Pharmacogenetics of tenofovir clearance among Southern Africans living with HIV
Metadata
Authors: Zinhle Cindi, Aida N Kawuma, Gary Maartens, Yuki Bradford, Simiso Sokhela, Nomathemba Chandiwana, Willem D Francois Venter, Roeland E Wasmann, Paolo Denti, Lubbe Wiesner, Marylyn D Ritchie, David W Haas, Phumla Sinxadi Journal: Pharmacogenetics and genomics Date: 2023 Mar 6 DOI: 10.1097/FPC.0000000000000495 PMID: 37098852 PMCID: PMC10154044 URL: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10154044/ PDF: https://pmc.ncbi.nlm.nih.gov/articles/PMC10154044/pdf/nihms-1869515.pdf
Abstract
Background:: Tenofovir is a component of preferred combination antiretroviral therapy (ART) regimens in Africa. Few pharmacogenetic studies have been conducted of tenofovir exposure in Africa, where genetic diversity is greatest.
Objective:: We characterized the pharmacogenetics of plasma tenofovir clearance in Southern Africans receiving tenofovir disoproxil fumarate (TDF) or tenofovir alafenamide (TAF).
Methods:: Adults randomised to TAF or TDF in dolutegravir-containing arms of the ADVANCE trial (NCT03122262) were studied. Linear regression models stratified by study arm examined associations with unexplained variability in tenofovir clearance. We investigated genetic associations with polymorphisms selected a priori followed by genome-wide associations.
Results:: A total of 268 participants (138 and 130 in the TAF and TDF arm, respectively) were evaluable for associations. Among polymorphisms previously associated with any drug-related phenotype, IFNL4 rs12979860 was associated with more rapid tenofovir clearance in both arms (TAF: P = 0.003; TDF: P = 0.003). Genome-wide, the lowest P-values for tenofovir clearance in TAF and TDF arms were LINC01684 rs9305223 (P = 3.0 × 10−8) and intergenic rs142693425 (P = 1.4 × 10−8), respectively.
Conclusion:: Among Southern Africans randomized to TAF or TDF in ADVANCE, unexplained variability in tenofovir clearance was associated with a polymorphism in IFNL4, an immune response gene. It is unclear how this gene would affect tenofovir disposition.
Keywords: antiretroviral therapy, HIV, tenofovir, tenofovir disoproxil fumarate, tenofovir alafenamide, pharmacokinetics, pharmacogenetics
Background:
Tenofovir is a component of preferred combination antiretroviral therapy (ART) regimens in Africa. Few pharmacogenetic studies have been conducted of tenofovir exposure in Africa, where genetic diversity is greatest.
Objective:
We characterized the pharmacogenetics of plasma tenofovir clearance in Southern Africans receiving tenofovir disoproxil fumarate (TDF) or tenofovir alafenamide (TAF).
Methods:
Adults randomised to TAF or TDF in dolutegravir-containing arms of the ADVANCE trial (NCT03122262NCT03122262) were studied. Linear regression models stratified by study arm examined associations with unexplained variability in tenofovir clearance. We investigated genetic associations with polymorphisms selected a prioria priori followed by genome-wide associations.
Results:
A total of 268 participants (138 and 130 in the TAF and TDF arm, respectively) were evaluable for associations. Among polymorphisms previously associated with any drug-related phenotype, IFNL4IFNL4 rs12979860 was associated with more rapid tenofovir clearance in both arms (TAF: P = 0.003; TDF: P = 0.003). Genome-wide, the lowest P-values for tenofovir clearance in TAF and TDF arms were LINC01684LINC01684 rs9305223 (P = 3.0 × 10^−8^−8) and intergenic rs142693425 (P = 1.4 × 10^−8^−8), respectively.
Conclusion:
Among Southern Africans randomized to TAF or TDF in ADVANCE, unexplained variability in tenofovir clearance was associated with a polymorphism in IFNL4IFNL4, an immune response gene. It is unclear how this gene would affect tenofovir disposition.
**Keywords:**Keywords: antiretroviral therapy, HIV, tenofovir, tenofovir disoproxil fumarate, tenofovir alafenamide, pharmacokinetics, pharmacogenetics
Introduction
South Africa is home to an estimated 8.2 million people living with HIV (PLWH) [11], including 5.2 million on antiretroviral therapy (ART) [22]. The World Health Organization recommends initiating ART using dolutegravir in combination with nucleoside reverse-transcriptase inhibitors (NRTI) [33].The preferred first-line regimen in South Africa is tenofovir disoproxil fumarate (TDF), lamivudine and dolutegravir [33], which is available as a fixed-dose formulation.
Tenofovir disoproxil fumarate, a prodrug of tenofovir, is a substrate for the efflux transporter P-glycoprotein (P-gp) [44]. After oral administration, TDF undergoes rapid metabolism in plasma to tenofovir, which is activated intracellularly [55, 66]. Tenofovir is excreted into urine via multidrug resistance protein 4 (MRP4) [77]. TDF is generally well tolerated but can cause nephrotoxicity and decreases in bone mineral density [88–1010].
Tenofovir alafenamide (TAF), a newer oral prodrug of tenofovir, has improved renal and bone safety outcomes. Unlike TDF, TAF is predominantly metabolized intracellularly to tenofovir by cathepsin A and is more stable in plasma [55, 1111, 1212]. This results in higher intracellular concentrations of the active metabolite, tenofovir diphosphate, and approximately tenfold lower plasma tenofovir concentrations than TDF [55, 1111, 1313], which is thought to explain the reduced risk of bone and renal toxicity [1414]. Transport of TDF through the basolateral membrane of renal cells involves organic anion transporter 1 (OAT1) and, to a lesser extent, OAT3 [1515], while TAF is not a substrate of either OAT1 or OAT3 but does undergo minor metabolism by CYP3A4 [1616]. Better understanding of the pharmacogenetics of tenofovir clearance may inform optimize prescribing in different populations and reduce tenofovir-associated toxicities.
In candidate gene studies, higher tenofovir concentrations have been associated with polymorphisms in SCL22A6SCL22A6, ABCC2ABCC2 and ABCC4ABCC4 [1717–1919]. In a Chinese cohort, SLCO1B3SLCO1B3 rs7311358 was independently associated with TAF AUC_0-t_0-t, while ABCC2ABCC2 rs3740066 was associated with tenofovir half-life. Higher TAF AUC and shorter tenofovir half-life with the ABCB1ABCB1 rs2032582 T allele and ABCC4ABCC4 rs3742106 CC genotype were noted though neither were statistically significant [2020]. Carriers of the ABCC4ABCC4 rs1751034 variant had slower tenofovir renal clearance [2121]. CYP3A4CYP3A4 rs35599367 was associated with higher plasma TAF AUC_0–24h_0–24h [2222]. Despite Africa having the world’s greatest genetic diversity, there are limited data on the pharmacogenetics of plasma tenofovir exposure in Africans. We hypothesized that polymorphisms in ABCB1, ABCC2, ABCC4, ABCG2, CYP3A4, SLC28A2ABCB1, ABCC2, ABCC4, ABCG2, CYP3A4, SLC28A2 and SLCO1B3SLCO1B3 may be associated with interindividual differences in plasma tenofovir clearance among Southern African PLWH receiving TAF or TDF. We also hypothesized that we would identify novel associations genome-wide.
Methods
Ethics
The present study was conducted in accordance with the Declaration of Helsinki and the ADVANCE protocol WRHI 060 (NCT03122262NCT03122262) received ethics and regulatory approvals from the Wits Human Research Ethics Committee (REF 160606B) and the South African Health Products Regulatory Authority (REF 20160620), respectively. Ethics approval for the pharmacogenetics sub-study was also granted by the University of Cape Town Health Sciences Human Research Ethics (REF 403/2019). Written informed consent for genetic research was obtained from study participants.
Study population
The ADVANCE study in South Africa was a phase 3 non-inferiority clinical trial in which 1053 HIV-positive, ART-naïve participants were randomly assigned to one of three treatment arms: 1) dolutegravir, TAF and emtricitabine; 2) dolutegravir, TDF and emtricitabine; or 3) efavirenz, TDF and emtricitabine [2323, 2424]. The present analyses included arm 1 and arm 2 participants who consented to genetic analysis.
Tenofovir assay
Plasma tenofovir concentrations were determined with a validated liquid chromatography tandem mass spectrometry assay developed at the Division of Clinical Pharmacology, University of Cape Town. The method utilised plasma protein precipitation followed by high performance liquid chromatography with tandem mass spectrometry detection. Chromatographic separation was achieved on a Waters Atlantis T3 column. The AB Sciex 5500 Qtrap mass spectrometer was used to monitor the transition of the protonated precursor ions 288.1 and 294.1 to the product ions 176.1 and 182.1 for tenofovir and tenofovir-d6 (internal standard), respectively. Electrospray ionisation was used for ion production. The calibration curve fitted a quadratic (weighted by 1/concentration) regression based on peak area ratios over the range 0.5 to 300 ng/mL. The combined accuracy (%Nom) of the limit of quantification, low, medium, and high-quality controls (3 validation batches, N=18) were between 93.8% and 103.8%, with precision (%CV) less than 13%.
Genetic polymorphisms
Whole blood was collected from consenting participants, and DNA extracted using the salting out method as described elsewhere [2525]. Samples were labelled with coded identifiers. Genotyping utilized the Illumina Infinium Multi-Ethnic Global BeadChip (MEGA^EX^EX) at Vanderbilt Technologies for Advanced Genomics (VANTAGE). Post-genotype quality control was performed by Vanderbilt Technologies for Advanced Genomics Analysis and Research Design (VANGARD), and included sex checks, call rates by marker and sample, identity by descent (IDB) plots to identify and remove related individuals, assessment for batch effects, concordance between duplicate samples, and HapMap controls.
All quality control steps were performed using PLINK version 1.9 [2626]. Genotyping efficiency per participant was > 95% for all samples. After quality control, data were imputed using the TOPMed reference panel after transforming to genome build 38 using liftOver and stratification by chromosome to parallelize the imputation process [2727, 2828]. We excluded polymorphisms with imputation scores < 0.3, genotyping call rates < 99%, minor allele frequency (MAF) < 0.05, or Hardy-Weinberg Equilibrium (HWE) P-values < 1.0×10^−8^−8. Linkage disequilibrium (LD) rr^2^22 values were determined using PLINK.
Pharmacokinetic sampling and analysis
In a subset of participants (equally divided between the TDF- and TAF-containing arms), intense pharmacokinetic sampling was done at steady state, with plasma samples collected pre-dose and 1, 2, 4, 6, 8 and 24 hours post dose. At the time of pharmacokinetic sampling, the median duration on ART was 19.9 months (IQR 19.2 to 20.7 months). Doses preceding intense sampling were observed and taken after a standard meal. For all other individuals, at least one plasma sample (sparse pharmacokinetic sampling) was collected at week 48 or 96 at random times post dose.
Deriving individual parameters and unexplained variability from a population pharmacokinetic model
We used a previously published pharmacokinetic model of tenofovir to estimate individual values of clearance and variability in clearance. Details about the population pharmacokinetic model have been presented elsewhere [2929]. Briefly, the model describing tenofovir pharmacokinetics was developed with data from the 41 intensively sampled individuals in a 1:1 ratio (21 on TDF, 20 on TAF) from the ADVANCE study. The model consists of two compartments with clearance, central volume, and peripheral volume estimated at 44.7 L/h, 378 L, and 356 L, respectively for a typical 70 kg individual. The model included between-subject variability in clearance of 20.1% (coefficient of variation). In addition, TDF and TAF absorption are described by two separate absorption processes. When given as TDF, tenofovir quickly appears in plasma with an estimated absorption rate constant of 3.04 (1/h). On the other hand, after TAF administration, tenofovir absorption was described using two pathways. A “fast” pathway with absorption rate constant of 1.45 1/h and a “slow” pathway in which tenofovir was first absorbed intracellularly and then transitioned from there to the plasma at a half-life of 6.8 days. Allometric scaling with weight was included as a predictor on all clearance and volume parameters. No other covariates were included in the model.
By employing a post-hoc Bayes estimation method and considering an individual’s pharmacokinetic data and characteristics (i.e., weight), the tenofovir model was used to generate individual estimates of clearance and unexplained variability in clearance for all individuals including those who underwent sparse sampling only. The formula below was applied where the individual clearance (Clearancei)Clearancei)ClearanceiClearanceCClleeaarraanncceeiii)) is defined by the typical population parameter value and individual influence (eBSVCLieBSVCLieBSVCLieeeBSVCLiBSVCLiBSVCLBBSSVVCCLLiii).
| Clearancei=Typicalclearancevalue.eBSVCLi |
|---|
Genetic association analyses
The outcome of interest was unexplained variability in plasma tenofovir clearance (CL_BSV_BSV) among individuals receiving TAF or TDF. To adjust for genetic ancestry, we estimated continuous axes of ancestry incorporating the intersection of common autosomal genotypes using EIGENSTRAT [3030]. We projected our individuals into a principal components analysis on the 1000Genomes. Principal component scree plots were visually inspected to determine how many components to include in analyses. Based on this, just the first two principal components were included. Other than principal components, no additional covariates were included in association analyses because they were already accounted for in the population pharmacokinetic model. We report regression coefficients (β) for additive associations with polymorphisms, where positive β values indicate an association with increased clearance. The Bonferroni method was used to correct of multiple testing, with significance thresholds of 0.05 divided by the number of polymorphisms tested in targeted polymorphism and gene analyses, and P < 5.0 ×10^−8^−8 for genome-wide analyses. We present nominal P-values uncorrected for multiple testing and indicate thresholds that would be significant with multiple testing.
We a prioria priori selected for analysis eight polymorphisms previously reported to affect tenofovir pharmacokinetics (ABCB1ABCB1 rs2032582; ABCC2ABCC2 rs3740066; ABCC4ABCC4 rs3742106 and rs1751034; ABCG2ABCG2 rs2231142; CYP3A4CYP3A4 rs35599367; SLC28A2SLC28A2 rs11854484; and SLCO1B3SLCO1B3 rs7311358). Beyond these polymorphisms, and to decrease the burden of multiple testing, we then used a stepwise approach to prioritize which sets of polymorphisms to interrogate, based on the rationale that polymorphisms that have previously been strongly associated with at least one drug-related phenotype, or that have been genome-wide significantly associated with any trait, are most likely to be true associations.
We used as references the Pharmacogenomics Knowledge Base (PharmGKB, accessed 5 April 2022) [3131] and the NHGRI-EBI GWAS Catalog (accessed 5 April 2022) [3232]. In PharmGKB, 141 polymorphisms were previously associated with at least one drug-related phenotype (pharmacokinetics, efficacy, or toxicity) with levels of evidence of 1 (the preponderance of evidence shows an association, which has been replicated in multiple cohorts, and preferably with strong effect size) or 2 (moderate evidence of association, which has been replicated but some studies may not show statistical significance, or the effect size is small). In the GWAS Catalog, 132 547 polymorphisms were previously associated with any trait at P < 5.0 × 10^−8^−8 in at least one published study. A subset of 22 polymorphisms were common to both PharmGKB and the GWAS Catalog. We prioritized polymorphisms common to both PharmGKB and the GWAS Catalog, considering these to have the most robust evidence for true drug-related associations. We secondarily explored all polymorphisms from PharmGKB and from the GWAS Catalog (based on criteria described above), and all polymorphisms in our imputed genome-wide genotype data.
Results
Study population
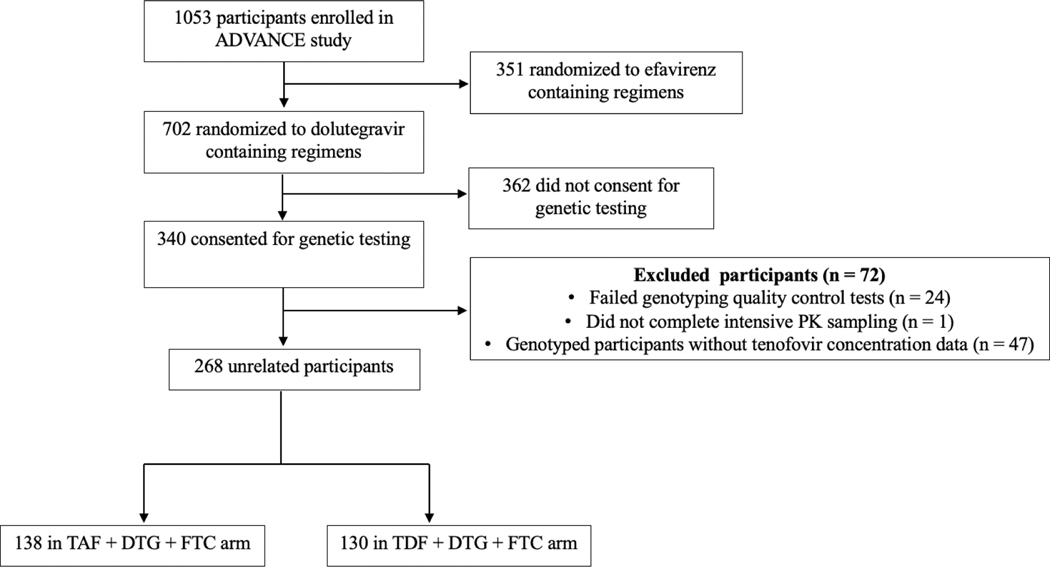
Among 340 ADVANCE participants who consented for genetic analyses, 268 were successfully genotyped and had pharmacokinetic data. Participant disposition is presented in Figure 1Figure 1. All participants were Black Africans and 62% were females. Study participant characteristics are shown in Table 1Table 1.
Figure 1. Disposition of study participants.
Of 1053 participants enrolled in the ADVANCE study, 268 randomized to TAF- and TDF-containing regimens had available pharmacokinetic sampling data and were evaluable for genetic associations.
Table 1:
Baseline characteristics for TAF and TDF recipients included in genetic analyses
| TAFa, DTG and FTC | TDF, DTG and FTC | |
|---|---|---|
| Characteristic | (n=138) | (n=130) |
| Age in years (IQR) | 32 (27 to 38) | 32 (27 to 37) |
| Sex | ||
| Female, n (%) | 86 (62) | 84 (65) |
| Male, n (%) | 52 (38) | 46 (35) |
| BMI in kg/m2, median (IQR) | 23.4 (20.5 to 26.5) | 22.9 (20.0 to 27.5) |
| CD4 T-cell count in cells/mm3, median (IQR) | 299 (164 to 491) | 283 (159 to 424) |
| CrCl in ml/min, median (IQR) | 125 (111 to 141) | 120 (104 to 148) |
Genetic associations with tenofovir clearance
Three of eight polymorphisms previously associated with tenofovir pharmacokinetics were not evaluable in our genetic data: CYP3A4CYP3A4 rs35599367 did not meet the imputation quality threshold, while ABCB1ABCB1 rs2032582 and ABCG2ABCG2 rs2231142 are very infrequent in Africans (MAF less than approximately 5%). The lowest P-value for association with tenofovir CL_BSV_BSV in the TAF arm was ABCC2ABCC2 rs3740066 (β = −0.1, P = 0.4). The lowest P-value for association with tenofovir CL_BSV_BSV in the TDF arm was ABCC2ABCC2 rs3740066 (β = −0.1, P = 0.3) (Table 2Table 2).
Table 2:
Associations with polymorphisms selected a priori
| Tenofovir prodrug | Polymorphism | Gene | MAFa | Beta | P-valueb |
|---|---|---|---|---|---|
| TAF | rs3740066 | ABCC2 | 0.2 | −0.1 | 0.4 |
| rs3742106 | ABCC4 | 0.4 | 0.03 | 0.5 | |
| rs11854484 | SLC28A2 | 0.2 | −0.03 | 0.6 | |
| rs7311358 | SLOC1B3 | 0.4 | 0.02 | 0.7 | |
| rs1751034 | ABCC4 | 0.4 | −0.001 | 1.0 | |
| TDF | rs3740066 | ABCC2 | 0.2 | −0.1 | 0.3 |
| rs1751034 | ABCC4 | 0.4 | −0.03 | 0.4 | |
| rs11854484 | SLC28A2 | 0.2 | 0.01 | 0.8 | |
| rs3742106 | ABCC4 | 0.4 | −0.002 | 1.0 | |
| Among polymorphisms from PharmGKB and the GWAS catalog, we report the five lowest P-value associations with TAF and TDF CL_BSV_BSV (Tables 3Tables 3 and 44). We were able to test for associations with 11 of 22 polymorphisms common to PharmGKB and the GWAS Catalog. Polymorphisms not included in our analyses did not meet imputation score, MAF, or HWE cutoffs. In the TAF arm the lowest P-value for association among these 11 polymorphisms was IFNL4IFNL4 rs12979860 (β = 0.1, P = 0.003) (Table 3Table 3), which withstood correction for multiple testing (cut-off P < 0.0045), and with the C allele being associated with increased tenofovir CL_BSV_BSV (Online supplemental material Figure S1Online supplemental material Figure S1). Considering PharmGKB polymorphisms that were not in the GWAS Catalog, we were able to test for associations with 15 (11%) of 141 polymorphisms. The lowest P-value for association among these 15 polymorphisms was SLC19A1SLC19A1 rs1051266 (β = 0.1, P = 0.1). Considering polymorphisms previously associated with any GWAS Catalog trait, we were able to test for associations with 82 785 (62%) of 132 547. The lowest P-value for association among these was LINC01414LINC01414 rs3850736 (β = 0.2, P = 3.5 × 10^−5^−5), which did not withstand correction for multiple testing. |
Table 3:
Lowest P-values for genetic association with unexplained variability in tenofovir clearance in the TAF arm
| Polymorphism | Gene | MAF | Beta | P-value | |
|---|---|---|---|---|---|
| PharmGKB and GWAS cataloga(n = 11 polymorphisms) | rs12979860 | IFNL4 | 0.4 | 0.1 | 0.003* |
| rs8099917 | Intergenic | 0.05 | −0.2 | 0.06 | |
| rs1801133 | MTHFR | 0.09 | −0.1 | 0.07 | |
| rs7412 | APOE | 0.2 | −0.05 | 0.27 | |
| rs396991 | FCGR3A | 0.3 | 0.04 | 0.4 | |
| PharmGKB but not in GWAS catalogb (n = 15 polymorphisms) | rs1051266 | SLC19A1 | 0.3 | 0.1 | 0.1 |
| rs4673993 | ATIC | 0.07 | −0.1 | 0.1 | |
| rs1042713 | ADRB2 | 0.5 | 0.1 | 0.1 | |
| rs7294 | VKORC1 | 0.4 | 0.04 | 0.3 | |
| rs20455 | KIF6 | 0.2 | −0.1 | 0.3 | |
| GWAS catalogc(n = 82 787 polymorphisms) | rs3850736 | LINC01414 | 0.2 | 0.2 | 3.5×10−5 |
| rs2356369 | LINC01414 | 0.2 | 0.2 | 4.6×10−5 | |
| rs7570090 | RPE | 0.4 | −0.2 | 8.2×10−5 | |
| rs7841320 | LINC01414 | 0.3 | 0.2 | 9.0×10−5 | |
| rs10032941 | C1QTNF7 | 0.3 | 0.2 | 1.0×10−4 | |
| Genome-wide genotype datad(n = 8 903 713 polymorphisms) | rs9305223e | LINC01684 | 0.3 | 0.2 | 3.0×10−8 |
| rs4816969 | LINC01684 | 0.3 | 0.2 | 3.7×10−8 | |
| rs2829163f | Intergenic | 0.4 | 0.2 | 4.5×10−8 | |
| rs7282679 | Intergenic | 0.2 | 0.2 | 5.9×10−8 | |
| rs2226443 | LINC01684 | 0.3 | 0.2 | 6.6×10−8 |
Table 4:
Lowest P-values for genetic association with unexplained variability in tenofovir clearance in the TDF arm
| Polymorphism | Gene | MAF | Beta | P-value | |
|---|---|---|---|---|---|
| PharmGKB and GWAS cataloga(n = 11 polymorphisms) | rs12979860 | IFNL4 | 0.4 | 0.1 | 0.003* |
| rs12777823 | Intergenic | 0.3 | −0.05 | 0.2 | |
| rs1800629 | TNF | 0.2 | −0.06 | 0.2 | |
| rs7412 | APOE | 0.2 | −0.05 | 0.25 | |
| rs10929302 | UGT1A | 0.3 | 0.02 | 0.6 | |
| PharmGKB but not in GWAS catalogb(n = 15 polymorphisms) | rs2297595 | DPYD | 0.1 | 0.1 | 0.03 |
| rs3745274 | CYP2B6 | 0.4 | 0.1 | 0.05 | |
| rs1042713 | ADRB2 | 0.5 | 0.04 | 0.2 | |
| rs28399499 | CYP2B6 | 0.1 | −0.1 | 0.3 | |
| rs1801159 | DPYD | 0.2 | 0.05 | 0.3 | |
| GWAS catalogc(n = 82 787 polymorphisms) | rs144511092 | Intergenic | 0.1 | 0.3 | 1.9×10−5 |
| rs7902657 | Intergenic | 0.5 | 0.1 | 2.5×10−5 | |
| rs4682844 | CCDC12 | 0.2 | 0.2 | 2.7×10−5 | |
| rs34940374 | GIMAP6 | 0.1 | 0.2 | 3.4×10−5 | |
| rs7970054 | LRIG3 | 0.05 | 0.3 | 5.0×10−5 | |
| Genome-wide genotype datad(n = 8 903 713 polymorphisms) | rs142693425 | Intergenic | 0.1 | 0.3 | 1.4×10−8 |
| rs112914324 | Intergenic | 0.3 | 0.2 | 1.6×10−8 | |
| rs11995962 | Intergenic | 0.1 | 0.3 | 2.3×10−8 | |
| rs73151902 | Intergenic | 0.05 | 0.4 | 1.1×10−7 | |
| rs866325353 | Intergenic | 0.05 | 0.4 | 2.1×10−7 | |
| In the TDF arm, among polymorphisms common to PharmGKB and the GWAS catalog, the lowest P-values was again IFNL4IFNL4 rs12979860 (β = 0.1, P = 0.003) (Table 4Table 4); which withstood correction for multiple testing (cut-off P < 0.0045), with the C allele associated with increased tenofovir CL_BSV_BSV (Online supplemental material Figure S2Online supplemental material Figure S2). Considering PharmGKB polymorphisms that were not in the GWAS Catalog, the lowest P-value was DPYDDPYD rs2297595 (β = 0.1, P = 0.03). Considering polymorphisms previously associated with any GWAS Catalog trait, the lowest P-value for association among these was rs144511092 (β = 0.3 P = 1.9 × 10^−5^−5), which did not withstand correction for multiple testing. In genome-wide associations combining both TAF and TDF arms (n = 268), IFNL4IFNL4 rs12979860 had a P-value of P = 2.1 × 10^−5^−5 (β = 0.12), which did not achieve genome-wide significance (Online supplemental material Figure S3Online supplemental material Figure S3). In both analyses that considered polymorphisms in PharmGKB only, and polymorphisms from the GWAS catalog, none of the polymorphisms with the lowest P-value withstood correction for multiple testing. |
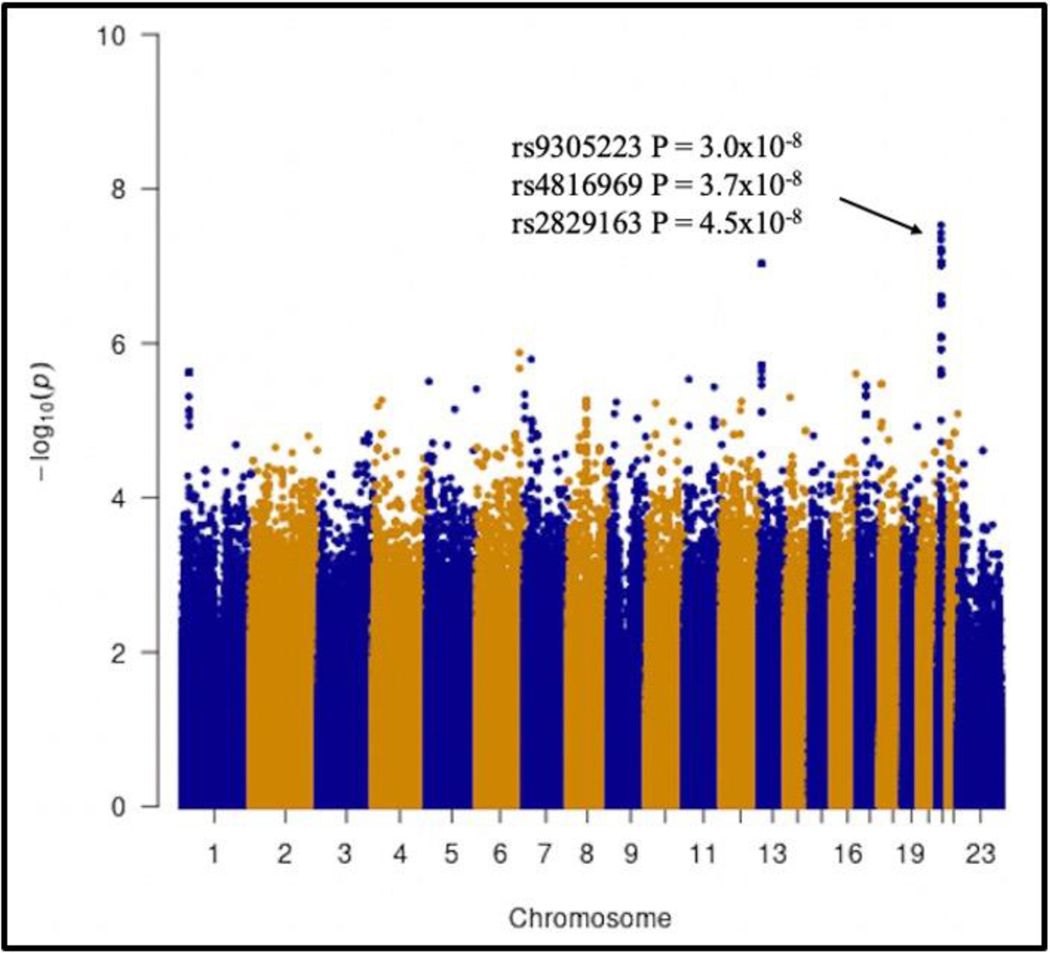
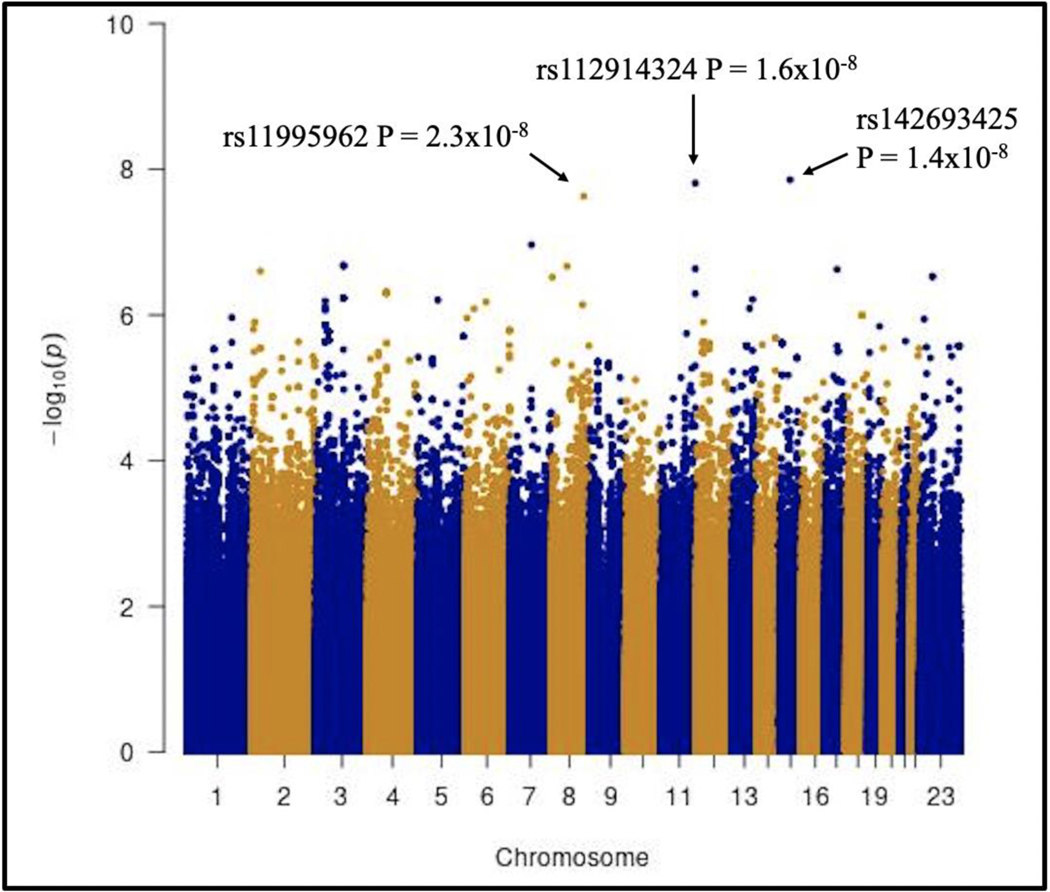
In analyses that explored genome-wide associations with tenofovir CL_BSV_BSV in the TAF arm, the lowest P-value was LINC01684LINC01684 rs9305223 (β = 0.2, P = 3.0 × 10^−8^−8), which was genome-wide significant. This polymorphism is in strong linkage disequilibrium (r^2^2 > 0.9) with LINC01684LINC01684 rs4816969 and rs2226443 on chromosome 21 (Figure 2Figure 2). Intergenic rs2829163 on chromosome 21 also reached genome-wide significance (β = 0.2, P = 4.5 × 10^−8^−8). The five lowest P-values for tenofovir CL_BSV_BSV in the TAF arm are presented in Table 3Table 3. In the TDF arm, the lowest P-value was for intergenic rs142693425 on chromosome 15 (β = 0.3, P = 1.4 × 10^−8^−8), and rs112914324 (β = 0.2, P = 1.6 × 10^−8^−8) on chromosome 11 and rs11995962 (β = 0.3, P = 2.3 × 10^−8^−8) on chromosome 8, which were each genome-wide significant (Figure 3Figure 3) and associated with increased tenofovir CL_BSV_BSV. The five lowest P-values for tenofovir CL_BSV_BSV in the TDF arm are presented in Table 4Table 4.
Figure 2. Manhattan plot of associations with unexplained variability in tenofovir clearance in the TAF arm.
The figure shows −log10 P-values for association among 138 TAF recipients who were evaluable for genetic associations. The black arrow indicates polymorphisms that reached genome-wide significance at P < 5.0×10−8.
Figure 3. Manhattan plot of associations with unexplained variability in tenofovir clearance in the TDF arm.
The figure shows −log10 P-values for association among 130 TDF recipients who were evaluable for genetic associations. The black arrows indicate polymorphisms that reached genome-wide significance at P < 5.0×10−8.
Discussion
We characterized genetic associations with unexplained variability in tenofovir clearance (CL_BSV_BSV) in ART-naïve participants randomized to TAF or TDF in the ADVANCE study in South Africa. We prioritized evaluation of polymorphisms from prior genetic association studies represented in PharmGKB and the GWAS Catalog, as present in our imputed genome-wide data. Among polymorphisms common to both PharmGKB and the GWAS catalog, the lowest P-value in both TDF and TAF arms was IFNL4IFNL4 rs12979860 (TAF P = 0.003, TDF P = 0.003), each of which withstood correction for multiple testing. This is a pseudogene which, in some humans, encodes the interferon lambda 4 (IFNL4) protein which is involved in immune response to viral infections. In prior reports, rs12979860 CC genotype was associated with lower hepatitis B virus plasma titers in Chinese and Moroccan patients treated with interferon-based therapy [3333, 3434]. In other studies, IFNL4IFNL4 rs12979860 CC genotype was independently associated with spontaneous HIV control in Caucasian Spanish individuals [3535], but not in African Americans [3636, 3737]. No association has been previously reported between IFNL4IFNL4 rs12979860 and tenofovir pharmacokinetics. While it is remarkable that IFNL4IFNL4 rs12979860 was associated with tenofovir clearance in both the TAF and TDF arms, it is unclear how this immune-response protein would mechanistically affect the pharmacokinetics of tenofovir, and there are no apparent drug metabolism or transport genes nearby on chromosome 19.
In genome-wide analyses for associations in the TAF arm, the lowest P-value was LINC01684LINC01684 rs9305223 (P = 3.0 × 10^−8^−8) on chromosome 21, which was in strong linkage disequilibrium with rs4816969 and rs2226443. Intergenic rs2829163 on chromosome 21 also reached genome-wide significance. In genome-wide analyses for associations in the TDF arm, the lowest P-value was intergenic rs142693425 (P = 1.4 × 10^−8^−8) which reached genome-wide significance, as did two additional intergenic polymorphisms, rs112914324 and rs11995962. It is unclear how these intergenic polymorphisms could affect tenofovir disposition. These associations warrant replication in independent cohorts.
We found no significant associations between polymorphisms selected a prioria priori (ABCC2ABCC2 rs3740066; ABCC4ABCC4 rs3742106 and rs1751034; SLC28A2SLC28A2 rs11854484 and SLOC1B3SLOC1B3 rs7311358) and CL_BSV._BSV. This may be due to low minor allele frequencies of these polymorphisms in our dataset. In a small American study, carriers of the ABCC4ABCC4 rs1751034 variant had slower tenofovir clearance [2121]. In contrast, in a Thai population, carriers of ABCC4ABCC4 rs1751034 had more rapid tenofovir clearance (CL/F) [3838]. In a Chinese population, ABCC2ABCC2 rs3740066, ABCC4ABCC4 rs3742106 and SLCO1B3SLCO1B3 rs7311358 were associated with TAF pharmacokinetics [2020]. In a study from Italy, SLC28A2SLC28A2 rs11854484 CT/TT genotypes were associated with plasma tenofovir exposure [3939].
Our study had limitations. Although our sample size was modest to detect genome-wide associations, this was the largest pharmacogenetic study of tenofovir clearance to date in Africa. In addition, the prioritized approach used in our analyses reduced the burden of multiple testing. A larger sample size may identify novel genome-wide significant associations with small effect sizes, or with infrequent polymorphisms. Several polymorphisms previously associated with tenofovir phenotypes could not be evaluated in our dataset.
In summary, unexplained variability in tenofovir clearance was associated with an IFNL4IFNL4 polymorphism that has previously been associated with response to hepatitis B virus treatment, and with immune control of HIV. In both TAF and TDF arms of ADVANCE study, we also identified genome-wide significant associations with intergenic polymorphisms. Further studies are needed to replicate these associations, as it is unclear mechanistically how these polymorphisms would affect tenofovir clearance.
Supplementary Material
Conflict of interest declaration and sources of funding
W. D. F. V. reports grants from ViiV; personal fees and nonfinancial support from ViiV Healthcare and Gilead Sciences, during the conduct of the study; and personal fees from Mylan, Merk, Adcock-Ingram, Aspen, Abbott, Roche, and Johnson and Johnson, outside the submitted work. S. S. reports grants from ViiV Healthcare and Gilead Sciences, during the conduct of the study. N. C. reports grants from ViiV Healthcare and Gilead Sciences, during the conduct of the study. S. S., N. C., and W. D. F. V. received research funding and drug donation for the ADVANCE trial through their institution from ViiV Healthcare and Gilead Sciences. NC declares personal fees and non-financial support from Johnson & Johnson, personal fees from Cipla, Frontiers Biotech, and Novo Nordisk outside the submitted work. All other authors have nothing to declare.
Research reported in this publication was funded, in part, by the Fogarty International Center of the National Institutes of Health (NIH), under Award Number D43 TW010559 (to Z. C. and D. W. H.), and the National Institute of Allergy and Infectious Diseases (NIAID) (Award Numbers UM1 AI068634, UM1 AI068636, and UM1 AI106701; to University of Cape Town’s Pharmacology laboratory and L. W.). NIH grant support also included AI110527, AI077505, and TR000445 (to D. W. H.). This work was also funded by the National Research Foundation through the Thuthuka Grant and Black Academic Advancement Programme (113983 and 120647 respectively; to P. S.). A. N. K. was supported by a doctoral training grant from Pharmacometrics Africa NPC. Grant support from USAID, Unitaid and the SAMRC was received for the ADVANCE clinical trial (to S. S., N. C., and W. D. F. V.).
Associated Data
*This section collects any data citations, data availability statements, or supplementary materials included in this article.*This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials
References
Stats SA. Statistical release P0302. 2021.
AIDSinfo | UNAIDS. http://aidsinfo.unaids.org/. Accessed 25 May 2021. http://aidsinfo.unaids.org/
Republic of South Africa National Department of Health. 2019 ART Clinical Guidelines for the Management of HIV in Adults, Pregnancy, Adolescents, Children, Infants and Neonates. 2019.
Mallants R, Van Oosterwyck K, Van Vaeck L, Mols R, De Clercq E, Augustijns P. Multidrug resistance-associated protein 2 (MRP2) affects hepatobiliary elimination but not the intestinal disposition of tenofovir disoproxil fumarate and its metabolites. Xenobiotica. 2005;35:1055–66. DOI | PubMed | Google Scholar
Lee WA, He GX, Eisenberg E, Cihlar T, Swaminathan S, Mulato A, et al. Selective intracellular activation of a novel prodrug of the human immunodeficiency virus reverse transcriptase inhibitor tenofovir leads to preferential distribution and accumulation in lymphatic tissue. Antimicrob Agents Chemother. 2005;49:1898–906. DOI | PMC free article | PubMed | Google Scholar
Sax PE, Zolopa A, Brar I, Elion R, Ortiz R, Post F, et al. Tenofovir alafenamide vs. tenofovir disoproxil fumarate in single tablet regimens for initial HIV-1 therapy: A randomized phase 2 study. J Acquir Immune Defic Syndr. 2014;67:52–8. DOI | PubMed | Google Scholar
Van Aubel RAMH Smeets PHE, Van Den Heuve JJMW, Russel FGM. Human organic anion transporter MRP4 (ABCC4) is an efflux pump for the purine end metabolite urate with multiple allosteric substrate binding sites. Am J Physiol - Ren Physiol. 2005;288:F327–33. DOI | PubMed | Google Scholar
Mocroft A, Kirk O, Reiss P, De Wit S, Sedlacek D, Beniowski M, et al. Estimated glomerular filtration rate, chronic kidney disease and antiretroviral drug use in HIV-positive patients. AIDS. 2010;24:1667–78. DOI | PubMed | Google Scholar
Morlat P, Vivot A, Vandenhende MA, Dauchy FA, Asselineau J, Déti E, et al. Role of Traditional Risk Factors and Antiretroviral Drugs in the Incidence of Chronic Kidney Disease, ANRS CO3 Aquitaine Cohort, France, 2004–2012. PLoS One. 2013;8:66223. DOI | PMC free article | PubMed | Google Scholar
McComsey GA, Kitch D, Daar ES, Tierney C, Jahed NC, Tebas P, et al. Bone mineral density and fractures in antiretroviral-naive persons randomized to receive abacavir-lamivudine or tenofovir disoproxil fumarate-emtricitabine along with efavirenz or atazanavir-ritonavir: AIDS Clinical Trials Group A5224s, a substudy of ACTG. J Infect Dis. 2011;203:1791–801. DOI | PMC free article | PubMed | Google Scholar
Birkus G, Kutty N, He GX, Mulato A, Lee W, McDermott M, et al. Activation of 9-[(R)-2-[[(S)-[[(S)-1-(Isopropoxycarbonyl)ethyl]amino] phenoxyphosphinyl]-methoxy]propyl]adenine (GS-7340) and other tenofovir phosphonoamidate prodrugs by human proteases. Mol Pharmacol. 2008;74:92–100. DOI | PubMed | Google Scholar
Birkus G, Wang R, Liu X, Kutty N, MacArthur H, Cihlar T, et al. Cathepsin a is the major hydrolase catalyzing the intracellular hydrolysis of the antiretroviral nucleotide phosphonoamidate prodrugs GS-7340 and GS-9131. Antimicrob Agents Chemother. 2007;51:543–50. DOI | PMC free article | PubMed | Google Scholar
Podany AT, Bares SH, Havens J, Dyavar SR, O’Neill J, Lee S, et al. Plasma and intracellular pharmacokinetics of tenofovir in patients switched from tenofovir disoproxil fumarate to tenofovir alafenamide. AIDS. 2018;32:761–5. DOI | PMC free article | PubMed | Google Scholar
Van Rompay KKA, Durand-Gasselin L, Brignolo LL, Ray AS, Abel K, Cihlar T, et al. Chronic administration of tenofovir to rhesus macaques from infancy through adulthood and pregnancy: Summary of pharmacokinetics and biological and virological effects. Antimicrob Agents Chemother. 2008;52:3144–60. DOI | PMC free article | PubMed | Google Scholar
Aceti A. Pharmacogenetics as a tool to tailor antiretroviral therapy: A review. World J Virol. 2015;4:198. DOI | PMC free article | PubMed | Google Scholar
Lazerwith SE, Siegel D, McFadden RM, Mish MR, Tse WC. New Antiretrovirals for HIV and Antivirals for HBV. In: Comprehensive Medicinal Chemistry III. Elsevier; 2017. p. 628–64. Google Scholar
Rungtivasuwan K, Avihingsanon A, Thammajaruk N, Mitruk S, Burger DM, Ruxrungtham K, et al. Influence of ABCC2 and ABCC4 polymorphisms on tenofovir plasma concentrations in Thai HIV-infected patients. Antimicrob Agents Chemother. 2015;59:3240–5. DOI | PMC free article | PubMed | Google Scholar
Manosuthi W, Sukasem C, Thongyen S, Nilkamhang S, Sungkanuparph S. ABCC2*1C and plasma tenofovir concentration are correlated to decreased glomerular filtration rate in patients receiving a tenofovir-containing antiretroviral regimen. J Antimicrob Chemother. 2014;69:2195–201. DOI | PubMed | Google Scholar
Bleasby K, Hall LA, Perry JL, Mohrenweiser HW, Pritchard JB. Functional consequences of single nucleotide polymorphisms in the human organic anion transporter hOAT1 (SLC22A6). J Pharmacol Exp Ther. 2005;314:923–31. DOI | PubMed | Google Scholar
Li X, Tan XY, Cui XJ, Yang M, Chen C, Chen XY. Pharmacokinetics of tenofovir alafenamide fumarate and tenofovir in the chinese people: Effects of non-genetic factors and genetic variations. Pharmgenomics Pers Med. 2021;14:1315–29. DOI | PMC free article | PubMed | Google Scholar
Kiser JJ, Aquilante CL, Anderson PL, King TM, Carten ML, Fletcher CV. Clinical and genetic determinants of intracellular tenofovir diphosphate concentrations in HIV-infected patients. J Acquir Immune Defic Syndr. 2008;47:298–303. DOI | PubMed | Google Scholar
Cerrone M, Alfarisi O, Neary M, Marzinke MA, Parsons TL, Owen A, et al. Rifampicin effect on intracellular and plasma pharmacokinetics of tenofovir alafenamide. J Antimicrob Chemother. 2019;74:1670–8. DOI | PMC free article | PubMed | Google Scholar
Venter WDF, Moorhouse M, Sokhela S, Fairlie L, Mashabane N, Masenya M, et al. Dolutegravir plus two different prodrugs of tenofovir to treat HIV. N Engl J Med. 2019;381:803–15. DOI | PubMed | Google Scholar
Venter WDF, Sokhela S, Simmons B, Moorhouse M, Fairlie L, Mashabane N, et al. Dolutegravir with emtricitabine and tenofovir alafenamide or tenofovir disoproxil fumarate versus efavirenz, emtricitabine, and tenofovir disoproxil fumarate for initial treatment of HIV-1 infection (ADVANCE): week 96 results from a randomised, phase 3, n. Lancet HIV. 2020;7:e666–76. DOI | PubMed | Google Scholar
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. DOI | PMC free article | PubMed | Google Scholar
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. DOI | PMC free article | PubMed | Google Scholar
Genomes Project C, Altshuler DL, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. DOI | PMC free article | PubMed | Google Scholar
Lift Genome Annotations. https://genome.ucsc.edu/cgi-bin/hgLiftOver. Accessed 17 Feb 2020. https://genome.ucsc.edu/cgi-bin/hgLiftOver
Kawuma A, Wasmann R, Sinxadi P, Sokhela S, Chandiwana N, Venter W, et al. 2022, ‘Population pharmacokinetics of tenofovir given as either tenofovir disoproxil fumarate (TDF) or tenofovir alafenamide (TAF)’, 30th Population Approach Group Europe (PAGE) 2022 conference, Ljubljana, Slovenia 28 June – 1 July Google Scholar
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. DOI | PubMed | Google Scholar
PharmGKB. https://www.pharmgkb.org/. Accessed 13 Jul 2021. https://www.pharmgkb.org/
Buniello A, Macarthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–12. DOI | PMC free article | PubMed | Google Scholar
Li W, Jiang Y, Jin Q, Shi X, Jin J, Gao Y, et al. Expression and gene polymorphisms of interleukin 28B and hepatitis B virus infection in a Chinese Han population. Liver Int. 2011;31:1118–26. DOI | PubMed | Google Scholar
Chihab H, Badre W, Tahiri M, Jadid FZ, Zaidane I, Elfihry R, et al. IFNL4 rs12979860 polymorphism influences HBV DNA viral loads but not the outcome of HBV infection in Moroccan patients. Microbes Infect. 2021;23:4–5. DOI | PubMed | Google Scholar
Machmach K, Abad-Molina C, Romero-Sánchez MC, Abad MA, Ferrando-Martínez S, Genebat M, et al. IL28B single-nucleotide polymorphism rs12979860 is associated with spontaneous HIV control in white subjects. J Infect Dis. 2013;207:651–5. DOI | PubMed | Google Scholar
Sajadi MM, Shakeri N, Talwani R, Howell CD, Pakyz R, Redfield RR, et al. IL28B Genotype Does Not Correlate with HIV Control in African Americans. Clin Transl Sci. 2011;4:282. DOI | PMC free article | PubMed | Google Scholar
Salgado M, Kirk GD, Cox A, Rutebemberwa A, Higgins Y, Astemborski J, et al. Protective interleukin-28B genotype affects hepatitis C virus clearance, but does not contribute to HIV-1 control in a cohort of African–American elite controllers/suppressors. AIDS. 2011;25:385. DOI | PMC free article | PubMed | Google Scholar
Rungtivasuwan K, Avihingsanon A, Thammajaruk N, Mitruk S, Burger DM, Ruxrungtham K, et al. Pharmacogenetics-based population pharmacokinetic analysis of tenofovir in Thai HIV-infected patients. Pharmacogenomics. 2017;18:1481–90. DOI | PubMed | Google Scholar
Calcagno A, Cusato J, Marinaro L, Trentini L, Alcantarini C, Mussa M, et al. Clinical pharmacology of tenofovir clearance: A pharmacokinetic/pharmacogenetic study on plasma and urines. Pharmacogenomics J. 2016;16:514–8. DOI | PubMed | Google Scholar