Pharmacogenetics of 6-mercaptopurine in a black Zimbabwean cohort treated for acute lymphoblastic leukaemia
Metadata
Authors: Pageneck Chikondowa, Derick Munkombwe, Zedias Chikwambi, Patience Kuona, Collen Masimirembwa Journal: Pharmacogenomics Date: 2023 May 30 DOI: 10.2217/pgs-2023-0026 PMID: 37248698 PMCID: PMC10463210 URL: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10463210/ PDF: https://pmc.ncbi.nlm.nih.gov/articles/PMC10463210/pdf/pgs-24-449.pdf
Abstract
Background:: 6-mercaptopurine usage is associated with myelotoxicity and increased risk in patients carrying metabolism-related genetic variations. This study aimed to determine the frequency of candidate gene polymorphisms and their association with 6-mercaptopurine intolerance.
Methods:: A total of 41 patients on acute lymphoblastic leukaemia treatment were genotyped for TPMT and NUDT15 (rs116855232) alleles, and their association with dose intensity was analyzed.
Results:: The defective TPMT3C allele frequency was 9.8%. The median maintenance dose intensity for TPMT1/3C participants was considerably lower (47%) when compared with the TPMT1/*1 wild-type (77%), although not statistically significant.
Conclusion:: This is the first pharmacogenetics study carried out in a black Zimbabwean leukemia patient cohort. The high defective TPMT*3C (9.8%) allele frequency points to the potential utility of pharmacogenetics testing for safe usage of 6-mercaptopurine in this population.
Keywords: 6-mercaptopurine, acute lymphoblastic leukaemia, pharmacogenetics, thiopurine methyltransferase, TPMT, Zimbabwe
Background:
6-mercaptopurine usage is associated with myelotoxicity and increased risk in patients carrying metabolism-related genetic variations. This study aimed to determine the frequency of candidate gene polymorphisms and their association with 6-mercaptopurine intolerance.
Methods:
A total of 41 patients on acute lymphoblastic leukaemia treatment were genotyped for TPMTTPMT and NUDT15NUDT15 (rs116855232) alleles, and their association with dose intensity was analyzed.
Results:
The defective TPMT3CTPMT3C allele frequency was 9.8%. The median maintenance dose intensity for TPMT1/3CTPMT*1/3C participants was considerably lower (47%) when compared with the TPMT1/1TPMT1/*1 wild-type (77%), although not statistically significant.
Conclusion:
This is the first pharmacogenetics study carried out in a black Zimbabwean leukemia patient cohort. The high defective TPMT3CTPMT3C (9.8%) allele frequency points to the potential utility of pharmacogenetics testing for safe usage of 6-mercaptopurine in this population.
**Keywords:**Keywords: 6-mercaptopurine, acute lymphoblastic leukaemia, pharmacogenetics, thiopurine methyltransferase, TPMTTPMT, Zimbabwe
Methodology
An observational, cross-sectional study was conducted at the Parirenyatwa Group of Hospitals (PGH) in Zimbabwe which recruited 41 patients receiving standard-of-care treatment for ALL. Study participants were recruited from PGH pediatric and adult hemato-oncology wards and clinics, while specimen storage, processing and analysis were carried out at the African Institute of Biomedical Science and Technology (Harare, Zimbabwe). A sample size of 28 was calculated using Dobson's formula, and convenience sampling was used to enrol both children (<18 years old) and adults (≥18 years old). We retrospectively identified as potential participants all patients with a confirmed diagnosis of ALL from January 2015 up to October 2022. Participants who had a bone marrow transplant were excluded from the study as it affects the determination of the patient's genotype. ALL was diagnosed using a full blood count and a peripheral film, whereas a bone marrow aspiration and trephine biopsy examination were used for confirmation. The clinical setting had limited access to advanced ALL diagnosis methods such as flow cytometry, cytogenetics, fluorescence in situin situ hybridization or molecular diagnosis.
A blood sample (3 ml) was collected from each enrolled participant for DNA extraction and genotyping. Participants' demographic information was gathered, and retrospective record reviews were carried out to look up baseline features and treatment data. All the adults (n = 4) were receiving induction therapy and none had started taking 6-MP; therefore, no 6-MP dosage information was available in their clinical records. The children were being treated using the Adapted Resource and Implementation Application (ARIA) guide protocol for ALL. The therapies given during the maintenance phase included weekly oral methotrexate (MTX), daily oral 6-MP, monthly intrathecal MTX, a monthly pulse of dexamethasone and monthly intravenous vincristine. For maintenance therapy, the starting dosages of 6-MP and MTX were 75 mg/m^2^2 per day and 20 mg/m^2^2 per week, respectively. At intervals of 4 weeks, a complete blood count was done, and 6-MP dosage was adjusted to keep the white blood cell level between 2.0 and 3.0 × 10^9^9/l. The 6-MP dose intensity (%) was defined as the ratio of the clinician-prescribed 6-MP dose to that of the protocol 6-MP dose during the maintenance stage of therapy to maintain a desirable absolute neutrophil count [1818]. Based on the ARIA guide protocol, the 6-MP dose was adjusted according to toxicities and infections; therefore, the 6-MP dose intensity directly indicated drug sensitivity or tolerance. The dose intensity as a measure of tolerance to 6-MP was included in the comparison analysis with TPMTTPMT and NUDT15NUDT15 polymorphisms.
All the participants and their legal guardians provided written informed consent and assent for children aged 7–17 years, where appropriate. The study was approved by the Joint Research Ethics Committee (JREC) of Parirenyatwa Hospital, the University of Zimbabwe, and the Medical Research Council of Zimbabwe (MRCZ).
Genotyping of TPMT & NUDT15 polymorphism
DNA were extracted from blood using the MagMax™ DNA Multi-Sample Ultra 2.0 Kit on an automated, high-throughput KingFisher Flex Magnetic Particle Processor system (Thermo Fisher Scientific, MA, USA). The extracted DNA was quantified using the Qubit 4 fluorometer (Thermo Fisher Scientific). TaqMan-based genotyping of TPMT2TPMT2 (rs1800462), TPMT3ATPMT3A (rs1800460, rs1142345), TPMT3BTPMT3B (rs1800460), TPMT3CTPMT3C (rs1142345), TPMT4TPMT4 (rs1800584) and NUDT152NUDT152 (rs116855232) was done using the Genopharm^®^® pharmacogenomics open array designed by Thermo Fisher Scientific [2121]. The QuantStudio™ 12 K Flex real-time PCR system and TaqMan PCR reagents from Applied Biosystems (CA, USA) were used.
Statistical analysis
Quantitative participants' demographic data were presented as mean and standard deviation (SD) if normally distributed; otherwise, median and interquartile range (IQR) were used. Respective frequencies and percentages were shown for the qualitative data. Normality was assessed using histograms and the Shapiro-Wilks test. Statistical analysis was performed using Stata v15 and the PRISM software (GraphPad6, CA, USA) was used for visualizations. TPMTTPMT and NUDT15NUDT15 allele and genotype frequencies were computed from the obtained results. Hardy-Weinberg equilibrium (HWE) of the observed and expected genotype frequencies was tested using the χ^2^2 test, with a p-value ≤ 0.05 indicating deviation from HWE. The Mann-Whitney nonparametric test was used for comparing the differences of TPMTTPMT and NUDT15NUDT15 genotypes with 6-MP dose intensity. Associations between other variables such as gender and 6-MP dose intensity were performed using the Mann-Whitney U test. A p-value < 0.05 was regarded as statistically significant for all analyses.
Results
General characteristics of study cohort
A total of 41 participants were enrolled in the study, of whom 37 were children (<18 years old) and 4 were adults (≥18 years old). Patients' baseline characteristics are shown in Table 1Table 1. For the children's group, the median age across ALL diagnoses was 4 years, with an IQR of 3–9 years. A total of 24 (65%) of the children were boys. The children's mean body surface area was 0.80 kg/m^2^2 with a SD of 0.18 kg/m^2^2. Four (11% of the 37 children) had B cells, four (11% of the 37 children) had T cells and 29 (78% of the 37 children) had a pre-B ALL diagnosis. Six children were in the induction phase, 32 had consolidation treatment records available and 23 were in the maintenance phase. A total of four adults were enrolled in the study. Three (75%) were males and the median age was 29 years, with an IQR of 20–49 years old. All the adults (n = 4) were receiving induction therapy and none had started taking 6-MP; therefore, no 6-MP dosage information was available in their clinical records.
Table 1. . General characteristics and demographics of the population (n = 41).
| Characteristics | n (%) | Median (IQR) |
|---|---|---|
| Children (n = 37) | ||
| Male | 24 (65) | |
| Age (years) | 6 (5–9) | |
| Age at diagnosis (years) | 4 (3–9) | |
| BMI | 15.9 (15.1–17.5) | |
| BSA | 0.8 (0.66–0.92) | |
| ALL diagnosis | ||
| B cell | 4 (10.8) | |
| T cell | 4 (10.8) | |
| Pre-B cell | 29 (78.4) | |
| ALL risk classification | ||
| Standard | 18 (48.7) | |
| High risk | 19 (51.30) | |
| Ethnicity | ||
| Ndebele | 3 (8.1) | |
| Shona | 34 (91.9) | |
| Adults (n = 4) | ||
| Male | 3 (75) | |
| Age (years) | 29 (20–49) | |
| Age at diagnosis (years) | 28.5 (19.5–49) | |
| Ethnicity | ||
| Shona | 4 (100) |
Frequency of TPMT & NUDT15 polymorphisms
Genotyping of TPMTTPMT and NUDT15NUDT15 was carried out for the 41 participants. A total of 33 (80%) participants had the wild-type TPMT1/1TPMT1/1 genotype. The remaining eight (20%) participants had a heterozygous TPMT1/3CTPMT1/3C genotype. The homozygous mutant TPMT3C/3CTPMT3C/3C genotype was not observed among the study participants. None of the 41 participants carried the NUDT15NUDT15 rs116855232 CT or TT variant genotypes as shown in Table 2Table 2. The observed and expected TPMTTPMT genotypes conformed with the HWE. The allele frequency for TPMT1TPMT1 was 90.2%, whereas it was 9.8% for TPMT3CTPMT3C. Other TPMTTPMT alleles 22, 3A3A and 3B3B were not detected in this study. The NUDT151NUDT151 allele was detected at a frequency of 100%.
Table 2. . TPMT and NUDT15 genotype frequencies.
| Gene | Genotype | n | Frequency | HWE |
|---|---|---|---|---|
| Observed | Expected | |||
| TPMT | *1/*1 | 33 | 0.805 | 0.814 |
| *1/*3C | 8 | 0.196 | 0.177 | |
| *3C/*3C | 0 | 0 | 0.009 | |
| NUDT15 (rs116855232) | CC | 41 | 1 | 1 |
Comparison between TPMT genotypes & 6-mercaptopurine dose intensity
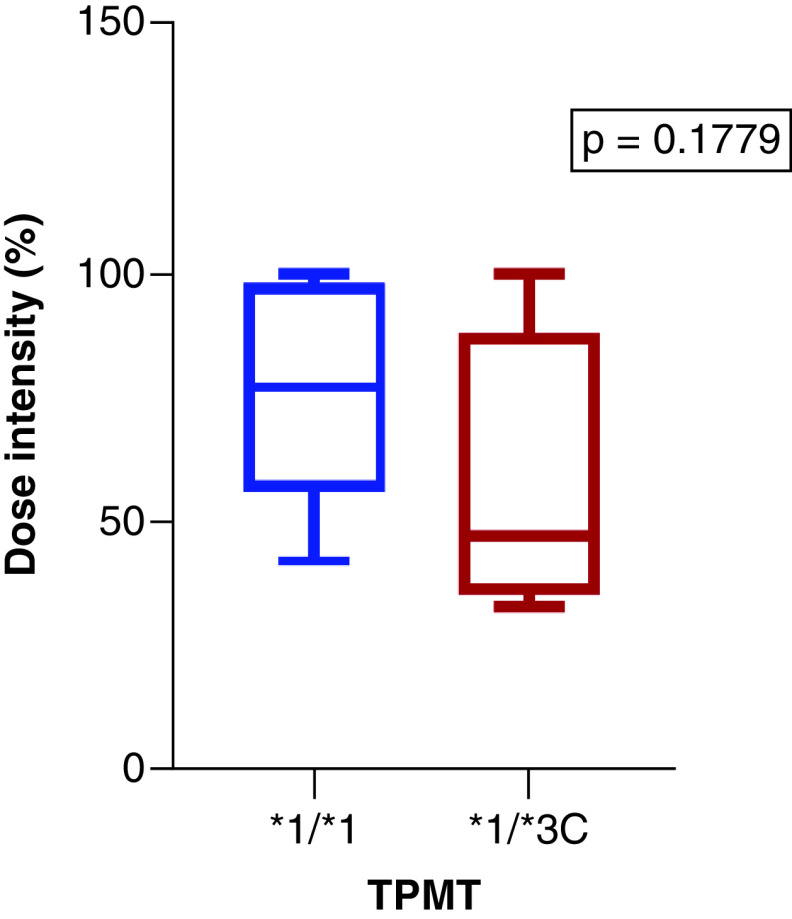
A total of 23 children had 6-MP maintenance records available (Table 1Table 1), whereas none of the adults had started taking 6-MP, so no 6-MP dosage information was available in their clinical records. The maintenance phase duration for the children ranged (IQR) from 6 to 16 months, and the median was 11 months. The median 6-MP dose intensity was 76%, ranging from 58 to 98%. The median 6-MP dose intensity was considerably low (47%) among TPMT1/3CTPMT*1/3C individuals (n = 4) compared with TPMT1/1TPMT1/1 (77%) individuals (n = 19) (Figure 1Figure 1). Similarly, the mean 6-MP dose intensity was lower (58%) for children with a TPMT1/3CTPMT1/3C genotype compared with TPMT1/1TPMT1/1 carriers (76%). However, comparison analysis using the Mann-Whitney test revealed no statistically significant difference (p = 0.1779) in dose intensity between TPMT1/1TPMT1/1 and TPMT1/3CTPMT1/*3C genotypes. Because no participant carried a variant for NUDT15NUDT15, no comparison analysis was performed with a 6-MP dose intensity.
Figure 1. . The comparison of 6-MP dose intensities among TPMT*1/1 (wild type) and TPMT1/*3C (heterozygous) individuals.
p-value was calculated using Mann-Whitney test.
Discussion
In this study, we determined the frequency of candidate genes involved in the metabolism of 6-MP and their association with 6-MP intolerance and toxicity. We replicated the findings from the current body of knowledge that the defective allele TPMT3CTPMT3C is the most frequent allele occurring in sub-Saharan Africans [22–2422–24]. This allele was reported at a frequency of 9.8% in our study cohort of Zimbabwean ALL patients. Similar to what has been reported from studies in Africans, none of the participants carried the NUDT15NUDT15 c.415C>T variant associated with a significantly higher risk of 6-MP-associated leukopenia [2525]. Comparison analysis showed that TPMT3CTPMT3C individuals had a lower 6-MP dose intensity when compared with TPMTTPMT wild-types.
TPMT3CTPMT3C was the only TPMTTPMT polymorphism reported in this study at a frequency of 9.8% (Table 3Table 3), which is considerably higher than the reported 5.3% for sub-Saharan Africans obtained from the PharmGKB database. However, it is comparable to the average global frequency of TPMTTPMT genetic variants, which is around 10% [77]. The comparison of allele frequencies from this study with those from other populations did not reveal any statistically significant difference. Of the fewer studies carried out in Africa so far, they have mostly looked at the frequency of TPMT2TPMT2, 3A3A, 3B3B, 3C3C and 44, and barely NUDT15NUDT15 polymorphisms. Closer to our finding, an allele frequency of 7.6% for TPMT3CTPMT3C was reported in 434 healthy Ghanaians [2323]. In other studies in Africans, allele frequencies of 5.3 and 5.6% were reported for TPMT3CTPMT3C in healthy Nigerians (n = 360) and neurological patients of Black admixed South Africans (n = 184), respectively [2626,2727]. A study carried out in Kenya that recruited 398 ALL patients reported an allele frequency of 5.4% [2828]. These differences in allele frequencies highlight the need to carry out ethnicity- or population-specific studies to guide the use of PGx in thiopurine dosing. All these findings support the uncontested claim that some ethnic groups exhibit considerably lower frequencies of these genetic variations than other groups, which warrants population-specific studies [77]. Based on the studies that have claimed considerable genetic diversity among African populations, it is somehow expected that these subpopulations have deviant allele frequencies [2929,3030]. In agreement with our study, these African studies did not report any TPMT2TPMT2, 3A3A, 3B3B or 44, emphasising the importance of TPMT3CTPMT3C in the PGx of thiopurines in Africans. Most importantly, none of the participants in our study had the NUDT15NUDT15 polymorphism, which agrees with previous studies [2525].
Table 3. . Comparison of allele frequencies from this study cohort with other populations.
| TPMT |
|---|
| Population |
| This study |
| African–American |
| South Asian |
| East Asian |
| European |
| Sub-Saharan Africa |
| The correlation of TPMTTPMT and NUDT15NUDT15 genotypes with 6-MP-induced toxic effects is usually performed during the maintenance phase of the therapy to exclude the influence of other drugs used in ALL treatment because it consists exclusively of 6-MP and low-dose MTX [3131]. However, comparison analysis employing the Mann-Whitney test did not reach statistical significance (p = 0.1779) (Figure 1Figure 1). Similar to this study, Correa-Jimenez *et al.*et al. also failed to demonstrate statistically significant correlations between TPMTTPMT and NUDT15NUDT15 genetic variations and toxicity outcomes [3232]. This lack of statistical significance is possibly due to the small sample size in our study [2525]. Eight out of 23 children (35%) had low-maintenance dose intensities (range: 42–67%), and they did not carry any of the tested TPMTTPMT or NUDT15NUDT15 variants. These patients could have a low-maintenance dose because of poor adherence to their medication; however, we did not measure adherence in this cohort. Furthermore, other genes or genetic variants that affect the activity of the TPMTTPMT gene could be involved since we did not perform TPMTTPMT phenotyping in this study. This probably suggests the need for TPMTTPMT enzymatic activity testing or whole-exome sequencing, which might reveal other TPMTTPMT and/or NUDT15NUDT15 loss-of-function variants not tested in this study, which lead to 6-MP intolerance. Because no NUDT15NUDT15 polymorphism was found in this study, no analysis of the relationship between 6-MP dose intensity and hematological toxicity was performed. Sequencing could have revealed novel and rare NUDT15NUDT15 polymorphisms, as shown in the study by Moriyama *et al.*et al., who discovered three novel NUDT15NUDT15 variants only seen in African and European patients. One of the variants exhibited extremely low thermostability and had no catalytic activity at all, with one of the patients only tolerating 43.5 mg/m^2^2/day of 75 mg/m^2^2/day [1717]. A genome-wide association study in children showed that patients with the homozygous NUDT15NUDT15 TT variant (NUDT15NUDT15 c.415C>T) could only tolerate 8.3% of their standard 6-MP dose, which was 75 mg/m^2^2/day [1818]. Similarly, 6-MP dose reductions of 42 and 82% in patients with the CT and TT genotypes, respectively, were observed in Japanese children with ALL [3333]. Another study of 124 ALL Uruguayan paediatrics discovered that the 6-MP dose was significantly lower in those with one or two TPMTTPMT and NUDT15NUDT15 risk alleles compared to those without risk alleles in all studied intervals [3434]. |
According to Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines, individuals heterozygous for TPMTTPMT should start at 50% and receive 30–70% of the standard dose (75 mg/m^2^2/day) to avoid toxicity as they cannot tolerate their full dose [1414]. From our study, eight (20%) of 41 participants had a TPMT1/3CTPMT*1/*3C heterozygous genotype, and they could have benefited from PGx testing and dose adjustments. The 3C3C allele occurred at a frequency of 9.8%, which is considerably high enough to consider using CPIC clinical guidelines in the Zimbabwean population. This implies that approximately 10 out of 100 individuals could benefit from TPMTTPMT screening. This would probably benefit both patients and the healthcare system by reducing the time needed to optimize the 6-MP dose by the clinicians, reducing unnecessary hospital visits due to frequent toxicity, and minimizing mortality and morbidity. Therefore, all patients diagnosed with ALL would need to be pre-emptively genotyped for TPMTTPMT before they are initiated on treatment. However, looking at resource-limited settings such as Zimbabwe, a reactive PGx approach could be considered. This means that only patients with severe, frequent toxicity and 6-MP intolerance would need to be genotyped for TPMTTPMT and NUDT15NUDT15, and the CPIC guidelines recommendations would be applied based on the results.
Limitations for this study include a shorter study duration and the lack of complete clinical records for some participants. In the future, we recommend a large multicentre, prospective study with longer follow-up, documenting the toxicity and intolerance of 6-MP and its association with TPMTTPMT and NUDT15NUDT15 genotypes. A larger sample size would allow us to make better inferences about the association between toxicity outcomes and generalize the findings to a larger population of patients in Zimbabwe. Future studies should also take either the sequencing approach to uncover novel TPMTTPMT and NUDT15NUDT15 variations that may explain intolerance and toxicity with thiopurines or the genome wide association study (GWAS) approach to identify risk variants in other genes.
Acknowledgments
The authors acknowledge the patients who volunteered to participate in the study and Parirenyatwa Group of Hospital, paediatrics department for helping with participant identification. Comfort Kanji and Marie Hidjo are sincerely acknowledged for providing technical support.
Footnotes
References
Franca R, Braidotti S, Stocco G, Decorti G. Understanding thiopurine methyltransferase polymorphisms for the targeted treatment of hematologic malignancies. Expert Opin. Drug Metab. Toxicol. 17(10), 1187–1198 (2021). DOI | PubMed | Google Scholar
Kotova ES, Gavrilina OA, Sudarikov AB. Significance of TPMT and NUDT15 variants in 6-mercaptopurine metabolism in acute lymphoblastic leukaemia/lymphoma patients. Russ. J. Hematol. Transfusiology 66(2), 253–262 (2021). Google Scholar
Jantararoungtong T, Wiwattanakul S, Tiyasirichokchai R et al. TPMT*3C as a predictor of 6-mercaptopurine-induced myelotoxicity in Thai children with acute lymphoblastic leukemia. J. Pers. Med. 11(8), 783 (2021). DOI | PMC free article | PubMed | Google Scholar
Zhou Y, Wang L, Zhai X-Y et al. Precision therapy of 6-mercaptopurine in Chinese children with acute lymphoblastic leukaemia. Br. J. Clin. Pharmacol. 86(8), 1519–1527 (2020). DOI | PMC free article | PubMed | Google Scholar
Inaba H, Pui C-H. Advances in the diagnosis and treatment of pediatric acute lymphoblastic leukemia. J. Clin. Med. 10(9), 1926 (2021). DOI | PMC free article | PubMed | Google Scholar
Liang D-C, Yang C-P, Liu H-C et al. NUDT15 gene polymorphism related to mercaptopurine intolerance in Taiwan Chinese children with acute lymphoblastic leukemia. Pharmacogenomics J. 16(6), 536–539 (2016). DOI | PubMed | Google Scholar
Moradveisi B, Muwakkit S, Zamani F, Ghaderi E, Mohammadi E, Zgheib NK. ITPA, TPMT, and NUDT15 genetic polymorphisms predict 6-mercaptopurine toxicity in Middle Eastern children with acute lymphoblastic leukemia. Front. Pharmacol. 10, 916 (2019). DOI | PMC free article | PubMed | Google Scholar
Buaboonnam J, Sripatanatadasakul P, Treesucon A et al. Effect of NUDT15 on incidence of neutropenia in children with acute lymphoblastic leukemia. Pediatr. Int. Off. J. Jpn. Pediatr. Soc. 61(8), 754–758 (2019). DOI | PubMed | Google Scholar
Tanaka Y, Yeoh AEJ, Moriyama T et al. An international retrospective study for tolerability of 6-mercaptopurine on NUDT15 bi-allelic variants in children with acute lymphoblastic leukemia. Haematologica 106(7), 2026–2029 (2021). DOI | PMC free article | PubMed | Google Scholar
Zarca K, Chansavang A, Loriot M-A, Durand-Zaleski I, Pallet N. Cost–effectiveness analysis of pretreatment screening for NUDT15 defective alleles. Pharmacogenet. Genomics 30(8), 175–183 (2020). DOI | PubMed | Google Scholar
Rudin S, Marable M, Huang RS. The promise of pharmacogenomics in reducing toxicity during acute lymphoblastic leukemia maintenance treatment. Genomics Proteomics Bioinformatics 15(2), 82–93 (2017). DOI | PMC free article | PubMed | Google Scholar
Koutsilieri S, Caudle KE, Alzghari SK, Monte AA, Relling MV, Patrinos GP. Optimizing thiopurine dosing based on TPMT and NUDT15 genotypes: it takes two to tango. Am. J. Hematol. 94(7), 737–740 (2019). DOI | PubMed | Google Scholar
Schaeffeler E, Jaeger SU, Klumpp V et al. Impact of NUDT15 genetics on severe thiopurine-related hematotoxicity in patients with European ancestry. Genet. Med. 21(9), 2145–2150 (2019). DOI | PMC free article | PubMed | Google Scholar
Relling MV, Schwab M, Whirl-Carrillo M et al. Clinical Pharmacogenetics Implementation Consortium Guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin. Pharmacol. Ther. 105(5), 1095–1105 (2019). DOI | PMC free article | PubMed | Google Scholar
Pratt VM, Wang WY, Boone EC et al. Characterization of reference materials for TPMT and NUDT15: a GeT-RM collaborative project. J. Mol. Diagn. 24(10), 1079–1088 (2022). DOI | PMC free article | PubMed | Google Scholar
Lennard L, Cartwright CS, Wade R, Vora A. Thiopurine dose intensity and treatment outcome in childhood lymphoblastic leukaemia: the influence of thiopurine methyltransferase pharmacogenetics. Br. J. Haematol. 169(2), 228–240 (2015). DOI | PMC free article | PubMed | Google Scholar
Moriyama T, Yang Y-L, Nishii R et al. Novel variants in NUDT15 and thiopurine intolerance in children with acute lymphoblastic leukemia from diverse ancestry. Blood 130(10), 1209–1212 (2017). DOI | PMC free article | PubMed | Google Scholar
Yang JJ, Landier W, Yang W et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J. Clin. Oncol. 33(11), 1235–1242 (2015). DOI | PMC free article | PubMed | Google Scholar
Yang JJ, Whirl-Carrillo M, Scott SA et al. Pharmacogene Variation Consortium gene introduction: NUDT15. Clin. Pharmacol. Ther. 105(5), 1091–1094 (2019). DOI | PMC free article | PubMed | Google Scholar
Zhou H, Li L, Yang P et al. Optimal predictor for 6-mercaptopurine intolerance in Chinese children with acute lymphoblastic leukemia: NUDT15, TPMT, or ITPA genetic variants? BMC Cancer 18(1), 516 (2018). DOI | PMC free article | PubMed | Google Scholar
Mbavha BT, Kanji CR, Stadler N et al. Population genetic polymorphisms of pharmacogenes in Zimbabwe, a potential guide for the safe and efficacious use of medicines in people of African ancestry. Pharmacogenet. Genomics 32(5), 173–182 (2022). DOI | PubMed | Google Scholar
Woillard J-B, Chouchana L, Picard N, Loriot M-A. Pharmacogenetics of immunosuppressants: State of the art and clinical implementation – recommendations from the French National Network of Pharmacogenetics (RNPGx). Therapies 72(2), 285–299 (2017). DOI | PubMed | Google Scholar
Ameyaw M. Thiopurine methyltransferase alleles in British and Ghanaian populations. Hum. Mol. Genet. 8(2), 367–370 (1999). DOI | PubMed | Google Scholar
Almoguera B, Vazquez L, Connolly JJ et al. Imputation of TPMT defective alleles for the identification of patients with high-risk phenotypes. Front. Genet. 5, 96 (2014). DOI | PMC free article | PubMed | Google Scholar
Zhang AL, Yang J, Wang H, Lu JL, Tang S, Zhang XJ. Association of NUDT15 c.415C>T allele and thiopurine-induced leukocytopenia in Asians: a systematic review and meta-analysis. Ir. J. Med. Sci. 187(1), 145–153 (2018). DOI | PubMed | Google Scholar
Adehin A, Bolaji OO, Kennedy MA, Adeagbo BA. Allele frequencies of thiopurine S-methyltransferase (TPMT) variants in the Nigerian population. Pol. Ann. Med. 24(2), 144–147 (2017). Google Scholar
Heckmann JM, Lambson EMT, Little F, Owen EP. Thiopurine methyltransferase (TPMT) heterozygosity and enzyme activity as predictive tests for the development of azathioprine-related adverse events. J. Neurol. Sci. 231(1-2), 71–80 (2005). DOI | PubMed | Google Scholar
McLeod HL, Pritchard SC, Githang J et al. Ethnic differences in thiopurine methyltransferase pharmacogenetics: evidence for allele specificity in Caucasian and Kenyan individuals. Pharmacogenetics 9(6), 773–776 (1999). DOI | PubMed | Google Scholar
Da Rocha JE, Othman H, Botha G et al. The extent and impact of variation in ADME genes in sub-Saharan African populations. Front. Pharmacol. 12, 634016 (2021). DOI | PMC free article | PubMed | Google Scholar
Rajman I, Knapp L, Morgan T, Masimirembwa C. African genetic diversity: implications for cytochrome P450-mediated drug metabolism and drug development. EBioMedicine 17, 67–74 (2017). DOI | PMC free article | PubMed | Google Scholar
Kodidela S, Dorababu P, Thakkar DN et al. Association of NUDT15 c.415C>T and FPGS 2572C>T variants with the risk of early hematologic toxicity during 6-MP and low-dose methotrexate-based maintenance therapy in indian patients with acute lymphoblastic leukemia. Genes 11(6), 594 (2020). DOI | PMC free article | PubMed | Google Scholar
Correa-Jimenez O, Yunis JJ, Linares-Ballesteros A, Sarmiento-Urbina I. Susceptibility to thiopurine toxicity by TPMT and NUDT15 variants in Colombian children with acute lymphoblastic leukemia. Colomb. Medica Cali Colomb. 52(3), e2074569 (2021). DOI | PMC free article | PubMed | Google Scholar
Tanaka Y, Kato M, Hasegawa D et al. Susceptibility to 6-MP toxicity conferred by a NUDT15 variant in Japanese children with acute lymphoblastic leukaemia. Br. J. Haematol. 171(1), 109–115 (2015). DOI | PubMed | Google Scholar
Soler AM, Olano N, Méndez Y et al. TPMT and NUDT15 genes are both related to mercaptopurine intolerance in acute lymphoblastic leukaemia patients from Uruguay. Br. J. Haematol. 181(2), 252–255 (2018). DOI | PubMed | Google Scholar