feat: setup Git LFS for large .jsonl files
Browse files🤖 Generated with [Claude Code](https://claude.ai/code)
Co-Authored-By: Claude <noreply@anthropic.com>
This view is limited to 50 files because it contains too many changes.
See raw diff
- .gitattributes +1 -0
- LICENSE +201 -0
- README.md +322 -0
- baseline_model.py +136 -0
- convert_to_json.py +79 -0
- dataset_infos.json +126 -0
- dataset_statistics.json +160 -0
- evaluate.py +71 -0
- test.jsonl +3 -0
- test/annotations.tsv +0 -0
- test/texts/PMC10179231.md +0 -0
- test/texts/PMC10278212.md +253 -0
- test/texts/PMC10418744.md +308 -0
- test/texts/PMC10499425.md +269 -0
- test/texts/PMC10502099.md +338 -0
- test/texts/PMC10566653.md +156 -0
- test/texts/PMC10618485.md +117 -0
- test/texts/PMC10957942.md +215 -0
- test/texts/PMC10995391.md +0 -0
- test/texts/PMC11102648.md +306 -0
- test/texts/PMC11106956.md +348 -0
- test/texts/PMC11120965.md +323 -0
- test/texts/PMC11134291.md +360 -0
- test/texts/PMC11141156.md +0 -0
- test/texts/PMC11148365.md +315 -0
- test/texts/PMC11252221.md +286 -0
- test/texts/PMC11257390.md +260 -0
- test/texts/PMC11315837.md +432 -0
- test/texts/PMC11507373.md +318 -0
- test/texts/PMC11528939.md +0 -0
- test/texts/PMC11531276.md +420 -0
- test/texts/PMC11628867.md +294 -0
- test/texts/PMC11666798.md +430 -0
- test/texts/PMC11677811.md +358 -0
- test/texts/PMC11787782.md +353 -0
- test/texts/PMC11803932.md +357 -0
- test/texts/PMC11852071.md +356 -0
- test/texts/PMC11887086.md +331 -0
- test/texts/PMC1365072.md +49 -0
- test/texts/PMC1365132.md +40 -0
- test/texts/PMC1474035.md +332 -0
- test/texts/PMC1762324.md +0 -0
- test/texts/PMC1873375.md +223 -0
- test/texts/PMC1885108.md +334 -0
- test/texts/PMC1974827.md +255 -0
- test/texts/PMC1975838.md +241 -0
- test/texts/PMC1978168.md +176 -0
- test/texts/PMC2014233.md +135 -0
- test/texts/PMC2564574.md +227 -0
- test/texts/PMC2596476.md +281 -0
.gitattributes
CHANGED
|
@@ -57,3 +57,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 57 |
# Video files - compressed
|
| 58 |
*.mp4 filter=lfs diff=lfs merge=lfs -text
|
| 59 |
*.webm filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
| 57 |
# Video files - compressed
|
| 58 |
*.mp4 filter=lfs diff=lfs merge=lfs -text
|
| 59 |
*.webm filter=lfs diff=lfs merge=lfs -text
|
| 60 |
+
*.jsonl filter=lfs diff=lfs merge=lfs -text
|
LICENSE
ADDED
|
@@ -0,0 +1,201 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Apache License
|
| 2 |
+
Version 2.0, January 2004
|
| 3 |
+
http://www.apache.org/licenses/
|
| 4 |
+
|
| 5 |
+
TERMS AND CONDITIONS FOR USE, REPRODUCTION, AND DISTRIBUTION
|
| 6 |
+
|
| 7 |
+
1. Definitions.
|
| 8 |
+
|
| 9 |
+
"License" shall mean the terms and conditions for use, reproduction,
|
| 10 |
+
and distribution as defined by Sections 1 through 9 of this document.
|
| 11 |
+
|
| 12 |
+
"Licensor" shall mean the copyright owner or entity authorized by
|
| 13 |
+
the copyright owner that is granting the License.
|
| 14 |
+
|
| 15 |
+
"Legal Entity" shall mean the union of the acting entity and all
|
| 16 |
+
other entities that control, are controlled by, or are under common
|
| 17 |
+
control with that entity. For the purposes of this definition,
|
| 18 |
+
"control" means (i) the power, direct or indirect, to cause the
|
| 19 |
+
direction or management of such entity, whether by contract or
|
| 20 |
+
otherwise, or (ii) ownership of fifty percent (50%) or more of the
|
| 21 |
+
outstanding shares, or (iii) beneficial ownership of such entity.
|
| 22 |
+
|
| 23 |
+
"You" (or "Your") shall mean an individual or Legal Entity
|
| 24 |
+
exercising permissions granted by this License.
|
| 25 |
+
|
| 26 |
+
"Source" form shall mean the preferred form for making modifications,
|
| 27 |
+
including but not limited to software source code, documentation
|
| 28 |
+
source, and configuration files.
|
| 29 |
+
|
| 30 |
+
"Object" form shall mean any form resulting from mechanical
|
| 31 |
+
transformation or translation of a Source form, including but
|
| 32 |
+
not limited to compiled object code, generated documentation,
|
| 33 |
+
and conversions to other media types.
|
| 34 |
+
|
| 35 |
+
"Work" shall mean the work of authorship, whether in Source or
|
| 36 |
+
Object form, made available under the License, as indicated by a
|
| 37 |
+
copyright notice that is included in or attached to the work
|
| 38 |
+
(an example is provided in the Appendix below).
|
| 39 |
+
|
| 40 |
+
"Derivative Works" shall mean any work, whether in Source or Object
|
| 41 |
+
form, that is based on (or derived from) the Work and for which the
|
| 42 |
+
editorial revisions, annotations, elaborations, or other modifications
|
| 43 |
+
represent, as a whole, an original work of authorship. For the purposes
|
| 44 |
+
of this License, Derivative Works shall not include works that remain
|
| 45 |
+
separable from, or merely link (or bind by name) to the interfaces of,
|
| 46 |
+
the Work and Derivative Works thereof.
|
| 47 |
+
|
| 48 |
+
"Contribution" shall mean any work of authorship, including
|
| 49 |
+
the original version of the Work and any modifications or additions
|
| 50 |
+
to that Work or Derivative Works thereof, that is intentionally
|
| 51 |
+
submitted to Licensor for inclusion in the Work by the copyright owner
|
| 52 |
+
or by an individual or Legal Entity authorized to submit on behalf of
|
| 53 |
+
the copyright owner. For the purposes of this definition, "submitted"
|
| 54 |
+
means any form of electronic, verbal, or written communication sent
|
| 55 |
+
to the Licensor or its representatives, including but not limited to
|
| 56 |
+
communication on electronic mailing lists, source code control systems,
|
| 57 |
+
and issue tracking systems that are managed by, or on behalf of, the
|
| 58 |
+
Licensor for the purpose of discussing and improving the Work, but
|
| 59 |
+
excluding communication that is conspicuously marked or otherwise
|
| 60 |
+
designated in writing by the copyright owner as "Not a Contribution."
|
| 61 |
+
|
| 62 |
+
"Contributor" shall mean Licensor and any individual or Legal Entity
|
| 63 |
+
on behalf of whom a Contribution has been received by Licensor and
|
| 64 |
+
subsequently incorporated within the Work.
|
| 65 |
+
|
| 66 |
+
2. Grant of Copyright License. Subject to the terms and conditions of
|
| 67 |
+
this License, each Contributor hereby grants to You a perpetual,
|
| 68 |
+
worldwide, non-exclusive, no-charge, royalty-free, irrevocable
|
| 69 |
+
copyright license to reproduce, prepare Derivative Works of,
|
| 70 |
+
publicly display, publicly perform, sublicense, and distribute the
|
| 71 |
+
Work and such Derivative Works in Source or Object form.
|
| 72 |
+
|
| 73 |
+
3. Grant of Patent License. Subject to the terms and conditions of
|
| 74 |
+
this License, each Contributor hereby grants to You a perpetual,
|
| 75 |
+
worldwide, non-exclusive, no-charge, royalty-free, irrevocable
|
| 76 |
+
(except as stated in this section) patent license to make, have made,
|
| 77 |
+
use, offer to sell, sell, import, and otherwise transfer the Work,
|
| 78 |
+
where such license applies only to those patent claims licensable
|
| 79 |
+
by such Contributor that are necessarily infringed by their
|
| 80 |
+
Contribution(s) alone or by combination of their Contribution(s)
|
| 81 |
+
with the Work to which such Contribution(s) was submitted. If You
|
| 82 |
+
institute patent litigation against any entity (including a
|
| 83 |
+
cross-claim or counterclaim in a lawsuit) alleging that the Work
|
| 84 |
+
or a Contribution incorporated within the Work constitutes direct
|
| 85 |
+
or contributory patent infringement, then any patent licenses
|
| 86 |
+
granted to You under this License for that Work shall terminate
|
| 87 |
+
as of the date such litigation is filed.
|
| 88 |
+
|
| 89 |
+
4. Redistribution. You may reproduce and distribute copies of the
|
| 90 |
+
Work or Derivative Works thereof in any medium, with or without
|
| 91 |
+
modifications, and in Source or Object form, provided that You
|
| 92 |
+
meet the following conditions:
|
| 93 |
+
|
| 94 |
+
(a) You must give any other recipients of the Work or
|
| 95 |
+
Derivative Works a copy of this License; and
|
| 96 |
+
|
| 97 |
+
(b) You must cause any modified files to carry prominent notices
|
| 98 |
+
stating that You changed the files; and
|
| 99 |
+
|
| 100 |
+
(c) You must retain, in the Source form of any Derivative Works
|
| 101 |
+
that You distribute, all copyright, patent, trademark, and
|
| 102 |
+
attribution notices from the Source form of the Work,
|
| 103 |
+
excluding those notices that do not pertain to any part of
|
| 104 |
+
the Derivative Works; and
|
| 105 |
+
|
| 106 |
+
(d) If the Work includes a "NOTICE" text file as part of its
|
| 107 |
+
distribution, then any Derivative Works that You distribute must
|
| 108 |
+
include a readable copy of the attribution notices contained
|
| 109 |
+
within such NOTICE file, excluding those notices that do not
|
| 110 |
+
pertain to any part of the Derivative Works, in at least one
|
| 111 |
+
of the following places: within a NOTICE text file distributed
|
| 112 |
+
as part of the Derivative Works; within the Source form or
|
| 113 |
+
documentation, if provided along with the Derivative Works; or,
|
| 114 |
+
within a display generated by the Derivative Works, if and
|
| 115 |
+
wherever such third-party notices normally appear. The contents
|
| 116 |
+
of the NOTICE file are for informational purposes only and
|
| 117 |
+
do not modify the License. You may add Your own attribution
|
| 118 |
+
notices within Derivative Works that You distribute, alongside
|
| 119 |
+
or as an addendum to the NOTICE text from the Work, provided
|
| 120 |
+
that such additional attribution notices cannot be construed
|
| 121 |
+
as modifying the License.
|
| 122 |
+
|
| 123 |
+
You may add Your own copyright statement to Your modifications and
|
| 124 |
+
may provide additional or different license terms and conditions
|
| 125 |
+
for use, reproduction, or distribution of Your modifications, or
|
| 126 |
+
for any such Derivative Works as a whole, provided Your use,
|
| 127 |
+
reproduction, and distribution of the Work otherwise complies with
|
| 128 |
+
the conditions stated in this License.
|
| 129 |
+
|
| 130 |
+
5. Submission of Contributions. Unless You explicitly state otherwise,
|
| 131 |
+
any Contribution intentionally submitted for inclusion in the Work
|
| 132 |
+
by You to the Licensor shall be under the terms and conditions of
|
| 133 |
+
this License, without any additional terms or conditions.
|
| 134 |
+
Notwithstanding the above, nothing herein shall supersede or modify
|
| 135 |
+
the terms of any separate license agreement you may have executed
|
| 136 |
+
with Licensor regarding such Contributions.
|
| 137 |
+
|
| 138 |
+
6. Trademarks. This License does not grant permission to use the trade
|
| 139 |
+
names, trademarks, service marks, or product names of the Licensor,
|
| 140 |
+
except as required for reasonable and customary use in describing the
|
| 141 |
+
origin of the Work and reproducing the content of the NOTICE file.
|
| 142 |
+
|
| 143 |
+
7. Disclaimer of Warranty. Unless required by applicable law or
|
| 144 |
+
agreed to in writing, Licensor provides the Work (and each
|
| 145 |
+
Contributor provides its Contributions) on an "AS IS" BASIS,
|
| 146 |
+
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or
|
| 147 |
+
implied, including, without limitation, any warranties or conditions
|
| 148 |
+
of TITLE, NON-INFRINGEMENT, MERCHANTABILITY, or FITNESS FOR A
|
| 149 |
+
PARTICULAR PURPOSE. You are solely responsible for determining the
|
| 150 |
+
appropriateness of using or redistributing the Work and assume any
|
| 151 |
+
risks associated with Your exercise of permissions under this License.
|
| 152 |
+
|
| 153 |
+
8. Limitation of Liability. In no event and under no legal theory,
|
| 154 |
+
whether in tort (including negligence), contract, or otherwise,
|
| 155 |
+
unless required by applicable law (such as deliberate and grossly
|
| 156 |
+
negligent acts) or agreed to in writing, shall any Contributor be
|
| 157 |
+
liable to You for damages, including any direct, indirect, special,
|
| 158 |
+
incidental, or consequential damages of any character arising as a
|
| 159 |
+
result of this License or out of the use or inability to use the
|
| 160 |
+
Work (including but not limited to damages for loss of goodwill,
|
| 161 |
+
work stoppage, computer failure or malfunction, or any and all
|
| 162 |
+
other commercial damages or losses), even if such Contributor
|
| 163 |
+
has been advised of the possibility of such damages.
|
| 164 |
+
|
| 165 |
+
9. Accepting Warranty or Additional Liability. While redistributing
|
| 166 |
+
the Work or Derivative Works thereof, You may choose to offer,
|
| 167 |
+
and charge a fee for, acceptance of support, warranty, indemnity,
|
| 168 |
+
or other liability obligations and/or rights consistent with this
|
| 169 |
+
License. However, in accepting such obligations, You may act only
|
| 170 |
+
on Your own behalf and on Your sole responsibility, not on behalf
|
| 171 |
+
of any other Contributor, and only if You agree to indemnify,
|
| 172 |
+
defend, and hold each Contributor harmless for any liability
|
| 173 |
+
incurred by, or claims asserted against, such Contributor by reason
|
| 174 |
+
of your accepting any such warranty or additional liability.
|
| 175 |
+
|
| 176 |
+
END OF TERMS AND CONDITIONS
|
| 177 |
+
|
| 178 |
+
APPENDIX: How to apply the Apache License to your work.
|
| 179 |
+
|
| 180 |
+
To apply the Apache License to your work, attach the following
|
| 181 |
+
boilerplate notice, with the fields enclosed by brackets "[]"
|
| 182 |
+
replaced with your own identifying information. (Don't include
|
| 183 |
+
the brackets!) The text should be enclosed in the appropriate
|
| 184 |
+
comment syntax for the file format. We also recommend that a
|
| 185 |
+
file or class name and description of purpose be included on the
|
| 186 |
+
same "printed page" as the copyright notice for easier
|
| 187 |
+
identification within third-party archives.
|
| 188 |
+
|
| 189 |
+
Copyright [yyyy] [name of copyright owner]
|
| 190 |
+
|
| 191 |
+
Licensed under the Apache License, Version 2.0 (the "License");
|
| 192 |
+
you may not use this file except in compliance with the License.
|
| 193 |
+
You may obtain a copy of the License at
|
| 194 |
+
|
| 195 |
+
http://www.apache.org/licenses/LICENSE-2.0
|
| 196 |
+
|
| 197 |
+
Unless required by applicable law or agreed to in writing, software
|
| 198 |
+
distributed under the License is distributed on an "AS IS" BASIS,
|
| 199 |
+
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 200 |
+
See the License for the specific language governing permissions and
|
| 201 |
+
limitations under the License.
|
README.md
ADDED
|
@@ -0,0 +1,322 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
license: apache-2.0
|
| 3 |
+
task_categories:
|
| 4 |
+
- text-classification
|
| 5 |
+
- information-extraction
|
| 6 |
+
- text-mining
|
| 7 |
+
language:
|
| 8 |
+
- en
|
| 9 |
+
tags:
|
| 10 |
+
- pharmacogenomics
|
| 11 |
+
- biomedical
|
| 12 |
+
- variant-drug-associations
|
| 13 |
+
- literature-mining
|
| 14 |
+
- genomics
|
| 15 |
+
size_categories:
|
| 16 |
+
- 1K<n<10K
|
| 17 |
+
source_datasets:
|
| 18 |
+
- original
|
| 19 |
+
multilinguality:
|
| 20 |
+
- monolingual
|
| 21 |
+
pretty_name: Variant-Drug Annotation Benchmark
|
| 22 |
+
dataset_info:
|
| 23 |
+
features:
|
| 24 |
+
- name: variant_annotation_id
|
| 25 |
+
dtype: string
|
| 26 |
+
- name: variant_haplotypes
|
| 27 |
+
dtype: string
|
| 28 |
+
- name: gene
|
| 29 |
+
dtype: string
|
| 30 |
+
- name: drugs
|
| 31 |
+
dtype: string
|
| 32 |
+
- name: pmid
|
| 33 |
+
dtype: string
|
| 34 |
+
- name: phenotype_category
|
| 35 |
+
dtype: string
|
| 36 |
+
- name: significance
|
| 37 |
+
dtype: string
|
| 38 |
+
- name: notes
|
| 39 |
+
dtype: string
|
| 40 |
+
- name: sentence
|
| 41 |
+
dtype: string
|
| 42 |
+
- name: alleles
|
| 43 |
+
dtype: string
|
| 44 |
+
- name: specialty_population
|
| 45 |
+
dtype: string
|
| 46 |
+
- name: metabolizer_types
|
| 47 |
+
dtype: string
|
| 48 |
+
- name: is_plural
|
| 49 |
+
dtype: string
|
| 50 |
+
- name: is_is_not_associated
|
| 51 |
+
dtype: string
|
| 52 |
+
- name: direction_of_effect
|
| 53 |
+
dtype: string
|
| 54 |
+
- name: pd_pk_terms
|
| 55 |
+
dtype: string
|
| 56 |
+
- name: multiple_drugs_and_or
|
| 57 |
+
dtype: string
|
| 58 |
+
- name: population_types
|
| 59 |
+
dtype: string
|
| 60 |
+
- name: population_phenotypes_or_diseases
|
| 61 |
+
dtype: string
|
| 62 |
+
- name: multiple_phenotypes_or_diseases_and_or
|
| 63 |
+
dtype: string
|
| 64 |
+
- name: comparison_alleles_or_genotypes
|
| 65 |
+
dtype: string
|
| 66 |
+
- name: comparison_metabolizer_types
|
| 67 |
+
dtype: string
|
| 68 |
+
- name: text
|
| 69 |
+
dtype: string
|
| 70 |
+
splits:
|
| 71 |
+
- name: train
|
| 72 |
+
num_examples: 3124
|
| 73 |
+
- name: validation
|
| 74 |
+
num_examples: 796
|
| 75 |
+
- name: test
|
| 76 |
+
num_examples: 596
|
| 77 |
+
---
|
| 78 |
+
|
| 79 |
+
# Variant-Drug Annotation Benchmark
|
| 80 |
+
|
| 81 |
+
## Dataset Description
|
| 82 |
+
|
| 83 |
+
The Variant-Drug Annotation Benchmark is a comprehensive dataset designed to evaluate models' ability to extract pharmacogenomic variant-drug associations from scientific literature. This benchmark addresses the critical need for automated systems that can identify genetic variants, associated drugs, and their clinical relationships from biomedical texts.
|
| 84 |
+
|
| 85 |
+
### Dataset Summary
|
| 86 |
+
|
| 87 |
+
This dataset contains **4,516 annotations** extracted from **1,431 unique scientific papers** (PMIDs), covering a wide range of pharmacogenomic relationships. Each annotation includes detailed information about genetic variants, drugs, phenotype categories, population specifics, and statistical associations.
|
| 88 |
+
|
| 89 |
+
The dataset is particularly valuable for:
|
| 90 |
+
- **Information Extraction**: Extracting structured pharmacogenomic data from unstructured text
|
| 91 |
+
- **Biomedical NLP**: Training models to understand genetic variant-drug relationships
|
| 92 |
+
- **Pharmacogenomics Research**: Supporting precision medicine applications
|
| 93 |
+
- **Literature Mining**: Automated knowledge extraction from scientific publications
|
| 94 |
+
|
| 95 |
+
### Supported Tasks
|
| 96 |
+
|
| 97 |
+
- **Named Entity Recognition**: Identifying variants, genes, drugs, and phenotypes
|
| 98 |
+
- **Relation Extraction**: Determining associations between variants and drug responses
|
| 99 |
+
- **Text Classification**: Categorizing phenotype types and significance levels
|
| 100 |
+
- **Information Extraction**: Structured data extraction from biomedical literature
|
| 101 |
+
|
| 102 |
+
### Languages
|
| 103 |
+
|
| 104 |
+
The dataset is in English (en).
|
| 105 |
+
|
| 106 |
+
## Dataset Structure
|
| 107 |
+
|
| 108 |
+
### Data Instances
|
| 109 |
+
|
| 110 |
+
Each example contains:
|
| 111 |
+
- **Core annotation fields**: Variant, gene, drug, PMID, phenotype category
|
| 112 |
+
- **Association details**: Significance, direction of effect, comparison data
|
| 113 |
+
- **Population information**: Specialty populations, metabolizer types
|
| 114 |
+
- **Full text**: Complete scientific article in markdown format
|
| 115 |
+
|
| 116 |
+
Example:
|
| 117 |
+
```json
|
| 118 |
+
{
|
| 119 |
+
"variant_annotation_id": "1450936460",
|
| 120 |
+
"variant_haplotypes": "rs28362731",
|
| 121 |
+
"gene": "AQP1",
|
| 122 |
+
"drugs": "cisplatin",
|
| 123 |
+
"pmid": "30840592",
|
| 124 |
+
"phenotype_category": "Efficacy",
|
| 125 |
+
"significance": "no",
|
| 126 |
+
"sentence": "Genotype AG is not associated with response to cisplatin in people with Mesothelioma as compared to genotype GG.",
|
| 127 |
+
"is_is_not_associated": "Not associated with",
|
| 128 |
+
"direction_of_effect": "",
|
| 129 |
+
"population_phenotypes_or_diseases": "Other:Mesothelioma",
|
| 130 |
+
"comparison_alleles_or_genotypes": "GG",
|
| 131 |
+
"text": "# Prediction of CYP2D6 poor metabolizers..."
|
| 132 |
+
}
|
| 133 |
+
```
|
| 134 |
+
|
| 135 |
+
### Data Fields
|
| 136 |
+
|
| 137 |
+
#### Core Fields
|
| 138 |
+
- `variant_annotation_id`: Unique identifier for each annotation
|
| 139 |
+
- `variant_haplotypes`: Genetic variant identifier (e.g., rs numbers, haplotypes)
|
| 140 |
+
- `gene`: Gene symbol (e.g., CYP2D6, ABCB1)
|
| 141 |
+
- `drugs`: Drug name(s) involved in the association
|
| 142 |
+
- `pmid`: PubMed identifier of the source article
|
| 143 |
+
|
| 144 |
+
#### Phenotype Information
|
| 145 |
+
- `phenotype_category`: Type of effect (Efficacy, Toxicity, Dosage, Metabolism/PK, etc.)
|
| 146 |
+
- `significance`: Statistical significance (yes/no/not stated)
|
| 147 |
+
- `sentence`: Key sentence describing the association
|
| 148 |
+
- `notes`: Additional context or study details
|
| 149 |
+
|
| 150 |
+
#### Association Details
|
| 151 |
+
- `is_is_not_associated`: Whether variant is associated with outcome
|
| 152 |
+
- `direction_of_effect`: Direction of association (increased/decreased)
|
| 153 |
+
- `pd_pk_terms`: Pharmacodynamic/pharmacokinetic terms
|
| 154 |
+
- `alleles`: Specific alleles involved
|
| 155 |
+
|
| 156 |
+
#### Population Context
|
| 157 |
+
- `specialty_population`: Specific patient populations
|
| 158 |
+
- `population_types`: General population categories
|
| 159 |
+
- `population_phenotypes_or_diseases`: Diseases or conditions
|
| 160 |
+
- `metabolizer_types`: CYP metabolizer classifications
|
| 161 |
+
|
| 162 |
+
#### Comparison Data
|
| 163 |
+
- `comparison_alleles_or_genotypes`: Reference genotypes for comparison
|
| 164 |
+
- `comparison_metabolizer_types`: Reference metabolizer types
|
| 165 |
+
|
| 166 |
+
#### Text Data
|
| 167 |
+
- `text`: Full text of the source scientific article in markdown format
|
| 168 |
+
|
| 169 |
+
### Data Splits
|
| 170 |
+
|
| 171 |
+
| Split | Annotations | Unique Papers |
|
| 172 |
+
|-------|-------------|---------------|
|
| 173 |
+
| Train | 3,124 | 1,001 |
|
| 174 |
+
| Validation | 796 | 215 |
|
| 175 |
+
| Test | 596 | 215 |
|
| 176 |
+
|
| 177 |
+
**Total**: 4,516 annotations across 1,431 papers
|
| 178 |
+
|
| 179 |
+
## Dataset Creation
|
| 180 |
+
|
| 181 |
+
### Curation Rationale
|
| 182 |
+
|
| 183 |
+
This benchmark was created to address the growing need for automated pharmacogenomic knowledge extraction. With the rapid expansion of pharmacogenomic literature, manual curation becomes increasingly challenging. This dataset provides a standardized evaluation framework for developing and comparing automated extraction systems.
|
| 184 |
+
|
| 185 |
+
### Source Data
|
| 186 |
+
|
| 187 |
+
#### Initial Data Collection and Normalization
|
| 188 |
+
|
| 189 |
+
The dataset is derived from peer-reviewed scientific publications in the pharmacogenomics domain. Articles were selected based on their content related to genetic variant-drug associations and clinical outcomes.
|
| 190 |
+
|
| 191 |
+
#### Who are the source language producers?
|
| 192 |
+
|
| 193 |
+
The source texts are scientific articles authored by researchers in pharmacogenomics, clinical pharmacology, and related biomedical fields, published in peer-reviewed journals.
|
| 194 |
+
|
| 195 |
+
### Annotations
|
| 196 |
+
|
| 197 |
+
#### Annotation process
|
| 198 |
+
|
| 199 |
+
Annotations were created by domain experts following a comprehensive schema covering:
|
| 200 |
+
- Genetic variant identification and standardization
|
| 201 |
+
- Drug name normalization
|
| 202 |
+
- Phenotype categorization using controlled vocabularies
|
| 203 |
+
- Population and study context extraction
|
| 204 |
+
- Statistical association characterization
|
| 205 |
+
|
| 206 |
+
#### Who are the annotators?
|
| 207 |
+
|
| 208 |
+
The annotations were created by experts in pharmacogenomics and biomedical informatics with specialized knowledge in genetic variant-drug associations.
|
| 209 |
+
|
| 210 |
+
### Personal and Sensitive Information
|
| 211 |
+
|
| 212 |
+
The dataset contains only published scientific literature and does not include personal or sensitive information about individuals.
|
| 213 |
+
|
| 214 |
+
## Considerations for Using the Data
|
| 215 |
+
|
| 216 |
+
### Social Impact of Dataset
|
| 217 |
+
|
| 218 |
+
This dataset supports the development of automated systems for pharmacogenomic knowledge extraction, which can:
|
| 219 |
+
- **Accelerate precision medicine**: Enable faster identification of clinically relevant variant-drug associations
|
| 220 |
+
- **Support clinical decision-making**: Facilitate evidence-based prescribing decisions
|
| 221 |
+
- **Advance research**: Enable large-scale analysis of pharmacogenomic literature
|
| 222 |
+
|
| 223 |
+
### Discussion of Biases
|
| 224 |
+
|
| 225 |
+
Potential biases in the dataset may include:
|
| 226 |
+
- **Publication bias**: Overrepresentation of statistically significant results
|
| 227 |
+
- **Population bias**: Uneven representation of different ethnic populations in source studies
|
| 228 |
+
- **Drug bias**: Focus on commonly studied drugs and variants
|
| 229 |
+
- **Temporal bias**: Emphasis on more recent research publications
|
| 230 |
+
|
| 231 |
+
### Other Known Limitations
|
| 232 |
+
|
| 233 |
+
- **Coverage**: Represents approximately 33.6% of original pharmacogenomic annotations from the source database
|
| 234 |
+
- **Language**: Limited to English-language publications
|
| 235 |
+
- **Domain scope**: Focused specifically on pharmacogenomics, may not generalize to other biomedical domains
|
| 236 |
+
- **Text quality**: Depends on the quality of PDF-to-text conversion for source articles
|
| 237 |
+
|
| 238 |
+
## Additional Information
|
| 239 |
+
|
| 240 |
+
### Dataset Curators
|
| 241 |
+
|
| 242 |
+
AutoGKB Team
|
| 243 |
+
|
| 244 |
+
### Licensing Information
|
| 245 |
+
|
| 246 |
+
This dataset is released under the Apache License 2.0.
|
| 247 |
+
|
| 248 |
+
### Citation Information
|
| 249 |
+
|
| 250 |
+
```bibtex
|
| 251 |
+
@misc{variant_drug_benchmark_2024,
|
| 252 |
+
title={Variant-Drug Annotation Benchmark},
|
| 253 |
+
author={AutoGKB Team},
|
| 254 |
+
year={2024},
|
| 255 |
+
note={A benchmark for pharmacogenomic variant-drug annotation extraction from scientific literature}
|
| 256 |
+
}
|
| 257 |
+
```
|
| 258 |
+
|
| 259 |
+
### Contributions
|
| 260 |
+
|
| 261 |
+
This dataset contributes to the biomedical NLP community by providing:
|
| 262 |
+
- A standardized benchmark for pharmacogenomic information extraction
|
| 263 |
+
- High-quality annotations with detailed schema
|
| 264 |
+
- Full-text articles paired with structured annotations
|
| 265 |
+
- Evaluation metrics and baseline models for comparison
|
| 266 |
+
|
| 267 |
+
## Usage Examples
|
| 268 |
+
|
| 269 |
+
### Loading the Dataset
|
| 270 |
+
|
| 271 |
+
```python
|
| 272 |
+
from datasets import load_dataset
|
| 273 |
+
|
| 274 |
+
dataset = load_dataset("variant_drug_benchmark")
|
| 275 |
+
|
| 276 |
+
# Access different splits
|
| 277 |
+
train_data = dataset["train"]
|
| 278 |
+
val_data = dataset["validation"]
|
| 279 |
+
test_data = dataset["test"]
|
| 280 |
+
|
| 281 |
+
# Example: Get all efficacy-related annotations
|
| 282 |
+
efficacy_examples = train_data.filter(
|
| 283 |
+
lambda x: "Efficacy" in x["phenotype_category"]
|
| 284 |
+
)
|
| 285 |
+
```
|
| 286 |
+
|
| 287 |
+
### Evaluation
|
| 288 |
+
|
| 289 |
+
The dataset includes evaluation scripts for measuring:
|
| 290 |
+
- Field-level exact match accuracy
|
| 291 |
+
- Overall accuracy across core fields
|
| 292 |
+
- Phenotype-specific performance
|
| 293 |
+
|
| 294 |
+
```bash
|
| 295 |
+
# Run baseline model
|
| 296 |
+
python baseline_model.py val baseline_predictions.tsv
|
| 297 |
+
|
| 298 |
+
# Evaluate predictions
|
| 299 |
+
python evaluate.py baseline_predictions.tsv val/annotations.tsv --output results.json
|
| 300 |
+
```
|
| 301 |
+
|
| 302 |
+
## File Structure
|
| 303 |
+
|
| 304 |
+
```
|
| 305 |
+
benchmark/
|
| 306 |
+
├── train/
|
| 307 |
+
│ ├── annotations.tsv # Training annotations
|
| 308 |
+
│ └── texts/ # Training text files (PMC*.md)
|
| 309 |
+
├── val/
|
| 310 |
+
│ ├── annotations.tsv # Validation annotations
|
| 311 |
+
│ └── texts/ # Validation text files
|
| 312 |
+
├── test/
|
| 313 |
+
│ ├── annotations.tsv # Test annotations
|
| 314 |
+
│ └── texts/ # Test text files
|
| 315 |
+
├── dataset_statistics.json
|
| 316 |
+
├── evaluate.py # Evaluation script
|
| 317 |
+
├── baseline_model.py # Rule-based baseline
|
| 318 |
+
├── variant_drug_benchmark.py # HuggingFace dataset script
|
| 319 |
+
├── dataset_infos.json # Dataset metadata
|
| 320 |
+
├── LICENSE # Apache 2.0 license
|
| 321 |
+
└── README.md # This file
|
| 322 |
+
```
|
baseline_model.py
ADDED
|
@@ -0,0 +1,136 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python3
|
| 2 |
+
"""
|
| 3 |
+
Rule-based baseline model for Variant-Drug Annotation Benchmark
|
| 4 |
+
"""
|
| 5 |
+
|
| 6 |
+
import pandas as pd
|
| 7 |
+
import re
|
| 8 |
+
from pathlib import Path
|
| 9 |
+
import argparse
|
| 10 |
+
|
| 11 |
+
class RuleBasedBaseline:
|
| 12 |
+
def __init__(self):
|
| 13 |
+
# Common drug patterns
|
| 14 |
+
self.drug_patterns = [
|
| 15 |
+
r'\b(?:warfarin|clopidogrel|codeine|tamoxifen|atorvastatin|simvastatin)\b',
|
| 16 |
+
r'\b\w+statin\b', # statins
|
| 17 |
+
r'\b\w+prazole\b', # proton pump inhibitors
|
| 18 |
+
]
|
| 19 |
+
|
| 20 |
+
# Variant patterns
|
| 21 |
+
self.variant_patterns = [
|
| 22 |
+
r'\b[A-Z]{2,}\d+[A-Z]\*\d+', # CYP2D6*1
|
| 23 |
+
r'\brs\d+', # rs123456
|
| 24 |
+
r'\b\*\d+', # *1, *2
|
| 25 |
+
r'\b[A-Z]+\*\d+', # CYP2D6*1 (simplified)
|
| 26 |
+
]
|
| 27 |
+
|
| 28 |
+
# Gene patterns
|
| 29 |
+
self.gene_patterns = [
|
| 30 |
+
r'\b[A-Z]{2,}\d[A-Z]\d', # CYP2D6
|
| 31 |
+
r'\bCYP\w+', # CYP genes
|
| 32 |
+
r'\bUGT\w+', # UGT genes
|
| 33 |
+
]
|
| 34 |
+
|
| 35 |
+
def extract_variants(self, text: str):
|
| 36 |
+
"""Extract variant mentions using regex patterns"""
|
| 37 |
+
variants = []
|
| 38 |
+
for pattern in self.variant_patterns:
|
| 39 |
+
matches = re.findall(pattern, text, re.IGNORECASE)
|
| 40 |
+
variants.extend(matches)
|
| 41 |
+
return list(set(variants))[:1] # Return first match
|
| 42 |
+
|
| 43 |
+
def extract_drugs(self, text: str):
|
| 44 |
+
"""Extract drug mentions using regex patterns"""
|
| 45 |
+
drugs = []
|
| 46 |
+
for pattern in self.drug_patterns:
|
| 47 |
+
matches = re.findall(pattern, text, re.IGNORECASE)
|
| 48 |
+
drugs.extend(matches)
|
| 49 |
+
return list(set(drugs))[:1] # Return first match
|
| 50 |
+
|
| 51 |
+
def extract_genes(self, text: str):
|
| 52 |
+
"""Extract gene mentions using regex patterns"""
|
| 53 |
+
genes = []
|
| 54 |
+
for pattern in self.gene_patterns:
|
| 55 |
+
matches = re.findall(pattern, text, re.IGNORECASE)
|
| 56 |
+
genes.extend(matches)
|
| 57 |
+
return list(set(genes))[:1] # Return first match
|
| 58 |
+
|
| 59 |
+
def predict_significance(self, text: str):
|
| 60 |
+
"""Simple heuristic for significance"""
|
| 61 |
+
if any(word in text.lower() for word in ['significant', 'p <', 'p=']):
|
| 62 |
+
return 'yes'
|
| 63 |
+
elif any(word in text.lower() for word in ['not significant', 'no association']):
|
| 64 |
+
return 'no'
|
| 65 |
+
return 'not stated'
|
| 66 |
+
|
| 67 |
+
def predict_direction(self, text: str):
|
| 68 |
+
"""Simple heuristic for direction"""
|
| 69 |
+
if any(word in text.lower() for word in ['increased', 'higher', 'elevated']):
|
| 70 |
+
return 'increased'
|
| 71 |
+
elif any(word in text.lower() for word in ['decreased', 'lower', 'reduced']):
|
| 72 |
+
return 'decreased'
|
| 73 |
+
return ''
|
| 74 |
+
|
| 75 |
+
def predict_annotations(self, text_file: str):
|
| 76 |
+
"""Predict annotations for a given text file"""
|
| 77 |
+
with open(text_file, 'r', encoding='utf-8') as f:
|
| 78 |
+
text = f.read()
|
| 79 |
+
|
| 80 |
+
variants = self.extract_variants(text)
|
| 81 |
+
drugs = self.extract_drugs(text)
|
| 82 |
+
genes = self.extract_genes(text)
|
| 83 |
+
|
| 84 |
+
# Simple prediction - one annotation per file
|
| 85 |
+
annotation = {
|
| 86 |
+
'Variant Annotation ID': 'baseline_1',
|
| 87 |
+
'Variant/Haplotypes': variants[0] if variants else '',
|
| 88 |
+
'Gene': genes[0] if genes else '',
|
| 89 |
+
'Drug(s)': drugs[0] if drugs else '',
|
| 90 |
+
'PMID': '', # Will be filled from filename
|
| 91 |
+
'Phenotype Category': 'Other',
|
| 92 |
+
'Significance': self.predict_significance(text),
|
| 93 |
+
'Notes': 'Generated by rule-based baseline',
|
| 94 |
+
'Sentence': '', # Would need more sophisticated extraction
|
| 95 |
+
'Alleles': '',
|
| 96 |
+
'Specialty Population': '',
|
| 97 |
+
'Metabolizer types': '',
|
| 98 |
+
'isPlural': '',
|
| 99 |
+
'Is/Is Not associated': 'Is',
|
| 100 |
+
'Direction of effect': self.predict_direction(text),
|
| 101 |
+
'PD/PK terms': '',
|
| 102 |
+
'Multiple drugs And/or': '',
|
| 103 |
+
'Population types': '',
|
| 104 |
+
'Population Phenotypes or diseases': '',
|
| 105 |
+
'Multiple phenotypes or diseases And/or': '',
|
| 106 |
+
'Comparison Allele(s) or Genotype(s)': '',
|
| 107 |
+
'Comparison Metabolizer types': ''
|
| 108 |
+
}
|
| 109 |
+
|
| 110 |
+
return [annotation]
|
| 111 |
+
|
| 112 |
+
def main():
|
| 113 |
+
parser = argparse.ArgumentParser(description="Run rule-based baseline")
|
| 114 |
+
parser.add_argument("input_dir", help="Directory containing text files")
|
| 115 |
+
parser.add_argument("output_file", help="Output TSV file")
|
| 116 |
+
|
| 117 |
+
args = parser.parse_args()
|
| 118 |
+
|
| 119 |
+
baseline = RuleBasedBaseline()
|
| 120 |
+
all_predictions = []
|
| 121 |
+
|
| 122 |
+
text_dir = Path(args.input_dir) / 'texts'
|
| 123 |
+
for text_file in text_dir.glob('*.md'):
|
| 124 |
+
predictions = baseline.predict_annotations(text_file)
|
| 125 |
+
for pred in predictions:
|
| 126 |
+
pred['PMID'] = text_file.stem.replace('PMC', '') # Extract PMCID
|
| 127 |
+
all_predictions.extend(predictions)
|
| 128 |
+
|
| 129 |
+
# Save predictions
|
| 130 |
+
df = pd.DataFrame(all_predictions)
|
| 131 |
+
df.to_csv(args.output_file, sep='\t', index=False)
|
| 132 |
+
|
| 133 |
+
print(f"Generated {len(all_predictions)} predictions in {args.output_file}")
|
| 134 |
+
|
| 135 |
+
if __name__ == "__main__":
|
| 136 |
+
main()
|
convert_to_json.py
ADDED
|
@@ -0,0 +1,79 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python3
|
| 2 |
+
"""
|
| 3 |
+
Convert TSV annotations to JSON/JSONL format for easier HuggingFace loading
|
| 4 |
+
"""
|
| 5 |
+
|
| 6 |
+
import csv
|
| 7 |
+
import json
|
| 8 |
+
import os
|
| 9 |
+
from pathlib import Path
|
| 10 |
+
from typing import Dict, List
|
| 11 |
+
|
| 12 |
+
def convert_tsv_to_jsonl(tsv_file: str, jsonl_file: str, texts_dir: str = None):
|
| 13 |
+
"""Convert TSV annotations to JSONL format with optional text inclusion"""
|
| 14 |
+
|
| 15 |
+
# Load text files if directory provided
|
| 16 |
+
text_cache = {}
|
| 17 |
+
if texts_dir and os.path.exists(texts_dir):
|
| 18 |
+
texts_path = Path(texts_dir)
|
| 19 |
+
for text_file in texts_path.glob("*.md"):
|
| 20 |
+
pmcid = text_file.stem
|
| 21 |
+
with open(text_file, 'r', encoding='utf-8') as f:
|
| 22 |
+
text_cache[pmcid] = f.read()
|
| 23 |
+
|
| 24 |
+
# Convert TSV to JSONL
|
| 25 |
+
with open(tsv_file, 'r', encoding='utf-8') as infile, \
|
| 26 |
+
open(jsonl_file, 'w', encoding='utf-8') as outfile:
|
| 27 |
+
|
| 28 |
+
reader = csv.DictReader(infile, delimiter='\t')
|
| 29 |
+
|
| 30 |
+
for row in reader:
|
| 31 |
+
# Clean field names (remove spaces, special characters)
|
| 32 |
+
clean_row = {}
|
| 33 |
+
for key, value in row.items():
|
| 34 |
+
# Convert field names to snake_case
|
| 35 |
+
clean_key = key.lower().replace(' ', '_').replace('/', '_').replace('(', '').replace(')', '')
|
| 36 |
+
clean_row[clean_key] = value if value else ""
|
| 37 |
+
|
| 38 |
+
# Add text if available
|
| 39 |
+
pmid = clean_row.get('pmid', '')
|
| 40 |
+
text_content = ""
|
| 41 |
+
|
| 42 |
+
# Try to find matching text file
|
| 43 |
+
for pmcid, content in text_cache.items():
|
| 44 |
+
if pmid in content or pmcid.replace('PMC', '') == pmid:
|
| 45 |
+
text_content = content
|
| 46 |
+
break
|
| 47 |
+
|
| 48 |
+
clean_row['text'] = text_content
|
| 49 |
+
|
| 50 |
+
# Write as JSON line
|
| 51 |
+
outfile.write(json.dumps(clean_row) + '\n')
|
| 52 |
+
|
| 53 |
+
def main():
|
| 54 |
+
"""Convert all splits to JSONL format"""
|
| 55 |
+
|
| 56 |
+
base_dir = Path(__file__).parent
|
| 57 |
+
|
| 58 |
+
splits = [
|
| 59 |
+
('train', 'train/annotations.tsv', 'train/texts'),
|
| 60 |
+
('val', 'val/annotations.tsv', 'val/texts'),
|
| 61 |
+
('test', 'test/annotations.tsv', 'test/texts')
|
| 62 |
+
]
|
| 63 |
+
|
| 64 |
+
for split_name, annotations_path, texts_path in splits:
|
| 65 |
+
tsv_file = base_dir / annotations_path
|
| 66 |
+
jsonl_file = base_dir / f"{split_name}.jsonl"
|
| 67 |
+
texts_dir = base_dir / texts_path if texts_path else None
|
| 68 |
+
|
| 69 |
+
if tsv_file.exists():
|
| 70 |
+
print(f"Converting {split_name} split...")
|
| 71 |
+
convert_tsv_to_jsonl(str(tsv_file), str(jsonl_file), str(texts_dir))
|
| 72 |
+
print(f"Created {jsonl_file}")
|
| 73 |
+
else:
|
| 74 |
+
print(f"Warning: {tsv_file} not found")
|
| 75 |
+
|
| 76 |
+
print("Conversion complete!")
|
| 77 |
+
|
| 78 |
+
if __name__ == "__main__":
|
| 79 |
+
main()
|
dataset_infos.json
ADDED
|
@@ -0,0 +1,126 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"default": {
|
| 3 |
+
"description": "Variant-Drug Annotation Benchmark: A dataset for evaluating models' ability to extract pharmacogenomic variant-drug associations from scientific literature. Contains 4,516 annotations across 1,431 papers with train/validation/test splits.",
|
| 4 |
+
"citation": "@misc{variant_drug_benchmark_2024,\n title={Variant-Drug Annotation Benchmark},\n author={AutoGKB Team},\n year={2024},\n note={A benchmark for pharmacogenomic variant-drug annotation extraction from scientific literature}\n}",
|
| 5 |
+
"homepage": "",
|
| 6 |
+
"license": "Apache-2.0",
|
| 7 |
+
"features": {
|
| 8 |
+
"variant_annotation_id": {
|
| 9 |
+
"dtype": "string",
|
| 10 |
+
"_type": "Value"
|
| 11 |
+
},
|
| 12 |
+
"variant_haplotypes": {
|
| 13 |
+
"dtype": "string",
|
| 14 |
+
"_type": "Value"
|
| 15 |
+
},
|
| 16 |
+
"gene": {
|
| 17 |
+
"dtype": "string",
|
| 18 |
+
"_type": "Value"
|
| 19 |
+
},
|
| 20 |
+
"drugs": {
|
| 21 |
+
"dtype": "string",
|
| 22 |
+
"_type": "Value"
|
| 23 |
+
},
|
| 24 |
+
"pmid": {
|
| 25 |
+
"dtype": "string",
|
| 26 |
+
"_type": "Value"
|
| 27 |
+
},
|
| 28 |
+
"phenotype_category": {
|
| 29 |
+
"dtype": "string",
|
| 30 |
+
"_type": "Value"
|
| 31 |
+
},
|
| 32 |
+
"significance": {
|
| 33 |
+
"dtype": "string",
|
| 34 |
+
"_type": "Value"
|
| 35 |
+
},
|
| 36 |
+
"notes": {
|
| 37 |
+
"dtype": "string",
|
| 38 |
+
"_type": "Value"
|
| 39 |
+
},
|
| 40 |
+
"sentence": {
|
| 41 |
+
"dtype": "string",
|
| 42 |
+
"_type": "Value"
|
| 43 |
+
},
|
| 44 |
+
"alleles": {
|
| 45 |
+
"dtype": "string",
|
| 46 |
+
"_type": "Value"
|
| 47 |
+
},
|
| 48 |
+
"specialty_population": {
|
| 49 |
+
"dtype": "string",
|
| 50 |
+
"_type": "Value"
|
| 51 |
+
},
|
| 52 |
+
"metabolizer_types": {
|
| 53 |
+
"dtype": "string",
|
| 54 |
+
"_type": "Value"
|
| 55 |
+
},
|
| 56 |
+
"is_plural": {
|
| 57 |
+
"dtype": "string",
|
| 58 |
+
"_type": "Value"
|
| 59 |
+
},
|
| 60 |
+
"is_is_not_associated": {
|
| 61 |
+
"dtype": "string",
|
| 62 |
+
"_type": "Value"
|
| 63 |
+
},
|
| 64 |
+
"direction_of_effect": {
|
| 65 |
+
"dtype": "string",
|
| 66 |
+
"_type": "Value"
|
| 67 |
+
},
|
| 68 |
+
"pd_pk_terms": {
|
| 69 |
+
"dtype": "string",
|
| 70 |
+
"_type": "Value"
|
| 71 |
+
},
|
| 72 |
+
"multiple_drugs_and_or": {
|
| 73 |
+
"dtype": "string",
|
| 74 |
+
"_type": "Value"
|
| 75 |
+
},
|
| 76 |
+
"population_types": {
|
| 77 |
+
"dtype": "string",
|
| 78 |
+
"_type": "Value"
|
| 79 |
+
},
|
| 80 |
+
"population_phenotypes_or_diseases": {
|
| 81 |
+
"dtype": "string",
|
| 82 |
+
"_type": "Value"
|
| 83 |
+
},
|
| 84 |
+
"multiple_phenotypes_or_diseases_and_or": {
|
| 85 |
+
"dtype": "string",
|
| 86 |
+
"_type": "Value"
|
| 87 |
+
},
|
| 88 |
+
"comparison_alleles_or_genotypes": {
|
| 89 |
+
"dtype": "string",
|
| 90 |
+
"_type": "Value"
|
| 91 |
+
},
|
| 92 |
+
"comparison_metabolizer_types": {
|
| 93 |
+
"dtype": "string",
|
| 94 |
+
"_type": "Value"
|
| 95 |
+
},
|
| 96 |
+
"text": {
|
| 97 |
+
"dtype": "string",
|
| 98 |
+
"_type": "Value"
|
| 99 |
+
}
|
| 100 |
+
},
|
| 101 |
+
"splits": {
|
| 102 |
+
"train": {
|
| 103 |
+
"name": "train",
|
| 104 |
+
"num_bytes": 0,
|
| 105 |
+
"num_examples": 3124,
|
| 106 |
+
"dataset_name": "variant_drug_benchmark"
|
| 107 |
+
},
|
| 108 |
+
"validation": {
|
| 109 |
+
"name": "validation",
|
| 110 |
+
"num_bytes": 0,
|
| 111 |
+
"num_examples": 796,
|
| 112 |
+
"dataset_name": "variant_drug_benchmark"
|
| 113 |
+
},
|
| 114 |
+
"test": {
|
| 115 |
+
"name": "test",
|
| 116 |
+
"num_bytes": 0,
|
| 117 |
+
"num_examples": 596,
|
| 118 |
+
"dataset_name": "variant_drug_benchmark"
|
| 119 |
+
}
|
| 120 |
+
},
|
| 121 |
+
"download_checksums": {},
|
| 122 |
+
"download_size": 0,
|
| 123 |
+
"dataset_size": 0,
|
| 124 |
+
"size_in_bytes": 0
|
| 125 |
+
}
|
| 126 |
+
}
|
dataset_statistics.json
ADDED
|
@@ -0,0 +1,160 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"train": {
|
| 3 |
+
"total_annotations": 3124,
|
| 4 |
+
"unique_pmids": 1001,
|
| 5 |
+
"phenotype_distribution": {
|
| 6 |
+
"Efficacy": 1420,
|
| 7 |
+
"Toxicity": 36,
|
| 8 |
+
"Dosage": 417,
|
| 9 |
+
"Metabolism/PK": 1029,
|
| 10 |
+
"Dosage, Metabolism/PK": 26,
|
| 11 |
+
"Toxicity, Metabolism/PK": 21,
|
| 12 |
+
"Dosage, Efficacy": 51,
|
| 13 |
+
"Dosage, Toxicity": 9,
|
| 14 |
+
"Efficacy, Metabolism/PK": 9,
|
| 15 |
+
"Other": 57,
|
| 16 |
+
"PD, Metabolism/PK": 1,
|
| 17 |
+
"Toxicity, Other": 1,
|
| 18 |
+
"Unknown": 4,
|
| 19 |
+
"Efficacy, Toxicity, Metabolism/PK": 6,
|
| 20 |
+
"Efficacy, Toxicity": 14,
|
| 21 |
+
"Other, Metabolism/PK": 12,
|
| 22 |
+
"Dosage, Efficacy, Toxicity, Metabolism/PK": 1,
|
| 23 |
+
"PD": 10
|
| 24 |
+
},
|
| 25 |
+
"significance_distribution": {
|
| 26 |
+
"no": 1471,
|
| 27 |
+
"not stated": 190,
|
| 28 |
+
"yes": 1463
|
| 29 |
+
},
|
| 30 |
+
"top_genes": {
|
| 31 |
+
"Unknown": 150,
|
| 32 |
+
"ABCB1": 143,
|
| 33 |
+
"CYP2B6": 130,
|
| 34 |
+
"CYP2C19": 129,
|
| 35 |
+
"CYP3A5": 125,
|
| 36 |
+
"CYP2C9": 120,
|
| 37 |
+
"CYP2D6": 118,
|
| 38 |
+
"OPRM1": 85,
|
| 39 |
+
"CYP3A4": 78,
|
| 40 |
+
"VKORC1": 78
|
| 41 |
+
},
|
| 42 |
+
"top_drugs": {
|
| 43 |
+
"warfarin": 233,
|
| 44 |
+
"tacrolimus": 166,
|
| 45 |
+
"methotrexate": 106,
|
| 46 |
+
"methadone": 90,
|
| 47 |
+
"methylphenidate": 79,
|
| 48 |
+
"efavirenz": 73,
|
| 49 |
+
"nicotine": 60,
|
| 50 |
+
"metformin": 53,
|
| 51 |
+
"fentanyl": 48,
|
| 52 |
+
"tenofovir": 46
|
| 53 |
+
}
|
| 54 |
+
},
|
| 55 |
+
"val": {
|
| 56 |
+
"total_annotations": 796,
|
| 57 |
+
"unique_pmids": 215,
|
| 58 |
+
"phenotype_distribution": {
|
| 59 |
+
"Efficacy": 386,
|
| 60 |
+
"Metabolism/PK": 302,
|
| 61 |
+
"Toxicity": 2,
|
| 62 |
+
"Dosage, Metabolism/PK": 12,
|
| 63 |
+
"Dosage, Efficacy, Toxicity": 2,
|
| 64 |
+
"Dosage": 73,
|
| 65 |
+
"Toxicity, Metabolism/PK": 2,
|
| 66 |
+
"Unknown": 1,
|
| 67 |
+
"Other": 6,
|
| 68 |
+
"Other, Metabolism/PK": 4,
|
| 69 |
+
"Dosage, Other": 1,
|
| 70 |
+
"Efficacy, Toxicity": 1,
|
| 71 |
+
"Dosage, Efficacy": 4
|
| 72 |
+
},
|
| 73 |
+
"significance_distribution": {
|
| 74 |
+
"no": 372,
|
| 75 |
+
"yes": 367,
|
| 76 |
+
"not stated": 57
|
| 77 |
+
},
|
| 78 |
+
"top_genes": {
|
| 79 |
+
"Unknown": 60,
|
| 80 |
+
"ABCB1": 43,
|
| 81 |
+
"CYP2C19": 37,
|
| 82 |
+
"CYP2D6": 36,
|
| 83 |
+
"CYP3A5": 35,
|
| 84 |
+
"CYP2B6": 33,
|
| 85 |
+
"CYP2C9": 25,
|
| 86 |
+
"SLCO1B1": 25,
|
| 87 |
+
"CYP3A4": 22,
|
| 88 |
+
"GLA": 20
|
| 89 |
+
},
|
| 90 |
+
"top_drugs": {
|
| 91 |
+
"warfarin": 53,
|
| 92 |
+
"efavirenz": 47,
|
| 93 |
+
"lithium": 38,
|
| 94 |
+
"vancomycin": 32,
|
| 95 |
+
"tacrolimus": 31,
|
| 96 |
+
"methotrexate": 24,
|
| 97 |
+
"botulinum toxin type a": 23,
|
| 98 |
+
"migalastat": 20,
|
| 99 |
+
"clopidogrel": 18,
|
| 100 |
+
"risperidone": 17
|
| 101 |
+
}
|
| 102 |
+
},
|
| 103 |
+
"test": {
|
| 104 |
+
"total_annotations": 596,
|
| 105 |
+
"unique_pmids": 215,
|
| 106 |
+
"phenotype_distribution": {
|
| 107 |
+
"Metabolism/PK": 206,
|
| 108 |
+
"Efficacy": 264,
|
| 109 |
+
"Dosage": 97,
|
| 110 |
+
"Dosage, Metabolism/PK": 5,
|
| 111 |
+
"Other": 11,
|
| 112 |
+
"Efficacy, Metabolism/PK": 4,
|
| 113 |
+
"Other, Metabolism/PK": 5,
|
| 114 |
+
"Unknown": 2,
|
| 115 |
+
"Toxicity, Metabolism/PK": 1,
|
| 116 |
+
"Toxicity": 1
|
| 117 |
+
},
|
| 118 |
+
"significance_distribution": {
|
| 119 |
+
"no": 227,
|
| 120 |
+
"yes": 310,
|
| 121 |
+
"not stated": 59
|
| 122 |
+
},
|
| 123 |
+
"top_genes": {
|
| 124 |
+
"ABCB1": 40,
|
| 125 |
+
"CYP2A6": 37,
|
| 126 |
+
"CYP2C19": 34,
|
| 127 |
+
"CYP2D6": 32,
|
| 128 |
+
"CYP2C9": 29,
|
| 129 |
+
"CYP3A5": 22,
|
| 130 |
+
"CYP2B6": 22,
|
| 131 |
+
"SLCO1B1": 21,
|
| 132 |
+
"VKORC1": 17,
|
| 133 |
+
"OPRM1": 16
|
| 134 |
+
},
|
| 135 |
+
"top_drugs": {
|
| 136 |
+
"warfarin": 48,
|
| 137 |
+
"nicotine": 45,
|
| 138 |
+
"methadone": 28,
|
| 139 |
+
"tacrolimus": 22,
|
| 140 |
+
"fenofibrate": 22,
|
| 141 |
+
"clopidogrel": 18,
|
| 142 |
+
"metformin": 14,
|
| 143 |
+
"fentanyl": 13,
|
| 144 |
+
"pioglitazone": 12,
|
| 145 |
+
"hydrochlorothiazide": 11
|
| 146 |
+
}
|
| 147 |
+
},
|
| 148 |
+
"overall": {
|
| 149 |
+
"total_annotations": 4516,
|
| 150 |
+
"unique_pmids": 1431,
|
| 151 |
+
"coverage_from_original": {
|
| 152 |
+
"total_original_annotations": 12474,
|
| 153 |
+
"total_unique_pmids": 4262,
|
| 154 |
+
"pmids_with_mapping": 1438,
|
| 155 |
+
"pmids_with_text": 1431,
|
| 156 |
+
"mapping_coverage": 0.337400281557954,
|
| 157 |
+
"text_coverage": 0.3357578601595495
|
| 158 |
+
}
|
| 159 |
+
}
|
| 160 |
+
}
|
evaluate.py
ADDED
|
@@ -0,0 +1,71 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python3
|
| 2 |
+
"""
|
| 3 |
+
Evaluation script for Variant-Drug Annotation Benchmark
|
| 4 |
+
"""
|
| 5 |
+
|
| 6 |
+
import pandas as pd
|
| 7 |
+
import json
|
| 8 |
+
from typing import Dict, Any
|
| 9 |
+
|
| 10 |
+
def exact_match_score(predicted, ground_truth):
|
| 11 |
+
"""Calculate exact match accuracy"""
|
| 12 |
+
if pd.isna(predicted) and pd.isna(ground_truth):
|
| 13 |
+
return 1.0
|
| 14 |
+
if pd.isna(predicted) or pd.isna(ground_truth):
|
| 15 |
+
return 0.0
|
| 16 |
+
return 1.0 if str(predicted).strip() == str(ground_truth).strip() else 0.0
|
| 17 |
+
|
| 18 |
+
def evaluate_predictions(predictions_file: str, ground_truth_file: str) -> Dict[str, Any]:
|
| 19 |
+
"""Evaluate model predictions against ground truth"""
|
| 20 |
+
|
| 21 |
+
predictions_df = pd.read_csv(predictions_file, sep='\t')
|
| 22 |
+
ground_truth_df = pd.read_csv(ground_truth_file, sep='\t')
|
| 23 |
+
|
| 24 |
+
# Core fields for evaluation
|
| 25 |
+
core_fields = [
|
| 26 |
+
'Variant/Haplotypes', 'Gene', 'Drug(s)',
|
| 27 |
+
'Phenotype Category', 'Significance',
|
| 28 |
+
'Is/Is Not associated', 'Direction of effect'
|
| 29 |
+
]
|
| 30 |
+
|
| 31 |
+
results = {}
|
| 32 |
+
|
| 33 |
+
for field in core_fields:
|
| 34 |
+
if field in predictions_df.columns and field in ground_truth_df.columns:
|
| 35 |
+
scores = []
|
| 36 |
+
for i in range(min(len(predictions_df), len(ground_truth_df))):
|
| 37 |
+
pred_val = predictions_df.iloc[i][field]
|
| 38 |
+
true_val = ground_truth_df.iloc[i][field]
|
| 39 |
+
scores.append(exact_match_score(pred_val, true_val))
|
| 40 |
+
|
| 41 |
+
results[field] = {
|
| 42 |
+
'accuracy': sum(scores) / len(scores) if scores else 0.0,
|
| 43 |
+
'total_examples': len(scores)
|
| 44 |
+
}
|
| 45 |
+
|
| 46 |
+
# Overall accuracy
|
| 47 |
+
field_accuracies = [result['accuracy'] for result in results.values()]
|
| 48 |
+
results['overall'] = {
|
| 49 |
+
'accuracy': sum(field_accuracies) / len(field_accuracies) if field_accuracies else 0.0,
|
| 50 |
+
'total_fields': len(results)
|
| 51 |
+
}
|
| 52 |
+
|
| 53 |
+
return results
|
| 54 |
+
|
| 55 |
+
if __name__ == "__main__":
|
| 56 |
+
import argparse
|
| 57 |
+
|
| 58 |
+
parser = argparse.ArgumentParser(description="Evaluate predictions")
|
| 59 |
+
parser.add_argument("predictions", help="Path to predictions TSV file")
|
| 60 |
+
parser.add_argument("ground_truth", help="Path to ground truth TSV file")
|
| 61 |
+
parser.add_argument("--output", help="Output JSON file for results")
|
| 62 |
+
|
| 63 |
+
args = parser.parse_args()
|
| 64 |
+
|
| 65 |
+
results = evaluate_predictions(args.predictions, args.ground_truth)
|
| 66 |
+
|
| 67 |
+
if args.output:
|
| 68 |
+
with open(args.output, 'w') as f:
|
| 69 |
+
json.dump(results, f, indent=2)
|
| 70 |
+
else:
|
| 71 |
+
print(json.dumps(results, indent=2))
|
test.jsonl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:841749055585002cc12a17982f020a48436e4f617848c7f7f6daadfaa4fe2b10
|
| 3 |
+
size 39851601
|
test/annotations.tsv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
test/texts/PMC10179231.md
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
test/texts/PMC10278212.md
ADDED
|
@@ -0,0 +1,253 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# The Impact of Single Nucleotide Polymorphisms on the Pharmacokinetics of Tacrolimus in Kidney Allograft Recipients of Northern-West, Iran
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Elaheh Jabbari Hagh, Ali Mousavi, Seyyedeh Mina Hejazian, Mehdi Haghi, Samaneh Esfahanian, Elham Ahmadian, Sepideh Zununi Vahed, Mohammadreza Ardalan
|
| 5 |
+
**Journal:** Advanced Pharmaceutical Bulletin
|
| 6 |
+
**Date:** 2022 Jan 2
|
| 7 |
+
**DOI:** [10.34172/apb.2023.038](https://doi.org/10.34172/apb.2023.038)
|
| 8 |
+
**PMID:** 37342387
|
| 9 |
+
**PMCID:** PMC10278212
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10278212/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC10278212/pdf/apb-13-393.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC10278212/pdf/apb-13-393.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
Purpose: Calcineurin inhibitors (CNIs) such as tacrolimus are a major immunosuppressive therapy after renal transplantation, which inhibit cytokine expression. The pharmacokinetics of such drugs is influenced by cytochrome P450 (CYP) enzymes, multi-drug resistance-1 (MDR-1), and C25385T pregnane X receptor (PXR). This study aimed to investigate the impact of single nucleotide polymorphisms (SNP) in these genes on the ratio of tacrolimus level per drug dosage (C/D ratio), acute graft rejection, and viral infections.
|
| 16 |
+
|
| 17 |
+
Methods: Kidney transplantation recipients (n=65) under similar immunosuppressive treatment were included. Amplification refractory mutation systempolymerase chain reaction (ARMS-PCR) method was applied to amplify the loci containing the SNPs of interest.
|
| 18 |
+
|
| 19 |
+
Results: Overall, 65 patients with a male/female ratio of 37/28 were included. The mean age was 38±1.75 years. The variant allele frequencies of CYP3A5*3, MDR-1 C3435T, and PXR C25385T were 95.38, 20.77, and 26.92%, respectively. No significant correlations were found between the studied SNPs and the tacrolimus C/D ratios. However, there was a significant difference in the C/D ratios at 2 and 8 weeks in homozygote CYP3A5 *3/*3 carriers (P=0.015). No significant association was found between the studied polymorphisms and viral infections and acute graft rejection (P>0.05).
|
| 20 |
+
|
| 21 |
+
Conclusion: Homozygote CYP3A5 *3/*3 genotype could influence the tacrolimus metabolism rate (C/D ratio).
|
| 22 |
+
|
| 23 |
+
Keywords: CYP 3A5 gene, Polymorphism, Tacrolimus, Kidney transplantation, PXR gene
|
| 24 |
+
|
| 25 |
+
**Keywords:**Keywords: CYP 3A5 gene, Polymorphism, Tacrolimus, Kidney transplantation, PXR gene
|
| 26 |
+
|
| 27 |
+
## Introduction
|
| 28 |
+
|
| 29 |
+
Calcineurin inhibitors (CNI) are administered in 96% of allograft recipients and provided significant benefits in short-term graft survival.^1^[1](#R1)1 However, it is shown that the long-term survival of the graft is limited by the nephrotoxicity of these drugs.^2^[2](#R2)2 In a 10-year histopathologic follow-up of the transplanted kidney grafts, the use of CNIs was associated with decreased early subclinical rejection while progressive arteriolar hyalinosis, glomerulosclerosis, and tubulointerstitial damage were observed.^3^[3](#R3)3 Although tacrolimus, in contrast to cyclosporine, causes less pathological changes in the transplanted allografts within the first year, its pathological insults were similar to those of cyclosporine’s after a long time.^4^[4](#R4)4 Since tacrolimus has therapeutic benefit with detrimental effects on graft survival, understanding its pharmacokinetics and interactions is important.^5^[5](#R5)5
|
| 30 |
+
|
| 31 |
+
Tacrolimus is metabolized by cytochrome P450 (CYP) 3A4 and 3A5 enzymes and variations in their coding genes were shown to be associated with altered drug clearance.^6^[6](#R6)6 Patients with high clearance of tacrolimus at first 90 days after transplantation are at an increased risk of acute graft rejection.^7^[7](#R7)7 CYP 3A4*22 T carriers require fewer doses of tacrolimus.^8^[8](#R8)8 The CYP 3A5*1 allele, which encodes the functional enzyme, causes a two-fold decrease in dose-normalized plasma levels of tacrolimus.^9^[9](#R9)9 This allele is more frequent among the patients with African-Americans ethnicity and associated with a 50% increase in total dosage requirements of tacrolimus to reach the first therapeutic trough level.^10^[10](#R10)10 In a study, among 337 renal allograft recipients, the presence of the CYP 3A4*22 allele and expression of CYP 3A5 were predictors of 20% and 160% clearance fluctuation, respectively.^11^[11](#R11)11 Thus, a personalized medicinal approach is necessary to determine the tacrolimus dosing to maintain therapeutic range in renal allograft recipients.
|
| 32 |
+
|
| 33 |
+
The A6986G (rs776746) single nucleotide polymorphism (SNP) in the *CYP 3A*CYP 3A5 gene has been a focus of researchers during recent years. This SNP is located on the 3^rd^rd intron of the gene and impacts the CYP 3A5 expression.^12^[12](#R12)12 However, there are conflicting findings among the studies. Additionally, two other SNPs including the C3435T of multi-drug resistance-1 (MDR-1) and C25385T pregnane X receptor (PXR, also known as nuclear receptor 112) genes were also investigated for their impact on the pharmacokinetics of tacrolimus. The obtained results are widely different across populations with different genetic backgrounds.
|
| 34 |
+
|
| 35 |
+
In the clinics, kidney recipients differently respond to a given pharmacological therapy, some of whom cannot reach the designated concentration(s) with recommended starting doses. The over-dosing of the drug increases the risks of nephrotoxicity, infection, and other severe drug-specific side effects, while its under-dosing may lead to the acute rejection. Hence, the CNIs management is a challenging issue in transplant patients. Considering that tacrolimus is widely used in transplant immunosuppression and there are inter-individual differences in respond to the drug, understanding the patients’ pharmacogenetics and personalized administration of the drug could be helpful in different populations. Therefore, in this study, the influence of the *CYP*CYP3A5, *MDR*MDR-1, and *PXR*PXR genes SNPs on the pharmacokinetics of tacrolimus and the ratio of concentration per dose of tacrolimus (C/D ratio) were evaluated in a group of kidney recipients in Northwest of Iran.
|
| 36 |
+
|
| 37 |
+
## Methods
|
| 38 |
+
|
| 39 |
+
### Patients
|
| 40 |
+
|
| 41 |
+
In this cross-sectional study, renal allograft recipients receiving tacrolimus were included. During the study period, renal transplant recipients with an age range of 20-60 years under triple therapy with tacrolimus (Prograf^®^® manufactured by Astellas Company, in 2 divided doses), mycophenolate mofetil (MMF), and prednisolone were included. Patients with ongoing acute allograft rejection, BK virus nephropathy, hepatitis, and cytomegalovirus (CMV) infections were excluded from the study.
|
| 42 |
+
|
| 43 |
+
### Laboratory examinations
|
| 44 |
+
|
| 45 |
+
A baseline laboratory examination including plasma creatinine, blood urea nitrogen, lipid profile, total bilirubin, and 25-hydroxy vitamin D levels were measured. The measurement of plasma levels of tacrolimus trough level was performed exactly 12 hours after the last dose of the drug and immediately before taking the next dose. Tacrolimus metabolism rate (C/D ratio) was calculated by dividing the blood concentration of the drug (C, ng/mL) to the daily dose of Tacrolimus (D, mg) at 2, 4 and 8 weeks after transplantation.
|
| 46 |
+
|
| 47 |
+
The genomic DNA was extracted from blood with the magnetic nanoparticle-based method using ZiAViZ^®^® DNA extraction kit (Itan). Amplification refractory mutation systempolymerase chain reaction (ARMS-PCR) method was performed using primers ([Table 1](#T1)Table 1) designed based on previous studies.^13,14^[13](#R13)13,[14](#R14)14 The commercial master mix kit (Sinaclon-Iran, #CatNo. MM2062) was used for the PCR procedure (Mastercycler Eppendorf, Germany).
|
| 48 |
+
|
| 49 |
+
### Table 1. The sequencing of the primers used in PCR .
|
| 50 |
+
|
| 51 |
+
| Primers | Type | Sequence (5’→3’) |
|
| 52 |
+
| ------- | ---- | ---------------- |
|
| 53 |
+
| CYP 3A5 (A6986G) | Forward | CACTTGATGATTTACCTGCCTTC |
|
| 54 |
+
| Wild-type reverse | GGTCCAAACAGGGAAGAGATAA | |
|
| 55 |
+
| Mutant reverse | GGTCCAAACAGGGAAGAGATAC | |
|
| 56 |
+
| MDR1 (C3435T) | Forward | ACTATAGGCCAGAGAGGCTGC |
|
| 57 |
+
| Wild-type reverse | GTGGTGTCACAGGAAGAGCTT | |
|
| 58 |
+
| Mutant reverse | GTGGTGTCACAGGAAGAGCTC | |
|
| 59 |
+
| NR1I2 (C25385T) | Forward | ACCACGATTGAGCAAACAGGTA |
|
| 60 |
+
| Wild-type reverse | TGGTCATTTTTTGGCAATCCCAGGTTC | |
|
| 61 |
+
| Mutant reverse | TGGTCATTTTTTGGCAATCCCAGGTTT | |
|
| 62 |
+
### Statistical analysis
|
| 63 |
+
|
| 64 |
+
The Hardy-Weinberg equilibrium was checked for each SNP. IBM SPSS Statistics version 24.0 was used for statistical analysis. Descriptive statistics demonstrates the frequency, percentage, mean, and standard deviation (SD). A logistic regression model with a confidence interval (CI) of 95% was applied for the evaluation of the correlations among the variables. ANOVA or Kruskal-Wallis tests were applied to compare the differences between tacrolimus C/D ratio at different time points (2, 4, and 8 weeks). The chi-square test was applied to check the correlation between the viral infections and graft rejection *vs.*vs. SNP genotypes. A *P*P value of < 0.05 was considered significant.
|
| 65 |
+
|
| 66 |
+
## Results and Discussion
|
| 67 |
+
|
| 68 |
+
In total, 65 renal recipients were included in the study with a male/female ratio of 37:28. The average age was 38 ± 14.17 years old. The demographic data of the patients are presented in [Table 2](#T2)Table 2. It is reported that in the CYP3A5 nonexpressers, genotyping of NR1I2/ MDR1/ CYP3A4 polymorphisms can be useful for controlling tacrolimus dosing.^15^[15](#R15)15 In the present study, the variant allele frequencies of CYP3A5*3, MDR-1 C3435T, and PXR C25385T were 95.38, 20.77, and 26.92%, respectively.
|
| 69 |
+
|
| 70 |
+
### Table 2. The baseline demographic and laboratory data .
|
| 71 |
+
|
| 72 |
+
| Variables | Values |
|
| 73 |
+
| --------- | ------ |
|
| 74 |
+
| Gender (male/female) | 37/28 |
|
| 75 |
+
| Age (y) | 38 ± 14.17 |
|
| 76 |
+
| Body mass index (kg/m2) | 24.23 ± 5.14 |
|
| 77 |
+
| Height (cm) | 162.47 ± 12.2 |
|
| 78 |
+
| Weight (kg) | 63.58 ± 14.83 |
|
| 79 |
+
| Blood urea nitrogen (mg/dL) | 56.38 ± 28.71 |
|
| 80 |
+
| Creatinine (mg/dL) | 1.11 (0.54-9.2) |
|
| 81 |
+
| Triglycerides (mg/dL) | 143 (29-592) |
|
| 82 |
+
| Total cholesterol (mg/dL) | 171.52 ± 49.39 |
|
| 83 |
+
| High-density lipoprotein (mg/dL) | 43.09 ± 7.38 |
|
| 84 |
+
| Low-density lipoprotein (mg/dL) | 92.5 (0-180) |
|
| 85 |
+
| 25-Hydroxy vitamin D3 (ng/ml) | 17.79 ± 13.22 |
|
| 86 |
+
### Allele frequency and genotype distribution of the CYP3A5*3
|
| 87 |
+
|
| 88 |
+
Most of the kidney recipients (90.8%, n = 59) were homozygotes for the *CYP3A5*3*CYP3A5*3 allele (*CYP3A5 *3/*CYP3A5 *3/*3, non-expressers CYP3A5), while 9.2% of the participants (n = 6) were heterozygous for the CYP3A5*3 (*CYP3A5 *1/*CYP3A5 *1/*3 genotype, functional CYP3A5 expressers). Across different populations, the frequency of CYP3A5*3 SNP differs widely. In our kidney recipient population, the frequency of the CYP3A5*3 allele was 95%. The result of a meta‐analysis has stated that the allele frequency of CYP3A5*3 is higher in European (94%), admixed American (80%), East Asian (71%), and South Asian (67%) subjects, while it is low (33%) in African subjects.^16^[16](#R16)16 Another meta-analysis (2015) concluded that the CYP3A5 A6986G SNP could affect the pharmacokinetics of tacrolimus.^17^[17](#R17)17 In the carriers of the wild-type allele (CYP3A5*1, expresser genotype, fast metabolizers), the expression of CYP3A5 leads to an elevated metabolism and clearance of tacrolimus and a lower C/D ratio compared to the other genotypes (*CYP3A5*1/*3*CYP3A5*1/*3 and *CYP3A5*3/*3*CYP3A5*3/*3 carriers). South African kidney transplant recipients in comparison with global transplant populations have a high rate of CYP3A5 expression that significantly affects the pharmacokinetics of tacrolimus.^18^[18](#R18)18 However, some studies did not find a statistically significant correlation between the A6986G SNP and higher clearance of tacrolimus^19^[19](#R19)19 being in agreement with our study.
|
| 89 |
+
|
| 90 |
+
### Allele frequency and genotype distribution of the MDR-1 C3435T
|
| 91 |
+
|
| 92 |
+
The frequency of the C and T alleles in *MDR*MDR-1 C3435T gene was 79.23 and 20.77%, respectively. The genotypes distribution and allele frequency of the studied polymorphism are presented in [Table 3](#T3)Table 3. The MDR-1 affects the pharmacodynamics of tacrolimus by active transporting of a wide variety of drugs to the outsides of the cells.^20^[20](#R20)20 Similar to the CYP3A5, wild-type carriers of at least one MDR-1 C allele are expected to express more P-gp. The MDR1 C3435T SNP is a synonymous mutation in which the ATC codon changes to ATT (Ile1145Ile), causing alteration in P-gp activity.^21-24^[21](#R21)21-[24](#R24)24 A higher activity of the MDR-1 limits the enteral absorption of tacrolimus. Similar to the results of Loh et al,^25^[25](#R25)25 67.7% of our recipients were homozygote C allele carriers in contrast to other reports.^26^[26](#R26)26 In addition, study by Tada et al^27^[27](#R27)27 revealed that the MDR1 C3435T polymorphism did not affect tacrolimus pharmacokinetics of renal transplanted patients. Among the Asian population, SNPs in the *MDR*MDR-1 and *CYP*CYP3A5 genes were shown to influence the plasma levels of tacrolimus but not cyclosporine. Moreover, it has been proposed that diltiazem, a non-dihydropyridine calcium channel blocker, may help achieve the optimum levels of tacrolimus by blocking the MDR-1 and CYP 3A4 proteins.^25^[25](#R25)25
|
| 93 |
+
|
| 94 |
+
### Table 3. Allele and genotype frequencies of the studied polymorphisms in kidney recipients .
|
| 95 |
+
|
| 96 |
+
| Polymorphisms | Genotype | Allelic status | Genotype frequencies, n (%) | Allelic frequencies, n (%)* |
|
| 97 |
+
| ------------- | -------- | -------------- | --------------------------- | --------------------------- |
|
| 98 |
+
| CYP 3A5 A6986G | *1/*1 (AA) | A | 0 (0%) | A | G |
|
| 99 |
+
| *1/*3 (AG) | A | 6 (9.2%) | 4.62% | 95.38% |
|
| 100 |
+
| *3/*3 (GG) | G | 59 (90.8%) | | |
|
| 101 |
+
| MDR-1 C3435T SNP | CC | C | 44 (67.7%) | C | T |
|
| 102 |
+
| CT | C | 15 (23.1%) | 79.23% | 20.77% |
|
| 103 |
+
| TT | T | 6 (9.2%) | | |
|
| 104 |
+
| PXR C25385T SNP | CC | C | 40 (61.5%) | C | T |
|
| 105 |
+
| CT | C | 15 (23.1%) | 73.08% | 26.92% |
|
| 106 |
+
| TT | T | 10 (15.4%) | | |
|
| 107 |
+
### Allele frequency and genotype distribution of the PXR C25385T SNP
|
| 108 |
+
|
| 109 |
+
The frequency of the C allele in the *PXR*PXR genes was 73.08%. The genotypes distribution and allele frequency of this SNP are presented in [Table 3](#T3)Table 3. The *PXR*PXR gene, encoding a nuclear receptor, is involved in the regulation of CYP3A enzymes, MDR-1, and other several proteins and therefore, plays a crucial role in the pharmacokinetics of tacrolimus.^28^[28](#R28)28 However, there are contradictions about its effect on the pharmacokinetics of tacrolimus.^29^[29](#R29)29 This study revealed that the frequency of the PXR C25385T allele was 26.92%.
|
| 110 |
+
|
| 111 |
+
### The correlation between clinical variables and gene polymorphisms
|
| 112 |
+
|
| 113 |
+
The median (min-max) C/D ratio (ng/mL/mg) for the kidney recipients at 2, 4, and 8 weeks were 1.2 (0.13-5), 1.68 (0.32-10.67), and 2 (0.33-13), respectively. There was a significant difference in C/D ratios at 2 and 8 weeks (P = 0.015). In addition, the C/D ratios of patients with different genotypes in the studied genes are given in [Table 4](#T4)Table 4. The mean of C/D ratios in the non-expresser group compared to the expresser group were 1.56 ± 1.1 *vs*vs. 0.83 ± 0.16 ng/mL/mg at 2 weeks, 2.01 ± 1.0 *vs*vs. 1.75 ± 0.91 ng/mL/mg at 4 weeks, and 2 *vs*vs. 1.2 at 8 weeks after renal transplantation, respectively. There was a significant difference when C/D ratios of homozygote *CYP3A5 *3/*CYP3A5 *3/*3 carriers were compared between 2 and 8 weeks (*P*P = 0.012).
|
| 114 |
+
|
| 115 |
+
### Table 4. The C/D ratios in 2nd, 4th and 8th weeks of tacrolimus administration in relation to SNP genotypes .
|
| 116 |
+
|
| 117 |
+
| SNPs | Genotype | Week 2 | Week 4 | Week 8 | P value* |
|
| 118 |
+
| ---- | -------- | ------ | ------ | ------ | -------- |
|
| 119 |
+
| Concentration | Daily dose | C/D ratio | Concentration | Daily dose | C/D ratio | Concentration | Daily dose | C/D ratio |
|
| 120 |
+
| CYP 3A5 A6986G | *1/*1 | - | - | - | - | - | - | - | - | - | 0.217a |
|
| 121 |
+
| *1/*3 | 5.66±0.57 | 7±1.73 | 0.83±0.16 | 10±3.6 | 6±1 | 1.75±0.91 | 9±4.35 | 5.33±0.57 | 1.2 (1.2-2.8) | 0.01 b |
|
| 122 |
+
| *3/*3 | 7.96±4.07 | 5.71±2.2 | 1.56±1.1 | 9.95±4.68 | 4.97±2.08 | 2.01±1.0 | 8.68±3.22 | 4.55±2.17 | 2 (0.33-13) | 0.99c |
|
| 123 |
+
| MDR-1 C3435T | CC | 7.71±3.68 | 5.66±2.38 | 1.57±1.03 | 9.07±4.88 | 5.03±2.24 | 1.66 (0.5-10.67) | 8.73±3.26 | 4.79±2.39 | 2.23±1.28 | 0.88a |
|
| 124 |
+
| CT | 9 (2-20) | 5.77±1.64 | 1.25 (0.29-5) | 8.11±4.45 | 4.88±1.83 | 1.69±0.6 | 8.77±3.19 | 3.83±1.62 | 2 (1-13) | 0.13b |
|
| 125 |
+
| TT | 4.8±1.09 | 6.6±2.07 | 0.8±0.4 | 6.32±2.83 | 5.4±1.34 | 1.28 (0.33-2) | 8.44±4.03 | 5±1.22 | 1.72±0.83 | 0.88c |
|
| 126 |
+
| PXR C25385T | CC | 6.46±2.79 | 5.48±1.76 | 1.25 (0.29-4.5) | 8.6±3.85 | 5.01±1.62 | 1.66 (0.33-10.6) | 8.67±3.31 | 4.59±1.42 | 2 (0.6-4.6) | 1.01a |
|
| 127 |
+
| CT | 9.8±5.67 | 6.5±3.13 | 1.35 (0.4-5) | 6.2 (3-22) | 4 (2-12) | 1.42 (0.5-4.86) | 7.99±3.45 | 3.25 (2-12) | 2 (0.33-4) | 0.13b |
|
| 128 |
+
| TT | 9.25±3.68 | 6.25±2.06 | 1.48±0.34 | 6.7±1.24 | 4.25±0.5 | 1.61±0.42 | 10.72±1.74 | 3.12±1.43 | 2.88 (2.2-13) | 0.13c |
|
| 129 |
+
| Total | 6 (1-23) | 5.66±2.07 | 1.2 (0.13-5) | 7.2 (1.9-32) | 4.95±1.95 | 1.68 (0.32-10.67) | 8.82±3.49 | 4.42±2.04 | 2 (0.33-13) | 0.19a |
|
| 130 |
+
| 0.01b | | | | | |
|
| 131 |
+
| 0.93c | | | | | |
|
| 132 |
+
Six patients experienced acute graft rejection, none of the SNPs seemed to have a significant impact on rejection rates (*P*P> 0.05). In addition, 10 patients experienced viral infections (CMV, BK virus, or both), however, none of the SNP genotypes had a significant predisposing influence on them (*P*P> 0.05).
|
| 133 |
+
|
| 134 |
+
### Pharmacogenetics and tacrolimus metabolism
|
| 135 |
+
|
| 136 |
+
Based on the studied genotypes and C/D ratios after 2, 4, and 8 weeks, renal transplant recipients were categorized into three groups ([Table 5](#T5)Table 5) based on the rate of tacrolimus metabolism. The groups were included C/D ratio < 1.05 (ng/mL/mg)/ (mg/kg/d) as fast metabolizers, 1.05 < C/D ratio < 1.55 (ng/mL/mg)/ (mg/kg/d) as intermediate metabolizers, and C/D ratio > 1.55 (ng/mL/mg)/ (mg/kg/d) as slow metabolizers, Thölking et al.^30^[30](#R30)30
|
| 137 |
+
|
| 138 |
+
### Table 5. Category of patients based on C/D ratio of Tacrolimus .
|
| 139 |
+
|
| 140 |
+
| Polymorphisms | Genotype | Metabolizers, n (mean±SD) |
|
| 141 |
+
| ------------- | -------- | ------------------------- |
|
| 142 |
+
| Fast metabolizers | Intermediate metabolizers | Slow metabolizers |
|
| 143 |
+
| CYP 3A5 A6986G SNP | *1/*1 | - | - | - |
|
| 144 |
+
| *1/*3 | 1 (1 ± 0.0) | 1 (1.06 ± 0.0) | 4 (2.29 ± 0.76) |
|
| 145 |
+
| *3/*3 | 17 (0.78 ± 0.18) | 7 (1.3 ± 0.12) | 30 (2.78 ± 1.28) |
|
| 146 |
+
| MDR-1 C3435T SNP | CC | 13 (0.76 ± 0.2) | 7 (1.28 ± 0.16) | 23 (2.62 ± 0.99) |
|
| 147 |
+
| CT | 2 (0.86 ± 0.12) | 2 (1.33 ± 0.007) | 9 (3.21 ± 1.74) |
|
| 148 |
+
| TT | 3 (0.91 ± 0.11) | 1 (1.33) | 2 (1.7 ± 0.16) |
|
| 149 |
+
| PXR C25385T SNP | CC | 12 (0.82 ± 0.18) | 7 (1.26 ± 0.11) | 20 (2.69 ± 1.4) |
|
| 150 |
+
| CT | 4 (0.69 ± 0.2) | 2 (1.44 ± 0.08) | 8 (2.81 ± 0.61) |
|
| 151 |
+
| TT | 2 (0.86 ± 0.19) | 1 (1.08) | 6 (2.74 ± 1.42) |
|
| 152 |
+
| Total | 19 (0.83 ± 0.22) | 8 (1.24 ± 0.12) | 34 (2.72 ± 1.23) |
|
| 153 |
+
It is reported that the expresser genotype (with a C/D ratio < 1.05) is correlated with a higher risk of chronic nephrotoxicity and acute rejection and a lower estimated glomerular filtration rate (eGFR) compared to slow and intermediate tacrolimus metabolizers.^17,31,32^[17](#R17)17,[31](#R31)31,[32](#R32)32 In the present study, about half of the kidney recipients with *CYP3A5*3/*3*CYP3A5*3/*3 genotype were slow-metabolizers, confirming the impact of other genes in the pharmacokinetics of tacrolimus.
|
| 154 |
+
|
| 155 |
+
It is shown that the 3435 C/C genotype of *MDR*MDR-1 gene is associated with 40% lower C/D ratios.^29^[29](#R29)29 Since drug concentrations accomplished with a standard dose will be lower for carriers of the wild-type C allele, C/D ratios is expected to be lower. Dissimilar to this expectation, our patients with the C allele had higher C/D ratios at week 2; 1.57 ± 1.03 for homozygotes and 1.25 (min-max 0.29-5) for heterozygotes versus 0.8 ± 0.4 T allele. The SNPs in *CYP*CYP3As and *MDR*MDR-1 genes are attracting attention in clinical practice.^33^[33](#R33)33 Our results showed that the frequency of the T allele was 20.77% among renal transplanted, 3 of the carriers were fast metabolizers with a 0.91 ± 0.11 C/D ratio and 2 patients were slow metabolizers with a 1.7 ± 0.16 C/D ratio. However, this polymorphism did no significant effect on the metabolism of tacrolimus.
|
| 156 |
+
|
| 157 |
+
Kurzawsk et al demonstrated that minor T allele carriers (CT or TT genotype) of the PXR C25385T require significantly low tacrolimus dose administration particularly in the first 6 months after renal transplantation compared to CC genotypes.^34^[34](#R34)34 In the present study, 2 of fast metabolizers with PXR SNP TT genotype had a 0.86 ± 0.19 C/D ratio and 6 patients were slow metabolizers with a 2.74 ± 1.42 C/D ratio. There were no significant differences between the studied polymorphism and fast/ intermediate/slow metabolizers ([Table 5](#T5)Table 5).
|
| 158 |
+
|
| 159 |
+
The nephrotoxicity of CNIs has led to the assumption that perhaps lowering the dosage of CNIs or utilizing a less toxic agent (e.g. belatacept, sirolimus) would be a safer option.^2,35^[2](#R2)2,[35](#R35)35 On the contrary, the triple regimen of tacrolimus, MMF, and a corticosteroid is ubiquitously used at present.^36^[36](#R36)36 It is shown that lower plasma levels of tacrolimus with a higher dosage of MMF are associated with better outcomes.^37^[37](#R37)37 Scholten et al developed a flexible method for accurate dosing of tacrolimus regardless of the genetic variables which is an ongoing field of research.^38^[38](#R38)38 There are somewhat controversial findings from other studies. We believe that the differences among the methodologies, tacrolimus release (immediate or extended), and perhaps ethnic differences may lead to such results.
|
| 160 |
+
|
| 161 |
+
## Conclusion
|
| 162 |
+
|
| 163 |
+
In this study, the frequency of the CYP3A5*3 allele was 95%. There was a significant difference in the C/D ratios at 2 and 8 weeks among homozygote *CYP3A5 *3/*CYP3A5 *3/*3 carriers, suggested that choosing the initial dosage according to the CYP3A5 genotype may result in a better outcome. No significant correlations were found between the *MDR*MDR-1, and *PXR*PXR gene SNPs and the tacrolimus C/D ratios. These result will be a step toward personalized medicine and may prolong the graft survival in renal allograft recipients. Pre-transplant genetic study of recipient could prevents low drug level rejection or high level nephrotoxicity to occur.
|
| 164 |
+
|
| 165 |
+
## Acknowledgments
|
| 166 |
+
|
| 167 |
+
The authors greatly appreciate the Kidney Research Center, Tabriz University of Medical Sciences, Tabriz, Iran for scientific and financial support (Grant number: 59763). This paper was extracted from the residential thesis of Elaheh Jabbari-Hagh, MD. Authors also thanks Transplantation Ward of Imam Reza Hospital of Tabriz University of Medical Science, Tabriz, Iran.
|
| 168 |
+
|
| 169 |
+
## Competing Interests
|
| 170 |
+
|
| 171 |
+
Authors declare no conflicts of interest regarding this manuscript.
|
| 172 |
+
|
| 173 |
+
## Ethical Approval
|
| 174 |
+
|
| 175 |
+
The Helsinki Declaration of ethics in medical research was honored in this study. The informed written consent was obtained from all patients. The study was approved by the ethics committee of Tabriz University of Medical Sciences, Tabriz, Iran and is registered with the following ethical code: IR.TBZMED.REC.1397.128.
|
| 176 |
+
|
| 177 |
+
## References
|
| 178 |
+
|
| 179 |
+
1. Rush D. The impact of calcineurin inhibitors on graft survival. Transplant Rev (Orlando) 2013;27(3):93–5. doi: 10.1016/j.trre.2013.04.003. [DOI](https://doi.org/10.1016/j.trre.2013.04.003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23743217/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplant%20Rev%20(Orlando)&title=The%20impact%20of%20calcineurin%20inhibitors%20on%20graft%20survival&author=D%20Rush&volume=27&issue=3&publication_year=2013&pages=93-5&pmid=23743217&doi=10.1016/j.trre.2013.04.003&)
|
| 180 |
+
|
| 181 |
+
2. Casey MJ, Meier-Kriesche HU. Calcineurin inhibitors in kidney transplantation: friend or foe? CurrOpin Nephrol Hypertens. 2011;20(6):610–5. doi: 10.1097/MNH.0b013e32834b4343. [DOI](https://doi.org/10.1097/MNH.0b013e32834b4343) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21885969/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=CurrOpin%20Nephrol%20Hypertens&title=Calcineurin%20inhibitors%20in%20kidney%20transplantation:%20friend%20or%20foe?&author=MJ%20Casey&author=HU%20Meier-Kriesche&volume=20&issue=6&publication_year=2011&pages=610-5&pmid=21885969&doi=10.1097/MNH.0b013e32834b4343&)
|
| 182 |
+
|
| 183 |
+
3. Nankivell BJ, Borrows RJ, Fung CL, O’Connell PJ, Allen RD, Chapman JR. The natural history of chronic allograft nephropathy. N Engl J Med. 2003;349(24):2326–33. doi: 10.1056/NEJMoa020009. [DOI](https://doi.org/10.1056/NEJMoa020009) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14668458/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=The%20natural%20history%20of%20chronic%20allograft%20nephropathy&author=BJ%20Nankivell&author=RJ%20Borrows&author=CL%20Fung&author=PJ%20O%E2%80%99Connell&author=RD%20Allen&volume=349&issue=24&publication_year=2003&pages=2326-33&pmid=14668458&doi=10.1056/NEJMoa020009&)
|
| 184 |
+
|
| 185 |
+
4. Nankivell BJ, PʼNg CH, OʼConnell PJ, Chapman JR. Calcineurin inhibitor nephrotoxicity through the lens of longitudinal histology: comparison of cyclosporine and tacrolimus eras. Transplantation. 2016;100(8):1723–31. doi: 10.1097/tp.0000000000001243. [DOI](https://doi.org/10.1097/tp.0000000000001243) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27306529/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplantation&title=Calcineurin%20inhibitor%20nephrotoxicity%20through%20the%20lens%20of%20longitudinal%20histology:%20comparison%20of%20cyclosporine%20and%20tacrolimus%20eras&author=BJ%20Nankivell&author=CH%20P%CA%BCNg&author=PJ%20O%CA%BCConnell&author=JR%20Chapman&volume=100&issue=8&publication_year=2016&pages=1723-31&pmid=27306529&doi=10.1097/tp.0000000000001243&)
|
| 186 |
+
|
| 187 |
+
5. Venkataramanan R, Swaminathan A, Prasad T, Jain A, Zuckerman S, Warty V, et al. Clinical pharmacokinetics of tacrolimus. Clin Pharmacokinet. 1995;29(6):404–30. doi: 10.2165/00003088-199529060-00003. [DOI](https://doi.org/10.2165/00003088-199529060-00003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8787947/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacokinet&title=Clinical%20pharmacokinetics%20of%20tacrolimus&author=R%20Venkataramanan&author=A%20Swaminathan&author=T%20Prasad&author=A%20Jain&author=S%20Zuckerman&volume=29&issue=6&publication_year=1995&pages=404-30&pmid=8787947&doi=10.2165/00003088-199529060-00003&)
|
| 188 |
+
|
| 189 |
+
6. Campagne O, Mager DE, Brazeau D, Venuto RC, Tornatore KM. Tacrolimus population pharmacokinetics and multiple CYP3A5 genotypes in black and white renal transplant recipients. J Clin Pharmacol. 2018;58(9):1184–95. doi: 10.1002/jcph.1118. [DOI](https://doi.org/10.1002/jcph.1118) | [PMC free article](/articles/PMC6105387/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29775201/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Clin%20Pharmacol&title=Tacrolimus%20population%20pharmacokinetics%20and%20multiple%20CYP3A5%20genotypes%20in%20black%20and%20white%20renal%20transplant%20recipients&author=O%20Campagne&author=DE%20Mager&author=D%20Brazeau&author=RC%20Venuto&author=KM%20Tornatore&volume=58&issue=9&publication_year=2018&pages=1184-95&pmid=29775201&doi=10.1002/jcph.1118&)
|
| 190 |
+
|
| 191 |
+
7. Egeland EJ, Robertsen I, Hermann M, Midtvedt K, Størset E, Gustavsen MT, et al. High tacrolimus clearance is a risk factor for acute rejection in the early phase after renal transplantation. Transplantation. 2017;101(8):e273–e9. doi: 10.1097/tp.0000000000001796. [DOI](https://doi.org/10.1097/tp.0000000000001796) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28452920/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplantation&title=High%20tacrolimus%20clearance%20is%20a%20risk%20factor%20for%20acute%20rejection%20in%20the%20early%20phase%20after%20renal%20transplantation&author=EJ%20Egeland&author=I%20Robertsen&author=M%20Hermann&author=K%20Midtvedt&author=E%20St%C3%B8rset&volume=101&issue=8&publication_year=2017&pages=e273-e9&pmid=28452920&doi=10.1097/tp.0000000000001796&)
|
| 192 |
+
|
| 193 |
+
8. Hesselink DA, Bouamar R, Elens L, van Schaik RH, van Gelder T. The role of pharmacogenetics in the disposition of and response to tacrolimus in solid organ transplantation. Clin Pharmacokinet. 2014;53(2):123–39. doi: 10.1007/s40262-013-0120-3. [DOI](https://doi.org/10.1007/s40262-013-0120-3) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24249597/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacokinet&title=The%20role%20of%20pharmacogenetics%20in%20the%20disposition%20of%20and%20response%20to%20tacrolimus%20in%20solid%20organ%20transplantation&author=DA%20Hesselink&author=R%20Bouamar&author=L%20Elens&author=RH%20van%20Schaik&author=T%20van%20Gelder&volume=53&issue=2&publication_year=2014&pages=123-39&pmid=24249597&doi=10.1007/s40262-013-0120-3&)
|
| 194 |
+
|
| 195 |
+
9. Macphee IA, Fredericks S, Mohamed M, Moreton M, Carter ND, Johnston A, et al. Tacrolimus pharmacogenetics: the CYP3A5*1 allele predicts low dose-normalized tacrolimus blood concentrations in whites and South Asians. Transplantation. 2005;79(4):499–502. doi: 10.1097/01.tp.0000151766.73249.12. [DOI](https://doi.org/10.1097/01.tp.0000151766.73249.12) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15729180/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplantation&title=Tacrolimus%20pharmacogenetics:%20the%20CYP3A5*1%20allele%20predicts%20low%20dose-normalized%20tacrolimus%20blood%20concentrations%20in%20whites%20and%20South%20Asians&author=IA%20Macphee&author=S%20Fredericks&author=M%20Mohamed&author=M%20Moreton&author=ND%20Carter&volume=79&issue=4&publication_year=2005&pages=499-502&pmid=15729180&doi=10.1097/01.tp.0000151766.73249.12&)
|
| 196 |
+
|
| 197 |
+
10. Maldonado AQ, Asempa T, Hudson S, Rebellato LM. Prevalence of CYP3A5 genomic variances and their impact on tacrolimus dosing requirements among kidney transplant recipients in eastern North Carolina. Pharmacotherapy. 2017;37(9):1081–8. doi: 10.1002/phar.1970. [DOI](https://doi.org/10.1002/phar.1970) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28605053/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacotherapy&title=Prevalence%20of%20CYP3A5%20genomic%20variances%20and%20their%20impact%20on%20tacrolimus%20dosing%20requirements%20among%20kidney%20transplant%20recipients%20in%20eastern%20North%20Carolina&author=AQ%20Maldonado&author=T%20Asempa&author=S%20Hudson&author=LM%20Rebellato&volume=37&issue=9&publication_year=2017&pages=1081-8&pmid=28605053&doi=10.1002/phar.1970&)
|
| 198 |
+
|
| 199 |
+
11. Andrews LM, Hesselink DA, van Schaik RHN, van Gelder T, de Fijter JW, Lloberas N, et al. A population pharmacokinetic model to predict the individual starting dose of tacrolimus in adult renal transplant recipients. Br J Clin Pharmacol. 2019;85(3):601–15. doi: 10.1111/bcp.13838. [DOI](https://doi.org/10.1111/bcp.13838) | [PMC free article](/articles/PMC6379219/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30552703/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=A%20population%20pharmacokinetic%20model%20to%20predict%20the%20individual%20starting%20dose%20of%20tacrolimus%20in%20adult%20renal%20transplant%20recipients&author=LM%20Andrews&author=DA%20Hesselink&author=RHN%20van%20Schaik&author=T%20van%20Gelder&author=JW%20de%20Fijter&volume=85&issue=3&publication_year=2019&pages=601-15&pmid=30552703&doi=10.1111/bcp.13838&)
|
| 200 |
+
|
| 201 |
+
12. Niioka T, Kagaya H, Saito M, Inoue T, Numakura K, Habuchi T, et al. Capability of utilizing CYP3A5 polymorphisms to predict therapeutic dosage of tacrolimus at early stage post-renal transplantation. Int J Mol Sci. 2015;16(1):1840–54. doi: 10.3390/ijms16011840. [DOI](https://doi.org/10.3390/ijms16011840) | [PMC free article](/articles/PMC4307337/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25594874/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int%20J%20Mol%20Sci&title=Capability%20of%20utilizing%20CYP3A5%20polymorphisms%20to%20predict%20therapeutic%20dosage%20of%20tacrolimus%20at%20early%20stage%20post-renal%20transplantation&author=T%20Niioka&author=H%20Kagaya&author=M%20Saito&author=T%20Inoue&author=K%20Numakura&volume=16&issue=1&publication_year=2015&pages=1840-54&pmid=25594874&doi=10.3390/ijms16011840&)
|
| 202 |
+
|
| 203 |
+
13. Fernando ME, Sellappan M, Srinivasa Prasad ND, Suren S, Thirumalvalavan K. Influence of CYP3A5 and ABCB1 polymorphism on tacrolimus drug dosing in South Indian renal allograft recipients. Indian J Nephrol. 2019;29(4):261–6. doi: 10.4103/ijn.IJN_97_18. [DOI](https://doi.org/10.4103/ijn.IJN_97_18) | [PMC free article](/articles/PMC6668310/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31423060/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Indian%20J%20Nephrol&title=Influence%20of%20CYP3A5%20and%20ABCB1%20polymorphism%20on%20tacrolimus%20drug%20dosing%20in%20South%20Indian%20renal%20allograft%20recipients&author=ME%20Fernando&author=M%20Sellappan&author=ND%20Srinivasa%20Prasad&author=S%20Suren&author=K%20Thirumalvalavan&volume=29&issue=4&publication_year=2019&pages=261-6&pmid=31423060&doi=10.4103/ijn.IJN_97_18&)
|
| 204 |
+
|
| 205 |
+
14. Kimura Y, Selmi C, Leung PS, Mao TK, Schauer J, Watnik M, et al. Genetic polymorphisms influencing xenobiotic metabolism and transport in patients with primary biliary cirrhosis. Hepatology. 2005;41(1):55–63. doi: 10.1002/hep.20516. [DOI](https://doi.org/10.1002/hep.20516) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15690482/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Hepatology&title=Genetic%20polymorphisms%20influencing%20xenobiotic%20metabolism%20and%20transport%20in%20patients%20with%20primary%20biliary%20cirrhosis&author=Y%20Kimura&author=C%20Selmi&author=PS%20Leung&author=TK%20Mao&author=J%20Schauer&volume=41&issue=1&publication_year=2005&pages=55-63&pmid=15690482&doi=10.1002/hep.20516&)
|
| 206 |
+
|
| 207 |
+
15. Li JL, Liu S, Fu Q, Zhang Y, Wang XD, Liu XM, et al. Interactive effects of CYP3A4, CYP3A5, MDR1 and NR1I2 polymorphisms on tracrolimus trough concentrations in early postrenal transplant recipients. Pharmacogenomics. 2015;16(12):1355–65. doi: 10.2217/pgs.15.78. [DOI](https://doi.org/10.2217/pgs.15.78) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26228923/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=Interactive%20effects%20of%20CYP3A4,%20CYP3A5,%20MDR1%20and%20NR1I2%20polymorphisms%20on%20tracrolimus%20trough%20concentrations%20in%20early%20postrenal%20transplant%20recipients&author=JL%20Li&author=S%20Liu&author=Q%20Fu&author=Y%20Zhang&author=XD%20Wang&volume=16&issue=12&publication_year=2015&pages=1355-65&pmid=26228923&doi=10.2217/pgs.15.78&)
|
| 208 |
+
|
| 209 |
+
16. Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin PharmacolTher. 2017;102(4):688–700. doi: 10.1002/cpt.690. [DOI](https://doi.org/10.1002/cpt.690) | [PMC free article](/articles/PMC5600063/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28378927/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20PharmacolTher&title=Worldwide%20distribution%20of%20cytochrome%20P450%20alleles:%20a%20meta-analysis%20of%20population-scale%20sequencing%20projects&author=Y%20Zhou&author=M%20Ingelman-Sundberg&author=VM%20Lauschke&volume=102&issue=4&publication_year=2017&pages=688-700&pmid=28378927&doi=10.1002/cpt.690&)
|
| 210 |
+
|
| 211 |
+
17. Rojas L, Neumann I, Herrero MJ, Bosó V, Reig J, Poveda JL, et al. Effect of CYP3A5*3 on kidney transplant recipients treated with tacrolimus: a systematic review and meta-analysis of observational studies. Pharmacogenomics J. 2015;15(1):38–48. doi: 10.1038/tpj.2014.38. [DOI](https://doi.org/10.1038/tpj.2014.38) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25201288/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J&title=Effect%20of%20CYP3A5*3%20on%20kidney%20transplant%20recipients%20treated%20with%20tacrolimus:%20a%20systematic%20review%20and%20meta-analysis%20of%20observational%20studies&author=L%20Rojas&author=I%20Neumann&author=MJ%20Herrero&author=V%20Bos%C3%B3&author=J%20Reig&volume=15&issue=1&publication_year=2015&pages=38-48&pmid=25201288&doi=10.1038/tpj.2014.38&)
|
| 212 |
+
|
| 213 |
+
18. Muller WK, Dandara C, Manning K, Mhandire D, Ensor J, Barday Z, et al. CYP3A5 polymorphisms and their effects on tacrolimus exposure in an ethnically diverse South African renal transplant population. S Afr Med J. 2020;110(2):159–66. doi: 10.7196/SAMJ.2020.v110i2.13969. [DOI](https://doi.org/10.7196/SAMJ.2020.v110i2.13969) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32657689/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=S%20Afr%20Med%20J&title=CYP3A5%20polymorphisms%20and%20their%20effects%20on%20tacrolimus%20exposure%20in%20an%20ethnically%20diverse%20South%20African%20renal%20transplant%20population&author=WK%20Muller&author=C%20Dandara&author=K%20Manning&author=D%20Mhandire&author=J%20Ensor&volume=110&issue=2&publication_year=2020&pages=159-66&pmid=32657689&doi=10.7196/SAMJ.2020.v110i2.13969&)
|
| 214 |
+
|
| 215 |
+
19. Shilbayeh S, Zmeili R, Almardini RI. The impact of CYP3A5 and MDR1 polymorphisms on tacrolimus dosage requirements and trough concentrations in pediatric renal transplant recipients. Saudi J Kidney Dis Transpl. 2013;24(6):1125–36. doi: 10.4103/1319-2442.121268. [DOI](https://doi.org/10.4103/1319-2442.121268) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24231473/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Saudi%20J%20Kidney%20Dis%20Transpl&title=The%20impact%20of%20CYP3A5%20and%20MDR1%20polymorphisms%20on%20tacrolimus%20dosage%20requirements%20and%20trough%20concentrations%20in%20pediatric%20renal%20transplant%20recipients&author=S%20Shilbayeh&author=R%20Zmeili&author=RI%20Almardini&volume=24&issue=6&publication_year=2013&pages=1125-36&pmid=24231473&doi=10.4103/1319-2442.121268&)
|
| 216 |
+
|
| 217 |
+
20. Mealey KL. Therapeutic implications of the MDR-1 gene. J Vet PharmacolTher. 2004;27(5):257–64. doi: 10.1111/j.1365-2885.2004.00607.x. [DOI](https://doi.org/10.1111/j.1365-2885.2004.00607.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15500562/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Vet%20PharmacolTher&title=Therapeutic%20implications%20of%20the%20MDR-1%20gene&author=KL%20Mealey&volume=27&issue=5&publication_year=2004&pages=257-64&pmid=15500562&doi=10.1111/j.1365-2885.2004.00607.x&)
|
| 218 |
+
|
| 219 |
+
21. Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmöller J, Johne A, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97(7):3473–8. doi: 10.1073/pnas.97.7.3473. [DOI](https://doi.org/10.1073/pnas.97.7.3473) | [PMC free article](/articles/PMC16264/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10716719/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Proc%20Natl%20Acad%20Sci%20U%20S%20A&title=Functional%20polymorphisms%20of%20the%20human%20multidrug-resistance%20gene:%20multiple%20sequence%20variations%20and%20correlation%20of%20one%20allele%20with%20P-glycoprotein%20expression%20and%20activity%20in%20vivo&author=S%20Hoffmeyer&author=O%20Burk&author=O%20von%20Richter&author=HP%20Arnold&author=J%20Brockm%C3%B6ller&volume=97&issue=7&publication_year=2000&pages=3473-8&pmid=10716719&doi=10.1073/pnas.97.7.3473&)
|
| 220 |
+
|
| 221 |
+
22. Drescher S, Schaeffeler E, Hitzl M, Hofmann U, Schwab M, Brinkmann U, et al. MDR1 gene polymorphisms and disposition of the P-glycoprotein substrate fexofenadine. Br J Clin Pharmacol. 2002;53(5):526–34. doi: 10.1046/j.1365-2125.2002.01591.x. [DOI](https://doi.org/10.1046/j.1365-2125.2002.01591.x) | [PMC free article](/articles/PMC1874364/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11994059/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=MDR1%20gene%20polymorphisms%20and%20disposition%20of%20the%20P-glycoprotein%20substrate%20fexofenadine&author=S%20Drescher&author=E%20Schaeffeler&author=M%20Hitzl&author=U%20Hofmann&author=M%20Schwab&volume=53&issue=5&publication_year=2002&pages=526-34&pmid=11994059&doi=10.1046/j.1365-2125.2002.01591.x&)
|
| 222 |
+
|
| 223 |
+
23. Goto M, Masuda S, Saito H, Uemoto S, Kiuchi T, Tanaka K, et al. C3435T polymorphism in the MDR1 gene affects the enterocyte expression level of CYP3A4 rather than Pgp in recipients of living-donor liver transplantation. Pharmacogenetics. 2002;12(6):451–7. doi: 10.1097/00008571-200208000-00005. [DOI](https://doi.org/10.1097/00008571-200208000-00005) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12172213/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=C3435T%20polymorphism%20in%20the%20MDR1%20gene%20affects%20the%20enterocyte%20expression%20level%20of%20CYP3A4%20rather%20than%20Pgp%20in%20recipients%20of%20living-donor%20liver%20transplantation&author=M%20Goto&author=S%20Masuda&author=H%20Saito&author=S%20Uemoto&author=T%20Kiuchi&volume=12&issue=6&publication_year=2002&pages=451-7&pmid=12172213&doi=10.1097/00008571-200208000-00005&)
|
| 224 |
+
|
| 225 |
+
24. Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, Gottesman MM. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22(47):7468–85. doi: 10.1038/sj.onc.1206948. [DOI](https://doi.org/10.1038/sj.onc.1206948) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14576852/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Oncogene&title=P-glycoprotein:%20from%20genomics%20to%20mechanism&author=SV%20Ambudkar&author=C%20Kimchi-Sarfaty&author=ZE%20Sauna&author=MM%20Gottesman&volume=22&issue=47&publication_year=2003&pages=7468-85&pmid=14576852&doi=10.1038/sj.onc.1206948&)
|
| 226 |
+
|
| 227 |
+
25. Loh PT, Lou HX, Zhao Y, Chin YM, Vathsala A. Significant impact of gene polymorphisms on tacrolimus but not cyclosporine dosing in Asian renal transplant recipients. Transplant Proc. 2008;40(5):1690–5. doi: 10.1016/j.transproceed.2008.04.010. [DOI](https://doi.org/10.1016/j.transproceed.2008.04.010) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18589174/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplant%20Proc&title=Significant%20impact%20of%20gene%20polymorphisms%20on%20tacrolimus%20but%20not%20cyclosporine%20dosing%20in%20Asian%20renal%20transplant%20recipients&author=PT%20Loh&author=HX%20Lou&author=Y%20Zhao&author=YM%20Chin&author=A%20Vathsala&volume=40&issue=5&publication_year=2008&pages=1690-5&pmid=18589174&doi=10.1016/j.transproceed.2008.04.010&)
|
| 228 |
+
|
| 229 |
+
26. Komoto C, Nakamura T, Sakaeda T, Kroetz DL, Yamada T, Omatsu H, et al. MDR1 haplotype frequencies in Japanese and Caucasian, and in Japanese patients with colorectal cancer and esophageal cancer. Drug MetabPharmacokinet. 2006;21(2):126–32. doi: 10.2133/dmpk.21.126. [DOI](https://doi.org/10.2133/dmpk.21.126) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16702732/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20MetabPharmacokinet&title=MDR1%20haplotype%20frequencies%20in%20Japanese%20and%20Caucasian,%20and%20in%20Japanese%20patients%20with%20colorectal%20cancer%20and%20esophageal%20cancer&author=C%20Komoto&author=T%20Nakamura&author=T%20Sakaeda&author=DL%20Kroetz&author=T%20Yamada&volume=21&issue=2&publication_year=2006&pages=126-32&pmid=16702732&doi=10.2133/dmpk.21.126&)
|
| 230 |
+
|
| 231 |
+
27. Tada H, Tsuchiya N, Satoh S, Kagaya H, Li Z, Sato K, et al. Impact of CYP3A5 and MDR1(ABCB1) C3435T polymorphisms on the pharmacokinetics of tacrolimus in renal transplant recipients. Transplant Proc. 2005;37(4):1730–2. doi: 10.1016/j.transproceed.2005.02.073. [DOI](https://doi.org/10.1016/j.transproceed.2005.02.073) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15919447/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplant%20Proc&title=Impact%20of%20CYP3A5%20and%20MDR1(ABCB1)%20C3435T%20polymorphisms%20on%20the%20pharmacokinetics%20of%20tacrolimus%20in%20renal%20transplant%20recipients&author=H%20Tada&author=N%20Tsuchiya&author=S%20Satoh&author=H%20Kagaya&author=Z%20Li&volume=37&issue=4&publication_year=2005&pages=1730-2&pmid=15919447&doi=10.1016/j.transproceed.2005.02.073&)
|
| 232 |
+
|
| 233 |
+
28. Barraclough KA, Isbel NM, Lee KJ, Bergmann TK, Johnson DW, McWhinney BC, et al. NR1I2 polymorphisms are related to tacrolimus dose-adjusted exposure and BK viremia in adult kidney transplantation. Transplantation. 2012;94(10):1025–32. doi: 10.1097/TP.0b013e31826c3985. [DOI](https://doi.org/10.1097/TP.0b013e31826c3985) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23095803/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplantation&title=NR1I2%20polymorphisms%20are%20related%20to%20tacrolimus%20dose-adjusted%20exposure%20and%20BK%20viremia%20in%20adult%20kidney%20transplantation&author=KA%20Barraclough&author=NM%20Isbel&author=KJ%20Lee&author=TK%20Bergmann&author=DW%20Johnson&volume=94&issue=10&publication_year=2012&pages=1025-32&pmid=23095803&doi=10.1097/TP.0b013e31826c3985&)
|
| 234 |
+
|
| 235 |
+
29. López-Montenegro Soria MA, Kanter Berga J, Beltrán Catalán S, Milara Payá J, Pallardó Mateu LM, Jiménez Torres NV. Genetic polymorphisms and individualized tacrolimus dosing. Transplant Proc. 2010;42(8):3031–3. doi: 10.1016/j.transproceed.2010.08.001. [DOI](https://doi.org/10.1016/j.transproceed.2010.08.001) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20970601/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplant%20Proc&title=Genetic%20polymorphisms%20and%20individualized%20tacrolimus%20dosing&author=MA%20L%C3%B3pez-Montenegro%20Soria&author=J%20Kanter%20Berga&author=S%20Beltr%C3%A1n%20Catal%C3%A1n&author=J%20Milara%20Pay%C3%A1&author=LM%20Pallard%C3%B3%20Mateu&volume=42&issue=8&publication_year=2010&pages=3031-3&pmid=20970601&doi=10.1016/j.transproceed.2010.08.001&)
|
| 236 |
+
|
| 237 |
+
30. Thölking G, Fortmann C, Koch R, Gerth HU, Pabst D, Pavenstädt H, et al. The tacrolimus metabolism rate influences renal function after kidney transplantation. PLoS One. 2014;9(10):e111128. doi: 10.1371/journal.pone.0111128. [DOI](https://doi.org/10.1371/journal.pone.0111128) | [PMC free article](/articles/PMC4207775/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25340655/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=PLoS%20One&title=The%20tacrolimus%20metabolism%20rate%20influences%20renal%20function%20after%20kidney%20transplantation&author=G%20Th%C3%B6lking&author=C%20Fortmann&author=R%20Koch&author=HU%20Gerth&author=D%20Pabst&volume=9&issue=10&publication_year=2014&pages=e111128&pmid=25340655&doi=10.1371/journal.pone.0111128&)
|
| 238 |
+
|
| 239 |
+
31. Kuypers DR, Naesens M, de Jonge H, Lerut E, Verbeke K, Vanrenterghem Y. Tacrolimus dose requirements and CYP3A5 genotype and the development of calcineurin inhibitor-associated nephrotoxicity in renal allograft recipients. Ther Drug Monit. 2010;32(4):394–404. doi: 10.1097/FTD.0b013e3181e06818. [DOI](https://doi.org/10.1097/FTD.0b013e3181e06818) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20526235/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ther%20Drug%20Monit&title=Tacrolimus%20dose%20requirements%20and%20CYP3A5%20genotype%20and%20the%20development%20of%20calcineurin%20inhibitor-associated%20nephrotoxicity%20in%20renal%20allograft%20recipients&author=DR%20Kuypers&author=M%20Naesens&author=H%20de%20Jonge&author=E%20Lerut&author=K%20Verbeke&volume=32&issue=4&publication_year=2010&pages=394-404&pmid=20526235&doi=10.1097/FTD.0b013e3181e06818&)
|
| 240 |
+
|
| 241 |
+
32. Thölking G, Gerth HU, Schuette-Nuetgen K, Reuter S. Influence of tacrolimus metabolism rate on renal function after solid organ transplantation. World J Transplant. 2017;7(1):26–33. doi: 10.5500/wjt.v7.i1.26. [DOI](https://doi.org/10.5500/wjt.v7.i1.26) | [PMC free article](/articles/PMC5324025/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28280692/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=World%20J%20Transplant&title=Influence%20of%20tacrolimus%20metabolism%20rate%20on%20renal%20function%20after%20solid%20organ%20transplantation&author=G%20Th%C3%B6lking&author=HU%20Gerth&author=K%20Schuette-Nuetgen&author=S%20Reuter&volume=7&issue=1&publication_year=2017&pages=26-33&pmid=28280692&doi=10.5500/wjt.v7.i1.26&)
|
| 242 |
+
|
| 243 |
+
33. Ashavaid TF, Raje HS, Shah BV, Shah SA. Design of allele specific PCR for rapid detection of CYP3A5 (A6986G) and Mdr-1 (C3435T) polymorphisms. Indian J Clin Biochem. 2011;26(1):18–21. doi: 10.1007/s12291-010-0085-z. [DOI](https://doi.org/10.1007/s12291-010-0085-z) | [PMC free article](/articles/PMC3068768/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22211008/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Indian%20J%20Clin%20Biochem&title=Design%20of%20allele%20specific%20PCR%20for%20rapid%20detection%20of%20CYP3A5%20(A6986G)%20and%20Mdr-1%20(C3435T)%20polymorphisms&author=TF%20Ashavaid&author=HS%20Raje&author=BV%20Shah&author=SA%20Shah&volume=26&issue=1&publication_year=2011&pages=18-21&pmid=22211008&doi=10.1007/s12291-010-0085-z&)
|
| 244 |
+
|
| 245 |
+
34. Kurzawski M, Malinowski D, Dziewanowski K, Droździk M. Analysis of common polymorphisms within NR1I2 and NR1I3 genes and tacrolimus dose-adjusted concentration in stable kidney transplant recipients. Pharmacogenet Genomics. 2017;27(10):372–7. doi: 10.1097/fpc.0000000000000301. [DOI](https://doi.org/10.1097/fpc.0000000000000301) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28777242/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenet%20Genomics&title=Analysis%20of%20common%20polymorphisms%20within%20NR1I2%20and%20NR1I3%20genes%20and%20tacrolimus%20dose-adjusted%20concentration%20in%20stable%20kidney%20transplant%20recipients&author=M%20Kurzawski&author=D%20Malinowski&author=K%20Dziewanowski&author=M%20Dro%C5%BAdzik&volume=27&issue=10&publication_year=2017&pages=372-7&pmid=28777242&doi=10.1097/fpc.0000000000000301&)
|
| 246 |
+
|
| 247 |
+
35. Leas BF, Uhl S, Sawinski DL, Trofe-Clark J, Tuteja S, Kaczmarek JL, et al. Calcineurin inhibitors for renal transplant. Rockville, MD: Agency for Healthcare Research and Quality (US); 2016. [PubMed](https://pubmed.ncbi.nlm.nih.gov/27123504/)
|
| 248 |
+
|
| 249 |
+
36. Meier-Kriesche HU, Li S, Gruessner RW, Fung JJ, Bustami RT, Barr ML, et al. Immunosuppression: evolution in practice and trends, 1994-2004. Am J Transplant. 2006;6(5 Pt 2):1111–31. doi: 10.1111/j.1600-6143.2006.01270.x. [DOI](https://doi.org/10.1111/j.1600-6143.2006.01270.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16613591/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Transplant&title=Immunosuppression:%20evolution%20in%20practice%20and%20trends,%201994-2004&author=HU%20Meier-Kriesche&author=S%20Li&author=RW%20Gruessner&author=JJ%20Fung&author=RT%20Bustami&volume=6&issue=5%20Pt%202&publication_year=2006&pages=1111-31&pmid=16613591&doi=10.1111/j.1600-6143.2006.01270.x&)
|
| 250 |
+
|
| 251 |
+
37. Ekberg H, van Gelder T, Kaplan B, Bernasconi C. Relationship of tacrolimus exposure and mycophenolate mofetil dose with renal function after renal transplantation. Transplantation. 2011;92(1):82–7. doi: 10.1097/TP.0b013e31821fad06. [DOI](https://doi.org/10.1097/TP.0b013e31821fad06) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21562449/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplantation&title=Relationship%20of%20tacrolimus%20exposure%20and%20mycophenolate%20mofetil%20dose%20with%20renal%20function%20after%20renal%20transplantation&author=H%20Ekberg&author=T%20van%20Gelder&author=B%20Kaplan&author=C%20Bernasconi&volume=92&issue=1&publication_year=2011&pages=82-7&pmid=21562449&doi=10.1097/TP.0b013e31821fad06&)
|
| 252 |
+
|
| 253 |
+
38. Scholten EM, Cremers SC, Schoemaker RC, Rowshani AT, van Kan EJ, den Hartigh J, et al. AUC-guided dosing of tacrolimus prevents progressive systemic overexposure in renal transplant recipients. Kidney Int. 2005;67(6):2440–7. doi: 10.1111/j.1523-1755.2005.00352.x. [DOI](https://doi.org/10.1111/j.1523-1755.2005.00352.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15882290/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Kidney%20Int&title=AUC-guided%20dosing%20of%20tacrolimus%20prevents%20progressive%20systemic%20overexposure%20in%20renal%20transplant%20recipients&author=EM%20Scholten&author=SC%20Cremers&author=RC%20Schoemaker&author=AT%20Rowshani&author=EJ%20van%20Kan&volume=67&issue=6&publication_year=2005&pages=2440-7&pmid=15882290&doi=10.1111/j.1523-1755.2005.00352.x&)
|
test/texts/PMC10418744.md
ADDED
|
@@ -0,0 +1,308 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Personalized CFTR Modulator Therapy for G85E and N1303K Homozygous Patients with Cystic Fibrosis
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Simon Y Graeber, Anita Balázs, Niklas Ziegahn, Tihomir Rubil, Constanze Vitzthum, Linus Piehler, Marika Drescher, Kathrin Seidel, Alexander Rohrbach, Jobst Röhmel, Stephanie Thee, Julia Duerr, Marcus A Mall, Mirjam Stahl
|
| 5 |
+
**Journal:** International Journal of Molecular Sciences
|
| 6 |
+
**Date:** 2023 Aug 2
|
| 7 |
+
**DOI:** [10.3390/ijms241512365](https://doi.org/10.3390/ijms241512365)
|
| 8 |
+
**PMID:** 37569738
|
| 9 |
+
**PMCID:** PMC10418744
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10418744/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC10418744/pdf/ijms-24-12365.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC10418744/pdf/ijms-24-12365.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
CFTR modulator therapy with elexacaftor/tezacaftor/ivacaftor (ETI) has been approved for people with CF and at least one F508del allele in Europe. In the US, the ETI label has been expanded to 177 rare CFTR mutations responsive in Fischer rat thyroid cells, including G85E, but not N1303K. However, knowledge on the effect of ETI on G85E or N1303K CFTR function remains limited. In vitro effects of ETI were measured in primary human nasal epithelial cultures (pHNECs) of a G85E homozygous patient and an N1303K homozygous patient. Effects of ETI therapy in vivo in these patients were assessed using clinical outcomes, including multiple breath washout and lung MRI, and the CFTR biomarkers sweat chloride concentration (SCC), nasal potential difference (NPD) and intestinal current measurement (ICM), before and after initiation of ETI. ETI increased CFTR-mediated chloride transport in G85E/G85E and N1303K/N1303K pHNECs. In the G85E/G85E and the N1303K/N1303K patient, we observed an improvement in lung function, SCC, and CFTR function in the respiratory and rectal epithelium after initiation of ETI. The approach of combining preclinical in vitro testing with subsequent in vivo verification can facilitate access to CFTR modulator therapy and enhance precision medicine for patients carrying rare CFTR mutations.
|
| 16 |
+
|
| 17 |
+
Keywords: cystic fibrosis, CFTR, CFTR modulator, G85E, N1303K, human nasal epithelial cells, intestinal current measurement, nasal potential difference
|
| 18 |
+
|
| 19 |
+
**Keywords:**Keywords: cystic fibrosis, CFTR, CFTR modulator, G85E, N1303K, human nasal epithelial cells, intestinal current measurement, nasal potential difference
|
| 20 |
+
|
| 21 |
+
## 1. Introduction
|
| 22 |
+
|
| 23 |
+
Cystic fibrosis (CF) is an autosomal recessive disease caused by mutations in the cystic fibrosis transmembrane conductance regulator (*CFTR*CFTR) gene [[1](#B1-ijms-24-12365)1]. As a consequence, the CFTR protein, which acts as an ion channel in various epithelia in the human body, is substantially reduced in quantity and/or quality, leading to an impaired function of the chloride channel [[2](#B2-ijms-24-12365)2]. This results in thickened secretions of multiple organs, including the airways, ultimately leading to damage of the affected organs and loss of their function. For a long time, therapeutic strategies aimed to reduce symptoms of organ dysfunction. The emergence of highly effective CFTR modulator therapies has transformed the clinical landscape of CF care. Novel triple combination elexacaftor/tezacaftor/ivacaftor (ETI) therapy is now available for ~90% of people with CF carrying at least one allele of the most common mutation *F508del*F508del [[3](#B3-ijms-24-12365)3,[4](#B4-ijms-24-12365)4,[5](#B5-ijms-24-12365)5,[6](#B6-ijms-24-12365)6,[7](#B7-ijms-24-12365)7,[8](#B8-ijms-24-12365)8,[9](#B9-ijms-24-12365)9]. ETI therapy leads to substantial clinical benefits and improvement in CFTR function to 40 to 50% of normal CFTR activity measured by nasal potential difference (NPD), intestinal current measurement (ICM), lung clearance index (LCI) and lung morphology in real world observational trials in patients with CF and at least one *F508del*F508del mutation [[10](#B10-ijms-24-12365)10,[11](#B11-ijms-24-12365)11]. Additionally, the FDA approved ETI for 177 rare *CFTR*CFTR mutations, based on in vitro data in Fischer rat thyroid cells. However, there is still an unmet need to address ~10% of people with non-approved mutations or ultra-rare mutations with unknown consequences, where personalized medicine approaches could become useful to enhance their accessibility to approved CFTR modulator therapies [[12](#B12-ijms-24-12365)12,[13](#B13-ijms-24-12365)13]. Here, we report an approach for individualized treatment for two patients with CF, homozygous for the *G85E*G85E and *N1303K*N1303K mutation and for whom CFTR modulator therapy is not available in Europe, as both conditions are not approved for ETI by the European Medicines Agency (EMA) (although *G85E*G85E has been approved by the FDA).
|
| 24 |
+
|
| 25 |
+
Both *N1303K*N1303K and *G85E*G85E are classified as processing mutations, which are refractory to lumacaftor [[14](#B14-ijms-24-12365)14,[15](#B15-ijms-24-12365)15]; with *N1303K*N1303K additionally being classified as a gating mutation [[15](#B15-ijms-24-12365)15,[16](#B16-ijms-24-12365)16]. However, recent reports from investigations of airway epithelial cells suggest that G85E and N1303K CFTR proteins are responsive to ETI [[17](#B17-ijms-24-12365)17,[18](#B18-ijms-24-12365)18]. Moreover, CF patients homozygous for rare *CFTR*CFTR mutations provide an opportunity to study the effect of specific mutations on CFTR function in a translational disease model system, which is not possible in compound heterozygous patients with CF. Furthermore, limited clinical data are available on the in vivo benefits of ETI for patients carrying these mutations [[13](#B13-ijms-24-12365)13,[19](#B19-ijms-24-12365)19,[20](#B20-ijms-24-12365)20,[21](#B21-ijms-24-12365)21].
|
| 26 |
+
|
| 27 |
+
In this study, we first utilized patient-derived primary human nasal epithelial cultures (pHNECs) from one *G85E*G85E homozygous patient and one *N1303K*N1303K homozygous patient as a tool to predict the clinical response to ETI therapy as a precision medicine model in vitro. In addition, we investigated the effect of ETI therapy in these patients on in vivo CFTR function with the CFTR biomarkers sweat chloride concentration (SCC), NPD, and ICM as well as clinical outcomes, including FEV_1_1 % predicted and sensitive endpoints for lung structure and function, such as LCI and magnetic resonance imaging (MRI) of the lung.
|
| 28 |
+
|
| 29 |
+
## 2. Results
|
| 30 |
+
|
| 31 |
+
### 2.1. Case Presentations
|
| 32 |
+
|
| 33 |
+
The *G85E*G85E/*G85E*G85E patient is a 15-year-old female diagnosed with CF at the age of two years secondary to recurrent respiratory infections and a failure to thrive. SCC at that time was clearly pathologic with 77 mmol/L and a genetic analysis that demonstrated homozygosity of *G85E*G85E in the *CFTR*CFTR gene. Symptomatic therapies were initiated according to the European Cystic Fibrosis Society guidelines, including bronchodilator and mucolytics inhalation with airway clearance, oral pancreatic enzyme replacement therapy, and fat-soluble vitamin supplementation. This led to an improvement in thriving, but the girl soon experienced a first and then recurrent episodes of allergic bronchopulmonary aspergillosis (ABPA). Therefore, she required extended symptomatic therapy including recurrent cycles of systemic and inhaled glucocorticosteroids during childhood and adolescence. Recurring ABPA episodes were treated with anti-IgE antibodies without clear response in clinical parameters. Under this treatment, the patient suffered mild-to-moderate obstructive lung disease at age 14 years and had already shown advanced structural CF lung disease in chest MRI ([Table 1](#ijms-24-12365-t001)Table 1). Continuous systemic antifungal therapy led to a deceleration of lung function decline; however, the patient still reported daily cough with both phases of persistent dry cough and extensive sputum production. In addition, bronchial tightness necessitated the frequent use of bronchodilators. In January 2021, our patient experienced a new significant decline in lung function during a pulmonary exacerbation that was only partly restored following intravenous antibiotic treatment.
|
| 34 |
+
|
| 35 |
+
### Table 1.
|
| 36 |
+
|
| 37 |
+
Clinical characetristics of the two patients at baseline.
|
| 38 |
+
|
| 39 |
+
| Clinical Characteristic | G85E/G85E | N1303K/N1303K |
|
| 40 |
+
| ----------------------- | --------- | ------------- |
|
| 41 |
+
| Age (years) | 15.0 | 19.8 |
|
| 42 |
+
| Sex | Female | Male |
|
| 43 |
+
| Pancreatic insufficiency | Yes | Yes |
|
| 44 |
+
| Sweat chloride (mmol/L) | 102.0 | 99.0 |
|
| 45 |
+
| FEV1 (% predicted) | 74.0 | 54.0 |
|
| 46 |
+
| LCI2.5 | 10.0 | 14.5 |
|
| 47 |
+
| BMI (kg/m2) | 20.9 | 19.0 |
|
| 48 |
+
The *N1303K*N1303K/*N1303K*N1303K patient is a 19-year-old male who was also diagnosed at preschool age due to recurrent pulmonary infections and failure to thrive. Issues with adherence to symptomatic therapy during adolescence added to the clear progression over time, especially of CF lung disease, with ultimately polymicrobial infection with Methicillin resistant Staphylococcus aureus (MRSA), Pseudomonas aeruginosa and Mycobacterium abscessus, among other pathogens. Repeated Pseudomonas-effective antibiotic therapy cycles did not lead to stabilization of lung disease ([Table 1](#ijms-24-12365-t001)Table 1). Besides the impaired pulmonary function, the *N1303K*N1303K/*N1303K*N1303K patient also suffered from an advanced CF liver disease with pronounced structural changes, but preserved function. However, infection with nontuberculous mycobacteria (NTM) could pose a problem if liver transplantation is required in the future. Therefore, we decided to start an antimycobacterial therapy in April 2021, taking into account the other pathogens present. The initial five-drug systemic NTM therapy with tigecycline, amikacin, cefoxitin, meropenem and clofazimine, in addition to colistin p.i., had to be adjusted in the first months due to limited tolerability, but was then carried out as a quadruple therapy over 9 months, which led to stabilization of the patient. However, with side effects increasing over time and a recurrence of pulmonary exacerbation under therapy, we evaluated the treatment with ETI for this patient.
|
| 49 |
+
|
| 50 |
+
### 2.2. Preclinical Testing of ETI Therapy In Vitro
|
| 51 |
+
|
| 52 |
+
To test the potential benefit of ETI therapy in vitro, we generated highly differentiated airway epithelial cultures from nasal brushings of both patients and performed transepithelial short-circuit current measurements in Ussing chambers ([Figure 1](#ijms-24-12365-f001)Figure 1). The basal short circuit current (I_sc_sc), amiloride-sensitive I_sc_sc and amiloride-insensitive I_sc_sc were not significantly different between vehicle control and ETI treatment in cultures derived from the *G85E*G85E/*G85E*G85E patient, although amiloride-sensitive I_sc_sc showed a trend towards a decrease with ETI treatment ([Figure 1](#ijms-24-12365-f001)Figure 1C,E–G). ETI treatment significantly enhanced CFTR-mediated chloride currents compared with vehicle-treated control, as indicated by forskolin peak response (*p*p < 0.05) and CFTRinhibitor-172 inhibited current (*p*p < 0.01) in *G85E*G85E/*G85E*G85E pHNECs ([Figure 1](#ijms-24-12365-f001)Figure 1H,J). Compared with our previously published data from pHNECs from healthy people (mean CFTRinhibitor-172 sensitive current −10.1 µA/mm^2^2, n = 31) [[22](#B22-ijms-24-12365)22], rescue of CFTR function by ETI treatment, as determined from CFTRinhibitor-172 sensitive current (−1.1 µA/mm^2^2), was 11.0% of the normal CFTR activity for pHNECs of the *G85E*G85E/*G85E*G85E patient. In pHNECs derived from the *N1303K*N1303K/*N1303K*N1303K patient, basal I_sc_sc and amiloride-insensitive I_sc_sc were unchanged, whereas amiloride-sensitive I_sc_sc was significantly reduced by ETI treatment compared with dimethyl sulfoxide (DMSO) control (*p*p < 0.05), suggesting decreased activity of the epithelial sodium channel (ENaC) ([Figure 1](#ijms-24-12365-f001)Figure 1D,K–M). Forskolin-stimulated I_sc_sc showed a trend towards an increase upon ETI treatment and acute ivacaftor response was significantly increased with ETI treatment compared with vehicle control ([Figure 1](#ijms-24-12365-f001)Figure 1D,N,O). Furthermore, ETI treatment significantly increased CFTRinhibitor-172 sensitive current (−1.1 µA/mm^2^2) compared with DMSO control (−0.44 µA/mm^2^2) in pHNECs from the *N1303K*N1303K/*N1303K*N1303K patient (*p*p < 0.05) corresponding to 11.0% of the normal CFTR activity ([Figure 1](#ijms-24-12365-f001)Figure 1D,P).
|
| 53 |
+
|
| 54 |
+
### Figure 1.
|
| 55 |
+
|
| 56 |
+

|
| 57 |
+
|
| 58 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=10418744_ijms-24-12365-g001.jpg)
|
| 59 |
+
|
| 60 |
+
Treatment with elexacaftor/tezacaftor/ivacaftor (ETI) rescues cystic fibrosis transmembrane conductance regulator (CFTR) function in nasal epithelial cultures from the G85E/G85E and the N1303K/N1303K patient. (A–D) Representative tracings of short-circuit transepithelial current (ISC) measurements in cultures from a healthy donor (A), a CF patient homozygous for F508del (B), the CF patient homozygous for G85E (C) and the CF patient homozygous for N1303K (D). CF cultures were incubated with elexacaftor/tezacaftor or dimethyl sulfoxide (DMSO) for 24 h. (E–J) Quantification of the basal ISC (E), amiloride-sensitive ISC (∆Amiloride) (F), amiloride-insensitive ISC (G), and effects of cAMP activation (∆Forskolin/IBMX) (H), ivacaftor (∆Ivacaftor) (I) and CFTR inhibitor-172 (∆CFTRinh-172) (J) on ISC in the G85E/G85E patient. Quantification of the basal ISC (K), amiloride-sensitive ISC (∆Amiloride) (L), amiloride-insensitive ISC (M), and effects of cAMP activation (∆Forskolin/IBMX) (N), ivacaftor (∆Ivacaftor) (O) and CFTR inhibitor-172 (∆CFTRinh-172) (P) on ISC in the N1303K/N1303K patient. n = 4–6 filters per individual per group, data are presented as mean ± SD. The black tracings and bars represent the vehicle control treated with DMSO and the blue tracings and bars represent the cultures treated with ETI. * p < 0.05, ** p < 0.01 ETI versus DMSO.
|
| 61 |
+
|
| 62 |
+
### 2.3. Improvements in CFTR Function In Vivo
|
| 63 |
+
|
| 64 |
+
Based on the in vitro evidence, ETI was started as an off-label therapy in both patients. To evaluate the in vivo effect of ETI therapy, we measured CFTR function in three different tissues using the established CFTR biomarkers SCC, NPD and ICM at baseline and after initiation of ETI therapy ([Figure 2](#ijms-24-12365-f002)Figure 2) [[11](#B11-ijms-24-12365)11,[23](#B23-ijms-24-12365)23,[24](#B24-ijms-24-12365)24].
|
| 65 |
+
|
| 66 |
+
### Figure 2.
|
| 67 |
+
|
| 68 |
+

|
| 69 |
+
|
| 70 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=10418744_ijms-24-12365-g002.jpg)
|
| 71 |
+
|
| 72 |
+
Elexacaftor/tezacaftor/ivacaftor (ETI) therapy improves mutant cystic fibrosis transmembrane conductance regulator (CFTR) function in both the G85E/G85E and the N1303K/N1303K patient. (A–F) Paired measurements of the CFTR biomarkers sweat chloride concentration (A), nasal potential difference (NPD) (B–D), and intestinal current measurement (ICM) (E–F) in a G85E/G85E (black) and an N1303K/N1303K (red) patient at baseline and 3 months (G85E/G85E) or 2 weeks (N1303K/N1303K) after initiation of ETI therapy. (A) Sweat chloride concentration. (B–D) NPD basal potential (B), NPD amiloride response (C), and NPD total chloride response (zero chloride plus isoproterenol (iso) solutions). (E,F) ICM cAMP response (E) and ICM total chloride response obtained by cAMP-dependent stimulation and cholinergic (calcium-dependent) co-activation (F). ICM studies were performed in the presence of amiloride and indomethacin. The dotted line represents median values for F508del/F508del patients treated with tezacaftor/ivacaftor (n = 41) and the dashed line represents median values for F508del/F508del patients treated with elexacaftor/tezacaftor/ivacaftor (n = 44) [11].
|
| 73 |
+
|
| 74 |
+
Both patients had a pathological SCC above 60 mmol/L and showed no CFTR function in the respiratory or rectal epithelium at baseline ([Figure 2](#ijms-24-12365-f002)Figure 2). In the *G85E*G85E/*G85E*G85E patient, SCC was reduced by 40 mmol/L after initiation of ETI therapy ([Figure 2](#ijms-24-12365-f002)Figure 2A). In the NPD, the basal potential was improved from –34.6 mV at baseline to –24.1 mV after initiation of ETI and the amiloride response improved from 20.8 mV to 13.9 mV ([Figure 2](#ijms-24-12365-f002)Figure 2B,C). Furthermore, NPD total chloride response improved from 0.0 mV to −6.9 mV corresponding to 32.4% of CFTR activity in the nasal epithelium of healthy people ([Figure 2](#ijms-24-12365-f002)Figure 2D). In the ICM, we observed a cAMP-dependent chloride secretory response of 32.0 µA/cm^2^22 after initiation of ETI therapy in the *G85E*G85E/*G85E*G85E patient, corresponding to rescue of CFTR function in the rectal mucosa to a level of 37.6% of normal ([Figure 2](#ijms-24-12365-f002)Figure 2E). The total chloride secretory response increased from 1.1 µA/cm^2^22 at baseline to 82.4 µA/cm^2^2 after initiation of ETI, corresponding to a level of 29.4% of normal CFTR activity ([Figure 2](#ijms-24-12365-f002)Figure 2F). In the *N1303K*N1303K/*N1303K*N1303K patient, SCC was reduced by 31 mmol/L after initiation of ETI therapy ([Figure 2](#ijms-24-12365-f002)Figure 2A). In the NPD, the basal potential was improved from −39.7 mV to –10.6 mV and the amiloride response from 14.1 mV to 3.9 mV ([Figure 2](#ijms-24-12365-f002)Figure 2B,C). NPD total chloride response improved from 0.8 mV to −1.2 mV, corresponding to 5.5% of normal CFTR activity in the nasal epithelium ([Figure 2](#ijms-24-12365-f002)Figure 2D). In the ICM, we observed a cAMP-dependent chloride secretory response of 14.0 µA/cm^2^22 after initiation of ETI therapy in the *N1303K*N1303K/*N1303K*N1303K patient, corresponding to rescue of CFTR function in the rectal mucosa to a level of 16.5% of normal ([Figure 2](#ijms-24-12365-f002)Figure 2E). The total chloride secretory response increased from 1.0 µA/cm^2^22 to 40.8 µA/cm^2^22, corresponding to a level of 14.6% of normal CFTR activity ([Figure 2](#ijms-24-12365-f002)Figure 2F).
|
| 75 |
+
|
| 76 |
+
### 2.4. Clinical Improvements following Treatment with ETI
|
| 77 |
+
|
| 78 |
+
The initiation of therapy with ETI resulted in prompt stabilization of the clinical status of both patients, with a particular improvement in FEV_1_1 % predicted and LCI ([Figure 3](#ijms-24-12365-f003)Figure 3A,B and [Figure 4](#ijms-24-12365-f004)Figure 4A,B). The *G85E*G85E/*G85E*G85E patient stayed in the normal range for BMI after initiation of ETI ([Figure 3](#ijms-24-12365-f003)Figure 3C). However, this patient showed a reduction in total serum IgE despite stopping all ABPA-related therapies ([Figure 3](#ijms-24-12365-f003)Figure 3D). Furthermore, we observed substantial improvements in mucus plugging and bronchial wall thickening in MRI images of the lung after initiation of ETI ([Figure 3](#ijms-24-12365-f003)Figure 3E).
|
| 79 |
+
|
| 80 |
+
### Figure 3.
|
| 81 |
+
|
| 82 |
+

|
| 83 |
+
|
| 84 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=10418744_ijms-24-12365-g003.jpg)
|
| 85 |
+
|
| 86 |
+
Elexacaftor/tezacaftor/ivacaftor (ETI) therapy improves clinical outcomes in the G85E/G85E patient. (A–D) Repeated measurements of FEV1% predicted (A), lung clearance index (LCI2.5) (B), BMI (C) and total immunoglobulin E (IgE) from 200 days before and 600 days after initiation of ETI. (E) Representative MRI image at baseline and after initiation of ETI. Structural airway abnormalities (wall thickening and/or bronchiectasis) are indicated by arrows and mucus plugging by white arrowheads. Perfusion abnormalities are indicated by black arrowheads.
|
| 87 |
+
|
| 88 |
+
### Figure 4.
|
| 89 |
+
|
| 90 |
+

|
| 91 |
+
|
| 92 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=10418744_ijms-24-12365-g004.jpg)
|
| 93 |
+
|
| 94 |
+
Elexacaftor/tezacaftor/ivacaftor (ETI) therapy improves clinical outcomes in the N1303K/N1303K patient. (A–D) Repeated measurements of FEV1% predicted (A), lung clearance index (LCI2.5) (B), BMI (C), aspartate transaminase (AST, GOT) and alanine transaminase (ALT, GPT) (D) from 200 days before and 200 days after initiation of ETI.
|
| 95 |
+
|
| 96 |
+
In the *N1303K*N1303K/*N1303K*N1303K patient, we were able to stop the NTM therapy after start of ETI without a decline in respiratory status ([Figure 4](#ijms-24-12365-f004)Figure 4A,B). Furthermore, an improvement and stabilization in weight into the normal BMI range appeared that was not achieved in previous years ([Figure 1](#ijms-24-12365-f001)Figure 1C). For safety monitoring, our patients received ophthalmology exams and routine hepatic function monitoring. In this context, transaminase levels showed improvements after initiation of ETI. Until now, neither the *G85E*G85E/*G85E*G85E patient who had been treated with ETI for 50 months, nor the *N1303K*N1303K/*N1303K*N1303K patient who had been treated for 16 months have shown signs of side effects and have reported no other adverse reactions after initiation of ETI.
|
| 97 |
+
|
| 98 |
+
## 3. Discussion
|
| 99 |
+
|
| 100 |
+
This study provides a comprehensive assessment of the effects of ETI on CFTR function in pHNECs of a *G85E*G85E homozygous patient and an *N1303K*N1303K homozygous patient in vitro as well as of the improvement of CFTR function in vivo after initiation of ETI in both patients. We were able to show that ETI increased CFTR-mediated currents in pHNECs of both patients ([Figure 1](#ijms-24-12365-f001)Figure 1). As predicted by the preclinical results, the in vivo CFTR function of the *G85E*G85E/*G85E*G85E patient improved to 30–40% of normal CFTR activity in the respiratory and rectal epithelium after initiation of ETI. The *N1303K*N1303K/*N1303K*N1303K patient showed an improvement to 5–20% in the respiratory and rectal epithelium after initiation of ETI ([Figure 2](#ijms-24-12365-f002)Figure 2). In both patients, partial restoration of CFTR function led to improvements in clinical outcomes after initiation of ETI ([Figure 3](#ijms-24-12365-f003)Figure 3 and [Figure 4](#ijms-24-12365-f004)Figure 4).
|
| 101 |
+
|
| 102 |
+
The *G85E*G85E mutation is classified as a class II missense mutation, resulting in decreased quantity and function of CFTR protein. In the US, therapy with ETI is approved for patients with CF carrying the *G85E*G85E mutation based on in vitro data in Fischer rat thyroid cells. In Europe, ETI is not approved for patients carrying the *G85E*G85E mutation (without *F508del*F508del on the other allele) due to the lack of clinical studies investigating efficacy and safety of this therapy in vivo. It has been previously shown that ETI improves CFTR function in rectal organoids of a *G85E*G85E heterozygous patient with a stop codon on the other allele [[25](#B25-ijms-24-12365)25]. We observed a consistent rescue of CFTR function by ETI in the pHNEC of the *G85E*G85E/*G85E*G85E patient. Of note, acute addition of ivacaftor did not further potentiate cAMP-stimulated chloride secretion in the *G85E*G85E/*G85E*G85E cultures and even tended to reduce the transepithelial current, indicating that the potentiator ivacaftor may not be essential for optimizing the chloride channel function of this specific trafficking mutation. However, to confirm this observation, the acute and chronic effects of ivacaftor on the G85E mutation need to be assessed in a larger sample size in future studies. Furthermore, by additionally measuring in vivo CFTR biomarkers, our study shows that ETI improves CFTR function in the *G85E*G85E homozygous patient to levels comparable to ETI in *F508del*F508del homozygous patients [[11](#B11-ijms-24-12365)11]. Overall, these data suggest that ETI is beneficial in patients with a *G85E*G85E mutation.
|
| 103 |
+
|
| 104 |
+
*N1303K*N1303K, the fourth most common *CFTR*CFTR mutation, is also classified as a class II missense mutation. Due to a lack of improvement above a threshold of 10% WT CFTR function after treatment with ETI in Fischer rat thyroid cells [[26](#B26-ijms-24-12365)26,[27](#B27-ijms-24-12365)27], ETI is not approved for treatment in patients carrying the *N1303K*N1303K mutation (without another responsive mutation on the other allele) in any country worldwide. Recent evidence suggests that, in addition to impaired CFTR trafficking, the *N1303K*N1303K mutation also causes a severe gating defect in CFTR [[16](#B16-ijms-24-12365)16]. Consistent with these data, our results in pHNEC suggest that potentiation by ivacaftor is an important mechanism in rescuing N1303K–CFTR function. This observation is supported by a study in *N1303K*N1303K/*N1303K*N1303K rectal organoids showing that potentiation by ivacaftor is important for reaching therapeutically relevant levels of organoid swelling after pre-treatment with elexacaftor and tezacaftor [[25](#B25-ijms-24-12365)25]. Recently, it has been suggested that patients with an *N1303K*N1303K mutation could clinically benefit from ETI therapy [[13](#B13-ijms-24-12365)13,[19](#B19-ijms-24-12365)19,[21](#B21-ijms-24-12365)21]. This is further supported by our study showing improvement of CFTR function in the *N1303K*N1303K/*N1303K*N1303K patient in the range of tezacaftor/ivacaftor in *F508del*F508del homozygous patients [[11](#B11-ijms-24-12365)11]. However, in previous studies, *N1303K*N1303K heterozygous and homozygous patients treated with ETI showed substantial improvements in lung function despite only a modest decrease in SCC [[13](#B13-ijms-24-12365)13,[19](#B19-ijms-24-12365)19,[21](#B21-ijms-24-12365)21]. The large decrease in SCC in the *N1303K*N1303K/*N1303K*N1303K patient in our study may partially be explained by the homozygous genotype. However, larger studies with *N1303K*N1303K homozygous and heterozygous patients are needed to address the question of a gene dosage effect for the *N1303K*N1303K mutation. Nevertheless, the relatively large improvement in lung function compared with the relatively modest improvement in sweat chloride in previous studies may suggest tissue-specific expression and therapeutic rescue of N1303K, indicating that SCC might not be the ideal biomarker to assess the response to therapy on the functional level therapy in patients carrying the *N1303K*N1303K mutation. This question needs to be addressed in future clinical trials and the in vivo CFTR biomarker NPD and ICM may help to solve this conundrum regarding N1303K. Interestingly, we further observed reduced amiloride-sensitive currents in ETI treated pHNECs of the *N1303K*N1303K/*N1303K*N1303K patient, suggesting decreased activity of ENaC. This observation may be explained by the restoration of a previously described functional interaction between CFTR and ENaC at the plasma membrane, leading to downregulation of ENaC-mediated sodium/fluid absorption and thus improved airway surface hydration and mucociliary clearance [[28](#B28-ijms-24-12365)28,[29](#B29-ijms-24-12365)29,[30](#B30-ijms-24-12365)30,[31](#B31-ijms-24-12365)31]. This is consistent with observations in the NPD of this patient showing a substantially lower basal transepithelial potential difference and a reduced amiloride response on ETI. In previous studies, improvements in basal potential difference and amiloride response were only observed for ETI in patients with at least one *F508del*F508del allele, but not for tezacaftor/ivacaftor or lumacaftor/ivacaftor in *F508del*F508del homozygous patients [[11](#B11-ijms-24-12365)11,[23](#B23-ijms-24-12365)23]. In pHNECs, we observed residual currents after inhibition of ENaC and CFTR at the end of the experiment. These residual currents were previously observed in freshly excised native nasal epithelium [[32](#B32-ijms-24-12365)32,[33](#B33-ijms-24-12365)33] and may be explained by (i) non-CFTR mediated anion conductance, such as SLC26A9 or TMEM16A; (ii) amiloride-insensitive sodium conductance; or (iii) passive chloride movement due to the chloride gradient applied under these experimental conditions (145 mM basolateral to 5 mM apical) [[34](#B34-ijms-24-12365)34]. Taken together, these data suggest that different preclinical models should be investigated, the cut-off in Fischer rat thyroid cells may need to be adjusted and the combination with in vivo testing may help to establish in vivo efficacy in patients with uncommon *CFTR*CFTR mutations.
|
| 105 |
+
|
| 106 |
+
In pHNECs from the *G85E*G85E/*G85E*G85E and the *N1303K*N1303K/*N1303K*N1303K patient, we observed an improvement of in vitro CFTR function to about 10% of CFTR activity of healthy controls. However, in vivo CFTR function was improved to 30–40% of normal CFTR activity in the *G85E*G85E/*G85E*G85E patient and to 5–20% in the *N1303K*N1303K/*N1303K*N1303K patient. This finding shows that patient-derived models such as pHNECs enable detection of functional restoration of CFTR variants in vitro but cannot predict the degree of the in vivo response in an individual patient. This has also previously been observed for other preclinical models, such as intestinal organoids, where no correlation between in vitro organoid swelling and in vivo improvement in CFTR function was observed in F508del homozygous patients treated with lumacaftor/ivacaftor [[35](#B35-ijms-24-12365)35]. We therefore suggest that in vivo biomarkers provide a valuable tool to confirm findings in preclinical in vitro models in individual patients.
|
| 107 |
+
|
| 108 |
+
Our study has several limitations: First, the small sample size may limit generalizability of the data. In n-of-1 studies, the variability of clinical endpoints could affect or disguise potential therapeutic effects. The intraindividual variability of repeated short-term measurements has been previously reported at 6.3% for FEV_1_1 % predicted and 7.4% for LCI [[36](#B36-ijms-24-12365)36,[37](#B37-ijms-24-12365)37]. The *G85E*G85E/*G85E*G85E patient showed an improvement in lung function in repeated measurements after initiation of ETI with a mean improvement of +6.6% in FEV_1_1 % predicted and −2.1 (20.2%) in LCI when comparing the mean values before and after initiation of ETI. In the *N1303K*N1303K/*N1303K*N1303K patient, FEV_1_1 % predicted improved initially by +10.3% and LCI improved by −2.3 (15.7%). Although FEV_1_1 % predicted decreased and LCI increased again, both parameters were still improved compared with the repeated measurements before initiation of ETI. Previous studies in *N1303K*N1303K patients have reported higher increases in FEV_1_1 % predicted after initiation of ETI treatment than observed in our study [[13](#B13-ijms-24-12365)13,[19](#B19-ijms-24-12365)19,[21](#B21-ijms-24-12365)21]. Clinical trials and real-world studies assessing the effects of ETI in patients with at least one F508del mutation have also observed a high heterogeneity in the response in FEV_1_1 % predicted, which may be explained by the fact that FEV_1_1 is influenced by numerous factors independent of CFTR function, such as fixed airflow limitation due to irreversible structural lung damage or adherence to concomitant therapies. We used multiple repeated clinical outcome measures and independent measurements of the in vitro and in vivo CFTR function to determine the response to ETI in both patients, suggesting that the improvement in lung function is related to the therapy rather than variability of the measurements. In addition, other recent reports support that ETI shows clinical benefit in patients with a *G85E*G85E and an *N1303K*N1303K mutation [[13](#B13-ijms-24-12365)13,[19](#B19-ijms-24-12365)19,[21](#B21-ijms-24-12365)21,[38](#B38-ijms-24-12365)38]. Second, pHNECs have not been validated in large patient cohorts receiving CFTR modulator therapy and it is unknown if a lack of response in pHNECs predicts a lack of clinical response. Third, for the in vitro experiments, we used a concentration of tezacaftor that was previously used in the pre-clinical development of the triple combination therapy [[39](#B39-ijms-24-12365)39], although it is unknown if these concentrations are reached in patients. Finally, the elexacaftor used in the in vitro experiments is a racemic mixture of two enantiomers which may have different effects than those of ETI in patients and therefore potentially underestimate the in vitro effects [[18](#B18-ijms-24-12365)18,[40](#B40-ijms-24-12365)40].
|
| 109 |
+
|
| 110 |
+
In summary, our results show an improvement of CFTR function and clinical benefit in patients homozygous for *G85E*G85E or *N1303K*N1303K after initiation of ETI therapy. Furthermore, these results support that patient-derived pHNECs may have the potential to indicate clinical response of rare *CFTR*CFTR mutations to ETI therapy. We suggest that the sequential use of in vitro testing of CFTR function in pHNECs and subsequent in vivo testing using biomarkers of CFTR function, in combination with clinical outcome measures, is a promising approach to facilitate access to CFTR modulator therapy and to enhance precision medicine for patients with CF carrying rare *CFTR*CFTR mutations.
|
| 111 |
+
|
| 112 |
+
## 4. Materials and Methods
|
| 113 |
+
|
| 114 |
+
### 4.1. Study Design and Participants
|
| 115 |
+
|
| 116 |
+
Patients participated in an ongoing multicenter longitudinal study on the effects of CFTR modulators (ClinicalTrials.gov Identifier: [NCT04732910](https://clinicaltrials.gov/ct2/show/NCT04732910)NCT04732910). The study was approved by the ethics committee of the Charité-Universitätsmedizin Berlin (EA2/220/18). Written informed consent was obtained from both patients included in the study and the parents of the *G85E/G85E*G85E/G85E patient. Patients provided nasal brushes for preclinical testing in primary nasal epithelial cell cultures. After proof of a significant preclinical response, the patients were treated with the standard dose of ELX 200 mg and TEZ 100 mg every 24 h in combination with IVA 150 mg every 12 h. Anthropometrics, spirometry, multiple-breath washout, SCC, NPD and ICM were assessed at baseline and after initiation of ETI. In the *G85E/G85E*G85E/G85E patient, total IgE levels and lung MRI were additionally performed at baseline and after initiation of ETI.
|
| 117 |
+
|
| 118 |
+
### 4.2. Nasal Epithelial Cell Cultures
|
| 119 |
+
|
| 120 |
+
Primary human nasal epithelial cells were cultured as previously described [[22](#B22-ijms-24-12365)22,[41](#B41-ijms-24-12365)41]. Briefly, cells were obtained by nasal brushing and expanded in co-culture with irradiated mouse 3T3 fibroblasts in the presence of RhoA kinase inhibitor Y-27632 (Stemcell Technologies, Cologne, Germany). Epithelial cells were seeded at passage 2 at a density of 200,000 cells/cm^2^2 on human placental type IV collagen-coated Snapwell supports (Corning, Glendale, NY, USA) in UNC–ALI medium and differentiated at the air–liquid interface (ALI) for at least 3 weeks.
|
| 121 |
+
|
| 122 |
+
### 4.3. Ussing Chamber Experiments in pHNECs
|
| 123 |
+
|
| 124 |
+
Cultures were treated with 3 µM elexacaftor (E) and 18 µM tezacaftor (T) or with 0.07% dimethyl sulfoxide (DMSO) (vehicle control) for 24 before Ussing chamber analysis. Ivacaftor was added acutely during transepithelial short-circuit current (I_sc_sc) measurements to ET pre-treated cultures, to provide conditions that maximally enhance CFTR function. CFTR modulators were obtained from Selleck Chemicals (Planegg, Germany), DMSO was obtained from Sigma-Aldrich (St Louis, MO, USA). I_sc_sc was measured by EasyMount Ussing chambers (Physiologic Instruments, San Diego, CA, USA) as previously described, in presence of a chloride gradient (basolateral 145 mM vs. apical 5 mM) [[22](#B22-ijms-24-12365)22,[42](#B42-ijms-24-12365)42]. After 10–20 min equilibration, amiloride (100 µM) was added to inhibit sodium absorption via the epithelial sodium channel (ENaC). To assess CFTR-mediated chloride secretion, forskolin (Fsk, 10 µM) and 3-isobutyl-1-methylxanthin (IBMX, 100 µM) were applied together, then ivacaftor (2.5 µM) was added, followed by CFTR inhibitor-172 (CFTRinh-172, 20 µM, TargetMol chemicals, Boston, MA, USA). All Ussing chamber chemicals and reagents, apart from CFTRinh-172 and ivacaftor, were obtained from Sigma-Aldrich (St Louis, MO, USA) at the highest level of available purity. Bioelectric responses were quantified by LabChart8 (AF Instruments, Oxfordshire, UK).
|
| 125 |
+
|
| 126 |
+
### 4.4. Lung Function and Multiple-Breath Washout (MBW) Measurements
|
| 127 |
+
|
| 128 |
+
Spirometry was performed according to the standards endorsed by the American Thoracic Society and European Respiratory Society and values were expressed as percent predicted according to published normative data [[43](#B43-ijms-24-12365)43,[44](#B44-ijms-24-12365)44]. MBW testing was performed with the Exhalyzer D system (Eco Medics, Duernten, Switzerland) and 100% oxygen was used to wash out resident nitrogen from the lungs with a mouthpiece as interface [[45](#B45-ijms-24-12365)45,[46](#B46-ijms-24-12365)46]. All measurements were evaluated using spiroware 3.3.1 (Eco Medics, Duernten, Switzerland) [[10](#B10-ijms-24-12365)10,[47](#B47-ijms-24-12365)47].
|
| 129 |
+
|
| 130 |
+
### 4.5. Morpho-Functional Chest Magnetic Resonance Imaging (MRI)
|
| 131 |
+
|
| 132 |
+
A standardized MRI protocol was used on a 1.5 Tesla MRI scanner (Magnetom Avanto, Magnetom Aera; Siemens Healthineers, Erlangen, Germany) at baseline and after initiation of therapy to acquire T1- and T2-weighted sequences. Lung perfusion was assessed by contrast-enhanced imaging [[48](#B48-ijms-24-12365)48]. Images were assessed for abnormalities in lung morphology and perfusion using a dedicated morpho-functional MRI score as previously described [[49](#B49-ijms-24-12365)49]. In this MRI score, structural findings are assessed by the MRI morphology score that comprises subscores for (i) bronchial wall abnormalities (wall thickening and/or bronchiectasis), (ii) mucus plugging, (iii) abscesses and/or sacculations, (iv) consolidations, and (v) special findings such as pleural effusion. The extent of these structural findings, as well as of changes in lung perfusion, are rated in each lobe (right upper lobe, right middle lobe, right lower lobe, left upper lobe, lingula, left lower lobe) as 0 (no abnormality), 1 (<50% of the lobe involved), or 2 (≥50% of the lobe involved) resulting in a range of the MRI global score from 0 (normal) to 72 (severely abnormal) with each score of ≥1 being rated as abnormal [[48](#B48-ijms-24-12365)48,[49](#B49-ijms-24-12365)49].
|
| 133 |
+
|
| 134 |
+
### 4.6. Sweat Chloride
|
| 135 |
+
|
| 136 |
+
Sweat tests were performed according to the German national diagnostic guideline [[50](#B50-ijms-24-12365)50] and the guidelines of the Clinical and Laboratory Standards Institute [[51](#B51-ijms-24-12365)51]. Sweating was stimulated by pilocarpine iontophoresis and samples were collected with the Macroduct^®^® system (Model 3700, Wescor, Logan, UT, USA). SCC was measured in a minimum volume of 30 μL using a chloridometer (KWM 20 Chloridometer, Kreienbaum, Langenfeld, Germany).
|
| 137 |
+
|
| 138 |
+
### 4.7. NPD Measurements
|
| 139 |
+
|
| 140 |
+
NPD measurements were performed as previously described [[23](#B23-ijms-24-12365)23,[52](#B52-ijms-24-12365)52,[53](#B53-ijms-24-12365)53]. The percentage of normal CFTR activity in the nasal epithelium was defined as the ratio of the total chloride conductance (TCC) of the individual CF patient to the median TCC in age-matched non-CF controls as previously described [[23](#B23-ijms-24-12365)23].
|
| 141 |
+
|
| 142 |
+
### 4.8. Intestinal Current Measurements
|
| 143 |
+
|
| 144 |
+
ICM was performed as previously described [[11](#B11-ijms-24-12365)11,[23](#B23-ijms-24-12365)23,[24](#B24-ijms-24-12365)24,[54](#B54-ijms-24-12365)54]. In brief, biopsies of the rectal mucosa were collected by endoscopic forceps biopsy, immediately stored in tissue medium (medium 199 containing Hank’s salts, L-glutamine and 25 mM HEPES complemented with 5 mM glycine and 0.5 mM sodium-DL-β-hydroxybutyrate or RPMI-1640 medium with L-glutamine and sodium bicarbonate) and mounted in custom-made perfused micro-Ussing chambers. The luminal and basolateral compartments were perfused continuously with a buffer solution of the following composition: 145 mM NaCl, 0.4 mM KH_2_2PO_4_4, 1.6 mM K_2_2HPO_4_4, 5 mM D-glucose, 1 mM MgCl_2_2, 1.3 mM Ca-gluconate, pH 7.4, at 37 °C. All reagents were obtained from Sigma-Aldrich at the highest level of available purity. Experiments were performed under open circuit conditions. Transepithelial voltage (V_te_te) was recorded and transepithelial resistance (R_te_te) was determined by applying intermittent (1 s) current pulses (ΔI = 0.5 µA). The equivalent short-circuit current (I_sc_sc) was determined from continuous V_te_te and R_te_te recordings according to Ohm’s law (I_sc_sc = V_te_te/R_te_te) after appropriate correction for fluid resistance [[34](#B34-ijms-24-12365)34,[55](#B55-ijms-24-12365)55,[56](#B56-ijms-24-12365)56,[57](#B57-ijms-24-12365)57]. The percentage of normal CFTR function for each CF patient was calculated by dividing the individual cAMP-induced Isc of the patient by the median cAMP-induced Isc of age-matched non-CF controls as previously described [[11](#B11-ijms-24-12365)11,[23](#B23-ijms-24-12365)23,[24](#B24-ijms-24-12365)24].
|
| 145 |
+
|
| 146 |
+
### 4.9. Statistical Analysis
|
| 147 |
+
|
| 148 |
+
Data were analyzed using GraphPad Prism version 9.4.1 (GraphPad Software, San Diego, CA, USA) and R 3.6.2 [[58](#B58-ijms-24-12365)58]. Data were normally distributed and are presented as mean and standard deviation. Differences between the ETI and vehicle group were tested by Student’s *t*t-test, *p*p < 0.05 was accepted to indicate statistical significance.
|
| 149 |
+
|
| 150 |
+
## Acknowledgments
|
| 151 |
+
|
| 152 |
+
The authors thank the patients for their participation in this study.
|
| 153 |
+
|
| 154 |
+
## Author Contributions
|
| 155 |
+
|
| 156 |
+
Conceptualization, S.Y.G., A.B., M.A.M. and M.S.; methodology, all authors; writing—original draft preparation, S.Y.G., A.B., T.R. and M.S.; writing—review and editing, all authors; visualization, S.Y.G. and A.B. All authors have read and agreed to the published version of the manuscript.
|
| 157 |
+
|
| 158 |
+
## Institutional Review Board Statement
|
| 159 |
+
|
| 160 |
+
The study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of the Charité – Universitätsmedizin Berlin (EA2/220/18).
|
| 161 |
+
|
| 162 |
+
## Informed Consent Statement
|
| 163 |
+
|
| 164 |
+
Informed consent was obtained from all subjects involved in the study.
|
| 165 |
+
|
| 166 |
+
## Data Availability Statement
|
| 167 |
+
|
| 168 |
+
The data that support the findings of this study are available on request from the corresponding author M.S. The data are not publicly available due to legal restrictions, e.g., they contain information that could compromise the privacy of research participants.
|
| 169 |
+
|
| 170 |
+
## Conflicts of Interest
|
| 171 |
+
|
| 172 |
+
S.Y.G. reports grants from Vertex Pharmaceuticals; and lecture honoraria from Chiesi and Vertex Pharmaceuticals; and advisory board participation for Chiesi and Vertex Pharmaceuticals, outside the submitted work. M.AM. reports grants from Vertex Pharmaceuticals; fees for advisory board participation or consulting from Abbvie, Antabio, Arrowhead Pharmaceuticals, Boehringer Ingelheim, Enterprise Therapeutics, Kither Biotech, Pari, Prieris, Recode, Santhera, Splisense, Vertex Pharmaceuticals; lecture honoraria from Vertex Pharmaceuticals; travel support from Boehringer Ingelheim and Vertex Pharmaceuticals outside the submitted work. J.R. received lecture honoraria from Vertex Pharmaceuticals outside the submitted work. M.S. reports grants from Vertex Pharmaceuticals; and lecture honoraria from Vertex Pharmaceuticals; and advisory board participation for Vertex Pharmaceuticals, outside the submitted work. All other authors declare no conflict of interest.
|
| 173 |
+
|
| 174 |
+
## Funding Statement
|
| 175 |
+
|
| 176 |
+
This research was supported by a financial grant from the Christiane Herzog Stiftung, Stuttgart, Germany and the Mukoviszidose Institut gGmbH, Bonn, the research and development arm of the German Cystic Fibrosis Association Mukoviszidose e.V. (2101 C-H-P), the German Federal Ministry of Education and Research (82DZL009B1) and the German Research Foundation (CRC 1449–project 431232613; sub-project Z02; and project 450557679). The funders had no role in the design, management, data collection, analyses, interpretation of the data, writing of the manuscript or the decision to submit for publication. S.Y.G., S.T. and M.S. are participants of the Berlin Institute of Health (BIH)-Charité Clinician Scientist Program funded by the Charité – Universitätsmedizin Berlin and the BIH.
|
| 177 |
+
|
| 178 |
+
## Footnotes
|
| 179 |
+
|
| 180 |
+
## Associated Data
|
| 181 |
+
|
| 182 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 183 |
+
|
| 184 |
+
### Data Availability Statement
|
| 185 |
+
|
| 186 |
+
The data that support the findings of this study are available on request from the corresponding author M.S. The data are not publicly available due to legal restrictions, e.g., they contain information that could compromise the privacy of research participants.
|
| 187 |
+
|
| 188 |
+
### Data Availability Statement
|
| 189 |
+
|
| 190 |
+
The data that support the findings of this study are available on request from the corresponding author M.S. The data are not publicly available due to legal restrictions, e.g., they contain information that could compromise the privacy of research participants.
|
| 191 |
+
|
| 192 |
+
## References
|
| 193 |
+
|
| 194 |
+
1. Elborn J.S. Cystic fibrosis. Lancet. 2016;388:2519–2531. doi: 10.1016/S0140-6736(16)00576-6. [DOI](https://doi.org/10.1016/S0140-6736(16)00576-6) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27140670/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Cystic%20fibrosis&author=J.S.%20Elborn&volume=388&publication_year=2016&pages=2519-2531&pmid=27140670&doi=10.1016/S0140-6736(16)00576-6&)
|
| 195 |
+
|
| 196 |
+
2. Mall M.A., Hartl D. CFTR: Cystic fibrosis and beyond. Eur. Respir. J. 2014;44:1042–1054. doi: 10.1183/09031936.00228013. [DOI](https://doi.org/10.1183/09031936.00228013) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24925916/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Respir.%20J.&title=CFTR:%20Cystic%20fibrosis%20and%20beyond&author=M.A.%20Mall&author=D.%20Hartl&volume=44&publication_year=2014&pages=1042-1054&pmid=24925916&doi=10.1183/09031936.00228013&)
|
| 197 |
+
|
| 198 |
+
3. Barry P.J., Mall M.A., Alvarez A., Colombo C., de Winter-de Groot K.M., Fajac I., McBennett K.A., McKone E.F., Ramsey B.W., Sutharsan S., et al. Triple Therapy for Cystic Fibrosis Phe508del-Gating and -Residual Function Genotypes. N. Engl. J. Med. 2021;385:815–825. doi: 10.1056/NEJMoa2100665. [DOI](https://doi.org/10.1056/NEJMoa2100665) | [PMC free article](/articles/PMC8982185/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34437784/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N.%20Engl.%20J.%20Med.&title=Triple%20Therapy%20for%20Cystic%20Fibrosis%20Phe508del-Gating%20and%20-Residual%20Function%20Genotypes&author=P.J.%20Barry&author=M.A.%20Mall&author=A.%20Alvarez&author=C.%20Colombo&author=K.M.%20de%20Winter-de%20Groot&volume=385&publication_year=2021&pages=815-825&pmid=34437784&doi=10.1056/NEJMoa2100665&)
|
| 199 |
+
|
| 200 |
+
4. Heijerman H.G.M., McKone E.F., Downey D.G., Van Braeckel E., Rowe S.M., Tullis E., Mall M.A., Welter J.J., Ramsey B.W., McKee C.M., et al. Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: A double-blind, randomised, phase 3 trial. Lancet. 2019;394:1940–1948. doi: 10.1016/S0140-6736(19)32597-8. [DOI](https://doi.org/10.1016/S0140-6736(19)32597-8) | [PMC free article](/articles/PMC7571408/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31679946/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Efficacy%20and%20safety%20of%20the%20elexacaftor%20plus%20tezacaftor%20plus%20ivacaftor%20combination%20regimen%20in%20people%20with%20cystic%20fibrosis%20homozygous%20for%20the%20F508del%20mutation:%20A%20double-blind,%20randomised,%20phase%203%20trial&author=H.G.M.%20Heijerman&author=E.F.%20McKone&author=D.G.%20Downey&author=E.%20Van%20Braeckel&author=S.M.%20Rowe&volume=394&publication_year=2019&pages=1940-1948&pmid=31679946&doi=10.1016/S0140-6736(19)32597-8&)
|
| 201 |
+
|
| 202 |
+
5. Mall M.A., Brugha R., Gartner S., Legg J., Moeller A., Mondejar-Lopez P., Prais D., Pressler T., Ratjen F., Reix P., et al. Efficacy and Safety of Elexacaftor/Tezacaftor/Ivacaftor in Children 6 Through 11 Years of Age with Cystic Fibrosis Heterozygous for F508del and a Minimal Function Mutation: A Phase 3b, Randomized, Placebo-controlled Study. Am. J. Respir. Crit. Care Med. 2022;206:1361–1369. doi: 10.1164/rccm.202202-0392OC. [DOI](https://doi.org/10.1164/rccm.202202-0392OC) | [PMC free article](/articles/PMC9746869/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35816621/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Efficacy%20and%20Safety%20of%20Elexacaftor/Tezacaftor/Ivacaftor%20in%20Children%206%20Through%2011%20Years%20of%20Age%20with%20Cystic%20Fibrosis%20Heterozygous%20for%20F508del%20and%20a%20Minimal%20Function%20Mutation:%20A%20Phase%203b,%20Randomized,%20Placebo-controlled%20Study&author=M.A.%20Mall&author=R.%20Brugha&author=S.%20Gartner&author=J.%20Legg&author=A.%20Moeller&volume=206&publication_year=2022&pages=1361-1369&pmid=35816621&doi=10.1164/rccm.202202-0392OC&)
|
| 203 |
+
|
| 204 |
+
6. Mall M.A., Mayer-Hamblett N., Rowe S.M. Cystic Fibrosis: Emergence of Highly Effective Targeted Therapeutics and Potential Clinical Implications. Am. J. Respir. Crit. Care Med. 2020;201:1193–1208. doi: 10.1164/rccm.201910-1943SO. [DOI](https://doi.org/10.1164/rccm.201910-1943SO) | [PMC free article](/articles/PMC7233349/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31860331/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Cystic%20Fibrosis:%20Emergence%20of%20Highly%20Effective%20Targeted%20Therapeutics%20and%20Potential%20Clinical%20Implications&author=M.A.%20Mall&author=N.%20Mayer-Hamblett&author=S.M.%20Rowe&volume=201&publication_year=2020&pages=1193-1208&pmid=31860331&doi=10.1164/rccm.201910-1943SO&)
|
| 205 |
+
|
| 206 |
+
7. Middleton P.G., Mall M.A., Drevinek P., Lands L.C., McKone E.F., Polineni D., Ramsey B.W., Taylor-Cousar J.L., Tullis E., Vermeulen F., et al. Elexacaftor-Tezacaftor-Ivacaftor for Cystic Fibrosis with a Single Phe508del Allele. N. Engl. J. Med. 2019;381:1809–1819. doi: 10.1056/NEJMoa1908639. [DOI](https://doi.org/10.1056/NEJMoa1908639) | [PMC free article](/articles/PMC7282384/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31697873/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N.%20Engl.%20J.%20Med.&title=Elexacaftor-Tezacaftor-Ivacaftor%20for%20Cystic%20Fibrosis%20with%20a%20Single%20Phe508del%20Allele&author=P.G.%20Middleton&author=M.A.%20Mall&author=P.%20Drevinek&author=L.C.%20Lands&author=E.F.%20McKone&volume=381&publication_year=2019&pages=1809-1819&pmid=31697873&doi=10.1056/NEJMoa1908639&)
|
| 207 |
+
|
| 208 |
+
8. Sutharsan S., McKone E.F., Downey D.G., Duckers J., MacGregor G., Tullis E., Van Braeckel E., Wainwright C.E., Watson D., Ahluwalia N., et al. Efficacy and safety of elexacaftor plus tezacaftor plus ivacaftor versus tezacaftor plus ivacaftor in people with cystic fibrosis homozygous for F508del-CFTR: A 24-week, multicentre, randomised, double-blind, active-controlled, phase 3b trial. Lancet Respir. Med. 2022;10:267–277. doi: 10.1016/S2213-2600(21)00454-9. [DOI](https://doi.org/10.1016/S2213-2600(21)00454-9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34942085/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet%20Respir.%20Med.&title=Efficacy%20and%20safety%20of%20elexacaftor%20plus%20tezacaftor%20plus%20ivacaftor%20versus%20tezacaftor%20plus%20ivacaftor%20in%20people%20with%20cystic%20fibrosis%20homozygous%20for%20F508del-CFTR:%20A%2024-week,%20multicentre,%20randomised,%20double-blind,%20active-controlled,%20phase%203b%20trial&author=S.%20Sutharsan&author=E.F.%20McKone&author=D.G.%20Downey&author=J.%20Duckers&author=G.%20MacGregor&volume=10&publication_year=2022&pages=267-277&pmid=34942085&doi=10.1016/S2213-2600(21)00454-9&)
|
| 209 |
+
|
| 210 |
+
9. Zemanick E.T., Taylor-Cousar J.L., Davies J., Gibson R.L., Mall M.A., McKone E.F., McNally P., Ramsey B.W., Rayment J.H., Rowe S.M., et al. A Phase 3 Open-Label Study of ELX/TEZ/IVA in Children 6 Through 11 Years of Age With CF and at Least One F508del Allele. Am. J. Respir. Crit. Care Med. 2021;203:1522–1532. doi: 10.1164/rccm.202102-0509OC. [DOI](https://doi.org/10.1164/rccm.202102-0509OC) | [PMC free article](/articles/PMC8483230/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33734030/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=A%20Phase%203%20Open-Label%20Study%20of%20ELX/TEZ/IVA%20in%20Children%206%20Through%2011%20Years%20of%20Age%20With%20CF%20and%20at%20Least%20One%20F508del%20Allele&author=E.T.%20Zemanick&author=J.L.%20Taylor-Cousar&author=J.%20Davies&author=R.L.%20Gibson&author=M.A.%20Mall&volume=203&publication_year=2021&pages=1522-1532&pmid=33734030&doi=10.1164/rccm.202102-0509OC&)
|
| 211 |
+
|
| 212 |
+
10. Graeber S.Y., Renz D.M., Stahl M., Pallenberg S.T., Sommerburg O., Naehrlich L., Berges J., Dohna M., Ringshausen F.C., Doellinger F., et al. Effects of Elexacaftor/Tezacaftor/Ivacaftor Therapy on Lung Clearance Index and Magnetic Resonance Imaging in Patients with Cystic Fibrosis and One or Two F508del Alleles. Am. J. Respir. Crit. Care Med. 2022;206:311–320. doi: 10.1164/rccm.202201-0219OC. [DOI](https://doi.org/10.1164/rccm.202201-0219OC) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35536314/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Effects%20of%20Elexacaftor/Tezacaftor/Ivacaftor%20Therapy%20on%20Lung%20Clearance%20Index%20and%20Magnetic%20Resonance%20Imaging%20in%20Patients%20with%20Cystic%20Fibrosis%20and%20One%20or%20Two%20F508del%20Alleles&author=S.Y.%20Graeber&author=D.M.%20Renz&author=M.%20Stahl&author=S.T.%20Pallenberg&author=O.%20Sommerburg&volume=206&publication_year=2022&pages=311-320&pmid=35536314&doi=10.1164/rccm.202201-0219OC&)
|
| 213 |
+
|
| 214 |
+
11. Graeber S.Y., Vitzthum C., Pallenberg S.T., Naehrlich L., Stahl M., Rohrbach A., Drescher M., Minso R., Ringshausen F.C., Rueckes-Nilges C., et al. Effects of Elexacaftor/Tezacaftor/Ivacaftor Therapy on CFTR Function in Patients with Cystic Fibrosis and One or Two F508del Alleles. Am. J. Respir. Crit. Care Med. 2022;205:540–549. doi: 10.1164/rccm.202110-2249OC. [DOI](https://doi.org/10.1164/rccm.202110-2249OC) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34936849/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Effects%20of%20Elexacaftor/Tezacaftor/Ivacaftor%20Therapy%20on%20CFTR%20Function%20in%20Patients%20with%20Cystic%20Fibrosis%20and%20One%20or%20Two%20F508del%20Alleles&author=S.Y.%20Graeber&author=C.%20Vitzthum&author=S.T.%20Pallenberg&author=L.%20Naehrlich&author=M.%20Stahl&volume=205&publication_year=2022&pages=540-549&pmid=34936849&doi=10.1164/rccm.202110-2249OC&)
|
| 215 |
+
|
| 216 |
+
12. Bell S.C., Mall M.A., Gutierrez H., Macek M., Madge S., Davies J.C., Burgel P.R., Tullis E., Castanos C., Castellani C., et al. The future of cystic fibrosis care: A global perspective. Lancet Respir. Med. 2020;8:65–124. doi: 10.1016/S2213-2600(19)30337-6. [DOI](https://doi.org/10.1016/S2213-2600(19)30337-6) | [PMC free article](/articles/PMC8862661/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31570318/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet%20Respir.%20Med.&title=The%20future%20of%20cystic%20fibrosis%20care:%20A%20global%20perspective&author=S.C.%20Bell&author=M.A.%20Mall&author=H.%20Gutierrez&author=M.%20Macek&author=S.%20Madge&volume=8&publication_year=2020&pages=65-124&pmid=31570318&doi=10.1016/S2213-2600(19)30337-6&)
|
| 217 |
+
|
| 218 |
+
13. Burgel P.R., Sermet-Gaudelus I., Durieu I., Kanaan R., Macey J., Grenet D., Porzio M., Coolen-Allou N., Chiron R., Marguet C., et al. The French Compassionate Program of elexacaftor-tezacaftor-ivacaftor in people with cystic fibrosis with advanced lung disease and no F508del CFTR variant. Eur. Respir. J. 2023;61:2202437. doi: 10.1183/13993003.02437-2022. [DOI](https://doi.org/10.1183/13993003.02437-2022) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36796836/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Respir.%20J.&title=The%20French%20Compassionate%20Program%20of%20elexacaftor-tezacaftor-ivacaftor%20in%20people%20with%20cystic%20fibrosis%20with%20advanced%20lung%20disease%20and%20no%20F508del%20CFTR%20variant&author=P.R.%20Burgel&author=I.%20Sermet-Gaudelus&author=I.%20Durieu&author=R.%20Kanaan&author=J.%20Macey&volume=61&publication_year=2023&pages=2202437&pmid=36796836&doi=10.1183/13993003.02437-2022&)
|
| 219 |
+
|
| 220 |
+
14. Awatade N.T., Uliyakina I., Farinha C.M., Clarke L.A., Mendes K., Solé A., Pastor J., Ramos M.M., Amaral M.D. Measurements of Functional Responses in Human Primary Lung Cells as a Basis for Personalized Therapy for Cystic Fibrosis. EBioMedicine. 2015;2:147–153. doi: 10.1016/j.ebiom.2014.12.005. [DOI](https://doi.org/10.1016/j.ebiom.2014.12.005) | [PMC free article](/articles/PMC4484512/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26137539/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=EBioMedicine&title=Measurements%20of%20Functional%20Responses%20in%20Human%20Primary%20Lung%20Cells%20as%20a%20Basis%20for%20Personalized%20Therapy%20for%20Cystic%20Fibrosis&author=N.T.%20Awatade&author=I.%20Uliyakina&author=C.M.%20Farinha&author=L.A.%20Clarke&author=K.%20Mendes&volume=2&publication_year=2015&pages=147-153&pmid=26137539&doi=10.1016/j.ebiom.2014.12.005&)
|
| 221 |
+
|
| 222 |
+
15. Ensinck M., De Keersmaecker L., Heylen L., Ramalho A.S., Gijsbers R., Farre R., De Boeck K., Christ F., Debyser Z., Carlon M.S. Phenotyping of Rare CFTR Mutations Reveals Distinct Trafficking and Functional Defects. Cells. 2020;9:754. doi: 10.3390/cells9030754. [DOI](https://doi.org/10.3390/cells9030754) | [PMC free article](/articles/PMC7140603/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32204475/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cells&title=Phenotyping%20of%20Rare%20CFTR%20Mutations%20Reveals%20Distinct%20Trafficking%20and%20Functional%20Defects&author=M.%20Ensinck&author=L.%20De%20Keersmaecker&author=L.%20Heylen&author=A.S.%20Ramalho&author=R.%20Gijsbers&volume=9&publication_year=2020&pages=754&pmid=32204475&doi=10.3390/cells9030754&)
|
| 223 |
+
|
| 224 |
+
16. DeStefano S., Gees M., Hwang T.C. Physiological and pharmacological characterization of the N1303K mutant CFTR. J. Cyst. Fibros. 2018;17:573–581. doi: 10.1016/j.jcf.2018.05.011. [DOI](https://doi.org/10.1016/j.jcf.2018.05.011) | [PMC free article](/articles/PMC7008954/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29887518/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Cyst.%20Fibros.&title=Physiological%20and%20pharmacological%20characterization%20of%20the%20N1303K%20mutant%20CFTR&author=S.%20DeStefano&author=M.%20Gees&author=T.C.%20Hwang&volume=17&publication_year=2018&pages=573-581&pmid=29887518&doi=10.1016/j.jcf.2018.05.011&)
|
| 225 |
+
|
| 226 |
+
17. Laselva O., Bartlett C., Gunawardena T.N.A., Ouyang H., Eckford P.D.W., Moraes T.J., Bear C.E., Gonska T. Rescue of multiple class II CFTR mutations by elexacaftor+ tezacaftor+ivacaftor mediated in part by the dual activities of Elexacaftor as both corrector and potentiator. Eur. Respir. J. 2020;57:2002774. doi: 10.1183/13993003.02774-2020. [DOI](https://doi.org/10.1183/13993003.02774-2020) | [PMC free article](/articles/PMC8209484/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33303536/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Respir.%20J.&title=Rescue%20of%20multiple%20class%20II%20CFTR%20mutations%20by%20elexacaftor+%20tezacaftor+ivacaftor%20mediated%20in%20part%20by%20the%20dual%20activities%20of%20Elexacaftor%20as%20both%20corrector%20and%20potentiator&author=O.%20Laselva&author=C.%20Bartlett&author=T.N.A.%20Gunawardena&author=H.%20Ouyang&author=P.D.W.%20Eckford&volume=57&publication_year=2020&pages=2002774&pmid=33303536&doi=10.1183/13993003.02774-2020&)
|
| 227 |
+
|
| 228 |
+
18. Veit G., Roldan A., Hancock M.A., Da Fonte D.F., Xu H., Hussein M., Frenkiel S., Matouk E., Velkov T., Lukacs G.L. Allosteric folding correction of F508del and rare CFTR mutants by elexacaftor-tezacaftor-ivacaftor (Trikafta) combination. JCI Insight. 2020;5:e139983. doi: 10.1172/jci.insight.139983. [DOI](https://doi.org/10.1172/jci.insight.139983) | [PMC free article](/articles/PMC7526550/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32853178/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=JCI%20Insight&title=Allosteric%20folding%20correction%20of%20F508del%20and%20rare%20CFTR%20mutants%20by%20elexacaftor-tezacaftor-ivacaftor%20(Trikafta)%20combination&author=G.%20Veit&author=A.%20Roldan&author=M.A.%20Hancock&author=D.F.%20Da%20Fonte&author=H.%20Xu&volume=5&publication_year=2020&pages=e139983&pmid=32853178&doi=10.1172/jci.insight.139983&)
|
| 229 |
+
|
| 230 |
+
19. Huang Y., Paul G., Lee J., Yarlagadda S., McCoy K., Naren A.P. Elexacaftor/Tezacaftor/Ivacaftor Improved Clinical Outcomes in a Patient with N1303K-CFTR Based on In Vitro Experimental Evidence. Am. J. Respir. Crit. Care Med. 2021;204:1231–1235. doi: 10.1164/rccm.202101-0090LE. [DOI](https://doi.org/10.1164/rccm.202101-0090LE) | [PMC free article](/articles/PMC8759307/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34379998/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Elexacaftor/Tezacaftor/Ivacaftor%20Improved%20Clinical%20Outcomes%20in%20a%20Patient%20with%20N1303K-CFTR%20Based%20on%20In%20Vitro%20Experimental%20Evidence&author=Y.%20Huang&author=G.%20Paul&author=J.%20Lee&author=S.%20Yarlagadda&author=K.%20McCoy&volume=204&publication_year=2021&pages=1231-1235&pmid=34379998&doi=10.1164/rccm.202101-0090LE&)
|
| 231 |
+
|
| 232 |
+
20. Stekolchik E., Saul D., Chidekel A. Clinical efficacy of elexacaftor-tezacaftor-ivacaftor in an adolescent with homozygous G85E cystic fibrosis. Respir. Med. Case Rep. 2022;40:101775. doi: 10.1016/j.rmcr.2022.101775. [DOI](https://doi.org/10.1016/j.rmcr.2022.101775) | [PMC free article](/articles/PMC9674891/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36411821/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Respir.%20Med.%20Case%20Rep.&title=Clinical%20efficacy%20of%20elexacaftor-tezacaftor-ivacaftor%20in%20an%20adolescent%20with%20homozygous%20G85E%20cystic%20fibrosis&author=E.%20Stekolchik&author=D.%20Saul&author=A.%20Chidekel&volume=40&publication_year=2022&pages=101775&pmid=36411821&doi=10.1016/j.rmcr.2022.101775&)
|
| 233 |
+
|
| 234 |
+
21. Sadras I., Kerem E., Livnat G., Sarouk I., Breuer O., Reiter J., Gileles-Hillel A., Inbar O., Cohen M., Gamliel A., et al. Clinical and functional efficacy of elexacaftor/tezacaftor/ivacaftor in people with cystic fibrosis carrying the N1303K mutation. J. Cyst. Fibros. 2023 doi: 10.1016/j.jcf.2023.06.001. in press . [DOI](https://doi.org/10.1016/j.jcf.2023.06.001) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37331863/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Cyst.%20Fibros.&title=Clinical%20and%20functional%20efficacy%20of%20elexacaftor/tezacaftor/ivacaftor%20in%20people%20with%20cystic%20fibrosis%20carrying%20the%20N1303K%20mutation&author=I.%20Sadras&author=E.%20Kerem&author=G.%20Livnat&author=I.%20Sarouk&author=O.%20Breuer&publication_year=2023&pmid=37331863&doi=10.1016/j.jcf.2023.06.001&)
|
| 235 |
+
|
| 236 |
+
22. Balazs A., Millar-Buchner P., Mulleder M., Farztdinov V., Szyrwiel L., Addante A., Kuppe A., Rubil T., Drescher M., Seidel K., et al. Age-Related Differences in Structure and Function of Nasal Epithelial Cultures From Healthy Children and Elderly People. Front. Immunol. 2022;13:822437. doi: 10.3389/fimmu.2022.822437. [DOI](https://doi.org/10.3389/fimmu.2022.822437) | [PMC free article](/articles/PMC8918506/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35296085/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front.%20Immunol.&title=Age-Related%20Differences%20in%20Structure%20and%20Function%20of%20Nasal%20Epithelial%20Cultures%20From%20Healthy%20Children%20and%20Elderly%20People&author=A.%20Balazs&author=P.%20Millar-Buchner&author=M.%20Mulleder&author=V.%20Farztdinov&author=L.%20Szyrwiel&volume=13&publication_year=2022&pages=822437&pmid=35296085&doi=10.3389/fimmu.2022.822437&)
|
| 237 |
+
|
| 238 |
+
23. Graeber S.Y., Dopfer C., Naehrlich L., Gyulumyan L., Scheuermann H., Hirtz S., Wege S., Mairbaurl H., Dorda M., Hyde R., et al. Effects of Lumacaftor-Ivacaftor Therapy on Cystic Fibrosis Transmembrane Conductance Regulator Function in Phe508del Homozygous Patients with Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2018;197:1433–1442. doi: 10.1164/rccm.201710-1983OC. [DOI](https://doi.org/10.1164/rccm.201710-1983OC) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29327948/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Effects%20of%20Lumacaftor-Ivacaftor%20Therapy%20on%20Cystic%20Fibrosis%20Transmembrane%20Conductance%20Regulator%20Function%20in%20Phe508del%20Homozygous%20Patients%20with%20Cystic%20Fibrosis&author=S.Y.%20Graeber&author=C.%20Dopfer&author=L.%20Naehrlich&author=L.%20Gyulumyan&author=H.%20Scheuermann&volume=197&publication_year=2018&pages=1433-1442&pmid=29327948&doi=10.1164/rccm.201710-1983OC&)
|
| 239 |
+
|
| 240 |
+
24. Graeber S.Y., Hug M.J., Sommerburg O., Hirtz S., Hentschel J., Heinzmann A., Dopfer C., Schulz A., Mainz J.G., Tummler B., et al. Intestinal Current Measurements Detect Activation of Mutant CFTR in Patients with Cystic Fibrosis with the G551D Mutation Treated with Ivacaftor. Am. J. Respir. Crit. Care Med. 2015;192:1252–1255. doi: 10.1164/rccm.201507-1271LE. [DOI](https://doi.org/10.1164/rccm.201507-1271LE) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26568242/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Intestinal%20Current%20Measurements%20Detect%20Activation%20of%20Mutant%20CFTR%20in%20Patients%20with%20Cystic%20Fibrosis%20with%20the%20G551D%20Mutation%20Treated%20with%20Ivacaftor&author=S.Y.%20Graeber&author=M.J.%20Hug&author=O.%20Sommerburg&author=S.%20Hirtz&author=J.%20Hentschel&volume=192&publication_year=2015&pages=1252-1255&pmid=26568242&doi=10.1164/rccm.201507-1271LE&)
|
| 241 |
+
|
| 242 |
+
25. Ensinck M.M., De Keersmaecker L., Ramalho A.S., Cuyx S., Van Biervliet S., Dupont L., Christ F., Debyser Z., Vermeulen F., Carlon M.S. Novel CFTR modulator combinations maximise rescue of G85E and N1303K in rectal organoids. ERJ Open Res. 2022;8:00716–2021. doi: 10.1183/23120541.00716-2021. [DOI](https://doi.org/10.1183/23120541.00716-2021) | [PMC free article](/articles/PMC9016267/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35449760/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=ERJ%20Open%20Res.&title=Novel%20CFTR%20modulator%20combinations%20maximise%20rescue%20of%20G85E%20and%20N1303K%20in%20rectal%20organoids&author=M.M.%20Ensinck&author=L.%20De%20Keersmaecker&author=A.S.%20Ramalho&author=S.%20Cuyx&author=S.%20Van%20Biervliet&volume=8&publication_year=2022&pages=00716-2021&pmid=35449760&doi=10.1183/23120541.00716-2021&)
|
| 243 |
+
|
| 244 |
+
26. US Food and Drug Administration Trikafta Label. [(accessed on 16 July 2023)]; Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/217660s000lbl.pdf. [https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/217660s000lbl.pdf](https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/217660s000lbl.pdf)
|
| 245 |
+
|
| 246 |
+
27. European Medical Agency Assessment Report Kaftrio. [(accessed on 16 July 2023)]. Available online: https://www.ema.europa.eu/en/documents/assessment-report/kaftrio-epar-public-assessment-report_en.pdf. [https://www.ema.europa.eu/en/documents/assessment-report/kaftrio-epar-public-assessment-report_en.pdf](https://www.ema.europa.eu/en/documents/assessment-report/kaftrio-epar-public-assessment-report_en.pdf)
|
| 247 |
+
|
| 248 |
+
28. Stutts M.J., Canessa C.M., Olsen J.C., Hamrick M., Cohn J.A., Rossier B.C., Boucher R.C. CFTR as a cAMP-dependent regulator of sodium channels. Science. 1995;269:847–850. doi: 10.1126/science.7543698. [DOI](https://doi.org/10.1126/science.7543698) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7543698/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Science&title=CFTR%20as%20a%20cAMP-dependent%20regulator%20of%20sodium%20channels&author=M.J.%20Stutts&author=C.M.%20Canessa&author=J.C.%20Olsen&author=M.%20Hamrick&author=J.A.%20Cohn&volume=269&publication_year=1995&pages=847-850&pmid=7543698&doi=10.1126/science.7543698&)
|
| 249 |
+
|
| 250 |
+
29. Mall M., Hipper A., Greger R., Kunzelmann K. Wild type but not Delta F508 CFTR inhibits Na+ conductance when coexpressed in Xenopus oocytes. FEBS Lett. 1996;381:47–52. doi: 10.1016/0014-5793(96)00079-8. [DOI](https://doi.org/10.1016/0014-5793(96)00079-8) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8641437/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=FEBS%20Lett.&title=Wild%20type%20but%20not%20Delta%20F508%20CFTR%20inhibits%20Na+%20conductance%20when%20coexpressed%20in%20Xenopus%20oocytes&author=M.%20Mall&author=A.%20Hipper&author=R.%20Greger&author=K.%20Kunzelmann&volume=381&publication_year=1996&pages=47-52&pmid=8641437&doi=10.1016/0014-5793(96)00079-8&)
|
| 251 |
+
|
| 252 |
+
30. Mall M., Bleich M., Schurlein M., Kuhr J., Seydewitz H.H., Brandis M., Greger R., Kunzelmann K. Cholinergic ion secretion in human colon requires coactivation by cAMP. Am. J. Physiol. 1998;275:G1274–G1281. doi: 10.1152/ajpgi.1998.275.6.G1274. [DOI](https://doi.org/10.1152/ajpgi.1998.275.6.G1274) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9843763/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Physiol.&title=Cholinergic%20ion%20secretion%20in%20human%20colon%20requires%20coactivation%20by%20cAMP&author=M.%20Mall&author=M.%20Bleich&author=M.%20Schurlein&author=J.%20Kuhr&author=H.H.%20Seydewitz&volume=275&publication_year=1998&pages=G1274-G1281&pmid=9843763&doi=10.1152/ajpgi.1998.275.6.G1274&)
|
| 253 |
+
|
| 254 |
+
31. Mall M.A. ENaC inhibition in cystic fibrosis: Potential role in the new era of CFTR modulator therapies. Eur. Respir. J. 2020;56:2000946. doi: 10.1183/13993003.00946-2020. [DOI](https://doi.org/10.1183/13993003.00946-2020) | [PMC free article](/articles/PMC7758539/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32732328/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Respir.%20J.&title=ENaC%20inhibition%20in%20cystic%20fibrosis:%20Potential%20role%20in%20the%20new%20era%20of%20CFTR%20modulator%20therapies&author=M.A.%20Mall&volume=56&publication_year=2020&pages=2000946&pmid=32732328&doi=10.1183/13993003.00946-2020&)
|
| 255 |
+
|
| 256 |
+
32. Mall M., Bleich M., Greger R., Schreiber R., Kunzelmann K. The amiloride-inhibitable Na+ conductance is reduced by the cystic fibrosis transmembrane conductance regulator in normal but not in cystic fibrosis airways. J. Clin. Investig. 1998;102:15–21. doi: 10.1172/JCI2729. [DOI](https://doi.org/10.1172/JCI2729) | [PMC free article](/articles/PMC509060/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9649552/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Investig.&title=The%20amiloride-inhibitable%20Na+%20conductance%20is%20reduced%20by%20the%20cystic%20fibrosis%20transmembrane%20conductance%20regulator%20in%20normal%20but%20not%20in%20cystic%20fibrosis%20airways&author=M.%20Mall&author=M.%20Bleich&author=R.%20Greger&author=R.%20Schreiber&author=K.%20Kunzelmann&volume=102&publication_year=1998&pages=15-21&pmid=9649552&doi=10.1172/JCI2729&)
|
| 257 |
+
|
| 258 |
+
33. Mall M., Wissner A., Gonska T., Calenborn D., Kuehr J., Brandis M., Kunzelmann K. Inhibition of amiloride-sensitive epithelial Na(+) absorption by extracellular nucleotides in human normal and cystic fibrosis airways. Am. J. Respir. Cell Mol. Biol. 2000;23:755–761. doi: 10.1165/ajrcmb.23.6.4207. [DOI](https://doi.org/10.1165/ajrcmb.23.6.4207) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11104728/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Cell%20Mol.%20Biol.&title=Inhibition%20of%20amiloride-sensitive%20epithelial%20Na(+)%20absorption%20by%20extracellular%20nucleotides%20in%20human%20normal%20and%20cystic%20fibrosis%20airways&author=M.%20Mall&author=A.%20Wissner&author=T.%20Gonska&author=D.%20Calenborn&author=J.%20Kuehr&volume=23&publication_year=2000&pages=755-761&pmid=11104728&doi=10.1165/ajrcmb.23.6.4207&)
|
| 259 |
+
|
| 260 |
+
34. O’Brodovich H., Yang P., Gandhi S., Otulakowski G. Amiloride-insensitive Na+ and fluid absorption in the mammalian distal lung. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;294:L401–L408. doi: 10.1152/ajplung.00431.2007. [DOI](https://doi.org/10.1152/ajplung.00431.2007) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18162600/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Physiol.%20Lung%20Cell%20Mol.%20Physiol.&title=Amiloride-insensitive%20Na+%20and%20fluid%20absorption%20in%20the%20mammalian%20distal%20lung&author=H.%20O%E2%80%99Brodovich&author=P.%20Yang&author=S.%20Gandhi&author=G.%20Otulakowski&volume=294&publication_year=2008&pages=L401-L408&pmid=18162600&doi=10.1152/ajplung.00431.2007&)
|
| 261 |
+
|
| 262 |
+
35. Graeber S.Y., van Mourik P., Vonk A.M., Kruisselbrink E., Hirtz S., van der Ent C.K., Mall M.A., Beekman J.M. Comparison of Organoid Swelling and In Vivo Biomarkers of CFTR Function to Determine Effects of Lumacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for the F508del Mutation. Am. J. Respir. Crit. Care Med. 2020;202:1589–1592. doi: 10.1164/rccm.202004-1200LE. [DOI](https://doi.org/10.1164/rccm.202004-1200LE) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32687398/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Comparison%20of%20Organoid%20Swelling%20and%20In%20Vivo%20Biomarkers%20of%20CFTR%20Function%20to%20Determine%20Effects%20of%20Lumacaftor-Ivacaftor%20in%20Patients%20with%20Cystic%20Fibrosis%20Homozygous%20for%20the%20F508del%20Mutation&author=S.Y.%20Graeber&author=P.%20van%20Mourik&author=A.M.%20Vonk&author=E.%20Kruisselbrink&author=S.%20Hirtz&volume=202&publication_year=2020&pages=1589-1592&pmid=32687398&doi=10.1164/rccm.202004-1200LE&)
|
| 263 |
+
|
| 264 |
+
36. Taylor-Robinson D., Whitehead M., Diderichsen F., Olesen H.V., Pressler T., Smyth R.L., Diggle P. Understanding the natural progression in %FEV1 decline in patients with cystic fibrosis: A longitudinal study. Thorax. 2012;67:860–866. doi: 10.1136/thoraxjnl-2011-200953. [DOI](https://doi.org/10.1136/thoraxjnl-2011-200953) | [PMC free article](/articles/PMC3446776/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22555277/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Thorax&title=Understanding%20the%20natural%20progression%20in%20%FEV1%20decline%20in%20patients%20with%20cystic%20fibrosis:%20A%20longitudinal%20study&author=D.%20Taylor-Robinson&author=M.%20Whitehead&author=F.%20Diderichsen&author=H.V.%20Olesen&author=T.%20Pressler&volume=67&publication_year=2012&pages=860-866&pmid=22555277&doi=10.1136/thoraxjnl-2011-200953&)
|
| 265 |
+
|
| 266 |
+
37. Frauchiger B.S., Ramsey K.A., Usemann J., Kieninger E., Casaulta C., Sirtes D., Yammine S., Spycher B., Moeller A., Latzin P. Variability of clinically measured lung clearance index in children with cystic fibrosis. Pediatr. Pulmonol. 2023;58:197–205. doi: 10.1002/ppul.26180. [DOI](https://doi.org/10.1002/ppul.26180) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36251441/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pediatr.%20Pulmonol.&title=Variability%20of%20clinically%20measured%20lung%20clearance%20index%20in%20children%20with%20cystic%20fibrosis&author=B.S.%20Frauchiger&author=K.A.%20Ramsey&author=J.%20Usemann&author=E.%20Kieninger&author=C.%20Casaulta&volume=58&publication_year=2023&pages=197-205&pmid=36251441&doi=10.1002/ppul.26180&)
|
| 267 |
+
|
| 268 |
+
38. Elson E.C., Capel P., Haynes J., Duehlmeyer S., Fischer M., Escobar H. CFTR Modulator Therapy in an Individual With Cystic Fibrosis Caused by a N1303K CFTR Variant and Infected With Mycobacterium abscessus. J. Pediatr. Pharmacol. Ther. 2022;27:396–399. doi: 10.5863/1551-6776-27.4.396. [DOI](https://doi.org/10.5863/1551-6776-27.4.396) | [PMC free article](/articles/PMC9088447/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35558347/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Pediatr.%20Pharmacol.%20Ther.&title=CFTR%20Modulator%20Therapy%20in%20an%20Individual%20With%20Cystic%20Fibrosis%20Caused%20by%20a%20N1303K%20CFTR%20Variant%20and%20Infected%20With%20Mycobacterium%20abscessus&author=E.C.%20Elson&author=P.%20Capel&author=J.%20Haynes&author=S.%20Duehlmeyer&author=M.%20Fischer&volume=27&publication_year=2022&pages=396-399&pmid=35558347&doi=10.5863/1551-6776-27.4.396&)
|
| 269 |
+
|
| 270 |
+
39. Keating D., Marigowda G., Burr L., Daines C., Mall M.A., McKone E.F., Ramsey B.W., Rowe S.M., Sass L.A., Tullis E., et al. VX-445-Tezacaftor-Ivacaftor in Patients with Cystic Fibrosis and One or Two Phe508del Alleles. N. Engl. J. Med. 2018;379:1612–1620. doi: 10.1056/NEJMoa1807120. [DOI](https://doi.org/10.1056/NEJMoa1807120) | [PMC free article](/articles/PMC6289290/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30334692/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N.%20Engl.%20J.%20Med.&title=VX-445-Tezacaftor-Ivacaftor%20in%20Patients%20with%20Cystic%20Fibrosis%20and%20One%20or%20Two%20Phe508del%20Alleles&author=D.%20Keating&author=G.%20Marigowda&author=L.%20Burr&author=C.%20Daines&author=M.A.%20Mall&volume=379&publication_year=2018&pages=1612-1620&pmid=30334692&doi=10.1056/NEJMoa1807120&)
|
| 271 |
+
|
| 272 |
+
40. Capurro V., Tomati V., Sondo E., Renda M., Borrelli A., Pastorino C., Guidone D., Venturini A., Giraudo A., Mandrup Bertozzi S., et al. Partial Rescue of F508del-CFTR Stability and Trafficking Defects by Double Corrector Treatment. Int. J. Mol. Sci. 2021;22:5262. doi: 10.3390/ijms22105262. [DOI](https://doi.org/10.3390/ijms22105262) | [PMC free article](/articles/PMC8156943/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34067708/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int.%20J.%20Mol.%20Sci.&title=Partial%20Rescue%20of%20F508del-CFTR%20Stability%20and%20Trafficking%20Defects%20by%20Double%20Corrector%20Treatment&author=V.%20Capurro&author=V.%20Tomati&author=E.%20Sondo&author=M.%20Renda&author=A.%20Borrelli&volume=22&publication_year=2021&pages=5262&pmid=34067708&doi=10.3390/ijms22105262&)
|
| 273 |
+
|
| 274 |
+
41. Gentzsch M., Boyles S.E., Cheluvaraju C., Chaudhry I.G., Quinney N.L., Cho C., Dang H., Liu X., Schlegel R., Randell S.H. Pharmacological Rescue of Conditionally Reprogrammed Cystic Fibrosis Bronchial Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2017;56:568–574. doi: 10.1165/rcmb.2016-0276MA. [DOI](https://doi.org/10.1165/rcmb.2016-0276MA) | [PMC free article](/articles/PMC5449492/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27983869/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Cell%20Mol.%20Biol.&title=Pharmacological%20Rescue%20of%20Conditionally%20Reprogrammed%20Cystic%20Fibrosis%20Bronchial%20Epithelial%20Cells&author=M.%20Gentzsch&author=S.E.%20Boyles&author=C.%20Cheluvaraju&author=I.G.%20Chaudhry&author=N.L.%20Quinney&volume=56&publication_year=2017&pages=568-574&pmid=27983869&doi=10.1165/rcmb.2016-0276MA&)
|
| 275 |
+
|
| 276 |
+
42. Salomon J.J., Albrecht T., Graeber S.Y., Scheuermann H., Butz S., Schatterny J., Mairbaurl H., Baumann I., Mall M.A. Chronic rhinosinusitis with nasal polyps is associated with impaired TMEM16A-mediated epithelial chloride secretion. J. Allergy Clin. Immunol. 2021;147:2191–2201.e2192. doi: 10.1016/j.jaci.2021.02.008. [DOI](https://doi.org/10.1016/j.jaci.2021.02.008) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33609628/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Allergy%20Clin.%20Immunol.&title=Chronic%20rhinosinusitis%20with%20nasal%20polyps%20is%20associated%20with%20impaired%20TMEM16A-mediated%20epithelial%20chloride%20secretion&author=J.J.%20Salomon&author=T.%20Albrecht&author=S.Y.%20Graeber&author=H.%20Scheuermann&author=S.%20Butz&volume=147&publication_year=2021&pages=2191-2201.e2192&pmid=33609628&doi=10.1016/j.jaci.2021.02.008&)
|
| 277 |
+
|
| 278 |
+
43. Beydon N., Davis S.D., Lombardi E., Allen J.L., Arets H.G., Aurora P., Bisgaard H., Davis G.M., Ducharme F.M., Eigen H., et al. An official American Thoracic Society/European Respiratory Society statement: Pulmonary function testing in preschool children. Am. J. Respir. Crit. Care Med. 2007;175:1304–1345. doi: 10.1164/rccm.200605-642ST. [DOI](https://doi.org/10.1164/rccm.200605-642ST) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17545458/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=An%20official%20American%20Thoracic%20Society/European%20Respiratory%20Society%20statement:%20Pulmonary%20function%20testing%20in%20preschool%20children&author=N.%20Beydon&author=S.D.%20Davis&author=E.%20Lombardi&author=J.L.%20Allen&author=H.G.%20Arets&volume=175&publication_year=2007&pages=1304-1345&pmid=17545458&doi=10.1164/rccm.200605-642ST&)
|
| 279 |
+
|
| 280 |
+
44. Quanjer P.H., Stanojevic S., Cole T.J., Baur X., Hall G.L., Culver B.H., Enright P.L., Hankinson J.L., Ip M.S., Zheng J., et al. Multi-ethnic reference values for spirometry for the 3-95-yr age range: The global lung function 2012 equations. Eur. Respir. J. 2012;40:1324–1343. doi: 10.1183/09031936.00080312. [DOI](https://doi.org/10.1183/09031936.00080312) | [PMC free article](/articles/PMC3786581/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22743675/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Respir.%20J.&title=Multi-ethnic%20reference%20values%20for%20spirometry%20for%20the%203-95-yr%20age%20range:%20The%20global%20lung%20function%202012%20equations&author=P.H.%20Quanjer&author=S.%20Stanojevic&author=T.J.%20Cole&author=X.%20Baur&author=G.L.%20Hall&volume=40&publication_year=2012&pages=1324-1343&pmid=22743675&doi=10.1183/09031936.00080312&)
|
| 281 |
+
|
| 282 |
+
45. Stahl M., Joachim C., Kirsch I., Uselmann T., Yu Y., Alfeis N., Berger C., Minso R., Rudolf I., Stolpe C., et al. Multicentre feasibility of multiple-breath washout in preschool children with cystic fibrosis and other lung diseases. ERJ Open Res. 2020;6:00408–2020. doi: 10.1183/23120541.00408-2020. [DOI](https://doi.org/10.1183/23120541.00408-2020) | [PMC free article](/articles/PMC7682699/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33263048/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=ERJ%20Open%20Res.&title=Multicentre%20feasibility%20of%20multiple-breath%20washout%20in%20preschool%20children%20with%20cystic%20fibrosis%20and%20other%20lung%20diseases&author=M.%20Stahl&author=C.%20Joachim&author=I.%20Kirsch&author=T.%20Uselmann&author=Y.%20Yu&volume=6&publication_year=2020&pages=00408-2020&pmid=33263048&doi=10.1183/23120541.00408-2020&)
|
| 283 |
+
|
| 284 |
+
46. Stahl M., Wielputz M.O., Graeber S.Y., Joachim C., Sommerburg O., Kauczor H.U., Puderbach M., Eichinger M., Mall M.A. Comparison of Lung Clearance Index and Magnetic Resonance Imaging for Assessment of Lung Disease in Children with Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2017;195:349–359. doi: 10.1164/rccm.201604-0893OC. [DOI](https://doi.org/10.1164/rccm.201604-0893OC) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27575911/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Comparison%20of%20Lung%20Clearance%20Index%20and%20Magnetic%20Resonance%20Imaging%20for%20Assessment%20of%20Lung%20Disease%20in%20Children%20with%20Cystic%20Fibrosis&author=M.%20Stahl&author=M.O.%20Wielputz&author=S.Y.%20Graeber&author=C.%20Joachim&author=O.%20Sommerburg&volume=195&publication_year=2017&pages=349-359&pmid=27575911&doi=10.1164/rccm.201604-0893OC&)
|
| 285 |
+
|
| 286 |
+
47. Wyler F., Oestreich M.A., Frauchiger B.S., Ramsey K.A., Latzin P. Correction of sensor crosstalk error in Exhalyzer D multiple-breath washout device significantly impacts outcomes in children with cystic fibrosis. J. Appl. Physiol. 2021;131:1148–1156. doi: 10.1152/japplphysiol.00338.2021. [DOI](https://doi.org/10.1152/japplphysiol.00338.2021) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34351818/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Appl.%20Physiol.&title=Correction%20of%20sensor%20crosstalk%20error%20in%20Exhalyzer%20D%20multiple-breath%20washout%20device%20significantly%20impacts%20outcomes%20in%20children%20with%20cystic%20fibrosis&author=F.%20Wyler&author=M.A.%20Oestreich&author=B.S.%20Frauchiger&author=K.A.%20Ramsey&author=P.%20Latzin&volume=131&publication_year=2021&pages=1148-1156&pmid=34351818&doi=10.1152/japplphysiol.00338.2021&)
|
| 287 |
+
|
| 288 |
+
48. Wielputz M.O., Puderbach M., Kopp-Schneider A., Stahl M., Fritzsching E., Sommerburg O., Ley S., Sumkauskaite M., Biederer J., Kauczor H.U., et al. Magnetic resonance imaging detects changes in structure and perfusion, and response to therapy in early cystic fibrosis lung disease. Am. J. Respir. Crit. Care Med. 2014;189:956–965. doi: 10.1164/rccm.201309-1659OC. [DOI](https://doi.org/10.1164/rccm.201309-1659OC) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24564281/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Magnetic%20resonance%20imaging%20detects%20changes%20in%20structure%20and%20perfusion,%20and%20response%20to%20therapy%20in%20early%20cystic%20fibrosis%20lung%20disease&author=M.O.%20Wielputz&author=M.%20Puderbach&author=A.%20Kopp-Schneider&author=M.%20Stahl&author=E.%20Fritzsching&volume=189&publication_year=2014&pages=956-965&pmid=24564281&doi=10.1164/rccm.201309-1659OC&)
|
| 289 |
+
|
| 290 |
+
49. Eichinger M., Optazaite D.E., Kopp-Schneider A., Hintze C., Biederer J., Niemann A., Mall M.A., Wielputz M.O., Kauczor H.U., Puderbach M. Morphologic and functional scoring of cystic fibrosis lung disease using MRI. Eur. J. Radiol. 2012;81:1321–1329. doi: 10.1016/j.ejrad.2011.02.045. [DOI](https://doi.org/10.1016/j.ejrad.2011.02.045) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21429685/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20J.%20Radiol.&title=Morphologic%20and%20functional%20scoring%20of%20cystic%20fibrosis%20lung%20disease%20using%20MRI&author=M.%20Eichinger&author=D.E.%20Optazaite&author=A.%20Kopp-Schneider&author=C.%20Hintze&author=J.%20Biederer&volume=81&publication_year=2012&pages=1321-1329&pmid=21429685&doi=10.1016/j.ejrad.2011.02.045&)
|
| 291 |
+
|
| 292 |
+
50. Naehrlich L., Stuhrmann-Spangenberg M., Barben J., Bargon J., Blankenstein O., Bremer W., Brunsmann F., Buchholz T., Ellemunter H., Fusch C., et al. S2-Konsensus-Leitlinie “Diagnose der Mukoviszidose” (AWMF 026-023) [(accessed on 21 June 2023)]. Available online: http://www.awmf.org/leitlinien/detail/ll/026-023.html. [http://www.awmf.org/leitlinien/detail/ll/026-023.html](http://www.awmf.org/leitlinien/detail/ll/026-023.html)
|
| 293 |
+
|
| 294 |
+
51. LeGrys V.A. Sweat Testing: Sample Collection and Quantitative Chloride Analysis, Approved Guideline. 4th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2019. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Sweat%20Testing:%20Sample%20Collection%20and%20Quantitative%20Chloride%20Analysis,%20Approved%20Guideline&author=V.A.%20LeGrys&publication_year=2019&)
|
| 295 |
+
|
| 296 |
+
52. Rowe S.M., Clancy J.P., Wilschanski M. Nasal potential difference measurements to assess CFTR ion channel activity. Methods Mol. Biol. 2011;741:69–86. doi: 10.1007/978-1-61779-117-8_6. [DOI](https://doi.org/10.1007/978-1-61779-117-8_6) | [PMC free article](/articles/PMC3760477/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21594779/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Methods%20Mol.%20Biol.&title=Nasal%20potential%20difference%20measurements%20to%20assess%20CFTR%20ion%20channel%20activity&author=S.M.%20Rowe&author=J.P.%20Clancy&author=M.%20Wilschanski&volume=741&publication_year=2011&pages=69-86&pmid=21594779&doi=10.1007/978-1-61779-117-8_6&)
|
| 297 |
+
|
| 298 |
+
53. Sermet-Gaudelus I., Girodon E., Sands D., Stremmler N., Vavrova V., Deneuville E., Reix P., Bui S., Huet F., Lebourgeois M., et al. Clinical phenotype and genotype of children with borderline sweat test and abnormal nasal epithelial chloride transport. Am. J. Respir. Crit. Care Med. 2010;182:929–936. doi: 10.1164/rccm.201003-0382OC. [DOI](https://doi.org/10.1164/rccm.201003-0382OC) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20538955/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Clinical%20phenotype%20and%20genotype%20of%20children%20with%20borderline%20sweat%20test%20and%20abnormal%20nasal%20epithelial%20chloride%20transport&author=I.%20Sermet-Gaudelus&author=E.%20Girodon&author=D.%20Sands&author=N.%20Stremmler&author=V.%20Vavrova&volume=182&publication_year=2010&pages=929-936&pmid=20538955&doi=10.1164/rccm.201003-0382OC&)
|
| 299 |
+
|
| 300 |
+
54. Graeber S.Y., Vitzthum C., Mall M.A. Potential of Intestinal Current Measurement for Personalized Treatment of Patients with Cystic Fibrosis. J. Pers. Med. 2021;11:384. doi: 10.3390/jpm11050384. [DOI](https://doi.org/10.3390/jpm11050384) | [PMC free article](/articles/PMC8151208/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34066648/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Pers.%20Med.&title=Potential%20of%20Intestinal%20Current%20Measurement%20for%20Personalized%20Treatment%20of%20Patients%20with%20Cystic%20Fibrosis&author=S.Y.%20Graeber&author=C.%20Vitzthum&author=M.A.%20Mall&volume=11&publication_year=2021&pages=384&pmid=34066648&doi=10.3390/jpm11050384&)
|
| 301 |
+
|
| 302 |
+
55. Mall M., Wissner A., Seydewitz H.H., Kuehr J., Brandis M., Greger R., Kunzelmann K. Defective cholinergic Cl(-) secretion and detection of K(+) secretion in rectal biopsies from cystic fibrosis patients. Am. J. Physiol. Gastrointest. Liver Physiol. 2000;278:G617–G624. doi: 10.1152/ajpgi.2000.278.4.G617. [DOI](https://doi.org/10.1152/ajpgi.2000.278.4.G617) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10762616/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Physiol.%20Gastrointest.%20Liver%20Physiol.&title=Defective%20cholinergic%20Cl(-)%20secretion%20and%20detection%20of%20K(+)%20secretion%20in%20rectal%20biopsies%20from%20cystic%20fibrosis%20patients&author=M.%20Mall&author=A.%20Wissner&author=H.H.%20Seydewitz&author=J.%20Kuehr&author=M.%20Brandis&volume=278&publication_year=2000&pages=G617-G624&pmid=10762616&doi=10.1152/ajpgi.2000.278.4.G617&)
|
| 303 |
+
|
| 304 |
+
56. Hirtz S., Gonska T., Seydewitz H.H., Thomas J., Greiner P., Kuehr J., Brandis M., Eichler I., Rocha H., Lopes A.I., et al. CFTR Cl- channel function in native human colon correlates with the genotype and phenotype in cystic fibrosis. Gastroenterology. 2004;127:1085–1095. doi: 10.1053/j.gastro.2004.07.006. [DOI](https://doi.org/10.1053/j.gastro.2004.07.006) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15480987/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Gastroenterology&title=CFTR%20Cl-%20channel%20function%20in%20native%20human%20colon%20correlates%20with%20the%20genotype%20and%20phenotype%20in%20cystic%20fibrosis&author=S.%20Hirtz&author=T.%20Gonska&author=H.H.%20Seydewitz&author=J.%20Thomas&author=P.%20Greiner&volume=127&publication_year=2004&pages=1085-1095&pmid=15480987&doi=10.1053/j.gastro.2004.07.006&)
|
| 305 |
+
|
| 306 |
+
57. Roth E.K., Hirtz S., Duerr J., Wenning D., Eichler I., Seydewitz H.H., Amaral M.D., Mall M.A. The K+ channel opener 1-EBIO potentiates residual function of mutant CFTR in rectal biopsies from cystic fibrosis patients. PLoS ONE. 2011;6:e24445. doi: 10.1371/journal.pone.0024445. [DOI](https://doi.org/10.1371/journal.pone.0024445) | [PMC free article](/articles/PMC3164200/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21909392/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=PLoS%20ONE&title=The%20K+%20channel%20opener%201-EBIO%20potentiates%20residual%20function%20of%20mutant%20CFTR%20in%20rectal%20biopsies%20from%20cystic%20fibrosis%20patients&author=E.K.%20Roth&author=S.%20Hirtz&author=J.%20Duerr&author=D.%20Wenning&author=I.%20Eichler&volume=6&publication_year=2011&pages=e24445&pmid=21909392&doi=10.1371/journal.pone.0024445&)
|
| 307 |
+
|
| 308 |
+
58. R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2018. [(accessed on 1 August 2022)]. Available online: https://www.R-project.org/ [https://www.R-project.org/](https://www.R-project.org/) | [Google Scholar](https://scholar.google.com/scholar_lookup?title=R:%20A%20Language%20and%20Environment%20for%20Statistical%20Computing&publication_year=2018&)
|
test/texts/PMC10499425.md
ADDED
|
@@ -0,0 +1,269 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Variant‐based heritability assessment of dexmedetomidine and fentanyl clearance in pediatric patients
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Morgan L Shannon, Ayesha Muhammad, Nathan T James, Michael L Williams, Joseph Breeyear, Todd Edwards, Jonathan D Mosley, Leena Choi, Prince Kannankeril, Sara Van Driest
|
| 5 |
+
**Journal:** Clinical and Translational Science
|
| 6 |
+
**Date:** 2023 Jun 26
|
| 7 |
+
**DOI:** [10.1111/cts.13574](https://doi.org/10.1111/cts.13574)
|
| 8 |
+
**PMID:** 37353859
|
| 9 |
+
**PMCID:** PMC10499425
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10499425/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC10499425/pdf/CTS-16-1628.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC10499425/pdf/CTS-16-1628.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
Despite complex pathways of drug disposition, clinical pharmacogenetic predictors currently rely on only a few high effect variants. Quantification of the polygenic contribution to variability in drug disposition is necessary to prioritize target drugs for pharmacogenomic approaches and guide analytic methods. Dexmedetomidine and fentanyl, often used in postoperative care of pediatric patients, have high rates of inter‐individual variability in dosing requirements. Analyzing previously generated population pharmacokinetic parameters, we used Bayesian hierarchical mixed modeling to measure narrow‐sense (additive) heritability (hSNP2) of dexmedetomidine and fentanyl clearance in children and identify relative contributions of small, moderate, and large effect‐size variants to hSNP2. We used genome‐wide association studies (GWAS) to identify variants contributing to variation in dexmedetomidine and fentanyl clearance, followed by functional analyses to identify associated pathways. For dexmedetomidine, median clearance was 33.0 L/h (interquartile range [IQR] 23.8–47.9 L/h) and hSNP2 was estimated to be 0.35 (90% credible interval 0.00–0.90), with 45% of hSNP2 attributed to large‐, 32% to moderate‐, and 23% to small‐effect variants. The fentanyl cohort had median clearance of 8.2 L/h (IQR 4.7–16.7 L/h), with estimated hSNP2 of 0.30 (90% credible interval 0.00–0.84). Large‐effect variants accounted for 30% of hSNP2, whereas moderate‐ and small‐effect variants accounted for 37% and 33%, respectively. As expected, given small sample sizes, no individual variants or pathways were significantly associated with dexmedetomidine or fentanyl clearance by GWAS. We conclude that clearance of both drugs is highly polygenic, motivating the future use of polygenic risk scores to guide appropriate dosing of dexmedetomidine and fentanyl.
|
| 16 |
+
|
| 17 |
+
There is wide inter‐individual variation in pharmacokinetics of dexmedetomidine and fentanyl. Genomic contribution to this inter‐individual pharmacokinetic variation for both drugs remains poorly understood, as candidate gene analyses have failed to identify specific genes explaining a significant portion of the variability.
|
| 18 |
+
|
| 19 |
+
How much of the inter‐individual variability in dexmedetomidine and fentanyl clearance can be attributed to common single nucleotide variation in the genome? In addition, within this heritable component, what proportion can be attributed to variants with individually small, medium, or large effects?
|
| 20 |
+
|
| 21 |
+
Dexmedetomidine and fentanyl clearance are moderately heritable and highly polygenic traits, with small‐ and moderate‐effect variants accounting for the majority of estimated heritability.
|
| 22 |
+
|
| 23 |
+
Our findings of polygenic contribution to dexmedetomidine and fentanyl clearance highlight the importance of using genome‐wide approaches for the investigation of pharmacologic phenotypes. Our study motivates the application of heritability analyses to additional pharmacokinetic datasets. Our findings demonstrate the need to assemble much larger cohorts that will enable the development, validation, and eventual implementation of polygenic risk scores for genetically‐informed drug dosing in clinical practice.
|
| 24 |
+
|
| 25 |
+
## INTRODUCTION
|
| 26 |
+
|
| 27 |
+
Precision medicine, and specifically the individualization of treatment decisions based on genetic variation (pharmacogenomics), is a rapidly evolving area of clinical care. Clinical pharmacogenomics is generally limited to the use of well‐studied genetic variants with high impact on drug outcomes. For example, the enzyme thiopurine S‐methyltransferase plays a vital role in metabolism of thiopurine drugs, and up to 10% of individuals in different populations are characterized as poor or intermediate metabolizers. In many clinical settings, genetic testing is performed to identify those with decreased enzyme activity prior to initiating therapy with a thiopurine agent, as alternate drugs or greatly reduced doses may be warranted to avoid severe toxicity.[1](#cts13574-bib-0001) ^1^1 , [2](#cts13574-bib-0002) ^2^2 , [3](#cts13574-bib-0003) ^3^3 More recently, a genome‐wide association study (GWAS) identified variants in a second gene, *NUDT15*NUDT15, contributing to thiopurine toxicity, leading to the addition of *NUDT15*NUDT15 variants to pharmacogenomic tests.[1](#cts13574-bib-0001) ^1^1 , [4](#cts13574-bib-0004) ^4^4 However, variants in these two genes are not fully predictive of thiopurine toxicity.[3](#cts13574-bib-0003) ^3^3 To more fully predict outcomes for drugs with residual variability or where no single pharmacogene has been identified, there is increasing focus on developing polygenic predictors of drug disposition and response.[5](#cts13574-bib-0005) ^5^5 , [6](#cts13574-bib-0006) ^6^6
|
| 28 |
+
|
| 29 |
+
Dexmedetomidine, an α_2_2‐agonist, and fentanyl, an opioid, are frequently used post‐operatively in pediatric intensive care units (ICUs) to achieve sedation and analgesia.[7](#cts13574-bib-0007) ^7^7 , [8](#cts13574-bib-0008) ^8^8 , [9](#cts13574-bib-0009) ^9^9 , [10](#cts13574-bib-0010) ^10^10 Both are commonly dosed via continuous intravenous infusions using fixed weight‐based rates, which is suitable for some patients but might require titration due to over‐ or undersedation for others. Importantly, both over‐ and undersedation can be harmful to pediatric ICU patients. Oversedation has been associated with bradycardia, respiratory depression, increased need for mechanical ventilation, and prolonged ICU stays, whereas undersedation increases the risk for agitation, patient self‐injury, and anxiety. Both over‐ and undersedation have been reported to lead to development of delirium, withdrawal syndromes, neuromuscular weakness and atrophy, and post‐traumatic stress disorder.[11](#cts13574-bib-0011) ^11^11
|
| 30 |
+
|
| 31 |
+
The need for dosing adjustments of dexmedetomidine and fentanyl to maintain appropriate sedation is due in part to well‐described inter‐individual variation in pharmacokinetics. For both drugs, previous studies have shown wide variation among individuals in drug absorption, bioavailability, and clearance.[8](#cts13574-bib-0008) ^8^8 , [12](#cts13574-bib-0012) ^12^12 , [13](#cts13574-bib-0013) ^13^13 , [14](#cts13574-bib-0014) ^14^14 , [15](#cts13574-bib-0015) ^15^15 Specifically, inter‐individual variations in drug clearance rates in pediatric ICU patients have been reported as high as three‐fold for dexmedetomidine[16](#cts13574-bib-0016) ^16^16 and 10‐fold for fentanyl.[17](#cts13574-bib-0017) ^17^17 Pharmacogenomics research on dexmedetomidine and fentanyl pharmacokinetics has focused primarily on genes encoding enzymes known to play important roles in drug metabolism; these include cytochrome P450 (CYP) 2A6 and glucuronidation enzymes UGT1A4 and UGT2B10 for dexmedetomidine and CYP3A4 and CYP3A5 for fentanyl. To date, genes that have been identified explain little of the inter‐individual variability observed for clearance of both drugs.[18](#cts13574-bib-0018) ^18^18 , [19](#cts13574-bib-0019) ^19^19 , [20](#cts13574-bib-0020) ^20^20 The broader polygenic contribution to dexmedetomidine and fentanyl clearance has not been explored.
|
| 32 |
+
|
| 33 |
+
Genome‐wide approaches, including GWAS and Bayesian nonlinear modeling, take a broader approach by looking at the contribution of variants across the genome to a phenotype of interest. GWAS analyses individually examine millions of common genetic variants to identify those associated with a given phenotype.[21](#cts13574-bib-0021) ^21^21 Bayesian nonlinear models can quantify the collective contributions of large numbers of single nucleotide polymorphisms (SNPs) and estimate narrow‐sense heritability (hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22), the proportion of phenotypic variation that can be attributed to additive influences of SNPs.[22](#cts13574-bib-0022) ^22^22 In contrast to GWAS‐based estimations of hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22, Bayesian modeling can account for linkage disequilibrium by assuming that not all variants will have a non‐zero effect on the phenotype. The estimated hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 can also be subdivided into relative contributions from genes with individually small, moderate, or large effects on the phenotype of interest. These genome‐wide approaches are complementary and frequently used in assessment of nonpharmacologic phenotypes, but their application within pharmacogenomics has been limited to date. We previously demonstrated validity of using one Bayesian method to explore pharmacologic outcomes, even when applied to small sample sizes,[23](#cts13574-bib-0023) ^23^23 and GWAS approaches have successfully detected pharmacogenomic signals despite small cohorts in a few examples where effect size of the variant(s) is very large,[24](#cts13574-bib-0024) ^24^24 , [25](#cts13574-bib-0025) ^25^25 including the *NUDT15*NUDT15 association to thiopurine toxicity, as described above.[1](#cts13574-bib-0001) ^1^1 , [4](#cts13574-bib-0004) ^4^4
|
| 34 |
+
|
| 35 |
+
In this study, we used the results of previously performed population pharmacokinetic analyses[16](#cts13574-bib-0016) ^16^16 , [17](#cts13574-bib-0017) ^17^17 and deployed polygenic approaches to assess genomic contribution to dexmedetomidine and fentanyl clearance in children after cardiac surgery. We used BayesR,[26](#cts13574-bib-0026) ^26^26 , [27](#cts13574-bib-0027) ^27^27 , [28](#cts13574-bib-0028) ^28^28 an established Bayesian hierarchical analysis method, to estimate hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 of dexmedetomidine and fentanyl clearance and assess the relative contribution of small‐, moderate‐, and large‐effect variants. We hypothesized that common genomic variation contributes substantially to inter‐individual variation in these phenotypes and that many variants with small effects on the phenotypes would cumulatively account for a greater proportion of heritability than few variants with large effects. We additionally assessed for specific variants that may be associated with dexmedetomidine or fentanyl clearance in our datasets using GWAS.
|
| 36 |
+
|
| 37 |
+
## METHODS
|
| 38 |
+
|
| 39 |
+
### Study design and data collection
|
| 40 |
+
|
| 41 |
+
The parent study supporting participant recruitment, specimen collection, and data collection was approved by the Vanderbilt University Medical Center Institutional Review Board and has been previously described in detail.[29](#cts13574-bib-0029) ^29^29 Pediatric patients undergoing surgery for congenital heart disease were voluntarily enrolled in this study via written parental informed consent and, when appropriate, informed assent from the patient. Enrollment of participants into the parent study began in 2007 and is ongoing; the genomic analyses presented here used data collected from April 2013 to October 2017. All participants were admitted to the pediatric cardiac ICU following surgery and underwent routine clinical care as determined by the primary clinical team, including selection and dosing of all medications. Study participants provided a blood or saliva sample for DNA extraction, and drug concentrations were measured from remnant blood samples collected during the course of clinical care. Participants were excluded from analysis if their surgery was canceled, if they had no genotype data available, if they did not survive to hospital discharge, or if they required extracorporeal membrane oxygenation postoperatively.
|
| 42 |
+
|
| 43 |
+
Study data were collected and stored using REDCap, a secure web application hosted at Vanderbilt University.[30](#cts13574-bib-0030) ^30^30 Medical history and demographic data were documented by the study team upon study enrollment. Surgical and clinical data were extracted from the electronic health record (EHR) by the study team during the hospital stay. Drug data, including dosing and times of administration, were extracted from the EHR and the Vanderbilt Enterprise Data Warehouse and standardized using the EHR2PKPD system.[31](#cts13574-bib-0031) ^31^31
|
| 44 |
+
|
| 45 |
+
This study was reviewed by the Institutional Review Board at Vanderbilt University Medical Center and determined to constitute non‐human subject research.
|
| 46 |
+
|
| 47 |
+
### Drug concentration measurement and clearance calculations
|
| 48 |
+
|
| 49 |
+
The primary outcomes used for analysis were the clearances estimated per individual for each of the two target drugs, as previously described.[16](#cts13574-bib-0016) ^16^16 , [17](#cts13574-bib-0017) ^17^17 Briefly, dexmedetomidine and fentanyl dosing data were extracted from the EHR and drug concentrations were measured in remnant plasma specimens collected for research purposes using high‐throughput tandem mass spectrometry. Measured drug concentrations, drug dosing data, and covariate data collected for the parent study were used to generate a population pharmacokinetic model to estimate clearance of each drug, using Monolix 2021R. A two‐compartment model was selected as the base model for both drugs, and covariate modeling was performed using prespecified covariates.
|
| 50 |
+
|
| 51 |
+
For dexmedetomidine, the following covariates were considered in the population pharmacokinetic modeling based on previous research and biologic plausibility: *UGT1A4*UGT1A4, *UGT2B10*UGT2B10, and *CYP2A6*CYP2A6 variants, body weight, postnatal age, postmenstrual age, sex, Society of Thoracic Surgeons‐European Association for Cardio‐Thoracic Surgery (STAT) Congenital Heart Surgery Mortality score, cardiac bypass time, length of ICU stay, and serum creatinine. Of these, only body weight and postmenstrual age significantly improved model fit and remained in the final covariate model.[16](#cts13574-bib-0016) ^16^16
|
| 52 |
+
|
| 53 |
+
For fentanyl, CYP3A metabolizer status (based on *CYP3A45*CYP3A45 genotype), body weight, postnatal age, postmenstrual age, sex, patient‐reported race, serum creatinine, STAT score, cardiac bypass time, concomitant CYP3A inducers and inhibitors, and the first five principal components of ancestry were considered in the pharmacokinetic model. Body weight, postnatal age, STAT score, and CYP3A metabolizer status were included in the final covariate model.[17](#cts13574-bib-0017) ^17^17
|
| 54 |
+
|
| 55 |
+
For the final model for each drug, patient covariates, estimated model parameters, and the conditional mode of the individual random effect on clearance were used to calculate the estimated clearances used in this analysis.
|
| 56 |
+
|
| 57 |
+
### Genotyping, quality control, and processing
|
| 58 |
+
|
| 59 |
+
Genome‐wide SNPs were derived from either the Affymetrix Axiom Precision Medicine Research Array or Precision Medicine Diversity Array (Thermo Fisher Scientific) using Genome Reference Consortium Human Build 37 (GRCh37/hg19). The BayesR method is sensitive to population substructure, meaning that inclusion of individuals across different ancestries could confound analyses and result in spurious findings. Therefore, analyses were restricted to individuals of European ancestry as this population was the most prominent in our cohorts and the only population for which sample size was sufficient for BayesR. To do this, within each cohort (dexmedetomidine and fentanyl), genotype data from participants was combined with that of Hapmap European (Utah residents with Northern and Western European ancestry from the CEPH collection), Asian (Han Chinese in Beijing, China and Japanese in Tokyo, Japan), and African (African ancestry in Southwest USA; Luhya in Webuye, Kenya; Maasai in Kinyawa, Kenya; and Yoruba in Ibadan, Nigeria) populations to assess ancestry. Principal component (PC) analyses were performed, and, because individuals with self‐reported white race overlapped heavily with the HapMap European population and segregated from Asian and African HapMap populations, final analyses were restricted to participants within three standard deviations of the mean of PC1 and PC2 of self‐reported white participants (Figure [S1](#cts13574-supitem-0001)S1). This approach effectively restricted our analyses to participants of European ancestry.
|
| 60 |
+
|
| 61 |
+
Genotype data were imputed to the TOPMed reference panel R2 using the TOPMed Imputation Server. Imputed gene dosages were converted to hard calls using PLINK2 and filtered by info scores greater than 0.8. SNPs were included if they met the following quality control (QC) parameters: minor allele frequency greater than 1% within the cohort, SNP genotyping rate greater than 98%, and Hardy–Weinberg equilibrium greater than 10^−6^−6. Sex chromosomes were excluded from analysis. Individuals were removed if they had a genotype call rate less than 98%, discrepancy between the genetically‐estimated sex and the sex assigned in the database, or a high degree of genetic relatedness to another sample using identity‐by‐descent analysis (Figure [S2](#cts13574-supitem-0002)S2).
|
| 62 |
+
|
| 63 |
+
For all individuals who passed QC, any clearance values greater than the median + 3 × interquartile range [IQR] or less than the median − 3 × IQR were removed as outliers from the data set for analysis.
|
| 64 |
+
|
| 65 |
+
### BayesR analysis
|
| 66 |
+
|
| 67 |
+
Following QC of data sets as described above, SNPs in high linkage disequilibrium (*r*r ^2^2 > 0.9) were removed. Residuals were then calculated from clearance values incorporating the first 20 PCs of ancestry. Residuals were used in the final Bayesian analysis because the BayesR software is not able to include covariates (Figure [S2](#cts13574-supitem-0002)S2).
|
| 68 |
+
|
| 69 |
+
For each cohort, BayesR (version 1), a hierarchical Bayesian mixture model using Markov Chain Monte Carlo estimation, was used to fit all SNPs simultaneously to a model predicting drug clearance in order to give unbiased estimates of SNP effect sizes.[26](#cts13574-bib-0026) ^26^26 , [27](#cts13574-bib-0027) ^27^27 BayesR uses *k*k different normal distributions to model the prior distribution of SNP effects, where the sum of the mixture proportions is constrained to unity. As in previous analyses, we set *k*k = 4,[27](#cts13574-bib-0027) ^27^27 where each component was modeled as a normal distribution with a mean of 0 and a variance of 0, 0.01%, 0.1%, and 1% of the additive genetic variance, respectively, as shown: pβ|π,σg2=π1N0,0×σg2+π2N0,0.0001×σg2+π3Npβ|π,σg2=π1N0,0×σg2+π2N0,0.0001×σg2+π3Npβ|π,σg2=π1N0,0×σg2+π2N0,0.0001×σg2+π3Npβ|π,σg2=π1N0,0×σg2+π2N0,0.0001×σg2+π3Npβ|π,σg2=π1N0,0×σg2+π2N0,0.0001×σg2+π3Npβ|π,σg2=π1N0,0×σg2+π2N0,0.0001×σg2+π3Npβ|π,σg2=π1N0,0×σg2+π2N0,0.0001×σg2+π3Npβ|π,σg2=π1N0,0×σg2+π2N0,0.0001×σg2+π3Nppβ|π,σg2ββ||π,σg2ππ,,σg2σσgg22==π1ππ11NN0,0×σg20,0×σg200,,00××σg2σσgg22++π2ππ22NN0,0.0001×σg20,0.0001×σg20,0.00010,0.0001××σg2σσgg22++π3ππ33NN 0,0.001×σg2+π4N0,0.01×σg20,0.001×σg2+π4N0,0.01×σg20,0.001×σg2+π4N0,0.01×σg20,0.001×σg2+π4N0,0.01×σg20,0.001×σg2+π4N0,0.01×σg20,0.001×σg2+π4N0,0.01×σg20,0.001×σg2+π4N0,0.01×σg20,0.001×σg2+π4N0,0.01×σg20,0.001×σg20,0.001×σg20,0.0010,0.001××σg2σσgg22++π4ππ44NN0,0.01×σg20,0.01×σg20,0.010,0.01××σg2σσgg22. In this model, β signifies a mixture of four zero‐mean normal distributions of SNP effects with a fixed relative variance for each mixture component, *π*π represents mixture proportions constrained to sum to unity, and σg2σg2σg2σg2σg2σσgg22 is additive genetic variance explained by SNPs. The mean and variance of the first component is set to zero to account for sparseness in the model. The components *k1*k1, *k2*k2, *k3*k3, and *k4*k4 are referred to as no‐effect, small‐effect (each SNP explaining 0.01% of the variance), moderate‐effect (each SNP explaining 0.1% of the variance), and large‐effect SNPs (each SNP explaining 1% of the variance), respectively. To estimate σg2σg2σg2σg2σg2σσgg22, the algorithm uses a Gibbs scheme to sample values from each unknown parameter's posterior distribution.[26](#cts13574-bib-0026) ^26^26 , [27](#cts13574-bib-0027) ^27^27
|
| 70 |
+
|
| 71 |
+
Narrow‐sense heritability (hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22) was calculated as hSNP2=σg2σg2+σe2hSNP2=σg2σg2+σe2hSNP2=σg2σg2+σe2hSNP2=σg2σg2+σe2hSNP2hhSNPSNP22==σg2σg2+σe2σg2σσgg22σg2+σe2σg2σσgg22++σe2σσee22 where σg2σg2σg2σg2σg2σσgg22 and σe2σe2σe2σe2σe2σσee22 (residual variance) were estimated by BayesR. The number of iterations was increased from the default 20,000 to 60,000 in our analyses to allow for convergence given the small sample sizes in our data sets. Default settings were used for all other prior distribution parameters. The 90% highest density credible intervals were calculated. Results were processed using custom R scripts.
|
| 72 |
+
|
| 73 |
+
### Genomewide association studies
|
| 74 |
+
|
| 75 |
+
Dexmedetomidine and fentanyl cohorts used for GWAS were identical to those input into BayesR, and the same quality control steps were performed (Figure [S2](#cts13574-supitem-0002)S2). SNPs in high linkage disequilibrium (*r*r ^2^2 > 0.7) were removed to reduce computational time, leaving 986,408 and 1,080,019 SNPs for final analysis for dexmedetomidine and fentanyl, respectively. PLINK software (versions 1.9 and 2.0) using linear regression was utilized to test each SNP for association with the continuous drug clearance phenotype. PLINK software was also used to extract the first 10 PCs, which were used as covariates.[32](#cts13574-bib-0032) ^32^32 Results were processed using custom R scripts, and Manhattan and quantile‐quantile (Q‐Q) plots were generated using the qqman package (version 0.1.8).
|
| 76 |
+
|
| 77 |
+
We performed post hoc functional analyses of variants with association *p*p values less than or equal to 1 × 10^−4^−4 by GWAS using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) functional annotation tool.[33](#cts13574-bib-0033) ^33^33 Gene lists for each data set (dexmedetomidine and fentanyl) included all genes within 100,000 base pairs of those GWAS SNPs with association *p*p values less than or equal to 1 × 10^−4^−4. Functional analyses using DAVID were restricted to gene ontology (molecular function, biological process, and cellular component), pathways (KEGG pathway, BioCarta, and biological biochemical image database), and diseases (OMIM and UniProt).
|
| 78 |
+
|
| 79 |
+
## RESULTS
|
| 80 |
+
|
| 81 |
+
### Study population characteristics
|
| 82 |
+
|
| 83 |
+
Characteristics of the dexmedetomidine and fentanyl cohorts are shown in Table [1](#cts13574-tbl-0001)1 and summaries of surgical procedures incurred by participants in each cohort are shown in Table [S1](#cts13574-supitem-0005)S1. The dexmedetomidine cohort included 354 participants, as described previously.[16](#cts13574-bib-0016) ^16^16 Due to sensitivity of the BayesR method to population substructure, 70 individuals were removed due to genotypes consistent with non‐European ancestry, classified as individuals with PCs outside three standard deviations of the mean PC1 or PC2 of self‐reported white participants (Figure [S1](#cts13574-supitem-0001)S1) and one individual was removed due to high relatedness. Three additional individuals were excluded from analysis due to identification of log‐transformed clearance values as outliers; therefore, a total of 280 subjects were included for final analysis of dexmedetomidine clearance. The median postnatal age at the time of surgery was 20.7 months (IQR 5.2–72.8) and the median body weight was 9.6 kg (IQR 6.1–19.6). Median length of ICU stay was 3 days (IQR 2–7), and median cardiac bypass time was 1.6 h (1.1–2.4). Median individual dexmedetomidine clearance, as estimated by population pharmacokinetic modeling,[16](#cts13574-bib-0016) ^16^16 was 33.4 L/h (IQR 23.5–47.8).
|
| 84 |
+
|
| 85 |
+
### TABLE 1.
|
| 86 |
+
|
| 87 |
+
Dexmedetomidine and fentanyl cohort characteristics.
|
| 88 |
+
|
| 89 |
+
| | Dexmedetomidine (n = 280) | Fentanyl (n = 346) |
|
| 90 |
+
| - | ------------------------- | ------------------ |
|
| 91 |
+
| Age (months) | 20.7 (5.2–72.8) | 8.5 (3.7–60.1) |
|
| 92 |
+
| Weight (kg) | 9.6 (6.1–19.6) | 7.7 (5.2–17.1) |
|
| 93 |
+
| Male sex | 145 (51.7%) | 184 (53.2%) |
|
| 94 |
+
| Self‐reported white race | 277 (98.9%) | 340 (98.3%) |
|
| 95 |
+
| Length of ICU stay (days) | 3 (2–7) | 4 (2–7) |
|
| 96 |
+
| Cardiac bypass time (h) | 1.6 (1.1–2.4) | 1.7 (1.1–2.5) |
|
| 97 |
+
| STAT score | 2 (1–3) | 2 (1–3) |
|
| 98 |
+
| Clearance (L/h) | 33.4 (23.5–47.8) | 8.1 (4.6–16.6) |
|
| 99 |
+
The previously described fentanyl cohort included 434 participants[17](#cts13574-bib-0017) ^17^17 ; 87 subjects were removed due to being outside of three standard deviations of the first two PCs of self‐reported white participants (Figure [S1](#cts13574-supitem-0001)S1) and one individual was removed due to high relatedness, leaving a total of 346 individuals for final analysis. The median postnatal age at the time of surgery was 8.5 months (IQR 3.7–60.1) and the median body weight was 7.7 kg (IQR 5.2–17.1). Median length of ICU stay was 4 days (IQR 2–7), and median cardiac bypass time was 1.7 h (1.1–2.5). Median fentanyl clearance, as estimated by population pharmacokinetic modeling,[17](#cts13574-bib-0017) ^17^17 was 8.1 L/h (IQR 4.6–16.6).
|
| 100 |
+
|
| 101 |
+
### Heritability estimates of dexmedetomidine and fentanyl clearance
|
| 102 |
+
|
| 103 |
+
Dexmedetomidine clearance values from 280 individuals were log‐transformed and adjusted for the first 20 PCs (Figure [S3](#cts13574-supitem-0003)S3). Covariates that were considered for inclusion in the population pharmacokinetic models used to calculate drug clearance values were not considered for inclusion in the BayesR model to estimate heritability. Residual values and 1,121,432 genetic variants were input into the BayesR model. The estimated additive genetic variance (σg2σg2σg2σg2σg2σσgg22) was 0.13 with a residual variance (σe2σe2σe2σe2σe2σσee22) of 0.24, resulting in a calculated hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 of 0.35 with a 90% highest density credible interval of 0.00–0.90 (Figure [1](#cts13574-fig-0001)1). As described above, the estimated hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 was further subdivided into relative contributions from small‐, moderate‐, and large‐effect variants, which account for 0.01%, 0.1%, and 1% of σg2σg2σg2σg2σg2σσgg22, respectively. For dexmedetomidine, 36% of the estimated hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 was accounted for by 2997 small‐effect variants, 26% was accounted for by 264 moderate‐effect variants, and 38% was accounted for by 57 large‐effect variants. Therefore, over half of the estimated hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 of dexmedetomidine clearance was attributed to small and moderate effect‐size variants (Figure [1](#cts13574-fig-0001)1).
|
| 104 |
+
|
| 105 |
+
### FIGURE 1.
|
| 106 |
+
|
| 107 |
+
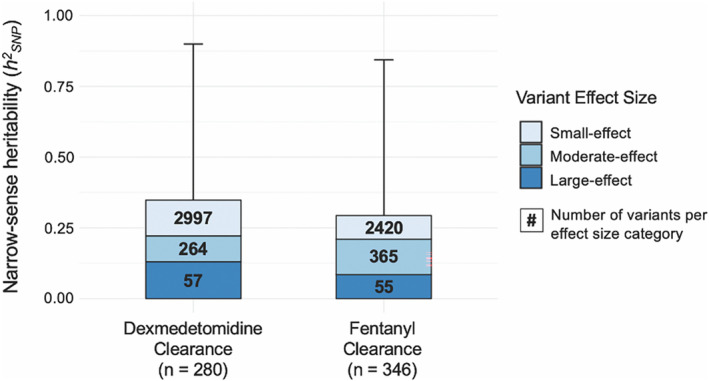
|
| 108 |
+
|
| 109 |
+
Narrow‐sense heritability (hSNP2) estimates of dexmedetomidine and fentanyl clearance. Total hSNP2 is further subdivided into relative contributions from large‐effect (dark blue), moderate‐effect (medium blue), and small‐effect (light blue) variants. Numerical values within each effect size box represent the number of variants that comprise each effect size category. Error bars represent 90% high‐density credible intervals for hSNP2.
|
| 110 |
+
|
| 111 |
+
For fentanyl, clearance values from 346 individuals were adjusted using the first 20 PCs (Figure [S3](#cts13574-supitem-0003)S3) and analyzed with 1,222,265 genetic variants using BayesR. The estimated σg2σg2σg2σg2σg2σσgg22 was 0.25, σe2σe2σe2σe2σe2σσee22 was 0.61, and calculated hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 was 0.29 with a 90% highest density credible interval of 0.00–0.84 (Figure [1](#cts13574-fig-0001)1). Relative contributions from small‐, moderate‐, and large‐effect variants were as follows: 29% of estimated hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 was accounted for by 2420 small‐effect variants, 42% was accounted for by 365 moderate‐effect variants, and 38% was accounted for by 55 large‐effect variants. As with dexmedetomidine, more than half of the estimated hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 of fentanyl clearance was attributed to small and moderate effect‐size variants (Figure [1](#cts13574-fig-0001)1).
|
| 112 |
+
|
| 113 |
+
### Genome‐wide association analysis of dexmedetomidine and fentanyl clearance
|
| 114 |
+
|
| 115 |
+
Given the substantial heritability of clearance for both drugs and small number of large‐effect variants for each drug, we performed GWAS to identify any variants significantly associated with the outcomes. To our knowledge, these are the first GWAS of dexmedetomidine and fentanyl clearance performed to date. Q‐Q plots demonstrated mild inflation of smaller *p*p values observed for dexmedetomidine and a relatively uniform distribution for fentanyl (Figure [S4](#cts13574-supitem-0004)S4). For both dexmedetomidine and fentanyl clearance, GWAS failed to identify any variants meeting the genome‐wide statistical significance threshold of 5 × 10^−8^−8 (Figure [2](#cts13574-fig-0002)2).
|
| 116 |
+
|
| 117 |
+
### FIGURE 2.
|
| 118 |
+
|
| 119 |
+
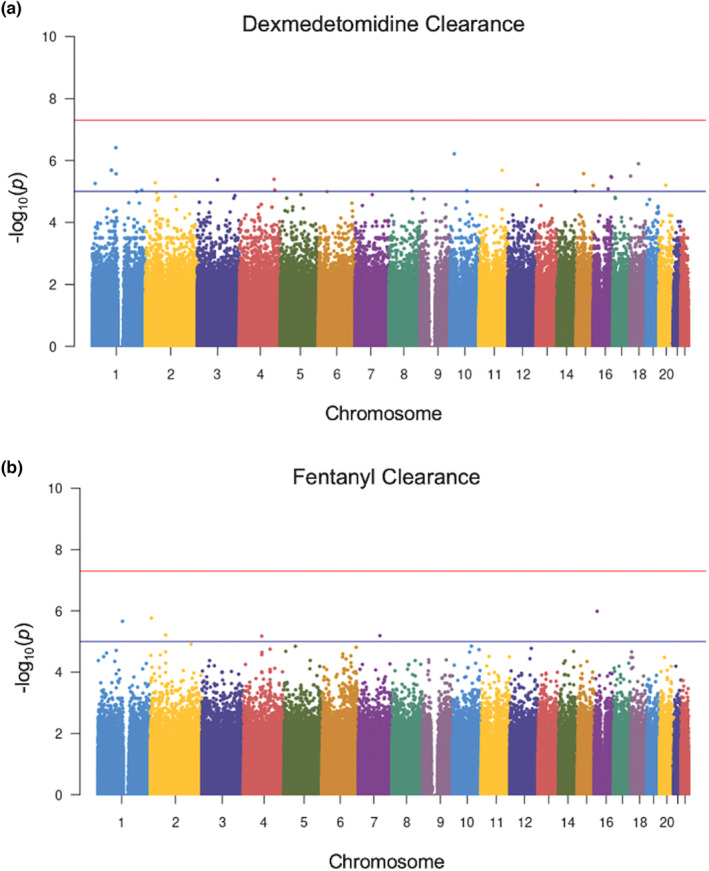
|
| 120 |
+
|
| 121 |
+
Manhattan plots showing genome‐wide association study results for dexmedetomidine (a) and fentanyl (b) clearance. For both plots, all genetic variants are shown as individual data points ordered by chromosome across the x‐axis. The y‐axis represents the negative log of the associated p value for each variant. The red line represents the standard genome‐wide significance threshold of 5 × 10−8. The blue line at 1 × 10−5 is suggestive of association. No variants in either the dexmedetomidine or fentanyl clearance analysis crossed this threshold to reach statistical significance.
|
| 122 |
+
|
| 123 |
+
Results of the GWAS analysis are reported in Tables [S2](#cts13574-supitem-0006)S2 and [S3](#cts13574-supitem-0007)S3. For dexmedetomidine clearance, the variants within 100,000 base pairs of coding regions with the strongest statistical associations (smallest *p*p values) were: *CACNB2*CACNB2, a voltage‐gated calcium channel; *LRRC8C*LRRC8C, which facilitates cAMP‐cGMP import across plasma membranes; and *RAP1A*RAP1A, a member of the RAS oncogene family (Table [S2](#cts13574-supitem-0006)S2). For fentanyl clearance, the variants within coding regions with the smallest *p*p values were: *CD58*CD58, involved in adhesion and activation of T cells; *DYSF*DYSF, which contributes to calcium‐mediated muscle contraction; and *ORAI2*ORAI2, which plays a role in store‐operated calcium channel activity (Table [S3](#cts13574-supitem-0007)S3).
|
| 124 |
+
|
| 125 |
+
We subsequently used the DAVID functional annotation tool to perform post hoc functional analyses on the variants with GWAS *p*p values less than or equal to 1 × 10^−4^−4 within 100,000 bp of known genes (Tables [S4](#cts13574-supitem-0008)S4 and [S5](#cts13574-supitem-0009)S5). These analyses did not identify any statistically significant pathways or functions enriched for clearance of either drug. For dexmedetomidine, the top three gene ontology categories were anchoring junction, specific granule membrane, and integral component of plasma membrane (Table [S6](#cts13574-supitem-0010)S6). Exogenous drug catabolism was among the top 10 pathways identified, which was associated with a total of four genes: *CYP2A13*CYP2A13, *CYP2A7*CYP2A7, *CYP2F1*CYP2F1, and *NOS1*NOS1. *CYP2A13*CYP2A13, *CYP2A7*CYP2A7, and *CYP2F1*CYP2F1 were all located within 100,000 base pairs of a single SNP on chromosome 19. For the fentanyl clearance data set, membrane, protein destabilization, and positive regulation of protein serine/threonine kinase activity were the top three gene ontology categories that emerged. No pathways related to drug metabolism were enriched for the fentanyl data set (Table [S7](#cts13574-supitem-0011)S7).
|
| 126 |
+
|
| 127 |
+
## DISCUSSION
|
| 128 |
+
|
| 129 |
+
In this study, we sought to explore the genomic contribution to variation among pediatric patients in clearance of two drugs commonly used after cardiac surgery: dexmedetomidine and fentanyl. Our data verify the clinical observation of large inter‐individual variability in drug clearance, as our pharmacokinetic analyses demonstrated an approximately two‐fold difference between the first and third quartile of estimated dexmedetomidine clearance and an almost four‐fold difference for fentanyl clearance, consistent with previous estimates.[16](#cts13574-bib-0016) ^16^16 , [17](#cts13574-bib-0017) ^17^17 We used the clearance data, coupled with genome‐wide genotype data, to employ a Bayesian hierarchical modeling methodology. Given that both hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 estimations were greater than 0.25, inter‐individual variation in clearance of dexmedetomidine and fentanyl in this population is moderately heritable. Further, the observation that less than 40% of hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 is attributable to large‐effect variants suggests that these traits are highly polygenic. The credible intervals for our observations are wide, which may be improved with larger sample sizes. These results suggest that, for both drugs, many genes with individually smaller effect sizes combine to comprise a substantial genomic contribution to drug clearance. Therefore, polygenic approaches combined with large sample sizes are necessary to gain a comprehensive understanding of genetic contribution to the individual pharmacokinetics of these drugs, which may ultimately be utilized, for example in the form of polygenic risk scores, in clinical practice to individualize therapy.
|
| 130 |
+
|
| 131 |
+
Although less than 40% of the estimated heritability for both dexmedetomidine and fentanyl clearance was attributed to variants with large effect sizes, their estimated contribution to heritability of both drugs was substantial. We therefore utilized GWAS in an attempt to identify any SNPs that were significantly associated with dexmedetomidine or fentanyl clearance. These analyses failed to identify any Bonferroni‐corrected statistically significant SNPs for clearance of either drug. Secondary functional analyses of genes with *p*p values smaller than 1 × 10^−4^−4 using the DAVID functional annotation tool also did not pinpoint specific pathways or processes that were enriched within our data sets. For dexmedetomidine clearance, exogenous drug catabolism and the epoxygenase P450 pathway were among the top functional hits. However, neither of these pathways reached statistical significance and each involved only a few genes, all of which were located near a single variant identified by GWAS. Furthermore, these enzymes have not been previously reported to play a role in dexmedetomidine metabolism. For fentanyl clearance, no pathways or processes related to drug metabolism were enriched. In agreement with the results of the Bayesian analysis, these findings also suggest a highly polygenic contribution of the genome to dexmedetomidine and fentanyl clearance and support continued efforts toward utilization of polygenic approaches to investigate the pharmacokinetics of both drugs. It is also important to consider that drug metabolism and distribution are dependent on pathways across many tissue types[34](#cts13574-bib-0034) ^34^34 , [35](#cts13574-bib-0035) ^35^35 and are highly variable across the lifespan.[36](#cts13574-bib-0036) ^36^36 , [37](#cts13574-bib-0037) ^37^37 Therefore, future efforts aimed at defining pathways and predictive models across different tissue types and different ages may aid in the identification of novel genetic associations with drug outcomes.
|
| 132 |
+
|
| 133 |
+
Taken together, our data motivate collection of larger and more ancestrally diverse cohorts of dexmedetomidine‐ and fentanyl‐exposed individuals to develop and validate polygenic predictive models for these drugs, incorporating effects of many genes with a spectrum of effect size. Such predictors are now available for nonpharmacologic phenotypes such as type 2 diabetes,[38](#cts13574-bib-0038) ^38^38 coronary artery disease,[39](#cts13574-bib-0039) ^39^39 , [40](#cts13574-bib-0040) ^40^40 neurodegenerative disease,[41](#cts13574-bib-0041) ^41^41 , [42](#cts13574-bib-0042) ^42^42 and some cancers.[43](#cts13574-bib-0043) ^43^43 , [44](#cts13574-bib-0044) ^44^44 , [45](#cts13574-bib-0045) ^45^45 Polygenic risk predictors have been demonstrated to be especially useful for those individuals at the tail ends of the distribution for a given phenotype.[46](#cts13574-bib-0046) ^46^46 Applied to dexmedetomidine and fentanyl clearance, polygenic risk scores may be most useful for identifying individuals at the highest risk for undersedation or oversedation with standard drug dosing regimens secondary to differences in drug clearance. These approaches may be useful across a wide range of drugs with complex pharmacokinetic and/or pharmacodynamic pathways. Estimations of hSNP2hSNP2hSNP2hSNP2hSNP2hhSNPSNP22 for drug traits may help identify the best target drugs for genomic approaches (i.e., those with high heritability).
|
| 134 |
+
|
| 135 |
+
Our study was limited by the application of modeling methodologies traditionally used with large sample sizes, such as Bayesian modeling and GWAS, to our small cohorts. This limitation likely resulted in decreased precision and wide credible intervals in our Bayesian analyses; however, Bayesian modeling has been previously used for pharmacogenomic analyses with similarly sized data sets.[23](#cts13574-bib-0023) ^23^23 Additionally, it has been demonstrated that in polygenic phenotypes accounted for by primarily small‐effect variants, as is true for both of our phenotypes, large sample sizes are required to detect significant variants with GWAS.[47](#cts13574-bib-0047) ^47^47 Therefore, given the small sample sizes, our GWAS analyses may be underpowered to detect the statistical association of some genes with our phenotypes and may have missed potentially significantly associated variants. Furthermore, development of clinically useful polygenic risk scores requires data sets much larger than those commonly available for pharmacologic outcomes. To combat this persistent limitation of pharmacogenomic research, we advocate for continued efforts toward curation of large data sets. To do so would require building scalable, sharable data structures and collaborative efforts both within and across international borders. In today's era, with the ubiquity of EHRs and advent of high throughput analytic techniques, such collaboration is both achievable and worthwhile.
|
| 136 |
+
|
| 137 |
+
Another limitation inherent in heritability studies is the need to restrict analyses to individuals of a single ancestry. Studies conducted in the United States and Europe often restrict analyses to European ancestry, and, as a result, the majority of publications in genomics include only individuals of European descent.[48](#cts13574-bib-0048) ^48^48 , [49](#cts13574-bib-0049) ^49^49 Our study was limited in a similar manner as our cohort was relatively small and currently lacks sufficient numbers of individuals of non‐European ancestries to perform analyses in other ancestries. It is paramount that we prioritize diversity within pharmacogenomics research; doing so will ensure applicability of findings across populations and foster new discoveries. We are continuously enrolling subjects in this cohort and hope to soon build large enough sample sizes to repeat these analyses for individuals of other ancestries. Additionally, methodological developments in datasets of diverse and complex ancestries would enable more ancestrally inclusive studies that may reveal important differences in inter‐individual drug variability.
|
| 138 |
+
|
| 139 |
+
Finally, both a limitation of our current study and an opportunity for future investigation is the application of these same methodologies to diverse pharmacodynamic outcomes. For dexmedetomidine and fentanyl, such outcomes may include standardized sedation scores. These analyses may identify a unique set of genes that influence the response to dexmedetomidine and fentanyl in this population. In order to be successful, these strategies must account for differences in pharmacokinetics, either through the use of robust modeling or through the measurement of drug concentrations.
|
| 140 |
+
|
| 141 |
+
In summary, our study demonstrates that the genome contributes substantially to inter‐individual variation in dexmedetomidine and fentanyl clearance in the pediatric cardiac surgery population. Clearance of both drugs was polygenic, with small and moderate effect‐size variants comprising the majority of estimated heritability, whereas no individual variant had a significant association with clearance of either drug. Taken together these findings motivate the development of polygenic predictors of drug responses with the ultimate goal of implementing genomically informed drug dosing into clinical practice.
|
| 142 |
+
|
| 143 |
+
## AUTHOR CONTRIBUTIONS
|
| 144 |
+
|
| 145 |
+
M.L.S., A.M., N.T.J., M.L.W., L.C., and S.L.V.D. wrote the manuscript. M.L.S., A.M., N.T.J., M.L.W., T.E., J.D.M., L.C., P.K., and S.L.V.D. designed the research. M.L.S., A.M., N.T.J., M.L.W., J.B., and S.L.V.D. performed the research. M.L.S. analyzed the data.
|
| 146 |
+
|
| 147 |
+
## FUNDING INFORMATION
|
| 148 |
+
|
| 149 |
+
This work is supported by the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) R01 GM132204 to S.L.V. This work is also supported in part by NIH/NIGMS R01 GM124109 to LC and R01 HD084461 from the NIH Eunice Kennedy Shriver National Institute of Child Health & Human Development to P.J.K. A.M. is supported by a grant from the American Heart Association (20PRE35180088) and from the Vanderbilt Medical Scientist Training Program (T32GM007347). S.L.V. and P.J.K. are supported by P50HD106446. This study used REDCap, supported by UL1 TR000445 from NIH National Center for Advancing Translational Sciences.
|
| 150 |
+
|
| 151 |
+
## CONFLICT OF INTEREST STATEMENT
|
| 152 |
+
|
| 153 |
+
The authors declared no competing interests for this work.
|
| 154 |
+
|
| 155 |
+
## Supporting information
|
| 156 |
+
|
| 157 |
+
## ACKNOWLEDGMENTS
|
| 158 |
+
|
| 159 |
+
The authors would like to thank the Research Immersion program at Vanderbilt University School of Medicine and Dr. Lea Davis for her advice and mentorship. The authors would also like to thank Dr. Sabrina Holley for her feedback and editing. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, Tennessee.
|
| 160 |
+
|
| 161 |
+
Shannon ML, Muhammad A, James NT, et al. Variant‐based heritability assessment of dexmedetomidine and fentanyl clearance in pediatric patients. Clin Transl Sci. 2023;16:1628‐1638. doi: 10.1111/cts.13574
|
| 162 |
+
|
| 163 |
+
## Associated Data
|
| 164 |
+
|
| 165 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 166 |
+
|
| 167 |
+
### Supplementary Materials
|
| 168 |
+
|
| 169 |
+
### Supplementary Materials
|
| 170 |
+
|
| 171 |
+
## References
|
| 172 |
+
|
| 173 |
+
1. Relling MV, Schwab M, Whirl‐Carrillo M, et al. Clinical pharmacogenetics implementation consortium guideline for thiopurine dosing based on tpmt and nudt 15 genotypes: 2018 update. Clin Pharmacol Ther. 2019;105(5):1095‐1105. doi: 10.1002/cpt.1304 [DOI](https://doi.org/10.1002/cpt.1304) | [PMC free article](/articles/PMC6576267/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30447069/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Clinical%20pharmacogenetics%20implementation%20consortium%20guideline%20for%20thiopurine%20dosing%20based%20on%20tpmt%20and%20nudt%2015%20genotypes:%202018%20update&author=MV%20Relling&author=M%20Schwab&author=M%20Whirl%E2%80%90Carrillo&volume=105&issue=5&publication_year=2019&pages=1095-1105&pmid=30447069&doi=10.1002/cpt.1304&)
|
| 174 |
+
|
| 175 |
+
2. Asadov C, Aliyeva G, Mustafayeva K. Thiopurine S‐methyltransferase as a pharmacogenetic biomarker: significance of testing and review of major methods. Cardiovasc Hematol Agents Med Chem. 2017;15(1):23‐30. doi: 10.2174/1871525715666170529091921 [DOI](https://doi.org/10.2174/1871525715666170529091921) | [PMC free article](/articles/PMC5740490/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28552060/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cardiovasc%20Hematol%20Agents%20Med%20Chem&title=Thiopurine%20S%E2%80%90methyltransferase%20as%20a%20pharmacogenetic%20biomarker:%20significance%20of%20testing%20and%20review%20of%20major%20methods&author=C%20Asadov&author=G%20Aliyeva&author=K%20Mustafayeva&volume=15&issue=1&publication_year=2017&pages=23-30&pmid=28552060&doi=10.2174/1871525715666170529091921&)
|
| 176 |
+
|
| 177 |
+
3. Lennard L. Implementation of TPMT testing: TPMT testing. Br J Clin Pharmacol. 2014;77(4):704‐714. doi: 10.1111/bcp.12226 [DOI](https://doi.org/10.1111/bcp.12226) | [PMC free article](/articles/PMC3971986/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23962279/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Implementation%20of%20TPMT%20testing:%20TPMT%20testing&author=L%20Lennard&volume=77&issue=4&publication_year=2014&pages=704-714&pmid=23962279&doi=10.1111/bcp.12226&)
|
| 178 |
+
|
| 179 |
+
4. Yang JJ, Landier W, Yang W, et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol. 2015;33(11):1235‐1242. doi: 10.1200/JCO.2014.59.4671 [DOI](https://doi.org/10.1200/JCO.2014.59.4671) | [PMC free article](/articles/PMC4375304/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25624441/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Clin%20Oncol&title=Inherited%20NUDT15%20variant%20is%20a%20genetic%20determinant%20of%20mercaptopurine%20intolerance%20in%20children%20with%20acute%20lymphoblastic%20leukemia&author=JJ%20Yang&author=W%20Landier&author=W%20Yang&volume=33&issue=11&publication_year=2015&pages=1235-1242&pmid=25624441&doi=10.1200/JCO.2014.59.4671&)
|
| 180 |
+
|
| 181 |
+
5. Johnson D, Wilke MAP, Lyle SM, et al. A systematic review and analysis of the use of polygenic scores in pharmacogenomics. Clin Pharmacol Ther. 2022;111(4):919‐930. doi: 10.1002/cpt.2520 [DOI](https://doi.org/10.1002/cpt.2520) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34953075/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=A%20systematic%20review%20and%20analysis%20of%20the%20use%20of%20polygenic%20scores%20in%20pharmacogenomics&author=D%20Johnson&author=MAP%20Wilke&author=SM%20Lyle&volume=111&issue=4&publication_year=2022&pages=919-930&pmid=34953075&doi=10.1002/cpt.2520&)
|
| 182 |
+
|
| 183 |
+
6. Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12(1):44. doi: 10.1186/s13073-020-00742-5 [DOI](https://doi.org/10.1186/s13073-020-00742-5) | [PMC free article](/articles/PMC7236300/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32423490/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genome%20Med&title=Polygenic%20risk%20scores:%20from%20research%20tools%20to%20clinical%20instruments&author=CM%20Lewis&author=E%20Vassos&volume=12&issue=1&publication_year=2020&pages=44&pmid=32423490&doi=10.1186/s13073-020-00742-5&)
|
| 184 |
+
|
| 185 |
+
7. Plambech MZ, Afshari A. Dexmedetomidine in the pediatric population: a review. Minerva Anestesiol. 2015;81(3):320‐332. [PubMed](https://pubmed.ncbi.nlm.nih.gov/24824958/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Minerva%20Anestesiol&title=Dexmedetomidine%20in%20the%20pediatric%20population:%20a%20review&author=MZ%20Plambech&author=A%20Afshari&volume=81&issue=3&publication_year=2015&pages=320-332&pmid=24824958&)
|
| 186 |
+
|
| 187 |
+
8. Weerink MAS, Struys MMRF, Hannivoort LN, Barends CRM, Absalom AR, Colin P. Clinical pharmacokinetics and pharmacodynamics of dexmedetomidine. Clin Pharmacokinet. 2017;56(8):893‐913. doi: 10.1007/s40262-017-0507-7 [DOI](https://doi.org/10.1007/s40262-017-0507-7) | [PMC free article](/articles/PMC5511603/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28105598/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacokinet&title=Clinical%20pharmacokinetics%20and%20pharmacodynamics%20of%20dexmedetomidine&author=MAS%20Weerink&author=MMRF%20Struys&author=LN%20Hannivoort&author=CRM%20Barends&author=AR%20Absalom&volume=56&issue=8&publication_year=2017&pages=893-913&pmid=28105598&doi=10.1007/s40262-017-0507-7&)
|
| 188 |
+
|
| 189 |
+
9. Lee S. Dexmedetomidine: present and future directions. Korean J Anesthesiol. 2019;72(4):323‐330. doi: 10.4097/kja.19259 [DOI](https://doi.org/10.4097/kja.19259) | [PMC free article](/articles/PMC6676029/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31220910/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Korean%20J%20Anesthesiol&title=Dexmedetomidine:%20present%20and%20future%20directions&author=S%20Lee&volume=72&issue=4&publication_year=2019&pages=323-330&pmid=31220910&doi=10.4097/kja.19259&)
|
| 190 |
+
|
| 191 |
+
10. Ziesenitz VC, Vaughns JD, Koch G, Mikus G, van den Anker JN. Pharmacokinetics of fentanyl and its derivatives in children: a comprehensive review. Clin Pharmacokinet. 2018;57(2):125‐149. doi: 10.1007/s40262-017-0569-6 [DOI](https://doi.org/10.1007/s40262-017-0569-6) | [PMC free article](/articles/PMC5756700/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28688027/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacokinet&title=Pharmacokinetics%20of%20fentanyl%20and%20its%20derivatives%20in%20children:%20a%20comprehensive%20review&author=VC%20Ziesenitz&author=JD%20Vaughns&author=G%20Koch&author=G%20Mikus&author=JN%20van%20den%20Anker&volume=57&issue=2&publication_year=2018&pages=125-149&pmid=28688027&doi=10.1007/s40262-017-0569-6&)
|
| 192 |
+
|
| 193 |
+
11. Egbuta C, Mason KP. Current state of analgesia and sedation in the pediatric intensive care unit. J Clin Med. 2021;10(9):1847. doi: 10.3390/jcm10091847 [DOI](https://doi.org/10.3390/jcm10091847) | [PMC free article](/articles/PMC8122992/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33922824/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Clin%20Med&title=Current%20state%20of%20analgesia%20and%20sedation%20in%20the%20pediatric%20intensive%20care%20unit&author=C%20Egbuta&author=KP%20Mason&volume=10&issue=9&publication_year=2021&pages=1847&pmid=33922824&doi=10.3390/jcm10091847&)
|
| 194 |
+
|
| 195 |
+
12. Oosten AW, Abrantes JA, Jönsson S, et al. Treatment with subcutaneous and transdermal fentanyl: results from a population pharmacokinetic study in cancer patients. Eur J Clin Pharmacol. 2016;72(4):459‐467. doi: 10.1007/s00228-015-2005-x [DOI](https://doi.org/10.1007/s00228-015-2005-x) | [PMC free article](/articles/PMC4792338/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26762381/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=Treatment%20with%20subcutaneous%20and%20transdermal%20fentanyl:%20results%20from%20a%20population%20pharmacokinetic%20study%20in%20cancer%20patients&author=AW%20Oosten&author=JA%20Abrantes&author=S%20J%C3%B6nsson&volume=72&issue=4&publication_year=2016&pages=459-467&pmid=26762381&doi=10.1007/s00228-015-2005-x&)
|
| 196 |
+
|
| 197 |
+
13. Iirola T, Aantaa R, Laitio R, et al. Pharmacokinetics of prolonged infusion of high‐dose dexmedetomidine in critically ill patients. Crit Care. 2011;15(5):R257. doi: 10.1186/cc10518 [DOI](https://doi.org/10.1186/cc10518) | [PMC free article](/articles/PMC3334808/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22030215/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Crit%20Care&title=Pharmacokinetics%20of%20prolonged%20infusion%20of%20high%E2%80%90dose%20dexmedetomidine%20in%20critically%20ill%20patients&author=T%20Iirola&author=R%20Aantaa&author=R%20Laitio&volume=15&issue=5&publication_year=2011&pages=R257&pmid=22030215&doi=10.1186/cc10518&)
|
| 198 |
+
|
| 199 |
+
14. Välitalo PA, Ahtola‐Sätilä T, Wighton A, Sarapohja T, Pohjanjousi P, Garratt C. Population pharmacokinetics of dexmedetomidine in critically ill patients. Clin Drug Investig. 2013;33(8):579‐587. doi: 10.1007/s40261-013-0101-1 [DOI](https://doi.org/10.1007/s40261-013-0101-1) | [PMC free article](/articles/PMC3717151/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23839483/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Drug%20Investig&title=Population%20pharmacokinetics%20of%20dexmedetomidine%20in%20critically%20ill%20patients&author=PA%20V%C3%A4litalo&author=T%20Ahtola%E2%80%90S%C3%A4til%C3%A4&author=A%20Wighton&author=T%20Sarapohja&author=P%20Pohjanjousi&volume=33&issue=8&publication_year=2013&pages=579-587&pmid=23839483&doi=10.1007/s40261-013-0101-1&)
|
| 200 |
+
|
| 201 |
+
15. Kuip EJM, Zandvliet ML, Koolen SLW, Mathijssen RHJ, van der Rijt CCD. A review of factors explaining variability in fentanyl pharmacokinetics; focus on implications for cancer patients: variability in fentanyl pharmacokinetics. Br J Clin Pharmacol. 2017;83(2):294‐313. doi: 10.1111/bcp.13129 [DOI](https://doi.org/10.1111/bcp.13129) | [PMC free article](/articles/PMC5237702/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27619152/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=A%20review%20of%20factors%20explaining%20variability%20in%20fentanyl%20pharmacokinetics;%20focus%20on%20implications%20for%20cancer%20patients:%20variability%20in%20fentanyl%20pharmacokinetics&author=EJM%20Kuip&author=ML%20Zandvliet&author=SLW%20Koolen&author=RHJ%20Mathijssen&author=CCD%20van%20der%20Rijt&volume=83&issue=2&publication_year=2017&pages=294-313&pmid=27619152&doi=10.1111/bcp.13129&)
|
| 202 |
+
|
| 203 |
+
16. James NT, Breeyear JH, Caprioli R, et al. Population pharmacokinetic analysis of dexmedetomidine in children using real‐world data from electronic health records and remnant specimens. Br J Clin Pharmacol. 2022;88:2885‐2898. doi: 10.1111/bcp.15194 [DOI](https://doi.org/10.1111/bcp.15194) | [PMC free article](/articles/PMC9106818/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34957589/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Population%20pharmacokinetic%20analysis%20of%20dexmedetomidine%20in%20children%20using%20real%E2%80%90world%20data%20from%20electronic%20health%20records%20and%20remnant%20specimens&author=NT%20James&author=JH%20Breeyear&author=R%20Caprioli&volume=88&publication_year=2022&pages=2885-2898&pmid=34957589&doi=10.1111/bcp.15194&)
|
| 204 |
+
|
| 205 |
+
17. Williams ML, Kannankeril PJ, Breeyear JH, Edwards TL, Van Driest SL, Choi L. Effect of CYP3A5 and CYP3A4 genetic variants on fentanyl pharmacokinetics in a pediatric population. Clin Pharmacol Ther. 2022;111(4):896‐908. doi: 10.1002/cpt.2506 [DOI](https://doi.org/10.1002/cpt.2506) | [PMC free article](/articles/PMC8940650/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34877660/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Effect%20of%20CYP3A5%20and%20CYP3A4%20genetic%20variants%20on%20fentanyl%20pharmacokinetics%20in%20a%20pediatric%20population&author=ML%20Williams&author=PJ%20Kannankeril&author=JH%20Breeyear&author=TL%20Edwards&author=SL%20Van%20Driest&volume=111&issue=4&publication_year=2022&pages=896-908&pmid=34877660&doi=10.1002/cpt.2506&)
|
| 206 |
+
|
| 207 |
+
18. Zhou S, Skaar DJ, Jacobson PA, Huang RS. Pharmacogenomics of medications commonly used in the intensive care unit. Front Pharmacol. 2018;9:1436. doi: 10.3389/fphar.2018.01436 [DOI](https://doi.org/10.3389/fphar.2018.01436) | [PMC free article](/articles/PMC6289166/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30564130/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front%20Pharmacol&title=Pharmacogenomics%20of%20medications%20commonly%20used%20in%20the%20intensive%20care%20unit&author=S%20Zhou&author=DJ%20Skaar&author=PA%20Jacobson&author=RS%20Huang&volume=9&publication_year=2018&pages=1436&pmid=30564130&doi=10.3389/fphar.2018.01436&)
|
| 208 |
+
|
| 209 |
+
19. Magarbeh L, Gorbovskaya I, Le Foll B, Jhirad R, Müller DJ. Reviewing pharmacogenetics to advance precision medicine for opioids. Biomed Pharmacother. 2021;142:112060. doi: 10.1016/j.biopha.2021.112060 [DOI](https://doi.org/10.1016/j.biopha.2021.112060) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34523422/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Biomed%20Pharmacother&title=Reviewing%20pharmacogenetics%20to%20advance%20precision%20medicine%20for%20opioids&author=L%20Magarbeh&author=I%20Gorbovskaya&author=B%20Le%20Foll&author=R%20Jhirad&author=DJ%20M%C3%BCller&volume=142&publication_year=2021&pages=112060&pmid=34523422&doi=10.1016/j.biopha.2021.112060&)
|
| 210 |
+
|
| 211 |
+
20. Kohli U, Pandharipande P, Muszkat M, et al. CYP2A6 genetic variation and dexmedetomidine disposition. Eur J Clin Pharmacol. 2012;68(6):937‐942. doi: 10.1007/s00228-011-1208-z [DOI](https://doi.org/10.1007/s00228-011-1208-z) | [PMC free article](/articles/PMC3352974/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22271297/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=CYP2A6%20genetic%20variation%20and%20dexmedetomidine%20disposition&author=U%20Kohli&author=P%20Pandharipande&author=M%20Muszkat&volume=68&issue=6&publication_year=2012&pages=937-942&pmid=22271297&doi=10.1007/s00228-011-1208-z&)
|
| 212 |
+
|
| 213 |
+
21. Uffelmann E, Huang QQ, Munung NS, et al. Genome‐wide association studies. Nat Rev Methods Primer. 2021;1(1):59. doi: 10.1038/s43586-021-00056-9 [DOI](https://doi.org/10.1038/s43586-021-00056-9) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat%20Rev%20Methods%20Primer&title=Genome%E2%80%90wide%20association%20studies&author=E%20Uffelmann&author=QQ%20Huang&author=NS%20Munung&volume=1&issue=1&publication_year=2021&pages=59&doi=10.1038/s43586-021-00056-9&)
|
| 214 |
+
|
| 215 |
+
22. Mayhew AJ, Meyre D. Assessing the heritability of complex traits in humans: methodological challenges and opportunities. Curr Genomics. 2017;18(4):332‐340. doi: 10.2174/1389202918666170307161450 [DOI](https://doi.org/10.2174/1389202918666170307161450) | [PMC free article](/articles/PMC5635617/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29081689/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Curr%20Genomics&title=Assessing%20the%20heritability%20of%20complex%20traits%20in%20humans:%20methodological%20challenges%20and%20opportunities&author=AJ%20Mayhew&author=D%20Meyre&volume=18&issue=4&publication_year=2017&pages=332-340&pmid=29081689&doi=10.2174/1389202918666170307161450&)
|
| 216 |
+
|
| 217 |
+
23. Muhammad A, Aka IT, Birdwell KA, et al. Genome‐wide approach to measure variant‐based heritability of drug outcome phenotypes. Clin Pharmacol Ther. 2021;110(3):714‐722. doi: 10.1002/cpt.2323 [DOI](https://doi.org/10.1002/cpt.2323) | [PMC free article](/articles/PMC8376753/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34151428/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Genome%E2%80%90wide%20approach%20to%20measure%20variant%E2%80%90based%20heritability%20of%20drug%20outcome%20phenotypes&author=A%20Muhammad&author=IT%20Aka&author=KA%20Birdwell&volume=110&issue=3&publication_year=2021&pages=714-722&pmid=34151428&doi=10.1002/cpt.2323&)
|
| 218 |
+
|
| 219 |
+
24. Ozeki T, Mushiroda T, Yowang A, et al. Genome‐wide association study identifies HLA‐A*3101 allele as a genetic risk factor for carbamazepine‐induced cutaneous adverse drug reactions in Japanese population. Hum Mol Genet. 2011;20(5):1034‐1041. doi: 10.1093/hmg/ddq537 [DOI](https://doi.org/10.1093/hmg/ddq537) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21149285/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Hum%20Mol%20Genet&title=Genome%E2%80%90wide%20association%20study%20identifies%20HLA%E2%80%90A*3101%20allele%20as%20a%20genetic%20risk%20factor%20for%20carbamazepine%E2%80%90induced%20cutaneous%20adverse%20drug%20reactions%20in%20Japanese%20population&author=T%20Ozeki&author=T%20Mushiroda&author=A%20Yowang&volume=20&issue=5&publication_year=2011&pages=1034-1041&pmid=21149285&doi=10.1093/hmg/ddq537&)
|
| 220 |
+
|
| 221 |
+
25. Ahlström S, Bergman P, Jokela R, et al. First genome‐wide association study on rocuronium dose requirements shows association with SLCO1A2. Br J Anaesth. 2021;126(5):949‐957. doi: 10.1016/j.bja.2021.01.029 [DOI](https://doi.org/10.1016/j.bja.2021.01.029) | [PMC free article](/articles/PMC8132880/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33676726/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Anaesth&title=First%20genome%E2%80%90wide%20association%20study%20on%20rocuronium%20dose%20requirements%20shows%20association%20with%20SLCO1A2&author=S%20Ahlstr%C3%B6m&author=P%20Bergman&author=R%20Jokela&volume=126&issue=5&publication_year=2021&pages=949-957&pmid=33676726&doi=10.1016/j.bja.2021.01.029&)
|
| 222 |
+
|
| 223 |
+
26. Erbe M, Hayes BJ, Matukumalli LK, et al. Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high‐density single nucleotide polymorphism panels. J Dairy Sci. 2012;95(7):4114‐4129. doi: 10.3168/jds.2011-5019 [DOI](https://doi.org/10.3168/jds.2011-5019) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22720968/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Dairy%20Sci&title=Improving%20accuracy%20of%20genomic%20predictions%20within%20and%20between%20dairy%20cattle%20breeds%20with%20imputed%20high%E2%80%90density%20single%20nucleotide%20polymorphism%20panels&author=M%20Erbe&author=BJ%20Hayes&author=LK%20Matukumalli&volume=95&issue=7&publication_year=2012&pages=4114-4129&pmid=22720968&doi=10.3168/jds.2011-5019&)
|
| 224 |
+
|
| 225 |
+
27. Moser G, Lee SH, Hayes BJ, Goddard ME, Wray NR, Visscher PM. Simultaneous discovery, estimation and prediction analysis of complex traits using a Bayesian mixture model. Haley C, ed. PLOS Genet. 2015;11(4):e1004969. doi: 10.1371/journal.pgen.1004969 [DOI](https://doi.org/10.1371/journal.pgen.1004969) | [PMC free article](/articles/PMC4388571/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25849665/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=PLOS%20Genet&title=Simultaneous%20discovery,%20estimation%20and%20prediction%20analysis%20of%20complex%20traits%20using%20a%20Bayesian%20mixture%20model.%20Haley%20C,%20ed&author=G%20Moser&author=SH%20Lee&author=BJ%20Hayes&author=ME%20Goddard&author=NR%20Wray&volume=11&issue=4&publication_year=2015&pages=e1004969&pmid=25849665&doi=10.1371/journal.pgen.1004969&)
|
| 226 |
+
|
| 227 |
+
28. Mollandin F, Rau A, Croiseau P. An evaluation of the predictive performance and mapping power of the BayesR model for genomic prediction. G3 (Bethesda). 2021;11(11):jkab225. doi: 10.1093/g3journal/jkab225 [DOI](https://doi.org/10.1093/g3journal/jkab225) | [PMC free article](/articles/PMC8527474/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34849780/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=G3%20(Bethesda)&title=An%20evaluation%20of%20the%20predictive%20performance%20and%20mapping%20power%20of%20the%20BayesR%20model%20for%20genomic%20prediction&author=F%20Mollandin&author=A%20Rau&author=P%20Croiseau&volume=11&issue=11&publication_year=2021&pages=jkab225&pmid=34849780&doi=10.1093/g3journal/jkab225&)
|
| 228 |
+
|
| 229 |
+
29. Van Driest SL, Marshall MD, Hachey B, et al. Pragmatic pharmacology: population pharmacokinetic analysis of fentanyl using remnant samples from children after cardiac surgery: population PK of fentanyl using remnant samples from children. Br J Clin Pharmacol. 2016;81(6):1165‐1174. doi: 10.1111/bcp.12903 [DOI](https://doi.org/10.1111/bcp.12903) | [PMC free article](/articles/PMC4876191/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26861166/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Pragmatic%20pharmacology:%20population%20pharmacokinetic%20analysis%20of%20fentanyl%20using%20remnant%20samples%20from%20children%20after%20cardiac%20surgery:%20population%20PK%20of%20fentanyl%20using%20remnant%20samples%20from%20children&author=SL%20Van%20Driest&author=MD%20Marshall&author=B%20Hachey&volume=81&issue=6&publication_year=2016&pages=1165-1174&pmid=26861166&doi=10.1111/bcp.12903&)
|
| 230 |
+
|
| 231 |
+
30. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata‐driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377‐381. doi: 10.1016/j.jbi.2008.08.010 [DOI](https://doi.org/10.1016/j.jbi.2008.08.010) | [PMC free article](/articles/PMC2700030/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18929686/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Biomed%20Inform&title=Research%20electronic%20data%20capture%20(REDCap)%E2%80%94a%20metadata%E2%80%90driven%20methodology%20and%20workflow%20process%20for%20providing%20translational%20research%20informatics%20support&author=PA%20Harris&author=R%20Taylor&author=R%20Thielke&author=J%20Payne&author=N%20Gonzalez&volume=42&issue=2&publication_year=2009&pages=377-381&pmid=18929686&doi=10.1016/j.jbi.2008.08.010&)
|
| 232 |
+
|
| 233 |
+
31. Choi L, Beck C, McNeer E, et al. Development of a system for Postmarketing population pharmacokinetic and pharmacodynamic studies using real‐world data from electronic health records. Clin Pharmacol Ther. 2020;107(4):934‐943. doi: 10.1002/cpt.1787 [DOI](https://doi.org/10.1002/cpt.1787) | [PMC free article](/articles/PMC7093250/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31957870/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Development%20of%20a%20system%20for%20Postmarketing%20population%20pharmacokinetic%20and%20pharmacodynamic%20studies%20using%20real%E2%80%90world%20data%20from%20electronic%20health%20records&author=L%20Choi&author=C%20Beck&author=E%20McNeer&volume=107&issue=4&publication_year=2020&pages=934-943&pmid=31957870&doi=10.1002/cpt.1787&)
|
| 234 |
+
|
| 235 |
+
32. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome‐wide association studies. Nat Genet. 2006;38(8):904‐909. doi: 10.1038/ng1847 [DOI](https://doi.org/10.1038/ng1847) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16862161/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat%20Genet&title=Principal%20components%20analysis%20corrects%20for%20stratification%20in%20genome%E2%80%90wide%20association%20studies&author=AL%20Price&author=NJ%20Patterson&author=RM%20Plenge&author=ME%20Weinblatt&author=NA%20Shadick&volume=38&issue=8&publication_year=2006&pages=904-909&pmid=16862161&doi=10.1038/ng1847&)
|
| 236 |
+
|
| 237 |
+
33. Sherman BT, Hao M, Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50(W1):W216‐W221. doi: 10.1093/nar/gkac194 [DOI](https://doi.org/10.1093/nar/gkac194) | [PMC free article](/articles/PMC9252805/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35325185/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nucleic%20Acids%20Res&title=DAVID:%20a%20web%20server%20for%20functional%20enrichment%20analysis%20and%20functional%20annotation%20of%20gene%20lists%20(2021%20update)&author=BT%20Sherman&author=M%20Hao&author=J%20Qiu&volume=50&issue=W1&publication_year=2022&pages=W216-W221&pmid=35325185&doi=10.1093/nar/gkac194&)
|
| 238 |
+
|
| 239 |
+
34. Piñero J, Gonzalez‐Perez A, Guney E, et al. Network, transcriptomic and genomic features differentiate genes relevant for drug response. Front Genet. 2018;9:412. doi: 10.3389/fgene.2018.00412 [DOI](https://doi.org/10.3389/fgene.2018.00412) | [PMC free article](/articles/PMC6168038/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30319692/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front%20Genet&title=Network,%20transcriptomic%20and%20genomic%20features%20differentiate%20genes%20relevant%20for%20drug%20response&author=J%20Pi%C3%B1ero&author=A%20Gonzalez%E2%80%90Perez&author=E%20Guney&volume=9&publication_year=2018&pages=412&pmid=30319692&doi=10.3389/fgene.2018.00412&)
|
| 240 |
+
|
| 241 |
+
35. Drozdzik M, Busch D, Lapczuk J, et al. Protein abundance of clinically relevant drug‐metabolizing enzymes in the human liver and intestine: a comparative analysis in paired tissue specimens. Clin Pharmacol Ther. 2018;104(3):515‐524. doi: 10.1002/cpt.967 [DOI](https://doi.org/10.1002/cpt.967) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29205295/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Protein%20abundance%20of%20clinically%20relevant%20drug%E2%80%90metabolizing%20enzymes%20in%20the%20human%20liver%20and%20intestine:%20a%20comparative%20analysis%20in%20paired%20tissue%20specimens&author=M%20Drozdzik&author=D%20Busch&author=J%20Lapczuk&volume=104&issue=3&publication_year=2018&pages=515-524&pmid=29205295&doi=10.1002/cpt.967&)
|
| 242 |
+
|
| 243 |
+
36. Mangoni AA, Jackson SHD. Age‐related changes in pharmacokinetics and pharmacodynamics: basic principles and practical applications. Br J Clin Pharmacol. 2004;57(1):6‐14. doi: 10.1046/j.1365-2125.2003.02007.x [DOI](https://doi.org/10.1046/j.1365-2125.2003.02007.x) | [PMC free article](/articles/PMC1884408/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14678335/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Age%E2%80%90related%20changes%20in%20pharmacokinetics%20and%20pharmacodynamics:%20basic%20principles%20and%20practical%20applications&author=AA%20Mangoni&author=SHD%20Jackson&volume=57&issue=1&publication_year=2004&pages=6-14&pmid=14678335&doi=10.1046/j.1365-2125.2003.02007.x&)
|
| 244 |
+
|
| 245 |
+
37. Batchelor HK, Marriott JF. Paediatric pharmacokinetics: key considerations. Br J Clin Pharmacol. 2015;79(3):395‐404. doi: 10.1111/bcp.12267 [DOI](https://doi.org/10.1111/bcp.12267) | [PMC free article](/articles/PMC4345950/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25855821/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Paediatric%20pharmacokinetics:%20key%20considerations&author=HK%20Batchelor&author=JF%20Marriott&volume=79&issue=3&publication_year=2015&pages=395-404&pmid=25855821&doi=10.1111/bcp.12267&)
|
| 246 |
+
|
| 247 |
+
38. Läll K, Mägi R, Morris A, Metspalu A, Fischer K. Personalized risk prediction for type 2 diabetes: the potential of genetic risk scores. Genet Med. 2017;19(3):322‐329. doi: 10.1038/gim.2016.103 [DOI](https://doi.org/10.1038/gim.2016.103) | [PMC free article](/articles/PMC5506454/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27513194/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genet%20Med&title=Personalized%20risk%20prediction%20for%20type%202%20diabetes:%20the%20potential%20of%20genetic%20risk%20scores&author=K%20L%C3%A4ll&author=R%20M%C3%A4gi&author=A%20Morris&author=A%20Metspalu&author=K%20Fischer&volume=19&issue=3&publication_year=2017&pages=322-329&pmid=27513194&doi=10.1038/gim.2016.103&)
|
| 248 |
+
|
| 249 |
+
39. Mega JL, Stitziel NO, Smith JG, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385(9984):2264‐2271. doi: 10.1016/S0140-6736(14)61730-X [DOI](https://doi.org/10.1016/S0140-6736(14)61730-X) | [PMC free article](/articles/PMC4608367/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25748612/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Genetic%20risk,%20coronary%20heart%20disease%20events,%20and%20the%20clinical%20benefit%20of%20statin%20therapy:%20an%20analysis%20of%20primary%20and%20secondary%20prevention%20trials&author=JL%20Mega&author=NO%20Stitziel&author=JG%20Smith&volume=385&issue=9984&publication_year=2015&pages=2264-2271&pmid=25748612&doi=10.1016/S0140-6736(14)61730-X&)
|
| 250 |
+
|
| 251 |
+
40. Inouye M, Abraham G, Nelson CP, et al. Genomic risk prediction of coronary artery disease in 480,000 adults. J Am Coll Cardiol. 2018;72(16):1883‐1893. doi: 10.1016/j.jacc.2018.07.079 [DOI](https://doi.org/10.1016/j.jacc.2018.07.079) | [PMC free article](/articles/PMC6176870/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30309464/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Am%20Coll%20Cardiol&title=Genomic%20risk%20prediction%20of%20coronary%20artery%20disease%20in%20480,000%20adults&author=M%20Inouye&author=G%20Abraham&author=CP%20Nelson&volume=72&issue=16&publication_year=2018&pages=1883-1893&pmid=30309464&doi=10.1016/j.jacc.2018.07.079&)
|
| 252 |
+
|
| 253 |
+
41. Escott‐Price V, Sims R, Bannister C, et al. Common polygenic variation enhances risk prediction for Alzheimer's disease. Brain. 2015;138(12):3673‐3684. doi: 10.1093/brain/awv268 [DOI](https://doi.org/10.1093/brain/awv268) | [PMC free article](/articles/PMC5006219/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26490334/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Brain&title=Common%20polygenic%20variation%20enhances%20risk%20prediction%20for%20Alzheimer's%20disease&author=V%20Escott%E2%80%90Price&author=R%20Sims&author=C%20Bannister&volume=138&issue=12&publication_year=2015&pages=3673-3684&pmid=26490334&doi=10.1093/brain/awv268&)
|
| 254 |
+
|
| 255 |
+
42. Escott‐Price V, Nalls MA, Morris HR, et al. Polygenic risk of p arkinson disease is correlated with disease age at onset. Ann Neurol. 2015;77(4):582‐591. doi: 10.1002/ana.24335 [DOI](https://doi.org/10.1002/ana.24335) | [PMC free article](/articles/PMC4737223/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25773351/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20Neurol&title=Polygenic%20risk%20of%20p%20arkinson%20disease%20is%20correlated%20with%20disease%20age%20at%20onset&author=V%20Escott%E2%80%90Price&author=MA%20Nalls&author=HR%20Morris&volume=77&issue=4&publication_year=2015&pages=582-591&pmid=25773351&doi=10.1002/ana.24335&)
|
| 256 |
+
|
| 257 |
+
43. The Breast and Prostate Cancer Cohort Consortium (BPC3), The PRACTICAL (Prostate Cancer Association Group to Investigate Cancer‐Associated Alterations in the Genome) Consortium, The COGS (Collaborative Oncological Gene‐environment Study) Consortium et al. A meta‐analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014;46(10):1103‐1109. doi: 10.1038/ng.3094 [DOI](https://doi.org/10.1038/ng.3094) | [PMC free article](/articles/PMC4383163/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25217961/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat%20Genet&title=A%20meta%E2%80%90analysis%20of%2087,040%20individuals%20identifies%2023%20new%20susceptibility%20loci%20for%20prostate%20cancer&volume=46&issue=10&publication_year=2014&pages=1103-1109&pmid=25217961&doi=10.1038/ng.3094&)
|
| 258 |
+
|
| 259 |
+
44. Mavaddat N, Pharoah PDP, Michailidou K, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015;107(5):djv036. doi: 10.1093/jnci/djv036 [DOI](https://doi.org/10.1093/jnci/djv036) | [PMC free article](/articles/PMC4754625/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25855707/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Natl%20Cancer%20Inst&title=Prediction%20of%20breast%20cancer%20risk%20based%20on%20profiling%20with%20common%20genetic%20variants&author=N%20Mavaddat&author=PDP%20Pharoah&author=K%20Michailidou&volume=107&issue=5&publication_year=2015&pages=djv036&pmid=25855707&doi=10.1093/jnci/djv036&)
|
| 260 |
+
|
| 261 |
+
45. Thomas M, Sakoda LC, Hoffmeister M, et al. Genome‐wide modeling of polygenic risk score in colorectal cancer risk. Am J Hum Genet. 2020;107(3):432‐444. doi: 10.1016/j.ajhg.2020.07.006 [DOI](https://doi.org/10.1016/j.ajhg.2020.07.006) | [PMC free article](/articles/PMC7477007/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32758450/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Hum%20Genet&title=Genome%E2%80%90wide%20modeling%20of%20polygenic%20risk%20score%20in%20colorectal%20cancer%20risk&author=M%20Thomas&author=LC%20Sakoda&author=M%20Hoffmeister&volume=107&issue=3&publication_year=2020&pages=432-444&pmid=32758450&doi=10.1016/j.ajhg.2020.07.006&)
|
| 262 |
+
|
| 263 |
+
46. Khera AV, Chaffin M, Aragam KG, et al. Genome‐wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219‐1224. doi: 10.1038/s41588-018-0183-z [DOI](https://doi.org/10.1038/s41588-018-0183-z) | [PMC free article](/articles/PMC6128408/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30104762/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat%20Genet&title=Genome%E2%80%90wide%20polygenic%20scores%20for%20common%20diseases%20identify%20individuals%20with%20risk%20equivalent%20to%20monogenic%20mutations&author=AV%20Khera&author=M%20Chaffin&author=KG%20Aragam&volume=50&issue=9&publication_year=2018&pages=1219-1224&pmid=30104762&doi=10.1038/s41588-018-0183-z&)
|
| 264 |
+
|
| 265 |
+
47. Nishino J, Ochi H, Kochi Y, Tsunoda T, Matsui S. Sample size for successful genome‐wide association study of major depressive disorder. Front Genet. 2018;9:227. doi: 10.3389/fgene.2018.00227 [DOI](https://doi.org/10.3389/fgene.2018.00227) | [PMC free article](/articles/PMC6032046/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30002671/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front%20Genet&title=Sample%20size%20for%20successful%20genome%E2%80%90wide%20association%20study%20of%20major%20depressive%20disorder&author=J%20Nishino&author=H%20Ochi&author=Y%20Kochi&author=T%20Tsunoda&author=S%20Matsui&volume=9&publication_year=2018&pages=227&pmid=30002671&doi=10.3389/fgene.2018.00227&)
|
| 266 |
+
|
| 267 |
+
48. Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177(1):26‐31. doi: 10.1016/j.cell.2019.02.048 [DOI](https://doi.org/10.1016/j.cell.2019.02.048) | [PMC free article](/articles/PMC7380073/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30901543/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cell&title=The%20missing%20diversity%20in%20human%20genetic%20studies&author=G%20Sirugo&author=SM%20Williams&author=SA%20Tishkoff&volume=177&issue=1&publication_year=2019&pages=26-31&pmid=30901543&doi=10.1016/j.cell.2019.02.048&)
|
| 268 |
+
|
| 269 |
+
49. Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538(7624):161‐164. doi: 10.1038/538161a [DOI](https://doi.org/10.1038/538161a) | [PMC free article](/articles/PMC5089703/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27734877/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nature&title=Genomics%20is%20failing%20on%20diversity&author=AB%20Popejoy&author=SM%20Fullerton&volume=538&issue=7624&publication_year=2016&pages=161-164&pmid=27734877&doi=10.1038/538161a&)
|
test/texts/PMC10502099.md
ADDED
|
@@ -0,0 +1,338 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Carriers of HLA-DRB1*04:05 have a better clinical response to abatacept in rheumatoid arthritis
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Mariko Inoue, Yasuo Nagafuchi, Mineto Ota, Haruka Tsuchiya, Shoko Tateishi, Hiroko Kanda, Keishi Fujio
|
| 5 |
+
**Journal:** Scientific Reports
|
| 6 |
+
**Date:** 2023 Sep 14
|
| 7 |
+
**DOI:** [10.1038/s41598-023-42324-6](https://doi.org/10.1038/s41598-023-42324-6)
|
| 8 |
+
**PMID:** 37709837
|
| 9 |
+
**PMCID:** PMC10502099
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10502099/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC10502099/pdf/41598_2023_Article_42324.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC10502099/pdf/41598_2023_Article_42324.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
HLA-DRB1 shared epitope risk alleles are the strongest genetic risk factors for rheumatoid arthritis (RA) and potential biomarkers for treatment response to biological disease-modifying antirheumatic drugs (bDMARDs). This study aimed to investigate the association between treatment response and individual HLA-DRB1 alleles in RA patients receiving different bDMARDs. We recruited 106 patients with active RA who had started abatacept, tocilizumab, or TNF inhibitors as a first-line bDMARDs. We examined the relationship between Simplified Disease Activity Index (SDAI) improvement at 3 months and HLA-DRB1 allele carriage. The results revealed that the HLA-DRB1*04:05 allele, a shared-epitope allele, was significantly associated with better SDAI improvement only after abatacept treatment (SDAI improvement 28.5% without the allele vs 59.8% with allele, p = 0.003). However, no significant association was found with other treatments. Both multivariate linear regression and mediation analysis confirmed that the HLA-DRB1*04:05 allele was independently associated with abatacept treatment response, regardless of anti-CCP antibody titers. The study concluded that in patients with RA receiving their first-line bDMARD treatment, carrying the HLA-DRB1*04:05 allele was associated with better SDAI improvement specifically in abatacept-treated patients. These disease-risk HLA alleles have the potential to serve as genomic biomarkers for predicting treatment response with co-stimulation blockage therapy.
|
| 16 |
+
|
| 17 |
+
Subject terms: Rheumatoid arthritis, Rheumatic diseases, Medical research, Rheumatology
|
| 18 |
+
|
| 19 |
+
**Subject terms:**Subject terms: Rheumatoid arthritis, Rheumatic diseases, Medical research, Rheumatology
|
| 20 |
+
|
| 21 |
+
## Introduction
|
| 22 |
+
|
| 23 |
+
Treatment of rheumatoid arthritis (RA) has improved dramatically with the development and approval of biologic agents (bDMARDs) and Janus kinase (JAK) inhibitors^1^[1](#CR1)1. Although many bDMARDs and JAK inhibitors are now available, treatment recommendations list the two options as equivalent due to the paucity of treatment-specific predictors of response for guiding drug selection^2,3^[2](#CR2)2,[3](#CR3)3. In practice, the choice of medication for RA is often based on the clinician's experience. In contrast, in the field of malignancy, genomic medicine has progressed, and drugs are routinely selected on a patient-by-patient basis according to the cancer genetic mutations present. In the field of autoimmune diseases, however, there is still a lack of evidence for personalized medicine. Among autoimmune diseases, RA presents a heavy burden, given the high cost of treatment and the progressive destruction of joints in the early stages of disease, and there is an urgent need to address it. Indeed, with the large number of treatment options available for RA, it seems like a plausible goal for the near future.
|
| 24 |
+
|
| 25 |
+
In predicting response to therapy for RA, the HLA gene has, to date, been the most examined genomic biomarker. The HLA-DRB1 gene is the most potent disease susceptibility gene, explaining 30–50% of the genetic risk of RA, and a specific sequence at positions 70–74 of the HLA-DRB1 allele is called a shared epitope (SE) and thought to be involved in the onset and pathology of RA^4,5^[4](#CR4)4,[5](#CR5)5. SE positivity is associated with progressive joint destruction^6^[6](#CR6)6, and it has been reported that bone destruction progresses more rapidly on X-ray when the amino acid at position 11 of HLA-DRB1 is valine^7^[7](#CR7)7. Regarding the association between HLA and the efficacy of bDMARDs, a study reported a significant improvement in disease activity with TNF inhibitors when the amino acids at positions 11, 71, and 74 of HLA-DRB1 were valine, lysine, and alanine, respectively, using RA cohort data from the United Kingdom^7^[7](#CR7)7. On the other hand, in 2016, an attempt was made to construct a predictive model for responsiveness to TNF inhibitors using genome-wide single nucleotide polymorphism information of RA patients, but no significant improvement in prediction accuracy was observed by adding genetic polymorphism information to the prediction model based on clinical information^8^[8](#CR8)8.
|
| 26 |
+
|
| 27 |
+
Abatacept (ABT, CTLA4-Ig) is a drug that inhibits T-cell co-stimulation. There are multiple reports of good response to treatment with ABT in HLA-DRB1 SE-positive cases. In patients with RA using ABT, the SDAI remission rate was 55.3% for SE-positive patients and 20.0% for SE-negative patients, demonstrating high efficacy of ABT in SE-positive RA patients^9^[9](#CR9)9. A study comparing the impact of SE on the efficacy of ABT and the JAK inhibitor tofacitinib also reported that SE was associated with DAS28 remission only in the ABT group^10^[10](#CR10)10. Furthermore, in the Early-AMPLE study, a prospective study directly comparing ABT with the TNF inhibitor adalimumab, ABT was more effective in SE-positive patients in terms of American College of Rheumatology core set and DAS28 remission^11^[11](#CR11)11. Although these reports suggest that SE alleles are strongly associated with treatment response, especially in ABT, details on the specific HLA-DRB1 allele associated with susceptibility are scarce.
|
| 28 |
+
|
| 29 |
+
In this study, we investigated the association between treatment response and individual HLA-DRB1 alleles in patients with RA using ABT, tocilizumab (TCZ, an IL-6 receptor inhibitor), or a TNF inhibitor as their first bDMARD, and attempted to identify HLA alleles associated with treatment outcome.
|
| 30 |
+
|
| 31 |
+
## Methods
|
| 32 |
+
|
| 33 |
+
### Patients
|
| 34 |
+
|
| 35 |
+
Japanese patients with RA who had received their first bDMARD between June 2012 and August 2018 in the Tokyo University Biologics Registry for RA (TOBIRA) and continued it for at least 3 months were included. All patients fulfilled the 1987 ACR^12^[12](#CR12)12 or 2010 ACR/EULAR classification criteria for RA^13^[13](#CR13)13. The selection of bDMARDs was made based on the judgment of both the physician and the patient. From the revised version in 2013 to the latest revised edition in 2022, EULAR recommendations for the management of RA have been listed TNF inhibitor, IL-6 inhibitor, and abatacept equally as bDMARDs^14,15^[14](#CR14)14,[15](#CR15)15, and treatment selection was made with reference to these recommendations. Patients who had been previously treated with bDMARDs or JAK inhibitors were excluded, and those who met SDAI remission (≤ 3.3) at the start of their first bDMARD regimen were excluded. The study was approved by the ethics committee of the University of Tokyo (no. 11592). All methods were carried out in accordance with Declaration of Helsinki and Japanese government’s Ethical Guidelines for Medical and Health Research Involving Human Subjects.
|
| 36 |
+
|
| 37 |
+
### Clinical data collection
|
| 38 |
+
|
| 39 |
+
We evaluated age, sex, disease duration, smoking history, Anti-CCP (cyclic citrullinated peptide) antibody, rheumatoid factor, and concomitant methotrexate (MTX) and oral prednisolone treatment at baseline. Anti-CCP antibody titer was measured using the STACIA MEBLux Test CCP, which is approved as an in vitro diagnostic medical device in Japan. The measurement range of this test kit is 0.6–500 U/mL. Oral glucocorticoid dose was converted to prednisolone equivalent dose. The following variables were evaluated at baseline and at 3 months of bDMARD treatment: tender joint count (TJC) and swollen joint count (SJC) in 28 joints, patient global assessment (PtGA), evaluator global assessment (EGA), and blood CRP level. The CRP level was measured as mg/dl. Global health (GH) was customarily replaced by the PtGA in millimeters on a visual analog scale. Scores for the PtGA and EGA were measured in centimeters on a 0–100 mm visual analog scale. We determined the SDAI score using the following equation^16^[16](#CR16)16:
|
| 40 |
+
|
| 41 |
+
| SDAI=SJC28+TJC28+PtGA+EGA+CRP |
|
| 42 |
+
| ----------------------------- |
|
| 43 |
+
Improvement in disease activity at 3 months was assessed based on change of SDAI value from baseline^17^[17](#CR17)17:
|
| 44 |
+
|
| 45 |
+
| SDAI at baseline-SDAI at 3months/SDAI at baseline×100[%]. |
|
| 46 |
+
| --------------------------------------------------------- |
|
| 47 |
+
DAS28-CRP was calculated following the definition:
|
| 48 |
+
|
| 49 |
+
| DAS28-CRP=0.56∗√TJC28+0.28∗√SJC28+0.014∗GH+0.36∗lnCRP+1+0.96 |
|
| 50 |
+
| ------------------------------------------------------------ |
|
| 51 |
+
### HLA allele and treatment responsiveness
|
| 52 |
+
|
| 53 |
+
HLA-DRB1 alleles were determined by next generation sequencing using peripheral blood of patients. We examined ten alleles with an allele frequency exceeding 0.03 based on the participants in this study. Of them, HLA-DRB1*01:01, *04:05, and *04:10 were defined as SE^18^[18](#CR18)18.
|
| 54 |
+
|
| 55 |
+
We used the percent change in SDAI from baseline to 3 months after each treatment as an assessment measure for treatment responsiveness for the following reasons. Tocilizumab blocks the IL-6 receptor and inhibits the production of acute-phase reactants, including ESR and CRP. Since the weight of ESR and CRP levels is quite high in the DAS28 formula^19,20^[19](#CR19)19,[20](#CR20)20, the remission rate of patients under tocilizumab treatment was reported to be higher when using DAS28 compared to SDAI^21^[21](#CR21)21. In this study, we evaluated the treatment responsiveness among bDMARDs, including tocilizumab, and therefore, we considered that using SDAI for evaluation was appropriate. The ACR20 has been validated as the best discriminator of efficacy in placebo-controlled trials. However, when assessing depth of response with respect to disease activity, continuous scales, such as SDAI is commonly employed. Smolen et al. investigated SDAI improvement rates in previous large-scale clinical trials and found that these rates serve as sensitive treatment response criteria. They also observed that optimal cutoffs for SDAI improvement rates vary across different trials^22^[22](#CR22)22. Given the limited number of cases in our study, we utilized the SDAI improvement rate as a more sensitive continuous variable to investigate the relationship between HLA haplotypes and treatment response.
|
| 56 |
+
|
| 57 |
+
The percent change in SDAI from baseline to 3 months after each treatment was examined, and the difference in percent change between carriers and non-carriers of these HLA alleles was analyzed with a Welch's *t*t test, followed by a multiple testing correction with the Benjamini–Hochberg method. We applied a significance threshold for multiple comparisons based on the Benjamini–Hochberg method with a false discovery rate (FDR) of < 0.05. Dose effects based on HLA-DRB1 alleles were not tested because of the limited number of study participants.
|
| 58 |
+
|
| 59 |
+
### Univariate and multivariate linear regression analysis of abatacept response
|
| 60 |
+
|
| 61 |
+
Thirty-seven patients who underwent ABT treatment were included in the analysis. A univariate linear regression analysis was performed using the SDAI improvement rate after 3 months as the objective variable and each clinical item and the HLA-DRB1*04:05 allele were the explanatory variables. To account for the varying scales of the explanatory variables, we normalized both the objective and explanatory variables. This normalization process aimed to ensure that all variables had a mean of zero and a variance of one. We further conducted multiple regression analysis using the HLA-DRB1*04:05 allele, disease duration, and the anti-CCP antibody titer as explanatory variables. These variables were selected based on their significance (*p*p < 0.20) in the univariate analyses. The response variable in this analysis was the SDAI improvement rate at 3 months.
|
| 62 |
+
|
| 63 |
+
### Mediation analysis
|
| 64 |
+
|
| 65 |
+
Thirty-seven ABT-treated patients were included in the mediation analysis. The relationships between the HLA-DRB1*04:05 allele (negative or positive), anti-CCP antibody titer, and SDAI improvement rate at 3 months were tested by mediation analysis. The SDAI improvement rate at 3 months was treated as the dependent variable of the linear regression model. The HLA-DRB1*04:05 allele was treated as the independent variable and anti-CCP antibody titer was treated as the mediator. To account for the varying scales of the variables, we normalized all variables. *p*p values were calculated via 1000-time bootstrapping.
|
| 66 |
+
|
| 67 |
+
### Statistical analysis
|
| 68 |
+
|
| 69 |
+
All statistical tests were performed using GraphPad Prism v9.3.1 (GraphPad Software) and R v4.1.3 (The R foundation). Mediation analysis was performed using the R mediation package v4.5.0.
|
| 70 |
+
|
| 71 |
+
### Ethics approval and consent to participate
|
| 72 |
+
|
| 73 |
+
This study was approved by the ethics committee of the University of Tokyo (11592), and informed consent was obtained from all participants included in the study.
|
| 74 |
+
|
| 75 |
+
## Results
|
| 76 |
+
|
| 77 |
+
### Patient characteristics and HLA alleles
|
| 78 |
+
|
| 79 |
+
The study included 106 patients with RA who had active disease and were initiating their first bDMARD (Supplementary Figure S1). Table [1](#Tab1)1 shows the patient characteristics. The bDMARDs used in this study included 37 patients with ABT, 28 with TCZ, and 41 with TNF inhibitors. There were no significant differences in SDAI disease activity at the start of bDMARD treatment or 3 months after treatment (Kruskal–Wallis test, *p*p = 0.63 at baseline, *p*p = 0.75 at 3 months). ABT patients were older (one-way analysis of variance, *p*p < 0.0001) and TNF inhibitors patients were more likely to use MTX (Chi-square test, *p*p = 0.014). This study includes patients who initiated bDMARDs with SDAI low disease activity. These patients began bDMARDs with the goal of tapering glucocorticoids, or their treatment was intensified with bDMARDs because their disease activity was considered moderate when evaluated using disease activity measures other than SDAI.
|
| 80 |
+
|
| 81 |
+
### Table 1.
|
| 82 |
+
|
| 83 |
+
Clinical characteristics of patients.
|
| 84 |
+
|
| 85 |
+
| Characteristic | Abatacept (n = 37) | Tocilizumab (n = 28) | TNF inhibitor (n = 41) | p |
|
| 86 |
+
| -------------- | ------------------ | -------------------- | ---------------------- | - |
|
| 87 |
+
| Age, years, mean ± SD | 68.0 ± 8.3 | 56.4 ± 12.6 | 59.2 ± 12.1 | < 0.0001§ |
|
| 88 |
+
| Sex, female, % | 86.5 | 82.1 | 82.9 | 0.87‡ |
|
| 89 |
+
| Disease duration, years | 6.4 (2.9–20.6) | 5.6 (1.3–12.3) | 4.4 (1.0–13.3) | 0.24† |
|
| 90 |
+
| Anti-CCP antibody positive, % | 86.5 | 67.9 | 80.5 | 0.18‡ |
|
| 91 |
+
| Anti-CCP antibody titer (U/mL) | 102 (17.2–333) | 25.9 (0.9–143) | 100 (10.8–230) | 0.20† |
|
| 92 |
+
| RF positive, % | 83.8 | 64.3 | 80.5 | 0.15‡ |
|
| 93 |
+
| RF titer (IU/mL) | 75.0 (21.5–131) | 38.0 (7.3–131) | 56.0 (20–148) | 0.44† |
|
| 94 |
+
| Smoking, % | 27.0 | 46.4 | 29.3 | 0.21‡ |
|
| 95 |
+
| Methotrexate user, % | 56.8 | 60.7 | 85.4 | 0.014‡ |
|
| 96 |
+
| Methotrexate dose (mg) | 6.0 (0.0–8.0) | 7.0 (0.0–10.0) | 10.0 (6.0–10.0) | 0.004† |
|
| 97 |
+
| Oral prednisolone user, % | 64.9 | 60.7 | 61.0 | 0.92‡ |
|
| 98 |
+
| Oral prednisolone dose (mg) | 4.0 (0.0–5.0) | 3.0 (0.0–5.0) | 3.0 (0.0–5.0) | 0.54† |
|
| 99 |
+
| Baseline DAS28-CRP, mean ± SD | 4.4 ± 1.5 | 4.0 ± 1.4 | 4.2 ± 1.1 | 0.52§ |
|
| 100 |
+
| Baseline SDAI | 18.7 (11.9–38.3) | 16.6 (11.7–25.5) | 20.5 (13.5–28.6) | 0.63† |
|
| 101 |
+
| Baseline SDAI Remission/LDA/MDA/ HDA | 0/5/19/13 | 0/5/17/6 | 0/8/21/12 | 0.80♦ |
|
| 102 |
+
| 3 months DAS28-CRP, mean ± SD | 3.3 ± 1.4 | 2.7 ± 1.1 | 2.9 ± 1.1 | 0.11§ |
|
| 103 |
+
| 3 months SDAI | 10.1 (4.8–16.9) | 8.5 (5.2–14.0) | 8.3 (4.8–17.0) | 0.75† |
|
| 104 |
+
| 3 months SDAI Remission/LDA/MDA/ HDA | 5/15/13/4 | 4/13/9/2 | 7/15/17/2 | 0.85♦ |
|
| 105 |
+
There were no significant differences in allele frequencies of the major HLA-DRB1 alleles between the ABT, TCZ, and TNF inhibitors groups (Table [2](#Tab2)2).
|
| 106 |
+
|
| 107 |
+
### Table 2.
|
| 108 |
+
|
| 109 |
+
HLA-DRB1 allele frequencies.
|
| 110 |
+
|
| 111 |
+
| HLA alleles | Abatacept (n = 37) | Tocilizumab (n = 28) | TNF inhibitor (n = 41) | p‡ |
|
| 112 |
+
| ----------- | ------------------ | -------------------- | ---------------------- | -- |
|
| 113 |
+
| Total | 74 (100) | 56 (100) | 82 (100) | |
|
| 114 |
+
| DRB1*01:01 # | 5 (6.8) | 3 (5.4) | 7 (8.5) | 0.77 |
|
| 115 |
+
| DRB1*04:05 # | 17 (23) | 9 (16.1) | 20 (24.4) | 0.48 |
|
| 116 |
+
| DRB1*04:06 | 2 (2.7) | 5 (8.9) | 3 (3.7) | 0.21 |
|
| 117 |
+
| DRB1*04:10 # | 3 (4.1) | 2 (3.6) | 4 (4.9) | 0.93 |
|
| 118 |
+
| DRB1*08:03 | 5 (6.8) | 4 (7.1) | 6 (7.3) | 0.99 |
|
| 119 |
+
| DRB1*09:01 | 10 (13.5) | 10 (17.9) | 15 (18.3) | 0.69 |
|
| 120 |
+
| DRB1*12:01 | 3 (4.1) | 2 (3.6) | 2 (2.4) | 0.85 |
|
| 121 |
+
| DRB1*13:02 | 3 (4.1) | 1 (1.8) | 4 (4.9) | 0.64 |
|
| 122 |
+
| DRB1*15:01 | 6 (8.1) | 0 (0) | 3 (3.7) | 0.072 |
|
| 123 |
+
| DRB1*15:02 | 3 (4.1) | 7 (12.5) | 9 (11) | 0.18 |
|
| 124 |
+
| Other DRB1 alleles | 17 (23.0) | 13 (23.2) | 9 (11.0) | 0.086 |
|
| 125 |
+
### Treatment response by HLA allele
|
| 126 |
+
|
| 127 |
+
In each of the 3 treatment groups, treatment response 3 months after bDMARD initiation was compared between carriers and non-carriers of the major HLA-DRB1 alleles (Table [3](#Tab3)3). Of all the HLA-DRB1 alleles, the only one that showed a significant difference in percent change in SDAI after 3 months was HLA-DRB1*04:05, one of the SE alleles in ABT use (28.5% SDAI improvement in HLA-DRB1*04:05 allele non-carriers and 59.8% SDAI improvement in HLA-DRB1*04:05 allele carriers, *p*p = 0.003, false discovery rate = 0.039). In other words, among the SE alleles, HLA-DRB1*04:05 was a clear predictor of good prognosis among those treated with ABT.
|
| 128 |
+
|
| 129 |
+
### Table 3.
|
| 130 |
+
|
| 131 |
+
Effects of HLA alleles on SDAI improvement after treatment.
|
| 132 |
+
|
| 133 |
+
| HLA alleles | Number of patients without allele | Number of patients with allele | Mean SDAI improvement without allele (%) | Mean SDAI improvement with allele (%) | p† | Benjamini–Hochberg adjusted p |
|
| 134 |
+
| ----------- | --------------------------------- | ------------------------------ | ---------------------------------------- | ------------------------------------- | -- | ----------------------------- |
|
| 135 |
+
| Abatacept (n = 37) | | | | | | |
|
| 136 |
+
| DRB1*01:01# | 32 | 5 | 41.2 | 53.4 | 0.449 | 0.662 |
|
| 137 |
+
| DRB1*04:05# | 20 | 17 | 28.5 | 59.8 | 0.003* | 0.039* |
|
| 138 |
+
| DRB1*04:06 | 35 | 2 | 45.0 | 5.5 | 0.654 | 0.662 |
|
| 139 |
+
| DRB1*04:10# | 34 | 3 | 44.3 | 27.0 | 0.662 | 0.662 |
|
| 140 |
+
| DRB1*08:03 | 33 | 4 | 41.4 | 55.3 | 0.320 | 0.662 |
|
| 141 |
+
| DRB1*09:01 | 30 | 7 | 47.0 | 25.0 | 0.185 | 0.662 |
|
| 142 |
+
| DRB1*12:01 | 34 | 3 | 43.5 | 35.3 | 0.558 | 0.662 |
|
| 143 |
+
| DRB1*13:02 | 34 | 3 | 43.4 | 36.7 | 0.657 | 0.662 |
|
| 144 |
+
| DRB1*15:01 | 31 | 6 | 47.7 | 17.8 | 0.237 | 0.662 |
|
| 145 |
+
| DRB1*15:02 | 34 | 3 | 44.7 | 22.0 | 0.446 | 0.662 |
|
| 146 |
+
| Tocilizumab (n = 28) | | | | | | |
|
| 147 |
+
| DRB1*01:01# | 25 | 3 | 43.1 | 42.3 | 0.974 | 0.974 |
|
| 148 |
+
| DRB1*04:05# | 19 | 9 | 47.2 | 34.2 | 0.521 | 0.759 |
|
| 149 |
+
| DRB1*04:06 | 23 | 5 | 47.5 | 22.4 | 0.447 | 0.759 |
|
| 150 |
+
| DRB1*04:10# | 26 | 2 | 41.7 | 60.5 | 0.552 | 0.759 |
|
| 151 |
+
| DRB1*08:03 | 24 | 4 | 42.3 | 47.5 | 0.853 | 0.938 |
|
| 152 |
+
| DRB1*09:01 | 19 | 9 | 47.8 | 33.0 | 0.244 | 0.759 |
|
| 153 |
+
| DRB1*12:01 | 26 | 2 | 41.4 | 64.5 | 0.292 | 0.759 |
|
| 154 |
+
| DRB1*13:02 | 27 | 1 | 42.7 | 52.0 | – | – |
|
| 155 |
+
| DRB1*15:01 | 28 | 0 | 43.0 | – | – | – |
|
| 156 |
+
| DRB1*15:02 | 21 | 7 | 40.8 | 49.9 | 0.541 | 0.759 |
|
| 157 |
+
| TNF inhibitors (n = 41) | | | | | | |
|
| 158 |
+
| DRB1*01:01# | 34 | 7 | 50.4 | 40.4 | 0.587 | 0.893 |
|
| 159 |
+
| DRB1*04:05# | 23 | 18 | 44.4 | 54.2 | 0.313 | 0.893 |
|
| 160 |
+
| DRB1*04:06 | 38 | 3 | 48.4 | 53.0 | 0.910 | 0.910 |
|
| 161 |
+
| DRB1*04:10# | 37 | 4 | 48.4 | 52.0 | 0.756 | 0.893 |
|
| 162 |
+
| DRB1*08:03 | 35 | 6 | 51.0 | 35.3 | 0.288 | 0.893 |
|
| 163 |
+
| DRB1*09:01 | 27 | 14 | 52.0 | 42.3 | 0.345 | 0.893 |
|
| 164 |
+
| DRB1*12:01 | 39 | 2 | 47.5 | 72.5 | 0.375 | 0.893 |
|
| 165 |
+
| DRB1*13:02 | 37 | 4 | 47.8 | 57.5 | 0.601 | 0.893 |
|
| 166 |
+
| DRB1*15:01 | 38 | 3 | 48.2 | 55.7 | 0.671 | 0.893 |
|
| 167 |
+
| DRB1*15:02 | 33 | 8 | 49.5 | 45.3 | 0.704 | 0.893 |
|
| 168 |
+
### HLA-DRB1*04:05 and treatment response in ABT use
|
| 169 |
+
|
| 170 |
+
In the Japanese ACPA-positive RA population, the HLA-DRB1*04:05 allele has a deep impact on RA pathogenesis, with an odds ratio of 5.0 compared to healthy individuals^23^[23](#CR23)23. Since our HLA allele analysis showed a specific prognostic effect of the HLA-DRB1*04:05 allele on ABT treatment (Table [3](#Tab3)3), we continued to focus on the effect of this allele on ABT treatment response.
|
| 171 |
+
|
| 172 |
+
Figure [1](#Fig1)1a shows the trend of SDAI between HLA-DRB1*04:05 carriers and non-carriers during ABT treatment. Three months after the start of treatment, SDAI significantly improved regardless of HLA-DRB1*04:05 status in the ABT group. However, the rate of improvement was significantly higher in HLA-DRB1*04:05 carriers (Fig. [1](#Fig1)1b,t test, *p*p = 0.0030).
|
| 173 |
+
|
| 174 |
+
### Figure 1.
|
| 175 |
+
|
| 176 |
+

|
| 177 |
+
|
| 178 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=10502099_41598_2023_42324_Fig1_HTML.jpg)
|
| 179 |
+
|
| 180 |
+
The HLA-DRB1*04:05 allele is associated with better response to abatacept treatment. (a) SDAI disease activity at baseline and 3 months after ABT treatment (HLA-DRB1*04:05 non-carrier, n = 20 and HLA-DRB1*04:05 carrier, n = 17; t test). (b) Comparison of SDAI improvement rate after 3 months of ABT treatment, stratified by the HLA-DRB1*04:05 carriage. t test.
|
| 181 |
+
|
| 182 |
+
Also, we investigated the association between the presence or absence of the HLA-DRB*04:05 haplotype and the achievement rate of SDAI50 in patients treated with ABT. SDAI 50 is defined as a 50% improvement in SDAI and was proposed by Alehata as a measure that correlates with ACR20, which is considered the gold standard for evaluating treatment response in clinical trials^24^[24](#CR24)24. It is worth noting that SDAI50 is also a measure that correlates with EULAR response, which is commonly used to assess treatment response in routine clinical practice.
|
| 183 |
+
|
| 184 |
+
The results revealed that the SDAI50 achievement rate was 70.6% in patients with the DRB1*04:05 haplotype, compared to 30.0% in those without the DRB1*04:05 haplotype (*p*p = 0.022, Fisher’s exact test).
|
| 185 |
+
|
| 186 |
+
Also, we investigated the differences in SDAI components based on the presence or absence of HLA-DRB1*04:05. We examined TJC28, SJC28, PtGA, EGA and CRP at baseline and after ABT treatment in relation to HLA-DRB1*04:05 carriers and non-carriers (Supplementary Figure S2). Regarding TJC28 and PtGA, we did not observe significant improvement after ABT treatment in HLA-DRB1*04:05 non-carriers. However, HLA-DRB1*04:05 carriers showed improvement in all SDAI components, including TJC and PtGA, after ABT treatment.
|
| 187 |
+
|
| 188 |
+
From these findings, it is concluded that the presence of the HLA-DRB1*04:05 haplotype is associated with better SDAI improvement in patients treated with ABT.
|
| 189 |
+
|
| 190 |
+
### Comparison of HLA-DRB1*04:05 and ACPA as predictors of ABT treatment response
|
| 191 |
+
|
| 192 |
+
To examine the factors involved in ABT response to treatment, linear regression analysis was performed on the rate of SDAI improvement after 3 months of treatment (Table [4](#Tab4)4). In the univariate analysis, being a carrier of HLA-DRB1*04:05 was significantly associated with an increased rate of SDAI improvement (standardized partial regression coefficient β = 0.46, *p*p = 0.0039), but other clinical parameters were not significant prognostic factors. We further conducted multiple regression analysis using the HLA-DRB1*04:05 allele, disease duration, and the anti-CCP antibody titer as explanatory variables. These variables were selected based on their significance (*p*p < 0.20) in the univariate analyses. As a result, carriage of HLA-DRB1*04:05 was associated with ABT efficacy independently of ACPA titer (β = 0.48, *p*p = 0.0052).
|
| 193 |
+
|
| 194 |
+
### Table 4.
|
| 195 |
+
|
| 196 |
+
Linear regression analysis of 3-month SDAI improvement rate after abatacept treatment.
|
| 197 |
+
|
| 198 |
+
| Variable | Univariate analysis | Multivariate analysis |
|
| 199 |
+
| -------- | ------------------- | --------------------- |
|
| 200 |
+
| Standardized β | 95%CI | p | Standardized β | 95%CI | p |
|
| 201 |
+
| Age (year) | − 0.04 | − 0.39, 0.3 | 0.8 | | | |
|
| 202 |
+
| Male sex | 0.22 | − 0.12, 0.55 | 0.2 | | | |
|
| 203 |
+
| Disease duration (year) | − 0.25 | − 0.58, 0.09 | 0.14 | − 0.28 | − 0.59, 0.02 | 0.06 |
|
| 204 |
+
| Anti− CCP antibody titer (U/mL) | 0.25 | − 0.09, 0.58 | 0.14 | 0.03 | − 0.29, 0.36 | 0.84 |
|
| 205 |
+
| RF titer (IU/mL) | 0.01 | − 0.33, 0.35 | 0.95 | | | |
|
| 206 |
+
| Smoking | − 0.13 | − 0.47, 0.21 | 0.43 | | | |
|
| 207 |
+
| Methotrexate dose (mg) | − 0.10 | − 0.44, 0.24 | 0.54 | | | |
|
| 208 |
+
| Prednisolone dose (mg) | − 0.16 | − 0.5, 0.17 | 0.33 | | | |
|
| 209 |
+
| Baseline DAS28-CRP | 0.08 | − 0.27, 0.42 | 0.65 | | | |
|
| 210 |
+
| Baseline SDAI | 0.07 | − 0.28, 0.41 | 0.69 | | | |
|
| 211 |
+
| HLA-DRB1*04:05 | 0.46 | 0.16, 0.77 | 0.0039* | 0.48 | 0.15, 0.8 | 0.0052* |
|
| 212 |
+
To further clarify the relationship between the HLA-DRB1*0405 allele, ACPA titer, and SDAI improvement rate, a causal mediation analysis was performed. The HLA-DRB1*04:05 allele was directly associated with the rate of SDAI improvement, rather than through an indirect effect on ACPA titer (Fig. [2](#Fig2)2). These results indicate that HLA-DRB1*04:05 is more directly associated with ABT prognosis than ACPA titer.
|
| 213 |
+
|
| 214 |
+
### Figure 2.
|
| 215 |
+
|
| 216 |
+

|
| 217 |
+
|
| 218 |
+
The HLA-DRB1*04:05 allele is directly linked to abatacept treatment response. A mediation analysis of the relationships between the HLA-DRB1*04:05 allele carriage, anti-CCP antibody titer, and the SDAI improvement rate 3 months after ABT treatment (n = 37). Solid lines represent significant associations, while the dashed line indicates a non-significant association.
|
| 219 |
+
|
| 220 |
+
## Discussion
|
| 221 |
+
|
| 222 |
+
In this study, we showed that among the SE alleles, HLA-DRB1*04:05 in particular was strongly associated with ABT treatment prognosis. The allele frequency of the HLA-DRB1*04:05 in Japanese patients with ACPA-positive RA is reported to be about 28%. Since each individual carries two HLA-DRB1 alleles, approximately half of the ACPA-positive RA patients have at least one copy of HLA-DRB1*04:05. And HLA-DRB1*04:05 is strongly associated with the development of ACPA-positive RA, having an odds ratio of 5.0^23^[23](#CR23)23. HLA, being innate and unchanging throughout a person's lifetime, suggests that the association between HLA and treatment prognosis is not merely coincidental. In other words, the HLA genotype is the cause, leading to favorable treatment outcomes. Although several associations between SE and ABT efficacy have been reported^9–11^[9](#CR9)9–[11](#CR11)11, details at the allele level are limited, even though the significance of the specific alleles as potential biomarkers is promising.
|
| 223 |
+
|
| 224 |
+
In this study, it was found that only HLA-DRB1*04:05 demonstrated an association with the responsiveness to ABT treatment, while HLA-DRB1*01:01 and 04:10, which share similar SE, did not show a significant association with treatment responsiveness. In addition to the effect of the small sample size, the following reasons can be considered. Amino acids at positions 11, 13, and 67 of HLA-DRB1, which are amino acid sequences other than SE, are also implicated in the risk of developing RA. Specifically, it has been found that in DRB1*04:05 and 04:10 the valine at position 11 is the amino acid most strongly associated with RA susceptibility, whereas DRB1*01:01 has a different amino acid, leucine, at position 11^25^[25](#CR25)25. Additionally, in a study reporting on the risk of developing RA among the Japanese population, it has been demonstrated that the risk of RA differs based on the variant of HLA-DRB1, even sharing the same HLA SE allele. It is suggested that HLA-DRB1*01:01, 04:05, and 04:10 are not biologically equivalent^23^[23](#CR23)23. Furthermore, it has been reported that HLA risk alleles for autoimmune diseases significantly impact the pattern of CDR3 sequences in T-cell receptors. Additionally, CDR3 sequences modified by HLA risk alleles have been associated with the recognition of citrullinated antigens. Therefore, it is believed that sequences other than SE are also associated with the development and progression of RA and other diseases^26^[26](#CR26)26.
|
| 225 |
+
|
| 226 |
+
SE and ACPA-positive RA are strongly associated, and ACPA is also associated with ABT treatment prognosis^27,28^[27](#CR27)27,[28](#CR28)28. Previous reports have also shown that SE is associated with ABT outcomes, even after adjusting for the effect of ACPA^9,10^[9](#CR9)9,[10](#CR10)10. In this study, both multiple regression analysis and mediation analysis suggested that the effect of the HLA-DRB1*04:05 allele was not an indirect effect mediated by ACPA (Table [4](#Tab4)4, Fig. [2](#Fig2)2). The impact of SE has been reported to be stronger in ACPA-positive RA than in ACPA-positive non-RA controls^29,30^[29](#CR29)29,[30](#CR30)30. In other words, SE may be involved in the onset of RA through mechanisms other than direct effects on ACPA positivity. RA-risk HLA is robustly associated with the T-cell receptor repertoire of CD4^+^+ T cells^26,31^[26](#CR26)26,[31](#CR31)31. RA-sensitive HLA alleles, such as HLA-DRB1*04:05, are associated with autoreactive CD4^+^+ T cells, which may be therapeutic targets for ABT.
|
| 227 |
+
|
| 228 |
+
In this investigation, the use of methotrexate was low in the abatacept group. Because, in general, it was reported that concomitant use of MTX may not augment the effectiveness of ABT. For example, in a phase III study, ABT did not elicit immunogenicity associated with loss of safety or efficacy, either with or without MTX^32^[32](#CR32)32. Also, in a retrospective cohort study, in RA patients with similar background characteristics undergoing abatacept treatment, concomitant MTX did not seem to affect clinical outcomes^33^[33](#CR33)33. Based on these findings, we believe that ABT would be a suitable treatment option in daily clinical practice in patients with contraindications to MTX.
|
| 229 |
+
|
| 230 |
+
In this study, the association between the HLA-DRB1*04:05 allele, an SE allele, and favorable treatment outcomes was significant only in ABT-treated patients, but not in those treated with the IL-6 receptor inhibitor TCZ or a TNF inhibitors. This is consistent with the association between the better prognosis with ABT and SE reported in the Early-AMPLE trial comparing ABT with the TNF inhibitor adalimumab^11^[11](#CR11)11. SE was not strongly associated with efficacy of the JAK inhibitor tofacitinib either^10^[10](#CR10)10. These findings may reflect the difference in mechanism of action between ABT, which inhibits co-stimulation of antigen-presenting cells and CD4^+^+ T cells, and IL-6 receptor inhibitors, TNF inhibitors, and JAK inhibitors, which are drugs that block inflammatory cytokine signaling.
|
| 231 |
+
|
| 232 |
+
There are several limitations to this study. First, because of the retrospective nature of this analysis, we cannot exclude the possibility of selection bias. Second, the number in each treatment group is small, so the effect of HLA alleles with a small frequency or small effect size may not have been fully realized. Third, since this study was conducted in a single Japanese cohort and there are ethnic differences in HLA-DRB1 allele frequencies, it is necessary to verify whether the results can be generalized to other cohorts, including other ethnic groups.
|
| 233 |
+
|
| 234 |
+
In conclusion, we analyzed the association between HLA-DRB1 alleles and prognosis in Japanese patients with RA who were starting ABT, TCZ, and TNF inhibitor treatment, and we showed that among SE alleles, the HLA- DRB1*04:05 allele was associated with better outcomes with ABT. This study demonstrates the possibility of stratifying RA patients by disease-risk HLA alleles, and supports the need for a larger prospective study.
|
| 235 |
+
|
| 236 |
+
## Acknowledgements
|
| 237 |
+
|
| 238 |
+
We would like to express our gratitude to all the study participants and the members of the Department of Allergy and Rheumatology for their cooperation in this study.
|
| 239 |
+
|
| 240 |
+
## Abbreviations
|
| 241 |
+
|
| 242 |
+
## Author contributions
|
| 243 |
+
|
| 244 |
+
M.I., Y.N., and K.F. conceived and designed the study concept. M.I., Y.N., M.O., H.T., S.T., H.K. contributed to clinical and H.L.A. data collection. M.I. and Y.N. analyzed the data. M.I. and Y.N. wrote the original manuscript draft. K.F. supervised and reviewed and edited the manuscript. All authors approved the final submitted version.
|
| 245 |
+
|
| 246 |
+
## Funding
|
| 247 |
+
|
| 248 |
+
This study was supported by the Ministry of Health, Labour and Welfare, Ministry of Education, Culture, Sports, Science and Technology KAKENHI Grant-in-Aid for Early-Career Scientists (18K16139) from the Japan Society for the Promotion of Science.
|
| 249 |
+
|
| 250 |
+
## Data availability
|
| 251 |
+
|
| 252 |
+
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. We cannot share raw HLA allele information of the patients publicly because it can be considered as personal information in Japanese regulations. Summary data of the clinical information and HLA allele frequencies are provided as Table [1](#Tab1)1 and Table [2](#Tab2)2. We used publicly available software for the analyses. Custom codes are available from the corresponding author on reasonable request.
|
| 253 |
+
|
| 254 |
+
## Competing interests
|
| 255 |
+
|
| 256 |
+
MI has received speaking fees and/or honoraria from Asahi Kasei and Daiichi Sankyo. YN belongs to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical, and has received financial support and/or speaking fees from Abbvie, BMS, Chugai, GlaxoSmithKline, Kissei, Mitsubishi Tanabe, Novartis, and Pfizer. MO belongs to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical. HT has received speaking fees and/or honoraria from Tanabe Mitsubishi, Janssen, Daiichi Sankyo, Amgen, Eisai, Gilead, Bristol-Myers Squibb, Asahi Kasei, Sanofi, UCB, Abbvie, and Eli Lilly, and has received grants from Abbvie, Mochida, and Takeda. ST declares no conflicts of interest. HK has received speaking fees and/or honoraria from AbbVie, Asahi Kasei, AstraZeneca, Astellas, Ayumi, Bristol Myers, Chugai, Eisai, Eli Lilly, Daiichi-Sankyo, Janssen, Novartis, Nihon Kayaku, Pfizer and Tanabe Mitsubishi and UCB, and has received research grants from AbbVie, Asahi Kasei, and Chugai. KF has received speaking fees and/or honoraria from Tanabe Mitsubishi, Bristol Myers, Eli Lilly, Chugai, Jansen, Pfizer, Ono, AbbVie, Ayumi, Astellas, Sanofi, Novartis, Daiichi Sankyo, Eisai, Asahi Kasei, Japan Blood Products Organization, and AstraZeneca, and has received research grants from Tanabe Mitsubishi, Bristol Myers, Eli Lilly, Chugai, AbbVie, Ayumi, Astellas, Sanofi, Eisai, Tsumura and Co., Asahi Kasei and AstraZeneca.
|
| 257 |
+
|
| 258 |
+
## Footnotes
|
| 259 |
+
|
| 260 |
+
## Associated Data
|
| 261 |
+
|
| 262 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 263 |
+
|
| 264 |
+
### Data Availability Statement
|
| 265 |
+
|
| 266 |
+
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. We cannot share raw HLA allele information of the patients publicly because it can be considered as personal information in Japanese regulations. Summary data of the clinical information and HLA allele frequencies are provided as Table [1](#Tab1)1 and Table [2](#Tab2)2. We used publicly available software for the analyses. Custom codes are available from the corresponding author on reasonable request.
|
| 267 |
+
|
| 268 |
+
### Data Availability Statement
|
| 269 |
+
|
| 270 |
+
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. We cannot share raw HLA allele information of the patients publicly because it can be considered as personal information in Japanese regulations. Summary data of the clinical information and HLA allele frequencies are provided as Table [1](#Tab1)1 and Table [2](#Tab2)2. We used publicly available software for the analyses. Custom codes are available from the corresponding author on reasonable request.
|
| 271 |
+
|
| 272 |
+
## References
|
| 273 |
+
|
| 274 |
+
1. Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. 2016;388(10055):2023–2038. doi: 10.1016/S0140-6736(16)30173-8. [DOI](https://doi.org/10.1016/S0140-6736(16)30173-8) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27156434/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Rheumatoid%20arthritis&author=JS%20Smolen&author=D%20Aletaha&author=IB%20McInnes&volume=388&issue=10055&publication_year=2016&pages=2023-2038&pmid=27156434&doi=10.1016/S0140-6736(16)30173-8&)
|
| 275 |
+
|
| 276 |
+
2. Smolen JS, Landewé RBM, Bijlsma JWJ, Burmester GR, Dougados M, Kerschbaumer A, McInnes IB, Sepriano A, van Vollenhoven RF, de Wit M, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann. Rheum. Dis. 2020;79(6):685–699. doi: 10.1136/annrheumdis-2019-216655. [DOI](https://doi.org/10.1136/annrheumdis-2019-216655) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31969328/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum.%20Dis.&title=EULAR%20recommendations%20for%20the%20management%20of%20rheumatoid%20arthritis%20with%20synthetic%20and%20biological%20disease-modifying%20antirheumatic%20drugs:%202019%20update&author=JS%20Smolen&author=RBM%20Landew%C3%A9&author=JWJ%20Bijlsma&author=GR%20Burmester&author=M%20Dougados&volume=79&issue=6&publication_year=2020&pages=685-699&pmid=31969328&doi=10.1136/annrheumdis-2019-216655&)
|
| 277 |
+
|
| 278 |
+
3. Fraenkel L, Bathon JM, England BR, St Clair EW, Arayssi T, Carandang K, Deane KD, Genovese M, Huston KK, Kerr G, et al. 2021 American College of Rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Care Res. (Hoboken) 2021;73(7):924–939. doi: 10.1002/acr.24596. [DOI](https://doi.org/10.1002/acr.24596) | [PMC free article](/articles/PMC9273041/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34101387/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Care%20Res.%20(Hoboken)&title=2021%20American%20College%20of%20Rheumatology%20guideline%20for%20the%20treatment%20of%20rheumatoid%20arthritis&author=L%20Fraenkel&author=JM%20Bathon&author=BR%20England&author=EW%20St%20Clair&author=T%20Arayssi&volume=73&issue=7&publication_year=2021&pages=924-939&pmid=34101387&doi=10.1002/acr.24596&)
|
| 279 |
+
|
| 280 |
+
4. Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30(11):1205–1213. doi: 10.1002/art.1780301102. [DOI](https://doi.org/10.1002/art.1780301102) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/2446635/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Rheum.&title=The%20shared%20epitope%20hypothesis.%20An%20approach%20to%20understanding%20the%20molecular%20genetics%20of%20susceptibility%20to%20rheumatoid%20arthritis&author=PK%20Gregersen&author=J%20Silver&author=RJ%20Winchester&volume=30&issue=11&publication_year=1987&pages=1205-1213&pmid=2446635&doi=10.1002/art.1780301102&)
|
| 281 |
+
|
| 282 |
+
5. Okada Y, Eyre S, Suzuki A, Kochi Y, Yamamoto K. Genetics of rheumatoid arthritis: 2018 status. Ann. Rheum. Dis. 2019;78(4):446–453. doi: 10.1136/annrheumdis-2018-213678. [DOI](https://doi.org/10.1136/annrheumdis-2018-213678) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30530827/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum.%20Dis.&title=Genetics%20of%20rheumatoid%20arthritis:%202018%20status&author=Y%20Okada&author=S%20Eyre&author=A%20Suzuki&author=Y%20Kochi&author=K%20Yamamoto&volume=78&issue=4&publication_year=2019&pages=446-453&pmid=30530827&doi=10.1136/annrheumdis-2018-213678&)
|
| 283 |
+
|
| 284 |
+
6. Kaltenhäuser S, Wagner U, Schuster E, Wassmuth R, Arnold S, Seidel W, Tröltzsch M, Loeffler M, Häntzschel H. Immunogenetic markers and seropositivity predict radiological progression in early rheumatoid arthritis independent of disease activity. J. Rheumatol. 2001;28(4):735–744. [PubMed](https://pubmed.ncbi.nlm.nih.gov/11327243/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Rheumatol.&title=Immunogenetic%20markers%20and%20seropositivity%20predict%20radiological%20progression%20in%20early%20rheumatoid%20arthritis%20independent%20of%20disease%20activity&author=S%20Kaltenh%C3%A4user&author=U%20Wagner&author=E%20Schuster&author=R%20Wassmuth&author=S%20Arnold&volume=28&issue=4&publication_year=2001&pages=735-744&pmid=11327243&)
|
| 285 |
+
|
| 286 |
+
7. Viatte S, Plant D, Han B, Fu B, Yarwood A, Thomson W, Symmons DP, Worthington J, Young A, Hyrich KL, et al. Association of HLA-DRB1 haplotypes with rheumatoid arthritis severity, mortality, and treatment response. JAMA. 2015;313(16):1645–1656. doi: 10.1001/jama.2015.3435. [DOI](https://doi.org/10.1001/jama.2015.3435) | [PMC free article](/articles/PMC4928097/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25919528/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=JAMA&title=Association%20of%20HLA-DRB1%20haplotypes%20with%20rheumatoid%20arthritis%20severity,%20mortality,%20and%20treatment%20response&author=S%20Viatte&author=D%20Plant&author=B%20Han&author=B%20Fu&author=A%20Yarwood&volume=313&issue=16&publication_year=2015&pages=1645-1656&pmid=25919528&doi=10.1001/jama.2015.3435&)
|
| 287 |
+
|
| 288 |
+
8. Sieberts SK, Zhu F, García-García J, Stahl E, Pratap A, Pandey G, Pappas D, Aguilar D, Anton B, Bonet J, et al. Crowdsourced assessment of common genetic contribution to predicting anti-TNF treatment response in rheumatoid arthritis. Nat. Commun. 2016;7:12460. doi: 10.1038/ncomms12460. [DOI](https://doi.org/10.1038/ncomms12460) | [PMC free article](/articles/PMC4996969/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27549343/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat.%20Commun.&title=Crowdsourced%20assessment%20of%20common%20genetic%20contribution%20to%20predicting%20anti-TNF%20treatment%20response%20in%20rheumatoid%20arthritis&author=SK%20Sieberts&author=F%20Zhu&author=J%20Garc%C3%ADa-Garc%C3%ADa&author=E%20Stahl&author=A%20Pratap&volume=7&publication_year=2016&pages=12460&pmid=27549343&doi=10.1038/ncomms12460&)
|
| 289 |
+
|
| 290 |
+
9. Oryoji K, Yoshida K, Kashiwado Y, Tanaka K, Mizuki SI, Tsukamoto H, Kamada K, Akashi K. Shared epitope positivity is related to efficacy of abatacept in rheumatoid arthritis. Ann. Rheum. Dis. 2018;77(8):1234–1236. doi: 10.1136/annrheumdis-2017-211430. [DOI](https://doi.org/10.1136/annrheumdis-2017-211430) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28830884/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum.%20Dis.&title=Shared%20epitope%20positivity%20is%20related%20to%20efficacy%20of%20abatacept%20in%20rheumatoid%20arthritis&author=K%20Oryoji&author=K%20Yoshida&author=Y%20Kashiwado&author=K%20Tanaka&author=SI%20Mizuki&volume=77&issue=8&publication_year=2018&pages=1234-1236&pmid=28830884&doi=10.1136/annrheumdis-2017-211430&)
|
| 291 |
+
|
| 292 |
+
10. Hirose W, Harigai M, Amano K, Hidaka T, Itoh K, Aoki K, Nakashima M, Nagasawa H, Komano Y, Nanki T, et al. Impact of the HLA-DRB1 shared epitope on responses to treatment with tofacitinib or abatacept in patients with rheumatoid arthritis. Arthritis Res. Ther. 2021;23(1):228. doi: 10.1186/s13075-021-02612-w. [DOI](https://doi.org/10.1186/s13075-021-02612-w) | [PMC free article](/articles/PMC8407060/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34465391/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Res.%20Ther.&title=Impact%20of%20the%20HLA-DRB1%20shared%20epitope%20on%20responses%20to%20treatment%20with%20tofacitinib%20or%20abatacept%20in%20patients%20with%20rheumatoid%20arthritis&author=W%20Hirose&author=M%20Harigai&author=K%20Amano&author=T%20Hidaka&author=K%20Itoh&volume=23&issue=1&publication_year=2021&pages=228&pmid=34465391&doi=10.1186/s13075-021-02612-w&)
|
| 293 |
+
|
| 294 |
+
11. Rigby W, Buckner JH, Louis Bridges S, Nys M, Gao S, Polinsky M, Ray N, Bykerk V. HLA-DRB1 risk alleles for RA are associated with differential clinical responsiveness to abatacept and adalimumab: Data from a head-to-head, randomized, single-blind study in autoantibody-positive early RA. Arthritis Res. Ther. 2021;23(1):245. doi: 10.1186/s13075-021-02607-7. [DOI](https://doi.org/10.1186/s13075-021-02607-7) | [PMC free article](/articles/PMC8449494/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34537057/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Res.%20Ther.&title=HLA-DRB1%20risk%20alleles%20for%20RA%20are%20associated%20with%20differential%20clinical%20responsiveness%20to%20abatacept%20and%20adalimumab:%20Data%20from%20a%20head-to-head,%20randomized,%20single-blind%20study%20in%20autoantibody-positive%20early%20RA&author=W%20Rigby&author=JH%20Buckner&author=S%20Louis%20Bridges&author=M%20Nys&author=S%20Gao&volume=23&issue=1&publication_year=2021&pages=245&pmid=34537057&doi=10.1186/s13075-021-02607-7&)
|
| 295 |
+
|
| 296 |
+
12. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31(3):315–324. doi: 10.1002/art.1780310302. [DOI](https://doi.org/10.1002/art.1780310302) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/3358796/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Rheum.&title=The%20American%20Rheumatism%20Association%201987%20revised%20criteria%20for%20the%20classification%20of%20rheumatoid%20arthritis&author=FC%20Arnett&author=SM%20Edworthy&author=DA%20Bloch&author=DJ%20McShane&author=JF%20Fries&volume=31&issue=3&publication_year=1988&pages=315-324&pmid=3358796&doi=10.1002/art.1780310302&)
|
| 297 |
+
|
| 298 |
+
13. Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, 3rd, Birnbaum NS, Burmester GR, Bykerk VP, Cohen MD, et al. 2010 Rheumatoid arthritis classification criteria: An American College of Rheumatology/European League against rheumatism collaborative initiative. Arthritis Rheum. 2010;62(9):2569–2581. doi: 10.1002/art.27584. [DOI](https://doi.org/10.1002/art.27584) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20872595/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Rheum.&title=2010%20Rheumatoid%20arthritis%20classification%20criteria:%20An%20American%20College%20of%20Rheumatology/European%20League%20against%20rheumatism%20collaborative%20initiative&author=D%20Aletaha&author=T%20Neogi&author=AJ%20Silman&author=J%20Funovits&author=DT%20Felson&volume=62&issue=9&publication_year=2010&pages=2569-2581&pmid=20872595&doi=10.1002/art.27584&)
|
| 299 |
+
|
| 300 |
+
14. Smolen JS, Landewe R, Breedveld FC, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2013 update. Ann. Rheum. Dis. 2014;73:492–509. doi: 10.1136/annrheumdis-2013-204573. [DOI](https://doi.org/10.1136/annrheumdis-2013-204573) | [PMC free article](/articles/PMC3933074/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24161836/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum.%20Dis.&title=EULAR%20recommendations%20for%20the%20management%20of%20rheumatoid%20arthritis%20with%20synthetic%20and%20biological%20disease-modifying%20antirheumatic%20drugs:%202013%20update&author=JS%20Smolen&author=R%20Landewe&author=FC%20Breedveld&volume=73&publication_year=2014&pages=492-509&pmid=24161836&doi=10.1136/annrheumdis-2013-204573&)
|
| 301 |
+
|
| 302 |
+
15. Smolen JS, Landewe R, Bergstra SA, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2022 update. Ann. Rheum. Dis. 2023;82:3–18. doi: 10.1136/ard-2022-223356. [DOI](https://doi.org/10.1136/ard-2022-223356) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36357155/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum.%20Dis.&title=EULAR%20recommendations%20for%20the%20management%20of%20rheumatoid%20arthritis%20with%20synthetic%20and%20biological%20disease-modifying%20antirheumatic%20drugs:%202022%20update&author=JS%20Smolen&author=R%20Landewe&author=SA%20Bergstra&volume=82&publication_year=2023&pages=3-18&pmid=36357155&doi=10.1136/ard-2022-223356&)
|
| 303 |
+
|
| 304 |
+
16. Smolen JS, Breedveld FC, Schiff MH, Kalden JR, Emery P, Eberl G, van Riel PL, Tugwell P. A simplified disease activity index for rheumatoid arthritis for use in clinical practice. Rheumatology (Oxford) 2003;42(2):244–257. doi: 10.1093/rheumatology/keg072. [DOI](https://doi.org/10.1093/rheumatology/keg072) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12595618/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Rheumatology%20(Oxford)&title=A%20simplified%20disease%20activity%20index%20for%20rheumatoid%20arthritis%20for%20use%20in%20clinical%20practice&author=JS%20Smolen&author=FC%20Breedveld&author=MH%20Schiff&author=JR%20Kalden&author=P%20Emery&volume=42&issue=2&publication_year=2003&pages=244-257&pmid=12595618&doi=10.1093/rheumatology/keg072&)
|
| 305 |
+
|
| 306 |
+
17. Inoue M, Kanda H, Tateishi S, Fujio K. Evaluation of response criteria in rheumatoid arthritis treated with biologic disease-modifying antirheumatic drugs. Arthritis Care Res. (Hoboken) 2020;72(7):942–949. doi: 10.1002/acr.23914. [DOI](https://doi.org/10.1002/acr.23914) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31058442/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Care%20Res.%20(Hoboken)&title=Evaluation%20of%20response%20criteria%20in%20rheumatoid%20arthritis%20treated%20with%20biologic%20disease-modifying%20antirheumatic%20drugs&author=M%20Inoue&author=H%20Kanda&author=S%20Tateishi&author=K%20Fujio&volume=72&issue=7&publication_year=2020&pages=942-949&pmid=31058442&doi=10.1002/acr.23914&)
|
| 307 |
+
|
| 308 |
+
18. Shimane K, Kochi Y, Suzuki A, Okada Y, Ishii T, Horita T, Saito K, Okamoto A, Nishimoto N, Myouzen K, et al. An association analysis of HLA-DRB1 with systemic lupus erythematosus and rheumatoid arthritis in a Japanese population: effects of *09:01 allele on disease phenotypes. Rheumatology (Oxford) 2013;52(7):1172–1182. doi: 10.1093/rheumatology/kes427. [DOI](https://doi.org/10.1093/rheumatology/kes427) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23407388/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Rheumatology%20(Oxford)&title=An%20association%20analysis%20of%20HLA-DRB1%20with%20systemic%20lupus%20erythematosus%20and%20rheumatoid%20arthritis%20in%20a%20Japanese%20population:%20effects%20of%20*09:01%20allele%20on%20disease%20phenotypes&author=K%20Shimane&author=Y%20Kochi&author=A%20Suzuki&author=Y%20Okada&author=T%20Ishii&volume=52&issue=7&publication_year=2013&pages=1172-1182&pmid=23407388&doi=10.1093/rheumatology/kes427&)
|
| 309 |
+
|
| 310 |
+
19. Bakker MF, Jacobs JW, Verstappen SM, Bijlsma JW. Tight control in the treatment of rheumatoid arthritis: efficacy and feasibility. Ann. Rheum. Dis. 2007;66(Suppl 3):iii56–60. doi: 10.1136/ard.2007.078360. [DOI](https://doi.org/10.1136/ard.2007.078360) | [PMC free article](/articles/PMC2095293/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17934098/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum.%20Dis.&title=Tight%20control%20in%20the%20treatment%20of%20rheumatoid%20arthritis:%20efficacy%20and%20feasibility&author=MF%20Bakker&author=JW%20Jacobs&author=SM%20Verstappen&author=JW%20Bijlsma&volume=66&issue=Suppl%203&publication_year=2007&pages=iii56-60&pmid=17934098&doi=10.1136/ard.2007.078360&)
|
| 311 |
+
|
| 312 |
+
20. Smolen JS, Aletaha D. The assessment of disease activity in rheumatoid arthritis. Clin. Exp. Rheumatol. 2010;28:S18–27. [PubMed](https://pubmed.ncbi.nlm.nih.gov/20576221/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Exp.%20Rheumatol.&title=The%20assessment%20of%20disease%20activity%20in%20rheumatoid%20arthritis&author=JS%20Smolen&author=D%20Aletaha&volume=28&publication_year=2010&pages=S18-27&pmid=20576221&)
|
| 313 |
+
|
| 314 |
+
21. Smolen JS, Aletaha D. Interleukin-6 receptor inhibition with tocilizumab and attainment of disease remission in rheumatoid arthritis: The role of acute-phase reactants. Arthritis Rheum. 2011;63(1):43–52. doi: 10.1002/art.27740. [DOI](https://doi.org/10.1002/art.27740) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21204103/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Rheum.&title=Interleukin-6%20receptor%20inhibition%20with%20tocilizumab%20and%20attainment%20of%20disease%20remission%20in%20rheumatoid%20arthritis:%20The%20role%20of%20acute-phase%20reactants&author=JS%20Smolen&author=D%20Aletaha&volume=63&issue=1&publication_year=2011&pages=43-52&pmid=21204103&doi=10.1002/art.27740&)
|
| 315 |
+
|
| 316 |
+
22. Smolen JS, Fleischimann R, Aletaha D, Li Y, Zhou Y, Sainsbury I, Galindo IL. Disease activity improvements with optimal discriminatory ability between treatment arms: Applicability in early and established rheumatoid arthritis clinical trials. Arthritis Res. Ther. 2019;21(1):231. doi: 10.1186/s13075-019-2005-9. [DOI](https://doi.org/10.1186/s13075-019-2005-9) | [PMC free article](/articles/PMC6842479/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31707982/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Res.%20Ther.&title=Disease%20activity%20improvements%20with%20optimal%20discriminatory%20ability%20between%20treatment%20arms:%20Applicability%20in%20early%20and%20established%20rheumatoid%20arthritis%20clinical%20trials&author=JS%20Smolen&author=R%20Fleischimann&author=D%20Aletaha&author=Y%20Li&author=Y%20Zhou&volume=21&issue=1&publication_year=2019&pages=231&pmid=31707982&doi=10.1186/s13075-019-2005-9&)
|
| 317 |
+
|
| 318 |
+
23. Okada Y, Suzuki A, Ikari K, Terao C, Kochi Y, Ohmura K, Higasa K, Akiyama M, Ashikawa K, Kanai M, et al. Contribution of a non-classical HLA Gene, HLA-DOA, to the risk of rheumatoid arthritis. Am. J. Hum. Genet. 2016;99(2):366–374. doi: 10.1016/j.ajhg.2016.06.019. [DOI](https://doi.org/10.1016/j.ajhg.2016.06.019) | [PMC free article](/articles/PMC4974094/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27486778/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Hum.%20Genet.&title=Contribution%20of%20a%20non-classical%20HLA%20Gene,%20HLA-DOA,%20to%20the%20risk%20of%20rheumatoid%20arthritis&author=Y%20Okada&author=A%20Suzuki&author=K%20Ikari&author=C%20Terao&author=Y%20Kochi&volume=99&issue=2&publication_year=2016&pages=366-374&pmid=27486778&doi=10.1016/j.ajhg.2016.06.019&)
|
| 319 |
+
|
| 320 |
+
24. Aletaha D, Martinez-Avila J, Kvien TK, Smolen JS. Definition of treatment response in rheumatoid arthritis based on the simplified and the clinical disease activity index. Ann. Rheum. Dis. 2012;71(7):1190–1196. doi: 10.1136/annrheumdis-2012-201491. [DOI](https://doi.org/10.1136/annrheumdis-2012-201491) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22454398/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum.%20Dis.&title=Definition%20of%20treatment%20response%20in%20rheumatoid%20arthritis%20based%20on%20the%20simplified%20and%20the%20clinical%20disease%20activity%20index&author=D%20Aletaha&author=J%20Martinez-Avila&author=TK%20Kvien&author=JS%20Smolen&volume=71&issue=7&publication_year=2012&pages=1190-1196&pmid=22454398&doi=10.1136/annrheumdis-2012-201491&)
|
| 321 |
+
|
| 322 |
+
25. Raychaudhuri S, Sandor C, Stahl EA, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 2012;44(3):291–296. doi: 10.1038/ng.1076. [DOI](https://doi.org/10.1038/ng.1076) | [PMC free article](/articles/PMC3288335/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22286218/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat.%20Genet.&title=Five%20amino%20acids%20in%20three%20HLA%20proteins%20explain%20most%20of%20the%20association%20between%20MHC%20and%20seropositive%20rheumatoid%20arthritis&author=S%20Raychaudhuri&author=C%20Sandor&author=EA%20Stahl&volume=44&issue=3&publication_year=2012&pages=291-296&pmid=22286218&doi=10.1038/ng.1076&)
|
| 323 |
+
|
| 324 |
+
26. Ishigaki K, Lagattuta KA, Luo Y, James EA, Buckner JH, Raychaudhuri S. HLA autoimmune risk alleles restrict the hypervariable region of T cell receptors. Nat. Genet. 2022;54(4):393–402. doi: 10.1038/s41588-022-01032-z. [DOI](https://doi.org/10.1038/s41588-022-01032-z) | [PMC free article](/articles/PMC9010379/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35332318/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat.%20Genet.&title=HLA%20autoimmune%20risk%20alleles%20restrict%20the%20hypervariable%20region%20of%20T%20cell%20receptors&author=K%20Ishigaki&author=KA%20Lagattuta&author=Y%20Luo&author=EA%20James&author=JH%20Buckner&volume=54&issue=4&publication_year=2022&pages=393-402&pmid=35332318&doi=10.1038/s41588-022-01032-z&)
|
| 325 |
+
|
| 326 |
+
27. Sokolove J, Schiff M, Fleischmann R, Weinblatt ME, Connolly SE, Johnsen A, Zhu J, Maldonado MA, Patel S, Robinson WH. Impact of baseline anti-cyclic citrullinated peptide-2 antibody concentration on efficacy outcomes following treatment with subcutaneous abatacept or adalimumab: 2-year results from the AMPLE trial. Ann. Rheum. Dis. 2015;75(4):709–714. doi: 10.1136/annrheumdis-2015-207942. [DOI](https://doi.org/10.1136/annrheumdis-2015-207942) | [PMC free article](/articles/PMC4819608/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26359449/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum.%20Dis.&title=Impact%20of%20baseline%20anti-cyclic%20citrullinated%20peptide-2%20antibody%20concentration%20on%20efficacy%20outcomes%20following%20treatment%20with%20subcutaneous%20abatacept%20or%20adalimumab:%202-year%20results%20from%20the%20AMPLE%20trial&author=J%20Sokolove&author=M%20Schiff&author=R%20Fleischmann&author=ME%20Weinblatt&author=SE%20Connolly&volume=75&issue=4&publication_year=2015&pages=709-714&pmid=26359449&doi=10.1136/annrheumdis-2015-207942&)
|
| 327 |
+
|
| 328 |
+
28. van Gaalen FA, van Aken J, Huizinga TW, Schreuder GM, Breedveld FC, Zanelli E, van Venrooij WJ, Verweij CL, Toes RE, de Vries RR. Association between HLA class II genes and autoantibodies to cyclic citrullinated peptides (CCPs) influences the severity of rheumatoid arthritis. Arthritis Rheum. 2004;50(7):2113–2121. doi: 10.1002/art.20316. [DOI](https://doi.org/10.1002/art.20316) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15248208/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Rheum.&title=Association%20between%20HLA%20class%20II%20genes%20and%20autoantibodies%20to%20cyclic%20citrullinated%20peptides%20(CCPs)%20influences%20the%20severity%20of%20rheumatoid%20arthritis&author=FA%20van%20Gaalen&author=J%20van%20Aken&author=TW%20Huizinga&author=GM%20Schreuder&author=FC%20Breedveld&volume=50&issue=7&publication_year=2004&pages=2113-2121&pmid=15248208&doi=10.1002/art.20316&)
|
| 329 |
+
|
| 330 |
+
29. Hensvold AH, Magnusson PK, Joshua V, Hansson M, Israelsson L, Ferreira R, Jakobsson PJ, Holmdahl R, Hammarström L, Malmström V, et al. Environmental and genetic factors in the development of anticitrullinated protein antibodies (ACPAs) and ACPA-positive rheumatoid arthritis: An epidemiological investigation in twins. Ann. Rheum. Dis. 2015;74(2):375–380. doi: 10.1136/annrheumdis-2013-203947. [DOI](https://doi.org/10.1136/annrheumdis-2013-203947) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24276366/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum.%20Dis.&title=Environmental%20and%20genetic%20factors%20in%20the%20development%20of%20anticitrullinated%20protein%20antibodies%20(ACPAs)%20and%20ACPA-positive%20rheumatoid%20arthritis:%20An%20epidemiological%20investigation%20in%20twins&author=AH%20Hensvold&author=PK%20Magnusson&author=V%20Joshua&author=M%20Hansson&author=L%20Israelsson&volume=74&issue=2&publication_year=2015&pages=375-380&pmid=24276366&doi=10.1136/annrheumdis-2013-203947&)
|
| 331 |
+
|
| 332 |
+
30. Wouters F, Maurits MP, van Boheemen L, Verstappen M, Mankia K, Matthijssen XME, Dorjée AL, Emery P, Knevel R, van Schaardenburg D, et al. Determining in which pre-arthritis stage HLA-shared epitope alleles and smoking exert their effect on the development of rheumatoid arthritis. Ann. Rheum. Dis. 2022;81(1):48–55. doi: 10.1136/annrheumdis-2021-220546. [DOI](https://doi.org/10.1136/annrheumdis-2021-220546) | [PMC free article](/articles/PMC7612192/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34285049/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum.%20Dis.&title=Determining%20in%20which%20pre-arthritis%20stage%20HLA-shared%20epitope%20alleles%20and%20smoking%20exert%20their%20effect%20on%20the%20development%20of%20rheumatoid%20arthritis&author=F%20Wouters&author=MP%20Maurits&author=L%20van%20Boheemen&author=M%20Verstappen&author=K%20Mankia&volume=81&issue=1&publication_year=2022&pages=48-55&pmid=34285049&doi=10.1136/annrheumdis-2021-220546&)
|
| 333 |
+
|
| 334 |
+
31. Nagafuchi Y, Ota M, Hatano H, Inoue M, Kobayashi S, Okubo M, Sugimori Y, Nakano M, Yamada S, Yoshida R, et al. Control of naive and effector CD4 T cell receptor repertoires by rheumatoid-arthritis-risk HLA alleles. J Autoimmun. 2022;133:102907. doi: 10.1016/j.jaut.2022.102907. [DOI](https://doi.org/10.1016/j.jaut.2022.102907) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36126366/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Autoimmun&title=Control%20of%20naive%20and%20effector%20CD4%20T%20cell%20receptor%20repertoires%20by%20rheumatoid-arthritis-risk%20HLA%20alleles&author=Y%20Nagafuchi&author=M%20Ota&author=H%20Hatano&author=M%20Inoue&author=S%20Kobayashi&volume=133&publication_year=2022&pages=102907&pmid=36126366&doi=10.1016/j.jaut.2022.102907&)
|
| 335 |
+
|
| 336 |
+
32. Nash P, Nayiager S, Genovese MC, et al. Immunogenicity, safety, and efficacy of abatacept administered subcutaneously with or without background methotrexate in patients with rheumatoid arthritis: Results from a phase III, international, multicenter, parallel-arm open-label study. Arthritis Care Res. 2013;65(5):718–728. doi: 10.1002/acr.21876. [DOI](https://doi.org/10.1002/acr.21876) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23097311/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Care%20Res.&title=Immunogenicity,%20safety,%20and%20efficacy%20of%20abatacept%20administered%20subcutaneously%20with%20or%20without%20background%20methotrexate%20in%20patients%20with%20rheumatoid%20arthritis:%20Results%20from%20a%20phase%20III,%20international,%20multicenter,%20parallel-arm%20open-label%20study&author=P%20Nash&author=S%20Nayiager&author=MC%20Genovese&volume=65&issue=5&publication_year=2013&pages=718-728&pmid=23097311&doi=10.1002/acr.21876&)
|
| 337 |
+
|
| 338 |
+
33. Takahashi N, Kojima T, Kida D, et al. Concomitant methotrexate has little effect on clinical outcomes of abatacept in rheumatoid arthritis: a propensity score matching analysis. Clin. Rheum. 2019;38(9):2451–2459. doi: 10.1007/s10067-019-04581-7. [DOI](https://doi.org/10.1007/s10067-019-04581-7) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31102087/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Rheum.&title=Concomitant%20methotrexate%20has%20little%20effect%20on%20clinical%20outcomes%20of%20abatacept%20in%20rheumatoid%20arthritis:%20a%20propensity%20score%20matching%20analysis&author=N%20Takahashi&author=T%20Kojima&author=D%20Kida&volume=38&issue=9&publication_year=2019&pages=2451-2459&pmid=31102087&doi=10.1007/s10067-019-04581-7&)
|
test/texts/PMC10566653.md
ADDED
|
@@ -0,0 +1,156 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Correlation of pain perception and fentanyl consumption after major abdominal surgery with CGRP 4218T/C polymorphism: A prospective interventional study
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** HT Prashant, Kirti N Saxena, Seema Kapoor, Bharti Wadhwa, Sukhyanti Kerai, Prachi Gaba
|
| 5 |
+
**Journal:** Indian Journal of Anaesthesia
|
| 6 |
+
**Date:** 2023 Sep 6
|
| 7 |
+
**DOI:** [10.4103/ija.ija_1033_22](https://doi.org/10.4103/ija.ija_1033_22)
|
| 8 |
+
**PMID:** 37829781
|
| 9 |
+
**PMCID:** PMC10566653
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10566653/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC10566653/pdf/IJA-67-796.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC10566653/pdf/IJA-67-796.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Background and Aims::**
|
| 16 |
+
Genetic polymorphisms contribute to patients’ variability in pain perception and response to opioid treatment. The present study evaluated the association of calcitonin gene-related peptide (CGRP) 4218T/C polymorphisms with fentanyl consumption over 24 h postoperatively in patients after major abdominal surgery.
|
| 17 |
+
|
| 18 |
+
**Methods::**
|
| 19 |
+
Eighty-five patients undergoing major abdominal surgery under general anaesthesia were recruited. For postoperative analgesia, epidural fentanyl and intravenous paracetamol were provided. The CGRP 4218T/C genotype was analysed, and the association between the genotype of the patient and the total consumption of fentanyl in the first 24 h after surgery was assessed. The association between different genotypes, the severity of postoperative pain and the side effects of opioids were also studied.
|
| 20 |
+
|
| 21 |
+
**Results::**
|
| 22 |
+
Our study population distribution included 52.9% of the T/T genotype (wild homozygote), 35.3% of the T/C genotype (heterozygote) and 11.8% of the C/C genotype (mutant homozygote). Mean (standard deviation) total fentanyl consumption in the first 24 h was found to be highest in the C/C group (212.0 [7.5] μg), followed by the T/T group (182.8 [9.9] μg) and was the least in the T/C group (159.6 [7.5] μg). The C/C group reported higher pain scores in all the study periods. There was no significant difference in the side effects of opioids, such as nausea, vomiting, sedation among different genotypes of CGRP 4218T/C.
|
| 23 |
+
|
| 24 |
+
**Conclusion::**
|
| 25 |
+
The polymorphism of CGRP 4218T/C affects postoperative pain perception and analgesic consumption. Patients with the C/C genotype had higher postoperative fentanyl consumption and pain scores.
|
| 26 |
+
|
| 27 |
+
Keywords: Analgesia, calcitonin gene-related peptide, CGRP 4218T/C, epidural analgesia, pharmacogenetics, postoperative analgesia, single-nucleotide polymorphisms
|
| 28 |
+
|
| 29 |
+
### Background and Aims:
|
| 30 |
+
|
| 31 |
+
Genetic polymorphisms contribute to patients’ variability in pain perception and response to opioid treatment. The present study evaluated the association of calcitonin gene-related peptide (CGRP) 4218T/C polymorphisms with fentanyl consumption over 24 h postoperatively in patients after major abdominal surgery.
|
| 32 |
+
|
| 33 |
+
### Methods:
|
| 34 |
+
|
| 35 |
+
Eighty-five patients undergoing major abdominal surgery under general anaesthesia were recruited. For postoperative analgesia, epidural fentanyl and intravenous paracetamol were provided. The CGRP 4218T/C genotype was analysed, and the association between the genotype of the patient and the total consumption of fentanyl in the first 24 h after surgery was assessed. The association between different genotypes, the severity of postoperative pain and the side effects of opioids were also studied.
|
| 36 |
+
|
| 37 |
+
### Results:
|
| 38 |
+
|
| 39 |
+
Our study population distribution included 52.9% of the T/T genotype (wild homozygote), 35.3% of the T/C genotype (heterozygote) and 11.8% of the C/C genotype (mutant homozygote). Mean (standard deviation) total fentanyl consumption in the first 24 h was found to be highest in the C/C group (212.0 [7.5] μg), followed by the T/T group (182.8 [9.9] μg) and was the least in the T/C group (159.6 [7.5] μg). The C/C group reported higher pain scores in all the study periods. There was no significant difference in the side effects of opioids, such as nausea, vomiting, sedation among different genotypes of CGRP 4218T/C.
|
| 40 |
+
|
| 41 |
+
### Conclusion:
|
| 42 |
+
|
| 43 |
+
The polymorphism of CGRP 4218T/C affects postoperative pain perception and analgesic consumption. Patients with the C/C genotype had higher postoperative fentanyl consumption and pain scores.
|
| 44 |
+
|
| 45 |
+
**Keywords:**Keywords: Analgesia, calcitonin gene-related peptide, CGRP 4218T/C, epidural analgesia, pharmacogenetics, postoperative analgesia, single-nucleotide polymorphisms
|
| 46 |
+
|
| 47 |
+
## INTRODUCTION
|
| 48 |
+
|
| 49 |
+
One of the important challenges in delivering satisfactory postoperative analgesia is interindividual variability in postoperative pain intensity.[[1](#R1)1] This variability has been attributed to complex genetic, environmental and social factors.[[2](#R2)2] Growing evidence shows that genetic factors are critical in pain sensitivity and susceptibility to developing chronic pain after surgery.[[3](#R3)3] The genetic variations in catechol-*O*O-methyltransferase (*COMT*COMT) and mu-opioid receptor gene (*OPRM1*OPRM1), brain-derived neurotrophic factor (*BDNF*BDNF) genes have previously been reported for interindividual differences in postoperative pain perception. Calcitonin gene-related peptide (CGRP) is a neuropeptide involved in the nociceptive pathway, and it is widely distributed in the central and peripheral nervous systems. It plays a crucial role in pain perception and modulation.[[4](#R4)4] The genetic polymorphism in the CGRP gene 4218T/C significantly influences postoperative fentanyl consumption.[[5](#R5)5,[6](#R6)6] Patients with the C/C genotype have been reported to have reduced sensitivity to fentanyl and increased pain perception. However, the influence of CGRP polymorphism on postoperative pain has not been evaluated in other ethnic populations.
|
| 50 |
+
|
| 51 |
+
The present study was designed to assess the role of different genotypes of CGRP on postoperative pain after major abdominal surgery. The primary objective was to evaluate the association of CGRP 4218T/C polymorphisms with fentanyl consumption over 24 h postoperatively following surgery. The secondary objectives were to assess the association of CGRP 4218T/C polymorphisms with the severity of postoperative pain, the incidence of nausea and vomiting, pruritus, sedation and respiratory depression over 24 h postoperatively. We hypothesise that there would be no difference in postoperative pain perception and analgesic consumption in patients with different CGRP genotypes.
|
| 52 |
+
|
| 53 |
+
## METHODS
|
| 54 |
+
|
| 55 |
+
This prospective interventional study was conducted on 85 patients from January 2019 to January 2020. The study was started after obtaining approval from the Institutional Ethical Committee (vide approval number 17/IEC/MAMC/2018/11, dated 26 October 2018) and was registered under the Clinical Trial Registry-India (vide registration number CTRI/2019/01/016986, [https://ctri.nic.in/](https://ctri.nic.in/)https://ctri.nic.in/). Adult patients aged between 20 and 65 years belonging to the American Society of Anesthesiologists (ASA) physical status I/II, who were scheduled to undergo elective major abdominal surgeries under general anaesthesia, who could comprehend and describe verbal or visual pain scale and who were without any history of drug dependence or recreational drug use were recruited in our study. Patients with a history of allergy to opioids or any other drugs to be given intraoperatively or with any contraindication to epidural block and pregnant patients were excluded from our study. Written informed consent was obtained from patients to participate in the study and use patient data for research and educational purposes. The study procedures followed the guidelines in the World Medical Association (WMA) Declaration of Helsinki-ethical Principles (2013) for medical research involving human subjects. Pre-anaesthetic checkup as per the institutional protocol was carried out in all patients. Standard anaesthetic monitors were applied in the operating room, including continuous electrocardiogram (ECG), noninvasive blood pressure and pulse oximeter, and baseline readings were noted. An intravenous (IV) access was secured, and a 3 ml venous blood sample was collected in an ethylenediaminetetraacetic acid (EDTA) vial; after proper labelling, the CGRP genetic analysis was performed using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method using HiPurA^™^™ blood genomic DNA miniprep purification kit (Himedia, Mumbai, India). Under all aseptic precautions, and after local infiltration with 2 ml of 2% lignocaine, an 18G Tuohy epidural needle was inserted in the desired space and an epidural catheter was threaded in up to 3–5 cm depth inside the epidural space. After proper fixation of the epidural catheter, the patients were premedicated with IV fentanyl 1.5 μg/kg lean body weight (LBW). Anaesthesia induction was done with IV thiopentone sodium 3–5 mg/kg titrated dose. After establishing the ability to ventilate via bag and mask, IV vecuronium 0.1 mg/kg was administered to facilitate tracheal intubation. Anaesthesia was maintained using a gaseous mixture of oxygen and nitrous oxide (50:50) in isoflurane. After 3 min of vecuronium administration, tracheal intubation was performed under direct laryngoscopy with an appropriately sized endotracheal tube. After successful tracheal intubation, controlled ventilation was established to target end-tidal carbon dioxide (EtCO_2_2) between 32 and 36 mmHg. Fentanyl IV infusion was started at 1 μg/kg/h 15 min after induction of anaesthesia. IV paracetamol 1 g was administered to all patients. Intraoperative titration of anaesthetic was left to the discretion of the attending anaesthesiologist. About 20–30 min before the anticipated conclusion of the surgery, fentanyl infusion was stopped, and IV ondansetron 4 mg was administered as prophylaxis for postoperative nausea and vomiting (PONV). At the end of the surgery, the neuromuscular blockade was antagonised with an appropriate dose IV neostigmine and glycopyrrolate. The patients were monitored at 0, 6, 12 and 24 h in the postoperative period. The severity of postoperative pain and side effects of opioids, such as nausea, vomiting, pruritus and respiratory depression, were assessed. Patients received IV paracetamol 1 g eight hourly for postoperative analgesia and epidural fentanyl infusion. For epidural analgesia, a background infusion of fentanyl at the rate of 5 μg/h was started as soon as the patients reached the postoperative care unit. The severity of postoperative pain was assessed using the visual analogue scale (VAS) with a rating of 0–10, where a score of 0 indicated no perceptible pain and a score of 10 indicated severe pain. If the patient complained of VAS more than 3, a rescue dose of IV fentanyl 5 μg was administered. The patient was re-evaluated 5–10 min after the rescue dose; if the patient still complained of VAS more than 3, the rescue dose was repeated. The rescue doses were repeated till VAS was less than 3. The total amount of fentanyl consumed over 24 h postoperatively was recorded. The side effects of opioids, such as PONV, pruritus, sedation and respiratory depression, were recorded at the study intervals. Vomiting was assessed by using a PONV impact scale. A PONV impact scale score of ≥5 defines clinically important PONV. Sedation was assessed using the Ramsay sedation scale, where a score of 1 meant anxious and agitated and 6 meant the patient exhibited no response. Respiratory depression was defined as a respiratory rate of fewer than 8 breaths per minute (bpm) or shallow breathing or oxygen saturation (SpO_2_2) value of less than 94%. The number of patients who had pruritus in the postoperative period was also recorded, and the severity was classified as mild, moderate and severe. The venous blood sample taken from the patients was analysed for CGRP 4218T/C polymorphisms. Based on the CGRP 4218T/C genotypes, we predict the influence of pain perception and analgesic consumption for developing effective postoperative pain management strategies. A study participant’s specific type of genotype was known only after performing the above-mentioned genotyping method. Based on this genotyping procedure, patients were categorised into respective genotypes, that is, C/C, C/T or T/T genotype. The investigator administering the anaesthesia and recording the parameter was blinded to the genotypes of the patients revealed at the time of data analysis.
|
| 56 |
+
|
| 57 |
+
For sample size calculation, a moderate correlation between CGRP 4218T/C polymorphisms with the total amount of fentanyl consumption over 24 h postoperatively was considered meaningful. To detect a moderate correlation (*r*r = 0.30), a sample of 85 subjects provides 80% power to discover that the correlation is significantly different from there being no correlation (i.e. the correlation is zero) at the 0.05 level. Statistical analysis was performed by the Statistical Package for the Social Sciences (SPSS) program for Windows, version 25.0 (IBM Corp., Armonk, NY). Continuous variables are presented as mean ± standard deviation (SD), and median (interquartile range [IQR]), and categorical variables are presented as absolute numbers and percentages. The genotype distribution was checked by Hardy–Weinberg Equilibrium (HWE) with the χ^2^2 test. Data were checked for normality before statistical analysis using the Shapiro–Wilk test. The Kruskal–Wallis test was used for those variables that were not normally distributed, and further comparisons were made using the Mann–Whitney U test. Categorical variables were analysed using the χ^2^2 test. A *P*P < 0.05 was taken for all statistical tests to indicate a significant difference. For measurement of the effect size of the difference between the genotypes, Cohen’s d was calculated.
|
| 58 |
+
|
| 59 |
+
## RESULTS
|
| 60 |
+
|
| 61 |
+
After the genetic analysis for CGRP 4218T/C polymorphisms, three different types of genotypes of CGRP4218T/C polymorphism was identified in the Indian population, C/C genotype, T/C genotype and T/T genotype, and the frequency of distribution of genotypes was 11.8% (10), 35.3% (30) and 52.9% (45), respectively. The patients’ demographic profiles were comparable among the three genotypes [[Table 1](#ija_1033_22-t001)Table 1], and the mean amount of fentanyl consumed in the first 24 h of the postoperative period, its effect size and precision were comparable [[Table 2](#ija_1033_22-t002)Table 2]. The mean total fentanyl consumption in the first 24 h was found to be highest in the C/C group, followed by the T/T group and was the least in the T/C group. In the paired comparison of C/C versus T/C and C/C versus T/T, the C/C group had a significantly higher requirement of mean total postoperative fentanyl in the first 24 h than T/T (*P*P < 0.001) and T/C (*P*P < 0.001). In a paired comparison of T/C versus T/T, the T/T group had a significantly higher requirement of mean total postoperative fentanyl in 24 h than T/C (*P*P < 0.001), with comparable effect size and precision [[Table 2](#ija_1033_22-t002)Table 2]. The mean VAS scores among genotypes and paired comparison showed that in the C/C genotype, the mean VAS score was highest in all periods. In the T/C genotype, the mean VAS score was least in all periods, and in the T/T genotype, the mean VAS scores were between the VAS of C/C and T/C in all periods. In the paired comparison of C/C versus T/C and C/C versus T/T, the C/C group had highly significant VAS or pain perception than T/C (*P*P < 0.001) and T/T (*P*P < 0.001) in all periods [[Table 3](#ija_1033_22-t003)Table 3]. The respiratory rate among genotypes and paired comparison showed that, in the paired comparison of respiratory rate in C/C versus T/C, statistically significant results were seen at the sixth hour (14.5 [1.3] vs. 12.5 [2.1] bpm) and the 12th-hour (13.3 [1.9] vs. 11.4 [2.2] bpm) assessment, with C/C having more respiratory rate compared to the T/T group. However, this data has no clinical relevance as the respiratory rate was more than 8 in all periods among genotypes. No significant results were observed with sedation scores, PONV scores and O_2_2 saturation. A negligible number of patients had complaints of pruritus (mild) at 0 h and 6-h assessment in all three genotypes. At 12-h (C/C [30%] and T/T [15.6%]) and 24-h (C/C [50%] and T/T [33.3%]) assessment, a significant number complained of pruritus. The frequency of pruritus in the T/C group was statistically insignificant at all periods.
|
| 62 |
+
|
| 63 |
+
### Table 1.
|
| 64 |
+
|
| 65 |
+
Demographic profile of the patients
|
| 66 |
+
|
| 67 |
+
| Parameters | Genotypes of CGRP 4218T/C |
|
| 68 |
+
| ---------- | ------------------------- |
|
| 69 |
+
| | |
|
| 70 |
+
| C/C (n=10) | T/C (n=30) | T/T (n=45) |
|
| 71 |
+
| Age (years) | 34.70 (9.15) | 39.00 (11.86) | 41.22 (9.05) |
|
| 72 |
+
| Gender (Female: Male) | 7:3 | 20:10 | 24:21 |
|
| 73 |
+
| Height (cm) | 165.00 (7.06) | 162.03 (7.29) | 163.76 (6.48) |
|
| 74 |
+
| Weight (kg) | 61.90 (8.35) | 58.13 (7.79) | 60.44 (6.57) |
|
| 75 |
+
| LBW (kg) | 46.20 (6.27) | 43.15 (6.29) | 46.41 (5.84) |
|
| 76 |
+
| ASA PS (I: II) | 9:1 | 22:8 | 31:14 |
|
| 77 |
+
| Duration of surgery (min) | 173.50 (26.57) | 157.67 (31.26) | 167.33 (32.34) |
|
| 78 |
+
### Table 2.
|
| 79 |
+
|
| 80 |
+
Comparison of the amount of IV fentanyl consumed postoperatively among genotypes and paired comparison of genotypes
|
| 81 |
+
|
| 82 |
+
| Amount of IV fentanyl consumed (µg) | Type of genotype | P | Effect size (95% CI) C/C versus T/C | Effect size (95% CI) C/C versus T/T | Effect size (95% CI) T/C versus T/T |
|
| 83 |
+
| ----------------------------------- | ---------------- | - | ----------------------------------- | ----------------------------------- | ----------------------------------- |
|
| 84 |
+
| | | | | | |
|
| 85 |
+
| C/C (n=10) | T/C (n=30) | T/T (n=45) | | | |
|
| 86 |
+
| 0–6 h | 32.0 (8.8) | 13.0 (3.8) | 27.9 (4.5) | <0.001 | 3.46 (2.78–4.25) | 0.75 (0.04–1.44) | 3.52 (2.78–4.25) |
|
| 87 |
+
| 6–12 h | 28.0 (6.3) | 11.8 (4.8) | 20.8 (3.9) | <0.001 | 3.09 (2.09–4.08) | 1.61 (0.85–2.36) | 2.06 (1.48–2.62) |
|
| 88 |
+
| 12–24 h | 27.5 (7.2) | 15.0 (4.9) | 14.1 (4.0) | <0.001 | 2.26 (1.37–3.12) | 2.84 (1.96–3.70) | 0.20 (0.66–3.81) |
|
| 89 |
+
| The total amount in the first 24 h | 212.0 (7.5) | 159.6 (7.5) | 182.9 (9.9) | <0.001 | 6.94 (5.22–8.65) | 3.04 (2.14–3.93) | 2.56 (1.94–3.18) |
|
| 90 |
+
### Table 3.
|
| 91 |
+
|
| 92 |
+
Mean pain scores among genotypes and paired comparison
|
| 93 |
+
|
| 94 |
+
| VAS score | Type of genotype | P | Effect size (95% CI) C/C versus T/C | Effect size (95% CI) C/C versus T/T | Effect size (95% CI) T/C versus T/T |
|
| 95 |
+
| --------- | ---------------- | - | ----------------------------------- | ----------------------------------- | ----------------------------------- |
|
| 96 |
+
| | | | | | |
|
| 97 |
+
| C/C | T/C | T/T | | | |
|
| 98 |
+
| 0 h | 5.9 (0.5) | 3.8 (0.8) | 5.3 (0.7) | <0.001 | 2.65 (1.71–3.57) | 1.03 (0.31–1.74) | 1.87 (1.32–2.42) |
|
| 99 |
+
| 6 h | 5.3 (0.5) | 3.2 (0.9) | 4.4 (0.6) | <0.001 | 2.49 (1.57–3.39) | 1.52 (0.77–2.25) | 1.50 (0.97–2.02) |
|
| 100 |
+
| 12 h | 4.8 (0.9) | 2.7 (1.1) | 3.9 (0.8) | <0.001 | 2.07 (1.21–2.91) | 1.18 (0.46–1.90) | 1.79 (1.19–2.37) |
|
| 101 |
+
| 24 h | 4.5 (0.8) | 2.6 (0.8) | 3.4 (1.0) | <0.001 | 2.19 (1.31–3.05) | 1.19 (0.46–1.91) | 0.74 (0.26–1.21) |
|
| 102 |
+
## DISCUSSION
|
| 103 |
+
|
| 104 |
+
The present study demonstrated that in the Indian population, CGRP4218T/C polymorphism was associated with variable fentanyl consumption in the postoperative period following major abdominal surgeries. The mean fentanyl consumption in the C/C group was significantly higher than in the T/T and T/C genotypes 24 h after surgery.
|
| 105 |
+
|
| 106 |
+
Yi *et al.*et al.,[[6](#R6)6] in a study involving ethnic Han Chinese patients undergoing open abdominal and lumbar surgeries, reported that the mean fentanyl consumption at 24 h after surgeries in T/T, T/C and C/C groups was 14.8 ± 6.1, 19.6 ± 6.4 and 23.4 ± 6.8 μg/kg, respectively. This is in contrast to our study and may be attributed to the difference in the postoperative analgesic protocol and the nature of the surgery included in their research. Yi *et al*et al. employed fentanyl with flurbiprofen axetil or fentanyl with propacetamol for patient-controlled analgesia (PCA), whereas in the present study, we used an epidural infusion of fentanyl and paracetamol IV. Another study by Xie *et al*et al.[[5](#R5)5] also supported the findings of our research. They reported that in parturients who underwent caesarean section, the C/C genotype was significantly linked to high consumption of patient-controlled epidural fentanyl.
|
| 107 |
+
|
| 108 |
+
CGRP is expressed ubiquitously in the body and has a well-documented role as a pronociceptive and proinflammatory neurotransmitter. It is released from the dorsal horn of the spinal cord in response to noxious stimuli. It amplifies the release of several other transmitters, such as substance P. Opioid medications act on the presynaptic terminals of afferent nociceptors and inhibit the release of these neurotransmitters.[[7](#R7)7,[8](#R8)8] It seems plausible that polymorphism in CGRP 4218 T/C leads to variable expression of these nociceptor transmitters in the pain signalling pathway, influencing the severity of pain.[[9](#R9)9] The present study noted that C/C genotype patients reported higher VAS scores during all study periods.
|
| 109 |
+
|
| 110 |
+
In agreement with previous studies, we found no association between the CGRP 4218T/C polymorphism and the side effects of opioids like pruritus, PONV, respiratory depression and sedation.[[5](#R5)5] The strength of the present study is that it is the first study done in the Indian population, as the previous studies were conducted in the Chinese Han ethnic group. There is limited data from our country regarding the effects of genetic variation on postoperative pain and the requirement for analgesics. A recent study by Kumar *et al*et al.[[10](#R10)10] on 257 South Indian women undergoing major breast surgery found that single nucleotide polymorphism (SNP) opioid receptor mu-1 (*OPRM1*OPRM1) (rs1799971) was associated with higher postoperative fentanyl requirement.
|
| 111 |
+
|
| 112 |
+
Our study adds another important evidence on the association of genetic polymorphisms with postoperative fentanyl consumption in patients undergoing surgery. In the future, incorporating genetics and epigenetic analysis into clinical practice will allow anaesthesiologists to tailor the analgesic dosage to maximise drug efficacy and minimise unnecessary adverse reactions. Our study has certain limitations; it was a single-centre study having a small sample size. In our research, intraoperatively, we used an infusion of IV fentanyl to provide analgesia. The epidural catheter was not activated, which would have confounded the analgesic assessment. Infusion of fentanyl without monitoring the depth of anaesthesia may raise concern for overdosing. However, it has been reported that bispectral index monitoring, commonly used to assess the depth of anaesthesia, fails to show the hypnotic-enhancing effect of fentanyl.[[11](#R11)11] We included patients of either gender with various major abdominal surgeries, which could have affected the patient-reported pain perception and not standardised the surgical procedure. The analgesics protocol used in the present study consisted of only fentanyl given through epidural and IV routes. As the combination of local anaesthetics and opioids is commonly used for postoperative analgesia, the study results need to be considered carefully. Future studies with larger sample sizes are required to support the current study’s findings.
|
| 113 |
+
|
| 114 |
+
## CONCLUSION
|
| 115 |
+
|
| 116 |
+
Our study concludes that the CGRP 4218T/C polymorphism is associated with variability in postoperative pain perception and fentanyl requirement. Patients with the C/C genotype experience more significant pain and have an increased demand for analgesics compared to those with T/C and T/T genotypes. However, there is no association between the polymorphism of CGRP4218T/C and the adverse effects of opioid analgesic medication.
|
| 117 |
+
|
| 118 |
+
### Study data availability
|
| 119 |
+
|
| 120 |
+
De-identified data may be requested with reasonable justification from the authors (email to the corresponding author) and shall be shared after approval as per the authors’ institution policy.
|
| 121 |
+
|
| 122 |
+
### Financial support and sponsorship
|
| 123 |
+
|
| 124 |
+
Nil.
|
| 125 |
+
|
| 126 |
+
### Conflicts of interest
|
| 127 |
+
|
| 128 |
+
There are no conflicts of interest.
|
| 129 |
+
|
| 130 |
+
## Acknowledgements
|
| 131 |
+
|
| 132 |
+
Mr. Sunil Kumar Polipalli, Mrs. Jyotsna Srivastava, Dr. Devika Mishra, Dr. Manisha Manohar and Dr. Mousumi Saha are acknowledged.
|
| 133 |
+
|
| 134 |
+
## References
|
| 135 |
+
|
| 136 |
+
1. Rawal N. Current issues in postoperative pain management. Eur J Anaesthesiol. 2016;33:160–71. doi: 10.1097/EJA.0000000000000366. [DOI](https://doi.org/10.1097/EJA.0000000000000366) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26509324/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Anaesthesiol&title=Current%20issues%20in%20postoperative%20pain%20management&author=N%20Rawal&volume=33&publication_year=2016&pages=160-71&pmid=26509324&doi=10.1097/EJA.0000000000000366&)
|
| 137 |
+
|
| 138 |
+
2. Fillingim RB. Individual differences in pain: Understanding the mosaic that makes pain perception. Pain. 2017;158:11–8. doi: 10.1097/j.pain.0000000000000775. [DOI](https://doi.org/10.1097/j.pain.0000000000000775) | [PMC free article](/articles/PMC5350021/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27902569/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pain&title=Individual%20differences%20in%20pain:%20Understanding%20the%20mosaic%20that%20makes%20pain%20perception&author=RB%20Fillingim&volume=158&publication_year=2017&pages=11-8&pmid=27902569&doi=10.1097/j.pain.0000000000000775&)
|
| 139 |
+
|
| 140 |
+
3. Palada V, Kaunisto MA, Kalso E. Genetics and genomics in postoperative pain and analgesia. Curr Opin Anaesthesiol. 2018;31:569–74. doi: 10.1097/ACO.0000000000000633. [DOI](https://doi.org/10.1097/ACO.0000000000000633) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29994939/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Curr%20Opin%20Anaesthesiol&title=Genetics%20and%20genomics%20in%20postoperative%20pain%20and%20analgesia&author=V%20Palada&author=MA%20Kaunisto&author=E%20Kalso&volume=31&publication_year=2018&pages=569-74&pmid=29994939&doi=10.1097/ACO.0000000000000633&)
|
| 141 |
+
|
| 142 |
+
4. Schou WS, Ashina S, Amin FM, Goadsby PJ, Ashina M. Calcitonin gene-related peptide and pain: A systematic review. J Headache Pain. 2017;18:34. doi: 10.1186/s10194-017-0741-2. [DOI](https://doi.org/10.1186/s10194-017-0741-2) | [PMC free article](/articles/PMC5355411/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28303458/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Headache%20Pain&title=Calcitonin%20gene-related%20peptide%20and%20pain:%20A%20systematic%20review&author=WS%20Schou&author=S%20Ashina&author=FM%20Amin&author=PJ%20Goadsby&author=M%20Ashina&volume=18&publication_year=2017&pages=34&pmid=28303458&doi=10.1186/s10194-017-0741-2&)
|
| 143 |
+
|
| 144 |
+
5. Xie W, Zhuang W, Chen L, Xie W, Jiang C, Liu N. 4218T/C polymorphism associations with post-cesarean patient-controlled epidural fentanyl consumption and pain perception. Acta Anaesthesiol Scand. 2018;62:376–83. doi: 10.1111/aas.13040. [DOI](https://doi.org/10.1111/aas.13040) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29148033/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Acta%20Anaesthesiol%20Scand&title=4218T/C%20polymorphism%20associations%20with%20post-cesarean%20patient-controlled%20epidural%20fentanyl%20consumption%20and%20pain%20perception&author=W%20Xie&author=W%20Zhuang&author=L%20Chen&author=W%20Xie&author=C%20Jiang&volume=62&publication_year=2018&pages=376-83&pmid=29148033&doi=10.1111/aas.13040&)
|
| 145 |
+
|
| 146 |
+
6. Yi Y, Zhao M, Xu F, Liu C, Yin Y, Yu J. CGRP4218T/C polymorphism correlated with the postoperative analgesic effect of fentanyl. Int J Clin Exp Pathol. 2015;8:5761–7. [PMC free article](/articles/PMC4503165/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26191294/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int%20J%20Clin%20Exp%20Pathol&title=CGRP4218T/C%20polymorphism%20correlated%20with%20the%20postoperative%20analgesic%20effect%20of%20fentanyl&author=Y%20Yi&author=M%20Zhao&author=F%20Xu&author=C%20Liu&author=Y%20Yin&volume=8&publication_year=2015&pages=5761-7&pmid=26191294&)
|
| 147 |
+
|
| 148 |
+
7. Frauenknecht J, Kirkham KR, Jacot-Guillarmod A, Albrecht E. Analgesic impact of intra-operative opioids vs opioid-free anaesthesia: A systemic review and meta-analysis. Anaesthesia. 2019;74:651–62. doi: 10.1111/anae.14582. [DOI](https://doi.org/10.1111/anae.14582) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30802933/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Anaesthesia&title=Analgesic%20impact%20of%20intra-operative%20opioids%20vs%20opioid-free%20anaesthesia:%20A%20systemic%20review%20and%20meta-analysis&author=J%20Frauenknecht&author=KR%20Kirkham&author=A%20Jacot-Guillarmod&author=E%20Albrecht&volume=74&publication_year=2019&pages=651-62&pmid=30802933&doi=10.1111/anae.14582&)
|
| 149 |
+
|
| 150 |
+
8. Friedrich S, Raub D, Teja BJ, Neves SE, Thevathasan T, Houle TT, et al. Effects of low-dose intraoperative fentanyl on postoperative respiratory complication rate: A pre-specified retrospective analysis. Br J Anaesth. 2019;122:180–8. doi: 10.1016/j.bja.2019.03.017. [DOI](https://doi.org/10.1016/j.bja.2019.03.017) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30982564/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Anaesth&title=Effects%20of%20low-dose%20intraoperative%20fentanyl%20on%20postoperative%20respiratory%20complication%20rate:%20A%20pre-specified%20retrospective%20analysis&author=S%20Friedrich&author=D%20Raub&author=BJ%20Teja&author=SE%20Neves&author=T%20Thevathasan&volume=122&publication_year=2019&pages=180-8&pmid=30982564&doi=10.1016/j.bja.2019.03.017&)
|
| 151 |
+
|
| 152 |
+
9. Leslie K, Myles P, Devereaux P, Williamson E, Rao-Melancini P, Forbes A, et al. Neuraxial block, death and serious cardiovascular morbidity and mortality in the POISE Trial. Br J Anaesth. 2013;111:382–90. doi: 10.1093/bja/aet120. [DOI](https://doi.org/10.1093/bja/aet120) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23611915/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Anaesth&title=Neuraxial%20block,%20death%20and%20serious%20cardiovascular%20morbidity%20and%20mortality%20in%20the%20POISE%20Trial&author=K%20Leslie&author=P%20Myles&author=P%20Devereaux&author=E%20Williamson&author=P%20Rao-Melancini&volume=111&publication_year=2013&pages=382-90&pmid=23611915&doi=10.1093/bja/aet120&)
|
| 153 |
+
|
| 154 |
+
10. Kumar S, Kesavan R, Sistla SC, Penumadu P, Natarajan S, Nair S, et al. Impact of genetic variation on postoperative pain and fentanyl dose requirement in patients undergoing major breast surgery: A candidate gene association study. Anesth Analg. 2023;137:409–17. doi: 10.1213/ANE.0000000000006330. [DOI](https://doi.org/10.1213/ANE.0000000000006330) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36538471/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Anesth%20Analg&title=Impact%20of%20genetic%20variation%20on%20postoperative%20pain%20and%20fentanyl%20dose%20requirement%20in%20patients%20undergoing%20major%20breast%20surgery:%20A%20candidate%20gene%20association%20study&author=S%20Kumar&author=R%20Kesavan&author=SC%20Sistla&author=P%20Penumadu&author=S%20Natarajan&volume=137&publication_year=2023&pages=409-17&pmid=36538471&doi=10.1213/ANE.0000000000006330&)
|
| 155 |
+
|
| 156 |
+
11. Lysakwoski C, Dumont L, Pellegrini M, Clergue F, Tassonyi E. Effects of fentanyl, alfentanil, remifentanil and sufentanil on loss of consciousness and Bispectral index during propofol induction of anaesthesia. Br J Anaesth. 2001;86:523–7. doi: 10.1093/bja/86.4.523. [DOI](https://doi.org/10.1093/bja/86.4.523) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11573626/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Anaesth&title=Effects%20of%20fentanyl,%20alfentanil,%20remifentanil%20and%20sufentanil%20on%20loss%20of%20consciousness%20and%20Bispectral%20index%20during%20propofol%20induction%20of%20anaesthesia&author=C%20Lysakwoski&author=L%20Dumont&author=M%20Pellegrini&author=F%20Clergue&author=E%20Tassonyi&volume=86&publication_year=2001&pages=523-7&pmid=11573626&doi=10.1093/bja/86.4.523&)
|
test/texts/PMC10618485.md
ADDED
|
@@ -0,0 +1,117 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Elexacaftor-Tezacaftor-Ivacaftor in 2 cystic fibrosis adults homozygous for M1101K with end-stage lung disease
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Winnie M Leung, Parastoo Molla Davoodi, Ashten Langevin, Clare Smith, Michael D Parkins
|
| 5 |
+
**Journal:** Respiratory Medicine Case Reports
|
| 6 |
+
**Date:** 2023 Oct 17
|
| 7 |
+
**DOI:** [10.1016/j.rmcr.2023.101938](https://doi.org/10.1016/j.rmcr.2023.101938)
|
| 8 |
+
**PMID:** 37920361
|
| 9 |
+
**PMCID:** PMC10618485
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10618485/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC10618485/pdf/main.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC10618485/pdf/main.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
Elexacaftor-tezacaftor-ivacaftor (ETI) therapy is shown to improve the health of individuals with cystic fibrosis (CF) who have the F508del variant. There are in vitro studies showing benefit with ETI for select rare CF variants. Limited data exists on the use of ETI in individuals with rare CF variants, particularly in those with advanced lung disease. We present 2 cases of CF individuals homozygous for the rare M1101K variant with end-stage lung disease who demonstrated sustained improvements in lung function, pulmonary exacerbation frequency, respiratory symptoms, and body mass index after 6 months of ETI treatment – similar to that expected with F508del.
|
| 16 |
+
|
| 17 |
+
Keywords: Cystic fibrosis, Cystic fibrosis transmembrane conductance regulator, CFTR modulator, Elexacaftor-tezacaftor-ivacaftor
|
| 18 |
+
|
| 19 |
+
**Keywords:**Keywords: Cystic fibrosis, Cystic fibrosis transmembrane conductance regulator, CFTR modulator, Elexacaftor-tezacaftor-ivacaftor
|
| 20 |
+
|
| 21 |
+
## 1. Introduction
|
| 22 |
+
|
| 23 |
+
Cystic Fibrosis (CF) is a progressive life-limiting disease of autosomal recessive inheritance, affecting approximately 105,000 individuals diagnosed with CF worldwide [[1](#bib1)1]. The basis for CF is from an abnormal cystic fibrosis transmembrane conductance regulator (CFTR) protein that regulates chloride and bicarbonate transport in epithelial cells [[2](#bib2)2]. Dysfunctional CFTR protein results in viscous and sticky mucus affecting many organs, most notably in the airways, gastrointestinal tract, pancreas and sweat glands, which cause impaired organ function that leads to morbidity and mortality in people with CF (pwCF) [[2](#bib2)2]. CFTR dysfunction is grouped into 6 classes based on the effect of the mutation on the CFTR protein production, trafficking, function, and stability [[2](#bib2)2,[3](#bib3)3]. CFTR modulators are small molecule therapies designed to restore the activity of mutant CFTR [[2](#bib2)2]. Clinical trials have demonstrated that the CFTR modulator elexacaftor-tezacaftor-ivacaftor (ETI) is effective in improving lung function, pulmonary exacerbation frequency, sweat chloride concentration, nutrition status and quality of life for pwCF with F508del, the most common CFTR variant worldwide [[4](#bib4)4,[5](#bib5)5]. Limited data is available for the clinical use of pwCF with non-F508del variants. However, pre-clinical studies examining the efficacy of ETI on non-F508del CFTR variants have shown *in vitro*in vitro benefit in selected CFTR variants, including c.3302T>A (M1101K) [[6](#bib6)6,[7](#bib7)7].
|
| 24 |
+
|
| 25 |
+
M1101K is a rare class II variant, found in 0.2% of CF patients registered in CFTR2 worldwide registry [[8](#bib8)8]. However, in Canada, M1101K is found in 1.7% of individuals with CF [[9](#bib9)9]. In particular, rates of M1101K in the Prairie provinces of Canada are disproportionally enriched because of the high prevalence of Hutterite colonies [[10](#bib10)10,[11](#bib11)11]. As such, the incidence of CF is 1:313 live births in these communities [[12](#bib12)12] compared to the national rate of 1:3850 [[9](#bib9)9].
|
| 26 |
+
|
| 27 |
+
Use of ETI in pwCF with M1101K is rarely reported. Whereas, approval for ETI use in those with M1101K is present in the United States [[13](#bib13)13], the majority of countries including Canada [[14](#bib14)14] have neither approval nor funding mechanisms for ETI use in non-F508del variants, and the lack of supporting clinical data is a barrier. Herein, we describe 2 cases of individuals homozygous for M1101K with end-stage lung disease who had marked clinical improvement following initiation of ETI through an Exceptional Drug Therapy funding program.
|
| 28 |
+
|
| 29 |
+
## 2. Case 1
|
| 30 |
+
|
| 31 |
+
ETI was started in a 28-year-old man with CF (genotype M1101K/M1101K) in December 2022. His past medical history included asthma, allergic bronchopulmonary aspergillosis, chronic pulmonary infection with *Pseudomonas aeruginosa,*Pseudomonas aeruginosa, intermittent respiratory cultures of methicillin-sensitive *Staphylococcus aureus*Staphylococcus aureus and *small colony variant S. aureus*small colony variant S. aureus, pancreatic sufficiency, CF-related diabetes, chronic abdominal pain, and osteopenia. Baseline percentage predicted forced expiratory volume in 1 second (ppFEV_1_1) 2 years prior to ETI initiation was 22%. In the 12 months preceding ETI initiation, there was progressive decline in lung health with development of chronic hypoxia, hypercapnia and pulmonary hypertension, new acquisition of methicillin-resistant *S. aureus*S. aureus, and 5 CF pulmonary exacerbation hospitalizations. Two weeks prior to ETI initiation, ppFEV_1_1 was 10% and there was a weight loss of 10% in the preceding month despite over 6 weeks of intravenous (IV) antibiotics. Arterial blood gas showed the partial pressure of carbon dioxide was 59 mm of mercury (mmHg), and partial pressure of oxygen was 50 mmHg, and he was on supplemental oxygen at 4 L per minute (lpm). Echocardiogram showed severely elevated pulmonary artery systolic pressure at 78 mmHg and mildly dilated right ventricle.
|
| 32 |
+
|
| 33 |
+
ETI was started as an outpatient on a reduced dose and escalated to the full dose by Day 7. Within the first week of ETI use, there was rapid improvement in sputum production, cough and dyspnea, there was a reduction in supplemental oxygen from 4 lpm to 1 lpm, and IV antibiotics were discontinued. Six weeks after ETI initiation, ppFEV_1_1 was 21%. At 6 months after ETI initiation, hypercapnia resolved. Depression and anxiety scores were measured by the Patient Health Questionnaire 9 (PHQ-9) and Generalized Anxiety Disorder 7-item (GAD-7) Scale, respectively. PHQ-9 score at baseline was 23 and after 6 months on ETI was 0. Baseline and 6-month post-ETI start GAD-7 scores were 18 and 2, respectively. No adverse effects were noted on initiation nor during the first 6 months of ETI therapy. There were sustained improvements in clinical status and sweat chloride comparing baseline to 6 months after ETI initiation ([Table 1](#tbl1)Table 1).
|
| 34 |
+
|
| 35 |
+
### Table 1.
|
| 36 |
+
|
| 37 |
+
Clinical characteristics before and after modulator therapy.
|
| 38 |
+
|
| 39 |
+
| Clinical characteristics | Case 1 | Case 2 |
|
| 40 |
+
| ------------------------ | ------ | ------ |
|
| 41 |
+
| Baseline | After 6 months on ETI | Baseline | After 6 months on ETI |
|
| 42 |
+
| FEV1, L | 0.44 | 0.84 | 0.85 | 1.19 |
|
| 43 |
+
| FEV1, % predicted | 10 | 21 | 27 | 38 |
|
| 44 |
+
| FVC, L | 1.68 | 2.62 | 1.81 | 2.34 |
|
| 45 |
+
| FVC, % predicted | 34 | 53 | 48 | 62 |
|
| 46 |
+
| Sweat chloride, mmol/L | 86 | 34 | 92 | 30 |
|
| 47 |
+
| BMI, kg/m2 | 16 | 22.2 | 22.2 | 23.8 |
|
| 48 |
+
| Number of CF pulmonary exacerbations | 5 in 12 months prior to ETI start | 0 | 6 in 12 months prior to ETI start | 1 |
|
| 49 |
+
| Days in hospital for CF pulmonary exacerbations | 49 in 12 months prior to ETI start | 0 | 33 in 12 months prior to ETI start | 0 |
|
| 50 |
+
| Days of IV antibiotics for CF pulmonary exacerbations | 149 in 12 months prior to ETI start | 0 | 33 in 12 months prior to ETI start | 0 |
|
| 51 |
+
## 3. Case 2
|
| 52 |
+
|
| 53 |
+
This is a 35-year-old female with CF (genotype M1101K/M1101K) who started ETI in September 2022. Her past medical history includes end-stage lung disease, chronic pulmonary infection with *P. aeruginosa*P. aeruginosa, chronic pulmonary infection with *Pseudallescheria boydii*Pseudallescheria boydii complex (and to a lower degree *Aspergillus fumigatus*Aspergillus fumigatus) requiring interventional bronchoscopy for debridement of mycetoma, hemoptysis, pancreatic sufficiency with repeated episodes of pancreatitis, depression and prior episodes of distal intestinal obstruction syndrome. Baseline ppFEV_1_1 2 years prior to ETI initiation was 46%. In the 12 months preceding ETI therapy, there was a progressive decline in lung health with development of chronic hypoxia requiring supplemental home oxygen, 6 CF pulmonary exacerbations treated with oral/IV antibiotics and a decrease in baseline ppFEV_1_1 to 27%. After starting ETI, there was a rapid decrease in sputum production, cough and dyspnea and cessation of episodes of hemoptysis. Her ppFEV_1_1 increased to 33% at 7 weeks and peaked at 38% at 6-months post initiation. This improvement in sputum production, cough and dyspnea has persisted with ongoing ETI therapy. There was a sustained improvement in overall health and sweat chloride after 6 months of ETI therapy ([Table 1](#tbl1)Table 1). No adverse effects were noted from initiation of ETI in this patient.
|
| 54 |
+
|
| 55 |
+
## 4. Discussion
|
| 56 |
+
|
| 57 |
+
To our knowledge, our cases are the first to demonstrate sustained clinical improvements with ETI therapy for pwCF and the rare M1101K variant for pulmonary and extra-pulmonary CF manifestations. Moreover, these cases demonstrate safety in initiating ETI for individuals with M1101K variant and end-stage lung disease. In both cases, ppFEV_1_1 improved after 1 month of ETI therapy, and they were sustained after 6 months with respect to ppFEV_1_1, pulmonary exacerbation rates and respiratory symptoms. There were associated improvements in nutrition as measured through body mass index (BMI), anxiety and depression. There were no reported adverse effects during ETI initiation nor with continued use during the observation period.
|
| 58 |
+
|
| 59 |
+
Those changes observed herein with the M1101K variant in our cases are similar clinical improvements compared with pwCF with F508del variant on ETI therapy. Our cases demonstrated increased ppFEV_1_1 after the first month of ETI therapy and maintained ppFEV_1_1 in the subsequent 6 months, with an absolute increase in ppFEV_1_1 at 11% for both cases, despite advanced lung disease at baseline. The rapid improvement that is sustained over time with reduction in pulmonary exacerbation rate in our cases is similar to results from clinical trials of ETI use in pwCF with F508del [[5](#bib5)5]. Similar improvements were shown in studies in pwCF with F508del with advanced lung disease on ETI [[15](#bib15)15,[16](#bib16)16]. A single center retrospective study of ETI therapy in pwCF with F508del variant and advanced lung disease with baseline median ppFEV_1_1 27.5% showed median ppFEV_1_1 increase of 10.5% at 6 months [[16](#bib16)16]. In the nutrition domain, BMI increased from 19.1 to 22.8 kg/m^2^2 after ETI therapy. Our cases similarly showed an increase in BMI to the normal weight category.
|
| 60 |
+
|
| 61 |
+
One case of a pwCF and M1101K with baseline ppFEV_1_1 37% showed improved ppFEV_1_1 and weight 4–6 weeks after treatment with ETI [[18](#bib18)18]. Our cases demonstrate that pulmonary and extra-pulmonary improvements from a much lower baseline ppFEV_1_1 are still achievable, and such improvements are sustained over 6 months.
|
| 62 |
+
|
| 63 |
+
*In vitro*In vitro studies using nasal epithelial cell cultures from pwCF homozygous for M1101K showed that ETI improved CFTR function [[6](#bib6)6,[7](#bib7)7]. Moreover, these improvements were superior to the CFTR function rescue by ETI in F508del homozygous nasal epithelial cell cultures [[6](#bib6)6]. Sweat chloride, a biomarker of CFTR function, can be used to assess the efficacy of treatments that influence CFTR function [[17](#bib17)17]. Our cases exhibited a reduction in sweat chloride to near normal CFTR function after ETI use, from baseline values within the CF diagnostic range [[19](#bib19)19]. The decrease in sweat chloride from baseline after ETI start in our cases exceeds the sweat chloride reduction reported in clinical trials of ETI in pwCF and F508del variant [[4](#bib4)4,[5](#bib5)5]. Our cases demonstrate improvement in CFTR function with ETI therapy, consistent with the *in vitro*in vitro data of CFTR function rescue.
|
| 64 |
+
|
| 65 |
+
More than 700 CFTR variants have been associated with CF [[8](#bib8)8]. The distribution of these alleles is often concentrated in specific communities [[20](#bib20)20]. Within Canada, those in Hutterite communities are disproportionally affected by CF and the M1101K homozygous genotype has been found in 39% of those with CF in this population [[11](#bib11)11]. Evaluation of the clinical effects for rare CFTR variants such as M1101K is challenging due to the paucity of cases available for enrollment in large-scale studies. Our cases not only demonstrate therapeutic benefit of ETI in individuals with non-F508del variants, but also provide supporting data in the worldwide efforts to access ETI for individuals who would benefit from this therapy.
|
| 66 |
+
|
| 67 |
+
## 5. Conclusion
|
| 68 |
+
|
| 69 |
+
We report 2 cases of pwCF with the rare M1101K variant who have improvements in lung function, pulmonary exacerbation frequency, respiratory symptoms and BMI following 6 months of ETI CFTR modulator therapy. CFTR function as measured by sweat chloride achieved near-normal values with ETI in our cases. These cases show sustained clinical efficacy in a rare but important Canadian variant, whose efficacy was previously shown in *in vitro*in vitro studies and a short-term study. Pulmonary and extra-pulmonary clinical improvements may still be safely achieved with ETI in end-stage lung disease.
|
| 70 |
+
|
| 71 |
+
## Declaration of competing interest
|
| 72 |
+
|
| 73 |
+
WML and MDP are site investigators for Vertex Pharmaceuticals clinical trials. PMD, AL and CS declare no competing interests associated with this manuscript.
|
| 74 |
+
|
| 75 |
+
Handling Editor: DR AC Amit Chopra
|
| 76 |
+
|
| 77 |
+
## References
|
| 78 |
+
|
| 79 |
+
1. Guo J., Garratt A., Hill A. Worldwide rates of diagnosis and effective treatment for cystic fibrosis. J. Cyst. Fibros. 2022;21:456–462. doi: 10.1016/j.jcf.2022.01.009. [DOI](https://doi.org/10.1016/j.jcf.2022.01.009) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35125294/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Cyst.%20Fibros.&title=Worldwide%20rates%20of%20diagnosis%20and%20effective%20treatment%20for%20cystic%20fibrosis&author=J.%20Guo&author=A.%20Garratt&author=A.%20Hill&volume=21&publication_year=2022&pages=456-462&pmid=35125294&doi=10.1016/j.jcf.2022.01.009&)
|
| 80 |
+
|
| 81 |
+
2. Gentzsch M., Mall M.A. Ion Channel modulators in cystic fibrosis. Chest. 2018;154:383–393. doi: 10.1016/j.chest.2018.04.036. [DOI](https://doi.org/10.1016/j.chest.2018.04.036) | [PMC free article](/articles/PMC6113631/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29750923/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chest&title=Ion%20Channel%20modulators%20in%20cystic%20fibrosis&author=M.%20Gentzsch&author=M.A.%20Mall&volume=154&publication_year=2018&pages=383-393&pmid=29750923&doi=10.1016/j.chest.2018.04.036&)
|
| 82 |
+
|
| 83 |
+
3. Boyle M.P., De Boeck K. A new era in the treatment of cystic fibrosis: correction of the underlying CFTR defect. Lancet Respir. Med. 2013;1:158–163. doi: 10.1016/S2213-2600(12)70057-7. [DOI](https://doi.org/10.1016/S2213-2600(12)70057-7) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24429096/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet%20Respir.%20Med.&title=A%20new%20era%20in%20the%20treatment%20of%20cystic%20fibrosis:%20correction%20of%20the%20underlying%20CFTR%20defect&author=M.P.%20Boyle&author=K.%20De%20Boeck&volume=1&publication_year=2013&pages=158-163&pmid=24429096&doi=10.1016/S2213-2600(12)70057-7&)
|
| 84 |
+
|
| 85 |
+
4. Heijerman H.G.M., McKone E.F., Downey D.G., et al. Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: a double-blind,randomised, phase 3 trial. Lancet. 2019;394:1940–1948. doi: 10.1016/S0140-6736(19)32597-8. [DOI](https://doi.org/10.1016/S0140-6736(19)32597-8) | [PMC free article](/articles/PMC7571408/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31679946/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Efficacy%20and%20safety%20of%20the%20elexacaftor%20plus%20tezacaftor%20plus%20ivacaftor%20combination%20regimen%20in%20people%20with%20cystic%20fibrosis%20homozygous%20for%20the%20F508del%20mutation:%20a%20double-blind,randomised,%20phase%203%20trial&author=H.G.M.%20Heijerman&author=E.F.%20McKone&author=D.G.%20Downey&volume=394&publication_year=2019&pages=1940-1948&pmid=31679946&doi=10.1016/S0140-6736(19)32597-8&)
|
| 86 |
+
|
| 87 |
+
5. Middleton P.G., Mall M.A., Dřevínek P., et al. Elexacaftor–Tezacaftor–Ivacaftor for Cystic Fibrosis with a Single Phe508del Allele. N. Engl. J. Med. 2019;381:1809–1819. doi: 10.1056/NEJMoa1908639. [DOI](https://doi.org/10.1056/NEJMoa1908639) | [PMC free article](/articles/PMC7282384/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31697873/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N.%20Engl.%20J.%20Med.&title=Elexacaftor%E2%80%93Tezacaftor%E2%80%93Ivacaftor%20for%20Cystic%20Fibrosis%20with%20a%20Single%20Phe508del%20Allele&author=P.G.%20Middleton&author=M.A.%20Mall&author=P.%20D%C5%99ev%C3%ADnek&volume=381&publication_year=2019&pages=1809-1819&pmid=31697873&doi=10.1056/NEJMoa1908639&)
|
| 88 |
+
|
| 89 |
+
6. Laselva O., Bartlett C., Gunawardena T.N., et al. Rescue of multiple class II CFTR mutations by elexacaftor+tezacaftor+ivacaftor mediated in part by the dual activities of elexacaftor as both corrector and potentiator. Eur. Respir. J. 2021;57 doi: 10.1183/13993003.02774-2020. [DOI](https://doi.org/10.1183/13993003.02774-2020) | [PMC free article](/articles/PMC8209484/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33303536/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Respir.%20J.&title=Rescue%20of%20multiple%20class%20II%20CFTR%20mutations%20by%20elexacaftor+tezacaftor+ivacaftor%20mediated%20in%20part%20by%20the%20dual%20activities%20of%20elexacaftor%20as%20both%20corrector%20and%20potentiator&author=O.%20Laselva&author=C.%20Bartlett&author=T.N.%20Gunawardena&volume=57&publication_year=2021&pmid=33303536&doi=10.1183/13993003.02774-2020&)
|
| 90 |
+
|
| 91 |
+
7. Veit G., Roldan A., Hancock M.A., et al. Allosteric folding correction of F508del and rare CFTR mutants by elexacaftor-tezacaftor-ivacaftor (Trikafta) combination. JCI Insight. 2020;5 doi: 10.1172/jci.insight.139983. [DOI](https://doi.org/10.1172/jci.insight.139983) | [PMC free article](/articles/PMC7526550/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32853178/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=JCI%20Insight&title=Allosteric%20folding%20correction%20of%20F508del%20and%20rare%20CFTR%20mutants%20by%20elexacaftor-tezacaftor-ivacaftor%20(Trikafta)%20combination&author=G.%20Veit&author=A.%20Roldan&author=M.A.%20Hancock&volume=5&publication_year=2020&pmid=32853178&doi=10.1172/jci.insight.139983&)
|
| 92 |
+
|
| 93 |
+
8. The clinical and functional TRanslation of CFTR (CFTR2) http://cftr2.org available at: [http://cftr2.org](http://cftr2.org)
|
| 94 |
+
|
| 95 |
+
9. Cystic Fibrosis Canada The Canadian cystic fibrosis registry 2021 annual data report. 2023. https://www.cysticfibrosis.ca/registry/2021AnnualDataReport.pdf Published February. [https://www.cysticfibrosis.ca/registry/2021AnnualDataReport.pdf](https://www.cysticfibrosis.ca/registry/2021AnnualDataReport.pdf)
|
| 96 |
+
|
| 97 |
+
10. Statistics Canada Table 98-10-0044-01 Type of collective dwelling and collective dwellings occupied by usual residents and population in collective dwellings: Canada, provinces and territories. 2022. Published 27 April. [DOI](https://doi.org/10.25318/9810004401-eng)
|
| 98 |
+
|
| 99 |
+
11. Pasterkamp H., Menzies K.J., Bayomi D.J. Cystic fibrosis in Canadian hutterites. Pediatr. Pulmonol. 2020;55:526–532. doi: 10.1002/ppul.24590. [DOI](https://doi.org/10.1002/ppul.24590) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31782915/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pediatr.%20Pulmonol.&title=Cystic%20fibrosis%20in%20Canadian%20hutterites&author=H.%20Pasterkamp&author=K.J.%20Menzies&author=D.J.%20Bayomi&volume=55&publication_year=2020&pages=526-532&pmid=31782915&doi=10.1002/ppul.24590&)
|
| 100 |
+
|
| 101 |
+
12. Mickle J.E., Cutting G.R. Clinical implications of cystic fibrosis transmembrane conductance regulator mutations. Clin. Chest Med. 1998;19:443–458. doi: 10.1016/S0272-5231(05)70092-7. [DOI](https://doi.org/10.1016/S0272-5231(05)70092-7) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9759548/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Chest%20Med.&title=Clinical%20implications%20of%20cystic%20fibrosis%20transmembrane%20conductance%20regulator%20mutations&author=J.E.%20Mickle&author=G.R.%20Cutting&volume=19&publication_year=1998&pages=443-458&pmid=9759548&doi=10.1016/S0272-5231(05)70092-7&)
|
| 102 |
+
|
| 103 |
+
13. Drugs@FDA: FDA Approved Drugs. U.S. Food & Drug Administration. 2023. https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/217660s000lbl.pdf Updated August. [https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/217660s000lbl.pdf](https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/217660s000lbl.pdf)
|
| 104 |
+
|
| 105 |
+
14. Drug product Database. Health Canada. 2022. https://pdf.hres.ca/dpd_pm/00065514.PDF Updated April. [https://pdf.hres.ca/dpd_pm/00065514.PDF](https://pdf.hres.ca/dpd_pm/00065514.PDF)
|
| 106 |
+
|
| 107 |
+
15. Burgel P.-R., Durieu I., Chiron R., et al. Rapid improvement after starting elexacaftor-tezacaftor-ivacaftor in patients with cystic fibrosis and advanced pulmonary disease. Am. J. Respir. Crit. Care Med. 2021;204:64–73. doi: 10.1164/rccm.202011-4153OC. [DOI](https://doi.org/10.1164/rccm.202011-4153OC) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33600738/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Rapid%20improvement%20after%20starting%20elexacaftor-tezacaftor-ivacaftor%20in%20patients%20with%20cystic%20fibrosis%20and%20advanced%20pulmonary%20disease&author=P.-R.%20Burgel&author=I.%20Durieu&author=R.%20Chiron&volume=204&publication_year=2021&pages=64-73&pmid=33600738&doi=10.1164/rccm.202011-4153OC&)
|
| 108 |
+
|
| 109 |
+
16. McCoy K.S., Blind J., Johnson T., et al. Clinical change 2 years from start of elexacaftor-tezacaftor-ivacaftor in severe cystic fibrosis. Pediatr. Pulmonol. 2023;58:1178–1184. doi: 10.1002/ppul.26318. [DOI](https://doi.org/10.1002/ppul.26318) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36650567/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pediatr.%20Pulmonol.&title=Clinical%20change%202%20years%20from%20start%20of%20elexacaftor-tezacaftor-ivacaftor%20in%20severe%20cystic%20fibrosis&author=K.S.%20McCoy&author=J.%20Blind&author=T.%20Johnson&volume=58&publication_year=2023&pages=1178-1184&pmid=36650567&doi=10.1002/ppul.26318&)
|
| 110 |
+
|
| 111 |
+
17. Collaco J.M., Blackman S.M., Raraigh K.S., et al. Sources of variation in sweat chloride measurements in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2016;194:1375–1382. doi: 10.1164/rccm.201603-0459OC. [DOI](https://doi.org/10.1164/rccm.201603-0459OC) | [PMC free article](/articles/PMC5148144/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27258095/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Respir.%20Crit.%20Care%20Med.&title=Sources%20of%20variation%20in%20sweat%20chloride%20measurements%20in%20cystic%20fibrosis&author=J.M.%20Collaco&author=S.M.%20Blackman&author=K.S.%20Raraigh&volume=194&publication_year=2016&pages=1375-1382&pmid=27258095&doi=10.1164/rccm.201603-0459OC&)
|
| 112 |
+
|
| 113 |
+
18. Burgel P.-R., Sermet-Gaudelus I., Durieu I., et al. The French compassionate programme of elexacaftor/tezacaftor/ivacaftor in people with cystic fibrosis with advanced lung disease and no F508del CFTR variant. Eur. Respir. J. 2023;61 doi: 10.1183/13993003.02437-2022. [DOI](https://doi.org/10.1183/13993003.02437-2022) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36796836/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Respir.%20J.&title=The%20French%20compassionate%20programme%20of%20elexacaftor/tezacaftor/ivacaftor%20in%20people%20with%20cystic%20fibrosis%20with%20advanced%20lung%20disease%20and%20no%20F508del%20CFTR%20variant&author=P.-R.%20Burgel&author=I.%20Sermet-Gaudelus&author=I.%20Durieu&volume=61&publication_year=2023&pmid=36796836&doi=10.1183/13993003.02437-2022&)
|
| 114 |
+
|
| 115 |
+
19. Farrell P.M., White T.B., Ren C.L., et al. Diagnosis of cystic fibrosis: consensus guidelines from the cystic fibrosis foundation. J. Pediatr. 2017;181S:S4–S15. doi: 10.1016/j.jpeds.2016.09.064. [DOI](https://doi.org/10.1016/j.jpeds.2016.09.064) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28129811/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Pediatr.&title=Diagnosis%20of%20cystic%20fibrosis:%20consensus%20guidelines%20from%20the%20cystic%20fibrosis%20foundation&author=P.M.%20Farrell&author=T.B.%20White&author=C.L.%20Ren&volume=181S&publication_year=2017&pages=S4-S15&pmid=28129811&doi=10.1016/j.jpeds.2016.09.064&)
|
| 116 |
+
|
| 117 |
+
20. Castellani C., Cuppens H., Macek M., Jr., et al. Consensus on the use and interpretation of cystic fibrosis mutation analysis in clinical practice. J. Cyst. Fibros. 2008;7:179–196. doi: 10.1016/j.jcf.2008.03.009. [DOI](https://doi.org/10.1016/j.jcf.2008.03.009) | [PMC free article](/articles/PMC2810954/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18456578/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Cyst.%20Fibros.&title=Consensus%20on%20the%20use%20and%20interpretation%20of%20cystic%20fibrosis%20mutation%20analysis%20in%20clinical%20practice&author=C.%20Castellani&author=H.%20Cuppens&author=M.%20Macek&volume=7&publication_year=2008&pages=179-196&pmid=18456578&doi=10.1016/j.jcf.2008.03.009&)
|
test/texts/PMC10957942.md
ADDED
|
@@ -0,0 +1,215 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Association of HLA-DRB1 locus with treatment response to abatacept or TNF inhibitors in patients with seropositive rheumatoid arthritis
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Soojin Cha, So-Young Bang, Young Bin Joo, Soo-Kyung Cho, Chan-Bum Choi, Yoon-Kyoung Sung, Tae-Hwan Kim, Jae-Bum Jun, Dae Hyun Yoo, Hye-Soon Lee, Sang-Cheol Bae
|
| 5 |
+
**Journal:** Scientific Reports
|
| 6 |
+
**Date:** 2024 Mar 21
|
| 7 |
+
**DOI:** [10.1038/s41598-024-56987-2](https://doi.org/10.1038/s41598-024-56987-2)
|
| 8 |
+
**PMID:** 38514707
|
| 9 |
+
**PMCID:** PMC10957942
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10957942/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC10957942/pdf/41598_2024_Article_56987.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC10957942/pdf/41598_2024_Article_56987.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
The strongest genetic risk factor for rheumatoid arthritis (RA) has been known as HLA-DRB1 based on amino acid positions 11, 71, and 74. This study analyzed the association between specific HLA-DRB1 locus and treatment response to abatacept or TNF inhibitors (TNFi) in patients with seropositive RA. A total of 374 Korean RA patients were treated with abatacept (n = 110) or TNFi (n = 264). Associations between HLA-DRB1 and treatment response after 6 months were analyzed using multivariable logistic regression. Seropositive RA patients with HLA-DRB1 shared epitope (SE) had a favorable response to abatacept (OR = 3.67, P = 0.067) and an inversely associated response to TNFi (OR 0.57, P = 0.058) based on EULAR response criteria, but the difference was not statistically significant in comparison to those without SE. In analyses using amino acid positions of HLA-DRB1, a significant association was found between valine at amino acid position 11 of SE and good response to abatacept (OR = 6.46, P = 5.4 × 10–3). The VRA haplotype also showed a good response to abatacept (OR = 4.56, P = 0.013), but not to TNFi. Our results suggest that treatment response to abatacept or TNFi may differ depending on HLA-DRB1 locus in seropositive RA, providing valuable insights for selecting optimal therapy.
|
| 16 |
+
|
| 17 |
+
Keywords: HLA-DRB1, Amino acid, Treatment response, Abatacept, TNF inhibitors, Seropositive rheumatoid arthritis
|
| 18 |
+
|
| 19 |
+
Subject terms: Rheumatic diseases, Rheumatoid arthritis, Genetics
|
| 20 |
+
|
| 21 |
+
**Keywords:**Keywords: HLA-DRB1, Amino acid, Treatment response, Abatacept, TNF inhibitors, Seropositive rheumatoid arthritis
|
| 22 |
+
|
| 23 |
+
**Subject terms:**Subject terms: Rheumatic diseases, Rheumatoid arthritis, Genetics
|
| 24 |
+
|
| 25 |
+
## Introduction
|
| 26 |
+
|
| 27 |
+
Rheumatoid arthritis (RA) is a systemic autoimmune disease caused by both genetic and environmental factors. Many studies have identified over 100 RA susceptibility loci across multiple ancestries^1,2^[1](#CR1)1,[2](#CR2)2. The shared epitope (SE) hypothesis suggests that HLA–DRB1 alleles, which share a common amino acid motif at positions 70–74 (QKRAA, QRRAA, and RRRAA), contribute to the susceptibility to RA^3^[3](#CR3)3. The SE has been identified to have a significant association with an increased RA risk and to influence the development of anti-cyclic citrullinated peptides (anti-CCP) antibodies in European and Asian populations^4–6^[4](#CR4)4–[6](#CR6)6. In addition, several studies have described associations between HLA-DRB1 non-SE alleles and seropositive RA. Amino acid position 11 of HLA-DRB1 (closely related to position 13), which is not traditionally associated with the SE, has shown the strongest association with seropositive RA risk in both Caucasian and Asian populations^7,8^[7](#CR7)7,[8](#CR8)8. These positions are located in the peptide-binding groove of the HLA class II beta chain, alongside positions 71 and 74 of HLA-DRB1, and play a role in antigen presentation to CD4 + T cells.
|
| 28 |
+
|
| 29 |
+
The use of biologic disease-modifying anti-rheumatic drugs (bDMARDs) has significantly improved RA patients’ clinical outcomes. However, since approximately one-third to half of RA patients still do not achieve a good response to bDMARDs, various efforts are focused on identifying biomarkers that can predict the therapeutic efficacy of each bDMARD. Several studies have investigated the relationship between the anti-CCP and the efficacy of abatacept or TNF inhibitors (TNFi)^9^[9](#CR9)9. A study on a European population found an association between a reduced response to TNFi and the presence of rheumatoid factor (RF) or anti-CCP^10^[10](#CR10)10. A meta-analysis showed that anti-CCP-positive patients are more likely to achieve a good response to abatacept, while not to TNFi, compared to anti-CCP-negative patients with RA^11^[11](#CR11)11.
|
| 30 |
+
|
| 31 |
+
Recent studies have suggested possible associations between HLA-DRB1 SE and the efficacy of abatacept, but conflicting findings regarding TNFi in RA patients. In Japanese observational studies, RA patients with SE receiving abatacept showed greater efficacy at 24 weeks based on European League Against Rheumatism (EULAR) response than SE-negative patients^12,13^[12](#CR12)12,[13](#CR13)13. In a head-to-head study in autoantibody-positive early RA (AMPLE study), SE-positive RA patients receiving abatacept showed greater efficacy compared to those receiving adalimumab^14^[14](#CR14)14. In a European study, SE-positive RA patients receiving adalimumab showed a significant association with low disease activity at week 26, while the efficacy of TNFi in a UK population of RA patients showed no association with SE status^10,15^[10](#CR10)10,[15](#CR15)15. Based on the understanding that amino acid positions 11, 71, and 74 of HLA-DRB1 have shown a stronger association with the risk of developing RA compared to SE^7,8^[7](#CR7)7,[8](#CR8)8, we hypothesized that treatment response to biologics could potentially be explained by the amino acids or their haplotypes at those specific positions. Therefore, in this study, we investigated the impact of HLA-DRB1 alleles specifically based on amino acid positions 11, 71, and 74 to predict treatment response to abatacept or TNFi in a prospective Korean RA cohort.
|
| 32 |
+
|
| 33 |
+
## Results
|
| 34 |
+
|
| 35 |
+
### Patient characteristics
|
| 36 |
+
|
| 37 |
+
In this study, 110 RA patients were treated with abatacept and 264 RA patients were treated with TNFi [including etanercept (n = 124), adalimumab (n = 92), golimumab (n = 30), and infliximab (n = 18)] due to moderate or high disease activity at enrollment, despite receiving conventional synthetic DMARDs for at least 6 months (Table [1](#Tab1)1). Among the patients receiving abatacept, the median age of RA onset was 47.0 years, and 58.2% of these patients had not previously received biologics. In the group receiving TNFi, the median age of RA onset was 41.0 years, and 76.9% of these patients had not previously received biologics. The change in DAS28 (ΔDAS28) was calculated as the difference between DAS28 at baseline and at 6 months after treatment. No significant differences were observed between the abatacept and TNFi treatment groups in the median baseline DAS28 [6.4 (5.8–7.0) vs. 6.2 (5.6–6.7), P > 0.05] or the median at 6 months [2.3 (1.6–2.9) in the abatacept-treatment group vs. 2.6 (1.8–3.4) in the TNFi-treatment group, P > 0.05].
|
| 38 |
+
|
| 39 |
+
### Table 1.
|
| 40 |
+
|
| 41 |
+
Clinical characteristics of seropositive RA patients receiving abatacept or TNF inhibitors.
|
| 42 |
+
|
| 43 |
+
| | | Abatacept (n = 110)a |
|
| 44 |
+
| - | - | -------------------- |
|
| 45 |
+
| Clinical | RA onset-age | 47.0 (35.0–56.8) |
|
| 46 |
+
| Biologics start age | 55.0 (48.0–63.0) | |
|
| 47 |
+
| Female | 90.0% | |
|
| 48 |
+
| BMI at baseline | 22.2 (19.9–23.7) | |
|
| 49 |
+
| Methotrexate | 83.6% | |
|
| 50 |
+
| Biologics naïve | 58.2% | |
|
| 51 |
+
| Anti-CCP | 93.6% | |
|
| 52 |
+
| RF | 89.1% | |
|
| 53 |
+
| DAS28 at baseline | 6.4 (5.8–7.0) | |
|
| 54 |
+
| SDAI at baseline | 36.4 (28.0–44.7) | |
|
| 55 |
+
| HLA-DRB1 | Shared epitopec | 70.9% |
|
| 56 |
+
| Valine at amino acid position 11 | 64.5% | |
|
| 57 |
+
The RA patients carrying HLA-DRB1 SE were associated with production of anti-CCP (*P*P = 1.1 × 10^–4^–4), but not RF, compared to those without SE (Supplementary Table [S1](#MOESM1)S1). Moreover, valine at amino acid position 11 (Val11) of HLA-DRB1 influenced production of anti-CCP (*P*P = 3.4 × 10^–4^–4) and high anti-CCP titers (> three times the upper limit of normal, *P*P = 3.2 × 10^–3^–3) compared to those without Val11.
|
| 58 |
+
|
| 59 |
+
### Clinical factors associated with treatment response to abatacept or TNFi in seropositive RA patients
|
| 60 |
+
|
| 61 |
+
To investigate the factors associated with treatment response to abatacept or TNFi, we divided seropositive RA patients treated with abatacept or TNFi into two groups based on EULAR response criteria: good responders (good response) and poor responders (moderate/non-response). We then analyzed clinical variables associated with good responders. In both abatacept and TNFi groups, treatment response was not associated with RA onset-age, biologics start age, sex, body mass index (BMI), co-treatment with methotrexate, or previous TNFi inefficacy (Table [2](#Tab2)2).
|
| 62 |
+
|
| 63 |
+
### Table 2.
|
| 64 |
+
|
| 65 |
+
Clinical factors associated with good responders to abatacept or TNF inhibitors in seropositive RA patients.
|
| 66 |
+
|
| 67 |
+
| | | Good respondersa | Poor respondersa | Pb |
|
| 68 |
+
| - | - | ---------------- | ---------------- | -- |
|
| 69 |
+
| Abatacept (n = 110) | Number of patients | 16 | 94 | |
|
| 70 |
+
| Female | 87.5% | 90.4% | 1.00 | |
|
| 71 |
+
| BMI at baseline | 22.2 (21.1–23.2) | 22.1 (19.6–24.0) | 0.98 | |
|
| 72 |
+
| RA onset-age | 41.0 (32.5–58.5) | 48.0 (37.2–56.0) | 0.36 | |
|
| 73 |
+
| Biologics start age | 47.5 (37.5–61.5) | 55.0 (49.0–63.0) | 0.06 | |
|
| 74 |
+
| Methotrexate | 93.8% | 81.9% | 0.41 | |
|
| 75 |
+
| Previous TNFi inefficacy | 18.8% | 25.5% | 0.79 | |
|
| 76 |
+
| TNF inhibitors (n = 264) | Number of patients | 88 | 176 | |
|
| 77 |
+
| Female | 83.0% | 88.1% | 0.34 | |
|
| 78 |
+
| BMI at baseline | 22.2 (19.7–23.9) | 22.0 (20.0–24.4) | 0.98 | |
|
| 79 |
+
| RA onset age | 43.0 (30.0–52.0) | 41.0 (31.0–54.0) | 0.63 | |
|
| 80 |
+
| Biologics start age | 51.0 (41.8–58.0) | 51.0 (41.8–60.0) | 0.30 | |
|
| 81 |
+
| Methotrexate | 93.2% | 89.2% | 0.41 | |
|
| 82 |
+
| Previous TNFi inefficacy | 3.4% | 2.8% | 1.00 | |
|
| 83 |
+
### Associations of HLA-DRB1 SE with treatment response to abatacept or TNFi
|
| 84 |
+
|
| 85 |
+
We conducted multivariable logistic regression analyses to explore the relationship between HLA-DRB1 SE and treatment response to TNFi or abatacept, while adjusting for RA onset-age and sex. In the TNFi group, we found that SE-positive patients were less likely to be classified as good responders compared to the SE-negative group (OR = 0.57 [0.32–1.02], *P*P = 0.058) (Table [3](#Tab3)3). Conversely, in the abatacept group, a higher proportion of good responders was observed among SE-positive patients, although this finding did not reach statistical significance (OR = 3.67 [0.92–24.71], *P*P = 0.067) (Table [4](#Tab4)4). However, the difference was not statistically significant in comparison to those without SE. We observed a negative trend in the association, although not statistically significant, between the HLA-DRB1 *09:01 allele, which is the second significant risk allele in Korean RA population, and treatment response to abatacept (OR = 0.19 [0.01–1.11], *P*P = 0.068) or TNFi (OR = 1.09 [0.61–1.92], *P*P = 0.78)^5^[5](#CR5)5.
|
| 86 |
+
|
| 87 |
+
### Table 3.
|
| 88 |
+
|
| 89 |
+
Association of shared epitope (SE) and amino acid positions 11, 71, and 74 of HLA-DRB1 with good responders in seropositive RA patients treated with TNF inhibitors.
|
| 90 |
+
|
| 91 |
+
| | HLA-DRB1a | ORb (95% CI) | Pb |
|
| 92 |
+
| - | --------- | ------------ | -- |
|
| 93 |
+
| TNF inhibitors (n = 264) | SE | 0.57 (0.32–1.02) | 0.058 |
|
| 94 |
+
| SE with Val11 | 0.82 (0.48–1.41) | 0.47 | |
|
| 95 |
+
| Val11 | 0.86 (0.49–1.51) | 0.60 | |
|
| 96 |
+
| VRA (positions 11, 71, and 74) | 0.96 (0.57–1.61) | 0.86 | |
|
| 97 |
+
### Table 4.
|
| 98 |
+
|
| 99 |
+
Association of shared epitope (SE) and amino acid positions 11, 71, and 74 of HLA-DRB1 with good responders in seropositive RA patients treated with abatacept.
|
| 100 |
+
|
| 101 |
+
| | HLA-DRB1a | ORb (95% CI) | Pb |
|
| 102 |
+
| - | --------- | ------------ | -- |
|
| 103 |
+
| Abatacept (n = 110) | SE | 3.67 (0.92–24.71) | 0.067 |
|
| 104 |
+
| SE with Val11 | 6.46 (1.65–43.17) | 5.4 × 10–3 | |
|
| 105 |
+
| Val11 | 5.17 (1.30–34.84) | 0.017 | |
|
| 106 |
+
| VRA (positions 11, 71, and 74) | 4.56 (1.35–21.01) | 0.013 | |
|
| 107 |
+
### Associations of amino acid positions 11, 71, and 74 of HLA-DRB1 with good responders in seropositive RA patients treated with abatacept or TNFi
|
| 108 |
+
|
| 109 |
+
We conducted further analyses to determine whether amino acid positions 11/13, 71, and 74 of HLA-DRB1, the strongest RA risk factor, influence treatment response to abatacept or TNFi among seropositive RA patients. We excluded consideration of amino acid position 13 in HLA-DRB1 due to its strong linkage disequilibrium (LD) with position 11^8^[8](#CR8)8. Intriguingly, patients with Val11 of HLA-DRB1 SE exhibited a more favorable response to abatacept (OR = 6.46 [1.65–43.17],* P* P = 5.4 × 10^–3^–3) (Table [4](#Tab4)4, Supplementary Table [S2](#MOESM1)S2). Interestingly, RA patients with Val11 also showed a good response to abatacept, regardless of SE (OR = 5.17 [1.30–34.84], *P*P = 0.017). However, no significant associations were observed between SE with Val11 or HLA-DRB1 Val11 and good responders in seropositive RA patients treated with TNFi (OR = 0.82 [0.48–1.41], *P*P = 0.47, OR = 0.86 [0.49–1.51], *P*P = 0.60, respectively) (Table [3](#Tab3)3).
|
| 110 |
+
|
| 111 |
+
Next, we investigated whether the haplotypes based on amino acid positions 11, 71, and 74 were associated with good responses. We observed that the valine arginine alanine (VRA) haplotype at amino acid positions 11, 71, and 74 showed a significant association with a good response in seropositive RA patients treated with abatacept (OR = 4.56 [1.35–21.01], *P*P = 0.013) (Table [4](#Tab4)4, Supplementary Table [S2](#MOESM1)S2). However, no significant association was observed between the VRA haplotype and treatment response to TNFi (OR = 0.96 [0.57–1.61], *P*P = 0.86) (Table [3](#Tab3)3). The association between the VRA haplotype and a good response was observed, not within the SE (Supplementary Fig. [S1](#MOESM1)S1) in patients treated with abatacept. This suggests that patients with the VRA haplotype were more likely to exhibit a favorable response compared to those with the SE.
|
| 112 |
+
|
| 113 |
+
## Discussion
|
| 114 |
+
|
| 115 |
+
This study investigated an effect of HLA-DRB1 on treatment response to abatacept or TNFi in seropositive RA patients. We demonstrated that Val11 of HLA-DRB1 might predict good treatment response to abatacept in seropositive RA patients. Our analysis further revealed a significant association between a good response to abatacept and SE with Val11 (*P*P = 5.4 × 10^–3^–3), as well as the VRA haplotype at amino acid positions 11, 71, and 74 of HLA-DRB1 (*P*P = 0.013).
|
| 116 |
+
|
| 117 |
+
Since a considerable proportion of patients with bDMARDs still do not experience a favorable response, the need for research on predictive biomarkers that can facilitate optimal bDMARDs selection has been consistently emphasized.
|
| 118 |
+
|
| 119 |
+
We demonstrated a positive association between SE and abatacept response in a Korean seropositive RA population, consistent with previous European and Japanese studies^12–14^[12](#CR12)12–[14](#CR14)14, and added a novel result by revealing the effect of HLA-DRB1 amino acid positions on treatment response, which offer greater explanatory power than SE.
|
| 120 |
+
|
| 121 |
+
In our study, RA patients with SE treated with TNFi showed a less favorable treatment response compared to those with SE-negative. At the amino acid level, we did not observe any significant association between Val11 or the VRA haplotype (at amino acid positions 11, 71, and 74) and treatment response to TNFi. Since a European study suggested a weak association between Val11 and a good EULAR response (OR = 1.14, *P*P = 0.04)^16^[16](#CR16)16 and previous studies regarding the association of SE with response to TNFi showed conflicting findings, further research is needed to investigate whether the specific HLA-DRB1 alleles or haplotypes could predict the response to TNFi or not.
|
| 122 |
+
|
| 123 |
+
Despite our study’s significant findings, there are several limitations. Although we can get statistically significant results even in a relatively small sample sized cohort, it is still necessary to conduct large-scaled studies to confirm our findings. The VKA haplotype (HLA-DRB1 *04:01) is uncommon in the Korean population but prevalent in European populations, necessitating additional studies to verify our findings across different populations. Lastly, both the abatacept and TNFi treatment groups in the study included patients who had previously failed on other bDMARDs. As a result, the proportion of good responders may be relatively lower compared to the group of patients who are naïve to bDMARDs. Also, we did not directly compare the effects of abatacept and TNFi treatments, as the two groups have distinct backgrounds. However, this may actually better reflect real-world data.
|
| 124 |
+
|
| 125 |
+
Our findings suggest that HLA-DRB1 alleles carrying Val11 may serve as predictive biomarkers for a favorable treatment response in seropositive RA patients receiving abatacept, but not in those receiving TNFi. These results indicate the potential clinical utility of using these biomarkers to guide the selection of optimal therapies for individual RA patients.
|
| 126 |
+
|
| 127 |
+
## Methods
|
| 128 |
+
|
| 129 |
+
### Patients
|
| 130 |
+
|
| 131 |
+
A total of 374 seropositive RA patients who were treated with abatacept (n = 110) or TNFi (n = 264) and fulfilled the 1987 revised American College of Rheumatology (ACR) or 2010 ACR/EULAR criteria were enrolled from Hanyang University Hospital for Rheumatic Diseases. We obtained clinical data including autoantibody profiles (RF, anti-CCP). The Disease Activity Score in 28 Joints (DAS28) was assessed by analyzing the 28-tender joint count (TJC), 28-swollen joint count (SJC), patient global assessment (PtGA) on a visual analogue scale (VAS), and erythrocyte sedimentation rate (ESR) at baseline and 6 months. Anti-CCP titers were measured using the ImmuLisa CCP ELISA test (normal range < 25.0 U/ml, IMMCO Diagnostics Inc., USA). All patients were categorized as good or moderate/non-responders based on the EULAR response criteria. The study obtained written informed consent from all RA patients and received approval from the Institutional Review Board of Hanyang University Hospital (HYG-14-032-14). This study was performed in accordance with the relevant guidelines and regulations.
|
| 132 |
+
|
| 133 |
+
### HLA-DRB1 genotyping
|
| 134 |
+
|
| 135 |
+
We extracted DNA from the blood of enrolled patients and sequenced for HLA-DRB1 using next-generation sequencing (NGS). HLA-DRB1 SE status was defined as *01:01, *04:01, *04:04, *04:05, *04:08, *04:10, *10:01, *14:02, or *14:06. Amino acids at positions 11, 71, and 74 of HLA-DRB1 were assigned according to the sequence information provided in the IMGT/HLA Database ([http://www.ebi.ac.uk/ipd/imgt/hla/](http://www.ebi.ac.uk/ipd/imgt/hla/)http://www.ebi.ac.uk/ipd/imgt/hla/), using the Sequence Alignment Tool, Release 3.53^17^[17](#CR17)17.
|
| 136 |
+
|
| 137 |
+
### Statistical analyses
|
| 138 |
+
|
| 139 |
+
In a univariable analysis, we used the Wilcoxon rank sum test and chi square test for continuous (numerical) and categorical variables, respectively. A logistic regression model was used in the multivariable analyses, and odds ratios (ORs), 95% confidence intervals, and p values were estimated by a likelihood ratio test (LRT). Individuals were defined as carriers of specific alleles, amino acids, or haplotypes if they had at least one copy of the allele, amino acid, or haplotype and were tested for an association with treatment response adjusting for RA onset-age and sex. All analyses were conducted in the R environment (R 4.1.0).
|
| 140 |
+
|
| 141 |
+
## Supplementary Information
|
| 142 |
+
|
| 143 |
+
## Author contributions
|
| 144 |
+
|
| 145 |
+
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. SC and SYB had full access to all data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study conception and design. SC, SYB, HSL, and SCB. Acquisition of data. SYB, YBJ, SKC, CBC, YKS, THK, JBJ, DHY, HSL, and SCB. Analysis of data. SC, SYB, HSL, and SCB. Interpretation of data. SC, SYB, HSL, and SCB.
|
| 146 |
+
|
| 147 |
+
## Funding
|
| 148 |
+
|
| 149 |
+
This study was funded by a Bristol-Myers Squibb grant (BMS_IM101-939) and by National Research Foundation of Korea (NRF) grants funded by the Korean government and Ministry of Education (NRF-2021R1A6A1A03038899, NRF-2022R1A2C2006073).
|
| 150 |
+
|
| 151 |
+
## Data availability
|
| 152 |
+
|
| 153 |
+
The datasets analyzed in this study are not publicly available but are available from the corresponding author on reasonable request.
|
| 154 |
+
|
| 155 |
+
## Competing interests
|
| 156 |
+
|
| 157 |
+
The authors declare no competing interests.
|
| 158 |
+
|
| 159 |
+
## Footnotes
|
| 160 |
+
|
| 161 |
+
## Supplementary Information
|
| 162 |
+
|
| 163 |
+
The online version contains supplementary material available at 10.1038/s41598-024-56987-2.
|
| 164 |
+
|
| 165 |
+
## Associated Data
|
| 166 |
+
|
| 167 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 168 |
+
|
| 169 |
+
### Supplementary Materials
|
| 170 |
+
|
| 171 |
+
### Data Availability Statement
|
| 172 |
+
|
| 173 |
+
The datasets analyzed in this study are not publicly available but are available from the corresponding author on reasonable request.
|
| 174 |
+
|
| 175 |
+
### Supplementary Materials
|
| 176 |
+
|
| 177 |
+
### Data Availability Statement
|
| 178 |
+
|
| 179 |
+
The datasets analyzed in this study are not publicly available but are available from the corresponding author on reasonable request.
|
| 180 |
+
|
| 181 |
+
## References
|
| 182 |
+
|
| 183 |
+
1. Ha E, Bae SC, Kim K. Large-scale meta-analysis across East Asian and European populations updated genetic architecture and variant-driven biology of rheumatoid arthritis, identifying 11 novel susceptibility loci. Ann. Rheum Dis. 2021;80:558–565. doi: 10.1136/annrheumdis-2020-219065. [DOI](https://doi.org/10.1136/annrheumdis-2020-219065) | [PMC free article](/articles/PMC8053349/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33310728/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum%20Dis.&title=Large-scale%20meta-analysis%20across%20East%20Asian%20and%20European%20populations%20updated%20genetic%20architecture%20and%20variant-driven%20biology%20of%20rheumatoid%20arthritis,%20identifying%2011%20novel%20susceptibility%20loci&author=E%20Ha&author=SC%20Bae&author=K%20Kim&volume=80&publication_year=2021&pages=558-565&pmid=33310728&doi=10.1136/annrheumdis-2020-219065&)
|
| 184 |
+
|
| 185 |
+
2. Kim K, Bang SY, Lee HS, Bae SC. Update on the genetic architecture of rheumatoid arthritis. Nat. Rev. Rheumatol. 2017;13:13–24. doi: 10.1038/nrrheum.2016.176. [DOI](https://doi.org/10.1038/nrrheum.2016.176) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27811914/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat.%20Rev.%20Rheumatol.&title=Update%20on%20the%20genetic%20architecture%20of%20rheumatoid%20arthritis&author=K%20Kim&author=SY%20Bang&author=HS%20Lee&author=SC%20Bae&volume=13&publication_year=2017&pages=13-24&pmid=27811914&doi=10.1038/nrrheum.2016.176&)
|
| 186 |
+
|
| 187 |
+
3. Gregersen, P. K., Silver, J. & Winchester, R. J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum30, 1205–1213. 10.1002/art.1780301102 (1987). [DOI](https://doi.org/10.1002/art.1780301102) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/2446635/)
|
| 188 |
+
|
| 189 |
+
4. Huizinga TW, et al. Refining the complex rheumatoid arthritis phenotype based on specificity of the HLA-DRB1 shared epitope for antibodies to citrullinated proteins. Arthritis Rheum. 2005;52:3433–3438. doi: 10.1002/art.21385. [DOI](https://doi.org/10.1002/art.21385) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16255021/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Rheum&title=Refining%20the%20complex%20rheumatoid%20arthritis%20phenotype%20based%20on%20specificity%20of%20the%20HLA-DRB1%20shared%20epitope%20for%20antibodies%20to%20citrullinated%20proteins&author=TW%20Huizinga&volume=52&publication_year=2005&pages=3433-3438&pmid=16255021&doi=10.1002/art.21385&)
|
| 190 |
+
|
| 191 |
+
5. Bang SY, Lee KH, Cho SK, Lee HS, Lee KW, Bae SC. Smoking increases rheumatoid arthritis susceptibility in individuals carrying the HLA-DRB1 shared epitope, regardless of rheumatoid factor or anti-cyclic citrullinated peptide antibody status. Arthritis Rheum. 2010;62:369–377. doi: 10.1002/art.27272. [DOI](https://doi.org/10.1002/art.27272) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20112396/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Rheum&title=Smoking%20increases%20rheumatoid%20arthritis%20susceptibility%20in%20individuals%20carrying%20the%20HLA-DRB1%20shared%20epitope,%20regardless%20of%20rheumatoid%20factor%20or%20anti-cyclic%20citrullinated%20peptide%20antibody%20status&author=SY%20Bang&author=KH%20Lee&author=SK%20Cho&author=HS%20Lee&author=KW%20Lee&volume=62&publication_year=2010&pages=369-377&pmid=20112396&doi=10.1002/art.27272&)
|
| 192 |
+
|
| 193 |
+
6. Freudenberg J, et al. Genome-wide association study of rheumatoid arthritis in Koreans: population-specific loci as well as overlap with European susceptibility loci. Arthritis Rheum. 2011;63:884–893. doi: 10.1002/art.30235. [DOI](https://doi.org/10.1002/art.30235) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21452313/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Rheum&title=Genome-wide%20association%20study%20of%20rheumatoid%20arthritis%20in%20Koreans:%20population-specific%20loci%20as%20well%20as%20overlap%20with%20European%20susceptibility%20loci&author=J%20Freudenberg&volume=63&publication_year=2011&pages=884-893&pmid=21452313&doi=10.1002/art.30235&)
|
| 194 |
+
|
| 195 |
+
7. Okada Y, et al. Risk for ACPA-positive rheumatoid arthritis is driven by shared HLA amino acid polymorphisms in Asian and European populations. Hum. Mol. Genet. 2014;23:6916–6926. doi: 10.1093/hmg/ddu387. [DOI](https://doi.org/10.1093/hmg/ddu387) | [PMC free article](/articles/PMC4245039/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25070946/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Hum.%20Mol.%20Genet.&title=Risk%20for%20ACPA-positive%20rheumatoid%20arthritis%20is%20driven%20by%20shared%20HLA%20amino%20acid%20polymorphisms%20in%20Asian%20and%20European%20populations&author=Y%20Okada&volume=23&publication_year=2014&pages=6916-6926&pmid=25070946&doi=10.1093/hmg/ddu387&)
|
| 196 |
+
|
| 197 |
+
8. Raychaudhuri S, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI](https://doi.org/10.1038/ng.1076) | [PMC free article](/articles/PMC3288335/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22286218/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat.%20Genet.&title=Five%20amino%20acids%20in%20three%20HLA%20proteins%20explain%20most%20of%20the%20association%20between%20MHC%20and%20seropositive%20rheumatoid%20arthritis&author=S%20Raychaudhuri&volume=44&publication_year=2012&pages=291-296&pmid=22286218&doi=10.1038/ng.1076&)
|
| 198 |
+
|
| 199 |
+
9. van der Helm-van Mil, A. H., Verpoort, K. N., Breedveld, F. C., Huizinga, T. W., Toes, R. E. & de Vries, R. R. The HLA-DRB1 shared epitope alleles are primarily a risk factor for anti-cyclic citrullinated peptide antibodies and are not an independent risk factor for development of rheumatoid arthritis. Arthritis Rheum54, 1117–1121. 10.1002/art.21739 (2006). [DOI](https://doi.org/10.1002/art.21739) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16572446/)
|
| 200 |
+
|
| 201 |
+
10. Potter C, et al. Association of rheumatoid factor and anti-cyclic citrullinated peptide positivity, but not carriage of shared epitope or PTPN22 susceptibility variants, with anti-tumour necrosis factor response in rheumatoid arthritis. Ann. Rheum. Dis. 2009;68:69–74. doi: 10.1136/ard.2007.084715. [DOI](https://doi.org/10.1136/ard.2007.084715) | [PMC free article](/articles/PMC2596303/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18375541/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum.%20Dis.&title=Association%20of%20rheumatoid%20factor%20and%20anti-cyclic%20citrullinated%20peptide%20positivity,%20but%20not%20carriage%20of%20shared%20epitope%20or%20PTPN22%20susceptibility%20variants,%20with%20anti-tumour%20necrosis%20factor%20response%20in%20rheumatoid%20arthritis&author=C%20Potter&volume=68&publication_year=2009&pages=69-74&pmid=18375541&doi=10.1136/ard.2007.084715&)
|
| 202 |
+
|
| 203 |
+
11. Alemao E, Postema R, Elbez Y, Mamane C, Finckh A. Presence of anti-cyclic citrullinated peptide antibodies is associated with better treatment response to abatacept but not to TNF inhibitors in patients with rheumatoid arthritis: a meta-analysis. Clin. Exp. Rheumatol. 2020;38:455–466. [PubMed](https://pubmed.ncbi.nlm.nih.gov/31770089/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Exp.%20Rheumatol.&title=Presence%20of%20anti-cyclic%20citrullinated%20peptide%20antibodies%20is%20associated%20with%20better%20treatment%20response%20to%20abatacept%20but%20not%20to%20TNF%20inhibitors%20in%20patients%20with%20rheumatoid%20arthritis:%20a%20meta-analysis&author=E%20Alemao&author=R%20Postema&author=Y%20Elbez&author=C%20Mamane&author=A%20Finckh&volume=38&publication_year=2020&pages=455-466&pmid=31770089&)
|
| 204 |
+
|
| 205 |
+
12. Oryoji K, et al. Shared epitope positivity is related to efficacy of abatacept in rheumatoid arthritis. Ann. Rheum Dis. 2018;77:1234–1236. doi: 10.1136/annrheumdis-2017-211430. [DOI](https://doi.org/10.1136/annrheumdis-2017-211430) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28830884/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Rheum%20Dis.&title=Shared%20epitope%20positivity%20is%20related%20to%20efficacy%20of%20abatacept%20in%20rheumatoid%20arthritis&author=K%20Oryoji&volume=77&publication_year=2018&pages=1234-1236&pmid=28830884&doi=10.1136/annrheumdis-2017-211430&)
|
| 206 |
+
|
| 207 |
+
13. Hirose W, et al. Impact of the HLA-DRB1 shared epitope on responses to treatment with tofacitinib or abatacept in patients with rheumatoid arthritis. Arthritis Res. Ther. 2021;23:228. doi: 10.1186/s13075-021-02612-w. [DOI](https://doi.org/10.1186/s13075-021-02612-w) | [PMC free article](/articles/PMC8407060/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34465391/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Res.%20Ther.&title=Impact%20of%20the%20HLA-DRB1%20shared%20epitope%20on%20responses%20to%20treatment%20with%20tofacitinib%20or%20abatacept%20in%20patients%20with%20rheumatoid%20arthritis&author=W%20Hirose&volume=23&publication_year=2021&pages=228&pmid=34465391&doi=10.1186/s13075-021-02612-w&)
|
| 208 |
+
|
| 209 |
+
14. Rigby W, et al. HLA-DRB1 risk alleles for RA are associated with differential clinical responsiveness to abatacept and adalimumab: data from a head-to-head, randomized, single-blind study in autoantibody-positive early RA. Arthritis Res. Ther. 2021;23:245. doi: 10.1186/s13075-021-02607-7. [DOI](https://doi.org/10.1186/s13075-021-02607-7) | [PMC free article](/articles/PMC8449494/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34537057/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arthritis%20Res.%20Ther.&title=HLA-DRB1%20risk%20alleles%20for%20RA%20are%20associated%20with%20differential%20clinical%20responsiveness%20to%20abatacept%20and%20adalimumab:%20data%20from%20a%20head-to-head,%20randomized,%20single-blind%20study%20in%20autoantibody-positive%20early%20RA&author=W%20Rigby&volume=23&publication_year=2021&pages=245&pmid=34537057&doi=10.1186/s13075-021-02607-7&)
|
| 210 |
+
|
| 211 |
+
15. Skapenko A, Smolen JS, Kavanaugh A, Arora V, Kupper H, Schulze-Koops H. Genetic markers associated with clinical and radiographic response in adalimumab plus methotrexate- or methotrexate-treated rheumatoid arthritis patients in OPTIMA. Clin. Exp. Rheumatol. 2019;37:783–790. [PubMed](https://pubmed.ncbi.nlm.nih.gov/30963994/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Exp.%20Rheumatol.&title=Genetic%20markers%20associated%20with%20clinical%20and%20radiographic%20response%20in%20adalimumab%20plus%20methotrexate-%20or%20methotrexate-treated%20rheumatoid%20arthritis%20patients%20in%20OPTIMA&author=A%20Skapenko&author=JS%20Smolen&author=A%20Kavanaugh&author=V%20Arora&author=H%20Kupper&volume=37&publication_year=2019&pages=783-790&pmid=30963994&)
|
| 212 |
+
|
| 213 |
+
16. Viatte S, et al. Association of HLA-DRB1 haplotypes with rheumatoid arthritis severity, mortality, and treatment response. JAMA. 2015;313:1645–1656. doi: 10.1001/jama.2015.3435. [DOI](https://doi.org/10.1001/jama.2015.3435) | [PMC free article](/articles/PMC4928097/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25919528/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=JAMA&title=Association%20of%20HLA-DRB1%20haplotypes%20with%20rheumatoid%20arthritis%20severity,%20mortality,%20and%20treatment%20response&author=S%20Viatte&volume=313&publication_year=2015&pages=1645-1656&pmid=25919528&doi=10.1001/jama.2015.3435&)
|
| 214 |
+
|
| 215 |
+
17. Robinson J, Halliwell JA, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2013;41:D1222–1227. doi: 10.1093/nar/gks949. [DOI](https://doi.org/10.1093/nar/gks949) | [PMC free article](/articles/PMC3531221/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23080122/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nucleic%20Acids%20Res.&title=The%20IMGT/HLA%20database&author=J%20Robinson&author=JA%20Halliwell&author=H%20McWilliam&author=R%20Lopez&author=P%20Parham&volume=41&publication_year=2013&pages=D1222-1227&pmid=23080122&doi=10.1093/nar/gks949&)
|
test/texts/PMC10995391.md
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
test/texts/PMC11102648.md
ADDED
|
@@ -0,0 +1,306 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# The pharmacokinetics and pharmacodynamics of ibogaine in opioid use disorder patients
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Thomas Knuijver, Rob ter Heine, Arnt F A Schellekens, Paniz Heydari, Luc Lucas, Sjoerd Westra, Maarten Belgers, Toon van Oosteren, Robbert Jan Verkes, Cornelis Kramers
|
| 5 |
+
**Journal:** Journal of Psychopharmacology (Oxford, England)
|
| 6 |
+
**Date:** 2024 Mar 22
|
| 7 |
+
**DOI:** [10.1177/02698811241237873](https://doi.org/10.1177/02698811241237873)
|
| 8 |
+
**PMID:** 38519421
|
| 9 |
+
**PMCID:** PMC11102648
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11102648/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11102648/pdf/10.1177_02698811241237873.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11102648/pdf/10.1177_02698811241237873.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Objective::**
|
| 16 |
+
Ibogaine is a hallucinogenic drug that may be used to treat opioid use disorder (OUD). The relationships between pharmacokinetics (PKs) of ibogaine and its metabolites and their clinical effects on side effects and opioid withdrawal severity are unknown. We aimed to study these relationships in patients with OUD undergoing detoxification supported by ibogaine.
|
| 17 |
+
|
| 18 |
+
**Methods::**
|
| 19 |
+
The study was performed in 14 subjects with OUD. They received a single dose of 10mg/kg ibogaine hydrochloride. Plasma PKs of ibogaine, noribogaine, and noribogaine glucuronide were obtained during 24 h. Cytochrome P450 isoenzyme 2D6 (CYP2D6) genotyping was performed. The PKs were analyzed by means of nonlinear mixed effects modeling and related with corrected QT interval (QTc) prolongation, cerebellar ataxia, and opioid withdrawal severity.
|
| 20 |
+
|
| 21 |
+
**Results::**
|
| 22 |
+
The PK of ibogaine were highly variable and significantly correlated to CYP2D6 genotype (p < 0.001). The basic clearance of ibogaine (at a CYP2D6 activity score (AS) of 0) was 0.82 L/h. This increased with 30.7 L/h for every point of AS. The relation between ibogaine plasma concentrations and QTc was best described by a sigmoid Emax model. Spearman correlations were significant (p < 0.03) for ibogaine but not noribogaine with QTc (p = 0.109) and cerebellar effects (p = 0.668); neither correlated with the severity of opioid withdrawal symptoms.
|
| 23 |
+
|
| 24 |
+
**Conclusions::**
|
| 25 |
+
The clearance of ibogaine is strongly related to CYPD2D6 genotype. Ibogaine cardiac side effects (QTc time) and cerebellar effects are most likely more driven by ibogaine rather than noribogaine. Future studies should aim at exploring lower doses and/or applying individualized dosing based on CYP2D6 genotype.
|
| 26 |
+
|
| 27 |
+
Keywords: Pharmacokinetics, pharmacodynamics, ibogaine, safety, opioid, CYP2D6, QTc
|
| 28 |
+
|
| 29 |
+
### Objective:
|
| 30 |
+
|
| 31 |
+
Ibogaine is a hallucinogenic drug that may be used to treat opioid use disorder (OUD). The relationships between pharmacokinetics (PKs) of ibogaine and its metabolites and their clinical effects on side effects and opioid withdrawal severity are unknown. We aimed to study these relationships in patients with OUD undergoing detoxification supported by ibogaine.
|
| 32 |
+
|
| 33 |
+
### Methods:
|
| 34 |
+
|
| 35 |
+
The study was performed in 14 subjects with OUD. They received a single dose of 10mg/kg ibogaine hydrochloride. Plasma PKs of ibogaine, noribogaine, and noribogaine glucuronide were obtained during 24 h. Cytochrome P450 isoenzyme 2D6 (CYP2D6) genotyping was performed. The PKs were analyzed by means of nonlinear mixed effects modeling and related with corrected QT interval (QTc) prolongation, cerebellar ataxia, and opioid withdrawal severity.
|
| 36 |
+
|
| 37 |
+
### Results:
|
| 38 |
+
|
| 39 |
+
The PK of ibogaine were highly variable and significantly correlated to CYP2D6 genotype (*p*p < 0.001). The basic clearance of ibogaine (at a CYP2D6 activity score (AS) of 0) was 0.82 L/h. This increased with 30.7 L/h for every point of AS. The relation between ibogaine plasma concentrations and QTc was best described by a sigmoid *E*E_max_max model. Spearman correlations were significant (*p*p < 0.03) for ibogaine but not noribogaine with QTc (*p*p = 0.109) and cerebellar effects (*p*p = 0.668); neither correlated with the severity of opioid withdrawal symptoms.
|
| 40 |
+
|
| 41 |
+
### Conclusions:
|
| 42 |
+
|
| 43 |
+
The clearance of ibogaine is strongly related to CYPD2D6 genotype. Ibogaine cardiac side effects (QTc time) and cerebellar effects are most likely more driven by ibogaine rather than noribogaine. Future studies should aim at exploring lower doses and/or applying individualized dosing based on CYP2D6 genotype.
|
| 44 |
+
|
| 45 |
+
**Keywords:**Keywords: Pharmacokinetics, pharmacodynamics, ibogaine, safety, opioid, CYP2D6, QTc
|
| 46 |
+
|
| 47 |
+
## Introduction
|
| 48 |
+
|
| 49 |
+
Ibogaine is one of the active alkaloid compounds found in the rootbark of the plant *Tabernanthe iboga*Tabernanthe iboga, a perennial shrub found in West Africa. It is considered an oneirophrenic and is used in treatment of various types of addiction, including opioid use disorder (OUD) in various settings from private clinics to home treatment ([Rodríguez-Cano et al., 2023](#bibr42-02698811241237873)Rodríguez-Cano et al., 2023; [Schenberg et al., 2014](#bibr43-02698811241237873)Schenberg et al., 2014, [Kim et al., 2023](#bibr19-02698811241237873)Kim et al., 2023). It is most often used in the form of iboga, a mixture of alkaloids extracted from the rootbark of the plant ([Mash et al., 2001](#bibr32-02698811241237873)Mash et al., 2001). Ibogaine has shown some promise in mitigating opioid withdrawal and decreasing opioid craving and relapse after detoxification in non-controlled studies ([Brown and Alper, 2017](#bibr6-02698811241237873)Brown and Alper, 2017; [Malcolm et al., 2018](#bibr29-02698811241237873)Malcolm et al., 2018; [Mash et al., 2018](#bibr31-02698811241237873)Mash et al., 2018; [Noller et al., 2017](#bibr35-02698811241237873)Noller et al., 2017). Dosing regimens in these studies vary greatly and currently no dose-effect studies of ibogaine exist.
|
| 50 |
+
|
| 51 |
+
Beyond the potential therapeutic effects of ibogaine, there are major concerns regarding its safety, especially because of cardiac side effects ([Knuijver et al., 2021](#bibr20-02698811241237873)Knuijver et al., 2021; [Koenig and Hilber, 2015](#bibr22-02698811241237873)Koenig and Hilber, 2015; [Koenig et al., 2013](#bibr24-02698811241237873)Koenig et al., 2013, [2014](#bibr23-02698811241237873)2014; [Litjens and Brunt, 2016](#bibr25-02698811241237873)Litjens and Brunt, 2016; [Paling et al., 2012](#bibr40-02698811241237873)Paling et al., 2012; [Thurner et al., 2014](#bibr47-02698811241237873)Thurner et al., 2014; [Vlaanderen et al., 2014](#bibr48-02698811241237873)Vlaanderen et al., 2014). Ibogaine and its active metabolite noribogaine are both known to prolong the corrected QT interval (QTc) interval, which is a risk factor for torsades des pointes, a life-threatening dysrhythmia ([Knuijver et al., 2021](#bibr20-02698811241237873)Knuijver et al., 2021; [Koenig and Hilber, 2015](#bibr22-02698811241237873)Koenig and Hilber, 2015; [Koenig et al., 2014](#bibr23-02698811241237873)Koenig et al., 2014; [Thurner et al., 2014](#bibr47-02698811241237873)Thurner et al., 2014). Data regarding ibogaine effects on QTc duration is currently limited to clinical observations of QTc prolongation after ibogaine ingestion without testing an exposure-response relationship([Grogan et al., 2019](#bibr15-02698811241237873)Grogan et al., 2019; [Knuijver et al., 2021](#bibr20-02698811241237873)Knuijver et al., 2021; [Koenig and Hilber, 2015](#bibr22-02698811241237873)Koenig and Hilber, 2015). Furthermore, animal studies have shown that ibogaine causes ataxia, and at very high doses is neurotoxic to cerebellar Purkinje cells ([Belgers et al., 2016](#bibr4-02698811241237873)Belgers et al., 2016; [Molinari et al., 1996](#bibr34-02698811241237873)Molinari et al., 1996; [O’Hearn and Molliver, 1993](#bibr36-02698811241237873)O’Hearn and Molliver, 1993). In vivo ibogaine has been shown to produce a reversible clinical cerebellar ataxia([Knuijver et al., 2021](#bibr20-02698811241237873)Knuijver et al., 2021; [Mash et al., 2018](#bibr31-02698811241237873)Mash et al., 2018). It is not known whether the cerebellar effects are due to ibogaine itself or its metabolites.
|
| 52 |
+
|
| 53 |
+
Ibogaine is metabolized into noribogaine by the cytochrome P450 isoenzyme 2D6 (CYP2D6). Noribogaine is then glucuronidated to noribogaineglucuronide (NIG) ([Obach et al., 1998](#bibr37-02698811241237873)Obach et al., 1998). The activity of CYPD2D6 is known to vary between individuals, in particular due to genetic polymorphisms ([Marez et al., 1997](#bibr30-02698811241237873)Marez et al., 1997). The impact of CYP2D6 genotype on the pharmacokinetics (PKs) of ibogaine and its metabolites has not yet been fully elucidated. This knowledge is, however, pivotal to help create safe dosing regimens in future trials for the development of ibogaine as a treatment for addiction.
|
| 54 |
+
|
| 55 |
+
This pharmacokinetic-pharmacodynamic (PKPD) study aimed to (1) investigate the effects of genetic variation in the CYP2D6 on ibogaine PKs, including effects on its main metabolites, and (2) explore the relationships between plasma levels of ibogaine and its main metabolites and the pharmacodynamic (PD) effects on opioid withdrawal severity, QTc prolongation, and ataxia.
|
| 56 |
+
|
| 57 |
+
## Methods
|
| 58 |
+
|
| 59 |
+
### Study design
|
| 60 |
+
|
| 61 |
+
We conducted an open-label study in patients with OUD in agonist treatment with methadone or buprenorphine to investigate the exposure-response relationship of ibogaine and its main metabolites. The study was conducted according to the World Medical Association Declaration of Helsinki and approved by the medical ethical committee “Commissie Mensgebonden Onderzoek” in Nijmegen (The Netherlands) under reference number 2014/081. All participants provided written informed consent. The EudraCT number of this study was 2014-000354-11 ([EudraCT, 2014](#bibr10-02698811241237873)EudraCT, 2014). Part of this study was to assess the safety profile of ibogaine in patients with OUD, which we previously reported. The details of the clinical study are described in detail elsewhere. For additional details on some of the procedures and outcomes used, for example, on withdrawal severity, we refer to this article. ([Knuijver et al., 2021](#bibr20-02698811241237873)Knuijver et al., 2021)
|
| 62 |
+
|
| 63 |
+
### Participants
|
| 64 |
+
|
| 65 |
+
The study was performed in 14 subjects with OUD. Exclusion criteria were a history of clinically significant cardiac disease (including ventricular fibrillation, long QT syndrome, history of syncope, QTc > 450 ms for men and >470 ms for women), serum potassium >5.0 mmol/l or <3.5 mmol/l, severe liver or renal dysfunction (estimated glomerular filtration rate < 30 ml/min/1.73 m^2^2), or pregnancy. Participants were not allowed to use QTc prolonging or CYP2D6-affecting medication, except for methadone prior to inclusion ([https://crediblemeds.org/](https://crediblemeds.org/)https://crediblemeds.org/; [Flockhart et al., 2014](#bibr11-02698811241237873)Flockhart et al., 2014). Patients with a history of major depressive or psychotic symptoms were also excluded.
|
| 66 |
+
|
| 67 |
+
### Measurements
|
| 68 |
+
|
| 69 |
+
Age, sex and opioid maintenance treatment, as well as the dose of morphine used in the 24 h prior to ibogaine administration were noted. Ibogaine and its metabolites noribogaine and NIG were quantified in plasma with a validated ultra-performance liquid chromatography-tandem mass spectrometry) method. This method was validated within a linear concentration range of 0.1–50 ng/mL for ibogaine and 0.1–250 ng/mL for noribogaine and NIG. CYPD2D6 polymorphisms were tested for by TaqMan® analysis ([Marez et al., 1997](#bibr30-02698811241237873)Marez et al., 1997). The determined genotype was used to calculate the CYP2D6 activity score (AS hereafter), according to [Gaedigk et al. (2008)](#bibr12-02698811241237873)Gaedigk et al. (2008) for further PK analysis. Automatic twelve lead ECG measurements were performed to assess QTc prolongation using a Philips Healthcare, multichannel TC50. QTc was calculated using Fridericia’s formula (RR/QT^1/3^1/3) to correct for the RR interval. In order to obtain a reliable estimation of the QT interval, the average of the QT durations in leads V5 and II was calculated, based on measures by two independent researchers by hand ([Knuijver et al., 2021](#bibr20-02698811241237873)Knuijver et al., 2021; [Postema and Wilde, 2014](#bibr41-02698811241237873)Postema and Wilde, 2014). Their findings were verified by an independent cardiologist ([Knuijver et al., 2021](#bibr20-02698811241237873)Knuijver et al., 2021; [Postema and Wilde, 2014](#bibr41-02698811241237873)Postema and Wilde, 2014). Cerebellar ataxia was assessed using the Scale for the Assessment and Rating of Ataxia (SARA), a structured clinical assessment of motor- and coordination skills related to the cerebellum, by a trained physician ([Schmitz-Hübsch et al., 2006](#bibr44-02698811241237873)Schmitz-Hübsch et al., 2006; [Weyer et al., 2007](#bibr49-02698811241237873)Weyer et al., 2007; [Yabe et al., 2008](#bibr50-02698811241237873)Yabe et al., 2008). The SARA has eight items with a maximum score of 40: gait (8), stance (6), sitting (4), speech (6), finger-chase test (4), nose-finger test (4), fast alternating movements (4), and a heel-to-shin test (4). The heel-to-shin test was performed while standing. Higher scores indicate worse performance ([Schmitz-Hübsch et al., 2006](#bibr44-02698811241237873)Schmitz-Hübsch et al., 2006, [2010](#bibr45-02698811241237873)2010; [Weyer et al., 2007](#bibr49-02698811241237873)Weyer et al., 2007; [Yabe et al., 2008](#bibr50-02698811241237873)Yabe et al., 2008). The SARA has been found reliable and consistent in scoring ataxia in several diseases([Schmitz-Hübsch et al., 2006](#bibr44-02698811241237873)Schmitz-Hübsch et al., 2006, [2010](#bibr45-02698811241237873)2010; [Weyer et al., 2007](#bibr49-02698811241237873)Weyer et al., 2007; [Yabe et al., 2008](#bibr50-02698811241237873)Yabe et al., 2008).
|
| 70 |
+
|
| 71 |
+
Withdrawal symptoms were measured using the Objective and Subjective Opioid Withdrawal Scales (OOWS and SOWS) ([Handelsman et al., 1987](#bibr16-02698811241237873)Handelsman et al., 1987). The OOWS is a reliable, standardized, and well-validated clinical observation tool that scores the presence of 12 opioid withdrawal symptoms over the last 10 min. The score represents the severity of withdrawal: 0–5 = none; 5–12 = mild; 13–24 = moderate; 25–36 = moderately severe; more than 36 = severe withdrawal. The SOWS lets subjects score 16 symptoms on a five-point scale, which then also gives a measure of the severity of opioid withdrawal: 1–10 = mild; 11–20 = moderate; 21–30 = severe.
|
| 72 |
+
|
| 73 |
+
### Study procedure
|
| 74 |
+
|
| 75 |
+
Subjects were admitted for 8 days during which they were detoxified from all other substances except tobacco. During these 8 days they were converted by titration to oral morphine in order to eliminate any QTc prolonging effects of methadone and to make the onset of withdrawal predictable. The last morphine administration was on the 9th day at 4:00 AM. Subjects then received ibogaine hydrochloride (denoted as ibogaine hereafter) 10 mg/kg orally, administered in a yoghurt mixture at 8:30 AM. For safety reasons, we chose a dosage in the lower range of doses administered in previous studies([Alper et al., 2000](#bibr1-02698811241237873)Alper et al., 2000; [Brown and Alper, 2018](#bibr7-02698811241237873)Brown and Alper, 2018; [Malcolm et al., 2018](#bibr29-02698811241237873)Malcolm et al., 2018; [Mash et al., 2001](#bibr32-02698811241237873)Mash et al., 2001, 2008; [Noller et al., 2017](#bibr35-02698811241237873)Noller et al., 2017; [Schenberg et al., 2014](#bibr43-02698811241237873)Schenberg et al., 2014; [Sheppard, 1994](#bibr46-02698811241237873)Sheppard, 1994).
|
| 76 |
+
|
| 77 |
+
GMP-grade ibogaine for human use (brand name Remogen) was obtained from Phytostan Enterprises (Montreal, Canada) ([inc. MefLEP, 2014](#bibr18-02698811241237873)inc. MefLEP, 2014). Purity was assessed by the manufacturer using a validated liquid chromatography assay with ultraviolet detection and confirmed by our pharmaceutical laboratory. Before ibogaine administration, subjects were administered 20mg of metoclopramide, to prevent nausea and vomiting and secure full ingestion. Patients were monitored for the next 24 h. Blood samples were obtained at 30 min before administration and then at 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, and 24 h after ingestion. An electrocardiogram was obtained every half hour for the first twelve hours. If prolongation of the QTc interval (>450 ms for men; >470 ms for women) continued after 24 h, ECG measurements were continued every hour; otherwise, an ECG measurement was performed every 4 h afterwards. After 24 h, a cardiologist assessed if cardiac monitoring should be continued. If during the first 24 h QTc exceeded 500 ms, participants received a magnesium bolus infusion of 10 mmol, followed by another 10 mmol of magnesium over the next 10 h, for myocardial stabilization. If withdrawal was too severe, opioid substitution therapy (OST) was resumed on request of the subject, with methadone only allowed after 24 h and with a QTc < 450 ms and OST was slowly titrated according to local treatment protocols.
|
| 78 |
+
|
| 79 |
+
The SARA, the OOWS, and SOWS were assessed at two, six, ten, and twenty-four hours after administration of ibogaine. They were not performed in the event a subject requested to return to OST, as the protocol allowed for subjects to return to OST unconditionally. Subjects were kept for observation for 5 days after ibogaine administration.
|
| 80 |
+
|
| 81 |
+
### Statistical analyses
|
| 82 |
+
|
| 83 |
+
#### Population PK analysis
|
| 84 |
+
|
| 85 |
+
The population PKPD analysis was performed by means of nonlinear mixed effects modeling using the software package NONMEM 7.4 (Icon, Dublin, Ireland). A sequential approach was employed. First, an integral population PK model for ibogaine, noribogaine, and NIG was developed. The CYP2D6 AS was investigated as a covariate for the clearance parameter describing the formation of noribogaine. Using the developed PK model, we derived the empirical Bayes estimates for ibogaine, noribogaine, and NIG plasma concentrations at the time of measurement of ECG, SARA, and OOWS/SOWS. Furthermore, the empirical Bayes individual estimates for time of maximum concentration (*T*T_max_max), maximum concentration (*C*C_max_max), and area under the concentration versus time curve (AUC) of ibogaine, noribogaine, and NIG were derived from the developed PK model.
|
| 86 |
+
|
| 87 |
+
#### PKPD analysis
|
| 88 |
+
|
| 89 |
+
To visually investigate the exposure-response relationship between the ibogaine and metabolite concentrations, individual hysteresis plots were inspected ([Louizos et al., 2014](#bibr27-02698811241237873)Louizos et al., 2014). The correlations of the PKs with the PDs were investigated using Spearman’s rank correlation coefficient. When a significant correlation was found, the interplay between PKs of ibogaine (metabolites) and QTc prolongation was analyzed by means of nonlinear mixed effects modeling. Details on the PKPD modeling can be found in the [Supplemental Material](https://journals.sagepub.com/doi/suppl/10.1177/20543581241228723)Supplemental Material.
|
| 90 |
+
|
| 91 |
+
## Results
|
| 92 |
+
|
| 93 |
+
Subject characteristics are shown in [Table 1](#table1-02698811241237873)Table 1. The observed PKs of ibogaine and its metabolites are presented in [Figure 1](#fig1-02698811241237873)Figure 1. The genotyping results for each individual in the study can be found in [Table S1 of the Supplemental Material](https://journals.sagepub.com/doi/suppl/10.1177/20543581241228723)Table S1 of the Supplemental Material, where the found CYP2D6 alleles, AS, and metabolizer status are presented per individual.
|
| 94 |
+
|
| 95 |
+
### Table 1.
|
| 96 |
+
|
| 97 |
+
Study population characteristics.
|
| 98 |
+
|
| 99 |
+
| Characteristics (n = 14) | |
|
| 100 |
+
| ------------------------ | - |
|
| 101 |
+
| Age (median; first and third percentile) | 48 (44–51) |
|
| 102 |
+
| Sex M/F | 12/2 |
|
| 103 |
+
| BMT/MMT | 2/12 |
|
| 104 |
+
| 24 h morphine need | 124 mg (60–240 mg) |
|
| 105 |
+
### Figure 1.
|
| 106 |
+
|
| 107 |
+

|
| 108 |
+
|
| 109 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11102648_10.1177_02698811241237873-fig1.jpg)
|
| 110 |
+
|
| 111 |
+
(a) Observed ibogaine PKs. (b) Observed noribogaine PKs. (c) Observed NIG PKs. NIG: noribogaineglucuronide; PK: pharmacokinetic.
|
| 112 |
+
|
| 113 |
+
### Population PK modeling
|
| 114 |
+
|
| 115 |
+
Empirical Bayes Estimates for the *C*C_max_max, *T*T_max_max, and AUC extrapolated to infinity of ibogaine, noribogaine and NIG are shown in [Table 2](#table2-02698811241237873)Table 2. As expected, the clearance of ibogaine to noribogaine was significantly (*p*p < 0.0001) associated with the CYP2D6 AS, shown in [Figure 2](#fig2-02698811241237873)Figure 2. The basic clearance (at an AS of 0) of ibogaine was estimated to be 0.82 L/h, but this increased to 30.7 L/h for every point of AS ([Figure 2](#fig2-02698811241237873)Figure 2 and [Supplemental Material](https://journals.sagepub.com/doi/suppl/10.1177/20543581241228723)Supplemental Material). Sex was not identified as a covariate for the PKs of ibogaine and its metabolites.
|
| 116 |
+
|
| 117 |
+
### Table 2.
|
| 118 |
+
|
| 119 |
+
PK parameters.
|
| 120 |
+
|
| 121 |
+
| | First quartile | Median | Third quartile |
|
| 122 |
+
| - | -------------- | ------ | -------------- |
|
| 123 |
+
| Cmax ibogaine (μM/L) | 4.12 | 4.77 | 6.14 |
|
| 124 |
+
| Tmax ibogaine (h) | 0.607 | 0.622 | 0.668 |
|
| 125 |
+
| Cmax noribogaine (μM/L) | 1.09 | 1.33 | 1.46 |
|
| 126 |
+
| Tmax noribogaine (h) | 5.93 | 7.57 | 10.4 |
|
| 127 |
+
| Cmax norbogaine glucuronide (μM/L) | 0.0173 | 0.0215 | 0.0350 |
|
| 128 |
+
| Tmax noribogaine glucuronide (h) | 8.22 | 8.90 | 11.7 |
|
| 129 |
+
| AUC ibogaine (μM × h/L) | 41.7 | 49.12 | 63.6 |
|
| 130 |
+
| AUC noribogaine (μM × h/L) | 36.3 | 60.4 | 73.4 |
|
| 131 |
+
| AUC noribogaine glucuronide (μM × h/L) | 0.772 | 0.917 | 1.50 |
|
| 132 |
+
### Figure 2.
|
| 133 |
+
|
| 134 |
+

|
| 135 |
+
|
| 136 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11102648_10.1177_02698811241237873-fig2.jpg)
|
| 137 |
+
|
| 138 |
+
CYP2D6 AS and ibogaine conversion to noribogaine. The dots represent the empirical Bayes estimates for clearance of ibogaine to noribogaine. The line represents the estimated relationship between CYP2D6 activity score and conversion of ibogaine to noribogaine. CYP2D6: Cytochrome P450 isoenzyme 2D6; AS: activity score.
|
| 139 |
+
|
| 140 |
+
### PKPD analysis
|
| 141 |
+
|
| 142 |
+
A total of 386 QTc measurements were performed. A modest, yet significant, Spearman rank correlation of 0.109 (*p*p < 0.05) between ibogaine and QTc was found ([Supplemental Table S4](https://journals.sagepub.com/doi/suppl/10.1177/20543581241228723)Supplemental Table S4). No hysteresis was observed for ibogaine; however, noribogaine and NIG showed a clockwise hysteresis curve (plots available in the [Supplemental Material; Figures S6–S8](https://journals.sagepub.com/doi/suppl/10.1177/20543581241228723)Supplemental Material; Figures S6–S8).
|
| 143 |
+
|
| 144 |
+
In the PKPD model describing the relationship between plasma concentrations of ibogaine and QTc prolongation, it was found that a sigmoid *E*E_max_max model best explained the relationship between QTc time and ibogaine concentrations. This means that the QTc prolongation reaches a plateau. The maximum QTc prolongation was estimated to be 67.9 ms, with a relative standard of estimate (RSE) of 10.9%. The ibogaine concentration where the half-maximum QTc prolongation was observed (EC50) was estimated to be 0.195 µM (RSE 64.1%). No effect of sex was found on this relationship between plasma concentrations and QTc prolongation. Notably, the majority of ibogaine concentrations were above this EC50, explaining why QTc prolongation quickly reached a plateau after administration. An in-depth description of the PKPD analysis can be found in the [Supplemental Material](https://journals.sagepub.com/doi/suppl/10.1177/20543581241228723)Supplemental Material.
|
| 145 |
+
|
| 146 |
+
Upon visual inspection of the SARA scores against plasma levels of ibogaine, noribogaine and NIG, we did not observe clear evidence of hysteresis. These SARA measurements showed a strong Spearman correlation of 0.67 (*p*p < 0.01, two-tailed; [Figure 3](#fig3-02698811241237873)Figure 3) with ibogaine concentrations but not with its metabolites. All correlations are shown in [Supplemental Table S4](https://journals.sagepub.com/doi/suppl/10.1177/20543581241228723)Supplemental Table S4.
|
| 147 |
+
|
| 148 |
+
### Figure 3.
|
| 149 |
+
|
| 150 |
+

|
| 151 |
+
|
| 152 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11102648_10.1177_02698811241237873-fig3.jpg)
|
| 153 |
+
|
| 154 |
+
SARA score versus ibogaine concentration. The correlation between ibogaine concentration and SARA score using Spearman’s rank correlation was significant (Spearman’s ρ = 0.668, p < 0.01). SARA: Scale for the Assessment and Rating of Ataxia.
|
| 155 |
+
|
| 156 |
+
A total of 54 SOWS and OOWS measurements were performed. No significant correlations were seen between plasma levels and both OOWS and SOWS ([Supplemental Table S4](https://journals.sagepub.com/doi/suppl/10.1177/20543581241228723)Supplemental Table S4). Again, no clear hysteresis was observed.
|
| 157 |
+
|
| 158 |
+
## Discussion
|
| 159 |
+
|
| 160 |
+
In this study, we set out to investigate the PKs of ibogaine, noribogaine, and NIG and quantify the effect of CYP2D6 status on these PKs after a single dose of 10 mg/kg ibogaine hydrochloride in OUD patients. Furthermore, we investigated the relationship between plasma levels of ibogaine and its metabolites with QTc prolongation, ataxia and withdrawal symptoms. We found that CYP2D6 genotype is a major determinant of ibogaine metabolism, with more than a 10-fold difference in clearance across the CYP2D6 ASs from 0 to 3. This indicates that CYP2D6-based dosing should be performed to assure equal exposure when dosing ibogaine. The *C*C_max_maxs of ibogaine and noribogaine were in agreement with previous studies ([Glue et al., 2015](#bibr14-02698811241237873)Glue et al., 2015; [Maciulaitis et al., 2008](#bibr28-02698811241237873)Maciulaitis et al., 2008; [Mash et al., 2000](#bibr33-02698811241237873)Mash et al., 2000, [2001](#bibr32-02698811241237873)2001) and more than ten-fold higher than the EC50. Consequently, to relevantly reduce the risk of QTc-prolongation, a more than ten-fold reduction in dose is required. As it stands, it is unknown whether such a low dose has any clinically meaningful effects.
|
| 161 |
+
|
| 162 |
+
QTc showed an anticlockwise hysteresis when plotted against ibogaine plasma concentrations, whereas noribogaine showed clockwise hysteresis. This implies that QTc prolongation occurs before noribogaine exposure increases. Furthermore, QTc correlated with ibogaine concentrations but not with noribogaine or NIG concentrations. We, therefore, postulate that QTc prolongation is mainly driven by ibogaine rather than its metabolites. This is in line with a previous clinical study of noribogaine, which exhibited a concentration-dependent QTc prolongation of 10–50 ms in the studied dose ([Glue et al., 2016](#bibr13-02698811241237873)Glue et al., 2016). However, the plasma noribogaine concentrations in this study were 10- to 100-fold higher than in our study, indicating that the low noribogaine concentrations in our study only marginally impact QTc. As shown by others ([Alper et al., 2012](#bibr2-02698811241237873)Alper et al., 2012; [Ona et al., 2022](#bibr38-02698811241237873)Ona et al., 2022), it should be noted that the low noribogaine concentrations in our population may still have contributed to QTc prolongation. However, as both the parent and metabolites are simultaneously present in the systemic circulation after administration of ibogaine, we could not distinguish the separate effects of both compounds on QTc prolongation. The suggested lower potential of noribogaine to cause cardiotoxicity may point to further clinical development of noribogaine for the same indication, As proposed by [Glue et al. (2016)](#bibr13-02698811241237873)Glue et al. (2016).
|
| 163 |
+
|
| 164 |
+
“It should be noted that tobacco smoking is likely to slightly prolong the QTc time ([Özdemir and Sökmen, 2020](#bibr39-02698811241237873)Özdemir and Sökmen, 2020). Considering the high incidence of tobacco smoking in OUD patients ([Clemmey et al., 1997](#bibr8-02698811241237873)Clemmey et al., 1997), it may be postulated that in patients who do not smoke, the QTc prolonging effects might be a little bit less pronounced. Smoking was allowed during the study. We did not observe an increase in cigarette consumption prior to or during ibogaine treatment, nor was a major increased QTc time at baseline observed. This makes a major confounding effect of nicotine use on the observed QTc prolongation after ibogaine ingestion unlikely. Our study investigated the effects of ibogaine in a real-world population where tobacco use is frequent, and we showed that ibogaine should be administered with caution in this population.
|
| 165 |
+
|
| 166 |
+
Ataxia was better correlated with ibogaine PKs than with noribogaine, suggesting ibogaine to be mainly responsible for the observed ataxia. This is in line with previous studies in rats, where it was observed that noribogaine does not induce ataxia in contrast to ibogaine ([Baumann et al., 2001](#bibr3-02698811241237873)Baumann et al., 2001). As morphine administration was ceased before treatment, withdrawal in the form of tremors may have had some effect on the ataxia measurement. However, only mild withdrawal was observed during ibogaine treatment.
|
| 167 |
+
|
| 168 |
+
We did not observe a correlation between the PKs of ibogaine or its metabolites and opioid withdrawal. Based on our dosing regimen, withdrawal is to be expected within 4–6 h after cessation of morphine use. We administered ibogaine 4 h after the last morphine and during the first 24 h withdrawal did not occur for most subjects. We, therefore, conclude that ibogaine is at least in part capable of mitigating withdrawal during the first 24 h and probably beyond. It may be postulated that in our study a plateau effect was reached and that a lower systemic exposure may have the potential to also prevent withdrawal symptoms. As it stands, the lowest effective exposure in relation to withdrawal is not yet known. This knowledge may facilitate the development of safer ibogaine dosing.
|
| 169 |
+
|
| 170 |
+
Our results should be seen in light of several considerations. It may be debated that the sample size of our study was relatively small, yet in line with most clinical studies with ibogaine ([Köck et al., 2022](#bibr21-02698811241237873)Köck et al., 2022). The sample size should be seen in context of the learning phase of our PKPD study. Although, from a statistical point of view, a larger sample size is always desirable, we were able to identify PKPD relationships in our study. If findings from our study are to be confirmed, our results may serve to design a trial and select an appropriate sample size.
|
| 171 |
+
|
| 172 |
+
Furthermore, reliably measuring the QTc interval is notoriously difficult. A 1 mm measuring error results in 40 ms of over- or underestimation of the QT interval. Current gold-standard research includes a full day of QTc measurement before administration to provide an individual baseline QTc correction, which we did not apply. A limited number of SARA, OOWS, and SOWS measurements were available, potentially reducing the likelihood of identifying hysteresis. This might have affected the ability to show the role of different compounds in our observations. Especially when it comes to mitigation of withdrawal symptoms, a different set-up with more frequent and longer measurement of OOWS/SOWS and measurement of morphine plasma levels might have produced a better understanding of the dose-effect relationship.
|
| 173 |
+
|
| 174 |
+
Some PKPD interactions with other drugs may have taken place in our study, which might have influenced our results. In theory, through competitive inhibition, metoclopramide might act as a CYP2D6 inhibitor. There is some in vitro evidence that metoclopramide is a reversible and competitive inhibitor, but not inactivator, of this metabolic enzyme ([Livezey et al., 2014](#bibr26-02698811241237873)Livezey et al., 2014), yet CYP2D6 inactivation was not observed in relevant concentrations. Concentrations at which only limited reversible CYP2D6 inhibition occurred were far above systemic concentrations that reached clinically relevant doses of metoclopramide of approximately 0.1–0.5 μM ([Bernardo-Escudero et al., 2011](#bibr5-02698811241237873)Bernardo-Escudero et al., 2011). The fact that a clear relationship was present between CYP2D6 AS and ibogaine PKs shows that at least CYP2D6 was not completely inhibited.
|
| 175 |
+
|
| 176 |
+
Both metoclopramide and methadone are listed in the Arizona QT-drugs list as QT-prolonging agents and may have influenced the duration of the QT ([https://crediblemeds.org/](https://crediblemeds.org/)https://crediblemeds.org/). The effect of metoclopramide on QT is not well known, as data suggest an effect on the QT variance and an increase in the length of the QT interval with increasing RR intervals.([Ellidokuz and Kaya, 2003](#bibr9-02698811241237873)Ellidokuz and Kaya, 2003) Higher plasma metoclopramide levels at the start versus end may confound the hysteresis plots and correlation. We do not consider methadone a relevant factor for QTc prolongation, given the elimination half-life of methadone, the fact that baseline QTc was within normal range and the clear prolongation of QTc after ibogaine ingestion.
|
| 177 |
+
|
| 178 |
+
The PKs of ibogaine are strongly related to CYPD2D6 genotype, expressed as an AS. Consequently, as plasma concentrations of ibogaine correlate with QTc time, reduced CYP2D6 activity increases exposure to ibogaine and induces more pronounced (with a maximum effect) and prolonged QTc prolongation. Cerebellar effects are most probably a result of ibogaine and not of noribogaine. Notably, no relationship between PKs and withdrawal severity was found. This might indicate that the studied dose was in the upper plateau of the PKPD effect curve. Significantly lower doses, preferably individualized using CYP2D6 genotyping, are required to increase cardiac safety, and clinical effects of such doses should be studied to understand the clinical potential of ibogaine to prevent withdrawal symptoms, craving and relapse in patients with OUD.
|
| 179 |
+
|
| 180 |
+
## Supplemental Material
|
| 181 |
+
|
| 182 |
+
Supplemental material, sj-docx-1-jop-10.1177_02698811241237873 for The pharmacokinetics and pharmacodynamics of ibogaine in opioid use disorder patients by Thomas Knuijver, Rob ter Heine, Arnt F. A. Schellekens, Paniz Heydari, Luc Lucas, Sjoerd Westra, Maarten Belgers, Toon van Oosteren, Robbert Jan Verkes and Cornelis Kramers in Journal of Psychopharmacology
|
| 183 |
+
|
| 184 |
+
Supplemental material, sj-docx-1-jop-10.1177_02698811241237873 for The pharmacokinetics and pharmacodynamics of ibogaine in opioid use disorder patients by Thomas Knuijver, Rob ter Heine, Arnt F. A. Schellekens, Paniz Heydari, Luc Lucas, Sjoerd Westra, Maarten Belgers, Toon van Oosteren, Robbert Jan Verkes and Cornelis Kramers in Journal of Psychopharmacology
|
| 185 |
+
|
| 186 |
+
## Acknowledgments
|
| 187 |
+
|
| 188 |
+
For aid in conducting our research, we would like to thank Sanders Dekkers, Jan Leijtens, Saskia Delis, Georghe Pop and Anne Loes In der Maur, NISPA and the nursing staff of IrisZorg and Radboudumc.
|
| 189 |
+
|
| 190 |
+
## Footnotes
|
| 191 |
+
|
| 192 |
+
## Associated Data
|
| 193 |
+
|
| 194 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 195 |
+
|
| 196 |
+
### Supplementary Materials
|
| 197 |
+
|
| 198 |
+
Supplemental material, sj-docx-1-jop-10.1177_02698811241237873 for The pharmacokinetics and pharmacodynamics of ibogaine in opioid use disorder patients by Thomas Knuijver, Rob ter Heine, Arnt F. A. Schellekens, Paniz Heydari, Luc Lucas, Sjoerd Westra, Maarten Belgers, Toon van Oosteren, Robbert Jan Verkes and Cornelis Kramers in Journal of Psychopharmacology
|
| 199 |
+
|
| 200 |
+
### Supplementary Materials
|
| 201 |
+
|
| 202 |
+
Supplemental material, sj-docx-1-jop-10.1177_02698811241237873 for The pharmacokinetics and pharmacodynamics of ibogaine in opioid use disorder patients by Thomas Knuijver, Rob ter Heine, Arnt F. A. Schellekens, Paniz Heydari, Luc Lucas, Sjoerd Westra, Maarten Belgers, Toon van Oosteren, Robbert Jan Verkes and Cornelis Kramers in Journal of Psychopharmacology
|
| 203 |
+
|
| 204 |
+
Supplemental material, sj-docx-1-jop-10.1177_02698811241237873 for The pharmacokinetics and pharmacodynamics of ibogaine in opioid use disorder patients by Thomas Knuijver, Rob ter Heine, Arnt F. A. Schellekens, Paniz Heydari, Luc Lucas, Sjoerd Westra, Maarten Belgers, Toon van Oosteren, Robbert Jan Verkes and Cornelis Kramers in Journal of Psychopharmacology
|
| 205 |
+
|
| 206 |
+
## References
|
| 207 |
+
|
| 208 |
+
1. Alper KR, Lotsof HS, Frenken GM, et al. (2000) Ibogaine in acute opioid withdrawal. An open label case series. Ann N Y Acad Sci 909: 257–259. [DOI](https://doi.org/10.1111/j.1749-6632.2000.tb06687.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10911935/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20N%20Y%20Acad%20Sci&title=Ibogaine%20in%20acute%20opioid%20withdrawal.%20An%20open%20label%20case%20series&author=KR%20Alper&author=HS%20Lotsof&author=GM%20Frenken&volume=909&publication_year=2000&pages=257-259&pmid=10911935&doi=10.1111/j.1749-6632.2000.tb06687.x&)
|
| 209 |
+
|
| 210 |
+
2. Alper KR, Stajić M, Gill JR. (2012) Fatalities temporally associated with the ingestion of ibogaine. Journal of Forensic Sciences 57: 398–412. [DOI](https://doi.org/10.1111/j.1556-4029.2011.02008.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22268458/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Journal%20of%20Forensic%20Sciences&title=Fatalities%20temporally%20associated%20with%20the%20ingestion%20of%20ibogaine&author=KR%20Alper&author=M%20Staji%C4%87&author=JR%20Gill&volume=57&publication_year=2012&pages=398-412&pmid=22268458&doi=10.1111/j.1556-4029.2011.02008.x&)
|
| 211 |
+
|
| 212 |
+
3. Baumann MH, Rothman RB, Pablo JP, et al. (2001) In vivo neurobiological effects of ibogaine and its O-desmethyl metabolite, 12-hydroxyibogamine (noribogaine), in rats. Journal of Pharmacology and Experimental Therapeutics 297: 531–539. [PubMed](https://pubmed.ncbi.nlm.nih.gov/11303040/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Journal%20of%20Pharmacology%20and%20Experimental%20Therapeutics&title=In%20vivo%20neurobiological%20effects%20of%20ibogaine%20and%20its%20O-desmethyl%20metabolite,%2012-hydroxyibogamine%20(noribogaine),%20in%20rats&author=MH%20Baumann&author=RB%20Rothman&author=JP%20Pablo&volume=297&publication_year=2001&pages=531-539&pmid=11303040&)
|
| 213 |
+
|
| 214 |
+
4. Belgers M, Leenaars M, Homberg JR, et al. (2016) Ibogaine and addiction in the animal model, a systematic review and meta-analysis. Transl Psychiatry 6: e826. [DOI](https://doi.org/10.1038/tp.2016.71) | [PMC free article](/articles/PMC5545647/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27244235/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transl%20Psychiatry&title=Ibogaine%20and%20addiction%20in%20the%20animal%20model,%20a%20systematic%20review%20and%20meta-analysis&author=M%20Belgers&author=M%20Leenaars&author=JR%20Homberg&volume=6&publication_year=2016&pmid=27244235&doi=10.1038/tp.2016.71&)
|
| 215 |
+
|
| 216 |
+
5. Bernardo-Escudero R, Alonso-Campero R, de Jesús Francisco-Doce MT, et al. (2011) Comparison of the pharmacokinetics of a new 15-mg modified-release tablet formulation of metoclopramide versus a 10-mg immediate-release tablet: A single-and multiple-dose, randomized, open-label, parallel-group study in healthy Mexican male volunteers. Clinical Therapeutics 33: 630–643. [DOI](https://doi.org/10.1016/j.clinthera.2011.04.016) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21665047/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clinical%20Therapeutics&title=Comparison%20of%20the%20pharmacokinetics%20of%20a%20new%2015-mg%20modified-release%20tablet%20formulation%20of%20metoclopramide%20versus%20a%2010-mg%20immediate-release%20tablet:%20A%20single-and%20multiple-dose,%20randomized,%20open-label,%20parallel-group%20study%20in%20healthy%20Mexican%20male%20volunteers&author=R%20Bernardo-Escudero&author=R%20Alonso-Campero&author=MT%20de%20Jes%C3%BAs%20Francisco-Doce&volume=33&publication_year=2011&pages=630-643&pmid=21665047&doi=10.1016/j.clinthera.2011.04.016&)
|
| 217 |
+
|
| 218 |
+
6. Brown TK, Alper K. (2017) Treatment of opioid use disorder with ibogaine: Detoxification and drug use outcomes. Am J Drug Alcohol Abuse 44: 24–36. DOI: 10.1080/00952990.2017.13208021-13. [DOI](https://doi.org/10.1080/00952990.2017.13208021-13.) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28541119/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Drug%20Alcohol%20Abuse&title=Treatment%20of%20opioid%20use%20disorder%20with%20ibogaine:%20Detoxification%20and%20drug%20use%20outcomes&author=TK%20Brown&author=K%20Alper&volume=44&publication_year=2017&pages=24-36&pmid=28541119&doi=10.1080/00952990.2017.13208021-13.&)
|
| 219 |
+
|
| 220 |
+
7. Brown TK, Alper K. (2018) Treatment of opioid use disorder with ibogaine: detoxification and drug use outcomes. Am J Drug Alcohol Abuse 44: 24–36. [DOI](https://doi.org/10.1080/00952990.2017.1320802) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28541119/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Drug%20Alcohol%20Abuse&title=Treatment%20of%20opioid%20use%20disorder%20with%20ibogaine:%20detoxification%20and%20drug%20use%20outcomes&author=TK%20Brown&author=K%20Alper&volume=44&publication_year=2018&pages=24-36&pmid=28541119&doi=10.1080/00952990.2017.1320802&)
|
| 221 |
+
|
| 222 |
+
8. Clemmey P, Brooner R, Chutuape MA, et al. (1997) Smoking habits and attitudes in a methadone maintenance treatment population. Drug Alcohol Depend 44: 123–132. [DOI](https://doi.org/10.1016/s0376-8716(96)01331-2) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9088784/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Alcohol%20Depend&title=Smoking%20habits%20and%20attitudes%20in%20a%20methadone%20maintenance%20treatment%20population&author=P%20Clemmey&author=R%20Brooner&author=MA%20Chutuape&volume=44&publication_year=1997&pages=123-132&pmid=9088784&doi=10.1016/s0376-8716(96)01331-2&)
|
| 223 |
+
|
| 224 |
+
9. Ellidokuz E, Kaya D. (2003) The effect of metoclopramide on QT dynamicity: double-blind, placebo-controlled, cross-over study in healthy male volunteers. Aliment Pharmacol Ther 18(1): 151-155. [DOI](https://doi.org/10.1046/j.1365-2036.2003.01641.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12848637/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Aliment%20Pharmacol%20Ther&title=The%20effect%20of%20metoclopramide%20on%20QT%20dynamicity:%20double-blind,%20placebo-controlled,%20cross-over%20study%20in%20healthy%20male%20volunteers&author=E%20Ellidokuz&author=D%20Kaya&volume=18&issue=1&publication_year=2003&pages=151-155&pmid=12848637&doi=10.1046/j.1365-2036.2003.01641.x&)
|
| 225 |
+
|
| 226 |
+
10. EudraCT (2014) The Efficacy of Ibogaine in the Treatment of Addiction; an open label, single fixed dose pilot-study of the efficacy of ibogaine in opioid-dependent subjects– EudraCT number 2014-000354-11. Available at: https://www.clinicaltrialsregister.eu/ctr-search/search?query=2014-000354-11. [https://www.clinicaltrialsregister.eu/ctr-search/search?query=2014-000354-11](https://www.clinicaltrialsregister.eu/ctr-search/search?query=2014-000354-11)
|
| 227 |
+
|
| 228 |
+
11. Flockhart DA, Thacker D, McDonald C, et al. (2014) The Flockhart Cytochrome P450 Drug-Drug Interaction Table. Division of Clinical Pharmacology, Indiana University School of Medicine. Updated 2021. Available at: https://drug-interactions.medicine.iu.edu (accessed 2014). [https://drug-interactions.medicine.iu.edu](https://drug-interactions.medicine.iu.edu)
|
| 229 |
+
|
| 230 |
+
12. Gaedigk A, Simon SD, Pearce RE, et al. (2008) The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther 83(2): 234-242. [DOI](https://doi.org/10.1038/sj.clpt.6100406) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17971818/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=The%20CYP2D6%20activity%20score:%20translating%20genotype%20information%20into%20a%20qualitative%20measure%20of%20phenotype&author=A%20Gaedigk&author=SD%20Simon&author=RE%20Pearce&volume=83&issue=2&publication_year=2008&pages=234-242&pmid=17971818&doi=10.1038/sj.clpt.6100406&)
|
| 231 |
+
|
| 232 |
+
13. Glue P, Cape G, Tunnicliff D, et al. (2016) Ascending single-dose, double-blind, placebo-controlled safety study of noribogaine in opioid-dependent patients. Clinical pharmacology in drug development 5(6): 460-468. [DOI](https://doi.org/10.1002/cpdd.254) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27870477/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clinical%20pharmacology%20in%20drug%20development&title=Ascending%20single-dose,%20double-blind,%20placebo-controlled%20safety%20study%20of%20noribogaine%20in%20opioid-dependent%20patients&author=P%20Glue&author=G%20Cape&author=D%20Tunnicliff&volume=5&issue=6&publication_year=2016&pages=460-468&pmid=27870477&doi=10.1002/cpdd.254&)
|
| 233 |
+
|
| 234 |
+
14. Glue P, Winter H, Garbe K, et al. (2015) Influence of CYP2D6 activity on the pharmacokinetics and pharmacodynamics of a single 20 mg dose of ibogaine in healthy volunteers. J Clin Pharmacol 55(6): 680-687. [DOI](https://doi.org/10.1002/jcph.471) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25651476/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Clin%20Pharmacol&title=Influence%20of%20CYP2D6%20activity%20on%20the%20pharmacokinetics%20and%20pharmacodynamics%20of%20a%20single%2020%20mg%20dose%20of%20ibogaine%20in%20healthy%20volunteers&author=P%20Glue&author=H%20Winter&author=K%20Garbe&volume=55&issue=6&publication_year=2015&pages=680-687&pmid=25651476&doi=10.1002/jcph.471&)
|
| 235 |
+
|
| 236 |
+
15. Grogan J, Gerona R, Snow JW, et al. (2019) Ibogaine Consumption With Seizure-Like Episodes, QTc-Prolongation, and Captured Cardiac Dysrhythmias. J Emerg Med 57(4): e99-e104. [DOI](https://doi.org/10.1016/j.jemermed.2019.06.052) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31630892/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Emerg%20Med&title=Ibogaine%20Consumption%20With%20Seizure-Like%20Episodes,%20QTc-Prolongation,%20and%20Captured%20Cardiac%20Dysrhythmias&author=J%20Grogan&author=R%20Gerona&author=JW%20Snow&volume=57&issue=4&publication_year=2019&pmid=31630892&doi=10.1016/j.jemermed.2019.06.052&)
|
| 237 |
+
|
| 238 |
+
16. Handelsman L, Cochrane KJ, Aronson MJ, et al. (1987) Two new rating scales for opiate withdrawal. Am J Drug Alcohol Abuse 13(3): 293-308. https://crediblemeds.org/TheArizonaQTdrugslist. [https://crediblemeds.org/TheArizonaQTdrugslist](https://crediblemeds.org/TheArizonaQTdrugslist) | [DOI](https://doi.org/10.3109/00952998709001515) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/3687892/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Drug%20Alcohol%20Abuse&title=Two%20new%20rating%20scales%20for%20opiate%20withdrawal&author=L%20Handelsman&author=KJ%20Cochrane&author=MJ%20Aronson&volume=13&issue=3&publication_year=1987&pages=293-308&pmid=3687892&doi=10.3109/00952998709001515&)
|
| 239 |
+
|
| 240 |
+
17. IBM Micromedex® (electronic version). IBM Watson Health, Greenwood Village, Colorado, USA. Available at: https://www.micromedexsolutions.com/ (cited: month/day/year). [https://www.micromedexsolutions.com/](https://www.micromedexsolutions.com/)
|
| 241 |
+
|
| 242 |
+
18. inc. MefLEP (2014) Ibogaïne Alliance website with information Remogen Ibogaine. https://www.ibogainealliance.org/conferences/vancouver-2012/archive/bob-sisko/. [https://www.ibogainealliance.org/conferences/vancouver-2012/archive/bob-sisko/](https://www.ibogainealliance.org/conferences/vancouver-2012/archive/bob-sisko/)
|
| 243 |
+
|
| 244 |
+
19. Kim S, Chen J, Cheng T, et al. (2023) PubChem 2023 update. Nucleic Acids Res. 51(D1):D1373–D1380. National Center for Biotechnology Information: PubChem Compound Database; CID=197060. Available at: https://pubchem.ncbi.nlm.nih.gov/compound/197060 (accessed 16 October 2018). [https://pubchem.ncbi.nlm.nih.gov/compound/197060](https://pubchem.ncbi.nlm.nih.gov/compound/197060) | [DOI](https://doi.org/10.1093/nar/gkac956) | [PMC free article](/articles/PMC9825602/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36305812/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nucleic%20Acids%20Res&title=PubChem%202023%20update&author=S%20Kim&author=J%20Chen&author=T%20Cheng&publication_year=2023&pmid=36305812&doi=10.1093/nar/gkac956&)
|
| 245 |
+
|
| 246 |
+
20. Knuijver T, Schellekens A, Belgers M, et al. (2021) Safety of ibogaine administration in detoxification of opioid-dependent individuals: a descriptive open-label observational study. Addiction. Epub ahead of print 20210223. DOI: 10.1111/add.15448. [DOI](https://doi.org/10.1111/add.15448.) | [PMC free article](/articles/PMC9292417/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33620733/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Addiction&title=Safety%20of%20ibogaine%20administration%20in%20detoxification%20of%20opioid-dependent%20individuals:%20a%20descriptive%20open-label%20observational%20study&author=T%20Knuijver&author=A%20Schellekens&author=M%20Belgers&publication_year=2021&pmid=33620733&doi=10.1111/add.15448.&)
|
| 247 |
+
|
| 248 |
+
21. Köck P, Froelich K, Walter M, et al. (2022) A systematic literature review of clinical trials and therapeutic applications of ibogaine. Journal of Substance Abuse Treatment 138: 108717. [DOI](https://doi.org/10.1016/j.jsat.2021.108717) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35012793/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Journal%20of%20Substance%20Abuse%20Treatment&title=A%20systematic%20literature%20review%20of%20clinical%20trials%20and%20therapeutic%20applications%20of%20ibogaine&author=P%20K%C3%B6ck&author=K%20Froelich&author=M%20Walter&volume=138&publication_year=2022&pages=108717&pmid=35012793&doi=10.1016/j.jsat.2021.108717&)
|
| 249 |
+
|
| 250 |
+
22. Koenig X, Hilber K. (2015) The anti-addiction drug ibogaine and the heart: a delicate relation. Molecules 20(2): 2208-2228. [DOI](https://doi.org/10.3390/molecules20022208) | [PMC free article](/articles/PMC4382526/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25642835/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Molecules&title=The%20anti-addiction%20drug%20ibogaine%20and%20the%20heart:%20a%20delicate%20relation&author=X%20Koenig&author=K%20Hilber&volume=20&issue=2&publication_year=2015&pages=2208-2228&pmid=25642835&doi=10.3390/molecules20022208&)
|
| 251 |
+
|
| 252 |
+
23. Koenig X, Kovar M, Boehm S, et al. (2014) Anti-addiction drug ibogaine inhibits hERG channels: a cardiac arrhythmia risk. Addict Biol 19(2): 237-239. [DOI](https://doi.org/10.1111/j.1369-1600.2012.00447.x) | [PMC free article](/articles/PMC4888945/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22458604/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Addict%20Biol&title=Anti-addiction%20drug%20ibogaine%20inhibits%20hERG%20channels:%20a%20cardiac%20arrhythmia%20risk&author=X%20Koenig&author=M%20Kovar&author=S%20Boehm&volume=19&issue=2&publication_year=2014&pages=237-239&pmid=22458604&doi=10.1111/j.1369-1600.2012.00447.x&)
|
| 253 |
+
|
| 254 |
+
24. Koenig X, Kovar M, Rubi L, et al. (2013) Anti-addiction drug ibogaine inhibits voltage-gated ionic currents: a study to assess the drug’s cardiac ion channel profile. Toxicol Appl Pharmacol 273(2): 259-268. [DOI](https://doi.org/10.1016/j.taap.2013.05.012) | [PMC free article](/articles/PMC3853361/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23707769/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Toxicol%20Appl%20Pharmacol&title=Anti-addiction%20drug%20ibogaine%20inhibits%20voltage-gated%20ionic%20currents:%20a%20study%20to%20assess%20the%20drug%E2%80%99s%20cardiac%20ion%20channel%20profile&author=X%20Koenig&author=M%20Kovar&author=L%20Rubi&volume=273&issue=2&publication_year=2013&pages=259-268&pmid=23707769&doi=10.1016/j.taap.2013.05.012&)
|
| 255 |
+
|
| 256 |
+
25. Litjens RP, Brunt TM. (2016) How toxic is ibogaine? Clin Toxicol (Phila) 54(4): 297-302. [DOI](https://doi.org/10.3109/15563650.2016.1138226) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26807959/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Toxicol%20(Phila)&title=How%20toxic%20is%20ibogaine?&author=RP%20Litjens&author=TM%20Brunt&volume=54&issue=4&publication_year=2016&pages=297-302&pmid=26807959&doi=10.3109/15563650.2016.1138226&)
|
| 257 |
+
|
| 258 |
+
26. Livezey MR, Briggs ED, Bolles AK, et al. (2014) Metoclopramide is metabolized by CYP2D6 and is a reversible inhibitor, but not inactivator, of CYP2D6. Xenobiotica 44(4): 309-319. [DOI](https://doi.org/10.3109/00498254.2013.835885) | [PMC free article](/articles/PMC4059401/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24010633/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Xenobiotica&title=Metoclopramide%20is%20metabolized%20by%20CYP2D6%20and%20is%20a%20reversible%20inhibitor,%20but%20not%20inactivator,%20of%20CYP2D6&author=MR%20Livezey&author=ED%20Briggs&author=AK%20Bolles&volume=44&issue=4&publication_year=2014&pages=309-319&pmid=24010633&doi=10.3109/00498254.2013.835885&)
|
| 259 |
+
|
| 260 |
+
27. Louizos C, Yáñez JA, Forrest L, et al. (2014) Understanding the hysteresis loop conundrum in pharmacokinetic/pharmacodynamic relationships. Journal of pharmacy & pharmaceutical sciences: a publication of the Canadian Society for Pharmaceutical Sciences, Societe canadienne des sciences pharmaceutiques 17(1): 34. [PMC free article](/articles/PMC4332569/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24735761/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Journal%20of%20pharmacy%20&%20pharmaceutical%20sciences:%20a%20publication%20of%20the%20Canadian%20Society%20for%20Pharmaceutical%20Sciences,%20Societe%20canadienne%20des%20sciences%20pharmaceutiques&title=Understanding%20the%20hysteresis%20loop%20conundrum%20in%20pharmacokinetic/pharmacodynamic%20relationships&author=C%20Louizos&author=JA%20Y%C3%A1%C3%B1ez&author=L%20Forrest&volume=17&issue=1&publication_year=2014&pages=34&pmid=24735761&)
|
| 261 |
+
|
| 262 |
+
28. Maciulaitis R, Kontrimaviciute V, Bressolle FM, et al. (2008) Ibogaine, an anti-addictive drug: pharmacology and time to go further in development. A narrative review. Hum Exp Toxicol 27(3): 181-194. [DOI](https://doi.org/10.1177/0960327107087802) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18650249/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Hum%20Exp%20Toxicol&title=Ibogaine,%20an%20anti-addictive%20drug:%20pharmacology%20and%20time%20to%20go%20further%20in%20development.%20A%20narrative%20review&author=R%20Maciulaitis&author=V%20Kontrimaviciute&author=FM%20Bressolle&volume=27&issue=3&publication_year=2008&pages=181-194&pmid=18650249&doi=10.1177/0960327107087802&)
|
| 263 |
+
|
| 264 |
+
29. Malcolm BJ, Polanco M, Barsuglia JP. (2018) Changes in Withdrawal and Craving Scores in Participants Undergoing Opioid Detoxification Utilizing Ibogaine. J Psychoactive Drugs. Epub ahead of print 2018/04/02. DOI: 10.1080/02791072.2018.1447175. 1-10. [DOI](https://doi.org/10.1080/02791072.2018.1447175.) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29608409/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Psychoactive%20Drugs&title=Changes%20in%20Withdrawal%20and%20Craving%20Scores%20in%20Participants%20Undergoing%20Opioid%20Detoxification%20Utilizing%20Ibogaine&author=BJ%20Malcolm&author=M%20Polanco&author=JP%20Barsuglia&publication_year=2018&pages=1-10&pmid=29608409&doi=10.1080/02791072.2018.1447175.&)
|
| 265 |
+
|
| 266 |
+
30. Marez D, Legrand M, Sabbagh N, et al. (1997) Polymorphism of the cytochrome P450 CYP2D6 gene in a European population: characterization of 48 mutations and 53 alleles, their frequencies and evolution. Pharmacogenetics 7(3): 193-202. [DOI](https://doi.org/10.1097/00008571-199706000-00004) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9241659/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Polymorphism%20of%20the%20cytochrome%20P450%20CYP2D6%20gene%20in%20a%20European%20population:%20characterization%20of%2048%20mutations%20and%2053%20alleles,%20their%20frequencies%20and%20evolution&author=D%20Marez&author=M%20Legrand&author=N%20Sabbagh&volume=7&issue=3&publication_year=1997&pages=193-202&pmid=9241659&doi=10.1097/00008571-199706000-00004&)
|
| 267 |
+
|
| 268 |
+
31. Mash DC, Duque L, Page B, et al. (2018) Ibogaine Detoxification Transitions Opioid and Cocaine Abusers Between Dependence and Abstinence: Clinical Observations and Treatment Outcomes. Front Pharmacol 9: 529. [DOI](https://doi.org/10.3389/fphar.2018.00529) | [PMC free article](/articles/PMC5996271/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29922156/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front%20Pharmacol&title=Ibogaine%20Detoxification%20Transitions%20Opioid%20and%20Cocaine%20Abusers%20Between%20Dependence%20and%20Abstinence:%20Clinical%20Observations%20and%20Treatment%20Outcomes&author=DC%20Mash&author=L%20Duque&author=B%20Page&volume=9&publication_year=2018&pages=529&pmid=29922156&doi=10.3389/fphar.2018.00529&)
|
| 269 |
+
|
| 270 |
+
32. Mash DC, Kovera CA, Pablo J, et al. (2001) Ibogaine in the treatment of heroin withdrawal. Alkaloids Chem Biol 56: 155-171. [DOI](https://doi.org/10.1016/s0099-9598(01)56012-5) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11705106/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Alkaloids%20Chem%20Biol&title=Ibogaine%20in%20the%20treatment%20of%20heroin%20withdrawal&author=DC%20Mash&author=CA%20Kovera&author=J%20Pablo&volume=56&publication_year=2001&pages=155-171&pmid=11705106&doi=10.1016/s0099-9598(01)56012-5&)
|
| 271 |
+
|
| 272 |
+
33. Mash DC, Kovera CA, Pablo J, et al. (2000) Ibogaine: complex pharmacokinetics, concerns for safety, and preliminary efficacy measures. Ann N Y Acad Sci 914: 394-401. [DOI](https://doi.org/10.1111/j.1749-6632.2000.tb05213.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11085338/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20N%20Y%20Acad%20Sci&title=Ibogaine:%20complex%20pharmacokinetics,%20concerns%20for%20safety,%20and%20preliminary%20efficacy%20measures&author=DC%20Mash&author=CA%20Kovera&author=J%20Pablo&volume=914&publication_year=2000&pages=394-401&pmid=11085338&doi=10.1111/j.1749-6632.2000.tb05213.x&)
|
| 273 |
+
|
| 274 |
+
34. Molinari HH, Maisonneuve IM, Glick SD. (1996) Ibogaine neurotoxicity: a re-evaluation. Brain Res 737(1-2): 255-262. [DOI](https://doi.org/10.1016/0006-8993(96)00739-1) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8930373/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Brain%20Res&title=Ibogaine%20neurotoxicity:%20a%20re-evaluation&author=HH%20Molinari&author=IM%20Maisonneuve&author=SD%20Glick&volume=737&issue=1-2&publication_year=1996&pages=255-262&pmid=8930373&doi=10.1016/0006-8993(96)00739-1&)
|
| 275 |
+
|
| 276 |
+
35. Noller GE, Frampton CM, Yazar-Klosinski B. (2017) Ibogaine treatment outcomes for opioid dependence from a twelve-month follow-up observational study. Am J Drug Alcohol Abuse. Epub ahead of print 2017/04/12. DOI: 10.1080/00952990.2017.1310218. 1-10. [DOI](https://doi.org/10.1080/00952990.2017.1310218.) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28402682/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Drug%20Alcohol%20Abuse&title=Ibogaine%20treatment%20outcomes%20for%20opioid%20dependence%20from%20a%20twelve-month%20follow-up%20observational%20study&author=GE%20Noller&author=CM%20Frampton&author=B%20Yazar-Klosinski&publication_year=2017&pages=1-10&pmid=28402682&doi=10.1080/00952990.2017.1310218.&)
|
| 277 |
+
|
| 278 |
+
36. O’Hearn E, Molliver ME. (1993) Degeneration of Purkinje cells in parasagittal zones of the cerebellar vermis after treatment with ibogaine or harmaline. Neuroscience 55(2): 303-310. [DOI](https://doi.org/10.1016/0306-4522(93)90500-f) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8377927/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Neuroscience&title=Degeneration%20of%20Purkinje%20cells%20in%20parasagittal%20zones%20of%20the%20cerebellar%20vermis%20after%20treatment%20with%20ibogaine%20or%20harmaline&author=E%20O%E2%80%99Hearn&author=ME%20Molliver&volume=55&issue=2&publication_year=1993&pages=303-310&pmid=8377927&doi=10.1016/0306-4522(93)90500-f&)
|
| 279 |
+
|
| 280 |
+
37. Obach RS, Pablo J, Mash DC. (1998) Cytochrome P4502D6 catalyzes the O-demethylation of the psychoactive alkaloid ibogaine to 12-hydroxyibogamine. Drug Metab Dispos 26(8): 764-768. [PubMed](https://pubmed.ncbi.nlm.nih.gov/9698290/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Metab%20Dispos&title=Cytochrome%20P4502D6%20catalyzes%20the%20O-demethylation%20of%20the%20psychoactive%20alkaloid%20ibogaine%20to%2012-hydroxyibogamine&author=RS%20Obach&author=J%20Pablo&author=DC%20Mash&volume=26&issue=8&publication_year=1998&pages=764-768&pmid=9698290&)
|
| 281 |
+
|
| 282 |
+
38. Ona G, Rocha JM, Bouso JC, et al. (2022) The adverse events of ibogaine in humans: an updated systematic review of the literature (2015–2020). Psychopharmacology 239(6): 1977-1987. [DOI](https://doi.org/10.1007/s00213-021-05964-y) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34406452/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Psychopharmacology&title=The%20adverse%20events%20of%20ibogaine%20in%20humans:%20an%20updated%20systematic%20review%20of%20the%20literature%20(2015%E2%80%932020)&author=G%20Ona&author=JM%20Rocha&author=JC%20Bouso&volume=239&issue=6&publication_year=2022&pages=1977-1987&pmid=34406452&doi=10.1007/s00213-021-05964-y&)
|
| 283 |
+
|
| 284 |
+
39. Özdemir L, Sökmen E. (2020) Effect of habitual cigarette smoking on the index of cardiac electrophysiological balance in apparently healthy individuals. Journal of Electrocardiology 59: 41-44. [DOI](https://doi.org/10.1016/j.jelectrocard.2020.01.003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31958651/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Journal%20of%20Electrocardiology&title=Effect%20of%20habitual%20cigarette%20smoking%20on%20the%20index%20of%20cardiac%20electrophysiological%20balance%20in%20apparently%20healthy%20individuals&author=L%20%C3%96zdemir&author=E%20S%C3%B6kmen&volume=59&publication_year=2020&pages=41-44&pmid=31958651&doi=10.1016/j.jelectrocard.2020.01.003&)
|
| 285 |
+
|
| 286 |
+
40. Paling FP, Andrews LM, Valk GD, et al. (2012) Life-threatening complications of ibogaine: three case reports. Neth J Med 70(9): 422-424. [PubMed](https://pubmed.ncbi.nlm.nih.gov/23123541/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Neth%20J%20Med&title=Life-threatening%20complications%20of%20ibogaine:%20three%20case%20reports&author=FP%20Paling&author=LM%20Andrews&author=GD%20Valk&volume=70&issue=9&publication_year=2012&pages=422-424&pmid=23123541&)
|
| 287 |
+
|
| 288 |
+
41. Postema PG, Wilde AA. (2014) The measurement of the QT interval. Curr Cardiol Rev 10(3): 287-294. [DOI](https://doi.org/10.2174/1573403X10666140514103612) | [PMC free article](/articles/PMC4040880/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24827793/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Curr%20Cardiol%20Rev&title=The%20measurement%20of%20the%20QT%20interval&author=PG%20Postema&author=AA%20Wilde&volume=10&issue=3&publication_year=2014&pages=287-294&pmid=24827793&doi=10.2174/1573403X10666140514103612&)
|
| 289 |
+
|
| 290 |
+
42. Rodríguez-Cano BJ, Kohek M, Ona G, et al. (2023) Underground ibogaine use for the treatment of substance use disorders: A qualitative analysis of subjective experiences. Drug and Alcohol Review 42(2): 401-414. [DOI](https://doi.org/10.1111/dar.13587) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36456173/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20and%20Alcohol%20Review&title=Underground%20ibogaine%20use%20for%20the%20treatment%20of%20substance%20use%20disorders:%20A%20qualitative%20analysis%20of%20subjective%20experiences&author=BJ%20Rodr%C3%ADguez-Cano&author=M%20Kohek&author=G%20Ona&volume=42&issue=2&publication_year=2023&pages=401-414&pmid=36456173&doi=10.1111/dar.13587&)
|
| 291 |
+
|
| 292 |
+
43. Schenberg EE, de Castro Comis MA, Chaves BR, et al. (2014) Treating drug dependence with the aid of ibogaine: a retrospective study. J Psychopharmacol 28(11): 993-1000. [DOI](https://doi.org/10.1177/0269881114552713) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25271214/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Psychopharmacol&title=Treating%20drug%20dependence%20with%20the%20aid%20of%20ibogaine:%20a%20retrospective%20study&author=EE%20Schenberg&author=MA%20de%20Castro%20Comis&author=BR%20Chaves&volume=28&issue=11&publication_year=2014&pages=993-1000&pmid=25271214&doi=10.1177/0269881114552713&)
|
| 293 |
+
|
| 294 |
+
44. Schmitz-Hübsch T, du Montcel ST, Baliko L, et al. (2006) Scale for the assessment and rating of ataxia: development of a new clinical scale. Neurology 66(11): 1717-1720. [DOI](https://doi.org/10.1212/01.wnl.0000219042.60538.92) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16769946/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Neurology&title=Scale%20for%20the%20assessment%20and%20rating%20of%20ataxia:%20development%20of%20a%20new%20clinical%20scale&author=T%20Schmitz-H%C3%BCbsch&author=ST%20du%20Montcel&author=L%20Baliko&volume=66&issue=11&publication_year=2006&pages=1717-1720&pmid=16769946&doi=10.1212/01.wnl.0000219042.60538.92&)
|
| 295 |
+
|
| 296 |
+
45. Schmitz-Hübsch T, Fimmers R, Rakowicz M, et al. (2010) Responsiveness of different rating instruments in spinocerebellar ataxia patients. Neurology 74(8): 678-684. [DOI](https://doi.org/10.1212/WNL.0b013e3181d1a6c9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20177122/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Neurology&title=Responsiveness%20of%20different%20rating%20instruments%20in%20spinocerebellar%20ataxia%20patients&author=T%20Schmitz-H%C3%BCbsch&author=R%20Fimmers&author=M%20Rakowicz&volume=74&issue=8&publication_year=2010&pages=678-684&pmid=20177122&doi=10.1212/WNL.0b013e3181d1a6c9&)
|
| 297 |
+
|
| 298 |
+
46. Sheppard SG. (1994) A preliminary investigation of ibogaine: case reports and recommendations for further study. J Subst Abuse Treat 11(4): 379-385. [DOI](https://doi.org/10.1016/0740-5472(94)90049-3) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7966509/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Subst%20Abuse%20Treat&title=A%20preliminary%20investigation%20of%20ibogaine:%20case%20reports%20and%20recommendations%20for%20further%20study&author=SG%20Sheppard&volume=11&issue=4&publication_year=1994&pages=379-385&pmid=7966509&doi=10.1016/0740-5472(94)90049-3&)
|
| 299 |
+
|
| 300 |
+
47. Thurner P, Stary-Weinzinger A, Gafar H, et al. (2014) Mechanism of hERG channel block by the psychoactive indole alkaloid ibogaine. J Pharmacol Exp Ther 348(2): 346-358. [DOI](https://doi.org/10.1124/jpet.113.209643) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24307198/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharmacol%20Exp%20Ther&title=Mechanism%20of%20hERG%20channel%20block%20by%20the%20psychoactive%20indole%20alkaloid%20ibogaine&author=P%20Thurner&author=A%20Stary-Weinzinger&author=H%20Gafar&volume=348&issue=2&publication_year=2014&pages=346-358&pmid=24307198&doi=10.1124/jpet.113.209643&)
|
| 301 |
+
|
| 302 |
+
48. Vlaanderen L, Martial LC, Franssen EJ, et al. (2014) Cardiac arrest after ibogaine ingestion. Clin Toxicol (Phila) 52(6): 642-643. [DOI](https://doi.org/10.3109/15563650.2014.927477) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24940646/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Toxicol%20(Phila)&title=Cardiac%20arrest%20after%20ibogaine%20ingestion&author=L%20Vlaanderen&author=LC%20Martial&author=EJ%20Franssen&volume=52&issue=6&publication_year=2014&pages=642-643&pmid=24940646&doi=10.3109/15563650.2014.927477&)
|
| 303 |
+
|
| 304 |
+
49. Weyer A, Abele M, Schmitz-Hübsch T, et al. (2007) Reliability and validity of the scale for the assessment and rating of ataxia: a study in 64 ataxia patients. Mov Disord 22(11): 1633-1637. [DOI](https://doi.org/10.1002/mds.21544) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17516493/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mov%20Disord&title=Reliability%20and%20validity%20of%20the%20scale%20for%20the%20assessment%20and%20rating%20of%20ataxia:%20a%20study%20in%2064%20ataxia%20patients&author=A%20Weyer&author=M%20Abele&author=T%20Schmitz-H%C3%BCbsch&volume=22&issue=11&publication_year=2007&pages=1633-1637&pmid=17516493&doi=10.1002/mds.21544&)
|
| 305 |
+
|
| 306 |
+
50. Yabe I, Matsushima M, Soma H, et al. (2008) Usefulness of the Scale for Assessment and Rating of Ataxia (SARA). J Neurol Sci 266(1-2): 164-166. [DOI](https://doi.org/10.1016/j.jns.2007.09.021) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17950753/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Neurol%20Sci&title=Usefulness%20of%20the%20Scale%20for%20Assessment%20and%20Rating%20of%20Ataxia%20(SARA)&author=I%20Yabe&author=M%20Matsushima&author=H%20Soma&volume=266&issue=1-2&publication_year=2008&pages=164-166&pmid=17950753&doi=10.1016/j.jns.2007.09.021&)
|
test/texts/PMC11106956.md
ADDED
|
@@ -0,0 +1,348 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Population pharmacokinetics of primaquine and its metabolites in African males
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Palang Chotsiri, Almahamoudou Mahamar, Halimatou Diawara, Pius S Fasinu, Kalifa Diarra, Koualy Sanogo, Teun Bousema, Larry A Walker, Joelle M Brown, Alassane Dicko, Roly Gosling, Ingrid Chen, Joel Tarning
|
| 5 |
+
**Journal:** Malaria Journal
|
| 6 |
+
**Date:** 2024 May 21
|
| 7 |
+
**DOI:** [10.1186/s12936-024-04979-y](https://doi.org/10.1186/s12936-024-04979-y)
|
| 8 |
+
**PMID:** 38773528
|
| 9 |
+
**PMCID:** PMC11106956
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11106956/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11106956/pdf/12936_2024_Article_4979.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11106956/pdf/12936_2024_Article_4979.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Background:**
|
| 16 |
+
Primaquine (PQ) is the prototype 8-aminoquinoline drug, a class which targets gametocytes and hypnozoites. The World Health Organization (WHO) recommends adding a single low dose of primaquine to the standard artemisinin-based combination therapy (ACT) in order to block malaria transmission in regions with low malaria transmission. However, the haemolytic toxicity is a major adverse outcome of primaquine in glucose-6-phosphate dehydrogenase (G6PD)-deficient subjects. This study aimed to characterize the pharmacokinetic properties of primaquine and its major metabolites in G6PD-deficient subjects.
|
| 17 |
+
|
| 18 |
+
**Methods:**
|
| 19 |
+
A single low-dose of primaquine (0.4–0.5 mg/kg) was administered in twenty-eight African males. Venous and capillary plasma were sampled up to 24 h after the drug administration. Haemoglobin levels were observed up to 28 days after drug administration. Only PQ, carboxy-primaquine (CPQ), and primaquine carbamoyl-glucuronide (PQCG) were present in plasma samples and measured using liquid chromatography mass spectrometry. Drug and metabolites’ pharmacokinetic properties were investigated using nonlinear mixed-effects modelling.
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
Population pharmacokinetic properties of PQ, CPQ, and PQCG can be described by one-compartment disposition kinetics with a transit-absorption model. Body weight was implemented as an allometric function on the clearance and volume parameters for all compounds. None of the covariates significantly affected the pharmacokinetic parameters. No significant correlations were detected between the exposures of the measured compounds and the change in haemoglobin or methaemoglobin levels. There was no significant haemoglobin drop in the G6PD-deficient patients after administration of a single low dose of PQ.
|
| 23 |
+
|
| 24 |
+
**Conclusions:**
|
| 25 |
+
A single low-dose of PQ was haematologically safe in this population of G6PD-normal and G6PD-deficient African males without malaria.
|
| 26 |
+
|
| 27 |
+
Trial registration NCT02535767
|
| 28 |
+
|
| 29 |
+
**Supplementary Information:**
|
| 30 |
+
The online version contains supplementary material available at 10.1186/s12936-024-04979-y.
|
| 31 |
+
|
| 32 |
+
Keywords: Primaquine, Carboxy-primaquine, Primaquine carbamoyl-glucuronide, Pharmacokinetics, Nonlinear mixed-effect model, G6PD-deficiency
|
| 33 |
+
|
| 34 |
+
### Background
|
| 35 |
+
|
| 36 |
+
Primaquine (PQ) is the prototype 8-aminoquinoline drug, a class which targets gametocytes and hypnozoites. The World Health Organization (WHO) recommends adding a single low dose of primaquine to the standard artemisinin-based combination therapy (ACT) in order to block malaria transmission in regions with low malaria transmission. However, the haemolytic toxicity is a major adverse outcome of primaquine in glucose-6-phosphate dehydrogenase (G6PD)-deficient subjects. This study aimed to characterize the pharmacokinetic properties of primaquine and its major metabolites in G6PD-deficient subjects.
|
| 37 |
+
|
| 38 |
+
### Methods
|
| 39 |
+
|
| 40 |
+
A single low-dose of primaquine (0.4–0.5 mg/kg) was administered in twenty-eight African males. Venous and capillary plasma were sampled up to 24 h after the drug administration. Haemoglobin levels were observed up to 28 days after drug administration. Only PQ, carboxy-primaquine (CPQ), and primaquine carbamoyl-glucuronide (PQCG) were present in plasma samples and measured using liquid chromatography mass spectrometry. Drug and metabolites’ pharmacokinetic properties were investigated using nonlinear mixed-effects modelling.
|
| 41 |
+
|
| 42 |
+
### Results
|
| 43 |
+
|
| 44 |
+
Population pharmacokinetic properties of PQ, CPQ, and PQCG can be described by one-compartment disposition kinetics with a transit-absorption model. Body weight was implemented as an allometric function on the clearance and volume parameters for all compounds. None of the covariates significantly affected the pharmacokinetic parameters. No significant correlations were detected between the exposures of the measured compounds and the change in haemoglobin or methaemoglobin levels. There was no significant haemoglobin drop in the G6PD-deficient patients after administration of a single low dose of PQ.
|
| 45 |
+
|
| 46 |
+
### Conclusions
|
| 47 |
+
|
| 48 |
+
A single low-dose of PQ was haematologically safe in this population of G6PD-normal and G6PD-deficient African males without malaria.
|
| 49 |
+
|
| 50 |
+
*Trial registration*Trial registration [NCT02535767](https://clinicaltrials.gov/ct2/show/NCT02535767)NCT02535767
|
| 51 |
+
|
| 52 |
+
### Supplementary Information
|
| 53 |
+
|
| 54 |
+
The online version contains supplementary material available at 10.1186/s12936-024-04979-y.
|
| 55 |
+
|
| 56 |
+
**Keywords:**Keywords: Primaquine, Carboxy-primaquine, Primaquine carbamoyl-glucuronide, Pharmacokinetics, Nonlinear mixed-effect model, G6PD-deficiency
|
| 57 |
+
|
| 58 |
+
## Background
|
| 59 |
+
|
| 60 |
+
Primaquine (PQ) is a widely available anti-malarial that kills the dormant liver stage (hypnozoites) of *Plasmodium vivax*Plasmodium vivax and *Plasmodium ovale*Plasmodium ovale [[1](#CR1)1]. The World Health Organization (WHO) suggests a daily treatment (i.e., 0.25 mg/kg daily for 14 days) for glucose-6-phosphate dehydrogenase (G6PD)-normal individuals and a weekly treatment (i.e., 0.75 mg/kg weekly for 8 weeks) for G6PD-deficient individuals. PQ also possesses a sterile effect against the sexual stage of the *Plasmodium*Plasmodium parasite. A single low dose (SLD) of PQ with a standard artemisinin-based combination therapy (ACT) has shown gametocyte reduction and transmission blocking properties [[2](#CR2)2–[4](#CR4)4]. Therefore, the WHO recommends a 0.25 mg/kg SLD-PQ in all *Plasmodium falciparum*Plasmodium falciparum-infected patients living in the area approaching malaria elimination and/or facing drug resistance [[1](#CR1)1, [5](#CR5)5].
|
| 61 |
+
|
| 62 |
+
Pharmacokinetic properties of PQ have been well characterized. PQ is almost completely absorbed (96% bioavailability) and rapidly eliminated (5–6 h terminal half-life). PQ can be metabolized via several pathways including the activities of monoamine oxidase (MAO-A), cytochrome P450 (CYP) isoenzyme, and uridine 5ʹ-diphospho-glucuronosyltransferase (UDP-glucuronosyltransferase, UGT). Also, its metabolism and pharmacokinetic profiles is enantioselective [[6](#CR6)6, [7](#CR7)7]. Carboxy-primaquine (CPQ) is the major metabolite found in human plasma and is generated via the MAO-A-mediated pathway. The pharmacological effects of PQ have been attributed to its metabolites, and although several metabolites of PQ can be detected in human plasma and urine, those responsible for the biologic activity are still poorly understood [[8](#CR8)8, [9](#CR9)9]. Presumably, the quinone-imine and orthoquinone metabolites were proposed as the active metabolites, because they can generate local reactive oxygen species (ROS) through a redox reaction which can results in oxidative damage to the parasites and potentially cause haemolysis of the red blood cell [[10](#CR10)10, [11](#CR11)11].
|
| 63 |
+
|
| 64 |
+
A major adverse effect of 8-aminoquinoline drug is the drug-induced haemolytic effect especially in G6PD-deficient individuals. Different variants of the G6PD genotypes are associated with different levels of haemolytic effect [[12](#CR12)12], i.e., individuals with South-East Asia variants or Middle East/West Asia variants have a stronger haemolytic response than individuals with the common African A- variants. Therefore, a long-term PQ dosing regimen for radical cure should be optimized according to patient’s G6PD activity status. For *P. falciparum*P. falciparum transmission-blocking regimen, the SLD-PQ is safe for G6PD-deficient individuals and it can be administered in all individuals without any G6PD testing [[5](#CR5)5].
|
| 65 |
+
|
| 66 |
+
This study evaluated the population pharmacokinetics of PQ, CPQ, and primaquine carbamoyl-glucuronide (PQCG) in G6PD-deficient African males. Also, the relationships between the pharmacokinetic parameters and the haemoglobin level in this population were evaluated.
|
| 67 |
+
|
| 68 |
+
## Methods
|
| 69 |
+
|
| 70 |
+
### Study design and ethical approval
|
| 71 |
+
|
| 72 |
+
This study was a part of an open-label, nonrandomized, dose-adjustment trial of the safety of SLD-PQ in G6PD-deficient and G6PD-normal males in Mali without microscopically detected malaria parasite infection. The main clinical trial was separated into two parts, i.e., Part I in adult males (aged 18–50) and Part II in children (aged 5–17). Full clinical details and results have been reported elsewhere [[13](#CR13)13]. Only 28 adult males from the main trial provided pharmacokinetic samples and were included in this study.
|
| 73 |
+
|
| 74 |
+
### Study procedure and blood sampling
|
| 75 |
+
|
| 76 |
+
After collection of day 0 samples, each participant received an oral dose of PQ according to his group assignment, after a fatty snack (biscuits) to minimized gastrointestinal symptoms. The study pharmacist prepared a dose by crushing a 15-mg tablet of PQ (Sanofi, Laval, Canada) in 15 mL of drinking water and administered the dose to the nearest 0.1 mL under direct observation. Venous samples (4 mL) were collected at pre-dose and 1, 4, 8, and 24 h post-dose. Capillary samples (0.5 mL) were collected at 2, 4, and 6 h post-dose. Both venous and capillary blood were collected using EDTA tubes, and centrifuged at 1100–1300×*g*g for 10–20 min to obtain plasma samples. The samples were store immediately at − 80 °C until analysed.
|
| 77 |
+
|
| 78 |
+
### Bioanalytical methods
|
| 79 |
+
|
| 80 |
+
Only PQ and its metabolites, CPQ and PQCG, were detectable in plasma samples collected here. All compounds were analysed using the liquid chromatography–mass spectrometry (LC–MS) method of Avula et al*.*. [[14](#CR14)14], modified to employ an ACQUITY UHPLC™, BEH Shield RP18 column (100 mm × 2.1 mm I.D., 1.7 mm) equipped with an LC-18 guard column (Vanguard 2.1 × 5 mm, Waters Corp, Milford, MA, USA). PQ, CPQ and PQCG were separated and eluted within 10-min retention time. The mobile phase, run at a flow rate of 0.25 ml/min, consisted of 0.05% formic acid in water (A) water and 0.05% formic acid in acetonitrile (B) and was applied in a linear gradient elution. The proportion of solvent A decreased from 90 to 63% during the first 5 min, then from 63 to 37% during minutes 5–8, then from 37 to 0% during minutes 8–10. A 3-min wash with 100% B and a 3.5 min equilibration period of 90% A followed each run. Samples were injected at 10 μL volume. The limits of quantification in plasma were 5 ng/mL for PQ and PQCG, and 1 ng/mL for CPQ. Quality control samples at 20 ng/mL were run at beginning and end of the batch analyses in duplicate. Intra-assay variability was less than 10%.
|
| 81 |
+
|
| 82 |
+
### Population pharmacokinetic model
|
| 83 |
+
|
| 84 |
+
PQ, CPQ, and PQCG concentrations were transformed into their natural logarithms and analysed using a nonlinear mixed-effects modelling approach in NONMEM version 7.4 (Icon Development Solution, Ellicott city, MD). Pirana version 3.0.0 [[15](#CR15)15], Perl-speaks-NONMEM version 5.3.0 (PsN) [[16](#CR16)16] and R version 4.2.0 were used for automation, model evaluation, and diagnostics during the model building process. The first-order conditional estimation method with interaction (FOCE-I) was applied for the estimation method. Each compound was modelled separately. Data below the LLOQ was either omitted (M1-method) or incorporated by imputing the first LLOQ data as half of LLOQ (M6-method) [[17](#CR17)17].
|
| 85 |
+
|
| 86 |
+
Pharmacokinetic parameters were implemented as a log-normal distribution as follows:
|
| 87 |
+
|
| 88 |
+
| θi=θTV×expηi,θ |
|
| 89 |
+
| -------------- |
|
| 90 |
+
where θiθiθiθθii is the individual iiiith pharmacokinetic parameter estimate, θTVθTVθTVθθTVTV is the typical value of the population mean estimate, and ηi,θηi,θηi,θηηi,θii,,θθ is the inter-individual variability of the parameter θθθθ for the iiiith individual, assumed to be normal distribution with zero mean and variance ω2ω2ω2ωωω22. Estimated inter-individual variability below 10% was fixed to zero.
|
| 91 |
+
|
| 92 |
+
A linear association between capillary and venous plasma concentrations of all compounds was assumed and modelled using an estimated conversion factor at the population level. Unexplained residual errors were modelled separately for capillary and venous plasma concentrations and implemented as an additive error model on the log-transformed concentrations, equivalent to an exponential error on arithmetic scale.
|
| 93 |
+
|
| 94 |
+
Individual body-weight (BWiBWiBWiBWBWii) was introduced into the pharmacokinetic model as a fixed allometric function on all clearance (n=0.75n=0.75n=0.75nn==0.750.75) and volume (n=1.00n=1.00n=1.00nn==1.001.00) parameters, centralized to 62.5 kg of body weight according to median body weight in the population.
|
| 95 |
+
|
| 96 |
+
| θi=θTV×expηi,θ×BWi62.5n |
|
| 97 |
+
| ----------------------- |
|
| 98 |
+
All other covariates of biological relevance (i.e. age, malaria status, G6PD genotypes, G6PD phenotypes, and CYP2D6 phenotype) were evaluated using a stepwise addition (*p*p < 0.05) followed by a stepwise elimination (*p*p < 0.001) approach.
|
| 99 |
+
|
| 100 |
+
### Model diagnostics and evaluations
|
| 101 |
+
|
| 102 |
+
The objective function value (OFV), calculated by NONMEM as proportional to − 2 × log-likelihood of the data, was used for evaluating the model fit. Any two hierarchical models were compared by a likelihood ratio test (LRT), based on the Chi-square distribution of OFV (i.e., p-value < 0.05 corresponding to ΔOFV > 3.48 at 1 degree of freedom difference). Potential model misspecification and systematic errors were determined by basic goodness-of-fit diagnostics. Eta and epsilon shrinkages were used to assess the ability of detecting model misspecification in the goodness-of-fit diagnostics [[18](#CR18)18]. Model robustness evaluation and generation of 95% confidence intervals were done using a sampling important resampling (SIR) procedure [[19](#CR19)19, [20](#CR20)20]. Predictive performances of the final pharmacokinetic models were assessed by prediction-corrected visual predictive checks (n = 1000) [[21](#CR21)21]. The 5th, 50th and 95th percentiles were overlaid with the 95% confidence interval of each simulated percentile.
|
| 103 |
+
|
| 104 |
+
### Pharmacokinetic effects on haemoglobin and methaemoglobin levels
|
| 105 |
+
|
| 106 |
+
The potential relationship between individual pharmacokinetic parameters (i.e., estimated maximum concentration; C_MAX_MAX, and 24-h exposure; AUC_24_24) of all compounds and the changes in haemoglobin and methaemoglobin levels were characterised using a simple linear regression.
|
| 107 |
+
|
| 108 |
+
## Results
|
| 109 |
+
|
| 110 |
+
This was an open-label, Phase 1, dose-adjustment clinical trial in adults and children, and the main clinical findings have been reported previously [[13](#CR13)13]. Adult participants (n = 28) provided pharmacokinetic samples and were included in this study. Participants enrolled in this study were divided into 3 dosing groups, i.e., 0.40 mg/kg (n = 7), 0.45 mg/kg (n = 7), and 0.50 mg/kg (n = 14). Full demographic characteristics are presented in Table [1](#Tab1)1.
|
| 111 |
+
|
| 112 |
+
### Table 1.
|
| 113 |
+
|
| 114 |
+
Baseline subject’s characteristics
|
| 115 |
+
|
| 116 |
+
| | Group 1 (n = 7)0.40 mg/kg | Group 2 (n = 7)0.45 mg/kg | Group 3 (n = 14)0.50 mg/kg |
|
| 117 |
+
| - | ------------------------- | ------------------------- | -------------------------- |
|
| 118 |
+
| Primaquine dose (mg) | 24.0 (17.6–28.0) | 26.1 (23.4–33.5) | 32.1 (28.4–41.5) |
|
| 119 |
+
| Weight (kg) | 60 (44.0–70.1) | 57.9 (52.0–74.4) | 64.1 (56.8–83.0) |
|
| 120 |
+
| Age (years) | 20 (18–50) | 32 (26–50) | 39 (25.47) |
|
| 121 |
+
| Temperature (°C) | 36.6 (36.5–37.4) | 36.5 (36.1–37.2) | 36.4 (36.1–37) |
|
| 122 |
+
| Haemoglobin (g/dL) | 13.3 (12.0–15.5) | 14.8 (14.4–17.5) | 14.9 (13.6–16.5) |
|
| 123 |
+
| Positive malaria PCR (n) | 5 | 5 | 4 |
|
| 124 |
+
| G6PD genotype (SNPs 202A and 376G) | | | |
|
| 125 |
+
| Wild-type | 3 | 2 | 7 |
|
| 126 |
+
| A− | 4 | 5 | 7 |
|
| 127 |
+
| G6PD semi-quantitativea | | | |
|
| 128 |
+
| Normal | 3 | 0 | 7 |
|
| 129 |
+
| Deficient | 4 | 6 | 7 |
|
| 130 |
+
| CYP2D6 metabolism | | | |
|
| 131 |
+
| Poor metabolizer | 1 | 0 | 0 |
|
| 132 |
+
| Intermediate metabolizer | 2 | 2 | 9 |
|
| 133 |
+
| Extensive metabolizer | 4 | 4 | 5 |
|
| 134 |
+
| Extensive/ultra-rapid metabolizer | 0 | 1 | 0 |
|
| 135 |
+
### Pharmacokinetic properties of PQ and its metabolites
|
| 136 |
+
|
| 137 |
+
Pharmacokinetic properties of PQ, CPQ, and PQCG were investigated separately. All 196 samples displayed concentrations above the LLOQ. Primaquine doses in molar units were used for metabolite dosing, under an assumption of 1:1 transformation factor.
|
| 138 |
+
|
| 139 |
+
Pharmacokinetic properties of PQ, CPQ, and PQCG were explained by one-disposition compartment models with transit absorption models. Adding an additional peripheral distribution compartment did not improve the model fits. Absorption of PQ and PQCG were best described by 5 transit compartment models, whereas the absorption of CPQ was best described by a 2 transit compartment model. Body-weight was added into the pharmacokinetic model as an allometric function. None of the other covariates were statistically significant. The G6PD genotype (A-variant and wild-type) and G6PD phenotype (determined by a semiquantitative test) were not statistically significant on any pharmacokinetic parameters of PQ or its metabolites. The final models of all compounds showed a satisfactory goodness-of-fit (Fig. [1](#Fig1)1) and predictive performance (Fig. [2](#Fig2)2). The venous-capillary conversion factors for all compounds were estimated using the final population pharmacokinetic models. Also, the estimated conversion factors agreed with a simple linear-regression model using measured concentrations of samples taken at the same time point (Figure S1). The final pharmacokinetic parameter estimates with its parameters’ uncertainty and the secondary parameter estimates are reported in Tables [2](#Tab2)2 and [3](#Tab3)3, respectively.
|
| 140 |
+
|
| 141 |
+
### Fig. 1.
|
| 142 |
+
|
| 143 |
+
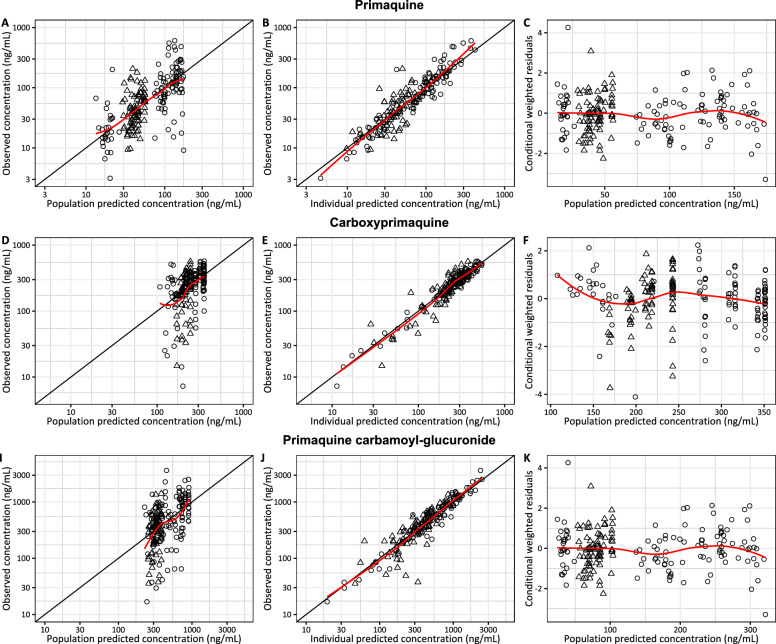
|
| 144 |
+
|
| 145 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11106956_12936_2024_4979_Fig1_HTML.jpg)
|
| 146 |
+
|
| 147 |
+
Goodness-of fit diagnostics of the final pharmacokinetic model of primaquine (A–C), carboxy-primaquine (D–F), and primaquine carbamoyl-glucuronide (I–K) stratified by biological matrix (i.e., circle = venous concentrations, and triangle = capillary concentrations). Red lines represent the locally weighted least-square regression based on the observations
|
| 148 |
+
|
| 149 |
+
### Fig. 2.
|
| 150 |
+
|
| 151 |
+
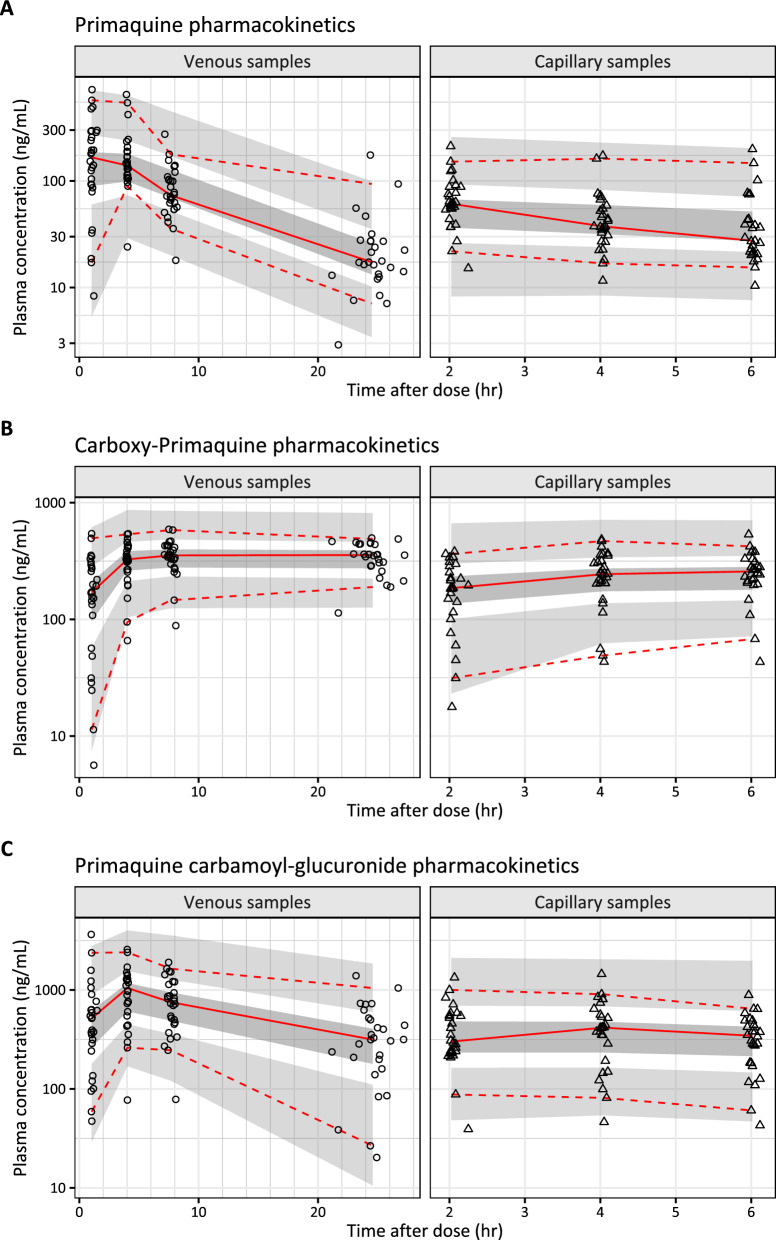
|
| 152 |
+
|
| 153 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11106956_12936_2024_4979_Fig2_HTML.jpg)
|
| 154 |
+
|
| 155 |
+
Visual predictive plots of the final pharmacokinetic model of primaquine (A), carboxy-primaquine (B), and primaquine carbamoyl-glucuronide (C). Solid and dashed lines represent the median, 5th, and 95th percentile of the observations. Shaded areas represent the simulated 95% confidence interval of each percentile
|
| 156 |
+
|
| 157 |
+
### Table 2.
|
| 158 |
+
|
| 159 |
+
Final pharmacokinetic parameter estimates
|
| 160 |
+
|
| 161 |
+
| Pharmacokinetic parameter | Population estimatea (%RSEb) | 95% CIb | IIVa (%CVb) | 95% CIb |
|
| 162 |
+
| ------------------------- | ---------------------------- | ------- | ----------- | ------- |
|
| 163 |
+
| Primaquine | | | | |
|
| 164 |
+
| F | 1 fixed | – | 52.9% (15.4%) | 39.7%–74.6% |
|
| 165 |
+
| MTT (h) | 0.563 (18.7%) | 0.329–0.754 | 63.3% (23.5%) | 36.8%–100% |
|
| 166 |
+
| CL/F (L/h) | 15.4 (9.57%) | 13.1–18.7 | 12% (12.4%) | 7.99%–14.3% |
|
| 167 |
+
| VC/F (L) | 163 (10.3%) | 134–201 | – | – |
|
| 168 |
+
| CF (%) | 32.9 (6.89%) | 28.7–37.5 | – | – |
|
| 169 |
+
| σVP | 0.173 (9.37%) | 0.125–0.245 | – | – |
|
| 170 |
+
| σCP | 0.226 (9.33%) | 0.157–0.320 | – | – |
|
| 171 |
+
| Carboxy-primaquine | | | | |
|
| 172 |
+
| MTT (h) | 1.24 (11.7%) | 0.962–0.746 | 65.3% (13.2%) | 50.6%–89.2% |
|
| 173 |
+
| CL/F (L/h) | 0.129 (28.5%) | 0.0765–0.216 | – | – |
|
| 174 |
+
| VC/F (L) | 93.3 (7.00%) | 81.8–108 | 37.4% (15.7%) | 29.5%–52.9% |
|
| 175 |
+
| CF (%) | 69.1 (3.83%) | 0.643–0.746 | – | – |
|
| 176 |
+
| σVP | 0.0328 (8.96%) | 0.0238–0.0474 | – | – |
|
| 177 |
+
| σCP | 0.101 (8.84%) | 0.0763–0.146 | – | – |
|
| 178 |
+
| Primaquine carbamoyl-glucuronide | | | | |
|
| 179 |
+
| F | 1 fixed | – | 63.5% (12.1%) | 48.7%–81.9% |
|
| 180 |
+
| MTT (h) | 1.13 (8.84%) | 0.929–1.33 | 34.4% (25.1%) | 24.2%–58.5% |
|
| 181 |
+
| CL/F (L/h) | 2.83 (15.8%) | 2.06–3.81 | 57.8% (12.1%) | 42.8%–75.1% |
|
| 182 |
+
| VC/F (L) | 55.4 (12.4%) | 44.1–72.2 | – | – |
|
| 183 |
+
| CF (%) | 40.1 (6.97%) | 0.353–0.465 | – | – |
|
| 184 |
+
| σVP | 0.108 (12.3%) | 0.0736–0.175 | – | – |
|
| 185 |
+
| σCP | 0.242 (8.00%) | 0.178–0.323 | – | – |
|
| 186 |
+
### Table 3.
|
| 187 |
+
|
| 188 |
+
Secondary pharmacokinetic parameter estimates
|
| 189 |
+
|
| 190 |
+
| Parameter | Primaquine | Carboxy-primaquine | Primaquine carbamoyl-glucuronide |
|
| 191 |
+
| --------- | ---------- | ------------------ | -------------------------------- |
|
| 192 |
+
| CMAX (ng/mL) | 259 (53.5–764) | 338 (74.3–546) | 1550 (246–7320) |
|
| 193 |
+
| TMAX (h) | 0.872 (0.237–1.77) | 4.57 (2.19–18.1) | 1.61 (0.572–1.78) |
|
| 194 |
+
| AUC24 (h × ng/mL) | 130 (59.2–491) | 8030 (1490–12,800) | 1240 (242–3100) |
|
| 195 |
+
| Half-life (h) | 7.42 (7.04–7.93) | 469 (325–1870) | 17.5 (11.1–28.0) |
|
| 196 |
+
### Effect of pharmacokinetic parameters on haemoglobin
|
| 197 |
+
|
| 198 |
+
Haemoglobin and methaemoglobin levels were observed for up to 28 days after PQ administration (Fig. [3](#Fig3)3, Figure S2). There were no significant haemoglobin declines in patients with G6PD deficient and G6PD-normal status. Suggestive trends were observed between the pharmacokinetic parameters of PQ and the small decreases observed in haemoglobin levels (Fig. [4](#Fig4)4, Table S1) or increases in methaemoglobin levels (Fig. [5](#Fig5)5, Table S2). However, these relationships were not statistically significant from the zero-correlation and not statistically significant different between the G6PD-deficient and G6PD-normal individuals.
|
| 199 |
+
|
| 200 |
+
### Fig. 3.
|
| 201 |
+
|
| 202 |
+
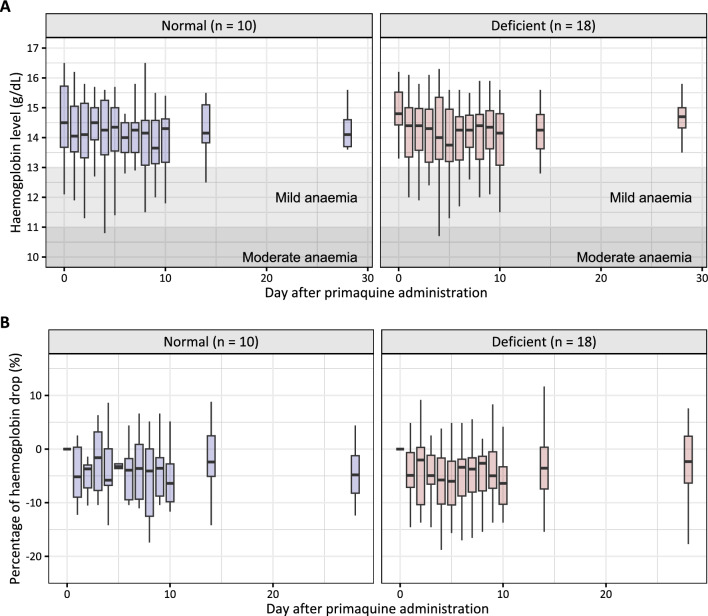
|
| 203 |
+
|
| 204 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11106956_12936_2024_4979_Fig3_HTML.jpg)
|
| 205 |
+
|
| 206 |
+
Observed haemoglobin concentration (A) and percentage of haemoglobin drop from the baseline (B) over 28 days of follow-up, stratified by G6PD status. Shaded areas represent a mild and moderate anaemia
|
| 207 |
+
|
| 208 |
+
### Fig. 4.
|
| 209 |
+
|
| 210 |
+
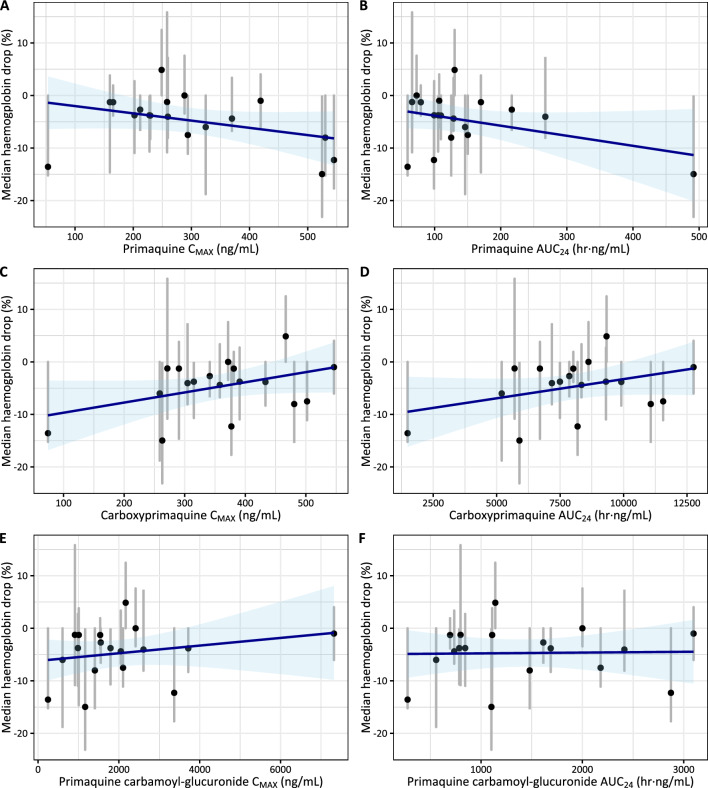
|
| 211 |
+
|
| 212 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11106956_12936_2024_4979_Fig4_HTML.jpg)
|
| 213 |
+
|
| 214 |
+
Correlation between pharmacokinetic parameters (CMAX and AUC24) and the median haemoglobin drop for primaquine (A, B), carboxy-primaquine (C, D), and primaquine cabarmoyl-glucuronide (E, F) in G6PD-deficient individuals (n = 18). Blue lines and shaded areas represent the linear regression with associated prediction intervals. Black circles and grey lines represent individual’s mean and range of observed haemoglobin drop. None of these correlations are significantly different from the zero-slope
|
| 215 |
+
|
| 216 |
+
### Fig. 5.
|
| 217 |
+
|
| 218 |
+
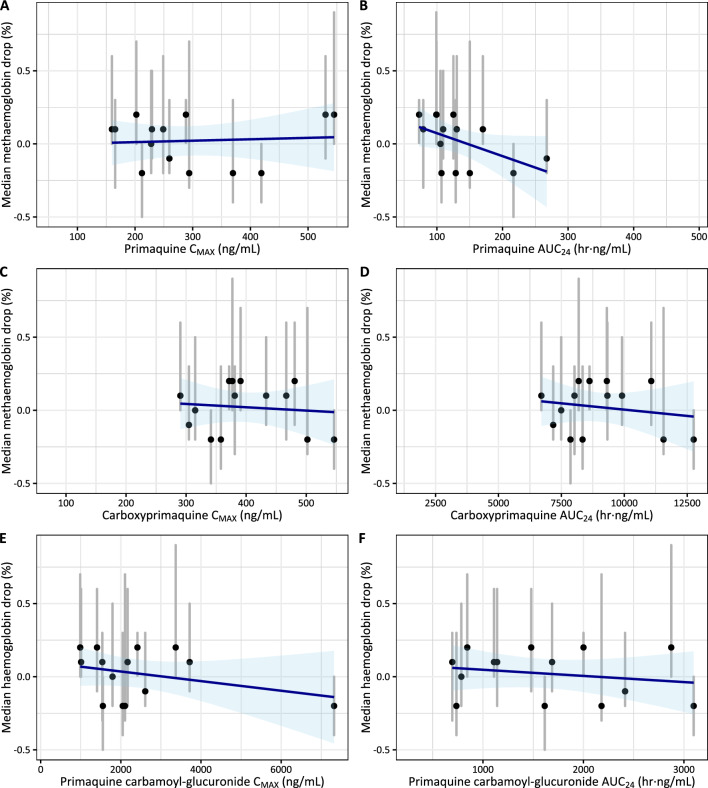
|
| 219 |
+
|
| 220 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11106956_12936_2024_4979_Fig5_HTML.jpg)
|
| 221 |
+
|
| 222 |
+
Correlation between pharmacokinetic parameters (CMAX and AUC24) and the median haemoglobin drop for primaquine (A, B), carboxy-primaquine (C, D), and primaquine cabarmoyl-glucuronide (E, F) in G6PD-deficient individuals (n = 18). Blue lines and shaded areas represent the linear regression with associated prediction intervals. Black circles and grey lines represent individual’s mean and range of observed methaemoglobin drop. None of these correlations are significantly different from the zero-slope
|
| 223 |
+
|
| 224 |
+
## Discussion
|
| 225 |
+
|
| 226 |
+
Pharmacokinetic properties of PQ and CPQ estimated in this study were similar to previous literature reports [[2](#CR2)2, [7](#CR7)7, [22](#CR22)22]. Pharmacokinetic properties of the carbamoyl-glucuronide metabolite are reported here for the first time.
|
| 227 |
+
|
| 228 |
+
Pharmacokinetic models of PQ and its metabolites were fitted separately due to the unknown fraction of total primaquine elimination resulting in each specific metabolic pathway. Metabolism of PQ is quite complex which several enzymes and intermediate metabolites involved [[8](#CR8)8]. CPQ is a stable abundant metabolite mediated by monoamine oxidase A (MAO-A) and it’s commonly measured together with the parent drug. On the other hand, PQ is also metabolized by the cytochrome P450 2D6 (CYP2D6) isoform. Quinone-imine and orthoquinone metabolites can generate a reactive oxygen species through redox cycling, and might be therefore responsible for antiparasitic activity and haemotoxicity. Drug-metabolite models of PQ and CPQ have been reported previously, both as empirical and mechanistic population pharmacokinetic models [[7](#CR7)7, [23](#CR23)23, [24](#CR24)24]. One-compartment disposition kinetic of PQ and CPQ reported here were congruent with previous studies. PQ was estimated to have a short terminal elimination half-life of approximately 7.42 h, while its carboxy metabolite had a longer terminal elimination half-life of approximately 19.5 days. This estimated terminal half-life of CPQ here was substantially longer than that previously reported in healthy volunteers (19.5 days vs 15.6 h) [[7](#CR7)7]. This might be explained by a short period of sampling times in previous studies (i.e. within 24 h); resulting in capturing mostly the absorption/distribution phase and not the terminal elimination phase, and thereby underestimating the true half-life of CPQ. Disposition kinetics of PQCG was explained best by using a one-compartment disposition model and resulted in an estimated terminal elimination half-life of 17.5 h. Implementation of CYP2D6 or G6PD status did not improve the pharmacokinetic model significantly.
|
| 229 |
+
|
| 230 |
+
As expected, it was a close correlation (1:1) between capillary plasma and venous plasma concentrations of PQ, CPQ and PQCG [[25](#CR25)25]. In this study, the conversion factor between capillary and venous plasma concentrations of PQ, CPQ, and PQCG were estimated at 32.9%, 69.1%, and 40.1%, respectively. This is not entirely consistent with literature, which report a capillary to venous plasma ratio of approximately 1:1 for PQ and CPQ [[25](#CR25)25]. This needs to be investigate further to elucidate potential differences in capillary vs venous plasma measurements in different populations and settings.
|
| 231 |
+
|
| 232 |
+
The WHO suggests to add PQ to the standard ACT for the treatment of uncomplicated falciparum malaria in order to block further malaria transmission [[1](#CR1)1]. This SLD-PQ (0.25 mg/kg) has been shown to be sufficient for reducing gametocyte carriage and preventing malaria transmission [[2](#CR2)2, [26](#CR26)26]. The same dose is also shown to be safe in G6PD-deficient individuals and does not require prior G6PD activity testing [[5](#CR5)5]. This study showed that the SLD-PQ is associated with a small increment of oxidative stress and low risk of haemolytic events in the G6PD-deficient individuals. However, a long-term PQ administration (15 mg daily for 14 days) in the G6PD-deficient individuals is associated with a high risk of acute haemolytic anaemia, and once weekly dosing for 8 weeks is recommended in these individuals. A mathematical modelling study proposed that a daily escalating PQ dose in G6PD-deficient individuals might be safer than the current weekly dosing regimen [[27](#CR27)27]. The predicted median reduction of haemoglobin after 0.25 and 0.40 mg/kg SLD-PQ in G6PD-deficient individual was 0.35 g/dL (90% CI 0.12–0.65 g/dL) and 0.56 g/dL (90% CI 0.21–1.00 g/dL), respectively [[28](#CR28)28].
|
| 233 |
+
|
| 234 |
+
This study has several limitations. Pharmacokinetic data were collected only for 24 h after drug administration, and the sampling schedule did not capture fully the elimination phase of CPQ. A pharmacokinetic study with longer duration of pharmacokinetic sampling and more data in the elimination phase of CPQ could improve the accuracy of the estimated parameters. PQ enantiomers have been shown to exhibit different pharmacokinetic profiles [[6](#CR6)6, [7](#CR7)7, [9](#CR9)9], but the bioanalytical assays used here could not distinguish between different enantiomers. Thus, the racemic mixture of PQ was modelled and no conclusions can be drawn on the contribution of individual enantiomers. The metabolism pathways of PQ are quite complex with several inactive compounds involved [[8](#CR8)8], and only few of these compounds can be quantified in clinical samples. More specifically 5,6-orthoquinoline, the metabolite believed to be responsible for adverse reactions and linked to clinical outcome [[11](#CR11)11], cannot be measured in human plasma. Therefore, the association between PQ and the active orthoquinone compound has not been well characterized.
|
| 235 |
+
|
| 236 |
+
## Conclusions
|
| 237 |
+
|
| 238 |
+
In summary, the population pharmacokinetic properties of PQ, CPQ, and PQCG have been characterized and reported here. No statistically significant relationships were seen between the pharmacokinetic parameters and the change in haemoglobin levels in G6PD-deficient patients after a single low dose of primaquine. A single low dose (0.50 mg/kg) of PQ was haematologically safe in this population of G6PD-deficient African males without malaria.
|
| 239 |
+
|
| 240 |
+
## Supplementary Information
|
| 241 |
+
|
| 242 |
+
## Acknowledgements
|
| 243 |
+
|
| 244 |
+
We are very thankful for all the participants, investigators, and the trial site staff who were involved in the conduct of this study.
|
| 245 |
+
|
| 246 |
+
## Abbreviations
|
| 247 |
+
|
| 248 |
+
## Author contributions
|
| 249 |
+
|
| 250 |
+
IC, RG, AD, JMB, and TB designed the clinical trial that collected samples for this study. IC and LAW conceived of this study. AM, KD, KS, and HD implemented the study. PC, PSF and JT contributed to the analysis and interpretation of the data. PC wrote the first draft of the manuscript. All authors critically revised the manuscript for content and approved the final version of the manuscript. All authors read and approved the final manuscript.
|
| 251 |
+
|
| 252 |
+
## Funding
|
| 253 |
+
|
| 254 |
+
This work was support by the Wellcome Trust [220211]; the Bill and Melinda Gates Foundation [INV-006052]; and was part of the Wellcome Trust-Mahidol University-Oxford Tropical Medicine Research Programme. The funding bodies did not have any role in the collection, analysis, interpretation of data, writing of the manuscript, or in the decision in submitting the manuscript for publication. A Creative Commons Attribution 4.0 Generic License has already been assigned to the Author Accepted Manuscript version.
|
| 255 |
+
|
| 256 |
+
## Data availability
|
| 257 |
+
|
| 258 |
+
Deidentified participant data will be available after publication to applicants who provide a sound proposal to the Mahidol Oxford Tropical Medicine Research Unit Data Access Committee. They can contact the corresponding author in the first instance.
|
| 259 |
+
|
| 260 |
+
## Declarations
|
| 261 |
+
|
| 262 |
+
### Ethics approval and consent to participate
|
| 263 |
+
|
| 264 |
+
This study was approved by the Ethics Committee of the Faculty of Medicine, Pharmacy, and Dentistry, University of Science, Techniques and Technologies of Bamako (approval no. 2015/89/CE/FMPOS), and by the Committee on Human Research at the University of California, San Francisco (institutional review board approval no. 14-14495). It also underwent human subjects review at the US Centers for Disease Control and Prevention and approved as non-engagement in human subject research. The trial was registered at ClinicalTrials.gov (registration no. [NCT02535767](https://clinicaltrials.gov/ct2/show/NCT02535767)NCT02535767).
|
| 265 |
+
|
| 266 |
+
### Consent for publication
|
| 267 |
+
|
| 268 |
+
The confidentiality of the participants’ records has been observed according to ethical regulations.
|
| 269 |
+
|
| 270 |
+
### Competing interests
|
| 271 |
+
|
| 272 |
+
All authors declare that they have no competing interests.
|
| 273 |
+
|
| 274 |
+
## Footnotes
|
| 275 |
+
|
| 276 |
+
## Associated Data
|
| 277 |
+
|
| 278 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 279 |
+
|
| 280 |
+
### Supplementary Materials
|
| 281 |
+
|
| 282 |
+
### Data Availability Statement
|
| 283 |
+
|
| 284 |
+
Deidentified participant data will be available after publication to applicants who provide a sound proposal to the Mahidol Oxford Tropical Medicine Research Unit Data Access Committee. They can contact the corresponding author in the first instance.
|
| 285 |
+
|
| 286 |
+
### Supplementary Materials
|
| 287 |
+
|
| 288 |
+
### Data Availability Statement
|
| 289 |
+
|
| 290 |
+
Deidentified participant data will be available after publication to applicants who provide a sound proposal to the Mahidol Oxford Tropical Medicine Research Unit Data Access Committee. They can contact the corresponding author in the first instance.
|
| 291 |
+
|
| 292 |
+
## References
|
| 293 |
+
|
| 294 |
+
1. WHO . Guidelines for the treatment of malaria. 3. Geneva: World Health Organization; 2015. [PubMed](https://pubmed.ncbi.nlm.nih.gov/26020088/) | [Google Scholar](https://scholar.google.com/scholar_lookup?title=Guidelines%20for%20the%20treatment%20of%20malaria&publication_year=2015&)
|
| 295 |
+
|
| 296 |
+
2. Chotsiri P, Mahamar A, Hoglund RM, Koita F, Sanogo K, Diawara H, et al. Mechanistic modeling of primaquine pharmacokinetics, gametocytocidal activity, and mosquito infectivity. Clin Pharmacol Ther. 2022;111:676–685. doi: 10.1002/cpt.2512. [DOI](https://doi.org/10.1002/cpt.2512) | [PMC free article](/articles/PMC9302630/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34905220/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Mechanistic%20modeling%20of%20primaquine%20pharmacokinetics,%20gametocytocidal%20activity,%20and%20mosquito%20infectivity&author=P%20Chotsiri&author=A%20Mahamar&author=RM%20Hoglund&author=F%20Koita&author=K%20Sanogo&volume=111&publication_year=2022&pages=676-685&pmid=34905220&doi=10.1002/cpt.2512&)
|
| 297 |
+
|
| 298 |
+
3. Bradley J, Soumare HM, Mahamar A, Diawara H, Roh M, Delves M, et al. Transmission-blocking effects of primaquine and methylene blue suggest Plasmodium falciparum gametocyte sterilization rather than effects on sex ratio. Clin Infect Dis. 2019;69:1436–1439. doi: 10.1093/cid/ciz134. [DOI](https://doi.org/10.1093/cid/ciz134) | [PMC free article](/articles/PMC6763632/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30753355/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Infect%20Dis&title=Transmission-blocking%20effects%20of%20primaquine%20and%20methylene%20blue%20suggest%20Plasmodium%20falciparum%20gametocyte%20sterilization%20rather%20than%20effects%20on%20sex%20ratio&author=J%20Bradley&author=HM%20Soumare&author=A%20Mahamar&author=H%20Diawara&author=M%20Roh&volume=69&publication_year=2019&pages=1436-1439&pmid=30753355&doi=10.1093/cid/ciz134&)
|
| 299 |
+
|
| 300 |
+
4. Dicko A, Brown JM, Diawara H, Baber I, Mahamar A, Soumare HM, et al. Primaquine to reduce transmission of Plasmodium falciparum malaria in Mali: a single-blind, dose-ranging, adaptive randomised phase 2 trial. Lancet Infect Dis. 2016;16:674–684. doi: 10.1016/S1473-3099(15)00479-X. [DOI](https://doi.org/10.1016/S1473-3099(15)00479-X) | [PMC free article](/articles/PMC10583596/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26906747/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet%20Infect%20Dis&title=Primaquine%20to%20reduce%20transmission%20of%20Plasmodium%20falciparum%20malaria%20in%20Mali:%20a%20single-blind,%20dose-ranging,%20adaptive%20randomised%20phase%202%20trial&author=A%20Dicko&author=JM%20Brown&author=H%20Diawara&author=I%20Baber&author=A%20Mahamar&volume=16&publication_year=2016&pages=674-684&pmid=26906747&doi=10.1016/S1473-3099(15)00479-X&)
|
| 301 |
+
|
| 302 |
+
5. WHO . Policy brief on single-dose primaquine as a gametocytocide in Plasmodium falciparum malaria. Geneva: World Health Organization; 2015. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Policy%20brief%20on%20single-dose%20primaquine%20as%20a%20gametocytocide%20in%20Plasmodium%20falciparum%20malaria&publication_year=2015&)
|
| 303 |
+
|
| 304 |
+
6. Tekwani BL, Avula B, Sahu R, Chaurasiya ND, Khan SI, Jain S, et al. Enantioselective pharmacokinetics of primaquine in healthy human volunteers. Drug Metab Dispos. 2015;43:571–577. doi: 10.1124/dmd.114.061127. [DOI](https://doi.org/10.1124/dmd.114.061127) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25637634/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Metab%20Dispos&title=Enantioselective%20pharmacokinetics%20of%20primaquine%20in%20healthy%20human%20volunteers&author=BL%20Tekwani&author=B%20Avula&author=R%20Sahu&author=ND%20Chaurasiya&author=SI%20Khan&volume=43&publication_year=2015&pages=571-577&pmid=25637634&doi=10.1124/dmd.114.061127&)
|
| 305 |
+
|
| 306 |
+
7. Chairat K, Jittamala P, Hanboonkunupakarn B, Pukrittayakamee S, Hanpithakpong W, Blessborn D, et al. Enantiospecific pharmacokinetics and drug-drug interactions of primaquine and blood-stage antimalarial drugs. J Antimicrob Chemother. 2018;73:3102–3113. doi: 10.1093/jac/dky297. [DOI](https://doi.org/10.1093/jac/dky297) | [PMC free article](/articles/PMC6198747/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30085149/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Antimicrob%20Chemother&title=Enantiospecific%20pharmacokinetics%20and%20drug-drug%20interactions%20of%20primaquine%20and%20blood-stage%20antimalarial%20drugs&author=K%20Chairat&author=P%20Jittamala&author=B%20Hanboonkunupakarn&author=S%20Pukrittayakamee&author=W%20Hanpithakpong&volume=73&publication_year=2018&pages=3102-3113&pmid=30085149&doi=10.1093/jac/dky297&)
|
| 307 |
+
|
| 308 |
+
8. Avula B, Tekwani BL, Chaurasiya ND, Fasinu P, Dhammika Nanayakkara NP, et al. Metabolism of primaquine in normal human volunteers: investigation of phase I and phase II metabolites from plasma and urine using ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry. Malar J. 2018;17:294. doi: 10.1186/s12936-018-2433-z. [DOI](https://doi.org/10.1186/s12936-018-2433-z) | [PMC free article](/articles/PMC6090659/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30103751/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Malar%20J&title=Metabolism%20of%20primaquine%20in%20normal%20human%20volunteers:%20investigation%20of%20phase%20I%20and%20phase%20II%20metabolites%20from%20plasma%20and%20urine%20using%20ultra-high%20performance%20liquid%20chromatography-quadrupole%20time-of-flight%20mass%20spectrometry&author=B%20Avula&author=BL%20Tekwani&author=ND%20Chaurasiya&author=P%20Fasinu&author=NP%20Dhammika%20Nanayakkara&volume=17&publication_year=2018&pages=294&pmid=30103751&doi=10.1186/s12936-018-2433-z&)
|
| 309 |
+
|
| 310 |
+
9. Fasinu PS, Avula B, Tekwani BL, Nanayakkara NP, Wang YH, Bandara Herath HM, et al. Differential kinetic profiles and metabolism of primaquine enantiomers by human hepatocytes. Malar J. 2016;15:224. doi: 10.1186/s12936-016-1270-1. [DOI](https://doi.org/10.1186/s12936-016-1270-1) | [PMC free article](/articles/PMC4837544/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27093859/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Malar%20J&title=Differential%20kinetic%20profiles%20and%20metabolism%20of%20primaquine%20enantiomers%20by%20human%20hepatocytes&author=PS%20Fasinu&author=B%20Avula&author=BL%20Tekwani&author=NP%20Nanayakkara&author=YH%20Wang&volume=15&publication_year=2016&pages=224&pmid=27093859&doi=10.1186/s12936-016-1270-1&)
|
| 311 |
+
|
| 312 |
+
10. Camarda G, Jirawatcharadech P, Priestley RS, Saif A, March S, Wong MHL, et al. Antimalarial activity of primaquine operates via a two-step biochemical relay. Nat Commun. 2019;10:3226. doi: 10.1038/s41467-019-11239-0. [DOI](https://doi.org/10.1038/s41467-019-11239-0) | [PMC free article](/articles/PMC6642103/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31324806/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat%20Commun&title=Antimalarial%20activity%20of%20primaquine%20operates%20via%20a%20two-step%20biochemical%20relay&author=G%20Camarda&author=P%20Jirawatcharadech&author=RS%20Priestley&author=A%20Saif&author=S%20March&volume=10&publication_year=2019&pages=3226&pmid=31324806&doi=10.1038/s41467-019-11239-0&)
|
| 313 |
+
|
| 314 |
+
11. Marcsisin SR, Reichard G, Pybus BS. Primaquine pharmacology in the context of CYP 2D6 pharmacogenomics: current state of the art. Pharmacol Ther. 2016;161:1–10. doi: 10.1016/j.pharmthera.2016.03.011. [DOI](https://doi.org/10.1016/j.pharmthera.2016.03.011) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27016470/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacol%20Ther&title=Primaquine%20pharmacology%20in%20the%20context%20of%20CYP%202D6%20pharmacogenomics:%20current%20state%20of%20the%20art&author=SR%20Marcsisin&author=G%20Reichard&author=BS%20Pybus&volume=161&publication_year=2016&pages=1-10&pmid=27016470&doi=10.1016/j.pharmthera.2016.03.011&)
|
| 315 |
+
|
| 316 |
+
12. Ashley EA, Recht J, White NJ. Primaquine: the risks and the benefits. Malar J. 2014;13:418. doi: 10.1186/1475-2875-13-418. [DOI](https://doi.org/10.1186/1475-2875-13-418) | [PMC free article](/articles/PMC4230503/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25363455/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Malar%20J&title=Primaquine:%20the%20risks%20and%20the%20benefits&author=EA%20Ashley&author=J%20Recht&author=NJ%20White&volume=13&publication_year=2014&pages=418&pmid=25363455&doi=10.1186/1475-2875-13-418&)
|
| 317 |
+
|
| 318 |
+
13. Chen I, Diawara H, Mahamar A, Sanogo K, Keita S, Kone D, et al. Safety of single-dose primaquine in G6PD-deficient and G6PD-normal males in mali without malaria: an open-label, phase 1, dose-adjustment trial. J Infect Dis. 2018;217:1298–1308. doi: 10.1093/infdis/jiy014. [DOI](https://doi.org/10.1093/infdis/jiy014) | [PMC free article](/articles/PMC5974787/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29342267/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Infect%20Dis&title=Safety%20of%20single-dose%20primaquine%20in%20G6PD-deficient%20and%20G6PD-normal%20males%20in%20mali%20without%20malaria:%20an%20open-label,%20phase%201,%20dose-adjustment%20trial&author=I%20Chen&author=H%20Diawara&author=A%20Mahamar&author=K%20Sanogo&author=S%20Keita&volume=217&publication_year=2018&pages=1298-1308&pmid=29342267&doi=10.1093/infdis/jiy014&)
|
| 319 |
+
|
| 320 |
+
14. Avula B, Khan SI, Tekwani BL, Nanayakkara NP, McChesney JD, Walker LA, et al. Analysis of primaquine and its metabolite carboxyprimaquine in biological samples: enantiomeric separation, method validation and quantification. Biomed Chromatogr. 2011;25:1010–1017. doi: 10.1002/bmc.1557. [DOI](https://doi.org/10.1002/bmc.1557) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21058417/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Biomed%20Chromatogr&title=Analysis%20of%20primaquine%20and%20its%20metabolite%20carboxyprimaquine%20in%20biological%20samples:%20enantiomeric%20separation,%20method%20validation%20and%20quantification&author=B%20Avula&author=SI%20Khan&author=BL%20Tekwani&author=NP%20Nanayakkara&author=JD%20McChesney&volume=25&publication_year=2011&pages=1010-1017&pmid=21058417&doi=10.1002/bmc.1557&)
|
| 321 |
+
|
| 322 |
+
15. Keizer RJ, van Benten M, Beijnen JH, Schellens JH, Huitema AD. Pirana and PCluster: a modeling environment and cluster infrastructure for NONMEM. Comput Methods Programs Biomed. 2011;101:72–79. doi: 10.1016/j.cmpb.2010.04.018. [DOI](https://doi.org/10.1016/j.cmpb.2010.04.018) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20627442/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Comput%20Methods%20Programs%20Biomed&title=Pirana%20and%20PCluster:%20a%20modeling%20environment%20and%20cluster%20infrastructure%20for%20NONMEM&author=RJ%20Keizer&author=M%20van%20Benten&author=JH%20Beijnen&author=JH%20Schellens&author=AD%20Huitema&volume=101&publication_year=2011&pages=72-79&pmid=20627442&doi=10.1016/j.cmpb.2010.04.018&)
|
| 323 |
+
|
| 324 |
+
16. Lindbom L, Ribbing J, Jonsson EN. Perl-speaks-NONMEM (PsN)–a Perl module for NONMEM related programming. Comput Methods Programs Biomed. 2004;75:85–94. doi: 10.1016/j.cmpb.2003.11.003. [DOI](https://doi.org/10.1016/j.cmpb.2003.11.003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15212851/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Comput%20Methods%20Programs%20Biomed&title=Perl-speaks-NONMEM%20(PsN)%E2%80%93a%20Perl%20module%20for%20NONMEM%20related%20programming&author=L%20Lindbom&author=J%20Ribbing&author=EN%20Jonsson&volume=75&publication_year=2004&pages=85-94&pmid=15212851&doi=10.1016/j.cmpb.2003.11.003&)
|
| 325 |
+
|
| 326 |
+
17. Ahn JE, Karlsson MO, Dunne A, Ludden TM. Likelihood based approaches to handling data below the quantification limit using NONMEM VI. J Pharmacokinet Pharmacodyn. 2008;35:401–421. doi: 10.1007/s10928-008-9094-4. [DOI](https://doi.org/10.1007/s10928-008-9094-4) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18686017/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharmacokinet%20Pharmacodyn&title=Likelihood%20based%20approaches%20to%20handling%20data%20below%20the%20quantification%20limit%20using%20NONMEM%20VI&author=JE%20Ahn&author=MO%20Karlsson&author=A%20Dunne&author=TM%20Ludden&volume=35&publication_year=2008&pages=401-421&pmid=18686017&doi=10.1007/s10928-008-9094-4&)
|
| 327 |
+
|
| 328 |
+
18. Karlsson MO, Savic RM. Diagnosing model diagnostics. Clin Pharmacol Ther. 2007;82:17–20. doi: 10.1038/sj.clpt.6100241. [DOI](https://doi.org/10.1038/sj.clpt.6100241) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17571070/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Diagnosing%20model%20diagnostics&author=MO%20Karlsson&author=RM%20Savic&volume=82&publication_year=2007&pages=17-20&pmid=17571070&doi=10.1038/sj.clpt.6100241&)
|
| 329 |
+
|
| 330 |
+
19. Dosne AG, Bergstrand M, Karlsson MO. An automated sampling importance resampling procedure for estimating parameter uncertainty. J Pharmacokinet Pharmacodyn. 2017;44:509–520. doi: 10.1007/s10928-017-9542-0. [DOI](https://doi.org/10.1007/s10928-017-9542-0) | [PMC free article](/articles/PMC5686280/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28887735/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharmacokinet%20Pharmacodyn&title=An%20automated%20sampling%20importance%20resampling%20procedure%20for%20estimating%20parameter%20uncertainty&author=AG%20Dosne&author=M%20Bergstrand&author=MO%20Karlsson&volume=44&publication_year=2017&pages=509-520&pmid=28887735&doi=10.1007/s10928-017-9542-0&)
|
| 331 |
+
|
| 332 |
+
20. Dosne AG, Bergstrand M, Harling K, Karlsson MO. Improving the estimation of parameter uncertainty distributions in nonlinear mixed effects models using sampling importance resampling. J Pharmacokinet Pharmacodyn. 2016;43:583–596. doi: 10.1007/s10928-016-9487-8. [DOI](https://doi.org/10.1007/s10928-016-9487-8) | [PMC free article](/articles/PMC5110709/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27730482/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharmacokinet%20Pharmacodyn&title=Improving%20the%20estimation%20of%20parameter%20uncertainty%20distributions%20in%20nonlinear%20mixed%20effects%20models%20using%20sampling%20importance%20resampling&author=AG%20Dosne&author=M%20Bergstrand&author=K%20Harling&author=MO%20Karlsson&volume=43&publication_year=2016&pages=583-596&pmid=27730482&doi=10.1007/s10928-016-9487-8&)
|
| 333 |
+
|
| 334 |
+
21. Bergstrand M, Hooker AC, Wallin JE, Karlsson MO. Prediction-corrected visual predictive checks for diagnosing nonlinear mixed-effects models. AAPS J. 2011;13:143–151. doi: 10.1208/s12248-011-9255-z. [DOI](https://doi.org/10.1208/s12248-011-9255-z) | [PMC free article](/articles/PMC3085712/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21302010/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=AAPS%20J&title=Prediction-corrected%20visual%20predictive%20checks%20for%20diagnosing%20nonlinear%20mixed-effects%20models&author=M%20Bergstrand&author=AC%20Hooker&author=JE%20Wallin&author=MO%20Karlsson&volume=13&publication_year=2011&pages=143-151&pmid=21302010&doi=10.1208/s12248-011-9255-z&)
|
| 335 |
+
|
| 336 |
+
22. Hanboonkunupakarn B, Ashley EA, Jittamala P, Tarning J, Pukrittayakamee S, Hanpithakpong W, et al. Open-label crossover study of primaquine and dihydroartemisinin-piperaquine pharmacokinetics in healthy adult thai subjects. Antimicrob Agents Chemother. 2014;58:7340–7346. doi: 10.1128/AAC.03704-14. [DOI](https://doi.org/10.1128/AAC.03704-14) | [PMC free article](/articles/PMC4249579/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25267661/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Open-label%20crossover%20study%20of%20primaquine%20and%20dihydroartemisinin-piperaquine%20pharmacokinetics%20in%20healthy%20adult%20thai%20subjects&author=B%20Hanboonkunupakarn&author=EA%20Ashley&author=P%20Jittamala&author=J%20Tarning&author=S%20Pukrittayakamee&volume=58&publication_year=2014&pages=7340-7346&pmid=25267661&doi=10.1128/AAC.03704-14&)
|
| 337 |
+
|
| 338 |
+
23. Moore BR, Salman S, Benjamin J, Page-Sharp M, Robinson LJ, Waita E, et al. Pharmacokinetic properties of single-dose primaquine in Papua New Guinean children: feasibility of abbreviated high-dose regimens for radical cure of vivax malaria. Antimicrob Agents Chemother. 2014;58:432–439. doi: 10.1128/AAC.01437-13. [DOI](https://doi.org/10.1128/AAC.01437-13) | [PMC free article](/articles/PMC3910777/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24189254/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Pharmacokinetic%20properties%20of%20single-dose%20primaquine%20in%20Papua%20New%20Guinean%20children:%20feasibility%20of%20abbreviated%20high-dose%20regimens%20for%20radical%20cure%20of%20vivax%20malaria&author=BR%20Moore&author=S%20Salman&author=J%20Benjamin&author=M%20Page-Sharp&author=LJ%20Robinson&volume=58&publication_year=2014&pages=432-439&pmid=24189254&doi=10.1128/AAC.01437-13&)
|
| 339 |
+
|
| 340 |
+
24. Goncalves BP, Pett H, Tiono AB, Murry D, Sirima SB, Niemi M, et al. Age, weight, and CYP2D6 genotype are major determinants of primaquine pharmacokinetics in African children. Antimicrob Agents Chemother. 2017;61:e02590–e2616. doi: 10.1128/AAC.02590-16. [DOI](https://doi.org/10.1128/AAC.02590-16) | [PMC free article](/articles/PMC5404566/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28289025/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Age,%20weight,%20and%20CYP2D6%20genotype%20are%20major%20determinants%20of%20primaquine%20pharmacokinetics%20in%20African%20children&author=BP%20Goncalves&author=H%20Pett&author=AB%20Tiono&author=D%20Murry&author=SB%20Sirima&volume=61&publication_year=2017&pages=e02590-e2616&pmid=28289025&doi=10.1128/AAC.02590-16&)
|
| 341 |
+
|
| 342 |
+
25. Gilder ME, Hanpithakphong W, Hoglund RM, Tarning J, Win HH, Hilda N, et al. Primaquine pharmacokinetics in lactating women and breastfed infant exposures. Clin Infect Dis. 2018;67:1000–1007. doi: 10.1093/cid/ciy235. [DOI](https://doi.org/10.1093/cid/ciy235) | [PMC free article](/articles/PMC6137118/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29590311/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Infect%20Dis&title=Primaquine%20pharmacokinetics%20in%20lactating%20women%20and%20breastfed%20infant%20exposures&author=ME%20Gilder&author=W%20Hanpithakphong&author=RM%20Hoglund&author=J%20Tarning&author=HH%20Win&volume=67&publication_year=2018&pages=1000-1007&pmid=29590311&doi=10.1093/cid/ciy235&)
|
| 343 |
+
|
| 344 |
+
26. Goncalves BP, Tiono AB, Ouedraogo A, Guelbeogo WM, Bradley J, Nebie I, et al. Single low dose primaquine to reduce gametocyte carriage and Plasmodium falciparum transmission after artemether-lumefantrine in children with asymptomatic infection: a randomised, double-blind, placebo-controlled trial. BMC Med. 2016;14:40. doi: 10.1186/s12916-016-0581-y. [DOI](https://doi.org/10.1186/s12916-016-0581-y) | [PMC free article](/articles/PMC4782330/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26952094/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=BMC%20Med&title=Single%20low%20dose%20primaquine%20to%20reduce%20gametocyte%20carriage%20and%20Plasmodium%20falciparum%20transmission%20after%20artemether-lumefantrine%20in%20children%20with%20asymptomatic%20infection:%20a%20randomised,%20double-blind,%20placebo-controlled%20trial&author=BP%20Goncalves&author=AB%20Tiono&author=A%20Ouedraogo&author=WM%20Guelbeogo&author=J%20Bradley&volume=14&publication_year=2016&pages=40&pmid=26952094&doi=10.1186/s12916-016-0581-y&)
|
| 345 |
+
|
| 346 |
+
27. Watson J, Taylor WR, Menard D, Kheng S, White NJ. Modelling primaquine-induced haemolysis in G6PD deficiency. Elife. 2017;6:e23061. doi: 10.7554/eLife.23061. [DOI](https://doi.org/10.7554/eLife.23061) | [PMC free article](/articles/PMC5330681/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28155819/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Elife&title=Modelling%20primaquine-induced%20haemolysis%20in%20G6PD%20deficiency&author=J%20Watson&author=WR%20Taylor&author=D%20Menard&author=S%20Kheng&author=NJ%20White&volume=6&publication_year=2017&pages=e23061&pmid=28155819&doi=10.7554/eLife.23061&)
|
| 347 |
+
|
| 348 |
+
28. van Beek SW, Svensson EM, Tiono AB, Okebe J, D'Alessandro U, Goncalves BP, et al. Model-based assessment of the safety of community interventions with primaquine in sub-Saharan Africa. Parasit Vectors. 2021;14:524. doi: 10.1186/s13071-021-05034-4. [DOI](https://doi.org/10.1186/s13071-021-05034-4) | [PMC free article](/articles/PMC8502297/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34627346/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Parasit%20Vectors&title=Model-based%20assessment%20of%20the%20safety%20of%20community%20interventions%20with%20primaquine%20in%20sub-Saharan%20Africa&author=SW%20van%20Beek&author=EM%20Svensson&author=AB%20Tiono&author=J%20Okebe&author=U%20D'Alessandro&volume=14&publication_year=2021&pages=524&pmid=34627346&doi=10.1186/s13071-021-05034-4&)
|
test/texts/PMC11120965.md
ADDED
|
@@ -0,0 +1,323 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Impact of CYP2C19 Gene Variants on Long-Term Treatment with Atorvastatin in Patients with Acute Coronary Syndromes
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Darius Čereškevičius, Vytautas Zabiela, Ali Aldujeli, Vaiva Lesauskaitė, Kristina Zubielienė, Vytautas Raškevičius, Ieva Čiapienė, Diana Žaliaduonytė, Agnė Giedraitienė, Vaidotas Žvikas, Valdas Jakštas, Vilius Skipskis, Olivija Dobilienė, Gintarė Šakalytė, Vacis Tatarūnas
|
| 5 |
+
**Journal:** International Journal of Molecular Sciences
|
| 6 |
+
**Date:** 2024 May 15
|
| 7 |
+
**DOI:** [10.3390/ijms25105385](https://doi.org/10.3390/ijms25105385)
|
| 8 |
+
**PMID:** 38791422
|
| 9 |
+
**PMCID:** PMC11120965
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11120965/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11120965/pdf/ijms-25-05385.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11120965/pdf/ijms-25-05385.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
The effectiveness of lipid-lowering therapies may be insufficient in high-risk cardiovascular patients and depends on the genetic variability of drug-metabolizing enzymes. Customizing statin therapy, including treatment with atorvastatin, may improve clinical outcomes. Currently, there is a lack of guidelines allowing the prediction of the therapeutic efficacy of lipid-lowering statin therapy. This study aimed to determine the effects of clinically significant gene variants of CYP2C19 on atorvastatin therapy in patients with acute coronary syndromes. In total, 92 patients with a confirmed diagnosis of ST-elevation myocardial infarction (STEMI) or non-ST-elevation myocardial infarction (NSTEMI) were sequenced for target regions within the CYP2C19 gene on the Illumina Miniseq system. The CYP2C19 poor metabolizer phenotype (carriers of CYP2C19*2, CYP2C19*4, and CYP2C19*8 alleles) was detected in 29% of patients. These patients had significantly lower responses to treatment with atorvastatin than patients with the normal metabolizer phenotype. CYP2C19-metabolizing phenotype, patient age, and smoking increased the odds of undertreatment in patients (∆LDL-C (mmol/L) < 1). These results revealed that the CYP2C19 phenotype may significantly impact atorvastatin therapy personalization in patients requiring LDL lipid-lowering therapy.
|
| 16 |
+
|
| 17 |
+
Keywords: statins, atorvastatin, LDL cholesterol, CYP2C19, STEMI, NSTEMI
|
| 18 |
+
|
| 19 |
+
**Keywords:**Keywords: statins, atorvastatin, LDL cholesterol, CYP2C19, STEMI, NSTEMI
|
| 20 |
+
|
| 21 |
+
## 1. Introduction
|
| 22 |
+
|
| 23 |
+
Hypercholesterolemia, or hyperlipidemia, is characterized by elevated levels of cholesterol in the blood and stands as a major risk factor for cardiovascular diseases, contributing significantly to the global burden of morbidity and mortality [[1](#B1-ijms-25-05385)1]. While lifestyle factors and dietary choices significantly impact cholesterol concentrations, genetic factors play a pivotal role in determining a predisposition to hypercholesterolemia [[2](#B2-ijms-25-05385)2]. A large study sponsored by the NIH (National Institutes of Health) discovered that a cholesterol-lowering diet and medical treatment decreased heart attacks in males with high cholesterol [[3](#B3-ijms-25-05385)3].
|
| 24 |
+
|
| 25 |
+
The discovery of compounds decreasing cholesterol levels in 1976 [[4](#B4-ijms-25-05385)4,[5](#B5-ijms-25-05385)5] has paved the way for a new class of drugs, 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMG-CoA reductase, HMGCR) inhibitors or statins. *Goldstein*Goldstein and *Brown*Brown were the first to describe that patients with familial hypercholesterolemia who lack low-density lipoprotein receptors (LDLr) and produce increased amounts of cholesterol in response to the absence of these receptors [[6](#B6-ijms-25-05385)6]. It was assumed that inhibiting cholesterol through HMG-CoA reductase and the upregulation of LDLr is a key tool in the prevention of atherosclerotic cardiovascular disease [[7](#B7-ijms-25-05385)7,[8](#B8-ijms-25-05385)8]. In addition, statins can have a positive effect by stabilizing the atherosclerotic plaque and reducing atherothrombotic events through this mechanism [[7](#B7-ijms-25-05385)7].
|
| 26 |
+
|
| 27 |
+
The current European Society of Cardiology guidelines for the management of dyslipidemias [[9](#B9-ijms-25-05385)9] and American Heart Association guidelines for the Management of Patients With Chronic Coronary Disease [[10](#B10-ijms-25-05385)10] agree that primary prevention with statins, according to the patients’ risk category, should be initiated when lifestyle modification does not work and the low-density lipoprotein–cholesterol (LDL-C) goal is not achieved. For chronic coronary syndrome (CCS), familial hypercholesterolemia (FH), severe diabetes mellitus (DM), and chronic kidney disease (CKD) patients’ treatment should be initiated immediately after the diagnosis [[10](#B10-ijms-25-05385)10,[11](#B11-ijms-25-05385)11].
|
| 28 |
+
|
| 29 |
+
The main focus of the current guidelines is on lowering LDL-C through the use of drug combinations along with other lipid-lowering therapies [[9](#B9-ijms-25-05385)9,[10](#B10-ijms-25-05385)10]. Studies show that lipid-lowering therapies are insufficiently effective in patients, especially in high cardiovascular disease (CVD) risk patients [[12](#B12-ijms-25-05385)12,[13](#B13-ijms-25-05385)13,[14](#B14-ijms-25-05385)14,[15](#B15-ijms-25-05385)15,[16](#B16-ijms-25-05385)16]. Atorvastatin is among the most commonly prescribed statins. However, studies show it in adherence in patients with dyslipidemia [[17](#B17-ijms-25-05385)17].
|
| 30 |
+
|
| 31 |
+
In this era of precision medicine, better-customized statin therapy can be used. Some authors show that genetic variants can influence statin therapy. According to *Cano-Corres*Cano-Corres, the *HMGCR*HMGCR c.1564-106A>G variant reduces the effect of statins [[18](#B18-ijms-25-05385)18] *Maxwell*Maxwell has examined a number of studies that have analyzed the influence of *ABCB1, APOE, KIF6, and TLR4*ABCB1, APOE, KIF6, and TLR4 on the improvement of clinical outcomes during statin therapy [[19](#B19-ijms-25-05385)19]. The first gene-based clinical guideline on the use of statins approved by the Clinical Pharmacogenetics Implementation Consortium (CPIC) was published in 2012. The gene-based prescription of simvastatin was based on *SLCO1B1*SLCO1B1 variants. In 2014, the document was updated [[20](#B20-ijms-25-05385)20]. The latest “CPIC^®^® Guideline for Statins and SLCO1B1, ABCG2 and CYP2C9“ summarizes data from genotyping studies that demonstrate the influence of *SLCO1B1, ABCG2*SLCO1B1, ABCG2, and *CYP2C9*CYP2C9 on safer statin therapy that may allow the avoidance of statin-associated effects [[21](#B21-ijms-25-05385)21]. The guidelines described here aim to reduce the risk of statin-associated musculoskeletal symptoms (SAMS), as research studies show that statins can cause muscle pain. Still, this pain is rare and usually manifests as mild symptoms [[22](#B22-ijms-25-05385)22,[23](#B23-ijms-25-05385)23].
|
| 32 |
+
|
| 33 |
+
Some studies have shown that variants in *CYP2C19*CYP2C19 may impact treatment effectiveness with statins [[24](#B24-ijms-25-05385)24]. Currently, no clear studies show that the effect of atorvastatin may depend on *CYP2C19*CYP2C19 gene variants. It is worth noting that an individual carrying a non-functional allele such as *CYP2C19*2, *3,*CYP2C19*2, *3, or **4**4 will have impaired drug metabolism and is considered an intermediate metabolizer. In contrast, an individual carrying two non-functional alleles will be regarded as a poor metabolizer [[25](#B25-ijms-25-05385)25]. Currently, no guidelines allow for the prediction of the therapeutic efficacy of lipid-lowering statin therapy. Various studies show that linoleic acid derivatives may be significant in lipid metabolism [[26](#B26-ijms-25-05385)26]. Thus, this study aimed to determine the effects of *CYP2C19*CYP2C19 gene variants and linoleic acid derivatives on atorvastatin therapy in patients with established cardiovascular disease.
|
| 34 |
+
|
| 35 |
+
## 2. Results
|
| 36 |
+
|
| 37 |
+
Among the patients, 30.4% were using statins before hospitalization, with 39.1% experiencing STEMI and 20.7% having undergone previous PCI ([Table 1](#ijms-25-05385-t001)Table 1).
|
| 38 |
+
|
| 39 |
+
### Table 1.
|
| 40 |
+
|
| 41 |
+
Basic characteristics of the represented patient’s group.
|
| 42 |
+
|
| 43 |
+
| Variable | n | % |
|
| 44 |
+
| -------- | - | - |
|
| 45 |
+
| Sex | | |
|
| 46 |
+
| Men | 49 | 53.3 |
|
| 47 |
+
| Women | 43 | 46.7 |
|
| 48 |
+
| Smoking | 54 | 58.7 |
|
| 49 |
+
| STEMI | 36 | 39.1 |
|
| 50 |
+
| Hypertension | 47 | 51.1 |
|
| 51 |
+
| Diabetes mellitus | 14 | 15.2 |
|
| 52 |
+
| Renal insufficiency | 11 | 12 |
|
| 53 |
+
| Statins at hospitalization | 28 | 30.4 |
|
| 54 |
+
| Family anamnesis of ischemic heart disease | 39 | 42.4 |
|
| 55 |
+
| Previous PCI | 19 | 20.7 |
|
| 56 |
+
Of the patients with STEMI, 28.6% were on statins, and 43.8% were non-users *prior*prior to hospitalization ([Table 2](#ijms-25-05385-t002)Table 2). No statistical significance was found between these two groups of patients.
|
| 57 |
+
|
| 58 |
+
### Table 2.
|
| 59 |
+
|
| 60 |
+
Patients with STEMI vs. NSTEMI according to statin therapy prior to hospitalization.
|
| 61 |
+
|
| 62 |
+
| STEMI | On Statins | No Statin | Fisher’s Exact Test p |
|
| 63 |
+
| ----- | ---------- | --------- | --------------------- |
|
| 64 |
+
| Yes n (%) | 8 (28.6) | 28 (43.8) | 0.245 |
|
| 65 |
+
| No n (%) | 20 (71.4) | 36 (56.3) | |
|
| 66 |
+
| Total n (%) | 28 (100) | 64 (100) | |
|
| 67 |
+
[Table 3](#ijms-25-05385-t003)Table 3 lists all variants detected in the *CYP2C19*CYP2C19 gene. Of the variants that impact CYP2C19 functionality, rs4244285 was the most common. Two other variants, rs28399504 and rs41291556, described in the PharmVar database [[27](#B27-ijms-25-05385)27], were also detected, leading to reduced enzyme activity. One patient was found to be carrying a rare variant of uncertain significance.
|
| 68 |
+
|
| 69 |
+
### Table 3.
|
| 70 |
+
|
| 71 |
+
CYP2C19 variants in the represented patient sample.
|
| 72 |
+
|
| 73 |
+
| dbSNP rs ID | c.DNA Position (NM_000771.4) | AA Change (NP_000762.2) | Pharmvar Allele | Impact on Function | No of Het | No of Hom |
|
| 74 |
+
| ----------- | ---------------------------- | ----------------------- | --------------- | ------------------ | --------- | --------- |
|
| 75 |
+
| rs3758581 | c.991A>G | p.Ile331Val | - | - | 7 | 0 |
|
| 76 |
+
| rs17885098 | c.99T>C | p.Pro33Pro | - | - | 8 | 0 |
|
| 77 |
+
| rs28399504 | c.1A>G | p.M1V | CYP2C19*4 | No function | 2 | 0 |
|
| 78 |
+
| rs58973490 | c.449G>A | p.Arg150His | CYP2C19*11 | Normal function | 1 | 0 |
|
| 79 |
+
| rs17878459 | c.276G>C | p.Glu92Asp | - | - | 12 | 0 |
|
| 80 |
+
| rs17882744 | c.1059C>T | p.His353= | - | - | 1 | 0 |
|
| 81 |
+
| rs3758580 | c.990C>T | p.Val330Val | - | - | 19 | 4 |
|
| 82 |
+
| rs142974781 | c.448C>T | p.Arg150Cys | - | - | 1 | 0 |
|
| 83 |
+
| rs41291556 | c.358T>C | p.Trp120Arg | CYP2C19*8 | No function | 1 | 0 |
|
| 84 |
+
| rs4244285 | c.681G>A | p.Pro227Pro | CYP2C19*2 | No function | 19 | 4 |
|
| 85 |
+
Patients were divided into two groups based on their CYP2C19 metabolism: poor metabolizers and normal metabolizers. Carriers of the CYP2C19*2, CYP2C19*4, and CYP2C19*8 alleles were considered poor metabolizers, while all other patients were considered normal metabolizers. Normal metabolizers had a higher reduction in LDL-C ([Table 4](#ijms-25-05385-t004)Table 4).
|
| 86 |
+
|
| 87 |
+
### Table 4.
|
| 88 |
+
|
| 89 |
+
Patient CYP2C19 phenotype in the represented samples.
|
| 90 |
+
|
| 91 |
+
| CYP2C19 Metabolizer Status | n (%) | LDL-C at Hospitalization (mmol/L) Median (Min-Max) | LDL-C 6 Months after Hospitalization (mmol/L) Median (Min-Max) | ∆LDL-C (mmol/L) Median (Min-Max) | p-Value *** |
|
| 92 |
+
| -------------------------- | ----- | -------------------------------------------------- | -------------------------------------------------------------- | -------------------------------- | ----------- |
|
| 93 |
+
| Normal metabolizer | 66 (71) | 3.88 (2.87–6.36) | 2.24 (1.17–5.41) ** | 1.69 (0.13–3.57) | 0.006 |
|
| 94 |
+
| Poor metabolizer * | 27 (29) | 3.78 (2.79–6.32) | 2.78 (1.19–5.97) ** | 0.49 (0.01–4.09) | |
|
| 95 |
+
| Total | 93 (100) | 3.87 (2.79–6.36) | 2.29 (1.17–5.97) | | |
|
| 96 |
+
### 2.1. Regression Model
|
| 97 |
+
|
| 98 |
+
The multivariable logistic regression model showed ([Table 5](#ijms-25-05385-t005)Table 5) that poor CYP2C19 metabolizing phenotype, patient age, and smoking increased the odds of undertreatment in patients (∆LDL-C (mmol/L) < 1) who received standard atorvastatin cholesterol-lowering therapy.
|
| 99 |
+
|
| 100 |
+
### Table 5.
|
| 101 |
+
|
| 102 |
+
Variables that may decrease the effect of atorvastatin lipid-lowering therapy.
|
| 103 |
+
|
| 104 |
+
| Variable | Odds Ratio | 95% CI | p-Value |
|
| 105 |
+
| -------- | ---------- | ------ | ------- |
|
| 106 |
+
| CYP2C19 poor metabolizer phenotype | 7.027 | (2.287–21.590) | 0.001 |
|
| 107 |
+
| Patient age in years | 1.064 | (1.017–1.113) | 0.007 |
|
| 108 |
+
| Smoking | 3.396 | (1.167–9.884) | 0.025 |
|
| 109 |
+
### 2.2. Analysis of Metabolite Data
|
| 110 |
+
|
| 111 |
+
Pairwise analysis of plasma metabolite concentrations ([Table 6](#ijms-25-05385-t006)Table 6) of atorvastatin, 4-OH-atorvastatin, 2-OH-atorvastatin, 9(10)-EpOME, and 12(13)-EpOME revealed that for normal metabolizers ([Figure 1](#ijms-25-05385-f001)Figure 1), 4-OH-atorvastatin concentration was dependent on atorvastatin concentration in blood plasma. Both 9,10-EpOME and 12,13-EpOME concentrations were associated with 4-OH-atorvastatin and 2-OH-atorvastatin plasma concentrations. There were different results in poor metabolizers ([Figure 2](#ijms-25-05385-f002)Figure 2): 4-OH-atorvastatin concentration was associated with atorvastatin and 9,10-EpOME and 12,13-EpOME concentrations. 12,13-EpOME concentration was associated with 2-OH-atorvastatin plasma concentrations.
|
| 112 |
+
|
| 113 |
+
### Table 6.
|
| 114 |
+
|
| 115 |
+
Atorvastatin, 4-OH-atorvastatin, 2-OH-atorvastatin, 9,10-EpOME, and 12,13-EpOME concentrations in the represented patient sample.
|
| 116 |
+
|
| 117 |
+
| CYP2C19 Metabolizer Status | Atorvastatin in ng, Median (Min-Max) | 4-OH-atorvastatin in ng, Median (Min-Max) | 2-OH-atorvastatin in ng, Median (Min-Max) | 9,10-EpOME, in ng, Median (Min-Max) | 12,13-EpOME in ng, Median (Min-Max) |
|
| 118 |
+
| -------------------------- | ------------------------------------ | ----------------------------------------- | ----------------------------------------- | ----------------------------------- | ----------------------------------- |
|
| 119 |
+
| Normal metabolizer (n = 42) | 6.5 (0–157.4) | 0.5 (0–32.1) | 4.1 (0–65.2) | 21.3 (6.9–167.3) | 37.4 (13–144.1) |
|
| 120 |
+
| Poor metabolizer(n = 21) | 5.3 (0–60.1) | 1.1 (0–8) | 5.9 (0–24.8) | 20 (0–148.7) | 29 (0–180) |
|
| 121 |
+
| Total (n = 63) | 5.3 (0–157.4) | 0.6 (0–32.1) | 4.5 (0–65.2) | 20.5 (0–167.3) | 34.8 (0–180) |
|
| 122 |
+
### Figure 1.
|
| 123 |
+
|
| 124 |
+
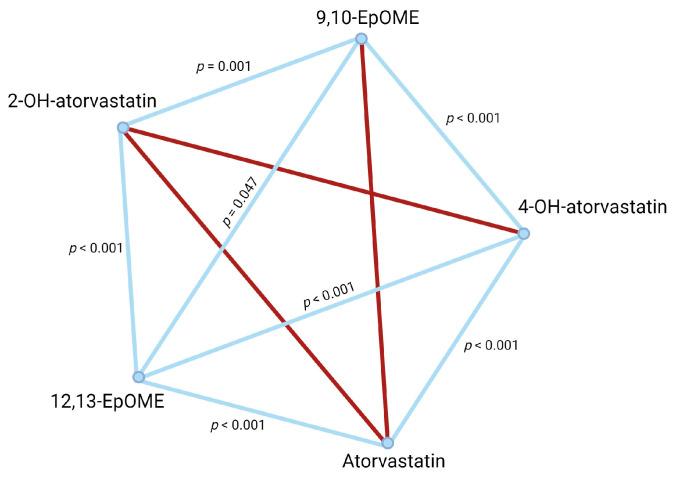
|
| 125 |
+
|
| 126 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11120965_ijms-25-05385-g001.jpg)
|
| 127 |
+
|
| 128 |
+
Pairwise analysis of compound concentrations in normal metabolizers. Significant associations are represented as blue lines. Non-significant associations are in red.
|
| 129 |
+
|
| 130 |
+
### Figure 2.
|
| 131 |
+
|
| 132 |
+
|
| 133 |
+
|
| 134 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11120965_ijms-25-05385-g002.jpg)
|
| 135 |
+
|
| 136 |
+
Pairwise analysis of compound concentrations in poor metabolizers. Significant associations are represented as blue lines. Non-significant associations are in red.
|
| 137 |
+
|
| 138 |
+
## 3. Discussion
|
| 139 |
+
|
| 140 |
+
The results of this study have shown for the first time that *CYP2C19*CYP2C19 variants may significantly affect atorvastatin lipid-lowering therapy. In addition, patient age and smoking were also associated with a decreased effect of atorvastatin therapy.
|
| 141 |
+
|
| 142 |
+
Statins are the cornerstone of lipid-lowering therapies for patients with cardiovascular disease. The European Society of Cardiology (ESC) guidelines divide patients into four risk categories: low, moderate, high, and very high risk. Lifestyle modification or drug therapy is considered according to the patient’s risk and LDL-C levels. If the primary (or secondary) prevention patient is without DM or FH but at very high risk, LDL-C reduction from baseline ≥ 50% and LDL-C goal of <1.4 mmol/L is recommended (I class, level C), and primary prevention for patients with high-risk LDL-C have a goal of <1.8 mmol/L (I class, level A). For individuals at moderate risk, the LDL-C goal is <2.6 mmol/L (IIa class, level A), and for low risk, <3 mmol/L (IIb class, level A). In most of the primary prevention cases, when lifestyle modification does not work and the desirable LDL-C-lowering effect is not achieved, statin therapy must be initiated [[9](#B9-ijms-25-05385)9]. Not only is the statin-induced LDL-C-lowering result relevant for secondary prevention patients, but its pleiotropic nature and their effect on CV morbidity and mortality are also vital for CVD prevention. The American Heart Association recommendations (2023) for chronic coronary syndrome (CCS) patients also support statin therapy to lower LDL-C to reduce the risk of major acute cardiovascular events (MACE) (class I, level A) [[10](#B10-ijms-25-05385)10].
|
| 143 |
+
|
| 144 |
+
Chronic oral statin treatment may have cardioprotective effects before STEMI [[28](#B28-ijms-25-05385)28]. Statins can prevent plaque rupture [[29](#B29-ijms-25-05385)29]. In a large study representing 5103 patients, 27% of the patients were low-intensity statin therapy users (LIST), and 17% used high-intensity statin therapy (HIST) before index acute coronary syndrome. In total, 56% were non-users (statin-naïve patients). The incidence of STEMI was lower in HIST patients than in LIST patients [[30](#B30-ijms-25-05385)30]. Our represented patients cannot confirm this effect because one of the limitations of this study was the relatively low patient number compared with the large-scale studies. However, the aim of this study was also different from the above-mentioned studies and was optimal for analyzing gene variants related to drug metabolism in the liver.
|
| 145 |
+
|
| 146 |
+
The most common loss-of-function variant in the represented patient group was rs4244285, also known as the *2 variant allele. *CYP2C19*CYP2C19 loss-of-function alleles are known to affect the effectiveness of clopidogrel antiplatelet therapy, residual platelet aggregation, and a higher rate of stent thrombosis [[31](#B31-ijms-25-05385)31]. The natural function of CYP2C19 in the liver is the metabolism of various substrates. Studies have shown that the *CYP2C9*CYP2C9 and *CYP2C19*CYP2C19 variant alleles are associated with a higher prevalence of atherosclerosis in cigarette smokers [[32](#B32-ijms-25-05385)32]. In patients following PCI, *CYP2C19*CYP2C19 loss-of-function alleles with peripheral endothelial dysfunction may predict future cardiovascular events [[33](#B33-ijms-25-05385)33]. Another study explained that a decline in epoxyeicosatrienoic acid concentrations due to *CYP2C19*CYP2C19 variants may be associated with coronary microvascular dysfunction [[34](#B34-ijms-25-05385)34]. This same study also shows that increased epoxyeicosatrienoic acid concentrations lead to lower inflammation in the microvascular system. Inflammation has been shown to suppress CYP-450, including CYP3A4 [[35](#B35-ijms-25-05385)35]. Our data show that patients with normal CYP2C19 function had different atorvastatin metabolites and EpOMEs profiles compared to poor metabolizers. Researchers have shown that EpOMEs (vernolic acid (12,13-EpOME) and coronaric acid (9,10-EpOME)) are produced by activated neutrophils and macrophages during inflammation and are known as leukotoxins [[36](#B36-ijms-25-05385)36]. These compounds are linoleic acid derivatives produced by CYP450 enzymes [[37](#B37-ijms-25-05385)37]. EpOMEs are further metabolized into 9,10-dihydroxy-octadecamonoenoate (9,10-DiHOME) and 12,13-dihydroxy-octadecamonoenoate (12,13-DiHOME). The later compounds suppress neutrophil respiratory (or oxidative) bursts, impair immunological signaling [[38](#B38-ijms-25-05385)38], and probably impact the cholesterol metabolism or cholesterol-lowering effect of statins.
|
| 147 |
+
|
| 148 |
+
Rosuvastatin undergoes metabolism through CYP2C9 and CYP2C19; however, according to the literature, the main atorvastatin-metabolizing enzyme remains as CYP3A4 [[39](#B39-ijms-25-05385)39,[40](#B40-ijms-25-05385)40]. One study showed an effect of rosuvastatin but not atorvastatin on P2Y_12_12 receptor reaction units (PRU) in patients with *CYP2C19*CYP2C19 variant alleles and concomitant use of clopidogrel and statin [[41](#B41-ijms-25-05385)41]. A recent study examined the levels of LDL cholesterol in patients who were taking statins. The authors found that *CYP2C19*CYP2C19 variants were associated with sdLDL-C levels and may predict the efficacy of statin therapy [[24](#B24-ijms-25-05385)24]. The results of our study revealed that reduction in LDL-C levels over six months was more effective in patients with normal metabolizer phenotypes of CYP2C19. Poor metabolizers had at least three times lower reduction in LDL-C blood plasma concentrations, despite the exact atorvastatin dosages as normal metabolizers (80 mg once daily). Our pioneering study describes the effect of the CYP2C19 phenotype on atorvastatin therapy in patients with acute coronary syndromes. Thus, the second limitation of this study is that functional studies were not performed to clarify the observations. However, one survey showed that CYP2C19 could metabolize atorvastatin lactone and 2-OH-atorvastatin lactone. The authors provide a more detailed description of how various statins can be metabolized. Atorvastatin is prescribed and used in the active acid form. Acid drugs are metabolized slower by liver cytochromes than more lipophilic lactone drugs. Under the action of certain enzymes, acid forms can be biotransformed into lactone forms and undergo metabolism through liver cytochromes [[42](#B42-ijms-25-05385)42]. Regarding the potentially inconsistent data on the impact of CYP2C19 on atorvastatin metabolism based on different studies, more detailed research is required to clarify the effect of atorvastatin metabolism in patients with acute coronary syndromes. The study utilized next-generation sequencing, a robust but time-consuming technique, to analyze variants of interest. However, alternative methods to sequencing can improve turnaround times while still detecting the most common variants in *CYP2C19*CYP2C19 (*CYP2C19*CYP2C19 *2, *4, *8 alleles) that have an impact. Real-time PCR or other alternative methods could be used for the fast detection of gene variants that may impact statin therapy, leading to precise cholesterol-reducing therapy with tailored treatments adopted for each patient individually.
|
| 149 |
+
|
| 150 |
+
## 4. Materials and Methods
|
| 151 |
+
|
| 152 |
+
### 4.1. Study Population and Inclusion Criteria
|
| 153 |
+
|
| 154 |
+
This study was a prospective, single-center investigation conducted at the cardiac intensive care unit of the Hospital of the Lithuanian University of Health Sciences Kaunas Clinics. The research included 92 consecutive patients admitted between January and November 2021, and all tested negative for COVID-19. These patients presented with either ST-Elevation Myocardial Infarction (STEMI) or Non-ST-Elevation Myocardial Infarction (NSTEMI). Each patient underwent invasive angiography followed by primary percutaneous coronary intervention (PCI). Patients were excluded from enrollment if they had a diagnosis of atrial fibrillation or pericardial diseases; a history of prior coronary artery bypass graft surgery; were pregnant; or had been diagnosed with significant structural heart diseases, including valvular heart diseases. Additionally, exclusion criteria encompassed patients with a documented history of hepatic, oncological, or lung diseases; allergy to contrast media; renal failure; and severe dementia.
|
| 155 |
+
|
| 156 |
+
Data collection included patient demographics, comorbidities, medications, and clinical course. STEMI, as per the 2023 ESC Guidelines, is marked by ST-segment elevation in two ECG leads, with specific thresholds for chest and limb leads and factoring in age and gender. A new left bundle branch block (LBBB) can also indicate STEMI. Diagnosis requires both ischemic symptoms and obstructive coronary artery disease confirmed by angiography. Without angiographic evidence of obstruction, a STEMI diagnosis is not made, even if ECG and clinical signs are present [[43](#B43-ijms-25-05385)43]. NSTEMI is identified through a 12-lead ECG showing a depressed ST-segment or T-wave inversion and a cardiac troponin rise exceeding five times the 99th percentile limit. Confirmation via coronary angiography is essential to diagnose the extent of arterial blockage [[43](#B43-ijms-25-05385)43]. All patients received standard treatment, including statins (80 mg of atorvastatin), angiotensin-converting enzyme inhibitors (or angiotensin receptor I blockers), β-adreno-blockers, and dual antiplatelet therapy (DAPT) comprising aspirin and a P2Y_12_12 inhibitor such as ticagrelor or clopidogrel.
|
| 157 |
+
|
| 158 |
+
### 4.2. Lipid Profile Assessment
|
| 159 |
+
|
| 160 |
+
Lipid levels in our study were assessed upon admission and again at six months. For most lipids, enzymatic hydrolysis was used to measure total cholesterol, triglycerides, and HDL cholesterol, with colorimetric analysis indicating their concentrations. LDL cholesterol was estimated using the Friedewald formula (LDL cholesterol = total cholesterol − HDL cholesterol − (triglycerides/2.2)), effective when triglycerides are below 4.5 mmol/L. For higher triglyceride levels, direct LDL measurement was employed.
|
| 161 |
+
|
| 162 |
+
### 4.3. DNA Extraction and Sequencing
|
| 163 |
+
|
| 164 |
+
Genomic DNA extraction was performed employing standard laboratory procedures, and subsequent library preparation utilized Illumina DNA Prep kits (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. Specifically, a DNA quantity ranging from 200 to 500 ng underwent bead-linked tagmentation. After tagmentation, samples were subjected to amplification through 9 cycles of polymerase chain reaction (PCR). A unique pair of indices was assigned to each during the amplification process to ensure sample distinction. After amplification and a subsequent clean-up step, samples were equitably pooled, each comprising 12 samples.
|
| 165 |
+
|
| 166 |
+
A customized panel was employed for target region enrichment, utilizing xGen Lock Down probes from Integrated DNA Technologies (IDT, Coralville, IA, USA). This panel comprehensively covered all exons of *CYP2C19*CYP2C19 included in the canonical transcript [NM_000769.4](https://www.ncbi.nlm.nih.gov/nuccore/NM_000769.4)NM_000769.4, as per the National Center for Biotechnology Information (NCBI) Reference Sequence Database ([https://www.ncbi.nlm.nih.gov/refseq](https://www.ncbi.nlm.nih.gov/refseq)https://www.ncbi.nlm.nih.gov/refseq, accessed on 1 October 2021). After enrichment, library pools underwent a sixteen-cycle PCR amplification. Subsequently, the prepared library pools were quantified using the Qubit High Sensitivity assay (Invitrogen, Carlsbad, CA, USA).
|
| 167 |
+
|
| 168 |
+
Sequencing was executed on an Illumina Miniseq system (Illumina, San Diego, CA, USA) utilizing the medium output 300 cycles kit. Data analysis was conducted with the Genomic Assembly Tool Kit (GATK) version 4.2.6.1, adhering to the best practice guidelines. This comprehensive methodology ensures robust and accurate interrogation of the genetic landscape, particularly focusing on the target region within the *CYP2C19*CYP2C19 gene.
|
| 169 |
+
|
| 170 |
+
### 4.4. Metabolite Analysis: UPLC-ESI-MS/MS Conditions
|
| 171 |
+
|
| 172 |
+
Analysis of targeted compounds in human plasma samples was carried out with an Acquity H-class UPLC system (Waters, Milford, MA, USA) equipped with a triple quadrupole tandem mass spectrometer (Xevo TQD, Waters, Milford, MA, USA) with an electrospray ionization source (ESI) working in both positive and negative modes. An Acquity UPLC BEH C18 (100 × 2.1 mm 1.7 µm) column was used for the separation of the targeted compounds. The column temperature was maintained at 40 °C. Gradient elution was performed with a mobile phase consisting of 0.1% acetic acid water solution (solvent A) and acetonitrile (solvent B) with the flow rate set to 0.5 mL/min. Linear gradient profile was applied with the following proportions of solvent A: 0 to 1 min–75%, 8.0 to 8.5 min.–5%, 8.51 min; 75% total analysis time—10 min. Electrospray ionization was applied for analysis with the following settings: capillary voltage: 2.5 kV for negative mode and 3.5 kV for positive mode, source temperature: –120 °C, desolvation temperature: –400 °C, desolvation gas flow: 650 L/h, cone gas flow: 10 L/h. Collision energy and cone voltage were optimized for each compound separately ([Table 7](#ijms-25-05385-t007)Table 7).
|
| 173 |
+
|
| 174 |
+
### Table 7.
|
| 175 |
+
|
| 176 |
+
MS condition and MRM transitions for compounds of interest.
|
| 177 |
+
|
| 178 |
+
| Compound | ESI Mode | Retention Time, min | Cone Voltage | Collision Energy | MRM Transition |
|
| 179 |
+
| -------- | -------- | ------------------- | ------------ | ---------------- | -------------- |
|
| 180 |
+
| Atorvastatin | Positive | 5.68 | 30 | 40 | 559 > 250 |
|
| 181 |
+
| Atorvastatin-d5 (IS) | Positive | 5.68 | 30 | 40 | 564 > 255 |
|
| 182 |
+
| 2-Hydroxyatorvastatin | Positive | 5.43 | 30 | 40 | 575 > 250 |
|
| 183 |
+
| Parahydroxyatorvastatin, or Atorvastatin-4OH | Positive | 4.42 | 30 | 40 | 575 > 250 |
|
| 184 |
+
| 20-HETE-d6 (IS) | Negative | 6.45 | 50 | 12 | 325 > 307 |
|
| 185 |
+
| 9,10-Epoxy-12Z-octadecenoic acid (9(10)-EpOME) | Negative | 6.79 | 30 | 20 | 295 > 171 |
|
| 186 |
+
| 12(13)Epoxy-9Z-octadecenoic acid (12(13)-EpOME) | Negative | 6.74 | 30 | 20 | 295 > 195 |
|
| 187 |
+
### 4.5. Statistical Analysis
|
| 188 |
+
|
| 189 |
+
Frequencies are presented in percentages. Quantitative clinical parameters were evaluated using a nonparametric Kruskal–Wallis test. Fisher’s exact test was used to assess the proportions of categorical variables. A *p*p-value < 0.05 was considered statistically significant. All variables were chosen for the multivariable model by backward selection, with the final model containing only those with *p*p < 0.05. According to Wang et al., 1 mmol/L reduction in LDL-C corresponds to a 19% lower risk of major vascular events and is independent of the baseline LDL cholesterol [[44](#B44-ijms-25-05385)44]. Therefore, a 1 mmol/L reduction in LDL cholesterol was used in this study to determine the relative efficacy of statin therapy.
|
| 190 |
+
|
| 191 |
+
Pairwise comparisons were used to evaluate the associations between atorvastatin, its metabolites (4-OH-atorvastatin, 2-OH-atorvastatin), and EPOME (9,10-epoxy-12Z-octadecenoic acid (9(10)-EpOME), as well as 12(13)epoxy-9Z-octadecenoic acid (12(13)-EpOME) concentrations in the represented patient sample.
|
| 192 |
+
|
| 193 |
+
## 5. Conclusions
|
| 194 |
+
|
| 195 |
+
The results revealed that the CYP2C19 phenotype may significantly impact atorvastatin therapy personalization in patients requiring LDL lipid-lowering therapy. However, more detailed studies are needed to show the exact role of CYP2C19 in determining the effect of atorvastatin.
|
| 196 |
+
|
| 197 |
+
## Author Contributions
|
| 198 |
+
|
| 199 |
+
Conceptualization, D.Č. and V.T.; methodology, A.A., D.Č., K.Z., V.Ž., V.J., V.S., A.G. and V.T.; validation, A.A., K.Z., D.Č., V.Ž., V.J., I.Č., V.S. and V.T.; formal analysis, D.Č. and V.T.; investigation, D.Č., I.Č., V.S. and V.Ž.; resources, V.Z., V.L., D.Ž., V.J., V.T., O.D. and G.Š.; data curation, D.Č., A.A., K.Z., V.R. and V.T.; writing—original draft preparation, D.Č., A.A., K.Z. and V.T.; writing—review and editing, all authors; visualization, D.Č. and I.Č.; supervision, V.T.; funding acquisition, V.L., D.Č. and V.T. All authors have read and agreed to the published version of the manuscript.
|
| 200 |
+
|
| 201 |
+
## Institutional Review Board Statement
|
| 202 |
+
|
| 203 |
+
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Regional Bioethics Committee of Kaunas, Lithuania (permission no. BE-2-5).
|
| 204 |
+
|
| 205 |
+
## Informed Consent Statement
|
| 206 |
+
|
| 207 |
+
Informed consent was obtained from all subjects involved in the study.
|
| 208 |
+
|
| 209 |
+
## Data Availability Statement
|
| 210 |
+
|
| 211 |
+
The datasets and resources generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
|
| 212 |
+
|
| 213 |
+
## Conflicts of Interest
|
| 214 |
+
|
| 215 |
+
The authors declare no conflicts of interest.
|
| 216 |
+
|
| 217 |
+
## Funding Statement
|
| 218 |
+
|
| 219 |
+
This work was supported by the funds of Lithuanian University of Health Sciences “Mokslo fondas”.
|
| 220 |
+
|
| 221 |
+
## Footnotes
|
| 222 |
+
|
| 223 |
+
## Associated Data
|
| 224 |
+
|
| 225 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 226 |
+
|
| 227 |
+
### Data Availability Statement
|
| 228 |
+
|
| 229 |
+
The datasets and resources generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
|
| 230 |
+
|
| 231 |
+
### Data Availability Statement
|
| 232 |
+
|
| 233 |
+
The datasets and resources generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
|
| 234 |
+
|
| 235 |
+
## References
|
| 236 |
+
|
| 237 |
+
1. Pirillo A., Norata G.D. The Burden of Hypercholesterolemia and Ischemic Heart Disease in an Ageing World. Pharmacol. Res. 2023;193:106814. doi: 10.1016/j.phrs.2023.106814. [DOI](https://doi.org/10.1016/j.phrs.2023.106814) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37271426/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacol.%20Res.&title=The%20Burden%20of%20Hypercholesterolemia%20and%20Ischemic%20Heart%20Disease%20in%20an%20Ageing%20World&author=A.%20Pirillo&author=G.D.%20Norata&volume=193&publication_year=2023&pages=106814&pmid=37271426&doi=10.1016/j.phrs.2023.106814&)
|
| 238 |
+
|
| 239 |
+
2. Jarauta E., Bea-Sanz A.M., Marco-Benedi V., Lamiquiz-Moneo I. Genetics of Hypercholesterolemia: Comparison Between Familial Hypercholesterolemia and Hypercholesterolemia Nonrelated to LDL Receptor. Front. Genet. 2020;11:554931. doi: 10.3389/fgene.2020.554931. [DOI](https://doi.org/10.3389/fgene.2020.554931) | [PMC free article](/articles/PMC7744656/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33343620/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front.%20Genet.&title=Genetics%20of%20Hypercholesterolemia:%20Comparison%20Between%20Familial%20Hypercholesterolemia%20and%20Hypercholesterolemia%20Nonrelated%20to%20LDL%20Receptor&author=E.%20Jarauta&author=A.M.%20Bea-Sanz&author=V.%20Marco-Benedi&author=I.%20Lamiquiz-Moneo&volume=11&publication_year=2020&pages=554931&pmid=33343620&doi=10.3389/fgene.2020.554931&)
|
| 240 |
+
|
| 241 |
+
3. Steinberg D. The Cholesterol Wars: The Cholesterol Skeptics vs the Preponderance of Evidence. Academic Press-Elsevier; San Diego, CA, USA: 2007. [Google Scholar](https://scholar.google.com/scholar_lookup?title=The%20Cholesterol%20Wars:%20The%20Cholesterol%20Skeptics%20vs%20the%20Preponderance%20of%20Evidence&author=D.%20Steinberg&publication_year=2007&)
|
| 242 |
+
|
| 243 |
+
4. Endo A., Kuroda M., Tsujita Y. ML-236A, ML-236B, and ML-236C, New Inhibitors of Cholesterogensis Produced by Penicillium Citrinum. J. Antibiot. 1976;29:1346–1348. doi: 10.7164/antibiotics.29.1346. [DOI](https://doi.org/10.7164/antibiotics.29.1346) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1010803/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Antibiot.&title=ML-236A,%20ML-236B,%20and%20ML-236C,%20New%20Inhibitors%20of%20Cholesterogensis%20Produced%20by%20Penicillium%20Citrinum&author=A.%20Endo&author=M.%20Kuroda&author=Y.%20Tsujita&volume=29&publication_year=1976&pages=1346-1348&pmid=1010803&doi=10.7164/antibiotics.29.1346&)
|
| 244 |
+
|
| 245 |
+
5. Endo A., Kuroda M., Tanzawa K. Competitive Inhibition of 3-hydroxy-3-methylglutaryl Coenzyme a Reductase by ML-236A and ML-236B Fungal Metabolites, Having Hypocholesterolemic Activity. FEBS Lett. 1976;72:323–326. doi: 10.1016/0014-5793(76)80996-9. [DOI](https://doi.org/10.1016/0014-5793(76)80996-9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16386050/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=FEBS%20Lett.&title=Competitive%20Inhibition%20of%203-hydroxy-3-methylglutaryl%20Coenzyme%20a%20Reductase%20by%20ML-236A%20and%20ML-236B%20Fungal%20Metabolites,%20Having%20Hypocholesterolemic%20Activity&author=A.%20Endo&author=M.%20Kuroda&author=K.%20Tanzawa&volume=72&publication_year=1976&pages=323-326&pmid=16386050&doi=10.1016/0014-5793(76)80996-9&)
|
| 246 |
+
|
| 247 |
+
6. Goldstein L.J., Brown S.M. The Low-Density Lipoprotein Pathway and Its Relation to Atherosclerosis. Annu. Rev. Biochem. 1977;46:897–930. doi: 10.1146/annurev.bi.46.070177.004341. [DOI](https://doi.org/10.1146/annurev.bi.46.070177.004341) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/197883/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Annu.%20Rev.%20Biochem.&title=The%20Low-Density%20Lipoprotein%20Pathway%20and%20Its%20Relation%20to%20Atherosclerosis&author=L.J.%20Goldstein&author=S.M.%20Brown&volume=46&publication_year=1977&pages=897-930&pmid=197883&doi=10.1146/annurev.bi.46.070177.004341&)
|
| 248 |
+
|
| 249 |
+
7. Razavi A.C., Mehta A., Sperling L.S. Statin Therapy for the Primary Prevention of Cardiovascular Disease: Pros. Atherosclerosis. 2022;356:41–45. doi: 10.1016/j.atherosclerosis.2022.07.004. [DOI](https://doi.org/10.1016/j.atherosclerosis.2022.07.004) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35945050/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Atherosclerosis&title=Statin%20Therapy%20for%20the%20Primary%20Prevention%20of%20Cardiovascular%20Disease:%20Pros&author=A.C.%20Razavi&author=A.%20Mehta&author=L.S.%20Sperling&volume=356&publication_year=2022&pages=41-45&pmid=35945050&doi=10.1016/j.atherosclerosis.2022.07.004&)
|
| 250 |
+
|
| 251 |
+
8. Li M., Wang X., Li X., Chen H., Hu Y., Zhang X., Tang X., Miao Y., Tian G., Shang H. Statins for the Primary Prevention of Coronary Heart Disease. BioMed Res. Int. 2019;2019:4870350. doi: 10.1155/2019/4870350. [DOI](https://doi.org/10.1155/2019/4870350) | [PMC free article](/articles/PMC6374814/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30834266/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=BioMed%20Res.%20Int.&title=Statins%20for%20the%20Primary%20Prevention%20of%20Coronary%20Heart%20Disease&author=M.%20Li&author=X.%20Wang&author=X.%20Li&author=H.%20Chen&author=Y.%20Hu&volume=2019&publication_year=2019&pages=4870350&pmid=30834266&doi=10.1155/2019/4870350&)
|
| 252 |
+
|
| 253 |
+
9. Mach F., Baigent C., Catapano A.L., Koskinas K.C., Casula M., Badimon L., Chapman M.J., De Backer G.G., Delgado V., Ference B.A., et al. 2019 ESC/EAS Guidelines for the Management of Dyslipidaemias: Lipid Modification to Reduce Cardiovascular Risk. Eur. Heart J. 2020;41:111–188. doi: 10.1093/eurheartj/ehz455. [DOI](https://doi.org/10.1093/eurheartj/ehz455) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31504418/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Heart%20J.&title=2019%20ESC/EAS%20Guidelines%20for%20the%20Management%20of%20Dyslipidaemias:%20Lipid%20Modification%20to%20Reduce%20Cardiovascular%20Risk&author=F.%20Mach&author=C.%20Baigent&author=A.L.%20Catapano&author=K.C.%20Koskinas&author=M.%20Casula&volume=41&publication_year=2020&pages=111-188&pmid=31504418&doi=10.1093/eurheartj/ehz455&)
|
| 254 |
+
|
| 255 |
+
10. Virani S.S., Newby L.K., Arnold S.V., Bittner V., Brewer L.C., Demeter S.H., Dixon D.L., Fearon W.F., Hess B., Johnson H.M., et al. 2023 AHA/ACC/ACCP/ASPC/NLA/PCNA Guideline for the Management of Patients with Chronic Coronary Disease: A Report of the American Heart Association/American College of Cardiology Joint Committee on Clinical Practice Guidelines. Circulation. 2023;148:E9–E119. doi: 10.1161/CIR.0000000000001168. [DOI](https://doi.org/10.1161/CIR.0000000000001168) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37471501/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circulation&title=2023%20AHA/ACC/ACCP/ASPC/NLA/PCNA%20Guideline%20for%20the%20Management%20of%20Patients%20with%20Chronic%20Coronary%20Disease:%20A%20Report%20of%20the%20American%20Heart%20Association/American%20College%20of%20Cardiology%20Joint%20Committee%20on%20Clinical%20Practice%20Guidelines&author=S.S.%20Virani&author=L.K.%20Newby&author=S.V.%20Arnold&author=V.%20Bittner&author=L.C.%20Brewer&volume=148&publication_year=2023&pages=E9-E119&pmid=37471501&doi=10.1161/CIR.0000000000001168&)
|
| 256 |
+
|
| 257 |
+
11. Marx N., Federici M., Schütt K., Müller-Wieland D., A Ajjan R., Antunes M.J., Christodorescu R.M., Crawford C., Di Angelantonio E., Eliasson B., et al. 2023 ESC Guidelines for the Management of Cardiovascular Disease in Patients with Diabetes. Eur. Heart J. 2023;44:4043–4140. doi: 10.1093/eurheartj/ehad192. [DOI](https://doi.org/10.1093/eurheartj/ehad192) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37622663/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Heart%20J.&title=2023%20ESC%20Guidelines%20for%20the%20Management%20of%20Cardiovascular%20Disease%20in%20Patients%20with%20Diabetes&author=N.%20Marx&author=M.%20Federici&author=K.%20Sch%C3%BCtt&author=D.%20M%C3%BCller-Wieland&author=R.%20A%20Ajjan&volume=44&publication_year=2023&pages=4043-4140&pmid=37622663&doi=10.1093/eurheartj/ehad192&)
|
| 258 |
+
|
| 259 |
+
12. Gitt A.K., Lautsch D., Ferrières J., De Ferrari G.M., Vyas A., Baxter C.A., Bash L.D., Ashton V., Horack M., Almahmeed W., et al. Cholesterol Target Value Attainment and Lipid-Lowering Therapy in Patients with Stable or Acute Coronary Heart Disease: Results from the Dyslipidemia International Study II. Atherosclerosis. 2017;266:158–166. doi: 10.1016/j.atherosclerosis.2017.08.013. [DOI](https://doi.org/10.1016/j.atherosclerosis.2017.08.013) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29028484/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Atherosclerosis&title=Cholesterol%20Target%20Value%20Attainment%20and%20Lipid-Lowering%20Therapy%20in%20Patients%20with%20Stable%20or%20Acute%20Coronary%20Heart%20Disease:%20Results%20from%20the%20Dyslipidemia%20International%20Study%20II&author=A.K.%20Gitt&author=D.%20Lautsch&author=J.%20Ferri%C3%A8res&author=G.M.%20De%20Ferrari&author=A.%20Vyas&volume=266&publication_year=2017&pages=158-166&pmid=29028484&doi=10.1016/j.atherosclerosis.2017.08.013&)
|
| 260 |
+
|
| 261 |
+
13. De Backer G., Jankowski P., Kotseva K., Mirrakhimov E., Reiner Ž., Rydén L., Tokgözoğlu L., Wood D., De Bacquer D., EUROASPIRE V collaborators et al. Management of Dyslipidaemia in Patients with Coronary Heart Disease: Results from the ESC-EORP EUROASPIRE V Survey in 27 Countries. Atherosclerosis. 2019;285:135–146. doi: 10.1016/j.atherosclerosis.2019.03.014. [DOI](https://doi.org/10.1016/j.atherosclerosis.2019.03.014) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31054483/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Atherosclerosis&title=Management%20of%20Dyslipidaemia%20in%20Patients%20with%20Coronary%20Heart%20Disease:%20Results%20from%20the%20ESC-EORP%20EUROASPIRE%20V%20Survey%20in%2027%20Countries&author=G.%20De%20Backer&author=P.%20Jankowski&author=K.%20Kotseva&author=E.%20Mirrakhimov&author=%C5%BD.%20Reiner&volume=285&publication_year=2019&pages=135-146&pmid=31054483&doi=10.1016/j.atherosclerosis.2019.03.014&)
|
| 262 |
+
|
| 263 |
+
14. Yao X., Shah N.D., Gersh B.J., Lopez-Jimenez F., Noseworthy P.A. Assessment of Trends in Statin Therapy for Secondary Prevention of Atherosclerotic Cardiovascular Disease in US Adults From 2007 to 2016. JAMA Netw. Open. 2020;3:e2025505. doi: 10.1001/jamanetworkopen.2020.25505. [DOI](https://doi.org/10.1001/jamanetworkopen.2020.25505) | [PMC free article](/articles/PMC7679951/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33216139/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=JAMA%20Netw.%20Open&title=Assessment%20of%20Trends%20in%20Statin%20Therapy%20for%20Secondary%20Prevention%20of%20Atherosclerotic%20Cardiovascular%20Disease%20in%20US%20Adults%20From%202007%20to%202016&author=X.%20Yao&author=N.D.%20Shah&author=B.J.%20Gersh&author=F.%20Lopez-Jimenez&author=P.A.%20Noseworthy&volume=3&publication_year=2020&pages=e2025505&pmid=33216139&doi=10.1001/jamanetworkopen.2020.25505&)
|
| 264 |
+
|
| 265 |
+
15. Shin S., Shin D.W., Cho I.Y., Jeong S.-M., Jung H. Status of Dyslipidemia Management and Statin Undertreatment in Korean Cancer Survivors: A Korean National Health and Nutrition Examination Survey Study. Eur. J. Prev. Cardiol. 2021;28:864–872. doi: 10.1177/2047487320905722. [DOI](https://doi.org/10.1177/2047487320905722) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34298552/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20J.%20Prev.%20Cardiol.&title=Status%20of%20Dyslipidemia%20Management%20and%20Statin%20Undertreatment%20in%20Korean%20Cancer%20Survivors:%20A%20Korean%20National%20Health%20and%20Nutrition%20Examination%20Survey%20Study&author=S.%20Shin&author=D.W.%20Shin&author=I.Y.%20Cho&author=S.-M.%20Jeong&author=H.%20Jung&volume=28&publication_year=2021&pages=864-872&pmid=34298552&doi=10.1177/2047487320905722&)
|
| 266 |
+
|
| 267 |
+
16. Danchin N., Almahmeed W., Al-Rasadi K., Azuri J., Berrah A., Cuneo C.A., Karpov Y., Kaul U., Kayıkçıoğlu M., Mitchenko O., et al. Achievement of Low-Density Lipoprotein Cholesterol Goals in 18 Countries Outside Western Europe: The International ChoLesterol Management Practice Study (ICLPS) Eur. J. Prev. Cardiol. 2018;25:1087–1094. doi: 10.1177/2047487318777079. [DOI](https://doi.org/10.1177/2047487318777079) | [PMC free article](/articles/PMC6039862/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29771156/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20J.%20Prev.%20Cardiol.&title=Achievement%20of%20Low-Density%20Lipoprotein%20Cholesterol%20Goals%20in%2018%20Countries%20Outside%20Western%20Europe:%20The%20International%20ChoLesterol%20Management%20Practice%20Study%20(ICLPS)&author=N.%20Danchin&author=W.%20Almahmeed&author=K.%20Al-Rasadi&author=J.%20Azuri&author=A.%20Berrah&volume=25&publication_year=2018&pages=1087-1094&pmid=29771156&doi=10.1177/2047487318777079&)
|
| 268 |
+
|
| 269 |
+
17. Tsioufis K., Vázquez J.M.C., Sykara G., Malvestiti F.M., van Vugt J. Real-World Evidence for Adherence and Persistence with Atorvastatin Therapy. Cardiol. Ther. 2021;10:445–464. doi: 10.1007/s40119-021-00240-8. [DOI](https://doi.org/10.1007/s40119-021-00240-8) | [PMC free article](/articles/PMC8555050/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34586613/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cardiol.%20Ther.&title=Real-World%20Evidence%20for%20Adherence%20and%20Persistence%20with%20Atorvastatin%20Therapy&author=K.%20Tsioufis&author=J.M.C.%20V%C3%A1zquez&author=G.%20Sykara&author=F.M.%20Malvestiti&author=J.%20van%20Vugt&volume=10&publication_year=2021&pages=445-464&pmid=34586613&doi=10.1007/s40119-021-00240-8&)
|
| 270 |
+
|
| 271 |
+
18. Cano-Corres R., Candás-Estébanez B., Padró-Miquel A., Fanlo-Maresma M., Pintó X., Alía-Ramos P. Influence of 6 Genetic Variants on the Efficacy of Statins in Patients with Dyslipidemia. J. Clin. Lab. Anal. 2018;32:e22566. doi: 10.1002/jcla.22566. [DOI](https://doi.org/10.1002/jcla.22566) | [PMC free article](/articles/PMC6817082/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29732606/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Lab.%20Anal.&title=Influence%20of%206%20Genetic%20Variants%20on%20the%20Efficacy%20of%20Statins%20in%20Patients%20with%20Dyslipidemia&author=R.%20Cano-Corres&author=B.%20Cand%C3%A1s-Est%C3%A9banez&author=A.%20Padr%C3%B3-Miquel&author=M.%20Fanlo-Maresma&author=X.%20Pint%C3%B3&volume=32&publication_year=2018&pages=e22566&pmid=29732606&doi=10.1002/jcla.22566&)
|
| 272 |
+
|
| 273 |
+
19. Maxwell W.D., Ramsey L.B., Johnson S.G., Moore K.G., Shtutman M., Schoonover J.H., Kawaguchi-Suzuki M. Impact of Pharmacogenetics on Efficacy and Safety of Statin Therapy for Dyslipidemia. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2017;37:1172–1190. doi: 10.1002/phar.1981. [DOI](https://doi.org/10.1002/phar.1981) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28672099/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacother.%20J.%20Hum.%20Pharmacol.%20Drug%20Ther.&title=Impact%20of%20Pharmacogenetics%20on%20Efficacy%20and%20Safety%20of%20Statin%20Therapy%20for%20Dyslipidemia&author=W.D.%20Maxwell&author=L.B.%20Ramsey&author=S.G.%20Johnson&author=K.G.%20Moore&author=M.%20Shtutman&volume=37&publication_year=2017&pages=1172-1190&pmid=28672099&doi=10.1002/phar.1981&)
|
| 274 |
+
|
| 275 |
+
20. Wilke R.A., Ramsey L.B., Johnson S.G., Maxwell W.D., McLeod H.L., Voora D., Krauss R.M., Roden D.M., Feng Q., Cooper-DeHoff R.M., et al. The Clinical Pharmacogenomics Implementation Consortium: CPIC Guideline for SLCO1B1 and Simvastatin-Induced Myopathy. Clin. Pharmacol. Ther. 2012;92:112–117. doi: 10.1038/clpt.2012.57. Updated in Clin. Pharmacol. Ther. 2014, 96, 423–428. [DOI](https://doi.org/10.1038/clpt.2012.57) | [PMC free article](/articles/PMC3384438/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22617227/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Pharmacol.%20Ther.&title=The%20Clinical%20Pharmacogenomics%20Implementation%20Consortium:%20CPIC%20Guideline%20for%20SLCO1B1%20and%20Simvastatin-Induced%20Myopathy&author=R.A.%20Wilke&author=L.B.%20Ramsey&author=S.G.%20Johnson&author=W.D.%20Maxwell&author=H.L.%20McLeod&volume=92&publication_year=2012&pages=112-117&pmid=22617227&doi=10.1038/clpt.2012.57&)
|
| 276 |
+
|
| 277 |
+
21. Cooper-DeHoff R.M., Niemi M., Ramsey L.B., Luzum J.A., Tarkiainen E.K., Straka R.J., Gong L., Tuteja S., Wilke R.A., Wadelius M., et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1, ABCG2, and CYP2C9 Genotypes and Statin-Associated Musculoskeletal Symptoms. Clin. Pharmacol. Ther. 2022;111:1007–1021. doi: 10.1002/cpt.2557. [DOI](https://doi.org/10.1002/cpt.2557) | [PMC free article](/articles/PMC9035072/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35152405/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Pharmacol.%20Ther.&title=The%20Clinical%20Pharmacogenetics%20Implementation%20Consortium%20Guideline%20for%20SLCO1B1,%20ABCG2,%20and%20CYP2C9%20Genotypes%20and%20Statin-Associated%20Musculoskeletal%20Symptoms&author=R.M.%20Cooper-DeHoff&author=M.%20Niemi&author=L.B.%20Ramsey&author=J.A.%20Luzum&author=E.K.%20Tarkiainen&volume=111&publication_year=2022&pages=1007-1021&pmid=35152405&doi=10.1002/cpt.2557&)
|
| 278 |
+
|
| 279 |
+
22. Reith C., Baigent C., Blackwell L., Emberson J., Spata E., Davies K., Halls H., Holland L., Wilson K., Armitage J., et al. Effect of Statin Therapy on Muscle Symptoms: An Individual Participant Data Meta-Analysis of Large-Scale, Randomised, Double-Blind Trials. Lancet. 2022;400:832–845. doi: 10.1016/S0140-6736(22)01545-8. [DOI](https://doi.org/10.1016/S0140-6736(22)01545-8) | [PMC free article](/articles/PMC7613583/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36049498/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Effect%20of%20Statin%20Therapy%20on%20Muscle%20Symptoms:%20An%20Individual%20Participant%20Data%20Meta-Analysis%20of%20Large-Scale,%20Randomised,%20Double-Blind%20Trials&author=C.%20Reith&author=C.%20Baigent&author=L.%20Blackwell&author=J.%20Emberson&author=E.%20Spata&volume=400&publication_year=2022&pages=832-845&pmid=36049498&doi=10.1016/S0140-6736(22)01545-8&)
|
| 280 |
+
|
| 281 |
+
23. Navar A.M., Peterson E.D., Li S., Robinson J.G., Roger V.L., Goldberg A.C., Virani S., Wilson P.W., Nanna M.G., Lee L.V., et al. Prevalence and Management of Symptoms Associated with Statin Therapy in Community Practice. Circ. Cardiovasc. Qual. Outcomes. 2018;11:e004249. doi: 10.1161/CIRCOUTCOMES.117.004249. [DOI](https://doi.org/10.1161/CIRCOUTCOMES.117.004249) | [PMC free article](/articles/PMC5858190/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29545393/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circ.%20Cardiovasc.%20Qual.%20Outcomes&title=Prevalence%20and%20Management%20of%20Symptoms%20Associated%20with%20Statin%20Therapy%20in%20Community%20Practice&author=A.M.%20Navar&author=E.D.%20Peterson&author=S.%20Li&author=J.G.%20Robinson&author=V.L.%20Roger&volume=11&publication_year=2018&pages=e004249&pmid=29545393&doi=10.1161/CIRCOUTCOMES.117.004249&)
|
| 282 |
+
|
| 283 |
+
24. Dai R., Zhao X., Zhuo H., Wang W., Xu Y., Hu Z., Zhang T., Zhao J. CYP2C19 Metabolizer Phenotypes May Affect the Efficacy of Statins on Lowering Small Dense Low-Density Lipoprotein Cholesterol of Patients with Coronary Artery Disease. Front. Cardiovasc. Med. 2022;9:1016126. doi: 10.3389/fcvm.2022.1016126. [DOI](https://doi.org/10.3389/fcvm.2022.1016126) | [PMC free article](/articles/PMC9806256/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36601065/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front.%20Cardiovasc.%20Med.&title=CYP2C19%20Metabolizer%20Phenotypes%20May%20Affect%20the%20Efficacy%20of%20Statins%20on%20Lowering%20Small%20Dense%20Low-Density%20Lipoprotein%20Cholesterol%20of%20Patients%20with%20Coronary%20Artery%20Disease&author=R.%20Dai&author=X.%20Zhao&author=H.%20Zhuo&author=W.%20Wang&author=Y.%20Xu&volume=9&publication_year=2022&pages=1016126&pmid=36601065&doi=10.3389/fcvm.2022.1016126&)
|
| 284 |
+
|
| 285 |
+
25. Caudle K.E., Dunnenberger H.M., Freimuth R.R., Peterson J.F., Burlison J.D., Whirl-Carrillo M., Scott S.A., Rehm H.L., Williams M.S., Klein T.E., et al. Standardizing Terms for Clinical Pharmacogenetic Test Results: Consensus Terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC) Genet. Med. 2017;19:215–223. doi: 10.1038/gim.2016.87. [DOI](https://doi.org/10.1038/gim.2016.87) | [PMC free article](/articles/PMC5253119/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27441996/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genet.%20Med.&title=Standardizing%20Terms%20for%20Clinical%20Pharmacogenetic%20Test%20Results:%20Consensus%20Terms%20from%20the%20Clinical%20Pharmacogenetics%20Implementation%20Consortium%20(CPIC)&author=K.E.%20Caudle&author=H.M.%20Dunnenberger&author=R.R.%20Freimuth&author=J.F.%20Peterson&author=J.D.%20Burlison&volume=19&publication_year=2017&pages=215-223&pmid=27441996&doi=10.1038/gim.2016.87&)
|
| 286 |
+
|
| 287 |
+
26. Hildreth K., Kodani S.D., Hammock B.D., Zhao L. Cytochrome P450-Derived Linoleic Acid Metabolites EpOMEs and DiHOMEs: A Review of Recent Studies. J. Nutr. Biochem. 2020;86:108484. doi: 10.1016/j.jnutbio.2020.108484. [DOI](https://doi.org/10.1016/j.jnutbio.2020.108484) | [PMC free article](/articles/PMC7606796/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32827665/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Nutr.%20Biochem.&title=Cytochrome%20P450-Derived%20Linoleic%20Acid%20Metabolites%20EpOMEs%20and%20DiHOMEs:%20A%20Review%20of%20Recent%20Studies&author=K.%20Hildreth&author=S.D.%20Kodani&author=B.D.%20Hammock&author=L.%20Zhao&volume=86&publication_year=2020&pages=108484&pmid=32827665&doi=10.1016/j.jnutbio.2020.108484&)
|
| 288 |
+
|
| 289 |
+
27. PharmVar Pharmacogene Variation Consortium. [(accessed on 17 January 2024)]. Available online: https://www.pharmvar.org/gene/CYP2C19. [https://www.pharmvar.org/gene/CYP2C19](https://www.pharmvar.org/gene/CYP2C19)
|
| 290 |
+
|
| 291 |
+
28. Badimon G.M., Calvo M., Guzman J., Perez P., Alamar M., Vilahur G., Gavara J., Vargas S., Rello P., Valente F., et al. CMR Analysis of the Cardioprotective Effects of Chronic Statin Therapy Prior to First STEMI: A Propensity Score Analysis. Eur. Heart J. 2021;42((Suppl. 1)):ehab724.1461. doi: 10.1093/eurheartj/ehab724.1461. [DOI](https://doi.org/10.1093/eurheartj/ehab724.1461) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Heart%20J.&title=CMR%20Analysis%20of%20the%20Cardioprotective%20Effects%20of%20Chronic%20Statin%20Therapy%20Prior%20to%20First%20STEMI:%20A%20Propensity%20Score%20Analysis&author=G.M.%20Badimon&author=M.%20Calvo&author=J.%20Guzman&author=P.%20Perez&author=M.%20Alamar&volume=42&issue=(Suppl.%201)&publication_year=2021&pages=ehab724.1461&doi=10.1093/eurheartj/ehab724.1461&)
|
| 292 |
+
|
| 293 |
+
29. Otsuka F., Hibi K., Kusama I., Endo M., Kosuge M., Iwahashi N., Okuda J., Tsukahara K., Ebina T., Kojima S., et al. Impact of Statin Pretreatment on the Incidence of Plaque Rupture in ST-Elevation Acute Myocardial Infarction. Atherosclerosis. 2010;213:505–511. doi: 10.1016/j.atherosclerosis.2010.09.005. [DOI](https://doi.org/10.1016/j.atherosclerosis.2010.09.005) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20926078/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Atherosclerosis&title=Impact%20of%20Statin%20Pretreatment%20on%20the%20Incidence%20of%20Plaque%20Rupture%20in%20ST-Elevation%20Acute%20Myocardial%20Infarction&author=F.%20Otsuka&author=K.%20Hibi&author=I.%20Kusama&author=M.%20Endo&author=M.%20Kosuge&volume=213&publication_year=2010&pages=505-511&pmid=20926078&doi=10.1016/j.atherosclerosis.2010.09.005&)
|
| 294 |
+
|
| 295 |
+
30. Dadon Z., Moriel M., Iakobishvili Z., Asher E., Samuel T.Y., Gavish D., Glikson M., Gottlieb S. Association of Contemporary Statin Pretreatment Intensity and LDL-C Levels on the Incidence of STEMI Presentation. Life. 2021;11:1268. doi: 10.3390/life11111268. [DOI](https://doi.org/10.3390/life11111268) | [PMC free article](/articles/PMC8625617/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34833144/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Life&title=Association%20of%20Contemporary%20Statin%20Pretreatment%20Intensity%20and%20LDL-C%20Levels%20on%20the%20Incidence%20of%20STEMI%20Presentation&author=Z.%20Dadon&author=M.%20Moriel&author=Z.%20Iakobishvili&author=E.%20Asher&author=T.Y.%20Samuel&volume=11&publication_year=2021&pages=1268&pmid=34833144&doi=10.3390/life11111268&)
|
| 296 |
+
|
| 297 |
+
31. Mega J.L., Close S.L., Wiviott S.D., Shen L., Hockett R.D., Brandt J.T., Walker J.R., Antman E.M., Macias W., Braunwald E., et al. Cytochrome P-450 Polymorphisms and Response to Clopidogrel. N. Engl. J. Med. 2009;360:354–362. doi: 10.1056/NEJMoa0809171. [DOI](https://doi.org/10.1056/NEJMoa0809171) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19106084/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N.%20Engl.%20J.%20Med.&title=Cytochrome%20P-450%20Polymorphisms%20and%20Response%20to%20Clopidogrel&author=J.L.%20Mega&author=S.L.%20Close&author=S.D.%20Wiviott&author=L.%20Shen&author=R.D.%20Hockett&volume=360&publication_year=2009&pages=354-362&pmid=19106084&doi=10.1056/NEJMoa0809171&)
|
| 298 |
+
|
| 299 |
+
32. Ercan B., Ayaz L., Çiçek D., Tamer L. Role of CYP2C9 and CYP2C19 Polymorphisms in Patients with Atherosclerosis. Cell Biochem. Funct. 2008;26:309–313. doi: 10.1002/cbf.1437. [DOI](https://doi.org/10.1002/cbf.1437) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17868191/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cell%20Biochem.%20Funct.&title=Role%20of%20CYP2C9%20and%20CYP2C19%20Polymorphisms%20in%20Patients%20with%20Atherosclerosis&author=B.%20Ercan&author=L.%20Ayaz&author=D.%20%C3%87i%C3%A7ek&author=L.%20Tamer&volume=26&publication_year=2008&pages=309-313&pmid=17868191&doi=10.1002/cbf.1437&)
|
| 300 |
+
|
| 301 |
+
33. Tabata N., Hokimoto S., Akasaka T., Arima Y., Sakamoto K., Yamamoto E., Tsujita K., Izumiya Y., Yamamuro M., Kojima S., et al. Patients with Both CYP2C19 Loss-of-Function Allele and Peripheral Endothelial Dysfunction Are Significantly Correlated with Adverse Cardiovascular Events Following Coronary Stent Implantation. J. Cardiol. 2016;67:104–109. doi: 10.1016/j.jjcc.2015.03.010. [DOI](https://doi.org/10.1016/j.jjcc.2015.03.010) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25851472/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Cardiol.&title=Patients%20with%20Both%20CYP2C19%20Loss-of-Function%20Allele%20and%20Peripheral%20Endothelial%20Dysfunction%20Are%20Significantly%20Correlated%20with%20Adverse%20Cardiovascular%20Events%20Following%20Coronary%20Stent%20Implantation&author=N.%20Tabata&author=S.%20Hokimoto&author=T.%20Akasaka&author=Y.%20Arima&author=K.%20Sakamoto&volume=67&publication_year=2016&pages=104-109&pmid=25851472&doi=10.1016/j.jjcc.2015.03.010&)
|
| 302 |
+
|
| 303 |
+
34. Akasaka T., Sueta D., Arima Y., Tabata N., Takashio S., Izumiya Y., Yamamoto E., Tsujita K., Kojima S., Kaikita K., et al. CYP2C19 Variants and Epoxyeicosatrienoic Acids in Patients with Microvascular Angina. IJC Heart Vasc. 2017;15:15–20. doi: 10.1016/j.ijcha.2017.03.001. [DOI](https://doi.org/10.1016/j.ijcha.2017.03.001) | [PMC free article](/articles/PMC5458130/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28616567/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=IJC%20Heart%20Vasc.&title=CYP2C19%20Variants%20and%20Epoxyeicosatrienoic%20Acids%20in%20Patients%20with%20Microvascular%20Angina&author=T.%20Akasaka&author=D.%20Sueta&author=Y.%20Arima&author=N.%20Tabata&author=S.%20Takashio&volume=15&publication_year=2017&pages=15-20&pmid=28616567&doi=10.1016/j.ijcha.2017.03.001&)
|
| 304 |
+
|
| 305 |
+
35. White C.M. Inflammation Suppresses Patients’ Ability to Metabolize Cytochrome P450 Substrate Drugs. Ann. Pharmacother. 2022;56:809–819. doi: 10.1177/10600280211047864. [DOI](https://doi.org/10.1177/10600280211047864) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34590872/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Pharmacother.&title=Inflammation%20Suppresses%20Patients%E2%80%99%20Ability%20to%20Metabolize%20Cytochrome%20P450%20Substrate%20Drugs&author=C.M.%20White&volume=56&publication_year=2022&pages=809-819&pmid=34590872&doi=10.1177/10600280211047864&)
|
| 306 |
+
|
| 307 |
+
36. Ozawa T., Sugiyama S., Hayakawa M., Satake T., Taki F., Iwata M., Taki K. Existence of Leukotoxin 9,10-Epoxy-12-0ctadecenoate in Lung Lavages from Rats Breathing Pure Oxygen and from Patients with the Adult Respiratory Distress Syndrome. Am. Rev. Respir. Dis. 1988;137:535–540. doi: 10.1164/ajrccm/137.3.535. [DOI](https://doi.org/10.1164/ajrccm/137.3.535) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/3345035/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20Rev.%20Respir.%20Dis.&title=Existence%20of%20Leukotoxin%209,10-Epoxy-12-0ctadecenoate%20in%20Lung%20Lavages%20from%20Rats%20Breathing%20Pure%20Oxygen%20and%20from%20Patients%20with%20the%20Adult%20Respiratory%20Distress%20Syndrome&author=T.%20Ozawa&author=S.%20Sugiyama&author=M.%20Hayakawa&author=T.%20Satake&author=F.%20Taki&volume=137&publication_year=1988&pages=535-540&pmid=3345035&doi=10.1164/ajrccm/137.3.535&)
|
| 308 |
+
|
| 309 |
+
37. Newman J.W., Morisseau C., Hammock B.D. Epoxide Hydrolases: Their Roles and Interactions with Lipid Metabolism. Prog. Lipid Res. 2005;44:1–51. doi: 10.1016/j.plipres.2004.10.001. [DOI](https://doi.org/10.1016/j.plipres.2004.10.001) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15748653/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Prog.%20Lipid%20Res.&title=Epoxide%20Hydrolases:%20Their%20Roles%20and%20Interactions%20with%20Lipid%20Metabolism&author=J.W.%20Newman&author=C.%20Morisseau&author=B.D.%20Hammock&volume=44&publication_year=2005&pages=1-51&pmid=15748653&doi=10.1016/j.plipres.2004.10.001&)
|
| 310 |
+
|
| 311 |
+
38. Thompson D.A., Hammock B.D. Dihydroxyoctadecamonoenoate esters inhibit the neutrophil respiratory burst. J. Biosci. 2007;32:279–291. doi: 10.1007/s12038-007-0028-x. [DOI](https://doi.org/10.1007/s12038-007-0028-x) | [PMC free article](/articles/PMC1904342/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17435320/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Biosci.&title=Dihydroxyoctadecamonoenoate%20esters%20inhibit%20the%20neutrophil%20respiratory%20burst&author=D.A.%20Thompson&author=B.D.%20Hammock&volume=32&publication_year=2007&pages=279-291&pmid=17435320&doi=10.1007/s12038-007-0028-x&)
|
| 312 |
+
|
| 313 |
+
39. Palleria C., Roberti R., Iannone L.F., Tallarico M., Barbieri M.A., Vero A., Manti A., De Sarro G., Spina E., Russo E. Clinically Relevant Drug Interactions between Statins and Antidepressants. J. Clin. Pharm. Ther. 2020;45:227–239. doi: 10.1111/jcpt.13058. [DOI](https://doi.org/10.1111/jcpt.13058) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31587356/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Pharm.%20Ther.&title=Clinically%20Relevant%20Drug%20Interactions%20between%20Statins%20and%20Antidepressants&author=C.%20Palleria&author=R.%20Roberti&author=L.F.%20Iannone&author=M.%20Tallarico&author=M.A.%20Barbieri&volume=45&publication_year=2020&pages=227-239&pmid=31587356&doi=10.1111/jcpt.13058&)
|
| 314 |
+
|
| 315 |
+
40. Kee P.S., Chin P.K.L., Kennedy M.A., Maggo S.D.S. Pharmacogenetics of Statin-Induced Myotoxicity. Front. Genet. 2020;11:575678. doi: 10.3389/fgene.2020.575678. [DOI](https://doi.org/10.3389/fgene.2020.575678) | [PMC free article](/articles/PMC7596698/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33193687/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front.%20Genet.&title=Pharmacogenetics%20of%20Statin-Induced%20Myotoxicity&author=P.S.%20Kee&author=P.K.L.%20Chin&author=M.A.%20Kennedy&author=S.D.S.%20Maggo&volume=11&publication_year=2020&pages=575678&pmid=33193687&doi=10.3389/fgene.2020.575678&)
|
| 316 |
+
|
| 317 |
+
41. Suh J.-W., Cha M.-J., Lee S.-P., Chae I.-H., Kwon T.-G., Bae J.-W., Cho M.-C., Rha S.-W., Kim H.-S. Relationship Between Statin Type and Responsiveness to Clopidogrel in Patients Treated with Percutaneous Coronary Intervention: A Subgroup Analysis of the CILON-T Trial. J. Atheroscler. Thromb. 2014;21:140–150. doi: 10.5551/jat.19265. [DOI](https://doi.org/10.5551/jat.19265) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24140730/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Atheroscler.%20Thromb.&title=Relationship%20Between%20Statin%20Type%20and%20Responsiveness%20to%20Clopidogrel%20in%20Patients%20Treated%20with%20Percutaneous%20Coronary%20Intervention:%20A%20Subgroup%20Analysis%20of%20the%20CILON-T%20Trial&author=J.-W.%20Suh&author=M.-J.%20Cha&author=S.-P.%20Lee&author=I.-H.%20Chae&author=T.-G.%20Kwon&volume=21&publication_year=2014&pages=140-150&pmid=24140730&doi=10.5551/jat.19265&)
|
| 318 |
+
|
| 319 |
+
42. Filppula A.M., Hirvensalo P., Parviainen H., Ivaska V.E., Lönnberg K.I., Deng F., Viinamäki J., Kurkela M., Neuvonen M., Niemi M. Comparative Hepatic and Intestinal Metabolism and Pharmacodynamics of Statins. Drug Metab. Dispos. 2021;49:658–667. doi: 10.1124/dmd.121.000406. [DOI](https://doi.org/10.1124/dmd.121.000406) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34045219/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Metab.%20Dispos.&title=Comparative%20Hepatic%20and%20Intestinal%20Metabolism%20and%20Pharmacodynamics%20of%20Statins&author=A.M.%20Filppula&author=P.%20Hirvensalo&author=H.%20Parviainen&author=V.E.%20Ivaska&author=K.I.%20L%C3%B6nnberg&volume=49&publication_year=2021&pages=658-667&pmid=34045219&doi=10.1124/dmd.121.000406&)
|
| 320 |
+
|
| 321 |
+
43. Byrne R.A., Rossello X., Coughlan J.J., Barbato E., Berry C., Chieffo A., Claeys M.J., Dan G.-A., Dweck M.R., Galbraith M., et al. 2023 ESC Guidelines for the Management of Acute Coronary Syndromes. Eur. Heart J. 2023;44:3720–3826. doi: 10.1093/eurheartj/ehad191. [DOI](https://doi.org/10.1093/eurheartj/ehad191) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37622654/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Heart%20J.&title=2023%20ESC%20Guidelines%20for%20the%20Management%20of%20Acute%20Coronary%20Syndromes&author=R.A.%20Byrne&author=X.%20Rossello&author=J.J.%20Coughlan&author=E.%20Barbato&author=C.%20Berry&volume=44&publication_year=2023&pages=3720-3826&pmid=37622654&doi=10.1093/eurheartj/ehad191&)
|
| 322 |
+
|
| 323 |
+
44. Wang N., Fulcher J., Abeysuriya N., Park L., Kumar S., Di Tanna G.L., Wilcox I., Keech A., Rodgers A., Lal S. Intensive LDL Cholesterol-Lowering Treatment beyond Current Recommendations for the Prevention of Major Vascular Events: A Systematic Review and Meta-Analysis of Randomised Trials Including 327 037 Participants. Lancet Diabetes Endocrinol. 2020;8:36–49. doi: 10.1016/S2213-8587(19)30388-2. [DOI](https://doi.org/10.1016/S2213-8587(19)30388-2) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31862150/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet%20Diabetes%20Endocrinol.&title=Intensive%20LDL%20Cholesterol-Lowering%20Treatment%20beyond%20Current%20Recommendations%20for%20the%20Prevention%20of%20Major%20Vascular%20Events:%20A%20Systematic%20Review%20and%20Meta-Analysis%20of%20Randomised%20Trials%20Including%20327%20037%20Participants&author=N.%20Wang&author=J.%20Fulcher&author=N.%20Abeysuriya&author=L.%20Park&author=S.%20Kumar&volume=8&publication_year=2020&pages=36-49&pmid=31862150&doi=10.1016/S2213-8587(19)30388-2&)
|
test/texts/PMC11134291.md
ADDED
|
@@ -0,0 +1,360 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Variability in plasma rifampicin concentrations and role of SLCO1B1, ABCB1, AADAC2 and CES2 genotypes in Ethiopian patients with tuberculosis
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Tesemma Sileshi, Eyasu Makonnen, Nigus Fikrie Telele, Victoria Barclay, Alimuddin Zumla, Eleni Aklillu
|
| 5 |
+
**Journal:** Infectious diseases (London, England)
|
| 6 |
+
**Date:** 2024 Feb 5
|
| 7 |
+
**DOI:** [10.1080/23744235.2024.2309348](https://doi.org/10.1080/23744235.2024.2309348)
|
| 8 |
+
**PMID:** 38315168
|
| 9 |
+
**PMCID:** PMC11134291
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11134291/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11134291/pdf/nihms-1991917.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11134291/pdf/nihms-1991917.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Background::**
|
| 16 |
+
Rifampicin, a key drug against tuberculosis (TB), displays wide between-patient pharmacokinetics variability and concentration-dependent antimicrobial effect. We investigated variability in plasma rifampicin concentrations and the role of SLCO1B1, ABCB1, arylacetamide deacetylase (AADAC) and carboxylesterase 2 (CES-2) genotypes in Ethiopian patients with TB.
|
| 17 |
+
|
| 18 |
+
**Methods::**
|
| 19 |
+
We enrolled adult patients with newly diagnosed TB (n = 119) who had received 2 weeks of rifampicin-based anti-TB therapy. Venous blood samples were obtained at three time points post-dose. Genotypes for SLCO1B1 (c.388A>G, c.521T>C), ABCB1 (c.3435C>T, c.4036A>G), AADACc.841G>A and CES-2 (c.269-965A>G) were determined. Rifampicin plasma concentration was quantified using LC-MS/MS. Predictors of rifampicin Cmax and AUC0–7h were analysed.
|
| 20 |
+
|
| 21 |
+
**Results::**
|
| 22 |
+
The median rifampicin Cmax and AUC0–7 were 6.76 μg/mL (IQR 5.37–8.48) and 17.05 μg·h/mL (IQR 13.87–22.26), respectively. Only 30.3% of patients achieved the therapeutic efficacy threshold (Cmax>8 μg/mL). The allele frequency for SLCO1B1* 1B (c.388A>G), SLCO1B1* 5 (c.521T>C), ABCB1 c.3435C>T, ABCB1c.4036A>G, AADAC c.841G>A and CES-2 c.269-965A>G were 2.2%, 20.2%, 24.4%, 14.6%, 86.1% and 30.6%, respectively. Sex, rifampicin dose and ABCB1c.4036A>G, genotypes were significant predictors of rifampicin Cmax and AUC0–7. AADACc.841G>A genotypes were significant predictors of rifampicin Cmax. There was no significant influence of SLCO1B1 (c.388A>G, c.521T>C), ABCB1c.3435C>T and CES-2 c.269-965A>G on rifampicin plasma exposure variability.
|
| 23 |
+
|
| 24 |
+
**Conclusions::**
|
| 25 |
+
Subtherapeutic rifampicin plasma concentrations occurred in two-thirds of Ethiopian TB patients. Rifampicin exposure varied with sex, dose and genotypes. AADACc.841G/G and ABCB1c.4036A/A genotypes and male patients are at higher risk of lower rifampicin plasma exposure. The impact on TB treatment outcomes and whether high-dose rifampicin is required to improve therapeutic efficacy requires further investigation.
|
| 26 |
+
|
| 27 |
+
Keywords: Rifampicin, pharmacokinetics, pharmacogenetics, SLCO1B1, ABCB1, AADAC, CES-2, genotype, Ethiopia, tuberculosis
|
| 28 |
+
|
| 29 |
+
### Background:
|
| 30 |
+
|
| 31 |
+
Rifampicin, a key drug against tuberculosis (TB), displays wide between-patient pharmacokinetics variability and concentration-dependent antimicrobial effect. We investigated variability in plasma rifampicin concentrations and the role of *SLCO1B1*SLCO1B1, *ABCB1*ABCB1, arylacetamide deacetylase (*AADAC*AADAC) and carboxylesterase 2 (*CES-2*CES-2) genotypes in Ethiopian patients with TB.
|
| 32 |
+
|
| 33 |
+
### Methods:
|
| 34 |
+
|
| 35 |
+
We enrolled adult patients with newly diagnosed TB (*n*n = 119) who had received 2 weeks of rifampicin-based anti-TB therapy. Venous blood samples were obtained at three time points post-dose. Genotypes for *SLCO1B1*SLCO1B1 (*c.388A>G*c.388A>G, *c.521T>C*c.521T>C), *ABCB1*ABCB1 (*c.3435C>T*c.3435C>T, c.4036A>G), *AADACc.841G>A*AADACc.841G>A and *CES-2*CES-2 (*c.269-965A>G*c.269-965A>G) were determined. Rifampicin plasma concentration was quantified using LC-MS/MS. Predictors of rifampicin *C*C_max_max and AUC_0–7h_0–7h were analysed.
|
| 36 |
+
|
| 37 |
+
### Results:
|
| 38 |
+
|
| 39 |
+
The median rifampicin *C*C_max_max and AUC_0–7_0–7 were 6.76 μg/mL (IQR 5.37–8.48) and 17.05 μg·h/mL (IQR 13.87–22.26), respectively. Only 30.3% of patients achieved the therapeutic efficacy threshold (*C*C_max_max>8 μg/mL). The allele frequency for *SLCO1B1* 1B*SLCO1B1* 1B (*c.388A>G*c.388A>G), *SLCO1B1* 5*SLCO1B1* 5 (*c.521T>C*c.521T>C), *ABCB1 c.3435C>T*ABCB1 c.3435C>T, *ABCB1c.4036A>G*ABCB1c.4036A>G, *AADAC c.841G>A*AADAC c.841G>A and *CES-2 c.269-965A>G*CES-2 c.269-965A>G were 2.2%, 20.2%, 24.4%, 14.6%, 86.1% and 30.6%, respectively. Sex, rifampicin dose and *ABCB1c.4036A>G*ABCB1c.4036A>G, genotypes were significant predictors of rifampicin *C*C_max_max and AUC_0–7_0–7. *AADACc.841G>A*AADACc.841G>A genotypes were significant predictors of rifampicin *C*C_max_max. There was no significant influence of *SLCO1B1*SLCO1B1 (*c.388A>G,*c.388A>G, c*.521T>C*.521T>C), *ABCB1c.3435C>T*ABCB1c.3435C>T and *CES-2 c.269-965A>G*CES-2 c.269-965A>G on rifampicin plasma exposure variability.
|
| 40 |
+
|
| 41 |
+
### Conclusions:
|
| 42 |
+
|
| 43 |
+
Subtherapeutic rifampicin plasma concentrations occurred in two-thirds of Ethiopian TB patients. Rifampicin exposure varied with sex, dose and genotypes. *AADACc.841G/G*AADACc.841G/G and *ABCB1c.4036A/A*ABCB1c.4036A/A genotypes and male patients are at higher risk of lower rifampicin plasma exposure. The impact on TB treatment outcomes and whether high-dose rifampicin is required to improve therapeutic efficacy requires further investigation.
|
| 44 |
+
|
| 45 |
+
**Keywords:**Keywords: Rifampicin, pharmacokinetics, pharmacogenetics, *SLCO1B1*SLCO1B1, *ABCB1*ABCB1, *AADAC*AADAC, *CES-*CES-2, genotype, Ethiopia, tuberculosis
|
| 46 |
+
|
| 47 |
+
## Introduction
|
| 48 |
+
|
| 49 |
+
Whilst effective tuberculosis (TB) treatment has been available for the past seven decades, the latest 2022 WHO Annual Global Tuberculosis Report highlights that TB remains a leading cause of death from an infectious disease worldwide [[1](#R1)1]. Considerable success have been achieved in treatment outcomes since the introduction of rifampicin in 1970. However, the global increase in HIV incidence, poor adherence to 6-month therapy and suboptimal drug concentrations due to interindividual pharmacokinetic variations of first-line antitubercular drugs have contributed to the emergence of resistance to TB drugs [[2](#R2)2–[4](#R4)4]. Drug-resistant TB is a concern in East African countries [[5](#R5)5]. Ethiopia is among the top 30 countries with the highest TB and TB-HIV burden with an incidence of 119 cases per 100,000 people in 2021 [[1](#R1)1].
|
| 50 |
+
|
| 51 |
+
A combination of rifampicin with isoniazid is the backbone of modern anti-TB therapy. Rifampicin has concentration-dependent bactericidal activity [[6](#R6)6]. The microbial killing of rifampicin was linked to the ratio of the area under the concentration-time curve and the minimum inhibitory concentration (AUC/MIC) and the maximum concentration (*C*C_max_max)/MIC (*C*C_max_max/MIC) ratio. Rifampicin prevents resistance to itself and attains sufficient bactericidal effect at a free *C*C_max_max/MIC ratio of ≥175 [[7](#R7)7,[8](#R8)8]. A rifampicin *C*C_max_max between 8 and 24 μg/mL is considered optimal and *C*C_max_max below 4 μg/mL is a risk factor for treatment failure [[9](#R9)9].
|
| 52 |
+
|
| 53 |
+
Rifampicin undergoes hepatic metabolism by genetically polymorphic carboxylesterases (CES) and arylacetamide deacetylase (AADAC), a serine esterase to 25-deacetylrifampicin [[10](#R10)10]. Rifampicin pharmacokinetics and treatment outcomes display wide between-patient variations [[11](#R11)11,[12](#R12)12]. Genetic variation in enzymes and transporter proteins relevant to rifampicin disposition may influence the variability of plasma rifampicin exposure. Previous studies in various populations investigated the impact of genetic variation in *AADAC*AADAC and *CES*CES on rifampicin plasma exposure with varying results [[13](#R13)13–[16](#R16)16]. Rifampicin is a substrate and inducer of the organic anion transporter polypeptide 1B1 (OAT1B1) encoded by the *SLCO1B1*SLCO1B1 gene [[17](#R17)17] and P-glycoprotein (P-gp) encoded by the *ABCB1*ABCB1 gene [[18](#R18)18]. OAT1B1 mediates hepatocellular uptake of rifampicin while P-gp mediates drug efflux. Both *SLCO1B1*SLCO1B1 and *ABCB1*ABCB1 genes are genetically polymorphic displaying wide between-population variation in enzyme activity and variant allele frequency distributions. In the few published studies investigating the effect of the *SLCO1B1*SLCO1B1 and *ABCB1*ABCB1 gene polymorphism on rifampicin pharmacokinetics, the result is inconclusive [[16](#R16)16,[19](#R19)19–[22](#R22)22].
|
| 54 |
+
|
| 55 |
+
The pharmacokinetics and pharmacogenetics of rifampicin display wide between-race and between-population variations, highlighting the need for investigation in different geographic locations where the burden of TB is high. The effect of pharmacogenetic variability in rifampicin pharmacokinetics using a targeted candidate gene approach has been explored in various Asian and Caucasian populations [[22](#R22)22–[24](#R24)24], but data from sub-Saharan Africa remain scarce. Ethiopia is the seventh top high-TB burden country globally [[1](#R1)1]and the second most populous nation in Africa. The pharmacogenetics of *SLCO1B1*SLCO1B1 and *ABCB1*ABCB1 in Ethiopians differs from that of other black African populations and inhabitants of European origin [[23](#R23)23,[25](#R25)25,[26](#R26)26]. In this study, we examined the variability in rifampicin *C*C_max_max and AUC_0–7_0–7 in Ethiopian TB patients in relation to the recommended target concentration for optimal therapeutic efficacy and the impact of common functional genetic variants in *SLCO1B1*SLCO1B1 (rs2306283 and rs4149056), *ABCB1*ABCB1 (rs1045642 and rs3842), *CES 2*CES 2 (rs4783745) and *AADAC*AADAC (rs1803155) on between-patient variability in rifampicin plasma concentration.
|
| 56 |
+
|
| 57 |
+
## Methods
|
| 58 |
+
|
| 59 |
+
### Study participants and settings
|
| 60 |
+
|
| 61 |
+
The study participants were newly diagnosed adults aged 18–65 years with either pulmonary or extrapulmonary drug-sensitive *Mycobacterium tuberculosis*Mycobacterium tuberculosis attending TB clinics in Addis Ababa (Beletshachew, Teklehymanote, Kazanchis, Woreda 2 and Areda Health Centre). The study was conducted from October 2019 to November 2021.
|
| 62 |
+
|
| 63 |
+
### Blood sample collection
|
| 64 |
+
|
| 65 |
+
Blood samples were obtained 2 weeks after treatment initiation during the intensive phase of TB therapy. Following overnight fasting, participants received drugs under direct observation in the morning. A total of 351 venous blood samples were collected in EDTA tubes, with three samples taken at different times from 113 subjects and two times from 6 subjects. The blood sampling points ranged from 1 to 7 h post-dose, with the majority of subjects sampled at 1, 2, 4, or 2, 4, or 6 h post-dose. Plasma was separated immediately and stored at −80 °C at the Department of Pharmacology and Clinical Pharmacy, Addis Ababa University until transported to Karolinska Institutet, Stockholm, Sweden for analysis.
|
| 66 |
+
|
| 67 |
+
### Ethical approval
|
| 68 |
+
|
| 69 |
+
Ethical approval was obtained from the Institutional Review Board of the College of Health Sciences at Addis Ababa University and the National Research Ethics Review Committee. All patients were informed about the purpose of the study and those willing to participate and who provided written informed consent were enrolled. The study was conducted following the ethical principle of the Helsinki Declaration.
|
| 70 |
+
|
| 71 |
+
### Treatment
|
| 72 |
+
|
| 73 |
+
Study participants received a standard daily dose of rifampicin in combination with isoniazid, pyrazinamide and ethambutol according to the Ethiopian treatment guidelines [[27](#R27)27]. Patients with a body weight above 55kg received four fixed-dose combinations (FDC) tablets daily. Patients with a body weight between 40 and 55 kg received three FDC tablets daily and those below 40 kg received two FDC tablets daily. Each FDC tablet contains 150, 75, 400 and 275 mg of rifampicin, isoniazid, pyrazinamide and ethambutol, respectively. Treatment was given as directly observed therapy at a primary health care facility in Addis Ababa, Ethiopia.
|
| 74 |
+
|
| 75 |
+
### DNA extraction and genotyping
|
| 76 |
+
|
| 77 |
+
Genomic DNA was extracted from whole blood samples using the QIAmp DNA Blood Midi Kit (QIAGEN GmbH, Hilden, Germany) following the manufacturer’s instructions. Common functional variant alleles in the black African population relevant to rifampicin disposition were selected for genotyping. Genotyping was performed using TaqMan^®^® drug metabolism assay reagents for allelic discrimination (Applied Biosystems Genotyping Assays) as described previously [[28](#R28)28] with the following ID numbers: C___8911003_1 for *AADAC2*AADAC2 (c.841G>A, rs1803155), C__31760486_10 for *CES2*CES2 (c.269-965A>G, rs4783745), C___7586657_20 for *ABCB1*ABCB1 (3435 C>T, rs1045642), C__11711730_20 for *ABCB1*ABCB1 (c.193A>G, rs3842), C___1901697_20 for *SLCO1B1*SLCO1B1 (c.388A>G, rs2306283) and C__30633906_10 for *SLCO1B1*SLCO1B1 (c.521T>C, rs4149056).
|
| 78 |
+
|
| 79 |
+
The final volume for each reaction was 10 μL, consisting of 1 μL genomic DNA and 9 μL of TaqMan^®^® fast advanced master mix (Applied Biosystems, Waltham, MA, United States), DNA/RNA free water, TaqMan 40X for *SLCO1B1*SLCO1B1, *ABCB1*ABCB1 and TaqMan 20 × for *AADAC2*AADAC2 and *CES2*CES2 drug metabolism genotyping assays mix (Applied Biosystems). Genotyping was performed by real-time qPCR (Applied Biosystems) equipped with 7500 software V2.3 (Life Technologies Corporation) for allelic discrimination.
|
| 80 |
+
|
| 81 |
+
### Quantification of rifampicin plasma concentrations
|
| 82 |
+
|
| 83 |
+
To determine rifampicin plasma concentrations, blood samples were collected 2 weeks after treatment initiation during the intensive phase of TB therapy. After overnight fasting, study participants received drugs under direct observation in the morning. Venous blood was taken in EDTA tubes at three time points from 1 to 7 h post-dose. Plasma was separated immediately and stored at −80° C at the Department of Pharmacology and Clinical Pharmacy, Addis Ababa University until transported to Karolinska Institutet, Stockholm, Sweden for analysis.
|
| 84 |
+
|
| 85 |
+
Rifampicin plasma concentrations were determined using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described previously [[11](#R11)11]. The method was validated according to the European Medicines Agency guidelines [[29](#R29)29]. The LC-MS/MS system consisted of an Acquity Ultra Performance LC-system coupled to a Xevo TQ-S Micro (Waters, Milford, MA, USA) and aYMC-ultraHT hydrosphere C18, 2 μm, 100 × 2 mm, reversed-phase column (Waters) was used. Sample preparation consisted of protein precipitation with acetonitrile containing deuterated rifampicin as an internal standard. In brief, 100 μL plasma samples were diluted with a 300 μL solution containing the internal standards dissolved in acetonitrile. After shaking for 30 s and 5 min of centrifugation, 150 μL of the supernatant was transferred to another plate. The supernatant dried for 30 min at 35° C and the dried sample was re-suspended with 15 μL methanol and 275 μL 0.1% formic acid for injection. The mobile phase gradient of 0.1% formic acid in Milli-Q pure water, 100% methanol:methanol/Milli-Q pure water:formic acid (10:90:0.1), methanol:Milli-Q pure water:isopropanol:formic acid (70:20:10:0.1), methanol:Milli-Q pure water (10:90). Rifampicin concentrations were calculated by linear regression from a six-point calibration curve. The limits of the quantitation range for rifampicin were 0.1 and 40 μg/mL.
|
| 86 |
+
|
| 87 |
+
### Pharmacokinetic and statistical analyses
|
| 88 |
+
|
| 89 |
+
Study participants’ sociodemographic and baseline clinical parameters are summarised as the median and interquartile range (IQR) or as frequency and percentages. The rifampicin *C*C_max_max was determined from the available plasma concentrations. The highest concentration observed was taken as *C*C_max_max. AUC_0–7 h_0–7 h calculation was performed using the trapezoidal rule. GraphPad Prism was used to calculate AUC_0–7 h_0–7 h.
|
| 90 |
+
|
| 91 |
+
The Shapiro–Wilk test was used to determine the normality of pharmacokinetics data. Non-normally distributed data are presented as median (IQR) and normally distributed as mean (standard deviations [*SD*SD]). The chi-square test was used to assess correlations between the observed and expected genotype frequencies according to the Hardy–Weinberg equilibrium. All plasma concentration data were log 10 transformed before conducting statistical analyses [[29](#R29)29]. The association of each genotype with between-patient variability in *C*C_max_max and AUC_0–7_0–7 was analyzed using a one-way analysis of variance, comparing the geometric mean of log-transformed concentration data [[30](#R30)30]. Predictors of *C*C_max_max and AUC_0–7 h_0–7 h of rifampicin were subjected to further analysis through univariate followed by multivariate regression analysis, incorporating study participant characteristics and genotypes as potential predictors. Variables with *p*p value <0.2 from the univariate analysis were included in the multivariate regression analysis. Data were analyzed using SPSS version 25 and a *p*p value ≤0.05 was considered to indicate statistical significance.
|
| 92 |
+
|
| 93 |
+
## Results
|
| 94 |
+
|
| 95 |
+
### Study participants characteristics
|
| 96 |
+
|
| 97 |
+
Of the 119 study participants, consisting of 62 males and 57 females, 78 were diagnosed with pulmonary TB and 41 had extrapulmonary TB. The median body weight was 54.8 kg (IQR, 48.0–61.7), and the median age was 28 years (IQR, 22 – 35). The mean dose of rifampicin was 9.39 mg/kg (*SD*SD = 0.98). The prevalence of cigarette, khat and alcohol use was 13.4%, 18.5% and 16.8%, respectively. Notably, a lower percentage of patients with extrapulmonary TB reported cigarette, khat and alcohol use compared to those with pulmonary TB. Furthermore, patients with extrapulmonary TB showed higher rifampicin *C*C_max_max (*p*p = 0.07) and AUC_0–7_0–7 (*p*p = 0.23) values but the differences were not statistically significant. The sociodemographic characteristics of the participants are presented in [Table 1](#T1)Table 1.
|
| 98 |
+
|
| 99 |
+
### Table 1.
|
| 100 |
+
|
| 101 |
+
Sociodemographic and clinical characteristics of 119 Ethiopian tuberculosis patients.
|
| 102 |
+
|
| 103 |
+
| Variables | | Pulmonary TB (n = 78) | Extrapulmonary TB (n = 41) | All patients (n = 119) |
|
| 104 |
+
| --------- | - | --------------------- | -------------------------- | ---------------------- |
|
| 105 |
+
| Sex (n) | Male | 45 | 17 | 62 (52.5%) |
|
| 106 |
+
| | Female | 33 | 24 | 57 (47.5%) |
|
| 107 |
+
| Smoking (n) | Yes | 15 | 1 | 16 (13.4%) |
|
| 108 |
+
| | No | 63 | 40 | 103 (86.6%) |
|
| 109 |
+
| Khat chewer (n) | Yes | 20 | 2 | 22 (18.5%) |
|
| 110 |
+
| | No | 58 | 39 | 97 (81.5%) |
|
| 111 |
+
| Alcohol (n) | Yes | 17 | 3 | 20 (16.8%) |
|
| 112 |
+
| | No | 61 | 38 | 99 (83.2%) |
|
| 113 |
+
| Age (years), median (IQR) | | 26 (21–35) | 28 (24.5–36) | 28 (22–35) |
|
| 114 |
+
| Median body weight in kg (IQR) | | 53 (45–60) | 58 (52.5–68.5) | 54.75 (48–61.75) |
|
| 115 |
+
| Drug dose (mg/kg, SD) | | 9.46 (0.99) | 9.26 (0.98) | 9.39 (0.98) |
|
| 116 |
+
| Median Cmax, μg/mL (IQR) | | 6.45 (5.13–8.54) | 7.46 (6.02–8.72) | 6.75 (5.39–8.58) |
|
| 117 |
+
| Median AUC0–7 μg.h/mL (IQR) | | 16.52 (13.81–21.98) | 17.55 (14.3–22.59) | 17.05 (13.87–22.26) |
|
| 118 |
+
### Genotype and allele frequency
|
| 119 |
+
|
| 120 |
+
Study participants were genotyped for *SLCO1B1c.388A>G*SLCO1B1c.388A>G, *SLCO1B1 c.521T>C*SLCO1B1 c.521T>C, *ABCB1 c.3435C>T*ABCB1 c.3435C>T, *ABCB1 c.4036A>G*ABCB1 c.4036A>G, *AADAC c.841G>A*AADAC c.841G>A and *CES-2 c.269-965A>G*CES-2 c.269-965A>G. The observed genotype and allele frequency distributions among patients are shown in [Table 2](#T2)Table 2. There were no significant differences between observed and expected genotype frequencies according to Hardy–Weinberg equilibrium. The variant allele *SLCO1B1c.388A>G*SLCO1B1c.388A>G was frequent (62.2%), while the defective variant allele *SLCO1B1c.521T>C*SLCO1B1c.521T>C (*5) was less frequent (20.2%). The minor variant allele frequency for *ABCB1 c.3435T*ABCB1 c.3435T and *ABCB1 c.4036G*ABCB1 c.4036G were 24, 4%, and 14.6%, respectively. The variant *AADAC c.841A*AADAC c.841A variant allele had a much higher frequency (86.1%), whereas the CES-2 c.269–965G allele occurred in 30.6%.
|
| 121 |
+
|
| 122 |
+
### Table 2.
|
| 123 |
+
|
| 124 |
+
Genotype and variant allele frequency of SLCO1B1, ABCB1, AADAC and CES-2.
|
| 125 |
+
|
| 126 |
+
| Variant allele | Protein | Genotype frequency (n, %) | Allele frequency (%) | χ2 | p value |
|
| 127 |
+
| -------------- | ------- | ------------------------- | -------------------- | -- | ------- |
|
| 128 |
+
| SLCO1B1*1B (c.388A>G) | Asn130Asp | A/A (15, 12.6) | A/G (60, 50.4) | G/G (37, 44) | A (37.7) | G (62.2) | 0.618 | 0.43 |
|
| 129 |
+
| SLCO1B1*5 (c.521T>C) | Val174Ala | T/T (76, 63.9) | T/C (38, 31.9) | C/C (5, 4.2) | T (79.8) | C (20.2) | 0.008 | 0.99 |
|
| 130 |
+
| ABCB1 c.3435C>T | Ile1145Ile | C/C (67, 56.3) | C/T (46, 38.7) | T/T (6, 5.0) | C (75.6) | T (24.4) | 0.28 | 0.59 |
|
| 131 |
+
| ABCB1c.4036A>G | Located in 3′-UTR | A/A (88, 73.8) | A/G (28, 23.5) | G/G (3, 2.5) | A (85.4) | G (14.6) | 0.183 | 0.67 |
|
| 132 |
+
| AADAC*2 (c.841G>A) | Val281Ile | G/G (3, 2.5) | G/A (26, 21.7) | A/A (90, 75) | G (13.9) | A (86.1) | 0.447 | 0.5 |
|
| 133 |
+
| CES-2 c.269-965A>G | Located in intron 1 | A/A (55, 46.2) | A/G (55, 46.2) | G/G (9, 7.6) | A (69.4) | G (30.6) | 0.896 | 0.34 |
|
| 134 |
+
### Rifampicin pharmacokinetics
|
| 135 |
+
|
| 136 |
+
There was high between-patient variability in rifampicin *C*C_max_max (range 1.90–18.57 μg/mL) and AUC_0–7_0–7 (range 3.61–47.1 μg × h/mL). The median rifampicin *C*C_max_max was 6.76 μg/mL (IQR 5.33–8.49). Only 30.3% (*n*n = 36) of participants achieved the target plasma concentration (> 8 μg/mL) for optimal therapeutic efficacy [[31](#R31)31]. *C*C_max_max <4 μg/mL, which is associated with risk for treatment failure, was observed in 5 (4.2%) patients. The median AUC_0–7 h_0–7 h was 17.1 μg × h/mL (IQR 13.9–22.3).
|
| 137 |
+
|
| 138 |
+
### Effect of genotype on rifampicin pharmacokinetics
|
| 139 |
+
|
| 140 |
+
A comparison of the median and geometric mean of rifampicin *C*C_max_max and AUC_0–7 h_0–7 h between the different genotypes using one-way analysis of variance is presented in [Table 3](#T3)Table 3. Although no significant influence of *SLCO1B1*SLCO1B1**1B*1B and *SLCO1B1*SLCO1B1**5*5 genotype on variation in rifampicin *C*C_max_max and AUC_0–7h_0–7h was found, patients homozygous for *SLCO1B*SLCO1B**5/*5/**5*5 (*C/C*C/C) had a *C*C_max_max below the target concentration. No significant difference in *C*C_max_max and AUC_0–7_0–7 was observed in *ABCB1 c.3435C>T*ABCB1 c.3435C>T and CES 2 c.269-965A>G genotype groups.
|
| 141 |
+
|
| 142 |
+
### Table 3.
|
| 143 |
+
|
| 144 |
+
Effects of SLCO1B1, ABCB1, AADAC and CES-2 genotype on rifampicin Cmax and AUC0–7 h in Ethiopian TB patients (n = 119).
|
| 145 |
+
|
| 146 |
+
| Genotype | | N | Cmax (μg/mL) | AUC0–7 (μg h/mL) |
|
| 147 |
+
| -------- | - | - | ------------ | ---------------- |
|
| 148 |
+
| Median (IQR) | Geometric mean ± SE | p Value* | Median (IQR) | Geometric mean ± SE | p value* |
|
| 149 |
+
| SLCO1B1*1B (c.388A>G) | A/A | 15 | 6.88 (5.83–9.36) | 7.08 ± 1.1 | 0.87 | 17.95 (16.59–20.93) | 17.78 ± 1.12 | 0.67 |
|
| 150 |
+
| | A/G | 60 | 6.62 (6.23–7.75) | 6.76 ± 1.05 | | 16.35 (14.36–18.22) | 16.6 ± 1.05 | |
|
| 151 |
+
| | G/G | 44 | 6.82 (6.1–7.4) | 6.92 ± 1.05 | | 17.14 (15.35–18.78) | 17.78 ± 1.07 | |
|
| 152 |
+
| SLCO1B1*5 (c.521T>C) | T/T | 76 | 6.59 (6.18–7.02) | 6.76 ± .05 | 0.15 | 16.65 (15.08–18.12) | 16.98 ± 1.05 | 0.18 |
|
| 153 |
+
| | T/C | 38 | 7.62 (6.76–8.12) | 7.24 ± 1.07 | | 18.49 (17.03–22.4) | 18.2 ± 1.07 | |
|
| 154 |
+
| | C/C | 5 | 5.1 (4.76–7.41) | 5.37 ± 1.1 | | 12.5 (12.44–19.61) | 13.18 ± 1.12 | |
|
| 155 |
+
| ABCB1 c.3435C>T | C/C | 67 | 6.42 (6.1–7.2) | 6.61 ± 1.05 | 0.70 | 16.7 (15.08–18.22) | 16.98 ± 1.05 | 0.87 |
|
| 156 |
+
| | C/T | 46 | 7.23 (6.63–7.95) | 6.92 ± 1.07 | | 17.85 (16.48–19.73) | 17.78 ± 1.07 | |
|
| 157 |
+
| | T/T | 6 | 6.76 (6.1–10.43) | 7.24 ± 1.15 | | 14.68 (13.4–23.99) | 16.6 ± 1.12 | |
|
| 158 |
+
| ABCB1 c.4036A>G | A/A | 88 | 6.53 (6.10–7.18) | 6.61 ± 1.05 | 0.018 | 16.7 (15.08–17.77) | 16.6 ± 1.05 | 0.02 |
|
| 159 |
+
| | A/G | 28 | 7.29 (6.59–8.85) | 7.41 ± 1.07 | | 18.31 (15.5–23.09) | 18.2 ± 1.07 | |
|
| 160 |
+
| | G/G | 3 | 9.35 (7.07–18.57) | 10.72 ± 1.32 | | 32.13 (17.3–47.05) | 29.51 ± 1.35 | |
|
| 161 |
+
| AADAC2 c.841G>A | A/A | 90 | 7.045 (6.49–7.82) | 7.08 ± 1.05 | 0.047 | 17.56 (16.48–18.58) | 17.78 ± 1.05 | 0.16 |
|
| 162 |
+
| | G/A | 26 | 6.21 (4.85–6.79) | 6.03 ± 1.07 | | 15.06 (14.11–20.29) | 15.49 ± 1.07 | |
|
| 163 |
+
| | G/G | 3 | 4.69 (4.27–6.63) | 5.13 ± 1.15 | | 13.99 (9.92–16.59) | 13.18 ± 1.17 | |
|
| 164 |
+
| CES-2 c.269-965A>G | A/A | 55 | 7.18 (6.59–8.04) | 7.08 ± 1.05 | 0.08 | 17.85 (15.5–19.06) | 17.38 ± 1.05 | 0.19 |
|
| 165 |
+
| | A/G | 55 | 6.42 (6.10–7.2) | 6.31 ± 1.05 | | 16.58 (14.82–17.3) | 16.22 ± 1.05 | |
|
| 166 |
+
| | G/G | 9 | 6.87 (6.18–13.94) | 8.13 ± 1.15 | | 18.32 (15.08–29.74) | 21.38 ± 1.15 | |
|
| 167 |
+
Significant variability in rifampicin *C*C_max_max (*p*p = 0.018) and AUC_0–7 h_0–7 h (0.02) between the *ABCB1 c.4036A>G*ABCB1 c.4036A>G genotype groups was observed. The geometric mean of *C*C_max_max and AUC_0–7 h_0–7 h was significantly higher among patients homozygous for the variant allele *ABCB1c.4036G/G*ABCB1c.4036G/G than heterozygous A/G or homozygous wild type (A/A)([Table 3](#T3)Table 3). A further post hoc analysis using Bonferroni correction indicated significant differences in *C*C_max_max (*p*p = 0.036) and AUC_0–7 h_0–7 h (*p*p = 0.023) between homozygous *ABCB1 c.4036 A/A*ABCB1 c.4036 A/A and homozygous wild-type (*G/G*G/G) groups. The comparison of *C*C_max_max and AUC_0–7 h_0–7 h between the different *ABCB1 c.4036A>G*ABCB1 c.4036A>G genotype groups is presented in [Figure 1(A)](#F1)Figure 1(A). No significant difference in *C*C_max_max and AUC_0–7_0–7 was observed in the different *ABCB1 c.3435C>T*ABCB1 c.3435C>T genotype groups.
|
| 168 |
+
|
| 169 |
+
### Figure 1.
|
| 170 |
+
|
| 171 |
+

|
| 172 |
+
|
| 173 |
+
Comparison of rifampicin Cmax and AUC0–7 h in the ABCB1 c.4036A>G (right) and AADAC2 c.841G>A (left) genotypes. The box plots show the median ± interquartile range, whereas whiskers denote the minimum and maximum values.
|
| 174 |
+
|
| 175 |
+
Furthermore, a significant association of *AADAC c.841G>A*AADAC c.841G>A genotype with rifampicin *C*C_max_max (*p*p = 0.047) and a similar trend for AUC_0–7_0–7 (*p*p = 0.16) was observed and was lower in the wild type (*G/G*G/G) genotype than heterozygous (*A/G*A/G) or homozygous for *A*A variant allele (*A/A*A/A) ([Figure 1(B)](#F1)Figure 1(B)). However, a post hoc test showed no significant variation for AUC_0–7_0–7 among the pairs of all three genotypes of *AADAC c.841G>A*AADAC c.841G>A. There was no significant association of *CES 2 c.269-965A>G*CES 2 c.269-965A>G genotype with rifampicin *C*C_max_max and AUC_0–7 h_0–7 h.
|
| 176 |
+
|
| 177 |
+
### Predictors of rifampicin pharmacokinetics
|
| 178 |
+
|
| 179 |
+
A univariate followed by a multivariate analysis was conducted to identify predictors of *C*C_max_max and AUC_0–7h_0–7h using log10 transformed concertation data. [Table 4](#T4)Table 4 shows the results of univariate and multivariate analyses of associations between variables and rifampicin *C*C_max_max and AUC_0–7 h_0–7 h. In univariate analysis, *ABCB1 c.4036A>G*ABCB1 c.4036A>G, *AADAC c.841G>A*AADAC c.841G>A genotypes and rifampicin dose were significant predictors of rifampicin *C*C_max_max (*p ≤*p ≤ 0.05), and a nearly significant effect was observed for sex (*p*p = 0.06). All variables with *p*p value <0.2 were further tested in the multivariate regression model. In multivariate analysis, sex, rifampicin dose, *ABCB1 c.4036A>G*ABCB1 c.4036A>G and *AADAC c.841G>A*AADAC c.841G>A genotypes remained independent predictors of rifampicin *C*C_max_max.
|
| 180 |
+
|
| 181 |
+
### Table 4.
|
| 182 |
+
|
| 183 |
+
Univariate and multivariate linear regression analysis of factors associated with rifampicin log10Cmax and log10AUC0–7 h in Ethiopian adult tuberculosis patients.
|
| 184 |
+
|
| 185 |
+
| Variable | C max | AUC |
|
| 186 |
+
| -------- | ----- | --- |
|
| 187 |
+
| Univariate | Multivariate | Univariate | Multivariate |
|
| 188 |
+
| Beta coefficients (95% CI) | p value | Adjusted beta coefficients (95% CI) | p value | Beta coefficients (95% CI) | p value | Adjusted beta coefficients (95% CI) | p value |
|
| 189 |
+
| Age | 0.002 (−0.001 to 0.006) | 0.12 | 0.002 (−0.048 to 0.004) | 0.24 | 0.002 (−0.001 to 0.006) | 0.19 | 0.002 (−0.002 to 0.005) | 0.31 |
|
| 190 |
+
| Sex (female vs. male) | −0.051 (−0.11 to 0.003) | 0.06 | −0.056 (−0.11 to 0.004) | 0.03 | −0.057 (−0.12 to 0.04) | 0.07 | −0.063 (−0.12 to 0.03) | 0.04 |
|
| 191 |
+
| Drug dose (mg) | 0.00 (0.00 to 0.001) | 0.05 | 0.000 (0.00 to 0.001) | 0.03 | 0.000 (0.00 to 0.01) | 0.05 | 0.000 (0.00 to 0.01) | 0.03 |
|
| 192 |
+
| Alcohol use (no vs. yes) | 0.00 (−0.066 to 0.8) | 0.84 | | | 0.002 (−0.08 to 0.083) | 0.97 | | |
|
| 193 |
+
| Khat chewing (no vs. yes) | 0.00 (−0.074 to 0.067) | 0.91 | | | 0.013 (−0.066 to 0.093) | 0.74 | | |
|
| 194 |
+
| Smoking (no vs. yes) | −0.012 (−0.093 to 0.067) | 0.75 | | | 0.016 (−0.076 to 0.11) | 0.73 | | |
|
| 195 |
+
| Days on drug therapy | −0.001 (−0.004 to 0.0012) | 0.43 | | | 0.001 (−0.005 to 0.003) | 0.59 | | |
|
| 196 |
+
| SLCO1B1c.388A>G | −001 (−0.043 to 0.04) | 0.96 | | | 0.007 (−0.04 to 0.054) | 0.76 | | |
|
| 197 |
+
| SLCO1B1c.521T>C | 0.002 (−0.05 to 0.046) | 0.94 | | | 0.002 (−0.052 to 0.056) | 0.95 | | |
|
| 198 |
+
| ABCB1 c.3435C>T | 0.02 (−0.026 to 0.066) | 0.4 | | | 0.006 (−0.046 to 0.059) | 0.81 | | |
|
| 199 |
+
| ABCB1 c.4036A>G | 0.071 (0.018 to 0.124) | 0.009 | 0.063 (0.013 to 0.114) | 0.015 | 0.071 (0.011 to 0.13) | 0.02 | 0.059 (0.001 to 0.13) | 0.048 |
|
| 200 |
+
| AADAC*2c.841G>A | −0.068 (−0.122 to −0.014) | 0.01 | −0.065 (−0.12 to −0.013) | 0.015 | −0.059 (−0.12 to 0.001) | 0.06 | −0.059 (−0.12 to 0.001) | 0.053 |
|
| 201 |
+
| CES-2 c.269-965A>G | −0.004 (−0.048 to0.04) | 0.86 | | | 0.008 (−0.043 to0.058) | 0.76 | | |
|
| 202 |
+
*ABCB1 c.4036A>G*ABCB1 c.4036A>G and drug dose were significant predictors for rifampicin AUC_0–7 h_0–7 h in both univariate and multivariate analysis. In multivariate analysis, sex was also a predictor of rifampicin AUC_0–7 h_0–7 h. Overall, females had higher exposure to rifampicin compared to males. Age, alcohol, cigarette and khat use, *SLCO1B1c.388A>G*SLCO1B1c.388A>G, *SLCO1B1c.521T>C*SLCO1B1c.521T>C, *ABCB1 c.3435C>T*ABCB1 c.3435C>T, *CES-2 c.269-965A>G*CES-2 c.269-965A>G genotypes and days on drug therapy did not predict rifampicin exposure (*C*C_max_max and AUC_0–7 h_0–7 h).
|
| 203 |
+
|
| 204 |
+
The stepwise multivariate regression analysis demonstrated that *ABCB1 c.4036A>G*ABCB1 c.4036A>G genotypes independently accounted for 5.8% of the variability in rifampicin *C*C_max_max. Combining *AADAC c.841G>A*AADAC c.841G>A and *ABCB1 c.4036A>G*ABCB1 c.4036A>G genotypes increased the explained variability to 10.8%. Additionally, 14% variability in rifampicin *C*C_max_max was observed when the drug dose (mg) was added to the two genotypes. The overall variability in rifampicin *C*C_max_max explained by the two genotypes, drug dose and sex was 17.2%. Similarly, *ABCB1 c.4036A>G*ABCB1 c.4036A>G genotypes explained 6.1% of the variability in rifampicin AUC_0–7_0–7 explained by. With the sequential addition of sex, drug dose and *AADAC c.841G>A*AADAC c.841G>A to the model, the variability in rifampicin AUC_0–7_0–7 increased to 10.1%, 15.8%, and 19.3%, respectively. These findings underscore the significant roles of *AADAC c.841G>A*AADAC c.841G>A and *ABCB1 c.4036A>G*ABCB1 c.4036A>G genotype, along with sex and drug dose in predicting rifampicin *C*C_max_max and AUC_0–7_0–7 among the variables examined.
|
| 205 |
+
|
| 206 |
+
## Discussion
|
| 207 |
+
|
| 208 |
+
The study is the first to examine the relationship between genetic polymorphism and rifampicin pharmacokinetics in the Ethiopian population. We investigated the between-patient variability of rifampicin pharmacokinetics parameters (*C*C_max_max and AUC_0–7 h_0–7 h) in Ethiopian adults commencing TB treatment and the role of pharmacogenetic variations in drug transporter proteins (*SLCO1B1*SLCO1B1 and *ABCB1*ABCB1) and metabolising enzymes relevant for rifampicin disposition (*AADAC2*AADAC2 and *CES2*CES2). There were several notable findings. First, there was substantial between-patient variability in rifampicin plasma concentrations. Second, a majority (70%) of patients had rifampicin plasma concentrations below the recommended target (≥8 μg/mL). Third, rifampicin dose, *ABCB1c.4036A>G*ABCB1c.4036A>G and *AADACc.841G>A*AADACc.841G>A genotypes and to some extent, sex were independent predictors of rifampicin *C*C_max_max and AUC_0–7 h_0–7 h.
|
| 209 |
+
|
| 210 |
+
Two weeks after treatment initiation, a 2-h post-dose plasma sample is recommended for therapeutic drug monitoring to predict TB treatment outcomes. Rifampicin *C*C_max_max should exceed 8 mg/L for optimal therapeutic efficacy [[32](#R32)32–[34](#R34)34]. This peak concentration was not attained in about 70% of our patients who received the standard rifampicin dose. Our finding is in line with previous studies reporting that many patients receiving first-line anti-TB therapy do not achieve the rifampicin *C*C_max_max target concentration, but the proportion varies between populations [[16](#R16)16,[34](#R34)34–[36](#R36)36]. To the best of our knowledge, the proportion of TB patients below the target 8 mg/mL in this study is one of the highest. This finding is of concern since subtherapeutic levels are associated with unfavorable outcomes and risk for development of drug resistance [[9](#R9)9,[37](#R37)37]. Indeed, drug-resistant TB is an increasing concern in Ethiopia [[5](#R5)5,[38](#R38)38,[39](#R39)39]. A higher dose of rifampicin or therapeutic drug monitoring in selected patients could be beneficial as suggested previously [[9](#R9)9,[40](#R40)40]. Whether high doses of rifampicin are safe and more effective than the standard dose is studied in clinical trials to shorten treatment duration and increase efficacy. The trial results indicated that a higher dose of rifampicin led to faster sputum sterilisation while maintaining a comparable level of toxicity to the standard dose [[41](#R41)41–[44](#R44)44]. Therefore, an increase in the dose of rifampicin in Ethiopian population may be warranted.
|
| 211 |
+
|
| 212 |
+
Several factors could contribute to the observed low rifampicin plasma concentrations in Ethiopian patients including genetic variations, malnutrition and HIV infection, which are quite prevalent in East Africa including Ethiopia [[1](#R1)1,[45](#R45)45,[46](#R46)46]. However, compared to the 70% observed in this study, only 35% of Tanzanian TB patients had a rifampicin *C*C_max_max below 8mg/L [[47](#R47)47]. The low rifampicin concentrations in Ethiopian TB patients could be due to either higher rifampicin metabolising enzyme activities or increased autoinduction due to pharmacogenetic variations [[23](#R23)23,[25](#R25)25,[26](#R26)26,[48](#R48)48,[49](#R49)49]. Lower plasma drug concentrations have been reported in earlier studies of antiretrovirals due to higher drug-metabolising enzyme activity and unique pharmacogenetic variation in Ethiopians compared to other populations, including Tanzanians [[23](#R23)23,[25](#R25)25,[26](#R26)26,[50](#R50)50]. Our study highlights the existence of substantial differences in rifampicin pharmacokinetics between populations in sub-Saharan Africa and findings from one population may not be directly extrapolated to others on the continent. Recently we reported high plasma isoniazid concentrations and a high prevalence of slow N-acetyltransferase 2 (NAT2) acetylators in Ethiopian TB patients [[51](#R51)51].
|
| 213 |
+
|
| 214 |
+
There have been inconsistent results about the effects of *SLCO1B1*SLCO1B1 genetic variation on rifampicin exposure. Previous studies in South African and Ugandan patients reported an association of the *SLCO1B1*SLCO1B1 genotype with variability in rifampicin pharmacokinetics [[21](#R21)21,[22](#R22)22,[52](#R52)52]. However, this finding was not replicated in many studies [[11](#R11)11,[15](#R15)15,[53](#R53)53,[54](#R54)54]. Likewise, we found no significant impact of *SLCO1B1 c.388A>G*SLCO1B1 c.388A>G and *SLCO1B1 c.521T>C*SLCO1B1 c.521T>C on rifampicin *C*C_max_max and AUC_0–7h_0–7h. *SLCO1B1*1B*SLCO1B1*1B and *SLCO1B1*5*SLCO1B1*5 are missense mutations, involving the change of asparagine to aspartic acid at position130 and valine to alanine at position 174, respectively ([Table 2](#T2)Table 2). The variant alleles *SLCO1B1*SLCO1B1**1B*1B and *SLCO1B1*SLCO1B1**5*5 were associated with increased and decreased transporter activity of OATP1B1, respectively. *SLCO1B1*SLCO1B1**1B*1B, which is associated with higher transporter activity, occurs at a higher frequency (62.2%) in Ethiopians and Tanzanians (86.8%) than in Europeans (34.2%) [[23](#R23)23]. On the other hand, the defective *SLCO1B1c.521T>C*SLCO1B1c.521T>C variant allele caused reduced enzyme activity occurs at a lower frequency among Ethiopians (2.8%) than Tanzanians (4.7%) or Europeans (8%) [[23](#R23)23,[25](#R25)25,[26](#R26)26].
|
| 215 |
+
|
| 216 |
+
Rifampicin is a substrate and inducer of P-gp which is a product of the *ABCB1*ABCB1 gene [[40](#R40)40,[55](#R55)55]. Few studies have evaluated the effect of *ABCB1*ABCB1 gene polymorphism on rifampicin pharmacokinetics. Huerta-García et al. reported that patients with *CC*CC or *CT*CT genotypes of *ABCB1*ABCB1 (*c.3435C>T*c.3435C>T) had lower *C*C_max_max and AUC_24_24 than those with a *TT*TT genotype [[56](#R56)56]. The *TT*TT homozygous genotype had significantly lower P-gp expression in the small intestine and showed the highest plasma concentrations of some drugs after oral administration [[24](#R24)24]. However, we found no significant variation in rifampicin *C*C_max_max and AUC_0–7 h_0–7 h for *ABCB1 c.3435C>T*ABCB1 c.3435C>T. The *ABCB1c.4036A>G*ABCB1c.4036A>G genotype, which is in linkage disequilibrium with *c.3435C>T,*c.3435C>T, significantly influenced between-patient variability of rifampicin *C*C_max_max and AUC_0–7 h_0–7 h. Rifampicin AUC_0–7 h_0–7 h was significantly higher in homozygous variant genotype (*GG*GG) carriers compared to the homozygous wild-type A/A ([Figure 1](#F1)Figure 1). Nevertheless, the homozygous variant genotype (*GG*GG) occurs at a low frequency in our study population, consistent with findings from a previous report [[49](#R49)49].
|
| 217 |
+
|
| 218 |
+
Few studies have investigated the impact of *AADAC*AADAC and *CES*CES genetic polymorphism on rifampicin pharmacokinetics. The association of *CES-2 c.−2263A>G*CES-2 c.−2263A>G (rs3759994) in the promotor region and closely linked to *c.269-965A>G*c.269-965A>G (rs4783745) and c.1612 + *136 G>A*136 G>A with increased rifampicin exposure is reported [[13](#R13)13]. Patients who carry the *CES2*CES2 (rs8192925) *G*G versus *A*A allele had a 17.2% increase in rifapentine AUC_0–24_0–24 (14). In our study, there was no significant association of *CES2 c.269-965A>G*CES2 c.269-965A>G genotypes with rifampicin *C*C_max_max and AUC_0–7 h_0–7 h. Likewise, no significant effect of *CES-2*CES-2 on rifampicin exposure variability was observed in Ghanaian children [[16](#R16)16]. *AADAC*AADAC and *CES-1*CES-1 genotypes were not associated with rifampicin pharmacokinetics in Malawian TB patients [[15](#R15)15].
|
| 219 |
+
|
| 220 |
+
We found a significant association between *AADAC c.841G>A*AADAC c.841G>A genotype and rifampicin *C*C_max_max, which was significantly higher in carriers of the mutant variant allele (A/A, G/A) than in those with wild-type G/G genotype ([Figure 1](#F1)Figure 1). Our result is consistent with previous reports [[3](#R3)3,[14](#R14)14]. Francis et al. reported that patients with *A/A*A/A genotype had a lower rifapentine clearance. Similarly, a previous study found an association of *AADAC c.841G*AADAC c.841G variant allele with low rifapentine AUC, particularly in black patients [[14](#R14)14]. However, this finding was not observed in Malawian adult TB patients [[15](#R15)15]. The low frequency of a wild-type (*GG*GG) genotype in Malawians may have contributed to the differing results. Indeed, the frequency distribution of *AADAC*AADAC**2*2 (c.841G>A) exhibits considerable variability across races and populations. Notably, the reported allele frequencies of *AADAC*AADAC**2*2 among European American, African American, Japanese and Korean populations were around 60%, contrasting with the 99.9% prevalence in Peruvian TB patients [[57](#R57)57] where the wild-type variant is almost missing. Our study among Ethiopian TB patients reveals *AADAC*AADAC**2*2 allele frequencies of 86%, and the wild-type *G*G variant was less prevalent with only three individuals exhibiting homozygosity for *G/G*G/G genotype. This underscores the need for further investigation in populations where the *AADAC c.841G*AADAC c.841G variant occurs at higher frequencies to replicate and confirm our findings.
|
| 221 |
+
|
| 222 |
+
In addition to genetic polymorphism, other predictors such as age, sex, duration of therapy with rifampicin, drug dose and substance use were tested in univariate followed by multivariate analyses. Sex and drug dose were significantly associated with rifampicin *C*C_max_max and AUC_0–7 h_0–7 h in multivariate analysis. Females had higher rifampicin exposure (higher *C*C_max_max and AUC_0–7 h_0–7 h) than males. This is consistent with previous studies where male sex was associated with lower rifampicin exposure [[35](#R35)35,[36](#R36)36,[52](#R52)52,[58](#R58)58].
|
| 223 |
+
|
| 224 |
+
Our study presents the first insight into the extent of variability in rifampicin exposure (*C*C_max_max and AUC_0–7_0–7) and the impact of genetic variation in drug transporters and metabolising enzymes in Ethiopian TB patients. However, it is imperative to acknowledge certain limitations in our study. The estimation of rifampicin pharmacokinetics in our study relied on three sampling time points within 7 h post-dose, adhering to the recommended approach for therapeutic drug monitoring [[31](#R31)31]. A 2-h post-dose sample approximates the *C*C_max_max for most TB drugs and adding a 6-h sample allows the clinician to distinguish between delayed absorption and malabsorption [[31](#R31)31,[32](#R32)32,[34](#R34)34]. Nevertheless, although the spare sampling strategy is useful for capturing the AUC_0–24 h_0–24 h [[59](#R59)59], the three time point concentration dataset in our study may not entirely capture the AUC accurately. Nevertheless, it is crucial to underscore that obtaining multiple blood samples solely for the study’s objectives from newly diagnosed TB-infected patients undergoing an intensive phase of treatment is impractical and raises ethical concerns.
|
| 225 |
+
|
| 226 |
+
Furthermore, in our study population, the occurrence of the wild-type *AADAC c.841 G/G*AADAC c.841 G/G and the variant *ABCB1 c.4036 G/G*ABCB1 c.4036 G/G genotype occurred at a lower frequency, potentially influencing the association of rifampicin *C*C_max_max and AUC_0–7 h_0–7 h with the investigated genotypes. It is note-worthy that globally, and particularly within Africa, G variant alleles exhibit lower frequencies for both *AADAC c.841G>A*AADAC c.841G>A and *ABCB1 c.4036A>G*ABCB1 c.4036A>G. The frequency of *ABCB1 c.4036A>G*ABCB1 c.4036A>G varies among black Africans, ranging from 29% in Tanzanians [[60](#R60)60] to 18% in Ethiopians [[28](#R28)28]. Considering these variations, future large-sample studies across diverse populations in high TB-burden areas, including Africa, where rifampicin is a cornerstone of TB therapy, are recommended to validate and replicate our findings.
|
| 227 |
+
|
| 228 |
+
In conclusion, we report low rifampicin exposure and high variability in rifampicin *C*C_max_max and AUC_0–7_0–7 in about two-thirds of Ethiopian TB patients. Rifampicin exposure varied with sex, dose, *ABCB1 c.4036A>G*ABCB1 c.4036A>G and *ADAC c.841G>A*ADAC c.841G>A genotypes. *AADAC c.841GG*AADAC c.841GG and *ABCB1 c.4036A>GAA*ABCB1 c.4036A>GAA genotype groups and male patients had a higher risk of low rifampicin plasma exposure than females. *SLCO1B1 c.388A>*SLCO1B1 c.388A>, *SLCO1B1 c.521T>C*SLCO1B1 c.521T>C, *ABCB1 c.3435C>T*ABCB1 c.3435C>T and *CES2 c.269-965A>G*CES2 c.269-965A>G genotypes did not affect rifampicin exposure. The impact of low rifampicin exposure on treatment outcomes needs further investigation in Ethiopian TB patients. Our findings may have important clinical implications and warrant studies on whether high-dose rifampicin improves therapeutic efficacy.
|
| 229 |
+
|
| 230 |
+
## Acknowledgments
|
| 231 |
+
|
| 232 |
+
The authors thank all study participants and staff of health centres involved in patient recruitment and sample collection. A.Z. acknowledges support from the EU-EDCTP-funded PANDORA-ID-NET program. A.Z. is in receipt of a UK National Institute for Health Research Senior Investigator Award.
|
| 233 |
+
|
| 234 |
+
### Funding
|
| 235 |
+
|
| 236 |
+
This study was supported by the Fogarty International Centre and the National Institute of Allergy and Infectious Disease of the National Institute of Health [Award No. D43 TW009127], Centre of Innovative Drug Development and Therapeutic Trial for Africa (CDT-Africa), Addis Ababa University, and The European & Developing Countries Clinical Trials Partnership (EDCTP2) [Grant Nos. CSA2016S-1618 and RIA2017MC-2009]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
|
| 237 |
+
|
| 238 |
+
## Footnotes
|
| 239 |
+
|
| 240 |
+
## References
|
| 241 |
+
|
| 242 |
+
1. WHO. Global Tuberculosis Report 2022. Geneva: World Health Organization; 2022. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Global%20Tuberculosis%20Report%202022&publication_year=2022&)
|
| 243 |
+
|
| 244 |
+
2. Sloan DJ, Davies GR, Khoo SH. Recent advances in tuberculosis: new drugs and treatment regimens. Curr Respir Med Rev. 2013; 9(3):200–210. doi: . [DOI](https://doi.org/10.2174/1573398×113099990017) | [PMC free article](/articles/PMC3968807/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24683386/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Curr%20Respir%20Med%20Rev&title=Recent%20advances%20in%20tuberculosis:%20new%20drugs%20and%20treatment%20regimens&author=DJ%20Sloan&author=GR%20Davies&author=SH%20Khoo&volume=9&issue=3&publication_year=2013&pages=200-210&pmid=24683386&doi=10.2174/1573398×113099990017&)
|
| 245 |
+
|
| 246 |
+
3. Francis J, Zvada SP, Denti P, et al. A population pharmacokinetic analysis shows that arylacetamide deacetylase (AADAC) gene polymorphism and HIV infection affect the exposure of rifapentine. Antimicrob Agents Chemother. 2019;63(4):e01964–18. doi: 10.1128/AAC.01964-18. [DOI](https://doi.org/10.1128/AAC.01964-18) | [PMC free article](/articles/PMC6437540/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30670438/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=A%20population%20pharmacokinetic%20analysis%20shows%20that%20arylacetamide%20deacetylase%20(AADAC)%20gene%20polymorphism%20and%20HIV%20infection%20affect%20the%20exposure%20of%20rifapentine&author=J%20Francis&author=SP%20Zvada&author=P%20Denti&volume=63&issue=4&publication_year=2019&pages=e01964-18&pmid=30670438&doi=10.1128/AAC.01964-18&)
|
| 247 |
+
|
| 248 |
+
4. Chakaya J, Khan M, Ntoumi F, et al. Global Tuberculosis Report 2020 – reflections on the global TB burden, treatment and prevention efforts. Int J Infect Dis. 2021; 113(Suppl 1):S7–S12. doi: 10.1016/j.ijid.2021.02.107. [DOI](https://doi.org/10.1016/j.ijid.2021.02.107) | [PMC free article](/articles/PMC8433257/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33716195/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int%20J%20Infect%20Dis&title=Global%20Tuberculosis%20Report%202020%20%E2%80%93%20reflections%20on%20the%20global%20TB%20burden,%20treatment%20and%20prevention%20efforts&author=J%20Chakaya&author=M%20Khan&author=F%20Ntoumi&volume=113&issue=Suppl%201&publication_year=2021&pages=S7-S12&pmid=33716195&doi=10.1016/j.ijid.2021.02.107&)
|
| 249 |
+
|
| 250 |
+
5. Molla KA, Reta MA, Ayene YY. Prevalence of multidrug-resistant tuberculosis in East Africa: a systematic review and meta-analysis. PLoS One. 2022;17(6):e0270272. doi: 10.1371/journal.pone.0270272. [DOI](https://doi.org/10.1371/journal.pone.0270272) | [PMC free article](/articles/PMC9246177/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35771884/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=PLoS%20One&title=Prevalence%20of%20multidrug-resistant%20tuberculosis%20in%20East%20Africa:%20a%20systematic%20review%20and%20meta-analysis&author=KA%20Molla&author=MA%20Reta&author=YY%20Ayene&volume=17&issue=6&publication_year=2022&pages=e0270272&pmid=35771884&doi=10.1371/journal.pone.0270272&)
|
| 251 |
+
|
| 252 |
+
6. Diacon AH, Patientia RF, Venter A, et al. Early bactericidal activity of high-dose rifampin in patients with pulmonary tuberculosis evidenced by positive sputum smears. Antimicrob Agents Chemother. 2007;51(8):2994–2996. doi: 10.1128/AAC.01474-06. [DOI](https://doi.org/10.1128/AAC.01474-06) | [PMC free article](/articles/PMC1932511/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17517849/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Early%20bactericidal%20activity%20of%20high-dose%20rifampin%20in%20patients%20with%20pulmonary%20tuberculosis%20evidenced%20by%20positive%20sputum%20smears&author=AH%20Diacon&author=RF%20Patientia&author=A%20Venter&volume=51&issue=8&publication_year=2007&pages=2994-2996&pmid=17517849&doi=10.1128/AAC.01474-06&)
|
| 253 |
+
|
| 254 |
+
7. Gumbo T, Louie A, Deziel MR, et al. Concentration-dependent Mycobacterium tuberculosis killing and prevention of resistance by rifampin. Antimicrob Agents Chemother. 2007;51(11):3781–3788. doi: 10.1128/AAC.01533-06. [DOI](https://doi.org/10.1128/AAC.01533-06) | [PMC free article](/articles/PMC2151424/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17724157/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Concentration-dependent%20Mycobacterium%20tuberculosis%20killing%20and%20prevention%20of%20resistance%20by%20rifampin&author=T%20Gumbo&author=A%20Louie&author=MR%20Deziel&volume=51&issue=11&publication_year=2007&pages=3781-3788&pmid=17724157&doi=10.1128/AAC.01533-06&)
|
| 255 |
+
|
| 256 |
+
8. Niward K, Davies Forsman L, Bruchfeld J, et al. Distribution of plasma concentrations of first-line anti-TB drugs and individual MICs: a prospective cohort study in a low endemic setting. J Antimicrob Chemother. 2018;73(10): 2838–2845. doi: 10.1093/jac/dky268. [DOI](https://doi.org/10.1093/jac/dky268) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30124844/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Antimicrob%20Chemother&title=Distribution%20of%20plasma%20concentrations%20of%20first-line%20anti-TB%20drugs%20and%20individual%20MICs:%20a%20prospective%20cohort%20study%20in%20a%20low%20endemic%20setting&author=K%20Niward&author=L%20Davies%20Forsman&author=J%20Bruchfeld&volume=73&issue=10&publication_year=2018&pages=2838-2845&pmid=30124844&doi=10.1093/jac/dky268&)
|
| 257 |
+
|
| 258 |
+
9. Ramachandran G, Chandrasekaran P, Gaikwad S, et al. Subtherapeutic rifampicin concentration is associated with unfavorable tuberculosis treatment outcomes. Clin Infect Dis. 2020;70(7):1463–1470. doi: 10.1093/cid/ciz380. [DOI](https://doi.org/10.1093/cid/ciz380) | [PMC free article](/articles/PMC7931830/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31075166/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Infect%20Dis&title=Subtherapeutic%20rifampicin%20concentration%20is%20associated%20with%20unfavorable%20tuberculosis%20treatment%20outcomes&author=G%20Ramachandran&author=P%20Chandrasekaran&author=S%20Gaikwad&volume=70&issue=7&publication_year=2020&pages=1463-1470&pmid=31075166&doi=10.1093/cid/ciz380&)
|
| 259 |
+
|
| 260 |
+
10. Nakajima A, Fukami T, Kobayashi Y, et al. Human arylacetamide deacetylase is responsible for deacetylation of rifamycins: rifampicin, rifabutin, and rifapentine. Biochem Pharmacol. 2011;82(11):1747–1756. doi: 10.1016/j.bcp.2011.08.003. [DOI](https://doi.org/10.1016/j.bcp.2011.08.003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21856291/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Biochem%20Pharmacol&title=Human%20arylacetamide%20deacetylase%20is%20responsible%20for%20deacetylation%20of%20rifamycins:%20rifampicin,%20rifabutin,%20and%20rifapentine&author=A%20Nakajima&author=T%20Fukami&author=Y%20Kobayashi&volume=82&issue=11&publication_year=2011&pages=1747-1756&pmid=21856291&doi=10.1016/j.bcp.2011.08.003&)
|
| 261 |
+
|
| 262 |
+
11. Mukonzo JK, Kengo A, Kutesa B, et al. Role of pharmacogenetics in rifampicin pharmacokinetics and the potential effect on TB-rifampicin sensitivity among Ugandan patients. Trans R Soc Trop Med Hyg. 2020;114(2):107–114. doi: 10.1093/trstmh/trz108. [DOI](https://doi.org/10.1093/trstmh/trz108) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31789383/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Trans%20R%20Soc%20Trop%20Med%20Hyg&title=Role%20of%20pharmacogenetics%20in%20rifampicin%20pharmacokinetics%20and%20the%20potential%20effect%20on%20TB-rifampicin%20sensitivity%20among%20Ugandan%20patients&author=JK%20Mukonzo&author=A%20Kengo&author=B%20Kutesa&volume=114&issue=2&publication_year=2020&pages=107-114&pmid=31789383&doi=10.1093/trstmh/trz108&)
|
| 263 |
+
|
| 264 |
+
12. Sileshi T, Tadesse E, Makonnen E, et al. The impact of first-line anti-tubercular drugs’ pharmacokinetics on treatment outcome: a systematic review. Clin Pharmacol. 2021;13:1–12. doi: 10.2147/CPAA.S289714. [DOI](https://doi.org/10.2147/CPAA.S289714) | [PMC free article](/articles/PMC7811439/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33469389/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol&title=The%20impact%20of%20first-line%20anti-tubercular%20drugs%E2%80%99%20pharmacokinetics%20on%20treatment%20outcome:%20a%20systematic%20review&author=T%20Sileshi&author=E%20Tadesse&author=E%20Makonnen&volume=13&publication_year=2021&pages=1-12&pmid=33469389&doi=10.2147/CPAA.S289714&)
|
| 265 |
+
|
| 266 |
+
13. Song SH, Chang HE, Jun SH, et al. Relationship between CES2 genetic variations and rifampicin metabolism. J Antimicrob Chemother. 2013;68(6):1281–1284. doi: 10.1093/jac/dkt036. [DOI](https://doi.org/10.1093/jac/dkt036) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23471941/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Antimicrob%20Chemother&title=Relationship%20between%20CES2%20genetic%20variations%20and%20rifampicin%20metabolism&author=SH%20Song&author=HE%20Chang&author=SH%20Jun&volume=68&issue=6&publication_year=2013&pages=1281-1284&pmid=23471941&doi=10.1093/jac/dkt036&)
|
| 267 |
+
|
| 268 |
+
14. Weiner M, Gelfond J, Johnson-Pais TL, et al. Decreased plasma rifapentine concentrations associated with AADAC single nucleotide polymorphism in adults with tuberculosis. J Antimicrob Chemother. 2021;76(3):582–586. doi: 10.1093/jac/dkaa490. [DOI](https://doi.org/10.1093/jac/dkaa490) | [PMC free article](/articles/PMC7879139/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33374006/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Antimicrob%20Chemother&title=Decreased%20plasma%20rifapentine%20concentrations%20associated%20with%20AADAC%20single%20nucleotide%20polymorphism%20in%20adults%20with%20tuberculosis&author=M%20Weiner&author=J%20Gelfond&author=TL%20Johnson-Pais&volume=76&issue=3&publication_year=2021&pages=582-586&pmid=33374006&doi=10.1093/jac/dkaa490&)
|
| 269 |
+
|
| 270 |
+
15. Sloan DJ, McCallum AD, Schipani A, et al. Genetic determinants of the pharmacokinetic variability of rifampin in Malawian adults with pulmonary tuberculosis. Antimicrob Agents Chemother. 2017;61(7):e00210–17. doi: 10.1128/AAC.00210-17. [DOI](https://doi.org/10.1128/AAC.00210-17) | [PMC free article](/articles/PMC5487625/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28461315/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Genetic%20determinants%20of%20the%20pharmacokinetic%20variability%20of%20rifampin%20in%20Malawian%20adults%20with%20pulmonary%20tuberculosis&author=DJ%20Sloan&author=AD%20McCallum&author=A%20Schipani&volume=61&issue=7&publication_year=2017&pages=e00210-17&pmid=28461315&doi=10.1128/AAC.00210-17&)
|
| 271 |
+
|
| 272 |
+
16. Dompreh A, Tang X, Zhou J, et al. Effect of genetic variation of NAT2 on isoniazid and SLCO1B1 and CES2 on rifampin pharmacokinetics in Ghanaian children with tuberculosis. Antimicrob Agents Chemother. 2018;62(3):e02099–17. doi: 10.1128/AAC.02099-17. [DOI](https://doi.org/10.1128/AAC.02099-17) | [PMC free article](/articles/PMC5826147/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29263072/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Effect%20of%20genetic%20variation%20of%20NAT2%20on%20isoniazid%20and%20SLCO1B1%20and%20CES2%20on%20rifampin%20pharmacokinetics%20in%20Ghanaian%20children%20with%20tuberculosis&author=A%20Dompreh&author=X%20Tang&author=J%20Zhou&volume=62&issue=3&publication_year=2018&pages=e02099-17&pmid=29263072&doi=10.1128/AAC.02099-17&)
|
| 273 |
+
|
| 274 |
+
17. Tirona RG, Leake BF, Wolkoff AW, et al. Human organic anion transporting polypeptide-C (SLC21A6) is a major determinant of rifampin-mediated pregnane X receptor activation. J Pharmacol Exp Ther. 2003;304(1):223–228. doi: 10.1124/jpet.102.043026. [DOI](https://doi.org/10.1124/jpet.102.043026) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12490595/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharmacol%20Exp%20Ther&title=Human%20organic%20anion%20transporting%20polypeptide-C%20(SLC21A6)%20is%20a%20major%20determinant%20of%20rifampin-mediated%20pregnane%20X%20receptor%20activation&author=RG%20Tirona&author=BF%20Leake&author=AW%20Wolkoff&volume=304&issue=1&publication_year=2003&pages=223-228&pmid=12490595&doi=10.1124/jpet.102.043026&)
|
| 275 |
+
|
| 276 |
+
18. Williamson B, Dooley KE, Zhang Y, et al. Induction of influx and efflux transporters and cytochrome P450 3A4 in primary human hepatocytes by rifampin, rifabutin, and rifapentine. Antimicrob Agents Chemother. 2013;57(12):6366–6369. doi: 10.1128/AAC.01124-13. [DOI](https://doi.org/10.1128/AAC.01124-13) | [PMC free article](/articles/PMC3837889/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24060875/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Induction%20of%20influx%20and%20efflux%20transporters%20and%20cytochrome%20P450%203A4%20in%20primary%20human%20hepatocytes%20by%20rifampin,%20rifabutin,%20and%20rifapentine&author=B%20Williamson&author=KE%20Dooley&author=Y%20Zhang&volume=57&issue=12&publication_year=2013&pages=6366-6369&pmid=24060875&doi=10.1128/AAC.01124-13&)
|
| 277 |
+
|
| 278 |
+
19. Thomas L, Sekhar Miraj S, Surulivelrajan M, et al. Influence of single nucleotide polymorphisms on rifampin pharmacokinetics in tuberculosis patients. Antibiotics (Basel). 2020; 9(6):307. doi: 10.3390/antibiotics9060307. [DOI](https://doi.org/10.3390/antibiotics9060307) | [PMC free article](/articles/PMC7344705/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32521634/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antibiotics%20(Basel)&title=Influence%20of%20single%20nucleotide%20polymorphisms%20on%20rifampin%20pharmacokinetics%20in%20tuberculosis%20patients&author=L%20Thomas&author=S%20Sekhar%20Miraj&author=M%20Surulivelrajan&volume=9&issue=6&publication_year=2020&pages=307&pmid=32521634&doi=10.3390/antibiotics9060307&)
|
| 279 |
+
|
| 280 |
+
20. Sileshi T, Mekonen G, Makonnen E, et al. Effect of genetic variations in drug-metabolizing enzymes and drug transporters on the pharmacokinetics of rifamycins: a systematic review. Pharmgenomics Pers Med. 2022;15:561–571. doi: 10.2147/PGPM.S363058. [DOI](https://doi.org/10.2147/PGPM.S363058) | [PMC free article](/articles/PMC9176238/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35693129/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmgenomics%20Pers%20Med&title=Effect%20of%20genetic%20variations%20in%20drug-metabolizing%20enzymes%20and%20drug%20transporters%20on%20the%20pharmacokinetics%20of%20rifamycins:%20a%20systematic%20review&author=T%20Sileshi&author=G%20Mekonen&author=E%20Makonnen&volume=15&publication_year=2022&pages=561-571&pmid=35693129&doi=10.2147/PGPM.S363058&)
|
| 281 |
+
|
| 282 |
+
21. Chigutsa E, Visser ME, Swart EC, et al. The SLCO1B1 rs4149032 polymorphism is highly prevalent in South Africans and is associated with reduced rifampin concentrations: dosing implications. Antimicrob Agents Chemother. 2011;55(9):4122–4127. doi: 10.1128/AAC.01833-10. [DOI](https://doi.org/10.1128/AAC.01833-10) | [PMC free article](/articles/PMC3165308/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21709081/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=The%20SLCO1B1%20rs4149032%20polymorphism%20is%20highly%20prevalent%20in%20South%20Africans%20and%20is%20associated%20with%20reduced%20rifampin%20concentrations:%20dosing%20implications&author=E%20Chigutsa&author=ME%20Visser&author=EC%20Swart&volume=55&issue=9&publication_year=2011&pages=4122-4127&pmid=21709081&doi=10.1128/AAC.01833-10&)
|
| 283 |
+
|
| 284 |
+
22. Weiner M, Peloquin C, Burman W, et al. Effects of tuberculosis, race, and human gene SLCO1B1 polymorphisms on rifampin concentrations. Antimicrob Agents Chemother. 2010;54(10):4192–4200. doi: 10.1128/AAC.00353-10. [DOI](https://doi.org/10.1128/AAC.00353-10) | [PMC free article](/articles/PMC2944564/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20660695/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Effects%20of%20tuberculosis,%20race,%20and%20human%20gene%20SLCO1B1%20polymorphisms%20on%20rifampin%20concentrations&author=M%20Weiner&author=C%20Peloquin&author=W%20Burman&volume=54&issue=10&publication_year=2010&pages=4192-4200&pmid=20660695&doi=10.1128/AAC.00353-10&)
|
| 285 |
+
|
| 286 |
+
23. Aklillu E, Habtewold A, Ngaimisi E, et al. SLCO1B1 gene variations among Tanzanians, Ethiopians, and Europeans: relevance for African and worldwide precision medicine. OMICS. 2016;20(9):538–545. doi: 10.1089/omi.2016.0119. [DOI](https://doi.org/10.1089/omi.2016.0119) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27631193/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=OMICS&title=SLCO1B1%20gene%20variations%20among%20Tanzanians,%20Ethiopians,%20and%20Europeans:%20relevance%20for%20African%20and%20worldwide%20precision%20medicine&author=E%20Aklillu&author=A%20Habtewold&author=E%20Ngaimisi&volume=20&issue=9&publication_year=2016&pages=538-545&pmid=27631193&doi=10.1089/omi.2016.0119&)
|
| 287 |
+
|
| 288 |
+
24. Ameyaw MM, Regateiro F, Li T, et al. MDR1 pharmacogenetics: frequency of the C3435T mutation in exon 26 is significantly influenced by ethnicity. Pharmacogenetics. 2001; 11(3):217–221. doi: 10.1097/00008571-200104000-00005. [DOI](https://doi.org/10.1097/00008571-200104000-00005) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11337937/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=MDR1%20pharmacogenetics:%20frequency%20of%20the%20C3435T%20mutation%20in%20exon%2026%20is%20significantly%20influenced%20by%20ethnicity&author=MM%20Ameyaw&author=F%20Regateiro&author=T%20Li&volume=11&issue=3&publication_year=2001&pages=217-221&pmid=11337937&doi=10.1097/00008571-200104000-00005&)
|
| 289 |
+
|
| 290 |
+
25. Mugusi S, Habtewold A, Ngaimisi E, et al. Impact of population and pharmacogenetics variations on efavirenz pharmacokinetics and immunologic outcomes during anti-tuberculosis Co-therapy: a parallel prospective cohort study in two Sub-Sahara African populations. Front Pharmacol. 2020;11:26. doi: 10.3389/fphar.2020.00026. [DOI](https://doi.org/10.3389/fphar.2020.00026) | [PMC free article](/articles/PMC7019112/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32116703/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front%20Pharmacol&title=Impact%20of%20population%20and%20pharmacogenetics%20variations%20on%20efavirenz%20pharmacokinetics%20and%20immunologic%20outcomes%20during%20anti-tuberculosis%20Co-therapy:%20a%20parallel%20prospective%20cohort%20study%20in%20two%20Sub-Sahara%20African%20populations&author=S%20Mugusi&author=A%20Habtewold&author=E%20Ngaimisi&volume=11&publication_year=2020&pages=26&pmid=32116703&doi=10.3389/fphar.2020.00026&)
|
| 291 |
+
|
| 292 |
+
26. Aklillu E, Mugusi S, Ngaimisi E, et al. Frequency of the SLCO1B1 388A>G and the 521T>C polymorphism in Tanzania genotyped by a new LightCycler®-based method. Eur J Clin Pharmacol. 2011;67(11):1139–1145. doi: 10.1007/s00228-011-1065-9. [DOI](https://doi.org/10.1007/s00228-011-1065-9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21630030/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=Frequency%20of%20the%20SLCO1B1%20388A>G%20and%20the%20521T>C%20polymorphism%20in%20Tanzania%20genotyped%20by%20a%20new%20LightCycler%C2%AE-based%20method&author=E%20Aklillu&author=S%20Mugusi&author=E%20Ngaimisi&volume=67&issue=11&publication_year=2011&pages=1139-1145&pmid=21630030&doi=10.1007/s00228-011-1065-9&)
|
| 293 |
+
|
| 294 |
+
27. EFMOH (Ethiopia Federal Ministry of Health). Guidelines for clinical and programmatic management of TB, TB/HIV, DR-TB and leprosy in Ethiopia. 2021. Available from: http://repository.iphce.org/xmlui/handle/123456789/1662 [http://repository.iphce.org/xmlui/handle/123456789/1662](http://repository.iphce.org/xmlui/handle/123456789/1662)
|
| 295 |
+
|
| 296 |
+
28. Chala A, Tadesse BT, Chaka TE, et al. Predictors of efavirenz plasma exposure, auto-induction profile, and effect of pharmacogenetic variations among HIV-Infected children in Ethiopia: a prospective cohort study. J Pers Med. 2021; 11(12):1303. doi: 10.3390/jpm11121303. [DOI](https://doi.org/10.3390/jpm11121303) | [PMC free article](/articles/PMC8707067/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34945777/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pers%20Med&title=Predictors%20of%20efavirenz%20plasma%20exposure,%20auto-induction%20profile,%20and%20effect%20of%20pharmacogenetic%20variations%20among%20HIV-Infected%20children%20in%20Ethiopia:%20a%20prospective%20cohort%20study&author=A%20Chala&author=BT%20Tadesse&author=TE%20Chaka&volume=11&issue=12&publication_year=2021&pages=1303&pmid=34945777&doi=10.3390/jpm11121303&)
|
| 297 |
+
|
| 298 |
+
29. FDA. Statistical approaches to establishing bioequivalence. Guidance for industry. 2001. Available from: https://www.fda.gov/media/70958/download [https://www.fda.gov/media/70958/download](https://www.fda.gov/media/70958/download) | [Google Scholar](https://scholar.google.com/scholar_lookup?title=Statistical%20approaches%20to%20establishing%20bioequivalence&publication_year=2001&)
|
| 299 |
+
|
| 300 |
+
30. Dunvald AD, Iversen DB, Svendsen ALO, et al. Tutorial: statistical analysis and reporting of clinical pharmacokinetic studies. Clin Transl Sci. 2022;15(8):1856–1866. doi: 10.1111/cts.13305. [DOI](https://doi.org/10.1111/cts.13305) | [PMC free article](/articles/PMC9372427/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35570335/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Transl%20Sci&title=Tutorial:%20statistical%20analysis%20and%20reporting%20of%20clinical%20pharmacokinetic%20studies&author=AD%20Dunvald&author=DB%20Iversen&author=ALO%20Svendsen&volume=15&issue=8&publication_year=2022&pages=1856-1866&pmid=35570335&doi=10.1111/cts.13305&)
|
| 301 |
+
|
| 302 |
+
31. Alsultan A, Peloquin CA. Therapeutic drug monitoring in the treatment of tuberculosis: an update. Drugs. 2014;74(8): 839–854. doi: 10.1007/s40265-014-0222-8. [DOI](https://doi.org/10.1007/s40265-014-0222-8) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24846578/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drugs&title=Therapeutic%20drug%20monitoring%20in%20the%20treatment%20of%20tuberculosis:%20an%20update&author=A%20Alsultan&author=CA%20Peloquin&volume=74&issue=8&publication_year=2014&pages=839-854&pmid=24846578&doi=10.1007/s40265-014-0222-8&)
|
| 303 |
+
|
| 304 |
+
32. Chawla PK, Udwadia ZF, Soman R, et al. Importance of therapeutic drug monitoring of rifampicin. J Assoc Physicians India. 2016;64(8):68–72. [PubMed](https://pubmed.ncbi.nlm.nih.gov/27762112/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Assoc%20Physicians%20India&title=Importance%20of%20therapeutic%20drug%20monitoring%20of%20rifampicin&author=PK%20Chawla&author=ZF%20Udwadia&author=R%20Soman&volume=64&issue=8&publication_year=2016&pages=68-72&pmid=27762112&)
|
| 305 |
+
|
| 306 |
+
33. Peloquin CA. Therapeutic drug monitoring in the treatment of tuberculosis. Drugs. 2002;62(15):2169–2183. doi: 10.2165/00003495-200262150-00001. [DOI](https://doi.org/10.2165/00003495-200262150-00001) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12381217/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drugs&title=Therapeutic%20drug%20monitoring%20in%20the%20treatment%20of%20tuberculosis&author=CA%20Peloquin&volume=62&issue=15&publication_year=2002&pages=2169-2183&pmid=12381217&doi=10.2165/00003495-200262150-00001&)
|
| 307 |
+
|
| 308 |
+
34. Prahl JB, Johansen IS, Cohen AS, et al. Clinical significance of 2 h plasma concentrations of first-line anti-tuberculosis drugs: a prospective observational study. J Antimicrob Chemother. 2014;69(10):2841–2847. doi: 10.1093/jac/dku210. [DOI](https://doi.org/10.1093/jac/dku210) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25140577/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Antimicrob%20Chemother&title=Clinical%20significance%20of%202%20h%20plasma%20concentrations%20of%20first-line%20anti-tuberculosis%20drugs:%20a%20prospective%20observational%20study&author=JB%20Prahl&author=IS%20Johansen&author=AS%20Cohen&volume=69&issue=10&publication_year=2014&pages=2841-2847&pmid=25140577&doi=10.1093/jac/dku210&)
|
| 309 |
+
|
| 310 |
+
35. Trentalange A, Borgogno E, Motta I, et al. Rifampicin and isoniazid maximal concentrations are below efficacy-associated thresholds in the majority of patients: time to increase the doses? Int J Antimicrob Agents. 2021;57(3):106297. doi: 10.1016/j.ijantimicag.2021.106297. [DOI](https://doi.org/10.1016/j.ijantimicag.2021.106297) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33539932/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int%20J%20Antimicrob%20Agents&title=Rifampicin%20and%20isoniazid%20maximal%20concentrations%20are%20below%20efficacy-associated%20thresholds%20in%20the%20majority%20of%20patients:%20time%20to%20increase%20the%20doses?&author=A%20Trentalange&author=E%20Borgogno&author=I%20Motta&volume=57&issue=3&publication_year=2021&pages=106297&pmid=33539932&doi=10.1016/j.ijantimicag.2021.106297&)
|
| 311 |
+
|
| 312 |
+
36. van Crevel R, Alisjahbana B, de Lange WC, et al. Low plasma concentrations of rifampicin in tuberculosis patients in Indonesia. Int J Tuberc Lung Dis. 2002;6(6):497–502. doi: 10.5588/09640569513002. [DOI](https://doi.org/10.5588/09640569513002) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12068982/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int%20J%20Tuberc%20Lung%20Dis&title=Low%20plasma%20concentrations%20of%20rifampicin%20in%20tuberculosis%20patients%20in%20Indonesia&author=R%20van%20Crevel&author=B%20Alisjahbana&author=WC%20de%20Lange&volume=6&issue=6&publication_year=2002&pages=497-502&pmid=12068982&doi=10.5588/09640569513002&)
|
| 313 |
+
|
| 314 |
+
37. Niward K, Ek Blom L, Davies Forsman L, et al. Plasma levels of rifampin correlate with the tuberculosis drug activity assay. Antimicrob Agents Chemother. 2018;62(5):e00218–18. doi: 10.1128/AAC.00218-18. [DOI](https://doi.org/10.1128/AAC.00218-18) | [PMC free article](/articles/PMC5923115/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29483112/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Plasma%20levels%20of%20rifampin%20correlate%20with%20the%20tuberculosis%20drug%20activity%20assay&author=K%20Niward&author=L%20Ek%20Blom&author=L%20Davies%20Forsman&volume=62&issue=5&publication_year=2018&pages=e00218-18&pmid=29483112&doi=10.1128/AAC.00218-18&)
|
| 315 |
+
|
| 316 |
+
38. Asgedom SW, Teweldemedhin M, Gebreyesus H. Prevalence of Multidrug-Resistant tuberculosis and associated factors in Ethiopia: a systematic review. J Pathog. 2018;2018:7104921–7104928. doi: 10.1155/2018/7104921. [DOI](https://doi.org/10.1155/2018/7104921) | [PMC free article](/articles/PMC5903304/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29850257/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pathog&title=Prevalence%20of%20Multidrug-Resistant%20tuberculosis%20and%20associated%20factors%20in%20Ethiopia:%20a%20systematic%20review&author=SW%20Asgedom&author=M%20Teweldemedhin&author=H%20Gebreyesus&volume=2018&publication_year=2018&pages=7104921-7104928&pmid=29850257&doi=10.1155/2018/7104921&)
|
| 317 |
+
|
| 318 |
+
39. Mehari K, Asmelash T, Hailekiros H, et al. Prevalence and factors associated with multidrug-resistant tuberculosis (MDR-TB) among presumptive MDR-TB patients in Tigray region, Northern Ethiopia. Can J Infect Dis Med Microbiol. 2019;2019:2923549. doi: 10.1155/2019/2923549. [DOI](https://doi.org/10.1155/2019/2923549) | [PMC free article](/articles/PMC6754863/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31583034/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Can%20J%20Infect%20Dis%20Med%20Microbiol&title=Prevalence%20and%20factors%20associated%20with%20multidrug-resistant%20tuberculosis%20(MDR-TB)%20among%20presumptive%20MDR-TB%20patients%20in%20Tigray%20region,%20Northern%20Ethiopia&author=K%20Mehari&author=T%20Asmelash&author=H%20Hailekiros&volume=2019&publication_year=2019&pages=2923549&pmid=31583034&doi=10.1155/2019/2923549&)
|
| 319 |
+
|
| 320 |
+
40. Stott KE, Pertinez H, Sturkenboom MGG, et al. Pharmacokinetics of rifampicin in adult TB patients and healthy volunteers: a systematic review and meta-analysis. J Antimicrob Chemother. 2018;73(9):2305–2313. doi: 10.1093/jac/dky152. [DOI](https://doi.org/10.1093/jac/dky152) | [PMC free article](/articles/PMC6105874/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29701775/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Antimicrob%20Chemother&title=Pharmacokinetics%20of%20rifampicin%20in%20adult%20TB%20patients%20and%20healthy%20volunteers:%20a%20systematic%20review%20and%20meta-analysis&author=KE%20Stott&author=H%20Pertinez&author=MGG%20Sturkenboom&volume=73&issue=9&publication_year=2018&pages=2305-2313&pmid=29701775&doi=10.1093/jac/dky152&)
|
| 321 |
+
|
| 322 |
+
41. Garcia-Prats AJ, Svensson EM, Winckler J, et al. Pharmacokinetics and safety of high-dose rifampicin in children with TB: the Opti-Rif trial. J Antimicrob Chemother. 2021;76(12):3237–3246. doi: 10.1093/jac/dkab336. [DOI](https://doi.org/10.1093/jac/dkab336) | [PMC free article](/articles/PMC8598292/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34529779/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Antimicrob%20Chemother&title=Pharmacokinetics%20and%20safety%20of%20high-dose%20rifampicin%20in%20children%20with%20TB:%20the%20Opti-Rif%20trial&author=AJ%20Garcia-Prats&author=EM%20Svensson&author=J%20Winckler&volume=76&issue=12&publication_year=2021&pages=3237-3246&pmid=34529779&doi=10.1093/jac/dkab336&)
|
| 323 |
+
|
| 324 |
+
42. Velásquez GE, Brooks MB, Coit JM, et al. Efficacy and safety of high-dose rifampin in pulmonary tuberculosis. A randomized controlled trial. Am J Respir Crit Care Med. 2018;198(5):657–666. doi: 10.1164/rccm.201712-2524OC. [DOI](https://doi.org/10.1164/rccm.201712-2524OC) | [PMC free article](/articles/PMC6118011/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29954183/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Respir%20Crit%20Care%20Med&title=Efficacy%20and%20safety%20of%20high-dose%20rifampin%20in%20pulmonary%20tuberculosis.%20A%20randomized%20controlled%20trial&author=GE%20Vel%C3%A1squez&author=MB%20Brooks&author=JM%20Coit&volume=198&issue=5&publication_year=2018&pages=657-666&pmid=29954183&doi=10.1164/rccm.201712-2524OC&)
|
| 325 |
+
|
| 326 |
+
43. Cao Y, Wang T, He K, et al. High-dose rifampicin for the treatment of tuberculous meningitis: a meta-analysis of randomized controlled trials. J Clin Pharm Ther. 2022;47(4): 445–454. doi: 10.1111/jcpt.13555. [DOI](https://doi.org/10.1111/jcpt.13555) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34897758/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Clin%20Pharm%20Ther&title=High-dose%20rifampicin%20for%20the%20treatment%20of%20tuberculous%20meningitis:%20a%20meta-analysis%20of%20randomized%20controlled%20trials&author=Y%20Cao&author=T%20Wang&author=K%20He&volume=47&issue=4&publication_year=2022&pages=445-454&pmid=34897758&doi=10.1111/jcpt.13555&)
|
| 327 |
+
|
| 328 |
+
44. Onorato L, Gentile V, Russo A, et al. Standard versus high dose of rifampicin in the treatment of pulmonary tuberculosis: a systematic review and meta-analysis. Clin Microbiol Infect. 2021;27(6):830–837. doi: 10.1016/j.cmi.2021.03.031. [DOI](https://doi.org/10.1016/j.cmi.2021.03.031) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33813119/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Microbiol%20Infect&title=Standard%20versus%20high%20dose%20of%20rifampicin%20in%20the%20treatment%20of%20pulmonary%20tuberculosis:%20a%20systematic%20review%20and%20meta-analysis&author=L%20Onorato&author=V%20Gentile&author=A%20Russo&volume=27&issue=6&publication_year=2021&pages=830-837&pmid=33813119&doi=10.1016/j.cmi.2021.03.031&)
|
| 329 |
+
|
| 330 |
+
45. Polasa K, Murthy KJ, Krishnaswamy K. Rifampicin kinetics in undernutrition. Br J Clin Pharmacol. 1984;17(4):481–484. doi: 10.1111/j.1365-2125.1984.tb02377.x. [DOI](https://doi.org/10.1111/j.1365-2125.1984.tb02377.x) | [PMC free article](/articles/PMC1463407/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/6721995/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Rifampicin%20kinetics%20in%20undernutrition&author=K%20Polasa&author=KJ%20Murthy&author=K%20Krishnaswamy&volume=17&issue=4&publication_year=1984&pages=481-484&pmid=6721995&doi=10.1111/j.1365-2125.1984.tb02377.x&)
|
| 331 |
+
|
| 332 |
+
46. Ramachandran G, Kumar AK, Kannan T, et al. Low serum concentrations of rifampicin and pyrazinamide associated with poor treatment outcomes in children with tuberculosis related to HIV status. Pediatr Infect Dis J 2016;35(5): 530–534. doi: 10.1097/INF.0000000000001069. [DOI](https://doi.org/10.1097/INF.0000000000001069) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26825153/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pediatr%20Infect%20Dis%20J&title=Low%20serum%20concentrations%20of%20rifampicin%20and%20pyrazinamide%20associated%20with%20poor%20treatment%20outcomes%20in%20children%20with%20tuberculosis%20related%20to%20HIV%20status&author=G%20Ramachandran&author=AK%20Kumar&author=T%20Kannan&volume=35&issue=5&publication_year=2016&pages=530-534&pmid=26825153&doi=10.1097/INF.0000000000001069&)
|
| 333 |
+
|
| 334 |
+
47. Tostmann A, Mtabho CM, Semvua HH, et al. Pharmacokinetics of first-line tuberculosis drugs in Tanzanian patients. Antimicrob Agents Chemother. 2013; 57(7):3208–3213. doi: 10.1128/AAC.02599-12. [DOI](https://doi.org/10.1128/AAC.02599-12) | [PMC free article](/articles/PMC3697325/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23629715/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Pharmacokinetics%20of%20first-line%20tuberculosis%20drugs%20in%20Tanzanian%20patients&author=A%20Tostmann&author=CM%20Mtabho&author=HH%20Semvua&volume=57&issue=7&publication_year=2013&pages=3208-3213&pmid=23629715&doi=10.1128/AAC.02599-12&)
|
| 335 |
+
|
| 336 |
+
48. Aklillu E, Zumla A, Habtewold A, et al. Early or deferred initiation of efavirenz during rifampicin-based TB therapy has no significant effect on CYP3A induction in TB-HIV infected patients. Br J Pharmacol. 2021;178(16):3294–3308. doi: 10.1111/bph.15309. [DOI](https://doi.org/10.1111/bph.15309) | [PMC free article](/articles/PMC8359173/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33155675/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Pharmacol&title=Early%20or%20deferred%20initiation%20of%20efavirenz%20during%20rifampicin-based%20TB%20therapy%20has%20no%20significant%20effect%20on%20CYP3A%20induction%20in%20TB-HIV%20infected%20patients&author=E%20Aklillu&author=A%20Zumla&author=A%20Habtewold&volume=178&issue=16&publication_year=2021&pages=3294-3308&pmid=33155675&doi=10.1111/bph.15309&)
|
| 337 |
+
|
| 338 |
+
49. Ngaimisi E, Habtewold A, Minzi O, et al. Importance of ethnicity, CYP2B6 and ABCB1 genotype for efavirenz pharmacokinetics and treatment outcomes: a parallel-group prospective cohort study in two sub-Saharan Africa populations. PLoS One. 2013;8(7):e67946. doi: 10.1371/journal.pone.0067946. [DOI](https://doi.org/10.1371/journal.pone.0067946) | [PMC free article](/articles/PMC3702506/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23861838/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=PLoS%20One&title=Importance%20of%20ethnicity,%20CYP2B6%20and%20ABCB1%20genotype%20for%20efavirenz%20pharmacokinetics%20and%20treatment%20outcomes:%20a%20parallel-group%20prospective%20cohort%20study%20in%20two%20sub-Saharan%20Africa%20populations&author=E%20Ngaimisi&author=A%20Habtewold&author=O%20Minzi&volume=8&issue=7&publication_year=2013&pages=e67946&pmid=23861838&doi=10.1371/journal.pone.0067946&)
|
| 339 |
+
|
| 340 |
+
50. Aklillu E, Djordjevic N, Carrillo JA, et al. High CYP2A6 enzyme activity as measured by a caffeine test and unique distribution of CYP2A6 variant alleles in Ethiopian population. OMICS. 2014;18(7):446–453. doi: 10.1089/omi.2013.0140. [DOI](https://doi.org/10.1089/omi.2013.0140) | [PMC free article](/articles/PMC4086241/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24380444/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=OMICS&title=High%20CYP2A6%20enzyme%20activity%20as%20measured%20by%20a%20caffeine%20test%20and%20unique%20distribution%20of%20CYP2A6%20variant%20alleles%20in%20Ethiopian%20population&author=E%20Aklillu&author=N%20Djordjevic&author=JA%20Carrillo&volume=18&issue=7&publication_year=2014&pages=446-453&pmid=24380444&doi=10.1089/omi.2013.0140&)
|
| 341 |
+
|
| 342 |
+
51. Sileshi T, Telele NF, Burkley V, et al. Correlation of N-acetyltransferase 2 genotype and acetylation status with plasma isoniazid concentration and its metabolic ratio in Ethiopian tuberculosis patients. Sci Rep. 2023;13(1):11438. doi: 10.1038/s41598-023-38716-3. [DOI](https://doi.org/10.1038/s41598-023-38716-3) | [PMC free article](/articles/PMC10349800/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37454203/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Sci%20Rep&title=Correlation%20of%20N-acetyltransferase%202%20genotype%20and%20acetylation%20status%20with%20plasma%20isoniazid%20concentration%20and%20its%20metabolic%20ratio%20in%20Ethiopian%20tuberculosis%20patients&author=T%20Sileshi&author=NF%20Telele&author=V%20Burkley&volume=13&issue=1&publication_year=2023&pages=11438&pmid=37454203&doi=10.1038/s41598-023-38716-3&)
|
| 343 |
+
|
| 344 |
+
52. Gengiah TN, Botha JH, Soowamber D, et al. Low rifampicin concentrations in tuberculosis patients with HIV infection. J Infect Dev Ctries. 2014;8(8):987–993. doi: 10.3855/jidc.4696. [DOI](https://doi.org/10.3855/jidc.4696) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25116663/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Infect%20Dev%20Ctries&title=Low%20rifampicin%20concentrations%20in%20tuberculosis%20patients%20with%20HIV%20infection&author=TN%20Gengiah&author=JH%20Botha&author=D%20Soowamber&volume=8&issue=8&publication_year=2014&pages=987-993&pmid=25116663&doi=10.3855/jidc.4696&)
|
| 345 |
+
|
| 346 |
+
53. Naidoo A, Chirehwa M, Ramsuran V, et al. Effects of genetic variability on rifampicin and isoniazid pharmacokinetics in South African patients with recurrent tuberculosis. Pharmacogenomics. 2019;20(4):225–240. doi: 10.2217/pgs-2018-0166. [DOI](https://doi.org/10.2217/pgs-2018-0166) | [PMC free article](/articles/PMC6562923/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30767706/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=Effects%20of%20genetic%20variability%20on%20rifampicin%20and%20isoniazid%20pharmacokinetics%20in%20South%20African%20patients%20with%20recurrent%20tuberculosis&author=A%20Naidoo&author=M%20Chirehwa&author=V%20Ramsuran&volume=20&issue=4&publication_year=2019&pages=225-240&pmid=30767706&doi=10.2217/pgs-2018-0166&)
|
| 347 |
+
|
| 348 |
+
54. Medellin-Garibay SE, Huerta-Garcia AP, Rodriguez-Baez AS, et al. A population approach of rifampicin pharmacogenetics and pharmacokinetics in Mexican patients with tuberculosis. Tuberculosis. 2020;124:101982. doi: 10.1016/j.tube.2020.101982. [DOI](https://doi.org/10.1016/j.tube.2020.101982) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32810723/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Tuberculosis&title=A%20population%20approach%20of%20rifampicin%20pharmacogenetics%20and%20pharmacokinetics%20in%20Mexican%20patients%20with%20tuberculosis&author=SE%20Medellin-Garibay&author=AP%20Huerta-Garcia&author=AS%20Rodriguez-Baez&volume=124&publication_year=2020&pages=101982&pmid=32810723&doi=10.1016/j.tube.2020.101982&)
|
| 349 |
+
|
| 350 |
+
55. Sissung TM, Baum CE, Kirkland CT, et al. Pharmacogenetics of membrane transporters: an update on current approaches. Mol Biotechnol. 2010;44(2):152–167. doi: 10.1007/s12033-009-9220-6. [DOI](https://doi.org/10.1007/s12033-009-9220-6) | [PMC free article](/articles/PMC6362991/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19950006/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mol%20Biotechnol&title=Pharmacogenetics%20of%20membrane%20transporters:%20an%20update%20on%20current%20approaches&author=TM%20Sissung&author=CE%20Baum&author=CT%20Kirkland&volume=44&issue=2&publication_year=2010&pages=152-167&pmid=19950006&doi=10.1007/s12033-009-9220-6&)
|
| 351 |
+
|
| 352 |
+
56. Huerta-García AP, Medell ın-Garibay SE, Salazar-González RA, et al. Anthropometric and genetic factors associated with the exposure of rifampicin and isoniazid in Mexican patients with tuberculosis. Ther Drug Monit. 2019;41(5): 648–656. doi: 10.1097/FTD.0000000000000631. [DOI](https://doi.org/10.1097/FTD.0000000000000631) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30939588/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ther%20Drug%20Monit&title=Anthropometric%20and%20genetic%20factors%20associated%20with%20the%20exposure%20of%20rifampicin%20and%20isoniazid%20in%20Mexican%20patients%20with%20tuberculosis&author=AP%20Huerta-Garc%C3%ADa&author=SE%20Medell%20%C4%B1n-Garibay&author=RA%20Salazar-Gonz%C3%A1lez&volume=41&issue=5&publication_year=2019&pages=648-656&pmid=30939588&doi=10.1097/FTD.0000000000000631&)
|
| 353 |
+
|
| 354 |
+
57. Levano KS, Jaramillo-Valverde L, Tarazona DD, et al. Allelic and genotypic frequencies of NAT2, CYP2E1, and AADAC genes in a cohort of Peruvian tuberculosis patients. Mol Genet Genomic Med. 2021;9(10):e1764. [DOI](https://doi.org/10.1002/mgg3.1764) | [PMC free article](/articles/PMC8580101/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34510815/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mol%20Genet%20Genomic%20Med&title=Allelic%20and%20genotypic%20frequencies%20of%20NAT2,%20CYP2E1,%20and%20AADAC%20genes%20in%20a%20cohort%20of%20Peruvian%20tuberculosis%20patients&author=KS%20Levano&author=L%20Jaramillo-Valverde&author=DD%20Tarazona&volume=9&issue=10&publication_year=2021&pages=e1764&pmid=34510815&doi=10.1002/mgg3.1764&)
|
| 355 |
+
|
| 356 |
+
58. McIlleron H, Rustomjee R, Vahedi M, et al. Reduced anti-tuberculosis drug concentrations in HIV-infected patients who are men or have low weight: implications for international dosing guidelines. Antimicrob Agents Chemother. 2012;56(6):3232–3238. doi: 10.1128/AAC.05526-11. [DOI](https://doi.org/10.1128/AAC.05526-11) | [PMC free article](/articles/PMC3370773/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22411614/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob%20Agents%20Chemother&title=Reduced%20anti-tuberculosis%20drug%20concentrations%20in%20HIV-infected%20patients%20who%20are%20men%20or%20have%20low%20weight:%20implications%20for%20international%20dosing%20guidelines&author=H%20McIlleron&author=R%20Rustomjee&author=M%20Vahedi&volume=56&issue=6&publication_year=2012&pages=3232-3238&pmid=22411614&doi=10.1128/AAC.05526-11&)
|
| 357 |
+
|
| 358 |
+
59. Cojutti P, Giangreco M, Isola M, et al. Limited sampling strategies for determining the area under the plasma concentration-time curve for isoniazid might be a valuable approach for optimizing treatment in adult patients with tuberculosis. Int J Antimicrob Agents. 2017;50(1):23–28. doi: 10.1016/j.ijantimicag.2017.01.036. [DOI](https://doi.org/10.1016/j.ijantimicag.2017.01.036) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28495479/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int%20J%20Antimicrob%20Agents&title=Limited%20sampling%20strategies%20for%20determining%20the%20area%20under%20the%20plasma%20concentration-time%20curve%20for%20isoniazid%20might%20be%20a%20valuable%20approach%20for%20optimizing%20treatment%20in%20adult%20patients%20with%20tuberculosis&author=P%20Cojutti&author=M%20Giangreco&author=M%20Isola&volume=50&issue=1&publication_year=2017&pages=23-28&pmid=28495479&doi=10.1016/j.ijantimicag.2017.01.036&)
|
| 359 |
+
|
| 360 |
+
60. Maganda BA, Minzi OM, Ngaimisi E, et al. CYP2B6*6 genotype and high efavirenz plasma concentration but not nevirapine are associated with low lumefantrine plasma exposure and poor treatment response in HIV-malaria-coinfected patients. Pharmacogenomics J 2016;16(1):88–95. doi: 10.1038/tpj.2015.37. [DOI](https://doi.org/10.1038/tpj.2015.37) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25963334/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J&title=CYP2B6*6%20genotype%20and%20high%20efavirenz%20plasma%20concentration%20but%20not%20nevirapine%20are%20associated%20with%20low%20lumefantrine%20plasma%20exposure%20and%20poor%20treatment%20response%20in%20HIV-malaria-coinfected%20patients&author=BA%20Maganda&author=OM%20Minzi&author=E%20Ngaimisi&volume=16&issue=1&publication_year=2016&pages=88-95&pmid=25963334&doi=10.1038/tpj.2015.37&)
|
test/texts/PMC11141156.md
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
test/texts/PMC11148365.md
ADDED
|
@@ -0,0 +1,315 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Meta-analysis of the effects of CYP3A5*3 gene polymorphisms on tacrolimus blood concentration and effectiveness in Chinese patients with membranous nephropathy
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Xiaona Dai, Fang Yuan, Lan Chai
|
| 5 |
+
**Journal:** Frontiers in Pharmacology
|
| 6 |
+
**Date:** 2024 May 21
|
| 7 |
+
**DOI:** [10.3389/fphar.2024.1385322](https://doi.org/10.3389/fphar.2024.1385322)
|
| 8 |
+
**PMID:** 38835664
|
| 9 |
+
**PMCID:** PMC11148365
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11148365/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11148365/pdf/fphar-15-1385322.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11148365/pdf/fphar-15-1385322.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Objective:**
|
| 16 |
+
The study aimed to systematically evaluate the relationship between CYP3A5*3 gene polymorphisms and the blood concentration and effectiveness of tacrolimus (TAC) in patients with membranous nephropathy (MN).
|
| 17 |
+
|
| 18 |
+
**Methods:**
|
| 19 |
+
PubMed, Cochrane Library, Embase, Web of Science, China Biomedical, China National Knowledge Infrastructure, Wanfang, Vipshop, ReadShow, Clinical Trials Registry, and other databases were searched. Studies on the relationship between CYP3A5*3 gene polymorphism and TAC blood concentration in MN patients were collected, and meta-analysis was performed using Stata 16 software.
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
A total of eight publications were included in the study, including 498 MN patients. CYP3A5*3 gene polymorphisms are associated with tacrolimus blood levels in patients with MN. The results of the relationship between CYP3A5*3 genotype polymorphisms and tacrolimus blood trough concentrations of the AA + AG genotype were lower than those of the GG genotype at ≤1 month [WMD = −2.08, 95% CI (−2.57, −1.59), p < 0.001] and 1–6 months [WMD = −0.63, 95% CI (−0.98, −0.27), p < 0.001]; however, they were not statistically significant at ≥6 months (p = 0.211). Furthermore, the subgroup analysis revealed that the dose-adjusted concentration of tacrolimus (C0/D) of the AA + AG genotype was lower than that of the GG genotype at ≤1 month [SMD = −1.93, 95% CI (−2.79, −1.08), p < 0.001], 1–6 months [SMD = −2.25, 95% CI (−2.71, −1.79), p < 0.001], and ≥6 months [SMD = −2.36, 95% CI (−2.86, −1.86), p < 0.001]. In addition, there was no statistically significant difference in effectiveness between the two groups at 3, 6, and 12 months of TAC administration (p > 0.05).
|
| 23 |
+
|
| 24 |
+
**Conclusion:**
|
| 25 |
+
Serum TAC concentrations in MN patients were correlated with CYP3A5*3 genotype polymorphisms. Detection of the CYP3A5*3 genotype before the administration of TAC may provide some clinical value for optimizing the treatment of MN patients.
|
| 26 |
+
|
| 27 |
+
**Systematic Review Registration::**
|
| 28 |
+
https://inplasy.com/, identifier [INPLASY202430083].
|
| 29 |
+
|
| 30 |
+
Keywords: gene polymorphism, CYP3A5, tacrolimus, meta-analysis, membranous nephropathy
|
| 31 |
+
|
| 32 |
+
### Objective
|
| 33 |
+
|
| 34 |
+
The study aimed to systematically evaluate the relationship between CYP3A5*3 gene polymorphisms and the blood concentration and effectiveness of tacrolimus (TAC) in patients with membranous nephropathy (MN).
|
| 35 |
+
|
| 36 |
+
### Methods
|
| 37 |
+
|
| 38 |
+
PubMed, Cochrane Library, Embase, Web of Science, China Biomedical, China National Knowledge Infrastructure, Wanfang, Vipshop, ReadShow, Clinical Trials Registry, and other databases were searched. Studies on the relationship between CYP3A5*3 gene polymorphism and TAC blood concentration in MN patients were collected, and meta-analysis was performed using Stata 16 software.
|
| 39 |
+
|
| 40 |
+
### Results
|
| 41 |
+
|
| 42 |
+
A total of eight publications were included in the study, including 498 MN patients. CYP3A5*3 gene polymorphisms are associated with tacrolimus blood levels in patients with MN. The results of the relationship between CYP3A5*3 genotype polymorphisms and tacrolimus blood trough concentrations of the AA + AG genotype were lower than those of the GG genotype at ≤1 month [WMD = −2.08, 95% CI (−2.57, −1.59), *p*p < 0.001] and 1–6 months [WMD = −0.63, 95% CI (−0.98, −0.27), *p*p < 0.001]; however, they were not statistically significant at ≥6 months (*p*p = 0.211). Furthermore, the subgroup analysis revealed that the dose-adjusted concentration of tacrolimus (C0/D) of the AA + AG genotype was lower than that of the GG genotype at ≤1 month [SMD = −1.93, 95% CI (−2.79, −1.08), *p*p < 0.001], 1–6 months [SMD = −2.25, 95% CI (−2.71, −1.79), *p*p < 0.001], and ≥6 months [SMD = −2.36, 95% CI (−2.86, −1.86), *p*p < 0.001]. In addition, there was no statistically significant difference in effectiveness between the two groups at 3, 6, and 12 months of TAC administration (*p*p > 0.05).
|
| 43 |
+
|
| 44 |
+
### Conclusion
|
| 45 |
+
|
| 46 |
+
Serum TAC concentrations in MN patients were correlated with CYP3A5*3 genotype polymorphisms. Detection of the CYP3A5*3 genotype before the administration of TAC may provide some clinical value for optimizing the treatment of MN patients.
|
| 47 |
+
|
| 48 |
+
### Systematic Review Registration:
|
| 49 |
+
|
| 50 |
+
[https://inplasy.com/](https://inplasy.com/)https://inplasy.com/, identifier [INPLASY202430083].
|
| 51 |
+
|
| 52 |
+
**Keywords:**Keywords: gene polymorphism, CYP3A5, tacrolimus, meta-analysis, membranous nephropathy
|
| 53 |
+
|
| 54 |
+
## 1 Introduction
|
| 55 |
+
|
| 56 |
+
Membranous nephropathy (MN) is one of the most common pathological types of nephrotic syndrome in adults and is characterized by the deposition of immune complexes under the epithelial cells of the glomerular basement membrane with diffuse thickening of the basement membrane ([Couser, 2017](#B5)Couser, 2017). In recent years, the incidence of membranous nephropathy has increased significantly in China, accounting for 23.4% of primary glomerular disease ([Wu et al., 2021](#B32)Wu et al., 2021). In idiopathic membranous nephropathy, approximately 20% of patients go into spontaneous complete or partial remission, while 30%–40% of idiopathic membranous nephropathy (IMN) patients progress to end-stage renal disease (ESRD) within 10–15 years and require renal replacement therapy.
|
| 57 |
+
|
| 58 |
+
Tacrolimus (TAC) is a potent macrolide immunosuppressive agent and is widely used for treating post-transplant rejection and autoimmune diseases. The 2021 KDIGO guideline recommends TAC for patients with intermediate and high-risk membranous nephropathy ([Beck et al., 2023](#B2)Beck et al., 2023). However, TAC is characterized by a narrow therapeutic window and variable oral bioavailability. Thus, therapeutic drug monitoring is required to enhance its effectiveness and avoid toxicity. The blood concentration of TAC is affected by many factors. In addition, it has been shown that individual differences in TAC pharmacokinetics are largely influenced by genetic factors ([Yang et al., 2020](#B34)Yang et al., 2020).
|
| 59 |
+
|
| 60 |
+
The cytochrome P450 (CYP) enzyme family is widely recognized as the primary catalyst responsible for the phase 1 metabolism of pharmaceuticals and other exogenous chemical agents ([Hakkola et al., 2020](#B9)Hakkola et al., 2020). CYP3A is a major subfamily affiliated with the CYP superfamily, which plays a crucial role in the metabolic processes of approximately 30%–60% of pharmaceutical medications presently available on the market ([Klyushova et al., 2022](#B17)Klyushova et al., 2022). Studies have demonstrated that the high intra-individual and inter-individual variability of tacrolimus is mostly attributed to the gene polymorphisms of CYP3A isozymes, predominantly CYP3A4 and CYP3A5 ([Barbarino et al., 2013](#B1)Barbarino et al., 2013). However, the CYP3A4 genotype has a very low frequency of mutation within the Chinese population, whereas the CYP3A5 genotype demonstrates a higher frequency of mutation ([Shao et al., 2020](#B25)Shao et al., 2020). Moreover, it has been observed that CYP3A5 exhibits a greater intrinsic clearance of tacrolimus compared to CYP3A4 ([Dai et al., 2006](#B6)Dai et al., 2006), which is widely recognized as the primary enzyme involved in the metabolic processes of tacrolimus.
|
| 61 |
+
|
| 62 |
+
CYP3A5*3 (6986A/G, rs776746) is a functional single-nucleotide polymorphism of CYP3A5 located in intron 3. Previous studies have shown a correlation between CYP3A5*3 gene polymorphisms and TAC blood concentrations in kidney transplant recipients. Specifically, CYP3A5 non-expressers (CYP3A5*3/*3) exhibit reduced metabolic functions and elevated TAC trough concentrations in comparison to CYP3A5 expressers (CYP3A5*1/*1 or CYP3A5*1/*3) ([van Gelder et al., 2020](#B27)van Gelder et al., 2020; [Hannachi et al., 2021](#B11)Hannachi et al., 2021; [He et al., 2022](#B12)He et al., 2022). Until now, research on the impact of CYP3A5 polymorphisms on TAC has primarily been limited to renal transplantation. However, it is important to note that the pharmacokinetic characteristics of TAC in individuals with organ transplants may not be generalizable to non-transplant patients ([Wang et al., 2023](#B29)Wang et al., 2023). In the context of liver transplant recipients, the post-operative day plays a significant role in affecting the clearance of tacrolimus, and this factor has the potential to enhance metabolic activity throughout the regeneration process of the transplanted liver, consequently leading to an increase in TAC clearance ([Kirubakaran et al., 2020](#B16)Kirubakaran et al., 2020). The occurrence of hypoproteinemia in primary nephrotic syndrome patients might lead to a decrease in the protein binding of TAC, consequently altering the metabolism and absorption of TAC ([Huang et al., 2019](#B14)Huang et al., 2019).
|
| 63 |
+
|
| 64 |
+
In recent times, many studies have focused their emphasis on investigating the correlation between CYP3A5*3 gene polymorphism and the plasma concentration and effectiveness of TAC in Chinese MN patients. Nevertheless, the findings of the studies are conflicting as a result of the limited sample size. Hence, we performed a comprehensive examination and statistical analysis of the existing research to ascertain the impact of CYP3A5*3 gene polymorphism on the blood concentration and effectiveness of TAC in Chinese patients with MN so as to provide clinicians with a reference on appropriate medication administration.
|
| 65 |
+
|
| 66 |
+
## 2 Materials and methods
|
| 67 |
+
|
| 68 |
+
### 2.1 Literature search strategy
|
| 69 |
+
|
| 70 |
+
The review protocol used for this study was pre-registered in INPLASY with the registration number INPLASY202430083. A search was performed to identify relevant studies in PubMed, Cochrane Library, Web of Science (WOS), China Biomedical (CBM), Embase, China Knowledge Network Infrastructure (CNKI), Wanfang, Vipshop, ReadShow, and [clinicaltrail.gov](http://clinicaltrail.gov)clinicaltrail.gov databases (updated on 28 October 2023). The search terms were tacrolimus, TAC, FK506, membranous nephropathy, membranous glomerulonephritis, CYP3A5, and cytochrome P4503A5. In order to expand the search scope of related articles, the references cited in the retrieved articles were further explored.
|
| 71 |
+
|
| 72 |
+
### 2.2 Inclusion and exclusion criteria
|
| 73 |
+
|
| 74 |
+
We included studies that met the following inclusion criteria: 1) all published domestic and international correlative studies on the effects of CYP3A5*3 gene polymorphisms and effectiveness on TAC blood concentrations in MN patients in any language. 2) The ethnicity in these studies must be Chinese population. 3) MN patients treated with TAC-based immunosuppressants, not using other drugs affecting TAC blood concentrations, and all tested for CYP3A5*3 gene polymorphisms, with no restrictions on patient age, gender, or test method. 4) Patients were classified into three different genotypes: AA (*1/*1), AG (*1/*3), and GG (*3/*3), or separately into GG (*3/*3) and AA + AG (*1/*3+*1/*1) genotypes. 5) Studies should provide either the TAC whole-blood trough concentration (C0) or dose-adjusted trough concentration (C0/D). Studies were excluded if they 1) were duplicate publications; 2) were not original studies (e.g., reviews, synthesis, case report-type articles, etc.); 3) have incomplete study outcomes; or 4) were irrelevant.
|
| 75 |
+
|
| 76 |
+
### 2.3 Literature screening and data extraction
|
| 77 |
+
|
| 78 |
+
The literature screening was done independently by two independent researchers and cross-checked, with any conflicting issues resolved through discussion and negotiation with a third author. The following information was extracted: first author, year of publication, sample size, gender, age, duration of dosing, CYP3A5*3 genotypes, tacrolimus blood concentration (C0), and dose-adjusted concentration of TAC (C0/D). In addition, the numbers of patients with complete remission (CR) and partial remission (PR) were extracted to measure the effectiveness of tacrolimus in MN patients at 3, 6, and 12 months after treatment.
|
| 79 |
+
|
| 80 |
+
### 2.4 Evaluation of the quality of the literature
|
| 81 |
+
|
| 82 |
+
STREGA ([Zeng et al., 2012](#B37)Zeng et al., 2012) was used to assess the quality of each included study in terms of 1) the adequacy of the sample size, 2) the clarity of the diagnostic criteria, 3) problems with the matching of subgroups, 4) whether study groups were comparable, 5) whether the genetic testing methods used were reasonable, and 6) the adequacy of the data. For each of the above-mentioned six items, one item should be met to obtain 1 point, with a total score of ≥3 being considered reliable in terms of quality.
|
| 83 |
+
|
| 84 |
+
### 2.5 Statistical analysis
|
| 85 |
+
|
| 86 |
+
Stata 16 software was used for forest plots. The relationship between genotypes and TAC blood concentrations was evaluated by standard mean differences (SMDs) and their corresponding 95% confidence intervals (95% CIs). The Mantel–Haenszel (M-H) method was used to analyze dichotomous data, and the strength of their association with effectiveness was determined by the odds ratio (OR). Heterogeneity assumptions were assessed using Q-tests based on chi-squared tests. If there was no statistical heterogeneity between studies (*p*p ≥ 0. 1, *I*I ^ 2 ^ *2*2 ≤ 50%), the fixed-effects model was used. On the other hand, if there was statistical heterogeneity between studies (*p*p < 0. 1, *I*I ^ 2 ^ *2*2 ≥ 50%), the data were analyzed by the random-effects model, and the causes of heterogeneity were analyzed, with subgroup analyses performed if necessary. The mean ± standard deviation was estimated using the formula of [McGrath et al. (2020)](#B21)McGrath et al. (2020) if the study reported only median and interquartile spacing. Publication bias was assessed by Begg’s test and Egger’s test. A *p*p-value ≤ 0.05 indicated significant publication bias.
|
| 87 |
+
|
| 88 |
+
## 3 Results
|
| 89 |
+
|
| 90 |
+
### 3.1 Study inclusion
|
| 91 |
+
|
| 92 |
+
A total of 71 studies were searched. After title and abstract screening, 14 studies were searched for their full texts. Then, two studies with no available data on reported outcomes and four studies without concerning outcomes were excluded. As a result, eight studies involving 498 Chinese membranous nephropathy patients were finally included through screening ([Ye et al., 2014](#B35)Ye et al., 2014; [Yang et al., 2015](#B33)Yang et al., 2015; [Wei et al., 2018](#B4)Wei et al., 2018; [Lin et al., 2019a](#B19)Lin et al., 2019a; [Wu, 2019](#B31)Wu, 2019; [Wang, 2020](#B30)Wang, 2020; [Zhang et al., 2020](#B38)Zhang et al., 2020; [Xu et al., 2021](#B13)Xu et al., 2021). The course of study selection is given in [Figure 1](#F1)Figure 1.
|
| 93 |
+
|
| 94 |
+
### FIGURE 1.
|
| 95 |
+
|
| 96 |
+
|
| 97 |
+
|
| 98 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11148365_fphar-15-1385322-g001.jpg)
|
| 99 |
+
|
| 100 |
+
Flow diagram of the studies included in this meta-analysis.
|
| 101 |
+
|
| 102 |
+
### 3.2 Basic characteristics and quality assessment
|
| 103 |
+
|
| 104 |
+
The basic characteristics of the included studies are shown in [Table 1](#T1)Table 1. We selected patients with three distinct CYP3A5 genotypes: GG (CYP3A5*3*3), AG (CYP3A5*1*3), and AA (CYP3A5*1*1). Of these, patients with the G genotype (CYP3A5*3) made up approximately 67.07% (668/996) of the Chinese population as a whole. The quality evaluation of the included literature is shown in [Table 2](#T2)Table 2.
|
| 105 |
+
|
| 106 |
+
### TABLE 1.
|
| 107 |
+
|
| 108 |
+
Basic characteristics of the included studies (“−”: no description).
|
| 109 |
+
|
| 110 |
+
| Study | Case number | Male/female | Age (y) | TAC testing time (weeks) | Cases in each group | |
|
| 111 |
+
| ----- | ----------- | ----------- | ------- | ------------------------ | ------------------- | - |
|
| 112 |
+
| AA (CYP3A5*1/*1) | AG (CYP3A5*1/*3) | GG (CYP3A5*3/*3) | G*3 (CYP3A5*3) | | | |
|
| 113 |
+
| Yang et al. (2015) | 60 | 38/22 | 34.97 ± 14.37 | 8, 12, 16, and 24 | 12 | 32 | 16 | 64 |
|
| 114 |
+
| Wu (2019) | 53 | 27/26 | 43.6 (19–69) | 24 | 2 | 22 | 29 | 80 |
|
| 115 |
+
| Xu et al., 2021 | 94 | 62/32 | 54.00 ± 13.38 | 1 | 17 | 32 | 45 | 122 |
|
| 116 |
+
| Ye et al. (2014) | 60 | − | − | 1, 2, 3, 4, 8, and 12 | 7 | 29 | 24 | 77 |
|
| 117 |
+
| Zhang et al. (2020) | 76 | 55/21 | 49.98 ± 15.64 | 4 | 0 | 35 | 41 | 117 |
|
| 118 |
+
| Lin et al. (2019a) | 66 | 36/30 | 37.2 ± 12.7 | 1 | 8 | 25 | 33 | 91 |
|
| 119 |
+
| Wei et al., 2018 | 44 | 27/17 | 35.9 ± 12.9 | 1, 2, 4, 12, 24, and 48 | 6 | 18 | 20 | 58 |
|
| 120 |
+
| Wang (2020) | 45 | 27/18 | 51.38 ± 14.47 | 1, 2, and 4 | 6 | 19 | 20 | 59 |
|
| 121 |
+
### TABLE 2.
|
| 122 |
+
|
| 123 |
+
Quality assessment of the included studies (Zeng et al., 2012).
|
| 124 |
+
|
| 125 |
+
| Study | Adequate sample size | Clear diagnostic criteria | Group match | Comparable between study groups | Genetic testing method | Sufficient data | Quality score (≥3) |
|
| 126 |
+
| ----- | -------------------- | ------------------------- | ----------- | ------------------------------- | ---------------------- | --------------- | ------------------ |
|
| 127 |
+
| Yang et al. (2015) | + | + | + | + | + | + | 6 |
|
| 128 |
+
| Wu (2019) | − | + | + | + | + | + | 5 |
|
| 129 |
+
| Xu et al., 2021 | + | + | + | + | + | + | 6 |
|
| 130 |
+
| Wang (2020) | − | + | + | + | + | − | 4 |
|
| 131 |
+
| Ye et al. (2014) | + | + | + | + | + | + | 6 |
|
| 132 |
+
| Zhang et al. (2020) | + | + | + | + | + | − | 5 |
|
| 133 |
+
| Lin et al. (2019a) | + | + | + | + | + | − | 5 |
|
| 134 |
+
| Wei et al., 2018 | − | + | + | + | + | − | 4 |
|
| 135 |
+
### 3.3 Relationship between CYP3A5*3 genotype polymorphisms and TAC blood trough concentrations
|
| 136 |
+
|
| 137 |
+
A total of seven studies ([Yang et al., 2015](#B33)Yang et al., 2015; [Wei et al., 2018](#B4)Wei et al., 2018; [Lin et al., 2019a](#B19)Lin et al., 2019a; [Wu, 2019](#B31)Wu, 2019; [Wang, 2020](#B30)Wang, 2020; [Zhang et al., 2020](#B38)Zhang et al., 2020; [Xu et al., 2021](#B13)Xu et al., 2021) reported the relationship between the AA + AG genotype (referred to as expressers) and the GG genotype (referred to as non-expressers) TAC blood trough concentrations in CYP3A5*3 genotypes. As the different durations of drug administration may have an impact on the pharmacokinetics and pharmacodynamics of TAC, the studies were divided into subgroups for analysis, according to the duration of TAC administration: ≤1 month, 1–6 months, and ≥6 months. The results of the heterogeneity test showed no statistical heterogeneity (*p*p > 0.1, *I*I ^ 2 ^ *2*2 < 50%) within the three subgroups, so the fixed-effects model was used for analysis. The results of the subgroup analysis showed that the duration of TAC administration was ≤1 month [WMD = −2.08, 95% CI (−2.57, −1.59), *p*p < 0.001] and 1–6 months [WMD = −0.63, 95% CI (−0.98, −0.27), *p*p < 0.001]. The TAC blood trough concentrations of the AA + AG genotype were significantly lower than those of the GG genotype. The difference in TAC blood trough concentrations between the AA + AG and GG groups was not statistically significant when taken for ≥6 months (*p*p = 0.211) ([Figure 2](#F2)Figure 2).
|
| 138 |
+
|
| 139 |
+
### FIGURE 2.
|
| 140 |
+
|
| 141 |
+
|
| 142 |
+
|
| 143 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11148365_fphar-15-1385322-g002.jpg)
|
| 144 |
+
|
| 145 |
+
Forest plot of a meta-analysis of the difference in tacrolimus blood trough concentration between AA + AG and GG genotypes. The studies were divided into subgroups for analysis according to the duration of tacrolimus administration as ≤1 month, 1–6 months, and ≥6 months. WMD, weighted mean difference; CI, confidence interval.
|
| 146 |
+
|
| 147 |
+
### 3.4 TAC C0/D levels between the AA + AG and GG genotypes
|
| 148 |
+
|
| 149 |
+
Five studies ([Yang et al., 2015](#B33)Yang et al., 2015; [Wu, 2019](#B31)Wu, 2019; [Wang, 2020](#B30)Wang, 2020; [Zhang et al., 2020](#B38)Zhang et al., 2020; [Xu et al., 2021](#B13)Xu et al., 2021) reported the TAC C0/D between the AA + AG and GG genotypes in MN patients. The heterogeneity results showed statistical heterogeneity between the two subgroups: ≤1 month (*p*p < 0.01, *I*I ^ 2 ^ *2*2 = 92.2%) and 1–6 months (*p*p = 0.064, *I*I ^ 2 ^ *2*2 = 55%). The random-effects model was used for meta-analysis. The meta-analysis results showed that at ≤1 month [SMD = −1.93, 95% CI (−2.79, −1.08), *p*p < 0.001], 1–6 months [SMD = −2.25, 95% CI (−2.71, −1.79), *p*p < 0.001], and ≥6 months [SMD = −2.36, 95% CI (−2.86, −1.86), *p*p < 0.001], the TAC C0/D levels of CYP3A5 expressers in MN patients were lower than those of CYP3A5 non-expressers ([Figure 3](#F3)Figure 3).
|
| 150 |
+
|
| 151 |
+
### FIGURE 3.
|
| 152 |
+
|
| 153 |
+
|
| 154 |
+
|
| 155 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11148365_fphar-15-1385322-g003.jpg)
|
| 156 |
+
|
| 157 |
+
Forest plot of the difference in tacrolimus C0/D between AA + AG and GG genotypes in MN patients. The studies were divided into subgroups for analysis according to the duration of tacrolimus administration as ≤1 month, 1–6 months, and ≥6 months. SMD, standard mean difference; CI, confidence interval.
|
| 158 |
+
|
| 159 |
+
### 3.5 Association of CYP3A5*3 gene polymorphisms with the effectiveness of TAC in the treatment of MN patients
|
| 160 |
+
|
| 161 |
+
Eight studies ([Ye et al., 2014](#B35)Ye et al., 2014; [Yang et al., 2015](#B33)Yang et al., 2015; [Wei et al., 2018](#B4)Wei et al., 2018; [Lin et al., 2019a](#B19)Lin et al., 2019a; [Wu, 2019](#B31)Wu, 2019; [Wang, 2020](#B30)Wang, 2020; [Zhang et al., 2020](#B38)Zhang et al., 2020; [Xu et al., 2021](#B13)Xu et al., 2021) reported the association between the AA + AG and GG genotypes and the effectiveness of TAC in treating MN patients. The heterogeneity results showed that there was no statistical heterogeneity among the studies in the subgroups of 3 months, 6 months, and 12 months after taking TAC, so the fixed-effects model was used for meta-analysis. The results showed that at 3 months [OR = 0.98, 95% CI (0.55, 1.76), *p*p = 0.949], 6 months [OR = 1.14, 95% CI (0.84, 1.56), *p*p = 0.401], and 12 months [OR = 1.20, 95% CI (0.66, 2.21), *p*p = 0.551], the remission rates of expressers were higher than those of non-expressers, but there was no statistically significant difference between the two groups (*p*p > 0.05) ([Figure 4](#F4)Figure 4).
|
| 162 |
+
|
| 163 |
+
### FIGURE 4.
|
| 164 |
+
|
| 165 |
+
|
| 166 |
+
|
| 167 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11148365_fphar-15-1385322-g004.jpg)
|
| 168 |
+
|
| 169 |
+
Forest plot of the relationship between CYP3A5*3 gene polymorphisms and the effectiveness of tacrolimus in treating MN patients. The studies were divided into subgroups for analysis according to the duration of tacrolimus administration as 3 months, 6 months, and 12 months. OR, odds ratio; CI, confidence interval.
|
| 170 |
+
|
| 171 |
+
### 3.6 Assessment of publication bias
|
| 172 |
+
|
| 173 |
+
Publication bias was assessed using the AA + AG genotype vs. GG genotype TAC blood concentrations in the CYP3A5*3 gene of MN patients at ≤1 month as an indicator. The funnel plot of this study was basically symmetrical, while the test of bias was performed, and the results of Begg’s test *p*p = 0.175 and Egger’s test *p*p = 0.124 were both greater than 0.05, so it can be judged that the possibility of publication bias in the study is low ([Figure 5](#F5)Figure 5).
|
| 174 |
+
|
| 175 |
+
### FIGURE 5.
|
| 176 |
+
|
| 177 |
+
|
| 178 |
+
|
| 179 |
+
Funnel plot of the publication bias assessment and Begg’s test.
|
| 180 |
+
|
| 181 |
+
## 4 Discussion
|
| 182 |
+
|
| 183 |
+
The main clinical manifestation of membranous nephropathy is nephrotic syndrome, which is characterized by severe proteinuria and hypoproteinemia. According to different etiologies, MN can be divided into IMN with unknown etiology and secondary membranous nephritis caused by systemic autoimmune diseases, drugs, infections, and malignant tumors ([Keri et al., 2019](#B15)Keri et al., 2019; [Hamilton et al., 2020](#B10)Hamilton et al., 2020). IMN accounts for approximately 80% of MN cases ([Couser, 2017](#B5)Couser, 2017). Due to the heterogeneity of prognosis, different drug responses, and high recurrence rate in patients with IMN, the treatment of IMN remains the focus of research.
|
| 184 |
+
|
| 185 |
+
Currently, the main treatment options include alkylating agents ([Fernandez-Juarez et al., 2021](#B8)Fernandez-Juarez et al., 2021), calcineurin inhibitors ([Lin S. et al., 2019](#B18)Lin S. et al., 2019; [Zou et al., 2019](#B39)Zou et al., 2019; [Ruo-Ji et al., 2022](#B24)Ruo-Ji et al., 2022), and rituximab ([Lu et al., 2020](#B20)Lu et al., 2020; [You et al., 2021](#B36)You et al., 2021). Previous studies have shown that alkylating agents are currently the only treatment proven to prevent patients from developing end-stage renal disease and reduce the risk of death ([von Groote et al., 2021](#B28)von Groote et al., 2021). However, the combination of alkylating agents and corticosteroids increases the risk of severe infections, advanced malignancies, infertility, and other serious adverse events ([van den Brand et al., 2017](#B26)van den Brand et al., 2017). Therefore, clinical applications are subject to many limitations. Because of the high cost of rituximab and other programs, it is necessary to comprehensively consider the economic level and condition of patients in clinical practice. Calmodulin inhibitors are still common in clinical applications as an alternative treatment method.
|
| 186 |
+
|
| 187 |
+
TAC is a calcineurin inhibitor that is mainly absorbed in the jejunum and ileum after oral administration and metabolized in the liver and intestine. It plays an immunosuppressive role mainly by inhibiting T-cell proliferation and reducing the production of cytokines, such as interleukin-2, thereby reducing proteinuria in MN patients ([Fang et al., 2019](#B7)Fang et al., 2019). However, due to the narrow therapeutic window of TAC and the large individual differences, it is important to closely monitor blood concentration during treatment. However, dose adjustment by monitoring blood levels has a time lag. Researchers are now focusing on how to assist clinicians in making a preliminary assessment before the detection of blood concentration.
|
| 188 |
+
|
| 189 |
+
The CYP3A5 isoenzyme is one of the important metabolic enzymes of TAC. One of the functional single-nucleotide polymorphisms in this gene is the mutation from guanine (G) to adenine (A) at site 6,986 in intron 3, which results in the appearance of the stop codon of the transcribed RNA at site 109, translating a non-functional protein fragment, reducing or even inactivating CYP3A5 isoenzyme activity, and TAC cannot be metabolized properly and accumulates in the body, producing adverse drug reactions ([Nair et al., 2015](#B23)Nair et al., 2015). The CYP3A5-A allele (also known as *1) encodes a functional CYP3A5 protein, and the pure heterozygous genotype AA and heterozygous genotype AG are known as gene expression types, while the pure heterozygous genotype GG is known as a non-expression type. Evidence-based studies on the relationship between CYP3A5*3 gene polymorphisms and TAC blood levels and effectiveness have been conducted in transplant patients. This study is the first systematic evaluation of CYP3A5 gene polymorphisms and TAC blood concentrations in MN patients.
|
| 190 |
+
|
| 191 |
+
The findings of the meta-analysis indicate that of the three CYP3A5 genotypes identified in Chinese patients, the CYP3A5*3 allele has a higher frequency. Out of the 498 patients included in eight studies, the percentage of CYP3A5*3 expressors in the Chinese population was found to be 67.07%. This finding aligns with a previous study, which also reported a high mutation rate of 72.7% for the CYP3A5*3 genotype in the Chinese population ([Cao et al., 2022](#B3)Cao et al., 2022). The meta-analysis revealed that tacrolimus blood concentrations of CYP3A5 expressers were significantly lower than those of CYP3A5 non-expressers in Chinese MN patients. The results of the subgroup analysis indicated that the plasma concentration of tacrolimus in type AA + AG was significantly lower than in type GG within 1 month and 1–6 months after administration. However, there was no significant difference between the two groups over 6 months, which may be attributable to factors such as the patient’s dosage, weight, and testing methodology. The adjustment of dosage holds significance in the interpretation of this outcome. However, due to the limitations to the included studies, specific dose-adjustment data were not recorded in detail and could not be obtained for different genotype groups at different time points. According to [Wu (2019)](#B31)Wu (2019), the tacrolimus daily dose was adjusted by target blood concentration, which was found to be higher in the AA and AG (CYP3A5 expressers) groups compared to the GG group (CYP3A5 non-expressers). The study conducted by [Wei et al., 2018](#B4)Wei et al., 2018 revealed that there was no statistically significant difference in the initial daily dosage between the two distinct groups. However, after 3, 6, and 12 months, the non-expressers exhibited a lower daily dosage compared to the expressers, and this difference was shown to be statistically significant. The variation in dosage between groups may potentially influence the disparity in blood concentrations of TAC. Therefore, in order to minimize the impact of dosage on blood concentrations, we additionally conducted a comparison of the dose-adjusted trough concentration. The results showed that in MN patients, TAC blood concentrations of CYP3A5 expressers are comparatively lower than those of CYP3A5 non-expressers at different time points (≤1 month, 1–6 months, and ≥6 months) after medication administration. The findings are consistent with the effect of different CYP3A5 * 3 genotypes on TAC in transplant patients ([Muraki et al., 2018](#B22)Muraki et al., 2018). However, the impact of the CYP3A5*3 genetic polymorphism on the effectiveness of TAC therapy in patients with MN remains uncertain. Therefore, when using TAC to treat MN, detecting the CYP3A5 * 3 gene polymorphism in patients can assist clinicians in determining the optimal initial dosage so as to achieve effective blood drug concentration in a shorter time and, thus, optimize the management of membranous nephropathy. However, it is important to note that this polymorphism cannot serve as a reliable indicator for predicting the clinical response of patients.
|
| 192 |
+
|
| 193 |
+
## 5 Limitations
|
| 194 |
+
|
| 195 |
+
Although statistical differences were eliminated through subgroup analysis among studies, heterogeneity still exists in some subgroups, which may be caused by the different times of taking TAC. In addition, the literature reporting specific dose adjustments was extremely scant, which made it impossible for us to compare the impact of dosage on TAC blood concentrations across genotype groups. Some of the data in the literature are expressed in quartiles, which we converted into the mean ± standard deviation form based on the formula of [McGrath et al. (2020)](#B21)McGrath et al. (2020), which may lead to errors. The subjects included in this study are all Chinese, and the sample size is small, which limits the generalizability of the findings. Therefore, more high-quality, large-sample, multicenter clinical studies are needed in the future to further evaluate the study in a comprehensive manner and draw more reliable conclusion.
|
| 196 |
+
|
| 197 |
+
## 6 Conclusion
|
| 198 |
+
|
| 199 |
+
Our meta-analysis indicated that there is a correlation between TAC blood levels and CYP3A5*3 gene polymorphisms in MN patients. However, there was no significant connection between CYP3A5*3 genetic polymorphisms and the effectiveness of TAC treatment based on the current available evidence. Detection of patients’ CYP3A5*3 genotypes before MN treatment might be useful for the administration of TAC and may contribute to individualized clinical treatment. However, regarding the mentioned limitations, further high-quality studies that are well-designed, multicentered, and have larger sample sizes are needed to better supplement and confirm our findings in the future.
|
| 200 |
+
|
| 201 |
+
## Acknowledgments
|
| 202 |
+
|
| 203 |
+
The authors sincerely thank the study participants for their contribution to the research.
|
| 204 |
+
|
| 205 |
+
## Funding Statement
|
| 206 |
+
|
| 207 |
+
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
|
| 208 |
+
|
| 209 |
+
## Data availability statement
|
| 210 |
+
|
| 211 |
+
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
|
| 212 |
+
|
| 213 |
+
## Author contributions
|
| 214 |
+
|
| 215 |
+
XD: writing–original draft. LC: writing–review and editing. FY: Formal analysis, Methodology, Supervision, Validation, Visualization, Writing – review & editing.
|
| 216 |
+
|
| 217 |
+
## Conflict of interest
|
| 218 |
+
|
| 219 |
+
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
|
| 220 |
+
|
| 221 |
+
## Publisher’s note
|
| 222 |
+
|
| 223 |
+
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
|
| 224 |
+
|
| 225 |
+
## Associated Data
|
| 226 |
+
|
| 227 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 228 |
+
|
| 229 |
+
### Data Availability Statement
|
| 230 |
+
|
| 231 |
+
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
|
| 232 |
+
|
| 233 |
+
### Data Availability Statement
|
| 234 |
+
|
| 235 |
+
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
|
| 236 |
+
|
| 237 |
+
## References
|
| 238 |
+
|
| 239 |
+
1. Barbarino J. M., Staatz C. E., Venkataramanan R., Klein T. E., Altman R. B. (2013). PharmGKB summary: cyclosporine and tacrolimus pathways. PHARMACOGENET GENOM 23 (10), 563–585. 10.1097/FPC.0b013e328364db84 [DOI](https://doi.org/10.1097/FPC.0b013e328364db84) | [PMC free article](/articles/PMC4119065/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23922006/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=PHARMACOGENET%20GENOM&title=PharmGKB%20summary:%20cyclosporine%20and%20tacrolimus%20pathways&author=J.%20M.%20Barbarino&author=C.%20E.%20Staatz&author=R.%20Venkataramanan&author=T.%20E.%20Klein&author=R.%20B.%20Altman&volume=23&issue=10&publication_year=2013&pages=563-585&pmid=23922006&doi=10.1097/FPC.0b013e328364db84&)
|
| 240 |
+
|
| 241 |
+
2. Beck L. H., Ayoub I., Caster D., Choi M. J., Cobb J., Geetha D., et al. (2023). KDOQI US commentary on the 2021 KDIGO clinical practice guideline for the management of glomerular diseases. Am. J. Kidney Dis. 82 (2), 121–175. 10.1053/j.ajkd.2023.02.003 [DOI](https://doi.org/10.1053/j.ajkd.2023.02.003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37341661/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Kidney%20Dis.&title=KDOQI%20US%20commentary%20on%20the%202021%20KDIGO%20clinical%20practice%20guideline%20for%20the%20management%20of%20glomerular%20diseases&author=L.%20H.%20Beck&author=I.%20Ayoub&author=D.%20Caster&author=M.%20J.%20Choi&author=J.%20Cobb&volume=82&issue=2&publication_year=2023&pages=121-175&pmid=37341661&doi=10.1053/j.ajkd.2023.02.003&)
|
| 242 |
+
|
| 243 |
+
3. Cao P., Zhang F., Zhang J., Zheng X., Sun Z., Yu B., et al. (2022). Cyp3a5 genetic polymorphism in Chinese population with renal transplantation: a meta-analysis review. Transpl. Proc. 54 (3), 638–644. 10.1016/j.transproceed.2021.10.031 [DOI](https://doi.org/10.1016/j.transproceed.2021.10.031) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35428510/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transpl.%20Proc.&title=Cyp3a5%20genetic%20polymorphism%20in%20Chinese%20population%20with%20renal%20transplantation:%20a%20meta-analysis%20review&author=P.%20Cao&author=F.%20Zhang&author=J.%20Zhang&author=X.%20Zheng&author=Z.%20Sun&volume=54&issue=3&publication_year=2022&pages=638-644&pmid=35428510&doi=10.1016/j.transproceed.2021.10.031&)
|
| 244 |
+
|
| 245 |
+
4. Couser W. G. (2017). Primary membranous nephropathy. Clin. J. Am. Soc. Nephrol. 12 (6), 983–997. 10.2215/CJN.11761116 [DOI](https://doi.org/10.2215/CJN.11761116) | [PMC free article](/articles/PMC5460716/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28550082/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20J.%20Am.%20Soc.%20Nephrol.&title=Primary%20membranous%20nephropathy&author=W.%20G.%20Couser&volume=12&issue=6&publication_year=2017&pages=983-997&pmid=28550082&doi=10.2215/CJN.11761116&)
|
| 246 |
+
|
| 247 |
+
5. Dai Y., Hebert M. F., Isoherranen N., Davis C. L., Marsh C., Shen D. D., et al. (2006). Effect of cyp3a5 polymorphism on tacrolimus metabolic clearance in vitro . Drug Metab. Dispos. 34 (5), 836–847. 10.1124/dmd.105.008680 [DOI](https://doi.org/10.1124/dmd.105.008680) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16501005/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Metab.%20Dispos.&title=Effect%20of%20cyp3a5%20polymorphism%20on%20tacrolimus%20metabolic%20clearance%20in%20vitro&author=Y.%20Dai&author=M.%20F.%20Hebert&author=N.%20Isoherranen&author=C.%20L.%20Davis&author=C.%20Marsh&volume=34&issue=5&publication_year=2006&pages=836-847&pmid=16501005&doi=10.1124/dmd.105.008680&)
|
| 248 |
+
|
| 249 |
+
6. Fang X., Qu Q., Xiao X. C. (2019). Research progress of tacrolimus in the treatment of idiopathic membranous nephropathy. Chin. Clin. Pharmacol. Ther. 24 (02), 228–234. 10.12092/j.issn.1009-2501.2019.02.018 [DOI](https://doi.org/10.12092/j.issn.1009-2501.2019.02.018) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chin.%20Clin.%20Pharmacol.%20Ther.&title=Research%20progress%20of%20tacrolimus%20in%20the%20treatment%20of%20idiopathic%20membranous%20nephropathy&author=X.%20Fang&author=Q.%20Qu&author=X.%20C.%20Xiao&volume=24&issue=02&publication_year=2019&pages=228-234&doi=10.12092/j.issn.1009-2501.2019.02.018&)
|
| 250 |
+
|
| 251 |
+
7. Fernandez-Juarez G., Rojas-Rivera J., Logt A. V., Justino J., Sevillano A., Caravaca- Fontán F., et al. (2021). The STARMEN trial indicates that alternating treatment with corticosteroids and cyclophosphamide is superior to sequential treatment with tacrolimus and rituximab in primary membranous nephropathy. Kidney Int. 99 (4), 986–998. 10.1016/j.kint.2020.10.014 [DOI](https://doi.org/10.1016/j.kint.2020.10.014) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33166580/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Kidney%20Int.&title=The%20STARMEN%20trial%20indicates%20that%20alternating%20treatment%20with%20corticosteroids%20and%20cyclophosphamide%20is%20superior%20to%20sequential%20treatment%20with%20tacrolimus%20and%20rituximab%20in%20primary%20membranous%20nephropathy&author=G.%20Fernandez-Juarez&author=J.%20Rojas-Rivera&author=A.%20V.%20Logt&author=J.%20Justino&author=A.%20Sevillano&volume=99&issue=4&publication_year=2021&pages=986-998&pmid=33166580&doi=10.1016/j.kint.2020.10.014&)
|
| 252 |
+
|
| 253 |
+
8. Hakkola J., Hukkanen J., Turpeinen M., Pelkonen O. (2020). Inhibition and induction of cyp enzymes in humans: an update. Arch. Toxicol. 94 (11), 3671–3722. 10.1007/s00204-020-02936-7 [DOI](https://doi.org/10.1007/s00204-020-02936-7) | [PMC free article](/articles/PMC7603454/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33111191/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arch.%20Toxicol.&title=Inhibition%20and%20induction%20of%20cyp%20enzymes%20in%20humans:%20an%20update&author=J.%20Hakkola&author=J.%20Hukkanen&author=M.%20Turpeinen&author=O.%20Pelkonen&volume=94&issue=11&publication_year=2020&pages=3671-3722&pmid=33111191&doi=10.1007/s00204-020-02936-7&)
|
| 254 |
+
|
| 255 |
+
9. Hamilton P., Wilson F., Chinnadurai R., Sinha S., Singh M., Ponnusamy A., et al. (2020). The investigative burden of membranous nephropathy in the UK. Clin. Kidney J. 13 (1), 27–34. 10.1093/ckj/sfz036 [DOI](https://doi.org/10.1093/ckj/sfz036) | [PMC free article](/articles/PMC7025364/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32082550/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Kidney%20J.&title=The%20investigative%20burden%20of%20membranous%20nephropathy%20in%20the%20UK&author=P.%20Hamilton&author=F.%20Wilson&author=R.%20Chinnadurai&author=S.%20Sinha&author=M.%20Singh&volume=13&issue=1&publication_year=2020&pages=27-34&pmid=32082550&doi=10.1093/ckj/sfz036&)
|
| 256 |
+
|
| 257 |
+
10. Hannachi I., Chadli Z., Kerkeni E., Kolsi A., Hammouda M., Chaabane A., et al. (2021). Influence of CYP3A polymorphisms on tacrolimus pharmacokinetics in kidney transplant recipients. Pharmacogenomics J. 21 (1), 69–77. 10.1038/s41397-020-00179-4 [DOI](https://doi.org/10.1038/s41397-020-00179-4) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32843687/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J.&title=Influence%20of%20CYP3A%20polymorphisms%20on%20tacrolimus%20pharmacokinetics%20in%20kidney%20transplant%20recipients&author=I.%20Hannachi&author=Z.%20Chadli&author=E.%20Kerkeni&author=A.%20Kolsi&author=M.%20Hammouda&volume=21&issue=1&publication_year=2021&pages=69-77&pmid=32843687&doi=10.1038/s41397-020-00179-4&)
|
| 258 |
+
|
| 259 |
+
11. He Y., Ma Y., Fu Q., Liang J., Yu X., Huang H., et al. (2022). The CYP3A5 and ABCB1 gene polymorphisms in kidney transplant patients and establishment of initial daily tacrolimus dosing formula. Ann. Pharmacother. 56 (4), 393–400. 10.1177/10600280211023495 [DOI](https://doi.org/10.1177/10600280211023495) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34362271/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Pharmacother.&title=The%20CYP3A5%20and%20ABCB1%20gene%20polymorphisms%20in%20kidney%20transplant%20patients%20and%20establishment%20of%20initial%20daily%20tacrolimus%20dosing%20formula&author=Y.%20He&author=Y.%20Ma&author=Q.%20Fu&author=J.%20Liang&author=X.%20Yu&volume=56&issue=4&publication_year=2022&pages=393-400&pmid=34362271&doi=10.1177/10600280211023495&)
|
| 260 |
+
|
| 261 |
+
12. Huang L., Wang J., Yang J., Zhang H., Ni Y., Zhu Z., et al. (2019). Impact of cyp3a4/5 and abcb1 polymorphisms on tacrolimus exposure and response in pediatric primary nephrotic syndrome. Pharmacogenomics 20 (15), 1071–1083. 10.2217/pgs-2019-0090 [DOI](https://doi.org/10.2217/pgs-2019-0090) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31588879/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=Impact%20of%20cyp3a4/5%20and%20abcb1%20polymorphisms%20on%20tacrolimus%20exposure%20and%20response%20in%20pediatric%20primary%20nephrotic%20syndrome&author=L.%20Huang&author=J.%20Wang&author=J.%20Yang&author=H.%20Zhang&author=Y.%20Ni&volume=20&issue=15&publication_year=2019&pages=1071-1083&pmid=31588879&doi=10.2217/pgs-2019-0090&)
|
| 262 |
+
|
| 263 |
+
13. Keri K. C., Blumenthal S., Kulkarni V., Beck L., Chongkrairatanakul T. (2019). Primary membranous nephropathy: comprehensive review and historical perspective. Postgrad. Med. J. 95 (1119), 23–31. 10.1136/postgradmedj-2018-135729 [DOI](https://doi.org/10.1136/postgradmedj-2018-135729) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30683678/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Postgrad.%20Med.%20J.&title=Primary%20membranous%20nephropathy:%20comprehensive%20review%20and%20historical%20perspective&author=K.%20C.%20Keri&author=S.%20Blumenthal&author=V.%20Kulkarni&author=L.%20Beck&author=T.%20Chongkrairatanakul&volume=95&issue=1119&publication_year=2019&pages=23-31&pmid=30683678&doi=10.1136/postgradmedj-2018-135729&)
|
| 264 |
+
|
| 265 |
+
14. Kirubakaran R., Stocker S. L., Hennig S., Day R. O., Carland J. E. (2020). Population pharmacokinetic models of tacrolimus in adult transplant recipients: a systematic review. Clin. Pharmacokinet. 59 (11), 1357–1392. 10.1007/s40262-020-00922-x [DOI](https://doi.org/10.1007/s40262-020-00922-x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32783100/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Pharmacokinet.&title=Population%20pharmacokinetic%20models%20of%20tacrolimus%20in%20adult%20transplant%20recipients:%20a%20systematic%20review&author=R.%20Kirubakaran&author=S.%20L.%20Stocker&author=S.%20Hennig&author=R.%20O.%20Day&author=J.%20E.%20Carland&volume=59&issue=11&publication_year=2020&pages=1357-1392&pmid=32783100&doi=10.1007/s40262-020-00922-x&)
|
| 266 |
+
|
| 267 |
+
15. Klyushova L. S., Perepechaeva M. L., Grishanova A. Y. (2022). The role of CYP3A in health and disease. BIOMEDICINES 10 (11), 2686. 10.3390/biomedicines10112686 [DOI](https://doi.org/10.3390/biomedicines10112686) | [PMC free article](/articles/PMC9687714/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36359206/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=BIOMEDICINES&title=The%20role%20of%20CYP3A%20in%20health%20and%20disease&author=L.%20S.%20Klyushova&author=M.%20L.%20Perepechaeva&author=A.%20Y.%20Grishanova&volume=10&issue=11&publication_year=2022&pages=2686&pmid=36359206&doi=10.3390/biomedicines10112686&)
|
| 268 |
+
|
| 269 |
+
16. Lin S., Li H. Y., Zhou T., Lin W., et al. (2019b). Efficacy and safety of cyclosporine A in the treatment of idiopathic membranous nephropathy in an Asian population. Drug Des. Devel Ther. 13, 2305–2330. 10.2147/DDDT.S204974 [DOI](https://doi.org/10.2147/DDDT.S204974) | [PMC free article](/articles/PMC6628962/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31371924/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Des.%20Devel%20Ther.&title=Efficacy%20and%20safety%20of%20cyclosporine%20A%20in%20the%20treatment%20of%20idiopathic%20membranous%20nephropathy%20in%20an%20Asian%20population&author=S.%20Lin&author=H.%20Y.%20Li&author=T.%20Zhou&author=W.%20Lin&volume=13&publication_year=2019b&pages=2305-2330&pmid=31371924&doi=10.2147/DDDT.S204974&)
|
| 270 |
+
|
| 271 |
+
17. Lin T.-T., Wei C., Sun S. (2019a). Study on the effect of CYP3A5 gene polymorphism on the blood concentration of tacrolimus in patients with membranous nephropathy. Strait Pharmacol. 31 (11), 115–118. [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Strait%20Pharmacol.&title=Study%20on%20the%20effect%20of%20CYP3A5%20gene%20polymorphism%20on%20the%20blood%20concentration%20of%20tacrolimus%20in%20patients%20with%20membranous%20nephropathy&author=T.-T.%20Lin&author=C.%20Wei&author=S.%20Sun&volume=31&issue=11&publication_year=2019a&pages=115-118&)
|
| 272 |
+
|
| 273 |
+
18. Lu W., Gong S., Li J., Luo H., Wang Y. (2020). Efficacy and safety of rituximab in the treatment of membranous nephropathy: a systematic review and meta-analysis. Med. Baltim. 99 (16), e19804. 10.1097/MD.0000000000019804 [DOI](https://doi.org/10.1097/MD.0000000000019804) | [PMC free article](/articles/PMC7440335/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32311997/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Med.%20Baltim.&title=Efficacy%20and%20safety%20of%20rituximab%20in%20the%20treatment%20of%20membranous%20nephropathy:%20a%20systematic%20review%20and%20meta-analysis&author=W.%20Lu&author=S.%20Gong&author=J.%20Li&author=H.%20Luo&author=Y.%20Wang&volume=99&issue=16&publication_year=2020&pages=e19804&pmid=32311997&doi=10.1097/MD.0000000000019804&)
|
| 274 |
+
|
| 275 |
+
19. McGrath S., Zhao X., Steele R., Thombs B. D., Benedetti A. (2020). Estimating the sample mean and standard deviation from commonly reported quantiles in meta-analysis. Stat. Methods Med. Res. 1387154872. 10.1177/0962280219889080 [DOI](https://doi.org/10.1177/0962280219889080) | [PMC free article](/articles/PMC7390706/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32292115/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Stat.%20Methods%20Med.%20Res.&title=Estimating%20the%20sample%20mean%20and%20standard%20deviation%20from%20commonly%20reported%20quantiles%20in%20meta-analysis&author=S.%20McGrath&author=X.%20Zhao&author=R.%20Steele&author=B.%20D.%20Thombs&author=A.%20Benedetti&publication_year=2020&pages=1387154872&pmid=32292115&doi=10.1177/0962280219889080&)
|
| 276 |
+
|
| 277 |
+
20. Muraki Y., Mizuno S., Nakatani K., Wakabayashi H., Ishikawa E., Araki T., et al. (2018). Monitoring of peripheral blood cluster of differentiation 4(+) adenosine triphosphate activity and CYP3A5 genotype to determine the pharmacokinetics, clinical effects and complications of tacrolimus in patients with autoimmune diseases. Exp. Ther. Med. 15 (1), 532–538. 10.3892/etm.2017.5364 [DOI](https://doi.org/10.3892/etm.2017.5364) | [PMC free article](/articles/PMC5763654/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29375701/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Exp.%20Ther.%20Med.&title=Monitoring%20of%20peripheral%20blood%20cluster%20of%20differentiation%204(+)%20adenosine%20triphosphate%20activity%20and%20CYP3A5%20genotype%20to%20determine%20the%20pharmacokinetics,%20clinical%20effects%20and%20complications%20of%20tacrolimus%20in%20patients%20with%20autoimmune%20diseases&author=Y.%20Muraki&author=S.%20Mizuno&author=K.%20Nakatani&author=H.%20Wakabayashi&author=E.%20Ishikawa&volume=15&issue=1&publication_year=2018&pages=532-538&pmid=29375701&doi=10.3892/etm.2017.5364&)
|
| 278 |
+
|
| 279 |
+
21. Nair S. S., Sarasamma S., Gracious N., George J., Anish T. S. N., Radhakrishnan R. (2015). Polymorphism of the CYP3A5 gene and its effect on tacrolimus blood level. Exp. Clin. Transpl. 13 (Suppl. 1), 197–200. 10.6002/ect.mesot2014.O167 [DOI](https://doi.org/10.6002/ect.mesot2014.O167) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25894154/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Exp.%20Clin.%20Transpl.&title=Polymorphism%20of%20the%20CYP3A5%20gene%20and%20its%20effect%20on%20tacrolimus%20blood%20level&author=S.%20S.%20Nair&author=S.%20Sarasamma&author=N.%20Gracious&author=J.%20George&author=T.%20S.%20N.%20Anish&volume=13&issue=Suppl.%201&publication_year=2015&pages=197-200&pmid=25894154&doi=10.6002/ect.mesot2014.O167&)
|
| 280 |
+
|
| 281 |
+
22. Ruo-Ji C., Fang X., Zhen-Shuang D., Yu-Lin Z., Zi-Li Z., Wei-Yuan L., et al. (2022). Comparative efficacy of three regimens (cyclosporine, tacrolimus, and cyclophosphamide) combined with steroids for the treatment of idiopathic membranous nephropathy. Nefrol. Engl. Ed. 42, 671–679. 10.1016/j.nefroe.2022.11.008 [DOI](https://doi.org/10.1016/j.nefroe.2022.11.008) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36402685/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nefrol.%20Engl.%20Ed.&title=Comparative%20efficacy%20of%20three%20regimens%20(cyclosporine,%20tacrolimus,%20and%20cyclophosphamide)%20combined%20with%20steroids%20for%20the%20treatment%20of%20idiopathic%20membranous%20nephropathy&author=C.%20Ruo-Ji&author=X.%20Fang&author=D.%20Zhen-Shuang&author=Z.%20Yu-Lin&author=Z.%20Zi-Li&volume=42&publication_year=2022&pages=671-679&pmid=36402685&doi=10.1016/j.nefroe.2022.11.008&)
|
| 282 |
+
|
| 283 |
+
23. Shao J., Wang C., Fu P., Chen F., Zhang Y., Wei J. (2020). Impact of donor and recipient cyp3a5*3 genotype on tacrolimus population pharmacokinetics in Chinese adult liver transplant recipients. Ann. Pharmacother. 54 (7), 652–661. 10.1177/1060028019897050 [DOI](https://doi.org/10.1177/1060028019897050) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31888346/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Pharmacother.&title=Impact%20of%20donor%20and%20recipient%20cyp3a5*3%20genotype%20on%20tacrolimus%20population%20pharmacokinetics%20in%20Chinese%20adult%20liver%20transplant%20recipients&author=J.%20Shao&author=C.%20Wang&author=P.%20Fu&author=F.%20Chen&author=Y.%20Zhang&volume=54&issue=7&publication_year=2020&pages=652-661&pmid=31888346&doi=10.1177/1060028019897050&)
|
| 284 |
+
|
| 285 |
+
24. van den Brand J., Ruggenenti P., Chianca A., Hofstra J., Perna A., Ruggiero B., et al. (2017). Safety of rituximab compared with steroids and cyclophosphamide for idiopathic membranous nephropathy. J. Am. Soc. Nephrol. 28 (9), 2729–2737. 10.1681/ASN.2016091022 [DOI](https://doi.org/10.1681/ASN.2016091022) | [PMC free article](/articles/PMC5576929/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28487395/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Am.%20Soc.%20Nephrol.&title=Safety%20of%20rituximab%20compared%20with%20steroids%20and%20cyclophosphamide%20for%20idiopathic%20membranous%20nephropathy&author=J.%20van%20den%20Brand&author=P.%20Ruggenenti&author=A.%20Chianca&author=J.%20Hofstra&author=A.%20Perna&volume=28&issue=9&publication_year=2017&pages=2729-2737&pmid=28487395&doi=10.1681/ASN.2016091022&)
|
| 286 |
+
|
| 287 |
+
25. van Gelder T., Meziyerh S., Swen J. J., de Vries A. J., Moes D. R., et al. (2020). The clinical impact of the C0/D ratio and the CYP3A5 genotype on outcome in tacrolimus treated kidney transplant recipients. Front. Pharmacol. 11, 1142. 10.3389/fphar.2020.01142 [DOI](https://doi.org/10.3389/fphar.2020.01142) | [PMC free article](/articles/PMC7411304/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32848756/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front.%20Pharmacol.&title=The%20clinical%20impact%20of%20the%20C0/D%20ratio%20and%20the%20CYP3A5%20genotype%20on%20outcome%20in%20tacrolimus%20treated%20kidney%20transplant%20recipients&author=T.%20van%20Gelder&author=S.%20Meziyerh&author=J.%20J.%20Swen&author=A.%20J.%20de%20Vries&author=D.%20R.%20Moes&volume=11&publication_year=2020&pages=1142&pmid=32848756&doi=10.3389/fphar.2020.01142&)
|
| 288 |
+
|
| 289 |
+
26. von Groote T. C., Williams G., Au E. H., Chen Y., Mathew A. T., Hodson E. M., et al. (2021). Immunosuppressive treatment for primary membranous nephropathy in adults with nephrotic syndrome. Cochrane Database Syst. Rev. 11 (11), D4293. 10.1002/14651858.cd004293.pub4 [DOI](https://doi.org/10.1002/14651858.cd004293.pub4) | [PMC free article](/articles/PMC8591447/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34778952/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cochrane%20Database%20Syst.%20Rev.&title=Immunosuppressive%20treatment%20for%20primary%20membranous%20nephropathy%20in%20adults%20with%20nephrotic%20syndrome&author=T.%20C.%20von%20Groote&author=G.%20Williams&author=E.%20H.%20Au&author=Y.%20Chen&author=A.%20T.%20Mathew&volume=11&issue=11&publication_year=2021&pages=D4293&pmid=34778952&doi=10.1002/14651858.cd004293.pub4&)
|
| 290 |
+
|
| 291 |
+
27. Wang C. B., Zhang Y. J., Zhao M. M., Zhao L. M. (2023). Population pharmacokinetic analyses of tacrolimus in non-transplant patients: a systematic review. Eur. J. Clin. Pharmacol. 79 (7), 897–913. 10.1007/s00228-023-03503-6 [DOI](https://doi.org/10.1007/s00228-023-03503-6) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37261481/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20J.%20Clin.%20Pharmacol.&title=Population%20pharmacokinetic%20analyses%20of%20tacrolimus%20in%20non-transplant%20patients:%20a%20systematic%20review&author=C.%20B.%20Wang&author=Y.%20J.%20Zhang&author=M.%20M.%20Zhao&author=L.%20M.%20Zhao&volume=79&issue=7&publication_year=2023&pages=897-913&pmid=37261481&doi=10.1007/s00228-023-03503-6&)
|
| 292 |
+
|
| 293 |
+
28. Wang M. (2020). Effect of CYP3A5 gene polymorphism on tacrolimus blood concentration, efficacy and adverse effects in patients with idiopathic membranous nephropathy. Hebei North College. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Effect%20of%20CYP3A5%20gene%20polymorphism%20on%20tacrolimus%20blood%20concentration,%20efficacy%20and%20adverse%20effects%20in%20patients%20with%20idiopathic%20membranous%20nephropathy&author=M.%20Wang&publication_year=2020&)
|
| 294 |
+
|
| 295 |
+
29. Wei C., Lin T., Dou M. (2018). Correlation of CYP3A5 gene polymorphisms with the efficacy and safety of tacrolimus in patients with membranous nephropathy. Chin. J. Hosp. Pharm. 38 (02), 161–164. 10.13286/j.cnki.chinhosppharmacyj.2018.02.11 [DOI](https://doi.org/10.13286/j.cnki.chinhosppharmacyj.2018.02.11) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chin.%20J.%20Hosp.%20Pharm.&title=Correlation%20of%20CYP3A5%20gene%20polymorphisms%20with%20the%20efficacy%20and%20safety%20of%20tacrolimus%20in%20patients%20with%20membranous%20nephropathy&author=C.%20Wei&author=T.%20Lin&author=M.%20Dou&volume=38&issue=02&publication_year=2018&pages=161-164&doi=10.13286/j.cnki.chinhosppharmacyj.2018.02.11&)
|
| 296 |
+
|
| 297 |
+
30. Wu F. (2019). Effect of CYP3A5 and MDR1 gene polymorphisms on the dose and efficacy of tacrolimus in patients with membranous nephropathy. Shandong University. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Effect%20of%20CYP3A5%20and%20MDR1%20gene%20polymorphisms%20on%20the%20dose%20and%20efficacy%20of%20tacrolimus%20in%20patients%20with%20membranous%20nephropathy&author=F.%20Wu&publication_year=2019&)
|
| 298 |
+
|
| 299 |
+
31. Wu L., Lai J., Ling Y., Weng Y., Zhou S., Wu S., et al. (2021). A review of the current practice of diagnosis and treatment of idiopathic membranous nephropathy in China. Med. Sci. Monit. 27, e930097. 10.12659/MSM.930097 [DOI](https://doi.org/10.12659/MSM.930097) | [PMC free article](/articles/PMC7876949/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33550324/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Med.%20Sci.%20Monit.&title=A%20review%20of%20the%20current%20practice%20of%20diagnosis%20and%20treatment%20of%20idiopathic%20membranous%20nephropathy%20in%20China&author=L.%20Wu&author=J.%20Lai&author=Y.%20Ling&author=Y.%20Weng&author=S.%20Zhou&volume=27&publication_year=2021&pages=e930097&pmid=33550324&doi=10.12659/MSM.930097&)
|
| 300 |
+
|
| 301 |
+
32. Xu H., Xu B., Wang S., Shen Z., Zhao Y., Wang Q., et al. (2021). Effect of CYP3A5*3 gene polymorphism on blood concentration and clinical efficacy of tacrolimus in the treatment of membranous nephropathy. China Ration. Drug Discov. 18 (03), 50–54. 10.3969/j.issn.2096-3327.2021.3.010 [DOI](https://doi.org/10.3969/j.issn.2096-3327.2021.3.010) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=China%20Ration.%20Drug%20Discov.&title=Effect%20of%20CYP3A5*3%20gene%20polymorphism%20on%20blood%20concentration%20and%20clinical%20efficacy%20of%20tacrolimus%20in%20the%20treatment%20of%20membranous%20nephropathy&author=H.%20Xu&author=B.%20Xu&author=S.%20Wang&author=Z.%20Shen&author=Y.%20Zhao&volume=18&issue=03&publication_year=2021&pages=50-54&doi=10.3969/j.issn.2096-3327.2021.3.010&)
|
| 302 |
+
|
| 303 |
+
33. Yang M., Wei C., Liu N., Liu Y. (2015). Predictive value of CYP3A5 gene polymorphisms on the initial dose of tacrolimus in patients with membranous nephropathy. Chin. J. Kidney Dis. 31 (10), 736–742. [DOI](https://doi.org/10.3760/cma.j.issn.1001—7097.2015.10.003) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chin.%20J.%20Kidney%20Dis.&title=Predictive%20value%20of%20CYP3A5%20gene%20polymorphisms%20on%20the%20initial%20dose%20of%20tacrolimus%20in%20patients%20with%20membranous%20nephropathy&author=M.%20Yang&author=C.%20Wei&author=N.%20Liu&author=Y.%20Liu&volume=31&issue=10&publication_year=2015&pages=736-742&doi=10.3760/cma.j.issn.1001—7097.2015.10.003&)
|
| 304 |
+
|
| 305 |
+
34. Yang T., Wu B., Li D., Xu T. (2020). Systematic evaluation of CYP3A5 gene polymorphisms and tacrolimus blood levels and efficacy and safety. Chin. J. Hosp. Pharm. 40 (03), 322–328. 10.13286/j.1001-5213.2020.03.16 [DOI](https://doi.org/10.13286/j.1001-5213.2020.03.16) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chin.%20J.%20Hosp.%20Pharm.&title=Systematic%20evaluation%20of%20CYP3A5%20gene%20polymorphisms%20and%20tacrolimus%20blood%20levels%20and%20efficacy%20and%20safety&author=T.%20Yang&author=B.%20Wu&author=D.%20Li&author=T.%20Xu&volume=40&issue=03&publication_year=2020&pages=322-328&doi=10.13286/j.1001-5213.2020.03.16&)
|
| 306 |
+
|
| 307 |
+
35. Ye D., Liu X., Lan S., Wang J. (2014). Effect of CYP3A5 genotype on the blood concentration to dose ratio of tacrolimus in patients with nephrotic syndrome. Chin. J. Hosp. Pharm. 34 (24), 2139–2143. 10.13286/j.cnki.chinhosppharmacyj.2014.24.15 [DOI](https://doi.org/10.13286/j.cnki.chinhosppharmacyj.2014.24.15) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chin.%20J.%20Hosp.%20Pharm.&title=Effect%20of%20CYP3A5%20genotype%20on%20the%20blood%20concentration%20to%20dose%20ratio%20of%20tacrolimus%20in%20patients%20with%20nephrotic%20syndrome&author=D.%20Ye&author=X.%20Liu&author=S.%20Lan&author=J.%20Wang&volume=34&issue=24&publication_year=2014&pages=2139-2143&doi=10.13286/j.cnki.chinhosppharmacyj.2014.24.15&)
|
| 308 |
+
|
| 309 |
+
36. You L., Ye P., Xiao G., Liang J., Kong Y. (2021). Rituximab for the treatment of idiopathic membranous nephropathy with nephrotic syndrome: a systematic review and meta-analysis. Turk J. Med. Sci. 51 (6), 2870–2880. 10.3906/sag-2104-177 [DOI](https://doi.org/10.3906/sag-2104-177) | [PMC free article](/articles/PMC10734821/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34391323/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Turk%20J.%20Med.%20Sci.&title=Rituximab%20for%20the%20treatment%20of%20idiopathic%20membranous%20nephropathy%20with%20nephrotic%20syndrome:%20a%20systematic%20review%20and%20meta-analysis&author=L.%20You&author=P.%20Ye&author=G.%20Xiao&author=J.%20Liang&author=Y.%20Kong&volume=51&issue=6&publication_year=2021&pages=2870-2880&pmid=34391323&doi=10.3906/sag-2104-177&)
|
| 310 |
+
|
| 311 |
+
37. Zeng X., Liu H., Chen X., Weidong L. (2012). Meta-analysis series No. 4: a quality assessment tool for observational studies. Chin. J. Evidence-Based Cardiovasc. Med. 4 (04), 297–299. 10.3969/j.1674-4055.2012.04.004 [DOI](https://doi.org/10.3969/j.1674-4055.2012.04.004) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chin.%20J.%20Evidence-Based%20Cardiovasc.%20Med.&title=Meta-analysis%20series%20No.%204:%20a%20quality%20assessment%20tool%20for%20observational%20studies&author=X.%20Zeng&author=H.%20Liu&author=X.%20Chen&author=L.%20Weidong&volume=4&issue=04&publication_year=2012&pages=297-299&doi=10.3969/j.1674-4055.2012.04.004&)
|
| 312 |
+
|
| 313 |
+
38. Zhang C., Duan S., Guo M., Yuan Y., Huang Z., Zhu J., et al. (2020). Effects of cyp3a5 polymorphisms on efficacy and safety of tacrolimus therapy in patients with idiopathic membranous nephropathy. Pharmacogenomics Personalized Med. 13, 141–149. 10.2147/PGPM.S247892 [DOI](https://doi.org/10.2147/PGPM.S247892) | [PMC free article](/articles/PMC7186213/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32368128/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20Personalized%20Med.&title=Effects%20of%20cyp3a5%20polymorphisms%20on%20efficacy%20and%20safety%20of%20tacrolimus%20therapy%20in%20patients%20with%20idiopathic%20membranous%20nephropathy&author=C.%20Zhang&author=S.%20Duan&author=M.%20Guo&author=Y.%20Yuan&author=Z.%20Huang&volume=13&publication_year=2020&pages=141-149&pmid=32368128&doi=10.2147/PGPM.S247892&)
|
| 314 |
+
|
| 315 |
+
39. Zou H., Jiang F., Xu G. (2019). Effectiveness and safety of cyclophosphamide or tacrolimus therapy for idiopathic membranous nephropathy. Ren. Fail 41 (1), 673–681. 10.1080/0886022X.2019.1637758 [DOI](https://doi.org/10.1080/0886022X.2019.1637758) | [PMC free article](/articles/PMC6711082/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31354007/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ren.%20Fail&title=Effectiveness%20and%20safety%20of%20cyclophosphamide%20or%20tacrolimus%20therapy%20for%20idiopathic%20membranous%20nephropathy&author=H.%20Zou&author=F.%20Jiang&author=G.%20Xu&volume=41&issue=1&publication_year=2019&pages=673-681&pmid=31354007&doi=10.1080/0886022X.2019.1637758&)
|
test/texts/PMC11252221.md
ADDED
|
@@ -0,0 +1,286 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Pharmacokinetics and safety of mavacamten in healthy Chinese participants with different CYP2C19 phenotypes
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Xiaojie Wu, Nanye Chen, Peiwen Hsu, Jing Sun, Wenting Li, Qi Wang, Merali Samira, Qiong Wei, Jicheng Yu, Guoying Cao, Haijing Yang, Lili Wang, Jingjing Wang, Yi Jin, Wei Liu, Jufang Wu, Jinjie He, Cheng Lyu, Jing Zhang
|
| 5 |
+
**Journal:** Clinical and Translational Science
|
| 6 |
+
**Date:** 2024 Jul 16
|
| 7 |
+
**DOI:** [10.1111/cts.13877](https://doi.org/10.1111/cts.13877)
|
| 8 |
+
**PMID:** 39014868
|
| 9 |
+
**PMCID:** PMC11252221
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11252221/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11252221/pdf/CTS-17-e13877.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11252221/pdf/CTS-17-e13877.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
Obstructive hypertrophic cardiomyopathy (oHCM) is a subtype of HCM characterized by left ventricular outflow tract obstruction resulting from cardiac muscle hypertrophy and anatomic alterations in the mitral valve and apparatus. Mavacamten, a cardiac myosin inhibitor metabolized primarily by CYP2C19 in the liver, is the first and only targeted medication approved for the treatment of symptomatic New York Heart Association (NYHA) class II–III oHCM. Previous pharmacokinetic (PK) results of mavacamten in healthy Caucasian, Japanese, and Asian participants demonstrated that mavacamten exposure was affected by CYP2C19 metabolism status. This open‐label, parallel‐group, phase I trial aimed to determine the PK and safety of mavacamten in healthy Chinese participants with different CYP2C19 genotypes. The primary outcome was to define the PK of mavacamten in healthy Chinese participants; the secondary outcome was to examine safety and tolerability. After a single oral dose of 15 or 25 mg mavacamten in fasted healthy adult Chinese individuals, C max was reached within a median T max of 0.6–1.5 h, indicating rapid absorption. Inter‐individual variability was moderate, and individuals carrying non‐functional CYP2C19 alleles (*2/*2, *3/*3, or *2/*3) exhibited longer half‐life and increased total exposure. After stratification of CYP2C19 genotypes, total mavacamten exposures were similar among different ethnic groups when compared with prior PK studies. No significant adverse events were observed in this study. Single oral administration of mavacamten at 15 mg was well tolerated across all CYP2C19 genotypes, and 25 mg dose was well tolerated in healthy participants with CYP2C19 genotypes UM/RM/NM. The PK profile of mavacamten in the healthy Chinese population was consistent with that in other healthy populations.
|
| 16 |
+
|
| 17 |
+
Mavacamten, a first‐in‐class cardiac myosin inhibitor, has been approved for the treatment of obstructive hypertrophic cardiomyopathy (oHCM) in 5 continents. This approval marks a significant breakthrough in oHCM treatment, as it provides a targeted approach to addressing the underlying mechanisms of oHCM. Prior studies conducted in Caucasian, Japanese, and Asian populations suggested that CYP2C19 genotype might influence mavacamten exposure.
|
| 18 |
+
|
| 19 |
+
This bridging study assessed the pharmacokinetics and safety of mavacamten in healthy adult Chinese participants with different genotypes of CYP2C19.
|
| 20 |
+
|
| 21 |
+
The pharmacokinetic profile of mavacamten in the Chinese population was found to be consistent with previous research, featuring rapid absorption and moderate inter‐individual variability. The total exposure of mavacamten varied based on the individual's CYP2C19 genotypes, with participants classified as poor metabolizers showing higher exposure and longer half‐life. Mavacamten was generally well tolerated, with no observed risk of cardiac toxicity in this study. No significant safety concerns emerged among participants classified as intermediate or poor metabolizers of CYP2C19.
|
| 22 |
+
|
| 23 |
+
This is the first study that provides the pharmacokinetic and safety profile of mavacamten in the Chinese population. These findings support an ongoing phase III trial of mavacamten in Chinese patients with oHCM.
|
| 24 |
+
|
| 25 |
+
## INTRODUCTION
|
| 26 |
+
|
| 27 |
+
Hypertrophic cardiomyopathy (HCM) is a primary myocardial disorder defined by left ventricular (LV) hypertrophy that cannot be explained by another cardiac or systemic disease. It is a chronic and often progressive disease of the cardiomyocyte. Two HCM phenotypes are recognized based on the presence or absence of obstruction of the left ventricular outflow tract (LVOT): obstructive HCM (oHCM) and non‐obstructive HCM. The disease often has a genetic basis and the pathophysiology is complex and not yet fully understood.[1](#cts13877-bib-0001) ^1^1 , [2](#cts13877-bib-0002) ^2^2 , [3](#cts13877-bib-0003) ^3^3 In patients with HCM, the myocardium becomes hypertrophied leading to cardiac dysfunction, resulting in significant comorbidities including heart failure, arrhythmia, and rarely, sudden death.[2](#cts13877-bib-0002) ^2^2 In oHCM, thickening of the heart muscle and anatomic alterations in the mitral valve and apparatus lead to LVOT obstruction,[4](#cts13877-bib-0004) ^4^4 which can cause symptoms such as shortness of breath, chest pain, and fainting, particularly during exercise.[5](#cts13877-bib-0005) ^5^5
|
| 28 |
+
|
| 29 |
+
Treatment for HCM depends on the severity of the disease and the presence of symptoms. In oHCM, medications such as beta‐blockers, calcium channel blockers, and disopyramide are used to relieve symptoms and improve blood flow, although they only have limited effectiveness and none of them have been specifically approved for the treatment of oHCM. In severe cases with LVOT gradient ≥50 mmHg, alcohol septal ablation and surgical septal myectomy may be needed to remove part of the thickened heart muscle. There is an increased risk of dysrhythmia, and a pacemaker or defibrillator may need to be implanted to regulate the heart's rhythm.[4](#cts13877-bib-0004) ^4^4 , [5](#cts13877-bib-0005) ^5^5 , [6](#cts13877-bib-0006) ^6^6
|
| 30 |
+
|
| 31 |
+
Mavacamten—a cardiac myosin inhibitor—is the first and only medication approved by the United States Food and Drug Administration, in 2022, for the treatment of adults with symptomatic New York Heart Association class II–III oHCM to improve functional capacity and symptoms.[7](#cts13877-bib-0007) ^7^7 The mechanism of action of mavacamten involves targeting the underlying pathophysiology of oHCM by reducing the number and activity of actin‐myosin bridges and alleviating hypercontractility. Mavacamten works by inhibiting the activity of the protein myosin, which is responsible for the contraction of heart muscle cells, and consequently helps to relax the heart muscle and improves diastolic function.[5](#cts13877-bib-0005) ^5^5 , [7](#cts13877-bib-0007) ^7^7 , [8](#cts13877-bib-0008) ^8^8 In prior phase III clinical studies, mavacamten significantly improved exercise capacity and LVOT obstruction, reduced important biomarkers of cardiac strain compared with placebo in patients with symptomatic oHCM, and also demonstrated a favorable safety profile.[9](#cts13877-bib-0009) ^9^9 , [10](#cts13877-bib-0010) ^10^10 , [11](#cts13877-bib-0011) ^11^11 The results of the phase III VALOR‐HCM trial ([NCT04349072](https://clinicaltrials.gov/ct2/show/NCT04349072)NCT04349072) demonstrated a statistically significant and clinically meaningful reduction for the primary end point, the composite of proceeding to septal reduction therapy (SRT) or remaining SRT Guideline–eligible after 16 weeks of treatment with mavacamten compared with placebo, among patients with symptomatic oHCM who met the 2011 American College of Cardiology Foundation/American Heart Association Guideline referral criteria for SRT.[12](#cts13877-bib-0012) ^12^12
|
| 32 |
+
|
| 33 |
+
Previous pharmacokinetic (PK) studies of mavacamten have provided important information about its absorption, distribution, metabolism, and elimination in humans. Mavacamten is rapidly absorbed following oral administration with peak plasma concentrations reached in around 1 h. The bioavailability of mavacamten is at least 85%, and it exhibits dose‐proportional PKs over the clinically relevant dose range of 1–15 mg. Mavacamten is extensively metabolized in the liver, primarily through CYP2C19 (74%), CYP3A4 (18%), and CYP2C9 (8%), and it is mainly excreted in urine (3% unchanged).[7](#cts13877-bib-0007) ^7^7 , [13](#cts13877-bib-0013) ^13^13 There is evidence indicating that the CYP2C19 genotype may influence mavacamten exposure. Normal metabolizers carry two functional alleles of CYP2C19, and poor metabolizers have two non‐functional alleles.[7](#cts13877-bib-0007) ^7^7 Higher plasma concentrations and longer half‐lives of mavacamten were observed in individuals who were poor metabolizers (PM) of CYP2C19 compared with normal metabolizers (NM).[7](#cts13877-bib-0007) ^7^7 , [13](#cts13877-bib-0013) ^13^13 This suggests that CYP2C19 genotyping may be helpful in dosing mavacamten. The prevalence of CYP2C19 PM varies depending on ancestry and is approximately 2% in Europeans, 4% in African Americans, and 14% in Chinese.[14](#cts13877-bib-0014) ^14^14
|
| 34 |
+
|
| 35 |
+
Currently, clinical studies of mavacamten in healthy participants and patients with oHCM have been conducted in the United States, the European Union, the United Kingdom, Australia, and Japan. The current bridging study (LB2001‐101) was designed to determine the PK and safety of mavacamten in healthy adult Chinese participants with different genotypes of CYP2C19.
|
| 36 |
+
|
| 37 |
+
## METHODS
|
| 38 |
+
|
| 39 |
+
### Study design and participants
|
| 40 |
+
|
| 41 |
+
This open‐label, parallel‐group, phase I trial (LB2001‐101) was conducted at a single center in China. Eligible patients were healthy Chinese adults (aged 18–60 years) who were CYP2C19 ultra‐rapid metabolizers (UM; *17/*17), CYP2C19 rapid metabolizers (RM; *1/*17), CYP2C19 NM (*1/*1), CYP2C19 intermediate metabolizers (IM; *1/*2 or *1/*3), or CYP2C19 PM (*2/*2, *3/*3, or *2/*3) as per central laboratory phenotype determination. Participants had a body mass index (BMI) of ≥18 and ≤30 kg/m^2^2 and a resting left ventricular ejection fraction (LVEF) ≥55% by echocardiography. Before dosing, each participant had no clinically significant abnormalities in their medical history, physical examinations, vital signs, clinical laboratory tests, or 12‐lead electrocardiograms (ECGs).
|
| 42 |
+
|
| 43 |
+
Key exclusion criteria included as follows: history of clinically significant arrhythmia, LV systolic dysfunction, or coronary artery disease; ECG showing corrected QT interval (QTc) >450 ms; positive results for human immunodeficiency virus test, hepatitis B virus, or hepatitis C virus; hypersensitivity to mavacamten or any of the components of its formulation; any known clinically significant abnormalities, conditions, or diseases that would pose a risk to participant's safety or interfere with study evaluation, procedures, or its completion. Details of the inclusion and exclusion criteria are provided in the Supplementary Information, and inclusion and exclusion criteria.
|
| 44 |
+
|
| 45 |
+
All prescribed medication was prohibited from 28 days before screening through the end of study, and all over‐the‐counter medication (including herbal preparations and nutritional supplements, but not including up to 1.5 g acetaminophen daily) was prohibited from 14 days before screening through the end of study.
|
| 46 |
+
|
| 47 |
+
The study was performed in accordance with the principles of the Declaration of Helsinki, the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use Guideline for Good Clinical Practice, and local applicable regulatory requirements for clinical studies. All participants provided written informed consent. The study protocol, amendments, and informed consent were approved by the institutional review board/ethics committee at the study center. The trial is registered at [ClinicalTrials.gov](http://clinicaltrials.gov)ClinicalTrials.gov, number: [NCT05135871](https://clinicaltrials.gov/ct2/show/NCT05135871)NCT05135871.
|
| 48 |
+
|
| 49 |
+
### End points
|
| 50 |
+
|
| 51 |
+
The primary objective was to characterize PK after a single, fasted, oral dose of mavacamten in healthy adult Chinese participants with different CYP2C19 genotypes. End points included area under the concentration–time curve from time zero to the last quantifiable concentration (AUC_0−last_0−last), area under the concentration–time curve from time zero extrapolated to infinity (AUC_0−inf_0−inf), maximum concentration (*C*C _max_max), time to maximum concentration (*T*T _max_max), apparent terminal elimination half‐life (*T*T _½_½), apparent volume of distribution (*V*V _ d _ *d*d /*F*F), and apparent clearance (CL/F). Secondary objectives were safety and tolerability, which were assessed via adverse events (AEs), vital signs, physical examination findings, ECG parameters, and clinical laboratory tests.
|
| 52 |
+
|
| 53 |
+
### Procedures
|
| 54 |
+
|
| 55 |
+
CYP2C19 genotypes were analyzed by the Beijing Prohealth Clinical Laboratory Co., Ltd (Beijing, China) using the ABI 7500 Real‐Time PCR System. Participants were divided into four cohorts according to CYP2C19 genotype (Cohort 1: CYP2C19 UM/RM/NM, 15 mg mavacamten; Cohort 2: CYP2C19 UM/RM/NM, 25 mg mavacamten; Cohort 3: CYP2C19 IM, 15 mg mavacamten; and Cohort 4: CYP2C19 PM, 15 mg mavacamten). For participants with CYP2C19 UM/RM/NM, enrollment in Cohort 1 was followed by Cohort 2. Participants could be enrolled in Cohort 3 and Cohort 4 in parallel with Cohort 1. The study period for each participant was up to 16 weeks (Figure [S1](#cts13877-supitem-0001)S1).
|
| 56 |
+
|
| 57 |
+
A single oral dose of mavacamten under fasted conditions was administered to each participant to evaluate the PK profile. Participants in Cohorts 1, 3, and 4 received 15 mg mavacamten, and participants in Cohort 2 received 25 mg mavacamten. Dose selection was based on findings from previous clinical or PK studies of mavacamten. To bridge PK data in the Chinese population with prior clinical or PK mavacamten studies, participants with CYP2C19 genotypes of UM/NM/RM received a single fasted dose of 25 mg mavacamten (Cohort 2), the same dose received by healthy Japanese and Caucasian participants with CYP2C19 UM/NM/RM; participants with CYP2C19 genotypes of IM or PM received a single fasted dose of 15 mg mavacamten (Cohorts 3 and 4, respectively), the same dose received by healthy Japanese participants with CYP2C19 IM and healthy Asian participants with CYP2C19 PM.[13](#cts13877-bib-0013) ^13^13 To evaluate the PK profiles of mavacamten in Chinese participants with different genotypes and make an ethnic comparison with PK findings for other populations, Cohorts 1, 3, and 4 received the same dose of 15 mg mavacamten.
|
| 58 |
+
|
| 59 |
+
### PK assessments
|
| 60 |
+
|
| 61 |
+
Blood samples (3 mL whole blood) were collected at each timepoint for PK analysis, stored at the study site at −20°C to −90°C, and shipped to a central laboratory for drug concentration analysis. The PK parameters of mavacamten were assessed pre‐dose (within 60 min before dosing), 10, 20, 30, 45 min, 1, 1.5, 2, 3, 4, 8, 12, 24 and 48 h post‐dose. Additional blood samples were also collected at Days 7, 10, 14, 21, 28, 35, 45, 60, and 75 (±1 day). The determination of mavacamten plasma concentrations was performed by Q Squared Solutions Co., Ltd (Beijing, China), using validated liquid chromatography with the tandem mass spectrometry method. The lower limit of quantification was set at 0.2 ng/mL, and the upper limit of quantification was 200 ng/mL. Calibration standards were prepared fresh for each analytical batch and used to generate a weighted (1/*x*x ^2^2) linear regression calibration curve with a dynamic range of 0.200–200 ng/mL. Quality control performance (accuracy and precision) was within ±15% (±20.0% at LLOQ) at four concentrations (0.2, 0.6, 75, and 150 ng/mL) across method qualification and sample analysis batches. A LC/MS/MS method for the quantitation of mavacamten in 50.0 μL of K_2_2EDTA human plasma has been validated over the concentration range of 0.200–200 ng/mL. The validity of preparing samples using a 20‐fold dilution factor was demonstrated. All stability experiments conducted during this validation showed no indication of instability.
|
| 62 |
+
|
| 63 |
+
### Safety assessment
|
| 64 |
+
|
| 65 |
+
AEs, vital signs, laboratory tests, and ECGs were recorded throughout the trial. AEs were coded according to the Medical Dictionary for Regulatory Activities Terminology version 24.1 and graded per the National Cancer Institute Common Terminology Criteria for Adverse Events version 5.0.
|
| 66 |
+
|
| 67 |
+
### Sample size estimation
|
| 68 |
+
|
| 69 |
+
Approximately 8 to 12 participants in each cohort were expected to provide sufficient data to allow the assessment of the safety, tolerability, and PK profile of mavacamten in Chinese participants based on Chinese regulatory considerations.
|
| 70 |
+
|
| 71 |
+
### Statistical analysis
|
| 72 |
+
|
| 73 |
+
The PK analysis set included all participants exposed to mavacamten with at least one plasma concentration result and no serious protocol violations affecting PK parameter results. The safety analysis set included all participants exposed to mavacamten, regardless of dose. The safety analysis period was defined as the time between study drug administration and the end‐of‐study visit.
|
| 74 |
+
|
| 75 |
+
The mean (± standard deviation [SD]) concentration–time curve was plotted for each cohort. The PK parameter data were listed and summarized descriptively. To examine the exposure to mavacamten for participants with the UM/RM/NM versus IM versus PM genotype, an analysis of variance model was constructed using log‐transformed AUC_0−last_0−last, AUC_0−inf_0−inf, and *C*C _max_max. The dependent variable was the ln‐transformed primary PK parameters (AUC_0−last_0−last, AUC_0−inf_0−inf, and *C*C _max_max), with the cohort as independent variable. The AUC_0−last_0−last, AUC_0−inf_0−inf, and *C*C _max_max were compared between Cohort 1 and Cohort 2 (CYP2C19 UM/RM/NM), Cohort 3 (CYP2C19 IM), and Cohort 4 (CYP2C19 PM), respectively. The geometric mean ratio between cohorts and 90% confidence intervals were calculated for each parameter (with Cohort 1 as the reference group for data analysis).
|
| 76 |
+
|
| 77 |
+
The PK parameters were calculated using a non‐compartmental model by WinNonlin version 8.3.4. Other statistical analyses were performed using SAS statistical software version 9.4.
|
| 78 |
+
|
| 79 |
+
## RESULTS
|
| 80 |
+
|
| 81 |
+
### Demographics
|
| 82 |
+
|
| 83 |
+
A total of 45 healthy Chinese participants were enrolled and assigned to four cohorts according to CYP2C19 genotype: Cohort 1 (12 participants), Cohort 2 (13 participants), Cohort 3 (12 participants), and Cohort 4 (eight participants).
|
| 84 |
+
|
| 85 |
+
A total of 44 participants received the study drug and completed the study. One participant in Cohort 2 withdrew from the study prior to the administration of the study drug due to an abnormal ECG and was excluded from the PK analysis set and safety analysis set. One additional participant was enrolled in Cohort 2 due to the withdrawal of the aforementioned participant, resulting in a total of 13 participants enrolled in Cohort 2. One participant in Cohort 3 was excluded from the PK analysis set due to a major protocol deviation of using the prohibited moderate CYP2C19 inhibitor omeprazole, which has the potential to affect the PK parameters. As a result, 44 participants were included in the safety analysis (Cohort 1, *n*n = 12; Cohort 2, *n*n = 12; Cohort 3, *n*n = 12; Cohort 4, *n*n = 8); 43 participants were included in the PK analysis (Cohort 1, *n*n = 12; Cohort 2, *n*n = 12; Cohort 3, *n*n = 11; Cohort 4, *n*n = 8).
|
| 86 |
+
|
| 87 |
+
The participants had a median age of 30 years, a median BMI of 23.8 kg/m^2^2, and a median LVEF of 67.5% (Table [1](#cts13877-tbl-0001)1). Most participants (*n*n = 33, 75%) were male. For Cohorts 1 and 2, one (8.3%) participant had CYP2C19 RM (*1/*17) genotype and 11 (91.7%) had CYP2C19 NM (*1/*1) genotype in each cohort. For Cohort 3, 10 (83.3%) participants had CYP2C19 IM (*1/*2) genotype and two (16.7%) had CYP2C19 IM (*1/*3) genotype. For Cohort 4, six (75.0%) participants had CYP2C19 PM (*2/*2) genotype and two (25.0%) had CYP2C19 PM (*2/*3) genotype.
|
| 88 |
+
|
| 89 |
+
### TABLE 1.
|
| 90 |
+
|
| 91 |
+
Participant demographics and baseline characteristics (safety analysis set).
|
| 92 |
+
|
| 93 |
+
| | Cohort 1 (N = 12) | Cohort 2 (N = 12) | Cohort 3 (N = 12) | Cohort 4 (N = 8) | Total (N = 44) |
|
| 94 |
+
| - | ----------------- | ----------------- | ----------------- | ---------------- | -------------- |
|
| 95 |
+
| Median age, years (range) | 26.0 (20–35) | 28.5 (21–45) | 30.5 (19–42) | 35.0 (30–43) | 30.0 (19–45) |
|
| 96 |
+
| Gender, n (%) | | | | | |
|
| 97 |
+
| Male | 9 (75.0) | 9 (75.0) | 9 (75.0) | 6 (75.0) | 33 (75.0) |
|
| 98 |
+
| Female | 3 (25.0) | 3 (25.0) | 3 (25.0) | 2 (25.0) | 11 (25.0) |
|
| 99 |
+
| Median body mass index a , kg/m2 (range) | 24.6 (20.9–28.4) | 24.4 (19.5–27.0) | 22.8 (20.7–27.4) | 22.3 (20.3–26.8) | 23.8 (19.5–28.4) |
|
| 100 |
+
| CYP2C19 genotype, n (%) | | | | | |
|
| 101 |
+
| UM | | | | | |
|
| 102 |
+
| *17/*17 | 0 | 0 | 0 | 0 | 0 |
|
| 103 |
+
| RM | | | | | |
|
| 104 |
+
| *1/*17 | 1 (8.3) | 1 (8.3) | 0 | 0 | 2 (4.5) |
|
| 105 |
+
| NM | | | | | |
|
| 106 |
+
| *1/*1 | 11 (91.7) | 11 (91.7) | 0 | 0 | 22 (50.0) |
|
| 107 |
+
| IM | | | | | |
|
| 108 |
+
| *1/*2 | 0 | 0 | 10 (83.3) | 0 | 10 (22.7) |
|
| 109 |
+
| *1/*3 | 0 | 0 | 2 (16.7) | 0 | 2 (4.5) |
|
| 110 |
+
| PM | | | | | |
|
| 111 |
+
| *2/*2 | 0 | 0 | 0 | 6 (75.0) | 6 (13.6) |
|
| 112 |
+
| *3/*3 | 0 | 0 | 0 | 0 | 0 |
|
| 113 |
+
| *2/*3 | 0 | 0 | 0 | 2 (25.0) | 2 (4.5) |
|
| 114 |
+
| Median LVEF, % (range) | 69.5 (61–74) | 66.5 (61–74) | 66.5 (61–72) | 66.0 (55–69) | 67.5 (55–74) |
|
| 115 |
+
### PK analysis
|
| 116 |
+
|
| 117 |
+
The mean plasma concentration–time curves of each cohort following a single oral dose of mavacamten are shown in Figure [1](#cts13877-fig-0001)1.
|
| 118 |
+
|
| 119 |
+
### FIGURE 1.
|
| 120 |
+
|
| 121 |
+

|
| 122 |
+
|
| 123 |
+
Mean (±SD) mavacamten plasma concentration–time data through 24 h (a) and 0–1776 h (b) per cohort in linear scale and semi‐logarithmic scale. Mavacamten was rapidly absorbed after a single oral dose. Datapoints represent mean ± standard deviation. Cohort 1: CYP2C19 UM/RM/NM, 15 mg mavacamten; Cohort 2: CYP2C19 UM/RM/NM, 25 mg mavacamten; Cohort 3: CYP2C19 IM, 15 mg mavacamten; Cohort 4: CYP2C19 PM, 15 mg mavacamten. CYP, cytochrome P450; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; RM, rapid metabolizer; UM, ultra‐rapid metabolizer.
|
| 124 |
+
|
| 125 |
+
After a single oral dose of mavacamten in fasted healthy adult Chinese individuals, the *C*C _max_max was reached within a median *T*T _max_max of 0.6–1.5 h, indicating rapid absorption. The inter‐individual variability was moderate, as the ranges of geometric coefficient of variation (CV%) for *C*C _max_max, AUC_0−last_0−last, AUC_0−inf_0−inf, and *T*T _½_½ were 25.7–40.0%, 21.1–50.5%, 21.2–52.5%, and 22.3–42.3%, respectively. The PK profiles of mavacamten varied among different cohorts, with a geometric mean *T*T _½_½ of 120.3, 143.6, 205.4, and 572.0 h; geometric mean AUC_0−last_0−last of 9956, 20,040, 18,270, and 39,980 h*ng/mL; and geometric mean AUC_0−inf_0−inf of 10,030, 20,280, 18,430, and 45,810 h*ng/mL for Cohorts 1 to 4, respectively. Individuals carrying non‐functional CYP2C19 alleles exhibited longer half‐life and increased total exposure (Table [2](#cts13877-tbl-0002)2).
|
| 126 |
+
|
| 127 |
+
### TABLE 2.
|
| 128 |
+
|
| 129 |
+
PK parameters of mavacamten following a single oral administration of mavacamten (PK analysis set).
|
| 130 |
+
|
| 131 |
+
| | Cohort 1 UM/RM/NM 15 mg (N = 12) | Cohort 2 UM/RM/NM 25 mg (N = 12) | Cohort 3 IM 15 mg (N = 11) | Cohort 4 PM 15 mg (N = 8) |
|
| 132 |
+
| - | -------------------------------- | -------------------------------- | -------------------------- | ------------------------- |
|
| 133 |
+
| C max (μg/mL), geometric mean (CV%) | 395.4 (40.0) | 571.8 (25.7) | 485.2 (35.0) | 435.7 (35.7) |
|
| 134 |
+
| T max (h), median (range) | 0.9 (0.5–3.0) | 1.5 (0.5–4.0) | 0.8 (0.5–3.0) | 0.6 (0.3–1.0) |
|
| 135 |
+
| T ½ (h), geometric mean (CV%) | 120.3 (23.4) | 143.6 (42.3) | 205.4 (22.3) | 572.0 (25.0) |
|
| 136 |
+
| AUC0−last (h*ng/mL), geometric mean (CV%) | 9956 (35.6) | 20,040 (50.5) | 18,270 (21.1) | 39,980 (23.6) |
|
| 137 |
+
| AUC0−inf (h*ng/mL), geometric mean (CV%) | 10,030 (35.4) | 20,280 (52.5) | 18,430 (21.2) | 45,810 (25.5) |
|
| 138 |
+
| %AUCex, geometric mean (CV%) | 0.7 (44.9) | 0.7 (112.2) | 0.8 (25.9) | 11.2 (53.6) |
|
| 139 |
+
| CL/F (mL/h), geometric mean (CV%) | 1495.0 (35.4) | 1233.0 (52.5) | 814.0 (21.2) | 327.5 (25.5) |
|
| 140 |
+
| V d /F (mL), geometric mean (CV%) | 259,500 (29.9) | 255,300 (16.6) | 241,200 (23.1) | 270,200 (27.0) |
|
| 141 |
+
Table [3](#cts13877-tbl-0003)3 shows the comparison of PK parameters of mavacamten between cohorts. The geometric mean ratio showed that total exposure in the IM group (Cohort 3) was approximately 1.8‐fold of that in the UM/RM/NM group (Cohort 1); total exposure in the PM group (Cohort 4) was approximately fourfold of that in the UM/RM/NM group. In the UM/RM/NM groups, total exposure of Cohort 2 (25 mg mavacamten) was around twofold of that in Cohort 1 (15 mg mavacamten). Increased exposure in the PM group versus the UM/RM/NM group is supported by the box plots of dose‐normalized *C*C _max_max, AUC_0−last_0−last, and AUC_0−inf_0−inf (Figure [2](#cts13877-fig-0002)2). Among individuals with the same CYP2C19 genotypes (UM/RM/NM), mavacamten exposure normalized by dose was comparable between groups regarding *C*C _max_max, AUC_0−last_0−last, and AUC_0−inf_0−inf after administering 25 mg and 15 mg doses (Cohort 2 and Cohort 1) (Figure [2](#cts13877-fig-0002)2).
|
| 142 |
+
|
| 143 |
+
### TABLE 3.
|
| 144 |
+
|
| 145 |
+
Comparison of PK parameters of mavacamten between cohorts (PK analysis set).
|
| 146 |
+
|
| 147 |
+
| Compared cohort | C max (μg/mL), geometric mean ratio (95% CI) | AUC0−last (h*ng/mL), geometric mean ratio (95% CI) | AUC0−inf (h*ng/mL), geometric mean ratio (95% CI) |
|
| 148 |
+
| --------------- | -------------------------------------------- | -------------------------------------------------- | ------------------------------------------------- |
|
| 149 |
+
| Cohort 2/Cohort 1 | 1.446 (1.15, 1.82) | 2.013 (1.59, 2.55) | 2.021 (1.59, 2.58) |
|
| 150 |
+
| Cohort 3/Cohort 1 | 1.227 (0.97, 1.55) | 1.835 (1.44, 2.34) | 1.837 (1.43, 2.35) |
|
| 151 |
+
| Cohort 4/Cohort 1 | 1.102 (0.85, 1.42) | 4.016 (3.08, 5.23) | 4.566 (3.48, 5.99) |
|
| 152 |
+
### FIGURE 2.
|
| 153 |
+
|
| 154 |
+

|
| 155 |
+
|
| 156 |
+
Box plots of dose‐normalized C max (a), AUC0−last (b), and AUC0−inf (c) of mavacamten (PK analysis set). Total exposures of mavacamten in the IM group (Cohort 3) and the PM group (Cohort 4) were higher than those in the UM/RM/NM groups (Cohorts 1 and 2). Datapoints represent minimum, first quartile, median, third quartile, and maximum. AUC0−inf, area under the concentration–time curve from time zero extrapolated to infinity; AUC0−last, area under the concentration–time curve from time zero to the last quantifiable concentration; C max, maximum concentration; IM, intermediate metabolizer; NM, normal metabolizer; PK, pharmacokinetics; PM, poor metabolizer; RM, rapid metabolizer; UM, ultra‐rapid metabolizer.
|
| 157 |
+
|
| 158 |
+
There was one outlier in Cohort 2 who met all the inclusion criteria and none of the exclusion criteria, with AUC parameters 3.8 to 6.4 times higher than other participants in the same cohort. After removing the outlier in Cohort 2, the geometric mean of inter‐individual *C*C _max_max, AUC_0−last_0−last, and AUC_0−inf_0−inf was 559.0 (CV%: 25.6%) ng/mL, 17,570 (CV%: 14.6%) h*ng/mL, and 17,690 (CV%: 14.5%) h*ng/mL, respectively. The geometric mean was 128.6 (CV%: 15.1%) h for T_½_½, 1413 (CV%: 14.5%) mL/h for CL/F, and 262,300 (CV%: 14.3%) mL for *V*V _ d _ *d*d /*F*F. The PK parameters of mavacamten in Cohort 2 with or without the outlier are presented in Table [S1](#cts13877-supitem-0001)S1. Comparing Cohort 2 without the outlier with Cohort 1, the geometric mean ratio of *C*C _max_max was approximately 1.4‐fold, and for AUC_0−last_0−last and AUC_0−inf_0−inf, approximately 1.8‐fold (Table [S2](#cts13877-supitem-0001)S2).
|
| 159 |
+
|
| 160 |
+
### Safety
|
| 161 |
+
|
| 162 |
+
The overview of treatment‐emergent AEs (TEAEs) is listed in Table [4](#cts13877-tbl-0004)4 and Table [S3](#cts13877-supitem-0001)S3. A total of 21 (47.7%) participants experienced at least one TEAE during the study. Only one participant in Cohort 3 reported one moderate TEAE (abdominal pain), while all other reported TEAEs were mild in severity. The one moderate TEAE was considered not related to the study drug. The most frequent TEAEs were upper respiratory tract infection (20.5%), blood fibrinogen increased (6.8%), and chest pain (6.8%). One participant from Cohort 2 (chest pain) and one participant from Cohort 4 (hemoglobin decreased) each had a single mild TEAE related to the study drug, whereas the TEAEs reported by the remaining participants were assessed as not related to the study drug.
|
| 163 |
+
|
| 164 |
+
### TABLE 4.
|
| 165 |
+
|
| 166 |
+
Overview of TEAEs (safety analysis set).
|
| 167 |
+
|
| 168 |
+
| n (%) | Cohort 1 (N = 12) | Cohort 2 (N = 12) | Cohort 3 (N = 12) | Cohort 4 (N = 8) | Total (N = 44) |
|
| 169 |
+
| ----- | ----------------- | ----------------- | ----------------- | ---------------- | -------------- |
|
| 170 |
+
| TEAEs | 3 (25.0) | 6 (50.0) | 9 (75.0) | 3 (37.5) | 21 (47.7) |
|
| 171 |
+
| Worst severity of TEAE | | | | | |
|
| 172 |
+
| Mild | 3 (25.0) | 6 (50.0) | 8 (66.7) | 3 (37.5) | 20 (45.5) |
|
| 173 |
+
| Moderate | 0 | 0 | 1 (8.3) | 0 | 1 (2.3) |
|
| 174 |
+
| Severe | 0 | 0 | 0 | 0 | 0 |
|
| 175 |
+
| TEAEs related to the study drug | 0 | 1 (8.3) | 0 | 1 (12.5) | 2 (4.5) |
|
| 176 |
+
| Serious TEAEs | 0 | 0 | 0 | 0 | 0 |
|
| 177 |
+
| TEAEs leading to treatment discontinuation | 0 | 0 | 0 | 0 | 0 |
|
| 178 |
+
| TEAEs leading to death | 0 | 0 | 0 | 0 | 0 |
|
| 179 |
+
| AESI | 0 | 0 | 0 | 0 | 0 |
|
| 180 |
+
| TEAEs occurred in ≥2 participants of total | | | | | |
|
| 181 |
+
| Upper respiratory tract infection | 2 (16.7) | 3 (25.0) | 3 (25.0) | 1 (12.5) | 9 (20.5) |
|
| 182 |
+
| Blood fibrinogen increased | 0 | 0 | 2 (16.7) | 1 (12.5) | 3 (6.8) |
|
| 183 |
+
| Chest pain | 0 | 2 (16.7) | 1 (8.3) | 0 | 3 (6.8) |
|
| 184 |
+
| White blood cell count decreased | 0 | 1 (8.3) | 0 | 1 (12.5) | 2 (4.5) |
|
| 185 |
+
| Headache | 0 | 1 (8.3) | 1 (8.3) | 0 | 2 (4.5) |
|
| 186 |
+
TEAEs related to the potential risk of mavacamten, symptomatic LVEF <50%, were not observed in this study. No serious TEAEs or AEs of special interest (including symptomatic overdose, and LVEF ≤30%) were reported in this study. No significant trends of shifts from baseline and no notable differences among the cohorts in terms of laboratory parameters, ECGs, vital signs, and physical examination results were observed.
|
| 187 |
+
|
| 188 |
+
## DISCUSSION
|
| 189 |
+
|
| 190 |
+
This study investigated the PK of single‐dose mavacamten in healthy adult Chinese participants with different phenotypes of CYP2C19, UM/RM/NM, IM, and PM. The results indicated rapid absorption after a single oral administration of 15 or 25 mg mavacamten, and the inter‐individual variability was moderate. Prolongations of T_½_½ and increased exposure for participants carrying non‐functional CYP2C19 alleles were observed. The total exposures for CYP2C19 IM and CYP2C19 PM were increased approximately 1.8‐ to 4‐fold when compared to CYP2C19 UM/RM/NM. Overall, the PK profile of mavacamten in the Chinese population was consistent with the previously known PK profile of mavacamten in other populations. No apparent difference in PK stratified by ethnicity was observed, once corrected for metabolizer status.
|
| 191 |
+
|
| 192 |
+
There was an increase in mavacamten exposure between 1056 and 1440 h in a semi‐log scale. This was due to an outlier in Cohort 2, who had the AUC parameters 3.8‐ to 6.4‐fold higher than other participants in the same cohort. In order to assess the impact of the outlier on PK evaluation, an ad hoc analysis was carried out by removing this particular participant from the PK analysis. The outlier in Cohort 2 did not experience TEAEs, took no concomitant medications, and no protocol deviations were recorded during the study. There were no clinically significant abnormalities in the safety tests for this outlier. All the PK parameters were similar with or without the outlier in Cohort 2. The total exposure of mavacamten in participants was approximately 1.4‐ to 1.8‐fold when the dose ratio of mavacamten was 1.67 times (25 mg/15 mg) among CYP2C19 UM/RM/NM genotypes, which was closer to a dose–concentration proportional relationship. The outlier did not significantly affect the conclusions and the titration plan for mavacamten in Chinese patients based on this study. The existence of an outlier showing higher AUC parameters suggested that the PK variability cannot be entirely accounted for by genotype alone. Therefore, an individualized titration scheme based on clinical status and echocardiographic assessment of patient response was adopted for mavacamten. Dose titration starting from 5 mg based on LVEF,, and Valsalva LVOT gradient is recommended for mavacamten per the FDA label.[13](#cts13877-bib-0013) ^13^13 While CYP2C19 genotyping for mavacamten initial dose determination is recommended per the EMA label,[15](#cts13877-bib-0015) ^15^15 individualized dosing based on CYP2C19 genotypes to reach the stable dose is not proposed.[16](#cts13877-bib-0016) ^16^16 Genotyping may be a helpful tool; however, it has not been routinely implemented in prospective trials and is not mandated for optimization globally.[13](#cts13877-bib-0013) ^13^13 In addition, no particular safety issues were observed in the IM/PM metabolizers. However, moderate to strong CYP2C19 inhibitors/inducers should be avoided when taking mavacamten.
|
| 193 |
+
|
| 194 |
+
PK parameters derived from this study (LB2001‐101) were compared with those obtained from prior studies in diverse ethnic populations who received a single dose of mavacamten. Mavacamten was quickly absorbed in this current study and was similar in the studies of healthy Japanese and Caucasian participants (*T*T _max_max: 1.0 h).[13](#cts13877-bib-0013) ^13^13 The PK of once‐daily oral mavacamten in healthy volunteers was approximately dose proportional within a dose range of 1–15 mg,[7](#cts13877-bib-0007) ^7^7 but less than dose proportional between 15 and 25 mg in Japanese participants. In Cohorts 1 and 2 of this study, the results confirmed that the exposure of mavacamten was approximately proportional between 15 and 25 mg mavacamten administered in Chinese participants with CYP2C19 UM/RM/NM. The findings of prolonged clearance and increased exposure in CYP2C19 IM and PM compared with CYP2C19 UM/RM/NM are consistent with results from previous studies.[13](#cts13877-bib-0013) ^13^13 Furthermore, after stratification by CYP2C19 genotype, mavacamten exposures were consistent between Chinese and Caucasians, consistent with the understanding based on previous clinical pharmacological studies of mavacamten. The PK profile for mavacamten was also consistent across various populations/ethnicities once corrected for CYP2C19 phenotype; therefore, dose adjustment based on ethnicity is not warranted.
|
| 195 |
+
|
| 196 |
+
Single oral administration of mavacamten was found to be safe and well tolerated in healthy participants. There were no serious TEAEs, TEAEs leading to study discontinuation, severe TEAEs, or AEs of special interest observed in this study. The most frequent TEAE was upper respiratory tract infection in the current study, all instances of which were found to be not related to the study drug. Among different CYP2C19 genotypes, there were no notable differences observed and safety results were similar. The risk of heart failure is a black box warning in the United States Prescribing Information for mavacamten.[13](#cts13877-bib-0013) ^13^13 No QT prolongation, symptomatic LVEF <50%, or cardiac failure were observed in the healthy participants in this study. This observation is consistent with prior studies, in which cardiac toxicity was not observed during single dosing.[13](#cts13877-bib-0013) ^13^13 It should be noted that the observation of cardiac adverse effect might be limited due to single dosing in the current study. None of the AEs of special interest, selected based on the phase III EXPLORER‐CN study,[17](#cts13877-bib-0017) ^17^17 were reported in our study. The reduction in LVEF is one safety risk associated with mavacamten, and dose interruption or adjustment is required based on LVEF assessment.[13](#cts13877-bib-0013) ^13^13 In a prior study, an exposure–response model was developed and the simulation suggested that the proportion of patients with LVEF <50% ranged 0.9%–2% at Week 40 across doses from 2.5 to 15 mg.[18](#cts13877-bib-0018) ^18^18 However, LVEF <50% was not observed in this study up to 25 mg. Again, this might be related to the single dosing. Additionally, prior clinical studies of mavacamten showed that the safety and tolerability were similar between mavacamten and placebo.[16](#cts13877-bib-0016) ^16^16 , [19](#cts13877-bib-0019) ^19^19 In our study, though the increase in exposure was fourfold and twofold in participants with CYP2C19 PM and IM, respectively, the incidence of TEAEs was not higher in CYP2C19 PM. Furthermore, only one TEAE each was reported in CYP2C19 UM/RM/NM (25 mg mavacamten) and CYP2C19 PM (15 mg mavacamten). This suggests that AEs do not increase with mavacamten dose, up to 25 mg in healthy participants. Therefore, individualized titration based on clinical status and echocardiographic assessment of patient response is recommended. Overall, single oral administration of mavacamten up to 25 mg in healthy Chinese participants showed a good safety and tolerability profile across all dose groups and genotypes, although the PK analysis showed different exposure levels of mavacamten in different dose groups and different CYP2C19 genotypes.
|
| 197 |
+
|
| 198 |
+
Mavacamten represents a significant advancement in the treatment of oHCM, offering a targeted approach to addressing the underlying pathophysiology of the disease. It is expected to have a major impact on the management of oHCM, improving oHCM symptoms and decreasing the need for surgical intervention.[1](#cts13877-bib-0001) ^1^1 , [8](#cts13877-bib-0008) ^8^8 , [12](#cts13877-bib-0012) ^12^12 Individual variation of PKs was observed in our study, indicating the importance of individualized titration according to the clinical response and safety monitoring for patients receiving mavacamten. The findings from this study support the individualized dose regimen based on clinical characteristics in a phase III trial that enrolled Chinese patients with symptomatic oHCM (EXPLORER‐CN; [NCT05174416](https://clinicaltrials.gov/ct2/show/NCT05174416)NCT05174416), which showed that mavacamten significantly improved Valsalva LVOT obstruction, New York Heart Association functional class, health status, and cardiac structure compared with placebo in these patients, regardless of CYP2C19 phenotype.[16](#cts13877-bib-0016) ^16^16
|
| 199 |
+
|
| 200 |
+
In conclusion, single‐dose PK of mavacamten in healthy Chinese participants in this study revealed rapid absorption and moderate inter‐individual variability. The total exposure of mavacamten varied depending on CYP2C19 genotypes, with higher exposure in participants with CYP2C19 PM, consistent with the understanding of mavacamten's disposition. Mavacamten was well tolerated overall. No cardiac AEs were observed in this study. No safety signals emerged in participants with CYP2C19 IM or PM. The PK profile of mavacamten in a healthy Chinese population was consistent with that in other healthy populations from prior studies in other ethnicities.
|
| 201 |
+
|
| 202 |
+
## AUTHOR CONTRIBUTIONS
|
| 203 |
+
|
| 204 |
+
P.H. wrote the manuscript. C.L. and J.Z. designed the study. X.W., N.C., Q.W., Q.W., J.Y., G.C., H.Y., L.W., J.W., Y.J., W.L., J.W., and J.H. performed the study. P.H., J.S., S.M., and W.L. analyzed the data.
|
| 205 |
+
|
| 206 |
+
## FUNDING INFORMATION
|
| 207 |
+
|
| 208 |
+
This work was supported by the Science and Technology Commission of Shanghai Municipality (22S11904701, 22S11904702), Shanghai LianBio Development Co., Ltd, and Bristol Myers Squibb.
|
| 209 |
+
|
| 210 |
+
## CONFLICT OF INTEREST STATEMENT
|
| 211 |
+
|
| 212 |
+
P. H. and C. L. are employees of Shanghai LianBio Development Co., Ltd. J. S., Q. W., and M. S. are employees of Bristol Myers Squibb. All other authors declared no competing interests for this work.
|
| 213 |
+
|
| 214 |
+
## Supporting information
|
| 215 |
+
|
| 216 |
+
## ACKNOWLEDGMENTS
|
| 217 |
+
|
| 218 |
+
We acknowledge funding from the Science and Technology Commission of Shanghai Municipality, and support from Manting Chiang, Shilpa Puli, and Vidya Perera of Bristol Myers Squibb. Vidya Perera was an employee of Bristol Myers Squibb at the time the study was conducted. Medical writing assistance was provided by Jing Yi Lee and Molly Yu from Parexel, which was funded by Shanghai LianBio Development Co., Ltd, and Bristol Myers Squibb.
|
| 219 |
+
|
| 220 |
+
Wu X, Chen N, Hsu P, et al. Pharmacokinetics and safety of mavacamten in healthy Chinese participants with different CYP2C19 phenotypes. Clin Transl Sci. 2024;17:e13877. doi: 10.1111/cts.13877
|
| 221 |
+
|
| 222 |
+
## Contributor Information
|
| 223 |
+
|
| 224 |
+
Cheng Lyu, Email: levvy.lv@aliyun.com.
|
| 225 |
+
|
| 226 |
+
Jing Zhang, Email: zhangj61@fudan.edu.cn.
|
| 227 |
+
|
| 228 |
+
## DATA AVAILABILITY STATEMENT
|
| 229 |
+
|
| 230 |
+
The data underlying this article will be shared upon a reasonable request.
|
| 231 |
+
|
| 232 |
+
## Associated Data
|
| 233 |
+
|
| 234 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 235 |
+
|
| 236 |
+
### Supplementary Materials
|
| 237 |
+
|
| 238 |
+
### Data Availability Statement
|
| 239 |
+
|
| 240 |
+
The data underlying this article will be shared upon a reasonable request.
|
| 241 |
+
|
| 242 |
+
### Supplementary Materials
|
| 243 |
+
|
| 244 |
+
### Data Availability Statement
|
| 245 |
+
|
| 246 |
+
The data underlying this article will be shared upon a reasonable request.
|
| 247 |
+
|
| 248 |
+
## References
|
| 249 |
+
|
| 250 |
+
1. Edelberg JM, Sehnert AJ, Mealiffe ME, Del Rio CL, McDowell R. The impact of Mavacamten on the pathophysiology of hypertrophic cardiomyopathy: a narrative review. Am J Cardiovasc Drugs. 2022;22(5):497‐510. doi: 10.1007/s40256-022-00532-x [DOI](https://doi.org/10.1007/s40256-022-00532-x) | [PMC free article](/articles/PMC9467968/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35435607/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Cardiovasc%20Drugs&title=The%20impact%20of%20Mavacamten%20on%20the%20pathophysiology%20of%20hypertrophic%20cardiomyopathy:%20a%20narrative%20review&author=JM%20Edelberg&author=AJ%20Sehnert&author=ME%20Mealiffe&author=CL%20Del%20Rio&author=R%20McDowell&volume=22&issue=5&publication_year=2022&pages=497-510&pmid=35435607&doi=10.1007/s40256-022-00532-x&)
|
| 251 |
+
|
| 252 |
+
2. Kawana M, Spudich JA, Ruppel KM. Hypertrophic cardiomyopathy: mutations to mechanisms to therapies. Front Physiol. 2022;13:975076. doi: 10.3389/fphys.2022.975076 [DOI](https://doi.org/10.3389/fphys.2022.975076) | [PMC free article](/articles/PMC9548533/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36225299/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front%20Physiol&title=Hypertrophic%20cardiomyopathy:%20mutations%20to%20mechanisms%20to%20therapies&author=M%20Kawana&author=JA%20Spudich&author=KM%20Ruppel&volume=13&publication_year=2022&pages=975076&pmid=36225299&doi=10.3389/fphys.2022.975076&)
|
| 253 |
+
|
| 254 |
+
3. Maron BA, Wang RS, Carnethon MR, et al. What causes hypertrophic cardiomyopathy? Am J Cardiol. 2022;179:74‐82. doi: 10.1016/j.amjcard.2022.06.017 [DOI](https://doi.org/10.1016/j.amjcard.2022.06.017) | [PMC free article](/articles/PMC9818026/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35843734/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Cardiol&title=What%20causes%20hypertrophic%20cardiomyopathy?&author=BA%20Maron&author=RS%20Wang&author=MR%20Carnethon&volume=179&publication_year=2022&pages=74-82&pmid=35843734&doi=10.1016/j.amjcard.2022.06.017&)
|
| 255 |
+
|
| 256 |
+
4. Ommen SR, Mital S, Burke MA, et al. 2020 AHA/ACC guideline for the diagnosis and treatment of patients with hypertrophic cardiomyopathy: a report of the American College of Cardiology/American Heart Association joint committee on clinical practice guidelines. Circulation. 2020;142(25):e558‐e631. doi: 10.1161/CIR.0000000000000937 [DOI](https://doi.org/10.1161/CIR.0000000000000937) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33215931/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circulation&title=2020%20AHA/ACC%20guideline%20for%20the%20diagnosis%20and%20treatment%20of%20patients%20with%20hypertrophic%20cardiomyopathy:%20a%20report%20of%20the%20American%20College%20of%20Cardiology/American%20Heart%20Association%20joint%20committee%20on%20clinical%20practice%20guidelines&author=SR%20Ommen&author=S%20Mital&author=MA%20Burke&volume=142&issue=25&publication_year=2020&pages=e558-e631&pmid=33215931&doi=10.1161/CIR.0000000000000937&)
|
| 257 |
+
|
| 258 |
+
5. Dong T, Alencherry B, Ospina S, Desai MY. Review of Mavacamten for obstructive hypertrophic cardiomyopathy and future directions. Drug Des Devel Ther. 2023;17:1097‐1106. doi: 10.2147/DDDT.S368590 [DOI](https://doi.org/10.2147/DDDT.S368590) | [PMC free article](/articles/PMC10094472/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37064432/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Des%20Devel%20Ther&title=Review%20of%20Mavacamten%20for%20obstructive%20hypertrophic%20cardiomyopathy%20and%20future%20directions&author=T%20Dong&author=B%20Alencherry&author=S%20Ospina&author=MY%20Desai&volume=17&publication_year=2023&pages=1097-1106&pmid=37064432&doi=10.2147/DDDT.S368590&)
|
| 259 |
+
|
| 260 |
+
6. Bayonas‐Ruiz A, Munoz‐Franco FM, Sabater‐Molina M, Oliva‐Sandoval MJ, Gimeno JR, Bonacasa B. Current therapies for hypertrophic cardiomyopathy: a systematic review and meta‐analysis of the literature. ESC Heart Fail. 2023;10(1):8‐23. doi: 10.1002/ehf2.14142 [DOI](https://doi.org/10.1002/ehf2.14142) | [PMC free article](/articles/PMC9871697/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36181355/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=ESC%20Heart%20Fail&title=Current%20therapies%20for%20hypertrophic%20cardiomyopathy:%20a%20systematic%20review%20and%20meta%E2%80%90analysis%20of%20the%20literature&author=A%20Bayonas%E2%80%90Ruiz&author=FM%20Munoz%E2%80%90Franco&author=M%20Sabater%E2%80%90Molina&author=MJ%20Oliva%E2%80%90Sandoval&author=JR%20Gimeno&volume=10&issue=1&publication_year=2023&pages=8-23&pmid=36181355&doi=10.1002/ehf2.14142&)
|
| 261 |
+
|
| 262 |
+
7. Keam SJ. Mavacamten: first approval. Drugs. 2022;82(10):1127‐1135. doi: 10.1007/s40265-022-01739-7 [DOI](https://doi.org/10.1007/s40265-022-01739-7) | [PMC free article](/articles/PMC9338109/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35802255/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drugs&title=Mavacamten:%20first%20approval&author=SJ%20Keam&volume=82&issue=10&publication_year=2022&pages=1127-1135&pmid=35802255&doi=10.1007/s40265-022-01739-7&)
|
| 263 |
+
|
| 264 |
+
8. Reyes KRL, Bilgili G, Rader F. Mavacamten: a first‐in‐class Oral modulator of cardiac myosin for the treatment of symptomatic hypertrophic obstructive cardiomyopathy. Heart Int. 2022;16(2):91‐98. doi: 10.17925/HI.2022.16.2.91 [DOI](https://doi.org/10.17925/HI.2022.16.2.91) | [PMC free article](/articles/PMC9872784/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36741099/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Heart%20Int&title=Mavacamten:%20a%20first%E2%80%90in%E2%80%90class%20Oral%20modulator%20of%20cardiac%20myosin%20for%20the%20treatment%20of%20symptomatic%20hypertrophic%20obstructive%20cardiomyopathy&author=KRL%20Reyes&author=G%20Bilgili&author=F%20Rader&volume=16&issue=2&publication_year=2022&pages=91-98&pmid=36741099&doi=10.17925/HI.2022.16.2.91&)
|
| 265 |
+
|
| 266 |
+
9. Olivotto I, Oreziak A, Barriales‐Villa R, et al. Mavacamten for treatment of symptomatic obstructive hypertrophic cardiomyopathy (EXPLORER‐HCM): a randomised, double‐blind, placebo‐controlled, phase 3 trial. Lancet. 2020;396(10253):759‐769. doi: 10.1016/S0140-6736(20)31792-X [DOI](https://doi.org/10.1016/S0140-6736(20)31792-X) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32871100/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Mavacamten%20for%20treatment%20of%20symptomatic%20obstructive%20hypertrophic%20cardiomyopathy%20(EXPLORER%E2%80%90HCM):%20a%20randomised,%20double%E2%80%90blind,%20placebo%E2%80%90controlled,%20phase%203%20trial&author=I%20Olivotto&author=A%20Oreziak&author=R%20Barriales%E2%80%90Villa&volume=396&issue=10253&publication_year=2020&pages=759-769&pmid=32871100&doi=10.1016/S0140-6736(20)31792-X&)
|
| 267 |
+
|
| 268 |
+
10. Spertus JA, Fine JT, Elliott P, et al. Mavacamten for treatment of symptomatic obstructive hypertrophic cardiomyopathy (EXPLORER‐HCM): health status analysis of a randomised, double‐blind, placebo‐controlled, phase 3 trial. Lancet. 2021;397(10293):2467‐2475. doi: 10.1016/S0140-6736(21)00763-7 [DOI](https://doi.org/10.1016/S0140-6736(21)00763-7) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34004177/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Mavacamten%20for%20treatment%20of%20symptomatic%20obstructive%20hypertrophic%20cardiomyopathy%20(EXPLORER%E2%80%90HCM):%20health%20status%20analysis%20of%20a%20randomised,%20double%E2%80%90blind,%20placebo%E2%80%90controlled,%20phase%203%20trial&author=JA%20Spertus&author=JT%20Fine&author=P%20Elliott&volume=397&issue=10293&publication_year=2021&pages=2467-2475&pmid=34004177&doi=10.1016/S0140-6736(21)00763-7&)
|
| 269 |
+
|
| 270 |
+
11. Hegde SM, Lester SJ, Solomon SD, et al. Effect of Mavacamten on echocardiographic features in symptomatic patients with obstructive hypertrophic cardiomyopathy. J Am Coll Cardiol. 2021;78(25):2518‐2532. doi: 10.1016/j.jacc.2021.09.1381 [DOI](https://doi.org/10.1016/j.jacc.2021.09.1381) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34915982/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Am%20Coll%20Cardiol&title=Effect%20of%20Mavacamten%20on%20echocardiographic%20features%20in%20symptomatic%20patients%20with%20obstructive%20hypertrophic%20cardiomyopathy&author=SM%20Hegde&author=SJ%20Lester&author=SD%20Solomon&volume=78&issue=25&publication_year=2021&pages=2518-2532&pmid=34915982&doi=10.1016/j.jacc.2021.09.1381&)
|
| 271 |
+
|
| 272 |
+
12. Desai MY, Owens A, Geske JB, et al. Myosin inhibition in patients with obstructive hypertrophic cardiomyopathy referred for septal reduction therapy. J Am Coll Cardiol. 2022;80(2):95‐108. doi: 10.1016/j.jacc.2022.04.048 [DOI](https://doi.org/10.1016/j.jacc.2022.04.048) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35798455/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Am%20Coll%20Cardiol&title=Myosin%20inhibition%20in%20patients%20with%20obstructive%20hypertrophic%20cardiomyopathy%20referred%20for%20septal%20reduction%20therapy&author=MY%20Desai&author=A%20Owens&author=JB%20Geske&volume=80&issue=2&publication_year=2022&pages=95-108&pmid=35798455&doi=10.1016/j.jacc.2022.04.048&)
|
| 273 |
+
|
| 274 |
+
13. MyoKardia, Inc., a wholly owned subsidiary of Bristol Myers Squibb. CAMZYOSTM (mavacamten): US prescribing information. 2022. Accessed April 24, 2023. https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/214998s000lbl.pdf [https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/214998s000lbl.pdf](https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/214998s000lbl.pdf)
|
| 275 |
+
|
| 276 |
+
14. Dean L, Kane M. Clopidogrel therapy and CYP2C19 genotype. In: Pratt VM, Scott SA, Pirmohamed M, Esquivel B, Kattman BL, Malheiro AJ, eds. Medical Genetics Summaries. National Center for Biotechnology Information; 2012. [PubMed](https://pubmed.ncbi.nlm.nih.gov/28520346/) | [Google Scholar](https://scholar.google.com/scholar_lookup?title=Medical%20Genetics%20Summaries&author=L%20Dean&author=M%20Kane&publication_year=2012&)
|
| 277 |
+
|
| 278 |
+
15. European Medicines Agency . Camzyos [product information]. Accessed April 08, 2024. https://www.ema.europa.eu/en/documents/product‐information/camzyos‐epar‐product‐information_en.pdf [https://www.ema.europa.eu/en/documents/product‐information/camzyos‐epar‐product‐information_en.pdf](https://www.ema.europa.eu/en/documents/product-information/camzyos-epar-product-information_en.pdf)
|
| 279 |
+
|
| 280 |
+
16. Tian Z, Li L, Li X, et al. Effect of Mavacamten on Chinese patients with symptomatic obstructive hypertrophic cardiomyopathy: the EXPLORER‐CN randomized clinical trial. JAMA Cardiol. 2023;8(10):957‐965. doi: 10.1001/jamacardio.2023.3030 [DOI](https://doi.org/10.1001/jamacardio.2023.3030) | [PMC free article](/articles/PMC10463173/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37639259/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=JAMA%20Cardiol&title=Effect%20of%20Mavacamten%20on%20Chinese%20patients%20with%20symptomatic%20obstructive%20hypertrophic%20cardiomyopathy:%20the%20EXPLORER%E2%80%90CN%20randomized%20clinical%20trial&author=Z%20Tian&author=L%20Li&author=X%20Li&volume=8&issue=10&publication_year=2023&pages=957-965&pmid=37639259&doi=10.1001/jamacardio.2023.3030&)
|
| 281 |
+
|
| 282 |
+
17. Tian Z, Wang F, Jin W, et al. Study design and rationale of EXPLORER‐CN: a phase III, randomised, double‐blind, placebo‐controlled clinical study to evaluate the efficacy and safety of mavacamten in Chinese adults with symptomatic obstructive hypertrophic cardiomyopathy. BMJ Open. 2023;13(6):e071473. doi: 10.1136/bmjopen-2022-071473 [DOI](https://doi.org/10.1136/bmjopen-2022-071473) | [PMC free article](/articles/PMC10314621/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37336533/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=BMJ%20Open&title=Study%20design%20and%20rationale%20of%20EXPLORER%E2%80%90CN:%20a%20phase%20III,%20randomised,%20double%E2%80%90blind,%20placebo%E2%80%90controlled%20clinical%20study%20to%20evaluate%20the%20efficacy%20and%20safety%20of%20mavacamten%20in%20Chinese%20adults%20with%20symptomatic%20obstructive%20hypertrophic%20cardiomyopathy&author=Z%20Tian&author=F%20Wang&author=W%20Jin&volume=13&issue=6&publication_year=2023&pages=e071473&pmid=37336533&doi=10.1136/bmjopen-2022-071473&)
|
| 283 |
+
|
| 284 |
+
18. Merali S, Salinger DH, Palmisano M, et al. Recommendation of mavacamten posology by model‐based analyses in adults with obstructive hypertrophic cardiomyopathy. CPT Pharmacometrics Syst Pharmacol. 2024. doi: 10.1002/psp4.13138. Online ahead of print. [DOI](https://doi.org/10.1002/psp4.13138) | [PMC free article](/articles/PMC11533099/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38695527/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=CPT%20Pharmacometrics%20Syst%20Pharmacol&title=Recommendation%20of%20mavacamten%20posology%20by%20model%E2%80%90based%20analyses%20in%20adults%20with%20obstructive%20hypertrophic%20cardiomyopathy&author=S%20Merali&author=DH%20Salinger&author=M%20Palmisano&publication_year=2024&pmid=38695527&doi=10.1002/psp4.13138&)
|
| 285 |
+
|
| 286 |
+
19. Rabiee Rad M, Ghasempour Dabaghi G, Habibi D. Safety and efficacy of mavacamten for treatment of hypertrophic cardiomyopathy: a systematic review and meta‐analysis of randomized clinical trials. Egypt Heart J. 2023;75(1):4. doi: 10.1186/s43044-023-00328-7 [DOI](https://doi.org/10.1186/s43044-023-00328-7) | [PMC free article](/articles/PMC9837360/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36633717/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Egypt%20Heart%20J&title=Safety%20and%20efficacy%20of%20mavacamten%20for%20treatment%20of%20hypertrophic%20cardiomyopathy:%20a%20systematic%20review%20and%20meta%E2%80%90analysis%20of%20randomized%20clinical%20trials&author=M%20Rabiee%20Rad&author=G%20Ghasempour%20Dabaghi&author=D%20Habibi&volume=75&issue=1&publication_year=2023&pages=4&pmid=36633717&doi=10.1186/s43044-023-00328-7&)
|
test/texts/PMC11257390.md
ADDED
|
@@ -0,0 +1,260 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# UGT1A4*3 polymorphism influences serum concentration and therapeutic effect of lamotrigine for epilepsy treatment: A meta-analysis
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Zhimei Jiang, Yuzhi Fu, Hongxin Shen
|
| 5 |
+
**Journal:** PLOS ONE
|
| 6 |
+
**Date:** 2024 Jul 18
|
| 7 |
+
**DOI:** [10.1371/journal.pone.0307377](https://doi.org/10.1371/journal.pone.0307377)
|
| 8 |
+
**PMID:** 39024362
|
| 9 |
+
**PMCID:** PMC11257390
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11257390/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11257390/pdf/pone.0307377.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11257390/pdf/pone.0307377.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Background:**
|
| 16 |
+
Lamotrigine as a broad-spectrum antiepileptic drug, is widely applied and its clinical efficacy is highly recognized. However, significant differences are observed in blood drug concentration of lamotrigine among individuals, which may have an impact on its efficacy. UGT1A4 is the main metabolic enzyme. However, it was inconsistent for the influence of UGT1A4 genetic polymorphism on concentration and efficacy of lamotrigine therapy. This study aimed to evaluate the influences of UGT1A4*3 genetic polymorphisms on lamotrigine concentration and therapeutic effect through meta-analysis.
|
| 17 |
+
|
| 18 |
+
**Methods:**
|
| 19 |
+
The literature search was conducted in Medline, Embase, PubMed, Web of Science, Wan Fang Database, China National Knowledge Infrastructure, China Science and Technology Journal Database until January 2024. The primary outcome included the mean serum concentration, concentration-to-dose-ratio by body weight (CDR), or efficacy related to different UGT1A4*3 genotype for lamotrigine therapy. Data were collected to access the Mean Difference or odds ratio with 95% confidence interval. Meta-analysis was performed by RevMan 5.2.
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
A total of eleven studies were enrolled. The meta-analysis for mean serum concentration of lamotrigine showed no significant difference between patients carrying TT genotypes and TG and GG genotypes group (MD: 0.12, 95% [-0.35, 0.58], P = 0.62). There was significant difference in CDR (MD: 0.49, 95% [0.03, 0.94], P = 0.04) and therapeutic efficacy (OR: 7.18, 95% [4.01, 12.83], P<0.00001) of lamotrigine, however no significant difference was found in subgroup analysis of CDR of children (MD: 0.03, 95% [-0.35, 0.42], P = 0.87) between patients carrying TT genotypes and TG and GG genotypes group.
|
| 23 |
+
|
| 24 |
+
**Conclusions:**
|
| 25 |
+
Polymorphism of UGT1A4*3 influenced the CDR and therapeutic efficacy of lamotrigine for antiepileptic therapy. Genotype analysis provided reference for personalized medication in the future. However, more high-quality evidences are necessary for precise and definitive conclusion.
|
| 26 |
+
|
| 27 |
+
### Background
|
| 28 |
+
|
| 29 |
+
Lamotrigine as a broad-spectrum antiepileptic drug, is widely applied and its clinical efficacy is highly recognized. However, significant differences are observed in blood drug concentration of lamotrigine among individuals, which may have an impact on its efficacy. UGT1A4 is the main metabolic enzyme. However, it was inconsistent for the influence of UGT1A4 genetic polymorphism on concentration and efficacy of lamotrigine therapy. This study aimed to evaluate the influences of UGT1A4*3 genetic polymorphisms on lamotrigine concentration and therapeutic effect through meta-analysis.
|
| 30 |
+
|
| 31 |
+
### Methods
|
| 32 |
+
|
| 33 |
+
The literature search was conducted in Medline, Embase, PubMed, Web of Science, Wan Fang Database, China National Knowledge Infrastructure, China Science and Technology Journal Database until January 2024. The primary outcome included the mean serum concentration, concentration-to-dose-ratio by body weight (CDR), or efficacy related to different UGT1A4*3 genotype for lamotrigine therapy. Data were collected to access the Mean Difference or odds ratio with 95% confidence interval. Meta-analysis was performed by RevMan 5.2.
|
| 34 |
+
|
| 35 |
+
### Results
|
| 36 |
+
|
| 37 |
+
A total of eleven studies were enrolled. The meta-analysis for mean serum concentration of lamotrigine showed no significant difference between patients carrying TT genotypes and TG and GG genotypes group (MD: 0.12, 95% [-0.35, 0.58], *P*P = 0.62). There was significant difference in CDR (MD: 0.49, 95% [0.03, 0.94], *P*P = 0.04) and therapeutic efficacy (OR: 7.18, 95% [4.01, 12.83], *P*P<0.00001) of lamotrigine, however no significant difference was found in subgroup analysis of CDR of children (MD: 0.03, 95% [-0.35, 0.42], P = 0.87) between patients carrying TT genotypes and TG and GG genotypes group.
|
| 38 |
+
|
| 39 |
+
### Conclusions
|
| 40 |
+
|
| 41 |
+
Polymorphism of UGT1A4*3 influenced the CDR and therapeutic efficacy of lamotrigine for antiepileptic therapy. Genotype analysis provided reference for personalized medication in the future. However, more high-quality evidences are necessary for precise and definitive conclusion.
|
| 42 |
+
|
| 43 |
+
## Introduction
|
| 44 |
+
|
| 45 |
+
Epilepsy is one of the most prevalent brain conditions in the world, it nearly affects over 70 million of people [[1](#pone.0307377.ref001)1]. It is a disease complex with multiple causes and strong genetic predisposition. Identification of causes has important therapeutic significance. The categories of causes are structural, metabolic, genetic, immune, infectious, and unknown [[2](#pone.0307377.ref002)2]. The characteristic of epileptic seizures is stereotyped behavioral alterations. Therefore, epilepsy is mostly diagnosed through detailed clinical history and observation of a seizure by reliable eyewitness. Surgery is an effective option for long-term seizure freedom. Antiseizure medications are the major choices for most people with epilepsy. Its aim is to stop seizures at the earliest moment which may reduce morbidity and the risk of premature death especially related with convulsions [[3](#pone.0307377.ref003)3, [4](#pone.0307377.ref004)4]. Over 25 medications are available worldwide for epilepsy treatment. However, it is effective for current drugs in only about 66% patients in high-income countries [[5](#pone.0307377.ref005)5]. Lamotrigine (LTG) is a novel antiepileptic drug and a mood stabilizer. It mainly exerts antiepileptic effects by blocking presynaptic membrane voltage sensitive sodium ion channels, and inhibiting the release of glutamate and aspartic acid. It is necessary to start with a small dose and gradually increase the dosage [[6](#pone.0307377.ref006)6]. However, the clinical efficacy of LTG varies greatly among individuals, the same for the occurrence of adverse reactions such as rash, nausea, vomiting, diarrhea, and headache. Taking the same dose of LTG may result in significant individual differences. For example, some patients are ineffective and others potentially experience adverse reactions. This is related to the significant individual differences in LTG steady-state blood drug valley concentration [[7](#pone.0307377.ref007)7]. Furthermore, plasma clearance of LTG could be increased by hepatic enzyme induced drugs (such as phenytoin sodium, carbamazepine, oxcarbazepine) through inducing hepatic glucuronic acid binding reactions [[8](#pone.0307377.ref008)8]. Valproate sodium reduces the plasma clearance rate of LTG through inhibiting the reactions [[9](#pone.0307377.ref009)9]. Therefore, application of therapeutic drug monitoring (TDM) is clinically needed to realize personalized treatment with LTG [[10](#pone.0307377.ref010)10]. Accordingly, TDM of LTG was considered as “recommended” and therapeutic window was suggested as 3–14 μg/mL for concentrations of LTG by the TDM expert group of the Arbeitsgemeinschaft für Neuropsychopharmakologie und Pharmakopsychiatrie consensus and ILAE Commission on Therapeutic Strategies [[11](#pone.0307377.ref011)11, [12](#pone.0307377.ref012)12]. At present, LTG dose-escalation regimens are mostly used by clinical physicians. Then the dosage regimens are adjusted empirically or based on TDM results which are time-consuming. Therefore, how to control epilepsy quickly and effectively, and enable LTG to reach therapeutic window is an urgent problem that needs to be solved in clinical practice.
|
| 46 |
+
|
| 47 |
+
Researches found that genetic factors such as mutations in metabolic enzymes and transporters might be one of the important reasons for the large individual differences in LTG serum concentration [[13](#pone.0307377.ref013)13, [14](#pone.0307377.ref014)14]. It is also a hot research topic in recent years. With the deepening research of pharmacogenomics, the impact of polymorphisms in drug metabolism enzymes and transporter on the serum concentration of LTG is increasingly being valued. LTG is mainly metabolized by UDP glycuronosyltransferase (UGT). It is metabolized into inactive glucuronic acid binding products, and then excreted from the body through the kidneys. UGT1A4 is the main metabolic enzyme [[15](#pone.0307377.ref015)15]. In its coding region, UGT1A4*3 is the most common single nucleotide gene polymorphism (SNP) [[16](#pone.0307377.ref016)16]. Zhou *et al*et al. [[16](#pone.0307377.ref016)16] found that mutations in UGT1A4 70C>A and 142T>G could lead to a decrease in UGT enzyme activity through in vitro studies, resulting in a decrease in LTG clearance rate. A study was conducted by Chang et al. [[17](#pone.0307377.ref017)17] on the correlation between LTG blood drug concentration and UGT1A4 70C>A and 142T>G genotypes in 106 Chinese epilepsy patients. It was found that the LTG standardized blood drug concentration/dose/normalized by body weight (CDR) of UGT1A4 142TT wild-type carriers was significantly higher than that of TG+GG carriers. However, some researches did not find the UGT1A4 142T>G gene mutation could affect the blood drug concentration of LTG. It is obviously that the conclusions about the influence of UGT1A4*3 polymorphism on the concentration and therapeutic efficacy of LTG is not entirely consistent. Furthermore, there is no systematic review discussing the collaborative impact by UGT1A4*3 polymorphism between serum concentration and therapeutic efficacy of LTG.
|
| 48 |
+
|
| 49 |
+
Therefore, our study systematically evaluated the effects of UGT1A4*3 polymorphisms on LTG serum concentration and therapeutic efficacy through meta-analysis. It was meaningful to provide a underlying mechanism of interindividual variation in the LTG treatment. The influence was useful to be considered as a factor when pharmacokinetic model was determined for LTG dose adjustment.
|
| 50 |
+
|
| 51 |
+
## Materials and methods
|
| 52 |
+
|
| 53 |
+
### Eligibility criteria
|
| 54 |
+
|
| 55 |
+
The present review included cohort researches investigating serum concentrations and effect of LTG influenced by UGT1A4*3 genetic polymorphisms during epilepsy. Types of prospective and retrospective both were included. The intervention was treated with LTG for epilepsy monotherapy. The exposed group showed TT genotype, whereas the control group was TG and GG genotype of UGT1A4*3. Eligible researches reported one of the following outcomes: mean serum concentration, CDR, or efficacy related to different UGT1A4*3 genotype. Studies published in English or Chinese were considered.
|
| 56 |
+
|
| 57 |
+
### Literature search
|
| 58 |
+
|
| 59 |
+
Four English language databases including Medline, Embase, PubMed, Web of Science were searched. Three Chinese databases including Wan Fang Database, China National Knowledge Infrastructure, China Science and Technology Journal Database were also searched.
|
| 60 |
+
|
| 61 |
+
The following principal search terms were used: “lamotrigine” and “gene” or “genetic” or “polymorphisms” and “UGT1A4”. They were combined to search for relevant studies. Furthermore, it was restricted to human studies. We would check additional studies in reference lists of included studies, and contact with authors to request missing data. The last retrieval was performed on January, 2024.
|
| 62 |
+
|
| 63 |
+
### Study selection and data extraction
|
| 64 |
+
|
| 65 |
+
Two trained reviewers independently screened potentially eligible studies based on title and abstract, and then read full texts for final eligibility. Disagreements were resolved by a third person. The information derived from included studies was collected and shown in a table including the following data: general characteristics of studies and patients, interventions and comparisons, outcomes (mean serum concentration, CDR, efficacy), assess of Hardy–Weinberg equilibrium. The MSC referred to the average steady-state valley concentration of LTG, and the CDR was derived through the formula: LTG steady-state valley plasma concentration/ LTG dose / weight. The efficacy of LTG was graded using Engel method [[18](#pone.0307377.ref018)18]. It was determined by monitoring the frequency of epileptic seizures in patients during a one-year follow-up.
|
| 66 |
+
|
| 67 |
+
### Quality assessment
|
| 68 |
+
|
| 69 |
+
The quality of included studies was evaluated according to Newcastle-Ottawa Scale (NOS). It includes three items. Selection of case and controls: 1) is the case definition adequate (Yes, with independent validation, score a star)? 2) Representativeness of the cases (consecutive or obviously representative series of cases, score a star); 3) Selection of controls (community controls, score a star); 4) Definition of controls (no history of disease (endpoint), score a star); Comparability of cases and controls: 1) Comparability of cases and controls on the basis of the design or analysis (study controls for (select the most important factor), score a star; study controls for any additional factor, score a star); Exposure: 1)Ascertainment of exposure (secure record, score a star; structured interview where blind to case/control status, score a star); 2)Same method of ascertainment for cases and controls (yes, score a star); 3)Non-response rate (same rate for both groups, score a star). It was not suitable to assess publication bias because of insufficient number of included studies.
|
| 70 |
+
|
| 71 |
+
### Statistical analysis
|
| 72 |
+
|
| 73 |
+
Statistical analyses were performed with Review Manager Version 5.2 (Copenhagen: The Nordic Cochrane Centre, 2012). It could perform meta-analysis of the data entered, and present the results graphically. The data of mean serum concentration and CDR outcomes were continuous data which were analyzed for Mean Difference (MD). The data of efficacy outcome was dichotomous data which were analyzed for odds ratio (OR) with 95% confidence interval (CI). Due to the fact that all included studies might not be highly consistent, i.e. there might be differences between subjects and interventions, and the resulting effect size might not be a fixed value, a random effects model was chosen for meta-analysis. Study heterogeneity was determined via statistical analyses, using the Q statistic for homogeneity and the I-squared (*I*I^2^*2*2) statistic. A P value of < 0.10 or *I*I^2^*2*2 >50% was considered to indicate significant heterogeneity. When only minimum (Min) and maximum (Max) were available for a specific sample, standard deviation (SD) was estimated (SD = (Max-Min)/4) [[19](#pone.0307377.ref019)19]. When Mean and SD in two groups were merged, formulas were as follows: mean (M) = (N_1_1M_1_1+N_2_2M_2_2)/(N_1_1+N_2_2), SD=(N1−1)SD12+(N2−1)SD22+N1N2N1+N2(M12+M22−2M1M2)N1+N2–1SD=(N1−1)SD12+(N2−1)SD22+N1N2N1+N2(M12+M22−2M1M2)N1+N2–1SSDD==(N1−1)SD12+(N2−1)SD22+N1N2N1+N2(M12+M22−2M1M2)N1+N2–1(N1−1)SD12+(N2−1)SD22+N1N2N1+N2(M12+M22−2M1M2)N1+N2–1(N1−1)SD12+(N2−1)SD22+N1N2N1+N2(M12+M22−2M1M2)(N1−1)SD12(N1−1)SD1((NN11−−11))SSDD11222++(N2−1)SD22(N2−1)SD2((NN22−−11))SSDD22222++N1N2N1+N2N1N2NN11NN22N1+N2NN11++NN22((M12+M1MM112+22++M22M2MM22222−−22MM11MM22))N1+N2–1NN1+1+NN22––11 (group A sample size N_1_1, mean M_1_1, standard deviation SD_1_1; group B sample sizeN2, mean M_2_2, standard deviation SD_2_2) [[20](#pone.0307377.ref020)20]. Sensitivity analysis was performed by individually excluding each study to assess the quality and consistency of the results.
|
| 74 |
+
|
| 75 |
+
## Results
|
| 76 |
+
|
| 77 |
+
### Search results and characteristics of enrolled studies
|
| 78 |
+
|
| 79 |
+
A total of 4268 records were identified by initial database search. After first screening of the title and abstract, 4219 articles were excluded. Following the full-text review, 11 studies were included ([Fig 1](#pone.0307377.g001)Fig 1). Of the 11 eligible studies, 7 studies were conducted for mean serum concentration for 541 patients; 8 studies enrolled 680 patients with the determination of CDR outcome; 3 studies shown efficacy outcome for 308 patients. General characteristic of included studies were shown in [Table 1](#pone.0307377.t001)Table 1. Detailed NOS results of quality assessment of included studies were shown in [Table 2](#pone.0307377.t002)Table 2.
|
| 80 |
+
|
| 81 |
+
### Fig 1. Follow diagram of selecting study.
|
| 82 |
+
|
| 83 |
+

|
| 84 |
+
|
| 85 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11257390_pone.0307377.g001.jpg)
|
| 86 |
+
|
| 87 |
+
### Table 1. Characteristics of included studies.
|
| 88 |
+
|
| 89 |
+
| Author(year) | Datasource/country | Number of patients (males) | Age, y | Outcome measurement of LTG | Administration dosage | Hardy–Weinberg equilibrium |
|
| 90 |
+
| ------------ | ------------------ | -------------------------- | ------ | -------------------------- | --------------------- | -------------------------- |
|
| 91 |
+
| Suzuki 2019 [21] | Japanese | 103(Unclear) | 42.2±16.2 | mean serum concentration | 100mg/d | Unclear |
|
| 92 |
+
| Petrenaite 2022 [22] | Danish | 195(87) | 18–67 | mean serum concentration | TT:335±168mg/d | Yes |
|
| 93 |
+
| TG:320±180mg/d | | | | | | |
|
| 94 |
+
| Gulcebi 2011 [23] | Turkish | 35(10) | 18–50 | mean serum concentration | 3.07±0.15mg/kg/d | Unclear |
|
| 95 |
+
| Zhou 2015 [24] | China | 119(57) | 29±12 | mean serum concentration; CDR | TT2.22:(1.46–3.12)mg/kg | Yes |
|
| 96 |
+
| GT+TT:2.27(1.96–3.08)mg/kg | | | | | | |
|
| 97 |
+
| Liu 2015 [25] | China | 56(31) | TT:7.74±2.53 | mean serum concentration; CDR | TT:68.82±36.81mg/d | Yes |
|
| 98 |
+
| TG+GG:46.02±12.43mg/d | | | | | | |
|
| 99 |
+
| TG+GG:8.55±3.14 | | | | | | |
|
| 100 |
+
| Lou 2021 [26] | China | 49(22) | 8.0(6.0–12.0) | mean serum concentration; CDR | 100mg/d | Yes |
|
| 101 |
+
| Liu 2014 [27] | China | 160(98) | 8.25±3.18 | mean serum concentration; CDR | 2.76±1.65mg/kg | Unclear |
|
| 102 |
+
| Yang 2013 [28] | China | 103(40) | 32.07±12.88 | CDR; efficacy | 0.41–4.30mg/Kg | Yes |
|
| 103 |
+
| Chang 2014 [17] | China | 106(40) | 44.55±11.79 | CDR; efficacy | 0.41–4.30 mg/kg | Yes |
|
| 104 |
+
| Du 2016[29] | China | 102(57) | 12.33±6.13 | CDR; efficacy | 2.74±1.17 mg/kg | Unclear |
|
| 105 |
+
| Reimers 2016 [29] | Norway | 127(Unclear) | 12–65 | CDR | 278.3(Mean) | Unclear |
|
| 106 |
+
### Table 2. Quality assessment of included studies using NOS.
|
| 107 |
+
|
| 108 |
+
| Included studies | Selection of case and controls | Comparability of cases and controls | Exposure |
|
| 109 |
+
| ---------------- | ------------------------------ | ----------------------------------- | -------- |
|
| 110 |
+
| is the case definition adequate | Representativeness of the cases | Selection of controls | Definition of controls | Comparability of cases and controls on the basis of the design or analysis | Ascertainment of exposure | Same method of ascertainment for cases and controls | Non-response rate |
|
| 111 |
+
| Suzuki 2019 [21] | ★ | - | - | ★ | ★ | ★★ | ★ | - |
|
| 112 |
+
| Petrenaite 2022 [22] | ★ | - | - | ★ | ★★ | ★★ | ★ | - |
|
| 113 |
+
| Gulcebi 2011 [23] | ★ | - | - | ★ | ★ | ★★ | ★ | - |
|
| 114 |
+
| Zhou 2015 [24] | ★ | - | - | ★ | ★★ | ★★ | ★ | - |
|
| 115 |
+
| Liu 2015 [25] | ★ | - | - | ★ | ★★ | ★★ | ★ | - |
|
| 116 |
+
| Lou 2021 [26] | ★ | - | - | ★ | ★★ | ★★ | ★ | - |
|
| 117 |
+
| Liu 2014 [27] | ★ | - | - | ★ | ★ | ★★ | ★ | - |
|
| 118 |
+
| Yang 2013 [28] | ★ | - | - | ★ | ★★ | ★★ | ★ | - |
|
| 119 |
+
| Chang 2014 [17] | ★ | - | - | ★ | ★★ | ★★ | ★ | - |
|
| 120 |
+
| Du 2016 [29] | ★ | - | - | ★ | ★★ | ★★ | ★ | - |
|
| 121 |
+
| Reimers 2016 [30] | ★ | - | - | ★ | ★★ | ★★ | ★ | - |
|
| 122 |
+
### Effect of UGT1A4*3 polymorphism on LTG mean serum concentration
|
| 123 |
+
|
| 124 |
+
Seven studies [[21](#pone.0307377.ref021)21–[27](#pone.0307377.ref027)27] involving 360 patients in the TT genotype group and 161 patients in the TG and GG genotype group reported mean serum concentration of LTG based on the UGT1A4*3 polymorphism during LTG monotherapy ([Fig 2](#pone.0307377.g002)Fig 2). Random-effects model was chosen because of statistical heterogeneity (*I*I^2^*2*2 = 82%). Pooled analysis of data showed no significant difference between the LTG serum concentration of subjects carrying TT genotype with TG and GG genotype (MD: 0.12, 95% [-0.35, 0.58], *P*P = 0.62).
|
| 125 |
+
|
| 126 |
+
### Fig 2. Forest plot of UGT1A4*3 polymorphism on mean serum concentration of LTG.
|
| 127 |
+
|
| 128 |
+

|
| 129 |
+
|
| 130 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11257390_pone.0307377.g002.jpg)
|
| 131 |
+
|
| 132 |
+
### Effect of UGT1A4*3 polymorphism on CDR
|
| 133 |
+
|
| 134 |
+
A total of 8 studies were included to pooled analysis CDR difference of LTG based on UGT1A4*3 polymorphism. 7 records [[17](#pone.0307377.ref017)17, [24](#pone.0307377.ref024)24–[29](#pone.0307377.ref029)29] reported appropriate data types for Meta-analysis ([Fig 3](#pone.0307377.g003)Fig 3). Random-effects model was used (*I*I ^2^*2*2 = 96%). Based on the data from those studies, patients with TT genotype yielded a significantly higher CDR of LTG than with TG and GG genotype (MD: 0.49, 95% [0.03, 0.94], *P*P = 0.04). One study [[30](#pone.0307377.ref030)30] only described individuals heterozygous for UGT1A4*3 had a significantly lower CDR than individuals with wild-type (TT) without detailed data. Clearly, it was consistent with our systematic Meta-analysis results. However, through subgroup analysis of CDR in children, there was no significant difference between patients carrying TT genotype group and TG and GG genotype group (MD: 0.03, 95% [-0.35, 0.42], P = 0.87).
|
| 135 |
+
|
| 136 |
+
### Fig 3. Forest plot of UGT1A4*3 polymorphism on CDR of LTG.
|
| 137 |
+
|
| 138 |
+

|
| 139 |
+
|
| 140 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11257390_pone.0307377.g003.jpg)
|
| 141 |
+
|
| 142 |
+
### Effect of UGT1A4*3 polymorphism on therapeutic efficacy of LTG
|
| 143 |
+
|
| 144 |
+
Only three studies [[28](#pone.0307377.ref028)28, [29](#pone.0307377.ref029)29], involving 308 patients, reported the therapeutic efficacy of LTG based on UGT1A4*3 polymorphism. No statistical heterogeneity existed among the article results (*I*I ^2^*2*2 = 0%). Random-effects model was used to perform the systematic analysis. There was significant difference between individuals with TT genotype and the control group in the therapeutic effect of LTG (OR: 7.18, 95% [4.01, 12.83], *P*P<0.00001) ([Fig 4](#pone.0307377.g004)Fig 4). The results indicated that the therapeutic effect of LTG with TT genotype was better than that with TG/GG.
|
| 145 |
+
|
| 146 |
+
### Fig 4. Forest plot of UGT1A4*3 polymorphism on therapeutic effect of LTG.
|
| 147 |
+
|
| 148 |
+

|
| 149 |
+
|
| 150 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11257390_pone.0307377.g004.jpg)
|
| 151 |
+
|
| 152 |
+
## Discussion
|
| 153 |
+
|
| 154 |
+
As reported, the therapeutic response of LTG therapy relied on its serum concentration. Unfortunately, there were significant interindividual differences in serum concentration or CDR among subjects [[31](#pone.0307377.ref031)31]. It was widely known that coadministration of valproate significantly increased LTG concentrations [[32](#pone.0307377.ref032)32, [33](#pone.0307377.ref033)33]. Therefore, we focused on the influence of UGT1A4 genetic polymorphisms based on LTG monotherpy or simultaneous medication of non-interacting antiepileptic drugs. UGT1A4 was the main metabolic enzymes of LTG into inactive glucuronic acid conjugates [[24](#pone.0307377.ref024)24]. Meanwhile, it was controversial for the correlation between genotype of UGT1A4*3 and serum or efficacy of LTG. This study systematically evaluated the influence of UGT1A4*3 on serum concentration and efficacy of LTG therapy through meta-analysis.
|
| 155 |
+
|
| 156 |
+
Mean serum and CDR were the most common parameters in eligible studies. It was interesting that there was no remarkable difference in mean serum concentration of LTG between patients carrying TT genotypes group and TG and GG genotypes group. However, significant difference was observed in CDR of LTG between patients carrying TT genotypes group and TG and GG genotypes group. Namely, the CDR level of LTG for patients carrying TT genotype was higher than that for patients carrying TG and GG genotype. Due to individual differences in weight and drug dosage among patients, CDR was adjusted based on daily dose and individual weight. It is standardized blood drug concentrations which excludes the influence of body weight and dosage on blood drug concentration. Therefore, it suggested that CDR parameter was to be a preferred detection indicator, to provide reference for personalized medication and achieve rational administration in different genotype of UGT1A4*3 patients. Through subgroup analysis of CDR in children, the meta-analysis suggested that there was no significant difference. It might be attributed to low activity of glucuronic acid transferase in young children [[34](#pone.0307377.ref034)34]. Therefore, adjusting dosage based on the overall population genetic polymorphism could not guarantee the safety of medication in children. It is necessary to consider organ maturation and individual development of drug metabolism enzymes in order to provide reasonable dosage.
|
| 157 |
+
|
| 158 |
+
Meta-analysis was performed on therapeutic efficacy of LTG in patients with different genotype of UGT1A4*3. It showed that the clinical efficacy of LTG therapy for TT genotype patients was better than that for TG and GG genotype patients. Reasonably, it was consistent with the CDR outcome results. Therefore, the conclusions may be helpful to understand the individual differences of LTG therapy. Patient’s genotype can be considered as a factor to establish pharmacokinetic model for reasonable dosage of LTG. On the other hand, its pharmacokinetic differences may be influenced by combined effects of multiple genes, and hepatic enzyme induced and inhibited drugs. Other significant factors are necessary to be further investigation for LTG individualized medication.
|
| 159 |
+
|
| 160 |
+
The present study still has limitations. First, because of the varied sample sizes of enrolled studies, inconsistency in the methods applied to determine LTG concentration and different dosage, the results might be biased due to clinical heterogeneity. The quality of the included studies was categorized as having a medium risk of bias (awarded six or seven stars). Five studies did not describe assess of Hardy-Weinberg equilibrium which was important for genetic analysis. Non-representative cases might make a risk of outcome bias. Secondly, multiple genetic polymorphism investigations are necessary to realize precision administration. UGT1A4*3 may be one of the genetic polymorphism which can influence the concentration and therapeutic effect of LTG. Zhou *et al*et al [[35](#pone.0307377.ref035)35] indicated that polymorphisms in HNF4α, ABCG2 and ABCB1 were associated with CDR of LTG. Therefore, much more single nucleotide polymorphisms involved in LTG pharmacokinetics can be further evaluated to provide factor when determine pharmacokinetic model for LTG dose adjustment. Finally, only three studies of Chinese reported the efficacy of LTG between different genotypes. It should be cautious with result of definitive correlation. Insufficient sample size in the meta-analysis might limit the reliability and accuracy.
|
| 161 |
+
|
| 162 |
+
In summary, current evidence indicated that UGT1A4*3 polymorphisms might influence CDR level and efficacy of LTG therapy. LTG administration is necessary to follow the principle of individualization.
|
| 163 |
+
|
| 164 |
+
## Supporting information
|
| 165 |
+
|
| 166 |
+
## Data Availability
|
| 167 |
+
|
| 168 |
+
All relevant data are within the manuscript and its [Supporting Information](#sec018)Supporting Information files.
|
| 169 |
+
|
| 170 |
+
## Funding Statement
|
| 171 |
+
|
| 172 |
+
The author(s) received no specific funding for this work.
|
| 173 |
+
|
| 174 |
+
## Associated Data
|
| 175 |
+
|
| 176 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 177 |
+
|
| 178 |
+
### Supplementary Materials
|
| 179 |
+
|
| 180 |
+
### Data Availability Statement
|
| 181 |
+
|
| 182 |
+
All relevant data are within the manuscript and its [Supporting Information](#sec018)Supporting Information files.
|
| 183 |
+
|
| 184 |
+
### Supplementary Materials
|
| 185 |
+
|
| 186 |
+
### Data Availability Statement
|
| 187 |
+
|
| 188 |
+
All relevant data are within the manuscript and its [Supporting Information](#sec018)Supporting Information files.
|
| 189 |
+
|
| 190 |
+
## References
|
| 191 |
+
|
| 192 |
+
1. Roland D Thijs, Rainer Surges, Prof Terence J O’Brien, Prof Josemir W Sander. Epilepsy in adults. The Lancet. 2019;393(10172):689–701. [DOI](https://doi.org/10.1016/S0140-6736(18)32596-0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30686584/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=The%20Lancet&title=Epilepsy%20in%20adults&author=D%20Thijs%20Roland&author=Surges%20Rainer&author=J%20O%E2%80%99Brien%20Prof%20Terence&author=W%20Sander%20Prof%20Josemir&volume=393&issue=10172&publication_year=2019&pages=689-701&pmid=30686584&doi=10.1016/S0140-6736(18)32596-0&)
|
| 193 |
+
|
| 194 |
+
2. Ingrid E Scheffer, Samuel Berkovic, Giuseppe Capovilla, Mary B Connolly, Jacqueline French, Laura Guilhoto, et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58(4):512–521. doi: 10.1111/epi.13709 [DOI](https://doi.org/10.1111/epi.13709) | [PMC free article](/articles/PMC5386840/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28276062/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Epilepsia&title=ILAE%20classification%20of%20the%20epilepsies:%20Position%20paper%20of%20the%20ILAE%20Commission%20for%20Classification%20and%20Terminology&author=E%20Scheffer%20Ingrid&author=Berkovic%20Samuel&author=Capovilla%20Giuseppe&author=B%20Connolly%20Mary&author=French%20Jacqueline&volume=58&issue=4&publication_year=2017&pages=512-521&pmid=28276062&doi=10.1111/epi.13709&)
|
| 195 |
+
|
| 196 |
+
3. Aidan Neligan, Willard A Hauser, Josemir W Sander. The epidemiology of the epilepsies. Handb Clin Neurol. 2012;107:113–33. doi: 10.1016/B978-0-444-52898-8.00006-9 [DOI](https://doi.org/10.1016/B978-0-444-52898-8.00006-9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22938966/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Handb%20Clin%20Neurol&title=The%20epidemiology%20of%20the%20epilepsies&author=Neligan%20Aidan&author=A%20Hauser%20Willard&author=W%20Sander%20Josemir&volume=107&publication_year=2012&pages=113-33&pmid=22938966&doi=10.1016/B978-0-444-52898-8.00006-9&)
|
| 197 |
+
|
| 198 |
+
4. Orrin Devinsky, Tanya Spruill, David Thurman, Daniel Friedman. Recognizing and preventing epilepsy-related mortality: A call for action. Neurology. 2016;86(8):779–86. doi: 10.1212/WNL.0000000000002253 [DOI](https://doi.org/10.1212/WNL.0000000000002253) | [PMC free article](/articles/PMC4763802/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26674330/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Neurology&title=Recognizing%20and%20preventing%20epilepsy-related%20mortality:%20A%20call%20for%20action&author=Devinsky%20Orrin&author=Spruill%20Tanya&author=Thurman%20David&author=Friedman%20Daniel&volume=86&issue=8&publication_year=2016&pages=779-86&pmid=26674330&doi=10.1212/WNL.0000000000002253&)
|
| 199 |
+
|
| 200 |
+
5. John S Duncan, Josemir W Sander, Sanjay M Sisodiya, Matthew C Walker. Adult epilepsy. The Lancet.2006;367(9516):1087–1100. [DOI](https://doi.org/10.1016/S0140-6736(06)68477-8) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16581409/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=The%20Lancet&title=Adult%20epilepsy&author=S%20Duncan%20John&author=W%20Sander%20Josemir&author=M%20Sisodiya%20Sanjay&author=C%20Walker%20Matthew&volume=367&issue=9516&publication_year=2006&pages=1087-1100&pmid=16581409&doi=10.1016/S0140-6736(06)68477-8&)
|
| 201 |
+
|
| 202 |
+
6. Song ZB, Liao WP, Yi YH, et al. The influence of lamotrigine on EEG at therapeutic serum concentration. Acta Acad Guangzhou Med Coll.2003;31(4): 16–18. [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Acta%20Acad%20Guangzhou%20Med%20Coll&title=The%20influence%20of%20lamotrigine%20on%20EEG%20at%20therapeutic%20serum%20concentration&author=ZB%20Song&author=WP%20Liao&author=YH%20Yi&volume=31&issue=4&publication_year=2003&pages=16-18&)
|
| 203 |
+
|
| 204 |
+
7. Schoretsanitis Georgios MD. Establishing and Extending the Use of Therapeutic Drug Monitoring in Neuropsychopharmacology. Therapeutic Drug Monitoring. 2024;46(2):141–142. doi: 10.1097/FTD.0000000000001171 [DOI](https://doi.org/10.1097/FTD.0000000000001171) | [PMC free article](/articles/PMC10930354/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38377175/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Therapeutic%20Drug%20Monitoring&title=Establishing%20and%20Extending%20the%20Use%20of%20Therapeutic%20Drug%20Monitoring%20in%20Neuropsychopharmacology&author=Georgios%20MD%20Schoretsanitis&volume=46&issue=2&publication_year=2024&pages=141-142&pmid=38377175&doi=10.1097/FTD.0000000000001171&)
|
| 205 |
+
|
| 206 |
+
8. May TW, Rambeck B, Jurgens U. Influence of oxcarbazepine and methsuximide on lamotrigine concentrations in epileptic patients with and without valproic acid comedication: results of a retrospective study. Ther Drug Monit.1999;21(2): 175–181. doi: 10.1097/00007691-199904000-00007 [DOI](https://doi.org/10.1097/00007691-199904000-00007) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10217337/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ther%20Drug%20Monit&title=Influence%20of%20oxcarbazepine%20and%20methsuximide%20on%20lamotrigine%20concentrations%20in%20epileptic%20patients%20with%20and%20without%20valproic%20acid%20comedication:%20results%20of%20a%20retrospective%20study&author=TW%20May&author=B%20Rambeck&author=U%20Jurgens&volume=21&issue=2&publication_year=1999&pages=175-181&pmid=10217337&doi=10.1097/00007691-199904000-00007&)
|
| 207 |
+
|
| 208 |
+
9. Yuen AW, Land G, Weatherley BC, Peck AW. Sodium valproate acutely inhibits lamotrigine metabolism. Br J Clin Pharmacol.1992;33(5):511–513. doi: 10.1111/j.1365-2125.1992.tb04079.x [DOI](https://doi.org/10.1111/j.1365-2125.1992.tb04079.x) | [PMC free article](/articles/PMC1381438/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1524964/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Sodium%20valproate%20acutely%20inhibits%20lamotrigine%20metabolism&author=AW%20Yuen&author=G%20Land&author=BC%20Weatherley&author=AW%20Peck&volume=33&issue=5&publication_year=1992&pages=511-513&pmid=1524964&doi=10.1111/j.1365-2125.1992.tb04079.x&)
|
| 209 |
+
|
| 210 |
+
10. Sara Baldelli 1, Simone Castoldi, Nitin Charbe, Valeria Cozzi, Serena Fucile, Dario Cattaneo, et al. Comparison of the QMS Analyzer With HPLC-UV for the Quantification of Lamotrigine Concentrations in Human Plasma Samples. Ther Drug Monit. 2015;37(5):689–94. doi: 10.1097/FTD.0000000000000202 [DOI](https://doi.org/10.1097/FTD.0000000000000202) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25730145/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ther%20Drug%20Monit&title=Comparison%20of%20the%20QMS%20Analyzer%20With%20HPLC-UV%20for%20the%20Quantification%20of%20Lamotrigine%20Concentrations%20in%20Human%20Plasma%20Samples&author=Baldelli%20Sara&author=Castoldi%20Simone&author=Charbe%20Nitin&author=Cozzi%20Valeria&author=Fucile%20Serena&volume=37&issue=5&publication_year=2015&pages=689-94&pmid=25730145&doi=10.1097/FTD.0000000000000202&)
|
| 211 |
+
|
| 212 |
+
11. Patsalos PN, Berry DJ, Bourgeois BF, et al. Antiepileptic drugs—best practice guidelines for therapeutic drug monitoring: a position paper by the sub commission on therapeutic drug monitoring, ILAE commission on therapeutic strategies. Epilepsia. 2008; 49:1239–1276. doi: 10.1111/j.1528-1167.2008.01561.x [DOI](https://doi.org/10.1111/j.1528-1167.2008.01561.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18397299/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Epilepsia&title=Antiepileptic%20drugs%E2%80%94best%20practice%20guidelines%20for%20therapeutic%20drug%20monitoring:%20a%20position%20paper%20by%20the%20sub%20commission%20on%20therapeutic%20drug%20monitoring,%20ILAE%20commission%20on%20therapeutic%20strategies&author=PN%20Patsalos&author=DJ%20Berry&author=BF%20Bourgeois&volume=49&publication_year=2008&pages=1239-1276&pmid=18397299&doi=10.1111/j.1528-1167.2008.01561.x&)
|
| 213 |
+
|
| 214 |
+
12. Hiemke C, Baumann P, Bergemann N, et al. AGNP consensus guidelines for therapeutic drug monitoring in psychiatry: update 2017. Pharmacopsychiatry. 2017; 51:9–62. [DOI](https://doi.org/10.1055/s-0031-1286287) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21969060/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacopsychiatry&title=AGNP%20consensus%20guidelines%20for%20therapeutic%20drug%20monitoring%20in%20psychiatry:%20update%202017&author=C%20Hiemke&author=P%20Baumann&author=N%20Bergemann&volume=51&publication_year=2017&pages=9-62&pmid=21969060&doi=10.1055/s-0031-1286287&)
|
| 215 |
+
|
| 216 |
+
13. ZHANG ZB, JI SM, HAN Y, et al. Population pharmacokinetic models of lamotrigine in different age groups of Chinese children with epilepsy. Eur J Clin Pharmacol. 2017; 73(4):1–9. doi: 10.1007/s00228-016-2190-2 [DOI](https://doi.org/10.1007/s00228-016-2190-2) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28064355/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=Population%20pharmacokinetic%20models%20of%20lamotrigine%20in%20different%20age%20groups%20of%20Chinese%20children%20with%20epilepsy&author=ZB%20ZHANG&author=SM%20JI&author=Y%20HAN&volume=73&issue=4&publication_year=2017&pages=1-9&pmid=28064355&doi=10.1007/s00228-016-2190-2&)
|
| 217 |
+
|
| 218 |
+
14. TAKEUCHI T, NATSUME J, KIDOKORO H, et al. The effects of co-medications on lamotrigine clearance in Japanese children with epilepsy. Brain Dev. 2016; 38(8):723–730. doi: 10.1016/j.braindev.2016.03.004 [DOI](https://doi.org/10.1016/j.braindev.2016.03.004) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27033151/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Brain%20Dev&title=The%20effects%20of%20co-medications%20on%20lamotrigine%20clearance%20in%20Japanese%20children%20with%20epilepsy&author=T%20TAKEUCHI&author=J%20NATSUME&author=H%20KIDOKORO&volume=38&issue=8&publication_year=2016&pages=723-730&pmid=27033151&doi=10.1016/j.braindev.2016.03.004&)
|
| 219 |
+
|
| 220 |
+
15. REIMERS A, SJURSEN W, HELDE G, et al. Frequencies of UGT1A4*2 (P24T) and *3 (L48V) and their effects on serum concentrations of lamotrigine. Eur J Dug Metab Pharmacokinet. 2016;41(2):149–155. doi: 10.1007/s13318-014-0247-0 [DOI](https://doi.org/10.1007/s13318-014-0247-0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25492569/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Dug%20Metab%20Pharmacokinet&title=Frequencies%20of%20UGT1A4*2%20(P24T)%20and%20*3%20(L48V)%20and%20their%20effects%20on%20serum%20concentrations%20of%20lamotrigine&author=A%20REIMERS&author=W%20SJURSEN&author=G%20HELDE&volume=41&issue=2&publication_year=2016&pages=149-155&pmid=25492569&doi=10.1007/s13318-014-0247-0&)
|
| 221 |
+
|
| 222 |
+
16. Jin Zhou, Upendra A Argikar, Rory P Remmel. Functional analysis of UGT1A4(P24T) and UGT1A4(L48V) variant enzymes. Pharmacogenomics. 2011;12(12):1671–9. doi: 10.2217/pgs.11.105 [DOI](https://doi.org/10.2217/pgs.11.105) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22047493/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=Functional%20analysis%20of%20UGT1A4(P24T)%20and%20UGT1A4(L48V)%20variant%20enzymes&author=Zhou%20Jin&author=A%20Argikar%20Upendra&author=P%20Remmel%20Rory&volume=12&issue=12&publication_year=2011&pages=1671-9&pmid=22047493&doi=10.2217/pgs.11.105&)
|
| 223 |
+
|
| 224 |
+
17. Ying Chang, Li-ya Yang, Meng-chao Zhang, Song-Yan Liu. Correlation of the UGT1A4 gene polymorphism with serum concentration and therapeutic efficacy of lamotrigine in Han Chinese of Northern China. Eur J Clin Pharmacol. 2014;70(8):941–6. doi: 10.1007/s00228-014-1690-1 [DOI](https://doi.org/10.1007/s00228-014-1690-1) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24820767/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=Correlation%20of%20the%20UGT1A4%20gene%20polymorphism%20with%20serum%20concentration%20and%20therapeutic%20efficacy%20of%20lamotrigine%20in%20Han%20Chinese%20of%20Northern%20China&author=Chang%20Ying&author=Yang%20Li-ya&author=Zhang%20Meng-chao&author=Liu%20Song-Yan&volume=70&issue=8&publication_year=2014&pages=941-6&pmid=24820767&doi=10.1007/s00228-014-1690-1&)
|
| 225 |
+
|
| 226 |
+
18. Engel J Jr, Van Ness PC, Rasmussen TB, Ojemann LM Outcome with respect to epileptic seizures. In: Engel J Jr (ed) Surgical treatment of the epilepsies, 2nd edn. Raven Press, New York;1993. pp 609–621. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Surgical%20treatment%20of%20the%20epilepsies&author=J%20Engel&author=PC%20Van%20Ness&author=TB%20Rasmussen&author=LM%20Ojemann&author=J%20Engel&publication_year=1993&)
|
| 227 |
+
|
| 228 |
+
19. Balram Chowbay, Huihua Li, Machin David, Yin Bun Cheung & Edmund J. D. Lee. Meta-analysis of the influence of MDR1 C3435T polymorphism on digoxin pharmacokinetics and MDR1 gene expression. British Journal of Clinical Pharmacology. 2004; 60(2):159–71. [DOI](https://doi.org/10.1111/j.1365-2125.2005.02392.x) | [PMC free article](/articles/PMC1884933/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16042669/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=British%20Journal%20of%20Clinical%20Pharmacology&title=Meta-analysis%20of%20the%20influence%20of%20MDR1%20C3435T%20polymorphism%20on%20digoxin%20pharmacokinetics%20and%20MDR1%20gene%20expression&author=Chowbay%20Balram&author=Li%20Huihua&author=David%20Machin&author=Cheung%20Yin%20Bun&author=J.%20D.%20Lee%20Edmund&volume=60&issue=2&publication_year=2004&pages=159-71&pmid=16042669&doi=10.1111/j.1365-2125.2005.02392.x&)
|
| 229 |
+
|
| 230 |
+
20. Bing Zhang, Jie Kang, Xiaoming Chen. Methods to Combine Standard Deviations of Different Subgroups in Meta-analysis. Chinese Journal of Evidence-Based Medicine.2016;7:851–854. [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chinese%20Journal%20of%20Evidence-Based%20Medicine&title=Methods%20to%20Combine%20Standard%20Deviations%20of%20Different%20Subgroups%20in%20Meta-analysis&author=Zhang%20Bing&author=Kang%20Jie&author=Chen%20Xiaoming&volume=7&publication_year=2016&pages=851-854&)
|
| 231 |
+
|
| 232 |
+
21. Takeshi Suzuki, Kazuo Mihara, Goyo Nagai, Shoko Kagawa, Akifumi Nakamura, et al. Relationship Between UGT1A4 and UGT2B7 Polymorphisms and the Steady-State Serum Concentrations of Lamotrigine in Patients With Treatment-Resistant Depressive Disorder Receiving Lamotrigine as Augmentation Therapy. Ther Drug Monit. 2019;41(1):86–90. [DOI](https://doi.org/10.1097/FTD.0000000000000577) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30489548/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ther%20Drug%20Monit&title=Relationship%20Between%20UGT1A4%20and%20UGT2B7%20Polymorphisms%20and%20the%20Steady-State%20Serum%20Concentrations%20of%20Lamotrigine%20in%20Patients%20With%20Treatment-Resistant%20Depressive%20Disorder%20Receiving%20Lamotrigine%20as%20Augmentation%20Therapy&author=Suzuki%20Takeshi&author=Mihara%20Kazuo&author=Nagai%20Goyo&author=Kagawa%20Shoko&author=Nakamura%20Akifumi&volume=41&issue=1&publication_year=2019&pages=86-90&pmid=30489548&doi=10.1097/FTD.0000000000000577&)
|
| 233 |
+
|
| 234 |
+
22. Vaiva Petrenaite, Inger Öhman, Frederik Peter Thal Jantzen, Lena Ekström. Effect of UGT1A4, UGT2B7, UGT2B15, UGT2B17 and ABC1B polymorphisms on lamotrigine metabolism in Danish patients. Epilepsy Res. 2022;182:106897. doi: 10.1016/j.eplepsyres.2022.106897 [DOI](https://doi.org/10.1016/j.eplepsyres.2022.106897) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35303539/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Epilepsy%20Res&title=Effect%20of%20UGT1A4,%20UGT2B7,%20UGT2B15,%20UGT2B17%20and%20ABC1B%20polymorphisms%20on%20lamotrigine%20metabolism%20in%20Danish%20patients&author=Petrenaite%20Vaiva&author=%C3%96hman%20Inger&author=Thal%20Jantzen%20Frederik%20Peter&author=Ekstr%C3%B6m%20Lena&volume=182&publication_year=2022&pages=106897&pmid=35303539&doi=10.1016/j.eplepsyres.2022.106897&)
|
| 235 |
+
|
| 236 |
+
23. Medine Idrizoglu Gulcebi 1, Aydan Ozkaynakcı, Mehmet Zafer Goren, Rezzan Gulhan Aker, Cigdem Ozkara, et al. The relationship between UGT1A4 polymorphism and serum concentration of lamotrigine in patients with epilepsy. Epilepsy Res. 2011;95(1–2):1–8. doi: 10.1016/j.eplepsyres.2011.01.016 [DOI](https://doi.org/10.1016/j.eplepsyres.2011.01.016) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21601426/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Epilepsy%20Res&title=The%20relationship%20between%20UGT1A4%20polymorphism%20and%20serum%20concentration%20of%20lamotrigine%20in%20patients%20with%20epilepsy&author=Gulcebi%20Medine%20Idrizoglu&author=Ozkaynakc%C4%B1%20Aydan&author=Zafer%20Goren%20Mehmet&author=Gulhan%20Aker%20Rezzan&author=Ozkara%20Cigdem&volume=95&issue=1%E2%80%932&publication_year=2011&pages=1-8&pmid=21601426&doi=10.1016/j.eplepsyres.2011.01.016&)
|
| 237 |
+
|
| 238 |
+
24. ZHOU Ya-fang, WANG Xue-ding, ZHOU Lie-min, ZHANG Jie, LI Hong-liang, et al. Interethnic differences in UGT1A4 genetic polymorphisms and its effect on lamotrigine trough concentrations. Chin J Clin Pharmacol.2015;31(6):439–442. [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chin%20J%20Clin%20Pharmacol&title=Interethnic%20differences%20in%20UGT1A4%20genetic%20polymorphisms%20and%20its%20effect%20on%20lamotrigine%20trough%20concentrations&author=Ya-fang%20ZHOU&author=Xue-ding%20WANG&author=Lie-min%20ZHOU&author=Jie%20ZHANG&author=Hong-liang%20LI&volume=31&issue=6&publication_year=2015&pages=439-442&)
|
| 239 |
+
|
| 240 |
+
25. Liu Limin, Zhao Limei, Wang Qiuning, Qiu Feng, Wu Xiujun, et al. Influence of valproic acid concentration and polymorphism of UGT1A4*3, UGT2B7 -161C>T and UGT2B7*2 on serum concentration of lamotrigine in Chinese epileptic children. Eur J Clin Pharmacol. 2015;71(11):1341–7. [DOI](https://doi.org/10.1007/s00228-015-1925-9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26303110/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=Influence%20of%20valproic%20acid%20concentration%20and%20polymorphism%20of%20UGT1A4*3,%20UGT2B7%20-161C>T%20and%20UGT2B7*2%20on%20serum%20concentration%20of%20lamotrigine%20in%20Chinese%20epileptic%20children&author=Limin%20Liu&author=Limei%20Zhao&author=Qiuning%20Wang&author=Feng%20Qiu&author=Xiujun%20Wu&volume=71&issue=11&publication_year=2015&pages=1341-7&pmid=26303110&doi=10.1007/s00228-015-1925-9&)
|
| 241 |
+
|
| 242 |
+
26. Jiang LOU, Neng-ming LIN, Ling CHEN, Zhan-li LIU, WANG Wei, et al. Correlation between multiple epilepsy related gene polymorphisms and serum concentrations of lamotrigine in the treatment of epilepsy in children. Chinese Journal of General Practice.2021;1(19):17–19,45. [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chinese%20Journal%20of%20General%20Practice&title=Correlation%20between%20multiple%20epilepsy%20related%20gene%20polymorphisms%20and%20serum%20concentrations%20of%20lamotrigine%20in%20the%20treatment%20of%20epilepsy%20in%20children&author=LOU%20Jiang&author=LIN%20Neng-ming&author=CHEN%20Ling&author=LIU%20Zhan-li&author=%20WANG%20Wei&volume=1&issue=19&publication_year=2021&pages=17-19,45&)
|
| 243 |
+
|
| 244 |
+
27. Liu Limin. The influence of UGT genetic polymorphisms on the drug interaction between lamotrigine and valproic in epileptic children. Liaoning, China Medical University.2014. [Google Scholar](https://scholar.google.com/scholar_lookup?title=The%20influence%20of%20UGT%20genetic%20polymorphisms%20on%20the%20drug%20interaction%20between%20lamotrigine%20and%20valproic%20in%20epileptic%20children&author=Limin%20Liu&publication_year=2014&)
|
| 245 |
+
|
| 246 |
+
28. Yang Liya. Association study of UGT1A4 genetic polymorphism with the serum concentration and clinical effects of lamotrigine. Jilin, Jilin University.2013. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Association%20study%20of%20UGT1A4%20genetic%20polymorphism%20with%20the%20serum%20concentration%20and%20clinical%20effects%20of%20lamotrigine&author=Liya%20Yang&publication_year=2013&)
|
| 247 |
+
|
| 248 |
+
29. Du Zhongliang, Jiao Yukun, Shi Lianting. Association of UGT2B7 and UGT1A4 Polymorphisms with Serum Concentration of Antiepileptic Drugs in Children. Med Sci Monit. 2016;22:4107–4113. doi: 10.12659/msm.897626 [DOI](https://doi.org/10.12659/msm.897626) | [PMC free article](/articles/PMC5100833/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27795544/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Med%20Sci%20Monit&title=Association%20of%20UGT2B7%20and%20UGT1A4%20Polymorphisms%20with%20Serum%20Concentration%20of%20Antiepileptic%20Drugs%20in%20Children&author=Zhongliang%20Du&author=Yukun%20Jiao&author=Lianting%20Shi&volume=22&publication_year=2016&pages=4107-4113&pmid=27795544&doi=10.12659/msm.897626&)
|
| 249 |
+
|
| 250 |
+
30. Reimers Arne, Sjursen Wenche, Helde Grethe, Brodtkorb Eylert. Frequencies of UGT1A4*2 (P24T) and *3 (L48V) and their effects on serum concentrations of lamotrigine. Eur J Drug Metab Pharmacokinet. 2016;41(2):149–55. doi: 10.1007/s13318-014-0247-0 [DOI](https://doi.org/10.1007/s13318-014-0247-0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25492569/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Drug%20Metab%20Pharmacokinet&title=Frequencies%20of%20UGT1A4*2%20(P24T)%20and%20*3%20(L48V)%20and%20their%20effects%20on%20serum%20concentrations%20of%20lamotrigine&author=Arne%20Reimers&author=Wenche%20Sjursen&author=Grethe%20Helde&author=Eylert%20Brodtkorb&volume=41&issue=2&publication_year=2016&pages=149-55&pmid=25492569&doi=10.1007/s13318-014-0247-0&)
|
| 251 |
+
|
| 252 |
+
31. Kagawa S, Mihara K, Nakamura A, et al. Relationship between serum concentrations of lamotrigine and its early therapeutic effect of lamotrigine augmentation therapy in treatment-resistant depressive disorder. Ther Drug Monit. 2014; 36:730–733. [DOI](https://doi.org/10.1097/FTD.0000000000000088) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24819973/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ther%20Drug%20Monit&title=Relationship%20between%20serum%20concentrations%20of%20lamotrigine%20and%20its%20early%20therapeutic%20effect%20of%20lamotrigine%20augmentation%20therapy%20in%20treatment-resistant%20depressive%20disorder&author=S%20Kagawa&author=K%20Mihara&author=A%20Nakamura&volume=36&publication_year=2014&pages=730-733&pmid=24819973&doi=10.1097/FTD.0000000000000088&)
|
| 253 |
+
|
| 254 |
+
32. Blanca Sánchez M, Herranz JL, Leno C, et al. UGT2B7 -161C.T polymorphism is associated with lamotrigine concentration-to-dose ratio in a multivariate study. Ther Drug Monit. 2010;32:177–184. [DOI](https://doi.org/10.1097/FTD.0b013e3181ceecc6) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20216122/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ther%20Drug%20Monit&title=UGT2B7%20-161C.T%20polymorphism%20is%20associated%20with%20lamotrigine%20concentration-to-dose%20ratio%20in%20a%20multivariate%20study&author=M%20Blanca%20S%C3%A1nchez&author=JL%20Herranz&author=C%20Leno&volume=32&publication_year=2010&pages=177-184&pmid=20216122&doi=10.1097/FTD.0b013e3181ceecc6&)
|
| 255 |
+
|
| 256 |
+
33. Milosheska D, Lorber B, Vovk T, et al. Pharmacokinetics of lamotrigine and its metabolite N-2-glucuronide: influence of polymorphism of UDPglucuronosyltransferases and drug transporters. Br J Clin Pharmacol. 2016;82:399–41. doi: 10.1111/bcp.12984 [DOI](https://doi.org/10.1111/bcp.12984) | [PMC free article](/articles/PMC4972156/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27096250/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Pharmacokinetics%20of%20lamotrigine%20and%20its%20metabolite%20N-2-glucuronide:%20influence%20of%20polymorphism%20of%20UDPglucuronosyltransferases%20and%20drug%20transporters&author=D%20Milosheska&author=B%20Lorber&author=T%20Vovk&volume=82&publication_year=2016&pages=399-41&pmid=27096250&doi=10.1111/bcp.12984&)
|
| 257 |
+
|
| 258 |
+
34. Yu Guo, Li Guofu, Zheng Qingshan, Wang Daxin. Pediatric physiologically based pharmacokinetic model and its applications in pediatric drug research. Chinese Journal of New Drugs and Clinical Remedies. 2014;33 (6):414–420. [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chinese%20Journal%20of%20New%20Drugs%20and%20Clinical%20Remedies&title=Pediatric%20physiologically%20based%20pharmacokinetic%20model%20and%20its%20applications%20in%20pediatric%20drug%20research&author=Guo%20Yu&author=Guofu%20Li&author=Qingshan%20Zheng&author=Daxin%20Wang&volume=33&issue=6&publication_year=2014&pages=414-420&)
|
| 259 |
+
|
| 260 |
+
35. Zhou Yafang, Wang Xueding, Li Hongliang, Zhang Jie, Chen Ziyi, et al. Polymorphisms of ABCG2, ABCB1 and HNF4α are associated with Lamotrigine trough concentrations in epilepsy patients. Drug Metab Pharmacokinet. 2015;30(4):282–7. [DOI](https://doi.org/10.1016/j.dmpk.2015.05.002) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26213157/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Metab%20Pharmacokinet&title=Polymorphisms%20of%20ABCG2,%20ABCB1%20and%20HNF4%CE%B1%20are%20associated%20with%20Lamotrigine%20trough%20concentrations%20in%20epilepsy%20patients&author=Yafang%20Zhou&author=Xueding%20Wang&author=Hongliang%20Li&author=Jie%20Zhang&author=Ziyi%20Chen&volume=30&issue=4&publication_year=2015&pages=282-7&pmid=26213157&doi=10.1016/j.dmpk.2015.05.002&)
|
test/texts/PMC11315837.md
ADDED
|
@@ -0,0 +1,432 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# A Semi-Mechanistic Population Pharmacokinetic Model of Noscapine in Healthy Subjects Considering Hepatic First-Pass Extraction and CYP2C9 Genotypes
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Zhendong Chen, Max Taubert, Chunli Chen, Jana Boland, Qian Dong, Muhammad Bilal, Charalambos Dokos, Bertil Wachall, Manfred Wargenau, Bernhard Scheidel, Martin H J Wiesen, Elke Schaeffeler, Roman Tremmel, Matthias Schwab, Uwe Fuhr
|
| 5 |
+
**Journal:** Drugs in R&D
|
| 6 |
+
**Date:** 2024 May 29
|
| 7 |
+
**DOI:** [10.1007/s40268-024-00466-6](https://doi.org/10.1007/s40268-024-00466-6)
|
| 8 |
+
**PMID:** 38809387
|
| 9 |
+
**PMCID:** PMC11315837
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11315837/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11315837/pdf/40268_2024_Article_466.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11315837/pdf/40268_2024_Article_466.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Introduction:**
|
| 16 |
+
Noscapine is a commonly used cough suppressant, with ongoing research on its anti-inflammatory and anti-tumor properties. The drug has a pronounced pharmacokinetic variability.
|
| 17 |
+
|
| 18 |
+
**Objective:**
|
| 19 |
+
This evaluation aims to describe the pharmacokinetics of noscapine using a semi-mechanistic population pharmacokinetic model and to identify covariates that could explain inter-individual pharmacokinetic variability.
|
| 20 |
+
|
| 21 |
+
**Methods:**
|
| 22 |
+
Forty-eight healthy volunteers (30 men and 18 women, mean age 33 years) were enrolled in a randomized, two-period, two-stage, crossover bioequivalence study of noscapine in two different liquid formulations. Noscapine plasma concentrations following oral administration of noscapine 50 mg were evaluated by a non-compartmental analysis and by a population pharmacokinetic model separately.
|
| 23 |
+
|
| 24 |
+
**Results:**
|
| 25 |
+
Compared to the reference formulation, the test formulation exhibited ratios (with 94.12% confidence intervals) of 0.784 (0.662–0.929) and 0.827 (0.762–0.925) for peak plasma concentrations and area under the plasma concentration–time curve, respectively. Significant differences in p values (< 0.01) were both observed when comparing peak plasma concentrations and area under the plasma concentration–time curve between CYP2C9 genotype-predicted phenotypes. A three-compartmental model with zero-order absorption and first-order elimination process best described the plasma data. The introduction of a liver compartment was able to describe the profound first-pass effect of noscapine. Total body weight and the CYP2C9 genotype-predicted phenotype were both identified as significant covariates on apparent clearance, which was estimated as 958 ± 548 L/h for extensive metabolizers (CYP2C9*1/*1 and *1/*9), 531 ± 304 L/h for intermediate metabolizers with an activity score of 1.5 (CYP2C9*1/*2), and 343 ± 197 L/h for poor metabolizers and intermediate metabolizers with an activity score of 1.0 (CYP2C9*1/*3, *2/*3, and*3/*3).
|
| 26 |
+
|
| 27 |
+
**Conclusion:**
|
| 28 |
+
The current work is expected to facilitate the future pharmacokinetic/pharmacodynamic development of noscapine. This study was registered prior to starting at “Deutsches Register Klinischer Studien” under registration no. DRKS00017760.
|
| 29 |
+
|
| 30 |
+
**Supplementary Information:**
|
| 31 |
+
The online version contains supplementary material available at 10.1007/s40268-024-00466-6.
|
| 32 |
+
|
| 33 |
+
### Introduction
|
| 34 |
+
|
| 35 |
+
Noscapine is a commonly used cough suppressant, with ongoing research on its anti-inflammatory and anti-tumor properties. The drug has a pronounced pharmacokinetic variability.
|
| 36 |
+
|
| 37 |
+
### Objective
|
| 38 |
+
|
| 39 |
+
This evaluation aims to describe the pharmacokinetics of noscapine using a semi-mechanistic population pharmacokinetic model and to identify covariates that could explain inter-individual pharmacokinetic variability.
|
| 40 |
+
|
| 41 |
+
### Methods
|
| 42 |
+
|
| 43 |
+
Forty-eight healthy volunteers (30 men and 18 women, mean age 33 years) were enrolled in a randomized, two-period, two-stage, crossover bioequivalence study of noscapine in two different liquid formulations. Noscapine plasma concentrations following oral administration of noscapine 50 mg were evaluated by a non-compartmental analysis and by a population pharmacokinetic model separately.
|
| 44 |
+
|
| 45 |
+
### Results
|
| 46 |
+
|
| 47 |
+
Compared to the reference formulation, the test formulation exhibited ratios (with 94.12% confidence intervals) of 0.784 (0.662–0.929) and 0.827 (0.762–0.925) for peak plasma concentrations and area under the plasma concentration–time curve, respectively. Significant differences in *p*p values (< 0.01) were both observed when comparing peak plasma concentrations and area under the plasma concentration–time curve between *CYP2C9*CYP2C9 genotype-predicted phenotypes. A three-compartmental model with zero-order absorption and first-order elimination process best described the plasma data. The introduction of a liver compartment was able to describe the profound first-pass effect of noscapine. Total body weight and the *CYP2C9*CYP2C9 genotype-predicted phenotype were both identified as significant covariates on apparent clearance, which was estimated as 958 ± 548 L/h for extensive metabolizers (*CYP2C9*1/*1*CYP2C9*1/*1 and **1/*9**1/*9), 531 ± 304 L/h for intermediate metabolizers with an activity score of 1.5 (*CYP2C9*1/*2*CYP2C9*1/*2), and 343 ± 197 L/h for poor metabolizers and intermediate metabolizers with an activity score of 1.0 (*CYP2C9*1/*3*CYP2C9*1/*3, **2/*3**2/*3, and**3/*3**3/*3).
|
| 48 |
+
|
| 49 |
+
### Conclusion
|
| 50 |
+
|
| 51 |
+
The current work is expected to facilitate the future pharmacokinetic/pharmacodynamic development of noscapine. This study was registered prior to starting at “Deutsches Register Klinischer Studien” under registration no. DRKS00017760.
|
| 52 |
+
|
| 53 |
+
### Supplementary Information
|
| 54 |
+
|
| 55 |
+
The online version contains supplementary material available at 10.1007/s40268-024-00466-6.
|
| 56 |
+
|
| 57 |
+
## Key Points
|
| 58 |
+
|
| 59 |
+
| A semi-physiological model of noscapine was established, effectively describing the first-pass hepatic metabolism through the inclusion of a liver compartment. |
|
| 60 |
+
| --------------------------------------------------------------------------------------------------------------------------------------------------------------- |
|
| 61 |
+
| The CYP2C9 genotype had a major significant impact on noscapine clearance, and differences in noscapine clearance between CYP2C9 genotypes were quantified. |
|
| 62 |
+
| Total body weight and age were identified as significant covariates on noscapine clearance and inter-compartmental clearance, respectively. |
|
| 63 |
+
## Introduction
|
| 64 |
+
|
| 65 |
+
Noscapine is a naturally occurring alkaloid derived from the opium poppy. It has a long history of medicinal use and is mainly used as an antitussive agent while showing minimal toxic effects [[1](#CR1)1]. The main mechanism of action of noscapine appears to be σ-receptor agonism [[2](#CR2)2]. Suppression of bradykinin production has also been suggested as a mode of action in successfully suppressing cough induced by angiotensin-converting enzyme inhibitors [[3](#CR3)3]. Furthermore, recent studies indicated its potential use in further therapeutic areas [[4](#CR4)4]. In addition to antitussive activity, noscapine showed anti-inflammatory effects and various anti-tumor properties, including inhibiting cell proliferation, inducing apoptosis, and disrupting microtubule dynamics in cancer cells [[5](#CR5)5]. These effects make noscapine a candidate for further investigation in cancer therapy, although more research is needed to fully understand its mechanisms and potential clinical applications.
|
| 66 |
+
|
| 67 |
+
The pharmacokinetic (PK) properties of noscapine have been investigated by previous studies. Following oral administration, noscapine is rapidly absorbed from the gastrointestinal tract with a peak plasma concentration (*C*C_max_max) reached within 1–2 h. However, noscapine undergoes an extensive first-pass metabolism in the liver, resulting in relatively low oral bioavailability of approximately 30% [[6](#CR6)6, [7](#CR7)7]. In addition, a disproportionate increase in the area under the plasma concentration–time curve (AUC) of noscapine was observed in the oral dose range of 100–300 mg, with a three-fold increase in dose resulting in a nine-fold increase in AUC due to saturable first-pass metabolism [[7](#CR7)7]. Previous clinical findings and in vitro studies demonstrated the involvement of cytochrome P450 (*CYP*CYP) *2C9*2C9, *2C19*2C19 and *3A4/5*3A4/5 in the noscapine metabolism, with *CYP2C9*CYP2C9 being identified as the most important enzyme [[8](#CR8)8–[11](#CR11)11]. After entering the circulatory system, noscapine exhibits wide distribution throughout the body with a large volume of distribution of 4.7 L/kg, indicating that it distributes well into tissues [[6](#CR6)6]. Noscapine exposure following oral administration showed significant variability among individuals, with a coefficient of variation (CV) of 73% observed for the AUC following administration of a 200-mg tablet [[7](#CR7)7]. The plasma elimination half-life of noscapine also varies between individuals but is generally around 4–5 h [[7](#CR7)7].
|
| 68 |
+
|
| 69 |
+
Genetic variations observed in CYP genes, which code for enzymes involved in drug metabolism, are known to result in altered enzyme activity. This alteration mainly leads to differences in the pharmacokinetics of drugs, which may also result in differences in pharmacodynamics [[12](#CR12)12]. Previous studies provided evidence that two most prevalent allelic variants in European and Asian populations, *CYP2C9*2*CYP2C9*2 and *CYP2C9*3*CYP2C9*3, exhibit enzyme activity decreased by approximately 30% and 80%, respectively, when compared with the normal-function allele, *CYP2C9*1*CYP2C9*1 [[13](#CR13)13]. The most common allele in Africans, *CYP2C9*9*CYP2C9*9, had no relevant effect on the pharmacokinetics of substrates including phenytoin and warfarin [[14](#CR14)14, [15](#CR15)15]. The *CYP2C19*CYP2C19 alleles *CYP2C19*2, CYP2C19*3, CYP3A5*3,*CYP2C19*2, CYP2C19*3, CYP3A5*3, and *CYP3A5*6*CYP3A5*6 are related to null activity, while *CYP2C19*17*CYP2C19*17 is related to increased activity as reflected by respective effects on the pharmacokinetics of substrates mainly eliminated by *CYP2C19*CYP2C19 [[16](#CR16)16, [17](#CR17)17]. In addition, *CYP3A4*22*CYP3A4*22 appears to be the most significant relevant genetic variants, which is associated with reduced enzyme activity compared with the wild-type allele [[18](#CR18)18]. To date, there has been no investigation into the correlation between genetic variations in these *CYP*CYPs and the variability observed in noscapine exposure after oral administration.
|
| 70 |
+
|
| 71 |
+
We conducted a bioequivalence study to primarily compare *C*C_max_max and AUC from timepoint zero up to the last timepoint (AUC_0–t_0–*t*t) between a reformulated noscapine suspension formulation relative to a respective reference formulation (Nipaxon^®^® 5-mg/mL oral suspension). The purpose of the present secondary evaluation is to develop a semi-physiological PK model that characterizes hepatic first-pass extraction of noscapine and identifies covariates that could explain inter-individual PK variability of noscapine. This model is expected to help future investigations of noscapine in different therapeutic indications by a PK/pharmacodynamic analysis.
|
| 72 |
+
|
| 73 |
+
## Material and Methods
|
| 74 |
+
|
| 75 |
+
### Study Design
|
| 76 |
+
|
| 77 |
+
Pharmacokinetic data were obtained from a randomized, two-period, two-stage, cross-over bioequivalence study with two different formulations of noscapine conducted in 30 male and 18 female healthy subjects. The participants were recruited from the volunteer panel of the Clinical Pharmacology Unit of the Institute for Pharmacology at the University Hospital of Cologne. Subjects aged ≥ 18 years with a body mass index between 18.5 and 30 kg/m^2^2 were enrolled for the study when all results of the screening examination were available and the subject was considered eligible. Key exclusion criteria were suspicion of hypersensitivity to noscapine or other excipients of the formulations, history of any severe disease, any relevant clinical or laboratory abnormality, any concomitant medication, smoking, drug addiction, and pregnancy or breast feeding. Because of the pandemic situation caused by severe acute respiratory syndrome coronavirus 2 during the study, a negative test result within 48 h prior to hospitalization on the ward was required, only one subject per room was allowed, and the subjects had to comply with certain infection control rules. The study was designed to include up to two stages, with 48 subjects in each stage receiving a single dose of a Nipaxon^®^® 5-mg/mL oral suspension (McNeil, Solna, Sweden) [as the reference] or a reformulated oral noscapine suspension (InfectoPharm, Heppenheim, Germany) [as the test]. In each stage, the subjects were randomly assigned to two treatment sequences (reference-test, or test-reference), with a wash-out phase of 6–14 days between each period. The two-stage design was used to account for the uncertainty of the true intra-individual variability for both *C*C_max_max and AUC_0–t_0–*t*t [[19](#CR19)19]. The second stage was to be conducted if bioequivalence was not shown in the first stage and if achieving bioequivalence was not futile. The sample size for the standard average bioavailability testing considered to allow an appropriate assessment of relative bioavailability of noscapine from the two formulations would be *n*n = 48 for alpha = 0.0294 (allowing for the two-stage design) and a power of 80% if the true *μ*μ_test_test/*μ*μ_reference_reference ratio was in the 0.95–1.05 range and intra-individual variability for both *C*C_max_max and AUC_0–t_0–*t*t did not exceed 30%. For the PK analysis, blood samples were collected using lithium heparin tubes at baseline, 0.17, 0.33, 0.5, 0.67, 0.83, 1, 1.25, 1.5, 1.75, 2, 2.5, 3, 3.5, 4, 6, 9, 12, 16, and 24 h after a single oral dose of noscapine 50 mg. Samples for laboratory tests were taken at the screening visit 2–21 days prior to the first administration of the study drug. Samples for genotyping were taken only from randomized subjects and just prior to the first administration of study drugs. A principal component analysis based on the genome-wide genetic data indicated that most individuals of the cohort were of European ancestry, while two subjects clustered to South Asian and American populations and two other individuals clustered to the African population (Fig. [S1](#MOESM1)S1 of the Electronic Supplementary Material [ESM]).
|
| 78 |
+
|
| 79 |
+
### Analytical Method
|
| 80 |
+
|
| 81 |
+
The quantification of noscapine in plasma employed a validated liquid chromatography–mass spectrometry method [[20](#CR20)20]. Noscapine was extracted from plasma by a liquid-liquid extraction method using diethyl ether. An LC-20AD liquid chromatography system (Shimadzu, Duisburg, Germany) with PAL HTC-xt autosampler (CTC Analytics AG, Zwingen, Switzerland) was used for chromatographic separation. The separation process was conducted using a Luna C_18_18 column (3 μm; 100 × 2.0 mm; Phenomenex, Aschaffenburg, Germany) coupled with a pre-column Security Guard C_18_18 (4 × 2.0 mm; Phenomenex) at a column temperature of 40 °C. The mobile phase consisted of acetonitrile/water (80/20, v/v) containing 0.4 μM of ammonium acetate and 0.1% ammonium hydroxide at a flow rate of 0.35 mL/min. For mass spectrometry analysis, a 5500 triple quadrupole mass spectrometer (AB Sciex, Concord, Canada) equipped with an electrospray ionization source (TurboIonSpray^®^®) was utilized. The settings for IonSpray voltage and temperature were 5500 V and 500 °C, respectively. The ion-pair transitions of 414.0→220.1 and 322.1→304.2 were used to monitor noscapine and oxycodone-d_6_6 (internal standard), respectively. A data analysis was carried out using Analyst software version 1.6.2 (AB Sciex). Linearity was demonstrated over a standard curve that ranges from 0.100 to 100 ng/mL. Accuracy and precision were evaluated across four concentrations: 0.250, 8.00, 40.0, and 80.0 ng/mL. The relative deviations for accuracy fell within the range of − 7.9 to 0.9%, while the coefficients of variations for precision fell within the range of 2.8–12.7%. An incurred sample reanalysis was performed to requirements of the European Medicines Agency guideline on bioanalytical method validation, and 100% of the repeated samples aligned with the acceptance criteria of ± 20% [[21](#CR21)21].
|
| 82 |
+
|
| 83 |
+
### Non-compartmental Pharmacokinetic Analysis
|
| 84 |
+
|
| 85 |
+
A non-compartmental analysis was conducted using SAS version 9.4 (SAS Institute, Cary, NC, USA). Plasma PK parameters were calculated including apparent elimination rate constants (*λ*λ_z_z), terminal plasma elimination half-life (*t*t_1/2_1/2), *C*C_max_max, time to reach maximum plasma concentration (*t*t_max_max), AUC_0–t_0–*t*t, and AUC from time 0 extrapolated to infinity (AUC_0–∞_0–∞). The *λ*λ_z_z was calculated by a linear regression analysis of the terminal log-linear portion of the individual plasma concentration versus time curves and *t*t_1/2_1/2 was then calculated as ln(2)/*λ*λ_z_z. Peak plasma concentration and *t*t_max_max were obtained directly from the actual sampling times and concentrations, respectively. Area under the plasma concentration–time curve from timepoint zero up to the last timepoint was determined according to the linear trapezoidal rule concerning the interval of t = 0 to *t*t_max_max. From there up to the last measured value above the lower limit of quantification (LLOQ), it was determined using the logarithmic trapezoidal rule. Area under the plasma concentration–time curve from time 0 extrapolated to infinity was calculated as AUC_0–t_0–*t*t + *C*C_last_last(fit)/*λ*λ_z_z, where the *C*C_last_last(fit) is the last fitted concentration according to regression analysis. Peak plasma concentration and AUC_0–t_0–*t*t were compared separately using the *t*t-test (*p*p < 0.01) between different genotype-predicted phenotype groups. To assess relative bioavailability, log-transformed *C*C_max_max, AUC_0–t_0–*t*t, and AUC_0–∞_0–∞ were submitted to a linear analysis of variance model with effects for sequence. The mean square error of the analysis of variance was used as the variance estimate to calculate 94.12% confidence intervals (CIs). Bioequivalence of two different formulations could be concluded if the respective 94.12% CIs for *μ*μ_test_test/*μ*μ_reference_reference estimates were completely within the respective boundaries of 0.80–1.25 for *C*C_max_max and AUC_0–t_0–*t*t.
|
| 86 |
+
|
| 87 |
+
### Population Pharmacokinetic Analysis
|
| 88 |
+
|
| 89 |
+
The population PK analysis was performed using a non-linear mixed-effects modeling software NONMEM version 7.4.0 (ICON Development Solutions, Omaha, NE, USA) [[22](#CR22)22]. Model execution and diagnostic procedures were performed using Perl-speaks-NONMEM (PsN) version 5.3.0 (Uppsala University, Uppsala, Sweden) [[23](#CR23)23]. The first-order conditional estimation with interaction method was used throughout model development. A statistical criterion based on a difference of > 3.84 in the objective function value (OFV) between two nested models (*p*p < 0.05) that differed by one parameter was employed for model selection [[24](#CR24)24]. Post-processing and plotting of NONMEM data were completed using R version 4.3.0 ([https://www.R-project.org/](https://www.R-project.org/)https://www.R-project.org/).
|
| 90 |
+
|
| 91 |
+
#### Structural Pharmacokinetic Model
|
| 92 |
+
|
| 93 |
+
The structural model was developed in a stepwise manner, initially starting with the simplest model, which was a one-compartment model with first-order absorption and linear elimination kinetics. Subsequently, following approaches were explored to improve the model, including: (i) evaluating the number of apparent distribution compartments; (ii) comparing first-order, zero-order, and combined (first-order plus zero-order) absorption kinetics; (iii) incorporating a liver compartment to describe first-pass metabolism; (iv) introducing a lag time or varying numbers of transit compartment between depot and central compartments; (v) separating the parameter estimation for absorption processes for the different formulations; (vi) comparing linear and non-linear elimination processes; and (vii) assessing the random effects including inter-individual variability (IIV) and inter-occasion variability (IOV).
|
| 94 |
+
|
| 95 |
+
The IIV for PK parameters was modeled exponentially as the following equation: θi=θ×eηiθi=θ×eηiθi=θ×eηiθiθθii==θθ××eηieeeηiηηii, where θiθiθi��θii represents the value of the individual parameter, θθθθ represents the population point estimate of the parameter, and ηiηiηiηηii represents a normally distributed random variable with a mean of 0 and variance of ω2ω2ω2ωωω22. An additive residual error model was applied to the log-transformed observations versus time data (equivalent to an exponential error model with untransformed data). The schematic of the structural model is shown in Fig. [1](#Fig1)1.
|
| 96 |
+
|
| 97 |
+
|
| 98 |
+
|
| 99 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11315837_40268_2024_466_Fig1_HTML.jpg)
|
| 100 |
+
|
| 101 |
+
Schematic of the structural model of noscapine. CL/F apparent clearance, F1 relative bioavailability for test formulation of noscapine, Qh liver plasma flow (fixed at 55 L/h), Qp1/F intercompartment clearance between central and first peripheral compartment, Qp2/F intercompartment clearance between central and second peripheral compartment, Rate apparent absorption constant rate (equivalent to dose/absorption duration), TR average transit rate, Vc/F central compartment volume, Vh liver volume (fixed at 1.5 L), Vp1/F first peripheral compartment volume, Vp2/F second peripheral compartment volume
|
| 102 |
+
|
| 103 |
+
#### Covariate Model
|
| 104 |
+
|
| 105 |
+
A covariate analysis was performed on the structural model using forward addition and backward elimination methods, applying significance levels of 0.05 (ΔOFV ≤ − 3.84) and 0.01 (ΔOFV ≤ − 6.63), respectively. The effects of age, total body weight (TBW), height, body mass index, alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, estimated glomerular filtration rate (CKD-EPI, 2009) [[25](#CR25)25], and plasma urea concentration from laboratory test results as well as genotype-predicted phenotypes of *CYP2C9*CYP2C9, *CYP2C19*CYP2C19, *CYP3A4*CYP3A4, *CYP3A5*CYP3A5, and *CYP2E1*CYP2E1 on appropriate PK parameters were investigated. For covariate relationships, continuous covariates were modeled with mean-centered power models and categorical covariates were modeled with conditional effects.
|
| 106 |
+
|
| 107 |
+
#### Model Evaluation
|
| 108 |
+
|
| 109 |
+
During development of the structural model, sensitivity analyses were performed to verify the impact of predefined values and equations for parameters related to the liver compartment on parameter estimates, especially on values of apparent clearance (CL/*F*F), relative bioavailability of test formulation (F1), IIV, and residual unexplained variability. Therefore, various values of liver volume and liver plasma flow (*Q*Q_h_h) as well as different approaches to encoding the relationship between the liver and central compartment were evaluated separately using reported values and equations [[26](#CR26)26–[30](#CR30)30].
|
| 110 |
+
|
| 111 |
+
Following the covariate analysis, the final model was evaluated by individual fit plots, goodness-of-fit plots, and visual predictive checks (*n*n = 1000) stratified by the covariates of interest. Model stability and parameter uncertainty were assessed by sampling importance resampling (SIR, *M*M/*m*m ratio = 5000/1000) [[31](#CR31)31].
|
| 112 |
+
|
| 113 |
+
## Results
|
| 114 |
+
|
| 115 |
+
### Demographics and Genetic Polymorphism Analysis
|
| 116 |
+
|
| 117 |
+
A total of 1920 plasma concentrations of noscapine were collected from 48 healthy subjects, all of whom completed the two-period treatment. Observations with concentrations below the LLOQ were treated as zero before *C*C_max_max (96 concentrations) and were discarded after *C*C_max_max (17 concentrations) for the analysis. The study population included 30 men and 18 women with a mean age of 33 years (range 19–65 years). Detailed demographic information and baseline covariate values of enrolled subjects are listed in Table [1](#Tab1)1.
|
| 118 |
+
|
| 119 |
+
### Table 1.
|
| 120 |
+
|
| 121 |
+
Demographics and covariates for enrolled subjects
|
| 122 |
+
|
| 123 |
+
| Characteristic | N | Mean | SD | Median | Range |
|
| 124 |
+
| -------------- | - | ---- | -- | ------ | ----- |
|
| 125 |
+
| Demographics | | | | | |
|
| 126 |
+
| Male | 30 | – | – | – | |
|
| 127 |
+
| Female | 18 | – | – | – | |
|
| 128 |
+
| Age (years) | 48 | 33 | 11 | 29 | 19.0–65.0 |
|
| 129 |
+
| Height (cm) | 48 | 177 | 9 | 178 | 159–197 |
|
| 130 |
+
| Total body weight (kg) | 48 | 77.3 | 12.7 | 77.0 | 54.3–107 |
|
| 131 |
+
| BMI (kg/m2) | 48 | 24.6 | 2.9 | 24.3 | 18.0–29.5 |
|
| 132 |
+
| Covariates | | | | | |
|
| 133 |
+
| ALT (U/L) | 48 | 24.1 | 14.0 | 22.0 | 7.0–96.0 |
|
| 134 |
+
| AST (U/L) | 48 | 23.5 | 5.7 | 22.0 | 14.0–39.0 |
|
| 135 |
+
| ALP (U/L) | 48 | 64.8 | 13.5 | 63.0 | 36.0–103 |
|
| 136 |
+
| eGFR (mL/min) | 48 | 107 | 14 | 76.0 | 76.0–132 |
|
| 137 |
+
| Urea (mg/dL) | 48 | 26.7 | 6.6 | 13.0 | 13.0–40.0 |
|
| 138 |
+
Genotyping of *CYP2C9*CYP2C9, *CYP2C19*CYP2C19, *CYP3A4*CYP3A4, *CYP3A5*CYP3A5, and *CYP2E1*CYP2E1 was performed for all 48 subjects in this study. Genotype frequencies and phenotype classifications are shown in Table [2](#Tab2)2. According to the reported relative enzyme functions of different genotypes, *CYP2C9*CYP2C9, *CYP2C19*CYP2C19, *CYP3A4,*CYP3A4, and *CYP3A5*CYP3A5 were classified into three, four, two, and three phenotypes, respectively. Because of limited predictive data support, the phenotype of *CYP2E1*CYP2E1 was not applicable in this study [[12](#CR12)12]. Extensive metabolizers (EMs) or normal metabolizers were defined as possessing two alleles with normal enzyme function (i.e., *CYP2C9*1*CYP2C9*1 and *CYP2C9*9*CYP2C9*9, *CYP2C19*1*CYP2C19*1, *CYP3A4*1*CYP3A4*1, and *CYP3A5*1*CYP3A5*1). Intermediate metabolizers (IMs) were individuals with one allele with relative dysfunction (i.e., *CYP2C9*2*CYP2C9*2, *CYP2C19*2*CYP2C19*2, and *CYP3A4*22*CYP3A4*22) or severe dysfunction (i.e., *CYP2C9*3, CYP3A5*3,*CYP2C9*3, CYP3A5*3, and *CYP3A5*6*CYP3A5*6) and no severe dysfunction in the other allele [[32](#CR32)32–[34](#CR34)34]. Poor metabolizers (PMs) were individuals with two alleles displaying severe dysfunction (i.e., *CYP2C9*3, CYP3A5*3,*CYP2C9*3, CYP3A5*3, and *CYP3A5*6*CYP3A5*6) [[32](#CR32)32]. Of note, *CYP2C9*1/*3*CYP2C9*1/*3 and *CYP2C9*2/*2*CYP2C9*2/*2 as IMs with an activity score (AS) of 1, were classified together with the PMs into one group in this study because of the limited sample size of homozygous carriers [[35](#CR35)35]. In contrast, rapid metabolizers (RMs) and ultrarapid metabolizers (UMs) were characterized by having one and two alleles of increased enzyme function (i.e., *CYP2C19*17*CYP2C19*17), respectively [[36](#CR36)36].
|
| 139 |
+
|
| 140 |
+
### Table 2.
|
| 141 |
+
|
| 142 |
+
Summary of genotype and classification of CYP2C9, CYP2C19, CYP3A4, CYP3A5, and CYP2E1 in enrolled subjects (n = 48)
|
| 143 |
+
|
| 144 |
+
| Predicted phenotype | Genotype | No. of subjects | Frequency (%) | Reported frequency in European population (PharmGKB, dbSNP) |
|
| 145 |
+
| ------------------- | -------- | --------------- | ------------- | ----------------------------------------------------------- |
|
| 146 |
+
| CYP2C9 | | | | |
|
| 147 |
+
| EM | *1/*1 | 31 | 64.6 | 62.8 |
|
| 148 |
+
| | *1/*9 | 1 | 2.1 | |
|
| 149 |
+
| IM with AS of 1.5 | *1/*2 | 10 | 20.8 | 20.2 |
|
| 150 |
+
| PM & IM with AS of 1 | *1/*3 | 3 | 6.3 | 12 |
|
| 151 |
+
| | *2/*3 | 2 | 4.2 | 1.9 |
|
| 152 |
+
| | *3/*3 | 1 | 2.1 | 0.5 |
|
| 153 |
+
| CYP2C19 | | | | |
|
| 154 |
+
| UM | *17/*17 | 2 | 4.2 | 4.6 |
|
| 155 |
+
| RM | *1/*17 | 9 | 18.8 | 26 |
|
| 156 |
+
| EM | *1/*1 | 24 | 50.0 | 39 |
|
| 157 |
+
| IM | *1/*2 | 10 | 20.8 | 18 |
|
| 158 |
+
| | *2/*17 | 3 | 6.3 | 6 |
|
| 159 |
+
| CYP3A4 | | | | |
|
| 160 |
+
| EM | *1/*1 | 39 | 81.3 | |
|
| 161 |
+
| IM | *1/*22 | 9 | 18.8 | MAF ~ 5, here 9% |
|
| 162 |
+
| CYP3A5 | | | | |
|
| 163 |
+
| EM | *1/*1 | 0 | 0 | |
|
| 164 |
+
| IM | *1/*3 | 12 | 25.0 | Frequency of *1 is 5.3%a |
|
| 165 |
+
| PM | *3/*3 | 35 | 72.9 | Frequency of *3 is 94.3%a |
|
| 166 |
+
| | *3/*6 | 1 | 2.1 | Frequency of *6 is 0.3%a |
|
| 167 |
+
| CYP2E1 | | | | |
|
| 168 |
+
| NAb | *1/*1 | 34 | 70.8 | |
|
| 169 |
+
| | *1/*5 | 6 | 12.5 | |
|
| 170 |
+
| | *1/*7 | 8 | 16.7 | |
|
| 171 |
+
### Non-compartmental Pharmacokinetic Analysis
|
| 172 |
+
|
| 173 |
+
The mean plasma concentration–time profiles of noscapine stratified by formulation and *CYP2C9*CYP2C9 genotype-predicted phenotype following a single oral dose of 50 mg are shown in Fig. S2 of the ESM. There were minor differences in *C*C_max_max and AUC_0–t_0–*t*t between the two formulations. However, it was evident that subjects with a dysfunctional *CYP2C9*CYP2C9 allele had a noticeable higher level in both *C*C_max_max and AUC_0–t_0–*t*t. The corresponding PK parameters of noscapine in both test and reference formulations are presented in Table [3](#Tab3)3. Noscapine was rapidly absorbed, with a median *t*t_max_max of 0.5 h in both liquid formulations. In both formulations, noscapine showed a similar *t*t_1/2_1/2 with 8.42 and 8.29 h. The inter-individual CV (coefficient of variation) of AUC_0–t_0–*t*t for test and reference formulations were 79.0 and 78.9%, respectively, which is comparable to the reported value of 73% [[7](#CR7)7]. In terms of *C*C_max_max, AUC_0–t_0–*t*t, and AUC_0–∞_0–∞ of noscapine, the test formulation exhibited ratios (with 94.12% CIs of 0.784 (0.662–0.929), 0.827 (0.748–0.915), and 0.840 (0.762–0.925) compared to the reference formulation, respectively. These values did not completely fall within the bioequivalence acceptability range of 0.8–1.25, indicating that the two formulations investigated in this study are not bioequivalent. Boxplots of the *C*C_max_max and AUC_0–t_0–*t*t stratified by different genotype-predicted phenotypes of *CYP2C9*CYP2C9, *CYP2C19*CYP2C19, *CYP3A4*CYP3A4, *CYP3A5*CYP3A5, and *CYP2E1*CYP2E1 are displayed in Fig. [2](#Fig2)2. Significant differences were observed only between genotype-predicted phenotype groups of *CYP2C9*CYP2C9 when comparing *C*C_max_max (*p*p values*,*, 0.00091 and 0.00013) and AUC_0–t_0–*t*t (*p*p values, 0.0047 and 0.00096) for EMs with IMs (AS of 1.5) and EMs with PMs & IMs (AS of 1.0), respectively.
|
| 174 |
+
|
| 175 |
+
### Table 3.
|
| 176 |
+
|
| 177 |
+
Pharmacokinetic parameters of noscapine 50 mg in test and reference formulations by a non-compartmental analysis (n = 48)
|
| 178 |
+
|
| 179 |
+
| Parametersa | Test | Reference | T/R ratio | 94.12% CIs | CVintra (%)c |
|
| 180 |
+
| ----------- | ---- | --------- | --------- | ---------- | ------------ |
|
| 181 |
+
| Geometric mean (SD) | CV (%) | Geometric mean (SD) | CV (%) | | |
|
| 182 |
+
| t1/2 (h) | 8.42 (4.00) | 47.5 | 8.29 (3.54) | 42.7 | – | – | – |
|
| 183 |
+
| tmax (h)b | 0.5 (0.17–2.0) | 40.5 | 0.5 (0.33–1.25) | 37.0 | – | – | – |
|
| 184 |
+
| Cmax (μg/L) | 36.8 (31.2) | 84.9 | 46.9 (41.4) | 88.2 | 0.784 (1.821) | 0.662–0.929 | 44.9 |
|
| 185 |
+
| AUC0–t (μg/L·h) | 53.1 (41.9) | 79.0 | 64.1 (50.6) | 78.9 | 0.827 (1.428) | 0.748–0.915 | 25.9 |
|
| 186 |
+
| AUC0–∞ (μg/L·h) | 57.0 (44.5) | 78.1 | 67.8 (53.1) | 78.3 | 0.840 (1.409) | 0.762–0.925 | 24.9 |
|
| 187 |
+
### Fig. 2.
|
| 188 |
+
|
| 189 |
+

|
| 190 |
+
|
| 191 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11315837_40268_2024_466_Fig2_HTML.jpg)
|
| 192 |
+
|
| 193 |
+
Peak plasma concentration (Cmax) and area under the plasma concentration–time curve from timepoint zero up to the last timepoint (AUC0–t) stratified by genotype-predicted phenotype groups of CYP2C9, CYP2C19, CYP3A4, CYP3A5, and CYP2E1. *Genotype-predicted phenotype of CYP2E1 is not available. AS activity score, EM extensive metabolizer, IM intermediate metabolizer, PM poor metabolizer, RM rapid metabolizer, UM ultrarapid metabolizer
|
| 194 |
+
|
| 195 |
+
### Population Pharmacokinetic Analysis
|
| 196 |
+
|
| 197 |
+
#### Structural Population Pharmacokinetic Model
|
| 198 |
+
|
| 199 |
+
A three-compartment model provided the most reliable fit for noscapine PK profiles, surpassing both one-compartment and two-compartment models. Further improvements and attempts to refine the three-compartment model were listed in Table [S1](#MOESM1)S1 of the ESM. Zero-order absorption described the absorption process best, while using transit compartments instead of simply introducing a lag time better described the delayed gastric emptying. Manually including four sequential transit compartments proved to be the most robust approach compared with estimating the number of transit compartments and transit time [[37](#CR37)37]. The introduction of an F1 for the test formulation and a liver compartment consistently improved the model fit, regardless of the absorption models used, and therefore, they were both retained in the structural model.
|
| 200 |
+
|
| 201 |
+
A non-linear elimination model was slightly better compared with the linear elimination model (ΔOFV = − 6.79), but the estimation precision (relative standard error) for the maximum rate of reaction (*V*V_max_max) and the concentration of substrate that allows the enzyme to achieve half *V*V_max_max (*K*K_m_m) were 37.0% and 45.0%, respectively. These values both further increased during subsequent model development, indicating that the data did not support precise estimation of *V*V_max_max and *K*K_m_m for the non-linear elimination of noscapine. Therefore, the linear elimination model was applied in the final model.
|
| 202 |
+
|
| 203 |
+
The IIVs of absorption duration, CL/*F*F, central compartment volume (*V*V_c_c/*F*F), and F1 were estimated at 209, 76.4, 29.3, and 34.1% in the base model, respectively. Incorporating IOVs of the transit rate and inter-compartment clearance (CL) between the central and first peripheral compartment (*Q*Q_p1_p1/*F*F) resulted in significant decreases of − 1360 and − 208 in ΔOFV, respectively. Consequently, the IOVs of the transit rate and *Q*Q_p1_p1/*F*F were estimated as 50.6 and 32.4%, respectively. The results of other attempts, such as separating the estimation of lag time or absorption duration for different formulations and incorporating a non-linear elimination process, are not presented because of their tendency to yield unstable models and parameter estimations lacking pharmacological significance.
|
| 204 |
+
|
| 205 |
+
#### Covariate Analysis
|
| 206 |
+
|
| 207 |
+
The impact of the type of preparation, subject covariates, and genotype-predicted phenotypes on post-hoc parameter estimates were explored based on the full structural PK model. Among the parameters describing drug liberation/absorption processes, only apparent bioavailability differed between test and reference preparations. In line with the findings from a non-compartmental analysis, the *CYP2C9*CYP2C9 genotype-predicted phenotype had a significant effect on CL/*F*F (ΔOFV = − 16.4 when incorporating two additional parameters). In addition, age (ΔOFV = − 10.1) and TBW (ΔOFV = − 7.63) were found to be significant covariates on *Q*Q_p1_p1/*F*F and CL/*F*F, respectively. Because of a lack of significant effects, other clinical characteristics and genotype-predicted phenotypes of *CYP*CYP enzymes were not incorporated into the analysis. Therefore, the final model equations for *Q*Q_p1_p1/*F*F and CL/*F*F are presented below:
|
| 208 |
+
|
| 209 |
+
| Qp1F=Age29.00.348×TVQp1F×eη1, |
|
| 210 |
+
| ----------------------------- |
|
| 211 |
+
| CLF=TBW77.31.34×TVCLphenotypeF×eη2, |
|
| 212 |
+
| ----------------------------------- |
|
| 213 |
+
where TVQ_p1_*p*p1/*F*F is the typical population value for *Q*Q_p1_p1/*F*F and TVCL/*F*F_phenotype is the typical population value for CL/*F*F classified according to different *CYP2C9*CYP2C9 genotype-predicted phenotypes. Point estimates for CL/*F*F were 958, 531, and 343 L/h for EMs, IMs (AS of 1.5), and PMs & IMs (AS of 1.0), respectively.
|
| 214 |
+
|
| 215 |
+
#### Model Evaluation
|
| 216 |
+
|
| 217 |
+
The comparison of parameter estimates in sensitivity analyses is displayed in Table S2 of the ESM. Schemes of different settings for predefined parameters and equations related to the liver compartment are shown in Fig. S3 of the ESM. Employing covariate-related values for liver volume and *Q*Q_h_h as well as incorporating the hepatic extraction rate using CL/*F*F and *Q*Q_h_h did not demonstrate any advantage in parameter estimates over settings in the final model. In contrast, estimates of essential parameters i.e., CL/*F*F, F1, IIVs, and residual unexplained variability, remained nearly unchanged in models with different settings of the liver compartment. Therefore, the simpler relationship between liver and central compartments, and predefined values of 1.5 L and 55 L/h for liver volume and *Q*Q_h_h, respectively, were applied in the structural model.
|
| 218 |
+
|
| 219 |
+
An overview of the goodness-of-fit plots for the final model is displayed in Fig. S4 of the ESM. As shown, a satisfactory fit was achieved between the observed and predicted values, without trends of conditional weighted residuals over time. The adequacy of the predictive properties of the final model was demonstrated in Fig. [3](#Fig3)3, which displays the visual predictive check plots with stratification based on the *CYP2C9*CYP2C9 genotype-predicted phenotype.
|
| 220 |
+
|
| 221 |
+
|
| 222 |
+
|
| 223 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11315837_40268_2024_466_Fig3_HTML.jpg)
|
| 224 |
+
|
| 225 |
+
Visual predictive check (n = 1000) stratified by predicted CYP2C9 genotype-predicted phenotype for the final model. Dots represent observed concentrations. Black solid lines represent the median values, while dashed lines show the 5th and 95th percentiles of observed concentrations. Shaded areas are the model-predicted 95% confidence intervals for the 5th (green), 50th (yellow), and 95th (green) percentiles from 1000 simulated data sets. AS activity score, EM extensive metabolizer, IM intermediate metabolizer, PM poor metabolizer
|
| 226 |
+
|
| 227 |
+
The final model parameter estimates and the 95% CI of the SIR result with an *M*M/*m*m ratio of 5000/1000 are displayed in Table [4](#Tab4)4. The model was demonstrated stable by the consistency observed between parameter estimates and the medians from the SIR method. The diagnostic plots of SIR including distributions of the differences in OFV, spatial trend plots, and temporal trend plots (shown in Figs. S5–S7, respectively) collectively suggested that a sufficient *M*M/*m*m ratio was used and no discernible trends of the resampling proportion were observed for all PK parameters.
|
| 228 |
+
|
| 229 |
+
Parameter estimates and SIR results from the final model
|
| 230 |
+
|
| 231 |
+
| Parameter | Final model | SIR with M/m ratio of 5000/1000 |
|
| 232 |
+
| --------- | ----------- | ------------------------------- |
|
| 233 |
+
| Estimate | RSE (%) | Median | 95% confidence interval |
|
| 234 |
+
| D1 (h) | 0.0624 | 28.6 | 0.0663 | 0.0353–0.105 |
|
| 235 |
+
| CL/F_EM (L/h) | 958 | 10.3 | 939 | 747–1144 |
|
| 236 |
+
| CL/F_IM_1.5 (L/h) | 531 | 23.0 | 549 | 347–809 |
|
| 237 |
+
| CL/F_PM_IM_1.0 (L/h) | 343 | 24.8 | 364 | 224–549 |
|
| 238 |
+
| Vc/F (L) | 49.7 | 5.9 | 49.6 | 43.7–55.2 |
|
| 239 |
+
| Qp1/F (L/h) | 18.9 | 4.0 | 18.8 | 17.3–20.3 |
|
| 240 |
+
| Vp1/F (L) | 243 | 3.6 | 243 | 227–261 |
|
| 241 |
+
| Qp2/F (L/h) | 25.8 | 4.3 | 25.8 | 23.6–27.8 |
|
| 242 |
+
| Vp2/F (L) | 31.2 | 4.5 | 31.2 | 28.4–33.9 |
|
| 243 |
+
| F1 (%) | 82.8 | 5.4 | 83.1 | 75.2–92.2 |
|
| 244 |
+
| TR (1/h) | 13.0 | 6.4 | 13.0 | 11.4–14.7 |
|
| 245 |
+
| Covariates | | |
|
| 246 |
+
| TBW on CL/F | 1.34 | 35.1 | 1.35 | 0.485–2.33 |
|
| 247 |
+
| Age on Qp1/F | 0.348 | 32.3 | 0.343 | 0.125–0.568 |
|
| 248 |
+
| Inter-individual variability (IIV, %)a | | |
|
| 249 |
+
| D1 | 226 (19.1%)c | 40.6 | 247 | 120–639 |
|
| 250 |
+
| CL/F | 57.3 (0.1%) | 18.8 | 58.3 | 45.5–75.6 |
|
| 251 |
+
| Vc/F | 28.5 (5.2%) | 15.5 | 29.2 | 24.0–36.2 |
|
| 252 |
+
| F1 | 34.1 (1.2%) | 16.4 | 34.5 | 27.9–40.6 |
|
| 253 |
+
| Correlation for IIVsb | | |
|
| 254 |
+
| D1 and CL/F | 0.109 | 225 | – | – |
|
| 255 |
+
| D1 and V/F | − 0.175 | 92.6 | – | – |
|
| 256 |
+
| D1 and F1 | 0.162 | 131 | – | – |
|
| 257 |
+
| Inter-occasion variability (%) | | |
|
| 258 |
+
| TR | 50.7 (2.7%)c | 16.5 | 51.0 | 41.3–62.4 |
|
| 259 |
+
| Qp1/F | 30.4 (14.5%) | 13.2 | 31.0 | 25.4–41.3 |
|
| 260 |
+
| Residual variability | | |
|
| 261 |
+
| Additive error | 0.0420d (9.5%)c | 2.4 | 0.0418 | 0.0397–0.438 |
|
| 262 |
+
## Discussion
|
| 263 |
+
|
| 264 |
+
A bioequivalence study was conducted on 48 healthy subjects to compare two different formulations of noscapine. Plasma data were collected after oral administration of noscapine 50 mg, which were then utilized to develop a population PK model. The established model facilitated the evaluation of how polymorphisms in *CYP2C9*CYP2C9, *CYP2C19*CYP2C19, *CYP3A4*CYP3A4, *CYP3A5*CYP3A5, and *CYP2E1*CYP2E1 affected noscapine CL. Additionally, the model also enabled the investigation into the influence of various covariates such as demographics (age, weight, height, and body mass index), liver function markers (aspartate aminotransferase, alanine aminotransferase, alkaline phosphatase), and kidney function indexes (estimated glomerular filtration rate, urea plasma concentration) on noscapine PK parameters.
|
| 265 |
+
|
| 266 |
+
Despite the complexity and influence of numerous physiological factors on the drug absorption process, the population PK models commonly used tend to be relatively simple and empirical. One of the challenges of this study was to find a stable model capable of simultaneously describing the absorption process of noscapine well in both formulations. In addition to the attempts listed in Table [S1](#MOESM1)S1 of the ESM, alternative approaches were explored, such as separate estimation of absorption duration, absorption rate constant, lag time, and the number of transit compartments. The results showed that none of above approaches can generate a stable model with pharmacologically reasonable estimates of PK parameters. Subsequently, introduction of four sequential transit compartments with IOVs of the transit rate following zero-order absorption was proven to be the best approach to describe the complicated absorption process of noscapine in this case for both preparations. As no difference was found between the reference and test formulations in noscapine PK parameters describing the rate of liberation/absorption processes, it is tempting to speculate that the lower bioavailability of the test formulation could be due to a lower amount of drug released from the suspension.
|
| 267 |
+
|
| 268 |
+
A three-compartmental model, consisting of one central compartment and two apparent distribution compartments, fits the noscapine data best and was consequently selected as the structural model. The volumes of distribution in central (*V*V_c_c), first peripheral (*V*V_p1_p1), and second peripheral (*V*V_p2_p2) compartments were estimated at 49.7, 243, and 31.2 L, respectively, which indicated that noscapine has a wide distribution in the human body. Two previous studies on noscapine distribution demonstrated that high concentrations of noscapine were observed in various organs, including the liver, spleen, kidney, lung, and brain, after intravenous administration in rats [[38](#CR38)38, [39](#CR39)39]. Therefore, considering its characteristics of lipophilicity (Log*P*P = 2.6), it is not surprising that a large volume of distribution was observed in previous studies and this study [[40](#CR40)40, [41](#CR41)41].
|
| 269 |
+
|
| 270 |
+
Saturable first-pass metabolism of noscapine was confirmed through the observation of non-linear pharmacokinetics following oral administration within the dose range of 100–300 mg [[7](#CR7)7]. In the present study, introduction of a liver compartment between depot and central compartments led to a significant decrease of 562 in ΔOFV, which well explained the first-pass metabolism effect. Although a non-linear elimination process was discovered during model development, the estimates of *V*V_max_max and *K*K_m_m lacked accuracy with a relative standard error above 30%. Therefore, an oral dose of noscapine 50 mg may not result in relevant saturation of the corresponding metabolism enzymes in humans. The apparent *t*t_1/2_1/2 observed in this study is approximately twice the previously reported value of 4.5 h [[6](#CR6)6]. The observed difference could be attributed to the utilization of a more sensitive liquid chromatography-mass spectrometry method with a LLOQ of 0.100 μg/L in this study, compared with the high-performance liquid chromatography-ultraviolet method with a LLOQ of 5.0 μg/L in the previous study.
|
| 271 |
+
|
| 272 |
+
A previous in vitro study demonstrated that that primary metabolism of noscapine is mainly mediated by *CYP2C9*CYP2C9, with some contribution of *CP2C19*CP2C19 and *CYP3A4/5*CYP3A4/5, while there is no in vivo information on the relative contribution of individual enzymes to overall noscapine CL [[11](#CR11)11]. As an over-the-counter drug, noscapine has been reported to interact with vitamin K antagonists such as acenocoumarol, phenprocoumon, and warfarin, all of which are known *CYP2C9*CYP2C9 substrates, thus *CYP2C9*CYP2C9 genotypes may also have an impact on such interactions [[42](#CR42)42]. In this study, genotypes of *CYP2C9*CYP2C9, *CYP2C19*CYP2C19, and *CYP3A4*CYP3A4 were investigated among all enrolled subjects. As shown in Table [2](#Tab2)2, genotype frequencies of *CYP2C9*CYP2C9 were close to reported genotype frequencies in Caucasian individuals [[43](#CR43)43]. The allele frequencies of *CYP2C19*2*CYP2C19*2_,_*,*,* CYP2C19*17, CYP3A5*3,* CYP2C19*17, CYP3A5*3, and *CYP2C19*6*CYP2C19*6 were found to be 13.5, 16.7, 86.5, and 1.0%, respectively, which closely align with the reported values [[44](#CR44)44]. However, the allele frequencies of *CYP3A4*22*CYP3A4*22 and *CYP3A5*1*CYP3A5*1 were 9.4 and 12.5%, respectively, which are both two-fold higher than reported values of 3–5%, and 5.3% [[34](#CR34)34, [45](#CR45)45]. Deviations from published data could be explained by the small sample size of 48 (and the genetically determined non-European ancestry of four samples [Fig. [S1](#MOESM1)S1 of the ESM]).
|
| 273 |
+
|
| 274 |
+
The impact of the *CYP2C9*CYP2C9 genotype on the pharmacokinetics of drugs metabolized by this enzyme has been described for a number of drugs. For instance, S-warfarin metabolism was demonstrated through in vitro experiments, which revealed that the *CYP2C9*2*CYP2C9*2 and **3**3 variants displayed 70% and 5% metabolic efficiency, in comparison to the wild-type enzyme, respectively [[46](#CR46)46]. Among 156 patients, oral plasma CL of warfarin was reported as 2.4 ± 1.2, 2.2 ± 0.9, and 1.5 ± 1.0 mL/min for *CYP2C9*1/*1*CYP2C9*1/*1, **1/*2**1/*2, and **1/*3**1/*3 carriers, respectively [[47](#CR47)47]. Additionally, the genotype effect on tolbutamide elimination was evident with plasma CLs of 0.85, 0.77, 0.60, and 0.57 L/h for *CYP2C9*1/*1*CYP2C9*1/*1, **1/*2**1/*2, **1/*3**1/*3, and **2/*2**2/*2 carriers, respectively [[48](#CR48)48]. However, a more pronounced effect of the *CYP2C9*CYP2C9 genotype was observed on noscapine elimination in this study, with apparent CL of 958, 531, and 343 L/h for EMs (**1/*1**1/*1 and **1/*9**1/*9), IMs with an AS of 1.5 (**1/*2**1/*2), and PMs & IMs with an AS of 1.0 (**1/*3**1/*3, **2/*3**2/*3, **3/*3**3/*3), respectively. Compared with the nearly 100% oral bioavailability of warfarin, the pronounced first-pass metabolism of noscapine by *CYP2C9*CYP2C9 results in a low oral bioavailability (~ 30%) [[49](#CR49)49]. Thus, the *CYP2C9*CYP2C9 genotype effects may influence oral bioavailability and thus contribute to large differences in apparent CL between the genotype groups.
|
| 275 |
+
|
| 276 |
+
Compared with the published PK studies of noscapine [[6](#CR6)6, [7](#CR7)7], we observed similar characteristics, including a fast absorption process, profound first-pass metabolism, a large volume of distribution, and high IIV in plasma exposure. However, no obvious saturable elimination process or enzyme inhibition was observed in noscapine elimination. This discrepancy may be attributed to the lower oral dose of 50 mg investigated in our study compared with the minimum oral dose of 100 mg investigated in previous PK studies. Furthermore, our findings indicate that genetic variations in *CYP2C9*CYP2C9 significantly influence noscapine metabolism following a single dose of 50 mg, while variations in *CYP3A4*CYP3A4 and *CYP2C19*CYP2C19 do not lead to PK differences between groups [[9](#CR9)9–[11](#CR11)11]. This observation could be also associated with the comparatively lower plasma concentrations of noscapine after a single dose of 50 mg.
|
| 277 |
+
|
| 278 |
+
Several limitations remained in this study. Only one dose group of noscapine 50 mg was investigated for pharmacokinetics, which failed to support the investigation of its non-linear metabolic behavior. The primary objective of this bioequivalence study was not focused on a genotype assessment, and thus the sample size was not specifically optimized for this purpose. The small sample size of the *CYP2C9*3*CYP2C9*3 carrier, with particularly only one *CYP2C9*3/*3*CYP2C9*3/*3 carrier, resulted in relatively high relative standard error for CL estimates of the *CYP2C9*3*CYP2C9*3 allele when attempting to separately estimate CL for each *CYP2C9*CYP2C9 allele. Consequently, to address this limitation, all six individuals with the *CYP2C9*3*CYP2C9*3 variant were grouped together as PMs & IMs with an AS of 1.0 and CL for each *CYP2C9*CYP2C9 genotype-predicted phenotype was estimated instead of the genotype. The high IIV of CL/*F*F, which remained at 57.3% even after inclusion of related covariates, may be linked to the unknown variability in absolute bioavailability. Unfortunately, there is a lack of intravenous administration data to enhance our understanding of first-pass metabolism, which would help determine the absolute CL of noscapine.
|
| 279 |
+
|
| 280 |
+
## Conclusions
|
| 281 |
+
|
| 282 |
+
Overall, a semi-physiological model of noscapine was successfully established, effectively describing the first-pass hepatic metabolism, and providing an explanation for the impact of genetic variations on noscapine metabolism. The *CYP2C9*CYP2C9 enzyme was confirmed to play the most important role at noscapine metabolism at oral dose of 50 mg. Additionally, total body weight and age were identified as significant covariates on CL and *Q*Q_p1_p1/*F*F, respectively. The current work has provided a stable PK model for noscapine, which is expected to facilitate its future PK/pharmacodynamic development.
|
| 283 |
+
|
| 284 |
+
## Supplementary Information
|
| 285 |
+
|
| 286 |
+
Below is the link to the electronic supplementary material.
|
| 287 |
+
|
| 288 |
+
## Acknowledgements
|
| 289 |
+
|
| 290 |
+
Zhendong Chen received a scholarship from the China Scholarship Council for support of his PhD studies. Roman Tremmel, Elke Schaeffeler, and Matthias Schwab were supported by the Robert Bosch Stiftung, Stuttgart, Germany. Chunli Chen is supported by the 2022 ESI International High Impact Research Article Cooperation Program (No. 212-54900112), the National Natural Science Foundation of Heilongjiang Province (No. YQ2022C017), and the International Postdoctoral Exchange Fellowship Program from the Office of China Postdoctoral Council (Nos. 2020106 and PC2020013). The excellent technical assistance concerning the genotyping of Thomas Hees is gratefully acknowledged. We thank Prof. Dr. Markus M. Nöthen and Dr. Per Hoffmann for providing the SNP microarray data used in this study.
|
| 291 |
+
|
| 292 |
+
## Declarations
|
| 293 |
+
|
| 294 |
+
### Funding
|
| 295 |
+
|
| 296 |
+
The clinical study was funded by InfectoPharm Arzneimittel und Consilium GmbH, 64646 Heppenheim, Germany.
|
| 297 |
+
|
| 298 |
+
### Conflict of interest
|
| 299 |
+
|
| 300 |
+
Beyond funding of the study, Zhendong Chen, Max Taubert, Chunli Chen, Jana Boland, Qian Dong, Muhammad Bilal, Charalambos Dokos, Bertil Wachall, Manfred Wargenau, Bernhard Scheidel, Martin H. J. Wiesen, Elke Schaeffeler, Roman Tremmel, Matthias Schwab, and Uwe Fuhr have no conflicts of interest that are directly relevant to the content of this article.
|
| 301 |
+
|
| 302 |
+
### Ethics approval
|
| 303 |
+
|
| 304 |
+
The study protocol was approved by the Ethics Committee (No. 21-1404-AMG-ff) of the Faculty of Medicine of the University of Cologne, Germany (EUDRA-CT No. 2019-002012-12), and by the pertinent authorities and was conducted in accordance with Good Clinical Practice guidelines and the Declaration of Helsinki (64th WMA General Assembly, Brazil, October 2013). This study was registered prior to starting at “Deutsches Register Klinischer Studien” under registration no. DRKS00017760.
|
| 305 |
+
|
| 306 |
+
### Consent to participate
|
| 307 |
+
|
| 308 |
+
All subjects provided informed written consent to participate in genotyping and pharmacokinetic studies.
|
| 309 |
+
|
| 310 |
+
### Consent for publication
|
| 311 |
+
|
| 312 |
+
Not applicable.
|
| 313 |
+
|
| 314 |
+
### Availability of data and material
|
| 315 |
+
|
| 316 |
+
The data that support the findings of this study are available on request from the corresponding author.
|
| 317 |
+
|
| 318 |
+
### Code availability
|
| 319 |
+
|
| 320 |
+
Not applicable.
|
| 321 |
+
|
| 322 |
+
### Author contributions
|
| 323 |
+
|
| 324 |
+
ZC and UF wrote the manuscript; UF and BW designed the research; ZC, QD, MB, CD, BW, BS, MHJW, ES, RT, MS, and UF performed the research; ZC, MT, CC, MW, and JB analyzed the data.
|
| 325 |
+
|
| 326 |
+
## Associated Data
|
| 327 |
+
|
| 328 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 329 |
+
|
| 330 |
+
### Supplementary Materials
|
| 331 |
+
|
| 332 |
+
### Supplementary Materials
|
| 333 |
+
|
| 334 |
+
## References
|
| 335 |
+
|
| 336 |
+
1. Empey DW, Laitinen LA, Young GA, Bye CE, Hughes DT. Comparison of the antitussive effects of codeine phosphate 20 mg, dextromethorphan 30 mg and noscapine 30 mg using citric acid-induced cough in normal subjects. Eur J Clin Pharmacol. 1979;16(6):393–7. 10.1007/BF00568199 [DOI](https://doi.org/10.1007/BF00568199) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/527635/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Empey%20DW,%20Laitinen%20LA,%20Young%20GA,%20Bye%20CE,%20Hughes%20DT.%20Comparison%20of%20the%20antitussive%20effects%20of%20codeine%20phosphate%2020%20mg,%20dextromethorphan%2030%20mg%20and%20noscapine%2030%20mg%20using%20citric%20acid-induced%20cough%20in%20normal%20subjects.%20Eur%20J%20Clin%20Pharmacol.%201979;16(6):393%E2%80%937.%2010.1007/BF00568199)
|
| 337 |
+
|
| 338 |
+
2. Kamei J. Role of opioidergic and serotonergic mechanisms in cough and antitussives. Pulm Pharmacol. 1996;9(5–6):349–56. 10.1006/pulp.1996.0046 [DOI](https://doi.org/10.1006/pulp.1996.0046) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9232674/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Kamei%20J.%20Role%20of%20opioidergic%20and%20serotonergic%20mechanisms%20in%20cough%20and%20antitussives.%20Pulm%20Pharmacol.%201996;9(5%E2%80%936):349%E2%80%9356.%2010.1006/pulp.1996.0046)
|
| 339 |
+
|
| 340 |
+
3. Mooraki A, Jenabi A, Jabbari M, Zolfaghari MI, Javanmardi SZ, Mahmoudian M, et al. Noscapine suppresses angiotensin converting enzyme inhibitors-induced cough. Nephrology (Carlton). 2005;10(4):348–50. 10.1111/j.1440-1797.2005.00429.x [DOI](https://doi.org/10.1111/j.1440-1797.2005.00429.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16109080/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Mooraki%20A,%20Jenabi%20A,%20Jabbari%20M,%20Zolfaghari%20MI,%20Javanmardi%20SZ,%20Mahmoudian%20M,%20et%20al.%20Noscapine%20suppresses%20angiotensin%20converting%20enzyme%20inhibitors-induced%20cough.%20Nephrology%20(Carlton).%202005;10(4):348%E2%80%9350.%2010.1111/j.1440-1797.2005.00429.x)
|
| 341 |
+
|
| 342 |
+
4. Karlsson MO, Dahlström B, Neil A. Characterization of high-affinity binding sites for the antitussive [3H]noscapine in guinea pig brain tissue. Eur J Pharmacol. 1988;145(2):195–203. 10.1016/0014-2999(88)90230-0 [DOI](https://doi.org/10.1016/0014-2999(88)90230-0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/3350041/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Karlsson%20MO,%20Dahlstr%C3%B6m%20B,%20Neil%20A.%20Characterization%20of%20high-affinity%20binding%20sites%20for%20the%20antitussive%20%5B3H%5Dnoscapine%20in%20guinea%20pig%20brain%20tissue.%20Eur%20J%20Pharmacol.%201988;145(2):195%E2%80%93203.%2010.1016/0014-2999(88)90230-0)
|
| 343 |
+
|
| 344 |
+
5. Rahmanian-Devin P, Baradaran Rahimi V, Jaafari MR, Golmohammadzadeh S, Sanei-Far Z, Askari VR. Noscapine, an emerging medication for different diseases: a mechanistic review. Evid Based Complement Altern Med. 2021;2021:8402517. 10.1155/2021/8402517 [DOI](https://doi.org/10.1155/2021/8402517) | [PMC free article](/articles/PMC8648453/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34880922/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Rahmanian-Devin%20P,%20Baradaran%20Rahimi%20V,%20Jaafari%20MR,%20Golmohammadzadeh%20S,%20Sanei-Far%20Z,%20Askari%20VR.%20Noscapine,%20an%20emerging%20medication%20for%20different%20diseases:%20a%20mechanistic%20review.%20Evid%20Based%20Complement%20Altern%20Med.%202021;2021:8402517.%2010.1155/2021/8402517) | [Retracted](/articles/PMC10732963/)
|
| 345 |
+
|
| 346 |
+
6. Dahlström B, Mellstrand T, Löfdahl CG, Johansson M. Pharmacokinetic properties of noscapine. Eur J Clin Pharmacol. 1982;22(6):535–9. 10.1007/BF00609627 [DOI](https://doi.org/10.1007/BF00609627) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7128665/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Dahlstr%C3%B6m%20B,%20Mellstrand%20T,%20L%C3%B6fdahl%20CG,%20Johansson%20M.%20Pharmacokinetic%20properties%20of%20noscapine.%20Eur%20J%20Clin%20Pharmacol.%201982;22(6):535%E2%80%939.%2010.1007/BF00609627)
|
| 347 |
+
|
| 348 |
+
7. Karlsson MO, Dahlström B, Eckernäs SA, Johansson M, Alm AT. Pharmacokinetics of oral noscapine. Eur J Clin Pharmacol. 1990;39(3):275–9. 10.1007/BF00315110 [DOI](https://doi.org/10.1007/BF00315110) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/2257866/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Karlsson%20MO,%20Dahlstr%C3%B6m%20B,%20Eckern%C3%A4s%20SA,%20Johansson%20M,%20Alm%20AT.%20Pharmacokinetics%20of%20oral%20noscapine.%20Eur%20J%20Clin%20Pharmacol.%201990;39(3):275%E2%80%939.%2010.1007/BF00315110)
|
| 349 |
+
|
| 350 |
+
8. Scordo MG, Melhus H, Stjernberg E, Edvardsson AM, Wadelius M. Warfarin-noscapine interaction: a series of four case reports. Ann Pharmacother. 2008;42(3):448–50. 10.1345/aph.1K544 [DOI](https://doi.org/10.1345/aph.1K544) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18303135/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Scordo%20MG,%20Melhus%20H,%20Stjernberg%20E,%20Edvardsson%20AM,%20Wadelius%20M.%20Warfarin-noscapine%20interaction:%20a%20series%20of%20four%20case%20reports.%20Ann%20Pharmacother.%202008;42(3):448%E2%80%9350.%2010.1345/aph.1K544)
|
| 351 |
+
|
| 352 |
+
9. Fang ZZ, Zhang YY, Ge GB, Huo H, Liang SC, Yang L. Time-dependent inhibition (TDI) of CYP3A4 and CYP2C9 by noscapine potentially explains clinical noscapine-warfarin interaction. Br J Clin Pharmacol. 2010;69(2):193–9. 10.1111/j.1365-2125.2009.03572.x [DOI](https://doi.org/10.1111/j.1365-2125.2009.03572.x) | [PMC free article](/articles/PMC2824481/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20233183/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Fang%20ZZ,%20Zhang%20YY,%20Ge%20GB,%20Huo%20H,%20Liang%20SC,%20Yang%20L.%20Time-dependent%20inhibition%20(TDI)%20of%20CYP3A4%20and%20CYP2C9%20by%20noscapine%20potentially%20explains%20clinical%20noscapine-warfarin%20interaction.%20Br%20J%20Clin%20Pharmacol.%202010;69(2):193%E2%80%939.%2010.1111/j.1365-2125.2009.03572.x)
|
| 353 |
+
|
| 354 |
+
10. Rosenborg S, Stenberg M, Otto S, Ostervall J, Masquelier M, Yue QY, et al. Clinically significant CYP2C inhibition by noscapine but not by glucosamine. Clin Pharmacol Ther. 2010;88(3):343–6. 10.1038/clpt.2010.107 [DOI](https://doi.org/10.1038/clpt.2010.107) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20668444/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Rosenborg%20S,%20Stenberg%20M,%20Otto%20S,%20Ostervall%20J,%20Masquelier%20M,%20Yue%20QY,%20et%20al.%20Clinically%20significant%20CYP2C%20inhibition%20by%20noscapine%20but%20not%20by%20glucosamine.%20Clin%20Pharmacol%20Ther.%202010;88(3):343%E2%80%936.%2010.1038/clpt.2010.107)
|
| 355 |
+
|
| 356 |
+
11. Fang ZZ, Krausz KW, Li F, Cheng J, Tanaka N, Gonzalez FJ. Metabolic map and bioactivation of the anti-tumour drug noscapine. Br J Pharmacol. 2012;167(6):1271–86. 10.1111/j.1476-5381.2012.02067.x [DOI](https://doi.org/10.1111/j.1476-5381.2012.02067.x) | [PMC free article](/articles/PMC3504993/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22671862/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Fang%20ZZ,%20Krausz%20KW,%20Li%20F,%20Cheng%20J,%20Tanaka%20N,%20Gonzalez%20FJ.%20Metabolic%20map%20and%20bioactivation%20of%20the%20anti-tumour%20drug%20noscapine.%20Br%20J%20Pharmacol.%202012;167(6):1271%E2%80%9386.%2010.1111/j.1476-5381.2012.02067.x)
|
| 357 |
+
|
| 358 |
+
12. Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–41. 10.1016/j.pharmthera.2012.12.007 [DOI](https://doi.org/10.1016/j.pharmthera.2012.12.007) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23333322/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Zanger%20UM,%20Schwab%20M.%20Cytochrome%20P450%20enzymes%20in%20drug%20metabolism:%20regulation%20of%20gene%20expression,%20enzyme%20activities,%20and%20impact%20of%20genetic%20variation.%20Pharmacol%20Ther.%202013;138(1):103%E2%80%9341.%2010.1016/j.pharmthera.2012.12.007)
|
| 359 |
+
|
| 360 |
+
13. Crespi CL, Miller VP. The R144C change in the CYP2C9*2 allele alters interaction of the cytochrome P450 with NADPH:cytochrome P450 oxidoreductase. Pharmacogenetics. 1997;7(3):203–10. 10.1097/00008571-199706000-00005 [DOI](https://doi.org/10.1097/00008571-199706000-00005) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9241660/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Crespi%20CL,%20Miller%20VP.%20The%20R144C%20change%20in%20the%20CYP2C9*2%20allele%20alters%20interaction%20of%20the%20cytochrome%20P450%20with%20NADPH:cytochrome%20P450%20oxidoreductase.%20Pharmacogenetics.%201997;7(3):203%E2%80%9310.%2010.1097/00008571-199706000-00005)
|
| 361 |
+
|
| 362 |
+
14. Blaisdell J, Jorge-Nebert LF, Coulter S, Ferguson SS, Lee SJ, Chanas B, et al. Discovery of new potentially defective alleles of human CYP2C9. Pharmacogenetics. 2004;14(8):527–37. 10.1097/01.fpc.0000114759.08559.51 [DOI](https://doi.org/10.1097/01.fpc.0000114759.08559.51) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15284535/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Blaisdell%20J,%20Jorge-Nebert%20LF,%20Coulter%20S,%20Ferguson%20SS,%20Lee%20SJ,%20Chanas%20B,%20et%20al.%20Discovery%20of%20new%20potentially%20defective%20alleles%20of%20human%20CYP2C9.%20Pharmacogenetics.%202004;14(8):527%E2%80%9337.%2010.1097/01.fpc.0000114759.08559.51)
|
| 363 |
+
|
| 364 |
+
15. Niinuma Y, Saito T, Takahashi M, Tsukada C, Ito M, Hirasawa N, et al. Functional characterization of 32 CYP2C9 allelic variants. Pharmacogenomics J. 2014;14(2):107–14. 10.1038/tpj.2013.22 [DOI](https://doi.org/10.1038/tpj.2013.22) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23752738/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Niinuma%20Y,%20Saito%20T,%20Takahashi%20M,%20Tsukada%20C,%20Ito%20M,%20Hirasawa%20N,%20et%20al.%20Functional%20characterization%20of%2032%20CYP2C9%20allelic%20variants.%20Pharmacogenomics%20J.%202014;14(2):107%E2%80%9314.%2010.1038/tpj.2013.22)
|
| 365 |
+
|
| 366 |
+
16. Lee SJ. Clinical application of CYP2C19 pharmacogenetics toward more Pprsonalized Medicine. Front Genet. 2013;3:318. 10.3389/fgene.2012.00318 [DOI](https://doi.org/10.3389/fgene.2012.00318) | [PMC free article](/articles/PMC3561709/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23378847/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Lee%20SJ.%20Clinical%20application%20of%20CYP2C19%20pharmacogenetics%20toward%20more%20Pprsonalized%20Medicine.%20Front%20Genet.%202013;3:318.%2010.3389/fgene.2012.00318)
|
| 367 |
+
|
| 368 |
+
17. Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27(4):383–91. 10.1038/86882 [DOI](https://doi.org/10.1038/86882) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11279519/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Kuehl%20P,%20Zhang%20J,%20Lin%20Y,%20Lamba%20J,%20Assem%20M,%20Schuetz%20J,%20et%20al.%20Sequence%20diversity%20in%20CYP3A%20promoters%20and%20characterization%20of%20the%20genetic%20basis%20of%20polymorphic%20CYP3A5%20expression.%20Nat%20Genet.%202001;27(4):383%E2%80%9391.%2010.1038/86882)
|
| 369 |
+
|
| 370 |
+
18. Werk AN, Cascorbi I. Functional gene variants of CYP3A4. Clin Pharmacol Ther. 2014;96(3):340–8. 10.1038/clpt.2014.129 [DOI](https://doi.org/10.1038/clpt.2014.129) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24926778/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Werk%20AN,%20Cascorbi%20I.%20Functional%20gene%20variants%20of%20CYP3A4.%20Clin%20Pharmacol%20Ther.%202014;96(3):340%E2%80%938.%2010.1038/clpt.2014.129)
|
| 371 |
+
|
| 372 |
+
19. European Medicine Agency. Committee for Medicinal Products for Human Use (CHMP) guideline on the investigation of bioequivalence. CPMP/EWP/QWP/1401/98 Rev 1/Corr **. 2010. Available from: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-investigation-bioequivalence-rev1_en.pdf. Accessed 16 May 2024. [https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-investigation-bioequivalence-rev1_en.pdf](https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-investigation-bioequivalence-rev1_en.pdf)
|
| 373 |
+
|
| 374 |
+
20. Zhu L, Chen X, Zhang Y, Yu H, Zhong D. Simultaneous determination of methylephedrine and noscapine in human plasma by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;820(2):175–82. 10.1016/j.jchromb.2005.03.010 [DOI](https://doi.org/10.1016/j.jchromb.2005.03.010) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15899371/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Zhu%20L,%20Chen%20X,%20Zhang%20Y,%20Yu%20H,%20Zhong%20D.%20Simultaneous%20determination%20of%20methylephedrine%20and%20noscapine%20in%20human%20plasma%20by%20liquid%20chromatography-tandem%20mass%20spectrometry.%20J%20Chromatogr%20B%20Analyt%20Technol%20Biomed%20Life%20Sci.%202005;820(2):175%E2%80%9382.%2010.1016/j.jchromb.2005.03.010)
|
| 375 |
+
|
| 376 |
+
21. Guideline on bioanalytical method validation (EMEA/CHMP/EWP/192217/2009 Rev.1 Corr. 2**); 2011. Available from: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-bioanalytical-method-validation_en.pdf. Accessed 16 May 2024. [https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-bioanalytical-method-validation_en.pdf](https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-bioanalytical-method-validation_en.pdf)
|
| 377 |
+
|
| 378 |
+
22. Boeckmann A, Sheiner L, Beal S. NONMEM users guide: part V. San Francisco: University of California; 2001. [Google Scholar](https://scholar.google.com/scholar_lookup?Boeckmann%20A,%20Sheiner%20L,%20Beal%20S.%20NONMEM%20users%20guide:%20part%20V.%20San%20Francisco:%20University%20of%20California;%202001.)
|
| 379 |
+
|
| 380 |
+
23. Lindbom L, Pihlgren P, Jonsson EN. PsN-Toolkit: a collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Comput Methods Progr Biomed. 2005;79(3):241–57. 10.1016/j.cmpb.2005.04.005 [DOI](https://doi.org/10.1016/j.cmpb.2005.04.005) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16023764/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Lindbom%20L,%20Pihlgren%20P,%20Jonsson%20EN.%20PsN-Toolkit:%20a%20collection%20of%20computer%20intensive%20statistical%20methods%20for%20non-linear%20mixed%20effect%20modeling%20using%20NONMEM.%20Comput%20Methods%20Progr%20Biomed.%202005;79(3):241%E2%80%9357.%2010.1016/j.cmpb.2005.04.005)
|
| 381 |
+
|
| 382 |
+
24. National Institute of Standards and Technology and Semiconductor Manufacturing Technology (NIST/SEMATECH). e-Handbook of statistical methods. Section 1.3.6.7.4. Critical values of the chi-square distribution. Available from: https://www.itl.nist.gov/div898/handbook/eda/section3/eda3674.htm. Accessed 14 June 2023. [https://www.itl.nist.gov/div898/handbook/eda/section3/eda3674.htm](https://www.itl.nist.gov/div898/handbook/eda/section3/eda3674.htm)
|
| 383 |
+
|
| 384 |
+
25. Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration), et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150(9):604–12. 10.7326/0003-4819-150-9-200905050-00006 [DOI](https://doi.org/10.7326/0003-4819-150-9-200905050-00006) | [PMC free article](/articles/PMC2763564/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19414839/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Levey%20AS,%20Stevens%20LA,%20Schmid%20CH,%20Zhang%20YL,%20Castro%20AF%203rd,%20Feldman%20HI,%20CKD-EPI%20(Chronic%20Kidney%20Disease%20Epidemiology%20Collaboration),%20et%20al.%20A%20new%20equation%20to%20estimate%20glomerular%20filtration%20rate.%20Ann%20Intern%20Med.%202009;150(9):604%E2%80%9312.%2010.7326/0003-4819-150-9-200905050-00006)
|
| 385 |
+
|
| 386 |
+
26. Brown RP, Delp MD, Lindstedt SL, Rhomberg LR, Beliles RP. Physiological parameter values for physiologically based pharmacokinetic models. Toxicol Ind Health. 1997;13(4):407–84. 10.1177/074823379701300401 [DOI](https://doi.org/10.1177/074823379701300401) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9249929/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Brown%20RP,%20Delp%20MD,%20Lindstedt%20SL,%20Rhomberg%20LR,%20Beliles%20RP.%20Physiological%20parameter%20values%20for%20physiologically%20based%20pharmacokinetic%20models.%20Toxicol%20Ind%20Health.%201997;13(4):407%E2%80%9384.%2010.1177/074823379701300401)
|
| 387 |
+
|
| 388 |
+
27. Johnson TN, Tucker GT, Tanner MS, Rostami-Hodjegan A. Changes in liver volume from birth to adulthood: a meta-analysis. Liver Transpl. 2005;11(12):1481–93. 10.1002/lt.20519 [DOI](https://doi.org/10.1002/lt.20519) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16315293/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Johnson%20TN,%20Tucker%20GT,%20Tanner%20MS,%20Rostami-Hodjegan%20A.%20Changes%20in%20liver%20volume%20from%20birth%20to%20adulthood:%20a%20meta-analysis.%20Liver%20Transpl.%202005;11(12):1481%E2%80%9393.%2010.1002/lt.20519)
|
| 389 |
+
|
| 390 |
+
28. Gordi T, Xie R, Huong NV, Huong DX, Karlsson MO, Ashton M. A semiphysiological pharmacokinetic model for artemisinin in healthy subjects incorporating autoinduction of metabolism and saturable first-pass hepatic extraction. Br J Clin Pharmacol. 2005;59(2):189–98. 10.1111/j.1365-2125.2004.02321.x [DOI](https://doi.org/10.1111/j.1365-2125.2004.02321.x) | [PMC free article](/articles/PMC1884742/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15676041/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Gordi%20T,%20Xie%20R,%20Huong%20NV,%20Huong%20DX,%20Karlsson%20MO,%20Ashton%20M.%20A%20semiphysiological%20pharmacokinetic%20model%20for%20artemisinin%20in%20healthy%20subjects%20incorporating%20autoinduction%20of%20metabolism%20and%20saturable%20first-pass%20hepatic%20extraction.%20Br%20J%20Clin%20Pharmacol.%202005;59(2):189%E2%80%9398.%2010.1111/j.1365-2125.2004.02321.x)
|
| 391 |
+
|
| 392 |
+
29. Quinney SK, Mohamed AN, Hebert MF, Haas DM, Clark S, Umans JG, et al. A semi-mechanistic metabolism model of CYP3A substrates in pregnancy: predicting changes in midazolam and nifedipine pharmacokinetics. CPT Pharmacometr Syst Pharmacol. 2012;1(9): e2. 10.1038/psp.2012.5 [DOI](https://doi.org/10.1038/psp.2012.5) | [PMC free article](/articles/PMC3603475/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23835882/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Quinney%20SK,%20Mohamed%20AN,%20Hebert%20MF,%20Haas%20DM,%20Clark%20S,%20Umans%20JG,%20et%20al.%20A%20semi-mechanistic%20metabolism%20model%20of%20CYP3A%20substrates%20in%20pregnancy:%20predicting%20changes%20in%20midazolam%20and%20nifedipine%20pharmacokinetics.%20CPT%20Pharmacometr%20Syst%20Pharmacol.%202012;1(9):%20e2.%2010.1038/psp.2012.5)
|
| 393 |
+
|
| 394 |
+
30. Krishnatry AS, Voelkner A, Dhar A, Prohn M, Ferron-Brady G. Population pharmacokinetic modeling of molibresib and its active metabolites in patients with solid tumors: a semimechanistic autoinduction model. CPT Pharmacometr Syst Pharmacol. 2021;10(7):709–22. 10.1002/psp4.12639 [DOI](https://doi.org/10.1002/psp4.12639) | [PMC free article](/articles/PMC8302244/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33955700/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Krishnatry%20AS,%20Voelkner%20A,%20Dhar%20A,%20Prohn%20M,%20Ferron-Brady%20G.%20Population%20pharmacokinetic%20modeling%20of%20molibresib%20and%20its%20active%20metabolites%20in%20patients%20with%20solid%20tumors:%20a%20semimechanistic%20autoinduction%20model.%20CPT%20Pharmacometr%20Syst%20Pharmacol.%202021;10(7):709%E2%80%9322.%2010.1002/psp4.12639)
|
| 395 |
+
|
| 396 |
+
31. Dosne AG, Bergstrand M, Harling K, Karlsson MO. Improving the estimation of parameter uncertainty distributions in nonlinear mixed effects models using sampling importance resampling. J Pharmacokinet Pharmacodyn. 2016;43(6):583–96. 10.1007/s10928-016-9487-8 [DOI](https://doi.org/10.1007/s10928-016-9487-8) | [PMC free article](/articles/PMC5110709/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27730482/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Dosne%20AG,%20Bergstrand%20M,%20Harling%20K,%20Karlsson%20MO.%20Improving%20the%20estimation%20of%20parameter%20uncertainty%20distributions%20in%20nonlinear%20mixed%20effects%20models%20using%20sampling%20importance%20resampling.%20J%20Pharmacokinet%20Pharmacodyn.%202016;43(6):583%E2%80%9396.%2010.1007/s10928-016-9487-8)
|
| 397 |
+
|
| 398 |
+
32. Kirchheiner J, Tsahuridu M, Jabrane W, Roots I, Brockmöller J. The CYP2C9 polymorphism: from enzyme kinetics to clinical dose recommendations. Per Med. 2004;1(1):63–84. 10.1517/17410541.1.1.63 [DOI](https://doi.org/10.1517/17410541.1.1.63) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29793229/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Kirchheiner%20J,%20Tsahuridu%20M,%20Jabrane%20W,%20Roots%20I,%20Brockm%C3%B6ller%20J.%20The%20CYP2C9%20polymorphism:%20from%20enzyme%20kinetics%20to%20clinical%20dose%20recommendations.%20Per%20Med.%202004;1(1):63%E2%80%9384.%2010.1517/17410541.1.1.63)
|
| 399 |
+
|
| 400 |
+
33. Hicks JK, Bishop JR, Sangkuhl K, Müller DJ, Ji Y, Leckband SG, Clinical Pharmacogenetics Implementation Consortium, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127–34. 10.1002/cpt.147 [DOI](https://doi.org/10.1002/cpt.147) | [PMC free article](/articles/PMC4512908/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25974703/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Hicks%20JK,%20Bishop%20JR,%20Sangkuhl%20K,%20M%C3%BCller%20DJ,%20Ji%20Y,%20Leckband%20SG,%20Clinical%20Pharmacogenetics%20Implementation%20Consortium,%20et%20al.%20Clinical%20Pharmacogenetics%20Implementation%20Consortium%20(CPIC)%20guideline%20for%20CYP2D6%20and%20CYP2C19%20genotypes%20and%20dosing%20of%20selective%20serotonin%20reuptake%20inhibitors.%20Clin%20Pharmacol%20Ther.%202015;98(2):127%E2%80%9334.%2010.1002/cpt.147)
|
| 401 |
+
|
| 402 |
+
34. Mulder TAM, van Eerden RAG, de With M, Elens L, Hesselink DA, Matic M, et al. CYP3A4∗22 genotyping in clinical practice: ready for implementation? Front Genet. 2021;12: 711943. 10.3389/fgene.2021.711943 [DOI](https://doi.org/10.3389/fgene.2021.711943) | [PMC free article](/articles/PMC8296839/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34306041/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Mulder%20TAM,%20van%20Eerden%20RAG,%20de%20With%20M,%20Elens%20L,%20Hesselink%20DA,%20Matic%20M,%20et%20al.%20CYP3A4%E2%88%9722%20genotyping%20in%20clinical%20practice:%20ready%20for%20implementation?%20Front%20Genet.%202021;12:%20711943.%2010.3389/fgene.2021.711943)
|
| 403 |
+
|
| 404 |
+
35. Theken KN, Lee CR, Gong L, Caudle KE, Formea CM, Gaedigk A, et al. Clinical Pharmacogenetics Implementation Consortium guideline (CPIC) for CYP2C9 and nonsteroidal anti-inflammatory drugs. Clin Pharmacol Ther. 2020;108(2):191–200. 10.1002/cpt.1830 [DOI](https://doi.org/10.1002/cpt.1830) | [PMC free article](/articles/PMC8080882/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32189324/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Theken%20KN,%20Lee%20CR,%20Gong%20L,%20Caudle%20KE,%20Formea%20CM,%20Gaedigk%20A,%20et%20al.%20Clinical%20Pharmacogenetics%20Implementation%20Consortium%20guideline%20(CPIC)%20for%20CYP2C9%20and%20nonsteroidal%20anti-inflammatory%20drugs.%20Clin%20Pharmacol%20Ther.%202020;108(2):191%E2%80%93200.%2010.1002/cpt.1830)
|
| 405 |
+
|
| 406 |
+
36. PharmGKB (2023) Gene-specific information tables for CYP2C19. Available from: https://www.pharmgkb.org/page/cyp2c19RefMaterials. [https://www.pharmgkb.org/page/cyp2c19RefMaterials](https://www.pharmgkb.org/page/cyp2c19RefMaterials)
|
| 407 |
+
|
| 408 |
+
37. Savic RM, Jonker DM, Kerbusch T, Karlsson MO. Implementation of a transit compartment model for describing drug absorption in pharmacokinetic studies. J Pharmacokinet Pharmacodyn. 2007;34(5):711–26. 10.1007/s10928-007-9066-0 [DOI](https://doi.org/10.1007/s10928-007-9066-0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17653836/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Savic%20RM,%20Jonker%20DM,%20Kerbusch%20T,%20Karlsson%20MO.%20Implementation%20of%20a%20transit%20compartment%20model%20for%20describing%20drug%20absorption%20in%20pharmacokinetic%20studies.%20J%20Pharmacokinet%20Pharmacodyn.%202007;34(5):711%E2%80%9326.%2010.1007/s10928-007-9066-0)
|
| 409 |
+
|
| 410 |
+
38. Nayak KP, Brochmann-hanssen E, Way EL. Biological disposition of noscapine. I: kinetics of metabolism, urinary excretion, and organ distribution. J Pharm Sci. 1965;54:191–4. 10.1002/jps.2600540206 [DOI](https://doi.org/10.1002/jps.2600540206) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14293802/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Nayak%20KP,%20Brochmann-hanssen%20E,%20Way%20EL.%20Biological%20disposition%20of%20noscapine.%20I:%20kinetics%20of%20metabolism,%20urinary%20excretion,%20and%20organ%20distribution.%20J%20Pharm%20Sci.%201965;54:191%E2%80%934.%2010.1002/jps.2600540206)
|
| 411 |
+
|
| 412 |
+
39. Priyadarshani A, Chuttani K, Mittal G, Bhatnagar A. Radiolabeling, biodistribution and gamma scintigraphy of noscapine hydrochloride in normal and polycystic ovary induced rats. J Ovarian Res. 2010;3:10. 10.1186/1757-2215-3-10 [DOI](https://doi.org/10.1186/1757-2215-3-10) | [PMC free article](/articles/PMC2877043/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20420718/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Priyadarshani%20A,%20Chuttani%20K,%20Mittal%20G,%20Bhatnagar%20A.%20Radiolabeling,%20biodistribution%20and%20gamma%20scintigraphy%20of%20noscapine%20hydrochloride%20in%20normal%20and%20polycystic%20ovary%20induced%20rats.%20J%20Ovarian%20Res.%202010;3:10.%2010.1186/1757-2215-3-10)
|
| 413 |
+
|
| 414 |
+
40. Nourbakhsh F, Askari VR. Biological and pharmacological activities of noscapine: focusing on its receptors and mechanisms. BioFactors. 2021;47(6):975–91. 10.1002/biof.1781 [DOI](https://doi.org/10.1002/biof.1781) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34534373/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Nourbakhsh%20F,%20Askari%20VR.%20Biological%20and%20pharmacological%20activities%20of%20noscapine:%20focusing%20on%20its%20receptors%20and%20mechanisms.%20BioFactors.%202021;47(6):975%E2%80%9391.%2010.1002/biof.1781)
|
| 415 |
+
|
| 416 |
+
41. Mansoor A, Mahabadi N. Volume of distribution [updated 2022 Jul 25]. In: StatPearls Treasure Island: StatPearls Publishing; 2023. Available from: https://www.ncbi.nlm.nih.gov/books/NBK545280/. Accessed 16 May 2024. [https://www.ncbi.nlm.nih.gov/books/NBK545280/](https://www.ncbi.nlm.nih.gov/books/NBK545280/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31424864/)
|
| 417 |
+
|
| 418 |
+
42. Lokhorst B, Rolfes L, Jessurun NT. Interaction of OTC drug noscapine and acenocoumarol and phenprocoumon. Br J Clin Pharmacol. 2019;85(5):1041–3. 10.1111/bcp.13887 [DOI](https://doi.org/10.1111/bcp.13887) | [PMC free article](/articles/PMC6475677/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30809820/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Lokhorst%20B,%20Rolfes%20L,%20Jessurun%20NT.%20Interaction%20of%20OTC%20drug%20noscapine%20and%20acenocoumarol%20and%20phenprocoumon.%20Br%20J%20Clin%20Pharmacol.%202019;85(5):1041%E2%80%933.%2010.1111/bcp.13887)
|
| 419 |
+
|
| 420 |
+
43. Siddiqi A, Khan DA, Khan FA, Naveed AK. Impact of CYP2C9 genetic polymorphism on warfarin dose requirements in Pakistani population. Pak J Pharm Sci. 2010;23(4):417–22. [PubMed](https://pubmed.ncbi.nlm.nih.gov/20884456/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Siddiqi%20A,%20Khan%20DA,%20Khan%20FA,%20Naveed%20AK.%20Impact%20of%20CYP2C9%20genetic%20polymorphism%20on%20warfarin%20dose%20requirements%20in%20Pakistani%20population.%20Pak%20J%20Pharm%20Sci.%202010;23(4):417%E2%80%9322.)
|
| 421 |
+
|
| 422 |
+
44. Petrović J, Pešić V, Lauschke VM. Frequencies of clinically important CYP2C19 and CYP2D6 alleles are graded across Europe. Eur J Hum Genet. 2020;28(1):88–94. 10.1038/s41431-019-0480-8. 10.1038/s41431-019-0480-8 [DOI](https://doi.org/10.1038/s41431-019-0480-8) | [PMC free article](/articles/PMC6906321/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31358955/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Petrovi%C4%87%20J,%20Pe%C5%A1i%C4%87%20V,%20Lauschke%20VM.%20Frequencies%20of%20clinically%20important%20CYP2C19%20and%20CYP2D6%20alleles%20are%20graded%20across%20Europe.%20Eur%20J%20Hum%20Genet.%202020;28(1):88%E2%80%9394.%2010.1038/s41431-019-0480-8.%2010.1038/s41431-019-0480-8)
|
| 423 |
+
|
| 424 |
+
45. Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther. 2017;102(4):688–700. 10.1002/cpt.690 [DOI](https://doi.org/10.1002/cpt.690) | [PMC free article](/articles/PMC5600063/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28378927/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Zhou%20Y,%20Ingelman-Sundberg%20M,%20Lauschke%20VM.%20Worldwide%20distribution%20of%20cytochrome%20P450%20alleles:%20a%20meta-analysis%20of%20population-scale%20sequencing%20projects.%20Clin%20Pharmacol%20Ther.%202017;102(4):688%E2%80%93700.%2010.1002/cpt.690)
|
| 425 |
+
|
| 426 |
+
46. Rettie AE, Korzekwa KR, Kunze KL, Lawrence RF, Eddy AC, Aoyama T, et al. Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: a role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chem Res Toxicol. 1992;5(1):54–9. 10.1021/tx00025a009 [DOI](https://doi.org/10.1021/tx00025a009) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1581537/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Rettie%20AE,%20Korzekwa%20KR,%20Kunze%20KL,%20Lawrence%20RF,%20Eddy%20AC,%20Aoyama%20T,%20et%20al.%20Hydroxylation%20of%20warfarin%20by%20human%20cDNA-expressed%20cytochrome%20P-450:%20a%20role%20for%20P-4502C9%20in%20the%20etiology%20of%20(S)-warfarin-drug%20interactions.%20Chem%20Res%20Toxicol.%201992;5(1):54%E2%80%939.%2010.1021/tx00025a009)
|
| 427 |
+
|
| 428 |
+
47. Loebstein R, Yonath H, Peleg D, Almog S, Rotenberg M, Lubetsky A, et al. Interindividual variability in sensitivity to warfarin: nature or nurture? Clin Pharmacol Ther. 2001;70(2):159–64. 10.1067/mcp.2001.117444 [DOI](https://doi.org/10.1067/mcp.2001.117444) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11503010/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Loebstein%20R,%20Yonath%20H,%20Peleg%20D,%20Almog%20S,%20Rotenberg%20M,%20Lubetsky%20A,%20et%20al.%20Interindividual%20variability%20in%20sensitivity%20to%20warfarin:%20nature%20or%20nurture?%20Clin%20Pharmacol%20Ther.%202001;70(2):159%E2%80%9364.%2010.1067/mcp.2001.117444)
|
| 429 |
+
|
| 430 |
+
48. Jetter A, Kinzig-Schippers M, Skott A, Lazar A, Tomalik-Scharte D, Kirchheiner J, et al. Cytochrome P450 2C9 phenotyping using low-dose tolbutamide. Eur J Clin Pharmacol. 2004;60(3):165–71. 10.1007/s00228-004-0754-z [DOI](https://doi.org/10.1007/s00228-004-0754-z) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15045499/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Jetter%20A,%20Kinzig-Schippers%20M,%20Skott%20A,%20Lazar%20A,%20Tomalik-Scharte%20D,%20Kirchheiner%20J,%20et%20al.%20Cytochrome%20P450%202C9%20phenotyping%20using%20low-dose%20tolbutamide.%20Eur%20J%20Clin%20Pharmacol.%202004;60(3):165%E2%80%9371.%2010.1007/s00228-004-0754-z)
|
| 431 |
+
|
| 432 |
+
49. Lv C, Liu C, Yao Z, Gao X, Sun L, Liu J, et al. The clinical pharmacokinetics and pharmacodynamics of warfarin when combined with compound Danshen: a case study for combined treatment of coronary heart diseases with atrial fibrillation. Front Pharmacol. 2017;8:826. 10.3389/fphar.2017.00826 [DOI](https://doi.org/10.3389/fphar.2017.00826) | [PMC free article](/articles/PMC5702344/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29209208/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Lv%20C,%20Liu%20C,%20Yao%20Z,%20Gao%20X,%20Sun%20L,%20Liu%20J,%20et%20al.%20The%20clinical%20pharmacokinetics%20and%20pharmacodynamics%20of%20warfarin%20when%20combined%20with%20compound%20Danshen:%20a%20case%20study%20for%20combined%20treatment%20of%20coronary%20heart%20diseases%20with%20atrial%20fibrillation.%20Front%20Pharmacol.%202017;8:826.%2010.3389/fphar.2017.00826)
|
test/texts/PMC11507373.md
ADDED
|
@@ -0,0 +1,318 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Analysis of ABCB1 Gene Polymorphisms and Their Impact on Tacrolimus Blood Levels in Kidney Transplant Recipients
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Corina Andreea Rotarescu, Ion Maruntelu, Ion Rotarescu, Alexandra-Elena Constantinescu, Ileana Constantinescu
|
| 5 |
+
**Journal:** International Journal of Molecular Sciences
|
| 6 |
+
**Date:** 2024 Oct 12
|
| 7 |
+
**DOI:** [10.3390/ijms252010999](https://doi.org/10.3390/ijms252010999)
|
| 8 |
+
**PMID:** 39456782
|
| 9 |
+
**PMCID:** PMC11507373
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11507373/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11507373/pdf/ijms-25-10999.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11507373/pdf/ijms-25-10999.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
Tacrolimus (Tc) is an immunosuppressant used in transplant patients, but its therapeutic range is narrow, making precise dosing essential. This study investigates the association of three single nucleotide polymorphisms (SNPs) (ABCB1 3435C>T, 1236C>T, 2677G>T/A) with Tc levels over time to gain better insights into their role in personalized medicine. We conducted the study over four distinct periods: 1–14 days, 15–30 days, 31–60 days, and beyond 60 days post-transplantation. The analysis included allele, genotype, haplotype, and diplotype frequencies of the three SNPs concerning Tc blood levels. Statistical significance was determined, and false discovery rate (PFDR) correction was applied where appropriate. Significant associations were found between the C (ABCB1 C1236T), A alleles (ABCB1 G2677T/A), the CAC haplotype and lower Tc levels. The CAC-TGT and TGT-TGT diplotypes significantly influence how patients metabolize the drug. The TGT haplotype and the AA genotype (ABCB1 G2677T/A) were associated with higher Tc levels, suggesting a long-term genetic influence. Genetic factors, specifically certain SNPs and diplotypes, significantly impact Tc blood levels, with their influence varying over time.
|
| 16 |
+
|
| 17 |
+
Keywords: kidney transplant, ABCB1, tacrolimus, haplotype, diplotype
|
| 18 |
+
|
| 19 |
+
**Keywords:**Keywords: kidney transplant, ABCB1, tacrolimus, haplotype, diplotype
|
| 20 |
+
|
| 21 |
+
## 1. Introduction
|
| 22 |
+
|
| 23 |
+
Chronic kidney disease is a progressive disorder characterized by a gradual decline in kidney function over months or years. Worldwide, an estimated 850 million people suffer from kidney disease, with the majority living in lower-middle-income nations, where they lack access to therapy, prevention and diagnosis [[1](#B1-ijms-25-10999)1]. End-stage renal disease is the most severe chronic kidney disease [[2](#B2-ijms-25-10999)2]. A glomerular filtration rate of fewer than 15 milliliters per minute is the hallmark of this disease, characterized by severe and irreversible kidney damage [[3](#B3-ijms-25-10999)3,[4](#B4-ijms-25-10999)4]. Treatments for end-stage renal disease primarily consist of renal replacement therapy or kidney transplants. Kidney transplantation is an indication of the implementation of immunosuppressive therapy. The most common immunosuppressive treatment involves Anti-thymocyte globuline or Basiliximab for initial induction. Maintenance includes Prednisone, Mycophenolate mofetil, Tacrolimus (Tc), or Cyclosporine.
|
| 24 |
+
|
| 25 |
+
Tc is one of the most studied ABCB1 substrates because it is frequently used to prevent transplant rejection [[5](#B5-ijms-25-10999)5]. Bioavailability changes significantly impact Tc pharmacokinetics. The drug’s oral bioavailability is poor and varies considerably between people, with an average of about 25% [[6](#B6-ijms-25-10999)6,[7](#B7-ijms-25-10999)7,[8](#B8-ijms-25-10999)8].
|
| 26 |
+
|
| 27 |
+
The ABCB1 gene, on chromosome 7 at position 7q21.12 is highly polymorphic and encodes the P-glycoprotein (a transmembrane protein) [[9](#B9-ijms-25-10999)9]. The protein’s molecular weight is 170 kDa [[10](#B10-ijms-25-10999)10], and it is present in the epithelia of numerous tissues, including the intestines, liver, kidneys, blood–brain barrier, testes, placenta, and lung [[11](#B11-ijms-25-10999)11,[12](#B12-ijms-25-10999)12,[13](#B13-ijms-25-10999)13]. ABCB1 gene polymorphisms are also linked to different pathologies, not only to drug metabolism. For instance, ABCB1 intron 3 C-rs3789243-T is associated with colorectal cancer (CRC) risk, as found in a prospective population-based study [[14](#B14-ijms-25-10999)14]. Similarly, a meta-analysis of case–control studies found an increased risk of CRC among carriers of the combined wild-type C3435T and G2677T/A alleles in Caucasians [[15](#B15-ijms-25-10999)15].
|
| 28 |
+
|
| 29 |
+
The ABCB1 gene polymorphisms can alter the structure and function of the P-glycoprotein, a key player in drug distribution and efficacy. These variations can affect the P-glycoprotein’s ability to expel drugs from cells, thereby influencing the distribution and efficacy of Tc in kidney transplantation.
|
| 30 |
+
|
| 31 |
+
Three polymorphic variants, ABCB1 3435C>T (exon 26), ABCB1 1236C>T (exon 12) and ABCB1 2677G>T/A (exon 21), have been extensively analyzed and demonstrated to hold functional significance in the context of kidney transplantation.
|
| 32 |
+
|
| 33 |
+
Our study aims to present findings from a retrospective single-center study on the impact of ABCB1 gene polymorphisms on Tc dosage in Romanian kidney transplant recipients. The goal is to contribute to optimizing Tc administration for kidney transplant recipients. Based on the findings of this study, individualized Tc administration may minimize the time required for optimal Tc levels with minimal adverse effects in clinical practice and reduce the risk of rejection in Romanian kidney transplant recipients.
|
| 34 |
+
|
| 35 |
+
## 2. Results
|
| 36 |
+
|
| 37 |
+
### 2.1. Demographic Data and Gene Frequencies
|
| 38 |
+
|
| 39 |
+
[Table 1](#ijms-25-10999-t001)Table 1 presents a detailed summary of 162 patients, including their medical history, demographics, and specific treatments. The average age of the patients is 40.68, with 65.4% being men. A significant finding is that over 40% of the patients have an unknown cause of their chronic kidney disease, highlighting the need for further investigation in this area. Regarding the donor type, 74.7% received kidneys from living donors, while 25.3% received kidneys from cadaveric donors.
|
| 40 |
+
|
| 41 |
+
### Table 1.
|
| 42 |
+
|
| 43 |
+
Characteristics of the patients.
|
| 44 |
+
|
| 45 |
+
| Number of Patients (n) | 162 |
|
| 46 |
+
| ---------------------- | --- |
|
| 47 |
+
| Age (mean ± standard deviation) | 40.68 ± 11.26 |
|
| 48 |
+
| Gender (n) | |
|
| 49 |
+
| Male | 106 (65.4%) |
|
| 50 |
+
| Female | 56 (34.6%) |
|
| 51 |
+
| Bodyweight (kg) (mean ± standard deviation) | 69.69 ± 14.85 |
|
| 52 |
+
| Blood groups (n) | |
|
| 53 |
+
| A | 74 (45.7%) |
|
| 54 |
+
| B | 32 (19.7%) |
|
| 55 |
+
| AB | 18 (11.1%) |
|
| 56 |
+
| O | 38 (23.5%) |
|
| 57 |
+
| Type of donor (n) | |
|
| 58 |
+
| Living donor | 121 (74.7%) |
|
| 59 |
+
| Cadaveric donor | 41 (25.3%) |
|
| 60 |
+
| Kidney disease causes (n) | |
|
| 61 |
+
| Unknown etiology | 67 (41.4%) |
|
| 62 |
+
| IgA nephropathy | 29 (17.9%) |
|
| 63 |
+
| Autosomal dominant polycystic kidney | 14 (8.6%) |
|
| 64 |
+
| Glomerulonephritis | 12 (7.4%) |
|
| 65 |
+
| Tubulointerstitial disease | 11 (6.8%) |
|
| 66 |
+
| Alport syndrome | 10 (6.2%) |
|
| 67 |
+
| Diabetic nephropathy | 8 (4.9%) |
|
| 68 |
+
| Hypertensive nephropathy | 3 (1.9%) |
|
| 69 |
+
| Others (LES, ANCA positive vasculitis, Fabry disease, Goodpasture syndrome) | 8 (4.9%) |
|
| 70 |
+
| Induction therapy (n) | |
|
| 71 |
+
| Anti-thymocyte globuline | 2 (1.2%) |
|
| 72 |
+
| Basiliximab | 160 (98.8%) |
|
| 73 |
+
| Characteristics over time (mean ± standard deviation) | 1–14 days | 15–30 days | 31–60 days | over 60 days |
|
| 74 |
+
| Tc C0 (ng/mL) | 12.18 ± 5.42 | 13.55 ± 4.42 | 11.53 ± 3.65 | 8.15 ± 2.91 |
|
| 75 |
+
| Tc dose (mg per day) | 12.92 ± 4.19 | 12.43 ± 4.89 | 8.89 ± 4.38 | 4.99 ± 2.56 |
|
| 76 |
+
| Tc C0/dose (ng/mL/mg per day) | 1.11 ± 1.20 | 1.29 ± 0.83 | 1.59 ± 0.87 | 1.98 ± 1.12 |
|
| 77 |
+
| Tc C0/dose/Bodyweight (ng/mL/mg/kg per day) | 0.016 ± 0.155 | 0.019 ± 0.151 | 0.024 ± 0.015 | 0.029 ± 0.016 |
|
| 78 |
+
| eGFR (mL/min/1.73 m2) | 37.87 ± 22.23 | 37.53 ± 21.49 | 43.98 ± 14.76 | 47.29 ± 15.81 |
|
| 79 |
+
[Table 2](#ijms-25-10999-t002)Table 2 presents the frequency distributions of genotypes for genetic variants in the ABCB1 gene. The allelic distribution of the G2677T/A variant was 70.4% for G, 29.6% for A, and 0% for T. The genotype frequencies for this variant were as follows: 28.5% (46/162) for homozygous wild-type, 41.9% (68/162) for heterozygous, and 29.6% (48/162) for homozygous mutant. Notably, no patients exhibited a GT/TA/TT genotype for this locus, highlighting the rarity of this genotype in the population under study. The C and T alleles were 75.3% and 24.7% for the C3435T SNP and 70.4% and 29.6% for the C1236T SNP, respectively. Regarding the genotypes ABCB1 3435C>T and ABCB1 1236C>T, the homozygous mutant was observed in 24.7% and 29.6% of patients. Notably, the genotype distribution adhered to the Hardy–Weinberg equilibrium, validating our research methodology and the reliability of our results.
|
| 80 |
+
|
| 81 |
+
### Table 2.
|
| 82 |
+
|
| 83 |
+
Genotype and allele frequencies of the patients.
|
| 84 |
+
|
| 85 |
+
| Genotype | Total (n = 162) | Allele | Total (n = 324) |
|
| 86 |
+
| -------- | --------------- | ------ | --------------- |
|
| 87 |
+
| ABCB1 3435C>T (rs1045642)CCCTTT | 38 (23.4%)84 (51.9%)40 (24.7%) | CT | 244 (75.3%)80 (24.7%) |
|
| 88 |
+
| ABCB1 1236C>T (rs1128503)CCCTTT | 38 (23.5%)76 (46.9%)48 (29.6%) | CT | 228 (70.4%)96 (29.6%) |
|
| 89 |
+
| ABCB1 2677G>T/A (rs2032582)GGGAAAGT/TA/TT | 46 (28.5%)68 (41.9%)48 (29.6%)0 (0%) | GAT | 228 (70.4%)96 (29.6%)0 (0%) |
|
| 90 |
+
### 2.2. ABCB1 SNP Alleles and Genotypes Analysis
|
| 91 |
+
|
| 92 |
+
The association between each SNP of the ABCB1 alleles and genotype and the Tc C0 was analyzed.
|
| 93 |
+
|
| 94 |
+
During the 1–14 day period, allele analysis revealed that the C and A alleles of ABCB1 C1236T and ABCB1 G2677A were significantly more frequent in the group with Tc levels outside the normal range, with *p*p-values of 0.0009 (OR = 0.39; 95% C.I. = 0.22–0.69) and 0.0015 (OR = 0.41; 95% C.I. = 0.23–0.72), respectively. The C allele of ABCB1 C3435T also showed a higher frequency in the outside range group (*p*p = 0.0215; OR = 0.53; 95% C.I. = 0.31–0.91) ([Figure 1](#ijms-25-10999-f001)Figure 1a)
|
| 95 |
+
|
| 96 |
+
### Figure 1.
|
| 97 |
+
|
| 98 |
+

|
| 99 |
+
|
| 100 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11507373_ijms-25-10999-g001.jpg)
|
| 101 |
+
|
| 102 |
+
Count of ABCB1 alleles and genotypes with Tc levels outside the normal range in the first two weeks: (a) Distribution of ABCB 1 with abnormal Tc levels. (b) Distribution of ABCB1 genotypes with abnormal Tc levels.
|
| 103 |
+
|
| 104 |
+
Patients homozygous on ABCB1 C1236T and GG from locus ABCB1 G2677T/A usually were outside of the normal range compared to heterozygotes status (ABCB1 1236CC with *p*p = 0.0061; *P*P_FDR_FDR = 0.0182; 95% C.I. = 0.01–0.04; ABCB1 1236TT with *p*p = 0.0102; *P*P_FDR_FDR = 0.0154; 95% C.I. = 0.01–0.09; ABCB1 2677GG with *p*p = 0.0010; *P*P_FDR_FDR = 0.0031; 95% C.I. = 0–0.01).
|
| 105 |
+
|
| 106 |
+
During the first two weeks after kidney transplantation, ABCB1 1236C, ABCB1 2677A and ABCB1 3435C alleles have been associated with low blood levels of Tc. This also applies to the CC and TT genotypes from the ABCB1 C1236T locus and the GG genotype from the ABCB1 G2677T/A locus ([Figure 1](#ijms-25-10999-f001)Figure 1b).
|
| 107 |
+
|
| 108 |
+
Beyond 60 days, significant associations re-emerged. The C allele of ABCB1 C1236T and the A allele of ABCB1 G2677A were associated with the standard range group, with *p*p-values of 0.0317 and 0.0106, respectively. Genotype analysis revealed that the AA genotype of ABCB1 G2677A was significantly associated with Tc levels within the normal range (*p*p = 0.0111; *P*P_FDR_FDR = 0.0334; 95% C.I. = 0.01–0.08).
|
| 109 |
+
|
| 110 |
+
Patients carrying the AA genotype from the ABCB1 2677 group tend to have lower concentrations than those without the AA genotype, as indicated by the lower mean rank (29.67 vs. 8.33) and *p*p-value = 0.005 ([Figure 2](#ijms-25-10999-f002)Figure 2). However, no other significant differences in Tc C0 were observed among ABCB1 1236C>T and 3435C>T genotype groups.
|
| 111 |
+
|
| 112 |
+
### Figure 2.
|
| 113 |
+
|
| 114 |
+

|
| 115 |
+
|
| 116 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11507373_ijms-25-10999-g002.jpg)
|
| 117 |
+
|
| 118 |
+
Blood concentration of Tc over 60 days for ABCB1 2677 AA genotype.
|
| 119 |
+
|
| 120 |
+
### 2.3. ABCB1 Haplotypes and Diplotypes Analysis
|
| 121 |
+
|
| 122 |
+
Patients who were homozygous for three variants or heterozygous for only one variant had their haplotypes (composed of alleles belonging to ABCB1 C1236T, ABCB1 G2677T/A and ABCB1 C3435T loci) unambiguously assigned, as reported by Lee et al. [[16](#B16-ijms-25-10999)16]. We used the Haploview software for the remaining patients to assign the haplotypes using the estimation-maximization algorithm.
|
| 123 |
+
|
| 124 |
+
A single haplotype block (formed by the G2677T/A, C3435T and C1236T) was identified in the Romanian group chromosome 7 region by the linkage disequilibrium (LD) analysis ([Figure 3](#ijms-25-10999-f003)Figure 3).
|
| 125 |
+
|
| 126 |
+
### Figure 3.
|
| 127 |
+
|
| 128 |
+

|
| 129 |
+
|
| 130 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11507373_ijms-25-10999-g003.jpg)
|
| 131 |
+
|
| 132 |
+
Linkage disequilibrium plots and haplotype analysis during periods of 1–14 days (A) and 31–60 days (B), respectively, over 60 days (C) with Tc levels in the outside range. For D’ < 1, LOD ≥ 2, use a shade of pink; for D’ = 1, LOD ≥ 2, use bright red.
|
| 133 |
+
|
| 134 |
+
Haplotype analysis indicated that the CAC haplotype was more prevalent in the abnormal range group (*p*p = 0.0006, *P*P_FDR_FDR = 0.0051) in the first two weeks.
|
| 135 |
+
|
| 136 |
+
We have observed no significant associations in allele or haplotype frequencies with the status range of Tc levels during the third and fourth weeks, following a kidney transplant. Genotype and diplotype analyses also showed no notable differences, except for the CAC-TGT diplotype, which had a marginal association with Tc levels. A marginal association indicates a weak but present relationship, with a *p*p-value of 0.0229 and *P*P_FDR_FDR = 0.3219. In other words, while the relationship is not strong enough to be considered significant, it is still worth noting as it may have some influence.
|
| 137 |
+
|
| 138 |
+
During the 31–60 day period, the analysis showed no significant differences in allele frequencies. However, the TGC haplotype was significantly more frequent in the standard range group (*p*p = 0.0048, *P*P_FDR_FDR = 0.0385). Diplotype analysis supported this finding, with the TGC-TGC diplotype being exclusive to the normal range group.
|
| 139 |
+
|
| 140 |
+
Beyond 60 days, the TGT haplotype was significantly more prevalent in the group with Tc levels in the normal range (*p*p = 0.0093, *P*P_FDR_FDR = 0.0467).
|
| 141 |
+
|
| 142 |
+
Out of 162 patients, 64.20% had heterozygous diplotypes ([Figure 4](#ijms-25-10999-f004)Figure 4). All the identified ABCB1 diplotypes were used to compare the Tc C0 levels at different periods since the transplant procedures.
|
| 143 |
+
|
| 144 |
+
### Figure 4.
|
| 145 |
+
|
| 146 |
+

|
| 147 |
+
|
| 148 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11507373_ijms-25-10999-g004.jpg)
|
| 149 |
+
|
| 150 |
+
Diplotype frequency of combined ABCB1 1236C>T, 2677G>T/A and 3435C>T.
|
| 151 |
+
|
| 152 |
+
Diplotype analysis found the TGT-TGT (*p*p = 0.0018, *P*P_FDR_FDR = 0.0213) and CAC-TGT (*p*p = 0.0070, *P*P_FDR_FDR = 0.0421) diplotypes more frequent in the abnormal range group, suggesting a potential impact on Tc levels.
|
| 153 |
+
|
| 154 |
+
### 2.4. Analysis of Tc Variability (C0/Dose Ratio) over Time through Different Mixed-Effects Models
|
| 155 |
+
|
| 156 |
+
A mixed-effects logistic regression model was employed to account for patient variability. This model considers the uniqueness of each patient’s baseline Tc C0/dose. Including patient-specific random effects in the model improved the AUC (Area Under the Curve) from 0.615 to 0.829, indicating a significantly better ability to predict which patients will have high Tc concentrations. This suggests that accounting for patient-specific variability is crucial in predicting Tc levels, emphasizing the need for individualized therapeutic monitoring.
|
| 157 |
+
|
| 158 |
+
Only during the 1 to 14 days were CAC-TGT and TGT-TGT diplotypes significantly associated with lower Tc blood levels. This suggests that patients with these specific genetic variations may metabolize Tc more efficiently, leading to lower drug concentrations.
|
| 159 |
+
|
| 160 |
+
Patients with the CAC-TGT diplotype had a 99% reduced likelihood of exhibiting high Tc concentrations, and those with the TGT-TGT diplotype had a 72% reduced likelihood. These findings were robust even after correcting for multiple comparisons using the False Discovery Rate (P_FDR_FDR) method, confirming the genetic variations as essential determinants of Tc pharmacokinetics.
|
| 161 |
+
|
| 162 |
+
Age showed a weak trend toward influencing Tc levels (*p*p = 0.07), with older patients slightly more likely to have higher concentrations. eGFR, an essential marker of kidney function, was not significantly associated with Tc levels in this model (*p*p = 0.11), suggesting that other factors, such as genetic variations, may play a more direct role in influencing drug concentration.
|
| 163 |
+
|
| 164 |
+
After 15 days of kidney transplantation, age and eGFR were significantly associated with Tc variability. Patients with higher eGFR, indicating better kidney function, were less likely to have elevated Tc levels (OR: 0.32, *p*p < 0.001), whereas older patients had increased odds of higher Tc C0 (OR: 0.31, *p*p = 0.005). Dose per weight was a critical factor, as patients receiving higher Tc C0/dose/bodyweight were much more likely to have higher blood levels (OR: 1.02, *p*p < 0.001).
|
| 165 |
+
|
| 166 |
+
## 3. Discussion
|
| 167 |
+
|
| 168 |
+
The impact of ABCB1 polymorphisms on drug response varies significantly across ethnic populations, influencing drug bioavailability and efficacy. Understanding these genetic differences is essential for developing personalized medicine strategies to optimize therapy based on an individual’s genotype.
|
| 169 |
+
|
| 170 |
+
Our study uniquely examines the relationship between alleles, genotypes, haplotypes, diplotypes, and clinical factors on Tc levels at different time intervals (1–14 days, 15–30 days, 31–60 days, and over 60 days) in Romanian kidney transplant recipients. The term ‘haplotype’ refers to a combination of alleles on specific chromosomes within a population [[17](#B17-ijms-25-10999)17], while ‘diplotype’ describes the combination of genotypes formed by homologous chromosome pairs [[17](#B17-ijms-25-10999)17].
|
| 171 |
+
|
| 172 |
+
In this study, the frequency of the variant C allele for ABCB1 C1236T and ABCB1 C3435T SNPs was 70.4% and 75.3%, differing from the Turkish population [[18](#B18-ijms-25-10999)18]. According to dbSNP [[19](#B19-ijms-25-10999)19], the ABCB1 1236C allele is the minor variant in Asians but the major variant in other populations, highlighting ethnic variability. The ABCB1 3435C allele also varies widely in frequency, being more common in some populations and less common in others [[20](#B20-ijms-25-10999)20,[21](#B21-ijms-25-10999)21,[22](#B22-ijms-25-10999)22]. The role of the ABCB1 1236C, ABCB1 2677A and ABCB1 3435C, along with the CAC haplotype, as potential risk factors for abnormal Tc levels aligns with findings from previous studies. In our research, beyond 60 days, kidney transplant recipients with the AA genotype in the ABCB1 2677 group tended to have a normal Tc concentration compared to patients without the AA genotype.
|
| 173 |
+
|
| 174 |
+
ABCB1 2677GG, homozygous ABCB1 C1236T is associated with abnormal Tc blood levels in the first two weeks.
|
| 175 |
+
|
| 176 |
+
In a study of Korean kidney transplant patients, carriers of the ABCB1 2677 GG genotype exhibited lower Tc dose-adjusted trough levels on day three after transplantation than non-GG carriers [[23](#B23-ijms-25-10999)23]. Fredericks et al. [[24](#B24-ijms-25-10999)24] examined the Tc dose requirements of 206 stable renal transplant patients. They discovered that ABCB1 G2677G and ABCB1 C3435C genotype patients had lower dose-normalized blood Tc concentrations than ABCB1 T2677T and ABCB1 T3435T patients [[24](#B24-ijms-25-10999)24]. Additionally, a research team led by Karim Akbas [[25](#B25-ijms-25-10999)25], conducted a study on the influence of ABCB1 polymorphism on Tc pharmacokinetics in Turkish individuals who had received a kidney transplant. The study findings showed that among the participants, 30.4% had the ABCB1 C3435C genotype, 47.8% had the ABCB1 C3435T genotype, and 21.7% had the ABCB1 T3435T genotype [[25](#B25-ijms-25-10999)25]. During the first six months after transplantation, patients with the 3435 TT genotype required significantly lower daily Tc [[25](#B25-ijms-25-10999)25]. However, in the following six months, individuals with the 3435 CC genotype had significantly lower trough Tc concentrations adjusted for dose than those with the 3435 TT and CT genotypes [[25](#B25-ijms-25-10999)25].
|
| 177 |
+
|
| 178 |
+
Nonetheless, a study by Provenzani et al. found that kidney transplant recipients with the wild-type genotype ABCB1 2677GG required significant doses of Tc [[26](#B26-ijms-25-10999)26]. Most scientists explored gene variants in ABCB1 and found no statistically significant association with dose-adjusted Tc trough levels [[27](#B27-ijms-25-10999)27,[28](#B28-ijms-25-10999)28]. This suggests that in the population studied, the ABCB1 genotype did not exert a notable impact on the pharmacokinetics of the medications transported by this gene. This could be attributed to several factors. These include the influence of other genes, environmental factors, and the limitations of the study design. Additional investigations are required to validate these findings and understand the complex interplay between genetics and transport.
|
| 179 |
+
|
| 180 |
+
Our findings indicate that the CAC haplotype (1–14 days) and TGC haplotype (31–60 days) are associated with abnormal Tc levels. The genetic influence on Tc metabolism diminishes over time, with non-genetic factors potentially playing a more significant role. As outlined in our hospital protocol, patients with the CAC haplotype required lower Tc doses during the first two weeks post-transplant to reach therapeutic levels.
|
| 181 |
+
|
| 182 |
+
The high frequency of CAC and TGT haplotypes may suggest evolutionary stability or a selective advantage in this population. Identifying CAC-TGT and TGT-TGT diplotypes as predictors of lower tacrolimus blood levels could aid in tailoring tacrolimus dosing. Due to enhanced drug metabolism, patients with these diplotypes may need higher doses to achieve therapeutic Tc concentrations. Genetic testing for ABCB1 polymorphisms could be integrated into routine post-transplant care, enabling more personalized dosing strategies.
|
| 183 |
+
|
| 184 |
+
The applied mixed-effects model highlights the importance of personalized medicine in managing kidney transplant recipients. While genetic factors significantly influence Tc metabolism, other factors, such as age, clinical status, and lifestyle, must be considered. Incorporating patient-specific variability into predictive models could improve dosing accuracy, preventing under- or over-dosing, which are associated with risks of transplant rejection or toxicity. The model also supports frequent therapeutic drug monitoring to ensure Tc levels remain within the target range.
|
| 185 |
+
|
| 186 |
+
Age significantly impacted Tc levels after 15 days, with older patients tending to have higher concentrations, likely due to reduced hepatic and renal clearance. Interestingly, despite being a key indicator of kidney function, eGFR did not significantly predict Tc concentration, possibly due to the dominant influence of genetic factors in this context.
|
| 187 |
+
|
| 188 |
+
Our findings suggest that genetic testing could inform personalized treatment protocols, especially regarding Tc dosing. Early dose adjustments based on genetic profiles may reduce the risk of acute rejection and tacrolimus-related nephrotoxicity. Routine monitoring should also account for individual variability, particularly in the early post-transplant months when Tc levels are more variable.
|
| 189 |
+
|
| 190 |
+
The small sample size for certain genetic variants can reduce the statistical power of the study, making it difficult to draw definitive conclusions about the impact of these genetic variants on Tacrolimus concentrations and dosing. Without randomization and a control group, it is challenging to establish causality. The observed effects might be influenced by confounding factors such as donor age and type that were not controlled for in the study design. Conducting the study at a single center limits the generalizability of our findings. The results may not apply to broader populations with different demographic or clinical characteristics. Retrospective studies are prone to various biases, which can affect the validity of the findings.
|
| 191 |
+
|
| 192 |
+
## 4. Materials and Methods
|
| 193 |
+
|
| 194 |
+
This study included 162 kidney transplant recipients treated with Tc (*Prograf or *Advagraf) for at least one year between January 2019 and May 2024 at Fundeni Clinical Institute, Bucharest, Romania (see [Figure 5](#ijms-25-10999-f005)Figure 5). Tc was administered orally once or twice daily with an initial dose of 0.15–0.20 mg/kg/day and then adjusted doses to achieve target concentrations: 12–15 ng/mL in the first 14 days, 10–12 ng/mL during 15–30 days, 8–10 ng/mL during 31–60 days, and 6–8 ng/mL after 60 days. All patients had normal liver function throughout the follow-up period. Therefore, we analyzed only: age, sex, renal function by eGFR, dose, concentration, and ABCB1 genes. The data on demographics, laboratory tests, underlying medical conditions, and medication during the perioperative period for this study were collected retrospectively from the electronic medical system (Hipocrate system). The study was conducted following the Declaration of Helsinki (2013) and was approved by the Commission of Ethics (24433/23.05.2024).
|
| 195 |
+
|
| 196 |
+
### Figure 5.
|
| 197 |
+
|
| 198 |
+

|
| 199 |
+
|
| 200 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11507373_ijms-25-10999-g005.jpg)
|
| 201 |
+
|
| 202 |
+
Design of the current study.
|
| 203 |
+
|
| 204 |
+
Tc concentrations (C0, ng/mL) were monitored biweekly for the first month following kidney transplantation and then monthly until the end of the first year. C0 refers to the Tc trough concentration, measured just before the next dose is administered (pre-dose). This measurement reflects the lowest concentration of the drug in the bloodstream, providing an indicator of drug exposure, and is commonly used to guide dosing adjustments in clinical practice. The chemiluminescent microparticle immunoassay method was used to measure Tc C0 in whole blood (on Architech System^®^® i2000 from Abbott Diagnostic, Lake Forest, IL, USA). In our laboratory, the measurement range (2–30 ng/mL) and detection limit (1 ng/mL) align with the manufacturer’s specifications [[29](#B29-ijms-25-10999)29].
|
| 205 |
+
|
| 206 |
+
DNA was isolated from 200 µL of whole blood using the QIAmp DNA Blood Mini Kit (Qiagen, Germany). The DNA was analyzed for the genes ABCB1 1236C>T (rs1128503), 2677G>T/A (rs2032582) and 3435C>T (rs1045642) using TaqMan^®^® Drug Metabolism Genotyping Assays. The TaqMan^®^® Drug Metabolism Genotyping Assays were chosen for their high specificity, sensitivity, and reliability, making them well-suited for analyzing the genetic influence on drug metabolism. The analysis was performed on an Applied Biosystems 7300 Real-Time PCR System.
|
| 207 |
+
|
| 208 |
+
Descriptive statistics were used to represent the demographic data and gene frequencies. Before using parametric or nonparametric tests, we checked for the normal distribution of our data using tests for normality such as the Shapiro–Wilk or Kolmogorov–Smirnov Test. Variables like C0/dose and estimated glomerular filtration rate (eGFR) were not normally distributed, so we log-transformed them for analysis.
|
| 209 |
+
|
| 210 |
+
We used T-student or Chi-square tests to study SNP alleles and Tc levels’ in/out range status as needed. Nonparametric tests were used to examine the impact of genotype, haplotype, and diplotype groups on Tc C0/dose. The groups were compared using the Mann–Whitney U test, and we utilized the SPSS software (v.20, IBM SPSS Statistics for Windows, Armonk, NY, USA) for analysis. GraphPad Prism version 9.3 for Windows (GraphPad Software, San Diego, CA, USA) was used for plotting graphs.
|
| 211 |
+
|
| 212 |
+
Hardy–Weinberg equilibrium, linkage disequilibrium and haplotype frequency analyses were determined using the Haploview 4.2 program (Broad Institute of Harvard and MIT, Cambridge, MA, USA).
|
| 213 |
+
|
| 214 |
+
A linear mixed-effects model was used to assess the impact of various covariates on Tc C0/dose over different time intervals (1–14 days, 15–30 days, 31–60 days, and over 61 days post-transplant). The model included age, gender, weight, time since transplant, and eGFR. The R package version 4.4.1 was used to analyze the mixed-effect model. The false discovery rate (P_FDR_FDR) method was used to reduce the likelihood of false positives in multiple comparisons [[30](#B30-ijms-25-10999)30]. The association was considered statistically significant if the *P*P_FDR_FDR was less than 0.05.
|
| 215 |
+
|
| 216 |
+
## 5. Conclusions
|
| 217 |
+
|
| 218 |
+
The study’s results underscore the importance of considering the temporal dynamics of genetic influence on Tc metabolism. The fluctuating significance of genetic markers over time suggests that personalized Tc therapy could benefit from a longitudinal approach, where genetic testing is not just a one-time assessment but is integrated into ongoing treatment evaluations. This approach can provide a more comprehensive understanding of the genetic influence on Tc levels, leading to more effective and personalized treatment strategies.
|
| 219 |
+
|
| 220 |
+
## Author Contributions
|
| 221 |
+
|
| 222 |
+
For Conceptualization and supervision, I.C.; methodology, C.A.R.; data curation, I.M.; writing—original draft preparation, I.M. and C.A.R.; writing—review and editing, I.C. and I.R.; visualization, A.-E.C. All authors have read and agreed to the published version of the manuscript.
|
| 223 |
+
|
| 224 |
+
## Institutional Review Board Statement
|
| 225 |
+
|
| 226 |
+
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Fundeni Clinical Institute, Bucharest, Romania (protocol code: 24433 and date of approval: 23 May 2024).
|
| 227 |
+
|
| 228 |
+
## Informed Consent Statement
|
| 229 |
+
|
| 230 |
+
Informed consent was obtained from all subjects involved in the study.
|
| 231 |
+
|
| 232 |
+
## Data Availability Statement
|
| 233 |
+
|
| 234 |
+
The dataset is available on request from the authors.
|
| 235 |
+
|
| 236 |
+
## Conflicts of Interest
|
| 237 |
+
|
| 238 |
+
The authors declare no conflicts of interest.
|
| 239 |
+
|
| 240 |
+
## Funding Statement
|
| 241 |
+
|
| 242 |
+
Publication of this paper was supported by the University of Medicine and Pharmacy Carol Davila, through the institutional program Publish not Perish.
|
| 243 |
+
|
| 244 |
+
## Footnotes
|
| 245 |
+
|
| 246 |
+
## Associated Data
|
| 247 |
+
|
| 248 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 249 |
+
|
| 250 |
+
### Data Availability Statement
|
| 251 |
+
|
| 252 |
+
The dataset is available on request from the authors.
|
| 253 |
+
|
| 254 |
+
### Data Availability Statement
|
| 255 |
+
|
| 256 |
+
The dataset is available on request from the authors.
|
| 257 |
+
|
| 258 |
+
## References
|
| 259 |
+
|
| 260 |
+
1. Jager K.J., Kovesdy C., Langham R., Rosenberg M., Jha V., Zoccali C. A Single Number for Advocacy and Communication-Worldwide More than 850 Million Individuals Have Kidney Diseases. Kidney Int. 2019;96:1048–1050. doi: 10.1016/j.kint.2019.07.012. [DOI](https://doi.org/10.1016/j.kint.2019.07.012) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31582227/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Kidney%20Int.&title=A%20Single%20Number%20for%20Advocacy%20and%20Communication-Worldwide%20More%20than%20850%20Million%20Individuals%20Have%20Kidney%20Diseases&author=K.J.%20Jager&author=C.%20Kovesdy&author=R.%20Langham&author=M.%20Rosenberg&author=V.%20Jha&volume=96&publication_year=2019&pages=1048-1050&pmid=31582227&doi=10.1016/j.kint.2019.07.012&)
|
| 261 |
+
|
| 262 |
+
2. Zoccali C., Kramer A., Jager K.J. Chronic Kidney Disease and End-Stage Renal Disease-a Review Produced to Contribute to the Report “the Status of Health in the European Union: Towards a Healthier Europe. ” NDT Plus. 2010;3:213–224. doi: 10.1093/ndtplus/sfp127. [DOI](https://doi.org/10.1093/ndtplus/sfp127) | [PMC free article](/articles/PMC5477935/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28657040/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=%E2%80%9D%20NDT%20Plus&title=Chronic%20Kidney%20Disease%20and%20End-Stage%20Renal%20Disease-a%20Review%20Produced%20to%20Contribute%20to%20the%20Report%20%E2%80%9Cthe%20Status%20of%20Health%20in%20the%20European%20Union:%20Towards%20a%20Healthier%20Europe&author=C.%20Zoccali&author=A.%20Kramer&author=K.J.%20Jager&volume=3&publication_year=2010&pages=213-224&pmid=28657040&doi=10.1093/ndtplus/sfp127&)
|
| 263 |
+
|
| 264 |
+
3. Hashmi M.F., Benjamin O., Lappin S.L. StatPearls. StatPearls Publishing; Treasure Island, FL, USA: 2024. End-Stage Renal Disease. [PubMed](https://pubmed.ncbi.nlm.nih.gov/29763036/) | [Google Scholar](https://scholar.google.com/scholar_lookup?title=StatPearls&author=M.F.%20Hashmi&author=O.%20Benjamin&author=S.L.%20Lappin&publication_year=2024&)
|
| 265 |
+
|
| 266 |
+
4. Stevens L.A., Li S., Wang C., Huang C., Becker B.N., Bomback A.S., Brown W.W., Burrows N.R., Jurkovitz C.T., McFarlane S.I., et al. Prevalence of CKD and Comorbid Illness in Elderly Patients in the United States: Results from the Kidney Early Evaluation Program (KEEP) Am. J. Kidney Dis. 2010;55:S23–S33. doi: 10.1053/j.ajkd.2009.09.035. [DOI](https://doi.org/10.1053/j.ajkd.2009.09.035) | [PMC free article](/articles/PMC4574484/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20172445/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Kidney%20Dis.&title=Prevalence%20of%20CKD%20and%20Comorbid%20Illness%20in%20Elderly%20Patients%20in%20the%20United%20States:%20Results%20from%20the%20Kidney%20Early%20Evaluation%20Program%20(KEEP)&author=L.A.%20Stevens&author=S.%20Li&author=C.%20Wang&author=C.%20Huang&author=B.N.%20Becker&volume=55&publication_year=2010&pages=S23-S33&pmid=20172445&doi=10.1053/j.ajkd.2009.09.035&)
|
| 267 |
+
|
| 268 |
+
5. Kasiske B.L., Zeier M.G., Chapman J.R., Craig J.C., Ekberg H., Garvey C.A., Green M.D., Jha V., Josephson M.A., Kiberd B.A., et al. KDIGO Clinical Practice Guideline for the Care of Kidney Transplant Recipients: A Summary. Kidney Int. 2010;77:299–311. doi: 10.1038/ki.2009.377. [DOI](https://doi.org/10.1038/ki.2009.377) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19847156/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Kidney%20Int.&title=KDIGO%20Clinical%20Practice%20Guideline%20for%20the%20Care%20of%20Kidney%20Transplant%20Recipients:%20A%20Summary&author=B.L.%20Kasiske&author=M.G.%20Zeier&author=J.R.%20Chapman&author=J.C.%20Craig&author=H.%20Ekberg&volume=77&publication_year=2010&pages=299-311&pmid=19847156&doi=10.1038/ki.2009.377&)
|
| 269 |
+
|
| 270 |
+
6. Tuteja S., Alloway R.R., Johnson J.A., Gaber A.O. The Effect of Gut Metabolism on Tacrolimus Bioavailability in Renal Transplant Recipients. Transplantation. 2001;71:1303–1307. doi: 10.1097/00007890-200105150-00021. [DOI](https://doi.org/10.1097/00007890-200105150-00021) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11397967/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplantation&title=The%20Effect%20of%20Gut%20Metabolism%20on%20Tacrolimus%20Bioavailability%20in%20Renal%20Transplant%20Recipients&author=S.%20Tuteja&author=R.R.%20Alloway&author=J.A.%20Johnson&author=A.O.%20Gaber&volume=71&publication_year=2001&pages=1303-1307&pmid=11397967&doi=10.1097/00007890-200105150-00021&)
|
| 271 |
+
|
| 272 |
+
7. Gruber S.A., Hewitt J.M., Sorenson A.L., Barber D.L., Bowers L., Rynders G., Arrazola L., Matas A.J., Rosenberg M.E., Canafax D.M. Pharmacokinetics of FK506 after Intravenous and Oral Administration in Patients Awaiting Renal Transplantation. J. Clin. Pharmacol. 1994;34:859–864. doi: 10.1002/j.1552-4604.1994.tb02052.x. [DOI](https://doi.org/10.1002/j.1552-4604.1994.tb02052.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7525661/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Pharmacol.&title=Pharmacokinetics%20of%20FK506%20after%20Intravenous%20and%20Oral%20Administration%20in%20Patients%20Awaiting%20Renal%20Transplantation&author=S.A.%20Gruber&author=J.M.%20Hewitt&author=A.L.%20Sorenson&author=D.L.%20Barber&author=L.%20Bowers&volume=34&publication_year=1994&pages=859-864&pmid=7525661&doi=10.1002/j.1552-4604.1994.tb02052.x&)
|
| 273 |
+
|
| 274 |
+
8. Masuda S., Goto M., Fukatsu S., Uesugi M., Ogura Y., Oike F., Kiuchi T., Takada Y., Tanaka K., Inui K.-I. Intestinal MDR1/ABCB1 Level at Surgery as a Risk Factor of Acute Cellular Rejection in Living-Donor Liver Transplant Patients. Clin. Pharmacol. Ther. 2006;79:90–102. doi: 10.1016/j.clpt.2005.09.013. [DOI](https://doi.org/10.1016/j.clpt.2005.09.013) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16413244/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Pharmacol.%20Ther.&title=Intestinal%20MDR1/ABCB1%20Level%20at%20Surgery%20as%20a%20Risk%20Factor%20of%20Acute%20Cellular%20Rejection%20in%20Living-Donor%20Liver%20Transplant%20Patients&author=S.%20Masuda&author=M.%20Goto&author=S.%20Fukatsu&author=M.%20Uesugi&author=Y.%20Ogura&volume=79&publication_year=2006&pages=90-102&pmid=16413244&doi=10.1016/j.clpt.2005.09.013&)
|
| 275 |
+
|
| 276 |
+
9. Ueda K., Clark D.P., Chen C.J., Roninson I.B., Gottesman M.M., Pastan I. The Human Multidrug Resistance (MDR1) Gene. cDNA Cloning and Transcription Initiation. J. Biol. Chem. 1987;262:505–508. doi: 10.1016/S0021-9258(19)75806-2. [DOI](https://doi.org/10.1016/S0021-9258(19)75806-2) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/3027054/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Biol.%20Chem.&title=The%20Human%20Multidrug%20Resistance%20(MDR1)%20Gene.%20cDNA%20Cloning%20and%20Transcription%20Initiation&author=K.%20Ueda&author=D.P.%20Clark&author=C.J.%20Chen&author=I.B.%20Roninson&author=M.M.%20Gottesman&volume=262&publication_year=1987&pages=505-508&pmid=3027054&doi=10.1016/S0021-9258(19)75806-2&)
|
| 277 |
+
|
| 278 |
+
10. Choudhuri S., Klaassen C.D. Structure, Function, Expression, Genomic Organization, and Single Nucleotide Polymorphisms of Human ABCB1 (MDR1), ABCC (MRP), and ABCG2 (BCRP) Efflux Transporters. Int. J. Toxicol. 2006;25:231–259. doi: 10.1080/10915810600746023. [DOI](https://doi.org/10.1080/10915810600746023) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16815813/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int.%20J.%20Toxicol.&title=Structure,%20Function,%20Expression,%20Genomic%20Organization,%20and%20Single%20Nucleotide%20Polymorphisms%20of%20Human%20ABCB1%20(MDR1),%20ABCC%20(MRP),%20and%20ABCG2%20(BCRP)%20Efflux%20Transporters&author=S.%20Choudhuri&author=C.D.%20Klaassen&volume=25&publication_year=2006&pages=231-259&pmid=16815813&doi=10.1080/10915810600746023&)
|
| 279 |
+
|
| 280 |
+
11. Thiebaut F., Tsuruo T., Hamada H., Gottesman M.M., Pastan I., Willingham M.C. Cellular Localization of the Multidrug-Resistance Gene Product P-Glycoprotein in Normal Human Tissues. Proc. Natl. Acad. Sci. USA. 1987;84:7735–7738. doi: 10.1073/pnas.84.21.7735. [DOI](https://doi.org/10.1073/pnas.84.21.7735) | [PMC free article](/articles/PMC299375/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/2444983/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Proc.%20Natl.%20Acad.%20Sci.%20USA&title=Cellular%20Localization%20of%20the%20Multidrug-Resistance%20Gene%20Product%20P-Glycoprotein%20in%20Normal%20Human%20Tissues&author=F.%20Thiebaut&author=T.%20Tsuruo&author=H.%20Hamada&author=M.M.%20Gottesman&author=I.%20Pastan&volume=84&publication_year=1987&pages=7735-7738&pmid=2444983&doi=10.1073/pnas.84.21.7735&)
|
| 281 |
+
|
| 282 |
+
12. Tatsuta T., Naito M., Oh-hara T., Sugawara I., Tsuruo T. Functional Involvement of P-Glycoprotein in Blood-Brain Barrier. J. Biol. Chem. 1992;267:20383–20391. doi: 10.1016/S0021-9258(19)88713-6. [DOI](https://doi.org/10.1016/S0021-9258(19)88713-6) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1356979/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Biol.%20Chem.&title=Functional%20Involvement%20of%20P-Glycoprotein%20in%20Blood-Brain%20Barrier&author=T.%20Tatsuta&author=M.%20Naito&author=T.%20Oh-hara&author=I.%20Sugawara&author=T.%20Tsuruo&volume=267&publication_year=1992&pages=20383-20391&pmid=1356979&doi=10.1016/S0021-9258(19)88713-6&)
|
| 283 |
+
|
| 284 |
+
13. Cordon-Cardo C., O’Brien J.P., Casals D., Rittman-Grauer L., Biedler J.L., Melamed M.R., Bertino J.R. Multidrug-Resistance Gene (P-Glycoprotein) Is Expressed by Endothelial Cells at Blood-Brain Barrier Sites. Proc. Natl. Acad. Sci. USA. 1989;86:695–698. doi: 10.1073/pnas.86.2.695. [DOI](https://doi.org/10.1073/pnas.86.2.695) | [PMC free article](/articles/PMC286540/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/2563168/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Proc.%20Natl.%20Acad.%20Sci.%20USA&title=Multidrug-Resistance%20Gene%20(P-Glycoprotein)%20Is%20Expressed%20by%20Endothelial%20Cells%20at%20Blood-Brain%20Barrier%20Sites&author=C.%20Cordon-Cardo&author=J.P.%20O%E2%80%99Brien&author=D.%20Casals&author=L.%20Rittman-Grauer&author=J.L.%20Biedler&volume=86&publication_year=1989&pages=695-698&pmid=2563168&doi=10.1073/pnas.86.2.695&)
|
| 285 |
+
|
| 286 |
+
14. Andersen V., Vogel U., Godiksen S., Frenzel F.B., Sæbø M., Hamfjord J., Kure E., Vogel L.K. Low ABCB1 Gene Expression Is an Early Event in Colorectal Carcinogenesis. PLoS ONE. 2013;8:e72119. doi: 10.1371/annotation/7cd6c6a6-b281-4bf3-b478-a289ed39b375. [DOI](https://doi.org/10.1371/annotation/7cd6c6a6-b281-4bf3-b478-a289ed39b375) | [PMC free article](/articles/PMC3747088/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23977225/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=PLoS%20ONE&title=Low%20ABCB1%20Gene%20Expression%20Is%20an%20Early%20Event%20in%20Colorectal%20Carcinogenesis&author=V.%20Andersen&author=U.%20Vogel&author=S.%20Godiksen&author=F.B.%20Frenzel&author=M.%20S%C3%A6b%C3%B8&volume=8&publication_year=2013&pages=e72119&pmid=23977225&doi=10.1371/annotation/7cd6c6a6-b281-4bf3-b478-a289ed39b375&)
|
| 287 |
+
|
| 288 |
+
15. He T., Mo A., Zhang K., Liu L. ABCB1/MDR1 Gene Polymorphism and Colorectal Cancer Risk: A Meta-Analysis of Case-Control Studies. Color. Dis. 2013;15:12–18. doi: 10.1111/j.1463-1318.2012.02919.x. [DOI](https://doi.org/10.1111/j.1463-1318.2012.02919.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23279665/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Color.%20Dis.&title=ABCB1/MDR1%20Gene%20Polymorphism%20and%20Colorectal%20Cancer%20Risk:%20A%20Meta-Analysis%20of%20Case-Control%20Studies&author=T.%20He&author=A.%20Mo&author=K.%20Zhang&author=L.%20Liu&volume=15&publication_year=2013&pages=12-18&pmid=23279665&doi=10.1111/j.1463-1318.2012.02919.x&)
|
| 289 |
+
|
| 290 |
+
16. Lee J., Huang H., Chen Y., Lu X. ABCB1 Haplotype Influences the Sirolimus Dose Requirements in Chinese Renal Transplant Recipients. Biopharm. Drug Dispos. 2014;35:164–172. doi: 10.1002/bdd.1881. [DOI](https://doi.org/10.1002/bdd.1881) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24285256/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Biopharm.%20Drug%20Dispos.&title=ABCB1%20Haplotype%20Influences%20the%20Sirolimus%20Dose%20Requirements%20in%20Chinese%20Renal%20Transplant%20Recipients&author=J.%20Lee&author=H.%20Huang&author=Y.%20Chen&author=X.%20Lu&volume=35&publication_year=2014&pages=164-172&pmid=24285256&doi=10.1002/bdd.1881&)
|
| 291 |
+
|
| 292 |
+
17. Zuo L., Wang K., Luo X. Use of Diplotypes—Matched Haplotype Pairs from Homologous Chromosomes—In Gene-Disease Association Studies. Shanghai Arch Psychiatry. 2014;26:165–170. doi: 10.3969/j.issn.1002-0829.2014.03.009. [DOI](https://doi.org/10.3969/j.issn.1002-0829.2014.03.009) | [PMC free article](/articles/PMC4118015/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25114493/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Shanghai%20Arch%20Psychiatry&title=Use%20of%20Diplotypes%E2%80%94Matched%20Haplotype%20Pairs%20from%20Homologous%20Chromosomes%E2%80%94In%20Gene-Disease%20Association%20Studies&author=L.%20Zuo&author=K.%20Wang&author=X.%20Luo&volume=26&publication_year=2014&pages=165-170&pmid=25114493&doi=10.3969/j.issn.1002-0829.2014.03.009&)
|
| 293 |
+
|
| 294 |
+
18. Gümüş-Akay G., Rüstemoğlu A., Karadağ A., Sunguroğlu A. Haplotype-Based Analysis of MDR1/ABCB1 Gene Polymorphisms in a Turkish Population. DNA Cell Biol. 2010;29:83–90. doi: 10.1089/dna.2009.0953. [DOI](https://doi.org/10.1089/dna.2009.0953) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20025534/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=DNA%20Cell%20Biol.&title=Haplotype-Based%20Analysis%20of%20MDR1/ABCB1%20Gene%20Polymorphisms%20in%20a%20Turkish%20Population&author=G.%20G%C3%BCm%C3%BC%C5%9F-Akay&author=A.%20R%C3%BCstemo%C4%9Flu&author=A.%20Karada%C4%9F&author=A.%20Sunguro%C4%9Flu&volume=29&publication_year=2010&pages=83-90&pmid=20025534&doi=10.1089/dna.2009.0953&)
|
| 295 |
+
|
| 296 |
+
19. Home—SNP—NCBI. [(accessed on 24 July 2024)]; Available online: https://www.ncbi.nlm.nih.gov/snp/ [https://www.ncbi.nlm.nih.gov/snp/](https://www.ncbi.nlm.nih.gov/snp/)
|
| 297 |
+
|
| 298 |
+
20. Schwab M., Eichelbaum M., Fromm M.F. Genetic Polymorphisms of the Human MDR1 Drug Transporter. Annu. Rev. Pharmacol. Toxicol. 2003;43:285–307. doi: 10.1146/annurev.pharmtox.43.100901.140233. [DOI](https://doi.org/10.1146/annurev.pharmtox.43.100901.140233) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12359865/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Annu.%20Rev.%20Pharmacol.%20Toxicol.&title=Genetic%20Polymorphisms%20of%20the%20Human%20MDR1%20Drug%20Transporter&author=M.%20Schwab&author=M.%20Eichelbaum&author=M.F.%20Fromm&volume=43&publication_year=2003&pages=285-307&pmid=12359865&doi=10.1146/annurev.pharmtox.43.100901.140233&)
|
| 299 |
+
|
| 300 |
+
21. Dey S. Single Nucleotide Polymorphisms in Human P-Glycoprotein: Its Impact on Drug Delivery and Disposition. Expert Opin. Drug Deliv. 2006;3:23–35. doi: 10.1517/17425247.3.1.23. [DOI](https://doi.org/10.1517/17425247.3.1.23) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16370938/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Expert%20Opin.%20Drug%20Deliv.&title=Single%20Nucleotide%20Polymorphisms%20in%20Human%20P-Glycoprotein:%20Its%20Impact%20on%20Drug%20Delivery%20and%20Disposition&author=S.%20Dey&volume=3&publication_year=2006&pages=23-35&pmid=16370938&doi=10.1517/17425247.3.1.23&)
|
| 301 |
+
|
| 302 |
+
22. Sakaeda T., Nakamura T., Okumura K. Pharmacogenetics of MDR1 and Its Impact on the Pharmacokinetics and Pharmacodynamics of Drugs. Pharmacogenomics. 2003;4:397–410. doi: 10.1517/phgs.4.4.397.22747. [DOI](https://doi.org/10.1517/phgs.4.4.397.22747) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12831320/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=Pharmacogenetics%20of%20MDR1%20and%20Its%20Impact%20on%20the%20Pharmacokinetics%20and%20Pharmacodynamics%20of%20Drugs&author=T.%20Sakaeda&author=T.%20Nakamura&author=K.%20Okumura&volume=4&publication_year=2003&pages=397-410&pmid=12831320&doi=10.1517/phgs.4.4.397.22747&)
|
| 303 |
+
|
| 304 |
+
23. Kim I.-W., Moon Y.J., Ji E., Kim K.I., Han N., Kim S.J., Shin W.G., Ha J., Yoon J.-H., Lee H.S., et al. Clinical and Genetic Factors Affecting Tacrolimus Trough Levels and Drug-Related Outcomes in Korean Kidney Transplant Recipients. Eur. J. Clin. Pharmacol. 2012;68:657–669. doi: 10.1007/s00228-011-1182-5. [DOI](https://doi.org/10.1007/s00228-011-1182-5) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22183771/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20J.%20Clin.%20Pharmacol.&title=Clinical%20and%20Genetic%20Factors%20Affecting%20Tacrolimus%20Trough%20Levels%20and%20Drug-Related%20Outcomes%20in%20Korean%20Kidney%20Transplant%20Recipients&author=I.-W.%20Kim&author=Y.J.%20Moon&author=E.%20Ji&author=K.I.%20Kim&author=N.%20Han&volume=68&publication_year=2012&pages=657-669&pmid=22183771&doi=10.1007/s00228-011-1182-5&)
|
| 305 |
+
|
| 306 |
+
24. Fredericks S., Moreton M., Reboux S., Carter N.D., Goldberg L., Holt D.W., MacPhee I.A.M. Multidrug Resistance Gene-1 (MDR-1) Haplotypes Have a Minor Influence on Tacrolimus Dose Requirements. Transplantation. 2006;82:705–708. doi: 10.1097/01.tp.0000234942.78716.c0. [DOI](https://doi.org/10.1097/01.tp.0000234942.78716.c0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16969296/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplantation&title=Multidrug%20Resistance%20Gene-1%20(MDR-1)%20Haplotypes%20Have%20a%20Minor%20Influence%20on%20Tacrolimus%20Dose%20Requirements&author=S.%20Fredericks&author=M.%20Moreton&author=S.%20Reboux&author=N.D.%20Carter&author=L.%20Goldberg&volume=82&publication_year=2006&pages=705-708&pmid=16969296&doi=10.1097/01.tp.0000234942.78716.c0&)
|
| 307 |
+
|
| 308 |
+
25. Akbas S.H., Bilgen T., Keser I., Tuncer M., Yucetin L., Tosun O., Gultekin M., Luleci G. The Effect of MDR1 (ABCB1) Polymorphism on the Pharmacokinetic of Tacrolimus in Turkish Renal Transplant Recipients. Transplant. Proc. 2006;38:1290–1292. doi: 10.1016/j.transproceed.2006.02.079. [DOI](https://doi.org/10.1016/j.transproceed.2006.02.079) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16797284/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplant.%20Proc.&title=The%20Effect%20of%20MDR1%20(ABCB1)%20Polymorphism%20on%20the%20Pharmacokinetic%20of%20Tacrolimus%20in%20Turkish%20Renal%20Transplant%20Recipients&author=S.H.%20Akbas&author=T.%20Bilgen&author=I.%20Keser&author=M.%20Tuncer&author=L.%20Yucetin&volume=38&publication_year=2006&pages=1290-1292&pmid=16797284&doi=10.1016/j.transproceed.2006.02.079&)
|
| 309 |
+
|
| 310 |
+
26. Provenzani A., Notarbartolo M., Labbozzetta M., Poma P., Vizzini G., Salis P., Caccamo C., Bertani T., Palazzo U., Polidori P., et al. Influence of CYP3A5 and ABCB1 Gene Polymorphisms and Other Factors on Tacrolimus Dosing in Caucasian Liver and Kidney Transplant Patients. Int. J. Mol. Med. 2011;28:1093–1102. doi: 10.3892/ijmm.2011.794. [DOI](https://doi.org/10.3892/ijmm.2011.794) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21922127/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int.%20J.%20Mol.%20Med.&title=Influence%20of%20CYP3A5%20and%20ABCB1%20Gene%20Polymorphisms%20and%20Other%20Factors%20on%20Tacrolimus%20Dosing%20in%20Caucasian%20Liver%20and%20Kidney%20Transplant%20Patients&author=A.%20Provenzani&author=M.%20Notarbartolo&author=M.%20Labbozzetta&author=P.%20Poma&author=G.%20Vizzini&volume=28&publication_year=2011&pages=1093-1102&pmid=21922127&doi=10.3892/ijmm.2011.794&)
|
| 311 |
+
|
| 312 |
+
27. Hamzah S., Teh L.K., Siew J.S.K., Ahmad G., Wong H.S., Zakaria Z.A., Salleh M.Z. Pharmacogenotyping of CYP3A5 in Predicting Dose-Adjusted Trough Levels of Tacrolimus among Malaysian Kidney-Transplant Patients. Can. J. Physiol. Pharmacol. 2014;92:50–57. doi: 10.1139/cjpp-2013-0128. [DOI](https://doi.org/10.1139/cjpp-2013-0128) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24383873/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Can.%20J.%20Physiol.%20Pharmacol.&title=Pharmacogenotyping%20of%20CYP3A5%20in%20Predicting%20Dose-Adjusted%20Trough%20Levels%20of%20Tacrolimus%20among%20Malaysian%20Kidney-Transplant%20Patients&author=S.%20Hamzah&author=L.K.%20Teh&author=J.S.K.%20Siew&author=G.%20Ahmad&author=H.S.%20Wong&volume=92&publication_year=2014&pages=50-57&pmid=24383873&doi=10.1139/cjpp-2013-0128&)
|
| 313 |
+
|
| 314 |
+
28. Cho J.-H., Yoon Y.-D., Park J.-Y., Song E.-J., Choi J.-Y., Yoon S.-H., Park S.-H., Kim Y.-L., Kim C.-D. Impact of Cytochrome P450 3A and ATP-Binding Cassette Subfamily B Member 1 Polymorphisms on Tacrolimus Dose-Adjusted Trough Concentrations among Korean Renal Transplant Recipients. Transplant. Proc. 2012;44:109–114. doi: 10.1016/j.transproceed.2011.11.004. [DOI](https://doi.org/10.1016/j.transproceed.2011.11.004) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22310591/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplant.%20Proc.&title=Impact%20of%20Cytochrome%20P450%203A%20and%20ATP-Binding%20Cassette%20Subfamily%20B%20Member%201%20Polymorphisms%20on%20Tacrolimus%20Dose-Adjusted%20Trough%20Concentrations%20among%20Korean%20Renal%20Transplant%20Recipients&author=J.-H.%20Cho&author=Y.-D.%20Yoon&author=J.-Y.%20Park&author=E.-J.%20Song&author=J.-Y.%20Choi&volume=44&publication_year=2012&pages=109-114&pmid=22310591&doi=10.1016/j.transproceed.2011.11.004&)
|
| 315 |
+
|
| 316 |
+
29. Wallemacq P., Goffinet J.-S., O’Morchoe S., Rosiere T., Maine G.T., Labalette M., Aimo G., Dickson D., Schmidt E., Schwinzer R., et al. Multi-Site Analytical Evaluation of the Abbott ARCHITECT Tacrolimus Assay. Ther. Drug Monit. 2009;31:198–204. doi: 10.1097/FTD.0b013e31819c6a37. [DOI](https://doi.org/10.1097/FTD.0b013e31819c6a37) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19258928/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ther.%20Drug%20Monit.&title=Multi-Site%20Analytical%20Evaluation%20of%20the%20Abbott%20ARCHITECT%20Tacrolimus%20Assay&author=P.%20Wallemacq&author=J.-S.%20Goffinet&author=S.%20O%E2%80%99Morchoe&author=T.%20Rosiere&author=G.T.%20Maine&volume=31&publication_year=2009&pages=198-204&pmid=19258928&doi=10.1097/FTD.0b013e31819c6a37&)
|
| 317 |
+
|
| 318 |
+
30. Glickman M.E., Rao S.R., Schultz M.R. False Discovery Rate Control Is a Recommended Alternative to Bonferroni-Type Adjustments in Health Studies. J. Clin. Epidemiol. 2014;67:850–857. doi: 10.1016/j.jclinepi.2014.03.012. [DOI](https://doi.org/10.1016/j.jclinepi.2014.03.012) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24831050/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Epidemiol.&title=False%20Discovery%20Rate%20Control%20Is%20a%20Recommended%20Alternative%20to%20Bonferroni-Type%20Adjustments%20in%20Health%20Studies&author=M.E.%20Glickman&author=S.R.%20Rao&author=M.R.%20Schultz&volume=67&publication_year=2014&pages=850-857&pmid=24831050&doi=10.1016/j.jclinepi.2014.03.012&)
|
test/texts/PMC11528939.md
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
test/texts/PMC11531276.md
ADDED
|
@@ -0,0 +1,420 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Pharmacogenomic Study of Selected Genes Affecting Amlodipine Blood Pressure Response in Patients with Hypertension
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Asif Jan, Abdullah R Alanzi, Ramzi A Mothana, Jun-Ya Kaimori, Syed Shaukat Ali, Tahir Muhammad, Muhammad Saeed, Rani Akbar, Mehtab Khan
|
| 5 |
+
**Journal:** Pharmacogenomics and Personalized Medicine
|
| 6 |
+
**Date:** 2024 Oct 29
|
| 7 |
+
**DOI:** [10.2147/PGPM.S481068](https://doi.org/10.2147/PGPM.S481068)
|
| 8 |
+
**PMID:** 39492848
|
| 9 |
+
**PMCID:** PMC11531276
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11531276/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11531276/pdf/pgpm-17-473.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11531276/pdf/pgpm-17-473.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Introduction:**
|
| 16 |
+
Despite the availability of various antihypertensive medications, the response to these medications varies among individuals. Understanding how individual genetic variations affect drugs treatment outcomes is a key area of focus in precision medicine. This study investigated the correlation between single nucleotide polymorphisms (SNPs) in selected genes (CACNA1C, CACNA1D, ABCB1, ACE, ADBR2, and NOS1AP) and the blood pressure (BP) control by amlodipine.
|
| 17 |
+
|
| 18 |
+
**Methods:**
|
| 19 |
+
Four hundred individuals of Pashtun ethnicity undergoing amlodipine treatment for hypertension were included in the present study and divided into the controlled (BP less than 140/90 mmHg) and uncontrolled (BP greater than 140/90 mmHg) hypertension groups. Blood samples (3 mL) were collected from each participant, and DNA was extracted using the Kit method. Ten SNPs in amlodipine pharmacogenes were selected and genotyped using real-time PCR with the TaqMan® system. Logistic regression model was used to determine the association between SNPs and the amlodipine BP response.
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
Notable association were observed between SNP rs2239050/CACNA1C and amlodipine blood pressure response, with GG genotype carriers demonstrating a better response (P=0.004) than individuals carrying CC or CG genotypes. SNP rs312481/CACNA1D also exhibited a positive pharmacogenetic association, Individuals with the GG genotype showing a considerable reduction in BP (P=0.021) compared to participants with AA or GA genotypes. In case of SNP rs429/ACE individuals carrying TA genotype were less likely to achieve BP control (P=0.002) than AA genotype carriers.
|
| 23 |
+
|
| 24 |
+
**Conclusion:**
|
| 25 |
+
Our finding suggests that the SNPs rs2239050/CACNA1C, rs312481/CACNA1D and rs429/ACE influence amlodipine blood pressure response in patients with hypertension. It is recommended that prior knowledge of amlodipine associated pharmacogenetic variants is important that could improve its treatment outcomes in hypertensive patients.
|
| 26 |
+
|
| 27 |
+
Keywords: pharmacogenomics, hypertension, genetic markers, personalized medicine, amlodipine, Pashtun, Pakistan
|
| 28 |
+
|
| 29 |
+
### Introduction
|
| 30 |
+
|
| 31 |
+
Despite the availability of various antihypertensive medications, the response to these medications varies among individuals. Understanding how individual genetic variations affect drugs treatment outcomes is a key area of focus in precision medicine. This study investigated the correlation between single nucleotide polymorphisms (SNPs) in selected genes (CACNA1C, CACNA1D, ABCB1, ACE, ADBR2, and NOS1AP) and the blood pressure (BP) control by amlodipine.
|
| 32 |
+
|
| 33 |
+
### Methods
|
| 34 |
+
|
| 35 |
+
Four hundred individuals of Pashtun ethnicity undergoing amlodipine treatment for hypertension were included in the present study and divided into the controlled (BP less than 140/90 mmHg) and uncontrolled (BP greater than 140/90 mmHg) hypertension groups. Blood samples (3 mL) were collected from each participant, and DNA was extracted using the Kit method. Ten SNPs in amlodipine pharmacogenes were selected and genotyped using real-time PCR with the TaqMan^®^® system. Logistic regression model was used to determine the association between SNPs and the amlodipine BP response.
|
| 36 |
+
|
| 37 |
+
### Results
|
| 38 |
+
|
| 39 |
+
Notable association were observed between SNP rs2239050/CACNA1C and amlodipine blood pressure response, with GG genotype carriers demonstrating a better response (P=0.004) than individuals carrying CC or CG genotypes. SNP rs312481/CACNA1D also exhibited a positive pharmacogenetic association, Individuals with the GG genotype showing a considerable reduction in BP (P=0.021) compared to participants with AA or GA genotypes. In case of SNP rs429/ACE individuals carrying TA genotype were less likely to achieve BP control (P=0.002) than AA genotype carriers.
|
| 40 |
+
|
| 41 |
+
### Conclusion
|
| 42 |
+
|
| 43 |
+
Our finding suggests that the SNPs rs2239050/CACNA1C, rs312481/CACNA1D and rs429/ACE influence amlodipine blood pressure response in patients with hypertension. It is recommended that prior knowledge of amlodipine associated pharmacogenetic variants is important that could improve its treatment outcomes in hypertensive patients.
|
| 44 |
+
|
| 45 |
+
**Keywords:**Keywords: pharmacogenomics, hypertension, genetic markers, personalized medicine, amlodipine, Pashtun, Pakistan
|
| 46 |
+
|
| 47 |
+
## Introduction
|
| 48 |
+
|
| 49 |
+
Hypertension (HTN) is a major public health issue, when left untreated leads to the development of various disorders like cardiovascular diseases, renal failure and premature death.[1](#cit0001)^1^1 The global burden of HTN has rapidly increased over the past decades, especially in low- and middle-income countries.[2](#cit0002)^2^2^,^,[3](#cit0003)^3^3 According to the World Health Organization (WHO) global report on hypertension (1990–2019) the HTN prevalence and incidence among adults has increased twofold, rising from 650 million in 1990 to 1.3 billion in 2019. The surge in the number of hypertensive cases is more frequent in the South Asian and Western Pacific regions than in the European and American populations.[4](#cit0004)^4^4^,^,[5](#cit0005)^5^5 The prevalence of hypertension in Pakistan, a developing, middle-income south Asian nation, is alarmingly high and requires immediate medical attention. National Health Survey of Pakistan on hypertension prevalence reported that 46.2% of adult Pakistani population is affected by hypertension and this number is escalating rapidly.[6](#cit0006)^6^6 Because of its growing mortality rate and lack of its initial sign and symptoms, it is recognized as one of the high prevalent chronic non-communicable diseases globally, earning the moniker of a “silent killer”.[7](#cit0007)^7^7 The key players contributing to high prevalence of hypertension are lack of exercise/physical activity, smoking, increased sodium intake, air pollution and rapid urbanization.[4](#cit0004)^4^4 In addition to factors such as poor lifestyle and various environmental factors, it has been documented that genetic factors (like Gene polymorphisms/ mutation in core genes) also play a significant role in both the onset of HTN and treatment response to anti-hypertensive medications.[8](#cit0008)^8^8^,^,[9](#cit0009)^9^9 Pharmacogenomic research studies as of the present study aim to elucidate how variations in different genes influence the response to antihypertensive medications. The ultimate goal is to develop pharmacotherapy based on individual genetic makeup aiming for personalized treatment strategies.
|
| 50 |
+
|
| 51 |
+
Different medications are used in blood pressure regulation; these include Diuretics, Beta Receptor Blocker (B-blockers), Calcium Channel Blockers (CCBs) Angiotensin Receptor Blockers (ARBs), direct vasodilators and Angiotensin-Converting Enzymes Inhibitors (ACEIs).[10](#cit0010)^10^10^,^,[11](#cit0011)^11^11 Calcium-Channel Blockers (CCBs) are commonly prescribed for hypertension treatment because CCBs controls high blood pressure effectively compared to other classes of antihypertensive medications.[12](#cit0012)^12^12 Amlodipine, a third-generation long-acting calcium channel blocker has demonstrated good efficacy in regulating elevated (high) blood pressure (BP) and has shown decreased risk of cardiovascular diseases in individuals with hypertension.[13](#cit0013)^13^13 Nonetheless, the BP lowering response to amlodipine varies significantly and several pharmacogenomic studies explored potential genetic polymorphisms that explain the observed differences among individuals and populations.[14–16](#cit0014)^14–16^14–16 These investigations analyzed polymorphisms in genes directly implicated in the Pharmacokinetics and Pharmacodynamics of amlodipine. Genetic polymorphisms in genes that encode ion channels (such as CACNA1C, CACNA1D, GNB3, TANC2, ADRB2, and ADRA1A) have been documented to alter the response to CCBs (such as amlodipine) by affecting their transport.[17](#cit0017)^17^17 For instance, a significant association between the CACNA1C variant rs2238032 and amlodipine treatment outcomes has been observed. Patients carrying the TT genotype showed positive treatment outcomes and those carrying the G allele showed negative treatment outcomes with amlodipine. Likewise in case rs2239050/ CACNA1C, individuals with GG genotype had better BP control with amlodipine compared to the individuals with CG genotype.[18](#cit0018)^18^18
|
| 52 |
+
|
| 53 |
+
Polymorphisms in the ABCB1 (a member of ATP-binding cassette (ABC) transporters) and CYP3A5 (a family member of cytochrome P450 enzyme) genes have been documented to influence the elimination and/or clearance of amlodipine.[19](#cit0019)^19^19 In the case of ABCB1, subjects carrying the TT genotype have an increased rate of amlodipine clearance compared to those carrying the CT or CC genotype.[20](#cit0020)^20^20 Whereas in case of CYP3A5 gene, CYP3A5*3/*3 genotype carriers showed reduced plasma amlodipine concentrations compared to CYP3A5*1 genotype carriers.[21](#cit0021)^21^21 Moreover, single nucleotide polymorphisms located within genes indirectly associated with the pharmacokinetics and antihypertensive action of amlodipine, such as AGT (Angiotensinogen) and ACE (angiotensin-converting enzyme) genes have been investigated.[22](#cit0022)^22^22 Both Angiotensinogen and angiotensin-converting enzyme play pivotal roles as an integral part of the renin-angiotensin system (RAS), which regulates BP by modulating fluid volume within the body.[23](#cit0023)^23^23 The variant rs4291 (of ACE gene) showed a significant correlation with the development of high BP;[22](#cit0022)^22^22 nevertheless, its direct link to the BP-lowering response of amlodipine remains unclear. A study conducted in an African American population documented that, in patients and subjects receiving amlodipine therapy, the presence of rs11122576 in the AGT gene demonstrated a lower risk of coronary heart disease. However, its association with BP regulation by amlodipine has not been documented/established.[24](#cit0024)^24^24 In addition, SNP rs10494366, located at the NOS1AP (nitric oxide synthase-1-adaptor protein) gene, was found to be associated with an increased risk of developing cardiovascular events in amlodipine users. Additionally, if rs1042713/ADRB2 SNP is present, cardiovascular drugs may show limited effectiveness.[25–27](#cit0025)^25–27^25–27
|
| 54 |
+
|
| 55 |
+
Given the multifaceted nature of HTN and the intricate physiological regulatory systems affecting its severity and management, it is crucial to study polymorphisms that are either directly or indirectly involved in pathways linked to the antihypertensive effects of pharmacological drugs. This will improve our understanding of the complex physiology of drug response outcomes in hypertensive individuals. Currently, there is limited information and data on specific SNPs that could affect the outcomes of hypertensive therapy in the Pakistani population. The present pharmacogenomic study in the Pashtun population of Pakistan aimed to explore polymorphisms in amlodipine-related genes and to evaluate their relationship with BP regulation. The Pakistani population is categorized into five major tribes/ethnic groups: the Sindhis, Baluchis, Punjabis, Muhajirs, and Pashtuns. Among these, Pashtuns have unique genetic makeup and distinct cultural practices, societal beliefs, and behaviors owing to these characteristics, making them the most suitable for such a pharmacogenomic study.
|
| 56 |
+
|
| 57 |
+
## Materials and Methods
|
| 58 |
+
|
| 59 |
+
### Study Subject Enrolment
|
| 60 |
+
|
| 61 |
+
A total of four hundred individuals of Pashtun ethnicity, taking amlodipine (for last 12 months) for hypertension treatment were included in the study. The enrolled participants were categorized into two groups: a) Un-controlled hypertensive patients (un-controlled HTN, n=200); and b) Controlled hypertensive patients (controlled HTN, n=200). Controlled hypertensive patients were marked/identified as individuals who were prescribed amlodipine (alone or in combination with any other antihypertensive medication) and maintained a mean arterial blood pressure of < 140/90 mmHg. Uncontrolled hypertensive patients included those who were prescribed amlodipine (alone or in combination with any other antihypertensive medication) but had a mean arterial blood pressure of ≥ 140/90 mmHg.
|
| 62 |
+
|
| 63 |
+
Study participants were selected from different districts in Khyber Pakhtunkhwa (KP), including Peshawar, Swabi, Charsadda, Bannu, Mardan, Nowshera, Dir, Swat, and Kohat. The study participants were enrolled from the cardiac care units of three large teaching- and research-based (tertiary care) hospitals (HMC), the Lady Reading Hospital (LRH), and Khyber Teaching Hospital (KTH). These hospitals offer special care and treatment for individuals with hypertension and other cardiovascular conditions. Written informed consent or an agreement form (that shows the wellness to be included in the study) was signed and obtained from all study subjects. For uneducated patients, the agreement form (or consent form) was verbally explained to them in local Pashtu language for ease of understanding. After agreeing to participate in the study and adhering to the terms and conditions, the agreement form was either signed by patients or by their attendant/relative on their behalf.
|
| 64 |
+
|
| 65 |
+
### Demographics and Clinical Data Collection
|
| 66 |
+
|
| 67 |
+
The demographic and detailed clinical data of the study participants were gathered using a proforma specially designed for this study. The information collected included: sex; age; district from which the patients belonged; smoking status; exercise level; presence or absence of comorbidities; socioeconomic status; drug and dietary compliance; prescribed medications for hypertension comprised amlodipine alone or in combination other antihypertensive medications like enalapril, losartan, and hydrochlorothiazide, or any other drug; glomerular filtration rate; triglyceride levels; BMI; urea and creatinine levels; blood glucose levels; and some other variables with potential influence on hypertension development.
|
| 68 |
+
|
| 69 |
+
### Inclusion and Exclusion Criteria
|
| 70 |
+
|
| 71 |
+
Study participants of Pashtun ethnicity, age between 20 to 80 years, using calcium channel blocker (the amlodipine) for blood pressure control from the past 12 months were included in the study whereas individuals who were bedridden, pregnant, or experiencing mental illness, chronic diseases such as HIV or Hepatitis were excluded from the study.
|
| 72 |
+
|
| 73 |
+
### Blood Samples Collection
|
| 74 |
+
|
| 75 |
+
With the help of a skilled and trained nurse, whole blood samples were collected from all the study participants using aseptic procedures. Blood samples were taken from the median cubital vein. Each participant contributed three millilitres (3 mL) of whole blood drawn into EDTA tubes that were carefully labeled to facilitate proper tracking and identification of the samples. Following blood collection and labelling, EDTA tubes (filled with 3 mL blood) were stored at −10 °C, until further testing or analysis.
|
| 76 |
+
|
| 77 |
+
### DNA Extraction and Quantification
|
| 78 |
+
|
| 79 |
+
For DNA extraction, two hundred (200) microliters (μL) of whole blood was used. The Wizard Genomic DNA Extraction Kit (model no. [W64120](https://www.ncbi.nlm.nih.gov/nuccore/W64120)W64120) was utilized for this purpose, carefully following the manufacturer’s instructions provided with the kit. Subsequent to the successful extraction of DNA, DNA was quantified using a Qubit™3 (Cat. No. [Q34860](https://www.ncbi.nlm.nih.gov/protein/Q34860)Q34860). Finally DNA concentration was adjusted to 5 ng/μL.
|
| 80 |
+
|
| 81 |
+
### Selection of Amlodipine Pharmacogenes
|
| 82 |
+
|
| 83 |
+
Ten known pharmacogenetic variants previously linked to blood pressure response to amlodipine, including rs2239050/CACNA1C, rs2238032/CACNA1C, rs312481/CACNA1D, rs3774425/CACNA1D, rs3774426/CACNA1D, rs2032582/ABCB1, rs4291/ACE, rs1799752/ACE, rs1042713/ADBR2, and rs10494366/NOS1AP, were carefully selected. The selection process involved consulting reliable sources, such as the Pharmacogenomics Knowledge Base (PharmGKB) and PharmacoGenomic Mutation Database (PGMD)[28](#cit0028)^28^28^,^,[29](#cit0029)^29^29 and conducting a comprehensive review of recent literature. Our focus was directed towards genes situated within pathways, either directly or indirectly, affecting the mechanism through which amlodipine lowers blood pressure.
|
| 84 |
+
|
| 85 |
+
### Genotyping
|
| 86 |
+
|
| 87 |
+
Genetic polymorphism analysis (genotyping) of the selected pharmacogenetic variants was carried out using Real-Time Polymerase Chain Reaction (real-time PCR) with the TaqMan^®^® SNP Genotyping Assay system (Applied Biosystems - Thermo Fisher Scientific). In brief, the reaction mixture consisted of 5 microlitre of TaqMan^®^® Master Mix and 0.5 microlitre of working reagent (primer/probe), resulting in a total of 5.5 microlitre of reagent per well. DNA samples along with controls were diluted with Nuclease-free water to attain a concentration of 10 ng/µL per well. Subsequently, 5.5 microlitre of the previously prepared reagent and 4.5 microlitre of the diluted sample were added to each well of the MicroAmp™ 96-well optical reaction plate, resulting in a total volume of 10 µL per well. Next the plate was sealed with adhesive tape and centrifuged at 1000 rpm before processing on real--time PCR machine. Allelic discrimination data analysis was conducted using Applied Bio-systems/Thermo Scientific 7500 v2.3 software.
|
| 88 |
+
|
| 89 |
+
### Statistical Analysis
|
| 90 |
+
|
| 91 |
+
All statistical analyses and tests were performed using the IBM SPSS version 25 (Statistical Package for Social Sciences, V25). The key variables considered for analysis included age, sex, triglyceride levels, BMI, co-administered or current medications, smoking status, districts from where the study subjects belonged, occupation, exercise level, lifestyle, diet, and specific genetic variants in CACNA1, CACNA1D, ABCB1, ACE, ADBR2, and NOS1AP. To identify genetic variants confirmatory of the Hardy-Weinberg equilibrium (HWE), the chi-square (χ^2^2) test was employed. To examine the allelic and genotypic frequencies differences between the uncontrolled and controlled AH groups, the χ^2^2 test was utilized. The correlation or association between alleles, genotypes, and BP response to amlodipine was tested using a logistic regression analysis. The effects of confounding factors on this association were assessed using an adjusted logistic regression model. Statistical significance was set at p < 0.05.
|
| 92 |
+
|
| 93 |
+
## Results
|
| 94 |
+
|
| 95 |
+
### Description of the Study Cohort
|
| 96 |
+
|
| 97 |
+
Sociodemographic characteristics, biochemical parameters of the study subjects, and the prevalence of comorbid conditions are given in [Tables 1–3](#t0001)Tables 1–3. The present study recruited 400 individuals age > 20 years, consisting of 200 individuals with controlled hypertension and 200 individuals with uncontrolled hypertension. Both the groups were age- and weight-matched.
|
| 98 |
+
|
| 99 |
+
### Table 1.
|
| 100 |
+
|
| 101 |
+
Sociodemographic Features of Study Subjects Under Investigation
|
| 102 |
+
|
| 103 |
+
| Variables | Controlled HTN n(f) | Uncontrolled HTN n(f) | P-value |
|
| 104 |
+
| --------- | ------------------- | --------------------- | ------- |
|
| 105 |
+
| Gender | | | 0.221 |
|
| 106 |
+
| Male (M) | 153 (76.5%) | 145 (72.5%) | |
|
| 107 |
+
| Female (F) | 47 (23.5%) | 55(27.5%) | |
|
| 108 |
+
| Mean age (yrs) | 54 ± 13:43 | 56 ± 13:40 | 0.805 |
|
| 109 |
+
| Mean weight (Kg) | 60.55 ± 8:32 | 62.64 ± 6:07 | 0.913 |
|
| 110 |
+
| Address | | | 0.328 |
|
| 111 |
+
| Peshawar | 50 (25.0%) | 55 (27.5%) | |
|
| 112 |
+
| Charsadda | 44 (22.0%) | 34 (17.0%) | |
|
| 113 |
+
| Mardan | 13 (6.5%) | 22 (11.0%) | |
|
| 114 |
+
| Kohat | 11 (5.5%) | 12 (6.0%) | |
|
| 115 |
+
| Swabi | 4 (2.0%) | 19 (9.5%) | |
|
| 116 |
+
| Nowshera | 35 (17.5%) | 17 (8.5%) | |
|
| 117 |
+
| Bannu | 10 (5.0%) | 18 (9.0%) | |
|
| 118 |
+
| Karak | 13 (6.5%) | 05 (2.5%) | |
|
| 119 |
+
| Dir | 10 (5.0%) | 06 (3.0%) | |
|
| 120 |
+
| Swat | 10 (5.0%) | 12 (6.0%) | |
|
| 121 |
+
| Occupation | | | 0.098 |
|
| 122 |
+
| Business | 16 (8.0%) | 20 (10.0%) | |
|
| 123 |
+
| Govt. servant | 28 (14.0%) | 37 (18.5%) | |
|
| 124 |
+
| Retired | 30 (15.0%) | 35 (17.5.0%) | |
|
| 125 |
+
| Farming | 55 (27.5%) | 35 (17.5%) | |
|
| 126 |
+
| House wife | 35 (17.5%) | 40 (20.0%) | |
|
| 127 |
+
| Labor | 36 (18.0%) | 33 (16.5%) | |
|
| 128 |
+
| Family Hx of HTN | | | 0.031 |
|
| 129 |
+
| Yes | 170 (85.0%) | 135 (67.5%) | |
|
| 130 |
+
| No | 30 (15.0%) | 65 (32.5%) | |
|
| 131 |
+
| Marital status | | | 0.138 |
|
| 132 |
+
| Single | 43 (21.5%) | 61 (30.5%) | |
|
| 133 |
+
| Married | 157 (78.6%) | 139 (69.5%) | |
|
| 134 |
+
| Smoking | | | 0.073 |
|
| 135 |
+
| Yes | 140 (70.0%) | 104 (52.0%) | |
|
| 136 |
+
| No | 60 (30.0%) | 96 (48.0%) | |
|
| 137 |
+
| Naswar (smokeless tobacco product) | | | 0.061 |
|
| 138 |
+
| Yes | 163 (81.5%) | 130 (65.0%) | |
|
| 139 |
+
| No | 47 (18.5%) | 70 (35.0% | |
|
| 140 |
+
| Socioeconomic status | | | 0.524 |
|
| 141 |
+
| Good | 24 (12.5%) | 22 (11.0%) | |
|
| 142 |
+
| Average | 153 (76.5%) | 132 (66.0%) | |
|
| 143 |
+
| Below | 23 (11.5%) | 46 (23%) | |
|
| 144 |
+
### Table 2.
|
| 145 |
+
|
| 146 |
+
Prevalence of Other Diseases Among Study Participants in Addition to HTN
|
| 147 |
+
|
| 148 |
+
| Name of Co Morbid Disease | Frequency (f) | P-value |
|
| 149 |
+
| ------------------------- | ------------- | ------- |
|
| 150 |
+
| Controlled HTN | Uncontrolled HTN | |
|
| 151 |
+
| Type 2 diabetes | 12.5% | 22.0% | 0.051 |
|
| 152 |
+
| IHD | 19.0% | 21.0% | 0.611 |
|
| 153 |
+
| Kidney Failure | 4.00% | 6.00% | 0.912 |
|
| 154 |
+
| Retinopathy | 9.1% | 23.0% | 0.012 |
|
| 155 |
+
| HBV | 0.00% | 0.00% | NA |
|
| 156 |
+
| HCV | 0.00% | 0.00% | NA |
|
| 157 |
+
### Table 3.
|
| 158 |
+
|
| 159 |
+
Clinical Features/Characteristics of the Participants Under Investigation
|
| 160 |
+
|
| 161 |
+
| Variables | Controlled HTN n(f) | Uncontrolled HTN n(f) |
|
| 162 |
+
| --------- | ------------------- | --------------------- |
|
| 163 |
+
| Total cholesterol (mg/dL) | | |
|
| 164 |
+
| Normal | 90 (45.0%) | 94 (47.0%) |
|
| 165 |
+
| Changed | 110 (55.0) | 106 (53.0%) |
|
| 166 |
+
| LDL- cholesterol (mg/dL) | | |
|
| 167 |
+
| Normal | 36 (18.0%) | 30 (15.0%) |
|
| 168 |
+
| Changed | 164 (82.5%) | 170(85.0%) |
|
| 169 |
+
| HDL- cholesterol (mg/dL) | | |
|
| 170 |
+
| Normal | 133 (66.5%) | 148 (74.0%) |
|
| 171 |
+
| Changed | 67 (33.5%) | 52 (26%) |
|
| 172 |
+
| Triglycerol (mg/dL) | | |
|
| 173 |
+
| Normal | 102 (51.0%) | 96 (48.0%) |
|
| 174 |
+
| Changed | 98 (49.0%) | 104 (52.0%) |
|
| 175 |
+
| Urea (mg/dL) | | |
|
| 176 |
+
| Normal | 183 (91.5%) | 173 (86.5%) |
|
| 177 |
+
| Changed | 17 (8.5%) | 27 (13.5%) |
|
| 178 |
+
| Creatinine (mg/dl) | | |
|
| 179 |
+
| Normal | 187 (93.5%) | 180 (90.0%) |
|
| 180 |
+
| Changed | 13 (6.5%) | 20 (10.0%) |
|
| 181 |
+
| HBA1C (%) | | |
|
| 182 |
+
| Normal | 179 (89.5%) | 182 (91.0%) |
|
| 183 |
+
| Changed | 21 (10.5%) | 18 (9.0%) |
|
| 184 |
+
| Amlodipine alone | 28 (14.0%) | 42 (21.0%) |
|
| 185 |
+
| Amlodipine + 1 drug | 44 (22.0%) | 58 (29.0%) |
|
| 186 |
+
| Amlodipine + 2 drugs | 91 (45.5%) | 62 (31.0%) |
|
| 187 |
+
| Amlodipine + 3 drugs | 36 (18.0%) | 22 (11.0%) |
|
| 188 |
+
In the controlled-HTN group, 75.5% of patients were males and 23.5% were females. District-wise, 25.5% of individuals were from district Peshawar, 22.5% from district Charsadda, 17.5% from district nowshera, 13 individuals (6.5%) from district Mardan, and a similar number of individuals from district Mardan were from district Karak. The occupation-wise majority (27.5%) was attached to farming. Moreover 85.0% showed family history of HTN whereas 15.0% replied “NO” when asked for family history of HTN. Seventy percent (70%) were smokers and 30% were non-smokers. Drug and diet compliance was good in the majority (71.0%) of individuals, whereas 29.0% of the study participants showed poor drug and dietary compliance. Considering the socioeconomic status majority (76.5%) were from average-income families and 11.5% were from poor families.
|
| 189 |
+
|
| 190 |
+
In case of uncontrolled-HTN group, 72.5% of the patients were males and 27.5% were females. Fifty-five participants (27.5%) were from Peshawar district, 17.0% were from Charsadda district, and a few participants (2.5%) were from Karak district. Occupation-wise, 18.5% were government servants, 20.0% among the female candidates were housewives, 10.0% were attached to business, and thirty five participants (67.5%) had a family history of hypertension, whereas the remaining 32.5% were documented to have no family history of hypertension. The majority (66.0%) of the patients belonged to middle-income families, whereas 23.0% had poor family backgrounds.
|
| 191 |
+
|
| 192 |
+
Comorbidities (Type 2 diabetes, renal failure, hypercholesterolemia, and retinopathy) were more prevalent in the un-controlled HTN group compared to control group ([Table 2](#t0002)Table 2). Similarly, lipid profiles (including triglycerides, total cholesterol, HDL-cholesterol, and LDL-cholesterol), renal function markers (urea and creatinine), and glycemic profiles were assessed in both groups. The detailed results are provided in [Table 3](#t0003)Table 3.
|
| 193 |
+
|
| 194 |
+
### Expression of Selected Pharmacogenetic Variants in the Study Participants
|
| 195 |
+
|
| 196 |
+
The study participants were screened for absence or presence of n=10 single nucleotide polymorphism associated with amlodipine response namely rs2239050, rs2238032, rs312481, rs3774425, rs3774426, rs2032582, rs4291, rs1799752, rs1042713, and rs10494366 using Real time PCR machine. Among the selected SNPs, seven variants (rs2239050, rs312481, rs3774425, rs3774426, rs4291, rs1042713, and rs10494366) were identified in the target population whereas the other three SNPs were not found in the study population. SNPs identified in the target population were in Hardy���Weinberg equilibrium (HWE). Details of the expressed and unexpressed SNPs in the study population are listed in [Table 4](#t0004)Table 4.
|
| 197 |
+
|
| 198 |
+
### Table 4.
|
| 199 |
+
|
| 200 |
+
Expressed/Un-Expressed Selected Single Nucleotide Polymorphisms (SNPs) in the Study Participants
|
| 201 |
+
|
| 202 |
+
| Gene | SNP | Status | Controlled HTN n(f) | Uncontrolled HTN n(f) |
|
| 203 |
+
| ---- | --- | ------ | ------------------- | --------------------- |
|
| 204 |
+
| CACNA1C | rs2239050 | Present | 175 (87.5%) | 135 (67.5%) |
|
| 205 |
+
| Absent | 35(17.5%) | 65(32.5%) | | |
|
| 206 |
+
| CACNA1C | rs2238032 | Present | – | – |
|
| 207 |
+
| Absent | 200(100%) | 200(100%) | | |
|
| 208 |
+
| CACNA1D | rs312481 | Present | 172(86%) | 180 (90%) |
|
| 209 |
+
| Absent | 28 (14%) | 20 (10%) | | |
|
| 210 |
+
| CACNA1D | rs3774425 | Present | 98 (49%) | 121 (60.5%) |
|
| 211 |
+
| Absent | 102 (51%) | 79(39.5%) | | |
|
| 212 |
+
| CACNA1D | rs3774426 | Present | 105(52.5%) | 112(56%) |
|
| 213 |
+
| Absent | 95(47.5%) | 88 (44%) | | |
|
| 214 |
+
| ABCB1 | rs2032582 | Present | – | – |
|
| 215 |
+
| Absent | 200 (100%) | 200 (100%) | | |
|
| 216 |
+
| ACE | rs4291 | Present | 165 (82.5%) | 146 (73%) |
|
| 217 |
+
| Absent | 35 (17.5%) | 54 (27%) | | |
|
| 218 |
+
| ACE | rs1799752 | Present | – | – |
|
| 219 |
+
| Absent | 200 (100%) | 200 (100%) | | |
|
| 220 |
+
| ADBR2 | rs1042713 | Present | 112(56%) | 132 (66%) |
|
| 221 |
+
| Absent | 88 (44%) | 68 (34%) | | |
|
| 222 |
+
| NOS1AP | rs10494366 | Present | 116(58%) | 98 (49%) |
|
| 223 |
+
| Absent | 84(42%) | 102 (51%) | | |
|
| 224 |
+
### SNPs and Their Association with Amlodipine Response
|
| 225 |
+
|
| 226 |
+
The influence of alleles and genotypes on the amlodipine-induced blood pressure control was observed using unadjusted and adjusted logistic regression models. A strong association between amlodipine response and the SNP rs2239050/CACNA1C was observed in the study participants. Participants carrying the GG genotype had better response/outcomes (P=0.004) when treated with amlodipine than carriers of CC or CG genotypes. After adjusting for confounding factors (age, sex, drug and diet compliance, etc)., no observable changes were noticed in the degree, level, or magnitude of the association. In the case of rs312481/CACNA1D, crude logistic regression analysis showed a positive pharmacogenetic association between SNP rs312481 and amlodipine response; individuals carrying the GG genotype showed a notable reduction in blood pressure (p=0.021). When confounding factors were considered, the association between SNP rs312481 and blood pressure regulation by amlodipine remained consistent, that is, the covariates/confounding factors showed no additive or opposing effect on the association. In the case of SNP rs429/ACE, individuals with the TA genotype were less likely to achieve blood pressure control (or have un-controlled hypertension) than those with the AA genotype. The remaining SNPs (rs3774425/ CACNA1D, rs3774426/ CACNA1D, rs1042713/ ADBR2, and rs10494366/ NOS1AP) showed no considerable influence on amlodipine response. The detailed results are presented in [Table 5](#t0005)Table 5.
|
| 227 |
+
|
| 228 |
+
### Table 5.
|
| 229 |
+
|
| 230 |
+
The Influence of Alleles and Genotypes on Amlodipine Produced Blood Pressure Response Using Unadjusted and Adjusted Regression Models
|
| 231 |
+
|
| 232 |
+
| Genotypes | Controlled HTN (n=200) n(f) | Un-controlled HTN (n=200) n(f) | Unadjusted Odds Ratios (95% CI) | p-value | Adjusted Odds Ratios (95% CI) | p-value |
|
| 233 |
+
| --------- | --------------------------- | ------------------------------ | ------------------------------- | ------- | ----------------------------- | ------- |
|
| 234 |
+
| rs2239050/ CACNA1C | | | | | | |
|
| 235 |
+
| CC | 67 (33.5%) | 70 (35%) | 1 | – | 1 | – |
|
| 236 |
+
| CG | 27 (13.5%) | 33 (16.5%) | 1.12 (0.61–2.96) | 0.163 | 1.37 (0.64–2.01) | 0.831 |
|
| 237 |
+
| GG | 106 (53%) | 97 (48.5%) | 4.69 (2.51–25.01) | 0.004 | 3.05 (2.76–20.08) | 0.006 |
|
| 238 |
+
| Allele C | 86 (43%) | 75(37.5%) | 1 | – | 1 | – |
|
| 239 |
+
| Allele G | 114(57%) | 125(62.5%) | 1.5 (0.60–3.76) | 0.431 | 1.5 (0.53–4.34) | 0.122 |
|
| 240 |
+
| rs312481/ CACNA1D | | | | | | |
|
| 241 |
+
| AA | 117(58.5%) | 129(64.5%) | 1 | – | 1 | – |
|
| 242 |
+
| AG | 26(13%) | 36(18%) | 1.3 (0.57–4.01) | 0.155 | 1.5 (0.71–4.96) | 0.093 |
|
| 243 |
+
| GG | 57(28.5%) | 35(17.5%) | 2.91(1.34–4.97) | 0.021 | 2.01 (1.12–5.01) | 0.024 |
|
| 244 |
+
| Allele A | 98(49%) | 105(52.5%) | | | | |
|
| 245 |
+
| Allele C | 102(51%) | 95(47.5%) | 0.32 (0.08–1.31) | 0.282 | 0.92 (0.42–1.44) | 0.227 |
|
| 246 |
+
| rs3774425/ CACNA1D | | | | | | |
|
| 247 |
+
| GG | 91(45.5%) | 89(44.5%) | 1 | – | 1 | – |
|
| 248 |
+
| AG | 44(22%) | 38(19%) | 2.4 (0.95–6.10) | 0.341 | 2.9 (1.96–7.08) | 0.812 |
|
| 249 |
+
| AA | 65(32.5%) | 73(36.5%) | 0.8 (0.31–1.81) | 0.213 | 0.7 (0.37–1.99) | 0.191 |
|
| 250 |
+
| Allele G | 102(51%) | 110(55%) | 1 | – | | – |
|
| 251 |
+
| Allele A | 98(49%) | 90(45%) | 0.9 (1.10–3.05) | 0.086 | 1.4 (0.52–2.13) | 0.761 |
|
| 252 |
+
| rs3774426/ CACNA1D | | | | | | |
|
| 253 |
+
| CC | 132(66%) | 140(70%) | 1 | – | 1 | – |
|
| 254 |
+
| CT | 22(11%) | 16(8.0%) | 0.3 (0.36–2.44) | 0.113 | 0.5 (0.33–2.74) | 0.101 |
|
| 255 |
+
| TT | 46(23%) | 44(22%) | 0.29 (0.19–1.92) | 0.314 | 0.82 (0.29–1.33) | 0.411 |
|
| 256 |
+
| Allele C | 135(67.5%) | 141(70.5%) | 1 | – | 1 | – |
|
| 257 |
+
| Allele T | 65(32.5%) | 59(29.5%) | 0.63 (0.16–2.06) | 0.112 | 0.57 (0.34–1.43) | 0.081 |
|
| 258 |
+
| rs4291/ACE | | | | | | |
|
| 259 |
+
| AA | 72(36%) | 65(32.5%) | 1 | – | 1 | – |
|
| 260 |
+
| TA | 82(41%) | 102(51%) | 3.27 (1.15–5.72) | 0.002 | 2.99 (1.17–5.81) | 0.005 |
|
| 261 |
+
| TT | 46(23%) | 33(16.5%) | 0.93 (0.46–1.09) | 0.272 | 0.35 (0.25–1.46) | 0.281 |
|
| 262 |
+
| Allele A | 122(61%) | 114(57%) | 1 | – | 1 | – |
|
| 263 |
+
| Allele T | 78(39%) | 86(43%) | 1.12 (0.73–2.41) | 0.601 | 1.02 (0.72–4.32) | 0.420 |
|
| 264 |
+
| rs1042713/ ADBR2 | | | | | | |
|
| 265 |
+
| GG | 88(44%) | 79(39.5%) | 1 | – | 1 | – |
|
| 266 |
+
| GA | 97(48.5%) | 91(45.5%) | 1.58 (0.91–4.01) | 0.514 | 1.26 (0.66–2.24) | 0.332 |
|
| 267 |
+
| AA | 15(7.5%) | 30(15%) | 1.44 (0.36–6.14 | 0.726 | 0.83 (0.36–3.91) | 0.644 |
|
| 268 |
+
| Allele G | 56(28%) | 76(38%) | 1 | – | 1 | – |
|
| 269 |
+
| Allele A | 144(72%) | 124(62%) | 1.71 (0.79–3.11) | 0.862 | 1.01 (0.27–3.42) | 0.566 |
|
| 270 |
+
| rs10494366/ NOS1AP | | | | | | |
|
| 271 |
+
| TT | 66(33%) | 70(5%) | 1 | – | 1 | – |
|
| 272 |
+
| GT | 102(51%) | 114(57%) | 1.55 (1.02–5.01) | 0.661 | 0.99 (0.52–4.28) | 0.472 |
|
| 273 |
+
| GG | 32(16%) | 16(8.0%) | 1.63 (0.250–3.44) | 0.540 | 1.24 (0.26–4.36) | 0.391 |
|
| 274 |
+
| T | 101(50.5%) | 110(55%) | 1 | – | 1 | – |
|
| 275 |
+
| G | 99(49.5%) | 90(45%) | 1.29 (0.55–2.72) | 0.326 | 1.09 (0.58–2.95) | 0.227 |
|
| 276 |
+
## Discussion
|
| 277 |
+
|
| 278 |
+
Amlodipine (a calcium channel blocker) is commonly prescribed for hypertension treatment. However, the outcomes of amlodipine treatment vary significantly from person to person, primarily due to genetic differences.[13](#cit0013)^13^13^,^,[30](#cit0030)^30^30 Considering genetic factors while prescribing anti-hypertensive medications would lead to the concept of personalized medicine that would help to address the rising incidence of hypertension. Pharmacogenes associated with amlodipine responses have been understudied in the Pakistani population. Owing to the high prevalence of HTN in Pakistan and the lack of pharmacogenomic data regarding anti-hypertensive drugs, the present study is carefully designed to examine the co-relation between ten selected genetic biomarkers/variants and blood pressure response to amlodipine therapy. Among the studies SNPs (n=10), seven SNPs (rs2239050, rs312481, rs3774425, rs3774426, rs4291, rs1042713, and rs10494366) were detected or reported or expressed in the Pashtun ethnic population whereas SNP rs1799752, rs1042713, and rs2238032 were not expressed within the study population. This may be attributed to the limited number of participants enrolled in the study. Studies with larger sample sizes tend to exhibit higher statistical power, which enables the detection of rare and common variants. The identified SNPs within the study population adhered to the Hardy–Weinberg equilibrium (HWE).
|
| 279 |
+
|
| 280 |
+
We reported a significant correlation between amlodipine response and SNP rs2239050 (located in the intron region of CACNA1C). Troy et al reported same association (SNP rs2239050×amlodipine treatment response) previously in Caucasian subjects.[18](#cit0018)^18^18 Our study findings suggest that individuals with GG (homozygous) genotype had significantly better response (un-adjusted odd-ratio (95% CI) = 4.69 (2.51–25.01 and crude P=0.004) to amlodipine treatment compared to the individuals with CC or CG genotypes. When adjusted for confounding factors/covariates, no observable changes (adjusted odds ratio (95% CI) = 3.05 (2.76–20.08), P=0.006) were observed in the degree, level, or magnitude of the association. No considerable changes in unadjusted, adjusted Odd ratios (ORs), confidence intervals (Cl), and P-values suggest that the study subjects (individuals with uncontrolled hypertension and individuals with controlled hypertension) are closely matched in terms of age, sex, and other related confounding factors. CACNA1C encodes a protein that forms the alpha-1C subunit of the L-type voltage-gated calcium channel, which plays an integral role in regulating calcium influx into cardiac and vascular smooth muscle cells.[31](#cit0031)^31^31^,^,[32](#cit0032)^32^32 Amlodipine (a calcium channel blocker), acts on this channel to lower blood pressure.[33](#cit0033)^33^33^,^,[34](#cit0034)^34^34 Studies have identified connections between CACNA1C gene variants and amlodipine response.[18](#cit0018)^18^18^,^,[35](#cit0035)^35^35 These findings imply that CACNA1C gene variants may affect an individual’s reaction to amlodipine, potentially influencing its effectiveness and side-effect profile. Thus, CACNA1C holds promise as a pharmacogenetic biomarker for tailored approaches to hypertension treatment.
|
| 281 |
+
|
| 282 |
+
Regarding rs312481/CACNA1D, the initial crude logistic regression analysis indicated a positive pharmacogenetic association between rs312481 and amlodipine treatment response. Individuals with the GG genotype demonstrated a significant independent reduction in blood pressure (unadjusted odds ratio (95% CI)=2.91(1.34–4.97), p=0.021). After adjusting for confounding variables, the association between SNP rs312481 and blood pressure regulation by amlodipine remained consistent (adjusted odds ratio (95% CI)= 2.01 (1.12–5.01), P=0.024). This suggests that the covariates and confounding factors have neither an additive nor opposing effect on the observed association. However, further research is needed to gain a comprehensive understanding of the relationship between rs312481 and amlodipine response. The SNP rs312481 is located within the CACNA1D gene, which encodes the alpha-1D subunit of the L-type voltage-gated calcium channel in humans.[36](#cit0036)^36^36^,^,[37](#cit0037)^37^37 Amlodipine, a calcium channel blocker, interacts with this subunit as a part of its mechanism of action. The rs312481 polymorphism involves a C to T substitution and is located in intron 3 of CACNA1D. Studies have identified the rs312481G>A polymorphism as being associated with variations in response to amlodipine, suggesting potential implications for pharmacokinetics or pharmacodynamics that could explain these response differences.[12](#cit0012)^12^12^,^,[16](#cit0016)^16^16
|
| 283 |
+
|
| 284 |
+
In the case of SNP rs4291/ACE, the TA genotype (rs4291) was independently and potentially linked to uncontrolled hypertension (unadjusted odds ratio(95% CI)= 3.27 (1.15–5.72, P=0.002) during amlodipine treatment. When adjusted for confounding factors (like age, gender and diet/drug compliance) the magnitude and direction of this association/correlation remained consistent (adjusted odd ratio (95% CI) = 2.99 (1.17–5.81), P=0.005). Individuals carrying the TA genotype (rs4291) showed a decreased response to amlodipine compared with individuals with the AA genotype. A study conducted in South African Adult population with Hypertension documented similar findings/results as we noticed in the present study.[35](#cit0035)^35^35 The ACE gene and renin-angiotensin-aldosterone system (RAAS) are intricately linked components of body regulatory mechanisms, particularly in the maintenance of blood pressure and electrolyte balance. ACE encodes the angiotensin-converting enzyme (ACE), which plays a pivotal role in the RAAS pathway. This pathway regulates diverse physiological processes; for example, it regulates the BP, inflow, and outflow of electrolytes, and maintains fluid homeostasis.[38](#cit0038)^38^38^,^,[39](#cit0039)^39^39 Overall, ACE is an important biomarker for hypertension and related cardiovascular diseases.[23](#cit0023)^23^23^,^,[40](#cit0040)^40^40^,^,[41](#cit0041)^41^41 Pharmacogenomic study of this ACE gene will help devise anti-hypertensive therapies according to the genetic makeup of the individual.
|
| 285 |
+
|
| 286 |
+
Furthermore, the present study finds no association between rs3774425/CACNA1D, rs3774426/CACNA1D, rs1042713/ADBR2, and rs10494366/NOS1AP and the blood pressure response to amlodipine. These nucleotide polymorphisms were investigated to determine their potential roles in the therapeutic outcomes of amlodipine. Despite the biological plausibility that variations in these genes may affect calcium channel function (CACNA1D), beta-adrenergic receptor activity (ADBR2), and nitric oxide signaling (NOS1AP), our findings did not demonstrate any significant correlation. This lack of association suggests that these genetic variants do not modify the efficacy of amlodipine in lowering blood pressure in hypertensive individuals. Our results indicate that the rs3774425, rs3774426, rs1042713, and rs10494366 genotypes are not reliable biomarkers for predicting patient responses to amlodipine therapy. This insight is crucial for clinicians and researchers focusing on personalized medicine and pharmacogenomics, as it highlights the need to explore other genetic factors or mechanisms that may contribute to variability in drug responses, thereby improving the precision and effectiveness of hypertension treatment strategies.
|
| 287 |
+
|
| 288 |
+
## Conclusion
|
| 289 |
+
|
| 290 |
+
In this Pharmacogenomic study, we examined the influence of various alleles and genotypes on blood pressure regulation by amlodipine using unadjusted and adjusted logistic regression models. Our findings revealed a potential postive association between the SNP rs2239050/CACNA1C and the efficacy of amlodipine in lowering blood pressure. Specifically, participants with the GG genotype demonstrated good response (P=0.004) compared with those carrying the CC or CG genotypes.
|
| 291 |
+
|
| 292 |
+
Similarly, the SNP rs312481/CACNA1D showed a positive pharmacogenetic association with the amlodipine response. Individuals with the GG genotype exhibited a lowering in blood pressure (P=0.021), and this association persisted after controlling for potential confounding variables, suggesting a stable genetic influence on drug response.
|
| 293 |
+
|
| 294 |
+
Conversely, for SNP rs429/ACE, individuals with the TA genotype were less likely to achieve blood pressure control than those with the AA genotype, highlighting a potential negative impact on treatment efficacy. In contrast, the SNPs rs3774425/CACNA1D, rs3774426/CACNA1D, rs1042713/ADBR2, and rs10494366/NOS1AP did not significantly affect the blood pressure response to amlodipine.
|
| 295 |
+
|
| 296 |
+
These findings underscore the importance of considering genetic biomarkers to predict patient response to amlodipine treatment. The observed associations suggested that genetic testing for these SNPs could potentially guide personalized hypertension treatment strategies and enhance therapeutic outcomes. Further research is warranted to validate these results and uncover the underlying pathways/mechanisms involved in these associations.
|
| 297 |
+
|
| 298 |
+
## Study Limitation
|
| 299 |
+
|
| 300 |
+
The study limitation in includes small sample size and targeting only Pashtun ethnic population. We adjusted the results for major confounding factors however other factors like lifestyle and non-hypertensive may have affect on the study outcomes. Future studies with larger cohorts and exploration of gene-environment interactions are needed to provide deeper insights into amlodipine pharmacogenomics.
|
| 301 |
+
|
| 302 |
+
## Acknowledgments
|
| 303 |
+
|
| 304 |
+
The authors would like to thank all volunteers who agreed to participate in this study. Authors would like to express their appreciation to Researchers Supporting Project Number (RSPD2024R885) at King Saud University Riyadh Saudi Arabia for supporting this research. We are also grateful to Prof. Dr. Johar Ali for the technical support in conducting this study.
|
| 305 |
+
|
| 306 |
+
## Funding Statement
|
| 307 |
+
|
| 308 |
+
The research was funded by the Researchers Supporting Project number (RSPD2024R885), King Saud University, Riyadh, Saudi Arabia.
|
| 309 |
+
|
| 310 |
+
## Data Sharing Statement
|
| 311 |
+
|
| 312 |
+
All the necessary data and information are provided in the manuscript. The corresponding author can be contacted for any additional data or information related to this article.
|
| 313 |
+
|
| 314 |
+
## Ethical Statement
|
| 315 |
+
|
| 316 |
+
This study was approved by the members of institutional research board District Head Quarter Hospital (approval number 247/Cl.Phar, dated 17/10/2021). All experiments and procedures were performed in accordance with the ethical guidelines established by the Declaration of Helsinki (1975).
|
| 317 |
+
|
| 318 |
+
## Informed Consent Statement
|
| 319 |
+
|
| 320 |
+
Informed consent was obtained from all the participants.
|
| 321 |
+
|
| 322 |
+
## Disclosure
|
| 323 |
+
|
| 324 |
+
The authors report no conflicts of interest in this work.
|
| 325 |
+
|
| 326 |
+
## Associated Data
|
| 327 |
+
|
| 328 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 329 |
+
|
| 330 |
+
### Data Availability Statement
|
| 331 |
+
|
| 332 |
+
All the necessary data and information are provided in the manuscript. The corresponding author can be contacted for any additional data or information related to this article.
|
| 333 |
+
|
| 334 |
+
### Data Availability Statement
|
| 335 |
+
|
| 336 |
+
All the necessary data and information are provided in the manuscript. The corresponding author can be contacted for any additional data or information related to this article.
|
| 337 |
+
|
| 338 |
+
## References
|
| 339 |
+
|
| 340 |
+
1. Kario K, Okura A, Hoshide S, Mogi M. The WHO Global report 2023 on hypertension warning the emerging hypertension burden in globe and its treatment strategy. Hypertens Res. 2024;47(5):1099–1102. doi: 10.1038/s41440-024-01622-w [DOI](https://doi.org/10.1038/s41440-024-01622-w) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38443614/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Hypertens%20Res&title=The%20WHO%20Global%20report%202023%20on%20hypertension%20warning%20the%20emerging%20hypertension%20burden%20in%20globe%20and%20its%20treatment%20strategy&author=K%20Kario&author=A%20Okura&author=S%20Hoshide&author=M%20Mogi&volume=47&issue=5&publication_year=2024&pages=1099-1102&pmid=38443614&doi=10.1038/s41440-024-01622-w&)
|
| 341 |
+
|
| 342 |
+
2. Mills KT, Stefanescu A, He J. The global epidemiology of hypertension. Nat Rev Nephrol. 2020;16(4):223–237. doi: 10.1038/s41581-019-0244-2 [DOI](https://doi.org/10.1038/s41581-019-0244-2) | [PMC free article](/articles/PMC7998524/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32024986/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat%20Rev%20Nephrol&title=The%20global%20epidemiology%20of%20hypertension&author=KT%20Mills&author=A%20Stefanescu&author=J%20He&volume=16&issue=4&publication_year=2020&pages=223-237&pmid=32024986&doi=10.1038/s41581-019-0244-2&)
|
| 343 |
+
|
| 344 |
+
3. Schutte AE, Srinivasapura Venkateshmurthy N, Mohan S, Prabhakaran D. Hypertension in Low- and Middle-Income Countries. Circ Res. 2021;128(7):808–826. doi: 10.1161/CIRCRESAHA.120.318729 [DOI](https://doi.org/10.1161/CIRCRESAHA.120.318729) | [PMC free article](/articles/PMC8091106/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33793340/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circ%20Res&title=Hypertension%20in%20Low-%20and%20Middle-Income%20Countries&author=AE%20Schutte&author=N%20Srinivasapura%20Venkateshmurthy&author=S%20Mohan&author=D%20Prabhakaran&volume=128&issue=7&publication_year=2021&pages=808-826&pmid=33793340&doi=10.1161/CIRCRESAHA.120.318729&)
|
| 345 |
+
|
| 346 |
+
4. World Health Organization. Global Report on Hypertension: The Race Against a Silent Killer. Geneva, Switzerland: World Health Organization; 2023:1–276. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Global%20Report%20on%20Hypertension:%20The%20Race%20Against%20a%20Silent%20Killer&publication_year=2023&)
|
| 347 |
+
|
| 348 |
+
5. NCD Risk Factor Collaboration (NCD-RisC). Worldwide trends in hypertension prevalence and progress in treatment and control from 1990 to 2019: a pooled analysis of 1201 population-representative studies with 104 million participants. Lancet. 2021;398(10304):957–980. doi: 10.1016/S0140-6736(21)01330-1 [DOI](https://doi.org/10.1016/S0140-6736(21)01330-1) | [PMC free article](/articles/PMC8446938/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34450083/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Worldwide%20trends%20in%20hypertension%20prevalence%20and%20progress%20in%20treatment%20and%20control%20from%201990%20to%202019:%20a%20pooled%20analysis%20of%201201%20population-representative%20studies%20with%20104%20million%20participants&volume=398&issue=10304&publication_year=2021&pages=957-980&pmid=34450083&doi=10.1016/S0140-6736(21)01330-1&)
|
| 349 |
+
|
| 350 |
+
6. Elahi A, Ali AA, Khan AH, et al. Challenges of managing hypertension in Pakistan - a review. Clin Hypertens. 2023;29(1):17. doi: 10.1186/s40885-023-00245-6 [DOI](https://doi.org/10.1186/s40885-023-00245-6) | [PMC free article](/articles/PMC10268336/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37316940/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Hypertens&title=Challenges%20of%20managing%20hypertension%20in%20Pakistan%20-%20a%20review&author=A%20Elahi&author=AA%20Ali&author=AH%20Khan&volume=29&issue=1&publication_year=2023&pages=17&pmid=37316940&doi=10.1186/s40885-023-00245-6&)
|
| 351 |
+
|
| 352 |
+
7. Shah WA, Jan A, Khan MA, et al. Association between Aldosterone Synthase (CYP11B2) gene polymorphism and hypertension in Pashtun ethnic population of Khyber Pakhtunkwha, Pakistan. Genes. 2023;14(6):1184. doi: 10.3390/genes14061184 [DOI](https://doi.org/10.3390/genes14061184) | [PMC free article](/articles/PMC10297898/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37372364/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genes&title=Association%20between%20Aldosterone%20Synthase%20(CYP11B2)%20gene%20polymorphism%20and%20hypertension%20in%20Pashtun%20ethnic%20population%20of%20Khyber%20Pakhtunkwha,%20Pakistan&author=WA%20Shah&author=A%20Jan&author=MA%20Khan&volume=14&issue=6&publication_year=2023&pages=1184&pmid=37372364&doi=10.3390/genes14061184&)
|
| 353 |
+
|
| 354 |
+
8. Oliveira-Paula GH, Pereira SC, Tanus-Santos JE, Lacchini R. Pharmacogenomics And Hypertension: current Insights. Pharmgenomics Pers Med. 2019;12:341–359. doi: 10.2147/PGPM.S230201 [DOI](https://doi.org/10.2147/PGPM.S230201) | [PMC free article](/articles/PMC6878918/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31819590/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmgenomics%20Pers%20Med&title=Pharmacogenomics%20And%20Hypertension:%20current%20Insights&author=GH%20Oliveira-Paula&author=SC%20Pereira&author=JE%20Tanus-Santos&author=R%20Lacchini&volume=12&publication_year=2019&pages=341-359&pmid=31819590&doi=10.2147/PGPM.S230201&)
|
| 355 |
+
|
| 356 |
+
9. Jan A, Saeed M, Mothana RA, et al. Association of CYP2C9*2 allele with sulphonylurea-induced hypoglycaemia in type 2 diabetes mellitus patients: a pharmacogenetic study in Pakistani pashtun population. Biomedicines. 2023;11(8):2282. doi: 10.3390/biomedicines11082282 [DOI](https://doi.org/10.3390/biomedicines11082282) | [PMC free article](/articles/PMC10452755/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37626778/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Biomedicines&title=Association%20of%20CYP2C9*2%20allele%20with%20sulphonylurea-induced%20hypoglycaemia%20in%20type%202%20diabetes%20mellitus%20patients:%20a%20pharmacogenetic%20study%20in%20Pakistani%20pashtun%20population&author=A%20Jan&author=M%20Saeed&author=RA%20Mothana&volume=11&issue=8&publication_year=2023&pages=2282&pmid=37626778&doi=10.3390/biomedicines11082282&)
|
| 357 |
+
|
| 358 |
+
10. Laurent S. Antihypertensive drugs. Pharmacol Res. 2017;124:116–125. doi: 10.1016/j.phrs.2017.07.026 [DOI](https://doi.org/10.1016/j.phrs.2017.07.026) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28780421/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacol%20Res&title=Antihypertensive%20drugs&author=S%20Laurent&volume=124&publication_year=2017&pages=116-125&pmid=28780421&doi=10.1016/j.phrs.2017.07.026&)
|
| 359 |
+
|
| 360 |
+
11. James PA, Oparil S, Carter BL, et al. 2014 evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8). JAMA. 2014;311(5):507–520. doi: 10.1001/jama.2013.284427 [DOI](https://doi.org/10.1001/jama.2013.284427) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24352797/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=JAMA&title=2014%20evidence-based%20guideline%20for%20the%20management%20of%20high%20blood%20pressure%20in%20adults:%20report%20from%20the%20panel%20members%20appointed%20to%20the%20Eighth%20Joint%20National%20Committee%20(JNC%208)&author=PA%20James&author=S%20Oparil&author=BL%20Carter&volume=311&issue=5&publication_year=2014&pages=507-520&pmid=24352797&doi=10.1001/jama.2013.284427&)
|
| 361 |
+
|
| 362 |
+
12. Kamide K, Yang J, Matayoshi T, et al. Genetic polymorphisms of l-type calcium channel.ALPHA.1C and.ALPHA.1D subunit genes are associated with sensitivity to the antihypertensive effects of l-type dihydropyridine calcium-channel blockers. Circ J. 2009;73(4):732–740. doi: 10.1253/circj.cj-08-0761 [DOI](https://doi.org/10.1253/circj.cj-08-0761) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19225208/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circ%20J&title=Genetic%20polymorphisms%20of%20l-type%20calcium%20channel.ALPHA.1C%20and.ALPHA.1D%20subunit%20genes%20are%20associated%20with%20sensitivity%20to%20the%20antihypertensive%20effects%20of%20l-type%20dihydropyridine%20calcium-channel%20blockers&author=K%20Kamide&author=J%20Yang&author=T%20Matayoshi&volume=73&issue=4&publication_year=2009&pages=732-740&pmid=19225208&doi=10.1253/circj.cj-08-0761&)
|
| 363 |
+
|
| 364 |
+
13. Fares H, DiNicolantonio JJ, O’Keefe JH, Lavie CJ. Amlodipine in hypertension: a first-line agent with efficacy for improving blood pressure and patient outcomes. Open Heart. 2016;3(2):e000473. doi: 10.1136/openhrt-2016-000473 [DOI](https://doi.org/10.1136/openhrt-2016-000473) | [PMC free article](/articles/PMC5051471/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27752334/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Open%20Heart&title=Amlodipine%20in%20hypertension:%20a%20first-line%20agent%20with%20efficacy%20for%20improving%20blood%20pressure%20and%20patient%20outcomes&author=H%20Fares&author=JJ%20DiNicolantonio&author=JH%20O%E2%80%99Keefe&author=CJ%20Lavie&volume=3&issue=2&publication_year=2016&pages=e000473&pmid=27752334&doi=10.1136/openhrt-2016-000473&)
|
| 365 |
+
|
| 366 |
+
14. Guo C, Pei QI, Tan H, Huang Z, Yuan H, Yang G. Effects of genetic factors on the pharmacokinetics and pharmacodynamics of amlodipine in primary hypertensive patients. Biomed Rep. 2015;3(2):195–200. doi: 10.3892/br.2014.395 [DOI](https://doi.org/10.3892/br.2014.395) | [PMC free article](/articles/PMC4448016/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26075072/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Biomed%20Rep&title=Effects%20of%20genetic%20factors%20on%20the%20pharmacokinetics%20and%20pharmacodynamics%20of%20amlodipine%20in%20primary%20hypertensive%20patients&author=C%20Guo&author=QI%20Pei&author=H%20Tan&author=Z%20Huang&author=H%20Yuan&volume=3&issue=2&publication_year=2015&pages=195-200&pmid=26075072&doi=10.3892/br.2014.395&)
|
| 367 |
+
|
| 368 |
+
15. Lynch AI, Irvin MR, Boerwinkle E, et al. RYR3 gene polymorphisms and cardiovascular disease outcomes in the context of antihypertensive treatment. Pharmacogenomics J. 2013;13(4):330–334. doi: 10.1038/tpj.2012.22 [DOI](https://doi.org/10.1038/tpj.2012.22) | [PMC free article](/articles/PMC3435442/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22664477/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J&title=RYR3%20gene%20polymorphisms%20and%20cardiovascular%20disease%20outcomes%20in%20the%20context%20of%20antihypertensive%20treatment&author=AI%20Lynch&author=MR%20Irvin&author=E%20Boerwinkle&volume=13&issue=4&publication_year=2013&pages=330-334&pmid=22664477&doi=10.1038/tpj.2012.22&)
|
| 369 |
+
|
| 370 |
+
16. Johnson R, Dludla P, Mabhida S, Benjeddou M, Louw J, February F. Pharmacogenomics of amlodipine and hydrochlorothiazide therapy and the quest for improved control of hypertension: a mini review. Heart Fail Rev. 2019;24(3):343–357. doi: 10.1007/s10741-018-09765-y [DOI](https://doi.org/10.1007/s10741-018-09765-y) | [PMC free article](/articles/PMC6476827/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30645721/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Heart%20Fail%20Rev&title=Pharmacogenomics%20of%20amlodipine%20and%20hydrochlorothiazide%20therapy%20and%20the%20quest%20for%20improved%20control%20of%20hypertension:%20a%20mini%20review&author=R%20Johnson&author=P%20Dludla&author=S%20Mabhida&author=M%20Benjeddou&author=J%20Louw&volume=24&issue=3&publication_year=2019&pages=343-357&pmid=30645721&doi=10.1007/s10741-018-09765-y&)
|
| 371 |
+
|
| 372 |
+
17. Türkmen D, Masoli JAH, Delgado J, et al. Calcium-channel blockers: clinical outcome associations with reported pharmacogenetics variants in 32 000 patients. Br J Clin Pharmacol. 2023;89(2):853–864. doi: 10.1111/bcp.15541 [DOI](https://doi.org/10.1111/bcp.15541) | [PMC free article](/articles/PMC10091789/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36134646/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Calcium-channel%20blockers:%20clinical%20outcome%20associations%20with%20reported%20pharmacogenetics%20variants%20in%2032%E2%80%89000%20patients&author=D%20T%C3%BCrkmen&author=JAH%20Masoli&author=J%20Delgado&volume=89&issue=2&publication_year=2023&pages=853-864&pmid=36134646&doi=10.1111/bcp.15541&)
|
| 373 |
+
|
| 374 |
+
18. Bremer T, Man A, Kask K, Diamond C. CACNA1C polymorphisms are associated with the efficacy of calcium channel blockers in the treatment of hypertension. Pharmacogenomics. 2006;7(3):271–279. doi: 10.2217/14622416.7.3.271 [DOI](https://doi.org/10.2217/14622416.7.3.271) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16610939/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=CACNA1C%20polymorphisms%20are%20associated%20with%20the%20efficacy%20of%20calcium%20channel%20blockers%20in%20the%20treatment%20of%20hypertension&author=T%20Bremer&author=A%20Man&author=K%20Kask&author=C%20Diamond&volume=7&issue=3&publication_year=2006&pages=271-279&pmid=16610939&doi=10.2217/14622416.7.3.271&)
|
| 375 |
+
|
| 376 |
+
19. PharmGKB. Amlodipine/clinical annotations. Available from: https://www.pharmgkb.org/chemical/PA448388/clinicalAnnotation. Accessed October 25, 2024. [https://www.pharmgkb.org/chemical/PA448388/clinicalAnnotation](https://www.pharmgkb.org/chemical/PA448388/clinicalAnnotation)
|
| 377 |
+
|
| 378 |
+
20. Kim K-A, Park P-W, Park J-Y. Effect of ABCB1 (MDR1) haplotypes derived from G2677T/C3435T on the pharmacokinetics of amlodipine in healthy subjects. Br J Clin Pharmacol. 2007;63(1):53–58. doi: 10.1111/j.1365-2125.2006.02733.x [DOI](https://doi.org/10.1111/j.1365-2125.2006.02733.x) | [PMC free article](/articles/PMC2000718/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16869811/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Effect%20of%20ABCB1%20(MDR1)%20haplotypes%20derived%20from%20G2677T/C3435T%20on%20the%20pharmacokinetics%20of%20amlodipine%20in%20healthy%20subjects&author=K-A%20Kim&author=P-W%20Park&author=J-Y%20Park&volume=63&issue=1&publication_year=2007&pages=53-58&pmid=16869811&doi=10.1111/j.1365-2125.2006.02733.x&)
|
| 379 |
+
|
| 380 |
+
21. Kim KA, Park PW, Lee OJ, et al. Effect of CYP3A53 genotype on the pharmacokinetics and pharmacodynamics of amlodipine in healthy Korean subjects. Clin Pharmacol Ther. 2006;80(6):646–656. doi: 10.1016/j.clpt.2006.09.009 [DOI](https://doi.org/10.1016/j.clpt.2006.09.009) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17178265/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Effect%20of%20CYP3A53%20genotype%20on%20the%20pharmacokinetics%20and%20pharmacodynamics%20of%20amlodipine%20in%20healthy%20Korean%20subjects&author=KA%20Kim&author=PW%20Park&author=OJ%20Lee&volume=80&issue=6&publication_year=2006&pages=646-656&pmid=17178265&doi=10.1016/j.clpt.2006.09.009&)
|
| 381 |
+
|
| 382 |
+
22. Martínez-Rodríguez N, Posadas-Romero C, Villarreal-Molina T, et al. Single nucleotide polymorphisms of the angiotensin-converting enzyme (ACE) gene are associated with essential hypertension and increased ACE enzyme levels in Mexican individuals. PLoS One. 2013;8(5):e65700. doi: 10.1371/journal.pone.0065700 [DOI](https://doi.org/10.1371/journal.pone.0065700) | [PMC free article](/articles/PMC3669228/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23741507/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=PLoS%20One&title=Single%20nucleotide%20polymorphisms%20of%20the%20angiotensin-converting%20enzyme%20(ACE)%20gene%20are%20associated%20with%20essential%20hypertension%20and%20increased%20ACE%20enzyme%20levels%20in%20Mexican%20individuals&author=N%20Mart%C3%ADnez-Rodr%C3%ADguez&author=C%20Posadas-Romero&author=T%20Villarreal-Molina&volume=8&issue=5&publication_year=2013&pages=e65700&pmid=23741507&doi=10.1371/journal.pone.0065700&)
|
| 383 |
+
|
| 384 |
+
23. Krishnan R, Sekar D, Karunanithy S, Subramanium S. Association of angiotensin converting enzyme gene insertion/deletion polymorphism with essential hypertension in south Indian population. Genes Dis. 2016;3(2):159–163. doi: 10.1016/j.gendis.2016.03.001 [DOI](https://doi.org/10.1016/j.gendis.2016.03.001) | [PMC free article](/articles/PMC6146176/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30258884/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genes%20Dis&title=Association%20of%20angiotensin%20converting%20enzyme%20gene%20insertion/deletion%20polymorphism%20with%20essential%20hypertension%20in%20south%20Indian%20population&author=R%20Krishnan&author=D%20Sekar&author=S%20Karunanithy&author=S%20Subramanium&volume=3&issue=2&publication_year=2016&pages=159-163&pmid=30258884&doi=10.1016/j.gendis.2016.03.001&)
|
| 385 |
+
|
| 386 |
+
24. Do AN, Irvin MR, Lynch AI, et al. The effects of angiotensinogen gene polymorphisms on cardiovascular disease outcomes during antihypertensive treatment in the GenHAT study. Front Pharmacol. 2014;5:210. doi: 10.3389/fphar.2014.00210 [DOI](https://doi.org/10.3389/fphar.2014.00210) | [PMC free article](/articles/PMC4165277/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25278896/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front%20Pharmacol&title=The%20effects%20of%20angiotensinogen%20gene%20polymorphisms%20on%20cardiovascular%20disease%20outcomes%20during%20antihypertensive%20treatment%20in%20the%20GenHAT%20study&author=AN%20Do&author=MR%20Irvin&author=AI%20Lynch&volume=5&publication_year=2014&pages=210&pmid=25278896&doi=10.3389/fphar.2014.00210&)
|
| 387 |
+
|
| 388 |
+
25. Becker ML, Visser LE, Newton‐Cheh C, et al. A common NOS1AP genetic polymorphism is associated with increased cardiovascular mortality in users of dihydropyridine calcium channel blockers. Br J Clin Pharmacol. 2009;67(1):61–67. doi: 10.1111/j.1365-2125.2008.03325.x [DOI](https://doi.org/10.1111/j.1365-2125.2008.03325.x) | [PMC free article](/articles/PMC2668085/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19076153/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=A%20common%20NOS1AP%20genetic%20polymorphism%20is%20associated%20with%20increased%20cardiovascular%20mortality%20in%20users%20of%20dihydropyridine%20calcium%20channel%20blockers&author=ML%20Becker&author=LE%20Visser&author=C%20Newton%E2%80%90Cheh&volume=67&issue=1&publication_year=2009&pages=61-67&pmid=19076153&doi=10.1111/j.1365-2125.2008.03325.x&)
|
| 389 |
+
|
| 390 |
+
26. Altoum SM, Al-Mahayri ZN, Ali BR. Antihypertensives associated adverse events: a review of mechanisms and pharmacogenomic biomarkers available evidence in multi-ethnic populations. Front Pharmacol. 2023;14:1286494. doi: 10.3389/fphar.2023.1286494 [DOI](https://doi.org/10.3389/fphar.2023.1286494) | [PMC free article](/articles/PMC10722273/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38108069/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front%20Pharmacol&title=Antihypertensives%20associated%20adverse%20events:%20a%20review%20of%20mechanisms%20and%20pharmacogenomic%20biomarkers%20available%20evidence%20in%20multi-ethnic%20populations&author=SM%20Altoum&author=ZN%20Al-Mahayri&author=BR%20Ali&volume=14&publication_year=2023&pages=1286494&pmid=38108069&doi=10.3389/fphar.2023.1286494&)
|
| 391 |
+
|
| 392 |
+
27. Kulminski AM, Culminskaya IV, Ukraintseva SV, et al. Polymorphisms in the ACE and ADRB2 genes and risks of aging-associated phenotypes: the case of myocardial infarction. Rejuvenation Res. 2010;13(1):13–21. doi: 10.1089/rej.2009.0905 [DOI](https://doi.org/10.1089/rej.2009.0905) | [PMC free article](/articles/PMC2944842/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20230274/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Rejuvenation%20Res&title=Polymorphisms%20in%20the%20ACE%20and%20ADRB2%20genes%20and%20risks%20of%20aging-associated%20phenotypes:%20the%20case%20of%20myocardial%20infarction&author=AM%20Kulminski&author=IV%20Culminskaya&author=SV%20Ukraintseva&volume=13&issue=1&publication_year=2010&pages=13-21&pmid=20230274&doi=10.1089/rej.2009.0905&)
|
| 393 |
+
|
| 394 |
+
28. Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenomics knowledge base. Methods Mol Biol. 2013;1015:311–320. doi: 10.1007/978-1-62703-435-7_20 [DOI](https://doi.org/10.1007/978-1-62703-435-7_20) | [PMC free article](/articles/PMC4084821/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23824865/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Methods%20Mol%20Biol&title=PharmGKB:%20the%20pharmacogenomics%20knowledge%20base&author=CF%20Thorn&author=TE%20Klein&author=RB%20Altman&volume=1015&publication_year=2013&pages=311-320&pmid=23824865&doi=10.1007/978-1-62703-435-7_20&)
|
| 395 |
+
|
| 396 |
+
29. Kaplun A, Hogan JD, Schacherer F, et al. PGMD: a comprehensive manually curated pharmacogenomic database. Pharmacogenomics J. 2016;16(2):124–128. doi: 10.1038/tpj.2015.32 [DOI](https://doi.org/10.1038/tpj.2015.32) | [PMC free article](/articles/PMC4819767/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25939485/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J&title=PGMD:%20a%20comprehensive%20manually%20curated%20pharmacogenomic%20database&author=A%20Kaplun&author=JD%20Hogan&author=F%20Schacherer&volume=16&issue=2&publication_year=2016&pages=124-128&pmid=25939485&doi=10.1038/tpj.2015.32&)
|
| 397 |
+
|
| 398 |
+
30. Cooper-DeHoff RM, Johnson JA. Hypertension pharmacogenomics: in search of personalized treatment approaches. Nat Rev Nephrol. 2016;12(2):110–122. doi: 10.1038/nrneph.2015.176 [DOI](https://doi.org/10.1038/nrneph.2015.176) | [PMC free article](/articles/PMC4778736/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26592190/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat%20Rev%20Nephrol&title=Hypertension%20pharmacogenomics:%20in%20search%20of%20personalized%20treatment%20approaches&author=RM%20Cooper-DeHoff&author=JA%20Johnson&volume=12&issue=2&publication_year=2016&pages=110-122&pmid=26592190&doi=10.1038/nrneph.2015.176&)
|
| 399 |
+
|
| 400 |
+
31. Eadon MT, Kanuri SH, Chapman AB. Pharmacogenomic studies of hypertension: paving the way for personalized antihypertensive treatment. Expert Rev Precis Med Drug. Dev. 2018;3(1):33–47. doi: 10.1080/23808993.2018.1420419 [DOI](https://doi.org/10.1080/23808993.2018.1420419) | [PMC free article](/articles/PMC5990020/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29888336/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Dev&title=Pharmacogenomic%20studies%20of%20hypertension:%20paving%20the%20way%20for%20personalized%20antihypertensive%20treatment.%20Expert%20Rev%20Precis%20Med%20Drug&author=MT%20Eadon&author=SH%20Kanuri&author=AB%20Chapman&volume=3&issue=1&publication_year=2018&pages=33-47&pmid=29888336&doi=10.1080/23808993.2018.1420419&)
|
| 401 |
+
|
| 402 |
+
32. Beitelshees AL, Navare H, Wang D, et al. CACNA1C gene polymorphisms, cardiovascular disease outcomes, and treatment response. Circ Cardiovasc Genet. 2009;2(4):362–370. doi: 10.1161/CIRCGENETICS.109.857839 [DOI](https://doi.org/10.1161/CIRCGENETICS.109.857839) | [PMC free article](/articles/PMC2761685/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20031608/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circ%20Cardiovasc%20Genet&title=CACNA1C%20gene%20polymorphisms,%20cardiovascular%20disease%20outcomes,%20and%20treatment%20response&author=AL%20Beitelshees&author=H%20Navare&author=D%20Wang&volume=2&issue=4&publication_year=2009&pages=362-370&pmid=20031608&doi=10.1161/CIRCGENETICS.109.857839&)
|
| 403 |
+
|
| 404 |
+
33. Viola HM, Macdonald WA, Tang H, Hool LC. The L-type Ca(2+) channel as a therapeutic target in heart disease. Curr Med Chem. 2009;16(26):3341–3358. doi: 10.2174/092986709789057671 [DOI](https://doi.org/10.2174/092986709789057671) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19548874/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Curr%20Med%20Chem&title=The%20L-type%20Ca(2+)%20channel%20as%20a%20therapeutic%20target%20in%20heart%20disease&author=HM%20Viola&author=WA%20Macdonald&author=H%20Tang&author=LC%20Hool&volume=16&issue=26&publication_year=2009&pages=3341-3358&pmid=19548874&doi=10.2174/092986709789057671&)
|
| 405 |
+
|
| 406 |
+
34. Tamargo J, Ruilope LM. Investigational calcium channel blockers for the treatment of hypertension. Expert Opin Investig Drugs. 2016;25(11):1295–1309. doi: 10.1080/13543784.2016.1241764 [DOI](https://doi.org/10.1080/13543784.2016.1241764) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27696904/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Expert%20Opin%20Investig%20Drugs&title=Investigational%20calcium%20channel%20blockers%20for%20the%20treatment%20of%20hypertension&author=J%20Tamargo&author=LM%20Ruilope&volume=25&issue=11&publication_year=2016&pages=1295-1309&pmid=27696904&doi=10.1080/13543784.2016.1241764&)
|
| 407 |
+
|
| 408 |
+
35. Masilela C, Adeniyi OV, Benjeddou M. Single nucleotide polymorphisms in amlodipine-associated genes and their correlation with blood pressure control among South African adults with hypertension. Genes (Basel). 2022;13(8):1394. doi: 10.3390/genes13081394 [DOI](https://doi.org/10.3390/genes13081394) | [PMC free article](/articles/PMC9407577/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36011305/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genes%20(Basel)&title=Single%20nucleotide%20polymorphisms%20in%20amlodipine-associated%20genes%20and%20their%20correlation%20with%20blood%20pressure%20control%20among%20South%20African%20adults%20with%20hypertension&author=C%20Masilela&author=OV%20Adeniyi&author=M%20Benjeddou&volume=13&issue=8&publication_year=2022&pages=1394&pmid=36011305&doi=10.3390/genes13081394&)
|
| 409 |
+
|
| 410 |
+
36. Reinbothe TM, Alkayyali S, Ahlqvist E, et al. The human L-type calcium channel Cav1.3 regulates insulin release and polymorphisms in CACNA1D associate with type 2 diabetes. Diabetologia. 2013;56(2):340–349. doi: 10.1007/s00125-012-2758-z [DOI](https://doi.org/10.1007/s00125-012-2758-z) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23229155/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Diabetologia&title=The%20human%20L-type%20calcium%20channel%20Cav1.3%20regulates%20insulin%20release%20and%20polymorphisms%20in%20CACNA1D%20associate%20with%20type%202%20diabetes&author=TM%20Reinbothe&author=S%20Alkayyali&author=E%20Ahlqvist&volume=56&issue=2&publication_year=2013&pages=340-349&pmid=23229155&doi=10.1007/s00125-012-2758-z&)
|
| 411 |
+
|
| 412 |
+
37. Szymanowicz O, Drużdż A, Słowikowski B, et al. A review of the CACNA gene family: its role in neurological disorders. Diseases. 2024;12(5):90. doi: 10.3390/diseases12050090 [DOI](https://doi.org/10.3390/diseases12050090) | [PMC free article](/articles/PMC11119137/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38785745/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Diseases&title=A%20review%20of%20the%20CACNA%20gene%20family:%20its%20role%20in%20neurological%20disorders&author=O%20Szymanowicz&author=A%20Dru%C5%BCd%C5%BC&author=B%20S%C5%82owikowski&volume=12&issue=5&publication_year=2024&pages=90&pmid=38785745&doi=10.3390/diseases12050090&)
|
| 413 |
+
|
| 414 |
+
38. Gelen V, Kükürt A, Şengül E. Role of the renin-angiotensin-aldosterone system in various disease processes: an overview. Renin Angiot Aldost Syst. 2021;12(1):87. doi: 10.5772/intechopen.97354 [DOI](https://doi.org/10.5772/intechopen.97354) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Renin%20Angiot%20Aldost%20Syst&title=Role%20of%20the%20renin-angiotensin-aldosterone%20system%20in%20various%20disease%20processes:%20an%20overview&author=V%20Gelen&author=A%20K%C3%BCk%C3%BCrt&author=E%20%C5%9Eeng%C3%BCl&volume=12&issue=1&publication_year=2021&pages=87&doi=10.5772/intechopen.97354&)
|
| 415 |
+
|
| 416 |
+
39. Fountain JH, Kaur J, Lappin SL. Physiology, Renin Angiotensin System. In:; StatPearls Treasure Island (FL): StatPearls; 2023. [PubMed](https://pubmed.ncbi.nlm.nih.gov/29261862/) | [Google Scholar](https://scholar.google.com/scholar_lookup?title=StatPearls&author=JH%20Fountain&author=J%20Kaur&author=SL%20Lappin&publication_year=2023&)
|
| 417 |
+
|
| 418 |
+
40. Razaq A, Khan A, Shah ST, Ullah S. Association of insertion /deletion polymorphism of ace gene with essential hypertension in patients of Khyber Pakhtunkhwa. Pak J Med Sci. 2024;40(3Part��II):461–466. doi: 10.12669/pjms.40.3.7354 [DOI](https://doi.org/10.12669/pjms.40.3.7354) | [PMC free article](/articles/PMC10862427/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38356813/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pak%20J%20Med%20Sci&title=Association%20of%20insertion%20/deletion%20polymorphism%20of%20ace%20gene%20with%20essential%20hypertension%20in%20patients%20of%20Khyber%20Pakhtunkhwa&author=A%20Razaq&author=A%20Khan&author=ST%20Shah&author=S%20Ullah&volume=40&issue=3Part%E2%80%93II&publication_year=2024&pages=461-466&pmid=38356813&doi=10.12669/pjms.40.3.7354&)
|
| 419 |
+
|
| 420 |
+
41. Moorthy N, Saligrama Ramegowda K, Jain S, et al. Role of angiotensin-converting enzyme (ACE) gene polymorphism and ACE activity in predicting outcome after acute myocardial infarction. Int J Cardiol Heart Vasc. 2021;32:100701. doi: 10.1016/j.ijcha.2020.100701 [DOI](https://doi.org/10.1016/j.ijcha.2020.100701) | [PMC free article](/articles/PMC7782316/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33426268/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int%20J%20Cardiol%20Heart%20Vasc&title=Role%20of%20angiotensin-converting%20enzyme%20(ACE)%20gene%20polymorphism%20and%20ACE%20activity%20in%20predicting%20outcome%20after%20acute%20myocardial%20infarction&author=N%20Moorthy&author=K%20Saligrama%20Ramegowda&author=S%20Jain&volume=32&publication_year=2021&pages=100701&pmid=33426268&doi=10.1016/j.ijcha.2020.100701&)
|
test/texts/PMC11628867.md
ADDED
|
@@ -0,0 +1,294 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Correlation of the DRD2 gene polymorphism with psychopathology and predictive antimanic responses in patients with bipolar mania
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Hejian Tao, Haiying Jin, Min Xu, Haihan Chen, Fengli Sun, Weidong Jin
|
| 5 |
+
**Journal:** Frontiers in Pharmacology
|
| 6 |
+
**Date:** 2024 Nov 26
|
| 7 |
+
**DOI:** [10.3389/fphar.2024.1465356](https://doi.org/10.3389/fphar.2024.1465356)
|
| 8 |
+
**PMID:** 39660002
|
| 9 |
+
**PMCID:** PMC11628867
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11628867/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11628867/pdf/fphar-15-1465356.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11628867/pdf/fphar-15-1465356.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
To explore the correlation of the DRD2 gene polymorphism with psychopathology and predict responses in patients with mania treated with lithium and olanzapine. Sixty patients with bipolar mania were treated with lithium combined with olanzapine for 8 weeks and assessed using YMRS, HAMD, and HAMA. The DRD2 gene polymorphism rs1800497 was tested. Eleven (24.4%) manic patients achieved an early effective response according to the reduction of the YMRS score of >20% in the 2nd week, with a lower HAMA score than the no early effective response group. Twenty-three (51.1%) manic patients achieved remission according to the reduction of the YMRS score of >75% at the 8th week with a higher dose of lithium at the 8th weekend (g/day) than in the no-remission group. Manic patients with genotype GG had lower YMRS scores and lower doses and serum concentrations of olanzapine than patients with genotype AA + AG from the 4th week to the 8th week. Manic patients with genotype GG had a higher relative change in the YMRS score than those with genotype AA + AG from the 2nd week to the 8th week. No differences in HAMA or HAMD were found between the groups with genotype GG and AA + AG. There were more patients who achieved an early effective response in the 2nd week and remission in the 8th in those with genotype GG compared to those with genotype AA + AG. Manic patients with genotype GG had a greater improvement in the YMRS score due to a greater early effective response and remission, which was not related to higher doses and serum concentrations of olanzapine and lithium.
|
| 16 |
+
|
| 17 |
+
Keywords: bipolar mania, DRD2 gene, gene polymorphism, lithium, olanzapine
|
| 18 |
+
|
| 19 |
+
**Keywords:**Keywords: bipolar mania, DRD2 gene, gene polymorphism, lithium, olanzapine
|
| 20 |
+
|
| 21 |
+
## 1 Introduction
|
| 22 |
+
|
| 23 |
+
Bipolar affective disorder is a common neuropsychiatric disorder. Although its neurobiological underpinnings are incompletely understood, the dopamine hypothesis has been a key theory of the pathophysiology of both manic and depressive phases of the disease for more than four decades ([Ashok et al., 2017](#B1)Ashok et al., 2017). Over the years, many genetic polymorphisms have been identified as having a greater risk of developing mood disorders. The countries with the highest activity and the most impactful research in the field were identified. Furthermore, a total of 13 main thematic clusters emerged in the literature. From the qualitative inspection of the clusters, it emerged that the research interest moved from a monogenic to a polygenic risk framework. Researchers have moved from studying single genes in the early 1990s to conducting genome-wide association studies around 2015 ([Bonacina et al., 2023](#B4)Bonacina et al., 2023). The increased use of antidopaminergics in the treatment of this disorder and new *in vivo*in vivo neuroimaging and postmortem studies make it timely to review this theory of dopamine hypothesis. The possible influence of dopamine receptor variants on drug response has not received as much attention. In contrast, there is some evidence that polymorphisms and mutations in dopamine receptors can alter functional activity and pharmacological profiles, but no conclusive data link these gene variants to drug response or disease. The lack of unequivocal findings may be related, in part, to the subtle changes in receptor pharmacology that these polymorphisms and mutations mediate. These subtle effects may be obscured by the influence of genes controlling drug metabolism and kinetics ([Wong et al., 2000](#B20)Wong et al., 2000).
|
| 24 |
+
|
| 25 |
+
In a systematic review, most antipsychotics, carbamazepine, lithium, tamoxifen, and valproate, were effective for acute mania, although only aripiprazole, olanzapine, quetiapine, and risperidone had better acceptability than placebo ([Kishi et al., 2022](#B11)Kishi et al., 2022). In particular, cariprazine (CAR) is an antipsychotic drug for the treatment of schizophrenia and bipolar disorder (BD) and acts as a partial agonist on dopamine receptors (DR), D2, and D3. The study found a significant association between *DRD2*DRD2 rs1800497 and rs6277 and the response to Cariprazine (CAR) treatment ([De Pieri et al., 2023](#B6)De Pieri et al., 2023). When genotypes were combined to obtain an arbitrary score, the receiver operating characteristic curve analysis showed, for the first time, a correlation between single nucleotide polymorphism (SNPs) in *DRD2*DRD2 and the response to CAR treatment. In several studies, *DRD2*DRD2 gene polymorphism was related to olanzapine effectiveness and safety variability ([Zubiaur et al., 2021](#B23)Zubiaur et al., 2021). These observations suggest that antipsychotics, such as risperidone and aripiprazole, can lead to the deposition of long-lasting epigenetic marks in addition to interacting with specific receptors, impairing the function of the nervous system ([Osuna-Luque et al., 2018](#B16)Osuna-Luque et al., 2018). There is reason to believe that the efficacy of olanzapine as an atypical antipsychotic in the treatment of mania is related to dopamine D2 receptor polymorphism.
|
| 26 |
+
|
| 27 |
+
This study aimed to find an association between *DRD2*DRD2 rs1800497 and olanzapine combined with lithium in the treatment of patients with bipolar mania.
|
| 28 |
+
|
| 29 |
+
## 2 Materials and methods
|
| 30 |
+
|
| 31 |
+
### 2.1 Research object
|
| 32 |
+
|
| 33 |
+
Patients with mania hospitalized at Tongde Hospital in Zhejiang Province, Jiaxing Kangci Hospital, Shaoxing Seventh Hospital, and Jinhua Second Hospital from 31 December 2020, to 1 January 2022, were selected according to the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition.
|
| 34 |
+
|
| 35 |
+
### 2.2 Research methods
|
| 36 |
+
|
| 37 |
+
Blood sample collection, DNA extraction, and gene polymorphism monitoring: 5 mL of peripheral venous blood was collected, and anticoagulated with ethylenediaminetetraacetic acid was also stored at −20°C. DNA extraction kit (TSINGKE) was used to extract genomic DNA, and the nucleic acid protein analyzer monitors its concentration and purity. A polymerase chain reaction was used to amplify the target gene containing the *DRD2*DRD2 gene rs1800497 polymorphism. The primer was designed and synthesized, the sequence of the SNP site was found at NCBI ([https://www.ncbi.nlm.nih.gov/](https://www.ncbi.nlm.nih.gov/)https://www.ncbi.nlm.nih.gov/), and the peripheral amplification primer was designed. The guiding primer and folding primer of rs1800497 were ATCCTCAAAGTGCTGGTC and AGGCAGGCGCCCAGCTGG, respectively. The amplification product was digested at 65°C by restriction endonuclease for 3 h. The genotype of the enzyme product was identified by 3% agarose gel electrophoresis and stained with bromophenol blue.
|
| 38 |
+
|
| 39 |
+
### 2.3 Drugs and management
|
| 40 |
+
|
| 41 |
+
The patients were treated with lithium carbonate combined with olanzapine from baseline for 8 weeks. The dose of lithium carbonate was 0.9–1.2 g/day and the dose of olanzapine was 15–25 mg/day. Other psychotropic drugs were not used in combination during treatment. If adverse reactions occur, benzodiazepines are administered in combination for symptomatic treatment.
|
| 42 |
+
|
| 43 |
+
### 2.4 Clinical assessment
|
| 44 |
+
|
| 45 |
+
#### 2.4.1 Scale assessment
|
| 46 |
+
|
| 47 |
+
Young manic rating scale (YMRS), Hamilton Anxiety Scale (HAMA), and Hamilton Depression Scale (HAMD) were used to assess clinical symptoms and changes by two psychiatrists on the 2nd, 4th, 6th, and 8th weekends before and after treatment with consistency ≥80%.
|
| 48 |
+
|
| 49 |
+
#### 2.4.2 Clinical efficacy
|
| 50 |
+
|
| 51 |
+
The main efficacy was defined as a decrease in the level of evaluation of YMRS. YMRS reduction score % = (total score before treatment - total score after treatment)/(total score before treatment) × 100%. Early effective therapy was expressed as an early effective response rate, which means that the YMRS reduction rate >20% on the 2nd week. Remission was expressed as a remission rate, which means a YMRS reduction rate ≥75% on the 8th week ([Jin et al., 2003](#B10)Jin et al., 2003).
|
| 52 |
+
|
| 53 |
+
### 2.5 Statistics analysis
|
| 54 |
+
|
| 55 |
+
Statistical analysis of the data was performed using SPSS 22.0 software. Measurement data were presented as mean ± standard deviation, and an independent sample *t*t-test was performed to compare differences between the two groups. The counting data were presented as composition ratio or frequency, and the chi-square test was used for comparison between groups. The statistical significance was set at P < 0.05.
|
| 56 |
+
|
| 57 |
+
This study was approved by the Medical Ethics Committee of Tongde Hospital of Zhejiang Province (2019–070) and registered for clinical research in China (CHICT2100054696). The research flowchart is shown in [Figure 1](#F1)Figure 1.
|
| 58 |
+
|
| 59 |
+
### FIGURE 1.
|
| 60 |
+
|
| 61 |
+

|
| 62 |
+
|
| 63 |
+
Research flowchart.
|
| 64 |
+
|
| 65 |
+
## 3 Results
|
| 66 |
+
|
| 67 |
+
### 3.1 Comparison of relative factors of patients and their therapeutic efficacy
|
| 68 |
+
|
| 69 |
+
We evaluated 45 patients with bipolar mania and observed changes in their YRMS score (reduction of >20% in the 2nd week and 75% in the 8th week) after 8 weeks of treatment with lithium combined with olanzapine. According to the reduction in the YRMS score, 11 patients were assigned to the early effective response group, with a reduction of a YMRS score >20% in the 2nd week, and 34 patients to the no early effective response group, with a reduction ≤20% in the 2nd week. No differences in sex, age, YMRS, HAMD, HAMA, drug dose, and concentration were found between the two groups, except for a relative reduction in YMRS in the 2nd week (see [Table 1](#T1)Table 1).
|
| 70 |
+
|
| 71 |
+
### TABLE 1.
|
| 72 |
+
|
| 73 |
+
The relative factors of 45 manic patients, early effective response group and no-early effective response group.
|
| 74 |
+
|
| 75 |
+
| Item | All | Early effective response group | No early effective response group | X 2 /t | P |
|
| 76 |
+
| ---- | --- | ------------------------------ | --------------------------------- | ------ | - |
|
| 77 |
+
| Number of cases | 45 | 11 (24.4%) | 34 (75.6%) | | |
|
| 78 |
+
| Age | 32.3 ± 9.9 | 32.1 ± 9.5 | 32.3 ± 10.2 | 0.057 | 0.953 |
|
| 79 |
+
| Age at first onset | 32.3 ± 9.9 | 21.6 ± 6.9 | 24.1 ± 9.9 | 0.758 | 0.374 |
|
| 80 |
+
| Sex (M/F) | 21/24 | 5/6 | 16/18 | | |
|
| 81 |
+
| Dosage of olanzapine at 2nd weekend (mg/d) | 11.5 ± 2.9 | 11.8 ± 2.5 | 11.4 ± 3.1 | 0.333 | 0.713 |
|
| 82 |
+
| Dosage of lithium at 2nd weekend (g/d) | 0.8 ± 0.2 | 0.8 ± 0.1 | 0.7 ± 0.2 | 0.593 | 0.526 |
|
| 83 |
+
| Concentration of olanzapine at 2nd weekend (ng/mL) | 36.2 ± 16.6 | 39.4 ± 18.6 | 35.1 ± 16.1 | 0.726 | 0.472 |
|
| 84 |
+
| Concentration of lithium at 2nd weekend (mmol/L) | 0.6 ± 0.1 | 0.6 ± 0.08 | 0.6 ± 0.1 | 0.622 | 0.537 |
|
| 85 |
+
| YMRS at 2nd weekend | 25.9 ± 2.6 | 25.5 ± 3.6 | 26.0 ± 2.2 | 0.527 | 0.601 |
|
| 86 |
+
| HAMD at 2nd weekend | 6.6 ± 1.1 | 6.3 ± 0.8 | 6.7 ± 1.2 | 0.824 | 0.308 |
|
| 87 |
+
| HAMA at 2nd weekend | 11.3 ± 1.3 | 10.6 ± 1.2 | 11.5 ± 1.3 | 2.081 | 0.049 |
|
| 88 |
+
| Relative reduction of YMRS at 2nd weekend (%) | 15.7 ± 6.7 | 25.3 ± 3.1 | 12.6 ± 3.7 | 10.199 | 0.000 |
|
| 89 |
+
Patients with a reduction of YMRS score ≥75% in the 8th week were assigned to the remission group; meanwhile, 22 patients with a reduction of YMRS score <75% in the 8th week were in the no-remission group. There were no differences in sex, age, HAMD, HAMA, drug dosage, and concentration of olanzapine between the two groups. The difference in dose of lithium, YMRS, and relative reduction of YMRS at the 8th week between the two groups was found (see [Table 2](#T2)Table 2).
|
| 90 |
+
|
| 91 |
+
### TABLE 2.
|
| 92 |
+
|
| 93 |
+
The relative factors of 45 manic patients, remission group and no-remission group.
|
| 94 |
+
|
| 95 |
+
| Item | All | Remission group | No remission group | X 2 /t | P |
|
| 96 |
+
| ---- | --- | --------------- | ------------------ | ------ | - |
|
| 97 |
+
| Number of cases | 45 | 23 (51.1%) | 22 (48.9%) | | |
|
| 98 |
+
| Age | 32.3 ± 9.9 | 33.8 ± 10.6 | 30.7 ± 9.1 | 1.058 | 0.294 |
|
| 99 |
+
| Age at first onset | 23.4 ± 9.2 | 23.3 ± 8.4 | 23.5 ± 10.2 | 0.071 | 0.943 |
|
| 100 |
+
| Sex (M/F) | 21/24 | 12/11 | 9/13 | | |
|
| 101 |
+
| Dosage of olanzapine at 8th weekend (mg/d) | 13.9 ± 2.6 | 14.3 ± 2.7 | 13.5 ± 2.5 | 1.050 | 0.299 |
|
| 102 |
+
| Dosage of lithium at 8th weekend (g/d) | 0.8 ± 0.1 | 0.8 ± 0.1 | 0.7 ± 0.1 | 2.247 | 0.003 |
|
| 103 |
+
| Concentration of olanzapine at 8th weekend (ng/mL) | 64.4 ± 18.0 | 69.3 ± 19.5 | 59.3 ± 15.2 | 1.896 | 0.054 |
|
| 104 |
+
| Concentration of lithium at 8th weekend (mmol/L) | 0.7 ± 0.1 | 0.7 ± 0.2 | 0.6 ± 0.1 | 1.985 | 0.052 |
|
| 105 |
+
| YMRS at 8th weekend | 8.0 ± 1.2 | 7.1 ± 1.0 | 8.8 ± 0.8 | 5.740 | 0.000 |
|
| 106 |
+
| HAMD at 8th weekend | 10.5 ± 0.6 | 10.9 ± 0.6 | 10.5 ± 0.5 | 0.857 | 0.380 |
|
| 107 |
+
| HAMA at 8th weekend | 6.3 ± 1.4 | 6.7 ± 1.5 | 6.4 ± 1.2 | 0.890 | 0.379 |
|
| 108 |
+
| Relative reduction of YMRS at 8th weekend (%) | 73.8 ± 4.9 | 77.8 ± 2.3 | 69.9 ± 2.9 | 10.407 | 0.000 |
|
| 109 |
+
### 3.2 Therapeutic efficacy and DRD2 gene polymorphism of rs1800497
|
| 110 |
+
|
| 111 |
+
The *DRD2*DRD2 gene polymorphism of rs1800497 in 45 patients with mania distributes 17 GG, 20 GA, and 8 AA. A previous study showed that the G allele may be related to more responses ([De Pieri et al., 2023](#B6)De Pieri et al., 2023). We compare the relative factors between the genotype GG groups and genotype AA + AG groups. There were no differences in sex, age, marriage, education, family history, clinical type, and dose and concentration of lithium between the two groups, except for the dose and concentration of olanzapine (see [Table 3](#T3)Table 3).
|
| 112 |
+
|
| 113 |
+
### TABLE 3.
|
| 114 |
+
|
| 115 |
+
Primary information between different groups of rs1800497 (AA + AG and GG genotype).
|
| 116 |
+
|
| 117 |
+
| Item | information | AA + AG | GG | X 2 | P |
|
| 118 |
+
| ---- | ----------- | ------- | -- | --- | - |
|
| 119 |
+
| Sex | Male | 13 | 8 | 0.002 | 0.967 |
|
| 120 |
+
| Female | 15 | 9 | | | |
|
| 121 |
+
| Age | 17–30 | 10 | 8 | 0.640 | 0.726 |
|
| 122 |
+
| 31–40 | 11 | 5 | | | |
|
| 123 |
+
| 41–60 | 7 | 4 | | | |
|
| 124 |
+
| Marriage | Unmarried | 11 | 11 | 2.917 | 0.233 |
|
| 125 |
+
| Married | 13 | 4 | | | |
|
| 126 |
+
| Divorce | 4 | 2 | | | |
|
| 127 |
+
| Education | Primary school | 0 | 1 | 5.578 | 0.233 |
|
| 128 |
+
| Junior school | 3 | 1 | | | |
|
| 129 |
+
| Senior school | 5 | 7 | | | |
|
| 130 |
+
| Junior college | 8 | 2 | | | |
|
| 131 |
+
| University | 12 | 6 | | | |
|
| 132 |
+
| Age of first onset | Less 17 | 7 | 5 | 3.876 | 0.275 |
|
| 133 |
+
| 17–30 | 18 | 9 | | | |
|
| 134 |
+
| 31–40 | 1 | 3 | | | |
|
| 135 |
+
| 41–60 | 2 | 0 | | | |
|
| 136 |
+
| Family history | Positive | 2 | 3 | 1.182 | 0.277 |
|
| 137 |
+
| Negative | 26 | 14 | | | |
|
| 138 |
+
| Clinical type | I type | 18 | 7 | 2.522 | 0.283 |
|
| 139 |
+
| II type | 6 | 3 | | | |
|
| 140 |
+
| Mixed episode | 10 | 7 | | | |
|
| 141 |
+
| Dosage of medicine | Olanzapine (mg/d) | 14.55 ± 2.81 | 12.95 ± 2.02 | 2.060 | 0.045 |
|
| 142 |
+
| Lithium carbonate (g/d) | 0.84 ± 0.15 | 0.78 ± 0.15 | 1.280 | 0.208 | |
|
| 143 |
+
| Concentration of medicine | Olanzapine (ng/mL) | 68.68 ± 18.58 | 57.51 ± 15.18 | 2.088 | 0.043 |
|
| 144 |
+
| Lithium carbonate (mmol/L) | 0.70 ± 0.13 | 0.67 ± 0.17 | 0.627 | 0.543 | |
|
| 145 |
+
The difference in YMRS and the relative change between the genotype GG groups and AA + AG groups were analyzed. The difference in YMRS appeared in the 4th week, and the relative change in YMRS appeared in the 2nd week between the genotype GG groups and genotype AA + AG groups (see [Table 4](#T4)Table 4).
|
| 146 |
+
|
| 147 |
+
### TABLE 4.
|
| 148 |
+
|
| 149 |
+
Symptom and Change between different between groups of rs1800497 (AA + AG and GG genotype).
|
| 150 |
+
|
| 151 |
+
| Scale/Relative change | Weeks | Group | t | P 1 | F | P 2 |
|
| 152 |
+
| --------------------- | ----- | ----- | - | --- | - | --- |
|
| 153 |
+
| AA + AG ( x¯ ±s) | GG ( x¯ ±s) | | | | | |
|
| 154 |
+
| YMRS | Week 0 | 30.64 ± 3.71 | 31.35 ± 4.24 | −0.59 | 0.56 | 2.580 | 0.115 |
|
| 155 |
+
| Weekend 2 | 26.29 ± 3.00 | 25.29 ± 1.76 | 1.40 | 0.17 | | |
|
| 156 |
+
| Weekend 4 | 20.86 ± 2.86 | 19.24 ± 2.02 | 2.05 | 0.05 | | |
|
| 157 |
+
| Weekend 6 | 15.79 ± 3.01 | 13.24 ± 2.17 | 3.04 | 0.00 | | |
|
| 158 |
+
| Weekend 8 | 8.39 ± 1.34 | 7.35 ± 0.93 | 2.80 | 0.00 | | |
|
| 159 |
+
| Relative change of YMRS (%) | Week 0 | — | — | — | — | | |
|
| 160 |
+
| Weekend 2 | 13.96 ± 6.46 | 18.63 ± 5.90 | −2.43 | 0.02 | 13.625 | 0.001 |
|
| 161 |
+
| Weekend 4 | 31.93 ± 4.77 | 38.11 ± 6.66 | −3.62 | 0.00 | | |
|
| 162 |
+
| Weekend 6 | 48.33 ± 9.24 | 57.53 ± 6.26 | −3.63 | 0.00 | | |
|
| 163 |
+
| Weekend 8 | 72.35 ± 5.04 | 76.27 ± 3.76 | −2.76 | 0.00 | | |
|
| 164 |
+
| HAMD | Week 0 | 6,29 ± 1.51 | 6 ± 1.23 | −0.66 | 0.51 | 1.137 | 0.292 |
|
| 165 |
+
| Weekend 2 | 6.55 ± 1.30 | 6.41 ± 1.00 | −0.92 | 0.36 | | |
|
| 166 |
+
| Weekend 4 | 8.145 ± 1.48 | 7.71 ± 0.92 | 1.22 | 0.23 | | |
|
| 167 |
+
| Weekend 6 | 9.18 ± 1.25 | 9.47 ± 0.87 | −0.85 | 0.36 | | |
|
| 168 |
+
| Weekend 8 | 10.54 ± 0.58 | 10.47 ± 0.62 | 0.36 | 0.72 | | |
|
| 169 |
+
| HAMA | Week 0 | 13.89 ± 1.69 | 13 ± 1.22 | 1.97 | 0.06 | 0.903 | 0.347 |
|
| 170 |
+
| Weekend 2 | 13.36 ± 1.77 | 13.41 ± 1.00 | −0.30 | 0.77 | | |
|
| 171 |
+
| Weekend 4 | 13.72 ± 2.63 | 12.94 ± 3.03 | −0.93 | 0.36 | | |
|
| 172 |
+
| Weekend 6 | 14.82 ± 3.18 | 15.53 ± 2.79 | −0.05 | 0.96 | | |
|
| 173 |
+
| Weekend 8 | 15.43 ± 3.814 | 16.29 ± 3.37 | −0.31 | 0.76 | | |
|
| 174 |
+
The correlation between therapeutic efficacy and gene polymorphism was also found. The early response in the 2nd week correlates with genotype GG (see [Table 5](#T5)Table 5). The higher early response in the 2nd week in patients with genotype GG of *DRD2*DRD2 gene polymorphism rs1800497 was found than that in patients with genotype AA + AG. Remission in the 8th week also correlates with genotyp GG (see [Table 6](#T6)Table 6). The higher remission in the 8th week in patients with genotype GG of *DRD2*DRD2 gene polymorphism rs1800497 was found than that in patients with genotype AA + AG.
|
| 175 |
+
|
| 176 |
+
### TABLE 5.
|
| 177 |
+
|
| 178 |
+
Comparison of response at second weekend between groups of rs1800497 (AA + AG and GG genotype).
|
| 179 |
+
|
| 180 |
+
| | response at 2nd week | Total |
|
| 181 |
+
| - | -------------------- | ----- |
|
| 182 |
+
| ≤20% | >20% | |
|
| 183 |
+
| RS1800497 | AA + AG | 24 | 4 | 28 |
|
| 184 |
+
| GG | 10 | 7 | 17 |
|
| 185 |
+
| Total | 34 | 11 | 45 |
|
| 186 |
+
| X 2 = 4.141 p = 0.042 | | |
|
| 187 |
+
### TABLE 6.
|
| 188 |
+
|
| 189 |
+
Comparison of remission at eighth weekend between groups of rs1800497 (AA + AG and GG genotype).
|
| 190 |
+
|
| 191 |
+
| | remission at 8th week | Total |
|
| 192 |
+
| - | --------------------- | ----- |
|
| 193 |
+
| <75% | ≥75% | |
|
| 194 |
+
| RS1800497 | AA + AG | 18 | 10 | 28 |
|
| 195 |
+
| GG | 4 | 13 | 17 |
|
| 196 |
+
| Total | 22 | 23 | 45 |
|
| 197 |
+
| X 2 = 7.032 p = 0.001 | | |
|
| 198 |
+
## 4 Discussion
|
| 199 |
+
|
| 200 |
+
Atypical antipsychotics, such as olanzapine combined with mood stabilizers, such as lithium carbonate, are commonly used and effective treatment methods for mania. Clinical studies and evidence-based medicine research have shown that olanzapine and lithium carbonate are effective drugs for treating mania. Treatment also suggest that the combination of lithium carbonate and olanzapine may yield better resultsfor treatment of mania ([Jin et al., 2004](#B9)Jin et al., 2004; [McKnight et al., 2019](#B14)McKnight et al., 2019; [Chen and Jin, 2022](#B5)Chen and Jin, 2022; [Yatham et al., 2021](#B22)Yatham et al., 2021; [Tang et al., 2007](#B17)Tang et al., 2007).Therefore, by the combination of lithium and olanzapine as a therapeutic treatment, 11 (24.4%) of the patients achieved early effective response in the 2nd week, and 23 (51.1%) achieved remission in the 8th week of 8-week treatment, which certainly suggested that the combination of lithium and olanzapine showed more improvement in manic patients (see [Tables 1](#T1)Tables 1, [2](#T2)2). Another finding of the study was that the remission group had higher doses of lithium carbonate, as well as possibly higher concentrations of lithium carbonate and olanzapine, which were clearly consistent with clinical practice ([Table 2](#T2)Table 2).
|
| 201 |
+
|
| 202 |
+
However, the next analysis had different results, which differed from the correlation between good efficacy and high doses or concentrations of the drug. The results show that patients with AA + AG of the *DRD2*DRD2 gene polymorphism have a higher dose and concentration of olanzapine than those with AA of the *DRD2*DRD2 gene polymorphism ([Table 3](#T3)Table 3), but patients with AA + AG of the *DRD2*DRD2 gene polymorphism achieve a greater improvement in symptoms compared to those with AA of the *DRD2*DRD2 gene polymorphism, which concludes a greater relative reduction in manic symptoms ([Table 4](#T4)Table 4), a higher early response rate and remission in the 2nd ([Table 5](#T5)Table 5) and 8th week ([Table 6](#T6)Table 6), respectively. These results suggest that therapeutic effects are related to the *DRD2*DRD2 gene polymorphism of rs1800497 rather than the higher dose and concentration of the drug. Personalized precision therapy should focus on polymorphisms, not drug concentrations or doses, although drug metabolism and concentration are also involved in the polymorphism of certain metabolic enzymes ([Milosavljevic et al., 2021](#B15)Milosavljevic et al., 2021). It also suggests that pharmacodynamics is very important in personalized precision therapy, at least in the treatment of mania ([De Pieri et al., 2023](#B6)De Pieri et al., 2023; [Urs et al., 2012](#B19)Urs et al., 2012). This also appeared to confirm the dopamine hypothesis that manic episodes are associated with dopamine hyperactivity, with dopamine receptor polymorphism more closely ([Bonacina et al., 2023](#B4)Bonacina et al., 2023; [Wong et al., 2000](#B20)Wong et al., 2000; [Azechi et al., 2019](#B2)Azechi et al., 2019).
|
| 203 |
+
|
| 204 |
+
Another finding of this study is equally significant since patients with genotype GG have early and late good treatment results, which appears to validate that “early effective response predicts late efficacy.” Early improvement in HAMD-17 and HAMD-6 scores was found to predict ultimate response and remission in depressed patients treated with fluoxetine or ECT ([Lin and Lin, 2019](#B13)Lin and Lin, 2019). Moreover, early improvement with vortioxetine was also found to predict response and remission in depressive patients ([Inoue et al., 2021](#B8)Inoue et al., 2021). This prediction pattern occurs not only in the treatment of depression, but in fact in the treatment of mania. Response and remission could be predicted by early improvement at week 2, while patients without early improvement were unlikely to reach response and remission at week 4 ([Li et al., 2017](#B12)Li et al., 2017). Early response at week 1 can predict treatment outcomes in adolescents with bipolar mania or mixed episodes treated with olanzapine ([Xiao et al., 2017](#B21)Xiao et al., 2017). We also found that an early improvement in more manic symptoms in the genotype GG also suggested a higher rate of remission. Therefore, it is reasonable to assume that this predictive pattern is also present in the treatment of patients with mania and that this predictive pattern is mediated by the gene polymorphism of rs1800497 with GG.
|
| 205 |
+
|
| 206 |
+
The shortcomings of this study are as follows. First, as a genetics and genetic polymorphism study, the sample size of the above studies is still small, and larger sample sizes and multi-center studies may lead to more reliable conclusions. Second, the treatment we used was a combination of lithium carbonate and olanzapine, making it difficult to distinguish between the therapeutic effects, which belong to lithium or olanzapine. It should be noted that combination therapy is more effective than monotherapy, especially lithium carbonate plus olanzapine, in the treatment of mania and even for refractory mania ([Fountoulakiskn et al., 2022](#B7)Fountoulakiskn et al., 2022; [Tohen et al., 2002](#B18)Tohen et al., 2002). Third, the scale of the study does not involve psychotic symptoms because these psychotic symptoms are relatively common in mania and are also important for therapeutic intervention ([Bjørklund et al., 2017](#B3)Bjørklund et al., 2017). Fourth, dopamine gene polymorphism can be involved in multiple sites, and we only chose rs1800497, but there may also be another gene that shares this characteristic.
|
| 207 |
+
|
| 208 |
+
## Acknowledgments
|
| 209 |
+
|
| 210 |
+
The authors thank Prof. Chen Wei (Zhejiang University) for giving us study ideas and Wang Zhiqiang (Tsinghua University) for helping us with literature retrieval. We thank Ren Xin (Zhejiang Province Mental Health Center) for helping us with part of the statistics. The authors thank Prof. Ma Yongchun (Zhejiang Province Mental Health Center) for the final revision of the article. We especially thank the laboratory of Di’an Biological Laboratory for the series of measures and tests of drug concentration in this study.
|
| 211 |
+
|
| 212 |
+
## Funding Statement
|
| 213 |
+
|
| 214 |
+
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Zhejiang Provincial Medical and Health Technology Project (2020KY088) and by the Peak Subject of Psychiatry, Tongde Hospital of Zhejiang Province (PSP2023-020) in design, data collection, statistics, and laboratory tests.
|
| 215 |
+
|
| 216 |
+
## Data availability statement
|
| 217 |
+
|
| 218 |
+
The original contributions presented in the study can be found here: [https://db.cngb.org/search/project/CNP0006542/](https://db.cngb.org/search/project/CNP0006542/)https://db.cngb.org/search/project/CNP0006542/. Further inquiries can be directed to the corresponding author.
|
| 219 |
+
|
| 220 |
+
## Ethics statement
|
| 221 |
+
|
| 222 |
+
The studies involving humans were approved by Medical Ethics Committee of Tongde Hospital of Zhejiang Province (2019-070). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
|
| 223 |
+
|
| 224 |
+
## Author contributions
|
| 225 |
+
|
| 226 |
+
HT: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. HJ: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing–original draft. MX: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing–original draft. HC: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing–original draft. FS: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing–original draft, Writing–review and editing. WJ: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
|
| 227 |
+
|
| 228 |
+
## Conflict of interest
|
| 229 |
+
|
| 230 |
+
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
|
| 231 |
+
|
| 232 |
+
## Publisher’s note
|
| 233 |
+
|
| 234 |
+
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
|
| 235 |
+
|
| 236 |
+
## Associated Data
|
| 237 |
+
|
| 238 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 239 |
+
|
| 240 |
+
### Data Availability Statement
|
| 241 |
+
|
| 242 |
+
The original contributions presented in the study can be found here: [https://db.cngb.org/search/project/CNP0006542/](https://db.cngb.org/search/project/CNP0006542/)https://db.cngb.org/search/project/CNP0006542/. Further inquiries can be directed to the corresponding author.
|
| 243 |
+
|
| 244 |
+
### Data Availability Statement
|
| 245 |
+
|
| 246 |
+
The original contributions presented in the study can be found here: [https://db.cngb.org/search/project/CNP0006542/](https://db.cngb.org/search/project/CNP0006542/)https://db.cngb.org/search/project/CNP0006542/. Further inquiries can be directed to the corresponding author.
|
| 247 |
+
|
| 248 |
+
## References
|
| 249 |
+
|
| 250 |
+
1. Ashok A. H., Marques T. R., Jauhar S., Nour M. M., Goodwin G. W., Young A. H., et al. (2017). The dopamine hypothesis of bipolar affective disorder: the state of the art and implications for treatment. Mol. Psychiatry 22 (5), 666–679. 10.1038/mp.2017.16 [DOI](https://doi.org/10.1038/mp.2017.16) | [PMC free article](/articles/PMC5401767/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28289283/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mol.%20Psychiatry&title=The%20dopamine%20hypothesis%20of%20bipolar%20affective%20disorder:%20the%20state%20of%20the%20art%20and%20implications%20for%20treatment&author=A.%20H.%20Ashok&author=T.%20R.%20Marques&author=S.%20Jauhar&author=M.%20M.%20Nour&author=G.%20W.%20Goodwin&volume=22&issue=5&publication_year=2017&pages=666-679&pmid=28289283&doi=10.1038/mp.2017.16&)
|
| 251 |
+
|
| 252 |
+
2. Azechi H., Hakamada K., Yamamoto T. (2019). A new inbred strain of Fawn-Hooded rats demonstrates mania-like behavioural and monoaminergic abnormalities. IBRO Rep. 7, 98–106. 10.1016/j.ibror.2019.11.001 [DOI](https://doi.org/10.1016/j.ibror.2019.11.001) | [PMC free article](/articles/PMC6861655/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31763490/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=IBRO%20Rep.&title=A%20new%20inbred%20strain%20of%20Fawn-Hooded%20rats%20demonstrates%20mania-like%20behavioural%20and%20monoaminergic%20abnormalities&author=H.%20Azechi&author=K.%20Hakamada&author=T.%20Yamamoto&volume=7&publication_year=2019&pages=98-106&pmid=31763490&doi=10.1016/j.ibror.2019.11.001&)
|
| 253 |
+
|
| 254 |
+
3. Bjørklund L. B., Horsdal H. T., Mors O., Gasse C., Østergaard S. D. (2017). Psychopharmacological treatment of psychotic mania and psychotic bipolar depression compared to non-psychotic mania and non-psychotic bipolar depression. Bipolar Disord. 19 (6), 505–512. 10.1111/bdi.12504 [DOI](https://doi.org/10.1111/bdi.12504) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28593691/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Bipolar%20Disord.&title=Psychopharmacological%20treatment%20of%20psychotic%20mania%20and%20psychotic%20bipolar%20depression%20compared%20to%20non-psychotic%20mania%20and%20non-psychotic%20bipolar%20depression&author=L.%20B.%20Bj%C3%B8rklund&author=H.%20T.%20Horsdal&author=O.%20Mors&author=C.%20Gasse&author=S.%20D.%20%C3%98stergaard&volume=19&issue=6&publication_year=2017&pages=505-512&pmid=28593691&doi=10.1111/bdi.12504&)
|
| 255 |
+
|
| 256 |
+
4. Bonacina G., Carollo A., Esposito G. (2023). The genetic side of the mood: a scientometric review of the genetic basis of mood disorders. Genes. (Basel) 14 (2), 352. 10.3390/genes14020352 [DOI](https://doi.org/10.3390/genes14020352) | [PMC free article](/articles/PMC9956267/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36833279/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genes.%20(Basel)&title=The%20genetic%20side%20of%20the%20mood:%20a%20scientometric%20review%20of%20the%20genetic%20basis%20of%20mood%20disorders&author=G.%20Bonacina&author=A.%20Carollo&author=G.%20Esposito&volume=14&issue=2&publication_year=2023&pages=352&pmid=36833279&doi=10.3390/genes14020352&)
|
| 257 |
+
|
| 258 |
+
5. Chen H. H., Jin W. D. (2022). Evaluation of effectiveness of antiepileptic drugs and lithium carbonate in the treatment of bipolar mania:network Meta-analysis of Chinese literature. J. Clin. Psychiatry 32 (6), 483–487. 10.3969/j.issn.1005-3220 [DOI](https://doi.org/10.3969/j.issn.1005-3220) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Psychiatry&title=Evaluation%20of%20effectiveness%20of%20antiepileptic%20drugs%20and%20lithium%20carbonate%20in%20the%20treatment%20of%20bipolar%20mania:network%20Meta-analysis%20of%20Chinese%20literature&author=H.%20H.%20Chen&author=W.%20D.%20Jin&volume=32&issue=6&publication_year=2022&pages=483-487&doi=10.3969/j.issn.1005-3220&)
|
| 259 |
+
|
| 260 |
+
6. De Pieri M., Ferrari M., Marino F., Traber R., Bolla E., Cosentino M. (2023). Functional single nucleotide polymorphisms in dopaminergic receptors D2 predict clinical response to Cariprazine. Front. Pharmacol. 14, 1182393. 10.3389/fphar.2023.1182393 [DOI](https://doi.org/10.3389/fphar.2023.1182393) | [PMC free article](/articles/PMC10203397/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37229261/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front.%20Pharmacol.&title=Functional%20single%20nucleotide%20polymorphisms%20in%20dopaminergic%20receptors%20D2%20predict%20clinical%20response%20to%20Cariprazine&author=M.%20De%20Pieri&author=M.%20Ferrari&author=F.%20Marino&author=R.%20Traber&author=E.%20Bolla&volume=14&publication_year=2023&pages=1182393&pmid=37229261&doi=10.3389/fphar.2023.1182393&)
|
| 261 |
+
|
| 262 |
+
7. Fountoulakiskn T. M., Zarate Jr C. A., Zarate C. A., Jr (2022). Lithium treatment of Bipolar disorder in adults: a systematic review of randomized trials and meta-analyses. Eur. Neuropsychopharmacol. 54, 100–115. 10.1016/j.euroneuro.2021.10.003 [DOI](https://doi.org/10.1016/j.euroneuro.2021.10.003) | [PMC free article](/articles/PMC8808297/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34980362/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20Neuropsychopharmacol.&title=Lithium%20treatment%20of%20Bipolar%20disorder%20in%20adults:%20a%20systematic%20review%20of%20randomized%20trials%20and%20meta-analyses&author=T.%20M.%20Fountoulakiskn&author=Jr%20C.%20A.%20Zarate&author=C.%20A.%20Zarate&volume=54&publication_year=2022&pages=100-115&pmid=34980362&doi=10.1016/j.euroneuro.2021.10.003&)
|
| 263 |
+
|
| 264 |
+
8. Inoue T., Fujimoto S., Marumoto T., Kitagawa T., Ishida K., Nakajima T., et al. (2021). Early improvement with vortioxetine predicts response and remission: a post hoc analysis of data from a clinical trial conducted in Japan. Neuropsychiatr. Dis. Treat. 17, 3735–3741. 10.2147/NDT.S340309 [DOI](https://doi.org/10.2147/NDT.S340309) | [PMC free article](/articles/PMC8694398/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34955641/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Neuropsychiatr.%20Dis.%20Treat.&title=Early%20improvement%20with%20vortioxetine%20predicts%20response%20and%20remission:%20a%20post%20hoc%20analysis%20of%20data%20from%20a%20clinical%20trial%20conducted%20in%20Japan&author=T.%20Inoue&author=S.%20Fujimoto&author=T.%20Marumoto&author=T.%20Kitagawa&author=K.%20Ishida&volume=17&publication_year=2021&pages=3735-3741&pmid=34955641&doi=10.2147/NDT.S340309&)
|
| 265 |
+
|
| 266 |
+
9. Jin W. D., Chen Z., Tang X. X., Feng B. (2004). Systematic review of olanzaping in treatment for mania in domestic index. J. Evidence-base Med. 4 (3), 145–157. 10.3969/j.issn.1671-5144.2004.03.012 [DOI](https://doi.org/10.3969/j.issn.1671-5144.2004.03.012) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Evidence-base%20Med.&title=Systematic%20review%20of%20olanzaping%20in%20treatment%20for%20mania%20in%20domestic%20index&author=W.%20D.%20Jin&author=Z.%20Chen&author=X.%20X.%20Tang&author=B.%20Feng&volume=4&issue=3&publication_year=2004&pages=145-157&doi=10.3969/j.issn.1671-5144.2004.03.012&)
|
| 267 |
+
|
| 268 |
+
10. Jin W. D., Shen Y., Chen H. (2003). Assessment of therapeutic effects and their difference in various antidepressants. Arch. Psychiatry 16 (4), 248–250. 10.3969/j.issn.1009-7201.2003.04.033 [DOI](https://doi.org/10.3969/j.issn.1009-7201.2003.04.033) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arch.%20Psychiatry&title=Assessment%20of%20therapeutic%20effects%20and%20their%20difference%20in%20various%20antidepressants&author=W.%20D.%20Jin&author=Y.%20Shen&author=H.%20Chen&volume=16&issue=4&publication_year=2003&pages=248-250&doi=10.3969/j.issn.1009-7201.2003.04.033&)
|
| 269 |
+
|
| 270 |
+
11. Kishi T., Ikuta T., Matsuda Y., Sakuma K., Okuya M., Nomura I., et al. (2022). Pharmacological treatment for bipolar mania: a systematic review and network meta-analysis of double-blind randomized controlled trials. Mol. Psychiatry 27 (2), 1136–1144. 10.1038/s41380-021-01334-4 [DOI](https://doi.org/10.1038/s41380-021-01334-4) | [PMC free article](/articles/PMC9054678/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34642461/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mol.%20Psychiatry&title=Pharmacological%20treatment%20for%20bipolar%20mania:%20a%20systematic%20review%20and%20network%20meta-analysis%20of%20double-blind%20randomized%20controlled%20trials&author=T.%20Kishi&author=T.%20Ikuta&author=Y.%20Matsuda&author=K.%20Sakuma&author=M.%20Okuya&volume=27&issue=2&publication_year=2022&pages=1136-1144&pmid=34642461&doi=10.1038/s41380-021-01334-4&)
|
| 271 |
+
|
| 272 |
+
12. Li D. J., Lin C. H., Lu M. J. (2017). Early improvement predicts treatment outcomes for patients with acute mania: a naturalistic study in taiwan. J. Clin. Psychopharmacol. 37 (4), 435–440. 10.1097/JCP.0000000000000728 [DOI](https://doi.org/10.1097/JCP.0000000000000728) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28590370/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Psychopharmacol.&title=Early%20improvement%20predicts%20treatment%20outcomes%20for%20patients%20with%20acute%20mania:%20a%20naturalistic%20study%20in%20taiwan&author=D.%20J.%20Li&author=C.%20H.%20Lin&author=M.%20J.%20Lu&volume=37&issue=4&publication_year=2017&pages=435-440&pmid=28590370&doi=10.1097/JCP.0000000000000728&)
|
| 273 |
+
|
| 274 |
+
13. Lin H. S., Lin C. H. (2019). Early improvement in HAMD-17 and HAMD-6 scores predicts ult,imate response and remission for depressed patients treated with fluoxetine or ECT. J. Affect Disord. 245, 91–97. 10.1016/j.jad.2018.10.105 [DOI](https://doi.org/10.1016/j.jad.2018.10.105) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30368075/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Affect%20Disord.&title=Early%20improvement%20in%20HAMD-17%20and%20HAMD-6%20scores%20predicts%20ult,imate%20response%20and%20remission%20for%20depressed%20patients%20treated%20with%20fluoxetine%20or%20ECT&author=H.%20S.%20Lin&author=C.%20H.%20Lin&volume=245&publication_year=2019&pages=91-97&pmid=30368075&doi=10.1016/j.jad.2018.10.105&)
|
| 275 |
+
|
| 276 |
+
14. McKnight R. F., de La Motte de Broöns de Vauvert SJGN, Chesney E., Amit B. H., Geddes J., Cipriani A. (2019). Lithium for acute mania. Cochrane Database Syst. Rev. 6 (6), CD004048. 10.1002/14651858.CD004048.pub4 [DOI](https://doi.org/10.1002/14651858.CD004048.pub4) | [PMC free article](/articles/PMC6544558/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31152444/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cochrane%20Database%20Syst.%20Rev.&title=Lithium%20for%20acute%20mania&author=R.%20F.%20McKnight&author=SJGN%20de%20La%20Motte%20de%20Bro%C3%B6ns%20de%20Vauvert&author=E.%20Chesney&author=B.%20H.%20Amit&author=J.%20Geddes&volume=6&issue=6&publication_year=2019&pages=CD004048&pmid=31152444&doi=10.1002/14651858.CD004048.pub4&)
|
| 277 |
+
|
| 278 |
+
15. Milosavljevic F., Bukvic N., Pavlovic N., Miljevic C., Pešic V., Molden E., et al. (2021). Association of CYP2C19 and CYP2D6 poor and intermediate metabolizer status with antidepressant and antipsychotic exposure: a systematic review and meta-analysis. JAMA Psychiatry 78 (3), 270–280. 10.1001/jamapsychiatry.2020.3643 [DOI](https://doi.org/10.1001/jamapsychiatry.2020.3643) | [PMC free article](/articles/PMC7702196/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33237321/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=JAMA%20Psychiatry&title=Association%20of%20CYP2C19%20and%20CYP2D6%20poor%20and%20intermediate%20metabolizer%20status%20with%20antidepressant%20and%20antipsychotic%20exposure:%20a%20systematic%20review%20and%20meta-analysis&author=F.%20Milosavljevic&author=N.%20Bukvic&author=N.%20Pavlovic&author=C.%20Miljevic&author=V.%20Pe%C5%A1ic&volume=78&issue=3&publication_year=2021&pages=270-280&pmid=33237321&doi=10.1001/jamapsychiatry.2020.3643&)
|
| 279 |
+
|
| 280 |
+
16. Osuna-Luque J., Rodríguez-Ramos A., Gámez-Del-Estal M. D. M., Ruiz-Rubio M. (2018). Behavioral mechanisms that depend on dopamine and serotonin in Caenorhabditis elegans interact with the antipsychotics risperidone and aripiprazole. J. Exp. Neurosci. 12, 1179069518798628. 10.1177/1179069518798628 [DOI](https://doi.org/10.1177/1179069518798628) | [PMC free article](/articles/PMC6144587/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30245571/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Exp.%20Neurosci.&title=Behavioral%20mechanisms%20that%20depend%20on%20dopamine%20and%20serotonin%20in%20Caenorhabditis%20elegans%20interact%20with%20the%20antipsychotics%20risperidone%20and%20aripiprazole&author=J.%20Osuna-Luque&author=A.%20Rodr%C3%ADguez-Ramos&author=M.%20D.%20M.%20G%C3%A1mez-Del-Estal&author=M.%20Ruiz-Rubio&volume=12&publication_year=2018&pages=1179069518798628&pmid=30245571&doi=10.1177/1179069518798628&)
|
| 281 |
+
|
| 282 |
+
17. Tang J. L., Jin W. D., Chen S. (2007). Evidence-base Medicine Process for an elderly patients with mania. Chin. General Pract. 10 (4), 290–291. 10.3969/j.issn.1007-9572.2007.04.016 [DOI](https://doi.org/10.3969/j.issn.1007-9572.2007.04.016) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chin.%20General%20Pract.&title=Evidence-base%20Medicine%20Process%20for%20an%20elderly%20patients%20with%20mania&author=J.%20L.%20Tang&author=W.%20D.%20Jin&author=S.%20Chen&volume=10&issue=4&publication_year=2007&pages=290-291&doi=10.3969/j.issn.1007-9572.2007.04.016&)
|
| 283 |
+
|
| 284 |
+
18. Tohen M., Chengappa K. N. R., Suppes T., Zarate Jr C. A., Calabrese J. R., Bowden C. L., et al. (2002). Efficacy of olanzapine in combination with valproate or lithium in the treatment of mania in patients partially nonresponsive to valproate or lithium monotherapy. Arch. Gen. Psychiatry 59 (1), 62–69. 10.1001/archpsyc.59.1.62 [DOI](https://doi.org/10.1001/archpsyc.59.1.62) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11779284/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arch.%20Gen.%20Psychiatry&title=Efficacy%20of%20olanzapine%20in%20combination%20with%20valproate%20or%20lithium%20in%20the%20treatment%20of%20mania%20in%20patients%20partially%20nonresponsive%20to%20valproate%20or%20lithium%20monotherapy&author=M.%20Tohen&author=K.%20N.%20R.%20Chengappa&author=T.%20Suppes&author=Jr%20C.%20A.%20Zarate&author=J.%20R.%20Calabrese&volume=59&issue=1&publication_year=2002&pages=62-69&pmid=11779284&doi=10.1001/archpsyc.59.1.62&)
|
| 285 |
+
|
| 286 |
+
19. Urs N. M., Snyder J. S., Jacobsen J. P. R., Peterson S. M., Caron M. G. (2012). Deletion of GSK3β in D2R-expressing neurons reveals distinct roles for β-arrestin signaling in antipsychotic and lithium action. Proc. Natl. Acad. Sci. U. S. A. 109 (50), 20732–20737. 10.1073/pnas.1215489109 [DOI](https://doi.org/10.1073/pnas.1215489109) | [PMC free article](/articles/PMC3528495/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23188793/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Proc.%20Natl.%20Acad.%20Sci.%20U.%20S.%20A.&title=Deletion%20of%20GSK3%CE%B2%20in%20D2R-expressing%20neurons%20reveals%20distinct%20roles%20for%20%CE%B2-arrestin%20signaling%20in%20antipsychotic%20and%20lithium%20action&author=N.%20M.%20Urs&author=J.%20S.%20Snyder&author=J.%20P.%20R.%20Jacobsen&author=S.%20M.%20Peterson&author=M.%20G.%20Caron&volume=109&issue=50&publication_year=2012&pages=20732-20737&pmid=23188793&doi=10.1073/pnas.1215489109&)
|
| 287 |
+
|
| 288 |
+
20. Wong A. H., Buckle C. E., Van Tol H. H. (2000). Polymorphisms in dopamine receptors: what do they tell us? Eur. J. Pharmacol. 410 (2-3), 183–203. 10.1016/s0014-2999(00)00815-3 [DOI](https://doi.org/10.1016/s0014-2999(00)00815-3) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11134669/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20J.%20Pharmacol.&title=Polymorphisms%20in%20dopamine%20receptors:%20what%20do%20they%20tell%20us?&author=A.%20H.%20Wong&author=C.%20E.%20Buckle&author=H.%20H.%20Van%20Tol&volume=410&issue=2-3&publication_year=2000&pages=183-203&pmid=11134669&doi=10.1016/s0014-2999(00)00815-3&)
|
| 289 |
+
|
| 290 |
+
21. Xiao L., Ganocy S. J., Findling R. L., Chang K., DelBello M. P., Kane J. M., et al. (2017). Baseline characteristics and early response at week 1 predict treatment outcome in adolescents with bipolar manic or mixed episode treated with olanzapine: results from a 3-week, randomized, placebo-controlled trial. J. Clin. Psychiatry 78 (9), e1158–e1166. 10.4088/JCP.16m10923 [DOI](https://doi.org/10.4088/JCP.16m10923) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28922591/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Psychiatry&title=Baseline%20characteristics%20and%20early%20response%20at%20week%201%20predict%20treatment%20outcome%20in%20adolescents%20with%20bipolar%20manic%20or%20mixed%20episode%20treated%20with%20olanzapine:%20results%20from%20a%203-week,%20randomized,%20placebo-controlled%20trial&author=L.%20Xiao&author=S.%20J.%20Ganocy&author=R.%20L.%20Findling&author=K.%20Chang&author=M.%20P.%20DelBello&volume=78&issue=9&publication_year=2017&pages=e1158-e1166&pmid=28922591&doi=10.4088/JCP.16m10923&)
|
| 291 |
+
|
| 292 |
+
22. Yatham L. N., Chakrabarty T., Bond D. J., Schaffer A., Beaulieu S., Parikh S. V., et al. (2021). Canadian network for mood and anxiety treatments (CANMAT) and international society for bipolar disorders (ISBD) recommendations for the management of patients with bipolar disorder with mixed presentations. Bipolar Disord. 23 (8), 767–788. 10.1111/bdi.13135 [DOI](https://doi.org/10.1111/bdi.13135) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34599629/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Bipolar%20Disord.&title=Canadian%20network%20for%20mood%20and%20anxiety%20treatments%20(CANMAT)%20and%20international%20society%20for%20bipolar%20disorders%20(ISBD)%20recommendations%20for%20the%20management%20of%20patients%20with%20bipolar%20disorder%20with%20mixed%20presentations&author=L.%20N.%20Yatham&author=T.%20Chakrabarty&author=D.%20J.%20Bond&author=A.%20Schaffer&author=S.%20Beaulieu&volume=23&issue=8&publication_year=2021&pages=767-788&pmid=34599629&doi=10.1111/bdi.13135&)
|
| 293 |
+
|
| 294 |
+
23. Zubiaur P., Soria-Chacartegui P., Villapalos-García G., Gordillo-Perdomo J. J., Abad-Santos F. (2021). The pharmacogenetics of treatment with olanzapine. Pharmacogenomics 22 (14), 939–958. 10.2217/pgs-2021-0051 [DOI](https://doi.org/10.2217/pgs-2021-0051) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34528455/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=The%20pharmacogenetics%20of%20treatment%20with%20olanzapine&author=P.%20Zubiaur&author=P.%20Soria-Chacartegui&author=G.%20Villapalos-Garc%C3%ADa&author=J.%20J.%20Gordillo-Perdomo&author=F.%20Abad-Santos&volume=22&issue=14&publication_year=2021&pages=939-958&pmid=34528455&doi=10.2217/pgs-2021-0051&)
|
test/texts/PMC11666798.md
ADDED
|
@@ -0,0 +1,430 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Influence of CYP2C8*3 and ABCG2 C421A genetic polymorphisms on trough concentration and molecular response of imatinib in Egyptian patients with chronic myeloid leukemia
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Safwat A Mangoura, Mahmoud H Abdel-Raheem, Hanan A Eltyb, Mohammed S Molla, Abeer M R Hussein
|
| 5 |
+
**Journal:** Cancer Chemotherapy and Pharmacology
|
| 6 |
+
**Date:** 2024 Dec 23
|
| 7 |
+
**DOI:** [10.1007/s00280-024-04723-y](https://doi.org/10.1007/s00280-024-04723-y)
|
| 8 |
+
**PMID:** 39714624
|
| 9 |
+
**PMCID:** PMC11666798
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11666798/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11666798/pdf/280_2024_Article_4723.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11666798/pdf/280_2024_Article_4723.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Purpose:**
|
| 16 |
+
The treatment landscape for chronic myeloid leukemia (CML) has been revolutionized by the introduction of imatinib, a tyrosine kinase inhibitor, which has transformed the disease from a fatal condition into a manageable chronic illness for a substantial number of patients. Despite this, some individuals do not respond adequately to the treatment, and others may experience disease progression even with continued therapy. This study examined how CYP2C8*3 (G416A; rs11572080) and ABCG2 C421A (rs2231142) single nucleotide polymorphisms (SNPs) affect the plasma trough concentration and therapeutic response of imatinib in Egyptian CML patients.
|
| 17 |
+
|
| 18 |
+
**Methods:**
|
| 19 |
+
The study included fifty patients with chronic-phase CML, who were categorized into two groups: responders (n = 26) and non-responders (n = 24), according to their BCR-ABL1 transcription levels after 12 months of imatinib treatment. Genotyping of the CYP2C8*3 and ABCG2 C421A polymorphisms was performed using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), while plasma trough concentrations were determined through high-performance liquid chromatography with ultraviolet-diode array detection (HPLC-UV/DAD).
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
Patients with the CA genotype of ABCG2 C421A showed significantly higher mean plasma trough concentrations of imatinib (CA: 1731 ± 424.7 ng/mL; CC: 1294 ± 381.3 ng/mL; p = 0.0132) and demonstrated a better molecular response compared to those with the CC genotype (p = 0.0395).
|
| 23 |
+
|
| 24 |
+
**Conclusion:**
|
| 25 |
+
The ABCG2 C421A polymorphism significantly influenced imatinib plasma trough concentrations and molecular responses in Egyptian chronic-phase CML patients. Genotyping of this polymorphism in these patients could assist in optimizing imatinib therapy, predicting more favorable treatment outcomes, and enabling the development of more personalized treatment plans.
|
| 26 |
+
|
| 27 |
+
**Supplementary Information:**
|
| 28 |
+
The online version contains supplementary material available at 10.1007/s00280-024-04723-y.
|
| 29 |
+
|
| 30 |
+
Keywords: CML, Imatinib, CYP2C8*3, ABCG2
|
| 31 |
+
|
| 32 |
+
### Purpose
|
| 33 |
+
|
| 34 |
+
The treatment landscape for chronic myeloid leukemia (CML) has been revolutionized by the introduction of imatinib, a tyrosine kinase inhibitor, which has transformed the disease from a fatal condition into a manageable chronic illness for a substantial number of patients. Despite this, some individuals do not respond adequately to the treatment, and others may experience disease progression even with continued therapy. This study examined how *CYP2C8*3*CYP2C8*3 (G416A; rs11572080) and *ABCG2*ABCG2 C421A (rs2231142) single nucleotide polymorphisms (SNPs) affect the plasma trough concentration and therapeutic response of imatinib in Egyptian CML patients.
|
| 35 |
+
|
| 36 |
+
### Methods
|
| 37 |
+
|
| 38 |
+
The study included fifty patients with chronic-phase CML, who were categorized into two groups: responders (*n*n = 26) and non-responders (*n*n = 24), according to their *BCR-ABL1*BCR-ABL1 transcription levels after 12 months of imatinib treatment. Genotyping of the *CYP2C8*3*CYP2C8*3 and *ABCG2*ABCG2 C421A polymorphisms was performed using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), while plasma trough concentrations were determined through high-performance liquid chromatography with ultraviolet-diode array detection (HPLC-UV/DAD).
|
| 39 |
+
|
| 40 |
+
### Results
|
| 41 |
+
|
| 42 |
+
Patients with the CA genotype of *ABCG2*ABCG2 C421A showed significantly higher mean plasma trough concentrations of imatinib (CA: 1731 ± 424.7 ng/mL; CC: 1294 ± 381.3 ng/mL; *p*p = 0.0132) and demonstrated a better molecular response compared to those with the CC genotype (*p*p = 0.0395).
|
| 43 |
+
|
| 44 |
+
### Conclusion
|
| 45 |
+
|
| 46 |
+
The *ABCG2*ABCG2 C421A polymorphism significantly influenced imatinib plasma trough concentrations and molecular responses in Egyptian chronic-phase CML patients. Genotyping of this polymorphism in these patients could assist in optimizing imatinib therapy, predicting more favorable treatment outcomes, and enabling the development of more personalized treatment plans.
|
| 47 |
+
|
| 48 |
+
### Supplementary Information
|
| 49 |
+
|
| 50 |
+
The online version contains supplementary material available at 10.1007/s00280-024-04723-y.
|
| 51 |
+
|
| 52 |
+
**Keywords:**Keywords: CML, Imatinib, *CYP2C8*3*CYP2C8*3, *ABCG2*ABCG2
|
| 53 |
+
|
| 54 |
+
## Introduction
|
| 55 |
+
|
| 56 |
+
Chronic myeloid leukemia (CML) is a malignant tumor that results from the abnormal proliferation of hematopoietic stem cells within the bone marrow. It constitutes approximately 15% of leukemia cases in adults [[1](#CR1)1].
|
| 57 |
+
|
| 58 |
+
A hallmark of this disease is the “Philadelphia (Ph) chromosome”, which is produced by a balanced reciprocal translocation between the long arms of chromosomes 9 and 22 [t (9; 22) (q34; q11)] [[2](#CR2)2].
|
| 59 |
+
|
| 60 |
+
This genetic translocation transposes the *Abelson murine leukemia 1 (ABL1)*Abelson murine leukemia 1 (ABL1) gene from chromosome 9 to the *breakpoint cluster region (BCR)*breakpoint cluster region (BCR) gene on chromosome 22, resulting in the generation of the *BCR-ABL1*BCR-ABL1 fusion oncogene that encodes the BCR-ABL1 oncoprotein [[3](#CR3)3].
|
| 61 |
+
|
| 62 |
+
The treatment of CML was revolutionized in 2001 with the introduction of imatinib, a tyrosine kinase inhibitor (TKI). This advancement transformed CML from a life-threatening illness into a manageable condition for most patients [[4](#CR4)4, [5](#CR5)5].
|
| 63 |
+
|
| 64 |
+
Despite this significant improvement in CML outcomes achieved with imatinib, some patients either experience disease progression during therapy or fail to respond adequately to the treatment [[6](#CR6)6].
|
| 65 |
+
|
| 66 |
+
Treatment failure with imatinib can be attributed to several mechanisms, including the upregulation of *BCR-ABL1*BCR-ABL1, the appearance of additional cytogenetic abnormalities, and mutations within the kinase domain of *BCR-ABL1*BCR-ABL1 [[7](#CR7)7]. Furthermore, variability in imatinib pharmacokinetics could influence its efficacy, as a correlation between imatinib exposure and clinical outcomes has been demonstrated [[8](#CR8)8–[10](#CR10)10].
|
| 67 |
+
|
| 68 |
+
Imatinib exhibits significant inter-patient variability in trough concentrations [[11](#CR11)11]. This variability may be attributed to differences in drug-metabolizing enzyme activity, the function of influx/efflux transporters [[12](#CR12)12], interaction with other drugs [[13](#CR13)13], and patient non-adherence [[14](#CR14)14].
|
| 69 |
+
|
| 70 |
+
It is predominantly metabolized into N-desmethyl imatinib (NDMI), by cytochrome P450 (CYP) 3A4 isozyme [[15](#CR15)15]. Furthermore, CYP2C8 is integral to imatinib metabolism, especially when CYP3A4 experiences auto-inhibition at imatinib’s steady state [[16](#CR16)16, [17](#CR17)17].
|
| 71 |
+
|
| 72 |
+
Other isozymes, including CYP1A2, CYP2D6, CYP2C9 and CYP2C19, also contribute to imatinib metabolism, but their roles are relatively minor [[18](#CR18)18].
|
| 73 |
+
|
| 74 |
+
Active efflux of imatinib from cells is carried out by adenosine triphosphate-binding cassette (ABC) transporters, notably ABCB1 and ABCG2. Conversely, the uptake of imatinib into cells is facilitated by the human organic cation transporter-1 (OCT1), also known as solute carrier family 22 member 1 (SLC22A1) [[19](#CR19)19].
|
| 75 |
+
|
| 76 |
+
Thus, genetic polymorphisms in the *CYP2C8*CYP2C8 and *ABCG2*ABCG2 genes are likely to affect intracellular or systemic concentrations of imatinib, potentially altering its therapeutic efficacy.
|
| 77 |
+
|
| 78 |
+
A notable gap in research existed regarding these polymorphisms in Egyptian CML patients. Therefore, this study aims to evaluate the influence of *CYP2C8*3*CYP2C8*3 (G416A; rs11572080) and *ABCG2*ABCG2 C421A (rs2231142) single nucleotide polymorphisms (SNPs) on plasma trough concentration and therapeutic response to imatinib in Egyptian patients with CML.
|
| 79 |
+
|
| 80 |
+
## Materials and methods
|
| 81 |
+
|
| 82 |
+
### Study design
|
| 83 |
+
|
| 84 |
+
This observational, cross-sectional study was conducted at the Medical Oncology Department of the South Egypt Cancer Institute (SECI), Assiut, Egypt.
|
| 85 |
+
|
| 86 |
+
The study protocol was reviewed and approved by the Ethics Committee of the Faculty of Medicine at Assuit University, Egypt (Institutional Review Board number: 17200117) and was registered in the ClinicalTrials.gov database (Identification Number: [NCT03262974](https://clinicaltrials.gov/ct2/show/NCT03262974)NCT03262974).
|
| 87 |
+
|
| 88 |
+
### Chemicals and kits
|
| 89 |
+
|
| 90 |
+
GeneJet Whole Blood Genomic DNA Purification Mini Kit (Thermo Scientific, Lithuania), COSMO PCR Master Mix (Willowfort, England), Forward and reverse primers (Macrogen, South Korea), Restriction enzymes: *Bse*BseRI and *Hpy*HpyCH4III (New England Biolabs, USA), Gel loading dye purple (6X) (New England Biolabs, USA), DNA ladder (GeneDireX, Taiwan) were purchased. Agarose powder (Bioline, USA), Tris base, Boric acid, ethylenediaminetetraacetic acid (EDTA) and ethidium bromide (Sigma-Aldrich, USA).
|
| 91 |
+
|
| 92 |
+
Imatinib mesylate (CAS No. 220127-57-1) and propranolol hydrochloride (CAS No. 318-98-9, the internal standard) were purchased from AK Scientific, Inc., USA. Acetonitrile and methanol (Sigma-Aldrich, Germany) were of HPLC grade. Water was purified by a Milli-Q Gradient A10 water purification system (Merck Millipore, USA).
|
| 93 |
+
|
| 94 |
+
### Patients
|
| 95 |
+
|
| 96 |
+
Patients were enrolled from the outpatient clinic of the Medical Oncology Department at the SECI, Egypt. The included patients had a diagnosis of Ph chromosome-positive CML, were aged over 18 years, and had been treated with imatinib for at least 12 months with good compliance to treatment. Exclusion criteria encompassed patients in accelerated or blastic phases, those taking medications that induce or inhibit liver microsomal enzymes (e.g., Ketoconazole, Phenytoin, and Valproic acid), individuals with poor compliance, and pregnant women. All patients provided informed consent before participation in the study.
|
| 97 |
+
|
| 98 |
+
Patient data, including age, sex, age at CML diagnosis, and *BCR-ABL1*BCR-ABL1 transcript levels at 12 months post-start of imatinib treatment, were collected during their regular follow-up visits.
|
| 99 |
+
|
| 100 |
+
The molecular response to imatinib was assessed using the International Scale, which quantifies the ratio of *BCR-ABL1*BCR-ABL1 transcripts to *ABL1*ABL1 transcripts, expressed as a percentage of *BCR-ABL1*BCR-ABL1 on a logarithmic scale. This scale corresponds to reductions of 2, 3, 4, 4.5, and 5 (equating to 1%, 0.1%, 0.01%, 0.0032%, and 0.001%, respectively) relative to the standardized baseline established in the International Randomized Study of Interferon versus STI571 (IRIS) [[20](#CR20)20].
|
| 101 |
+
|
| 102 |
+
In accordance with the European LeukemiaNet recommendations for CML treatment, treatment failure is defined as *BCR-ABL1*BCR-ABL1 levels exceeding 1% after 12 months of imatinib therapy [[21](#CR21)21].
|
| 103 |
+
|
| 104 |
+
Accordingly, patients in this study were classified into two groups based on their *BCR-ABL1*BCR-ABL1 transcript levels 12 months after initiating imatinib treatment: responders, with transcript levels of 1% or below, who continued imatinib therapy, and non-responders, with transcript levels above 1%, who were transitioned to second-generation TKIs such as nilotinib or dasatinib.
|
| 105 |
+
|
| 106 |
+
### Blood sampling
|
| 107 |
+
|
| 108 |
+
Peripheral blood samples were collected from patients into EDTA tubes during their routine follow-up visits at the outpatient clinic, concurrent with their regular laboratory tests.
|
| 109 |
+
|
| 110 |
+
Two milliliters were collected from all patients for DNA extraction and subsequent genotyping. An additional two milliliters were collected from patients receiving imatinib for at least 12 months, approximately 24 ± 3 h after their last dose but before the next dose (trough sample), following 28 consecutive days of administration [[9](#CR9)9].
|
| 111 |
+
|
| 112 |
+
Samples were then centrifuged at 2500 RPM (1000 ×*g*g) for 15 min [[22](#CR22)22] using a benchtop centrifuge (Rotofix 32 A, Hettich, Germany). Plasma samples obtained were stored at -80 °C until subsequent measurement of trough concentrations.
|
| 113 |
+
|
| 114 |
+
All blood samples were collected within the same timeframe; however, there was a lapse between the molecular response evaluation and the blood sample collection for each patient, which depended on the duration of their imatinib treatment.
|
| 115 |
+
|
| 116 |
+
### DNA extraction and genotyping
|
| 117 |
+
|
| 118 |
+
Whole blood DNA was extracted using a commercial extraction kit following the manufacturer’s instructions. The concentration and purity of the extracted DNA were assessed using a microplate spectrophotometer (Epoch, BioTek Instruments Inc., USA). Ratios of A_260_260/A_280_280 ranging from 1.7 to 1.9 indicate pure template DNA, optimal for PCR [[23](#CR23)23]. Extracted DNA samples were stored at -80 °C until genotyping analysis.
|
| 119 |
+
|
| 120 |
+
The *CYP2C8*3*CYP2C8*3 (G416A; rs11572080) [[24](#CR24)24] and *ABCG2*ABCG2 C421A (rs2231142) [[25](#CR25)25] polymorphisms were genotyped using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method, as previously described. Detailed primer sequences, PCR thermal conditions, and restriction enzymes used are provided in the Online Resource 1.
|
| 121 |
+
|
| 122 |
+
The PCR was performed using a thermal cycler (Veriti 96 Well Thermal Cycler, Applied Biosystems, USA). The resulting PCR products were then digested with appropriate restriction enzymes according to the manufacturer’s protocol.
|
| 123 |
+
|
| 124 |
+
The digested products were separated by electrophoresis on 3% agarose gel using a horizontal electrophoresis unit (Biometra Compact M, Analytik Jena, Germany) and visualized under ultraviolet (UV) light at 312 nm wavelength using a UV transilluminator (UVstar, Analytik Jena, Germany). Gel images were captured and documented using a gel documentation system (BioDocAnalyze, Analytik Jena, Germany). DNA extraction and PCR-RFLP analyses were carried out at the Medical Research Center, Assiut University, Assiut, Egypt.
|
| 125 |
+
|
| 126 |
+
### Measurement of plasma trough concentration of imatinib
|
| 127 |
+
|
| 128 |
+
Plasma trough concentrations were measured using the high-performance liquid chromatography with ultraviolet-diode array detection (HPLC-UV/DAD) method described previously [[26](#CR26)26]. The analysis was performed using a Waters 2695 Alliance Separations Module equipped with a 996 PDA detector (Waters Corporation, USA), with the detector wavelength set at 265 nm. Chromatographic separation was achieved on a C18 reversed-phase analytical column (Inertsil ODS 4 analytical column, 250 mm × 4.6 mm internal diameter, five µm particle size; GL Sciences, Japan) using isocratic elution. Chromatogram processing, data generation, and concentration calculations were conducted using Empower 3 chromatography data software (Waters Corporation, USA). These measurements were carried out at Nawah Scientific Inc., Cairo, Egypt.
|
| 129 |
+
|
| 130 |
+
### Statistical analysis
|
| 131 |
+
|
| 132 |
+
Possible deviation from Hardy–Weinberg equilibrium (HWE) of various genotypes was assessed using Chi-square test.
|
| 133 |
+
|
| 134 |
+
The Shapiro–Wilk test was employed to evaluate the normality of variable distributions. Continuous variables that followed a normal distribution were reported as mean ± standard deviation (SD), whereas those deviating from normality were presented as median and range. Categorical variables were reported in terms of frequency and percentage.
|
| 135 |
+
|
| 136 |
+
Fisher’s exact test was utilized to compare the frequencies of various genotypes between imatinib responders and non-responders. Additionally, the unpaired Student’s *t*t-test was applied to assess differences in plasma trough concentrations of imatinib across patients with different genotypes.
|
| 137 |
+
|
| 138 |
+
GraphPad Prism software version 9.5.1 (GraphPad Software Inc., USA) was used to perform statistical analyses and create graphical representations. Statistically significant differences were indicated by a *p*p-value of less than 0.05.
|
| 139 |
+
|
| 140 |
+
## Results
|
| 141 |
+
|
| 142 |
+
### Patients’ characteristics
|
| 143 |
+
|
| 144 |
+
Table [1](#Tab1)1 presents the demographic and clinical characteristics of the patients. A total of 50 Egyptian patients with Ph chromosome-positive CML in the chronic phase were included in the study. Of whom, 30 (60%) were females. The mean age of the patients at enrollment was 43.84 ± 13.30 years. At the time of CML diagnosis, the mean age was 38.48 ± 13.25 years. Twenty-six patients (52%) were responders to imatinib, with a mean treatment duration of 5.58 ± 2.16 years. The remaining 24 patients (48%) were non-responders.
|
| 145 |
+
|
| 146 |
+
### Table 1.
|
| 147 |
+
|
| 148 |
+
Demographic and clinical characteristics of Egyptian patients with CML (n = 50)
|
| 149 |
+
|
| 150 |
+
| Sex: | |
|
| 151 |
+
| ---- | - |
|
| 152 |
+
| - Male, n (%) - Female, n (%) | 20 (40%) 30 (60%) |
|
| 153 |
+
| Age (years), mean ± SD | 43.84 ± 13.30 |
|
| 154 |
+
| Age at diagnosis (years), mean ± SD | 38.48 ± 13.25 |
|
| 155 |
+
| Molecular response to imatinib at 12 months: | |
|
| 156 |
+
| - Responders (BCR-ABL1 transcript level ≤ 1%), n (%) - Non-responders (BCR-ABL1 transcript level > 1%), n (%) | 26 (52%) 24 (48%) |
|
| 157 |
+
| Duration of treatment with imatinib (years), mean ± SD | 5.58 ± 2.16 |
|
| 158 |
+
| BCR-ABL1transcript level at 12 month (%): | |
|
| 159 |
+
| - Responders, median (range) - Non-responders, median (range) | 0.2 (0-0.9) 30 (2-100) |
|
| 160 |
+
### Genotype and allele frequency
|
| 161 |
+
|
| 162 |
+
Table [2](#Tab2)2 presents the frequency of different genotypes and alleles of *CYP2C8*3*CYP2C8*3 G416A and *ABCG2*ABCG2 C421A polymorphisms in our patients. For the *CYP2C8*3*CYP2C8*3 G416A polymorphism, the frequencies of the GG (*CYP2C8*1/*1*CYP2C8*1/*1, homozygous wild type) and GA (*CYP2C8*1/*3*CYP2C8*1/*3, heterozygous type) genotypes were 76% and 24%, respectively. The frequency of the variant allele (A allele) was 12%. For the *ABCG2*ABCG2 C421A polymorphism, the frequencies of the CC (homozygous wild) and CA (heterozygous) genotypes was 78% and 22%, respectively. The frequency of the variant allele (A allele) was 11%. In the present study, we did not detect the AA (homozygous variant) genotype for either *CYP2C8*3*CYP2C8*3 G416A or *ABCG2*ABCG2 C421A polymorphisms. All reported frequencies did not significantly deviate from the HWE (*p >*p > 0.05).
|
| 163 |
+
|
| 164 |
+
### Table 2.
|
| 165 |
+
|
| 166 |
+
Genotype and allele frequencies of CYP2C8*3 and ABCG2 C421A polymorphisms among Egyptian patients with CML
|
| 167 |
+
|
| 168 |
+
| SNP | Genotype | Frequency (%) | Allele | Frequency (%) | HWEp-value |
|
| 169 |
+
| --- | -------- | ------------- | ------ | ------------- | ---------- |
|
| 170 |
+
| CYP2C8*3 (G416A; rs11572080) | GG | 76 | G | 88 | 0.3349 |
|
| 171 |
+
| GA | 24 | A | 12 | | |
|
| 172 |
+
| ABCG2 C421A (rs2231142) | CC | 78 | C | 89 | 0.3821 |
|
| 173 |
+
| CA | 22 | A | 11 | | |
|
| 174 |
+
### Influence of different genotypes on imatinib trough concentrations
|
| 175 |
+
|
| 176 |
+
It was observed that there was no statistically significant difference in the mean plasma trough concentration of imatinib in patients carrying different genotypes of the *CYP2C8*3*CYP2C8*3 polymorphism (GG: 1477 ± 459.8 vs. GA: 1270 ± 321.5; *p*p = 0.3995) (Fig. [1](#Fig1)1a).
|
| 177 |
+
|
| 178 |
+
### Fig. 1.
|
| 179 |
+
|
| 180 |
+
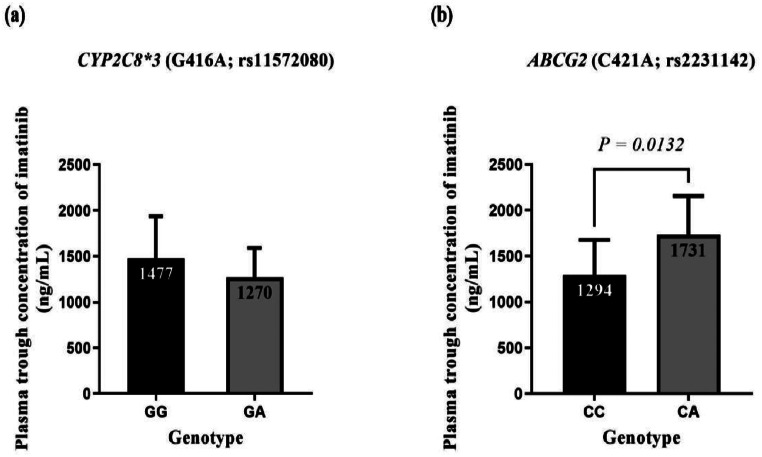
|
| 181 |
+
|
| 182 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11666798_280_2024_4723_Fig1_HTML.jpg)
|
| 183 |
+
|
| 184 |
+
Plasma trough concentrations of imatinib in different genotypes of CYP2C8*3 (a) and ABCG2 C421A (b) polymorphisms in CML patients
|
| 185 |
+
|
| 186 |
+
On the other hand, patients carrying the CA genotype of the *ABCG2*ABCG2 C421A polymorphism had a statistically significant higher mean plasma trough concentration of imatinib compared to those carrying the CC genotype (CA: 1731 ± 424.7 ng/mL vs. CC: 1294 ± 381.3 ng/mL; *p*p = 0.0132) (Fig. [1](#Fig1)1b).
|
| 187 |
+
|
| 188 |
+
Data are presented as mean ± SD. No statistically significant difference was found in the mean plasma trough concentration of imatinib between the different genotypes of *CYP2C8*3*CYP2C8*3 polymorphism (*p*p = 0.3995). A statistically significant difference was observed in the mean plasma trough concentration of imatinib between the CC and CA genotypes of *ABCG2*ABCG2 C421A polymorphism (*p*p = 0.0132). Results were compared using the unpaired Student’s *t*t-test.
|
| 189 |
+
|
| 190 |
+
### Influence of different genotypes on molecular response of imatinib
|
| 191 |
+
|
| 192 |
+
Regarding the genotypes of the *CYP2C8*3*CYP2C8*3 polymorphism, no statistically significant difference was observed in their distribution between responder and non-responder patients (*p*p = 0.1902) (Fig. [2](#Fig2)2a).
|
| 193 |
+
|
| 194 |
+
### Fig. 2.
|
| 195 |
+
|
| 196 |
+
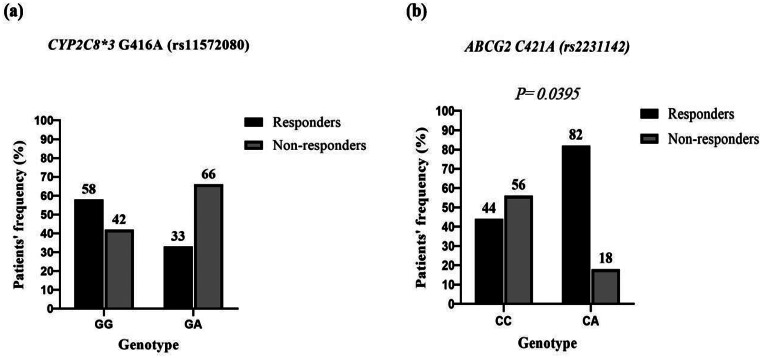
|
| 197 |
+
|
| 198 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11666798_280_2024_4723_Fig2_HTML.jpg)
|
| 199 |
+
|
| 200 |
+
Frequency of different genotypes of CYP2C8*3 (a) and ABCG2 C421A (b) polymorphisms in imatinib responders and non-responders
|
| 201 |
+
|
| 202 |
+
However, a statistically significant difference was noted between the two patient groups regarding the distribution of different genotypes of the *ABCG2*ABCG2 C421A polymorphism with predominance of the CA genotype in responder patients (*p*p = 0.0395) (Fig. [2](#Fig2)2b).
|
| 203 |
+
|
| 204 |
+
Assessment of molecular response to imatinib based on the *BCR-ABL1*BCR-ABL1 transcript level at 12 months. Responders (*n*n = 26); Non-responders (*n*n = 24). No statistically significant difference was found between the frequency of GG and GA genotypes of *CYP2C8*3*CYP2C8*3 in the two groups (*p*p = 0.1902). However, There was a statistically significant difference between the frequency of CC and CA genotypes of *ABCG2*ABCG2 C421A in the two groups (*p*p = 0.0395). Results were compared using Fisher’s exact test.
|
| 205 |
+
|
| 206 |
+
## Discussion
|
| 207 |
+
|
| 208 |
+
Despite the transformative impact of imatinib on the treatment of CML, a substantial proportion of CML patients (30–35%) exhibit inadequate response, either as suboptimal responders or as resistant to the treatment [[27](#CR27)27]. Various factors contribute to these outcomes [[28](#CR28)28], including genetic polymorphisms that impact drug metabolism and transport [[29](#CR29)29].
|
| 209 |
+
|
| 210 |
+
In this study, we focused on two key SNPs, *CYP2C8*3*CYP2C8*3 (G416A; rs11572080) and *ABCG2*ABCG2 C421A (rs2231142), which may influence imatinib pharmacokinetics. These genetic variations can alter plasma trough concentrations of imatinib and impact treatment outcomes in Egyptian patients with chronic-phase CML.
|
| 211 |
+
|
| 212 |
+
Over the past three decades, many methods have been developed for SNP genotyping, with PCR-RFLP being a widely used and cost-effective option [[30](#CR30)30]. In this study, we utilized PCR-RFLP to genotype target SNPs in all 50 patients. However, this technique has limitations, including its dependence on specific restriction enzymes to distinguish alleles, which can restrict its effectiveness when suitable enzymes are unavailable or show non-specific binding. Additionally, the multi-step process is time-intensive, increasing the risk of contamination and error, and is less efficient than high-throughput methods like real-time PCR and next-generation sequencing [[31](#CR31)31].
|
| 213 |
+
|
| 214 |
+
Many authors recommend HPLC with UV detection for measuring imatinib plasma concentrations, as it is cost-effective, straightforward, and suitable for routine laboratory use. Studies have also shown that HPLC-UV provides results comparable to those obtained with LC-MS/MS [[32](#CR32)32, [33](#CR33)33]. Additionally, incorporating a diode-array detector (DAD) into HPLC-UV systems improves specificity by allowing spectral comparisons and peak purity verification, making HPLC-UV/DAD sufficiently sensitive and specific for imatinib analysis [[34](#CR34)34].
|
| 215 |
+
|
| 216 |
+
The CYP2C8 enzyme, comprising 7% of the liver’s CYP content, is crucial for metabolizing about 20% of commonly prescribed medications. It is encoded by the *CYP2C8*CYP2C8 gene, situated on chromosome 10q24 [[35](#CR35)35].
|
| 217 |
+
|
| 218 |
+
This isozyme shows significant genetic diversity, with various SNPs identified, including *CYP2C8*2*CYP2C8*2, **3**3, and **4**4, which account for most of the non-synonymous variability of *CYP2C8*CYP2C8 in humans [[36](#CR36)36, [37](#CR37)37].
|
| 219 |
+
|
| 220 |
+
Among these SNPs, *CYP2C8*3*CYP2C8*3 stands out as the most extensively studied functional polymorphism. It encompasses two non-synonymous variants, G416A (Arg139Lys; rs11572080) and A1196G (Lys399Arg; rs10509681), often found in complete or near-complete linkage disequilibrium [[38](#CR38)38].
|
| 221 |
+
|
| 222 |
+
In vitro studies suggest that *CYP2C8*3*CYP2C8*3 is linked to enhanced enzyme activity, which may expedite the conversion of imatinib to NDMI [[11](#CR11)11, [39](#CR39)39].
|
| 223 |
+
|
| 224 |
+
However, in vivo studies indicate substrate-specific effects, including enhanced metabolism of drugs like pioglitazone and repaglinide, and reduced metabolism of ibuprofen [[40](#CR40)40].
|
| 225 |
+
|
| 226 |
+
In the context of our study on Egyptian CML patients, we did not observe a significant influence of the *CYP2C8*3*CYP2C8*3 polymorphism on imatinib’s plasma trough concentration or molecular response.
|
| 227 |
+
|
| 228 |
+
Nonetheless, we noted a trend towards lower imatinib concentrations in patients carrying the GA (*CYP2C*1/*3*CYP2C*1/*3) genotype, particularly prevalent among non-responders. These findings suggest a potential detrimental impact of this genotype on imatinib pharmacokinetics and clinical efficacy.
|
| 229 |
+
|
| 230 |
+
To our knowledge, this study is the first to investigate the impact of *CYP2C8*3*CYP2C8*3 (G416A; rs11572080) on imatinib therapy outcomes in Egyptian CML patients, highlighting the need for further validation through larger studies.
|
| 231 |
+
|
| 232 |
+
Results of the present study align with those of Verboom et al. [[41](#CR41)41], who similarly found no significant effect of the *CYP2C8*3*CYP2C8*3 on imatinib pharmacokinetics in Dutch patients with GIST or CML.
|
| 233 |
+
|
| 234 |
+
Conversely, studies by Barratt et al. [[42](#CR42)42] and Dalle Fratte et al. [[43](#CR43)43] reported increased metabolic ratios of NDMI/imatinib in CML and GIST patients carrying the *CYP2C8*3*CYP2C8*3 allele, contrasting with our findings. Further research in diverse populations and larger cohorts is crucial to elucidate the full spectrum of *CYP2C8*3’*CYP2C8*3’s impact on imatinib metabolism and treatment response.
|
| 235 |
+
|
| 236 |
+
The ABCG2, also known as the breast cancer resistance protein, belongs to the subfamily G of the ABC efflux transporter superfamily and mediates ATP-dependent efflux of diverse molecules across cell membranes [[44](#CR44)44]. This protein is encoded by the *ABCG2*ABCG2 gene, which is situated on chromosome 4q22.1 [[45](#CR45)45].
|
| 237 |
+
|
| 238 |
+
This transporter is extensively distributed in the human body. It operates in the apical membrane of enterocytes, where it restricts intestinal absorption; in the sinusoidal membrane of hepatocytes, where it facilitates hepatobiliary excretion; and in the apical membrane of proximal tubular cells in the kidney, where it contributes to uric acid elimination [[46](#CR46)46]. Furthermore, it significantly affects the pharmacokinetics of diverse compounds including anticancer drugs, antibiotics, antivirals and analgesics [[47](#CR47)47].
|
| 239 |
+
|
| 240 |
+
Numerous SNPs have been identified in the *ABCG2*ABCG2 gene, with C421A (rs2231142) in exon 5 being extensively studied. This SNP involves a substitution of glutamine with lysine at codon 141 (Q141K) [[48](#CR48)48].
|
| 241 |
+
|
| 242 |
+
In both in vitro and in vivo studies, this polymorphism was reported to generally reduce ABCG2 protein expression. Additionally, some studies indicate that it may also diminish ATPase activity, leading to compromised transport function [[49](#CR49)49].
|
| 243 |
+
|
| 244 |
+
Given its location on the apical membrane of hepatocytes, ABCG2 is likely essential for the excretion of imatinib. Consequently, genetic variations in the *ABCG2*ABCG2 gene may affect the pharmacokinetics and clinical response to imatinib [[18](#CR18)18]. However, the precise effect of the *ABCG2*ABCG2 C421A SNP on imatinib’s plasma trough concentration and therapeutic response remains a subject of debate.
|
| 245 |
+
|
| 246 |
+
In this study, we found that patients with the CA genotype of the *ABCG2*ABCG2 C421A polymorphism showed significantly higher plasma trough concentrations of imatinib compared to those with the CC genotype. Additionally, the CA genotype was more prevalent among responder patients, suggesting a potential role of this polymorphism in enhancing the therapeutic efficacy of imatinib.
|
| 247 |
+
|
| 248 |
+
Results of the present study align with those of Takahashi et al. [[50](#CR50)50], who reported higher imatinib trough concentrations in Japanese CML patients with the CA or AA genotypes compared to those with the CC genotype. However, they did not find a significant association between this SNP and treatment response.
|
| 249 |
+
|
| 250 |
+
Seong and his colleagues [[18](#CR18)18] identified a potential association between the ABCG2 C421A variant and an increased rate of major molecular response in Korean CML patients. However, they did not observe a significant effect on imatinib trough concentrations.
|
| 251 |
+
|
| 252 |
+
Jiang et al. [[51](#CR51)51] conducted a meta-analysis that included seven studies with nearly 2,200 patients, which further reinforced the association between the ABCG2 C421A variant allele and a higher clinical response rate.
|
| 253 |
+
|
| 254 |
+
Similarly, Alves et al. [[52](#CR52)52] observed that the CC genotype was associated with imatinib resistance, while the CA genotype was linked to a favorable response in Portuguese CML patients.
|
| 255 |
+
|
| 256 |
+
Findings of the current study are comparable with those of Cheng et al. [[53](#CR53)53], who examined the influence of the *ABCG2*ABCG2 C421A polymorphism on imatinib plasma concentration and response in 190 Chinese CML patients. They found that individuals with the CA or AA genotype exhibited higher imatinib concentrations and more favorable cytogenetic and molecular responses compared to those with the CC genotype. This finding supports our observation that the CA genotype is linked to higher plasma trough concentrations of imatinib and an improved therapeutic response.
|
| 257 |
+
|
| 258 |
+
In contrast, several studies have found no significant association between the *ABCG2*ABCG2 C421A SNP and imatinib response. For instance, Francis et al. [[54](#CR54)54] reported no significant impact of this SNP on imatinib trough concentration in Indian CML patients.
|
| 259 |
+
|
| 260 |
+
Similarly, Omran et al. [[55](#CR55)55] and Rajamani et al. [[5](#CR5)5] reported no significant influence of the ABCG2 C421A SNP on imatinib response in Egyptian and Indian CML patients, respectively. Additionally, Sabri et al. [[56](#CR56)56] found no significant association between ABCG2 gene expression and response to imatinib in their study on Egyptian CML patients.
|
| 261 |
+
|
| 262 |
+
Recent studies by Nouri et al. [[57](#CR57)57] and Mohammadi et al. [[58](#CR58)58] also concluded that the ABCG2 C421A SNP had no impact on imatinib response in Iranian CML patients.
|
| 263 |
+
|
| 264 |
+
The present study faced several limitations. The primary constraint was the small sample size, which might have hindered the identification of some genotypes within the patient population. Thus, large and multicenter studies are necessary. Furthermore, financial restrictions limited our investigation to a selected number of polymorphisms.
|
| 265 |
+
|
| 266 |
+
## Conclusion
|
| 267 |
+
|
| 268 |
+
The current study revealed that the *ABCG2*ABCG2 C421A (rs2231142) polymorphism significantly impacted both the plasma trough concentration and molecular response to imatinib in Egyptian patients with chronic-phase CML. Patients carrying the CA genotype showed higher plasma imatinib concentrations and more favorable treatment outcomes compared to those with the CC genotype. In contrast, the *CYP2C8*3*CYP2C8*3 (G416A; rs11572080) polymorphism did not significantly affect imatinib pharmacokinetics or clinical outcomes in the study population. These findings suggest that genotyping for the *ABCG2*ABCG2 C421A SNP could be a valuable tool in optimizing imatinib therapy for CML patients, allowing for more personalized treatment strategies.
|
| 269 |
+
|
| 270 |
+
## Electronic supplementary material
|
| 271 |
+
|
| 272 |
+
Below is the link to the electronic supplementary material.
|
| 273 |
+
|
| 274 |
+
## Author contributions
|
| 275 |
+
|
| 276 |
+
S.A.M., M.H.A., H.A.E., and A.M.R.H. conceptualized the study and supervised the work. H.A.E. and M.S.M. collected data and recruited patients. M.S.M. conducted genetic and HPLC analyses, performed statistical analysis, and wrote the original draft. All authors reviewed and approved the final manuscript and agreed to be accountable for all aspects of the work, ensuring its accuracy and integrity.
|
| 277 |
+
|
| 278 |
+
## Funding
|
| 279 |
+
|
| 280 |
+
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). Financial support for this study was partially provided by the Grant Office of the Faculty of Medicine at Assiut University, Egypt, under Grant Number 2017-08-30-024-R2.
|
| 281 |
+
|
| 282 |
+
## Data availability
|
| 283 |
+
|
| 284 |
+
No datasets were generated or analysed during the current study.
|
| 285 |
+
|
| 286 |
+
## Declarations
|
| 287 |
+
|
| 288 |
+
### Declaration of generative AI and AI-assisted technologies in the writing process
|
| 289 |
+
|
| 290 |
+
In the preparation of this manuscript, generative AI, specifically OpenAI’s ChatGPT, was utilized to support the writing process. ChatGPT was employed to assist with language refinement, editing, and the restructuring of certain passages to ensure clarity and adherence to scientific writing standards. The use of AI was guided by the authors, who provided all the intellectual content and made final decisions on the presentation and interpretation of the research findings. All AI-generated text was carefully reviewed and edited by the authors to align with the scientific rigor and standards expected of the manuscript. The final content of the manuscript is the result of the authors’ original work and expertise.
|
| 291 |
+
|
| 292 |
+
### Competing interests
|
| 293 |
+
|
| 294 |
+
The authors declare no competing interests.
|
| 295 |
+
|
| 296 |
+
## Footnotes
|
| 297 |
+
|
| 298 |
+
## Associated Data
|
| 299 |
+
|
| 300 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 301 |
+
|
| 302 |
+
### Supplementary Materials
|
| 303 |
+
|
| 304 |
+
### Data Availability Statement
|
| 305 |
+
|
| 306 |
+
No datasets were generated or analysed during the current study.
|
| 307 |
+
|
| 308 |
+
### Supplementary Materials
|
| 309 |
+
|
| 310 |
+
### Data Availability Statement
|
| 311 |
+
|
| 312 |
+
No datasets were generated or analysed during the current study.
|
| 313 |
+
|
| 314 |
+
## References
|
| 315 |
+
|
| 316 |
+
1. Cortes J, Pavlovsky C, Saußele S (2021) Chronic myeloid leukaemia. Lancet 398(10314):1914–1926. 10.1016/S0140-6736(21)01891-8 [DOI](https://doi.org/10.1016/S0140-6736(21)01204-6) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34425075/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Cortes%20J,%20Pavlovsky%20C,%20Sau%C3%9Fele%20S%20(2021)%20Chronic%20myeloid%20leukaemia.%20Lancet%20398(10314):1914%E2%80%931926.%2010.1016/S0140-6736(21)01891-8)
|
| 317 |
+
|
| 318 |
+
2. Kang ZJ, Liu YF, Xu LZ, Long ZJ, Huang D, Yang Y, Liu B, Feng JX, Pan YJ, Yan JS, Liu Q (2016) The Philadelphia chromosome in leukemogenesis. Chin J Cancer 35:48. 10.1186/s40880-016-0108-0 [DOI](https://doi.org/10.1186/s40880-016-0108-0) | [PMC free article](/articles/PMC4896164/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27233483/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Kang%20ZJ,%20Liu%20YF,%20Xu%20LZ,%20Long%20ZJ,%20Huang%20D,%20Yang%20Y,%20Liu%20B,%20Feng%20JX,%20Pan%20YJ,%20Yan%20JS,%20Liu%20Q%20(2016)%20The%20Philadelphia%20chromosome%20in%20leukemogenesis.%20Chin%20J%20Cancer%2035:48.%2010.1186/s40880-016-0108-0)
|
| 319 |
+
|
| 320 |
+
3. Kumar V, Singh P, Gupta SK, Ali V, Verma M (2022) Transport and metabolism of tyrosine kinase inhibitors associated with chronic myeloid leukemia therapy: a review. Mol Cell Biochem 477(4):1261–1279. 10.1007/s11010-022-04474-0 [DOI](https://doi.org/10.1007/s11010-022-04376-6) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35129779/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Kumar%20V,%20Singh%20P,%20Gupta%20SK,%20Ali%20V,%20Verma%20M%20(2022)%20Transport%20and%20metabolism%20of%20tyrosine%20kinase%20inhibitors%20associated%20with%20chronic%20myeloid%20leukemia%20therapy:%20a%20review.%20Mol%20Cell%20Biochem%20477(4):1261%E2%80%931279.%2010.1007/s11010-022-04474-0)
|
| 321 |
+
|
| 322 |
+
4. Harivenkatesh N, Kumar L, Bakhshi S, Sharma A, Kabra M, Velpandian T, Gogia A, Shastri SS, Biswas NR, Gupta YK (2017) Influence of MDR1 and CYP3A5 genetic polymorphisms on trough levels and therapeutic response of imatinib in newly diagnosed patients with chronic myeloid leukemia. Pharmacol Res 120:138–145. 10.1016/j.phrs.2017.03.003 [DOI](https://doi.org/10.1016/j.phrs.2017.03.011) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28330783/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Harivenkatesh%20N,%20Kumar%20L,%20Bakhshi%20S,%20Sharma%20A,%20Kabra%20M,%20Velpandian%20T,%20Gogia%20A,%20Shastri%20SS,%20Biswas%20NR,%20Gupta%20YK%20(2017)%20Influence%20of%20MDR1%20and%20CYP3A5%20genetic%20polymorphisms%20on%20trough%20levels%20and%20therapeutic%20response%20of%20imatinib%20in%20newly%20diagnosed%20patients%20with%20chronic%20myeloid%20leukemia.%20Pharmacol%20Res%20120:138%E2%80%93145.%2010.1016/j.phrs.2017.03.003)
|
| 323 |
+
|
| 324 |
+
5. Rajamani BM, Benjamin ES, Abraham A, Ganesan S, Lakshmi KM, Anandan S, Karathedath S, Varatharajan S, Mohanan E, Janet NB, Srivastava VM (2020) Plasma imatinib levels and ABCB1 polymorphism influences early molecular response and failure-free survival in newly diagnosed chronic phase CML patients. Sci Rep 10(1):20640. 10.1038/s41598-020-77691-6 [DOI](https://doi.org/10.1038/s41598-020-77140-9) | [PMC free article](/articles/PMC7691501/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33244077/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Rajamani%20BM,%20Benjamin%20ES,%20Abraham%20A,%20Ganesan%20S,%20Lakshmi%20KM,%20Anandan%20S,%20Karathedath%20S,%20Varatharajan%20S,%20Mohanan%20E,%20Janet%20NB,%20Srivastava%20VM%20(2020)%20Plasma%20imatinib%20levels%20and%20ABCB1%20polymorphism%20influences%20early%20molecular%20response%20and%20failure-free%20survival%20in%20newly%20diagnosed%20chronic%20phase%20CML%20patients.%20Sci%20Rep%2010(1):20640.%2010.1038/s41598-020-77691-6)
|
| 325 |
+
|
| 326 |
+
6. Kavanagh S, Nee A, Lipton JH (2018) Emerging alternatives to tyrosine kinase inhibitors for treating chronic myeloid leukemia. Expert Opin Emerg Drugs 23(1):51–62. 10.1080/14728214.2018.1400461 [DOI](https://doi.org/10.1080/14728214.2018.1445717) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29480034/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Kavanagh%20S,%20Nee%20A,%20Lipton%20JH%20(2018)%20Emerging%20alternatives%20to%20tyrosine%20kinase%20inhibitors%20for%20treating%20chronic%20myeloid%20leukemia.%20Expert%20Opin%20Emerg%20Drugs%2023(1):51%E2%80%9362.%2010.1080/14728214.2018.1400461)
|
| 327 |
+
|
| 328 |
+
7. Soverini S, Mancini M, Bavaro L, Cavo M, Martinelli G (2018) Chronic myeloid leukemia: the paradigm of targeting oncogenic tyrosine kinase signaling and counteracting resistance for successful cancer therapy. Mol Cancer 17:49. 10.1186/s12943-018-0808-x [DOI](https://doi.org/10.1186/s12943-018-0780-6) | [PMC free article](/articles/PMC5817796/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29455643/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Soverini%20S,%20Mancini%20M,%20Bavaro%20L,%20Cavo%20M,%20Martinelli%20G%20(2018)%20Chronic%20myeloid%20leukemia:%20the%20paradigm%20of%20targeting%20oncogenic%20tyrosine%20kinase%20signaling%20and%20counteracting%20resistance%20for%20successful%20cancer%20therapy.%20Mol%20Cancer%2017:49.%2010.1186/s12943-018-0808-x)
|
| 329 |
+
|
| 330 |
+
8. Larson RA, Druker BJ, Guilhot F, O’Brien SG, Riviere GJ, Krahnke T, Gathmann I, Wang Y (2008) Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia: a subanalysis of the IRIS study. Blood 111(8):4022–4028. 10.1182/blood-2007-10-116475 [DOI](https://doi.org/10.1182/blood-2007-10-116475) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18256322/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Larson%20RA,%20Druker%20BJ,%20Guilhot%20F,%20O%E2%80%99Brien%20SG,%20Riviere%20GJ,%20Krahnke%20T,%20Gathmann%20I,%20Wang%20Y%20(2008)%20Imatinib%20pharmacokinetics%20and%20its%20correlation%20with%20response%20and%20safety%20in%20chronic-phase%20chronic%20myeloid%20leukemia:%20a%20subanalysis%20of%20the%20IRIS%20study.%20Blood%20111(8):4022%E2%80%934028.%2010.1182/blood-2007-10-116475)
|
| 331 |
+
|
| 332 |
+
9. Guilhot F, Hughes TP, Cortes J, Druker BJ, Baccarani M, Gathmann I, Hayes M, Granvil C, Wang Y (2012) Plasma exposure of imatinib and its correlation with clinical response in the Tyrosine Kinase Inhibitor Optimization and selectivity trial. Haematologica 97(5):731–738. 10.3324/haematol.2011.056200 [DOI](https://doi.org/10.3324/haematol.2011.045666) | [PMC free article](/articles/PMC3342976/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22315495/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Guilhot%20F,%20Hughes%20TP,%20Cortes%20J,%20Druker%20BJ,%20Baccarani%20M,%20Gathmann%20I,%20Hayes%20M,%20Granvil%20C,%20Wang%20Y%20(2012)%20Plasma%20exposure%20of%20imatinib%20and%20its%20correlation%20with%20clinical%20response%20in%20the%20Tyrosine%20Kinase%20Inhibitor%20Optimization%20and%20selectivity%20trial.%20Haematologica%2097(5):731%E2%80%93738.%2010.3324/haematol.2011.056200)
|
| 333 |
+
|
| 334 |
+
10. Verheijen RB, Yu H, Schellens JH, Beijnen JH, Steeghs N, Huitema AD (2017) Practical recommendations for therapeutic drug monitoring of kinase inhibitors in oncology. Clin Pharmacol Ther 102(5):765–776. 10.1002/cpt.693 [DOI](https://doi.org/10.1002/cpt.787) | [PMC free article](/articles/PMC5656880/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28699160/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Verheijen%20RB,%20Yu%20H,%20Schellens%20JH,%20Beijnen%20JH,%20Steeghs%20N,%20Huitema%20AD%20(2017)%20Practical%20recommendations%20for%20therapeutic%20drug%20monitoring%20of%20kinase%20inhibitors%20in%20oncology.%20Clin%20Pharmacol%20Ther%20102(5):765%E2%80%93776.%2010.1002/cpt.693)
|
| 335 |
+
|
| 336 |
+
11. Farag S, Verheijen RB, Martijn Kerst J, Cats A, Huitema AD, Steeghs N (2017) Imatinib pharmacokinetics in a large observational cohort of gastrointestinal stromal tumour patients. Clin Pharmacokinet 56:287–292. 10.1007/s40262-016-0442-2 [DOI](https://doi.org/10.1007/s40262-016-0439-7) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27435281/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Farag%20S,%20Verheijen%20RB,%20Martijn%20Kerst%20J,%20Cats%20A,%20Huitema%20AD,%20Steeghs%20N%20(2017)%20Imatinib%20pharmacokinetics%20in%20a%20large%20observational%20cohort%20of%20gastrointestinal%20stromal%20tumour%20patients.%20Clin%20Pharmacokinet%2056:287%E2%80%93292.%2010.1007/s40262-016-0442-2)
|
| 337 |
+
|
| 338 |
+
12. Barratt DT, Somogyi AA (2017) Role of pharmacogenetics in personalised imatinib dosing. Transl Cancer Res 6(Suppl 10). 10.21037/tcr.2017.06.01. S1541-S1557
|
| 339 |
+
|
| 340 |
+
13. Shao J, Markowitz JS, Bei D, An G (2014) Enzyme-transporter-mediated drug interactions with small molecule tyrosine kinase inhibitors. J Pharm Sci 103(12):3810–3833. 10.1002/jps.24126 [DOI](https://doi.org/10.1002/jps.24113) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25308414/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Shao%20J,%20Markowitz%20JS,%20Bei%20D,%20An%20G%20(2014)%20Enzyme-transporter-mediated%20drug%20interactions%20with%20small%20molecule%20tyrosine%20kinase%20inhibitors.%20J%20Pharm%20Sci%20103(12):3810%E2%80%933833.%2010.1002/jps.24126)
|
| 341 |
+
|
| 342 |
+
14. Lankheet NA, Desar IM, Mulder SF, Burger DM, Kweekel DM, van Herpen CM, van der Graaf WT, van Erp NP (2017) Optimizing the dose in cancer patients treated with imatinib, sunitinib and pazopanib. Br J Clin Pharmacol 83(10):2195–2204. 10.1111/bcp.13328 [DOI](https://doi.org/10.1111/bcp.13327) | [PMC free article](/articles/PMC5595974/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28500677/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Lankheet%20NA,%20Desar%20IM,%20Mulder%20SF,%20Burger%20DM,%20Kweekel%20DM,%20van%20Herpen%20CM,%20van%20der%20Graaf%20WT,%20van%20Erp%20NP%20(2017)%20Optimizing%20the%20dose%20in%20cancer%20patients%20treated%20with%20imatinib,%20sunitinib%20and%20pazopanib.%20Br%20J%20Clin%20Pharmacol%2083(10):2195%E2%80%932204.%2010.1111/bcp.13328)
|
| 343 |
+
|
| 344 |
+
15. Filppula AM, Laitila J, Neuvonen PJ, Backman JT (2012) Potent mechanism-based inhibition of CYP3A4 by Imatinib explains its liability to interact with CYP3A4 substrates. Br J Pharmacol 165(8):2787–2798. 10.1111/j.1476-5381.2011.01717.x [DOI](https://doi.org/10.1111/j.1476-5381.2011.01732.x) | [PMC free article](/articles/PMC3423228/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22014153/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Filppula%20AM,%20Laitila%20J,%20Neuvonen%20PJ,%20Backman%20JT%20(2012)%20Potent%20mechanism-based%20inhibition%20of%20CYP3A4%20by%20Imatinib%20explains%20its%20liability%20to%20interact%20with%20CYP3A4%20substrates.%20Br%20J%20Pharmacol%20165(8):2787%E2%80%932798.%2010.1111/j.1476-5381.2011.01717.x)
|
| 345 |
+
|
| 346 |
+
16. Nebot N, Crettol S, d’Esposito F, Tattam B, Hibbs DE, Murray M (2010) Participation of CYP2C8 and CYP3A4 in the N-demethylation of imatinib in human hepatic microsomes. Br J Pharmacol 161(5):1059–1069. 10.1111/j.1476-5381.2010.00924.x [DOI](https://doi.org/10.1111/j.1476-5381.2010.00946.x) | [PMC free article](/articles/PMC2998687/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20977456/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Nebot%20N,%20Crettol%20S,%20d%E2%80%99Esposito%20F,%20Tattam%20B,%20Hibbs%20DE,%20Murray%20M%20(2010)%20Participation%20of%20CYP2C8%20and%20CYP3A4%20in%20the%20N-demethylation%20of%20imatinib%20in%20human%20hepatic%20microsomes.%20Br%20J%20Pharmacol%20161(5):1059%E2%80%931069.%2010.1111/j.1476-5381.2010.00924.x)
|
| 347 |
+
|
| 348 |
+
17. Filppula AM, Neuvonen M, Laitila J, Neuvonen PJ, Backman JT (2013) Autoinhibition of CYP3A4 leads to important role of CYP2C8 in imatinib metabolism: variability in CYP2C8 activity may alter plasma concentrations and response. Drug Metab Dispos 41(1):50–59. 10.1124/dmd.112.046094 [DOI](https://doi.org/10.1124/dmd.112.048017) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23028140/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Filppula%20AM,%20Neuvonen%20M,%20Laitila%20J,%20Neuvonen%20PJ,%20Backman%20JT%20(2013)%20Autoinhibition%20of%20CYP3A4%20leads%20to%20important%20role%20of%20CYP2C8%20in%20imatinib%20metabolism:%20variability%20in%20CYP2C8%20activity%20may%20alter%20plasma%20concentrations%20and%20response.%20Drug%20Metab%20Dispos%2041(1):50%E2%80%9359.%2010.1124/dmd.112.046094)
|
| 349 |
+
|
| 350 |
+
18. Seong SJ, Lim M, Sohn SK, Moon JH, Oh SJ, Kim BS, Ryoo HM, Chung JS, Joo YD, Bang SM, Jung CW (2013) Influence of enzyme and transporter polymorphisms on trough imatinib concentration and clinical response in chronic myeloid leukemia patients. Ann Oncol 24(3):756–760. 10.1093/annonc/mds501 [DOI](https://doi.org/10.1093/annonc/mds532) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23117072/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Seong%20SJ,%20Lim%20M,%20Sohn%20SK,%20Moon%20JH,%20Oh%20SJ,%20Kim%20BS,%20Ryoo%20HM,%20Chung%20JS,%20Joo%20YD,%20Bang%20SM,%20Jung%20CW%20(2013)%20Influence%20of%20enzyme%20and%20transporter%20polymorphisms%20on%20trough%20imatinib%20concentration%20and%20clinical%20response%20in%20chronic%20myeloid%20leukemia%20patients.%20Ann%20Oncol%2024(3):756%E2%80%93760.%2010.1093/annonc/mds501)
|
| 351 |
+
|
| 352 |
+
19. Peng B, Lloyd P, Schran H (2005) Clinical pharmacokinetics of imatinib. Clin Pharmacokinet 44:879–894. 10.2165/00003088-200544080-00002 [DOI](https://doi.org/10.2165/00003088-200544090-00001) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16122278/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Peng%20B,%20Lloyd%20P,%20Schran%20H%20(2005)%20Clinical%20pharmacokinetics%20of%20imatinib.%20Clin%20Pharmacokinet%2044:879%E2%80%93894.%2010.2165/00003088-200544080-00002)
|
| 353 |
+
|
| 354 |
+
20. Cross NC, White HE, Müller MC, Saglio G, Hochhaus A (2012) Standardized definitions of molecular response in chronic myeloid leukemia. Leukemia 26(10):2172–2175. 10.1038/leu.2012.104 [DOI](https://doi.org/10.1038/leu.2012.104) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22504141/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Cross%20NC,%20White%20HE,%20M%C3%BCller%20MC,%20Saglio%20G,%20Hochhaus%20A%20(2012)%20Standardized%20definitions%20of%20molecular%20response%20in%20chronic%20myeloid%20leukemia.%20Leukemia%2026(10):2172%E2%80%932175.%2010.1038/leu.2012.104)
|
| 355 |
+
|
| 356 |
+
21. Hochhaus A, Baccarani M, Silver RT, Schiffer C, Apperley JF, Cervantes F, Clark RE, Cortes JE, Deininger MW, Guilhot F, Hjorth-Hansen H (2020) European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 34(4):966–984. 10.1038/s41375-020-0776-2 [DOI](https://doi.org/10.1038/s41375-020-0776-2) | [PMC free article](/articles/PMC7214240/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32127639/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Hochhaus%20A,%20Baccarani%20M,%20Silver%20RT,%20Schiffer%20C,%20Apperley%20JF,%20Cervantes%20F,%20Clark%20RE,%20Cortes%20JE,%20Deininger%20MW,%20Guilhot%20F,%20Hjorth-Hansen%20H%20(2020)%20European%20LeukemiaNet%202020%20recommendations%20for%20treating%20chronic%20myeloid%20leukemia.%20Leukemia%2034(4):966%E2%80%93984.%2010.1038/s41375-020-0776-2)
|
| 357 |
+
|
| 358 |
+
22. Malhotra H, Sharma P, Bhargava S, Rathore OS, Malhotra B, Kumar M (2014) Correlation of plasma trough levels of imatinib with molecular response in patients with chronic myeloid leukemia. Leuk Lymphoma 55(11):2614–2619. 10.3109/10428194.2014.898748 [DOI](https://doi.org/10.3109/10428194.2014.885515) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24446903/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Malhotra%20H,%20Sharma%20P,%20Bhargava%20S,%20Rathore%20OS,%20Malhotra%20B,%20Kumar%20M%20(2014)%20Correlation%20of%20plasma%20trough%20levels%20of%20imatinib%20with%20molecular%20response%20in%20patients%20with%20chronic%20myeloid%20leukemia.%20Leuk%20Lymphoma%2055(11):2614%E2%80%932619.%2010.3109/10428194.2014.898748)
|
| 359 |
+
|
| 360 |
+
23. Bruijns B, Hoekema T, Oomens L, Tiggelaar R, Gardeniers H (2022) Performance of spectrophotometric and fluorometric DNA quantification methods. Analytica 3(3):371–384. 10.3390/analytica3030031 [Google Scholar](https://scholar.google.com/scholar_lookup?Bruijns%20B,%20Hoekema%20T,%20Oomens%20L,%20Tiggelaar%20R,%20Gardeniers%20H%20(2022)%20Performance%20of%20spectrophotometric%20and%20fluorometric%20DNA%20quantification%20methods.%20Analytica%203(3):371%E2%80%93384.%2010.3390/analytica3030031)
|
| 361 |
+
|
| 362 |
+
24. Pechandova K, Buzkova H, Matouskova O, Perlik F, Slanar O (2012) Genetic polymorphisms of CYP2C8 in the Czech Republic. Genet Test Mol Biomarkers 16(7):812–816. 10.1089/gtmb.2011.0332 [DOI](https://doi.org/10.1089/gtmb.2011.0275) | [PMC free article](/articles/PMC3396005/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22313047/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Pechandova%20K,%20Buzkova%20H,%20Matouskova%20O,%20Perlik%20F,%20Slanar%20O%20(2012)%20Genetic%20polymorphisms%20of%20CYP2C8%20in%20the%20Czech%20Republic.%20Genet%20Test%20Mol%20Biomarkers%2016(7):812%E2%80%93816.%2010.1089/gtmb.2011.0332)
|
| 363 |
+
|
| 364 |
+
25. El Mesallamy HO, Rashed WM, Hamdy NM, Hamdy N (2014) High-dose methotrexate in Egyptian pediatric acute lymphoblastic leukemia: the impact of ABCG2 C421A genetic polymorphism on plasma levels, what is next? J Cancer Res Clin Oncol 140:1359–1365. 10.1007/s00432-014-1695-1 [DOI](https://doi.org/10.1007/s00432-014-1670-y) | [PMC free article](/articles/PMC11823488/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24718721/) | [Google Scholar](https://scholar.google.com/scholar_lookup?El%20Mesallamy%20HO,%20Rashed%20WM,%20Hamdy%20NM,%20Hamdy%20N%20(2014)%20High-dose%20methotrexate%20in%20Egyptian%20pediatric%20acute%20lymphoblastic%20leukemia:%20the%20impact%20of%20ABCG2%20C421A%20genetic%20polymorphism%20on%20plasma%20levels,%20what%20is%20next?%20J%20Cancer%20Res%20Clin%20Oncol%20140:1359%E2%80%931365.%2010.1007/s00432-014-1695-1)
|
| 365 |
+
|
| 366 |
+
26. Kaza M, Piorkowska E, Filist M, Rudzki PJ (2016) HPLC-UV assay of imatinib in human plasma optimized for bioequivalence studies. Acta Pol Pharm 73(6):1495–1503 [PubMed](https://pubmed.ncbi.nlm.nih.gov/29634103/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Kaza%20M,%20Piorkowska%20E,%20Filist%20M,%20Rudzki%20PJ%20(2016)%20HPLC-UV%20assay%20of%20imatinib%20in%20human%20plasma%20optimized%20for%20bioequivalence%20studies.%20Acta%20Pol%20Pharm%2073(6):1495%E2%80%931503)
|
| 367 |
+
|
| 368 |
+
27. Ankathil R, Azlan H, Dzarr AA, Baba AA (2018) Pharmacogenetics and the treatment of chronic myeloid leukemia: how relevant clinically? An update. Pharmacogenomics 19(5):475–493. 10.2217/pgs-2018-0017 [DOI](https://doi.org/10.2217/pgs-2017-0193) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29569526/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Ankathil%20R,%20Azlan%20H,%20Dzarr%20AA,%20Baba%20AA%20(2018)%20Pharmacogenetics%20and%20the%20treatment%20of%20chronic%20myeloid%20leukemia:%20how%20relevant%20clinically?%20An%20update.%20Pharmacogenomics%2019(5):475%E2%80%93493.%2010.2217/pgs-2018-0017)
|
| 369 |
+
|
| 370 |
+
28. Volpe G, Panuzzo C, Ulisciani S, Cilloni D (2009) Imatinib resistance in CML. Cancer Lett 274(1):1–9. 10.1016/j.canlet.2008.08.002 [DOI](https://doi.org/10.1016/j.canlet.2008.06.003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18653275/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Volpe%20G,%20Panuzzo%20C,%20Ulisciani%20S,%20Cilloni%20D%20(2009)%20Imatinib%20resistance%20in%20CML.%20Cancer%20Lett%20274(1):1%E2%80%939.%2010.1016/j.canlet.2008.08.002)
|
| 371 |
+
|
| 372 |
+
29. Angelini S, Soverini S, Ravegnini G, Barnett M, Turrini E, Thornquist M, Pane F, Hughes TP, White DL, Radich J, Kim DW (2013) Association between Imatinib transporters and metabolizing enzymes genotype and response in newly diagnosed chronic myeloid leukemia patients receiving imatinib therapy. Haematologica 98(2):193–200. 10.3324/haematol.2012.068676 [DOI](https://doi.org/10.3324/haematol.2012.066480) | [PMC free article](/articles/PMC3561425/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22875622/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Angelini%20S,%20Soverini%20S,%20Ravegnini%20G,%20Barnett%20M,%20Turrini%20E,%20Thornquist%20M,%20Pane%20F,%20Hughes%20TP,%20White%20DL,%20Radich%20J,%20Kim%20DW%20(2013)%20Association%20between%20Imatinib%20transporters%20and%20metabolizing%20enzymes%20genotype%20and%20response%20in%20newly%20diagnosed%20chronic%20myeloid%20leukemia%20patients%20receiving%20imatinib%20therapy.%20Haematologica%2098(2):193%E2%80%93200.%2010.3324/haematol.2012.068676)
|
| 373 |
+
|
| 374 |
+
30. Kratochwil CF, Kautt AF, Rometsch SJ, Meyer A (2022) Benefits and limitations of a new genome-based PCR‐RFLP genotyping assay (GB‐RFLP): a SNP‐based detection method for identification of species in extremely young adaptive radiations. Ecol Evol 12(3):e8751. 10.1002/ece3.8751 [DOI](https://doi.org/10.1002/ece3.8751) | [PMC free article](/articles/PMC8941502/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35356554/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Kratochwil%20CF,%20Kautt%20AF,%20Rometsch%20SJ,%20Meyer%20A%20(2022)%20Benefits%20and%20limitations%20of%20a%20new%20genome-based%20PCR%E2%80%90RFLP%20genotyping%20assay%20(GB%E2%80%90RFLP):%20a%20SNP%E2%80%90based%20detection%20method%20for%20identification%20of%20species%20in%20extremely%20young%20adaptive%20radiations.%20Ecol%20Evol%2012(3):e8751.%2010.1002/ece3.8751)
|
| 375 |
+
|
| 376 |
+
31. Jordan D, Mills D (2021) Past, present, and future of DNA typing for analyzing human and non-human forensic samples. Front Ecol Evol 9:646130. 10.3389/fevo.2021.646130 [Google Scholar](https://scholar.google.com/scholar_lookup?Jordan%20D,%20Mills%20D%20(2021)%20Past,%20present,%20and%20future%20of%20DNA%20typing%20for%20analyzing%20human%20and%20non-human%20forensic%20samples.%20Front%20Ecol%20Evol%209:646130.%2010.3389/fevo.2021.646130)
|
| 377 |
+
|
| 378 |
+
32. Caterino M, Casadei GM, Arvonio R, De Francia S, Pirro E, Piccione FM, Pane F, Ruoppolo M (2013) Quantification of imatinib plasma levels in patients with chronic myeloid leukemia: comparison between HPLC–UV and LC–MS/MS. Int J Pept Res Ther 19:109–116. 10.1007/s10989-012-9321-0 [Google Scholar](https://scholar.google.com/scholar_lookup?Caterino%20M,%20Casadei%20GM,%20Arvonio%20R,%20De%20Francia%20S,%20Pirro%20E,%20Piccione%20FM,%20Pane%20F,%20Ruoppolo%20M%20(2013)%20Quantification%20of%20imatinib%20plasma%20levels%20in%20patients%20with%20chronic%20myeloid%20leukemia:%20comparison%20between%20HPLC%E2%80%93UV%20and%20LC%E2%80%93MS/MS.%20Int%20J%20Pept%20Res%20Ther%2019:109%E2%80%93116.%2010.1007/s10989-012-9321-0)
|
| 379 |
+
|
| 380 |
+
33. Miura M, Takahashi N (2016) Routine therapeutic drug monitoring of tyrosine kinase inhibitors by HPLC–UV or LC–MS/MS methods. Drug Metab Pharmacokinet 31(1):12–20. 10.1016/j.dmpk.2015.09.002 [DOI](https://doi.org/10.1016/j.dmpk.2015.09.002) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26732608/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Miura%20M,%20Takahashi%20N%20(2016)%20Routine%20therapeutic%20drug%20monitoring%20of%20tyrosine%20kinase%20inhibitors%20by%20HPLC%E2%80%93UV%20or%20LC%E2%80%93MS/MS%20methods.%20Drug%20Metab%20Pharmacokinet%2031(1):12%E2%80%9320.%2010.1016/j.dmpk.2015.09.002)
|
| 381 |
+
|
| 382 |
+
34. Roth O, Spreux-Varoquaux O, Bouchet S, Rousselot P, Castaigne S, Rigaudeau S, Raggueneau V, Therond P, Devillier P, Molimard M, Maneglier B (2010) Imatinib assay by HPLC with photodiode-array UV detection in plasma from patients with chronic myeloid leukemia: comparison with LC-MS/MS. Clin Chim Acta 411(3–4):140–146. 10.1016/j.cca.2009.10.007 [DOI](https://doi.org/10.1016/j.cca.2009.10.007) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19853594/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Roth%20O,%20Spreux-Varoquaux%20O,%20Bouchet%20S,%20Rousselot%20P,%20Castaigne%20S,%20Rigaudeau%20S,%20Raggueneau%20V,%20Therond%20P,%20Devillier%20P,%20Molimard%20M,%20Maneglier%20B%20(2010)%20Imatinib%20assay%20by%20HPLC%20with%20photodiode-array%20UV%20detection%20in%20plasma%20from%20patients%20with%20chronic%20myeloid%20leukemia:%20comparison%20with%20LC-MS/MS.%20Clin%20Chim%20Acta%20411(3%E2%80%934):140%E2%80%93146.%2010.1016/j.cca.2009.10.007)
|
| 383 |
+
|
| 384 |
+
35. Aquilante CL, Niemi M, Gong L, Altman RB, Klein TE (2013) PharmGKB summary: very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 8. Pharmacogenet Genomics 23(12):721–728. 10.1097/FPC.0000000000000004 [DOI](https://doi.org/10.1097/FPC.0b013e3283653b27) | [PMC free article](/articles/PMC4038626/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23962911/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Aquilante%20CL,%20Niemi%20M,%20Gong%20L,%20Altman%20RB,%20Klein%20TE%20(2013)%20PharmGKB%20summary:%20very%20important%20pharmacogene%20information%20for%20cytochrome%20P450,%20family%202,%20subfamily%20C,%20polypeptide%208.%20Pharmacogenet%20Genomics%2023(12):721%E2%80%93728.%2010.1097/FPC.0000000000000004)
|
| 385 |
+
|
| 386 |
+
36. Zhou Y, Ingelman-Sundberg M, Lauschke VM (2017) Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther 102(4):688–700. 10.1002/cpt.685 [DOI](https://doi.org/10.1002/cpt.690) | [PMC free article](/articles/PMC5600063/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28378927/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Zhou%20Y,%20Ingelman-Sundberg%20M,%20Lauschke%20VM%20(2017)%20Worldwide%20distribution%20of%20cytochrome%20P450%20alleles:%20a%20meta-analysis%20of%20population-scale%20sequencing%20projects.%20Clin%20Pharmacol%20Ther%20102(4):688%E2%80%93700.%2010.1002/cpt.685)
|
| 387 |
+
|
| 388 |
+
37. Abudahab S, Hakooz N, Tobeh N, Gogazeh E, Gharaibeh M, Al-Eitan L, Zihlif M, Dajani R (2022) Variability of CYP2C8 polymorphisms in three Jordanian populations: circassians, chechens, and Jordanian-Arabs. J Immigr Minor Health 24(5):1167–1176. 10.1007/s10903-021-01275-2 [DOI](https://doi.org/10.1007/s10903-021-01264-x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34448113/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Abudahab%20S,%20Hakooz%20N,%20Tobeh%20N,%20Gogazeh%20E,%20Gharaibeh%20M,%20Al-Eitan%20L,%20Zihlif%20M,%20Dajani%20R%20(2022)%20Variability%20of%20CYP2C8%20polymorphisms%20in%20three%20Jordanian%20populations:%20circassians,%20chechens,%20and%20Jordanian-Arabs.%20J%20Immigr%20Minor%20Health%2024(5):1167%E2%80%931176.%2010.1007/s10903-021-01275-2)
|
| 389 |
+
|
| 390 |
+
38. Tornio A, Backman JT (2018) Cytochrome P450 in pharmacogenetics: an update. Adv Pharmacol 83:3–32. 10.1016/bs.apha.2018.03.001 [DOI](https://doi.org/10.1016/bs.apha.2018.04.007) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29801580/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Tornio%20A,%20Backman%20JT%20(2018)%20Cytochrome%20P450%20in%20pharmacogenetics:%20an%20update.%20Adv%20Pharmacol%2083:3%E2%80%9332.%2010.1016/bs.apha.2018.03.001)
|
| 391 |
+
|
| 392 |
+
39. Khan MS, Barratt DT, Somogyi AA (2016) Impact of CYP2C8*3 polymorphism on in vitro metabolism of imatinib to N-desmethyl imatinib. Xenobiotica 46(3):278–287. 10.3109/00498254.2015.1060367 [DOI](https://doi.org/10.3109/00498254.2015.1060649) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26161459/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Khan%20MS,%20Barratt%20DT,%20Somogyi%20AA%20(2016)%20Impact%20of%20CYP2C8*3%20polymorphism%20on%20in%20vitro%20metabolism%20of%20imatinib%20to%20N-desmethyl%20imatinib.%20Xenobiotica%2046(3):278%E2%80%93287.%2010.3109/00498254.2015.1060367)
|
| 393 |
+
|
| 394 |
+
40. Aquilante CL, Kosmiski LA, Bourne DW, Bushman LR, Daily EB, Hammond KP, Hopley CW, Kadam RS, Kanack AT, Kompella UB, Le M (2013) Impact of the CYP2C8*3 polymorphism on the drug–drug interaction between gemfibrozil and pioglitazone. Br J Clin Pharmacol 75(1):217–226. 10.1111/j.1365-2125.2012.04328.x [DOI](https://doi.org/10.1111/j.1365-2125.2012.04343.x) | [PMC free article](/articles/PMC3555061/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22625877/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Aquilante%20CL,%20Kosmiski%20LA,%20Bourne%20DW,%20Bushman%20LR,%20Daily%20EB,%20Hammond%20KP,%20Hopley%20CW,%20Kadam%20RS,%20Kanack%20AT,%20Kompella%20UB,%20Le%20M%20(2013)%20Impact%20of%20the%20CYP2C8*3%20polymorphism%20on%20the%20drug%E2%80%93drug%20interaction%20between%20gemfibrozil%20and%20pioglitazone.%20Br%20J%20Clin%20Pharmacol%2075(1):217%E2%80%93226.%2010.1111/j.1365-2125.2012.04328.x)
|
| 395 |
+
|
| 396 |
+
41. Verboom MC, Visser L, Kouwen S, Swen JJ, Diepstraten J, Posthuma WF, Gelderblom H, Van Lammeren D, Wilms EB (2017) Influence of CYP2C8 polymorphisms on imatinib steady-state trough level in chronic myeloid leukemia and gastrointestinal stromal tumor patients. Pharmacogenet Genomics 27(6):223–226. 10.1097/FPC.0000000000000282 [DOI](https://doi.org/10.1097/FPC.0000000000000278) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28383355/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Verboom%20MC,%20Visser%20L,%20Kouwen%20S,%20Swen%20JJ,%20Diepstraten%20J,%20Posthuma%20WF,%20Gelderblom%20H,%20Van%20Lammeren%20D,%20Wilms%20EB%20(2017)%20Influence%20of%20CYP2C8%20polymorphisms%20on%20imatinib%20steady-state%20trough%20level%20in%20chronic%20myeloid%20leukemia%20and%20gastrointestinal%20stromal%20tumor%20patients.%20Pharmacogenet%20Genomics%2027(6):223%E2%80%93226.%2010.1097/FPC.0000000000000282)
|
| 397 |
+
|
| 398 |
+
42. Barratt DT, Cox HK, Menelaou A, Yeung DT, White DL, Hughes TP, Somogyi AA (2017) CYP2C8 genotype significantly alters imatinib metabolism in chronic myeloid leukemia patients. Clin Pharmacokinet 56:977–985. 10.1007/s40262-016-0505-y [DOI](https://doi.org/10.1007/s40262-016-0494-0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27995529/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Barratt%20DT,%20Cox%20HK,%20Menelaou%20A,%20Yeung%20DT,%20White%20DL,%20Hughes%20TP,%20Somogyi%20AA%20(2017)%20CYP2C8%20genotype%20significantly%20alters%20imatinib%20metabolism%20in%20chronic%20myeloid%20leukemia%20patients.%20Clin%20Pharmacokinet%2056:977%E2%80%93985.%2010.1007/s40262-016-0505-y)
|
| 399 |
+
|
| 400 |
+
43. Dalle Fratte C, Gagno S, Roncato R, Polesel J, Zanchetta M, Buzzo M, Posocco B, De Mattia E, Borsatti R, Puglisi F, Foltran L (2023) CYP2D6 and CYP2C8 pharmacogenetics and pharmacological interactions to predict imatinib plasmatic exposure in GIST patients. Br J Clin Pharmacol 89(3):1089–1098. 10.1111/bcp.15572 [DOI](https://doi.org/10.1111/bcp.15551) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36178950/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Dalle%20Fratte%20C,%20Gagno%20S,%20Roncato%20R,%20Polesel%20J,%20Zanchetta%20M,%20Buzzo%20M,%20Posocco%20B,%20De%20Mattia%20E,%20Borsatti%20R,%20Puglisi%20F,%20Foltran%20L%20(2023)%20CYP2D6%20and%20CYP2C8%20pharmacogenetics%20and%20pharmacological%20interactions%20to%20predict%20imatinib%20plasmatic%20exposure%20in%20GIST%20patients.%20Br%20J%20Clin%20Pharmacol%2089(3):1089%E2%80%931098.%2010.1111/bcp.15572)
|
| 401 |
+
|
| 402 |
+
44. Kukal S, Guin D, Rawat C, Bora S, Mishra MK, Sharma P, Paul PR, Kanojia N, Grewal GK, Kukreti S, Saso L (2021) Multidrug efflux transporter ABCG2: expression and regulation. Cell Mol Life Sci 78:6887–6939. 10.1007/s00018-021-03910-x [DOI](https://doi.org/10.1007/s00018-021-03901-y) | [PMC free article](/articles/PMC11072723/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34586444/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Kukal%20S,%20Guin%20D,%20Rawat%20C,%20Bora%20S,%20Mishra%20MK,%20Sharma%20P,%20Paul%20PR,%20Kanojia%20N,%20Grewal%20GK,%20Kukreti%20S,%20Saso%20L%20(2021)%20Multidrug%20efflux%20transporter%20ABCG2:%20expression%20and%20regulation.%20Cell%20Mol%20Life%20Sci%2078:6887%E2%80%936939.%2010.1007/s00018-021-03910-x)
|
| 403 |
+
|
| 404 |
+
45. Bruckmueller H, Cascorbi I (2021) ABCB1, ABCG2, ABCC1, ABCC2, and ABCC3 drug transporter polymorphisms and their impact on drug bioavailability: what is our current understanding? Expert Opin Drug Metab Toxicol 17(4):369–396. 10.1080/17425255.2021.1896652 [DOI](https://doi.org/10.1080/17425255.2021.1876661) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33459081/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Bruckmueller%20H,%20Cascorbi%20I%20(2021)%20ABCB1,%20ABCG2,%20ABCC1,%20ABCC2,%20and%20ABCC3%20drug%20transporter%20polymorphisms%20and%20their%20impact%20on%20drug%20bioavailability:%20what%20is%20our%20current%20understanding?%20Expert%20Opin%20Drug%20Metab%20Toxicol%2017(4):369%E2%80%93396.%2010.1080/17425255.2021.1896652)
|
| 405 |
+
|
| 406 |
+
46. Woodward OM, Köttgen A, Coresh J, Boerwinkle E, Guggino WB, Köttgen M (2009) Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci U S A 106(25):10338–11042. 10.1073/pnas.0902308106 [DOI](https://doi.org/10.1073/pnas.0901249106) | [PMC free article](/articles/PMC2700910/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19506252/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Woodward%20OM,%20K%C3%B6ttgen%20A,%20Coresh%20J,%20Boerwinkle%20E,%20Guggino%20WB,%20K%C3%B6ttgen%20M%20(2009)%20Identification%20of%20a%20urate%20transporter,%20ABCG2,%20with%20a%20common%20functional%20polymorphism%20causing%20gout.%20Proc%20Natl%20Acad%20Sci%20U%20S%20A%20106(25):10338%E2%80%9311042.%2010.1073/pnas.0902308106)
|
| 407 |
+
|
| 408 |
+
47. Safar Z, Kis E, Erdo F, Zolnerciks JK, Krajcsi P (2019) ABCG2/BCRP: variants, transporter interaction profile of substrates and inhibitors. Expert Opin Drug Metab Toxicol 15(4):313–328. 10.1080/17425255.2019.1574854 [DOI](https://doi.org/10.1080/17425255.2019.1591373) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30856014/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Safar%20Z,%20Kis%20E,%20Erdo%20F,%20Zolnerciks%20JK,%20Krajcsi%20P%20(2019)%20ABCG2/BCRP:%20variants,%20transporter%20interaction%20profile%20of%20substrates%20and%20inhibitors.%20Expert%20Opin%20Drug%20Metab%20Toxicol%2015(4):313%E2%80%93328.%2010.1080/17425255.2019.1574854)
|
| 409 |
+
|
| 410 |
+
48. Fohner AE, Brackman DJ, Giacomini KM, Altman RB, Klein TE (2017) PharmGKB summary: very important pharmacogene information for: ABCG2. Pharmacogenet Genomics 27(11):420–427. 10.1097/FPC.0000000000000348 [DOI](https://doi.org/10.1097/FPC.0000000000000305) | [PMC free article](/articles/PMC5788016/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28858993/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Fohner%20AE,%20Brackman%20DJ,%20Giacomini%20KM,%20Altman%20RB,%20Klein%20TE%20(2017)%20PharmGKB%20summary:%20very%20important%20pharmacogene%20information%20for:%20ABCG2.%20Pharmacogenet%20Genomics%2027(11):420%E2%80%93427.%2010.1097/FPC.0000000000000348)
|
| 411 |
+
|
| 412 |
+
49. Woodward OM, Tukaye DN, Cui J, Greenwell P, Constantoulakis LM, Parker BS, Rao A, Köttgen M, Maloney PC, Guggino WB (2013) Gout-causing Q141K mutation in ABCG2 leads to instability of the nucleotide-binding domain and can be corrected with small molecules. Proc Natl Acad Sci U S A 110(13):5223–5228. 10.1073/pnas.1219808110 [DOI](https://doi.org/10.1073/pnas.1214530110) | [PMC free article](/articles/PMC3612674/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23493553/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Woodward%20OM,%20Tukaye%20DN,%20Cui%20J,%20Greenwell%20P,%20Constantoulakis%20LM,%20Parker%20BS,%20Rao%20A,%20K%C3%B6ttgen%20M,%20Maloney%20PC,%20Guggino%20WB%20(2013)%20Gout-causing%20Q141K%20mutation%20in%20ABCG2%20leads%20to%20instability%20of%20the%20nucleotide-binding%20domain%20and%20can%20be%20corrected%20with%20small%20molecules.%20Proc%20Natl%20Acad%20Sci%20U%20S%20A%20110(13):5223%E2%80%935228.%2010.1073/pnas.1219808110)
|
| 413 |
+
|
| 414 |
+
50. Takahashi N, Miura M, Scott SA, Kagaya H, Kameoka Y, Tagawa H, Saitoh H, Fujishima N, Yoshioka T, Hirokawa M, Sawada K (2010) Influence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemia. J Hum Genet 55(11):731–737. 10.1038/jhg.2010.108 [DOI](https://doi.org/10.1038/jhg.2010.98) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20720558/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Takahashi%20N,%20Miura%20M,%20Scott%20SA,%20Kagaya%20H,%20Kameoka%20Y,%20Tagawa%20H,%20Saitoh%20H,%20Fujishima%20N,%20Yoshioka%20T,%20Hirokawa%20M,%20Sawada%20K%20(2010)%20Influence%20of%20CYP3A5%20and%20drug%20transporter%20polymorphisms%20on%20imatinib%20trough%20concentration%20and%20clinical%20response%20among%20patients%20with%20chronic%20phase%20chronic%20myeloid%20leukemia.%20J%20Hum%20Genet%2055(11):731%E2%80%93737.%2010.1038/jhg.2010.108)
|
| 415 |
+
|
| 416 |
+
51. Jiang ZP, Zhao XL, Takahashi N, Angelini S, Dubashi B, Sun L, Xu P (2017) Trough concentration and ABCG2 polymorphism are better to predict imatinib response in chronic myeloid leukemia: a meta-analysis. Pharmacogenomics 18(1):35–56. 10.2217/pgs-2016-0047 [DOI](https://doi.org/10.2217/pgs-2016-0103) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27991849/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Jiang%20ZP,%20Zhao%20XL,%20Takahashi%20N,%20Angelini%20S,%20Dubashi%20B,%20Sun%20L,%20Xu%20P%20(2017)%20Trough%20concentration%20and%20ABCG2%20polymorphism%20are%20better%20to%20predict%20imatinib%20response%20in%20chronic%20myeloid%20leukemia:%20a%20meta-analysis.%20Pharmacogenomics%2018(1):35%E2%80%9356.%2010.2217/pgs-2016-0047)
|
| 417 |
+
|
| 418 |
+
52. Alves R, Gonçalves AC, Jorge J, Marques G, Ribeiro AB, Tenreiro R, Coucelo M, Diamond J, Oliveiros B, Pereira A, Freitas-Tavares P (2022) Genetic variants of ABC and SLC transporter genes and chronic myeloid leukemia: impact on susceptibility and prognosis. Int J Mol Sci 23(17):9815. 10.3390/ijms23179815 [DOI](https://doi.org/10.3390/ijms23179815) | [PMC free article](/articles/PMC9456284/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36077209/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Alves%20R,%20Gon%C3%A7alves%20AC,%20Jorge%20J,%20Marques%20G,%20Ribeiro%20AB,%20Tenreiro%20R,%20Coucelo%20M,%20Diamond%20J,%20Oliveiros%20B,%20Pereira%20A,%20Freitas-Tavares%20P%20(2022)%20Genetic%20variants%20of%20ABC%20and%20SLC%20transporter%20genes%20and%20chronic%20myeloid%20leukemia:%20impact%20on%20susceptibility%20and%20prognosis.%20Int%20J%20Mol%20Sci%2023(17):9815.%2010.3390/ijms23179815)
|
| 419 |
+
|
| 420 |
+
53. Cheng F, Cui Z, Li Q, Chen S, Li W, Zhang Y (2024) Influence of genetic polymorphisms on imatinib concentration and therapeutic response in patients with chronic-phase chronic myeloid leukemia. Int Immunopharmacol 133:112090. 10.1016/j.intimp.2024.112090 [DOI](https://doi.org/10.1016/j.intimp.2024.112090) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38640718/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Cheng%20F,%20Cui%20Z,%20Li%20Q,%20Chen%20S,%20Li%20W,%20Zhang%20Y%20(2024)%20Influence%20of%20genetic%20polymorphisms%20on%20imatinib%20concentration%20and%20therapeutic%20response%20in%20patients%20with%20chronic-phase%20chronic%20myeloid%20leukemia.%20Int%20Immunopharmacol%20133:112090.%2010.1016/j.intimp.2024.112090)
|
| 421 |
+
|
| 422 |
+
54. Francis J, Dubashi B, Sundaram R, Pradhan SC, Chandrasekaran A (2015) A study to explore the correlation of ABCB1, ABCG2, OCT1 genetic polymorphisms and trough level concentration with imatinib mesylate-induced thrombocytopenia in chronic myeloid leukemia patients. Cancer Chemother Pharmacol 76:1185–1189. 10.1007/s00280-015-2871-8 [DOI](https://doi.org/10.1007/s00280-015-2905-6) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26546461/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Francis%20J,%20Dubashi%20B,%20Sundaram%20R,%20Pradhan%20SC,%20Chandrasekaran%20A%20(2015)%20A%20study%20to%20explore%20the%20correlation%20of%20ABCB1,%20ABCG2,%20OCT1%20genetic%20polymorphisms%20and%20trough%20level%20concentration%20with%20imatinib%20mesylate-induced%20thrombocytopenia%20in%20chronic%20myeloid%20leukemia%20patients.%20Cancer%20Chemother%20Pharmacol%2076:1185%E2%80%931189.%2010.1007/s00280-015-2871-8)
|
| 423 |
+
|
| 424 |
+
55. Omran MM, Abdelfattah R, Moussa HS, Alieldin N, Shouman SA (2020) Association of the trough, peak/trough ratio of imatinib, pyridine–N-oxide imatinib and ABCG2 SNPs 34 G > A and SLCO1B3 334 T > G with imatinib response in Egyptian chronic myeloid leukemia patients. Front Oncol 10:1348. 10.3389/fonc.2020.01348 [DOI](https://doi.org/10.3389/fonc.2020.01348) | [PMC free article](/articles/PMC7466443/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32974132/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Omran%20MM,%20Abdelfattah%20R,%20Moussa%20HS,%20Alieldin%20N,%20Shouman%20SA%20(2020)%20Association%20of%20the%20trough,%20peak/trough%20ratio%20of%20imatinib,%20pyridine%E2%80%93N-oxide%20imatinib%20and%20ABCG2%20SNPs%2034%20G%E2%80%89>%E2%80%89A%20and%20SLCO1B3%20334%20T%E2%80%89>%E2%80%89G%20with%20imatinib%20response%20in%20Egyptian%20chronic%20myeloid%20leukemia%20patients.%20Front%20Oncol%2010:1348.%2010.3389/fonc.2020.01348)
|
| 425 |
+
|
| 426 |
+
56. Sabri A, Omran MM, Azim SA, Abdelfattah R, Allam RM, Shouman SA (2023) Role of IDH1 (R132) mutation on imatinib toxicity and effect of ABCG2/OCT1 expression on N-desmethyl imatinib plasma level in Egyptian chronic myeloid leukemia patients. Drug Res (Stuttg) 73(3):146–155. 10.1055/a-1924-7746 [DOI](https://doi.org/10.1055/a-1924-7746) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36630991/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Sabri%20A,%20Omran%20MM,%20Azim%20SA,%20Abdelfattah%20R,%20Allam%20RM,%20Shouman%20SA%20(2023)%20Role%20of%20IDH1%20(R132)%20mutation%20on%20imatinib%20toxicity%20and%20effect%20of%20ABCG2/OCT1%20expression%20on%20N-desmethyl%20imatinib%20plasma%20level%20in%20Egyptian%20chronic%20myeloid%20leukemia%20patients.%20Drug%20Res%20(Stuttg)%2073(3):146%E2%80%93155.%2010.1055/a-1924-7746)
|
| 427 |
+
|
| 428 |
+
57. Nouri N, Mehrzad V, Khalaj Z, Zaker E, Zare F, Abbasi E, Khosravi M, Kalantar SM, Salehi M (2023) Effects of ABCG2 C421A and ABCG2 G34A genetic polymorphisms on clinical outcome and response to imatinib mesylate, in Iranian chronic myeloid leukemia patients. Egypt J Med Hum Genet 24:1. 10.1186/s43042-023-00467-0 [Google Scholar](https://scholar.google.com/scholar_lookup?Nouri%20N,%20Mehrzad%20V,%20Khalaj%20Z,%20Zaker%20E,%20Zare%20F,%20Abbasi%20E,%20Khosravi%20M,%20Kalantar%20SM,%20Salehi%20M%20(2023)%20Effects%20of%20ABCG2%20C421A%20and%20ABCG2%20G34A%20genetic%20polymorphisms%20on%20clinical%20outcome%20and%20response%20to%20imatinib%20mesylate,%20in%20Iranian%20chronic%20myeloid%20leukemia%20patients.%20Egypt%20J%20Med%20Hum%20Genet%2024:1.%2010.1186/s43042-023-00467-0)
|
| 429 |
+
|
| 430 |
+
58. Mohammadi F, Rostami G, Hamid M, Shafiei M, Azizi M, Bahmani H (2023) Association of ABCB1, ABCG2 drug transporter polymorphisms and smoking with disease risk and cytogenetic response to imatinib in chronic myeloid leukemia patients. Leuk Res 126:107021. 10.1016/j.leukres.2023.107021 [DOI](https://doi.org/10.1016/j.leukres.2023.107021) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36696828/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Mohammadi%20F,%20Rostami%20G,%20Hamid%20M,%20Shafiei%20M,%20Azizi%20M,%20Bahmani%20H%20(2023)%20Association%20of%20ABCB1,%20ABCG2%20drug%20transporter%20polymorphisms%20and%20smoking%20with%20disease%20risk%20and%20cytogenetic%20response%20to%20imatinib%20in%20chronic%20myeloid%20leukemia%20patients.%20Leuk%20Res%20126:107021.%2010.1016/j.leukres.2023.107021)
|
test/texts/PMC11677811.md
ADDED
|
@@ -0,0 +1,358 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Pharmacogenetics of Neoadjuvant MAP Chemotherapy in Localized Osteosarcoma: A Study Based on Data from the GEIS-33 Protocol
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Juliana Salazar, María J Arranz, Javier Martin-Broto, Francisco Bautista, Jerónimo Martínez-García, Javier Martínez-Trufero, Yolanda Vidal-Insua, Aizpea Echebarria-Barona, Roberto Díaz-Beveridge, Claudia Valverde, Pablo Luna, María A Vaz-Salgado, Pilar Blay, Rosa Álvarez, Ana Sebio
|
| 5 |
+
**Journal:** Pharmaceutics
|
| 6 |
+
**Date:** 2024 Dec 12
|
| 7 |
+
**DOI:** [10.3390/pharmaceutics16121585](https://doi.org/10.3390/pharmaceutics16121585)
|
| 8 |
+
**PMID:** 39771563
|
| 9 |
+
**PMCID:** PMC11677811
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11677811/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11677811/pdf/pharmaceutics-16-01585.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11677811/pdf/pharmaceutics-16-01585.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
Background: Osteosarcoma is a rare disease, but it is the most frequent malignant bone tumor. Primary treatment consists of preoperative MAP (methotrexate (MTX), doxorubicin and cisplatin) chemotherapy followed by surgery and adjuvant chemotherapy. Pathological response to preoperative chemotherapy is one of the most important prognostic factors, but molecular biomarkers are lacking. Additionally, chemotherapy-induced toxicity might jeopardize treatment completion. We evaluated variants in genes involved in DNA repair and drug metabolism pathways as predictors of response to MAP-based treatment. Material and Methods: Germline polymorphisms in MTHFR, SLC19A1, ABCB1, ABCC2, ABCC3, ERCC1, ERCC2 and GSTP1 genes were determined for association studies in 69 patients diagnosed with localized osteosarcoma who enrolled in the prospective GEIS-33 trial. P-glycoprotein expression in tumor tissue was also analyzed. Results: In the multivariate analysis, the ABCC2 rs2273697 (odds ratio [OR] 12.3, 95% CI 2.3–66.2; p = 0.003) and ERCC2 rs1799793 (OR 9.6, 95% CI 2.1–43.2; p = 0.003) variants were associated with poor pathological response. P-glycoprotein expression did not correlate with pathological response. The ABCB1 rs1128503 (OR 11.4, 95% CI 2.2–58.0; p = 0.003) and ABCC3 rs4793665 (OR 12.0, 95% CI 2.1–70.2; p = 0.006) variants were associated with MTX grade 3–4 hepatotoxicity. Conclusions: Our findings add to the evidence that genetic variants in the ABC transporters and DNA-repair genes may serve as predictive biomarkers for MAP chemotherapy and contribute to treatment personalization.
|
| 16 |
+
|
| 17 |
+
Keywords: osteosarcoma, neoadjuvant chemotherapy, pharmacogenomics, personalized medicine
|
| 18 |
+
|
| 19 |
+
**Keywords:**Keywords: osteosarcoma, neoadjuvant chemotherapy, pharmacogenomics, personalized medicine
|
| 20 |
+
|
| 21 |
+
## 1. Introduction
|
| 22 |
+
|
| 23 |
+
Standard first-line treatment for localized high-grade osteosarcoma consist of neoadjuvant chemotherapy based on high-dose methotrexate (MTX), doxorubicin and cisplatin (the so-called MAP regimen), followed by complete surgical resection of the primary tumor and subsequent adjuvant chemotherapy [[1](#B1-pharmaceutics-16-01585)1,[2](#B2-pharmaceutics-16-01585)2,[3](#B3-pharmaceutics-16-01585)3]. However, despite multimodality treatment, the 5-year survival rate is around 70%. Risk stratification of patients is based on pathological response to neoadjuvant MAP chemotherapy that correlates with prognosis [[4](#B4-pharmaceutics-16-01585)4,[5](#B5-pharmaceutics-16-01585)5]. Patients with a pathological response ≥90% are considered good responders. However, more than 40% of the patients do not achieve a good response [[6](#B6-pharmaceutics-16-01585)6]. Strategies to improve survival in poor responders, such as the addition of chemotherapy agents such as ifosfamide plus etoposide to adjuvant chemotherapy, have been evaluated in a large-scale clinical trial, but with negative results [[7](#B7-pharmaceutics-16-01585)7,[8](#B8-pharmaceutics-16-01585)8]. Identification of poor responders at diagnosis could improve clinical outcomes by treatment escalation, or by introducing alternative therapeutic agents or targeted therapies to the neoadjuvant setting. Additionally, in a subset of patients, multidrug chemotherapy regimens could lead to early severe adverse effects such as hepatotoxicity and myelotoxicity [[9](#B9-pharmaceutics-16-01585)9], that cannot be anticipated due to the lack of predictive biomarkers of chemotherapy toxicity.
|
| 24 |
+
|
| 25 |
+
Cytostatic drugs used in the treatment of osteosarcoma exert their antitumoral activity by interfering with cell proliferation processes that ultimately lead to apoptosis. High-dose MTX acts through the folate cycle by affecting de novo synthesis of pyrimidine and purine nucleotides, both of which are essential for DNA and RNA syntheses [[10](#B10-pharmaceutics-16-01585)10]. Cisplatin and doxorubicin bind to DNA, inhibiting DNA replication [[11](#B11-pharmaceutics-16-01585)11]. The nucleotide excision repair (NER) pathway is involved in the removal of platinum adducts [[12](#B12-pharmaceutics-16-01585)12] and the glutathione-S-transferase (GST) enzymes in the detoxification processes [[13](#B13-pharmaceutics-16-01585)13]. The ATP-binding cassette (ABC) family includes the P-glycoprotein encoded by the *ABCB1*ABCB1 gene. The P-glycoprotein effluxes the chemotherapeutic agents back into the intestinal lumen, affecting their exposure and clearance.
|
| 26 |
+
|
| 27 |
+
Polymorphisms in genes involved in these processes may alter protein functionality and explain, at least partially, individual variation in response to MAP chemotherapy. In osteosarcomas, most studies investigating genetic variants as predictors of pathological response or toxicity are based on pathways related to MTX [[14](#B14-pharmaceutics-16-01585)14,[15](#B15-pharmaceutics-16-01585)15,[16](#B16-pharmaceutics-16-01585)16], cisplatin [[17](#B17-pharmaceutics-16-01585)17,[18](#B18-pharmaceutics-16-01585)18] or multidrug chemotherapy regimens [[19](#B19-pharmaceutics-16-01585)19,[20](#B20-pharmaceutics-16-01585)20,[21](#B21-pharmaceutics-16-01585)21]. Other strategies focused on testing a larger number of single-nucleotide polymorphisms (SNPs) have identified new genes associated with treatment outcomes, but the evidence of their causality remains low [[22](#B22-pharmaceutics-16-01585)22,[23](#B23-pharmaceutics-16-01585)23]. Thus, the genetic variants found to be associated in these studies need further confirmation in larger and more homogeneous cohorts before they can be used in treatment planification. Prospective pharmacogenetic studies in large clinical trials offer the perfect setting for evaluating and validating these genetic variants as predictive biomarkers of efficacy and toxicity. In the present study, we conducted a multicenter pharmacogenetic association study embedded within a prospective osteosarcoma study of the Spanish Group of Sarcoma Research (GEIS, by its Spanish acronym). Germline SNPs in genes relevant to the pharmacokinetics or pharmacodynamics of MTX, doxorubicin and cisplatin were analyzed as predictive biomarkers of response to chemotherapy in localized high-grade osteosarcomas. In the analysis, we also included P-glycoprotein expression values, determined centrally in the GEIS-33 protocol.
|
| 28 |
+
|
| 29 |
+
## 2. Materials and Methods
|
| 30 |
+
|
| 31 |
+
### 2.1. Study Design
|
| 32 |
+
|
| 33 |
+
This was a multicenter study, embedded in the GEIS-33 protocol, a prospective observational study for patients with newly diagnosed high-grade osteosarcoma localized in the extremities. Patients included in this study were enrolled in 13 tertiary hospitals in Spain.
|
| 34 |
+
|
| 35 |
+
The GEIS-33 protocol was conducted according to the provisions of the Declaration of Helsinki, and it was approved by the ethics committees of all participating centers (ISG-GEIS-OS-2). All patients or, in the case of children, their parents or guardians, provided written informed consent to participate.
|
| 36 |
+
|
| 37 |
+
### 2.2. Patients’ Characteristics
|
| 38 |
+
|
| 39 |
+
Between July 2016 and November 2020, 69 patients with non-metastatic high-grade osteosarcoma localized in the extremities, enrolled in the GEIS-33 protocol and with an available DNA sample, were included in this study. [Table 1](#pharmaceutics-16-01585-t001)Table 1 shows the characteristics of the patients and tumors.
|
| 40 |
+
|
| 41 |
+
### Table 1.
|
| 42 |
+
|
| 43 |
+
Clinical and pathological characteristics of osteosarcoma patients.
|
| 44 |
+
|
| 45 |
+
| Characteristic | n (%) |
|
| 46 |
+
| -------------- | ----- |
|
| 47 |
+
| Age at diagnosis (years), median (range) | 14 (4–32) |
|
| 48 |
+
| Sex | |
|
| 49 |
+
| Female | 32 (46.4) |
|
| 50 |
+
| Male | 37 (53.6) |
|
| 51 |
+
| Primary tumor site | |
|
| 52 |
+
| Femur | 45 (65.2) |
|
| 53 |
+
| Tibia/fibula | 12 (17.4) |
|
| 54 |
+
| Humerus/radius | 8 (11.6) |
|
| 55 |
+
| Other | 4 (5.8) |
|
| 56 |
+
| Surgical margins | |
|
| 57 |
+
| Wide | 44 (63.8) |
|
| 58 |
+
| Radical | 7 (10.1) |
|
| 59 |
+
| Marginal | 15 (21.7) |
|
| 60 |
+
| Not available | 3 (4.3) |
|
| 61 |
+
| Pathological response | |
|
| 62 |
+
| ≥90 | 26 (37.7) |
|
| 63 |
+
| <90 | 40 (58) |
|
| 64 |
+
| Not available | 3 (4.3) |
|
| 65 |
+
| Death (Yes) | 14 (21.2) |
|
| 66 |
+
| Progression (Yes) | 16 (23.9) |
|
| 67 |
+
All patients received 2 cycles over 8 weeks of standard preoperative chemotherapy consisting of high-dose MTX 12 g/m^2^2, doxorubicin 90 mg/m^2^2 (Adriamycin) and cisplatin 120 mg/m^2^2. After neoadjuvant chemotherapy, patients underwent surgery with curative intent. After surgery, patients were stratified according to P-glycoprotein expression and histological response. Patients with negative tumor expression of P-glycoprotein received conventional adjuvant MAP chemotherapy. Patients with positive tumor expression of P-glycoprotein were stratified according to histological response to receive mifamurtide (tumor necrosis ≥ 90%; good responders) or high-dose ifosfamide plus mifamurtide (tumor necrosis < 90%; poor responders).
|
| 68 |
+
|
| 69 |
+
### 2.3. Outcome Measures
|
| 70 |
+
|
| 71 |
+
Pathological response to neoadjuvant MAP chemotherapy was evaluated histologically in the resected surgical specimen. Pathological response classification was dichotomized into good response (tumor necrosis ≥ 90%) and poor response (tumor necrosis (<90%). Pathological response was not available for 3 patients.
|
| 72 |
+
|
| 73 |
+
Chemotherapy-induced toxicities were recorded prospectively for each drug and treatment cycle. Hepatotoxicity related to high-dose MTX treatment was evaluated based on alanine transaminase (ALT) and aspartate transaminase (AST) enzymes levels. Hematological toxicities related to doxorubicin and cisplatin were anemia, neutropenia and thrombocytopenia. Toxicities were graded according to Common Terminology Criteria for Adverse Events (CTCAE) Version 4.0. [[24](#B24-pharmaceutics-16-01585)24]. The highest grade of each toxicity was used for the analyses and dichotomized into grades 0/1–2 versus grades 3–4. Toxicity data were not available for 9 patients (n = 60), and not all toxicities were available for all 60 patients (from 58 patients for high-dose MTX hepatotoxicity and from 57 patients for hematological toxicities induced by cisplatin and/or doxorubicin).
|
| 74 |
+
|
| 75 |
+
Overall survival (OS) was calculated from the date of diagnosis to death from any cause or last clinical follow-up. Recurrence-free survival (RFS) was defined as the time from the initiation of MAP chemotherapy until the date of local or distant recurrence, whichever occurred first. Data for survival analyses were not available for all the patients (from 65 patients for OS and from 63 patients for RFS).
|
| 76 |
+
|
| 77 |
+
### 2.4. SNPs Selection and Genotyping
|
| 78 |
+
|
| 79 |
+
We selected 13 SNPs in eight genes (*MTHFR*MTHFR, *SLC19A1*SLC19A1, *ABCB1*ABCB1, *ABCC2*ABCC2, *ABCC3*ABCC3, *ERCC1*ERCC1, *ERCC2*ERCC2 and *GSTP1*GSTP1) related to the DNA-repair and folic acid pathways, and to the transport and detoxification of the cytostatic drugs used in high-grade osteosarcoma treatment. These polymorphisms have previously been associated with clinical outcomes in patients with osteosarcoma [[14](#B14-pharmaceutics-16-01585)14,[16](#B16-pharmaceutics-16-01585)16,[20](#B20-pharmaceutics-16-01585)20,[25](#B25-pharmaceutics-16-01585)25,[26](#B26-pharmaceutics-16-01585)26,[27](#B27-pharmaceutics-16-01585)27,[28](#B28-pharmaceutics-16-01585)28,[29](#B29-pharmaceutics-16-01585)29,[30](#B30-pharmaceutics-16-01585)30,[31](#B31-pharmaceutics-16-01585)31] (see [Table 2](#pharmaceutics-16-01585-t002)Table 2).
|
| 80 |
+
|
| 81 |
+
### Table 2.
|
| 82 |
+
|
| 83 |
+
Main characteristics of the genetic variants analyzed.
|
| 84 |
+
|
| 85 |
+
| Gene Symbol | Reference SNP | Molecular Consequence | Identifiers |
|
| 86 |
+
| ----------- | ------------- | --------------------- | ----------- |
|
| 87 |
+
| ERCC1 | rs11615 | Synonymous | NM_001983.4:c.354T>C (p.Asn118=) |
|
| 88 |
+
| ERCC2 | rs13181 | Missense | NM_000400.4:c.2251A>C (p.Lys751Gln) |
|
| 89 |
+
| ERCC2 | rs1799793 | Missense | NM_000400.4:c.934G>A (p.Asp312Asn) |
|
| 90 |
+
| GSTP1 | rs1695 | Missense | NM_000852.4:c.313A>G (p.Ile105Val) |
|
| 91 |
+
| ABCC3 | rs4793665 | 2KB upstream | NC_000017.11:g.50634726C>T |
|
| 92 |
+
| ABCB1 | rs1045642 | Synonymous | NM_001348944.2:c.3435T>C (p.Ile1145=) |
|
| 93 |
+
| ABCB1 | rs2032582 | Missense | NM_001348944.2:c.2677T>G (p.Ser893Ala); c.2677T>A (p.Ser893Thr) |
|
| 94 |
+
| ABCB1 | rs1128503 | Synonymous | NM_001348944.2:c.1236T>C (p.Gly412=) |
|
| 95 |
+
| ABCC2 | rs2273697 | Missense | NM_000392.5:c.1249G>A (p.Val417Ile) |
|
| 96 |
+
| ABCC2 | rs3740066 | synonymous | NM_000392.5:c.3972C>T (p.Ile1324=) |
|
| 97 |
+
| MTHFR | rs1801133 | Missense | NM_005957.4:c.665C>T (p.Ala222Val) a |
|
| 98 |
+
| MTHFR | rs1801131 | Missense | NM_005957.4:c.1286A>C (p.Glu429Ala) a |
|
| 99 |
+
| SLC19A1 | rs1051266 | Missense | NM_194255.4(SLC19A1):c.80A>G (p.His27Arg) |
|
| 100 |
+
DNA from peripheral blood samples was isolated by automatic extraction (Autopure, Qiagen, Hilden, Germany), and DNA concentration was measured using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). Samples were processed by real-time PCR using TaqMan^®^® SNP genotyping assays on a 7900 HT Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). All the methods were performed following the manufacturers’ recommendations. The call rates were higher than 99% for SNPs and samples. The allele frequencies of the SNPs did not differ from those reported in European populations [[32](#B32-pharmaceutics-16-01585)32]. All the SNPs were in Hardy–Weinberg equilibrium (*p*p > 0.05).
|
| 101 |
+
|
| 102 |
+
### 2.5. Immunohistochemical Studies
|
| 103 |
+
|
| 104 |
+
P-glycoprotein expression was evaluated in the biopsy at diagnosis and was prospectively recorded. P-glycoprotein determination was performed according to the GEIS-33 protocol and centralized at the Instituto Ortopedico Rizzoli (IRCCS), Italy. In brief, immunohistochemistry was performed in 4 to 6 μm thick formalin-fixed paraffin-embedded (FFPE) tissue sections using an avidin–biotin peroxidase complex method (Vectastain ABC kit; Vector Laboratories, Inc, Burlingame, CA, USA) and three monoclonal antibodies (JSB-1 (Monosan Sanbio, Uden, The Netherlands), MRK16 (MyBioSource Aurogene Srl, Rome, Italy) and C494 (Invitrogen, Ltd., Paisley, UK)) [[33](#B33-pharmaceutics-16-01585)33]. P-glycoprotein expression was classified as positive, negative or not evaluable.
|
| 105 |
+
|
| 106 |
+
### 2.6. Statistical Analysis
|
| 107 |
+
|
| 108 |
+
The study sample size had more than 80% statistical power with two-sided 95% confidence intervals (CIs) to detect genetic effect sizes of moderate magnitude (odds ratios (OR) ≤ 3), assuming 40% of poor responders to neoadjuvant MAP chemotherapy (calculated by Epi Info 7TM version 7.2.5.0 ([https://www.cdc.gov/epiinfo](https://www.cdc.gov/epiinfo)https://www.cdc.gov/epiinfo); accessed on 9 January 2024). Chi-square was used to detect statistical differences between categorical variables. Logistic regression analyses were performed including as covariates age (4–10 years versus 10–32 years), gender and tumor site (femur/humerus versus other) for pathological response, and age and gender for toxicity. Kaplan–Meier curves and a log-rank test were used for OS and RFS analyses. Significant associations were presented with the ORs and 95% CIs. Statistical significance was defined as a *p*p value < 0.05. Bonferroni correction for multiple comparisons was set at *p*p < 0.001. Statistical analyses were performed using IBM SPSS Statistics version 26.0, and the statistical package PLINK version 1.07.2 [[34](#B34-pharmaceutics-16-01585)34].
|
| 109 |
+
|
| 110 |
+
## 3. Results
|
| 111 |
+
|
| 112 |
+
### 3.1. Clinical Results
|
| 113 |
+
|
| 114 |
+
The median follow-up was 62.4 (interquartile range [IQR], 38.4–80.3] months, and the median age at diagnosis was 14 (IQR, 4–32) years. Twenty-six (37.7%) patients achieved a good pathological response (≥90%) after two cycles of neoadjuvant MAP chemotherapy. During the study follow up, the disease progressed in 16 (23.9%) patients and 14 (21.2%) patients died. None of the clinicopathological variables analyzed regarding the pathological response showed statistical significance: age (*p*p = 0.08), gender (*p*p = 0.42) and tumor site (*p*p = 0.33).
|
| 115 |
+
|
| 116 |
+
### 3.2. P-Glycoprotein Expression and ABCB1 Genetic Variants
|
| 117 |
+
|
| 118 |
+
P-glycoprotein expression in tumor samples was positive in 35 (50.7%) patients, negative in 28 (40.6%) patients and not evaluable in 6 (8.7%) patients. We analyzed the correlation between P-glycoprotein expression and the rs1045642, rs2032582 and rs1128503 *ABCB1*ABCB1 genetic variants determined in germline DNA. We found that the *ABCB1*ABCB1 rs1045642-A allele was marginally correlated with positive P-glycoprotein expression (*p*p = 0.049): 51% of tumors had positive expression and 34% of tumors had negative expression ([Figure 1](#pharmaceutics-16-01585-f001)Figure 1). P-glycoprotein expression was not correlated with the other two genetic variants, rs2032582 (*p*p = 0.28) or rs1128503 (*p*p = 0.24).
|
| 119 |
+
|
| 120 |
+
### Figure 1.
|
| 121 |
+
|
| 122 |
+

|
| 123 |
+
|
| 124 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11677811_pharmaceutics-16-01585-g001.jpg)
|
| 125 |
+
|
| 126 |
+
(A) P-glycoprotein expression in tumor samples (number of patients; %). (B) Distribution of ABCB1 rs1045642 alleles according to P-glycoprotein expression (number of alleles; %).
|
| 127 |
+
|
| 128 |
+
### 3.3. Association Analyses and Pathological Response
|
| 129 |
+
|
| 130 |
+
P-glycoprotein expression was not associated with pathological response (*p*p = 0.52). [Table 3](#pharmaceutics-16-01585-t003)Table 3 shows univariate analyses for genetic variants and pathological response. Univariate analyses showed marginal associations between the *ABCC2*ABCC2 rs2273697 and *ERCC2*ERCC2 rs1799793 variants and pathological response. Patients carrying the *ABCC2*ABCC2 rs2273697-A allele (*p*p = 0.04) and patients carrying the *ERCC2*ERCC2 rs1799793-A allele (*p*p = 0.047) had a higher risk of poor pathological response. For *ABCC2*ABCC2 rs2273697, 50% of patients with the GG genotype presented poor pathological response, compared to 81.8% of patients with GA or AA genotypes (*p*p = 0.02 in a dominant model). For *ERCC2*ERCC2 rs1799793, 45.2% of patients with the GG genotype presented poor pathological response, compared to 74.3% of patients with the GA or AA genotypes (*p*p = 0.02 in a dominant model). Multivariate analyses including these two SNPs and age, gender and tumor site as covariates showed significant associations for both genetic variants: *ABCC2*ABCC2 rs2273697 (OR 12.3, 95% CI 2.3–66.2; *p*p = 0.003) and *ERCC2*ERCC2 rs1799793 (OR 9.6, 95% CI 2.1–43.2; *p*p = 0.003). However, these associations were not statistically significant after the Bonferroni test.
|
| 131 |
+
|
| 132 |
+
### Table 3.
|
| 133 |
+
|
| 134 |
+
Non-corrected univariate analyses for genetic variants and pathological response.
|
| 135 |
+
|
| 136 |
+
| Pathological Response (n = 66) |
|
| 137 |
+
| ------------------------------ |
|
| 138 |
+
| Gene | Allele | Poor Responders Frequency | Good Responders Frequency | p-Value | OR |
|
| 139 |
+
| MTHFR | rs1801131-G | 0.35 | 0.33 | 0.78 | 1.11 |
|
| 140 |
+
| MTHFR | rs1801133-A | 0.31 | 0.33 | 0.86 | 0.94 |
|
| 141 |
+
| SLC19A1 | rs1051266-A | 0.43 | 0.48 | 0.53 | 0.80 |
|
| 142 |
+
| ABCB1 | rs1045642-A | 0.48 | 0.37 | 0.21 | 1.57 |
|
| 143 |
+
| ABCB1 | rs2032582-A | 0.41 | 0.25 | 0.06 | 2.11 |
|
| 144 |
+
| ABCB1 | rs1128503-A | 0.4 | 0.33 | 0.40 | 1.37 |
|
| 145 |
+
| ABCC2 | rs2273697-A | 0.24 | 0.1 | 0.04 | 2.93 |
|
| 146 |
+
| ABCC2 | rs3740066-T | 0.5 | 0.35 | 0.08 | 1.89 |
|
| 147 |
+
| ABCC3 | rs4793665-C | 0.45 | 0.33 | 0.16 | 1.68 |
|
| 148 |
+
| ERCC1 | rs11615-G | 0.45 | 0.33 | 0.16 | 1.68 |
|
| 149 |
+
| ERCC2 | rs13181-G | 0.39 | 0.31 | 0.35 | 1.42 |
|
| 150 |
+
| ERCC2 | rs1799793-A | 0.39 | 0.21 | 0.047 | 2.24 |
|
| 151 |
+
| GSTP1 | rs1695-G | 0.43 | 0.48 | 0.53 | 0.80 |
|
| 152 |
+
### 3.4. Association Analyses and Toxicity
|
| 153 |
+
|
| 154 |
+
[Table 4](#pharmaceutics-16-01585-t004)Table 4 shows univariate analyses for high-dose MTX hepatotoxicity and SNPs in genes related to the folic acid pathway and drug transport. Univariate analyses showed statistically significant associations between the *ABCB1*ABCB1 rs1128503 and *ABCC3*ABCC3 rs4793665 variants and severe hepatotoxicity. Patients carrying the *ABCB1*ABCB1 rs1128503-G allele (*p*p = 0.009) and patients carrying the *ABCC3*ABCC3 rs4793665-T allele (*p*p = 0.04) presented a higher risk of developing grade 3–4 hepatotoxicity because of MTX treatment. For *ABCB1*ABCB1 rs1128503, 86.4% of patients with the GG genotype developed grade 3–4 hepatotoxicity, compared to 44.4% of patients with GA or AA genotypes (*p*p = 0.002 in a dominant model). For *ABCC3*ABCC3 rs4793665, 90.5% of patients with the TT genotype developed grade 3–4 hepatotoxicity, compared to 43.2% of patients with TC or CC genotypes (*p*p < 0.001 in a dominant model). Multivariate analyses including these two SNPs and age and gender as covariates showed significant associations for *ABCB1*ABCB1 rs1128503 (OR 11.4, 95% CI 2.2–58.0; *p*p = 0.003) and *ABCC3*ABCC3 rs4793665 (OR 12.0, 95% CI 2.1–70.2; *p*p = 0.006) variants. However, these associations were not statistically significant after the Bonferroni test.
|
| 155 |
+
|
| 156 |
+
### Table 4.
|
| 157 |
+
|
| 158 |
+
Non-corrected univariate analyses for high-dose MTX hepatotoxicity and variants in genes related to the folic acid pathway and drug transport.
|
| 159 |
+
|
| 160 |
+
| High-Dose Methotrexate Hepatotoxicity (n = 58) |
|
| 161 |
+
| ---------------------------------------------- |
|
| 162 |
+
| Gene | Allele | Grade 3–4 Frequency | Grade 0–2 Frequency | p-Value | OR |
|
| 163 |
+
| MTHFR | rs1801131-G | 0.43 | 0.26 | 0.07 | 2.13 |
|
| 164 |
+
| MTHFR | rs1801133-A | 0.24 | 0.41 | 0.05 | 0.46 |
|
| 165 |
+
| SLC19A1 | rs1051266-A | 0.46 | 0.41 | 0.64 | 1.2 |
|
| 166 |
+
| ABCB1 | rs1045642-A | 0.44 | 0.46 | 0.88 | 0.95 |
|
| 167 |
+
| ABCB1 | rs2032582-A | 0.33 | 0.43 | 0.25 | 0.64 |
|
| 168 |
+
| ABCB1 | rs1128503-G | 0.7 | 0.46 | 0.009 | 2.78 |
|
| 169 |
+
| ABCC2 | rs2273697-A | 0.21 | 0.13 | 0.25 | 1.82 |
|
| 170 |
+
| ABCC2 | rs3740066-T | 0.41 | 0.43 | 0.83 | 0.92 |
|
| 171 |
+
| ABCC3 | rs4793665-T | 0.69 | 0.5 | 0.04 | 2.17 |
|
| 172 |
+
The effect of a possible gene–gene interaction between these two genetic variants on MTX-induced hepatotoxicity was also investigated considering age and gender as covariates. Analyses showed statistically significant interactions between the *ABCB1*ABCB1 rs1128503 and *ABCC3*ABCC3 rs4793665 polymorphisms (*p*p = 0.01).
|
| 173 |
+
|
| 174 |
+
Univariate analyses for hematological toxicities induced by cisplatin and doxorubicin treatment and SNPs in genes related to the DNA-repair pathway and drug transport showed non-significant associations ([Supplementary Table S1](#app1-pharmaceutics-16-01585)Supplementary Table S1).
|
| 175 |
+
|
| 176 |
+
### 3.5. Survival Analyses
|
| 177 |
+
|
| 178 |
+
Univariate analyses for the *ABCC2*ABCC2 rs2273697 and *ERCC2*ERCC2 rs1799793 variants associated with pathological response, and for the *ABCB1*ABCB1 rs1128503 and *ABCC3*ABCC3 rs4793665 variants associated with MTX hepatotoxicity, showed no statistically significant associations with OS and RFS ([Supplementary Table S2](#app1-pharmaceutics-16-01585)Supplementary Table S2).
|
| 179 |
+
|
| 180 |
+
## 4. Discussion
|
| 181 |
+
|
| 182 |
+
One of the most important prognostic criteria in high-grade osteosarcoma is the assessment of the pathological response, but validated molecular biomarkers for treatment stratification at the time of diagnosis are lacking. In addition, there are as of yet no molecular biomarkers to predict treatment-derived toxicity that may delay treatment or lead to serious clinical complications. In this study, we found that the *ABCC2*ABCC2 rs2273697 and *ERCC2*ERCC2 rs1799793 germline variants were associated with poor pathological response. Moreover, we found significant associations of the *ABCB1*ABCB1 rs1128503 and *ABCC3*ABCC3 rs4793665 variants with MTX-induced hepatotoxicity.
|
| 183 |
+
|
| 184 |
+
As optimization of risk-stratification is a challenge in the systemic treatment strategy in non-metastatic osteosarcoma, there is a need to identify new biomarkers. The *ABCB1*ABCB1 gene encodes P-glycoprotein, an efflux transporter involved in the reduction in the intracellular concentration of many toxic compounds. Overexpression of this protein in tumor tissue has been associated with a worse response to MAP chemotherapy in patients with osteosarcoma [[35](#B35-pharmaceutics-16-01585)35]. Based on this observation, a prospective trial that stratified patients according to P-glycoprotein expression was conducted in Italy (ISG/OS-2) and Spain (GEIS-33). However, the Italian group [[36](#B36-pharmaceutics-16-01585)36] reported that P-glycoprotein expression was not a predictor of pathological response in induction chemotherapy. Here, we analyzed the effect of P-glycoprotein expression in tumor tissue on pathological response and also found no association. This finding is in line with a previous retrospective study that had the limitation of small sample size [[37](#B37-pharmaceutics-16-01585)37], but also with a prospective study conducted in 685 patients with localized high-grade osteosarcoma [[38](#B38-pharmaceutics-16-01585)38]. However, it should be noted that the appropriate assessment of P-glycoprotein expression may depend on tumor heterogeneity, which could influence the observed results by underestimating possible subclonal expression of P-glycoprotein.
|
| 185 |
+
|
| 186 |
+
We also analyzed whether there was a correlation between germline variants in the *ABCB1*ABCB1 gene and P-glycoprotein expression in the tumor, with marginal results. This observation suggests that tumor protein expression would be a more informative biomarker than genetic variants for those proteins whose regulation can be modified by the tumor microenvironment.
|
| 187 |
+
|
| 188 |
+
Other research has focused on the characterization of genetic biomarkers that may be useful in therapeutic guidance in osteosarcoma. Most of these are association studies that have analyzed candidate SNPs for certain pathways related to MAP chemotherapy [[14](#B14-pharmaceutics-16-01585)14,[15](#B15-pharmaceutics-16-01585)15,[16](#B16-pharmaceutics-16-01585)16,[17](#B17-pharmaceutics-16-01585)17,[18](#B18-pharmaceutics-16-01585)18,[19](#B19-pharmaceutics-16-01585)19,[20](#B20-pharmaceutics-16-01585)20,[21](#B21-pharmaceutics-16-01585)21]. The significant genetic alterations revealed in these studies may have an impact on the efficacy and safety of MAP chemotherapy, but more evidence is needed before their clinical use can be considered.
|
| 189 |
+
|
| 190 |
+
In our study, we found significant associations between the *ABCC2*ABCC2 rs2273697-A and *ERCC2*ERCC2 rs1799793-A alleles and poor pathological response to neoadjuvant chemotherapy. The ABCC2 protein mediates the efflux of xenobiotic compounds such as MTX, cisplatin and doxorubicin. In vitro studies showed that a haplotype including the A-allele of the *ABCC2*ABCC2 rs2273697 (p.Val417Ile) variant was associated with increased protein expression [[39](#B39-pharmaceutics-16-01585)39]. We thus speculate that it could have a negative effect on the response to MAP chemotherapy. Along similar lines, other studies analyzing the *ABCC2*ABCC2 rs2273697 variant and the pharmacokinetics of some drugs showed associations of the A-allele with reduced oral bioavailability of talinolol [[40](#B40-pharmaceutics-16-01585)40] and with reduced dose-normalized concentration of tacrolimus [[41](#B41-pharmaceutics-16-01585)41]. However, it is noteworthy that the studies mentioned above were performed on orally administered drugs, whereas chemotherapy is administered intravenously.
|
| 191 |
+
|
| 192 |
+
*ERCC2*ERCC2 rs1799793 (p.Asp312Asn) is a missense variant that may reduce the DNA repair capacity of the enzyme and enhance the cytotoxic effect of cisplatin [[42](#B42-pharmaceutics-16-01585)42]. Liu et al. [[17](#B17-pharmaceutics-16-01585)17] described that patients with osteosarcoma carrying the AA genotype for *ERCC2*ERCC2 rs1799793 presented better response. However, other studies that analyzed the variant in relation to the pathological response did not find significant associations [[14](#B14-pharmaceutics-16-01585)14,[18](#B18-pharmaceutics-16-01585)18,[43](#B43-pharmaceutics-16-01585)43]. These data contrast with our findings, indicating that the *ERCC2*ERCC2 rs1799793 variant warrants further investigation in the context of MAP chemotherapy.
|
| 193 |
+
|
| 194 |
+
Hepatotoxicity is a limiting complication of high-dose MTX treatment in patients with localized osteosarcoma, leading to dose adjustment and treatment delays. We found that the *ABCB1*ABCB1 rs1128503-G and *ABCC3*ABCC3 rs4793665-T alleles were associated with grade 3–4 hepatotoxicity. However, we did not find a gene–gene interaction between the two variants on hepatotoxicity.
|
| 195 |
+
|
| 196 |
+
ABCC3 transport protein is actively involved in the removal of MTX from the hepatocytes into the blood circulation and ABCB1 in its excretion into the bile [[44](#B44-pharmaceutics-16-01585)44]. *ABCB1*ABCB1 rs1128503 (p.Gly412=) is a synonymous variant encoding the amino acid glycine. It is located near amino acid residues that are critical for ATP binding and ATP hydrolysis [[45](#B45-pharmaceutics-16-01585)45]. We did not find, however, a correlation between the rs1128503 variant and protein expression in the tumor, although P-glycoprotein expression in the tumor would not be representative of its activity in the liver, as P-glycoprotein is overexpressed in numerous cancer-transformed tissues [[46](#B46-pharmaceutics-16-01585)46]. In line with our results, Hattinger et al. [[21](#B21-pharmaceutics-16-01585)21] found an association between the *ABCB1*ABCB1 rs1128503-G allele and hepatotoxicity, defined as transaminases grade 4 in 57 high-grade osteosarcoma patients.
|
| 197 |
+
|
| 198 |
+
*ABCC3*ABCC3 rs4793665 is a promoter variant that has a maximum score of 1a according to the RegulomeDB database [[47](#B47-pharmaceutics-16-01585)47], suggesting it would be a functional variant. Accordingly, the *ABCC3*ABCC3 rs4793665-T allele has been associated with low levels of hepatic mRNA and with reduced binding affinity of nuclear factors to this promoter region [[48](#B48-pharmaceutics-16-01585)48]. The variant was also found to be associated with MTX pharmacokinetic parameters, such as the area under the concentration–time curve and the maximum concentration [[16](#B16-pharmaceutics-16-01585)16].
|
| 199 |
+
|
| 200 |
+
Since ABCB1 and ABCC3 proteins are relevant for preserving liver integrity at high doses of MTX, these variants are promising biomarkers for MTX-induced hepatotoxicity in localized osteosarcoma patients. If validated, the determination of the *ABCB1*ABCB1 rs1128503 and *ABCC3*ABCC3 rs4793665 variants before neoadjuvant MAP chemotherapy may enhance the benefits of high-dose MTX by reducing or preventing the risk of toxicity in some patients. In vitro research is warranted to elucidate the exact mechanism involving these polymorphisms in the drug-induced hepatotoxicity.
|
| 201 |
+
|
| 202 |
+
We note that the associations observed may have been conditioned by the fact that some ABC transporters show binding and transport affinity for more than one anticancer agent used in MAP chemotherapy, so their efflux activity may not only depend on germline polymorphisms, but also on drug–drug interactions.
|
| 203 |
+
|
| 204 |
+
In addition, survival analyses were also performed to explore the long-term effect of the genetic variants found to be associated with pathological response and hepatotoxicity to MTX in this study. However, we found no significant associations, probably due to the moderate sample size.
|
| 205 |
+
|
| 206 |
+
Currently, most patients diagnosed with localized disease are treated with the classical regimen of neoadjuvant and adjuvant MAP chemotherapy as clinical trials testing new therapies have reported limited improvements in survival rates. In addition to the evaluation of emerging therapies, the incorporation of predictive biomarkers into clinical trials may contribute to the achievement of better outcomes in these patients. In this sense, our observations support the utility of pharmacogenetic markers in explaining part of the variation in response to MAP chemotherapy among patients with osteosarcoma, but there are limitations. First, although this is a national multicenter study, the sample size is small. This is because osteosarcoma is a rare disease and access to study samples of patients treated with the same regimen is limited. In addition, safety data were not available for all the patients included in the study. However, these data are valuable because, despite the multidrug combination chemotherapy regimen, toxicities were prospectively recorded for each chemotherapy cycle and were analyzed for each drug. Second, our selection of genes has focused only on those harboring SNPs previously associated with osteosarcoma treatment in an attempt to validate them in a homogenous prospective cohort. Although there is currently no strong evidence of their contribution to osteosarcomas treatment, other candidate genes such as *DHFR*DHFR, *ABCC1*ABCC1 or *ABCG2*ABCG2 should be investigated in future studies with large samples. Third, additional studies on the effect of the *ABCC2*ABCC2 rs2273697 and *ERCC2*ERCC2 rs1799793 variants on protein expression in the tumor may contribute further evidence on the clinical utility of these variants as treatment predictors. Finally, this study should be considered exploratory, as none of the associations described survived Bonferroni corrections for multiple comparisons. Bonferroni corrections are, however, too conservative in candidate gene studies due to the high correlation between SNPs in the same chromosomal region.
|
| 207 |
+
|
| 208 |
+
## 5. Conclusions
|
| 209 |
+
|
| 210 |
+
Our prospective pharmacogenetic study conducted in patients diagnosed with non-metastatic high-grade osteosarcoma of the extremities and treated with neoadjuvant MAP chemotherapy shows variants in ABC transporter genes that may identify patients with poor response and patients at risk of hepatic toxicity at diagnosis. These genetic variants may help personalize treatment and select a more effective and safer neoadjuvant therapy in localized osteosarcoma. Additional validation in clinical trials will be required before these genetic variants can be incorporated into clinical practice.
|
| 211 |
+
|
| 212 |
+
## Acknowledgments
|
| 213 |
+
|
| 214 |
+
We thank Carolyn Newey for language editing and Olga Bell for technical support.
|
| 215 |
+
|
| 216 |
+
## Supplementary Materials
|
| 217 |
+
|
| 218 |
+
The following supporting information can be downloaded at: [https://www.mdpi.com/article/10.3390/pharmaceutics16121585/s1](https://www.mdpi.com/article/10.3390/pharmaceutics16121585/s1)https://www.mdpi.com/article/10.3390/pharmaceutics16121585/s1, Table S1: Non-significant associations for cisplatin- and doxorubicin-induced hematological toxicities and SNPs in genes related to the DNA-repair pathway and drug transport; Table S2: Non-significant associations between genetic variants and survival.
|
| 219 |
+
|
| 220 |
+
## Author Contributions
|
| 221 |
+
|
| 222 |
+
Conceptualization, A.S.; Methodology, J.S., M.J.A. and A.S.; Formal analysis, J.S. and M.J.A.; Resources, J.S., J.M.-B., F.B., J.M.-G., J.M.-T., Y.V.-I., A.E.-B., R.D.-B., C.V., P.L., M.A.V.-S., P.B., R.Á. and A.S.; Data curation, J.M.-B., F.B., J.M.-G., J.M.-T., Y.V.-I., A.E.-B., R.D.-B., C.V., P.L., M.A.V.-S., P.B., R.Á. and A.S.; Writing––original draft, J.S.; Writing-review and editing, J.S., M.J.A., J.M.-B., F.B., J.M.-G., J.M.-T., Y.V.-I., A.E.-B., R.D.-B., C.V., P.L., M.A.V.-S., P.B., R.Á. and A.S.; Investigation, J.S., M.J.A. and A.S.; Funding acquisition, A.S.; Supervision, J.S. and A.S.; Project administration, J.S. and A.S. All authors have read and agreed to the published version of the manuscript.
|
| 223 |
+
|
| 224 |
+
## Institutional Review Board Statement
|
| 225 |
+
|
| 226 |
+
This study was conducted following the guidelines of the Declaration of Helsinki and approved by the ethics committee of each participating center (ISG-GEIS-OS-2; 12 May 2016).
|
| 227 |
+
|
| 228 |
+
## Informed Consent Statement
|
| 229 |
+
|
| 230 |
+
Informed consent was obtained from all subjects or, in the case of children, from their parents or guardians, who participated in this study.
|
| 231 |
+
|
| 232 |
+
## Data Availability Statement
|
| 233 |
+
|
| 234 |
+
The data presented in this study are not publicly available due to ethical committee regulations but are available upon request from the corresponding authors.
|
| 235 |
+
|
| 236 |
+
## Conflicts of Interest
|
| 237 |
+
|
| 238 |
+
The authors declare no conflicts of interest.
|
| 239 |
+
|
| 240 |
+
## Funding Statement
|
| 241 |
+
|
| 242 |
+
This research was funded by the Spanish Group of Sarcoma Research (GEIS). Funding identifier: Translational research projects by young researchers 2017.
|
| 243 |
+
|
| 244 |
+
## Footnotes
|
| 245 |
+
|
| 246 |
+
## Associated Data
|
| 247 |
+
|
| 248 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 249 |
+
|
| 250 |
+
### Supplementary Materials
|
| 251 |
+
|
| 252 |
+
### Data Availability Statement
|
| 253 |
+
|
| 254 |
+
The data presented in this study are not publicly available due to ethical committee regulations but are available upon request from the corresponding authors.
|
| 255 |
+
|
| 256 |
+
### Supplementary Materials
|
| 257 |
+
|
| 258 |
+
### Data Availability Statement
|
| 259 |
+
|
| 260 |
+
The data presented in this study are not publicly available due to ethical committee regulations but are available upon request from the corresponding authors.
|
| 261 |
+
|
| 262 |
+
## References
|
| 263 |
+
|
| 264 |
+
1. Bacci G., Briccoli A., Ferrari S., Longhi A., Mercuri M., Capanna R., Donati D., Lari S., Forni C., DePaolis M. Neoadjuvant Chemotherapy for Osteosarcoma of the Extremity: Long-Term Results of the Rizzoli’s 4th Protocol. Eur. J. Cancer. 2001;37:2030–2039. doi: 10.1016/S0959-8049(01)00229-5. [DOI](https://doi.org/10.1016/S0959-8049(01)00229-5) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11597381/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20J.%20Cancer&title=Neoadjuvant%20Chemotherapy%20for%20Osteosarcoma%20of%20the%20Extremity:%20Long-Term%20Results%20of%20the%20Rizzoli%E2%80%99s%204th%20Protocol&author=G.%20Bacci&author=A.%20Briccoli&author=S.%20Ferrari&author=A.%20Longhi&author=M.%20Mercuri&volume=37&publication_year=2001&pages=2030-2039&pmid=11597381&doi=10.1016/S0959-8049(01)00229-5&)
|
| 265 |
+
|
| 266 |
+
2. Ferrari S., Smeland S., Mercuri M., Bertoni F., Longhi A., Ruggieri P., Alvegard T.A., Picci P., Capanna R., Bernini G., et al. Neoadjuvant Chemotherapy with High-Dose Ifosfamide, High-Dose Methotrexate, Cisplatin, and Doxorubicin for Patients with Localized Osteosarcoma of the Extremity: A Joint Study by the Italian and Scandinavian Sarcoma Groups. J. Clin. Oncol. 2005;23:8845–8852. doi: 10.1200/JCO.2004.00.5785. [DOI](https://doi.org/10.1200/JCO.2004.00.5785) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16246977/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Oncol.&title=Neoadjuvant%20Chemotherapy%20with%20High-Dose%20Ifosfamide,%20High-Dose%20Methotrexate,%20Cisplatin,%20and%20Doxorubicin%20for%20Patients%20with%20Localized%20Osteosarcoma%20of%20the%20Extremity:%20A%20Joint%20Study%20by%20the%20Italian%20and%20Scandinavian%20Sarcoma%20Groups&author=S.%20Ferrari&author=S.%20Smeland&author=M.%20Mercuri&author=F.%20Bertoni&author=A.%20Longhi&volume=23&publication_year=2005&pages=8845-8852&pmid=16246977&doi=10.1200/JCO.2004.00.5785&)
|
| 267 |
+
|
| 268 |
+
3. Meyers P.A., Schwartz C.L., Krailo M.D., Healey J.H., Bernstein M.L., Betcher D., Ferguson W.S., Gebhardt M.C., Goorin A.M., Harris M., et al. Osteosarcoma: The Addition of Muramyl Tripeptide to Chemotherapy Improves Overall Survival—A Report from the Children’s Oncology Group. J. Clin. Oncol. 2008;26:633–638. doi: 10.1200/JCO.2008.14.0095. [DOI](https://doi.org/10.1200/JCO.2008.14.0095) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18235123/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Oncol.&title=Osteosarcoma:%20The%20Addition%20of%20Muramyl%20Tripeptide%20to%20Chemotherapy%20Improves%20Overall%20Survival%E2%80%94A%20Report%20from%20the%20Children%E2%80%99s%20Oncology%20Group&author=P.A.%20Meyers&author=C.L.%20Schwartz&author=M.D.%20Krailo&author=J.H.%20Healey&author=M.L.%20Bernstein&volume=26&publication_year=2008&pages=633-638&pmid=18235123&doi=10.1200/JCO.2008.14.0095&)
|
| 269 |
+
|
| 270 |
+
4. Provisor A.J., Ettinger L.J., Nachman J.B., Krailo M.D., Makley J.T., Yunis E.J., Huvos A.G., Betcher D.L., Baum E.S., Kisker C.T., et al. Treatment of Nonmetastatic Osteosarcoma of the Extremity with Preoperative and Postoperative Chemotherapy: A Report from the Children’s Cancer Group. J. Clin. Oncol. 1997;15:76–84. doi: 10.1200/JCO.1997.15.1.76. [DOI](https://doi.org/10.1200/JCO.1997.15.1.76) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8996127/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Oncol.&title=Treatment%20of%20Nonmetastatic%20Osteosarcoma%20of%20the%20Extremity%20with%20Preoperative%20and%20Postoperative%20Chemotherapy:%20A%20Report%20from%20the%20Children%E2%80%99s%20Cancer%20Group&author=A.J.%20Provisor&author=L.J.%20Ettinger&author=J.B.%20Nachman&author=M.D.%20Krailo&author=J.T.%20Makley&volume=15&publication_year=1997&pages=76-84&pmid=8996127&doi=10.1200/JCO.1997.15.1.76&)
|
| 271 |
+
|
| 272 |
+
5. Bacci G., Mercuri M., Longhi A., Ferrari S., Bertoni F., Versari M., Picci P. Grade of Chemotherapy-Induced Necrosis as a Predictor of Local and Systemic Control in 881 Patients with Non-Metastatic Osteosarcoma of the Extremities Treated with Neoadjuvant Chemotherapy in a Single Institution. Eur. J. Cancer. 2005;41:2079–2085. doi: 10.1016/j.ejca.2005.03.036. [DOI](https://doi.org/10.1016/j.ejca.2005.03.036) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16115755/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur.%20J.%20Cancer&title=Grade%20of%20Chemotherapy-Induced%20Necrosis%20as%20a%20Predictor%20of%20Local%20and%20Systemic%20Control%20in%20881%20Patients%20with%20Non-Metastatic%20Osteosarcoma%20of%20the%20Extremities%20Treated%20with%20Neoadjuvant%20Chemotherapy%20in%20a%20Single%20Institution&author=G.%20Bacci&author=M.%20Mercuri&author=A.%20Longhi&author=S.%20Ferrari&author=F.%20Bertoni&volume=41&publication_year=2005&pages=2079-2085&pmid=16115755&doi=10.1016/j.ejca.2005.03.036&)
|
| 273 |
+
|
| 274 |
+
6. Bielack S.S., Kempf-Bielack B., Delling G., Exner G.U., Flege S., Helmke K., Kotz R., Salzer-Kuntschik M., Werner M., Winkelmann W., et al. Prognostic Factors in High-Grade Osteosarcoma of the Extremities or Trunk: An Analysis of 1,702 Patients Treated on Neoadjuvant Cooperative Osteosarcoma Study Group Protocols. J. Clin. Oncol. 2002;20:776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI](https://doi.org/10.1200/JCO.2002.20.3.776) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11821461/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Oncol.&title=Prognostic%20Factors%20in%20High-Grade%20Osteosarcoma%20of%20the%20Extremities%20or%20Trunk:%20An%20Analysis%20of%201,702%20Patients%20Treated%20on%20Neoadjuvant%20Cooperative%20Osteosarcoma%20Study%20Group%20Protocols&author=S.S.%20Bielack&author=B.%20Kempf-Bielack&author=G.%20Delling&author=G.U.%20Exner&author=S.%20Flege&volume=20&publication_year=2002&pages=776-790&pmid=11821461&doi=10.1200/JCO.2002.20.3.776&)
|
| 275 |
+
|
| 276 |
+
7. Whelan J.S., Bielack S.S., Marina N., Smeland S., Jovic G., Hook J.M., Krailo M., Anninga J., Butterfass-Bahloul T., Böhling T., et al. EURAMOS-1, an International Randomised Study for Osteosarcoma: Results from Pre-Randomisation Treatment. Ann. Oncol. 2015;26:407–414. doi: 10.1093/annonc/mdu526. [DOI](https://doi.org/10.1093/annonc/mdu526) | [PMC free article](/articles/PMC4304379/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25421877/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann.%20Oncol.&title=EURAMOS-1,%20an%20International%20Randomised%20Study%20for%20Osteosarcoma:%20Results%20from%20Pre-Randomisation%20Treatment&author=J.S.%20Whelan&author=S.S.%20Bielack&author=N.%20Marina&author=S.%20Smeland&author=G.%20Jovic&volume=26&publication_year=2015&pages=407-414&pmid=25421877&doi=10.1093/annonc/mdu526&)
|
| 277 |
+
|
| 278 |
+
8. Marina N.M., Smeland S., Bielack S.S., Bernstein M., Jovic G., Krailo M.D., Hook J.M., Arndt C., van den Berg H., Brennan B., et al. Comparison of MAPIE versus MAP in Patients with a Poor Response to Preoperative Chemotherapy for Newly Diagnosed High-Grade Osteosarcoma (EURAMOS-1): An Open-Label, International, Randomised Controlled Trial. Lancet Oncol. 2016;17:1396–1408. doi: 10.1016/S1470-2045(16)30214-5. [DOI](https://doi.org/10.1016/S1470-2045(16)30214-5) | [PMC free article](/articles/PMC5052459/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27569442/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet%20Oncol.&title=Comparison%20of%20MAPIE%20versus%20MAP%20in%20Patients%20with%20a%20Poor%20Response%20to%20Preoperative%20Chemotherapy%20for%20Newly%20Diagnosed%20High-Grade%20Osteosarcoma%20(EURAMOS-1):%20An%20Open-Label,%20International,%20Randomised%20Controlled%20Trial&author=N.M.%20Marina&author=S.%20Smeland&author=S.S.%20Bielack&author=M.%20Bernstein&author=G.%20Jovic&volume=17&publication_year=2016&pages=1396-1408&pmid=27569442&doi=10.1016/S1470-2045(16)30214-5&)
|
| 279 |
+
|
| 280 |
+
9. Janeway K.A., Grier H.E. Sequelae of Osteosarcoma Medical Therapy: A Review of Rare Acute Toxicities and Late Effects. Lancet Oncol. 2010;11:670–678. doi: 10.1016/S1470-2045(10)70062-0. [DOI](https://doi.org/10.1016/S1470-2045(10)70062-0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20347613/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet%20Oncol.&title=Sequelae%20of%20Osteosarcoma%20Medical%20Therapy:%20A%20Review%20of%20Rare%20Acute%20Toxicities%20and%20Late%20Effects&author=K.A.%20Janeway&author=H.E.%20Grier&volume=11&publication_year=2010&pages=670-678&pmid=20347613&doi=10.1016/S1470-2045(10)70062-0&)
|
| 281 |
+
|
| 282 |
+
10. Chan E.S.L., Cronstein B.N. Mechanisms of Action of Methotrexate. Bull. Hosp. Joint Dis. 2013;71:S5–S8. [PubMed](https://pubmed.ncbi.nlm.nih.gov/24219035/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Bull.%20Hosp.%20Joint%20Dis.&title=Mechanisms%20of%20Action%20of%20Methotrexate&author=E.S.L.%20Chan&author=B.N.%20Cronstein&volume=71&publication_year=2013&pages=S5-S8&pmid=24219035&)
|
| 283 |
+
|
| 284 |
+
11. Rabik C.A., Dolan M.E. Molecular Mechanisms of Resistance and Toxicity Associated with Platinating Agents. Cancer Treat. Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI](https://doi.org/10.1016/j.ctrv.2006.09.006) | [PMC free article](/articles/PMC1855222/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17084534/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cancer%20Treat.%20Rev.&title=Molecular%20Mechanisms%20of%20Resistance%20and%20Toxicity%20Associated%20with%20Platinating%20Agents&author=C.A.%20Rabik&author=M.E.%20Dolan&volume=33&publication_year=2007&pages=9-23&pmid=17084534&doi=10.1016/j.ctrv.2006.09.006&)
|
| 285 |
+
|
| 286 |
+
12. Zamble D.B., Mu D., Reardon J.T., Sancar A., Lippard S.J. Repair of Cisplatin-DNA Adducts by the Mammalian Excision Nuclease. Biochemistry. 1996;35:10004–10013. doi: 10.1021/bi960453+. [DOI](https://doi.org/10.1021/bi960453+) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8756462/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Biochemistry&title=Repair%20of%20Cisplatin-DNA%20Adducts%20by%20the%20Mammalian%20Excision%20Nuclease&author=D.B.%20Zamble&author=D.%20Mu&author=J.T.%20Reardon&author=A.%20Sancar&author=S.J.%20Lippard&volume=35&publication_year=1996&pages=10004-10013&pmid=8756462&doi=10.1021/bi960453+&)
|
| 287 |
+
|
| 288 |
+
13. Singh R.R., Reindl K.M. Glutathione S-Transferases in Cancer. Antioxidants. 2021;10:701. doi: 10.3390/antiox10050701. [DOI](https://doi.org/10.3390/antiox10050701) | [PMC free article](/articles/PMC8146591/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33946704/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antioxidants&title=Glutathione%20S-Transferases%20in%20Cancer&author=R.R.%20Singh&author=K.M.%20Reindl&volume=10&publication_year=2021&pages=701&pmid=33946704&doi=10.3390/antiox10050701&)
|
| 289 |
+
|
| 290 |
+
14. Caronia D., Patiño-García A., Milne R.L., Zalacain-Díez M., Pita G., Alonso M.R., Moreno L.T., Sierrasesumaga-Ariznabarreta L., Benítez J., Gonzáles-Neira A. Common Variations in ERCC2 Are Associated with Response to Cisplatin Chemotherapy and Clinical Outcome in Osteosarcoma Patients. Pharmacogenomics J. 2009;9:347–353. doi: 10.1038/tpj.2009.19. [DOI](https://doi.org/10.1038/tpj.2009.19) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19434073/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J.&title=Common%20Variations%20in%20ERCC2%20Are%20Associated%20with%20Response%20to%20Cisplatin%20Chemotherapy%20and%20Clinical%20Outcome%20in%20Osteosarcoma%20Patients&author=D.%20Caronia&author=A.%20Pati%C3%B1o-Garc%C3%ADa&author=R.L.%20Milne&author=M.%20Zalacain-D%C3%ADez&author=G.%20Pita&volume=9&publication_year=2009&pages=347-353&pmid=19434073&doi=10.1038/tpj.2009.19&)
|
| 291 |
+
|
| 292 |
+
15. Jabeen S., Holmboe L., Alnæs G.I.G., Andersen A.M., Hall K.S., Kristensen V.N. Impact of Genetic Variants of RFC1, DHFR and MTHFR in Osteosarcoma Patients Treated with High-Dose Methotrexate. Pharmacogenomics J. 2015;15:385–390. doi: 10.1038/tpj.2015.11. [DOI](https://doi.org/10.1038/tpj.2015.11) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25778468/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J.&title=Impact%20of%20Genetic%20Variants%20of%20RFC1,%20DHFR%20and%20MTHFR%20in%20Osteosarcoma%20Patients%20Treated%20with%20High-Dose%20Methotrexate&author=S.%20Jabeen&author=L.%20Holmboe&author=G.I.G.%20Aln%C3%A6s&author=A.M.%20Andersen&author=K.S.%20Hall&volume=15&publication_year=2015&pages=385-390&pmid=25778468&doi=10.1038/tpj.2015.11&)
|
| 293 |
+
|
| 294 |
+
16. Hegyi M., Arany A., Semsei A.F., Csordas K., Eipel O., Gezsi A., Kutszegi N., Csoka M., Muller J., Erdelyi D.J., et al. Pharmacogenetic Analysis of High-Dose Methotrexate Treatment in Children with Osteosarcoma. Oncotarget. 2017;8:9388–9398. doi: 10.18632/oncotarget.11543. [DOI](https://doi.org/10.18632/oncotarget.11543) | [PMC free article](/articles/PMC5354739/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27566582/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Oncotarget&title=Pharmacogenetic%20Analysis%20of%20High-Dose%20Methotrexate%20Treatment%20in%20Children%20with%20Osteosarcoma&author=M.%20Hegyi&author=A.%20Arany&author=A.F.%20Semsei&author=K.%20Csordas&author=O.%20Eipel&volume=8&publication_year=2017&pages=9388-9398&pmid=27566582&doi=10.18632/oncotarget.11543&)
|
| 295 |
+
|
| 296 |
+
17. Liu Z.F., Asila A.L.J., Aikenmu K., Zhao J., Meng Q.C., Fang R. Influence of ERCC2 Gene Polymorphisms on the Treatment Outcome of Osteosarcoma. Genet. Mol. Res. 2015;14:12967–12972. doi: 10.4238/2015.October.21.17. [DOI](https://doi.org/10.4238/2015.October.21.17) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26505449/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genet.%20Mol.%20Res.&title=Influence%20of%20ERCC2%20Gene%20Polymorphisms%20on%20the%20Treatment%20Outcome%20of%20Osteosarcoma&author=Z.F.%20Liu&author=A.L.J.%20Asila&author=K.%20Aikenmu&author=J.%20Zhao&author=Q.C.%20Meng&volume=14&publication_year=2015&pages=12967-12972&pmid=26505449&doi=10.4238/2015.October.21.17&)
|
| 297 |
+
|
| 298 |
+
18. Ji W.P., He N. Bin Investigation on the DNA Repaired Gene Polymorphisms and Response to Chemotherapy and Overall Survival of Osteosarcoma. Int. J. Clin. Exp. Pathol. 2015;8:894–899. [PMC free article](/articles/PMC4348835/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25755792/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int.%20J.%20Clin.%20Exp.%20Pathol.&title=Bin%20Investigation%20on%20the%20DNA%20Repaired%20Gene%20Polymorphisms%20and%20Response%20to%20Chemotherapy%20and%20Overall%20Survival%20of%20Osteosarcoma&author=W.P.%20Ji&author=N.%20He&volume=8&publication_year=2015&pages=894-899&pmid=25755792&)
|
| 299 |
+
|
| 300 |
+
19. Caronia D., Patiño-Garcia A., Peréz-Martínez A., Pita G., Moreno L.T., Zalacain-Díez M., Molina B., Colmenero I., Sierrasesúmaga L., Benítez J., et al. Effect of ABCB1 and ABCC3 Polymorphisms on Osteosarcoma Survival after Chemotherapy: A Pharmacogenetic Study. PLoS ONE. 2011;6:e26091. doi: 10.1371/journal.pone.0026091. [DOI](https://doi.org/10.1371/journal.pone.0026091) | [PMC free article](/articles/PMC3189235/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22016816/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=PLoS%20ONE&title=Effect%20of%20ABCB1%20and%20ABCC3%20Polymorphisms%20on%20Osteosarcoma%20Survival%20after%20Chemotherapy:%20A%20Pharmacogenetic%20Study&author=D.%20Caronia&author=A.%20Pati%C3%B1o-Garcia&author=A.%20Per%C3%A9z-Mart%C3%ADnez&author=G.%20Pita&author=L.T.%20Moreno&volume=6&publication_year=2011&pages=e26091&pmid=22016816&doi=10.1371/journal.pone.0026091&)
|
| 301 |
+
|
| 302 |
+
20. Windsor R.E., Strauss S.J., Kallis C., Wood N.E., Whelan J.S. Germline Genetic Polymorphisms May Influence Chemotherapy Response and Disease Outcome in Osteosarcoma: A Pilot Study. Cancer. 2012;118:1856–1867. doi: 10.1002/cncr.26472. [DOI](https://doi.org/10.1002/cncr.26472) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21887680/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cancer&title=Germline%20Genetic%20Polymorphisms%20May%20Influence%20Chemotherapy%20Response%20and%20Disease%20Outcome%20in%20Osteosarcoma:%20A%20Pilot%20Study&author=R.E.%20Windsor&author=S.J.%20Strauss&author=C.%20Kallis&author=N.E.%20Wood&author=J.S.%20Whelan&volume=118&publication_year=2012&pages=1856-1867&pmid=21887680&doi=10.1002/cncr.26472&)
|
| 303 |
+
|
| 304 |
+
21. Hattinger C.M., Biason P., Iacoboni E., Gagno S., Fanelli M., Tavanti E., Vella S., Ferrari S., Roli A., Roncato R., et al. Candidate Germline Polymorphisms of Genes Belonging to the Pathways of Four Drugs Used in Osteosarcoma Standard Chemotherapy Associated with Risk, Survival and Toxicity in Non-Metastatic High-Grade Osteosarcoma. Oncotarget. 2016;7:61970–61987. doi: 10.18632/oncotarget.11486. [DOI](https://doi.org/10.18632/oncotarget.11486) | [PMC free article](/articles/PMC5308704/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27566557/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Oncotarget&title=Candidate%20Germline%20Polymorphisms%20of%20Genes%20Belonging%20to%20the%20Pathways%20of%20Four%20Drugs%20Used%20in%20Osteosarcoma%20Standard%20Chemotherapy%20Associated%20with%20Risk,%20Survival%20and%20Toxicity%20in%20Non-Metastatic%20High-Grade%20Osteosarcoma&author=C.M.%20Hattinger&author=P.%20Biason&author=E.%20Iacoboni&author=S.%20Gagno&author=M.%20Fanelli&volume=7&publication_year=2016&pages=61970-61987&pmid=27566557&doi=10.18632/oncotarget.11486&)
|
| 305 |
+
|
| 306 |
+
22. Bhuvaneshwar K., Harris M., Gusev Y., Madhavan S., Iyer R., Vilboux T., Deeken J., Yang E., Shankar S. Genome Sequencing Analysis of Blood Cells Identifies Germline Haplotypes Strongly Associated with Drug Resistance in Osteosarcoma Patients. BMC Cancer. 2019;19:357. doi: 10.1186/s12885-019-5474-y. [DOI](https://doi.org/10.1186/s12885-019-5474-y) | [PMC free article](/articles/PMC6466653/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30991985/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=BMC%20Cancer&title=Genome%20Sequencing%20Analysis%20of%20Blood%20Cells%20Identifies%20Germline%20Haplotypes%20Strongly%20Associated%20with%20Drug%20Resistance%20in%20Osteosarcoma%20Patients&author=K.%20Bhuvaneshwar&author=M.%20Harris&author=Y.%20Gusev&author=S.%20Madhavan&author=R.%20Iyer&volume=19&publication_year=2019&pages=357&pmid=30991985&doi=10.1186/s12885-019-5474-y&)
|
| 307 |
+
|
| 308 |
+
23. Hurkmans E.G.E., Klumpers M.J., Vermeulen S.H., Hagleitner M.M., Flucke U., Schreuder H.W.B., Gelderblom H., Bras J., Guchelaar H.J., Coenen M.J.H., et al. Analysis of Drug Metabolizing Gene Panel in Osteosarcoma Patients Identifies Association Between Variants in SULT1E1, CYP2B6 and CYP4F8 and Methotrexate Levels and Toxicities. Front. Pharmacol. 2020;11:1241. doi: 10.3389/fphar.2020.01241. [DOI](https://doi.org/10.3389/fphar.2020.01241) | [PMC free article](/articles/PMC7435008/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32903464/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front.%20Pharmacol.&title=Analysis%20of%20Drug%20Metabolizing%20Gene%20Panel%20in%20Osteosarcoma%20Patients%20Identifies%20Association%20Between%20Variants%20in%20SULT1E1,%20CYP2B6%20and%20CYP4F8%20and%20Methotrexate%20Levels%20and%20Toxicities&author=E.G.E.%20Hurkmans&author=M.J.%20Klumpers&author=S.H.%20Vermeulen&author=M.M.%20Hagleitner&author=U.%20Flucke&volume=11&publication_year=2020&pages=1241&pmid=32903464&doi=10.3389/fphar.2020.01241&)
|
| 309 |
+
|
| 310 |
+
24. National Cancer Institute Common Terminology Criteria for Adverse Events (CTCAE) Version 4.0. U.S. Department of Health and Human Services; Washington, DC, USA: 2009. [Google Scholar](https://scholar.google.com/scholar_lookup?title=National%20Cancer%20Institute%20Common%20Terminology%20Criteria%20for%20Adverse%20Events%20(CTCAE)%20Version%204.0&publication_year=2009&)
|
| 311 |
+
|
| 312 |
+
25. Xie L., Guo W., Yang Y., Ji T., Xu J. More Severe Toxicity of Genetic Polymorphisms on MTHFR Activity in Osteosarcoma Patients Treated with High-Dose Methotrexate. Oncotarget. 2018;9:11465–11476. doi: 10.18632/oncotarget.23222. [DOI](https://doi.org/10.18632/oncotarget.23222) | [PMC free article](/articles/PMC5837742/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29545912/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Oncotarget&title=More%20Severe%20Toxicity%20of%20Genetic%20Polymorphisms%20on%20MTHFR%20Activity%20in%20Osteosarcoma%20Patients%20Treated%20with%20High-Dose%20Methotrexate&author=L.%20Xie&author=W.%20Guo&author=Y.%20Yang&author=T.%20Ji&author=J.%20Xu&volume=9&publication_year=2018&pages=11465-11476&pmid=29545912&doi=10.18632/oncotarget.23222&)
|
| 313 |
+
|
| 314 |
+
26. Hattinger C.M., Patrizio M.P., Luppi S., Serra M. Pharmacogenomics and Pharmacogenetics in Osteosarcoma: Translational Studies and Clinical Impact. Int. J. Mol. Sci. 2020;21:4659. doi: 10.3390/ijms21134659. [DOI](https://doi.org/10.3390/ijms21134659) | [PMC free article](/articles/PMC7369799/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32629971/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int.%20J.%20Mol.%20Sci.&title=Pharmacogenomics%20and%20Pharmacogenetics%20in%20Osteosarcoma:%20Translational%20Studies%20and%20Clinical%20Impact&author=C.M.%20Hattinger&author=M.P.%20Patrizio&author=S.%20Luppi&author=M.%20Serra&volume=21&publication_year=2020&pages=4659&pmid=32629971&doi=10.3390/ijms21134659&)
|
| 315 |
+
|
| 316 |
+
27. Zhang W., Liu Z., Yang Z., Feng C., Zhou X., Tu C., Li Z. MTHFR Polymorphism Is Associated With Severe Methotrexate-Induced Toxicity in Osteosarcoma Treatment. Front. Oncol. 2021;11:781386. doi: 10.3389/fonc.2021.781386. [DOI](https://doi.org/10.3389/fonc.2021.781386) | [PMC free article](/articles/PMC8714641/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34976820/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front.%20Oncol.&title=MTHFR%20Polymorphism%20Is%20Associated%20With%20Severe%20Methotrexate-Induced%20Toxicity%20in%20Osteosarcoma%20Treatment&author=W.%20Zhang&author=Z.%20Liu&author=Z.%20Yang&author=C.%20Feng&author=X.%20Zhou&volume=11&publication_year=2021&pages=781386&pmid=34976820&doi=10.3389/fonc.2021.781386&)
|
| 317 |
+
|
| 318 |
+
28. Biason P., Hattinger C.M., Innocenti F., Talamini R., Alberghini M., Scotlandi K., Zanusso C., Serra M., Toffoli G. Nucleotide Excision Repair Gene Variants and Association with Survival in Osteosarcoma Patients Treated with Neoadjuvant Chemotherapy. Pharmacogenomics J. 2012;12:476–483. doi: 10.1038/tpj.2011.33. [DOI](https://doi.org/10.1038/tpj.2011.33) | [PMC free article](/articles/PMC3935514/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21826087/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J.&title=Nucleotide%20Excision%20Repair%20Gene%20Variants%20and%20Association%20with%20Survival%20in%20Osteosarcoma%20Patients%20Treated%20with%20Neoadjuvant%20Chemotherapy&author=P.%20Biason&author=C.M.%20Hattinger&author=F.%20Innocenti&author=R.%20Talamini&author=M.%20Alberghini&volume=12&publication_year=2012&pages=476-483&pmid=21826087&doi=10.1038/tpj.2011.33&)
|
| 319 |
+
|
| 320 |
+
29. Hattinger C.M., Casotti C., Patrizio M.P., Luppi S., Fantoni L., Scotlandi K., Ibrahim T., Serra M. Pharmacogenomic Profiling of Cisplatin-Resistant and -Sensitive Human Osteosarcoma Cell Lines by Multimodal Targeted Next Generation Sequencing. Int. J. Mol. Sci. 2022;23:11787. doi: 10.3390/ijms231911787. [DOI](https://doi.org/10.3390/ijms231911787) | [PMC free article](/articles/PMC9570120/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36233089/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int.%20J.%20Mol.%20Sci.&title=Pharmacogenomic%20Profiling%20of%20Cisplatin-Resistant%20and%20-Sensitive%20Human%20Osteosarcoma%20Cell%20Lines%20by%20Multimodal%20Targeted%20Next%20Generation%20Sequencing&author=C.M.%20Hattinger&author=C.%20Casotti&author=M.P.%20Patrizio&author=S.%20Luppi&author=L.%20Fantoni&volume=23&publication_year=2022&pages=11787&pmid=36233089&doi=10.3390/ijms231911787&)
|
| 321 |
+
|
| 322 |
+
30. Hurkmans E.G.E., Brand A.C.A.M., Verdonschot J.A.J., te Loo D.M.W.M., Coenen M.J.H. Pharmacogenetics of Chemotherapy Treatment Response and -Toxicities in Patients with Osteosarcoma: A Systematic Review. BMC Cancer. 2022;22:1326. doi: 10.1186/s12885-022-10434-5. [DOI](https://doi.org/10.1186/s12885-022-10434-5) | [PMC free article](/articles/PMC9761983/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36536332/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=BMC%20Cancer&title=Pharmacogenetics%20of%20Chemotherapy%20Treatment%20Response%20and%20-Toxicities%20in%20Patients%20with%20Osteosarcoma:%20A%20Systematic%20Review&author=E.G.E.%20Hurkmans&author=A.C.A.M.%20Brand&author=J.A.J.%20Verdonschot&author=D.M.W.M.%20te%20Loo&author=M.J.H.%20Coenen&volume=22&publication_year=2022&pages=1326&pmid=36536332&doi=10.1186/s12885-022-10434-5&)
|
| 323 |
+
|
| 324 |
+
31. Liu B., Liu G., Liu B., Guo Y., Peng N., Li T. Correlation between Gene Polymorphism and Adverse Reactions of High-Dose Methotrexate in Osteosarcoma Patients: A Systematic Review and Meta-Analysis. World J. Surg. Oncol. 2024;22:19. doi: 10.1186/s12957-023-03287-0. [DOI](https://doi.org/10.1186/s12957-023-03287-0) | [PMC free article](/articles/PMC10782754/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38212758/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=World%20J.%20Surg.%20Oncol.&title=Correlation%20between%20Gene%20Polymorphism%20and%20Adverse%20Reactions%20of%20High-Dose%20Methotrexate%20in%20Osteosarcoma%20Patients:%20A%20Systematic%20Review%20and%20Meta-Analysis&author=B.%20Liu&author=G.%20Liu&author=B.%20Liu&author=Y.%20Guo&author=N.%20Peng&volume=22&publication_year=2024&pages=19&pmid=38212758&doi=10.1186/s12957-023-03287-0&)
|
| 325 |
+
|
| 326 |
+
32. Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E., Flicek P., et al. A Global Reference for Human Genetic Variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI](https://doi.org/10.1038/nature15393) | [PMC free article](/articles/PMC4750478/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26432245/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nature&title=A%20Global%20Reference%20for%20Human%20Genetic%20Variation&author=A.%20Auton&author=G.R.%20Abecasis&author=D.M.%20Altshuler&author=R.M.%20Durbin&author=D.R.%20Bentley&volume=526&publication_year=2015&pages=68-74&pmid=26432245&doi=10.1038/nature15393&)
|
| 327 |
+
|
| 328 |
+
33. Serra M., Scotlandi K., Reverter-Branchat G., Ferrari S., Manara M.C., Benini S., Incaprera M., Bertoni F., Mercuri M., Briccoli A., et al. Value of P-Glycoprotein and Clinicopathologic Factors as the Basis for New Treatment Strategies in High-Grade Osteosarcoma of the Extremities. J. Clin. Oncol. 2003;21:536–542. doi: 10.1200/JCO.2003.03.144. [DOI](https://doi.org/10.1200/JCO.2003.03.144) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12560446/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Oncol.&title=Value%20of%20P-Glycoprotein%20and%20Clinicopathologic%20Factors%20as%20the%20Basis%20for%20New%20Treatment%20Strategies%20in%20High-Grade%20Osteosarcoma%20of%20the%20Extremities&author=M.%20Serra&author=K.%20Scotlandi&author=G.%20Reverter-Branchat&author=S.%20Ferrari&author=M.C.%20Manara&volume=21&publication_year=2003&pages=536-542&pmid=12560446&doi=10.1200/JCO.2003.03.144&)
|
| 329 |
+
|
| 330 |
+
34. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., De Bakker P.I.W., Daly M.J., et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI](https://doi.org/10.1086/519795) | [PMC free article](/articles/PMC1950838/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17701901/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Hum.%20Genet.&title=PLINK:%20A%20Tool%20Set%20for%20Whole-Genome%20Association%20and%20Population-Based%20Linkage%20Analyses&author=S.%20Purcell&author=B.%20Neale&author=K.%20Todd-Brown&author=L.%20Thomas&author=M.A.R.%20Ferreira&volume=81&publication_year=2007&pages=559-575&pmid=17701901&doi=10.1086/519795&)
|
| 331 |
+
|
| 332 |
+
35. Serra M., Hattinger C.M. The Pharmacogenomics of Osteosarcoma. Pharmacogenomics J. 2017;17:11–20. doi: 10.1038/tpj.2016.45. [DOI](https://doi.org/10.1038/tpj.2016.45) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27241064/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J.&title=The%20Pharmacogenomics%20of%20Osteosarcoma&author=M.%20Serra&author=C.M.%20Hattinger&volume=17&publication_year=2017&pages=11-20&pmid=27241064&doi=10.1038/tpj.2016.45&)
|
| 333 |
+
|
| 334 |
+
36. Palmerini E., Meazza C., Tamburini A., Bisogno G., Ferraresi V., Asaftei S.D., Milano G.M., Coccoli L., Manzitti C., Luksch R., et al. Phase 2 Study for Nonmetastatic Extremity High-Grade Osteosarcoma in Pediatric and Adolescent and Young Adult Patients with a Risk-Adapted Strategy Based on ABCB1/P-Glycoprotein Expression: An Italian Sarcoma Group Trial (ISG/OS-2) Cancer. 2022;128:1958–1966. doi: 10.1002/cncr.34131. [DOI](https://doi.org/10.1002/cncr.34131) | [PMC free article](/articles/PMC9305236/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35201621/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cancer&title=Phase%202%20Study%20for%20Nonmetastatic%20Extremity%20High-Grade%20Osteosarcoma%20in%20Pediatric%20and%20Adolescent%20and%20Young%20Adult%20Patients%20with%20a%20Risk-Adapted%20Strategy%20Based%20on%20ABCB1/P-Glycoprotein%20Expression:%20An%20Italian%20Sarcoma%20Group%20Trial%20(ISG/OS-2)&author=E.%20Palmerini&author=C.%20Meazza&author=A.%20Tamburini&author=G.%20Bisogno&author=V.%20Ferraresi&volume=128&publication_year=2022&pages=1958-1966&pmid=35201621&doi=10.1002/cncr.34131&)
|
| 335 |
+
|
| 336 |
+
37. Zhao Z.G., Ding F., Liu M., Ma D.Z., Zheng C.K., Kan W.S. Association between P-Glycoprotein Expression and Response to Chemotherapy in Patients with Osteosarcoma: A Systematic and Meta-Analysis. J. Cancer Res. Ther. 2014;10:C206–C209. doi: 10.4103/0973-1482.145874. [DOI](https://doi.org/10.4103/0973-1482.145874) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25450283/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Cancer%20Res.%20Ther.&title=Association%20between%20P-Glycoprotein%20Expression%20and%20Response%20to%20Chemotherapy%20in%20Patients%20with%20Osteosarcoma:%20A%20Systematic%20and%20Meta-Analysis&author=Z.G.%20Zhao&author=F.%20Ding&author=M.%20Liu&author=D.Z.%20Ma&author=C.K.%20Zheng&volume=10&publication_year=2014&pages=C206-C209&pmid=25450283&doi=10.4103/0973-1482.145874&)
|
| 337 |
+
|
| 338 |
+
38. Schwartz C.L., Gorlick R., Teot L., Krailo M., Chen Z., Goorin A., Grier H.E., Bernstein M.L., Meyers P. Multiple Drug Resistance in Osteogenic Sarcoma: INT0133 from the Children’s Oncology Group. J. Clin. Oncol. 2007;25:2057–2062. doi: 10.1200/JCO.2006.07.7776. [DOI](https://doi.org/10.1200/JCO.2006.07.7776) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17513810/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Clin.%20Oncol.&title=Multiple%20Drug%20Resistance%20in%20Osteogenic%20Sarcoma:%20INT0133%20from%20the%20Children%E2%80%99s%20Oncology%20Group&author=C.L.%20Schwartz&author=R.%20Gorlick&author=L.%20Teot&author=M.%20Krailo&author=Z.%20Chen&volume=25&publication_year=2007&pages=2057-2062&pmid=17513810&doi=10.1200/JCO.2006.07.7776&)
|
| 339 |
+
|
| 340 |
+
39. Laechelt S., Turrini E., Ruehmkorf A., Siegmund W., Cascorbi I., Haenisch S. Impact of ABCC2 Haplotypes on Transcriptional and Posttranscriptional Gene Regulation and Function. Pharmacogenomics J. 2011;11:25–34. doi: 10.1038/tpj.2010.20. [DOI](https://doi.org/10.1038/tpj.2010.20) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20351751/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J.&title=Impact%20of%20ABCC2%20Haplotypes%20on%20Transcriptional%20and%20Posttranscriptional%20Gene%20Regulation%20and%20Function&author=S.%20Laechelt&author=E.%20Turrini&author=A.%20Ruehmkorf&author=W.%20Siegmund&author=I.%20Cascorbi&volume=11&publication_year=2011&pages=25-34&pmid=20351751&doi=10.1038/tpj.2010.20&)
|
| 341 |
+
|
| 342 |
+
40. Haenisch S., May K., Wegner D., Caliebe A., Cascorbi I., Siegmund W. Influence of Genetic Polymorphisms on Intestinal Expression and Rifampicin-Type Induction of ABCC2 and on Bioavailability of Talinolol. Pharmacogenet. Genomics. 2008;18:357–365. doi: 10.1097/FPC.0b013e3282f974b7. [DOI](https://doi.org/10.1097/FPC.0b013e3282f974b7) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18334920/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenet.%20Genomics&title=Influence%20of%20Genetic%20Polymorphisms%20on%20Intestinal%20Expression%20and%20Rifampicin-Type%20Induction%20of%20ABCC2%20and%20on%20Bioavailability%20of%20Talinolol&author=S.%20Haenisch&author=K.%20May&author=D.%20Wegner&author=A.%20Caliebe&author=I.%20Cascorbi&volume=18&publication_year=2008&pages=357-365&pmid=18334920&doi=10.1097/FPC.0b013e3282f974b7&)
|
| 343 |
+
|
| 344 |
+
41. Ogasawara K., Chitnis S.D., Gohh R.Y., Christians U., Akhlaghi F. Multidrug Resistance-Associated Protein 2 (MRP2/ABCC2) Haplotypes Significantly Affect the Pharmacokinetics of Tacrolimus in Kidney Transplant Recipients. Clin. Pharmacokinet. 2013;52:751–762. doi: 10.1007/s40262-013-0069-2. [DOI](https://doi.org/10.1007/s40262-013-0069-2) | [PMC free article](/articles/PMC3755037/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23633119/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Pharmacokinet.&title=Multidrug%20Resistance-Associated%20Protein%202%20(MRP2/ABCC2)%20Haplotypes%20Significantly%20Affect%20the%20Pharmacokinetics%20of%20Tacrolimus%20in%20Kidney%20Transplant%20Recipients&author=K.%20Ogasawara&author=S.D.%20Chitnis&author=R.Y.%20Gohh&author=U.%20Christians&author=F.%20Akhlaghi&volume=52&publication_year=2013&pages=751-762&pmid=23633119&doi=10.1007/s40262-013-0069-2&)
|
| 345 |
+
|
| 346 |
+
42. Benhamou S., Sarasin A. ERCC2/XPD Gene Polymorphisms and Lung Cancer: A HuGE Review. Am. J. Epidemiol. 2005;161:1–14. doi: 10.1093/aje/kwi018. [DOI](https://doi.org/10.1093/aje/kwi018) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15615908/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am.%20J.%20Epidemiol.&title=ERCC2/XPD%20Gene%20Polymorphisms%20and%20Lung%20Cancer:%20A%20HuGE%20Review&author=S.%20Benhamou&author=A.%20Sarasin&volume=161&publication_year=2005&pages=1-14&pmid=15615908&doi=10.1093/aje/kwi018&)
|
| 347 |
+
|
| 348 |
+
43. Goričar K., Kovač V., Jazbec J., Zakotnik B., Lamovec J., Dolžan V. Genetic Variability of DNA Repair Mechanisms and Glutathione-S-Transferase Genes Influences Treatment Outcome in Osteosarcoma. Cancer Epidemiol. 2015;39:182–188. doi: 10.1016/j.canep.2014.12.009. [DOI](https://doi.org/10.1016/j.canep.2014.12.009) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25592234/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cancer%20Epidemiol.&title=Genetic%20Variability%20of%20DNA%20Repair%20Mechanisms%20and%20Glutathione-S-Transferase%20Genes%20Influences%20Treatment%20Outcome%20in%20Osteosarcoma&author=K.%20Gori%C4%8Dar&author=V.%20Kova%C4%8D&author=J.%20Jazbec&author=B.%20Zakotnik&author=J.%20Lamovec&volume=39&publication_year=2015&pages=182-188&pmid=25592234&doi=10.1016/j.canep.2014.12.009&)
|
| 349 |
+
|
| 350 |
+
44. Mikkelsen T.S., Thorn C.F., Yang J.J., Ulrich C.M., French D., Zaza G., Dunnenberger H.M., Marsh S., McLeod H.L., Giacomini K., et al. PharmGKB Summary: Methotrexate Pathway. Pharmacogenet. Genomics. 2011;21:679–686. doi: 10.1097/FPC.0b013e328343dd93. [DOI](https://doi.org/10.1097/FPC.0b013e328343dd93) | [PMC free article](/articles/PMC3139712/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21317831/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenet.%20Genomics&title=PharmGKB%20Summary:%20Methotrexate%20Pathway&author=T.S.%20Mikkelsen&author=C.F.%20Thorn&author=J.J.%20Yang&author=C.M.%20Ulrich&author=D.%20French&volume=21&publication_year=2011&pages=679-686&pmid=21317831&doi=10.1097/FPC.0b013e328343dd93&)
|
| 351 |
+
|
| 352 |
+
45. Fung K.L., Gottesman M.M. A Synonymous Polymorphism in a Common MDR1 (ABCB1) Haplotype Shapes Protein Function. Biochim. Biophys. Acta. 2009;1794:860–871. doi: 10.1016/j.bbapap.2009.02.014. [DOI](https://doi.org/10.1016/j.bbapap.2009.02.014) | [PMC free article](/articles/PMC2810319/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19285158/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Biochim.%20Biophys.%20Acta&title=A%20Synonymous%20Polymorphism%20in%20a%20Common%20MDR1%20(ABCB1)%20Haplotype%20Shapes%20Protein%20Function&author=K.L.%20Fung&author=M.M.%20Gottesman&volume=1794&publication_year=2009&pages=860-871&pmid=19285158&doi=10.1016/j.bbapap.2009.02.014&)
|
| 353 |
+
|
| 354 |
+
46. Gottesman M.M., Fojo T., Bates S.E. Multidrug Resistance in Cancer: Role of ATP-Dependent Transporters. Nat. Rev. Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI](https://doi.org/10.1038/nrc706) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11902585/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat.%20Rev.%20Cancer&title=Multidrug%20Resistance%20in%20Cancer:%20Role%20of%20ATP-Dependent%20Transporters&author=M.M.%20Gottesman&author=T.%20Fojo&author=S.E.%20Bates&volume=2&publication_year=2002&pages=48-58&pmid=11902585&doi=10.1038/nrc706&)
|
| 355 |
+
|
| 356 |
+
47. Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. Annotation of Functional Variation in Personal Genomes Using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI](https://doi.org/10.1101/gr.137323.112) | [PMC free article](/articles/PMC3431494/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22955989/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genome%20Res.&title=Annotation%20of%20Functional%20Variation%20in%20Personal%20Genomes%20Using%20RegulomeDB&author=A.P.%20Boyle&author=E.L.%20Hong&author=M.%20Hariharan&author=Y.%20Cheng&author=M.A.%20Schaub&volume=22&publication_year=2012&pages=1790-1797&pmid=22955989&doi=10.1101/gr.137323.112&)
|
| 357 |
+
|
| 358 |
+
48. Lang T., Hitzi M., Burk O., Mornhinweg E., Keil A., Kerb R., Klein K., Zanger U.M., Eichelbaum M., Fromm M.F. Genetic Polymorphisms in the Multidrug Resistance-Associated Protein 3 (ABCC3, MRP3) Gene and Relationship to Its MRNA and Protein Expression in Human Liver. Pharmacogenetics. 2004;14:155–164. doi: 10.1097/00008571-200403000-00003. [DOI](https://doi.org/10.1097/00008571-200403000-00003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15167703/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Genetic%20Polymorphisms%20in%20the%20Multidrug%20Resistance-Associated%20Protein%203%20(ABCC3,%20MRP3)%20Gene%20and%20Relationship%20to%20Its%20MRNA%20and%20Protein%20Expression%20in%20Human%20Liver&author=T.%20Lang&author=M.%20Hitzi&author=O.%20Burk&author=E.%20Mornhinweg&author=A.%20Keil&volume=14&publication_year=2004&pages=155-164&pmid=15167703&doi=10.1097/00008571-200403000-00003&)
|
test/texts/PMC11787782.md
ADDED
|
@@ -0,0 +1,353 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Pharmacodynamic Modeling of Warfarin Dosing Algorithm for Cardiovascular Patients in Indonesia: A Tailored Method to Anticoagulation Therapy
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Norisca Aliza Putriana, Irma Rahayu Latarissa, Taofik Rusdiana, Tina Rostinawati, Mohammad Rizki Akbar
|
| 5 |
+
**Journal:** Drug Design, Development and Therapy
|
| 6 |
+
**Date:** 2025 Jan 29
|
| 7 |
+
**DOI:** [10.2147/DDDT.S497738](https://doi.org/10.2147/DDDT.S497738)
|
| 8 |
+
**PMID:** 39896937
|
| 9 |
+
**PMCID:** PMC11787782
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11787782/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11787782/pdf/dddt-19-671.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11787782/pdf/dddt-19-671.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Purpose:**
|
| 16 |
+
Warfarin is an anticoagulant drug widely used for treating thromboembolism-related conditions. The main challenge with this drug is the high variability in patients response, which is influenced by both clinical, non-clinical, and genetic factors, such as VKORC1, CYP2C9, and CYP4F2. Therefore, this research aimed to evaluate the impact of clinical and genetic factors on warfarin dose adjustment and to develop a dosing algorithm for patients with cardiovascular disease.
|
| 17 |
+
|
| 18 |
+
**Patients and Methods:**
|
| 19 |
+
A total of 77 research subjects were selected using consecutive sampling based on the inclusion criteria of cardiac outpatients on warfarin for ≥3 months with PT-INR data, complete medical records, and willingness to participate. Exclusion criteria included vitamin K use and inability to follow up. Patients demographic data and clinical characteristics were collected from medical records. Blood samples were obtained for genetic testing of CYP4F2 rs2108622 (sequencing). Statistical analyses included both bivariate and multivariate analyses (logistic regression) with a significance level set at <0.05.
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
Statistical analysis using the Kruskal–Wallis test showed that the CC, CT, and TT genotypes were significantly associated with warfarin dose (p = 0.02). Furthermore, the Mann–Whitney test results showed that gender did not have a significant relationship with warfarin dose (p = 0.16). The Spearman Rank correlation test showed that age (p = 0.02) and BMI (p = 0.03) had significant relationships with warfarin dose (p < 0.05). However, gender (p = 0.89) had no effect, while age (p = 0.01), BMI (p = 0.01), and genotype (p = 0.01) significantly influenced warfarin dose determination.
|
| 23 |
+
|
| 24 |
+
**Conclusion:**
|
| 25 |
+
In conclusion, the combined contribution of age (8.76%), BMI (7.95%), and CYP4F2 genotype (8.29%) to warfarin dose adjustment was 25%. The linear regression model for predicting warfarin dose was determined to be y = 12.736–0.16*age + 0.55*BMI + 3.55*genotype, where 1 = CC, 2 = CT, and 3 = TT.
|
| 26 |
+
|
| 27 |
+
Keywords: warfarin dosing, cardiovascular disease, CYP4F2 genotype, anticoagulation therapy, genetic polymorphism
|
| 28 |
+
|
| 29 |
+
### Purpose
|
| 30 |
+
|
| 31 |
+
Warfarin is an anticoagulant drug widely used for treating thromboembolism-related conditions. The main challenge with this drug is the high variability in patients response, which is influenced by both clinical, non-clinical, and genetic factors, such as *VKORC1, CYP2C9*VKORC1, CYP2C9, and *CYP4F2*CYP4F2. Therefore, this research aimed to evaluate the impact of clinical and genetic factors on warfarin dose adjustment and to develop a dosing algorithm for patients with cardiovascular disease.
|
| 32 |
+
|
| 33 |
+
### Patients and Methods
|
| 34 |
+
|
| 35 |
+
A total of 77 research subjects were selected using consecutive sampling based on the inclusion criteria of cardiac outpatients on warfarin for ≥3 months with PT-INR data, complete medical records, and willingness to participate. Exclusion criteria included vitamin K use and inability to follow up. Patients demographic data and clinical characteristics were collected from medical records. Blood samples were obtained for genetic testing of *CYP4F2 rs2108622*CYP4F2 rs2108622 (sequencing). Statistical analyses included both bivariate and multivariate analyses (logistic regression) with a significance level set at <0.05.
|
| 36 |
+
|
| 37 |
+
### Results
|
| 38 |
+
|
| 39 |
+
Statistical analysis using the Kruskal–Wallis test showed that the CC, CT, and TT genotypes were significantly associated with warfarin dose (p = 0.02). Furthermore, the Mann–Whitney test results showed that gender did not have a significant relationship with warfarin dose (p = 0.16). The Spearman Rank correlation test showed that age (p = 0.02) and BMI (p = 0.03) had significant relationships with warfarin dose (p < 0.05). However, gender (p = 0.89) had no effect, while age (p = 0.01), BMI (p = 0.01), and genotype (p = 0.01) significantly influenced warfarin dose determination.
|
| 40 |
+
|
| 41 |
+
### Conclusion
|
| 42 |
+
|
| 43 |
+
In conclusion, the combined contribution of age (8.76%), BMI (7.95%), and *CYP4F2*CYP4F2 genotype (8.29%) to warfarin dose adjustment was 25%. The linear regression model for predicting warfarin dose was determined to be y = 12.736–0.16*age + 0.55*BMI + 3.55*genotype, where 1 = CC, 2 = CT, and 3 = TT.
|
| 44 |
+
|
| 45 |
+
**Keywords:**Keywords: warfarin dosing, cardiovascular disease, CYP4F2 genotype, anticoagulation therapy, genetic polymorphism
|
| 46 |
+
|
| 47 |
+
## Introduction
|
| 48 |
+
|
| 49 |
+
Warfarin is a class of anticoagulant drugs that are often used to treat diseases associated with thromboembolism, such as atrial fibrillation, venous thrombosis, and pulmonary thrombosis.[1](#cit0001)1,[2](#cit0002)2 The main problem with the use of warfarin is that the variation in response between patients is very high.[3](#cit0003)3 This causes difficulty in determining the initial dose of each patients appropriately, which will then result in the occurrence of DRP (drug-related problem) cases in the form of adverse drug reactions.[3–5](#cit0003)3–5 The high variation occurs due to the uniqueness of the drugs, which has the characteristics of a narrow therapeutic index. Therefore, underdose condition results in inadequate treatment or complications, while overdose leads to bleeding phenomena, ranging from severe instances such as cerebral hemorrhage to minor cases, namely ocular bleeding.[6–9](#cit0006)6–9
|
| 50 |
+
|
| 51 |
+
During the COVID-19 pandemic, the use of anticoagulants, including warfarin, gained significant attention due to the increased risk of thromboembolic complications in infected patients.[10–13](#cit0010)10–13 This highlights the critical need for precise warfarin dosing, as mismanagement could exacerbate complications related to both thromboembolism and bleeding. A previous study showed that 44% of patients who experienced bleeding had an INR value >3.0, whereas 48% of patients with thromboembolic events had an INR value <2.15.[14](#cit0014)14 These findings highlight the significant risks associated with improper dosing and the need for careful monitoring of INR values in warfarin therapy.
|
| 52 |
+
|
| 53 |
+
Some of the factors that cause significant variations in response to warfarin use include clinical/demographic (age, weight, gender, body surface area, disease), non-clinical, and genetic factors (*VKORC1, CYP2C9, CYP4F2*VKORC1, CYP2C9, CYP4F2).[15](#cit0015)15,[16](#cit0016)16 Previous research has shown that genetic factors *VKORC1*VKORC1 and *CYP2C9*CYP2C9 significantly influence variations in the pharmacokinetic and pharmacodynamic responses of warfarin.[17](#cit0017)17 Patients carrying the homomutant *VKORC1*VKORC1 gene type carrier (AA) show a low warfarin dose requirement, while the *VKORC1*VKORC1 gene type (GG) tends to require a higher dose. Meanwhile, patients with homomutant (*3/*3) type carriers of *CYP2C9*CYP2C9 are at great risk of side effects in the form of bleeding. This condition necessitates the administration of warfarin at low doses. *CYP2C9*CYP2C9 wildtype (*1/*1) tends to require higher doses and risk disease complications when given standard doses.[18](#cit0018)18
|
| 54 |
+
|
| 55 |
+
In recent research, another SNPs that could potentially influence warfarin therapy was found, namely *CYP4F2 rs2108622. CYP4F2*CYP4F2 rs2108622. CYP4F2 catalyzes the conversion of vitamin K to its inactive metabolite, hydroxyvitamin K.[19](#cit0019)19 The *rs2108622*rs2108622 V433M variant results from a C > T nucleotide substitution, where the T allele replaces valine with methionine at position 433, reducing catalytic activity and potentially affecting blood clotting and warfarin response.[17](#cit0017)17
|
| 56 |
+
|
| 57 |
+
A dosing algorithm model was needed to determine the appropriate initial and maintenance doses for patients receiving warfarin therapy. Several countries have developed algorithmic models to determine warfarin doses that are influenced by clinical, non-clinical, and genetic factors. Some of these models include Japan (Dose = 2.263 + 4.248 x (*VKORC1*VKORC1 G/G) + 1.067 x (*VKOCR1*VKOCR1 A/G) − 2.416 x (*CYP2C9*CYP2C9*3/*3) − 0.864 (x*CYP2C9*CYP2C9*1/*3) + 1.308 x *BSA*BSA + 0.025 x age), in China (Dose = 0.727–0.007 x age + 0.384 x *BSA*BSA + 0.403 x (*VKORC1*VKORC1 G/A) + 0.554 x (*VKORC1*VKORC1 G/G) − 0.482 x (*CYP2C9*CYP2C9*1/*3) − 1.583 x (*CYP2C9*CYP2C9*3/*3), in Italy (Dose = 7.39764–0.02734 x age + 1.06287 x *BSA*BSA − 1.04468 x *VKORC1*VKORC1 A/G − 2.12117 x *VKORC1*VKORC1), and USA (Dose = 3.52–0.006 x age + 0.38 x BSA − 0.15 x *hypertension*hypertension − 0.23 x (*CYP2C9*CYP2C9*1/*3 or *3/*3) − 0.24 x (*VKORC1*VKORC1 A/G) − 0.48 x (*VKORC1*VKORC1).[20–22](#cit0020)20–22 In Indonesia, there is still no development of this warfarin dosing algorithm model. Therefore, this research aimed to obtain a model of warfarin dosing algorithm or pattern according to the condition of each patient. The results can be applied as a guide in warfarin therapy in cardiac hospitals or clinics where cardiologists treat patients using warfarin.
|
| 58 |
+
|
| 59 |
+
## Materials and Methods
|
| 60 |
+
|
| 61 |
+
### Ethics Statement
|
| 62 |
+
|
| 63 |
+
This research complies with the principles outlined in the Declaration of Helsinki. Ethical approval was obtained from the West Java Health Ethics Commission-Faculty of Medicine, Universitas Padjadjaran with registration number 1342/UN6.KEP/EC/2019.
|
| 64 |
+
|
| 65 |
+
### Subjects
|
| 66 |
+
|
| 67 |
+
The inclusion criteria were outpatients of the cardiac clinic who had been on warfarin therapy for ≥ 3 months, had Prothrombin Time-International Normalized Ratio (PT-INR) laboratory data available, had complete medical records, made routine medical visits, and were willing to participate. Similarly, the exclusion criteria were patients who took supplements containing vitamin K, and those who could not be followed up due to death, relocation of treatment, or inability to be contacted.
|
| 68 |
+
|
| 69 |
+
The sample size required for this study was calculated using the Lemeshow formula based on the allele prevalence:
|
| 70 |
+
|
| 71 |
+
| \documentclass[12pt]{minimal} \usepackage{wasysym} \usepackage[substack]{amsmath} \usepackage{amsfonts} \usepackage{amssymb} \usepackage{amsbsy} \usepackage[mathscr]{eucal} \usepackage{mathrsfs} \DeclareFontFamily{T1}{linotext}{} \DeclareFontShape{T1}{linotext}{m}{n} {linotext }{} \DeclareSymbolFont{linotext}{T1}{linotext}{m}{n} \DeclareSymbolFontAlphabet{\mathLINOTEXT}{linotext} \begin{document}$$n{\left({1 + x} \right)^n} = {{N{\mathrm{}}Prev{\mathrm{}}\left({1 - Prev} \right)} \over {\left({n - 1} \right){{{d^2}} \over {{{\left({Z1 - {a \over 2}} \right)}^n}{\mathrm{}}}} + Prev{\mathrm{}}\left({1 - Prev} \right)}}$$\end{document} |
|
| 72 |
+
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
|
| 73 |
+
Explanation of variables:
|
| 74 |
+
|
| 75 |
+
n: required sample size
|
| 76 |
+
|
| 77 |
+
d: margin of error (5%)
|
| 78 |
+
|
| 79 |
+
N: population size
|
| 80 |
+
|
| 81 |
+
Prev: prevalence of the CYP4F2 polymorphism (31.45% in the Asian population, as reported by Singh et al, 2011)[17](#cit0017)17
|
| 82 |
+
|
| 83 |
+
Z: confidence level (95%, corresponding to 1.96)
|
| 84 |
+
|
| 85 |
+
Given that the population of warfarin therapy patients at Hasan Sadikin Hospital, Bandung, was 100, and the polymorphism prevalence (C > T) was 31.45%, the calculation is as follows:
|
| 86 |
+
|
| 87 |
+
| \documentclass[12pt]{minimal} \usepackage{wasysym} \usepackage[substack]{amsmath} \usepackage{amsfonts} \usepackage{amssymb} \usepackage{amsbsy} \usepackage[mathscr]{eucal} \usepackage{mathrsfs} \DeclareFontFamily{T1}{linotext}{} \DeclareFontShape{T1}{linotext}{m}{n} {linotext }{} \DeclareSymbolFont{linotext}{T1}{linotext}{m}{n} \DeclareSymbolFontAlphabet{\mathLINOTEXT}{linotext} \begin{document}$$n{\left({1 + x} \right)^n} = {{100{\mathrm{ }}x{\mathrm{ }}0.3145{\mathrm{ }}\left({1 - 0.3145} \right)} \over {\left({100 - 1} \right)x{\mathrm{ }}{{{{0.05}^2}} \over {{{1.96}^2}{\mathrm{ }}}} + 0.3145{\mathrm{ }}\left({1 - 0.3145} \right)}} = 76.99 = 77sample$$\end{document} |
|
| 88 |
+
| ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
|
| 89 |
+
All patients provided informed consent, then clinical characteristics, medical history, medications used, and daily warfarin doses were recorded. Clinical data were collected by reviewing medical records and direct inquiry during regular scheduled clinic visits. The clinical data included age, height, weight, gender, target INR, concomitant diseases, combined medications, and warfarin dosage.
|
| 90 |
+
|
| 91 |
+
### Blood Sampling
|
| 92 |
+
|
| 93 |
+
A 3 mL blood sample was collected into marked EDTA tubes and stored at −20°C. The design of gene-specific primers for *CYP4F2 rs2108622*CYP4F2 rs2108622 was carried out by downloading the gene sequence from the National Center for Biotechnology Information (NCBI). After obtaining the sequence, the nitrogenous base sequence was input into the Primer-BLAST tool on the NCBI website (www.ncbi.nlm.nih.gov/tools/primer-blast/[www.ncbi.nlm.nih.gov/tools/primer-blast/](http://www.ncbi.nlm.nih.gov/tools/primer-blast/)www.ncbi.nlm.nih.gov/tools/primer-blast/). The primers were then verified using the online OligoCalc software (http://biotools.nubic.northwestern.edu/OligoCalc.html[http://biotools.nubic.northwestern.edu/OligoCalc.html](http://biotools.nubic.northwestern.edu/OligoCalc.html)http://biotools.nubic.northwestern.edu/OligoCalc.html). The primers are shown in [Table 1](#t0001)Table 1, respectively.
|
| 94 |
+
|
| 95 |
+
### Table 1.
|
| 96 |
+
|
| 97 |
+
Primer
|
| 98 |
+
|
| 99 |
+
| Gen CYP4F2 | Primer | Fragment Size |
|
| 100 |
+
| ---------- | ------ | ------------- |
|
| 101 |
+
| Rs2108622 | Forward 5’ TACTCCTGATCAAAACCCTGCC 3’ | 170 pb |
|
| 102 |
+
| Reverse 5’CTTCTCCTGACTGCTCCCTT 3’ | | |
|
| 103 |
+
### Deoxyribonucleic Acid (DNA) Extraction and Genotyping
|
| 104 |
+
|
| 105 |
+
A total of 200 μL of blood was placed in a 1.5 mL Eppendorf tube and 20 μL of proteinase K and 20 μL of Ribonuclease (RNAse) A solution were added. The mixture was homogenized by vortexing, then 200 μL of lysis solution C was added to the Eppendorf tube, and the tube was vortexed again for 15 seconds. The mixture was then incubated for 10 minutes at 55°C. After incubation, 200 μL of 95% ethanol was added to the lysate, and the mixture was homogenized by vortexing for 10 seconds.
|
| 106 |
+
|
| 107 |
+
DNA purification was performed using GenElute™ miniprep binding columns. The lysates, previously mixed with 95% ethanol, were transferred into the columns and centrifuged at 6,500 x g for one minute. The liquid in the collection tubes (2.0 mL) was discarded and replaced. The next step in the DNA purification process was the washing stage, using a wash solution concentrate that had been diluted with 95% ethanol. The DNA extraction process was concluded with the elution stage, where 100 μL of elution solution was added to the column and centrifuged at 6,500 x g for one minute, and the process was repeated twice.
|
| 108 |
+
|
| 109 |
+
The Polymerase Chain Reaction (PCR) process consists of three stages, namely denaturation, annealing, and extension. Several temperature variations were used to determine the optimal primer annealing temperature, including 55.4°C, 56.4°C, 57.4°C, 58°C, 59°C, 60°C, 61°C, 62°C, 63.4°C, and 64.4°C. The total reaction volume was 25 μL, comprising 2 μL of DNA template, 1 μL of forward primer, 1 μL of reverse primer, 12.5 μL of PCR Master Mix, and 8.5 μL of nuclease-free water. The PCR product was then electrophoresed on a 2% agarose gel at 80 volts for 90 minutes. The electrophoresis results were visualized under UV light at 312 nm using a fluorescence scanner. The PCR products were then sent to Humanizing Genomics Macrogen (https://www.macrogen.com/en/main/index.php[https://www.macrogen.com/en/main/index.php](https://www.macrogen.com/en/main/index.php)https://www.macrogen.com/en/main/index.php), Korea, for sequencing. Sequencing was performed using the Sanger method, which relied on DNA synthesis with chain termination.
|
| 110 |
+
|
| 111 |
+
### Statistical Analysis
|
| 112 |
+
|
| 113 |
+
The characteristics of the data were assessed to determine the normality using the D’Agostino or Kolmogorov–Smirnov tests. Based on the results, appropriate statistical test methods were applied. For normally distributed data, ANOVA or Student’s *t*t-test was used for analysis, at a significance level of α = 0.05. Otherwise, the Kruskal–Wallis or Mann–Whitney *U*U-test was applied.
|
| 114 |
+
|
| 115 |
+
Univariate analysis was conducted for descriptive analysis to determine the characteristics of each research variable, presented as number and percentage (n, %). Bivariate analysis was conducted to identify variables that could be included in the multivariate model, with a p-value < 0.05. Furthermore, the multivariate regression analysis (logistic regression) was used to examine the correlation and develop warfarin dosing model, considering both clinical and non-clinical factors, with a p-value < 0.05.
|
| 116 |
+
|
| 117 |
+
## Results
|
| 118 |
+
|
| 119 |
+
A total of 77 patients participated in this research from March to December 2021. Demographic data and clinical characteristics of patients were obtained by reviewing medical records. [Table 2](#t0002)Table 2 shows the description of patients demographic characteristics.
|
| 120 |
+
|
| 121 |
+
### Table 2.
|
| 122 |
+
|
| 123 |
+
Baseline Demographic, Clinical Characteristic and Mean INR Value
|
| 124 |
+
|
| 125 |
+
| Variables | Value | Mean INR Value |
|
| 126 |
+
| --------- | ----- | -------------- |
|
| 127 |
+
| Median age (range) (year) | 54 (28–80) | – |
|
| 128 |
+
| Male/female N (%) | 37/40 (48/52) | 2.57/2.96 |
|
| 129 |
+
| Mean BMI (range) (kg/m2) | 23.63 (17–35) | – |
|
| 130 |
+
| Mean warfarin dose (range) (mg/week) | 20.25 (7–46) | – |
|
| 131 |
+
| Mean INR < 2 N (%) | 29 (37) | – |
|
| 132 |
+
| Mean INR 2–3 N (%) | 41 (53) | – |
|
| 133 |
+
| Mean INR > 3 N (%) | 7 (9) CC: 4 CT: 3 TT: 0 | – |
|
| 134 |
+
| Primary indication N (%) | | |
|
| 135 |
+
| Rheumatic Heart Disease | 32 (41.60) | 2.04 |
|
| 136 |
+
| Atrial Fibrillation | 25 (32.50) | 2.7 |
|
| 137 |
+
| Mitral Valve Prolapse | 8 (10.40) | 2.43 |
|
| 138 |
+
| Coronary Artery Disease | 7 (9.10) | 2.12 |
|
| 139 |
+
| Hypertensive Heart Disease | 5 (6.50) | 2.3 |
|
| 140 |
+
| Concomitant medication N (%) | | |
|
| 141 |
+
| Erythromycin | 35 (46.10) | 2.31 |
|
| 142 |
+
| Spironolactone | 15 (19.70) | 2.09 |
|
| 143 |
+
| Sucralfate | 3 (3.90) | 2.66 |
|
| 144 |
+
| Lansoprazole | 11 (14.50) | 2.01 |
|
| 145 |
+
| Simvastatin | 3 (3.90) | 2.36 |
|
| 146 |
+
| Allopurinol | 2 (2.60) | 2.71 |
|
| 147 |
+
| Diltiazem | 1 (1.30) | 1.84 |
|
| 148 |
+
| Comorbidities | | |
|
| 149 |
+
| Hypertension | 7 (9.2) | 2.26 |
|
| 150 |
+
| Diabetes Mellitus | 2 (2) | 2.47 |
|
| 151 |
+
| Turner Syndrome | 1 (1) | 2.91 |
|
| 152 |
+
| Epilepsy | 1 (1) | 2.57 |
|
| 153 |
+
| Hyperthyroidism | 2 (2) | 2.36 |
|
| 154 |
+
| Tuberculosis | 1 (1) | 2.85 |
|
| 155 |
+
| Hyperlipidemia | 2 (2) | 3.01 |
|
| 156 |
+
| Gout | 3 (3) | 2.07 |
|
| 157 |
+
| CY4F2 (rs2108622) N (%) | | |
|
| 158 |
+
| CC | 47 (61) | 2.2 |
|
| 159 |
+
| CT | 27 (35) | 2.4 |
|
| 160 |
+
| TT | 3 (4) | 2.7 |
|
| 161 |
+
The average weekly dose based on age, Body Mass Index (BMI), and *CYP4F2*CYP4F2 rs 2108622 genotype are shown in [Table 3](#t0003)Table 3. The results showed that the required dose decreases with increasing age. Specifically, patients aged 70–79 required a weekly dose of 16.17 mg, which is 27.33% lower than the highest average dose for patients aged 30–39, while patients aged 80–89 required a significantly lower dose of 7 mg (3 times smaller than the largest dose).
|
| 162 |
+
|
| 163 |
+
### Table 3.
|
| 164 |
+
|
| 165 |
+
Mean Weekly Doses (in Mg) for Age, BMI, and CYP4F2 Rs 2108622 Genotype
|
| 166 |
+
|
| 167 |
+
| Variable | Total | Mean ± SD |
|
| 168 |
+
| -------- | ----- | --------- |
|
| 169 |
+
| Age (yr) | | |
|
| 170 |
+
| 20–29 | 4 | 21.13 ± 3.07 |
|
| 171 |
+
| 30–39 | 12 | 22.25 ± 7.79 |
|
| 172 |
+
| 40–49 | 13 | 22.08 ± 7.93 |
|
| 173 |
+
| 50–59 | 26 | 20.96 ± 7.08 |
|
| 174 |
+
| 60–69 | 15 | 18.10 ± 6.45 |
|
| 175 |
+
| 70–79 | 6 | 16.17 ± 5.23 |
|
| 176 |
+
| 80–89 | 1 | 7 ± 0.00 |
|
| 177 |
+
| BMI (kg/m) | | |
|
| 178 |
+
| Underweight (<18.50) | 6 | 17.67 ± 7.23 |
|
| 179 |
+
| Normal (18.50–24.90) | 48 | 19.05 ± 5.21 |
|
| 180 |
+
| Overweight (>25) | 20 | 23.33 ± 10.15 |
|
| 181 |
+
| Obesity (>30) | 3 | 24.00 ± 8.72 |
|
| 182 |
+
| CYP4F2 rs 2108622 genotype | | |
|
| 183 |
+
| CT | 47 | 19 ± 6.42 |
|
| 184 |
+
| CC | 27 | 21 ± 7.35 |
|
| 185 |
+
| TT | 3 | 33 ± 6.78 |
|
| 186 |
+
### Bivariate Analysis
|
| 187 |
+
|
| 188 |
+
The results of the bivariate analysis between patients demographics and genotypes on warfarin dose are shown in [Table 4](#t0004)Table 4. Variables with a p-value <0.25 in the bivariate analysis are eligible to enter the multivariate model.
|
| 189 |
+
|
| 190 |
+
### Table 4.
|
| 191 |
+
|
| 192 |
+
Results of Bivariate Analysis Between Patients Demographics and Genotype on Warfarin Dose
|
| 193 |
+
|
| 194 |
+
| Patients Demographics | Bivariate Analysis | p-value | Correlation Coefficient | Multivariate Analysis |
|
| 195 |
+
| --------------------- | ------------------ | ------- | ----------------------- | --------------------- |
|
| 196 |
+
| Genotype | CC | Kruskal–Wallis | 0.02* | - | Yes |
|
| 197 |
+
| CT | | | | |
|
| 198 |
+
| TT | | | | |
|
| 199 |
+
| Sex | Male | Mann–Whitney | 0.16 | - | Yes |
|
| 200 |
+
| Female | | | | |
|
| 201 |
+
| Age (year) | 28–80 | Spearman’s Rank | 0.02* | −0.28 | Yes |
|
| 202 |
+
| BMI | 17–35 | Spearman’s Rank | 0.03* | 0.25 | Yes |
|
| 203 |
+
The Kruskal–Wallis test on genotype showed a p-value of 0.02 (<0.05), suggesting that the CC, CT, and TT genotypes have a significant association with warfarin dosage. Meanwhile, the Mann–Whitney test on gender had a p-value of 0.16 (>0.05). This result showed that gender does not have a significant relationship with warfarin dosage. However, gender was included in the multivariate analysis (p < 0.25) as a confounding factor.
|
| 204 |
+
|
| 205 |
+
The results of the Spearman Rank correlation analysis for age (p = 0.02) and BMI (p = 0.03) showed p-values <0.05. This implies that age and BMI have a significant relationship with warfarin dosage. The correlation coefficient values from this analysis were −0.28 for age and 0.25 for BMI. These results suggest that the strength of the relationship between age, BMI, and warfarin dosage is very weak (correlation coefficient: 0.00–0.30).[23](#cit0023)23 Specifically, as age increases, the required dose of warfarin decreases. Conversely, as BMI increases, the required dose of warfarin also increases.
|
| 206 |
+
|
| 207 |
+
### Multivariate Analysis
|
| 208 |
+
|
| 209 |
+
Multivariate analysis aimed to determine the factors associated with warfarin dosing. Multiple linear regression was used to select age, BMI, sex, and *CYP4F2*CYP4F2 genotype for the creation of warfarin dosing formula. The results of the multiple linear regression analysis are shown in [Table 5](#t0005)Table 5.
|
| 210 |
+
|
| 211 |
+
### Table 5.
|
| 212 |
+
|
| 213 |
+
Multiple Linear Regression Analysis Between Age, BMI, Gender, Genotype, and Warfarin Dose
|
| 214 |
+
|
| 215 |
+
| Variable | Beta coefficient | SE (B) | t-value | p-value | Description |
|
| 216 |
+
| -------- | ---------------- | ------ | ------- | ------- | ----------- |
|
| 217 |
+
| Initial Model: Sex | 0.21 | 1.49 | 0.14 | 0.89 | Non-significant |
|
| 218 |
+
| Age | −0.16 | 0.06 | −2.83 | 0.01* | Significant |
|
| 219 |
+
| BMI | 0.54 | 0.20 | 2.67 | 0.01* | Significant |
|
| 220 |
+
| Genotype | 3.51 | 1.30 | 2.71 | 0.01* | Significant |
|
| 221 |
+
| Final Model: Age | −0.16 | 0.06 | −2.88 | 0.01* | Significant |
|
| 222 |
+
| BMI | 0.54 | 0.20 | 2.70 | 0.01* | Significant |
|
| 223 |
+
| Genotype | 3.55 | 1.27 | 2.8 | 0.01* | Significant |
|
| 224 |
+
| Konstanta | 12.736 | - | 0 | - | |
|
| 225 |
+
### Quality of Life
|
| 226 |
+
|
| 227 |
+
Quality of life of warfarin therapy patients in Dr. Hasan Sadikin Central General Hospital is presented in [Table 4](#t0004)Table 4, with categories. The lower score showed a better quality of life and the higher score showed worse conditions. In addition, the results showed that the highest percentage score was included in the category < 56,266. This showed that most patients on warfarin therapy had a better quality of life.
|
| 228 |
+
|
| 229 |
+
The principle of multiple linear regression analysis used was backward elimination. In the initial model, all variables were entered simultaneously, and those with a significance value >0.05 were excluded. The final model of this regression analysis included three variables, namely age, BMI, and genotype. [Table 5](#t0005)Table 5 shows that the final model analysis has a significance value of <0.01 for each variable. This result suggests that age (p = 0.01), BMI (p = 0.01), and genotype (p = 0.01) have a significant influence on the determination of warfarin dose.
|
| 230 |
+
|
| 231 |
+
Based on [Table 5](#t0005)Table 5, the regression model can be expressed as y = 12.736–0.160×1 + 0.540×2 + 3.545X3, or dose = 12.736–0.16**age + 0.54**age + 0.54*BMI + 3.55**CYP4F2*CYP4F2 genotype, where 1 = CC, 2 = CT, and 3 = TT. The constant 12.736 represents warfarin dose in mg/week when age, BMI, and genotype are not considered. The regression coefficient of −0.16 (β1) shows that for every decrease in age, warfarin dose increases by 0.16 mg/week. The regression coefficient of 0.54 (β2) shows that each unit increase in BMI will raise warfarin dose by 0.54 mg/week. Finally, the regression coefficient of 3.55 (β3) suggests that the presence of the *CYP4F2*CYP4F2 C > T polymorphism increases warfarin dose by 3.55 mg/week.
|
| 232 |
+
|
| 233 |
+
The result in [Table 5](#t0005)Table 5 showed an R-squared value of 0.25, showing that 25% of the variance in warfarin dose was explained by age, BMI, and *CYP4F2*CYP4F2 genotype, while the remaining 75% was determined by other factors not included in this research. The effective contribution of each variable was 8.76%, 8.29%, and 7.95% for age, *CYP4F2*CYP4F2 gene polymorphism, and BMI. The effective contribution can be calculated using the formula SE% = βx × rxy × 100%.
|
| 234 |
+
|
| 235 |
+
## Discussion
|
| 236 |
+
|
| 237 |
+
In this research, 77 patients met the inclusion criteria, consisting of 37 men and 40 women, with an average BMI of 23.63 kg/m². The *CYP4F2 rs2108622*CYP4F2 rs2108622 gene polymorphism profile included 47 patients with the CC genotype, 27 with CT, and 3 with the TT. [Table 3](#t0003)Table 3 shows that the older patients, the lower the dose required. The results of this research are consistent with previous reports that patients with middle and old age require warfarin doses 10.60% lower than young age, as the age of patients decreases the weekly dose by 0.40 mg per year of age.[24](#cit0024)24 In addition, in old age, there are many hemorrhagic events due to the use of drugs that can increase the risk of bleeding, such as antiplatelets, anticoagulants, statins, and amiodarone.[25](#cit0025)25 The low dose of warfarin in elderly patients was attributed to decreased activity of the vitamin K redox recycling system, which was affected by age-related physiological changes. These changes included alterations in body composition, an increase in fat tissue (leading to an increased volume of distribution for fat-soluble drugs), slowing of metabolic processes, and reduced blood perfusion to the intestinal region.[26](#cit0026)26,[27](#cit0027)27
|
| 238 |
+
|
| 239 |
+
Dosing based on BMI classification showed that the higher the BMI index, the greater the weekly dose required. The average weekly dose for obese patients was 24 mg, which was 26.38% greater than the underweight and 5 mg higher than normal-weight patients ([Table 3](#t0003)Table 3). This result was consistent with previous research showing a correlation between weekly dose and BMI. Research by Alshammari et al (2020) and Mueller et al (2014) showed significant results that obese patients require weekly doses 20% higher than those of normal and overweight.[28](#cit0028)28,[29](#cit0029)29 According to Yoo et al (2012), an increase in body weight was directly proportional to the required warfarin dose and INR value. Patients over 80 years old and weighing less than 55 kg needed a maintenance dose of 3 mg. Meanwhile, those under 55 years old and weighing more than 50 kg required a dose of 10 mg. Patients within these two age and weight ranges needed a dose of 3–7 mg.[30](#cit0030)30 This is due to differences in pharmacokinetics in obese patients, specifically, in drug distribution within tissues, volume of distribution (Vd), blood flow, plasma protein binding, and drug elimination. The absorption process remains similar to that of normal-weight patients. Obese patients have greater absolute body and fat mass, and the hemodynamic conditions can enhance drug kinetics. Changes in plasma protein-binding concentrations can impact the movement of drugs into tissue compartments, influencing therapeutic effects. Furthermore, the need for larger weekly doses in obese patients was attributed to increased body weight, which affected the volume of distribution and clearance of warfarin, leading to elevated coagulation factors.[31](#cit0031)31
|
| 240 |
+
|
| 241 |
+
Dosing based on the *CYP4F2 rs2108622*CYP4F2 rs2108622 genetic polymorphism showed that patients with CC, CT, and TT genotypes required doses of 19 mg, 21 mg, and 33 mg, respectively. The weekly dose for TT patients was significantly greater than CC and CT, as shown in [Table 3](#t0003)Table 3. Several countries have conducted research on *CYP4F2*CYP4F2 polymorphism and the effect on warfarin dosing. Research in China,[32](#cit0032)32 Iran,[33](#cit0033)33 Italy,[34](#cit0034)34 and India[17](#cit0017)17 showed that patients with the *CYP4F2*CYP4F2 polymorphism required higher warfarin doses. However, research conducted on populations in the UK,[35](#cit0035)35 Japan,[36](#cit0036)36 and Norway[37](#cit0037)37 suggested that *CYP4F2*CYP4F2 polymorphism had no significant influence on warfarin dosing.
|
| 242 |
+
|
| 243 |
+
The *CYP4F2*CYP4F2 gene expression catalyzes the hydroxylation of vitamin K1 (VK1) into an inactive form, hydroxyvitamin K. This gene served as an important negative regulator of vitamin K levels, thereby affecting blood clotting.[38](#cit0038)38 The *CYP4F2 rs2108622*CYP4F2 rs2108622 V433M variant arises from a polymorphism including the C > T nucleotide substitution. The T allele in *rs2108622*rs2108622 replaced a valine residue with a methionine residue at position 433 in the coding region. This change impacted enzyme activity, and drug metabolism, as well as physiological and pathophysiological processes. The increase in warfarin dose for CT and TT genotypes was consistent with the observed rise in plasma concentration.
|
| 244 |
+
|
| 245 |
+
Molecular dynamics (MD) research showed that the *CYP4F2*CYP4F2 V433M variant was associated with a decrease in protein stability, as evident by free energy values. Free energy values below zero suggested low stability. Destabilization of the protein structure could alter biological function and disrupt signal cascades and normal protein pathways. The V433M variant impacted the physicochemical characteristics, intermolecular interactions, as well as functional and structural properties of the protein. Furthermore, the mutant amino acid (methionine) was larger than the wild-type (valine), leading to structural mismatches within the protein. The wild-type amino acid was located in a critical position for interacting with other molecules that are essential for protein activity. Mutations could disrupt these interactions, affecting the signaling cascade from the binding to the activity domain.[19](#cit0019)19
|
| 246 |
+
|
| 247 |
+
Research by McDonald et al in 2009 showed the participation of *CYP4F2*CYP4F2 in the oxidative degradation of vitamin K and oxidative activity. The protein encoded by the *rs2108622*rs2108622 T allele had reduced activity compared to the wild-type in the genotyping of liver microsomal enzymes, with the TT phenotype showing a 75% reduction in vitamin K oxidative activity. The *CYP4F2 rs2108622*CYP4F2 rs2108622 V433M variant had a diminished ability to metabolize VK1 to hydroxyvitamin K1, resulting in reduced steady-state hepatic enzyme concentration. Consequently, patients with the *rs2108622*rs2108622 polymorphism tend to have elevated hepatic VK1 levels, leading to a requirement for higher warfarin doses to achieve the same anticoagulant response.[19](#cit0019)19
|
| 248 |
+
|
| 249 |
+
Based on the INR values obtained in this study, the majority of patients with CYP4F2 genotypes CC, CT, and TT had INR values within the target therapeutic range of 2–3. Among the CC genotype group, only 4 patients had INR values exceeding 3, while 3 patients in the CT group exhibited similar results. Notably, no patients with the TT genotype had INR values above 3. These findings suggest that most patients across all genotypes were effectively managed within the desired therapeutic range, reducing the risk of adverse outcomes such as bleeding. Furthermore, there were no reports of major bleeding events among the study participants, further supporting the safety of the dosing regimens utilized in this population ([Table 2](#t0002)Table 2).
|
| 250 |
+
|
| 251 |
+
The algorithm model obtained was y = 12.736–0.160×1 + 0.540×2 + 3.545X3, or dose = 12.736–0.16**age + 0.54**age + 0.54*BMI + 3.55**CYP4F2*CYP4F2 genotype, where 1 = CC, 2 = CT, and 3 = TT. The results of this algorithm are consistent with several models developed in various countries, such as in Japan (Dose = 2.263 + 4.248 x (*VKORC1*VKORC1 G/G) + 1.067 x (*VKOCR1*VKOCR1 A/G) − 2.416 x (*CYP2C9*CYP2C9*3/*3) − 0.864 (x*CYP2C9*CYP2C9*1/*3) + 1.308 x *BSA*BSA + 0.025 x age), China (Dose = 0.727–0.007 x age + 0.384 x *BSA*BSA + 0.403 x (*VKORC1*VKORC1 G/A) + 0.554 x (*VKORC1*VKORC1 G/G) − 0.482 x (*CYP2C9*CYP2C9*1/*3) − 1.583 x (*CYP2C9*CYP2C9*3/*3), Italia (Dose = 7.39764–0.02734 x age + 1.06287 x *BSA*BSA − 1.04468 x *VKORC1*VKORC1 A/G − 2.12117 x *VKORC1*VKORC1), and USA (Dose = 3.52–0.006 x age + 0.38 x BSA − 0.15 x *hypertension*hypertension − 0.23 x (*CYP2C9*CYP2C9*1/*3 or *3/*3) − 0.24 x (*VKORC1*VKORC1 A/G) − 0.48 x (*VKORC1*VKORC1).[22](#cit0022)22,[39](#cit0039)39,[40](#cit0040)40
|
| 252 |
+
|
| 253 |
+
The similarity of the algorithm obtained in this research with those from several other countries was in the inclusion of age and BMI or BSA as factors in the dosing model. The correlation between age and dose was negative across research, namely Japan (+0.025 x age), China (−0.007 x age), Italy (−0.02734 x age), America (−0.006 x age), and Indonesia (−0.16 x age). This result showed that as age increases, the required dose tends to decrease. In contrast, BMI showed a positive correlation, suggesting that the higher the BMI, the greater the required dose. A key difference between the algorithm developed in this research and models from other countries was the genetic factors. While previous investigation focused on *VKORC1*VKORC1 and *CYP2C9*CYP2C9, this research emphasized *CYP4F2*CYP4F2, due to its crucial role in the vitamin K cycle, which was directly related to the vitamin K intake.
|
| 254 |
+
|
| 255 |
+
The results of this study align with previous findings indicating that age and BMI significantly influence warfarin dosing. For example, Khoury et al (2014) demonstrated that warfarin dosage decreases with age, consistent with our findings.[41](#cit0041)41 Similarly, the observed correlation between higher BMI and increased warfarin requirements corresponds with results reported by Alshammari et al (2020) and Mueller et al (2014).[28](#cit0028)28,[29](#cit0029)29 However, our study highlights CYP4F2 as a genetic factor in warfarin dosing, diverging from studies in other countries that emphasize VKORC1 and CYP2C9. This underscores the importance of considering population-specific genetic variations, such as CYP4F2 in Indonesia, in developing dosing algorithms.
|
| 256 |
+
|
| 257 |
+
The limitations of this research include the relatively small sample size, which may not accurately represent the broader population, thereby limiting the generalizability of the results to all patients with similar conditions. Future research with larger sample sizes is needed to validate these results. Additionally, this research was conducted at only one hospital within a specific geographical area, which could introduce location and population bias, as patients from other regions or hospitals may exhibit different characteristics. Comprehensive analyses that incorporate more genetic factors, as well as other non-clinical variables, are necessary for a more thorough understanding of these issues.
|
| 258 |
+
|
| 259 |
+
## Conclusion
|
| 260 |
+
|
| 261 |
+
In conclusion, the factors that influenced warfarin dose adjustment in cardiovascular patients in Indonesia were age, BMI, and the *CYP4F2*CYP4F2 gene polymorphism *rs2108622*rs2108622. Specifically, as age increased, the required dose decreased. The *CYP4F2 rs2108622*CYP4F2 rs2108622 gene polymorphism also affected warfarin dose variation, with patients carrying the TT polymorphism requiring higher doses. The percentage contributions of each factor to warfarin dose adjustment included 8.76%, 7.95%, and 8.29% for age, BMI, and gene polymorphism, respectively. The total contribution of age, BMI, and *CYP4F2*CYP4F2 genotype to warfarin dose adjustment was 25%. Finally, the linear regression model for predicting warfarin dose was represented by the equation y = 12.736–0.16*Age + 0.54*Age + 0.54 BMI + 3.55*Genotype. In addition, further exploration of International Normalized Ratio (INR) data could provide more insights into the warfarin response, as INR is a key parameter for monitoring warfarin therapy. The relationship between INR levels and the influencing factors identified in this study may help optimize dosing strategies for cardiovascular patients in Indonesia.
|
| 262 |
+
|
| 263 |
+
## Funding Statement
|
| 264 |
+
|
| 265 |
+
The authors are grateful to the Rector of Universitas Padjadjaran for funding this study (RKDU grant No 1918/UN6.3.1/PT.00/2024).
|
| 266 |
+
|
| 267 |
+
## Disclosure
|
| 268 |
+
|
| 269 |
+
The authors report no conflicts of interest in this work.
|
| 270 |
+
|
| 271 |
+
## References
|
| 272 |
+
|
| 273 |
+
1. Martin J, Somogyi A. Pharmacogenomics and warfarin therapy. therapeutic drug monitoring: newer drugs and biomarkers. Therape Drug Monitor Newer Drugs Biomark. 2012:161–173. doi: 10.1016/B978-0-12-385467-4.00008-7 [DOI](https://doi.org/10.1016/B978-0-12-385467-4.00008-7) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Therape%20Drug%20Monitor%20Newer%20Drugs%20Biomark&title=Pharmacogenomics%20and%20warfarin%20therapy.%20therapeutic%20drug%20monitoring:%20newer%20drugs%20and%20biomarkers&author=J%20Martin&author=A%20Somogyi&publication_year=2012&pages=161-173&doi=10.1016/B978-0-12-385467-4.00008-7&)
|
| 274 |
+
|
| 275 |
+
2. Daly AK. Pharmacogenomics of Warfarin. Handbook Pharmacogenomics Strat Med. 2014;497–507. doi: 10.1016/B978-0-12-386882-4.00024-4 [DOI](https://doi.org/10.1016/B978-0-12-386882-4.00024-4) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Handbook%20Pharmacogenomics%20Strat%20Med&title=Pharmacogenomics%20of%20Warfarin&author=AK%20Daly&publication_year=2014&pages=497-507&doi=10.1016/B978-0-12-386882-4.00024-4&)
|
| 276 |
+
|
| 277 |
+
3. Putriana NA, Destiani DP, Putri AN, Latarissa IR. Quality of life of patients receiving warfarin therapy at a tertiary care centre in Indonesia using DASS (duke anticoagulation satisfaction scale). Vasc Health Risk Manag. 2024;20:403–413. doi: 10.2147/VHRM.S467656 [DOI](https://doi.org/10.2147/VHRM.S467656) | [PMC free article](/articles/PMC11352524/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/39206433/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Vasc%20Health%20Risk%20Manag&title=Quality%20of%20life%20of%20patients%20receiving%20warfarin%20therapy%20at%20a%20tertiary%20care%20centre%20in%20Indonesia%20using%20DASS%20(duke%20anticoagulation%20satisfaction%20scale)&author=NA%20Putriana&author=DP%20Destiani&author=AN%20Putri&author=IR%20Latarissa&volume=20&publication_year=2024&pages=403-413&pmid=39206433&doi=10.2147/VHRM.S467656&)
|
| 278 |
+
|
| 279 |
+
4. Loebstein R, Yonath H, Peleg D, et al. Interindividual variability in sensitivity to warfarin--Nature or nurture? Clin Pharmacol Ther. 2001;70(2):159–164. doi: 10.1067/MCP.2001.117444 [DOI](https://doi.org/10.1067/MCP.2001.117444) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11503010/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Interindividual%20variability%20in%20sensitivity%20to%20warfarin--Nature%20or%20nurture?&author=R%20Loebstein&author=H%20Yonath&author=D%20Peleg&volume=70&issue=2&publication_year=2001&pages=159-164&pmid=11503010&doi=10.1067/MCP.2001.117444&)
|
| 280 |
+
|
| 281 |
+
5. Van Spall HGC, Wallentin L, Yusuf S, et al. Variation in warfarin dose adjustment practice is responsible for differences in the quality of anticoagulation control between centers and countries: an analysis of patients receiving warfarin in the randomized evaluation of long-term anticoagulation therapy (RE-LY) trial. Circulation. 2012;126(19):2309–2316. doi: 10.1161/CIRCULATIONAHA.112.101808/ASSET/2B596CB7-59AE-4100-A2E8-97818129888B/ASSETS/GRAPHIC/ZHC0441213230001.JPEG [DOI](https://doi.org/10.1161/CIRCULATIONAHA.112.101808/ASSET/2B596CB7-59AE-4100-A2E8-97818129888B/ASSETS/GRAPHIC/ZHC0441213230001.JPEG) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23027801/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circulation&title=Variation%20in%20warfarin%20dose%20adjustment%20practice%20is%20responsible%20for%20differences%20in%20the%20quality%20of%20anticoagulation%20control%20between%20centers%20and%20countries:%20an%20analysis%20of%20patients%20receiving%20warfarin%20in%20the%20randomized%20evaluation%20of%20long-term%20anticoagulation%20therapy%20(RE-LY)%20trial&author=HGC%20Van%20Spall&author=L%20Wallentin&author=S%20Yusuf&volume=126&issue=19&publication_year=2012&pages=2309-2316&pmid=23027801&doi=10.1161/CIRCULATIONAHA.112.101808/ASSET/2B596CB7-59AE-4100-A2E8-97818129888B/ASSETS/GRAPHIC/ZHC0441213230001.JPEG&)
|
| 282 |
+
|
| 283 |
+
6. Cross B, Turner RM, Zhang JE, Pirmohamed M. Being precise with anticoagulation to reduce adverse drug reactions: are we there yet? Pharmacogenomics J. 2024;24(2):1–23. doi: 10.1038/s41397-024-00329-y [DOI](https://doi.org/10.1038/s41397-024-00329-y) | [PMC free article](/articles/PMC10914631/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38443337/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J&title=Being%20precise%20with%20anticoagulation%20to%20reduce%20adverse%20drug%20reactions:%20are%20we%20there%20yet?&author=B%20Cross&author=RM%20Turner&author=JE%20Zhang&author=M%20Pirmohamed&volume=24&issue=2&publication_year=2024&pages=1-23&pmid=38443337&doi=10.1038/s41397-024-00329-y&)
|
| 284 |
+
|
| 285 |
+
7. Petty GW, Brown RD, Whisnant JP, Sicks JRD, O’Fallon WM, Wiebers DO. Frequency of major complications of aspirin, warfarin, and intravenous heparin for secondary stroke prevention. A population-based study. Ann Intern Med. 1999;130(1):14–22. doi: 10.7326/0003-4819-130-1-199901050-00004 [DOI](https://doi.org/10.7326/0003-4819-130-1-199901050-00004) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9890845/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20Intern%20Med&title=Frequency%20of%20major%20complications%20of%20aspirin,%20warfarin,%20and%20intravenous%20heparin%20for%20secondary%20stroke%20prevention.%20A%20population-based%20study&author=GW%20Petty&author=RD%20Brown&author=JP%20Whisnant&author=JRD%20Sicks&author=WM%20O%E2%80%99Fallon&volume=130&issue=1&publication_year=1999&pages=14-22&pmid=9890845&doi=10.7326/0003-4819-130-1-199901050-00004&)
|
| 286 |
+
|
| 287 |
+
8. Gulløv AL, Koefoed BG, Petersen P. Bleeding during warfarin and aspirin therapy in patients with atrial fibrillation: the AFASAK 2 study. Atrial fibrillation aspirin and anticoagulation. Arch Intern Med. 1999;159(12):1322–1328. doi: 10.1001/ARCHINTE.159.12.1322 [DOI](https://doi.org/10.1001/ARCHINTE.159.12.1322) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10386508/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arch%20Intern%20Med&title=Bleeding%20during%20warfarin%20and%20aspirin%20therapy%20in%20patients%20with%20atrial%20fibrillation:%20the%20AFASAK%202%20study.%20Atrial%20fibrillation%20aspirin%20and%20anticoagulation&author=AL%20Gull%C3%B8v&author=BG%20Koefoed&author=P%20Petersen&volume=159&issue=12&publication_year=1999&pages=1322-1328&pmid=10386508&doi=10.1001/ARCHINTE.159.12.1322&)
|
| 288 |
+
|
| 289 |
+
9. Kimmel SE. Warfarin therapy: in need of improvement after all these years. Expert Opin Pharmacother. 2008;9(5):677. doi: 10.1517/14656566.9.5.677 [DOI](https://doi.org/10.1517/14656566.9.5.677) | [PMC free article](/articles/PMC2855533/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18345947/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Expert%20Opin%20Pharmacother&title=Warfarin%20therapy:%20in%20need%20of%20improvement%20after%20all%20these%20years&author=SE%20Kimmel&volume=9&issue=5&publication_year=2008&pages=677&pmid=18345947&doi=10.1517/14656566.9.5.677&)
|
| 290 |
+
|
| 291 |
+
10. Barnes GD, Burnett A, Allen A, et al. Thromboembolism and anticoagulant therapy during the COVID-19 pandemic: interim clinical guidance from the anticoagulation forum. J Thromb Thrombolysis. 2020;50(1):72. doi: 10.1007/S11239-020-02138-Z [DOI](https://doi.org/10.1007/S11239-020-02138-Z) | [PMC free article](/articles/PMC7241581/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32440883/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Thromb%20Thrombolysis&title=Thromboembolism%20and%20anticoagulant%20therapy%20during%20the%20COVID-19%20pandemic:%20interim%20clinical%20guidance%20from%20the%20anticoagulation%20forum&author=GD%20Barnes&author=A%20Burnett&author=A%20Allen&volume=50&issue=1&publication_year=2020&pages=72&pmid=32440883&doi=10.1007/S11239-020-02138-Z&)
|
| 292 |
+
|
| 293 |
+
11. Latarissa IR, Barliana MI, Meiliana A, et al. Efficacy of quinine sulfate in patients with mild-to-moderate COVID-19:A randomized controlled trial. Indones Biomedl J. 2023;15(6):366–374. doi: 10.18585/INABJ.V15I6.2543 [DOI](https://doi.org/10.18585/INABJ.V15I6.2543) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Indones%20Biomedl%20J&title=Efficacy%20of%20quinine%20sulfate%20in%20patients%20with%20mild-to-moderate%20COVID-19:A%20randomized%20controlled%20trial&author=IR%20Latarissa&author=MI%20Barliana&author=A%20Meiliana&volume=15&issue=6&publication_year=2023&pages=366-374&doi=10.18585/INABJ.V15I6.2543&)
|
| 294 |
+
|
| 295 |
+
12. Latarissa IR, Meiliana A, Sormin IP, et al. The efficacy of herbal medicines on the length of stay and negative conversion time/rate outcomes in patients with COVID-19: a systematic review. Front Pharmacol. 2024;15:1383359. doi: 10.3389/FPHAR.2024.1383359 [DOI](https://doi.org/10.3389/FPHAR.2024.1383359) | [PMC free article](/articles/PMC11169809/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38873430/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front%20Pharmacol&title=The%20efficacy%20of%20herbal%20medicines%20on%20the%20length%20of%20stay%20and%20negative%20conversion%20time/rate%20outcomes%20in%20patients%20with%20COVID-19:%20a%20systematic%20review&author=IR%20Latarissa&author=A%20Meiliana&author=IP%20Sormin&volume=15&publication_year=2024&pages=1383359&pmid=38873430&doi=10.3389/FPHAR.2024.1383359&)
|
| 296 |
+
|
| 297 |
+
13. Latarissa IR, Rendrayani F, Iftinan GN, et al. The efficacy of oral/intravenous corticosteroid use in COVID-19 patients: a systematic review. J Exp Pharmacol. 2024;16:321–337. doi: 10.2147/JEP.S484596 [DOI](https://doi.org/10.2147/JEP.S484596) | [PMC free article](/articles/PMC11453156/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/39371262/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Exp%20Pharmacol&title=The%20efficacy%20of%20oral/intravenous%20corticosteroid%20use%20in%20COVID-19%20patients:%20a%20systematic%20review&author=IR%20Latarissa&author=F%20Rendrayani&author=GN%20Iftinan&volume=16&publication_year=2024&pages=321-337&pmid=39371262&doi=10.2147/JEP.S484596&)
|
| 298 |
+
|
| 299 |
+
14. Putriana NA, Rusdiana T, Rostinawati T, Akbar MR, Destiani DP. Evaluation of adverse drug reaction in patients warfarin therapy. J Adv Pharm Technol Res. 2022;13(4):291–295. doi: 10.4103/JAPTR.JAPTR_439_22 [DOI](https://doi.org/10.4103/JAPTR.JAPTR_439_22) | [PMC free article](/articles/PMC9784044/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36568047/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Adv%20Pharm%20Technol%20Res&title=Evaluation%20of%20adverse%20drug%20reaction%20in%20patients%20warfarin%20therapy&author=NA%20Putriana&author=T%20Rusdiana&author=T%20Rostinawati&author=MR%20Akbar&author=DP%20Destiani&volume=13&issue=4&publication_year=2022&pages=291-295&pmid=36568047&doi=10.4103/JAPTR.JAPTR_439_22&)
|
| 300 |
+
|
| 301 |
+
15. Yoshizawa M, Hayashi H, Tashiro Y, et al. Effect of VKORC1-1639 G>A polymorphism, body weight, age, and serum albumin alterations on warfarin response in Japanese patients. Thromb Res. 2009;124(2):161–166. doi: 10.1016/J.THROMRES.2008.11.011 [DOI](https://doi.org/10.1016/J.THROMRES.2008.11.011) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19135231/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Thromb%20Res&title=Effect%20of%20VKORC1-1639%20G>A%20polymorphism,%20body%20weight,%20age,%20and%20serum%20albumin%20alterations%20on%20warfarin%20response%20in%20Japanese%20patients&author=M%20Yoshizawa&author=H%20Hayashi&author=Y%20Tashiro&volume=124&issue=2&publication_year=2009&pages=161-166&pmid=19135231&doi=10.1016/J.THROMRES.2008.11.011&)
|
| 302 |
+
|
| 303 |
+
16. Patel S, Singh R, Preuss CV, Patel NW. Hemostasis and thrombosis: fourth edition. 2023. Published online March 24.
|
| 304 |
+
|
| 305 |
+
17. Singh O, Sandanaraj E, Subramanian K, Lee LH, Chowbay B. Influence of CYP4F2 rs2108622 (V433M) on warfarin dose requirement in Asian patients. Drug Metab Pharmacokinet. 2011;26(2):130–136. doi: 10.2133/DMPK.DMPK-10-RG-080 [DOI](https://doi.org/10.2133/DMPK.DMPK-10-RG-080) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21084764/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Metab%20Pharmacokinet&title=Influence%20of%20CYP4F2%20rs2108622%20(V433M)%20on%20warfarin%20dose%20requirement%20in%20Asian%20patients&author=O%20Singh&author=E%20Sandanaraj&author=K%20Subramanian&author=LH%20Lee&author=B%20Chowbay&volume=26&issue=2&publication_year=2011&pages=130-136&pmid=21084764&doi=10.2133/DMPK.DMPK-10-RG-080&)
|
| 306 |
+
|
| 307 |
+
18. Rusdiana T, Araki T, Nakamura T, Subarnas A, Yamamoto K. Responsiveness to low-dose warfarin associated with genetic variants of VKORC1, CYP2C9, CYP2C19, and CYP4F2 in an Indonesian population. Eur J Clin Pharmacol. 2013;69(3):395–405. doi: 10.1007/S00228-012-1356-9 [DOI](https://doi.org/10.1007/S00228-012-1356-9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22855348/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=Responsiveness%20to%20low-dose%20warfarin%20associated%20with%20genetic%20variants%20of%20VKORC1,%20CYP2C9,%20CYP2C19,%20and%20CYP4F2%20in%20an%20Indonesian%20population&author=T%20Rusdiana&author=T%20Araki&author=T%20Nakamura&author=A%20Subarnas&author=K%20Yamamoto&volume=69&issue=3&publication_year=2013&pages=395-405&pmid=22855348&doi=10.1007/S00228-012-1356-9&)
|
| 308 |
+
|
| 309 |
+
19. McDonald MG, Rieder MJ, Nakano M, Hsia CK, Rettie AE. CYP4F2 is a vitamin K1 oxidase: an explanation for altered warfarin dose in carriers of the V433M variant. Mol Pharmacol. 2009;75(6):1337–1346. doi: 10.1124/MOL.109.054833 [DOI](https://doi.org/10.1124/MOL.109.054833) | [PMC free article](/articles/PMC2684883/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19297519/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mol%20Pharmacol&title=CYP4F2%20is%20a%20vitamin%20K1%20oxidase:%20an%20explanation%20for%20altered%20warfarin%20dose%20in%20carriers%20of%20the%20V433M%20variant&author=MG%20McDonald&author=MJ%20Rieder&author=M%20Nakano&author=CK%20Hsia&author=AE%20Rettie&volume=75&issue=6&publication_year=2009&pages=1337-1346&pmid=19297519&doi=10.1124/MOL.109.054833&)
|
| 310 |
+
|
| 311 |
+
20. Pei L, Tian X, Long Y, et al. Establishment of a Han Chinese-specific pharmacogenetic-guided warfarin dosing algorithm. Medicine. 2018;97(36):e12178. doi: 10.1097/MD.0000000000012178 [DOI](https://doi.org/10.1097/MD.0000000000012178) | [PMC free article](/articles/PMC6133597/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/30200121/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Medicine&title=Establishment%20of%20a%20Han%20Chinese-specific%20pharmacogenetic-guided%20warfarin%20dosing%20algorithm&author=L%20Pei&author=X%20Tian&author=Y%20Long&volume=97&issue=36&publication_year=2018&pages=e12178&pmid=30200121&doi=10.1097/MD.0000000000012178&)
|
| 312 |
+
|
| 313 |
+
21. Cho EH, Lee K, Yang M, et al. Development and validation of a novel warfarin dosing algorithm for Korean patients with VKORC1 1173C. Ann Lab Med. 2020;40(3):216. doi: 10.3343/ALM.2020.40.3.216 [DOI](https://doi.org/10.3343/ALM.2020.40.3.216) | [PMC free article](/articles/PMC6933054/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31858761/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20Lab%20Med&title=Development%20and%20validation%20of%20a%20novel%20warfarin%20dosing%20algorithm%20for%20Korean%20patients%20with%20VKORC1%201173C&author=EH%20Cho&author=K%20Lee&author=M%20Yang&volume=40&issue=3&publication_year=2020&pages=216&pmid=31858761&doi=10.3343/ALM.2020.40.3.216&)
|
| 314 |
+
|
| 315 |
+
22. Ramirez AH, Shi Y, Schildcrout JS, et al. Predicting warfarin dosage in European–Americans and African–Americans using DNA samples linked to an electronic health record. Pharmacogenomics. 2012;13(4):407. doi: 10.2217/PGS.11.164 [DOI](https://doi.org/10.2217/PGS.11.164) | [PMC free article](/articles/PMC3361510/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22329724/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=Predicting%20warfarin%20dosage%20in%20European%E2%80%93Americans%20and%20African%E2%80%93Americans%20using%20DNA%20samples%20linked%20to%20an%20electronic%20health%20record&author=AH%20Ramirez&author=Y%20Shi&author=JS%20Schildcrout&volume=13&issue=4&publication_year=2012&pages=407&pmid=22329724&doi=10.2217/PGS.11.164&)
|
| 316 |
+
|
| 317 |
+
23. Mukaka MM. A guide to appropriate use of correlation coefficient in medical research. Malawi Med J. 2012;24(3):69. [PMC free article](/articles/PMC3576830/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23638278/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Malawi%20Med%20J&title=A%20guide%20to%20appropriate%20use%20of%20correlation%20coefficient%20in%20medical%20research&author=MM%20Mukaka&volume=24&issue=3&publication_year=2012&pages=69&pmid=23638278&)
|
| 318 |
+
|
| 319 |
+
24. Garcia D, Regan S, Crowther M, Hughes RA, Hylek EM. Warfarin maintenance dosing patterns in clinical practice: implications for safer anticoagulation in the elderly population. Chest. 2005;127(6):2049–2056. doi: 10.1378/CHEST.127.6.2049 [DOI](https://doi.org/10.1378/CHEST.127.6.2049) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15947319/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chest&title=Warfarin%20maintenance%20dosing%20patterns%20in%20clinical%20practice:%20implications%20for%20safer%20anticoagulation%20in%20the%20elderly%20population&author=D%20Garcia&author=S%20Regan&author=M%20Crowther&author=RA%20Hughes&author=EM%20Hylek&volume=127&issue=6&publication_year=2005&pages=2049-2056&pmid=15947319&doi=10.1378/CHEST.127.6.2049&)
|
| 320 |
+
|
| 321 |
+
25. Shendre A, Parmar GM, Dillon C, Beasley TM, Limdi NA. Influence of age on warfarin dose, anticoagulation control, and risk of hemorrhage. Pharmacotherapy. 2018;38(6):588–596. doi: 10.1002/PHAR.2089 [DOI](https://doi.org/10.1002/PHAR.2089) | [PMC free article](/articles/PMC6014885/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29393514/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacotherapy&title=Influence%20of%20age%20on%20warfarin%20dose,%20anticoagulation%20control,%20and%20risk%20of%20hemorrhage&author=A%20Shendre&author=GM%20Parmar&author=C%20Dillon&author=TM%20Beasley&author=NA%20Limdi&volume=38&issue=6&publication_year=2018&pages=588-596&pmid=29393514&doi=10.1002/PHAR.2089&)
|
| 322 |
+
|
| 323 |
+
26. Miura T, Nishinaka T, Terada T, Yonezawa K. Relationship between aging and dosage of warfarin: the current status of warfarin anticoagulant therapy for Japanese outpatients in a department of cardiovascular medicine. J Cardiol. 2009;53(3):355–360. doi: 10.1016/J.JJCC.2008.12.003 [DOI](https://doi.org/10.1016/J.JJCC.2008.12.003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19477376/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Cardiol&title=Relationship%20between%20aging%20and%20dosage%20of%20warfarin:%20the%20current%20status%20of%20warfarin%20anticoagulant%20therapy%20for%20Japanese%20outpatients%20in%20a%20department%20of%20cardiovascular%20medicine&author=T%20Miura&author=T%20Nishinaka&author=T%20Terada&author=K%20Yonezawa&volume=53&issue=3&publication_year=2009&pages=355-360&pmid=19477376&doi=10.1016/J.JJCC.2008.12.003&)
|
| 324 |
+
|
| 325 |
+
27. Aktan A, Güzel T, Aslan B, et al. Comparison of the real-life clinical outcomes of warfarin with effective time in therapeutic range and non-vitamin K antagonist oral anticoagulants: insight from the AFTER-2 trial. Kardiol Pol. 2023;81(2):132–140. doi: 10.33963/KP.A2022.0287 [DOI](https://doi.org/10.33963/KP.A2022.0287) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36594528/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Kardiol%20Pol&title=Comparison%20of%20the%20real-life%20clinical%20outcomes%20of%20warfarin%20with%20effective%20time%20in%20therapeutic%20range%20and%20non-vitamin%20K%20antagonist%20oral%20anticoagulants:%20insight%20from%20the%20AFTER-2%20trial&author=A%20Aktan&author=T%20G%C3%BCzel&author=B%20Aslan&volume=81&issue=2&publication_year=2023&pages=132-140&pmid=36594528&doi=10.33963/KP.A2022.0287&)
|
| 326 |
+
|
| 327 |
+
28. Alshammari A, Altuwayjiri A, Alshaharani Z, Bustami R, Almodaimegh HS. Warfarin dosing requirement according to body mass index. Cureus. 2020;12(10). doi: 10.7759/CUREUS.11047 [DOI](https://doi.org/10.7759/CUREUS.11047) | [PMC free article](/articles/PMC7676436/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33224644/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cureus&title=Warfarin%20dosing%20requirement%20according%20to%20body%20mass%20index&author=A%20Alshammari&author=A%20Altuwayjiri&author=Z%20Alshaharani&author=R%20Bustami&author=HS%20Almodaimegh&volume=12&issue=10&publication_year=2020&pmid=33224644&doi=10.7759/CUREUS.11047&)
|
| 328 |
+
|
| 329 |
+
29. Mueller JA, Patel T, Halawa A, Dumitrascu A, Dawson NL. Warfarin dosing and body mass index. Ann Pharmacother. 2014;48(5):584–588. doi: 10.1177/1060028013517541 [DOI](https://doi.org/10.1177/1060028013517541) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24558184/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20Pharmacother&title=Warfarin%20dosing%20and%20body%20mass%20index&author=JA%20Mueller&author=T%20Patel&author=A%20Halawa&author=A%20Dumitrascu&author=NL%20Dawson&volume=48&issue=5&publication_year=2014&pages=584-588&pmid=24558184&doi=10.1177/1060028013517541&)
|
| 330 |
+
|
| 331 |
+
30. Yoo SH, Kwon SU, Jo MW, Kang DW, Kim JS. Age- and weight-adjusted warfarin initiation nomogram for ischaemic stroke patients. Eur J Neurol. 2012;19(12):1547–1553. doi: 10.1111/J.1468-1331.2012.03772.X [DOI](https://doi.org/10.1111/J.1468-1331.2012.03772.X) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22672718/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Neurol&title=Age-%20and%20weight-adjusted%20warfarin%20initiation%20nomogram%20for%20ischaemic%20stroke%20patients&author=SH%20Yoo&author=SU%20Kwon&author=MW%20Jo&author=DW%20Kang&author=JS%20Kim&volume=19&issue=12&publication_year=2012&pages=1547-1553&pmid=22672718&doi=10.1111/J.1468-1331.2012.03772.X&)
|
| 332 |
+
|
| 333 |
+
31. Cheymol G. Effects of obesity on pharmacokinetics implications for drug therapy. Clin Pharmacokinet. 2000;39(3):215–231. doi: 10.2165/00003088-200039030-00004 [DOI](https://doi.org/10.2165/00003088-200039030-00004) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11020136/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacokinet&title=Effects%20of%20obesity%20on%20pharmacokinetics%20implications%20for%20drug%20therapy&author=G%20Cheymol&volume=39&issue=3&publication_year=2000&pages=215-231&pmid=11020136&doi=10.2165/00003088-200039030-00004&)
|
| 334 |
+
|
| 335 |
+
32. Li JH, Ma GG, Zhu SQ, Yan H, Wu YB, Xu JJ. Correlation between single nucleotide polymorphisms in CYP4F2 and warfarin dosing in Chinese valve replacement patients. J Cardiothorac Surg. 2012;7(1):97. doi: 10.1186/1749-8090-7-97 [DOI](https://doi.org/10.1186/1749-8090-7-97) | [PMC free article](/articles/PMC3487995/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23013706/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Cardiothorac%20Surg&title=Correlation%20between%20single%20nucleotide%20polymorphisms%20in%20CYP4F2%20and%20warfarin%20dosing%20in%20Chinese%20valve%20replacement%20patients&author=JH%20Li&author=GG%20Ma&author=SQ%20Zhu&author=H%20Yan&author=YB%20Wu&volume=7&issue=1&publication_year=2012&pages=97&pmid=23013706&doi=10.1186/1749-8090-7-97&)
|
| 336 |
+
|
| 337 |
+
33. Khosropanah S, Faraji SN, Habibi H, Yavarian M, Mansoori R, Haghpanah S. Correlation between Rs2108622 locus of CYP4F2 gene single nucleotide polymorphism and warfarin dosage in Iranian cardiovascular patients. Iran J Pharm Res. 2017;16(3):1238. [PMC free article](/articles/PMC5610780/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29201113/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Iran%20J%20Pharm%20Res&title=Correlation%20between%20Rs2108622%20locus%20of%20CYP4F2%20gene%20single%20nucleotide%20polymorphism%20and%20warfarin%20dosage%20in%20Iranian%20cardiovascular%20patients&author=S%20Khosropanah&author=SN%20Faraji&author=H%20Habibi&author=M%20Yavarian&author=R%20Mansoori&volume=16&issue=3&publication_year=2017&pages=1238&pmid=29201113&)
|
| 338 |
+
|
| 339 |
+
34. Borgiani P, Ciccacci C, Forte V, et al. CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics. 2009;10(2):261–266. doi: 10.2217/14622416.10.2.261 [DOI](https://doi.org/10.2217/14622416.10.2.261) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19207028/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=CYP4F2%20genetic%20variant%20(rs2108622)%20significantly%20contributes%20to%20warfarin%20dosing%20variability%20in%20the%20Italian%20population&author=P%20Borgiani&author=C%20Ciccacci&author=V%20Forte&volume=10&issue=2&publication_year=2009&pages=261-266&pmid=19207028&doi=10.2217/14622416.10.2.261&)
|
| 340 |
+
|
| 341 |
+
35. Zhang JE, Jorgensen AL, Alfirevic A, et al. Effects of CYP4F2 genetic polymorphisms and haplotypes on clinical outcomes in patients initiated on warfarin therapy. Pharmacogenet Genomics. 2009;19(10):781–789. doi: 10.1097/FPC.0B013E3283311347 [DOI](https://doi.org/10.1097/FPC.0B013E3283311347) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19741565/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenet%20Genomics&title=Effects%20of%20CYP4F2%20genetic%20polymorphisms%20and%20haplotypes%20on%20clinical%20outcomes%20in%20patients%20initiated%20on%20warfarin%20therapy&author=JE%20Zhang&author=AL%20Jorgensen&author=A%20Alfirevic&volume=19&issue=10&publication_year=2009&pages=781-789&pmid=19741565&doi=10.1097/FPC.0B013E3283311347&)
|
| 342 |
+
|
| 343 |
+
36. Harada T, Ariyoshi N, Shimura H, et al. Application of Akaike information criterion to evaluate warfarin dosing algorithm. Thromb Res. 2010;126(3):183–190. doi: 10.1016/J.THROMRES.2010.05.016 [DOI](https://doi.org/10.1016/J.THROMRES.2010.05.016) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20553802/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Thromb%20Res&title=Application%20of%20Akaike%20information%20criterion%20to%20evaluate%20warfarin%20dosing%20algorithm&author=T%20Harada&author=N%20Ariyoshi&author=H%20Shimura&volume=126&issue=3&publication_year=2010&pages=183-190&pmid=20553802&doi=10.1016/J.THROMRES.2010.05.016&)
|
| 344 |
+
|
| 345 |
+
37. Kringen MK, Haug KBF, Grimholt RM, et al. Genetic variation of VKORC1 and CYP4F2 genes related to warfarin maintenance dose in patients with myocardial infarction. J Biomed Biotechnol. 2011;2011. doi: 10.1155/2011/739751 [DOI](https://doi.org/10.1155/2011/739751) | [PMC free article](/articles/PMC2992873/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21127708/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Biomed%20Biotechnol&title=Genetic%20variation%20of%20VKORC1%20and%20CYP4F2%20genes%20related%20to%20warfarin%20maintenance%20dose%20in%20patients%20with%20myocardial%20infarction&author=MK%20Kringen&author=KBF%20Haug&author=RM%20Grimholt&volume=2011&publication_year=2011&pmid=21127708&doi=10.1155/2011/739751&)
|
| 346 |
+
|
| 347 |
+
38. Antman EM. Cardiovascular therapeutics: a companion to braunwald’s heart disease: fourth edition. Elsevier. 1–807.
|
| 348 |
+
|
| 349 |
+
39. Lei X, Guo Y, Sun J, et al. Accuracy assessment of pharmacogenetic algorithms for warfarin dose prediction in Chinese patients. Am J Hematol. 2012;87(5):541–544. doi: 10.1002/AJH.23151 [DOI](https://doi.org/10.1002/AJH.23151) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22460248/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Hematol&title=Accuracy%20assessment%20of%20pharmacogenetic%20algorithms%20for%20warfarin%20dose%20prediction%20in%20Chinese%20patients&author=X%20Lei&author=Y%20Guo&author=J%20Sun&volume=87&issue=5&publication_year=2012&pages=541-544&pmid=22460248&doi=10.1002/AJH.23151&)
|
| 350 |
+
|
| 351 |
+
40. Cho HJ, On YK, Bang OY, et al. Development and comparison of a warfarin-dosing algorithm for Korean patients with atrial fibrillation. Clin Ther. 2011;33(10):1371–1380. doi: 10.1016/J.CLINTHERA.2011.09.004 [DOI](https://doi.org/10.1016/J.CLINTHERA.2011.09.004) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21981797/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Ther&title=Development%20and%20comparison%20of%20a%20warfarin-dosing%20algorithm%20for%20Korean%20patients%20with%20atrial%20fibrillation&author=HJ%20Cho&author=YK%20On&author=OY%20Bang&volume=33&issue=10&publication_year=2011&pages=1371-1380&pmid=21981797&doi=10.1016/J.CLINTHERA.2011.09.004&)
|
| 352 |
+
|
| 353 |
+
41. Khoury G, Sheikh-Taha M. Effect of age and sex on warfarin dosing. Clin Pharmacol. 2014;6(1):103–106. doi: 10.2147/CPAA.S66776 [DOI](https://doi.org/10.2147/CPAA.S66776) | [PMC free article](/articles/PMC4103915/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25050078/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol&title=Effect%20of%20age%20and%20sex%20on%20warfarin%20dosing&author=G%20Khoury&author=M%20Sheikh-Taha&volume=6&issue=1&publication_year=2014&pages=103-106&pmid=25050078&doi=10.2147/CPAA.S66776&)
|
test/texts/PMC11803932.md
ADDED
|
@@ -0,0 +1,357 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Drug response is related to NR3C1 and FAAH polymorphism in Chinese pediatric epilepsy patients
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Hongli Wang, Chu Li, Qian Li, Ning Li, Huiling Qin
|
| 5 |
+
**Journal:** Italian Journal of Pediatrics
|
| 6 |
+
**Date:** 2025 Feb 7
|
| 7 |
+
**DOI:** [10.1186/s13052-025-01870-7](https://doi.org/10.1186/s13052-025-01870-7)
|
| 8 |
+
**PMID:** 39920787
|
| 9 |
+
**PMCID:** PMC11803932
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11803932/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11803932/pdf/13052_2025_Article_1870.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11803932/pdf/13052_2025_Article_1870.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Background:**
|
| 16 |
+
Childhood epilepsy is a common neurological syndrome with complex etiology and recurrent seizures. It seriously affects the growth and development of child patients.
|
| 17 |
+
|
| 18 |
+
**Methods:**
|
| 19 |
+
NR3C1 rs41423247 and FAAH rs324420 polymorphisms were detected by the polymerase chain reaction in 105 pediatric epilepsy patients. Patients were divided into the good response group and the poor response group after anti-seizure medications (ASMs) treatment. According to the results of the liver function test (LFT), patients were divided into the no LFT disturbance group and the LFT disturbance group. Hardy–Weinberg balance was applied to analyze the population representation. The correlations were calculated by logistic regression analysis.
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
NR3C1 rs41423247 genotype and allele frequencies differed significantly between good response and poor response groups, while FAAH rs324420 did not. The CG genotype and C allele of NR3C1 rs41423247 were associated with good drug response, and the association was also detected in the dominant model. In addition, polymorphisms in NR3C1 and FAAH were not associated with liver damage induced by epilepsy medication.
|
| 23 |
+
|
| 24 |
+
**Conclusion:**
|
| 25 |
+
The polymorphism of NR3C1 rs41423247 might influence the drug response of epilepsy children.
|
| 26 |
+
|
| 27 |
+
Keywords: NR3C1 rs41423247, FAAH rs324420, Drug response, Epilepsy, Liver dysfunction
|
| 28 |
+
|
| 29 |
+
### Background
|
| 30 |
+
|
| 31 |
+
Childhood epilepsy is a common neurological syndrome with complex etiology and recurrent seizures. It seriously affects the growth and development of child patients.
|
| 32 |
+
|
| 33 |
+
### Methods
|
| 34 |
+
|
| 35 |
+
NR3C1 rs41423247 and FAAH rs324420 polymorphisms were detected by the polymerase chain reaction in 105 pediatric epilepsy patients. Patients were divided into the good response group and the poor response group after anti-seizure medications (ASMs) treatment. According to the results of the liver function test (LFT), patients were divided into the no LFT disturbance group and the LFT disturbance group. Hardy–Weinberg balance was applied to analyze the population representation. The correlations were calculated by logistic regression analysis.
|
| 36 |
+
|
| 37 |
+
### Results
|
| 38 |
+
|
| 39 |
+
NR3C1 rs41423247 genotype and allele frequencies differed significantly between good response and poor response groups, while FAAH rs324420 did not. The CG genotype and C allele of NR3C1 rs41423247 were associated with good drug response, and the association was also detected in the dominant model. In addition, polymorphisms in NR3C1 and FAAH were not associated with liver damage induced by epilepsy medication.
|
| 40 |
+
|
| 41 |
+
### Conclusion
|
| 42 |
+
|
| 43 |
+
The polymorphism of NR3C1 rs41423247 might influence the drug response of epilepsy children.
|
| 44 |
+
|
| 45 |
+
**Keywords:**Keywords: NR3C1 rs41423247, FAAH rs324420, Drug response, Epilepsy, Liver dysfunction
|
| 46 |
+
|
| 47 |
+
## Introduction
|
| 48 |
+
|
| 49 |
+
Epilepsy is a disorder in which patients experience sudden, brief, and recurrent seizures of symptoms and/or signs, caused by a variety of etiological factors that result in abnormal or excessive firing activity of neurons in the brain [[1](#CR1)1]. Seizures, in the form of altered consciousness, involuntary motor, sensory, or psychiatric events, are a common chronic disease of the nervous system and are a significant cause of disability and death [[2](#CR2)2]. Since there is still a lack of effective preventive measures and cures for epileptic disorders at this stage, most patients require regular long-term drug therapy. Antiepileptic drugs are used to control the frequency of epileptic seizures and can be used as a single drug, but they are usually administered in combination with multiple drugs [[3](#CR3)3]. Most anti-seizure medications (ASMs) have different degrees of adverse effects, with dose-related adverse effects being the most common, including tremors, anorexia, nausea, vomiting, and drowsiness [[4](#CR4)4]. Long-term use is difficult for patients to accept, and therefore is prone to interruptions in treatment and recurrence of the disease. The ineffectiveness of antiepileptic treatment affects as many as 30% of epileptic patients, which is particularly important in the clinical treatment of epilepsy [[5](#CR5)5, [6](#CR6)6]. The role of genetic diversity in the pathogenesis of epilepsy drug resistance seems indisputable [[7](#CR7)7]. Accordingly, this research spotlighted the roles of genetic polymorphism in treating epilepsy.
|
| 50 |
+
|
| 51 |
+
Single-nucleotide polymorphisms (SNPs) refer to single-nucleotide variations in a genetic sequence among people, which are the most frequent nucleotide variations in the human genome. SNPs are associated with epilepsy, which can influence the occurrence and therapy efficacy of epilepsy. The last decade has observed a significant improvement in unraveling several mutations in the genes that are responsible for drug resistance and associated adverse drug reactions in the treatment of epilepsy [[8](#CR8)8]. For example, the G allele of rs1491974 or rs6798347 of microglial P2Y12 receptors may elevate the risk of the frequency of seizure [[9](#CR9)9]. The rs57095329 SNP of miR-146a and rs3789243 of ATP-binding cassette subfamily B member 1 influence the drug response and resistance of epilepsy, which may guide the clinical treatment [[10](#CR10)10, [11](#CR11)11]. These investigations unveil that SNPs play crucial roles in epilepsy. These findings would help the concerned neurologists to prescribe the medicine in epilepsy patients with more accuracy and to achieve the maximum therapeutic benefit. This would also help to improve the quality of life of patients with epilepsy and will avoid the recurrence of seizures.
|
| 52 |
+
|
| 53 |
+
Glucocorticoid receptor (NR3C1) is a gene encoded glucocorticoid receptor. The rs41423247 locus of NR3C1 carries a significantly higher frequency of the G allele in patients with functional seizures and major depression [[12](#CR12)12]. Mutations in fatty acid amide hydrolase (FAAH) (C385A; rs324420) have been associated with alterations in fronto-amygdala function, which may be associated with anxiety and fear symptoms [[13](#CR13)13]. COMT rs4680, FAAH rs324420, and OPRM1 rs1799971 models are related to the response of a placebo to nerve pain and can predict the effect of a placebo [[14](#CR14)14]. FAAH C384A genotype is associated with the risk of generalized epilepsy in Iranians [[15](#CR15)15]. Nowadays, the SNPs in NR3C1 and FAAH on the drug response of epilepsy remain unclear.
|
| 54 |
+
|
| 55 |
+
In this observation, the polymorphism of the rs41423247 locus on the NR3C1 gene and the rs324420 locus on the FAAH gene in children with epilepsy were detected, and the relationship between them and drug response in children with epilepsy was analyzed, so as to understand the clinical significance of these genetic polymorphisms in improving treatment outcomes for children with epilepsy.
|
| 56 |
+
|
| 57 |
+
## Materials and methods
|
| 58 |
+
|
| 59 |
+
### Participants
|
| 60 |
+
|
| 61 |
+
105 epileptic patients who were diagnosed and treated by Dongying People’s Hospital from June 2021 to October 2023 and met the criteria for admission and discharge were selected. This study has been approved by the Ethics Committee of Dongying People’s Hospital. All subjects signed informed consent before entering the group to collect blood samples. To protect patient privacy, only authorized medical personnel are allowed to access patient data. And the medical staff strictly follow the privacy policies established by the hospital.
|
| 62 |
+
|
| 63 |
+
Epilepsy is diagnosed by electroencephalography (EEG) examination results or typical seizure history. The diagnostic criteria refer to the Practical Clinical Definition of Epilepsy published by ILAE in 2014 The included criteria were: (1) receiving ASM treatment over 12 months; (2) aged from 2 years old to 17 years old, and (3) no consanguineous relationship between the subjects and no history of intermarriage. Patients with the following characteristics were excluded from the study: 1) comorbidity with other psychiatric disorders, 2) pseudoseizures, 3) unreliable seizure frequency without continuous electroencephalographic (CEEG) monitoring, 4) comorbidity with liver disease that can affect our detection indicators and failure of vital organs that can affect the life safety, 5) non-compliance with ASM treatment, and 6) incomplete clinical records.
|
| 64 |
+
|
| 65 |
+
Drug response to ASM therapy in patients with epilepsy was determined according to the International League Against Epilepsy (ILAE) definitions. A good drug response indicated completely seizure-free patients based on regular follow-up for at least 1 year during monotherapy or combination therapy at the best tolerated therapeutic dose [[16](#CR16)16, [17](#CR17)17]. Patients with an adverse response are those who have been correctly medicated at the maximum tolerated dose for at least 12 months after monotherapy or combination therapy, with ineffectiveness and persistent seizures.
|
| 66 |
+
|
| 67 |
+
Liver function tests (LFT), including alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, total bilirubin, direct bilirubin, and so on, were performed to assess liver function at baseline and outpatient clinic follow-ups at 12 months after ASM treatments.
|
| 68 |
+
|
| 69 |
+
### DNA isolation from blood samples
|
| 70 |
+
|
| 71 |
+
DNA was extracted using a blood genome column rapid extraction kit (CWBIO, Taizhou, China) with an additional isopropanol precipitation step for optimal DNA quality. 200 μl of blood was added to a centrifuge tube, and 20 μl of Proteinase K and 200 μl Buffer GL was added successively. After incubating at 80℃ for 10 min, 200 μl isopropanol and 20 μl magnetic beads were added and mixed upside down. The centrifuge tube was fixed on a magnetic rack and left to stand for 1 min and the supernatant was aspirated and discarded. Then, buffer GW1 was added to the centrifuge tube for repeated washes of the magnetic beads. The wash solution was discarded. Finally, the eluent was added, mixed, and transferred to a new centrifuge tube. The desired DNA solution was collected and stored at −20°C.
|
| 72 |
+
|
| 73 |
+
The DNA sample quality was described by NanoDrop (Thermo Scientific, Willmington, USA), and the DNA sample with A260/A280 between 1.7 and 1.9 met the experimental requirements.
|
| 74 |
+
|
| 75 |
+
### Detection of target genotype
|
| 76 |
+
|
| 77 |
+
Specific site amplification primers were designed according to the target site sequence, and the polymorphisms of FAAH and NR3C1 genes were analyzed by the dideoxy terminal termination method (Sanger method). PCR amplification reaction system included: PCR master mix (Takara, Shiga, Japan) 25 μl, Forward Primer 1 μl, Reverse Primer 1 μl, template 1 μl, and ddH_2_2O 22 μl. PCR reaction procedure was: pre-denaturation at 94℃ for 5 min; 28 circles of denaturation at 94℃ for 0.5 min, annealing at 58℃ for 0.5 min, extension at 72℃ for 1 min; and finally, 72°C extended for another 10 min. The primers of rs41423247 and rs324420 are listed in Table [1](#Tab1)1. The specificity of primers was verified by showing a single band of the correct amplified fragment size via agarose gel electrophoresis of the PCR products. All reagents, except for the template DNA, were prepared in an isolated pre-PCR room to prevent contamination. The PCR production was purified and put on the 3730 xl sequencer (Applied Biosystems, Foster City, USA) was used for sequencing. The Sequencing results will be analyzed by the DNA Chromas Analysis software (Technelysium, South Brisbane, Queensland).
|
| 78 |
+
|
| 79 |
+
### Table 1.
|
| 80 |
+
|
| 81 |
+
Primer sequences of FAAH and NR3C1
|
| 82 |
+
|
| 83 |
+
| SNP ID | Gene Symbol | Nucleotide Change Location | Forward Primer | Reverse Primer |
|
| 84 |
+
| ------ | ----------- | -------------------------- | -------------- | -------------- |
|
| 85 |
+
| rs41423247 | NR3C1 | 646 C > G | 5′TGCTGCCTTATTTGTAAATTCGT 3′ | 5′ AAGCTTAACAATTTTGGCCATC 3′ |
|
| 86 |
+
| rs324420 | FAAH | 385 C > A | 5′ TGTTGCTGGTTACCCCTCTC3′ | 5′ CCCAAAATGACCCAAGATGC3′ |
|
| 87 |
+
### Statistical analysis
|
| 88 |
+
|
| 89 |
+
SPSS 25.0 statistical software was applied to the study data for statistical processing (testing level *P*P < 0.05 was considered statistically significant). Measurement data with normal distribution were presented as mean ± standard deviation (SD). The Categorical variables were expressed as number or percentage, and the χ2 test was used for comparison between groups, and the genetic specific risk was estimated as odds ratios (ORs) with associated 95% confidence intervals (CIs). Hardy–Weinberg balance was used to verify the population representation of target genes. And HWE deviations in control cohorts are frequently caused by a relatively small sample size, and expanding the sample size is desirable.
|
| 90 |
+
|
| 91 |
+
## Results
|
| 92 |
+
|
| 93 |
+
### General clinicopathological characteristics
|
| 94 |
+
|
| 95 |
+
A total of 105 patients were included in the analysis, including 50 males and 55 females, aged (9.54 ± 3.04) years (Table [2](#Tab2)2). There were 13 patients with epilepsy history, accounting for 12.4% (Table [2](#Tab2)2). There were 59 patients with good drug reactions, accounting for 56.2, and 46 patients with poor drug reactions, accounting for 43.8% (Table [2](#Tab2)2).
|
| 96 |
+
|
| 97 |
+
### Table 2.
|
| 98 |
+
|
| 99 |
+
Clinical Characteristics of Patients with Epilepsy
|
| 100 |
+
|
| 101 |
+
| Items | Patients with Epilepsy (N = 105) |
|
| 102 |
+
| ----- | -------------------------------- |
|
| 103 |
+
| Age (years) | 9.54 ± 3.04 |
|
| 104 |
+
| Gender | |
|
| 105 |
+
| Male (N, %) | 50, 47.6 |
|
| 106 |
+
| Female (N, %) | 55, 52.4 |
|
| 107 |
+
| Family History of Epilepsy | |
|
| 108 |
+
| Yes (N, %) | 13, 12.4 |
|
| 109 |
+
| No (N, %) | 92, 87.6 |
|
| 110 |
+
| Drug Response | |
|
| 111 |
+
| Good (N, %) | 59, 56.2 |
|
| 112 |
+
| Poor (N, %) | 46, 43.8 |
|
| 113 |
+
### Polymorphism detection results
|
| 114 |
+
|
| 115 |
+
Sequencing of target gene loci in 105 epilepsy patients showed that 8 cases of NR3C1 rs41423247 were wild type (CC) and 97 cases had gene mutations, including 32 cases of heterozygous mutations (CG) and pure 65 cases of combined mutation (GG). The NR3C1 646 C > G genotype frequency was tested to be consistent with Hard-Weinberg equilibrium (*P*P = 0.10, Table [3](#Tab3)3).
|
| 116 |
+
|
| 117 |
+
### Table 3.
|
| 118 |
+
|
| 119 |
+
Distribution of rs41423247 and rs324420 in Patients with Epilepsy
|
| 120 |
+
|
| 121 |
+
| SNP ID | N, % |
|
| 122 |
+
| ------ | ---- |
|
| 123 |
+
| rs41423247 | |
|
| 124 |
+
| CC | 8, 7.6 |
|
| 125 |
+
| CG | 32, 30.5 |
|
| 126 |
+
| GG | 65, 61.9 |
|
| 127 |
+
| C | 44, 21.0 |
|
| 128 |
+
| G | 166, 79.0 |
|
| 129 |
+
| PHWE | 0.10 |
|
| 130 |
+
| rs324420 | |
|
| 131 |
+
| AA | 4, 3.8 |
|
| 132 |
+
| AC | 20, 19.1 |
|
| 133 |
+
| CC | 81, 77.1 |
|
| 134 |
+
| A | 28, 13.3 |
|
| 135 |
+
| C | 182, 86.7 |
|
| 136 |
+
| PHWE | 0.07 |
|
| 137 |
+
In all patients with epilepsy, 81 cases of FAAH rs324420 were wild type (CC), and 24 cases had gene mutations, including 20 cases of heterozygous mutations (CA) and 4 cases of homozygous mutations (AA). The rs324420 site complies with Hardy–Weinberg equilibrium (*P*P = 0.07, Table [3](#Tab3)3).
|
| 138 |
+
|
| 139 |
+
### Correlation between target SNP and drug response
|
| 140 |
+
|
| 141 |
+
There was a statistically significant difference in the comparison of rs41423247 genotype frequencies between the good response and poor response groups (*P*P < 0.001, Table [4](#Tab4)4). The frequency of the C allele was significantly higher in the good response group than in the poor response group (*P*P < 0.001, Table [4](#Tab4)4). Significantly, χ^2^2 analysis found that GG genotypes (OR = 6.023, 95% CI = 1.022–35.509, *P*P = 0.047, Table [4](#Tab4)4) and C allele (OR = 4.822, 95% CI = 1.363–17.064, *P*P = 0.015, Table [4](#Tab4)4) were associated with increased adverse drug reactions. The CG genotype increased the risk of poor response by over six times and the G allele elevated the risk of poor response by almost fivefold (Table [4](#Tab4)4).
|
| 142 |
+
|
| 143 |
+
### Table 4.
|
| 144 |
+
|
| 145 |
+
Distribution of rs41423247 and rs324420 in Patients with Epilepsy
|
| 146 |
+
|
| 147 |
+
| SNP ID | Good response(N = 59) | Poor response(N = 46) | χ2 | P value | OR (95%CI) | P value |
|
| 148 |
+
| ------ | --------------------- | --------------------- | -- | ------- | ---------- | ------- |
|
| 149 |
+
| rs41423247 | | | | | | |
|
| 150 |
+
| CC | 6, 10.2 | 2, 4.3 | / | / | 1.00 | |
|
| 151 |
+
| CG | 27, 45.8 | 5, 10.9 | / | / | 0.721 (0.101–5.122) | 0.744 |
|
| 152 |
+
| GG | 26, 44.1 | 39, 84.8 | 18.397 | < 0.001 | 6.023 (1.022–35.509) | 0.047 |
|
| 153 |
+
| C | 36, 30.5 | 8, 8.7 | / | / | 1.00 | |
|
| 154 |
+
| G | 82, 69.5 | 84, 91.3 | 7.425 | 0.008 | 4.822 (1.363–17.064) | 0.015 |
|
| 155 |
+
| rs324420 | | | | | | |
|
| 156 |
+
| AA | 3, 5.1 | 1, 2.2 | / | / | 1.00 | |
|
| 157 |
+
| AC | 10, 16.9 | 10, 21.7 | / | / | 0.581 (0.046–7.263) | 0.674 |
|
| 158 |
+
| CC | 46, 78.0 | 35, 76.1 | 0.898 | 0.638 | 1.655 (0.536–5.114) | 0.381 |
|
| 159 |
+
| A | 12, 10.2 | 16, 17.4 | / | / | 1.00 | |
|
| 160 |
+
| C | 106, 89.8 | 76, 82.6 | 1.167 | 0.280 | 1.902 (0.503–7.189) | 0.343 |
|
| 161 |
+
No difference in rs324420 genotypes and alleles was found between the good response and poor response groups and no correlations were observed between this polymorphism and drug response (All *P*P > 0.05, Table [4](#Tab4)4).
|
| 162 |
+
|
| 163 |
+
### Inheritance genotype models of different drug responses
|
| 164 |
+
|
| 165 |
+
The genotype model of inheritance of rs41423247 and rs324420 was exhibited in Table [5](#Tab5)5. In genetic model analysis, the dominant model of rs41423247 showed a difference between the good response group and the poor response group (*P*P < 0.001, Table [5](#Tab5)5) and it was related to the good drug response (OR = 8.253, 95% CI = 2.784–24.467, *P*P < 0.001, Table [5](#Tab5)5). The recessive model of rs41423247 together with the dominant and recessive models of rs324420 represented no correlation with the drug response of ASMs (All *P*P > 0.05, Table [5](#Tab5)5).
|
| 166 |
+
|
| 167 |
+
### Table 5.
|
| 168 |
+
|
| 169 |
+
Distribution of genetic model of rs41423247 and rs324420 in Patients with Epilepsy
|
| 170 |
+
|
| 171 |
+
| Model | Genotype | Good response(N = 59) | Poor response(N = 46) | χ2 | P value | OR (95%CI) | P value |
|
| 172 |
+
| ----- | -------- | --------------------- | --------------------- | -- | ------- | ---------- | ------- |
|
| 173 |
+
| | rs41423247 | | | | | | |
|
| 174 |
+
| Dominant | CC-CG | 33, 55.9 | 7, 15.2 | / | / | 1.00 | |
|
| 175 |
+
| | GG | 26, 44.1 | 39, 84.8 | 18.169 | < 0.001 | 8.253 (2.784–24.467) | < 0.001 |
|
| 176 |
+
| Recessive | CC | 6, 10.2 | 2, 4.3 | / | / | 1.00 | |
|
| 177 |
+
| | CG-GG | 53, 89.8 | 44, 95.7 | 1.245 | 0.461 | 1.826 (0.280–11.921) | 0.529 |
|
| 178 |
+
| | rs324420 | | | | | | |
|
| 179 |
+
| Dominant | AA-AC | 13, 22.0 | 11, 23.9 | / | / | 1.00 | |
|
| 180 |
+
| | CC | 46, 78.0 | 35, 76.1 | 0.052 | 0.820 | 1.527 (0.507–4.605) | 0.452 |
|
| 181 |
+
| Recessive | AA | 3, 5.1 | 1, 2.2 | / | / | 1.00 | |
|
| 182 |
+
| | AC-CC | 56, 94.9 | 45, 97.8 | 0.598 | 0.630 | 0.333 (0.023–4.717) | 0.416 |
|
| 183 |
+
### Relationship between polymorphisms and the risk of liver dysfunction
|
| 184 |
+
|
| 185 |
+
Of the genotyped patients, 39 cases experienced hepatotoxicity while 66 did not. Individuals with the variant of the rs41423247 and rs324420 represented no statistical significance of LFT impairment (All *P*P > 0.05, Table [6](#Tab6)6).
|
| 186 |
+
|
| 187 |
+
### Table 6.
|
| 188 |
+
|
| 189 |
+
Distribution of rs41423247 and rs324420 in patients with epilepsy
|
| 190 |
+
|
| 191 |
+
| SNP ID | No LFT disturbance(N = 66) | LFT disturbance(N = 39) | χ2 | P value | OR (95%CI) | P value |
|
| 192 |
+
| ------ | -------------------------- | ----------------------- | -- | ------- | ---------- | ------- |
|
| 193 |
+
| rs41423247 | | | | | | |
|
| 194 |
+
| CC | 3, 4.5 | 5, 12.8 | / | / | 1.00 | |
|
| 195 |
+
| CG | 19, 28.8 | 13, 33.3 | / | / | 3.950 (0.832–18.750) | 0.084 |
|
| 196 |
+
| GG | 44, 66.7 | 21, 53.8 | 3.020 | 0.221 | 1.418 (0.572–33.519) | 0.451 |
|
| 197 |
+
| rs324420 | | | | | | |
|
| 198 |
+
| AA | 3, 4.5 | 1, 2.6 | / | / | 1.00 | |
|
| 199 |
+
| AC | 9, 13.6 | 12, 30.8 | / | / | 0.886 (0.085–9.223) | 0.920 |
|
| 200 |
+
| CC | 54, 81.8 | 26, 66.7 | 4.589 | 0.101 | 2.990 (1.099–8.131) | 0.032 |
|
| 201 |
+
## Discussion
|
| 202 |
+
|
| 203 |
+
This study investigated the relationship between drug response to epilepsy treatment and the rs41423247 polymorphism on the NR3C1 gene or the SNP rs324420 on the FAAH gene. The genotypes and allele frequencies of the rs41423247 locus on the NR3C1 gene and the rs324420 locus on the FAAH gene were found to be in accordance with the Hardy–Weinberg balance in 105 epileptic patients. Of the 105 patients who completed ASM treatment in this study, 59 had a good response to the ASMs, and 46 responded negatively. In the good response group, the CG genotype frequency, the C allele frequency, dominant genotype at the rs41423247 locus on the NR3C1 gene were significantly higher than in the poor response group. However, there was no significant difference in rs324420 polymorphism of the FAAH gene between the two groups. Rs41423247 polymorphism on the NR3C1 gene might be associated with drug response to ASM treatment, and antiepileptic efficacy was better in patients with the C allele and CG genotype. In addition, the rs41423247 polymorphism on the NR3C1 gene and the rs324420 on the FAAH gene were not associated with liver injury after epilepsy drug therapy. The findings indicated that the liver function of epilepsy cases after drug therapy might not be influenced by different genotypes of rs41423247 and rs324420 polymorphisms. However, other factors, such as medication type and duration of treatment, may be confounding factors that were not considered in the present study, which should be verified in future studies.
|
| 204 |
+
|
| 205 |
+
Epilepsy is a chronic disease caused by abnormal discharges of neurons in the brain. Patients may experience sudden loss of consciousness, foaming at the mouth, convulsions, muscle stiffness, or tremors during an attack [[18](#CR18)18]. The causes of epilepsy are varied, including genetic factors, brain infections, traumatic brain injuries, brain tumors, and so on [[19](#CR19)19]. Non-pharmacological treatments such as epilepsy surgery, neuromodulation, and ketogenic diets have made great strides, but pharmacological treatments are still the mainstay [[20](#CR20)20]. Antiepileptic drugs can control seizures by modulating neuronal excitability and inhibiting neuronal over-discharge [[21](#CR21)21]. Phenobarbital is the oldest antiepileptic drug still in wide clinical use. These drugs successfully suppress seizures in the majority of patients. However, in approximately 20–40% of patients, epilepsy is drug resistant [[22](#CR22)22].
|
| 206 |
+
|
| 207 |
+
The therapeutic response to ASMs varies significantly in different individuals, and blood concentrations at conventional doses can exceed the range of effective therapeutic concentrations, leading to therapeutic failure, reduced tolerance, and adverse effects. Genetic polymorphisms are one of the main reasons for these differences. Therefore, this article focused on the polymorphism in the drug response of treatment in epilepsy in order to explore the mechanism of drug response in ASM, so as to assist personalized treatment strategies. Many genetic polymorphisms play a role in the pathogenesis and treatment of epilepsy, e.g., SCN1A ABCG2, SCN1A, CYP3A5, and SCN2A [[23](#CR23)23–[25](#CR25)25]. The rs211037 polymorphism on the GABRG2 gene is associated with valproic acid-induced adverse drug reactions, and the CC genotype was associated with the absence of seizures after treatment [[26](#CR26)26]. The rs2556375 of BCL11A increases the seizure susceptibility and risk of drug resistance [[27](#CR27)27]. The SNP of NR3C1 is correlated with difficult-to-treat rhinosinusitis, glucose metabolism type 2 diabetes, and IgA nephropathies [[28](#CR28)28–[30](#CR30)30]. Notably, NR3C1 rs41423247 polymorphism is related to functional seizures of Iranian [[12](#CR12)12]. In our study, we found a correlation between NR3C1 and drug response to ASM treatment in Chinese pediatric epilepsy patients, mainly in the form of CG genotype, and C allele is associated with a good response to the drug, suggesting that the NR3C1 rs41423247 polymorphism may affect the therapeutic effect of ASM in epilepsy patients. The role of glucocorticoid receptors in the nervous system may influence epileptic drug response. Glucocorticoids can regulate the release of neurotransmitters, neuronal excitability and synaptic plasticity. The functional status of the glucocorticoid receptor encoded by the NR3C1 gene may affect the regulatory effect of glucocorticoids on the nervous system, and thus affect the efficacy of epilepsy drugs. In addition, the polymorphism of NR3C1 gene may affect the metabolism and transport of epilepsy drugs. Different NR3C1 gene polymorphisms may lead to changes in the structure and function of glucocorticoid receptors, thereby affecting drug metabolism and transport. Some studies have suggested that polymorphisms in the NR3C1 gene may indirectly affect the metabolism and transport of epilepsy drugs by affecting the function of the hypothalamic–pituitary–adrenal axis (HPA) [[31](#CR31)31]. However, the mechanism should be verified in future studies. In the clinical treatment of patients with epilepsy, understanding the genotype of the NR3C1 rs41423247 site can help doctors develop a more personalized treatment plan. For carriers of the CG genotype, doctors may be more inclined to choose conventional antiepileptic drugs during initial treatment. Because these patients may have better anti-epileptic efficacy according to the conclusion of the study, they can avoid overuse of some powerful drugs that may have more side effects. For carriers of the CC gene, doctors can consider adjusting treatment options, such as changing drug types, adjusting drug dosages, or combining other drugs. In addition, accumulating data supports an autoimmune basis in patients with antiepileptic drug-resistant seizures [[32](#CR32)32, [33](#CR33)33]. NR3C1 has been reported to serve as potential immune-related biomarkers [[34](#CR34)34]. And NR3C1 is linked with a deregulated hypothalamus–pituitary–adrenal (HPA) axis and psychopathology [[35](#CR35)35]. Therefore, the role of NR3C1-mediated immune response in epilepsy drug response is worth exploring in depth. Besides, there is a close correlation between probiotic-mediated immunity and drug resistance in epileptic patients [[36](#CR36)36]. Probiotics may have a positive impact on drug resistance in patients with epilepsy by regulating the immune system, enhancing the intestinal barrier, regulating intestinal microbiota, and other mechanisms [[37](#CR37)37]. In the clinical treatment of epilepsy patients, probiotics supplement therapy has important research significance.
|
| 208 |
+
|
| 209 |
+
FAAH rs324420 is widely researched in several diseases or physiological processes, such as motor performance, memory fading, and susceptibility to methamphetamine dependence [[38](#CR38)38–[40](#CR40)40]. In the Iranian population, FAAH rs324420 genotype and allele distribution were shown to be associated with generalized epilepsy and not with focal epilepsy [[15](#CR15)15]. In our study, the FAAH rs324420 polymorphism did not correlate with drug response after ASM treatment in patients with epilepsy These results suggest that polymorphisms in FAAH do not correlate with drug response to ASM therapy for epilepsy. However, the sample size of this study is relatively small, and the comparison of polycentric large samples of homogenous populations is lacking. Although relatively few direct studies have been conducted on the role of FAAH in patients with different types of epilepsy, its role in epilepsy warrants further investigation because it is a key enzyme in the endocannabinoid system [[41](#CR41)41]. And the endocannabinoid system plays an important role in neuropsychiatric diseases [[42](#CR42)42]. The study showed that FAAH polymorphism is not associated with epilepsy resistance, which may mean that FAAH gene polymorphism is not involved in these key drug action links in the mechanism of epilepsy resistance. On the other hand, it is also possible that our study sample is small and there is a certain result bias. Therefore, in the future, large-scale clinical studies can be conducted to collect NR3C1 and FAAH gene information and epilepsy drug treatment response data, and analyze the relationship between NR3C1 and FAAH gene polymorphisms and epilepsy drug response. In addition, the relationship of NR3C1 and FAAH with epileptic drug response can be comprehensively evaluated in combination with other biological indicators or genome interactions. Then explore the personalized epilepsy treatment strategy based on NR3C1 and FAAH. In addition, other external factors such as diet and environmental exposures may influence genetic expression and drug metabolism, potentially interacting with the polymorphisms studied. But they were not included in the current study. Thus, future studies should take into account external factors and observe the relationship between NR3C1 and FAAH genetic polymorphisms and epilepsy drug response after controlling for these factors.
|
| 210 |
+
|
| 211 |
+
Interestingly, the present results also demonstrated that both NR3C1 and FAAH polymorphisms showed no significant correlation with liver injury associated with ASM treatment. Antiepileptic drugs are mainly metabolized by cytochrome P450 enzyme series (CYP) or glucosylation reaction. The polymorphism of NR3C1 and FAAH genes may not affect the metabolic process of antiepileptic drugs in the liver, and thus have nothing to do with liver injury. Some current studies have found that probiotics can improve liver function in patients with epilepsy [[43](#CR43)43]. This may be because probiotics improve gut microbiota outcomes and reduce the production of inflammatory mediators, thereby reducing the burden of inflammation on the liver. Signaling molecules produced by probiotics may affect NR3C1 gene expression [[44](#CR44)44]. This regulatory effect of probiotics may vary in individuals with different polymorphisms of the NR3C1 gene. FAAH gene polymorphism may also affect the metabolism of probiotics. Different FAAH genotypes may lead to differences in the endocannabinoid system, which in turn affect the physiological function of the gut. In addition, NR3C1 gene polymorphism may affect the body's ability to regulate inflammation, and thus affect liver function [[45](#CR45)45]. Therefore, inflammatory responses may mask the potential effects of genetic polymorphisms on liver function. Therefore, exploring the role of systemic inflammation in the relationship between gene polymorphism and liver function is helpful to understand the influencing factors of liver function more comprehensively.
|
| 212 |
+
|
| 213 |
+
## Conclusion
|
| 214 |
+
|
| 215 |
+
In consequence, NR3C1 rs41423247 polymorphism may be related to the response to anti-epileptic drugs, and G allele and CG genotype carriers have better anti-epileptic efficacy. FAAH rs324420 polymorphism was not associated with drug response to epilepsy treatment. At the same time, NR3C1 rs41423247 and FAAH rs324420 polymorphisms were not associated with liver injury in epilepsy treatment. These findings provide new ideas and methods for personalized treatment of epilepsy.
|
| 216 |
+
|
| 217 |
+
## Acknowledgements
|
| 218 |
+
|
| 219 |
+
Not Applicable.
|
| 220 |
+
|
| 221 |
+
## Authors’ contributions
|
| 222 |
+
|
| 223 |
+
Q.L and N. L designed the research study. H.L. W, C. L, Q. L, N. L and H.L. Q performed the research and analyzed the data. Q.L and N. L wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
|
| 224 |
+
|
| 225 |
+
## Funding
|
| 226 |
+
|
| 227 |
+
No funding was received to assist with the preparation of this work.
|
| 228 |
+
|
| 229 |
+
## Data availability
|
| 230 |
+
|
| 231 |
+
Corresponding authors may provide data and materials.
|
| 232 |
+
|
| 233 |
+
## Declarations
|
| 234 |
+
|
| 235 |
+
### Ethics approval and consent to participate
|
| 236 |
+
|
| 237 |
+
The study protocol was approved by The Ethics Committee of Dongying People’s Hospital and followed the principles outlined in the Declaration of Helsinki. In addition, informed consent has been obtained from the participants involved.
|
| 238 |
+
|
| 239 |
+
### Consent for publication
|
| 240 |
+
|
| 241 |
+
Not applicable.
|
| 242 |
+
|
| 243 |
+
### Competing interests
|
| 244 |
+
|
| 245 |
+
The authors declare that they have no competing interests.
|
| 246 |
+
|
| 247 |
+
## Footnotes
|
| 248 |
+
|
| 249 |
+
## Contributor Information
|
| 250 |
+
|
| 251 |
+
Qian Li, Email: liqiandr@163.com.
|
| 252 |
+
|
| 253 |
+
Huiling Qin, Email: qinhuiling533000@163.com.
|
| 254 |
+
|
| 255 |
+
## Associated Data
|
| 256 |
+
|
| 257 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 258 |
+
|
| 259 |
+
### Data Availability Statement
|
| 260 |
+
|
| 261 |
+
Corresponding authors may provide data and materials.
|
| 262 |
+
|
| 263 |
+
### Data Availability Statement
|
| 264 |
+
|
| 265 |
+
Corresponding authors may provide data and materials.
|
| 266 |
+
|
| 267 |
+
## References
|
| 268 |
+
|
| 269 |
+
1. Žuvela T, Filipović-Grčić B, Rušić D, Leskur D, Modun D, Čohadžić T, et al. Knowledge and Attitudes towards Epilepsy of Croatian General Student Population and Biomedical Students: A Cross-Sectional Study. Healthcare (Basel, Switzerland). 2023;11(18):2550. Epub 2023/09/28. [DOI](https://doi.org/10.3390/healthcare11182550) | [PMC free article](/articles/PMC10531231/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37761747/)
|
| 270 |
+
|
| 271 |
+
2. Gyaase D, Gyaase TI, Tawiah R, Atta-Osei G, Owusu I, Mprah WK, et al. Perceived causes and management of epilepsy among rural community dwellers in Ghana: a qualitative synthesis. Front Neurol. 2023;14:1230336. Epub 2023/10/20. [DOI](https://doi.org/10.3389/fneur.2023.1230336) | [PMC free article](/articles/PMC10583556/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37859650/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Gyaase%20D,%20Gyaase%20TI,%20Tawiah%20R,%20Atta-Osei%20G,%20Owusu%20I,%20Mprah%20WK,%20et%20al.%20Perceived%20causes%20and%20management%20of%20epilepsy%20among%20rural%20community%20dwellers%20in%20Ghana:%20a%20qualitative%20synthesis.%20Front%20Neurol.%202023;14:1230336.%20Epub%202023/10/20.)
|
| 272 |
+
|
| 273 |
+
3. Bodor GS, Rands AJ. Quantitative LC-MS/MS Method for the Simultaneous Measurement of Six Antiepileptics and Pentobarbital in Human Serum. Methods Mol Biol (Clifton, NJ). 2024;2737:43–54. Epub 2023/12/01. [DOI](https://doi.org/10.1007/978-1-0716-3541-4_5) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38036809/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Bodor%20GS,%20Rands%20AJ.%20Quantitative%20LC-MS/MS%20Method%20for%20the%20Simultaneous%20Measurement%20of%20Six%20Antiepileptics%20and%20Pentobarbital%20in%20Human%20Serum.%20Methods%20Mol%20Biol%20(Clifton,%20NJ).%202024;2737:43%E2%80%9354.%20Epub%202023/12/01.)
|
| 274 |
+
|
| 275 |
+
4. Hoxhaj P, Habiya SK, Sayabugari R, Balaji R, Xavier R, Ahmad A, et al. Investigating the Impact of Epilepsy on Cognitive Function: A Narrative Review. Cureus. 2023;15(6):e41223 Epub 2023/08/01. [DOI](https://doi.org/10.7759/cureus.41223) | [PMC free article](/articles/PMC10387362/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37525802/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Hoxhaj%20P,%20Habiya%20SK,%20Sayabugari%20R,%20Balaji%20R,%20Xavier%20R,%20Ahmad%20A,%20et%20al.%20Investigating%20the%20Impact%20of%20Epilepsy%20on%20Cognitive%20Function:%20A%20Narrative%20Review.%20Cureus.%202023;15(6):e41223%20Epub%202023/08/01.)
|
| 276 |
+
|
| 277 |
+
5. Smolarz B, Makowska M, Romanowicz H. Pharmacogenetics of Drug-Resistant Epilepsy (review of literature). Int J Mol Sci. 2021;22(21):11696. Epub 2021/11/14. [DOI](https://doi.org/10.3390/ijms222111696) | [PMC free article](/articles/PMC8584095/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34769124/)
|
| 278 |
+
|
| 279 |
+
6. Monteagudo-Gimeno E, Sánchez-González R, Raduà-Castaño J, Fortea-González L, Boget-Llucià T, Carreño-Martínez M, et al. Association between depressive and anxious symptoms with cognitive function and quality of life in drug-resistant epilepsy. Heliyon. 2023;9(10):e20903 Epub 2023/10/27. [DOI](https://doi.org/10.1016/j.heliyon.2023.e20903) | [PMC free article](/articles/PMC10597766/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37886767/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Monteagudo-Gimeno%20E,%20S%C3%A1nchez-Gonz%C3%A1lez%20R,%20Radu%C3%A0-Casta%C3%B1o%20J,%20Fortea-Gonz%C3%A1lez%20L,%20Boget-Lluci%C3%A0%20T,%20Carre%C3%B1o-Mart%C3%ADnez%20M,%20et%20al.%20Association%20between%20depressive%20and%20anxious%20symptoms%20with%20cognitive%20function%20and%20quality%20of%20life%20in%20drug-resistant%20epilepsy.%20Heliyon.%202023;9(10):e20903%20Epub%202023/10/27.)
|
| 280 |
+
|
| 281 |
+
7. Shevlyakov AD, Kolesnikova TO, de Abreu MS, Petersen EV, Yenkoyan KB, Demin KA, et al. Forward genetics-based approaches to understanding the systems biology and molecular mechanisms of epilepsy. Int J Mol Sci. 2023;24(6):5280. Epub 2023/03/30. [DOI](https://doi.org/10.3390/ijms24065280) | [PMC free article](/articles/PMC10049737/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36982355/)
|
| 282 |
+
|
| 283 |
+
8. Shaheen U, Prasad DK, Sharma V, Suryaprabha T, Ahuja YR, Jyothy A, et al. Significance of MDR1 gene polymorphism C3435T in predicting drug response in epilepsy. Epilepsy Res. 2014;108(2):251–6. [DOI](https://doi.org/10.1016/j.eplepsyres.2013.11.009) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24300029/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Shaheen%20U,%20Prasad%20DK,%20Sharma%20V,%20Suryaprabha%20T,%20Ahuja%20YR,%20Jyothy%20A,%20et%20al.%20Significance%20of%20MDR1%20gene%20polymorphism%20C3435T%20in%20predicting%20drug%20response%20in%20epilepsy.%20Epilepsy%20Res.%202014;108(2):251%E2%80%936.)
|
| 284 |
+
|
| 285 |
+
9. Wang Q, Shi NR, Lv P, Liu J, Zhang JZ, Deng BL, et al. P2Y12 receptor gene polymorphisms are associated with epilepsy. Purinergic Signalling. 2023;19(1):155–62. Epub 2022/02/18. [DOI](https://doi.org/10.1007/s11302-022-09848-4) | [PMC free article](/articles/PMC9984642/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35175489/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Wang%20Q,%20Shi%20NR,%20Lv%20P,%20Liu%20J,%20Zhang%20JZ,%20Deng%20BL,%20et%20al.%20P2Y12%20receptor%20gene%20polymorphisms%20are%20associated%20with%20epilepsy.%20Purinergic%20Signalling.%202023;19(1):155%E2%80%9362.%20Epub%202022/02/18.)
|
| 286 |
+
|
| 287 |
+
10. Abdel-Rasol HA, Abdel Ghaffar H, Mohamed MS, Jad RW, Abelaleem OO, Abdelghaffar NK. A functional SNP in miR-146a and genetic susceptibility to drug-resistant epilepsy. Neurol Res. 2023;45(8):765–72. Epub 2023/05/05. [DOI](https://doi.org/10.1080/01616412.2023.2203617) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37142567/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Abdel-Rasol%20HA,%20Abdel%20Ghaffar%20H,%20Mohamed%20MS,%20Jad%20RW,%20Abelaleem%20OO,%20Abdelghaffar%20NK.%20A%20functional%20SNP%20in%20miR-146a%20and%20genetic%20susceptibility%20to%20drug-resistant%20epilepsy.%20Neurol%20Res.%202023;45(8):765%E2%80%9372.%20Epub%202023/05/05.)
|
| 288 |
+
|
| 289 |
+
11. Zhu J, Lu J, He Y, Shen X, Xia H, Li W, et al. Association of ABCB1 polymorphisms with efficacy and adverse drug reactions of valproic acid in children with epilepsy. Pharmaceuticals (Basel, Switzerland). 2023;16(11):1536. Epub 2023/11/25. [DOI](https://doi.org/10.3390/ph16111536) | [PMC free article](/articles/PMC10675623/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/38004402/)
|
| 290 |
+
|
| 291 |
+
12. Firouzabadi N, Asadi-Pooya AA, Alimoradi N, Simani L, Asadollahi M. Polymorphism of glucocorticoid receptor gene (rs41423247) in functional seizures (psychogenic nonepileptic seizures/attacks). Epilepsia Open. 2023;8(4):1425–31 Epub 2023/08/18. [DOI](https://doi.org/10.1002/epi4.12816) | [PMC free article](/articles/PMC10690659/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37593891/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Firouzabadi%20N,%20Asadi-Pooya%20AA,%20Alimoradi%20N,%20Simani%20L,%20Asadollahi%20M.%20Polymorphism%20of%20glucocorticoid%20receptor%20gene%20(rs41423247)%20in%20functional%20seizures%20(psychogenic%20nonepileptic%20seizures/attacks).%20Epilepsia%20Open.%202023;8(4):1425%E2%80%9331%20Epub%202023/08/18.)
|
| 292 |
+
|
| 293 |
+
13. Dincheva I, Drysdale AT, Hartley CA, Johnson DC, Jing D, King EC, et al. FAAH genetic variation enhances fronto-amygdala function in mouse and human. Nat Commun. 2015;6:6395. Epub 2015/03/04. [DOI](https://doi.org/10.1038/ncomms7395) | [PMC free article](/articles/PMC4351757/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25731744/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Dincheva%20I,%20Drysdale%20AT,%20Hartley%20CA,%20Johnson%20DC,%20Jing%20D,%20King%20EC,%20et%20al.%20FAAH%20genetic%20variation%20enhances%20fronto-amygdala%20function%20in%20mouse%20and%20human.%20Nat%20Commun.%202015;6:6395.%20Epub%202015/03/04.)
|
| 294 |
+
|
| 295 |
+
14. Colloca L, Wang Y, Martinez PE, Chang YC, Ryan KA, Hodgkinson C, et al. OPRM1 rs1799971, COMT rs4680, and FAAH rs324420 genes interact with placebo procedures to induce hypoalgesia. Pain. 2019;160(8):1824–34. Epub 2019/07/25. [DOI](https://doi.org/10.1097/j.pain.0000000000001578) | [PMC free article](/articles/PMC6668362/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31335650/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Colloca%20L,%20Wang%20Y,%20Martinez%20PE,%20Chang%20YC,%20Ryan%20KA,%20Hodgkinson%20C,%20et%20al.%20OPRM1%20rs1799971,%20COMT%20rs4680,%20and%20FAAH%20rs324420%20genes%20interact%20with%20placebo%20procedures%20to%20induce%20hypoalgesia.%20Pain.%202019;160(8):1824%E2%80%9334.%20Epub%202019/07/25.)
|
| 296 |
+
|
| 297 |
+
15. Anvar LH, Alejafar A, Moosavi SE, Charsouei S, Zeynalzadeh N, Fanid LM, et al. The study of rs324420 (C385A) polymorphism of the FAAH gene of the endocannabinoid system in patients with epilepsy and ADHD. Epilepsy Res. 2023;192: 107100. Epub 2023/04/06. [DOI](https://doi.org/10.1016/j.eplepsyres.2023.107100) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37018974/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Anvar%20LH,%20Alejafar%20A,%20Moosavi%20SE,%20Charsouei%20S,%20Zeynalzadeh%20N,%20Fanid%20LM,%20et%20al.%20The%20study%20of%20rs324420%20(C385A)%20polymorphism%20of%20the%20FAAH%20gene%20of%20the%20endocannabinoid%20system%20in%20patients%20with%20epilepsy%20and%20ADHD.%20Epilepsy%20Res.%202023;192:%20107100.%20Epub%202023/04/06.)
|
| 298 |
+
|
| 299 |
+
16. Zahra MA, Kamha ES, Abdelaziz HK, Nounou HA, Deeb HME. Aberrant Expression of Serum MicroRNA-153 and -199a in Generalized Epilepsy and its Correlation with Drug Resistance. Ann Neurosci. 2022;29(4):203–8. [DOI](https://doi.org/10.1177/09727531221077667) | [PMC free article](/articles/PMC10101161/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37064282/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Zahra%20MA,%20Kamha%20ES,%20Abdelaziz%20HK,%20Nounou%20HA,%20Deeb%20HME.%20Aberrant%20Expression%20of%20Serum%20MicroRNA-153%20and%20-199a%20in%20Generalized%20Epilepsy%20and%20its%20Correlation%20with%20Drug%20Resistance.%20Ann%20Neurosci.%202022;29(4):203%E2%80%938.)
|
| 300 |
+
|
| 301 |
+
17. Sterjev Z, Trencevska GK, Cvetkovska E, Petrov I, Kuzmanovski I, Ribarska JT, et al. The association of C3435T single-nucleotide polymorphism, Pgp-glycoprotein gene expression levels and carbamazepine maintenance dose in patients with epilepsy. Neuropsychiatr Dis Treat. 2012;8:191–6. [DOI](https://doi.org/10.2147/NDT.S28285) | [PMC free article](/articles/PMC3346059/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22570551/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Sterjev%20Z,%20Trencevska%20GK,%20Cvetkovska%20E,%20Petrov%20I,%20Kuzmanovski%20I,%20Ribarska%20JT,%20et%20al.%20The%20association%20of%20C3435T%20single-nucleotide%20polymorphism,%20Pgp-glycoprotein%20gene%20expression%20levels%20and%20carbamazepine%20maintenance%20dose%20in%20patients%20with%20epilepsy.%20Neuropsychiatr%20Dis%20Treat.%202012;8:191%E2%80%936.)
|
| 302 |
+
|
| 303 |
+
18. Huang D, Wen X, Lu C, Zhang B, Fu Z, Huang Y, et al. Investigating the molecular mechanism of Compound Danshen Dropping Pills for the treatment of epilepsy by utilizing network pharmacology and molecular docking technology. Ann Transl Med. 2022;10(4):216 Epub 2022/03/15. [DOI](https://doi.org/10.21037/atm-22-195) | [PMC free article](/articles/PMC8908140/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35280369/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Huang%20D,%20Wen%20X,%20Lu%20C,%20Zhang%20B,%20Fu%20Z,%20Huang%20Y,%20et%20al.%20Investigating%20the%20molecular%20mechanism%20of%20Compound%20Danshen%20Dropping%20Pills%20for%20the%20treatment%20of%20epilepsy%20by%20utilizing%20network%20pharmacology%20and%20molecular%20docking%20technology.%20Ann%20Transl%20Med.%202022;10(4):216%20Epub%202022/03/15.)
|
| 304 |
+
|
| 305 |
+
19. Luo X, Ruan Z, Liu L. Causal relationship between telomere length and epilepsy: A bidirectional Mendelian randomization study. Epilepsia Open. 2023;8(4):1432–9 Epub 2023/08/18. [DOI](https://doi.org/10.1002/epi4.12817) | [PMC free article](/articles/PMC10690705/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37593897/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Luo%20X,%20Ruan%20Z,%20Liu%20L.%20Causal%20relationship%20between%20telomere%20length%20and%20epilepsy:%20A%20bidirectional%20Mendelian%20randomization%20study.%20Epilepsia%20Open.%202023;8(4):1432%E2%80%939%20Epub%202023/08/18.)
|
| 306 |
+
|
| 307 |
+
20. van Hezik-Wester V, de Groot S, Kanters T, Versteegh M, Wagner L, Ardesch J, et al. Burden of illness in people with medically refractory epilepsy who suffer from daily to weekly seizures: 12-month follow-up of participants in the EPISODE study. Front Neurol. 2022;13:1012486. Epub 2022/11/18. [DOI](https://doi.org/10.3389/fneur.2022.1012486) | [PMC free article](/articles/PMC9650114/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36388190/) | [Google Scholar](https://scholar.google.com/scholar_lookup?van%20Hezik-Wester%20V,%20de%20Groot%20S,%20Kanters%20T,%20Versteegh%20M,%20Wagner%20L,%20Ardesch%20J,%20et%20al.%20Burden%20of%20illness%20in%20people%20with%20medically%20refractory%20epilepsy%20who%20suffer%20from%20daily%20to%20weekly%20seizures:%2012-month%20follow-up%20of%20participants%20in%20the%20EPISODE%20study.%20Front%20Neurol.%202022;13:1012486.%20Epub%202022/11/18.)
|
| 308 |
+
|
| 309 |
+
21. Acharya AR, Larsen LE, Delbeke J, Wadman WJ, Vonck K, Meurs A, et al. In vivo inhibition of epileptiform afterdischarges in rat hippocampus by light-activated chloride channel, stGtACR2. CNS Neurosci Ther. 2023;29(3):907–16. Epub 2022/12/10. [DOI](https://doi.org/10.1111/cns.14029) | [PMC free article](/articles/PMC9928558/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36482869/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Acharya%20AR,%20Larsen%20LE,%20Delbeke%20J,%20Wadman%20WJ,%20Vonck%20K,%20Meurs%20A,%20et%20al.%20In%20vivo%20inhibition%20of%20epileptiform%20afterdischarges%20in%20rat%20hippocampus%20by%20light-activated%20chloride%20channel,%20stGtACR2.%20CNS%20Neurosci%20Ther.%202023;29(3):907%E2%80%9316.%20Epub%202022/12/10.)
|
| 310 |
+
|
| 311 |
+
22. Baum L, Kwan P. Antiepileptic drug delivery. Adv Drug Deliv Rev. 2012;64(10):885–6. [DOI](https://doi.org/10.1016/j.addr.2012.04.007) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/22575859/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Baum%20L,%20Kwan%20P.%20Antiepileptic%20drug%20delivery.%20Adv%20Drug%20Deliv%20Rev.%202012;64(10):885%E2%80%936.)
|
| 312 |
+
|
| 313 |
+
23. Berseem NF, Khattab E, Saad DS, Abd Elnaby SA. Role of SCN2A c.56G/A Gene Polymorphism in Egyptian Children with Genetic Epilepsy with Febrile Seizure Plus. CNS & Neurol Disorders Drug Targets. 2022;21(5):450–7 Epub 2021/10/06. [DOI](https://doi.org/10.2174/1871527320666211004123731) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34607551/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Berseem%20NF,%20Khattab%20E,%20Saad%20DS,%20Abd%20Elnaby%20SA.%20Role%20of%20SCN2A%20c.56G/A%20Gene%20Polymorphism%20in%20Egyptian%20Children%20with%20Genetic%20Epilepsy%20with%20Febrile%20Seizure%20Plus.%20CNS%20&%20Neurol%20Disorders%20Drug%20Targets.%202022;21(5):450%E2%80%937%20Epub%202021/10/06.)
|
| 314 |
+
|
| 315 |
+
24. Mousavi SF, Hasanpour K, Nazarzadeh M, Adli A, Bazghandi MS, Asadi A, et al. ABCG2, SCN1A and CYP3A5 genes polymorphism and drug-resistant epilepsy in children: A case-control study. Seizure. 2022;97:58–62. Epub 2022/03/27. [DOI](https://doi.org/10.1016/j.seizure.2022.03.009) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35338956/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Mousavi%20SF,%20Hasanpour%20K,%20Nazarzadeh%20M,%20Adli%20A,%20Bazghandi%20MS,%20Asadi%20A,%20et%20al.%20ABCG2,%20SCN1A%20and%20CYP3A5%20genes%20polymorphism%20and%20drug-resistant%20epilepsy%20in%20children:%20A%20case-control%20study.%20Seizure.%202022;97:58%E2%80%9362.%20Epub%202022/03/27.)
|
| 316 |
+
|
| 317 |
+
25. Zhou Z, Wu S, Zou X, Gu S. Association between SCN1A polymorphism and risk of epilepsy in children: A systematic review and meta-analysis. Seizure. 2023;112:40–7. Epub 2023/09/24. [DOI](https://doi.org/10.1016/j.seizure.2023.09.012) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37741152/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Zhou%20Z,%20Wu%20S,%20Zou%20X,%20Gu%20S.%20Association%20between%20SCN1A%20polymorphism%20and%20risk%20of%20epilepsy%20in%20children:%20A%20systematic%20review%20and%20meta-analysis.%20Seizure.%202023;112:40%E2%80%937.%20Epub%202023/09/24.)
|
| 318 |
+
|
| 319 |
+
26. Lu J, Xia H, Li W, Shen X, Guo H, Zhang J, et al. Genetic Polymorphism of GABRG2 rs211037 is Associated with Drug Response and Adverse Drug Reactions to Valproic Acid in Chinese Southern Children with Epilepsy. Pharmacogenom Personalized Med. 2021;14:1141–50 Epub 2021/09/24. [DOI](https://doi.org/10.2147/PGPM.S329594) | [PMC free article](/articles/PMC8450188/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34552348/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Lu%20J,%20Xia%20H,%20Li%20W,%20Shen%20X,%20Guo%20H,%20Zhang%20J,%20et%20al.%20Genetic%20Polymorphism%20of%20GABRG2%20rs211037%20is%20Associated%20with%20Drug%20Response%20and%20Adverse%20Drug%20Reactions%20to%20Valproic%20Acid%20in%20Chinese%20Southern%20Children%20with%20Epilepsy.%20Pharmacogenom%20Personalized%20Med.%202021;14:1141%E2%80%9350%20Epub%202021/09/24.)
|
| 320 |
+
|
| 321 |
+
27. Wang S, Cai X, Liu S, Zhou Q, Wang T, Du S, et al. A novel BCL11A polymorphism influences gene expression, therapeutic response and epilepsy risk: A multicenter study. Front Mol Neurosci. 2022;15:1010101. Epub 2022/12/27. [DOI](https://doi.org/10.3389/fnmol.2022.1010101) | [PMC free article](/articles/PMC9780294/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36568279/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Wang%20S,%20Cai%20X,%20Liu%20S,%20Zhou%20Q,%20Wang%20T,%20Du%20S,%20et%20al.%20A%20novel%20BCL11A%20polymorphism%20influences%20gene%20expression,%20therapeutic%20response%20and%20epilepsy%20risk:%20A%20multicenter%20study.%20Front%20Mol%20Neurosci.%202022;15:1010101.%20Epub%202022/12/27.)
|
| 322 |
+
|
| 323 |
+
28. Wu C, Fang F, Zhan X, Wei Y. The association between glucocorticoid receptor (NR3C1) gene polymorphism and difficult-to-treat rhinosinusitis. Eur Arch Oto-Rhino-Laryngology. 2022;279(8):3981–7 Epub 2022/01/31. [DOI](https://doi.org/10.1007/s00405-021-07228-z) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35094121/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Wu%20C,%20Fang%20F,%20Zhan%20X,%20Wei%20Y.%20The%20association%20between%20glucocorticoid%20receptor%20(NR3C1)%20gene%20polymorphism%20and%20difficult-to-treat%20rhinosinusitis.%20Eur%20Arch%20Oto-Rhino-Laryngology.%202022;279(8):3981%E2%80%937%20Epub%202022/01/31.)
|
| 324 |
+
|
| 325 |
+
29. Wei D, Liu X, Huo W, Yu S, Li L, Wang C, et al. Serum cortisone and glucocorticoid receptor gene (NR3C1) polymorphism in human dysglycemia. Hormones (Athens). 2020;19(3):385–93. Epub 2020/04/19. [DOI](https://doi.org/10.1007/s42000-020-00196-9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32304041/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Wei%20D,%20Liu%20X,%20Huo%20W,%20Yu%20S,%20Li%20L,%20Wang%20C,%20et%20al.%20Serum%20cortisone%20and%20glucocorticoid%20receptor%20gene%20(NR3C1)%20polymorphism%20in%20human%20dysglycemia.%20Hormones%20(Athens).%202020;19(3):385%E2%80%9393.%20Epub%202020/04/19.)
|
| 326 |
+
|
| 327 |
+
30. Pac M, Krata N, Moszczuk B, Wyczałkowska-Tomasik A, Kaleta B, Foroncewicz B, et al. NR3C1 glucocorticoid receptor gene polymorphisms are associated with Membranous and IgA Nephropathies. Cells. 2021;10(11):3186. Epub 2021/11/28. [DOI](https://doi.org/10.3390/cells10113186) | [PMC free article](/articles/PMC8625873/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34831409/)
|
| 328 |
+
|
| 329 |
+
31. Bakusic J, Ghosh M, Polli A, Bekaert B, Schaufeli W, Claes S, et al. Role of NR3C1 and SLC6A4 methylation in the HPA axis regulation in burnout. J Affect Disord. 2021;295:505–12. [DOI](https://doi.org/10.1016/j.jad.2021.08.081) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34509065/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Bakusic%20J,%20Ghosh%20M,%20Polli%20A,%20Bekaert%20B,%20Schaufeli%20W,%20Claes%20S,%20et%20al.%20Role%20of%20NR3C1%20and%20SLC6A4%20methylation%20in%20the%20HPA%20axis%20regulation%20in%20burnout.%20J%20Affect%20Disord.%202021;295:505%E2%80%9312.)
|
| 330 |
+
|
| 331 |
+
32. Greco A, Rizzo MI, De Virgilio A, Conte M, Gallo A, Attanasio G, et al. Autoimmune epilepsy. Autoimmun Rev. 2016;15(3):221–5. [DOI](https://doi.org/10.1016/j.autrev.2015.11.007) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26626229/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Greco%20A,%20Rizzo%20MI,%20De%20Virgilio%20A,%20Conte%20M,%20Gallo%20A,%20Attanasio%20G,%20et%20al.%20Autoimmune%20epilepsy.%20Autoimmun%20Rev.%202016;15(3):221%E2%80%935.)
|
| 332 |
+
|
| 333 |
+
33. Vitaliti G, Pavone P, Guglielmo F, Spataro G, Falsaperla R. The immunomodulatory effect of probiotics beyond atopy: an update. J Asthma. 2014;51(3):320–32. [DOI](https://doi.org/10.3109/02770903.2013.862259) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24256057/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Vitaliti%20G,%20Pavone%20P,%20Guglielmo%20F,%20Spataro%20G,%20Falsaperla%20R.%20The%20immunomodulatory%20effect%20of%20probiotics%20beyond%20atopy:%20an%20update.%20J%20Asthma.%202014;51(3):320%E2%80%9332.)
|
| 334 |
+
|
| 335 |
+
34. Luo D, Gao X, Zhu X, Wu J, Yang Q, Xu Y, et al. Identification of steroid-induced osteonecrosis of the femoral head biomarkers based on immunization and animal experiments. BMC Musculoskelet Disord. 2024;25(1):596. [DOI](https://doi.org/10.1186/s12891-024-07707-4) | [PMC free article](/articles/PMC11285486/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/39069636/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Luo%20D,%20Gao%20X,%20Zhu%20X,%20Wu%20J,%20Yang%20Q,%20Xu%20Y,%20et%20al.%20Identification%20of%20steroid-induced%20osteonecrosis%20of%20the%20femoral%20head%20biomarkers%20based%20on%20immunization%20and%20animal%20experiments.%20BMC%20Musculoskelet%20Disord.%202024;25(1):596.)
|
| 336 |
+
|
| 337 |
+
35. Wadji DL, Tandon T, Ketcha Wanda GJM, Wicky C, Dentz A, Hasler G, et al. Child maltreatment and NR3C1 exon 1(F) methylation, link with deregulated hypothalamus-pituitary-adrenal axis and psychopathology: A systematic review. Child Abuse Negl. 2021;122: 105304. [DOI](https://doi.org/10.1016/j.chiabu.2021.105304) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34488052/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Wadji%20DL,%20Tandon%20T,%20Ketcha%20Wanda%20GJM,%20Wicky%20C,%20Dentz%20A,%20Hasler%20G,%20et%20al.%20Child%20maltreatment%20and%20NR3C1%20exon%201(F)%20methylation,%20link%20with%20deregulated%20hypothalamus-pituitary-adrenal%20axis%20and%20psychopathology:%20A%20systematic%20review.%20Child%20Abuse%20Negl.%202021;122:%20105304.)
|
| 338 |
+
|
| 339 |
+
36. Yue Q, Cai M, Xiao B, Zhan Q, Zeng C. The Microbiota-Gut-Brain Axis and Epilepsy. Cell Mol Neurobiol. 2022;42(2):439–53. [DOI](https://doi.org/10.1007/s10571-021-01130-2) | [PMC free article](/articles/PMC11441249/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34279746/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Yue%20Q,%20Cai%20M,%20Xiao%20B,%20Zhan%20Q,%20Zeng%20C.%20The%20Microbiota-Gut-Brain%20Axis%20and%20Epilepsy.%20Cell%20Mol%20Neurobiol.%202022;42(2):439%E2%80%9353.)
|
| 340 |
+
|
| 341 |
+
37. Zubareva OE, Dyomina AV, Kovalenko AA, Roginskaya AI, Melik-Kasumov TB, Korneeva MA, et al. Beneficial effects of probiotic bifidobacterium longum in a lithium-pilocarpine model of temporal lobe epilepsy in rats. Int J Mol Sci. 2023;24(9):8451. Epub 2023/05/05. [DOI](https://doi.org/10.3390/ijms24098451) | [PMC free article](/articles/PMC10179354/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37176158/)
|
| 342 |
+
|
| 343 |
+
38. Silva HH, Tavares V, Neto BV, Cerqueira F, Medeiros R, Silva MG. FAAH rs324420 polymorphism: biological pathways, impact on elite athletic performance and insights for sport medicine. Genes. 2023;14(10):1946. Epub 2023/10/28. [DOI](https://doi.org/10.3390/genes14101946) | [PMC free article](/articles/PMC10606937/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37895295/)
|
| 344 |
+
|
| 345 |
+
39. Spohrs J, Ulrich M, Grön G, Plener PL, Abler B. FAAH polymorphism (rs324420) modulates extinction recall in healthy humans: an fMRI study. Eur Arch Psychiatry Clin Neurosci. 2022;272(8):1495–504. Epub 2021/12/12. [DOI](https://doi.org/10.1007/s00406-021-01367-4) | [PMC free article](/articles/PMC9653364/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34893921/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Spohrs%20J,%20Ulrich%20M,%20Gr%C3%B6n%20G,%20Plener%20PL,%20Abler%20B.%20FAAH%20polymorphism%20(rs324420)%20modulates%20extinction%20recall%20in%20healthy%20humans:%20an%20fMRI%20study.%20Eur%20Arch%20Psychiatry%20Clin%20Neurosci.%202022;272(8):1495%E2%80%93504.%20Epub%202021/12/12.)
|
| 346 |
+
|
| 347 |
+
40. Zhang W, Liu H, Deng XD, Ma Y, Liu Y. FAAH levels and its genetic polymorphism association with susceptibility to methamphetamine dependence. Ann Hum Genet. 2020;84(3):259–70. Epub 2019/12/04. [DOI](https://doi.org/10.1111/ahg.12368) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31789429/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Zhang%20W,%20Liu%20H,%20Deng%20XD,%20Ma%20Y,%20Liu%20Y.%20FAAH%20levels%20and%20its%20genetic%20polymorphism%20association%20with%20susceptibility%20to%20methamphetamine%20dependence.%20Ann%20Hum%20Genet.%202020;84(3):259%E2%80%9370.%20Epub%202019/12/04.)
|
| 348 |
+
|
| 349 |
+
41. Ren SY, Wang ZZ, Zhang Y, Chen NH. Potential application of endocannabinoid system agents in neuropsychiatric and neurodegenerative diseases-focusing on FAAH/MAGL inhibitors. Acta Pharmacol Sin. 2020;41(10):1263–71. [DOI](https://doi.org/10.1038/s41401-020-0385-7) | [PMC free article](/articles/PMC7608191/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32203086/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Ren%20SY,%20Wang%20ZZ,%20Zhang%20Y,%20Chen%20NH.%20Potential%20application%20of%20endocannabinoid%20system%20agents%20in%20neuropsychiatric%20and%20neurodegenerative%20diseases-focusing%20on%20FAAH/MAGL%20inhibitors.%20Acta%20Pharmacol%20Sin.%202020;41(10):1263%E2%80%9371.)
|
| 350 |
+
|
| 351 |
+
42. Martinez-Aguirre C, Cinar R, Rocha L. Targeting Endocannabinoid System in Epilepsy: For Good or for Bad. Neuroscience. 2022;482:172–85. [DOI](https://doi.org/10.1016/j.neuroscience.2021.12.013) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/34923038/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Martinez-Aguirre%20C,%20Cinar%20R,%20Rocha%20L.%20Targeting%20Endocannabinoid%20System%20in%20Epilepsy:%20For%20Good%20or%20for%20Bad.%20Neuroscience.%202022;482:172%E2%80%9385.)
|
| 352 |
+
|
| 353 |
+
43. Mu C, Nikpoor N, Tompkins TA, Rho JM, Scantlebury MH, Shearer J. Probiotics counteract hepatic steatosis caused by ketogenic diet and upregulate AMPK signaling in a model of infantile epilepsy. EBioMedicine. 2022;76: 103838. [DOI](https://doi.org/10.1016/j.ebiom.2022.103838) | [PMC free article](/articles/PMC8882998/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/35148983/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Mu%20C,%20Nikpoor%20N,%20Tompkins%20TA,%20Rho%20JM,%20Scantlebury%20MH,%20Shearer%20J.%20Probiotics%20counteract%20hepatic%20steatosis%20caused%20by%20ketogenic%20diet%20and%20upregulate%20AMPK%20signaling%20in%20a%20model%20of%20infantile%20epilepsy.%20EBioMedicine.%202022;76:%20103838.)
|
| 354 |
+
|
| 355 |
+
44. Tian P, O’Riordan KJ, Lee YK, Wang G, Zhao J, Zhang H, et al. Towards a psychobiotic therapy for depression: Bifidobacterium breve CCFM1025 reverses chronic stress-induced depressive symptoms and gut microbial abnormalities in mice. Neurobiol Stress. 2020;12: 100216. [DOI](https://doi.org/10.1016/j.ynstr.2020.100216) | [PMC free article](/articles/PMC7109524/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32258258/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Tian%20P,%20O%E2%80%99Riordan%20KJ,%20Lee%20YK,%20Wang%20G,%20Zhao%20J,%20Zhang%20H,%20et%20al.%20Towards%20a%20psychobiotic%20therapy%20for%20depression:%20Bifidobacterium%20breve%20CCFM1025%20reverses%20chronic%20stress-induced%20depressive%20symptoms%20and%20gut%20microbial%20abnormalities%20in%20mice.%20Neurobiol%20Stress.%202020;12:%20100216.)
|
| 356 |
+
|
| 357 |
+
45. Newton R. Anti-inflammatory glucocorticoids: changing concepts. Eur J Pharmacol. 2014;724:231–6. [DOI](https://doi.org/10.1016/j.ejphar.2013.05.035) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23747654/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Newton%20R.%20Anti-inflammatory%20glucocorticoids:%20changing%20concepts.%20Eur%20J%20Pharmacol.%202014;724:231%E2%80%936.)
|
test/texts/PMC11852071.md
ADDED
|
@@ -0,0 +1,356 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Pharmacogenetics and Pharmacokinetics of Moxifloxacin in MDR-TB Patients in Indonesia: Analysis for ABCB1 and SLCO1B1
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Nurul Annisa, Nadiya N Afifah, Prayudi Santoso, Vycke Yunivita, Lindsey H M te Brake, Rob E Aarnoutse, Melisa I Barliana, Rovina Ruslami
|
| 5 |
+
**Journal:** Antibiotics
|
| 6 |
+
**Date:** 2025 Feb 16
|
| 7 |
+
**DOI:** [10.3390/antibiotics14020204](https://doi.org/10.3390/antibiotics14020204)
|
| 8 |
+
**PMID:** 40001447
|
| 9 |
+
**PMCID:** PMC11852071
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11852071/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11852071/pdf/antibiotics-14-00204.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11852071/pdf/antibiotics-14-00204.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
Background/Objectives: Studies show that SNPs in ABCB1 rs2032582 and SLCO1B1 rs4149015 affect the PK profile of moxifloxacin, a key drug for MDR-TB. This study aimed to assess the genotype frequencies of ABCB1 rs2032582 and SLCO1B1 rs4149015; describe moxifloxacin AUC0–24 and Cmax; and evaluate the association between genotype variations and moxifloxacin AUC0–24 and Cmax, corrected for the effect of other determinants in MDR-TB patients in Indonesia. Methods: The genotypes were identified using DNA sequencing. Plasma samples for PK analysis were collected at either two or four timepoints post-dose, at steady state. AUC0–24 values were assessed with a limited sampling formula. A multivariate linear regression analysis identified the determinants for moxifloxacin AUC0–24 and Cmax. Results: We recruited 204 MDR-TB patients for PG analysis, with 80 providing PK samples. The majority of the ABCB1 and SLCO1B1 genotypes were wildtype (GG), 41.7% and 93.6%, respectively. The geometric mean AUC0–24 for moxifloxacin was 78.6 mg·h/L and that for Cmax was 6.1 mg/L. No statistically significant difference in exposure to moxifloxacin could be shown between the genotypes. Sex, age, and dose in mg/kg/body weight were significant determinants of the AUC0–24 of moxifloxacin. Conclusions: The major genotype of ABCB1 rs2032582 and SLCO1B1 rs4149015 was wildtype, and the exposure to moxifloxacin was high but not related to the studied genotype in an Indonesian population.
|
| 16 |
+
|
| 17 |
+
Keywords: pharmacogenetics, pharmacokinetics, AUC0–24, Cmax, moxifloxacin, ABCB1 rs2032582, SLCO1B1 rs4149015, MDR-TB
|
| 18 |
+
|
| 19 |
+
**Keywords:**Keywords: pharmacogenetics, pharmacokinetics, AUC_0–24_0–24, C_max_max, moxifloxacin, *ABCB1*ABCB1 rs2032582, *SLCO1B1*SLCO1B1 rs4149015, MDR-TB
|
| 20 |
+
|
| 21 |
+
## 1. Introduction
|
| 22 |
+
|
| 23 |
+
Multidrug-resistant tuberculosis (MDR-TB) poses a significant challenge, necessitating the optimization of treatment strategies [[1](#B1-antibiotics-14-00204)1]. The fluoroquinolone antibiotics moxifloxacin and levofloxacin are crucial for treating MDR-TB and are designated as group A MDR-TB drugs. In so-called shorter (9–11 months) multi-drug MDR-TB treatment regimens, moxifloxacin or levofloxacin are part of the initial phase (4–6 months) as well as the continuation phase (5 months) of treatment [[2](#B2-antibiotics-14-00204)2]. Most recently, an even shorter (6 month) regimen was introduced that comprises bedaquiline, pretomanid, linezolid, and moxifloxacin (BPaLM) [[3](#B3-antibiotics-14-00204)3].
|
| 24 |
+
|
| 25 |
+
Moxifloxacin exerts its bactericidal effect on *Mycobacterium tuberculosis*Mycobacterium tuberculosis by inhibiting topoisomerase II (deoxyribonucleic acid (DNA)-gyrase) and topoisomerase IV, essential enzymes for bacterial DNA processes [[4](#B4-antibiotics-14-00204)4,[5](#B5-antibiotics-14-00204)5]. The efficacy of moxifloxacin relies on achieving a target area under the plasma concentration for the 0–24 h (AUC_0–24_0–24)/MIC ratio, in which AUC represents the total exposure to a drug in the plasma [[6](#B6-antibiotics-14-00204)6]. The total exposure to a drug can be influenced by various factors, including variations in genes involved in pharmacokinetic (PK) mechanisms. These variations include single-nucleotide polymorphisms (SNPs) in transporter genes. Indeed, ATP-binding cassette (ABC) proteins and solute carrier (SLC) transporters exhibit polymorphisms that impact the AUC_0–24_0–24 and C_max_max of moxifloxacin. Moxifloxacin is a substrate for p-glycoprotein (P-gp), a transporter encoded by the ATP-binding cassette subfamily B member 1 (*ABCB1)*ABCB1) gene. Studies have indicated that a polymorphism in *ABCB1*ABCB1 can lead to a decrease in the AUC_0–24_0–24 and an increase in the time to reach the maximum concentration (C_max_max) of moxifloxacin (t_max_max) [[7](#B7-antibiotics-14-00204)7]. Furthermore, research involving pulmonary TB patients from Africa and the USA revealed that a variant g.—11187G>A in solute carrier organic anion transporter family member 1B1 (*SLCO1B1*SLCO1B1) rs4149015 was associated with an increase in both the AUC_0–24_0–24 and C_max_max of moxifloxacin [[8](#B8-antibiotics-14-00204)8]. However, no studies on the association between pharmacogenetics (PG) and the PK of moxifloxacin have been conducted in an Indonesian population, particularly in treating MDR-TB, although Indonesia ranks number two for tuberculosis prevalence [[1](#B1-antibiotics-14-00204)1].
|
| 26 |
+
|
| 27 |
+
Therefore, the objectives of this study were (1) to describe the *ABCB1*ABCB1 rs2032582 and *SLCO1B1*SLCO1B1 rs4149015 genotype distribution in Indonesian MDR-TB patients; (2) to describe the AUC_0–24_0–24 and C_max_max of moxifloxacin in Indonesian MDR-TB patients; and (3) to assess the association between *ABCB1*ABCB1 rs2032582 and *SLCO1B1*SLCO1B1 rs4149015 SNPs and moxifloxacin AUC_0–24_0–24 and C_max_max, corrected for the effect of other predictors.
|
| 28 |
+
|
| 29 |
+
## 2. Results
|
| 30 |
+
|
| 31 |
+
From June 2020 to May 2022, 204 patients treated at the Hasan Sadikin Hospital in Bandung were recruited to participate in the PG study. Of these, 168 were eligible for inclusion in the PK study ([Figure 1](#antibiotics-14-00204-f001)Figure 1).
|
| 32 |
+
|
| 33 |
+
### Figure 1.
|
| 34 |
+
|
| 35 |
+

|
| 36 |
+
|
| 37 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11852071_antibiotics-14-00204-g001.jpg)
|
| 38 |
+
|
| 39 |
+
Patient tree.
|
| 40 |
+
|
| 41 |
+
Of the 168 eligible individuals, 83 were enrolled in the PK substudy. Three subjects were eliminated due to outlier data. The patients in the PK substudy were categorized into two groups, based on the daily dosage of moxifloxacin received (400 mg or 600 mg) based on their body weight.
|
| 42 |
+
|
| 43 |
+
The patient characteristics, data on drug regimens, and laboratory results of the patients are summarized in [Table 1](#antibiotics-14-00204-t001)Table 1a,b. The majority of patients were male with a median age of 39–40 years and predominantly an underweight status.
|
| 44 |
+
|
| 45 |
+
### Table 1.
|
| 46 |
+
|
| 47 |
+
(a). Patient characteristics. (b). Laboratory results and drug regimens of patients in the PK study.
|
| 48 |
+
|
| 49 |
+
| (a) |
|
| 50 |
+
| --- |
|
| 51 |
+
| Variable | PG Study(N = 204) | PK Study(N = 80) |
|
| 52 |
+
| Sex | | |
|
| 53 |
+
| Male | 150 (58.8) | 45 (56.3) |
|
| 54 |
+
| Age (years), median (min–max) | 39 (18–81) | 40 (18–66) |
|
| 55 |
+
| Body weight (kg), median (min–max) | 46.0 (29–84) | 44.5 (34–73) |
|
| 56 |
+
| BMI | | |
|
| 57 |
+
| Overweight | 9 (4.4) | 5 (6.3) |
|
| 58 |
+
| Normal | 84 (41.2) | 24 (30.0) |
|
| 59 |
+
| Underweight | 111 (54.4) | 51 (63.7) |
|
| 60 |
+
| Comorbidity | | |
|
| 61 |
+
| Diabetes mellitus | 44 (21.6) | 14 (17.5) |
|
| 62 |
+
| Hypertension | 14 (6.9) | 5 (6.3) |
|
| 63 |
+
| (b) |
|
| 64 |
+
| Variable | N = 80 |
|
| 65 |
+
| Drug regimen | |
|
| 66 |
+
| Injection containing | 11 (13.8) |
|
| 67 |
+
| All oral | 69 (86.2) |
|
| 68 |
+
| Moxifloxacin daily dose | |
|
| 69 |
+
| 400 mg | 41 (51.2) |
|
| 70 |
+
| 600 mg | 39 (48.8) |
|
| 71 |
+
| Dose/body weight (mg/kg), median (min–max) | 10.3 (8.2–14.0) |
|
| 72 |
+
| Time of taking medicine before sampling (hours), median (min–max) | 23 (11–27) |
|
| 73 |
+
| Number of days taking medicine, median (min–max) | 31 (14–126) |
|
| 74 |
+
| Taking with meal | |
|
| 75 |
+
| Yes | 33 (41.3) |
|
| 76 |
+
| Laboratory results | |
|
| 77 |
+
| Albumin (g/dL), median (min–max) | 3.4 (2.2–4.6) |
|
| 78 |
+
| Abnormal | 35 (43.7) |
|
| 79 |
+
| Normal (3.5–5 g/dL) | 45 (56.3) |
|
| 80 |
+
| ALT (U/L), median (min–max) | 25 (11–721) |
|
| 81 |
+
| Low level | 4 (5.3) |
|
| 82 |
+
| Normal level (14–59 U/L) | 63 (82.9) |
|
| 83 |
+
| High level | 9 (11.8) |
|
| 84 |
+
| eGFR (mL/min), median (min–max) | 110 (49–330) |
|
| 85 |
+
| eGFR criteria, n (%) | n = 76 |
|
| 86 |
+
| Abnormal | 1 (1.3) |
|
| 87 |
+
| Normal (>60 mL/min) | 75 (98.7) |
|
| 88 |
+
The data show that patients, on average, displayed normal laboratory parameter values, including liver and renal parameters.
|
| 89 |
+
|
| 90 |
+
The distribution of the genotype frequencies (n = 204) is summarized in [Table 2](#antibiotics-14-00204-t002)Table 2. For *ABCB1*ABCB1 rs2032582, the majority showed a wildtype GG (41.7%), while, for *SLCO1B1*SLCO1B1 rs4149015, the majority genotype was wildtype GG (93.6%).
|
| 91 |
+
|
| 92 |
+
### Table 2.
|
| 93 |
+
|
| 94 |
+
Genotype frequencies of ABCB1 rs2032582 and SLCO1B1 rs4149015.
|
| 95 |
+
|
| 96 |
+
| Variable | PG StudyN = 204 | PK StudyN = 80 |
|
| 97 |
+
| -------- | --------------- | -------------- |
|
| 98 |
+
| ABCB1 rs2032582, n (%) | | |
|
| 99 |
+
| Genotype | | |
|
| 100 |
+
| GG | 85 (41.7) | 38 (47.5) |
|
| 101 |
+
| GT | 77 (37.7) | 34 (42.5) |
|
| 102 |
+
| TT | 26 (12.7) | 4 (5.0) |
|
| 103 |
+
| GA | 11 (5.4) | 2 (2.5) |
|
| 104 |
+
| AT | 5 (2.5) | 2 (2.5) |
|
| 105 |
+
| SLCO1B1 rs4149015, n (%) | | |
|
| 106 |
+
| Genotype | | |
|
| 107 |
+
| GG | 191 (93.6) | 73 (91.3) |
|
| 108 |
+
| GA | 13 (6.4) | 7 (8.7) |
|
| 109 |
+
In patients with PK data (n = 80, [Table 3](#antibiotics-14-00204-t003)Table 3), the geometric mean of the moxifloxacin AUC_0–24_0–24 was 78.6 mg·h/L, while, for C_max_max, it was 6.2 mg/L.
|
| 110 |
+
|
| 111 |
+
### Table 3.
|
| 112 |
+
|
| 113 |
+
Moxifloxacin AUC0–24 and Cmax with determinants.
|
| 114 |
+
|
| 115 |
+
| Variable | n | AUC0–24 (mg·h/L) | p-Value | Cmax (mg/L) | p-Value |
|
| 116 |
+
| -------- | - | ---------------- | ------- | ----------- | ------- |
|
| 117 |
+
| All | 80 | 78.6 (12.0–656.8) | - | 6.1 (1.6–21.0) | - |
|
| 118 |
+
| Sex | | | | | |
|
| 119 |
+
| Male | 45 | 60.5 (12.0–387.6) | <0.001 * | 5.2 (41.6–21.0) | <0.001 * |
|
| 120 |
+
| Female | 35 | 110.0 (37.0–656.8) | | 7.7 (3.2–20.9) | |
|
| 121 |
+
| Age (year) | | | | | |
|
| 122 |
+
| 18–34 | 31 | 63.9 (12.0–339.4) | 0.049 * | 5.8 (1.6–20.9) | 0.359 |
|
| 123 |
+
| 35–49 | 30 | 78.0 (18.9–335.2) | | 5.9 (3.1–18.9) | |
|
| 124 |
+
| 50–66 | 19 | 111.5 (45.0–656.8) | | 7.2 (3.3–21.0) | |
|
| 125 |
+
| BMI | | | | | |
|
| 126 |
+
| Overweight | 5 | 84.1 (45.0–335.21) | 0.660 | 6.0 (3.3–18.9) | 0.815 |
|
| 127 |
+
| Normal | 24 | 88.1 (24.9–656.8) | | 6.5 (3.1–17.5) | |
|
| 128 |
+
| Underweight | 51 | 74.0 (12.0–387.6) | | 6.0 (1.6–21.0) | |
|
| 129 |
+
| Intake with meal | | | | | |
|
| 130 |
+
| Yes | 47 | 87.3 (12.0–387.6) | 0.154 | 6.7 (1.6–21.0) | 0.131 |
|
| 131 |
+
| No | 33 | 67.7 (17.4–656.8) | | 5.5 (2.8–17.5) | |
|
| 132 |
+
| Comorbidity | | | | | |
|
| 133 |
+
| DM status | | | | | |
|
| 134 |
+
| Yes | 14 | 113.2 (45.0–263.6) | 0.054 | 6.8 (3.3–15.4) | 0.457 |
|
| 135 |
+
| No | 66 | 72.7 (12.0–656.8) | | 6.0 (1.6–21.0) | |
|
| 136 |
+
| Hypertension | | | | | |
|
| 137 |
+
| Yes | 5 | 87.0 (46.4–656.8) | 0.765 | 5.2 (3.2–17.5) | 0.502 |
|
| 138 |
+
| No | 75 | 78.0 (12.0–387.6) | | 6.2 (1.6–21.0) | |
|
| 139 |
+
| Moxifloxacin dose | | | | | |
|
| 140 |
+
| 400 mg | 41 | 71.4 (17.4–339.4) | 0.262 | 5.7 (2.6–20.9) | 0.274 |
|
| 141 |
+
| 600 mg | 39 | 86.9 (12.0–656.8) | | 6.6 (1.6–21.0) | |
|
| 142 |
+
| Drug regimen | | | | | |
|
| 143 |
+
| Injection containing | 11 | 67.7 (37.0–108.7) | 0.501 | 4.4 (3.1–6.5) | 0.030 * |
|
| 144 |
+
| All oral | 69 | 80.5 (12.0–656.8) | | 6.5 (1.6–21.0) | |
|
| 145 |
+
| Dose per body weight ** | 80 | R = 0.3 | 0.004 * | R = 0.361 | 0.001 * |
|
| 146 |
+
Sex demonstrated significant associations with AUC_0–24_0–24 and C_max_max. Female patients showed higher exposure to moxifloxacin, either AUC_0–24_0–24 (110.0 vs. 60.5 mg·h/L, *p*p < 0.001) or C_max_max (7.7 vs. 5.2 mg/L, *p*p < 0.001) than male patients, while patients with a higher age showed higher AUC_0–24_0–24 values (*p*p = 0.049; see [Table 3](#antibiotics-14-00204-t003)Table 3). In addition, a significant difference was observed in moxifloxacin C_max_max between patients on all-oral regimens and those on a regimen with a kanamycin injection (6.5 vs. 4.4 mg/L, *p*p = 0.030). Both the AUC_0–24_0–24 and the C_max_max of moxifloxacin were also correlated with dose per body weight (r = 0.292, *p*p = 0.004 and r = 0.361, *p*p = 0.001).
|
| 147 |
+
|
| 148 |
+
The effect of *ABCB1*ABCB1 rs2032582 and *SLCO1B1*SLCO1B1 rs4149015 genotype variation on moxifloxacin AUC_0–24_0–24 and C_max_max is shown in [Table 4](#antibiotics-14-00204-t004)Table 4 and [Figure 2](#antibiotics-14-00204-f002)Figure 2. No statistically significant difference in exposure to moxifloxacin could be shown between the genotypes, although the geometric mean AUC_0–24_0–24 and C_max_max were lower in the TT genotype in *ABCB1*ABCB1 rs2032582 than that in the other genotypes, while the geometric mean AUC_0–24_0–24 (128.2 h.mg/L) and C_max_max (8.4 mg/L) were higher in the GA genotype of *SLCO1B1*SLCO1B1 rs4149015 ([Table 4](#antibiotics-14-00204-t004)Table 4).
|
| 149 |
+
|
| 150 |
+
### Table 4.
|
| 151 |
+
|
| 152 |
+
The effect of ABCB1 rs2032582 and SLCO1B1 rs4149015 gene polymorphisms on the moxifloxacin AUC0–24 and Cmax.
|
| 153 |
+
|
| 154 |
+
| Variable | n | AUC0–24 (mg·h/L) | p-Value | Cmax (mg/L) | p-Value |
|
| 155 |
+
| -------- | - | ---------------- | ------- | ----------- | ------- |
|
| 156 |
+
| Geometric Mean (Min–Max) | Geometric Mean (Min–Max) | | | | |
|
| 157 |
+
| ABCB1 rs2032582 | | | | | |
|
| 158 |
+
| GG | 38 | 79.5 (12.0–656.8) | 0.883 | 6.1 (2.9–17.5) | 0.651 |
|
| 159 |
+
| GT | 34 | 80.5 (17.4–387.6) | | 6.5 (1.6–21.0) | |
|
| 160 |
+
| TT | 4 | 53.3 (37.0–77.6) | | 4.3 (3.1–7.7) | |
|
| 161 |
+
| GA | 2 | 72.2 (57.0–91.4) | | 4.9 (4.1–5.9) | |
|
| 162 |
+
| AT | 2 | 97.6 (28.0–339.4) | | 7.4 (2.6–20.9) | |
|
| 163 |
+
| SLCO1B1 rs4149015 | | | | | |
|
| 164 |
+
| GG | 73 | 75.0 (12.0–656.8) | 0.083 | 6.0 (1.6–21.0) | 0.064 |
|
| 165 |
+
| GA | 7 | 128.2 (47.2–339.4) | | 8.4 (3.1–20.9) | |
|
| 166 |
+
### Figure 2.
|
| 167 |
+
|
| 168 |
+

|
| 169 |
+
|
| 170 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11852071_antibiotics-14-00204-g002.jpg)
|
| 171 |
+
|
| 172 |
+
(a) Boxplot ABCB1 rs2032582 genotype with moxifloxacin geometric mean AUC0–24; (b) boxplot ABCB1 rs2032582 genotype with moxifloxacin geometric mean Cmax.
|
| 173 |
+
|
| 174 |
+
[Figure 2](#antibiotics-14-00204-f002)Figure 2 depicts a boxplot comparing the *ABCB1*ABCB1 rs2032582 genotypes (GG, GT, TT, GA, and AT) with the moxifloxacin geometric mean AUC_0–24_0–24 and C_max_max values.
|
| 175 |
+
|
| 176 |
+
[Figure 3](#antibiotics-14-00204-f003)Figure 3 depicts a boxplot comparing the *SLCO1B1*SLCO1B1 rs4149015 genotypes (GG and GA) with the moxifloxacin geometric mean AUC_0–24_0–24 and C_max_max values.
|
| 177 |
+
|
| 178 |
+
### Figure 3.
|
| 179 |
+
|
| 180 |
+

|
| 181 |
+
|
| 182 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=11852071_antibiotics-14-00204-g003.jpg)
|
| 183 |
+
|
| 184 |
+
(a) Boxplot SLCO1B1 rs4149015 genotype with moxifloxacin geometric mean AUC0–24; (b) boxplot SLCO1B1 rs4149015 genotype with moxifloxacin geometric mean Cmax.
|
| 185 |
+
|
| 186 |
+
The subsequent multiple linear regression analysis revealed that sex and dose in mg/kg predicted the AUC_0–24_0–24 and C_max_max of moxifloxacin, whereas age also affected the AUC_0–24_0–24 ([Table 5](#antibiotics-14-00204-t005)Table 5).
|
| 187 |
+
|
| 188 |
+
### Table 5.
|
| 189 |
+
|
| 190 |
+
Predictors of moxifloxacin AUC0–24 and Cmax.
|
| 191 |
+
|
| 192 |
+
| Variable | Unstandardized Coefficients | Standardized Coefficients | p-Value |
|
| 193 |
+
| -------- | --------------------------- | ------------------------- | ------- |
|
| 194 |
+
| B | Std. Error | Beta | |
|
| 195 |
+
| Moxifloxacin AUC0–24 | | | | |
|
| 196 |
+
| (Constant) | 0.927 | 0.274 | | 0.001 * |
|
| 197 |
+
| Female sex | 0.221 | 0.071 | 0.324 | 0.003 * |
|
| 198 |
+
| Age | 0.007 | 0.003 | 0.278 | 0.014 * |
|
| 199 |
+
| Dose per body weight (mg/kg) | 0.052 | 0.024 | 0.221 | 0.033 * |
|
| 200 |
+
| Dependent variable: Log AUC0–24 moxifloxacin, R2 = 0.282Moxifloxacin Cmax | | | |
|
| 201 |
+
| (Constant) | −0.034 | 0.235 | | 0.887 |
|
| 202 |
+
| Female sex | 0.125 | 0.052 | 0.256 | 0.020 * |
|
| 203 |
+
| Dose per body weight (mg/kg) | 0.040 | 0.018 | 0.236 | 0.027 * |
|
| 204 |
+
| Dependent variable: Log Cmax, R2 = 0.180 | | | |
|
| 205 |
+
## 3. Discussion
|
| 206 |
+
|
| 207 |
+
This is the first study on a PG analysis concerning the PK of moxifloxacin within the Indonesian population. The first objective of this study was to assess the genotypes of *ABCB1*ABCB1 rs2032582 and *SLCO1B1*SLCO1B1 rs4149015 in MDR-TB patients in Indonesia (PG study). Secondly, the moxifloxacin AUC_0–24_0–24 and C_max_max were described in eligible participants (PK study). The third objective was to evaluate the association between genotype variants and the AUC_0–24_0–24 and C_max_max of moxifloxacin.
|
| 208 |
+
|
| 209 |
+
Within the dataset of the PG substudy, the most frequently observed *ABCB1*ABCB1 rs2032582 genotypes were GG, GT, TT, GA, and AT, with the major genotype being the wildtype GG. These findings align with genotype patterns identified in the Chinese Han population, characterized by GG, GT, and TT [[9](#B9-antibiotics-14-00204)9]. Additionally, the genotype variations observed in our study were consistent with the results of another study conducted in the Polish population [[10](#B10-antibiotics-14-00204)10]. Meanwhile, other studies within a South African population showed a predominantly CC genotype [[11](#B11-antibiotics-14-00204)11]. As for *SLCO1B1*SLCO1B1 rs4149015, genotype GG was shown to be the major genotype, which is also the wildtype, with GA as a variant. This is consistent with another study conducted in white and black populations, which revealed the same genotype distribution and frequency [[8](#B8-antibiotics-14-00204)8].
|
| 210 |
+
|
| 211 |
+
When only considering the moxifloxacin dose of 400 mg once daily, the exposure to moxifloxacin in MDR-TB patients in this study was higher than that in drug-sensitive pulmonary TB patients [[12](#B12-antibiotics-14-00204)12] or TB meningitis patients [[13](#B13-antibiotics-14-00204)13] from the same Indonesian setting. The AUC_0–24_0–24 values were 71.4, 48.2, and 33.6, while those for C_max_max were 5.7, 4.7, and 7.4, respectively. The AUC_0–24_0–24 of moxifloxacin in the MDR-TB patients in this study was also higher than that of the TB patients in another heterogeneous population (78.6 vs. 33.6 mg·h/L) [[14](#B14-antibiotics-14-00204)14]. A higher exposure to 400 mg of moxifloxacin in this study may partly be explained by the (absent) effect of rifampicin on the metabolism of moxifloxacin [[12](#B12-antibiotics-14-00204)12]. Furthermore, considering both the 400 and 600 mg moxifloxacin doses, it should be considered that over 60% of patients in our study had a low body weight, and the average moxifloxacin dose received was high at 10.3 mg/kg of body weight ([Table 1](#antibiotics-14-00204-t001)Table 1b).
|
| 212 |
+
|
| 213 |
+
This study found no association between genotype variations in *ABCB1*ABCB1 and *SLCO1B1*SLCO1B1 and the AUC_0–24_0–24 and C_max_max of moxifloxacin. Still, the analysis on *SLCO1B1*SLCO1B1 rs4149015 revealed a trend that patients with the GA genotype exhibited a higher moxifloxacin AUC_0–24_0–24 and C_max_max than those with the GG genotype ([Table 4](#antibiotics-14-00204-t004)Table 4). This result is consistent with a previous study conducted in Africa and the United States. In the study of *SLCO1B1*SLCO1B1 g.11187, the variant genotype AG had a median moxifloxacin AUC_0–24_0–24 that was 46% higher and a median C_max_max that was 30% higher than that for the GG genotype (wildtype) [[8](#B8-antibiotics-14-00204)8].
|
| 214 |
+
|
| 215 |
+
In this study, a multiple linear regression analysis revealed that sex, age, and dose/kg body weight were significant predictors for moxifloxacin AUC_0–24_0–24, while sex and dose per body weight were predictors for C_max_max. One possible reason for the sex-related differences in the PK might be due to variations in the regulation of drug metabolism through endogenous hormonal influences. It may involve a combination of genetics and physiological factors [[15](#B15-antibiotics-14-00204)15]. The differential tissue expression of P-gp between the genders has been reported as a significant contributor to gender differences in both the PK and pharmacodynamic (PD) response observed between the genders for many of its drug substrates [[16](#B16-antibiotics-14-00204)16]. As for organic anion transporter polypeptide (OATP) expression (encoded by the *SLCO1B1*SLCO1B1 gene) in age and gender, these variations can influence drug disposition and efficacy and may be the basis for drug interactions, especially in children and the elderly [[17](#B17-antibiotics-14-00204)17]. Another study noted a higher exposure to moxifloxacin in elderly females than that in elderly men. However, no significant differences were found when normalized to body weight, and no serious adverse effects were associated [[18](#B18-antibiotics-14-00204)18]. Age or gender had no effect on the bioavailability of levofloxacin, another fluoroquinolone, as reported by another study [[19](#B19-antibiotics-14-00204)19].
|
| 216 |
+
|
| 217 |
+
In addition to *ABCB1*ABCB1 and *SLCO1B1*SLCO1B1, other genes such as uridine 5′-diphosphate-glucuronosyltransferase family 1 member A1 (*UGT1A1*UGT1A1) and member A9 (*UGT1A9*UGT1A9) also influence the PK parameters of moxifloxacin, but they were not analyzed in this study [[7](#B7-antibiotics-14-00204)7,[11](#B11-antibiotics-14-00204)11,[20](#B20-antibiotics-14-00204)20], which is a notable limitation. Another limitation is the relatively small sample size in our PG and PK studies. A larger sample size is essential for a more comprehensive understanding of the relationship between genotype variations and moxifloxacin PK in Indonesian MDR-TB patients.
|
| 218 |
+
|
| 219 |
+
## 4. Materials and Methods
|
| 220 |
+
|
| 221 |
+
### 4.1. Patients and Study Design
|
| 222 |
+
|
| 223 |
+
This study was part of the MDR-TB cohort study conducted between 2020 and 2022 at the Hasan Sadikin Hospital, Bandung, a referral hospital for the West Java province in Indonesia. The diagnosis of MDR-TB was based on a rapid molecular test (Xpert^®^® MTB-RIF Assay G4) for the identification of rifampicin resistance in *M. tuberculosis*M. tuberculosis from sputum samples. The MDR-TB therapeutic regimen to be given by the clinician was based on the line probe assay (LPA) results. LPA is a DNA strip-based rapid test designed to determine the drug resistance profile by detecting the most common mutations associated with resistance to first- and second-line anti-TB agents, as well as specific *M. tuberculosis*M. tuberculosis wildtype DNA sequences.
|
| 224 |
+
|
| 225 |
+
All the MDR-TB patients were sampled for the PG study. Adult (>18 years old) MDR-TB patients who received a short-course (9–11 months) regimen containing moxifloxacin were recruited for the PK study. Pregnant and breastfeeding females were excluded from this PK study. This study was approved by the Research Ethical Committee of Universitas Padjadjaran (No. 643/UN6.KEP/EC/2020). Eligible patients were asked for written informed consent.
|
| 226 |
+
|
| 227 |
+
### 4.2. PG and PK Study
|
| 228 |
+
|
| 229 |
+
Samples for the PG study were taken from the leftovers during routine laboratory examinations at baseline by taking the patient’s whole blood. The DNA-isolation process involved the use of the Gene Elute™ Mammalian Genomic DNA Miniprep kit (Sigma Aldrich, Merck, Darmstadt, Germany, catalog number G1N10). Subsequently, the isolated DNA was quantified with a spectrophotometer and stored at −20 °C until further analysis. Gene sequences were obtained from the National Center for Biotechnology Information ([https://www.ncbi.nlm.nih.gov](https://www.ncbi.nlm.nih.gov)https://www.ncbi.nlm.nih.gov) (accessed on 16 September 2021). For *ABCB1*ABCB1 rs2032582, the specific primers used were as follows: forward primer for *ABCB1*ABCB1 rs2032582: 5′-GAGCATAGTAAGCAGTAGGGAGT-3′ and reverse primer: 5′-GCAGGCTATAGGTTCCAGGC-3′; forward primer for *SLCO1B1*SLCO1B1: 5′-GGCCTTGGGTCTACATTTCTCA-3′ and reverse primer: 5′-AGTACAGACCCTTCTCTCACA-3′ (Macrogen, Singapore, Singapore). A polymerase chain reaction (PCR) was carried out using the GoTaq™ Green Master Mix (Promega, Madison, WI, USA). The PCR conditions included denaturation at 95 °C for 2 min, annealing at 61 °C for *ABCB1*ABCB1 rs2032582 and 59 °C for *SLCO1B1*SLCO1B1 rs4149015, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min, with a total of 30 cycles. The PCR products underwent 2% agarose gel electrophoresis and were visualized at a 312 nm wavelength using a Biometra instrument. The expected amplicon size was 298 bp for *ABCB1*ABCB1 rs2032582 and 365 bp for *SLCO1B1*SLCO1B1 rs4149015. Genotyping was further confirmed through a DNA sequencing/capillary electrophoresis method.
|
| 230 |
+
|
| 231 |
+
PK sampling. PK sampling for moxifloxacin was conducted at the patient’s scheduled follow-up, typically one month after initiating drug treatment, when a steady state for moxifloxacin can be expected. Serial blood samples (2 mL each) were collected to assess the plasma concentrations of moxifloxacin at specific time points (at 0, 2, 4, and 6 h post-dose). If this was not possible (because patients could not stay longer at the clinic), the blood samples were collected at 0 and 2 h post-dose. Blood sampling was performed using a single insertion of a venous catheter. The blood samples were centrifuged at 3000 rpm for 15 min, and plasma was separated and stored at −80 °C within 30 min after sampling.
|
| 232 |
+
|
| 233 |
+
Bioanalysis of PK samples. The plasma concentrations of moxifloxacin were measured using high-performance liquid chromatography (HPLC) with UV detection. The mobile phase consisted of triethanolamine (TEA) (0.4%) in Milli-Q^®^® water (pH ±3) and acetonitrile 100% with an eluent ratio of 75:25%. A Sunfire™ Column C-18 (4.6 × 100 mm, 5 µL: Waters™: Ireland) served as the stationary phase, and detection was achieved using a Waters detector 2998 photodiode array (PDA) at 296 nm. The calibration range was 0.20–10.2 mg/L for human plasma. The intra-day and inter-day imprecision, expressed as the coefficient of variation (%CV), were shown to be lower than 7.2% and 5.0%, respectively, at all the concentrations tested, and the accuracies were between 95.5% and 103.4%.
|
| 234 |
+
|
| 235 |
+
PK data analysis. The AUC_0–24_0–24 of moxifloxacin was calculated using the limited sampling formula developed by Magis et al. [[14](#B14-antibiotics-14-00204)14]. The formula to calculate AUC_0–24_0–24 was [AUC_0–24_0–24 = −4.35 + (3.97 × C2) − (6.49 × C4) + (20.05 × C6)] for subjects who had concentration data at 0, 2, 4, and 6 h after drug administration [[14](#B14-antibiotics-14-00204)14]. For subjects who had concentration data at 0 and 2 h post-dose, moxifloxacin exposures were calculated using the formula: AUC_0–24_0–24 = [5.056 + (31.687 × C0) + (4.413 × C2)]. The C_max_max value was the highest measured concentration in a patient.
|
| 236 |
+
|
| 237 |
+
Statistical analysis. The patient characteristics in the PG and PK studies as well as the genotype distributions in the PG study were presented descriptively. Moxifloxacin AUC_0–24_0–24 and C_max_max values obtained in the PK study were presented as geometric mean and minimum–maximum values. Differences in the AUC_0–24_0–24 and C_max_max values between genotypes or between patient subgroups based on age, sex, body mass index (BMI), comorbidity, intake of drugs with or without food, moxifloxacin dose, and drug regimen were assessed using an unpaired t-test for 2 groups or a one-way ANOVA in the case of more than 2 groups, after log-transformation of the PK data. The moxifloxacin AUC_0–24_0–24 and C_max_max were correlated with dose in mg/kg by using Spearman rank correlation.
|
| 238 |
+
|
| 239 |
+
After these univariate analyses, possible predictors with *p*p < 0.20 were included in a multiple linear regression to identify moxifloxacin AUC_0–24_0–24 and C_max_max predictors. Statistical significance was achieved with *p*p < 0.05. All the statistical analyses were performed in SPSS software version 22. The graphs were generated using GraphPad Prism software (version 9.2.0; GraphPad Software, San Diego, CA, USA).
|
| 240 |
+
|
| 241 |
+
## 5. Conclusions
|
| 242 |
+
|
| 243 |
+
In conclusion, our observation found that the major genotype of *ABCB1*ABCB1 rs2032582 and *SLCO1B1*SLCO1B1 rs4149015 was GG. The average AUC_0–24_0–24 and C_max_max values of moxifloxacin were high at 78.6 mg·h/L and 6.1 mg/L, respectively. There was no association between genotype frequencies with AUC_0–24_0–24 and C_max_max. Multivariate analysis revealed that sex, age, and dose per body weight were significant determinants for AUC_0–24_0–24, while sex and dose per body weight were predictors for C_max_max.
|
| 244 |
+
|
| 245 |
+
## Acknowledgments
|
| 246 |
+
|
| 247 |
+
We acknowledge the support from Claudia Selvyanti, Septiwi Rizkayani, Rhea V. Nugraha, Agnesya Gunawan, Irawati N. Hidayah, and Zakiyyatul Aimmah (ALG MDR-TB field team); Harold Atmaja, Triana Nurul Meirina, and Atu Purnama Dewi (Pharmacokinetics Laboratory, Faculty of Medicine Universitas Padjadjaran); Hanny Nugrahani (Biotechnology Laboratory, Faculty of Pharmacy Universitas Padjadjaran); and Evan Susandi (Statistician, Faculty of Medicine Universitas Padjadjaran).
|
| 248 |
+
|
| 249 |
+
## Abbreviations
|
| 250 |
+
|
| 251 |
+
| MDR-TB | Multidrug-resistant tuberculosis |
|
| 252 |
+
| ------ | -------------------------------- |
|
| 253 |
+
| PK | Pharmacokinetics |
|
| 254 |
+
| PG | Pharmacogenetics |
|
| 255 |
+
| BPaLM | Bedaquiline, pretomanid, linezolid, and moxifloxacin |
|
| 256 |
+
| DNA | Deoxyribonucleic acid |
|
| 257 |
+
| SNPs | Single-nucleotide polymorphisms |
|
| 258 |
+
| ABC | ATP-binding cassette |
|
| 259 |
+
| SLC | Solute carrier |
|
| 260 |
+
| P-gp | p-Glycoprotein |
|
| 261 |
+
| Cmax | The maximum concentration |
|
| 262 |
+
| Tmax | The time to reach the maximum concentration |
|
| 263 |
+
| AUC0–24 | Area under the plasma concentration |
|
| 264 |
+
| ABCB1 | ATP-Binding Cassette Subfamily B Member 1 |
|
| 265 |
+
| SLCO1B1 | Solute Carrier Organic Anion Transporter Family Member 1B1 |
|
| 266 |
+
| UGT1A1 | Uridine 5′-diphosphate-glucuronosyltransferase family 1 member A1 |
|
| 267 |
+
| LPA | Line probe assay |
|
| 268 |
+
| PCR | Polymerase chain reaction |
|
| 269 |
+
| HPLC | High-performance liquid chromatography |
|
| 270 |
+
| TEA | Triethanolamine |
|
| 271 |
+
| PDA | Photodiode array |
|
| 272 |
+
| CV | Coefficient of variation |
|
| 273 |
+
| BMI | Body mass index |
|
| 274 |
+
| ALT | Alanine aminotransferase |
|
| 275 |
+
| eGFR | Estimated glomerular filtration rate |
|
| 276 |
+
| PD | Pharmacodynamic |
|
| 277 |
+
| OATPs | Organic anion transporter polypeptides |
|
| 278 |
+
## Author Contributions
|
| 279 |
+
|
| 280 |
+
N.A. and M.I.B. performed the PG analyses. P.S. and V.Y. led the field team in sampling and were responsible for clinical data. R.R. and V.Y. were responsible for PK measurements. N.A., V.Y., L.H.M.t.B., R.E.A. and R.R. were responsible for PK data analysis. N.A. performed the statistical analysis. N.A., N.N.A. and M.I.B. wrote the first complete draft of the report. R.R., R.E.A. and M.I.B. supervised this study. All authors provided contributions and suggestions. All authors have read and agreed to the published version of the manuscript.
|
| 281 |
+
|
| 282 |
+
## Institutional Review Board Statement
|
| 283 |
+
|
| 284 |
+
This study was conducted in accordance with the Declaration of Helsinki and approved by the Research Ethics Committee of Universitas Padjadjaran (No. 643/UN6.KEP/EC/2020; 21 July 2020).
|
| 285 |
+
|
| 286 |
+
## Informed Consent Statement
|
| 287 |
+
|
| 288 |
+
Informed consent was obtained from all subjects involved in this study.
|
| 289 |
+
|
| 290 |
+
## Data Availability Statement
|
| 291 |
+
|
| 292 |
+
Dataset available on request from the authors.
|
| 293 |
+
|
| 294 |
+
## Conflicts of Interest
|
| 295 |
+
|
| 296 |
+
The authors declare no conflicts of interest.
|
| 297 |
+
|
| 298 |
+
## Funding Statement
|
| 299 |
+
|
| 300 |
+
This research was funded partially by the Indonesian Ministry of Education, Culture, Research, and Technology for MIB (grant number: 1827/UN6.3.1/LT/2020) and grants-in-aid from Universitas Padjadjaran (Academic Leadership Grant) for RR (grant number: 2476/UN6.C/LT/2018).
|
| 301 |
+
|
| 302 |
+
## Footnotes
|
| 303 |
+
|
| 304 |
+
## Associated Data
|
| 305 |
+
|
| 306 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 307 |
+
|
| 308 |
+
### Data Availability Statement
|
| 309 |
+
|
| 310 |
+
Dataset available on request from the authors.
|
| 311 |
+
|
| 312 |
+
### Data Availability Statement
|
| 313 |
+
|
| 314 |
+
Dataset available on request from the authors.
|
| 315 |
+
|
| 316 |
+
## References
|
| 317 |
+
|
| 318 |
+
1. World Health Organization . Global Tuberculosis Report 2022. WHO; Geneva, Switzerland: 2022. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Global%20Tuberculosis%20Report%202022&publication_year=2022&)
|
| 319 |
+
|
| 320 |
+
2. Kemenkes R.I. Penatalaksanaan Tuberkulosis Resisten Obat di Indonesia. Kementerian Kesehatan RI; Jakarta, Indonesia: 2020. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Penatalaksanaan%20Tuberkulosis%20Resisten%20Obat%20di%20Indonesia&author=R.I.%20Kemenkes&publication_year=2020&)
|
| 321 |
+
|
| 322 |
+
3. World Health Organization . WHO Consolidated Guidelines on Tuberculosis. Module 4: Treatment. WHO; Geneva, Switzerland: 2022. Drug-Resistant Tuberculosis Treatment, 2022 Update. [Google Scholar](https://scholar.google.com/scholar_lookup?title=WHO%20Consolidated%20Guidelines%20on%20Tuberculosis.%20Module%204:%20Treatment&publication_year=2022&)
|
| 323 |
+
|
| 324 |
+
4. FDA . Highlights of Prescribing Information. Avelox® (Moxifloxacin Hydrocloride) FDA; Silver Spring, MD, USA: 2016. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Highlights%20of%20Prescribing%20Information.%20Avelox%C2%AE%20(Moxifloxacin%20Hydrocloride)&publication_year=2016&)
|
| 325 |
+
|
| 326 |
+
5. Fàbrega A., Madurga S., Giralt E., Vila J. Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2009;2:40–61. doi: 10.1111/j.1751-7915.2008.00063.x. [DOI](https://doi.org/10.1111/j.1751-7915.2008.00063.x) | [PMC free article](/articles/PMC3815421/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21261881/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Microb.%20Biotechnol.&title=Mechanism%20of%20action%20of%20and%20resistance%20to%20quinolones&author=A.%20F%C3%A0brega&author=S.%20Madurga&author=E.%20Giralt&author=J.%20Vila&volume=2&publication_year=2009&pages=40-61&pmid=21261881&doi=10.1111/j.1751-7915.2008.00063.x&)
|
| 327 |
+
|
| 328 |
+
6. Gumbo T., Angulo-Barturen I., Ferrer-Bazaga S. Pharmacokinetic-Pharmacodynamic and Dose-Response Relationships of Antituberculosis Drugs: Recommendations and Standards for Industry and Academia. J. Infect. Dis. 2015;211((Suppl. 3)):S96–S106. doi: 10.1093/infdis/jiu610. [DOI](https://doi.org/10.1093/infdis/jiu610) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26009618/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J.%20Infect.%20Dis.&title=Pharmacokinetic-Pharmacodynamic%20and%20Dose-Response%20Relationships%20of%20Antituberculosis%20Drugs:%20Recommendations%20and%20Standards%20for%20Industry%20and%20Academia&author=T.%20Gumbo&author=I.%20Angulo-Barturen&author=S.%20Ferrer-Bazaga&volume=211&issue=(Suppl.%203)&publication_year=2015&pages=S96-S106&pmid=26009618&doi=10.1093/infdis/jiu610&)
|
| 329 |
+
|
| 330 |
+
7. Annisa N., Barliana M.I., Santoso P., Ruslami R. Transporter and metabolizer gene polymorphisms affect fluoroquinolone pharmacokinetic parameters. Front. Pharmacol. 2022;13:1063413. doi: 10.3389/fphar.2022.1063413. [DOI](https://doi.org/10.3389/fphar.2022.1063413) | [PMC free article](/articles/PMC9798452/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/36588725/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front.%20Pharmacol.&title=Transporter%20and%20metabolizer%20gene%20polymorphisms%20affect%20fluoroquinolone%20pharmacokinetic%20parameters&author=N.%20Annisa&author=M.I.%20Barliana&author=P.%20Santoso&author=R.%20Ruslami&volume=13&publication_year=2022&pages=1063413&pmid=36588725&doi=10.3389/fphar.2022.1063413&)
|
| 331 |
+
|
| 332 |
+
8. Weiner M., Gelfond J., Johnson-Pais T.L., Engle M., Peloquin C.A., Johnson J.L., Sizemore E.E., Mac Kenzie W.R. Elevated plasma moxifloxacin concentrations and SLCO1B1 g.11187G>A polymorphism in adults with pulmonary tuberculosis. Antimicrob. Agents Chemother. 2018;62:1–11. doi: 10.1128/AAC.01802-17. [DOI](https://doi.org/10.1128/AAC.01802-17) | [PMC free article](/articles/PMC5923103/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29463526/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Antimicrob.%20Agents%20Chemother.&title=Elevated%20plasma%20moxifloxacin%20concentrations%20and%20SLCO1B1%20g.11187G>A%20polymorphism%20in%20adults%20with%20pulmonary%20tuberculosis&author=M.%20Weiner&author=J.%20Gelfond&author=T.L.%20Johnson-Pais&author=M.%20Engle&author=C.A.%20Peloquin&volume=62&publication_year=2018&pages=1-11&pmid=29463526&doi=10.1128/AAC.01802-17&)
|
| 333 |
+
|
| 334 |
+
9. Shan X.X., Qiu Y., Xie W.W., Wu R.R., Yu Y., Wu H.S., Li L.H. ABCB1 Gene Is Associated with Clinical Response to SNRIs in a Local Chinese Han Population. Front. Pharmacol. 2019;10:761. doi: 10.3389/fphar.2019.00761. [DOI](https://doi.org/10.3389/fphar.2019.00761) | [PMC free article](/articles/PMC6620233/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31333472/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Front.%20Pharmacol.&title=ABCB1%20Gene%20Is%20Associated%20with%20Clinical%20Response%20to%20SNRIs%20in%20a%20Local%20Chinese%20Han%20Population&author=X.X.%20Shan&author=Y.%20Qiu&author=W.W.%20Xie&author=R.R.%20Wu&author=Y.%20Yu&volume=10&publication_year=2019&pages=761&pmid=31333472&doi=10.3389/fphar.2019.00761&)
|
| 335 |
+
|
| 336 |
+
10. Rychlik-Sych M., Barańska M., Dudarewicz M., Skrętkowicz J., Żebrowska A., Woźniacka A., Owczarek J., Orszulak-Michalak D., Waszczykowska E. Haplotypes of ABCB1 1236C >T (rs1128503), 2677G >T/A (rs2032582), and 3435C >T (rs1045642) in patients with bullous pemphigoid. Arch. Dermatol. Res. 2018;310:515–522. doi: 10.1007/s00403-018-1842-8. [DOI](https://doi.org/10.1007/s00403-018-1842-8) | [PMC free article](/articles/PMC6060767/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29948283/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arch.%20Dermatol.%20Res.&title=Haplotypes%20of%20ABCB1%201236C%20>T%20(rs1128503),%202677G%20>T/A%20(rs2032582),%20and%203435C%20>T%20(rs1045642)%20in%20patients%20with%20bullous%20pemphigoid&author=M.%20Rychlik-Sych&author=M.%20Bara%C5%84ska&author=M.%20Dudarewicz&author=J.%20Skr%C4%99tkowicz&author=A.%20%C5%BBebrowska&volume=310&publication_year=2018&pages=515-522&pmid=29948283&doi=10.1007/s00403-018-1842-8&)
|
| 337 |
+
|
| 338 |
+
11. Naidoo A., Ramsuran V., Chirehwa M., Denti P., McIlleron H., Naidoo K., Yende-Zuma N., Singh R., Ngcapu S., Chaudhry M., et al. Effect of genetic variation in UGT1A and ABCB1 on moxifloxacin pharmacokinetics in South African patients with tuberculosis. Pharmacogenomics. 2017;19:17–29. doi: 10.2217/pgs-2017-0144. [DOI](https://doi.org/10.2217/pgs-2017-0144) | [PMC free article](/articles/PMC5753622/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29210323/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=Effect%20of%20genetic%20variation%20in%20UGT1A%20and%20ABCB1%20on%20moxifloxacin%20pharmacokinetics%20in%20South%20African%20patients%20with%20tuberculosis&author=A.%20Naidoo&author=V.%20Ramsuran&author=M.%20Chirehwa&author=P.%20Denti&author=H.%20McIlleron&volume=19&publication_year=2017&pages=17-29&pmid=29210323&doi=10.2217/pgs-2017-0144&)
|
| 339 |
+
|
| 340 |
+
12. Nijland H.M.J., Ruslami R., Suroto A.J., Burger D.M., Alisjahbana B., Van Crevel R., Aarnoutse R.E. Rifampicin reduces plasma concentrations of moxifloxacin in patients with tuberculosis. Clin. Infect. Dis. 2007;45:1001–1007. doi: 10.1086/521894. [DOI](https://doi.org/10.1086/521894) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17879915/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Infect.%20Dis.&title=Rifampicin%20reduces%20plasma%20concentrations%20of%20moxifloxacin%20in%20patients%20with%20tuberculosis&author=H.M.J.%20Nijland&author=R.%20Ruslami&author=A.J.%20Suroto&author=D.M.%20Burger&author=B.%20Alisjahbana&volume=45&publication_year=2007&pages=1001-1007&pmid=17879915&doi=10.1086/521894&)
|
| 341 |
+
|
| 342 |
+
13. Ruslami R., Ganiem A.R., Dian S., Apriani L., Achmad T.H., van der Ven A.J., Borm G., Aarnoutse R.E., van Crevel R. Intensified regimen containing rifampicin and moxifloxacin for tuberculous meningitis: An open-label, randomised controlled phase 2 trial. Lancet Infect. Dis. 2013;13:27–35. doi: 10.1016/S1473-3099(12)70264-5. [DOI](https://doi.org/10.1016/S1473-3099(12)70264-5) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/23103177/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet%20Infect.%20Dis.&title=Intensified%20regimen%20containing%20rifampicin%20and%20moxifloxacin%20for%20tuberculous%20meningitis:%20An%20open-label,%20randomised%20controlled%20phase%202%20trial&author=R.%20Ruslami&author=A.R.%20Ganiem&author=S.%20Dian&author=L.%20Apriani&author=T.H.%20Achmad&volume=13&publication_year=2013&pages=27-35&pmid=23103177&doi=10.1016/S1473-3099(12)70264-5&)
|
| 343 |
+
|
| 344 |
+
14. Magis-Escurra C., Later-Nijland H.M.J., Alffenaar J.W.C., Broeders J., Burger D.M., Van Crevel R., Boeree M.J., Donders A.R.T., van Altena R., van Der Werf T.S., et al. Population pharmacokinetics and limited sampling strategy for first-line tuberculosis drugs and moxifloxacin. Int. J. Antimicrob. Agents. 2014;44:229–234. doi: 10.1016/j.ijantimicag.2014.04.019. [DOI](https://doi.org/10.1016/j.ijantimicag.2014.04.019) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/24985091/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int.%20J.%20Antimicrob.%20Agents&title=Population%20pharmacokinetics%20and%20limited%20sampling%20strategy%20for%20first-line%20tuberculosis%20drugs%20and%20moxifloxacin&author=C.%20Magis-Escurra&author=H.M.J.%20Later-Nijland&author=J.W.C.%20Alffenaar&author=J.%20Broeders&author=D.M.%20Burger&volume=44&publication_year=2014&pages=229-234&pmid=24985091&doi=10.1016/j.ijantimicag.2014.04.019&)
|
| 345 |
+
|
| 346 |
+
15. Soldin O.P., Mattison D.R. Sex differences in pharmacokinetics and pharmacodynamics. Clin. Pharmacokinet. 2009;48:143–157. doi: 10.2165/00003088-200948030-00001. [DOI](https://doi.org/10.2165/00003088-200948030-00001) | [PMC free article](/articles/PMC3644551/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19385708/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Pharmacokinet.&title=Sex%20differences%20in%20pharmacokinetics%20and%20pharmacodynamics&author=O.P.%20Soldin&author=D.R.%20Mattison&volume=48&publication_year=2009&pages=143-157&pmid=19385708&doi=10.2165/00003088-200948030-00001&)
|
| 347 |
+
|
| 348 |
+
16. Bebawy M., Chetty M. Gender Differences in P-Glycoprotein Expression and Function: Effects on Drug Disposition and Outcome. Curr. Drug Metab. 2009;10:322–328. doi: 10.2174/138920009788498996. [DOI](https://doi.org/10.2174/138920009788498996) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/19519340/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Curr.%20Drug%20Metab.&title=Gender%20Differences%20in%20P-Glycoprotein%20Expression%20and%20Function:%20Effects%20on%20Drug%20Disposition%20and%20Outcome&author=M.%20Bebawy&author=M.%20Chetty&volume=10&publication_year=2009&pages=322-328&pmid=19519340&doi=10.2174/138920009788498996&)
|
| 349 |
+
|
| 350 |
+
17. Hou W.Y., Xu S.F., Zhu Q.N., Lu Y.F., Cheng X.G., Liu J. Age- and sex-related differences of organic anion-transporting polypeptide gene expression in livers of rats. Toxicol. Appl. Pharmacol. 2014;280:370–377. doi: 10.1016/j.taap.2014.08.020. [DOI](https://doi.org/10.1016/j.taap.2014.08.020) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25168429/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Toxicol.%20Appl.%20Pharmacol.&title=Age-%20and%20sex-related%20differences%20of%20organic%20anion-transporting%20polypeptide%20gene%20expression%20in%20livers%20of%20rats&author=W.Y.%20Hou&author=S.F.%20Xu&author=Q.N.%20Zhu&author=Y.F.%20Lu&author=X.G.%20Cheng&volume=280&publication_year=2014&pages=370-377&pmid=25168429&doi=10.1016/j.taap.2014.08.020&)
|
| 351 |
+
|
| 352 |
+
18. Sullivan J.T., Lettieri J.T., Liu P., Heller A.H., Watson S.J. The influence of age and gender on the pharmacokinetics of moxifloxacin. Clin. Pharmacokinet. 2001;40((Suppl. 1)):11–18. doi: 10.2165/00003088-200140001-00002. [DOI](https://doi.org/10.2165/00003088-200140001-00002) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11352437/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin.%20Pharmacokinet.&title=The%20influence%20of%20age%20and%20gender%20on%20the%20pharmacokinetics%20of%20moxifloxacin&author=J.T.%20Sullivan&author=J.T.%20Lettieri&author=P.%20Liu&author=A.H.%20Heller&author=S.J.%20Watson&volume=40&issue=(Suppl.%201)&publication_year=2001&pages=11-18&pmid=11352437&doi=10.2165/00003088-200140001-00002&)
|
| 353 |
+
|
| 354 |
+
19. Chien S.C., Chow A.T., Natarajan J., Williams R.R., Wong F.A., Rogge M.C., Nayak R.K. Absence of age and gender effects on the pharmacokinetics of a single 500-milligram oral dose of levofloxacin in healthy subjects, Antimicrob. Agents Chemother. 1997;41:1562–1565. doi: 10.1128/AAC.41.7.1562. [DOI](https://doi.org/10.1128/AAC.41.7.1562) | [PMC free article](/articles/PMC163959/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9210685/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Agents%20Chemother.&title=Absence%20of%20age%20and%20gender%20effects%20on%20the%20pharmacokinetics%20of%20a%20single%20500-milligram%20oral%20dose%20of%20levofloxacin%20in%20healthy%20subjects,%20Antimicrob&author=S.C.%20Chien&author=A.T.%20Chow&author=J.%20Natarajan&author=R.R.%20Williams&author=F.A.%20Wong&volume=41&publication_year=1997&pages=1562-1565&pmid=9210685&doi=10.1128/AAC.41.7.1562&)
|
| 355 |
+
|
| 356 |
+
20. Hasunuma T., Tohkin M., Kaniwa N., Jang I.J., Yimin C., Kaneko M., Saito Y., Takeuchi M., Watanabe H., Yamazoe Y., et al. Absence of ethnic differences in the pharmacokinetics of moxifloxacin, simvastatin, and meloxicam among three East Asian populations and Caucasians. Br. J. Clin. Pharmacol. 2016;81:1078–1090. doi: 10.1111/bcp.12884. [DOI](https://doi.org/10.1111/bcp.12884) | [PMC free article](/articles/PMC4876172/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26774055/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br.%20J.%20Clin.%20Pharmacol.&title=Absence%20of%20ethnic%20differences%20in%20the%20pharmacokinetics%20of%20moxifloxacin,%20simvastatin,%20and%20meloxicam%20among%20three%20East%20Asian%20populations%20and%20Caucasians&author=T.%20Hasunuma&author=M.%20Tohkin&author=N.%20Kaniwa&author=I.J.%20Jang&author=C.%20Yimin&volume=81&publication_year=2016&pages=1078-1090&pmid=26774055&doi=10.1111/bcp.12884&)
|
test/texts/PMC11887086.md
ADDED
|
@@ -0,0 +1,331 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Valproic acid levels in neurodevelopmental disorders: correlation with CYP and SULT genes using LC-MS/MS
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Shada Abutaleb, Eyad Mallah, Luay Abu-Qatouseh, Ahmad Abu-awwad, Kenza Mansoor, Sarah Khallad, Khaled W Omari, Omar Mouhtady, Tawfiq Arafat
|
| 5 |
+
**Journal:** BMC Neurology
|
| 6 |
+
**Date:** 2025 Mar 7
|
| 7 |
+
**DOI:** [10.1186/s12883-025-04065-z](https://doi.org/10.1186/s12883-025-04065-z)
|
| 8 |
+
**PMID:** 40055599
|
| 9 |
+
**PMCID:** PMC11887086
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11887086/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC11887086/pdf/12883_2025_Article_4065.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC11887086/pdf/12883_2025_Article_4065.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Background:**
|
| 16 |
+
Valproic acid (VPA) is one of the most widely prescribed antiepileptic drugs worldwide, which is used to treat migraines, bipolar disorder, and anxiety. However, VPA is associated with a wide range of side effects. This study evaluates therapeutic drug monitoring (TDM) in individuals with neurodevelopmental disorders. It explores the correlation between valproic acid (VPA) plasma levels and genetic polymorphisms in cytochrome P450 (CYP) enzymes and cytosolic sulfotransferase (SULT) genes.
|
| 17 |
+
|
| 18 |
+
**Methods:**
|
| 19 |
+
A simple and accurate LC-MS/MS method was developed, validated, and applied in the TDM of 14 individuals on VPA therapy. Plasma VPA levels were measured, and genotyped genes SULT1A1, CYP2D64, CYP2D610, CYP3A5, and CYP2C19*2. Statistical analyses were conducted using SPSS.
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
Of the fourteen participants, two had toxic plasma VPA levels (≥ 100 µg/mL), one had a sub-therapeutic level (< 50 µg/mL), and eleven were within or slightly above the therapeutic range (50–100 µg/mL). No significant correlation was observed between VPA plasma concentrations and genotypes of SULT1A1 (p = 0.522), CYP2C192 (p = 0.288), CYP2D64 (p = 0.895), or CYP2D6*10 (p = 0.067). While no direct associations were found, genotype-guided drug therapy remains a promising strategy for optimizing drug efficacy and minimizing toxicity.
|
| 23 |
+
|
| 24 |
+
**Conclusions:**
|
| 25 |
+
This study highlights the complexity of valproic acid (VPA) therapy in individuals with neurodevelopmental disorders and the limited influence of common genetic polymorphisms in CYP and SULT genes on VPA plasma levels. While therapeutic drug monitoring (TDM) remains an invaluable tool for optimizing VPA therapy, the lack of significant correlations between genetic variants and VPA concentrations suggests that routine pharmacogenetic testing for these specific variants may not be warranted in clinical practice. However, the observed toxic and sub-therapeutic VPA levels emphasize the importance of regular TDM to mitigate risks associated with overdose or insufficient dosing.
|
| 26 |
+
|
| 27 |
+
**Supplementary Information:**
|
| 28 |
+
The online version contains supplementary material available at 10.1186/s12883-025-04065-z.
|
| 29 |
+
|
| 30 |
+
Keywords: Therapeutic drug monitoring, Valproic acid, Neurodevelopmental disorders, Autism spectrum disorder (ASD), LC-MS/MS, CYP and SULT genes
|
| 31 |
+
|
| 32 |
+
### Background
|
| 33 |
+
|
| 34 |
+
Valproic acid (VPA) is one of the most widely prescribed antiepileptic drugs worldwide, which is used to treat migraines, bipolar disorder, and anxiety. However, VPA is associated with a wide range of side effects. This study evaluates therapeutic drug monitoring (TDM) in individuals with neurodevelopmental disorders. It explores the correlation between valproic acid (VPA) plasma levels and genetic polymorphisms in cytochrome P450 (CYP) enzymes and cytosolic sulfotransferase (SULT) genes.
|
| 35 |
+
|
| 36 |
+
### Methods
|
| 37 |
+
|
| 38 |
+
A simple and accurate LC-MS/MS method was developed, validated, and applied in the TDM of 14 individuals on VPA therapy. Plasma VPA levels were measured, and genotyped genes SULT1A1, CYP2D64, CYP2D610, CYP3A5, and CYP2C19*2. Statistical analyses were conducted using SPSS.
|
| 39 |
+
|
| 40 |
+
### Results
|
| 41 |
+
|
| 42 |
+
Of the fourteen participants, two had toxic plasma VPA levels (≥ 100 µg/mL), one had a sub-therapeutic level (< 50 µg/mL), and eleven were within or slightly above the therapeutic range (50–100 µg/mL). No significant correlation was observed between VPA plasma concentrations and genotypes of SULT1A1 (*p*p = 0.522), CYP2C192 (*p*p = 0.288), *CYP2D64*CYP2D64 (*p*p = 0.895), or CYP2D6*10 (*p*p = 0.067). While no direct associations were found, genotype-guided drug therapy remains a promising strategy for optimizing drug efficacy and minimizing toxicity.
|
| 43 |
+
|
| 44 |
+
### Conclusions
|
| 45 |
+
|
| 46 |
+
This study highlights the complexity of valproic acid (VPA) therapy in individuals with neurodevelopmental disorders and the limited influence of common genetic polymorphisms in CYP and SULT genes on VPA plasma levels. While therapeutic drug monitoring (TDM) remains an invaluable tool for optimizing VPA therapy, the lack of significant correlations between genetic variants and VPA concentrations suggests that routine pharmacogenetic testing for these specific variants may not be warranted in clinical practice. However, the observed toxic and sub-therapeutic VPA levels emphasize the importance of regular TDM to mitigate risks associated with overdose or insufficient dosing.
|
| 47 |
+
|
| 48 |
+
### Supplementary Information
|
| 49 |
+
|
| 50 |
+
The online version contains supplementary material available at 10.1186/s12883-025-04065-z.
|
| 51 |
+
|
| 52 |
+
**Keywords:**Keywords: Therapeutic drug monitoring, Valproic acid, Neurodevelopmental disorders, Autism spectrum disorder (ASD), LC-MS/MS, CYP and SULT genes
|
| 53 |
+
|
| 54 |
+
## Background
|
| 55 |
+
|
| 56 |
+
Intellectual disability (ID) is characterized by deficiencies in general mental skills such as reasoning, problem-solving, planning, logical thought, judgment, academic learning, and learning from experience. The stated global prevalence of intellectual disability is 1%, ranging from 1–3% per country, with a male-to-female ratio of 2:1 [[1](#CR1)1]. An intelligence quotient (IQ) of 70 or lower indicates an ID diagnosis. Intellectual disorder is diagnosed before the age of 18, where the intensity of the condition has been described using the phrases “mild,” “moderate,” severe,” and “profound” [[2](#CR2)2]. Additionally, data from the United States between 2019 and 2021 showed that boys were more than three times as likely as girls to be diagnosed with autism spectrum disorder, which often co-occurs with ID. These variations highlight the influence of diagnostic criteria, healthcare access, and reporting methods on prevalence estimates [[3](#CR3)3]. It is always a good idea to use inclusive language for neurodivergent conditions, such as autism spectrum disorder (ASD), which aims to employ respectful and accurate terminology; readers may refer to recent literature [[4](#CR4)4].
|
| 57 |
+
|
| 58 |
+
Autism Spectrum Disorder (ASD) is a neurodevelopmental disorder characterized by challenges in social communication and interaction, along with restricted, repetitive patterns of behavior, interests, or activities. According to the DSM-5 (2013), ASD now encompasses previous diagnoses, including autism, Asperger’s syndrome, and pervasive developmental disorders not otherwise specified (PDD-NOS), under a single diagnostic category. In the past ten years, the number of individuals has risen dramatically, and it currently affects 1 to 1.5% of the population [[2](#CR2)2]. ASD is diagnosed more frequently in males than in females, with a reported diagnostic ratio of 4:1. The new diagnostic definition of ASD focuses on two central domains: social contact disorder and restricted interests and repetitive behaviors [[5](#CR5)5]. ASD can be detected as early as the age of 2 years [[6](#CR6)6]. In extreme cases, individuals with autism may show specific developmental disabilities, including cognitive impairment [[7](#CR7)7]. There is some proof of an elevated risk of epilepsy correlated with other causes, such as ASD etiology, seriousness of autistic traits, developmental regression, and family background [[8](#CR8)8]. Epilepsy is common in autism spectrum disorder (ASD), with prevalence estimates ranging from 5 to 46% depending on the study population, age group, and severity of ASD symptoms. A meta-analysis study reported an overall pooled prevalence of epilepsy in ASD to be approximately 21.4%, with higher rates observed in individuals with co-occurring intellectual disabilities (up to 46%) compared to those without intellectual disabilities (approximately 8%) [[9](#CR9)9].
|
| 59 |
+
|
| 60 |
+
Valproic acid (VPA) was used in clinical practice nearly 50 years ago. Its effectiveness and tolerability profiles have been well-documented in experimental and clinical trials. Because of its broad scope of efficacy over a wide variety of seizures and epileptic syndromes, it is a mainstay of anticonvulsant treatment [[10](#CR10)10]. VPA is available in various pharmaceutical formulations, including sodium valproate, valproic acid on its own, and valproic acid combined with valproic acid. A modified release formulation containing a 2.3:1 sodium valproate and VPA ratio is also provided. Valproate semisodium (divalproex semisodium in the United States) is a medication sold in the United Kingdom by Sanofi-Synthélabo under the brand name Depakote^®^® [[11](#CR11)11]. It is an enteric-coated compound of sodium valproate and VPA in a 1:1 molar ratio, dissociating to release valproate ions in the gastrointestinal system.
|
| 61 |
+
|
| 62 |
+
VPA comes in several forms: immediate-release, enteric-coated, delayed-release (12 h), and extended-release (24 h). It may also be administered as an intravenous solution. Children’s therapeutic regular doses vary from 15 to 60 mg/kg/day, while adults’ doses range from 500 mg to 2 g a day [[12](#CR12)12]. VPA is associated with a wide range of side effects that are particularly concerning in polytherapy and long-term treatment. Gastrointestinal intolerance symptoms may be reduced when using an enteric-coated formulation or taking the medication at mealtime. Nervous system stimulation, such as nervousness, insomnia, and tremor, can occur regularly in some individuals. VPA has been linked to Parkinsonism and cognitive dysfunction. It can cause hyperammonaemia, encephalopathy, thrombocytopenia, and other blood diseases documented infrequently [[13](#CR13)13]. VPA has also been linked to toxic epidermal necrolysis (TEN), Stevens-Johnson syndrome (SJS), and drug reactions with eosinophilia and systemic symptoms (DRESS), among other delayed hypersensitivity reactions [[14](#CR14)14]. Pancreatitis has been recorded in children and adults who have taken valproate; most cases have been identified in epileptic children or adults with kidney failure [[15](#CR15)15]. VPA has been linked to severe congenital malformations, most notably neural tube defects (e.g., spina bifida). Lower IQ scores in utero have been related to VPA exposure.
|
| 63 |
+
|
| 64 |
+
Drugs that increase the expression of hepatic enzymes can increase valproate clearance. Phenytoin, carbamazepine, and phenobarbital (or primidone) can, for instance, double VPA clearance. Since cytochrome P450 microsomal mediated oxidation is a minor secondary metabolic pathway compared to glucuronidation and beta-oxidation, drugs that inhibit cytochrome P450 isozymes, such as antidepressants, may have little impact valproate clearance. Valproate weakly inhibits some P450 isozymes, epoxide hydrase, and glucuronosyltransferases.
|
| 65 |
+
|
| 66 |
+
VPA’s pharmacokinetics are non-linear due to their saturable binding to plasma proteins. The suggested VPA reference range for the treatment of epilepsy is 50–100 µg/mL. For bipolar disorder treatment, a higher dosage range of 50–125 µg/mL is recommended [[16](#CR16)16]. VPA’s time to steady state is 2–4 days [[17](#CR17)17]. The T_max_max for valproate semisodium was 3.6 h, and for enteric-coated valproate, it was 3.8 h [[11](#CR11)11]. The maximum concentration reached for a medication, C_max_max, was 103 µg/mL for valproate semisodium at a dosage of 500 mg (the VPA equivalent of 500 mg) twice daily. In contrast, enteric-coated sodium valproate was 91.33 µg/mL at a dose of 500 mg (VPA equivalent: 433 mg) twice a day. In a study, depending on the dosage given (400–1500 mg/day), C_max_max after around 4 h ranged from 60 to 330 µg/mL [[18](#CR18)18]. In another study, following regular doses of 250–1000 mg of Depakote ER (equivalent to 250 mg of VPA), C_max_max for total VPA in children (ages 8–11) ranged from 55.7 to 132.7 µg/mL. Following regular doses of 500–1750 mg Depakote ER, C_max_max for total VPA in teenagers (ages 12–17) ranged from (31.7–120.6) µg/mL [[19](#CR19)19].
|
| 67 |
+
|
| 68 |
+
Therapeutic drug monitoring (TDM) of antiepileptic drugs (AEDs) has had a significant effect on epilepsy treatment for the last 10–20 years because of increased understanding of AED pharmacokinetics. VPA’s ability to treat a variety of seizures with a single anticonvulsant has led to its widespread use, especially among children. Moreover, it’s also increasingly used in the treatment of psychotic and schizoaffective conditions, neuropathic pain, and migraine headache prophylaxis. The clearance of VPA is stated to be concentration- or dose-dependent due to substantial inter-individual pharmacokinetic variation and concentration-dependent plasma protein binding pharmacokinetics of VPA [[20](#CR20)20]. Furthermore, the therapeutic effects of VPA are closely related to the drug’s serum concentration. Its proper serum concentration is determined by various factors, including age, total body weight, VPA dosage, and co-administration of other medications that impair VPA’s pharmacokinetics. Many methods of analysis for quantitating VPA in human plasma have been developed. Fluorescence polarization immunoassay (FPIA) has been used to monitor VPA in most of them. Additionally, other techniques, such as High-Performance Liquid Chromatography (HPLC) assays combined with either ultraviolet or fluorescence detection [[21](#CR21)21] and Gas Chromatography (GC) with mass spectrometric (MS) detection, have also shown satisfactory results. Most of these methods necessitate time-consuming analysis or running time. Analytical procedures requiring evaporation steps are undesirable due to the volatility of VPA. On the other hand, VPA could need a liquid-liquid extraction to be observed by ultraviolet or fluorescence, leading to a high processing time and poor recovery. So many methods have been used, including liquid chromatography-tandem mass spectrometric detection (LC-MS/MS). The primary benefit of such techniques would be that no prior chemical alteration of VPA is necessary.
|
| 69 |
+
|
| 70 |
+
Sulfonation has typically been regarded as a detoxifying pathway leading to even more water-soluble compounds and assisting their elimination through the kidneys or bile, as in the case of most xenobiotics and relatively insignificant endogenous substrates. Baumann originally discovered sulfonate conjugation in 1876, and it has subsequently been found to be a key process in the biotransformation of a wide range of xenobiotics and endobiotics, including medicines, chemical carcinogens, hormones, bile acids, neurotransmitters, peptides, and lipids. 3′-phosphoadenosine-5′-phosphosulfate (PAPS) is the widespread sulfonate donor for such processes. Thus, the transference of sulfonate (SO-3) toward a hydroxyl or amino group would be catalyzed via a supergene family of enzymes known as sulfotransferases (SULTs) [[22](#CR22)22]. SULTs seem genetically polymorphic and indicated in a diverse range of organs. Four leading SULT families are SULT1, SULT2, SULT4, and SULT6, with 13 human cytosolic SULT isoforms. The SULTs alloenzymes are encoded by this gene; SULT1A (SULT1A1*1 wild-type, SULT1A1*2, SULT1A1*3, and SULT1A1*4) were localized to chromosome 16p12.1–p11.2, with considerable biochemical changes in their activities. This polymorphism is particularly significant in the case of a mutation in exon seven at nucleotide 638 (codon 213), which results in a replacement of histidine by arginine (SULT1A1*2 allele), which is linked with lower enzymatic activity and heat resistance when compared to the wild-type allele (SULT1A1*1 allele) [[23](#CR23)23].
|
| 71 |
+
|
| 72 |
+
The current study aims to develop a simple and accurate LC-MS/MS method for determining VPA concentration in human plasma. The objective is to determine concentrations of VPA in individuals with neurodevelopmental disorders (intellectual disability and/or autism spectrum disorder), to evaluate TDM, and study the relationship between common genotypes of CYPs and SULTs genes and levels of VPA in these individuals by restriction fragment length polymorphism.
|
| 73 |
+
|
| 74 |
+
## Methods
|
| 75 |
+
|
| 76 |
+
### Human participants and study design
|
| 77 |
+
|
| 78 |
+
This research was carried out after receiving ethical approval from the Jordan Center of Pharmaceutical Sciences’ inter-regulatory board on October 3, 2020. The Arabic Village for Special Education Center obtained parental approval separately. Fourteen participants were registered for the study. Blood samples were obtained from the Arabic Village for Special Education Center with the help of nurses. The selection criteria were based on individuals suffering from neurodevelopmental disorders (ID and/or ASD) and taking VPA at a steady state. In October 2020, blood chemistry data from the Arabic Village for Special Education Center was collected, including kidney, liver, and thyroid function tests and hematologic analysis. A total of 1–5 mL of blood was obtained from each participant. Blood samples were collected at 12 h post-administration and centrifuged to give blood plasma samples using citrate tubes to analyze VPA. The samples were transferred within 30 min to the Jordan Center of Pharmaceutical Research for LC-MS/MS analysis. Half a milliliter of whole blood from each participant was withdrawn into a plain tube and frozen at -80 °C at the University of Petra until assay.
|
| 79 |
+
|
| 80 |
+
### LC-MS/MS
|
| 81 |
+
|
| 82 |
+
The study used an LC-MS/MS system composed of high-pressure liquid chromatography (HPLC) (DIONEX Ultimate 3000) coupled with a mass spectrometer and detector (Thermo LCQ Fleet). Genotyping was carried out at the biotechnology laboratory of the Pharmaceutical Center of Petra University. The instruments used include: a UV spectrophotometry apparatus NanoDrop™ Thermo Fisher Scientific, Inc. (USA), a gel electrophoresis apparatus, nano PAC-300 from Cleaver Scientific (UK), a gel imaging instrument Bio-Rad gel Doc™ EZ imager (USA), and for DNA amplification, GeneAMP^®^® PCR system 9700.
|
| 83 |
+
|
| 84 |
+
Under chromatographic conditions, the mobile phase consisted of an isocratic mixture of 0.1% acetic acid and acetonitrile 1:1 v/v. The pH was adjusted by adding 0.1% triethylamine. The internal standard (IS) was Ondansetron, and the chromatographic separation was achieved using analytical column C18 ACE (2.1 mm, 5 cm, 5 μm). The mobile phase flow rate was 25 mL/min, and the injection volume was 2 µL.
|
| 85 |
+
|
| 86 |
+
MS Conditions, the mass spectrometry detection mode for VPA was unfavorable, the VPA’s mass transition (m/z) was 143, and the collision energy was zero. The mass spectrometry detection mode for Ondansetron was positive, the mass transition (m/z) was 294, and the collision energy was 25. Ion source temperature was maintained at 300 °C, Ionization voltage was kept at 5 Kv, and Sheath gas, auxiliary gas, and sweep gas flow rates were 50, 25, and 0 L/min, respectively.
|
| 87 |
+
|
| 88 |
+
### Sample preparation: plasma extraction
|
| 89 |
+
|
| 90 |
+
Acetonitrile (precipitating agent) containing 10 µg/mL IS (Ondansetron) was added to a spiked plasma in a ratio of 3:2 v/v. The mixture was vortexed and centrifuged for 5 min at 14,000 rpm; the supernatant was transferred into an auto-sampler vial.
|
| 91 |
+
|
| 92 |
+
### Standard calibration points and quality control sample preparation
|
| 93 |
+
|
| 94 |
+
100 mg of VPA raw material was weighed and dissolved in 5 mL methanol. The stock solution was mixed in a ratio of 1:1 v/v to prepare 20,000 µg/mL from VPA as a mixture solution. A serial dilution was prepared as calibrators of (50, 100, 200, 400, 700, 1000, and 1500) µg/mL, and quality control (QC) samples as follows: 150 low, 750 mid, and 1500 high µg/mL QC concentrations.
|
| 95 |
+
|
| 96 |
+
The validation fulfilled the guidelines set out by the European Medicines Agency [[24](#CR24)24]. The method was validated in accuracy, precision, sensitivity, specificity, and linearity.
|
| 97 |
+
|
| 98 |
+
The LC-MS/MS method used in this study to determine valproic acid (VPA) concentrations in plasma was thoroughly validated per EMA guidelines. The technique demonstrated excellent accuracy, with within-run and between-run accuracy values ranging from 97.5 to 102.9% across all quality control (QC) levels. Precision was confirmed with a coefficient of variation (CV) below 5.25%, meeting the acceptance criteria of ≤ 15% for QC samples and ≤ 20% for the lower limit of quantification (LLOQ). Linearity was established over a 5–150 µg/mL concentration range, with a correlation coefficient (R²) of 0.9998. Sensitivity was defined at the LLOQ of 5 µg/mL, ensuring reliable quantification at low concentrations. Specificity tests showed no interference from endogenous or exogenous substances in the matrix. These robust validation parameters ensure that the LC-MS/MS method provides accurate, precise, and reliable results for therapeutic drug monitoring of VPA in clinical settings.
|
| 99 |
+
|
| 100 |
+
There were no interferences at the VPA and IS retention times. The peaks were well-formed and well-resolved from the plasma constituents. The matrix peak was less than 5% of the internal standard’s peak area, deemed acceptable by the European Medicines Agency [[25](#CR25)25].
|
| 101 |
+
|
| 102 |
+
For genotyping, blood samples were collected from each participant using EDTA tubes. A total of 0.3 mL of each sample was utilized, with the remainder stored at -80 °C in case further testing is needed. QIAamp Blood mini kit was used to extract DNA from blood samples (Qiagen, Germany).
|
| 103 |
+
|
| 104 |
+
## Results
|
| 105 |
+
|
| 106 |
+
Demography and VPA administration, the 14 individuals, all of whom were males, were diagnosed with a neurodevelopmental disorder (intellectual disability and/or autism spectrum disorder) within their first two years. The participants’ actual weight ranged from 31.5 Kg to 87 Kg, their height ranged from 1.4 m to 1.7 m, and their ages ranged from 12 to 36 years old. Table [1](#Tab1)1 Shows the daily doses for every individual based on the sample number.
|
| 107 |
+
|
| 108 |
+
### Table 1.
|
| 109 |
+
|
| 110 |
+
VPA dose regimens
|
| 111 |
+
|
| 112 |
+
| Sample number | The total daily dose of VPA |
|
| 113 |
+
| ------------- | --------------------------- |
|
| 114 |
+
| 1 | 250 mg |
|
| 115 |
+
| 2 | 300 mg |
|
| 116 |
+
| 3 | 500 mg |
|
| 117 |
+
| 4, 5, 6, 7, 8 and 9 | 1000 mg |
|
| 118 |
+
| 10, 11 and 12 | 1250 mg |
|
| 119 |
+
| 13 | 1500 mg |
|
| 120 |
+
| 14 | 2000 mg |
|
| 121 |
+
For blood screening, fourteen whole blood samples were withdrawn and centrifuged into plasma and serum. Blood serum samples were analyzed for blood chemistry analysis described in Supplementary Materials S1, S2, and S3. When the CBC results were compared to normal ranges, the RBC, Hb, and HCT were lower than usual limits, as shown in Supplementary Materials S1. Some of the hematological adverse effects of VPA treatment that have been described include aplastic anemia, pure red cell aplasia, macrocytosis, leukopenia, and thrombocytopenia. This might contribute to the low RBC, Hb, and HCT levels in the sample numbers (2, 3, 4, 5, 9, 10, 11, and 14) [[26](#CR26)26]. ASD individuals with intellectual impairment had lower hemoglobin and hematocrit levels than ASD individuals with standard intellectual capability. Iron deficiency anemia was found to be higher in children with intellectual disabilities [[27](#CR27)27], and this might explain the low levels of RBC, Hb, and HCT. In sample numbers (5, 9, 10, 12, and 14), there is a decrease in neutrophil values that could be described as neutropenia. Neutropenia can also be caused by VPA treatment, which is reported in the literature. Numbers (1, 3, and 14) showed a decrease in their MCV values, which could be explained by iron deficiency anemia. Meanwhile, sample numbers (1, 4, 5, 9, 13, and 14) show decreased MCH values. A change in MCH usually corresponds to a change in MCV. Microcytic anemias are associated with a reduction in hemoglobin levels, whereas macrocytic anemias are associated with an elevation in hemoglobin levels. As a result, the MCH provides minimal information that is not already included in the MCV. There was a decrease in MCHC value in sample numbers (1, 3, 8, 9, and 10), indicating that RBCs were hypochromic, pale, and contained less Hemoglobin. Sample numbers (5 and 12) showed a decrease in WBCs, which is called leukopenia. Leukopenia is caused by a reduction in overall WBC production or an increase in WBC breakdown throughout the bone marrow [[28](#CR28)28].
|
| 122 |
+
|
| 123 |
+
In Supplementary Materials S2, sample numbers (1, 2, and 12) showed a low uric acid level. Long-term antiepileptic therapy, including VPA, has been shown to lower blood uric acid levels [[29](#CR29)29]. In sample number (11), there was an increase in uric acid blood and creatinine concentrations. Increased uric Acid production reported in some gout patients may be related to accelerated creatinine synthesis [[30](#CR30)30]. In sample numbers (7, 9, 11, and 13), there was a slight elevation of serum creatinine, reported in the literature as seen in individuals treated with VPA, and could be a sign that kidneys begin to dysfunction [[31](#CR31)31]. Sample number (2) had increased serum potassium, a condition called hyperkalemia. Hyperkalemia caused by VPA has rarely been documented in the literature. Also, in the sample, there was an increase in chloride serum levels. No correlation was mentioned between high chloride serum levels and VPA in the literature. Sample numbers (13 and 14) showed an increase in GGT levels, as shown in Supplementary Materials S2. Both samples are taking VPA along with carbamazepine. VPA and carbamazepine increase ALT, AST, and GGT, but carbamazepine can potentially increase GGT [[32](#CR32)32] more than VPA because of the enzyme induction [[19](#CR19)19]. Sample numbers (3 and 8) had an increase in globulin levels. There is no relation between globulin and VPA found in the literature. Sample number (5) had an increased level of direct bilirubin, which could be explained via chronic VPA treatment, alone or in combination. Acute hepatic impairment associated with prolonged VPA treatment has also been linked to an increase in plasma bilirubin levels. It is recommended for individuals on long-term VPA treatment to undergo frequent liver function testing [[33](#CR33)33]. Sample numbers (5 and 6) showed increased direct bilirubin level, also called conjugated bilirubin. Hepatocellular illness or cholestasis is the most common cause of conjugated hyperbilirubinemia (intrahepatic and extrahepatic) [[34](#CR34)34].
|
| 124 |
+
|
| 125 |
+
As shown in Supplementary Materials S3, sample numbers (1, 2, 6, and 13) had an increased TSH value. It was reported that VPA might cause changes in thyroid hormones [[35](#CR35)35]. Sample number (10) had a very low ferritin level. In children with ASD, there has been a high frequency of iron deficiency. However, research on the link between iron deficiency characteristics and ASD clinical symptoms is limited [[27](#CR27)27]. Sample numbers (4, 5, 6, 7, 8, 11, and 14) had a decreased level of vitamin D3. Vitamin D is a neurosteroid hormone that plays a vital role in brain development. As a result, its lack throughout pregnancy and early childhood might have a significant influence on the development of the brain, potentially leading to negative neuropsychological consequences such as autism spectrum disorder [[36](#CR36)36]. A study has revealed that there is a higher prevalence of vitamin D3 insufficiency in epileptic children under valproate therapy compared to healthy children [[37](#CR37)37].
|
| 126 |
+
|
| 127 |
+
Plasma Valproic acid (VPA) concentration and plasma concentrations of VPA were determined using LC-MS/MS, as shown in Table [2](#Tab2)2. When we Compare the results to the steady state mean concentration of VPA in the literature, we can classify them as sub-therapeutic, slightly high, high, or toxic [[16](#CR16)16].
|
| 128 |
+
|
| 129 |
+
### Table 2.
|
| 130 |
+
|
| 131 |
+
VPA plasma concentration
|
| 132 |
+
|
| 133 |
+
| Sample # | VPA Measured concentration (µg/mL) by LC-MS/MS |
|
| 134 |
+
| -------- | ---------------------------------------------- |
|
| 135 |
+
| 1 | 25.9 |
|
| 136 |
+
| 2 | 61.4 |
|
| 137 |
+
| 3 | 68.9 |
|
| 138 |
+
| 4 | 96.5 |
|
| 139 |
+
| 5 | 90.0 |
|
| 140 |
+
| 6 | 90.1 |
|
| 141 |
+
| 7 | 58.9 |
|
| 142 |
+
| 8 | 107.9 |
|
| 143 |
+
| 9 | 75.3 |
|
| 144 |
+
| 10 | 91.2 |
|
| 145 |
+
| 11 | 80.5 |
|
| 146 |
+
| 12 | 60.6 |
|
| 147 |
+
| 13 | 93.2 |
|
| 148 |
+
| 14 | 105.4 |
|
| 149 |
+
| Average | 79.0 |
|
| 150 |
+
| CV% | 28% |
|
| 151 |
+
The study identified various abnormal laboratory findings and VPA levels among the participants. Two participants (samples 8 and 14) exhibited toxic plasma VPA concentrations, exceeding 100 µg/mL, with levels recorded at 107.9 µg/mL and 105.4 µg/mL, respectively. These toxic levels were accompanied by elevated gamma-glutamyl transferase (GGT) levels, suggesting potential liver damage or bile duct dysfunction. One participant (sample 1) had a sub-therapeutic VPA level of 25.9 µg/mL, which was significantly below the recommended therapeutic range of 50–100 µg/mL. Several participants showed hematological abnormalities: samples 3, 4, 5, and 9 demonstrated low platelet counts when VPA concentrations exceeded 80 µg/mL, consistent with thrombocytopenia observed in VPA treatment. Hypochromic and microcytic red blood cells were indicated by decreased mean corpuscular hemoglobin (MCH) and mean corpuscular hemoglobin concentration (MCHC) values in samples 1, 3, and 9. Additionally, neutropenia was observed in sample 14, aligning with VPA’s documented effects on white blood cells. Low serum vitamin D3 levels were recorded in multiple participants, with sample 10 showing particularly low levels, which could have implications for bone health and neurodevelopment. These results emphasize the complex interplay between VPA levels and associated biochemical abnormalities, reinforcing the need for routine laboratory monitoring in patients receiving VPA therapy.
|
| 152 |
+
|
| 153 |
+
## Discussion
|
| 154 |
+
|
| 155 |
+
Individuals 3, 4, 5, 9, 10, 11, 12, and 14 showed signs of a potential correlation between VPA and haematological parameters, and sample numbers 4, 5, 6, and 13 showed signs of low platelet count when the plasma VPA level rose above 80 µg/mL, as documented in the literature [[38](#CR38)38]. In addition, there is a possible correlation between creatinine serum level and VPA serum level in individuals’ numbers (7, 9, 11, and 13). Sample number (8) has normal liver enzyme levels, but these levels may surge as the individual’s VPA level approaches toxic levels. Sample numbers (13 and 14) have elevated GGT levels, and there is a link between VPA plasma levels and GGT [[32](#CR32)32]. Sample numbers (3, 4, 5, 6, 8, 11, and 14) have low vitamin D3 levels, which could be linked to VPA plasma concentration.
|
| 156 |
+
|
| 157 |
+
Genotyping, as shown in Supplementary Materials S4, all participants were homozygous for SULT1A1 and CYP2D6-4, except for sample numbers (3 and 8) that were heterozygous abnormal. Sample number (8) has a toxic level of VPA, but sample number (3) has a therapeutic VPA level. Sample numbers (2 and 8) have heterozygous abnormal CYP 2C19*2; sample number (2) has a therapeutic VPA level, but sample number (8) has a toxic level of VPA. Sample number (3) has an abnormal heterozygosity for CYP6-10 and a therapeutic drug level. The classification of intellectual disability (ID) levels used in this study is based on established diagnostic criteria, categorizing ID into four severity levels: mild, moderate, severe, and profound. These classifications are determined by deficits in intellectual functioning, such as reasoning, problem-solving, and judgment, and adaptive functioning in conceptual, social, and practical domains. An IQ score below 70 is indicative of ID, with specific ranges defining the severity: mild (IQ 50–69), moderate (IQ 35–49), severe (IQ 20–34), and profound (IQ below 20). In this study, individuals were classified into these categories based on their clinical diagnoses, as shown in Supplementary Table S4. For example, participants 1, 2, and 5 were categorized as having moderate ID, while participants 3 and 4 were classified with mild ID. This classification provides a framework for understanding the variability in therapeutic outcomes and drug response among individuals with neurodevelopmental disorders.
|
| 158 |
+
|
| 159 |
+
Statistical analyses were conducted using SPSS version 28.0.0.0. Descriptive statistics were used to summarize demographic, clinical, and laboratory data, while inferential statistics were applied to evaluate the relationships between VPA plasma levels and genetic polymorphisms. Associations between categorical variables (e.g., genotype combinations and therapeutic drug monitoring (TDM) categories) were assessed using the chi-square (χ²) test, with statistical significance set at a p-value of < 0.05. Continuous variables (e.g., VPA plasma concentrations) were analyzed using independent samples t-tests or one-way ANOVA, as appropriate.
|
| 160 |
+
|
| 161 |
+
Power calculations were performed retrospectively to assess the adequacy of the sample size (*n*n = 14) for detecting significant associations. Given the small sample size, the study was underpowered for detecting small-to-moderate effect sizes (e.g., Cohen’s d < 0.5 or odds ratios < 2.0). For example, a sample size of 14 provides only 30% power to detect a moderate effect size (d = 0.5) at a significance level of 0.05. This limitation highlights the exploratory nature of the study, with findings intended to generate hypotheses for future research rather than provide definitive conclusions.
|
| 162 |
+
|
| 163 |
+
The small sample size was determined by the availability of participants meeting the inclusion criteria at the study site, specifically individuals with neurodevelopmental disorders receiving stable VPA therapy. While this limited sample size restricts the generalizability of the findings, the study offers valuable preliminary data that can inform the design of larger, more adequately powered studies in the future.
|
| 164 |
+
|
| 165 |
+
Pearson’s χ2 test was used to compare genotype and TDM category. The χ2 test was used to examine the statistical significance of the variations in SULT1A1, CYP3A5 + D, CYP2C19*2, CYP2D6*4, and 2D6*10 between individuals. Statistical significance was defined as a probability value of less than 0.05.
|
| 166 |
+
|
| 167 |
+
No statistically significant correlations were identified between VPA plasma concentrations and the genetic variants SULT1A1 (*p*p = 0.522), CYP2C192 (*p*p = 0.288), CYP2D64 (*p*p = 0.895), or CYP2D6*10 (*p*p = 0.067). Specifically, the p-values for SULT1A1 and CYP2D64 were 0.522 and 0.895, respectively, indicating no significant relationship as shown in Table [3](#Tab3)3. Similarly, the CYP2C192 variant showed a p-value of 0.288, and the CYP2D610 variant approached significance (*p*p = 0.067) but did not reach the threshold for statistical significance. Confidence intervals for the observed associations further demonstrated the lack of a strong relationship, as the intervals overlapped extensively with the null value. The χ² test for the genotype groups and VPA levels did not reveal any significant deviations, confirming that these genetic variants are unlikely to contribute to the variability in VPA plasma levels within this cohort. These results align with previous studies suggesting minimal impact of these genotypes on VPA pharmacokinetics.
|
| 168 |
+
|
| 169 |
+
### Table 3.
|
| 170 |
+
|
| 171 |
+
Statistical analysis for the association between the genotype and TDM category
|
| 172 |
+
|
| 173 |
+
| | SULT1A1 | CYP3A5 + D | CYP2C19*2 | CYP2D6*4 | CYP2D6*10 |
|
| 174 |
+
| - | ------- | ---------- | --------- | -------- | --------- |
|
| 175 |
+
| P-value | 0.522 | Constant | 0.067 | 0.288 | 0.895 |
|
| 176 |
+
| χ2 | 3.217 | Constant | 8.782 | 2.492 | 1.098 |
|
| 177 |
+
This investigation aimed to determine whether VPA and drug-metabolizing enzymes, the most significant cytochrome and sulfotransferases, were related in individuals with nervous system impairments or diseases. Our findings showed that specific genotypes’ proportions were increasing in certain groups. Compared to previous studies’ outcomes, these percentages from the harmful and regular TDM reference groups happened by accident rather than being statistically related. It was found that individualized VPA dosage regimens can be helpful for CYP2C19 genetic variations [[39](#CR39)39]. Allele CYP2C19*2 and CYP2C19*3 carriers exhibit greater trough plasma VPA concentrations than CYP2C19 wild-type patients, indicating that VPA plasma concentrations are strongly affected by CYP2C19 genetic variants and that the dose of VPA for intermediate and poor metabolizers may be lower than for extensive ones. Our study disagrees with them, and there are several explanations for this. One of these explanations could be the difference in the study community. They have females in their study, their mean age group is different, they have a larger sample size, and their nationality is various. Like our results, the CYP2C19 genotype was not clinically significant for VPA pharmacokinetic variability [[40](#CR40)40].
|
| 178 |
+
|
| 179 |
+
Therapeutic drug monitoring (TDM) of valproate (VPA) is a critical tool in clinical practice, enhancing the precision of treatment regimens and improving patient outcomes in neuropsychiatric disorders. A retrospective study in Saudi Arabia highlighted TDM’s effectiveness in managing mood stabilizers like VPA, ensuring therapeutic levels, and minimizing adverse effects, particularly in bipolar disorder patients [[41](#CR41)41]. Similarly, a 5-year analysis conducted in Italy demonstrated the frequent underdosing of VPA in clinical settings, underscoring the role of TDM in preventing subtherapeutic levels and optimizing treatment efficacy [[42](#CR42)42]. Furthermore, the AGNP Consensus Guidelines strongly recommend TDM for valproate, recognizing its value in individualizing treatment and reducing the risk of toxicity [[43](#CR43)43]. These findings emphasize the indispensable role of TDM in ensuring the safe and effective use of valproate in diverse clinical scenarios.
|
| 180 |
+
|
| 181 |
+
The study has limitations that should be acknowledged to contextualize the findings. First, the sample size (*n*n = 14) is small, which limits the statistical power to detect genetic associations and reduces the generalizability of the results to broader populations. The small sample size may increase the risk of type II errors, where significant associations could be missed. Second, the absence of female participants restricts the study’s applicability to only male populations, neglecting potential sex-specific differences in the pharmacokinetics and pharmacodynamics of valproate. Additionally, the study does not account for potential confounding variables such as comorbid conditions, co-administered medications, or environmental factors that could influence drug metabolism and therapeutic outcomes. These limitations highlight the need for larger, more diverse studies to validate the findings and explore the broader implications of genotype-guided therapy for valproate in neurodevelopmental disorders.
|
| 182 |
+
|
| 183 |
+
## Conclusions
|
| 184 |
+
|
| 185 |
+
This study highlights the complexity of valproic acid (VPA) therapy in individuals with neurodevelopmental disorders and the limited influence of common genetic polymorphisms in CYP and SULT genes on VPA plasma levels. While therapeutic drug monitoring (TDM) remains an invaluable tool for optimizing VPA therapy, the lack of significant correlations between genetic variants and VPA concentrations suggests that routine pharmacogenetic testing for these specific variants may not be warranted in clinical practice. However, the observed toxic and sub-therapeutic VPA levels emphasize the importance of regular TDM to mitigate risks associated with overdose or insufficient dosing.
|
| 186 |
+
|
| 187 |
+
Based on these findings, healthcare providers should prioritize individualized TDM and integrate laboratory assessments, such as liver function and hematological profiles, into routine monitoring for patients on VPA therapy. Future studies should explore larger, more diverse populations, including females and individuals with co-existing medical conditions, to validate these findings. Additionally, further research should focus on identifying other genetic, epigenetic, or environmental factors that might influence VPA pharmacokinetics and therapeutic outcomes. Finally, studies assessing the cost-effectiveness and clinical utility of genotype-guided VPA therapy would provide valuable insights for integrating pharmacogenomics into standard care.
|
| 188 |
+
|
| 189 |
+
## Electronic supplementary material
|
| 190 |
+
|
| 191 |
+
Below is the link to the electronic supplementary material.
|
| 192 |
+
|
| 193 |
+
## Acknowledgements
|
| 194 |
+
|
| 195 |
+
We thank the University of Petra and the Jordan Center for Pharmaceutical Research for their support.
|
| 196 |
+
|
| 197 |
+
## Abbreviations
|
| 198 |
+
|
| 199 |
+
## Author contributions
|
| 200 |
+
|
| 201 |
+
Each author contributed to the analysis and interpretation of the findings. Additionally, the authors consented to accept the work in its submitted form. Conceptualization, E.M., and T.A.; methodology, A.A. and K.M.; software, A.A., E.M., L.A., and S.A.; validation, S.A., S.K., E.M., and A.A.; formal analysis, S.A., and S.K.; resources, T.A.; data curation, E.M., A.A., and S.A.; writing—original draft preparation, E.M., L.A., and K.W.O.; writing—review and editing, T.A., A.A., E.M., K.W.O., and O.M.; Verification, K.W.O. and O.M.; supervision, E.M., and L.A; project administration, T.A.; funding acquisition, T.A. The authors reviewed and approved the final version of the published manuscript for significant intellectual content.
|
| 202 |
+
|
| 203 |
+
## Data availability
|
| 204 |
+
|
| 205 |
+
No datasets were generated or analysed during the current study.
|
| 206 |
+
|
| 207 |
+
## Declarations
|
| 208 |
+
|
| 209 |
+
### Ethics approval and consent to participate
|
| 210 |
+
|
| 211 |
+
The Helsinki Declaration was followed when conducting the study, the ICH HARMONISED GUIDELINE: INTEGRATED ADDENDUM TO ICH E6(R1): GUIDELINE FOR GOOD CLINICAL PRACTICE E6(R2) dated November 9, 2016. (ICH 2016). The Jordan Center of Pharmaceutical Research’s Institutional Review Board (IRB) also approved it locally. All individuals and/or their legal representatives gave written informed consent before entering the study. All procedures, interventions, and laboratory tests were conducted per the Jordanian Ministry of Health’s recommendations.
|
| 212 |
+
|
| 213 |
+
### Consent for publication
|
| 214 |
+
|
| 215 |
+
Not applicable.
|
| 216 |
+
|
| 217 |
+
### Competing interests
|
| 218 |
+
|
| 219 |
+
The authors declare no competing interests.
|
| 220 |
+
|
| 221 |
+
## Footnotes
|
| 222 |
+
|
| 223 |
+
## Contributor Information
|
| 224 |
+
|
| 225 |
+
Eyad Mallah, Email: emallah@uop.edu.jo.
|
| 226 |
+
|
| 227 |
+
Khaled W. Omari, Email: khaled.omari@aum.edu.kw
|
| 228 |
+
|
| 229 |
+
## Associated Data
|
| 230 |
+
|
| 231 |
+
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
|
| 232 |
+
|
| 233 |
+
### Supplementary Materials
|
| 234 |
+
|
| 235 |
+
### Data Availability Statement
|
| 236 |
+
|
| 237 |
+
No datasets were generated or analysed during the current study.
|
| 238 |
+
|
| 239 |
+
### Supplementary Materials
|
| 240 |
+
|
| 241 |
+
### Data Availability Statement
|
| 242 |
+
|
| 243 |
+
No datasets were generated or analysed during the current study.
|
| 244 |
+
|
| 245 |
+
## References
|
| 246 |
+
|
| 247 |
+
1. Patel DR, Cabral MD, Ho A, Merrick J. A clinical primer on intellectual disability. Transl Pediatr. 2020;9(Suppl 1):S23–35. [DOI](https://doi.org/10.21037/tp.2020.02.02) | [PMC free article](/articles/PMC7082244/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32206581/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Patel%20DR,%20Cabral%20MD,%20Ho%20A,%20Merrick%20J.%20A%20clinical%20primer%20on%20intellectual%20disability.%20Transl%20Pediatr.%202020;9(Suppl%201):S23%E2%80%9335.)
|
| 248 |
+
|
| 249 |
+
2. Committee to Evaluate the Supplemental Security Income Disability Program for Children with Mental Disorders; Board on the Health of Select Populations; Board on Children, Youth, and Families; Institute of Medicine; Division of Behavioral and Social Sciences and Education; The National Academies of Sciences, Engineering, and Medicine; Boat TF, Wu JT, editors. Mental Disorders and Disabilities Among Low-Income Children. Washington (DC): National Academies Press (US). 2015 Oct 28. 14, Prevalence of Autism Spectrum Disorder. https://nap.nationalacademies.org/initiative/committee-to-evaluate-the-supplemental-security-income-disability-program-for-children-with-mental-disorders. Accessed 22 Oct 2021. [https://nap.nationalacademies.org/initiative/committee-to-evaluate-the-supplemental-security-income-disability-program-for-children-with-mental-disorders](https://nap.nationalacademies.org/initiative/committee-to-evaluate-the-supplemental-security-income-disability-program-for-children-with-mental-disorders) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/26632628/)
|
| 250 |
+
|
| 251 |
+
3. Zablotsky B, Ng AE, Black LI, Blumberg SJ. Diagnosed developmental disabilities in children aged 3–17 years: United States, 2019–2021. Natural Center for Health Statistics. 2023;473. [PubMed](https://pubmed.ncbi.nlm.nih.gov/37440277/)
|
| 252 |
+
|
| 253 |
+
4. Bottini SB, Morton HE, Buchanan KA, Gould K. Moving from disorder to difference: a systematic review of recent language use in autism research. Autism Adulthood. 2024;6(2):128–40. [DOI](https://doi.org/10.1089/aut.2023.0030) | [PMC free article](/articles/PMC11319857/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/39144072/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Bottini%20SB,%20Morton%20HE,%20Buchanan%20KA,%20Gould%20K.%20Moving%20from%20disorder%20to%20difference:%20a%20systematic%20review%20of%20recent%20language%20use%20in%20autism%20research.%20Autism%20Adulthood.%202024;6(2):128%E2%80%9340.)
|
| 254 |
+
|
| 255 |
+
5. Sharma S, Gonda X, Tarazi F. Autism spectrum disorder: classification, diagnosis and therapy. Pharmacol Ther. 2018;190:91–104. [DOI](https://doi.org/10.1016/j.pharmthera.2018.05.007) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29763648/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Sharma%20S,%20Gonda%20X,%20Tarazi%20F.%20Autism%20spectrum%20disorder:%20classification,%20diagnosis%20and%20therapy.%20Pharmacol%20Ther.%202018;190:91%E2%80%93104.)
|
| 256 |
+
|
| 257 |
+
6. Lord C, Risi S, DiLavore P, Shulman C, Thurm A, Pickles A. Autism from 2 to 9 years of age. Arch Gen Psychiatry. 2006;63(6):694. [DOI](https://doi.org/10.1001/archpsyc.63.6.694) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16754843/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Lord%20C,%20Risi%20S,%20DiLavore%20P,%20Shulman%20C,%20Thurm%20A,%20Pickles%20A.%20Autism%20from%202%20to%209%20years%20of%20age.%20Arch%20Gen%20Psychiatry.%202006;63(6):694.)
|
| 258 |
+
|
| 259 |
+
7. Geschwind D, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17(1):103–11. [DOI](https://doi.org/10.1016/j.conb.2007.01.009) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17275283/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Geschwind%20D,%20Levitt%20P.%20Autism%20spectrum%20disorders:%20developmental%20disconnection%20syndromes.%20Curr%20Opin%20Neurobiol.%202007;17(1):103%E2%80%9311.)
|
| 260 |
+
|
| 261 |
+
8. El Achkar C, Spence S. Clinical characteristics of children and young adults with co-occurring autism spectrum disorder and epilepsy. Epilepsy Behav. 2015;47:183–90. [DOI](https://doi.org/10.1016/j.yebeh.2014.12.022) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/25599987/) | [Google Scholar](https://scholar.google.com/scholar_lookup?El%20Achkar%20C,%20Spence%20S.%20Clinical%20characteristics%20of%20children%20and%20young%20adults%20with%20co-occurring%20autism%20spectrum%20disorder%20and%20epilepsy.%20Epilepsy%20Behav.%202015;47:183%E2%80%9390.)
|
| 262 |
+
|
| 263 |
+
9. Lukmanji S, Manji SA, Kadhim S, Sauro KM, Wirrell EC, Kwon CS, Jette N. The co-occurrence of epilepsy and autism: a systematic review. Epilepsy Behav. 2019;98:238–48. [DOI](https://doi.org/10.1016/j.yebeh.2019.07.037) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31398688/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Lukmanji%20S,%20Manji%20SA,%20Kadhim%20S,%20Sauro%20KM,%20Wirrell%20EC,%20Kwon%20CS,%20Jette%20N.%20The%20co-occurrence%20of%20epilepsy%20and%20autism:%20a%20systematic%20review.%20Epilepsy%20Behav.%202019;98:238%E2%80%9348.)
|
| 264 |
+
|
| 265 |
+
10. Löscher W. Basic Pharmacology of Valproate. CNS Drugs. 2002;16(10):669–94. [DOI](https://doi.org/10.2165/00023210-200216100-00003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12269861/) | [Google Scholar](https://scholar.google.com/scholar_lookup?L%C3%B6scher%20W.%20Basic%20Pharmacology%20of%20Valproate.%20CNS%20Drugs.%202002;16(10):669%E2%80%9394.)
|
| 266 |
+
|
| 267 |
+
11. Fisher C, Broderick W. Sodium valproate or valproate semisodium: is there a difference in the treatment of bipolar disorder. Psychiatr Bull. 2003;27(12):446–8. [Google Scholar](https://scholar.google.com/scholar_lookup?Fisher%20C,%20Broderick%20W.%20Sodium%20valproate%20or%20valproate%20semisodium:%20is%20there%20a%20difference%20in%20the%20treatment%20of%20bipolar%20disorder.%20Psychiatr%20Bull.%202003;27(12):446%E2%80%938.)
|
| 268 |
+
|
| 269 |
+
12. Citrome L. Valproate: do formulations matter? J Clin Pharm Ther. 2008;33(4):457–457. [DOI](https://doi.org/10.1111/j.1365-2710.2008.00919_1.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18613865/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Citrome%20L.%20Valproate:%20do%20formulations%20matter?%20J%20Clin%20Pharm%20Ther.%202008;33(4):457%E2%80%93457.)
|
| 270 |
+
|
| 271 |
+
13. Acharya S, Bussel J. Hematologic toxicity of Sodium Valproate. J Pediatr Hematol Oncol. 2000;22(1):62–5. [DOI](https://doi.org/10.1097/00043426-200001000-00012) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10695824/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Acharya%20S,%20Bussel%20J.%20Hematologic%20toxicity%20of%20Sodium%20Valproate.%20J%20Pediatr%20Hematol%20Oncol.%202000;22(1):62%E2%80%935.)
|
| 272 |
+
|
| 273 |
+
14. Gupta T. Valproate-induced drug Rash Eosinophilia with systemic symptoms syndrome: an unknown hepatotoxicity. Euroasian J Hepatogastroenterol. 2019;9(2):102–3. [DOI](https://doi.org/10.5005/jp-journals-10018-1298) | [PMC free article](/articles/PMC7047308/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32117699/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Gupta%20T.%20Valproate-induced%20drug%20Rash%20Eosinophilia%20with%20systemic%20symptoms%20syndrome:%20an%20unknown%20hepatotoxicity.%20Euroasian%20J%20Hepatogastroenterol.%202019;9(2):102%E2%80%933.)
|
| 274 |
+
|
| 275 |
+
15. Huang W, Ren X, Shen F, Xing B. Sodium valproate induced acute pancreatitis in a bipolar disorder patient: a case report. BMC Pharmacol Toxicol. 2019;20(1):71. [DOI](https://doi.org/10.1186/s40360-019-0373-z) | [PMC free article](/articles/PMC6884746/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/31783774/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Huang%20W,%20Ren%20X,%20Shen%20F,%20Xing%20B.%20Sodium%20valproate%20induced%20acute%20pancreatitis%20in%20a%20bipolar%20disorder%20patient:%20a%20case%20report.%20BMC%20Pharmacol%20Toxicol.%202019;20(1):71.)
|
| 276 |
+
|
| 277 |
+
16. Methaneethorn J. A systematic review of population pharmacokinetics of valproic acid. Br Br J Clin Pharmacol. 2018;84(5):816–34. [DOI](https://doi.org/10.1111/bcp.13510) | [PMC free article](/articles/PMC5903263/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29328514/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Methaneethorn%20J.%20A%20systematic%20review%20of%20population%20pharmacokinetics%20of%20valproic%20acid.%20Br%20Br%20J%20Clin%20Pharmacol.%202018;84(5):816%E2%80%9334.)
|
| 278 |
+
|
| 279 |
+
17. Patsalos P, Berry D, Bourgeois B, Cloyd J, Glauser T, Johannessen S, Leppik I, Tomson T, Perucca E. Antiepileptic drugsbest practice guidelines for therapeutic drug monitoring: a position paper by the subcommission on therapeutic drug monitoring, ILAE Commission on therapeutic strategies. Epilepsia. 2008;49(7):1239–76. [DOI](https://doi.org/10.1111/j.1528-1167.2008.01561.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/18397299/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Patsalos%20P,%20Berry%20D,%20Bourgeois%20B,%20Cloyd%20J,%20Glauser%20T,%20Johannessen%20S,%20Leppik%20I,%20Tomson%20T,%20Perucca%20E.%20Antiepileptic%20drugsbest%20practice%20guidelines%20for%20therapeutic%20drug%20monitoring:%20a%20position%20paper%20by%20the%20subcommission%20on%20therapeutic%20drug%20monitoring,%20ILAE%20Commission%20on%20therapeutic%20strategies.%20Epilepsia.%202008;49(7):1239%E2%80%9376.)
|
| 280 |
+
|
| 281 |
+
18. Vasudev K, Das S, Goswami U, Tayal G. Pharmacokinetics of valproic acid in patients with bipolar disorder. J Psychopharmacol. 2001;15(3):187–90. [DOI](https://doi.org/10.1177/026988110101500305) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11565626/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Vasudev%20K,%20Das%20S,%20Goswami%20U,%20Tayal%20G.%20Pharmacokinetics%20of%20valproic%20acid%20in%20patients%20with%20bipolar%20disorder.%20J%20Psychopharmacol.%202001;15(3):187%E2%80%9390.)
|
| 282 |
+
|
| 283 |
+
19. Morris P. Compliance Profile of Depakote ER compared to Depakote DR and Valproic Acid in Bipolar patients. J Correctional Health Care. 2008;14(4):311–7. [Google Scholar](https://scholar.google.com/scholar_lookup?Morris%20P.%20Compliance%20Profile%20of%20Depakote%20ER%20compared%20to%20Depakote%20DR%20and%20Valproic%20Acid%20in%20Bipolar%20patients.%20J%20Correctional%20Health%20Care.%202008;14(4):311%E2%80%937.)
|
| 284 |
+
|
| 285 |
+
20. Patsalos P, Spencer E. Therapeutic drug monitoring of antiepileptic drugs in Epilepsy: a 2018 Update. Ther Drug Monit. 2018;40(5):526–48. [DOI](https://doi.org/10.1097/FTD.0000000000000546) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/29957667/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Patsalos%20P,%20Spencer%20E.%20Therapeutic%20drug%20monitoring%20of%20antiepileptic%20drugs%20in%20Epilepsy:%20a%202018%20Update.%20Ther%20Drug%20Monit.%202018;40(5):526%E2%80%9348.)
|
| 286 |
+
|
| 287 |
+
21. Fernández-Campos F, Calpena A, Soy D, Colom H. Determination of total and unbound valproic acid concentrations in human plasma by liquid chromatography-tandem mass spectrometry. J Liq Chromatogr Relat Technol. 2012;35(9):1171–83. [Google Scholar](https://scholar.google.com/scholar_lookup?Fern%C3%A1ndez-Campos%20F,%20Calpena%20A,%20Soy%20D,%20Colom%20H.%20Determination%20of%20total%20and%20unbound%20valproic%20acid%20concentrations%20in%20human%20plasma%20by%20liquid%20chromatography-tandem%20mass%20spectrometry.%20J%20Liq%20Chromatogr%20Relat%20Technol.%202012;35(9):1171%E2%80%9383.)
|
| 288 |
+
|
| 289 |
+
22. Gamage N, Barnett A, Hempel N, Duggleby R, Windmill K, Martin J, McManus M. Human sulfotransferases and their role in Chemical metabolism. Toxicol Sci. 2005;90(1):5–22. [DOI](https://doi.org/10.1093/toxsci/kfj061) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16322073/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Gamage%20N,%20Barnett%20A,%20Hempel%20N,%20Duggleby%20R,%20Windmill%20K,%20Martin%20J,%20McManus%20M.%20Human%20sulfotransferases%20and%20their%20role%20in%20Chemical%20metabolism.%20Toxicol%20Sci.%202005;90(1):5%E2%80%9322.)
|
| 290 |
+
|
| 291 |
+
23. Bustami M, Al-Shudifat A, Hussein N, Yacoub M, Atwa E, Sabri I, Abu-Hamdah R, Abu-Rayyan W, Arafat T, Badran A, Abu-Qatouseh L. Impact of genetic polymorphism of Sulpha transferase genes (SULT1A) genes on the risk of females with breast Cancer in Jordan. Preprints. 2018. 10.20944/preprints201710.0050.v2. Accessed 22 Oct 2021. [Google Scholar](https://scholar.google.com/scholar_lookup?Bustami%20M,%20Al-Shudifat%20A,%20Hussein%20N,%20Yacoub%20M,%20Atwa%20E,%20Sabri%20I,%20Abu-Hamdah%20R,%20Abu-Rayyan%20W,%20Arafat%20T,%20Badran%20A,%20Abu-Qatouseh%20L.%20Impact%20of%20genetic%20polymorphism%20of%20Sulpha%20transferase%20genes%20(SULT1A)%20genes%20on%20the%20risk%20of%20females%20with%20breast%20Cancer%20in%20Jordan.%20Preprints.%202018.%2010.20944/preprints201710.0050.v2.%20Accessed%2022%20Oct%202021.)
|
| 292 |
+
|
| 293 |
+
24. European Medicines Agency. Guideline on process validation for finished products - information and data to be provided in regulatory submissions. 2016. https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-process-validation-finished-products-information-data-be-provided-regulatory-submissions_en.pdf. Accessed 22 Oct 2021. [https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-process-validation-finished-products-information-data-be-provided-regulatory-submissions_en.pdf](https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-process-validation-finished-products-information-data-be-provided-regulatory-submissions_en.pdf)
|
| 294 |
+
|
| 295 |
+
25. European Medicines Agency. Overview of the Agency’s contribution to science, medicines and health in the European Union. 2011. chromehttps://www.ema.europa.eu/en/documents/annual-report/annual-report-european-medicines-agency-2011_en.pdf. Accessed 22 Oct 2021. [https://www.ema.europa.eu/en/documents/annual-report/annual-report-european-medicines-agency-2011_en.pdf](https://www.ema.europa.eu/en/documents/annual-report/annual-report-european-medicines-agency-2011_en.pdf)
|
| 296 |
+
|
| 297 |
+
26. Johnson-Wimbley T, Graham D. Diagnosis and management of iron deficiency anemia in the 21st century. Th Adv Gastroenterol. 2011;4(3):177–84. [DOI](https://doi.org/10.1177/1756283X11398736) | [PMC free article](/articles/PMC3105608/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/21694802/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Johnson-Wimbley%20T,%20Graham%20D.%20Diagnosis%20and%20management%20of%20iron%20deficiency%20anemia%20in%20the%2021st%20century.%20Th%20Adv%20Gastroenterol.%202011;4(3):177%E2%80%9384.)
|
| 298 |
+
|
| 299 |
+
27. Gunes S, Ekinci O, Celik T. Iron deficiency parameters in autism spectrum disorder: clinical correlates and associated factors. Ital J Pediatr. 2017;43(1):86. [DOI](https://doi.org/10.1186/s13052-017-0407-3) | [PMC free article](/articles/PMC5609017/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28934988/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Gunes%20S,%20Ekinci%20O,%20Celik%20T.%20Iron%20deficiency%20parameters%20in%20autism%20spectrum%20disorder:%20clinical%20correlates%20and%20associated%20factors.%20Ital%20J%20Pediatr.%202017;43(1):86.)
|
| 300 |
+
|
| 301 |
+
28. George-Gay B, Parker K. Understanding the complete blood count with differential. J Perianesth Nurs. 2003;18(2):96–117. [DOI](https://doi.org/10.1053/jpan.2003.50013) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12710004/) | [Google Scholar](https://scholar.google.com/scholar_lookup?George-Gay%20B,%20Parker%20K.%20Understanding%20the%20complete%20blood%20count%20with%20differential.%20J%20Perianesth%20Nurs.%202003;18(2):96%E2%80%93117.)
|
| 302 |
+
|
| 303 |
+
29. Krause K, Berlit P, Schmidt-Gayk H, Schellenberg B. Antiepileptic drugs reduce serum uric acid. Epilepsy Res. 1987;1(5):306–7. [DOI](https://doi.org/10.1016/0920-1211(87)90007-6) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/3143553/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Krause%20K,%20Berlit%20P,%20Schmidt-Gayk%20H,%20Schellenberg%20B.%20Antiepileptic%20drugs%20reduce%20serum%20uric%20acid.%20Epilepsy%20Res.%201987;1(5):306%E2%80%937.)
|
| 304 |
+
|
| 305 |
+
30. Nishida Y. Relation between creatinine and uric acid excretion. Ann Rheum Dis. 1992;51(1):101–2. [DOI](https://doi.org/10.1136/ard.51.1.101) | [PMC free article](/articles/PMC1004629/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1540011/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Nishida%20Y.%20Relation%20between%20creatinine%20and%20uric%20acid%20excretion.%20Ann%20Rheum%20Dis.%201992;51(1):101%E2%80%932.)
|
| 306 |
+
|
| 307 |
+
31. Maneenin C, Lapyuneyong N, Tongpan S, Yannasithinon S, Burawat J, Maneenin N, Sukhorum W, Arun S, Iamsaard S. The alterations of microvasculature, tyrosine phosphorylation, and lipid peroxidation in kidney of rats treated with valproic acid. Int J Morphol. 2019;37(1):65–70. [Google Scholar](https://scholar.google.com/scholar_lookup?Maneenin%20C,%20Lapyuneyong%20N,%20Tongpan%20S,%20Yannasithinon%20S,%20Burawat%20J,%20Maneenin%20N,%20Sukhorum%20W,%20Arun%20S,%20Iamsaard%20S.%20The%20alterations%20of%20microvasculature,%20tyrosine%20phosphorylation,%20and%20lipid%20peroxidation%20in%20kidney%20of%20rats%20treated%20with%20valproic%20acid.%20Int%20J%20Morphol.%202019;37(1):65%E2%80%9370.)
|
| 308 |
+
|
| 309 |
+
32. HadzagicCatibusic F, Hasanbegovic E, Melunovic M, Zubcevic S, Uzicanin S. Effects of Carbamazepine and Valproate on serum aspartate aminotransferase, Alanine Aminotransferase and Gamma - Glutamyltransferase in Children. Med Arch. 2017;71(4):239. [DOI](https://doi.org/10.5455/medarh.2017.71.239-242) | [PMC free article](/articles/PMC5585805/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28974841/) | [Google Scholar](https://scholar.google.com/scholar_lookup?HadzagicCatibusic%20F,%20Hasanbegovic%20E,%20Melunovic%20M,%20Zubcevic%20S,%20Uzicanin%20S.%20Effects%20of%20Carbamazepine%20and%20Valproate%20on%20serum%20aspartate%20aminotransferase,%20Alanine%20Aminotransferase%20and%20Gamma%20-%20Glutamyltransferase%20in%20Children.%20Med%20Arch.%202017;71(4):239.)
|
| 310 |
+
|
| 311 |
+
33. Ratnaike RN, Schapel GJ, Purdie G, Rischbieth RH, Hoffmann S. Hyperammonaemia and hepatotoxicity during chronic valproate therapy: enhancement by combination with other antiepileptic drugs. Br J Clin Pharmacol. 1986;22(1):100–3. [PMC free article](/articles/PMC1401093/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/3091053/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Ratnaike%20RN,%20Schapel%20GJ,%20Purdie%20G,%20Rischbieth%20RH,%20Hoffmann%20S.%20Hyperammonaemia%20and%20hepatotoxicity%20during%20chronic%20valproate%20therapy:%20enhancement%20by%20combination%20with%20other%20antiepileptic%20drugs.%20Br%20J%20Clin%20Pharmacol.%201986;22(1):100%E2%80%933.)
|
| 312 |
+
|
| 313 |
+
34. Tripathi N, Jialal I, Conjugated H. 2023 Jul 24. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan–. PMID: 32965843. https://pubmed.ncbi.nlm.nih.gov/32965843/#:~:text=Pathologic%20elevation%20of%20conjugated%20or,is%20derived%20from%20hemoglobin%20metabolism. Accessed 22 Oct 2021. [PubMed](https://pubmed.ncbi.nlm.nih.gov/32965843/#:~:text=Pathologic%20elevation%20of%20conjugated%20or,is%20derived%20from%20hemoglobin%20metabolism)
|
| 314 |
+
|
| 315 |
+
35. Vainionpää L, Mikkonen K, Rättyä J, Knip M, Pakarinen A, Myllylä V, Isojärvi J. Thyroid function in girls with Epilepsy with Carbamazepine, Oxcarbazepine, or Valproate Monotherapy and after Withdrawal of Medication. Epilepsia. 2004;45(3):197–203. [DOI](https://doi.org/10.1111/j.0013-9580.2004.26003.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15009219/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Vainionp%C3%A4%C3%A4%20L,%20Mikkonen%20K,%20R%C3%A4tty%C3%A4%20J,%20Knip%20M,%20Pakarinen%20A,%20Myllyl%C3%A4%20V,%20Isoj%C3%A4rvi%20J.%20Thyroid%20function%20in%20girls%20with%20Epilepsy%20with%20Carbamazepine,%20Oxcarbazepine,%20or%20Valproate%20Monotherapy%20and%20after%20Withdrawal%20of%20Medication.%20Epilepsia.%202004;45(3):197%E2%80%93203.)
|
| 316 |
+
|
| 317 |
+
36. Siracusano M, Riccioni A, Abate R, Benvenuto A, Curatolo P, Mazzone L. Vitamin D Deficiency and Autism Spectrum Disorder. Curr Pharm Des. 2020;26(21):2460–74. [DOI](https://doi.org/10.2174/1381612826666200415174311) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32294031/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Siracusano%20M,%20Riccioni%20A,%20Abate%20R,%20Benvenuto%20A,%20Curatolo%20P,%20Mazzone%20L.%20Vitamin%20D%20Deficiency%20and%20Autism%20Spectrum%20Disorder.%20Curr%20Pharm%20Des.%202020;26(21):2460%E2%80%9374.)
|
| 318 |
+
|
| 319 |
+
37. Ameena AT, Zaher TM. Vitamin D status in epileptic children on Valproic Acid; a case-control study. Archives Acad Emerg Med. 2020;8(1):e13. [PMC free article](/articles/PMC7130439/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32259112/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Ameena%20AT,%20Zaher%20TM.%20Vitamin%20D%20status%20in%20epileptic%20children%20on%20Valproic%20Acid;%20a%20case-control%20study.%20Archives%20Acad%20Emerg%20Med.%202020;8(1):e13.)
|
| 320 |
+
|
| 321 |
+
38. Vasudev K, Keown P, Gibb I, McAllister-Williams R. Hematological effects of valproate in psychiatric patients: what are the risk factors? J Clin Psychopharmacol. 2010;30(3):282–5. [DOI](https://doi.org/10.1097/JCP.0b013e3181db2684) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/20473063/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Vasudev%20K,%20Keown%20P,%20Gibb%20I,%20McAllister-Williams%20R.%20Hematological%20effects%20of%20valproate%20in%20psychiatric%20patients:%20what%20are%20the%20risk%20factors?%20J%20Clin%20Psychopharmacol.%202010;30(3):282%E2%80%935.)
|
| 322 |
+
|
| 323 |
+
39. Song C, Li X, Mao P, Song W, Liu L, Zhang Y. Impact of CYP2C19 and CYP2C9 gene polymorphisms on sodium valproate plasma concentration in patients with epilepsy. Eur J Hosp Pharm. 2022;29(4):198–201. [DOI](https://doi.org/10.1136/ejhpharm-2020-002367) | [PMC free article](/articles/PMC9251156/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/32868386/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Song%20C,%20Li%20X,%20Mao%20P,%20Song%20W,%20Liu%20L,%20Zhang%20Y.%20Impact%20of%20CYP2C19%20and%20CYP2C9%20gene%20polymorphisms%20on%20sodium%20valproate%20plasma%20concentration%20in%20patients%20with%20epilepsy.%20Eur%20J%20Hosp%20Pharm.%202022;29(4):198%E2%80%93201.)
|
| 324 |
+
|
| 325 |
+
40. Smith R, Haslemo T, Refsum H, Molden E. Impact of age, gender and CYP2C9/2C19 genotypes on dose-adjusted steady-state serum concentrations of valproic acid—a large-scale study based on naturalistic therapeutic drug monitoring data. Eur J Clin Pharmacol. 2016;72(9):1099–104. [DOI](https://doi.org/10.1007/s00228-016-2087-0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27353638/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Smith%20R,%20Haslemo%20T,%20Refsum%20H,%20Molden%20E.%20Impact%20of%20age,%20gender%20and%20CYP2C9/2C19%20genotypes%20on%20dose-adjusted%20steady-state%20serum%20concentrations%20of%20valproic%20acid%E2%80%94a%20large-scale%20study%20based%20on%20naturalistic%20therapeutic%20drug%20monitoring%20data.%20Eur%20J%20Clin%20Pharmacol.%202016;72(9):1099%E2%80%93104.)
|
| 326 |
+
|
| 327 |
+
41. Abu-Qurain H, Almashhad F, Lucca JM, Abumadini MS. A retrospective study on therapeutic drug monitoring of mood stabilizers in real-life clinical scenario. J Pharm Bioallied Sci. 2020;12(3):351–5. [DOI](https://doi.org/10.4103/jpbs.JPBS_368_19) | [PMC free article](/articles/PMC7574742/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/33100796/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Abu-Qurain%20H,%20Almashhad%20F,%20Lucca%20JM,%20Abumadini%20MS.%20A%20retrospective%20study%20on%20therapeutic%20drug%20monitoring%20of%20mood%20stabilizers%20in%20real-life%20clinical%20scenario.%20J%20Pharm%20Bioallied%20Sci.%202020;12(3):351%E2%80%935.)
|
| 328 |
+
|
| 329 |
+
42. Biso L, Carli M, Kolachalam S, Monticelli G, Calabrò PF, di Paolo A, Giorgi FS, Bocci G, Scarselli M. A 5-Year study of antiseizure medications (ASMs) monitoring in patients with neuropsychiatric disorders in an Italian clinical Center. Pharmaceuticals. 2023;16(7):945. [DOI](https://doi.org/10.3390/ph16070945) | [PMC free article](/articles/PMC10383891/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/37513857/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Biso%20L,%20Carli%20M,%20Kolachalam%20S,%20Monticelli%20G,%20Calabr%C3%B2%20PF,%20di%20Paolo%20A,%20Giorgi%20FS,%20Bocci%20G,%20Scarselli%20M.%20A%205-Year%20study%20of%20antiseizure%20medications%20(ASMs)%20monitoring%20in%20patients%20with%20neuropsychiatric%20disorders%20in%20an%20Italian%20clinical%20Center.%20Pharmaceuticals.%202023;16(7):945.)
|
| 330 |
+
|
| 331 |
+
43. Hiemke C, Bergemann N, Clement HW, Conca A, Deckert J, Domschke K, Eckermann G, Egberts K, Gerlach M, Greiner C, Gründer G. Consensus guidelines for therapeutic drug monitoring in neuropsychopharmacology: update 2017. Pharmacopsychiatry. 2018;51(01/02):9–62. [DOI](https://doi.org/10.1055/s-0043-116492) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/28910830/) | [Google Scholar](https://scholar.google.com/scholar_lookup?Hiemke%20C,%20Bergemann%20N,%20Clement%20HW,%20Conca%20A,%20Deckert%20J,%20Domschke%20K,%20Eckermann%20G,%20Egberts%20K,%20Gerlach%20M,%20Greiner%20C,%20Gr%C3%BCnder%20G.%20Consensus%20guidelines%20for%20therapeutic%20drug%20monitoring%20in%20neuropsychopharmacology:%20update%202017.%20Pharmacopsychiatry.%202018;51(01/02):9%E2%80%9362.)
|
test/texts/PMC1365072.md
ADDED
|
@@ -0,0 +1,49 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Biotransformation and pharmacokinetics of ethylmorphine after a single oral dose
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** T A Aasmundstad, B Q Xu, I Johansson, A Ripel, A Bjørneboe, A S Christophersen, E Bodd, J Mørland
|
| 5 |
+
**Journal:** British Journal of Clinical Pharmacology
|
| 6 |
+
**Date:** 1995 Jun
|
| 7 |
+
**DOI:** [10.1111/j.1365-2125.1995.tb05720.x](https://doi.org/10.1111/j.1365-2125.1995.tb05720.x)
|
| 8 |
+
**PMID:** 7654478
|
| 9 |
+
**PMCID:** PMC1365072
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1365072/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC1365072/pdf/brjclinpharm00008-0026.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC1365072/pdf/brjclinpharm00008-0026.pdf)
|
| 12 |
+
|
| 13 |
+
*Note: This is a scanned document with limited structured text. Full content available in PDF.*
|
| 14 |
+
|
| 15 |
+
## Abstract
|
| 16 |
+
|
| 17 |
+
1. The pharmacokinetics of ethylmorphine after administration of a single dose of the cough mixture Cosylan were investigated in 10 healthy subjects. 2. The median urinary recovery of ethylmorphine and measured metabolites was 77% over 48 h. The median tmax of unchanged ethylmorphine was 45 min, and the terminal elimination t1/2 was 2 h. Ethylmorphine-6-glucuronide was found to be the major metabolite. 3. Two subjects had significantly lower urinary recovery (0.48 h) of morphine and morphine-glucuronides than the remainder. Furthermore, these two had urinary metabolic ratios (MRO) and partial metabolic clearances (CLmO) for O-deethylation of ethylmorphine tentatively classifying them phenotypically as poor metabolisers of the debrisoquine/sparteine type. 4. Genotyping for cytochrome P450 (CYP) 2D6 alleles revealed five homozygote (wt/wt) and five heterozygote subjects. Two subjects phenotypically classified as poor metabolisers were genotypically CYP2D6A/wt and CYP2D6D/wt, respectively. 5. Serum and urine samples taken more than 8 and 24 h after administration of ethyl-morphine respectively, contained morphine and morphine-glucuronides, but no ethylmorphine, ethylmorphine-6-glucuronide or (serum only) norethylmorphine. Norethylmorphine could be detected after hydrolysis of urine samples in all subjects. The urinary recovery of the active metabolites morphine and morphine-6-glucuronide after administration of ethylmorphine varied by a factor of 9 between individuals. 6. The wide variation in recovery of morphine and morphine-glucuronides after oral administration of ethylmorphine could not be explained simply by a difference in CYP2D6 genotype. Constitutional variation in other enzymatic pathways involved in ethylmorphine metabolism is probably crucial. Ratios of morphine to parent drug cannot be used to distinguish the source of morphine after administration of ethylmorphine. Norethylmorphine should be included in urine assays for opiates in forensic toxicology, and no firm conclusions about the source of morphine are possible based on serum samples obtained more than 24 h after drug administration.
|
| 18 |
+
|
| 19 |
+
## Full Text (Scanned Pages)
|
| 20 |
+
|
| 21 |
+
### Page 1
|
| 22 |
+

|
| 23 |
+
|
| 24 |
+
### Page 2
|
| 25 |
+
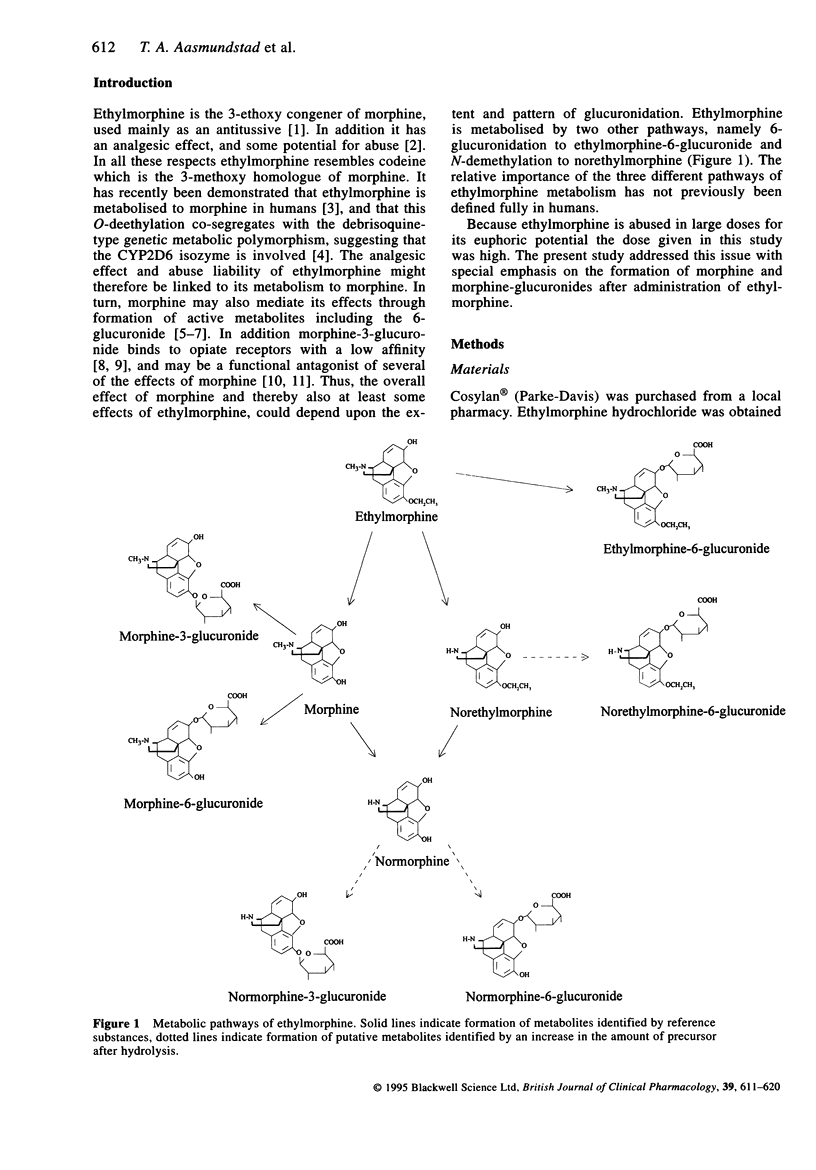
|
| 26 |
+
|
| 27 |
+
### Page 3
|
| 28 |
+

|
| 29 |
+
|
| 30 |
+
### Page 4
|
| 31 |
+

|
| 32 |
+
|
| 33 |
+
### Page 5
|
| 34 |
+
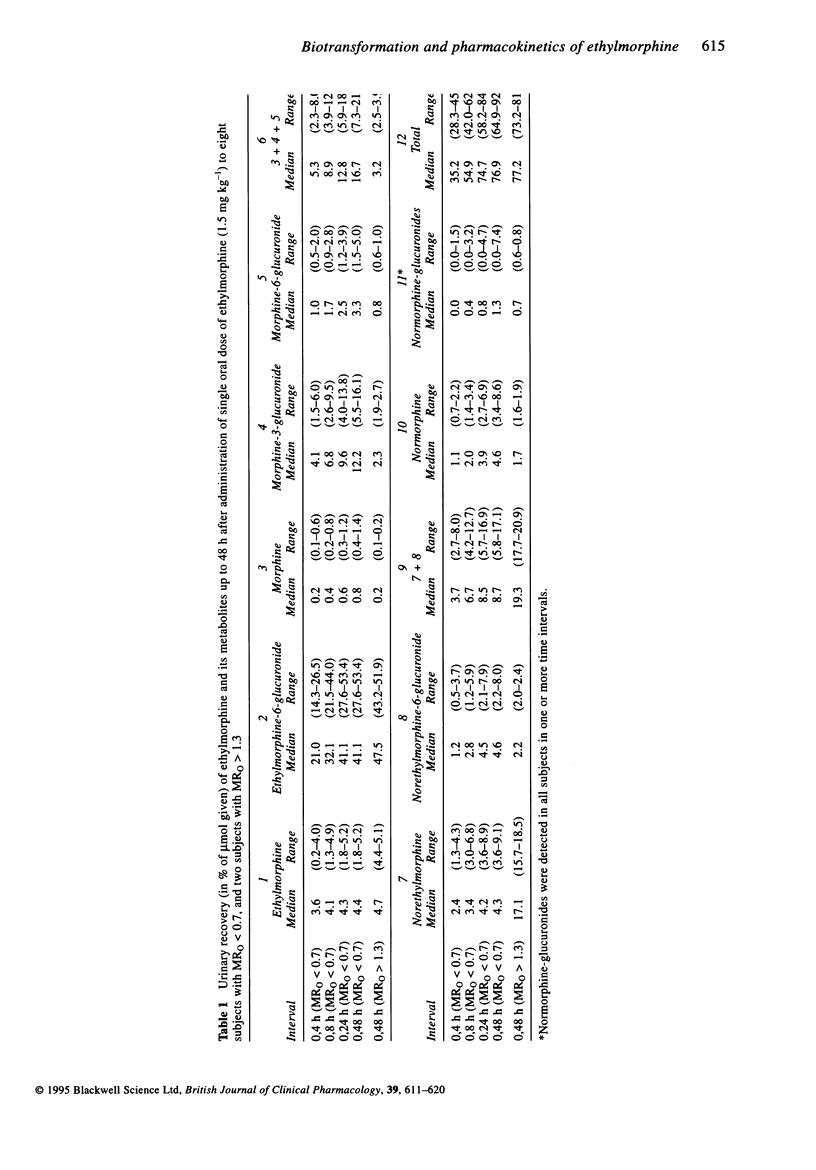
|
| 35 |
+
|
| 36 |
+
### Page 6
|
| 37 |
+
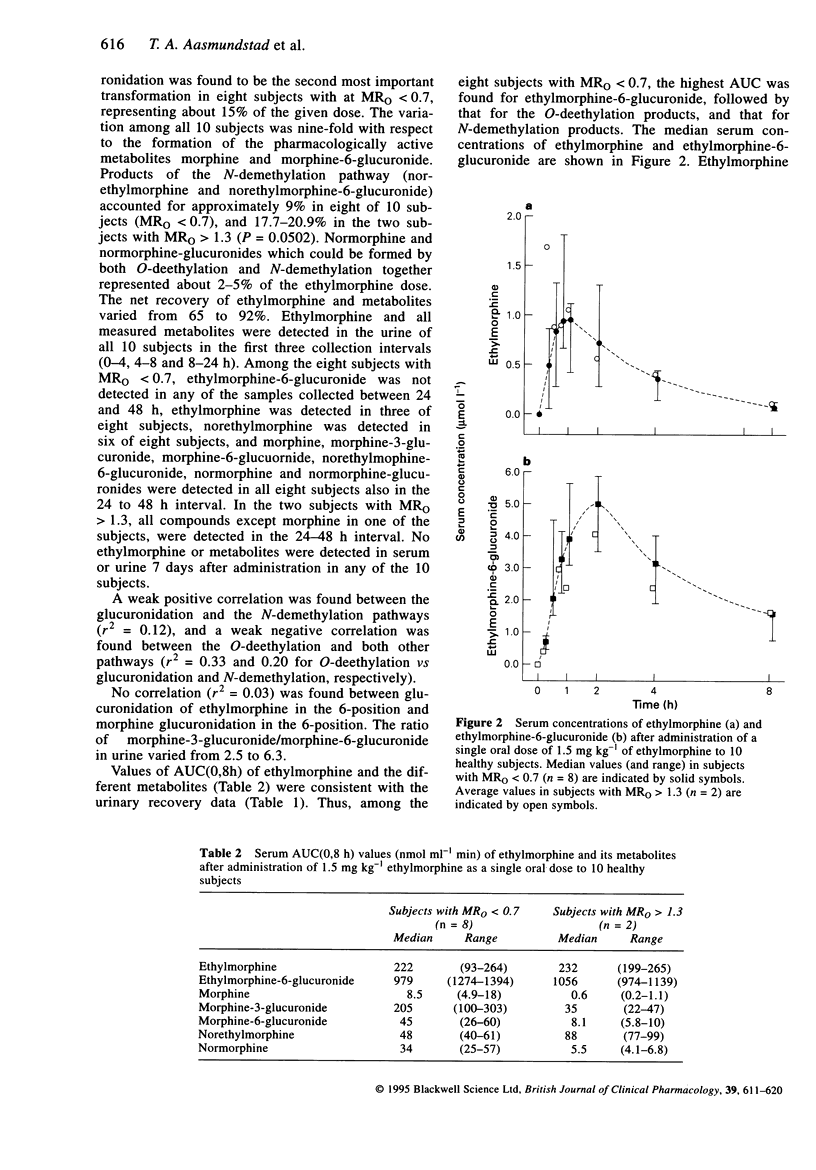
|
| 38 |
+
|
| 39 |
+
### Page 7
|
| 40 |
+
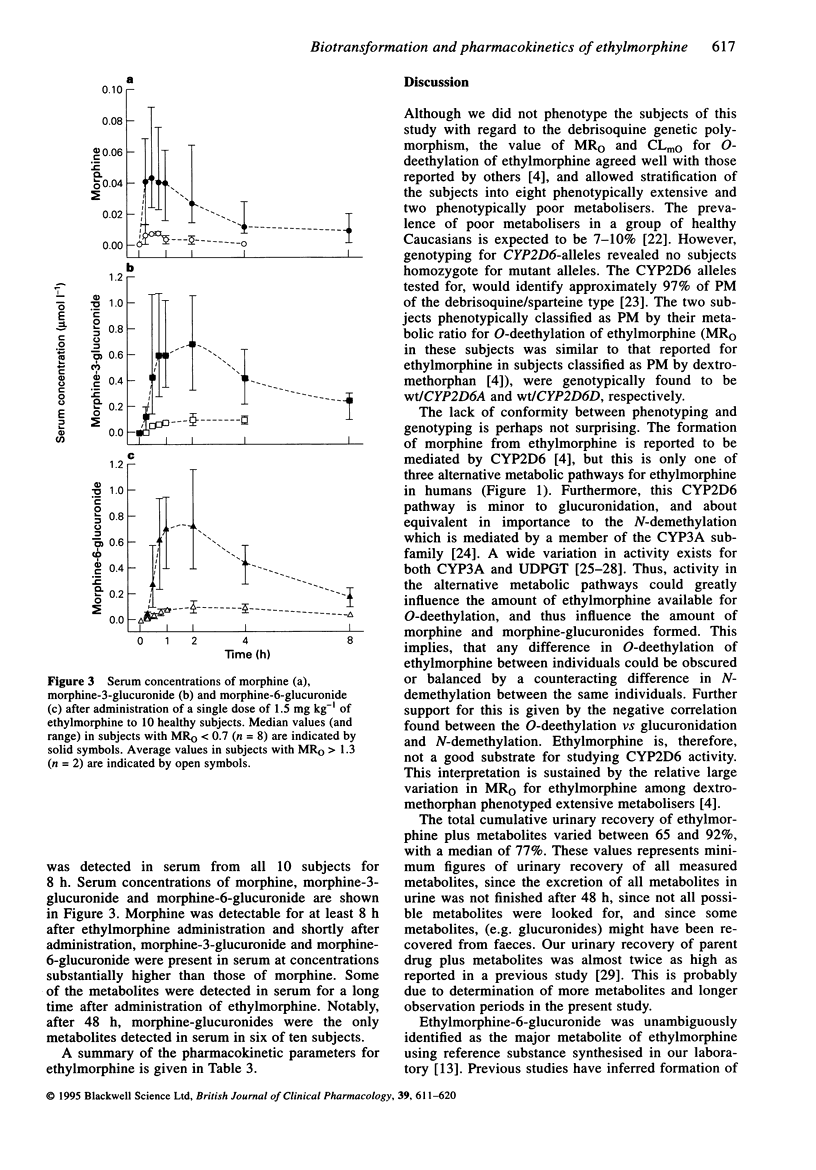
|
| 41 |
+
|
| 42 |
+
### Page 8
|
| 43 |
+

|
| 44 |
+
|
| 45 |
+
### Page 9
|
| 46 |
+

|
| 47 |
+
|
| 48 |
+
### Page 10
|
| 49 |
+

|
test/texts/PMC1365132.md
ADDED
|
@@ -0,0 +1,40 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Imipramine metabolism in relation to the sparteine and mephenytoin oxidation polymorphisms--a population study
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** H Madsen, K K Nielsen, K Brøsen
|
| 5 |
+
**Journal:** British Journal of Clinical Pharmacology
|
| 6 |
+
**Date:** 1995 Apr
|
| 7 |
+
**DOI:** [10.1111/j.1365-2125.1995.tb04473.x](https://doi.org/10.1111/j.1365-2125.1995.tb04473.x)
|
| 8 |
+
**PMID:** 7640151
|
| 9 |
+
**PMCID:** PMC1365132
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1365132/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC1365132/pdf/brjclinpharm00010-0092.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC1365132/pdf/brjclinpharm00010-0092.pdf)
|
| 12 |
+
|
| 13 |
+
*Note: This is a scanned document with limited structured text. Full content available in PDF.*
|
| 14 |
+
|
| 15 |
+
## Abstract
|
| 16 |
+
|
| 17 |
+
1. Sparteine and mephenytoin phenotyping tests were carried out in 327 healthy Danish subjects. Two weeks later each subject took 25 mg imipramine followed by urine collection for 24 h. The urinary content of imipramine, desipramine, 2-hydroxy-imipramine and 2-hydroxy-desipramine was assayed by h.p.l.c. 2. The medians of the hydroxylation ratios (i.e. 2-hydroxy-metabolite over parent compound) were 6 to 14 times higher in 300 extensive metabolizers of sparteine (EMs) as compared with 27 poor metabolizers (PMs), but none of the ratios separated the two phenotypes completely. 3. There were 324 EM of mephenytoin (EMM) and three PM (PMM) in the sample. The demethylation ratios between desipramine, 2-hydroxy-desipramine and their corresponding tertiary amines showed statistically significant correlations with the mephenytoin S/R isomer ratio (Spearman's rs: -0.20 and -0.27, P < 0.05). 4. The demethylation ratios were higher in 80 smokers than in 245 non-smokers. This indicates that CYP1A2, which is induced by cigarette smoking, also catalyzes the N-demethylation of imipramine. 5. CYP2D6 genotyping was carried out by PCR in 325 of the subjects, and the D6-wt allele was amplified in 298 EMs, meaning that they were genotyped correctly. One PMs was D6-wt/D6-B, another PMs had the genotype D6-wt/ and hence both were misclassified as EMs. The remaining 25 PMs were D6-A/D6-B (n = 5), D6-B/ (n = 18) or D6-D/D6-D (no PCR amplification, n = 2).(ABSTRACT TRUNCATED AT 250 WORDS)
|
| 18 |
+
|
| 19 |
+
## Full Text (Scanned Pages)
|
| 20 |
+
|
| 21 |
+
### Page 1
|
| 22 |
+

|
| 23 |
+
|
| 24 |
+
### Page 2
|
| 25 |
+

|
| 26 |
+
|
| 27 |
+
### Page 3
|
| 28 |
+
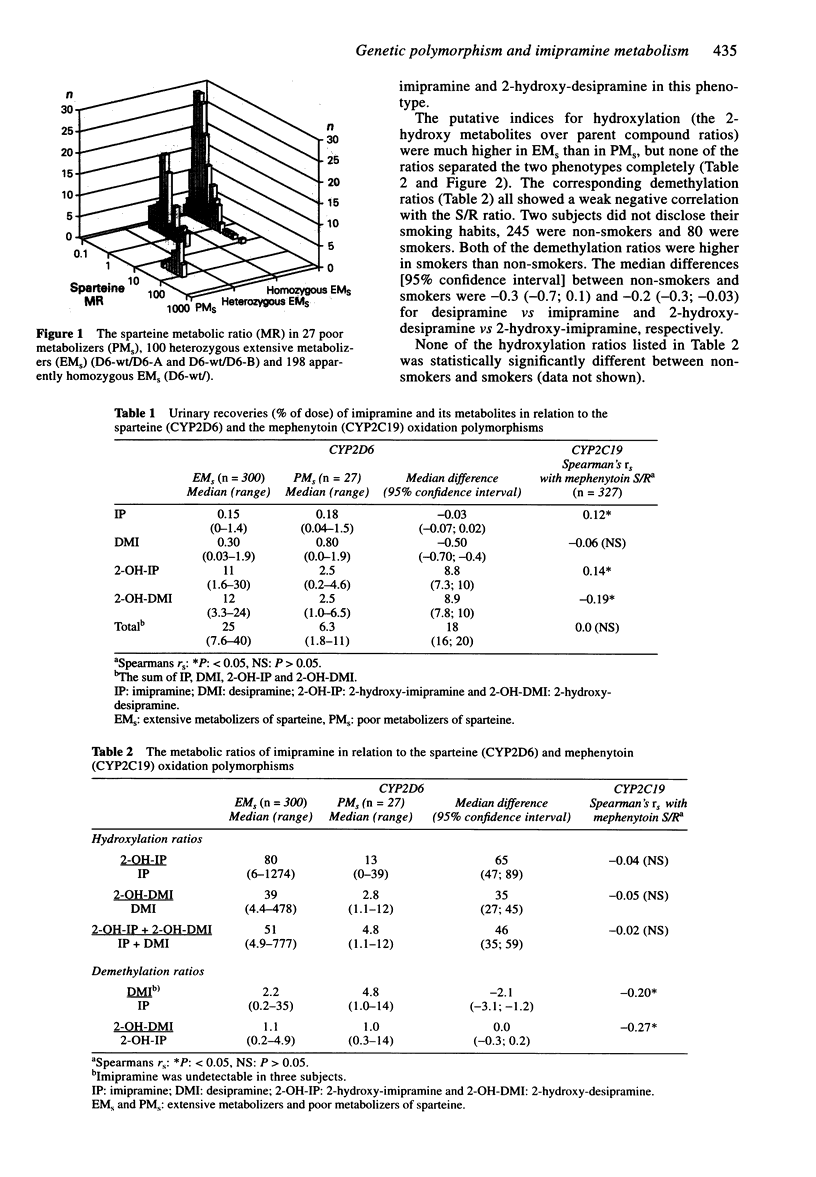
|
| 29 |
+
|
| 30 |
+
### Page 4
|
| 31 |
+
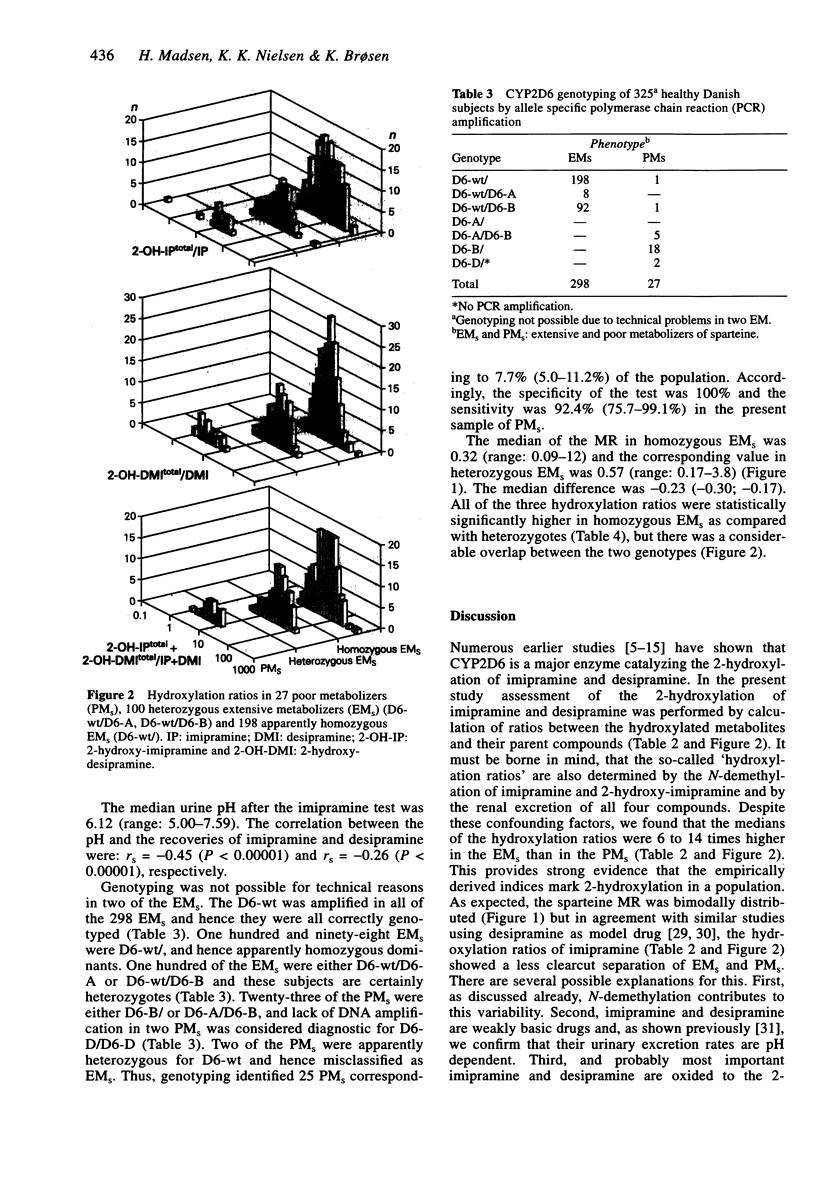
|
| 32 |
+
|
| 33 |
+
### Page 5
|
| 34 |
+

|
| 35 |
+
|
| 36 |
+
### Page 6
|
| 37 |
+

|
| 38 |
+
|
| 39 |
+
### Page 7
|
| 40 |
+

|
test/texts/PMC1474035.md
ADDED
|
@@ -0,0 +1,332 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Variation in the Gene Encoding the Serotonin 2A Receptor Is Associated with Outcome of Antidepressant Treatment
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Francis J McMahon, Silvia Buervenich, Dennis Charney, Robert Lipsky, A John Rush, Alexander F Wilson, Alexa J M Sorant, George J Papanicolaou, Gonzalo Laje, Maurizio Fava, Madhukar H Trivedi, Stephen R Wisniewski, Husseini Manji
|
| 5 |
+
**Journal:** American Journal of Human Genetics
|
| 6 |
+
**Date:** 2006 Mar 20
|
| 7 |
+
**DOI:** [10.1086/503820](https://doi.org/10.1086/503820)
|
| 8 |
+
**PMID:** 16642436
|
| 9 |
+
**PMCID:** PMC1474035
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1474035/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC1474035/pdf/AJHGv78p804.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC1474035/pdf/AJHGv78p804.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
Depressive disorders account for a large and increasing global burden of disease. Although the condition of many patients improves with medication, only a minority experience full remission, and patients whose condition responds to one medication may not have a response to others. Individual variation in antidepressant treatment outcome is, at present, unpredictable but may have a partial genetic basis. We searched for genetic predictors of treatment outcome in 1,953 patients with major depressive disorder who were treated with the antidepressant citalopram in the Sequenced Treatment Alternatives for Depression (STAR*D) study and were prospectively assessed. In a split-sample design, a selection of 68 candidate genes was genotyped, with 768 single-nucleotide–polymorphism markers chosen to detect common genetic variation. We detected significant and reproducible association between treatment outcome and a marker in HTR2A (P range 1×10-6 to 3.7×10-5 in the total sample). Other markers in HTR2A also showed evidence of association with treatment outcome in the total sample. HTR2A encodes the serotonin 2A receptor, which is downregulated by citalopram. Participants who were homozygous for the A allele had an 18% reduction in absolute risk of having no response to treatment, compared with those homozygous for the other allele. The A allele was over six times more frequent in white than in black participants, and treatment was less effective among black participants. The A allele may contribute to racial differences in outcomes of antidepressant treatment. Taken together with prior neurobiological findings, these new genetic data make a compelling case for a key role of HTR2A in the mechanism of antidepressant action.
|
| 16 |
+
|
| 17 |
+
## Methods
|
| 18 |
+
|
| 19 |
+
### Sample
|
| 20 |
+
|
| 21 |
+
The rationale, methods, and design of the STAR*D study have been detailed elsewhere.[13](#RF16)^13^13 In brief, investigators at 14 regional centers across the United States implemented a standard study protocol at 41 clinical sites.
|
| 22 |
+
|
| 23 |
+
Subjects provided separate written informed consent for study participation and for the collection of blood samples for genetic studies. Outpatients aged 18–75 years with a baseline Hamilton Depression Rating Scale score[14](#RF17)^14^14^,^,[15](#RF18)^15^15 of ⩾14 who met DSM-IV[16](#RF19)^16^16 criteria for nonpsychotic MDD were eligible. Patients with bipolar, psychotic, or obsessive-compulsive disorders were excluded, as were those with primary eating disorders, general medical conditions that contraindicated study medications, substance dependence requiring inpatient detoxification, and clear nonresponse or intolerance to any protocol antidepressant during current episode or those who were pregnant or breast-feeding.
|
| 24 |
+
|
| 25 |
+
The 16-item Quick Inventory of Depressive Symptomatology–Clinician-rated (QIDS-C_16_16)[12](#RF15)^12^12^,^,[13](#RF16)^13^13^,^,[17](#RF20)^17^17^,^,[18](#RF21)^18^18 was obtained at baseline and at each treatment visit, to measure symptom severity. The intraclass correlation coefficient for the QIDS-C_16_16, repeated across raters over 4 years, was 0.96 (A.J.R.'s unpublished data). Patients with a baseline QIDS-C_16_16 >10 were eligible if the treating clinician determined that outpatient treatment with an antidepressant medication was indicated and safe. At level 1, the protocol required an adequate dose of citalopram for a sufficient time to maximize the likelihood of treatment success, to ensure that those who did not improve were most likely unresponsive to the medication, not just underdosed.[12](#RF15)^12^12 No concomitant medications were allowed, aside from benzodiazepines and hypnotics if needed. A CONSORT (ConConsolidated SStandards oof RReporting TTrials) diagram of the current study sample is shown in [figure 1](#FG1)figure 1.
|
| 26 |
+
|
| 27 |
+
### Figure 1.
|
| 28 |
+
|
| 29 |
+
|
| 30 |
+
|
| 31 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=1474035_AJHGv78p804fg1.jpg)
|
| 32 |
+
|
| 33 |
+
CONSORT chart of genotyping and analysis of STAR*D sample. Samples dropped from analysis comprised 34 subjects who were noncompliant with medications, 19 with initial QIDS-C16 <10 or missing, 5 with missing clinical data, 4 whose molecular and recorded sex did not match, and 3 duplicate samples.
|
| 34 |
+
|
| 35 |
+
### DNA Samples
|
| 36 |
+
|
| 37 |
+
DNA samples were collected from 1,953 participants. A sample of 20 ml of whole blood was collected in citrate-treated vacuum tubes and was shipped overnight to the Rutgers Cell Repository, where lymphocytes were extracted and cryopreserved using standard methods. DNA was extracted using GenePure chemistry (Qiagen) and was shipped on dry ice to the NIH laboratories. Samples were arrayed using a Tecan Genesis robot (Lipsky Lab), then sex-verified with a set of three X-linked and two Y-linked markers (McMahon Lab). Four sex discrepancies were identified and were excluded before samples were genotyped further.
|
| 38 |
+
|
| 39 |
+
A summary of the sample characteristics is shown in [table 1](#TB1)table 1. Those who consented to have blood drawn were similar to those in the full study sample but showed slight differences in several variables that reached statistical significance because of the large sample size. Subjects who consented to have blood drawn were older and better educated, with higher household income, and were more likely to be married, to be retired, and to describe themselves as white. These subjects were also more likely to come from a primary-care setting and to report more time elapsed since their first major depressive episode (MDE), more episodes, and greater comorbidity. These differences cannot affect the genetic association results, which derive from comparisons among the genotyped subjects. However, these differences may limit the generalizability of our findings, and clinical outcomes in the genotyped sample may differ somewhat from those in the full STAR*D sample.[12](#RF15)^12^12
|
| 40 |
+
|
| 41 |
+
### Table 1.
|
| 42 |
+
|
| 43 |
+
Selected Demographic and Clinical Characteristics of the STAR*D Sample[Note]
|
| 44 |
+
|
| 45 |
+
| | | Patients with Blood Drawn | Comparison |
|
| 46 |
+
| - | - | ------------------------- | ---------- |
|
| 47 |
+
| Characteristic | Complete Sample(N=4,041) | Yes(n=1,953) | No(n=2,088) | Test Statistica | df | Pb |
|
| 48 |
+
| Sociodemographic: | | | | | | |
|
| 49 |
+
| Mean (±SD) age (years) | 40.5 ± 13.3 | 42.7 ± 13.4 | 38.4 ± 12.9 | t = 10.31 | 4,037 | <.0001 |
|
| 50 |
+
| Sex: | | | | χ2 = 1.48 | 1 | NS |
|
| 51 |
+
| Male | 1,509 (37.3) | 748 (38.3) | 761 (36.4) | | | |
|
| 52 |
+
| Female | 2,532 (62.7) | 1,205 (61.7) | 1,327 (63.6) | | | |
|
| 53 |
+
| Race: | | | | χ2 = 13.20 | 2 | .0014 |
|
| 54 |
+
| White | 3,055 (75.7) | 1,526 (78.2) | 1,529 (73.4) | | | |
|
| 55 |
+
| Black | 709 (17.6) | 313 (16.0) | 396 (19.0) | | | |
|
| 56 |
+
| Other/mixed | 272 (6.7) | 113 (5.8) | 159 (7.6) | | | |
|
| 57 |
+
| Mean (±SD) no. of years of education | 13.4 ± 3.2 | 13.6 ± 3.2 | 13.3 ± 3.2 | χ2 = 13.01 | 1 | .0003 |
|
| 58 |
+
| Employment: | | | | χ2 = 19.65 | 2 | <.0001 |
|
| 59 |
+
| Employed | 2,311 (57.3) | 1,092 (55.9) | 1,219 (58.6) | | | |
|
| 60 |
+
| Unemployed | 1,489 (36.9) | 715 (36.6) | 774 (37.2) | | | |
|
| 61 |
+
| Retired | 234 (5.8) | 146 (7.5) | 88 (4.2) | | | |
|
| 62 |
+
| Mean (±SD) monthly household income (U.S. $) | 2,419 ± 3,143 | 2,521 ± 3,202 | 2,318 ± 3,082 | χ2 = 6.23 | 1 | .0125 |
|
| 63 |
+
| Medical insurance: | | | | χ2 = .68 | 2 | NS |
|
| 64 |
+
| Private | 2,022 (51.8) | 998 (52.4) | 1,024 (51.2) | | | |
|
| 65 |
+
| Public | 553 (14.2) | 270 (14.2) | 283 (14.1) | | | |
|
| 66 |
+
| None | 1,332 (34.1) | 638 (33.5) | 694 (34.7) | | | |
|
| 67 |
+
| Marital status: | | | | χ2 = 15.12 | 3 | .0017 |
|
| 68 |
+
| Single | 1,207 (29.9) | 545 (27.9) | 662 (31.8) | | | |
|
| 69 |
+
| Married/cohabiting | 1,663 (41.2) | 838 (42.9) | 825 (39.6) | | | |
|
| 70 |
+
| Divorced/separated | 1,037 (25.7) | 493 (25.2) | 544 (26.1) | | | |
|
| 71 |
+
| Widowed | 128 (3.2) | 77 (3.9) | 51 (2.4) | | | |
|
| 72 |
+
| Clinical: | | | | | | |
|
| 73 |
+
| Mean (±SD) age at first MDE (years) | 25.5 ± 14.4 | 26.1 ± 14.9 | 24.9 ± 13.9 | χ2 = 3.40 | 1 | NS |
|
| 74 |
+
| Mean (±SD) time since first MDE (years) | 15.0 ± 13.1 | 16.6 ± 13.9 | 13.5 ± 12.1 | χ2 = 42.07 | 1 | <.0001 |
|
| 75 |
+
| Mean (±SD) no. of MDEs | 5.9 ± 11.4 | 6.4 ± 12.5 | 5.4 ± 10.2 | χ2 = 11.09 | 1 | .0009 |
|
| 76 |
+
| Suicide ever attempted: | | | | χ2 = 6.26 | 1 | .0123 |
|
| 77 |
+
| Yes | 667 (16.5) | 293 (15.0) | 374 (17.9) | | | |
|
| 78 |
+
| No | 3,370 (83.5) | 1,659 (85.0) | 1,711 (82.1) | | | |
|
| 79 |
+
| No. of psychiatric comorbidities: | | | | χ2 = 24.39 | 4 | <.0001 |
|
| 80 |
+
| 0 | 1,510 (38.2) | 781 (40.9) | 729 (35.7) | | | |
|
| 81 |
+
| 1 | 1,028 (26.0) | 510 (26.7) | 518 (25.4) | | | |
|
| 82 |
+
| 2 | 607 (15.4) | 282 (14.8) | 325 (15.9) | | | |
|
| 83 |
+
| 3 | 342 (8.7) | 133 (7.0) | 209 (10.2) | | | |
|
| 84 |
+
| ⩾4 | 465 (11.8) | 204 (10.7) | 261 (12.8) | | | |
|
| 85 |
+
| Current episode: | | | | | | |
|
| 86 |
+
| Clinical setting: | | | | χ2 = 29.97 | 1 | <.0001 |
|
| 87 |
+
| Primary | 1,575 (39) | 846 (43.3) | 729 (34.9) | | | |
|
| 88 |
+
| Specialty | 2,466 (61) | 1,107 (56.7) | 1,359 (65.1) | | | |
|
| 89 |
+
| Mean (±SD) duration of current episode (mo) | 24.5 ± 52.0 | 24.8 ± 53.1 | 24.3 ± 51.0 | χ2 = .78 | 1 | .3764 |
|
| 90 |
+
| HDRS-17c | 18.8 ± 6.5 | 18.4 ± 6.2 | 19.6 ± 6.9 | t = 1.65 | 330 | .099 |
|
| 91 |
+
| QID-S16 | 13.8 ± 4.2 | 13.4 ± 4.1 | 14.5 ± 4.5 | t = 2.20 | 373 | .0283 |
|
| 92 |
+
### Phenotypes
|
| 93 |
+
|
| 94 |
+
All phenotype definitions and assignments were settled in advance and were assigned before genotyping. Patients were scored for treatment outcome in two ways: designated remission and response ([fig. 2](#FG2)fig. 2). In the absence of external validators, our choice of categorical phenotypes was guided (1) by careful work with the STAR*D clinicians—in advance of the genotyping—to develop distinctions that had face validity and took advantage of the large body of data available from the STAR*D trial; (2) by ensuring maximal contrast between the outcome groups, to improve power, and creating “probable” groups that approximated the more narrowly defined categories, to test their robustness; (3) and by paying special attention to full remission of symptoms, since this was the primary target outcome of treatment.
|
| 95 |
+
|
| 96 |
+
### Figure 2.
|
| 97 |
+
|
| 98 |
+
|
| 99 |
+
|
| 100 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=1474035_AJHGv78p804fg2.jpg)
|
| 101 |
+
|
| 102 |
+
Treatment-outcome phenotypes. Subjects who completed at least 6 wk of treatment with citalopram were assigned a remission and response phenotype that was based on the QIDS-C16 score at the last treatment visit. Those who met score criteria for remission or response after 3–6 wk of treatment were grouped with probable remitters or probable responders, respectively.
|
| 103 |
+
|
| 104 |
+
*Remitters*Remitters achieved a QIDS-C_16_16 score of ⩽5 at the last treatment visit; probable remitters achieved a score of 6 or 7. Nonremitters had a QIDS-C_16_16 score of ⩾10 at the last visit. Those with a final QIDS-C_16_16 score in the borderline range of 8 and 9 were excluded from analysis.
|
| 105 |
+
|
| 106 |
+
*Responders*Responders achieved at least a 50% reduction in baseline QIDS-C_16_16 at the last treatment visit; probable responders achieved a 45%–50% reduction. Nonresponders did not achieve even a 40% reduction in baseline QIDS-C_16_16 score at the last treatment visit. Those with a reduction in QIDS-C_16_16 in the borderline range of 40%–45% were excluded from analysis.
|
| 107 |
+
|
| 108 |
+
Only patients who completed at least 6 wk of treatment were included in the primary analysis. Patients who achieved the required QIDS-C_16_16 scores after <6 wk of treatment but who received at least 3 wk of treatment were assigned to the appropriate outcome group but were classified as “probable.” Those who did not complete at least 3 wk of treatment were excluded from analysis. Similarly, subjects who were classified as “intolerant” or “probably intolerant” were removed from the nonremitter and nonresponder groups but were retained in the remitter and responder groups, since intolerant subjects were probably not able to take the full effective dose of citalopram but might have responded if they had. Assessment of tolerability is discussed below. Subjects who did not adhere to the treatment regimen were excluded from analysis.
|
| 109 |
+
|
| 110 |
+
As a secondary test, relative change in QIDS-C_16_16 score at the last visit (expressed as percentage change from initial score) was tested as a quantitative trait, after removal of intolerant and nonadherent subjects.
|
| 111 |
+
|
| 112 |
+
### Tolerability
|
| 113 |
+
|
| 114 |
+
Medication tolerability comprises an individual’s objective and perceived side-effect burden and typically increases over time and with response to treatment.[19](#RF22)^19^19 Since failure to consider tolerability could lead to misclassification of intolerant patients as nonresponders, we scored all subjects as tolerant, probably tolerant, intolerant, or probably intolerant on the basis of an algorithm that considered study exit data and the Global Rating of Side Effect Burden (GRSEB).[13](#RF16)^13^13 In brief, all subjects who elected to continue citalopram at the end of the level 1 treatment period were considered tolerant, whereas subjects who refused to continue citalopram or who left the study because of side effects were considered intolerant. The remaining subjects were classified on the basis of GRSEB score into probably tolerant (no more than moderate side effects) or probably intolerant (more than moderate side effects). A small number of subjects with missing GRSEB scores were classified according to whether they took citalopram for <4 wk (probably intolerant) or ⩾4 wk (probably tolerant).
|
| 115 |
+
|
| 116 |
+
### Candidate Genes
|
| 117 |
+
|
| 118 |
+
Sixty-eight genes were chosen for study from among a larger list of plausible candidates. Genes primarily involved in drug metabolism were excluded, by prior agreement, since these will be studied by another group using the same set of DNA samples. Genes were scored by an expert panel (D.C., W. Drevets, H.M., and F.J.M.) on the basis of (1) prior evidence of association with antidepressant outcome (1–3 points), (2) prior evidence of association with major mood disorders (1–3 points), and (3) known functional variant(s) (0–1 points). Under this scoring system, candidate genes could receive 0–7 points; higher scores conferred higher priority for study. Genes with a score of ⩾4 were used to seed sets of related genes broadly encompassing five main pathways: serotonin related (n=20*n*n=20), glutamate related (n=16*n*n=16), dopamine related (n=3*n*n=3), adrenergic (n=4*n*n=4), and neurotrophic (n=4*n*n=4), along with selected genes in other pathways (n=21*n*n=21). The complete list of genes studied is shown in [table 2](#TB2)table 2.
|
| 119 |
+
|
| 120 |
+
### Table 2.
|
| 121 |
+
|
| 122 |
+
List of Genes Screened
|
| 123 |
+
|
| 124 |
+
| Hypothesis and Gene Symbol | Gene Name |
|
| 125 |
+
| -------------------------- | --------- |
|
| 126 |
+
| Dopamine hypothesis: | |
|
| 127 |
+
| TH | Tyrosine hydroxylase |
|
| 128 |
+
| COMT | Catechol-O-methyltransferase |
|
| 129 |
+
| MAOA | Monoamine-oxidase A |
|
| 130 |
+
| Adrenergic hypothesis: | |
|
| 131 |
+
| ADRA2A | alpha-2a adrenergic receptor |
|
| 132 |
+
| ADRA2C | alpha-2c adrenergic receptor |
|
| 133 |
+
| DBH | Dopamine beta-hydroxylase |
|
| 134 |
+
| SLC6A2 | Norepinephrin transporter |
|
| 135 |
+
| Serotonin hypothesis: | |
|
| 136 |
+
| SLC6A4 | Serotonin transporter |
|
| 137 |
+
| TPH1 | Tryptophane hydroxylase-1 |
|
| 138 |
+
| TPH2 | Tryptophane hydroxylase-2 |
|
| 139 |
+
| HTR1A | Serotonin receptors |
|
| 140 |
+
| HTR1B | Serotonin receptors |
|
| 141 |
+
| HTR1D | Serotonin receptors |
|
| 142 |
+
| HTR1E | Serotonin receptors |
|
| 143 |
+
| HTR1F | Serotonin receptors |
|
| 144 |
+
| HTR2A | Serotonin receptors |
|
| 145 |
+
| HTR2B/PSMD1 | Serotonin receptors |
|
| 146 |
+
| HTR2C | Serotonin receptors |
|
| 147 |
+
| HTR3A | Serotonin receptors |
|
| 148 |
+
| HTR3B | Serotonin receptors |
|
| 149 |
+
| HTR3C | Serotonin receptors |
|
| 150 |
+
| HTR3D | Serotonin receptors |
|
| 151 |
+
| HTR3E | Serotonin receptors |
|
| 152 |
+
| HTR4 | Serotonin receptors |
|
| 153 |
+
| HTR5A | Serotonin receptors |
|
| 154 |
+
| HTR6 | Serotonin receptors |
|
| 155 |
+
| HTR7 | Serotonin receptors |
|
| 156 |
+
| Glutamate hypothesis: | |
|
| 157 |
+
| GRIA1 | AMPA receptors |
|
| 158 |
+
| GRIA2 | AMPA receptors |
|
| 159 |
+
| GRIA3 | AMPA receptors |
|
| 160 |
+
| GRIA4 | AMPA receptors |
|
| 161 |
+
| GRIN1 | NMDA receptors |
|
| 162 |
+
| GRIN2A | NMDA receptors |
|
| 163 |
+
| GRIN2B | NMDA receptors |
|
| 164 |
+
| GRIN2C | NMDA receptors |
|
| 165 |
+
| GRIN2D | NMDA receptors |
|
| 166 |
+
| GRIN3A | NMDA receptors |
|
| 167 |
+
| GRIK1 | Kainate receptors |
|
| 168 |
+
| GRIK2 | Kainate receptors |
|
| 169 |
+
| GRIK3 | Kainate receptors |
|
| 170 |
+
| GRIK4 | Kainate receptors |
|
| 171 |
+
| GRIK5 | Kainate receptors |
|
| 172 |
+
| SLC1A1 | Glutamate/aspartate transporter |
|
| 173 |
+
| Neurotrophin hypothesis: | |
|
| 174 |
+
| BDNF | BDNF |
|
| 175 |
+
| NTRK2 | Trk-B |
|
| 176 |
+
| BCL2 | B-cell CLL/lymphoma 2 |
|
| 177 |
+
| BAG1 | BCL2-associated athanogene |
|
| 178 |
+
| Other signaling pathways: | |
|
| 179 |
+
| PPP1R1B | DARP-32 |
|
| 180 |
+
| NR3C2 | Mineralocorticoid receptor |
|
| 181 |
+
| CREB1 | CREB |
|
| 182 |
+
| MAPK1 | Mitogen-activated protein kinase 1 |
|
| 183 |
+
| GSK3B | Glycogen synthase kinase 3 beta |
|
| 184 |
+
| CAMK1 | Calcium/calmodulin-dependent protein kinase I |
|
| 185 |
+
| PPP3R2 | Calcineurin B (located within GRIN3A) |
|
| 186 |
+
| Other genes: | |
|
| 187 |
+
| FKBP5 | FK506 binding protein 5 |
|
| 188 |
+
| LAMA4 | Laminin alpha-4 |
|
| 189 |
+
| GNB3 | Guanine nucleotide binding protein |
|
| 190 |
+
| OGG1 | 8-Oxoguanine DNA glycosylase |
|
| 191 |
+
| NET-5 | Tetraspan NET-5 |
|
| 192 |
+
| NBL1 | Neuroblastoma, suppression of tumorigenicity 1 |
|
| 193 |
+
| GRWD1 | Glutamate-rich WD repeat containing 1 |
|
| 194 |
+
| RPP30 | Ribonuclease P (30 kDa) |
|
| 195 |
+
| RNF20 | Hypothetical protein FLJ20690 |
|
| 196 |
+
| FBXO38 | F-box only protein 38 |
|
| 197 |
+
| ARHGAP10 | rho GTPase activating protein 21 |
|
| 198 |
+
| NR1I2 | Orphan nuclear receptor PAR2 |
|
| 199 |
+
| KDELR1 | KDELR1 protein |
|
| 200 |
+
| ATP1A3 | ATPase, Na+/K+ transporting, alpha-3 polypeptide |
|
| 201 |
+
### Selection of SNP Markers
|
| 202 |
+
|
| 203 |
+
For each candidate gene, genotype data spanning the coding region and up to 2 kb of flanking sequence were downloaded from the [International HapMap Project](#RF2)International HapMap Project, accessed November 2004.[20](#RF23)^20^20 Since the STAR*D sample is mostly white, data from the CEPH sample (Utah residents with northern and western European ancestry) were used. The program LDSelect[21](#RF24)^21^21 was used to select an optimal set of available SNPs to genotype, at an r2*r*r^2^2 threshold of ⩾0.8. From the remaining SNPs, we further excluded those with a minor-allele frequency <7.5%, since we expected the alleles that contribute to treatment outcome in this data set to be common. Six nonsynonymous SNPs and four SNPs reported elsewhere[22](#RF25)^22^22^,^,[23](#RF26)^23^23 to be associated with treatment outcome were added to the set, which brought the total to 768. Illumina then performed a bioinformatic screen that identified 12 markers that would likely fail in their BeadArray assay. Predicted failures were replaced by a nearby marker that was in strong linkage disequilibrium (LD) with the excluded SNP, if available. Absent this, a nearby marker with an allele frequency similar to that of the excluded marker was selected. The complete list of SNPs genotyped, along with flanking sequence and expected alleles, is available on request.
|
| 204 |
+
|
| 205 |
+
### Genotyping Methods
|
| 206 |
+
|
| 207 |
+
Since all of the collected samples were not available at the start of the experiment, only the first 1,380 samples were shipped to Illumina, where they were genotyped on the Illumina BeadArray platform, a highly accurate, high-throughput assay.[24](#RF27)^24^24 At Illumina, 99.78% of samples were successfully genotyped, and 97.92% of SNPs produced usable data, so that a total of 1,034,602 of a possible 1,035,504 genotypes were returned, including 11,280 blind duplicate genotypes, all of which matched exactly.
|
| 208 |
+
|
| 209 |
+
On the basis of the results of the first 1,380 samples, five SNPs in the remaining samples were genotyped using Taqman chemistry, were scored on a fluorescent plate reader (Molecular Dynamics) without regard to phenotype. By design, 394 samples were genotyped at *rs7997012*rs7997012 and *rs1928040*rs1928040 both by Illumina and in-house (McMahon Lab). No discrepancies were detected.
|
| 210 |
+
|
| 211 |
+
### Analysis Plan
|
| 212 |
+
|
| 213 |
+
The primary experiment was based on comparison of allele and genotype frequencies between subjects who benefited or did not benefit from citalopram therapy. Because the number of tests in this experiment was large, a split sample design was employed. The 1,380 samples genotyped for all SNPs were divided, a priori, into a discovery sample and a replication sample. The discovery sample consisted of two-thirds of the total sample genotyped at Illumina; the replication sample consisted of the remaining one-third. The choice of asymmetric sample sizes for the discovery and test samples was based on the large overall sample size. Splitting the sample in this fashion allowed for the detection of genetic effects of equal size but with a more conservative nominal significance level in the discovery sample than in the test sample. The split samples were matched on sex and reported race (collapsed into “white,” “black,” and “other/mixed”). Any SNPs meeting the a priori significance levels in both the discovery and test samples (see below) were tested for robustness in the total data set.
|
| 214 |
+
|
| 215 |
+
### Power Analysis
|
| 216 |
+
|
| 217 |
+
The power to detect association was estimated in two ways. In the first, standard statistical methods[25](#RF28)^25^25 were used to determine the effect size for approximate sample sizes for the discovery and replication samples and for a third sample that included the discovery, replication, and all remaining samples. The discovery sample was screened with a nominal significance level of .01, which had a power of 0.9 to detect an allelic effect size of 0.16. The power to detect a similar effect size in the smaller replication sample was ∼0.8 at the P=.05*P*P=.05 level.
|
| 218 |
+
|
| 219 |
+
In addition, the power to detect allelic and genotypic (additive) effects was estimated for logistic regression analysis for a variety of genetic models (Quanto v. 1.0 beta [[Gene×Environment, Gene×Gene Interaction home page](#RF1)Gene×E*Gene*Gene×*E*Environment, Gene×Gene*Gene*Gene×*Gene*Gene Interaction home page]).[26](#RF29)^26^26 There are a large number of possible underlying genetic models. Estimates of power under a specified model are appropriate only when the specified model is, in fact, the true underlying model. For example, in an unmatched case-control design (case:control ratio of 1:0.5), for a log-additive model with allele frequency of .3, a significance level of .01, and a power of .90, the detectable genetic relative risks for sample sizes of 600 and 300 were ∼1.5 and ∼1.7, respectively. At a power of .80 and a significance level of .05, corresponding genetic relative risks were ∼1.4 and ∼1.6, respectively.
|
| 220 |
+
|
| 221 |
+
### Race and Ethnicity
|
| 222 |
+
|
| 223 |
+
Subjects were enrolled into the STAR*D study without regard to race or ethnicity. Detailed data on ethnicity were not collected, but most subjects identified themselves as “white,” “black,” or “other.” We attempted to verify this self-reported race using STRUCTURE,[27](#RF30)^27^27 which assigns a probability of group membership to individuals on the basis of marker-allele frequencies. Since the number of subjects of “other” ethnicity in the sample was small, they were excluded from the analysis. We prepared a test data set of 1,284 individuals containing 57 unlinked loci selected from among those genotyped at Illumina, and we ran this in STRUCTURE under an admixture model with correlated allele frequencies, performing 20,000 burn-ins and 20,000 repetitions. This set of markers robustly distinguished between black and white subjects in this sample, with median posterior probabilities of group membership of 0.8 for the black subjects and 0.6 for the white subjects. We then divided the sample into self-identified whites and self-identified blacks and ran STRUCTURE again with the same markers. In each subject, STRUCTURE identified only one major cluster, and a one-population model gave the best fit to the data (data not shown).
|
| 224 |
+
|
| 225 |
+
### Statistical Methods
|
| 226 |
+
|
| 227 |
+
All pairs of individuals were compared using RELCHECK,[28](#RF31)^28^28 which verified that individuals were unrelated. Because the sample consisted entirely of patients with major depression, a marker that increased risk for major depression in many of these individuals would not necessarily be in Hardy-Weinberg equilibrium.[29](#RF32)^29^29 Thus, SNPs were not removed from analysis because of deviation from Hardy-Weinberg expectations.
|
| 228 |
+
|
| 229 |
+
Tests of association included the Pearson χ^2^2 test (2×22×2 for allelic and 2×32×3 for genotypic associations), likelihood-ratio χ^2^2 test (2×22×2 for allelic and 2×32×3 for genotypic associations), and Fisher’s exact test (2×22×2 for allelic associations only). Since each test has complementary advantages, all tests were used to make decisions about replication. This strategy might increase false-positive results due to multiple testing, but the high correlation between the tests makes this unlikely. Some of the SNPs tested had relatively rare alleles, which led to contingency tables with small cell sizes. The Pearson χ^2^2 test is only an approximation for these SNPs but makes no assumptions about the underlying model. Fisher’s exact test is robust to small cells but assumes that the marginal distributions of the contingency table are fixed, which may not hold in this sample. When small cell size warnings were ignored, correlations among *P*P values from the Pearson χ^2^2, likelihood-ratio χ^2^2, and Fisher’s exact test were high (> 0.9) for the main SNP of interest. Fisher’s exact test was the most conservative of the tests considered, so those are the findings presented in the “[Results](#SC2)Results” section. For the quantitative outcome phenotype, genotypic means were compared with the *F*F test. All *P*P values were from two-tailed tests. LD between adjacent SNPs was estimated using Haploview 3.2,[30](#RF33)^30^30 which generates estimates of *D′*D′ and r2*r*r^2^2 that are based on the input genotypes.
|
| 230 |
+
|
| 231 |
+
## Results
|
| 232 |
+
|
| 233 |
+
### Marker Coverage
|
| 234 |
+
|
| 235 |
+
The genotyped SNPs sampled the common variation within the genes selected for study at a median pairwise *D′*D′ value of 0.81. Overall, 75% of adjacent marker pairs were in LD at a D′>0.73*D*D^′^′>0.73. Only 5% of SNP pairs were associated with each other at an r2*r*r^2^2 value of >0.8, demonstrating that our SNP-selection strategy effectively excluded redundant markers.
|
| 236 |
+
|
| 237 |
+
### Allelic Association Results
|
| 238 |
+
|
| 239 |
+
Each SNP was tested for association with treatment response and remission in the discovery sample (n=1,380*n*n=1,380). A total of 12 SNPs met or exceeded the nominal significance level of .01 for one or both phenotypes. Of these, only one SNP met or exceeded the nominal significance level of .05 in the replication sample for the same allele and phenotype ([fig. 3](#FG3)fig. 3). This SNP, *rs7997012,*rs7997012, resides in the second intron of the gene *HTR2A,*HTR2A, which encodes the serotonin 2A receptor. None of the other 767 SNPs met these strict criteria for association and replication in this experiment.
|
| 240 |
+
|
| 241 |
+
### Figure 3.
|
| 242 |
+
|
| 243 |
+
|
| 244 |
+
|
| 245 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=1474035_AJHGv78p804fg3.jpg)
|
| 246 |
+
|
| 247 |
+
Allelic association between treatment outcome and each of 768 SNPs representing 68 candidate genes. Allelic association P values were ranked in the discovery and replication samples, then the ranks were summed across samples. For visual clarity, the inverse of the sum of ranks is shown on the Y-axis. The remission phenotype is indicated by the unblackened circles, the response phenotype by the blackened circles. Only rs7997012 met the a priori P value thresholds in both samples. Another SNP, rs2178865, ranked highly in both samples but fell short of the a priori significance level in the replication sample.
|
| 248 |
+
|
| 249 |
+
In *HTR2A,*HTR2A, significant association was detected, in both samples, between the same (A) allele of *rs7997012*rs7997012 and treatment response ([table 3](#TB3)table 3). (Evidence of association was also detected between this allele and the remission phenotype, but this did not meet our a priori significance levels in both samples.) An additional SNP in *HTR2A*HTR2A (*rs1928040*rs1928040) showed evidence of association with response and remission in the discovery sample but not in the replication sample. LD analysis demonstrated that these two SNPs were not in strong LD with each other ([fig. 4](#FG4)fig. 4).
|
| 250 |
+
|
| 251 |
+
### Table 3.
|
| 252 |
+
|
| 253 |
+
Results of Association Analysis of Genotyped HTR2A SNPs, Stratified by Race[Note]
|
| 254 |
+
|
| 255 |
+
| | All | White | Black |
|
| 256 |
+
| - | --- | ----- | ----- |
|
| 257 |
+
| | | P | | P | | P |
|
| 258 |
+
| Phenotype and SNP | N | Allelewise | Genotypewise | n | Allelewise | Genotypewise | n | Allelewise | Genotypewise |
|
| 259 |
+
| Remission: | | | | | | | | | |
|
| 260 |
+
| rs7997012 | 1,149 | .000024 | .000035 | 911 | .00107 | .00183 | 170 | NS | NS |
|
| 261 |
+
| rs1928040 | 1,148 | .0446 | .0701 | 910 | .0626 | NS | 170 | NS | NS |
|
| 262 |
+
| rs6313 | 1,183 | NS | NS | 942 | NS | NS | 172 | NS | NS |
|
| 263 |
+
| rs6311 | 1,180 | NS | NS | 939 | NS | NS | 172 | .0431 | .0874 |
|
| 264 |
+
| Response: | | | | | | | | | |
|
| 265 |
+
| rs7997012 | 1,329 | .000037 | .000002 | 1,049 | .00183 | .000157 | 199 | NS | NS |
|
| 266 |
+
| rs1928040 | 1,327 | .0709 | NS | 1,048 | NS | NS | 199 | NS | NS |
|
| 267 |
+
| rs6313 | 1,372 | NS | NS | 1,086 | NS | NS | 202 | NS | NS |
|
| 268 |
+
| rs6311 | 1,371 | NS | NS | 1,084 | NS | NS | 203 | .0918 | .0149 |
|
| 269 |
+
| Change in QIDS-C16: | | | | | | | | | |
|
| 270 |
+
| rs7997012 | 1,749 | .000007 | .00000146 | 1,380 | .00123 | .000516 | 261 | NS | NS |
|
| 271 |
+
| rs1928040 | 1,747 | .0214 | .0072 | 1,378 | .0738 | .0887 | 261 | NS | NS |
|
| 272 |
+
| rs6313 | 1,802 | NS | .0878 | 1,425 | NS | NS | 264 | NS | .0353 |
|
| 273 |
+
| rs6311 | 1,804 | .0599 | .0494 | 1,426 | NS | NS | 265 | .0094 | .0261 |
|
| 274 |
+
### Figure 4.
|
| 275 |
+
|
| 276 |
+
|
| 277 |
+
|
| 278 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=1474035_AJHGv78p804fg4.jpg)
|
| 279 |
+
|
| 280 |
+
Physical positions and LD relationships among SNP markers genotyped in the STAR*D sample. This figure shows the Refseq gene model for HTR2A taken from the UCSC Genome Browser (build 43), juxtaposed on a graphical representation of intermarker r2 values produced by Haploview 3.2.30
|
| 281 |
+
|
| 282 |
+
On the basis of these results, we genotyped *rs7997012*rs7997012 and *rs1928040*rs1928040 in the remaining subjects and tested association with treatment outcome in the total sample. Significant evidence of association was again observed between *rs7997012*rs7997012 and treatment outcome ([table 3](#TB3)table 3), with *P*P values on the order of 10-610^-6^-6 for the treatment response phenotype. Evidence of association between *rs1928040*rs1928040 and treatment outcome was not substantially stronger in the total sample.
|
| 283 |
+
|
| 284 |
+
Since *rs7997012*rs7997012 and *rs1928040*rs1928040 are both intronic SNPs with no known function, we also genotyped two SNPs in *HTR2A*HTR2A (*rs6313*rs6313 and *rs6311*rs6311) that may have functional importance.[31](#RF34)^31^31^^^–^–[33](#RF36)^33^33 Both SNPs were in tight LD with each other and with *rs1928040,*rs1928040, but they were not in significant LD with *rs7997012.*rs7997012. Neither *rs6316*rs6316 nor *rs6311*rs6311 showed significant association with treatment response or remission in the total sample ([table 3](#TB3)table 3).
|
| 285 |
+
|
| 286 |
+
Further support for association with *rs7997012*rs7997012 was obtained in the quantitative-trait analysis ([table 3](#TB3)table 3). The marker *rs7997012*rs7997012 was significantly associated with relative change in initial QIDS-C_16_16 score, with a *P*P value of 7.0×10-57.0×10^-5^-5 for allelic and 1.0×10-61.0×10^-6^-6 for genotypic association. Less-significant evidence of association was also observed at other *HTR2A*HTR2A SNPs ([table 3](#TB3)table 3).
|
| 287 |
+
|
| 288 |
+
### Stratified Analyses
|
| 289 |
+
|
| 290 |
+
Having established an association between treatment outcome and *HTR2A*HTR2A in this sample, we next sought to characterize the association with respect to medication tolerability and race, using data sets stratified by the variable of interest. We used *rs7997012*rs7997012 as the test SNP and remission as the outcome variable, since they had yielded the strongest evidence of association in the earlier analyses.
|
| 291 |
+
|
| 292 |
+
In the primary analysis, medication-intolerant subjects were dropped from the *nonremitter*nonremitter and *nonresponder*nonresponder groups. However, this clinically justified decision created an imbalance with the *remitter*remitter and *responder*responder groups, which included medication-intolerant subjects. To rule out any role of this imbalance in the primary association finding, we dropped the medication-intolerant subjects from the *remitter*remitter and *responder*responder groups, then reanalyzed the data. Significant association between *rs7997012*rs7997012 and treatment outcome remained in the sample of medication-tolerant subjects (P=2.3×10-5*P*P=2.3×10^-5^-5, n=1,140*n*n=1,140 for the remission; P=2.3×10-5*P*P=2.3×10^-5^-5, n=1,303*n*n=1,303 for the response phenotypes).
|
| 293 |
+
|
| 294 |
+
It has been suggested that black patients respond less well than white patients to antidepressant medications, particularly SSRIs, but firm data are lacking. When we divided this sample by race into “white” and “black” strata, two important differences emerged. First, we noted that the A allele of *rs7997012*rs7997012 that was associated with better treatment outcome in the mixed-race sample was more than six times more frequent in the white than in the black participants (allele frequency in whites=0.42*whites*whites=0.42, in blacks, 0.06). Second, it was apparent that the association between *HTR2A*HTR2A and treatment outcome was largely confined to the white participants. No significant association between *rs7997012*rs7997012 and either treatment response or remission was detected in the black participants, although nominally significant results appeared for *rs6313*rs6313 and *rs6311*rs6311 in some analyses ([table 3](#TB3)table 3).
|
| 295 |
+
|
| 296 |
+
### Potential Confounders
|
| 297 |
+
|
| 298 |
+
Carriers of the A allele of *rs7997012*rs7997012 were compared with the other cases by means of several variables that could themselves be associated with treatment outcome: maximum citalopram dose (in mg), tolerability, sex, and initial QIDS-C_16_16 score. Carriers of the A allele did not differ from the other cases for any of these variables (data not shown).
|
| 299 |
+
|
| 300 |
+
### Strength of Association
|
| 301 |
+
|
| 302 |
+
For treatment-outcome phenotypes, the usual measures of strength of association, on the basis of odds ratios, are perhaps less informative than are measures that directly capture differences in clinical outcome.[34](#RF37)^34^34 Thus, we compared the rates of treatment response among participants grouped by genotype at *rs7997012.*rs7997012. Overall, 79.9% of those homozygous for the A allele were classified as “responders,” compared with 62.4% of those homozygous for the G allele. Among white participants, the values were 79.7% and 63.5%, respectively. Thus, the AA genotype at *rs7997013*rs7997013 confers a 16%–18% reduction in absolute risk of being a nonresponder in this sample.
|
| 303 |
+
|
| 304 |
+
## Discussion
|
| 305 |
+
|
| 306 |
+
To our knowledge, this is the first demonstration of significant, reproducible association between genetic variation and outcome of antidepressant treatment. The association signal reproduces in both our discovery and our replication samples and increases in significance with the inclusion of additional subjects. We evaluated three related definitions of treatment effectiveness: two categorical outcomes (response and remission) and a quantitative outcome based on relative change in the final symptom score. The categorical outcomes were defined in a way that created gaps between responders/remitters and nonresponders/nonremitters, thus increasing the contrast between these groups. The quantitative-trait analysis (which included all patients) supported the results of the categorical phenotypes and demonstrated that the observed association was not a consequence of the arbitrary outcome categories. Furthermore, the *HTR2A*HTR2A allele that was associated with better treatment outcome was more than six times more frequent in white than in black participants, who had an overall less favorable treatment outcome in this sample.[12](#RF15)^12^12
|
| 307 |
+
|
| 308 |
+
This study has several limitations and strengths. We did not perform a complete genomewide association study, so we cannot conclude that we have found all the genes that may be relevant to antidepressant-treatment outcome. Instead, we screened 68 genes selected by an expert panel as the best candidates. Given the available marker coverage, we did not sample all common variation in these genes, but pairwise LD analysis indicated that much of the common variation was sampled. The large sample size allowed detection of weak effects with high statistical confidence, along with replication testing. The association results, however, do not necessarily point to functional variants in *HTR2A.*HTR2A. Our strongest results are not attributable to either of the suspected functional variants in *HTR2A.*HTR2A. Instead, these results point to intronic variants whose functional relevance is unknown. (Indeed, given the local patterns of LD, we cannot fully exclude the possibility that the association signal actually reflects another, nearby gene.) However, our results set the stage for a focused search for functional variants in the 3′ end of *HTR2A*HTR2A in future studies. We employed phenotypes on the basis of a uniform rating of clinical symptoms that allowed for comparatively simple scoring of treatment outcome. Alternative methods for scoring treatment outcome exist and could point to different genes. The STAR*D study employed a naturalistic ascertainment and treatment protocol. Thus, these results should have general relevance in the clinic population but may differ from those obtained in more narrowly ascertained study groups.
|
| 309 |
+
|
| 310 |
+
As expected for a single gene, the clinical impact of *HTR2A*HTR2A on treatment outcome is modest. In the total sample, white subjects homozygous for the allele associated with better treatment outcome had an 18% decrease in absolute risk of nonresponse to citalopram, compared with subjects homozygous for the other allele. Additional alleles predictive of treatment outcome will need to be discovered before clinically more-relevant effect sizes are obtainable. In this regard, one pertinent negative finding should be highlighted. Outcome after treatment with SSRIs has been most often associated with variation in the gene encoding the serotonin transporter, *SLC6A4,*SLC6A4, the primary target of SSRI action (reviewed by Anguelova et al.[35](#RF38)^35^35). We found no evidence of association among any of the four genotyped *SLC6A4*SLC6A4 markers and treatment outcome in these data. Our split sample design, with a requirement that the same allele show association with the same phenotype by the same test in both samples, was implemented as a way to control for multiple testing but may have reduced the power to detect modest association signals.[36](#RF39)^36^36
|
| 311 |
+
|
| 312 |
+
We found an interesting two-way relationship between race, *HTR2A*HTR2A variation, and treatment outcomes in this sample. First, significantly fewer black patients had favorable treatment outcomes in this study, partly because of more early dropouts.[12](#RF15)^12^12 This is consistent with the few published reports that black patients respond less often than do whites to antidepressant medications,[37](#RF40)^37^37 although the differences have generally been attributed to psychosocial factors. Second, the association found between *HTR2A*HTR2A and antidepressant outcome in this study was largely confined to the white subjects. Although the sample size of the black population was smaller, no trend of association with *rs7997012*rs7997012 was detected for any treatment-outcome phenotype. Third, the allele that was associated with better treatment outcome was more than six times more frequent in whites than blacks in this sample. These results suggest that genetic variation in *HTR2A*HTR2A should be considered along with psychosocial factors in attempts to explain racial differences in antidepressant-treatment outcomes.
|
| 313 |
+
|
| 314 |
+
Genetic variation in *HTR2A*HTR2A has been widely implicated in a variety of neuropsychiatric disorders (reviewed by Norton and Owen[38](#RF41)^38^38), but it has not been convincingly demonstrated that it affects antidepressant-treatment outcome. One previous study found suggestive evidence that *HTR2A*HTR2A was associated with a delayed and sustained pattern of treatment outcome in a small sample.[39](#RF42)^39^39 Another small study found suggestive evidence that a different SNP in *HTR2A*HTR2A was associated with short-term treatment outcome.[40](#RF43)^40^40 One study of 443 depressed inpatients detected a marginally significant association between *HTR2A*HTR2A variants and outcome.[41](#RF44)^41^41 Finally, one small study observed differential treatment response in patients with one or two C alleles of the T102C polymorphism *rs6311*rs6311.[42](#RF45)^42^42
|
| 315 |
+
|
| 316 |
+
Although the precise molecular mechanisms by which antidepressants exert their beneficial effects remain to be fully elucidated, considerable data implicate the serotonergic system.[43](#RF46)^43^43^,^,[44](#RF47)^44^44 On the basis of radioligand binding, signal transduction, and amino acid sequences, 5-HT effectors currently comprise seven distinct receptors (5HT1–7). The 5-HT2A, B, and C subtypes are positively coupled with the enzyme phospholipase C (PLC).[45](#RF48)^45^45 The 5-HT2A receptors are postsynaptic receptors that are highly enriched in neocortex and regulate the function of prefrontal-subcortical circuits implicated in the pathophysiology of depression.[46](#RF49)^46^46^,^,[47](#RF50)^47^47 The 5-HT2A receptors interact with Gq/G11 guanine nucleotide binding proteins (G proteins) and thereby stimulate PLC to produce the intracellular second messengers sn-1,2-DAG (an endogenous activator of protein kinase C) and inositol-1,4,5-trisphosphate (IP3), which stimulates the release of Ca^++^++ from intracellular stores.
|
| 317 |
+
|
| 318 |
+
Considerable neurobiological data suggest that the 5-HT2A receptor plays an important role in antidepressant-drug action. Different classes of antidepressants, including citalopram, downregulate 5-HT2A receptors in rodent and primate forebrain within a time frame paralleling therapeutic effects in humans.[44](#RF47)^44^44^,^,[46](#RF49)^46^46^,^,[48](#RF51)^48^48^,^,[49](#RF52)^49^49 Selective 5-HT2A antagonists are effective in animal models of depression, and antisense oligonucleotides directed against 5-HT2A receptors regulate the development of depressivelike behavior in the learned-helplessness model.[50](#RF53)^50^50 Although citalopram does not directly bind to 5-HT2A receptors, several antidepressants do bind 5-HT2A receptors as antagonists, which likely plays an important role in their therapeutic action.[43](#RF46)^43^43 Finally, a growing body of work suggests that antidepressants bring about their delayed therapeutic effects by the induction of neuronal plasticity mediated by increased expression of brain-derived neurotrophic factor (BDNF).[47](#RF50)^47^47^,^,[51](#RF54)^51^51^,^,[52](#RF55)^52^52 In this context, it is noteworthy that 5-HT2A antagonists block stress-induced downregulation of BDNF mRNA in rodents.[53](#RF56)^53^53 Together, these results suggest that many of the neuroplastic events believed to underlie the efficacy of SSRIs are mediated, in part, via 5HT2A receptors.[53](#RF56)^53^53 Although future studies are needed to delineate the precise cellular mechanisms by which the *HTR2A*HTR2A SNP described herein affects response to the therapeutic effects of citalopram, the new genetic data presented here, taken together with the existing neurobiologic findings, make a compelling case for a key role of *HTR2A*HTR2A in the mechanism of antidepressant action.
|
| 319 |
+
|
| 320 |
+
In conclusion, this study demonstrates that genetic variation in *HTR2A*HTR2A is reproducibly associated with outcome of citalopram treatment in a large sample of outpatients with MDD. This same variation may contribute to racial differences in outcomes with SSRI treatment. Further studies are needed to define the functional changes in *HTR2A*HTR2A that account for the association signal in this sample.
|
| 321 |
+
|
| 322 |
+
## Acknowledgments
|
| 323 |
+
|
| 324 |
+
This research was supported in part by the Intramural Research Programs of the NIMH, the National Institute on Alcohol Abuse and Alcoholism, and the National Human Genome Research Institute, NIH. The authors appreciate the efforts of the STAR*D research team in performing the clinical study and gathering the DNA samples. Data and sample collection was funded with federal funds from the NIMH, NIH, under contract N01MH90003 to University of Texas Southwestern Medical Center at Dallas (principal investigator, A. J. Rush). We thank Nirmala Akula and Jo Steele, for technical advice, and the Rutgers Cell and DNA Repository, for extracting DNA and providing samples to our laboratories. The content of this publication does not necessarily reflect the views or policies of the DHHS, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. We appreciate the support of Forest Laboratories for providing citalopram at no cost for the STAR*D study. Most important, we thank the study participants, without whom this study would not be possible.
|
| 325 |
+
|
| 326 |
+
## References
|
| 327 |
+
|
| 328 |
+
1. Gene×Environment, Gene×Gene Interaction home page, http://hydra.usc.edu/gxe/ (for Quanto, v. 1.0 beta) [http://hydra.usc.edu/gxe/](http://hydra.usc.edu/gxe/)
|
| 329 |
+
|
| 330 |
+
2. International HapMap Project, http://www.hapmap.org/ [http://www.hapmap.org/](http://www.hapmap.org/)
|
| 331 |
+
|
| 332 |
+
3. UCSC Genome Browser, http://genome.ucsc.edu/ [http://genome.ucsc.edu/](http://genome.ucsc.edu/)
|
test/texts/PMC1762324.md
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
test/texts/PMC1873375.md
ADDED
|
@@ -0,0 +1,223 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Non-linear fluvoxamine disposition
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Olav Spigset, Kerstin Granberg, Staffan Hägg, Emma Söderström, Rune Dahlqvist
|
| 5 |
+
**Journal:** British Journal of Clinical Pharmacology
|
| 6 |
+
**Date:** 1998 Mar
|
| 7 |
+
**DOI:** [10.1046/j.1365-2125.1998.00670.x](https://doi.org/10.1046/j.1365-2125.1998.00670.x)
|
| 8 |
+
**PMID:** 9517369
|
| 9 |
+
**PMCID:** PMC1873375
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1873375/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC1873375/pdf/bcp0045-0257.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC1873375/pdf/bcp0045-0257.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Aims:**
|
| 16 |
+
To study the pharmacokinetics of fluvoxamine when given in increasing doses to healthy volunteers.
|
| 17 |
+
|
| 18 |
+
**Methods:**
|
| 19 |
+
Ten healthy, non-smoking men were given maintenance treatment with fluvoxamine for 4 weeks. Eight subjects were CYP2D6 extensive metabolisers (EMs) and two were CYP2D6 poor metabolisers (PMs). As a measure of the CYP1A2 phenotype, the paraxanthine/caffeine ratio in saliva after intake of caffeine was studied. The fluvoxamine doses given were 25 mg day−1 the first week, 50 mg day−1 the second week, 100 mg day−1 the third week and 200 mg day−1 the fourth week, divided in two daily doses. On the seventh day every week, serum concentrations of fluvoxamine were followed for a dose interval of 12 h. After discontinuation of treatment, fluvoxamine concentrations were followed for 1 week.
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
For each of the three two-fold increases in given dose, the mean AUC increased 3.25-fold, 3.17-fold and 3.14-fold, respectively (P<0.0001), indicating a decrease in oral clearance with increasing dose. The elimination half-life based upon the serum concentrations 12–48 h after discontinuation of fluvoxamine was 32.1±11.0 h whereas the half-life based upon the concentrations 3–7 days after discontinuation was significantly shorter, 15.8±4.2 h (means±s.d.; P<0.001). There were no significant correlations between the CYP1A2 phenotype and fluvoxamine AUCs at different doses (r =−0.56; P =0.095 for the correlation between the paraxanthine/caffeine ratio in saliva and fluvoxamine AUC at a dose of 50 mg day−1). The two CYP2D6 PMs had AUC values in the same range as the EMs.
|
| 23 |
+
|
| 24 |
+
**Conclusions:**
|
| 25 |
+
The present study conclusively demonstrates that fluvoxamine exhibits non-linear kinetics within the therapeutic dose interval. The reason for non-linearity is not Michaelis-Menten saturation kinetics of a single metabolic pathway, but rather a complex involvement of multiple parallel pathways.
|
| 26 |
+
|
| 27 |
+
Keywords: CYP1A2, CYP2D6, fluvoxamine, non-linear, disposition, pharmacokinetics, adverse drug reactions
|
| 28 |
+
|
| 29 |
+
### Aims
|
| 30 |
+
|
| 31 |
+
To study the pharmacokinetics of fluvoxamine when given in increasing doses to healthy volunteers.
|
| 32 |
+
|
| 33 |
+
### Methods
|
| 34 |
+
|
| 35 |
+
Ten healthy, non-smoking men were given maintenance treatment with fluvoxamine for 4 weeks. Eight subjects were CYP2D6 extensive metabolisers (EMs) and two were CYP2D6 poor metabolisers (PMs). As a measure of the CYP1A2 phenotype, the paraxanthine/caffeine ratio in saliva after intake of caffeine was studied. The fluvoxamine doses given were 25 mg day^−1^−1 the first week, 50 mg day^−1^−1 the second week, 100 mg day^−1^−1 the third week and 200 mg day^−1^−1 the fourth week, divided in two daily doses. On the seventh day every week, serum concentrations of fluvoxamine were followed for a dose interval of 12 h. After discontinuation of treatment, fluvoxamine concentrations were followed for 1 week.
|
| 36 |
+
|
| 37 |
+
### Results
|
| 38 |
+
|
| 39 |
+
For each of the three two-fold increases in given dose, the mean AUC increased 3.25-fold, 3.17-fold and 3.14-fold, respectively (*P*P<0.0001), indicating a decrease in oral clearance with increasing dose. The elimination half-life based upon the serum concentrations 12–48 h after discontinuation of fluvoxamine was 32.1±11.0 h whereas the half-life based upon the concentrations 3–7 days after discontinuation was significantly shorter, 15.8±4.2 h (means±s.d.; *P*P<0.001). There were no significant correlations between the CYP1A2 phenotype and fluvoxamine AUCs at different doses (*r*r =−0.56; *P*P =0.095 for the correlation between the paraxanthine/caffeine ratio in saliva and fluvoxamine AUC at a dose of 50 mg day^−1^−1). The two CYP2D6 PMs had AUC values in the same range as the EMs.
|
| 40 |
+
|
| 41 |
+
### Conclusions
|
| 42 |
+
|
| 43 |
+
The present study conclusively demonstrates that fluvoxamine exhibits non-linear kinetics within the therapeutic dose interval. The reason for non-linearity is not Michaelis-Menten saturation kinetics of a single metabolic pathway, but rather a complex involvement of multiple parallel pathways.
|
| 44 |
+
|
| 45 |
+
**Keywords:**Keywords: CYP1A2, CYP2D6, fluvoxamine, non-linear, disposition, pharmacokinetics, adverse drug reactions
|
| 46 |
+
|
| 47 |
+
## Introduction
|
| 48 |
+
|
| 49 |
+
The selective serotonin reuptake inhibitor fluvoxamine is used in the treatment of depression and obsessive compulsive disorder, and is currently evaluated in the treatment of a number of other psychiatric diseases [[1](#b1)1, [2](#b2)2]. Fluvoxamine is extensively metabolised in the liver [[3](#b3)3], and the limited data available indicate that it is a high clearance drug [[4](#b4)4]. There are some data to indicate that the isozymes CYP1A2 [[5](#b5)5] and CYP2D6 [[6](#b6)6], but not CYP2C19 [[6](#b6)6], are important for its metabolism. Fluvoxamine is a potent inhibitor of CYP1A2 *in vitro*in vitro [[7](#b7)7, [8](#b8)8] as well as *in vivo*in vivo [[9](#b9)9–[13](#b13)13]. Saturation kinetics have been demonstrated for other drugs metabolised by CYP1A2, such as theophylline and caffeine [[14](#b14)14–[16](#b16)16], and drug metabolism by the polymorphic isozyme CYP2D6 is also characterised by saturation kinetics [[17](#b17)17].
|
| 50 |
+
|
| 51 |
+
There is evidence to suggest that fluvoxamine may exhibit non-linear kinetics. In a crossover study in six healthy volunteers [[18](#b18)18], area under the concentration-time curve (AUC) over a dose interval at steady-state (dose 50 mg ×2) was 30% higher than predicted from AUC from 0 to infinity after a single dose of 50 mg. Moreover, the terminal half-lives were 20–50% longer at steady state than in the same subjects after a single dose [[18](#b18)18]. In a study of nine patients with depression, doubling the fluvoxamine dose from 100 to 200 mg day^−1^−1 caused a 3.3-fold increase in the mean steady state plasma concentration [[19](#b19)19]. On the other hand, no indications of non-linearity were found in the dose range 25–100 mg after single oral doses [[20](#b20)20], and according to some unpublished data (cited in [[21](#b21)21]), linearity has been demonstrated in the 100 to 300 mg day^−1^−1 dose range in two small clinical studies. In a recent case report of fluvoxamine intoxication [[22](#b22)22], in which serum concentrations of fluvoxamine were followed for 1 week, non-linearity was apparently present at serum levels over 150 ng ml^−1^−1 (approximately 500 nmol l^−1^−1). The half-life of the first part of the elimination phase was 38 h, which is longer than other reports in the literature, except among some patients with liver cirrhosis [[23](#b23)23]. In contrast, the half-life of the terminal elimination phase was 19 h.
|
| 52 |
+
|
| 53 |
+
In order to study the non-linear kinetics of fluvoxamine more thoroughly and to elucidate the role of different enzymes involved, we performed a pharmacokinetic study in which fluvoxamine was given in increasing doses to healthy volunteers.
|
| 54 |
+
|
| 55 |
+
## Methods
|
| 56 |
+
|
| 57 |
+
### Subjects
|
| 58 |
+
|
| 59 |
+
After giving their informed consent, 10 non-smoking men took part in the investigation, which was approved by the Ethics Committee at Umeå University. Their age (mean±s.d.) was 28.9±5.2 years, and their body weight was 85.6±7.6 kg. All subjects were healthy, as assessed by medical history, physical examination, and routine blood chemistry tests. They had been entirely drug-free for at least 2 weeks prior to study start. Drugs taken as needed during the study period included single doses of bromhexine, chlorzoxazone, codeine, dextropropoxyphene, ephedrine, ibuprofen and paracetamol. These drugs were never ingested less than 3 days before the study days.
|
| 60 |
+
|
| 61 |
+
### Study protocol
|
| 62 |
+
|
| 63 |
+
Fluvoxamine (Fevarin; enteric-coated fluvoxamine maleate, Solvay Duphar B.V., Veesp, The Netherlands) was given to the subjects in increasing doses for a total of 4 weeks. The doses were 25 mg day^−1^−1 the first week, 50 mg day^−1^−1 the second week, 100 mg day^−1^−1 the third week, and 200 mg day^−1^−1 the fourth week. Half of the daily dose was ingested at 08.00 h and the other half at 20.00 h.
|
| 64 |
+
|
| 65 |
+
On the seventh day at each dose level, venous blood samples (10 ml) were drawn at 08.00 h, and 1, 2, 3, 4, 6, 8, 10, and 12 h later. Serum was separated within 30 min and stored frozen at −80° C until analysis. On these study days, the tablet intake at 08.00 h was followed by a standardized, light breakfast meal at 08.30 h, a standardized lunch at noon, and a standardized dinner at 17.00 h. Intake of alcohol and of food or beverages containing caffeine or other methylxanthines was not allowed or the study days and was otherwise registered daily throughout the study period. After discontinuation of fluvoxamine at the end of week 4, single blood samples were, in addition to the samples obtained during the first 12 h after tablet intake, also obtained after 24, 32 and 48 h, and thereafter once daily except Sundays for a total period of 1 week. Throughout the study, the subjects daily registered possible adverse effects of fluvoxamine by open reporting on a registration sheet.
|
| 66 |
+
|
| 67 |
+
### CYP2D6 phenotyping
|
| 68 |
+
|
| 69 |
+
The CYP2D6 phenotype was determined after intake of 50 mg dextromethorphan hydrobromide (Tussidyl mixture, 2 mg ml^−1^−1; Tika, Lund, Sweden) and collection of urine for 10 h. Dextromethorphan and its *O*O-demethylated metabolite dextrorphan were analysed by a capillary gas chromatography method described in detail elsewhere [[6](#b6)6]. Using this method, dextromethorphan/dextrorphan ratios >0.9 defined the poor metabolisers, whereas ratios <0.9 defined the extensive metabolisers. The subjects were chosen to include eight CYP2D6 extensive metabolisers (EMs) and two CYP2D6 poor metabolisers (PMs). The EMs had metabolic ratios ranging from 0.03 to 0.47, and the PMs had metabolic ratios of 1.9 and 48, respectively.
|
| 70 |
+
|
| 71 |
+
### CYP2C19 phenotyping
|
| 72 |
+
|
| 73 |
+
The CYP2C19 phenotype was determined after intake of 100 mg racemic mephenytoin (Mesantoin; Sandoz, Basel, Switzerland) and collection of urine for 10 h [[6](#b6)6]. The analytical method used is described in detail elsewhere [[24](#b24)24]. Using this method, mephenytoin S/R enantiomeric ratios /0.8 defined the poor metabolisers, whereas ratios <0.8 defined the extensive metabolisers. All subjects were CYP2C19 extensive metabolisers with metabolic ratios ranging from 0.02 to 0.18.
|
| 74 |
+
|
| 75 |
+
### CYP1A2 phenotyping
|
| 76 |
+
|
| 77 |
+
As a measure of CYP1A2 metabolic capacity, the paraxanthine/caffeine ratio in saliva was studied [[25](#b25)25]. After methylxanthine abstinence for at least 30 h, 200 mg caffeine (2 tablets of 100 mg Koffein ACO; ACO AB, Helsingborg, Sweden) was ingested. Seven hours later, 1.5 ml saliva was collected into polyethylene tubes by the spit method without chemical stimulation. All samples were stored at −20° C until analysis.
|
| 78 |
+
|
| 79 |
+
Concentrations of caffeine and paraxanthine were analysed by a high performance liquid chromatography (h.p.l.c.) method developed in our laboratory. After thawing, the saliva samples were centrifuged at 11 000 *g*g for 2 min. Solid-phase extraction cartridges (Isolute MF C18; International Sorbent Technology, Mid Glamorgan, UK) were prepared by prewashing with 2×1 ml methanol and 2×1 ml water. 240 μl saliva, 60 μl (14.39 ng) of the internal standard β-hydroxyethyltheophylline, and 900 μl water was gently mixed, and 1 ml of the mixture was applied to the column. Thereafter, the column was washed with 2×1 ml water and 350 μl acetone and the methylxanthines were eluted with 800 μl acetone. After the eluate was evaporated to dryness under nitrogen and redissolved in 100 μl of the mobile phase by ultrasound, 40 μl were injected onto the chromatograph. The mobile phase consisted of 3 mm sodium acetate (pH 4.0) with 1.4% acetonitrile, 1% methanol, and 1.6% tetrahydrofurane stabilised with 0.025% butylated hydroxytoluene. The separation was performed on a Nucleosil ODS 5 μm 25 cm column (Jones Chromatography, Mid Glamorgan, UK) with a flow of 1.2 ml min^−1^−1. The ultraviolet detector was set at a wavelength of 273 nm. Mean (±s.d.) recoveries after solid-phase extraction were 97.6% (±3.2%) for caffeine, 99.8% (±4.5%) for paraxanthine, and 94.8% (±2.4%) for β-hydroxyethyltheophylline. At a concentration of 10 μmol l^−1^−1, the intraassay coefficients of variation were 2.7% for paraxanthine and 1.8% for caffeine, and the interassay coefficients of variation were 2.9% and 4.0%, respectively. The limit of quantification was 0.5 μmol l^−1^−1, and the method was linear at least up to 250 μmol l^−1^−1. The salivary concentrations of caffeine and paraxanthine in the volunteers ranged from 3.4 to 9.6 μmol l^−1^−1 and from 3.3 to 4.5 μmol l^−1^−1, respectively, and the paraxanthine/caffeine ratios ranged from 0.36 to 1.14. Unfortunately, the ratio from one CYP2D6 PM was not available for administrative reasons.
|
| 80 |
+
|
| 81 |
+
### Fluvoxamine analysis
|
| 82 |
+
|
| 83 |
+
The serum concentrations of fluvoxamine were determined by a h.p.l.c. method described in detail elsewhere [[5](#b5)5]. In brief, to 2 ml serum were added 5 ml 0.3 m Na_3_3PO_4_4 and 400 μl di-isopropylether. After mixing and centrifuging, the organic layer was separated on a straight phase 150×4.6 mm Apex Silica 3 μm column and analysed with the ultraviolet detector set at a wavelength of 254 nm. The limit of quantification was 0.5 nmol l^−1^−1 and the method was linear at least up to 3000 nmol l^−1^−1.
|
| 84 |
+
|
| 85 |
+
### Pharmacokinetic and statistical analyses
|
| 86 |
+
|
| 87 |
+
Trough serum concentrations of fluvoxamine at steady state (*C*C_0_0), peak serum concentrations of fluvoxamine at steady state (*C*C_max_max) and time of peak concentrations (*t*t_max_max) were derived directly from the measured values. Other pharmacokinetic parameters were calculated by use of the pharmacokinetic program package Siphar/Win, version 1.13 (SIMED S.A., Creteil, France). Areas under the serum concentration-time curves during the dose intervals of 12 h at steady state (AUC) were calculated by use of the linear trapezoidal rule. Average serum concentrations at steady-state (*C*C_av_av) were calculated as AUC/12 h. Oral clearance (CL/*F*F, in which *F*F is the oral bioavailability) was calculated as Dose (half of the daily dose)/AUC. The parameter estimates describing the linear slopes of the log-concentration of fluvoxamine were calculated by means of the peeling procedure in the program package. For each subject, two slopes were calculated: λ_12–48_12–48, which is the slope represented by the serum concentrations 12, 24, 32 and 48 h after discontinuation of fluvoxamine, and λ_z_z, which is the slope represented by the four last measurable serum concentrations. The corresponding half-lives (*t*t_1/2,12–48_1/2,12–48 and *t*t_1/2,z_1/2,z) were calculated as ln2/λ_12–48_12–48, and ln2/λ_z_z, respectively. In addition, the presence of non-linear kinetics was evaluated by plotting the daily dose/*C*C_av_av ratio against daily dose (Eadie-Hofstee plot with interchanged axes).
|
| 88 |
+
|
| 89 |
+
For comparisons of pharmacokinetic parameters at different dose levels, two-way analysis of variance was used. For comparisons of elimination half-lives, paired Student's *t*t-test was used. For the study of correlations between variables, Pearson's correlation test was used. Metabolic ratios were log-transformed prior to the inclusion in the analyses. All calculations were carried out by the SYSTAT program package, version 5.2.1. (Systat Inc., Evanston, IL, USA). *P*P values of less than 0.05 were regarded as statistically significant.
|
| 90 |
+
|
| 91 |
+
## Results
|
| 92 |
+
|
| 93 |
+
The pharmacokinetic parameters for the four different maintenance doses of fluvoxamine are presented in [Table 1](#tbl1)Table 1, and the increases in fluvoxamine average steady-state concentrations with increasing doses are illustrated in [Figure 1](#fig01)Figure 1. There was a consistent and disproportionate increase in fluvoxamine AUC values throughout the dose range studied (*P*P<0.0001). Relative to the AUC/dose ratio at 25 mg day^−1^−1, the mean increase was 58% at 50 mg day^−1^−1, 153% at 100 mg day^−1^−1, and 299% at 200 mg day^−1^−1. Mean AUC ratios for the doses 50 mg *vs*vs 25 mg, 100 mg *vs*vs 50 mg, and 200 mg *vs*vs 100 mg were fairly constant, 3.25, 3.17 and 3.14, respectively. All subjects showed disproportionate increases in *C*C_0_0 and *C*C_max_max, and a marked decrease in CL/*F*F with increasing doses (*P*P<0.0001 for all parameters). There were no changes in *t*t_max_max with increasing doses. Mean *C*C_max_max/*C*C_0_0 ratios decreased significantly with increasing dose and were 1.96±0.78 at a dose of 25 mg day^−1^−1, 1.62±0.32 at a dose of 50 mg day^−1^−1, 1.51±0.21 at a dose of 100 mg day^−1^−1, and 1.48±0.41 at a dose of 200 mg day^−1^−1 (*P*P =0.008).
|
| 94 |
+
|
| 95 |
+
### Table 1.
|
| 96 |
+
|
| 97 |
+
Pharmacokinetic parameters for fluvoxamine at steady-state in 10 healthy volunteers given fluvoxamine in increasing doses for 4 weeks
|
| 98 |
+
|
| 99 |
+
### Figure 1.
|
| 100 |
+
|
| 101 |
+

|
| 102 |
+
|
| 103 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=1873375_bcp0045-0257-f1.jpg)
|
| 104 |
+
|
| 105 |
+
Plot of fluvoxamine dose against average steady-state serum concentration in eight CYP2D6 extensive metabolisers (whole lines) and two CYP2D6 poor metabolisers (dotted lines). Identical symbols in Figures 1–3 represent the same subject
|
| 106 |
+
|
| 107 |
+
Individual elimination rates after discontinuation of fluvoxamine are illustrated in [Figure 2](#fig02)Figure 2. Mean *t*t_1/2,12–48_1/2,12–48 was 32.1±11.1 h (range 11.5–45.5 h) whereas mean *t*t_1/2,z_1/2,z was 15.8±4.2 h (range 7.9–19.4 h) (*P*P<0.001). There was a considerable interindividual variation in the average steady-state concentrations ([Table 1](#tbl1)Table 1, [Figure 1](#fig01)Figure 1). For example, at a dose of 100 mg day^−1^−1, a 6.8-fold variation in average steady-state serum concentrations was encountered. Moreover, the subject with the lowest serum concentrations had an average steady-state concentration at a dose of 200 mg day^−1^−1 that the subject with the highest serum concentrations would be expected to have at a dose of approximately 75 mg day^−1^−1.
|
| 108 |
+
|
| 109 |
+
### Figure 2.
|
| 110 |
+
|
| 111 |
+

|
| 112 |
+
|
| 113 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=1873375_bcp0045-0257-f2.jpg)
|
| 114 |
+
|
| 115 |
+
Elimination rates of fluvoxamine in eight CYP2D6 extensive metabolisers (whole lines) and two CYP2D6 poor metabolisers (dotted lines) after discontinuation of fluvoxamine. The subjects had been treated with fluvoxamine 200 mg day−1, and steady-state conditions were achieved in all subjects. The abscissa represents the time after the last dose of fluvoxamine was ingested. Identical symbols in Figures 1–3 represent the same subject
|
| 116 |
+
|
| 117 |
+
There were no correlations between age, body weight, or CYP2C19 metabolic ratio and AUC at different dose levels. There was a non-significant, negative correlation between paraxanthine/caffeine ratio in saliva and fluvoxamine AUC (*r*r =−0.48 at 25 mg day^−1^−1; *r*r =−0.56 (*P*P =0.095) at 50 mg day^−1^−1, *r*r =−0.33 at 100 mg day^−1^−1; *r*r =−0.13 at 200 mg day^−1^−1). The two CYP2D6 PMs had fluvoxamine concentrations ([Figure 1](#fig01)Figure 1) and AUC values ([Table 2](#tbl2)Table 2) within the range of the eight CYP2D6 EMs. In both the EMs and the PMs, the plots of daily dose/*C*C_av_av against daily dose revealed downward convex curves of essentially the same shape ([Figure 3](#fig03)Figure 3).
|
| 118 |
+
|
| 119 |
+
### Table 2.
|
| 120 |
+
|
| 121 |
+
AUC (nmol l−1 h) for CYP2D6 extensive metabolisers (n =8) and CYP2D6 poor metabolisers (n =2) given fluvoxamine in increasing doses for 4 weeks
|
| 122 |
+
|
| 123 |
+
### Figure 3.
|
| 124 |
+
|
| 125 |
+

|
| 126 |
+
|
| 127 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=1873375_bcp0045-0257-f3.jpg)
|
| 128 |
+
|
| 129 |
+
Plot of fluvoxamine daily dose/average steady-state serum concentration ratio against daily dose in eight CYP2D6 extensive metabolisers (whole lines) and two CYP2D6 poor metabolisers (dotted lines). Identical symbols in Figures 1–3 represent the same subject
|
| 130 |
+
|
| 131 |
+
At the doses 25 and 50 mg day^−1^−1, fluvoxamine was well tolerated. During the 2 weeks at these doses, one subject had transitory light headache, one had intermittent diarrhoea, and two experienced transient nausea. At a dose of 100 mg day^−1^−1, five of the volunteers experienced more long-lasting adverse drug reactions, including nausea and drowsiness (two subjects), nausea, headache, and excessive yawning. At a dose of 200 mg day^−1^−1, all but one subject reported adverse drug reactions. These were drowsiness (*n*n =7), nervousness/restlessness (*n*n =6), nausea, particularly after sports and training (*n*n =4), excessive yawning (*n*n =4), headache (*n*n =3), insomnia (*n*n =3), tremor (*n*n =2), dyspepsia (*n*n =1), decreased libido (*n*n =1), paraesthesias (*n*n =1), diarrhoea (*n*n =1) and dry mouth (*n*n =1). No subjects interrupted the treatment due to adverse effects. There was a close to significant positive correlation between the number of suspected adverse drug reactions reported during week 4 and the trough fluvoxamine serum concentration at the end of the same week ([Figure 4](#fig04)Figure 4) (*r*r =0.62; *P*P =0.057).
|
| 132 |
+
|
| 133 |
+
### Figure 4.
|
| 134 |
+
|
| 135 |
+

|
| 136 |
+
|
| 137 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=1873375_bcp0045-0257-f4.jpg)
|
| 138 |
+
|
| 139 |
+
Correlation between number of suspected adverse drug reactions reported during 1 week of treatment with fluvoxamine 200 mg day−1 and trough steady-state serum fluvoxamine concentrations at the end of the same week (r =0.62; P =0.057)
|
| 140 |
+
|
| 141 |
+
## Discussion
|
| 142 |
+
|
| 143 |
+
The present study clearly demonstrates that fluvoxamine AUC disproportionally increases with increasing doses within the clinically used dose range. The evidence of non-linear kinetics of fluvoxamine confirms the results from some earlier studies [[18](#b18)18, [19](#b19)19, [22](#b22)22].
|
| 144 |
+
|
| 145 |
+
For practical reasons as well as for safety reasons, the order of the different maintenance doses of fluvoxamine were not randomised, but were given in order of increasing dose. In the statistical analysis, a possible effect of the order of treatment can therefore not be separated from that due to the different doses. However, the shape of the concentration-time curves after discontinuation of fluvoxamine clearly supports that our findings are dose-dependent rather that time-dependent.
|
| 146 |
+
|
| 147 |
+
The design of the study does not allow us to state with certainty that steady-state conditions have been achieved at all dose levels. Based on the elimination half-lives reported in the literature [[3](#b3)3–[6](#b6)6, [18](#b18)18, [20](#b20)20, [21](#b21)21, [26](#b26)26], it seems reasonable that steady-state conditions have been achieved at doses of 25 and 50 mg day^−1^−1. At doses of 100 and 200 mg day^−1^−1, mean increases in concentrations from 0 to 12 h post-dose day 7 were 2.6% and 0.1%, respectively, indicating that conditions close to steady-state have been achieved. Moreover, initial half-lives after discontinuation of fluvoxamine ([Figure 2](#fig02)Figure 2) can roughly be estimated to be less than 50 h in most cases, although it cannot be excluded that a few subjects might have initial half-lives up to 100 h. Nevertheless, if steady-state conditions have not been achieved at the highest dose levels, the real differences will, in fact, be larger than found in the current study.
|
| 148 |
+
|
| 149 |
+
In the only study where fluvoxamine has been given intravenously [[4](#b4)4], mean oral bioavailability was 53%. Based on the mean AUC value in that study, clearance can be estimated to be approximately 2100 ml min^−1^−1 after a single intravenous dose of 30 mg. Thus, fluvoxamine is a high clearance drug, at least at low doses. The high clearance values compared with liver blood flow are probably mainly caused by the fact that the concentration of fluvoxamine is 50–70% higher in erythrocytes than in serum (unpublished observations from our laboratory), although the possibility of extrahepatic metabolism cannot be excluded. As shown in the present study, oral clearance will clearly decrease with increasing fluvoxamine dosage and serum concentrations. Moreover, in subjects lacking functional CYP2D6 and having a low metabolic capacity of other major enzymes involved in fluvoxamine metabolism, fluvoxamine behaves as a low-clearance rather than a high-clearance drug [[26](#b26)26].
|
| 150 |
+
|
| 151 |
+
In most other studies, mean elimination half-lives of fluvoxamine in healthy, young volunteers have been 15–20 h after single oral doses of 25–100 mg [[3](#b3)3, [18](#b18)18, [20](#b20)20, [21](#b21)21], although several recent studies have reported mean half-lives of 10–14 h [[4](#b4)4–[6](#b6)6, [26](#b26)26]. The terminal elimination half-lives (*t*t_1/2,z_1/2,z) seen in the present study correspond well with these values. The half-lives based on the serum concentrations 12–48 h after the last dose (*t*t_1/2,12–48_1/2,12–48) are longer than ever reported earlier, except among patients with liver cirrhosis [[23](#b23)23] and after fluvoxamine overdose [[22](#b22)22]. Moreover, as seen in [Figure 2](#fig02)Figure 2, these half-lives most probably are an underestimation of the real initial half-lives.
|
| 152 |
+
|
| 153 |
+
Plots of daily dose/*C*C_av_av ratio against daily dose should yield a horizontal line (slope=0) if the elimination pathway involved exhibits linear kinetics at the concentrations studied, and would have a negative slope if the pathway is saturable at the concentrations studied. A curvilinear plot with a downwards convex slope (negative slope decreasing), as here, implies the coexistence of multiple parallel pathways. In both the eight CYP2D6 EMs and the two CYP2D6 PMs, the plots revealed downward convex curves. Thus, our results indicate that several parallel metabolic pathways exist in CYP2D6 PMs as well as in CYP2D6 EMs. Most probably, one of these is CYP1A2. CYP1A2 is one of the major enzymes involved in the metabolism of fluvoxamine [[5](#b5)5], and saturation kinetics have been demonstrated for other drugs metabolised by this enzyme, such as theophylline and caffeine [[14](#b14)14, [15](#b15)15]. The finding that the *r*r values for the correlation between CYP1A2 metabolic ratio and fluvoxamine AUC at the doses of 100 and 200 mg day^−1^−1 were closer to zero than the corresponding *r*r values at lower does might also indicate that CYP1A2 gradually becomes saturated with increasing fluvoxamine doses.
|
| 154 |
+
|
| 155 |
+
In CYP2D6 EMs, the other enzyme involved might be CYP2D6 [[6](#b6)6], which is also characterized by saturation kinetics [[17](#b17)17]. However, the finding that CYP2D6 PMs also had curvilinear plots with the negative slope decreasing indicates that at least one other, non-CYP1A2, non-CYP2D6 pathway, is involved in the metabolism of fluvoxamine. This pathway is most likely not CYP2C19 [[6](#b6)6]. The reason why the two CYP2D6 PMs had AUC values in the same range as the EMs might be that they underwent a faster metabolism via other enzyme systems; in fact, the PM tested was the second fastest caffeine metaboliser. Model fitting with the Michaelis-Menten equation to ascertain *V*V_max_max and *K*K_m_*m*m values was considered not meaningful due to the small number of data points available from each subject and due to the complex situation when two different saturable processes (CYP1A2 and CYP2D6) with different *V*V_max_max and *K*K_m_*m*m values might occur concomitantly in addition to a presumably non-saturable low-affinity metabolic pathway.
|
| 156 |
+
|
| 157 |
+
We found a close to significant positive correlation between the number of suspected adverse effects reported during week 4 and trough fluvoxamine serum concentrations at the end of the same week, indicating that a concentration-adverse drug reaction relationship might exist. At a dose of 200 mg day^−1^−1, there was no clear cut-off serum concentration below which adverse effects were not present. However, on the other hand, the relative lack of adverse effects at the doses 25–50 mg day^−1^−1 strongly indicate that the occurrence of adverse drug reactions is dose and concentration dependent within the individuals, although the influence of time factors cannot be completely ruled out. A concentration-adverse drug reaction relationship has also been observed in a study of 18 patients with major depression treated with fluvoxamine 100–300 mg day^−1^−1 for 4 weeks [[27](#b27)27]. In contrast, no concentration-adverse drug reaction effect relationships were revealed in two other studies [[28](#b28)28, [29](#b29)29], but the methodology for the detection of adverse effects in these studies is insufficiently described.
|
| 158 |
+
|
| 159 |
+
In conclusion, the present study clearly demonstrates that fluvoxamine exhibits non-linear kinetics within the therapeutic dose interval. The reason for non-linearity is not Michaelis-Menten saturation kinetics of a single metabolic pathway, but rather a more complex metabolic picture with multiple pathways involved. The clinical implications of these findings are two-fold: First, changes in fluvoxamine dosage cause disproportionate changes in the serum levels of fluvoxamine. Although fluvoxamine has a broad therapeutic range [[1](#b1)1, [2](#b2)2], the results from the present study as well as from another study [[27](#b27)27] indicate that higher serum concentrations are associated with more adverse effects. Therefore, large and sudden increases in maintenance doses of fluvoxamine should preferably be avoided. Second, the present study confirms the finding from a case report [[20](#b20)20], indicating that a disproportionate extension of the recovery time may occur in cases of fluvoxamine intoxications.
|
| 160 |
+
|
| 161 |
+
## Acknowledgments
|
| 162 |
+
|
| 163 |
+
We acknowledge Martin Bäckström, Lena Carleborg and Åke Norström for excellent clinical and technical assistance. Olav Spigset was the recipient of a fellowship in clinical pharmacology, funded by Merck Sharp & Dohme (Sweden). Financial support was achieved from research funds from the Medical Faculty, Umeå University.
|
| 164 |
+
|
| 165 |
+
## References
|
| 166 |
+
|
| 167 |
+
1. Wilde MI, Plosker GL, Benfield P. Fluvoxamine. Drugs. 1993;46:895–924. doi: 10.2165/00003495-199346050-00008. [DOI](https://doi.org/10.2165/00003495-199346050-00008) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7507038/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drugs&title=Fluvoxamine&author=MI%20Wilde&author=GL%20Plosker&author=P%20Benfield&volume=46&publication_year=1993&pages=895-924&pmid=7507038&doi=10.2165/00003495-199346050-00008&)
|
| 168 |
+
|
| 169 |
+
2. Palmer KH, Benfield P. Fluvoxamine. CNS Drugs. 1994;1:57–87. [Google Scholar](https://scholar.google.com/scholar_lookup?journal=CNS%20Drugs&title=Fluvoxamine&author=KH%20Palmer&author=P%20Benfield&volume=1&publication_year=1994&pages=57-87&)
|
| 170 |
+
|
| 171 |
+
3. de Bree H, van der Schoot JB, Post LC. Fluvoxamine maleate: disposition in man. Eur J Drug Metabolism Pharmacokinet. 1983;8:175–179. doi: 10.1007/BF03188757. [DOI](https://doi.org/10.1007/BF03188757) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/6418548/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Drug%20Metabolism%20Pharmacokinet&title=Fluvoxamine%20maleate:%20disposition%20in%20man&author=H%20de%20Bree&author=JB%20van%20der%20Schoot&author=LC%20Post&volume=8&publication_year=1983&pages=175-179&pmid=6418548&doi=10.1007/BF03188757&)
|
| 172 |
+
|
| 173 |
+
4. van Harten J, Lönnebo A, Grahnén A. Pharmacokinetics of fluvoxamine after intravenous and oral administration [abstract] Neuropsychopharmacology. 1994;10 [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Neuropsychopharmacology&title=Pharmacokinetics%20of%20fluvoxamine%20after%20intravenous%20and%20oral%20administration%20%5Babstract%5D&author=J%20van%20Harten&author=A%20L%C3%B6nnebo&author=A%20Grahn%C3%A9n&volume=10&publication_year=1994&)
|
| 174 |
+
|
| 175 |
+
5. Spigset O, Carleborg L, Hedenmalm K, Dahlqvist R. Effect of cigarette smoking on fluvoxamine pharmacokinetics in humans. Clin Pharmacol Ther. 1995;58:399–403. doi: 10.1016/0009-9236(95)90052-7. [DOI](https://doi.org/10.1016/0009-9236(95)90052-7) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7586931/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Effect%20of%20cigarette%20smoking%20on%20fluvoxamine%20pharmacokinetics%20in%20humans&author=O%20Spigset&author=L%20Carleborg&author=K%20Hedenmalm&author=R%20Dahlqvist&volume=58&publication_year=1995&pages=399-403&pmid=7586931&doi=10.1016/0009-9236(95)90052-7&)
|
| 176 |
+
|
| 177 |
+
6. Spigset O, Granberg K, Hägg S, Norström Å, Dahlqvist R. Relationship between fluvoxamine pharmacokinetics and CYP2D6/CYP2C19 polymorphisms. Eur J Clin Pharmacol. 1997;52:129–133. doi: 10.1007/s002280050261. [DOI](https://doi.org/10.1007/s002280050261) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9174682/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=Relationship%20between%20fluvoxamine%20pharmacokinetics%20and%20CYP2D6/CYP2C19%20polymorphisms&author=O%20Spigset&author=K%20Granberg&author=S%20H%C3%A4gg&author=%C3%85%20Norstr%C3%B6m&author=R%20Dahlqvist&volume=52&publication_year=1997&pages=129-133&pmid=9174682&doi=10.1007/s002280050261&)
|
| 178 |
+
|
| 179 |
+
7. Brøsen K, Skjelbo E, Rasmussen BB, Poulsen HE, Loft S. Fluvoxamine is a potent inhibitor of cytochrome P4501A2. Biochem Pharmacol. 1993;45:1211–1214. doi: 10.1016/0006-2952(93)90272-x. [DOI](https://doi.org/10.1016/0006-2952(93)90272-x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8466541/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Biochem%20Pharmacol&title=Fluvoxamine%20is%20a%20potent%20inhibitor%20of%20cytochrome%20P4501A2&author=K%20Br%C3%B8sen&author=E%20Skjelbo&author=BB%20Rasmussen&author=HE%20Poulsen&author=S%20Loft&volume=45&publication_year=1993&pages=1211-1214&pmid=8466541&doi=10.1016/0006-2952(93)90272-x&)
|
| 180 |
+
|
| 181 |
+
8. Skjelbo E, Brøsen K. Inhibitors of imipramine metabolism by human liver microsomes. Br J Clin Pharmacol. 1992;34:256–261. doi: 10.1111/j.1365-2125.1992.tb04133.x. [DOI](https://doi.org/10.1111/j.1365-2125.1992.tb04133.x) | [PMC free article](/articles/PMC1381397/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1389950/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Inhibitors%20of%20imipramine%20metabolism%20by%20human%20liver%20microsomes&author=E%20Skjelbo&author=K%20Br%C3%B8sen&volume=34&publication_year=1992&pages=256-261&pmid=1389950&doi=10.1111/j.1365-2125.1992.tb04133.x&)
|
| 182 |
+
|
| 183 |
+
9. Spina E, Pollicino AM, Avenoso A, Campo GM, Perucca E, Caputi AP. Effect of fluvoxamine on the pharmacokinetics of imipramine and desipramine in healthy subjects. Ther Drug Monit. 1993;15:243–246. doi: 10.1097/00007691-199306000-00011. [DOI](https://doi.org/10.1097/00007691-199306000-00011) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8333005/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ther%20Drug%20Monit&title=Effect%20of%20fluvoxamine%20on%20the%20pharmacokinetics%20of%20imipramine%20and%20desipramine%20in%20healthy%20subjects&author=E%20Spina&author=AM%20Pollicino&author=A%20Avenoso&author=GM%20Campo&author=E%20Perucca&volume=15&publication_year=1993&pages=243-246&pmid=8333005&doi=10.1097/00007691-199306000-00011&)
|
| 184 |
+
|
| 185 |
+
10. Sperber AD. Toxic interaction between fluvoxamine and sustained release theophylline in an 11-year-old boy. Drug Saf. 1991;6:460–462. doi: 10.2165/00002018-199106060-00006. [DOI](https://doi.org/10.2165/00002018-199106060-00006) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1793525/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Saf&title=Toxic%20interaction%20between%20fluvoxamine%20and%20sustained%20release%20theophylline%20in%20an%2011-year-old%20boy&author=AD%20Sperber&volume=6&publication_year=1991&pages=460-462&pmid=1793525&doi=10.2165/00002018-199106060-00006&)
|
| 186 |
+
|
| 187 |
+
11. Donaldsson KM, Wright DM, Mathlener IS, Harry JD. The effect of fluvoxamine at steady state on the pharmacokinetics of theophylline after a single dose in healthy male volunteers. Br J Clin Pharmacol. 1994;37 [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=The%20effect%20of%20fluvoxamine%20at%20steady%20state%20on%20the%20pharmacokinetics%20of%20theophylline%20after%20a%20single%20dose%20in%20healthy%20male%20volunteers&author=KM%20Donaldsson&author=DM%20Wright&author=IS%20Mathlener&author=JD%20Harry&volume=37&publication_year=1994&)
|
| 188 |
+
|
| 189 |
+
12. Jeppesen U, Gram LF, Vistisen K, Loft S, Poulsen HE, Brøsen K. Dose-dependent inhibition of CYP1A2, CYP2C19 and CYP2D6 by citalopram, fluoxetine, fluvoxamine and paroxetine. Eur J Clin Pharmacol. 1996;51:73–78. doi: 10.1007/s002280050163. [DOI](https://doi.org/10.1007/s002280050163) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8880055/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=Dose-dependent%20inhibition%20of%20CYP1A2,%20CYP2C19%20and%20CYP2D6%20by%20citalopram,%20fluoxetine,%20fluvoxamine%20and%20paroxetine&author=U%20Jeppesen&author=LF%20Gram&author=K%20Vistisen&author=S%20Loft&author=HE%20Poulsen&volume=51&publication_year=1996&pages=73-78&pmid=8880055&doi=10.1007/s002280050163&)
|
| 190 |
+
|
| 191 |
+
13. Jeppesen U, Loft S, Poulsen HE, Brøsen K. A fluvoxamine-caffeine interaction study. Pharmacogenetics. 1996;6:213–222. doi: 10.1097/00008571-199606000-00003. [DOI](https://doi.org/10.1097/00008571-199606000-00003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8807660/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=A%20fluvoxamine-caffeine%20interaction%20study&author=U%20Jeppesen&author=S%20Loft&author=HE%20Poulsen&author=K%20Br%C3%B8sen&volume=6&publication_year=1996&pages=213-222&pmid=8807660&doi=10.1097/00008571-199606000-00003&)
|
| 192 |
+
|
| 193 |
+
14. Dahlqvist R, Billing B, Miners JO, Birkett DJ. Nonlinear metabolic disposition of theophylline. Ther Drug Monit. 1984;6:290–297. doi: 10.1097/00007691-198409000-00006. [DOI](https://doi.org/10.1097/00007691-198409000-00006) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/6506136/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ther%20Drug%20Monit&title=Nonlinear%20metabolic%20disposition%20of%20theophylline&author=R%20Dahlqvist&author=B%20Billing&author=JO%20Miners&author=DJ%20Birkett&volume=6&publication_year=1984&pages=290-297&pmid=6506136&doi=10.1097/00007691-198409000-00006&)
|
| 194 |
+
|
| 195 |
+
15. Kamimori GH, Lugo SI, Penetar DM, et al. Dose-dependent caffeine pharmacokinetics during severe sleep deprivation in humans. Int J Clin Pharmacol Ther. 1995;33:182–186. [PubMed](https://pubmed.ncbi.nlm.nih.gov/7599918/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int%20J%20Clin%20Pharmacol%20Ther&title=Dose-dependent%20caffeine%20pharmacokinetics%20during%20severe%20sleep%20deprivation%20in%20humans&author=GH%20Kamimori&author=SI%20Lugo&author=DM%20Penetar&volume=33&publication_year=1995&pages=182-186&pmid=7599918&)
|
| 196 |
+
|
| 197 |
+
16. Campbell ME, Grant DM, Inaba T, Kalow W. Biotransformation of caffeine, theophylline and theobromine by polycyclic aromatic hydrocarbon-inducible cytochrome(s) P-450 in human liver microsomes. Drug Metab Dispos. 1987;15:237–248. [PubMed](https://pubmed.ncbi.nlm.nih.gov/2882985/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Metab%20Dispos&title=Biotransformation%20of%20caffeine,%20theophylline%20and%20theobromine%20by%20polycyclic%20aromatic%20hydrocarbon-inducible%20cytochrome(s)%20P-450%20in%20human%20liver%20microsomes&author=ME%20Campbell&author=DM%20Grant&author=T%20Inaba&author=W%20Kalow&volume=15&publication_year=1987&pages=237-248&pmid=2882985&)
|
| 198 |
+
|
| 199 |
+
17. Sindrup SH, Brøsen K, Gram LF. Pharmacokinetics of the selective serotonin reuptake inhibitor paroxetine: nonlinearity and relation to the sparteine oxidation polymorphism. Clin Pharmacol Ther. 1992;51:288–295. doi: 10.1038/clpt.1992.24. [DOI](https://doi.org/10.1038/clpt.1992.24) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1531951/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Pharmacokinetics%20of%20the%20selective%20serotonin%20reuptake%20inhibitor%20paroxetine:%20nonlinearity%20and%20relation%20to%20the%20sparteine%20oxidation%20polymorphism&author=SH%20Sindrup&author=K%20Br%C3%B8sen&author=LF%20Gram&volume=51&publication_year=1992&pages=288-295&pmid=1531951&doi=10.1038/clpt.1992.24&)
|
| 200 |
+
|
| 201 |
+
18. de Vries MH, Raghoebar M, Mathlener IS, van Harten J. Single and multiple oral dose fluvoxamine kinetics in young and elderly subjects. Ther Drug Monit. 1992;14:493–598. doi: 10.1097/00007691-199212000-00010. [DOI](https://doi.org/10.1097/00007691-199212000-00010) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1485372/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ther%20Drug%20Monit&title=Single%20and%20multiple%20oral%20dose%20fluvoxamine%20kinetics%20in%20young%20and%20elderly%20subjects&author=MH%20de%20Vries&author=M%20Raghoebar&author=IS%20Mathlener&author=J%20van%20Harten&volume=14&publication_year=1992&pages=493-598&pmid=1485372&doi=10.1097/00007691-199212000-00010&)
|
| 202 |
+
|
| 203 |
+
19. Härtter S, Wetzel H, Hiemke C. Automated determination of fluvoxamine in plasma by column-switching high-performance liquid chromatography. Clin Chem. 1992;38:2082–2086. [PubMed](https://pubmed.ncbi.nlm.nih.gov/1394994/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Chem&title=Automated%20determination%20of%20fluvoxamine%20in%20plasma%20by%20column-switching%20high-performance%20liquid%20chromatography&author=S%20H%C3%A4rtter&author=H%20Wetzel&author=C%20Hiemke&volume=38&publication_year=1992&pages=2082-2086&pmid=1394994&)
|
| 204 |
+
|
| 205 |
+
20. de Vries MH, van Harten J, van Bemmel P, Raghoebar M. Pharmacokinetics of fluvoxamine maleate after single oral doses in healthy subjects. Biopharm Drug Dispos. 1993;14:291–296. doi: 10.1002/bdd.2510140403. [DOI](https://doi.org/10.1002/bdd.2510140403) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8499580/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Biopharm%20Drug%20Dispos&title=Pharmacokinetics%20of%20fluvoxamine%20maleate%20after%20single%20oral%20doses%20in%20healthy%20subjects&author=MH%20de%20Vries&author=J%20van%20Harten&author=P%20van%20Bemmel&author=M%20Raghoebar&volume=14&publication_year=1993&pages=291-296&pmid=8499580&doi=10.1002/bdd.2510140403&)
|
| 206 |
+
|
| 207 |
+
21. Perucca E, Gatti G, Spina E. Clinical pharmacokinetics of fluvoxamine. Clin Pharmacokinet. 1994;27:175–190. doi: 10.2165/00003088-199427030-00002. [DOI](https://doi.org/10.2165/00003088-199427030-00002) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7988100/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacokinet&title=Clinical%20pharmacokinetics%20of%20fluvoxamine&author=E%20Perucca&author=G%20Gatti&author=E%20Spina&volume=27&publication_year=1994&pages=175-190&pmid=7988100&doi=10.2165/00003088-199427030-00002&)
|
| 208 |
+
|
| 209 |
+
22. Spigset O, Öhman R. Case of fluvoxamine intoxication demonstrating non-linear elimination pharmacokinetics. J Clin Psychopharmacol. 1996;16:255–256. doi: 10.1097/00004714-199606000-00012. [DOI](https://doi.org/10.1097/00004714-199606000-00012) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8784660/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Clin%20Psychopharmacol&title=Case%20of%20fluvoxamine%20intoxication%20demonstrating%20non-linear%20elimination%20pharmacokinetics&author=O%20Spigset&author=R%20%C3%96hman&volume=16&publication_year=1996&pages=255-256&pmid=8784660&doi=10.1097/00004714-199606000-00012&)
|
| 210 |
+
|
| 211 |
+
23. van Harten J, Duchier J, Devissaguet J-P, van Bemmel P, de Vries MH, Raghoebar M. Pharmacokinetics of fluvoxamine maleate in patients with liver cirrhosis after single-dose oral administration. Clin Pharmacokinet. 1993;24:177–182. doi: 10.2165/00003088-199324020-00006. [DOI](https://doi.org/10.2165/00003088-199324020-00006) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8453824/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacokinet&title=Pharmacokinetics%20of%20fluvoxamine%20maleate%20in%20patients%20with%20liver%20cirrhosis%20after%20single-dose%20oral%20administration&author=J%20van%20Harten&author=J%20Duchier&author=J-P%20Devissaguet&author=P%20van%20Bemmel&author=MH%20de%20Vries&volume=24&publication_year=1993&pages=177-182&pmid=8453824&doi=10.2165/00003088-199324020-00006&)
|
| 212 |
+
|
| 213 |
+
24. Sanz EJ, Villén T, Alm C, Bertilsson L. S-mephenytoin hydroxylation phenotypes in a Swedish population determined after coadministration with debrisoquin. Clin Pharmacol Ther. 1989;45:495–499. doi: 10.1038/clpt.1989.63. [DOI](https://doi.org/10.1038/clpt.1989.63) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/2721104/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=S-mephenytoin%20hydroxylation%20phenotypes%20in%20a%20Swedish%20population%20determined%20after%20coadministration%20with%20debrisoquin&author=EJ%20Sanz&author=T%20Vill%C3%A9n&author=C%20Alm&author=L%20Bertilsson&volume=45&publication_year=1989&pages=495-499&pmid=2721104&doi=10.1038/clpt.1989.63&)
|
| 214 |
+
|
| 215 |
+
25. Fuhr U, Rost KL. Simple and reliable CYP1A2 phenotyping by the paraxanthine/caffeine ratio in plasma and in saliva. Pharmacogenetics. 1994;4:109–116. doi: 10.1097/00008571-199406000-00001. [DOI](https://doi.org/10.1097/00008571-199406000-00001) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7920690/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Simple%20and%20reliable%20CYP1A2%20phenotyping%20by%20the%20paraxanthine/caffeine%20ratio%20in%20plasma%20and%20in%20saliva&author=U%20Fuhr&author=KL%20Rost&volume=4&publication_year=1994&pages=109-116&pmid=7920690&doi=10.1097/00008571-199406000-00001&)
|
| 216 |
+
|
| 217 |
+
26. Carrillo JA, Dahl M-L, Svensson J-O, Alm C, Rodríguez I, Bertilsson L. Disposition of fluvoxamine in humans is determined by the polymorphic CYP2D6 and also by the CYP1A2 activity. Clin Pharmacol Ther. 1996;60:183–190. doi: 10.1016/S0009-9236(96)90134-4. [DOI](https://doi.org/10.1016/S0009-9236(96)90134-4) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8823236/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Disposition%20of%20fluvoxamine%20in%20humans%20is%20determined%20by%20the%20polymorphic%20CYP2D6%20and%20also%20by%20the%20CYP1A2%20activity&author=JA%20Carrillo&author=M-L%20Dahl&author=J-O%20Svensson&author=C%20Alm&author=I%20Rodr%C3%ADguez&volume=60&publication_year=1996&pages=183-190&pmid=8823236&doi=10.1016/S0009-9236(96)90134-4&)
|
| 218 |
+
|
| 219 |
+
27. Kasper S, Dötsch M, Kick H, Vieira A, Möller H-J. Plasma concentrations of fluvoxamine and maprotiline in major depression: implications on therapeutic efficacy and side effects. Eur Neuropsychopharmacol. 1993;3:13–21. doi: 10.1016/0924-977x(93)90290-3. [DOI](https://doi.org/10.1016/0924-977x(93)90290-3) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8471827/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20Neuropsychopharmacol&title=Plasma%20concentrations%20of%20fluvoxamine%20and%20maprotiline%20in%20major%20depression:%20implications%20on%20therapeutic%20efficacy%20and%20side%20effects&author=S%20Kasper&author=M%20D%C3%B6tsch&author=H%20Kick&author=A%20Vieira&author=H-J%20M%C3%B6ller&volume=3&publication_year=1993&pages=13-21&pmid=8471827&doi=10.1016/0924-977x(93)90290-3&)
|
| 220 |
+
|
| 221 |
+
28. Klok CJ, Brouwer GJ, van Praag HM, Doogan D. Fluvoxamine and clomipramine in depressed patients. Acta Psychiatr Scand. 1981;64:1–11. doi: 10.1111/j.1600-0447.1981.tb00756.x. [DOI](https://doi.org/10.1111/j.1600-0447.1981.tb00756.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/6172005/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Acta%20Psychiatr%20Scand&title=Fluvoxamine%20and%20clomipramine%20in%20depressed%20patients&author=CJ%20Klok&author=GJ%20Brouwer&author=HM%20van%20Praag&author=D%20Doogan&volume=64&publication_year=1981&pages=1-11&pmid=6172005&doi=10.1111/j.1600-0447.1981.tb00756.x&)
|
| 222 |
+
|
| 223 |
+
29. de Wilde JEM, Doogan DP. Fluvoxamine and chlorimipramine in endogenous depression. J Affect Disord. 1982;4:249–259. doi: 10.1016/0165-0327(82)90009-x. [DOI](https://doi.org/10.1016/0165-0327(82)90009-x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/6215443/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Affect%20Disord&title=Fluvoxamine%20and%20chlorimipramine%20in%20endogenous%20depression&author=JEM%20de%20Wilde&author=DP%20Doogan&volume=4&publication_year=1982&pages=249-259&pmid=6215443&doi=10.1016/0165-0327(82)90009-x&)
|
test/texts/PMC1885108.md
ADDED
|
@@ -0,0 +1,334 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Pharmacokinetics and response to pravastatin in paediatric patients with familial hypercholesterolaemia and in paediatric cardiac transplant recipients in relation to polymorphisms of the SLCO1B1 and ABCB1 genes
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Mia Hedman, Marjatta Antikainen, Christer Holmberg, Mikko Neuvonen, Michel Eichelbaum, Kari T Kivistö, Pertti J Neuvonen, Mikko Niemi
|
| 5 |
+
**Journal:** British Journal of Clinical Pharmacology
|
| 6 |
+
**Date:** 2006 Apr 21
|
| 7 |
+
**DOI:** [10.1111/j.1365-2125.2006.02643.x](https://doi.org/10.1111/j.1365-2125.2006.02643.x)
|
| 8 |
+
**PMID:** 16722833
|
| 9 |
+
**PMCID:** PMC1885108
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1885108/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC1885108/pdf/bcp0061-0706.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC1885108/pdf/bcp0061-0706.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Aims:**
|
| 16 |
+
Our aim was to investigate associations between the single nucleotide polymorphisms (SNPs) in the SLCO1B1 (encoding OATP1B1) and ABCB1 (encoding P-glycoprotein) genes with the pharmacokinetics and efficacy of pravastatin in children with heterozygous familial hypercholesterolaemia (HeFH) and in paediatric cardiac transplant recipients.
|
| 17 |
+
|
| 18 |
+
**Methods:**
|
| 19 |
+
Twenty children with HeFH (aged 4.9–15.6 years) and 12 cardiac transplant recipients (aged 4.4–18.7 years and receiving triple immunosuppressive medication) who had participated in previous pharmacokinetic and pharmacodynamic studies with pravastatin were genotyped for the −11187G > A and 521T > C SNPs in the SLCO1B1 gene and for the 2677G > T/A and 3435C > T SNPs in the ABCB1 gene.
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
Two HeFH patients with the −11187GA genotype had a 81% lower peak plasma pravastatin concentration (Cmax) (difference in means −13.9 ng ml−1, 95% CI −21.1, −6.7; P < 0.001) and a 74% smaller area under the plasma concentration-time curve (AUC(0, ∞)) (−25.3 ng ml−1 h, 95% CI −35.6, −15.0; P < 0.0001) and significantly greater increase in high density lipoprotein (HDL) cholesterol after 2 months treatment with pravastatin than patients with the reference genotype. No significant differences were seen in the pharmacokinetics or effects of pravastatin between HeFH patients with the SLCO1B1 521TC and 521TT genotypes. The cardiac transplant recipients with the SLCO1B1 521TC genotype (n = 3) had a 46% lower Cmax (−67.7 ng ml−1, 95% CI −135.7, 0.3; P = 0.055) and 62% lower AUC(0,24 h) (−228.5 ng ml−1 h, 95% CI −402.7, −54.3; P = 0.016) and a shorter half-life (t1/2) (0.9 ± 0.1 vs. 1.3 ± 0.4 h, P = 0.015) of pravastatin than those with the reference genotype. Decreases in total and low-density lipoprotein cholesterol by pravastatin were significantly smaller, and the increase in HDL-cholesterol was greater in the transplant recipients with the 521TC genotype compared with patients with the 521TT reference genotype.
|
| 23 |
+
|
| 24 |
+
**Conclusions:**
|
| 25 |
+
In children with HeFH and in paediatric cardiac transplant recipients receiving immunosuppressive medication, the −11187G > A and SLCO1B1 521T > C SNPs were associated with decreased plasma concentrations of pravastatin. These differences are opposite to those seen previously in healthy adults. The mechanisms underlying these phenomena are unclear and warrant further study.
|
| 26 |
+
|
| 27 |
+
Keywords: children, cyclosporin, OATP1B1, pravastatin, SLCO1B1
|
| 28 |
+
|
| 29 |
+
### Aims
|
| 30 |
+
|
| 31 |
+
Our aim was to investigate associations between the single nucleotide polymorphisms (SNPs) in the *SLCO1B1*SLCO1B1 (encoding OATP1B1) and *ABCB1*ABCB1 (encoding P-glycoprotein) genes with the pharmacokinetics and efficacy of pravastatin in children with heterozygous familial hypercholesterolaemia (HeFH) and in paediatric cardiac transplant recipients.
|
| 32 |
+
|
| 33 |
+
### Methods
|
| 34 |
+
|
| 35 |
+
Twenty children with HeFH (aged 4.9–15.6 years) and 12 cardiac transplant recipients (aged 4.4–18.7 years and receiving triple immunosuppressive medication) who had participated in previous pharmacokinetic and pharmacodynamic studies with pravastatin were genotyped for the −11187G > A and 521T > C SNPs in the *SLCO1B1*SLCO1B1 gene and for the 2677G > T/A and 3435C > T SNPs in the *ABCB1*ABCB1 gene.
|
| 36 |
+
|
| 37 |
+
### Results
|
| 38 |
+
|
| 39 |
+
Two HeFH patients with the −11187GA genotype had a 81% lower peak plasma pravastatin concentration (*C*C_max_max) (difference in means −13.9 ng ml^−1^−1, 95% CI −21.1, −6.7; *P*P < 0.001) and a 74% smaller area under the plasma concentration-time curve (AUC(0, ∞)) (−25.3 ng ml^−1^−1 h, 95% CI −35.6, −15.0; *P*P < 0.0001) and significantly greater increase in high density lipoprotein (HDL) cholesterol after 2 months treatment with pravastatin than patients with the reference genotype. No significant differences were seen in the pharmacokinetics or effects of pravastatin between HeFH patients with the *SLCO1B1*SLCO1B1 521TC and 521TT genotypes. The cardiac transplant recipients with the *SLCO1B1*SLCO1B1 521TC genotype (*n*n = 3) had a 46% lower *C*C_max_max (−67.7 ng ml^−1^−1, 95% CI −135.7, 0.3; *P*P = 0.055) and 62% lower AUC(0,24 h) (−228.5 ng ml^−1^−1 h, 95% CI −402.7, −54.3; *P*P = 0.016) and a shorter half-life (*t*t_1/2_1/2) (0.9 ± 0.1 *vs.*vs. 1.3 ± 0.4 h, *P*P = 0.015) of pravastatin than those with the reference genotype. Decreases in total and low-density lipoprotein cholesterol by pravastatin were significantly smaller, and the increase in HDL-cholesterol was greater in the transplant recipients with the 521TC genotype compared with patients with the 521TT reference genotype.
|
| 40 |
+
|
| 41 |
+
### Conclusions
|
| 42 |
+
|
| 43 |
+
In children with HeFH and in paediatric cardiac transplant recipients receiving immunosuppressive medication, the −11187G > A and *SLCO1B1*SLCO1B1 521T > C SNPs were associated with decreased plasma concentrations of pravastatin. These differences are opposite to those seen previously in healthy adults. The mechanisms underlying these phenomena are unclear and warrant further study.
|
| 44 |
+
|
| 45 |
+
**Keywords:**Keywords: children, cyclosporin, OATP1B1, pravastatin, SLCO1B1
|
| 46 |
+
|
| 47 |
+
## Introduction
|
| 48 |
+
|
| 49 |
+
The plasma concentrations and the cholesterol-lowering efficacy of pravastatin, a hydrophilic, semisynthetic inhibitor of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase, vary considerably between individuals, both adults [[1](#b1)1] and children [[2](#b2)2]. However, no clear difference in the pharmacokinetics of pravastatin exists between hypercholesterolaemic and normocholesterolaemic adults [[3](#b3)3–[5](#b5)5]. Pravastatin is currently approved by the US Food and Drug Administration for the treatment of children over 8 years of age with familial hypercholesterolaemia (FH). In adult cardiac transplant recipients, pravastatin slows the development of accelerated coronary artery disease, and decreases the rates of cardiovascular morbidity and mortality [[6](#b6)6]. Because progressive coronary artery disease is also the main cause of poor long-term survival in paediatric cardiac transplant recipients [[7](#b7)7, [8](#b8)8], the tolerability, safety and efficacy of pravastatin in this patient group is currently being evaluated [[9](#b9)9, [10](#b10)10].
|
| 50 |
+
|
| 51 |
+
The oral bioavailability of pravastatin is about 20% and it is cleared by both renal and non-renal routes (47% and 53%, respectively) [[5](#b5)5, [11](#b11)11]. The metabolites of pravastatin include 3α-iso-pravastatin and 6-epi-pravastatin, which are formed by nonenzymatic acid-catalyzed isomerization in the stomach or produced in the cytosol of liver and small intestinal mucosa cells, and 3α, 5β-dihydroxy-pravastatin and 3-hydroxypravastatin, which are the major oxidized metabolites [[5](#b5)5]. Unlike many other statins, pravastatin is not significantly biotransformed by cytochrome P450 (CYP) enzymes [[12](#b12)12], and thus it is not susceptible to CYP3A4- [[1](#b1)1], CYP2C9- [[13](#b13)13] or CYP2C19-mediated [[13](#b13)13] drug interactions. Cyclosporin is a known inhibitor/substrate of CYP3A4 [[14](#b14)14], P-glycoprotein [[15](#b15)15], organic anion transporting polypeptide 1B1 (OATP1B1; previously known as OATP-C, OATP2 and LST-1) [[16](#b16)16] and multidrug resistance-associated protein 2 (MRP2) [[17](#b17)17], and it greatly elevates the plasma concentrations of many statins, including pravastatin [[10](#b10)10, [18](#b18)18–[20](#b20)20], and increases the risk of myopathy and rhabdomyolysis during statin therapy [[21](#b21)21, [22](#b22)22].
|
| 52 |
+
|
| 53 |
+
OATP1B1 is responsible for the hepatic uptake of pravastatin [[23](#b23)23, [24](#b24)24]. OATP2B1 (OATP-B) transports pravastatin from gut lumen into the cytosol of intestinal epithelial cells and may facilitate its absorption [[25](#b25)25]. MRP2 transports absorbed pravastatin back into the gut lumen and also mediates the biliary excretion of pravastatin from hepatocytes [[26](#b26)26]. The organic anion transporter 3 (OAT3), a member of the *SLC22*SLC22 superfamily, may affect the urinary excretion of pravastatin [[27](#b27)27]. In adults, the 521T > C (Val174Ala) and −11187G > A single nucleotide polymorphisms (SNPs) in the *SLCO1B1*SLCO1B1 gene encoding for OATP1B1 have been associated with increased plasma concentrations of pravastatin [[28](#b28)28–[30](#b30)30], whereas SNPs in *SLCO2B1*SLCO2B1 (encoding OATP2B1) [[29](#b29)29], *SLC22A8*SLC22A8 (encoding OAT3) [[28](#b28)28], *ABCC2*ABCC2 (encoding MRP2) [[29](#b29)29] or *ABCB1*ABCB1 (encoding P-glycoprotein) genes [[29](#b29)29], have not affected its pharmacokinetics.
|
| 54 |
+
|
| 55 |
+
We have recently studied the pharmacokinetics and efficacy of pravastatin in paediatric patients with heterozygous familial hypercholesterolaemia (HeFH) who received no other concomitant medication [[2](#b2)2] and in paediatric cardiac transplant recipients on a regimen of triple immunosuppression [[10](#b10)10]. As the *SLCO1B1*SLCO1B1 521T > C and −11187G > A SNPs are associated with increased plasma pravastatin concentrations in adults, we investigated possible associations of these SNPs with the pharmacokinetics and efficacy of pravastatin in children. Moreover, as cyclosporin is a substrate of P-glycoprotein, we hypothesized that *ABCB1*ABCB1 SNPs might modulate the effects of cyclosporin on pravastatin. Thus, associations between the *ABCB1*ABCB1 3435C > T and 2677G > T/A SNPs and pravastatin pharmacokinetics were also evaluated.
|
| 56 |
+
|
| 57 |
+
## Methods
|
| 58 |
+
|
| 59 |
+
### Patients
|
| 60 |
+
|
| 61 |
+
Twenty children (13 girls and seven boys) with HeFH, who had participated in our previous pravastatin study [[2](#b2)2], were included. Their mean age was 10.3 ± 2.9 years and their other characteristics are listed in [Table 1](#tbl1)Table 1. The diagnosis of HeFH had been verified by LDL receptor mutation analysis [[31](#b31)31] or by a lymphocyte test [[32](#b32)32], as previously described [[2](#b2)2]. Ten patients tested were positive for the FH-Helsinki LDL receptor mutation, six patients for the FH-North Karelia LDL receptor mutation, and four patients had defective cholesterol intake in the lymphocyte test. Other than HeFH, all patients were healthy and none had received daily medication previously.
|
| 62 |
+
|
| 63 |
+
### Table 1.
|
| 64 |
+
|
| 65 |
+
Characteristics of 20 children with familial hypercholesterolaemia (HeFH) and 12 paediatric cardiac transplant recipients
|
| 66 |
+
|
| 67 |
+
| | Sex | Age (years) | Weight (kg) | Height (cm) | Pravastatin dose (mg kg−1) |
|
| 68 |
+
| - | --- | ----------- | ----------- | ----------- | -------------------------- |
|
| 69 |
+
| Patients with HeFH | | | | | |
|
| 70 |
+
| 1 | Female | 13.0 | 42.0 | 153.8 | 0.24 |
|
| 71 |
+
| 2 | Female | 7.9 | 29.4 | 125.9 | 0.34 |
|
| 72 |
+
| 3 | Female | 6.9 | 26.1 | 125.1 | 0.38 |
|
| 73 |
+
| 4 | Female | 9.7 | 28.4 | 139.7 | 0.35 |
|
| 74 |
+
| 5 | Female | 9.8 | 33.0 | 141.0 | 0.30 |
|
| 75 |
+
| 6 | Female | 13.4 | 45.3 | 160.5 | 0.22 |
|
| 76 |
+
| 7 | Female | 11.9 | 43.5 | 152.5 | 0.23 |
|
| 77 |
+
| 8 | Female | 6.0 | 19.2 | 114.1 | 0.52 |
|
| 78 |
+
| 9 | Female | 14.0 | 63.0 | 161.9 | 0.16 |
|
| 79 |
+
| 10 | Female | 15.6 | 57.8 | 168.4 | 0.17 |
|
| 80 |
+
| 11 | Female | 11.7 | 45.0 | 157.8 | 0.22 |
|
| 81 |
+
| 12 | female | 12.0 | 44.5 | 161.4 | 0.22 |
|
| 82 |
+
| 13 | Female | 12.0 | 50.3 | 152.7 | 0.20 |
|
| 83 |
+
| 14 | Female | 10.3 | 27.5 | 134.5 | 0.36 |
|
| 84 |
+
| 15 | Male | 6.3 | 20.5 | 119.2 | 0.49 |
|
| 85 |
+
| 16 | Male | 7.7 | 23.1 | 120.8 | 0.43 |
|
| 86 |
+
| 17 | Male | 10.3 | 48.8 | 151.8 | 0.20 |
|
| 87 |
+
| 18 | Male | 4.9 | 16.4 | 108.9 | 0.61 |
|
| 88 |
+
| 19 | Male | 10.7 | 56.6 | 145.2 | 0.18 |
|
| 89 |
+
| 20 | Male | 10.7 | 42.5 | 142.3 | 0.24 |
|
| 90 |
+
| Mean ± SD | | 10.3 ± 2.9 | 38.1 ± 13.8 | 141.9 ± 17.7 | 0.30 ± 0.13 |
|
| 91 |
+
| Transplant recipients | | | | | |
|
| 92 |
+
| 21 | Female | 8.0 | 20.0 | 127.0 | 0.50 |
|
| 93 |
+
| 22 | Female | 18.7 | 46.3 | 160.0 | 0.22 |
|
| 94 |
+
| 23 | Female | 8.0 | 24.1 | 127.5 | 0.41 |
|
| 95 |
+
| 24 | Male | 11.6 | 42.5 | 146.0 | 0.24 |
|
| 96 |
+
| 25 | Female | 6.1 | 23.0 | 115.0 | 0.43 |
|
| 97 |
+
| 26 | Male | 13.8 | 40.3 | 143.0 | 0.25 |
|
| 98 |
+
| 27 | Female | 16.1 | 55.6 | 150.0 | 0.18 |
|
| 99 |
+
| 28 | Female | 4.4 | 15.5 | 103.0 | 0.65 |
|
| 100 |
+
| 29 | Female | 11.4 | 61.0 | 154.0 | 0.16 |
|
| 101 |
+
| 30 | Female | 16.9 | 71.0 | 163.0 | 0.14 |
|
| 102 |
+
| 31 | Female | 13.9 | 42.5 | 154.0 | 0.24 |
|
| 103 |
+
| 32 | Fale | 8.8 | 25.0 | 130.5 | 0.40 |
|
| 104 |
+
| Mean ± SD | | 11.5 ± 4.5 | 38.9 ± 17.7 | 139.0 ± 18.8 | 0.32 ± 0.16 |
|
| 105 |
+
Of the 19 paediatric cardiac transplant recipients who had participated in our previous pravastatin study [[10](#b10)10], 12 (nine girls and three boys) were studied again. Their mean age was 11.5 ± 4.5 years and their other characteristics are listed in [Table 1](#tbl1)Table 1. The immunosuppressive protocols included triple therapy with cyclosporin in microemulsion composition (12 patients), methylprednisolone (12 patients), and azathioprine (11 patients) or mycophenolate mofetil (one patient). Detailed information on the immunosuppressive regimen has been published previously [[10](#b10)10]. Additional medication on the day of the pravastatin pharmacokinetic study was the following: diltiazem (*n*n = 1), felodipine (*n*n = 4), nifedipine (*n*n = 1), furosemide (*n*n = 4), propranolol (*n*n = 2), atenolol (*n*n = 1), bisoprolol (*n*n = 1), valganciclovir (*n*n = 2), aciclovir (*n*n = 1), and omeprazole (*n*n = 1).
|
| 106 |
+
|
| 107 |
+
### Study design
|
| 108 |
+
|
| 109 |
+
The protocol was approved by the ethics committee for paediatrics, adolescent medicine and psychiatry of the Helsinki and Uusimaa Hospital District. Written informed consent was obtained from the parents.
|
| 110 |
+
|
| 111 |
+
The patients in the two previous pharmacokinetic studies [[2](#b2)2, [10](#b10)10] had ingested a single 10 mg dose of pravastatin (Pravachol, Bristol-Myers Squibb, Epernon, France) with 150 ml water after an overnight fast and did not eat earlier than 1.5 h after administration of pravastatin. The mean pravastatin doses kg^−1^−1 bodyweight in the children with HeFH and in the transplant recipients were 0.30 ± 0.13 mg kg ^−1^−1 and 0.32 ± 0.16 mg kg^−1^−1, respectively ([Table 1](#tbl1)Table 1). The transplant recipients ingested their morning medication with pravastatin. Plasma pravastatin concentrations had been determined by liquid chromatography-ionspray tandem mass spectrometry with use of the PE SCIEX API 3000 LC/MS/MS system (Sciex Division of MDS Inc, Toronto, Canada) [[33](#b33)33] from samples taken before pravastatin and 0.5, 1, 1.5, 2, 3, 4, 8 and 10 h later. In the transplant recipients additional samples were taken at 12 and 24 h [[2](#b2)2, [10](#b10)10]. The ion transition monitored was m/z 442 to m/z 269, and the limit of quantification was 0.25 ng ml^−1^−1 for pravastatin. The day-to-day coefficient of variation (CV) was 7.8% at 1 ng ml^−1^−1 (*n*n = 6). After the pharmacokinetic study, all patients were treated with pravastatin 10 mg day^−1^−1 and blood lipids and safety parameters were monitored as described earlier [[2](#b2)2, [10](#b10)10]. In the present genetic study, a 1 ml EDTA blood sample was drawn from each subject and stored at −20 °C. DNA was extracted using standard methods (QIAamp DNA Blood Mini Kit, Qiagen, Hilden, Germany).
|
| 112 |
+
|
| 113 |
+
### Pharmacokinetics and pharmacodynamics analysis
|
| 114 |
+
|
| 115 |
+
The pharmacokinetics of pravastatin were characterized by the peak concentration in plasma (*C*C_max_max), the time to peak concentration (*t*t_max_max), the elimination half-life (*t*t_1/2_1/2), and the area under the plasma concentration-time curve from time 0 to infinity (AUC(0, ∞)) (in HeFH patients) [[2](#b2)2] and from 0 to 24 h (AUC(0,24 h)) (in cardiac transplant recipients) [[10](#b10)10]. The pharmacokinetic software used was the MK Model (Biosoft, Cambridge, UK). The pharmacokinetics of pravastatin and the effect of 2 months daily pravastatin treatment on serum cholesterol and triglycerides in HeFH and transplant patients [[2](#b2)2, [10](#b10)10] were evaluated in relation to the current genotyping results.
|
| 116 |
+
|
| 117 |
+
### SLCO1B1 genotyping
|
| 118 |
+
|
| 119 |
+
All patients were genotyped for the −11187G > A SNP in the promoter region and the 521T > C (Val174Ala) SNP in exon 5 of the *SLCO1B1*SLCO1B1 gene by allelic discrimination with TaqMan® 5′nuclease assays, using the ABI Prism 7700 Sequence Detection System (Applied Biosystems, Weiterstadt, Germany). The primers used in 521T > C genotyping were 5′-GAAACACTCTCTTA TCTACATAGGTTGTTTA-3′ (forward) and 5′-CCCC TATTCCACGAAGCAT-3′ (reverse). The TaqMan® MGB probes were VIC-TACCCATGAACACATATA and FAM-TACCCATGAACGCATATA. The primers used for −11187G > A were 5′-CATATATGCATCCTC ACATTACCACAT-3′ (forward) and 5′-AATAAAGTAC AGACCCTTCTCTCACATAAA-3′ (reverse), and the TaqMan® MGB probes were VIC-TGTATACAGGT AAAAGTG and FAM-TGTGTATACAAGTAAAAG. The PCR conditions were one cycle at 50 °C for 2 min and at 95 °C for 10 min, followed by 40 cycles at 92 °C for 15 s and at 60 °C for 1 min, as recommended by the manufacturer. The accuracy of genotyping was confirmed by sequencing.
|
| 120 |
+
|
| 121 |
+
### ABCB1 genotyping
|
| 122 |
+
|
| 123 |
+
All patients were genotyped for the 3435C > T SNP in exon 26 and the 2677G > T/A SNP in exon 21 of the *ABCB1*ABCB1 gene, by denaturing high-performance liquid chromatography [[34](#b34)34]. *ABCB1*ABCB1 haplotype analysis was performed as described previously [[35](#b35)35].
|
| 124 |
+
|
| 125 |
+
### Statistical analysis
|
| 126 |
+
|
| 127 |
+
Results are expressed as mean values (± SD) in the text and tables and, for clarity, as mean (± SEM) in the figures. Statistical comparisons between two groups were made with Student’s *t*t-test for unpaired values. Differences in continuous variables between more than two groups were compared by one-way anovaanova with *a posteriori*a posteriori testing using the Tukey test. *t*t_max_max values were compared with the Mann–Whitney U-test or Kruskall-Wallis anovaanova with *a posteriori*a posteriori testing using Dunn’s test. StatsDirect (StatsDirect Ltd, Cheshire, UK) and SPSS 11.0 for Windows (SPSS Inc., Chicago, Illinois, USA) were used for the statistical analyses. *P*P values less than 0.05 were considered statistically significant.
|
| 128 |
+
|
| 129 |
+
## Results
|
| 130 |
+
|
| 131 |
+
The *SLCO1B1*SLCO1B1 and *ABCB1*ABCB1 alleles and genotypes found in children with HeFH and in the cardiac transplant recipients are shown in [Table 2](#tbl2)Table 2. All observed genotype frequencies were consistent with the Hardy–Weinberg equilibrium.
|
| 132 |
+
|
| 133 |
+
### Table 2.
|
| 134 |
+
|
| 135 |
+
SLCO1B1 and ABCB1 genotypes in 20 children with heterozygous familial hypercholesterolaemia (HeFH) and in 12 paediatric cardiac transplant recipients
|
| 136 |
+
|
| 137 |
+
| Gene | Region | Position | Allele | Patients with HeFH n (%) | Transplant recipients n (%) | Genotype | Patients with HeFH n (%) | Transplant recipients n (%) |
|
| 138 |
+
| ---- | ------ | -------- | ------ | ------------------------ | --------------------------- | -------- | ------------------------ | --------------------------- |
|
| 139 |
+
| SLCO1B1 | Promoter | −11187 | G | 38 (95%) | 24 (100%) | GG | 18 (90%) | 12 (100%) |
|
| 140 |
+
| | | | A | 2 (5%) | 0 (0%) | GA | 2 (10%) | 0 (0%) |
|
| 141 |
+
| SLCO1B1 | Exon 5 | 521 | T | 34 (85%) | 21 (88%) | TT | 14 (70%) | 9 (75%) |
|
| 142 |
+
| | | | C | 6 (15%) | 3 (13%) | TC | 6 (30%) | 3 (25%) |
|
| 143 |
+
| ABCB1 | Exon 21 | 2677 | G | 17 (43%) | 15 (63%) | GG | 4 (20%) | 5 (42%) |
|
| 144 |
+
| | | | T | 21 (53%) | 8 (33%) | GT | 8 (40%) | 5 (42%) |
|
| 145 |
+
| | | | A | 2 (5%) | 1 (4%) | TT | 6 (30%) | 1 (8%) |
|
| 146 |
+
| | | | | | | GA | 1 (5%) | 0 (0%) |
|
| 147 |
+
| | | | | | | TA | 1 (5%) | 1 (8%) |
|
| 148 |
+
| ABCB1 | Exon 26 | 3435 | C | 20 (50%) | 13 (54%) | CC | 5 (25%) | 3 (25%) |
|
| 149 |
+
| | | | T | 20 (50%) | 11 (46%) | CT | 10 (50%) | 7 (58%) |
|
| 150 |
+
| | | | | | | TT | 5 (25%) | 2 (17%) |
|
| 151 |
+
Compared with 18 patients with the reference (wild type) genotype, two patients with the *SLCO1B1*SLCO1B1−11187GA genotype (who also had the 521TC genotype) had an 81% lower mean *C*C_max_max (*P*P< 0.001) and a 74% lower AUC(0, ∞) (*P*P< 0.0001), and significantly greater increase in HDL-cholesterol after 2 months of pravastatin 10 mg day^−1^−1 (26.7% ± 2.9%*vs.*vs. 3.2% ± 14.0%, *P*P = 0.0002) ([Table 3](#tbl3)Table 3, [Figure 1](#fig01)Figure 1). Children with the *SLCO1B1*SLCO1B1 521TC genotype (*n*n = 6) had a 49% lower mean *C*C_max_max (*P*P= 0.136) and 26% lower AUC(0, ∞) (*P*P= 0.409) of pravastatin than those with the reference genotype, but these differences were not statistically significant. Patient characteristics were similar in subjects with different *SLCO1B1*SLCO1B1 genotypes ([Table 3](#tbl3)Table 3). No significant differences existed in the lipid-lowering efficacy of pravastatin. Furthermore, there were no significant differences in the pharmacokinetics or effects of pravastatin between subjects with different *ABCB1*ABCB1 3435C > T or 2677G > T/A genotypes or haplotypes (data not shown).
|
| 152 |
+
|
| 153 |
+
### Table 3.
|
| 154 |
+
|
| 155 |
+
Comparison of the baseline characteristics and the pravastatin pharmacokinetic and pharmacodynamic data in relation to the SLCO1B1 521T > C and −11187G > A single nucleotide polymorphisms in children with heterozygous familial hypercholesterolaemia
|
| 156 |
+
|
| 157 |
+
| Variable | 521TT (n = 14) | 521TC (n = 6) | Difference in means (95% CI) | P | −11187GG (n = 18) | −11187GA (n = 2) | Difference in means (95% CI) | P |
|
| 158 |
+
| -------- | -------------- | ------------- | ---------------------------- | - | ----------------- | ---------------- | ---------------------------- | - |
|
| 159 |
+
| Age (years) | 10.1 ± 3.2 | 10.6 ± 2.2 | 0.5 (−2.6, 3.5) | | 10.2 ± 2.8 | 10.1 ± 4.6 | −0.1 (−4.7, 4.5) | |
|
| 160 |
+
| Weight (kg) | 37.1 ± 15.0 | 40.6 ± 11.5 | 3.5 (−11.0, 17.9) | | 38.4 ± 14.2 | 35.7 ± 13.6 | −0.7 (−24.9, 19.5) | |
|
| 161 |
+
| Height (cm) | 140.2 ± 19.5 | 145.7 ± 13.7 | 5.5 (−13.0, 24.0) | | 141.8 ± 17.7 | 142.8 ± 25.0 | 1.0 (−27.5, 29.6) | |
|
| 162 |
+
| Body mass index (kg m−2) | 17.9 ± 3.1 | 19.0 ± 4.4 | 1.1 (−2.5, 4.6) | | 18.4 ± 3.6 | 17.2 ± 0.6 | −1.2 (−6.7, 4.3) | |
|
| 163 |
+
| Pravastatin | | | | | | | | |
|
| 164 |
+
| dose (mg kg−1) | 0.32 ± 0.14 | 0.27 ± 0.08 | −0.05 (−0.19, 0.08) | 0.403 | 0.30 ± 0.13 | 0.30 ± 0.11 | 0.0 (−0.21, 0.20) | 0.974 |
|
| 165 |
+
| Cmax (ng ml−1) | 18.4 ± 15.6 | 9.4 ± 9.6 | −9.0 (−21.0, 3.0) | 0.136 | 17.1 ± 14.6 | 3.2 ± 0.1 | −13.9 (−21.1, −6.7) | <0.001 |
|
| 166 |
+
| AUC(0,∞) (ng ml−1 h) | 34.5 ± 21.7 | 25.7 ± 20.0 | −8.8 (−30.6, 13.0) | 0.409 | 34.4 ± 20.7 | 9.1 ± 0.4 | −25.3 (−35.6, −15.0) | <0.0001 |
|
| 167 |
+
| tmax (h) | 1.3 (0.5–4.0) | 1.3 (1.0–1.5) | | 0.785 | 1.3 (0.5–4) | 1.3 (1–1.5) | | 0.568 |
|
| 168 |
+
| t1/2 (h) | 1.6 ± 0.8 | 1.5 ± 0.2 | −0.1 (−0.6, 0.4) | 0.731 | 1.5 ± 0.8 | 1.7 ± 0.2 | 0.2 (−1.0, 1.3) | 0.814 |
|
| 169 |
+
| Change in cholesterol (mmol l−1)* | −1.3 ± 1.0 | −1.4 ± 0.8 | −0.1 (−1.1, 0.9) | 0.869 | −1.4 ± 1.0 | −0.8 ± 0.8 | 0.6 (−0.9, 2.1) | 0.401 |
|
| 170 |
+
| Change in cholesterol (%)* | −15.3 ± 8.7 | −18.6 ± 10.1 | −3.3 (−12.5, 6.0) | 0.466 | −16.7 ± 8.7 | −13.3 ± 14.8 | 3.4 (−11.0, 17.7) | 0.631 |
|
| 171 |
+
| Change in LDL-cholesterol (mmol l−1)* | −1.4 ± 1.0 | −1.3 ± 0.6 | 0.1 (−0.9, 1.0) | 0.908 | −1.4 ± 1.0 | −1.1 ± 0.7 | 0.3 (−1.2, 1.7) | 0.702 |
|
| 172 |
+
| Change in LDL-cholesterol (%)* | −20.1 ± 10.2 | −23.2 ± 11.6 | −3.1 (−14.0, 7.8) | 0.554 | −20.7 ± 9.8 | −24.8 ± 20.4 | −4.1 (−20.8, 12.5) | 0.610 |
|
| 173 |
+
| Change in HDL-cholesterol (mmol l−1)* | 0.1 ± 0.1 | 0.0 ± 0.3 | −0.1 (−0.3, 0.2) | 0.548 | 0.0 ± 0.2 | 0.3 ± 0.0 | 0.3 (0.2, 0.4) | 0.0001 |
|
| 174 |
+
| Change in HDL-cholesterol (%)* | 7.2 ± 12.5 | 1.7 ± 20.9 | −5.5 (−24.7, 13.8) | 0.572 | 3.1 ± 14.0 | 26.7 ± 3.0 | 23.6 (15.3, 31.7) | 0.0002 |
|
| 175 |
+
| Change in triglycerides (mmol l−1)* | −0.1 ± 0.6 | −0.2 ± 0.4 | −0.1 (−0.6, 0.5) | 0.728 | −0.2 ± 0.5 | −0.1 ± 0.1 | 0.1 (−0.7, 0.9) | 0.801 |
|
| 176 |
+
| Change in triglycerides (%)* | −2.6 ± 35.7 | −14.5 ± 24.4 | −11.9 (−40.9, 17.0) | 0.401 | −6.1 ± 34.2 | −6.2 ± 14.1 | −0.1 (−52.4, 52.3) | 0.999 |
|
| 177 |
+
### Figure 1.
|
| 178 |
+
|
| 179 |
+

|
| 180 |
+
|
| 181 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=1885108_bcp0061-0706-f1.jpg)
|
| 182 |
+
|
| 183 |
+
Plasma pravastatin concentrations (mean ± SEM) in relation to the SLCO1B1 521T > C (A) SLCO1B1 521TT subjects (n = 14) (○); SLCO1B1 521TC subjects (n = 6) (•) and −11187 G > A (B) SLCO1B1−11187GG subjects (n = 18) (○); SLCO1B1−11187GA subjects (n = 2) (•) single nucleotide polymorphisms in children with heterozygous familial hypercholesterolaemia
|
| 184 |
+
|
| 185 |
+
Cardiac transplant recipients with the *SLCO1B1*SLCO1B1 521TC genotype (*n*n = 3) had a 46% lower *C*C_max_max (*P*P= 0.055), a 62% smaller AUC(0,24 h) (*P*P= 0.016) and a shorter *t*t_1/2_1/2 (0.9 ± 0.1 *vs.*vs. 1.3 ± 0.4 h, *P*P = 0.015) of pravastatin than those with the reference genotype ([Table 4](#tbl4)Table 4, [Figure 2](#fig02)Figure 2). In transplant recipients with the 521TC genotype, the decreases in total (−1.2% ± 5.8%*vs.*vs.−18.9% ± 17.6%, *P*P = 0.031) and LDL cholesterol (−7.7% ± 7.7%*vs.*vs.−33.5% ± 20.8%, *P*P = 0.011) were smaller and the increase in HDL cholesterol was greater (36.0% ± 15.6%*vs.*vs. 0.4% ± 23.0%, *P*P = 0.026) than in those with the reference genotype ([Table 4](#tbl4)Table 4). No differences were apparent in patient characteristics between subjects with different *SLCO1B1*SLCO1B1 521T > C genotypes ([Table 4](#tbl4)Table 4). There were no significant differences in the pharmacokinetics or effects of pravastatin between subjects with different *ABCB1*ABCB1 3435C > T and 2677G > T/A genotypes or haplotypes (data not shown).
|
| 186 |
+
|
| 187 |
+
### Table 4.
|
| 188 |
+
|
| 189 |
+
Comparison of the baseline characteristics and the pravastatin pharmacokinetic and pharmacodynamic data in relation to the SLCO1B1 521T > C single nucleotide polymorphism in paediatric cardiac transplant recipients on a regimen of triple immunosuppression
|
| 190 |
+
|
| 191 |
+
| Variable | SLCO1B1 521TT (n = 9) | SLCO1B1 521TC (n = 3) | Difference in means (95% CI) | P |
|
| 192 |
+
| -------- | --------------------- | --------------------- | ---------------------------- | - |
|
| 193 |
+
| Age (years) | 11.6 ± 4.1 | 11.2 ± 6.6 | −0.4 (−7.4, 6.7) | |
|
| 194 |
+
| Weight (kg) | 41.4 ± 19.0 | 31.4 ± 12.9 | −10.0 (−36.6, 16.7) | |
|
| 195 |
+
| Height (cm) | 140.8 ± 18.6 | 135.2 ± 22.9 | −5.6 (−34.6, 23.3) | |
|
| 196 |
+
| Body mass index (mg m−2) | 19.6 ± 5.2 | 16.7 ± 1.8 | −2.9 (−9.9, 4.1) | |
|
| 197 |
+
| Cyclosporin dose (mg) | 160 (135–300) | 250 (150–300) | | |
|
| 198 |
+
| Cyclosporin dose (mg kg−1) | 5.4 ± 2.0 | 7.6 ± 2.1 | 2.2 (−0.8, 5.2) | |
|
| 199 |
+
| Azathioprine dose (mg) | 46.3 (18.8–75) | 25 (25–75) | | |
|
| 200 |
+
| Azathioprine dose (mg kg−1) | 1.2 ± 0.1 | 1.2 ± 0.3 | 0.0 (−0.4, 0.5) | |
|
| 201 |
+
| Azathioprine morning dose (mg) | 21.88 (12.5–37.5) | 12.5 (12.5–50) | | |
|
| 202 |
+
| Pravastatin | | | | |
|
| 203 |
+
| dose (mg kg−1) | 0.31 ± 0.17 | 0.35 ± 0.12 | 0.04 (−0.20, 0.29) | 0.701 |
|
| 204 |
+
| Cmax (ng ml−1) | 145.7 ± 89.8 | 78.0 ± 10.3 | −67.7 (−135.7, 0.3) | 0.055 |
|
| 205 |
+
| AUC(0,24 h) (ng ml−1 h) | 366.3 ± 223.5 | 137.8 ± 41.0 | −228.5 (−402.7, −54.3) | 0.016 |
|
| 206 |
+
| tmax (h) | 1.0 (0.5–2.0) | 1.0 (1.0–1.5) | | 0.981 |
|
| 207 |
+
| t1/2 (h) | 1.3 ± 0.4 | 0.9 ± 0.1 | −0.4 (−0.7, −0.1) | 0.015 |
|
| 208 |
+
| Change in cholesterol (mmol l−1)* | −1.1 ± 1.2 | −0.1 ± 0.3 | 1.0 (0.1, 2.0) | 0.037 |
|
| 209 |
+
| Change in cholesterol (%)* | −18.9 ± 17.6 | −1.2 ± 5.8 | 17.7 (1.9, 32.1) | 0.031 |
|
| 210 |
+
| Change in LDL-cholesterol (mmol l−1)* | −1.0 ± 0.8 | −0.2 ± 0.3 | 0.8 (0.1, 1.5) | 0.022 |
|
| 211 |
+
| Change in LDL-cholesterol (%)* | −33.5 ± 20.8 | −7.7 ± 7.7 | 25.8 (7.4, 44.2) | 0.011 |
|
| 212 |
+
| Change in HDL-cholesterol (mmol l−1)* | 0.0 ± 0.4 | 0.4 ± 0.2 | 0.4 (0.1, 0.7) | 0.032 |
|
| 213 |
+
| Change in HDL-cholesterol (%)* | 0.4 ± 23.0 | 36.0 ± 15.6 | 35.6 (10.0, 62.7) | 0.026 |
|
| 214 |
+
| Change in triglycerides (mmol l−1)* | −0.1 ± 0.8 | −0.6 ± 0.6 | −0.5 (−1.6, 0.7) | 0.358 |
|
| 215 |
+
| Change in triglycerides (%)* | −2.5 ± 39.0 | −27.1 ± 15.1 | −24.6 (−59.5, 10.2) | 0.149 |
|
| 216 |
+
### Figure 2.
|
| 217 |
+
|
| 218 |
+

|
| 219 |
+
|
| 220 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=1885108_bcp0061-0706-f2.jpg)
|
| 221 |
+
|
| 222 |
+
Plasma pravastatin concentrations (mean ± SEM) in relation to the SLCO1B1 521T > C single nucleotide polymorphism in paediatric cardiac transplant recipients on a regimen of triple immunosuppression, SLCO1B1 521TT subjects (n = 9) (○); SLCO1B1 521TC subjects (n = 3) (•)
|
| 223 |
+
|
| 224 |
+
## Discussion
|
| 225 |
+
|
| 226 |
+
To our knowledge, this is the first study to explore associations between the pharmacokinetics and efficacy of pravastatin (with and without immunosuppressive medication) and polymorphisms of the *SLCO1B1*SLCO1B1 and *ABCB1*ABCB1 genes in children. In patients with HeFH, the 521T > C SNP had no significant effect on the pharmacokinetics or efficacy of pravastatin, but two patients with the −11187GA genotype had significantly lower plasma concentrations of pravastatin than those with the reference genotype. Furthermore, the AUC(0,24 h) of pravastatin was markedly lower and the *t*t_1/2_1/2 shorter in paediatric cardiac transplant recipients with the *SLCO1B1*SLCO1B1 521TC genotype than in patients with the reference genotype. Decreases in total and LDL cholesterol by pravastatin were smaller, and increases in HDL cholesterol greater in association with the 521T > C SNP.
|
| 227 |
+
|
| 228 |
+
Recently, a number of SNPs in the *SLCO1B1*SLCO1B1 gene have been characterized. Increasing evidence suggests that some of these SNPs are functionally significant and decrease the activity of the encoded transporter OATP1B1 *in vitro*in vitro[[36](#b36)36–[39](#b39)39]. For example, Tirona et al. observed decreased uptake of oestrone sulphate, oestradiol 17β-D-glucuronide and rifampicin by OATP1B1 in association with several *SLCO1B1*SLCO1B1 SNPs, including 521T > C [[36](#b36)36, [37](#b37)37]. In addition, Kameyama et al. found reduced transport of pravastatin and atorvastatin acid in association with the 521T > C SNP [[38](#b38)38]. Overall, decreased transport activity of OATP1B1 is thought to lower the hepatic uptake of pravastatin from blood, and thus to impair its elimination and increase its plasma concentrations.
|
| 229 |
+
|
| 230 |
+
Recent *in vivo*in vivo studies in healthy adults have reported altered pharmacokinetics of pravastatin in association with polymorphisms in *SLCO1B1*SLCO1B1[[28](#b28)28–[30](#b30)30]. One study observed decreased total and nonrenal clearance of pravastatin in healthy volunteers carrying the *SLCO1B1*SLCO1B1 haplotype *15 (388G and 521C) compared with those homozygous for *SLCO1B1*1B*SLCO1B1*1B (388G and 521T) [[28](#b28)28]. Another study reported significantly higher pravastatin AUC(0,12 h) in subjects with the −11187GA or 521TC genotypes or *15B or *17 (−11187 A, 388G and 521C) haplotypes [[29](#b29)29], and a further study found increased pravastatin AUC(0,6 h) in heterozygous carriers of *SLCO1B1*5*SLCO1B1*5 (388 A and 521C), and decreased amounts of pravastatin excreted into urine in subjects heterozygous or homozygous for *SLCO1B1*1B*SLCO1B1*1B, compared with subjects with the reference genotype [[30](#b30)30]. As statins inhibit cholesterol synthesis in the liver, decreased uptake of pravastatin into hepatocytes due to defective transport might decrease its cholesterol-lowering efficacy. In support of this, a recent study showed that the inhibitory effect of pravastatin on cholesterol synthesis, evaluated by changes in lathosterol concentrations and lathosterol : cholesterol ratios, was smaller in patients with the *SLCO1B1*SLCO1B1 haplotype *17 [[40](#b40)40]. Another investigation found significantly lower decreases in serum cholesterol during treatment with statins (pravastatin, atorvastatin or simvastatin) in 20 hypercholesterolaemic adults with the 521TC genotype, compared with 44 patients with the 521TT genotype (−16.5%*vs.*vs.−22.3%) [[41](#b41)41].
|
| 231 |
+
|
| 232 |
+
In contrast to a previous study in healthy adults [[29](#b29)29], the mean pravastatin AUC(0, ∞) in two children with HeFH having the −11187GA genotype was approximately 74% lower than the corresponding value observed in patients with the reference genotype. Notably, no differences existed in the baseline characteristics between patients with different *SLCO1B1*SLCO1B1 genotypes. The mechanisms underlying these results are unclear and warrant further study. Very little is currently known about the potential role of developmental characteristics affecting the pharmacokinetics of statins in childhood or about age-associated differences in the expression and function of drug transporters [[42](#b42)42]. The present results suggest that the effects of *SLCO1B1*SLCO1B1 polymorphism on OATP1B1 phenotype may be modulated by age.
|
| 233 |
+
|
| 234 |
+
In paediatric cardiac transplant recipients, the 521T > C SNP was associated with significantly decreased plasma concentrations of pravastatin and with a shorter half-life, despite similar baseline characteristics among the patients. It is unclear why the plasma concentrations of pravastatin were decreased. Along with the low plasma concentrations of pravastatin in our cardiac transplant recipients with the 521TC genotype, the total and LDL cholesterol-lowering effects in these patients were less, suggesting that hepatocellular pravastatin concentrations were also decreased.
|
| 235 |
+
|
| 236 |
+
Cyclosporin is known to cause substantial increases in the plasma concentrations of pravastatin [[10](#b10)10, [18](#b18)18, [19](#b19)19], through mechanisms that are not completely understood. As cyclosporin inhibits drug transporters such as P-glycoprotein [[43](#b43)43], MRP2 [[17](#b17)17] and OATP1B1 [[16](#b16)16], the interaction could be due to inhibition of pravastatin transport. In our previous pharmacokinetic study in paediatric cardiac transplant patients, immunosuppressive medication increased pravastatin *C*C_max_max 8-fold and AUC 10-fold, but left the *t*t_1/2_1/2 unaffected [[10](#b10)10].
|
| 237 |
+
|
| 238 |
+
The allelic frequencies of the *SLCO1B1*SLCO1B1 521C variant (15% and 13%) in children with HeFH and paediatric cardiac transplant recipients were similar to those found in Caucasian and Japanese adult populations (11–18%) [[28](#b28)28,[29](#b29)29]. The respective frequencies of the promoter variant (−11187 A) were also similar, though slightly smaller than those previously reported in Caucasians (5% and 0%*vs.*vs. 7.5%) [[29](#b29)29]. The allelic frequencies of the ABCB1 3435T (50% and 46%), 2677T (53% and 33%) and 2677 A (5% and 4%) variants were also comparable with those previously reported in adult Finnish Caucasians (55%, 48% and 1%, respectively) [[29](#b29)29].
|
| 239 |
+
|
| 240 |
+
The present results suggest that differences may exist in the pharmacogenetics of pravastatin between children and adults. Larger trials are required to assess the underlying mechanisms and clinical significance of these findings. Because undertreated hypercholesterolaemic patients are likely to remain at an increased risk of cardiovascular morbidity and mortality, further characterization of the factors affecting the pharmacokinetics of statins is important in both adults and children.
|
| 241 |
+
|
| 242 |
+
In conclusion, in children with HeFH, the *SLCO1B1*SLCO1B1−11187GA genotype was unexpectedly associated with markedly decreased plasma concentrations of pravastatin. Similarly, cardiac transplant recipients undergoing triple immunosuppressive therapy had lower plasma concentrations and a shorter half-life of pravastatin in association with the *SLCO1B1*SLCO1B1 521T > C SNP. Owing to the small number of subjects included, these results require substantiation in larger trials.
|
| 243 |
+
|
| 244 |
+
## Acknowledgments
|
| 245 |
+
|
| 246 |
+
This study was supported by grants from the Helsinki University Central Hospital Research Fund (Helsinki, Finland), the Robert Bosch Foundation (Stuttgart, Germany) and the Alexander von Humboldt Foundation (MN) (Bonn, Germany).
|
| 247 |
+
|
| 248 |
+
## References
|
| 249 |
+
|
| 250 |
+
1. Neuvonen PJ, Kantola T, Kivistö KT. Simvastatin but not pravastatin is very susceptible to interaction with the CYP3A4 inhibitor irtaconazole. Clin Pharmacol Ther. 1998;63:332–41. doi: 10.1016/S0009-9236(98)90165-5. [DOI](https://doi.org/10.1016/S0009-9236(98)90165-5) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9542477/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Simvastatin%20but%20not%20pravastatin%20is%20very%20susceptible%20to%20interaction%20with%20the%20CYP3A4%20inhibitor%20irtaconazole&author=PJ%20Neuvonen&author=T%20Kantola&author=KT%20Kivist%C3%B6&volume=63&publication_year=1998&pages=332-41&pmid=9542477&doi=10.1016/S0009-9236(98)90165-5&)
|
| 251 |
+
|
| 252 |
+
2. Hedman M, Neuvonen PJ, Neuvonen M, Antikainen M. Pharmacokinetics and pharmacodynamics of pravastatin in children with familial hypercholesterolemia. Clin Pharmacol Ther. 2003;74:178–85. doi: 10.1016/S0009-9236(03)00153-X. [DOI](https://doi.org/10.1016/S0009-9236(03)00153-X) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12891228/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Pharmacokinetics%20and%20pharmacodynamics%20of%20pravastatin%20in%20children%20with%20familial%20hypercholesterolemia&author=M%20Hedman&author=PJ%20Neuvonen&author=M%20Neuvonen&author=M%20Antikainen&volume=74&publication_year=2003&pages=178-85&pmid=12891228&doi=10.1016/S0009-9236(03)00153-X&)
|
| 253 |
+
|
| 254 |
+
3. Pan HY, De Vault AR, Swites BJ, Whigan D, Ivashkiv E, Willard DA, Brescia D. Pharmacokinetics and pharmacodynamics of pravastatin alone and with cholestyramine in hypercholesterolemia. Clin Pharmacol Ther. 1990;48:201–7. doi: 10.1038/clpt.1990.136. [DOI](https://doi.org/10.1038/clpt.1990.136) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/2116260/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Pharmacokinetics%20and%20pharmacodynamics%20of%20pravastatin%20alone%20and%20with%20cholestyramine%20in%20hypercholesterolemia&author=HY%20Pan&author=AR%20De%20Vault&author=BJ%20Swites&author=D%20Whigan&author=E%20Ivashkiv&volume=48&publication_year=1990&pages=201-7&pmid=2116260&doi=10.1038/clpt.1990.136&)
|
| 255 |
+
|
| 256 |
+
4. Jones PH, Farmer JA, Cressman DO, McKenney JM, Wright JT, Proctor JD, Berkson DM, Farnham DJ, Wolfson PM, Colfer HT. Once-daily pravastatin in patients with primary hypercholesterolemia: a dose–response study. Clin Cardiol. 1991;14:146–51. doi: 10.1002/clc.4960140211. [DOI](https://doi.org/10.1002/clc.4960140211) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1904333/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Cardiol&title=Once-daily%20pravastatin%20in%20patients%20with%20primary%20hypercholesterolemia:%20a%20dose%E2%80%93response%20study&author=PH%20Jones&author=JA%20Farmer&author=DO%20Cressman&author=JM%20McKenney&author=JT%20Wright&volume=14&publication_year=1991&pages=146-51&pmid=1904333&doi=10.1002/clc.4960140211&)
|
| 257 |
+
|
| 258 |
+
5. Hatanaka T. Clinical pharmacokinetics of pravastatin. Mechanisms of pharmacokinetic events. Clin Pharmacokinet. 2000;39:397–412. doi: 10.2165/00003088-200039060-00002. [DOI](https://doi.org/10.2165/00003088-200039060-00002) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11192473/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacokinet&title=Clinical%20pharmacokinetics%20of%20pravastatin.%20Mechanisms%20of%20pharmacokinetic%20events&author=T%20Hatanaka&volume=39&publication_year=2000&pages=397-412&pmid=11192473&doi=10.2165/00003088-200039060-00002&)
|
| 259 |
+
|
| 260 |
+
6. Kobashigawa JA, Katznelson S, Laks H, Johnson JA, Yeatman L, Wang XM, Chia D, Terasaki PI, Sabad A, Cogert GA. Effect of pravastatin on outcomes after cardiac transplantation. N Engl J Med. 1995;333:621–7. doi: 10.1056/NEJM199509073331003. [DOI](https://doi.org/10.1056/NEJM199509073331003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7637722/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Effect%20of%20pravastatin%20on%20outcomes%20after%20cardiac%20transplantation&author=JA%20Kobashigawa&author=S%20Katznelson&author=H%20Laks&author=JA%20Johnson&author=L%20Yeatman&volume=333&publication_year=1995&pages=621-7&pmid=7637722&doi=10.1056/NEJM199509073331003&)
|
| 261 |
+
|
| 262 |
+
7. Hertz MI, Taylor DO, Trulock EP, Boucek MM, Mohacsi PJ, Edwards LB, Keck BM. The registry of the international society for heart and lung transplantation: nineteenth official report-2002. J Heart Lung Transplant. 2002;21:950–70. doi: 10.1016/s1053-2498(02)00498-9. [DOI](https://doi.org/10.1016/s1053-2498(02)00498-9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12231366/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Heart%20Lung%20Transplant&title=The%20registry%20of%20the%20international%20society%20for%20heart%20and%20lung%20transplantation:%20nineteenth%20official%20report-2002&author=MI%20Hertz&author=DO%20Taylor&author=EP%20Trulock&author=MM%20Boucek&author=PJ%20Mohacsi&volume=21&publication_year=2002&pages=950-70&pmid=12231366&doi=10.1016/s1053-2498(02)00498-9&)
|
| 263 |
+
|
| 264 |
+
8. Boucek MM, Edwards LB, Keck BM, Trulock EP, Taylor DO, Mohacsi PJ, Hertz MI. The registry of the international society for heart and lung transplantation: fifth official pediatric report-2001–02. J Heart Lung Transplant. 2002;21:827–40. doi: 10.1016/s1053-2498(02)00496-5. [DOI](https://doi.org/10.1016/s1053-2498(02)00496-5) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12163082/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Heart%20Lung%20Transplant&title=The%20registry%20of%20the%20international%20society%20for%20heart%20and%20lung%20transplantation:%20fifth%20official%20pediatric%20report-2001%E2%80%9302&author=MM%20Boucek&author=LB%20Edwards&author=BM%20Keck&author=EP%20Trulock&author=DO%20Taylor&volume=21&publication_year=2002&pages=827-40&pmid=12163082&doi=10.1016/s1053-2498(02)00496-5&)
|
| 265 |
+
|
| 266 |
+
9. Penson MG, Fricker FJ, Thompson JR, Harker K, Kahler DA, Schowengerdt KO. Safety and efficacy of pravastatin therapy for the prevention of hyperlipidemia in pediatric and adolescent cardiac transplant recipients. J Heart Lung Transplant. 2001;20:611–8. doi: 10.1016/s1053-2498(01)00251-0. [DOI](https://doi.org/10.1016/s1053-2498(01)00251-0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11404165/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Heart%20Lung%20Transplant&title=Safety%20and%20efficacy%20of%20pravastatin%20therapy%20for%20the%20prevention%20of%20hyperlipidemia%20in%20pediatric%20and%20adolescent%20cardiac%20transplant%20recipients&author=MG%20Penson&author=FJ%20Fricker&author=JR%20Thompson&author=K%20Harker&author=DA%20Kahler&volume=20&publication_year=2001&pages=611-8&pmid=11404165&doi=10.1016/s1053-2498(01)00251-0&)
|
| 267 |
+
|
| 268 |
+
10. Hedman M, Neuvonen PJ, Neuvonen M, Holmberg C, Antikainen M. Pharmacokinetics and pharmacodynamics of pravastatin in pediatric and adolescent cardiac transplant recipients on a regimen of triple immunosuppression. Clin Pharmacol Ther. 2004;75:101–9. doi: 10.1016/j.clpt.2003.09.011. [DOI](https://doi.org/10.1016/j.clpt.2003.09.011) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14749696/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Pharmacokinetics%20and%20pharmacodynamics%20of%20pravastatin%20in%20pediatric%20and%20adolescent%20cardiac%20transplant%20recipients%20on%20a%20regimen%20of%20triple%20immunosuppression&author=M%20Hedman&author=PJ%20Neuvonen&author=M%20Neuvonen&author=C%20Holmberg&author=M%20Antikainen&volume=75&publication_year=2004&pages=101-9&pmid=14749696&doi=10.1016/j.clpt.2003.09.011&)
|
| 269 |
+
|
| 270 |
+
11. Singhvi SM, Pan HY, Morrison RA, Willard DA. Disposition of pravastatin sodium, a tissue-selective HMG-CoA reductase inhibitor, in healthy subjects. Br J Clin Pharmacol. 1990;29:239–43. doi: 10.1111/j.1365-2125.1990.tb03626.x. [DOI](https://doi.org/10.1111/j.1365-2125.1990.tb03626.x) | [PMC free article](/articles/PMC1380090/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/2106337/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Disposition%20of%20pravastatin%20sodium,%20a%20tissue-selective%20HMG-CoA%20reductase%20inhibitor,%20in%20healthy%20subjects&author=SM%20Singhvi&author=HY%20Pan&author=RA%20Morrison&author=DA%20Willard&volume=29&publication_year=1990&pages=239-43&pmid=2106337&doi=10.1111/j.1365-2125.1990.tb03626.x&)
|
| 271 |
+
|
| 272 |
+
12. Everett DW, Chando TJ, Didonato GC, Singhvi SM, Pan HY, Weinstein SH. Biotransformation of pravastatin sodium in humans. Drug Metab Dispos. 1991;19:740–8. [PubMed](https://pubmed.ncbi.nlm.nih.gov/1680649/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Metab%20Dispos&title=Biotransformation%20of%20pravastatin%20sodium%20in%20humans&author=DW%20Everett&author=TJ%20Chando&author=GC%20Didonato&author=SM%20Singhvi&author=HY%20Pan&volume=19&publication_year=1991&pages=740-8&pmid=1680649&)
|
| 273 |
+
|
| 274 |
+
13. Kantola T, Backman JT, Niemi M, Kivistö KT, Neuvonen PJ. Effect of fluconazole on plasma fluvastatin and pravastatin concentrations. Eur J Clin Pharmacol. 2000;56:225–9. doi: 10.1007/s002280000127. [DOI](https://doi.org/10.1007/s002280000127) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10952477/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=Effect%20of%20fluconazole%20on%20plasma%20fluvastatin%20and%20pravastatin%20concentrations&author=T%20Kantola&author=JT%20Backman&author=M%20Niemi&author=KT%20Kivist%C3%B6&author=PJ%20Neuvonen&volume=56&publication_year=2000&pages=225-9&pmid=10952477&doi=10.1007/s002280000127&)
|
| 275 |
+
|
| 276 |
+
14. Yee GC. Pharmacokinetic interactions between cyclosporine and other drugs. Transplant Proc. 1990;22:1203–7. [PubMed](https://pubmed.ncbi.nlm.nih.gov/2190382/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplant%20Proc&title=Pharmacokinetic%20interactions%20between%20cyclosporine%20and%20other%20drugs&author=GC%20Yee&volume=22&publication_year=1990&pages=1203-7&pmid=2190382&)
|
| 277 |
+
|
| 278 |
+
15. Saeki T, Ueda K, Tanigawara Y, Hori R, Komano T. Human P-glycoprotein transports cyclosporine A and Fk506. J Biol Chem. 1993;268:6077–80. [PubMed](https://pubmed.ncbi.nlm.nih.gov/7681059/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Biol%20Chem&title=Human%20P-glycoprotein%20transports%20cyclosporine%20A%20and%20Fk506&author=T%20Saeki&author=K%20Ueda&author=Y%20Tanigawara&author=R%20Hori&author=T%20Komano&volume=268&publication_year=1993&pages=6077-80&pmid=7681059&)
|
| 279 |
+
|
| 280 |
+
16. Shitara Y, Sugiyama D, Kusuhara H, Kato Y, Abe T, Meier PJ, Ithoh T, Sugiyama Y. Comparative inhibitory effects of different compounds on rat Oatp1 (Slc21a1)- and Oatp2 (Slc21a5)-mediated transport. Pharm Res. 2002;19:147–53. doi: 10.1023/a:1014264614637. [DOI](https://doi.org/10.1023/a:1014264614637) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11883641/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharm%20Res&title=Comparative%20inhibitory%20effects%20of%20different%20compounds%20on%20rat%20Oatp1%20(Slc21a1)-%20and%20Oatp2%20(Slc21a5)-mediated%20transport&author=Y%20Shitara&author=D%20Sugiyama&author=H%20Kusuhara&author=Y%20Kato&author=T%20Abe&volume=19&publication_year=2002&pages=147-53&pmid=11883641&doi=10.1023/a:1014264614637&)
|
| 281 |
+
|
| 282 |
+
17. Chen ZS, Kawabe T, Ono M, Aoki S, Sunizawa T, Ferukawa T, Uchiumi T, Wada M, Kuwano M, Akiyama SI. Effects of multidrug resistance-reversing agents on transporting activity of human canalicular multispecific organic anion transporter. Mol Pharmacol. 1999;56:1219–28. doi: 10.1124/mol.56.6.1219. [DOI](https://doi.org/10.1124/mol.56.6.1219) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10570049/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mol%20Pharmacol&title=Effects%20of%20multidrug%20resistance-reversing%20agents%20on%20transporting%20activity%20of%20human%20canalicular%20multispecific%20organic%20anion%20transporter&author=ZS%20Chen&author=T%20Kawabe&author=M%20Ono&author=S%20Aoki&author=T%20Sunizawa&volume=56&publication_year=1999&pages=1219-28&pmid=10570049&doi=10.1124/mol.56.6.1219&)
|
| 283 |
+
|
| 284 |
+
18. Park J-W, Siekmeier R, Merz M, Krell B, Harder S, Marz W, Seidel D, Schuler S, Gross W. Pharmacokinetics of pravastatin in heart-transplant patients taking cyclosporin A. Int J Clin Pharm Ther. 2002;40:439–50. doi: 10.5414/cpp40439. [DOI](https://doi.org/10.5414/cpp40439) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12395976/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int%20J%20Clin%20Pharm%20Ther&title=Pharmacokinetics%20of%20pravastatin%20in%20heart-transplant%20patients%20taking%20cyclosporin%20A&author=J-W%20Park&author=R%20Siekmeier&author=M%20Merz&author=B%20Krell&author=S%20Harder&volume=40&publication_year=2002&pages=439-50&pmid=12395976&doi=10.5414/cpp40439&)
|
| 285 |
+
|
| 286 |
+
19. Regazzi MB, Iacona I, Campana C, Raddato V, Lesi C, Perani G, Gavazzi A, Vigano M. Altered disposition of pravastatin following concomitant drug therapy with cyclosporin A in transplant recipients. Transplant Proc. 1993;25:2732–4. [PubMed](https://pubmed.ncbi.nlm.nih.gov/8356729/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplant%20Proc&title=Altered%20disposition%20of%20pravastatin%20following%20concomitant%20drug%20therapy%20with%20cyclosporin%20A%20in%20transplant%20recipients&author=MB%20Regazzi&author=I%20Iacona&author=C%20Campana&author=V%20Raddato&author=C%20Lesi&volume=25&publication_year=1993&pages=2732-4&pmid=8356729&)
|
| 287 |
+
|
| 288 |
+
20. Olbricht C, Wanner C, Eisenhauer T, Kliem V, Doll R, Boddaert M, O’Grady P, Krekler M, Mangold B, Chritians U. Accumulation of lovastatin, but not pravastatin, in the blood of cyclosporin-treated kidney graft patients after multiple doses. Clin Pharmacol Ther. 1997;62:311–21. doi: 10.1016/S0009-9236(97)90034-5. [DOI](https://doi.org/10.1016/S0009-9236(97)90034-5) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9333107/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Accumulation%20of%20lovastatin,%20but%20not%20pravastatin,%20in%20the%20blood%20of%20cyclosporin-treated%20kidney%20graft%20patients%20after%20multiple%20doses&author=C%20Olbricht&author=C%20Wanner&author=T%20Eisenhauer&author=V%20Kliem&author=R%20Doll&volume=62&publication_year=1997&pages=311-21&pmid=9333107&doi=10.1016/S0009-9236(97)90034-5&)
|
| 289 |
+
|
| 290 |
+
21. Corpier CL, Jones PH, Suki WN, Lederer ED, Quinones MA, Schmidt SW, Young JB. Rhabdomyolysis and renal injury with lovastatin use: report of two cases in cardiac transplant recipients. JAMA. 1988;260:239–41. [PubMed](https://pubmed.ncbi.nlm.nih.gov/3290520/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=JAMA&title=Rhabdomyolysis%20and%20renal%20injury%20with%20lovastatin%20use:%20report%20of%20two%20cases%20in%20cardiac%20transplant%20recipients&author=CL%20Corpier&author=PH%20Jones&author=WN%20Suki&author=ED%20Lederer&author=MA%20Quinones&volume=260&publication_year=1988&pages=239-41&pmid=3290520&)
|
| 291 |
+
|
| 292 |
+
22. Kobashigawa JA, Murphy FL, Stevenson LW, Moriguchi JD, Kawata N, Kamjoo P, Brownfield E, Leonard L, Chuck C. Low-dose lovastatin safely lowers cholesterol after cardiac transplantation. Circulation. 1990;82(Suppl):IV–281–IV–283. [PubMed](https://pubmed.ncbi.nlm.nih.gov/2225417/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circulation&title=Low-dose%20lovastatin%20safely%20lowers%20cholesterol%20after%20cardiac%20transplantation&author=JA%20Kobashigawa&author=FL%20Murphy&author=LW%20Stevenson&author=JD%20Moriguchi&author=N%20Kawata&volume=82&issue=Suppl&publication_year=1990&pages=IV%E2%80%93281-IV%E2%80%93283&pmid=2225417&)
|
| 293 |
+
|
| 294 |
+
23. Hsiang B, Zhu Y, Wang Z, Wu Y, Sasseville V, Yang WP, Kirchgessner TG. A novel human hepatic organic anion transporting polypeptide (OATP2). Identification of a liver-specific human organic anion transporting polypeptide and identification of rat and human hydroxymethylglutaryl-CoA reductase inhibitor transporters. J Biol Chem. 1999;274:37161–8. doi: 10.1074/jbc.274.52.37161. [DOI](https://doi.org/10.1074/jbc.274.52.37161) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10601278/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Biol%20Chem&title=A%20novel%20human%20hepatic%20organic%20anion%20transporting%20polypeptide%20(OATP2).%20Identification%20of%20a%20liver-specific%20human%20organic%20anion%20transporting%20polypeptide%20and%20identification%20of%20rat%20and%20human%20hydroxymethylglutaryl-CoA%20reductase%20inhibitor%20transporters&author=B%20Hsiang&author=Y%20Zhu&author=Z%20Wang&author=Y%20Wu&author=V%20Sasseville&volume=274&publication_year=1999&pages=37161-8&pmid=10601278&doi=10.1074/jbc.274.52.37161&)
|
| 295 |
+
|
| 296 |
+
24. Nakai D, Nakagomi R, Furuta Y, Tokui T, Abe T, Ikeda T, Nishimura K. Human liver-specific organic anion transporter, LST-1, mediates uptake of pravastatin by human hepatocytes. J Pharmacol Exp Ther. 2001;297:861–7. [PubMed](https://pubmed.ncbi.nlm.nih.gov/11356905/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharmacol%20Exp%20Ther&title=Human%20liver-specific%20organic%20anion%20transporter,%20LST-1,%20mediates%20uptake%20of%20pravastatin%20by%20human%20hepatocytes&author=D%20Nakai&author=R%20Nakagomi&author=Y%20Furuta&author=T%20Tokui&author=T%20Abe&volume=297&publication_year=2001&pages=861-7&pmid=11356905&)
|
| 297 |
+
|
| 298 |
+
25. Kobayashi D, Nozawa T, Imai K, Nezu JI, Tsuji A, Tamai I. Involvement of human organic anion transporting polypeptide OATP-B (SLC21A9) in pH-dependent transport across intestinal apical membrane. J Pharmacol Exp Ther. 2003;306:703–8. doi: 10.1124/jpet.103.051300. [DOI](https://doi.org/10.1124/jpet.103.051300) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12724351/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharmacol%20Exp%20Ther&title=Involvement%20of%20human%20organic%20anion%20transporting%20polypeptide%20OATP-B%20(SLC21A9)%20in%20pH-dependent%20transport%20across%20intestinal%20apical%20membrane&author=D%20Kobayashi&author=T%20Nozawa&author=K%20Imai&author=JI%20Nezu&author=A%20Tsuji&volume=306&publication_year=2003&pages=703-8&pmid=12724351&doi=10.1124/jpet.103.051300&)
|
| 299 |
+
|
| 300 |
+
26. Yamazaki M, Akiyama S, Ni’Inuma K, Nishigaki R, Sugiyama Y. Biliary excretion of pravastatin in rats: contribution of the excretion pathway mediated by canalicular multispecific organic anion transporter (cMOAT) Drug Metab Dispos. 1997;25:1123–9. [PubMed](https://pubmed.ncbi.nlm.nih.gov/9321514/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Metab%20Dispos&title=Biliary%20excretion%20of%20pravastatin%20in%20rats:%20contribution%20of%20the%20excretion%20pathway%20mediated%20by%20canalicular%20multispecific%20organic%20anion%20transporter%20(cMOAT)&author=M%20Yamazaki&author=S%20Akiyama&author=K%20Ni%E2%80%99Inuma&author=R%20Nishigaki&author=Y%20Sugiyama&volume=25&publication_year=1997&pages=1123-9&pmid=9321514&)
|
| 301 |
+
|
| 302 |
+
27. Cha SH, Sekine T, Fukushima JI, Kanai Y, Kobayashi Y, Goya T, Endou H. Identification and characterization of human organic anion transporter 3 expressing predominantly in the kidney. Mol Pharmacol. 2001;59:1277–86. doi: 10.1124/mol.59.5.1277. [DOI](https://doi.org/10.1124/mol.59.5.1277) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11306713/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mol%20Pharmacol&title=Identification%20and%20characterization%20of%20human%20organic%20anion%20transporter%203%20expressing%20predominantly%20in%20the%20kidney&author=SH%20Cha&author=T%20Sekine&author=JI%20Fukushima&author=Y%20Kanai&author=Y%20Kobayashi&volume=59&publication_year=2001&pages=1277-86&pmid=11306713&doi=10.1124/mol.59.5.1277&)
|
| 303 |
+
|
| 304 |
+
28. Nishizato Y, Ieiri I, Suzuki H, Kimura M, Kawabata K, Hirota T, Takane H, Irie S, Kusuhara H, Urasaki Y, Urae A, Higuchi S, Otsubo K, Sugiyama Y. Polymorphisms of OATP-C (SLC21A6) and OAT3 (SLC22A8) genes: consequences for pravastatin pharmacokinetics. Clin Pharmacol Ther. 2003;73:554–65. doi: 10.1016/S0009-9236(03)00060-2. [DOI](https://doi.org/10.1016/S0009-9236(03)00060-2) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12811365/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Polymorphisms%20of%20OATP-C%20(SLC21A6)%20and%20OAT3%20(SLC22A8)%20genes:%20consequences%20for%20pravastatin%20pharmacokinetics&author=Y%20Nishizato&author=I%20Ieiri&author=H%20Suzuki&author=M%20Kimura&author=K%20Kawabata&volume=73&publication_year=2003&pages=554-65&pmid=12811365&doi=10.1016/S0009-9236(03)00060-2&)
|
| 305 |
+
|
| 306 |
+
29. Niemi M, Schaeffeler E, Lang T, Fromm MF, Neuvonen M, Kyrklund C, Backman JT, Kerb R, Schwab M, Neuvonen PJ, Eichelbaum M, Kivistö KT. High plasma pravastatin concentrations are associated with single nucleotide polymorphisms and haplotypes of organic anion transporting polypeptide-C (OATP-C, SLCO1B1) Pharmacogenetics. 2004;14:429–40. doi: 10.1097/01.fpc.0000114750.08559.32. [DOI](https://doi.org/10.1097/01.fpc.0000114750.08559.32) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15226675/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=High%20plasma%20pravastatin%20concentrations%20are%20associated%20with%20single%20nucleotide%20polymorphisms%20and%20haplotypes%20of%20organic%20anion%20transporting%20polypeptide-C%20(OATP-C,%20SLCO1B1)&author=M%20Niemi&author=E%20Schaeffeler&author=T%20Lang&author=MF%20Fromm&author=M%20Neuvonen&volume=14&publication_year=2004&pages=429-40&pmid=15226675&doi=10.1097/01.fpc.0000114750.08559.32&)
|
| 307 |
+
|
| 308 |
+
30. Mwinyi J, Johne A, Bauer S, Roots I, Gerloff T. Evidence for inverse effects of OATP-C (SLC21A6) *5 and *1b haplotypes on pravastatin kinetics. Clin Pharmacol Ther. 2004;75:415–21. doi: 10.1016/j.clpt.2003.12.016. [DOI](https://doi.org/10.1016/j.clpt.2003.12.016) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15116054/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Evidence%20for%20inverse%20effects%20of%20OATP-C%20(SLC21A6)%20*5%20and%20*1b%20haplotypes%20on%20pravastatin%20kinetics&author=J%20Mwinyi&author=A%20Johne&author=S%20Bauer&author=I%20Roots&author=T%20Gerloff&volume=75&publication_year=2004&pages=415-21&pmid=15116054&doi=10.1016/j.clpt.2003.12.016&)
|
| 309 |
+
|
| 310 |
+
31. Vuorio AF, Aalto-Setälä K, Koivisto U-M, Turtola H, Nissen H, Kovanen PT, Miettinen T, Gylling H, Oksanen H, Kontula K Finnish FH-group. Familial hypercholesterolemia in Finland. Common, rare and mild mutations of the LDL receptor and their clinical consequences. Ann Med. 2001;33:410–21. doi: 10.3109/07853890108995954. [DOI](https://doi.org/10.3109/07853890108995954) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11585102/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20Med&title=Familial%20hypercholesterolemia%20in%20Finland.%20Common,%20rare%20and%20mild%20mutations%20of%20the%20LDL%20receptor%20and%20their%20clinical%20consequences&author=AF%20Vuorio&author=K%20Aalto-Set%C3%A4l%C3%A4&author=U-M%20Koivisto&author=H%20Turtola&author=H%20Nissen&volume=33&publication_year=2001&pages=410-21&pmid=11585102&doi=10.3109/07853890108995954&)
|
| 311 |
+
|
| 312 |
+
32. Cuthbert JA, East CA, Bilheimer DW, Lipsky PE. Detection of familial hypercholesterolemia by assaying functional low-density-lipoprotein receptors on lymphocytes. N Engl J Med. 1986;314:879–83. doi: 10.1056/NEJM198604033141404. [DOI](https://doi.org/10.1056/NEJM198604033141404) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/3633381/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Detection%20of%20familial%20hypercholesterolemia%20by%20assaying%20functional%20low-density-lipoprotein%20receptors%20on%20lymphocytes&author=JA%20Cuthbert&author=CA%20East&author=DW%20Bilheimer&author=PE%20Lipsky&volume=314&publication_year=1986&pages=879-83&pmid=3633381&doi=10.1056/NEJM198604033141404&)
|
| 313 |
+
|
| 314 |
+
33. Mulvana D, Jemal M, Pulver SC. Quantitative determination of pravastatin and its biotransformation products in human serum by turbo ion spray LC/MS/MS. J Pharm Biomed Anal. 2000;23:851–66. doi: 10.1016/s0731-7085(00)00372-1. [DOI](https://doi.org/10.1016/s0731-7085(00)00372-1) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11022911/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharm%20Biomed%20Anal&title=Quantitative%20determination%20of%20pravastatin%20and%20its%20biotransformation%20products%20in%20human%20serum%20by%20turbo%20ion%20spray%20LC/MS/MS&author=D%20Mulvana&author=M%20Jemal&author=SC%20Pulver&volume=23&publication_year=2000&pages=851-66&pmid=11022911&doi=10.1016/s0731-7085(00)00372-1&)
|
| 315 |
+
|
| 316 |
+
34. Furuno T, Landi MT, Ceroni M, Caporaso N, Bernucci I, Nappi G, Martignoni E, Schaeffeler E, Eichelbaum M, Schwab M, Zanger UM. Expression polymorphism of the blood–brain barrier component P-glycoprotein (MDR1) in relation to Parkinson’s disease. Pharmacogenetics. 2002;12:529–34. doi: 10.1097/00008571-200210000-00004. [DOI](https://doi.org/10.1097/00008571-200210000-00004) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12360103/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Expression%20polymorphism%20of%20the%20blood%E2%80%93brain%20barrier%20component%20P-glycoprotein%20(MDR1)%20in%20relation%20to%20Parkinson%E2%80%99s%20disease&author=T%20Furuno&author=MT%20Landi&author=M%20Ceroni&author=N%20Caporaso&author=I%20Bernucci&volume=12&publication_year=2002&pages=529-34&pmid=12360103&doi=10.1097/00008571-200210000-00004&)
|
| 317 |
+
|
| 318 |
+
35. Johne A, Köpke K, Gerloff T, Mai I, Rietbrock S, Meisel C, Hoffmeyer S, Kerb R, Fromm MF, Brinkmann U, Eichelbaum M, Brockmoller J, Cascorbi I, Roots I. Modulation of steady-state kinetics of digoxin by haplotypes of the P-glycoprotein MDR1 gene. Clin Pharmacol Ther. 2002;72:584–94. doi: 10.1067/mcp.2002.129196. [DOI](https://doi.org/10.1067/mcp.2002.129196) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12426522/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Modulation%20of%20steady-state%20kinetics%20of%20digoxin%20by%20haplotypes%20of%20the%20P-glycoprotein%20MDR1%20gene&author=A%20Johne&author=K%20K%C3%B6pke&author=T%20Gerloff&author=I%20Mai&author=S%20Rietbrock&volume=72&publication_year=2002&pages=584-94&pmid=12426522&doi=10.1067/mcp.2002.129196&)
|
| 319 |
+
|
| 320 |
+
36. Tirona RG, Leake BF, Merino G, Kim RB. Polymorphisms in OATP-C. Identification of multiple allelic variants associated with altered transport activity among European and African Americans. J Biol Chem. 2001;276:35669–75. doi: 10.1074/jbc.M103792200. [DOI](https://doi.org/10.1074/jbc.M103792200) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11477075/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Biol%20Chem&title=Polymorphisms%20in%20OATP-C.%20Identification%20of%20multiple%20allelic%20variants%20associated%20with%20altered%20transport%20activity%20among%20European%20and%20African%20Americans&author=RG%20Tirona&author=BF%20Leake&author=G%20Merino&author=RB%20Kim&volume=276&publication_year=2001&pages=35669-75&pmid=11477075&doi=10.1074/jbc.M103792200&)
|
| 321 |
+
|
| 322 |
+
37. Tirona RG, Leake BF, Wolkoff AW, Kim RB. Human organic anion transporting polypeptide-C (SLC21A6) is a major determinant of rifampin-mediated pregnane X receptor activation. J Pharmacol Exp Ther. 2003;304:223–8. doi: 10.1124/jpet.102.043026. [DOI](https://doi.org/10.1124/jpet.102.043026) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12490595/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharmacol%20Exp%20Ther&title=Human%20organic%20anion%20transporting%20polypeptide-C%20(SLC21A6)%20is%20a%20major%20determinant%20of%20rifampin-mediated%20pregnane%20X%20receptor%20activation&author=RG%20Tirona&author=BF%20Leake&author=AW%20Wolkoff&author=RB%20Kim&volume=304&publication_year=2003&pages=223-8&pmid=12490595&doi=10.1124/jpet.102.043026&)
|
| 323 |
+
|
| 324 |
+
38. Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics. 2005;15:513–22. doi: 10.1097/01.fpc.0000170913.73780.5f. [DOI](https://doi.org/10.1097/01.fpc.0000170913.73780.5f) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15970799/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenet%20Genomics&title=Functional%20characterization%20of%20SLCO1B1%20(OATP-C)%20variants,%20SLCO1B1*5,%20SLCO1B1*15%20and%20SLCO1B1*15+C1007G,%20by%20using%20transient%20expression%20systems%20of%20HeLa%20and%20HEK293%20cells&author=Y%20Kameyama&author=K%20Yamashita&author=K%20Kobayashi&author=M%20Hosokawa&author=K%20Chiba&volume=15&publication_year=2005&pages=513-22&pmid=15970799&doi=10.1097/01.fpc.0000170913.73780.5f&)
|
| 325 |
+
|
| 326 |
+
39. Michalski C, Cui Y, Nies AT, Nuessler AK, Neuhaus P, Zanger UM, Klein K, Eichelbaum M, Keppler D, Konig J. A naturally occurring mutation in the SLC21A6 gene causing impaired membrane localization of the hepatocyte uptake transporter. J Biol Chem. 2002;277:43058–63. doi: 10.1074/jbc.M207735200. [DOI](https://doi.org/10.1074/jbc.M207735200) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12196548/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Biol%20Chem&title=A%20naturally%20occurring%20mutation%20in%20the%20SLC21A6%20gene%20causing%20impaired%20membrane%20localization%20of%20the%20hepatocyte%20uptake%20transporter&author=C%20Michalski&author=Y%20Cui&author=AT%20Nies&author=AK%20Nuessler&author=P%20Neuhaus&volume=277&publication_year=2002&pages=43058-63&pmid=12196548&doi=10.1074/jbc.M207735200&)
|
| 327 |
+
|
| 328 |
+
40. Niemi M, Neuvonen PJ, Hofmann U, Backman JT, Schwab M, Lutjohann D, von Bergmann K, Eichelbaum M, Kivistö KT. Acute effects of pravastatin on cholesterol synthesis are associated with SLCO1B1 (encoding OATP1B1) haplotype * 17. Pharmacogenet Genomics. 2005;15:303–9. doi: 10.1097/01213011-200505000-00005. [DOI](https://doi.org/10.1097/01213011-200505000-00005) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15864131/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenet%20Genomics&title=Acute%20effects%20of%20pravastatin%20on%20cholesterol%20synthesis%20are%20associated%20with%20SLCO1B1%20(encoding%20OATP1B1)%20haplotype%20*%2017&author=M%20Niemi&author=PJ%20Neuvonen&author=U%20Hofmann&author=JT%20Backman&author=M%20Schwab&volume=15&publication_year=2005&pages=303-9&pmid=15864131&doi=10.1097/01213011-200505000-00005&)
|
| 329 |
+
|
| 330 |
+
41. Tachibana-Iimori R, Tabara Y, Kusuhara H, Kohara K, Kawamoto R, Nakura J, Tokunaga K, Kondo I, Sugiyama Y, Miki T. Effect of genetic polymorphism of OATP-C (SLCO1B1) on lipid-lowering response to HMG-CoA reductase inhibitors. Drug Metab Pharmacokin. 2004;19:375–80. doi: 10.2133/dmpk.19.375. [DOI](https://doi.org/10.2133/dmpk.19.375) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15548849/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drug%20Metab%20Pharmacokin&title=Effect%20of%20genetic%20polymorphism%20of%20OATP-C%20(SLCO1B1)%20on%20lipid-lowering%20response%20to%20HMG-CoA%20reductase%20inhibitors&author=R%20Tachibana-Iimori&author=Y%20Tabara&author=H%20Kusuhara&author=K%20Kohara&author=R%20Kawamoto&volume=19&publication_year=2004&pages=375-80&pmid=15548849&doi=10.2133/dmpk.19.375&)
|
| 331 |
+
|
| 332 |
+
42. Kearns GL, Abdel-Rahman SM, Alander SW, Blowey DL, Leeder JS, Kauffman RE. Drug therapy, developmental pharmacology – drug disposition, action, and therapy in infants and children. N Engl J Med. 2003;349:1157–67. doi: 10.1056/NEJMra035092. [DOI](https://doi.org/10.1056/NEJMra035092) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/13679531/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Drug%20therapy,%20developmental%20pharmacology%20%E2%80%93%20drug%20disposition,%20action,%20and%20therapy%20in%20infants%20and%20children&author=GL%20Kearns&author=SM%20Abdel-Rahman&author=SW%20Alander&author=DL%20Blowey&author=JS%20Leeder&volume=349&publication_year=2003&pages=1157-67&pmid=13679531&doi=10.1056/NEJMra035092&)
|
| 333 |
+
|
| 334 |
+
43. Tamai I, Safa AR. Competetive interaction of cyclosporins with the Vinca alkaloid-binding site of P-glycoprotein in multidrug-resistant cells. J Biol Chem. 1990;265:16509–13. [PubMed](https://pubmed.ncbi.nlm.nih.gov/1975813/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Biol%20Chem&title=Competetive%20interaction%20of%20cyclosporins%20with%20the%20Vinca%20alkaloid-binding%20site%20of%20P-glycoprotein%20in%20multidrug-resistant%20cells&author=I%20Tamai&author=AR%20Safa&volume=265&publication_year=1990&pages=16509-13&pmid=1975813&)
|
test/texts/PMC1974827.md
ADDED
|
@@ -0,0 +1,255 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Influence of the CYP3A5 and MDR1 genetic polymorphisms on the pharmacokinetics of tacrolimus in healthy Korean subjects
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Ji H Choi, Yoon J Lee, Seong B Jang, Jong-Eun Lee, Kyung H Kim, Kyungsoo Park
|
| 5 |
+
**Journal:** British Journal of Clinical Pharmacology
|
| 6 |
+
**Date:** 2007 Mar 28
|
| 7 |
+
**DOI:** [10.1111/j.1365-2125.2007.02874.x](https://doi.org/10.1111/j.1365-2125.2007.02874.x)
|
| 8 |
+
**PMID:** 17391324
|
| 9 |
+
**PMCID:** PMC1974827
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1974827/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC1974827/pdf/bcp0064-0185.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC1974827/pdf/bcp0064-0185.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**What is already known about this subject:**
|
| 16 |
+
It was found that the genetic polymorphisms of CYP3A5, CYP3A4 and MDR1 could affect the pharmacokinetics of tacrolimus.
|
| 17 |
+
|
| 18 |
+
This study was conducted to find such a possibility in the Korean population.
|
| 19 |
+
|
| 20 |
+
**What this study adds:**
|
| 21 |
+
CYP3A5 polymorphisms are likely to be associated with altered pharmacokinetics of tacrolimus in Koreans.
|
| 22 |
+
|
| 23 |
+
MDR1 polymorphisms have no important role in the pharmacokinetics of tacrolimus.
|
| 24 |
+
|
| 25 |
+
**Aims:**
|
| 26 |
+
To determine the frequencies of the genotypes of CYP3A5 and MDR1 and to examine the influence of the polymorphisms of these genes on tacrolimus pharmacokinetics in the Korean population.
|
| 27 |
+
|
| 28 |
+
**Methods:**
|
| 29 |
+
Twenty-nine healthy Koreans who participated in the previous tacrolimus pharmacokinetic study were genotyped for CYP3A4*1B, CYP3A5*3, MDR1 c.1236C→T, MDR1 c.2677G→A/T and MDR1 c.3435C→T. The relationship between the genotypes so obtained and tacrolimus pharmacokinetics observed in the previous study was examined.
|
| 30 |
+
|
| 31 |
+
**Results:**
|
| 32 |
+
No subject in this study had the CYP3A4*1B variant. The observed frequencies of CYP3A5*1/*1, *1/*3, and *3/*3 were 0.069 [confidence interval (CI) −0.023, 0.161], 0.483 (CI 0.301, 0.665) and 0.448 (CI 0.267, 0.629), respectively. AUC0–∞ for the CYP3A5*1/*1 or *1/*3 genotype was 131.5 ± 44.8 ng h ml−1 (CI 109.6, 153.5), which was much lower compared with the CYP3A5*3/*3 genotype of 323.8 ± 129.3 ng h ml−1 (CI 253.5, 394.1) (P = 2.063E−07). Similarly, Cmax for the CYP3A5*1/*1 or *1/*3 genotype was 11.8 ± 3.4 ng ml−1 (CI 10.1, 13.5), which was also much lower compared with the CYP3A5*3/*3 genotype of 24.4 ± 12.3 ng ml−1 (CI 17.8, 31.1) (P = 0.0001). However, there was no significant difference in tacrolimus pharmacokinetics among the MDR1 diplotypes of CGC-CGC, CGC-TTT, CGC-TGC, TTT-TGC or TTT-TTT (P = 0.2486).
|
| 33 |
+
|
| 34 |
+
**Conclusions:**
|
| 35 |
+
This study shows that the CYP3A5*3 genetic polymorphisms may be associated with the individual difference in tacrolimus pharmacokinetics. An individualized dosage regimen design incorporating such genetic information would help increase clinical efficacy of the drug while reducing adverse drug reactions.
|
| 36 |
+
|
| 37 |
+
Keywords: CYP3A5, MDR1, pharmacokinetics, polymorphism, tacrolimus
|
| 38 |
+
|
| 39 |
+
### What is already known about this subject
|
| 40 |
+
|
| 41 |
+
### What this study adds
|
| 42 |
+
|
| 43 |
+
### Aims
|
| 44 |
+
|
| 45 |
+
To determine the frequencies of the genotypes of *CYP3A5*CYP3A5 and *MDR1*MDR1 and to examine the influence of the polymorphisms of these genes on tacrolimus pharmacokinetics in the Korean population.
|
| 46 |
+
|
| 47 |
+
### Methods
|
| 48 |
+
|
| 49 |
+
Twenty-nine healthy Koreans who participated in the previous tacrolimus pharmacokinetic study were genotyped for *CYP3A4*CYP3A4**1B*1B, *CYP3A5*CYP3A5**3*3, *MDR1*MDR1 c.1236C→T, *MDR1*MDR1 c.2677G→A/T and *MDR1*MDR1 c.3435C→T. The relationship between the genotypes so obtained and tacrolimus pharmacokinetics observed in the previous study was examined.
|
| 50 |
+
|
| 51 |
+
### Results
|
| 52 |
+
|
| 53 |
+
No subject in this study had the *CYP3A4*CYP3A4**1B*1B variant. The observed frequencies of *CYP3A5*CYP3A5**1/*1/**1*1, **1/*1/**3*3, and **3/*3/**3*3 were 0.069 [confidence interval (CI) −0.023, 0.161], 0.483 (CI 0.301, 0.665) and 0.448 (CI 0.267, 0.629), respectively. AUC_0–∞_0–∞ for the *CYP3A5*CYP3A5**1/*1/**1*1 or **1/*1/**3*3 genotype was 131.5 ± 44.8 ng h ml^−1^−1 (CI 109.6, 153.5), which was much lower compared with the *CYP3A5*CYP3A5**3/*3/**3*3 genotype of 323.8 ± 129.3 ng h ml^−1^−1 (CI 253.5, 394.1) (*P*P = 2.063E−07). Similarly, *C*C_max_max for the *CYP3A5*CYP3A5**1/*1/**1*1 or **1/*1/**3*3 genotype was 11.8 ± 3.4 ng ml^−1^−1 (CI 10.1, 13.5), which was also much lower compared with the *CYP3A5*CYP3A5**3/*3/**3*3 genotype of 24.4 ± 12.3 ng ml^−1^−1 (CI 17.8, 31.1) (*P*P = 0.0001). However, there was no significant difference in tacrolimus pharmacokinetics among the *MDR1*MDR1 diplotypes of CGC-CGC, CGC-TTT, CGC-TGC, TTT-TGC or TTT-TTT (*P*P = 0.2486).
|
| 54 |
+
|
| 55 |
+
### Conclusions
|
| 56 |
+
|
| 57 |
+
This study shows that the *CYP3A5*CYP3A5**3*3 genetic polymorphisms may be associated with the individual difference in tacrolimus pharmacokinetics. An individualized dosage regimen design incorporating such genetic information would help increase clinical efficacy of the drug while reducing adverse drug reactions.
|
| 58 |
+
|
| 59 |
+
**Keywords:**Keywords: CYP3A5, MDR1, pharmacokinetics, polymorphism, tacrolimus
|
| 60 |
+
|
| 61 |
+
## Introduction
|
| 62 |
+
|
| 63 |
+
Tacrolimus, one of the calcineurin inhibitors, is highly effective in preventing acute rejection after transplantation of solid organs, including the liver and kidney [[1](#b1)1]. However, tacrolimus shows high between- and within-subject variability in pharmacokinetics, with a narrow therapeutic index, necessitating therapeutic drug monitoring to optimize treatment [[2](#b2)2–[4](#b4)4].
|
| 64 |
+
|
| 65 |
+
It has become clear that the variability in the pharmacokinetics of tacrolimus is largely determined by differences in oral bioavailability [[5](#b5)5]. Tacrolimus is a substrate of P-glycoprotein, the product of the multidrug resistance (*MDR1*MDR1) gene and has been known to undergo extensive hepatic metabolism by cytochrome P450 3A4 (CYP3A4) and CYP3A5 [[6](#b6)6–[8](#b8)8]. The variability in oral bioavailability of tacrolimus thus has been attributed to individual differences in expression of CYP3A4, CYP3A5 and P-glycoprotein [[9](#b9)9].
|
| 66 |
+
|
| 67 |
+
A limited number of studies have reported on the association between *CYP3A4*CYP3A4 variants and the pharmacokinetics of tacrolimus. Hesselink *et al.*et al. found that *CYP3A4*CYP3A4**1B*1B carriers require larger dose of tacrolimus to reach target trough concentrations compared with *CYP3A4*CYP3A4**1*1 homozygotes [[5](#b5)5]. In their work, it was established that the dose requirement for tacrolimus could be affected by *CYP3A5*CYP3A5 variants. Several studies have reported that the dose-adjusted trough concentration of tacrolimus is much higher in *CYP3A5*CYP3A5**3/*3/**3*3 subjects than in **1/*1/**1*1 or **1/*1/**3*3 subjects [[10](#b10)10–[15](#b15)15]. The *CYP3A5*CYP3A5**3*3 variant creates an alternative splice in pre-mRNA and produces aberrant mRNA that does not translate into functional CYP3A5 protein [[16](#b16)16]. In contrast, numerous studies have investigated whether the genetic polymorphisms in *MDR1*MDR1, including c.1236C→T, c.2677G→A/T and c.3435C→T, could affect the pharmacokinetics of tacrolimus, yielding controversial results [[9](#b9)9].
|
| 68 |
+
|
| 69 |
+
In a previous pharmacokinetic study, we compared the pharmacokinetics of twice-daily dosing of 1-mg tacrolimus capsules of two different formulations in healthy Korean volunteers: Prograf (Astellas Pharma Korea, Inc.), the reference, or conventional formulation, and TacroBell® (Chong Kun Dang Pharmaceutical Corp., Korea), the test, or newly developed formulation. While no significant difference in formulation was observed, we found high interindividual variability [51–76%, coefficient of variation (CV)] in the pharmacokinetics of tacrolimus and suspected a possible genetic polymorphism as a source of pharmacokinetic variation [[17](#b17)17]. In this regard, this study was intended to see if such a possibility existed in the Korean population. Subjects who had participated in the previous tacrolimus pharmacokinetic study were therefore first examined for the frequencies of *CYP3A4*CYP3A4**1B*1B, *CYP3A5*CYP3A5**3*3 and three single nucleotide polymorphisms (SNPs) of *MDR1*MDR1, c.1236C→T, c.2677G→A/T and c.3435C→ T, and then for an association between the resulting genotypes and tacrolimus pharmacokinetics obtained in the previous study.
|
| 70 |
+
|
| 71 |
+
## Materials and methods
|
| 72 |
+
|
| 73 |
+
### Subjects
|
| 74 |
+
|
| 75 |
+
This study was approved by the institutional review board of Yonsei University Medical Centre, Seoul, Korea. A total of 29 unrelated healthy Korean volunteers participated in this study after giving written informed consent. They were recruited from the volunteers who had participated in the previous pharmacokinetic study of two oral formulations of tacrolimus [[17](#b17)17].
|
| 76 |
+
|
| 77 |
+
### Genetic analysis
|
| 78 |
+
|
| 79 |
+
We investigated the genotype frequencies of *CYP3A4*CYP3A4**1B*1B (GenBank accession number [AC069294](https://www.ncbi.nlm.nih.gov/nuccore/AC069294)AC069294), *CYP3A5*CYP3A5**3*3 (GenBank accession number [AC005020](https://www.ncbi.nlm.nih.gov/nuccore/AC005020)AC005020) and three SNPs of *MDR1*MDR1, c.1236C→T, c.2677G→A/T and c.3435C→T (GenBank accession numbers NC000007, NM000927) for the subjects by direct sequencing using an automated genetic analyser (Model 3700; Applied Biosystems, Foster City, CA, USA). The nomenclature of *CYP3A4*CYP3A4 and *CYP3A5*CYP3A5 was based on the website [http://www.cypalleles.ki.se/](http://www.cypalleles.ki.se/)http://www.cypalleles.ki.se/. For each subject, one blood sample was collected and DNA was extracted using a purification kit (Qiagen, Hilden, Germany). Haplotype assembly was performed using the Haploview 3.2 program (Broad Institute of Harvard and MIT, Cambridge, MA, USA), based on a standard expectation-maximization algorithm to reconstruct individual haplotypes from population genotype data [[18](#b18)18].
|
| 80 |
+
|
| 81 |
+
### Pharmacokinetic data
|
| 82 |
+
|
| 83 |
+
For pharmacokinetic data, we used data obtained from the previous pharmacokinetic study, which can be summarized as follows [[17](#b17)17]. It was as an open-label, randomized, two-period, cross-over study with a 3-week wash-out period in 29 healthy Korean volunteers. Each subject received two 1-mg capsules of a conventional (reference) or a newly developed (test) oral tacrolimus formulation twice a day, morning and evening (total daily dose of 4 mg). Blood samples were collected before dosing and at 0.5, 1, 1.5, 2, 3, 4, 7, 12, 12.5, 13, 13.5, 14, 15, 16, 19, 24, 36, 48, 72 and 96 h after dosing. Whole blood concentrations were analysed by LC/MS/MS. Pharmacokinetic parameters such as area under the concentration curve from time zero to infinity (AUC_0–∞_0–∞), maximum concentration (*C*C_max_max), time to *C*C_max_max (*t*t_max_max) and half-life (*t*t_1/2_1/2) for tacrolimus were estimated by noncompartmental analysis using WinNonlin Professional 4.1. In results, the two formulations were not significantly different and showed similarly large interindividual variation in tacrolimus pharmacokinetics [59.3–61.1% (test) and 50.7–75.6% (reference) in CV]. Therefore, in this study, without loss of generality, only the conventional formulation's pharmacokinetic data were used in examining genetic influences on tacrolimus pharmacokinetics.
|
| 84 |
+
|
| 85 |
+
### Statistical analysis
|
| 86 |
+
|
| 87 |
+
Differences in pharmacokinetic parameters between the genotype groups were tested using the Wilcoxon rank sum test or the Kruskal–Wallis test. The frequency of *CYP3A5*CYP3A5**3/*3/**3*3 genotype was compared with that of the other ethnic groups, using χ^2^2 test. *P*P-values < 0.05 were considered to be statistically significant.
|
| 88 |
+
|
| 89 |
+
## Results
|
| 90 |
+
|
| 91 |
+
### Polymorphisms of CYP3A5 and MDR1 in Koreans
|
| 92 |
+
|
| 93 |
+
[Table 1](#tbl1)Table 1 shows frequency distributions of *CYP3A5*CYP3A5 and *MDR1*MDR1 genotypes in 29 Korean subjects examined in this study. Each genotype did not deviate from Hardy–Weinberg equilibrium. Among the *CYP3A4*CYP3A4**1B*1B, *CYP3A5*CYP3A5**3*3 and *MDR1*MDR1 genotypes examined in our study, the *CYP3A4*CYP3A4**1B*1B genotype was not observed in any subject and the *CYP3A5*CYP3A5**1/*1/**1*1 genotype was observed in only two subjects, yielding a frequency of 0.069 [confidence interval (CI) −0.023, 0.161), with CI indicating a 95% CI. In contrast, for the *CYP3A5*CYP3A5**3*3 variant, the heterozygous allele was observed in 14 subjects with a frequency of 0.483 (CI 0.301, 0.665), while the homozygous allele was observed in 13 subjects with a frequency of 0.448 (CI 0.267, 0.629). The frequency of *CYP3A5*CYP3A5**3/*3/**3*3 observed with our subjects was similar to that of 0.400 (CI 0.268, 0.532) with Chinese (*P*P = 0.5313) or 0.583 (CI 0.512, 0.654) with Japanese (*P*P = 0.1737) [[19](#b19)19, [20](#b20)20]. However, some significant differences were also found in the frequency of this genotype. That is, the frequency of *CYP3A5*CYP3A5**3/*3/**3*3 genotype was higher in Koreans compared with the frequency of 0.029 (CI −0.027, 0.085) in African-Americans (*P*P = 0.0001), but was much lower compared with the frequency of 0.830 (CI 0.756, 0.904) in Whites (*P*P = 0.0001) [[21](#b21)21, [22](#b22)22]. These results suggest that the frequency of *CYP3A5*CYP3A5**3/*3/**3*3 genotype is dependent on ethnicity.
|
| 94 |
+
|
| 95 |
+
### Table 1.
|
| 96 |
+
|
| 97 |
+
Genotype frequencies for CYP3A5 and MDR1 in healthy Koreans (n = 29)
|
| 98 |
+
|
| 99 |
+
| Variant | Genotype | No. (frequency) | 95% CI |
|
| 100 |
+
| ------- | -------- | --------------- | ------ |
|
| 101 |
+
| CYP3A5*3 | *1/*1 | 2 (0.069) | (−0.023, 0.161) |
|
| 102 |
+
| | *1/*3 | 14 (0.483) | (0.301, 0.665) |
|
| 103 |
+
| | *3/*3 | 13 (0.448) | (0.267, 0.629) |
|
| 104 |
+
| MDR1 c.1236C→T | CC | 3 (0.103) | (−0.008, 0.214) |
|
| 105 |
+
| | CT | 12 (0.414) | (0.235, 0.593) |
|
| 106 |
+
| | TT | 14 (0.483) | (0.301, 0.665) |
|
| 107 |
+
| MDR1 c.2677G→A/T | GG | 4 (0.138) | (0.012, 0.264) |
|
| 108 |
+
| | GA | 2 (0.069) | (−0.023, 0.161) |
|
| 109 |
+
| | GT | 15 (0.517) | (0.335, 0.699) |
|
| 110 |
+
| | AT | 1 (0.034) | (−0.032, 0.100) |
|
| 111 |
+
| | AA | 2 (0.069) | (−0.023, 0.161) |
|
| 112 |
+
| | TT | 5 (0.172) | (0.035, 0.309) |
|
| 113 |
+
| MDR1 c.3435C→T | CC | 8 (0.276) | (0.113, 0.439) |
|
| 114 |
+
| | CT | 17 (0.586) | (0.407, 0.765) |
|
| 115 |
+
| | TT | 4 (0.138) | (0.012, 0.264) |
|
| 116 |
+
The *MDR1*MDR1 haplotypes were estimated from the three *MDR1*MDR1 genotypes at c.1236C→T, c.2677G→A/T and c.3435C→T using a standard expectation-maximization algorithm ([Table 2](#tbl2)Table 2). Three major haplotypes that were relatively commonly observed were TTT, CGC and TGC, accounting for 41.4%, 20.7% and 20.7% of the total haplotype diversity, respectively. The combinations of these three predominant haplotypes constituted the five diplotypes examined in this study, CGC-CGC (3.4%, *n*n = 1), CGC-TTT (17.2%, *n*n = 5), CGC-TGC (10.3%, *n*n = 3), TTT-TGC (31.0%, *n*n = 9) and TTT-TTT (10.3%, *n*n = 3) ([Table 3](#tbl3)Table 3).
|
| 117 |
+
|
| 118 |
+
### Table 2.
|
| 119 |
+
|
| 120 |
+
Haplotype frequencies of MDR1 c.1236C→T, c.2677G→A/T, c.3435C→T in healthy Koreans (n = 29)
|
| 121 |
+
|
| 122 |
+
| Haplotype | c.1236C→T | c.2677G→A/T | c.3435C→T | Frequency (95% CI) |
|
| 123 |
+
| --------- | --------- | ----------- | --------- | ------------------ |
|
| 124 |
+
| TTT | T | T | T | 0.414 (0.287, 0.541) |
|
| 125 |
+
| CGC | C | G | C | 0.207 (0.103, 0.311) |
|
| 126 |
+
| TGC | T | G | C | 0.207 (0.103, 0.311) |
|
| 127 |
+
| CAC | C | A | C | 0.069 (0.004, 0.134) |
|
| 128 |
+
| TAC | T | A | C | 0.034 (−0.013, 0.081) |
|
| 129 |
+
| TTC | T | T | C | 0.034 (−0.013, 0.081) |
|
| 130 |
+
| CGT | C | G | T | 0.017 (−0.016, 0.050) |
|
| 131 |
+
### Table 3.
|
| 132 |
+
|
| 133 |
+
Tacrolimus pharmacokinetic parameters against major MDR1 genotypes in healthy Koreans (n = 21)
|
| 134 |
+
|
| 135 |
+
| Diplotype (no.) | AUC0–∞ (ng h ml−1) | Cmax (ng ml−1) | tmax (h) | t1/2 (h) |
|
| 136 |
+
| --------------- | ------------------ | -------------- | -------- | -------- |
|
| 137 |
+
| CGC-CGC (1) | 565.7 | 30.9 | 2.0 | 31.2 |
|
| 138 |
+
| CGC-TTT (5) | 138.1 ± 38.9 (104.0, 172.2) | 11.7 ± 3.5 (8.6, 14.8) | 1.2 ± 0.3 (1.0, 1.4) | 23.3 ± 12.6 (12.3, 34.4) |
|
| 139 |
+
| CGC-TGC (3) | 233.0 ± 106.0 (113.1, 353.0) | 18.3 ± 4.4 (13.3, 23.3) | 1.7 ± 0.3 (1.3, 2.0) | 23.4 ± 3.1 (19.9, 26.9) |
|
| 140 |
+
| TTT-TGC (9) | 250.9 ± 157.8 (147.9, 354.0) | 19.0 ± 13.4 (10.3, 27.7) | 1.6 ± 0.9 (1.0, 2.2) | 31.6 ± 12.0 (23.8, 39.4) |
|
| 141 |
+
| TTT-TTT (3) | 200.3 ± 69.2 (122.0, 278.6) | 16.3 ± 5.6 (9.9, 22.6) | 1.3 ± 0.3 (1.0, 1.7) | 33.9 ± 11.4 (21.0, 46.9) |
|
| 142 |
+
| P-value* | 0.2486 | 0.3926 | 0.4733 | 0.4609 |
|
| 143 |
+
### Pharmacokinetic parameters of tacrolimus
|
| 144 |
+
|
| 145 |
+
In this study, pharmacokinetic values were given as mean ± SD with 95% CI. [Table 4](#tbl4)Table 4 shows that AUC_0–∞_0–∞ for the subject with the *CYP3A5*CYP3A5**1/*1/**1*1 or **1/*1/**3*3 genotype was 131.5 ± 44.8 ng h ml^−1^−1 (CI 109.6, 153.5), which was much lower compared with the *CYP3A5*CYP3A5**3/*3/**3*3 genotype of 323.8 ± 129.3 ng h ml^−1^−1 (CI 253.5, 394.1) (*P*P = 2.063E−07). [Table 4](#tbl4)Table 4 also shows that *C*C_max_max for the subject with the *CYP3A5*CYP3A5**1/*1/**1*1 or **1/*1/**3*3 genotype was 11.8 ± 3.4 ng ml^−1^−1 (CI 10.1, 13.5), which was also much lower compared with the *CYP3A5*CYP3A5**3/*3/**3*3 genotype of 24.4 ± 12.3 ng ml^−1^−1 (CI 17.8, 31.1) (*P*P = 0.0001). The values of *t*t_max_max and *t*t_1/2_1/2 were similar between the two genotype groups (*P*P = 0.1160, 0.1103, respectively). The genotype differences in AUC_0–∞_0–∞ and *C*C_max_max of tacrolimus are visually depicted in [Figure 1](#fig01)Figure 1.
|
| 146 |
+
|
| 147 |
+
### Table 4.
|
| 148 |
+
|
| 149 |
+
Tacrolimus pharmacokinetic parameters vs. CYP3A5 genotypes in healthy Koreans (n = 29)
|
| 150 |
+
|
| 151 |
+
| Genotype | AUC0–∞ (ng h ml−1) | Cmax (ng ml−1) | tmax (h) | t1/2 (h) |
|
| 152 |
+
| -------- | ------------------ | -------------- | -------- | -------- |
|
| 153 |
+
| CYP3A5 | 131.5 ± 44.8 | 11.8 ± 3.4 | 1.4 ± 0.7 | 23.9 ± 11.9 |
|
| 154 |
+
| *1/*1 or *1/*3 | (109.6, 153.5) | (10.1, 13.5) | (1.1, 1.7) | (18.0, 29.7) |
|
| 155 |
+
| CYP3A5 | 323.8 ± 129.3 | 24.4 ± 12.3 | 1.6 ± 0.4 | 30.6 ± 8.7 |
|
| 156 |
+
| *3/*3 | (253.5, 394.1) | (17.8, 31.1) | (1.4, 1.8) | (25.9, 35.3) |
|
| 157 |
+
| P-value* | 2.0630E−07 | 0.0001 | 0.1160 | 0.1103 |
|
| 158 |
+
### Figure 1.
|
| 159 |
+
|
| 160 |
+

|
| 161 |
+
|
| 162 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=2000625_bcp0064-0185-f1.jpg)
|
| 163 |
+
|
| 164 |
+
AUC0–∞ (A) and Cmax (B) of tacrolimus illustrated according to the CYP3A5 genotype. Triangles, CYP3A5*1/*1 or *1/*3 (n = 16); circles, CYP3A5*3/*3 (n = 13). A thick bar indicates the mean value of the group
|
| 165 |
+
|
| 166 |
+
There was no significant difference in pharmacokinetics of tacrolimus among the five *MDR1*MDR1 diplotype groups (*P*P = 0.2486) ([Table 3](#tbl3)Table 3).
|
| 167 |
+
|
| 168 |
+
## Discussion
|
| 169 |
+
|
| 170 |
+
In the present study, we observed a significant association between the blood concentration of tacrolimus and the *CYP3A5*CYP3A5**3*3 variant. Our results show that subjects with the *CYP3A5*CYP3A5**1/*1/**1*1 or **1/*1/**3*3 genotype yield much lower AUC_0–∞_0–∞ and *C*C_max_max values than the *CYP3A5*CYP3A5**3/*3/**3*3 genotype. To investigate other possibilities that may contribute to such pharmacokinetic differences, demographic factors including age, sex, height and body weight were compared between the two genotype groups. No demographic factor showed a significant difference between the two groups other than age, which showed distributions (mean ± SD) of 31.4 ± 8.3 years for the *CYP3A5*CYP3A5**1/*1/**1*1 or **1/*1/**3*3 genotype *vs.*vs. 25.6 ± 2.1 years for the *CYP3A5*CYP3A5**3/*3/**3*3 genotype (*P*P = 0.0153) ([Table 5](#tbl5)Table 5). However, it has been reported that CYP3A activity is not significantly influenced by age [[23](#b23)23].
|
| 171 |
+
|
| 172 |
+
### Table 5.
|
| 173 |
+
|
| 174 |
+
Demographic characteristics of the CYP3A5 genotype in healthy Koreans (n = 29)
|
| 175 |
+
|
| 176 |
+
| Genotype | Sex (male:female) | Age*(years) | Weight*(kg) | Height*(cm) |
|
| 177 |
+
| -------- | ----------------- | ----------- | ----------- | ----------- |
|
| 178 |
+
| CYP3A5 | 12 : 4 | 31.4 ± 8.3 | 64.4 ± 9.0 | 170.5 ± 5.4 |
|
| 179 |
+
| *1/*1 or *1/*3 | | (20–51) | (48–78) | (160–182) |
|
| 180 |
+
| CYP3A5 | 10 : 3 | 25.6 ± 2.1 | 68.7 ± 11.1 | 172.2 ± 8.5 |
|
| 181 |
+
| *3/*3 | | (23–30) | (48–87) | (155–181) |
|
| 182 |
+
| P-value | >0.9999† | 0.0153‡ | 0.2760‡ | 0.5314‡ |
|
| 183 |
+
Several studies have reported the interethnic variation in the pharmacokinetics of tacrolimus [[24](#b24)24, [25](#b25)25]. In relation to interethnic differences in the frequency of the *CYP3A5*CYP3A5**3*3 allele, Thompson *et al.*et al. found that the frequency is lowest in sub-Saharan Africa and highest in European and East Asian populations, showing an increasing trend with distance from the equator [[26](#b26)26]. Similarly in this work, we found that the *CYP3A5*CYP3A5**3/*3/**3*3 genotype was more common in Koreans and Whites than in African-Americans. The interethnic difference in the frequency of *CYP3A5*CYP3A5**3/*3/**3*3 genotype may contribute to the interethnic variation in the pharmacokinetics of tacrolimus.
|
| 184 |
+
|
| 185 |
+
In our subjects, no one had the *CYP3A4*CYP3A4**1B*1B variant. According to the work by Lamba *et al.*et al., this genotype was not found in Japanese-Americans or in Chinese-Americans, while frequencies of 2.0–9.6% and 35–67% were observed in Whites and African-Americans, respectively [[16](#b16)16]. These findings indicate that the frequency of the *CYP3A4*CYP3A4**1B*1B allele may vary among ethnic groups.
|
| 186 |
+
|
| 187 |
+
In our results, no significant difference was found in tacrolimus pharmacokinetics against *MDR1*MDR1 polymorphisms in c.1236C→T, c.2677G→A/T or c.3435C→T, indicating a nonsignificant association between the blood concentration of tacrolimus and these three genotype groups (data not shown). Although many researchers have tried to examine whether these three SNPs could affect the expression and function of MDR1, the results are still controversial [[27](#b27)27]. Recently, it has been confirmed that *MDR1*MDR1 c.3435C→T is in significant linkage disequilibrium with c.1236C→T and c.2677G→A/T and the analysis of *MDR1*MDR1 haplotypes may be superior to that of SNPs in revealing genotype–phenotype associations in pharmacokinetic studies [[8](#b8)8, [27](#b27)27–[29](#b29)29].
|
| 188 |
+
|
| 189 |
+
In our work, we estimated the *MDR1*MDR1 haplotypes from the three *MDR1*MDR1 genotypes at c.1236C→T, c.2677G→A/T and c.3435C→T using a standard expectation-maximization algorithm. Three predominant haplotypes were observed and the combinations of these three haplotypes constituted the five diplotypes, with no significant difference observed in tacrolimus pharmacokinetics among these five *MDR1*MDR1 diplotypes. Thus, although it was found that the subject with CGC-CGC diplotype showed much higher AUC_0–∞_0–∞ and *C*C_max_max compared with the other subjects ([Table 3](#tbl3)Table 3), this may be mainly due to the influence of the *CYP3A5*CYP3A5**3/*3/**3*3 genotype of this subject ([Table 4](#tbl4)Table 4).
|
| 190 |
+
|
| 191 |
+
In conclusion, our study demonstrates that the *CYP3A5*CYP3A5 genetic polymorphisms may be associated with the pharmacokinetic variation of tacrolimus in Korean populations. Therefore, dosage regimen design incorporating genetic polymorphisms in *CYP3A5*CYP3A5 may be of help in identifying the optimal dose for the individual patient.
|
| 192 |
+
|
| 193 |
+
## Acknowledgments
|
| 194 |
+
|
| 195 |
+
This study was supported by the grant 03-PJ10-PG13-GD01-0002 from the Korea Health 21 R&D Project, Ministry of Health & Welfare, Korea, by the grant A050001 from the Korean Ministry of Health and Welfare Public Health and Medical Technology Infrastructure Development Plan, and by Chong Kun Dang Pharmaceutical Corp., Seoul, Korea. [Correction added after online publication 10 July 2007: acknowledgement omitted].
|
| 196 |
+
|
| 197 |
+
## References
|
| 198 |
+
|
| 199 |
+
1. First MR. Tacrolimus based immunosuppression. J Nephrol. 2004;17(Suppl. 8):S25–31. [PubMed](https://pubmed.ncbi.nlm.nih.gov/15599882/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Nephrol&title=Tacrolimus%20based%20immunosuppression&author=MR%20First&volume=17&issue=Suppl.%208&publication_year=2004&pages=S25-31&pmid=15599882&)
|
| 200 |
+
|
| 201 |
+
2. Felipe CR, Silva HT, Machado PG, Garcia R, da Silva Moreira SR, Pestana JO. The impact of ethnic miscegenation on tacrolimus clinical pharmacokinetics and therapeutic drug monitoring. Clin Transplant. 2002;16:262–72. doi: 10.1034/j.1399-0012.2002.01103.x. [DOI](https://doi.org/10.1034/j.1399-0012.2002.01103.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12099982/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Transplant&title=The%20impact%20of%20ethnic%20miscegenation%20on%20tacrolimus%20clinical%20pharmacokinetics%20and%20therapeutic%20drug%20monitoring&author=CR%20Felipe&author=HT%20Silva&author=PG%20Machado&author=R%20Garcia&author=SR%20da%20Silva%20Moreira&volume=16&publication_year=2002&pages=262-72&pmid=12099982&doi=10.1034/j.1399-0012.2002.01103.x&)
|
| 202 |
+
|
| 203 |
+
3. Taylor AL, Watson CJ, Bradley JA. Immunosuppressive agents in solid organ transplantation: mechanisms of action and therapeutic efficacy. Crit Rev Oncol Hematol. 2005;56:23–46. doi: 10.1016/j.critrevonc.2005.03.012. [DOI](https://doi.org/10.1016/j.critrevonc.2005.03.012) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16039869/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Crit%20Rev%20Oncol%20Hematol&title=Immunosuppressive%20agents%20in%20solid%20organ%20transplantation:%20mechanisms%20of%20action%20and%20therapeutic%20efficacy&author=AL%20Taylor&author=CJ%20Watson&author=JA%20Bradley&volume=56&publication_year=2005&pages=23-46&pmid=16039869&doi=10.1016/j.critrevonc.2005.03.012&)
|
| 204 |
+
|
| 205 |
+
4. Scott LJ, McKeage K, Keam SJ, Plosker GL. Tacrolimus: a further update of its use in the management of organ transplantation. Drugs. 2003;63:1247–97. doi: 10.2165/00003495-200363120-00006. [DOI](https://doi.org/10.2165/00003495-200363120-00006) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12790696/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Drugs&title=Tacrolimus:%20a%20further%20update%20of%20its%20use%20in%20the%20management%20of%20organ%20transplantation&author=LJ%20Scott&author=K%20McKeage&author=SJ%20Keam&author=GL%20Plosker&volume=63&publication_year=2003&pages=1247-97&pmid=12790696&doi=10.2165/00003495-200363120-00006&)
|
| 206 |
+
|
| 207 |
+
5. Hesselink DA, van Schaik RH, van der Heiden IP, van der Werf M, Gregoor PJ, Lindemans J, Weimar W, van Gelder T. Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR-1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin Pharmacol Ther. 2003;74:245–54. doi: 10.1016/S0009-9236(03)00168-1. [DOI](https://doi.org/10.1016/S0009-9236(03)00168-1) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12966368/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Genetic%20polymorphisms%20of%20the%20CYP3A4,%20CYP3A5,%20and%20MDR-1%20genes%20and%20pharmacokinetics%20of%20the%20calcineurin%20inhibitors%20cyclosporine%20and%20tacrolimus&author=DA%20Hesselink&author=RH%20van%20Schaik&author=IP%20van%20der%20Heiden&author=M%20van%20der%20Werf&author=PJ%20Gregoor&volume=74&publication_year=2003&pages=245-54&pmid=12966368&doi=10.1016/S0009-9236(03)00168-1&)
|
| 208 |
+
|
| 209 |
+
6. Roy JN, Barama A, Poirier C, Vinet B, Roger M. Cyp3A4, Cyp3A5, and MDR-1 genetic influences on tacrolimus pharmacokinetics in renal transplant recipients. Pharmacogenet Genomics. 2006;16:659–65. doi: 10.1097/01.fpc.0000220571.20961.dd. [DOI](https://doi.org/10.1097/01.fpc.0000220571.20961.dd) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16906020/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenet%20Genomics&title=Cyp3A4,%20Cyp3A5,%20and%20MDR-1%20genetic%20influences%20on%20tacrolimus%20pharmacokinetics%20in%20renal%20transplant%20recipients&author=JN%20Roy&author=A%20Barama&author=C%20Poirier&author=B%20Vinet&author=M%20Roger&volume=16&publication_year=2006&pages=659-65&pmid=16906020&doi=10.1097/01.fpc.0000220571.20961.dd&)
|
| 210 |
+
|
| 211 |
+
7. Anglicheau D, Verstuyft C, Laurent-Puig P, Becquemont L, Schlageter MH, Cassinat B, Beaune P, Legendre C, Thervet E. Association of the multidrug resistance-1 gene single-nucleotide polymorphisms with the tacrolimus dose requirements in renal transplant recipients. J Am Soc Nephrol. 2003;14:1889–96. doi: 10.1097/01.asn.0000073901.94759.36. [DOI](https://doi.org/10.1097/01.asn.0000073901.94759.36) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12819250/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Am%20Soc%20Nephrol&title=Association%20of%20the%20multidrug%20resistance-1%20gene%20single-nucleotide%20polymorphisms%20with%20the%20tacrolimus%20dose%20requirements%20in%20renal%20transplant%20recipients&author=D%20Anglicheau&author=C%20Verstuyft&author=P%20Laurent-Puig&author=L%20Becquemont&author=MH%20Schlageter&volume=14&publication_year=2003&pages=1889-96&pmid=12819250&doi=10.1097/01.asn.0000073901.94759.36&)
|
| 212 |
+
|
| 213 |
+
8. Mai I, Perloff ES, Bauer S, Goldammer M, Johne A, Filler G, Budde K, Roots I. MDR1 haplotypes derived from exons 21 and 26 do not affect the steady-state pharmacokinetics of tacrolimus in renal transplant patients. Br J Clin Pharmacol. 2004;58:548–53. doi: 10.1111/j.1365-2125.2004.02182.x. [DOI](https://doi.org/10.1111/j.1365-2125.2004.02182.x) | [PMC free article](/articles/PMC1884628/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15521904/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=MDR1%20haplotypes%20derived%20from%20exons%2021%20and%2026%20do%20not%20affect%20the%20steady-state%20pharmacokinetics%20of%20tacrolimus%20in%20renal%20transplant%20patients&author=I%20Mai&author=ES%20Perloff&author=S%20Bauer&author=M%20Goldammer&author=A%20Johne&volume=58&publication_year=2004&pages=548-53&pmid=15521904&doi=10.1111/j.1365-2125.2004.02182.x&)
|
| 214 |
+
|
| 215 |
+
9. Hesselink DA, van Gelder T, van Schaik RH. The pharmacogenetics of calcineurin inhibitors: one step closer toward individualized immunosuppression? Pharmacogenomics. 2005;6:323–37. doi: 10.1517/14622416.6.4.323. [DOI](https://doi.org/10.1517/14622416.6.4.323) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16004552/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=The%20pharmacogenetics%20of%20calcineurin%20inhibitors:%20one%20step%20closer%20toward%20individualized%20immunosuppression?&author=DA%20Hesselink&author=T%20van%20Gelder&author=RH%20van%20Schaik&volume=6&publication_year=2005&pages=323-37&pmid=16004552&doi=10.1517/14622416.6.4.323&)
|
| 216 |
+
|
| 217 |
+
10. Macphee IA, Fredericks S, Mohamed M, Moreton M, Carter ND, Johnston A, Goldberg L, Holt DW. Tacrolimus pharmacogenetics: the CYP3A5*1 allele predicts low dose-normalized tacrolimus blood concentrations in whites and South Asians. Transplantation. 2005;79:499–502. doi: 10.1097/01.tp.0000151766.73249.12. [DOI](https://doi.org/10.1097/01.tp.0000151766.73249.12) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15729180/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplantation&title=Tacrolimus%20pharmacogenetics:%20the%20CYP3A5*1%20allele%20predicts%20low%20dose-normalized%20tacrolimus%20blood%20concentrations%20in%20whites%20and%20South%20Asians&author=IA%20Macphee&author=S%20Fredericks&author=M%20Mohamed&author=M%20Moreton&author=ND%20Carter&volume=79&publication_year=2005&pages=499-502&pmid=15729180&doi=10.1097/01.tp.0000151766.73249.12&)
|
| 218 |
+
|
| 219 |
+
11. Tsuchiya N, Satoh S, Tada H, Li Z, Ohyama C, Sato K, Suzuki T, Habuchi T, Kato T. Influence of CYP3A5 and MDR1 (ABCB1) polymorphisms on the pharmacokinetics of tacrolimus in renal transplant recipients. Transplantation. 2004;78:1182–7. doi: 10.1097/01.tp.0000137789.58694.b4. [DOI](https://doi.org/10.1097/01.tp.0000137789.58694.b4) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15502717/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplantation&title=Influence%20of%20CYP3A5%20and%20MDR1%20(ABCB1)%20polymorphisms%20on%20the%20pharmacokinetics%20of%20tacrolimus%20in%20renal%20transplant%20recipients&author=N%20Tsuchiya&author=S%20Satoh&author=H%20Tada&author=Z%20Li&author=C%20Ohyama&volume=78&publication_year=2004&pages=1182-7&pmid=15502717&doi=10.1097/01.tp.0000137789.58694.b4&)
|
| 220 |
+
|
| 221 |
+
12. Zheng H, Webber S, Zeevi A, Schuetz E, Zhang J, Bowman P, Boyle G, Law Y, Miller S, Lamba J, Burckart GJ. Tacrolimus dosing in pediatric heart transplant patients is related to CYP3A5 and MDR1 gene polymorphisms. Am J Transplant. 2003;3:477–83. doi: 10.1034/j.1600-6143.2003.00077.x. [DOI](https://doi.org/10.1034/j.1600-6143.2003.00077.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12694072/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Transplant&title=Tacrolimus%20dosing%20in%20pediatric%20heart%20transplant%20patients%20is%20related%20to%20CYP3A5%20and%20MDR1%20gene%20polymorphisms&author=H%20Zheng&author=S%20Webber&author=A%20Zeevi&author=E%20Schuetz&author=J%20Zhang&volume=3&publication_year=2003&pages=477-83&pmid=12694072&doi=10.1034/j.1600-6143.2003.00077.x&)
|
| 222 |
+
|
| 223 |
+
13. Goto M, Masuda S, Kiuchi T, Ogura Y, Oike F, Okuda M, Tanaka K, Inui K. CYP3A5*1-carrying graft liver reduces the concentration/oral dose ratio of tacrolimus in recipients of living-donor liver transplantation. Pharmacogenetics. 2004;14:471–8. doi: 10.1097/01.fpc.0000114747.08559.49. [DOI](https://doi.org/10.1097/01.fpc.0000114747.08559.49) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15226679/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=CYP3A5*1-carrying%20graft%20liver%20reduces%20the%20concentration/oral%20dose%20ratio%20of%20tacrolimus%20in%20recipients%20of%20living-donor%20liver%20transplantation&author=M%20Goto&author=S%20Masuda&author=T%20Kiuchi&author=Y%20Ogura&author=F%20Oike&volume=14&publication_year=2004&pages=471-8&pmid=15226679&doi=10.1097/01.fpc.0000114747.08559.49&)
|
| 224 |
+
|
| 225 |
+
14. Zheng H, Zeevi A, Schuetz E, Lamba J, McCurry K, Griffith BP, Webber S, Ristich J, Dauber J, Iacono A, Grgurich W, Zaldonis D, McDade K, Zhang J, Burckart GJ. Tacrolimus dosing in adult lung transplant patients is related to cytochrome P4503A5 gene polymorphism. J Clin Pharmacol. 2004;44:135–40. doi: 10.1177/0091270003262108. [DOI](https://doi.org/10.1177/0091270003262108) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14747421/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Clin%20Pharmacol&title=Tacrolimus%20dosing%20in%20adult%20lung%20transplant%20patients%20is%20related%20to%20cytochrome%20P4503A5%20gene%20polymorphism&author=H%20Zheng&author=A%20Zeevi&author=E%20Schuetz&author=J%20Lamba&author=K%20McCurry&volume=44&publication_year=2004&pages=135-40&pmid=14747421&doi=10.1177/0091270003262108&)
|
| 226 |
+
|
| 227 |
+
15. Thervet E, Anglicheau D, King B, Schlageter MH, Cassinat B, Beaune P, Legendre C, Daly AK. Impact of cytochrome p450 3A5 genetic polymorphism on tacrolimus doses and concentration-to-dose ratio in renal transplant recipients. Transplantation. 2003;76:1233–5. doi: 10.1097/01.TP.0000090753.99170.89. [DOI](https://doi.org/10.1097/01.TP.0000090753.99170.89) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14578760/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplantation&title=Impact%20of%20cytochrome%20p450%203A5%20genetic%20polymorphism%20on%20tacrolimus%20doses%20and%20concentration-to-dose%20ratio%20in%20renal%20transplant%20recipients&author=E%20Thervet&author=D%20Anglicheau&author=B%20King&author=MH%20Schlageter&author=B%20Cassinat&volume=76&publication_year=2003&pages=1233-5&pmid=14578760&doi=10.1097/01.TP.0000090753.99170.89&)
|
| 228 |
+
|
| 229 |
+
16. Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–94. doi: 10.1016/s0169-409x(02)00066-2. [DOI](https://doi.org/10.1016/s0169-409x(02)00066-2) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12406645/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Adv%20Drug%20Deliv%20Rev&title=Genetic%20contribution%20to%20variable%20human%20CYP3A-mediated%20metabolism&author=JK%20Lamba&author=YS%20Lin&author=EG%20Schuetz&author=KE%20Thummel&volume=54&publication_year=2002&pages=1271-94&pmid=12406645&doi=10.1016/s0169-409x(02)00066-2&)
|
| 230 |
+
|
| 231 |
+
17. Park K, Kim YS, Kwon K, Park MS, Lee YJ, Kim KH. A randomized, open-label, two-period cross-over bioavailability study of two oral formualtions of tacrolimus in healthy Korean adults. Clin Ther. 2007;29:154–62. doi: 10.1016/j.clinthera.2007.01.016. [DOI](https://doi.org/10.1016/j.clinthera.2007.01.016) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17379055/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Ther&title=A%20randomized,%20open-label,%20two-period%20cross-over%20bioavailability%20study%20of%20two%20oral%20formualtions%20of%20tacrolimus%20in%20healthy%20Korean%20adults&author=K%20Park&author=YS%20Kim&author=K%20Kwon&author=MS%20Park&author=YJ%20Lee&volume=29&publication_year=2007&pages=154-62&pmid=17379055&doi=10.1016/j.clinthera.2007.01.016&)
|
| 232 |
+
|
| 233 |
+
18. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI](https://doi.org/10.1093/bioinformatics/bth457) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15297300/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Bioinformatics&title=Haploview:%20analysis%20and%20visualization%20of%20LD%20and%20haplotype%20maps&author=JC%20Barrett&author=B%20Fry&author=J%20Maller&author=MJ%20Daly&volume=21&publication_year=2005&pages=263-5&pmid=15297300&doi=10.1093/bioinformatics/bth457&)
|
| 234 |
+
|
| 235 |
+
19. Yu S, Wu L, Jin J, Yan S, Jiang G, Xie H, Zheng S. Influence of CYP3A5 gene polymorphisms of donor rather than recipient to tacrolimus individual dose requirement in liver transplantation. Transplantation. 2006;81:46–51. doi: 10.1097/01.tp.0000188118.34633.bf. [DOI](https://doi.org/10.1097/01.tp.0000188118.34633.bf) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16421475/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Transplantation&title=Influence%20of%20CYP3A5%20gene%20polymorphisms%20of%20donor%20rather%20than%20recipient%20to%20tacrolimus%20individual%20dose%20requirement%20in%20liver%20transplantation&author=S%20Yu&author=L%20Wu&author=J%20Jin&author=S%20Yan&author=G%20Jiang&volume=81&publication_year=2006&pages=46-51&pmid=16421475&doi=10.1097/01.tp.0000188118.34633.bf&)
|
| 236 |
+
|
| 237 |
+
20. Saeki M, Saito Y, Nakamura T, Murayama N, Kim SR, Ozawa S, Komamura K, Ueno K, Kamakura S, Nakajima T, Saito H, Kitamura Y, Kamatani N, Sawada J. Single nucleotide polymorphisms and haplotype frequencies of CYP3A5 in a Japanese population. Hum Mutat. 2003;21:653. doi: 10.1002/humu.9147. [DOI](https://doi.org/10.1002/humu.9147) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14961555/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Hum%20Mutat&title=Single%20nucleotide%20polymorphisms%20and%20haplotype%20frequencies%20of%20CYP3A5%20in%20a%20Japanese%20population&author=M%20Saeki&author=Y%20Saito&author=T%20Nakamura&author=N%20Murayama&author=SR%20Kim&volume=21&publication_year=2003&pages=653&pmid=14961555&doi=10.1002/humu.9147&)
|
| 238 |
+
|
| 239 |
+
21. Floyd MD, Gervasini G, Masica AL, Mayo G, George AL, Jr, Bhat K, Kim RB, Wilkinson GR. Genotype–phenotype associations for common CYP3A4 and CYP3A5 variants in the basal and induced metabolism of midazolam in European- and African-American men and women. Pharmacogenetics. 2003;13:595–606. doi: 10.1097/00008571-200310000-00003. [DOI](https://doi.org/10.1097/00008571-200310000-00003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14515058/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Genotype%E2%80%93phenotype%20associations%20for%20common%20CYP3A4%20and%20CYP3A5%20variants%20in%20the%20basal%20and%20induced%20metabolism%20of%20midazolam%20in%20European-%20and%20African-American%20men%20and%20women&author=MD%20Floyd&author=G%20Gervasini&author=AL%20Masica&author=G%20Mayo&author=AL%20George&volume=13&publication_year=2003&pages=595-606&pmid=14515058&doi=10.1097/00008571-200310000-00003&)
|
| 240 |
+
|
| 241 |
+
22. Haufroid V, Mourad M, Van Kerckhove V, Wawrzyniak J, De Meyer M, Eddour DC, Malaise J, Lison D, Squifflet JP, Wallemacq P. The effect of CYP3A5 and MDR1 (ABCB1) polymorphisms on cyclosporine and tacrolimus dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenetics. 2004;14:147–54. doi: 10.1097/00008571-200403000-00002. [DOI](https://doi.org/10.1097/00008571-200403000-00002) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15167702/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=The%20effect%20of%20CYP3A5%20and%20MDR1%20(ABCB1)%20polymorphisms%20on%20cyclosporine%20and%20tacrolimus%20dose%20requirements%20and%20trough%20blood%20levels%20in%20stable%20renal%20transplant%20patients&author=V%20Haufroid&author=M%20Mourad&author=V%20Van%20Kerckhove&author=J%20Wawrzyniak&author=M%20De%20Meyer&volume=14&publication_year=2004&pages=147-54&pmid=15167702&doi=10.1097/00008571-200403000-00002&)
|
| 242 |
+
|
| 243 |
+
23. Baker SD, van Schaik RH, Rivory LP, Ten Tije AJ, Dinh K, Graveland WJ, Schenk PW, Charles KA, Clarke SJ, Carducci MA, McGuire WP, Dawkins F, Gelderblom H, Verweij J, Sparreboom A. Factors affecting cytochrome P-450 3A activity in cancer patients. Clin Cancer Res. 2004;10:8341–50. doi: 10.1158/1078-0432.CCR-04-1371. [DOI](https://doi.org/10.1158/1078-0432.CCR-04-1371) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15623611/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Cancer%20Res&title=Factors%20affecting%20cytochrome%20P-450%203A%20activity%20in%20cancer%20patients&author=SD%20Baker&author=RH%20van%20Schaik&author=LP%20Rivory&author=AJ%20Ten%20Tije&author=K%20Dinh&volume=10&publication_year=2004&pages=8341-50&pmid=15623611&doi=10.1158/1078-0432.CCR-04-1371&)
|
| 244 |
+
|
| 245 |
+
24. Mancinelli LM, Frassetto L, Floren LC, Dressler D, Carrier S, Bekersky I, Benet LZ, Christians U. The pharmacokinetics and metabolic disposition of tacrolimus: a comparison across ethnic groups. Clin Pharmacol Ther. 2001;69:24–31. doi: 10.1067/mcp.2001.113183. [DOI](https://doi.org/10.1067/mcp.2001.113183) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11180035/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=The%20pharmacokinetics%20and%20metabolic%20disposition%20of%20tacrolimus:%20a%20comparison%20across%20ethnic%20groups&author=LM%20Mancinelli&author=L%20Frassetto&author=LC%20Floren&author=D%20Dressler&author=S%20Carrier&volume=69&publication_year=2001&pages=24-31&pmid=11180035&doi=10.1067/mcp.2001.113183&)
|
| 246 |
+
|
| 247 |
+
25. Dirks NL, Huth B, Yates CR, Meibohm B. Pharmacokinetics of immunosuppressants: a perspective on ethnic differences. Int J Clin Pharmacol Ther. 2004;42:701–18. doi: 10.5414/cpp42701. [DOI](https://doi.org/10.5414/cpp42701) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15624287/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Int%20J%20Clin%20Pharmacol%20Ther&title=Pharmacokinetics%20of%20immunosuppressants:%20a%20perspective%20on%20ethnic%20differences&author=NL%20Dirks&author=B%20Huth&author=CR%20Yates&author=B%20Meibohm&volume=42&publication_year=2004&pages=701-18&pmid=15624287&doi=10.5414/cpp42701&)
|
| 248 |
+
|
| 249 |
+
26. Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet. 2004;75:1059–69. doi: 10.1086/426406. [DOI](https://doi.org/10.1086/426406) | [PMC free article](/articles/PMC1182141/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15492926/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Hum%20Genet&title=CYP3A%20variation%20and%20the%20evolution%20of%20salt-sensitivity%20variants&author=EE%20Thompson&author=H%20Kuttab-Boulos&author=D%20Witonsky&author=L%20Yang&author=BA%20Roe&volume=75&publication_year=2004&pages=1059-69&pmid=15492926&doi=10.1086/426406&)
|
| 250 |
+
|
| 251 |
+
27. Sakaeda T, Nakamura T, Okumura K. Pharmacogenetics of MDR1 and its impact on the pharmacokinetics and pharmacodynamics of drugs. Pharmacogenomics. 2003;4:397–410. doi: 10.1517/phgs.4.4.397.22747. [DOI](https://doi.org/10.1517/phgs.4.4.397.22747) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12831320/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=Pharmacogenetics%20of%20MDR1%20and%20its%20impact%20on%20the%20pharmacokinetics%20and%20pharmacodynamics%20of%20drugs&author=T%20Sakaeda&author=T%20Nakamura&author=K%20Okumura&volume=4&publication_year=2003&pages=397-410&pmid=12831320&doi=10.1517/phgs.4.4.397.22747&)
|
| 252 |
+
|
| 253 |
+
28. Johne A, Kopke K, Gerloff T, Mai I, Rietbrock S, Meisel C, Hoffmeyer S, Kerb R, Fromm MF, Brinkmann U, Eichelbaum M, Brockmoller J, Cascorbi I, Roots I. Modulation of steady-state kinetics of digoxin by haplotypes of the P-glycoprotein MDR1 gene. Clin Pharmacol Ther. 2002;72:584–94. doi: 10.1067/mcp.2002.129196. [DOI](https://doi.org/10.1067/mcp.2002.129196) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12426522/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Modulation%20of%20steady-state%20kinetics%20of%20digoxin%20by%20haplotypes%20of%20the%20P-glycoprotein%20MDR1%20gene&author=A%20Johne&author=K%20Kopke&author=T%20Gerloff&author=I%20Mai&author=S%20Rietbrock&volume=72&publication_year=2002&pages=584-94&pmid=12426522&doi=10.1067/mcp.2002.129196&)
|
| 254 |
+
|
| 255 |
+
29. Woodahl EL, Ho RJ. The role of MDR1 genetic polymorphisms in interindividual variability in P-glycoprotein expression and function. Curr Drug Metab. 2004;5:11–9. doi: 10.2174/1389200043489108. [DOI](https://doi.org/10.2174/1389200043489108) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14965248/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Curr%20Drug%20Metab&title=The%20role%20of%20MDR1%20genetic%20polymorphisms%20in%20interindividual%20variability%20in%20P-glycoprotein%20expression%20and%20function&author=EL%20Woodahl&author=RJ%20Ho&volume=5&publication_year=2004&pages=11-9&pmid=14965248&doi=10.2174/1389200043489108&)
|
test/texts/PMC1975838.md
ADDED
|
@@ -0,0 +1,241 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Genetic-based dosing in orthopedic patients beginning warfarin therapy
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Eric A Millican, Petra A Lenzini, Paul E Milligan, Leonard Grosso, Charles Eby, Elena Deych, Gloria Grice, John C Clohisy, Robert L Barrack, R Stephen J Burnett, Deepak Voora, Susan Gatchel, Amy Tiemeier, Brian F Gage
|
| 5 |
+
**Journal:** Blood
|
| 6 |
+
**Date:** 2007 Mar 26
|
| 7 |
+
**DOI:** [10.1182/blood-2007-01-069609](https://doi.org/10.1182/blood-2007-01-069609)
|
| 8 |
+
**PMID:** 17387222
|
| 9 |
+
**PMCID:** PMC1975838
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1975838/
|
| 11 |
+
|
| 12 |
+
## Abstract
|
| 13 |
+
|
| 14 |
+
High variability in drug response and a narrow therapeutic index complicate warfarin therapy initiation. No existing algorithm provides recommendations on refining the initial warfarin dose based on genetic variables, clinical data, and international normalized ratio (INR) values. Our goal was to develop such an algorithm. We studied 92 patients undergoing primary or revision total hip or knee replacement. From each patient we collected a blood sample, clinical variables, current medications, and preoperative and postoperative laboratory values. We genotyped for polymorphisms in the cytochrome P450 (CYP) 2C9 and vitamin K epoxide reductase (VKORC1) genes. Using stepwise regression, we developed a model for refining the warfarin dose after the third warfarin dose. The algorithm explained four fifths of the variability in therapeutic dose (R2adj of 79%). Significant (P > .05) predictors were INR value after 3 doses (47% reduction per 0.25-unit rise), first warfarin dose (+7% per 1 mg), CYP2C9*3 and CYP2C9*2 genotype (−38% and −17% per allele), estimated blood loss (interacting with INR3), smoking status (+20% in current smokers), and VKORC1 (−11% per copy of haplotype A). If validated, this model should provide a safer, more effective process for initiating warfarin therapy.
|
| 15 |
+
|
| 16 |
+
## Introduction
|
| 17 |
+
|
| 18 |
+
Warfarin sodium is characterized by a narrow therapeutic range (eg, an international normalized ratio [INR]) of 2.0–3.0), a marked interindividual variation in dosing requirements, and an increased risk of adverse events when the dose is too high or low.^1,2^[1](#B1)1,[2](#B2)2 To minimize the high incidence of such events,^3–5^[3](#B3)3–[5](#B5)5 particularly during the first few weeks of initiating therapy,^1,6^[1](#B1)1,[6](#B6)6 most guidelines recommend prescribing warfarin at or near the anticipated maintenance dose and then adjusting the dose by trial and error.^1,7,8^[1](#B1)1,[7](#B7)7,[8](#B8)8 While algorithms for predicting this maintenance dose a priori have improved,^9–16^[9](#B9)9–[16](#B16)16 there remains little guidance on how this starting dose should be adjusted a posteriori based on the subsequent INR values. We hypothesized that use of genetic markers could help optimize these dose refinements.
|
| 19 |
+
|
| 20 |
+
Two common single nucleotide polymorphisms (SNPs) in the cytochrome P450 *(CYP)*(CYP) 2C9 system are associated with impaired metabolism of warfarin,^3–6,11,17^[3](#B3)3–[6](#B6)6,[11](#B11)11,[17](#B17)17 while SNPs in the gene for vitamin K epoxide reductase complex 1 *(VKORC1)*(VKORC1) correlate with warfarin sensitivity and resistance.^2,18–20^[2](#B2)2,[18](#B18)18–[20](#B20)20 No prior study has examined the impact of these SNPs on warfarin-dose adjustments. Given the current knowledge about these markers, we hypothesize that for a given INR, a patient who is a slow metabolizer of warfarin may need a more cautious adjustment in their dose than a similar patient who is a normal metabolizer. Failure to tailor dose refinements during warfarin induction in poor metabolizers may have contributed to the 3-fold increased risk of (laboratory or clinical) adverse events among poor metabolizers in our initial prospective study of pharmacogenetic-based warfarin therapy.^4^[4](#B4)4
|
| 21 |
+
|
| 22 |
+
The purpose of this study was to develop a dose-refinement nomogram to guide clinicians in adjusting warfarin doses. This nomogram would be similar to prior algorithms,^21,22^[21](#B21)21,[22](#B22)22 but will have 2 advantages: (1) it will allow for, but not require, a first dose that is tailored to clinical and/or genetic factors and (2) it will incorporate genetics and clinical factors that are independent predictors of how much the dose should be refined.^1,11^[1](#B1)1,[11](#B11)11 If successful, the proposed warfarin nomogram would simplify and standardize warfarin initiation.
|
| 23 |
+
|
| 24 |
+
## Patients, materials, and methods
|
| 25 |
+
|
| 26 |
+
The study was a retrospective analysis of 2 cohorts of orthopedic surgery patients who had participated in 2 prospective studies of pharmacogenetic-based warfarin therapy. The Human Research Protection Office at Washington University Medical Center approved these studies.
|
| 27 |
+
|
| 28 |
+
### Patients
|
| 29 |
+
|
| 30 |
+
For patients in both cohorts, we offered participation if they were scheduled for primary or revision total knee or hip arthroplasty at Washington University Medical Center and if they were 18 years or older. We excluded patients who had previously taken warfarin or who had contraindications to warfarin treatment. To allow time for genotyping, we also excluded patients scheduled for surgery fewer than 7 days following referral to our anticoagulation service. As previously described,^4^[4](#B4)4 we recruited the first cohort (n = 46) between August 2003 and March 2004. We recruited the second cohort (n = 72) between February 2006 and July 2006. We excluded patients who stopped their warfarin therapy prior to becoming therapeutic (n = 26), leaving a final cohort of 92 participants. All patients provided written informed consent in accordance with the Declaration of Helsinki.
|
| 31 |
+
|
| 32 |
+
### Data collection
|
| 33 |
+
|
| 34 |
+
We prospectively collected the following information: age, sex, race, ethnicity, height, weight, smoking status, current medications and history of hypertension, high blood pressure, cancer, gastrointestinal bleeding, and liver disease. We defined liver disease as cirrhosis, chronic hepatitis, or a 2-fold elevation in aspartate or alanine transaminase in the last 24 months. We estimated body surface area (BSA) from the classic formula.^23^[23](#B23)23 We used electronic records to obtain baseline INR values, preoperative and postoperative levels of creatinine, hematocrit, and platelets, estimated blood loss during surgery (EBL), and hemovac drainage following surgery. When EBL was recorded as “minimal,” we estimated a value of 50 mL.
|
| 35 |
+
|
| 36 |
+
### Genotyping
|
| 37 |
+
|
| 38 |
+
We collected a 5-mL anticoagulated blood sample from each patient, centrifuged it, and isolated genomic DNA from buffy coat. We genotyped each patient for SNPs in the *CYP2C9*CYP2C9 and *VKORC1*VKORC1 genes (listed with dbSNP reference SNP identifiers, as follows). Each sample was genotyped for the *CYP2C9*CYP2C9**2*2 (rs1799853) and *CYP2C9*CYP2C9**3*3 (rs1057910) alleles using the restriction fragment length polymorphism technique described previously.^11^[11](#B11)11 We tested each sample for *VKORC1*VKORC1 SNPs 3673G>A (rs9923231; also designated −1639 G > A) and 6853G>C (rs8050894) using Pyrosequencing^18^[18](#B18)18 or 50 cycles of asymmetric polymerase chain reaction (PCR) using DNA isolated from 200 μL peripheral blood (0.5% of isolated DNA per genotype) with fluorescent minor groove binding probes (Nanogen, San Diego, CA; forward primer final concentration = 100 nM AA*AGA*CTCCTGTTA*GTTACCTC; reverse primer final concentration = 2 μM CCA*CTCCATGCAATCTTGGTGA; probe 1 final concentration = 400 nM Minor Groove Binder Eclipse Dark Quencher-CGCTCCGTGATGA-6-carboxyfluorescein; probe 2 final concentration = 400 nM Minor Groove Binder Eclipse Dark Quencher-ACGCTCGGTGA*T-2′,4′,1,4,-tetrachlorofluorescein, where each G* or A* is a modified proprietary base created by Nanogen that reduces G-G self association or improve stability of A-T bonding, respectively^24^[24](#B24)24) and melt curve analysis on a SmartCycler (Cepheid, Sunnyvale, CA). While blinded to clinical variables, we performed and interpreted genotyping in a clinical DNA diagnostic laboratory following CLIA guidelines.
|
| 39 |
+
|
| 40 |
+
### Treatment
|
| 41 |
+
|
| 42 |
+
For both cohorts, the first warfarin dose was taken approximately 24 hours preoperatively and was tailored to clinical factors and some genetic factors. In the first cohort, we prescribed the first 2 warfarin doses based on *CYP2C9*CYP2C9 genotype and clinical factors, but rounded the first dose up to the nearest 5 mg, as previously described.^4^[4](#B4)4 In the second cohort, we prescribed the first dose (rounded to the nearest 0.5 or 1 mg) based on *VKORC1*VKORC1 genotype and clinical factors using a pharmacogenetic algorithm that estimated initial dose ([www.WarfarinDosing.org](http://www.WarfarinDosing.org)www.WarfarinDosing.org). In this cohort, we reduced the warfarin dose in patients with *CYP2C9*CYP2C9 variants beginning with the second dose. For inpatients, warfarin was taken at 5:00 pmpm and laboratory tests were drawn after midnight. After discharge, we recommended that warfarin be taken in the evening and commonly ordered morning INR draws by the home care nurses. Outpatients had INR tests drawn on Mondays and Thursdays and as clinically necessary. For most patients, we prescribed warfarin for 5 to 6 weeks if they were in the first cohort and for 4 to 5 weeks if in the second.
|
| 43 |
+
|
| 44 |
+
### End point
|
| 45 |
+
|
| 46 |
+
The end point for this study was the therapeutic warfarin dose. We defined therapeutic dose as a dose that gave an INR in the target therapeutic range after 7 consecutive days or, where that never occurred (n = 7), after 6 consecutive days. In general, the target therapeutic range was 2 to 3 in cohort 1 and 1.7 to 2.7 in cohort 2.
|
| 47 |
+
|
| 48 |
+
### Statistical analysis
|
| 49 |
+
|
| 50 |
+
To select candidate variables for the multiple regression model, we tested all variables for collinearity. Noncollinear variables and biologically plausible interaction terms were then tested for association with logarithmically transformed therapeutic warfarin dose using the stepwise regression procedure (PROC REG) in SAS (Version 9.0 for Windows; SAS Institute; Cary, NC). The natural logarithm (ln) was used for all logarithmic transforms.
|
| 51 |
+
|
| 52 |
+
We coded *CYP2C9*CYP2C9 SNPs as 0 (if wild type), 1 (heterozygous), and 2 (homozygous) to model additive allelic effects on warfarin dose. We coded 2 copies of the warfarin-sensitive *VKORC1*VKORC1 haplotype A as 2, 1 copy as 1, and 2 copies of *VKORC1*VKORC1 B haplotype as 0. Where haplotype-distinguishing SNP 3673G>A was missing (n = 4), we used the genotype at position 6853G>C on the basis of high pairwise linkage disequilibrium (D′ = .97) and our prior work with this SNP.^18^[18](#B18)18 Dummy variables were used to code for demographic factors (sex, race, ethnicity, and cohort), clinical variables (history of liver disease, hypertension, cancer, or gastrointestinal bleeding), specific habits (current smoking), and individual medications. We used continuous variables for the other factors (age, BSA, creatinine, preoperative INR, INR after 3 warfarin doses [INR_3_3], preoperative warfarin dose, hemovac drainage, estimated blood loss [EBL], platelet count, and change in hematocrit). We logarithmically transformed EBL and INR_3_3 because of their positively skewed distributions. Where EBL was missing from the electronic record (n = 19), we imputed it as a decreasing linear function of the ratio of postoperative (day 2) platelet count to preoperative platelet count. We also tested for INR_3_3 interactions with *CYP2C9*CYP2C9*2, *CYP2C9*CYP2C9*3, and EBL.
|
| 53 |
+
|
| 54 |
+
While we set *P*P > .3 thresholds for entry and exit into the stepwise regression, we considered statistically significant only those variables with *P*P > .05 and a type II sum of squares contribution greater than .5. To prevent overfitting, the number of variables in the model, including the intercept, was constrained to be fewer than or equal to 10. This number included a variable representing the second warfarin dose, a variable representing an individual's target INR, and a variable representing *VKORC1*VKORC1, all of which we forced into the regression model for clinical reasons. Because R^2^2 can be inflated by the number of independent variables, we also present adjusted R^2^2 (R^2^2_adj_adj) for the model. In the final model, variance inflation factors were less than 3 for noninteracting terms, and less than 8 for interactions.
|
| 55 |
+
|
| 56 |
+
The authors had full access to the data and take responsibility for its integrity. All authors have read and agree to the paper as written.
|
| 57 |
+
|
| 58 |
+
## Results
|
| 59 |
+
|
| 60 |
+
The mean age was 58 years, with a range of 21 to 83 years and median of 59 years ([Table 1](#T1)Table 1). The therapeutic daily warfarin dose ranged from 1.36 to 13.75 mg. The arithmetic mean dose was 5.4 mg/day and the geometric mean was 4.9 mg/day. The mean INR_3_3 was 1.7 (SD = 0.5). EBL ranged from 50 mL to 4000 mL with an arithmetic mean of 564 mL and a geometric mean of 374 mL. Two percent and 22% of participants were *CYP2C9*CYP2C9*2 homozygotes and heterozygotes, respectively ([Table 2](#T2)Table 2), and 15% were *CYP2C9*CYP2C9*3 heterozygotes. Individuals with 1 or 2 copies of the *VKORC1*VKORC1 haplotype A, represented 40% and 10% of the sample, respectively.
|
| 61 |
+
|
| 62 |
+
### Table 1.
|
| 63 |
+
|
| 64 |
+
Demographic and clinical factors in the 92 participants reaching therapeutic dose
|
| 65 |
+
|
| 66 |
+
| Variable | Value |
|
| 67 |
+
| -------- | ----- |
|
| 68 |
+
| Age, mean (SD) | 58.2 (15.5) |
|
| 69 |
+
| BSA, m2, mean (SD) | 2 (0.3) |
|
| 70 |
+
| White, no. (%) | 79 (86) |
|
| 71 |
+
| African-American, no. (%) | 13 (14) |
|
| 72 |
+
| Female, no. (%) | 44 (48) |
|
| 73 |
+
| Therapeutic warfarin dose, geometric mean (SD) | 4.9 (2.5) |
|
| 74 |
+
| EBL, geometric mean (SD) | 376 (670) |
|
| 75 |
+
| INR0, geometric mean (SD) | 1.0 (0.1) |
|
| 76 |
+
| INR3, geometric mean (SD) | 1.6 (0.5) |
|
| 77 |
+
| Target INR, mean (SD) | 2.3 (0.2) |
|
| 78 |
+
| Warfarin dose, day before surgery, mean (SD) | 6.5 (2.3) |
|
| 79 |
+
| Warfarin dose, day of surgery, mean (SD) | 4.6 (1.7) |
|
| 80 |
+
| History of liver disease, no. (%) | 2 (2) |
|
| 81 |
+
| Smokes, no. (%) | 17 (18) |
|
| 82 |
+
| Takes amiodarone, no. (%) | 1 (1) |
|
| 83 |
+
| Takes simvastatin, no. (%) | 10 (11) |
|
| 84 |
+
| Takes fluvastatin, no. (%) | 7 (8) |
|
| 85 |
+
### Table 2.
|
| 86 |
+
|
| 87 |
+
Genotype-based frequencies and therapeutic doses
|
| 88 |
+
|
| 89 |
+
| SNP alleles | No. (%) | Mean therapeutic dose (SD), mg/day |
|
| 90 |
+
| ----------- | ------- | ---------------------------------- |
|
| 91 |
+
| CYP2C9, WT / WT | 58 (63) | 6.1 (2.5) |
|
| 92 |
+
| CYP2C9*2 / WT | 17 (18) | 5.3 (2.3) |
|
| 93 |
+
| CYP2C9*2 /CYP2C9*2 | 2 (2) | 4.3 (2.4) |
|
| 94 |
+
| CYP2C9*3 / WT | 11 (12) | 3.3 (1.0) |
|
| 95 |
+
| CYP2C9*2 / CYP2C9*3 | 4 (4) | 2.6 (0.8) |
|
| 96 |
+
| CYP2C9*3 / CYP2C9*3 | 0 | — |
|
| 97 |
+
| VKORC1 | | |
|
| 98 |
+
| VKORC1 B / B | 45 (49) | 6.6 (2.5) |
|
| 99 |
+
| VKORC1 A / B | 37 (40) | 4.5 (2.0) |
|
| 100 |
+
| VKORC1 A / A | 10 (11) | 3.8 (1.7) |
|
| 101 |
+
The final model consisted of 8 factors significantly associated with higher therapeutic warfarin dose: lower INR_3_3, higher first dose, fewer *CYP2C9*CYP2C9*2 and *CYP2C9*CYP2C9*3 copies, fewer *VKORC1*VKORC1 haplotype A copies, an increase in the value of the product of EBL and INR_3_3, and current smoking status ([Table 3](#T3)Table 3). Creatinine clearance was not significant in the multivariate model.
|
| 102 |
+
|
| 103 |
+
### Table 3.
|
| 104 |
+
|
| 105 |
+
Multivariate analysis: independent predictors of therapeutic warfarin dose at day 3
|
| 106 |
+
|
| 107 |
+
| Entry into model | Variable | % Change in therapeutic warfarin dose (95% CI)* | R2 after entry, % | P in final model |
|
| 108 |
+
| ---------------- | -------- | ----------------------------------------------- | ----------------- | ---------------- |
|
| 109 |
+
| — | Intercept | — | — | < .001 |
|
| 110 |
+
| 1 | INR3 | −46.5 (−33.3 to −57.1) | 34.4 | < .001 |
|
| 111 |
+
| 2 | 1st warfarin dose, per mg | +7.1 (+4.0 to +10.4) | 54.7 | < .001 |
|
| 112 |
+
| 3 | CYP2C9*3, per allele | −38.1(−29.3 to −45.7) | 69.8 | < .001 |
|
| 113 |
+
| 4 | 2nd warfarin dose, per mg | +3.9 (+0.0 to +8.0) | 73.6 | .051 |
|
| 114 |
+
| 5 | CYP2C9*2, per allele | −17.4 (−8.3 to −25.6) | 75.6 | .001 |
|
| 115 |
+
| 6 | EBL × INR3 | +44.9 (+16.6 to +80.2) | 78.1 | .001 |
|
| 116 |
+
| 7 | Smokes | +20.1 (+6.0 to +36.2) | 79.9 | .005 |
|
| 117 |
+
| 8 | VKORC1 haplotype A, per copy | −10.7 (−2.0 to −18.6) | 81.1 | .018 |
|
| 118 |
+
| 9 | Target INR | +14.6 (−7.9 to +42.5) | 81.5 | .218 |
|
| 119 |
+
After including forced variables, the pharmacogenetic algorithm that best estimated therapeutic dose (mg/day) from data available on the third day after surgery was as follows: Exp[1.0138 − 2.5047 × ln(INR_3_3) + 0.0690 × 1st warfarin dose + 0.0385 × 2nd warfarin dose + 0.2474 × ln(EBL) × ln(INR_3_3) − 0.1912 × CYP2C9*2 − 0.4793 × CYP2C9*3 + 0.1835 × Smokes − 0.1132 × VKORC1 + 0.2724 × TargetINR], where Exp is the exponential function. In total, the pharmacogenetic model explained 81.5% of the variation in therapeutic warfarin dose (R^2^2 = 81.5%; R^2^2_adj_adj = 79.3%) ([Table 3](#T3)Table 3; [Figure 1](#F1)Figure 1). History of liver disease was found nominally significant (*P*P = .067) and was not included in the final model. Removing *VKORC1*VKORC1, however, resulted in an alternative model capable of explaining 81.2% of the variation (R^2^2 = 81.2%; R^2^2_adj_adj = 79.0%) in which history of liver disease was statistically significant (*P*P = .033): EXP[1.2091 − 0.1575 × *CYP2C9*CYP2C9**2*2− 0.4814 × *CYP2C9*CYP2C9**3*3 − 0.3610 × History of Liver Disease + 0.1939 × Smokes + 0.1084 × Target INR − 2.5682 × ln(INR_3_3) + 0.0906 × 1st Warfarin Dose + 0.0405 × 2nd Warfarin dose + 0.2452 × ln(INR_3_3) × ln(EBL)].
|
| 120 |
+
|
| 121 |
+
### Figure 1.
|
| 122 |
+
|
| 123 |
+
|
| 124 |
+
|
| 125 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=1975838_zh80130703470001.jpg)
|
| 126 |
+
|
| 127 |
+
Predicted vs therapeutic warfarin dose (mg/day) and line of identity.
|
| 128 |
+
|
| 129 |
+
When the data were analyzed without the 19 patients for which EBL was imputed, the interaction of INR_3_3 and blood loss remained significant (*P*P = .012) with a similar impact on the dose estimation.
|
| 130 |
+
|
| 131 |
+
## Discussion
|
| 132 |
+
|
| 133 |
+
The initiation of warfarin therapy carries a high risk of adverse events, particularly during the first weeks of therapy as the proper dose for each patient is determined.^1,5,25–27^[1](#B1)1,[5](#B5)5,[25](#B25)25–[27](#B27)27 Current dosing algorithms attempt to minimize this risk by prescribing either a standard dose or an estimate of the maintenance dose. For either strategy, the actual maintenance dose is eventually found through a process of trial and error as the starting dose is adjusted according to the INR value—often starting on the third or fourth day of therapy.^9,21,22,28^[9](#B9)9,[21](#B21)21,[22](#B22)22,[28](#B28)28 Our results demonstrate that the INR response is only one of several factors that should be used to estimate the therapeutic dose in orthopedic patients: the accuracy of dose refinements based solely on INR is inherently limited, heightening the chance of excessive or inadequate warfarin dose adjustments.
|
| 134 |
+
|
| 135 |
+
After INR response, the next variable to enter the stepwise regression was the initial warfarin dose, which explained 20.3% of the variability in therapeutic dose. This finding is interesting because 34 (37%) of the included patients were given initial doses that did not take *VKORC1*VKORC1 genotype into account, while the remainder received genetically tailored doses. This observation suggests that our model should be appropriate for orthopedic patients, regardless of whether their genotype is known before the first warfarin dose. It is also important to note that the first dose was adjusted for age, BSA, and amiodarone in both cohorts. This adjustment helps explain the statistical significance of the first dose as well as the absence of these variables elsewhere in our model, given their known correlation with therapeutic dose.^11,29–32^[11](#B11)11,[29](#B29)29–[32](#B32)32
|
| 136 |
+
|
| 137 |
+
Consistent with previous studies,^3–6,11–17^[3](#B3)3–[6](#B6)6,[11](#B11)11–[17](#B17)17 we also found that the *CYP2C9*CYP2C9*2 and *3 alleles were significant predictors of therapeutic dose. The therapeutic dose was 17.4% lower per *2 allele and 38.1% lower per *3 allele ([Table 3](#T3)Table 3). The significance of *VKORC1*VKORC1 is also consistent with recent studies, which suggest an increased sensitivity to warfarin per copy of haplotype A.^2,12–16,19,20^[2](#B2)2,[12](#B12)12–[16](#B16)16,[19](#B19)19,[20](#B20)20 However, an alternative model that did not include *VKORC1*VKORC1 was nearly as predictive as the full model, because much of the predictive power of *VKORC1*VKORC1 genotype was captured in INR_3_3. This raises the interesting question of whether genotyping for *VKORC1*VKORC1 status is a necessary and cost-effective step when initiating warfarin therapy. We did not analyze any of the additional *CYP2C9*CYP2C9 alleles that may be associated with warfarin metabolism. However, a recent review found that of 30 genes possibly related to warfarin action and metabolism, *CYP2C9*CYP2C9 and *VKORC1*VKORC1 are the ones that are clearly important.^16^[16](#B16)16 Thus, the inclusion of additional genes is unlikely to increase the predictive value of the model significantly.
|
| 138 |
+
|
| 139 |
+
Of interest, we found that the interaction between INR_3_3 and EBL during surgery was significantly related to the eventual therapeutic dose, with the dose increasing 44.9% for every 1.5-point increase in the interaction term. This result must be interpreted carefully: rather than affecting the patient's long-term warfarin requirements, we profess that blood loss affects the value and interpretation of INR_3_3. With blood loss during surgery, patients experience an acute loss of clotting factors that are not replenished via transfusions of packed red blood cells. This loss temporarily inflates the INR values commensurate with EBL. Indeed, though our current study focuses on orthopedic patients, this correlation may explain why postoperative patients are particularly sensitive to warfarin after valve-replacement surgery.^33^[33](#B33)33 This observation should be validated because we had to impute EBL from the change in platelets for 19 patients. Nevertheless, the association is unlikely to be spurious, as excluding those with imputed EBL values did not significantly alter the predictive weight of this variable.
|
| 140 |
+
|
| 141 |
+
Two clinically relevant variables were exposed as potential indicators of therapeutic dose. First, we found a significant impact on the therapeutic dose from smoking. The effect of smoking (+ 20.1%) was larger than expected, since hepatic enzyme induction from smoking results in only a 10% increase in warfarin clearance.^29^[29](#B29)29 Directionally, however, this is consistent with other studies that have shown a significant relationship between smoking status and warfarin dose.^15^[15](#B15)15 While nominally significant, the effect of a history of liver disease on therapeutic dose is clinically relevant and is consistent with previous studies linking liver disease to a decrease in warfarin requirements.^34^[34](#B34)34 This impact should be investigated further, as only 2 patients in our sample had liver disease.
|
| 142 |
+
|
| 143 |
+
In addition to requiring a cautious interpretation of the effects of EBL and liver disease variables, this study has other limitations. First, our study population consisted entirely of patients initiating warfarin for deep vein thrombosis prophylaxis following total hip or knee arthroplasty. The ability to generalize our model for other indications is unknown and should be studied in a broad population. In particular, the appropriate starting doses and the ability to safely initiate warfarin without genetic information need to be examined in other patient groups—including nonsurgical populations. Moreover, like any data-driven model, it might reflect peculiarities within our data rather than causal relationships between the variables and the therapeutic dose.^35^[35](#B35)35 In addition, our definition of therapeutic dose was ad hoc as definitions vary among studies.^4,8,11,21,22^[4](#B4)4,[8](#B8)8,[11](#B11)11,[21](#B21)21,[22](#B22)22 Our definition was appropriate for orthopedic patients who tend to use warfarin for a limited timeframe. A final minor limitation was that only 19 patients were taking a statin and only 1 was taking amiodarone, so we had inadequate statistical power to determine how these, and other drugs that interact with warfarin, affected warfarin dose.
|
| 144 |
+
|
| 145 |
+
Despite these limitations, this research has important implications about the timing and effectiveness of pharmacogenetics-based warfarin therapy in orthopedic patients, and potentially additional populations as well. Prior pharmacogenetic dosing algorithms explain 51% to 60% of the variability (R^2^2) in warfarin dose,^2,15,32,36,37^[2](#B2)2,[15](#B15)15,[32](#B32)32,[36](#B36)36,[37](#B37)37 but implicitly expect the genotype to be available rapidly. Same-day genotyping is unlikely to be available in most settings, but may be unnecessary: even in poor metabolizers who carry one copy of the *CYP2C9*CYP2C9*2 or *CYP2C9*CYP2C9*3 alleles, S-warfarin plasma concentrations are subtherapeutic for at least 3 days.^3^[3](#B3)3 Moreover, while INR values rise more rapidly in patients with 2 variant *CYP2C9*CYP2C9 alleles, the rise may be slow enough to avoid excessive INR_3_3 values (ie, INR_3_3 < 3).^38^[38](#B38)38 However, additional study is needed to quantify how very poor warfarin metabolism affects the rate of INR rise. With frequent monitoring of INR values, a patient with extremely poor warfarin metabolism should be identifiable well before he/she reaches a dangerously supratherapeutic INR. Thus, provided the initial warfarin doses are not excessive and the INR is closely monitored, physicians may have a 3-day window between the time that they start warfarin therapy and the time when they need to know genotype. Although the efficacy of this approach should be demonstrated in a prospective cohort of nonorthopedic patients, the principle of a moderate initial dose followed by genetically tailored dose refinement should be broadly applicable. This knowledge, and the approach developed here, should allow for more effective and less expensive pharmacogenetics-based warfarin therapy.
|
| 146 |
+
|
| 147 |
+
The next steps are to make the new pharmacogenetics refinement algorithm accessible to clinicians and to validate it in a diverse population, including patients with nonsurgical indications for warfarin therapy. In pursuit of these goals, we have made an interactive and free version of this refinement algorithm available online at [www.WarfarinDosing.org](http://www.WarfarinDosing.org)www.WarfarinDosing.org. Provided that the variables in [Table 3](#T3)Table 3 are available, the website uses the algorithm developed here. The website can estimate warfarin dose without INR_3_3 and genotype, but the accuracy of these estimates is much less than that of the full model. The website also solicits information about key interacting drugs to provide relevant warnings about warfarin dose (and to quantify the relevance of these drugs in future research). In summary, [www.WarfarinDosing.org](http://www.WarfarinDosing.org)www.WarfarinDosing.org allows clinicians to incorporate pharmacogenetics into their dosing plans—bearing in mind the experimental nature of this algorithm and its unproven applicability to nonorthopedic patients—and allows us to validate this pharmacogenetic approach in a diverse, anonymous population.
|
| 148 |
+
|
| 149 |
+
The results of this study contain a number of interesting possibilities for safe, effective, and efficient warfarin initiation. Incorporating clinical and genetic factors with the INR in determining warfarin dose refinements could substantially improve trial-and-error dose adjustments and reduce the risks of initiating warfarin therapy. Moreover, allowing physicians to initiate warfarin therapy while the genotype is being processed and then incorporating genetic information into a refinement after 3 doses would allow for offsite genotyping, thereby facilitating this approach in any setting. Ultimately, with further validation and refinement, this pharmacogenetic model should yield a streamlined approach to refining the dose and improving the safety and efficiency of warfarin initiation.
|
| 150 |
+
|
| 151 |
+
## Acknowledgment
|
| 152 |
+
|
| 153 |
+
This research was funded by the NIH, R01 HL074724 and T35 HL007815.
|
| 154 |
+
|
| 155 |
+
## Footnotes
|
| 156 |
+
|
| 157 |
+
## Authorship
|
| 158 |
+
|
| 159 |
+
Contribution: C.E. and B.F.G. designed research; E.A.M., P.E.M., C.E., G.G., J.C.C., R.L.B., R.S.J.B., D.V., S.G., A.T., and B.F.G. collected data; E.A.M., P.A.L., L.G., E.D., G.G., and B.F.G. analyzed and interpreted data; P.A.L. and E.D. performed statistical analysis; E.A.M., P.A.L., and B.F.G. drafted the paper.
|
| 160 |
+
|
| 161 |
+
Conflict-of-interest disclosure: The authors declare no competing financial interests.
|
| 162 |
+
|
| 163 |
+
Correspondence: Brian F. Gage, Campus Box 8005, Dept of Medicine, 660 South Euclid, St Louis, MO 63110; e-mail: bgage@im.wustl.edubgage@im.wustl.edu.
|
| 164 |
+
|
| 165 |
+
## References
|
| 166 |
+
|
| 167 |
+
1. Hirsh J, Fuster V, Ansell J, Halperin JL. American Heart Association/American College of Cardiology Foundation guide to warfarin therapy. Circulation. 2003;107:1692–1711. doi: 10.1161/01.CIR.0000063575.17904.4E. [DOI](https://doi.org/10.1161/01.CIR.0000063575.17904.4E) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12668507/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circulation&title=American%20Heart%20Association/American%20College%20of%20Cardiology%20Foundation%20guide%20to%20warfarin%20therapy.&author=J%20Hirsh&author=V%20Fuster&author=J%20Ansell&author=JL%20Halperin&volume=107&publication_year=2003&pages=1692-1711&pmid=12668507&doi=10.1161/01.CIR.0000063575.17904.4E&)
|
| 168 |
+
|
| 169 |
+
2. Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI](https://doi.org/10.1038/sj.tpj.6500313) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15883587/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J&title=Common%20VKORC1%20and%20GGCX%20polymorphisms%20associated%20with%20warfarin%20dose.&author=M%20Wadelius&author=LY%20Chen&author=K%20Downes&volume=5&publication_year=2005&pages=262-270&pmid=15883587&doi=10.1038/sj.tpj.6500313&)
|
| 170 |
+
|
| 171 |
+
3. Linder MW, Looney S, Adams JE, III, et al. Warfarin dose adjustments based on CYP2C9 genetic polymorphisms. J Thromb Thrombolysis. 2002;14:227–232. doi: 10.1023/a:1025052827305. [DOI](https://doi.org/10.1023/a:1025052827305) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12913403/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Thromb%20Thrombolysis&title=Warfarin%20dose%20adjustments%20based%20on%20CYP2C9%20genetic%20polymorphisms.&author=MW%20Linder&author=S%20Looney&author=JE%20Adams&volume=14&publication_year=2002&pages=227-232&pmid=12913403&doi=10.1023/a:1025052827305&)
|
| 172 |
+
|
| 173 |
+
4. Voora D, Eby C, Linder MW, et al. Prospective dosing of warfarin based on cytochrome P-450 2C9 genotype. Thromb Haemost. 2005;93:700–705. doi: 10.1160/TH04-08-0542. [DOI](https://doi.org/10.1160/TH04-08-0542) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15841315/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Thromb%20Haemost&title=Prospective%20dosing%20of%20warfarin%20based%20on%20cytochrome%20P-450%202C9%20genotype.&author=D%20Voora&author=C%20Eby&author=MW%20Linder&volume=93&publication_year=2005&pages=700-705&pmid=15841315&doi=10.1160/TH04-08-0542&)
|
| 174 |
+
|
| 175 |
+
5. Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–1698. doi: 10.1001/jama.287.13.1690. [DOI](https://doi.org/10.1001/jama.287.13.1690) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11926893/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=JAMA&title=Association%20between%20CYP2C9%20genetic%20variants%20and%20anticoagulation-related%20outcomes%20during%20warfarin%20therapy.&author=MK%20Higashi&author=DL%20Veenstra&author=LM%20Kondo&volume=287&publication_year=2002&pages=1690-1698&pmid=11926893&doi=10.1001/jama.287.13.1690&)
|
| 176 |
+
|
| 177 |
+
6. Aithal GP, Day CP, Kesteven PJL, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI](https://doi.org/10.1016/S0140-6736(98)04474-2) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10073515/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Association%20of%20polymorphisms%20in%20the%20cytochrome%20P450%20CYP2C9%20with%20warfarin%20dose%20requirement%20and%20risk%20of%20bleeding%20complications.&author=GP%20Aithal&author=CP%20Day&author=PJL%20Kesteven&author=AK%20Daly&volume=353&publication_year=1999&pages=717-719&pmid=10073515&doi=10.1016/S0140-6736(98)04474-2&)
|
| 178 |
+
|
| 179 |
+
7. Harrison L, Johnston M, Massicotte MP, Crowther M, Moffat K, Hirsh J. Comparison of 5-mg and 10-mg loading doses in initiation of warfarin therapy. Ann Intern Med. 1997;126:133–136. doi: 10.7326/0003-4819-126-2-199701150-00006. [DOI](https://doi.org/10.7326/0003-4819-126-2-199701150-00006) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9005747/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20Intern%20Med&title=Comparison%20of%205-mg%20and%2010-mg%20loading%20doses%20in%20initiation%20of%20warfarin%20therapy.&author=L%20Harrison&author=M%20Johnston&author=MP%20Massicotte&author=M%20Crowther&author=K%20Moffat&volume=126&publication_year=1997&pages=133-136&pmid=9005747&doi=10.7326/0003-4819-126-2-199701150-00006&)
|
| 180 |
+
|
| 181 |
+
8. Crowther MA, Ginsberg JB, Kearon C, et al. A randomized trial comparing 5-mg and 10-mg warfarin loading doses. Arch Intern Med. 1999;159:46–48. doi: 10.1001/archinte.159.1.46. [DOI](https://doi.org/10.1001/archinte.159.1.46) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9892329/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arch%20Intern%20Med&title=A%20randomized%20trial%20comparing%205-mg%20and%2010-mg%20warfarin%20loading%20doses.&author=MA%20Crowther&author=JB%20Ginsberg&author=C%20Kearon&volume=159&publication_year=1999&pages=46-48&pmid=9892329&doi=10.1001/archinte.159.1.46&)
|
| 182 |
+
|
| 183 |
+
9. Gedge J, Orme S, Hampton KK, Channer KS, Hendra TJ. A comparison of a low-dose warfarin induction regimen with the modified Fennerty regimen in elderly inpatients. Age Ageing. 2000;29:31–34. doi: 10.1093/ageing/29.1.31. [DOI](https://doi.org/10.1093/ageing/29.1.31) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10690692/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Age%20Ageing&title=A%20comparison%20of%20a%20low-dose%20warfarin%20induction%20regimen%20with%20the%20modified%20Fennerty%20regimen%20in%20elderly%20inpatients.&author=J%20Gedge&author=S%20Orme&author=KK%20Hampton&author=KS%20Channer&author=TJ%20Hendra&volume=29&publication_year=2000&pages=31-34&pmid=10690692&doi=10.1093/ageing/29.1.31&)
|
| 184 |
+
|
| 185 |
+
10. Pengo V, Biasiolo A, Pegoraro C. A simple scheme to initiate oral anticoagulant treatment in outpatients with nonrheumatic atrial fibrillation. Am J Cardiol. 2001;88:1214–1216. doi: 10.1016/s0002-9149(01)02069-0. [DOI](https://doi.org/10.1016/s0002-9149(01)02069-0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11703979/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Cardiol&title=A%20simple%20scheme%20to%20initiate%20oral%20anticoagulant%20treatment%20in%20outpatients%20with%20nonrheumatic%20atrial%20fibrillation.&author=V%20Pengo&author=A%20Biasiolo&author=C%20Pegoraro&volume=88&publication_year=2001&pages=1214-1216&pmid=11703979&doi=10.1016/s0002-9149(01)02069-0&)
|
| 186 |
+
|
| 187 |
+
11. Gage BF, Eby C, Milligan PE, Banet GA, Duncan JR, McLeod HL. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb Haemost. 2004;91:87–94. doi: 10.1160/TH03-06-0379. [DOI](https://doi.org/10.1160/TH03-06-0379) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14691573/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Thromb%20Haemost&title=Use%20of%20pharmacogenetics%20and%20clinical%20factors%20to%20predict%20the%20maintenance%20dose%20of%20warfarin.&author=BF%20Gage&author=C%20Eby&author=PE%20Milligan&author=GA%20Banet&author=JR%20Duncan&volume=91&publication_year=2004&pages=87-94&pmid=14691573&doi=10.1160/TH03-06-0379&)
|
| 188 |
+
|
| 189 |
+
12. Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI](https://doi.org/10.1182/blood-2005-03-1108) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15947090/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Blood&title=The%20impact%20of%20CYP2C9%20and%20VKORC1%20genetic%20polymorphism%20and%20patient%20characteristics%20upon%20warfarin%20dose%20requirements:%20proposal%20for%20a%20new%20dosing%20regimen.&author=EA%20Sconce&author=TI%20Khan&author=HA%20Wynne&volume=106&publication_year=2005&pages=2329-2333&pmid=15947090&doi=10.1182/blood-2005-03-1108&)
|
| 190 |
+
|
| 191 |
+
13. Takahashi H, Wilkinson GR, Nutescu EA, et al. Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African-Americans. Pharmacogenet Genomics. 2006;16:101–110. doi: 10.1097/01.fpc.0000184955.08453.a8. [DOI](https://doi.org/10.1097/01.fpc.0000184955.08453.a8) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16424822/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenet%20Genomics&title=Different%20contributions%20of%20polymorphisms%20in%20VKORC1%20and%20CYP2C9%20to%20intra-%20and%20inter-population%20differences%20in%20maintenance%20dose%20of%20warfarin%20in%20Japanese,%20Caucasians%20and%20African-Americans.&author=H%20Takahashi&author=GR%20Wilkinson&author=EA%20Nutescu&volume=16&publication_year=2006&pages=101-110&pmid=16424822&doi=10.1097/01.fpc.0000184955.08453.a8&)
|
| 192 |
+
|
| 193 |
+
14. Carlquist JF, Horne BD, Muhlestein JB, et al. Genotypes of the cytochrome p450 isoform, CYP2C9, and the vitamin K epoxide reductase complex subunit 1 conjointly determine stable warfarin dose: a prospective study. J Thromb Thrombolysis. 2006;22:191–197. doi: 10.1007/s11239-006-9030-7. [DOI](https://doi.org/10.1007/s11239-006-9030-7) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17111199/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Thromb%20Thrombolysis&title=Genotypes%20of%20the%20cytochrome%20p450%20isoform,%20CYP2C9,%20and%20the%20vitamin%20K%20epoxide%20reductase%20complex%20subunit%201%20conjointly%20determine%20stable%20warfarin%20dose:%20a%20prospective%20study.&author=JF%20Carlquist&author=BD%20Horne&author=JB%20Muhlestein&volume=22&publication_year=2006&pages=191-197&pmid=17111199&doi=10.1007/s11239-006-9030-7&)
|
| 194 |
+
|
| 195 |
+
15. Aquilante CL, Lobmeyer MT, Langaee TY, Johnson JA. Comparison of cytochrome P450 2C9 genotyping methods and implications for the clinical laboratory. Pharmacotherapy. 2004;24:720–726. doi: 10.1592/phco.24.8.720.36074. [DOI](https://doi.org/10.1592/phco.24.8.720.36074) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15222661/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacotherapy&title=Comparison%20of%20cytochrome%20P450%202C9%20genotyping%20methods%20and%20implications%20for%20the%20clinical%20laboratory.&author=CL%20Aquilante&author=MT%20Lobmeyer&author=TY%20Langaee&author=JA%20Johnson&volume=24&publication_year=2004&pages=720-726&pmid=15222661&doi=10.1592/phco.24.8.720.36074&)
|
| 196 |
+
|
| 197 |
+
16. Wadelius M, Chen LY, Eriksson N, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007;121:23–34. doi: 10.1007/s00439-006-0260-8. [DOI](https://doi.org/10.1007/s00439-006-0260-8) | [PMC free article](/articles/PMC1797064/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17048007/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Hum%20Genet&title=Association%20of%20warfarin%20dose%20with%20genes%20involved%20in%20its%20action%20and%20metabolism.&author=M%20Wadelius&author=LY%20Chen&author=N%20Eriksson&volume=121&publication_year=2007&pages=23-34&pmid=17048007&doi=10.1007/s00439-006-0260-8&)
|
| 198 |
+
|
| 199 |
+
17. Margaglione M, Colaizzo D, D'Andrea G, et al. Genetic modulation of oral anticoagulation with warfarin. Thromb Haemost. 2000;84:775–778. [PubMed](https://pubmed.ncbi.nlm.nih.gov/11127854/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Thromb%20Haemost&title=Genetic%20modulation%20of%20oral%20anticoagulation%20with%20warfarin.&author=M%20Margaglione&author=D%20Colaizzo&author=G%20D'Andrea&volume=84&publication_year=2000&pages=775-778&pmid=11127854&)
|
| 200 |
+
|
| 201 |
+
18. Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI](https://doi.org/10.1056/NEJMoa044503) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15930419/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Effect%20of%20VKORC1%20haplotypes%20on%20transcriptional%20regulation%20and%20warfarin%20dose.&author=MJ%20Rieder&author=AP%20Reiner&author=BF%20Gage&volume=352&publication_year=2005&pages=2285-2293&pmid=15930419&doi=10.1056/NEJMoa044503&)
|
| 202 |
+
|
| 203 |
+
19. D'Andrea G, D'Ambrosio RL, Di Perna P, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105:645–649. doi: 10.1182/blood-2004-06-2111. [DOI](https://doi.org/10.1182/blood-2004-06-2111) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15358623/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Blood&title=A%20polymorphism%20in%20the%20VKORC1%20gene%20is%20associated%20with%20an%20interindividual%20variability%20in%20the%20dose-anticoagulant%20effect%20of%20warfarin.&author=G%20D'Andrea&author=RL%20D'Ambrosio&author=P%20Di%20Perna&volume=105&publication_year=2005&pages=645-649&pmid=15358623&doi=10.1182/blood-2004-06-2111&)
|
| 204 |
+
|
| 205 |
+
20. Yuan HY, Chen JJ, Lee MT, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet. 2005;14:1745–1751. doi: 10.1093/hmg/ddi180. [DOI](https://doi.org/10.1093/hmg/ddi180) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15888487/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Hum%20Mol%20Genet&title=A%20novel%20functional%20VKORC1%20promoter%20polymorphism%20is%20associated%20with%20inter-individual%20and%20inter-ethnic%20differences%20in%20warfarin%20sensitivity.&author=HY%20Yuan&author=JJ%20Chen&author=MT%20Lee&volume=14&publication_year=2005&pages=1745-1751&pmid=15888487&doi=10.1093/hmg/ddi180&)
|
| 206 |
+
|
| 207 |
+
21. Siguret V, Gouin I, Debray M, et al. Initiation of warfarin therapy in elderly medical inpatients: a safe and accurate regimen. Am J Med. 2005;118:137–142. doi: 10.1016/j.amjmed.2004.07.053. [DOI](https://doi.org/10.1016/j.amjmed.2004.07.053) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15694897/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Med&title=Initiation%20of%20warfarin%20therapy%20in%20elderly%20medical%20inpatients:%20a%20safe%20and%20accurate%20regimen.&author=V%20Siguret&author=I%20Gouin&author=M%20Debray&volume=118&publication_year=2005&pages=137-142&pmid=15694897&doi=10.1016/j.amjmed.2004.07.053&)
|
| 208 |
+
|
| 209 |
+
22. Fennerty A, Dolben J, Thomas P, et al. Flexible induction dose regimen for warfarin and prediction of maintenance dose. Br Med J (Clin Res Ed) 1984;288:1268–1270. doi: 10.1136/bmj.288.6426.1268. [DOI](https://doi.org/10.1136/bmj.288.6426.1268) | [PMC free article](/articles/PMC1441080/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/6424820/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20Med%20J%20(Clin%20Res%20Ed)&title=Flexible%20induction%20dose%20regimen%20for%20warfarin%20and%20prediction%20of%20maintenance%20dose.&author=A%20Fennerty&author=J%20Dolben&author=P%20Thomas&volume=288&publication_year=1984&pages=1268-1270&pmid=6424820&doi=10.1136/bmj.288.6426.1268&)
|
| 210 |
+
|
| 211 |
+
23. DuBois D, DuBois E. Clinical Calorimetry; a formula to estimate the approximate surface area if height and weight be known. Arch Int med. 1916;17:863–871. [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Arch%20Int%20med&title=Clinical%20Calorimetry;%20a%20formula%20to%20estimate%20the%20approximate%20surface%20area%20if%20height%20and%20weight%20be%20known.&author=D%20DuBois&author=E%20DuBois&volume=17&publication_year=1916&pages=863-871&)
|
| 212 |
+
|
| 213 |
+
24. Belousov YS, Welch RA, Sanders S, et al. Single nucleotide polymorphism genotyping by two colour melting curve analysis using the MGB Eclipse Probe System in challenging sequence environment. Hum Genomics. 2004;1:209–217. doi: 10.1186/1479-7364-1-3-209. [DOI](https://doi.org/10.1186/1479-7364-1-3-209) | [PMC free article](/articles/PMC3525082/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15588480/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Hum%20Genomics&title=Single%20nucleotide%20polymorphism%20genotyping%20by%20two%20colour%20melting%20curve%20analysis%20using%20the%20MGB%20Eclipse%20Probe%20System%20in%20challenging%20sequence%20environment.&author=YS%20Belousov&author=RA%20Welch&author=S%20Sanders&volume=1&publication_year=2004&pages=209-217&pmid=15588480&doi=10.1186/1479-7364-1-3-209&)
|
| 214 |
+
|
| 215 |
+
25. White RH, Beyth RJ, Zhou H, Romano PS. Major bleeding after hospitalization for deep-venous thrombosis. Am J Med. 1999;107:414–424. doi: 10.1016/s0002-9343(99)00267-3. [DOI](https://doi.org/10.1016/s0002-9343(99)00267-3) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10569295/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Med&title=Major%20bleeding%20after%20hospitalization%20for%20deep-venous%20thrombosis.&author=RH%20White&author=RJ%20Beyth&author=H%20Zhou&author=PS%20Romano&volume=107&publication_year=1999&pages=414-424&pmid=10569295&doi=10.1016/s0002-9343(99)00267-3&)
|
| 216 |
+
|
| 217 |
+
26. Beyth RJ, Quinn L, Landefeld CS. A Multicomponent Intervention To Prevent Major Bleeding Complications in Older Patients Receiving Warfarin: a Randomized, Controlled Trial. Ann Intern Med. 2000;133:687–695. doi: 10.7326/0003-4819-133-9-200011070-00010. [DOI](https://doi.org/10.7326/0003-4819-133-9-200011070-00010) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11074901/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20Intern%20Med&title=A%20Multicomponent%20Intervention%20To%20Prevent%20Major%20Bleeding%20Complications%20in%20Older%20Patients%20Receiving%20Warfarin:%20a%20Randomized,%20Controlled%20Trial.&author=RJ%20Beyth&author=L%20Quinn&author=CS%20Landefeld&volume=133&publication_year=2000&pages=687-695&pmid=11074901&doi=10.7326/0003-4819-133-9-200011070-00010&)
|
| 218 |
+
|
| 219 |
+
27. Ezekowitz MD, James KE, Radford MJ, Rickles FR, Redmond N. Initiating and maintaining patients on warfarin anticoagulation: the importance of monitoring. J Cardiovasc Pharmacol Ther. 1999;4:3–8. doi: 10.1177/107424849900400102. [DOI](https://doi.org/10.1177/107424849900400102) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10684518/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Cardiovasc%20Pharmacol%20Ther&title=Initiating%20and%20maintaining%20patients%20on%20warfarin%20anticoagulation:%20the%20importance%20of%20monitoring.&author=MD%20Ezekowitz&author=KE%20James&author=MJ%20Radford&author=FR%20Rickles&author=N%20Redmond&volume=4&publication_year=1999&pages=3-8&pmid=10684518&doi=10.1177/107424849900400102&)
|
| 220 |
+
|
| 221 |
+
28. Kovacs MJ, Rodger M, Anderson DR, et al. Comparison of 10-mg and 5-mg warfarin initiation nomograms together with low-molecular-weight heparin for outpatient treatment of acute venous thromboembolism: a randomized, double-blind, controlled trial. Ann Intern Med. 2003;138:714–719. doi: 10.7326/0003-4819-138-9-200305060-00007. [DOI](https://doi.org/10.7326/0003-4819-138-9-200305060-00007) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12729425/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20Intern%20Med&title=Comparison%20of%2010-mg%20and%205-mg%20warfarin%20initiation%20nomograms%20together%20with%20low-molecular-weight%20heparin%20for%20outpatient%20treatment%20of%20acute%20venous%20thromboembolism:%20a%20randomized,%20double-blind,%20controlled%20trial.&author=MJ%20Kovacs&author=M%20Rodger&author=DR%20Anderson&volume=138&publication_year=2003&pages=714-719&pmid=12729425&doi=10.7326/0003-4819-138-9-200305060-00007&)
|
| 222 |
+
|
| 223 |
+
29. Mungall DR, Ludden TM, Marshall J, Hawkins DW, Talbert RL, Crawford MH. Population kinetics of racemic warfarin. J Pharmacokinetics Biopharmaceutics. 1985;13:213–227. doi: 10.1007/BF01065653. [DOI](https://doi.org/10.1007/BF01065653) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/3841364/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharmacokinetics%20Biopharmaceutics&title=Population%20kinetics%20of%20racemic%20warfarin.&author=DR%20Mungall&author=TM%20Ludden&author=J%20Marshall&author=DW%20Hawkins&author=RL%20Talbert&volume=13&publication_year=1985&pages=213-227&pmid=3841364&doi=10.1007/BF01065653&)
|
| 224 |
+
|
| 225 |
+
30. Wynne H, Cope L, Kelly P, Whittingham T, Edwards C, Kamali F. The influence of age, liver size and enantiomer concentrations on warfarin requirements. Br J Clin Pharmacol. 1995;40:203–207. [PMC free article](/articles/PMC1365098/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8527280/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=The%20influence%20of%20age,%20liver%20size%20and%20enantiomer%20concentrations%20on%20warfarin%20requirements.&author=H%20Wynne&author=L%20Cope&author=P%20Kelly&author=T%20Whittingham&author=C%20Edwards&volume=40&publication_year=1995&pages=203-207&pmid=8527280&)
|
| 226 |
+
|
| 227 |
+
31. Gurwitz J, Avorn J, Ross-Degnan D, Choodnovskiy I, Ansell Aging and the anticoagulant response to warfarin therapy. Ann Intern Med. 1992;116:901–904. doi: 10.7326/0003-4819-116-11-901. [DOI](https://doi.org/10.7326/0003-4819-116-11-901) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1580446/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20Intern%20Med&title=Aging%20and%20the%20anticoagulant%20response%20to%20warfarin%20therapy.&author=J%20Gurwitz&author=J%20Avorn&author=D%20Ross-Degnan&author=I%20Choodnovskiy&author=%20Ansell&volume=116&publication_year=1992&pages=901-904&pmid=1580446&doi=10.7326/0003-4819-116-11-901&)
|
| 228 |
+
|
| 229 |
+
32. Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI](https://doi.org/10.1182/blood-2005-03-1108) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15947090/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Blood&title=The%20impact%20of%20CYP2C9%20and%20VKORC1%20genetic%20polymorphism%20and%20patient%20characteristics%20upon%20warfarin%20dose%20requirements:%20proposal%20for%20a%20new%20dosing%20regimen.&author=EA%20Sconce&author=TI%20Khan&author=HA%20Wynne&volume=106&publication_year=2005&pages=2329-2333&pmid=15947090&doi=10.1182/blood-2005-03-1108&)
|
| 230 |
+
|
| 231 |
+
33. Rahman M, Binesmael TM, Payne N, Butchart EG. Increased sensitivity to warfarin after heart valve replacement. Ann Pharmacother. 2006;40:397–401. doi: 10.1345/aph.1G407. [DOI](https://doi.org/10.1345/aph.1G407) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16507614/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20Pharmacother&title=Increased%20sensitivity%20to%20warfarin%20after%20heart%20valve%20replacement.&author=M%20Rahman&author=TM%20Binesmael&author=N%20Payne&author=EG%20Butchart&volume=40&publication_year=2006&pages=397-401&pmid=16507614&doi=10.1345/aph.1G407&)
|
| 232 |
+
|
| 233 |
+
34. Fergusson RJ, Eade OE, Logie AW, Gaddie J. A flexible loading dose schedule for warfarin therapy. Scott Med J. 1987;32:169–171. doi: 10.1177/003693308703200604. [DOI](https://doi.org/10.1177/003693308703200604) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/3329767/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Scott%20Med%20J&title=A%20flexible%20loading%20dose%20schedule%20for%20warfarin%20therapy.&author=RJ%20Fergusson&author=OE%20Eade&author=AW%20Logie&author=J%20Gaddie&volume=32&publication_year=1987&pages=169-171&pmid=3329767&doi=10.1177/003693308703200604&)
|
| 234 |
+
|
| 235 |
+
35. Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI](https://doi.org/10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8668867/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Stat%20Med&title=Multivariable%20prognostic%20models:%20issues%20in%20developing%20models,%20evaluating%20assumptions%20and%20adequacy,%20and%20measuring%20and%20reducing%20errors.&author=FE%20Harrell&author=KL%20Lee&author=DB%20Mark&volume=15&publication_year=1996&pages=361-387&pmid=8668867&doi=10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4&)
|
| 236 |
+
|
| 237 |
+
36. Takahashi H, Wilkinson GR, Nutescu EA, et al. Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African-Americans. Pharmacogenet Genomics. 2006;16:101–110. doi: 10.1097/01.fpc.0000184955.08453.a8. [DOI](https://doi.org/10.1097/01.fpc.0000184955.08453.a8) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16424822/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenet%20Genomics&title=Different%20contributions%20of%20polymorphisms%20in%20VKORC1%20and%20CYP2C9%20to%20intra-%20and%20inter-population%20differences%20in%20maintenance%20dose%20of%20warfarin%20in%20Japanese,%20Caucasians%20and%20African-Americans.&author=H%20Takahashi&author=GR%20Wilkinson&author=EA%20Nutescu&volume=16&publication_year=2006&pages=101-110&pmid=16424822&doi=10.1097/01.fpc.0000184955.08453.a8&)
|
| 238 |
+
|
| 239 |
+
37. Hermans C, Claeys D. Review of the rebound phenomenon in new anticoagulant treatments. Curr Med Res Opin. 2006;22:471–481. doi: 10.1185/030079906X89801. [DOI](https://doi.org/10.1185/030079906X89801) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16574031/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Curr%20Med%20Res%20Opin&title=Review%20of%20the%20rebound%20phenomenon%20in%20new%20anticoagulant%20treatments.&author=C%20Hermans&author=D%20Claeys&volume=22&publication_year=2006&pages=471-481&pmid=16574031&doi=10.1185/030079906X89801&)
|
| 240 |
+
|
| 241 |
+
38. Veenstra DL, You JH, Rieder MJ, et al. Association of vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet Genomics. 2005;15:687–691. doi: 10.1097/01.fpc.0000174789.77614.68. [DOI](https://doi.org/10.1097/01.fpc.0000174789.77614.68) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16141794/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenet%20Genomics&title=Association%20of%20vitamin%20K%20epoxide%20reductase%20complex%201%20(VKORC1)%20variants%20with%20warfarin%20dose%20in%20a%20Hong%20Kong%20Chinese%20patient%20population.&author=DL%20Veenstra&author=JH%20You&author=MJ%20Rieder&volume=15&publication_year=2005&pages=687-691&pmid=16141794&doi=10.1097/01.fpc.0000174789.77614.68&)
|
test/texts/PMC1978168.md
ADDED
|
@@ -0,0 +1,176 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# The Effect of CYP2D6 polymorphisms on the Response to Pain Treatment for Pediatric Sickle Cell Pain Crisis
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** David C Brousseau, D Gail McCarver, Amy L Drendel, Karthika Divakaran, Julie A Panepinto
|
| 5 |
+
**Journal:** The Journal of pediatrics
|
| 6 |
+
**Date:** 2007 Jun
|
| 7 |
+
**DOI:** [10.1016/j.jpeds.2007.01.049](https://doi.org/10.1016/j.jpeds.2007.01.049)
|
| 8 |
+
**PMID:** 17517247
|
| 9 |
+
**PMCID:** PMC1978168
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1978168/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC1978168/pdf/nihms24354.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC1978168/pdf/nihms24354.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Objectives:**
|
| 16 |
+
To test the hypothesis that children taking hydroxyurea who fail codeine therapy would have reduced functioning CYP2D6 alleles.
|
| 17 |
+
|
| 18 |
+
**Study design:**
|
| 19 |
+
Children with sickle cell disease presenting to an emergency department with a pain crisis unresponsive to codeine were genotyped. The proportion of children with reduced functioning alleles, and CYP2D6 enzyme activity scores ≤ 1.5, were compared, by chi-square analysis, between children taking hydroxyurea and those with mild disease.
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
73 children completed the study; 42 possessed reduced functioning alleles. 82% of 27 children taking hydroxyurea had reduced functioning alleles versus 47% of 36 mild children (p < 0.05). 78% of children taking hydroxyurea had decreased activity scores versus 44% of mild children (p<0.05). The odds ratios (95% CI), of children taking hydroxyurea, were 4.9 (1.5 – 15.9) for having reduced functioning alleles, and 4.4 (1.4 – 13.4) for having a low activity score.
|
| 23 |
+
|
| 24 |
+
**Conclusions:**
|
| 25 |
+
Failing codeine therapy for a pain crisis while taking hydroxyurea is associated with an increase in reduced functioning CYP2D6 alleles. We recommend genetic analysis or trial of a non-CYP2D6 analgesic for these children.
|
| 26 |
+
|
| 27 |
+
Keywords: genetics, hydroxyurea
|
| 28 |
+
|
| 29 |
+
### Objectives
|
| 30 |
+
|
| 31 |
+
To test the hypothesis that children taking hydroxyurea who fail codeine therapy would have reduced functioning CYP2D6 alleles.
|
| 32 |
+
|
| 33 |
+
### Study design
|
| 34 |
+
|
| 35 |
+
Children with sickle cell disease presenting to an emergency department with a pain crisis unresponsive to codeine were genotyped. The proportion of children with reduced functioning alleles, and CYP2D6 enzyme activity scores ≤ 1.5, were compared, by chi-square analysis, between children taking hydroxyurea and those with mild disease.
|
| 36 |
+
|
| 37 |
+
### Results
|
| 38 |
+
|
| 39 |
+
73 children completed the study; 42 possessed reduced functioning alleles. 82% of 27 children taking hydroxyurea had reduced functioning alleles versus 47% of 36 mild children (p < 0.05). 78% of children taking hydroxyurea had decreased activity scores versus 44% of mild children (p<0.05). The odds ratios (95% CI), of children taking hydroxyurea, were 4.9 (1.5 – 15.9) for having reduced functioning alleles, and 4.4 (1.4 – 13.4) for having a low activity score.
|
| 40 |
+
|
| 41 |
+
### Conclusions
|
| 42 |
+
|
| 43 |
+
Failing codeine therapy for a pain crisis while taking hydroxyurea is associated with an increase in reduced functioning CYP2D6 alleles. We recommend genetic analysis or trial of a non-CYP2D6 analgesic for these children.
|
| 44 |
+
|
| 45 |
+
**Keywords:**Keywords: genetics, hydroxyurea
|
| 46 |
+
|
| 47 |
+
## Methods
|
| 48 |
+
|
| 49 |
+
### Population/setting
|
| 50 |
+
|
| 51 |
+
Children with sickle cell disease (4–18 years) presenting to a Children’s Hospital ED for continued painful crisis after taking outpatient oral codeine or acetaminophen with codeine therapy were eligible. There was no threshold dose of codeine required to be eligible; any child who reported taking codeine was eligible. Those taking other CYP2D6 substrates (e.g. tramadol, paroxetine, or dextromethorphan) were excluded as they would potentially interfere with the activation of codeine even with normal functioning alleles. The study was approved by the hospital Institutional Review Board.
|
| 52 |
+
|
| 53 |
+
Children who met study criteria, were accompanied by a legal guardian, and presented for care when a research assistant was present in the ED (generally 8am to 10 pm weekdays and 8 am to 6 pm weekends) were approached for participation. Enrollment occurred between January, 2004 and June, 2005. There were seven refusals. After written informed consent was obtained from parents, and assent from children greater than or equal to seven years of age, blood was drawn for *CYP2D6*CYP2D6 genotyping.
|
| 54 |
+
|
| 55 |
+
### Severity
|
| 56 |
+
|
| 57 |
+
Children were classified as having “severe” sickle cell disease based strictly on the clinical criteria of having a history of three or more hospitalizations for vaso-occlusive crises in the last three years. In addition, any child placed on hydroxyurea for frequent painful crises was classified as severe.[2](#R2)^2^2 All other children were considered “mild”. Most severe children/families had agreed to take hydroxyurea; these children were classified as “severe, taking hydroxyurea.” Severity was determined by one of the investigators (DB) through chart review of all enrolled children, excluding the study visit and this investigator worked independently of the personnel who determined the CYP2D6 genotyping. Severity classification was determined before the blood samples were genotyped.
|
| 58 |
+
|
| 59 |
+
### DNA analysis
|
| 60 |
+
|
| 61 |
+
Blood samples were analyzed for 11 *CYP2D6*CYP2D6 SNPs, and *CYP2D6*CYP2D6 gene deletion and duplication. The combination of differences in these 11 SNPs accounts for the greater than 40 polymorphisms described in the literature.[18](#R18)^18^18 As an example, CYP2D6*4 was determined by detecting the following three SNPs in the gene: 100C>T, 1846G>A, and 4180G>C. Similarly, other alleles were assigned as defined previously.[18](#R18)^18^18 Genomic DNA was PCR-amplified to produce a 5.1 kb CYP2D6 amplicon. Nine SNPs, 100 C>T, 1023 C>T, 1707 G>A, 1846 G>A, 2549 delA, 2613 delAGA, 2850 C>T, 3183 G>A, and 4180 G>C were identified using this product. A second aliquot was used in a PCR reaction to generate the 418 bp region containing the SNPs 1659 G>A and 1758 G>A, which was sequenced using amplification primers by Dye Terminator Cycle Sequencing following the Quick Start Kit protocol (Beckman Coulter, Fullerton, CA). Gene duplications and deletions were detected by long-range PCR. [13](#R13)^13^13^, ^, [19](#R19)^19^19 The DNA analysis was performed by a single investigator (KD) who was blinded to the identity and severity status of the children.
|
| 62 |
+
|
| 63 |
+
### Outcome measures
|
| 64 |
+
|
| 65 |
+
The main outcome measure was the proportion of children possessing at least one reduced functioning allele, defined as: *CYP2D6*CYP2D6 *4, *5 (deletion allele), *6, *10, *17 and *40.[18](#R18)^18^18 A secondary outcome was the *CYP2D6*CYP2D6 activity score.[20](#R20)^20^20^, ^, [21](#R21)^21^21 The enzymatic activity of CYP2D6 was not measured in our study subjects, rather the activity score, based on extensive genotype/phenotype comparisons and measurements of enzymatic activity, is used to approximate the phenotype for a given genotype in an individual. The scoring system assigns a value of one for normal functioning alleles, and values of 0, 0.5, or 0.75 for reduced functioning alleles, based on enzymatic activity. A child with a normal functioning allele and an allele with an activity score of 0.5 would therefore have an overall activity score of 1.5. Duplicated alleles have their values counted twice, as determined to be valid in the derivation of the activity score.[20](#R20)^20^20^, ^, [21](#R21)^21^21 Phenotypic extensive metabolizers are classified as “high metabolizers” if their activity score is greater than 1.5.[21](#R21)^21^21 “High metabolizers” would more readily convert codeine to its active form, resulting in improved pain control, although children with activity scores ≤1.5 would have decreased activation of codeine and thus decreased pain control.
|
| 66 |
+
|
| 67 |
+
### Statistical analysis
|
| 68 |
+
|
| 69 |
+
The proportion of children with reduced functioning alleles, and the proportion with activity scores ≤ 1.5, were compared by Chi-square analysis between children with severe disease and those with mild disease. The analysis was repeated in children taking hydroxyurea, as they should have less severe crises, but reduced activation of codeine could cause continued visits for pain crises. Significant interaction terms existed between sickle cell genotype and severity/hydroxyurea status; a subset analysis was therefore performed in children with hemoglobin (Hgb) SS, the only genotype with a significant number of children classified as severe or severe taking hydroxyurea. Odds ratios for the presence of a reduced functioning allele based on severity status, and the odds ratio of being classified as severe given the presence of a reduced functioning allele were calculated, along with 95% confidence intervals.
|
| 70 |
+
|
| 71 |
+
## Results
|
| 72 |
+
|
| 73 |
+
78 children were enrolled; two children were excluded because of insufficient DNA. Six of the remaining 76 children came from three families; one child from each family was randomly withdrawn to obtain independent allelic frequencies, leaving 73 children. All of these children had reported taking codeine or acetaminophen with codeine. No other opioids had been used in the outpatient setting prior to arrival. The mean age was 11.5 years (s.d. 4.3); 42 (58%) were female. The majority were Hgb SS (47/73, 64%), 15 (21%) were Hgb SC, 3 (4%) were HgbSβ° and 8 (11%) were HgbSβ^+^+. Approximately one-half of children (37/73) were classified as severe, and 27 of those 37 children were taking hydroxyurea.
|
| 74 |
+
|
| 75 |
+
Within the entire sample, 42/73 children (58%) had a reduced functioning allele, a proportion similar to African American norms.[12](#R12)^12^12^, ^, [14](#R14)^14^14 Ten children (14%) had *CYP2D6*CYP2D6 duplications and 15 (21%) had deletions. Using univariate testing, children classified as having severe disease were older, more likely to Hgb SS, and more likely to have a reduced functioning allele and an activity score ≤ 1.5 ([Table I](#T1)Table I). A child with severe disease had twice the odds of having a reduced functioning allele and twice the odds of having an activity score ≤ 1.5, although the difference was only statistically significant for the activity score. Among the children taking hydroxyurea, 82% possessed a reduced functioning allele. Children taking hydroxyurea had 4.9 times the odds of possessing a reduced functioning allele and 4.4 times the odds of having an activity score ≤ 1.5 compared to those with mild disease.
|
| 76 |
+
|
| 77 |
+
### Table 1.
|
| 78 |
+
|
| 79 |
+
Differences in characteristics between children with mild disease, severe disease and severe disease taking hydroxyurea. Results are odds ratios (OR) and 95% Confidence Intervals for having a reduced functioning allele or activity score ≤ 1.5 compared to mild disease.
|
| 80 |
+
|
| 81 |
+
| | Mild (n = 36) | Severe (n = 37) | Severe taking Hydroxyurea (n=27) |
|
| 82 |
+
| - | ------------- | --------------- | -------------------------------- |
|
| 83 |
+
| Mean age (s.d.) | 10.1 (4.3) | 12.9 (3.9)* | 12.9 (4.0)* |
|
| 84 |
+
| Female | 18 (50%) | 24 (65%) | 17 (63%) |
|
| 85 |
+
| Sickle cell type Hgb SS Hgb Sβ° Hgb SC Hgb Sβ+ | 17 (47%) 1 (3%) 12 (33%) 6 (17%) | 30 (81%)* 2 (5%) 3 (8%) 2 (5%) | 22 (82%)* 2 (7%) 2 (7%) 1 (4%) |
|
| 86 |
+
| Possessed a reduced functioning allele | 17 (47%) | 25 (68%) | 22 (82%)* |
|
| 87 |
+
| 2D6 Activity score ≤ 1.5 | 16 (44%) | 25 (68%)* | 21 (78%)* |
|
| 88 |
+
| O.R. (95% CI) for having reduced functioning allele | referent | 2.3 (0.9 – 6.0) | 4.9 (1.5 – 15.9)* |
|
| 89 |
+
| O.R. (95% CI) for activity score ≤ 1.5 | referent | 2.6 (1.0 – 6.7)* | 4.4 (1.4 – 13.4)* |
|
| 90 |
+
Within the subset of children with HgbSS ([Table II](#T2)Table II), 30/47 (64%) were classified as severe and 22 (47%) of the 47 children were taking hydroxyurea. HgbSS children with severe disease again had twice the odds of having a reduced functioning allele and having an activity score ≤ 1.5 compared to children with mild disease, although in this smaller sample, neither odds ratio was statistically significant. HgbSS children taking hydroxyurea had over five times the odds of having a reduced functioning allele and an activity score ≤ 1.5, both significantly increased over children with mild disease.
|
| 91 |
+
|
| 92 |
+
### Table 2.
|
| 93 |
+
|
| 94 |
+
Differences in characteristics between children with mild disease, severe disease and severe disease taking hydroxyurea, in subset of children with HgbSS. Results are odds ratios (OR) and 95% Confidence Intervals for having a reduced functioning allele or activity score ≤ 1.5 compared to mild disease.
|
| 95 |
+
|
| 96 |
+
| | Mild (n = 17) | Severe (n = 30) | Severe taking Hydroxyurea (n=22) |
|
| 97 |
+
| - | ------------- | --------------- | -------------------------------- |
|
| 98 |
+
| Mean age (s.d.) | 10.1 (4.1) | 12.6 (3.9)* | 12.6 (3.9)* |
|
| 99 |
+
| Female | 8 (47%) | 21 (70%) | 15 (68%) |
|
| 100 |
+
| Possessed a reduced functioning allele | 9 (53%) | 21 (70%) | 19 (86%)* |
|
| 101 |
+
| 2D6 Activity score ≤ 1.5 | 16/36 (44%) | 20 (67%) | 18 (82%)* |
|
| 102 |
+
| O.R. (95% CI) for having reduced functioning allele | referent | 2.1 (0.6 – 7.1) | 5.6 (1.2 – 26.4)* |
|
| 103 |
+
| O.R. (95% CI) for activity score ≤ 1.5 | referent | 2.3 (0.7 – 7.6) | 5.1 (1.2 – 21.4)* |
|
| 104 |
+
Alternatively, for the entire population, a child having a reduced functioning allele had 4.9 times the odds of being on hydroxuyrea, and a child with an activity score ≤ 1.5 had 4.4 times the odds of being on hydroxyurea. Limited to children with HgbSS disease, a child with a reduced functioning allele had 5.6 times the odds of being on hydroxyurea, and a child with an activity score ≤ 1.5 had 5.1 times the odds of being on hydroxyurea.
|
| 105 |
+
|
| 106 |
+
## Discussion
|
| 107 |
+
|
| 108 |
+
Children taking hydroxyurea who present to the ED with sickle cell pain crises after failing outpatient codeine are significantly more likely to possess a reduced functioning CYP2D6 allele, and to have reduced CYP2D6 activity. Although some children may be classified as severe based on more severe underlying pathology, this study suggests that some children may have more frequent healthcare utilization for sickle cell pain crises due to an inadequate pain response to oral codeine.
|
| 109 |
+
|
| 110 |
+
Although it may be true that all children with severe disease have an increased risk of reduced functioning alleles, we were only able to show a significant difference for children who failed codeine while taking hydroxyurea. Since hydroxyurea lessens the pain of a sickle cell crisis, those children with an inadequate pain response to oral codeine would continue to present to the ED, whereas children whose now reduced pain is controlled with codeine would not require ED management.
|
| 111 |
+
|
| 112 |
+
For our study, we chose to analyze children who continued to have pain after oral codeine or acetaminophen with codeine. We chose to analyze codeine metabolism because: 1) codeine is the most commonly prescribed oral opioid for outpatient sickle cell pain at our institution, and 2) the important clinical effects of CYP2D6 polymorphisms have been shown with codeine activation, as well as the activation or metabolism of many commonly used drugs, including tramadol, and many cardiovascular or psychoactive drugs. [9](#R9)^9^9^, ^, [20](#R20)^20^20^–^–[22](#R22)^22^22
|
| 113 |
+
|
| 114 |
+
There are study limitations that need to be addressed. First, we did not measure enzyme activity; however, the calculated activity scores closely approximate phenotype, and account for duplications, providing an accurate assessment of the enzyme activity. A second limitation is the definition of sickle cell severity. We used adapted criteria established to determine eligibility for bone marrow transplant or hydroxyurea. There are other markers of severity in sickle cell disease, including elevated white blood cell count, low fetal hemoglobin, and elevated pulmonary artery pressures. [1](#R1)^1^1^, ^, [23](#R23)^23^23^, ^, [24](#R24)^24^24 We used previously published clinical criteria because treatment interventions for children with frequent complications from their sickle cell disease has become widely accepted. [2](#R2)^2^2^, ^, [4](#R4)^4^4 Finally, we did not ensure that children took the maximum allowable dose of codeine. Although it is possible that there may have been underdosing of codeine, this underdosing would have to be systematically different between the two groups to explain the differences we found in our study.
|
| 115 |
+
|
| 116 |
+
In conclusion, we believe that children taking hydroxyurea who continue to present to the ED for sickle cell pain crisis after failing oral codeine warrant CYP2D6 genotyping. Alternatively, these children could be given a non-CYP2D6 dependent analgesic such as oxycodone or hydrocodone for pain.
|
| 117 |
+
|
| 118 |
+
## Acknowledgments
|
| 119 |
+
|
| 120 |
+
The authors would like to thank Andrea Gaedigk PhD for her assistance in analyzing DNA sample duplications. We would also like to thank the Research Assistants in the Emergency Department of the Children’s Hospital of Wisconsin for their help in recruiting patients.
|
| 121 |
+
|
| 122 |
+
Financial support: This study was supported by the General Clinical Research Center Grant M01-RR00058 from the National Institutes of Health
|
| 123 |
+
|
| 124 |
+
## List of abbreviations
|
| 125 |
+
|
| 126 |
+
## Footnotes
|
| 127 |
+
|
| 128 |
+
## References
|
| 129 |
+
|
| 130 |
+
1. Platt OS, Thorington BD, Brambilla DJ, Milner PF, Rosse WF, Vichinsky E, et al. Pain in sickle cell disease. Rates and risk factors. N Engl J Med. 1991;325:11–16. doi: 10.1056/NEJM199107043250103. [DOI](https://doi.org/10.1056/NEJM199107043250103) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1710777/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Pain%20in%20sickle%20cell%20disease.%20Rates%20and%20risk%20factors&author=OS%20Platt&author=BD%20Thorington&author=DJ%20Brambilla&author=PF%20Milner&author=WF%20Rosse&volume=325&publication_year=1991&pages=11-16&pmid=1710777&doi=10.1056/NEJM199107043250103&)
|
| 131 |
+
|
| 132 |
+
2. Panepinto JA, O’Mahar KM, DeBaun MR, Loberiza FR, Scott JP. Health-related quality of life in children with sickle cell disease: child and parent perception. Br J Haematol. 2005;130:437–444. doi: 10.1111/j.1365-2141.2005.05622.x. [DOI](https://doi.org/10.1111/j.1365-2141.2005.05622.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16042695/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Haematol&title=Health-related%20quality%20of%20life%20in%20children%20with%20sickle%20cell%20disease:%20child%20and%20parent%20perception&author=JA%20Panepinto&author=KM%20O%E2%80%99Mahar&author=MR%20DeBaun&author=FR%20Loberiza&author=JP%20Scott&volume=130&publication_year=2005&pages=437-444&pmid=16042695&doi=10.1111/j.1365-2141.2005.05622.x&)
|
| 133 |
+
|
| 134 |
+
3. Scott JP, Hillery CA, Brown ER, Misiewicz V, Labotka RJ. Hydroxyurea therapy in children severely affected with sickle cell disease. J Pediatr. 1996;128:820–828. doi: 10.1016/s0022-3476(96)70335-9. [DOI](https://doi.org/10.1016/s0022-3476(96)70335-9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8648542/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pediatr&title=Hydroxyurea%20therapy%20in%20children%20severely%20affected%20with%20sickle%20cell%20disease&author=JP%20Scott&author=CA%20Hillery&author=ER%20Brown&author=V%20Misiewicz&author=RJ%20Labotka&volume=128&publication_year=1996&pages=820-828&pmid=8648542&doi=10.1016/s0022-3476(96)70335-9&)
|
| 135 |
+
|
| 136 |
+
4. Charache S, Terrin ML, Moore RD, Dover GJ, Barton FB, Eckert SV, et al. Effect of hydroxyurea on the frequency of painful crises in sickle cell anemia. Investigators of the Multicenter Study of Hydroxyurea in Sickle Cell Anemia. N Engl J Med. 1995;332:1317–1322. doi: 10.1056/NEJM199505183322001. [DOI](https://doi.org/10.1056/NEJM199505183322001) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7715639/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Effect%20of%20hydroxyurea%20on%20the%20frequency%20of%20painful%20crises%20in%20sickle%20cell%20anemia.%20Investigators%20of%20the%20Multicenter%20Study%20of%20Hydroxyurea%20in%20Sickle%20Cell%20Anemia&author=S%20Charache&author=ML%20Terrin&author=RD%20Moore&author=GJ%20Dover&author=FB%20Barton&volume=332&publication_year=1995&pages=1317-1322&pmid=7715639&doi=10.1056/NEJM199505183322001&)
|
| 137 |
+
|
| 138 |
+
5. Taylor JGt, Tang DC, Savage SA, Leitman SF, Heller SI, Serjeant GR, et al. Variants in the VCAM1 gene and risk for symptomatic stroke in sickle cell disease. Blood. 2002;100:4303–4309. doi: 10.1182/blood-2001-12-0306. [DOI](https://doi.org/10.1182/blood-2001-12-0306) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12393616/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Blood&title=Variants%20in%20the%20VCAM1%20gene%20and%20risk%20for%20symptomatic%20stroke%20in%20sickle%20cell%20disease&author=JGt%20Taylor&author=DC%20Tang&author=SA%20Savage&author=SF%20Leitman&author=SI%20Heller&volume=100&publication_year=2002&pages=4303-4309&pmid=12393616&doi=10.1182/blood-2001-12-0306&)
|
| 139 |
+
|
| 140 |
+
6. Hoppe C, Cheng S, Grow M, Silbergleit A, Klitz W, Trachtenberg E, et al. A novel multilocus genotyping assay to identify genetic predictors of stroke in sickle cell anaemia. Br J Haematol. 2001;114:718–720. doi: 10.1046/j.1365-2141.2001.02997.x. [DOI](https://doi.org/10.1046/j.1365-2141.2001.02997.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11553004/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Haematol&title=A%20novel%20multilocus%20genotyping%20assay%20to%20identify%20genetic%20predictors%20of%20stroke%20in%20sickle%20cell%20anaemia&author=C%20Hoppe&author=S%20Cheng&author=M%20Grow&author=A%20Silbergleit&author=W%20Klitz&volume=114&publication_year=2001&pages=718-720&pmid=11553004&doi=10.1046/j.1365-2141.2001.02997.x&)
|
| 141 |
+
|
| 142 |
+
7. Sullivan KJ, Kissoon N, Duckworth LJ, Sandler E, Freeman B, Bayne E, et al. Low exhaled nitric oxide and a polymorphism in the NOS I gene is associated with acute chest syndrome. Am J Respir Crit Care Med. 2001;164:2186–2190. doi: 10.1164/ajrccm.164.12.2012090. [DOI](https://doi.org/10.1164/ajrccm.164.12.2012090) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11751185/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Respir%20Crit%20Care%20Med&title=Low%20exhaled%20nitric%20oxide%20and%20a%20polymorphism%20in%20the%20NOS%20I%20gene%20is%20associated%20with%20acute%20chest%20syndrome&author=KJ%20Sullivan&author=N%20Kissoon&author=LJ%20Duckworth&author=E%20Sandler&author=B%20Freeman&volume=164&publication_year=2001&pages=2186-2190&pmid=11751185&doi=10.1164/ajrccm.164.12.2012090&)
|
| 143 |
+
|
| 144 |
+
8. Passon RG, Howard TA, Zimmerman SA, Schultz WH, Ware RE. Influence of bilirubin uridine diphosphate-glucuronosyltransferase 1A promoter polymorphisms on serum bilirubin levels and cholelithiasis in children with sickle cell anemia. J Pediatr Hematol Oncol. 2001;23:448–451. doi: 10.1097/00043426-200110000-00011. [DOI](https://doi.org/10.1097/00043426-200110000-00011) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11878580/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pediatr%20Hematol%20Oncol&title=Influence%20of%20bilirubin%20uridine%20diphosphate-glucuronosyltransferase%201A%20promoter%20polymorphisms%20on%20serum%20bilirubin%20levels%20and%20cholelithiasis%20in%20children%20with%20sickle%20cell%20anemia&author=RG%20Passon&author=TA%20Howard&author=SA%20Zimmerman&author=WH%20Schultz&author=RE%20Ware&volume=23&publication_year=2001&pages=448-451&pmid=11878580&doi=10.1097/00043426-200110000-00011&)
|
| 145 |
+
|
| 146 |
+
9. Wuttke H, Rau T, Heidi R, Bergmann K, Bohm M, Weil J, et al. Increased Frequency of cytochrome P450 2D6 poor metabolizers among patients with metoprolol-associated adverse events. Clin Pharmacol Ther. 2002;72:429–437. doi: 10.1067/mcp.2002.127111. [DOI](https://doi.org/10.1067/mcp.2002.127111) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12386645/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Increased%20Frequency%20of%20cytochrome%20P450%202D6%20poor%20metabolizers%20among%20patients%20with%20metoprolol-associated%20adverse%20events&author=H%20Wuttke&author=T%20Rau&author=R%20Heidi&author=K%20Bergmann&author=M%20Bohm&volume=72&publication_year=2002&pages=429-437&pmid=12386645&doi=10.1067/mcp.2002.127111&)
|
| 147 |
+
|
| 148 |
+
10. Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348:529–537. doi: 10.1056/NEJMra020021. [DOI](https://doi.org/10.1056/NEJMra020021) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12571261/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Inheritance%20and%20drug%20response&author=R%20Weinshilboum&volume=348&publication_year=2003&pages=529-537&pmid=12571261&doi=10.1056/NEJMra020021&)
|
| 149 |
+
|
| 150 |
+
11. Evans W, McLeod H. Pharmacogenomics - drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI](https://doi.org/10.1056/NEJMra020526) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12571262/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Pharmacogenomics%20-%20drug%20disposition,%20drug%20targets,%20and%20side%20effects&author=W%20Evans&author=H%20McLeod&volume=348&publication_year=2003&pages=538-549&pmid=12571262&doi=10.1056/NEJMra020526&)
|
| 151 |
+
|
| 152 |
+
12. Bradford L. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229–243. doi: 10.1517/14622416.3.2.229. [DOI](https://doi.org/10.1517/14622416.3.2.229) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11972444/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics&title=CYP2D6%20allele%20frequency%20in%20European%20Caucasians,%20Asians,%20Africans%20and%20their%20descendants&author=L%20Bradford&volume=3&publication_year=2002&pages=229-243&pmid=11972444&doi=10.1517/14622416.3.2.229&)
|
| 153 |
+
|
| 154 |
+
13. Gaedigk A, Bradford L, Marcucci K, Leeder J. Unique CYP2D6 activity distribution and genotype-phenotype discordance in black Americans. Clinical Pharmacology and Therapeutics. 2002;72:76–89. doi: 10.1067/mcp.2002.125783. [DOI](https://doi.org/10.1067/mcp.2002.125783) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12152006/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clinical%20Pharmacology%20and%20Therapeutics&title=Unique%20CYP2D6%20activity%20distribution%20and%20genotype-phenotype%20discordance%20in%20black%20Americans&author=A%20Gaedigk&author=L%20Bradford&author=K%20Marcucci&author=J%20Leeder&volume=72&publication_year=2002&pages=76-89&pmid=12152006&doi=10.1067/mcp.2002.125783&)
|
| 155 |
+
|
| 156 |
+
14. Wan Y-J, Poland R, Han G. Analysis of CYP2D6 polymorphism and enzyme activity in African-Americans in Southen California. Pharmacogenetics. 2001;11:489–499. doi: 10.1097/00008571-200108000-00004. [DOI](https://doi.org/10.1097/00008571-200108000-00004) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11505219/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Analysis%20of%20CYP2D6%20polymorphism%20and%20enzyme%20activity%20in%20African-Americans%20in%20Southen%20California&author=Y-J%20Wan&author=R%20Poland&author=G%20Han&volume=11&publication_year=2001&pages=489-499&pmid=11505219&doi=10.1097/00008571-200108000-00004&)
|
| 157 |
+
|
| 158 |
+
15. Eckhardt K, Li S, Ammon S, Schanzle G, Mikus G, Eichelbaum M. Same incidence of adverse drug events after codeine administration irrespective of the genetically determined differences in morphine formation. Pain. 1998;76:27–33. doi: 10.1016/s0304-3959(98)00021-9. [DOI](https://doi.org/10.1016/s0304-3959(98)00021-9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9696456/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pain&title=Same%20incidence%20of%20adverse%20drug%20events%20after%20codeine%20administration%20irrespective%20of%20the%20genetically%20determined%20differences%20in%20morphine%20formation&author=K%20Eckhardt&author=S%20Li&author=S%20Ammon&author=G%20Schanzle&author=G%20Mikus&volume=76&publication_year=1998&pages=27-33&pmid=9696456&doi=10.1016/s0304-3959(98)00021-9&)
|
| 159 |
+
|
| 160 |
+
16. Poulsen L, Brosen K, Arendt-Nielsen L, Gram LF, Elbaek K, Sindrup SH. Codeine and morphine in extensive and poor metabolizers of sparteine: pharmacokinetics, analgesic effect and side effects. Eur J Clin Pharmacol. 1996;51:289–295. doi: 10.1007/s002280050200. [DOI](https://doi.org/10.1007/s002280050200) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9010701/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=Codeine%20and%20morphine%20in%20extensive%20and%20poor%20metabolizers%20of%20sparteine:%20pharmacokinetics,%20analgesic%20effect%20and%20side%20effects&author=L%20Poulsen&author=K%20Brosen&author=L%20Arendt-Nielsen&author=LF%20Gram&author=K%20Elbaek&volume=51&publication_year=1996&pages=289-295&pmid=9010701&doi=10.1007/s002280050200&)
|
| 161 |
+
|
| 162 |
+
17. Caraco Y, Sheller J, Wood AJ. Pharmacogenetic determination of the effects of codeine and prediction of drug interactions. J Pharmacol Exp Ther. 1996;278:1165–1174. [PubMed](https://pubmed.ncbi.nlm.nih.gov/8819499/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharmacol%20Exp%20Ther&title=Pharmacogenetic%20determination%20of%20the%20effects%20of%20codeine%20and%20prediction%20of%20drug%20interactions&author=Y%20Caraco&author=J%20Sheller&author=AJ%20Wood&volume=278&publication_year=1996&pages=1165-1174&pmid=8819499&)
|
| 163 |
+
|
| 164 |
+
18. [Accessed October 25, 2006]; http://www.imm.ki.se/CYPalleles/cyp2D6.htm. [http://www.imm.ki.se/CYPalleles/cyp2D6.htm](http://www.imm.ki.se/CYPalleles/cyp2D6.htm)
|
| 165 |
+
|
| 166 |
+
19. Gaedigk A, Gotschall R, Forbes N, Simon S, Kearns G, Leeder J. Optimization of cytochrome P4502D6 (CYP2D6) phenotype assignment using a genotyping algorithm based on allele frequency data. Pharmacogenetics. 1999;9:669–682. doi: 10.1097/01213011-199912000-00002. [DOI](https://doi.org/10.1097/01213011-199912000-00002) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10634130/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Optimization%20of%20cytochrome%20P4502D6%20(CYP2D6)%20phenotype%20assignment%20using%20a%20genotyping%20algorithm%20based%20on%20allele%20frequency%20data&author=A%20Gaedigk&author=R%20Gotschall&author=N%20Forbes&author=S%20Simon&author=G%20Kearns&volume=9&publication_year=1999&pages=669-682&pmid=10634130&doi=10.1097/01213011-199912000-00002&)
|
| 167 |
+
|
| 168 |
+
20. Findling RL, Nucci G, Piergies AA, et al. Multiple Dose Pharmacokinetics of Paroxetine in Children and Adolescents with Major Depressive Disorder or Obsessive-Compulsive Disorder. Neuropsychopharmacology. 2005 Nov 23; doi: 10.1038/sj.npp.1300960. [DOI](https://doi.org/10.1038/sj.npp.1300960) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16319918/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Neuropsychopharmacology&title=Multiple%20Dose%20Pharmacokinetics%20of%20Paroxetine%20in%20Children%20and%20Adolescents%20with%20Major%20Depressive%20Disorder%20or%20Obsessive-Compulsive%20Disorder&author=RL%20Findling&author=G%20Nucci&author=AA%20Piergies&publication_year=2005&pmid=16319918&doi=10.1038/sj.npp.1300960&)
|
| 169 |
+
|
| 170 |
+
21. Zineh I, Beitelshees AL, Gaedigk A, Walker JR, Pauly DF, Eberst K, et al. Pharmacokinetics and CYP2D6 genotypes do not predict metoprolol adverse events or efficacy in hypertension. Clin Pharmacol Ther. 2004;76:536–544. doi: 10.1016/j.clpt.2004.08.020. [DOI](https://doi.org/10.1016/j.clpt.2004.08.020) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15592325/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Pharmacokinetics%20and%20CYP2D6%20genotypes%20do%20not%20predict%20metoprolol%20adverse%20events%20or%20efficacy%20in%20hypertension&author=I%20Zineh&author=AL%20Beitelshees&author=A%20Gaedigk&author=JR%20Walker&author=DF%20Pauly&volume=76&publication_year=2004&pages=536-544&pmid=15592325&doi=10.1016/j.clpt.2004.08.020&)
|
| 171 |
+
|
| 172 |
+
22. Tirkkonen T, Laine K. Drug interactions with the potential to prevent prodrug activation as a common source of irrational prescribing in hospital inpatients. Clin Pharmacol Ther. 2004;76(6):639–647. doi: 10.1016/j.clpt.2004.08.017. [DOI](https://doi.org/10.1016/j.clpt.2004.08.017) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15592335/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Drug%20interactions%20with%20the%20potential%20to%20prevent%20prodrug%20activation%20as%20a%20common%20source%20of%20irrational%20prescribing%20in%20hospital%20inpatients&author=T%20Tirkkonen&author=K%20Laine&volume=76&issue=6&publication_year=2004&pages=639-647&pmid=15592335&doi=10.1016/j.clpt.2004.08.017&)
|
| 173 |
+
|
| 174 |
+
23. Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O, Steinberg MH, et al. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med. 1994;330:1639–1644. doi: 10.1056/NEJM199406093302303. [DOI](https://doi.org/10.1056/NEJM199406093302303) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7993409/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Mortality%20in%20sickle%20cell%20disease.%20Life%20expectancy%20and%20risk%20factors%20for%20early%20death&author=OS%20Platt&author=DJ%20Brambilla&author=WF%20Rosse&author=PF%20Milner&author=O%20Castro&volume=330&publication_year=1994&pages=1639-1644&pmid=7993409&doi=10.1056/NEJM199406093302303&)
|
| 175 |
+
|
| 176 |
+
24. Gladwin MT, Sachdev V, Jison ML, Shizukuda Y, Plehn JF, Minter K, et al. Pulmonary hypertension as a risk factor for death in patients with sickle cell disease. N Engl J Med. 2004;350:886–895. doi: 10.1056/NEJMoa035477. [DOI](https://doi.org/10.1056/NEJMoa035477) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14985486/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Pulmonary%20hypertension%20as%20a%20risk%20factor%20for%20death%20in%20patients%20with%20sickle%20cell%20disease&author=MT%20Gladwin&author=V%20Sachdev&author=ML%20Jison&author=Y%20Shizukuda&author=JF%20Plehn&volume=350&publication_year=2004&pages=886-895&pmid=14985486&doi=10.1056/NEJMoa035477&)
|
test/texts/PMC2014233.md
ADDED
|
@@ -0,0 +1,135 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Functional significance of a C→A polymorphism in intron 1 of the cytochrome P450 CYP1A2 gene tested with caffeine
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Christoph Sachse, Jürgen Brockmöller, Steffen Bauer, Ivar Roots
|
| 5 |
+
**Journal:** British Journal of Clinical Pharmacology
|
| 6 |
+
**Date:** 1999 Apr
|
| 7 |
+
**DOI:** [10.1046/j.1365-2125.1999.00898.x](https://doi.org/10.1046/j.1365-2125.1999.00898.x)
|
| 8 |
+
**PMID:** 10233211
|
| 9 |
+
**PMCID:** PMC2014233
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2014233/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC2014233/pdf/bcp0047-0445.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC2014233/pdf/bcp0047-0445.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Aims:**
|
| 16 |
+
The cytochrome P450 enzyme CYP1A2 metabolises several drugs and carcinogens. We wanted to determine how much of the variability of CYP1A2 activity is explained by a newly discovered gene polymorphism in intron 1.
|
| 17 |
+
|
| 18 |
+
**Methods:**
|
| 19 |
+
A single nucleotide polymorphism in intron 1 of the CYP1A2 gene at position 734 downstream of the first transcribed nucleotide was identified by DNA sequence analysis. The functional significance of this C/A polymorphism was assessed in 185 healthy Caucasian non-smokers and in 51 smokers by genotyping and phenotyping using caffeine (100 mg oral dose).
|
| 20 |
+
|
| 21 |
+
**Results:**
|
| 22 |
+
Out of the total sample, 46% were homozygous for the variant A, 44% were heterozygous, and 10% were homozygous for the variant C. The ratio of 1,7-dimethylxanthine (17X) plus 1,7-dimethyluric acid divided by caffeine in 0–5 h urine samples from 185 non-smokers did not differ significantly between the three CYP1A2 genotypes. In the 51 smokers, analysis of variance revealed significant differences in the 5 h plasma 17X/caffeine ratios between the genotypes (P=0.008, F-test). The mean ratio was 1.37 in carriers of the A/A genotype, 0.88 in heterozygotes and 0.82 in carriers of C/C. The mean difference between the A/A and C/A groups was 0.48 (95% confidence interval 0.15–0.81; P=0.01).
|
| 23 |
+
|
| 24 |
+
**Conclusions:**
|
| 25 |
+
The A/A genotype, which may represent a CYP1A2 high inducibility genotype, may either be a direct cause of increased CYP1A2activity, or be genetically linked to polymorphisms conferring high inducibility. Further studies are needed to define the role of this polymorphism on the pharmacokinetics of drugs metabolised by CYP1A2 and in the activation of carcinogens.
|
| 26 |
+
|
| 27 |
+
Keywords: caffeine test, cytochrome P450 1A2, CYP1A2, gene polymorphism, enzyme induction
|
| 28 |
+
|
| 29 |
+
### Aims
|
| 30 |
+
|
| 31 |
+
The cytochrome P450 enzyme CYP1A2 metabolises several drugs and carcinogens. We wanted to determine how much of the variability of CYP1A2 activity is explained by a newly discovered gene polymorphism in intron 1.
|
| 32 |
+
|
| 33 |
+
### Methods
|
| 34 |
+
|
| 35 |
+
A single nucleotide polymorphism in intron 1 of the *CYP1A2*CYP1A2 gene at position 734 downstream of the first transcribed nucleotide was identified by DNA sequence analysis. The functional significance of this C/A polymorphism was assessed in 185 healthy Caucasian non-smokers and in 51 smokers by genotyping and phenotyping using caffeine (100 mg oral dose).
|
| 36 |
+
|
| 37 |
+
### Results
|
| 38 |
+
|
| 39 |
+
Out of the total sample, 46% were homozygous for the variant A, 44% were heterozygous, and 10% were homozygous for the variant C. The ratio of 1,7-dimethylxanthine (17X) plus 1,7-dimethyluric acid divided by caffeine in 0–5 h urine samples from 185 non-smokers did not differ significantly between the three *CYP1A2*CYP1A2 genotypes. In the 51 smokers, analysis of variance revealed significant differences in the 5 h plasma 17X/caffeine ratios between the genotypes (*P*P=0.008, *F*F-test). The mean ratio was 1.37 in carriers of the A/A genotype, 0.88 in heterozygotes and 0.82 in carriers of C/C. The mean difference between the A/A and C/A groups was 0.48 (95% confidence interval 0.15–0.81; *P*P=0.01).
|
| 40 |
+
|
| 41 |
+
### Conclusions
|
| 42 |
+
|
| 43 |
+
The A/A genotype, which may represent a *CYP1A2*CYP1A2 high inducibility genotype, may either be a direct cause of increased *CYP1A2*CYP1A2activity, or be genetically linked to polymorphisms conferring high inducibility. Further studies are needed to define the role of this polymorphism on the pharmacokinetics of drugs metabolised by CYP1A2 and in the activation of carcinogens.
|
| 44 |
+
|
| 45 |
+
**Keywords:**Keywords: caffeine test, cytochrome P450 1A2, *CYP1A2*CYP1A2, gene polymorphism, enzyme induction
|
| 46 |
+
|
| 47 |
+
## Introduction
|
| 48 |
+
|
| 49 |
+
The cytochrome P450 enzyme CYP1A2 plays a major role in metabolism of many commonly used drugs including clozapine, imipramine, caffeine, paracetamol, phenacetin, theophylline, tacrine [[1](#b1)1], and some neurotoxins [[2](#b2)2]. Furthermore, CYP1A2 activates several aromatic amines and thus is a key enzyme in chemical carcinogenesis [[3](#b3)3]. CYP1A2 activity is induced by the binding of aromatic hydrocarbons to the Ah-receptor. *In vivo*In vivo measurement of CYP1A2 activity in several human populations has shown wide interindividual variability, and population studies have reported either unimodal [[4](#b4)4], bimodal or trimodal distributions of CYP1A2 activity [[5](#b5)5–[7](#b7)7]. Also, so-called metabolic crosstalk with polymorphisms of *CYP2C19*CYP2C19 [[8](#b8)8] or *GSTM1*GSTM1 [[9](#b9)9] appears to modify CYP1A2 activity in humans. The wide interindividual variation in and possible polymodal distribution of CYP1A2 activity are suggestive of polymorphic control of enzyme activity. Very recently, sequencing of genomic DNA has revealed putative polymorphisms in exons 2 and 7 and in intron 1 of *CYP1A2*CYP1A2 [[10](#b10)10–[12](#b12)12]. Furthermore, the intron 1 polymorphism appears to affect the inducibility of *CYP1A2*CYP1A2 [[12](#b12)12]. By sequencing intron 1 of *CYP1A2*CYP1A2 in Caucasian healthy volunteers, we have confirmed the presence of a polymorphism occurring at high frequency and have studied its functional relevance using caffeine as a test drug in populations of non-smokers and smokers.
|
| 50 |
+
|
| 51 |
+
## Methods
|
| 52 |
+
|
| 53 |
+
The functional effect of the *CYP1A2*CYP1A2 polymorphism was studied in two groups. First, 185 non-smoking healthy Caucasian volunteers were phenotyped for their CYP1A2 metabolic capacity, using a single oral dose of 100 mg caffeine (Coffeinum purum^TM^TM, Berlin-Chemie). Immediately prior to taking the tablet, the volunteers were asked to empty their bladder and urine was collected for the subsequent 5 h. These subjects were recruited from a pool of healthy non-smoking volunteers regularly participating in phase I studies. Non-smoking status was confirmed by inquiry on the day of the test. All subjects had given written informed consent to the phenotyping procedures and to genotype analyses for heritable polymorphisms in foreign compound metabolising enzymes. The volunteers did not participate in drug trials for at least four weeks prior to phenotyping. Urine concentrations of caffeine (137X) and the CYP1A2 catalysed metabolites 1,7-dimethylxanthine (17X) and 1,7-dimethyluric acid (17U) were quantified by h.p.l.c. essentially as described by Grant *et al*et al. [[13](#b13)13]. The interassay coefficient of variation (5 mg l^−1^−1) of each of the caffeine metabolites was 8.4% for 137X, 4.7% for 17U, and 4.8% for 17X (*n*n=13). The urine 17X+17U/137X ratio, calculated on a molar basis, was used as the index of CYP1A2 activity [[14](#b14)14].
|
| 54 |
+
|
| 55 |
+
The second study was performed in 51 healthy volunteers who were current smokers (1 to 50 [mean 15] cigarettes per day for at least 2 weeks prior to the test). They received an oral dose of 100 mg caffeine and a venous blood sample was taken 5 h after administration. The molar metabolic concentration ratio (MR) 17X/137X was used as the index of CYP1A2 activity [[15](#b15)15]. 17X and 137X were quantified in plasma as described above but using 10% methanol in the mobile phase. The lower limit of quantification was 0.02 mg l^−1^−1 for 17X and 0.03 mg l^−1^−1 for 137X. Interassay coefficients of variation were 8.8% for 17X and 8.0% for 137X at a concentration of 5 mg l^−1^−1 for both compounds (*n*n=20).
|
| 56 |
+
|
| 57 |
+
For the determination of genotype, 5–10 ml of whole blood were collected in EDTA tubes. DNA was extracted using a standard phenol/chloroform extraction procedure [[16](#b16)16], and DNA samples were dissolved in 10 mm Tris/1 mm EDTA, pH 8.0 and stored at 4° C. Intron 1 of *CYP1A2*CYP1A2 was amplified by PCR using forward primer P1f (5′-CAACCCTGCCAATCTCAAGCAC, located in exon 1) and reverse primer P4r (5′-AGAAGCTCTGTGGCCGAGAAGG, located in exon 2). A 25 μl PCR mix comprised 10 mm Tris-HCl pH 8.3, 1.25 mm MgCl_2_2, 50 mm KCl, 200 μm dNTPs, 0.2 μm of each of the primers, 1.25 U *Taq*Taq polymerase (AmpliTaq^TM^TM, Perkin Elmer), and 1 μl of genomic DNA. PCR was performed with an initial denaturation for 2 min at 94° C followed by 35 cycles of 30 s at 94° C, 10 s at 60° C, 1 min at 72° C, and a terminal extension for 7 min at 72° C. For sequencing the *CYP1A2*CYP1A2 intron 1 region from the P1f/P4r PCR product, two shorter fragments were amplified in a nested PCR to increase the specificity ([Figure 1](#fig01)Figure 1). One fragment was amplified with primers P1f (see above) and P2r (5′-AAGAGTCCCTGCCAGTGCTGGC, located in intron 1), and the other fragment was flanked by P3f (5′-GGAACTCCTGGTCCCTTGGGTA, located in intron 1) and P4r (see above). DNA sequence analysis was performed on an ABI 373A instrument using a Taq DyeDeoxy Cycle Sequencing Kit^TM^TM (Perkin Elmer, Weiterstadt, Germany).
|
| 58 |
+
|
| 59 |
+
### Figure 1.
|
| 60 |
+
|
| 61 |
+
|
| 62 |
+
|
| 63 |
+
Structure of the CYP1A2 gene. Exons are shown as filled boxes, the dashed lines indicating incompletely characterised intron sequences, and the arrows showing the positions of the primers used for identification and routine genotyping of the intron 1 polymorphism. The boxes below show the DNA sequencing signals of the four nucleotides around the CYP1A2734 polymorphism for the three genotypes C/C, C/A and A/A. Number 734 refers to the position downstream of the first transcribed CYP1A2 nucleotide.
|
| 64 |
+
|
| 65 |
+
For routine genotyping, a PCR-RFLP test specific for the C/A polymorphism was established using the reaction with primers P1f and P4r as described above followed by a digestion with *Bsp*Bsp120I (MBI Fermentas). Wild-type alleles were cut into two fragments of 709 and 211 bp by the enzyme *Bsp*Bsp120l, whereas the mutant alleles remained uncleaved. Cleavage products were analyzed by 3% agarose gel electrophoresis.
|
| 66 |
+
|
| 67 |
+
Differences between the genetically defined subgroups were tested by one-way analysis of variance with the SAS statistical program (SAS institute, Cary, USA) with the procedure GLM using the logarithmically (base 10) transformed metabolic ratios. The coefficient of determination (R^2^2) was also calculated using the SAS procedure GLM. Analysis of variance was applied after normal distribution of the logarithmically transformed data from all groups was confirmed using the Shapiro Wilk test. In addition, the prerequisite of homoscedasticity was fulfilled after logarithmic transformation. Means, confidence limits both for absolute values and differences between means were determined from logarithmically transformed values.
|
| 68 |
+
|
| 69 |
+
## Results
|
| 70 |
+
|
| 71 |
+
We sequenced intron 1 of the *CYP1A2*CYP1A2 gene in DNA from eight volunteers. Only one polymorphism was identified, namely a C to A transversion at position 734 downstream of the first transcribed nucleotide of *CYP1A2*CYP1A2 (based on the *CYP1A2*CYP1A2 sequence published at GenBank accession no. [M31664](https://www.ncbi.nlm.nih.gov/nuccore/M31664)M31664; [Figure 1](#fig01)Figure 1). A mutation-specific PCR-RFLP test was then developed and used to genotype 236 subjects. Only 24 subjects had the homozygous C/C genotype (10%, 95% confidence limits 6.6–15%), whereas 104 subjects had the A/C genotype (44%, 95% CL 38–51%) and 108 were homozygous A/A (46%, 95% CL 39–52%). This distribution is in accordance with the frequencies expected, namely 43% for A/C and 46% for A/A, when applying the Hardy-Weinberg principle based on the experimentally determined frequency of the C/C genotype.
|
| 72 |
+
|
| 73 |
+
The molar urinary ratios of (17X+17U)/137X following the oral administration of 100 mg caffeine to 185 non-smoking subjects are depicted on the left hand side of [Figure 2](#fig02)Figure 2. Mean values and their 95% confidence limits were 2.54 (2.19–2.99) for the A/A genotype, 2.28 (1.97–2.65) for the A/C genotype, and 3.26 (2.36–4.50) for the C/C genotype. No significant differences between *CYP1A2*CYP1A2 genotypes were observed according to one way analysis of variance.
|
| 74 |
+
|
| 75 |
+
### Figure 2.
|
| 76 |
+
|
| 77 |
+
|
| 78 |
+
|
| 79 |
+
Frequency distribution of caffeine metabolic ratios in the different genotypes with respect to the CYP1A2 intron 1 polymorphism. On the left, logarithmically transformed molar urinary (17X+17U)/137X ratios from 185 non-smoking subjects are shown. There were no significant differences in the mean ratios between the three groups. On the right, the logarithmically transformed plasma 17X/137X ratios from 51 smokers are shown. In this group, the C/A polymorphism had a significant effect on the caffeine metabolic ratios (P=0.008). Dotted lines indicate the mean values in each group. The frequency distribution of each of the subgroups was not significantly different from a normal distribution according to the Shapiro-Wilk test.
|
| 80 |
+
|
| 81 |
+
In contrast, the mean molar ratios of 17X/137X in plasma from 51 smokers differed significantly between the *CYP1A2*CYP1A2 genotypes ([Figure 2](#fig02)Figure 2). Mean values and their 95% confidence intervals were 1.37 (1.07–1.72) for the A/A genotype, 0.88 (0.73–1.06) for the A/C, genotype, and 0.82 (0.46–1.46) for the C/C genotype. These differences were significant according to one way analysis of variance (*F*F-test, *P*P=0.008) with a coefficient of determination (R^2^2) of 0.18, indicating that 18% of the phenotypic variability might be explained by the C/A polymorphism. The distribution of the logarithmically transformed data was not significantly different from that of a normal distribution according to the Shapiro Wilk test. However, the metabolic ratios of the A/C group did not appear to be normally distributed on visual inspection of the histogram ([Figure 2](#fig02)Figure 2). Thus, the nonparametric Kruskal-Wallis test was applied to confirm that the differences between the groups were significant (*P*P=0.02 with 2 degrees of freedom). *Post-hoc*Post-hoc tests using a Bonferroni correction showed a significant difference between the A/A and the A/C group (*P*P=0.01; mean difference: 0.48; 95% confidence limits: 0.15–0.81). In contrast, no significant differences were seen between the C/C and A/C group (mean difference: 0.06, 95% confidence limits: -0.42–0.55) and the C/C and A/A group (mean difference: 0.55, 95% confidence limits:-0.06–1.19).
|
| 82 |
+
|
| 83 |
+
The smokers were stratified into equal sized groups of moderate smokers (<15 cigarettes per day) and heavy smokers (≥15 cigarettes per day). In the heavy smoker subgroup (*n*n=26), mean (95% confidence limits) of the metabolic ratios were 0.77 (0.34–1.76) for C/C, 0.81 (0.55–1.20) for A/C and 1.40 (1.02–1.92) for A/A, and there was a significant *P*P difference of 0.59 (95% confidence limits 0.17–1.07) between the A/C and A/A groups.
|
| 84 |
+
|
| 85 |
+
## Discussion
|
| 86 |
+
|
| 87 |
+
In this work we have confirmed the preliminary data of McLeod *et al*et al. [[12](#b12)12] showing the presence of a polymorphism in intron 1 of the *CYP1A2*CYP1A2 gene. A single base change, namely a C to A transversion downstream of the first transcribed nucleotide, was identified. The previously published sequence of the wild type *CYP1A2*CYP1A2 gene contained C at position 734 [[17](#b17)17]. However, in the present large population study we have found that the A variant is more frequent and suggest that it be termed *CYP1A2*CYP1A2**1A*1A and the C variant *CYP1A2*CYP1A2**1B*1B in accordance with the standard nomenclature for polymorphic drug metabolising enzymes [[18](#b18)18].
|
| 88 |
+
|
| 89 |
+
There were no significant differences in CYP1A2 metabolic activity between the genotypes in non-smoking individuals (i.e., with uninduced CYP1A2 levels) with respect to the intron 1 polymorphism. However, in smokers a 1.6-fold higher metabolic activity was observed in subjects homozygous for the A allele compared with the other genotypes, a difference that was statistically significant. These data are in accordance with those published by McLeod *et al*et al. [[12](#b12)12] in a smaller study. We observed a significant difference between the A/A and A/C genotypes but not between the A/C and the C/C genotypes. This might indicate that the A allele is a recessive factor for high inducibility but with only 5 subjects in the C/C group the power to establish the mode of inheritance for this gene was too small.
|
| 90 |
+
|
| 91 |
+
One shortcoming of this study was the different caffeine test ratios used in the non-smoker group and smoker groups. Although it has been shown that both urinary and plasma caffeine ratios reflect CYP1A2 activity [[14](#b14)14], plasma ratios may be less biased and more sensitive parameters of CYP1A2 activity [[6](#b6)6, [15](#b15)15]. For practical reasons only the urinary (17X+17U)/caffeine ratios could be measured in the study of non-smokers. Therefore, the small effect of this *CYP1A2*CYP1A2 polymorphism on enzyme induction seen in smokers could also be present in non-smokers, but may not have been detected because a less sensitive index of *CYP1A2*CYP1A2activity was used in the latter group. Alternatively, in non-smokers other segments in the *CYP1A2*CYP1A2 gene may be more important for regulation of the CYP1A2 activity.
|
| 92 |
+
|
| 93 |
+
Mechanistically, this single nucleotide polymorphism in the non-coding region of the *CYP1A2*CYP1A2 gene may either result in differential binding of putative regulatory proteins as suggested by McLeod *et al*et al. [[12](#b12)12], or it may be in linkage disequilibrium with other mutations affecting CYP1A2 inducibility. The C/A polymorphism could contribute to interindividual differences in the metabolism of other CYP1A2 substrates. It may also be a risk factor in cancers associated with exposure to arylamines or heterocyclic amines, such as urinary bladder or colon cancer.
|
| 94 |
+
|
| 95 |
+
## Acknowledgments
|
| 96 |
+
|
| 97 |
+
The study was supported by grants 01EC9408 and 01ZZ9511 of the German Federal Ministry of Education, Science, Research, and Technology. Furthermore, we thank Mr. A. Becker for his contribution to this study.
|
| 98 |
+
|
| 99 |
+
## References
|
| 100 |
+
|
| 101 |
+
1. Bertz RJ, Grannemann GR. Use of in vitro data and in vivo data to estimate the likelihood of metabolic pharmacokinetic interactions. Clin Pharmacokinet. 1997;32:210–258. doi: 10.2165/00003088-199732030-00004. [DOI](https://doi.org/10.2165/00003088-199732030-00004) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9084960/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacokinet&title=Use%20of%20in%20vitro%20data%20and%20in%20vivo%20data%20to%20estimate%20the%20likelihood%20of%20metabolic%20pharmacokinetic%20interactions&author=RJ%20Bertz&author=GR%20Grannemann&volume=32&publication_year=1997&pages=210-258&pmid=9084960&doi=10.2165/00003088-199732030-00004&)
|
| 102 |
+
|
| 103 |
+
2. Coleman T, Ellis SW, Martin IJ, Lennard MS, Tucker GT. 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) is N-demethylated by cytochromes P450 2D6, 1A2 and 3A4—implications for susceptibility to Parkinson’s disease. J Pharmacol Exp Ther. 1996;277:685–690. [PubMed](https://pubmed.ncbi.nlm.nih.gov/8627546/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Pharmacol%20Exp%20Ther&title=1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine%20(MPTP)%20is%20N-demethylated%20by%20cytochromes%20P450%202D6,%201A2%20and%203A4%E2%80%94implications%20for%20susceptibility%20to%20Parkinson%E2%80%99s%20disease&author=T%20Coleman&author=SW%20Ellis&author=IJ%20Martin&author=MS%20Lennard&author=GT%20Tucker&volume=277&publication_year=1996&pages=685-690&pmid=8627546&)
|
| 104 |
+
|
| 105 |
+
3. Gooderham NJ, Murray S, Lynch AM, et al. Heterocyclic amines: evaluation of their role in diet associated human cancer. Br J Clin Pharmacol. 1996;42:91–98. doi: 10.1046/j.1365-2125.1996.37513.x. [DOI](https://doi.org/10.1046/j.1365-2125.1996.37513.x) | [PMC free article](/articles/PMC2042638/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8807149/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=Heterocyclic%20amines:%20evaluation%20of%20their%20role%20in%20diet%20associated%20human%20cancer&author=NJ%20Gooderham&author=S%20Murray&author=AM%20Lynch&volume=42&publication_year=1996&pages=91-98&pmid=8807149&doi=10.1046/j.1365-2125.1996.37513.x&)
|
| 106 |
+
|
| 107 |
+
4. Catteau A, Bechtel YC, Poisson N, Bechtel PR, Bonaiti-Pellie C. A population and family study of CYP1A2 using caffeine urinary metabolites. Eur J Clin Pharmacol. 1995;47:423–430. doi: 10.1007/BF00196856. [DOI](https://doi.org/10.1007/BF00196856) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7720764/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Eur%20J%20Clin%20Pharmacol&title=A%20population%20and%20family%20study%20of%20CYP1A2%20using%20caffeine%20urinary%20metabolites&author=A%20Catteau&author=YC%20Bechtel&author=N%20Poisson&author=PR%20Bechtel&author=C%20Bonaiti-Pellie&volume=47&publication_year=1995&pages=423-430&pmid=7720764&doi=10.1007/BF00196856&)
|
| 108 |
+
|
| 109 |
+
5. Kalow W, Tang BK. Use of caffeine metabolite ratios to explore CYP1A2 and xanthine oxidase activities. Clin Pharmacol Ther. 1991;50:508–519. doi: 10.1038/clpt.1991.176. [DOI](https://doi.org/10.1038/clpt.1991.176) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1934864/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Use%20of%20caffeine%20metabolite%20ratios%20to%20explore%20CYP1A2%20and%20xanthine%20oxidase%20activities&author=W%20Kalow&author=BK%20Tang&volume=50&publication_year=1991&pages=508-519&pmid=1934864&doi=10.1038/clpt.1991.176&)
|
| 110 |
+
|
| 111 |
+
6. Fuhr U, Rost KL. Simple and reliable CYP1A2 phenotyping by the paraxanthine/caffeine ratio in plasma and in saliva. Pharmacogenetics. 1994;4:109–116. doi: 10.1097/00008571-199406000-00001. [DOI](https://doi.org/10.1097/00008571-199406000-00001) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7920690/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Simple%20and%20reliable%20CYP1A2%20phenotyping%20by%20the%20paraxanthine/caffeine%20ratio%20in%20plasma%20and%20in%20saliva&author=U%20Fuhr&author=KL%20Rost&volume=4&publication_year=1994&pages=109-116&pmid=7920690&doi=10.1097/00008571-199406000-00001&)
|
| 112 |
+
|
| 113 |
+
7. Eaton DL, Gallagher EP, Bammler TK, Kunze KL. Role of cytochrome P4501A2 in chemical carcinogenesis: implications for human variability in expression and enzyme activity. Pharmacogenetics. 1995;5:259–274. doi: 10.1097/00008571-199510000-00001. [DOI](https://doi.org/10.1097/00008571-199510000-00001) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8563766/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Role%20of%20cytochrome%20P4501A2%20in%20chemical%20carcinogenesis:%20implications%20for%20human%20variability%20in%20expression%20and%20enzyme%20activity&author=DL%20Eaton&author=EP%20Gallagher&author=TK%20Bammler&author=KL%20Kunze&volume=5&publication_year=1995&pages=259-274&pmid=8563766&doi=10.1097/00008571-199510000-00001&)
|
| 114 |
+
|
| 115 |
+
8. Rost KL, Brösicke H, Brockmöller J, Scheffler M, Helge H, Roots I. Increase of cytochrome P450IA2 activity by omeprazole: evidence by the 13C-[N-3-methyl]-caffeine breath test in poor and extensive metabolizers of S-mephenytoin. Clin Pharmacol Ther. 1992;52:170–180. doi: 10.1038/clpt.1992.126. [DOI](https://doi.org/10.1038/clpt.1992.126) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1505152/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&title=Increase%20of%20cytochrome%20P450IA2%20activity%20by%20omeprazole:%20evidence%20by%20the%2013C-%5BN-3-methyl%5D-caffeine%20breath%20test%20in%20poor%20and%20extensive%20metabolizers%20of%20S-mephenytoin&author=KL%20Rost&author=H%20Br%C3%B6sicke&author=J%20Brockm%C3%B6ller&author=M%20Scheffler&author=H%20Helge&volume=52&publication_year=1992&pages=170-180&pmid=1505152&doi=10.1038/clpt.1992.126&)
|
| 116 |
+
|
| 117 |
+
9. MacLeod S, Sinha R, Kadlubar FF, Lang NP. Polymorphisms of CYP1A1 and GSTM1 influence the in vivo function of CYP1A2. Mutat Res. 1997;376:135–142. doi: 10.1016/s0027-5107(97)00036-5. [DOI](https://doi.org/10.1016/s0027-5107(97)00036-5) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9202749/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mutat%20Res&title=Polymorphisms%20of%20CYP1A1%20and%20GSTM1%20influence%20the%20in%20vivo%20function%20of%20CYP1A2&author=S%20MacLeod&author=R%20Sinha&author=FF%20Kadlubar&author=NP%20Lang&volume=376&publication_year=1997&pages=135-142&pmid=9202749&doi=10.1016/s0027-5107(97)00036-5&)
|
| 118 |
+
|
| 119 |
+
10. Nakajima M, Yokoi T, Mizutani M, Shin S, Kadlubar FF, Kamataki T. Phenotyping of CYP1A2 in Japanese population by analysis of caffeine urinary metabolites: absence of mutation prescribing the phenotype in the CYP1A2 gene. Cancer Epidemiol Biomarkers Prev. 1994;3:413–421. [PubMed](https://pubmed.ncbi.nlm.nih.gov/7920209/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cancer%20Epidemiol%20Biomarkers%20Prev&title=Phenotyping%20of%20CYP1A2%20in%20Japanese%20population%20by%20analysis%20of%20caffeine%20urinary%20metabolites:%20absence%20of%20mutation%20prescribing%20the%20phenotype%20in%20the%20CYP1A2%20gene&author=M%20Nakajima&author=T%20Yokoi&author=M%20Mizutani&author=S%20Shin&author=FF%20Kadlubar&volume=3&publication_year=1994&pages=413-421&pmid=7920209&)
|
| 120 |
+
|
| 121 |
+
11. Yokoi T, Sawada M, Kamataki T. Polymorphic drug metabolism: studies with recombinant Chinese hamster cells and analyses in human populations. Pharmacogenetics. 1995;5:S65–S69. doi: 10.1097/00008571-199512001-00003. [DOI](https://doi.org/10.1097/00008571-199512001-00003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7581492/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Polymorphic%20drug%20metabolism:%20studies%20with%20recombinant%20Chinese%20hamster%20cells%20and%20analyses%20in%20human%20populations&author=T%20Yokoi&author=M%20Sawada&author=T%20Kamataki&volume=5&publication_year=1995&pages=S65-S69&pmid=7581492&doi=10.1097/00008571-199512001-00003&)
|
| 122 |
+
|
| 123 |
+
12. MacLeod SL, Tang Y-M, Yokoi Yokoi, et al. The role of recently discovered genetic polymorphism in the regulation of the human CYP1A2 gene. Proceedings of the American Association for Cancer Research. 1998;39:396. [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Proceedings%20of%20the%20American%20Association%20for%20Cancer%20Research&title=The%20role%20of%20recently%20discovered%20genetic%20polymorphism%20in%20the%20regulation%20of%20the%20human%20CYP1A2%20gene&author=SL%20MacLeod&author=Y-M%20Tang&author=Yokoi%20Yokoi&volume=39&publication_year=1998&pages=396&)
|
| 124 |
+
|
| 125 |
+
13. Grant DM, Tang BK, Kalow W. A simple test for acetylator phenotype using caffeine. Br J Clin Pharmacol. 1984;52:459–464. doi: 10.1111/j.1365-2125.1984.tb02372.x. [DOI](https://doi.org/10.1111/j.1365-2125.1984.tb02372.x) | [PMC free article](/articles/PMC1463406/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/6721992/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Br%20J%20Clin%20Pharmacol&title=A%20simple%20test%20for%20acetylator%20phenotype%20using%20caffeine&author=DM%20Grant&author=BK%20Tang&author=W%20Kalow&volume=52&publication_year=1984&pages=459-464&pmid=6721992&doi=10.1111/j.1365-2125.1984.tb02372.x&)
|
| 126 |
+
|
| 127 |
+
14. Butler MA, Lang NP, Young JF, et al. Determination of CYP1A2 and NAT2 phenotypes in human populations by analysis of caffeine urinary metabolites. Pharmacogenetics. 1992;2:116–127. doi: 10.1097/00008571-199206000-00003. [DOI](https://doi.org/10.1097/00008571-199206000-00003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1306111/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Determination%20of%20CYP1A2%20and%20NAT2%20phenotypes%20in%20human%20populations%20by%20analysis%20of%20caffeine%20urinary%20metabolites&author=MA%20Butler&author=NP%20Lang&author=JF%20Young&volume=2&publication_year=1992&pages=116-127&pmid=1306111&doi=10.1097/00008571-199206000-00003&)
|
| 128 |
+
|
| 129 |
+
15. Rostami-Hodjegan A, Nurminen S, Jackson PR, Tucker GT. Caffeine urinary metabolite ratios as markers of enzyme activity: a theoretical assessment. Pharmacogenetics. 1996;6:121–149. doi: 10.1097/00008571-199604000-00001. [DOI](https://doi.org/10.1097/00008571-199604000-00001) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9156692/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Caffeine%20urinary%20metabolite%20ratios%20as%20markers%20of%20enzyme%20activity:%20a%20theoretical%20assessment&author=A%20Rostami-Hodjegan&author=S%20Nurminen&author=PR%20Jackson&author=GT%20Tucker&volume=6&publication_year=1996&pages=121-149&pmid=9156692&doi=10.1097/00008571-199604000-00001&)
|
| 130 |
+
|
| 131 |
+
16. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Molecular%20cloning:%20a%20laboratory%20manual&author=J%20Sambrook&author=EF%20Fritsch&author=T%20Maniatis&publication_year=1989&)
|
| 132 |
+
|
| 133 |
+
17. Ikeya K, Jaiswal AK, Owens RA, Jones JE, Nebert DW, Kimura S. Human CYP1A2: sequence, gene structure, comparison with the mouse and rat orthologous gene, and differences in liver 1A2 mRNA expression. Mol Endocrinol. 1989;3:1399–1408. doi: 10.1210/mend-3-9-1399. [DOI](https://doi.org/10.1210/mend-3-9-1399) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/2575218/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mol%20Endocrinol&title=Human%20CYP1A2:%20sequence,%20gene%20structure,%20comparison%20with%20the%20mouse%20and%20rat%20orthologous%20gene,%20and%20differences%20in%20liver%201A2%20mRNA%20expression&author=K%20Ikeya&author=AK%20Jaiswal&author=RA%20Owens&author=JE%20Jones&author=DW%20Nebert&volume=3&publication_year=1989&pages=1399-1408&pmid=2575218&doi=10.1210/mend-3-9-1399&)
|
| 134 |
+
|
| 135 |
+
18. Daly AK, Brockmöller J, Broly F, et al. Nomenclature for human CYP2D6 alleles. Pharmacogenetics. 1996;6:193–201. doi: 10.1097/00008571-199606000-00001. [DOI](https://doi.org/10.1097/00008571-199606000-00001) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8807658/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenetics&title=Nomenclature%20for%20human%20CYP2D6%20alleles&author=AK%20Daly&author=J%20Brockm%C3%B6ller&author=F%20Broly&volume=6&publication_year=1996&pages=193-201&pmid=8807658&doi=10.1097/00008571-199606000-00001&)
|
test/texts/PMC2564574.md
ADDED
|
@@ -0,0 +1,227 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Polymorphisms in the VKORC1 gene are strongly associated with warfarin dosage requirements in patients receiving anticoagulation
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** T Li, L A Lange, X Li, L Susswein, B Bryant, R Malone, E M Lange, T‐Y Huang, D W Stafford, J P Evans
|
| 5 |
+
**Journal:** Journal of Medical Genetics
|
| 6 |
+
**Date:** 2006 Apr 12
|
| 7 |
+
**DOI:** [10.1136/jmg.2005.040410](https://doi.org/10.1136/jmg.2005.040410)
|
| 8 |
+
**PMID:** 16611750
|
| 9 |
+
**PMCID:** PMC2564574
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2564574/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC2564574/pdf/740.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC2564574/pdf/740.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
**Background:**
|
| 16 |
+
Warfarin is a mainstay of therapy for conditions associated with an increased risk of thromboembolic events. However, the use of this common agent is fraught with complications and little is known regarding inter‐individual variation in warfarin response.
|
| 17 |
+
|
| 18 |
+
**Objective:**
|
| 19 |
+
We tested for association between single nucleotide polymorphisms (SNPs) in VKORC1 and CYP2C9 and average weekly warfarin dose required to maintain patients at their desired anticoagulation target.
|
| 20 |
+
|
| 21 |
+
**Methods:**
|
| 22 |
+
The sample consisted of 93 European‐American patients from anticoagulation clinics at the University of North Carolina at Chapel Hill. Data on mean weekly warfarin dose were collected over a mean treatment period of 20.6 months. ANCOVA models were used and haplotype analysis was performed.
|
| 23 |
+
|
| 24 |
+
**Results:**
|
| 25 |
+
Three of six VKORC1 SNPs were found to be very strongly associated with the average warfarin dose required to achieve the target international normalised ratio (INR; p<0.0001). The mean weekly dose by genotype ranged from approximately 27 to 47 mg. There was no evidence for an association between either of the two CYP2C9 polymorphisms studied, CYP2C9*2 and CYP2C9*3. CYP2C9*3 was significantly (p = 0.05) associated with average warfarin dosage after adjustment for VKORC1*1173.
|
| 26 |
+
|
| 27 |
+
**Conclusions:**
|
| 28 |
+
These results are of considerable clinical interest and confirm recently published results regarding the role of these two genes in modifying warfarin metabolism and maintenance dosage. The consistent findings regarding the role of VKORC1 and CYP2C9 in warfarin metabolism and maintenance dosage represent a clinically useful proof of principal for the use of pharmacogenomic information in medicine and may lead to improved understanding of warfarin's actions.
|
| 29 |
+
|
| 30 |
+
Keywords: anticoagulation, CYP2C9, pharmacogenomics, VKORC1, warfarin
|
| 31 |
+
|
| 32 |
+
### Background
|
| 33 |
+
|
| 34 |
+
Warfarin is a mainstay of therapy for conditions associated with an increased risk of thromboembolic events. However, the use of this common agent is fraught with complications and little is known regarding inter‐individual variation in warfarin response.
|
| 35 |
+
|
| 36 |
+
### Objective
|
| 37 |
+
|
| 38 |
+
We tested for association between single nucleotide polymorphisms (SNPs) in *VKORC1*VKORC1 and *CYP2C9*CYP2C9 and average weekly warfarin dose required to maintain patients at their desired anticoagulation target.
|
| 39 |
+
|
| 40 |
+
### Methods
|
| 41 |
+
|
| 42 |
+
The sample consisted of 93 European‐American patients from anticoagulation clinics at the University of North Carolina at Chapel Hill. Data on mean weekly warfarin dose were collected over a mean treatment period of 20.6 months. ANCOVA models were used and haplotype analysis was performed.
|
| 43 |
+
|
| 44 |
+
### Results
|
| 45 |
+
|
| 46 |
+
Three of six *VKORC1*VKORC1 SNPs were found to be very strongly associated with the average warfarin dose required to achieve the target international normalised ratio (INR; p<0.0001). The mean weekly dose by genotype ranged from approximately 27 to 47 mg. There was no evidence for an association between either of the two *CYP2C9*CYP2C9 polymorphisms studied, *CYP2C9*2*CYP2C9*2 and *CYP2C9*3*CYP2C9*3. *CYP2C9*3*CYP2C9*3 was significantly (p = 0.05) associated with average warfarin dosage after adjustment for *VKORC1**VKORC1*1173.
|
| 47 |
+
|
| 48 |
+
### Conclusions
|
| 49 |
+
|
| 50 |
+
These results are of considerable clinical interest and confirm recently published results regarding the role of these two genes in modifying warfarin metabolism and maintenance dosage. The consistent findings regarding the role of *VKORC1*VKORC1 and *CYP2C9*CYP2C9 in warfarin metabolism and maintenance dosage represent a clinically useful proof of principal for the use of pharmacogenomic information in medicine and may lead to improved understanding of warfarin's actions.
|
| 51 |
+
|
| 52 |
+
**Keywords:**Keywords: anticoagulation, CYP2C9, pharmacogenomics, VKORC1, warfarin
|
| 53 |
+
|
| 54 |
+
## Methods
|
| 55 |
+
|
| 56 |
+
This study was approved by the Biomedical Institutional Review Board at the University of North Carolina at Chapel Hill. Patients attending anticoagulation clinics at UNC who were being treated with warfarin were approached for participation in the current study. After obtaining informed consent, blood was obtained from 93 European‐American patients for DNA analysis. An extensive database is maintained on these patients that includes the indication for treatment, duration of treatment, average dose, dosage adjustments, and international normalised ratio (INR). Mean weekly warfarin dose required to achieve each patient's target INR was gathered from the database over a mean period of treatment of 20.6 months. Genomic DNAs were extracted from whole blood using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA).
|
| 57 |
+
|
| 58 |
+
### Genomic sequencing
|
| 59 |
+
|
| 60 |
+
Direct sequencing of 60 patients identified six single nucleotide polymorphisms (SNPs) in the *VKORC1*VKORC1 gene. Approximately 10 ng DNA was used for PCR reactions. The primers used to amplify the 5′‐UTR and exon 1 region were: CCAATCGCCGAGTCAGAGG and CCCAGTCCCCAGCACTGTCT; primers for exon 2 and flanking region were: AGGGGAGGATAGGGTCAGTG and CCTGTTAGTTACCTCCCCACA; and primers for exon 3 and the 3′‐UTR were: ATACGTGCGTAAGCCACCAC and ACCCAGATATGCCCCCTTAG. Sequencing was via high throughput capillary electrophoresis. Genotyping for these SNPs was then performed in the remaining patients.
|
| 61 |
+
|
| 62 |
+
Assay of known SNPs was via real time PCR. The assay reagents for SNP genotyping were obtained from the Assay‐by‐Design service (Applied Biosystems, Foster City, CA). The primers and probes (FAM and VIC dye‐labelled) were designed using Primer Express software and were synthesised in Applied Biosystems (table 1). PCR reactions used 2× TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems). The real time PCR reactions were performed in an Opticon II system (Bio‐Rad Laboratories/MJ Research, Waltham, MA). Conditions were as follows: 95°C 10 min preheat, 92°C 15 s, 60°C 1 min followed by a plate reading, 40 cycles. The results were read according to the signal value of FAM and VIC dye.
|
| 63 |
+
|
| 64 |
+
### Table 1 The primers and probes used in real time PCR genotyping.
|
| 65 |
+
|
| 66 |
+
| VKOR SNPs | VIC probe sequence | FAM probe sequence | Forward primer | Reverse primer |
|
| 67 |
+
| --------- | ------------------ | ------------------ | -------------- | -------------- |
|
| 68 |
+
| VKOR 497 T>G | CCCCTTCaCCTGCGC | CCCCTTCcCCTGCGC | GCGGTAGAGATTGACGATGGT | GCAGCCATCGCCAACAC |
|
| 69 |
+
| VKOR 698 C>T | CAAGGCTgGTATAACG | CAAGGCTaGTATAACG | CTCTGATGCAAAACCGAGTGAAC | GCCCGGCCCTTAAGTAATTCTT |
|
| 70 |
+
| VKOR 1173 C>T | CCTAGTCCAAGgGTCGAT | CTAGTCCAAGaGTCGAT | CCCGGTGCCAGGAGATC | CACCTGGGCTATCCTCTGTTC |
|
| 71 |
+
| VKOR 1542 G>C | TCATCACgGAGCGTC | TCATCACcGAGCGTC | GGTGATCCACACAGCTGACA | CCTGTTAGTTACCTCCCCACATC |
|
| 72 |
+
| VKOR 2255 T>C | CCAGGACCaTGGTGC | CCAGGACCgTGGTGC | GCTCCAGAGAAGGCATCACT | GCCAAGTCTGAACCATGTGTCA |
|
| 73 |
+
| VKOR 3730 G>A | ATACCCgCACATGAC | CATACCCaCACATGAC | GTCCCTAGAAGGCCCTAGATGT | GTGTGGCACATTTGGTCCATT |
|
| 74 |
+
### Statistical methods
|
| 75 |
+
|
| 76 |
+
Each of the SNPs was first assessed to determine if the observed genotype frequencies were consistent with expected Hardy‐Weinberg proportions using Pearson's χ^2^2 tests. Pair‐wise marker‐marker linkage disequilibrium was assessed using Lewontin's D′ statistic[14](#ref14)^14^14 and Devlin and Risch's Δ^2^2 statistic[15](#ref15)^15^15 as implemented in the computer program GOLD ([www.sph.umich.edu/csg/abecasis/GOLD](http://www.sph.umich.edu/csg/abecasis/GOLD)www.sph.umich.edu/csg/abecasis/GOLD).
|
| 77 |
+
|
| 78 |
+
To test for association between individual SNPs and the continuously distributed outcomes, average warfarin dose, and average INR, we performed analysis of covariance models (ANCOVA) using PROC GLM within the SAS software system (version 8.0, SAS Institute, Cary, NC). The outcomes were first examined for adherence to distributional assumptions (including approximate normality of error terms conditional on covariates and homoscedasticity); the natural log transformation was subsequently applied to average dosage to adhere to these assumptions. Covariate adjustment was made for age, gender, and target INR in all analytic models. Genotype was tested for general association (no mode of inheritance assumption) using a 2 df F test. A 1 df F test was used to test the null hypotheses of no association for the *CYP2C9*CYP2C9 genotypes due to a lack of observed homozygotes for the *CYP2C9*2*CYP2C9*2 or **3**3 alleles. We additionally examined a two locus model, in which we selected the most statistically significant polymorphism from each of the two genes and tested the effects of each while controlling for the other. Finally, a model testing the interaction effects between the two polymorphisms was performed.
|
| 79 |
+
|
| 80 |
+
Tests of haplotype effects were performed using a score test developed by Schaid, as implemented in the computer program HAPLO.STAT ([http://mayoresearch.mayo.edu/mayo/research/biostat/schaid.cfm](http://mayoresearch.mayo.edu/mayo/research/biostat/schaid.cfm)http://mayoresearch.mayo.edu/mayo/research/biostat/schaid.cfm). HAPLO.STAT utilises a weighting scheme based on EM derived haplotype frequency estimates and weights every haplotype rather than assigning a “most likely” haplotype to an individual. Statistical differences in overall haplotype frequencies (excluding haplotypes with extremely low frequencies, for example 0.01) were tested, after adjustment for covariates (age, gender, and target INR), for association with the outcomes. Specific individual haplotype effects were also tested.
|
| 81 |
+
|
| 82 |
+
## Results
|
| 83 |
+
|
| 84 |
+
The sample consisted of 93 European‐American subjects with both genotype and phenotype data. The mean age was 63 years (SD 16, range 24–90 years) and 31% (n = 29) of the subjects were female. Mean treatment period for this study was 20.6 months (SD 11.0, range 12 days to 37.3 months). Indications for warfarin were predominantly atrial fibrillation or atrial flutter (45%, n = 47), with another large group (26%, n = 24) being treated or prophylaxed for venous thromboembolic events (VTE). Thirteen (14%) of the patients were receiving warfarin due to prosthetic mitral or aortic valves. The target INR for the majority of patients (83%, n = 77) was 2–3. Three of the prophylaxed VTE patients had a goal of 1.5–2.0. Thirteen patients had a goal of 2.5–3.5; this group included those with a prosthestic mitral valve as well as those with both a prosthetic atrial valve and atrial fibrillation. The positions and the corresponding observed allele frequencies of the identified SNPs in all 93 patients are given in table 2. The observed distributions of genotype data for the *VKORC1*VKORC1 and *CYP2C9*CYP2C9 SNPs were consistent with expected Hardy‐Weinberg proportions (data not shown).
|
| 85 |
+
|
| 86 |
+
### Table 2 VKORC1 and CYP2C9 SNP major allele frequencies.
|
| 87 |
+
|
| 88 |
+
| SNPs | Position | NCBI dbSNP rs# | Frequency (%) of wild type allele |
|
| 89 |
+
| ---- | -------- | -------------- | --------------------------------- |
|
| 90 |
+
| VKORC1*497 T>G | Intron 1 | Rs2884737 | 68.8 |
|
| 91 |
+
| VKORC1*698 C>T | Intron 1 | Rs17708472 | 79.6 |
|
| 92 |
+
| VKORC1*1173 C>T | Intron 1 | Rs9934438 | 54.8 |
|
| 93 |
+
| VKORC1*1542 G>C | Intron 2 | Rs8050894 | 56.5 |
|
| 94 |
+
| VKORC1*2255 T>C | Intron 2 | Rs2359612 | 44.1 |
|
| 95 |
+
| VKORC1*3730 G>A | 3′‐UTR | Rs7294 | 64.5 |
|
| 96 |
+
| CYP2C9*2 | Exon 3, C144R | Rs1799853 | 92.5 |
|
| 97 |
+
| CYP2C9*3 | Exon 7, L359I | Rs1057910 | 93.0 |
|
| 98 |
+
Table 3 displays the association results of the six *VKORC1*VKORC1 polymorphisms with mean weekly warfarin dose and INR in these patients. As expected, the observed INRs cluster tightly around 2.5 since warfarin doses were continually adjusted clinically in order to achieve an appropriate level of anticoagulation, with an INR of 2–3 being the most common target. Five of the six *VKORC1*VKORC1 SNPs were found to be statistically associated with average warfarin dose required to achieve the target INR (p ranging from <0.0001 to 0.021). The *VKORC1*VKORC1*1173, *VKORC1*VKORC1*1542, and *VKORC1*VKORC1*2255 SNPs were found to be the most strongly associated with the average warfarin dose required to achieve the target INR (p<0.0001). The trend in mean warfarin dosage by all three genotypes is consistent with an additive mode of inheritance, where the mean for the heterozygote is roughly midway between the two homozygotes
|
| 99 |
+
|
| 100 |
+
### Table 3 Individual VKORC1 gene SNP association analysis results.
|
| 101 |
+
|
| 102 |
+
| VKORC1 gene SNPs | n | Average (least square means) weekly warfarin dose (mg) | INR |
|
| 103 |
+
| ---------------- | - | ------------------------------------------------------ | --- |
|
| 104 |
+
| 497 T>G | | | |
|
| 105 |
+
| TT | 43 | 43.6 | 2.5 |
|
| 106 |
+
| TG | 42 | 34.4 | 2.7 |
|
| 107 |
+
| GG | 8 | 24.9 | 2.7 |
|
| 108 |
+
| p value | | 0.0015 | 0.12 |
|
| 109 |
+
| 698 C>T | | | |
|
| 110 |
+
| CC | 59 | 35.2 | 2.7 |
|
| 111 |
+
| CT | 30 | 40.0 | 2.5 |
|
| 112 |
+
| TT | 4 | 54.7 | 2.4 |
|
| 113 |
+
| p value | | 0.056 | 0.18 |
|
| 114 |
+
| 1173 C>T | | | |
|
| 115 |
+
| CC | 31 | 47.1 | 2.5 |
|
| 116 |
+
| CT | 40 | 35.8 | 2.6 |
|
| 117 |
+
| TT | 22 | 26.9 | 2.7 |
|
| 118 |
+
| p value | | <0.0001 | 0.10 |
|
| 119 |
+
| 1542 G>C | | | |
|
| 120 |
+
| GG | 32 | 46.4 | 2.5 |
|
| 121 |
+
| GC | 41 | 36.0 | 2.6 |
|
| 122 |
+
| CC | 20 | 26.5 | 2.7 |
|
| 123 |
+
| p value | | <0.0001 | 0.11 |
|
| 124 |
+
| 2255 T>C | | | |
|
| 125 |
+
| TT | 20 | 26.5 | 2.7 |
|
| 126 |
+
| TC | 42 | 35.7 | 2.6 |
|
| 127 |
+
| CC | 31 | 47.1 | 2.5 |
|
| 128 |
+
| p value | | <0.0001 | 0.11 |
|
| 129 |
+
| 3730 G>A | | | |
|
| 130 |
+
| GG | 40 | 33.3 | 2.6 |
|
| 131 |
+
| GA | 40 | 39.7 | 2.6 |
|
| 132 |
+
| AA | 13 | 46.4 | 2.5 |
|
| 133 |
+
| p value | | 0.021 | 0.63 |
|
| 134 |
+
The *VKORC1**VKORC1*1173, *1542, and *2255 SNPs were found to be in very strong linkage disequilibrium (LD) with each other, with estimated D′ values of 1.00 and estimated Δ^2^2 values ranging from 0.94 to 0.98. Estimated D′ values ranged from 0.84 to 1.00 and estimated Δ^2 ^2 values ranged from 0.11 to 0.56 for all other SNP pair combinations. Haplotype analysis results are reported in table 4. Four haplotypes were observed with an estimated frequency greater than 3%. The permutation based global p value for any difference between haplotypes was 0.0004. All four of the individual haplotype specific tests were found to be statistically significant (p<0.05). The two haplotypes (EA1 and EA2) characterised by the T allele for *VKOR*VKOR 1173, the C allele for *VKOR*VKOR 1542, and the T allele for *VKOR*VKOR 2255 were associated with negative score statistics (indicating lower mean dosage), while the other two haplotypes were associated with positive score statistics (indicating higher mean dosage). Overall, the haplotype results are consistent with the single SNP results and suggest that the causal polymorphism is one of the identified SNPs (*VKORC1*VKORC1*1173, *1542, or ****2255) or another polymorphism in LD with these three SNPs.
|
| 135 |
+
|
| 136 |
+
### Table 4 Association between VKORC1 gene haplotypes and log average dose.
|
| 137 |
+
|
| 138 |
+
| Haplotype | VKORC1 gene SNPs | | Score* | p† |
|
| 139 |
+
| --------- | ---------------- | - | ------ | -- |
|
| 140 |
+
| 497 | 698 | 1173 | 1542 | 2255 | 3730 | Estimated frequency (%) |
|
| 141 |
+
| EA1 | G | C | T | C | T | G | 30.1 | −3.42 | 0.00063 |
|
| 142 |
+
| EA2 | T | C | T | C | T | G | 12.9 | −2.26 | 0.024 |
|
| 143 |
+
| EA3 | T | T | C | G | C | G | 19.3 | 2.47 | 0.013 |
|
| 144 |
+
| EA4 | T | C | C | G | C | A | 34.9 | 2.73 | 0.0063 |
|
| 145 |
+
| Global‡ | | | | | | | | | 0.0004 |
|
| 146 |
+
The *CYP2C9*CYP2C9 SNP association results are presented in table 5. Only 14 and 13 heterozygotes were observed for *CYP2C9*2*CYP2C9*2 and *CYP2C9*3*CYP2C9*3 polymorphisms, respectively. There was no evidence for an association between *CYP2C9*CYP2C9 genotypes and average warfarin weekly dose before additional adjustment for *VKORC1*VKORC1 genotype. *CYP2C9*3*CYP2C9*3 became marginally statistically significant (p = 0.050) after adjustment for *VKOR 1173*VKOR 1173. The *CYP2C9*2*CYP2C9*2 and *CYP2C9*3*CYP2C9*3 SNPs were not found to be in significant LD with one another (D′ = 0.04, Δ^2^2 = 0.00, p = 0.97). Given the lack of LD between the two SNPs, haplotype analyses were not performed.
|
| 147 |
+
|
| 148 |
+
### Table 5 Association between CYP2C9 genotypes and log average dose in European‐American subjects.
|
| 149 |
+
|
| 150 |
+
| CYP2C9 gene SNPs | n | Least square means | INR |
|
| 151 |
+
| ---------------- | - | ------------------ | --- |
|
| 152 |
+
| Average weekly dose (mg) | Average weekly dose (mg) after adjustment for VKORC1*1173 | | |
|
| 153 |
+
| CYP2C9*2 | | | | |
|
| 154 |
+
| 11 | 79 | 38.8 | 37.3 | 2.6 |
|
| 155 |
+
| 12 | 14 | 32.1 | 31.0 | 2.6 |
|
| 156 |
+
| 22 | 0 | – | – | – |
|
| 157 |
+
| p value | | 0.19 | 0.16 | 0.93 |
|
| 158 |
+
| CYP2C9*3 | | | | |
|
| 159 |
+
| 11 | 80 | 38.5 | 37.1 | 2.6 |
|
| 160 |
+
| 12 | 13 | 33.0 | 31.5 | 2.7 |
|
| 161 |
+
| 22 | 0 | – | – | – |
|
| 162 |
+
| p value | | 0.24 | 0.050 | 0.12 |
|
| 163 |
+
## Discussion
|
| 164 |
+
|
| 165 |
+
We have here shown that variants within *VKORC1*VKORC1 are strongly associated with the mean weekly dose of warfarin required to maintain a desired target INR in a sample of European‐American outpatients undergoing anticoagulation treatment. In addition to their robust statistical significance, the results presented here are potentially clinically meaningful given the notoriously narrow therapeutic window[16](#ref16)^16^16 of the drug in question. The mean weekly dose of warfarin required to maintain the desired target INR is almost doubled (from 27 to 47 mg) for those who are homozygous C/C at position 1173, G/G at position 1542, or C/C at position 2255, as compared with the homozygotes for the opposite allele at each site. Also bolstering the significance of these results is the dose‐response curve observed in heterozygotes, who demonstrate intermediate warfarin sensitivity. Moreover, these alleles are common in this unselected sample with an estimated prevalence of 54.2% for the *VKORC1*VKORC1*1173 C allele, 55.7% for the *VKORC1*VKORC1*1542 G allele, and 54.7% for the *VKORC1*VKORC1*2255 C allele. These results are strongly consistent with recent findings regarding the effects of *VKORC1*VKORC1 genotype on warfarin dosage levels required to reach desired INR both in terms of overall statistical significance and mean warfarin dosage levels by genotype.[7](#ref7)^7^7,[8](#ref8)^8^8,[9](#ref9)^9^9,[10](#ref10)^10^10,[11](#ref11)^11^11,[12](#ref12)^12^12 For example, D'Andrea *et al*et al[7](#ref7)^7^7 report an observed warfarin mean weekly dosage of 49.0, 35.7, and 25.9 (calculated by multiplying their reported mean daily dosage by 7) for *VKORC1*VKORC1*1173 genotypes C/C, C/T, and T/T, respectively, as compared with our observed weekly mean dosages of 47.1, 35.8, and 26.9, respectively, for the same genotypes. In addition, there is considerable agreement with the direction of the haplotype based associations. In our study, EA1 and EA2 are associated with lower mean levels of warfarin dosage and haplotypes EA3 and EA4 are associated with higher mean levels of warfarin dosage. Our haplotyes EA1 and EA2 have identical alleles as do haplotypes H3a and H3b in Wadelius *et al*et al[10](#ref10)^10^10 for *VKORC1**VKORC1*1173, *2255, and *3730. Our haplotype EA3 corresponds to H2 and our haplotype EA4 corresponds to H1 in Wadelius *et al*et al,[10](#ref10)^10^10 where H1 and H2 are also associated with higher warfarin dosage. Finally, our haplotypes EA1, EA2, EA3, and EA4 correspond to the sets of haplotypes {H2, H5}, {H1, H3}, {H4, H6, H9}, and {H7, H8}, respectively, in Rieder *et al*et al[8](#ref8)^8^8; the direction of results between the two studies with respect to which haplotypes are associated with higher or lower warfarin dosage levels is also entirely consistent.
|
| 166 |
+
|
| 167 |
+
In addition to our *VKORC1*VKORC1 results, we did not detect a statistically significant association between the two *CYP2C9*CYP2C9 polymorphisms and warfarin dosage before adjustment for *VKORC1*VKORC1; however, results for *CYP2C9*3*CYP2C9*3 did reach a marginal level of statistical significance (p = 0.05) after adjustment for *VKORC1**VKORC1*1173. Because the *CYP2C9*CYP2C9 SNPs have lower minor allele frequencies than the *VKORC1*VKORC1 SNPs examined, the power to detect significant departures from the null hypothesis of no association for *CYP2C9*CYP2C9 was less than that for *VKORC1*VKORC1.
|
| 168 |
+
|
| 169 |
+
In previously reported work, Sconce *et al*et al[9](#ref9)^9^9 demonstrated the best multivariable regression model for fitting warfarin dosage included both *CYP2C9*CYP2C9 and *VKORC1*VKORC1. Our multivariable model including age, gender, target INR, and *VKORC1*VKORC1*1173 genotype explained approximately 30% (R^2^2 = 0.30) of our total observed variation. Our multivariable model including age, gender, target INR, and *CYP2C9*3*CYP2C9*3 explained only 12% of the total variation. Including both *CYP2C9*3*CYP2C9*3 and *VKORC1*VKORC1*1173 along with our covariates explained 34% of the total variation. Thus, our multivariable model appeared to explain less of the total variation in the mean warfarin dosage required to reach the target INR than the multivariable regression model of Sconce *et al*et al[9](#ref9)^9^9 (R^2^2 = 0.55). It should be noted that their model included height, a measure that we did not have available to us for this study. In addition, our results suggest, consistent with the findings of Wadelius *et al*et al[10](#ref10)^10^10, that *VKORC1*VKORC1 polymorphisms explain a larger amount of the observed variation in mean warfarin dosage required to reach the target INR than do *CYP2C9*CYP2C9 polymorphisms in the European‐American population.
|
| 170 |
+
|
| 171 |
+
During the course of our study, we also collected data on warfarin dosage requirements to reach the target INR for 18 African‐Americans. Of these 18 patients, one had a target INR of 1.5–2.0, 14 had a target INR of 2.0–3.0, and three had a target INR of 2.5–3.5. Marginally statistically significant results were observed for *VKORC1*VKORC1*1173 (p = 0.03) and *2255 (p = 0.03) and *CYP2C9*3*CYP2C9*3 (p = 0.01) in this limited data set. These results could be entirely explained by the low observed warfarin dosages (average weekly dosage of 9.3 mg) of a single individual who was homozygous T/T at both *VKORC1**VKORC1*1173 and *2255 and heterozygous at *CYP2C9*3*CYP2C9*3. This individual's genotypes at these three markers were consistent with the genotypes that were shown to be associated with low warfarin dosage in the European‐American population. In addition, this individual had the lowest target INR (1.75) and was also the only African‐American individual in our sample with these aforementioned genotypes. We note that in our limited sample, there were strong differences in allele frequencies (data not shown) between the European‐American and African‐American samples. For example, *VKORC1**VKORC1*698 and *CYP2C9*2*CYP2C9*2 were uninformative (completely homozygous for the wild type allele) in this small African‐American sample. We also note that we observed weaker marker‐marker LD and a larger diversity in haplotypes in this limited African‐American sample (data not shown). These findings are all consistent with the comparative results between the European and African‐Yoruban samples genotyped at *VKORC1*VKORC1 as part of the International HapMap Project ([www.hapmap.org](http://www.hapmap.org)www.hapmap.org). While these results based on 18 African‐American patients are limited and inconclusive, they do suggest that it is important to assess the role of *VKORC1*VKORC1 SNPs in warfarin response in other populations and that studying different populations with different haplotype patterns may be critical in ultimately identifying the functionally important variants.
|
| 172 |
+
|
| 173 |
+
It remains undetermined whether one or more of the SNPs identified here or elsewhere are causal in producing differential warfarin sensitivity or whether they are in LD with the actual causative SNP. Strong LD between *VKORC1*VKORC1 polymorphisms in the European‐American population hinders our ability to fine map/identify the functional loci. Further analysis of warfarin related gene(s) and surrounding sequence and the inclusion of larger numbers of patients, ideally in different populations, may lead to elucidation of the specific genotypic factors which influence warfarin sensitivity. In addition, prospective studies will ultimately be critical to establishing the clinical utility of assessing *VKORC1*VKORC1 genotype for determining optimal warfarin dosage. Nonetheless, combining genotype data from SNPs of the *VKORC1*VKORC1 gene with SNPs of the cytochrome *CYP2C9*CYP2C9 gene in a clinical setting promises to result in a more robust assessment of warfarin sensitivity.
|
| 174 |
+
|
| 175 |
+
The results presented here support those from other recent reports[7](#ref7)^7^7,[8](#ref8)^8^8,[9](#ref9)^9^9,[10](#ref10)^10^10,[11](#ref11)^11^11,[12](#ref12)^12^12 and further demonstrate the need for prospective studies to evaluate the utility of *VKORC1*VKORC1 and *CYP2C9*CYP2C9 genotype information with respect to outcomes such as length of time until the target INR is reached and adverse events. This current demonstration of common polymorphisms that predict warfarin sensitivity represents a potentially clinically useful proof of principal for the use of pharmacogenomic information in medicine and will likely play a critical role in future predictive models for warfarin dosage. Such models will have strong potential to guide physicians in quickly prescribing appropriate dosages and, ultimately, avoid complications.
|
| 176 |
+
|
| 177 |
+
## Electronic‐database information
|
| 178 |
+
|
| 179 |
+
Information about the computer program GOLD is at [www.sph.umich.edu/csg/abecasis/GOLD,](http://www.sph.umich.edu/csg/abecasis/GOLD,)www.sph.umich.edu/csg/abecasis/GOLD, the computer program HAPLO.STAT at [http://mayoresearch.mayo.edu/mayo/research/biostat/schaid.cfm](http://mayoresearch.mayo.edu/mayo/research/biostat/schaid.cfm)http://mayoresearch.mayo.edu/mayo/research/biostat/schaid.cfm, and the International HapMap Project at [www.hapmap.org](http://www.hapmap.org)www.hapmap.org.
|
| 180 |
+
|
| 181 |
+
## Abbreviations
|
| 182 |
+
|
| 183 |
+
ANCOVA - analysis of covariance
|
| 184 |
+
|
| 185 |
+
INR - international normalised ratio
|
| 186 |
+
|
| 187 |
+
LD - linkage disequilibrium
|
| 188 |
+
|
| 189 |
+
SNP - single nucleotide polymorphism
|
| 190 |
+
|
| 191 |
+
VTE - venous thromboembolic events
|
| 192 |
+
|
| 193 |
+
## Footnotes
|
| 194 |
+
|
| 195 |
+
## References
|
| 196 |
+
|
| 197 |
+
1. Shulman S. Oral anticoagulation. In: Beutler E, Lichtman MA, Coller BS, Kipps TJ, Seligsohn U, eds. Williams hematology. 6th ed. New York: McGraw‐Hill, 20011777–1792.
|
| 198 |
+
|
| 199 |
+
2. Levine M N, Raskob G, Landefeld S, Kearon C. Hemorrhagic complications of anticoagulant treatment. Chest 2001119108S–21S. [DOI](https://doi.org/10.1378/chest.119.1_suppl.108s) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11157645/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Chest&volume=119&publication_year=2001&pages=108S-21S&pmid=11157645&doi=10.1378/chest.119.1_suppl.108s&)
|
| 200 |
+
|
| 201 |
+
3. Poller L, Shiach C R, MacCallum P K, Johansen A M, Munster A M, Magalhaes A, Jespersen J. Multicentre randomised study of computerised anticoagulant dosage. European Concerted Action on Anticoagulation. Lancet 19983521505–1509. [DOI](https://doi.org/10.1016/s0140-6736(98)04147-6) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9820298/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&volume=352&publication_year=1998&pages=1505-1509&pmid=9820298&doi=10.1016/s0140-6736(98)04147-6&)
|
| 202 |
+
|
| 203 |
+
4. Doecke C J, Cosh D G, Gallus A S. Standardised initial warfarin treatment: evaluation of initial treatment response and maintenance dose prediction by randomised trial, and risk factors for an excessive warfarin response. Aust N Z J Med 199121319–324. [DOI](https://doi.org/10.1111/j.1445-5994.1991.tb04697.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/1953510/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Aust%20N%20Z%20J%20Med&volume=21&publication_year=1991&pages=319-324&pmid=1953510&doi=10.1111/j.1445-5994.1991.tb04697.x&)
|
| 204 |
+
|
| 205 |
+
5. Li T, Chang C Y, Jin D Y, Lin P J, Khvorova A, Stafford D W. Identification of the gene for vitamin K epoxide reductase. Nature 2004427541–544. [DOI](https://doi.org/10.1038/nature02254) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14765195/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nature&volume=427&publication_year=2004&pages=541-544&pmid=14765195&doi=10.1038/nature02254&)
|
| 206 |
+
|
| 207 |
+
6. Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hortnagel K, Pelz H J, Lappegard K, Seifried E, Scharrer I, Tuddenham E G, Muller C R, Strom T M, Oldenburg J. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 2004427537–541. [DOI](https://doi.org/10.1038/nature02214) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/14765194/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nature&volume=427&publication_year=2004&pages=537-541&pmid=14765194&doi=10.1038/nature02214&)
|
| 208 |
+
|
| 209 |
+
7. D'Andrea G, D'Ambrosio R L, Di Perna P, Chetta M, Santacroce R, Brancaccio V, Grandone E, Margaglione M. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose‐anticoagulant effect of warfarin. Blood 2005105645–649. [DOI](https://doi.org/10.1182/blood-2004-06-2111) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15358623/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Blood&volume=105&publication_year=2005&pages=645-649&pmid=15358623&doi=10.1182/blood-2004-06-2111&)
|
| 210 |
+
|
| 211 |
+
8. Rieder M J, Reiner A P, Gage B F, Nickerson D A, Eby C S, McLeod H L, Blough D K, Thummel K E, Veenstra D L, Rettie A E. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med 20053522285–2293. [DOI](https://doi.org/10.1056/NEJMoa044503) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15930419/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&volume=352&publication_year=2005&pages=2285-2293&pmid=15930419&doi=10.1056/NEJMoa044503&)
|
| 212 |
+
|
| 213 |
+
9. Sconce E A, Khan T I, Wynne H A, Avery P, Monkhouse L, King B P, Wood P, Kesteven P, Daly A K, Kamali F. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood 2005106(7)2329–2333. [DOI](https://doi.org/10.1182/blood-2005-03-1108) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15947090/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Blood&volume=106&publication_year=2005&pages=2329-2333&pmid=15947090&doi=10.1182/blood-2005-03-1108&)
|
| 214 |
+
|
| 215 |
+
10. Wadelius M, Chen L Y, Downes K, Ghori J, Hunt S, Eriksson N, Wallerman O, Melhus H, Wadelius C, Bentley D, Deloukas P. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J 20055(4)262–270. [DOI](https://doi.org/10.1038/sj.tpj.6500313) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15883587/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Pharmacogenomics%20J&volume=5&publication_year=2005&pages=262-270&pmid=15883587&doi=10.1038/sj.tpj.6500313&)
|
| 216 |
+
|
| 217 |
+
11. Yuan H Y, Chen J J, Lee M T, Wung J C, Chen Y F, Charng M J, Lu M J, Hung C R, Wei C Y, Chen C H, Wu J Y, Chen Y T. A novel functional VKORC1 promoter polymorphism is associated with inter‐individual and inter‐ethnic differences in warfarin sensitivity. Hum Mol Genet 2005141745–1751. [DOI](https://doi.org/10.1093/hmg/ddi180) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15888487/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Hum%20Mol%20Genet&volume=14&publication_year=2005&pages=1745-1751&pmid=15888487&doi=10.1093/hmg/ddi180&)
|
| 218 |
+
|
| 219 |
+
12. Lee S C, Ng S S, Oldenburg J, Chong P Y, Rost S, Guo J Y, Yap H L, Rankin S C, Khor H B, Yeo T C, Ng K S, Soong R, Goh B C. Interethnic variability of warfarin maintenance requirement is explained by VKORC1 genotype in an Asian population. Clin Pharmacol Ther 200679197–205. [DOI](https://doi.org/10.1016/j.clpt.2005.11.006) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16513444/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Pharmacol%20Ther&volume=79&publication_year=2006&pages=197-205&pmid=16513444&doi=10.1016/j.clpt.2005.11.006&)
|
| 220 |
+
|
| 221 |
+
13. Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin‐treated patients: a HuGEnet systematic review and meta‐analysis. Genet Med 2005797–104. [DOI](https://doi.org/10.1097/01.gim.0000153664.65759.cf) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15714076/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genet%20Med&volume=7&publication_year=2005&pages=97-104&pmid=15714076&doi=10.1097/01.gim.0000153664.65759.cf&)
|
| 222 |
+
|
| 223 |
+
14. Lewontin R C. The interaction of selection and linkage. I. General considerations. Genetics 19644949–67. [DOI](https://doi.org/10.1093/genetics/49.1.49) | [PMC free article](/articles/PMC1210557/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17248194/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genetics&volume=49&publication_year=1964&pages=49-67&pmid=17248194&doi=10.1093/genetics/49.1.49&)
|
| 224 |
+
|
| 225 |
+
15. Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine‐scale mapping. Genomics 199529311–322. [DOI](https://doi.org/10.1006/geno.1995.9003) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8666377/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genomics&volume=29&publication_year=1995&pages=311-322&pmid=8666377&doi=10.1006/geno.1995.9003&)
|
| 226 |
+
|
| 227 |
+
16. Schulman S. Clinical practice. Care of patients receiving long‐term anticoagulant therapy. N Engl J Med 2003349675–683. [DOI](https://doi.org/10.1056/NEJMcp025373) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12917305/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&volume=349&publication_year=2003&pages=675-683&pmid=12917305&doi=10.1056/NEJMcp025373&)
|
test/texts/PMC2596476.md
ADDED
|
@@ -0,0 +1,281 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# A GRK5 Polymorphism that Inhibits β-Adrenergic Receptor Signaling is Protective in Heart Failure
|
| 2 |
+
|
| 3 |
+
## Metadata
|
| 4 |
+
**Authors:** Stephen B Liggett, Sharon Cresci, Reagan J Kelly, Faisal M Syed, Scot J Matkovich, Harvey S Hahn, Abhinav Diwan, Jeffrey S Martini, Li Sparks, Rohan R Parekh, John A Spertus, Walter J Koch, Sharon L R Kardia, Gerald W Dorn, II
|
| 5 |
+
**Journal:** Nature medicine
|
| 6 |
+
**Date:** 2008 Apr 20
|
| 7 |
+
**DOI:** [10.1038/nm1750](https://doi.org/10.1038/nm1750)
|
| 8 |
+
**PMID:** 18425130
|
| 9 |
+
**PMCID:** PMC2596476
|
| 10 |
+
**URL:** https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2596476/
|
| 11 |
+
**PDF:** [https://pmc.ncbi.nlm.nih.gov/articles/PMC2596476/pdf/nihms65652.pdf](https://pmc.ncbi.nlm.nih.gov/articles/PMC2596476/pdf/nihms65652.pdf)
|
| 12 |
+
|
| 13 |
+
## Abstract
|
| 14 |
+
|
| 15 |
+
β-adrenergic receptor (βAR) blockade is standard therapy for cardiac failure and ischemia. G-protein coupled receptor kinases (GRKs) desensitize βAR, suggesting that genetic GRK variants might modify outcomes in these syndromes. Re-sequencing of GRK2 and GRK5 revealed a non-synonymous polymorphism of GRK5, common in African Americans (AA), substituting leucine (L) for glutamine (Q) at position 41. GRK5-L41 more effectively uncoupled isoproterenol-stimulated responses than GRK5-Q41 in transfected cells and transgenic mice, and like pharmacological βAR blockade, GRK5-L41 protected against experimental catecholamine-induced cardiomyopathy. Human association studies showed a pharmacogenomic interaction between GRK5-L41 and β-blocker treatment on mortality outcome in independent cohorts of AA cardiac failure (P=0.036) and ischemia (P=0.023). In 375 prospectively followed AA heart failure subjects, GRK5-L41 was protective against death/cardiac transplant (single allele: RR=0.28, 95% CI=0.12-0.66; two alleles: RR=0.08, 95% CI=0.04-0.19; P=0.004). The gain-of-function GRK5-L41 polymorphism facilitates βAR desensitization during catecholamine excess, imparting “genetic β-blockade” and improving survival in heart failure.
|
| 16 |
+
|
| 17 |
+
## INTRODUCTION
|
| 18 |
+
|
| 19 |
+
Heart failure is an incurable syndrome of multiple causes that will affect one in five adults, conferring a ~25% chance of dying within a year of diagnosis and a mortality rate of ~50% at five years [1](#R1)^1^1^,^,[2](#R2)^2^2. Heart failure management is complicated by disease heterogeneity in both inherited genetic cardiomyopathies [3](#R3)^3^3 and the more common non-familial dilated and ischemic cardiomyopathies [4](#R4)^4^4^–^–[7](#R7)^7^7. We and others have proposed that inter-individual differences in genetic polymorphisms involving catecholamine signaling pathways can modify heart failure risk, prognosis, or response to treatment. Especially relevant would be pharmacogenomic interactions between genetic variants of catecholamine receptors or their effectors and β-adrenergic receptor (βAR) antagonism (β-blockade), which is a standard therapy for heart failure and myocardial ischemia [8](#R8)^8^8. Indeed, pharmacogenomics of β-blockade might have special importance as this therapy prolongs life and ameliorates symptoms, but concomitantly impairs a critical mechanism for acutely increasing cardiac output in response to physiological stress. Maintaining the proper balance between preservation of βAR signaling and avoidance of βAR-mediated catecholamine toxicity may therefore be critical to outcome.
|
| 20 |
+
|
| 21 |
+
An important mechanism downregulating βAR signaling in heart failure is increased expression of myocardial G-protein receptor kinase (GRK)-2, which phosphorylates cardiac βAR, leading to recruitment of β-arrestin and receptor uncoupling from G-proteins and downstream signaling effectors [9](#R9)^9^9. While multiple studies have shown that expression of a GRK2-inhibiting mini-gene can improve cardiac function in experimental models of heart failure [10](#R10)^10^10^,^,[11](#R11)^11^11, cardiac-specific ablation of GRK2 in mice actually accelerates catecholamine-induced heart failure [12](#R12)^12^12. Thus, the effects of GRK2 on heart function appear to depend both upon expression level and pathophysiological context. The function of the other dominant GRK in the heart, GRK5 [13](#R13)^13^13, has not been as well defined: Genetic GRK5 ablation was not associated with a cardiac phenotype in mice [14](#R14)^14^14, but massive cardiac overexpression of bovine GRK5 depressed cardiac βAR responsiveness [15](#R15)^15^15^,^,[16](#R16)^16^16. Differences between GRK5 and GRK2 in sub-cellular localization, mechanism of activation, and receptor specificity suggest that these two cardiac GRKs may have non-redundant modulatory roles in the heart. Of particular interest is the rapid up- and down-regulation of GRK2 that correlates with ventricular function [17](#R17)^17^17^,^,[18](#R18)^18^18, which implies that the role of this GRK in the heart may be acute regulation. On the other hand, GRK5 expression appears to less dynamic and may therefore be more important in chronic regulation [19](#R19)^19^19^,^,[20](#R20)^20^20. As such, GRK5-mediated βAR desensitization could provide adaptive, beneficial effects during early ventricular decompensation, and prior to frank failure.
|
| 22 |
+
|
| 23 |
+
To examine the above notion, we searched for human genetic variants of cardiac-expressed GRK2 and GRK5 that might impart risk, modify the course, or alter the response to therapy of heart failure. We identified a non-synonymous polymorphism of GRK5 that changes amino acid 41 in the non-catalytic regulatory domain from glutamine (Q, the most common allele) to leucine (L). In levels of increasing complexity, we have defined the phenotype of this polymorphism in transfected cells, transgenic mice, and in two independent cohorts of cardiac disease among African Americans, in whom the polymorphism is common. We show that the GRK5-L41 variant augments βAR desensitization and represents a form of “genetic β-blockade” that diminishes βAR signaling, confers resistance to experimental catecholamine-induced cardiomyopathy, and protects against early death in African Americans with heart failure.
|
| 24 |
+
|
| 25 |
+
## RESULTS
|
| 26 |
+
|
| 27 |
+
### GRK5, but not GRK2, exhibits genetic variability
|
| 28 |
+
|
| 29 |
+
Of the seven human GRKs, GRK5 and GRK2 predominate in myocardium [10](#R10)^10^10^,^,[12](#R12)^12^12^,^,[13](#R13)^13^13. Polymorphism discovery in the 16 exons of GRK5 and 21 exons of GRK2 was performed by resequencing 96 DNA samples (Human Variation Collection of the Coriell Institute [[http://ccr.coriell.org/nigms/cells/humdiv.html](http://ccr.coriell.org/nigms/cells/humdiv.html)http://ccr.coriell.org/nigms/cells/humdiv.html]) from individuals of diverse ethnicity (40 Caucasians, 40 African American, 16 Asians), providing a 98% probability of detecting polymorphisms with allele frequencies as low as 0.02. Four non-synonymous polymorphisms were detected for GRK5, at cDNA nucleic acid positions 122 (A/T), 840 (G/A), 1274 (C/T) and 1624 (C/G), resulting in amino acid changes at residues 41 (Q to L) (rs17098707), 304 (R to H) (rs12718341), 425 (T to M) and 542 (P to A). The GRK5-Q41L variant was the only one with an allele frequency greater than 2% in any ethnic group, and was therefore studied further. In contrast to GRK5, we found no non-synonymous polymorphisms of the GRK2 coding exons, including the D457V and K465M variants reported in dbSNP (NCBI SNP Cluster ID # rs1977983 and rs1977982). To further confirm this, templates from the Whitehead Institute with the reported polymorphisms were sent to the University of Cincinnati and sequenced; these two SNPs were not confirmed.
|
| 30 |
+
|
| 31 |
+
### GRK5-Q41 and -L41 differentially affect β-adrenergic receptor desensitization in transfected CHO cells
|
| 32 |
+
|
| 33 |
+
Amino acid 41 of GRK5 is adjacent to a lipid and calmodulin binding domain. To determine the significance of the Q to L substitution at this position on the canonical desensitization function of GRK5, the effects of recombinantly expressed GRK5-Q41 and -L41 on β_1_1AR desensitization were examined during continuous agonist exposure. Chinese Hamster Ovary (CHO) cells were co-transfected with human β_1_1AR (Arg389 variant, which is the most common) and either GRK5-Q41 or -L41. Receptor expression (determined by radioligand binding, data not shown) and GRK expression as determined by Western blotting were equivalent (not shown). The rate and maximal level of cAMP accumulation over time in response to 10 μM isoproterenol in these cells is a measure of receptor coupling to Gαs/adenylyl cyclase, and is inversely related to receptor desensitization. As shown in [Figure 1a](#F1)Figure 1a, GRK5-L41 cells had a different desensitization pattern compared to GRK-Q41 (*P*P < 0.001 by ANOVA), with a ~25% decrease in the rate of cAMP accumulation (4.7±1.2 vs 6.4±1.9, *P*P<0.05) and a ~33% decrease in maximal response (2.6±0.2 vs 3.8±0.4% conversion, *P*P<0.05), compared to GRK5-Q41. Thus, the GRK5-L41 polymorphism decreases β_1_1AR signaling by enhancing agonist-promoted desensitization.
|
| 34 |
+
|
| 35 |
+
### Figure 1. Characteristics of mouse hearts expressing GRK5-Q41 and GRK5-L41.
|
| 36 |
+
|
| 37 |
+
|
| 38 |
+
|
| 39 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=2596476_nihms65652f1.jpg)
|
| 40 |
+
|
| 41 |
+
a. Time dependent accumulation of cAMP in GRK5-transfected cells stimulated with isoproterenol, 10 μM. b. (top) Immunoblot analysis of cardiac GRK5 from multiple lines of GRK5-Q41 and GRK5-L41 transgenic mice. (bottom) Comparative subcellular localization of GRK5-Q41 and –L41. c. Rhodopsin phosphorylation (bottom panel) by buffer (b) and cardiac membranes from nontransgenic (ntg), Q41, and L41 hearts. Top panel shows immunoreactive GRK5 from same fractions. d. Comparative immunoblot analysis of GRK5 and GRK2 in mouse hearts. e. Representative M-mode echocardiograms. ef In vivo contractile dose-response (+dP/dt) to isoproterenol in GRK5-Q41 (black squares) and L41 (black triangles) mice. Nontransgenic (NTG, open circles) are shown for comparison. *P<0.05 vs ntg for Q41 and L41; #P<0.05 for L41. g. Mean data for time-dependent desensitization of dP/dt response to infused isoproterenol in closed chest in vivo catheterization studies. n=12/group. *P<0.05 vs ntg and Q41; # P=0.081 vs ntg and Q41.
|
| 42 |
+
|
| 43 |
+
### Identification of a pharmacogenomic interaction between GRK5 –L41 and β-blockade in human cardiac ischemia and failure
|
| 44 |
+
|
| 45 |
+
Diminished isoproterenol-stimulated βAR signaling by GRK5-L41 resembles receptor antagonism by pharmacological β-blockers, suggesting that this polymorphism might interact with or mimic β-blockade in human cardiac syndromes wherein β-blockers are standard therapy. Accordingly, we genotyped for GRK5-Q41 or –L41 in two independent closely matched and highly phenotyped cohorts of cardiac disease subjects, 810 individuals (568 Caucasians and 242 African Americans) from Cincinnati with New York Heart Association class II – IV heart failure, and 822 individuals (580 Caucasians and 242 African Americans) from Kansas City and Atlanta with acute cardiac ischemia. Clinical characteristics of the two study groups are in [Table 1](#T1)Table 1. A non-affected control group consisted of 513 subjects (406 Caucasians and 107 African Americans) with negative histories and physical examinations for cardiac disease, normal ECGs, and normal echocardiograms. Among Caucasians, L41 allele frequencies were 0.013 (unaffected), 0.024 (heart failure), and 0.010 (acute ischemia), with no association between Q41L genotype and either cardiac disease. Among African Americans, L41 allele frequencies were ~10-fold higher than Caucasians, but again did not differ among unaffected (0.23), heart failure (0.24), and acute ischemia (0.28) subjects. Heterozygous L41 genotype frequency among African Americans was 0.35 and homozygous frequency was 0.062, consistent with predictions from Hardy-Weinberg equilibrium (*P*P=0.57). Thus, the GRK5-L41 allele is rare in Caucasians, but common in African Americans, and is not disproportionately represented in cardiac disease.
|
| 46 |
+
|
| 47 |
+
### Table 1.
|
| 48 |
+
|
| 49 |
+
Descriptive Statistics of Heart Failure and Coronary Ischemia Cohorts in Gene Association Studies.
|
| 50 |
+
|
| 51 |
+
| Variable | Heart Failure | Acute Coronary Ischemia |
|
| 52 |
+
| -------- | ------------- | ----------------------- |
|
| 53 |
+
| Caucasian n=568 | AA n=242 | Caucasian n=580 | AA n=242 |
|
| 54 |
+
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD |
|
| 55 |
+
| Age at enrollment (yrs) | 53 ± 13 | 51 ± 13 | 62 ± 12 | 55 ± 11 |
|
| 56 |
+
| Males (%) | 69.3 | 55.0 | 65.7 | 57.4 |
|
| 57 |
+
| Follow-up (yrs) | 2.3 ± 2.2 | 2.3 ± 2.1 | 3.8 ± 1.1 | 3.3 ± 1.4 |
|
| 58 |
+
| Height (cm) | 172 ± 10 | 172 ± 10 | 171 ± 10 | 169 ± 10 |
|
| 59 |
+
| Weight (kg) | 86 ± 21 | 90 ± 26 | 86 ± 18 | 83±21 |
|
| 60 |
+
| Left Ventricular EF (%) | 28 ± 14 | 33 ± 15 | 48 ± 12 | 44 ± 16 |
|
| 61 |
+
| Hypertension (%) | 45.1 | 80.2 | 62.8 | 81.0 |
|
| 62 |
+
| βblocker use (%) | 69.2 | 80.2 | 82.2 | 73.4 |
|
| 63 |
+
| Diagnosis (%) | | |
|
| 64 |
+
| Heart Failure | Acute Coronary Ischemia | |
|
| 65 |
+
| | Caucasian | AA | | Caucasian | AA |
|
| 66 |
+
| Non-ischemic CHF | 54.8 | 71.1 | STEMI | 30.9 | 15.7 |
|
| 67 |
+
| Ischemic CHF | 43.1 | 26.8 | NSTEMI | 33.1 | 54.1 |
|
| 68 |
+
| Other | 2.2 | 1.5 | UA | 36.0 | 30.2 |
|
| 69 |
+
Since GRKs exhibit activity only for ligand-occupied receptors, and functional differences between GRK5-Q41 and –L41 were observed only after catecholamine challenge (see [Figure 1](#F1)Figure 1), it is not surprising that the GRK5 polymorphism does not alter the risk of developing either heart failure or ischemia. In these syndromes, catecholamine excess and chronic βAR stimulation occur during the course of the disease, and GRK5 effects (like protection afforded by β-blockade) may therefore only be detectable after the disease and accompanying catecholamine excess develop. Accordingly, we examined the heart failure and acute coronary ischemia cohorts for any interaction between GRK5-L41 and β-blocker usage that influenced survival.
|
| 70 |
+
|
| 71 |
+
Among African Americans either with heart failure or acute coronary ischemic syndromes, age- and gender-adjusted Cox proportional hazards modeling showed significant interactions between GRK5-L41 and β-blocker usage on the endpoint of death (Likelihood Ratio P=0.036 and 0.023, respectively). In Caucasians, wherein the GRK5-L41 variant is rare, there was no significant interaction in either the heart failure (P=0.46)or acute coronary ischemia (P=0.72) cohorts. These results reveal pharmacogenomic interactions between the GRK5-L41 allele and β-blocker therapy for heart failure and acute ischemic syndromes in African Americans only.
|
| 72 |
+
|
| 73 |
+
### GRK5-L41 enhances β1-adrenergic receptor desensitization in transgenic mice
|
| 74 |
+
|
| 75 |
+
The above studies show that the GRK5-L41 variant decreases β_1_1AR signaling in transfected CHO cells much like partial β-blockade, and that there is a significant interaction between GRK5-L41 and β-blocker use that affects long-term outcome in human heart failure and acute myocardial ischemia. As human gene-association studies do not address molecular mechanisms and are subject to epistatic effects from other polymorphisms, we further characterized the effects of GRK5-Q41 and -L41 using mice in which each allele was specifically expressed only in cardiac myocytes, so phenotypes would only reflect cardiac effects of the GRK5s. Since prior 30-fold overexpression of bovine GRK5 in transgenic mouse hearts significantly depressed basal and catecholamine-stimulated cardiac function [15](#R15)^15^15^,^,[16](#R16)^16^16, we created a large number of founder lines to identify pairs of human GRK5-Q41 and -L41 expressing mice with comparable, low levels of expression ([Figure 1b](#F1)Figure 1b, upper panel). The selected lines (Q41A and L41B) each exhibited 4 to 6-fold increases in myocardial GRK activity assessed by rhodopsin kinase assay ([Figure 1c](#F1)Figure 1c) and immunoblotting ([Figures 1b-d](#F1)Figures 1b-d), with no counter-regulation of GRK2 ([Figure 1d](#F1)Figure 1d). Importantly, no abnormalities of cardiac size, histological appearance, disease-related gene expression, or basal contractile function (M-mode echocardiography) were detected in GRK5-Q41 or -L41mice up to 6 months of age ([Fig. 1e](#F1)Fig. 1e and data not shown), demonstrating that the presence of transgenic GRK5 at these levels does not cause dysfunction, and revealing no apparent effect of the L41 variant in normal hearts.
|
| 76 |
+
|
| 77 |
+
To determine the consequences of GRK5-Q41 and -L41 on βAR stimulation of cardiac contractility, mice underwent cardiac catheterization to measure left ventricular peak positive dP/dt at baseline and in response to increasing doses of intravenous isoproterenol. Both GRK5-Q41 and -L41 produced rightward shifts of the dose-response curves in comparison with nontransgenic mice ([Figure 1f](#F1)Figure 1f; EC_50_50 for GRK5-Q41 transgenics =0.135±0.041 ng/g/min, and GRK5-L41 transgenics =0.086±0.025 ng/g/min, compared to 0.046±0.011 ng/g/min for nontransgenics, P=0.02), reproducing previous findings with transgenic GRK2 and GRK5 [10](#R10)^10^10^,^,[15](#R15)^15^15^,^,[16](#R16)^16^16. In contrast, desensitization of βAR-stimulated contraction, measured during continuous high-dose isoproterenol infusion, was greater (e.g., peak positive dP/dt response was lower) after 10 minutes in GRK5-L41 mice than either -Q41 mice or nontransgenic controls (P<0.001, n=12/group) ([Figure 1g](#F1)Figure 1g). This trend continued at 20 min (P=0.081), but after 30 minutes of continuous isoproterenol infusion desensitization in all groups had achieved similar levels (P=0.133). These data demonstrate that GRK5-L41 is more effective than -Q41 in desensitizingcardiac βAR under conditions of acute catecholamine excess.
|
| 78 |
+
|
| 79 |
+
### GRK5-L41 protects against experimental heart failure caused by catecholamine excess
|
| 80 |
+
|
| 81 |
+
In experimental mouse models, overexpression of βARs [21](#R21)^21^21 or their Gαs G-protein signaling transducer [22](#R22)^22^22 causes cardiac dilation and failure. Conversely, genetic ablation of the βAR/Gαs downstream effector, adenylyl cyclase, preserves myocardial function after physiological stress [23](#R23)^23^23. The finding that GRK5-L41 accelerates isoproterenol-promoted βAR desensitization suggested that, like beta-blockers [22](#R22)^22^22, it might protect hearts from the effects of persistent βAR stimulation, i.e. cardiac dilation, ventricular hypertrophy, and heart failure ([Fig. 2a-c](#F2)Fig. 2a-c)[12](#R12)^12^12. To directly examine this possibility we chronically administered isoproterenol to GRK5-Q41 and -L41 transgenic mice via implanted osmotic mini-pump. Whereas GRK5-Q41 did not protect against isoproterenol-mediated increases in left ventricular chamber size (LVEDD; [Fig. 2d](#F2)Fig. 2d) and mass (LVM; [Figs 2e and f](#F2)Figs 2e and f), GRK5-L41 expression at the same levels was protective. Whereas β-blockade ameliorated the effects of catecholamine cardiomyopathy in NTG and GRK-Q41 mice (note LVEDD and LVM effects), in –L41 hearts β-blockade had no additional protective effect ([Figs. 2d-f](#F2)Figs. 2d-f). Both GRK5-Q41 and -L41 blunted the isoproterenol-mediated deterioration in cardiac contractility (cVcf), while propranolol administration had relatively little effect on this parameter ([Fig. 2g](#F2)Fig. 2g). These results demonstrate that enhanced βAR desensitization by GRK5-L41 can modulate catecholamine cardiotoxicity, protecting against left ventricular remodeling and cardiomyopathy development similar to pharmacological β-blockade.
|
| 82 |
+
|
| 83 |
+
### Figure 2. Cardiac expression of GRK5-L41, but not -Q41, confers resistance to catecholamine-induced cardiomyopathy.
|
| 84 |
+
|
| 85 |
+
|
| 86 |
+
|
| 87 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=2596476_nihms65652f2.jpg)
|
| 88 |
+
|
| 89 |
+
a–c. Time course for development of catecholamine cardiomyopathy in normal mice (closed triangles) and effect of β-blockade with propanolol (open triangles). LVEDD, left ventricular end diastolic dimension; cVcf, velocity of circumferential shortening corrected for heart rate. (n=6/group, *P<0.05 vs no propanolol). d–g. % change in cardiac parameters before and after 8 days of chronic isoproterenol treatment in vehicle (black, n=12/group) and propanolol (grey, n=6/group) treated mice.. P values compare GRK5-Q41 and –L41 responses. *P<0.05 for propanolol vs vehicle at day eight. ntg is shown for comparison.
|
| 90 |
+
|
| 91 |
+
### GRK5-L41 prolongs survival in β-blocker naive human heart failure
|
| 92 |
+
|
| 93 |
+
In our human case-control gene association studies and transgenic mouse experiments we found noevidence that the GRK5-L41 polymorphism could cause heart disease. However, we identified a pharmacogenomic interaction between GRK5-L41 and β-blockers in African American heart failure and coronary ischemia, and enhanced β-adrenergic receptor desensitization in GRK5-L41 transgenic mice likewise altered the response to β-blocker treatment under conditions of catecholamine excess. To further define the magnitude of the clinical benefit of the GRK5 polymorphism, we undertook a prospective analysis of 375 African American heart failure subjects, comparing time to death or cardiac transplantation as a function of GRK5-Q41L genotype and β-blocker treatment status. Characteristics of the prospective study group, classified by GRK5 genotype and β-blocker use, are in [Table 2](#T2)Table 2. Kaplan-Meyer analysis and Cox proportional modeling were utilized to assess potential effects on outcomes. In addition, race-specific genotyping at short-tandem repeats was undertaken to address potential population stratification. Validation techniques were then employed to show internal reproducibility and predictiveness of the model.
|
| 94 |
+
|
| 95 |
+
### Table 2.
|
| 96 |
+
|
| 97 |
+
Descriptive Statistics of AA Heart Failure Subjects in the Prospective Study
|
| 98 |
+
|
| 99 |
+
| | Only Q Alleles | ≥ 1 L Allele |
|
| 100 |
+
| - | -------------- | ------------ |
|
| 101 |
+
| Variable | No BB Use (N=34) | BB Use (N=182) | No BB Use (N=27) | BB Use (N=132) |
|
| 102 |
+
| Fractional Shortening (%) | 26±12 | 22±11 | 24±10 | 23±12 |
|
| 103 |
+
| LV Ejection Fraction (%) | 34±25 | 34±14 | 31±14 | 34±14 |
|
| 104 |
+
| Non-ischemic Cardiomyopathy (%) | 76 | 66 | 69 | 71 |
|
| 105 |
+
| Ischemic Cardiomyopathy (%) | 21 | 31 | 27 | 29 |
|
| 106 |
+
| LV Mass Indexed to BSA (g) | 160±51 | 178±58 | 180±72 | 186±69 |
|
| 107 |
+
| Percent Predicted LV Mass (%) | 175±55 | 178±56 | 179±55 | 198±69 |
|
| 108 |
+
| Hypertension (%) | 85 | 80 | 67 | 81 |
|
| 109 |
+
| Female (%) | 56 | 43 | 48 | 46 |
|
| 110 |
+
Consistent with therapeutic benefits of β-blocker treatment in heart failure [4](#R4)^4^4^,^,[24](#R24)^24^24^–^–[26](#R26)^26^26, individuals homozygous for “wild-type” GRK5-Q41 who received β-blockers had longer transplant-free survival times than those of the same genotype who were β-blocker naïve (controls) (HR=0.22, 95% CI=0.12 – 0.40, *P*P<0.001; [Figure 3a](#F3)Figure 3a). In contrast, there was no difference in outcome among L41 heart failure subjects regardless of β-blocker treatment (HR=0.78, 95% CI= 0.35 – 1.7, *P*P=0.53; [Figure 3b](#F3)Figure 3b). To formally evaluate the interaction effect of β-blockers and GRK5-L41 on time to death/transplant, we compared Cox Proportional Hazards models that included age, sex, β-blocker usage, and genotype status, with and without an interaction term between β-blocker usage and Q41L status. The model with the interaction term was significantly better than the reduced model (likelihood ratio test P=0.005). The β-blocker-L41 carrier status interaction term was significant at P=0.004, demonstrating an interaction between GRK5 genotype and β-blocker usage for the endpoint of death/transplant. β-blocker naïve L41 subjects had transplant-free survival times significantly greater than β-blocker naïve Q41 subjects ([Figure 3c](#F3)Figure 3c), but β-blocker treatment fully mimicked the survival advantage of L41 ([Figure 3d](#F3)Figure 3d). Similar results were obtained when all-cause mortality alone was considered ([Figures 3e, f](#F3)Figures 3e, f). When the homozygous Q41 β-blocker naïve group was set as the reference to derive age- and sex-adjusted hazard ratios for genotype and β-blocker treatment, β-blocker naïve L41 carriers were protected against death/transplantation ([Table 3](#T3)Table 3) and death alone. In β-blocker naïve subjects with two L41 alleles, the hazard ratio was 0.081 (95% CI 0.035 to 0.19), suggesting a gene-dose response.
|
| 111 |
+
|
| 112 |
+
### Figure 3. Prospective analysis of GRK5 polymorphism interaction with β-blockade as a determinant of heart failure outcome in African Americans.
|
| 113 |
+
|
| 114 |
+
|
| 115 |
+
|
| 116 |
+
[View larger image](https://www.ncbi.nlm.nih.gov/core/lw/2.0/html/tileshop_pmc/tileshop_pmc_inline.html?title=Click%20on%20image%20to%20zoom&p=PMC3&id=2596476_nihms65652f3.jpg)
|
| 117 |
+
|
| 118 |
+
Kaplan-Meier curves for time from diagnosis of heart failure to death or cardiac transplantation (a–d) or death alone (e,f). a. GRK5-Q41 by β-blocker usage. b. GRK5-L41 by β-blocker usage. c. GRK5-Q41 no β-blocker use vs. GRK5-L41 no β-blocker use. d. GRK5-Q41 β-blocker users vs. GRK5-L41 β-blocker users. e. GRK5-Q41 by β-blocker usage. f. GRK5-L41 by β-blocker usage.
|
| 119 |
+
|
| 120 |
+
### Table 3.
|
| 121 |
+
|
| 122 |
+
Cox proportional hazards of GRK5-Q41L polymorphism, β-blocker use, and death/transplant, adjusted for age at heart failure diagnosis and sex (n=375).
|
| 123 |
+
|
| 124 |
+
| Group | Hazard ratio | 95% CI | P-value |
|
| 125 |
+
| ----- | ------------ | ------ | ------- |
|
| 126 |
+
| Genotype† | β-blocker | N (# of Events) | | | |
|
| 127 |
+
| Without adjustment for % African ancestry * | | | |
|
| 128 |
+
| Q41 | − | 34 (17) | 1.0 (reference) | -- | -- |
|
| 129 |
+
| Q41 | + | 182 (37) | 0.19 | 0.10 to 0.34 | <0.001 |
|
| 130 |
+
| L41 | − | 27 (8) | 0.28 | 0.12 to 0.66 | 0.004 |
|
| 131 |
+
| L41 | + | 132 (27) | 0.20 | 0.05 to 0.80 | 0.02 |
|
| 132 |
+
| With adjustment for % African ancestry ** | | | |
|
| 133 |
+
| Q41 | − | 34 (17) | 1.0 (reference) | -- | -- |
|
| 134 |
+
| Q41 | + | 182 (37) | 0.19 | 0.10 to 0.37 | <0.001 |
|
| 135 |
+
| L41 | − | 27 (8) | 0.31 | 0.13 to 0.73 | 0.007 |
|
| 136 |
+
| L41 | + | 132 (27) | 0.20 | 0.05 to 0.84 | 0.03 |
|
| 137 |
+
Although not a prospective study, a cohort analysis within the case-control study of acute coronary ischemia also showed an interaction between GRK5 genotype and β-blocker use. The specific effect of genotype was most evident in those receiving β-blockers, where GRK5-L41 carriers exhibited improvement in survival (HR = 0.45, 95% CI = 0.238 – 0.853, P = 0.01).
|
| 138 |
+
|
| 139 |
+
Given the reported associations between the β_1_1AR Arg389 polymorphism and the response to β-blocker treatment in heart failure [27](#R27)^27^27, and between β_1_1AR Arg-389 and α2c del 322-325 polymorphisms and the risk of having heart failure [28](#R28)^28^28, we examined whether an interaction between these functionally-related polymorphisms and GRK5-L41 might provide for a more predictive model. We observed no significant interaction affecting time to death or cardiac transplant between GRK5-L41 and β1AR Arg389 (P=0.46), between GRK5-L41 and α2c del322-325 (P=0.21), or between all three polymorphisms (P=0.87).
|
| 140 |
+
|
| 141 |
+
Population stratification and significant differences in admixture within the African American cohort have the potential to result in spurious associations. Therefore, we estimated % African ancestry based on genotyping of 13 race-informative short tandem repeats [29](#R29)^29^29^,^,[30](#R30)^30^30. No difference in racial admixture was found between the β-blocker treated (African ancestry = 73.4%) and β-blocker naïve groups (African ancestry = 76.8 %; *P*P=0.22). We then utilized the estimated African ancestry as an additional term to adjust the Cox Proportional Hazards model, which did not significantly change the hazard ratios and P-values ([Table 3](#T3)Table 3).
|
| 142 |
+
|
| 143 |
+
Internal reproducibility of the β-blocker-GRK5 interaction was assessed using a sequential analysis procedure [31](#R31)^31^31. We rejected the null hypothesis of no interaction at P<0.05 using 50 randomly chosen subjects (Stage I) and found that the remaining sample had a P=0.01 (Stage II). When randomization and re-analysis were repeated 100 times, in 86% of the instances it was rejected at least twice, considered a very high degree of internal reproducibility [32](#R32)^32^32. To assess the interaction’s predictive utility, we performed a leave-one-out cross-validation, demonstrating that relative risks calculated from the β-blocker-GRK5 allele interaction Cox Proportional Hazards model were significant predictors of transplant-free survival (P<0.001) with an R^2^2 of 0.039 (Supplemental Table S1). The predictive ability of the GRK5-L41 allele compares favorably to clinical predictors of heart failure outcome (age, sex, hypertension), which had an R^2^2 of 0.023. The R^2^2 increased to 0.05 when these clinical predictors were included in the β-blocker-GRK5 allele interaction model.
|
| 144 |
+
|
| 145 |
+
## DISCUSSION
|
| 146 |
+
|
| 147 |
+
Multiple polymorphisms within the βAR signaling pathway have been proposed as modifiers of heart failure risk [7](#R7)^7^7^,^,[8](#R8)^8^8^,^,[27](#R27)^27^27^,^,[33](#R33)^33^33. Given the morbidity, mortality and health-care costs of heart failure [34](#R34)^34^34^,^,[35](#R35)^35^35, efforts are underway to identify additional genetic markers that will indicate prognosis and guide patient management. Here, we examined the genes encoding GRK5 and GRK2 because they constitute a critical regulatory node for the pathophysiologically important cardiac βAR signaling pathway that has not previously been explored for genotype-phenotype interactions in human heart disease. Thesecandidate genes were particularly attractive since their role in signaling is to modify receptor coupling to G-proteins and downstream adenylyl cyclase, i.e., the important parameters of βAR function that are perturbed in heart failure. Furthermore, GRK5 and GRK2 have the potential to modify signaling through both β_1_1- and β_2_2AR receptor subtypes, as well as other critical receptors in heart failure [16](#R16)^16^16^,^,[36](#R36)^36^36. Compared to the highly polymorphic receptors they regulate, we found these two human GRKs to be highly conserved, with only one common non-synonymous polymorphism in GRK5 identified by screening the complete coding sequences of both genes in 192 chromosomes. Interestingly, there is also striking cross-species similarity in amino acid sequence for GRKs among mammals, with 96% identity between human and mouse GRK5, and absolute conservation of Q at position 41 or its analog from humans to zebrafish (Supplemental Figure 1). The rarity of polymorphic variations in human GRK2 and GRK5 and the high degree of sequence conservation between species for GRK5 suggest that minor changes can have significant functional and physiological consequences, as we found for GRK5-L41.
|
| 148 |
+
|
| 149 |
+
A major function of GRKs is uncoupling of ligand-occupied receptors from signaling effectors, resulting in decreased cellular responsiveness (desensitization) and a time-dependent loss of agonist promoted function at the organ level [9](#R9)^9^9. In human heart failure, increases in myocardial GRK2 activity [17](#R17)^17^17^,^,[18](#R18)^18^18 and marked desensitization of cardiac β_1_1- and β_2_2-AR [37](#R37)^37^37 represent endogenous processes that may be acting to protect the heart from high circulating catecholamine levels [38](#R38)^38^38^,^,[39](#R39)^39^39. Here, we found a gain-of-function genetic polymorphism of GRK5 that augments βAR desensitization. As expected for a kinase that specifically modulates the agonist-occupied form of G-protein coupled receptors, the GRK5-Q41 and –L41 variants could not be distinguished in the absence of agonist. Indeed, within the African American population where it is fairly common (~40% carry at least one allele), the GRK5-L41 polymorphism also did not alter the risk for developing heart failure. However, GRK5-L41 was markedly more effective than –Q41 in promoting isoproterenol-mediated β-receptor desensitization in transfected cells and transgenic mice, and was associated with prolonged survival in clinical heart failure. The transgenic mouse studies with GRK5-Q41 and -L41, which also showed a β-blocker-like protective effect in the context of chronic catecholamine excess, indicate a major role for direct cardioprotection by this polymorphism since expression was targeted specifically to cardiac myocytes. Taken together, these data suggest that GRK5-L41 acts to attenuate β_1_1AR signaling in a manner similar to partial β_1_1AR antagonism with β-blockers, favoring protection against remodeling and improving survival.
|
| 150 |
+
|
| 151 |
+
An important feature of the current work is the mechanistic experiments in transfected cells and transgenic mice. These experiments revealed a phenotype that aided in the design of our human studies. Indeed, had analysis been restricted to conventional assessments of risk or survival, the important pharmacogenomic phenotype might have been overlooked. Such mechanistic studies are also crucial in making the case that the polymorphism is the basis of the human phenotype, rather than another locus that is in linkage disequilibrium with GRK5-L41 [8](#R8)^8^8. Our findings with this polymorphism in heart failure and acute ischemia, and the observation that its actions are consistent with genetic β-blockade, provide additional insight into inter-individual variation in outcome in these diseases, and suggest alternate monitoring and treatment strategies in individuals with GRK5-L41. For example, in African-Americans with heart failure who carry the GRK5-L41 allele it may be more prudent to maximize treatment focused on other pathways rather than continue to treat aggressively with drugs that may provide less additional benefit in these individuals. Since ~40% of African-Americans carry GRK5-L41, personalization of medical care could impact a large number of individuals suffering from cardiac disease.
|
| 152 |
+
|
| 153 |
+
## METHODS
|
| 154 |
+
|
| 155 |
+
### Study Subjects
|
| 156 |
+
|
| 157 |
+
Human study protocols were approved by the Institutional Review Boards of the University of Cincinnati and Washington University. Subjects provided written informed consent. Enrollment criteria for the heart failure observational study were: age of 18 to 80 years, left ventricular ejection fraction of less than 40%, and New York Heart Association heart failure class II–IV. Enrollment criteria for the acute coronary ischemia observational study were: age of 18 to 80 years and hospital admission with the confirmed diagnosis of acute myocardial infarction or unstable coronary syndrome. Non-affected controls were recruited from the greater Cincinnati area. Racial classification as Caucasian or African American was self-reported.
|
| 158 |
+
|
| 159 |
+
In the prospective GRK5-β-blocker interaction study, 402 African American heart failure subjects were consented between May 1, 2000 and June 1, 2006, 242 of whom were also in the observational study. Of these, 383 subjects completed input studies and had blood draw for DNA. GRK5 genotypes were not obtained on five subjects (success rate of 98.7%), and three subjects (0.8%) were lost to follow-up. Genotypes of these three subjects were Q/Q. The primary study endpoint was death or transplantation, with secondary endpoints of death or cardiac transplantation only, with an average follow-up period of 30 months. β-blocker use was defined as continuous therapy for at least 6 months. Decisions regarding β-blocker treatment (73%-carvedilol, 22%-metoprolol, and 5%-others) were made by the subjects’ physicians.
|
| 160 |
+
|
| 161 |
+
### Sequencing and Genotyping
|
| 162 |
+
|
| 163 |
+
Based on sequences of the human genes (accession numbers [NC_000011](https://www.ncbi.nlm.nih.gov/nuccore/NC_000011)NC_000011 and [NC_000010](https://www.ncbi.nlm.nih.gov/nuccore/NC_000010)NC_000010) the exons for GRK2 and GRK5 were amplified from genomic DNA using PCR. (primers for GRK5 coding exons are in Supplemental Table S2). Polymorphism discovery and GRK5 genotyping used bi-directional automated sequencing and outputs were aligned with reference sequence using SeqScape v2.5, and variants individually verified by an investigator (RRP). African American heart failure subjects were further genotyped at 13 race-informative short tandem repeat loci [29](#R29)^29^29.
|
| 164 |
+
|
| 165 |
+
### In vitro βAR desensitization studies
|
| 166 |
+
|
| 167 |
+
Chinese Hamster Ovary (CHO) cells were transfected with cDNAs encoding the human β_1_1AR, and either empty vector, GRK5-Q41 or GRK5-L41. Cell monolayers were treated with isoproterenol for the indicated times at 37°, and cAMP quantitated as previously described [40](#R40)^40^40.
|
| 168 |
+
|
| 169 |
+
### Experimental Heart Failure
|
| 170 |
+
|
| 171 |
+
Transgenic mice (FVB/N background) were generated using the α–myosin heavy chain (MHC) promoter to express human GRK5-Q41 and L41 using methods similar to those previously described [41](#R41)^41^41. Multiple founders were identified by genomic Southern analysis of tail clip DNA. F1 or F2 mice were sacrificed and myocardial GRK5 content analyzed by immunoblotting with GRK5-specific antisera (Santa Cruz). Mouse lines with equivalent cardiac GRK5-Q41 and GRK5-L41 protein expression were propagated for study. Animals were treated in accordance with approved University of Cincinnati Animal Care and Use Committee protocols.
|
| 172 |
+
|
| 173 |
+
To assess βAR responsiveness and desensitization, mice underwent left ventricular catheterization [41](#R41)^41^41 during graded infusions of the nonselective β-agonist isoproterenol, 0.01 to 0.32 ng/g/min., and sustained 30 minute infusion of 20 ng/g/min to evoke desensitization [42](#R42)^42^42. Heart failure was induced by chronic isoproterenol infusion by osmotic mini-pump [12](#R12)^12^12. Cardiac remodeling was assessed by transthoracic echocardiography.
|
| 174 |
+
|
| 175 |
+
### Statistical Analysis
|
| 176 |
+
|
| 177 |
+
Student’s t-tests and chi-square tests were used to assess significant differences in variables between ethnic groups and between genotype classes within ethnic groups. Hardy-Weinberg Equilibrium (HWE) was assessed in each ethnic group separately. The primary clinical endpoint was combined all-cause mortality or cardiac transplant; secondary endpoints were all-cause mortality and cardiac transplantation. Differences in time from diagnosis to endpoint were assessed using Kaplan-Meier curves and Log Rank tests [43](#R43)^43^43. Hazards ratios were obtained by Cox Proportional Hazards modeling [44](#R44)^44^44 using an additive genetic model [45](#R45)^45^45 after adjustment for age at diagnosis, β-blocker usage, hypertension status, and sex. To assess internal reproducibility of the association between GRK5 allele and survival we used an analytical strategy that reduces the probability of type I errors through sequential hypothesis testing [31](#R31)^31^31. The smallest possible sample sizes were used to reject the null hypothesis of no GRK5-β-blocker interaction, and the remaining samples were used to confirm those findings [46](#R46)^46^46. The type I error (α), type II error (β), and effect size D were preset to α=0.05, β =0.8, and D=0.5 to create stopping rules for the procedure where the sample size N is considered a random variable. Starting with n=50, the null hypothesis of no GRK5-β-blocker interaction was tested using the Wald test and then sequentially another individual was added and retested.
|
| 178 |
+
|
| 179 |
+
To assess the predictive value of Cox proportional hazards models, we utilized leave-one-out cross-validation. Each individual was sequentially left out and a Cox proportional hazards model for time to death or transplant were fitted. Using the coefficients estimated with the *n*n-1 individuals, an overall relative risk was calculated for the individual left out. These relative risks were then used as the predictor in a new Cox proportional hazards model. Because each individual is omitted from the model used to calculate his relative risk, the performance of a model using these relative risks as predictors approximates the predictive ability of the association in an independent sample drawn from the same population. The performance of each model was assessed using overall model significance and Cox Snell R^2^2, a measure of the model’s predictive ability [47](#R47)^47^47. All analyses were carried out using the R Statistical Language [32](#R32)^32^32. Two-tailed tests and an alpha level of 0.05 was used to assess significance. Percent African ancestry was estimated with the program Structure [30](#R30)^30^30.
|
| 180 |
+
|
| 181 |
+
## Acknowledgments
|
| 182 |
+
|
| 183 |
+
Supported by NHLBI Special Clinical Centers of Research in Heart Failure, P50 HL77101 and HL77113, and by HL87871.
|
| 184 |
+
|
| 185 |
+
## Footnotes
|
| 186 |
+
|
| 187 |
+
## References
|
| 188 |
+
|
| 189 |
+
1. Levy D, et al. Long-term trends in the incidence of and survival with heart failure. N Engl J Med. 2002;347:1397–1402. doi: 10.1056/NEJMoa020265. [DOI](https://doi.org/10.1056/NEJMoa020265) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12409541/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Long-term%20trends%20in%20the%20incidence%20of%20and%20survival%20with%20heart%20failure&author=D%20Levy&volume=347&publication_year=2002&pages=1397-1402&pmid=12409541&doi=10.1056/NEJMoa020265&)
|
| 190 |
+
|
| 191 |
+
2. Roger VL, et al. Trends in heart failure incidence and survival in a community-based population. JAMA. 2004;292:344–350. doi: 10.1001/jama.292.3.344. [DOI](https://doi.org/10.1001/jama.292.3.344) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15265849/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=JAMA&title=Trends%20in%20heart%20failure%20incidence%20and%20survival%20in%20a%20community-based%20population&author=VL%20Roger&volume=292&publication_year=2004&pages=344-350&pmid=15265849&doi=10.1001/jama.292.3.344&)
|
| 192 |
+
|
| 193 |
+
3. Franz WM, Muller OJ, Katus HA. Cardiomyopathies: from genetics to the prospect of treatment. Lancet. 2001;358:1627–1637. doi: 10.1016/S0140-6736(01)06657-0. [DOI](https://doi.org/10.1016/S0140-6736(01)06657-0) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11716909/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Cardiomyopathies:%20from%20genetics%20to%20the%20prospect%20of%20treatment&author=WM%20Franz&author=OJ%20Muller&author=HA%20Katus&volume=358&publication_year=2001&pages=1627-1637&pmid=11716909&doi=10.1016/S0140-6736(01)06657-0&)
|
| 194 |
+
|
| 195 |
+
4. The Merit HF Investigators. Effect of metoprolol CR/XL in chronic heart failure: Metoprolol CR/XL Randomised Intervention Trial in Congestive Heart Failure (MERIT-HF) Lancet. 1999;353:2001–2007. [PubMed](https://pubmed.ncbi.nlm.nih.gov/10376614/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Effect%20of%20metoprolol%20CR/XL%20in%20chronic%20heart%20failure:%20Metoprolol%20CR/XL%20Randomised%20Intervention%20Trial%20in%20Congestive%20Heart%20Failure%20(MERIT-HF)&volume=353&publication_year=1999&pages=2001-2007&pmid=10376614&)
|
| 196 |
+
|
| 197 |
+
5. BEST Trial Investigators. A trial of the beta-blocker bucindolol in patients with advanced chronic heart failure. N Engl J Med. 2001;344:1659–1667. doi: 10.1056/NEJM200105313442202. [DOI](https://doi.org/10.1056/NEJM200105313442202) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11386264/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=A%20trial%20of%20the%20beta-blocker%20bucindolol%20in%20patients%20with%20advanced%20chronic%20heart%20failure&volume=344&publication_year=2001&pages=1659-1667&pmid=11386264&doi=10.1056/NEJM200105313442202&)
|
| 198 |
+
|
| 199 |
+
6. van Campen LC, Visser FC, Visser CA. Ejection fraction improvement by beta-blocker treatment in patients with heart failure: an analysis of studies published in the literature. J Cardiovasc Pharmacol. 1998;32(Suppl 1):S31–S35. doi: 10.1097/00005344-199800003-00006. [DOI](https://doi.org/10.1097/00005344-199800003-00006) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9731693/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Cardiovasc%20Pharmacol&title=Ejection%20fraction%20improvement%20by%20beta-blocker%20treatment%20in%20patients%20with%20heart%20failure:%20an%20analysis%20of%20studies%20published%20in%20the%20literature&author=LC%20van%20Campen&author=FC%20Visser&author=CA%20Visser&volume=32&issue=Suppl%201&publication_year=1998&pages=S31-S35&pmid=9731693&doi=10.1097/00005344-199800003-00006&)
|
| 200 |
+
|
| 201 |
+
7. Wagoner LE, et al. Polymorphisms of the beta(2)-adrenergic receptor determine exercise capacity in patients with heart failure. Circ Res. 2000;86:834–840. doi: 10.1161/01.res.86.8.834. [DOI](https://doi.org/10.1161/01.res.86.8.834) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10785504/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circ%20Res&title=Polymorphisms%20of%20the%20beta(2)-adrenergic%20receptor%20determine%20exercise%20capacity%20in%20patients%20with%20heart%20failure&author=LE%20Wagoner&volume=86&publication_year=2000&pages=834-840&pmid=10785504&doi=10.1161/01.res.86.8.834&)
|
| 202 |
+
|
| 203 |
+
8. Liggett SB. Pharmacogenetic applications of the Human Genome project. Nat Med. 2001;7:281–283. doi: 10.1038/85411. [DOI](https://doi.org/10.1038/85411) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11231618/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Nat%20Med&title=Pharmacogenetic%20applications%20of%20the%20Human%20Genome%20project&author=SB%20Liggett&volume=7&publication_year=2001&pages=281-283&pmid=11231618&doi=10.1038/85411&)
|
| 204 |
+
|
| 205 |
+
9. Kohout TA, Lefkowitz RJ. Regulation of G protein-coupled receptor kinases and arrestins during receptor desensitization. Mol Pharmacol. 2003;63:9–18. doi: 10.1124/mol.63.1.9. [DOI](https://doi.org/10.1124/mol.63.1.9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12488531/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mol%20Pharmacol&title=Regulation%20of%20G%20protein-coupled%20receptor%20kinases%20and%20arrestins%20during%20receptor%20desensitization&author=TA%20Kohout&author=RJ%20Lefkowitz&volume=63&publication_year=2003&pages=9-18&pmid=12488531&doi=10.1124/mol.63.1.9&)
|
| 206 |
+
|
| 207 |
+
10. Koch WJ, et al. Cardiac function in mice overexpressing the beta-adrenergic receptor kinase or a beta ARK inhibitor. Science. 1995;268:1350–1353. doi: 10.1126/science.7761854. [DOI](https://doi.org/10.1126/science.7761854) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7761854/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Science&title=Cardiac%20function%20in%20mice%20overexpressing%20the%20beta-adrenergic%20receptor%20kinase%20or%20a%20beta%20ARK%20inhibitor&author=WJ%20Koch&volume=268&publication_year=1995&pages=1350-1353&pmid=7761854&doi=10.1126/science.7761854&)
|
| 208 |
+
|
| 209 |
+
11. Koch WJ. Genetic and phenotypic targeting of beta-adrenergic signaling in heart failure. Mol Cell Biochem. 2004;263:5–9. doi: 10.1023/B:MCBI.0000041843.64809.48. [DOI](https://doi.org/10.1023/B:MCBI.0000041843.64809.48) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/27520660/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mol%20Cell%20Biochem&title=Genetic%20and%20phenotypic%20targeting%20of%20beta-adrenergic%20signaling%20in%20heart%20failure&author=WJ%20Koch&volume=263&publication_year=2004&pages=5-9&pmid=27520660&doi=10.1023/B:MCBI.0000041843.64809.48&)
|
| 210 |
+
|
| 211 |
+
12. Matkovich SJ, et al. Cardiac-specific ablation of G-protein receptor kinase 2 redefines its roles in heart development and beta-adrenergic signaling. Circ Res. 2006;99:996–1003. doi: 10.1161/01.RES.0000247932.71270.2c. [DOI](https://doi.org/10.1161/01.RES.0000247932.71270.2c) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/17008600/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circ%20Res&title=Cardiac-specific%20ablation%20of%20G-protein%20receptor%20kinase%202%20redefines%20its%20roles%20in%20heart%20development%20and%20beta-adrenergic%20signaling&author=SJ%20Matkovich&volume=99&publication_year=2006&pages=996-1003&pmid=17008600&doi=10.1161/01.RES.0000247932.71270.2c&)
|
| 212 |
+
|
| 213 |
+
13. Premont RT, Koch WJ, Inglese J, Lefkowitz RJ. Identification, purification, and characterization of GRK5, a member of the family of G protein-coupled receptor kinases. J Biol Chem. 1994;269:6832–6841. [PubMed](https://pubmed.ncbi.nlm.nih.gov/8120045/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Biol%20Chem&title=Identification,%20purification,%20and%20characterization%20of%20GRK5,%20a%20member%20of%20the%20family%20of%20G%20protein-coupled%20receptor%20kinases&author=RT%20Premont&author=WJ%20Koch&author=J%20Inglese&author=RJ%20Lefkowitz&volume=269&publication_year=1994&pages=6832-6841&pmid=8120045&)
|
| 214 |
+
|
| 215 |
+
14. Gainetdinov RR, et al. Muscarinic supersensitivity and impaired receptor desensitization in G protein-coupled receptor kinase 5-deficient mice. Neuron. 1999;24:1029–1036. doi: 10.1016/s0896-6273(00)81048-x. [DOI](https://doi.org/10.1016/s0896-6273(00)81048-x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10624964/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Neuron&title=Muscarinic%20supersensitivity%20and%20impaired%20receptor%20desensitization%20in%20G%20protein-coupled%20receptor%20kinase%205-deficient%20mice&author=RR%20Gainetdinov&volume=24&publication_year=1999&pages=1029-1036&pmid=10624964&doi=10.1016/s0896-6273(00)81048-x&)
|
| 216 |
+
|
| 217 |
+
15. Chen EP, Bittner HB, Akhter SA, Koch WJ, Davis RD. Myocardial function in hearts with transgenic overexpression of the G protein-coupled receptor kinase 5. Ann Thorac Surg. 2001;71:1320–1324. doi: 10.1016/s0003-4975(00)01754-9. [DOI](https://doi.org/10.1016/s0003-4975(00)01754-9) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11308180/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Ann%20Thorac%20Surg&title=Myocardial%20function%20in%20hearts%20with%20transgenic%20overexpression%20of%20the%20G%20protein-coupled%20receptor%20kinase%205&author=EP%20Chen&author=HB%20Bittner&author=SA%20Akhter&author=WJ%20Koch&author=RD%20Davis&volume=71&publication_year=2001&pages=1320-1324&pmid=11308180&doi=10.1016/s0003-4975(00)01754-9&)
|
| 218 |
+
|
| 219 |
+
16. Rockman HA, et al. Receptor-specific in vivo desensitization by the G protein-coupled receptor kinase-5 in transgenic mice. Proc Natl Acad Sci U S A. 1996;93:9954–9959. doi: 10.1073/pnas.93.18.9954. [DOI](https://doi.org/10.1073/pnas.93.18.9954) | [PMC free article](/articles/PMC38536/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8790438/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Proc%20Natl%20Acad%20Sci%20U%20S%20A&title=Receptor-specific%20in%20vivo%20desensitization%20by%20the%20G%20protein-coupled%20receptor%20kinase-5%20in%20transgenic%20mice&author=HA%20Rockman&volume=93&publication_year=1996&pages=9954-9959&pmid=8790438&doi=10.1073/pnas.93.18.9954&)
|
| 220 |
+
|
| 221 |
+
17. Ungerer M, Bohm M, Elce JS, Erdmann E, Lohse MJ. Altered expression of beta-adrenergic receptor kinase and beta 1-adrenergic receptors in the failing human heart. Circulation. 1993;87:454–463. doi: 10.1161/01.cir.87.2.454. [DOI](https://doi.org/10.1161/01.cir.87.2.454) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8381058/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circulation&title=Altered%20expression%20of%20beta-adrenergic%20receptor%20kinase%20and%20beta%201-adrenergic%20receptors%20in%20the%20failing%20human%20heart&author=M%20Ungerer&author=M%20Bohm&author=JS%20Elce&author=E%20Erdmann&author=MJ%20Lohse&volume=87&publication_year=1993&pages=454-463&pmid=8381058&doi=10.1161/01.cir.87.2.454&)
|
| 222 |
+
|
| 223 |
+
18. Ungerer M, et al. Expression of beta-arrestins and beta-adrenergic receptor kinases in the failing human heart. Circ Res. 1994;74:206–213. doi: 10.1161/01.res.74.2.206. [DOI](https://doi.org/10.1161/01.res.74.2.206) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/8293560/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circ%20Res&title=Expression%20of%20beta-arrestins%20and%20beta-adrenergic%20receptor%20kinases%20in%20the%20failing%20human%20heart&author=M%20Ungerer&volume=74&publication_year=1994&pages=206-213&pmid=8293560&doi=10.1161/01.res.74.2.206&)
|
| 224 |
+
|
| 225 |
+
19. Oyama N, et al. Angiotensin converting enzyme inhibitors attenuated the expression of G-protein coupled receptor kinases in heart failure patients. Circ J. 2006;70:362–363. doi: 10.1253/circj.70.362. [DOI](https://doi.org/10.1253/circj.70.362) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16501306/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circ%20J&title=Angiotensin%20converting%20enzyme%20inhibitors%20attenuated%20the%20expression%20of%20G-protein%20coupled%20receptor%20kinases%20in%20heart%20failure%20patients&author=N%20Oyama&volume=70&publication_year=2006&pages=362-363&pmid=16501306&doi=10.1253/circj.70.362&)
|
| 226 |
+
|
| 227 |
+
20. Dzimiri N, Basco C, Moorji A, Afrane B, Al Halees Z. Characterization of lymphocyte beta 2-adrenoceptor signalling in patients with left ventricular volume overload disease. Clin Exp Pharmacol Physiol. 2002;29:181–188. doi: 10.1046/j.1440-1681.2002.03625.x. [DOI](https://doi.org/10.1046/j.1440-1681.2002.03625.x) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11906480/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Clin%20Exp%20Pharmacol%20Physiol&title=Characterization%20of%20lymphocyte%20beta%202-adrenoceptor%20signalling%20in%20patients%20with%20left%20ventricular%20volume%20overload%20disease&author=N%20Dzimiri&author=C%20Basco&author=A%20Moorji&author=B%20Afrane&author=Z%20Al%20Halees&volume=29&publication_year=2002&pages=181-188&pmid=11906480&doi=10.1046/j.1440-1681.2002.03625.x&)
|
| 228 |
+
|
| 229 |
+
21. Liggett SB, et al. Early and delayed consequences of beta(2)-adrenergic receptor overexpression in mouse hearts: critical role for expression level. Circulation. 2000;101:1707–1714. doi: 10.1161/01.cir.101.14.1707. [DOI](https://doi.org/10.1161/01.cir.101.14.1707) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10758054/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circulation&title=Early%20and%20delayed%20consequences%20of%20beta(2)-adrenergic%20receptor%20overexpression%20in%20mouse%20hearts:%20critical%20role%20for%20expression%20level&author=SB%20Liggett&volume=101&publication_year=2000&pages=1707-1714&pmid=10758054&doi=10.1161/01.cir.101.14.1707&)
|
| 230 |
+
|
| 231 |
+
22. Asai K, et al. Beta-adrenergic receptor blockade arrests myocyte damage and preserves cardiac function in the transgenic G(salpha) mouse. J Clin Invest. 1999;104:551–558. doi: 10.1172/JCI7418. [DOI](https://doi.org/10.1172/JCI7418) | [PMC free article](/articles/PMC408547/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10487769/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Clin%20Invest&title=Beta-adrenergic%20receptor%20blockade%20arrests%20myocyte%20damage%20and%20preserves%20cardiac%20function%20in%20the%20transgenic%20G(salpha)%20mouse&author=K%20Asai&volume=104&publication_year=1999&pages=551-558&pmid=10487769&doi=10.1172/JCI7418&)
|
| 232 |
+
|
| 233 |
+
23. Okumura S, et al. Disruption of type 5 adenylyl cyclase gene preserves cardiac function against pressure overload. Proc Natl Acad Sci U S A. 2003;100:9986–9990. doi: 10.1073/pnas.1733772100. [DOI](https://doi.org/10.1073/pnas.1733772100) | [PMC free article](/articles/PMC187910/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12904575/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Proc%20Natl%20Acad%20Sci%20U%20S%20A&title=Disruption%20of%20type%205%20adenylyl%20cyclase%20gene%20preserves%20cardiac%20function%20against%20pressure%20overload&author=S%20Okumura&volume=100&publication_year=2003&pages=9986-9990&pmid=12904575&doi=10.1073/pnas.1733772100&)
|
| 234 |
+
|
| 235 |
+
24. Bristow MR. beta-adrenergic receptor blockade in chronic heart failure. Circulation. 2000;101:558–569. doi: 10.1161/01.cir.101.5.558. [DOI](https://doi.org/10.1161/01.cir.101.5.558) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10662755/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circulation&title=beta-adrenergic%20receptor%20blockade%20in%20chronic%20heart%20failure&author=MR%20Bristow&volume=101&publication_year=2000&pages=558-569&pmid=10662755&doi=10.1161/01.cir.101.5.558&)
|
| 236 |
+
|
| 237 |
+
25. Packer M. Current role of beta-adrenergic blockers in the management of chronic heart failure. Am J Med. 2001;110(Suppl 7A):81S–94S. doi: 10.1016/s0002-9343(01)00676-3. [DOI](https://doi.org/10.1016/s0002-9343(01)00676-3) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11334782/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Am%20J%20Med&title=Current%20role%20of%20beta-adrenergic%20blockers%20in%20the%20management%20of%20chronic%20heart%20failure&author=M%20Packer&volume=110&issue=Suppl%207A&publication_year=2001&pages=81S-94S&pmid=11334782&doi=10.1016/s0002-9343(01)00676-3&)
|
| 238 |
+
|
| 239 |
+
26. Waagstein F, et al. Beneficial effects of metoprolol in idiopathic dilated cardiomyopathy. Metoprolol in Dilated Cardiomyopathy (MDC) Trial Study Group. Lancet. 1993;342:1441–1446. doi: 10.1016/0140-6736(93)92930-r. [DOI](https://doi.org/10.1016/0140-6736(93)92930-r) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/7902479/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Beneficial%20effects%20of%20metoprolol%20in%20idiopathic%20dilated%20cardiomyopathy.%20Metoprolol%20in%20Dilated%20Cardiomyopathy%20(MDC)%20Trial%20Study%20Group&author=F%20Waagstein&volume=342&publication_year=1993&pages=1441-1446&pmid=7902479&doi=10.1016/0140-6736(93)92930-r&)
|
| 240 |
+
|
| 241 |
+
27. Liggett SB, et al. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci U S A. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI](https://doi.org/10.1073/pnas.0509937103) | [PMC free article](/articles/PMC1523317/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16844790/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Proc%20Natl%20Acad%20Sci%20U%20S%20A&title=A%20polymorphism%20within%20a%20conserved%20beta(1)-adrenergic%20receptor%20motif%20alters%20cardiac%20function%20and%20beta-blocker%20response%20in%20human%20heart%20failure&author=SB%20Liggett&volume=103&publication_year=2006&pages=11288-11293&pmid=16844790&doi=10.1073/pnas.0509937103&)
|
| 242 |
+
|
| 243 |
+
28. Small KM, Wagoner LE, Levin AM, Kardia SL, Liggett SB. Synergistic polymorphisms of beta1- and alpha2C-adrenergic receptors and the risk of congestive heart failure. N Engl J Med. 2002;347:1135–1142. doi: 10.1056/NEJMoa020803. [DOI](https://doi.org/10.1056/NEJMoa020803) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12374873/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Synergistic%20polymorphisms%20of%20beta1-%20and%20alpha2C-adrenergic%20receptors%20and%20the%20risk%20of%20congestive%20heart%20failure&author=KM%20Small&author=LE%20Wagoner&author=AM%20Levin&author=SL%20Kardia&author=SB%20Liggett&volume=347&publication_year=2002&pages=1135-1142&pmid=12374873&doi=10.1056/NEJMoa020803&)
|
| 244 |
+
|
| 245 |
+
29. Barnholtz-Sloan JS, Chakraborty R, Sellers TA, Schwartz AG. Examining population stratification via individual ancestry estimates versus self-reported race. Cancer Epidemiol Biomarkers Prev. 2005;14:1545–1551. doi: 10.1158/1055-9965.EPI-04-0832. [DOI](https://doi.org/10.1158/1055-9965.EPI-04-0832) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15941970/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Cancer%20Epidemiol%20Biomarkers%20Prev&title=Examining%20population%20stratification%20via%20individual%20ancestry%20estimates%20versus%20self-reported%20race&author=JS%20Barnholtz-Sloan&author=R%20Chakraborty&author=TA%20Sellers&author=AG%20Schwartz&volume=14&publication_year=2005&pages=1545-1551&pmid=15941970&doi=10.1158/1055-9965.EPI-04-0832&)
|
| 246 |
+
|
| 247 |
+
30. Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI](https://doi.org/10.1093/genetics/155.2.945) | [PMC free article](/articles/PMC1461096/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/10835412/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genetics&title=Inference%20of%20population%20structure%20using%20multilocus%20genotype%20data&author=JK%20Pritchard&author=M%20Stephens&author=P%20Donnelly&volume=155&publication_year=2000&pages=945-959&pmid=10835412&doi=10.1093/genetics/155.2.945&)
|
| 248 |
+
|
| 249 |
+
31. Bechhoffer RE, Kiefer J, Sobel M. Sequential Identification and Ranking Proceedures. University of Chicago Press; Chicago: 1986. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Sequential%20Identification%20and%20Ranking%20Proceedures&author=RE%20Bechhoffer&author=J%20Kiefer&author=M%20Sobel&publication_year=1986&)
|
| 250 |
+
|
| 251 |
+
32. R Develpoment Core Team. R: A language and environment for statistical computing. (2.3.0). 2005. R foundation for statistical computing. Ref Type: Computer Program
|
| 252 |
+
|
| 253 |
+
33. Liggett SB. Beta-adrenergic receptors in the failing heart: the good, the bad, and the unknown. J Clin Invest. 2001;107:947–948. doi: 10.1172/JCI12774. [DOI](https://doi.org/10.1172/JCI12774) | [PMC free article](/articles/PMC199564/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11306597/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Clin%20Invest&title=Beta-adrenergic%20receptors%20in%20the%20failing%20heart:%20the%20good,%20the%20bad,%20and%20the%20unknown&author=SB%20Liggett&volume=107&publication_year=2001&pages=947-948&pmid=11306597&doi=10.1172/JCI12774&)
|
| 254 |
+
|
| 255 |
+
34. Lloyd-Jones DM, et al. Lifetime risk for developing congestive heart failure: the Framingham Heart Study. Circulation. 2002;106:3068–3072. doi: 10.1161/01.cir.0000039105.49749.6f. [DOI](https://doi.org/10.1161/01.cir.0000039105.49749.6f) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/12473553/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circulation&title=Lifetime%20risk%20for%20developing%20congestive%20heart%20failure:%20the%20Framingham%20Heart%20Study&author=DM%20Lloyd-Jones&volume=106&publication_year=2002&pages=3068-3072&pmid=12473553&doi=10.1161/01.cir.0000039105.49749.6f&)
|
| 256 |
+
|
| 257 |
+
35. Thom T, et al. Heart disease and stroke statistics--2006 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2006;113:e85–151. doi: 10.1161/CIRCULATIONAHA.105.171600. [DOI](https://doi.org/10.1161/CIRCULATIONAHA.105.171600) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/16407573/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Circulation&title=Heart%20disease%20and%20stroke%20statistics--2006%20update:%20a%20report%20from%20the%20American%20Heart%20Association%20Statistics%20Committee%20and%20Stroke%20Statistics%20Subcommittee&author=T%20Thom&volume=113&publication_year=2006&pages=e85-151&pmid=16407573&doi=10.1161/CIRCULATIONAHA.105.171600&)
|
| 258 |
+
|
| 259 |
+
36. Kim J, et al. Functional antagonism of different G protein-coupled receptor kinases for beta-arrestin-mediated angiotensin II receptor signaling. Proc Natl Acad Sci U S A. 2005;102:1442–1447. doi: 10.1073/pnas.0409532102. [DOI](https://doi.org/10.1073/pnas.0409532102) | [PMC free article](/articles/PMC547874/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15671181/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Proc%20Natl%20Acad%20Sci%20U%20S%20A&title=Functional%20antagonism%20of%20different%20G%20protein-coupled%20receptor%20kinases%20for%20beta-arrestin-mediated%20angiotensin%20II%20receptor%20signaling&author=J%20Kim&volume=102&publication_year=2005&pages=1442-1447&pmid=15671181&doi=10.1073/pnas.0409532102&)
|
| 260 |
+
|
| 261 |
+
37. Port JD, Bristow MR. Altered beta-adrenergic receptor gene regulation and signaling in chronic heart failure. J Mol Cell Cardiol. 2001;33:887–905. doi: 10.1006/jmcc.2001.1358. [DOI](https://doi.org/10.1006/jmcc.2001.1358) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11343413/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=J%20Mol%20Cell%20Cardiol&title=Altered%20beta-adrenergic%20receptor%20gene%20regulation%20and%20signaling%20in%20chronic%20heart%20failure&author=JD%20Port&author=MR%20Bristow&volume=33&publication_year=2001&pages=887-905&pmid=11343413&doi=10.1006/jmcc.2001.1358&)
|
| 262 |
+
|
| 263 |
+
38. Cohn JN, et al. Plasma norepinephrine as a guide to prognosis in patients with chronic congestive heart failure. N Engl J Med. 1984;311:819–823. doi: 10.1056/NEJM198409273111303. [DOI](https://doi.org/10.1056/NEJM198409273111303) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/6382011/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=N%20Engl%20J%20Med&title=Plasma%20norepinephrine%20as%20a%20guide%20to%20prognosis%20in%20patients%20with%20chronic%20congestive%20heart%20failure&author=JN%20Cohn&volume=311&publication_year=1984&pages=819-823&pmid=6382011&doi=10.1056/NEJM198409273111303&)
|
| 264 |
+
|
| 265 |
+
39. Bristow MR. Why does the myocardium fail? Insights from basic science. Lancet. 1998;352(Suppl 1):SI8–14. doi: 10.1016/s0140-6736(98)90311-7. [DOI](https://doi.org/10.1016/s0140-6736(98)90311-7) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9736474/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Lancet&title=Why%20does%20the%20myocardium%20fail?%20Insights%20from%20basic%20science&author=MR%20Bristow&volume=352&issue=Suppl%201&publication_year=1998&pages=SI8-14&pmid=9736474&doi=10.1016/s0140-6736(98)90311-7&)
|
| 266 |
+
|
| 267 |
+
40. Liggett SB, et al. Altered patterns of agonist-stimulated cAMP accumulation in cells expressing mutant beta 2-adrenergic receptors lacking phosphorylation sites. Mol Pharmacol. 1989;36:641–646. [PubMed](https://pubmed.ncbi.nlm.nih.gov/2554115/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Mol%20Pharmacol&title=Altered%20patterns%20of%20agonist-stimulated%20cAMP%20accumulation%20in%20cells%20expressing%20mutant%20beta%202-adrenergic%20receptors%20lacking%20phosphorylation%20sites&author=SB%20Liggett&volume=36&publication_year=1989&pages=641-646&pmid=2554115&)
|
| 268 |
+
|
| 269 |
+
41. D’Angelo DD, et al. Transgenic Galphaq overexpression induces cardiac contractile failure in mice. Proc Natl Acad Sci U S A. 1997;94:8121–8126. doi: 10.1073/pnas.94.15.8121. [DOI](https://doi.org/10.1073/pnas.94.15.8121) | [PMC free article](/articles/PMC21567/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/9223325/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Proc%20Natl%20Acad%20Sci%20U%20S%20A&title=Transgenic%20Galphaq%20overexpression%20induces%20cardiac%20contractile%20failure%20in%20mice&author=DD%20D%E2%80%99Angelo&volume=94&publication_year=1997&pages=8121-8126&pmid=9223325&doi=10.1073/pnas.94.15.8121&)
|
| 270 |
+
|
| 271 |
+
42. Odley A, et al. Regulation of cardiac contractility by Rab4-modulated beta2-adrenergic receptor recycling. Proc Natl Acad Sci U S A. 2004;101:7082–7087. doi: 10.1073/pnas.0308335101. [DOI](https://doi.org/10.1073/pnas.0308335101) | [PMC free article](/articles/PMC406469/) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/15105445/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Proc%20Natl%20Acad%20Sci%20U%20S%20A&title=Regulation%20of%20cardiac%20contractility%20by%20Rab4-modulated%20beta2-adrenergic%20receptor%20recycling&author=A%20Odley&volume=101&publication_year=2004&pages=7082-7087&pmid=15105445&doi=10.1073/pnas.0308335101&)
|
| 272 |
+
|
| 273 |
+
43. Kaplan EL, Meier Paul. Nonparametric Estimation from Incomplete Observations. Journal of the American Statistical Association. 2006;53:457–481. [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Journal%20of%20the%20American%20Statistical%20Association&title=Nonparametric%20Estimation%20from%20Incomplete%20Observations&author=EL%20Kaplan&author=Paul%20Meier&volume=53&publication_year=2006&pages=457-481&)
|
| 274 |
+
|
| 275 |
+
44. Parmar M, Machin D. Survival Analysis: A practical approach. John Wiley & Sons; 1995. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Survival%20Analysis:%20A%20practical%20approach&author=M%20Parmar&author=D%20Machin&publication_year=1995&)
|
| 276 |
+
|
| 277 |
+
45. Lynch M, Walsh B. Genetics and Analysis of Quantative Traits. Sinauer Associates; 1998. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Genetics%20and%20Analysis%20of%20Quantative%20Traits&author=M%20Lynch&author=B%20Walsh&publication_year=1998&)
|
| 278 |
+
|
| 279 |
+
46. Province MA. A single, sequential, genome-wide test to identify simultaneously all promising areas in a linkage scan. Genet Epidemiol. 2000;19:301–322. doi: 10.1002/1098-2272(200012)19:4<301::AID-GEPI3>3.0.CO;2-G. [DOI](https://doi.org/10.1002/1098-2272(200012)19:4<301::AID-GEPI3>3.0.CO;2-G) | [PubMed](https://pubmed.ncbi.nlm.nih.gov/11108641/) | [Google Scholar](https://scholar.google.com/scholar_lookup?journal=Genet%20Epidemiol&title=A%20single,%20sequential,%20genome-wide%20test%20to%20identify%20simultaneously%20all%20promising%20areas%20in%20a%20linkage%20scan&author=MA%20Province&volume=19&publication_year=2000&pages=301-322&pmid=11108641&doi=10.1002/1098-2272(200012)19:4<301::AID-GEPI3>3.0.CO;2-G&)
|
| 280 |
+
|
| 281 |
+
47. Cox DR, snell EJ. Analysis of Binary Data. Chapman & Hall; 1989. [Google Scholar](https://scholar.google.com/scholar_lookup?title=Analysis%20of%20Binary%20Data&author=DR%20Cox&author=EJ%20snell&publication_year=1989&)
|