code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
<img src="https://rhyme.com/assets/img/logo-dark.png" align="center">
<h2 align="center">Linear Regression</h2>
Linear Regression is a useful tool for predicting a quantitative response.
We have an input vector $X^T = (X_1, X_2,...,X_p)$, and want to predict a real-valued output $Y$. The linear regression model has the form
<h4 align="center"> $f(x) = \beta_0 + \sum_{j=1}^p X_j \beta_j$. </h4>
The linear model either assumes that the regression function $E(Y|X)$ is linear, or that the linear model is a reasonable approximation.Here the $\beta_j$'s are unknown parameters or coefficients, and the variables $X_j$ can come from different sources. No matter the source of $X_j$, the model is linear in the parameters.
### Task 1: Import Libraries
---
```
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
%matplotlib inline
```
### Task 2: Load the Data
The adverstiting dataset captures sales revenue generated with respect to advertisement spends across multiple channles like radio, tv and newspaper. [Source](http://www-bcf.usc.edu/~gareth/ISL/Advertising.csv)
```
advert = pd.read_csv('Advertising.csv')
advert.head()
advert.info()
```
### Task 3: Remove the index column
```
advert.columns
advert.drop(['Unnamed: 0'], axis = 1, inplace = True)
advert.head()
```
### Task 4: Exploratory Analysis
```
import seaborn as sns
sns.distplot(advert.sales);
sns.distplot(advert.newspaper);
sns.distplot(advert.radio);
sns.distplot(advert.TV);
```
### Task 5: Exploring Relationships between Predictors and Response
```
sns.pairplot(advert, x_vars=['TV', 'radio', 'newspaper'], y_vars='sales', height=7, aspect=0.7, kind='reg');
advert.TV.corr(advert.sales)
advert.corr()
sns.heatmap( advert.corr(), annot=True );
```
### Task 6: Creating the Simple Linear Regression Model
General linear regression model:
$y=\beta_{0}+\beta_{1}x_{1}+\beta_{2}x_{2}+...+\beta_{n}x_{n}$
- $y$ is the response
- $\beta_{0}$ is the intercept
- $\beta_{1}$ is the coefficient for x1 (the first feature)
- $\beta_{n}$ is the coefficient for xn (the nth feature)
- $\beta_1$ is the feature coefficient and $y$ is the y-intercept
In our case: $y=\beta_{0}+\beta_{1}×TV+\beta_{2}×Radio+\beta_{3}×Newspaper$
The $\beta$ values are called the **model coefficients*:
- These values are "learned" during the model fitting step using the "least squares" criterion
- The fitted model is then used to make predictions
```
X = advert[['TV']]
X.head()
# check the type and shape of X
print(type(X))
print(X.shape)
y = advert.sales
y.head()
print(type(y))
print(y.shape)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
from sklearn.linear_model import LinearRegression
linreg = LinearRegression()
linreg.fit(X_train, y_train)
```
### Task 7: Interpreting Model Coefficients
```
# print the intercept and coefficients
print(linreg.intercept_)
print(linreg.coef_)
zip(advert.TV, linreg.coef_)
```
### Task 8: Making Predictions with our Model
```
# make predictions on the testing set
y_pred = linreg.predict(X_test)
```
### Task 9: Model Evaluation Metrics
```
# define true and predicted response values
true = [100, 50, 30, 20]
pred = [90, 50, 50, 30]
```
**Mean Absolute Error** (MAE) is the mean of the absolute value of the errors:;
$$ \frac{1}{n} \sum_{i=1}^{n} \left |y_i - \hat{y}_i \right |$$
```
# calculate MAE by hand
print((10 + 0 + 20 + 10) / 4)
# calculate MAE using scikit-learn
from sklearn import metrics
print(metrics.mean_absolute_error(true, pred))
```
**Mean Squared Error** (MSE) is the mean of the squared errors:
$$\frac{1}{n} \sum_{i=1}^{n} (y_i - \hat{y}_i)^2$$
```
# calculate MSE by hand
print((10**2 + 0**2 + 20**2 + 10**2) / 4)
# calculate MSE using scikit-learn
print(metrics.mean_squared_error(true, pred))
```
**Root Mean Squared Error** (RMSE) is the square root of the mean of the squared errors:
$$\sqrt{\frac{1}{n} \sum_{i=1}^{n} (y_i - \hat{y}_i)^2}$$
```
# calculate RMSE by hand
print(np.sqrt(((10**2 + 0**2 + 20**2 + 10**2) / 4)))
# calculate RMSE using scikit-learn
print(np.sqrt(metrics.mean_squared_error(true, pred)))
print(np.sqrt(metrics.mean_squared_error(y_test, y_pred)))
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
%matplotlib inline
advert = pd.read_csv('Advertising.csv')
advert.head()
advert.info()
advert.columns
advert.drop(['Unnamed: 0'], axis = 1, inplace = True)
advert.head()
import seaborn as sns
sns.distplot(advert.sales);
sns.distplot(advert.newspaper);
sns.distplot(advert.radio);
sns.distplot(advert.TV);
sns.pairplot(advert, x_vars=['TV', 'radio', 'newspaper'], y_vars='sales', height=7, aspect=0.7, kind='reg');
advert.TV.corr(advert.sales)
advert.corr()
sns.heatmap( advert.corr(), annot=True );
X = advert[['TV']]
X.head()
# check the type and shape of X
print(type(X))
print(X.shape)
y = advert.sales
y.head()
print(type(y))
print(y.shape)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
from sklearn.linear_model import LinearRegression
linreg = LinearRegression()
linreg.fit(X_train, y_train)
# print the intercept and coefficients
print(linreg.intercept_)
print(linreg.coef_)
zip(advert.TV, linreg.coef_)
# make predictions on the testing set
y_pred = linreg.predict(X_test)
# define true and predicted response values
true = [100, 50, 30, 20]
pred = [90, 50, 50, 30]
# calculate MAE by hand
print((10 + 0 + 20 + 10) / 4)
# calculate MAE using scikit-learn
from sklearn import metrics
print(metrics.mean_absolute_error(true, pred))
# calculate MSE by hand
print((10**2 + 0**2 + 20**2 + 10**2) / 4)
# calculate MSE using scikit-learn
print(metrics.mean_squared_error(true, pred))
# calculate RMSE by hand
print(np.sqrt(((10**2 + 0**2 + 20**2 + 10**2) / 4)))
# calculate RMSE using scikit-learn
print(np.sqrt(metrics.mean_squared_error(true, pred)))
print(np.sqrt(metrics.mean_squared_error(y_test, y_pred)))
| 0.424889 | 0.978135 |
# Concept Drift
In the context of data streams, it is assumed that data can change over time. The change in the relationship between the data (features) and the target to learn is known as **Concept Drift**. As examples we can mention, the electricity demand across the year, the stock market, and the likelihood of a new movie to be successful. Let's consider the movie example: Two movies can have similar features such as popular actors/directors, storyline, production budget, marketing campaigns, etc. yet it is not certain that both will be similarly successful. What the target audience *considers* worth watching (and their money) is constantly changing and production companies must adapt accordingly to avoid "box office flops".
## Impact of drift on learning
Concept drift can have a significant impact on predictive performance if not handled properly. Most batch learning models will fail in the presence of concept drift as they are essentially trained on different data. On the other hand, stream learning methods continuously update themselves and adapt to new concepts. Furthermore, drift-aware methods use change detection methods (a.k.a. drift detectors) to trigger *mitigation mechanisms* if a change in performance is detected.
## Detecting concept drift
Multiple drift detection methods have been proposed. The goal of a drift detector is to signal an alarm in the presence of drift. A good drift detector maximizes the number of true positives while keeping the number of false positives to a minimum. It must also be resource-wise efficient to work in the context of infinite data streams.
For this example, we will generate a synthetic data stream by concatenating 3 distributions of 1000 samples each:
- $dist_a$: $\mu=0.8$, $\sigma=0.05$
- $dist_b$: $\mu=0.4$, $\sigma=0.02$
- $dist_c$: $\mu=0.6$, $\sigma=0.1$.
```
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import gridspec
# Generate data for 3 distributions
random_state = np.random.RandomState(seed=42)
dist_a = random_state.normal(0.8, 0.05, 1000)
dist_b = random_state.normal(0.4, 0.02, 1000)
dist_c = random_state.normal(0.6, 0.1, 1000)
# Concatenate data to simulate a data stream with 2 drifts
stream = np.concatenate((dist_a, dist_b, dist_c))
# Auxiliary function to plot the data
def plot_data(dist_a, dist_b, dist_c, drifts=None):
fig = plt.figure(figsize=(7,3), tight_layout=True)
gs = gridspec.GridSpec(1, 2, width_ratios=[3, 1])
ax1, ax2 = plt.subplot(gs[0]), plt.subplot(gs[1])
ax1.grid()
ax1.plot(stream, label='Stream')
ax2.grid(axis='y')
ax2.hist(dist_a, label=r'$dist_a$')
ax2.hist(dist_b, label=r'$dist_b$')
ax2.hist(dist_c, label=r'$dist_c$')
if drifts is not None:
for drift_detected in drifts:
ax1.axvline(drift_detected, color='red')
plt.show()
plot_data(dist_a, dist_b, dist_c)
```
### Drift detection test
We will use the ADaptive WINdowing (`ADWIN`) drift detection method. Remember that the goal is to indicate that drift has occurred after samples **1000** and **2000** in the synthetic data stream.
```
from river import drift
drift_detector = drift.ADWIN()
drifts = []
for i, val in enumerate(stream):
drift_detector.update(val) # Data is processed one sample at a time
if drift_detector.change_detected:
# The drift detector indicates after each sample if there is a drift in the data
print(f'Change detected at index {i}')
drifts.append(i)
drift_detector.reset() # As a best practice, we reset the detector
plot_data(dist_a, dist_b, dist_c, drifts)
```
We see that `ADWIN` successfully indicates the presence of drift (red vertical lines) close to the begining of a new data distribution.
---
We conclude this example with some remarks regarding concept drift detectors and their usage:
- In practice, drift detectors provide stream learning methods with robustness against concept drift. Drift detectors monitor the model usually through a performance metric.
- Drift detectors work on univariate data. This is why they are used to monitor a model's performance and not the data itself. Remember that concept drift is defined as a change in the relationship between data and the target to learn (in supervised learning).
- Drift detectors define their expectations regarding input data. It is important to know these expectations to feed a given drift detector with the correct data.
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import gridspec
# Generate data for 3 distributions
random_state = np.random.RandomState(seed=42)
dist_a = random_state.normal(0.8, 0.05, 1000)
dist_b = random_state.normal(0.4, 0.02, 1000)
dist_c = random_state.normal(0.6, 0.1, 1000)
# Concatenate data to simulate a data stream with 2 drifts
stream = np.concatenate((dist_a, dist_b, dist_c))
# Auxiliary function to plot the data
def plot_data(dist_a, dist_b, dist_c, drifts=None):
fig = plt.figure(figsize=(7,3), tight_layout=True)
gs = gridspec.GridSpec(1, 2, width_ratios=[3, 1])
ax1, ax2 = plt.subplot(gs[0]), plt.subplot(gs[1])
ax1.grid()
ax1.plot(stream, label='Stream')
ax2.grid(axis='y')
ax2.hist(dist_a, label=r'$dist_a$')
ax2.hist(dist_b, label=r'$dist_b$')
ax2.hist(dist_c, label=r'$dist_c$')
if drifts is not None:
for drift_detected in drifts:
ax1.axvline(drift_detected, color='red')
plt.show()
plot_data(dist_a, dist_b, dist_c)
from river import drift
drift_detector = drift.ADWIN()
drifts = []
for i, val in enumerate(stream):
drift_detector.update(val) # Data is processed one sample at a time
if drift_detector.change_detected:
# The drift detector indicates after each sample if there is a drift in the data
print(f'Change detected at index {i}')
drifts.append(i)
drift_detector.reset() # As a best practice, we reset the detector
plot_data(dist_a, dist_b, dist_c, drifts)
| 0.647464 | 0.990216 |
```
from pyspark.sql import *
from pyspark.sql.types import Row
from pyspark.sql import functions as f
from pyspark.sql.functions import unix_timestamp, from_unixtime
from pyspark.sql.functions import *
from pyspark.sql.types import *
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
from pyspark.ml.fpm import FPGrowth
import plotly.express as px
import plotly.graph_objects as go
from plotly.subplots import make_subplots
import plotly.io as pio
from datetime import datetime, timedelta
df = spark.read.csv("superstore.csv", inferSchema = True, header = True)
print(df.count())
df.printSchema()
data = df.drop("Row ID", "City", "State", "Postal Code", "Region", "Customer Name", "Order Priority", "Discount", "Shipping Cost", "Ship Mode", "Ship Date")
data = data.withColumn("Order Date", f.regexp_replace("Order Date", "/", "-"))
data = data.withColumn("Order Date",to_date(data["Order Date"], 'dd-MM-yyyy'))
data = data.withColumn("Order Date", data["Order Date"].cast(StringType()))
data = data.withColumn("Month", data["Order Date"].substr(0, 7))
data = data.withColumn("Quantity", data["Quantity"].cast("long"))
data = data.withColumn("Sales", data["Sales"].cast("double"))
print(data.printSchema())
```
# Most ordered products
```
prod = data.select(data["Product ID"], data["Quantity"])
most_prod = prod.groupby("Product ID").sum().withColumnRenamed("sum(Quantity)", "Total Ordered")
topten=most_prod.orderBy("Total Ordered" ,ascending = False).limit(20)
topten.show()
print(prod.count())
print(most_prod.count())
p = topten.select(topten["Product ID"])
o = topten.select(topten["Total Ordered"])
l1 = [row["Product ID"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
plt.bar(l1, l2)
plt.xticks(l1,rotation="vertical")
plt.xlabel("Product ID")
plt.ylabel("Total Quantity Ordered")
plt.savefig('product_orders.png', dpi=300, bbox_inches='tight')
plt.show()
```
# Most Ordered Category
### Most Ordered Sub-Category
```
subcatg = data.select(data["Sub-Category"], data["Quantity"])
most_subcatg = subcatg.groupBy(subcatg["Sub-Category"]).sum().withColumnRenamed("sum(Quantity)", "Total Ordered")
most_subcatg.orderBy(most_subcatg["Total Ordered"], ascending = False).show()
p = most_subcatg.select(most_subcatg["Sub-Category"])
o = most_subcatg.select(most_subcatg["Total Ordered"])
l1 = [row["Sub-Category"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
plt.bar(l1, l2)
plt.xticks(l1,rotation="vertical")
plt.xlabel("Sub-Category")
plt.ylabel("Total Quantity Ordered")
plt.show()
```
### Most Ordered Category
```
catg = data.select(data["Category"], data["Quantity"])
most_catg = catg.groupBy(catg["Category"]).sum().withColumnRenamed("sum(Quantity)", "Total Ordered")
most_catg.orderBy(most_catg["Total Ordered"], ascending = False).show()
p = most_catg.select(most_catg["Category"])
o = most_catg.select(most_catg["Total Ordered"])
l1 = [row["Category"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
sizes=[122015,41011,35388]
labels="Office Supplies","Furniture","Technology"
plt.pie(sizes,labels=labels,shadow=True,autopct='%1.1f%%')
plt.savefig('category_orders.png', dpi=300, bbox_inches='tight')
plt.show()
```
# Country with the most orders
```
country = data.select(data["Country"], data["Order ID"])
most_country = country.distinct().groupBy(country["Country"]).count().withColumnRenamed("count", "Total Ordered")
most_country.orderBy(most_country["Total Ordered"], ascending = False).show()
countries=most_country.rdd.map(lambda x: x[0]).collect()
vs=most_country.rdd.map(lambda x: x[1]).collect()
dt=[dict(type="choropleth", autocolorscale=True, locations=countries, z=vs, locationmode="country names", text=countries, colorbar= dict(title='Orders'))]
layout=dict(geo=dict(scope="world", projection=dict(type="natural earth"), showlakes=True, lakecolor='rgb(0,0,255)'))
fig=dict(data=dt, layout=layout)
pio.show(fig)
```
# Market with the most orders
```
market = data.select(data["Market"], data["Quantity"])
most_market = market.groupBy(market["Market"]).sum().withColumnRenamed("sum(Quantity)", "Total Ordered")
most_market.orderBy(most_market["Total Ordered"], ascending = False).show()
p = most_market.select(most_market["Market"])
o = most_market.select(most_market["Total Ordered"])
l1 = [row["Market"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
plt.bar(l1, l2)
plt.xticks(l1,rotation="vertical")
plt.xlabel("Market")
plt.ylabel("Total Quantity Ordered")
plt.show()
```
# Segment with most orders
```
segm = data.select(data["Segment"], data["Quantity"])
most_segm = segm.groupBy(segm["Segment"]).sum().withColumnRenamed("sum(Quantity)", "Total Ordered")
most_segm.orderBy(most_segm["Total Ordered"], ascending = False).show()
p = most_segm.select(most_segm["Segment"])
o = most_segm.select(most_segm["Total Ordered"])
l1 = [row["Segment"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
sizes=[100554, 61548, 36312]
labels= "Consumer", "Corporate", "Home Office"
plt.pie(sizes,labels=labels,shadow=True,autopct='%1.1f%%')
plt.savefig('segment_orders.png', dpi=300, bbox_inches='tight')
plt.show()
```
# Most Profitable Products
```
profit = data.select(data["Product ID"], data["Profit"])
prod_profit = profit.groupBy(profit["Product ID"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
top = prod_profit.orderBy(prod_profit["Total Profit"], ascending = False).limit(20)
#print(top.show())
mix = topten.join(top, "Product ID", "full")
print(mix.orderBy(mix["Total Ordered"], ascending = False).show())
print(mix.orderBy(mix["Total Profit"], ascending = False).show())
p = top.select(top["Product ID"])
o = top.select(top["Total Profit"])
l1 = [row["Product ID"] for row in p.collect()]
l2 = [row["Total Profit"] for row in o.collect()]
plt.bar(l1, l2)
plt.xticks(l1,rotation="vertical")
plt.xlabel("Product ID")
plt.ylabel("Total Profit")
plt.savefig('prod_profit.png', dpi=300, bbox_inches='tight')
plt.show()
```
# Most Profit per Category
### Sub-Category
```
sub_profit = data.select(data["Sub-Category"], data["Profit"])
most_sub_porfit = sub_profit.groupBy(sub_profit["Sub-Category"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_sub_porfit.orderBy(most_sub_porfit["Total Profit"], ascending = False).show()
# p = most_sub_porfit.select(most_sub_porfit["Sub-Category"])
# o = most_sub_porfit.select(most_sub_porfit["Total Profit"])
# l1 = [row["Sub-Category"] for row in p.collect()]
# l2 = [row["Total Profit"] for row in o.collect()]
# plt.bar(l1, l2)
# plt.xticks(l1,rotation="vertical")
# plt.xlabel("Sub-Category")
# plt.ylabel("Total Profit")
# plt.show()
p = most_subcatg.select(most_subcatg["Sub-Category"])
o = most_subcatg.select(most_subcatg["Total Ordered"])
l1 = [row["Sub-Category"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
m = most_sub_porfit.select(most_sub_porfit["Sub-Category"])
n = most_sub_porfit.select(most_sub_porfit["Total Profit"])
l3 = [row["Sub-Category"] for row in m.collect()]
l4 = [row["Total Profit"] for row in n.collect()]
fig,ax2=plt.subplots()
ax1=ax2.twinx()
ax2.bar(l1, l2)
ax1.plot(l3, l4, 'r')
ax2.set_xlabel("Sub-Category")
ax2.set_ylabel("Number of Orders")
ax1.set_ylabel("Total Profit", color = "r")
ax2.set_xticklabels(l1,rotation='vertical')
plt.savefig('Sub-Category.png', dpi=300, bbox_inches='tight')
plt.show()
```
### Category
```
cat_profit = data.select(data["Category"], data["Profit"])
most_cat_profit = cat_profit.groupBy(cat_profit["Category"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_cat_profit.orderBy(most_cat_profit["Total Profit"], ascending = False).show()
p = most_cat_profit.select(most_cat_profit["Category"])
o = most_cat_profit.select(most_cat_profit["Total Profit"])
l1 = [row["Category"] for row in p.collect()]
l2 = [row["Total Profit"] for row in o.collect()]
sizes=[663712.0816800011, 516615.9118999997, 286439.87820000004]
labels= "Technology", "Office Supplies", "Furniture"
plt.pie(sizes,labels=labels,shadow=True,autopct='%1.1f%%')
plt.savefig('category_profit.png', dpi=300, bbox_inches='tight')
plt.show()
```
# Most profit per country
```
country_prof = data.select(data["Country"], data["Profit"])
most_country_prof = country_prof.groupBy(country_prof["Country"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_country_prof.orderBy(most_country_prof["Total Profit"], ascending = False).show()
prcountries=most_country_prof.rdd.map(lambda x: x[0]).collect()
prvs=most_country_prof.rdd.map(lambda x: x[1]).collect()
pdt=[dict(type="choropleth", autocolorscale=True, locations=prcountries, z=prvs, locationmode="country names", text=prcountries, colorbar= dict(title='Profit'))]
playout=dict(geo=dict(scope="world", projection=dict(type="natural earth"), showlakes=True, lakecolor='rgb(0,0,255)'))
fig=dict(data=pdt, layout=playout)
pio.show(fig)
```
# Most profit per market
```
mark_prof = data.select(data["Market"], data["Profit"])
most_mark_prof = mark_prof.groupBy(mark_prof["Market"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_mark_prof.orderBy(most_mark_prof["Total Profit"], ascending = False).show()
# p = most_mark_prof.select(most_mark_prof["Market"])
# o = most_mark_prof.select(most_mark_prof["Total Profit"])
# l1 = [row["Market"] for row in p.collect()]
# l2 = [row["Total Profit"] for row in o.collect()]
# plt.bar(l1, l2)
# plt.xticks(l1,rotation="vertical")
# plt.xlabel("Market")
# plt.ylabel("Total Profit")
# plt.show()
p = most_market.select(most_market["Market"])
o = most_market.select(most_market["Total Ordered"])
l1 = [row["Market"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
m = most_mark_prof.select(most_mark_prof["Market"])
n = most_mark_prof.select(most_mark_prof["Total Profit"])
l3 = [row["Market"] for row in m.collect()]
l4 = [row["Total Profit"] for row in n.collect()]
fig,ax2=plt.subplots()
ax1=ax2.twinx()
ax2.bar(l1, l2)
ax1.plot(l3, l4, 'r')
ax2.set_xlabel("Market")
ax2.set_ylabel("Number of Orders")
ax1.set_ylabel("Total Profit", color = "r")
ax2.set_xticklabels(l1,rotation='vertical')
plt.savefig('market.png', dpi=300, bbox_inches='tight')
plt.show()
```
# Most profit per segment
```
seg_prof = data.select(data["Segment"], data["Profit"])
most_seg_prof = seg_prof.groupBy(seg_prof["Segment"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_seg_prof.orderBy(most_seg_prof["Total Profit"], ascending = False).show()
p = most_seg_prof.select(most_seg_prof["Segment"])
o = most_seg_prof.select(most_seg_prof["Total Profit"])
l1 = [row["Segment"] for row in p.collect()]
l2 = [row["Total Profit"] for row in o.collect()]
sizes=[749564.4320600019, 440659.1595600009, 276544.2801599999]
labels= "Consumer", "Corporate", "Home Office"
plt.pie(sizes,labels=labels,shadow=True,autopct='%1.1f%%')
plt.savefig('segment_profit.png', dpi=300, bbox_inches='tight')
plt.show()
```
# Month with most orders
```
date = data.select(data["Month"], data["Order ID"])
date.show()
# split_date=pyspark.sql.functions.split(df['Date'], '[-/]')
month_order = date.groupBy(date["Month"]).count()
month_orders = month_order.orderBy(month_order["Month"], ascending = True)
month_orders.show()
```
# Monthly Profit
```
month_profit = data.select(data["Month"], data["Profit"])
most_month_profit = month_profit.groupBy(month_profit["Month"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_month_profits = most_month_profit.orderBy(most_month_profit["Month"])
most_month_profits.show()
p = month_orders.select(month_orders["Month"])
o = month_orders.select(month_orders["count"])
l1 = [row["Month"] for row in p.collect()]
l2 = [row["count"] for row in o.collect()]
m = most_month_profits.select(most_month_profits["Month"])
n = most_month_profits.select(most_month_profits["Total Profit"])
l3 = [row["Month"] for row in m.collect()]
l4 = [row["Total Profit"] for row in n.collect()]
fig,ax2=plt.subplots()
ax1=ax2.twinx()
ax2.bar(l1, l2)
ax1.plot(l3, l4, 'r')
ax2.set_xlabel("Month")
ax2.set_ylabel("Number of Orders")
ax1.set_ylabel("Total Profit", color = "r")
ax2.set_xticklabels(l1,rotation='vertical')
fig.set_size_inches(15.5,7.5)
plt.savefig('month.png', dpi=300, bbox_inches='tight')
plt.show()
```
# Customer Orders
```
cust_order = data.select(data["Order ID"], data["Customer ID"])
most_cust_order = cust_order.distinct()
most_cust_order = most_cust_order.groupBy(most_cust_order["Customer ID"]).count()
top_cust = most_cust_order.orderBy(most_cust_order["count"], ascending = False).limit(20)
top_cust.show()
q = top_cust.select(top_cust["Customer ID"])
t = top_cust.select(top_cust["count"])
l5 = [row["Customer ID"] for row in q.collect()]
l6 = [row["count"] for row in t.collect()]
plt.bar(l5, l6)
plt.xticks(l5,rotation="vertical")
plt.xlabel("Customer ID")
plt.ylabel("Total Orders")
plt.savefig('customers.png', dpi=300, bbox_inches='tight')
plt.show()
```
# Days with most orders
```
dwmo=data.rdd.map(lambda x: (((datetime(int(x[1][:4]),int(x[1][5:7]),int(x[1][8:10])).strftime("%A")),datetime(int(x[1][:4]),int(x[1][5:7]),int(x[1][8:10])).weekday()),1)).\
reduceByKey(lambda x,y:x+y).sortBy(lambda x: x[0][1])
dwmo.collect()
days=dwmo.map(lambda x: x[0][0]).collect()
dvls=dwmo.map(lambda x:x[1]).collect()
plt.bar(days, dvls)
plt.xticks(days, rotation=70,size=10)
plt.title("Number Of Orders By Day",size=18)
plt.show()
```
# Frequent Patterns
```
orders = data.select(data["Order ID"], data["Product ID"])
orders = orders.distinct()
rdd = orders.rdd.map(lambda x: (x[0], [x[1]]))
rdd.take(5)
rdd1 = rdd.reduceByKey(lambda x,y: x+y)
rdd1.take(5)
order = sqlContext.createDataFrame(rdd1, ['Order ID', 'Product ID'])
fpGrowth = FPGrowth(itemsCol="Product ID", minSupport=0.00001, minConfidence=0.05)
model = fpGrowth.fit(order)
model.associationRules.show()
model.freqItemsets.show()
```
|
github_jupyter
|
from pyspark.sql import *
from pyspark.sql.types import Row
from pyspark.sql import functions as f
from pyspark.sql.functions import unix_timestamp, from_unixtime
from pyspark.sql.functions import *
from pyspark.sql.types import *
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
from pyspark.ml.fpm import FPGrowth
import plotly.express as px
import plotly.graph_objects as go
from plotly.subplots import make_subplots
import plotly.io as pio
from datetime import datetime, timedelta
df = spark.read.csv("superstore.csv", inferSchema = True, header = True)
print(df.count())
df.printSchema()
data = df.drop("Row ID", "City", "State", "Postal Code", "Region", "Customer Name", "Order Priority", "Discount", "Shipping Cost", "Ship Mode", "Ship Date")
data = data.withColumn("Order Date", f.regexp_replace("Order Date", "/", "-"))
data = data.withColumn("Order Date",to_date(data["Order Date"], 'dd-MM-yyyy'))
data = data.withColumn("Order Date", data["Order Date"].cast(StringType()))
data = data.withColumn("Month", data["Order Date"].substr(0, 7))
data = data.withColumn("Quantity", data["Quantity"].cast("long"))
data = data.withColumn("Sales", data["Sales"].cast("double"))
print(data.printSchema())
prod = data.select(data["Product ID"], data["Quantity"])
most_prod = prod.groupby("Product ID").sum().withColumnRenamed("sum(Quantity)", "Total Ordered")
topten=most_prod.orderBy("Total Ordered" ,ascending = False).limit(20)
topten.show()
print(prod.count())
print(most_prod.count())
p = topten.select(topten["Product ID"])
o = topten.select(topten["Total Ordered"])
l1 = [row["Product ID"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
plt.bar(l1, l2)
plt.xticks(l1,rotation="vertical")
plt.xlabel("Product ID")
plt.ylabel("Total Quantity Ordered")
plt.savefig('product_orders.png', dpi=300, bbox_inches='tight')
plt.show()
subcatg = data.select(data["Sub-Category"], data["Quantity"])
most_subcatg = subcatg.groupBy(subcatg["Sub-Category"]).sum().withColumnRenamed("sum(Quantity)", "Total Ordered")
most_subcatg.orderBy(most_subcatg["Total Ordered"], ascending = False).show()
p = most_subcatg.select(most_subcatg["Sub-Category"])
o = most_subcatg.select(most_subcatg["Total Ordered"])
l1 = [row["Sub-Category"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
plt.bar(l1, l2)
plt.xticks(l1,rotation="vertical")
plt.xlabel("Sub-Category")
plt.ylabel("Total Quantity Ordered")
plt.show()
catg = data.select(data["Category"], data["Quantity"])
most_catg = catg.groupBy(catg["Category"]).sum().withColumnRenamed("sum(Quantity)", "Total Ordered")
most_catg.orderBy(most_catg["Total Ordered"], ascending = False).show()
p = most_catg.select(most_catg["Category"])
o = most_catg.select(most_catg["Total Ordered"])
l1 = [row["Category"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
sizes=[122015,41011,35388]
labels="Office Supplies","Furniture","Technology"
plt.pie(sizes,labels=labels,shadow=True,autopct='%1.1f%%')
plt.savefig('category_orders.png', dpi=300, bbox_inches='tight')
plt.show()
country = data.select(data["Country"], data["Order ID"])
most_country = country.distinct().groupBy(country["Country"]).count().withColumnRenamed("count", "Total Ordered")
most_country.orderBy(most_country["Total Ordered"], ascending = False).show()
countries=most_country.rdd.map(lambda x: x[0]).collect()
vs=most_country.rdd.map(lambda x: x[1]).collect()
dt=[dict(type="choropleth", autocolorscale=True, locations=countries, z=vs, locationmode="country names", text=countries, colorbar= dict(title='Orders'))]
layout=dict(geo=dict(scope="world", projection=dict(type="natural earth"), showlakes=True, lakecolor='rgb(0,0,255)'))
fig=dict(data=dt, layout=layout)
pio.show(fig)
market = data.select(data["Market"], data["Quantity"])
most_market = market.groupBy(market["Market"]).sum().withColumnRenamed("sum(Quantity)", "Total Ordered")
most_market.orderBy(most_market["Total Ordered"], ascending = False).show()
p = most_market.select(most_market["Market"])
o = most_market.select(most_market["Total Ordered"])
l1 = [row["Market"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
plt.bar(l1, l2)
plt.xticks(l1,rotation="vertical")
plt.xlabel("Market")
plt.ylabel("Total Quantity Ordered")
plt.show()
segm = data.select(data["Segment"], data["Quantity"])
most_segm = segm.groupBy(segm["Segment"]).sum().withColumnRenamed("sum(Quantity)", "Total Ordered")
most_segm.orderBy(most_segm["Total Ordered"], ascending = False).show()
p = most_segm.select(most_segm["Segment"])
o = most_segm.select(most_segm["Total Ordered"])
l1 = [row["Segment"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
sizes=[100554, 61548, 36312]
labels= "Consumer", "Corporate", "Home Office"
plt.pie(sizes,labels=labels,shadow=True,autopct='%1.1f%%')
plt.savefig('segment_orders.png', dpi=300, bbox_inches='tight')
plt.show()
profit = data.select(data["Product ID"], data["Profit"])
prod_profit = profit.groupBy(profit["Product ID"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
top = prod_profit.orderBy(prod_profit["Total Profit"], ascending = False).limit(20)
#print(top.show())
mix = topten.join(top, "Product ID", "full")
print(mix.orderBy(mix["Total Ordered"], ascending = False).show())
print(mix.orderBy(mix["Total Profit"], ascending = False).show())
p = top.select(top["Product ID"])
o = top.select(top["Total Profit"])
l1 = [row["Product ID"] for row in p.collect()]
l2 = [row["Total Profit"] for row in o.collect()]
plt.bar(l1, l2)
plt.xticks(l1,rotation="vertical")
plt.xlabel("Product ID")
plt.ylabel("Total Profit")
plt.savefig('prod_profit.png', dpi=300, bbox_inches='tight')
plt.show()
sub_profit = data.select(data["Sub-Category"], data["Profit"])
most_sub_porfit = sub_profit.groupBy(sub_profit["Sub-Category"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_sub_porfit.orderBy(most_sub_porfit["Total Profit"], ascending = False).show()
# p = most_sub_porfit.select(most_sub_porfit["Sub-Category"])
# o = most_sub_porfit.select(most_sub_porfit["Total Profit"])
# l1 = [row["Sub-Category"] for row in p.collect()]
# l2 = [row["Total Profit"] for row in o.collect()]
# plt.bar(l1, l2)
# plt.xticks(l1,rotation="vertical")
# plt.xlabel("Sub-Category")
# plt.ylabel("Total Profit")
# plt.show()
p = most_subcatg.select(most_subcatg["Sub-Category"])
o = most_subcatg.select(most_subcatg["Total Ordered"])
l1 = [row["Sub-Category"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
m = most_sub_porfit.select(most_sub_porfit["Sub-Category"])
n = most_sub_porfit.select(most_sub_porfit["Total Profit"])
l3 = [row["Sub-Category"] for row in m.collect()]
l4 = [row["Total Profit"] for row in n.collect()]
fig,ax2=plt.subplots()
ax1=ax2.twinx()
ax2.bar(l1, l2)
ax1.plot(l3, l4, 'r')
ax2.set_xlabel("Sub-Category")
ax2.set_ylabel("Number of Orders")
ax1.set_ylabel("Total Profit", color = "r")
ax2.set_xticklabels(l1,rotation='vertical')
plt.savefig('Sub-Category.png', dpi=300, bbox_inches='tight')
plt.show()
cat_profit = data.select(data["Category"], data["Profit"])
most_cat_profit = cat_profit.groupBy(cat_profit["Category"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_cat_profit.orderBy(most_cat_profit["Total Profit"], ascending = False).show()
p = most_cat_profit.select(most_cat_profit["Category"])
o = most_cat_profit.select(most_cat_profit["Total Profit"])
l1 = [row["Category"] for row in p.collect()]
l2 = [row["Total Profit"] for row in o.collect()]
sizes=[663712.0816800011, 516615.9118999997, 286439.87820000004]
labels= "Technology", "Office Supplies", "Furniture"
plt.pie(sizes,labels=labels,shadow=True,autopct='%1.1f%%')
plt.savefig('category_profit.png', dpi=300, bbox_inches='tight')
plt.show()
country_prof = data.select(data["Country"], data["Profit"])
most_country_prof = country_prof.groupBy(country_prof["Country"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_country_prof.orderBy(most_country_prof["Total Profit"], ascending = False).show()
prcountries=most_country_prof.rdd.map(lambda x: x[0]).collect()
prvs=most_country_prof.rdd.map(lambda x: x[1]).collect()
pdt=[dict(type="choropleth", autocolorscale=True, locations=prcountries, z=prvs, locationmode="country names", text=prcountries, colorbar= dict(title='Profit'))]
playout=dict(geo=dict(scope="world", projection=dict(type="natural earth"), showlakes=True, lakecolor='rgb(0,0,255)'))
fig=dict(data=pdt, layout=playout)
pio.show(fig)
mark_prof = data.select(data["Market"], data["Profit"])
most_mark_prof = mark_prof.groupBy(mark_prof["Market"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_mark_prof.orderBy(most_mark_prof["Total Profit"], ascending = False).show()
# p = most_mark_prof.select(most_mark_prof["Market"])
# o = most_mark_prof.select(most_mark_prof["Total Profit"])
# l1 = [row["Market"] for row in p.collect()]
# l2 = [row["Total Profit"] for row in o.collect()]
# plt.bar(l1, l2)
# plt.xticks(l1,rotation="vertical")
# plt.xlabel("Market")
# plt.ylabel("Total Profit")
# plt.show()
p = most_market.select(most_market["Market"])
o = most_market.select(most_market["Total Ordered"])
l1 = [row["Market"] for row in p.collect()]
l2 = [row["Total Ordered"] for row in o.collect()]
m = most_mark_prof.select(most_mark_prof["Market"])
n = most_mark_prof.select(most_mark_prof["Total Profit"])
l3 = [row["Market"] for row in m.collect()]
l4 = [row["Total Profit"] for row in n.collect()]
fig,ax2=plt.subplots()
ax1=ax2.twinx()
ax2.bar(l1, l2)
ax1.plot(l3, l4, 'r')
ax2.set_xlabel("Market")
ax2.set_ylabel("Number of Orders")
ax1.set_ylabel("Total Profit", color = "r")
ax2.set_xticklabels(l1,rotation='vertical')
plt.savefig('market.png', dpi=300, bbox_inches='tight')
plt.show()
seg_prof = data.select(data["Segment"], data["Profit"])
most_seg_prof = seg_prof.groupBy(seg_prof["Segment"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_seg_prof.orderBy(most_seg_prof["Total Profit"], ascending = False).show()
p = most_seg_prof.select(most_seg_prof["Segment"])
o = most_seg_prof.select(most_seg_prof["Total Profit"])
l1 = [row["Segment"] for row in p.collect()]
l2 = [row["Total Profit"] for row in o.collect()]
sizes=[749564.4320600019, 440659.1595600009, 276544.2801599999]
labels= "Consumer", "Corporate", "Home Office"
plt.pie(sizes,labels=labels,shadow=True,autopct='%1.1f%%')
plt.savefig('segment_profit.png', dpi=300, bbox_inches='tight')
plt.show()
date = data.select(data["Month"], data["Order ID"])
date.show()
# split_date=pyspark.sql.functions.split(df['Date'], '[-/]')
month_order = date.groupBy(date["Month"]).count()
month_orders = month_order.orderBy(month_order["Month"], ascending = True)
month_orders.show()
month_profit = data.select(data["Month"], data["Profit"])
most_month_profit = month_profit.groupBy(month_profit["Month"]).sum().withColumnRenamed("sum(Profit)", "Total Profit")
most_month_profits = most_month_profit.orderBy(most_month_profit["Month"])
most_month_profits.show()
p = month_orders.select(month_orders["Month"])
o = month_orders.select(month_orders["count"])
l1 = [row["Month"] for row in p.collect()]
l2 = [row["count"] for row in o.collect()]
m = most_month_profits.select(most_month_profits["Month"])
n = most_month_profits.select(most_month_profits["Total Profit"])
l3 = [row["Month"] for row in m.collect()]
l4 = [row["Total Profit"] for row in n.collect()]
fig,ax2=plt.subplots()
ax1=ax2.twinx()
ax2.bar(l1, l2)
ax1.plot(l3, l4, 'r')
ax2.set_xlabel("Month")
ax2.set_ylabel("Number of Orders")
ax1.set_ylabel("Total Profit", color = "r")
ax2.set_xticklabels(l1,rotation='vertical')
fig.set_size_inches(15.5,7.5)
plt.savefig('month.png', dpi=300, bbox_inches='tight')
plt.show()
cust_order = data.select(data["Order ID"], data["Customer ID"])
most_cust_order = cust_order.distinct()
most_cust_order = most_cust_order.groupBy(most_cust_order["Customer ID"]).count()
top_cust = most_cust_order.orderBy(most_cust_order["count"], ascending = False).limit(20)
top_cust.show()
q = top_cust.select(top_cust["Customer ID"])
t = top_cust.select(top_cust["count"])
l5 = [row["Customer ID"] for row in q.collect()]
l6 = [row["count"] for row in t.collect()]
plt.bar(l5, l6)
plt.xticks(l5,rotation="vertical")
plt.xlabel("Customer ID")
plt.ylabel("Total Orders")
plt.savefig('customers.png', dpi=300, bbox_inches='tight')
plt.show()
dwmo=data.rdd.map(lambda x: (((datetime(int(x[1][:4]),int(x[1][5:7]),int(x[1][8:10])).strftime("%A")),datetime(int(x[1][:4]),int(x[1][5:7]),int(x[1][8:10])).weekday()),1)).\
reduceByKey(lambda x,y:x+y).sortBy(lambda x: x[0][1])
dwmo.collect()
days=dwmo.map(lambda x: x[0][0]).collect()
dvls=dwmo.map(lambda x:x[1]).collect()
plt.bar(days, dvls)
plt.xticks(days, rotation=70,size=10)
plt.title("Number Of Orders By Day",size=18)
plt.show()
orders = data.select(data["Order ID"], data["Product ID"])
orders = orders.distinct()
rdd = orders.rdd.map(lambda x: (x[0], [x[1]]))
rdd.take(5)
rdd1 = rdd.reduceByKey(lambda x,y: x+y)
rdd1.take(5)
order = sqlContext.createDataFrame(rdd1, ['Order ID', 'Product ID'])
fpGrowth = FPGrowth(itemsCol="Product ID", minSupport=0.00001, minConfidence=0.05)
model = fpGrowth.fit(order)
model.associationRules.show()
model.freqItemsets.show()
| 0.359364 | 0.755005 |
# <img style="float: left; padding-right: 10px; width: 45px" src="https://github.com/Harvard-IACS/2018-CS109A/blob/master/content/styles/iacs.png?raw=true"> CS109A Introduction to Data Science
## Lecture 14 (PCA and High Dimensionality)
**Harvard University**<br>
**Fall 2019**<br>
**Instructors:** Pavlos Protopapas, Kevin Rader, and Chris Tanner<br>
---
```
import pandas as pd
import sys
import numpy as np
import scipy as sp
import sklearn as sk
import matplotlib.pyplot as plt
# import statsmodels.api as sm
# from statsmodels.tools import add_constant
# from statsmodels.regression.linear_model import RegressionResults
# import seaborn as sns
# from sklearn.preprocessing import MinMaxScaler
# from sklearn.model_selection import KFold
# from sklearn.linear_model import LinearRegression
# from sklearn.linear_model import Ridge
# from sklearn.linear_model import Lasso
# from sklearn.preprocessing import PolynomialFeatures
# from sklearn.neighbors import KNeighborsRegressor
from sklearn.linear_model import LogisticRegression
from sklearn.decomposition import PCA
# sns.set(style="ticks")
# %matplotlib inline
heart_df = pd.read_csv('../data/Heart.csv')
print(heart_df.shape)
heart_df.head()
heart_df.describe()
# For pedagogical purposes, let's simplify our lives and use just 4 predictors
X = heart_df[['Age','RestBP','Chol','MaxHR']]
y = 1*(heart_df['AHD']=='Yes')
#fit the 'full' model on the 4 predictors. and print out the coefficients
logit_full = LogisticRegression(C=1000000,solver="lbfgs").fit(X,y)
beta = logit_full.coef_[0]
print(beta)
# investigating what happens when two identical predictors are used
logit1 = LogisticRegression(C=1000000,solver="lbfgs").fit(heart_df[['Age']],y)
logit2 = LogisticRegression(C=1000000,solver="lbfgs").fit(heart_df[['Age','Age']],y)
print("The coef estimate for Age (when in the model once):",logit1.coef_)
print("The coef estimates for Age (when in the model twice):",logit2.coef_)
X = heart_df[['Age','RestBP','Chol','MaxHR']]
# create/fit the 'full' pca transformation
pca = PCA().fit(X)
# apply the pca transformation to the full predictor set
pcaX = pca.transform(X)
# convert to a data frame
pcaX_df = pd.DataFrame(pcaX, columns=[['PCA1' , 'PCA2', 'PCA3', 'PCA4']])
# here are the weighting (eigen-vectors) of the variables (first 2 at least)
print("First PCA Component (w1):",pca.components_[0,:])
print("Second PCA Component (w2):",pca.components_[1,:])
# here is the variance explained:
print("Variance explained by each component:",pca.explained_variance_ratio_)
# Plot the response over the first 2 PCA component vectors
plt.scatter(pcaX_df['PCA1'][y==0],pcaX_df['PCA2'][y==0])
plt.scatter(pcaX_df['PCA1'][y==1],pcaX_df['PCA2'][y==1])
plt.legend(["AHD = No","AHD = Yes"])
plt.xlabel("First PCA Component Vector (Z1)")
plt.ylabel("Second PCA Component Vector (Z2)");
logit_pcr1 = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaX_df[['PCA1']],y)
print("Intercept from simple PCR-Logistic:",logit_pcr1.intercept_)
print("'Slope' from simple PCR-Logistic:", logit_pcr1.coef_)
print("First PCA Component (w1):",pca.components_[0,:])
# Fit the other 3 PCRs on the rest of the 4 predictors
logit_pcr2 = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaX_df[['PCA1','PCA2']],y)
logit_pcr3 = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaX_df[['PCA1','PCA2','PCA3']],y)
logit_pcr4 = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaX_df[['PCA1','PCA2','PCA3','PCA4']],y)
pcr1=(logit_pcr1.coef_*np.transpose(pca.components_[0:1,:])).sum(axis=1)
pcr2=(logit_pcr2.coef_*np.transpose(pca.components_[0:2,:])).sum(axis=1)
pcr3=(logit_pcr3.coef_*np.transpose(pca.components_[0:3,:])).sum(axis=1)
pcr4=(logit_pcr4.coef_*np.transpose(pca.components_[0:4,:])).sum(axis=1)
print(pcr1)
print(pcr2)
print(pcr3)
print(pcr4)
results = np.vstack((pcr1,pcr2,pcr3,pcr4,beta))
plt.plot(['PCR1' , 'PCR2', 'PCR3', 'PCR4', 'Logistic'],results)
plt.ylabel("Back-calculated Beta Coefficients");
plt.legend(X.columns);
scaler = sk.preprocessing.StandardScaler()
scaler.fit(X)
Z = scaler.transform(X)
pca = PCA(n_components=4).fit(Z)
pcaZ = pca.transform(Z)
pcaZ_df = pd.DataFrame(pcaZ, columns=[['PCA1' , 'PCA2', 'PCA3', 'PCA4']])
print("First PCA Component (w1):",pca.components_[0,:])
print("Second PCA Component (w2):",pca.components_[1,:])
#fit the 'full' model on the 4 predictors. and print out the coefficients
logit_full = LogisticRegression(C=1000000,solver="lbfgs").fit(Z,y)
betaZ = logit_full.coef_[0]
print("Logistic coef. on standardized predictors:",betaZ)
# Fit the PCR
logit_pcr1Z = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaZ_df[['PCA1']],y)
logit_pcr2Z = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaZ_df[['PCA1','PCA2']],y)
logit_pcr3Z = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaZ_df[['PCA1','PCA2','PCA3']],y)
logit_pcr4Z = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaZ_df[['PCA1','PCA2','PCA3','PCA4']],y)
pcr1Z=(logit_pcr1Z.coef_*np.transpose(pca.components_[0:1,:])).sum(axis=1)
pcr2Z=(logit_pcr2Z.coef_*np.transpose(pca.components_[0:2,:])).sum(axis=1)
pcr3Z=(logit_pcr3Z.coef_*np.transpose(pca.components_[0:3,:])).sum(axis=1)
pcr4Z=(logit_pcr4Z.coef_*np.transpose(pca.components_[0:4,:])).sum(axis=1)
resultsZ = np.vstack((pcr1Z,pcr2Z,pcr3Z,pcr4Z,betaZ))
print(resultsZ)
plt.plot(['PCR1-Z' , 'PCR2-Z', 'PCR3-Z', 'PCR4-Z', 'Logistic'],resultsZ)
plt.ylabel("Back-calculated Beta Coefficients");
plt.legend(X.columns);
```
---
|
github_jupyter
|
import pandas as pd
import sys
import numpy as np
import scipy as sp
import sklearn as sk
import matplotlib.pyplot as plt
# import statsmodels.api as sm
# from statsmodels.tools import add_constant
# from statsmodels.regression.linear_model import RegressionResults
# import seaborn as sns
# from sklearn.preprocessing import MinMaxScaler
# from sklearn.model_selection import KFold
# from sklearn.linear_model import LinearRegression
# from sklearn.linear_model import Ridge
# from sklearn.linear_model import Lasso
# from sklearn.preprocessing import PolynomialFeatures
# from sklearn.neighbors import KNeighborsRegressor
from sklearn.linear_model import LogisticRegression
from sklearn.decomposition import PCA
# sns.set(style="ticks")
# %matplotlib inline
heart_df = pd.read_csv('../data/Heart.csv')
print(heart_df.shape)
heart_df.head()
heart_df.describe()
# For pedagogical purposes, let's simplify our lives and use just 4 predictors
X = heart_df[['Age','RestBP','Chol','MaxHR']]
y = 1*(heart_df['AHD']=='Yes')
#fit the 'full' model on the 4 predictors. and print out the coefficients
logit_full = LogisticRegression(C=1000000,solver="lbfgs").fit(X,y)
beta = logit_full.coef_[0]
print(beta)
# investigating what happens when two identical predictors are used
logit1 = LogisticRegression(C=1000000,solver="lbfgs").fit(heart_df[['Age']],y)
logit2 = LogisticRegression(C=1000000,solver="lbfgs").fit(heart_df[['Age','Age']],y)
print("The coef estimate for Age (when in the model once):",logit1.coef_)
print("The coef estimates for Age (when in the model twice):",logit2.coef_)
X = heart_df[['Age','RestBP','Chol','MaxHR']]
# create/fit the 'full' pca transformation
pca = PCA().fit(X)
# apply the pca transformation to the full predictor set
pcaX = pca.transform(X)
# convert to a data frame
pcaX_df = pd.DataFrame(pcaX, columns=[['PCA1' , 'PCA2', 'PCA3', 'PCA4']])
# here are the weighting (eigen-vectors) of the variables (first 2 at least)
print("First PCA Component (w1):",pca.components_[0,:])
print("Second PCA Component (w2):",pca.components_[1,:])
# here is the variance explained:
print("Variance explained by each component:",pca.explained_variance_ratio_)
# Plot the response over the first 2 PCA component vectors
plt.scatter(pcaX_df['PCA1'][y==0],pcaX_df['PCA2'][y==0])
plt.scatter(pcaX_df['PCA1'][y==1],pcaX_df['PCA2'][y==1])
plt.legend(["AHD = No","AHD = Yes"])
plt.xlabel("First PCA Component Vector (Z1)")
plt.ylabel("Second PCA Component Vector (Z2)");
logit_pcr1 = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaX_df[['PCA1']],y)
print("Intercept from simple PCR-Logistic:",logit_pcr1.intercept_)
print("'Slope' from simple PCR-Logistic:", logit_pcr1.coef_)
print("First PCA Component (w1):",pca.components_[0,:])
# Fit the other 3 PCRs on the rest of the 4 predictors
logit_pcr2 = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaX_df[['PCA1','PCA2']],y)
logit_pcr3 = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaX_df[['PCA1','PCA2','PCA3']],y)
logit_pcr4 = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaX_df[['PCA1','PCA2','PCA3','PCA4']],y)
pcr1=(logit_pcr1.coef_*np.transpose(pca.components_[0:1,:])).sum(axis=1)
pcr2=(logit_pcr2.coef_*np.transpose(pca.components_[0:2,:])).sum(axis=1)
pcr3=(logit_pcr3.coef_*np.transpose(pca.components_[0:3,:])).sum(axis=1)
pcr4=(logit_pcr4.coef_*np.transpose(pca.components_[0:4,:])).sum(axis=1)
print(pcr1)
print(pcr2)
print(pcr3)
print(pcr4)
results = np.vstack((pcr1,pcr2,pcr3,pcr4,beta))
plt.plot(['PCR1' , 'PCR2', 'PCR3', 'PCR4', 'Logistic'],results)
plt.ylabel("Back-calculated Beta Coefficients");
plt.legend(X.columns);
scaler = sk.preprocessing.StandardScaler()
scaler.fit(X)
Z = scaler.transform(X)
pca = PCA(n_components=4).fit(Z)
pcaZ = pca.transform(Z)
pcaZ_df = pd.DataFrame(pcaZ, columns=[['PCA1' , 'PCA2', 'PCA3', 'PCA4']])
print("First PCA Component (w1):",pca.components_[0,:])
print("Second PCA Component (w2):",pca.components_[1,:])
#fit the 'full' model on the 4 predictors. and print out the coefficients
logit_full = LogisticRegression(C=1000000,solver="lbfgs").fit(Z,y)
betaZ = logit_full.coef_[0]
print("Logistic coef. on standardized predictors:",betaZ)
# Fit the PCR
logit_pcr1Z = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaZ_df[['PCA1']],y)
logit_pcr2Z = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaZ_df[['PCA1','PCA2']],y)
logit_pcr3Z = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaZ_df[['PCA1','PCA2','PCA3']],y)
logit_pcr4Z = LogisticRegression(C=1000000,solver="lbfgs").fit(pcaZ_df[['PCA1','PCA2','PCA3','PCA4']],y)
pcr1Z=(logit_pcr1Z.coef_*np.transpose(pca.components_[0:1,:])).sum(axis=1)
pcr2Z=(logit_pcr2Z.coef_*np.transpose(pca.components_[0:2,:])).sum(axis=1)
pcr3Z=(logit_pcr3Z.coef_*np.transpose(pca.components_[0:3,:])).sum(axis=1)
pcr4Z=(logit_pcr4Z.coef_*np.transpose(pca.components_[0:4,:])).sum(axis=1)
resultsZ = np.vstack((pcr1Z,pcr2Z,pcr3Z,pcr4Z,betaZ))
print(resultsZ)
plt.plot(['PCR1-Z' , 'PCR2-Z', 'PCR3-Z', 'PCR4-Z', 'Logistic'],resultsZ)
plt.ylabel("Back-calculated Beta Coefficients");
plt.legend(X.columns);
| 0.501221 | 0.889337 |
Nama : Ghozy Ghulamul Afif
NIM : 1301170379
Judul TA : Implementasi Information Gain (IG) dan Genetic Algorithm (GA) untuk Reduksi Dimensi pada Klasifikasi Data Microarray Menggunakan Functional Link Neural Network (FLNN)
Pembimbing 1 : Widi Astuti, S.T., M.Kom.
Pembimbing 2 : Prof. Dr. Adiwijaya
# 1. Preprocessing
## 1.1. Import Library
```
import pandas as pd
from sklearn.preprocessing import MinMaxScaler
from pandas import DataFrame
from scipy.special import legendre
import numpy as np
from sklearn.model_selection import StratifiedKFold
import matplotlib.pyplot as plt
import keras
from tensorflow.python.keras.layers import Dense
from keras.optimizers import Adam
from tensorflow.python.keras import Sequential
from sklearn.metrics import accuracy_score, f1_score
from sklearn.metrics import classification_report
import random
import timeit
from sklearn.feature_selection import mutual_info_classif
from sklearn.feature_selection import SelectKBest
```
## 1.2. Import Dataset
```
# data colon
url = "https://raw.githubusercontent.com/jamessaldo/final-task/master/colonTumor.data"
data_colon = pd.read_csv(url, header=None)
```
## 1.3. Check Missing Value
```
print('Total Missing Value pada Data colon Tumor:',data_colon.isnull().sum().sum())
```
## 1.4. Normalization
```
# Melakukan normalisasi
# data colon
data_new_colon = data_colon.drop([2000],axis=1)
scaler = MinMaxScaler()
data_new_colon = scaler.fit_transform(data_new_colon)
data_new_colon = DataFrame(data_new_colon)
data_new_colon['label'] = list(data_colon[2000])
dic = {'negative':1,'positive':0}
data_new_colon.replace(dic,inplace=True)
```
# 2. Define Reusable Function
## FLNN Classifier
```
def FLNN_Classifier(data_train, data_test, orde, lr):
start = timeit.default_timer()
x_data_train = data_train.drop(['label'],axis=1)
y_data_train = data_train['label']
x_data_test = data_test.drop(['label'],axis=1)
y_data_test = data_test['label']
df_train = pd.DataFrame()
df_test = pd.DataFrame()
for x in range(1, orde+1):
pn = legendre(x)
y_orde = pn(x_data_train)
df_train = pd.concat([df_train, y_orde], axis=1)
pn = legendre(x)
y_orde = pn(x_data_test)
df_test = pd.concat([df_test, y_orde], axis=1)
df_train.columns = ["Attribut"+str(i) for i in range(len(df_train.columns))]
df_train['label'] = y_data_train.reset_index().label
X_train = df_train.iloc[:, 0:len(df_train.columns)-1].values
y_train = df_train.iloc[:, len(df_train.columns)-1].values
df_test.columns = ["Attribut"+str(i) for i in range(len(df_test.columns))]
df_test['label'] = y_data_test.reset_index().label
X_test = df_test.iloc[:, 0:len(df_test.columns)-1].values
y_test = df_test.iloc[:, len(df_test.columns)-1].values
# Melakukan proses klasifikasi FLNN
# Inisialisasi FLNN
Model = Sequential()
# Menambah input layer dan hidden layer pertama
Model.add(Dense(units = len(df_train.columns)-1, kernel_initializer = 'uniform', input_dim = len(df_train.columns)-1))
# Menambah output layer
Model.add(Dense(units = 1, kernel_initializer = 'uniform', activation = 'sigmoid'))
# Menjalankan ANN
Model.compile(optimizer = Adam(learning_rate=lr), loss = 'mean_squared_error', metrics = ['accuracy'])
# Fitting ANN ke training set
history = Model.fit(X_train, y_train, batch_size = 50, epochs = 100, validation_split = 0.2, verbose=False)
#Memprediksi hasil test set
y_pred = Model(X_test)
y_pred =(y_pred >= 0.5)
#print("X_Train :", X_train)
print("Y_Train :", y_train)
#print("X_Test :", X_test)
print("Y_Test :", y_test)
akurasi = accuracy_score(y_test,y_pred)
F1 = f1_score(y_test, y_pred, average='macro')
print("Akurasi : ", akurasi)
print("F1_Score : ", F1)
print(classification_report(y_test,y_pred))
# Membuat confusion matrix
from sklearn.metrics import confusion_matrix
from mlxtend.plotting import plot_confusion_matrix
cm = confusion_matrix(y_test, y_pred)
fig, ax = plot_confusion_matrix(conf_mat = cm, figsize=(5,5))
plt.show()
stop = timeit.default_timer()
print('Running Time: ', stop - start)
return akurasi, F1, stop-start
```
# 3. Classification
```
start = timeit.default_timer()
akurasi_IG_2_v1,f1_IG_2_v1,rt_IG_2_v1 = [],[],[]
akurasi_IG_3_v1,f1_IG_3_v1,rt_IG_3_v1 = [],[],[]
akurasi_IG_4_v1,f1_IG_4_v1,rt_IG_4_v1 = [],[],[]
akurasi_IG_2_v2,f1_IG_2_v2,rt_IG_2_v2 = [],[],[]
akurasi_IG_3_v2,f1_IG_3_v2,rt_IG_3_v2 = [],[],[]
akurasi_IG_4_v2,f1_IG_4_v2,rt_IG_4_v2 = [],[],[]
#Melakukan proses K-Fold
kf = StratifiedKFold(n_splits=5, shuffle=True, random_state=10)
kf.get_n_splits(data_new_colon)
X = data_new_colon.copy().iloc[:, 0:len(data_new_colon.columns)-1].values
Y = data_new_colon.copy().iloc[:, len(data_new_colon.columns)-1].values
for train_index, test_index in kf.split(X,Y):
print("Train : ", train_index, "Test : ", test_index)
data_train, data_test, y_train, y_test = pd.DataFrame(X[train_index]), pd.DataFrame(X[test_index]), Y[train_index], Y[test_index]
data_train['label'] = y_train
data_test['label'] = y_test
print("colon Orde 2 v1")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 2, 0.6)
akurasi_IG_2_v1.append(acc)
f1_IG_2_v1.append(f1)
rt_IG_2_v1.append(rt)
print("colon Orde 3 v1")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 3, 0.6)
akurasi_IG_3_v1.append(acc)
f1_IG_3_v1.append(f1)
rt_IG_3_v1.append(rt)
print("colon Orde 4 v1")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 4, 0.6)
akurasi_IG_4_v1.append(acc)
f1_IG_4_v1.append(f1)
rt_IG_4_v1.append(rt)
print("colon Orde 2 v2")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 2, 0.001)
akurasi_IG_2_v2.append(acc)
f1_IG_2_v2.append(f1)
rt_IG_2_v2.append(rt)
print("colon Orde 3 v2")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 3, 0.001)
akurasi_IG_3_v2.append(acc)
f1_IG_3_v2.append(f1)
rt_IG_3_v2.append(rt)
print("colon Orde 4 v1")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 4, 0.001)
akurasi_IG_4_v2.append(acc)
f1_IG_4_v2.append(f1)
rt_IG_4_v2.append(rt)
akurasi_IG_2_v1,f1_IG_2_v1,rt_IG_2_v1 = np.array(akurasi_IG_2_v1),np.array(f1_IG_2_v1),np.array(rt_IG_2_v1)
akurasi_IG_3_v1,f1_IG_3_v1,rt_IG_3_v1 = np.array(akurasi_IG_3_v1),np.array(f1_IG_3_v1),np.array(rt_IG_3_v1)
akurasi_IG_4_v1,f1_IG_4_v1,rt_IG_4_v1 = np.array(akurasi_IG_4_v1),np.array(f1_IG_4_v1),np.array(rt_IG_4_v1)
akurasi_IG_2_v2,f1_IG_2_v2,rt_IG_2_v2 = np.array(akurasi_IG_2_v2),np.array(f1_IG_2_v2),np.array(rt_IG_2_v2)
akurasi_IG_3_v2,f1_IG_3_v2,rt_IG_3_v2 = np.array(akurasi_IG_3_v2),np.array(f1_IG_3_v2),np.array(rt_IG_3_v2)
akurasi_IG_4_v2,f1_IG_4_v2,rt_IG_4_v2 = np.array(akurasi_IG_4_v2),np.array(f1_IG_4_v2),np.array(rt_IG_4_v2)
#Print Result
print('===============================================================================================================================================================================================================')
print('Avg accuracy colon cancer orde 2 v1 : ', akurasi_IG_2_v1.mean())
print('Avg F1 score colon cancer orde 2 v1 : ', f1_IG_2_v1.mean())
print('Avg running time colon cancer orde 2 v1 : ', rt_IG_2_v1.mean())
print('Avg accuracy colon cancer orde 3 v1 : ', akurasi_IG_3_v1.mean())
print('Avg F1 score colon cancer orde 3 v1 : ', f1_IG_3_v1.mean())
print('Avg running time colon cancer orde 3 v1 : ', rt_IG_3_v1.mean())
print('Avg accuracy colon cancer orde 4 v1 : ', akurasi_IG_4_v1.mean())
print('Avg F1 score colon cancer orde 4 v1 : ', f1_IG_4_v1.mean())
print('Avg running time colon cancer orde 4 v1 : ', rt_IG_4_v1.mean())
print('===============================================================================================================================================================================================================')
print('Avg accuracy colon cancer orde 2 v2 : ', akurasi_IG_2_v2.mean())
print('Avg F1 score colon cancer orde 2 v2 : ', f1_IG_2_v2.mean())
print('Avg running time colon cancer orde 2 v2 : ', rt_IG_2_v2.mean())
print('Avg accuracy colon cancer orde 3 v2 : ', akurasi_IG_3_v2.mean())
print('Avg F1 score colon cancer orde 3 v2 : ', f1_IG_3_v2.mean())
print('Avg running time colon cancer orde 3 v2 : ', rt_IG_3_v2.mean())
print('Avg accuracy colon cancer orde 4 v2 : ', akurasi_IG_4_v2.mean())
print('Avg F1 score colon cancer orde 4 v2 : ', f1_IG_4_v2.mean())
print('Avg running time colon cancer orde 4 v2 : ', rt_IG_4_v2.mean())
print()
stop = timeit.default_timer()
print("Overall Running Time : ", stop-start)
```
|
github_jupyter
|
import pandas as pd
from sklearn.preprocessing import MinMaxScaler
from pandas import DataFrame
from scipy.special import legendre
import numpy as np
from sklearn.model_selection import StratifiedKFold
import matplotlib.pyplot as plt
import keras
from tensorflow.python.keras.layers import Dense
from keras.optimizers import Adam
from tensorflow.python.keras import Sequential
from sklearn.metrics import accuracy_score, f1_score
from sklearn.metrics import classification_report
import random
import timeit
from sklearn.feature_selection import mutual_info_classif
from sklearn.feature_selection import SelectKBest
# data colon
url = "https://raw.githubusercontent.com/jamessaldo/final-task/master/colonTumor.data"
data_colon = pd.read_csv(url, header=None)
print('Total Missing Value pada Data colon Tumor:',data_colon.isnull().sum().sum())
# Melakukan normalisasi
# data colon
data_new_colon = data_colon.drop([2000],axis=1)
scaler = MinMaxScaler()
data_new_colon = scaler.fit_transform(data_new_colon)
data_new_colon = DataFrame(data_new_colon)
data_new_colon['label'] = list(data_colon[2000])
dic = {'negative':1,'positive':0}
data_new_colon.replace(dic,inplace=True)
def FLNN_Classifier(data_train, data_test, orde, lr):
start = timeit.default_timer()
x_data_train = data_train.drop(['label'],axis=1)
y_data_train = data_train['label']
x_data_test = data_test.drop(['label'],axis=1)
y_data_test = data_test['label']
df_train = pd.DataFrame()
df_test = pd.DataFrame()
for x in range(1, orde+1):
pn = legendre(x)
y_orde = pn(x_data_train)
df_train = pd.concat([df_train, y_orde], axis=1)
pn = legendre(x)
y_orde = pn(x_data_test)
df_test = pd.concat([df_test, y_orde], axis=1)
df_train.columns = ["Attribut"+str(i) for i in range(len(df_train.columns))]
df_train['label'] = y_data_train.reset_index().label
X_train = df_train.iloc[:, 0:len(df_train.columns)-1].values
y_train = df_train.iloc[:, len(df_train.columns)-1].values
df_test.columns = ["Attribut"+str(i) for i in range(len(df_test.columns))]
df_test['label'] = y_data_test.reset_index().label
X_test = df_test.iloc[:, 0:len(df_test.columns)-1].values
y_test = df_test.iloc[:, len(df_test.columns)-1].values
# Melakukan proses klasifikasi FLNN
# Inisialisasi FLNN
Model = Sequential()
# Menambah input layer dan hidden layer pertama
Model.add(Dense(units = len(df_train.columns)-1, kernel_initializer = 'uniform', input_dim = len(df_train.columns)-1))
# Menambah output layer
Model.add(Dense(units = 1, kernel_initializer = 'uniform', activation = 'sigmoid'))
# Menjalankan ANN
Model.compile(optimizer = Adam(learning_rate=lr), loss = 'mean_squared_error', metrics = ['accuracy'])
# Fitting ANN ke training set
history = Model.fit(X_train, y_train, batch_size = 50, epochs = 100, validation_split = 0.2, verbose=False)
#Memprediksi hasil test set
y_pred = Model(X_test)
y_pred =(y_pred >= 0.5)
#print("X_Train :", X_train)
print("Y_Train :", y_train)
#print("X_Test :", X_test)
print("Y_Test :", y_test)
akurasi = accuracy_score(y_test,y_pred)
F1 = f1_score(y_test, y_pred, average='macro')
print("Akurasi : ", akurasi)
print("F1_Score : ", F1)
print(classification_report(y_test,y_pred))
# Membuat confusion matrix
from sklearn.metrics import confusion_matrix
from mlxtend.plotting import plot_confusion_matrix
cm = confusion_matrix(y_test, y_pred)
fig, ax = plot_confusion_matrix(conf_mat = cm, figsize=(5,5))
plt.show()
stop = timeit.default_timer()
print('Running Time: ', stop - start)
return akurasi, F1, stop-start
start = timeit.default_timer()
akurasi_IG_2_v1,f1_IG_2_v1,rt_IG_2_v1 = [],[],[]
akurasi_IG_3_v1,f1_IG_3_v1,rt_IG_3_v1 = [],[],[]
akurasi_IG_4_v1,f1_IG_4_v1,rt_IG_4_v1 = [],[],[]
akurasi_IG_2_v2,f1_IG_2_v2,rt_IG_2_v2 = [],[],[]
akurasi_IG_3_v2,f1_IG_3_v2,rt_IG_3_v2 = [],[],[]
akurasi_IG_4_v2,f1_IG_4_v2,rt_IG_4_v2 = [],[],[]
#Melakukan proses K-Fold
kf = StratifiedKFold(n_splits=5, shuffle=True, random_state=10)
kf.get_n_splits(data_new_colon)
X = data_new_colon.copy().iloc[:, 0:len(data_new_colon.columns)-1].values
Y = data_new_colon.copy().iloc[:, len(data_new_colon.columns)-1].values
for train_index, test_index in kf.split(X,Y):
print("Train : ", train_index, "Test : ", test_index)
data_train, data_test, y_train, y_test = pd.DataFrame(X[train_index]), pd.DataFrame(X[test_index]), Y[train_index], Y[test_index]
data_train['label'] = y_train
data_test['label'] = y_test
print("colon Orde 2 v1")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 2, 0.6)
akurasi_IG_2_v1.append(acc)
f1_IG_2_v1.append(f1)
rt_IG_2_v1.append(rt)
print("colon Orde 3 v1")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 3, 0.6)
akurasi_IG_3_v1.append(acc)
f1_IG_3_v1.append(f1)
rt_IG_3_v1.append(rt)
print("colon Orde 4 v1")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 4, 0.6)
akurasi_IG_4_v1.append(acc)
f1_IG_4_v1.append(f1)
rt_IG_4_v1.append(rt)
print("colon Orde 2 v2")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 2, 0.001)
akurasi_IG_2_v2.append(acc)
f1_IG_2_v2.append(f1)
rt_IG_2_v2.append(rt)
print("colon Orde 3 v2")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 3, 0.001)
akurasi_IG_3_v2.append(acc)
f1_IG_3_v2.append(f1)
rt_IG_3_v2.append(rt)
print("colon Orde 4 v1")
acc,f1,rt = FLNN_Classifier(data_train.copy(), data_test.copy(), 4, 0.001)
akurasi_IG_4_v2.append(acc)
f1_IG_4_v2.append(f1)
rt_IG_4_v2.append(rt)
akurasi_IG_2_v1,f1_IG_2_v1,rt_IG_2_v1 = np.array(akurasi_IG_2_v1),np.array(f1_IG_2_v1),np.array(rt_IG_2_v1)
akurasi_IG_3_v1,f1_IG_3_v1,rt_IG_3_v1 = np.array(akurasi_IG_3_v1),np.array(f1_IG_3_v1),np.array(rt_IG_3_v1)
akurasi_IG_4_v1,f1_IG_4_v1,rt_IG_4_v1 = np.array(akurasi_IG_4_v1),np.array(f1_IG_4_v1),np.array(rt_IG_4_v1)
akurasi_IG_2_v2,f1_IG_2_v2,rt_IG_2_v2 = np.array(akurasi_IG_2_v2),np.array(f1_IG_2_v2),np.array(rt_IG_2_v2)
akurasi_IG_3_v2,f1_IG_3_v2,rt_IG_3_v2 = np.array(akurasi_IG_3_v2),np.array(f1_IG_3_v2),np.array(rt_IG_3_v2)
akurasi_IG_4_v2,f1_IG_4_v2,rt_IG_4_v2 = np.array(akurasi_IG_4_v2),np.array(f1_IG_4_v2),np.array(rt_IG_4_v2)
#Print Result
print('===============================================================================================================================================================================================================')
print('Avg accuracy colon cancer orde 2 v1 : ', akurasi_IG_2_v1.mean())
print('Avg F1 score colon cancer orde 2 v1 : ', f1_IG_2_v1.mean())
print('Avg running time colon cancer orde 2 v1 : ', rt_IG_2_v1.mean())
print('Avg accuracy colon cancer orde 3 v1 : ', akurasi_IG_3_v1.mean())
print('Avg F1 score colon cancer orde 3 v1 : ', f1_IG_3_v1.mean())
print('Avg running time colon cancer orde 3 v1 : ', rt_IG_3_v1.mean())
print('Avg accuracy colon cancer orde 4 v1 : ', akurasi_IG_4_v1.mean())
print('Avg F1 score colon cancer orde 4 v1 : ', f1_IG_4_v1.mean())
print('Avg running time colon cancer orde 4 v1 : ', rt_IG_4_v1.mean())
print('===============================================================================================================================================================================================================')
print('Avg accuracy colon cancer orde 2 v2 : ', akurasi_IG_2_v2.mean())
print('Avg F1 score colon cancer orde 2 v2 : ', f1_IG_2_v2.mean())
print('Avg running time colon cancer orde 2 v2 : ', rt_IG_2_v2.mean())
print('Avg accuracy colon cancer orde 3 v2 : ', akurasi_IG_3_v2.mean())
print('Avg F1 score colon cancer orde 3 v2 : ', f1_IG_3_v2.mean())
print('Avg running time colon cancer orde 3 v2 : ', rt_IG_3_v2.mean())
print('Avg accuracy colon cancer orde 4 v2 : ', akurasi_IG_4_v2.mean())
print('Avg F1 score colon cancer orde 4 v2 : ', f1_IG_4_v2.mean())
print('Avg running time colon cancer orde 4 v2 : ', rt_IG_4_v2.mean())
print()
stop = timeit.default_timer()
print("Overall Running Time : ", stop-start)
| 0.388038 | 0.873701 |
```
import numpy as np
import pandas as pd
import scipy.stats as stats
import matplotlib.pyplot as plt
import seaborn as sms
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error, mean_squared_error, r2_score
from IPython.display import display
import math
%matplotlib inline
datafMultivariable = pd.read_csv('weatherHistory.csv')
datafMultivariable.head()
datafMultivariable.columns
datafMultivariable.info()
nan_df = datafMultivariable[datafMultivariable.isna().any(axis=1)]
nan_df.count()
datafMultivariable.shape
proporcionNaN = 517/datafMultivariable.shape[0]*100
print('La proporción de datos NaN o perdidos en el dataframe es:', proporcionNaN, '%')
datafMultivariable.dropna(inplace=True)
datafMultivariable.shape
dfAUX = datafMultivariable[['Temperature (C)',
'Apparent Temperature (C)',
'Humidity',
'Wind Speed (km/h)',
'Wind Bearing (degrees)',
'Visibility (km)',
'Loud Cover',
'Pressure (millibars)']]
dfAUX.head()
dfAUX.info()
# Parece que no se debería dejar por fuera de nuestro modelo
datafMultivariable['Precip Type'].unique()
# Para variables tipo objeto que representan categorías, existe una función "dummy"
# la cual permite transformar a variables "categoricas numéricas"
dfDummy = pd.get_dummies(datafMultivariable['Precip Type'])
dfDummy.head()
dfAUX = dfAUX.merge(dfDummy, left_index= True, right_index=True)
dfAUX.head()
dfAUX.describe().T
sms.heatmap(dfAUX.corr())
# Matriz de correlación
matrixCorrelacion = dfAUX.corr()
matrixCorrelacion
def relacionFeatures(corrMatrix, umbral):
feature = []
valores = []
for i, index in enumerate(corrMatrix.index):
if abs(corrMatrix[index]) > umbral:
feature.append(index)
valores.append(corrMatrix[index])
df = pd.DataFrame(data = valores, index = feature, columns=['Valor de Correlación'])
return df
```
**Funcion de entrenamiento y de Obtencion de metricas**
Paso 1: Identificar las variables del umbral de correlacion <br>
Dividir en los conjuntos entrenamiento y prueba<br>
Seleccionar el modelo<br>
Entrenar<br>
Obtener las metricas<br>
Paso 2: Identificar la MULTICOLINEALIDAD
```
def training(X, y):
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=0)
modelo = LinearRegression()
modelo.fit(X_train, y_train)
y_predict_train = modelo.predict(X_train)
y_predict = modelo.predict(X_test)
return y_predict, y_test
def metricas(umbral):
valorCorrelacion = relacionFeatures(matrixCorrelacion['Apparent Temperature (C)'], umbral)
dataCorrelacion = dfAUX[valorCorrelacion.index]
X = dataCorrelacion.drop('Apparent Temperature (C)', axis=1)
y = dataCorrelacion['Apparent Temperature (C)']
y_predict, y_test = training(X, y)
puntuacion = r2_score(y_test, y_predict)
meanabsoluteerror = mean_absolute_error(y_test, y_predict)
mse = mean_squared_error(y_test, y_predict)
valorCorrelacion = valorCorrelacion.T
valorCorrelacion['r2_score'] = puntuacion
valorCorrelacion['MAE'] = meanabsoluteerror
valorCorrelacion['MSE'] = mse
valorCorrelacion.reset_index(inplace=True, drop=True)
print('Metricas\n')
return valorCorrelacion
umbral=0.2
print('Ejemplo Umbral de 0.2')
metricas(umbral)
```
**EJEMPLOS**
```
# ejemplo -> umbral = 0.4
print('\033[1mCon un umbral de 0.4 \033[0m \n')
metricas(umbral=0.4)
# ejemplo -> umbral = 0.6
print('\033[1mCon un umbral de 0.6 \033[0m \n')
metricas(umbral=0.6)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import scipy.stats as stats
import matplotlib.pyplot as plt
import seaborn as sms
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error, mean_squared_error, r2_score
from IPython.display import display
import math
%matplotlib inline
datafMultivariable = pd.read_csv('weatherHistory.csv')
datafMultivariable.head()
datafMultivariable.columns
datafMultivariable.info()
nan_df = datafMultivariable[datafMultivariable.isna().any(axis=1)]
nan_df.count()
datafMultivariable.shape
proporcionNaN = 517/datafMultivariable.shape[0]*100
print('La proporción de datos NaN o perdidos en el dataframe es:', proporcionNaN, '%')
datafMultivariable.dropna(inplace=True)
datafMultivariable.shape
dfAUX = datafMultivariable[['Temperature (C)',
'Apparent Temperature (C)',
'Humidity',
'Wind Speed (km/h)',
'Wind Bearing (degrees)',
'Visibility (km)',
'Loud Cover',
'Pressure (millibars)']]
dfAUX.head()
dfAUX.info()
# Parece que no se debería dejar por fuera de nuestro modelo
datafMultivariable['Precip Type'].unique()
# Para variables tipo objeto que representan categorías, existe una función "dummy"
# la cual permite transformar a variables "categoricas numéricas"
dfDummy = pd.get_dummies(datafMultivariable['Precip Type'])
dfDummy.head()
dfAUX = dfAUX.merge(dfDummy, left_index= True, right_index=True)
dfAUX.head()
dfAUX.describe().T
sms.heatmap(dfAUX.corr())
# Matriz de correlación
matrixCorrelacion = dfAUX.corr()
matrixCorrelacion
def relacionFeatures(corrMatrix, umbral):
feature = []
valores = []
for i, index in enumerate(corrMatrix.index):
if abs(corrMatrix[index]) > umbral:
feature.append(index)
valores.append(corrMatrix[index])
df = pd.DataFrame(data = valores, index = feature, columns=['Valor de Correlación'])
return df
def training(X, y):
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=0)
modelo = LinearRegression()
modelo.fit(X_train, y_train)
y_predict_train = modelo.predict(X_train)
y_predict = modelo.predict(X_test)
return y_predict, y_test
def metricas(umbral):
valorCorrelacion = relacionFeatures(matrixCorrelacion['Apparent Temperature (C)'], umbral)
dataCorrelacion = dfAUX[valorCorrelacion.index]
X = dataCorrelacion.drop('Apparent Temperature (C)', axis=1)
y = dataCorrelacion['Apparent Temperature (C)']
y_predict, y_test = training(X, y)
puntuacion = r2_score(y_test, y_predict)
meanabsoluteerror = mean_absolute_error(y_test, y_predict)
mse = mean_squared_error(y_test, y_predict)
valorCorrelacion = valorCorrelacion.T
valorCorrelacion['r2_score'] = puntuacion
valorCorrelacion['MAE'] = meanabsoluteerror
valorCorrelacion['MSE'] = mse
valorCorrelacion.reset_index(inplace=True, drop=True)
print('Metricas\n')
return valorCorrelacion
umbral=0.2
print('Ejemplo Umbral de 0.2')
metricas(umbral)
# ejemplo -> umbral = 0.4
print('\033[1mCon un umbral de 0.4 \033[0m \n')
metricas(umbral=0.4)
# ejemplo -> umbral = 0.6
print('\033[1mCon un umbral de 0.6 \033[0m \n')
metricas(umbral=0.6)
| 0.552298 | 0.741627 |
## 9.4 设置主题样式
使用Beamer制作幻灯片的一道特色就是有现成的主题样式可供选择和直接使用,其中,主题样式对于幻灯片的演示效果而言十分重要,简言之,主题样式就是幻灯片的“外观”,改变幻灯片最简单的方式就是变换不同的主题样式。Beamer中提供的每种主题样式都具有良好的可用性和可读性,这也使得Beamer制作出来的幻灯片看起来十分专业,同时,反复使用的难度也不大。
在英文中,主题对应的英文单词为theme。狭义来看,幻灯片主题是指幻灯片的主题样式;但从广义来看,其实幻灯片主题包括了包括主题样式、颜色主题、字体主题、内部主题、外部主题。
### 9.4.1 基本介绍
使用Beamer制作幻灯片时,我们可以选择很多已经封装好的幻灯片主题样式,不同样式可以达到不同的视觉效果。其实,使用这些主题样式的方法非常简单。通常来说,在前导代码中插入`\usetheme{}`命令即可,例如使用`Copenhagen`(哥本哈根主题样式)只需要在前导代码中申明`\usetheme{Copenhagen}`,这种方式调用主题样式是非常省事。
<p align="center">
<img align="middle" src="tikz_graphics/beamer_theme.png" width="800" />
</p>
<center><b>图9.4.1</b> Beamer文档类型中的主题样式</center>
在Beamer文档类型中,有几十种主题样式可供选择和使用,比较常用的主题样式包括以下这些:
- `Berlin`:柏林主题样式,默认样式为蓝色调。
- `Copenhagen`: 哥本哈根主题样式,默认样式为蓝色调。
- `CambridgeUS`:美国剑桥主题样式,默认样式为红色调。
- `Berkeley`:伯克利主题样式,默认样式为蓝色调。
- `Singapore`:新加坡主题样式。
- `Warsaw`:默认样式为蓝色调。
【**例37**】在`beamer`文档类型中使用`CambridgeUS`主题样式制作一个简单的幻灯片。
```tex
\documentclass{beamer}
\usetheme{CambridgeUS}
\begin{document}
\begin{frame}{Example}
This is a simple example for the CambridgeUS theme.
\end{frame}
\end{document}
```
编译上述代码,得到幻灯片如图9.4.1所示。
<p align="center">
<img align="middle" src="graphics/example_sec2_1.png" width="450" />
</p>
<center><b>图9.4.1</b> 编译后的幻灯片效果</center>
当然,在这些主题样式基础上,我们也能够使用一些特定的主题样式如颜色主题、字体主题、内部主题、外部主题对幻灯片的显示效果进行调整。
<p align="center">
<img align="middle" src="tikz_graphics/other_themes.png" width="600" />
</p>
<center><b>图9.4.2</b> Beamer文档类型中的其他几种主题设置</center>
### 9.4.2 颜色主题
使用Beamer制作幻灯片时,我们能够自行设置幻灯片主题样式的色调,使用`\usecolortheme{}`命令即可,这些色调包括`beetle`、`beaver`、`orchid`、`whale`、`dolphin`等。这里的色调又被称为颜色主题,它定义了幻灯片各部分的颜色搭配,设置特定的颜色主题后,我们能够得到不同的组合样式,具体可参考[https://hartwork.org/beamer-theme-matrix/](https://hartwork.org/beamer-theme-matrix/)网站提供的组合样式矩阵。
【**例38**】在`beamer`文档类型中使用`CambridgeUS`主题样式和`dolphin`色调制作一个简单的幻灯片。
```tex
\documentclass{beamer}
\usetheme{CambridgeUS}
\usecolortheme{dolphin}
\begin{document}
\begin{frame}{Example}
This is a simple example for the CambridgeUS theme with dolphin (color theme).
\end{frame}
\end{document}
```
编译上述代码,得到幻灯片如图9.4.3所示。
<p align="center">
<img align="middle" src="graphics/example_sec2_2.png" width="450" />
</p>
<center><b>图9.4.3</b> 编译后的幻灯片效果</center>
### 9.4.3 字体主题
实际上,对于幻灯片的文本字体,我们可以调用字体样式对其进行调整。在Beamer中,字体样式被称为字体主题,它定义了幻灯片的字体搭配。具体使用方法是:在前导代码中要用到的命令为`\usefonttheme{A}`,位置A填写的一般是字体类型,例如`serif`。
【**例39**】使用`beamer`文档类型创建一个简单的幻灯片,并在前导代码中申明使用`serif`对应的字体样式。
```tex
\documentclass{beamer}
\usefonttheme{serif}
\begin{document}
\begin{frame}
This is a simple example for using \alert{serif} font theme.
\end{frame}
\end{document}
```
编译上述代码,得到幻灯片如图9.4.4所示。
<p align="center">
<img align="middle" src="graphics/example_sec2_3.png" width="450" />
</p>
<center><b>图9.4.4</b> 编译后的幻灯片效果</center>
我们知道:在常规文档中,可以使用各种字体对应的宏包达到调用字体的作用,使用规则为`\usepackage{A}`,位置A填写的一般是字体类型,包括serif、avant、bookman、chancery、charter、euler、helvet、mathtime、mathptm、mathptmx、newcent、palatino、pifont、utopia等。
【**例40**】使用`beamer`文档类型创建一个简单的幻灯片,并在前导代码中申明使用字体`palatino`对应的宏包。
```tex
\documentclass{beamer}
\usepackage{palatino}
\begin{document}
\begin{frame}
This is a simple example for using \alert{palatino} font.
\end{frame}
\end{document}
```
编译上述代码,得到幻灯片如图9.4.5所示。
<p align="center">
<img align="middle" src="graphics/example_sec2_4.png" width="450" />
</p>
<center><b>图9.4.5</b> 编译后的幻灯片效果</center>
### 9.4.4 内部主题
内部主题定义了幻灯片展示区域的样式,如列表、定理等,内部主题不包括页眉、页脚、导航栏等部分。每一种主题样式都有默认的内部主题,更换内部主题需使用`\useinnertheme{A}`命令,位置A可供选择的内部主题包括`circles`、`rectangles`、`rounded`和`inmargin`。
【**例41**】在`beamer`文档类型中分别使用`circles`和`inmargin`两种内部主题制作幻灯片。
- 使用`circles`内部主题:
```tex
\documentclass{beamer}
\usetheme{CambridgeUS}
\usefonttheme{professionalfonts}
\useinnertheme{circles}
\begin{document}
\begin{frame}
\frametitle{Parent function}
\framesubtitle{A short list}
Please check out the following parent function list.
\begin{enumerate}
\item $y=x$
\item $y=|x|$
\item $y=x^{2}$
\item $y=x^{3}$
\item $y=x^{b}$
\end{enumerate}
\end{frame}
\end{document}
```
编译上述代码,得到幻灯片如图9.4.6所示。
<p align="center">
<img align="middle" src="graphics/example_innertheme_circles.png" width="450" />
</p>
<center><b>图9.4.6</b> 编译后的幻灯片效果</center>
- 使用`inmargin`内部主题:
```tex
\documentclass{beamer}
\usetheme{CambridgeUS}
\usefonttheme{professionalfonts}
\useinnertheme{inmargin}
\begin{document}
\begin{frame}
\frametitle{Parent function}
\framesubtitle{A short list}
Please check out the following parent function list.
\begin{enumerate}
\item $y=x$
\item $y=|x|$
\item $y=x^{2}$
\item $y=x^{3}$
\item $y=x^{b}$
\end{enumerate}
\end{frame}
\end{document}
```
编译上述代码,得到幻灯片如图9.4.7所示。
<p align="center">
<img align="middle" src="graphics/example_innertheme_inmargin.png" width="450" />
</p>
<center><b>图9.4.7</b> 编译后的幻灯片效果</center>
### 9.4.5 外部主题
外部主题定义了幻灯片的边框、页眉、页脚、导航栏等部分的样式。更换外部主题需使用`\useoutertheme{A}`,位置A可供选择的外部主题包括`infolines`、`smoothbars`、`sidebar`、`split`和`tree`。
### 9.4.6 表格字体大小
在Beamer中制作表格,当我们想对表头或者表格内容文字大小进行调整时,可以使用在前导代码中申明使用`caption`宏包,即`\usepackage{caption}`,然后设置具体的字体大小即可,如`\captionsetup{font = scriptsize, labelfont = scriptsize}`可以将表头和表格内容字体大小调整为`scriptsize`。
【**例9-41**】使用`\begin{table} \end{table}`环境创建一个简单表格,并使用`caption`宏包将表头字体大小设置为`Large`、将表格内容字体大小设置为`large`。
```tex
\documentclass{beamer}
\usepackage{booktabs}
\usepackage{caption}
\captionsetup{font = large, labelfont = Large}
\begin{document}
\begin{frame}
\begin{table}
\caption{A simple table.}
\begin{tabular}{l|ccc}
\toprule
& \textbf{header3} & \textbf{header4} & \textbf{header5} \\
\midrule
\textbf{header1} & cell1 & cell2 & cell3 \\
\midrule
\textbf{header2} & cell4 & cell5 & cell6 \\
\bottomrule
\end{tabular}
\end{table}
\end{frame}
\end{document}
```
编译上述代码,得到幻灯片如图9.4.6所示。
<p align="center">
<img align="middle" src="graphics/example_sec2_4_0.png" width="450" />
</p>
<center><b>图9.4.6</b> 编译后的幻灯片效果</center>
其中,单就设置表头字体大小而言,除了使用`caption`宏包之外,还可以通过对幻灯片设置全局参数达到调整字体大小的效果,例如`\setbeamerfont{caption}{size = \Large}`。
### 9.5.4 样式调整
在Beamer文档类型中,除了可以使用各种主题样式,另外也可以根据幻灯片组成部分,分别对侧边栏、导航栏以及Logo等进行调整。其中,侧边栏是由所选幻灯片主题样式自动生成的,主要用于显示幻灯片目录。有时为了显示幻灯片的层次,使用侧边栏进行目录索引。
【**例9-42**】使用`Berkeley`主题样式,并将侧边栏显示在右侧。
```tex
\documentclass{beamer}
\PassOptionsToPackage{right}{beamerouterthemesidebar}
\usetheme{Berkeley}
\usefonttheme{professionalfonts}
\begin{document}
\begin{frame}
\frametitle{Parent function}
\framesubtitle{A short list}
Please check out the following parent function list.
\begin{enumerate}
\item $y=x$
\item $y=|x|$
\item $y=x^{2}$
\item $y=x^{3}$
\item $y=x^{b}$
\end{enumerate}
\end{frame}
\end{document}
```
编译上述代码,得到幻灯片如图9.4.7所示。
<p align="center">
<img align="middle" src="graphics/example_sec2_6.png" width="450" />
</p>
<center><b>图9.4.7</b> 编译后的幻灯片效果</center>
很多时候我们会发现,在各类学术汇报中,幻灯片的首页通常会有主讲人所在的研究机构Logo。在Beamer文档类型中,有`\logo`和`\titlegraphic`两个命令可供使用,使用`\logo`命令添加的Logo会在每一页幻灯片中都显示,而使用`\titlegraphic`命令添加的Logo只出现在标题页。
【**例9-43**】使用`\logo`命令在幻灯片中添加Logo。
```tex
\documentclass{beamer}
\usefonttheme{professionalfonts}
\title{A Simple Beamer Example}
\author{Author's Name}
\institute{Author's Institute}
\logo{\includegraphics[width=2cm]{logopolito}}
\begin{document}
\begin{frame}
\titlepage
\end{frame}
\begin{frame}{Parent function}{A short list}
Please check out the following parent function list.
\begin{enumerate}
\item $y=x$
\item $y=|x|$
\item $y=x^{2}$
\item $y=x^{3}$
\item $y=x^{b}$
\end{enumerate}
\end{frame}
\end{document}
```
编译上述代码,得到幻灯片如图9.4.8所示。
<p align="center">
<table>
<tr>
<td><img align="middle" src="graphics/example_sec2_7_0.png" width="450"></td>
<td><img align="middle" src="graphics/example_sec2_7_1.png" width="450"></td>
</tr>
</table>
</p>
<center><b>图9.4.8</b> 编译后的幻灯片效果</center>
【**例9-44**】使用`\titlegraphic`命令在幻灯片的标题页添加Logo。
```tex
\documentclass{beamer}
\usefonttheme{professionalfonts}
\title{A Simple Beamer Example}
\author{Author's Name}
\institute{Author's Institute}
\titlegraphic{\includegraphics[width=2cm]{logopolito}\hspace*{4.75cm}~
\includegraphics[width=2cm]{logopolito}
}
\begin{document}
\begin{frame}
\titlepage
\end{frame}
\begin{frame}{Parent function}{A short list}
Please check out the following parent function list.
\begin{enumerate}
\item $y=x$
\item $y=|x|$
\item $y=x^{2}$
\item $y=x^{3}$
\item $y=x^{b}$
\end{enumerate}
\end{frame}
\end{document}
```
编译上述代码,得到幻灯片如图9.4.9所示。
<p align="center">
<table>
<tr>
<td><img align="middle" src="graphics/example_sec2_8_0.png" width="450"></td>
<td><img align="middle" src="graphics/example_sec2_8_1.png" width="450"></td>
</tr>
</table>
</p>
<center><b>图9.4.9</b> 编译后的幻灯片效果</center>
### 参考资料
- Prathik Naidu, Adam Pahlavan. [Fun with Beamer: An Epic Quest To Create the Perfect Presentation](http://web.mit.edu/rsi/www/pdfs/beamer-tutorial.pdf), June 28, 2017.
- [Beamer: change size of figure caption](https://tex.stackexchange.com/questions/52132).
- [logo in the first page only](https://tex.stackexchange.com/questions/61051).
【回放】[**9.3 块与盒子——添加框元素**](https://nbviewer.jupyter.org/github/xinychen/latex-cookbook/blob/main/chapter-9/section3.ipynb)
【继续】[**9.5 插入程序源代码**](https://nbviewer.jupyter.org/github/xinychen/latex-cookbook/blob/main/chapter-9/section5.ipynb)
### License
<div class="alert alert-block alert-danger">
<b>This work is released under the MIT license.</b>
</div>
|
github_jupyter
|
\documentclass{beamer}
\usetheme{CambridgeUS}
\begin{document}
\begin{frame}{Example}
This is a simple example for the CambridgeUS theme.
\end{frame}
\end{document}
\documentclass{beamer}
\usetheme{CambridgeUS}
\usecolortheme{dolphin}
\begin{document}
\begin{frame}{Example}
This is a simple example for the CambridgeUS theme with dolphin (color theme).
\end{frame}
\end{document}
\documentclass{beamer}
\usefonttheme{serif}
\begin{document}
\begin{frame}
This is a simple example for using \alert{serif} font theme.
\end{frame}
\end{document}
\documentclass{beamer}
\usepackage{palatino}
\begin{document}
\begin{frame}
This is a simple example for using \alert{palatino} font.
\end{frame}
\end{document}
\documentclass{beamer}
\usetheme{CambridgeUS}
\usefonttheme{professionalfonts}
\useinnertheme{circles}
\begin{document}
\begin{frame}
\frametitle{Parent function}
\framesubtitle{A short list}
Please check out the following parent function list.
\begin{enumerate}
\item $y=x$
\item $y=|x|$
\item $y=x^{2}$
\item $y=x^{3}$
\item $y=x^{b}$
\end{enumerate}
\end{frame}
\end{document}
\documentclass{beamer}
\usetheme{CambridgeUS}
\usefonttheme{professionalfonts}
\useinnertheme{inmargin}
\begin{document}
\begin{frame}
\frametitle{Parent function}
\framesubtitle{A short list}
Please check out the following parent function list.
\begin{enumerate}
\item $y=x$
\item $y=|x|$
\item $y=x^{2}$
\item $y=x^{3}$
\item $y=x^{b}$
\end{enumerate}
\end{frame}
\end{document}
\documentclass{beamer}
\usepackage{booktabs}
\usepackage{caption}
\captionsetup{font = large, labelfont = Large}
\begin{document}
\begin{frame}
\begin{table}
\caption{A simple table.}
\begin{tabular}{l|ccc}
\toprule
& \textbf{header3} & \textbf{header4} & \textbf{header5} \\
\midrule
\textbf{header1} & cell1 & cell2 & cell3 \\
\midrule
\textbf{header2} & cell4 & cell5 & cell6 \\
\bottomrule
\end{tabular}
\end{table}
\end{frame}
\end{document}
\documentclass{beamer}
\PassOptionsToPackage{right}{beamerouterthemesidebar}
\usetheme{Berkeley}
\usefonttheme{professionalfonts}
\begin{document}
\begin{frame}
\frametitle{Parent function}
\framesubtitle{A short list}
Please check out the following parent function list.
\begin{enumerate}
\item $y=x$
\item $y=|x|$
\item $y=x^{2}$
\item $y=x^{3}$
\item $y=x^{b}$
\end{enumerate}
\end{frame}
\end{document}
\documentclass{beamer}
\usefonttheme{professionalfonts}
\title{A Simple Beamer Example}
\author{Author's Name}
\institute{Author's Institute}
\logo{\includegraphics[width=2cm]{logopolito}}
\begin{document}
\begin{frame}
\titlepage
\end{frame}
\begin{frame}{Parent function}{A short list}
Please check out the following parent function list.
\begin{enumerate}
\item $y=x$
\item $y=|x|$
\item $y=x^{2}$
\item $y=x^{3}$
\item $y=x^{b}$
\end{enumerate}
\end{frame}
\end{document}
\documentclass{beamer}
\usefonttheme{professionalfonts}
\title{A Simple Beamer Example}
\author{Author's Name}
\institute{Author's Institute}
\titlegraphic{\includegraphics[width=2cm]{logopolito}\hspace*{4.75cm}~
\includegraphics[width=2cm]{logopolito}
}
\begin{document}
\begin{frame}
\titlepage
\end{frame}
\begin{frame}{Parent function}{A short list}
Please check out the following parent function list.
\begin{enumerate}
\item $y=x$
\item $y=|x|$
\item $y=x^{2}$
\item $y=x^{3}$
\item $y=x^{b}$
\end{enumerate}
\end{frame}
\end{document}
| 0.494873 | 0.934515 |
```
import os
import matplotlib.pyplot as plt
import csv
import pickle
import math
# Don't edit
done_load=0
load_dest=""
import time
def deleteDB(db='ycsb', host='vmtest3.westus.cloudapp.azure.com:27017', mongo_dir=r"C:\Program Files\MongoDB\Server\3.6\bin"):
curr_dir=os.getcwd()
os.chdir(mongo_dir)
delete_string=r'mongo ycsb --host "' + host + '" --eval "db.usertable.drop()"'
print(delete_string)
status = os.system(delete_string)
os.chdir(curr_dir)
return status
def deleteDBMongo():
deleteDB(host='mongotcoa.westus.cloudapp.azure.com:27017')
def deleteDBAtlas(mongo_dir=r"C:\Program Files\MongoDB\Server\3.6\bin"):
curr_dir=os.getcwd()
os.chdir(mongo_dir)
u=r"anfeldma"
p=r"O!curmt0"
host=r"mongodb+srv://atlas36shard1ncaluswest.fr0to.mongodb.net/ycsb"
run_str=r'mongo "' + host + r'" --username anfeldma --password O!curmt0' + r' --eval "db.usertable.drop()"'
print(run_str)
status = os.system(run_str)
# create_cmd=r'mongo ycsb --host ' + host + r' -u ' + u + r' -p ' + p + r' --ssl < inp.txt'
# status = os.system(create_cmd)
os.chdir(curr_dir)
time.sleep(2)
def deleteDBCosmos(mongo_dir=r"C:\Program Files\MongoDB\Server\3.6\bin"):
curr_dir=os.getcwd()
os.chdir(mongo_dir)
u=r"mongo-api-benchmark"
p=r"KiYRdcJp41NN268oTcyeM2ilpLwYUAo8tsX9sYoBNTd6DzjXuJHtcaSylh5VJNGs2wg1FVGExRC0m5Z6pEk7ow=="
host=r"mongo-api-benchmark.mongo.cosmos.azure.com:10255"
run_str=r'mongo ycsb --host ' + host + r' -u ' + u + r' -p ' + p + r' --ssl --eval "db.usertable.drop()"'
print(run_str)
status = os.system(run_str)
os.chdir(curr_dir)
time.sleep(2)
return status
# deleteDB(host=r'mongo-api-benchmark:KiYRdcJp41NN268oTcyeM2ilpLwYUAo8tsX9sYoBNTd6DzjXuJHtcaSylh5VJNGs2wg1FVGExRC0m5Z6pEk7ow^=^=@mongo-api-benchmark.mongo.cosmos.azure.com:10255/?ssl^=true^&replicaSet^=globaldb^&retrywrites^=false^&maxIdleTimeMS^=120000^&appName^=@mongo-api-benchmark@')
# deleteDB(host=r'mongo-api-benchmark:KiYRdcJp41NN268oTcyeM2ilpLwYUAo8tsX9sYoBNTd6DzjXuJHtcaSylh5VJNGs2wg1FVGExRC0m5Z6pEk7ow==@mongo-api-benchmark.mongo.cosmos.azure.com:10255/?ssl=true&replicaSet=globaldb&retrywrites=false&maxIdleTimeMS=120000&appName=@mongo-api-benchmark@')
def runYCSB(cmd="run", ycsb_dir=r'C:\Users\anfeldma\codeHome\YCSB\bin',workload_dir=r'C:\Users\anfeldma\codeHome\YCSB\workloads',workload='workloadw', \
mongo_endpoint=r'mongodb://vmtest3.westus.cloudapp.azure.com:27017/',operation_count=1000,record_count=100, \
nthreads=1,logdir=".\\",logfn="log.csv"):
curr_dir=os.getcwd()
os.chdir(ycsb_dir)
ycsb_str=r'ycsb ' + cmd + ' mongodb -s -P "' + workload_dir + "\\" + workload + r'" -p mongodb.url="' + mongo_endpoint + \
r'" -p operationcount=' + str(operation_count) + r' -p recordcount=' + str(record_count) + r' -threads ' + str(nthreads) + \
r" " + \
' > ' + logdir + logfn
# r"^&maxPoolSize^=" + str(10*nthreads) + \
print(ycsb_str)
#status=0
os.system(ycsb_str)
os.chdir(curr_dir)
return ycsb_str
def runYCSBMongo36(execmd="run", op_count=10000, rec_count=10000, nthr=1, wkld="workloadw"):
return runYCSB(cmd=execmd, operation_count=op_count, record_count=rec_count, nthreads=nthr, workload=wkld, mongo_endpoint=r"mongodb://mongotcoa.westus.cloudapp.azure.com:27017/")
def runYCSBCosmos36(execmd="run", op_count=10000, rec_count=10000, nthr=1, wkld="workloadw"):
return runYCSB(cmd=execmd, mongo_endpoint=r'mongodb://mongo-api-benchmark:KiYRdcJp41NN268oTcyeM2ilpLwYUAo8tsX9sYoBNTd6DzjXuJHtcaSylh5VJNGs2wg1FVGExRC0m5Z6pEk7ow^=^=@mongo-api-benchmark.mongo.cosmos.azure.com:10255/?ssl^=true^&replicaSet^=globaldb^&retrywrites^=false^&maxIdleTimeMS^=120000^&appName^=@mongo-api-benchmark@', \
operation_count=op_count, record_count=rec_count, nthreads=nthr, workload=wkld)
def runYCSBAtlas36(execmd="run", op_count=10000, rec_count=10000, nthr=1, wkld="workloadw"):
return runYCSB(cmd=execmd, mongo_endpoint=r'mongodb+srv://anfeldma:O%21curmt0@atlas36shard1ncaluswest.fr0to.mongodb.net/ycsb?authSource^=admin^&retryWrites^=true^&w^=majority', \
operation_count=op_count, record_count=rec_count, nthreads=nthr, workload=wkld)
def parseLog(logdir=r'C:\Users\anfeldma\codeHome\YCSB\bin', logfn='log.csv'):
metrics_dict={}
with open(logdir + '\\' + logfn, newline='') as csvfile:
csvrdr = csv.reader(csvfile)#csv.reader(csvfile, delimiter='', quotechar='|')
for row in csvrdr:
if len(row) > 0 and row[0][0] == "[":
arg0 = row[0].lstrip().rstrip()
arg1 = row[1].lstrip().rstrip()
met_val = row[2].lstrip().rstrip()
if not(arg0 in metrics_dict):
metrics_dict[arg0] = {}
metrics_dict[arg0][arg1] = float(met_val)
return metrics_dict
def getIndividualMetrics(met_thrpt_dict_array):
# Plot response curve
thrpt_list=[]
metric_list=[]
max_thrpt=0
for idx in range(len(met_thrpt_dict_array)):
thrpt_list.append(met_thrpt_dict_array[idx][rt_thrpt_field][thrpt_field])
metric_list.append(met_thrpt_dict_array[idx][optype_field][metric_field])
return thrpt_list, metric_list, max_thrpt
def plotResponseCurve(thrpt_list, metric_list, max_thrpt, optype_field):
plt.plot(thrpt_list, metric_list, marker="x")
ax = plt.gca()
for idx in range(len(met_thrpt_dict_array)):
ax.annotate(str(thrpt_list[idx]),
xy=(thrpt_list[idx], metric_list[idx]))
plt.grid(True)
plt.title(optype_field)
plt.xlabel(thrpt_field)
plt.ylabel(metric_field)
fig=plt.gcf()
plt.show()
return fig
def saveResult(met_thrpt_dict_array,thrpt_list,metric_list,nthread_list,max_thrpt,optype_field,ycsb_str,fig):
print("Making " + optype_field + " dir.")
os.makedirs(optype_field, exist_ok=True)
print("Saving result data...")
dumpObj={}
with open(optype_field + "\\pickle.obj", "wb") as fileObj:
dumpObj["met_thrpt_dict_array"]=met_thrpt_dict_array
dumpObj["thrpt_list"]=thrpt_list
dumpObj["metric_list"]=metric_list
dumpObj["nthread_list"]=nthread_list
dumpObj["max_thrpt"]=max_thrpt
dumpObj["optype_field"]=optype_field
dumpObj["ycsb_str"]=max_thrpt
pickle.dump(dumpObj,fileObj)
print("Saving plot...")
fig.savefig(optype_field + "\\" + optype_field + ".png")
def saveComparison(op_max_rate):
print("Making " + "ycsb_op_comparison" + " dir.")
os.makedirs("ycsb_op_comparison", exist_ok=True)
print("Saving comparison data...")
dumpObj={}
with open(optype_field + "\\pickle.obj", "wb") as fileObj:
dumpObj["op_max_rate"]=op_max_rate
pickle.dump(dumpObj,fileObj)
op_mapping={"insert":{"optype_field":"[INSERT]","workload_name":"workloadw"}, \
"read":{"optype_field":"[READ]","workload_name":"workloadr"}, \
"update":{"optype_field":"[UPDATE]","workload_name":"workloadu"} \
}
db_type="atlas" #"cosmos", "mongo", "atlas"
rt_thrpt_field="[OVERALL]"
rt_field="RunTime(ms)"
thrpt_field="Throughput(ops/sec)"
ops_list=["read"] #["insert","read","update"]
opname=""
optype_field=""
workload_name=""
metric_field="99thPercentileLatency(us)"
doc_count=10000000#4000000
nthread_list=[100,150,200]#range(65,73,1)#[20,50,64,100] #[10,12,14,16,18,20] # [1,2,5,10,20,50,64,100]
print(str(range(65,129,7)[-1]))
print(str(len(range(100,129,7))))
met_thrpt_dict_array = []
os.chdir(r"C:\Users\anfeldma\codeHome\YCSB")
op_max_rate={}
for jdx in range(len(ops_list)):
opname = ops_list[jdx]
optype_field=op_mapping[opname]["optype_field"]
workload_name=op_mapping[opname]["workload_name"]
if opname != "insert":
if True or (done_load>=doc_count and load_dest==db_type):
print("Already loaded data.")
else:
print("Deleting existing data.")
if db_type=="mongo":
deleteDBMongo()
print("Starting YCSB load using max thread count...")
runYCSBMongo36(execmd="load",op_count=doc_count, rec_count=doc_count, nthr=max(nthread_list), wkld=workload_name)
elif db_type=="atlas":
deleteDBAtlas()
print("Starting YCSB load using max thread count...")
runYCSBAtlas36(execmd="load",op_count=doc_count, rec_count=doc_count, nthr=max(nthread_list), wkld=workload_name)
elif db_type=="cosmos":
deleteDBCosmos()
print("Starting YCSB load using max thread count...")
runYCSBCosmos36(execmd="load",op_count=doc_count, rec_count=doc_count, nthr=max(nthread_list), wkld=workload_name)
done_load=doc_count
load_dest=db_type
print("Finished YCSB load.")
for idx in range(len(nthread_list)):
print("Starting YCSB " + db_type + " run, opname " + opname + ", workload " + workload_name + ", thread count " + str(nthread_list[idx]))
if opname=="insert":
if db_type=="mongo":
deleteDBMongo()
elif db_type=="atlas":
deleteDBAtlas()
elif db_type=="cosmos":
deleteDBCosmos()
print("Done deleting existing YCSB dataset.")
done_load=0
operation_count=doc_count
if opname=="read" or opname=="update":
print(opname)
#operation_count=int(doc_count)
operation_count=int(doc_count/7)
elif opname=="insert":
print(opname)
operation_count=int(doc_count)
operation_count=int(doc_count/3)
if db_type=="mongo":
ycsb_str=runYCSBMongo36(op_count=operation_count, rec_count=doc_count, nthr=nthread_list[idx], wkld=workload_name)
elif db_type=="atlas":
ycsb_str=runYCSBAtlas36(op_count=operation_count, rec_count=doc_count, nthr=nthread_list[idx], wkld=workload_name)
elif db_type=="cosmos":
ycsb_str=runYCSBCosmos36(op_count=operation_count, rec_count=doc_count, nthr=nthread_list[idx], wkld=workload_name)
met_thrpt_dict_array.append(parseLog())
print("Finished YCSB run, thread count " + str(nthread_list[idx]))
thrpt_list, metric_list, max_thrpt = getIndividualMetrics(met_thrpt_dict_array)
max_thrpt=max(thrpt_list)
met_thrpt_dict_array=[]
fig=plotResponseCurve(thrpt_list, metric_list, max_thrpt, opname)
saveResult(met_thrpt_dict_array,thrpt_list,metric_list,nthread_list,max_thrpt,optype_field,ycsb_str,fig)
print("Max throughput: " + str(max_thrpt))
op_max_rate[opname]=max_thrpt
saveComparison(op_max_rate)
print(op_max_rate)
r'mongodb+srv://anfeldma:O%21curmt0@atlas36shard1ncaluswest.fr0to.mongodb.net/ycsb?authSource=admin&retryWrites=true&w=majority'
if opname=="insert":
if db_type=="mongo":
deleteDBMongo()
elif db_type=="atlas":
deleteDBAtlas()
elif db_type=="cosmos":
deleteDBCosmos()
print("Done deleting existing YCSB dataset.")
done_load=0
plt.bar(op_max_rate.keys(),op_max_rate.values())
os.getcwd()
```
|
github_jupyter
|
import os
import matplotlib.pyplot as plt
import csv
import pickle
import math
# Don't edit
done_load=0
load_dest=""
import time
def deleteDB(db='ycsb', host='vmtest3.westus.cloudapp.azure.com:27017', mongo_dir=r"C:\Program Files\MongoDB\Server\3.6\bin"):
curr_dir=os.getcwd()
os.chdir(mongo_dir)
delete_string=r'mongo ycsb --host "' + host + '" --eval "db.usertable.drop()"'
print(delete_string)
status = os.system(delete_string)
os.chdir(curr_dir)
return status
def deleteDBMongo():
deleteDB(host='mongotcoa.westus.cloudapp.azure.com:27017')
def deleteDBAtlas(mongo_dir=r"C:\Program Files\MongoDB\Server\3.6\bin"):
curr_dir=os.getcwd()
os.chdir(mongo_dir)
u=r"anfeldma"
p=r"O!curmt0"
host=r"mongodb+srv://atlas36shard1ncaluswest.fr0to.mongodb.net/ycsb"
run_str=r'mongo "' + host + r'" --username anfeldma --password O!curmt0' + r' --eval "db.usertable.drop()"'
print(run_str)
status = os.system(run_str)
# create_cmd=r'mongo ycsb --host ' + host + r' -u ' + u + r' -p ' + p + r' --ssl < inp.txt'
# status = os.system(create_cmd)
os.chdir(curr_dir)
time.sleep(2)
def deleteDBCosmos(mongo_dir=r"C:\Program Files\MongoDB\Server\3.6\bin"):
curr_dir=os.getcwd()
os.chdir(mongo_dir)
u=r"mongo-api-benchmark"
p=r"KiYRdcJp41NN268oTcyeM2ilpLwYUAo8tsX9sYoBNTd6DzjXuJHtcaSylh5VJNGs2wg1FVGExRC0m5Z6pEk7ow=="
host=r"mongo-api-benchmark.mongo.cosmos.azure.com:10255"
run_str=r'mongo ycsb --host ' + host + r' -u ' + u + r' -p ' + p + r' --ssl --eval "db.usertable.drop()"'
print(run_str)
status = os.system(run_str)
os.chdir(curr_dir)
time.sleep(2)
return status
# deleteDB(host=r'mongo-api-benchmark:KiYRdcJp41NN268oTcyeM2ilpLwYUAo8tsX9sYoBNTd6DzjXuJHtcaSylh5VJNGs2wg1FVGExRC0m5Z6pEk7ow^=^=@mongo-api-benchmark.mongo.cosmos.azure.com:10255/?ssl^=true^&replicaSet^=globaldb^&retrywrites^=false^&maxIdleTimeMS^=120000^&appName^=@mongo-api-benchmark@')
# deleteDB(host=r'mongo-api-benchmark:KiYRdcJp41NN268oTcyeM2ilpLwYUAo8tsX9sYoBNTd6DzjXuJHtcaSylh5VJNGs2wg1FVGExRC0m5Z6pEk7ow==@mongo-api-benchmark.mongo.cosmos.azure.com:10255/?ssl=true&replicaSet=globaldb&retrywrites=false&maxIdleTimeMS=120000&appName=@mongo-api-benchmark@')
def runYCSB(cmd="run", ycsb_dir=r'C:\Users\anfeldma\codeHome\YCSB\bin',workload_dir=r'C:\Users\anfeldma\codeHome\YCSB\workloads',workload='workloadw', \
mongo_endpoint=r'mongodb://vmtest3.westus.cloudapp.azure.com:27017/',operation_count=1000,record_count=100, \
nthreads=1,logdir=".\\",logfn="log.csv"):
curr_dir=os.getcwd()
os.chdir(ycsb_dir)
ycsb_str=r'ycsb ' + cmd + ' mongodb -s -P "' + workload_dir + "\\" + workload + r'" -p mongodb.url="' + mongo_endpoint + \
r'" -p operationcount=' + str(operation_count) + r' -p recordcount=' + str(record_count) + r' -threads ' + str(nthreads) + \
r" " + \
' > ' + logdir + logfn
# r"^&maxPoolSize^=" + str(10*nthreads) + \
print(ycsb_str)
#status=0
os.system(ycsb_str)
os.chdir(curr_dir)
return ycsb_str
def runYCSBMongo36(execmd="run", op_count=10000, rec_count=10000, nthr=1, wkld="workloadw"):
return runYCSB(cmd=execmd, operation_count=op_count, record_count=rec_count, nthreads=nthr, workload=wkld, mongo_endpoint=r"mongodb://mongotcoa.westus.cloudapp.azure.com:27017/")
def runYCSBCosmos36(execmd="run", op_count=10000, rec_count=10000, nthr=1, wkld="workloadw"):
return runYCSB(cmd=execmd, mongo_endpoint=r'mongodb://mongo-api-benchmark:KiYRdcJp41NN268oTcyeM2ilpLwYUAo8tsX9sYoBNTd6DzjXuJHtcaSylh5VJNGs2wg1FVGExRC0m5Z6pEk7ow^=^=@mongo-api-benchmark.mongo.cosmos.azure.com:10255/?ssl^=true^&replicaSet^=globaldb^&retrywrites^=false^&maxIdleTimeMS^=120000^&appName^=@mongo-api-benchmark@', \
operation_count=op_count, record_count=rec_count, nthreads=nthr, workload=wkld)
def runYCSBAtlas36(execmd="run", op_count=10000, rec_count=10000, nthr=1, wkld="workloadw"):
return runYCSB(cmd=execmd, mongo_endpoint=r'mongodb+srv://anfeldma:O%21curmt0@atlas36shard1ncaluswest.fr0to.mongodb.net/ycsb?authSource^=admin^&retryWrites^=true^&w^=majority', \
operation_count=op_count, record_count=rec_count, nthreads=nthr, workload=wkld)
def parseLog(logdir=r'C:\Users\anfeldma\codeHome\YCSB\bin', logfn='log.csv'):
metrics_dict={}
with open(logdir + '\\' + logfn, newline='') as csvfile:
csvrdr = csv.reader(csvfile)#csv.reader(csvfile, delimiter='', quotechar='|')
for row in csvrdr:
if len(row) > 0 and row[0][0] == "[":
arg0 = row[0].lstrip().rstrip()
arg1 = row[1].lstrip().rstrip()
met_val = row[2].lstrip().rstrip()
if not(arg0 in metrics_dict):
metrics_dict[arg0] = {}
metrics_dict[arg0][arg1] = float(met_val)
return metrics_dict
def getIndividualMetrics(met_thrpt_dict_array):
# Plot response curve
thrpt_list=[]
metric_list=[]
max_thrpt=0
for idx in range(len(met_thrpt_dict_array)):
thrpt_list.append(met_thrpt_dict_array[idx][rt_thrpt_field][thrpt_field])
metric_list.append(met_thrpt_dict_array[idx][optype_field][metric_field])
return thrpt_list, metric_list, max_thrpt
def plotResponseCurve(thrpt_list, metric_list, max_thrpt, optype_field):
plt.plot(thrpt_list, metric_list, marker="x")
ax = plt.gca()
for idx in range(len(met_thrpt_dict_array)):
ax.annotate(str(thrpt_list[idx]),
xy=(thrpt_list[idx], metric_list[idx]))
plt.grid(True)
plt.title(optype_field)
plt.xlabel(thrpt_field)
plt.ylabel(metric_field)
fig=plt.gcf()
plt.show()
return fig
def saveResult(met_thrpt_dict_array,thrpt_list,metric_list,nthread_list,max_thrpt,optype_field,ycsb_str,fig):
print("Making " + optype_field + " dir.")
os.makedirs(optype_field, exist_ok=True)
print("Saving result data...")
dumpObj={}
with open(optype_field + "\\pickle.obj", "wb") as fileObj:
dumpObj["met_thrpt_dict_array"]=met_thrpt_dict_array
dumpObj["thrpt_list"]=thrpt_list
dumpObj["metric_list"]=metric_list
dumpObj["nthread_list"]=nthread_list
dumpObj["max_thrpt"]=max_thrpt
dumpObj["optype_field"]=optype_field
dumpObj["ycsb_str"]=max_thrpt
pickle.dump(dumpObj,fileObj)
print("Saving plot...")
fig.savefig(optype_field + "\\" + optype_field + ".png")
def saveComparison(op_max_rate):
print("Making " + "ycsb_op_comparison" + " dir.")
os.makedirs("ycsb_op_comparison", exist_ok=True)
print("Saving comparison data...")
dumpObj={}
with open(optype_field + "\\pickle.obj", "wb") as fileObj:
dumpObj["op_max_rate"]=op_max_rate
pickle.dump(dumpObj,fileObj)
op_mapping={"insert":{"optype_field":"[INSERT]","workload_name":"workloadw"}, \
"read":{"optype_field":"[READ]","workload_name":"workloadr"}, \
"update":{"optype_field":"[UPDATE]","workload_name":"workloadu"} \
}
db_type="atlas" #"cosmos", "mongo", "atlas"
rt_thrpt_field="[OVERALL]"
rt_field="RunTime(ms)"
thrpt_field="Throughput(ops/sec)"
ops_list=["read"] #["insert","read","update"]
opname=""
optype_field=""
workload_name=""
metric_field="99thPercentileLatency(us)"
doc_count=10000000#4000000
nthread_list=[100,150,200]#range(65,73,1)#[20,50,64,100] #[10,12,14,16,18,20] # [1,2,5,10,20,50,64,100]
print(str(range(65,129,7)[-1]))
print(str(len(range(100,129,7))))
met_thrpt_dict_array = []
os.chdir(r"C:\Users\anfeldma\codeHome\YCSB")
op_max_rate={}
for jdx in range(len(ops_list)):
opname = ops_list[jdx]
optype_field=op_mapping[opname]["optype_field"]
workload_name=op_mapping[opname]["workload_name"]
if opname != "insert":
if True or (done_load>=doc_count and load_dest==db_type):
print("Already loaded data.")
else:
print("Deleting existing data.")
if db_type=="mongo":
deleteDBMongo()
print("Starting YCSB load using max thread count...")
runYCSBMongo36(execmd="load",op_count=doc_count, rec_count=doc_count, nthr=max(nthread_list), wkld=workload_name)
elif db_type=="atlas":
deleteDBAtlas()
print("Starting YCSB load using max thread count...")
runYCSBAtlas36(execmd="load",op_count=doc_count, rec_count=doc_count, nthr=max(nthread_list), wkld=workload_name)
elif db_type=="cosmos":
deleteDBCosmos()
print("Starting YCSB load using max thread count...")
runYCSBCosmos36(execmd="load",op_count=doc_count, rec_count=doc_count, nthr=max(nthread_list), wkld=workload_name)
done_load=doc_count
load_dest=db_type
print("Finished YCSB load.")
for idx in range(len(nthread_list)):
print("Starting YCSB " + db_type + " run, opname " + opname + ", workload " + workload_name + ", thread count " + str(nthread_list[idx]))
if opname=="insert":
if db_type=="mongo":
deleteDBMongo()
elif db_type=="atlas":
deleteDBAtlas()
elif db_type=="cosmos":
deleteDBCosmos()
print("Done deleting existing YCSB dataset.")
done_load=0
operation_count=doc_count
if opname=="read" or opname=="update":
print(opname)
#operation_count=int(doc_count)
operation_count=int(doc_count/7)
elif opname=="insert":
print(opname)
operation_count=int(doc_count)
operation_count=int(doc_count/3)
if db_type=="mongo":
ycsb_str=runYCSBMongo36(op_count=operation_count, rec_count=doc_count, nthr=nthread_list[idx], wkld=workload_name)
elif db_type=="atlas":
ycsb_str=runYCSBAtlas36(op_count=operation_count, rec_count=doc_count, nthr=nthread_list[idx], wkld=workload_name)
elif db_type=="cosmos":
ycsb_str=runYCSBCosmos36(op_count=operation_count, rec_count=doc_count, nthr=nthread_list[idx], wkld=workload_name)
met_thrpt_dict_array.append(parseLog())
print("Finished YCSB run, thread count " + str(nthread_list[idx]))
thrpt_list, metric_list, max_thrpt = getIndividualMetrics(met_thrpt_dict_array)
max_thrpt=max(thrpt_list)
met_thrpt_dict_array=[]
fig=plotResponseCurve(thrpt_list, metric_list, max_thrpt, opname)
saveResult(met_thrpt_dict_array,thrpt_list,metric_list,nthread_list,max_thrpt,optype_field,ycsb_str,fig)
print("Max throughput: " + str(max_thrpt))
op_max_rate[opname]=max_thrpt
saveComparison(op_max_rate)
print(op_max_rate)
r'mongodb+srv://anfeldma:O%21curmt0@atlas36shard1ncaluswest.fr0to.mongodb.net/ycsb?authSource=admin&retryWrites=true&w=majority'
if opname=="insert":
if db_type=="mongo":
deleteDBMongo()
elif db_type=="atlas":
deleteDBAtlas()
elif db_type=="cosmos":
deleteDBCosmos()
print("Done deleting existing YCSB dataset.")
done_load=0
plt.bar(op_max_rate.keys(),op_max_rate.values())
os.getcwd()
| 0.202049 | 0.146942 |
<a href="https://colab.research.google.com/github/yuritpinheiro/deep-learning-v2-pytorch/blob/master/intro-to-pytorch/Part%203%20-%20Training%20Neural%20Networks%20(Exercises).ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Training Neural Networks
The network we built in the previous part isn't so smart, it doesn't know anything about our handwritten digits. Neural networks with non-linear activations work like universal function approximators. There is some function that maps your input to the output. For example, images of handwritten digits to class probabilities. The power of neural networks is that we can train them to approximate this function, and basically any function given enough data and compute time.
<img src="https://github.com/yuritpinheiro/deep-learning-v2-pytorch/blob/master/intro-to-pytorch/assets/function_approx.png?raw=1" width=500px>
At first the network is naive, it doesn't know the function mapping the inputs to the outputs. We train the network by showing it examples of real data, then adjusting the network parameters such that it approximates this function.
To find these parameters, we need to know how poorly the network is predicting the real outputs. For this we calculate a **loss function** (also called the cost), a measure of our prediction error. For example, the mean squared loss is often used in regression and binary classification problems
$$
\large \ell = \frac{1}{2n}\sum_i^n{\left(y_i - \hat{y}_i\right)^2}
$$
where $n$ is the number of training examples, $y_i$ are the true labels, and $\hat{y}_i$ are the predicted labels.
By minimizing this loss with respect to the network parameters, we can find configurations where the loss is at a minimum and the network is able to predict the correct labels with high accuracy. We find this minimum using a process called **gradient descent**. The gradient is the slope of the loss function and points in the direction of fastest change. To get to the minimum in the least amount of time, we then want to follow the gradient (downwards). You can think of this like descending a mountain by following the steepest slope to the base.
<img src='https://github.com/yuritpinheiro/deep-learning-v2-pytorch/blob/master/intro-to-pytorch/assets/gradient_descent.png?raw=1' width=350px>
## Backpropagation
For single layer networks, gradient descent is straightforward to implement. However, it's more complicated for deeper, multilayer neural networks like the one we've built. Complicated enough that it took about 30 years before researchers figured out how to train multilayer networks.
Training multilayer networks is done through **backpropagation** which is really just an application of the chain rule from calculus. It's easiest to understand if we convert a two layer network into a graph representation.
<img src='https://github.com/yuritpinheiro/deep-learning-v2-pytorch/blob/master/intro-to-pytorch/assets/backprop_diagram.png?raw=1' width=550px>
In the forward pass through the network, our data and operations go from bottom to top here. We pass the input $x$ through a linear transformation $L_1$ with weights $W_1$ and biases $b_1$. The output then goes through the sigmoid operation $S$ and another linear transformation $L_2$. Finally we calculate the loss $\ell$. We use the loss as a measure of how bad the network's predictions are. The goal then is to adjust the weights and biases to minimize the loss.
To train the weights with gradient descent, we propagate the gradient of the loss backwards through the network. Each operation has some gradient between the inputs and outputs. As we send the gradients backwards, we multiply the incoming gradient with the gradient for the operation. Mathematically, this is really just calculating the gradient of the loss with respect to the weights using the chain rule.
$$
\large \frac{\partial \ell}{\partial W_1} = \frac{\partial L_1}{\partial W_1} \frac{\partial S}{\partial L_1} \frac{\partial L_2}{\partial S} \frac{\partial \ell}{\partial L_2}
$$
**Note:** I'm glossing over a few details here that require some knowledge of vector calculus, but they aren't necessary to understand what's going on.
We update our weights using this gradient with some learning rate $\alpha$.
$$
\large W^\prime_1 = W_1 - \alpha \frac{\partial \ell}{\partial W_1}
$$
The learning rate $\alpha$ is set such that the weight update steps are small enough that the iterative method settles in a minimum.
## Losses in PyTorch
Let's start by seeing how we calculate the loss with PyTorch. Through the `nn` module, PyTorch provides losses such as the cross-entropy loss (`nn.CrossEntropyLoss`). You'll usually see the loss assigned to `criterion`. As noted in the last part, with a classification problem such as MNIST, we're using the softmax function to predict class probabilities. With a softmax output, you want to use cross-entropy as the loss. To actually calculate the loss, you first define the criterion then pass in the output of your network and the correct labels.
Something really important to note here. Looking at [the documentation for `nn.CrossEntropyLoss`](https://pytorch.org/docs/stable/nn.html#torch.nn.CrossEntropyLoss),
> This criterion combines `nn.LogSoftmax()` and `nn.NLLLoss()` in one single class.
>
> The input is expected to contain scores for each class.
This means we need to pass in the raw output of our network into the loss, not the output of the softmax function. This raw output is usually called the *logits* or *scores*. We use the logits because softmax gives you probabilities which will often be very close to zero or one but floating-point numbers can't accurately represent values near zero or one ([read more here](https://docs.python.org/3/tutorial/floatingpoint.html)). It's usually best to avoid doing calculations with probabilities, typically we use log-probabilities.
```
import torch
from torch import nn
import torch.nn.functional as F
from torchvision import datasets, transforms
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5,), (0.5,)),
])
# Download and load the training data
trainset = datasets.MNIST('~/.pytorch/MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
```
### Note
If you haven't seen `nn.Sequential` yet, please finish the end of the Part 2 notebook.
```
# Build a feed-forward network
model = nn.Sequential(nn.Linear(784, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 10))
# Define the loss
criterion = nn.CrossEntropyLoss()
# Get our data
images, labels = next(iter(trainloader))
# Flatten images
images = images.view(images.shape[0], -1)
# Forward pass, get our logits
logits = model(images)
# Calculate the loss with the logits and the labels
loss = criterion(logits, labels)
print(loss)
```
In my experience it's more convenient to build the model with a log-softmax output using `nn.LogSoftmax` or `F.log_softmax` ([documentation](https://pytorch.org/docs/stable/nn.html#torch.nn.LogSoftmax)). Then you can get the actual probabilities by taking the exponential `torch.exp(output)`. With a log-softmax output, you want to use the negative log likelihood loss, `nn.NLLLoss` ([documentation](https://pytorch.org/docs/stable/nn.html#torch.nn.NLLLoss)).
>**Exercise:** Build a model that returns the log-softmax as the output and calculate the loss using the negative log likelihood loss. Note that for `nn.LogSoftmax` and `F.log_softmax` you'll need to set the `dim` keyword argument appropriately. `dim=0` calculates softmax across the rows, so each column sums to 1, while `dim=1` calculates across the columns so each row sums to 1. Think about what you want the output to be and choose `dim` appropriately.
```
# TODO: Build a feed-forward network
model = nn.Sequential(nn.Linear(784, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 10),
nn.LogSoftmax(dim=1))
# TODO: Define the loss
criterion = nn.NLLLoss()
### Run this to check your work
# Get our data
images, labels = next(iter(trainloader))
# Flatten images
images = images.view(images.shape[0], -1)
# Forward pass, get our logits
logits = model(images)
# Calculate the loss with the logits and the labels
loss = criterion(logits, labels)
print(loss)
```
## Autograd
Now that we know how to calculate a loss, how do we use it to perform backpropagation? Torch provides a module, `autograd`, for automatically calculating the gradients of tensors. We can use it to calculate the gradients of all our parameters with respect to the loss. Autograd works by keeping track of operations performed on tensors, then going backwards through those operations, calculating gradients along the way. To make sure PyTorch keeps track of operations on a tensor and calculates the gradients, you need to set `requires_grad = True` on a tensor. You can do this at creation with the `requires_grad` keyword, or at any time with `x.requires_grad_(True)`.
You can turn off gradients for a block of code with the `torch.no_grad()` content:
```python
x = torch.zeros(1, requires_grad=True)
>>> with torch.no_grad():
... y = x * 2
>>> y.requires_grad
False
```
Also, you can turn on or off gradients altogether with `torch.set_grad_enabled(True|False)`.
The gradients are computed with respect to some variable `z` with `z.backward()`. This does a backward pass through the operations that created `z`.
```
x = torch.randn(2,2, requires_grad=True)
print(x)
y = x**2
print(y)
```
Below we can see the operation that created `y`, a power operation `PowBackward0`.
```
## grad_fn shows the function that generated this variable
print(y.grad_fn)
```
The autograd module keeps track of these operations and knows how to calculate the gradient for each one. In this way, it's able to calculate the gradients for a chain of operations, with respect to any one tensor. Let's reduce the tensor `y` to a scalar value, the mean.
```
z = y.mean()
print(z)
```
You can check the gradients for `x` and `y` but they are empty currently.
```
print(x.grad)
```
To calculate the gradients, you need to run the `.backward` method on a Variable, `z` for example. This will calculate the gradient for `z` with respect to `x`
$$
\frac{\partial z}{\partial x} = \frac{\partial}{\partial x}\left[\frac{1}{n}\sum_i^n x_i^2\right] = \frac{x}{2}
$$
```
z.backward()
print(x.grad)
print(x/2)
```
These gradients calculations are particularly useful for neural networks. For training we need the gradients of the cost with respect to the weights. With PyTorch, we run data forward through the network to calculate the loss, then, go backwards to calculate the gradients with respect to the loss. Once we have the gradients we can make a gradient descent step.
## Loss and Autograd together
When we create a network with PyTorch, all of the parameters are initialized with `requires_grad = True`. This means that when we calculate the loss and call `loss.backward()`, the gradients for the parameters are calculated. These gradients are used to update the weights with gradient descent. Below you can see an example of calculating the gradients using a backwards pass.
```
# Build a feed-forward network
model = nn.Sequential(nn.Linear(784, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 10),
nn.LogSoftmax(dim=1))
criterion = nn.NLLLoss()
images, labels = next(iter(trainloader))
images = images.view(images.shape[0], -1)
logits = model(images)
loss = criterion(logits, labels)
print('Before backward pass: \n', model[0].weight.grad)
loss.backward()
print('After backward pass: \n', model[0].weight.grad)
```
## Training the network!
There's one last piece we need to start training, an optimizer that we'll use to update the weights with the gradients. We get these from PyTorch's [`optim` package](https://pytorch.org/docs/stable/optim.html). For example we can use stochastic gradient descent with `optim.SGD`. You can see how to define an optimizer below.
```
from torch import optim
# Optimizers require the parameters to optimize and a learning rate
optimizer = optim.SGD(model.parameters(), lr=0.01)
```
Now we know how to use all the individual parts so it's time to see how they work together. Let's consider just one learning step before looping through all the data. The general process with PyTorch:
* Make a forward pass through the network
* Use the network output to calculate the loss
* Perform a backward pass through the network with `loss.backward()` to calculate the gradients
* Take a step with the optimizer to update the weights
Below I'll go through one training step and print out the weights and gradients so you can see how it changes. Note that I have a line of code `optimizer.zero_grad()`. When you do multiple backwards passes with the same parameters, the gradients are accumulated. This means that you need to zero the gradients on each training pass or you'll retain gradients from previous training batches.
```
print('Initial weights - ', model[0].weight)
images, labels = next(iter(trainloader))
images.resize_(64, 784)
# Clear the gradients, do this because gradients are accumulated
optimizer.zero_grad()
# Forward pass, then backward pass, then update weights
output = model(images)
loss = criterion(output, labels)
loss.backward()
print('Gradient -', model[0].weight.grad)
# Take an update step and few the new weights
optimizer.step()
print('Updated weights - ', model[0].weight)
```
### Training for real
Now we'll put this algorithm into a loop so we can go through all the images. Some nomenclature, one pass through the entire dataset is called an *epoch*. So here we're going to loop through `trainloader` to get our training batches. For each batch, we'll doing a training pass where we calculate the loss, do a backwards pass, and update the weights.
>**Exercise:** Implement the training pass for our network. If you implemented it correctly, you should see the training loss drop with each epoch.
```
## Your solution here
model = nn.Sequential(nn.Linear(784, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 10),
nn.LogSoftmax(dim=1))
criterion = nn.NLLLoss()
optimizer = optim.SGD(model.parameters(), lr=0.003)
epochs = 5
for e in range(epochs):
running_loss = 0
for images, labels in trainloader:
# Flatten MNIST images into a 784 long vector
images = images.view(images.shape[0], -1)
# TODO: Training pass
optimizer.zero_grad()
out = model(images)
loss = criterion(out, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
else:
print(f"Training loss: {running_loss/len(trainloader)}")
```
With the network trained, we can check out it's predictions.
```
%matplotlib inline
import helper
import matplotlib.pyplot as plt
images, labels = next(iter(trainloader))
img = images[0].view(1, 784)
# Turn off gradients to speed up this part
with torch.no_grad():
logps = model(img)
# Output of the network are log-probabilities, need to take exponential for probabilities
ps = torch.exp(logps)
# helper.view_classify(img.view(1, 28, 28), ps)
plt.imshow(img.view(28,28))
plt.show()
plt.barh([0,1,2,3,4,5,6,7,8,9], ps[0])
plt.show()
```
Now our network is brilliant. It can accurately predict the digits in our images. Next up you'll write the code for training a neural network on a more complex dataset.
|
github_jupyter
|
import torch
from torch import nn
import torch.nn.functional as F
from torchvision import datasets, transforms
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5,), (0.5,)),
])
# Download and load the training data
trainset = datasets.MNIST('~/.pytorch/MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
# Build a feed-forward network
model = nn.Sequential(nn.Linear(784, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 10))
# Define the loss
criterion = nn.CrossEntropyLoss()
# Get our data
images, labels = next(iter(trainloader))
# Flatten images
images = images.view(images.shape[0], -1)
# Forward pass, get our logits
logits = model(images)
# Calculate the loss with the logits and the labels
loss = criterion(logits, labels)
print(loss)
# TODO: Build a feed-forward network
model = nn.Sequential(nn.Linear(784, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 10),
nn.LogSoftmax(dim=1))
# TODO: Define the loss
criterion = nn.NLLLoss()
### Run this to check your work
# Get our data
images, labels = next(iter(trainloader))
# Flatten images
images = images.view(images.shape[0], -1)
# Forward pass, get our logits
logits = model(images)
# Calculate the loss with the logits and the labels
loss = criterion(logits, labels)
print(loss)
x = torch.zeros(1, requires_grad=True)
>>> with torch.no_grad():
... y = x * 2
>>> y.requires_grad
False
x = torch.randn(2,2, requires_grad=True)
print(x)
y = x**2
print(y)
## grad_fn shows the function that generated this variable
print(y.grad_fn)
z = y.mean()
print(z)
print(x.grad)
z.backward()
print(x.grad)
print(x/2)
# Build a feed-forward network
model = nn.Sequential(nn.Linear(784, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 10),
nn.LogSoftmax(dim=1))
criterion = nn.NLLLoss()
images, labels = next(iter(trainloader))
images = images.view(images.shape[0], -1)
logits = model(images)
loss = criterion(logits, labels)
print('Before backward pass: \n', model[0].weight.grad)
loss.backward()
print('After backward pass: \n', model[0].weight.grad)
from torch import optim
# Optimizers require the parameters to optimize and a learning rate
optimizer = optim.SGD(model.parameters(), lr=0.01)
print('Initial weights - ', model[0].weight)
images, labels = next(iter(trainloader))
images.resize_(64, 784)
# Clear the gradients, do this because gradients are accumulated
optimizer.zero_grad()
# Forward pass, then backward pass, then update weights
output = model(images)
loss = criterion(output, labels)
loss.backward()
print('Gradient -', model[0].weight.grad)
# Take an update step and few the new weights
optimizer.step()
print('Updated weights - ', model[0].weight)
## Your solution here
model = nn.Sequential(nn.Linear(784, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 10),
nn.LogSoftmax(dim=1))
criterion = nn.NLLLoss()
optimizer = optim.SGD(model.parameters(), lr=0.003)
epochs = 5
for e in range(epochs):
running_loss = 0
for images, labels in trainloader:
# Flatten MNIST images into a 784 long vector
images = images.view(images.shape[0], -1)
# TODO: Training pass
optimizer.zero_grad()
out = model(images)
loss = criterion(out, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
else:
print(f"Training loss: {running_loss/len(trainloader)}")
%matplotlib inline
import helper
import matplotlib.pyplot as plt
images, labels = next(iter(trainloader))
img = images[0].view(1, 784)
# Turn off gradients to speed up this part
with torch.no_grad():
logps = model(img)
# Output of the network are log-probabilities, need to take exponential for probabilities
ps = torch.exp(logps)
# helper.view_classify(img.view(1, 28, 28), ps)
plt.imshow(img.view(28,28))
plt.show()
plt.barh([0,1,2,3,4,5,6,7,8,9], ps[0])
plt.show()
| 0.87362 | 0.993636 |
```
import networkx as nx
import matplotlib.pyplot as plt
import numpy as np
import random
random.seed(0)
np.random.seed(0)
V = 4039
T = 1000*V
InitNode = 0
nodes = list(range(V)) # Get a list of only the node names
edges = np.loadtxt('facebook_combined.txt',dtype=int)
G = nx.Graph()
G.add_nodes_from(nodes)
G.add_edges_from(edges)
print(nx.info(G))
z = list(G.degree([n for n in G]))
y = [y[1] for y in z]
b = np.linspace(0,max(y))
plt.hist(y,bins=b)
plt.show()
nx.set_edge_attributes(G, 0,'visits')
H = G.to_directed()
nx.set_node_attributes(G, 0,'visits')
pi = np.array([x[1] for x in list(G.degree())])
pi = pi/np.sum(pi)
t = 0
v = InitNode
freq = np.empty(V)
err = []
ferr = []
while t < T:
v_next = random.choice(list(H.adj[v]))
H[v][v_next]['visits'] += 1
G.nodes[v_next]['visits'] += 1
t += 1
v = v_next
if t%(V//10) == 0:
for i in range(V):
freq[i] = G.nodes[i]['visits']
pi_hat = freq/np.sum(freq)
err.append(np.mean(abs(pi-pi_hat)))
ferr.append(np.mean(abs(pi-pi_hat)/pi))
G = nx.Graph()
G.add_nodes_from(nodes)
G.add_edges_from(edges)
nx.set_edge_attributes(G, 0,'visits')
H = G.to_directed()
nx.set_node_attributes(G, 0,'visits')
Gf = G.copy()
t = 0
v = InitNode
freq1 = np.empty(V)
freq2 = np.empty(V)
err1 = []
err2 = []
ferr1 = []
ferr2 = []
explore = np.arange(1,T,T//(100*np.log(T)))
while t < T:
if t in explore:
v_next = np.random.randint(0,V)
else:
v_next = random.choice(list(H.adj[v]))
G.nodes[v_next]['visits'] += 1
H[v][v_next]['visits'] += 1
Gf.nodes[v_next]['visits'] += 1
t += 1
v = v_next
if t%(V//10) == 0:
for i in range(V):
freq1[i] = G.nodes[i]['visits']
freq2[i] = G.nodes[i]['visits']
pi_hat1 = freq1/np.sum(freq1)
pi_hat2 = freq2/np.sum(freq2)
err1.append(np.mean(abs(pi-pi_hat1)))
ferr1.append(np.mean(abs(pi-pi_hat1)/pi))
err2.append(np.mean(abs(pi-pi_hat2)))
ferr2.append(np.mean(abs(pi-pi_hat2)/pi))
for i in range(V):
if G.nodes[i] != Gf.nodes[i]:
print(G.nodes[i],Gf.nodes[i])
plt.figure(2,figsize=(8,5))
plt.plot(np.array(list(range(len(err)//5)))*(V//10),np.log(err[:len(err)//5]),color='black')
plt.plot(np.array(list(range(len(err)//5)))*(V//10),np.log(err1[:len(err)//5]),color='blue')
plt.grid()
G = nx.Graph()
G.add_nodes_from(nodes)
G.add_edges_from(edges)
nx.set_edge_attributes(G, 0,'visits')
H = G.to_directed()
nx.set_node_attributes(G, 0,'visits')
Gf = G.copy()
t = 0
v = InitNode
freq3 = np.empty(V)
err3 = []
h_len = 100
threshold = 10
history = [None]*h_len
pointer = 0
while t < T:
temp = max(history,key=history.count)
if temp != None and history.count(temp) > threshold:
v_next = np.random.randint(0,V)
print('Improvising')
else:
v_next = random.choice(list(H.adj[v]))
G.nodes[v_next]['visits'] += 1
t += 1
history[pointer] = v
v = v_next
pointer = (pointer + 1) % h_len
if t%(V//10) == 0:
for i in range(V):
freq3[i] = G.nodes[i]['visits']
pi_hat3 = freq3/np.sum(freq3)
err3.append(np.mean(abs(pi-pi_hat3)))
plt.figure(2,figsize=(8,5))
plt.plot(np.array(list(range(len(err))))*(V//10),np.log(err),color='black')
plt.plot(np.array(list(range(len(err))))*(V//10),np.log(err3),color='blue')
plt.grid()
```
|
github_jupyter
|
import networkx as nx
import matplotlib.pyplot as plt
import numpy as np
import random
random.seed(0)
np.random.seed(0)
V = 4039
T = 1000*V
InitNode = 0
nodes = list(range(V)) # Get a list of only the node names
edges = np.loadtxt('facebook_combined.txt',dtype=int)
G = nx.Graph()
G.add_nodes_from(nodes)
G.add_edges_from(edges)
print(nx.info(G))
z = list(G.degree([n for n in G]))
y = [y[1] for y in z]
b = np.linspace(0,max(y))
plt.hist(y,bins=b)
plt.show()
nx.set_edge_attributes(G, 0,'visits')
H = G.to_directed()
nx.set_node_attributes(G, 0,'visits')
pi = np.array([x[1] for x in list(G.degree())])
pi = pi/np.sum(pi)
t = 0
v = InitNode
freq = np.empty(V)
err = []
ferr = []
while t < T:
v_next = random.choice(list(H.adj[v]))
H[v][v_next]['visits'] += 1
G.nodes[v_next]['visits'] += 1
t += 1
v = v_next
if t%(V//10) == 0:
for i in range(V):
freq[i] = G.nodes[i]['visits']
pi_hat = freq/np.sum(freq)
err.append(np.mean(abs(pi-pi_hat)))
ferr.append(np.mean(abs(pi-pi_hat)/pi))
G = nx.Graph()
G.add_nodes_from(nodes)
G.add_edges_from(edges)
nx.set_edge_attributes(G, 0,'visits')
H = G.to_directed()
nx.set_node_attributes(G, 0,'visits')
Gf = G.copy()
t = 0
v = InitNode
freq1 = np.empty(V)
freq2 = np.empty(V)
err1 = []
err2 = []
ferr1 = []
ferr2 = []
explore = np.arange(1,T,T//(100*np.log(T)))
while t < T:
if t in explore:
v_next = np.random.randint(0,V)
else:
v_next = random.choice(list(H.adj[v]))
G.nodes[v_next]['visits'] += 1
H[v][v_next]['visits'] += 1
Gf.nodes[v_next]['visits'] += 1
t += 1
v = v_next
if t%(V//10) == 0:
for i in range(V):
freq1[i] = G.nodes[i]['visits']
freq2[i] = G.nodes[i]['visits']
pi_hat1 = freq1/np.sum(freq1)
pi_hat2 = freq2/np.sum(freq2)
err1.append(np.mean(abs(pi-pi_hat1)))
ferr1.append(np.mean(abs(pi-pi_hat1)/pi))
err2.append(np.mean(abs(pi-pi_hat2)))
ferr2.append(np.mean(abs(pi-pi_hat2)/pi))
for i in range(V):
if G.nodes[i] != Gf.nodes[i]:
print(G.nodes[i],Gf.nodes[i])
plt.figure(2,figsize=(8,5))
plt.plot(np.array(list(range(len(err)//5)))*(V//10),np.log(err[:len(err)//5]),color='black')
plt.plot(np.array(list(range(len(err)//5)))*(V//10),np.log(err1[:len(err)//5]),color='blue')
plt.grid()
G = nx.Graph()
G.add_nodes_from(nodes)
G.add_edges_from(edges)
nx.set_edge_attributes(G, 0,'visits')
H = G.to_directed()
nx.set_node_attributes(G, 0,'visits')
Gf = G.copy()
t = 0
v = InitNode
freq3 = np.empty(V)
err3 = []
h_len = 100
threshold = 10
history = [None]*h_len
pointer = 0
while t < T:
temp = max(history,key=history.count)
if temp != None and history.count(temp) > threshold:
v_next = np.random.randint(0,V)
print('Improvising')
else:
v_next = random.choice(list(H.adj[v]))
G.nodes[v_next]['visits'] += 1
t += 1
history[pointer] = v
v = v_next
pointer = (pointer + 1) % h_len
if t%(V//10) == 0:
for i in range(V):
freq3[i] = G.nodes[i]['visits']
pi_hat3 = freq3/np.sum(freq3)
err3.append(np.mean(abs(pi-pi_hat3)))
plt.figure(2,figsize=(8,5))
plt.plot(np.array(list(range(len(err))))*(V//10),np.log(err),color='black')
plt.plot(np.array(list(range(len(err))))*(V//10),np.log(err3),color='blue')
plt.grid()
| 0.057315 | 0.57335 |
# Duel of sorcerers
You are witnessing an epic battle between two powerful sorcerers: Gandalf and Saruman. Each sorcerer has 10 spells of variable power in their mind and they are going to throw them one after the other. The winner of the duel will be the one who wins more of those clashes between spells. Spells are represented as a list of 10 integers whose value equals the power of the spell.
```
gandalf = [10, 11, 13, 30, 22, 11, 10, 33, 22, 22]
saruman = [23, 66, 12, 43, 12, 10, 44, 23, 12, 17]
```
For example:
1. The first clash is won by Saruman: 10 against 23, wins 23
2. The second clash wins Saruman: 11 against 66, wins 66
3. etc.
You will create two variables, one for each sorcerer, where the sum of clashes won will be stored. Depending on which variable is greater at the end of the duel, you will show one of the following three results on the screen:
* Gandalf wins
* Saruman wins
* Tie
<img src="images/content_lightning_bolt_big.jpg" width="400">
## Solution
```
# Assign spell power lists to variables
gandalf = [10, 11, 13, 30, 22, 11, 10, 33, 22, 22]
saruman = [23, 66, 12, 43, 12, 10, 44, 23, 12, 17]
# Assign 0 to each variable that stores the victories
g_win = 0
s_win = 0
for i in range(len(gandalf)):
if gandalf[i] > saruman[i]:
g_win += 1
else:
s_win += 1
# We check who has won, do not forget the possibility of a draw.
# Print the result based on the winner.
if g_win > s_win:
print("Gandalf wins with a total of:", g_win, "victories!")
elif s_win > g_win:
print("Saruman wins with a total of:", s_win, "victories!")
else:
print("Tie")
```
## Goals
1. Treatment of lists
2. Use of **for loop**
3. Use of conditional **if-elif-else**
4. Use of the functions **range(), len()**
5. Print
## Bonus
1. Spells now have a name and there is a dictionary that relates that name to a power.
2. A sorcerer wins if he succeeds in winning 3 spell clashes in a row.
3. Average of each of the spell lists.
4. Standard deviation of each of the spell lists.
```
POWER = {
'Fireball': 50,
'Lightning bolt': 40,
'Magic arrow': 10,
'Black Tentacles': 25,
'Contagion': 45
}
gandalf = ['Fireball', 'Lightning bolt', 'Lightning bolt', 'Magic arrow', 'Fireball',
'Magic arrow', 'Lightning bolt', 'Fireball', 'Fireball', 'Fireball']
saruman = ['Contagion', 'Contagion', 'Black Tentacles', 'Fireball', 'Black Tentacles',
'Lightning bolt', 'Magic arrow', 'Contagion', 'Magic arrow', 'Magic arrow']
```
Good luck!
```
# 1. Spells now have a name and there is a dictionary that relates that name to a power.
# variables
POWER = {
'Fireball': 50,
'Lightning bolt': 40,
'Magic arrow': 10,
'Black Tentacles': 25,
'Contagion': 45
}
gandalf = ['Fireball', 'Lightning bolt', 'Lightning bolt', 'Magic arrow', 'Fireball',
'Magic arrow', 'Lightning bolt', 'Fireball', 'Magic arrow', 'Fireball']
saruman = ['Contagion', 'Contagion', 'Black Tentacles', 'Fireball', 'Black Tentacles',
'Lightning bolt', 'Magic arrow', 'Contagion', 'Magic arrow', 'Magic arrow']
# Assign spell power lists to variables
for i in gandalf:
i = POWER.values()
print(i)
Fireball = 50
Lightning_bolt = 40
Magic_arrow = 10
Black_Tentacles = 25
Contagion = 45
g_total = 0
s_total = 0
# 2. A sorcerer wins if he succeeds in winning 3 spell clashes in a row.
# Execution of spell clashes
for i in gandalf:
if gandalf [i] == power.keys():
g_total += power.values
print(g_total)
# check for 3 wins in a row
# check the winner
# 3. Average of each of the spell lists.
# 4. Standard deviation of each of the spell lists.
```
|
github_jupyter
|
gandalf = [10, 11, 13, 30, 22, 11, 10, 33, 22, 22]
saruman = [23, 66, 12, 43, 12, 10, 44, 23, 12, 17]
# Assign spell power lists to variables
gandalf = [10, 11, 13, 30, 22, 11, 10, 33, 22, 22]
saruman = [23, 66, 12, 43, 12, 10, 44, 23, 12, 17]
# Assign 0 to each variable that stores the victories
g_win = 0
s_win = 0
for i in range(len(gandalf)):
if gandalf[i] > saruman[i]:
g_win += 1
else:
s_win += 1
# We check who has won, do not forget the possibility of a draw.
# Print the result based on the winner.
if g_win > s_win:
print("Gandalf wins with a total of:", g_win, "victories!")
elif s_win > g_win:
print("Saruman wins with a total of:", s_win, "victories!")
else:
print("Tie")
POWER = {
'Fireball': 50,
'Lightning bolt': 40,
'Magic arrow': 10,
'Black Tentacles': 25,
'Contagion': 45
}
gandalf = ['Fireball', 'Lightning bolt', 'Lightning bolt', 'Magic arrow', 'Fireball',
'Magic arrow', 'Lightning bolt', 'Fireball', 'Fireball', 'Fireball']
saruman = ['Contagion', 'Contagion', 'Black Tentacles', 'Fireball', 'Black Tentacles',
'Lightning bolt', 'Magic arrow', 'Contagion', 'Magic arrow', 'Magic arrow']
# 1. Spells now have a name and there is a dictionary that relates that name to a power.
# variables
POWER = {
'Fireball': 50,
'Lightning bolt': 40,
'Magic arrow': 10,
'Black Tentacles': 25,
'Contagion': 45
}
gandalf = ['Fireball', 'Lightning bolt', 'Lightning bolt', 'Magic arrow', 'Fireball',
'Magic arrow', 'Lightning bolt', 'Fireball', 'Magic arrow', 'Fireball']
saruman = ['Contagion', 'Contagion', 'Black Tentacles', 'Fireball', 'Black Tentacles',
'Lightning bolt', 'Magic arrow', 'Contagion', 'Magic arrow', 'Magic arrow']
# Assign spell power lists to variables
for i in gandalf:
i = POWER.values()
print(i)
Fireball = 50
Lightning_bolt = 40
Magic_arrow = 10
Black_Tentacles = 25
Contagion = 45
g_total = 0
s_total = 0
# 2. A sorcerer wins if he succeeds in winning 3 spell clashes in a row.
# Execution of spell clashes
for i in gandalf:
if gandalf [i] == power.keys():
g_total += power.values
print(g_total)
# check for 3 wins in a row
# check the winner
# 3. Average of each of the spell lists.
# 4. Standard deviation of each of the spell lists.
| 0.208582 | 0.959307 |
# Spectroscopy of a three cavity - two qubit system, with thermal losses: <mark>Solving the Master Equation</mark>
1. **Introduction**
2. **Problem parameters**
3. **Setting up operators and Hamiltonian's**
4. **Frequency spectrum of the coupled system**
5. **Evolving qubit 1 in the system with time**
<u>Author</u> : Soumya Shreeram (shreeramsoumya@gmail.com)<br>
<u>Supervisor</u> : Yu-Chin Chao (ychao@fnal.gov) <br>
<u>Date</u>$\ \ \ \$: 7th July 2019<br>
This script was coded as part of the Helen Edwards Summer Internship program at Fermilab.
## 1. Introduction
A multi-mode QED architecture is explored as described in by [McKay et *al*](http://schusterlab.uchicago.edu/static/pdfs/McKay2015.pdf). The hamiltonian for such a system with two qubits with frequencies $v_{Q,1}$, $v_{Q,2}$, and $n$ mode filter can be described as the sum of the qubit Hamiltonian, $\hat{H}_Q$, the filter Hamiltonian, $\hat{H}_F$, and the qubit-filter coupling Hamiltonian, $\hat{H}_{Q-F},$
$$ \hat{H} = \hat{H_Q} + \hat{H_F} + \hat{H}_{Q-F} $$
$$ \hat{H_Q} = h\ v_{Q,1}\ \frac{\hat{ \sigma}^z_1}{2} + h\ v_{Q,2}\ \frac{\hat{ \sigma}^z_2}{2}$$
$$ \hat{H}_{F} = \sum_{i=1}^{n}h\ v_{F}\ \hat{a}^{\dagger}_i \hat{a}_i + \sum_{i=2}^{n}h\ g_{F}\ (\hat{a}^{\dagger}_i \hat{a}_{i-1} + \hat{a}^{\dagger}_{i-1} \hat{a}_i)$$
$$ \hat{H}_{Q-F} = h\ g_{Q1,F}\ (\hat{a}^{\dagger}_1 \hat{\sigma}^-_1 + \hat{a}_1 \hat{\sigma}^+_1) + h\ g_{Q2,F}\ (\hat{a}^{\dagger}_n \hat{\sigma}^-_2 + \hat{a}_n \hat{\sigma}^+_2)$$
where $\hat{\sigma}^{+(-)}$ is the raising and lowering operator for the qubit, $\hat{a}_i$ creates a photon in the $i^{th}$ resonantor, $g_F$ is the filter-filter coupling, and $g_{Q,F}$ is the qubit-filter coupling.
Here we must also account for the interaction of the quantum state with it's environment. This can be represented by a non-Hermitian term in the Hamiltonian such that,
$$\displaystyle H_{\rm eff}(t) = H(t) - \frac{i\hbar}{2}\sum_n c_n^\dagger c_n$$
where $c_n$ is the collapse operator
The code calculates the eigen modes for such a system when the qubit 1 frequency is changed.
```
%matplotlib inline
import matplotlib.pyplot as plt
plt.rcParams.update({'font.size': 16})
import numpy as np
from math import pi
from qutip import *
```
## 2. Problem parameters
Here we use $\hbar=1$; the coupling terms are redefined with a multiple of $2\pi$ before them for convinience.
```
"""------------- FREQUENCIES --------------------"""
w_q1 = 2*pi*6.5; # Qubit 1 frequency
w_q2 = 2*pi*6.8; # Qubit 2 frequency: range from 1-9 GHz
w_f = 2*pi*7.1 # Resonator/ Filter frequency
"""------------- COUPLING --------------------"""
g_f1 = 2*pi*1.18 # Filter-filter coupling
g_f2 = 2*pi*3.44
g_q1f = 2*pi*1.35 # qubit 1-fitler coupling
g_q2f = 2*pi*4.15 # qubit 2-fitler coupling
numF = 3 # number of filters
N = 2 # number of fock states
kappa = 1.0/0.129 # cavity dissipation rate
n_th_a = 0.063 # avg. no. of thermal bath excitation
r1 = 0.0075 # qubit relaxation rate
r2 = 0.0025 # qubit dephasing rate
```
## 3. Setting up operators and the Hamiltonian's
For every qubit: <br> <br>
**sm** $\ \rightarrow \ \hat{\sigma}^{+(-)}$ is the raising and lowering operator of the *qubit* <br>
**sz** $\ \ \rightarrow \ \sigma_z $ is the Pauli-z matrix of the *qubit* <br>
**n** $\ \ \ \rightarrow \ n$ is the number operator
```
def numOp(m):
"""
Computes the number operator
@param loweringMat :: lowering matrix operator for a system
"""
return m.dag()*m
def rwaCoupling(m1, m2):
return m1.dag()*m2 + m1*m2.dag()
```
### 3.1 Qubit-cavity system operators and Hamiltonian's
```
# cavity 1, 2, 3 destruction operators
a1 = tensor(destroy(N), qeye(N), qeye(N), qeye(2), qeye(2))
a2 = tensor(qeye(N), destroy(N), qeye(N), qeye(2), qeye(2))
a3 = tensor(qeye(N), qeye(N), destroy(N), qeye(2), qeye(2))
# operators for qubit 1
sm1 = tensor(qeye(N), qeye(N), qeye(N), sigmam(), qeye(2))
sz1 = tensor(qeye(N), qeye(N), qeye(N), sigmaz(), qeye(2))
n1 = sm1.dag() * sm1
# operators for qubit 2
sm2 = tensor(qeye(N), qeye(N), qeye(N), qeye(2), sigmam())
sz2 = tensor(qeye(N), qeye(N), qeye(N), qeye(2), sigmaz())
n2 = sm2.dag() * sm2
# collapse operators
c_ops = []
# Qubit Hamiltonians (Hq1+Hq2)
Hq1 = 0.5*sz1
Hq2 = 0.5*sz2
# Filter Hamiltonians (refer formula in the Introduction)
Hf = numOp(a1) + numOp(a2) + numOp(a3)
H_f12 = g_f1*(rwaCoupling(a1, a2) + rwaCoupling(a2, a3))
# Qubit-Filter Hamiltonian
Hqf = g_q1f*(rwaCoupling(a1, sm1) + rwaCoupling(a3, sm2))
# Qubit 1 -independent Hamiltonian (see later)
H0 = H_f12 + Hqf + w_f*Hf + w_q2*Hq2
H = w_f*Hf + H_f12 + Hqf + w_q1*Hq1 + w_q2*Hq2 # Resultant Hamiltonian
```
### 3.2 Collapse operator used to describe dissipation
```
# collapse operator list
c_ops = []
# cavity relaxation
rate = kappa * (1 + n_th_a)
c_ops.append(np.sqrt(rate) * a1)
# cavity excitation
# qubit 1 relaxation
c_ops.append(np.sqrt(r1 * (1+n_th_a)) * sm1)
c_ops.append(np.sqrt(r1 * n_th_a) * sm1.dag())
c_ops.append(np.sqrt(r2) * sz1)
# qubit 2 relaxation
c_ops.append(np.sqrt(r1 * (1+n_th_a)) * sm2)
c_ops.append(np.sqrt(r1 * n_th_a) * sm2.dag())
c_ops.append(np.sqrt(r2) * sz2)
# initial state of the system. Qubit 1: excited, Qubit 2: ground st.
psi0 = tensor(basis(N,0), basis(N,0), basis(N,0), basis(2,0), basis(2,1))
times = np.linspace(0.0,6,500)
output = mesolve(H, psi0, times, c_ops, [n1, n2, numOp(a1), numOp(a2), numOp(a3)])
fig, ax = plt.subplots(2, 2, figsize=(14,12))
# qubit 1 - Cavity 1
ax[0, 0].plot(times, output.expect[0], 'r:', linewidth=1.8, label="Qubit 1")
ax[0, 0].plot(times, output.expect[0], 'b:', linewidth=1.8, label="Cavity 1")
ax[0, 0].set_title('Qubit 1 - Cavities 1');
ax[0, 0].legend(loc="upper right")
# qubit 1 - Cavity 2, 3
ax[0, 1].plot(times, output.expect[0], 'r-', label="Qubit 1")
ax[0, 1].plot(times, output.expect[3], 'k--', linewidth=1.8, label="Cavity 2")
ax[0, 1].plot(times, output.expect[4], 'y--', linewidth=1.5, label="Cavity 3")
ax[0, 1].set_title('Qubit 1 - Cavities 2,3');
ax[0, 1].legend(loc="upper right")
# qubit 2 - Cavity 1
ax[1, 0].plot(times, output.expect[1], 'g--', label="Qubit 2")
ax[1, 0].plot(times, output.expect[0], 'b', label="Cavity 1")
ax[1, 0].set_xlabel('Time (ns)');
ax[1, 0].set_ylabel('Occupation probability');
ax[1, 0].set_title('Qubits 2 - Cavity 1');
ax[1, 0].legend(loc="upper right")
# qubit 2 - Cavity 2, 3
ax[1,1].plot(times, output.expect[1], 'g:', linewidth=1.5, label="Qubit 2")
ax[1,1].plot(times, output.expect[3], 'k--', linewidth=1.8, label="Cavity 2")
ax[1, 1].plot(times, output.expect[4], 'y--', linewidth=1.5, label="Cavity 3")
ax[1,1].set_xlabel('Time (ns)')
ax[1,1].set_title('Qubit 2 - Cavities 2,3');
ax[1,1].legend(loc="upper right")
fig, axes = plt.subplots(1, 1, figsize=(12,7))
axes.plot(times, output.expect[0] + output.expect[1] + output.expect[2] + output.expect[3] + output.expect[4], 'k--', linewidth=1.5, label="Total Probability")
axes.plot(times, output.expect[2], 'r--', linewidth=1.5, label="cavity 1")
axes.plot(times, output.expect[3], 'r:', linewidth=1.5, label="cavity 2")
axes.plot(times, output.expect[4], 'r', linewidth=1, label="cavity 3")
axes.plot(times, output.expect[0], 'b', linewidth=0.9, label="qubit 1")
axes.plot(times, output.expect[1], '#b0ed3e', linewidth=1.8, label="qubit 2")
axes.set_xlabel("Time (ns)")
axes.set_ylabel("Occupation \n probability")
axes.legend(loc = 'center left', bbox_to_anchor = (1.0, 0.5))
fig.tight_layout()
```
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
plt.rcParams.update({'font.size': 16})
import numpy as np
from math import pi
from qutip import *
"""------------- FREQUENCIES --------------------"""
w_q1 = 2*pi*6.5; # Qubit 1 frequency
w_q2 = 2*pi*6.8; # Qubit 2 frequency: range from 1-9 GHz
w_f = 2*pi*7.1 # Resonator/ Filter frequency
"""------------- COUPLING --------------------"""
g_f1 = 2*pi*1.18 # Filter-filter coupling
g_f2 = 2*pi*3.44
g_q1f = 2*pi*1.35 # qubit 1-fitler coupling
g_q2f = 2*pi*4.15 # qubit 2-fitler coupling
numF = 3 # number of filters
N = 2 # number of fock states
kappa = 1.0/0.129 # cavity dissipation rate
n_th_a = 0.063 # avg. no. of thermal bath excitation
r1 = 0.0075 # qubit relaxation rate
r2 = 0.0025 # qubit dephasing rate
def numOp(m):
"""
Computes the number operator
@param loweringMat :: lowering matrix operator for a system
"""
return m.dag()*m
def rwaCoupling(m1, m2):
return m1.dag()*m2 + m1*m2.dag()
# cavity 1, 2, 3 destruction operators
a1 = tensor(destroy(N), qeye(N), qeye(N), qeye(2), qeye(2))
a2 = tensor(qeye(N), destroy(N), qeye(N), qeye(2), qeye(2))
a3 = tensor(qeye(N), qeye(N), destroy(N), qeye(2), qeye(2))
# operators for qubit 1
sm1 = tensor(qeye(N), qeye(N), qeye(N), sigmam(), qeye(2))
sz1 = tensor(qeye(N), qeye(N), qeye(N), sigmaz(), qeye(2))
n1 = sm1.dag() * sm1
# operators for qubit 2
sm2 = tensor(qeye(N), qeye(N), qeye(N), qeye(2), sigmam())
sz2 = tensor(qeye(N), qeye(N), qeye(N), qeye(2), sigmaz())
n2 = sm2.dag() * sm2
# collapse operators
c_ops = []
# Qubit Hamiltonians (Hq1+Hq2)
Hq1 = 0.5*sz1
Hq2 = 0.5*sz2
# Filter Hamiltonians (refer formula in the Introduction)
Hf = numOp(a1) + numOp(a2) + numOp(a3)
H_f12 = g_f1*(rwaCoupling(a1, a2) + rwaCoupling(a2, a3))
# Qubit-Filter Hamiltonian
Hqf = g_q1f*(rwaCoupling(a1, sm1) + rwaCoupling(a3, sm2))
# Qubit 1 -independent Hamiltonian (see later)
H0 = H_f12 + Hqf + w_f*Hf + w_q2*Hq2
H = w_f*Hf + H_f12 + Hqf + w_q1*Hq1 + w_q2*Hq2 # Resultant Hamiltonian
# collapse operator list
c_ops = []
# cavity relaxation
rate = kappa * (1 + n_th_a)
c_ops.append(np.sqrt(rate) * a1)
# cavity excitation
# qubit 1 relaxation
c_ops.append(np.sqrt(r1 * (1+n_th_a)) * sm1)
c_ops.append(np.sqrt(r1 * n_th_a) * sm1.dag())
c_ops.append(np.sqrt(r2) * sz1)
# qubit 2 relaxation
c_ops.append(np.sqrt(r1 * (1+n_th_a)) * sm2)
c_ops.append(np.sqrt(r1 * n_th_a) * sm2.dag())
c_ops.append(np.sqrt(r2) * sz2)
# initial state of the system. Qubit 1: excited, Qubit 2: ground st.
psi0 = tensor(basis(N,0), basis(N,0), basis(N,0), basis(2,0), basis(2,1))
times = np.linspace(0.0,6,500)
output = mesolve(H, psi0, times, c_ops, [n1, n2, numOp(a1), numOp(a2), numOp(a3)])
fig, ax = plt.subplots(2, 2, figsize=(14,12))
# qubit 1 - Cavity 1
ax[0, 0].plot(times, output.expect[0], 'r:', linewidth=1.8, label="Qubit 1")
ax[0, 0].plot(times, output.expect[0], 'b:', linewidth=1.8, label="Cavity 1")
ax[0, 0].set_title('Qubit 1 - Cavities 1');
ax[0, 0].legend(loc="upper right")
# qubit 1 - Cavity 2, 3
ax[0, 1].plot(times, output.expect[0], 'r-', label="Qubit 1")
ax[0, 1].plot(times, output.expect[3], 'k--', linewidth=1.8, label="Cavity 2")
ax[0, 1].plot(times, output.expect[4], 'y--', linewidth=1.5, label="Cavity 3")
ax[0, 1].set_title('Qubit 1 - Cavities 2,3');
ax[0, 1].legend(loc="upper right")
# qubit 2 - Cavity 1
ax[1, 0].plot(times, output.expect[1], 'g--', label="Qubit 2")
ax[1, 0].plot(times, output.expect[0], 'b', label="Cavity 1")
ax[1, 0].set_xlabel('Time (ns)');
ax[1, 0].set_ylabel('Occupation probability');
ax[1, 0].set_title('Qubits 2 - Cavity 1');
ax[1, 0].legend(loc="upper right")
# qubit 2 - Cavity 2, 3
ax[1,1].plot(times, output.expect[1], 'g:', linewidth=1.5, label="Qubit 2")
ax[1,1].plot(times, output.expect[3], 'k--', linewidth=1.8, label="Cavity 2")
ax[1, 1].plot(times, output.expect[4], 'y--', linewidth=1.5, label="Cavity 3")
ax[1,1].set_xlabel('Time (ns)')
ax[1,1].set_title('Qubit 2 - Cavities 2,3');
ax[1,1].legend(loc="upper right")
fig, axes = plt.subplots(1, 1, figsize=(12,7))
axes.plot(times, output.expect[0] + output.expect[1] + output.expect[2] + output.expect[3] + output.expect[4], 'k--', linewidth=1.5, label="Total Probability")
axes.plot(times, output.expect[2], 'r--', linewidth=1.5, label="cavity 1")
axes.plot(times, output.expect[3], 'r:', linewidth=1.5, label="cavity 2")
axes.plot(times, output.expect[4], 'r', linewidth=1, label="cavity 3")
axes.plot(times, output.expect[0], 'b', linewidth=0.9, label="qubit 1")
axes.plot(times, output.expect[1], '#b0ed3e', linewidth=1.8, label="qubit 2")
axes.set_xlabel("Time (ns)")
axes.set_ylabel("Occupation \n probability")
axes.legend(loc = 'center left', bbox_to_anchor = (1.0, 0.5))
fig.tight_layout()
| 0.750644 | 0.986891 |
```
import sys
import numpy as np
from numpy import genfromtxt
import tkinter as tk
from tkinter import filedialog
import os
import pandas as pd
import matplotlib.pyplot as plt
import scipy.signal as signal
from scipy import interpolate
from scipy.optimize import curve_fit
from scipy.interpolate import UnivariateSpline
from scipy import stats
from ipfx import feature_extractor
from ipfx import subthresh_features as subt
from ipfx import feature_vectors as fv
from ipfx.sweep import Sweep
from sklearn.preprocessing import minmax_scale
from pyAPisolation.loadABF import loadABF
import sklearn.preprocessing
import pyabf
import logging
import glob
method='trf'
import autograd.numpy as np
from autograd import grad
def exp_grow(t, a, b, alpha):
return a - b * np.exp(-alpha * t)
def exp_grow_2p(t, a, b1, alphaFast, b2, alphaSlow):
return a - b1 * np.exp(-alphaFast * t) - b2*np.exp(-alphaSlow*t)
def exp_grow_clampfit(t, a, b1, alphaFast, b2, alphaSlow):
return b1 * np.exp(-alphaFast * t) + b2*np.exp(-alphaSlow*t)
f1 = grad(exp_grow_2p) # 1st derivative of f
f2 = grad(f1) # 2nd derivative of f
def curvature(x, a, b1, alphaFast, b2, alphaSlow):
return np.abs(f2(x, a, b1, alphaFast, b2, alphaSlow))*(1 + f1(x, a, b1, alphaFast, b2, alphaSlow)**2)**-1.5
def curvature_real(dy, ddy):
return abs(dy)*(1 + ddy**2)**-1.5
def curvature_splines(x, y=None, error=0.1, smoothing=None):
"""Calculate the signed curvature of a 2D curve at each point
using interpolating splines.
Parameters
----------
x,y: numpy.array(dtype=float) shape (n_points, )
or
y=None and
x is a numpy.array(dtype=complex) shape (n_points, )
In the second case the curve is represented as a np.array
of complex numbers.
error : float
The admisible error when interpolating the splines
Returns
-------
curvature: numpy.array shape (n_points, )
Note: This is 2-3x slower (1.8 ms for 2000 points) than `curvature_gradient`
but more accurate, especially at the borders.
"""
# handle list of complex case
if y is None:
x, y = x.real, x.imag
t = np.arange(x.shape[0])
std = error * np.ones_like(x)
fx = UnivariateSpline(t, x, k=4, w=1 / np.sqrt(std), s=smoothing)
fy = UnivariateSpline(t, y, k=4, w=1 / np.sqrt(std), s=smoothing)
xˈ = fx.derivative(1)(t)
xˈˈ = fx.derivative(2)(t)
yˈ = fy.derivative(1)(t)
yˈˈ = fy.derivative(2)(t)
curvature = (xˈ* yˈˈ - yˈ* xˈˈ) / np.power(xˈ** 2 + yˈ** 2, 3 / 2)
return curvature
def derivative(x,y):
return np.diff(y)/np.diff(x)
def curve_detrend(x,y, curve2):
test = curvature_splines(x, signal.savgol_filter(y, 51, 1), error=1, smoothing=25)
cy = np.array([curvature(xi, *curve2) for xi in x])
#detrend using first and last point
lin_res = stats.linregress([x[0], x[-1]], [cy[0], cy[-1]])
trend = x*lin_res.slope + lin_res.intercept
#plt.plot(x,trend)
detrended_data = cy - trend
return detrended_data
def exp_growth_factor(dataT,dataV,dataI, end_index=300):
#try:
diff_I = np.diff(dataI)
upwardinfl = np.argmax(diff_I)
#Compute out -50 ms from threshold
dt = dataT[1] - dataT[0]
offset = 0.01/ dt
end_index = int(end_index - offset)
upperC = np.amax(dataV[upwardinfl:end_index])
lowerC = np.amin(dataV[upwardinfl:end_index])
diffC = np.abs(lowerC - upperC)
t1 = dataT[upwardinfl:end_index] - dataT[upwardinfl]
curve = curve_fit(exp_grow, t1, dataV[upwardinfl:end_index], maxfev=50000, bounds=([-np.inf, -np.inf, -np.inf], [np.inf, np.inf, np.inf]))[0]
curve = np.hstack((curve, np.full(2,1)))
curve2 = curve_fit(exp_grow_2p, t1, dataV[upwardinfl:end_index], maxfev=50000,method='trf', p0=curve, bounds=([upperC-5, 0, 10, 0, -np.inf], [upperC+5, diffC, np.inf, diffC,np.inf]), xtol=None, gtol=None, ftol=1e-12, jac='3-point')[0]
tau = curve[2]
tau1 = 1/curve2[2]
tau2 = 1/curve2[4]
tau_idx = [2, 4]
fast = tau_idx[np.argmin([tau1, tau2])]
slow = tau_idx[np.argmax([tau1, tau2])]
curve_out = [curve2[0], curve2[fast-1], curve2[fast], curve2[slow-1], curve2[slow]]
#plt.subplot(1,2,1)
plt.plot(t1, dataV[upwardinfl:end_index], c='k', alpha=0.5)
plt.plot(t1, exp_grow_2p(t1, *curve2), label=f'2 phase fit', c='r', alpha=0.5)
plt.plot(t1, exp_grow(t1, *curve_out[:3]), label=f'Fast phase', c='g', alpha=0.5)
plt.plot(t1, exp_grow(t1, curve_out[0], *curve_out[3:]), label=f'slow phase', c='b', alpha=0.5)
plt.title(f" CELL will tau1 {1/curve2[fast]} and tau2 {1/curve2[slow]}")
#plt.subplot(1,2,2)
plt.legend()
#plt.twinx()
#plt.subplot(1,2,2)
dy = curve_detrend(t1, dataV[upwardinfl:end_index], curve2)
#signal.savgol_filter(nt1p.diff(dataV[upwardinfl:end_index])/np.diff(t1), 71, 2, mode='mirror')
#plt.plot(t1,dy)
curve_out = [curve2[0], curve2[fast-1], 1/curve2[fast], curve2[slow-1], 1/curve2[slow]]
return curve_out, np.amax(dy)
#except:
return [np.nan, np.nan, np.nan, np.nan, np.nan]
files = glob.glob('C:\\Users\\SMest\\Documents\\clustering-data\\\All IC1s\\*.abf', recursive=True)
cell_type_df = pd.read_csv("C:\\Users\\SMest\\Documents\\clustering-data\\MARM_PVN_IC1\\spike_count_sort_out.csv")
print(cell_type_df.head)
file_names = cell_type_df['filename'].to_numpy()
cell_type_label = cell_type_df['cell_label'].to_numpy()
curves = []
label = []
ids = []
max_curve = []
for i, f in enumerate(files[-20:]):
print(i)
#try:
base = os.path.basename(f)
base = base.split(".")[0]
if base in file_names:
x, y, c = loadABF(f)
temp_curves =[]
#plt.clf()
iterd = 0
for sweepX, sweepY, sweepC in zip(x,y,c):
spikext = feature_extractor.SpikeFeatureExtractor(filter=0, end=1.25)
res = spikext.process(sweepX, sweepY, sweepC)
if res.empty==False and iterd < 3:
iterd += 1
spike_time = res['threshold_index'].to_numpy()[0]
#plt.figure(num=2)
curve, max_dy = exp_growth_factor(sweepX, sweepY, sweepC, spike_time)
max_curve.append(max_dy)
temp_curves.append(curve)
temp_curves = np.vstack(temp_curves)
div = np.ravel((temp_curves[:,2]) / (temp_curves[:,4])).reshape(-1,1)
sum_height= (temp_curves[:,1] + temp_curves[:,3])
ratio = (temp_curves[:,2] / (temp_curves[:,1] / sum_height)) / (temp_curves[:,4] / (temp_curves[:,3] / sum_height))
ratio = np.ravel(ratio).reshape(-1,1)
temp_curves = np.hstack([temp_curves, div, ratio])
print(temp_curves)
meanC = np.nanmean(temp_curves, axis=0)
print(meanC.shape)
curves.append(meanC)
label_idx = np.argwhere(file_names==base)
label.append(cell_type_label[label_idx])
ids.append(base)
plt.savefig(f+".png")
plt.show()
plt.close()
#except:
#print("fail")
curves = np.vstack(curves)
#lab = sklearn.preprocessing.LabelEncoder()
#int_lab = lab.fit_transform(label)
print(curves)
label = np.ravel(label).reshape(-1,1)
div = np.ravel((curves[:,2]) / (curves[:,4])).reshape(-1,1)
print(div)
sum_height= (curves[:,1] + curves[:,3])
ratio = (curves[:,2] / (curves[:,1]/sum_height)) / (curves[:,4] / (curves[:,3]/sum_height))
ratio = np.ravel(ratio).reshape(-1,1)
curves_out = np.hstack([curves, div, ratio, label])
np.savetxt('curves.csv', curves_out, fmt='%.8f', delimiter=',')
np.savetxt('curves_id.csv', ids, fmt='%s', delimiter=',')
print(curves)
means = []
plt.figure(figsize=(10,10))
plt.clf()
for x in np.unique(label).astype(np.int32):
idx = np.argwhere(label[:,0]==x).astype(np.int32)
mcur = curves[idx]
plt.scatter(np.full(len(idx), x), (curves[idx,2]) / (curves[idx,4]), label=label[x])
means.append(np.nanmean((curves[idx,2]) / (curves[idx,4])))
plt.legend()
plt.yscale('log')
#plt.ylim(0,1)
print(means)
1=1
curves = []
label = []
ids = []
for i, f in enumerate(files[:38]):
print(i)
x, y, c = loadABF(f)
d_name = os.path.dirname(f)
base = os.path.basename(f)
ids.append(base)
label.append(d_name)
dfs = []
temp_curves
plt.clf()
for sweepX, sweepY, sweepC in zip(x,y,c):
spikext = feature_extractor.SpikeFeatureExtractor(filter=0)
res = spikext.process(sweepX, sweepY, sweepC)
dfs.append(res)
if res.empty==False:
if len(non_empty_df) > 1:
sweep_to_use = non_empty_df[1]
else:
sweep_to_use = non_empty_df[-1]
non_empty_df = np.nonzero(np.invert([df.empty for df in dfs]))[0]
try:
spike_time = dfs[sweep_to_use]['threshold_index'].to_numpy()[0]
curve = exp_growth_factor(x[sweep_to_use,:], y[sweep_to_use,:], c[sweep_to_use,:], spike_time)
curves.append(curve)
except:
curves.append([np.nan, np.nan, np.nan, np.nan, np.nan])
plt.show()
print(non_empty_df)
```
|
github_jupyter
|
import sys
import numpy as np
from numpy import genfromtxt
import tkinter as tk
from tkinter import filedialog
import os
import pandas as pd
import matplotlib.pyplot as plt
import scipy.signal as signal
from scipy import interpolate
from scipy.optimize import curve_fit
from scipy.interpolate import UnivariateSpline
from scipy import stats
from ipfx import feature_extractor
from ipfx import subthresh_features as subt
from ipfx import feature_vectors as fv
from ipfx.sweep import Sweep
from sklearn.preprocessing import minmax_scale
from pyAPisolation.loadABF import loadABF
import sklearn.preprocessing
import pyabf
import logging
import glob
method='trf'
import autograd.numpy as np
from autograd import grad
def exp_grow(t, a, b, alpha):
return a - b * np.exp(-alpha * t)
def exp_grow_2p(t, a, b1, alphaFast, b2, alphaSlow):
return a - b1 * np.exp(-alphaFast * t) - b2*np.exp(-alphaSlow*t)
def exp_grow_clampfit(t, a, b1, alphaFast, b2, alphaSlow):
return b1 * np.exp(-alphaFast * t) + b2*np.exp(-alphaSlow*t)
f1 = grad(exp_grow_2p) # 1st derivative of f
f2 = grad(f1) # 2nd derivative of f
def curvature(x, a, b1, alphaFast, b2, alphaSlow):
return np.abs(f2(x, a, b1, alphaFast, b2, alphaSlow))*(1 + f1(x, a, b1, alphaFast, b2, alphaSlow)**2)**-1.5
def curvature_real(dy, ddy):
return abs(dy)*(1 + ddy**2)**-1.5
def curvature_splines(x, y=None, error=0.1, smoothing=None):
"""Calculate the signed curvature of a 2D curve at each point
using interpolating splines.
Parameters
----------
x,y: numpy.array(dtype=float) shape (n_points, )
or
y=None and
x is a numpy.array(dtype=complex) shape (n_points, )
In the second case the curve is represented as a np.array
of complex numbers.
error : float
The admisible error when interpolating the splines
Returns
-------
curvature: numpy.array shape (n_points, )
Note: This is 2-3x slower (1.8 ms for 2000 points) than `curvature_gradient`
but more accurate, especially at the borders.
"""
# handle list of complex case
if y is None:
x, y = x.real, x.imag
t = np.arange(x.shape[0])
std = error * np.ones_like(x)
fx = UnivariateSpline(t, x, k=4, w=1 / np.sqrt(std), s=smoothing)
fy = UnivariateSpline(t, y, k=4, w=1 / np.sqrt(std), s=smoothing)
xˈ = fx.derivative(1)(t)
xˈˈ = fx.derivative(2)(t)
yˈ = fy.derivative(1)(t)
yˈˈ = fy.derivative(2)(t)
curvature = (xˈ* yˈˈ - yˈ* xˈˈ) / np.power(xˈ** 2 + yˈ** 2, 3 / 2)
return curvature
def derivative(x,y):
return np.diff(y)/np.diff(x)
def curve_detrend(x,y, curve2):
test = curvature_splines(x, signal.savgol_filter(y, 51, 1), error=1, smoothing=25)
cy = np.array([curvature(xi, *curve2) for xi in x])
#detrend using first and last point
lin_res = stats.linregress([x[0], x[-1]], [cy[0], cy[-1]])
trend = x*lin_res.slope + lin_res.intercept
#plt.plot(x,trend)
detrended_data = cy - trend
return detrended_data
def exp_growth_factor(dataT,dataV,dataI, end_index=300):
#try:
diff_I = np.diff(dataI)
upwardinfl = np.argmax(diff_I)
#Compute out -50 ms from threshold
dt = dataT[1] - dataT[0]
offset = 0.01/ dt
end_index = int(end_index - offset)
upperC = np.amax(dataV[upwardinfl:end_index])
lowerC = np.amin(dataV[upwardinfl:end_index])
diffC = np.abs(lowerC - upperC)
t1 = dataT[upwardinfl:end_index] - dataT[upwardinfl]
curve = curve_fit(exp_grow, t1, dataV[upwardinfl:end_index], maxfev=50000, bounds=([-np.inf, -np.inf, -np.inf], [np.inf, np.inf, np.inf]))[0]
curve = np.hstack((curve, np.full(2,1)))
curve2 = curve_fit(exp_grow_2p, t1, dataV[upwardinfl:end_index], maxfev=50000,method='trf', p0=curve, bounds=([upperC-5, 0, 10, 0, -np.inf], [upperC+5, diffC, np.inf, diffC,np.inf]), xtol=None, gtol=None, ftol=1e-12, jac='3-point')[0]
tau = curve[2]
tau1 = 1/curve2[2]
tau2 = 1/curve2[4]
tau_idx = [2, 4]
fast = tau_idx[np.argmin([tau1, tau2])]
slow = tau_idx[np.argmax([tau1, tau2])]
curve_out = [curve2[0], curve2[fast-1], curve2[fast], curve2[slow-1], curve2[slow]]
#plt.subplot(1,2,1)
plt.plot(t1, dataV[upwardinfl:end_index], c='k', alpha=0.5)
plt.plot(t1, exp_grow_2p(t1, *curve2), label=f'2 phase fit', c='r', alpha=0.5)
plt.plot(t1, exp_grow(t1, *curve_out[:3]), label=f'Fast phase', c='g', alpha=0.5)
plt.plot(t1, exp_grow(t1, curve_out[0], *curve_out[3:]), label=f'slow phase', c='b', alpha=0.5)
plt.title(f" CELL will tau1 {1/curve2[fast]} and tau2 {1/curve2[slow]}")
#plt.subplot(1,2,2)
plt.legend()
#plt.twinx()
#plt.subplot(1,2,2)
dy = curve_detrend(t1, dataV[upwardinfl:end_index], curve2)
#signal.savgol_filter(nt1p.diff(dataV[upwardinfl:end_index])/np.diff(t1), 71, 2, mode='mirror')
#plt.plot(t1,dy)
curve_out = [curve2[0], curve2[fast-1], 1/curve2[fast], curve2[slow-1], 1/curve2[slow]]
return curve_out, np.amax(dy)
#except:
return [np.nan, np.nan, np.nan, np.nan, np.nan]
files = glob.glob('C:\\Users\\SMest\\Documents\\clustering-data\\\All IC1s\\*.abf', recursive=True)
cell_type_df = pd.read_csv("C:\\Users\\SMest\\Documents\\clustering-data\\MARM_PVN_IC1\\spike_count_sort_out.csv")
print(cell_type_df.head)
file_names = cell_type_df['filename'].to_numpy()
cell_type_label = cell_type_df['cell_label'].to_numpy()
curves = []
label = []
ids = []
max_curve = []
for i, f in enumerate(files[-20:]):
print(i)
#try:
base = os.path.basename(f)
base = base.split(".")[0]
if base in file_names:
x, y, c = loadABF(f)
temp_curves =[]
#plt.clf()
iterd = 0
for sweepX, sweepY, sweepC in zip(x,y,c):
spikext = feature_extractor.SpikeFeatureExtractor(filter=0, end=1.25)
res = spikext.process(sweepX, sweepY, sweepC)
if res.empty==False and iterd < 3:
iterd += 1
spike_time = res['threshold_index'].to_numpy()[0]
#plt.figure(num=2)
curve, max_dy = exp_growth_factor(sweepX, sweepY, sweepC, spike_time)
max_curve.append(max_dy)
temp_curves.append(curve)
temp_curves = np.vstack(temp_curves)
div = np.ravel((temp_curves[:,2]) / (temp_curves[:,4])).reshape(-1,1)
sum_height= (temp_curves[:,1] + temp_curves[:,3])
ratio = (temp_curves[:,2] / (temp_curves[:,1] / sum_height)) / (temp_curves[:,4] / (temp_curves[:,3] / sum_height))
ratio = np.ravel(ratio).reshape(-1,1)
temp_curves = np.hstack([temp_curves, div, ratio])
print(temp_curves)
meanC = np.nanmean(temp_curves, axis=0)
print(meanC.shape)
curves.append(meanC)
label_idx = np.argwhere(file_names==base)
label.append(cell_type_label[label_idx])
ids.append(base)
plt.savefig(f+".png")
plt.show()
plt.close()
#except:
#print("fail")
curves = np.vstack(curves)
#lab = sklearn.preprocessing.LabelEncoder()
#int_lab = lab.fit_transform(label)
print(curves)
label = np.ravel(label).reshape(-1,1)
div = np.ravel((curves[:,2]) / (curves[:,4])).reshape(-1,1)
print(div)
sum_height= (curves[:,1] + curves[:,3])
ratio = (curves[:,2] / (curves[:,1]/sum_height)) / (curves[:,4] / (curves[:,3]/sum_height))
ratio = np.ravel(ratio).reshape(-1,1)
curves_out = np.hstack([curves, div, ratio, label])
np.savetxt('curves.csv', curves_out, fmt='%.8f', delimiter=',')
np.savetxt('curves_id.csv', ids, fmt='%s', delimiter=',')
print(curves)
means = []
plt.figure(figsize=(10,10))
plt.clf()
for x in np.unique(label).astype(np.int32):
idx = np.argwhere(label[:,0]==x).astype(np.int32)
mcur = curves[idx]
plt.scatter(np.full(len(idx), x), (curves[idx,2]) / (curves[idx,4]), label=label[x])
means.append(np.nanmean((curves[idx,2]) / (curves[idx,4])))
plt.legend()
plt.yscale('log')
#plt.ylim(0,1)
print(means)
1=1
curves = []
label = []
ids = []
for i, f in enumerate(files[:38]):
print(i)
x, y, c = loadABF(f)
d_name = os.path.dirname(f)
base = os.path.basename(f)
ids.append(base)
label.append(d_name)
dfs = []
temp_curves
plt.clf()
for sweepX, sweepY, sweepC in zip(x,y,c):
spikext = feature_extractor.SpikeFeatureExtractor(filter=0)
res = spikext.process(sweepX, sweepY, sweepC)
dfs.append(res)
if res.empty==False:
if len(non_empty_df) > 1:
sweep_to_use = non_empty_df[1]
else:
sweep_to_use = non_empty_df[-1]
non_empty_df = np.nonzero(np.invert([df.empty for df in dfs]))[0]
try:
spike_time = dfs[sweep_to_use]['threshold_index'].to_numpy()[0]
curve = exp_growth_factor(x[sweep_to_use,:], y[sweep_to_use,:], c[sweep_to_use,:], spike_time)
curves.append(curve)
except:
curves.append([np.nan, np.nan, np.nan, np.nan, np.nan])
plt.show()
print(non_empty_df)
| 0.559531 | 0.530662 |
```
cd ..
#source: https://www.kaggle.com/bhaveshsk/getting-started-with-titanic-dataset/data
#data analysis and wrangling
import pandas as pd
import numpy as np
import random as rnd
#data visualization
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
#machine learning packages
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import Perceptron
from sklearn.linear_model import SGDClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn import metrics
train_df = pd.read_csv("./input/train.csv")
test_df = pd.read_csv("./input/test.csv")
df = pd.concat([train_df,test_df], sort=True)
df.head()
from src.preprocessing import add_derived_title
df = add_derived_title(df)
df['Title'] = df['Title'].map({"Mr": 1, "Miss": 2, "Mrs": 3, "Master": 4, "Rare": 5}).fillna(0)
freq_port = df.Embarked.dropna().mode()[0]
df['Embarked'] = df['Embarked'].fillna(freq_port)
# EXERCISE 2: Write a unit test and extract the following implementation into a function:
# df = impute_nans(df, columns)
# 'Fare' column
df['Fare'] = df['Fare'].fillna(df['Fare'].dropna().median())
# 'Age' column
df['Age'] = df['Age'].fillna(df['Age'].dropna().median())
df['Sex'] = df['Sex'].map( {'female': 1, 'male': 0} ).astype(int)
df['AgeBand'] = pd.cut(df['Age'], 5)
df.loc[ df['Age'] <= 16, 'Age'] = 0
df.loc[(df['Age'] > 16) & (df['Age'] <= 32), 'Age'] = 1
df.loc[(df['Age'] > 32) & (df['Age'] <= 48), 'Age'] = 2
df.loc[(df['Age'] > 48) & (df['Age'] <= 64), 'Age'] = 3
df = df.drop(['AgeBand'], axis=1)
# EXERCISE 3: Write a unit test and extract the following implementation into a function: add_is_alone_column(df)
df['FamilySize'] = df['SibSp'] + df['Parch'] + 1
df['IsAlone'] = 0
df.loc[df['FamilySize'] == 1, 'IsAlone'] = 1
# drop unused columns
df = df.drop(['Parch', 'SibSp', 'FamilySize'], axis=1)
df = df.drop(['Ticket', 'Cabin'], axis=1)
df = df.drop(['Name', 'PassengerId'], axis=1)
df['Age*Class'] = df.Age * df.Pclass
df['Embarked'] = df['Embarked'].map( {'S': 0, 'C': 1, 'Q': 2} ).astype(int)
df['FareBand'] = pd.qcut(df['Fare'], 4)
df.loc[ df['Fare'] <= 7.91, 'Fare'] = 0
df.loc[(df['Fare'] > 7.91) & (df['Fare'] <= 14.454), 'Fare'] = 1
df.loc[(df['Fare'] > 14.454) & (df['Fare'] <= 31), 'Fare'] = 2
df.loc[ df['Fare'] > 31, 'Fare'] = 3
df['Fare'] = df['Fare'].astype(int)
df = df.drop(['FareBand'], axis=1)
train_df = df[-df['Survived'].isna()]
test_df = df[df['Survived'].isna()]
test_df = test_df.drop('Survived', axis=1)
X_train = train_df.drop("Survived", axis=1)
Y_train = train_df["Survived"]
X_test = test_df.copy()
# EXERCISE 1: Create a function, train_model(...), to eliminate the duplication in the next few cells.
def train_model(ModelClass, X, Y, **kwargs):
model = ModelClass(**kwargs)
model.fit(X, Y)
acc_score = round(model.score(X, Y) * 100, 2)
return model, acc_score
_, svc_acc = train_model(SVC, X_train, Y_train)
_, knn_acc = train_model(KNeighborsClassifier, X_train, Y_train)
_, gauss_acc = train_model(GaussianNB, X_train, Y_train)
_, percept_acc = train_model(Perceptron, X_train, Y_train)
_, sgd_acc = train_model(SGDClassifier, X_train, Y_train)
_, decision_tree_acc = train_model(DecisionTreeClassifier, X_train, Y_train)
_, random_forest_acc = train_model(RandomForestClassifier, X_train, Y_train, n_estimators=100)
models = pd.DataFrame({
'Model': ['Support Vector Machines', 'KNN',
'Random Forest', 'Naive Bayes', 'Perceptron',
'Stochastic Gradient Decent',
'Decision Tree'],
'Score': [acc_svc, acc_knn,
acc_random_forest, acc_gaussian, acc_perceptron,
acc_sgd, acc_decision_tree]})
models.sort_values(by='Score', ascending=False)
```
|
github_jupyter
|
cd ..
#source: https://www.kaggle.com/bhaveshsk/getting-started-with-titanic-dataset/data
#data analysis and wrangling
import pandas as pd
import numpy as np
import random as rnd
#data visualization
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
#machine learning packages
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import Perceptron
from sklearn.linear_model import SGDClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn import metrics
train_df = pd.read_csv("./input/train.csv")
test_df = pd.read_csv("./input/test.csv")
df = pd.concat([train_df,test_df], sort=True)
df.head()
from src.preprocessing import add_derived_title
df = add_derived_title(df)
df['Title'] = df['Title'].map({"Mr": 1, "Miss": 2, "Mrs": 3, "Master": 4, "Rare": 5}).fillna(0)
freq_port = df.Embarked.dropna().mode()[0]
df['Embarked'] = df['Embarked'].fillna(freq_port)
# EXERCISE 2: Write a unit test and extract the following implementation into a function:
# df = impute_nans(df, columns)
# 'Fare' column
df['Fare'] = df['Fare'].fillna(df['Fare'].dropna().median())
# 'Age' column
df['Age'] = df['Age'].fillna(df['Age'].dropna().median())
df['Sex'] = df['Sex'].map( {'female': 1, 'male': 0} ).astype(int)
df['AgeBand'] = pd.cut(df['Age'], 5)
df.loc[ df['Age'] <= 16, 'Age'] = 0
df.loc[(df['Age'] > 16) & (df['Age'] <= 32), 'Age'] = 1
df.loc[(df['Age'] > 32) & (df['Age'] <= 48), 'Age'] = 2
df.loc[(df['Age'] > 48) & (df['Age'] <= 64), 'Age'] = 3
df = df.drop(['AgeBand'], axis=1)
# EXERCISE 3: Write a unit test and extract the following implementation into a function: add_is_alone_column(df)
df['FamilySize'] = df['SibSp'] + df['Parch'] + 1
df['IsAlone'] = 0
df.loc[df['FamilySize'] == 1, 'IsAlone'] = 1
# drop unused columns
df = df.drop(['Parch', 'SibSp', 'FamilySize'], axis=1)
df = df.drop(['Ticket', 'Cabin'], axis=1)
df = df.drop(['Name', 'PassengerId'], axis=1)
df['Age*Class'] = df.Age * df.Pclass
df['Embarked'] = df['Embarked'].map( {'S': 0, 'C': 1, 'Q': 2} ).astype(int)
df['FareBand'] = pd.qcut(df['Fare'], 4)
df.loc[ df['Fare'] <= 7.91, 'Fare'] = 0
df.loc[(df['Fare'] > 7.91) & (df['Fare'] <= 14.454), 'Fare'] = 1
df.loc[(df['Fare'] > 14.454) & (df['Fare'] <= 31), 'Fare'] = 2
df.loc[ df['Fare'] > 31, 'Fare'] = 3
df['Fare'] = df['Fare'].astype(int)
df = df.drop(['FareBand'], axis=1)
train_df = df[-df['Survived'].isna()]
test_df = df[df['Survived'].isna()]
test_df = test_df.drop('Survived', axis=1)
X_train = train_df.drop("Survived", axis=1)
Y_train = train_df["Survived"]
X_test = test_df.copy()
# EXERCISE 1: Create a function, train_model(...), to eliminate the duplication in the next few cells.
def train_model(ModelClass, X, Y, **kwargs):
model = ModelClass(**kwargs)
model.fit(X, Y)
acc_score = round(model.score(X, Y) * 100, 2)
return model, acc_score
_, svc_acc = train_model(SVC, X_train, Y_train)
_, knn_acc = train_model(KNeighborsClassifier, X_train, Y_train)
_, gauss_acc = train_model(GaussianNB, X_train, Y_train)
_, percept_acc = train_model(Perceptron, X_train, Y_train)
_, sgd_acc = train_model(SGDClassifier, X_train, Y_train)
_, decision_tree_acc = train_model(DecisionTreeClassifier, X_train, Y_train)
_, random_forest_acc = train_model(RandomForestClassifier, X_train, Y_train, n_estimators=100)
models = pd.DataFrame({
'Model': ['Support Vector Machines', 'KNN',
'Random Forest', 'Naive Bayes', 'Perceptron',
'Stochastic Gradient Decent',
'Decision Tree'],
'Score': [acc_svc, acc_knn,
acc_random_forest, acc_gaussian, acc_perceptron,
acc_sgd, acc_decision_tree]})
models.sort_values(by='Score', ascending=False)
| 0.568536 | 0.521471 |
## Follow-Up using `soynlp`
- `soynlp`를 이용, 띄어쓰기 교정 한 후 동일한 모델에 학습시키기 위함
- 띄어쓰기 학습 데이터로 4,030건의 뉴스 기사 22,039 문장을 사용했으나, 리뷰 교정엔 적용하기 어려웠음 (영화 리뷰에서 학습용 데이터 선정 요망)
```
from pprint import pprint
from soyspacing.countbase import RuleDict, CountSpace
import soyspacing
from words_preprocessing import *
from file_io import *
train = load_pickle('../train_data.pkl')
test = load_pickle('../test_data.pkl')
corpus_fname = '../134963_norm.txt'
model = CountSpace()
model.train(corpus_fname)
model_fname = '../model/spacing.model'
model.save_model(model_fname, json_format=False)
model = CountSpace()
model.load_model('../model/spacing.model', json_format=False)
```
### 띄어쓰기 교정 함수 적용
- arguments
- 4개의 parameter
- `force_abs_threshold` : 점수의 절대값이 이 수준 이상이면 최고점이 아니더라도 즉각 태깅
- `nonspace_threshold` : 이 점수 이하일 때만 0으로 태깅
- `space_threshold` : 이 점수 이상일 때만 1로 태깅
- `min_count` : L, C, R 각각의 feature 빈도수가 min_count 이하이면 불확실한 정보로 판단, 띄어쓰기 계산 시 무시
- `verbose`: iteration 마다 띄어쓰기가 어떻게 되고 있는지 확인
rules : 점수와 관계없이 반드시 태깅을 먼저 할 (chars, tags)
```
verbose=False
mc = 10 # min_count
ft = 0.3 # force_abs_threshold
nt =-0.3 # nonspace_threshold
st = 0.3 # space_threshold
N = 24
pprint(train[N][0][0])
sent_corrected, tags = model.correct(doc=train[N][0][0],
verbose=verbose,
force_abs_threshold=ft,
nonspace_threshold=nt,
space_threshold=st,
min_count=mc)
pprint(sent_corrected)
%%time
train_spaced = [(model.correct(row[0][0],
verbose=verbose,
force_abs_threshold=ft,
nonspace_threshold=nt,
space_threshold=st,
min_count=mc)[0],
row[1]) for row in train]
test_spaced = [(model.correct(row[0][0],
verbose=verbose,
force_abs_threshold=ft,
nonspace_threshold=nt,
space_threshold=st,
min_count=mc)[0],
row[1]) for row in test]
pprint(train_spaced[20])
pprint(test_spaced[20])
from soynlp.word import WordExtractor
word_extractor = WordExtractor(min_count=10,
min_cohesion_forward=0.05,
min_right_branching_entropy=0.0)
sentences_spaced = [row[0] for row in train_spaced]
word_extractor.train(sentences_spaced) # list of str or like
words = word_extractor.extract()
from soynlp.tokenizer import MaxScoreTokenizer
scores = {items[0]: items[1][0] for items in list(words.items())}
tokenizer = MaxScoreTokenizer(scores=scores)
train_tokenized = [(tokenizer.tokenize(row[0][0]), row[1]) for row in train]
test_tokenized = [(tokenizer.tokenize(row[0][0]), row[1]) for row in test]
train_tokenized[N]
save_pickle('../train_space_tokenized.pkl' , train_tokenized)
save_pickle('../test_space_tokenized.pkl' , test_tokenized)
save_pickle('../train_space_corrected.pkl', train_spaced)
save_pickle('../test_space_corrected.pkl', test_spaced)
```
|
github_jupyter
|
from pprint import pprint
from soyspacing.countbase import RuleDict, CountSpace
import soyspacing
from words_preprocessing import *
from file_io import *
train = load_pickle('../train_data.pkl')
test = load_pickle('../test_data.pkl')
corpus_fname = '../134963_norm.txt'
model = CountSpace()
model.train(corpus_fname)
model_fname = '../model/spacing.model'
model.save_model(model_fname, json_format=False)
model = CountSpace()
model.load_model('../model/spacing.model', json_format=False)
verbose=False
mc = 10 # min_count
ft = 0.3 # force_abs_threshold
nt =-0.3 # nonspace_threshold
st = 0.3 # space_threshold
N = 24
pprint(train[N][0][0])
sent_corrected, tags = model.correct(doc=train[N][0][0],
verbose=verbose,
force_abs_threshold=ft,
nonspace_threshold=nt,
space_threshold=st,
min_count=mc)
pprint(sent_corrected)
%%time
train_spaced = [(model.correct(row[0][0],
verbose=verbose,
force_abs_threshold=ft,
nonspace_threshold=nt,
space_threshold=st,
min_count=mc)[0],
row[1]) for row in train]
test_spaced = [(model.correct(row[0][0],
verbose=verbose,
force_abs_threshold=ft,
nonspace_threshold=nt,
space_threshold=st,
min_count=mc)[0],
row[1]) for row in test]
pprint(train_spaced[20])
pprint(test_spaced[20])
from soynlp.word import WordExtractor
word_extractor = WordExtractor(min_count=10,
min_cohesion_forward=0.05,
min_right_branching_entropy=0.0)
sentences_spaced = [row[0] for row in train_spaced]
word_extractor.train(sentences_spaced) # list of str or like
words = word_extractor.extract()
from soynlp.tokenizer import MaxScoreTokenizer
scores = {items[0]: items[1][0] for items in list(words.items())}
tokenizer = MaxScoreTokenizer(scores=scores)
train_tokenized = [(tokenizer.tokenize(row[0][0]), row[1]) for row in train]
test_tokenized = [(tokenizer.tokenize(row[0][0]), row[1]) for row in test]
train_tokenized[N]
save_pickle('../train_space_tokenized.pkl' , train_tokenized)
save_pickle('../test_space_tokenized.pkl' , test_tokenized)
save_pickle('../train_space_corrected.pkl', train_spaced)
save_pickle('../test_space_corrected.pkl', test_spaced)
| 0.206894 | 0.863392 |
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from pandas import Series, DataFrame
import numpy.random as rnd
import os
from datetime import datetime
aadata = pd.read_csv('data/AviationData.txt',
delimiter='|',
skiprows=1,
names=['id', 'type', 'number', 'date',
'location', 'country', 'lat', 'long', 'airport_code',
'airport_name', 'injury_severity', 'aircraft_damage',
'aircraft_cat', 'reg_no', 'make', 'model',
'amateur_built', 'no_engines', 'engine_type', 'FAR_desc',
'schedule', 'purpose', 'air_carrier', 'fatal',
'serious', 'minor', 'uninjured',
'weather', 'broad_phase', 'report_status',
'pub_date', 'none'])
aadata.columns
selection = aadata['date'] != ' '
#2space
aadata = aadata[selection]
aadata['datetime'] = [datetime.strptime(x, ' %m/%d/%Y ') for x in aadata['date']]
aadata['month'] = [int(x.month) for x in aadata['datetime']]
aadata['year'] = [int(x.year) for x in aadata['datetime']]
def decyear(date):
start = datetime(year=date.year, month=1, day=1)
end = datetime(year=date.year+1, month=1, day=1)
decimal = (date - start)/(end - start)
return date.year + decimal
aadata['decyear'] = aadata['datetime'].apply(decyear)
cols = ['lat', 'long',
'fatal',
'serious',
'minor',
'uninjured']
aadata[cols] = aadata[cols].applymap(
lambda x: np.nan if isinstance(x, str) and x.isspace() else float(x))
plt.figure(figsize=(9, 4.5))
plt.step(aadata['decyear'], aadata['fatal'],
lw=1.75, where='mid', alpha=0.5, label='Fatal')
plt.step(aadata['decyear'], aadata['minor']+200,
lw=1.75, where='mid', label='Minor')
plt.step(aadata['decyear'], aadata['serious']+2000*2,
lw=1.75, where='mid', label='Serious')
plt.xticks(rotation=45)
plt.legend(loc=(0.01, .4), fontsize=15)
plt.ylim(-10, 600)
plt.grid(axis='y')
plt.title('Accident injuries {0}-{1}'.format(aadata['year'].min(), aadata['year'].max()))
plt.text(0.15, 0.92, 'source: NTSB', size=12, transform=plt.gca().transAxes, ha='right')
plt.yticks(np.arange(0, 600, 100), [0, 100, 0, 100, 0, 100])
plt.xlablel('Year')
plt.ylabel('No injuries recorded')
plt.xlim((aadata['decyear'].min()-0.5, aadata['decyear'].max()+0.5))
plt.figure(figsize=(9,3))
plt.subplot(121)
year_selection = (aadata['year']>=1975) & (aadata['year']<=2016)
plt.hist(aadata[year_selection]['year'].values, bins=np.arange(1975, 2016+2, 1), align='mid')
plt.xlabel('Year')
plt.xticks(rotation=45)
plt.ylabel('Accident recorded')
plt.subplot(122)
year_selection = (aadata['year'] >=1976) & (aadata['year']<=1986)
plt.hist(aadata[year_selection]['year'].values, bins=np.arange(1976, 1986+2, 1), align='mid')
plt.xlabel('Year')
plt.xticks(rotation=45)
aadata[aadata['year']<=1981]
aadata = aadata[aadata['year']>1981]
plt.figure(figsize=(9,4.5))
plt.step(aadata['decyear'], aadata['fatal'],
lw=1.75, where='mid', alpha=0.5, label='Fatal')
plt.step(aadata['decyear'], aadata['minor']+200,
lw=1.75, where='mid', label='Minor')
plt.step(aadata['decyear'], aadata['serious']+200*2,
lw=1.75, where='mid', label='Serious')
plt.xticks(rotation=45)
plt.legend(loc=(0.8, 0.74), fontsize=15)
plt.ylim(-10, 600)
plt.grid(axis='y')
plt.title('Accidents {0}-{1}'.format(aadata['year'].min(), aadata['year'].max()))
plt.text(0.16, 0.92, 'source: NTSB', size=12, transform=plt.gca().transAxes, ha='right')
plt.yticks(np.arange(0, 600, 100), [0, 100, 0, 100, 0, 100])
plt.xlabel('Year')
plt.ylabel('No injuries recorded')
plt.xlim((aadata['decyear'].min()-0.5, aadata['decyear'].max()+0.5))
bins = np.arange(aadata.year.min(), aadata.year.max()+1, 1)
yearly_dig = aadata.groupby(np.digitize(aadata.year, bins))
yearly_dig.mean().head()
np.floor(yearly_dig['year'].mean()).values
def plot_trend(groups, fields=['Fatal'], which='year', what='max'):
fig, ax = plt.subplots(1,1, figsize=(9, 3.5))
x = np.floor(groups.mean()[which.lower()]).values
width = 0.9
colors = ['LightSalmon', 'SteelBlue', 'Green']
bottom = np.zeros(len(groups.max()[fields[0].lower()]))
for i in range(len(fields)):
if what == 'max':
ax.bar(x, groups.max()[fields[int(i)].lower()],
width, color=colors[int(i)], label=fields[int(i)],
align='center', bottom=bottom, zorder=4)
bottom += groups.max()[fields[int(i)].lower()].values
elif what == 'mean':
ax.bar(x, groups.mean()[fields[int(i)].lower()],
align='center', bottom=bottom, zorder=4)
bottom += groups.mean([fileds[int(i)].lower()].values)
ax.legend(loc=2, ncol=2, frameon=False)
ax.grid(b=True, which='major', axis='y', color='0.65', linestyle='-', zorder=-1)
ax.yaxis.set_ticks_position('left')
ax.xaxis.set_ticks_position('bottom')
for tic1, tic2 in zip(ax.xaxis.get_major_ticks(), ax.yaxis.get_major_ticks()):
tic1.tick10n = tic1.tick20n = False
tic2.tick10n = tic2.tick20n = False
for spine in ['left', 'right', 'top', 'bottom']:
ax.spines[spine].set_color('w')
xticks = np.arange(x.min(), x.max()+1, 1)
ax.set_xticks(xticks)
ax.set_xticklabels([str(int(x)) for x in xticks])
fig.autofmt_xdate(rotation=90, ha='center')
ax.set_xlim((xticks.min()-1.5, xticks.max()+0.5))
ax.set_ylim(0, bottom.max()*1.15)
if what == 'max':
ax.set_title('Plane accidents maximu injuries')
ax.set_ylabel('Max value')
elif waht == 'mean':
ax.set_title('Plane accdients mean injuries')
ax.set_ylabel('Mean value')
ax.set_xlabel(str(which))
return ax
ax = plot_trend(yearly_dig, fields=['Fatal', 'Serious', 'Minor'], which='year')
import pymc
from pymc import Matplot as mcplt
x = np.floor(yearly_dig.mean()['year']).values
y = yearly_dig.max()['fatal'].values
def model_fatalities(y=y):
s = pymc.DiscreteUniform('s', lower=5, upper=18, value=14)
e = pymc.Exponential('e', beta=1.)
l = pymc.Exponential('l', beta=1.)
@pymc.deterministic(plot=False)
def m(s=s, e=e, l=l):
meanval = np.empty(len(y))
meanval[:s] = e
meanval[s:] = l
return meanval
D = pymc.Poisson('D', mu=m, value=y, observed=True)
return locals()
np.random.seed(1234)
MDL = pymc.MCMC(model_fatalities(y=y))
MDL.sample(5e4, 5e3, 2)
MDL.step_method_dict
early = MDL.stats()['e']['mean']
earlyerr = MDL.stats()['e']['standard deviation']
late = MDL.stats()['l']['mean']
lateerr = MDL.stats()['l']['standard deviation']
spt = MDL.stats()['s']['mean']
spterr = MDL.stats()['s']['standard deviation']
mcplt.plot(MDL)
mcplt.autocorrelation(MDL.l)
s = int(np.floor(spt))
print(spt, spterr, x[s])
ax = plot_trend(yearly_dig, fields=['Fatal'], which='Year')
ax.plot([x[0]-1.5, x[s]], [early, early], 'k', lw=2, zorder=5)
ax.fill_between([x[0]-1.5, x[s]], [early-3*earlyerr, early-3*earlyerr], [early+3*earlyerr, early+3*earlyerr], color='0.3', alpha=0.5, zorder=5)
ax.plot([x[s], x[-1]+0.5], [late, late], 'k', lw=2, zorder=5)
ax.fill_between([x[s], x[-1]+0.5],
[late-3*lateerr, late-3*lateerr],
[late+3*lateerr, late+3*lateerr],
color ='0.3', alpha=0.5, zorder=5)
ax.axvline(int(x[s]), color='0.4', dashes=(3,3), lw=2)
bbox_args = dict(boxstyle='round', fc='w', alpha=0.85)
ax.annotate('{0:.1f}$\pm${1:.1f}'.format(early, earlyerr),
xy = (x[s]-1, early),
bbox = bbox_args, ha='right', va='center', zorder=5)
ax.annotate('{0:.1f}$\pm${1:.1f}'.format(late, lateerr),
xy = (x[s]+1, late),
bbox = bbox_args, ha='right', va='center', zorder=5)
ax.annotate('{0}'.format(int(x[s])), xy = (int(x[s]), 300), bbox = bbox_args, ha='center', va='center', zorder=5)
bins = np.arange(1, 12+1, 1)
monthly_dig = aadata.groupby(np.digitize(aadata.month, bins))
monthly_dig.mean().head()
ax = plot_trend(monthly_dig, fields=['Fatal', 'Serious', 'Minor'], which='Month')
ax.set_xlim(0.5, 12.5)
lats, lons = aadata['lat'].values, aadata['long'].values
import cartopy.crs as ccrs
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
import matplotlib.ticker as mticker
fig = plt.figure(figsize=(12, 10))
ax = fig.add_axes([0,0,1,1], projection=ccrs.PlateCarree())
ax.stock_img()
ax.scatter(aadata['long'], aadata['lat'], color='IndianRed', s=aadata['fatal']*2, transform=ccrs.PlateCarree())
gl = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels = True, linewidth=2, color='gray', alpha=0.5, linestyle='--')
gl.xlabels_top = False
gl.ylabels_right = False
gl.xlocator = mticker.FixedLocator(np.arange(-180, 180+1, 60))
gl.xformatter = LONGITUDE_FORMATTER
gl.yformatter = LATITUDE_FORMATTER
import os
conda_file_dir = conda.__file__
conda_dir = conda_file_dir.split('lib')[0]
proj_lib = os.path.join(os.path.join(conda_dir, 'share'), 'proj')
os.environ["PROJ_LIB"] = proj_lib
from mpl_toolkits.basemap import Basemap
co2_gr = pd.read_csv('data/co2_gr_gl.txt',
delim_whitespace=True,
skiprows=62,
names=['year', 'rate', 'err'])
co2_now = pd.read_csv('data/co2_annmean_gl.txt',
delim_whitespace=True,
skiprows=57,
names=['year', 'co2', 'err'])
co2_200 = pd.read_csv('data/siple2.013.dat',
delim_whitespace=True,
skiprows=36,
names=['depth', 'year', 'co2'])
co2_1000 = pd.read_csv('data/lawdome.smoothed.yr75.dat',
delim_whitespace=True,
skiprows=22,
names=['year', 'co2'])
co2_200.tail()
co2_200 = co2_200[:-3]
print(co2_200['year'].dtype, co2_1000['co2'].dtype, co2_now['co2'].dtype, co2_gr['rate'].dtype)
co2_200['year'] = pd.to_numeric(co2_200['year'])
co2_200['co2'] = pd.to_numeric(co2_200['co2'])
co2_200['co2'].dtype, co2_200['year'].dtype
fig,axs = plt.subplots(1,2, figsize=(9, 3.5))
ax2 = axs[0]
ax2.errorbar(co2_now['year'], co2_now['co2'],
color='Coral',
ls='None',
elinewidth=1,
capthick=1.5,
marker='.',
ms=6)
ax2.plot(co2_1000['year'], co2_1000['co2'],
color='Green',
ls='None',
marker='s', mew=0,
ms=5)
ax2.plot(co2_200['year'], co2_200['co2'],
color='0.3',
ls='None',
marker='x', mew=2,
ms=8)
ax2.legend(['Recent', 'LAW ice core', 'SIPLE ice core'], fontsize=15, loc=2)
ax2.axvline(1800, lw=2, color='Gray', dashes=(6,5))
ax2.axvline(co2_gr['year'][0], lw=2, color='SteelBlue', dashes=(6,5))
print(co2_gr['year'][0])
labels = ax2.get_xticklabels()
plt.setp(labels, rotation=33, ha='right')
ax2.set_ylabel('CO$_2$ (ppm)')
ax2.set_xlabel('Year')
ax2.set_title('Past CO$_2$')
ax1 = axs[1]
ax1.errorbar(co2_gr['year'], co2_gr['rate'],
yerr=co2_gr['err'],
color='SteelBlue',
ls='None',
elinewidth=1.5,
capthick=1.5,
marker='.',
ms=8)
labels = ax1.get_xticklabels()
plt.setp(labels, rotation=33, ha='right')
ax1.set_ylabel('CO$_2$ growth (ppm/yr)')
ax1.set_xlabel('Year')
ax1.set_xlim((1957, 2016))
ax1.set_title('Growth rate since 1960')
_ = plt.hist(co2_gr['err'], bins=20)
plt.xlabel('Uncertainty')
plt.ylabel('Count')
x = co2_gr['year'].values
y = co2_gr['rate'].values
y_error = co2_gr['err'].values
def model(x, y):
slope = pymc.Normal('slope', 0.1, 1.)
intercept = pymc.Normal('intercept', -50., 10.)
@pymc.deterministic(plot=False)
def linear(x=x, slope=slope, intercept=intercept):
return x * slope + intercept
f = pymc.Normal('f', mu=linear, tau=1.0/y_error, value=y, observed=True)
return locals()
MDL = pymc.MCMC(model(x,y))
MDL.sample(5e5, 5e4, 100)
y_min = MDL.stats()['linear']['quantiles'][2.5]
y_max = MDL.stats()['linear']['quantiles'][97.5]
y_fit = MDL.stats()['linear']['mean']
slope = MDL.stats()['slope']['mean']
slope_err = MDL.stats()['slope']['standard deviation']
intercept = MDL.stats()['intercept']['mean']
intercept_err = MDL.stats()['intercept']['standard deviation']
mcplt.plot(MDL)
import statsmodels.formula.api as smf
from statsmodels.sandbox.regression.predstd import wls_prediction_std
ols_results = smf.ols("rate ~ year", co2_gr).fit()
prstd, iv_l , iv_u = wls_prediction_std(ols_results)
ols_params = np.flipud(ols_results.params)
ols_err = np.flipud(np.diag(ols_results.cov_params())**.5)
print ('OLS: slope:{0:.3f}, intercept:{1:.2f}'.format(*ols_params))
print('Bay: slope:{0:.3f}, intercept:{1:.2f}'.format(slope, intercept))
ols_results.conf_int(alpha=0.05)
MDL.stats(['intercept', 'slope'])
plt.figure(figsize=(8,4))
plt.title('Growth rate since 1960')
plt.errorbar(x, y, yerr=y_error,
color='SteelBlue', ls='None',
elinewidth=1.5, capthick=1.5,
marker='.', ms=8,
label='Observed')
plt.xlabel('Year')
plt.ylabel('CO$_2$ growth rate (ppm/yr)')
plt.plot(x, y_fit, 'k', lw=2, label='pymc')
plt.fill_between(x, y_min, y_max,
color='0.5', alpha=0.5,
label='Uncertainty')
plt.plot([x.min(), x.max()], [ols_results.fittedvalues.min(), ols_results.fittedvalues.max()],
'r', dashes=(13,2), lw=1.5, label='OLS', zorder=32)
plt.legend(loc=2, numpoints=1, fontsize=12)
```
|
github_jupyter
|
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from pandas import Series, DataFrame
import numpy.random as rnd
import os
from datetime import datetime
aadata = pd.read_csv('data/AviationData.txt',
delimiter='|',
skiprows=1,
names=['id', 'type', 'number', 'date',
'location', 'country', 'lat', 'long', 'airport_code',
'airport_name', 'injury_severity', 'aircraft_damage',
'aircraft_cat', 'reg_no', 'make', 'model',
'amateur_built', 'no_engines', 'engine_type', 'FAR_desc',
'schedule', 'purpose', 'air_carrier', 'fatal',
'serious', 'minor', 'uninjured',
'weather', 'broad_phase', 'report_status',
'pub_date', 'none'])
aadata.columns
selection = aadata['date'] != ' '
#2space
aadata = aadata[selection]
aadata['datetime'] = [datetime.strptime(x, ' %m/%d/%Y ') for x in aadata['date']]
aadata['month'] = [int(x.month) for x in aadata['datetime']]
aadata['year'] = [int(x.year) for x in aadata['datetime']]
def decyear(date):
start = datetime(year=date.year, month=1, day=1)
end = datetime(year=date.year+1, month=1, day=1)
decimal = (date - start)/(end - start)
return date.year + decimal
aadata['decyear'] = aadata['datetime'].apply(decyear)
cols = ['lat', 'long',
'fatal',
'serious',
'minor',
'uninjured']
aadata[cols] = aadata[cols].applymap(
lambda x: np.nan if isinstance(x, str) and x.isspace() else float(x))
plt.figure(figsize=(9, 4.5))
plt.step(aadata['decyear'], aadata['fatal'],
lw=1.75, where='mid', alpha=0.5, label='Fatal')
plt.step(aadata['decyear'], aadata['minor']+200,
lw=1.75, where='mid', label='Minor')
plt.step(aadata['decyear'], aadata['serious']+2000*2,
lw=1.75, where='mid', label='Serious')
plt.xticks(rotation=45)
plt.legend(loc=(0.01, .4), fontsize=15)
plt.ylim(-10, 600)
plt.grid(axis='y')
plt.title('Accident injuries {0}-{1}'.format(aadata['year'].min(), aadata['year'].max()))
plt.text(0.15, 0.92, 'source: NTSB', size=12, transform=plt.gca().transAxes, ha='right')
plt.yticks(np.arange(0, 600, 100), [0, 100, 0, 100, 0, 100])
plt.xlablel('Year')
plt.ylabel('No injuries recorded')
plt.xlim((aadata['decyear'].min()-0.5, aadata['decyear'].max()+0.5))
plt.figure(figsize=(9,3))
plt.subplot(121)
year_selection = (aadata['year']>=1975) & (aadata['year']<=2016)
plt.hist(aadata[year_selection]['year'].values, bins=np.arange(1975, 2016+2, 1), align='mid')
plt.xlabel('Year')
plt.xticks(rotation=45)
plt.ylabel('Accident recorded')
plt.subplot(122)
year_selection = (aadata['year'] >=1976) & (aadata['year']<=1986)
plt.hist(aadata[year_selection]['year'].values, bins=np.arange(1976, 1986+2, 1), align='mid')
plt.xlabel('Year')
plt.xticks(rotation=45)
aadata[aadata['year']<=1981]
aadata = aadata[aadata['year']>1981]
plt.figure(figsize=(9,4.5))
plt.step(aadata['decyear'], aadata['fatal'],
lw=1.75, where='mid', alpha=0.5, label='Fatal')
plt.step(aadata['decyear'], aadata['minor']+200,
lw=1.75, where='mid', label='Minor')
plt.step(aadata['decyear'], aadata['serious']+200*2,
lw=1.75, where='mid', label='Serious')
plt.xticks(rotation=45)
plt.legend(loc=(0.8, 0.74), fontsize=15)
plt.ylim(-10, 600)
plt.grid(axis='y')
plt.title('Accidents {0}-{1}'.format(aadata['year'].min(), aadata['year'].max()))
plt.text(0.16, 0.92, 'source: NTSB', size=12, transform=plt.gca().transAxes, ha='right')
plt.yticks(np.arange(0, 600, 100), [0, 100, 0, 100, 0, 100])
plt.xlabel('Year')
plt.ylabel('No injuries recorded')
plt.xlim((aadata['decyear'].min()-0.5, aadata['decyear'].max()+0.5))
bins = np.arange(aadata.year.min(), aadata.year.max()+1, 1)
yearly_dig = aadata.groupby(np.digitize(aadata.year, bins))
yearly_dig.mean().head()
np.floor(yearly_dig['year'].mean()).values
def plot_trend(groups, fields=['Fatal'], which='year', what='max'):
fig, ax = plt.subplots(1,1, figsize=(9, 3.5))
x = np.floor(groups.mean()[which.lower()]).values
width = 0.9
colors = ['LightSalmon', 'SteelBlue', 'Green']
bottom = np.zeros(len(groups.max()[fields[0].lower()]))
for i in range(len(fields)):
if what == 'max':
ax.bar(x, groups.max()[fields[int(i)].lower()],
width, color=colors[int(i)], label=fields[int(i)],
align='center', bottom=bottom, zorder=4)
bottom += groups.max()[fields[int(i)].lower()].values
elif what == 'mean':
ax.bar(x, groups.mean()[fields[int(i)].lower()],
align='center', bottom=bottom, zorder=4)
bottom += groups.mean([fileds[int(i)].lower()].values)
ax.legend(loc=2, ncol=2, frameon=False)
ax.grid(b=True, which='major', axis='y', color='0.65', linestyle='-', zorder=-1)
ax.yaxis.set_ticks_position('left')
ax.xaxis.set_ticks_position('bottom')
for tic1, tic2 in zip(ax.xaxis.get_major_ticks(), ax.yaxis.get_major_ticks()):
tic1.tick10n = tic1.tick20n = False
tic2.tick10n = tic2.tick20n = False
for spine in ['left', 'right', 'top', 'bottom']:
ax.spines[spine].set_color('w')
xticks = np.arange(x.min(), x.max()+1, 1)
ax.set_xticks(xticks)
ax.set_xticklabels([str(int(x)) for x in xticks])
fig.autofmt_xdate(rotation=90, ha='center')
ax.set_xlim((xticks.min()-1.5, xticks.max()+0.5))
ax.set_ylim(0, bottom.max()*1.15)
if what == 'max':
ax.set_title('Plane accidents maximu injuries')
ax.set_ylabel('Max value')
elif waht == 'mean':
ax.set_title('Plane accdients mean injuries')
ax.set_ylabel('Mean value')
ax.set_xlabel(str(which))
return ax
ax = plot_trend(yearly_dig, fields=['Fatal', 'Serious', 'Minor'], which='year')
import pymc
from pymc import Matplot as mcplt
x = np.floor(yearly_dig.mean()['year']).values
y = yearly_dig.max()['fatal'].values
def model_fatalities(y=y):
s = pymc.DiscreteUniform('s', lower=5, upper=18, value=14)
e = pymc.Exponential('e', beta=1.)
l = pymc.Exponential('l', beta=1.)
@pymc.deterministic(plot=False)
def m(s=s, e=e, l=l):
meanval = np.empty(len(y))
meanval[:s] = e
meanval[s:] = l
return meanval
D = pymc.Poisson('D', mu=m, value=y, observed=True)
return locals()
np.random.seed(1234)
MDL = pymc.MCMC(model_fatalities(y=y))
MDL.sample(5e4, 5e3, 2)
MDL.step_method_dict
early = MDL.stats()['e']['mean']
earlyerr = MDL.stats()['e']['standard deviation']
late = MDL.stats()['l']['mean']
lateerr = MDL.stats()['l']['standard deviation']
spt = MDL.stats()['s']['mean']
spterr = MDL.stats()['s']['standard deviation']
mcplt.plot(MDL)
mcplt.autocorrelation(MDL.l)
s = int(np.floor(spt))
print(spt, spterr, x[s])
ax = plot_trend(yearly_dig, fields=['Fatal'], which='Year')
ax.plot([x[0]-1.5, x[s]], [early, early], 'k', lw=2, zorder=5)
ax.fill_between([x[0]-1.5, x[s]], [early-3*earlyerr, early-3*earlyerr], [early+3*earlyerr, early+3*earlyerr], color='0.3', alpha=0.5, zorder=5)
ax.plot([x[s], x[-1]+0.5], [late, late], 'k', lw=2, zorder=5)
ax.fill_between([x[s], x[-1]+0.5],
[late-3*lateerr, late-3*lateerr],
[late+3*lateerr, late+3*lateerr],
color ='0.3', alpha=0.5, zorder=5)
ax.axvline(int(x[s]), color='0.4', dashes=(3,3), lw=2)
bbox_args = dict(boxstyle='round', fc='w', alpha=0.85)
ax.annotate('{0:.1f}$\pm${1:.1f}'.format(early, earlyerr),
xy = (x[s]-1, early),
bbox = bbox_args, ha='right', va='center', zorder=5)
ax.annotate('{0:.1f}$\pm${1:.1f}'.format(late, lateerr),
xy = (x[s]+1, late),
bbox = bbox_args, ha='right', va='center', zorder=5)
ax.annotate('{0}'.format(int(x[s])), xy = (int(x[s]), 300), bbox = bbox_args, ha='center', va='center', zorder=5)
bins = np.arange(1, 12+1, 1)
monthly_dig = aadata.groupby(np.digitize(aadata.month, bins))
monthly_dig.mean().head()
ax = plot_trend(monthly_dig, fields=['Fatal', 'Serious', 'Minor'], which='Month')
ax.set_xlim(0.5, 12.5)
lats, lons = aadata['lat'].values, aadata['long'].values
import cartopy.crs as ccrs
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
import matplotlib.ticker as mticker
fig = plt.figure(figsize=(12, 10))
ax = fig.add_axes([0,0,1,1], projection=ccrs.PlateCarree())
ax.stock_img()
ax.scatter(aadata['long'], aadata['lat'], color='IndianRed', s=aadata['fatal']*2, transform=ccrs.PlateCarree())
gl = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels = True, linewidth=2, color='gray', alpha=0.5, linestyle='--')
gl.xlabels_top = False
gl.ylabels_right = False
gl.xlocator = mticker.FixedLocator(np.arange(-180, 180+1, 60))
gl.xformatter = LONGITUDE_FORMATTER
gl.yformatter = LATITUDE_FORMATTER
import os
conda_file_dir = conda.__file__
conda_dir = conda_file_dir.split('lib')[0]
proj_lib = os.path.join(os.path.join(conda_dir, 'share'), 'proj')
os.environ["PROJ_LIB"] = proj_lib
from mpl_toolkits.basemap import Basemap
co2_gr = pd.read_csv('data/co2_gr_gl.txt',
delim_whitespace=True,
skiprows=62,
names=['year', 'rate', 'err'])
co2_now = pd.read_csv('data/co2_annmean_gl.txt',
delim_whitespace=True,
skiprows=57,
names=['year', 'co2', 'err'])
co2_200 = pd.read_csv('data/siple2.013.dat',
delim_whitespace=True,
skiprows=36,
names=['depth', 'year', 'co2'])
co2_1000 = pd.read_csv('data/lawdome.smoothed.yr75.dat',
delim_whitespace=True,
skiprows=22,
names=['year', 'co2'])
co2_200.tail()
co2_200 = co2_200[:-3]
print(co2_200['year'].dtype, co2_1000['co2'].dtype, co2_now['co2'].dtype, co2_gr['rate'].dtype)
co2_200['year'] = pd.to_numeric(co2_200['year'])
co2_200['co2'] = pd.to_numeric(co2_200['co2'])
co2_200['co2'].dtype, co2_200['year'].dtype
fig,axs = plt.subplots(1,2, figsize=(9, 3.5))
ax2 = axs[0]
ax2.errorbar(co2_now['year'], co2_now['co2'],
color='Coral',
ls='None',
elinewidth=1,
capthick=1.5,
marker='.',
ms=6)
ax2.plot(co2_1000['year'], co2_1000['co2'],
color='Green',
ls='None',
marker='s', mew=0,
ms=5)
ax2.plot(co2_200['year'], co2_200['co2'],
color='0.3',
ls='None',
marker='x', mew=2,
ms=8)
ax2.legend(['Recent', 'LAW ice core', 'SIPLE ice core'], fontsize=15, loc=2)
ax2.axvline(1800, lw=2, color='Gray', dashes=(6,5))
ax2.axvline(co2_gr['year'][0], lw=2, color='SteelBlue', dashes=(6,5))
print(co2_gr['year'][0])
labels = ax2.get_xticklabels()
plt.setp(labels, rotation=33, ha='right')
ax2.set_ylabel('CO$_2$ (ppm)')
ax2.set_xlabel('Year')
ax2.set_title('Past CO$_2$')
ax1 = axs[1]
ax1.errorbar(co2_gr['year'], co2_gr['rate'],
yerr=co2_gr['err'],
color='SteelBlue',
ls='None',
elinewidth=1.5,
capthick=1.5,
marker='.',
ms=8)
labels = ax1.get_xticklabels()
plt.setp(labels, rotation=33, ha='right')
ax1.set_ylabel('CO$_2$ growth (ppm/yr)')
ax1.set_xlabel('Year')
ax1.set_xlim((1957, 2016))
ax1.set_title('Growth rate since 1960')
_ = plt.hist(co2_gr['err'], bins=20)
plt.xlabel('Uncertainty')
plt.ylabel('Count')
x = co2_gr['year'].values
y = co2_gr['rate'].values
y_error = co2_gr['err'].values
def model(x, y):
slope = pymc.Normal('slope', 0.1, 1.)
intercept = pymc.Normal('intercept', -50., 10.)
@pymc.deterministic(plot=False)
def linear(x=x, slope=slope, intercept=intercept):
return x * slope + intercept
f = pymc.Normal('f', mu=linear, tau=1.0/y_error, value=y, observed=True)
return locals()
MDL = pymc.MCMC(model(x,y))
MDL.sample(5e5, 5e4, 100)
y_min = MDL.stats()['linear']['quantiles'][2.5]
y_max = MDL.stats()['linear']['quantiles'][97.5]
y_fit = MDL.stats()['linear']['mean']
slope = MDL.stats()['slope']['mean']
slope_err = MDL.stats()['slope']['standard deviation']
intercept = MDL.stats()['intercept']['mean']
intercept_err = MDL.stats()['intercept']['standard deviation']
mcplt.plot(MDL)
import statsmodels.formula.api as smf
from statsmodels.sandbox.regression.predstd import wls_prediction_std
ols_results = smf.ols("rate ~ year", co2_gr).fit()
prstd, iv_l , iv_u = wls_prediction_std(ols_results)
ols_params = np.flipud(ols_results.params)
ols_err = np.flipud(np.diag(ols_results.cov_params())**.5)
print ('OLS: slope:{0:.3f}, intercept:{1:.2f}'.format(*ols_params))
print('Bay: slope:{0:.3f}, intercept:{1:.2f}'.format(slope, intercept))
ols_results.conf_int(alpha=0.05)
MDL.stats(['intercept', 'slope'])
plt.figure(figsize=(8,4))
plt.title('Growth rate since 1960')
plt.errorbar(x, y, yerr=y_error,
color='SteelBlue', ls='None',
elinewidth=1.5, capthick=1.5,
marker='.', ms=8,
label='Observed')
plt.xlabel('Year')
plt.ylabel('CO$_2$ growth rate (ppm/yr)')
plt.plot(x, y_fit, 'k', lw=2, label='pymc')
plt.fill_between(x, y_min, y_max,
color='0.5', alpha=0.5,
label='Uncertainty')
plt.plot([x.min(), x.max()], [ols_results.fittedvalues.min(), ols_results.fittedvalues.max()],
'r', dashes=(13,2), lw=1.5, label='OLS', zorder=32)
plt.legend(loc=2, numpoints=1, fontsize=12)
| 0.381911 | 0.587144 |
$$Exercise:1$$
```
import pandas as pd
df = pd.read_csv('cleveland_heart_attr.csv')
```
$$Q:1$$
```
count_rows = len(df.axes[0])
count_cols = len(df.axes[1])
print('No. of rows: ',count_rows, '\nNo. of columns: ',count_cols)
df.info()
```
$$Q:2$$
**Here**, the num_major_vessels_fluroscopy column contains the character '?' in the 167, 193, 288, 303 rows and thal column contains the character '?' in the 88, 267 rows. Since, the above mentioned columns contains characters, that's why they have been considered as 'objects'.
```
df.describe()
df.hist(column='age')
```
$$Q:3$$
Here, the no. of bins = 10
And bin size = (77-29)/10 = 4.8
$$Q:4$$
```
df.hist(column='age',bins=50)
```
Here, Bin size = (77-29)/50 = 0.96
**Observations:** As we increase the no. of bins, the height of the bars decreases because, the sum of age in each bin decreases due to the decrease in bin size.
```
import seaborn as sns
sns.set(rc={'figure.figsize':(13,7)})
#[R] What is the KDE option useful for in histplot()? Explain the details.
#[R] Plot pandas based histogram and seaborn based histogram for serum_cholesterol attribute. Use bin sizes from {default, 20, 50, 100, 200, 500}. For seaborn, use KDE. Report the observations.
```
$$Q-5: $$
A kernel density estimate (KDE) plot is a method for visualizing the distribution of observations in a dataset, analagous to a histogram. KDE represents the data using a continuous probability density curve in one or more dimensions.
$$Q:6$$
```
df.serum_cholesterol.plot.hist(color='blue')
sns.histplot(df.serum_cholesterol,color='red',kde=True)
df.serum_cholesterol.plot.hist(bins=20, color='blue')
sns.histplot(df.serum_cholesterol,bins=20,color='red',kde=True)
df.serum_cholesterol.plot.hist(bins=50, color='blue')
sns.histplot(df.serum_cholesterol,bins=50,color='red',kde=True)
df.serum_cholesterol.plot.hist(bins=100, color='blue')
sns.histplot(df.serum_cholesterol,bins=100,color='red',kde=True)
df.serum_cholesterol.plot.hist(bins=200, color='blue')
sns.histplot(df.serum_cholesterol,bins=200,color='red',kde=True)
df.serum_cholesterol.plot.hist(bins=500, color='blue')
sns.histplot(df.serum_cholesterol,bins=500,color='red',kde=True)
```
Observations:
Since, number of bins are inversenly proportional to bin size. So, increasing the no. of bins reduces the bin size significintly. Although the shape of the graph remains same but the values of the bars decreases.
$$Q:7$$
```
import matplotlib.pyplot as plt
import numpy as np
sns.histplot(df.serum_cholesterol, bins=100)
plt.axvline(x=np.mean(df.serum_cholesterol),color='red',label='mean')
plt.axvline(x=np.median(df.serum_cholesterol),color='magenta',label='median')
plt.axvline(x=np.percentile(df.serum_cholesterol, 25),color='green',label='25 percentile')
plt.axvline(x=np.percentile(df.serum_cholesterol, 75),color='yellow',label='75 percentile')
plt.legend(loc='upper right')
df['gender'] = np.where(df['sex']==1.0,'male','female')
sns.barplot(x="gender",y="serum_cholesterol",data=df)
```
$$Q:8$$
```
sns.barplot(x="gender",y="serum_cholesterol",data=df, order=['female', 'male'])
```
Plotting the bar using median estimator:
```
sns.barplot(x="gender",y="serum_cholesterol",data=df,estimator=np.median)
```
$$Q:9$$
There is slight change in the size and also in the position of the error bar for male in between the bar plot obtained using the median estimator for
gender vs serum cholesterol and the bar plot obtained before.
```
#We can create bar plots with even more fine-grained grouping
#Let us group according to chest_pain_type
sns.barplot(x="gender",y="serum_cholesterol",hue="chest_pain_type",data=df)
```
$$Q:10$$
Males with chest pain type 3 and 4 have the minimum and maximum serum cholesterol respectively.
Females with chest pain type 1 and 4 have the minimum and maximum serum cholesterol respectively.
Females are likely to have more serum cholesterol than males.
$$Q:11$$
```
conditions=[df['chest_pain_type']==1,df['chest_pain_type']==2,df['chest_pain_type']==3]
choice=['typical_angina','atypical_anginae','non_anginal_pain']
df['chest_pain_type_description']=np.select(conditions,choice,default='asymptomatic')
ax = sns.barplot(x="gender",y="serum_cholesterol",hue="chest_pain_type_description",data=df)
ax.legend(loc='lower right', ncol=1)
ax.set_xlabel("Gender",fontsize=15)
ax.set_ylabel("Serum Cholesterol",fontsize=15)
ax.set_xticklabels(ax.get_xticklabels(),fontsize=13)
```
$$Q:12$$
```
plot = sns.barplot(x="gender",y="serum_cholesterol",hue="chest_pain_type",data=df)
for p,line in zip(plot.patches, plot.lines):
plot.annotate(format(p.get_height(),'.1f'),
(p.get_x() + p.get_width() / 2., line.get_ydata()[1]),
ha = 'center', va = 'top',
xytext = (0, 9),
textcoords = 'offset points')
```
$$Q:13$$
```
import matplotlib.pyplot as plt
plot = sns.barplot(x="gender",y="serum_cholesterol",hue="chest_pain_type",data=df)
x=plot.patches[7].get_x() + plot.patches[4].get_width() / 2.
y=plot.lines[1].get_ydata()[1]
plt.annotate('The average serum cholesterol\n for female of chest pain type 4 is the highest!', xy=(x,y), xytext=(x+0.1,y+25.5), arrowprops=dict(facecolor='indigo'))
sns.scatterplot(x="age",y="serum_cholesterol",data=df)
```
$$Q:14$$
**Observations:**
The serum cholesterol level tends to increase as the age increases.
The 40-70 age group have most no. of serum cholesterol data.
The average serum cholesterol value lies in between 200-300.
Only 50-70 age group have samples with serum cholesterol above 400.
```
sns.lineplot(x="age",y="serum_cholesterol",data=df)
```
$$Q:15$$
The light-coloured bands and the dark line indicates the 95% confidence interval and the mean value of the serum cholesterol corresponding to the age respectively.
```
sns.boxplot(x="chest_pain_type",y="serum_cholesterol",data=df)
```
$$Q:16$$
The upper and the lower boundaries of the box of chest pain type and serum cholesterol indicates the 75 and 25 percentile value of the serum cholesterol corresponding to the chest pain type for majority of samples.
The line inside the box indicates the median value of serum cholestero corresponding to the chest pain type.
The points marked beyond
the error bars indicates the outliers.
```
sns.boxplot(x="chest_pain_type",y="serum_cholesterol",hue="gender",data=df)
```
$$Q:17$$
The 75 percentile value of serum cholesterol is greater in female than male in all chest pain type except for the 1st type.
The 25 percentile value of serum cholesterol is lower in male than female in all chest pain type except for the 3rd type. But, lowest exists for male in 3rd type too.
The median value of serum cholesterol is greater in females compared to males in all chest pain type.
Females have some serum cholesterol value greater than 400 for 3rd type of chest pain.
$$Q:18$$
Using violin plot to plot the relationship between chest pain type and serum cholesterol
```
sns.violinplot(x="chest_pain_type",y="serum_cholesterol",data=df)
```
**Observations:**
The no. of outliers is minimum for 1st type of chest pain.
The no. of outliers is maximum for 3rd type of chest pain.
Grouping the violinplots based on gender information
```
sns.violinplot(x="chest_pain_type",y="serum_cholesterol",hue="gender",data=df)
```
Observations:
The serum cholesterol values of females is higher compared to males.
The no. of outliers is minimum for type 1 chest pain in females and maximun for type 3 chest pain in females.
|
github_jupyter
|
import pandas as pd
df = pd.read_csv('cleveland_heart_attr.csv')
count_rows = len(df.axes[0])
count_cols = len(df.axes[1])
print('No. of rows: ',count_rows, '\nNo. of columns: ',count_cols)
df.info()
df.describe()
df.hist(column='age')
df.hist(column='age',bins=50)
import seaborn as sns
sns.set(rc={'figure.figsize':(13,7)})
#[R] What is the KDE option useful for in histplot()? Explain the details.
#[R] Plot pandas based histogram and seaborn based histogram for serum_cholesterol attribute. Use bin sizes from {default, 20, 50, 100, 200, 500}. For seaborn, use KDE. Report the observations.
df.serum_cholesterol.plot.hist(color='blue')
sns.histplot(df.serum_cholesterol,color='red',kde=True)
df.serum_cholesterol.plot.hist(bins=20, color='blue')
sns.histplot(df.serum_cholesterol,bins=20,color='red',kde=True)
df.serum_cholesterol.plot.hist(bins=50, color='blue')
sns.histplot(df.serum_cholesterol,bins=50,color='red',kde=True)
df.serum_cholesterol.plot.hist(bins=100, color='blue')
sns.histplot(df.serum_cholesterol,bins=100,color='red',kde=True)
df.serum_cholesterol.plot.hist(bins=200, color='blue')
sns.histplot(df.serum_cholesterol,bins=200,color='red',kde=True)
df.serum_cholesterol.plot.hist(bins=500, color='blue')
sns.histplot(df.serum_cholesterol,bins=500,color='red',kde=True)
import matplotlib.pyplot as plt
import numpy as np
sns.histplot(df.serum_cholesterol, bins=100)
plt.axvline(x=np.mean(df.serum_cholesterol),color='red',label='mean')
plt.axvline(x=np.median(df.serum_cholesterol),color='magenta',label='median')
plt.axvline(x=np.percentile(df.serum_cholesterol, 25),color='green',label='25 percentile')
plt.axvline(x=np.percentile(df.serum_cholesterol, 75),color='yellow',label='75 percentile')
plt.legend(loc='upper right')
df['gender'] = np.where(df['sex']==1.0,'male','female')
sns.barplot(x="gender",y="serum_cholesterol",data=df)
sns.barplot(x="gender",y="serum_cholesterol",data=df, order=['female', 'male'])
sns.barplot(x="gender",y="serum_cholesterol",data=df,estimator=np.median)
#We can create bar plots with even more fine-grained grouping
#Let us group according to chest_pain_type
sns.barplot(x="gender",y="serum_cholesterol",hue="chest_pain_type",data=df)
conditions=[df['chest_pain_type']==1,df['chest_pain_type']==2,df['chest_pain_type']==3]
choice=['typical_angina','atypical_anginae','non_anginal_pain']
df['chest_pain_type_description']=np.select(conditions,choice,default='asymptomatic')
ax = sns.barplot(x="gender",y="serum_cholesterol",hue="chest_pain_type_description",data=df)
ax.legend(loc='lower right', ncol=1)
ax.set_xlabel("Gender",fontsize=15)
ax.set_ylabel("Serum Cholesterol",fontsize=15)
ax.set_xticklabels(ax.get_xticklabels(),fontsize=13)
plot = sns.barplot(x="gender",y="serum_cholesterol",hue="chest_pain_type",data=df)
for p,line in zip(plot.patches, plot.lines):
plot.annotate(format(p.get_height(),'.1f'),
(p.get_x() + p.get_width() / 2., line.get_ydata()[1]),
ha = 'center', va = 'top',
xytext = (0, 9),
textcoords = 'offset points')
import matplotlib.pyplot as plt
plot = sns.barplot(x="gender",y="serum_cholesterol",hue="chest_pain_type",data=df)
x=plot.patches[7].get_x() + plot.patches[4].get_width() / 2.
y=plot.lines[1].get_ydata()[1]
plt.annotate('The average serum cholesterol\n for female of chest pain type 4 is the highest!', xy=(x,y), xytext=(x+0.1,y+25.5), arrowprops=dict(facecolor='indigo'))
sns.scatterplot(x="age",y="serum_cholesterol",data=df)
sns.lineplot(x="age",y="serum_cholesterol",data=df)
sns.boxplot(x="chest_pain_type",y="serum_cholesterol",data=df)
sns.boxplot(x="chest_pain_type",y="serum_cholesterol",hue="gender",data=df)
sns.violinplot(x="chest_pain_type",y="serum_cholesterol",data=df)
sns.violinplot(x="chest_pain_type",y="serum_cholesterol",hue="gender",data=df)
| 0.558809 | 0.978529 |
# RadarCOVID-Report
## Data Extraction
```
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import pycountry
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
```
### Constants
```
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
```
### Parameters
```
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
environment_enable_multi_backend_download = \
os.environ.get("RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD")
if environment_enable_multi_backend_download:
report_backend_identifiers = None
else:
report_backend_identifiers = [report_backend_identifier]
report_backend_identifiers
environment_invalid_shared_diagnoses_dates = \
os.environ.get("RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES")
if environment_invalid_shared_diagnoses_dates:
invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(",")
else:
invalid_shared_diagnoses_dates = []
invalid_shared_diagnoses_dates
```
### COVID-19 Cases
```
report_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=report_backend_identifier)
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe():
return pd.read_csv("https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv")
confirmed_df_ = download_cases_dataframe()
confirmed_df_.iloc[0]
confirmed_df = confirmed_df_.copy()
confirmed_df = confirmed_df[["date", "new_cases", "iso_code"]]
confirmed_df.rename(
columns={
"date": "sample_date",
"iso_code": "country_code",
},
inplace=True)
def convert_iso_alpha_3_to_alpha_2(x):
try:
return pycountry.countries.get(alpha_3=x).alpha_2
except Exception as e:
logging.info(f"Error converting country ISO Alpha 3 code '{x}': {repr(e)}")
return None
confirmed_df["country_code"] = confirmed_df.country_code.apply(convert_iso_alpha_3_to_alpha_2)
confirmed_df.dropna(inplace=True)
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_days_df["sample_date_string"] = \
confirmed_days_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_days_df.tail()
def sort_source_regions_for_display(source_regions: list) -> list:
if report_backend_identifier in source_regions:
source_regions = [report_backend_identifier] + \
list(sorted(set(source_regions).difference([report_backend_identifier])))
else:
source_regions = list(sorted(source_regions))
return source_regions
report_source_regions = report_backend_client.source_regions_for_date(
date=extraction_datetime.date())
report_source_regions = sort_source_regions_for_display(
source_regions=report_source_regions)
report_source_regions
def get_cases_dataframe(source_regions_for_date_function, columns_suffix=None):
source_regions_at_date_df = confirmed_days_df.copy()
source_regions_at_date_df["source_regions_at_date"] = \
source_regions_at_date_df.sample_date.apply(
lambda x: source_regions_for_date_function(date=x))
source_regions_at_date_df.sort_values("sample_date", inplace=True)
source_regions_at_date_df["_source_regions_group"] = source_regions_at_date_df. \
source_regions_at_date.apply(lambda x: ",".join(sort_source_regions_for_display(x)))
source_regions_at_date_df.tail()
#%%
source_regions_for_summary_df_ = \
source_regions_at_date_df[["sample_date", "_source_regions_group"]].copy()
source_regions_for_summary_df_.rename(columns={"_source_regions_group": "source_regions"}, inplace=True)
source_regions_for_summary_df_.tail()
#%%
confirmed_output_columns = ["sample_date", "new_cases", "covid_cases"]
confirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)
for source_regions_group, source_regions_group_series in \
source_regions_at_date_df.groupby("_source_regions_group"):
source_regions_set = set(source_regions_group.split(","))
confirmed_source_regions_set_df = \
confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()
confirmed_source_regions_group_df = \
confirmed_source_regions_set_df.groupby("sample_date").new_cases.sum() \
.reset_index().sort_values("sample_date")
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df.merge(
confirmed_days_df[["sample_date_string"]].rename(
columns={"sample_date_string": "sample_date"}),
how="right")
confirmed_source_regions_group_df["new_cases"] = \
confirmed_source_regions_group_df["new_cases"].clip(lower=0)
confirmed_source_regions_group_df["covid_cases"] = \
confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[confirmed_output_columns]
confirmed_source_regions_group_df = confirmed_source_regions_group_df.replace(0, np.nan)
confirmed_source_regions_group_df.fillna(method="ffill", inplace=True)
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[
confirmed_source_regions_group_df.sample_date.isin(
source_regions_group_series.sample_date_string)]
confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)
result_df = confirmed_output_df.copy()
result_df.tail()
#%%
result_df.rename(columns={"sample_date": "sample_date_string"}, inplace=True)
result_df = confirmed_days_df[["sample_date_string"]].merge(result_df, how="left")
result_df.sort_values("sample_date_string", inplace=True)
result_df.fillna(method="ffill", inplace=True)
result_df.tail()
#%%
result_df[["new_cases", "covid_cases"]].plot()
if columns_suffix:
result_df.rename(
columns={
"new_cases": "new_cases_" + columns_suffix,
"covid_cases": "covid_cases_" + columns_suffix},
inplace=True)
return result_df, source_regions_for_summary_df_
confirmed_eu_df, source_regions_for_summary_df = get_cases_dataframe(
report_backend_client.source_regions_for_date)
confirmed_es_df, _ = get_cases_dataframe(
lambda date: [spain_region_country_code],
columns_suffix=spain_region_country_code.lower())
```
### Extract API TEKs
```
raw_zip_path_prefix = "Data/TEKs/Raw/"
base_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=base_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
```
### Dump API TEKs
```
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_base_df = tek_list_df[tek_list_df.region == report_backend_identifier]
tek_list_base_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_base_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_base_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_base_df.head()
```
### Load TEK Dumps
```
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
```
### Daily New TEKs
```
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \
.groupby(["upload_date"]).shared_teks.max().reset_index() \
.sort_values(["upload_date"], ascending=False) \
.rename(columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
invalid_shared_diagnoses_dates_mask = \
estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)
estimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0
estimated_shared_diagnoses_df.head()
```
### Hourly New TEKs
```
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
```
### Official Statistics
```
import requests
import pandas.io.json
official_stats_response = requests.get("https://radarcovid.covid19.gob.es/kpi/statistics/basics")
official_stats_response.raise_for_status()
official_stats_df_ = pandas.io.json.json_normalize(official_stats_response.json())
official_stats_df = official_stats_df_.copy()
official_stats_df["date"] = pd.to_datetime(official_stats_df["date"], dayfirst=True)
official_stats_df.head()
official_stats_column_map = {
"date": "sample_date",
"applicationsDownloads.totalAcummulated": "app_downloads_es_accumulated",
"communicatedContagions.totalAcummulated": "shared_diagnoses_es_accumulated",
}
accumulated_suffix = "_accumulated"
accumulated_values_columns = \
list(filter(lambda x: x.endswith(accumulated_suffix), official_stats_column_map.values()))
interpolated_values_columns = \
list(map(lambda x: x[:-len(accumulated_suffix)], accumulated_values_columns))
official_stats_df = \
official_stats_df[official_stats_column_map.keys()] \
.rename(columns=official_stats_column_map)
official_stats_df["extraction_date"] = extraction_date
official_stats_df.head()
official_stats_path = "Data/Statistics/Current/RadarCOVID-Statistics.json"
previous_official_stats_df = pd.read_json(official_stats_path, orient="records", lines=True)
previous_official_stats_df["sample_date"] = pd.to_datetime(previous_official_stats_df["sample_date"], dayfirst=True)
official_stats_df = official_stats_df.append(previous_official_stats_df)
official_stats_df.head()
official_stats_df = official_stats_df[~(official_stats_df.shared_diagnoses_es_accumulated == 0)]
official_stats_df.sort_values("extraction_date", ascending=False, inplace=True)
official_stats_df.drop_duplicates(subset=["sample_date"], keep="first", inplace=True)
official_stats_df.head()
official_stats_stored_df = official_stats_df.copy()
official_stats_stored_df["sample_date"] = official_stats_stored_df.sample_date.dt.strftime("%Y-%m-%d")
official_stats_stored_df.to_json(official_stats_path, orient="records", lines=True)
official_stats_df.drop(columns=["extraction_date"], inplace=True)
official_stats_df = confirmed_days_df.merge(official_stats_df, how="left")
official_stats_df.sort_values("sample_date", ascending=False, inplace=True)
official_stats_df.head()
official_stats_df[accumulated_values_columns] = \
official_stats_df[accumulated_values_columns] \
.astype(float).interpolate(limit_area="inside")
official_stats_df[interpolated_values_columns] = \
official_stats_df[accumulated_values_columns].diff(periods=-1)
official_stats_df.drop(columns="sample_date", inplace=True)
official_stats_df.head()
```
### Data Merge
```
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
official_stats_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_eu_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df = confirmed_es_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df = result_summary_df.merge(source_regions_for_summary_df, how="left")
result_summary_df.set_index(["sample_date", "source_regions"], inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(result_summary_df.shared_diagnoses_es / result_summary_df.covid_cases_es).fillna(0)
result_summary_df.head(daily_plot_days)
def compute_aggregated_results_summary(days) -> pd.DataFrame:
aggregated_result_summary_df = result_summary_df.copy()
aggregated_result_summary_df["covid_cases_for_ratio"] = \
aggregated_result_summary_df.covid_cases.mask(
aggregated_result_summary_df.shared_diagnoses == 0, 0)
aggregated_result_summary_df["covid_cases_for_ratio_es"] = \
aggregated_result_summary_df.covid_cases_es.mask(
aggregated_result_summary_df.shared_diagnoses_es == 0, 0)
aggregated_result_summary_df = aggregated_result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(days).agg({
"covid_cases": "sum",
"covid_cases_es": "sum",
"covid_cases_for_ratio": "sum",
"covid_cases_for_ratio_es": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum",
"shared_diagnoses_es": "sum",
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
aggregated_result_summary_df = aggregated_result_summary_df.fillna(0).astype(int)
aggregated_result_summary_df["teks_per_shared_diagnosis"] = \
(aggregated_result_summary_df.shared_teks_by_upload_date /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case"] = \
(aggregated_result_summary_df.shared_diagnoses /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(aggregated_result_summary_df.shared_diagnoses_es /
aggregated_result_summary_df.covid_cases_for_ratio_es).fillna(0)
return aggregated_result_summary_df
aggregated_result_with_7_days_window_summary_df = compute_aggregated_results_summary(days=7)
aggregated_result_with_7_days_window_summary_df.head()
last_7_days_summary = aggregated_result_with_7_days_window_summary_df.to_dict(orient="records")[1]
last_7_days_summary
aggregated_result_with_14_days_window_summary_df = compute_aggregated_results_summary(days=13)
last_14_days_summary = aggregated_result_with_14_days_window_summary_df.to_dict(orient="records")[1]
last_14_days_summary
```
## Report Results
```
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"source_regions": "Source Countries",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases (Source Countries)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date (Source Countries)",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date (Source Countries)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date (Source Countries)",
"shared_diagnoses": "Shared Diagnoses (Source Countries – Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis (Source Countries)",
"shared_diagnoses_per_covid_case": "Usage Ratio (Source Countries)",
"covid_cases_es": "COVID-19 Cases (Spain)",
"app_downloads_es": "App Downloads (Spain – Official)",
"shared_diagnoses_es": "Shared Diagnoses (Spain – Official)",
"shared_diagnoses_per_covid_case_es": "Usage Ratio (Spain)",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
"covid_cases_es",
"app_downloads_es",
"shared_diagnoses_es",
"shared_diagnoses_per_covid_case_es",
]
summary_percentage_columns= [
"shared_diagnoses_per_covid_case_es",
"shared_diagnoses_per_covid_case",
]
```
### Daily Summary Table
```
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
```
### Daily Summary Plots
```
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.droplevel(level=["source_regions"]) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 30), legend=False)
ax_ = summary_ax_list[0]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
for percentage_column in summary_percentage_columns:
percentage_column_index = summary_columns.index(percentage_column)
summary_ax_list[percentage_column_index].yaxis \
.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
```
### Daily Generation to Upload Period Table
```
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
```
### Hourly Summary Plots
```
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
```
### Publish Results
```
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case_es"]: lambda x: f"{x:.2%}" if x != 0 else "",
}
general_columns = \
list(filter(lambda x: x not in display_formatters, display_column_name_mapping.values()))
general_formatter = lambda x: f"{x}" if x != 0 else ""
display_formatters.update(dict(map(lambda x: (x, general_formatter), general_columns)))
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index.get_level_values("sample_date") == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.item()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.item()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.item()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.item()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.item()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
if len(report_source_regions) == 1:
display_brief_source_regions = report_source_regions[0]
else:
display_brief_source_regions = f"{len(report_source_regions)} 🇪🇺"
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
df = df.copy()
df_styler = df.style.format(display_formatters)
media_path = get_temporary_image_path()
dfi.export(df_styler, media_path)
return media_path
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
```
### Save Results
```
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
```
### Publish Results as JSON
```
def generate_summary_api_results(df: pd.DataFrame) -> list:
api_df = df.reset_index().copy()
api_df["sample_date_string"] = \
api_df["sample_date"].dt.strftime("%Y-%m-%d")
api_df["source_regions"] = \
api_df["source_regions"].apply(lambda x: x.split(","))
return api_df.to_dict(orient="records")
summary_api_results = \
generate_summary_api_results(df=result_summary_df)
today_summary_api_results = \
generate_summary_api_results(df=extraction_date_result_summary_df)[0]
summary_results = dict(
backend_identifier=report_backend_identifier,
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=today_summary_api_results,
last_7_days=last_7_days_summary,
last_14_days=last_14_days_summary,
daily_results=summary_api_results)
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
```
### Publish on README
```
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
```
### Publish on Twitter
```
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
def format_shared_diagnoses_per_covid_case(value) -> str:
if value == 0:
return "–"
return f"≤{value:.2%}"
display_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=shared_diagnoses_per_covid_case)
display_last_14_days_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case"])
display_last_14_days_shared_diagnoses_per_covid_case_es = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case_es"])
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: {display_shared_diagnoses_per_covid_case}
Last 14 Days:
- Usage Ratio (Estimation): {display_last_14_days_shared_diagnoses_per_covid_case}
- Usage Ratio (Official): {display_last_14_days_shared_diagnoses_per_covid_case_es}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
```
|
github_jupyter
|
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import pycountry
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
environment_enable_multi_backend_download = \
os.environ.get("RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD")
if environment_enable_multi_backend_download:
report_backend_identifiers = None
else:
report_backend_identifiers = [report_backend_identifier]
report_backend_identifiers
environment_invalid_shared_diagnoses_dates = \
os.environ.get("RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES")
if environment_invalid_shared_diagnoses_dates:
invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(",")
else:
invalid_shared_diagnoses_dates = []
invalid_shared_diagnoses_dates
report_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=report_backend_identifier)
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe():
return pd.read_csv("https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv")
confirmed_df_ = download_cases_dataframe()
confirmed_df_.iloc[0]
confirmed_df = confirmed_df_.copy()
confirmed_df = confirmed_df[["date", "new_cases", "iso_code"]]
confirmed_df.rename(
columns={
"date": "sample_date",
"iso_code": "country_code",
},
inplace=True)
def convert_iso_alpha_3_to_alpha_2(x):
try:
return pycountry.countries.get(alpha_3=x).alpha_2
except Exception as e:
logging.info(f"Error converting country ISO Alpha 3 code '{x}': {repr(e)}")
return None
confirmed_df["country_code"] = confirmed_df.country_code.apply(convert_iso_alpha_3_to_alpha_2)
confirmed_df.dropna(inplace=True)
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_days_df["sample_date_string"] = \
confirmed_days_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_days_df.tail()
def sort_source_regions_for_display(source_regions: list) -> list:
if report_backend_identifier in source_regions:
source_regions = [report_backend_identifier] + \
list(sorted(set(source_regions).difference([report_backend_identifier])))
else:
source_regions = list(sorted(source_regions))
return source_regions
report_source_regions = report_backend_client.source_regions_for_date(
date=extraction_datetime.date())
report_source_regions = sort_source_regions_for_display(
source_regions=report_source_regions)
report_source_regions
def get_cases_dataframe(source_regions_for_date_function, columns_suffix=None):
source_regions_at_date_df = confirmed_days_df.copy()
source_regions_at_date_df["source_regions_at_date"] = \
source_regions_at_date_df.sample_date.apply(
lambda x: source_regions_for_date_function(date=x))
source_regions_at_date_df.sort_values("sample_date", inplace=True)
source_regions_at_date_df["_source_regions_group"] = source_regions_at_date_df. \
source_regions_at_date.apply(lambda x: ",".join(sort_source_regions_for_display(x)))
source_regions_at_date_df.tail()
#%%
source_regions_for_summary_df_ = \
source_regions_at_date_df[["sample_date", "_source_regions_group"]].copy()
source_regions_for_summary_df_.rename(columns={"_source_regions_group": "source_regions"}, inplace=True)
source_regions_for_summary_df_.tail()
#%%
confirmed_output_columns = ["sample_date", "new_cases", "covid_cases"]
confirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)
for source_regions_group, source_regions_group_series in \
source_regions_at_date_df.groupby("_source_regions_group"):
source_regions_set = set(source_regions_group.split(","))
confirmed_source_regions_set_df = \
confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()
confirmed_source_regions_group_df = \
confirmed_source_regions_set_df.groupby("sample_date").new_cases.sum() \
.reset_index().sort_values("sample_date")
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df.merge(
confirmed_days_df[["sample_date_string"]].rename(
columns={"sample_date_string": "sample_date"}),
how="right")
confirmed_source_regions_group_df["new_cases"] = \
confirmed_source_regions_group_df["new_cases"].clip(lower=0)
confirmed_source_regions_group_df["covid_cases"] = \
confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[confirmed_output_columns]
confirmed_source_regions_group_df = confirmed_source_regions_group_df.replace(0, np.nan)
confirmed_source_regions_group_df.fillna(method="ffill", inplace=True)
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[
confirmed_source_regions_group_df.sample_date.isin(
source_regions_group_series.sample_date_string)]
confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)
result_df = confirmed_output_df.copy()
result_df.tail()
#%%
result_df.rename(columns={"sample_date": "sample_date_string"}, inplace=True)
result_df = confirmed_days_df[["sample_date_string"]].merge(result_df, how="left")
result_df.sort_values("sample_date_string", inplace=True)
result_df.fillna(method="ffill", inplace=True)
result_df.tail()
#%%
result_df[["new_cases", "covid_cases"]].plot()
if columns_suffix:
result_df.rename(
columns={
"new_cases": "new_cases_" + columns_suffix,
"covid_cases": "covid_cases_" + columns_suffix},
inplace=True)
return result_df, source_regions_for_summary_df_
confirmed_eu_df, source_regions_for_summary_df = get_cases_dataframe(
report_backend_client.source_regions_for_date)
confirmed_es_df, _ = get_cases_dataframe(
lambda date: [spain_region_country_code],
columns_suffix=spain_region_country_code.lower())
raw_zip_path_prefix = "Data/TEKs/Raw/"
base_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=base_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_base_df = tek_list_df[tek_list_df.region == report_backend_identifier]
tek_list_base_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_base_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_base_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_base_df.head()
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \
.groupby(["upload_date"]).shared_teks.max().reset_index() \
.sort_values(["upload_date"], ascending=False) \
.rename(columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
invalid_shared_diagnoses_dates_mask = \
estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)
estimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0
estimated_shared_diagnoses_df.head()
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
import requests
import pandas.io.json
official_stats_response = requests.get("https://radarcovid.covid19.gob.es/kpi/statistics/basics")
official_stats_response.raise_for_status()
official_stats_df_ = pandas.io.json.json_normalize(official_stats_response.json())
official_stats_df = official_stats_df_.copy()
official_stats_df["date"] = pd.to_datetime(official_stats_df["date"], dayfirst=True)
official_stats_df.head()
official_stats_column_map = {
"date": "sample_date",
"applicationsDownloads.totalAcummulated": "app_downloads_es_accumulated",
"communicatedContagions.totalAcummulated": "shared_diagnoses_es_accumulated",
}
accumulated_suffix = "_accumulated"
accumulated_values_columns = \
list(filter(lambda x: x.endswith(accumulated_suffix), official_stats_column_map.values()))
interpolated_values_columns = \
list(map(lambda x: x[:-len(accumulated_suffix)], accumulated_values_columns))
official_stats_df = \
official_stats_df[official_stats_column_map.keys()] \
.rename(columns=official_stats_column_map)
official_stats_df["extraction_date"] = extraction_date
official_stats_df.head()
official_stats_path = "Data/Statistics/Current/RadarCOVID-Statistics.json"
previous_official_stats_df = pd.read_json(official_stats_path, orient="records", lines=True)
previous_official_stats_df["sample_date"] = pd.to_datetime(previous_official_stats_df["sample_date"], dayfirst=True)
official_stats_df = official_stats_df.append(previous_official_stats_df)
official_stats_df.head()
official_stats_df = official_stats_df[~(official_stats_df.shared_diagnoses_es_accumulated == 0)]
official_stats_df.sort_values("extraction_date", ascending=False, inplace=True)
official_stats_df.drop_duplicates(subset=["sample_date"], keep="first", inplace=True)
official_stats_df.head()
official_stats_stored_df = official_stats_df.copy()
official_stats_stored_df["sample_date"] = official_stats_stored_df.sample_date.dt.strftime("%Y-%m-%d")
official_stats_stored_df.to_json(official_stats_path, orient="records", lines=True)
official_stats_df.drop(columns=["extraction_date"], inplace=True)
official_stats_df = confirmed_days_df.merge(official_stats_df, how="left")
official_stats_df.sort_values("sample_date", ascending=False, inplace=True)
official_stats_df.head()
official_stats_df[accumulated_values_columns] = \
official_stats_df[accumulated_values_columns] \
.astype(float).interpolate(limit_area="inside")
official_stats_df[interpolated_values_columns] = \
official_stats_df[accumulated_values_columns].diff(periods=-1)
official_stats_df.drop(columns="sample_date", inplace=True)
official_stats_df.head()
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
official_stats_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_eu_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df = confirmed_es_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df = result_summary_df.merge(source_regions_for_summary_df, how="left")
result_summary_df.set_index(["sample_date", "source_regions"], inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(result_summary_df.shared_diagnoses_es / result_summary_df.covid_cases_es).fillna(0)
result_summary_df.head(daily_plot_days)
def compute_aggregated_results_summary(days) -> pd.DataFrame:
aggregated_result_summary_df = result_summary_df.copy()
aggregated_result_summary_df["covid_cases_for_ratio"] = \
aggregated_result_summary_df.covid_cases.mask(
aggregated_result_summary_df.shared_diagnoses == 0, 0)
aggregated_result_summary_df["covid_cases_for_ratio_es"] = \
aggregated_result_summary_df.covid_cases_es.mask(
aggregated_result_summary_df.shared_diagnoses_es == 0, 0)
aggregated_result_summary_df = aggregated_result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(days).agg({
"covid_cases": "sum",
"covid_cases_es": "sum",
"covid_cases_for_ratio": "sum",
"covid_cases_for_ratio_es": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum",
"shared_diagnoses_es": "sum",
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
aggregated_result_summary_df = aggregated_result_summary_df.fillna(0).astype(int)
aggregated_result_summary_df["teks_per_shared_diagnosis"] = \
(aggregated_result_summary_df.shared_teks_by_upload_date /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case"] = \
(aggregated_result_summary_df.shared_diagnoses /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(aggregated_result_summary_df.shared_diagnoses_es /
aggregated_result_summary_df.covid_cases_for_ratio_es).fillna(0)
return aggregated_result_summary_df
aggregated_result_with_7_days_window_summary_df = compute_aggregated_results_summary(days=7)
aggregated_result_with_7_days_window_summary_df.head()
last_7_days_summary = aggregated_result_with_7_days_window_summary_df.to_dict(orient="records")[1]
last_7_days_summary
aggregated_result_with_14_days_window_summary_df = compute_aggregated_results_summary(days=13)
last_14_days_summary = aggregated_result_with_14_days_window_summary_df.to_dict(orient="records")[1]
last_14_days_summary
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"source_regions": "Source Countries",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases (Source Countries)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date (Source Countries)",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date (Source Countries)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date (Source Countries)",
"shared_diagnoses": "Shared Diagnoses (Source Countries – Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis (Source Countries)",
"shared_diagnoses_per_covid_case": "Usage Ratio (Source Countries)",
"covid_cases_es": "COVID-19 Cases (Spain)",
"app_downloads_es": "App Downloads (Spain – Official)",
"shared_diagnoses_es": "Shared Diagnoses (Spain – Official)",
"shared_diagnoses_per_covid_case_es": "Usage Ratio (Spain)",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
"covid_cases_es",
"app_downloads_es",
"shared_diagnoses_es",
"shared_diagnoses_per_covid_case_es",
]
summary_percentage_columns= [
"shared_diagnoses_per_covid_case_es",
"shared_diagnoses_per_covid_case",
]
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.droplevel(level=["source_regions"]) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 30), legend=False)
ax_ = summary_ax_list[0]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
for percentage_column in summary_percentage_columns:
percentage_column_index = summary_columns.index(percentage_column)
summary_ax_list[percentage_column_index].yaxis \
.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case_es"]: lambda x: f"{x:.2%}" if x != 0 else "",
}
general_columns = \
list(filter(lambda x: x not in display_formatters, display_column_name_mapping.values()))
general_formatter = lambda x: f"{x}" if x != 0 else ""
display_formatters.update(dict(map(lambda x: (x, general_formatter), general_columns)))
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index.get_level_values("sample_date") == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.item()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.item()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.item()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.item()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.item()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
if len(report_source_regions) == 1:
display_brief_source_regions = report_source_regions[0]
else:
display_brief_source_regions = f"{len(report_source_regions)} 🇪🇺"
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
df = df.copy()
df_styler = df.style.format(display_formatters)
media_path = get_temporary_image_path()
dfi.export(df_styler, media_path)
return media_path
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
def generate_summary_api_results(df: pd.DataFrame) -> list:
api_df = df.reset_index().copy()
api_df["sample_date_string"] = \
api_df["sample_date"].dt.strftime("%Y-%m-%d")
api_df["source_regions"] = \
api_df["source_regions"].apply(lambda x: x.split(","))
return api_df.to_dict(orient="records")
summary_api_results = \
generate_summary_api_results(df=result_summary_df)
today_summary_api_results = \
generate_summary_api_results(df=extraction_date_result_summary_df)[0]
summary_results = dict(
backend_identifier=report_backend_identifier,
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=today_summary_api_results,
last_7_days=last_7_days_summary,
last_14_days=last_14_days_summary,
daily_results=summary_api_results)
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
def format_shared_diagnoses_per_covid_case(value) -> str:
if value == 0:
return "–"
return f"≤{value:.2%}"
display_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=shared_diagnoses_per_covid_case)
display_last_14_days_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case"])
display_last_14_days_shared_diagnoses_per_covid_case_es = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case_es"])
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: {display_shared_diagnoses_per_covid_case}
Last 14 Days:
- Usage Ratio (Estimation): {display_last_14_days_shared_diagnoses_per_covid_case}
- Usage Ratio (Official): {display_last_14_days_shared_diagnoses_per_covid_case_es}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
| 0.268749 | 0.215464 |
# Confidence Interval Test for Confined Aquifers
**Synthetic data**
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from ttim import *
```
Set basic parameters for the model:
```
H = 7 #aquifer thickness
k = 70 #hydraulic conductivity
S = 1e-4 #specific storage
Q = 788 #constant discharge
d1 = 30 #observation well 1
d2 = 90 #observation well 2 (positions same as for Oude Korendijk)
```
Load data of test site 'Oude Korendijk':
```
data1 = np.loadtxt('data/piezometer_h30.txt', skiprows = 1)
t = data1[:, 0] / 60 / 24 # convert min to days
```
Create conceptual model:
```
ml = ModelMaq(kaq=70, z =[-18, -25], Saq=1e-4, tmin=1e-5, tmax=1)
w = Well(ml, xw=0, yw=0, rw=0.1, tsandQ=[(0, 788)])
ml.solve(silent='True')
h1 = ml.head(d1, 0, t)
h2 = ml.head(d2, 0, t)
```
Add noises:
```
np.savetxt('data/syn_30_0.0.txt', h1[0])
np.savetxt('data/syn_90_0.0.txt', h2[0])
#print(h2[0])
np.random.seed(5)
he12 = h1[0] - np.random.randn(len(t)) * 0.02
he22 = h2[0] - np.random.randn(len(t)) * 0.02
np.savetxt('data/syn_p30_0.02.txt', he12)
np.savetxt('data/syn_p90_0.02.txt', he22)
np.random.seed(4)
he15 = h1[0] - np.random.randn(len(t)) * 0.05
he25 = h2[0] - np.random.randn(len(t)) * 0.05
np.savetxt('data/syn_p30_0.05.txt', he15)
np.savetxt('data/syn_p90_0.05.txt', he25)
plt.figure(figsize=(8, 5))
plt.semilogx(t, he12, '.', label='obs at 30 m with sig=0.02')
plt.semilogx(t, h1[0], label='ttim at 30 m')
plt.semilogx(t, he22, '.', label='obs at 90 m with sig=0.02')
plt.semilogx(t, h2[0], label='ttim at 90 m')
plt.legend()
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)');
plt.figure(figsize=(8, 5))
plt.semilogx(t, he15, '.', label='obs at 30 m with sig=0.05')
plt.semilogx(t, h1[0], label='ttim at 30 m')
plt.semilogx(t, he25, '.', label='obs at 90 m with sig=0.05')
plt.semilogx(t, h2[0], label='ttim at 90 m')
plt.legend()
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)');
```
#### Test if TTim finds the parameters back
Calibrate with two datasets respectively (0.02):
```
ca23 = Calibrate(ml)
ca23.set_parameter(name='kaq0', initial=10)
ca23.set_parameter(name='Saq0', initial=1e-3)
ca23.series(name='obs1', x=d1, y=0, t=t, h=he12, layer=0)
ca23.fit(report=True)
display(ca23.parameters)
print('rmse:', ca23.rmse())
h123 = ml.head(d1, 0, t)
h223 = ml.head(d2, 0 ,t)
plt.figure(figsize = (8, 5))
plt.semilogx(t, he12, '.', label='obs at 30 m with sig=0.02')
plt.semilogx(t, h123[0], label='ttim at 30 m')
plt.semilogx(t, he22, '.', label='obs at 90 m with sig=0.02')
plt.semilogx(t, h223[0], label='ttim at 90 m')
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)')
plt.title('ttim analysis with synthetic data, sig=0.02 errors at 30 m.')
plt.legend();
ca29 = Calibrate(ml)
ca29.set_parameter(name='kaq0', initial=10)
ca29.set_parameter(name='Saq0', initial=1e-3)
ca29.series(name='obs2', x=d2, y=0, t=t, h=he22, layer=0)
ca29.fit(report=True)
display(ca29.parameters)
print('rmse:', ca29.rmse())
h129 = ml.head(d1, 0, t)
h229 = ml.head(d2, 0, t)
plt.figure(figsize = (8, 5))
plt.semilogx(t, he15, '.', label='obs at 30 m with sig=0.02')
plt.semilogx(t, h129[0], label='ttim at 30 m')
plt.semilogx(t, he25, '.', label='obs at 90 m with sig=0.02')
plt.semilogx(t, h229[0], label='ttim at 90 m')
plt.legend()
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)');
plt.title('ttim analysis with synthetic data, sig=0.02 errors at 90 m.')
plt.legend(loc = 'best');
```
#### Calibrate with two datasets simultaneously
Drawdown without errors:
```
ca0 = Calibrate(ml)
ca0.set_parameter(name='kaq0', initial=10)
ca0.set_parameter(name='Saq0', initial=1e-3)
ca0.series(name='obs1', x=d1, y=0, t=t, h=h1[0], layer=0)
ca0.series(name='obs2', x=d2, y=0, t=t, h=h2[0], layer=0)
ca0.fit(report=True)
display(ca0.parameters)
print('rmse:', ca0.rmse())
h1n = ml.head(d1, 0, t)
h2n = ml.head(d2, 0, t)
plt.figure(figsize = (7, 4))
plt.semilogx(t, h1[0], 'b.', label='obs at 30 m no errors')
plt.semilogx(t, h1n[0], color = 'r', label = 'ttim at 30 m')
plt.semilogx(t, h2[0], 'g.', label='obs at 90 m no errors')
plt.semilogx(t, h2n[0], color='orange', label = 'ttim at 90 m')
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)')
plt.title('ttim analysis with synthetic data without errors.')
plt.legend();
```
Drawdowns with errors with $\sigma=0.02$.
```
ca2 = Calibrate(ml)
ca2.set_parameter(name='kaq0', initial=10)
ca2.set_parameter(name='Saq0', initial=1e-3)
ca2.series(name='obs1', x=d1, y=0, t=t, h=he12, layer=0)
ca2.series(name='obs2', x=d2, y=0, t=t, h=he22, layer=0)
ca2.fit()
display(ca2.parameters)
print('rmse:', ca2.rmse())
h12 = ml.head(d1, 0, t)
h22 = ml.head(d2, 0, t)
plt.figure(figsize = (8, 5))
plt.semilogx(t, he12, 'b.', label='obs at 30 m, sig=0.02')
plt.semilogx(t, h12[0], color = 'r', label = 'ttim at 30 m')
plt.semilogx(t, he22, 'g.', label='obs at 90 m, sig=0.02')
plt.semilogx(t, h22[0], color='orange', label = 'ttim at 90 m')
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)')
plt.title('ttim analysis with synthetic data and errors with sig=0.02')
plt.legend();
```
Drawdowns with errors with $\sigma=0.05$.
```
ca5 = Calibrate(ml)
ca5.set_parameter(name='kaq0', initial=10)
ca5.set_parameter(name='Saq0', initial=1e-3)
ca5.series(name='obs1', x=d1, y=0, t=t, h=he15, layer=0)
ca5.series(name='obs2', x=d2, y=0, t=t, h=he25, layer=0)
ca5.fit()
display(ca5.parameters)
print('rmse:', ca5.rmse())
h15 = ml.head(d1, 0, t)
h25 = ml.head(d2, 0, t)
plt.figure(figsize=(8, 5))
plt.semilogx(t, he15, 'b.', label='obs at 30 m')
plt.semilogx(t, h15[0], color = 'r', label = 'ttim at 30 m')
plt.semilogx(t, he25, 'g.', label='obs at 90 m')
plt.semilogx(t, h25[0], color='orange', label = 'ttim at 90 m')
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)')
plt.title('ttim analysis with synthetic data and errors with sig=0.05')
plt.legend();
```
|
github_jupyter
|
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from ttim import *
H = 7 #aquifer thickness
k = 70 #hydraulic conductivity
S = 1e-4 #specific storage
Q = 788 #constant discharge
d1 = 30 #observation well 1
d2 = 90 #observation well 2 (positions same as for Oude Korendijk)
data1 = np.loadtxt('data/piezometer_h30.txt', skiprows = 1)
t = data1[:, 0] / 60 / 24 # convert min to days
ml = ModelMaq(kaq=70, z =[-18, -25], Saq=1e-4, tmin=1e-5, tmax=1)
w = Well(ml, xw=0, yw=0, rw=0.1, tsandQ=[(0, 788)])
ml.solve(silent='True')
h1 = ml.head(d1, 0, t)
h2 = ml.head(d2, 0, t)
np.savetxt('data/syn_30_0.0.txt', h1[0])
np.savetxt('data/syn_90_0.0.txt', h2[0])
#print(h2[0])
np.random.seed(5)
he12 = h1[0] - np.random.randn(len(t)) * 0.02
he22 = h2[0] - np.random.randn(len(t)) * 0.02
np.savetxt('data/syn_p30_0.02.txt', he12)
np.savetxt('data/syn_p90_0.02.txt', he22)
np.random.seed(4)
he15 = h1[0] - np.random.randn(len(t)) * 0.05
he25 = h2[0] - np.random.randn(len(t)) * 0.05
np.savetxt('data/syn_p30_0.05.txt', he15)
np.savetxt('data/syn_p90_0.05.txt', he25)
plt.figure(figsize=(8, 5))
plt.semilogx(t, he12, '.', label='obs at 30 m with sig=0.02')
plt.semilogx(t, h1[0], label='ttim at 30 m')
plt.semilogx(t, he22, '.', label='obs at 90 m with sig=0.02')
plt.semilogx(t, h2[0], label='ttim at 90 m')
plt.legend()
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)');
plt.figure(figsize=(8, 5))
plt.semilogx(t, he15, '.', label='obs at 30 m with sig=0.05')
plt.semilogx(t, h1[0], label='ttim at 30 m')
plt.semilogx(t, he25, '.', label='obs at 90 m with sig=0.05')
plt.semilogx(t, h2[0], label='ttim at 90 m')
plt.legend()
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)');
ca23 = Calibrate(ml)
ca23.set_parameter(name='kaq0', initial=10)
ca23.set_parameter(name='Saq0', initial=1e-3)
ca23.series(name='obs1', x=d1, y=0, t=t, h=he12, layer=0)
ca23.fit(report=True)
display(ca23.parameters)
print('rmse:', ca23.rmse())
h123 = ml.head(d1, 0, t)
h223 = ml.head(d2, 0 ,t)
plt.figure(figsize = (8, 5))
plt.semilogx(t, he12, '.', label='obs at 30 m with sig=0.02')
plt.semilogx(t, h123[0], label='ttim at 30 m')
plt.semilogx(t, he22, '.', label='obs at 90 m with sig=0.02')
plt.semilogx(t, h223[0], label='ttim at 90 m')
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)')
plt.title('ttim analysis with synthetic data, sig=0.02 errors at 30 m.')
plt.legend();
ca29 = Calibrate(ml)
ca29.set_parameter(name='kaq0', initial=10)
ca29.set_parameter(name='Saq0', initial=1e-3)
ca29.series(name='obs2', x=d2, y=0, t=t, h=he22, layer=0)
ca29.fit(report=True)
display(ca29.parameters)
print('rmse:', ca29.rmse())
h129 = ml.head(d1, 0, t)
h229 = ml.head(d2, 0, t)
plt.figure(figsize = (8, 5))
plt.semilogx(t, he15, '.', label='obs at 30 m with sig=0.02')
plt.semilogx(t, h129[0], label='ttim at 30 m')
plt.semilogx(t, he25, '.', label='obs at 90 m with sig=0.02')
plt.semilogx(t, h229[0], label='ttim at 90 m')
plt.legend()
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)');
plt.title('ttim analysis with synthetic data, sig=0.02 errors at 90 m.')
plt.legend(loc = 'best');
ca0 = Calibrate(ml)
ca0.set_parameter(name='kaq0', initial=10)
ca0.set_parameter(name='Saq0', initial=1e-3)
ca0.series(name='obs1', x=d1, y=0, t=t, h=h1[0], layer=0)
ca0.series(name='obs2', x=d2, y=0, t=t, h=h2[0], layer=0)
ca0.fit(report=True)
display(ca0.parameters)
print('rmse:', ca0.rmse())
h1n = ml.head(d1, 0, t)
h2n = ml.head(d2, 0, t)
plt.figure(figsize = (7, 4))
plt.semilogx(t, h1[0], 'b.', label='obs at 30 m no errors')
plt.semilogx(t, h1n[0], color = 'r', label = 'ttim at 30 m')
plt.semilogx(t, h2[0], 'g.', label='obs at 90 m no errors')
plt.semilogx(t, h2n[0], color='orange', label = 'ttim at 90 m')
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)')
plt.title('ttim analysis with synthetic data without errors.')
plt.legend();
ca2 = Calibrate(ml)
ca2.set_parameter(name='kaq0', initial=10)
ca2.set_parameter(name='Saq0', initial=1e-3)
ca2.series(name='obs1', x=d1, y=0, t=t, h=he12, layer=0)
ca2.series(name='obs2', x=d2, y=0, t=t, h=he22, layer=0)
ca2.fit()
display(ca2.parameters)
print('rmse:', ca2.rmse())
h12 = ml.head(d1, 0, t)
h22 = ml.head(d2, 0, t)
plt.figure(figsize = (8, 5))
plt.semilogx(t, he12, 'b.', label='obs at 30 m, sig=0.02')
plt.semilogx(t, h12[0], color = 'r', label = 'ttim at 30 m')
plt.semilogx(t, he22, 'g.', label='obs at 90 m, sig=0.02')
plt.semilogx(t, h22[0], color='orange', label = 'ttim at 90 m')
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)')
plt.title('ttim analysis with synthetic data and errors with sig=0.02')
plt.legend();
ca5 = Calibrate(ml)
ca5.set_parameter(name='kaq0', initial=10)
ca5.set_parameter(name='Saq0', initial=1e-3)
ca5.series(name='obs1', x=d1, y=0, t=t, h=he15, layer=0)
ca5.series(name='obs2', x=d2, y=0, t=t, h=he25, layer=0)
ca5.fit()
display(ca5.parameters)
print('rmse:', ca5.rmse())
h15 = ml.head(d1, 0, t)
h25 = ml.head(d2, 0, t)
plt.figure(figsize=(8, 5))
plt.semilogx(t, he15, 'b.', label='obs at 30 m')
plt.semilogx(t, h15[0], color = 'r', label = 'ttim at 30 m')
plt.semilogx(t, he25, 'g.', label='obs at 90 m')
plt.semilogx(t, h25[0], color='orange', label = 'ttim at 90 m')
plt.xlabel('time (d)')
plt.ylabel('drawdown (m)')
plt.title('ttim analysis with synthetic data and errors with sig=0.05')
plt.legend();
| 0.246896 | 0.928991 |
## Exploration of information gain and feature importance on a healthcare dataset
Example of using information gain (Gini impurity) to perform EDA on a tabular dataset:
**Heart Disease Data Set - UCI Machine Learning Repository** (https://archive.ics.uci.edu/ml/datasets/heart+disease)
```
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
np.random.seed(42)
names=['age','sex','cp','restbps','chol','fbs','restecg','thalac','exang','oldpeak','slope','ca','thal','outcome']
df=pd.read_csv('processed.cleveland.data.csv', header=None, names=names )
print(df.shape)
df.head(5)
df=df.apply(lambda x: pd.to_numeric(x, errors='coerce'))
df[df.isna().any(axis=1)]
df=df.dropna(axis='rows')
df["outcome"][df["outcome"]>1]=1
df.head(5)
```
### Brute force calculation of information gain for each feature
```
from information_gain import gini_calc
from information_gain import plot_gini_hist
y=df['outcome']
data=df['age']
threshold,ig = gini_calc(data,y)
plot_gini_hist(data,y,threshold,ig,'Hearth Disease',data.name)
```
In the case of **age** the optimal split is at 54.5 years. Information gain is ~0.05 (max 0.5) so we are far from a pure node. However it's easy to see how people with heart disease cluster at ages above the treshold.
### Compare with sklearn
Check if my implementation of Gini Purity is matched sklearn.
```
y=df['outcome']
data=df['age'].to_numpy().reshape(-1, 1)
from sklearn.tree import DecisionTreeClassifier
clf = DecisionTreeClassifier(max_depth=1)
clf.fit(data, y)
feat_importance = clf.tree_.compute_feature_importances(normalize=False)
print("information gain = " + str(feat_importance))
from sklearn import tree
fig = plt.figure(figsize=(5,5))
_ = tree.plot_tree(clf, feature_names=df['age'].name, class_names=['0','1'],filled=True)
```
### Rank Feature Importance by information gain
Note that this would be the first step generarting a decision tree. Howver I find it useful to plot feature importance at this stage because information theory does not assume anything about the feature distribution
```
new_df = df.apply(lambda x: gini_calc(x,df['outcome']) if not x.name == 'outcome' else x)
new_df=new_df[:-1]
new_df=new_df.to_frame()
new_df=pd.DataFrame(new_df[0].values.tolist(), index=new_df.index, columns=['threshold','ig'])
new_df=new_df.sort_values(by=['ig'],ascending=True)
new_df=new_df['ig']
new_df.plot(kind='barh', color=['red'],rot=0,legend=True,figsize=(12,4))
```
Note how no feature is near 0.5 (which would correspond to a perfect separation between heart disease and no heart disease). Moreover:
1. To decide what feature are important one should inspect how much features are correlated
2. Check if there are outliers that bias the information gain score
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
np.random.seed(42)
names=['age','sex','cp','restbps','chol','fbs','restecg','thalac','exang','oldpeak','slope','ca','thal','outcome']
df=pd.read_csv('processed.cleveland.data.csv', header=None, names=names )
print(df.shape)
df.head(5)
df=df.apply(lambda x: pd.to_numeric(x, errors='coerce'))
df[df.isna().any(axis=1)]
df=df.dropna(axis='rows')
df["outcome"][df["outcome"]>1]=1
df.head(5)
from information_gain import gini_calc
from information_gain import plot_gini_hist
y=df['outcome']
data=df['age']
threshold,ig = gini_calc(data,y)
plot_gini_hist(data,y,threshold,ig,'Hearth Disease',data.name)
y=df['outcome']
data=df['age'].to_numpy().reshape(-1, 1)
from sklearn.tree import DecisionTreeClassifier
clf = DecisionTreeClassifier(max_depth=1)
clf.fit(data, y)
feat_importance = clf.tree_.compute_feature_importances(normalize=False)
print("information gain = " + str(feat_importance))
from sklearn import tree
fig = plt.figure(figsize=(5,5))
_ = tree.plot_tree(clf, feature_names=df['age'].name, class_names=['0','1'],filled=True)
new_df = df.apply(lambda x: gini_calc(x,df['outcome']) if not x.name == 'outcome' else x)
new_df=new_df[:-1]
new_df=new_df.to_frame()
new_df=pd.DataFrame(new_df[0].values.tolist(), index=new_df.index, columns=['threshold','ig'])
new_df=new_df.sort_values(by=['ig'],ascending=True)
new_df=new_df['ig']
new_df.plot(kind='barh', color=['red'],rot=0,legend=True,figsize=(12,4))
| 0.293202 | 0.923039 |
```
from IPython.display import HTML
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
The raw code for this IPython notebook is by default hidden for easier reading.
To toggle on/off the raw code, click <a href="javascript:code_toggle()">here</a>.''')
```
<a id="top-title"></a>
# From Ptolemy to Kepler
Ptolemy, Copernicus, Brahe, Kepler
<a id="CC-2.1"></a>
# Cosmic Calculations 2.1: Kepler’s Third Law
First, let's review the three laws discovered by Kepler from the careful measurements of [Tycho Brache](https://physicsworld.com/a/kepler-and-tycho-brahe-the-odd-couple/). This is one of the most ineteresting stories of scientific collaboration that transformed years of observations into laws about the universe. I recommend to read/watch [***The Character of Physical Law***](https://www.youtube.com/watch?v=j3mhkYbznBk) by Richard Feynman if you want to indulge in the details.
<p><a href="https://commons.wikimedia.org/wiki/File:Tycho-Kepler-Statue-Prague.jpg#/media/File:Tycho-Kepler-Statue-Prague.jpg"><img src="https://upload.wikimedia.org/wikipedia/commons/7/73/Tycho-Kepler-Statue-Prague.jpg" alt="Tycho-Kepler-Statue-Prague.jpg" width="360" height="480"></a><br>By <a href="https://en.wikipedia.org/wiki/hu:User:Both_El%C5%91d" class="extiw" title="w:hu:User:Both Előd">Both Előd</a> at <a href="https://en.wikipedia.org/wiki/hu:" class="extiw" title="w:hu:">Hungarian Wikipedia</a>, <a href="https://creativecommons.org/licenses/by-sa/2.5" title="Creative Commons Attribution-Share Alike 2.5">CC BY-SA 2.5</a>, <a href="https://commons.wikimedia.org/w/index.php?curid=47229075">Link</a></p>
## Kepler's laws:
***1. The orbit of every planet is an ellipse with the Sun at one of the two foci.***
In the figure below, you can imagine the yellow dot as the sun, and a planet would be moving on the blue curve a certain distance away from it. The elliptical orbit can be described by the semi-major and semi-minor axes, which define the eccentricity of the orbit.
Look for the *perihelion* and *aphelion* of Earth's orbit. From those values, what's the flattening of Earth's orbit and its eccentricity?
```
import numpy as np
import matplotlib.pyplot as plt
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
# Set default font size for plots:
font = {'size' : 18}
plt.rc('font',**font)
def elliptic_orbit(a,b,t):
'''Plot an elliptical orbit and see the radial distance from one focal point
a= semi-major axis
b=semi-minor axis
t=location at an angle between 0 and 360'''
p=np.linspace(0,2*np.pi,360)
x = a*np.cos(p)
y = b*np.sin(p)
plt.figure('Ellipse2',figsize=(10,5))
plt.plot(x,y,'-')
plt.axis('equal')
plt.grid(True)
#t is the angle varying from 0 to 360 degrees
X = a*np.cos(t*np.pi/180)
Y = b*np.sin(t*np.pi/180)
#Conditionals in case of changing length of largest semi-major axis
if a>=b:
c=np.sqrt(a**2-b**2)
plt.scatter(c,0,s=200,c='y')
plt.scatter(-c, 0, s=200, facecolors='none', edgecolors='y')
plt.arrow(c, 0, X-c, Y, head_width=0.1, head_length=0.1, fc='red', ec='red')
plt.scatter(X,Y,s=50,c='b')
f=(a-b)/a
e=c/a
#print('Orbital flattening : ',f)
print('Orbital eccentricity : ',e)
#plt.show()
else:
c=np.sqrt(b**2-a**2)
plt.scatter(0,c,s=200,c='y')
plt.scatter(0, -c, s=200, facecolors='none', edgecolors='y')
plt.arrow(0, c, X, Y-c, head_width=0.1, head_length=0.1, fc='red', ec='red')
plt.scatter(X,Y,s=50,c='b')
f=(b-a)/b
e=c/b
#print('Orbital flattening : ',f)
print('Orbital eccentricity : ',e)
plt.show()
return
interactive(elliptic_orbit, a = (0,20,1),b=(0,20,1),t=(0,360,20),continuous_update=False)
```
The reason the orbits are ellipses, and not circles, would not be understood until the arrival of Newton's equations on Gravitation.
***2. A line joining a planet and the Sun sweeps out equal areas during equal intervals of time.***
<p><a href="https://commons.wikimedia.org/wiki/File:Kepler-second-law.gif#/media/File:Kepler-second-law.gif"><img src="https://upload.wikimedia.org/wikipedia/commons/6/69/Kepler-second-law.gif" alt="Kepler-second-law.gif"></a><br>By <a href="https://en.wikipedia.org/wiki/User:Gonfer" class="extiw" title="en:User:Gonfer">Gonfer</a> (<a href="//commons.wikimedia.org/wiki/User_talk:Gonfer" title="User talk:Gonfer">talk</a>) - <a href="https://en.wikipedia.org/wiki/User:Gonfer" class="extiw" title="en:User:Gonfer">Gonfer</a>, <a href="https://creativecommons.org/licenses/by-sa/3.0" title="Creative Commons Attribution-Share Alike 3.0">CC BY-SA 3.0</a>, <a href="https://commons.wikimedia.org/w/index.php?curid=24871608">Link</a></p>
When Kepler discovered his third law ($p^2 = a^3$), he knew only that it applied to the orbits of planets about the Sun. In fact, it applies to any orbiting object as long as the following two conditions are met:
1. The object orbits the Sun or another star of precisely the same mass.
2. We use units of years for the orbital period and AU for the orbital distance. (Newton extended the law to all orbiting objects; see [Cosmic Calculations 7.1](#CC-7.1).)
In other words, these two conditions make the relationship a perfect equality.
**Example 1:** The largest asteroid, Ceres, orbits the Sun at an average distance (semimajor axis) of 2.77 AU. What is its orbital period?
***Solution:*** Both conditions are met, so we solve Kepler’s third law for the orbital period $p$ and substitute the given orbital distance, $a = 2.77~AU$.
$$p^2 = a^3$$
$$ p = \sqrt{a^3} = \sqrt{2.77^3} \approx 4.6~y$$
Ceres has an orbital period of 4.6 years.
**Example 2:** A planet is discovered orbiting every three months around a star of the same mass as our Sun. What is the planet’s average orbital distance?
***Solution:*** The first condition is met, and we can satisfy the second by converting the orbital period from months to years: $p = 3$ months = 0.25 year. We now solve Kepler’s third law for the average distance a:
$a = \sqrt[3]{p^2}$
$a = \sqrt[3]{0.25^2} \approx 0.40~AU$
The planet orbits its star at an average distance of $0.40~AU$, which is nearly the same as Mercury’s average distance from the Sun.
These observations offered clear proof that Earth is not the center of everything.* Although we now recognize that Galileo won the day, the story was more complex in his own time, when Catholic Church doctrine still held Earth to be the center of the universe. On June 22, 1633, Galileo was brought before a Church inquisition in Rome and ordered to recant his claim that Earth orbits the Sun. Nearly 70 years old and fearing for his life, Galileo did as ordered and his life was spared. However, legend has it that as he rose from his knees, he whispered under his breath, Eppur si muove— Italian for “And yet it moves.” (Given the likely consequences if Church officials had heard him say this, most historians doubt the legend.) The Church did not formally vindicate Galileo until 1992, but the Church had given up the argument long before that. Today, Catholic scientists are at the forefront of much astronomical research, and official Church teachings are compatible not only with Earth’s planetary status but also with the theories of the Big Bang and the subsequent evolution of the cosmos and of life.
<object classid="clsid:D27CDB6E-AE6D-11cf-96B8-444553540000" width="750" height="400"><param name="movie" value="KeplerFirstLaw.swf" /><!--[if !IE]>--><object type="application/x-shockwave-flash" data="KeplerFirstLaw.swf" width="750" height="400"><!--<![endif]--><p>flash animation</p><!--[if !IE]>--></object><!--<![endif]--></object>
[Go back to the top of the page](#top-title)
|
github_jupyter
|
from IPython.display import HTML
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
The raw code for this IPython notebook is by default hidden for easier reading.
To toggle on/off the raw code, click <a href="javascript:code_toggle()">here</a>.''')
import numpy as np
import matplotlib.pyplot as plt
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
# Set default font size for plots:
font = {'size' : 18}
plt.rc('font',**font)
def elliptic_orbit(a,b,t):
'''Plot an elliptical orbit and see the radial distance from one focal point
a= semi-major axis
b=semi-minor axis
t=location at an angle between 0 and 360'''
p=np.linspace(0,2*np.pi,360)
x = a*np.cos(p)
y = b*np.sin(p)
plt.figure('Ellipse2',figsize=(10,5))
plt.plot(x,y,'-')
plt.axis('equal')
plt.grid(True)
#t is the angle varying from 0 to 360 degrees
X = a*np.cos(t*np.pi/180)
Y = b*np.sin(t*np.pi/180)
#Conditionals in case of changing length of largest semi-major axis
if a>=b:
c=np.sqrt(a**2-b**2)
plt.scatter(c,0,s=200,c='y')
plt.scatter(-c, 0, s=200, facecolors='none', edgecolors='y')
plt.arrow(c, 0, X-c, Y, head_width=0.1, head_length=0.1, fc='red', ec='red')
plt.scatter(X,Y,s=50,c='b')
f=(a-b)/a
e=c/a
#print('Orbital flattening : ',f)
print('Orbital eccentricity : ',e)
#plt.show()
else:
c=np.sqrt(b**2-a**2)
plt.scatter(0,c,s=200,c='y')
plt.scatter(0, -c, s=200, facecolors='none', edgecolors='y')
plt.arrow(0, c, X, Y-c, head_width=0.1, head_length=0.1, fc='red', ec='red')
plt.scatter(X,Y,s=50,c='b')
f=(b-a)/b
e=c/b
#print('Orbital flattening : ',f)
print('Orbital eccentricity : ',e)
plt.show()
return
interactive(elliptic_orbit, a = (0,20,1),b=(0,20,1),t=(0,360,20),continuous_update=False)
| 0.281801 | 0.858244 |
# Prophet - predição de vendas no varejo.
<img src="https://i.imgur.com/Hi7XNrM.png" />
## Facebook PROPHET
A ferramenta visa contribuir para problemas de geração de previsões e cenários futuros para séries temporais. Veja as características do Prophet (em que ele brilha, segundo o site):
* Observações horárias, diárias ou semanais com pelo menos alguns meses (preferivelmente um ano) de histórico.
* Feriados importantes que ocorrem em intervalos irregulares que são conhecidos antecipadamente (por exemplo, o Super Bowl).
* Um número razoável de observações ausentes ou grandes outliers,
* mudanças históricas de tendência, por exemplo, devido a lançamentos de produtos ou alterações no registro.
* Tendências que são curvas de crescimento não lineares, em que uma tendência atinge um limite natural ou satura.
A ideia de oferecer uma ferramenta para forecast com menor esforço está tornando o Prophet largamente investigado.
Nesse projeto vamos testa a eficácia dele, usaremos um dataset simples para tal:
```
import pandas as pd
import numpy as np
from fbprophet import Prophet
import matplotlib.pyplot as plt
pd.plotting.register_matplotlib_converters()
%matplotlib inline
plt.rcParams['figure.figsize']=(20,10)
plt.style.use('ggplot')
%time sales_df = pd.read_csv('C:/Users/skite/OneDrive/Documentos/GitHub/Projeto_TimeSires_Phophet/example_retail_sales.csv', sep = ',', encoding = 'ISO-8859-1', index_col='Date', parse_dates=True)
sales_df.head()
sales_df.info()
```
**Esse dataset e bem pequeno e foi ajustado propositalmente testas modelos preditores que usem séries temporais.**
## Preparação dos dados para o Prophet
```
df = sales_df.reset_index()
df.head()
```
*Vamos renomear as colunas.*
*O Prophet exige que os dados estajão organizados como 'ds' (datas) e 'y' (valor).*
```
df=df.rename(columns={'Date':'ds', 'Sales':'y'})
df.head()
```
*Plot dos dados*
```
df.set_index('ds').y.plot()
```
*Transformação dos dados aplicando a função log do numpy*
```
df['y'] = np.log(df['y'])
df.tail()
df.set_index('ds').y.plot()
```
# Execução do Prophet
*A ferramenta automaticamente seleciona uma sazonalidade adequada*
```
model = Prophet()
model.fit(df);
```
*Montagem do dataframe que irá receber os dados das predições.*
```
future = model.make_future_dataframe(periods=24, freq = 'm')
future.head()
future.tail()
```
*Execução da predição do modelo instanciado.*
```
forecast = model.predict(future)
```
*Resultado da execução:*
```
forecast.tail()
```
*Seleção dos dados específicos das predições da série temporal:*
```
forecast[['ds', 'yhat', 'yhat_lower', 'yhat_upper']].head()
forecast[['ds', 'yhat', 'yhat_lower', 'yhat_upper']].tail()
```
# Plot dos resultados
*Prophet tem uma função embarcada que já faz o desenho do gráfico - plot.*
```
model.plot(forecast);
```
*Sazonalidade do dados.*
# Montando a visualização no formato original dos dados
*Equalização dos índices para```ds```*
```
df.set_index('ds', inplace=True)
forecast.set_index('ds', inplace=True)
```
*Junção dos dados originais e do forecast*
```
viz_df = sales_df.join(forecast[['yhat', 'yhat_lower','yhat_upper']], how = 'outer')
```
*Observação: os dados da predição ainda estão em valores relativos à transformação logarítimica que realizamos.*
```
viz_df.head()
```
*retornando os dados de log para a escala original*
```
viz_df['yhat_rescaled'] = np.exp(viz_df['yhat'])
viz_df.head()
```
*Visualização dos dados das vendas e do yhat restaurado ao valor original*
```
viz_df[['Sales', 'yhat_rescaled']].plot()
```
*Vamos utilizar a data como índice:*
```
sales_df.index = pd.to_datetime(sales_df.index)
```
*Seleciona a penúltima data:*
```
connect_date = sales_df.index[-2]
```
*Critério com os índices maiores que a penúltima:*
```
mask = (forecast.index > connect_date)
```
*Seleção dos dados:*
```
predict_df = forecast.loc[mask]
```
*O predict_df terá somente os valores abaixo que forem True:*
```
mask
predict_df
```
*Construção do DataFrame para a visualização.*
```
viz_df = sales_df.join(predict_df[['yhat', 'yhat_lower','yhat_upper']], how = 'outer')
viz_df
```
*Adição da coluna com o yhat original:*
```
viz_df['yhat_scaled']=np.exp(viz_df['yhat'])
```
## Visualizaçao Final
```
fig, ax1 = plt.subplots()
ax1.plot(viz_df.Sales)
ax1.plot(viz_df.yhat_scaled, color='black', linestyle=':')
ax1.fill_between(viz_df.index, np.exp(viz_df['yhat_upper']), np.exp(viz_df['yhat_lower']), alpha=0.5, color='darkgray')
ax1.set_title('Vendas (Laranja) vs Previsão de vendas (Preto)')
ax1.set_ylabel('Vendas em Dólares')
ax1.set_xlabel('Data')
```
**A previsão realizada pelo modelo reforça a tendência de crescimento das vendas para os próximos meses.Eantretanto, os intervalos de confiança tem momentos em que se elevam rapidamente, isso indica que outros fatores podem levar a uma rápida aumento ou quedas das vendas.**
*O Prophoet se mostrou uma ferramenta de forecasting muito boa.*
|
github_jupyter
|
import pandas as pd
import numpy as np
from fbprophet import Prophet
import matplotlib.pyplot as plt
pd.plotting.register_matplotlib_converters()
%matplotlib inline
plt.rcParams['figure.figsize']=(20,10)
plt.style.use('ggplot')
%time sales_df = pd.read_csv('C:/Users/skite/OneDrive/Documentos/GitHub/Projeto_TimeSires_Phophet/example_retail_sales.csv', sep = ',', encoding = 'ISO-8859-1', index_col='Date', parse_dates=True)
sales_df.head()
sales_df.info()
df = sales_df.reset_index()
df.head()
df=df.rename(columns={'Date':'ds', 'Sales':'y'})
df.head()
df.set_index('ds').y.plot()
df['y'] = np.log(df['y'])
df.tail()
df.set_index('ds').y.plot()
model = Prophet()
model.fit(df);
future = model.make_future_dataframe(periods=24, freq = 'm')
future.head()
future.tail()
forecast = model.predict(future)
forecast.tail()
forecast[['ds', 'yhat', 'yhat_lower', 'yhat_upper']].head()
forecast[['ds', 'yhat', 'yhat_lower', 'yhat_upper']].tail()
model.plot(forecast);
df.set_index('ds', inplace=True)
forecast.set_index('ds', inplace=True)
viz_df = sales_df.join(forecast[['yhat', 'yhat_lower','yhat_upper']], how = 'outer')
viz_df.head()
viz_df['yhat_rescaled'] = np.exp(viz_df['yhat'])
viz_df.head()
viz_df[['Sales', 'yhat_rescaled']].plot()
sales_df.index = pd.to_datetime(sales_df.index)
connect_date = sales_df.index[-2]
mask = (forecast.index > connect_date)
predict_df = forecast.loc[mask]
mask
predict_df
viz_df = sales_df.join(predict_df[['yhat', 'yhat_lower','yhat_upper']], how = 'outer')
viz_df
viz_df['yhat_scaled']=np.exp(viz_df['yhat'])
fig, ax1 = plt.subplots()
ax1.plot(viz_df.Sales)
ax1.plot(viz_df.yhat_scaled, color='black', linestyle=':')
ax1.fill_between(viz_df.index, np.exp(viz_df['yhat_upper']), np.exp(viz_df['yhat_lower']), alpha=0.5, color='darkgray')
ax1.set_title('Vendas (Laranja) vs Previsão de vendas (Preto)')
ax1.set_ylabel('Vendas em Dólares')
ax1.set_xlabel('Data')
| 0.351422 | 0.918187 |
```
# https://www.reddit.com/r/financialindependence/comments/9w8h2j/the_4_rule_is_there_some_builtin_flexibility/
%matplotlib inline
from pprint import pprint
from plot import plot_two
from simulate import simulate_withdrawals
from harvesting import N_60_RebalanceHarvesting, N_100_RebalanceHarvesting
import harvesting
import itertools
from decimal import Decimal
from montecarlo import conservative
from matplotlib import pyplot as plt
import matplotlib
import plot
import math
from market import Returns_US_1871
import withdrawal
class EAH(withdrawal.WithdrawalStrategy):
def __init__(self, portfolio, harvest):
super().__init__(portfolio, harvest)
self.last_withdrawal = Decimal('.04') * portfolio.value
self.portfolio_highwater = portfolio.value
def start(self):
withdraw = self.last_withdrawal
return withdraw
def next(self):
self.portfolio_highwater *= (1 + self.current_inflation)
if self.portfolio.value > self.portfolio_highwater:
withdraw = Decimal('.04') * self.portfolio.value
self.portfolio_highwater = self.portfolio.value
else:
withdraw = self.last_withdrawal * (1 + self.current_inflation)
self.last_withdrawal = withdraw
return withdraw
def compare(series, years=30):
(r1, r2) = itertools.tee(series)
portfolio = (600000, 400000)
x = simulate_withdrawals(r1, years=years, harvesting=N_60_RebalanceHarvesting, withdraw=withdrawal.ConstantDollar, portfolio=portfolio)
y = simulate_withdrawals(r2, years=years, harvesting=N_60_RebalanceHarvesting, withdraw=EAH, portfolio=portfolio)
s1 = [n.withdraw_r for n in x]
s2 = [n.withdraw_r for n in y]
plot.plot_n({'4%': s1, 'EAH': s2}, 'Years', '4% vs /u/ElephantsAreHeavy')
compare(Returns_US_1871().iter_from(1928))
c_cd = 0
c_eah = 0
count = 0
for year in range(1871, 2017-30):
series = Returns_US_1871().iter_from(year)
(r1, r2) = itertools.tee(series)
portfolio = (600000, 400000)
x = simulate_withdrawals(r1, years=30, harvesting=N_60_RebalanceHarvesting, withdraw=withdrawal.ConstantDollar, portfolio=portfolio)
y = simulate_withdrawals(r2, years=30, harvesting=N_60_RebalanceHarvesting, withdraw=EAH, portfolio=portfolio)
cd = x[-1].portfolio_n
eah = y[-1].portfolio_n
count += 1
if cd > 0:
c_cd += 1
if eah > 0:
c_eah += 1
print(c_cd/count)
print(c_eah/count)
print(count, c_cd, c_eah)
```
|
github_jupyter
|
# https://www.reddit.com/r/financialindependence/comments/9w8h2j/the_4_rule_is_there_some_builtin_flexibility/
%matplotlib inline
from pprint import pprint
from plot import plot_two
from simulate import simulate_withdrawals
from harvesting import N_60_RebalanceHarvesting, N_100_RebalanceHarvesting
import harvesting
import itertools
from decimal import Decimal
from montecarlo import conservative
from matplotlib import pyplot as plt
import matplotlib
import plot
import math
from market import Returns_US_1871
import withdrawal
class EAH(withdrawal.WithdrawalStrategy):
def __init__(self, portfolio, harvest):
super().__init__(portfolio, harvest)
self.last_withdrawal = Decimal('.04') * portfolio.value
self.portfolio_highwater = portfolio.value
def start(self):
withdraw = self.last_withdrawal
return withdraw
def next(self):
self.portfolio_highwater *= (1 + self.current_inflation)
if self.portfolio.value > self.portfolio_highwater:
withdraw = Decimal('.04') * self.portfolio.value
self.portfolio_highwater = self.portfolio.value
else:
withdraw = self.last_withdrawal * (1 + self.current_inflation)
self.last_withdrawal = withdraw
return withdraw
def compare(series, years=30):
(r1, r2) = itertools.tee(series)
portfolio = (600000, 400000)
x = simulate_withdrawals(r1, years=years, harvesting=N_60_RebalanceHarvesting, withdraw=withdrawal.ConstantDollar, portfolio=portfolio)
y = simulate_withdrawals(r2, years=years, harvesting=N_60_RebalanceHarvesting, withdraw=EAH, portfolio=portfolio)
s1 = [n.withdraw_r for n in x]
s2 = [n.withdraw_r for n in y]
plot.plot_n({'4%': s1, 'EAH': s2}, 'Years', '4% vs /u/ElephantsAreHeavy')
compare(Returns_US_1871().iter_from(1928))
c_cd = 0
c_eah = 0
count = 0
for year in range(1871, 2017-30):
series = Returns_US_1871().iter_from(year)
(r1, r2) = itertools.tee(series)
portfolio = (600000, 400000)
x = simulate_withdrawals(r1, years=30, harvesting=N_60_RebalanceHarvesting, withdraw=withdrawal.ConstantDollar, portfolio=portfolio)
y = simulate_withdrawals(r2, years=30, harvesting=N_60_RebalanceHarvesting, withdraw=EAH, portfolio=portfolio)
cd = x[-1].portfolio_n
eah = y[-1].portfolio_n
count += 1
if cd > 0:
c_cd += 1
if eah > 0:
c_eah += 1
print(c_cd/count)
print(c_eah/count)
print(count, c_cd, c_eah)
| 0.447943 | 0.355691 |
```
import os
import json
import numpy as np
import pandas as pd
import pyproj
import matplotlib.pyplot as plt
from matplotlib import gridspec
from matplotlib import cm
from pysheds.grid import Grid
from matplotlib import colors
import seaborn as sns
import warnings
from swmmtoolbox import swmmtoolbox
import matplotlib.patches as mpatches
from matplotlib.lines import Line2D
warnings.filterwarnings('ignore')
sns.set()
sns.set_palette('husl', 7)
%matplotlib inline
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
var = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_11.2mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('11.2mm' in fn) and (not 'under' in fn):
if 'k30' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('11.2mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
var = pd.Series(var).sort_values()
colors = pd.Series(var.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
var.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Hydrograph variance $(m^3/s)^2$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title('Outlet discharge, small storm', size=14)
ax[1].set_title('Flashiness (hydrograph variance)', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_var_small.png', bbox_inches='tight', dpi=200)
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
var = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_16.87mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('16.87mm' in fn) and (not 'under' in fn):
if 'k30' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('16.87mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
var = pd.Series(var).sort_values()
colors = pd.Series(var.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
var.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Hydrograph variance $(m^3/s)^2$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title('Outlet discharge, medium storm', size=14)
ax[1].set_title('Flashiness (hydrograph variance)', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_var_med.png', bbox_inches='tight', dpi=200)
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
var = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_23.38mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('23.38mm' in fn) and (not 'under' in fn):
if 'k30' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('23.38mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
var = pd.Series(var).sort_values()
colors = pd.Series(var.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
var.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Hydrograph variance $(m^3/s)^2$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title('Outlet discharge, large storm', size=14)
ax[1].set_title('Flashiness (hydrograph variance)', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_var_large.png', bbox_inches='tight', dpi=200)
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
maxes = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_11.2mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('11.2mm' in fn) and (not 'under' in fn):
if 'k30' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('11.2mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
maxes = pd.Series(maxes).sort_values()
colors = pd.Series(maxes.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
maxes.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Peak discharge $(m^3/s)$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title(r'Outlet discharge, small storm', size=14)
ax[1].set_title(r'Peak discharge', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_max_small.png', bbox_inches='tight', dpi=200)
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
maxes = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_16.87mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('16.87mm' in fn) and (not 'under' in fn):
if 'k18' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('16.87mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
maxes = pd.Series(maxes).sort_values()
colors = pd.Series(maxes.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
maxes.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Peak discharge $(m^3/s)$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title(r'Outlet discharge, medium storm', size=14)
ax[1].set_title(r'Peak discharge', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_max_med.png', bbox_inches='tight', dpi=200)
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
maxes = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_23.38mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('23.38mm' in fn) and (not 'under' in fn):
if 'k18' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('23.38mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
maxes = pd.Series(maxes).sort_values()
colors = pd.Series(maxes.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
maxes.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Peak discharge $(m^3/s)$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title(r'Outlet discharge, large storm', size=14)
ax[1].set_title(r'Peak discharge', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_max_large.png', bbox_inches='tight', dpi=200)
maxes
var[0] / var[-1]
var[1:-1].mean() / var[-1]
maxes[1] / maxes[-1]
maxes[2:-1].mean() / maxes[-1]
```
|
github_jupyter
|
import os
import json
import numpy as np
import pandas as pd
import pyproj
import matplotlib.pyplot as plt
from matplotlib import gridspec
from matplotlib import cm
from pysheds.grid import Grid
from matplotlib import colors
import seaborn as sns
import warnings
from swmmtoolbox import swmmtoolbox
import matplotlib.patches as mpatches
from matplotlib.lines import Line2D
warnings.filterwarnings('ignore')
sns.set()
sns.set_palette('husl', 7)
%matplotlib inline
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
var = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_11.2mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('11.2mm' in fn) and (not 'under' in fn):
if 'k30' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('11.2mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
var = pd.Series(var).sort_values()
colors = pd.Series(var.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
var.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Hydrograph variance $(m^3/s)^2$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title('Outlet discharge, small storm', size=14)
ax[1].set_title('Flashiness (hydrograph variance)', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_var_small.png', bbox_inches='tight', dpi=200)
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
var = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_16.87mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('16.87mm' in fn) and (not 'under' in fn):
if 'k30' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('16.87mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
var = pd.Series(var).sort_values()
colors = pd.Series(var.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
var.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Hydrograph variance $(m^3/s)^2$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title('Outlet discharge, medium storm', size=14)
ax[1].set_title('Flashiness (hydrograph variance)', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_var_med.png', bbox_inches='tight', dpi=200)
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
var = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_23.38mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('23.38mm' in fn) and (not 'under' in fn):
if 'k30' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('23.38mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
var[basename] = outfall.var()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
var = pd.Series(var).sort_values()
colors = pd.Series(var.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
var.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Hydrograph variance $(m^3/s)^2$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title('Outlet discharge, large storm', size=14)
ax[1].set_title('Flashiness (hydrograph variance)', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_var_large.png', bbox_inches='tight', dpi=200)
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
maxes = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_11.2mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('11.2mm' in fn) and (not 'under' in fn):
if 'k30' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('11.2mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
maxes = pd.Series(maxes).sort_values()
colors = pd.Series(maxes.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
maxes.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Peak discharge $(m^3/s)$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title(r'Outlet discharge, small storm', size=14)
ax[1].set_title(r'Peak discharge', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_max_small.png', bbox_inches='tight', dpi=200)
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
maxes = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_16.87mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('16.87mm' in fn) and (not 'under' in fn):
if 'k18' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('16.87mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
maxes = pd.Series(maxes).sort_values()
colors = pd.Series(maxes.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
maxes.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Peak discharge $(m^3/s)$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title(r'Outlet discharge, medium storm', size=14)
ax[1].set_title(r'Peak discharge', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_max_med.png', bbox_inches='tight', dpi=200)
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
output_dir = '../data/out'
maxes = {}
for fn in os.listdir(output_dir):
basename = fn.split('_diff')[0]
if (('50pct' in fn)) or ('uncontrolled' in fn):
if fn == 'uncontrolled_23.38mm.out':
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='k', zorder=1)
elif fn.startswith('linear') and ('23.38mm' in fn) and (not 'under' in fn):
if 'k18' in fn:
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='r', zorder=2)
elif fn.startswith('naive') and ('23.38mm' in fn) and (not 'under' in fn):
outfall = swmmtoolbox.extract('../data/out/{0}'.format(fn), 'system,Flow_leaving_outfalls,11')
maxes[basename] = outfall.max()[0]
outfall.plot(ax=ax[0], legend=False, color='0.75', alpha=0.3, zorder=0)
maxes = pd.Series(maxes).sort_values()
colors = pd.Series(maxes.index.str.split('_').str[0]).map({'linear' : 'r', 'naive' : '0.75', 'uncontrolled' : 'k'})
maxes.plot(ax=ax[1], kind='bar', colors=colors)
ax[0].set_ylabel('Outlet discharge $(m^3/s)$', size=13)
ax[1].set_ylabel('Peak discharge $(m^3/s)$', size=13)
plt.tight_layout()
ax[0].get_xaxis().set_ticklabels([])
ax[0].minorticks_off()
ax[1].get_xaxis().set_ticks([])
ax[0].set_title(r'Outlet discharge, large storm', size=14)
ax[1].set_title(r'Peak discharge', size=14)
custom_lines = [Line2D([0], [0], color='k', lw=2),
Line2D([0], [0], color='0.75', lw=2),
Line2D([0], [0], color='r', lw=2)]
ax[0].legend(custom_lines, ['Uncontrolled', 'Randomized', 'Optimized'], loc=1, fontsize=13)
ax[0].set_xlabel('Time', size=13)
ax[1].set_xlabel('Trials', size=13, labelpad=8)
red_patch = mpatches.Patch(facecolor='r', label='Optimized trial', linewidth=1.2, edgecolor='k')
white_patch = mpatches.Patch(facecolor='0.85', label='Randomized trial', linewidth=1.2, edgecolor='k')
black_patch = mpatches.Patch(facecolor='k', label='Uncontrolled trial', linewidth=1.2, edgecolor='k')
leg1 = ax[1].legend(handles=[red_patch, white_patch, black_patch],
frameon=True, fontsize=13, loc=2)
ax[1].get_legend().get_title().set_fontsize(13)
ax[1].get_legend().get_title().set_fontweight('bold')
ax[1].set_xlim(-1, 52)
plt.tight_layout()
#plt.savefig('../img/performance_max_large.png', bbox_inches='tight', dpi=200)
maxes
var[0] / var[-1]
var[1:-1].mean() / var[-1]
maxes[1] / maxes[-1]
maxes[2:-1].mean() / maxes[-1]
| 0.332202 | 0.349921 |
```
import math, sys, random
import os
import pygame
from pygame.locals import *
from pygame.color import *
import pymunk
from pymunk import Vec2d
import pymunk.pygame_util
width, height = 600,600
collision_types = {
"ball": 1,
"brick": 2,
"bottom": 3,
"player": 4,
}
def spawn_ball(space, position, direction):
ball_body = pymunk.Body(1, pymunk.inf)
ball_body.position = position
ball_shape = pymunk.Circle(ball_body, 5)
ball_shape.color = THECOLORS["green"]
ball_shape.elasticity = 1.0
ball_shape.collision_type = collision_types["ball"]
ball_body.apply_impulse_at_local_point(Vec2d(direction))
# Keep ball velocity at a static value
def constant_velocity(body, gravity, damping, dt):
body.velocity = body.velocity.normalized() * 400
ball_body.velocity_func = constant_velocity
space.add(ball_body, ball_shape)
def setup_level(space, player_body):
# Remove balls and bricks
for s in space.shapes[:]:
if s.body.body_type == pymunk.Body.DYNAMIC and s.body not in [player_body]:
space.remove(s.body, s)
# Spawn a ball for the player to have something to play with
spawn_ball(space, player_body.position + (0,40), random.choice([(1,10),(-1,10)]))
# Spawn bricks
for x in range(0,21):
x = x * 20 + 100
for y in range(0,5):
y = y * 10 + 400
brick_body = pymunk.Body(body_type=pymunk.Body.KINEMATIC)
brick_body.position = x, y
brick_shape = pymunk.Poly.create_box(brick_body, (20,10))
brick_shape.elasticity = 1.0
brick_shape.color = THECOLORS['blue']
brick_shape.group = 1
brick_shape.collision_type = collision_types["brick"]
space.add(brick_body, brick_shape)
# Make bricks be removed when hit by ball
def remove_brick(arbiter, space, data):
brick_shape = arbiter.shapes[0]
space.remove(brick_shape, brick_shape.body)
h = space.add_collision_handler(
collision_types["brick"],
collision_types["ball"])
h.separate = remove_brick
def main():
### PyGame init
pygame.init()
screen = pygame.display.set_mode((width,height))
clock = pygame.time.Clock()
running = True
font = pygame.font.SysFont("Arial", 16)
### Physics stuff
space = pymunk.Space()
draw_options = pymunk.pygame_util.DrawOptions(screen)
### Game area
# walls - the left-top-right walls
static_lines = [pymunk.Segment(space.static_body, (50, 50), (50, 550), 2)
,pymunk.Segment(space.static_body, (50, 550), (550, 550), 2)
,pymunk.Segment(space.static_body, (550, 550), (550, 50), 2)
]
for line in static_lines:
line.color = THECOLORS['lightgray']
line.elasticity = 1.0
space.add(static_lines)
# bottom - a sensor that removes anything touching it
bottom = pymunk.Segment(space.static_body, (50, 50), (550, 50), 2)
bottom.sensor = True
bottom.collision_type = collision_types["bottom"]
bottom.color = THECOLORS['red']
def remove_first(arbiter, space, data):
ball_shape = arbiter.shapes[0]
space.remove(ball_shape, ball_shape.body)
return True
h = space.add_collision_handler(
collision_types["ball"],
collision_types["bottom"])
h.begin = remove_first
space.add(bottom)
### Player ship
player_body = pymunk.Body(500, pymunk.inf)
player_body.position = 300,100
player_shape = pymunk.Segment(player_body, (-50,0), (50,0), 8)
player_shape.color = THECOLORS["red"]
player_shape.elasticity = 1.0
player_shape.collision_type = collision_types["player"]
def pre_solve(arbiter, space, data):
# We want to update the collision normal to make the bounce direction
# dependent of where on the paddle the ball hits. Note that this
# calculation isn't perfect, but just a quick example.
set_ = arbiter.contact_point_set
if len(set_.points) > 0:
player_shape = arbiter.shapes[0]
width = (player_shape.b - player_shape.a).x
delta = (player_shape.body.position - set_.points[0].point_a.x).x
normal = Vec2d(0, 1).rotated(delta / width / 2)
set_.normal = normal
set_.points[0].distance = 0
arbiter.contact_point_set = set_
return True
h = space.add_collision_handler(
collision_types["player"],
collision_types["ball"])
h.pre_solve = pre_solve
# restrict movement of player to a straigt line
move_joint = pymunk.GrooveJoint(space.static_body, player_body, (100,100), (500,100), (0,0))
space.add(player_body, player_shape, move_joint)
global state
# Start game
setup_level(space, player_body)
while running:
for event in pygame.event.get():
if event.type == QUIT:
running = False
elif event.type == KEYDOWN and (event.key in [K_ESCAPE, K_q]):
running = False
elif event.type == KEYDOWN and event.key == K_p:
pygame.image.save(screen, "breakout.png")
elif event.type == KEYDOWN and event.key == K_LEFT:
player_body.velocity = (-600,0)
elif event.type == KEYUP and event.key == K_LEFT:
player_body.velocity = 0,0
elif event.type == KEYDOWN and event.key == K_RIGHT:
player_body.velocity = (600,0)
elif event.type == KEYUP and event.key == K_RIGHT:
player_body.velocity = 0,0
elif event.type == KEYDOWN and event.key == K_r:
setup_level(space, player_body)
elif event.type == KEYDOWN and event.key == K_SPACE:
spawn_ball(space, player_body.position + (0,40), random.choice([(1,10),(-1,10)]))
### Clear screen
screen.fill(THECOLORS["black"])
### Draw stuff
space.debug_draw(draw_options)
state = []
for x in space.shapes:
s = "%s %s %s" % (x, x.body.position, x.body.velocity)
state.append(s)
### Update physics
fps = 60
dt = 1./fps
space.step(dt)
### Info and flip screen
screen.blit(font.render("fps: " + str(clock.get_fps()), 1, THECOLORS["white"]), (0,0))
screen.blit(font.render("Move with left/right arrows, space to spawn a ball", 1, THECOLORS["darkgrey"]), (5,height - 35))
screen.blit(font.render("Press R to reset, ESC or Q to quit", 1, THECOLORS["darkgrey"]), (5,height - 20))
pygame.display.flip()
clock.tick(fps)
if __name__ == '__main__':
sys.exit(main())
```
|
github_jupyter
|
import math, sys, random
import os
import pygame
from pygame.locals import *
from pygame.color import *
import pymunk
from pymunk import Vec2d
import pymunk.pygame_util
width, height = 600,600
collision_types = {
"ball": 1,
"brick": 2,
"bottom": 3,
"player": 4,
}
def spawn_ball(space, position, direction):
ball_body = pymunk.Body(1, pymunk.inf)
ball_body.position = position
ball_shape = pymunk.Circle(ball_body, 5)
ball_shape.color = THECOLORS["green"]
ball_shape.elasticity = 1.0
ball_shape.collision_type = collision_types["ball"]
ball_body.apply_impulse_at_local_point(Vec2d(direction))
# Keep ball velocity at a static value
def constant_velocity(body, gravity, damping, dt):
body.velocity = body.velocity.normalized() * 400
ball_body.velocity_func = constant_velocity
space.add(ball_body, ball_shape)
def setup_level(space, player_body):
# Remove balls and bricks
for s in space.shapes[:]:
if s.body.body_type == pymunk.Body.DYNAMIC and s.body not in [player_body]:
space.remove(s.body, s)
# Spawn a ball for the player to have something to play with
spawn_ball(space, player_body.position + (0,40), random.choice([(1,10),(-1,10)]))
# Spawn bricks
for x in range(0,21):
x = x * 20 + 100
for y in range(0,5):
y = y * 10 + 400
brick_body = pymunk.Body(body_type=pymunk.Body.KINEMATIC)
brick_body.position = x, y
brick_shape = pymunk.Poly.create_box(brick_body, (20,10))
brick_shape.elasticity = 1.0
brick_shape.color = THECOLORS['blue']
brick_shape.group = 1
brick_shape.collision_type = collision_types["brick"]
space.add(brick_body, brick_shape)
# Make bricks be removed when hit by ball
def remove_brick(arbiter, space, data):
brick_shape = arbiter.shapes[0]
space.remove(brick_shape, brick_shape.body)
h = space.add_collision_handler(
collision_types["brick"],
collision_types["ball"])
h.separate = remove_brick
def main():
### PyGame init
pygame.init()
screen = pygame.display.set_mode((width,height))
clock = pygame.time.Clock()
running = True
font = pygame.font.SysFont("Arial", 16)
### Physics stuff
space = pymunk.Space()
draw_options = pymunk.pygame_util.DrawOptions(screen)
### Game area
# walls - the left-top-right walls
static_lines = [pymunk.Segment(space.static_body, (50, 50), (50, 550), 2)
,pymunk.Segment(space.static_body, (50, 550), (550, 550), 2)
,pymunk.Segment(space.static_body, (550, 550), (550, 50), 2)
]
for line in static_lines:
line.color = THECOLORS['lightgray']
line.elasticity = 1.0
space.add(static_lines)
# bottom - a sensor that removes anything touching it
bottom = pymunk.Segment(space.static_body, (50, 50), (550, 50), 2)
bottom.sensor = True
bottom.collision_type = collision_types["bottom"]
bottom.color = THECOLORS['red']
def remove_first(arbiter, space, data):
ball_shape = arbiter.shapes[0]
space.remove(ball_shape, ball_shape.body)
return True
h = space.add_collision_handler(
collision_types["ball"],
collision_types["bottom"])
h.begin = remove_first
space.add(bottom)
### Player ship
player_body = pymunk.Body(500, pymunk.inf)
player_body.position = 300,100
player_shape = pymunk.Segment(player_body, (-50,0), (50,0), 8)
player_shape.color = THECOLORS["red"]
player_shape.elasticity = 1.0
player_shape.collision_type = collision_types["player"]
def pre_solve(arbiter, space, data):
# We want to update the collision normal to make the bounce direction
# dependent of where on the paddle the ball hits. Note that this
# calculation isn't perfect, but just a quick example.
set_ = arbiter.contact_point_set
if len(set_.points) > 0:
player_shape = arbiter.shapes[0]
width = (player_shape.b - player_shape.a).x
delta = (player_shape.body.position - set_.points[0].point_a.x).x
normal = Vec2d(0, 1).rotated(delta / width / 2)
set_.normal = normal
set_.points[0].distance = 0
arbiter.contact_point_set = set_
return True
h = space.add_collision_handler(
collision_types["player"],
collision_types["ball"])
h.pre_solve = pre_solve
# restrict movement of player to a straigt line
move_joint = pymunk.GrooveJoint(space.static_body, player_body, (100,100), (500,100), (0,0))
space.add(player_body, player_shape, move_joint)
global state
# Start game
setup_level(space, player_body)
while running:
for event in pygame.event.get():
if event.type == QUIT:
running = False
elif event.type == KEYDOWN and (event.key in [K_ESCAPE, K_q]):
running = False
elif event.type == KEYDOWN and event.key == K_p:
pygame.image.save(screen, "breakout.png")
elif event.type == KEYDOWN and event.key == K_LEFT:
player_body.velocity = (-600,0)
elif event.type == KEYUP and event.key == K_LEFT:
player_body.velocity = 0,0
elif event.type == KEYDOWN and event.key == K_RIGHT:
player_body.velocity = (600,0)
elif event.type == KEYUP and event.key == K_RIGHT:
player_body.velocity = 0,0
elif event.type == KEYDOWN and event.key == K_r:
setup_level(space, player_body)
elif event.type == KEYDOWN and event.key == K_SPACE:
spawn_ball(space, player_body.position + (0,40), random.choice([(1,10),(-1,10)]))
### Clear screen
screen.fill(THECOLORS["black"])
### Draw stuff
space.debug_draw(draw_options)
state = []
for x in space.shapes:
s = "%s %s %s" % (x, x.body.position, x.body.velocity)
state.append(s)
### Update physics
fps = 60
dt = 1./fps
space.step(dt)
### Info and flip screen
screen.blit(font.render("fps: " + str(clock.get_fps()), 1, THECOLORS["white"]), (0,0))
screen.blit(font.render("Move with left/right arrows, space to spawn a ball", 1, THECOLORS["darkgrey"]), (5,height - 35))
screen.blit(font.render("Press R to reset, ESC or Q to quit", 1, THECOLORS["darkgrey"]), (5,height - 20))
pygame.display.flip()
clock.tick(fps)
if __name__ == '__main__':
sys.exit(main())
| 0.391988 | 0.468791 |
```
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import seaborn as sns
import requests
import time
from pprint import pprint
# Import API key
from config import api_key
# Incorporated citipy to determine city based on latitude and longitude
from citipy import citipy
# Output File (CSV)
output_data_file = "output_data/cities.csv"
# Range of latitudes and longitudes
lat_range = (-90, 90)
lng_range = (-180, 180)
```
## Generate Cities List
```
# List for holding lat_lngs and cities
lat_lngs = []
cities = []
# Create a set of random lat and lng combinations
lats = np.random.uniform(low=-90.000, high=90.000, size=1500)
lngs = np.random.uniform(low=-180.000, high=180.000, size=1500)
lat_lngs = zip(lats, lngs)
# Identify nearest city for each lat, lng combination
for lat_lng in lat_lngs:
city = citipy.nearest_city(lat_lng[0], lat_lng[1]).city_name
# If the city is unique, then add it to a our cities list
if city not in cities:
cities.append(city)
# Print the city count to confirm sufficient count
len(cities)
```
## Perform API Calls
```
# OpenWeatherMap API Key
# Starting URL for Weather Map API Call
def call_city(city, request, api_key):
try:
url = "http://api.openweathermap.org/data/2.5/weather?units=Imperial&appid=" + api_key + "&q=" + city
data = requests.get(url).json()
latitude = data['coord']['lat']
if (request == 'temperature'):
answer = data['main']['temp_max']
elif(request == 'humidity'):
answer = data['main']['humidity']
elif request == 'clouds':
answer = data['clouds']['all']
elif request == 'wind':
answer = data['wind']['speed']
else:
print('That request is not valid!\nUse "temperature" , "humidity", "clouds", or "wind"')
return latitude, answer
except:
return 'NA', "errorhere" + city
def create_df(cities, request, api_key):
df = pd.DataFrame(columns = ['Latitude', request], index = range(len(cities)))
df['Latitude'] = [call_city(city, request, api_key)[0] for city in cities]
df[request] = [call_city(city, request, api_key)[1] for city in cities]
return df
```
## Temperature (F) vs. Latitude
```
temp_df = create_df(cities,'temperature',api_key)
temp_df.head()
temp_df.to_csv('temperatures.csv')
temp_error_indexes = temp_df['Latitude'][temp_df['Latitude'] == 'NA'].index
cleaned_temp = temp_df.drop(temp_df.index[temp_error_indexes])
plt.scatter(cleaned_temp['Latitude'], cleaned_temp['temperature'])
plt.xlabel('Latitude')
plt.ylabel('Temperature (F)')
plt.title('Temperature at Different Latitudes')
sns.set_style("dark")
```
There's clearly a very strong inverse correlation between distance from the equator and temperature. As in, the closer you get to the equator, the hotter it gets. It seems that this trend is a bit stronger in the southern hemisphere.
## Humidity (%) vs. Latitude
```
hum_df = create_df(cities,'humidity',api_key)
hum_df.head()
hum_df.to_csv('humidity.csv')
hum_error_indexes = hum_df['Latitude'][hum_df['Latitude'] == 'NA'].index
cleaned_hum = hum_df.drop(hum_df.index[hum_error_indexes])
plt.scatter(cleaned_hum['Latitude'], cleaned_hum['humidity'])
plt.xlabel('Latitude')
plt.ylabel('Humidity (%)')
plt.title('Humidity at Different Latitudes')
sns.set_style("dark")
```
It appears that there is a slight correlation with higher humidity the closer you get to the equator, albeit far less strong that temperature correlation with latitude. Mostly it appears that this correlation starts around +/- 20 latitude. The closer you get from that point, the more humid it will be. However, further out than that, the humidity appears to have no correlation with latitude.
## Cloudiness (%) vs. Latitude
```
clouds_df = create_df(cities,'clouds',api_key)
clouds_df.head()
clouds_df.to_csv('clouds.csv')
clouds_error_indexes = clouds_df['Latitude'][clouds_df['Latitude'] == 'NA'].index
cleaned_clouds = clouds_df.drop(clouds_df.index[clouds_error_indexes])
plt.scatter(cleaned_clouds['Latitude'], cleaned_clouds['clouds'])
plt.xlabel('Latitude')
plt.ylabel('Clouds (%)')
plt.title('Cloud Pecentage at Different Latitudes')
sns.set_style("dark")
```
There is clearly no correlation between cloud percentage and different latitudes. However, the data looks discrete, as evidenced by the neat rows. The cloud density is probably estimated by percentages of 5 or 2.5.
## Wind Speed (mph) vs. Latitude
```
wind_df = create_df(cities,'wind',api_key)
wind_df.head()
wind_df.to_csv('wind.csv')
wind_error_indexes = wind_df['Latitude'][wind_df['Latitude'] == 'NA'].index
cleaned_wind = wind_df.drop(wind_df.index[wind_error_indexes])
plt.scatter(cleaned_wind['Latitude'], cleaned_wind['wind'])
plt.xlabel('Latitude')
plt.ylabel('Wind Speed (mph)')
plt.title('Wind Speed at Different Latitudes')
sns.set_style("dark")
```
Clearly here, there is absolutely no correlation.
|
github_jupyter
|
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import seaborn as sns
import requests
import time
from pprint import pprint
# Import API key
from config import api_key
# Incorporated citipy to determine city based on latitude and longitude
from citipy import citipy
# Output File (CSV)
output_data_file = "output_data/cities.csv"
# Range of latitudes and longitudes
lat_range = (-90, 90)
lng_range = (-180, 180)
# List for holding lat_lngs and cities
lat_lngs = []
cities = []
# Create a set of random lat and lng combinations
lats = np.random.uniform(low=-90.000, high=90.000, size=1500)
lngs = np.random.uniform(low=-180.000, high=180.000, size=1500)
lat_lngs = zip(lats, lngs)
# Identify nearest city for each lat, lng combination
for lat_lng in lat_lngs:
city = citipy.nearest_city(lat_lng[0], lat_lng[1]).city_name
# If the city is unique, then add it to a our cities list
if city not in cities:
cities.append(city)
# Print the city count to confirm sufficient count
len(cities)
# OpenWeatherMap API Key
# Starting URL for Weather Map API Call
def call_city(city, request, api_key):
try:
url = "http://api.openweathermap.org/data/2.5/weather?units=Imperial&appid=" + api_key + "&q=" + city
data = requests.get(url).json()
latitude = data['coord']['lat']
if (request == 'temperature'):
answer = data['main']['temp_max']
elif(request == 'humidity'):
answer = data['main']['humidity']
elif request == 'clouds':
answer = data['clouds']['all']
elif request == 'wind':
answer = data['wind']['speed']
else:
print('That request is not valid!\nUse "temperature" , "humidity", "clouds", or "wind"')
return latitude, answer
except:
return 'NA', "errorhere" + city
def create_df(cities, request, api_key):
df = pd.DataFrame(columns = ['Latitude', request], index = range(len(cities)))
df['Latitude'] = [call_city(city, request, api_key)[0] for city in cities]
df[request] = [call_city(city, request, api_key)[1] for city in cities]
return df
temp_df = create_df(cities,'temperature',api_key)
temp_df.head()
temp_df.to_csv('temperatures.csv')
temp_error_indexes = temp_df['Latitude'][temp_df['Latitude'] == 'NA'].index
cleaned_temp = temp_df.drop(temp_df.index[temp_error_indexes])
plt.scatter(cleaned_temp['Latitude'], cleaned_temp['temperature'])
plt.xlabel('Latitude')
plt.ylabel('Temperature (F)')
plt.title('Temperature at Different Latitudes')
sns.set_style("dark")
hum_df = create_df(cities,'humidity',api_key)
hum_df.head()
hum_df.to_csv('humidity.csv')
hum_error_indexes = hum_df['Latitude'][hum_df['Latitude'] == 'NA'].index
cleaned_hum = hum_df.drop(hum_df.index[hum_error_indexes])
plt.scatter(cleaned_hum['Latitude'], cleaned_hum['humidity'])
plt.xlabel('Latitude')
plt.ylabel('Humidity (%)')
plt.title('Humidity at Different Latitudes')
sns.set_style("dark")
clouds_df = create_df(cities,'clouds',api_key)
clouds_df.head()
clouds_df.to_csv('clouds.csv')
clouds_error_indexes = clouds_df['Latitude'][clouds_df['Latitude'] == 'NA'].index
cleaned_clouds = clouds_df.drop(clouds_df.index[clouds_error_indexes])
plt.scatter(cleaned_clouds['Latitude'], cleaned_clouds['clouds'])
plt.xlabel('Latitude')
plt.ylabel('Clouds (%)')
plt.title('Cloud Pecentage at Different Latitudes')
sns.set_style("dark")
wind_df = create_df(cities,'wind',api_key)
wind_df.head()
wind_df.to_csv('wind.csv')
wind_error_indexes = wind_df['Latitude'][wind_df['Latitude'] == 'NA'].index
cleaned_wind = wind_df.drop(wind_df.index[wind_error_indexes])
plt.scatter(cleaned_wind['Latitude'], cleaned_wind['wind'])
plt.xlabel('Latitude')
plt.ylabel('Wind Speed (mph)')
plt.title('Wind Speed at Different Latitudes')
sns.set_style("dark")
| 0.378574 | 0.783699 |
```
library(rdmc)
library(tidyverse)
library(ape)
theme_set(cowplot::theme_cowplot(15))
library(patchwork)
options(repr.plot.width = 10, repr.plot.height = 7, repr.plot.res = 200)
gen_map_all_chr <- read_delim("../data/map/ogut_v5.map.txt", delim = "\t") %>%
drop_na() %>%
mutate(cm = cm + abs(min(cm))) %>%
group_by(chr) %>%
group_modify(~{
df1 <- slice(.x, -nrow(.x))
df2 <- slice(.x, -1)
to_keep <- df2$cm > df1$cm & df2$pos > df1$pos
df1 <- df1[to_keep, ]
df2 <- df2[to_keep, ]
cm_mb <- tibble(cm_mb = 1e6*(df2$cm - df1$cm)/(df2$pos - df1$pos))
cm_bp <- tibble(rr = (df2$cm - df1$cm)/(df2$pos - df1$pos)/100)
bind_cols(df2, cm_mb, cm_bp)
}) %>%
mutate(chr = paste0("chr", chr))
median(gen_map_all_chr$rr)
get_rr <- function(genetic_df, sweep_chr, sweep_positions){
chr_df <- filter(genetic_df, chr == sweep_chr)
median(approx(x = chr_df$pos, y = chr_df$rr, xout = sweep_positions)$y)
}
get_cm <- function(genetic_df, sweep_chr, sweep_start, sweep_end){
chr_df <- filter(genetic_df, chr == sweep_chr)
cm_start <- approx(x = chr_df$pos, y = chr_df$cm, xout = sweep_start)$y
cm_end <- approx(x = chr_df$pos, y = chr_df$cm, xout = sweep_end)$y
cm_end - cm_start
}
gmap <- "../data/map/ogut_v5.map.txt"
gen_map_all_chr <- vroom::vroom(gmap, delim = "\t") %>%
drop_na() %>%
mutate(cm = cm + abs(min(cm))) %>%
group_by(chr) %>%
group_modify(~{
df1 <- slice(.x, -nrow(.x))
df2 <- slice(.x, -1)
to_keep <- df2$cm > df1$cm & df2$pos > df1$pos
df1 <- df1[to_keep, ]
df2 <- df2[to_keep, ]
cm_mb <- tibble(cm_mb = 1e6*(df2$cm - df1$cm)/(df2$pos - df1$pos))
cm_bp <- tibble(rr = (df2$cm - df1$cm)/(df2$pos - df1$pos)/100)
bind_cols(df2, cm_mb, cm_bp)
}) %>%
ungroup() %>%
mutate(chr = paste0("chr", chr))
median(gen_map_all_chr$rr)
#how to get a constant density of samples over sweeps that vary in their **genetic** size (not physical)
#get_cm(gen_map_all_chr, "chr1", sweep_freqs$end[1], tail(sweep_freqs$end, 1))
MIN_FREQ <- 1/20
DEFAULT_SITES <- 1e4
MAX_SITES <- 1e5
MIN_SITES <- 1e3
SNP_K <- 250000
s_file <- "../data/rdmc/sweep_freq/v5--sweep--chr1--0--308452471_start31723921_end31727430_pops3-4.txt"
start <- str_split(s_file, "start", simplify = TRUE) %>%
`[`(2) %>%
str_split("_", simplify = TRUE) %>%
`[`(1) %>%
as.numeric(c)
end <- str_split(s_file, "end", simplify = TRUE) %>%
`[`(2) %>%
str_split("_", simplify = TRUE) %>%
`[`(1) %>%
as.numeric(c)
sweep_cM <- get_cm(gen_map_all_chr, "chr1", start, end)
n_snps <- round(SNP_K*sweep_cM)
if(is.na(n_snps)) n_snps <- DEFAULT_SITES
n_sites <- case_when(
is.na(n_snps) ~ NA_real_,
n_snps >= MIN_SITES && n_snps <= MAX_SITES ~ n_snps,
n_snps < MIN_SITES ~ MIN_SITES,
n_snps > MAX_SITES ~ MAX_SITES,
)
n_snps
print("here")
vroom::vroom(file = s_file,
delim = "\t",
col_names = FREQ_POPS) %>% head()
FREQ_POPS = c(
"chrom",
"start",
"end",
"v5--LR--Amatlan_de_Canas",
"v5--LR--Crucero_Lagunitas",
"v5--LR--Los_Guajes",
"v5--LR--random1_Palmar_Chico",
"v5--LR--San_Lorenzo",
"v5--Teo--Amatlan_de_Canas",
"v5--Teo--Crucero_Lagunitas",
"v5--Teo--El_Rodeo",
"v5--Teo--Los_Guajes",
"v5--Teo--random1_Palmar_Chico",
"v5--Teo--San_Lorenzo"
)
MIN_FREQ <- 1/20
neutral_freqs <- vroom::vroom(file = "../data/rdmc/v5--neutral_freqs.txt",
delim = "\t",
col_names = FREQ_POPS) %>%
mutate(varz = apply(select(., -c(chrom, start, end)), 1, max)) %>%
filter(varz >= MIN_FREQ) %>%
sample_n(50000) %>%
select(-varz)
s_file <- "../data/rdmc/sweep_freq/v5--sweep--chr10--0--152435371_start100234619_end100464359_pops1-4-10.txt"
#s_file <- "../test_chr_start7110396592_end110877024_1MB_buffer_sweep_pops1-2-3-4-5.txt"
#s_file <- "../sweep_test.txt"
#str_split(s_file, "pops", simplify = FALSE)
sel_vec <- str_split(s_file, "pops", simplify = TRUE)[,2] %>%
str_remove(".txt") %>%
str_split("-", simplify = TRUE) %>%
as_vector() %>%
as.numeric()
#sel_vec <- c(1,4,8)
sel_vec
sweep_freqs <- vroom::vroom(file = s_file,
delim = "\t",
col_names = FREQ_POPS) %>%
mutate(varz = apply(select(., -c(chrom, start, end)), 1, max)) %>%
filter(varz >= MIN_FREQ) %>%
select(-varz) %>%
sample_n(min(nrow(.), n_sites)) %>%
arrange(start)
nrow(neutral_freqs)
nrow(sweep_freqs)
sweep_freqs[0:10, ]
#sweep_freqs %>%
# apply(select(., -c(chrom, start, end)), 1, var)
# apply(., 1, var)
pop_types <- (names(neutral_freqs)[-c(1:3)] %>% str_remove_all('v5--') %>% str_split('--', simplify = TRUE))[,1]
pops <- (names(neutral_freqs)[-c(1:3)] %>% str_remove_all('v5--') %>% str_split('--', simplify = TRUE))[,2]
pop_pc <- neutral_freqs %>%
select(-c(chrom, start, end)) %>%
t() %>%
prcomp()
plot(pop_pc$x[,1], pop_pc$x[,2], bg = factor(pops), pch = 21, cex = 2)
text(pop_pc$x[,1], pop_pc$x[,2], pops)
dist_mat <- neutral_freqs %>%
select(-c(chrom, start, end)) %>%
t() %>%
dist()
plot.phylo(nj(dist_mat), type = "unrooted")
pos_vec <- select(sweep_freqs, end) %>% pull(end)
sweep_mat <- sweep_freqs %>%
select(-c(chrom, start, end)) %>%
t()
neut_mat <-
neutral_freqs %>%
select(-c(chrom, start, end)) %>%
t()
rr <- get_rr(gen_map_all_chr, "chr1", sweep_freqs$end)
param_list <-
parameter_barge(
Ne = 50000,
rec = rr,
neutral_freqs = neut_mat,
selected_freqs = sweep_mat,
selected_pops = sel_vec,
positions = pos_vec,
n_sites = 20,
sample_sizes = rep(10, nrow(neut_mat)),
num_bins = 1000,
sels = 10^seq(-5, -1, length.out = 15),
times = c(1e2, 1e3, 1e4, 1e5),
gs = 10^seq(-3, -1, length.out = 3),
migs = 10^(seq(-3, -1, length.out = 2)),
sources = sel_vec,
locus_name = s_file,
cholesky = TRUE
)
rep(mean(apply(param_list$allFreqs, 1, mean)), 11)
quantile(apply(mvtnorm::rmvnorm(n = 10000, mean = rep(mean(apply(param_list$allFreqs, 1, mean)), 11), sigma = param_list$F_estimate), 2, sd), 0.01)
mode_wrapper <- function(barge, mode) {
cle_out <- try(mode_cle(barge, mode))
if(class(cle_out)[1] == 'try-error'){
barge$cholesky <- FALSE
cle_out <- suppressWarnings(mode_cle(barge, mode))
barge$cholesky <- TRUE
}
return(cle_out)
}
t <- Sys.time()
#fit composite likelihood models
print("neutral")
neut_cle <- mode_wrapper(param_list, mode = "neutral")
print("ind")
ind_cle <- mode_wrapper(param_list, mode = "independent")
print("standing")
sv_cle <- mode_wrapper(param_list, mode = "standing")
print("mig")
mig_cle <- mode_wrapper(param_list, mode = "migration")
Sys.time() - t
head(neut_cle)
#neut <- unique(neut_cle$cle)
#merge data frame of all fit models
all_mods <-
bind_rows(
ind_cle,
mig_cle,
sv_cle
) %>%
mutate(sel_pop_ids = paste(FREQ_POPS[sel_vec+3], collapse = "; "),
neut_cle = unique(neut_cle$cle))
#max composite likelihood estimate
#of all params over all models
all_mods %>%
group_by(model) %>%
filter(cle == max(cle,na.rm=T)) %>%
mutate(mcle = cle - neut_cle) %>%
ungroup() %>%
mutate(mcle_delta = mcle - max(mcle)) %>%
arrange(desc(mcle))
best_mcle <- all_mods %>%
group_by(model) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ungroup() %>%
arrange(desc(mcle))
(best_mod <- best_mcle %>% slice(1) %>% pull(model))
#best_mod <- "standing"
dim(neut_cle)
all_mods %>%
group_by(selected_sites, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(selected_sites, mcle, colour = model)) +
geom_line() +
geom_point() +
xlab("Position") +
ylab("Composite likelihood") +
scale_color_brewer(palette = "Set1")
#visualize likelihood surface wrt selection coefficients
all_mods %>%
group_by(sels, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(log10(sels), mcle, colour = model)) +
geom_line() +
geom_point() +
ylab("Composite likelihood") +
xlab("Selection coefficient") +
scale_color_brewer(palette = "Set1")
if (best_mod == "standing"){
#visualize likelihood surface wrt age
a <- all_mods %>%
group_by(times, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(log10(times), mcle, colour = model)) +
geom_line() +
geom_point() +
ylab("Composite likelihood") +
xlab("Age") +
scale_color_brewer(palette = "Set1")
#visualize likelihood surface wrt age
b <- all_mods %>%
group_by(gs, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(gs, mcle, colour = model)) +
geom_line() +
geom_point() +
ylab("Composite likelihood") +
xlab("Initial allele frequency") +
scale_color_brewer(palette = "Set1")
a / b
} else if(best_mod == "migration"){
a <- all_mods %>%
group_by(migs, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(log10(migs), mcle, colour = model)) +
geom_line() +
geom_point() +
ylab("Composite likelihood") +
xlab("Migration rate") +
scale_color_brewer(palette = "Set1")
b <- all_mods %>%
group_by(sources, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(factor(sources), mcle, colour = model)) +
geom_point(size = 3) +
ylab("Composite likelihood") +
xlab("Source pop") +
scale_color_brewer(palette = "Set1")
a+b
}
sel_vec
paste(FREQ_POPS[sel_vec+3], collapse = "; ")
```
|
github_jupyter
|
library(rdmc)
library(tidyverse)
library(ape)
theme_set(cowplot::theme_cowplot(15))
library(patchwork)
options(repr.plot.width = 10, repr.plot.height = 7, repr.plot.res = 200)
gen_map_all_chr <- read_delim("../data/map/ogut_v5.map.txt", delim = "\t") %>%
drop_na() %>%
mutate(cm = cm + abs(min(cm))) %>%
group_by(chr) %>%
group_modify(~{
df1 <- slice(.x, -nrow(.x))
df2 <- slice(.x, -1)
to_keep <- df2$cm > df1$cm & df2$pos > df1$pos
df1 <- df1[to_keep, ]
df2 <- df2[to_keep, ]
cm_mb <- tibble(cm_mb = 1e6*(df2$cm - df1$cm)/(df2$pos - df1$pos))
cm_bp <- tibble(rr = (df2$cm - df1$cm)/(df2$pos - df1$pos)/100)
bind_cols(df2, cm_mb, cm_bp)
}) %>%
mutate(chr = paste0("chr", chr))
median(gen_map_all_chr$rr)
get_rr <- function(genetic_df, sweep_chr, sweep_positions){
chr_df <- filter(genetic_df, chr == sweep_chr)
median(approx(x = chr_df$pos, y = chr_df$rr, xout = sweep_positions)$y)
}
get_cm <- function(genetic_df, sweep_chr, sweep_start, sweep_end){
chr_df <- filter(genetic_df, chr == sweep_chr)
cm_start <- approx(x = chr_df$pos, y = chr_df$cm, xout = sweep_start)$y
cm_end <- approx(x = chr_df$pos, y = chr_df$cm, xout = sweep_end)$y
cm_end - cm_start
}
gmap <- "../data/map/ogut_v5.map.txt"
gen_map_all_chr <- vroom::vroom(gmap, delim = "\t") %>%
drop_na() %>%
mutate(cm = cm + abs(min(cm))) %>%
group_by(chr) %>%
group_modify(~{
df1 <- slice(.x, -nrow(.x))
df2 <- slice(.x, -1)
to_keep <- df2$cm > df1$cm & df2$pos > df1$pos
df1 <- df1[to_keep, ]
df2 <- df2[to_keep, ]
cm_mb <- tibble(cm_mb = 1e6*(df2$cm - df1$cm)/(df2$pos - df1$pos))
cm_bp <- tibble(rr = (df2$cm - df1$cm)/(df2$pos - df1$pos)/100)
bind_cols(df2, cm_mb, cm_bp)
}) %>%
ungroup() %>%
mutate(chr = paste0("chr", chr))
median(gen_map_all_chr$rr)
#how to get a constant density of samples over sweeps that vary in their **genetic** size (not physical)
#get_cm(gen_map_all_chr, "chr1", sweep_freqs$end[1], tail(sweep_freqs$end, 1))
MIN_FREQ <- 1/20
DEFAULT_SITES <- 1e4
MAX_SITES <- 1e5
MIN_SITES <- 1e3
SNP_K <- 250000
s_file <- "../data/rdmc/sweep_freq/v5--sweep--chr1--0--308452471_start31723921_end31727430_pops3-4.txt"
start <- str_split(s_file, "start", simplify = TRUE) %>%
`[`(2) %>%
str_split("_", simplify = TRUE) %>%
`[`(1) %>%
as.numeric(c)
end <- str_split(s_file, "end", simplify = TRUE) %>%
`[`(2) %>%
str_split("_", simplify = TRUE) %>%
`[`(1) %>%
as.numeric(c)
sweep_cM <- get_cm(gen_map_all_chr, "chr1", start, end)
n_snps <- round(SNP_K*sweep_cM)
if(is.na(n_snps)) n_snps <- DEFAULT_SITES
n_sites <- case_when(
is.na(n_snps) ~ NA_real_,
n_snps >= MIN_SITES && n_snps <= MAX_SITES ~ n_snps,
n_snps < MIN_SITES ~ MIN_SITES,
n_snps > MAX_SITES ~ MAX_SITES,
)
n_snps
print("here")
vroom::vroom(file = s_file,
delim = "\t",
col_names = FREQ_POPS) %>% head()
FREQ_POPS = c(
"chrom",
"start",
"end",
"v5--LR--Amatlan_de_Canas",
"v5--LR--Crucero_Lagunitas",
"v5--LR--Los_Guajes",
"v5--LR--random1_Palmar_Chico",
"v5--LR--San_Lorenzo",
"v5--Teo--Amatlan_de_Canas",
"v5--Teo--Crucero_Lagunitas",
"v5--Teo--El_Rodeo",
"v5--Teo--Los_Guajes",
"v5--Teo--random1_Palmar_Chico",
"v5--Teo--San_Lorenzo"
)
MIN_FREQ <- 1/20
neutral_freqs <- vroom::vroom(file = "../data/rdmc/v5--neutral_freqs.txt",
delim = "\t",
col_names = FREQ_POPS) %>%
mutate(varz = apply(select(., -c(chrom, start, end)), 1, max)) %>%
filter(varz >= MIN_FREQ) %>%
sample_n(50000) %>%
select(-varz)
s_file <- "../data/rdmc/sweep_freq/v5--sweep--chr10--0--152435371_start100234619_end100464359_pops1-4-10.txt"
#s_file <- "../test_chr_start7110396592_end110877024_1MB_buffer_sweep_pops1-2-3-4-5.txt"
#s_file <- "../sweep_test.txt"
#str_split(s_file, "pops", simplify = FALSE)
sel_vec <- str_split(s_file, "pops", simplify = TRUE)[,2] %>%
str_remove(".txt") %>%
str_split("-", simplify = TRUE) %>%
as_vector() %>%
as.numeric()
#sel_vec <- c(1,4,8)
sel_vec
sweep_freqs <- vroom::vroom(file = s_file,
delim = "\t",
col_names = FREQ_POPS) %>%
mutate(varz = apply(select(., -c(chrom, start, end)), 1, max)) %>%
filter(varz >= MIN_FREQ) %>%
select(-varz) %>%
sample_n(min(nrow(.), n_sites)) %>%
arrange(start)
nrow(neutral_freqs)
nrow(sweep_freqs)
sweep_freqs[0:10, ]
#sweep_freqs %>%
# apply(select(., -c(chrom, start, end)), 1, var)
# apply(., 1, var)
pop_types <- (names(neutral_freqs)[-c(1:3)] %>% str_remove_all('v5--') %>% str_split('--', simplify = TRUE))[,1]
pops <- (names(neutral_freqs)[-c(1:3)] %>% str_remove_all('v5--') %>% str_split('--', simplify = TRUE))[,2]
pop_pc <- neutral_freqs %>%
select(-c(chrom, start, end)) %>%
t() %>%
prcomp()
plot(pop_pc$x[,1], pop_pc$x[,2], bg = factor(pops), pch = 21, cex = 2)
text(pop_pc$x[,1], pop_pc$x[,2], pops)
dist_mat <- neutral_freqs %>%
select(-c(chrom, start, end)) %>%
t() %>%
dist()
plot.phylo(nj(dist_mat), type = "unrooted")
pos_vec <- select(sweep_freqs, end) %>% pull(end)
sweep_mat <- sweep_freqs %>%
select(-c(chrom, start, end)) %>%
t()
neut_mat <-
neutral_freqs %>%
select(-c(chrom, start, end)) %>%
t()
rr <- get_rr(gen_map_all_chr, "chr1", sweep_freqs$end)
param_list <-
parameter_barge(
Ne = 50000,
rec = rr,
neutral_freqs = neut_mat,
selected_freqs = sweep_mat,
selected_pops = sel_vec,
positions = pos_vec,
n_sites = 20,
sample_sizes = rep(10, nrow(neut_mat)),
num_bins = 1000,
sels = 10^seq(-5, -1, length.out = 15),
times = c(1e2, 1e3, 1e4, 1e5),
gs = 10^seq(-3, -1, length.out = 3),
migs = 10^(seq(-3, -1, length.out = 2)),
sources = sel_vec,
locus_name = s_file,
cholesky = TRUE
)
rep(mean(apply(param_list$allFreqs, 1, mean)), 11)
quantile(apply(mvtnorm::rmvnorm(n = 10000, mean = rep(mean(apply(param_list$allFreqs, 1, mean)), 11), sigma = param_list$F_estimate), 2, sd), 0.01)
mode_wrapper <- function(barge, mode) {
cle_out <- try(mode_cle(barge, mode))
if(class(cle_out)[1] == 'try-error'){
barge$cholesky <- FALSE
cle_out <- suppressWarnings(mode_cle(barge, mode))
barge$cholesky <- TRUE
}
return(cle_out)
}
t <- Sys.time()
#fit composite likelihood models
print("neutral")
neut_cle <- mode_wrapper(param_list, mode = "neutral")
print("ind")
ind_cle <- mode_wrapper(param_list, mode = "independent")
print("standing")
sv_cle <- mode_wrapper(param_list, mode = "standing")
print("mig")
mig_cle <- mode_wrapper(param_list, mode = "migration")
Sys.time() - t
head(neut_cle)
#neut <- unique(neut_cle$cle)
#merge data frame of all fit models
all_mods <-
bind_rows(
ind_cle,
mig_cle,
sv_cle
) %>%
mutate(sel_pop_ids = paste(FREQ_POPS[sel_vec+3], collapse = "; "),
neut_cle = unique(neut_cle$cle))
#max composite likelihood estimate
#of all params over all models
all_mods %>%
group_by(model) %>%
filter(cle == max(cle,na.rm=T)) %>%
mutate(mcle = cle - neut_cle) %>%
ungroup() %>%
mutate(mcle_delta = mcle - max(mcle)) %>%
arrange(desc(mcle))
best_mcle <- all_mods %>%
group_by(model) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ungroup() %>%
arrange(desc(mcle))
(best_mod <- best_mcle %>% slice(1) %>% pull(model))
#best_mod <- "standing"
dim(neut_cle)
all_mods %>%
group_by(selected_sites, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(selected_sites, mcle, colour = model)) +
geom_line() +
geom_point() +
xlab("Position") +
ylab("Composite likelihood") +
scale_color_brewer(palette = "Set1")
#visualize likelihood surface wrt selection coefficients
all_mods %>%
group_by(sels, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(log10(sels), mcle, colour = model)) +
geom_line() +
geom_point() +
ylab("Composite likelihood") +
xlab("Selection coefficient") +
scale_color_brewer(palette = "Set1")
if (best_mod == "standing"){
#visualize likelihood surface wrt age
a <- all_mods %>%
group_by(times, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(log10(times), mcle, colour = model)) +
geom_line() +
geom_point() +
ylab("Composite likelihood") +
xlab("Age") +
scale_color_brewer(palette = "Set1")
#visualize likelihood surface wrt age
b <- all_mods %>%
group_by(gs, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(gs, mcle, colour = model)) +
geom_line() +
geom_point() +
ylab("Composite likelihood") +
xlab("Initial allele frequency") +
scale_color_brewer(palette = "Set1")
a / b
} else if(best_mod == "migration"){
a <- all_mods %>%
group_by(migs, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(log10(migs), mcle, colour = model)) +
geom_line() +
geom_point() +
ylab("Composite likelihood") +
xlab("Migration rate") +
scale_color_brewer(palette = "Set1")
b <- all_mods %>%
group_by(sources, model) %>%
filter(model == best_mod) %>%
summarise(mcle = max(cle, na.rm = T) - neut_cle) %>%
ggplot(aes(factor(sources), mcle, colour = model)) +
geom_point(size = 3) +
ylab("Composite likelihood") +
xlab("Source pop") +
scale_color_brewer(palette = "Set1")
a+b
}
sel_vec
paste(FREQ_POPS[sel_vec+3], collapse = "; ")
| 0.2819 | 0.311859 |
# 字符串
## 1. 字符串的定义
- Python 中字符串被定义为引号之间的字符集合。
- Python 支持使用成对的 单引号 或 双引号。
```
t1 = 'i love Python!'
print(t1, type(t1))
# i love Python! <class 'str'>
t2 = "I love Python!"
print(t2, type(t2))
# I love Python! <class 'str'>
print(5 + 8) # 13
print('5' + '8') # 58
```
- Python 的常用转义字符
**转义字符** | **描述**
- \\ | 反斜杠符号
- \' | 单引号
- \" | 双引号
- \n | 换行
- \t | 横向制表符(TAB)
- \r | 回车
如果字符串中需要出现单引号或双引号,可以使用转义符号\对字符串中的符号进行转义。
```
print('let\'s go') # let's go
print("let's go") # let's go
print('C:\\now') # C:\now
print("C:\\Program Files\\Intel\\Wifi\\Help")
# C:\Program Files\Intel\Wifi\Help
```
原始字符串只需要在字符串前边加一个英文字母 r 即可。
```
print(r'C:\Program Files\Intel\Wifi\Help')
# C:\Program Files\Intel\Wifi\Help
print(r'C:\Program Files\Intel\Wifi\Help')
```
三引号允许一个字符串跨多行,字符串中可以包含换行符、制表符以及其他特殊字符。
```
para_str = """这是一个多行字符串的实例
多行字符串可以使用制表符
TAB ( \t )。
也可以使用换行符 [ \n ]。
"""
print(para_str)
# 这是一个多行字符串的实例
# 多行字符串可以使用制表符
# TAB ( )。
# 也可以使用换行符 [
# ]。
para_str = '''这是一个多行字符串的实例
多行字符串可以使用制表符
TAB ( \t )。
也可以使用换行符 [ \n ]。
'''
print(para_str)
# 这是一个多行字符串的实例
# 多行字符串可以使用制表符
# TAB ( )。
# 也可以使用换行符 [
# ]。
```
## 2. 字符串的切片与拼接
- 类似于元组具有不可修改性
- 从 0 开始 (和 Java 一样)
- 切片通常写成 start:end 这种形式,包括「start 索引」对应的元素,不包括「end索引」对应的元素。
- 索引值可正可负,正索引从 0 开始,从左往右;负索引从 -1 开始,从右往左。使用负数索引时,会从最后一个元素开始计数。最后一个元素的位置编号是 -1。
**左开右闭**
```
str1 = 'I Love LsgoGroup'
print(str1[:6]) # I Love
print(str1[5]) # e
print(str1[:6] + " 插入的字符串 " + str1[6:])
# I Love 插入的字符串 LsgoGroup
s = 'Python'
print(s) # Python
print(s[2:4]) # th
print(s[-5:-2]) # yth
print(s[2]) # t
print(s[-1]) # n
```
## 3. 字符串的常用内置方法
capitalize() 将字符串的第一个字符转换为大写。
```
str2 = 'xiaoxie'
print(str2.capitalize()) # Xiaoxie
```
- ower() 转换字符串中所有大写字符为小写。
- upper() 转换字符串中的小写字母为大写。
- swapcase() 将字符串中大写转换为小写,小写转换为大写。
```
str2 = "DAXIExiaoxie"
print(str2.lower()) # daxiexiaoxie
print(str2.upper()) # DAXIEXIAOXIE
print(str2.swapcase()) # daxieXIAOXIE
```
count(str, beg= 0,end=len(string)) 返回str在 string 里面出现的次数,如果beg或者end指定则返回指定范围内str出现的次数。
```
str2 = "DAXIExiaoxie"
print(str2.count('xi')) # 2
```
- endswith(suffix, beg=0, end=len(string)) 检查字符串是否以指定子字符串 suffix 结束,如果是,返回 True,否则返回 False。如果 beg 和 end 指定值,则在指定范围内检查。
- startswith(substr, beg=0,end=len(string)) 检查字符串是否以指定子字符串 substr 开头,如果是,返回 True,否则返回 False。如果 beg 和 end 指定值,则在指定范围内检查。
```
str2 = "DAXIExiaoxie"
print(str2.endswith('ie')) # True
print(str2.endswith('xi')) # False
print(str2.startswith('Da')) # False
print(str2.startswith('DA')) # True
```
- find(str, beg=0, end=len(string)) 检测 str 是否包含在字符串中,如果指定范围 beg 和 end,则检查是否包含在指定范围内,如果包含,返回开始的索引值,否则返回 -1。
- rfind(str, beg=0,end=len(string)) 类似于 find() 函数,不过是从右边开始查找
```
str2 = "DAXIExiaoxie"
print(str2.find('xi')) # 5
print(str2.find('ix')) # -1
print(str2.rfind('xi')) # 9
```
isnumeric() 如果字符串中只包含数字字符,则返回 True,否则返回 False。
```
str3 = '12345'
print(str3.isnumeric()) # True
str3 += 'a'
print(str3.isnumeric()) # False
```
- ljust(width[, fillchar])返回一个原字符串左对齐,并使用fillchar(默认空格)填充至长度width的新字符串。
- rjust(width[, fillchar])返回一个原字符串右对齐,并使用fillchar(默认空格)填充至长度width的新字符串。
```
str4 = '1101'
print(str4.ljust(8, '0')) # 11010000
print(str4.rjust(8, '0')) # 00001101
```
- lstrip([chars]) 截掉字符串左边的空格或指定字符。
- rstrip([chars]) 删除字符串末尾的空格或指定字符。
- strip([chars]) 在字符串上执行lstrip()和rstrip()。
```
str5 = ' I Love LsgoGroup '
print(str5.lstrip()) # 'I Love LsgoGroup '
print(str5.lstrip().strip('I')) # ' Love LsgoGroup '
print(str5.rstrip()) # ' I Love LsgoGroup'
print(str5.strip()) # 'I Love LsgoGroup'
print(str5.strip().strip('p')) # 'I Love LsgoGrou'
```
- partition(sub) 找到子字符串sub,把字符串分为一个三元组(pre_sub,sub,fol_sub),如果字符串中不包含sub则返回('原字符串','','')。
- rpartition(sub)类似于partition()方法,不过是从右边开始查找。
```
str5 = ' I Love LsgoGroup '
print(str5.strip().partition('o')) # ('I L', 'o', 've LsgoGroup')
print(str5.strip().partition('m')) # ('I Love LsgoGroup', '', '')
print(str5.strip().rpartition('o')) # ('I Love LsgoGr', 'o', 'up')
```
- replace(old, new [, max]) 把 将字符串中的old替换成new,如果max指定,则替换不超过max次。
```
str5 = ' I Love LsgoGroup '
print(str5.strip().replace('I', 'We')) # We Love LsgoGroup
```
- **split**(str="", num) 不带参数默认是以空格为分隔符切片字符串,如果num参数有设置,则仅分隔num个子字符串,返回切片后的子字符串拼接的列表。
```
str5 = ' I Love LsgoGroup '
print(str5.strip().split()) # ['I', 'Love', 'LsgoGroup']
print(str5.strip().split('o')) # ['I L', 've Lsg', 'Gr', 'up']
u = "www.baidu.com.cn"
# 使用默认分隔符
print(u.split()) # ['www.baidu.com.cn']
# 以"."为分隔符
print((u.split('.'))) # ['www', 'baidu', 'com', 'cn']
# 分割0次
print((u.split(".", 0))) # ['www.baidu.com.cn']
# 分割一次
print((u.split(".", 1))) # ['www', 'baidu.com.cn']
# 分割两次
print(u.split(".", 2)) # ['www', 'baidu', 'com.cn']
# 分割两次,并取序列为1的项
print((u.split(".", 2)[1])) # baidu
# 分割两次,并把分割后的三个部分保存到三个变量
u1, u2, u3 = u.split(".", 2)
print(u1) # www
print(u2) # baidu
print(u3) # com.cn
c = '''say
hello
baby'''
print(c)
# say
# hello
# baby
print(c.split('\n')) # ['say', 'hello', 'baby']
string = "hello boy<[www.baidu.com]>byebye"
# 先分割成"hello boy<", "www.baidu.com]>byebye",取1, 即为www.baidu.com]>byebye,
# 再根据]分割成"www.baidu.com>","byebye", 取0, 即为www.baidu.com
print(string.split('[')[1].split(']')[0]) # www.baidu.com
# 在第一题的基础上再根据.进行分割,得到三个部分
print(string.split('[')[1].split(']')[0].split('.')) # ['www', 'baidu', 'com']
```
- splitlines([keepends]) 按照行('\r', '\r\n', \n')分隔,返回一个包含各行作为元素的列表,如果参数keepends为 False,不包含换行符,如果为 True,则保留换行符。
```
str6 = 'I \n Love \n LsgoGroup'
print(str6.splitlines()) # ['I ', ' Love ', ' LsgoGroup']
print(str6.splitlines(True)) # ['I \n', ' Love \n', ' LsgoGroup']
```
- maketrans(intab, outtab) 创建字符映射的转换表,第一个参数是字符串,表示需要转换的字符,第二个参数也是字符串表示转换的目标。
- translate(table, deletechars="") 根据参数table给出的表,转换字符串的字符,要过滤掉的字符放到deletechars参数中。
```
str7 = 'this is string example....wow!!!'
intab = 'aeiou'
outtab = '12345'
trantab = str7.maketrans(intab, outtab)
print(trantab) # {97: 49, 111: 52, 117: 53, 101: 50, 105: 51}
print(str7.translate(trantab)) # th3s 3s str3ng 2x1mpl2....w4w!!!
```
## 4. 字符串格式化
- format 格式化函数
```
str8 = "{0} Love {1}".format('I', 'Lsgogroup') # 位置参数
print(str8) # I Love Lsgogroup
str8 = "{a} Love {b}".format(a='I', b='Lsgogroup') # 关键字参数
print(str8) # I Love Lsgogroup
str8 = "{0} Love {b}".format('I', b='Lsgogroup') # 位置参数要在关键字参数之前
print(str8) # I Love Lsgogroup
str8 = '{0:.2f}{1}'.format(27.658, 'GB') # 保留小数点后两位
print(str8) # 27.66GB
```
- Python 字符串格式化符号
**符 号** **描述**
- %c 格式化字符及其ASCII码
- %s 格式化字符串,用str()方法处理对象
- %r 格式化字符串,用rper()方法处理对象
- %d 格式化整数
- %o 格式化无符号八进制数
- %x 格式化无符号十六进制数
- %X 格式化无符号十六进制数(大写)
- %f 格式化浮点数字,可指定小数点后的精度
- %e 用科学计数法格式化浮点数
- %E 作用同%e,用科学计数法格式化浮点数
- %g 根据值的大小决定使用%f或%e
- %G 作用同%g,根据值的大小决定使用%f或%E
```
print('%c' % 97) # a
print('%c %c %c' % (97, 98, 99)) # a b c
print('%d + %d = %d' % (4, 5, 9)) # 4 + 5 = 9
print("我叫 %s 今年 %d 岁!" % ('小明', 10)) # 我叫 小明 今年 10 岁!
print('%o' % 10) # 12
print('%x' % 10) # a
print('%X' % 10) # A
print('%f' % 27.658) # 27.658000
print('%e' % 27.658) # 2.765800e+01
print('%E' % 27.658) # 2.765800E+01
print('%g' % 27.658) # 27.658
text = "I am %d years old." % 22
print("I said: %s." % text) # I said: I am 22 years old..
print("I said: %r." % text) # I said: 'I am 22 years old.'
```
## 练习题
1.
- replace函数
- spilt(' ')
- lstrip(' ') 截掉字符串左边的空格或指定字符
2. 实现函数isdigit, 判断字符串里是否只包含数字0~9
```
def isdigit(string):
if(string.isnumeric()):
return True
else:
return False
def longestPalindrome(s) -> str:
pass
```
|
github_jupyter
|
t1 = 'i love Python!'
print(t1, type(t1))
# i love Python! <class 'str'>
t2 = "I love Python!"
print(t2, type(t2))
# I love Python! <class 'str'>
print(5 + 8) # 13
print('5' + '8') # 58
print('let\'s go') # let's go
print("let's go") # let's go
print('C:\\now') # C:\now
print("C:\\Program Files\\Intel\\Wifi\\Help")
# C:\Program Files\Intel\Wifi\Help
print(r'C:\Program Files\Intel\Wifi\Help')
# C:\Program Files\Intel\Wifi\Help
print(r'C:\Program Files\Intel\Wifi\Help')
para_str = """这是一个多行字符串的实例
多行字符串可以使用制表符
TAB ( \t )。
也可以使用换行符 [ \n ]。
"""
print(para_str)
# 这是一个多行字符串的实例
# 多行字符串可以使用制表符
# TAB ( )。
# 也可以使用换行符 [
# ]。
para_str = '''这是一个多行字符串的实例
多行字符串可以使用制表符
TAB ( \t )。
也可以使用换行符 [ \n ]。
'''
print(para_str)
# 这是一个多行字符串的实例
# 多行字符串可以使用制表符
# TAB ( )。
# 也可以使用换行符 [
# ]。
str1 = 'I Love LsgoGroup'
print(str1[:6]) # I Love
print(str1[5]) # e
print(str1[:6] + " 插入的字符串 " + str1[6:])
# I Love 插入的字符串 LsgoGroup
s = 'Python'
print(s) # Python
print(s[2:4]) # th
print(s[-5:-2]) # yth
print(s[2]) # t
print(s[-1]) # n
str2 = 'xiaoxie'
print(str2.capitalize()) # Xiaoxie
str2 = "DAXIExiaoxie"
print(str2.lower()) # daxiexiaoxie
print(str2.upper()) # DAXIEXIAOXIE
print(str2.swapcase()) # daxieXIAOXIE
str2 = "DAXIExiaoxie"
print(str2.count('xi')) # 2
str2 = "DAXIExiaoxie"
print(str2.endswith('ie')) # True
print(str2.endswith('xi')) # False
print(str2.startswith('Da')) # False
print(str2.startswith('DA')) # True
str2 = "DAXIExiaoxie"
print(str2.find('xi')) # 5
print(str2.find('ix')) # -1
print(str2.rfind('xi')) # 9
str3 = '12345'
print(str3.isnumeric()) # True
str3 += 'a'
print(str3.isnumeric()) # False
str4 = '1101'
print(str4.ljust(8, '0')) # 11010000
print(str4.rjust(8, '0')) # 00001101
str5 = ' I Love LsgoGroup '
print(str5.lstrip()) # 'I Love LsgoGroup '
print(str5.lstrip().strip('I')) # ' Love LsgoGroup '
print(str5.rstrip()) # ' I Love LsgoGroup'
print(str5.strip()) # 'I Love LsgoGroup'
print(str5.strip().strip('p')) # 'I Love LsgoGrou'
str5 = ' I Love LsgoGroup '
print(str5.strip().partition('o')) # ('I L', 'o', 've LsgoGroup')
print(str5.strip().partition('m')) # ('I Love LsgoGroup', '', '')
print(str5.strip().rpartition('o')) # ('I Love LsgoGr', 'o', 'up')
str5 = ' I Love LsgoGroup '
print(str5.strip().replace('I', 'We')) # We Love LsgoGroup
str5 = ' I Love LsgoGroup '
print(str5.strip().split()) # ['I', 'Love', 'LsgoGroup']
print(str5.strip().split('o')) # ['I L', 've Lsg', 'Gr', 'up']
u = "www.baidu.com.cn"
# 使用默认分隔符
print(u.split()) # ['www.baidu.com.cn']
# 以"."为分隔符
print((u.split('.'))) # ['www', 'baidu', 'com', 'cn']
# 分割0次
print((u.split(".", 0))) # ['www.baidu.com.cn']
# 分割一次
print((u.split(".", 1))) # ['www', 'baidu.com.cn']
# 分割两次
print(u.split(".", 2)) # ['www', 'baidu', 'com.cn']
# 分割两次,并取序列为1的项
print((u.split(".", 2)[1])) # baidu
# 分割两次,并把分割后的三个部分保存到三个变量
u1, u2, u3 = u.split(".", 2)
print(u1) # www
print(u2) # baidu
print(u3) # com.cn
c = '''say
hello
baby'''
print(c)
# say
# hello
# baby
print(c.split('\n')) # ['say', 'hello', 'baby']
string = "hello boy<[www.baidu.com]>byebye"
# 先分割成"hello boy<", "www.baidu.com]>byebye",取1, 即为www.baidu.com]>byebye,
# 再根据]分割成"www.baidu.com>","byebye", 取0, 即为www.baidu.com
print(string.split('[')[1].split(']')[0]) # www.baidu.com
# 在第一题的基础上再根据.进行分割,得到三个部分
print(string.split('[')[1].split(']')[0].split('.')) # ['www', 'baidu', 'com']
str6 = 'I \n Love \n LsgoGroup'
print(str6.splitlines()) # ['I ', ' Love ', ' LsgoGroup']
print(str6.splitlines(True)) # ['I \n', ' Love \n', ' LsgoGroup']
str7 = 'this is string example....wow!!!'
intab = 'aeiou'
outtab = '12345'
trantab = str7.maketrans(intab, outtab)
print(trantab) # {97: 49, 111: 52, 117: 53, 101: 50, 105: 51}
print(str7.translate(trantab)) # th3s 3s str3ng 2x1mpl2....w4w!!!
str8 = "{0} Love {1}".format('I', 'Lsgogroup') # 位置参数
print(str8) # I Love Lsgogroup
str8 = "{a} Love {b}".format(a='I', b='Lsgogroup') # 关键字参数
print(str8) # I Love Lsgogroup
str8 = "{0} Love {b}".format('I', b='Lsgogroup') # 位置参数要在关键字参数之前
print(str8) # I Love Lsgogroup
str8 = '{0:.2f}{1}'.format(27.658, 'GB') # 保留小数点后两位
print(str8) # 27.66GB
print('%c' % 97) # a
print('%c %c %c' % (97, 98, 99)) # a b c
print('%d + %d = %d' % (4, 5, 9)) # 4 + 5 = 9
print("我叫 %s 今年 %d 岁!" % ('小明', 10)) # 我叫 小明 今年 10 岁!
print('%o' % 10) # 12
print('%x' % 10) # a
print('%X' % 10) # A
print('%f' % 27.658) # 27.658000
print('%e' % 27.658) # 2.765800e+01
print('%E' % 27.658) # 2.765800E+01
print('%g' % 27.658) # 27.658
text = "I am %d years old." % 22
print("I said: %s." % text) # I said: I am 22 years old..
print("I said: %r." % text) # I said: 'I am 22 years old.'
def isdigit(string):
if(string.isnumeric()):
return True
else:
return False
def longestPalindrome(s) -> str:
pass
| 0.069946 | 0.771757 |
# Week3-InteractiveVisualization
This notebook loads a multi-site dataset of subcortical nucleus (thalamus, globus pallidus, and striatum) volumes, performs site harmonization on these volumes using ComBat, then displays a number of interactive figures to explore the harmonized data.
```
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import neuroCombat as nc
```
## Load and Prepare Data
```
data = pd.read_csv("data_trimmed.csv", index_col=0)
# Map site IDs to integers, as required by neuroCombat()
sites = data['Site'].unique()
site_dict = dict(zip(sites, range(1, len(sites)+1)))
data['Site_no'] = data['Site'].map(site_dict)
#data['Site'].map(site_dict)
```
## ComBat Site Harmonization
```
# The list of feature names
features = ['L_str_vol', 'L_GP_vol', 'L_thal_vol', 'R_str_vol', 'R_GP_vol', 'R_thal_vol']
# Perform harmonization with ComBat
harmonized_features = nc.neuroCombat(data[features].transpose(), covars=data[['Site_no', 'Age', 'DX']], batch_col='Site_no', categorical_cols=[], continuous_cols=[]).transpose()
harmonized_features = pd.DataFrame(harmonized_features)
harmonized_features.columns = features
# Add back site data and measures that were not harmonized.
harmonized_features['Site'] = data['Site']
harmonized_features['Age'] = data['Age']
harmonized_features['TBV'] = data['TBV']
harmonized_features['DX'] = data['DX'] # I sure hope that ComBat doesn't change the order of the rows!!!
```
## Compute linear regression models on the harmonized data.
Regress nucleus volume against diagnosis and age.
```
# Linear regression on ComBat harmonized data
from statsmodels.formula.api import ols
models = pd.Series(dtype='object')
model_params = pd.Series(dtype = 'object')
for nucleus in harmonized_features.columns[:-4]: # Don't include last (covariate) columns
models[nucleus] = ols(formula = nucleus + ' ~ DX + Age', data = harmonized_features).fit()
print(models[nucleus].summary())
model_params[nucleus] = models[nucleus].params
print(model_params[nucleus])
```
## Compute Linear Regression on Unharmonized Data
```
# Linear regression on unharmonized data
models_unh = pd.Series(dtype='object')
model_unh_params = pd.Series(dtype = 'object')
for nucleus in features:
models_unh[nucleus] = ols(formula = nucleus + ' ~ DX + Age', data = data).fit()
print(models_unh[nucleus].summary())
model_unh_params[nucleus] = models_unh[nucleus].params
print(model_unh_params[nucleus])
```
## Interactive Visualizations
```
# Set up ipywidgets for interactive chart.
!jupyter nbextension enable --py widgetsnbextension
import ipywidgets as widgets
from ipywidgets import interact, fixed
```
### Scatterplots of nucleus volume against Age and Total Brain Volume
### Split violin plot showing original and harmonized distributions for each site
```
# Function to plot original and harmonized distributions together, by site.
def plot_dual_distributions(x, y, data1, data2, scale = 'count', ax=None):
# Plots two distibutions (e.g. harmonized and unharmonized data) as the two halves of stacked violin plots.
plt.figure()
vp_data = pd.DataFrame({x: data1[x], y: data1[y], 'Legend': 'Unharmonized'})
vp_data = vp_data.append(pd.DataFrame({x: data2[x], y: data2[y], 'Legend': 'Harmonized'}))
ax_h_uh = sns.violinplot(y = y, x = x, data = vp_data, hue = 'Legend', split = True, scale = scale, ax = ax)
#ax_h_uh.legend().remove()
plt.show()
# Plot a panel of figures useful for quick data exploration and verification.
def plot_panel(nucleus, data1, data2):
fig, axes = plt.subplots(ncols=2)
plt.subplots_adjust(wspace = 0.5)
sns.scatterplot('Age', nucleus, data = data, hue = "DX", ax=axes[0])
sns.scatterplot('TBV', nucleus, data = data, hue = "DX", ax=axes[1])
plot_dual_distributions(x=nucleus, y = 'Site', data1 = data1, data2 = data2, scale = 'count')
interact(plot_panel, nucleus = features, data1 = fixed(data), data2=fixed(harmonized_features))
```
### Plot fitted values against residuals for the ComBat-harmonized data
```
# Function to plot fitted values against residuals
def plot_fitted_vs_residual(models, ex_data, x_fitted_col, y_resid_col, display_col, hue_col, title_add = None):
plt.figure()
fitted_y = models[display_col].fittedvalues
resid = models[display_col].resid
# The following is not ideal. Need to revisit this solution.
fit_resid_df = pd.DataFrame({'Residual': resid, 'Fitted_Value': fitted_y, 'Site': ex_data['Site'], 'DX': ex_data['DX']})
ax = sns.scatterplot(fitted_y, resid, hue = hue_col, data=fit_resid_df)
ax.set_title('Fitted vs. Residual Plot for ' + display_col + title_add)
ax.set(xlabel = 'Fitted', ylabel = 'Residual')
ax.legend().remove()
plt.show()
# Plot fitted values vs. residuals of ComBat harmonized data
interact(plot_fitted_vs_residual,
models = fixed(models),
ex_data = fixed(harmonized_features),
x_fitted_col = fixed('Fitted_Value'),
y_resid_col = fixed('Residual'),
display_col = features,
hue_col = ['DX', 'Site'],
title_add = fixed(' ComBat Harmonized'))
# Plot fitted values vs. residuals of unharmonized data
interact(plot_fitted_vs_residual,
models = fixed(models_unh),
ex_data = fixed(data),
x_fitted_col = fixed('Fitted_Value'),
y_resid_col = fixed('Residual'),
display_col = features,
hue_col = ['DX', 'Site'],
title_add = fixed(' Unharmonized'))
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import neuroCombat as nc
data = pd.read_csv("data_trimmed.csv", index_col=0)
# Map site IDs to integers, as required by neuroCombat()
sites = data['Site'].unique()
site_dict = dict(zip(sites, range(1, len(sites)+1)))
data['Site_no'] = data['Site'].map(site_dict)
#data['Site'].map(site_dict)
# The list of feature names
features = ['L_str_vol', 'L_GP_vol', 'L_thal_vol', 'R_str_vol', 'R_GP_vol', 'R_thal_vol']
# Perform harmonization with ComBat
harmonized_features = nc.neuroCombat(data[features].transpose(), covars=data[['Site_no', 'Age', 'DX']], batch_col='Site_no', categorical_cols=[], continuous_cols=[]).transpose()
harmonized_features = pd.DataFrame(harmonized_features)
harmonized_features.columns = features
# Add back site data and measures that were not harmonized.
harmonized_features['Site'] = data['Site']
harmonized_features['Age'] = data['Age']
harmonized_features['TBV'] = data['TBV']
harmonized_features['DX'] = data['DX'] # I sure hope that ComBat doesn't change the order of the rows!!!
# Linear regression on ComBat harmonized data
from statsmodels.formula.api import ols
models = pd.Series(dtype='object')
model_params = pd.Series(dtype = 'object')
for nucleus in harmonized_features.columns[:-4]: # Don't include last (covariate) columns
models[nucleus] = ols(formula = nucleus + ' ~ DX + Age', data = harmonized_features).fit()
print(models[nucleus].summary())
model_params[nucleus] = models[nucleus].params
print(model_params[nucleus])
# Linear regression on unharmonized data
models_unh = pd.Series(dtype='object')
model_unh_params = pd.Series(dtype = 'object')
for nucleus in features:
models_unh[nucleus] = ols(formula = nucleus + ' ~ DX + Age', data = data).fit()
print(models_unh[nucleus].summary())
model_unh_params[nucleus] = models_unh[nucleus].params
print(model_unh_params[nucleus])
# Set up ipywidgets for interactive chart.
!jupyter nbextension enable --py widgetsnbextension
import ipywidgets as widgets
from ipywidgets import interact, fixed
# Function to plot original and harmonized distributions together, by site.
def plot_dual_distributions(x, y, data1, data2, scale = 'count', ax=None):
# Plots two distibutions (e.g. harmonized and unharmonized data) as the two halves of stacked violin plots.
plt.figure()
vp_data = pd.DataFrame({x: data1[x], y: data1[y], 'Legend': 'Unharmonized'})
vp_data = vp_data.append(pd.DataFrame({x: data2[x], y: data2[y], 'Legend': 'Harmonized'}))
ax_h_uh = sns.violinplot(y = y, x = x, data = vp_data, hue = 'Legend', split = True, scale = scale, ax = ax)
#ax_h_uh.legend().remove()
plt.show()
# Plot a panel of figures useful for quick data exploration and verification.
def plot_panel(nucleus, data1, data2):
fig, axes = plt.subplots(ncols=2)
plt.subplots_adjust(wspace = 0.5)
sns.scatterplot('Age', nucleus, data = data, hue = "DX", ax=axes[0])
sns.scatterplot('TBV', nucleus, data = data, hue = "DX", ax=axes[1])
plot_dual_distributions(x=nucleus, y = 'Site', data1 = data1, data2 = data2, scale = 'count')
interact(plot_panel, nucleus = features, data1 = fixed(data), data2=fixed(harmonized_features))
# Function to plot fitted values against residuals
def plot_fitted_vs_residual(models, ex_data, x_fitted_col, y_resid_col, display_col, hue_col, title_add = None):
plt.figure()
fitted_y = models[display_col].fittedvalues
resid = models[display_col].resid
# The following is not ideal. Need to revisit this solution.
fit_resid_df = pd.DataFrame({'Residual': resid, 'Fitted_Value': fitted_y, 'Site': ex_data['Site'], 'DX': ex_data['DX']})
ax = sns.scatterplot(fitted_y, resid, hue = hue_col, data=fit_resid_df)
ax.set_title('Fitted vs. Residual Plot for ' + display_col + title_add)
ax.set(xlabel = 'Fitted', ylabel = 'Residual')
ax.legend().remove()
plt.show()
# Plot fitted values vs. residuals of ComBat harmonized data
interact(plot_fitted_vs_residual,
models = fixed(models),
ex_data = fixed(harmonized_features),
x_fitted_col = fixed('Fitted_Value'),
y_resid_col = fixed('Residual'),
display_col = features,
hue_col = ['DX', 'Site'],
title_add = fixed(' ComBat Harmonized'))
# Plot fitted values vs. residuals of unharmonized data
interact(plot_fitted_vs_residual,
models = fixed(models_unh),
ex_data = fixed(data),
x_fitted_col = fixed('Fitted_Value'),
y_resid_col = fixed('Residual'),
display_col = features,
hue_col = ['DX', 'Site'],
title_add = fixed(' Unharmonized'))
| 0.671147 | 0.946399 |
## Model-Based Collaborative Filtering Systems
### SVD Matrix Factorization
```
# Import Dependencies
import numpy as np
import pandas as pd
import sklearn
from sklearn.decomposition import TruncatedSVD
```
The MovieLens dataset was collected by the GroupLens Research Project at the University of Minnesota. You can download the dataset for this demostration at the following URL: https://grouplens.org/datasets/movielens/100k/
### Preparing the data
```
# Loading the movie rating data set
columns = ['user_id', 'item_id', 'rating', 'timestamp']
rating_df = pd.read_csv('./data/u.data', sep='\t', names=columns)
rating_df.head()
# Loading the movie data set
columns = ['item_id', 'movie title', 'release date', 'video release date', 'IMDb URL', 'unknown', 'Action', 'Adventure',
'Animation', 'Childrens', 'Comedy', 'Crime', 'Documentary', 'Drama', 'Fantasy', 'Film-Noir', 'Horror',
'Musical', 'Mystery', 'Romance', 'Sci-Fi', 'Thriller', 'War', 'Western']
movies_df = pd.read_csv('./data/u.item', sep='|', names=columns, encoding='latin-1')
movie_names = movies_df[['item_id', 'movie title']]
movie_names.head()
# Join the two dataframes by item_id
joined_df = pd.merge(rating_df, movie_names, on='item_id')
joined_df.head()
# Check the number of times an item is rated
joined_df.groupby('item_id')['rating'].count().sort_values(ascending=False).head()
# Check the title of the most rated movie in the data
most_counted_movie = joined_df['item_id']==50
joined_df[most_counted_movie]['movie title'].unique()
```
### Building a Utility Matrix
```
# Create the rating crosstab to track user rating for each movie
rating_crosstab = joined_df.pivot_table(values='rating', index='user_id',
columns='movie title', fill_value=0)
rating_crosstab.head()
```
### Transposing the Matrix
```
# Check the dimension of the crosstab matrix
rating_crosstab.shape
# Transposing the crosstable matrix
X = rating_crosstab.T
X.shape
```
### Decomposing the Matrix
```
# Using the SVD model to fit the data
SVD = TruncatedSVD(n_components=12, random_state=17)
resultant_matrix = SVD.fit_transform(X)
resultant_matrix.shape
```
### Generating a Correlation Matrix
```
# Creating a correlation matrix
corr_mat = np.corrcoef(resultant_matrix)
corr_mat.shape
```
### Isolating Star Wars from the Correlation Matrix
```
# Create a list of movie names from rating crosstab
movie_names = rating_crosstab.columns
movie_list = list(movie_names)
# Find and assign the index of Star Wars (1977)
star_wars = movie_list.index('Star Wars (1977)')
star_wars
# Calculate the correlation of all movies to Star Wars (1977)
corr_star_wars = corr_mat[1398]
corr_star_wars.shape
```
### Recommending a Highly Correlated Movie
```
# List all movie titles that is 90% correlated with Star Wars (1977)
list(movie_names[(corr_star_wars < 1.0) & (corr_star_wars > 0.9)])
# List all movie titles that is 95% correlated with Star Wars (1977)
list(movie_names[(corr_star_wars < 1.0) & (cor_star_wars > 9.5)])
```
|
github_jupyter
|
# Import Dependencies
import numpy as np
import pandas as pd
import sklearn
from sklearn.decomposition import TruncatedSVD
# Loading the movie rating data set
columns = ['user_id', 'item_id', 'rating', 'timestamp']
rating_df = pd.read_csv('./data/u.data', sep='\t', names=columns)
rating_df.head()
# Loading the movie data set
columns = ['item_id', 'movie title', 'release date', 'video release date', 'IMDb URL', 'unknown', 'Action', 'Adventure',
'Animation', 'Childrens', 'Comedy', 'Crime', 'Documentary', 'Drama', 'Fantasy', 'Film-Noir', 'Horror',
'Musical', 'Mystery', 'Romance', 'Sci-Fi', 'Thriller', 'War', 'Western']
movies_df = pd.read_csv('./data/u.item', sep='|', names=columns, encoding='latin-1')
movie_names = movies_df[['item_id', 'movie title']]
movie_names.head()
# Join the two dataframes by item_id
joined_df = pd.merge(rating_df, movie_names, on='item_id')
joined_df.head()
# Check the number of times an item is rated
joined_df.groupby('item_id')['rating'].count().sort_values(ascending=False).head()
# Check the title of the most rated movie in the data
most_counted_movie = joined_df['item_id']==50
joined_df[most_counted_movie]['movie title'].unique()
# Create the rating crosstab to track user rating for each movie
rating_crosstab = joined_df.pivot_table(values='rating', index='user_id',
columns='movie title', fill_value=0)
rating_crosstab.head()
# Check the dimension of the crosstab matrix
rating_crosstab.shape
# Transposing the crosstable matrix
X = rating_crosstab.T
X.shape
# Using the SVD model to fit the data
SVD = TruncatedSVD(n_components=12, random_state=17)
resultant_matrix = SVD.fit_transform(X)
resultant_matrix.shape
# Creating a correlation matrix
corr_mat = np.corrcoef(resultant_matrix)
corr_mat.shape
# Create a list of movie names from rating crosstab
movie_names = rating_crosstab.columns
movie_list = list(movie_names)
# Find and assign the index of Star Wars (1977)
star_wars = movie_list.index('Star Wars (1977)')
star_wars
# Calculate the correlation of all movies to Star Wars (1977)
corr_star_wars = corr_mat[1398]
corr_star_wars.shape
# List all movie titles that is 90% correlated with Star Wars (1977)
list(movie_names[(corr_star_wars < 1.0) & (corr_star_wars > 0.9)])
# List all movie titles that is 95% correlated with Star Wars (1977)
list(movie_names[(corr_star_wars < 1.0) & (cor_star_wars > 9.5)])
| 0.599251 | 0.915091 |
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib notebook
```
# This Laptop Is Inadequate:
# An Aperitif for DSFP Session 8
**Version 0.1**
By AA Miller 2019 Mar 24
When I think about LSST there are a few numbers that always stick in my head:
- 37 billion (the total number of sources that will be detected by LSST)
- 10 (the number of years for the baseline survey)
- 1000 (~the number of observations per source)
- 37 trillion ($37 \times 10^9 \times 10^4$ = the total number of source observations)
These numbers are *eye-popping*, though the truth is that there are now several astronomical databases that have $\sim{10^9}$ sources (e.g., PanSTARRS-1, which we will hear more about later today).
A pressing question, for current and future surveys, is: how are we going to deal with all that data?
If you're anything like me - then, you love your laptop.
And if you had it your way, you wouldn't need anything but your laptop... ever.
But is that practical?
## Problem 1) The Inadequacy of Laptops
**Problem 1a**
Suppose you could describe every source detected by LSST with a single number. Assuming you are on a computer with a 64 bit architecture, to within an order of magnitude, how much RAM would you need to store every LSST source within your laptop's memory?
*Bonus question* - can you think of a single number to describe every source in LSST that could produce a meaningful science result?
*Take a minute to discuss with your partner*
**Solution 1a**
As for a single number to perform useful science, I can think of two.
First - you could generate a [heirarchical triangular mesh](http://www.skyserver.org/HTM/) with enough trixels to characterize every LSST resolution element on the night sky. Then you could assign a number to each trixel, and describe the position of every source in LSST with a single number. Under the assumption that every source detected by LSST is a galaxy, this is not a terrible assumption, you could look at the clustering of these positions to (potentially) learn things about structure formation or galaxy formation (though without redshifts you may not learn all that much).
The other number is the flux (or magnitude) of every source in a single filter. Again, under the assumption that everything is a galaxy, the number counts (i.e. a histogram) of the flux measurements tells you a bit about the Universe.
It probably isn't a shock that you won't be able to analyze every individual LSST source on your laptop.
But that raises the question - how should you analyze LSST data?
- By buying a large desktop?
- On a local or national supercomputer?
- In the cloud?
- On computers that LSST hosts/maintains?
We will discuss some of these issues a bit later in the week...
## Problem 2) Laptop or Not You Should Be Worried About the Data
### Pop quiz
We will now re-visit a question from a previous session:
**Problem 2a**
What is data?
*Take a minute to discuss with your partner*
**Solution 2a**
This leads to another question:
Q - What is the defining property of a constant?
A - They don't change.
If data are constants, and constants don't change, then we should probably be sure that our data storage solutions do not alter the data in any way.
Within the data science community, the python [`pandas`](https://pandas.pydata.org/) package is particularly popular for reading, writing, and manipulating data (we will talk more about the utility of `pandas` later).
The `pandas` docs state the `read_csv()` method is the [workhorse function for reading text files](http://pandas.pydata.org/pandas-docs/stable/user_guide/io.html#csv-text-files). Let's now take a look at how well this workhorse "maintains the constant nature of data".
**Problem 2b**
Create a `numpy` array, called `nums`, of length 10000 filled with random numbers. Create a `pandas` `Series` object, called `s`, based on that array, and then write the `Series` to a file called `tmp.txt` using the `to_csv()` method.
*Hint* - you'll need to name the `Series` and add the `header=True` option to the `to_csv()` call.
```
from numpy.random import rand
import pandas as pd
nums = rand(10000)
s = pd.Series(nums)
s.to_csv(path='tmp.txt', header=['nums'])
```
**Problem 2c**
Using the `pandas` `read_csv()` method, read in the data to a new variable, called `s_read`. Do you expect `s_read` and `nums` to be the same? Check whether or not your expectations are correct.
*Hint* - take the sum of the difference not equal to zero to identify if any elements are not the same.
```
import numpy as np
s_read = pd.read_csv('tmp.txt')
sum(nums - s_read['nums'].values != 0)
```
So, it turns out that $\sim{23}\%$ of the time, `pandas` does not in fact read in the same number that it wrote to disk.
The truth is that these differences are quite small (see next slide), but there are many mathematical operations (e.g., subtraction of very similar numbers) that may lead these tiny differences to compound over time such that your data are not, in fact, constant.
```
print(np.max(np.abs(nums - s_read['nums'].values)))
```
So, what is going on?
Sometimes, when you convert a number to ASCII (i.e. text) format, there is some precision that is lost in that conversion.
How do you avoid this?
One way is to directly write your files in binary. To do so has serveral advantages: it is possible to reproduce byte level accuracy, and, binary storage is almost always more efficient than text storage (the same number can be written in binary with less space than in ascii).
The downside is that developing your own procedure to write data in binary is a pain, and it places strong constraints on where and how you can interact with the data once it has been written to disk.
Fortuantely, we live in a world with `pandas`. All this hard work has been done for you, as `pandas` naturally interfaces with the [`hdf5`](https://www.hdfgroup.org/solutions/hdf5/) binary table format. (You may want to also take a look at [`pyTables`](https://www.pytables.org/))
(Historically astronomers have used [FITS](https://fits.gsfc.nasa.gov/fits_primer.html) files as a binary storage solution)
**Problem 2d**
Repeat your procedure from above, but instead of writing to a csv file, use the `pandas` `to_hdf()` and `read_df()` method to see if there are any differences in `s` and `s_read`.
*Hint* - You will need to specify a name for the table that you have written to the `hdf5` file in the call to `to_hdf()` as a required argument. Any string will do.
*Hint 2* - Use `s_read.values` instead of `s_read['nums'].values`.
```
s2 = pd.Series(nums)
s2.to_hdf(path_or_buf='tmp2.h5', key='nums')
s2_read = pd.read_hdf('tmp2.h5')
print(np.sum(nums-s2_read.values))
```
# Noiccuh
So, if you are using `pandas` anyway, and if you aren't using `pandas` –– check it out!, then I strongly suggest removing csv files from your workflow to instead focus on binary hdf5 files. This requires typing the same number of characters, but it ensures byte level reproducibility.
And reproducibiliy is the pillar upon which the scientific method is built.
Is that the end of the story? ... No.
In the previous example, I was being a little tricky in order to make a point. It *is* in fact possible to create reproducible csv files with `pandas`. By default, `pandas` sacrifices a little bit of precision in order to gain a lot more speed. If you want to ensure reproducibility then you can specify that the `float_precision` should be `round_trip`, meaning you get the same thing back after reading from a file that you wrote to disk.
```
s.to_csv('tmp.txt', header=['nums'])
s_read = pd.read_csv('tmp.txt', float_precision='round_trip')
sum(nums - s_read['nums'].values != 0)
```
So was all of this in service of a lie?
No. What I said before remains true - text files do not guarantee byte level precision, and they take more space on disk. Text files have some advantages:
- anyone, anywhere, on any platform can easily manipulate text files
- text files can be easily inspected (and corrected) if necessary
- special packages are needed to read/write in binary
- binary files, which are not easily interpretable, are difficult to use in version control (and banned by some version control platforms)
To summarize, here is my advice: think of binary as your (new?) default for storing data.
But, as with all things, consider your audience: if you are sharing/working with people that won't be able to deal with binary data, or, you have an incredibly small amount of data, csv (or other text files) should be fine.
## Problem 3) Binary or ASCII - Doesn't Matter if You Aren't Organized
While the reproducibility of data is essential, ultimately, concerns about binary vs ascii are useless if you cannot access the data you need *when* you need it.
Your data are valuable (though cheaper to acquire than ever before), which means you need a good solution for managing that data, or else you are going to run into a significant loss of time and money.
**Problem 3a**
How would you organize the following: (a) 3 very deep images of a galaxy, (b) 4 nights of optical observations ($\sim$50 images night$^{-1}$) of a galaxy cluster in the $ugrizY$ filters, (c) images from a 50 night time-domain survey ($\sim$250 images night$^{-1}$) covering 1000 deg$^2$?
Similarly, how would you organize: (a) photometric information for your galaxy observations, (b) photometry for all the galaxies in your cluster field, (c) the observations/light curves from your survey?
*Take a minute to discuss with your partner*
**Solution 3a**
...
Keeping in mind that there are several suitable answers to each of these questions, here are a few thoughts: (a) the 3 images should be kept together, probably in a single file directory. (b) With 200 images taken over the course of 4 nights, I would create a directory structure that includes every night (top level), with sub-directories based on the individual filters. (c) Similar to (b), I'd create a tree-based file structure, though given that the primary science is time variability, I would likely organize the observations by fieldID at the top level, then by filter and date after that.
As a final note - for each of these data sets, backups are essential! There should be no risk of a single point failure wiping away all that information for good.
The photometric data requires more than just a file structure. In all three cases I would want to store everything in a single file (so directories are not necessary).
For 3 observations of a single galaxy, I would use... a text file (not worth the trouble for binary storage)
Assuming there are 5000 galaxies in the cluster field, I would store the photometric information that I extract for those galaxies in a *table*. In this table, each row would represent a single galaxy, while the columns would include brightness/shape measurements for the galaxies in each of the observed filters. I would organize this table as a `pandas` DataFrame (and write it to an hdf5 file).
For the time-domain survey, the organization of all the photometric information is far less straight forward.
Could you use a single table? Yes. Though this would be highly inefficient given that not all sources were observed at the same time. The table would then need columns like `obs1_JD`, `obs1_flux`, `obs1_flux_unc`, `obs2_JD`, `obs2_flux`, `obs2_flux_unc`, ..., all the way up to $N$, the maximum number of observations of any individual source. This will lead to several columns that are empty for several sources.
I would instead use a collection of tables. First, a master source table:
|objID|RA|Dec|mean_mag|mean_mag_unc|
|:--:|:--:|:--:|:--:|:--:|
|0001|246.98756|-12.06547|18.35|0.08|
|0002|246.98853|-12.04325|19.98|0.21|
|.|.|.|.|.|
|.|.|.|.|.|
|.|.|.|.|.|
Coupled with a table holding the individual flux measurements:
|objID|JD|filt|mag|mag_unc|
|:--:|:--:|:--:|:--:|:--:|
|0001|2456785.23465|r|18.21|0.07|
|0001|2456785.23469|z|17.81|0.12|
|.|.|.|.|.|
|.|.|.|.|.|
|.|.|.|.|.|
|0547|2456821.36900|g|16.04|0.02|
|0547|2456821.36906|i|17.12|0.05|
|.|.|.|.|.|
|.|.|.|.|.|
|.|.|.|.|.|
The critical thing to notice about these tables is that they both contain `objID`. That information allows us to connect the tables via a "join". This table, or relational, structure allows us to easily connect subsets of the data as way to minimize storage (relative to having everything in a single table) while also maintaining computational speed.
Typically, when astronomers (or data scientists) need to organize data into several connected tables capable of performing fast relational algebra operations they use a database. We will hear a lot more about databases over the next few days, so I won't provide a detailed introduction now.
One very nice property of (many) database systems is that provide an efficient means for searching large volumes of data that cannot be stored in memory (recall problem **1a**). Whereas, your laptop, or even a specialized high-memory computer, would not be able to open a csv file with all the LSST observations in it.
Another quick aside –– `pandas` can deal with files that are too large to fit in memory by loading a portion of the file at a time:
light_curves = pd.read_csv(lc_csv_file, chunksize=100000)
If you are building a data structure where loading the data in "chunks" is necessary, I would strongly advise considering an alternative to storing the data in a csv file.
A question you may currently be wondering is: why has there been such an intense focus on `pandas` today?
The short answer: the developers of `pandas` wanted to create a product that is good at relational algebra (like traditional database tools) but with lower overhead in construction, and a lot more flexibility (which is essential in a world of heterogeneous data storage and management, see Tuesday's lecture on Data Wrangling).
(You'll get several chances to practice working with databases throughout the week)
We will now run through a few examples that highlight how `pandas` can be used in a manner similar to a relational database. Throughout the week, as you think about your own data management needs, I think the critical thing to consider is scope. Can my data be organized into something that is smaller than a full-on database?
**Problem 3b**
Download the [SDSS data set](https://northwestern.box.com/s/sjegm0tx62l2i8dkzqw22s4gmq1a9sg1) that will be used in the exercises for tomorrow.
Read that data, stored in a csv file, into a `pandas` DataFrame called `sdss_spec`.
In a lecture where I have spent a great deal of time describing the value of binary data storage, does the fact that I am now providing a (moderate) amount of data as a plain ascii file mean that I am a bad teacher...
*probably*
```
sdss_spec = pd.read_csv("DSFP_SDSS_spec_train.csv")
sdss_spec.head()
```
`pandas` provides many different methods for selecting columns from the DataFrame. Supposing you wanted `psfMag`, you could use any of the following:
sdss_spec['psfMag_g']
sdss_spec[['psfMag_r', 'psfMag_z']]
sdss_spec.psfMag_g
(notice that selecting multiple columns requires a list within `[]`)
**Problem 3c**
Plot a histogram of the `psfMag_g` - `modelMag_g` distribution for the data set (which requires a selection of those two columns).
Do you notice anything interesting?
*Hint* - you may want to use more than the default number of bins (=10).
```
mag_diff = sdss_spec['psfMag_g'] - sdss_spec['modelMag_g']
plt.hist(mag_diff,bins=100)
# complete
```
Pandas can also be used to aggregate the results of a search.
**Problem 3d**
How many extended sources (`type` = `ext`) have `modelMag_i` between 19 and 20? Use as few lines as possible.
```
len(sdss_spec[(sdss_spec['type']=='ext') & (sdss_spec['modelMag_i'] < 20) & (sdss_spec['modelMag_i'] > 19)])
# aww yiss
```
## WORKING RIGHT HERE NOW!!!|
`pandas` also enables [`GROUP BY`](http://pandas.pydata.org/pandas-docs/stable/user_guide/groupby.html) operations, where the data are split based on some criterion, a function is then applied to the groups, and the results are then combined back into a data structure.
**Problem 3e**
Group the data by their `type` and then report the minimum, median, and maximum redshift of each group. Can you immediately tell anything about these sources based on these results?
*Hint* - just execute the cell below.
```
grouped = sdss_spec.groupby([sdss_spec.type])
print(grouped['z'].min())
print(grouped['z'].median())
print(grouped['z'].max())
```
Finally, we have only briefly discussed joining tables, but this is where relational databases really shine.
For this example we only have a single table, so we will exclude any examples of a `pandas` join, but there is functionality to [join or merge](https://pandas.pydata.org/pandas-docs/stable/user_guide/merging.html) dataframes in a fashion that is fully analogous to databases.
In summary, there are many different possible solutions for data storage and management.
For "medium" to "big" data that won't easily fit into memory ($\sim$16 GB), it is likely that a database is your best solution. For slightly smaller problems `pandas` provides a really nice, lightweight alternative to a full blown database that nevertheless maintains a lot of the same functionality and power.
## Problem 4) We Aren't Done Talking About Your Laptop's Inadequacies
So far we have been focused on only a single aspect of computing: storage (and your laptop sucks at that).
But here's the thing - your laptop is also incredibly slow.
Supposing for a moment that you could hold all (or even a significant fraction) of the information from LSST in memory on your laptop, you would still be out of luck, as you would die before you could actually process the data and make any meaningful calculations.
(we argued that it would take [$\sim$200 yr to process LSST on your laptop](https://github.com/LSSTC-DSFP/LSSTC-DSFP-Sessions/blob/master/Session5/Day1/PhotonsArentScienceSolutions.ipynb) in Session 5)
You are in luck, however, as you need not limit yourself to your laptop. You can take advantage of multiple computers, also known as parallel processing.
At a previous session, I asked Robert Lupton, one of the primary developers of the LSST photometric pipeline, "How many CPUs are being used to process LSST data?" To which he replied, "However many are needed to process everything within 1 month."
The critical point here is that if you can figure out how to split a calculation over multiple computers, then you can finish any calculation arbitrarily fast with enough processors (to within some limits, like the speed of light, etc)
We will spend a lot more time talking about both efficient algorithm design and parallel processing later this week, but I want to close with a quick example that touches on each of these things.
Suppose that you have some 2k x 2k detector (i.e. 4 million pixels), and you need to manipulate the data in that array. For instance, the detector will report the number of counts per pixel, but this number is larger than the actual number of detected photons by a factor $g$, the gain of the telescope.
How long does it take divide every pixel by the gain?
(This is where I spend a moment telling you that - if you are going to time portions of your code as a means of measuring performance it is essential that you turn off *everything else* that may be running on your computer, as background processes can mess up your timing results)
```
import time
pixel_data = np.random.rand(4000000)
photons = np.empty_like(pixel_data)
tstart = time.time()
for pix_num, pixel in enumerate(pixel_data):
photons[pix_num] = pixel/8
trun = time.time() - tstart
print('It takes {:.6f} s to correct for the gain'.format(trun))
```
1.5 s isn't too bad in the grand scheme of things.
Except that this example should make you cringe. There is absolutely no need to use a for loop for these operations.
This brings us to fast coding lesson number 1 - **vectorize everything**.
```
photons = np.empty_like(pixel_data)
tstart = time.time()
photons = pixel_data/8
trun = time.time() - tstart
print('It takes {:.6f} s to correct for the gain'.format(trun))
```
By removing the for loop we improve the speed of this particular calculation by a factor of $\sim$125. That is a massive win.
Alternatively, we could have sped up the operations via the use of parallel programing. The [`multiprocessing`](https://docs.python.org/2/library/multiprocessing.html) library in python makes it relatively easy to implement parallel operations. There are many different ways to implement parallel processing in python, here we will just use one simple example.
(again, we will go over multiprocessing in far more detail later this week)
```
from multiprocessing import Pool
def divide_by_gain(number, gain=8):
return number/gain
pool = Pool()
tstart = time.time()
photons = pool.map(divide_by_gain, pixel_data)
trun = time.time() - tstart
print('It takes {:.6f} s to correct for the gain'.format(trun))
```
Wait, parallel processing slows this down by a factor $\sim$7? What's going on here?
It turns out there is some overhead in copying the data and spawning multiple processes, and in this case that overhead is enormous. Of course, for more complex functions/operations (here we were only doing simple division), that overhead is tiny compared to the individual calculations and using multiple processors can lead to almost an $N$x gain in speed, where $N$ is the number of processors available.
This brings me to fast coding lesson number 2 - **before you parallelize, profile**. (We will talk more about software profiling later in the week), but in short - there is no point to parallelizing inefficient operations. Even if the parallel gain calculation provided a factor of $\sim$4 (the number of CPUS on my machine) speed up relative to our initial for loop, that factor of 4 is a waste compared to the factor of 125 gained by vectorizing the code.
## Conclusions
As we go through this week - think about the types of problems that you encounter in your own workflow, and consider how you might be able to improve that workflow by moving off of your laptop.
This may come in several forms, including: superior data organization, better data structures (`pandas` or databases), more efficient algorithms, and finally parallel processing.
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib notebook
from numpy.random import rand
import pandas as pd
nums = rand(10000)
s = pd.Series(nums)
s.to_csv(path='tmp.txt', header=['nums'])
import numpy as np
s_read = pd.read_csv('tmp.txt')
sum(nums - s_read['nums'].values != 0)
print(np.max(np.abs(nums - s_read['nums'].values)))
s2 = pd.Series(nums)
s2.to_hdf(path_or_buf='tmp2.h5', key='nums')
s2_read = pd.read_hdf('tmp2.h5')
print(np.sum(nums-s2_read.values))
s.to_csv('tmp.txt', header=['nums'])
s_read = pd.read_csv('tmp.txt', float_precision='round_trip')
sum(nums - s_read['nums'].values != 0)
sdss_spec = pd.read_csv("DSFP_SDSS_spec_train.csv")
sdss_spec.head()
mag_diff = sdss_spec['psfMag_g'] - sdss_spec['modelMag_g']
plt.hist(mag_diff,bins=100)
# complete
len(sdss_spec[(sdss_spec['type']=='ext') & (sdss_spec['modelMag_i'] < 20) & (sdss_spec['modelMag_i'] > 19)])
# aww yiss
grouped = sdss_spec.groupby([sdss_spec.type])
print(grouped['z'].min())
print(grouped['z'].median())
print(grouped['z'].max())
import time
pixel_data = np.random.rand(4000000)
photons = np.empty_like(pixel_data)
tstart = time.time()
for pix_num, pixel in enumerate(pixel_data):
photons[pix_num] = pixel/8
trun = time.time() - tstart
print('It takes {:.6f} s to correct for the gain'.format(trun))
photons = np.empty_like(pixel_data)
tstart = time.time()
photons = pixel_data/8
trun = time.time() - tstart
print('It takes {:.6f} s to correct for the gain'.format(trun))
from multiprocessing import Pool
def divide_by_gain(number, gain=8):
return number/gain
pool = Pool()
tstart = time.time()
photons = pool.map(divide_by_gain, pixel_data)
trun = time.time() - tstart
print('It takes {:.6f} s to correct for the gain'.format(trun))
| 0.284377 | 0.933127 |
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import pywt
df = pd.read_excel("Data/df_100.xlsx")
df.head()
df.info()
df["Avg_Soil_TP100_TempC_Plus_1"] = df["Avg_Soil_TP100_TempC"].shift(-1)
df["Avg_Soil_TP100_TempC_Plus_2"] = df["Avg_Soil_TP100_TempC"].shift(-2)
df["Max_Soil_TP100_TempC_Plus_1"] = df["Max_Soil_TP100_TempC"].shift(-1)
df["Max_Soil_TP100_TempC_Plus_2"] = df["Max_Soil_TP100_TempC"].shift(-2)
df["Min_Soil_TP100_TempC_Plus_1"] = df["Min_Soil_TP100_TempC"].shift(-1)
df["Min_Soil_TP100_TempC_Plus_2"] = df["Min_Soil_TP100_TempC"].shift(-2)
df["AvgAir_T_TempC_Minus_6"] = df["AvgAir_T_TempC"].shift(6)
df["AvgAir_T_TempC_Minus_5"] = df["AvgAir_T_TempC"].shift(5)
df["AvgAir_T_TempC_Minus_4"] = df["AvgAir_T_TempC"].shift(4)
df["AvgAir_T_TempC_Minus_3"] = df["AvgAir_T_TempC"].shift(3)
df["AvgAir_T_TempC_Minus_2"] = df["AvgAir_T_TempC"].shift(2)
df["AvgAir_T_TempC_Minus_1"] = df["AvgAir_T_TempC"].shift(1)
df["MaxAir_T_TempC_Minus_6"] = df["MaxAir_T_TempC"].shift(6)
df["MaxAir_T_TempC_Minus_5"] = df["MaxAir_T_TempC"].shift(5)
df["MaxAir_T_TempC_Minus_4"] = df["MaxAir_T_TempC"].shift(4)
df["MaxAir_T_TempC_Minus_3"] = df["MaxAir_T_TempC"].shift(3)
df["MaxAir_T_TempC_Minus_2"] = df["MaxAir_T_TempC"].shift(2)
df["MaxAir_T_TempC_Minus_1"] = df["MaxAir_T_TempC"].shift(1)
df["MinAir_T_TempC_Minus_6"] = df["MinAir_T_TempC"].shift(6)
df["MinAir_T_TempC_Minus_5"] = df["MinAir_T_TempC"].shift(5)
df["MinAir_T_TempC_Minus_4"] = df["MinAir_T_TempC"].shift(4)
df["MinAir_T_TempC_Minus_3"] = df["MinAir_T_TempC"].shift(3)
df["MinAir_T_TempC_Minus_2"] = df["MinAir_T_TempC"].shift(2)
df["MinAir_T_TempC_Minus_1"] = df["MinAir_T_TempC"].shift(1)
df = df[["AvgAir_T_TempC_Minus_6","MaxAir_T_TempC_Minus_6","MinAir_T_TempC_Minus_6",
"AvgAir_T_TempC_Minus_5","MaxAir_T_TempC_Minus_5","MinAir_T_TempC_Minus_5",
"AvgAir_T_TempC_Minus_4","MaxAir_T_TempC_Minus_4","MinAir_T_TempC_Minus_4",
"AvgAir_T_TempC_Minus_3","MaxAir_T_TempC_Minus_3","MinAir_T_TempC_Minus_3",
"AvgAir_T_TempC_Minus_2","MaxAir_T_TempC_Minus_2","MinAir_T_TempC_Minus_2",
"AvgAir_T_TempC_Minus_1","MaxAir_T_TempC_Minus_1","MinAir_T_TempC_Minus_1",
"AvgAir_T_TempC","MaxAir_T_TempC","MinAir_T_TempC",
"Avg_Soil_TP100_TempC","Max_Soil_TP100_TempC","Min_Soil_TP100_TempC",
"Avg_Soil_TP100_TempC_Plus_1","Max_Soil_TP100_TempC_Plus_1","Min_Soil_TP100_TempC_Plus_1",
"Avg_Soil_TP100_TempC_Plus_2","Max_Soil_TP100_TempC_Plus_2","Min_Soil_TP100_TempC_Plus_2"]]
df.dropna(inplace=True)
df.reset_index(drop="True", inplace=True)
def WaveletTransform(x,walvelettype,level):
size = np.shape(x)
length = (size[0] // (2**level)) * (2 ** level)
x = x.iloc[0:length]
bigmats = []
for i in x.columns:
vec = x[i].values
cAs_cDs = pywt.swt(vec, wavelet=walvelettype, level=level,axis=0)
for j in cAs_cDs:
bigmats.append(j[0])
bigmats.append(j[1])
features_num = len(bigmats)
x = np.zeros((length, features_num))
for i in range(0 ,len(bigmats)):
for j in range(0,len(bigmats[0])):
x[j,i] = x[j,i] + bigmats[i][j]
return(x)
x = df.drop(["Avg_Soil_TP100_TempC","Max_Soil_TP100_TempC","Min_Soil_TP100_TempC",
"Avg_Soil_TP100_TempC_Plus_1","Max_Soil_TP100_TempC_Plus_1","Min_Soil_TP100_TempC_Plus_1",
"Avg_Soil_TP100_TempC_Plus_2","Max_Soil_TP100_TempC_Plus_2","Min_Soil_TP100_TempC_Plus_2"],axis=1)
y = df[["Avg_Soil_TP100_TempC","Max_Soil_TP100_TempC","Min_Soil_TP100_TempC",
"Avg_Soil_TP100_TempC_Plus_1","Max_Soil_TP100_TempC_Plus_1","Min_Soil_TP100_TempC_Plus_1",
"Avg_Soil_TP100_TempC_Plus_2","Max_Soil_TP100_TempC_Plus_2","Min_Soil_TP100_TempC_Plus_2"]].values
x = WaveletTransform(x,"db5",5)
x.shape
y = df[0:x.shape[0]]
x = pd.DataFrame(x)
df = pd.concat([x,y],axis=1)
df
final_result_table_100cm_with_wavelet = pd.DataFrame(columns=["output_variable", "MLAlgo", "real_value",
"predicted_value", "mse_list", "mae_list", "r_sqaured"])
final_result_table_100cm_with_wavelet
```
# 2 - Feature Selection
```
# df --> Dataframe
#Ys --> a list of all target variables
#y --> the main target variable - string
def FeatureSelection(df,Ys,y,corr):
X = df.drop(Ys,axis=1)
Y = df[y]
df_final = pd.concat([X,Y],axis=1)
indexes = df_final.corr()[df_final.corr()[y] > corr].index
df_final = df_final[indexes]
return df_final
Ys = ["Avg_Soil_TP100_TempC", "Max_Soil_TP100_TempC", "Min_Soil_TP100_TempC","Avg_Soil_TP100_TempC_Plus_1",
"Max_Soil_TP100_TempC_Plus_1","Min_Soil_TP100_TempC_Plus_1", "Avg_Soil_TP100_TempC_Plus_2",
"Max_Soil_TP100_TempC_Plus_2", "Min_Soil_TP100_TempC_Plus_2"]
df_avg = FeatureSelection(df,Ys,y = "Avg_Soil_TP100_TempC",corr=0.5)
df_max = FeatureSelection(df,Ys,y = "Max_Soil_TP100_TempC",corr=0.5)
df_min = FeatureSelection(df,Ys,y = "Min_Soil_TP100_TempC",corr=0.5)
df_avg_plus1 = FeatureSelection(df,Ys,y = "Avg_Soil_TP100_TempC_Plus_1",corr=0.5)
df_max_plus1 = FeatureSelection(df,Ys,y = "Max_Soil_TP100_TempC_Plus_1",corr=0.5)
df_min_plus1 = FeatureSelection(df,Ys,y = "Min_Soil_TP100_TempC_Plus_1",corr=0.5)
df_avg_plus2 = FeatureSelection(df,Ys,y = "Avg_Soil_TP100_TempC_Plus_2",corr=0.5)
df_max_plus2 = FeatureSelection(df,Ys,y = "Max_Soil_TP100_TempC_Plus_2",corr=0.5)
df_min_plus2 = FeatureSelection(df,Ys,y = "Min_Soil_TP100_TempC_Plus_2",corr=0.5)
```
# 3 - SVR Support vector machines for regression Problem
### Train test split and data transform
```
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler, MinMaxScaler
#df: data frame
#y: target variable --> string
def SplitTransform(df,y,test_size, scaler):
X = df.drop(y,axis=1).values
y = df[y].values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=test_size, random_state=101)
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
data = [X_train, X_test,y_train,y_test]
return data
from sklearn.svm import SVR
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import mean_absolute_error, mean_squared_error, explained_variance_score
model = SVR(gamma="auto")
param_grid = {"kernel" : ["rbf"],
"C" : [40]}
grid = GridSearchCV(model,param_grid, cv = 5)
Ys = {"Avg_Soil_TP100_TempC":df_avg, "Max_Soil_TP100_TempC":df_max, "Min_Soil_TP100_TempC":df_min,"Avg_Soil_TP100_TempC_Plus_1":df_avg_plus1,
"Max_Soil_TP100_TempC_Plus_1":df_max_plus1,"Min_Soil_TP100_TempC_Plus_1":df_min_plus1, "Avg_Soil_TP100_TempC_Plus_2":df_avg_plus2,
"Max_Soil_TP100_TempC_Plus_2":df_max_plus2, "Min_Soil_TP100_TempC_Plus_2":df_min_plus2}
SVM_results = []
for i in (Ys):
print(i)
X_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[0]
X_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[1]
y_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[2]
y_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[3]
grid.fit(X_train, y_train)
best_params = grid.best_params_
score = grid.score(X_test,y_test)
prediction = grid.predict(X_test)
MSE = mean_squared_error(y_test,prediction)
MAE = mean_absolute_error(y_test,prediction)
dictinary = {"Target Variable" : i, "Best Parameters" : best_params, "R Squared" : score, "MAE": MAE,
"MSE": MSE}
SVM_results.append(dictinary)
length = len(prediction)
output_variable = [i for j in range(length)]
MLAlgo = ["WSVM" for j in range(length)]
mse_list = [MSE for j in range(length)]
mae_list = [MAE for j in range(length)]
r_squared = [score for j in range(length)]
predicted_value = prediction.tolist()
real_value = y_test.tolist()
result = pd.DataFrame(list(zip(output_variable,MLAlgo,real_value,predicted_value,mse_list,mae_list,r_squared)),
columns=["output_variable", "MLAlgo", "real_value","predicted_value", "mse_list", "mae_list", "r_sqaured"])
final_result_table_100cm_with_wavelet = pd.concat([final_result_table_100cm_with_wavelet,result])
final_result_table_100cm_with_wavelet
```
# 4 - XGBoost (Extreme Gradient Boosting Method) for regression problem
```
import xgboost as xg
model = xg.XGBRegressor(objective ='reg:squarederror')
param_grid = {
'max_depth': [7],
'learning_rate': [0.05],
'gamma': [0],
'reg_lambda': [1.0],
'n_estimators' : [150]
}
grid = GridSearchCV(model,param_grid, cv = 5, verbose=3)
XGBoost_results = []
for i in (Ys):
print(i)
X_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[0]
X_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[1]
y_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[2]
y_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[3]
grid.fit(X_train, y_train)
best_params = grid.best_params_
score = grid.score(X_test,y_test)
prediction = grid.predict(X_test)
MSE = mean_squared_error(y_test,prediction)
MAE = mean_absolute_error(y_test,prediction)
dictinary = {"Target Variable" : i, "Best Parameters" : best_params, "R Squared" : score, "MAE": MAE,
"MSE": MSE}
XGBoost_results.append(dictinary)
length = len(prediction)
output_variable = [i for j in range(length)]
MLAlgo = ["WXGBoost" for j in range(length)]
mse_list = [MSE for j in range(length)]
mae_list = [MAE for j in range(length)]
r_squared = [score for j in range(length)]
predicted_value = prediction.tolist()
real_value = y_test.tolist()
result = pd.DataFrame(list(zip(output_variable,MLAlgo,real_value,predicted_value,mse_list,mae_list,r_squared)),
columns=["output_variable", "MLAlgo", "real_value","predicted_value", "mse_list", "mae_list", "r_sqaured"])
final_result_table_100cm_with_wavelet = pd.concat([final_result_table_100cm_with_wavelet,result])
final_result_table_100cm_with_wavelet
```
# 5 - ANN
### Building the Model
```
from keras.models import Sequential
from keras.layers import Dense
from keras.wrappers.scikit_learn import KerasRegressor
def create_model(neurons,hidden_layers, optimizer, activation):
model = Sequential()
# Add aninput layer
model.add(Dense(X_train.shape[1], activation = activation))
for i in range(hidden_layers):
# Add one hidden layer
model.add(Dense(neurons, activation=activation))
# Add an output layer
model.add(Dense(1))
# Compile model
model.compile(optimizer = optimizer, loss="mse")
return model
model = KerasRegressor(build_fn=create_model, verbose = 3)
param_grid = {
'batch_size': [128],
'epochs': [400],
'neurons': [350],
'hidden_layers': [4],
'optimizer' : ['Adam'],
"activation" : ["relu"]
}
grid = GridSearchCV(model,param_grid,n_jobs=-1, verbose=3)
ANN_results = []
for i in (Ys):
print(i)
X_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[0]
X_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[1]
y_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[2]
y_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[3]
grid.fit(X_train, y_train)
best_params = grid.best_params_
score = grid.score(X_test,y_test)
prediction = grid.predict(X_test)
MSE = mean_squared_error(y_test,prediction)
MAE = mean_absolute_error(y_test,prediction)
dictinary = {"Target Variable" : i, "Best Parameters" : best_params, "R Squared" : score, "MAE": MAE,
"MSE": MSE}
ANN_results.append(dictinary)
length = len(prediction)
output_variable = [i for j in range(length)]
MLAlgo = ["WANN" for j in range(length)]
mse_list = [MSE for j in range(length)]
mae_list = [MAE for j in range(length)]
r_squared = [score for j in range(length)]
predicted_value = prediction.tolist()
real_value = y_test.tolist()
result = pd.DataFrame(list(zip(output_variable,MLAlgo,real_value,predicted_value,mse_list,mae_list,r_squared)),
columns=["output_variable", "MLAlgo", "real_value","predicted_value", "mse_list", "mae_list", "r_sqaured"])
final_result_table_100cm_with_wavelet = pd.concat([final_result_table_100cm_with_wavelet,result])
final_result_table_100cm_with_wavelet.reset_index(drop = True)
final_result_table_100cm_with_wavelet.to_csv(r'C:\Users\siava\OneDrive - University of Manitoba\D\Computer science\Data Science Projects\Soil-temperature\Results\100 cm\final_result_table_100cm_with_wavelet.csv'
,index=False)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import pywt
df = pd.read_excel("Data/df_100.xlsx")
df.head()
df.info()
df["Avg_Soil_TP100_TempC_Plus_1"] = df["Avg_Soil_TP100_TempC"].shift(-1)
df["Avg_Soil_TP100_TempC_Plus_2"] = df["Avg_Soil_TP100_TempC"].shift(-2)
df["Max_Soil_TP100_TempC_Plus_1"] = df["Max_Soil_TP100_TempC"].shift(-1)
df["Max_Soil_TP100_TempC_Plus_2"] = df["Max_Soil_TP100_TempC"].shift(-2)
df["Min_Soil_TP100_TempC_Plus_1"] = df["Min_Soil_TP100_TempC"].shift(-1)
df["Min_Soil_TP100_TempC_Plus_2"] = df["Min_Soil_TP100_TempC"].shift(-2)
df["AvgAir_T_TempC_Minus_6"] = df["AvgAir_T_TempC"].shift(6)
df["AvgAir_T_TempC_Minus_5"] = df["AvgAir_T_TempC"].shift(5)
df["AvgAir_T_TempC_Minus_4"] = df["AvgAir_T_TempC"].shift(4)
df["AvgAir_T_TempC_Minus_3"] = df["AvgAir_T_TempC"].shift(3)
df["AvgAir_T_TempC_Minus_2"] = df["AvgAir_T_TempC"].shift(2)
df["AvgAir_T_TempC_Minus_1"] = df["AvgAir_T_TempC"].shift(1)
df["MaxAir_T_TempC_Minus_6"] = df["MaxAir_T_TempC"].shift(6)
df["MaxAir_T_TempC_Minus_5"] = df["MaxAir_T_TempC"].shift(5)
df["MaxAir_T_TempC_Minus_4"] = df["MaxAir_T_TempC"].shift(4)
df["MaxAir_T_TempC_Minus_3"] = df["MaxAir_T_TempC"].shift(3)
df["MaxAir_T_TempC_Minus_2"] = df["MaxAir_T_TempC"].shift(2)
df["MaxAir_T_TempC_Minus_1"] = df["MaxAir_T_TempC"].shift(1)
df["MinAir_T_TempC_Minus_6"] = df["MinAir_T_TempC"].shift(6)
df["MinAir_T_TempC_Minus_5"] = df["MinAir_T_TempC"].shift(5)
df["MinAir_T_TempC_Minus_4"] = df["MinAir_T_TempC"].shift(4)
df["MinAir_T_TempC_Minus_3"] = df["MinAir_T_TempC"].shift(3)
df["MinAir_T_TempC_Minus_2"] = df["MinAir_T_TempC"].shift(2)
df["MinAir_T_TempC_Minus_1"] = df["MinAir_T_TempC"].shift(1)
df = df[["AvgAir_T_TempC_Minus_6","MaxAir_T_TempC_Minus_6","MinAir_T_TempC_Minus_6",
"AvgAir_T_TempC_Minus_5","MaxAir_T_TempC_Minus_5","MinAir_T_TempC_Minus_5",
"AvgAir_T_TempC_Minus_4","MaxAir_T_TempC_Minus_4","MinAir_T_TempC_Minus_4",
"AvgAir_T_TempC_Minus_3","MaxAir_T_TempC_Minus_3","MinAir_T_TempC_Minus_3",
"AvgAir_T_TempC_Minus_2","MaxAir_T_TempC_Minus_2","MinAir_T_TempC_Minus_2",
"AvgAir_T_TempC_Minus_1","MaxAir_T_TempC_Minus_1","MinAir_T_TempC_Minus_1",
"AvgAir_T_TempC","MaxAir_T_TempC","MinAir_T_TempC",
"Avg_Soil_TP100_TempC","Max_Soil_TP100_TempC","Min_Soil_TP100_TempC",
"Avg_Soil_TP100_TempC_Plus_1","Max_Soil_TP100_TempC_Plus_1","Min_Soil_TP100_TempC_Plus_1",
"Avg_Soil_TP100_TempC_Plus_2","Max_Soil_TP100_TempC_Plus_2","Min_Soil_TP100_TempC_Plus_2"]]
df.dropna(inplace=True)
df.reset_index(drop="True", inplace=True)
def WaveletTransform(x,walvelettype,level):
size = np.shape(x)
length = (size[0] // (2**level)) * (2 ** level)
x = x.iloc[0:length]
bigmats = []
for i in x.columns:
vec = x[i].values
cAs_cDs = pywt.swt(vec, wavelet=walvelettype, level=level,axis=0)
for j in cAs_cDs:
bigmats.append(j[0])
bigmats.append(j[1])
features_num = len(bigmats)
x = np.zeros((length, features_num))
for i in range(0 ,len(bigmats)):
for j in range(0,len(bigmats[0])):
x[j,i] = x[j,i] + bigmats[i][j]
return(x)
x = df.drop(["Avg_Soil_TP100_TempC","Max_Soil_TP100_TempC","Min_Soil_TP100_TempC",
"Avg_Soil_TP100_TempC_Plus_1","Max_Soil_TP100_TempC_Plus_1","Min_Soil_TP100_TempC_Plus_1",
"Avg_Soil_TP100_TempC_Plus_2","Max_Soil_TP100_TempC_Plus_2","Min_Soil_TP100_TempC_Plus_2"],axis=1)
y = df[["Avg_Soil_TP100_TempC","Max_Soil_TP100_TempC","Min_Soil_TP100_TempC",
"Avg_Soil_TP100_TempC_Plus_1","Max_Soil_TP100_TempC_Plus_1","Min_Soil_TP100_TempC_Plus_1",
"Avg_Soil_TP100_TempC_Plus_2","Max_Soil_TP100_TempC_Plus_2","Min_Soil_TP100_TempC_Plus_2"]].values
x = WaveletTransform(x,"db5",5)
x.shape
y = df[0:x.shape[0]]
x = pd.DataFrame(x)
df = pd.concat([x,y],axis=1)
df
final_result_table_100cm_with_wavelet = pd.DataFrame(columns=["output_variable", "MLAlgo", "real_value",
"predicted_value", "mse_list", "mae_list", "r_sqaured"])
final_result_table_100cm_with_wavelet
# df --> Dataframe
#Ys --> a list of all target variables
#y --> the main target variable - string
def FeatureSelection(df,Ys,y,corr):
X = df.drop(Ys,axis=1)
Y = df[y]
df_final = pd.concat([X,Y],axis=1)
indexes = df_final.corr()[df_final.corr()[y] > corr].index
df_final = df_final[indexes]
return df_final
Ys = ["Avg_Soil_TP100_TempC", "Max_Soil_TP100_TempC", "Min_Soil_TP100_TempC","Avg_Soil_TP100_TempC_Plus_1",
"Max_Soil_TP100_TempC_Plus_1","Min_Soil_TP100_TempC_Plus_1", "Avg_Soil_TP100_TempC_Plus_2",
"Max_Soil_TP100_TempC_Plus_2", "Min_Soil_TP100_TempC_Plus_2"]
df_avg = FeatureSelection(df,Ys,y = "Avg_Soil_TP100_TempC",corr=0.5)
df_max = FeatureSelection(df,Ys,y = "Max_Soil_TP100_TempC",corr=0.5)
df_min = FeatureSelection(df,Ys,y = "Min_Soil_TP100_TempC",corr=0.5)
df_avg_plus1 = FeatureSelection(df,Ys,y = "Avg_Soil_TP100_TempC_Plus_1",corr=0.5)
df_max_plus1 = FeatureSelection(df,Ys,y = "Max_Soil_TP100_TempC_Plus_1",corr=0.5)
df_min_plus1 = FeatureSelection(df,Ys,y = "Min_Soil_TP100_TempC_Plus_1",corr=0.5)
df_avg_plus2 = FeatureSelection(df,Ys,y = "Avg_Soil_TP100_TempC_Plus_2",corr=0.5)
df_max_plus2 = FeatureSelection(df,Ys,y = "Max_Soil_TP100_TempC_Plus_2",corr=0.5)
df_min_plus2 = FeatureSelection(df,Ys,y = "Min_Soil_TP100_TempC_Plus_2",corr=0.5)
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler, MinMaxScaler
#df: data frame
#y: target variable --> string
def SplitTransform(df,y,test_size, scaler):
X = df.drop(y,axis=1).values
y = df[y].values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=test_size, random_state=101)
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
data = [X_train, X_test,y_train,y_test]
return data
from sklearn.svm import SVR
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import mean_absolute_error, mean_squared_error, explained_variance_score
model = SVR(gamma="auto")
param_grid = {"kernel" : ["rbf"],
"C" : [40]}
grid = GridSearchCV(model,param_grid, cv = 5)
Ys = {"Avg_Soil_TP100_TempC":df_avg, "Max_Soil_TP100_TempC":df_max, "Min_Soil_TP100_TempC":df_min,"Avg_Soil_TP100_TempC_Plus_1":df_avg_plus1,
"Max_Soil_TP100_TempC_Plus_1":df_max_plus1,"Min_Soil_TP100_TempC_Plus_1":df_min_plus1, "Avg_Soil_TP100_TempC_Plus_2":df_avg_plus2,
"Max_Soil_TP100_TempC_Plus_2":df_max_plus2, "Min_Soil_TP100_TempC_Plus_2":df_min_plus2}
SVM_results = []
for i in (Ys):
print(i)
X_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[0]
X_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[1]
y_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[2]
y_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[3]
grid.fit(X_train, y_train)
best_params = grid.best_params_
score = grid.score(X_test,y_test)
prediction = grid.predict(X_test)
MSE = mean_squared_error(y_test,prediction)
MAE = mean_absolute_error(y_test,prediction)
dictinary = {"Target Variable" : i, "Best Parameters" : best_params, "R Squared" : score, "MAE": MAE,
"MSE": MSE}
SVM_results.append(dictinary)
length = len(prediction)
output_variable = [i for j in range(length)]
MLAlgo = ["WSVM" for j in range(length)]
mse_list = [MSE for j in range(length)]
mae_list = [MAE for j in range(length)]
r_squared = [score for j in range(length)]
predicted_value = prediction.tolist()
real_value = y_test.tolist()
result = pd.DataFrame(list(zip(output_variable,MLAlgo,real_value,predicted_value,mse_list,mae_list,r_squared)),
columns=["output_variable", "MLAlgo", "real_value","predicted_value", "mse_list", "mae_list", "r_sqaured"])
final_result_table_100cm_with_wavelet = pd.concat([final_result_table_100cm_with_wavelet,result])
final_result_table_100cm_with_wavelet
import xgboost as xg
model = xg.XGBRegressor(objective ='reg:squarederror')
param_grid = {
'max_depth': [7],
'learning_rate': [0.05],
'gamma': [0],
'reg_lambda': [1.0],
'n_estimators' : [150]
}
grid = GridSearchCV(model,param_grid, cv = 5, verbose=3)
XGBoost_results = []
for i in (Ys):
print(i)
X_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[0]
X_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[1]
y_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[2]
y_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[3]
grid.fit(X_train, y_train)
best_params = grid.best_params_
score = grid.score(X_test,y_test)
prediction = grid.predict(X_test)
MSE = mean_squared_error(y_test,prediction)
MAE = mean_absolute_error(y_test,prediction)
dictinary = {"Target Variable" : i, "Best Parameters" : best_params, "R Squared" : score, "MAE": MAE,
"MSE": MSE}
XGBoost_results.append(dictinary)
length = len(prediction)
output_variable = [i for j in range(length)]
MLAlgo = ["WXGBoost" for j in range(length)]
mse_list = [MSE for j in range(length)]
mae_list = [MAE for j in range(length)]
r_squared = [score for j in range(length)]
predicted_value = prediction.tolist()
real_value = y_test.tolist()
result = pd.DataFrame(list(zip(output_variable,MLAlgo,real_value,predicted_value,mse_list,mae_list,r_squared)),
columns=["output_variable", "MLAlgo", "real_value","predicted_value", "mse_list", "mae_list", "r_sqaured"])
final_result_table_100cm_with_wavelet = pd.concat([final_result_table_100cm_with_wavelet,result])
final_result_table_100cm_with_wavelet
from keras.models import Sequential
from keras.layers import Dense
from keras.wrappers.scikit_learn import KerasRegressor
def create_model(neurons,hidden_layers, optimizer, activation):
model = Sequential()
# Add aninput layer
model.add(Dense(X_train.shape[1], activation = activation))
for i in range(hidden_layers):
# Add one hidden layer
model.add(Dense(neurons, activation=activation))
# Add an output layer
model.add(Dense(1))
# Compile model
model.compile(optimizer = optimizer, loss="mse")
return model
model = KerasRegressor(build_fn=create_model, verbose = 3)
param_grid = {
'batch_size': [128],
'epochs': [400],
'neurons': [350],
'hidden_layers': [4],
'optimizer' : ['Adam'],
"activation" : ["relu"]
}
grid = GridSearchCV(model,param_grid,n_jobs=-1, verbose=3)
ANN_results = []
for i in (Ys):
print(i)
X_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[0]
X_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[1]
y_train = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[2]
y_test = SplitTransform(df = Ys[i], y = i, test_size= 0.3, scaler = StandardScaler())[3]
grid.fit(X_train, y_train)
best_params = grid.best_params_
score = grid.score(X_test,y_test)
prediction = grid.predict(X_test)
MSE = mean_squared_error(y_test,prediction)
MAE = mean_absolute_error(y_test,prediction)
dictinary = {"Target Variable" : i, "Best Parameters" : best_params, "R Squared" : score, "MAE": MAE,
"MSE": MSE}
ANN_results.append(dictinary)
length = len(prediction)
output_variable = [i for j in range(length)]
MLAlgo = ["WANN" for j in range(length)]
mse_list = [MSE for j in range(length)]
mae_list = [MAE for j in range(length)]
r_squared = [score for j in range(length)]
predicted_value = prediction.tolist()
real_value = y_test.tolist()
result = pd.DataFrame(list(zip(output_variable,MLAlgo,real_value,predicted_value,mse_list,mae_list,r_squared)),
columns=["output_variable", "MLAlgo", "real_value","predicted_value", "mse_list", "mae_list", "r_sqaured"])
final_result_table_100cm_with_wavelet = pd.concat([final_result_table_100cm_with_wavelet,result])
final_result_table_100cm_with_wavelet.reset_index(drop = True)
final_result_table_100cm_with_wavelet.to_csv(r'C:\Users\siava\OneDrive - University of Manitoba\D\Computer science\Data Science Projects\Soil-temperature\Results\100 cm\final_result_table_100cm_with_wavelet.csv'
,index=False)
| 0.15772 | 0.334943 |
# 실험 실행
Azure Machine Learning SDK를 사용하여 메트릭을 기록하고 출력을 생성하는 코드 실험을 실행할 수 있습니다. 이 실행 과정은 Azure Machine Learning에서 수행하는 대다수 기계 학습 작업의 핵심 요소입니다.
## 작업 영역에 연결
Azure Machine Learning 작업 영역 내에서 모든 실험 및 관련 리소스를 관리합니다. 대부분의 경우에는 JSON 구성 파일에 작업 영역 구성을 저장해야 합니다. 이렇게 하면 Azure 구독 ID 등의 세부 정보를 기억할 필요 없이 더욱 쉽게 다시 연결할 수 있습니다. Azure Portal의 작업 영역 블레이드에서 JSON 구성 파일을 다운로드할 수 있습니다. 그러나 작업 영역 내에서 컴퓨팅 인스턴스를 사용 중이라면 구성 파일은 루트 폴더에 이미 다운로드된 상태입니다.
아래 코드는 구성 파일을 사용하여 작업 영역에 연결합니다.
> **참고**: Azure 구독에 인증된 세션을 아직 설정하지 않은 경우에는 링크를 클릭하고 인증 코드를 입력한 다음 Azure에 로그인하여 인증하라는 메시지가 표시됩니다.
```
import azureml.core
from azureml.core import Workspace
# Load the workspace from the saved config file
ws = Workspace.from_config()
print('Ready to use Azure ML {} to work with {}'.format(azureml.core.VERSION, ws.name))
```
## 실험 실행
데이터 과학자가 수행해야 하는 가장 기본적인 작업 중 하나는 데이터를 처리하고 분석하는 실험을 만들고 실행하는 것입니다. 이 연습에서는 Azure ML *실험*을 사용하여 Python 코드를 실행하고 데이터에서 추출된 값을 기록하는 방법을 알아봅니다. 여기서는 당뇨병 진단을 받은 환자의 세부 정보가 포함된 간단한 데이터 세트를 사용합니다. 실험을 실행하여 데이터를 살펴본 다음 통계, 시각화 및 데이터 샘플을 추출합니다. 여기서 사용할 대다수 코드는 데이터 탐색 프로세스에서 실행할 수 있는 코드와 같은 매우 일반적인 Python 코드입니다. 그러나 여기에 코드를 몇 줄만 추가하면 Azure ML *실험*을 사용하여 실행 세부 정보를 기록하도록 수정할 수 있습니다.
```
from azureml.core import Experiment
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
# Create an Azure ML experiment in your workspace
experiment = Experiment(workspace=ws, name="mslearn-diabetes")
# Start logging data from the experiment, obtaining a reference to the experiment run
run = experiment.start_logging()
print("Starting experiment:", experiment.name)
# load the data from a local file
data = pd.read_csv('data/diabetes.csv')
# Count the rows and log the result
row_count = (len(data))
run.log('observations', row_count)
print('Analyzing {} rows of data'.format(row_count))
# Plot and log the count of diabetic vs non-diabetic patients
diabetic_counts = data['Diabetic'].value_counts()
fig = plt.figure(figsize=(6,6))
ax = fig.gca()
diabetic_counts.plot.bar(ax = ax)
ax.set_title('Patients with Diabetes')
ax.set_xlabel('Diagnosis')
ax.set_ylabel('Patients')
plt.show()
run.log_image(name='label distribution', plot=fig)
# log distinct pregnancy counts
pregnancies = data.Pregnancies.unique()
run.log_list('pregnancy categories', pregnancies)
# Log summary statistics for numeric columns
med_columns = ['PlasmaGlucose', 'DiastolicBloodPressure', 'TricepsThickness', 'SerumInsulin', 'BMI']
summary_stats = data[med_columns].describe().to_dict()
for col in summary_stats:
keys = list(summary_stats[col].keys())
values = list(summary_stats[col].values())
for index in range(len(keys)):
run.log_row(col, stat=keys[index], value = values[index])
# Save a sample of the data and upload it to the experiment output
data.sample(100).to_csv('sample.csv', index=False, header=True)
run.upload_file(name='outputs/sample.csv', path_or_stream='./sample.csv')
# Complete the run
run.complete()
```
## 실행 세부 정보 확인
Jupyter Notebook에서 **RunDetails** 위젯을 사용하면 실행 세부 정보의 시각화를 확인할 수 있습니다.
```
from azureml.widgets import RunDetails
RunDetails(run).show()
```
### Azure Machine Learning Studio에서 추가 세부 정보 확인
**RunDetails** 위젯에는 Azure Machine Learning Studio에서 **실행 세부 정보를 확인**할 수 있는 링크가 포함되어 있습니다. 이 링크를 클릭하면 새 브라우저 탭이 열리고 실행 세부 정보가 표시됩니다. [Azure Machine Learning Studio](https://ml.azure.com)를 열고 **실험** 페이지에서 실행을 찾아도 됩니다. Azure Machine Learning Studio에서 실행을 확인할 때는 다음 사항에 유의하세요.
- **세부 정보** 탭에는 실험 실행의 일반 속성이 포함되어 있습니다.
- **메트릭** 탭에서는 기록된 메트릭을 선택하여 표나 차트로 표시할 수 있습니다.
- **이미지** 탭에서는 실험에서 기록된 이미지나 그림(여기서는 *레이블 분포* 그림)을 선택하여 확인할 수 있습니다.
- **자식 실행** 탭에는 자식 실행의 목록이 표시됩니다. 이 실험에서는 자식 실행이 없습니다.
- **출력 + 로그** 탭에는 실험에서 생성된 출력 파일이나 로그 파일이 표시됩니다.
- **스냅샷** 탭에는 실험 코드가 실행된 폴더의 모든 파일(여기서는 이 Notebook과 같은 폴더에 있는 모든 파일)이 포함되어 있습니다.
- **설명** 탭은 실험에서 생성된 모델 설명을 표시하는 데 사용됩니다. 여기서는 생성된 설명이 없습니다.
- **공정성** 탭은 기계 학습 모델의 공정성을 평가하는 데 도움이 되는 예측 성능 차이(여기서는 없음)의 시각화에 사용됩니다.
### SDK를 사용하여 실험 세부 정보 검색
앞에서 실행한 코드의 **run** 변수는 **Run** 개체의 인스턴스입니다. Run 개체는 Azure Machine Learning에서 진행하는 실험의 개별 실행에 대한 참조입니다. 이 참조를 사용하여 실험 및 해당 출력 관련 정보를 가져올 수 있습니다.
```
import json
# Get logged metrics
print("Metrics:")
metrics = run.get_metrics()
for metric_name in metrics:
print(metric_name, ":", metrics[metric_name])
# Get output files
print("\nFiles:")
files = run.get_file_names()
for file in files:
print(file)
```
실험에서 생성된 파일은 **download_file** 메서드를 사용해 개별적으로 다운로드할 수도 있고 **download_files** 메서드를 사용해 여러 파일을 검색한 다음 다운로드할 수도 있습니다. 다음 코드는 실행의 **output** 폴더에 있는 모든 파일을 다운로드합니다.
```
import os
download_folder = 'downloaded-files'
# Download files in the "outputs" folder
run.download_files(prefix='outputs', output_directory=download_folder)
# Verify the files have been downloaded
for root, directories, filenames in os.walk(download_folder):
for filename in filenames:
print (os.path.join(root,filename))
```
실험 실행의 문제를 해결해야 하는 경우 **get_details** 메서드를 사용해 실행의 기본 세부 정보를 검색할 수도 있고, **get_details_with_logs** 메서드를 사용하여 실행 세부 정보와 실행에서 생성된 로그 파일의 내용을 검색할 수도 있습니다.
```
run.get_details_with_logs()
```
세부 정보에는 실험을 실행한 컴퓨팅 대상 관련 정보, 그리고 실험을 시작 및 종료한 날짜와 시간이 포함됩니다. 또한 실험 코드가 포함된 Notebook(이 Notebook)이 복제된 Git 리포지토리에 있으므로 해당 리포지토리, 분기, 상태 관련 세부 정보도 실행 기록에 기록됩니다.
여기서 세부 정보의 **logFiles** 항목은 로그 파일이 생성되지 않았음을 나타냅니다. 방금 실행한 실험과 같은 인라인 실험에서는 대개 로그 파일이 생성되지 않습니다. 하지만 스크립트를 실험으로 실행하는 경우에는 결과가 달라집니다. 다음에는 이러한 실험에 대해 살펴보겠습니다.
## 실험 스크립트 실행
이전 예제에서는 이 Notebook에서 실험을 인라인으로 실행했습니다. 실험을 더 유동적으로 실행하려는 경우 별도의 실험용 스크립트를 작성하여 이 스크립트에 필요한 다른 파일과 함께 폴더에 저장한 다음, Azure ML을 사용하여 해당 폴더의 스크립트를 기준으로 실험을 실행하면 됩니다.
먼저 실험 파일용 폴더를 만들고 이 폴더에 데이터를 복사해 보겠습니다.
```
import os, shutil
# Create a folder for the experiment files
folder_name = 'diabetes-experiment-files'
experiment_folder = './' + folder_name
os.makedirs(folder_name, exist_ok=True)
# Copy the data file into the experiment folder
shutil.copy('data/diabetes.csv', os.path.join(folder_name, "diabetes.csv"))
```
실험용 코드가 포함된 Python 스크립트를 만들어 실험 폴더에 저장합니다.
> **참고**: 다음 셀을 실행하면 스크립트 파일이 *작성*만 되며 실행되지는 않습니다.
```
%%writefile $folder_name/diabetes_experiment.py
from azureml.core import Run
import pandas as pd
import os
# Get the experiment run context
run = Run.get_context()
# load the diabetes dataset
data = pd.read_csv('diabetes.csv')
# Count the rows and log the result
row_count = (len(data))
run.log('observations', row_count)
print('Analyzing {} rows of data'.format(row_count))
# Count and log the label counts
diabetic_counts = data['Diabetic'].value_counts()
print(diabetic_counts)
for k, v in diabetic_counts.items():
run.log('Label:' + str(k), v)
# Save a sample of the data in the outputs folder (which gets uploaded automatically)
os.makedirs('outputs', exist_ok=True)
data.sample(100).to_csv("outputs/sample.csv", index=False, header=True)
# Complete the run
run.complete()
```
이 코드는 이전에 사용했던 인라인 코드의 간단한 버전이며 다음과 같은 작업을 수행합니다.
- `Run.get_context()` 메서드를 사용하여 스크립트 실행 시에 실험 실행 컨텍스트를 검색합니다.
- 스크립트가 있는 폴더에서 당뇨병 데이터를 로드합니다.
- **outputs** 폴더를 만들고 이 폴더에 샘플 파일을 작성합니다. 이 폴더는 실험 실행에 자동으로 업로드됩니다.
이제 실험을 실행할 준비가 거의 완료되었습니다. 스크립트를 실행하려면 실험에서 실행할 Python 스크립트 파일을 식별하는 **ScriptRunConfig**를 만든 다음, 이를 기반으로 실험을 실행해야 합니다.
> **참고**: 또한 ScriptRunConfig는 컴퓨팅 대상 및 Python 환경을 결정합니다. 이 경우 Python 환경은 일부 Conda 및 pip 패키지를 포함하도록 정의되지만 컴퓨팅 대상은 생략되므로 기본 로컬 컴퓨터가 사용됩니다.
다음 셀은 스크립트 기반 실험을 구성하고 제출합니다.
```
from azureml.core import Experiment, ScriptRunConfig, Environment
from azureml.widgets import RunDetails
# Create a Python environment for the experiment (from a .yml file)
env = Environment.from_conda_specification("experiment_env", "environment.yml")
# Create a script config
script_config = ScriptRunConfig(source_directory=experiment_folder,
script='diabetes_experiment.py',
environment=env)
# submit the experiment
experiment = Experiment(workspace=ws, name='mslearn-diabetes')
run = experiment.submit(config=script_config)
RunDetails(run).show()
run.wait_for_completion()
```
이전과 마찬가지로 [Azure Machine Learning Studio](https://ml.azure.com)의 실험 링크나 위젯을 사용해 실험에서 생성된 출력을 확인할 수 있습니다. 또한 실험에서 생성된 파일과 메트릭을 검색하는 코드를 작성할 수도 있습니다.
```
# Get logged metrics
metrics = run.get_metrics()
for key in metrics.keys():
print(key, metrics.get(key))
print('\n')
for file in run.get_file_names():
print(file)
```
이번에는 실행에서 로그 파일이 생성되었습니다. 이러한 로그 파일은 위젯에서 확인할 수도 있고 앞에서 사용했던 것과 같은 **get_details_with_logs** 메서드를 사용할 수도 있습니다. 이번에는 이 메서드를 사용하면 출력에 로그 데이터가 포함됩니다.
```
run.get_details_with_logs()
```
위의 출력에서도 로그 세부 정보를 확인할 수는 있지만 일반적으로는 로그 파일을 다운로드하여 텍스트 편집기에서 확인하는 방식이 더 쉽습니다.
```
import os
log_folder = 'downloaded-logs'
# Download all files
run.get_all_logs(destination=log_folder)
# Verify the files have been downloaded
for root, directories, filenames in os.walk(log_folder):
for filename in filenames:
print (os.path.join(root,filename))
```
## 실험 실행 기록 확인
같은 실험을 여러 번 실행했으므로 [Azure Machine Learning Studio](https://ml.azure.com)에서 기록을 확인할 수 있으며 기록된 각 실행을 살펴볼 수도 있습니다. 작업 영역에서 이름으로 실험을 검색하고 SDK를 사용하여 해당 실행을 반복할 수도 있습니다.
```
from azureml.core import Experiment, Run
diabetes_experiment = ws.experiments['mslearn-diabetes']
for logged_run in diabetes_experiment.get_runs():
print('Run ID:', logged_run.id)
metrics = logged_run.get_metrics()
for key in metrics.keys():
print('-', key, metrics.get(key))
```
## MLflow 사용
MLflow는 기계 학습 프로세스를 관리하기 위한 오픈 소스 플랫폼입니다. MLflow는 전적으로는 아니지만 일반적으로 Databricks 환경에서 실험을 조정하고 메트릭을 추적하는 데 사용됩니다. Azure Machine Learning 실험에서는 기본 로그 기능 대신 MLflow를 사용하여 메트릭을 추적할 수 있습니다.
이 기능을 활용하려면 **azureml-mlflow** 패키지가 필요합니다. 이 패키지가 설치되어 있는지 확인해 보겠습니다.
```
!pip show azureml-mlflow
```
### 인라인 실험에 MLflow 사용
MLflow를 사용하여 인라인 실험의 메트릭을 추적하려면 MLflow *추적 URI*를 실험이 실행 중인 작업 공간으로 설정해야 합니다. 이를 통해 **mlflow** 추적 방법을 사용하여 실험 실행에 데이터를 로그할 수 있습니다.
```
from azureml.core import Experiment
import pandas as pd
import mlflow
# Set the MLflow tracking URI to the workspace
mlflow.set_tracking_uri(ws.get_mlflow_tracking_uri())
# Create an Azure ML experiment in your workspace
experiment = Experiment(workspace=ws, name='mslearn-diabetes-mlflow')
mlflow.set_experiment(experiment.name)
# start the MLflow experiment
with mlflow.start_run():
print("Starting experiment:", experiment.name)
# Load data
data = pd.read_csv('data/diabetes.csv')
# Count the rows and log the result
row_count = (len(data))
mlflow.log_metric('observations', row_count)
print("Run complete")
```
이제 실행 중에 로깅된 메트릭을 확인해 보겠습니다.
```
# Get the latest run of the experiment
run = list(experiment.get_runs())[0]
# Get logged metrics
print("\nMetrics:")
metrics = run.get_metrics()
for key in metrics.keys():
print(key, metrics.get(key))
# Get a link to the experiment in Azure ML studio
experiment_url = experiment.get_portal_url()
print('See details at', experiment_url)
```
위의 코드를 실행한 후 표시되는 링크를 사용하여 Azure Machine Learning Studio에서 실험을 볼 수 있습니다. 그런 다음 실험의 최신 실행을 선택하여 **메트릭** 탭을 보고 로그된 메트릭을 확인합니다.
### 실험 스크립트에서 MLflow 사용
MLflow를 사용하여 실험 스크립트의 메트릭을 추적할 수도 있습니다.
다음 두 셀을 실행하여 MLflow를 사용하는 실험을 위한 폴더와 스크립트를 만듭니다.
```
import os, shutil
# Create a folder for the experiment files
folder_name = 'mlflow-experiment-files'
experiment_folder = './' + folder_name
os.makedirs(folder_name, exist_ok=True)
# Copy the data file into the experiment folder
shutil.copy('data/diabetes.csv', os.path.join(folder_name, "diabetes.csv"))
%%writefile $folder_name/mlflow_diabetes.py
from azureml.core import Run
import pandas as pd
import mlflow
# start the MLflow experiment
with mlflow.start_run():
# Load data
data = pd.read_csv('diabetes.csv')
# Count the rows and log the result
row_count = (len(data))
print('observations:', row_count)
mlflow.log_metric('observations', row_count)
```
Azure ML 실험 스크립트에서 MLflow 추적을 사용하면 실험 실행을 시작할 때 MLflow 추적 URI가 자동으로 설정됩니다. 그러나 스크립트를 실행해야 하는 환경에는 필수 **mlflow** 패키지가 포함되어 있어야 합니다.
```
from azureml.core import Experiment, ScriptRunConfig, Environment
from azureml.widgets import RunDetails
# Create a Python environment for the experiment (from a .yml file)
env = Environment.from_conda_specification("experiment_env", "environment.yml")
# Create a script config
script_mlflow = ScriptRunConfig(source_directory=experiment_folder,
script='mlflow_diabetes.py',
environment=env)
# submit the experiment
experiment = Experiment(workspace=ws, name='mslearn-diabetes-mlflow')
run = experiment.submit(config=script_mlflow)
RunDetails(run).show()
run.wait_for_completion()
```
평소와 같이 실험이 완료되면 로그된 메트릭을 실험 실행에서 얻을 수 있습니다.
```
# Get logged metrics
metrics = run.get_metrics()
for key in metrics.keys():
print(key, metrics.get(key))
```
> **추가 정보**: 실험 실행에 대해 자세히 알아보려면 Azure ML 설명서에서 [이 토픽](https://docs.microsoft.com/azure/machine-learning/how-to-manage-runs)을 참조하세요. 실행에서 메트릭을 기록하는 방법에 대한 자세한 내용은 [이 토픽](https://docs.microsoft.com/azure/machine-learning/how-to-track-experiments)을 참조하세요. Azure ML 실험과 MLflow의 통합에 대한 자세한 내용은 [이 토픽](https://docs.microsoft.com/en-us/azure/machine-learning/how-to-use-mlflow)을 참조하세요.
|
github_jupyter
|
import azureml.core
from azureml.core import Workspace
# Load the workspace from the saved config file
ws = Workspace.from_config()
print('Ready to use Azure ML {} to work with {}'.format(azureml.core.VERSION, ws.name))
from azureml.core import Experiment
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
# Create an Azure ML experiment in your workspace
experiment = Experiment(workspace=ws, name="mslearn-diabetes")
# Start logging data from the experiment, obtaining a reference to the experiment run
run = experiment.start_logging()
print("Starting experiment:", experiment.name)
# load the data from a local file
data = pd.read_csv('data/diabetes.csv')
# Count the rows and log the result
row_count = (len(data))
run.log('observations', row_count)
print('Analyzing {} rows of data'.format(row_count))
# Plot and log the count of diabetic vs non-diabetic patients
diabetic_counts = data['Diabetic'].value_counts()
fig = plt.figure(figsize=(6,6))
ax = fig.gca()
diabetic_counts.plot.bar(ax = ax)
ax.set_title('Patients with Diabetes')
ax.set_xlabel('Diagnosis')
ax.set_ylabel('Patients')
plt.show()
run.log_image(name='label distribution', plot=fig)
# log distinct pregnancy counts
pregnancies = data.Pregnancies.unique()
run.log_list('pregnancy categories', pregnancies)
# Log summary statistics for numeric columns
med_columns = ['PlasmaGlucose', 'DiastolicBloodPressure', 'TricepsThickness', 'SerumInsulin', 'BMI']
summary_stats = data[med_columns].describe().to_dict()
for col in summary_stats:
keys = list(summary_stats[col].keys())
values = list(summary_stats[col].values())
for index in range(len(keys)):
run.log_row(col, stat=keys[index], value = values[index])
# Save a sample of the data and upload it to the experiment output
data.sample(100).to_csv('sample.csv', index=False, header=True)
run.upload_file(name='outputs/sample.csv', path_or_stream='./sample.csv')
# Complete the run
run.complete()
from azureml.widgets import RunDetails
RunDetails(run).show()
import json
# Get logged metrics
print("Metrics:")
metrics = run.get_metrics()
for metric_name in metrics:
print(metric_name, ":", metrics[metric_name])
# Get output files
print("\nFiles:")
files = run.get_file_names()
for file in files:
print(file)
import os
download_folder = 'downloaded-files'
# Download files in the "outputs" folder
run.download_files(prefix='outputs', output_directory=download_folder)
# Verify the files have been downloaded
for root, directories, filenames in os.walk(download_folder):
for filename in filenames:
print (os.path.join(root,filename))
run.get_details_with_logs()
import os, shutil
# Create a folder for the experiment files
folder_name = 'diabetes-experiment-files'
experiment_folder = './' + folder_name
os.makedirs(folder_name, exist_ok=True)
# Copy the data file into the experiment folder
shutil.copy('data/diabetes.csv', os.path.join(folder_name, "diabetes.csv"))
%%writefile $folder_name/diabetes_experiment.py
from azureml.core import Run
import pandas as pd
import os
# Get the experiment run context
run = Run.get_context()
# load the diabetes dataset
data = pd.read_csv('diabetes.csv')
# Count the rows and log the result
row_count = (len(data))
run.log('observations', row_count)
print('Analyzing {} rows of data'.format(row_count))
# Count and log the label counts
diabetic_counts = data['Diabetic'].value_counts()
print(diabetic_counts)
for k, v in diabetic_counts.items():
run.log('Label:' + str(k), v)
# Save a sample of the data in the outputs folder (which gets uploaded automatically)
os.makedirs('outputs', exist_ok=True)
data.sample(100).to_csv("outputs/sample.csv", index=False, header=True)
# Complete the run
run.complete()
from azureml.core import Experiment, ScriptRunConfig, Environment
from azureml.widgets import RunDetails
# Create a Python environment for the experiment (from a .yml file)
env = Environment.from_conda_specification("experiment_env", "environment.yml")
# Create a script config
script_config = ScriptRunConfig(source_directory=experiment_folder,
script='diabetes_experiment.py',
environment=env)
# submit the experiment
experiment = Experiment(workspace=ws, name='mslearn-diabetes')
run = experiment.submit(config=script_config)
RunDetails(run).show()
run.wait_for_completion()
# Get logged metrics
metrics = run.get_metrics()
for key in metrics.keys():
print(key, metrics.get(key))
print('\n')
for file in run.get_file_names():
print(file)
run.get_details_with_logs()
import os
log_folder = 'downloaded-logs'
# Download all files
run.get_all_logs(destination=log_folder)
# Verify the files have been downloaded
for root, directories, filenames in os.walk(log_folder):
for filename in filenames:
print (os.path.join(root,filename))
from azureml.core import Experiment, Run
diabetes_experiment = ws.experiments['mslearn-diabetes']
for logged_run in diabetes_experiment.get_runs():
print('Run ID:', logged_run.id)
metrics = logged_run.get_metrics()
for key in metrics.keys():
print('-', key, metrics.get(key))
!pip show azureml-mlflow
from azureml.core import Experiment
import pandas as pd
import mlflow
# Set the MLflow tracking URI to the workspace
mlflow.set_tracking_uri(ws.get_mlflow_tracking_uri())
# Create an Azure ML experiment in your workspace
experiment = Experiment(workspace=ws, name='mslearn-diabetes-mlflow')
mlflow.set_experiment(experiment.name)
# start the MLflow experiment
with mlflow.start_run():
print("Starting experiment:", experiment.name)
# Load data
data = pd.read_csv('data/diabetes.csv')
# Count the rows and log the result
row_count = (len(data))
mlflow.log_metric('observations', row_count)
print("Run complete")
# Get the latest run of the experiment
run = list(experiment.get_runs())[0]
# Get logged metrics
print("\nMetrics:")
metrics = run.get_metrics()
for key in metrics.keys():
print(key, metrics.get(key))
# Get a link to the experiment in Azure ML studio
experiment_url = experiment.get_portal_url()
print('See details at', experiment_url)
import os, shutil
# Create a folder for the experiment files
folder_name = 'mlflow-experiment-files'
experiment_folder = './' + folder_name
os.makedirs(folder_name, exist_ok=True)
# Copy the data file into the experiment folder
shutil.copy('data/diabetes.csv', os.path.join(folder_name, "diabetes.csv"))
%%writefile $folder_name/mlflow_diabetes.py
from azureml.core import Run
import pandas as pd
import mlflow
# start the MLflow experiment
with mlflow.start_run():
# Load data
data = pd.read_csv('diabetes.csv')
# Count the rows and log the result
row_count = (len(data))
print('observations:', row_count)
mlflow.log_metric('observations', row_count)
from azureml.core import Experiment, ScriptRunConfig, Environment
from azureml.widgets import RunDetails
# Create a Python environment for the experiment (from a .yml file)
env = Environment.from_conda_specification("experiment_env", "environment.yml")
# Create a script config
script_mlflow = ScriptRunConfig(source_directory=experiment_folder,
script='mlflow_diabetes.py',
environment=env)
# submit the experiment
experiment = Experiment(workspace=ws, name='mslearn-diabetes-mlflow')
run = experiment.submit(config=script_mlflow)
RunDetails(run).show()
run.wait_for_completion()
# Get logged metrics
metrics = run.get_metrics()
for key in metrics.keys():
print(key, metrics.get(key))
| 0.595257 | 0.943191 |
# Partial Least Squares Regression (PLSR) on Near Infrared Spectroscopy (NIR) data and octane data
This notebook illustrates how to use the **hoggorm** package to carry out partial least squares regression (PLSR) on multivariate data. Furthermore, we will learn how to visualise the results of the PLSR using the **hoggormPlot** package.
---
### Import packages and prepare data
First import **hoggorm** for analysis of the data and **hoggormPlot** for plotting of the analysis results. We'll also import **pandas** such that we can read the data into a data frame. **numpy** is needed for checking dimensions of the data.
```
import hoggorm as ho
import hoggormplot as hop
import pandas as pd
import numpy as np
```
Next, load the data that we are going to analyse using **hoggorm**. After the data has been loaded into the pandas data frame, we'll display it in the notebook.
```
# Load fluorescence data
X_df = pd.read_csv('gasoline_NIR.txt', header=None, sep='\s+')
X_df
# Load response data, that is octane measurements
y_df = pd.read_csv('gasoline_octane.txt', header=None, sep='\s+')
y_df
```
The ``nipalsPLS1`` class in hoggorm accepts only **numpy** arrays with numerical values and not pandas data frames. Therefore, the pandas data frames holding the imported data need to be "taken apart" into three parts:
* two numpy array holding the numeric values
* two Python list holding variable (column) names
* two Python list holding object (row) names.
The numpy arrays with values will be used as input for the ``nipalsPLS2`` class for analysis. The Python lists holding the variable and row names will be used later in the plotting function from the **hoggormPlot** package when visualising the results of the analysis. Below is the code needed to access both data, variable names and object names.
```
# Get the values from the data frame
X = X_df.values
y = y_df.values
# Get the variable or columns names
X_varNames = list(X_df.columns)
y_varNames = list(y_df.columns)
# Get the object or row names
X_objNames = list(X_df.index)
y_objNames = list(y_df.index)
```
---
### Apply PLSR to our data
Now, let's run PLSR on the data using the ``nipalsPLS1`` class, since we have a univariate response. The documentation provides a [description of the input parameters](https://hoggorm.readthedocs.io/en/latest/plsr.html). Using input paramter ``arrX`` and ``vecy`` we define which numpy array we would like to analyse. ``vecy`` is what typically is considered to be the response vector, while the measurements are typically defined as ``arrX``. By setting input parameter ``Xstand=False`` we make sure that the variables are only mean centered, not scaled to unit variance, if this is what you want. This is the default setting and actually doesn't need to expressed explicitly. Setting paramter ``cvType=["loo"]`` we make sure that we compute the PLS2 model using full cross validation. ``"loo"`` means "Leave One Out". By setting paramter ``numpComp=10`` we ask for four components to be computed.
```
model = ho.nipalsPLS1(arrX=X, Xstand=False,
vecy=Y,
cvType=["loo"],
numComp=10)
```
That's it, the PLS2 model has been computed. Now we would like to inspect the results by visualising them. We can do this using plotting functions of the separate [**hoggormPlot** package](https://hoggormplot.readthedocs.io/en/latest/). If we wish to plot the results for component 1 and component 2, we can do this by setting the input argument ``comp=[1, 2]``. The input argument ``plots=[1, 6]`` lets the user define which plots are to be plotted. If this list for example contains value ``1``, the function will generate the scores plot for the model. If the list contains value ``6`` the explained variance plot for y will be plotted. The hoggormPlot documentation provides a [description of input paramters](https://hoggormplot.readthedocs.io/en/latest/mainPlot.html).
```
hop.plot(model, comp=[1, 2],
plots=[1, 6],
objNames=X_objNames,
XvarNames=X_varNames,
YvarNames=Y_varNames)
```
Plots can also be called separately.
```
# Plot cumulative explained variance (both calibrated and validated) using a specific function for that.
hop.explainedVariance(model)
# Plot cumulative validated explained variance in X.
hop.explainedVariance(model, which='X')
hop.scores(model)
# Plot X loadings in line plot
hop.loadings(model, weights=True, line=True)
# Plot regression coefficients
hop.coefficients(model, comp=3)
```
---
### Accessing numerical results
Now that we have visualised the PLSR results, we may also want to access the numerical results. Below are some examples. For a complete list of accessible results, please see this part of the documentation.
```
# Get X scores and store in numpy array
X_scores = model.X_scores()
# Get scores and store in pandas dataframe with row and column names
X_scores_df = pd.DataFrame(model.X_scores())
X_scores_df.index = X_objNames
X_scores_df.columns = ['Comp {0}'.format(x+1) for x in range(model.X_scores().shape[1])]
X_scores_df
help(ho.nipalsPLS1.X_scores)
# Dimension of the X_scores
np.shape(model.X_scores())
```
We see that the numpy array holds the scores for all countries and OECD (35 in total) for four components as required when computing the PCA model.
```
# Get X loadings and store in numpy array
X_loadings = model.X_loadings()
# Get X loadings and store in pandas dataframe with row and column names
X_loadings_df = pd.DataFrame(model.X_loadings())
X_loadings_df.index = X_varNames
X_loadings_df.columns = ['Comp {0}'.format(x+1) for x in range(model.X_loadings().shape[1])]
X_loadings_df
help(ho.nipalsPLS1.X_loadings)
np.shape(model.X_loadings())
```
Here we see that the array holds the loadings for the 10 variables in the data across four components.
```
# Get Y loadings and store in numpy array
Y_loadings = model.Y_loadings()
# Get Y loadings and store in pandas dataframe with row and column names
Y_loadings_df = pd.DataFrame(model.Y_loadings())
Y_loadings_df.index = Y_varNames
Y_loadings_df.columns = ['Comp {0}'.format(x+1) for x in range(model.Y_loadings().shape[1])]
Y_loadings_df
# Get X correlation loadings and store in numpy array
X_corrloadings = model.X_corrLoadings()
# Get X correlation loadings and store in pandas dataframe with row and column names
X_corrloadings_df = pd.DataFrame(model.X_corrLoadings())
X_corrloadings_df.index = X_varNames
X_corrloadings_df.columns = ['Comp {0}'.format(x+1) for x in range(model.X_corrLoadings().shape[1])]
X_corrloadings_df
help(ho.nipalsPLS1.X_corrLoadings)
# Get Y loadings and store in numpy array
Y_corrloadings = model.X_corrLoadings()
# Get Y loadings and store in pandas dataframe with row and column names
Y_corrloadings_df = pd.DataFrame(model.Y_corrLoadings())
Y_corrloadings_df.index = Y_varNames
Y_corrloadings_df.columns = ['Comp {0}'.format(x+1) for x in range(model.Y_corrLoadings().shape[1])]
Y_corrloadings_df
help(ho.nipalsPLS1.Y_corrLoadings)
# Get calibrated explained variance of each component in X
X_calExplVar = model.X_calExplVar()
# Get calibrated explained variance in X and store in pandas dataframe with row and column names
X_calExplVar_df = pd.DataFrame(model.X_calExplVar())
X_calExplVar_df.columns = ['calibrated explained variance in X']
X_calExplVar_df.index = ['Comp {0}'.format(x+1) for x in range(model.X_loadings().shape[1])]
X_calExplVar_df
help(ho.nipalsPLS1.X_calExplVar)
# Get calibrated explained variance of each component in Y
Y_calExplVar = model.Y_calExplVar()
# Get calibrated explained variance in Y and store in pandas dataframe with row and column names
Y_calExplVar_df = pd.DataFrame(model.Y_calExplVar())
Y_calExplVar_df.columns = ['calibrated explained variance in Y']
Y_calExplVar_df.index = ['Comp {0}'.format(x+1) for x in range(model.Y_loadings().shape[1])]
Y_calExplVar_df
help(ho.nipalsPLS1.Y_calExplVar)
# Get cumulative calibrated explained variance in X
X_cumCalExplVar = model.X_cumCalExplVar()
# Get cumulative calibrated explained variance in X and store in pandas dataframe with row and column names
X_cumCalExplVar_df = pd.DataFrame(model.X_cumCalExplVar())
X_cumCalExplVar_df.columns = ['cumulative calibrated explained variance in X']
X_cumCalExplVar_df.index = ['Comp {0}'.format(x) for x in range(model.X_loadings().shape[1] + 1)]
X_cumCalExplVar_df
help(ho.nipalsPLS1.X_cumCalExplVar)
# Get cumulative calibrated explained variance in Y
Y_cumCalExplVar = model.Y_cumCalExplVar()
# Get cumulative calibrated explained variance in Y and store in pandas dataframe with row and column names
Y_cumCalExplVar_df = pd.DataFrame(model.Y_cumCalExplVar())
Y_cumCalExplVar_df.columns = ['cumulative calibrated explained variance in Y']
Y_cumCalExplVar_df.index = ['Comp {0}'.format(x) for x in range(model.Y_loadings().shape[1] + 1)]
Y_cumCalExplVar_df
help(ho.nipalsPLS1.Y_cumCalExplVar)
# Get cumulative calibrated explained variance for each variable in X
X_cumCalExplVar_ind = model.X_cumCalExplVar_indVar()
# Get cumulative calibrated explained variance for each variable in X and store in pandas dataframe with row and column names
X_cumCalExplVar_ind_df = pd.DataFrame(model.X_cumCalExplVar_indVar())
X_cumCalExplVar_ind_df.columns = X_varNames
X_cumCalExplVar_ind_df.index = ['Comp {0}'.format(x) for x in range(model.X_loadings().shape[1] + 1)]
X_cumCalExplVar_ind_df
help(ho.nipalsPLS1.X_cumCalExplVar_indVar)
# Get calibrated predicted Y for a given number of components
# Predicted Y from calibration using 1 component
Y_from_1_component = model.Y_predCal()[1]
# Predicted Y from calibration using 1 component stored in pandas data frame with row and columns names
Y_from_1_component_df = pd.DataFrame(model.Y_predCal()[1])
Y_from_1_component_df.index = Y_objNames
Y_from_1_component_df.columns = Y_varNames
Y_from_1_component_df
# Get calibrated predicted Y for a given number of components
# Predicted Y from calibration using 4 component
Y_from_4_component = model.Y_predCal()[4]
# Predicted Y from calibration using 1 component stored in pandas data frame with row and columns names
Y_from_4_component_df = pd.DataFrame(model.Y_predCal()[4])
Y_from_4_component_df.index = Y_objNames
Y_from_4_component_df.columns = Y_varNames
Y_from_4_component_df
help(ho.nipalsPLS1.X_predCal)
# Get validated explained variance of each component X
X_valExplVar = model.X_valExplVar()
# Get calibrated explained variance in X and store in pandas dataframe with row and column names
X_valExplVar_df = pd.DataFrame(model.X_valExplVar())
X_valExplVar_df.columns = ['validated explained variance in X']
X_valExplVar_df.index = ['Comp {0}'.format(x+1) for x in range(model.X_loadings().shape[1])]
X_valExplVar_df
help(ho.nipalsPLS1.X_valExplVar)
# Get validated explained variance of each component Y
Y_valExplVar = model.Y_valExplVar()
# Get calibrated explained variance in X and store in pandas dataframe with row and column names
Y_valExplVar_df = pd.DataFrame(model.Y_valExplVar())
Y_valExplVar_df.columns = ['validated explained variance in Y']
Y_valExplVar_df.index = ['Comp {0}'.format(x+1) for x in range(model.Y_loadings().shape[1])]
Y_valExplVar_df
help(ho.nipalsPLS1.Y_valExplVar)
# Get cumulative validated explained variance in X
X_cumValExplVar = model.X_cumValExplVar()
# Get cumulative validated explained variance in X and store in pandas dataframe with row and column names
X_cumValExplVar_df = pd.DataFrame(model.X_cumValExplVar())
X_cumValExplVar_df.columns = ['cumulative validated explained variance in X']
X_cumValExplVar_df.index = ['Comp {0}'.format(x) for x in range(model.X_loadings().shape[1] + 1)]
X_cumValExplVar_df
help(ho.nipalsPLS1.X_cumValExplVar)
# Get cumulative validated explained variance in Y
Y_cumValExplVar = model.Y_cumValExplVar()
# Get cumulative validated explained variance in Y and store in pandas dataframe with row and column names
Y_cumValExplVar_df = pd.DataFrame(model.Y_cumValExplVar())
Y_cumValExplVar_df.columns = ['cumulative validated explained variance in Y']
Y_cumValExplVar_df.index = ['Comp {0}'.format(x) for x in range(model.Y_loadings().shape[1] + 1)]
Y_cumValExplVar_df
help(ho.nipalsPLS1.Y_cumValExplVar)
help(ho.nipalsPLS1.X_cumValExplVar_indVar)
# Get validated predicted Y for a given number of components
# Predicted Y from validation using 1 component
Y_from_1_component_val = model.Y_predVal()[1]
# Predicted Y from calibration using 1 component stored in pandas data frame with row and columns names
Y_from_1_component_val_df = pd.DataFrame(model.Y_predVal()[1])
Y_from_1_component_val_df.index = Y_objNames
Y_from_1_component_val_df.columns = Y_varNames
Y_from_1_component_val_df
# Get validated predicted Y for a given number of components
# Predicted Y from validation using 3 components
Y_from_3_component_val = model.Y_predVal()[3]
# Predicted Y from calibration using 3 components stored in pandas data frame with row and columns names
Y_from_3_component_val_df = pd.DataFrame(model.Y_predVal()[3])
Y_from_3_component_val_df.index = Y_objNames
Y_from_3_component_val_df.columns = Y_varNames
Y_from_3_component_val_df
help(ho.nipalsPLS1.Y_predVal)
# Get predicted scores for new measurements (objects) of X
# First pretend that we acquired new X data by using part of the existing data and overlaying some noise
import numpy.random as npr
new_X = X[0:4, :] + npr.rand(4, np.shape(X)[1])
np.shape(X)
# Now insert the new data into the existing model and compute scores for two components (numComp=2)
pred_X_scores = model.X_scores_predict(new_X, numComp=2)
# Same as above, but results stored in a pandas dataframe with row names and column names
pred_X_scores_df = pd.DataFrame(model.X_scores_predict(new_X, numComp=2))
pred_X_scores_df.columns = ['Comp {0}'.format(x+1) for x in range(2)]
pred_X_scores_df.index = ['new object {0}'.format(x+1) for x in range(np.shape(new_X)[0])]
pred_X_scores_df
help(ho.nipalsPLS1.X_scores_predict)
# Predict Y from new X data
pred_Y = model.Y_predict(new_X, numComp=2)
# Predict Y from nex X data and store results in a pandas dataframe with row names and column names
pred_Y_df = pd.DataFrame(model.Y_predict(new_X, numComp=2))
pred_Y_df.columns = Y_varNames
pred_Y_df.index = ['new object {0}'.format(x+1) for x in range(np.shape(new_X)[0])]
pred_Y_df
```
|
github_jupyter
|
import hoggorm as ho
import hoggormplot as hop
import pandas as pd
import numpy as np
# Load fluorescence data
X_df = pd.read_csv('gasoline_NIR.txt', header=None, sep='\s+')
X_df
# Load response data, that is octane measurements
y_df = pd.read_csv('gasoline_octane.txt', header=None, sep='\s+')
y_df
# Get the values from the data frame
X = X_df.values
y = y_df.values
# Get the variable or columns names
X_varNames = list(X_df.columns)
y_varNames = list(y_df.columns)
# Get the object or row names
X_objNames = list(X_df.index)
y_objNames = list(y_df.index)
model = ho.nipalsPLS1(arrX=X, Xstand=False,
vecy=Y,
cvType=["loo"],
numComp=10)
hop.plot(model, comp=[1, 2],
plots=[1, 6],
objNames=X_objNames,
XvarNames=X_varNames,
YvarNames=Y_varNames)
# Plot cumulative explained variance (both calibrated and validated) using a specific function for that.
hop.explainedVariance(model)
# Plot cumulative validated explained variance in X.
hop.explainedVariance(model, which='X')
hop.scores(model)
# Plot X loadings in line plot
hop.loadings(model, weights=True, line=True)
# Plot regression coefficients
hop.coefficients(model, comp=3)
# Get X scores and store in numpy array
X_scores = model.X_scores()
# Get scores and store in pandas dataframe with row and column names
X_scores_df = pd.DataFrame(model.X_scores())
X_scores_df.index = X_objNames
X_scores_df.columns = ['Comp {0}'.format(x+1) for x in range(model.X_scores().shape[1])]
X_scores_df
help(ho.nipalsPLS1.X_scores)
# Dimension of the X_scores
np.shape(model.X_scores())
# Get X loadings and store in numpy array
X_loadings = model.X_loadings()
# Get X loadings and store in pandas dataframe with row and column names
X_loadings_df = pd.DataFrame(model.X_loadings())
X_loadings_df.index = X_varNames
X_loadings_df.columns = ['Comp {0}'.format(x+1) for x in range(model.X_loadings().shape[1])]
X_loadings_df
help(ho.nipalsPLS1.X_loadings)
np.shape(model.X_loadings())
# Get Y loadings and store in numpy array
Y_loadings = model.Y_loadings()
# Get Y loadings and store in pandas dataframe with row and column names
Y_loadings_df = pd.DataFrame(model.Y_loadings())
Y_loadings_df.index = Y_varNames
Y_loadings_df.columns = ['Comp {0}'.format(x+1) for x in range(model.Y_loadings().shape[1])]
Y_loadings_df
# Get X correlation loadings and store in numpy array
X_corrloadings = model.X_corrLoadings()
# Get X correlation loadings and store in pandas dataframe with row and column names
X_corrloadings_df = pd.DataFrame(model.X_corrLoadings())
X_corrloadings_df.index = X_varNames
X_corrloadings_df.columns = ['Comp {0}'.format(x+1) for x in range(model.X_corrLoadings().shape[1])]
X_corrloadings_df
help(ho.nipalsPLS1.X_corrLoadings)
# Get Y loadings and store in numpy array
Y_corrloadings = model.X_corrLoadings()
# Get Y loadings and store in pandas dataframe with row and column names
Y_corrloadings_df = pd.DataFrame(model.Y_corrLoadings())
Y_corrloadings_df.index = Y_varNames
Y_corrloadings_df.columns = ['Comp {0}'.format(x+1) for x in range(model.Y_corrLoadings().shape[1])]
Y_corrloadings_df
help(ho.nipalsPLS1.Y_corrLoadings)
# Get calibrated explained variance of each component in X
X_calExplVar = model.X_calExplVar()
# Get calibrated explained variance in X and store in pandas dataframe with row and column names
X_calExplVar_df = pd.DataFrame(model.X_calExplVar())
X_calExplVar_df.columns = ['calibrated explained variance in X']
X_calExplVar_df.index = ['Comp {0}'.format(x+1) for x in range(model.X_loadings().shape[1])]
X_calExplVar_df
help(ho.nipalsPLS1.X_calExplVar)
# Get calibrated explained variance of each component in Y
Y_calExplVar = model.Y_calExplVar()
# Get calibrated explained variance in Y and store in pandas dataframe with row and column names
Y_calExplVar_df = pd.DataFrame(model.Y_calExplVar())
Y_calExplVar_df.columns = ['calibrated explained variance in Y']
Y_calExplVar_df.index = ['Comp {0}'.format(x+1) for x in range(model.Y_loadings().shape[1])]
Y_calExplVar_df
help(ho.nipalsPLS1.Y_calExplVar)
# Get cumulative calibrated explained variance in X
X_cumCalExplVar = model.X_cumCalExplVar()
# Get cumulative calibrated explained variance in X and store in pandas dataframe with row and column names
X_cumCalExplVar_df = pd.DataFrame(model.X_cumCalExplVar())
X_cumCalExplVar_df.columns = ['cumulative calibrated explained variance in X']
X_cumCalExplVar_df.index = ['Comp {0}'.format(x) for x in range(model.X_loadings().shape[1] + 1)]
X_cumCalExplVar_df
help(ho.nipalsPLS1.X_cumCalExplVar)
# Get cumulative calibrated explained variance in Y
Y_cumCalExplVar = model.Y_cumCalExplVar()
# Get cumulative calibrated explained variance in Y and store in pandas dataframe with row and column names
Y_cumCalExplVar_df = pd.DataFrame(model.Y_cumCalExplVar())
Y_cumCalExplVar_df.columns = ['cumulative calibrated explained variance in Y']
Y_cumCalExplVar_df.index = ['Comp {0}'.format(x) for x in range(model.Y_loadings().shape[1] + 1)]
Y_cumCalExplVar_df
help(ho.nipalsPLS1.Y_cumCalExplVar)
# Get cumulative calibrated explained variance for each variable in X
X_cumCalExplVar_ind = model.X_cumCalExplVar_indVar()
# Get cumulative calibrated explained variance for each variable in X and store in pandas dataframe with row and column names
X_cumCalExplVar_ind_df = pd.DataFrame(model.X_cumCalExplVar_indVar())
X_cumCalExplVar_ind_df.columns = X_varNames
X_cumCalExplVar_ind_df.index = ['Comp {0}'.format(x) for x in range(model.X_loadings().shape[1] + 1)]
X_cumCalExplVar_ind_df
help(ho.nipalsPLS1.X_cumCalExplVar_indVar)
# Get calibrated predicted Y for a given number of components
# Predicted Y from calibration using 1 component
Y_from_1_component = model.Y_predCal()[1]
# Predicted Y from calibration using 1 component stored in pandas data frame with row and columns names
Y_from_1_component_df = pd.DataFrame(model.Y_predCal()[1])
Y_from_1_component_df.index = Y_objNames
Y_from_1_component_df.columns = Y_varNames
Y_from_1_component_df
# Get calibrated predicted Y for a given number of components
# Predicted Y from calibration using 4 component
Y_from_4_component = model.Y_predCal()[4]
# Predicted Y from calibration using 1 component stored in pandas data frame with row and columns names
Y_from_4_component_df = pd.DataFrame(model.Y_predCal()[4])
Y_from_4_component_df.index = Y_objNames
Y_from_4_component_df.columns = Y_varNames
Y_from_4_component_df
help(ho.nipalsPLS1.X_predCal)
# Get validated explained variance of each component X
X_valExplVar = model.X_valExplVar()
# Get calibrated explained variance in X and store in pandas dataframe with row and column names
X_valExplVar_df = pd.DataFrame(model.X_valExplVar())
X_valExplVar_df.columns = ['validated explained variance in X']
X_valExplVar_df.index = ['Comp {0}'.format(x+1) for x in range(model.X_loadings().shape[1])]
X_valExplVar_df
help(ho.nipalsPLS1.X_valExplVar)
# Get validated explained variance of each component Y
Y_valExplVar = model.Y_valExplVar()
# Get calibrated explained variance in X and store in pandas dataframe with row and column names
Y_valExplVar_df = pd.DataFrame(model.Y_valExplVar())
Y_valExplVar_df.columns = ['validated explained variance in Y']
Y_valExplVar_df.index = ['Comp {0}'.format(x+1) for x in range(model.Y_loadings().shape[1])]
Y_valExplVar_df
help(ho.nipalsPLS1.Y_valExplVar)
# Get cumulative validated explained variance in X
X_cumValExplVar = model.X_cumValExplVar()
# Get cumulative validated explained variance in X and store in pandas dataframe with row and column names
X_cumValExplVar_df = pd.DataFrame(model.X_cumValExplVar())
X_cumValExplVar_df.columns = ['cumulative validated explained variance in X']
X_cumValExplVar_df.index = ['Comp {0}'.format(x) for x in range(model.X_loadings().shape[1] + 1)]
X_cumValExplVar_df
help(ho.nipalsPLS1.X_cumValExplVar)
# Get cumulative validated explained variance in Y
Y_cumValExplVar = model.Y_cumValExplVar()
# Get cumulative validated explained variance in Y and store in pandas dataframe with row and column names
Y_cumValExplVar_df = pd.DataFrame(model.Y_cumValExplVar())
Y_cumValExplVar_df.columns = ['cumulative validated explained variance in Y']
Y_cumValExplVar_df.index = ['Comp {0}'.format(x) for x in range(model.Y_loadings().shape[1] + 1)]
Y_cumValExplVar_df
help(ho.nipalsPLS1.Y_cumValExplVar)
help(ho.nipalsPLS1.X_cumValExplVar_indVar)
# Get validated predicted Y for a given number of components
# Predicted Y from validation using 1 component
Y_from_1_component_val = model.Y_predVal()[1]
# Predicted Y from calibration using 1 component stored in pandas data frame with row and columns names
Y_from_1_component_val_df = pd.DataFrame(model.Y_predVal()[1])
Y_from_1_component_val_df.index = Y_objNames
Y_from_1_component_val_df.columns = Y_varNames
Y_from_1_component_val_df
# Get validated predicted Y for a given number of components
# Predicted Y from validation using 3 components
Y_from_3_component_val = model.Y_predVal()[3]
# Predicted Y from calibration using 3 components stored in pandas data frame with row and columns names
Y_from_3_component_val_df = pd.DataFrame(model.Y_predVal()[3])
Y_from_3_component_val_df.index = Y_objNames
Y_from_3_component_val_df.columns = Y_varNames
Y_from_3_component_val_df
help(ho.nipalsPLS1.Y_predVal)
# Get predicted scores for new measurements (objects) of X
# First pretend that we acquired new X data by using part of the existing data and overlaying some noise
import numpy.random as npr
new_X = X[0:4, :] + npr.rand(4, np.shape(X)[1])
np.shape(X)
# Now insert the new data into the existing model and compute scores for two components (numComp=2)
pred_X_scores = model.X_scores_predict(new_X, numComp=2)
# Same as above, but results stored in a pandas dataframe with row names and column names
pred_X_scores_df = pd.DataFrame(model.X_scores_predict(new_X, numComp=2))
pred_X_scores_df.columns = ['Comp {0}'.format(x+1) for x in range(2)]
pred_X_scores_df.index = ['new object {0}'.format(x+1) for x in range(np.shape(new_X)[0])]
pred_X_scores_df
help(ho.nipalsPLS1.X_scores_predict)
# Predict Y from new X data
pred_Y = model.Y_predict(new_X, numComp=2)
# Predict Y from nex X data and store results in a pandas dataframe with row names and column names
pred_Y_df = pd.DataFrame(model.Y_predict(new_X, numComp=2))
pred_Y_df.columns = Y_varNames
pred_Y_df.index = ['new object {0}'.format(x+1) for x in range(np.shape(new_X)[0])]
pred_Y_df
| 0.816955 | 0.987228 |
### This is Example 4.3. Gambler’s Problem from Sutton's book.
A gambler has the opportunity to make bets on the outcomes of a sequence of coin flips.
If the coin comes up heads, he wins as many dollars as he has staked on that flip;
if it is tails, he loses his stake. The game ends when the gambler wins by reaching his goal of $100,
or loses by running out of money.
On each flip, the gambler must decide what portion of his capital to stake, in integer numbers of dollars.
This problem can be formulated as an undiscounted, episodic, finite MDP.
The state is the gambler’s capital, s ∈ {1, 2, . . . , 99}.
The actions are stakes, a ∈ {0, 1, . . . , min(s, 100 − s)}.
The reward is zero on all transitions except those on which the gambler reaches his goal, when it is +1.
The state-value function then gives the probability of winning from each state. A policy is a mapping from levels of capital to stakes. The optimal policy maximizes the probability of reaching the goal. Let p_h denote the probability of the coin coming up heads. If p_h is known, then the entire problem is known and it can be solved, for instance, by value iteration.
```
import numpy as np
import sys
import matplotlib.pyplot as plt
if "../" not in sys.path:
sys.path.append("../")
```
### Exercise 4.9 (programming)
Implement value iteration for the gambler’s problem and solve it for p_h = 0.25 and p_h = 0.55.
```
def value_iteration_for_gamblers(p_h, theta=0.0001, discount_factor=1.0):
"""
Args:
p_h: Probability of the coin coming up heads
"""
def one_step_lookahead(s, V, rewards):
"""
Helper function to calculate the value for all action in a given state.
Args:
s: The gambler’s capital. Integer.
V: The vector that contains values at each state.
rewards: The reward vector.
Returns:
A vector containing the expected value of each action.
Its length equals to the number of actions.
"""
A = np.zeros(101)
for a in range(1, min(s, 100-s)+1):
A[a] = p_h * (rewards[s+a] + discount_factor*V[s+a]) + (1-p_h) * (rewards[s-a] + discount_factor*V[s-a])
return A
rewards = np.zeros(101)
rewards[100] = 1
V = np.zeros(101)
policy = np.zeros(101)
while True:
delta = 0
for s in range(1, 101):
A = one_step_lookahead(s, V, rewards)
v = np.max(A)
delta = max(delta, np.abs(V[s]-v))
V[s] = v
if delta < theta:
break
for s in range(1, 101):
A = one_step_lookahead(s, V, rewards)
policy[s] = np.argmax(A)
return policy, V
policy, v = value_iteration_for_gamblers(0.25)
print("Optimized Policy:")
print(policy)
print("")
print("Optimized Value Function:")
print(v)
print("")
# Plotting Final Policy (action stake) vs State (Capital)
# Implement!
# Plotting Capital vs Final Policy
# Implement!
```
|
github_jupyter
|
import numpy as np
import sys
import matplotlib.pyplot as plt
if "../" not in sys.path:
sys.path.append("../")
def value_iteration_for_gamblers(p_h, theta=0.0001, discount_factor=1.0):
"""
Args:
p_h: Probability of the coin coming up heads
"""
def one_step_lookahead(s, V, rewards):
"""
Helper function to calculate the value for all action in a given state.
Args:
s: The gambler’s capital. Integer.
V: The vector that contains values at each state.
rewards: The reward vector.
Returns:
A vector containing the expected value of each action.
Its length equals to the number of actions.
"""
A = np.zeros(101)
for a in range(1, min(s, 100-s)+1):
A[a] = p_h * (rewards[s+a] + discount_factor*V[s+a]) + (1-p_h) * (rewards[s-a] + discount_factor*V[s-a])
return A
rewards = np.zeros(101)
rewards[100] = 1
V = np.zeros(101)
policy = np.zeros(101)
while True:
delta = 0
for s in range(1, 101):
A = one_step_lookahead(s, V, rewards)
v = np.max(A)
delta = max(delta, np.abs(V[s]-v))
V[s] = v
if delta < theta:
break
for s in range(1, 101):
A = one_step_lookahead(s, V, rewards)
policy[s] = np.argmax(A)
return policy, V
policy, v = value_iteration_for_gamblers(0.25)
print("Optimized Policy:")
print(policy)
print("")
print("Optimized Value Function:")
print(v)
print("")
# Plotting Final Policy (action stake) vs State (Capital)
# Implement!
# Plotting Capital vs Final Policy
# Implement!
| 0.469763 | 0.931898 |
# Full Python Course
## By: Dr. Amin Oroji
## Chief Data Scientist
## Prata Technology
# Python
- An open source
- cross-platform programming language
- become increasingly popular over the last ten years
- It was first released in 1991
- A multi-purpose programming languages (due to its many extensions)
- examples are **scientific computing** and **calculations**, **simulations**, **web development** (using, e.g., the Django Web framework), **Data Analytics**, etc.
- Interpreted Programming language
## ***WHY Python?***
- open source and free.
- highly extendable due to its high number of free available Python Packaged and Libraries.
- simple, flexible code structure and easy to learn.
- can be used on all platforms (Windows, macOS and Linux).
- The popularity of Python is growing fast.
- Multi-purpose programming language.
- It has efficient high-level data structures.
- has simple but effective approach to object-oriented programming (OOP).
## Chapter1: Getting Started
for the main source please click [here](https://github.com/AminOroji/Elementary-Python-suf)
### Python Installation on Windows
**Step 1: Select Version of Python to Install**\
**Step 2: Download Python Executable Installer**\
Open your web browser and navigate to the [Downloads for Windows section](https://www.python.org/downloads/windows/) of the [official Python website](https://www.python.org/).\
**Step 3: Run Executable Installer**\
- Run the Python Installer once downloaded. (In this example, we have downloaded Python 3.7.3.)
- Make sure you select the Install launcher for all users and Add Python 3.7 to PATH checkboxes.\
The latter places the interpreter in the execution path. For older versions of Python that do not support the Add Python to Path checkbox, see Step 6.
- Select Install Now – the recommended installation options.\
For all recent versions of Python, the recommended installation options include Pip and IDLE. Older versions might not include such additional features.
- The next dialog will prompt you to select whether to Disable path length limit. Choosing this option will allow Python to bypass the 260-character MAX_PATH limit. Effectively, it will enable Python to use long path names.
**Step 4: Verify Python Was Installed On Windows**
- Navigate to the directory in which Python was installed on the system. In our case, it is C:\Users\Username\AppData\Local\Programs\Python\Python37 since we have installed the latest version.
- Double-click python.exe.
**Step 5: Verify Pip Was Installed**
If you opted to install an older version of Python, it is possible that it did not come with Pip preinstalled. Pip is a powerful package management system for Python software packages. Thus, make sure that you have it installed.
We recommend using Pip for most Python packages, especially when working in virtual environments.
To verify whether Pip was installed:
- Open the Start menu and type "cmd."
- Select the Command Prompt application.
- Enter pip -V in the console.
Pip has not been installed yet if you get the following output:
```
'pip' is not recognized as an internal or external command,
Operable program or batch file
```
**Step 6: Add Python Path to Environment Variables (Optional)**
We recommend you go through this step if your version of the Python installer does not include the Add Python to PATH checkbox or if you have not selected that option.
Setting up the Python path to system variables alleviates the need for using full paths. It instructs Windows to look through all the PATH folders for “python” and find the install folder that contains the python.exe file.
- Open the Start menu and start the Run app.
- Type sysdm.cpl and click OK. This opens the System Properties window.
- Navigate to the Advanced tab and select Environment Variables.
- Under System Variables, find and select the Path variable.
- Click Edit.
- Select the Variable value field. Add the path to the python.exe file preceded with a semicolon (;). For example, ```;C:\Python34.```
- Click OK and close all windows.
By setting this up, you can execute Python scripts like this: ```Python script.py```
Instead of this: ```C:/Python34/Python script.py```
As you can see, it is cleaner and more manageable.
**Step 7: Install virtualnv (Optional)**
You have Python, and you have Pip to manage packages. Now, you need one last software package - virtualnv. Virtualnv enables you to create isolated local virtual environments for your Python projects.
***Why use virtualnv?***
Python software packages are installed system-wide by default. Consequently, whenever a single project-specific package is changed, it changes for all your Python projects. You would want to avoid this, and having separate virtual environments for each project is the easiest solution.
To install virtualnv:
1. Open the Start menu and type "cmd."
2. Select the Command Prompt application.
3. Type the following pip command in the console:
```C:\Users\Username> pip install virtualenv```
Upon completion, virtualnv is installed on your system.
### Python IDEs
- [vscode](https://code.visualstudio.com/download)
- [sublim](https://www.sublimetext.com/download)
### Jupyter Notebook
Jupyter Notebook is an open-source web application that allows you to create and share documents that contain live code, equations, visualizations, and narrative text. Uses include data cleaning and transformation, numerical simulation, statistical modeling, data visualization, machine learning, and much more.
Jupyter has support for over 40 different programming languages and Python is one of them. Python is a requirement (Python 3.3 or greater, or Python 2.7) for installing the Jupyter Notebook itself.
#### Installing Jupyter Notebook using pip:
PIP is a package management system used to install and manage software packages/libraries written in Python. These files are stored in a large **on-line repository** termed as Python Package Index (PyPI).
pip uses PyPI as the default source for packages and their dependencies.
To install Jupyter using pip, we need to first check if pip is updated in our system.
- Use the following command to update pip:
```python -m pip install --upgrade pip```
After updating the pip version, follow the instructions provided below to install Jupyter:
- Command to install Jupyter:
```python -m pip install jupyter```
- Launching Jupyter:
Use the following command to launch Jupyter using command-line:
```jupyter notebook```
### Google Colab
### Let's Play (Using Python as a Calculator)
```
2 + 2
3 * 4
50 - 5*6
(50 - 5*6) / 4
8 / 5
print(17 / 3) # classic division returns a float
print(17 // 3) # floor division discards the fractional part
print(17 % 3) # the % operator returns the remainder of the division
print(5 * 3 + 2)
print(2 ** 10)
width = 20
height = 5 * 9
width * height
2++2
2++++++++++2
02
print('Hello World!')
print 'Hello World!'
print('Hello World!'
print('Hello World!)
```
In interactive mode, the last printed expression is assigned to the variable _. This means that when you are using Python as a desk calculator, it is somewhat easier to continue calculations, for example:
```
tax = 12.5 / 100
price = 100.50
price * tax
price + _
round(_, 2)
'spam eggs' # single quotes
'doesn\'t' # use \' to escape the single quote...
"doesn't" # ...or use double quotes instead
'"Yes," they said.'
"\"Yes,\" they said."
'"Isn\'t," they said.'
print('"Isn\'t," they said.')
s = 'First line.\nSecond line.' # \n means newline
s # without print(), \n is included in the output
print(s)
print("""\
Usage: thingy [OPTIONS]
-h Display this usage message
-H hostname Hostname to connect to
""")
print(3 * 'un' + 'ium')
print(3 * ('un' + 'ium'))
```
|
github_jupyter
|
'pip' is not recognized as an internal or external command,
Operable program or batch file
- Click OK and close all windows.
By setting this up, you can execute Python scripts like this: ```Python script.py```
Instead of this: ```C:/Python34/Python script.py```
As you can see, it is cleaner and more manageable.
**Step 7: Install virtualnv (Optional)**
You have Python, and you have Pip to manage packages. Now, you need one last software package - virtualnv. Virtualnv enables you to create isolated local virtual environments for your Python projects.
***Why use virtualnv?***
Python software packages are installed system-wide by default. Consequently, whenever a single project-specific package is changed, it changes for all your Python projects. You would want to avoid this, and having separate virtual environments for each project is the easiest solution.
To install virtualnv:
1. Open the Start menu and type "cmd."
2. Select the Command Prompt application.
3. Type the following pip command in the console:
Upon completion, virtualnv is installed on your system.
### Python IDEs
- [vscode](https://code.visualstudio.com/download)
- [sublim](https://www.sublimetext.com/download)
### Jupyter Notebook
Jupyter Notebook is an open-source web application that allows you to create and share documents that contain live code, equations, visualizations, and narrative text. Uses include data cleaning and transformation, numerical simulation, statistical modeling, data visualization, machine learning, and much more.
Jupyter has support for over 40 different programming languages and Python is one of them. Python is a requirement (Python 3.3 or greater, or Python 2.7) for installing the Jupyter Notebook itself.
#### Installing Jupyter Notebook using pip:
PIP is a package management system used to install and manage software packages/libraries written in Python. These files are stored in a large **on-line repository** termed as Python Package Index (PyPI).
pip uses PyPI as the default source for packages and their dependencies.
To install Jupyter using pip, we need to first check if pip is updated in our system.
- Use the following command to update pip:
After updating the pip version, follow the instructions provided below to install Jupyter:
- Command to install Jupyter:
- Launching Jupyter:
Use the following command to launch Jupyter using command-line:
### Google Colab
### Let's Play (Using Python as a Calculator)
In interactive mode, the last printed expression is assigned to the variable _. This means that when you are using Python as a desk calculator, it is somewhat easier to continue calculations, for example:
| 0.754373 | 0.9455 |
```
!wget "https://raw.githubusercontent.com/udacity/deep-learning-v2-pytorch/master/convolutional-neural-networks/conv-visualization/data/udacity_sdc.png"
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.image as mpimg
import cv2
import numpy as np
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import torch
import torch.nn.functional as F
import torch.nn as nn
# Any results you write to the current directory are saved as output.
```
CNNs bases their principles in Image feature extraction using Convolutional kernels let's see how this kernels works on and Image and then we will see how to contruct a CNN network using Pytorch
```
from torchvision import datasets
import torchvision.transforms as transforms
from torch.utils.data.sampler import SubsetRandomSampler
# number of subprocesses to use for data loading
num_workers = 3 # lets try to parallelize the data loading
# how many samples per batch to load
batch_size = 20
# percentage of training set to use as validation
valid_size = 0.2
# convert data to a normalized torch.FloatTensor
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))
])
# choose the training and test datasets
train_data = datasets.CIFAR10('input/CIFAR', train=True,
download=True, transform=transform)
test_data = datasets.CIFAR10('input/CIFAR', train=False,
download=True, transform=transform)
# obtain training indices that will be used for validation
num_train = len(train_data)
indices = list(range(num_train))
np.random.shuffle(indices)
split = int(np.floor(valid_size * num_train))
train_idx, valid_idx = indices[split:], indices[:split]
# define samplers for obtaining training and validation batches
train_sampler = SubsetRandomSampler(train_idx)
valid_sampler = SubsetRandomSampler(valid_idx)
# prepare data loaders (combine dataset and sampler)
train_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=train_sampler, num_workers=num_workers)
valid_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=valid_sampler, num_workers=num_workers)
test_loader = torch.utils.data.DataLoader(test_data, batch_size=batch_size,
num_workers=num_workers)
# specify the image classes
classes = ['airplane', 'automobile', 'bird', 'cat', 'deer',
'dog', 'frog', 'horse', 'ship', 'truck']
import matplotlib.pyplot as plt
# helper function to un-normalize and display an image
def imshow(img,label=None):
img = img / 2 + 0.5 # unnormalize
if label:
plt.title(classes[label])
plt.imshow(np.transpose(img, (1, 2, 0))) # convert from Tensor image
```
## Visualizing Kernel effects
let's se how convolutions affects our Images
```
dataiter = iter(train_loader)
images, labels = dataiter.next()
imshow(images[0],labels[0])
```
In the following cell you can be able to see how applying a particular filter (Convolutional kernel) will affect an image filtering out unrelevant features or emphasizing particular ones, in this case applying the sobel_y filter will emphasize horizontal lines and filtersout other features.
for edge detection it's important that all te elements of the kernel sum 0 in order to don't alterate the original brightness of the image, since we just want to detect the edges (high frequency regions in image (high-pass filter))
- 0 no change
- positive - more brighter
- negative - less brighter
```
# 3x3 array for edge detection
sobel_y = np.array([[ -1, -2, -1],
[ 0, 0, 0],
[ 1, 2, 1]])
# Filter the image using filter2D, which has inputs: (grayscale image, bit-depth, kernel)
filtered_image = cv2.filter2D(images[0].numpy()[0], -1, sobel_y)
plt.imshow(filtered_image, cmap='gray')
```
To see it clear let's actually write a convolutional layer to see the outputs it's produce and also how the activation function affect the results
So let's say we have the following images and filters gray scale image
```
bgr_img = cv2.imread("udacity_sdc.png") # we normalize the image to ly into 0-1 range so tat our filters and sGD will work better
gray_img = cv2.cvtColor(bgr_img, cv2.COLOR_BGR2GRAY).astype("float32")/255
plt.imshow(gray_img,cmap='gray')
gray_img_tensor = torch.from_numpy(gray_img).unsqueeze(0).unsqueeze(1).float()
```
Then we have 4 filters one for left vertical lines, other for right vertical lines and others 2 for horizontal lines
```
right_V = np.array([[-1, -1, 1, 1],
[-1, -1, 1, 1],
[-1, -1, 1, 1],
[-1, -1, 1, 1]])
left_V = -right_V
down_H = right_V.T
up_H = -down_H
filters = np.array([right_V, left_V, down_H, up_H])
"""
Cite: taken from udacity/deep-learning-v2-pytorch
"""
def viz_filters(filters):
fig = plt.figure(figsize=(10, 5))
fig.subplots_adjust(left=0, right=1.5, bottom=0.8, top=1, hspace=0.05, wspace=0.05)
for i in range(4):
ax = fig.add_subplot(1, 4, i+1, xticks=[], yticks=[])
ax.imshow(filters[i], cmap='gray')
ax.set_title('Filter %s' % str(i+1))
width, height = filters[i].shape
for x in range(width):
for y in range(height):
ax.annotate(str(filters[i][x][y]), xy=(y,x),
horizontalalignment='center',
verticalalignment='center',
color='white' if filters[i][x][y]<0 else 'black')
viz_filters(filters)
```
then we will create a conv layer that uses this filters as conv kernels
```
import torch.nn.functional as F
class convtest(torch.nn.Module):
def __init__(self,weights):
super().__init__()
kernel_height,kernels_width=weights.shape[-2:] #the last two are the rows and columns of the kernels
#we input 1 "feature map(image) 1 dimension", we have 4 filters so we output 4 feature maps
self.convLayer1=nn.Conv2d(1, 4, kernel_size=(kernel_height, kernels_width), bias=False)
#we replace the weights with custom filters
self.convLayer1.weight = torch.nn.Parameter(weights)
# define a pooling layer
self.maxPool = nn.MaxPool2d(2, 2)#maxpooling with stride 2 and window of size 2x2 we speact to have the a half of the size o the input
self.averagePool=nn.AvgPool2d(2,2)#Avgpooling with stride 2 and window of size 2x2 we speact to have the a half of the size o the input
def forward(self,inputs):
convolved=self.convLayer1(inputs)
withActivation=F.relu(convolved)
maxPooled=self.maxPool(withActivation)
avgPooled=self.averagePool(withActivation)
return convolved,withActivation,maxPooled,avgPooled
```
we will create the layers, cast our filters to
```
weights = torch.from_numpy(filters).unsqueeze(1).type(torch.FloatTensor)
model = convtest(weights)
model
# helper function for visualizing the output of a given layer
# default number of filters is 4
def viz_layer(layer, n_filters= 4,title=''):
fig = plt.figure(figsize=(20, 20))
fig.suptitle(title,y=0.6)
for i in range(n_filters):
ax = fig.add_subplot(1, n_filters, i+1, xticks=[], yticks=[])
# grab layer outputs
ax.imshow(np.squeeze(layer[0,i].data.numpy()), cmap='gray')
ax.set_title(f"Output {i+1}")
# plot original image
plt.imshow(gray_img, cmap='gray')
viz_filters(model.convLayer1.weight.squeeze(1).detach().int().numpy())
gray_img_tensor = torch.from_numpy(gray_img).unsqueeze(0).unsqueeze(1)
# get the convolutional layer (pre and post activation)
conv_layer, activated_layer,maxPooled,avgPooled = model(gray_img_tensor)
# visualize the output of a conv layer
viz_layer(conv_layer,title='Conv Layer No Activation Function')
viz_layer(activated_layer,title='Conv Layer with Activation Function')
viz_layer(maxPooled,title='Max Pooling results')
viz_layer(avgPooled,title='Avg Pooling results')
print(f"Conv Layer Output Shape: {str(conv_layer.shape):>40}")
print(f"Activated Conv Output Shape: {str(activated_layer.shape):>36}")
print(f"MaxPooling Output Shape: {str(maxPooled.shape):>40}")
print(f"AveragePooling Output Shape: {str(avgPooled.shape):>36}")
```
## Building a CNN for the CIFAR dataset
Now let's build a network to classify the images in the CIFAR dataset and see the kernels that our network will learn
For image classification tasks we usually want to increase the depth of our features but to decrease its size to create a bootleneck for compressing information obtaining a hicherical structure of the features going from general to particular
- In our case we are going to **increase** by the double the number of kernels to get more feature maps trought each convLayer **16 features--> 32 features--> 64 features**
- then we will **downsample** the features **HxW** by a half each layer using MaxPooling to extract the most important features for classification **size(32x32) --> size (16x16) --> size (8x8) --> size (4x4)**
- At the end we will perfom the classification flattening our output feature maps and passing it throught a fully connected network so the input of the fc network will be
$$n_{features} = depth*height*width$$
where depth is the number of feature maps coming and usually height and weight are the same value and could be calculated as
$$(height | width)=\frac{input_{(height|width)}}{P_{1}.stride*P_{2}.stride....*P_{n}.stride}$$
**only when you mantain the same input-output size in each conv layer**
where P refers to the pooling layers applied until the desired output sequencially , remember pooling layers each time their are applied downsample our height|weight by a factor of the stride size (2 a half,1 maintain the same size, etc...)
```
from tqdm.auto import tqdm,trange
class metrics():
def __init__(self):
self.loss=[]
self.accuracy=[]
def append(self,loss,accuracy):
self.loss.append(loss)
self.accuracy.append(accuracy)
class CIFARNET(torch.nn.Module):
def __init__(self,input_d,size):#size assuming H and W are the same (32x32)
super().__init__()
"""
the First Layer will have 16 kernels with size 3
an stride of 1 to preserve te size and a padding of 1 to complete the missing pixels of the 3x3 kernels
"""
self.conv1=nn.Conv2d(input_d,16,kernel_size=3,stride=1,padding=1)#sees (32x32)x3
self.conv2=nn.Conv2d(self.conv1.out_channels,32,kernel_size=3,stride=1,padding=1)#sees (16x16)x16 image tensor we've downsample the HxW with maxpooling a half
self.conv3=nn.Conv2d(self.conv2.out_channels,64,kernel_size=3,stride=1,padding=1)#sees (8x8)x32 image tensor we've downsample the HxW with maxpooling a half
self.maxPooling=nn.MaxPool2d(2,2)#window size 2,stride 2 downsample the features height and weight to a half of their size
"""The fc network recieves a flatten tensor of size 64(feature maps)*height*width
where this H and w has been downsample by a factor of 2 each time we applied MaxPooling with stride 2
so the input image has a size of 32x32 pixels then the feature smaps of conv1 16x16, then 8x8 and finaly 4*4
"""
fc_input_f=self.conv3.out_channels*((size//(self.maxPooling.stride**3))**2)
self.fc=nn.Sequential(#sees (4x4)x64 image tensor we've downsample the HxW with maxpooling a half
nn.Linear(fc_input_f,512),
nn.ReLU(),
nn.Dropout(0.25),
nn.Linear(512,10),
nn.LogSoftmax(dim=1)
)
""" we can also don't use activation an use crossentropy loss
but cross entropy loss applies logsoftmax and Nlloss so is more efficient to use Logsofmax
and NLLoss, and then just use torch.exp to get the actual probabilities"""
def forward(self,inputs):
h_1=self.maxPooling(F.relu(self.conv1(inputs)))#feature maps by the conv1 max pooled (16x16)x16
h_2=self.maxPooling(F.relu(self.conv2(h_1))) #feature maps by the conv2 max pooled (8x8)x32
h_3=self.maxPooling(F.relu(self.conv3(h_2))) #feature maps by the conv3 max pooled (4x4)x64
h_3_flatten=h_3.view(-1,self.fc._modules['0'].in_features)# conv3 feature maps flattened (64 * 4 * 4) , we have already calculated in is the in-features of fc layer
return self.fc(h_3_flatten) #log probabilities for each class (we must use exp to get the actual probabilities
def fit(self,train_generator,val_generator,criterion,Optimizer,faccuracy,Epochs=10,device='cuda'):
self.to(device)
train_batches=len(train_generator)
val_batches=len(val_generator)
val_metrics,train_metrics = metrics(),metrics()
for epoch in trange(Epochs,desc='Epochs:'):
#Train steps
self.train()
train_accuracy,train_loss=0,0
for images,labels in tqdm(train_generator,desc='Train Steps:',leave=False):
images,labels=images.to(device),labels.to(device)
Optimizer.zero_grad()#clean the gradients of optimizer
logProbs=self.forward(images)#calculate logProbabilities
loss=criterion(logProbs,labels)#calculating loss
loss.backward()#Calculating loss gradient with respect the parameters
Optimizer.step()#Optimization step (backpropagation)
train_loss+=loss.item()
train_accuracy+=faccuracy(torch.exp(logProbs),labels).item()
train_metrics.append(train_loss/train_batches,train_accuracy/train_batches)
#Validation steps
self.eval()#turns off dropout
val_accuracy,val_loss=0,0
for images,labels in tqdm(val_generator,desc='Val Steps:',leave=False):
with torch.no_grad():
images,labels=images.to(device),labels.to(device)
logProbs=self.forward(images)
val_loss+=criterion(logProbs,labels).item()
val_accuracy+=faccuracy(torch.exp(logProbs),labels).item()
val_metrics.append(val_loss/val_batches,val_accuracy/val_batches)
print(f"EPOCH: {epoch}"
f"\nTrain loss: {train_metrics.loss[-1]:.4f} Train accuracy: {train_metrics.accuracy[-1]:.4f}"
f"\nVal loss: {val_metrics.loss[-1]:.4f} Val accuracy: {val_metrics.accuracy[-1]:.4f}")
return train_metrics,val_metrics
images.shape
model=CIFARNET(images.shape[1],images.shape[2])
print("Model Description: ",model,"\nTest model\n",torch.sum(torch.exp(model(images)),dim=1))
```
Let's train and test our mode
```
import matplotlib.pyplot as plt
def plot_train_history(train_metrics,val_metrics):
train_loss,train_accuracy = train_metrics.loss,train_metrics.accuracy
val_loss,val_accuracy = val_metrics.loss,val_metrics.accuracy
plt.plot(train_loss, label='Training loss')
plt.plot(val_loss, label='Validation loss')
plt.legend(frameon=False)
plt.show()
plt.plot(train_accuracy, label='Training accuracy')
plt.plot(val_accuracy, label='Validation accuracy')
plt.legend(frameon=False)
plt.show()
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
def f_accuracy(predictions,labels):
top_p, top_class = predictions.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
return torch.mean(equals.type(torch.FloatTensor))
criterion=nn.NLLLoss()
Optimizer=torch.optim.SGD(model.parameters(), lr=0.01)
train_metrics,val_metrics=model.fit(train_loader,valid_loader,criterion,Optimizer,f_accuracy)
plot_train_history(train_metrics,val_metrics)
```
## Data Augmentation
We have several problems in this model the first one is that our maximum accuracy archieved whitout overfitting the model is around 70% that is not too god for this data set so there are several problems that could be happening in our.
First we know that convNets have some properties by themselves for example they provide translation invariance (in a some way by the kernels working) so they can found a pattern in an image (matrix) wherever they are but there are a lack of propierties that we need to correctly perfom our task (detect a kind of object and classify it) such as
- Rotation Invariance (recognizes patterns no matter it rotation in the image)
- Scale Invariance (recognizes patterns no metter it's size in the image)
So in order to produce better results and give our model rotation,scale and translation invariences propierties we can do several things one of the simplest way is just adding images that train our model to detect this patter in dificult situations so basically we perfom a preprocessing in our training set such as:
- Image rotations: to give examples with diferent rotation
- Shift images: to give examples where the object is not in a typical location
- Scale the image: to give our image examples where the patterns appear in different scale(this is more harder so there is other ways to perfom this than Data Augmentation )
Data augmentation is considered a resampling techinique which helps to deal a lot of problems in ML algorithms in this case give our images better examples for classfication creating sintetic data.
**we only apply data augmentation in our training set we didn't modify our test set cause we want ur test data to be the more similar to our real cases**
```
from torchvision import datasets
import torchvision.transforms as transforms
from torch.utils.data.sampler import SubsetRandomSampler
# number of subprocesses to use for data loading
num_workers = 3 # lets try to parallelize the data loading
# how many samples per batch to load
batch_size = 20
# percentage of training set to use as validation
valid_size = 0.2
# let's perom data augmentation (sintetized samples)
transform_Augmented = transforms.Compose([
transforms.RandomRotation(10),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))
])
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))
])
# choose the training and test datasets
train_data = datasets.CIFAR10('input/CIFAR', train=True,
download=True, transform=transform_Augmented)
#we only apply data augmentation in our training set we didn't modify our test set cause we want ur test data to be the more similar to our real cases
test_data = datasets.CIFAR10('input/CIFAR', train=False,
download=True, transform=transform)
# obtain training indices that will be used for validation
num_train = len(train_data)
indices = list(range(num_train))
np.random.shuffle(indices)
split = int(np.floor(valid_size * num_train))
train_idx, valid_idx = indices[split:], indices[:split]
# define samplers for obtaining training and validation batches
train_sampler = SubsetRandomSampler(train_idx)
valid_sampler = SubsetRandomSampler(valid_idx)
# prepare data loaders (combine dataset and sampler)
train_loader_aug = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=train_sampler, num_workers=num_workers)
valid_loader_aug = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=valid_sampler, num_workers=num_workers)
test_loader_aug = torch.utils.data.DataLoader(test_data, batch_size=batch_size,
num_workers=num_workers)
```
Then this is how our data looks now
```
classes = ['airplane', 'automobile', 'bird', 'cat', 'deer',
'dog', 'frog', 'horse', 'ship', 'truck']
dataiter = iter(train_loader_aug)
images, labels = dataiter.next()
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(25, 4))
# display 20 images
for idx in np.arange(20):
ax = fig.add_subplot(2, 20/2, idx+1, xticks=[], yticks=[])
imshow(images[idx])
ax.set_title(classes[labels[idx]])
```
then let's see if our model valid loss and accuracy could be improved by using this resample technique
```
model_aug=CIFARNET(images.shape[1],images.shape[2])
Optimizer=torch.optim.SGD(model_aug.parameters(), lr=0.01)
train_metrics_aug,val_metrics_aug=model_aug.fit(train_loader_aug,valid_loader_aug,criterion,Optimizer,f_accuracy,Epochs=28)
plot_train_history(train_metrics_aug,val_metrics_aug)
```
|
github_jupyter
|
!wget "https://raw.githubusercontent.com/udacity/deep-learning-v2-pytorch/master/convolutional-neural-networks/conv-visualization/data/udacity_sdc.png"
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.image as mpimg
import cv2
import numpy as np
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import torch
import torch.nn.functional as F
import torch.nn as nn
# Any results you write to the current directory are saved as output.
from torchvision import datasets
import torchvision.transforms as transforms
from torch.utils.data.sampler import SubsetRandomSampler
# number of subprocesses to use for data loading
num_workers = 3 # lets try to parallelize the data loading
# how many samples per batch to load
batch_size = 20
# percentage of training set to use as validation
valid_size = 0.2
# convert data to a normalized torch.FloatTensor
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))
])
# choose the training and test datasets
train_data = datasets.CIFAR10('input/CIFAR', train=True,
download=True, transform=transform)
test_data = datasets.CIFAR10('input/CIFAR', train=False,
download=True, transform=transform)
# obtain training indices that will be used for validation
num_train = len(train_data)
indices = list(range(num_train))
np.random.shuffle(indices)
split = int(np.floor(valid_size * num_train))
train_idx, valid_idx = indices[split:], indices[:split]
# define samplers for obtaining training and validation batches
train_sampler = SubsetRandomSampler(train_idx)
valid_sampler = SubsetRandomSampler(valid_idx)
# prepare data loaders (combine dataset and sampler)
train_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=train_sampler, num_workers=num_workers)
valid_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=valid_sampler, num_workers=num_workers)
test_loader = torch.utils.data.DataLoader(test_data, batch_size=batch_size,
num_workers=num_workers)
# specify the image classes
classes = ['airplane', 'automobile', 'bird', 'cat', 'deer',
'dog', 'frog', 'horse', 'ship', 'truck']
import matplotlib.pyplot as plt
# helper function to un-normalize and display an image
def imshow(img,label=None):
img = img / 2 + 0.5 # unnormalize
if label:
plt.title(classes[label])
plt.imshow(np.transpose(img, (1, 2, 0))) # convert from Tensor image
dataiter = iter(train_loader)
images, labels = dataiter.next()
imshow(images[0],labels[0])
# 3x3 array for edge detection
sobel_y = np.array([[ -1, -2, -1],
[ 0, 0, 0],
[ 1, 2, 1]])
# Filter the image using filter2D, which has inputs: (grayscale image, bit-depth, kernel)
filtered_image = cv2.filter2D(images[0].numpy()[0], -1, sobel_y)
plt.imshow(filtered_image, cmap='gray')
bgr_img = cv2.imread("udacity_sdc.png") # we normalize the image to ly into 0-1 range so tat our filters and sGD will work better
gray_img = cv2.cvtColor(bgr_img, cv2.COLOR_BGR2GRAY).astype("float32")/255
plt.imshow(gray_img,cmap='gray')
gray_img_tensor = torch.from_numpy(gray_img).unsqueeze(0).unsqueeze(1).float()
right_V = np.array([[-1, -1, 1, 1],
[-1, -1, 1, 1],
[-1, -1, 1, 1],
[-1, -1, 1, 1]])
left_V = -right_V
down_H = right_V.T
up_H = -down_H
filters = np.array([right_V, left_V, down_H, up_H])
"""
Cite: taken from udacity/deep-learning-v2-pytorch
"""
def viz_filters(filters):
fig = plt.figure(figsize=(10, 5))
fig.subplots_adjust(left=0, right=1.5, bottom=0.8, top=1, hspace=0.05, wspace=0.05)
for i in range(4):
ax = fig.add_subplot(1, 4, i+1, xticks=[], yticks=[])
ax.imshow(filters[i], cmap='gray')
ax.set_title('Filter %s' % str(i+1))
width, height = filters[i].shape
for x in range(width):
for y in range(height):
ax.annotate(str(filters[i][x][y]), xy=(y,x),
horizontalalignment='center',
verticalalignment='center',
color='white' if filters[i][x][y]<0 else 'black')
viz_filters(filters)
import torch.nn.functional as F
class convtest(torch.nn.Module):
def __init__(self,weights):
super().__init__()
kernel_height,kernels_width=weights.shape[-2:] #the last two are the rows and columns of the kernels
#we input 1 "feature map(image) 1 dimension", we have 4 filters so we output 4 feature maps
self.convLayer1=nn.Conv2d(1, 4, kernel_size=(kernel_height, kernels_width), bias=False)
#we replace the weights with custom filters
self.convLayer1.weight = torch.nn.Parameter(weights)
# define a pooling layer
self.maxPool = nn.MaxPool2d(2, 2)#maxpooling with stride 2 and window of size 2x2 we speact to have the a half of the size o the input
self.averagePool=nn.AvgPool2d(2,2)#Avgpooling with stride 2 and window of size 2x2 we speact to have the a half of the size o the input
def forward(self,inputs):
convolved=self.convLayer1(inputs)
withActivation=F.relu(convolved)
maxPooled=self.maxPool(withActivation)
avgPooled=self.averagePool(withActivation)
return convolved,withActivation,maxPooled,avgPooled
weights = torch.from_numpy(filters).unsqueeze(1).type(torch.FloatTensor)
model = convtest(weights)
model
# helper function for visualizing the output of a given layer
# default number of filters is 4
def viz_layer(layer, n_filters= 4,title=''):
fig = plt.figure(figsize=(20, 20))
fig.suptitle(title,y=0.6)
for i in range(n_filters):
ax = fig.add_subplot(1, n_filters, i+1, xticks=[], yticks=[])
# grab layer outputs
ax.imshow(np.squeeze(layer[0,i].data.numpy()), cmap='gray')
ax.set_title(f"Output {i+1}")
# plot original image
plt.imshow(gray_img, cmap='gray')
viz_filters(model.convLayer1.weight.squeeze(1).detach().int().numpy())
gray_img_tensor = torch.from_numpy(gray_img).unsqueeze(0).unsqueeze(1)
# get the convolutional layer (pre and post activation)
conv_layer, activated_layer,maxPooled,avgPooled = model(gray_img_tensor)
# visualize the output of a conv layer
viz_layer(conv_layer,title='Conv Layer No Activation Function')
viz_layer(activated_layer,title='Conv Layer with Activation Function')
viz_layer(maxPooled,title='Max Pooling results')
viz_layer(avgPooled,title='Avg Pooling results')
print(f"Conv Layer Output Shape: {str(conv_layer.shape):>40}")
print(f"Activated Conv Output Shape: {str(activated_layer.shape):>36}")
print(f"MaxPooling Output Shape: {str(maxPooled.shape):>40}")
print(f"AveragePooling Output Shape: {str(avgPooled.shape):>36}")
from tqdm.auto import tqdm,trange
class metrics():
def __init__(self):
self.loss=[]
self.accuracy=[]
def append(self,loss,accuracy):
self.loss.append(loss)
self.accuracy.append(accuracy)
class CIFARNET(torch.nn.Module):
def __init__(self,input_d,size):#size assuming H and W are the same (32x32)
super().__init__()
"""
the First Layer will have 16 kernels with size 3
an stride of 1 to preserve te size and a padding of 1 to complete the missing pixels of the 3x3 kernels
"""
self.conv1=nn.Conv2d(input_d,16,kernel_size=3,stride=1,padding=1)#sees (32x32)x3
self.conv2=nn.Conv2d(self.conv1.out_channels,32,kernel_size=3,stride=1,padding=1)#sees (16x16)x16 image tensor we've downsample the HxW with maxpooling a half
self.conv3=nn.Conv2d(self.conv2.out_channels,64,kernel_size=3,stride=1,padding=1)#sees (8x8)x32 image tensor we've downsample the HxW with maxpooling a half
self.maxPooling=nn.MaxPool2d(2,2)#window size 2,stride 2 downsample the features height and weight to a half of their size
"""The fc network recieves a flatten tensor of size 64(feature maps)*height*width
where this H and w has been downsample by a factor of 2 each time we applied MaxPooling with stride 2
so the input image has a size of 32x32 pixels then the feature smaps of conv1 16x16, then 8x8 and finaly 4*4
"""
fc_input_f=self.conv3.out_channels*((size//(self.maxPooling.stride**3))**2)
self.fc=nn.Sequential(#sees (4x4)x64 image tensor we've downsample the HxW with maxpooling a half
nn.Linear(fc_input_f,512),
nn.ReLU(),
nn.Dropout(0.25),
nn.Linear(512,10),
nn.LogSoftmax(dim=1)
)
""" we can also don't use activation an use crossentropy loss
but cross entropy loss applies logsoftmax and Nlloss so is more efficient to use Logsofmax
and NLLoss, and then just use torch.exp to get the actual probabilities"""
def forward(self,inputs):
h_1=self.maxPooling(F.relu(self.conv1(inputs)))#feature maps by the conv1 max pooled (16x16)x16
h_2=self.maxPooling(F.relu(self.conv2(h_1))) #feature maps by the conv2 max pooled (8x8)x32
h_3=self.maxPooling(F.relu(self.conv3(h_2))) #feature maps by the conv3 max pooled (4x4)x64
h_3_flatten=h_3.view(-1,self.fc._modules['0'].in_features)# conv3 feature maps flattened (64 * 4 * 4) , we have already calculated in is the in-features of fc layer
return self.fc(h_3_flatten) #log probabilities for each class (we must use exp to get the actual probabilities
def fit(self,train_generator,val_generator,criterion,Optimizer,faccuracy,Epochs=10,device='cuda'):
self.to(device)
train_batches=len(train_generator)
val_batches=len(val_generator)
val_metrics,train_metrics = metrics(),metrics()
for epoch in trange(Epochs,desc='Epochs:'):
#Train steps
self.train()
train_accuracy,train_loss=0,0
for images,labels in tqdm(train_generator,desc='Train Steps:',leave=False):
images,labels=images.to(device),labels.to(device)
Optimizer.zero_grad()#clean the gradients of optimizer
logProbs=self.forward(images)#calculate logProbabilities
loss=criterion(logProbs,labels)#calculating loss
loss.backward()#Calculating loss gradient with respect the parameters
Optimizer.step()#Optimization step (backpropagation)
train_loss+=loss.item()
train_accuracy+=faccuracy(torch.exp(logProbs),labels).item()
train_metrics.append(train_loss/train_batches,train_accuracy/train_batches)
#Validation steps
self.eval()#turns off dropout
val_accuracy,val_loss=0,0
for images,labels in tqdm(val_generator,desc='Val Steps:',leave=False):
with torch.no_grad():
images,labels=images.to(device),labels.to(device)
logProbs=self.forward(images)
val_loss+=criterion(logProbs,labels).item()
val_accuracy+=faccuracy(torch.exp(logProbs),labels).item()
val_metrics.append(val_loss/val_batches,val_accuracy/val_batches)
print(f"EPOCH: {epoch}"
f"\nTrain loss: {train_metrics.loss[-1]:.4f} Train accuracy: {train_metrics.accuracy[-1]:.4f}"
f"\nVal loss: {val_metrics.loss[-1]:.4f} Val accuracy: {val_metrics.accuracy[-1]:.4f}")
return train_metrics,val_metrics
images.shape
model=CIFARNET(images.shape[1],images.shape[2])
print("Model Description: ",model,"\nTest model\n",torch.sum(torch.exp(model(images)),dim=1))
import matplotlib.pyplot as plt
def plot_train_history(train_metrics,val_metrics):
train_loss,train_accuracy = train_metrics.loss,train_metrics.accuracy
val_loss,val_accuracy = val_metrics.loss,val_metrics.accuracy
plt.plot(train_loss, label='Training loss')
plt.plot(val_loss, label='Validation loss')
plt.legend(frameon=False)
plt.show()
plt.plot(train_accuracy, label='Training accuracy')
plt.plot(val_accuracy, label='Validation accuracy')
plt.legend(frameon=False)
plt.show()
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
def f_accuracy(predictions,labels):
top_p, top_class = predictions.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
return torch.mean(equals.type(torch.FloatTensor))
criterion=nn.NLLLoss()
Optimizer=torch.optim.SGD(model.parameters(), lr=0.01)
train_metrics,val_metrics=model.fit(train_loader,valid_loader,criterion,Optimizer,f_accuracy)
plot_train_history(train_metrics,val_metrics)
from torchvision import datasets
import torchvision.transforms as transforms
from torch.utils.data.sampler import SubsetRandomSampler
# number of subprocesses to use for data loading
num_workers = 3 # lets try to parallelize the data loading
# how many samples per batch to load
batch_size = 20
# percentage of training set to use as validation
valid_size = 0.2
# let's perom data augmentation (sintetized samples)
transform_Augmented = transforms.Compose([
transforms.RandomRotation(10),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))
])
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))
])
# choose the training and test datasets
train_data = datasets.CIFAR10('input/CIFAR', train=True,
download=True, transform=transform_Augmented)
#we only apply data augmentation in our training set we didn't modify our test set cause we want ur test data to be the more similar to our real cases
test_data = datasets.CIFAR10('input/CIFAR', train=False,
download=True, transform=transform)
# obtain training indices that will be used for validation
num_train = len(train_data)
indices = list(range(num_train))
np.random.shuffle(indices)
split = int(np.floor(valid_size * num_train))
train_idx, valid_idx = indices[split:], indices[:split]
# define samplers for obtaining training and validation batches
train_sampler = SubsetRandomSampler(train_idx)
valid_sampler = SubsetRandomSampler(valid_idx)
# prepare data loaders (combine dataset and sampler)
train_loader_aug = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=train_sampler, num_workers=num_workers)
valid_loader_aug = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=valid_sampler, num_workers=num_workers)
test_loader_aug = torch.utils.data.DataLoader(test_data, batch_size=batch_size,
num_workers=num_workers)
classes = ['airplane', 'automobile', 'bird', 'cat', 'deer',
'dog', 'frog', 'horse', 'ship', 'truck']
dataiter = iter(train_loader_aug)
images, labels = dataiter.next()
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(25, 4))
# display 20 images
for idx in np.arange(20):
ax = fig.add_subplot(2, 20/2, idx+1, xticks=[], yticks=[])
imshow(images[idx])
ax.set_title(classes[labels[idx]])
model_aug=CIFARNET(images.shape[1],images.shape[2])
Optimizer=torch.optim.SGD(model_aug.parameters(), lr=0.01)
train_metrics_aug,val_metrics_aug=model_aug.fit(train_loader_aug,valid_loader_aug,criterion,Optimizer,f_accuracy,Epochs=28)
plot_train_history(train_metrics_aug,val_metrics_aug)
| 0.8398 | 0.882326 |
# "Recreating the BBC style graphic in Python - `plotnine` and `altair`"
> Want to make awesome chart with bbc style? This is an attempt to reproduce https://bbc.github.io/rcookbook/ in python.
- toc: true
- badges: true
- comments: true
- categories: [python]
- hide: trues
# Todo
- [ ] Missing Subtitle (plotnine)
- [ ] Missing Style
`plotnine` is an implementation of `ggplot` in python, while `altair` is base on `vega-lite`. Pick one that sparks joy.
# Difference between plotnine and ggplot
90% of them are the same, except that in python you have to wrap column names in `''`, otherwise it will be treated as variable and caused error. Most of the time you just need to wrap a `''` or replaced with `_` depends on the function.
Some of the features are missing in plotnine, for example, subtitle is not possible.
I tried to produce the same chart with `plotnine` and `altair`, and hopefully you will see their difference. `plotnine` covers 99% of `ggplot2`, so if you are coming from R, just go ahead with `plotnine`! `altair` is another interesting visualization library that base on vega-lite, therefore it can be integrated with website easily. In addition, it can also produce interactive chart with very simple function, which is a big plus!
# Setup
```
# collapse-hide
# !pip install plotnine[all]
# !pip install altair
# !pip install gapminder
from gapminder import gapminder
from plotnine.data import mtcars
from plotnine import *
from plotnine import ggplot, geom_point, aes, stat_smooth, facet_wrap, geom_line
from plotnine import ggplot # https://plotnine.readthedocs.io/en/stable/
import altair as alt
import pandas as pd
import plotnine
from bbc_plot.bbc_plot import bbc_style
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
alt.renderers.enable('html')
print(f'altair version: {alt.__version__}')
print(f'plotnine version: {plotnine.__version__}')
print(f'pandas version: {pd.__version__}')
```
# Make a Line Chart
```r
# ggplot
line_df <- gapminder %>%
filter(country == "Malawi")
#Make plot
line <- ggplot(line_df, aes(x = year, y = lifeExp)) +
geom_line(colour = "#1380A1", size = 1) +
geom_hline(yintercept = 0, size = 1, colour="#333333") +
bbc_style() +
labs(title="Living longer",
subtitle = "Life expectancy in Malawi 1952-2007")
```
```
#hide
line_df = gapminder.query(" country == 'Malawi' ")
(ggplot(line_df, aes(x='year', y='lifeExp')) +
geom_line(colour='#1380A1', size=1) +
geom_hline(yintercept = 0, size = 1, colour='#333333') +
labs(title='Living longer',
subtitle = 'Life expectancy in Malawi 1952-2007')
)
## altair
line = (alt.Chart(line_df).mark_line().encode(
x='year',
y='lifeExp')
.properties(title={'text': 'Living Longer',
'subtitle': 'Life expectancy in Malawi 1952-2007'})
)
# hline
overlay = overlay = pd.DataFrame({'y': [0]})
hline = alt.Chart(overlay).mark_rule(color='#333333', strokeWidth=3).encode(y='y:Q')
line + hline
```
# The BBC style
```R
function ()
{
font <- "Helvetica"
ggplot2::theme(plot.title = ggplot2::element_text(family = font,
size = 28, face = "bold", color = "#222222"), plot.subtitle = ggplot2::element_text(family = font,
size = 22, margin = ggplot2::margin(9, 0, 9, 0)), plot.caption = ggplot2::element_blank(),
legend.position = "top", legend.text.align = 0, legend.background = ggplot2::element_blank(),
legend.title = ggplot2::element_blank(), legend.key = ggplot2::element_blank(),
legend.text = ggplot2::element_text(family = font, size = 18,
color = "#222222"), axis.title = ggplot2::element_blank(),
axis.text = ggplot2::element_text(family = font, size = 18,
color = "#222222"), axis.text.x = ggplot2::element_text(margin = ggplot2::margin(5,
b = 10)), axis.ticks = ggplot2::element_blank(),
axis.line = ggplot2::element_blank(), panel.grid.minor = ggplot2::element_blank(),
panel.grid.major.y = ggplot2::element_line(color = "#cbcbcb"),
panel.grid.major.x = ggplot2::element_blank(), panel.background = ggplot2::element_blank(),
strip.background = ggplot2::element_rect(fill = "white"),
strip.text = ggplot2::element_text(size = 22, hjust = 0))
}
<environment: namespace:bbplot>
```
```
font = "Helvetica"
theme(plot_title=element_text(family=font,
size=28, face="bold", color="#222222"),
# plot_subtitle=element_text(family=font,
# size=22, plot_margin=(9, 0, 9, 0)), plot_caption=element_blank(),
legend_position="top", legend_title_align=0, legend_background=element_blank(),
legend_title=element_blank(), legend_key=element_blank(),
legend_text=element_text(family=font, size=18,
color="#222222"), axis_title=element_blank(),
axis_text=element_text(family=font, size=18,
color="#222222"),
axis_text_x=element_text(margin={'t': 5, 'b': 10}),
axis_ticks=element_blank(),
axis_line=element_blank(), panel_grid_minor=element_blank(),
panel_grid_major_y=element_line(color="#cbcbcb"),
panel_grid_major_x=element_blank(), panel_background=element_blank(),
strip_background=element_rect(fill="white"),
strip_text=element_text(size=22, hjust=0)
)
```
The `finalise_plot()` function does more than just save out your chart, it also left-aligns the title and subtitle as is standard for BBC graphics, adds a footer with the logo on the right side and lets you input source text on the left side.
```
# altair
line = (alt.Chart(line_df).mark_line().encode(
x='year',
y='lifeExp')
.properties(title={'text': 'Living Longer',
'subtitle': 'Life expectancy in China 1952-2007'})
)
# hline
overlay = overlay = pd.DataFrame({'lifeExp': [0]})
hline = alt.Chart(overlay).mark_rule(color='#333333', strokeWidth=3).encode(y='lifeExp:Q')
line + hline
```
# Make a multiple line chart
```
# hide
# Prepare data
multiline_df = gapminder.query(
'country == "China" | country =="United States" ')
```
```r
# ggplot
#Prepare data
multiple_line_df <- gapminder %>%
filter(country == "China" | country == "United States")
#Make plot
multiple_line <- ggplot(multiple_line_df, aes(x = year, y = lifeExp, colour = country)) +
geom_line(size = 1) +
geom_hline(yintercept = 0, size = 1, colour="#333333") +
scale_colour_manual(values = c("#FAAB18", "#1380A1")) +
bbc_style() +
labs(title="Living longer",
subtitle = "Life expectancy in China and the US")
```
```
# Make plot
multiline = (
ggplot(multiline_df, aes(x='year', y='lifeExp', colour='country')) +
geom_line(colour="#1380A1", size=1) +
geom_hline(yintercept=0, size=1, color="#333333") +
scale_colour_manual(values=["#FAAB18", "#1380A1"]) +
bbc_style() +
labs(title="Living longer",
subtitle="Life expectancy in China 1952-2007"))
multiline
multiline_altair = (alt.Chart(multiline_df).mark_line().encode(
x='year',
y='lifeExp',
color='country')
.properties(title={'text': 'Living Longer',
'subtitle': 'Life expectancy in China 1952-2007'})
)
# hline
overlay = overlay = pd.DataFrame({'lifeExp': [0]})
hline = alt.Chart(overlay).mark_rule(color='#333333', strokeWidth=3).encode(y='lifeExp:Q')
multiline_altair + hline
```
# Make a bar chart
```r
# ggplot
#Prepare data
bar_df <- gapminder %>%
filter(year == 2007 & continent == "Africa") %>%
arrange(desc(lifeExp)) %>%
head(5)
#Make plot
bars <- ggplot(bar_df, aes(x = country, y = lifeExp)) +
geom_bar(stat="identity",
position="identity",
fill="#1380A1") +
geom_hline(yintercept = 0, size = 1, colour="#333333") +
bbc_style() +
labs(title="Reunion is highest",
subtitle = "Highest African life expectancy, 2007")
```
```
## hide
bar_df = gapminder.query(' year == 2007 & continent == "Africa" ').nlargest(5, 'lifeExp')
bars_ggplot = (ggplot(bar_df, aes(x='country', y='lifeExp')) +
geom_bar(stat="identity",
position="identity",
fill="#1380A1") +
geom_hline(yintercept=0, size=1, colour="#333333") +
# bbc_style() +
labs(title="Reunion is highest",
subtitle="Highest African life expectancy, 2007"))
bars_ggplot
bars_altair = (alt.Chart(bar_df).mark_bar().encode(
x='country',
y='lifeExp',
# color='country'
)
.properties(title={'text': 'Reunion is highest',
'subtitle': 'Highest African life expectancy, 2007'})
)
bars_altair
```
# Make a stacked bar chart
```
## collapse-hide
stacked_bar_df = (
gapminder.query(' year == 2007')
.assign(
lifeExpGrouped=lambda x: pd.cut(
x['lifeExp'],
bins=[0, 50, 65, 80, 90],
labels=["under 50", "50-65", "65-80", "80+"]))
.groupby(
['continent', 'lifeExpGrouped'], as_index=True)
.agg({'pop': 'sum'})
.rename(columns={'pop': 'continentPop'})
.reset_index()
)
stacked_bar_df['lifeExpGrouped'] = pd.Categorical(stacked_bar_df['lifeExpGrouped'], ordered=True)
stacked_bar_df.head(6)
```
```r
# ggplot
#prepare data
stacked_df <- gapminder %>%
filter(year == 2007) %>%
mutate(lifeExpGrouped = cut(lifeExp,
breaks = c(0, 50, 65, 80, 90),
labels = c("Under 50", "50-65", "65-80", "80+"))) %>%
group_by(continent, lifeExpGrouped) %>%
summarise(continentPop = sum(as.numeric(pop)))
#set order of stacks by changing factor levels
stacked_df$lifeExpGrouped = factor(stacked_df$lifeExpGrouped, levels = rev(levels(stacked_df$lifeExpGrouped)))
#create plot
stacked_bars <- ggplot(data = stacked_df,
aes(x = continent,
y = continentPop,
fill = lifeExpGrouped)) +
geom_bar(stat = "identity",
position = "fill") +
bbc_style() +
scale_y_continuous(labels = scales::percent) +
scale_fill_viridis_d(direction = -1) +
geom_hline(yintercept = 0, size = 1, colour = "#333333") +
labs(title = "How life expectancy varies",
subtitle = "% of population by life expectancy band, 2007") +
theme(legend.position = "top",
legend.justification = "left") +
guides(fill = guide_legend(reverse = TRUE))
```
```
# altair
stacked_bar_altair = (
alt.Chart(stacked_bar_df)
.mark_bar()
.encode(x='continent:N',
y=alt.Y('continentPop', stack='normalize',
axis=alt.Axis(format='%'),
sort=['80+', '65-80', '50-65', 'under 50']),
# order=alt.Order(
# # Sort the segments of the bars by this field
# 'lifeExpGrouped',
# sort='descending'),
fill=alt.Fill('lifeExpGrouped:O',
scale=alt.Scale(scheme='viridis',
reverse=True,
domain=['under 50','50-65', '65-80', '80+', ],
range=['rgb(253, 231, 37)',
'rgb(53, 183, 121)',
'rgb(49, 104, 142)',
'rgb(68, 1, 84)']),
legend=alt.Legend(title="Life Expectancy")
)
)
.properties(title={'text': 'How life expectancy varies',
'subtitle': '% of population by life expectancy band, 2007'},
)
)
overay = overlay = pd.DataFrame({'continentPop': [0]})
hline = alt.Chart(overlay).mark_rule(
color='#333333', strokeWidth=2).encode(y='continentPop:Q')
(stacked_bar_altair + hline).configure_legend(orient ='right')
```
# Make a grouped bar chart
```
# hide
grouped_bar_df = (
gapminder[[
'country', 'year', 'lifeExp'
]].query(' year == 1967 | year == 2007 ')
.pivot_table(
index=['country'], columns='year',
values='lifeExp')
.assign(gap=lambda x: x[2007] - x[1967])
.nlargest(5, 'gap')
.reset_index()
.melt(value_vars=[1967, 2007],
id_vars=['country', 'gap'],
value_name='lifeExp')
)
grouped_bar_df
```
```r
# ggplot
#Prepare data
grouped_bar_df <- gapminder %>%
filter(year == 1967 | year == 2007) %>%
select(country, year, lifeExp) %>%
spread(year, lifeExp) %>%
mutate(gap = `2007` - `1967`) %>%
arrange(desc(gap)) %>%
head(5) %>%
gather(key = year,
value = lifeExp,
-country,
-gap)
#Make plot
grouped_bars <- ggplot(grouped_bar_df,
aes(x = country,
y = lifeExp,
fill = as.factor(year))) +
geom_bar(stat="identity", position="dodge") +
geom_hline(yintercept = 0, size = 1, colour="#333333") +
bbc_style() +
scale_fill_manual(values = c("#1380A1", "#FAAB18")) +
labs(title="We're living longer",
subtitle = "Biggest life expectancy rise, 1967-2007")
```
```
# plotnine
grouped_bars_ggplot = (ggplot(grouped_bar_df,
aes(x='country',
y='lifeExp',
fill='year')) +
geom_bar(stat="identity", position="dodge") +
geom_hline(yintercept=0, size=1, colour="#333333") +
# bbc_style() +
scale_fill_manual(values=("#1380A1", "#FAAB18")) +
labs(title="We're living longer",
subtitle="Biggest life expectancy rise, 1967-2007"))
grouped_bars_ggplot
# altair
grouped_bars_altair = (
alt.Chart(grouped_bar_df)
.mark_bar(size=42)
.encode(x='year:N',
y='lifeExp',
color=alt.Color('year:N', scale=alt.Scale(
range=["#1380A1", "#FAAB18"])),
column='country:N')
.properties(title={'text': "We're living longer",
'subtitle': 'Biggest life expectancy rise, 1967-2007'},
width=100
).configure_facet(
spacing=0.5,
# strokeWidth=1.0,
).configure_scale(
bandPaddingInner=0.4,
bandPaddingOuter=0.4
).configure_header(labelOrient='bottom',
labelPadding=6,
titleOrient='bottom').configure_axisX(
ticks=False,
labels=False,
title=None,
).configure_view(
stroke=None
)
)
grouped_bars_altair
```
|
github_jupyter
|
# collapse-hide
# !pip install plotnine[all]
# !pip install altair
# !pip install gapminder
from gapminder import gapminder
from plotnine.data import mtcars
from plotnine import *
from plotnine import ggplot, geom_point, aes, stat_smooth, facet_wrap, geom_line
from plotnine import ggplot # https://plotnine.readthedocs.io/en/stable/
import altair as alt
import pandas as pd
import plotnine
from bbc_plot.bbc_plot import bbc_style
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
alt.renderers.enable('html')
print(f'altair version: {alt.__version__}')
print(f'plotnine version: {plotnine.__version__}')
print(f'pandas version: {pd.__version__}')
# ggplot
line_df <- gapminder %>%
filter(country == "Malawi")
#Make plot
line <- ggplot(line_df, aes(x = year, y = lifeExp)) +
geom_line(colour = "#1380A1", size = 1) +
geom_hline(yintercept = 0, size = 1, colour="#333333") +
bbc_style() +
labs(title="Living longer",
subtitle = "Life expectancy in Malawi 1952-2007")
#hide
line_df = gapminder.query(" country == 'Malawi' ")
(ggplot(line_df, aes(x='year', y='lifeExp')) +
geom_line(colour='#1380A1', size=1) +
geom_hline(yintercept = 0, size = 1, colour='#333333') +
labs(title='Living longer',
subtitle = 'Life expectancy in Malawi 1952-2007')
)
## altair
line = (alt.Chart(line_df).mark_line().encode(
x='year',
y='lifeExp')
.properties(title={'text': 'Living Longer',
'subtitle': 'Life expectancy in Malawi 1952-2007'})
)
# hline
overlay = overlay = pd.DataFrame({'y': [0]})
hline = alt.Chart(overlay).mark_rule(color='#333333', strokeWidth=3).encode(y='y:Q')
line + hline
function ()
{
font <- "Helvetica"
ggplot2::theme(plot.title = ggplot2::element_text(family = font,
size = 28, face = "bold", color = "#222222"), plot.subtitle = ggplot2::element_text(family = font,
size = 22, margin = ggplot2::margin(9, 0, 9, 0)), plot.caption = ggplot2::element_blank(),
legend.position = "top", legend.text.align = 0, legend.background = ggplot2::element_blank(),
legend.title = ggplot2::element_blank(), legend.key = ggplot2::element_blank(),
legend.text = ggplot2::element_text(family = font, size = 18,
color = "#222222"), axis.title = ggplot2::element_blank(),
axis.text = ggplot2::element_text(family = font, size = 18,
color = "#222222"), axis.text.x = ggplot2::element_text(margin = ggplot2::margin(5,
b = 10)), axis.ticks = ggplot2::element_blank(),
axis.line = ggplot2::element_blank(), panel.grid.minor = ggplot2::element_blank(),
panel.grid.major.y = ggplot2::element_line(color = "#cbcbcb"),
panel.grid.major.x = ggplot2::element_blank(), panel.background = ggplot2::element_blank(),
strip.background = ggplot2::element_rect(fill = "white"),
strip.text = ggplot2::element_text(size = 22, hjust = 0))
}
<environment: namespace:bbplot>
font = "Helvetica"
theme(plot_title=element_text(family=font,
size=28, face="bold", color="#222222"),
# plot_subtitle=element_text(family=font,
# size=22, plot_margin=(9, 0, 9, 0)), plot_caption=element_blank(),
legend_position="top", legend_title_align=0, legend_background=element_blank(),
legend_title=element_blank(), legend_key=element_blank(),
legend_text=element_text(family=font, size=18,
color="#222222"), axis_title=element_blank(),
axis_text=element_text(family=font, size=18,
color="#222222"),
axis_text_x=element_text(margin={'t': 5, 'b': 10}),
axis_ticks=element_blank(),
axis_line=element_blank(), panel_grid_minor=element_blank(),
panel_grid_major_y=element_line(color="#cbcbcb"),
panel_grid_major_x=element_blank(), panel_background=element_blank(),
strip_background=element_rect(fill="white"),
strip_text=element_text(size=22, hjust=0)
)
# altair
line = (alt.Chart(line_df).mark_line().encode(
x='year',
y='lifeExp')
.properties(title={'text': 'Living Longer',
'subtitle': 'Life expectancy in China 1952-2007'})
)
# hline
overlay = overlay = pd.DataFrame({'lifeExp': [0]})
hline = alt.Chart(overlay).mark_rule(color='#333333', strokeWidth=3).encode(y='lifeExp:Q')
line + hline
# hide
# Prepare data
multiline_df = gapminder.query(
'country == "China" | country =="United States" ')
# ggplot
#Prepare data
multiple_line_df <- gapminder %>%
filter(country == "China" | country == "United States")
#Make plot
multiple_line <- ggplot(multiple_line_df, aes(x = year, y = lifeExp, colour = country)) +
geom_line(size = 1) +
geom_hline(yintercept = 0, size = 1, colour="#333333") +
scale_colour_manual(values = c("#FAAB18", "#1380A1")) +
bbc_style() +
labs(title="Living longer",
subtitle = "Life expectancy in China and the US")
# Make plot
multiline = (
ggplot(multiline_df, aes(x='year', y='lifeExp', colour='country')) +
geom_line(colour="#1380A1", size=1) +
geom_hline(yintercept=0, size=1, color="#333333") +
scale_colour_manual(values=["#FAAB18", "#1380A1"]) +
bbc_style() +
labs(title="Living longer",
subtitle="Life expectancy in China 1952-2007"))
multiline
multiline_altair = (alt.Chart(multiline_df).mark_line().encode(
x='year',
y='lifeExp',
color='country')
.properties(title={'text': 'Living Longer',
'subtitle': 'Life expectancy in China 1952-2007'})
)
# hline
overlay = overlay = pd.DataFrame({'lifeExp': [0]})
hline = alt.Chart(overlay).mark_rule(color='#333333', strokeWidth=3).encode(y='lifeExp:Q')
multiline_altair + hline
# ggplot
#Prepare data
bar_df <- gapminder %>%
filter(year == 2007 & continent == "Africa") %>%
arrange(desc(lifeExp)) %>%
head(5)
#Make plot
bars <- ggplot(bar_df, aes(x = country, y = lifeExp)) +
geom_bar(stat="identity",
position="identity",
fill="#1380A1") +
geom_hline(yintercept = 0, size = 1, colour="#333333") +
bbc_style() +
labs(title="Reunion is highest",
subtitle = "Highest African life expectancy, 2007")
## hide
bar_df = gapminder.query(' year == 2007 & continent == "Africa" ').nlargest(5, 'lifeExp')
bars_ggplot = (ggplot(bar_df, aes(x='country', y='lifeExp')) +
geom_bar(stat="identity",
position="identity",
fill="#1380A1") +
geom_hline(yintercept=0, size=1, colour="#333333") +
# bbc_style() +
labs(title="Reunion is highest",
subtitle="Highest African life expectancy, 2007"))
bars_ggplot
bars_altair = (alt.Chart(bar_df).mark_bar().encode(
x='country',
y='lifeExp',
# color='country'
)
.properties(title={'text': 'Reunion is highest',
'subtitle': 'Highest African life expectancy, 2007'})
)
bars_altair
## collapse-hide
stacked_bar_df = (
gapminder.query(' year == 2007')
.assign(
lifeExpGrouped=lambda x: pd.cut(
x['lifeExp'],
bins=[0, 50, 65, 80, 90],
labels=["under 50", "50-65", "65-80", "80+"]))
.groupby(
['continent', 'lifeExpGrouped'], as_index=True)
.agg({'pop': 'sum'})
.rename(columns={'pop': 'continentPop'})
.reset_index()
)
stacked_bar_df['lifeExpGrouped'] = pd.Categorical(stacked_bar_df['lifeExpGrouped'], ordered=True)
stacked_bar_df.head(6)
# ggplot
#prepare data
stacked_df <- gapminder %>%
filter(year == 2007) %>%
mutate(lifeExpGrouped = cut(lifeExp,
breaks = c(0, 50, 65, 80, 90),
labels = c("Under 50", "50-65", "65-80", "80+"))) %>%
group_by(continent, lifeExpGrouped) %>%
summarise(continentPop = sum(as.numeric(pop)))
#set order of stacks by changing factor levels
stacked_df$lifeExpGrouped = factor(stacked_df$lifeExpGrouped, levels = rev(levels(stacked_df$lifeExpGrouped)))
#create plot
stacked_bars <- ggplot(data = stacked_df,
aes(x = continent,
y = continentPop,
fill = lifeExpGrouped)) +
geom_bar(stat = "identity",
position = "fill") +
bbc_style() +
scale_y_continuous(labels = scales::percent) +
scale_fill_viridis_d(direction = -1) +
geom_hline(yintercept = 0, size = 1, colour = "#333333") +
labs(title = "How life expectancy varies",
subtitle = "% of population by life expectancy band, 2007") +
theme(legend.position = "top",
legend.justification = "left") +
guides(fill = guide_legend(reverse = TRUE))
# altair
stacked_bar_altair = (
alt.Chart(stacked_bar_df)
.mark_bar()
.encode(x='continent:N',
y=alt.Y('continentPop', stack='normalize',
axis=alt.Axis(format='%'),
sort=['80+', '65-80', '50-65', 'under 50']),
# order=alt.Order(
# # Sort the segments of the bars by this field
# 'lifeExpGrouped',
# sort='descending'),
fill=alt.Fill('lifeExpGrouped:O',
scale=alt.Scale(scheme='viridis',
reverse=True,
domain=['under 50','50-65', '65-80', '80+', ],
range=['rgb(253, 231, 37)',
'rgb(53, 183, 121)',
'rgb(49, 104, 142)',
'rgb(68, 1, 84)']),
legend=alt.Legend(title="Life Expectancy")
)
)
.properties(title={'text': 'How life expectancy varies',
'subtitle': '% of population by life expectancy band, 2007'},
)
)
overay = overlay = pd.DataFrame({'continentPop': [0]})
hline = alt.Chart(overlay).mark_rule(
color='#333333', strokeWidth=2).encode(y='continentPop:Q')
(stacked_bar_altair + hline).configure_legend(orient ='right')
# hide
grouped_bar_df = (
gapminder[[
'country', 'year', 'lifeExp'
]].query(' year == 1967 | year == 2007 ')
.pivot_table(
index=['country'], columns='year',
values='lifeExp')
.assign(gap=lambda x: x[2007] - x[1967])
.nlargest(5, 'gap')
.reset_index()
.melt(value_vars=[1967, 2007],
id_vars=['country', 'gap'],
value_name='lifeExp')
)
grouped_bar_df
# ggplot
#Prepare data
grouped_bar_df <- gapminder %>%
filter(year == 1967 | year == 2007) %>%
select(country, year, lifeExp) %>%
spread(year, lifeExp) %>%
mutate(gap = `2007` - `1967`) %>%
arrange(desc(gap)) %>%
head(5) %>%
gather(key = year,
value = lifeExp,
-country,
-gap)
#Make plot
grouped_bars <- ggplot(grouped_bar_df,
aes(x = country,
y = lifeExp,
fill = as.factor(year))) +
geom_bar(stat="identity", position="dodge") +
geom_hline(yintercept = 0, size = 1, colour="#333333") +
bbc_style() +
scale_fill_manual(values = c("#1380A1", "#FAAB18")) +
labs(title="We're living longer",
subtitle = "Biggest life expectancy rise, 1967-2007")
# plotnine
grouped_bars_ggplot = (ggplot(grouped_bar_df,
aes(x='country',
y='lifeExp',
fill='year')) +
geom_bar(stat="identity", position="dodge") +
geom_hline(yintercept=0, size=1, colour="#333333") +
# bbc_style() +
scale_fill_manual(values=("#1380A1", "#FAAB18")) +
labs(title="We're living longer",
subtitle="Biggest life expectancy rise, 1967-2007"))
grouped_bars_ggplot
# altair
grouped_bars_altair = (
alt.Chart(grouped_bar_df)
.mark_bar(size=42)
.encode(x='year:N',
y='lifeExp',
color=alt.Color('year:N', scale=alt.Scale(
range=["#1380A1", "#FAAB18"])),
column='country:N')
.properties(title={'text': "We're living longer",
'subtitle': 'Biggest life expectancy rise, 1967-2007'},
width=100
).configure_facet(
spacing=0.5,
# strokeWidth=1.0,
).configure_scale(
bandPaddingInner=0.4,
bandPaddingOuter=0.4
).configure_header(labelOrient='bottom',
labelPadding=6,
titleOrient='bottom').configure_axisX(
ticks=False,
labels=False,
title=None,
).configure_view(
stroke=None
)
)
grouped_bars_altair
| 0.73848 | 0.91302 |
# Immoscout24.de Scraper
Ein Script zum dumpen (in `.csv` schreiben) von Immobilien, welche auf [immoscout24.de](http://immoscout24.de) angeboten werden
```
from bs4 import BeautifulSoup
import json
import urllib.request as urllib2
import random
from random import choice
import time
# urlquery from Achim Tack. Thank you!
# https://github.com/ATack/GoogleTrafficParser/blob/master/google_traffic_parser.py
def urlquery(url):
# function cycles randomly through different user agents and time intervals to simulate more natural queries
try:
sleeptime = float(random.randint(1,6))/5
time.sleep(sleeptime)
agents = ['Mozilla/5.0 (Macintosh; Intel Mac OS X 10_8_2) AppleWebKit/537.17 (KHTML, like Gecko) Chrome/24.0.1309.0 Safari/537.17',
'Mozilla/5.0 (compatible; MSIE 10.6; Windows NT 6.1; Trident/5.0; InfoPath.2; SLCC1; .NET CLR 3.0.4506.2152; .NET CLR 3.5.30729; .NET CLR 2.0.50727) 3gpp-gba UNTRUSTED/1.0',
'Opera/12.80 (Windows NT 5.1; U; en) Presto/2.10.289 Version/12.02',
'Mozilla/4.0 (compatible; MSIE 5.5; Windows NT)',
'Mozilla/3.0',
'Mozilla/5.0 (iPhone; U; CPU like Mac OS X; en) AppleWebKit/420+ (KHTML, like Gecko) Version/3.0 Mobile/1A543a Safari/419.3',
'Mozilla/5.0 (Linux; U; Android 0.5; en-us) AppleWebKit/522+ (KHTML, like Gecko) Safari/419.3',
'Opera/9.00 (Windows NT 5.1; U; en)']
agent = choice(agents)
opener = urllib2.build_opener()
opener.addheaders = [('User-agent', agent)]
html = opener.open(url).read()
time.sleep(sleeptime)
return html
except Exception as e:
print('Something went wrong with Crawling:\n%s' % e)
def immoscout24parser(url):
''' Parser holt aus Immoscout24.de Suchergebnisseiten die Immobilien '''
try:
soup = BeautifulSoup(urlquery(url), 'html.parser')
scripts = soup.findAll('script')
for script in scripts:
#print script.text.strip()
if 'IS24.resultList' in script.text.strip():
s = script.string.split('\n')
for line in s:
#print('\n\n\'%s\'' % line)
if line.strip().startswith('resultListModel'):
resultListModel = line.strip('resultListModel: ')
immo_json = json.loads(resultListModel[:-1])
searchResponseModel = immo_json[u'searchResponseModel']
resultlist_json = searchResponseModel[u'resultlist.resultlist']
return resultlist_json
except Exception as e:
print("Fehler in immoscout24 parser: %s" % e)
```
## Main Loop
Geht Wohnungen und Häuser, jeweils zum Kauf und Miete durch und sammelt die Daten
```
immos = {}
# See immoscout24.de URL in Browser!
b = 'Baden-Wuerttemberg' # Bundesland
s = 'Stuttgart' # Stadt
k = 'Wohnung' # Wohnung oder Haus
w = 'Miete' # Miete oder Kauf
page = 0
print('Suche %s / %s' % (k, w))
while True:
page+=1
url = 'http://www.immobilienscout24.de/Suche/S-T/P-%s/%s-%s/%s/%s?pagerReporting=true' % (page, k, w, b, s)
# Because of some timeout or immoscout24.de errors,
# we try until it works \o/
resultlist_json = None
while resultlist_json is None:
try:
resultlist_json = immoscout24parser(url)
numberOfPages = int(resultlist_json[u'paging'][u'numberOfPages'])
pageNumber = int(resultlist_json[u'paging'][u'pageNumber'])
except:
pass
if page>numberOfPages:
break
# Get the data
for resultlistEntry in resultlist_json['resultlistEntries'][0][u'resultlistEntry']:
realEstate_json = resultlistEntry[u'resultlist.realEstate']
realEstate = {}
realEstate[u'Miete/Kauf'] = w
realEstate[u'Haus/Wohnung'] = k
realEstate['address'] = realEstate_json['address']['description']['text']
realEstate['city'] = realEstate_json['address']['city']
realEstate['postcode'] = realEstate_json['address']['postcode']
realEstate['quarter'] = realEstate_json['address']['quarter']
try:
realEstate['lat'] = realEstate_json['address'][u'wgs84Coordinate']['latitude']
realEstate['lon'] = realEstate_json['address'][u'wgs84Coordinate']['longitude']
except:
realEstate['lat'] = None
realEstate['lon'] = None
realEstate['title'] = realEstate_json['title']
realEstate['numberOfRooms'] = realEstate_json['numberOfRooms']
realEstate['livingSpace'] = realEstate_json['livingSpace']
if k=='Wohnung':
realEstate['balcony'] = realEstate_json['balcony']
realEstate['builtInKitchen'] = realEstate_json['builtInKitchen']
realEstate['garden'] = realEstate_json['garden']
realEstate['price'] = realEstate_json['price']['value']
realEstate['privateOffer'] = realEstate_json['privateOffer']
elif k=='Haus':
realEstate['isBarrierFree'] = realEstate_json['isBarrierFree']
realEstate['cellar'] = realEstate_json['cellar']
realEstate['plotArea'] = realEstate_json['plotArea']
realEstate['price'] = realEstate_json['price']['value']
realEstate['privateOffer'] = realEstate_json['privateOffer']
realEstate['energyPerformanceCertificate'] = realEstate_json['energyPerformanceCertificate']
realEstate['floorplan'] = realEstate_json['floorplan']
realEstate['from'] = realEstate_json['companyWideCustomerId']
realEstate['ID'] = realEstate_json[u'@id']
realEstate['url'] = u'https://www.immobilienscout24.de/expose/%s' % realEstate['ID']
immos[realEstate['ID']] = realEstate
print('Scrape Page %i/%i (%i Immobilien %s %s gefunden)' % (page, numberOfPages, len(immos), k, w))
print("Scraped %i Immos" % len(immos))
```
## Datenaufbereitung & Cleaning
Die gesammelten Daten werden in ein sauberes Datenformat konvertiert, welches z.B. auch mit Excel gelesen werden kann. Weiterhin werden die Ergebnisse pseudonymisiert, d.h. die Anbieter bekommen eindeutige Nummern statt Klarnamen.
```
from datetime import datetime
timestamp = datetime.strftime(datetime.now(), '%Y-%m-%d')
import pandas as pd
df = pd.DataFrame(immos).T
df.index.name = 'ID'
df.livingSpace[df.livingSpace==0] = None
df['EUR/qm'] = df.price / df.livingSpace
df.sort_values(by='EUR/qm', inplace=True)
len(df)
df.head()
```
## Alles Dumpen
```
f = open('%s-%s-%s-%s-%s.csv' % (timestamp, b, s, k, w), 'w')
f.write('# %s %s from immoscout24.de on %s\n' % (k,w,timestamp))
df[(df['Haus/Wohnung']==k) & (df['Miete/Kauf']==w)].to_csv(f, encoding='utf-8')
f.close()
df.to_excel('%s-%s-%s-%s-%s.xlsx' % (timestamp, b, s, k, w))
```
Fragen? [@Balzer82](https://twitter.com/Balzer82)
|
github_jupyter
|
from bs4 import BeautifulSoup
import json
import urllib.request as urllib2
import random
from random import choice
import time
# urlquery from Achim Tack. Thank you!
# https://github.com/ATack/GoogleTrafficParser/blob/master/google_traffic_parser.py
def urlquery(url):
# function cycles randomly through different user agents and time intervals to simulate more natural queries
try:
sleeptime = float(random.randint(1,6))/5
time.sleep(sleeptime)
agents = ['Mozilla/5.0 (Macintosh; Intel Mac OS X 10_8_2) AppleWebKit/537.17 (KHTML, like Gecko) Chrome/24.0.1309.0 Safari/537.17',
'Mozilla/5.0 (compatible; MSIE 10.6; Windows NT 6.1; Trident/5.0; InfoPath.2; SLCC1; .NET CLR 3.0.4506.2152; .NET CLR 3.5.30729; .NET CLR 2.0.50727) 3gpp-gba UNTRUSTED/1.0',
'Opera/12.80 (Windows NT 5.1; U; en) Presto/2.10.289 Version/12.02',
'Mozilla/4.0 (compatible; MSIE 5.5; Windows NT)',
'Mozilla/3.0',
'Mozilla/5.0 (iPhone; U; CPU like Mac OS X; en) AppleWebKit/420+ (KHTML, like Gecko) Version/3.0 Mobile/1A543a Safari/419.3',
'Mozilla/5.0 (Linux; U; Android 0.5; en-us) AppleWebKit/522+ (KHTML, like Gecko) Safari/419.3',
'Opera/9.00 (Windows NT 5.1; U; en)']
agent = choice(agents)
opener = urllib2.build_opener()
opener.addheaders = [('User-agent', agent)]
html = opener.open(url).read()
time.sleep(sleeptime)
return html
except Exception as e:
print('Something went wrong with Crawling:\n%s' % e)
def immoscout24parser(url):
''' Parser holt aus Immoscout24.de Suchergebnisseiten die Immobilien '''
try:
soup = BeautifulSoup(urlquery(url), 'html.parser')
scripts = soup.findAll('script')
for script in scripts:
#print script.text.strip()
if 'IS24.resultList' in script.text.strip():
s = script.string.split('\n')
for line in s:
#print('\n\n\'%s\'' % line)
if line.strip().startswith('resultListModel'):
resultListModel = line.strip('resultListModel: ')
immo_json = json.loads(resultListModel[:-1])
searchResponseModel = immo_json[u'searchResponseModel']
resultlist_json = searchResponseModel[u'resultlist.resultlist']
return resultlist_json
except Exception as e:
print("Fehler in immoscout24 parser: %s" % e)
immos = {}
# See immoscout24.de URL in Browser!
b = 'Baden-Wuerttemberg' # Bundesland
s = 'Stuttgart' # Stadt
k = 'Wohnung' # Wohnung oder Haus
w = 'Miete' # Miete oder Kauf
page = 0
print('Suche %s / %s' % (k, w))
while True:
page+=1
url = 'http://www.immobilienscout24.de/Suche/S-T/P-%s/%s-%s/%s/%s?pagerReporting=true' % (page, k, w, b, s)
# Because of some timeout or immoscout24.de errors,
# we try until it works \o/
resultlist_json = None
while resultlist_json is None:
try:
resultlist_json = immoscout24parser(url)
numberOfPages = int(resultlist_json[u'paging'][u'numberOfPages'])
pageNumber = int(resultlist_json[u'paging'][u'pageNumber'])
except:
pass
if page>numberOfPages:
break
# Get the data
for resultlistEntry in resultlist_json['resultlistEntries'][0][u'resultlistEntry']:
realEstate_json = resultlistEntry[u'resultlist.realEstate']
realEstate = {}
realEstate[u'Miete/Kauf'] = w
realEstate[u'Haus/Wohnung'] = k
realEstate['address'] = realEstate_json['address']['description']['text']
realEstate['city'] = realEstate_json['address']['city']
realEstate['postcode'] = realEstate_json['address']['postcode']
realEstate['quarter'] = realEstate_json['address']['quarter']
try:
realEstate['lat'] = realEstate_json['address'][u'wgs84Coordinate']['latitude']
realEstate['lon'] = realEstate_json['address'][u'wgs84Coordinate']['longitude']
except:
realEstate['lat'] = None
realEstate['lon'] = None
realEstate['title'] = realEstate_json['title']
realEstate['numberOfRooms'] = realEstate_json['numberOfRooms']
realEstate['livingSpace'] = realEstate_json['livingSpace']
if k=='Wohnung':
realEstate['balcony'] = realEstate_json['balcony']
realEstate['builtInKitchen'] = realEstate_json['builtInKitchen']
realEstate['garden'] = realEstate_json['garden']
realEstate['price'] = realEstate_json['price']['value']
realEstate['privateOffer'] = realEstate_json['privateOffer']
elif k=='Haus':
realEstate['isBarrierFree'] = realEstate_json['isBarrierFree']
realEstate['cellar'] = realEstate_json['cellar']
realEstate['plotArea'] = realEstate_json['plotArea']
realEstate['price'] = realEstate_json['price']['value']
realEstate['privateOffer'] = realEstate_json['privateOffer']
realEstate['energyPerformanceCertificate'] = realEstate_json['energyPerformanceCertificate']
realEstate['floorplan'] = realEstate_json['floorplan']
realEstate['from'] = realEstate_json['companyWideCustomerId']
realEstate['ID'] = realEstate_json[u'@id']
realEstate['url'] = u'https://www.immobilienscout24.de/expose/%s' % realEstate['ID']
immos[realEstate['ID']] = realEstate
print('Scrape Page %i/%i (%i Immobilien %s %s gefunden)' % (page, numberOfPages, len(immos), k, w))
print("Scraped %i Immos" % len(immos))
from datetime import datetime
timestamp = datetime.strftime(datetime.now(), '%Y-%m-%d')
import pandas as pd
df = pd.DataFrame(immos).T
df.index.name = 'ID'
df.livingSpace[df.livingSpace==0] = None
df['EUR/qm'] = df.price / df.livingSpace
df.sort_values(by='EUR/qm', inplace=True)
len(df)
df.head()
f = open('%s-%s-%s-%s-%s.csv' % (timestamp, b, s, k, w), 'w')
f.write('# %s %s from immoscout24.de on %s\n' % (k,w,timestamp))
df[(df['Haus/Wohnung']==k) & (df['Miete/Kauf']==w)].to_csv(f, encoding='utf-8')
f.close()
df.to_excel('%s-%s-%s-%s-%s.xlsx' % (timestamp, b, s, k, w))
| 0.316369 | 0.545407 |
### 使用 CART 算法来创建分类树
```
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.datasets import load_iris
# 准备数据集
iris = load_iris()
# 获取数据集合分类标识
features = iris.data
labels = iris.target
# 随机抽取33%的数据作为测试集,其余为训练集
train_features, test_features, train_labels, test_labels = train_test_split(
features, labels, test_size=0.33, random_state=0)
# 创建CART分类树
clf = DecisionTreeClassifier(criterion='gini')
# 拟合构造CART分类树
clf = clf.fit(train_features, train_labels)
# 用CART分类树做预测
test_predict = clf.predict(test_features)
# 预测结果与测试结果作对比
score = accuracy_score(test_labels, test_predict)
print('CART分类树准确率 %.4f' % score)
```
#### 画决策树
Mac用户安装 graphviz
```
pip install graphviz
brew install graphviz
```
```
from sklearn import tree
import graphviz
dot_data = tree.export_graphviz(clf, out_file=None)
graph = graphviz.Source(dot_data)
graph
```
### 使用 CART 回归树做预测
```
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import train_test_split
from sklearn.datasets import load_boston
from sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error
from sklearn.tree import DecisionTreeRegressor
# 准备数据集
boston = load_boston()
# 探索数据
boston.feature_names
# 获取特征集和房价
features = boston.data
prices = boston.target
# 随机抽取33%的数据作为测试集,其余为训练集
train_features, test_features, train_price, test_price = train_test_split(
features, prices, test_size=0.33, random_state=0)
# 创建CART回归树
dtr = DecisionTreeRegressor()
# 拟合构造CART回归树
dtr.fit(train_features, train_price)
# 预测测试集中的房价
predict_price = dtr.predict(test_features)
# 测试集的结果评价
print('回归树二乘偏差均值:', mean_squared_error(test_price, predict_price))
print('回归树绝对值偏差均值:', mean_absolute_error(test_price, predict_price))
```
#### 画决策树
```
from sklearn import tree
import graphviz
dot_data = tree.export_graphviz(dtr, out_file=None)
graph = graphviz.Source(dot_data)
graph.render('./CART_Regressor_boston')
```
### 对手写数据集做CART建模
```
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.datasets import load_digits
# 准备数据集
digits = load_digits()
# 获取数据集合分类标识
features = digits.data
digits = digits.target
# 随机抽取33%的数据作为测试集,其余为训练集
train_features, test_features, train_digits, test_digits = train_test_split(
features, digits, test_size=0.33, random_state=0)
# 创建CART分类树
clf = DecisionTreeClassifier(criterion='gini')
# 拟合构造CART分类树
clf.fit(train_features, train_digits)
# 用CART分类树做预测
predict_digits = clf.predict(test_features)
# 预测结果与测试结果作对比
score = accuracy_score(test_digits, predict_digits)
print('CART分类树准确率 %.4f' % score)
from sklearn import tree
import graphviz
dot_data = tree.export_graphviz(clf, out_file=None)
graph = graphviz.Source(dot_data)
graph.render('./CART_Classifier_digits')
```
|
github_jupyter
|
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.datasets import load_iris
# 准备数据集
iris = load_iris()
# 获取数据集合分类标识
features = iris.data
labels = iris.target
# 随机抽取33%的数据作为测试集,其余为训练集
train_features, test_features, train_labels, test_labels = train_test_split(
features, labels, test_size=0.33, random_state=0)
# 创建CART分类树
clf = DecisionTreeClassifier(criterion='gini')
# 拟合构造CART分类树
clf = clf.fit(train_features, train_labels)
# 用CART分类树做预测
test_predict = clf.predict(test_features)
# 预测结果与测试结果作对比
score = accuracy_score(test_labels, test_predict)
print('CART分类树准确率 %.4f' % score)
pip install graphviz
brew install graphviz
from sklearn import tree
import graphviz
dot_data = tree.export_graphviz(clf, out_file=None)
graph = graphviz.Source(dot_data)
graph
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import train_test_split
from sklearn.datasets import load_boston
from sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error
from sklearn.tree import DecisionTreeRegressor
# 准备数据集
boston = load_boston()
# 探索数据
boston.feature_names
# 获取特征集和房价
features = boston.data
prices = boston.target
# 随机抽取33%的数据作为测试集,其余为训练集
train_features, test_features, train_price, test_price = train_test_split(
features, prices, test_size=0.33, random_state=0)
# 创建CART回归树
dtr = DecisionTreeRegressor()
# 拟合构造CART回归树
dtr.fit(train_features, train_price)
# 预测测试集中的房价
predict_price = dtr.predict(test_features)
# 测试集的结果评价
print('回归树二乘偏差均值:', mean_squared_error(test_price, predict_price))
print('回归树绝对值偏差均值:', mean_absolute_error(test_price, predict_price))
from sklearn import tree
import graphviz
dot_data = tree.export_graphviz(dtr, out_file=None)
graph = graphviz.Source(dot_data)
graph.render('./CART_Regressor_boston')
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.datasets import load_digits
# 准备数据集
digits = load_digits()
# 获取数据集合分类标识
features = digits.data
digits = digits.target
# 随机抽取33%的数据作为测试集,其余为训练集
train_features, test_features, train_digits, test_digits = train_test_split(
features, digits, test_size=0.33, random_state=0)
# 创建CART分类树
clf = DecisionTreeClassifier(criterion='gini')
# 拟合构造CART分类树
clf.fit(train_features, train_digits)
# 用CART分类树做预测
predict_digits = clf.predict(test_features)
# 预测结果与测试结果作对比
score = accuracy_score(test_digits, predict_digits)
print('CART分类树准确率 %.4f' % score)
from sklearn import tree
import graphviz
dot_data = tree.export_graphviz(clf, out_file=None)
graph = graphviz.Source(dot_data)
graph.render('./CART_Classifier_digits')
| 0.585931 | 0.88054 |
# Hands-on example (Azure ML)
```
!python --version
import os
import warnings
import sys
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
#Sklearn
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score
from sklearn.model_selection import train_test_split
from sklearn.linear_model import ElasticNet
#Azure
import azureml.core
import azureml.dataprep
from azureml.core import Workspace, Dataset
from azure.storage.blob import BlobServiceClient
from azureml.core.authentication import ServicePrincipalAuthentication
#MLFlow
import mlflow
import mlflow.sklearn
from mlflow.entities import ViewType
#Temporarily filter all warning for demo
warnings.filterwarnings("ignore")
# Check core SDK version number
print("SDK version:", azureml.core.VERSION)
```
### Set Up Authentication
```
from settings import (AZURE_SUBSCRIPTION_ID,AZURE_RESOURCE_GROUP,
AZURE_TENANT_ID)
subscription_id = AZURE_SUBSCRIPTION_ID
resource_group = AZURE_RESOURCE_GROUP
workspace_name = 'mlflow_tutorial'
```
## 0. The data
* The data set used in this example is from http://archive.ics.uci.edu/ml/datasets/Wine+Quality
* P. Cortez, A. Cerdeira, F. Almeida, T. Matos and J. Reis.
* Modeling wine preferences by data mining from physicochemical properties. In Decision Support Systems, Elsevier, 47(4):547-553, 2009.
```
##Use this if inside AML Studio
# ws = Workspace.from_config()
ws = Workspace(subscription_id, resource_group, workspace_name)
dataset = Dataset.get_by_name(ws, name='wine-quality')
dataset.to_pandas_dataframe().head()
```
## 1. Tracking experiments
### Set Tracking URI to Azure ML Workspace
```
mlflow.set_tracking_uri(ws.get_mlflow_tracking_uri())
mlflow.tracking.get_tracking_uri()
exp_name = "ElasticNet_wine_AML"
mlflow.set_experiment(exp_name)
```
### What do we track?
- **Code Version**: Git commit hash used for the run (if it was run from an MLflow Project)
- **Start & End Time**: Start and end time of the run
- **Source**: what code run?
- **Parameters**: Key-value input parameters.
- **Metrics**: Key-value metrics, where the value is numeric (can be updated over the run)
- **Artifacts**: Output files in any format.
```
def eval_metrics(actual, pred):
# compute relevant metrics
rmse = np.sqrt(mean_squared_error(actual, pred))
mae = mean_absolute_error(actual, pred)
r2 = r2_score(actual, pred)
return rmse, mae, r2
def load_data(dataset):
data = dataset.to_pandas_dataframe()
# Split the data into training and test sets. (0.75, 0.25) split.
train, test = train_test_split(data)
# The predicted column is "quality" which is a scalar from [3, 9]
train_x = train.drop(["quality"], axis=1)
test_x = test.drop(["quality"], axis=1)
train_y = train[["quality"]]
test_y = test[["quality"]]
return train_x, train_y, test_x, test_y
def train(alpha=0.5, l1_ratio=0.5,full_view=False):
# train a model with given parameters
warnings.filterwarnings("ignore")
np.random.seed(40)
# Read the wine-quality csv file (make sure you're running this from the root of MLflow!)
data_path = "data/wine-quality.csv"
train_x, train_y, test_x, test_y = load_data(dataset)
# Useful for multiple runs (only doing one run in this sample notebook)
with mlflow.start_run():
# Execute ElasticNet
lr = ElasticNet(alpha=alpha, l1_ratio=l1_ratio, random_state=42)
lr.fit(train_x, train_y)
# Evaluate Metrics
predicted_qualities = lr.predict(test_x)
(rmse, mae, r2) = eval_metrics(test_y, predicted_qualities)
# Print out metrics
print("Elasticnet model (alpha=%f, l1_ratio=%f):" % (alpha, l1_ratio))
print(" RMSE: %s" % rmse)
print(" MAE: %s" % mae)
print(" R2: %s" % r2)
# Log parameter, metrics, and model to MLflow
mlflow.log_param(key="alpha", value=alpha)
mlflow.log_param(key="l1_ratio", value=l1_ratio)
mlflow.log_metric(key="rmse", value=rmse)
mlflow.log_metrics({"mae": mae, "r2": r2})
print("Save to: {}".format(mlflow.get_artifact_uri()))
if full_view:
print("Run IDs: \n{}".format(mlflow.search_runs(ViewType.ACTIVE_ONLY)))
else:
pass
mlflow.sklearn.log_model(lr, "model")
train(0.5, 0.5)
train(0.2, 0.2)
train(0.1, 0.1,full_view=True)
```
### 1.1 Comparing runs
Connect to Azure Portal to view experiments and run metrics.
```
ws.experiments[exp_name]
```
## 2. Tagging runs
```
from datetime import datetime
from mlflow.tracking import MlflowClient
client = MlflowClient()
experiments = client.list_experiments() # returns a list of mlflow.entities.Experiment
print(experiments)
# get the run
_run = client.get_run(run_id="53254795-c368-4155-a04e-b570ca7d9e60")
print(_run)
# add a tag to the run
dt = datetime.now().strftime("%d-%m-%Y (%H:%M:%S.%f)")
client.set_tag(_run.info.run_id, "deployed", dt)
```
|
github_jupyter
|
!python --version
import os
import warnings
import sys
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
#Sklearn
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score
from sklearn.model_selection import train_test_split
from sklearn.linear_model import ElasticNet
#Azure
import azureml.core
import azureml.dataprep
from azureml.core import Workspace, Dataset
from azure.storage.blob import BlobServiceClient
from azureml.core.authentication import ServicePrincipalAuthentication
#MLFlow
import mlflow
import mlflow.sklearn
from mlflow.entities import ViewType
#Temporarily filter all warning for demo
warnings.filterwarnings("ignore")
# Check core SDK version number
print("SDK version:", azureml.core.VERSION)
from settings import (AZURE_SUBSCRIPTION_ID,AZURE_RESOURCE_GROUP,
AZURE_TENANT_ID)
subscription_id = AZURE_SUBSCRIPTION_ID
resource_group = AZURE_RESOURCE_GROUP
workspace_name = 'mlflow_tutorial'
##Use this if inside AML Studio
# ws = Workspace.from_config()
ws = Workspace(subscription_id, resource_group, workspace_name)
dataset = Dataset.get_by_name(ws, name='wine-quality')
dataset.to_pandas_dataframe().head()
mlflow.set_tracking_uri(ws.get_mlflow_tracking_uri())
mlflow.tracking.get_tracking_uri()
exp_name = "ElasticNet_wine_AML"
mlflow.set_experiment(exp_name)
def eval_metrics(actual, pred):
# compute relevant metrics
rmse = np.sqrt(mean_squared_error(actual, pred))
mae = mean_absolute_error(actual, pred)
r2 = r2_score(actual, pred)
return rmse, mae, r2
def load_data(dataset):
data = dataset.to_pandas_dataframe()
# Split the data into training and test sets. (0.75, 0.25) split.
train, test = train_test_split(data)
# The predicted column is "quality" which is a scalar from [3, 9]
train_x = train.drop(["quality"], axis=1)
test_x = test.drop(["quality"], axis=1)
train_y = train[["quality"]]
test_y = test[["quality"]]
return train_x, train_y, test_x, test_y
def train(alpha=0.5, l1_ratio=0.5,full_view=False):
# train a model with given parameters
warnings.filterwarnings("ignore")
np.random.seed(40)
# Read the wine-quality csv file (make sure you're running this from the root of MLflow!)
data_path = "data/wine-quality.csv"
train_x, train_y, test_x, test_y = load_data(dataset)
# Useful for multiple runs (only doing one run in this sample notebook)
with mlflow.start_run():
# Execute ElasticNet
lr = ElasticNet(alpha=alpha, l1_ratio=l1_ratio, random_state=42)
lr.fit(train_x, train_y)
# Evaluate Metrics
predicted_qualities = lr.predict(test_x)
(rmse, mae, r2) = eval_metrics(test_y, predicted_qualities)
# Print out metrics
print("Elasticnet model (alpha=%f, l1_ratio=%f):" % (alpha, l1_ratio))
print(" RMSE: %s" % rmse)
print(" MAE: %s" % mae)
print(" R2: %s" % r2)
# Log parameter, metrics, and model to MLflow
mlflow.log_param(key="alpha", value=alpha)
mlflow.log_param(key="l1_ratio", value=l1_ratio)
mlflow.log_metric(key="rmse", value=rmse)
mlflow.log_metrics({"mae": mae, "r2": r2})
print("Save to: {}".format(mlflow.get_artifact_uri()))
if full_view:
print("Run IDs: \n{}".format(mlflow.search_runs(ViewType.ACTIVE_ONLY)))
else:
pass
mlflow.sklearn.log_model(lr, "model")
train(0.5, 0.5)
train(0.2, 0.2)
train(0.1, 0.1,full_view=True)
ws.experiments[exp_name]
from datetime import datetime
from mlflow.tracking import MlflowClient
client = MlflowClient()
experiments = client.list_experiments() # returns a list of mlflow.entities.Experiment
print(experiments)
# get the run
_run = client.get_run(run_id="53254795-c368-4155-a04e-b570ca7d9e60")
print(_run)
# add a tag to the run
dt = datetime.now().strftime("%d-%m-%Y (%H:%M:%S.%f)")
client.set_tag(_run.info.run_id, "deployed", dt)
| 0.476823 | 0.811116 |
# Image Embedding Similarity Search
## Creating image embeddings with features extracted using PyTorch
```
import faiss
import numpy as np
from tqdm import tqdm
from PIL import Image
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import torch
from torch.utils.data import DataLoader
from torchvision import models, transforms, datasets
# Check for gpu and initalise model
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
model = models.resnet101(pretrained=True, progress=True)
print("[INFO] Using Device:", DEVICE)
print(f"[INFO] Loading model: {model.__class__.__name__}")
class ImageFolderWithPaths(datasets.ImageFolder):
"""Custom dataset class that includes image file paths. Extends
torchvision.datasets.ImageFolder
Source: https://gist.github.com/andrewjong/6b02ff237533b3b2c554701fb53d5c4d
"""
# override the __getitem__ method. this is the method that dataloader calls
def __getitem__(self, index):
# this is what ImageFolder normally returns
original_tuple = super(ImageFolderWithPaths, self).__getitem__(index)
# the image file path
path = self.imgs[index][0]
# make a new tuple that includes original and the path
tuple_with_path = original_tuple + (path,)
return tuple_with_path
def pooling_output(x, model):
for layer_name, layer in model._modules.items():
x = layer(x)
if layer_name == "avgpool":
break
return x
transform = transforms.Compose(
[
transforms.Resize(size=[224, 224], interpolation=2),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
def extract_features_paths(data_directory):
dataset = ImageFolderWithPaths(
data_directory, transform=transform
) # our custom dataset
# strip away unnecessary info and store path to each image
img_paths = [dataset.imgs[i][0] for i in range(len(dataset.imgs))]
# initialize the dataloaders
dataloader = DataLoader(dataset)
features = []
print(f"[INFO][STARTED] Feature Extraction using {model.__class__.__name__}")
model.to(DEVICE)
with torch.no_grad():
model.eval()
for inputs, labels, paths in tqdm(dataloader):
result = pooling_output(inputs.to(DEVICE), model)
features.append(result.cpu().view(1, -1).numpy())
torch.cuda.empty_cache()
print(f"[INFO][DONE] Feature Extraction using {model.__class__.__name__}")
features = np.vstack(features)
print("[DEBUG]: Preview Features:", features[0])
print("[DEBUG]: Preview Image Paths:", img_paths[0])
return features, img_paths
# TODO: Provide the path to a folder of image files
data_directory = "/home/umar-musashi/Documents/Musashi Part Images/lobe_dummy"
features, img_paths = extract_features_paths(data_directory)
# The feature length that resnet outputs
dimension = 2048
# Add trained features to FAISS index
index = faiss.IndexFlatL2(dimension)
index.add(features)
my_index = 3400
query_image = img_paths[my_index]
# query_image = "/home/umar-musashi/Downloads/black-triangle-shape.png"
print("file path: ", query_image)
PIL_img = Image.open(query_image)
# PIL_img = PIL_img.transpose(Image.FLIP_TOP_BOTTOM)
# PIL_img = PIL_img.transpose(Image.FLIP_LEFT_RIGHT)
PIL_img = PIL_img.convert('RGB')
plt.imshow(PIL_img)
input_tensor = transform(PIL_img)
input_tensor = input_tensor.view(1, *input_tensor.shape)
# Search index for neighbor embeddings
with torch.no_grad():
model.eval()
result = pooling_output(input_tensor.to(DEVICE), model)
query_descriptors = result.cpu().view(1, -1).numpy()
torch.cuda.empty_cache()
print("query_descriptors shape", query_descriptors.shape)
distances, indices = index.search(query_descriptors, 9)
print("input:", query_descriptors)
print("input shape:", query_descriptors.shape)
print("input type:", type(query_descriptors))
fig, ax = plt.subplots(3, 3, figsize=(15,15))
i = 0
for file_index, ax_i in zip(indices[0], np.array(ax).flatten()):
ax_i.imshow(plt.imread(img_paths[file_index]))
distance = str(float("{0:.2f}".format(distances[0][i])))
ax_i.set_title(f"Distance: {distance} \nFile: {img_paths[file_index].split('/')[-1][0:25]}")
i+=1
plt.show()
```
|
github_jupyter
|
import faiss
import numpy as np
from tqdm import tqdm
from PIL import Image
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import torch
from torch.utils.data import DataLoader
from torchvision import models, transforms, datasets
# Check for gpu and initalise model
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
model = models.resnet101(pretrained=True, progress=True)
print("[INFO] Using Device:", DEVICE)
print(f"[INFO] Loading model: {model.__class__.__name__}")
class ImageFolderWithPaths(datasets.ImageFolder):
"""Custom dataset class that includes image file paths. Extends
torchvision.datasets.ImageFolder
Source: https://gist.github.com/andrewjong/6b02ff237533b3b2c554701fb53d5c4d
"""
# override the __getitem__ method. this is the method that dataloader calls
def __getitem__(self, index):
# this is what ImageFolder normally returns
original_tuple = super(ImageFolderWithPaths, self).__getitem__(index)
# the image file path
path = self.imgs[index][0]
# make a new tuple that includes original and the path
tuple_with_path = original_tuple + (path,)
return tuple_with_path
def pooling_output(x, model):
for layer_name, layer in model._modules.items():
x = layer(x)
if layer_name == "avgpool":
break
return x
transform = transforms.Compose(
[
transforms.Resize(size=[224, 224], interpolation=2),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
def extract_features_paths(data_directory):
dataset = ImageFolderWithPaths(
data_directory, transform=transform
) # our custom dataset
# strip away unnecessary info and store path to each image
img_paths = [dataset.imgs[i][0] for i in range(len(dataset.imgs))]
# initialize the dataloaders
dataloader = DataLoader(dataset)
features = []
print(f"[INFO][STARTED] Feature Extraction using {model.__class__.__name__}")
model.to(DEVICE)
with torch.no_grad():
model.eval()
for inputs, labels, paths in tqdm(dataloader):
result = pooling_output(inputs.to(DEVICE), model)
features.append(result.cpu().view(1, -1).numpy())
torch.cuda.empty_cache()
print(f"[INFO][DONE] Feature Extraction using {model.__class__.__name__}")
features = np.vstack(features)
print("[DEBUG]: Preview Features:", features[0])
print("[DEBUG]: Preview Image Paths:", img_paths[0])
return features, img_paths
# TODO: Provide the path to a folder of image files
data_directory = "/home/umar-musashi/Documents/Musashi Part Images/lobe_dummy"
features, img_paths = extract_features_paths(data_directory)
# The feature length that resnet outputs
dimension = 2048
# Add trained features to FAISS index
index = faiss.IndexFlatL2(dimension)
index.add(features)
my_index = 3400
query_image = img_paths[my_index]
# query_image = "/home/umar-musashi/Downloads/black-triangle-shape.png"
print("file path: ", query_image)
PIL_img = Image.open(query_image)
# PIL_img = PIL_img.transpose(Image.FLIP_TOP_BOTTOM)
# PIL_img = PIL_img.transpose(Image.FLIP_LEFT_RIGHT)
PIL_img = PIL_img.convert('RGB')
plt.imshow(PIL_img)
input_tensor = transform(PIL_img)
input_tensor = input_tensor.view(1, *input_tensor.shape)
# Search index for neighbor embeddings
with torch.no_grad():
model.eval()
result = pooling_output(input_tensor.to(DEVICE), model)
query_descriptors = result.cpu().view(1, -1).numpy()
torch.cuda.empty_cache()
print("query_descriptors shape", query_descriptors.shape)
distances, indices = index.search(query_descriptors, 9)
print("input:", query_descriptors)
print("input shape:", query_descriptors.shape)
print("input type:", type(query_descriptors))
fig, ax = plt.subplots(3, 3, figsize=(15,15))
i = 0
for file_index, ax_i in zip(indices[0], np.array(ax).flatten()):
ax_i.imshow(plt.imread(img_paths[file_index]))
distance = str(float("{0:.2f}".format(distances[0][i])))
ax_i.set_title(f"Distance: {distance} \nFile: {img_paths[file_index].split('/')[-1][0:25]}")
i+=1
plt.show()
| 0.552057 | 0.864024 |
# TME sur les données blablacar
**Ce fichier est le fichier de travail**, l'autre fichier blablacar est donné pour information et pour montrer comment les données ont été collectées.
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# %matplotlib notebook
import pickle as pkl
```
## Chargement des données
Les données sont stockées au format pickle (code fourni ci-dessous):
1. Importer le module : import `pickle as pkl`
1. Charger les données avec `load`
1. La structure est un dictionnaire, les données sont dans le champ `data`
1. La description des colonnes est dans `indexcol`
```
import pickle as pkl
# chargement des données
fich = pkl.load( open('donnees_blablacar.pkl', 'rb'))
# {'indexcol': cols , 'data':pp2db, 'villes': villes, 'marques':marques }
titles_col = fich['indexcol']
print(len(titles_col), titles_col)
data = fich['data']
print(data.shape)
dico_villes = fich['villes']
dico_marques = fich['marques']
print(dico_marques)
```
## Discrétisation et histogramme
Nous nous intéressons à la variable `distance` (dernière colonne). Nous allons procéder de la manière suivante:
1. Analyse rapide de la variable aléatoire: calcul de la moyenne et de l'écart-type
1. Analyse plus fine (1): affichage des 10 quantiles
1. Analyse plus fine (2): discrétisation de la variable en 10 intervalles de largeur constante & comptage des effectifs dans chaque catégorie (= construction d'un histogramme)
- Construire l'histogramme à la main. (1) calculer les bornes des intervalles puis (2) utiliser `np.where` pour déterminer les effectifs dans chaque classe. Utiliser `plt.bar` pour l'affichage.
- Vérifier vos résultats avec `np.histogram` et `plt.hist` pour l'affichage
- Comparer les quantiles et les bornes des intervalles discrets
1. Discuter le nombre d'intervalles pour l'histogramme et trouver une valeur satisfaisante
**Notes** :
- dans `np.where`, il faut mettre des parenthèses s'il y a plusieurs clause
> `np.where((x>a) & (x<b))` : tous les indices de x qui satisfont la clause
>
> `np.where((x>a) & (x<b), 1, 0).sum()` : le comptage associé
- Dans `plt.bar`, il faut donner une largeur importante aux bar, sinon on ne voit rien
```
# Analyse rapide : moyenne, écart-type, calcul des quantiles pour faire la synthèse de cette variable aléatoire
d = data[:,-1] # extraction de la variable distance
# A vous de jouer pour calculer tous les descripteurs = recherche dans la doc numpy !
d = np.sort(d)
quantile_values = np.array([])
range_quantile = 10
print("mean:", np.mean(d))
print("std:", np.std(d))
for i in range(range_quantile):
quantile_value = np.quantile(d, i/range_quantile)
print("quantile ", i, "/", range_quantile, ":", quantile_value)
quantile_values = np.append(quantile_values, quantile_value)
print("quantile_values:", quantile_values)
# Discrétisation des distances & histogramme avec np.histogram
np.histogram(d, bins = np.linspace(0, d.shape[0])
)
hist, bins = np.histogram(d, bins = np.linspace(0, d.shape[0]))
print(hist)
print(bins)
plt.hist(d, bins = np.linspace(0, d.shape[0]), width=100)
plt.title("histogramme")
plt.show()
# Discrétisation des distances & histogramme a la main
n = 30 # nb intervalle
inter = np.size(d)//n
bornes = np.array([])
for i in range(n):
bornes = np.append(bornes, i*inter)
# calcul des effectifs avec np.where
effectifs = np.array([])
for i in range(n - 1):
values = np.where((d >= bornes[i]) & (d <= bornes[i+1]))
effectif = np.size(values)
# print("borne1 :",bornes[i])
# print("borne2 :",bornes[i+1])
# print("effectif :",effectif)
# print("------------")
effectifs = np.append(effectifs, effectif)
bornes = bornes[:-1]
# print(bornes.shape)
# print(effectifs.shape)
# print(bornes)
# print(effectifs)
# affichage avec plt.bar = histogramme à la main
fig = plt.figure()
ax = fig.add_axes([0,0,1,1])
ax.bar(bornes, effectifs, width = 200)
plt.title("histogramme")
plt.show()
```
## histogramme (bis)
Tracer l'histogramme des prix au km
```
# histogramme des prix au km : construction de la variable puis utilisation de np.histogramme
prix = data[:,10]
km = data[:,13]
# print(prix.shape)
# print(km.shape)
values = prix/km
# print(value.shape)
bins = np.linspace(0, values[0], n)
np.histogram(values, bins = bins)
hist, bins = np.histogram(values, bins = bins)
plt.hist(values, bins = bins, width=0.002)
plt.title("histogramme")
plt.show()
```
# Distributions jointes, distributions conditionnelles
Nous voulons maintenant étudier la distribution jointe entre la distance et la marque de la voiture. Partir des distributions discrètes ou discétisées et construire le tableau d'effectif puis normaliser par les effectifs de l'échantillon pour estimer la loi jointe.
Il est diffile d'analyser cette probabilité jointe (cf ci-dessous pour l'affichage)... Nous allons donc passer à la loi conditionnelle: nous voulons donc calculer la probabilité de la distance conditionnellement à la marque de la voiture.
1. Proposer un critère rapide pour vérifier que votre distribution conditionnelle respecte bien les propriétés de base
1. Cette distribution conditionnelle fait apparaitre des pics très marqués: pouvons-nous tirer parti de ces informations?
**Note:**
- pour afficher une matrice `p_dm`, la meilleure solution est la suivante:
> `plt.imshow(p_dm, interpolation='nearest')`
>
> `plt.show()`
- la variable `marque` est bruitée. Vous pourrez vous amuser à éliminer ou fusionner certaines catégories
- les indices dans une matrice doivent toujours être entiers. `int(...)`
- pour ajouter une description sur l'axe des x:
```python
fig, ax = plt.subplots(1,1)
plt.imshow(p_dsm, interpolation='nearest')
ax.set_xticks(np.arange(len(dico_marques)))
ax.set_xticklabels(dico_marques.keys(),rotation=90,fontsize=8)
plt.show()
```
- Si l'image est trop petite pour voir quelque chose: solution = sauvegarde en pdf (ie vectorielle) + ouverture avec un logiciel de lecture pdf
```python
plt.savefig('mafigure.pdf')
```
```
distances = data[:,13]
marques = data[:,11]
n = 10 # nb intervalle
inter = np.size(data[:,13])//n
bornes = np.array([])
for i in range(n):
bornes = np.append(bornes, i*inter)
print("nombre de bornes:\n",np.size(bornes)-1)
print("bornes distance:\n",bornes)
print("nombre de marque de voiture:\n",np.size(unique_marque))
tab_marques_dist = np.array([])
for i in range(n - 1):
values = np.array([])
for m in unique_marque:
values = np.append(values, np.size(np.where((distances > bornes[i]) & (distances < bornes[i+1]) & (marques == m)))/np.size(unique_marque))
tab_marques_dist = np.append(tab_marques_dist, values)
print("shape de nos valeurs:\n",tab_marques_dist.shape)
tab_marques_dist = tab_marques_dist.reshape(np.size(bornes)-1, np.size(unique_marque))
print("shape de nos valeurs après reshape en matrice:\n",tab_marques_dist.shape)
# construction de la distance discrétisée
# Dimensions : = (Nind x 1) = mêmes dimensions que d
# contenu = catégorie de distance (entre 0 et 29 par exemple si on a discrétisé en 30 catégories)
# dd = np.zeros(d.shape)
# remplissage avec np.where
# p_dm = np.zeros((len(bornes)-1, len(dico_marques)))
# remplissage de la matrice p_dm = double boucle + comptage
# p_dm /= p_dm.sum() # normalisation
# affichage du résultat
print("affichage resultat:")
plt.imshow(tab_marques_dist, interpolation='nearest')
plt.show()
# loi conditionnelle distance | marque
# calcul d'une marginale
# p_m =
# calcul de la conditionnelle
# p_dsm =
# affichage
# fig, ax = plt.subplots(1,1)
# plt.imshow(p_dsm, interpolation='nearest')
# ax.set_xticks(np.arange(len(dico_marques)))
# ax.set_xticklabels(dico_marques.keys(),rotation=90,fontsize=8)
# plt.show()
# proposition d'un critère très rapide pour vérifier qu'il s'agit bien d'une distribution conditionnelle
```
## Tracé de l'ensemble de l'échantillon avec des codes couleurs
Nous proposons ensuite de tracer toutes les trajectoires des voitures blablacar. Pour cela, il faut utiliser la commande `plt.plot`.
Vous devez optenir des étoiles à partir des 7 villes requêtes: `['Paris', 'Marseille', 'Grenoble', 'Lille', 'Strasbourg', 'Nantes', 'Bordeaux']`.
Mais on ne voit pas grand chose... Et ça prend beaucoup de temps à tracer avec une boucle for. On propose donc une série d'exercice pour mieux comprendre ce qui se passe.
1. Attention à l'ordre des arguments dans le plot:
```plt.plot(tous_les_x, tous_les_y)```
Afin de tracer des trajectoires, il faut envoyer les x et les y 2 par 2 dans une boucle `for`
1. Pour éviter les boucles, il existe une méthode `quiver` dédiée au tracé de champs de vecteurs: ça ira beaucoup plus vite qu'avec plot. Il faut juste bien comprendre les mécanismes d'échelles. Pour utiliser l'échelle 1, la commande est la suivante:
```python
plt.quiver(x_dep, y_dep, delta_x, delta_y,\
angles='xy', scale_units='xy', scale=1)
```
1. Isoler les trajets proposés à partir de chacune des villes sachant les coordonnées sont:
```python
coord = np.array([[45.18721767, 5.72345183],
[47.22572172, -1.56558993],
[50.63010695, 3.07071992],
[48.5782548, 7.74078742],
[44.83848889, -0.58156509],
[43.2991509, 5.38925024],
[48.8477201, 2.34607889]])
```
Chaque trajectoire (point de départ) sera rattachée à la ville la plus proche.
Une fois la distance calculée pour chaque origine de trajectoire, vous pourrez avoir besoin de `argmin`
1. Tracer les trajets d'une couleur spéciale en fonction des origines.
Les commandes matplotlib attendent des instructions de couleur au format RGB ou avec des lettres. Je vous propose une solution élégante pour distinguer les villes.
- soit l'index de la ville la plus proche sotcké dans `ville_or` (0,...,7)
- construire le dictionnaire: `dict({0:'b', 1:'r', 2:'k', 3:'y', 4:'c', 5:'m', 6:'g'})`
- transformer `ville_or` en `ville_c` en vectorisant l'appel à la table de hash:
```python
ville_c = np.vectorize(dico.get)(ville_or)
```
```
# tracé de l'ensemble des trajectoires avec un code couleur en fonction de la ville de départ
# trouver l'information sur la ville la plus proche
coord = np.array([[45.18721767, 5.72345183],
[47.22572172, -1.56558993],
[50.63010695, 3.07071992],
[48.5782548, 7.74078742],
[44.83848889, -0.58156509],
[43.2991509, 5.38925024],
[48.8477201, 2.34607889]])
# calcul de la matrice de distance
dist =
# indice de la ville d'origine du trajet (plus petite distance dans le tableau ci-dessus)
ville_or =
# astuce pour construire une correspondance indice => code couleur
dico = dict({0:'b', 1:'r', 2:'k', 3:'y', 4:'c', 5:'m', 6:'g'})
ville_c = np.vectorize(dico.get)(ville_or)
plt.figure()
plt.quiver([... coordonnées des vecteurs ...]\
color= ville_c, angles='xy', scale_units='xy', scale=1)
plt.show()
```
## Etude de la corrélation entre variables
On propose d'étudier la corrélation entre la distance du trajet et le nombre d'étoiles de confort. Attention, les étoiles ne sont pas toujours renseignées (-1 = inconnu). On fera aussi ces opérations entre la distance et le prix.
1. Tracer dans le plan les coordonnées (distance,etoile) pour les points concernés
Vous utiliserez la commande `scatter` pour réaliser l'opération
1. Calculer le coefficient de corrélation entre les deux variables aléatoires
```
# test de corrélation entre la distance et le confort de la voiture
plt.figure()
# affichage du nuage de points correspondant au deux variables
plt.show()
# calcul du coefficient de corrélation pour tous les points admissibles
# (ceux pour lesquels les étoiles sont renseignées)
# test de corrélation entre la distance et le prix
plt.figure()
# affichage du nuage de points correspondant au deux variables
plt.show()
# calcul du coefficient de corrélation pour tous les points admissibles
```
## Quelques questions supplémentaires
### prix au kilomètre en fonction de l'origine
On s'intérroge sur le prix des courses en fonction des villes de départ. On ne veut pas tomber dans des pièges liés à des résumés simplistes, nous allons donc calculer la distribution jointe (prix_km, ville_origine).
1. Mettre au propre le code de discretisation en construisant une méthode de signature:
```python
def discretisation(x, nintervalles):
#[...]
return bornes, effectifs, xd
```
1. Mettre au propre le code de calcul de la loi jointe entre deux échantillons de variables aléatoires discrètes
```python
def pjointe(xd, yd):
#[...]
return pj
```
1. En l'état, nous avons du mal à analyser les données. Ceci est du aux valeurs extrêmes (notamment hautes). Afin de rendre l'analyse robuste, seuiller le prix au km au 99ème percentile (toutes les valeurs supérieures sont ramenées à cette valeur limite).
1. Proposer quelques analyses.
### même analyse pour voir dans quelle ville les gens sont plus matinaux, s'ils partent plus vers le sud ou le nord, l'est ou l'ouest...
Si vous étiez un journaliste en manque de sujet de reportage, quel(s) graphique(s) calculeriez vous à partir de ces données?
```
def discretisation(x, nintervalles, eps = 0.0000001) :
# [...]
return bornes, effectifs, xd
def pjointe(xd, yd): # variable codées de 0 à valmax
# [...]
return pj
# prix km
pkm = data[:,-4]/data[:,-1]
n = 30 # nb catégories
# robustesse : calcul du 99è percentile et seuillage des valeurs
bornes, effectifs, pkmd = discretisation(pkm, n)
pj = pjointe(pkmd,ville_or)
pv = # calcul de la marginale
p_km_s_v = # calcul de la conditionnelle
fig, ax = plt.subplots(1,1)
plt.imshow(p_km_s_v, interpolation='nearest')
plt.show()
# analyses
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# %matplotlib notebook
import pickle as pkl
import pickle as pkl
# chargement des données
fich = pkl.load( open('donnees_blablacar.pkl', 'rb'))
# {'indexcol': cols , 'data':pp2db, 'villes': villes, 'marques':marques }
titles_col = fich['indexcol']
print(len(titles_col), titles_col)
data = fich['data']
print(data.shape)
dico_villes = fich['villes']
dico_marques = fich['marques']
print(dico_marques)
# Analyse rapide : moyenne, écart-type, calcul des quantiles pour faire la synthèse de cette variable aléatoire
d = data[:,-1] # extraction de la variable distance
# A vous de jouer pour calculer tous les descripteurs = recherche dans la doc numpy !
d = np.sort(d)
quantile_values = np.array([])
range_quantile = 10
print("mean:", np.mean(d))
print("std:", np.std(d))
for i in range(range_quantile):
quantile_value = np.quantile(d, i/range_quantile)
print("quantile ", i, "/", range_quantile, ":", quantile_value)
quantile_values = np.append(quantile_values, quantile_value)
print("quantile_values:", quantile_values)
# Discrétisation des distances & histogramme avec np.histogram
np.histogram(d, bins = np.linspace(0, d.shape[0])
)
hist, bins = np.histogram(d, bins = np.linspace(0, d.shape[0]))
print(hist)
print(bins)
plt.hist(d, bins = np.linspace(0, d.shape[0]), width=100)
plt.title("histogramme")
plt.show()
# Discrétisation des distances & histogramme a la main
n = 30 # nb intervalle
inter = np.size(d)//n
bornes = np.array([])
for i in range(n):
bornes = np.append(bornes, i*inter)
# calcul des effectifs avec np.where
effectifs = np.array([])
for i in range(n - 1):
values = np.where((d >= bornes[i]) & (d <= bornes[i+1]))
effectif = np.size(values)
# print("borne1 :",bornes[i])
# print("borne2 :",bornes[i+1])
# print("effectif :",effectif)
# print("------------")
effectifs = np.append(effectifs, effectif)
bornes = bornes[:-1]
# print(bornes.shape)
# print(effectifs.shape)
# print(bornes)
# print(effectifs)
# affichage avec plt.bar = histogramme à la main
fig = plt.figure()
ax = fig.add_axes([0,0,1,1])
ax.bar(bornes, effectifs, width = 200)
plt.title("histogramme")
plt.show()
# histogramme des prix au km : construction de la variable puis utilisation de np.histogramme
prix = data[:,10]
km = data[:,13]
# print(prix.shape)
# print(km.shape)
values = prix/km
# print(value.shape)
bins = np.linspace(0, values[0], n)
np.histogram(values, bins = bins)
hist, bins = np.histogram(values, bins = bins)
plt.hist(values, bins = bins, width=0.002)
plt.title("histogramme")
plt.show()
fig, ax = plt.subplots(1,1)
plt.imshow(p_dsm, interpolation='nearest')
ax.set_xticks(np.arange(len(dico_marques)))
ax.set_xticklabels(dico_marques.keys(),rotation=90,fontsize=8)
plt.show()
plt.savefig('mafigure.pdf')
distances = data[:,13]
marques = data[:,11]
n = 10 # nb intervalle
inter = np.size(data[:,13])//n
bornes = np.array([])
for i in range(n):
bornes = np.append(bornes, i*inter)
print("nombre de bornes:\n",np.size(bornes)-1)
print("bornes distance:\n",bornes)
print("nombre de marque de voiture:\n",np.size(unique_marque))
tab_marques_dist = np.array([])
for i in range(n - 1):
values = np.array([])
for m in unique_marque:
values = np.append(values, np.size(np.where((distances > bornes[i]) & (distances < bornes[i+1]) & (marques == m)))/np.size(unique_marque))
tab_marques_dist = np.append(tab_marques_dist, values)
print("shape de nos valeurs:\n",tab_marques_dist.shape)
tab_marques_dist = tab_marques_dist.reshape(np.size(bornes)-1, np.size(unique_marque))
print("shape de nos valeurs après reshape en matrice:\n",tab_marques_dist.shape)
# construction de la distance discrétisée
# Dimensions : = (Nind x 1) = mêmes dimensions que d
# contenu = catégorie de distance (entre 0 et 29 par exemple si on a discrétisé en 30 catégories)
# dd = np.zeros(d.shape)
# remplissage avec np.where
# p_dm = np.zeros((len(bornes)-1, len(dico_marques)))
# remplissage de la matrice p_dm = double boucle + comptage
# p_dm /= p_dm.sum() # normalisation
# affichage du résultat
print("affichage resultat:")
plt.imshow(tab_marques_dist, interpolation='nearest')
plt.show()
# loi conditionnelle distance | marque
# calcul d'une marginale
# p_m =
# calcul de la conditionnelle
# p_dsm =
# affichage
# fig, ax = plt.subplots(1,1)
# plt.imshow(p_dsm, interpolation='nearest')
# ax.set_xticks(np.arange(len(dico_marques)))
# ax.set_xticklabels(dico_marques.keys(),rotation=90,fontsize=8)
# plt.show()
# proposition d'un critère très rapide pour vérifier qu'il s'agit bien d'une distribution conditionnelle
Afin de tracer des trajectoires, il faut envoyer les x et les y 2 par 2 dans une boucle `for`
1. Pour éviter les boucles, il existe une méthode `quiver` dédiée au tracé de champs de vecteurs: ça ira beaucoup plus vite qu'avec plot. Il faut juste bien comprendre les mécanismes d'échelles. Pour utiliser l'échelle 1, la commande est la suivante:
1. Isoler les trajets proposés à partir de chacune des villes sachant les coordonnées sont:
Chaque trajectoire (point de départ) sera rattachée à la ville la plus proche.
Une fois la distance calculée pour chaque origine de trajectoire, vous pourrez avoir besoin de `argmin`
1. Tracer les trajets d'une couleur spéciale en fonction des origines.
Les commandes matplotlib attendent des instructions de couleur au format RGB ou avec des lettres. Je vous propose une solution élégante pour distinguer les villes.
- soit l'index de la ville la plus proche sotcké dans `ville_or` (0,...,7)
- construire le dictionnaire: `dict({0:'b', 1:'r', 2:'k', 3:'y', 4:'c', 5:'m', 6:'g'})`
- transformer `ville_or` en `ville_c` en vectorisant l'appel à la table de hash:
## Etude de la corrélation entre variables
On propose d'étudier la corrélation entre la distance du trajet et le nombre d'étoiles de confort. Attention, les étoiles ne sont pas toujours renseignées (-1 = inconnu). On fera aussi ces opérations entre la distance et le prix.
1. Tracer dans le plan les coordonnées (distance,etoile) pour les points concernés
Vous utiliserez la commande `scatter` pour réaliser l'opération
1. Calculer le coefficient de corrélation entre les deux variables aléatoires
## Quelques questions supplémentaires
### prix au kilomètre en fonction de l'origine
On s'intérroge sur le prix des courses en fonction des villes de départ. On ne veut pas tomber dans des pièges liés à des résumés simplistes, nous allons donc calculer la distribution jointe (prix_km, ville_origine).
1. Mettre au propre le code de discretisation en construisant une méthode de signature:
1. Mettre au propre le code de calcul de la loi jointe entre deux échantillons de variables aléatoires discrètes
1. En l'état, nous avons du mal à analyser les données. Ceci est du aux valeurs extrêmes (notamment hautes). Afin de rendre l'analyse robuste, seuiller le prix au km au 99ème percentile (toutes les valeurs supérieures sont ramenées à cette valeur limite).
1. Proposer quelques analyses.
### même analyse pour voir dans quelle ville les gens sont plus matinaux, s'ils partent plus vers le sud ou le nord, l'est ou l'ouest...
Si vous étiez un journaliste en manque de sujet de reportage, quel(s) graphique(s) calculeriez vous à partir de ces données?
| 0.132515 | 0.954265 |
# Library Spectra Export process results
Read spectra files exported from the Bruker Spectra Library. All spectra files shall end with ".spectrum" and be located in one folder. Only one spectrum per file.
Please, specify the folder containing \*.spectrum files and the name for the results file:
```
folder = 'D:\data\Libraries\Example_Xpec'
archive = 'all_spectra.json'
```
Load modules and define functions.
```
import sys
import os.path
import codecs
import json
def dpstr2dict(filename, data_points_string, meta_count):
data_points = {} # using a dict to store the spectrum
# TODO: maybe use numpy ndarray instead
points = data_points_string.strip().split()
count = len(points)
if (count / 2) != int(meta_count):
print "Could not convert string to dict! Data point mismatch in spectrum: " + filename
return data_points_string # preserve original data
for i in xrange(0, count, 2):
data_points[float(points[i])] = int(points[i + 1])
#print (meta_count)
#print len(data_points)
return data_points
def readspectrum(filename, filecontent):
spectrum = {'SpecFile': filename}
values = ""
for line in filecontent.splitlines():
contents = line.split(':')
if len(contents) == 2:
key, value = contents
spectrum[key.strip()] = value.strip()
# 'Date', 'AnalName' and probably comments have multiple ':'
elif len(contents) > 2:
key = contents[0]
value = ':'.join(contents[1:])
spectrum[key.strip()] = value.strip()
# spectra do not contain ":"
else:
values += contents[0]
values = dpstr2dict(filename, values, spectrum['Num Peaks'])
spectrum['Values'] = values
return spectrum
```
Run the main script, which uses the functions above to collect all spectra in a list of dicts.
```
folder = os.path.abspath(folder)
if not os.path.exists(folder) or not os.path.isdir(folder):
print "Folder not found!"
sys.exit(0)
library = []
for spectrum in os.listdir(folder):
if not spectrum.endswith('.spectrum'):
print "Skipped file: " + spectrum
continue
spectrum = os.path.join(folder, spectrum)
#print spectrum
with codecs.open(spectrum, 'r', 'cp1252') as s:
data = readspectrum(spectrum, s.read())
library.append(data)
print 'The library contains ' + str(len(library)) + ' spectra.'
```
Store all spectra in a single *JSON* file.
```
archive = os.path.join(folder, archive)
with open(archive, 'w') as out:
json.dump(library, out, indent = 4, sort_keys = True)
```
|
github_jupyter
|
folder = 'D:\data\Libraries\Example_Xpec'
archive = 'all_spectra.json'
import sys
import os.path
import codecs
import json
def dpstr2dict(filename, data_points_string, meta_count):
data_points = {} # using a dict to store the spectrum
# TODO: maybe use numpy ndarray instead
points = data_points_string.strip().split()
count = len(points)
if (count / 2) != int(meta_count):
print "Could not convert string to dict! Data point mismatch in spectrum: " + filename
return data_points_string # preserve original data
for i in xrange(0, count, 2):
data_points[float(points[i])] = int(points[i + 1])
#print (meta_count)
#print len(data_points)
return data_points
def readspectrum(filename, filecontent):
spectrum = {'SpecFile': filename}
values = ""
for line in filecontent.splitlines():
contents = line.split(':')
if len(contents) == 2:
key, value = contents
spectrum[key.strip()] = value.strip()
# 'Date', 'AnalName' and probably comments have multiple ':'
elif len(contents) > 2:
key = contents[0]
value = ':'.join(contents[1:])
spectrum[key.strip()] = value.strip()
# spectra do not contain ":"
else:
values += contents[0]
values = dpstr2dict(filename, values, spectrum['Num Peaks'])
spectrum['Values'] = values
return spectrum
folder = os.path.abspath(folder)
if not os.path.exists(folder) or not os.path.isdir(folder):
print "Folder not found!"
sys.exit(0)
library = []
for spectrum in os.listdir(folder):
if not spectrum.endswith('.spectrum'):
print "Skipped file: " + spectrum
continue
spectrum = os.path.join(folder, spectrum)
#print spectrum
with codecs.open(spectrum, 'r', 'cp1252') as s:
data = readspectrum(spectrum, s.read())
library.append(data)
print 'The library contains ' + str(len(library)) + ' spectra.'
archive = os.path.join(folder, archive)
with open(archive, 'w') as out:
json.dump(library, out, indent = 4, sort_keys = True)
| 0.162015 | 0.798344 |
```
import pandas as pd
import numpy as np
from sklearn.feature_extraction.text import CountVectorizer
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.models import Sequential
from keras.layers import Dense, Embedding, LSTM, Bidirectional, Dropout
from sklearn.model_selection import train_test_split
from keras.utils.np_utils import to_categorical
import re
import matplotlib.pyplot as plt
from google.colab import drive
drive.mount('/content/drive')
train = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/kannada_offensive_train (1).csv', delimiter='\t', names=['sentence','classes','nan'])
train = train.drop(columns=['nan'])
train.head()
val = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/kannada_offensive_dev.csv', delimiter='\t', names=['sentence','classes','nan'])
val = val.drop(columns=['nan'])
test = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/kannada_offensive_test.csv',delimiter='\t',names=['sentence'])
test.head(4)
train.count()
train['classes'].apply(len).max()
train['sentence'].apply(len).max()
set(train['classes'])
encode_dict = {}
def encode_cat(x):
if x not in encode_dict.keys():
encode_dict[x] = len(encode_dict)
return encode_dict[x]
train['encode_cat'] = train['classes'].apply(lambda x: encode_cat(x))
val['encode_cat'] = val['classes'].apply(lambda x: encode_cat(x))
train.head(9)
y_train = train['encode_cat']
y_val = val['encode_cat']
def clean(df):
df['sentence'] = df['sentence'].apply(lambda x: x.lower())
df['sentence'] = df['sentence'].apply(lambda x: re.sub(r' +', ' ',x))
df['sentence'] = df['sentence'].apply(lambda x: re.sub("[!@#$+%*:()'-]", ' ',x))
df['sentence'] = df['sentence'].str.replace('\d+', '')
clean(train)
clean(val)
clean(test)
max_features = 2000
max_len = 512
tokenizer = Tokenizer(num_words=max_features, split=' ')
tokenizer.fit_on_texts(train['sentence'].values)
X_train = tokenizer.texts_to_sequences(train['sentence'].values)
# vocab_size = len(tokenizer.word_index) + 1
X_train = pad_sequences(X_train,padding = 'post', maxlen=max_len)
tokenizer.fit_on_texts(val['sentence'].values)
X_val = tokenizer.texts_to_sequences(val['sentence'].values)
# vocab_size = len(tokenizer.word_index) + 1
X_val = pad_sequences(X_val,padding = 'post', maxlen=max_len)
tokenizer.fit_on_texts(test['sentence'].values)
X_test = tokenizer.texts_to_sequences(test['sentence'].values)
# vocab_size = len(tokenizer.word_index) + 1
X_test = pad_sequences(X_test,padding = 'post', maxlen=max_len)
train['sentence'].apply(len).max()
train.describe()
Y_train = pd.get_dummies(y_train).values
Y_val = pd.get_dummies(y_val).values
print(X_train.shape,Y_train.shape)
print(X_val.shape,Y_val.shape)
print(X_test.shape)
from keras.layers import Layer
from keras.layers import Input
from keras.models import Model
from tensorflow.keras import backend as K
class attention(Layer):
def __init__(self):
super(attention,self).__init__()
def build(self,input_shape):
self.W=self.add_weight(name="att_weight",shape=(input_shape[-1],1),initializer="normal")
self.b=self.add_weight(name="att_bias",shape=(input_shape[1],1),initializer="zeros")
super(attention, self).build(input_shape)
def call(self,x):
et=K.squeeze(K.tanh(K.dot(x,self.W)+self.b),axis=-1)
at=K.softmax(et)
at=K.expand_dims(at,axis=-1)
output=x*at
return K.sum(output,axis=1)
def compute_output_shape(self,input_shape):
return (input_shape[0],input_shape[-1])
def get_config(self):
return super(attention,self).get_config()
!wget --header="Host: nlp.stanford.edu" --header="User-Agent: Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/69.0.3497.100 Safari/537.36" --header="Accept: text/html,application/xhtml+xml,application/xml;q=0.9,image/webp,image/apng,*/*;q=0.8" --header="Accept-Language: en-US,en;q=0.9" --header="Cookie: _ga=GA1.2.456156586.1539718115; _gid=GA1.2.491677602.1539718115; _gat=1" --header="Connection: keep-alive" "https://nlp.stanford.edu/data/glove.6B.zip" -O "glove.6B.zip" -c
!unzip glove.6B.zip
from numpy import array
from numpy import asarray
from numpy import zeros
embeddings_index = dict()
glove_file = open('glove.6B.100d.txt', encoding="utf8")
for line in glove_file:
records = line.split()
word = records[0]
vector_dimensions = asarray(records[1:], dtype='float32')
embeddings_index[word] = vector_dimensions
glove_file.close()
print('Found %s word vectors.' %len(embeddings_index))
word_index = tokenizer.word_index
print(len(word_index))
num_words = min(max_features, len(word_index)) + 1
print(num_words)
embedding_dim = 100
embedding_matrix = np.zeros((num_words, embedding_dim))
for word, i in word_index.items():
if i > max_features:
continue
embedding_vector = embeddings_index.get(word)
if embedding_vector is not None:
embedding_matrix[i] = embedding_vector
else:
embedding_matrix[i] = np.random.randn(embedding_dim)
K.clear_session()
from keras.regularizers import l2
from keras.initializers import Constant
embed_dim = 100
lstm_out = 128
# model = Sequential()
inputs = Input(shape=(512,))
x = Embedding(num_words, embed_dim,embeddings_initializer=Constant(embedding_matrix),input_length = X_train.shape[1])(inputs)
att_in = Bidirectional(LSTM(lstm_out,return_sequences=True, dropout=0.2))(x)
att_out = attention()(att_in)
d = Dropout(0.2)(att_out)
outputs = Dense(6, activation='softmax')(d)
model = Model(inputs,outputs)
print(model.summary())
import numpy as np
from sklearn.utils import class_weight
class_weights = class_weight.compute_class_weight('balanced',
np.unique(train.encode_cat.values),
train.encode_cat.values)
class_weights = dict(enumerate(class_weights))
model.compile(loss = 'categorical_crossentropy', optimizer='adam',metrics = ['accuracy'])
history = model.fit(X_train, Y_train,batch_size = 128, validation_data=(X_val,Y_val), epochs=10, verbose=2)
import matplotlib.pyplot as plt
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
plt.title('Model Accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train','val'])
#plt.show()
plt.savefig('Model_accuracy.png', dpi=600)
# score = model.evaluate(X_test,verbose=1)
predictions = np.argmax(model.predict(X_test),axis = -1)
# print("val score is {}".format(score[0]))
# print("val Accuracy is {}".format(score[1]))
_, train_acc = model.evaluate(X_train, Y_train, verbose=0)
_, val_acc = model.evaluate(X_val, Y_val, verbose=0)
print(val_acc)
print(train_acc)
rounded_predictions = np.argmax(model.predict(X_test, batch_size=128, verbose=0),axis = -1)
print(rounded_predictions)
a = {'id':[i for i in range(778)]}
a = pd.DataFrame(a)
df = pd.DataFrame({'id':a.id,'labels':rounded_predictions})
df.labels = df.labels.apply({0:'Not_offensive',5:'Offensive_Untargetede',3:'Offensive_Targeted_Insult_Other',
4:'Offensive_Targeted_Insult_Individual',1:'not-Kannada',2:'Offensive_Targeted_Insult_Group'}.get)
df
df.to_csv('LSTM_with_attention_Kannada_submission.csv',index=False)
from google.colab import files
files.download("LSTM_with_attention_Kannada_submission.csv")
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from sklearn.feature_extraction.text import CountVectorizer
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.models import Sequential
from keras.layers import Dense, Embedding, LSTM, Bidirectional, Dropout
from sklearn.model_selection import train_test_split
from keras.utils.np_utils import to_categorical
import re
import matplotlib.pyplot as plt
from google.colab import drive
drive.mount('/content/drive')
train = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/kannada_offensive_train (1).csv', delimiter='\t', names=['sentence','classes','nan'])
train = train.drop(columns=['nan'])
train.head()
val = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/kannada_offensive_dev.csv', delimiter='\t', names=['sentence','classes','nan'])
val = val.drop(columns=['nan'])
test = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/kannada_offensive_test.csv',delimiter='\t',names=['sentence'])
test.head(4)
train.count()
train['classes'].apply(len).max()
train['sentence'].apply(len).max()
set(train['classes'])
encode_dict = {}
def encode_cat(x):
if x not in encode_dict.keys():
encode_dict[x] = len(encode_dict)
return encode_dict[x]
train['encode_cat'] = train['classes'].apply(lambda x: encode_cat(x))
val['encode_cat'] = val['classes'].apply(lambda x: encode_cat(x))
train.head(9)
y_train = train['encode_cat']
y_val = val['encode_cat']
def clean(df):
df['sentence'] = df['sentence'].apply(lambda x: x.lower())
df['sentence'] = df['sentence'].apply(lambda x: re.sub(r' +', ' ',x))
df['sentence'] = df['sentence'].apply(lambda x: re.sub("[!@#$+%*:()'-]", ' ',x))
df['sentence'] = df['sentence'].str.replace('\d+', '')
clean(train)
clean(val)
clean(test)
max_features = 2000
max_len = 512
tokenizer = Tokenizer(num_words=max_features, split=' ')
tokenizer.fit_on_texts(train['sentence'].values)
X_train = tokenizer.texts_to_sequences(train['sentence'].values)
# vocab_size = len(tokenizer.word_index) + 1
X_train = pad_sequences(X_train,padding = 'post', maxlen=max_len)
tokenizer.fit_on_texts(val['sentence'].values)
X_val = tokenizer.texts_to_sequences(val['sentence'].values)
# vocab_size = len(tokenizer.word_index) + 1
X_val = pad_sequences(X_val,padding = 'post', maxlen=max_len)
tokenizer.fit_on_texts(test['sentence'].values)
X_test = tokenizer.texts_to_sequences(test['sentence'].values)
# vocab_size = len(tokenizer.word_index) + 1
X_test = pad_sequences(X_test,padding = 'post', maxlen=max_len)
train['sentence'].apply(len).max()
train.describe()
Y_train = pd.get_dummies(y_train).values
Y_val = pd.get_dummies(y_val).values
print(X_train.shape,Y_train.shape)
print(X_val.shape,Y_val.shape)
print(X_test.shape)
from keras.layers import Layer
from keras.layers import Input
from keras.models import Model
from tensorflow.keras import backend as K
class attention(Layer):
def __init__(self):
super(attention,self).__init__()
def build(self,input_shape):
self.W=self.add_weight(name="att_weight",shape=(input_shape[-1],1),initializer="normal")
self.b=self.add_weight(name="att_bias",shape=(input_shape[1],1),initializer="zeros")
super(attention, self).build(input_shape)
def call(self,x):
et=K.squeeze(K.tanh(K.dot(x,self.W)+self.b),axis=-1)
at=K.softmax(et)
at=K.expand_dims(at,axis=-1)
output=x*at
return K.sum(output,axis=1)
def compute_output_shape(self,input_shape):
return (input_shape[0],input_shape[-1])
def get_config(self):
return super(attention,self).get_config()
!wget --header="Host: nlp.stanford.edu" --header="User-Agent: Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/69.0.3497.100 Safari/537.36" --header="Accept: text/html,application/xhtml+xml,application/xml;q=0.9,image/webp,image/apng,*/*;q=0.8" --header="Accept-Language: en-US,en;q=0.9" --header="Cookie: _ga=GA1.2.456156586.1539718115; _gid=GA1.2.491677602.1539718115; _gat=1" --header="Connection: keep-alive" "https://nlp.stanford.edu/data/glove.6B.zip" -O "glove.6B.zip" -c
!unzip glove.6B.zip
from numpy import array
from numpy import asarray
from numpy import zeros
embeddings_index = dict()
glove_file = open('glove.6B.100d.txt', encoding="utf8")
for line in glove_file:
records = line.split()
word = records[0]
vector_dimensions = asarray(records[1:], dtype='float32')
embeddings_index[word] = vector_dimensions
glove_file.close()
print('Found %s word vectors.' %len(embeddings_index))
word_index = tokenizer.word_index
print(len(word_index))
num_words = min(max_features, len(word_index)) + 1
print(num_words)
embedding_dim = 100
embedding_matrix = np.zeros((num_words, embedding_dim))
for word, i in word_index.items():
if i > max_features:
continue
embedding_vector = embeddings_index.get(word)
if embedding_vector is not None:
embedding_matrix[i] = embedding_vector
else:
embedding_matrix[i] = np.random.randn(embedding_dim)
K.clear_session()
from keras.regularizers import l2
from keras.initializers import Constant
embed_dim = 100
lstm_out = 128
# model = Sequential()
inputs = Input(shape=(512,))
x = Embedding(num_words, embed_dim,embeddings_initializer=Constant(embedding_matrix),input_length = X_train.shape[1])(inputs)
att_in = Bidirectional(LSTM(lstm_out,return_sequences=True, dropout=0.2))(x)
att_out = attention()(att_in)
d = Dropout(0.2)(att_out)
outputs = Dense(6, activation='softmax')(d)
model = Model(inputs,outputs)
print(model.summary())
import numpy as np
from sklearn.utils import class_weight
class_weights = class_weight.compute_class_weight('balanced',
np.unique(train.encode_cat.values),
train.encode_cat.values)
class_weights = dict(enumerate(class_weights))
model.compile(loss = 'categorical_crossentropy', optimizer='adam',metrics = ['accuracy'])
history = model.fit(X_train, Y_train,batch_size = 128, validation_data=(X_val,Y_val), epochs=10, verbose=2)
import matplotlib.pyplot as plt
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
plt.title('Model Accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train','val'])
#plt.show()
plt.savefig('Model_accuracy.png', dpi=600)
# score = model.evaluate(X_test,verbose=1)
predictions = np.argmax(model.predict(X_test),axis = -1)
# print("val score is {}".format(score[0]))
# print("val Accuracy is {}".format(score[1]))
_, train_acc = model.evaluate(X_train, Y_train, verbose=0)
_, val_acc = model.evaluate(X_val, Y_val, verbose=0)
print(val_acc)
print(train_acc)
rounded_predictions = np.argmax(model.predict(X_test, batch_size=128, verbose=0),axis = -1)
print(rounded_predictions)
a = {'id':[i for i in range(778)]}
a = pd.DataFrame(a)
df = pd.DataFrame({'id':a.id,'labels':rounded_predictions})
df.labels = df.labels.apply({0:'Not_offensive',5:'Offensive_Untargetede',3:'Offensive_Targeted_Insult_Other',
4:'Offensive_Targeted_Insult_Individual',1:'not-Kannada',2:'Offensive_Targeted_Insult_Group'}.get)
df
df.to_csv('LSTM_with_attention_Kannada_submission.csv',index=False)
from google.colab import files
files.download("LSTM_with_attention_Kannada_submission.csv")
| 0.46563 | 0.301298 |
```
%load_ext autoreload
%autoreload 2
```
This notebook contains basic usage of the `FunctionClassifier`.
```
import numpy as np
import pandas as pd
from hulearn.datasets import load_titanic
from hulearn.classification import FunctionClassifier
from sklearn.model_selection import GridSearchCV
df = load_titanic(as_frame=True)
X, y = df.drop(columns=['survived']), df['survived']
def last_name(dataf, sex='male', pclass=1):
predicate = (dataf['sex'] == sex) & (dataf['pclass'] == pclass)
return np.array(predicate).astype(int)
def fare_based(dataf, threshold=10):
return np.array(dataf['fare'] > threshold).astype(int)
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import precision_score, recall_score, accuracy_score, make_scorer
mod = FunctionClassifier(fare_based, threshold=10)
grid = GridSearchCV(mod,
cv=2,
param_grid={'threshold': np.linspace(0, 100, 30)},
scoring={'accuracy': make_scorer(accuracy_score),
'precision': make_scorer(precision_score),
'recall': make_scorer(recall_score)},
refit='accuracy'
)
grid.fit(X, y)
score_df = (pd.DataFrame(grid.cv_results_)
.set_index('param_threshold')
[['mean_test_accuracy', 'mean_test_precision', 'mean_test_recall']])
score_df.plot(figsize=(12, 5), title="scores vs. fare-threshold");
score_df.sort_values("mean_test_precision", ascending=False).head()
```
We seem to be able to achieve 78.5% precision given the right threshold! What about a `RandomForest`?
```
from sklearn.ensemble import RandomForestClassifier
X_subset = X.assign(sex=lambda d: d['sex'] == 'female')[['pclass', 'sex', 'age', 'fare']]
grid = GridSearchCV(RandomForestClassifier(),
cv=10,
param_grid={},
scoring={'accuracy': make_scorer(accuracy_score),
'precision': make_scorer(precision_score),
'recall': make_scorer(recall_score)},
refit='accuracy'
)
pd.DataFrame(grid.fit(X_subset, y).cv_results_)[['mean_test_accuracy', 'mean_test_precision', 'mean_test_recall']]
```
Our simple threhold model seems competative on precision. This is interesting.
# Bigger Grid
We're not limited to just one parameter for our grid. With the `FunctionClassifier` you can define many.
```
def custom_predict(dataf, sex='male', pclass=1):
predicate = (dataf['sex'] == sex) & (dataf['pclass'] == pclass)
return np.array(predicate).astype(int)
mod = FunctionClassifier(custom_predict, pclass=10, sex='male')
grid = GridSearchCV(mod,
cv=2,
param_grid={'pclass': [1, 2, 3], 'sex': ['male', 'female']},
scoring={'accuracy': make_scorer(accuracy_score),
'precision': make_scorer(precision_score),
'recall': make_scorer(recall_score)},
refit='accuracy')
grid.fit(X, y)
pd.DataFrame(grid.cv_results_)[['param_pclass', 'param_sex', 'mean_test_accuracy', 'mean_test_precision', 'mean_test_recall']]
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
import numpy as np
import pandas as pd
from hulearn.datasets import load_titanic
from hulearn.classification import FunctionClassifier
from sklearn.model_selection import GridSearchCV
df = load_titanic(as_frame=True)
X, y = df.drop(columns=['survived']), df['survived']
def last_name(dataf, sex='male', pclass=1):
predicate = (dataf['sex'] == sex) & (dataf['pclass'] == pclass)
return np.array(predicate).astype(int)
def fare_based(dataf, threshold=10):
return np.array(dataf['fare'] > threshold).astype(int)
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import precision_score, recall_score, accuracy_score, make_scorer
mod = FunctionClassifier(fare_based, threshold=10)
grid = GridSearchCV(mod,
cv=2,
param_grid={'threshold': np.linspace(0, 100, 30)},
scoring={'accuracy': make_scorer(accuracy_score),
'precision': make_scorer(precision_score),
'recall': make_scorer(recall_score)},
refit='accuracy'
)
grid.fit(X, y)
score_df = (pd.DataFrame(grid.cv_results_)
.set_index('param_threshold')
[['mean_test_accuracy', 'mean_test_precision', 'mean_test_recall']])
score_df.plot(figsize=(12, 5), title="scores vs. fare-threshold");
score_df.sort_values("mean_test_precision", ascending=False).head()
from sklearn.ensemble import RandomForestClassifier
X_subset = X.assign(sex=lambda d: d['sex'] == 'female')[['pclass', 'sex', 'age', 'fare']]
grid = GridSearchCV(RandomForestClassifier(),
cv=10,
param_grid={},
scoring={'accuracy': make_scorer(accuracy_score),
'precision': make_scorer(precision_score),
'recall': make_scorer(recall_score)},
refit='accuracy'
)
pd.DataFrame(grid.fit(X_subset, y).cv_results_)[['mean_test_accuracy', 'mean_test_precision', 'mean_test_recall']]
def custom_predict(dataf, sex='male', pclass=1):
predicate = (dataf['sex'] == sex) & (dataf['pclass'] == pclass)
return np.array(predicate).astype(int)
mod = FunctionClassifier(custom_predict, pclass=10, sex='male')
grid = GridSearchCV(mod,
cv=2,
param_grid={'pclass': [1, 2, 3], 'sex': ['male', 'female']},
scoring={'accuracy': make_scorer(accuracy_score),
'precision': make_scorer(precision_score),
'recall': make_scorer(recall_score)},
refit='accuracy')
grid.fit(X, y)
pd.DataFrame(grid.cv_results_)[['param_pclass', 'param_sex', 'mean_test_accuracy', 'mean_test_precision', 'mean_test_recall']]
| 0.677474 | 0.859605 |
# Project 2
##### Team member: Zhihong Zhang.
## Part 1: Introduction ##
The dataset of this project is based on the salaries package provided in the R programming language Package.
#### The setting up of the project
```
#starting point:import the library and data set
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
salaries ='https://raw.githubusercontent.com/steinszzh/DAV-5400/master/project2/Salaries.csv'
salary = pd.read_csv(salaries,index_col=0)
# data set check - make sure data was read in as expected
salary.head()
```
As the data base mentioned, the data set contains the 2008-2009 nine-month academic salary for Assistant Professors, Associate Professors and Professors in a college in the U.S. The data were collected as part of the on-going effort of the college's administration to monitor salary differences between male and female faculty members.
After checking the data, it is clear to see some potential correlation between salaries and some other attributes. I would like to explore the difference on salaries between the genders.
#### research question
To better serve the goal of exploring the data set on gender difference, one foundamental question is seeked to ask :
- Is the pay fair for both genders?
According to this goal of comparision on performance, the research questions are created and I decide to see the salary difference based on area of study, years of service,rank and gender.
## Part 2: Data Summary
The dataset was acquired from the R data set repository[https://vincentarelbundock.github.io/Rdatasets/csv/carData/Salaries.csv]. The dataset contains 397 rows and 6 attributes. These attributes are:
- rank: a factor with levels (values are AssocProf, AsstProf,and Prof).It is categorical/ordinal.
- discipline: a factor with levels A (“theoretical” departments) or B (“applied” departments). It is categorical/regular.
- yrs.since.phd: years since PhD. It is numerical/continuous.
- yrs.service: years of service. It is numerical/continuous.
- Sex: a factor with levels Female and Male. It is categorical/regular.
- salary : nine-month salary, in dollars. It is numerical/continuous.
## Part 3: Exploratory Data Analysis (EDA)
To explore the data, I firstly need to make sure how dimensions are distributed in the data set. Due to the possibility that data set is not evenly distributed into two genders (which means that one gender will have more people) To make comparision, this situation of sample size needs to be taken into account. To get start, I use the describe function to know the quantative stats for the numerical variables.
```
#create a copy for first section to make sure not messing up data
salary1 = salary.copy()
#displays descriptive stats for all numeric variables
salary1.describe()
```
There are 397 rows among these data set. And it showed descriptive stats for all numeric variables. mean of yrs since phd is 22.314861 and mean of years service is 17.614610. They are both greater than median value of 21 and 16. It means that both two values are skewed to the right. Similarly, the salary is also skewed to the right.
Firstly, for the rank attribute, I choose bar chart to explore this attribute.
#### Attribute: rank
```
# side-by-side barplot Matplotlib and Seaborn for rank based on the number of the distribution.
# define a new figure for plotting
plt.figure(figsize=(10,5))
# define the left plot which is created by matplotlib
plt.subplot(121)
# matplotlib graph of rank
rank_count = salary1.groupby('rank').count()['salary']
rank_count.plot.bar();
# give the plot a title
plt.title('Rank Bar Chart (Matplotlib)')
plt.xlabel('Rank')
plt.ylabel('Count')
#Then using seaborn to create graph
# define the right graph
plt.subplot(122)
# Seaborn graph of rank
sns.countplot(x='rank',data=salary1, order=['AssocProf','AsstProf','Prof'])
# give the plot a title
plt.title('Rank Bar Chart (Seaborn)')
plt.xlabel('Rank')
plt.ylabel('Count')
plt.show()
```
Based on the graph, we know that in this data set, majority is professor rank and there is more assistant prof than asscociated prof but they are basically the same level.
Then I choose bar plot for discipline attributes.
#### Attribute: discipline
```
# plot barplot by using Matplotlib and Seaborn for discipline based on the number of the distribution.
# make the appropriate size of figure
plt.figure(figsize=(10,5))
# make matplotlib plot on the left
plt.subplot(121)
# matplotlib graph of discipline
discipline_count = salary1.groupby('discipline').count()['salary']
discipline_count.plot.bar();
# give the plot a title
plt.title('discipline Bar Chart (Matplotlib)')
plt.xlabel('discipline')
plt.ylabel('Count')
#Then using seaborn to create graph
# create the right side graph
plt.subplot(122)
# make Seaborn graph of discpline
sns.countplot(x='discipline',data=salary1, order=['A','B'])
# give the plot a title
plt.title('discipline Bar Chart (Seaborn)')
plt.xlabel('discipline')
plt.ylabel('Count')
plt.show()
#drawing scatterplots by Matplotlib and Seaborn
#plot matploblib on the left
plt.figure(figsize=(20,20))
#plots discipline vs salary using matplotlib
plt.subplot(121)
plt.scatter(x='discipline', y='salary', data=salary1)
plt.title('Scatterplot discipline vs. salary(Matplotlib)')
plt.xlabel('discipline')
plt.ylabel('salary')
```
By checking the spread of discipline vs salary, we can tell that in general, applied field has a smaller standard deviation (narrow spread). Also two bar charts showed that applied and theoritical has the similar level of quantity but there is more samples in applied disciplines.
For years since phd attribute, box plot was made.
#### Attribute: yrs.since.phd
```
# make boxplot for years since phd by two method
salary_yrs_phd = salary1['yrs.since.phd']
# define a new figure for plotting
plt.figure(figsize=(10,10))
# define the plot that will appear on the lefthand side of the subplot:
plt.subplot(121)
# Matplotlib boxplot for years since phd
plt.boxplot(x=salary_yrs_phd)
# give the plot a title
plt.title('yrs.since.phd Boxplot (Matplotlib)')
#make right side plot
plt.subplot(122)
# create Seaborn boxplot for years since phd
sns.boxplot(tuple(salary_yrs_phd),orient='v')
# give the plot a title
plt.title('years since phd Boxplot (Seaborn)')
```
Based on the graph, we know that the median of years since phd is around 22, up bound and low bound is roughly 33 and 14.
Scatterplot was used to demonstrate the years of service attributes.
#### Attribute: yrs.service
```
#drawing scatterplots by Matplotlib and Seaborn
#plot matploblib on the left
plt.figure(figsize=(20,20))
#plots years service vs salary using matplotlib
plt.subplot(121)
plt.scatter(x='yrs.service', y='salary', data=salary1)
plt.title('Scatterplot yrs.service vs. salary(Matplotlib)')
plt.xlabel('yrs.service')
plt.ylabel('salary')
#plots years service vs salary using seaborn
plt.subplot(122)
sns.regplot(x='yrs.service', y='salary', data=salary1)
plt.title('Scatterplot yrs.service vs. salary(Seaborn)')
plt.xlabel('yrs.service')
plt.ylabel('salary')
```
As the result showed, in general, the years of service is positively correlated to the salary. The correlation does not show a direct linear regression and it can be contributed to the fact that other factors such as rank or discipline impacts the salary
For the sex attribue, bar chart was used to check the frequency of the distribution.
#### Attribute: Sex
```
# side-by-side barplot Matplotlib and Seaborn for Sex based on the number of the distribution.
# define a new figure for plotting
plt.figure(figsize=(10,5))
# define the left plot which is created by matplotlib
plt.subplot(121)
# matplotlib graph of Sex
gender_count = salary1.groupby('sex').count()['salary']
gender_count.plot.bar();
# give the plot a title
plt.title('Gender Bar Chart (Matplotlib)')
plt.xlabel('Sex')
plt.ylabel('Count')
#Then using seaborn to create graph
# define the right graph
plt.subplot(122)
# Seaborn graph of Sex
sns.countplot(x='sex',data=salary1, order=['Female','Male'])
# give the plot a title
plt.title('Gender Bar Chart (Seaborn)')
plt.xlabel('Sex')
plt.ylabel('Count')
plt.show()
```
Based on this bar chart, we know that the majority of the data set is male. And it has more than 1:8 gender ratio between male and female.
#### Attribute: Salary
```
# plot histogram by using Matplotlib and Seaborn
salary_hist = salary1['salary']
#plot matplotlib graph on the left
plt.subplot(121)
#Matlplotlib histogram for salary
plt.hist(tuple(salary_hist))
plt.title('Histogram of salary (Matplotlib)')
#plot seaborn on the right side
plt.subplot(122)
#Seaborn histogram for salary
sns.distplot(tuple(salary_hist))
plt.title('Histogram of salary (Seaborn)')
plt.show()
```
Based on the histogram of salary, it is clear that majority of the salary level is between 80,000 to 11,000 for professor.
## Part 4: Inference
Based on the EDA section, I have better understanding of the patterns of each attributes. Firstly, I decide to make a new dataframe.
```
#create a copy for salary to make sure not messing up data
salary2 = salary.copy()
```
To test whether there is a salary ineuality, I test the data of salary abover the average. Since there is more than half of data below the mean , it means that the data has more high salary outliers to drag mean to the right.
```
# Calculate the salary mean
salary_mean = salary2.salary.mean()
salary_mean
# Create a column "special_salary" that contains all of salary values less than the salary mean
special_salary = salary2.salary[salary2.salary<salary_mean]
special_salary.count()
special_salary.describe()
```
After getting the data, I then created box charts of salary based on gender. For this general plot, I firstly includes all attributesto see the patterns of salary plot.
```
#print boxplot by using matplotlib
salary2.boxplot(by ='sex', column =['salary'], grid = False)
# print boxplot by using seaborns
sns.boxplot(x="sex", y="salary", data=salary2, palette="Set1")
#sns.plt.show()
```
Through these two bar charts, it is easy to tell that for both two genders, the mean is about the same. Compared with the male faculty, the female has a narrow spread. Also the minimum salary level for the female is higher than the male. Due to the fact that the male has some outlier drags the average to higher value. Since there are other factors impacting on the average(such as the rank or years of service), other steps need to be used.
To further explore the data, I decide to make the controlled group. Based on that the rank of professor occupies the majority of the samples, I decided to compare salary when both genders have the same rank in the applied field.
```
#create subset of rank, sex,discipline and salary
salary_select = salary2.loc[:,['rank','discipline','sex','salary']]
#filter data to be prof in applied field only
salary_select_prof = salary_select[(salary_select['rank']=='Prof')& (salary_select['discipline']=='B')]
#get most frequent salary
print(salary_select_prof.describe())
#print boxplot by using matplotlib
salary_select_boxplot=salary_select_prof.boxplot(by ='sex', column =['salary'], grid = False)
salary_select_boxplot;
# print boxplot by using seaborns
sns.boxplot(x="sex", y="salary", data=salary_select_prof, palette="Set1")
#sns.plt.show()
```
After this controlled group we can tell that female has a even narrowed spread. To dig further, I tried to import another variable, employer who worked for more than 18 years and in applied field. Similarily,I make plot as well.
```
#create subset of rank, sex,discipline and salary
salary_select_yrs = salary2.loc[:,['rank','discipline','yrs.service','sex','salary']]
#filter data to be prof in applied field work for more than 18 years
salary_select_yrs = salary_select_yrs[(salary_select_yrs['rank']=='Prof')& (salary_select_yrs['discipline']=='B')&(salary_select_yrs['yrs.service']>=18)]
#get most frequent salary
print(salary_select_prof.describe())
#print boxplot by using matplotlib
salary_select_boxplot=salary_select_yrs.boxplot(by ='sex', column =['salary'], grid = False)
salary_select_boxplot;
# print boxplot by using seaborns as well
sns.boxplot(x="sex", y="salary", data=salary_select_yrs, palette="Set1")
#sns.plt.show()
```
After this controlled group in the apllied field, the female controlled group has a narrower spread. Interestingly, now the female faculty has a higher average salary value than the male.
##### Result:
Based on these printed graph on different control groups, although it has a very similar shape and spread, that as we filtered more , the size of spread on female professor becomes smaller. The main reason of this phenomenon is that there is not enough samples for the selected group(female) the filter dramistically changed the possible output.
## Part 5: Conclusion
By answering the research question and explore various type of data, in my opinion,the research question provides the part of view how the the salary level differ from each individuals within a college.
Based on the information we have, we could not say that there is any direct evidence that the female employee did not get a fair pay or not. Contributing to the limited sample size of the data(Especially there is a relatively small group of female professors), the samples does not spread a lot, and it does not provide enough data for making the right judgement on salary inquality.
Also, with some research of this question, the reference does mention the salary inquality near 2006, while it cannot help to explain this data by this college since this data set may be one of the special cases in the educational system.
Therefore, Compared the huge difference between the size of samples between two genders, we still lacked the envidence to confidently say that there is a significant difference between male and female on salary in this college.
## References ##
1. https://www.insidehighered.com/news/2018/04/11/aaups-annual-report-faculty-compensation-takes-salary-compression-and-more
2. https://ed.psu.edu/cshe/working-papers/wp-11
|
github_jupyter
|
#starting point:import the library and data set
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
salaries ='https://raw.githubusercontent.com/steinszzh/DAV-5400/master/project2/Salaries.csv'
salary = pd.read_csv(salaries,index_col=0)
# data set check - make sure data was read in as expected
salary.head()
#create a copy for first section to make sure not messing up data
salary1 = salary.copy()
#displays descriptive stats for all numeric variables
salary1.describe()
# side-by-side barplot Matplotlib and Seaborn for rank based on the number of the distribution.
# define a new figure for plotting
plt.figure(figsize=(10,5))
# define the left plot which is created by matplotlib
plt.subplot(121)
# matplotlib graph of rank
rank_count = salary1.groupby('rank').count()['salary']
rank_count.plot.bar();
# give the plot a title
plt.title('Rank Bar Chart (Matplotlib)')
plt.xlabel('Rank')
plt.ylabel('Count')
#Then using seaborn to create graph
# define the right graph
plt.subplot(122)
# Seaborn graph of rank
sns.countplot(x='rank',data=salary1, order=['AssocProf','AsstProf','Prof'])
# give the plot a title
plt.title('Rank Bar Chart (Seaborn)')
plt.xlabel('Rank')
plt.ylabel('Count')
plt.show()
# plot barplot by using Matplotlib and Seaborn for discipline based on the number of the distribution.
# make the appropriate size of figure
plt.figure(figsize=(10,5))
# make matplotlib plot on the left
plt.subplot(121)
# matplotlib graph of discipline
discipline_count = salary1.groupby('discipline').count()['salary']
discipline_count.plot.bar();
# give the plot a title
plt.title('discipline Bar Chart (Matplotlib)')
plt.xlabel('discipline')
plt.ylabel('Count')
#Then using seaborn to create graph
# create the right side graph
plt.subplot(122)
# make Seaborn graph of discpline
sns.countplot(x='discipline',data=salary1, order=['A','B'])
# give the plot a title
plt.title('discipline Bar Chart (Seaborn)')
plt.xlabel('discipline')
plt.ylabel('Count')
plt.show()
#drawing scatterplots by Matplotlib and Seaborn
#plot matploblib on the left
plt.figure(figsize=(20,20))
#plots discipline vs salary using matplotlib
plt.subplot(121)
plt.scatter(x='discipline', y='salary', data=salary1)
plt.title('Scatterplot discipline vs. salary(Matplotlib)')
plt.xlabel('discipline')
plt.ylabel('salary')
# make boxplot for years since phd by two method
salary_yrs_phd = salary1['yrs.since.phd']
# define a new figure for plotting
plt.figure(figsize=(10,10))
# define the plot that will appear on the lefthand side of the subplot:
plt.subplot(121)
# Matplotlib boxplot for years since phd
plt.boxplot(x=salary_yrs_phd)
# give the plot a title
plt.title('yrs.since.phd Boxplot (Matplotlib)')
#make right side plot
plt.subplot(122)
# create Seaborn boxplot for years since phd
sns.boxplot(tuple(salary_yrs_phd),orient='v')
# give the plot a title
plt.title('years since phd Boxplot (Seaborn)')
#drawing scatterplots by Matplotlib and Seaborn
#plot matploblib on the left
plt.figure(figsize=(20,20))
#plots years service vs salary using matplotlib
plt.subplot(121)
plt.scatter(x='yrs.service', y='salary', data=salary1)
plt.title('Scatterplot yrs.service vs. salary(Matplotlib)')
plt.xlabel('yrs.service')
plt.ylabel('salary')
#plots years service vs salary using seaborn
plt.subplot(122)
sns.regplot(x='yrs.service', y='salary', data=salary1)
plt.title('Scatterplot yrs.service vs. salary(Seaborn)')
plt.xlabel('yrs.service')
plt.ylabel('salary')
# side-by-side barplot Matplotlib and Seaborn for Sex based on the number of the distribution.
# define a new figure for plotting
plt.figure(figsize=(10,5))
# define the left plot which is created by matplotlib
plt.subplot(121)
# matplotlib graph of Sex
gender_count = salary1.groupby('sex').count()['salary']
gender_count.plot.bar();
# give the plot a title
plt.title('Gender Bar Chart (Matplotlib)')
plt.xlabel('Sex')
plt.ylabel('Count')
#Then using seaborn to create graph
# define the right graph
plt.subplot(122)
# Seaborn graph of Sex
sns.countplot(x='sex',data=salary1, order=['Female','Male'])
# give the plot a title
plt.title('Gender Bar Chart (Seaborn)')
plt.xlabel('Sex')
plt.ylabel('Count')
plt.show()
# plot histogram by using Matplotlib and Seaborn
salary_hist = salary1['salary']
#plot matplotlib graph on the left
plt.subplot(121)
#Matlplotlib histogram for salary
plt.hist(tuple(salary_hist))
plt.title('Histogram of salary (Matplotlib)')
#plot seaborn on the right side
plt.subplot(122)
#Seaborn histogram for salary
sns.distplot(tuple(salary_hist))
plt.title('Histogram of salary (Seaborn)')
plt.show()
#create a copy for salary to make sure not messing up data
salary2 = salary.copy()
# Calculate the salary mean
salary_mean = salary2.salary.mean()
salary_mean
# Create a column "special_salary" that contains all of salary values less than the salary mean
special_salary = salary2.salary[salary2.salary<salary_mean]
special_salary.count()
special_salary.describe()
#print boxplot by using matplotlib
salary2.boxplot(by ='sex', column =['salary'], grid = False)
# print boxplot by using seaborns
sns.boxplot(x="sex", y="salary", data=salary2, palette="Set1")
#sns.plt.show()
#create subset of rank, sex,discipline and salary
salary_select = salary2.loc[:,['rank','discipline','sex','salary']]
#filter data to be prof in applied field only
salary_select_prof = salary_select[(salary_select['rank']=='Prof')& (salary_select['discipline']=='B')]
#get most frequent salary
print(salary_select_prof.describe())
#print boxplot by using matplotlib
salary_select_boxplot=salary_select_prof.boxplot(by ='sex', column =['salary'], grid = False)
salary_select_boxplot;
# print boxplot by using seaborns
sns.boxplot(x="sex", y="salary", data=salary_select_prof, palette="Set1")
#sns.plt.show()
#create subset of rank, sex,discipline and salary
salary_select_yrs = salary2.loc[:,['rank','discipline','yrs.service','sex','salary']]
#filter data to be prof in applied field work for more than 18 years
salary_select_yrs = salary_select_yrs[(salary_select_yrs['rank']=='Prof')& (salary_select_yrs['discipline']=='B')&(salary_select_yrs['yrs.service']>=18)]
#get most frequent salary
print(salary_select_prof.describe())
#print boxplot by using matplotlib
salary_select_boxplot=salary_select_yrs.boxplot(by ='sex', column =['salary'], grid = False)
salary_select_boxplot;
# print boxplot by using seaborns as well
sns.boxplot(x="sex", y="salary", data=salary_select_yrs, palette="Set1")
#sns.plt.show()
| 0.438545 | 0.979823 |
<a href="https://colab.research.google.com/github/mittshah2/Music-Genre-Classification/blob/main/music_classif.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import os
import librosa,librosa.display
import matplotlib.pyplot as plt
import numpy as np
import math
import json
import tensorflow as tf
import sklearn
from tqdm import tqdm
import shutil
import warnings
warnings.filterwarnings('ignore')
SAMPLE_RATE=22050
DURATION=30
num_segment=6
hop_length=512
n_fft=2048
n_mfcc=15
sample_per_segment=int((SAMPLE_RATE*DURATION)/num_segment)
expected_mfcc_vector_per_segment=math.ceil(sample_per_segment/hop_length)
audio_path='/content/drive/My Drive/audio classification/Data/genres_original/blues/blues.00000.wav'
signal,sr=librosa.load(audio_path,sr=22050) #length of signal=time_of_audio*sr
signal.shape
librosa.display.waveplot(signal,sr=sr)
plt.xlabel('time')
plt.ylabel('amplitude')
fft=np.fft.fft(signal)
fft.shape
magnitude=np.abs(fft)
freq=np.linspace(0,sr,len(magnitude))
freq=freq[:int(len(freq)/2)]
magnitude=magnitude[:int(len(magnitude)/2)]
plt.xlabel('freq')
plt.ylabel('magnitude')
plt.title('fft plot')
plt.plot(freq,magnitude)
n_fft=2048 #frame size
hop_length=512 #strides while shifting in fft
stft=librosa.core.stft(signal,hop_length=hop_length,n_fft=n_fft)
spectogram=np.abs(stft)
librosa.display.specshow(spectogram,sr=sr,hop_length=hop_length)
plt.xlabel('time')
plt.ylabel('freq')
plt.colorbar()
n_fft=2048 #frame size
hop_length=512 #strides while shifting in fft
stft=librosa.core.stft(signal,hop_length=hop_length,n_fft=n_fft)
spectrogram=np.abs(stft)
spectrogram=librosa.amplitude_to_db(spectrogram)
librosa.display.specshow(spectrogram,sr=sr,hop_length=hop_length)
plt.xlabel('time')
plt.ylabel('freq')
plt.colorbar()
#MFCC
mfcc=librosa.feature.mfcc(signal,n_fft=n_fft,hop_length=hop_length,n_mfcc=n_mfcc)
librosa.display.specshow(mfcc,sr=sr,hop_length=hop_length)
plt.xlabel('time')
plt.ylabel('mfcc')
plt.colorbar()
def save_mfcc(data_path,json_path,n_mfcc=n_mfcc,hop_length=hop_length,n_fft=n_fft,num_segments=num_segment,c=0):
for i,name in enumerate(os.listdir(data_path)):
if name=='jazz':
continue
print('Processing',name)
for f in os.listdir(os.path.join(data_path,name)):
signal,sr=librosa.load(os.path.join(os.path.join(data_path,name),f),sr=SAMPLE_RATE)
for j in range(num_segments):
start=j*sample_per_segment
end=start+sample_per_segment
mfcc=librosa.feature.mfcc(signal[start:end],sr=sr,n_fft=n_fft,hop_length=hop_length,n_mfcc=n_mfcc)
# mfcc=sklearn.preprocessing.scale(mfcc,axis=-1)
# mfcc=librosa.power_to_db(mfcc**2,ref=np.max)
mfcc=mfcc.T
if len(mfcc)==expected_mfcc_vector_per_segment:
data={}
data['labels']=i
data['mfcc']=mfcc.tolist()
with open(os.path.join(json_path,'data_'+str(c)+'.json'),'w') as fp:
json.dump(data,fp,indent=4)
c=c+1
save_mfcc('/content/drive/My Drive/audio classification/Data/genres_original','/content/drive/My Drive/audio classification/json_files')
train_path='/content/drive/My Drive/audio classification/json_files'
val_path='/content/drive/My Drive/audio classification/val'
val_size=int(0.15*len(os.listdir(train_path)))
for i in range(val_size):
rand=np.random.choice(os.listdir(train_path))
shutil.move(os.path.join(train_path,rand),'/content/drive/My Drive/audio classification/val/'+rand)
x_train=[]
y_train=[]
for f in tqdm(os.listdir(train_path)):
with open(os.path.join(train_path,f),'r') as fp:
data=json.load(fp)
img=np.array(data['mfcc'])
a=np.zeros((9,))
a[data['labels']]=1
y_train.append(a)
img=img[...,np.newaxis]
x_train.append(img)
x_train=np.array(x_train)
y_train=np.array(y_train)
x_test=[]
y_test=[]
for f in tqdm(os.listdir(val_path)):
with open(os.path.join(val_path,f),'r') as fp:
data=json.load(fp)
img=np.array(data['mfcc'])
a=np.zeros((9,))
a[data['labels']]=1
y_test.append(a)
img=img[...,np.newaxis]
x_test.append(img)
x_test=np.array(x_test)
y_test=np.array(y_test)
np.save('/content/drive/My Drive/audio classification/x_train.npy',x_train)
np.save('/content/drive/My Drive/audio classification/y_train.npy',y_train)
np.save('/content/drive/My Drive/audio classification/x_test.npy',x_test)
np.save('/content/drive/My Drive/audio classification/y_test.npy',y_test)
x_train=np.load('/content/drive/My Drive/audio classification/x_train.npy')
y_train=np.load('/content/drive/My Drive/audio classification/y_train.npy')
x_test=np.load('/content/drive/My Drive/audio classification/x_test.npy')
y_test=np.load('/content/drive/My Drive/audio classification/y_test.npy')
model=tf.keras.models.Sequential()
model.add(tf.keras.layers.Conv2D(32,3,activation='relu',input_shape=[x_train.shape[1],x_train.shape[2],1]))
model.add(tf.keras.layers.MaxPool2D(3,(2,2),padding='same'))
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.Conv2D(32,3,activation='relu'))
model.add(tf.keras.layers.MaxPool2D(3,(2,2),padding='same'))
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.Conv2D(32,2,activation='relu'))
model.add(tf.keras.layers.MaxPool2D(3,padding='same'))
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.Flatten())
model.add(tf.keras.layers.Dense(64,activation='relu'))
model.add(tf.keras.layers.Dropout(0.5))
model.add(tf.keras.layers.Dense(9,activation='softmax'))
model.compile('adam',loss='categorical_crossentropy',metrics=['accuracy'])
model.summary()
early=tf.keras.callbacks.EarlyStopping(patience=4)
reducelr=tf.keras.callbacks.ReduceLROnPlateau(patience=5)
model.fit(x_train,y_train,batch_size=16,epochs=100,validation_data=(x_test,y_test),validation_batch_size=8,callbacks=[early,reducelr])
import pandas as pd
import seaborn as sns
sns.set_style('whitegrid')
loss=pd.DataFrame(model.history.history)
loss[['accuracy','val_accuracy']].plot()
loss[['loss','val_loss']].plot()
decode={
0:'blues',
1:'classical',
2:'country',
3:'disco',
4:'hiphop',
5:'metal',
6:'pop',
7:'reggae',
8:'rock'
}
plt.figure(figsize=(10,8))
pred=model.predict(x_test)
from sklearn.metrics import roc_curve,auc
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(9):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], pred[:, i])
roc_auc[i] = auc(fpr[i], tpr[i])
for i in range(9):
plt.plot(fpr[i],tpr[i],label=decode[i],linewidth=2)
plt.plot([0, 1], [0, 1], 'k--', lw=2,label='random guess')
plt.legend(loc="lower right")
from sklearn.metrics import classification_report,confusion_matrix
predictions=model.predict_classes(x_test)
check=[]
for i in range(len(y_test)):
for j in range(9):
if(y_test[i][j]==1):
check.append(j)
check=np.asarray(check)
print(classification_report(check,predictions))
print(confusion_matrix(check,predictions))
```
|
github_jupyter
|
import os
import librosa,librosa.display
import matplotlib.pyplot as plt
import numpy as np
import math
import json
import tensorflow as tf
import sklearn
from tqdm import tqdm
import shutil
import warnings
warnings.filterwarnings('ignore')
SAMPLE_RATE=22050
DURATION=30
num_segment=6
hop_length=512
n_fft=2048
n_mfcc=15
sample_per_segment=int((SAMPLE_RATE*DURATION)/num_segment)
expected_mfcc_vector_per_segment=math.ceil(sample_per_segment/hop_length)
audio_path='/content/drive/My Drive/audio classification/Data/genres_original/blues/blues.00000.wav'
signal,sr=librosa.load(audio_path,sr=22050) #length of signal=time_of_audio*sr
signal.shape
librosa.display.waveplot(signal,sr=sr)
plt.xlabel('time')
plt.ylabel('amplitude')
fft=np.fft.fft(signal)
fft.shape
magnitude=np.abs(fft)
freq=np.linspace(0,sr,len(magnitude))
freq=freq[:int(len(freq)/2)]
magnitude=magnitude[:int(len(magnitude)/2)]
plt.xlabel('freq')
plt.ylabel('magnitude')
plt.title('fft plot')
plt.plot(freq,magnitude)
n_fft=2048 #frame size
hop_length=512 #strides while shifting in fft
stft=librosa.core.stft(signal,hop_length=hop_length,n_fft=n_fft)
spectogram=np.abs(stft)
librosa.display.specshow(spectogram,sr=sr,hop_length=hop_length)
plt.xlabel('time')
plt.ylabel('freq')
plt.colorbar()
n_fft=2048 #frame size
hop_length=512 #strides while shifting in fft
stft=librosa.core.stft(signal,hop_length=hop_length,n_fft=n_fft)
spectrogram=np.abs(stft)
spectrogram=librosa.amplitude_to_db(spectrogram)
librosa.display.specshow(spectrogram,sr=sr,hop_length=hop_length)
plt.xlabel('time')
plt.ylabel('freq')
plt.colorbar()
#MFCC
mfcc=librosa.feature.mfcc(signal,n_fft=n_fft,hop_length=hop_length,n_mfcc=n_mfcc)
librosa.display.specshow(mfcc,sr=sr,hop_length=hop_length)
plt.xlabel('time')
plt.ylabel('mfcc')
plt.colorbar()
def save_mfcc(data_path,json_path,n_mfcc=n_mfcc,hop_length=hop_length,n_fft=n_fft,num_segments=num_segment,c=0):
for i,name in enumerate(os.listdir(data_path)):
if name=='jazz':
continue
print('Processing',name)
for f in os.listdir(os.path.join(data_path,name)):
signal,sr=librosa.load(os.path.join(os.path.join(data_path,name),f),sr=SAMPLE_RATE)
for j in range(num_segments):
start=j*sample_per_segment
end=start+sample_per_segment
mfcc=librosa.feature.mfcc(signal[start:end],sr=sr,n_fft=n_fft,hop_length=hop_length,n_mfcc=n_mfcc)
# mfcc=sklearn.preprocessing.scale(mfcc,axis=-1)
# mfcc=librosa.power_to_db(mfcc**2,ref=np.max)
mfcc=mfcc.T
if len(mfcc)==expected_mfcc_vector_per_segment:
data={}
data['labels']=i
data['mfcc']=mfcc.tolist()
with open(os.path.join(json_path,'data_'+str(c)+'.json'),'w') as fp:
json.dump(data,fp,indent=4)
c=c+1
save_mfcc('/content/drive/My Drive/audio classification/Data/genres_original','/content/drive/My Drive/audio classification/json_files')
train_path='/content/drive/My Drive/audio classification/json_files'
val_path='/content/drive/My Drive/audio classification/val'
val_size=int(0.15*len(os.listdir(train_path)))
for i in range(val_size):
rand=np.random.choice(os.listdir(train_path))
shutil.move(os.path.join(train_path,rand),'/content/drive/My Drive/audio classification/val/'+rand)
x_train=[]
y_train=[]
for f in tqdm(os.listdir(train_path)):
with open(os.path.join(train_path,f),'r') as fp:
data=json.load(fp)
img=np.array(data['mfcc'])
a=np.zeros((9,))
a[data['labels']]=1
y_train.append(a)
img=img[...,np.newaxis]
x_train.append(img)
x_train=np.array(x_train)
y_train=np.array(y_train)
x_test=[]
y_test=[]
for f in tqdm(os.listdir(val_path)):
with open(os.path.join(val_path,f),'r') as fp:
data=json.load(fp)
img=np.array(data['mfcc'])
a=np.zeros((9,))
a[data['labels']]=1
y_test.append(a)
img=img[...,np.newaxis]
x_test.append(img)
x_test=np.array(x_test)
y_test=np.array(y_test)
np.save('/content/drive/My Drive/audio classification/x_train.npy',x_train)
np.save('/content/drive/My Drive/audio classification/y_train.npy',y_train)
np.save('/content/drive/My Drive/audio classification/x_test.npy',x_test)
np.save('/content/drive/My Drive/audio classification/y_test.npy',y_test)
x_train=np.load('/content/drive/My Drive/audio classification/x_train.npy')
y_train=np.load('/content/drive/My Drive/audio classification/y_train.npy')
x_test=np.load('/content/drive/My Drive/audio classification/x_test.npy')
y_test=np.load('/content/drive/My Drive/audio classification/y_test.npy')
model=tf.keras.models.Sequential()
model.add(tf.keras.layers.Conv2D(32,3,activation='relu',input_shape=[x_train.shape[1],x_train.shape[2],1]))
model.add(tf.keras.layers.MaxPool2D(3,(2,2),padding='same'))
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.Conv2D(32,3,activation='relu'))
model.add(tf.keras.layers.MaxPool2D(3,(2,2),padding='same'))
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.Conv2D(32,2,activation='relu'))
model.add(tf.keras.layers.MaxPool2D(3,padding='same'))
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.Flatten())
model.add(tf.keras.layers.Dense(64,activation='relu'))
model.add(tf.keras.layers.Dropout(0.5))
model.add(tf.keras.layers.Dense(9,activation='softmax'))
model.compile('adam',loss='categorical_crossentropy',metrics=['accuracy'])
model.summary()
early=tf.keras.callbacks.EarlyStopping(patience=4)
reducelr=tf.keras.callbacks.ReduceLROnPlateau(patience=5)
model.fit(x_train,y_train,batch_size=16,epochs=100,validation_data=(x_test,y_test),validation_batch_size=8,callbacks=[early,reducelr])
import pandas as pd
import seaborn as sns
sns.set_style('whitegrid')
loss=pd.DataFrame(model.history.history)
loss[['accuracy','val_accuracy']].plot()
loss[['loss','val_loss']].plot()
decode={
0:'blues',
1:'classical',
2:'country',
3:'disco',
4:'hiphop',
5:'metal',
6:'pop',
7:'reggae',
8:'rock'
}
plt.figure(figsize=(10,8))
pred=model.predict(x_test)
from sklearn.metrics import roc_curve,auc
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(9):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], pred[:, i])
roc_auc[i] = auc(fpr[i], tpr[i])
for i in range(9):
plt.plot(fpr[i],tpr[i],label=decode[i],linewidth=2)
plt.plot([0, 1], [0, 1], 'k--', lw=2,label='random guess')
plt.legend(loc="lower right")
from sklearn.metrics import classification_report,confusion_matrix
predictions=model.predict_classes(x_test)
check=[]
for i in range(len(y_test)):
for j in range(9):
if(y_test[i][j]==1):
check.append(j)
check=np.asarray(check)
print(classification_report(check,predictions))
print(confusion_matrix(check,predictions))
| 0.302082 | 0.70416 |
```
import pandas as pd
import numpy as np
from pathlib import Path
%matplotlib inline
# read csv files for each playoff teams
bills = pd.read_csv(Path("resources/Buf_Bills2021.csv"), header=1)
rams = pd.read_csv(Path("resources/LA_Rams2021.csv"), header=1)
bengals = pd.read_csv(Path("resources/Cin_Bengals.csv"), header=1)
chiefs = pd.read_csv(Path("resources/KC_Chiefs2021.csv"), header=1)
cowboys = pd.read_csv(Path("resources/Dallas_Cowboys2021.csv"), header=1)
titans = pd.read_csv(Path("resources/Ten_Titans.csv"), header=1)
packers = pd.read_csv(Path("resources/GB_Packers2021.csv"), header=1)
buccaneers = pd.read_csv(Path("resources/Tampa_Buccaneers2021.csv"), header=1)
cardinals = pd.read_csv(Path("resources/Arizona_Cardinals.csv"), header=1)
patriots = pd.read_csv(Path("resources/NE_Patriots.csv"), header=1)
eagles = pd.read_csv(Path("resources/Philadelphia_Eagles.csv"), header=1)
fortyniners = pd.read_csv(Path("resources/SanFrancisco_49ers.csv"), header=1)
steelers = pd.read_csv(Path("resources/Pittsburgh_Steelers.csv"), header=1)
raiders = pd.read_csv(Path("resources/LasVegas_Raiders.csv"), header=1)
fortyniners.head()
bills = bills[0:16].drop(columns=['Date'])
rams = rams[0:16].drop(columns=['Date'])
bengals = bengals[0:16].drop(columns=['Date'])
chiefs = chiefs[0:16].drop(columns=['Date'])
cowboys = cowboys[0:16].drop(columns=['Date'])
titans = titans[0:16].drop(columns=['Date'])
packers = packers[0:16].drop(columns=['Date'])
buccaneers = buccaneers[0:16].drop(columns=['Date'])
cardinals = cardinals[0:16].drop(columns=['Date'])
patriots = patriots[0:16].drop(columns=['Date'])
eagles = eagles[0:16].drop(columns=['Date'])
fortyniners = fortyniners[0:16].drop(columns=['Date'])
steelers = steelers[0:16].drop(columns=['Date'])
raiders = raiders[0:16].drop(columns=['Date'])
bills.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
rams.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
bengals.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
chiefs.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
cowboys.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
titans.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
packers.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
buccaneers.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
cardinals.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
patriots.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
eagles.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
fortyniners.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
steelers.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
raiders.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
steelers
bills.insert(0,"Team",'Buffalo Bills')
rams.insert(0,"Team",'Los Angeles Rams')
bengals.insert(0,"Team",'Cincinnati Bengals')
chiefs.insert(0,"Team",'Kansas City Chiefs')
cowboys.insert(0,"Team",'Dallas Cowboys')
titans.insert(0,"Team",'Tennessee Titans')
packers.insert(0,"Team",'Green Bay Packers')
buccaneers.insert(0,"Team",'Tampa Bay Buccaneers')
cardinals.insert(0,"Team",'Arizona Cardinals')
patriots.insert(0,"Team",'New England Patriots')
eagles.insert(0,"Team",'Philadelphia Eagles')
fortyniners.insert(0,"Team",'San Francisco 49ers')
steelers.insert(0,"Team",'Pittsburgh Steelers')
raiders.insert(0,"Team",'Las Vegas Raiders')
rams
Team_df = [bills, rams, bengals, chiefs, cowboys, titans, packers, buccaneers, cardinals, patriots, eagles, fortyniners
, steelers, raiders]
total_team_df = pd.concat(Team_df)
total_team_df
total_team_df = total_team_df.fillna(1)
total_team_df = total_team_df.replace('@', 0)
total_team_df.head()
total_team_df = total_team_df.replace('W', 1)
total_team_df = total_team_df.replace('L', 0)
total_team_df.head()
LR = pd.read_csv(Path("resources/Prediction_LR.csv"), header=0)
ADA = pd.read_csv(Path("resources/Prediction_ada.csv"), header=0)
LR.head()
LR_prediction = LR[['Team','Prediction']]
ADA_prediction = ADA[['Team', 'Prediction']]
LR_prediction.columns = ['Team','Prediction_LR']
ADA_prediction.columns = ['Team','Prediction_ADA']
LR_prediction = LR_prediction.drop([7,8,12,13,14,16,17,19,20,21,22,25,26,28,29,30,31,6], axis=0)
ADA_prediction = ADA_prediction.drop([4,12,13,15,16,17,18,19,21,22,23,24,25,27,28,29,30,31], axis=0)
ADA_prediction
total_team_pred_df = pd.merge(total_team_df,
LR_prediction,
# ADA_prediction,
left_on = 'Team', right_on ='Team', how = 'left')
total_team_pred_df = pd.merge(total_team_pred_df, ADA_prediction, left_on= 'Team', right_on= 'Team', how='left')
total_team_pred_df.head(20)
total_team_pred_df.to_csv('./resources/Team_df.csv', index=False)
bills
rams
bengals
chiefs
cowboys
titans
packers
buccaneers
cardinals
patriots
eagles
fortyniners
steelers
raiders
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from pathlib import Path
%matplotlib inline
# read csv files for each playoff teams
bills = pd.read_csv(Path("resources/Buf_Bills2021.csv"), header=1)
rams = pd.read_csv(Path("resources/LA_Rams2021.csv"), header=1)
bengals = pd.read_csv(Path("resources/Cin_Bengals.csv"), header=1)
chiefs = pd.read_csv(Path("resources/KC_Chiefs2021.csv"), header=1)
cowboys = pd.read_csv(Path("resources/Dallas_Cowboys2021.csv"), header=1)
titans = pd.read_csv(Path("resources/Ten_Titans.csv"), header=1)
packers = pd.read_csv(Path("resources/GB_Packers2021.csv"), header=1)
buccaneers = pd.read_csv(Path("resources/Tampa_Buccaneers2021.csv"), header=1)
cardinals = pd.read_csv(Path("resources/Arizona_Cardinals.csv"), header=1)
patriots = pd.read_csv(Path("resources/NE_Patriots.csv"), header=1)
eagles = pd.read_csv(Path("resources/Philadelphia_Eagles.csv"), header=1)
fortyniners = pd.read_csv(Path("resources/SanFrancisco_49ers.csv"), header=1)
steelers = pd.read_csv(Path("resources/Pittsburgh_Steelers.csv"), header=1)
raiders = pd.read_csv(Path("resources/LasVegas_Raiders.csv"), header=1)
fortyniners.head()
bills = bills[0:16].drop(columns=['Date'])
rams = rams[0:16].drop(columns=['Date'])
bengals = bengals[0:16].drop(columns=['Date'])
chiefs = chiefs[0:16].drop(columns=['Date'])
cowboys = cowboys[0:16].drop(columns=['Date'])
titans = titans[0:16].drop(columns=['Date'])
packers = packers[0:16].drop(columns=['Date'])
buccaneers = buccaneers[0:16].drop(columns=['Date'])
cardinals = cardinals[0:16].drop(columns=['Date'])
patriots = patriots[0:16].drop(columns=['Date'])
eagles = eagles[0:16].drop(columns=['Date'])
fortyniners = fortyniners[0:16].drop(columns=['Date'])
steelers = steelers[0:16].drop(columns=['Date'])
raiders = raiders[0:16].drop(columns=['Date'])
bills.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
rams.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
bengals.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
chiefs.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
cowboys.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
titans.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
packers.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
buccaneers.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
cardinals.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
patriots.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
eagles.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
fortyniners.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
steelers.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
raiders.columns = ['Week','Result','Rec','Home','Opp','TmScore','OppScore',
'O_1stD','O_Tot_yd','O_P_Yd','O_R_Yd','O_TO',
'D_1stD','D_Tot_Yd','D_P_Yd','D_R_Yd','D_TO','Offence','Defense','Sp.Tms']
steelers
bills.insert(0,"Team",'Buffalo Bills')
rams.insert(0,"Team",'Los Angeles Rams')
bengals.insert(0,"Team",'Cincinnati Bengals')
chiefs.insert(0,"Team",'Kansas City Chiefs')
cowboys.insert(0,"Team",'Dallas Cowboys')
titans.insert(0,"Team",'Tennessee Titans')
packers.insert(0,"Team",'Green Bay Packers')
buccaneers.insert(0,"Team",'Tampa Bay Buccaneers')
cardinals.insert(0,"Team",'Arizona Cardinals')
patriots.insert(0,"Team",'New England Patriots')
eagles.insert(0,"Team",'Philadelphia Eagles')
fortyniners.insert(0,"Team",'San Francisco 49ers')
steelers.insert(0,"Team",'Pittsburgh Steelers')
raiders.insert(0,"Team",'Las Vegas Raiders')
rams
Team_df = [bills, rams, bengals, chiefs, cowboys, titans, packers, buccaneers, cardinals, patriots, eagles, fortyniners
, steelers, raiders]
total_team_df = pd.concat(Team_df)
total_team_df
total_team_df = total_team_df.fillna(1)
total_team_df = total_team_df.replace('@', 0)
total_team_df.head()
total_team_df = total_team_df.replace('W', 1)
total_team_df = total_team_df.replace('L', 0)
total_team_df.head()
LR = pd.read_csv(Path("resources/Prediction_LR.csv"), header=0)
ADA = pd.read_csv(Path("resources/Prediction_ada.csv"), header=0)
LR.head()
LR_prediction = LR[['Team','Prediction']]
ADA_prediction = ADA[['Team', 'Prediction']]
LR_prediction.columns = ['Team','Prediction_LR']
ADA_prediction.columns = ['Team','Prediction_ADA']
LR_prediction = LR_prediction.drop([7,8,12,13,14,16,17,19,20,21,22,25,26,28,29,30,31,6], axis=0)
ADA_prediction = ADA_prediction.drop([4,12,13,15,16,17,18,19,21,22,23,24,25,27,28,29,30,31], axis=0)
ADA_prediction
total_team_pred_df = pd.merge(total_team_df,
LR_prediction,
# ADA_prediction,
left_on = 'Team', right_on ='Team', how = 'left')
total_team_pred_df = pd.merge(total_team_pred_df, ADA_prediction, left_on= 'Team', right_on= 'Team', how='left')
total_team_pred_df.head(20)
total_team_pred_df.to_csv('./resources/Team_df.csv', index=False)
bills
rams
bengals
chiefs
cowboys
titans
packers
buccaneers
cardinals
patriots
eagles
fortyniners
steelers
raiders
| 0.12661 | 0.196267 |
```
%matplotlib inline
morange = u'#ff7f0e'
mblue = u'#1f77b4'
mgreen = u'#2ca02c'
mred = u'#d62728'
mpurple = u'#9467bd'
from cosmodc2.sdss_colors import load_umachine_processed_sdss_catalog
sdss = load_umachine_processed_sdss_catalog()
print(sdss.keys())
import os
from astropy.table import Table
# MDPL2-based mock
dirname = "/Users/aphearin/work/random/ARCHIVES/2018/March2018/0331"
basename = "cutmock_1e9.hdf5"
fname = os.path.join(dirname, basename)
mock = Table.read(fname, path='data')
mock.Lbox = 500.
```
## Assign black hole mass
```
from cosmodc2.black_hole_modeling import monte_carlo_black_hole_mass
mock['bt'] = np.random.rand(len(mock))
mock['bulge_mass'] = mock['obs_sm']*mock['bt']
mock['bh_mass'] = monte_carlo_black_hole_mass(mock['bulge_mass'])
from cosmodc2.black_hole_modeling import monte_carlo_bh_acc_rate
eddington_ratio_z0, accretion_rate_z0 = monte_carlo_bh_acc_rate(
0., mock['bh_mass'], mock['sfr_percentile'])
eddington_ratio_z1, accretion_rate_z1 = monte_carlo_bh_acc_rate(
1., mock['bh_mass'], mock['sfr_percentile'])
eddington_ratio_z2, accretion_rate_z2 = monte_carlo_bh_acc_rate(
2., mock['bh_mass'], mock['sfr_percentile'])
fig, ax = plt.subplots(1, 1)
__=ax.hist(np.log10(eddington_ratio_z0), bins=100, normed=True,
alpha=0.8, color=mblue, label=r'${\rm z=0}$')
__=ax.hist(np.log10(eddington_ratio_z1), bins=100, normed=True,
alpha=0.8, color=mgreen, label=r'${\rm z=1}$')
__=ax.hist(np.log10(eddington_ratio_z2), bins=100, normed=True,
alpha=0.8, color=mred, label=r'${\rm z=2}$')
legend = ax.legend()
xlabel = ax.set_xlabel(r'${\rm \log \lambda_{Edd}}$')
xlim = ax.set_xlim(-4.5, 0)
figname = 'black_hole_eddington_ratios_redshift_evolution.pdf'
fig.savefig(figname, bbox_extra_artists=[xlabel], bbox_inches='tight')
from cosmodc2.black_hole_modeling import monte_carlo_bh_acc_rate
redshift = 0.
eddington_ratio, accretion_rate = monte_carlo_bh_acc_rate(
redshift, mock['bh_mass'], mock['sfr_percentile'])
mock['eddington_ratio'] = eddington_ratio
mock['bh_acc_rate'] = accretion_rate
```
### Plot redshift-dependence
```
sm_mask = mock['obs_sm'] > 10**10
sm_mask *= mock['obs_sm'] < 10**10.5
eddington_ratio_z0, accretion_rate_z0 = monte_carlo_bh_acc_rate(
0., mock['bh_mass'], mock['sfr_percentile'])
eddington_ratio_z1, accretion_rate_z1 = monte_carlo_bh_acc_rate(
1., mock['bh_mass'], mock['sfr_percentile'])
eddington_ratio_z2, accretion_rate_z2 = monte_carlo_bh_acc_rate(
2., mock['bh_mass'], mock['sfr_percentile'])
fig, ax = plt.subplots(1, 1)
# __=ax.hist(np.log10(accretion_rate_z0[sm_mask]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm z=0}$', color=mblue)
# __=ax.hist(np.log10(accretion_rate_z1[sm_mask]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm z=1}$', color=mgreen)
# __=ax.hist(np.log10(accretion_rate_z2[sm_mask]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm z=2}$', color=mred)
from scipy.stats import gaussian_kde
kde_z0 = gaussian_kde(np.log10(accretion_rate_z0[sm_mask]))
kde_z1 = gaussian_kde(np.log10(accretion_rate_z1[sm_mask]))
kde_z2 = gaussian_kde(np.log10(accretion_rate_z2[sm_mask]))
x = np.linspace(-7, 0, 150)
pdf_z0 = kde_z0.evaluate(x)
pdf_z1 = kde_z1.evaluate(x)
pdf_z2 = kde_z2.evaluate(x)
__=ax.fill(x, pdf_z0, alpha=0.8, label=r'${\rm z=0}$', color=mblue)
__=ax.fill(x, pdf_z1, alpha=0.8, label=r'${\rm z=1}$', color=mgreen)
__=ax.fill(x, pdf_z2, alpha=0.8, label=r'${\rm z=2}$', color=mred)
# title = ax.set_title(r'${\rm M_{\ast}\approx10^{10.5}M_{\odot}$}')
title = ax.set_title(r'${\rm DC2\ black\ hole\ model}$')
xlabel = ax.set_xlabel(r'${\rm dM_{BH}/dt\ [M_{\odot}/yr]}$')
legend = ax.legend()
xlim = ax.set_xlim(-6, 0)
ylim = ax.set_ylim(ymin=0)
__=ax.set_xticks((-6, -4, -2, 0))
xticklabels = (r'$10^{-6}$', r'$10^{-4}$', r'$10^{-2}$', r'$10^{0}$')
__=ax.set_xticklabels(xticklabels)
figname = 'black_hole_accretion_rates_v4_redshift_evolution.pdf'
fig.savefig(figname, bbox_extra_artists=[xlabel], bbox_inches='tight')
```
### Plot color-dependence
```
sm_mask = mock['obs_sm'] > 10**10
sm_mask *= mock['obs_sm'] < 10**10.5
median_sfr = np.median(mock['obs_sfr'][sm_mask])
sfr_mask1 = mock['obs_sfr'] < median_sfr
mask1 = sm_mask & sfr_mask1
sfr_mask2 = mock['obs_sfr'] > median_sfr
mask2 = sm_mask & sfr_mask2
eddington_ratio_z0, accretion_rate_z0 = monte_carlo_bh_acc_rate(
0., mock['bh_mass'], mock['sfr_percentile'])
fig, ax = plt.subplots(1, 1)
# __=ax.hist(np.log10(accretion_rate_z0[mask1]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm z=0\ red\ galaxies}$', color=mred)
# __=ax.hist(np.log10(accretion_rate_z0[mask2]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm z=0\ blue\ galaxies}$', color=mblue)
from scipy.stats import gaussian_kde
kde_red = gaussian_kde(np.log10(accretion_rate_z0[mask1]))
kde_blue = gaussian_kde(np.log10(accretion_rate_z0[mask2]))
x = np.linspace(-7, 0, 150)
pdf_red = kde_red.evaluate(x)
pdf_blue = kde_blue.evaluate(x)
__=ax.fill(x, pdf_red, alpha=0.8, label=r'${\rm z=0\ red\ galaxies}$', color=mred)
__=ax.fill(x, pdf_blue, alpha=0.8, label=r'${\rm z=0\ blue\ galaxies}$', color=mblue)
ylim = ax.set_ylim(0, 0.8)
title = ax.set_title(r'${\rm M_{\ast}\approx10^{10.5}M_{\odot}$}')
xlabel = ax.set_xlabel(r'${\rm dM_{BH}/dt\ [M_{\odot}/yr]}$')
legend = ax.legend(loc='upper left')
xlim = ax.set_xlim(-7, -2)
__=ax.set_xticks((-6, -4, -2))
xticklabels = (r'$10^{-6}$', r'$10^{-4}$', r'$10^{-2}$')
__=ax.set_xticklabels(xticklabels)
figname = 'black_hole_accretion_rates_v4_sfr_dependence.pdf'
fig.savefig(figname, bbox_extra_artists=[xlabel], bbox_inches='tight')
```
### Plot mass-dependence
```
mask1 = (mock['obs_sm'] > 10**9.75) & (mock['obs_sm'] < 10**10)
mask2 = (mock['obs_sm'] > 10**10.5) & (mock['obs_sm'] < 10**10.75)
mask3 = (mock['obs_sm'] > 10**11.25) #& (mock['obs_sm'] < 10**10)
eddington_ratio_z0, accretion_rate_z0 = monte_carlo_bh_acc_rate(
0., mock['bh_mass'], mock['sfr_percentile'])
fig, ax = plt.subplots(1, 1)
# __=ax.hist(np.log10(accretion_rate_z0[mask1]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm M_{\ast}\approx10^{9.75}M_{\odot}}$')
# __=ax.hist(np.log10(accretion_rate_z0[mask2]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm M_{\ast}\approx10^{10.5}M_{\odot}}$')
# __=ax.hist(np.log10(accretion_rate_z0[mask3]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm M_{\ast}\approx10^{11.25}M_{\odot}}$')
from scipy.stats import gaussian_kde
kde_red = gaussian_kde(np.log10(accretion_rate_z0[mask1]))
kde_blue = gaussian_kde(np.log10(accretion_rate_z0[mask2]))
kde_orange = gaussian_kde(np.log10(accretion_rate_z0[mask3]))
x = np.linspace(-7, 0, 150)
pdf_red = kde_red.evaluate(x)
pdf_blue = kde_blue.evaluate(x)
pdf_orange = kde_orange.evaluate(x)
__=ax.fill(x, pdf_red, alpha=0.8, label=r'${\rm M_{\ast}\approx10^{9.75}M_{\odot}}$')
__=ax.fill(x, pdf_blue, alpha=0.8, label=r'${\rm M_{\ast}\approx10^{10.5}M_{\odot}}$')
__=ax.fill(x, pdf_orange, alpha=0.8, label=r'${\rm M_{\ast}\approx10^{11.25}M_{\odot}}$')
title = ax.set_title(r'${\rm z=0}$')
xlabel = ax.set_xlabel(r'${\rm dM_{BH}/dt\ [M_{\odot}/yr]}$')
legend = ax.legend()
ylim = ax.set_ylim(0, 1.25)
xlim = ax.set_xlim(-7, 0)
__=ax.set_xticks((-6, -4, -2, 0))
xticklabels = (r'$10^{-6}$', r'$10^{-4}$', r'$10^{-2}$', r'$10^{0}$')
__=ax.set_xticklabels(xticklabels)
figname = 'black_hole_accretion_rates_v4_mstar_dependence.pdf'
fig.savefig(figname, bbox_extra_artists=[xlabel], bbox_inches='tight')
```
|
github_jupyter
|
%matplotlib inline
morange = u'#ff7f0e'
mblue = u'#1f77b4'
mgreen = u'#2ca02c'
mred = u'#d62728'
mpurple = u'#9467bd'
from cosmodc2.sdss_colors import load_umachine_processed_sdss_catalog
sdss = load_umachine_processed_sdss_catalog()
print(sdss.keys())
import os
from astropy.table import Table
# MDPL2-based mock
dirname = "/Users/aphearin/work/random/ARCHIVES/2018/March2018/0331"
basename = "cutmock_1e9.hdf5"
fname = os.path.join(dirname, basename)
mock = Table.read(fname, path='data')
mock.Lbox = 500.
from cosmodc2.black_hole_modeling import monte_carlo_black_hole_mass
mock['bt'] = np.random.rand(len(mock))
mock['bulge_mass'] = mock['obs_sm']*mock['bt']
mock['bh_mass'] = monte_carlo_black_hole_mass(mock['bulge_mass'])
from cosmodc2.black_hole_modeling import monte_carlo_bh_acc_rate
eddington_ratio_z0, accretion_rate_z0 = monte_carlo_bh_acc_rate(
0., mock['bh_mass'], mock['sfr_percentile'])
eddington_ratio_z1, accretion_rate_z1 = monte_carlo_bh_acc_rate(
1., mock['bh_mass'], mock['sfr_percentile'])
eddington_ratio_z2, accretion_rate_z2 = monte_carlo_bh_acc_rate(
2., mock['bh_mass'], mock['sfr_percentile'])
fig, ax = plt.subplots(1, 1)
__=ax.hist(np.log10(eddington_ratio_z0), bins=100, normed=True,
alpha=0.8, color=mblue, label=r'${\rm z=0}$')
__=ax.hist(np.log10(eddington_ratio_z1), bins=100, normed=True,
alpha=0.8, color=mgreen, label=r'${\rm z=1}$')
__=ax.hist(np.log10(eddington_ratio_z2), bins=100, normed=True,
alpha=0.8, color=mred, label=r'${\rm z=2}$')
legend = ax.legend()
xlabel = ax.set_xlabel(r'${\rm \log \lambda_{Edd}}$')
xlim = ax.set_xlim(-4.5, 0)
figname = 'black_hole_eddington_ratios_redshift_evolution.pdf'
fig.savefig(figname, bbox_extra_artists=[xlabel], bbox_inches='tight')
from cosmodc2.black_hole_modeling import monte_carlo_bh_acc_rate
redshift = 0.
eddington_ratio, accretion_rate = monte_carlo_bh_acc_rate(
redshift, mock['bh_mass'], mock['sfr_percentile'])
mock['eddington_ratio'] = eddington_ratio
mock['bh_acc_rate'] = accretion_rate
sm_mask = mock['obs_sm'] > 10**10
sm_mask *= mock['obs_sm'] < 10**10.5
eddington_ratio_z0, accretion_rate_z0 = monte_carlo_bh_acc_rate(
0., mock['bh_mass'], mock['sfr_percentile'])
eddington_ratio_z1, accretion_rate_z1 = monte_carlo_bh_acc_rate(
1., mock['bh_mass'], mock['sfr_percentile'])
eddington_ratio_z2, accretion_rate_z2 = monte_carlo_bh_acc_rate(
2., mock['bh_mass'], mock['sfr_percentile'])
fig, ax = plt.subplots(1, 1)
# __=ax.hist(np.log10(accretion_rate_z0[sm_mask]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm z=0}$', color=mblue)
# __=ax.hist(np.log10(accretion_rate_z1[sm_mask]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm z=1}$', color=mgreen)
# __=ax.hist(np.log10(accretion_rate_z2[sm_mask]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm z=2}$', color=mred)
from scipy.stats import gaussian_kde
kde_z0 = gaussian_kde(np.log10(accretion_rate_z0[sm_mask]))
kde_z1 = gaussian_kde(np.log10(accretion_rate_z1[sm_mask]))
kde_z2 = gaussian_kde(np.log10(accretion_rate_z2[sm_mask]))
x = np.linspace(-7, 0, 150)
pdf_z0 = kde_z0.evaluate(x)
pdf_z1 = kde_z1.evaluate(x)
pdf_z2 = kde_z2.evaluate(x)
__=ax.fill(x, pdf_z0, alpha=0.8, label=r'${\rm z=0}$', color=mblue)
__=ax.fill(x, pdf_z1, alpha=0.8, label=r'${\rm z=1}$', color=mgreen)
__=ax.fill(x, pdf_z2, alpha=0.8, label=r'${\rm z=2}$', color=mred)
# title = ax.set_title(r'${\rm M_{\ast}\approx10^{10.5}M_{\odot}$}')
title = ax.set_title(r'${\rm DC2\ black\ hole\ model}$')
xlabel = ax.set_xlabel(r'${\rm dM_{BH}/dt\ [M_{\odot}/yr]}$')
legend = ax.legend()
xlim = ax.set_xlim(-6, 0)
ylim = ax.set_ylim(ymin=0)
__=ax.set_xticks((-6, -4, -2, 0))
xticklabels = (r'$10^{-6}$', r'$10^{-4}$', r'$10^{-2}$', r'$10^{0}$')
__=ax.set_xticklabels(xticklabels)
figname = 'black_hole_accretion_rates_v4_redshift_evolution.pdf'
fig.savefig(figname, bbox_extra_artists=[xlabel], bbox_inches='tight')
sm_mask = mock['obs_sm'] > 10**10
sm_mask *= mock['obs_sm'] < 10**10.5
median_sfr = np.median(mock['obs_sfr'][sm_mask])
sfr_mask1 = mock['obs_sfr'] < median_sfr
mask1 = sm_mask & sfr_mask1
sfr_mask2 = mock['obs_sfr'] > median_sfr
mask2 = sm_mask & sfr_mask2
eddington_ratio_z0, accretion_rate_z0 = monte_carlo_bh_acc_rate(
0., mock['bh_mass'], mock['sfr_percentile'])
fig, ax = plt.subplots(1, 1)
# __=ax.hist(np.log10(accretion_rate_z0[mask1]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm z=0\ red\ galaxies}$', color=mred)
# __=ax.hist(np.log10(accretion_rate_z0[mask2]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm z=0\ blue\ galaxies}$', color=mblue)
from scipy.stats import gaussian_kde
kde_red = gaussian_kde(np.log10(accretion_rate_z0[mask1]))
kde_blue = gaussian_kde(np.log10(accretion_rate_z0[mask2]))
x = np.linspace(-7, 0, 150)
pdf_red = kde_red.evaluate(x)
pdf_blue = kde_blue.evaluate(x)
__=ax.fill(x, pdf_red, alpha=0.8, label=r'${\rm z=0\ red\ galaxies}$', color=mred)
__=ax.fill(x, pdf_blue, alpha=0.8, label=r'${\rm z=0\ blue\ galaxies}$', color=mblue)
ylim = ax.set_ylim(0, 0.8)
title = ax.set_title(r'${\rm M_{\ast}\approx10^{10.5}M_{\odot}$}')
xlabel = ax.set_xlabel(r'${\rm dM_{BH}/dt\ [M_{\odot}/yr]}$')
legend = ax.legend(loc='upper left')
xlim = ax.set_xlim(-7, -2)
__=ax.set_xticks((-6, -4, -2))
xticklabels = (r'$10^{-6}$', r'$10^{-4}$', r'$10^{-2}$')
__=ax.set_xticklabels(xticklabels)
figname = 'black_hole_accretion_rates_v4_sfr_dependence.pdf'
fig.savefig(figname, bbox_extra_artists=[xlabel], bbox_inches='tight')
mask1 = (mock['obs_sm'] > 10**9.75) & (mock['obs_sm'] < 10**10)
mask2 = (mock['obs_sm'] > 10**10.5) & (mock['obs_sm'] < 10**10.75)
mask3 = (mock['obs_sm'] > 10**11.25) #& (mock['obs_sm'] < 10**10)
eddington_ratio_z0, accretion_rate_z0 = monte_carlo_bh_acc_rate(
0., mock['bh_mass'], mock['sfr_percentile'])
fig, ax = plt.subplots(1, 1)
# __=ax.hist(np.log10(accretion_rate_z0[mask1]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm M_{\ast}\approx10^{9.75}M_{\odot}}$')
# __=ax.hist(np.log10(accretion_rate_z0[mask2]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm M_{\ast}\approx10^{10.5}M_{\odot}}$')
# __=ax.hist(np.log10(accretion_rate_z0[mask3]), bins=100, normed=True,
# alpha=0.8, label=r'${\rm M_{\ast}\approx10^{11.25}M_{\odot}}$')
from scipy.stats import gaussian_kde
kde_red = gaussian_kde(np.log10(accretion_rate_z0[mask1]))
kde_blue = gaussian_kde(np.log10(accretion_rate_z0[mask2]))
kde_orange = gaussian_kde(np.log10(accretion_rate_z0[mask3]))
x = np.linspace(-7, 0, 150)
pdf_red = kde_red.evaluate(x)
pdf_blue = kde_blue.evaluate(x)
pdf_orange = kde_orange.evaluate(x)
__=ax.fill(x, pdf_red, alpha=0.8, label=r'${\rm M_{\ast}\approx10^{9.75}M_{\odot}}$')
__=ax.fill(x, pdf_blue, alpha=0.8, label=r'${\rm M_{\ast}\approx10^{10.5}M_{\odot}}$')
__=ax.fill(x, pdf_orange, alpha=0.8, label=r'${\rm M_{\ast}\approx10^{11.25}M_{\odot}}$')
title = ax.set_title(r'${\rm z=0}$')
xlabel = ax.set_xlabel(r'${\rm dM_{BH}/dt\ [M_{\odot}/yr]}$')
legend = ax.legend()
ylim = ax.set_ylim(0, 1.25)
xlim = ax.set_xlim(-7, 0)
__=ax.set_xticks((-6, -4, -2, 0))
xticklabels = (r'$10^{-6}$', r'$10^{-4}$', r'$10^{-2}$', r'$10^{0}$')
__=ax.set_xticklabels(xticklabels)
figname = 'black_hole_accretion_rates_v4_mstar_dependence.pdf'
fig.savefig(figname, bbox_extra_artists=[xlabel], bbox_inches='tight')
| 0.448185 | 0.606877 |
```
# define logging and working directory
from ProjectRoot import change_wd_to_project_root
change_wd_to_project_root()
import SimpleITK as sitk
import ipywidgets as widgets
import matplotlib.pyplot as plt
import pandas as pd
import os
import glob
import shutil
import numpy as np
from scipy.spatial.transform import Rotation as R
from src.data.Preprocess import *
from src.utils.Notebook_imports import *
```
# Resample to isotrop
This method creates strange artefacts, so far I dont know how to solve
```
ax_src = '/mnt/ssd/data/gcn/ax_sax_from_flo/ax3d/'
sax_src = '/mnt/ssd/data/gcn/ax_sax_from_flo/sax3d/'
ax_iso_dst = '/mnt/ssd/data/gcn/ax_sax_from_flo/ax3d_iso_linear/'
sax_iso_dst = '/mnt/ssd/data/gcn/ax_sax_from_flo/sax3d_iso_linear/'
# resample 3D image / mask into a fake isotrop resolution, use multithreadding, combine iso and orientation resample, everything in one step
import concurrent.futures
from concurrent.futures import as_completed
slice_first = 2
workers = 16
futures_ax_iso = set()
futures_sax_iso = set()
futures_ax2sax_iso = set()
futures_sax2ax_iso = set()
ax_files = sorted(glob.glob(ax_src+'*.nrrd', recursive = True))
sax_files = sorted(glob.glob(sax_src+'*.nrrd', recursive = True))
ensure_dir(ax_iso_dst)
ensure_dir(sax_iso_dst)
target_spacing = (1.5,1.5,1.5)
# spawn multiple threads or processes
with concurrent.futures.ThreadPoolExecutor(max_workers=workers) as executor:# resample ax towards sax and vize versa
for ax_file, sax_file in zip(ax_files, sax_files):
print('processing file: {}'.format(ax_file))
reader1 = sitk.ImageFileReader()
reader1.SetFileName(ax_file)
ax_img = reader1.Execute()
reader2 = sitk.ImageFileReader()
reader2.SetFileName(sax_file)
sax_img = reader2.Execute()
# make a pseudo isotrop volume from both input images
if 'img' in os.path.basename(ax_file):
futures_ax_iso.add(executor.submit(transform_to_isotrop_voxels, ax_img, sitk.sitkLinear,target_spacing, ax_file))
futures_sax_iso.add(executor.submit(transform_to_isotrop_voxels, sax_img, sitk.sitkLinear,target_spacing, sax_file))
else:
futures_ax_iso.add(executor.submit(max_thres_resample2_iso_label_img, ax_img,50, target_spacing, ax_file, sitk.sitkLinear))
futures_sax_iso.add(executor.submit(max_thres_resample2_iso_label_img, sax_img,50, target_spacing, sax_file, sitk.sitkLinear))
# this part is sequential, we need only one writer
writer = sitk.ImageFileWriter()
for future in as_completed(futures_ax_iso):
try:
res = future.result()
iso_resampled, file_path = res
writer.SetFileName(os.path.join(ax_iso_dst,os.path.basename(file_path)))
writer.Execute(iso_resampled)
except Exception as e:
logging.error(
'Exception {} in datagenerator with: image: {} or mask: {}'.format(str(e)))
for future in as_completed(futures_sax_iso):
try:
res = future.result()
iso_resampled, file_path = res
writer.SetFileName(os.path.join(sax_iso_dst,os.path.basename(file_path)))
writer.Execute(iso_resampled)
except Exception as e:
logging.error(
'Exception {} in datagenerator with: image: {} or mask: {}'.format(str(e)))
ax_src = '/mnt/ssd/data/gcn/ax_sax_from_flo/ax3d_iso_linear/'
sax_src = '/mnt/ssd/data/gcn/ax_sax_from_flo/sax3d_iso_linear/'
dst_ax2sax = '/mnt/ssd/data/gcn/ax_sax_from_flo/sax2ax3d_iso/'
ax2sax_iso_dst = '/mnt/ssd/data/gcn/ax_sax_from_flo/ax2sax3d_iso_linear/'
sax2ax_iso_dst = '/mnt/ssd/data/gcn/ax_sax_from_flo/sax2ax3d_iso_linear/'
```
# Resample to other orientation
```
# resample 3D image / mask from axial view into sax view or vize versa, extend the transformed vol by 10 voxel along the target z axis
import concurrent.futures
from concurrent.futures import as_completed
workers = 16
futures_ax2sax = set()
futures_sax2ax = set()
ensure_dir(ax2sax_iso_dst)
ensure_dir(sax2ax_iso_dst)
# spawn multiple threads or processes
# transform the image of file_path with according to the direction of
ax_files = sorted(glob.glob(ax_src+'*.nrrd', recursive = True))
sax_files = sorted(glob.glob(sax_src+'*.nrrd', recursive = True))
print('axial files: {}'.format(len(ax_files)))
print('sax files: {}'.format(len(sax_files)))
with concurrent.futures.ThreadPoolExecutor(max_workers=workers) as executor:
for ax_file_path, sax_file_path in zip(ax_files, sax_files):
reader1 = sitk.ImageFileReader()
reader1.SetFileName(ax_file_path)
ax_img = reader1.Execute()
reader2 = sitk.ImageFileReader()
reader2.SetFileName(sax_file_path)
sax_img = reader2.Execute()
if 'img' in os.path.basename(ax_file_path):
futures_ax2sax.add(executor.submit(resample_direcion_origin_spacing_shift, ax_img, sax_img, (0,0,-10), sitk.sitkLinear, ax_file_path))
futures_sax2ax.add(executor.submit(resample_direcion_origin_spacing_shift, sax_img, ax_img, (0,0,-10), sitk.sitkLinear, sax_file_path))
else:
futures_ax2sax.add(executor.submit(max_thres_resample2_label_img_shift, ax_img, sax_img, 50, (0,0,-10), ax_file_path))
futures_sax2ax.add(executor.submit(max_thres_resample2_label_img_shift, sax_img, ax_img, 50, (0,0,-10), sax_file_path))
writer = sitk.ImageFileWriter()
for i, future in enumerate(as_completed(futures_ax2sax)):
try:
res = future.result()
resampled, file_path = res
writer.SetFileName(os.path.join(ax2sax_iso_dst,os.path.basename(file_path)))
writer.Execute(resampled)
except Exception as e:
logging.error(
'Exception {} in datagenerator with: image: {} or mask: {}'.format(str(e)))
for i, future in enumerate(as_completed(futures_sax2ax)):
try:
res = future.result()
resampled, file_path = res
writer.SetFileName(os.path.join(sax2ax_iso_dst,os.path.basename(file_path)))
writer.Execute(resampled)
except Exception as e:
logging.error(
'Exception {} in datagenerator with: image: {} or mask: {}'.format(str(e)))
```
# Manual tests, not needed currently
```
ax_src = '/home/rflo/master/all_with_7_worst_regi/ax3d/'
sax_src = '/home/rflo/master/all_with_7_worst_regi/sax3d/'
dst = '/home/rflo/master/all_with_7_worst_regi/testground/'
ensure_dir(dst)
ax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/ax3d/'+'*.nrrd', recursive = True))[0]
sax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/sax3d/'+'*.nrrd', recursive = True))[0]
ax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/ax3d_iso_linear/'+'*.nrrd', recursive = True))[0]
sax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/sax3d_iso_linear/'+'*.nrrd', recursive = True))[0]
ax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/ax3d_iso_linear/'+'*.nrrd', recursive = True))[28]
sax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/sax3d_iso_linear/'+'*.nrrd', recursive = True))[28]
print(ax_src)
print(sax_src)
from scipy.ndimage import affine_transform
#testing single pictures
ax_file_path = ax_src
sax_file_path = sax_src
reader1 = sitk.ImageFileReader()
reader1.SetFileName(ax_file_path)
ax_img = reader1.Execute()
reader2 = sitk.ImageFileReader()
reader2.SetFileName(sax_file_path)
sax_img = reader2.Execute()
for shift in range(-20,1,2):
resampled = resample_direcion_origin_spacing_shift(ax_img, sax_img, shift = (0,0,shift), interpolate=sitk.sitkLinear)
writer = sitk.ImageFileWriter()
writer.SetFileName(os.path.join(dst,'shift'+str((0,0,shift))+'.nrrd'))
writer.Execute(resampled)
for shift in range(-20,21,5):
resampled = resample_direcion_origin_spacing_shift(ax_img, sax_img, shift = (shift,0,0), interpolate=sitk.sitkLinear)
writer = sitk.ImageFileWriter()
writer.SetFileName(os.path.join(dst,'shift'+str((shift,0,0))+'.nrrd'))
writer.Execute(resampled)
resampled = resample_direcion_origin_spacing_shift(ax_img, sax_img, shift = (0,shift,0), interpolate=sitk.sitkLinear)
writer = sitk.ImageFileWriter()
writer.SetFileName(os.path.join(dst,'shift'+str((0,shift,0))+'.nrrd'))
writer.Execute(resampled)
resampled = resample_direcion_origin_spacing_shift(ax_img, sax_img, shift = (0,0,shift), interpolate=sitk.sitkLinear)
writer = sitk.ImageFileWriter()
writer.SetFileName(os.path.join(dst,'shift'+str((0,0,shift))+'.nrrd'))
writer.Execute(resampled)
"""
sitk_img1 = ax_img
sitk_img2 = sax_ixmg
label1_img1 = get_single_label_img(sitk_img1,1)
label2_img1 = get_single_label_img(sitk_img1,2)
label3_img1 = get_single_label_img(sitk_img1,3)
resampled_label1 = resample_img(label1_img1, sitk_img2)
resampled_label2 = resample_img(label2_img1, sitk_img2)
resampled_label3 = resample_img(label3_img1, sitk_img2)
"""
size = ax_img.GetSize()
spacing = ax_img.GetSpacing()
size_new = [int(s*p) for s,p in zip(size,spacing)]
size_new = tuple(size_new)
resampler = sitk.ResampleImageFilter()
resampler.SetSize(size_new)
resampler.SetOutputSpacing((1,1,1))
resampler.SetOutputOrigin(ax_img.GetOrigin())
#resampler.SetInterpolator(sitk.sitkNearestNeighbor)
new_img = resampler.Execute(ax_img)
#sitk_img, size=(12, 256, 256), spacing=(8, 1.25, 1.25), interpolate=sitk.sitkNearestNeighbor
ax_array = sitk.GetArrayFromImage(new_img)
#ax_img.SetDirection(sax_img.GetDirection())
direction = sax_img.GetDirection()
direction = np.reshape(direction,(3,3))
#r = R.from_matrix(direction)
new_array = affine_transform(ax_array,direction)
new_img = sitk.GetImageFromArray(new_array)
new_img.SetOrigin(ax_img.GetOrigin())
new_img.SetDirection(sax_img.GetDirection())
#resampled = max_thres_resample_label_img(ax_img, sax_img,50)
#resampled = percentage_resample_label_img(ax_img, sax_img,80)
writer = sitk.ImageFileWriter()
writer.SetFileName(os.path.join(dst,os.path.basename('img_ax_new.nrrd')))
writer.Execute(new_img)
#writer = sitk.ImageFileWriter()
#writer.SetFileName(os.path.join(dst,os.path.basename('sax_0.nrrd')))
#writer.Execute(sax_img)
#writer = sitk.ImageFileWriter()
#writer.SetFileName(os.path.join(dst,os.path.basename(ax_file_path)))
#writer.Execute(resampled)
png_path = '/home/rflo/master/data_search/ax'
png_path = '/home/rflo/master/data_search/ax_to_sax/'
from IPython.display import Image
from ipywidgets import interact
path_to_png = png_path
@interact
def show_png(i = (0, len(sorted(glob.glob(os.path.join(path_to_png,'*.png'))))-1)):
f_name = sorted(glob.glob(os.path.join(path_to_png,'*.png')))[i]
print(f_name)
return Image(filename=f_name)
ax_src = '/home/rflo/master/all_with_7_worst_regi/ax3d/'
sax_src = '/home/rflo/master/all_with_7_worst_regi/sax3d/'
dst = '/home/rflo/master/all_with_7_worst_regi/testground/'
ax_src = '/home/rflo/master/all_with_7_worst_regi/ax3d_iso_linear/'
sax_src = '/home/rflo/master/all_with_7_worst_regi/sax3d_iso_linear/'
dst = '/home/rflo/master/all_with_7_worst_regi/testground/'
ensure_dir(dst)
src = ax_src
#copy all msk into dst
for file_path in sorted(glob.glob(src+'*.nrrd', recursive = True)):
if 'msk' in os.path.basename(file_path):
shutil.copyfile(file_path,os.path.join(dst,os.path.basename(file_path)))
```
|
github_jupyter
|
# define logging and working directory
from ProjectRoot import change_wd_to_project_root
change_wd_to_project_root()
import SimpleITK as sitk
import ipywidgets as widgets
import matplotlib.pyplot as plt
import pandas as pd
import os
import glob
import shutil
import numpy as np
from scipy.spatial.transform import Rotation as R
from src.data.Preprocess import *
from src.utils.Notebook_imports import *
ax_src = '/mnt/ssd/data/gcn/ax_sax_from_flo/ax3d/'
sax_src = '/mnt/ssd/data/gcn/ax_sax_from_flo/sax3d/'
ax_iso_dst = '/mnt/ssd/data/gcn/ax_sax_from_flo/ax3d_iso_linear/'
sax_iso_dst = '/mnt/ssd/data/gcn/ax_sax_from_flo/sax3d_iso_linear/'
# resample 3D image / mask into a fake isotrop resolution, use multithreadding, combine iso and orientation resample, everything in one step
import concurrent.futures
from concurrent.futures import as_completed
slice_first = 2
workers = 16
futures_ax_iso = set()
futures_sax_iso = set()
futures_ax2sax_iso = set()
futures_sax2ax_iso = set()
ax_files = sorted(glob.glob(ax_src+'*.nrrd', recursive = True))
sax_files = sorted(glob.glob(sax_src+'*.nrrd', recursive = True))
ensure_dir(ax_iso_dst)
ensure_dir(sax_iso_dst)
target_spacing = (1.5,1.5,1.5)
# spawn multiple threads or processes
with concurrent.futures.ThreadPoolExecutor(max_workers=workers) as executor:# resample ax towards sax and vize versa
for ax_file, sax_file in zip(ax_files, sax_files):
print('processing file: {}'.format(ax_file))
reader1 = sitk.ImageFileReader()
reader1.SetFileName(ax_file)
ax_img = reader1.Execute()
reader2 = sitk.ImageFileReader()
reader2.SetFileName(sax_file)
sax_img = reader2.Execute()
# make a pseudo isotrop volume from both input images
if 'img' in os.path.basename(ax_file):
futures_ax_iso.add(executor.submit(transform_to_isotrop_voxels, ax_img, sitk.sitkLinear,target_spacing, ax_file))
futures_sax_iso.add(executor.submit(transform_to_isotrop_voxels, sax_img, sitk.sitkLinear,target_spacing, sax_file))
else:
futures_ax_iso.add(executor.submit(max_thres_resample2_iso_label_img, ax_img,50, target_spacing, ax_file, sitk.sitkLinear))
futures_sax_iso.add(executor.submit(max_thres_resample2_iso_label_img, sax_img,50, target_spacing, sax_file, sitk.sitkLinear))
# this part is sequential, we need only one writer
writer = sitk.ImageFileWriter()
for future in as_completed(futures_ax_iso):
try:
res = future.result()
iso_resampled, file_path = res
writer.SetFileName(os.path.join(ax_iso_dst,os.path.basename(file_path)))
writer.Execute(iso_resampled)
except Exception as e:
logging.error(
'Exception {} in datagenerator with: image: {} or mask: {}'.format(str(e)))
for future in as_completed(futures_sax_iso):
try:
res = future.result()
iso_resampled, file_path = res
writer.SetFileName(os.path.join(sax_iso_dst,os.path.basename(file_path)))
writer.Execute(iso_resampled)
except Exception as e:
logging.error(
'Exception {} in datagenerator with: image: {} or mask: {}'.format(str(e)))
ax_src = '/mnt/ssd/data/gcn/ax_sax_from_flo/ax3d_iso_linear/'
sax_src = '/mnt/ssd/data/gcn/ax_sax_from_flo/sax3d_iso_linear/'
dst_ax2sax = '/mnt/ssd/data/gcn/ax_sax_from_flo/sax2ax3d_iso/'
ax2sax_iso_dst = '/mnt/ssd/data/gcn/ax_sax_from_flo/ax2sax3d_iso_linear/'
sax2ax_iso_dst = '/mnt/ssd/data/gcn/ax_sax_from_flo/sax2ax3d_iso_linear/'
# resample 3D image / mask from axial view into sax view or vize versa, extend the transformed vol by 10 voxel along the target z axis
import concurrent.futures
from concurrent.futures import as_completed
workers = 16
futures_ax2sax = set()
futures_sax2ax = set()
ensure_dir(ax2sax_iso_dst)
ensure_dir(sax2ax_iso_dst)
# spawn multiple threads or processes
# transform the image of file_path with according to the direction of
ax_files = sorted(glob.glob(ax_src+'*.nrrd', recursive = True))
sax_files = sorted(glob.glob(sax_src+'*.nrrd', recursive = True))
print('axial files: {}'.format(len(ax_files)))
print('sax files: {}'.format(len(sax_files)))
with concurrent.futures.ThreadPoolExecutor(max_workers=workers) as executor:
for ax_file_path, sax_file_path in zip(ax_files, sax_files):
reader1 = sitk.ImageFileReader()
reader1.SetFileName(ax_file_path)
ax_img = reader1.Execute()
reader2 = sitk.ImageFileReader()
reader2.SetFileName(sax_file_path)
sax_img = reader2.Execute()
if 'img' in os.path.basename(ax_file_path):
futures_ax2sax.add(executor.submit(resample_direcion_origin_spacing_shift, ax_img, sax_img, (0,0,-10), sitk.sitkLinear, ax_file_path))
futures_sax2ax.add(executor.submit(resample_direcion_origin_spacing_shift, sax_img, ax_img, (0,0,-10), sitk.sitkLinear, sax_file_path))
else:
futures_ax2sax.add(executor.submit(max_thres_resample2_label_img_shift, ax_img, sax_img, 50, (0,0,-10), ax_file_path))
futures_sax2ax.add(executor.submit(max_thres_resample2_label_img_shift, sax_img, ax_img, 50, (0,0,-10), sax_file_path))
writer = sitk.ImageFileWriter()
for i, future in enumerate(as_completed(futures_ax2sax)):
try:
res = future.result()
resampled, file_path = res
writer.SetFileName(os.path.join(ax2sax_iso_dst,os.path.basename(file_path)))
writer.Execute(resampled)
except Exception as e:
logging.error(
'Exception {} in datagenerator with: image: {} or mask: {}'.format(str(e)))
for i, future in enumerate(as_completed(futures_sax2ax)):
try:
res = future.result()
resampled, file_path = res
writer.SetFileName(os.path.join(sax2ax_iso_dst,os.path.basename(file_path)))
writer.Execute(resampled)
except Exception as e:
logging.error(
'Exception {} in datagenerator with: image: {} or mask: {}'.format(str(e)))
ax_src = '/home/rflo/master/all_with_7_worst_regi/ax3d/'
sax_src = '/home/rflo/master/all_with_7_worst_regi/sax3d/'
dst = '/home/rflo/master/all_with_7_worst_regi/testground/'
ensure_dir(dst)
ax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/ax3d/'+'*.nrrd', recursive = True))[0]
sax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/sax3d/'+'*.nrrd', recursive = True))[0]
ax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/ax3d_iso_linear/'+'*.nrrd', recursive = True))[0]
sax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/sax3d_iso_linear/'+'*.nrrd', recursive = True))[0]
ax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/ax3d_iso_linear/'+'*.nrrd', recursive = True))[28]
sax_src = sorted(glob.glob('/home/rflo/master/all_with_7_worst_regi/sax3d_iso_linear/'+'*.nrrd', recursive = True))[28]
print(ax_src)
print(sax_src)
from scipy.ndimage import affine_transform
#testing single pictures
ax_file_path = ax_src
sax_file_path = sax_src
reader1 = sitk.ImageFileReader()
reader1.SetFileName(ax_file_path)
ax_img = reader1.Execute()
reader2 = sitk.ImageFileReader()
reader2.SetFileName(sax_file_path)
sax_img = reader2.Execute()
for shift in range(-20,1,2):
resampled = resample_direcion_origin_spacing_shift(ax_img, sax_img, shift = (0,0,shift), interpolate=sitk.sitkLinear)
writer = sitk.ImageFileWriter()
writer.SetFileName(os.path.join(dst,'shift'+str((0,0,shift))+'.nrrd'))
writer.Execute(resampled)
for shift in range(-20,21,5):
resampled = resample_direcion_origin_spacing_shift(ax_img, sax_img, shift = (shift,0,0), interpolate=sitk.sitkLinear)
writer = sitk.ImageFileWriter()
writer.SetFileName(os.path.join(dst,'shift'+str((shift,0,0))+'.nrrd'))
writer.Execute(resampled)
resampled = resample_direcion_origin_spacing_shift(ax_img, sax_img, shift = (0,shift,0), interpolate=sitk.sitkLinear)
writer = sitk.ImageFileWriter()
writer.SetFileName(os.path.join(dst,'shift'+str((0,shift,0))+'.nrrd'))
writer.Execute(resampled)
resampled = resample_direcion_origin_spacing_shift(ax_img, sax_img, shift = (0,0,shift), interpolate=sitk.sitkLinear)
writer = sitk.ImageFileWriter()
writer.SetFileName(os.path.join(dst,'shift'+str((0,0,shift))+'.nrrd'))
writer.Execute(resampled)
"""
sitk_img1 = ax_img
sitk_img2 = sax_ixmg
label1_img1 = get_single_label_img(sitk_img1,1)
label2_img1 = get_single_label_img(sitk_img1,2)
label3_img1 = get_single_label_img(sitk_img1,3)
resampled_label1 = resample_img(label1_img1, sitk_img2)
resampled_label2 = resample_img(label2_img1, sitk_img2)
resampled_label3 = resample_img(label3_img1, sitk_img2)
"""
size = ax_img.GetSize()
spacing = ax_img.GetSpacing()
size_new = [int(s*p) for s,p in zip(size,spacing)]
size_new = tuple(size_new)
resampler = sitk.ResampleImageFilter()
resampler.SetSize(size_new)
resampler.SetOutputSpacing((1,1,1))
resampler.SetOutputOrigin(ax_img.GetOrigin())
#resampler.SetInterpolator(sitk.sitkNearestNeighbor)
new_img = resampler.Execute(ax_img)
#sitk_img, size=(12, 256, 256), spacing=(8, 1.25, 1.25), interpolate=sitk.sitkNearestNeighbor
ax_array = sitk.GetArrayFromImage(new_img)
#ax_img.SetDirection(sax_img.GetDirection())
direction = sax_img.GetDirection()
direction = np.reshape(direction,(3,3))
#r = R.from_matrix(direction)
new_array = affine_transform(ax_array,direction)
new_img = sitk.GetImageFromArray(new_array)
new_img.SetOrigin(ax_img.GetOrigin())
new_img.SetDirection(sax_img.GetDirection())
#resampled = max_thres_resample_label_img(ax_img, sax_img,50)
#resampled = percentage_resample_label_img(ax_img, sax_img,80)
writer = sitk.ImageFileWriter()
writer.SetFileName(os.path.join(dst,os.path.basename('img_ax_new.nrrd')))
writer.Execute(new_img)
#writer = sitk.ImageFileWriter()
#writer.SetFileName(os.path.join(dst,os.path.basename('sax_0.nrrd')))
#writer.Execute(sax_img)
#writer = sitk.ImageFileWriter()
#writer.SetFileName(os.path.join(dst,os.path.basename(ax_file_path)))
#writer.Execute(resampled)
png_path = '/home/rflo/master/data_search/ax'
png_path = '/home/rflo/master/data_search/ax_to_sax/'
from IPython.display import Image
from ipywidgets import interact
path_to_png = png_path
@interact
def show_png(i = (0, len(sorted(glob.glob(os.path.join(path_to_png,'*.png'))))-1)):
f_name = sorted(glob.glob(os.path.join(path_to_png,'*.png')))[i]
print(f_name)
return Image(filename=f_name)
ax_src = '/home/rflo/master/all_with_7_worst_regi/ax3d/'
sax_src = '/home/rflo/master/all_with_7_worst_regi/sax3d/'
dst = '/home/rflo/master/all_with_7_worst_regi/testground/'
ax_src = '/home/rflo/master/all_with_7_worst_regi/ax3d_iso_linear/'
sax_src = '/home/rflo/master/all_with_7_worst_regi/sax3d_iso_linear/'
dst = '/home/rflo/master/all_with_7_worst_regi/testground/'
ensure_dir(dst)
src = ax_src
#copy all msk into dst
for file_path in sorted(glob.glob(src+'*.nrrd', recursive = True)):
if 'msk' in os.path.basename(file_path):
shutil.copyfile(file_path,os.path.join(dst,os.path.basename(file_path)))
| 0.358129 | 0.594198 |
```
import pandas as pd
import matplotlib.pyplot as plt
import ipywidgets as widgets
from IPython.display import HTML
plt.ioff()
DATA_URL = 'https://gist.githubusercontent.com/chriddyp/cb5392c35661370d95f300086accea51/raw/8e0768211f6b747c0db42a9ce9a0937dafcbd8b2/indicators.csv'
EXPLANATION = """\
<div class="app-sidebar">
<p><em>Compare different development indicators.</em><p>
<p>Select what indicators to plot in the dropdowns, and use the slider
to sub-select a fraction of years to include in the plot.</p>
<p>Data and idea copied from the <a href="https://dash.plot.ly/getting-started-part-2">
Plotly Dash documentation</a>.</p>
<p>This example demonstrates combining matplotlib with Jupyter widgets. For more interactive plots,
consider using <a href="https://github.com/bloomberg/bqplot">bqplot</a>.
</div>
"""
HTML("""\
<style>
.app-subtitle {
font-size: 1.5em;
}
.app-subtitle a {
color: #106ba3;
}
.app-subtitle a:hover {
text-decoration: underline;
}
.app-sidebar p {
margin-bottom: 1em;
line-height: 1.7;
}
.app-sidebar a {
color: #106ba3;
}
.app-sidebar a:hover {
text-decoration: underline;
}
</style>
""")
class App:
def __init__(self, df):
self._df = df
available_indicators = self._df['Indicator Name'].unique()
self._x_dropdown = self._create_indicator_dropdown(available_indicators, 0)
self._y_dropdown = self._create_indicator_dropdown(available_indicators, 1)
self._plot_container = widgets.Output()
self._year_slider, year_slider_box = self._create_year_slider(
min(df['Year']), max(df['Year'])
)
_app_container = widgets.VBox([
widgets.HBox([self._x_dropdown, self._y_dropdown]),
self._plot_container,
year_slider_box
], layout=widgets.Layout(align_items='center', flex='3 0 auto'))
self.container = widgets.VBox([
widgets.HTML(
(
'<h1>Development indicators</h1>'
'<h2 class="app-subtitle"><a href="https://github.com/pbugnion/voila-gallery/blob/master/country-indicators/index.ipynb">Link to code</a></h2>'
),
layout=widgets.Layout(margin='0 0 5em 0')
),
widgets.HBox([
_app_container,
widgets.HTML(EXPLANATION, layout=widgets.Layout(margin='0 0 0 2em'))
])
], layout=widgets.Layout(flex='1 1 auto', margin='0 auto 0 auto', max_width='1024px'))
self._update_app()
@classmethod
def from_url(cls, url):
df = pd.read_csv(url)
return cls(df)
def _create_indicator_dropdown(self, indicators, initial_index):
dropdown = widgets.Dropdown(options=indicators, value=indicators[initial_index])
dropdown.observe(self._on_change, names=['value'])
return dropdown
def _create_year_slider(self, min_year, max_year):
year_slider_label = widgets.Label('Year range: ')
year_slider = widgets.IntRangeSlider(
min=min_year, max=max_year,
layout=widgets.Layout(width='500px')
)
year_slider.observe(self._on_change, names=['value'])
year_slider_box = widgets.HBox([year_slider_label, year_slider])
return year_slider, year_slider_box
def _create_plot(self, x_indicator, y_indicator, year_range):
df = self._df[self._df['Year'].between(*year_range)]
xs = df[df['Indicator Name'] == x_indicator]['Value']
ys = df[df['Indicator Name'] == y_indicator]['Value']
plt.figure(figsize=(10, 8))
plt.xlabel(x_indicator, size=16)
plt.ylabel(y_indicator, size=16)
plt.gca().tick_params(axis='both', which='major', labelsize=16)
plt.plot(xs, ys, 'o', alpha=0.3)
def _on_change(self, _):
self._update_app()
def _update_app(self):
x_indicator = self._x_dropdown.value
y_indicator = self._y_dropdown.value
year_range = self._year_slider.value
self._plot_container.clear_output(wait=True)
with self._plot_container:
self._create_plot(x_indicator, y_indicator, year_range)
plt.show()
app = App.from_url(DATA_URL)
app.container
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import ipywidgets as widgets
from IPython.display import HTML
plt.ioff()
DATA_URL = 'https://gist.githubusercontent.com/chriddyp/cb5392c35661370d95f300086accea51/raw/8e0768211f6b747c0db42a9ce9a0937dafcbd8b2/indicators.csv'
EXPLANATION = """\
<div class="app-sidebar">
<p><em>Compare different development indicators.</em><p>
<p>Select what indicators to plot in the dropdowns, and use the slider
to sub-select a fraction of years to include in the plot.</p>
<p>Data and idea copied from the <a href="https://dash.plot.ly/getting-started-part-2">
Plotly Dash documentation</a>.</p>
<p>This example demonstrates combining matplotlib with Jupyter widgets. For more interactive plots,
consider using <a href="https://github.com/bloomberg/bqplot">bqplot</a>.
</div>
"""
HTML("""\
<style>
.app-subtitle {
font-size: 1.5em;
}
.app-subtitle a {
color: #106ba3;
}
.app-subtitle a:hover {
text-decoration: underline;
}
.app-sidebar p {
margin-bottom: 1em;
line-height: 1.7;
}
.app-sidebar a {
color: #106ba3;
}
.app-sidebar a:hover {
text-decoration: underline;
}
</style>
""")
class App:
def __init__(self, df):
self._df = df
available_indicators = self._df['Indicator Name'].unique()
self._x_dropdown = self._create_indicator_dropdown(available_indicators, 0)
self._y_dropdown = self._create_indicator_dropdown(available_indicators, 1)
self._plot_container = widgets.Output()
self._year_slider, year_slider_box = self._create_year_slider(
min(df['Year']), max(df['Year'])
)
_app_container = widgets.VBox([
widgets.HBox([self._x_dropdown, self._y_dropdown]),
self._plot_container,
year_slider_box
], layout=widgets.Layout(align_items='center', flex='3 0 auto'))
self.container = widgets.VBox([
widgets.HTML(
(
'<h1>Development indicators</h1>'
'<h2 class="app-subtitle"><a href="https://github.com/pbugnion/voila-gallery/blob/master/country-indicators/index.ipynb">Link to code</a></h2>'
),
layout=widgets.Layout(margin='0 0 5em 0')
),
widgets.HBox([
_app_container,
widgets.HTML(EXPLANATION, layout=widgets.Layout(margin='0 0 0 2em'))
])
], layout=widgets.Layout(flex='1 1 auto', margin='0 auto 0 auto', max_width='1024px'))
self._update_app()
@classmethod
def from_url(cls, url):
df = pd.read_csv(url)
return cls(df)
def _create_indicator_dropdown(self, indicators, initial_index):
dropdown = widgets.Dropdown(options=indicators, value=indicators[initial_index])
dropdown.observe(self._on_change, names=['value'])
return dropdown
def _create_year_slider(self, min_year, max_year):
year_slider_label = widgets.Label('Year range: ')
year_slider = widgets.IntRangeSlider(
min=min_year, max=max_year,
layout=widgets.Layout(width='500px')
)
year_slider.observe(self._on_change, names=['value'])
year_slider_box = widgets.HBox([year_slider_label, year_slider])
return year_slider, year_slider_box
def _create_plot(self, x_indicator, y_indicator, year_range):
df = self._df[self._df['Year'].between(*year_range)]
xs = df[df['Indicator Name'] == x_indicator]['Value']
ys = df[df['Indicator Name'] == y_indicator]['Value']
plt.figure(figsize=(10, 8))
plt.xlabel(x_indicator, size=16)
plt.ylabel(y_indicator, size=16)
plt.gca().tick_params(axis='both', which='major', labelsize=16)
plt.plot(xs, ys, 'o', alpha=0.3)
def _on_change(self, _):
self._update_app()
def _update_app(self):
x_indicator = self._x_dropdown.value
y_indicator = self._y_dropdown.value
year_range = self._year_slider.value
self._plot_container.clear_output(wait=True)
with self._plot_container:
self._create_plot(x_indicator, y_indicator, year_range)
plt.show()
app = App.from_url(DATA_URL)
app.container
| 0.683631 | 0.369088 |
Deep Learning
=============
Assignment 4
------------
Previously in `2_fullyconnected.ipynb` and `3_regularization.ipynb`, we trained fully connected networks to classify [notMNIST](http://yaroslavvb.blogspot.com/2011/09/notmnist-dataset.html) characters.
The goal of this assignment is make the neural network convolutional.
```
# These are all the modules we'll be using later. Make sure you can import them
# before proceeding further.
from __future__ import print_function
import numpy as np
import tensorflow as tf
from six.moves import cPickle as pickle
from six.moves import range
pickle_file = 'notMNIST.pickle'
with open(pickle_file, 'rb') as f:
save = pickle.load(f)
train_dataset = save['train_dataset']
train_labels = save['train_labels']
valid_dataset = save['valid_dataset']
valid_labels = save['valid_labels']
test_dataset = save['test_dataset']
test_labels = save['test_labels']
del save # hint to help gc free up memory
print('Training set', train_dataset.shape, train_labels.shape)
print('Validation set', valid_dataset.shape, valid_labels.shape)
print('Test set', test_dataset.shape, test_labels.shape)
```
Reformat into a TensorFlow-friendly shape:
- convolutions need the image data formatted as a cube (width by height by #channels)
- labels as float 1-hot encodings.
```
image_size = 28
num_labels = 10
num_channels = 1 # grayscale
import numpy as np
def reformat(dataset, labels):
dataset = dataset.reshape(
(-1, image_size, image_size, num_channels)).astype(np.float32)
labels = (np.arange(num_labels) == labels[:,None]).astype(np.float32)
return dataset, labels
train_dataset, train_labels = reformat(train_dataset, train_labels)
valid_dataset, valid_labels = reformat(valid_dataset, valid_labels)
test_dataset, test_labels = reformat(test_dataset, test_labels)
print('Training set', train_dataset.shape, train_labels.shape)
print('Validation set', valid_dataset.shape, valid_labels.shape)
print('Test set', test_dataset.shape, test_labels.shape)
def accuracy(predictions, labels):
return (100.0 * np.sum(np.argmax(predictions, 1) == np.argmax(labels, 1))
/ predictions.shape[0])
```
Let's build a small network with two convolutional layers, followed by one fully connected layer. Convolutional networks are more expensive computationally, so we'll limit its depth and number of fully connected nodes.
```
batch_size = 16
patch_size = 5
depth = 16
num_hidden = 64
graph = tf.Graph()
with graph.as_default():
# Input data.
tf_train_dataset = tf.placeholder(
tf.float32, shape=(batch_size, image_size, image_size, num_channels))
tf_train_labels = tf.placeholder(tf.float32, shape=(batch_size, num_labels))
tf_valid_dataset = tf.constant(valid_dataset)
tf_test_dataset = tf.constant(test_dataset)
# Variables.
layer1_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, num_channels, depth], stddev=0.1))
layer1_biases = tf.Variable(tf.zeros([depth]))
layer2_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, depth, depth], stddev=0.1))
layer2_biases = tf.Variable(tf.constant(1.0, shape=[depth]))
layer3_weights = tf.Variable(tf.truncated_normal(
[image_size // 4 * image_size // 4 * depth, num_hidden], stddev=0.1))
layer3_biases = tf.Variable(tf.constant(1.0, shape=[num_hidden]))
layer4_weights = tf.Variable(tf.truncated_normal(
[num_hidden, num_labels], stddev=0.1))
layer4_biases = tf.Variable(tf.constant(1.0, shape=[num_labels]))
# Model.
def model(data):
conv = tf.nn.conv2d(data, layer1_weights, [1, 2, 2, 1], padding='SAME')
hidden = tf.nn.relu(conv + layer1_biases)
conv = tf.nn.conv2d(hidden, layer2_weights, [1, 2, 2, 1], padding='SAME')
hidden = tf.nn.relu(conv + layer2_biases)
shape = hidden.get_shape().as_list()
reshape = tf.reshape(hidden, [shape[0], shape[1] * shape[2] * shape[3]])
hidden = tf.nn.relu(tf.matmul(reshape, layer3_weights) + layer3_biases)
return tf.matmul(hidden, layer4_weights) + layer4_biases
# Training computation.
logits = model(tf_train_dataset)
loss = tf.reduce_mean(
tf.nn.softmax_cross_entropy_with_logits(logits, tf_train_labels))
# Optimizer.
optimizer = tf.train.GradientDescentOptimizer(0.05).minimize(loss)
# Predictions for the training, validation, and test data.
train_prediction = tf.nn.softmax(logits)
valid_prediction = tf.nn.softmax(model(tf_valid_dataset))
test_prediction = tf.nn.softmax(model(tf_test_dataset))
num_steps = 1001
with tf.Session(graph=graph) as session:
tf.initialize_all_variables().run()
print('Initialized')
for step in range(num_steps):
offset = (step * batch_size) % (train_labels.shape[0] - batch_size)
batch_data = train_dataset[offset:(offset + batch_size), :, :, :]
batch_labels = train_labels[offset:(offset + batch_size), :]
feed_dict = {tf_train_dataset : batch_data, tf_train_labels : batch_labels}
_, l, predictions = session.run(
[optimizer, loss, train_prediction], feed_dict=feed_dict)
if (step % 50 == 0):
print('Minibatch loss at step %d: %f' % (step, l))
print('Minibatch accuracy: %.1f%%' % accuracy(predictions, batch_labels))
print('Validation accuracy: %.1f%%' % accuracy(
valid_prediction.eval(), valid_labels))
print('Test accuracy: %.1f%%' % accuracy(test_prediction.eval(), test_labels))
```
---
Problem 1
---------
The convolutional model above uses convolutions with stride 2 to reduce the dimensionality. Replace the strides by a max pooling operation (`nn.max_pool()`) of stride 2 and kernel size 2.
---
```
batch_size = 16
patch_size = 5
depth = 16
num_hidden = 64
graph = tf.Graph()
with graph.as_default():
# Input data.
tf_train_dataset = tf.placeholder(
tf.float32, shape=(batch_size, image_size, image_size, num_channels))
tf_train_labels = tf.placeholder(tf.float32, shape=(batch_size, num_labels))
tf_valid_dataset = tf.constant(valid_dataset)
tf_test_dataset = tf.constant(test_dataset)
# Variables.
layer1_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, num_channels, depth], stddev=0.1))
layer1_biases = tf.Variable(tf.zeros([depth]))
layer2_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, depth, depth], stddev=0.1))
layer2_biases = tf.Variable(tf.constant(1.0, shape=[depth]))
layer3_weights = tf.Variable(tf.truncated_normal(
[image_size // 4 * image_size // 4 * depth, num_hidden], stddev=0.1))
layer3_biases = tf.Variable(tf.constant(1.0, shape=[num_hidden]))
layer4_weights = tf.Variable(tf.truncated_normal(
[num_hidden, num_labels], stddev=0.1))
layer4_biases = tf.Variable(tf.constant(1.0, shape=[num_labels]))
# Model.
def model(data):
conv = tf.nn.conv2d(data, layer1_weights, [1, 1, 1, 1], padding='SAME')
hidden = tf.nn.relu(conv + layer1_biases)
conv = tf.nn.conv2d(tf.nn.max_pool(hidden, [1,2,2,1], [1,2,2,1], 'SAME'),\
layer2_weights, [1, 1, 1, 1], padding='SAME')
hidden = tf.nn.relu(conv + layer2_biases)
shape = tf.nn.max_pool(hidden, [1,2,2,1], [1,2,2,1], 'SAME').get_shape().as_list()
reshape = tf.reshape(tf.nn.max_pool(hidden, [1,2,2,1], [1,2,2,1], 'SAME'),\
[shape[0], shape[1] * shape[2] * shape[3]])
hidden = tf.nn.relu(tf.matmul(reshape, layer3_weights) + layer3_biases)
return tf.matmul(hidden, layer4_weights) + layer4_biases
# Training computation.
logits = model(tf_train_dataset)
loss = tf.reduce_mean(
tf.nn.softmax_cross_entropy_with_logits(logits, tf_train_labels))
# Optimizer.
optimizer = tf.train.GradientDescentOptimizer(0.05).minimize(loss)
# Predictions for the training, validation, and test data.
train_prediction = tf.nn.softmax(logits)
valid_prediction = tf.nn.softmax(model(tf_valid_dataset))
test_prediction = tf.nn.softmax(model(tf_test_dataset))
num_steps = 1001
with tf.Session(graph=graph) as session:
tf.initialize_all_variables().run()
print('Initialized')
for step in range(num_steps):
offset = (step * batch_size) % (train_labels.shape[0] - batch_size)
batch_data = train_dataset[offset:(offset + batch_size), :, :, :]
batch_labels = train_labels[offset:(offset + batch_size), :]
feed_dict = {tf_train_dataset : batch_data, tf_train_labels : batch_labels}
_, l, predictions = session.run(
[optimizer, loss, train_prediction], feed_dict=feed_dict)
if (step % 50 == 0):
print('Minibatch loss at step %d: %f' % (step, l))
print('Minibatch accuracy: %.1f%%' % accuracy(predictions, batch_labels))
print('Validation accuracy: %.1f%%' % accuracy(
valid_prediction.eval(), valid_labels))
print('Test accuracy: %.1f%%' % accuracy(test_prediction.eval(), test_labels))
```
---
Problem 2
---------
Try to get the best performance you can using a convolutional net. Look for example at the classic [LeNet5](http://yann.lecun.com/exdb/lenet/) architecture, adding Dropout, and/or adding learning rate decay.
---
```
batch_size = 128
patch_size = 5
depth1 = 16
depth2 = 32
num_hidden1 = 128
num_hidden2 = 64
# Construct a 7-layer CNN.
# C1: convolutional layer, batch_size x 28 x 28 x 16, convolution size: 5 x 5 x 1 x 16
# S2: sub-sampling layer, batch_size x 14 x 14 x 16
# C3: convolutional layer, batch_size x 10 x 10 x 32, convolution size: 5 x 5 x 6 x 32
# S4: sub-sampling layer, batch_size x 5 x 5 x 32
# C5: convolutional layer, batch_size x 1 x 1 x 128, convolution size: 5 x 5 x 16 x 128
# F6: fully-connected layer, weight size: 128 x 64
# Output layer, weight size: 64 x 10
graph = tf.Graph()
with graph.as_default():
# Input data.
tf_train_dataset = tf.placeholder(
tf.float32, shape=(batch_size, image_size, image_size, num_channels))
tf_train_labels = tf.placeholder(tf.float32, shape=(batch_size, num_labels))
tf_valid_dataset = tf.constant(valid_dataset)
tf_test_dataset = tf.constant(test_dataset)
# Variables.
layer1_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, num_channels, depth1], stddev=0.1))
layer1_biases = tf.Variable(tf.zeros([depth1]))
layer2_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, depth1, depth2], stddev=0.1))
layer2_biases = tf.Variable(tf.constant(1.0, shape=[depth2]))
layer3_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, depth2, num_hidden1], stddev=0.1))
layer3_biases = tf.Variable(tf.constant(1.0, shape=[num_hidden1]))
layer4_weights = tf.Variable(tf.truncated_normal(
[num_hidden1, num_hidden2], stddev=0.1))
layer4_biases = tf.Variable(tf.constant(1.0, shape=[num_hidden2]))
layer5_weights = tf.Variable(tf.truncated_normal(
[num_hidden2, num_labels], stddev=0.1))
layer5_biases = tf.Variable(tf.constant(1.0, shape=[num_labels]))
# Model.
def model(data, keep_prob):
conv = tf.nn.conv2d(data, layer1_weights, [1,1,1,1], 'SAME', name='C1')
hidden = tf.nn.relu(conv + layer1_biases)
hidden = tf.nn.dropout(hidden, keep_prob)
sub = tf.nn.max_pool(hidden, [1,2,2,1], [1,2,2,1], 'SAME', name='S2')
conv = tf.nn.conv2d(sub, layer2_weights, [1,1,1,1], padding='VALID', name='C3')
hidden = tf.nn.relu(conv + layer2_biases)
hidden = tf.nn.dropout(hidden, keep_prob)
sub = tf.nn.max_pool(hidden, [1,2,2,1], [1,2,2,1], 'SAME', name='S4')
conv = tf.nn.conv2d(sub, layer3_weights, [1,1,1,1], padding='VALID', name='C5')
hidden = tf.nn.relu(conv + layer3_biases)
hidden = tf.nn.dropout(hidden, keep_prob)
shape = hidden.get_shape().as_list()
reshape = tf.reshape(hidden, [shape[0], shape[1] * shape[2] * shape[3]])
hidden = tf.nn.relu(tf.matmul(reshape, layer4_weights) + layer4_biases)
hidden = tf.nn.dropout(hidden, keep_prob)
return tf.matmul(hidden, layer5_weights) + layer5_biases
# Training computation.
logits = model(tf_train_dataset, 0.5)
loss = tf.reduce_mean(
tf.nn.softmax_cross_entropy_with_logits(logits, tf_train_labels))
# Optimizer.
#optimizer = tf.train.AdagradOptimizer(0.05).minimize(loss)
global_step = tf.Variable(0)
learning_rate = tf.train.exponential_decay(0.05, global_step, 2000, 0.95)
optimizer = tf.train.AdagradOptimizer(learning_rate).minimize(loss, global_step=global_step)
# Predictions for the training, validation, and test data.
train_prediction = tf.nn.softmax(model(tf_train_dataset, 1.0))
valid_prediction = tf.nn.softmax(model(tf_valid_dataset, 1.0))
test_prediction = tf.nn.softmax(model(tf_test_dataset, 1.0))
num_steps = 2001
with tf.Session(graph=graph) as session:
tf.initialize_all_variables().run()
print('Initialized')
for step in range(num_steps):
offset = (step * batch_size) % (train_labels.shape[0] - batch_size)
batch_data = train_dataset[offset:(offset + batch_size), :, :, :]
batch_labels = train_labels[offset:(offset + batch_size), :]
feed_dict = {tf_train_dataset : batch_data, tf_train_labels : batch_labels}
_, l, predictions = session.run(
[optimizer, loss, train_prediction], feed_dict=feed_dict)
if (step % 100 == 0):
print('Minibatch loss at step %d: %f' % (step, l))
print('Minibatch accuracy: %.1f%%' % accuracy(predictions, batch_labels))
print('Validation accuracy: %.1f%%' % accuracy(valid_prediction.eval(), valid_labels))
print('Test accuracy: %.1f%%' % accuracy(test_prediction.eval(), test_labels))
```
|
github_jupyter
|
# These are all the modules we'll be using later. Make sure you can import them
# before proceeding further.
from __future__ import print_function
import numpy as np
import tensorflow as tf
from six.moves import cPickle as pickle
from six.moves import range
pickle_file = 'notMNIST.pickle'
with open(pickle_file, 'rb') as f:
save = pickle.load(f)
train_dataset = save['train_dataset']
train_labels = save['train_labels']
valid_dataset = save['valid_dataset']
valid_labels = save['valid_labels']
test_dataset = save['test_dataset']
test_labels = save['test_labels']
del save # hint to help gc free up memory
print('Training set', train_dataset.shape, train_labels.shape)
print('Validation set', valid_dataset.shape, valid_labels.shape)
print('Test set', test_dataset.shape, test_labels.shape)
image_size = 28
num_labels = 10
num_channels = 1 # grayscale
import numpy as np
def reformat(dataset, labels):
dataset = dataset.reshape(
(-1, image_size, image_size, num_channels)).astype(np.float32)
labels = (np.arange(num_labels) == labels[:,None]).astype(np.float32)
return dataset, labels
train_dataset, train_labels = reformat(train_dataset, train_labels)
valid_dataset, valid_labels = reformat(valid_dataset, valid_labels)
test_dataset, test_labels = reformat(test_dataset, test_labels)
print('Training set', train_dataset.shape, train_labels.shape)
print('Validation set', valid_dataset.shape, valid_labels.shape)
print('Test set', test_dataset.shape, test_labels.shape)
def accuracy(predictions, labels):
return (100.0 * np.sum(np.argmax(predictions, 1) == np.argmax(labels, 1))
/ predictions.shape[0])
batch_size = 16
patch_size = 5
depth = 16
num_hidden = 64
graph = tf.Graph()
with graph.as_default():
# Input data.
tf_train_dataset = tf.placeholder(
tf.float32, shape=(batch_size, image_size, image_size, num_channels))
tf_train_labels = tf.placeholder(tf.float32, shape=(batch_size, num_labels))
tf_valid_dataset = tf.constant(valid_dataset)
tf_test_dataset = tf.constant(test_dataset)
# Variables.
layer1_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, num_channels, depth], stddev=0.1))
layer1_biases = tf.Variable(tf.zeros([depth]))
layer2_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, depth, depth], stddev=0.1))
layer2_biases = tf.Variable(tf.constant(1.0, shape=[depth]))
layer3_weights = tf.Variable(tf.truncated_normal(
[image_size // 4 * image_size // 4 * depth, num_hidden], stddev=0.1))
layer3_biases = tf.Variable(tf.constant(1.0, shape=[num_hidden]))
layer4_weights = tf.Variable(tf.truncated_normal(
[num_hidden, num_labels], stddev=0.1))
layer4_biases = tf.Variable(tf.constant(1.0, shape=[num_labels]))
# Model.
def model(data):
conv = tf.nn.conv2d(data, layer1_weights, [1, 2, 2, 1], padding='SAME')
hidden = tf.nn.relu(conv + layer1_biases)
conv = tf.nn.conv2d(hidden, layer2_weights, [1, 2, 2, 1], padding='SAME')
hidden = tf.nn.relu(conv + layer2_biases)
shape = hidden.get_shape().as_list()
reshape = tf.reshape(hidden, [shape[0], shape[1] * shape[2] * shape[3]])
hidden = tf.nn.relu(tf.matmul(reshape, layer3_weights) + layer3_biases)
return tf.matmul(hidden, layer4_weights) + layer4_biases
# Training computation.
logits = model(tf_train_dataset)
loss = tf.reduce_mean(
tf.nn.softmax_cross_entropy_with_logits(logits, tf_train_labels))
# Optimizer.
optimizer = tf.train.GradientDescentOptimizer(0.05).minimize(loss)
# Predictions for the training, validation, and test data.
train_prediction = tf.nn.softmax(logits)
valid_prediction = tf.nn.softmax(model(tf_valid_dataset))
test_prediction = tf.nn.softmax(model(tf_test_dataset))
num_steps = 1001
with tf.Session(graph=graph) as session:
tf.initialize_all_variables().run()
print('Initialized')
for step in range(num_steps):
offset = (step * batch_size) % (train_labels.shape[0] - batch_size)
batch_data = train_dataset[offset:(offset + batch_size), :, :, :]
batch_labels = train_labels[offset:(offset + batch_size), :]
feed_dict = {tf_train_dataset : batch_data, tf_train_labels : batch_labels}
_, l, predictions = session.run(
[optimizer, loss, train_prediction], feed_dict=feed_dict)
if (step % 50 == 0):
print('Minibatch loss at step %d: %f' % (step, l))
print('Minibatch accuracy: %.1f%%' % accuracy(predictions, batch_labels))
print('Validation accuracy: %.1f%%' % accuracy(
valid_prediction.eval(), valid_labels))
print('Test accuracy: %.1f%%' % accuracy(test_prediction.eval(), test_labels))
batch_size = 16
patch_size = 5
depth = 16
num_hidden = 64
graph = tf.Graph()
with graph.as_default():
# Input data.
tf_train_dataset = tf.placeholder(
tf.float32, shape=(batch_size, image_size, image_size, num_channels))
tf_train_labels = tf.placeholder(tf.float32, shape=(batch_size, num_labels))
tf_valid_dataset = tf.constant(valid_dataset)
tf_test_dataset = tf.constant(test_dataset)
# Variables.
layer1_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, num_channels, depth], stddev=0.1))
layer1_biases = tf.Variable(tf.zeros([depth]))
layer2_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, depth, depth], stddev=0.1))
layer2_biases = tf.Variable(tf.constant(1.0, shape=[depth]))
layer3_weights = tf.Variable(tf.truncated_normal(
[image_size // 4 * image_size // 4 * depth, num_hidden], stddev=0.1))
layer3_biases = tf.Variable(tf.constant(1.0, shape=[num_hidden]))
layer4_weights = tf.Variable(tf.truncated_normal(
[num_hidden, num_labels], stddev=0.1))
layer4_biases = tf.Variable(tf.constant(1.0, shape=[num_labels]))
# Model.
def model(data):
conv = tf.nn.conv2d(data, layer1_weights, [1, 1, 1, 1], padding='SAME')
hidden = tf.nn.relu(conv + layer1_biases)
conv = tf.nn.conv2d(tf.nn.max_pool(hidden, [1,2,2,1], [1,2,2,1], 'SAME'),\
layer2_weights, [1, 1, 1, 1], padding='SAME')
hidden = tf.nn.relu(conv + layer2_biases)
shape = tf.nn.max_pool(hidden, [1,2,2,1], [1,2,2,1], 'SAME').get_shape().as_list()
reshape = tf.reshape(tf.nn.max_pool(hidden, [1,2,2,1], [1,2,2,1], 'SAME'),\
[shape[0], shape[1] * shape[2] * shape[3]])
hidden = tf.nn.relu(tf.matmul(reshape, layer3_weights) + layer3_biases)
return tf.matmul(hidden, layer4_weights) + layer4_biases
# Training computation.
logits = model(tf_train_dataset)
loss = tf.reduce_mean(
tf.nn.softmax_cross_entropy_with_logits(logits, tf_train_labels))
# Optimizer.
optimizer = tf.train.GradientDescentOptimizer(0.05).minimize(loss)
# Predictions for the training, validation, and test data.
train_prediction = tf.nn.softmax(logits)
valid_prediction = tf.nn.softmax(model(tf_valid_dataset))
test_prediction = tf.nn.softmax(model(tf_test_dataset))
num_steps = 1001
with tf.Session(graph=graph) as session:
tf.initialize_all_variables().run()
print('Initialized')
for step in range(num_steps):
offset = (step * batch_size) % (train_labels.shape[0] - batch_size)
batch_data = train_dataset[offset:(offset + batch_size), :, :, :]
batch_labels = train_labels[offset:(offset + batch_size), :]
feed_dict = {tf_train_dataset : batch_data, tf_train_labels : batch_labels}
_, l, predictions = session.run(
[optimizer, loss, train_prediction], feed_dict=feed_dict)
if (step % 50 == 0):
print('Minibatch loss at step %d: %f' % (step, l))
print('Minibatch accuracy: %.1f%%' % accuracy(predictions, batch_labels))
print('Validation accuracy: %.1f%%' % accuracy(
valid_prediction.eval(), valid_labels))
print('Test accuracy: %.1f%%' % accuracy(test_prediction.eval(), test_labels))
batch_size = 128
patch_size = 5
depth1 = 16
depth2 = 32
num_hidden1 = 128
num_hidden2 = 64
# Construct a 7-layer CNN.
# C1: convolutional layer, batch_size x 28 x 28 x 16, convolution size: 5 x 5 x 1 x 16
# S2: sub-sampling layer, batch_size x 14 x 14 x 16
# C3: convolutional layer, batch_size x 10 x 10 x 32, convolution size: 5 x 5 x 6 x 32
# S4: sub-sampling layer, batch_size x 5 x 5 x 32
# C5: convolutional layer, batch_size x 1 x 1 x 128, convolution size: 5 x 5 x 16 x 128
# F6: fully-connected layer, weight size: 128 x 64
# Output layer, weight size: 64 x 10
graph = tf.Graph()
with graph.as_default():
# Input data.
tf_train_dataset = tf.placeholder(
tf.float32, shape=(batch_size, image_size, image_size, num_channels))
tf_train_labels = tf.placeholder(tf.float32, shape=(batch_size, num_labels))
tf_valid_dataset = tf.constant(valid_dataset)
tf_test_dataset = tf.constant(test_dataset)
# Variables.
layer1_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, num_channels, depth1], stddev=0.1))
layer1_biases = tf.Variable(tf.zeros([depth1]))
layer2_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, depth1, depth2], stddev=0.1))
layer2_biases = tf.Variable(tf.constant(1.0, shape=[depth2]))
layer3_weights = tf.Variable(tf.truncated_normal(
[patch_size, patch_size, depth2, num_hidden1], stddev=0.1))
layer3_biases = tf.Variable(tf.constant(1.0, shape=[num_hidden1]))
layer4_weights = tf.Variable(tf.truncated_normal(
[num_hidden1, num_hidden2], stddev=0.1))
layer4_biases = tf.Variable(tf.constant(1.0, shape=[num_hidden2]))
layer5_weights = tf.Variable(tf.truncated_normal(
[num_hidden2, num_labels], stddev=0.1))
layer5_biases = tf.Variable(tf.constant(1.0, shape=[num_labels]))
# Model.
def model(data, keep_prob):
conv = tf.nn.conv2d(data, layer1_weights, [1,1,1,1], 'SAME', name='C1')
hidden = tf.nn.relu(conv + layer1_biases)
hidden = tf.nn.dropout(hidden, keep_prob)
sub = tf.nn.max_pool(hidden, [1,2,2,1], [1,2,2,1], 'SAME', name='S2')
conv = tf.nn.conv2d(sub, layer2_weights, [1,1,1,1], padding='VALID', name='C3')
hidden = tf.nn.relu(conv + layer2_biases)
hidden = tf.nn.dropout(hidden, keep_prob)
sub = tf.nn.max_pool(hidden, [1,2,2,1], [1,2,2,1], 'SAME', name='S4')
conv = tf.nn.conv2d(sub, layer3_weights, [1,1,1,1], padding='VALID', name='C5')
hidden = tf.nn.relu(conv + layer3_biases)
hidden = tf.nn.dropout(hidden, keep_prob)
shape = hidden.get_shape().as_list()
reshape = tf.reshape(hidden, [shape[0], shape[1] * shape[2] * shape[3]])
hidden = tf.nn.relu(tf.matmul(reshape, layer4_weights) + layer4_biases)
hidden = tf.nn.dropout(hidden, keep_prob)
return tf.matmul(hidden, layer5_weights) + layer5_biases
# Training computation.
logits = model(tf_train_dataset, 0.5)
loss = tf.reduce_mean(
tf.nn.softmax_cross_entropy_with_logits(logits, tf_train_labels))
# Optimizer.
#optimizer = tf.train.AdagradOptimizer(0.05).minimize(loss)
global_step = tf.Variable(0)
learning_rate = tf.train.exponential_decay(0.05, global_step, 2000, 0.95)
optimizer = tf.train.AdagradOptimizer(learning_rate).minimize(loss, global_step=global_step)
# Predictions for the training, validation, and test data.
train_prediction = tf.nn.softmax(model(tf_train_dataset, 1.0))
valid_prediction = tf.nn.softmax(model(tf_valid_dataset, 1.0))
test_prediction = tf.nn.softmax(model(tf_test_dataset, 1.0))
num_steps = 2001
with tf.Session(graph=graph) as session:
tf.initialize_all_variables().run()
print('Initialized')
for step in range(num_steps):
offset = (step * batch_size) % (train_labels.shape[0] - batch_size)
batch_data = train_dataset[offset:(offset + batch_size), :, :, :]
batch_labels = train_labels[offset:(offset + batch_size), :]
feed_dict = {tf_train_dataset : batch_data, tf_train_labels : batch_labels}
_, l, predictions = session.run(
[optimizer, loss, train_prediction], feed_dict=feed_dict)
if (step % 100 == 0):
print('Minibatch loss at step %d: %f' % (step, l))
print('Minibatch accuracy: %.1f%%' % accuracy(predictions, batch_labels))
print('Validation accuracy: %.1f%%' % accuracy(valid_prediction.eval(), valid_labels))
print('Test accuracy: %.1f%%' % accuracy(test_prediction.eval(), test_labels))
| 0.779783 | 0.934574 |
```
import numpy as np
import cv2
import misc
from RL_networks import Stand_alone_net
import pickle
```
### Naive Random Walk, judt push the images to the networks and see if they can learn to extract information fro m a random combination of images.
In this implimintation we have the Syclop run a random walk on the image, we will see if a basic CNN or RNN network can work with a changing path over the images.
<br> For each image a new, random, path will be initialized.
<br> This does not work at all, the networks run poorly compared to the case when the same random path is used for all images.
<br>
```
import importlib
importlib.reload(misc)
from __future__ import division, print_function, absolute_import
# PyTorch libraries and modules
import torchvision
import torch
import torch.nn as nn
from torch.utils.data import Dataset, DataLoader
from torch.autograd import Variable
from torch.nn import Linear, ReLU, CrossEntropyLoss, Sequential, Conv2d, MaxPool2d, Module, Softmax, BatchNorm2d, Dropout
from torch.optim import Adam, SGD
import torch.nn as nn
from mnist import MNIST
mnist = MNIST('/home/labs/ahissarlab/orra/datasets/mnist')
images, labels = mnist.load_training()
# Training Parameters
learning_rate = 0.001
num_steps = 1000
batch_size = 128
validation_index=-5000
# Network Parameters
size=None
padding_size=(128,128)
# num_input = padding_size[0]*padding_size[1] # MNIST data input (img shape: 28*28)
num_classes = None
# dropout = 0.25 # Dropout, probability to drop a unit
import matplotlib.pyplot as plt
%matplotlib notebook
import SYCLOP_env as syc
#Define function for low resolution lens on syclop
def bad_res101(img,res):
sh=np.shape(img)
dwnsmp=cv2.resize(img,res, interpolation = cv2.INTER_CUBIC)
upsmp = cv2.resize(dwnsmp,sh, interpolation = cv2.INTER_CUBIC)
return upsmp
plt.figure()
plt.imshow(misc.build_mnist_padded(1./256*np.reshape(images[0],[1,28,28])))
img=misc.build_mnist_padded(1./256*np.reshape(images[45],[1,28,28]))
scene = syc.Scene(image_matrix=img)
sensor = syc.Sensor(winx=56,winy=56,centralwinx=28,centralwiny=28)
agent = syc.Agent(max_q = [scene.maxx-sensor.hp.winx,scene.maxy-sensor.hp.winy])
starting_point = np.array([agent.max_q[0]//2,agent.max_q[1]//2])
steps = []
for j in range(5):
steps.append(starting_point*1)
starting_point += np.random.randint(-5,5,2)
q_sequence = np.array(steps).astype(int)
q_sequence
sensor.hp.resolution_fun = lambda x: bad_res101(x,(28,28))
imim=[]
dimim=[]
agent.set_manual_trajectory(manual_q_sequence=q_sequence)
for t in range(10):
agent.manual_act()
sensor.update(scene, agent)
imim.append(sensor.central_frame_view)
dimim.append(sensor.central_dvs_view)
for i in range(5):
plt.figure()
plt.imshow(imim[i])
```
### Create a Dataset from the Syclops visual inputs
We are starting with a simple time series where the syclop starts from the same starting point, at the middle of the img on the x axis and the middle - 10 pixles on the y axis - (middle_point, middle_point - 10)
<br> Each time step the syclop will move one pixle up on the y axis, to a final point at (middle_point, middle_point + 10) and make a circular movement in the x direction.
<br>
```
def create_dataset(images, labels, res, sample = 5, mixed_state = True, dvs = False):
'''
Creates a torch dataloader object of syclop outputs
from a list of images and labels.
Parameters
----------
images : List object holding the images to proces
labels : List object holding the labels
res : resolution dawnsampling factor - to be used in cv.resize(orig_img, res)
sample: the number of samples to have in syclop
mixed_state : if False, use the same trajectory on every image.
Returns
-------
train_dataloader, test_dataloader - torch DataLoader class objects
'''
count = 0
ts_images = []
dvs_images = []
count = 0
#create subplot to hold examples from the dataset
fig, ax = plt.subplots(2,5)
i = 0 #indexises for the subplot for image and for syclop vision
for img in images:
orig_img = np.reshape(img,[28,28])
#Set the padded image
img=misc.build_mnist_padded(1./256*np.reshape(img,[1,28,28]))
if count < 5:
ax[0,i].imshow(orig_img)
plt.title(labels[count])
#Set the sensor and the agent
scene = syc.Scene(image_matrix=img)
sensor = syc.Sensor(winx=56,winy=56,centralwinx=28,centralwiny=28)
agent = syc.Agent(max_q = [scene.maxx-sensor.hp.winx,scene.maxy-sensor.hp.winy])
#Setting the coordinates to visit
starting_point = np.array([agent.max_q[0]//2,agent.max_q[1]//2])
steps = []
for j in range(5):
steps.append(starting_point*1)
starting_point += np.random.randint(-5,5,2)
if mixed_state:
q_sequence = np.array(steps).astype(int)
else:
if count == 0:
q_sequence = np.array(steps).astype(int)
#Setting the resolution function - starting with the regular resolution
sensor.hp.resolution_fun = lambda x: bad_res101(x,(res,res))
#Create empty lists to store the syclops outputs
imim=[]
dimim=[]
agent.set_manual_trajectory(manual_q_sequence=q_sequence)
#Run Syclop for 20 time steps
for t in range(5):
agent.manual_act()
sensor.update(scene, agent)
imim.append(sensor.central_frame_view)
dimim.append(sensor.central_dvs_view)
#Create a unified matrix from the list
if count < 5:
ax[1,i].imshow(imim[0])
plt.title(labels[count])
i+=1
imim = np.array(imim)
dimim = np.array(dimim)
#Add current proccessed image to lists
ts_images.append(imim)
dvs_images.append(dimim)
count += 1
ts_train = ts_images[:55_000]
train_labels = labels[:55_000]
ts_val = ts_images[55_000:]
val_labels = labels[55_000:]
dvs_train = dvs_images[:55_000]
dvs_val = dvs_images[55_000:]
class mnist_dataset(Dataset):
def __init__(self, data, labels, transform = None):
self.data = data
self.labels = labels
self.transform = transform
def __len__(self):
return len(self.data)
def __getitem__(self, idx):
'''
args idx (int) : index
returns: tuple(data, label)
'''
data = self.data[idx]
label = self.labels[idx]
if self.transform:
data = self.transform(data)
return data, label
else:
return data, label
def dataset(self):
return self.data
def labels(self):
return self.labels
if dvs:
train_dataset = mnist_dataset(dvs_train, train_labels)
test_dataset = mnist_dataset(dvs_val, val_labels)
else:
train_dataset = mnist_dataset(ts_train, train_labels)
test_dataset = mnist_dataset(ts_val, val_labels)
batch = 64
train_dataloader = torch.utils.data.DataLoader(train_dataset, batch_size = batch, shuffle = True)
test_dataloader = torch.utils.data.DataLoader(test_dataset, batch_size = batch, shuffle = False)
return train_dataloader, test_dataloader
class CNN(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(5,16,3,stride=1, padding=1)
self.bn1 = nn.BatchNorm2d(16)
self.conv2 = nn.Conv2d(16,32,3, stride=1, padding=2)
self.bn2 = nn.BatchNorm2d(32)
self.conv3 = nn.Conv2d(32,16,3,stride=1, padding=1)
self.bn3 = nn.BatchNorm2d(16)
self.pool = nn.MaxPool2d(2)
#After the layers and pooling the first two we should get
# 16,3,3
#Flatting it we get:
# 144
self.fc1 = nn.Linear(8*8*16,64)
self.fc2 = nn.Linear(64,10)
self.relu = nn.ReLU()
def forward(self, img):
img = self.pool(self.relu(self.bn1(self.conv1(img.double()))))
img = self.pool(self.relu(self.bn2(self.conv2(img))))
img = self.relu(self.bn3(self.conv3(img)))
#print(img.shape)
img = img.view(img.shape[0],8*8*16)
img = self.relu(self.fc1(img))
img = self.fc2(img)
return img
class CNN_one_layer(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1,16,3,stride=1, padding=1)
self.bn1 = nn.BatchNorm2d(16)
self.conv2 = nn.Conv2d(16,32,3, stride=1, padding=2)
self.bn2 = nn.BatchNorm2d(32)
self.conv3 = nn.Conv2d(32,16,3,stride=1, padding=1)
self.bn3 = nn.BatchNorm2d(16)
self.pool = nn.MaxPool2d(2)
#After the layers and pooling the first two we should get
# 16,3,3
#Flatting it we get:
# 144
self.fc1 = nn.Linear(8*8*16,64)
self.fc2 = nn.Linear(64,10)
self.relu = nn.ReLU()
def forward(self, img):
img = img[:,0,:,:]
img = img.unsqueeze(1)
img = self.pool(self.relu(self.bn1(self.conv1(img.double()))))
img = self.pool(self.relu(self.bn2(self.conv2(img))))
img = self.relu(self.bn3(self.conv3(img)))
#print(img.shape)
img = img.view(img.shape[0],8*8*16)
img = self.relu(self.fc1(img))
img = self.fc2(img)
return img
def train(train_dataloader, test_dataloader, net, epochs = 10):
lr = 3e-3
#net = CNN().double()
optimizer = Adam(net.parameters(), lr = lr)
loss_func = nn.CrossEntropyLoss()
if torch.cuda.is_available():
net = net.cuda()
train_loss = []
test_loss = []
test_accur = []
for epoch in range(epochs):
batch_loss = []
for batch_idx, (data,targets) in enumerate(train_dataloader):
if net.__class__.__name__ == 'RNN_Net':
data = data.unsqueeze(2)
if torch.cuda.is_available():
data = data.to('cuda', non_blocking=True)
targets = targets.to('cuda', non_blocking = True)
#print(batch_idx, data.shape, targets.shape)
optimizer.zero_grad()
output = net(data.double())
loss = loss_func(output, targets)
loss.backward()
optimizer.step()
batch_loss.append(loss.item())
train_loss.append(np.mean(batch_loss))
if epoch%1 == 0:
correct = 0
test_batch_loss = []
test_accuracy = []
for batch_idx, (test_data,test_targets) in enumerate(test_dataloader):
if net.__class__.__name__ == 'RNN_Net':
test_data = test_data.unsqueeze(2)
if torch.cuda.is_available():
test_data = test_data.to('cuda', non_blocking=True)
test_targets = test_targets.to('cuda', non_blocking = True)
#print(batch_idx, data.shape, targets.shape)
test_output = net(test_data)
loss = loss_func(test_output, test_targets)
test_batch_loss.append(loss.item())
test_pred = test_output.data.max(1, keepdim = True)[1]
correct = test_pred.eq(test_targets.data.view_as(test_pred)).sum()
test_accuracy.append(100.*correct.to('cpu')/len(test_targets))
print('Net',net.__class__.__name__,'Epoch : ',epoch+1, '\t', 'loss :', loss.to('cpu').item(), 'accuracy :',np.mean(test_accuracy) )
test_loss.append(np.mean(test_batch_loss))
test_accur.append(np.mean(test_accuracy))
return train_loss, test_loss, test_accur
class RNN_Net(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1,16,3,stride=1, padding=1)
self.bn1 = nn.BatchNorm2d(16)
self.conv2 = nn.Conv2d(16,32,3, stride=1, padding=2)
self.bn2 = nn.BatchNorm2d(32)
self.conv3 = nn.Conv2d(32,16,3,stride=1, padding=1)
self.bn3 = nn.BatchNorm2d(16)
self.pool = nn.MaxPool2d(2)
#After the layers and pooling the first two we should get
# 16,3,3
#Flatting it we get:
# 144
self.gru = nn.GRU(8*8*16,100)
self.fc1 = nn.Linear(100,10)
#self.fc2 = nn.Linear(6,10)
self.relu = nn.ReLU()
def forward(self, data):
hn = torch.zeros([1,data.shape[0],100]).double()
if torch.cuda.is_available():
hn = hn.to('cuda')
#print(data.shape)
for i in range(data.shape[1]):
img = data[:,i,:,:,:]
#print(img.shape)
#plt.figure()
#plt.imshow(img[0][0])
img = self.pool(self.relu(self.bn1(self.conv1(img.double()))))
img = self.pool(self.relu(self.bn2(self.conv2(img))))
img = self.relu(self.bn3(self.conv3(img)))
img = img.view(img.shape[0],8*8*16)
out, hn = self.gru(img.unsqueeze(0),hn)
#print(out.shape)
output = self.fc1(out[0, :, :])
return output
class RNN_Net2(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1,4,3,stride=1, padding=1)
self.bn1 = nn.BatchNorm2d(16)
self.conv2 = nn.Conv2d(16,32,3, stride=1, padding=2)
self.bn2 = nn.BatchNorm2d(32)
self.conv3 = nn.Conv2d(32,16,3,stride=1, padding=1)
self.bn3 = nn.BatchNorm2d(16)
self.pool = nn.MaxPool2d(2)
#After the layers and pooling the first two we should get
# 16,3,3
#Flatting it we get:
# 144
self.gru = nn.GRU(8*8*16,100, batch_first=True)
self.fc1 = nn.Linear(100,10)
#self.fc2 = nn.Linear(6,10)
self.relu = nn.ReLU()
def forward(self, data):
batch_size, timesteps, C, H, W = data.size()
img = datax.view(batch_size * timesteps, C, H, W)
img = self.pool(self.relu(self.bn1(self.conv1(img.double()))))
img = self.pool(self.relu(self.bn2(self.conv2(img))))
img = self.relu(self.bn3(self.conv3(img)))
print(img.shape)
img = img.view(img.shape[0],img.shape[1],8*8*16)
out, hn = self.gru(img)
output = self.fc1(hn.squeeze(0))
return output
```
### Random Walk: Drew a new random walk for each image
```
#Load the training data
train_dataloader, test_dataloader = create_dataset(images, labels,res = 6)
#Run the CNN over one image only
train_loss, test_loss, test_accur = train(train_dataloader, test_dataloader,net = CNN_one_layer().double(), epochs = 10)
#Run the CNN over the stacked images 47-54.7
train_loss, test_loss, test_accur = train(train_dataloader, test_dataloader,net = CNN().double(), epochs = 10)
#Run the CNN+RNN over the stacked images
train_loss, test_loss, test_accur = train(train_dataloader, test_dataloader,net = RNN_Net().double(), epochs = 10)
```
### Random Walk: The same random trajectory on all images
```
#Run with a constant trajectory - test with a few trajectories to see is there is a change
cnn_one_image = []
cnn_lasagna = []
cnn_rnn = []
cnn_one_image_train_loss = []
cnn_lasagna_train_loss = []
cnn_rnn_train_loss = []
for i in range(4):
#Load the training data
train_dataloader, test_dataloader = create_dataset(images, labels,res = 6,mixed_state = False)
print('Try {} Run the CNN over one image only'.format(i))
cnn1_train_loss, cnn1_test_loss, cnn1_test_accur = train(train_dataloader, test_dataloader,net = CNN_one_layer().double(), epochs = 4)
cnn_one_image.append(cnn1_test_accur[-1])
cnn_one_image_train_loss.append(cnn1_train_loss[-1])
print('Try {} Run the CNN over the stacked images'.format(i))
cnn_train_loss, cnn_test_loss, cnn_test_accur = train(train_dataloader, test_dataloader,net = CNN().double(), epochs = 4)
cnn_lasagna.append(cnn_test_accur[-1])
cnn_lasagna_train_loss.append(cnn_train_loss[-1])
print('Try {} Run the CNN+RNN over the stacked images'.format(i))
rnn_train_loss, rnn_test_loss, rnn_test_accur = train(train_dataloader, test_dataloader,net = RNN_Net().double(), epochs = 4)
cnn_rnn.append(rnn_test_accur[-1])
cnn_rnn_train_loss.append(rnn_train_loss[-1])
```
|
github_jupyter
|
import numpy as np
import cv2
import misc
from RL_networks import Stand_alone_net
import pickle
import importlib
importlib.reload(misc)
from __future__ import division, print_function, absolute_import
# PyTorch libraries and modules
import torchvision
import torch
import torch.nn as nn
from torch.utils.data import Dataset, DataLoader
from torch.autograd import Variable
from torch.nn import Linear, ReLU, CrossEntropyLoss, Sequential, Conv2d, MaxPool2d, Module, Softmax, BatchNorm2d, Dropout
from torch.optim import Adam, SGD
import torch.nn as nn
from mnist import MNIST
mnist = MNIST('/home/labs/ahissarlab/orra/datasets/mnist')
images, labels = mnist.load_training()
# Training Parameters
learning_rate = 0.001
num_steps = 1000
batch_size = 128
validation_index=-5000
# Network Parameters
size=None
padding_size=(128,128)
# num_input = padding_size[0]*padding_size[1] # MNIST data input (img shape: 28*28)
num_classes = None
# dropout = 0.25 # Dropout, probability to drop a unit
import matplotlib.pyplot as plt
%matplotlib notebook
import SYCLOP_env as syc
#Define function for low resolution lens on syclop
def bad_res101(img,res):
sh=np.shape(img)
dwnsmp=cv2.resize(img,res, interpolation = cv2.INTER_CUBIC)
upsmp = cv2.resize(dwnsmp,sh, interpolation = cv2.INTER_CUBIC)
return upsmp
plt.figure()
plt.imshow(misc.build_mnist_padded(1./256*np.reshape(images[0],[1,28,28])))
img=misc.build_mnist_padded(1./256*np.reshape(images[45],[1,28,28]))
scene = syc.Scene(image_matrix=img)
sensor = syc.Sensor(winx=56,winy=56,centralwinx=28,centralwiny=28)
agent = syc.Agent(max_q = [scene.maxx-sensor.hp.winx,scene.maxy-sensor.hp.winy])
starting_point = np.array([agent.max_q[0]//2,agent.max_q[1]//2])
steps = []
for j in range(5):
steps.append(starting_point*1)
starting_point += np.random.randint(-5,5,2)
q_sequence = np.array(steps).astype(int)
q_sequence
sensor.hp.resolution_fun = lambda x: bad_res101(x,(28,28))
imim=[]
dimim=[]
agent.set_manual_trajectory(manual_q_sequence=q_sequence)
for t in range(10):
agent.manual_act()
sensor.update(scene, agent)
imim.append(sensor.central_frame_view)
dimim.append(sensor.central_dvs_view)
for i in range(5):
plt.figure()
plt.imshow(imim[i])
def create_dataset(images, labels, res, sample = 5, mixed_state = True, dvs = False):
'''
Creates a torch dataloader object of syclop outputs
from a list of images and labels.
Parameters
----------
images : List object holding the images to proces
labels : List object holding the labels
res : resolution dawnsampling factor - to be used in cv.resize(orig_img, res)
sample: the number of samples to have in syclop
mixed_state : if False, use the same trajectory on every image.
Returns
-------
train_dataloader, test_dataloader - torch DataLoader class objects
'''
count = 0
ts_images = []
dvs_images = []
count = 0
#create subplot to hold examples from the dataset
fig, ax = plt.subplots(2,5)
i = 0 #indexises for the subplot for image and for syclop vision
for img in images:
orig_img = np.reshape(img,[28,28])
#Set the padded image
img=misc.build_mnist_padded(1./256*np.reshape(img,[1,28,28]))
if count < 5:
ax[0,i].imshow(orig_img)
plt.title(labels[count])
#Set the sensor and the agent
scene = syc.Scene(image_matrix=img)
sensor = syc.Sensor(winx=56,winy=56,centralwinx=28,centralwiny=28)
agent = syc.Agent(max_q = [scene.maxx-sensor.hp.winx,scene.maxy-sensor.hp.winy])
#Setting the coordinates to visit
starting_point = np.array([agent.max_q[0]//2,agent.max_q[1]//2])
steps = []
for j in range(5):
steps.append(starting_point*1)
starting_point += np.random.randint(-5,5,2)
if mixed_state:
q_sequence = np.array(steps).astype(int)
else:
if count == 0:
q_sequence = np.array(steps).astype(int)
#Setting the resolution function - starting with the regular resolution
sensor.hp.resolution_fun = lambda x: bad_res101(x,(res,res))
#Create empty lists to store the syclops outputs
imim=[]
dimim=[]
agent.set_manual_trajectory(manual_q_sequence=q_sequence)
#Run Syclop for 20 time steps
for t in range(5):
agent.manual_act()
sensor.update(scene, agent)
imim.append(sensor.central_frame_view)
dimim.append(sensor.central_dvs_view)
#Create a unified matrix from the list
if count < 5:
ax[1,i].imshow(imim[0])
plt.title(labels[count])
i+=1
imim = np.array(imim)
dimim = np.array(dimim)
#Add current proccessed image to lists
ts_images.append(imim)
dvs_images.append(dimim)
count += 1
ts_train = ts_images[:55_000]
train_labels = labels[:55_000]
ts_val = ts_images[55_000:]
val_labels = labels[55_000:]
dvs_train = dvs_images[:55_000]
dvs_val = dvs_images[55_000:]
class mnist_dataset(Dataset):
def __init__(self, data, labels, transform = None):
self.data = data
self.labels = labels
self.transform = transform
def __len__(self):
return len(self.data)
def __getitem__(self, idx):
'''
args idx (int) : index
returns: tuple(data, label)
'''
data = self.data[idx]
label = self.labels[idx]
if self.transform:
data = self.transform(data)
return data, label
else:
return data, label
def dataset(self):
return self.data
def labels(self):
return self.labels
if dvs:
train_dataset = mnist_dataset(dvs_train, train_labels)
test_dataset = mnist_dataset(dvs_val, val_labels)
else:
train_dataset = mnist_dataset(ts_train, train_labels)
test_dataset = mnist_dataset(ts_val, val_labels)
batch = 64
train_dataloader = torch.utils.data.DataLoader(train_dataset, batch_size = batch, shuffle = True)
test_dataloader = torch.utils.data.DataLoader(test_dataset, batch_size = batch, shuffle = False)
return train_dataloader, test_dataloader
class CNN(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(5,16,3,stride=1, padding=1)
self.bn1 = nn.BatchNorm2d(16)
self.conv2 = nn.Conv2d(16,32,3, stride=1, padding=2)
self.bn2 = nn.BatchNorm2d(32)
self.conv3 = nn.Conv2d(32,16,3,stride=1, padding=1)
self.bn3 = nn.BatchNorm2d(16)
self.pool = nn.MaxPool2d(2)
#After the layers and pooling the first two we should get
# 16,3,3
#Flatting it we get:
# 144
self.fc1 = nn.Linear(8*8*16,64)
self.fc2 = nn.Linear(64,10)
self.relu = nn.ReLU()
def forward(self, img):
img = self.pool(self.relu(self.bn1(self.conv1(img.double()))))
img = self.pool(self.relu(self.bn2(self.conv2(img))))
img = self.relu(self.bn3(self.conv3(img)))
#print(img.shape)
img = img.view(img.shape[0],8*8*16)
img = self.relu(self.fc1(img))
img = self.fc2(img)
return img
class CNN_one_layer(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1,16,3,stride=1, padding=1)
self.bn1 = nn.BatchNorm2d(16)
self.conv2 = nn.Conv2d(16,32,3, stride=1, padding=2)
self.bn2 = nn.BatchNorm2d(32)
self.conv3 = nn.Conv2d(32,16,3,stride=1, padding=1)
self.bn3 = nn.BatchNorm2d(16)
self.pool = nn.MaxPool2d(2)
#After the layers and pooling the first two we should get
# 16,3,3
#Flatting it we get:
# 144
self.fc1 = nn.Linear(8*8*16,64)
self.fc2 = nn.Linear(64,10)
self.relu = nn.ReLU()
def forward(self, img):
img = img[:,0,:,:]
img = img.unsqueeze(1)
img = self.pool(self.relu(self.bn1(self.conv1(img.double()))))
img = self.pool(self.relu(self.bn2(self.conv2(img))))
img = self.relu(self.bn3(self.conv3(img)))
#print(img.shape)
img = img.view(img.shape[0],8*8*16)
img = self.relu(self.fc1(img))
img = self.fc2(img)
return img
def train(train_dataloader, test_dataloader, net, epochs = 10):
lr = 3e-3
#net = CNN().double()
optimizer = Adam(net.parameters(), lr = lr)
loss_func = nn.CrossEntropyLoss()
if torch.cuda.is_available():
net = net.cuda()
train_loss = []
test_loss = []
test_accur = []
for epoch in range(epochs):
batch_loss = []
for batch_idx, (data,targets) in enumerate(train_dataloader):
if net.__class__.__name__ == 'RNN_Net':
data = data.unsqueeze(2)
if torch.cuda.is_available():
data = data.to('cuda', non_blocking=True)
targets = targets.to('cuda', non_blocking = True)
#print(batch_idx, data.shape, targets.shape)
optimizer.zero_grad()
output = net(data.double())
loss = loss_func(output, targets)
loss.backward()
optimizer.step()
batch_loss.append(loss.item())
train_loss.append(np.mean(batch_loss))
if epoch%1 == 0:
correct = 0
test_batch_loss = []
test_accuracy = []
for batch_idx, (test_data,test_targets) in enumerate(test_dataloader):
if net.__class__.__name__ == 'RNN_Net':
test_data = test_data.unsqueeze(2)
if torch.cuda.is_available():
test_data = test_data.to('cuda', non_blocking=True)
test_targets = test_targets.to('cuda', non_blocking = True)
#print(batch_idx, data.shape, targets.shape)
test_output = net(test_data)
loss = loss_func(test_output, test_targets)
test_batch_loss.append(loss.item())
test_pred = test_output.data.max(1, keepdim = True)[1]
correct = test_pred.eq(test_targets.data.view_as(test_pred)).sum()
test_accuracy.append(100.*correct.to('cpu')/len(test_targets))
print('Net',net.__class__.__name__,'Epoch : ',epoch+1, '\t', 'loss :', loss.to('cpu').item(), 'accuracy :',np.mean(test_accuracy) )
test_loss.append(np.mean(test_batch_loss))
test_accur.append(np.mean(test_accuracy))
return train_loss, test_loss, test_accur
class RNN_Net(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1,16,3,stride=1, padding=1)
self.bn1 = nn.BatchNorm2d(16)
self.conv2 = nn.Conv2d(16,32,3, stride=1, padding=2)
self.bn2 = nn.BatchNorm2d(32)
self.conv3 = nn.Conv2d(32,16,3,stride=1, padding=1)
self.bn3 = nn.BatchNorm2d(16)
self.pool = nn.MaxPool2d(2)
#After the layers and pooling the first two we should get
# 16,3,3
#Flatting it we get:
# 144
self.gru = nn.GRU(8*8*16,100)
self.fc1 = nn.Linear(100,10)
#self.fc2 = nn.Linear(6,10)
self.relu = nn.ReLU()
def forward(self, data):
hn = torch.zeros([1,data.shape[0],100]).double()
if torch.cuda.is_available():
hn = hn.to('cuda')
#print(data.shape)
for i in range(data.shape[1]):
img = data[:,i,:,:,:]
#print(img.shape)
#plt.figure()
#plt.imshow(img[0][0])
img = self.pool(self.relu(self.bn1(self.conv1(img.double()))))
img = self.pool(self.relu(self.bn2(self.conv2(img))))
img = self.relu(self.bn3(self.conv3(img)))
img = img.view(img.shape[0],8*8*16)
out, hn = self.gru(img.unsqueeze(0),hn)
#print(out.shape)
output = self.fc1(out[0, :, :])
return output
class RNN_Net2(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1,4,3,stride=1, padding=1)
self.bn1 = nn.BatchNorm2d(16)
self.conv2 = nn.Conv2d(16,32,3, stride=1, padding=2)
self.bn2 = nn.BatchNorm2d(32)
self.conv3 = nn.Conv2d(32,16,3,stride=1, padding=1)
self.bn3 = nn.BatchNorm2d(16)
self.pool = nn.MaxPool2d(2)
#After the layers and pooling the first two we should get
# 16,3,3
#Flatting it we get:
# 144
self.gru = nn.GRU(8*8*16,100, batch_first=True)
self.fc1 = nn.Linear(100,10)
#self.fc2 = nn.Linear(6,10)
self.relu = nn.ReLU()
def forward(self, data):
batch_size, timesteps, C, H, W = data.size()
img = datax.view(batch_size * timesteps, C, H, W)
img = self.pool(self.relu(self.bn1(self.conv1(img.double()))))
img = self.pool(self.relu(self.bn2(self.conv2(img))))
img = self.relu(self.bn3(self.conv3(img)))
print(img.shape)
img = img.view(img.shape[0],img.shape[1],8*8*16)
out, hn = self.gru(img)
output = self.fc1(hn.squeeze(0))
return output
#Load the training data
train_dataloader, test_dataloader = create_dataset(images, labels,res = 6)
#Run the CNN over one image only
train_loss, test_loss, test_accur = train(train_dataloader, test_dataloader,net = CNN_one_layer().double(), epochs = 10)
#Run the CNN over the stacked images 47-54.7
train_loss, test_loss, test_accur = train(train_dataloader, test_dataloader,net = CNN().double(), epochs = 10)
#Run the CNN+RNN over the stacked images
train_loss, test_loss, test_accur = train(train_dataloader, test_dataloader,net = RNN_Net().double(), epochs = 10)
#Run with a constant trajectory - test with a few trajectories to see is there is a change
cnn_one_image = []
cnn_lasagna = []
cnn_rnn = []
cnn_one_image_train_loss = []
cnn_lasagna_train_loss = []
cnn_rnn_train_loss = []
for i in range(4):
#Load the training data
train_dataloader, test_dataloader = create_dataset(images, labels,res = 6,mixed_state = False)
print('Try {} Run the CNN over one image only'.format(i))
cnn1_train_loss, cnn1_test_loss, cnn1_test_accur = train(train_dataloader, test_dataloader,net = CNN_one_layer().double(), epochs = 4)
cnn_one_image.append(cnn1_test_accur[-1])
cnn_one_image_train_loss.append(cnn1_train_loss[-1])
print('Try {} Run the CNN over the stacked images'.format(i))
cnn_train_loss, cnn_test_loss, cnn_test_accur = train(train_dataloader, test_dataloader,net = CNN().double(), epochs = 4)
cnn_lasagna.append(cnn_test_accur[-1])
cnn_lasagna_train_loss.append(cnn_train_loss[-1])
print('Try {} Run the CNN+RNN over the stacked images'.format(i))
rnn_train_loss, rnn_test_loss, rnn_test_accur = train(train_dataloader, test_dataloader,net = RNN_Net().double(), epochs = 4)
cnn_rnn.append(rnn_test_accur[-1])
cnn_rnn_train_loss.append(rnn_train_loss[-1])
| 0.635222 | 0.859664 |
<h1>Using pre-trained embeddings with TensorFlow Hub</h1>
This notebook illustrates:
<ol>
<li>How to instantiate a TensorFlow Hub module</li>
<li>How to find pre-trained TensorFlow Hub modules for a variety of purposes</li>
<li>How to examine the embeddings of a Hub module</li>
<li>How one Hub module composes representations of sentences from individual words</li>
<li>How to assess word embeddings using a semantic similarity test</li>
</ol>
```
# change these to try this notebook out# chang
BUCKET = 'cloud-training-demos-ml'
PROJECT = 'cloud-training-demos'
REGION = 'us-central1'
```
Install the TensorFlow Hub library
```
!pip install -q tensorflow-hub
import tensorflow as tf
import tensorflow_hub as hub
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import re
import seaborn as sns
import scipy
import math
os.environ['BUCKET'] = BUCKET
os.environ['PROJECT'] = PROJECT
os.environ['REGION'] = REGION
os.environ['TFVERSION'] = '1.8'
import tensorflow as tf
print(tf.__version__)
```
<h2>TensorFlow Hub Concepts</h2>
TensorFlow Hub is a library for the publication, discovery, and consumption of reusable parts of machine learning models. A module is a self-contained piece of a TensorFlow graph, along with its weights and assets, that can be reused across different tasks in a process known as transfer learning, which we covered as part of the course on Image Models.
To download and use a module, it's as easy as:
However, because modules are self-contained parts of a TensorFlow graph, in order to actually collect values from a module, you'll need to evaluate it in the context of a session.
First, let's explore what hub modules there are. Go to https://www.tensorflow.org/hub/modules/ and explore a bit.
Note that TensorFlow Hub has modules for Images, Text, and Other. In this case, we're interested in a Text module, so navigate to the Text section.
Within the Text section, there are a number of modules. If you click on a link, you'll be taken to a page that describes the module and links to the original paper where the model was proposed. Click on a model in the Word2Vec section of the page.
Note the details section, which describes what the module expects as input, how it preprocesses data, what it does when it encounters a word it hasn't seen before (OOV means "out of vocabulary") and in this case, how word embeddings can be composed to form sentence embeddings.
Finally, note the URL of the page. This is the URL you can copy to instantiate your module.
<h2>Task 1: Create an embedding using the NNLM model</h2>
To complete this task:
<ol>
<li>Find the module URL for the NNLM 50 dimensional English model</li>
<li>Use it to instantiate a module as 'embed'</li>
<li>Print the embedded representation of "cat"</li>
</ol>
NOTE: downloading hub modules requires downloading a lot of data. Instantiating the module will take a few minutes.
```
# Task 1
import tensorflow as tf
import tensorflow_hub as hub
module_url = "https://tfhub.dev/google/nnlm-en-dim50/1"
embed = hub.Module(module_url)
embeddings = embed(["cat"])
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
sess.run(tf.tables_initializer())
print(sess.run(embeddings))
```
When I completed this exercise, I got a vector that looked like `[[ 0.11233182 -0.3176392 -0.0166118...]]`
<h2>Task 2: Assess the Embeddings Informally</h2>
<ol>
<li>Identify some words to test</li>
<li>Retrieve the embeddings for each word</li>
<li>Determine what method to use to compare each pair of embeddings</li>
</ol>
So, now we have some vectors but the question is, are they any good? One way of testing whether they are any good is to try them for your task. But, first, let's just take a peak.
For our test, we'll need three common words such that two of the words are much closer in meaning than the third.
```
word_1 = "cat"
word_2 = "dog"
word_3 = "potato"
```
Now, we'll use the same process of using our Hub module to generate embeddings but instead of printing the embeddings, capture them in a variable called 'my_embeddings'.
```
# Task 2b
# Reduce logging output.
tf.logging.set_verbosity(tf.logging.ERROR)
messages = [word_1, word_2, word_3]
def create_embeddings(messages, embed):
my_embeddings = None
with tf.Session() as session:
session.run([tf.global_variables_initializer(), tf.tables_initializer()])
message_embeddings = session.run(embed(messages))
my_embeddings = np.array(message_embeddings)
for i, message_embedding in enumerate(np.array(my_embeddings).tolist()):
print("Message: {}".format(messages[i]))
print("Embedding size: {}".format(len(message_embedding)))
message_embedding_snippet = ", ".join(
(str(x) for x in message_embedding[:3]))
print("Embedding: [{}, ...]\n".format(message_embedding_snippet))
return my_embeddings
my_embeddings = create_embeddings(messages, embed)
```
Now, we'll use Seaborn's heatmap function to see how the vectors compare to each other. I've written the shell of a function that you'll need to complete that will generate a heatmap. The one piece that's missing is how we'll compare each pair of vectors. Note that because we are computing a score for every pair of vectors, we should have len(my_embeddings)^2 scores. There are many valid ways of comparing vectors. Generality, similarity scores are symmetric. The simplest is to take their dot product. For extra credit, implement a more complicated vector comparison function.
```
def plot_similarity(labels, embeddings):
corr = np.inner(embeddings, embeddings)
sns.set(font_scale=1.2)
g = sns.heatmap(
corr,
xticklabels=labels,
yticklabels=labels,
vmin=0,
vmax=1,
cmap="YlOrRd")
g.set_xticklabels(labels, rotation=90)
g.set_title("Semantic Textual Similarity")
plot_similarity([word_1, word_2, word_3], my_embeddings)
```
What you should observe is that, trivially, all words are identical to themselves, and, more interestingly, that the two more similar words have more similar embeddings than the third word.
<h2>Task 3: From Words to Sentences</h2>
Up until now, we've used our module to produce representations of words. But, in fact, if we want to, we can also use it to construct representations of sentences. The methods used by the module to compose a representation of a sentence won't be as nuanced as what an RNN might do, but they are still worth examining because they are so convenient.
<ol>
<li> Examine the documentation for our hub module and determine how to ask it to construct a representation of a sentence</li>
<li> Figure out how the module takes word embeddings and uses them to construct sentence embeddings </li>
<li> Construct a embeddings of a "cat", "The cat sat on the mat", "dog" and "The cat sat on the dog" and plot their similarity
</ol>
```
messages = ["cat", "The cat sat on the mat", "dog", "The cat sat on the dog"]
my_embeddings = create_embeddings(messages, embed)
plot_similarity(messages, my_embeddings)
```
Which is cat more similar to, "The cat sat on the mat" or "dog"? Is this desireable?
Think back to how an RNN scans a sequence and maintains its state. Naive methods of embedding composition (mapping many to one) can't possibly compete with a network trained for this very purpose!
<h2>Task 4: Assessing the Embeddings Formally</h2>
Of course, it's great to know that our embeddings match our intuitions to an extent, but it'd be better to have a formal, data-driven measure of the quality of the representation.
Researchers have
The STS Benchmark http://ixa2.si.ehu.es/stswiki/index.php/STSbenchmark provides an intristic evaluation of the degree to which similarity scores computed using sentence embeddings align with human judgements. The benchmark requires systems to return similarity scores for a diverse selection of sentence pairs. Pearson correlation is then used to evaluate the quality of the machine similarity scores against human judgements.
```
def load_sts_dataset(filename):
# Loads a subset of the STS dataset into a DataFrame. In particular both
# sentences and their human rated similarity score.
sent_pairs = []
with tf.gfile.GFile(filename, "r") as f:
for line in f:
ts = line.strip().split("\t")
# (sent_1, sent_2, similarity_score)
sent_pairs.append((ts[5], ts[6], float(ts[4])))
return pd.DataFrame(sent_pairs, columns=["sent_1", "sent_2", "sim"])
def download_and_load_sts_data():
sts_dataset = tf.keras.utils.get_file(
fname="Stsbenchmark.tar.gz",
origin="http://ixa2.si.ehu.es/stswiki/images/4/48/Stsbenchmark.tar.gz",
extract=True)
sts_dev = load_sts_dataset(
os.path.join(os.path.dirname(sts_dataset), "stsbenchmark", "sts-dev.csv"))
sts_test = load_sts_dataset(
os.path.join(
os.path.dirname(sts_dataset), "stsbenchmark", "sts-test.csv"))
return sts_dev, sts_test
sts_dev, sts_test = download_and_load_sts_data()
```
Take a look at the data. The sim score is higher when the sentences are more similar and lower when they are not.
```
sts_dev.head()
```
<h3>Build the Evaluation Graph</h3>
Next, we need to build the evaluation graph.
```
sts_input1 = tf.placeholder(tf.string, shape=(None))
sts_input2 = tf.placeholder(tf.string, shape=(None))
# For evaluation we use exactly normalized rather than
# approximately normalized.
sts_encode1 = tf.nn.l2_normalize(embed(sts_input1), axis=1)
sts_encode2 = tf.nn.l2_normalize(embed(sts_input2), axis=1)
cosine_similarities = tf.reduce_sum(tf.multiply(sts_encode1, sts_encode2), axis=1)
clip_cosine_similarities = tf.clip_by_value(cosine_similarities, -1.0, 1.0)
sim_scores = 1.0 - tf.acos(clip_cosine_similarities)
```
<h3>Evaluate Sentence Embeddings</h3>
Finally, we need to create a session and run our evaluation.
```
sts_data = sts_dev #@param ["sts_dev", "sts_test"] {type:"raw"}
text_a = sts_data['sent_1'].tolist()
text_b = sts_data['sent_2'].tolist()
dev_scores = sts_data['sim'].tolist()
def run_sts_benchmark(session):
"""Returns the similarity scores"""
emba, embb, scores = session.run(
[sts_encode1, sts_encode2, sim_scores],
feed_dict={
sts_input1: text_a,
sts_input2: text_b
})
return scores
with tf.Session() as session:
session.run(tf.global_variables_initializer())
session.run(tf.tables_initializer())
scores = run_sts_benchmark(session)
pearson_correlation = scipy.stats.pearsonr(scores, dev_scores)
print('Pearson correlation coefficient = {0}\np-value = {1}'.format(
pearson_correlation[0], pearson_correlation[1]))
```
<h3>Extra Credit</h3>
For extra credit, re-run this analysis with a different Hub module. Are the results different? If so, how?
<h2>Further Reading</h2>
We published a [blog post](https://developers.googleblog.com/2018/04/text-embedding-models-contain-bias.html) on how bias can affect text embeddings. It's worth a read!
|
github_jupyter
|
# change these to try this notebook out# chang
BUCKET = 'cloud-training-demos-ml'
PROJECT = 'cloud-training-demos'
REGION = 'us-central1'
!pip install -q tensorflow-hub
import tensorflow as tf
import tensorflow_hub as hub
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import re
import seaborn as sns
import scipy
import math
os.environ['BUCKET'] = BUCKET
os.environ['PROJECT'] = PROJECT
os.environ['REGION'] = REGION
os.environ['TFVERSION'] = '1.8'
import tensorflow as tf
print(tf.__version__)
# Task 1
import tensorflow as tf
import tensorflow_hub as hub
module_url = "https://tfhub.dev/google/nnlm-en-dim50/1"
embed = hub.Module(module_url)
embeddings = embed(["cat"])
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
sess.run(tf.tables_initializer())
print(sess.run(embeddings))
word_1 = "cat"
word_2 = "dog"
word_3 = "potato"
# Task 2b
# Reduce logging output.
tf.logging.set_verbosity(tf.logging.ERROR)
messages = [word_1, word_2, word_3]
def create_embeddings(messages, embed):
my_embeddings = None
with tf.Session() as session:
session.run([tf.global_variables_initializer(), tf.tables_initializer()])
message_embeddings = session.run(embed(messages))
my_embeddings = np.array(message_embeddings)
for i, message_embedding in enumerate(np.array(my_embeddings).tolist()):
print("Message: {}".format(messages[i]))
print("Embedding size: {}".format(len(message_embedding)))
message_embedding_snippet = ", ".join(
(str(x) for x in message_embedding[:3]))
print("Embedding: [{}, ...]\n".format(message_embedding_snippet))
return my_embeddings
my_embeddings = create_embeddings(messages, embed)
def plot_similarity(labels, embeddings):
corr = np.inner(embeddings, embeddings)
sns.set(font_scale=1.2)
g = sns.heatmap(
corr,
xticklabels=labels,
yticklabels=labels,
vmin=0,
vmax=1,
cmap="YlOrRd")
g.set_xticklabels(labels, rotation=90)
g.set_title("Semantic Textual Similarity")
plot_similarity([word_1, word_2, word_3], my_embeddings)
messages = ["cat", "The cat sat on the mat", "dog", "The cat sat on the dog"]
my_embeddings = create_embeddings(messages, embed)
plot_similarity(messages, my_embeddings)
def load_sts_dataset(filename):
# Loads a subset of the STS dataset into a DataFrame. In particular both
# sentences and their human rated similarity score.
sent_pairs = []
with tf.gfile.GFile(filename, "r") as f:
for line in f:
ts = line.strip().split("\t")
# (sent_1, sent_2, similarity_score)
sent_pairs.append((ts[5], ts[6], float(ts[4])))
return pd.DataFrame(sent_pairs, columns=["sent_1", "sent_2", "sim"])
def download_and_load_sts_data():
sts_dataset = tf.keras.utils.get_file(
fname="Stsbenchmark.tar.gz",
origin="http://ixa2.si.ehu.es/stswiki/images/4/48/Stsbenchmark.tar.gz",
extract=True)
sts_dev = load_sts_dataset(
os.path.join(os.path.dirname(sts_dataset), "stsbenchmark", "sts-dev.csv"))
sts_test = load_sts_dataset(
os.path.join(
os.path.dirname(sts_dataset), "stsbenchmark", "sts-test.csv"))
return sts_dev, sts_test
sts_dev, sts_test = download_and_load_sts_data()
sts_dev.head()
sts_input1 = tf.placeholder(tf.string, shape=(None))
sts_input2 = tf.placeholder(tf.string, shape=(None))
# For evaluation we use exactly normalized rather than
# approximately normalized.
sts_encode1 = tf.nn.l2_normalize(embed(sts_input1), axis=1)
sts_encode2 = tf.nn.l2_normalize(embed(sts_input2), axis=1)
cosine_similarities = tf.reduce_sum(tf.multiply(sts_encode1, sts_encode2), axis=1)
clip_cosine_similarities = tf.clip_by_value(cosine_similarities, -1.0, 1.0)
sim_scores = 1.0 - tf.acos(clip_cosine_similarities)
sts_data = sts_dev #@param ["sts_dev", "sts_test"] {type:"raw"}
text_a = sts_data['sent_1'].tolist()
text_b = sts_data['sent_2'].tolist()
dev_scores = sts_data['sim'].tolist()
def run_sts_benchmark(session):
"""Returns the similarity scores"""
emba, embb, scores = session.run(
[sts_encode1, sts_encode2, sim_scores],
feed_dict={
sts_input1: text_a,
sts_input2: text_b
})
return scores
with tf.Session() as session:
session.run(tf.global_variables_initializer())
session.run(tf.tables_initializer())
scores = run_sts_benchmark(session)
pearson_correlation = scipy.stats.pearsonr(scores, dev_scores)
print('Pearson correlation coefficient = {0}\np-value = {1}'.format(
pearson_correlation[0], pearson_correlation[1]))
| 0.78233 | 0.97382 |
# Introduction to Analysis of Algorithms
## What is an algorithm?
- unambiguous, finite sequence of instructions to solve a problem
- Keywords:
- unambiguous
- finite
- problem
- sequence (semantically "sequential" steps; loops are allowed)
## What is a problem?
- Given all possible inputs, how do we achieve the desired output?
- Keywords:
- desired
- inputs/outputs
- We will be studying the 'best' algorithms for a host of problems
## What does best mean?
- Fastest
- Smallest amount of memory
- Easiest to implement and to read
- Most general
## Why study algorithms?
- Saves time, resources and therefore money
- Without an efficient algorithm, regardless of how fast the hardware is, some problems are unsolvable
>We should forget about small efficiencies, say about 97% of the time: premature optimization is the root of all evil. >Yet we should not pass up our opportunities in that critical 3%. - Donald Knuth
## Parallel Algorithms
- Warmup:
- Given an array, will a `in` operation be faster if we have more than one CPU
- Will two cores for an array of n elements be computed more quickly?
- How about n cores for n elements?
- `O(1)` time for checking each element, but doing a boolean && may take n operations
- How to synchronize and what about overhead?
## Scientific Method applied to Algorithm Design
1. Outline specification - input, output, "best"
2. Design an algorithm with the goal in mind
3. Prove that the algorithm is correct
4. Does it satisfy the spec?
5. If not, go back to 2. and make iterative improvements
6. Otherwise, code the algorithm
## Miscellaneous Insights/Notes
### Sieve of Erastothenes
```
Input: an integer n > 1.
Let A be an array of Boolean values, indexed by integers 2 to n,
initially all set to true.
for i = 2, 3, 4, ..., not exceeding √n:
if A[i] is true:
for j = i2, i2+i, i2+2i, i2+3i, ..., not exceeding n:
A[j] := false.
Output: all i such that A[i] is true.
```
Note that we start from $i^2$ because all of the factors of $i$ from $2i$, $3i$ etc. to $(i-1)i$ have already been eliminated.
### Amortized Analysis
Best illustrated by an example
- Resizing array
- To resize, when the array is full, we double the size. Adding elements is `O(1)`, but doubling the size involves copying n elements across, which takes n operations depending on n.
- This would indicate that adding n elements to a resizing array is `O(log n)`
- However, this is unfair, as after each double, we get to add many more elements.
- For example, doubling from 8 to 16 allows 8 more additional elements at `O(1)` to be added.
- If we divide number of cumulative operations / number of elements, we find that this number is constant (if we ignore minimal array overhead)
- This is amortized analysis: in practical application, doubling an array is so infrequent and also opens up more potential operations that it makes more sense to evaluate this operation as having **constant amortized time**
|
github_jupyter
| 0.096376 | 0.967686 |
|
```
from scipy.io import wavfile
import wave
import webrtcvad
import numpy as np
import librosa
import webrtcvad
import IPython.display as ipd
import glob
def find_silences(filename):
global args
blend_duration = 0.005
with wave.open(filename) as wav:
size = wav.getnframes()
channels = wav.getnchannels()
sample_width = wav.getsampwidth()
frame_rate = wav.getframerate()
max_value = 1 << (8 * sample_width - 1)
half_blend_frames = int(blend_duration * frame_rate / 2)
blend_frames = half_blend_frames * 2
assert size > blend_frames > 0
square_threshold = max_value ** 2 * 10 ** (args.threshold_level / 10)
blend_squares = collections.deque()
blend = 0
def get_values():
frames_read = 0
while frames_read < size:
frames = wav.readframes(min(0x1000, size - frames_read))
frames_count = len(frames) // sample_width // channels
for frame_index in range(frames_count):
yield frames[frame_index*channels*sample_width:(frame_index+1)*channels*sample_width]
frames_read += frames_count
def get_is_silence(blend):
results = 0
frames = get_values()
for index in range(half_blend_frames):
frame = next(frames)
square = 0
for channel in range(channels):
value = int.from_bytes(frame[sample_width*channel:sample_width*channel+sample_width], 'little', signed=True)
square += value*value
blend_squares.append(square)
blend += square
for index in range(size-half_blend_frames):
frame = next(frames)
square = 0
for channel in range(channels):
value = int.from_bytes(frame[sample_width*channel:sample_width*channel+sample_width], 'little', signed=True)
square += value*value
blend_squares.append(square)
blend += square
if index < half_blend_frames:
yield blend < square_threshold * channels * (half_blend_frames + index + 1)
else:
result = blend < square_threshold * channels * (blend_frames + 1)
if result:
results += 1
yield result
blend -= blend_squares.popleft()
for index in range(half_blend_frames):
blend -= blend_squares.popleft()
yield blend < square_threshold * channels * (blend_frames - index)
is_silence = get_is_silence(blend)
def to_regions(iterable):
iterator = enumerate(iterable)
while True:
try:
index, value = next(iterator)
except StopIteration:
return
if value:
start = index
while True:
try:
index, value = next(iterator)
if not value:
yield start, index
break
except StopIteration:
yield start, index+1
return
threshold_frames = int(args.threshold_duration * frame_rate)
silence_regions = ( (start, end) for start, end in to_regions(is_silence) if end-start >= blend_duration )
silence_regions = ( (start + (half_blend_frames if start > 0 else 0), end - (half_blend_frames if end < size else 0)) for start, end in silence_regions )
silence_regions = [ (start, end) for start, end in silence_regions if end-start >= threshold_frames ]
including_end = len(silence_regions) == 0 or silence_regions[-1][1] == size
silence_regions = [ (start/frame_rate, end/frame_rate) for start, end in silence_regions ]
# print(args.save_silence)
if args.save_silence:
with wave.open(args.save_silence, 'wb') as out_wav:
out_wav.setnchannels(channels)
out_wav.setsampwidth(sample_width)
out_wav.setframerate(frame_rate)
for start, end in silence_regions:
wav.setpos(start)
frames = wav.readframes(end-start)
out_wav.writeframes(frames)
return silence_regions, including_end
def transform_duration(duration):
global args
return args.constant + args.sublinear * math.log(duration + 1) + args.linear * duration
def format_offset(offset):
return '{}:{}:{}'.format(int(offset) // 3600, int(offset) % 3600 // 60, offset % 60)
def closest_frames(duration, frame_rate):
return int((duration + 1 / frame_rate / 2) // (1 / frame_rate))
def compress_audio(wav, start_frame, end_frame, result_frames):
# print(start_frame, end_frame, result_frames)
if result_frames == 0:
return b''
elif result_frames == end_frame - start_frame:
# print("same")
wav.setpos(start_frame)
return wav.readframes(result_frames)
else:
channels = wav.getnchannels()
sample_width = wav.getsampwidth()
frame_width = sample_width*channels
if result_frames*2 <= end_frame - start_frame:
left_length = result_frames
right_length = result_frames
else:
left_length = (end_frame - start_frame + 1) // 2
right_length = end_frame - start_frame - left_length
crossfade_length = right_length + left_length - result_frames
crossfade_start = (result_frames - crossfade_length) // 2
wav.setpos(start_frame)
left_frames = wav.readframes(left_length)
wav.setpos(end_frame - right_length)
right_frames = wav.readframes(right_length)
result = bytearray(b'\x00'*result_frames*frame_width)
result[:(left_length-crossfade_length)*frame_width] = left_frames[:-crossfade_length*frame_width]
result[-(right_length-crossfade_length)*frame_width:] = right_frames[crossfade_length*frame_width:]
for i in range(crossfade_length):
r = i / (crossfade_length - 1)
l = 1 - r
for channel in range(channels):
signal_left = int.from_bytes(left_frames[(left_length-crossfade_length+i)*frame_width+channel*sample_width:(left_length-crossfade_length+i)*frame_width+(channel+1)*sample_width], 'little', signed=True)
signal_right = int.from_bytes(right_frames[i*frame_width+channel*sample_width:i*frame_width+(channel+1)*sample_width], 'little', signed=True)
result[(left_length-crossfade_length+i)*frame_width+channel*sample_width:(left_length-crossfade_length+i)*frame_width+(channel+1)*sample_width] = int(signal_left*l + signal_right*r).to_bytes(sample_width, 'little', signed=True)
return result
class Frame(object):
"""Represents a "frame" of audio data."""
def __init__(self, bytes, timestamp, duration):
self.bytes = bytes
self.timestamp = timestamp
self.duration = duration
def frame_generator(frame_duration_ms, audio, sample_rate):
frames = []
n = int(sample_rate * (frame_duration_ms / 1000.0) * 2)
offset = 0
timestamp = 0.0
duration = (float(n) / sample_rate) / 2.0
while offset + n < len(audio):
frames.append(Frame(audio[offset:offset + n], timestamp, duration))
timestamp += duration
offset += n
return frames
import argparse
parser = argparse.ArgumentParser()
parser.add_argument('-path', type=str, default='files/', help='path to video')
parser.add_argument('-threshold-duration', type=float, default=0.2, help='threshold duration in seconds')
parser.add_argument('-check', type=bool, default=True, help='path to text file')
parser.add_argument('-p', type=str, default='results/', help='path to video')
parser.add_argument('--threshold-level', type=float, default=-35, help='threshold level in dB')
parser.add_argument('--constant', type=float, default=0, help='duration constant transform value')
parser.add_argument('--sublinear', type=float, default=0, help='duration sublinear transform factor')
parser.add_argument('--linear', type=float, default=0.1, help='duration linear transform factor')
parser.add_argument('--save-silence', type=str, help='filename for saving silence')
parser.add_argument('--recalculate-time-in-description', type=str, help='path to text file')
parser.add_argument('-f')
args = parser.parse_args()
from scipy.io.wavfile import write
import os
import wave
from pydub import AudioSegment
import collections
dir_path = args.path +'*.wav'
paths = glob.glob(dir_path)
for path in paths:
# print(path)
sample_rate, samples = wavfile.read(path)
# print('sample rate : {}, samples.shape : {}'.format(sample_rate, samples.shape))
vad = webrtcvad.Vad()
vad.set_mode(3)
# 10, 20, or 30
frame_duration = 10 # ms
frames = frame_generator(frame_duration, samples, sample_rate)
flag = True
for i, frame in enumerate(frames):
if vad.is_speech(frame.bytes, sample_rate):
if flag:
start_idx = i
flag = False
else:
end_idx = i
if start_idx > 1:
start_idx -= 1
if end_idx < len(frames):
end_idx += 1
audio_start_frame = int(start_idx/100.0*sample_rate*2)
audio_end_frame = int(end_idx/100.0*sample_rate*2)
audio_result_frames = audio_end_frame - audio_start_frame
dst = args.p + path.split(args.path)[-1]
if not os.path.isdir(args.p):
os.mkdir(args.p)
wav = wave.open(path, mode='rb')
out_wav = wave.open(dst, mode='wb')
channels = wav.getnchannels()
sample_width = wav.getsampwidth()
audio_frame_rate = wav.getframerate()
out_wav.setnchannels(channels)
out_wav.setsampwidth(sample_width)
out_wav.setframerate(audio_frame_rate)
out_wav.writeframes(compress_audio(wav, audio_start_frame, audio_end_frame, audio_result_frames))
# write(dst, sample_rate, samples_cut)
out_wav.close()
silences, including_end = find_silences(dst)
if silences[0][0] == 0.0:
start_gap = silences[0][1] - silences[0][0]
else:
start_gap = 0
if including_end:
end_gap = silences[-1][1] - silences[-1][0]
else:
end_gap = 0
seg = AudioSegment.silent(duration=200)
song = AudioSegment.from_wav(dst)
if start_gap < 0.2:
start_gap += 0.2
song = seg + song
if end_gap < 0.2:
end_gap += 0.2
song = song + seg
song.export(dst, format="wav")
# print(start_gap, end_gap)
import librosa
sample_rate, samples = wavfile.read(path)
print(sample_rate)
y, sr = librosa.load(path, sr=sample_rate)
resample = librosa.resample(y, sr, 16000)
```
|
github_jupyter
|
from scipy.io import wavfile
import wave
import webrtcvad
import numpy as np
import librosa
import webrtcvad
import IPython.display as ipd
import glob
def find_silences(filename):
global args
blend_duration = 0.005
with wave.open(filename) as wav:
size = wav.getnframes()
channels = wav.getnchannels()
sample_width = wav.getsampwidth()
frame_rate = wav.getframerate()
max_value = 1 << (8 * sample_width - 1)
half_blend_frames = int(blend_duration * frame_rate / 2)
blend_frames = half_blend_frames * 2
assert size > blend_frames > 0
square_threshold = max_value ** 2 * 10 ** (args.threshold_level / 10)
blend_squares = collections.deque()
blend = 0
def get_values():
frames_read = 0
while frames_read < size:
frames = wav.readframes(min(0x1000, size - frames_read))
frames_count = len(frames) // sample_width // channels
for frame_index in range(frames_count):
yield frames[frame_index*channels*sample_width:(frame_index+1)*channels*sample_width]
frames_read += frames_count
def get_is_silence(blend):
results = 0
frames = get_values()
for index in range(half_blend_frames):
frame = next(frames)
square = 0
for channel in range(channels):
value = int.from_bytes(frame[sample_width*channel:sample_width*channel+sample_width], 'little', signed=True)
square += value*value
blend_squares.append(square)
blend += square
for index in range(size-half_blend_frames):
frame = next(frames)
square = 0
for channel in range(channels):
value = int.from_bytes(frame[sample_width*channel:sample_width*channel+sample_width], 'little', signed=True)
square += value*value
blend_squares.append(square)
blend += square
if index < half_blend_frames:
yield blend < square_threshold * channels * (half_blend_frames + index + 1)
else:
result = blend < square_threshold * channels * (blend_frames + 1)
if result:
results += 1
yield result
blend -= blend_squares.popleft()
for index in range(half_blend_frames):
blend -= blend_squares.popleft()
yield blend < square_threshold * channels * (blend_frames - index)
is_silence = get_is_silence(blend)
def to_regions(iterable):
iterator = enumerate(iterable)
while True:
try:
index, value = next(iterator)
except StopIteration:
return
if value:
start = index
while True:
try:
index, value = next(iterator)
if not value:
yield start, index
break
except StopIteration:
yield start, index+1
return
threshold_frames = int(args.threshold_duration * frame_rate)
silence_regions = ( (start, end) for start, end in to_regions(is_silence) if end-start >= blend_duration )
silence_regions = ( (start + (half_blend_frames if start > 0 else 0), end - (half_blend_frames if end < size else 0)) for start, end in silence_regions )
silence_regions = [ (start, end) for start, end in silence_regions if end-start >= threshold_frames ]
including_end = len(silence_regions) == 0 or silence_regions[-1][1] == size
silence_regions = [ (start/frame_rate, end/frame_rate) for start, end in silence_regions ]
# print(args.save_silence)
if args.save_silence:
with wave.open(args.save_silence, 'wb') as out_wav:
out_wav.setnchannels(channels)
out_wav.setsampwidth(sample_width)
out_wav.setframerate(frame_rate)
for start, end in silence_regions:
wav.setpos(start)
frames = wav.readframes(end-start)
out_wav.writeframes(frames)
return silence_regions, including_end
def transform_duration(duration):
global args
return args.constant + args.sublinear * math.log(duration + 1) + args.linear * duration
def format_offset(offset):
return '{}:{}:{}'.format(int(offset) // 3600, int(offset) % 3600 // 60, offset % 60)
def closest_frames(duration, frame_rate):
return int((duration + 1 / frame_rate / 2) // (1 / frame_rate))
def compress_audio(wav, start_frame, end_frame, result_frames):
# print(start_frame, end_frame, result_frames)
if result_frames == 0:
return b''
elif result_frames == end_frame - start_frame:
# print("same")
wav.setpos(start_frame)
return wav.readframes(result_frames)
else:
channels = wav.getnchannels()
sample_width = wav.getsampwidth()
frame_width = sample_width*channels
if result_frames*2 <= end_frame - start_frame:
left_length = result_frames
right_length = result_frames
else:
left_length = (end_frame - start_frame + 1) // 2
right_length = end_frame - start_frame - left_length
crossfade_length = right_length + left_length - result_frames
crossfade_start = (result_frames - crossfade_length) // 2
wav.setpos(start_frame)
left_frames = wav.readframes(left_length)
wav.setpos(end_frame - right_length)
right_frames = wav.readframes(right_length)
result = bytearray(b'\x00'*result_frames*frame_width)
result[:(left_length-crossfade_length)*frame_width] = left_frames[:-crossfade_length*frame_width]
result[-(right_length-crossfade_length)*frame_width:] = right_frames[crossfade_length*frame_width:]
for i in range(crossfade_length):
r = i / (crossfade_length - 1)
l = 1 - r
for channel in range(channels):
signal_left = int.from_bytes(left_frames[(left_length-crossfade_length+i)*frame_width+channel*sample_width:(left_length-crossfade_length+i)*frame_width+(channel+1)*sample_width], 'little', signed=True)
signal_right = int.from_bytes(right_frames[i*frame_width+channel*sample_width:i*frame_width+(channel+1)*sample_width], 'little', signed=True)
result[(left_length-crossfade_length+i)*frame_width+channel*sample_width:(left_length-crossfade_length+i)*frame_width+(channel+1)*sample_width] = int(signal_left*l + signal_right*r).to_bytes(sample_width, 'little', signed=True)
return result
class Frame(object):
"""Represents a "frame" of audio data."""
def __init__(self, bytes, timestamp, duration):
self.bytes = bytes
self.timestamp = timestamp
self.duration = duration
def frame_generator(frame_duration_ms, audio, sample_rate):
frames = []
n = int(sample_rate * (frame_duration_ms / 1000.0) * 2)
offset = 0
timestamp = 0.0
duration = (float(n) / sample_rate) / 2.0
while offset + n < len(audio):
frames.append(Frame(audio[offset:offset + n], timestamp, duration))
timestamp += duration
offset += n
return frames
import argparse
parser = argparse.ArgumentParser()
parser.add_argument('-path', type=str, default='files/', help='path to video')
parser.add_argument('-threshold-duration', type=float, default=0.2, help='threshold duration in seconds')
parser.add_argument('-check', type=bool, default=True, help='path to text file')
parser.add_argument('-p', type=str, default='results/', help='path to video')
parser.add_argument('--threshold-level', type=float, default=-35, help='threshold level in dB')
parser.add_argument('--constant', type=float, default=0, help='duration constant transform value')
parser.add_argument('--sublinear', type=float, default=0, help='duration sublinear transform factor')
parser.add_argument('--linear', type=float, default=0.1, help='duration linear transform factor')
parser.add_argument('--save-silence', type=str, help='filename for saving silence')
parser.add_argument('--recalculate-time-in-description', type=str, help='path to text file')
parser.add_argument('-f')
args = parser.parse_args()
from scipy.io.wavfile import write
import os
import wave
from pydub import AudioSegment
import collections
dir_path = args.path +'*.wav'
paths = glob.glob(dir_path)
for path in paths:
# print(path)
sample_rate, samples = wavfile.read(path)
# print('sample rate : {}, samples.shape : {}'.format(sample_rate, samples.shape))
vad = webrtcvad.Vad()
vad.set_mode(3)
# 10, 20, or 30
frame_duration = 10 # ms
frames = frame_generator(frame_duration, samples, sample_rate)
flag = True
for i, frame in enumerate(frames):
if vad.is_speech(frame.bytes, sample_rate):
if flag:
start_idx = i
flag = False
else:
end_idx = i
if start_idx > 1:
start_idx -= 1
if end_idx < len(frames):
end_idx += 1
audio_start_frame = int(start_idx/100.0*sample_rate*2)
audio_end_frame = int(end_idx/100.0*sample_rate*2)
audio_result_frames = audio_end_frame - audio_start_frame
dst = args.p + path.split(args.path)[-1]
if not os.path.isdir(args.p):
os.mkdir(args.p)
wav = wave.open(path, mode='rb')
out_wav = wave.open(dst, mode='wb')
channels = wav.getnchannels()
sample_width = wav.getsampwidth()
audio_frame_rate = wav.getframerate()
out_wav.setnchannels(channels)
out_wav.setsampwidth(sample_width)
out_wav.setframerate(audio_frame_rate)
out_wav.writeframes(compress_audio(wav, audio_start_frame, audio_end_frame, audio_result_frames))
# write(dst, sample_rate, samples_cut)
out_wav.close()
silences, including_end = find_silences(dst)
if silences[0][0] == 0.0:
start_gap = silences[0][1] - silences[0][0]
else:
start_gap = 0
if including_end:
end_gap = silences[-1][1] - silences[-1][0]
else:
end_gap = 0
seg = AudioSegment.silent(duration=200)
song = AudioSegment.from_wav(dst)
if start_gap < 0.2:
start_gap += 0.2
song = seg + song
if end_gap < 0.2:
end_gap += 0.2
song = song + seg
song.export(dst, format="wav")
# print(start_gap, end_gap)
import librosa
sample_rate, samples = wavfile.read(path)
print(sample_rate)
y, sr = librosa.load(path, sr=sample_rate)
resample = librosa.resample(y, sr, 16000)
| 0.479504 | 0.369315 |
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#Case-1---Coronary-Heart-Disease-Classification" data-toc-modified-id="Case-1---Coronary-Heart-Disease-Classification-1"><span class="toc-item-num">1 </span>Case 1 - Coronary Heart Disease Classification</a></span></li><li><span><a href="#Background" data-toc-modified-id="Background-2"><span class="toc-item-num">2 </span>Background</a></span></li><li><span><a href="#Model-1:-Imputing-the-missing-values-with-Zero-and-preprocessing-data-using-StandardScaler" data-toc-modified-id="Model-1:-Imputing-the-missing-values-with-Zero-and-preprocessing-data-using-StandardScaler-3"><span class="toc-item-num">3 </span>Model 1: Imputing the missing values with Zero and preprocessing data using StandardScaler</a></span><ul class="toc-item"><li><span><a href="#Data" data-toc-modified-id="Data-3.1"><span class="toc-item-num">3.1 </span>Data</a></span><ul class="toc-item"><li><span><a href="#Read-and-prepare-the-dataset" data-toc-modified-id="Read-and-prepare-the-dataset-3.1.1"><span class="toc-item-num">3.1.1 </span>Read and prepare the dataset</a></span><ul class="toc-item"><li><span><a href="#Check-missing-values-in-the-dataset" data-toc-modified-id="Check-missing-values-in-the-dataset-3.1.1.1"><span class="toc-item-num">3.1.1.1 </span>Check missing values in the dataset</a></span></li><li><span><a href="#Fill-the-missing-value-with-zero" data-toc-modified-id="Fill-the-missing-value-with-zero-3.1.1.2"><span class="toc-item-num">3.1.1.2 </span>Fill the missing value with zero</a></span></li><li><span><a href="#Separate-Data-and-Label" data-toc-modified-id="Separate-Data-and-Label-3.1.1.3"><span class="toc-item-num">3.1.1.3 </span>Separate Data and Label</a></span></li><li><span><a href="#Treat-any-value-higher-than-one-as-one" data-toc-modified-id="Treat-any-value-higher-than-one-as-one-3.1.1.4"><span class="toc-item-num">3.1.1.4 </span>Treat any value higher than one as one</a></span></li></ul></li><li><span><a href="#Data-Preprocessing" data-toc-modified-id="Data-Preprocessing-3.1.2"><span class="toc-item-num">3.1.2 </span>Data Preprocessing</a></span><ul class="toc-item"><li><span><a href="#Convert-Dataframe-to-Numpy-array" data-toc-modified-id="Convert-Dataframe-to-Numpy-array-3.1.2.1"><span class="toc-item-num">3.1.2.1 </span>Convert Dataframe to Numpy array</a></span></li><li><span><a href="#Preprocessing-data-using-StandardScaler" data-toc-modified-id="Preprocessing-data-using-StandardScaler-3.1.2.2"><span class="toc-item-num">3.1.2.2 </span>Preprocessing data using StandardScaler</a></span></li></ul></li></ul></li><li><span><a href="#Models-and-Training" data-toc-modified-id="Models-and-Training-3.2"><span class="toc-item-num">3.2 </span>Models and Training</a></span><ul class="toc-item"><li><span><a href="#Split-data-into-Training-set,-Validation-set-and-Testing-set" data-toc-modified-id="Split-data-into-Training-set,-Validation-set-and-Testing-set-3.2.1"><span class="toc-item-num">3.2.1 </span>Split data into Training set, Validation set and Testing set</a></span></li><li><span><a href="#Construct-the-Model-Architecture" data-toc-modified-id="Construct-the-Model-Architecture-3.2.2"><span class="toc-item-num">3.2.2 </span>Construct the Model Architecture</a></span></li><li><span><a href="#Build-the-Model" data-toc-modified-id="Build-the-Model-3.2.3"><span class="toc-item-num">3.2.3 </span>Build the Model</a></span></li></ul></li><li><span><a href="#Results" data-toc-modified-id="Results-3.3"><span class="toc-item-num">3.3 </span>Results</a></span><ul class="toc-item"><li><span><a href="#Plot-the-training-and-validation-Loss-and-Accuracy-Graph" data-toc-modified-id="Plot-the-training-and-validation-Loss-and-Accuracy-Graph-3.3.1"><span class="toc-item-num">3.3.1 </span>Plot the training and validation Loss and Accuracy Graph</a></span></li><li><span><a href="#Cut-off-the-first-few-data-points" data-toc-modified-id="Cut-off-the-first-few-data-points-3.3.2"><span class="toc-item-num">3.3.2 </span>Cut off the first few data points</a></span></li><li><span><a href="#Smoothen-the-previous-curve-for-clarity" data-toc-modified-id="Smoothen-the-previous-curve-for-clarity-3.3.3"><span class="toc-item-num">3.3.3 </span>Smoothen the previous curve for clarity</a></span></li><li><span><a href="#Rebuild-the-Model" data-toc-modified-id="Rebuild-the-Model-3.3.4"><span class="toc-item-num">3.3.4 </span>Rebuild the Model</a></span></li><li><span><a href="#Test-the-Model" data-toc-modified-id="Test-the-Model-3.3.5"><span class="toc-item-num">3.3.5 </span>Test the Model</a></span></li></ul></li></ul></li><li><span><a href="#Model-2:-Imputing-the-missing-values-with-Zero-and-preprocessing-data-using-Normalization" data-toc-modified-id="Model-2:-Imputing-the-missing-values-with-Zero-and-preprocessing-data-using-Normalization-4"><span class="toc-item-num">4 </span>Model 2: Imputing the missing values with Zero and preprocessing data using Normalization</a></span><ul class="toc-item"><li><span><a href="#Data" data-toc-modified-id="Data-4.1"><span class="toc-item-num">4.1 </span>Data</a></span><ul class="toc-item"><li><span><a href="#Data-preprocessing" data-toc-modified-id="Data-preprocessing-4.1.1"><span class="toc-item-num">4.1.1 </span>Data preprocessing</a></span><ul class="toc-item"><li><span><a href="#Preprocessing-data-using-Normalization" data-toc-modified-id="Preprocessing-data-using-Normalization-4.1.1.1"><span class="toc-item-num">4.1.1.1 </span>Preprocessing data using Normalization</a></span></li></ul></li></ul></li><li><span><a href="#Models-and-Training" data-toc-modified-id="Models-and-Training-4.2"><span class="toc-item-num">4.2 </span>Models and Training</a></span><ul class="toc-item"><li><span><a href="#Split-data-into-Training-set,-Validation-set-and-Testing-set" data-toc-modified-id="Split-data-into-Training-set,-Validation-set-and-Testing-set-4.2.1"><span class="toc-item-num">4.2.1 </span>Split data into Training set, Validation set and Testing set</a></span></li><li><span><a href="#Construct-the-Model-Architecture" data-toc-modified-id="Construct-the-Model-Architecture-4.2.2"><span class="toc-item-num">4.2.2 </span>Construct the Model Architecture</a></span></li><li><span><a href="#Build-the-Model" data-toc-modified-id="Build-the-Model-4.2.3"><span class="toc-item-num">4.2.3 </span>Build the Model</a></span></li></ul></li><li><span><a href="#Results" data-toc-modified-id="Results-4.3"><span class="toc-item-num">4.3 </span>Results</a></span><ul class="toc-item"><li><span><a href="#Plot-the-training-and-validation-Loss-and-Accuracy-Graph" data-toc-modified-id="Plot-the-training-and-validation-Loss-and-Accuracy-Graph-4.3.1"><span class="toc-item-num">4.3.1 </span>Plot the training and validation Loss and Accuracy Graph</a></span></li><li><span><a href="#Cut-off-the-first-few-data-points" data-toc-modified-id="Cut-off-the-first-few-data-points-4.3.2"><span class="toc-item-num">4.3.2 </span>Cut off the first few data points</a></span></li><li><span><a href="#Smoothen-the-previous-curve-for-clarity" data-toc-modified-id="Smoothen-the-previous-curve-for-clarity-4.3.3"><span class="toc-item-num">4.3.3 </span>Smoothen the previous curve for clarity</a></span></li><li><span><a href="#Rebuild-the-Model" data-toc-modified-id="Rebuild-the-Model-4.3.4"><span class="toc-item-num">4.3.4 </span>Rebuild the Model</a></span></li><li><span><a href="#Test-the-Model" data-toc-modified-id="Test-the-Model-4.3.5"><span class="toc-item-num">4.3.5 </span>Test the Model</a></span></li></ul></li></ul></li><li><span><a href="#Model-3:-Imputing-the-missing-values-with-Mean-and-Preprocessing-data-using-StandardScaler" data-toc-modified-id="Model-3:-Imputing-the-missing-values-with-Mean-and-Preprocessing-data-using-StandardScaler-5"><span class="toc-item-num">5 </span>Model 3: Imputing the missing values with Mean and Preprocessing data using StandardScaler</a></span><ul class="toc-item"><li><span><a href="#Data" data-toc-modified-id="Data-5.1"><span class="toc-item-num">5.1 </span>Data</a></span><ul class="toc-item"><li><span><a href="#Read-in-the-dataset-from-.csv-file" data-toc-modified-id="Read-in-the-dataset-from-.csv-file-5.1.1"><span class="toc-item-num">5.1.1 </span>Read in the dataset from .csv file</a></span><ul class="toc-item"><li><span><a href="#Fill-the-missing-value-with-mean" data-toc-modified-id="Fill-the-missing-value-with-mean-5.1.1.1"><span class="toc-item-num">5.1.1.1 </span>Fill the missing value with mean</a></span></li><li><span><a href="#Separate-Data-and-Label" data-toc-modified-id="Separate-Data-and-Label-5.1.1.2"><span class="toc-item-num">5.1.1.2 </span>Separate Data and Label</a></span></li><li><span><a href="#Treat-any-value-higher-than-one-as-one" data-toc-modified-id="Treat-any-value-higher-than-one-as-one-5.1.1.3"><span class="toc-item-num">5.1.1.3 </span>Treat any value higher than one as one</a></span></li></ul></li><li><span><a href="#Data-Preprocessing" data-toc-modified-id="Data-Preprocessing-5.1.2"><span class="toc-item-num">5.1.2 </span>Data Preprocessing</a></span><ul class="toc-item"><li><span><a href="#Convert-Dataframe-to-Numpy-Array" data-toc-modified-id="Convert-Dataframe-to-Numpy-Array-5.1.2.1"><span class="toc-item-num">5.1.2.1 </span>Convert Dataframe to Numpy Array</a></span></li><li><span><a href="#Preprocessing-data-using-StandardScaler" data-toc-modified-id="Preprocessing-data-using-StandardScaler-5.1.2.2"><span class="toc-item-num">5.1.2.2 </span>Preprocessing data using StandardScaler</a></span></li></ul></li></ul></li><li><span><a href="#Models-and-Training" data-toc-modified-id="Models-and-Training-5.2"><span class="toc-item-num">5.2 </span>Models and Training</a></span><ul class="toc-item"><li><span><a href="#Split-data-into-Training-set,-Validation-set-and-Testing-set" data-toc-modified-id="Split-data-into-Training-set,-Validation-set-and-Testing-set-5.2.1"><span class="toc-item-num">5.2.1 </span>Split data into Training set, Validation set and Testing set</a></span></li><li><span><a href="#Construct-the-Model-Architecture" data-toc-modified-id="Construct-the-Model-Architecture-5.2.2"><span class="toc-item-num">5.2.2 </span>Construct the Model Architecture</a></span></li><li><span><a href="#Build-the-Model" data-toc-modified-id="Build-the-Model-5.2.3"><span class="toc-item-num">5.2.3 </span>Build the Model</a></span></li></ul></li><li><span><a href="#Results" data-toc-modified-id="Results-5.3"><span class="toc-item-num">5.3 </span>Results</a></span><ul class="toc-item"><li><span><a href="#Plot-the-training-and-validation-Loss-and-Accuracy-Graph" data-toc-modified-id="Plot-the-training-and-validation-Loss-and-Accuracy-Graph-5.3.1"><span class="toc-item-num">5.3.1 </span>Plot the training and validation Loss and Accuracy Graph</a></span></li><li><span><a href="#Cut-off-the-first-few-data-points" data-toc-modified-id="Cut-off-the-first-few-data-points-5.3.2"><span class="toc-item-num">5.3.2 </span>Cut off the first few data points</a></span></li><li><span><a href="#Smoothen-the-previous-curve-for-clarity" data-toc-modified-id="Smoothen-the-previous-curve-for-clarity-5.3.3"><span class="toc-item-num">5.3.3 </span>Smoothen the previous curve for clarity</a></span></li><li><span><a href="#Rebuild-the-Model" data-toc-modified-id="Rebuild-the-Model-5.3.4"><span class="toc-item-num">5.3.4 </span>Rebuild the Model</a></span></li><li><span><a href="#Test-the-Model" data-toc-modified-id="Test-the-Model-5.3.5"><span class="toc-item-num">5.3.5 </span>Test the Model</a></span></li></ul></li></ul></li><li><span><a href="#Model-4:-Imputing-the-missing-values-with-Mean-and-preprocessing-data-using-Normalization" data-toc-modified-id="Model-4:-Imputing-the-missing-values-with-Mean-and-preprocessing-data-using-Normalization-6"><span class="toc-item-num">6 </span>Model 4: Imputing the missing values with Mean and preprocessing data using Normalization</a></span><ul class="toc-item"><li><span><a href="#Data" data-toc-modified-id="Data-6.1"><span class="toc-item-num">6.1 </span>Data</a></span><ul class="toc-item"><li><span><a href="#Data-preprocessing" data-toc-modified-id="Data-preprocessing-6.1.1"><span class="toc-item-num">6.1.1 </span>Data preprocessing</a></span><ul class="toc-item"><li><span><a href="#Preprocessing-data-using-Normalization" data-toc-modified-id="Preprocessing-data-using-Normalization-6.1.1.1"><span class="toc-item-num">6.1.1.1 </span>Preprocessing data using Normalization</a></span></li></ul></li></ul></li><li><span><a href="#Models-and-Training" data-toc-modified-id="Models-and-Training-6.2"><span class="toc-item-num">6.2 </span>Models and Training</a></span><ul class="toc-item"><li><span><a href="#Split-data-into-Training-set,-Validation-set-and-Testing-set" data-toc-modified-id="Split-data-into-Training-set,-Validation-set-and-Testing-set-6.2.1"><span class="toc-item-num">6.2.1 </span>Split data into Training set, Validation set and Testing set</a></span></li><li><span><a href="#Construct-the-Model-Architecture" data-toc-modified-id="Construct-the-Model-Architecture-6.2.2"><span class="toc-item-num">6.2.2 </span>Construct the Model Architecture</a></span></li><li><span><a href="#Build-the-Model" data-toc-modified-id="Build-the-Model-6.2.3"><span class="toc-item-num">6.2.3 </span>Build the Model</a></span></li></ul></li><li><span><a href="#Results" data-toc-modified-id="Results-6.3"><span class="toc-item-num">6.3 </span>Results</a></span><ul class="toc-item"><li><span><a href="#Plot-the-training-and-validation-Loss-and-Accuracy-Graph" data-toc-modified-id="Plot-the-training-and-validation-Loss-and-Accuracy-Graph-6.3.1"><span class="toc-item-num">6.3.1 </span>Plot the training and validation Loss and Accuracy Graph</a></span></li><li><span><a href="#Cut-off-the-first-few-data-points" data-toc-modified-id="Cut-off-the-first-few-data-points-6.3.2"><span class="toc-item-num">6.3.2 </span>Cut off the first few data points</a></span></li><li><span><a href="#Smoothen-the-previous-curve-for-clarity" data-toc-modified-id="Smoothen-the-previous-curve-for-clarity-6.3.3"><span class="toc-item-num">6.3.3 </span>Smoothen the previous curve for clarity</a></span></li><li><span><a href="#Rebuild-the-Model" data-toc-modified-id="Rebuild-the-Model-6.3.4"><span class="toc-item-num">6.3.4 </span>Rebuild the Model</a></span></li><li><span><a href="#Test-the-Model" data-toc-modified-id="Test-the-Model-6.3.5"><span class="toc-item-num">6.3.5 </span>Test the Model</a></span></li></ul></li></ul></li><li><span><a href="#Model-5:-Imputing-the-missing-values-with-Median-and-preprocessing-data-using-StandardScaler" data-toc-modified-id="Model-5:-Imputing-the-missing-values-with-Median-and-preprocessing-data-using-StandardScaler-7"><span class="toc-item-num">7 </span>Model 5: Imputing the missing values with Median and preprocessing data using StandardScaler</a></span><ul class="toc-item"><li><span><a href="#Data" data-toc-modified-id="Data-7.1"><span class="toc-item-num">7.1 </span>Data</a></span><ul class="toc-item"><li><span><a href="#Read-in-the-dataset-from-.csv-file" data-toc-modified-id="Read-in-the-dataset-from-.csv-file-7.1.1"><span class="toc-item-num">7.1.1 </span>Read in the dataset from .csv file</a></span><ul class="toc-item"><li><span><a href="#Fill-the-missing-value-with-mean" data-toc-modified-id="Fill-the-missing-value-with-mean-7.1.1.1"><span class="toc-item-num">7.1.1.1 </span>Fill the missing value with mean</a></span></li><li><span><a href="#Separate-Data-and-Label" data-toc-modified-id="Separate-Data-and-Label-7.1.1.2"><span class="toc-item-num">7.1.1.2 </span>Separate Data and Label</a></span></li><li><span><a href="#Treat-any-value-higher-than-one-as-one" data-toc-modified-id="Treat-any-value-higher-than-one-as-one-7.1.1.3"><span class="toc-item-num">7.1.1.3 </span>Treat any value higher than one as one</a></span></li></ul></li><li><span><a href="#Data-Preprocessing" data-toc-modified-id="Data-Preprocessing-7.1.2"><span class="toc-item-num">7.1.2 </span>Data Preprocessing</a></span><ul class="toc-item"><li><span><a href="#Convert-Dataframe-to-Numpy-Array" data-toc-modified-id="Convert-Dataframe-to-Numpy-Array-7.1.2.1"><span class="toc-item-num">7.1.2.1 </span>Convert Dataframe to Numpy Array</a></span></li><li><span><a href="#Preprocessing-data-using-StandardScaler" data-toc-modified-id="Preprocessing-data-using-StandardScaler-7.1.2.2"><span class="toc-item-num">7.1.2.2 </span>Preprocessing data using StandardScaler</a></span></li></ul></li></ul></li><li><span><a href="#Models-and-Training" data-toc-modified-id="Models-and-Training-7.2"><span class="toc-item-num">7.2 </span>Models and Training</a></span><ul class="toc-item"><li><span><a href="#Split-data-into-Training-set,-Validation-set-and-Testing-set" data-toc-modified-id="Split-data-into-Training-set,-Validation-set-and-Testing-set-7.2.1"><span class="toc-item-num">7.2.1 </span>Split data into Training set, Validation set and Testing set</a></span></li><li><span><a href="#Construct-the-Model-Architecture" data-toc-modified-id="Construct-the-Model-Architecture-7.2.2"><span class="toc-item-num">7.2.2 </span>Construct the Model Architecture</a></span></li><li><span><a href="#Build-the-Model" data-toc-modified-id="Build-the-Model-7.2.3"><span class="toc-item-num">7.2.3 </span>Build the Model</a></span></li></ul></li><li><span><a href="#Results" data-toc-modified-id="Results-7.3"><span class="toc-item-num">7.3 </span>Results</a></span><ul class="toc-item"><li><span><a href="#Plot-the-training-and-validation-Loss-and-Accuracy-Graph" data-toc-modified-id="Plot-the-training-and-validation-Loss-and-Accuracy-Graph-7.3.1"><span class="toc-item-num">7.3.1 </span>Plot the training and validation Loss and Accuracy Graph</a></span></li><li><span><a href="#Cut-off-the-first-few-data-points" data-toc-modified-id="Cut-off-the-first-few-data-points-7.3.2"><span class="toc-item-num">7.3.2 </span>Cut off the first few data points</a></span></li><li><span><a href="#Smoothen-the-previous-curve-for-clarity" data-toc-modified-id="Smoothen-the-previous-curve-for-clarity-7.3.3"><span class="toc-item-num">7.3.3 </span>Smoothen the previous curve for clarity</a></span></li><li><span><a href="#Rebuild-the-Model" data-toc-modified-id="Rebuild-the-Model-7.3.4"><span class="toc-item-num">7.3.4 </span>Rebuild the Model</a></span></li><li><span><a href="#Test-the-Model" data-toc-modified-id="Test-the-Model-7.3.5"><span class="toc-item-num">7.3.5 </span>Test the Model</a></span></li></ul></li></ul></li><li><span><a href="#Model-6:-Imputing-the-missing-values-with-Median-and-preprocessing-data-using-Normalization" data-toc-modified-id="Model-6:-Imputing-the-missing-values-with-Median-and-preprocessing-data-using-Normalization-8"><span class="toc-item-num">8 </span>Model 6: Imputing the missing values with Median and preprocessing data using Normalization</a></span><ul class="toc-item"><li><span><a href="#Data" data-toc-modified-id="Data-8.1"><span class="toc-item-num">8.1 </span>Data</a></span><ul class="toc-item"><li><span><a href="#Data-preprocessing" data-toc-modified-id="Data-preprocessing-8.1.1"><span class="toc-item-num">8.1.1 </span>Data preprocessing</a></span><ul class="toc-item"><li><span><a href="#Preprocessing-data-using-Normalization" data-toc-modified-id="Preprocessing-data-using-Normalization-8.1.1.1"><span class="toc-item-num">8.1.1.1 </span>Preprocessing data using Normalization</a></span></li></ul></li></ul></li><li><span><a href="#Models-and-Training" data-toc-modified-id="Models-and-Training-8.2"><span class="toc-item-num">8.2 </span>Models and Training</a></span><ul class="toc-item"><li><span><a href="#Split-data-into-Training-set,-Validation-set-and-Testing-set" data-toc-modified-id="Split-data-into-Training-set,-Validation-set-and-Testing-set-8.2.1"><span class="toc-item-num">8.2.1 </span>Split data into Training set, Validation set and Testing set</a></span></li><li><span><a href="#Construct-the-Model-Architecture" data-toc-modified-id="Construct-the-Model-Architecture-8.2.2"><span class="toc-item-num">8.2.2 </span>Construct the Model Architecture</a></span></li><li><span><a href="#Build-the-Model" data-toc-modified-id="Build-the-Model-8.2.3"><span class="toc-item-num">8.2.3 </span>Build the Model</a></span></li></ul></li><li><span><a href="#Results" data-toc-modified-id="Results-8.3"><span class="toc-item-num">8.3 </span>Results</a></span><ul class="toc-item"><li><span><a href="#Plot-the-training-and-validation-Loss-and-Accuracy-Graph" data-toc-modified-id="Plot-the-training-and-validation-Loss-and-Accuracy-Graph-8.3.1"><span class="toc-item-num">8.3.1 </span>Plot the training and validation Loss and Accuracy Graph</a></span></li><li><span><a href="#Cut-off-the-first-few-data-points" data-toc-modified-id="Cut-off-the-first-few-data-points-8.3.2"><span class="toc-item-num">8.3.2 </span>Cut off the first few data points</a></span></li><li><span><a href="#Smoothen-the-previous-curve-for-clarity" data-toc-modified-id="Smoothen-the-previous-curve-for-clarity-8.3.3"><span class="toc-item-num">8.3.3 </span>Smoothen the previous curve for clarity</a></span></li><li><span><a href="#Rebuild-the-Model" data-toc-modified-id="Rebuild-the-Model-8.3.4"><span class="toc-item-num">8.3.4 </span>Rebuild the Model</a></span></li><li><span><a href="#Test-the-Model" data-toc-modified-id="Test-the-Model-8.3.5"><span class="toc-item-num">8.3.5 </span>Test the Model</a></span></li></ul></li></ul></li><li><span><a href="#Finalized-Result-and-Discussion" data-toc-modified-id="Finalized-Result-and-Discussion-9"><span class="toc-item-num">9 </span>Finalized Result and Discussion</a></span></li><li><span><a href="#Conclusion" data-toc-modified-id="Conclusion-10"><span class="toc-item-num">10 </span>Conclusion</a></span></li></ul></div>
# Case 1 - Coronary Heart Disease Classification
<br>*Matit Leesathapron . Sithan Janetunyalux . Yanisa Nitisaropas*<br>
Last Edited : 1st February 2020 Sat 11:45 UTC+2<br>
Neural Networks for Health Technology Applications<br>
[Helsinki Metropolia University of Applied Sciences](http://www.metropolia.fi/en/)<br>
# Background
Diagnostic decision is one of the fields in health technology application which can be implemented by using machine learning algorithms. This document<br> aims to investigate heart disease classification.
The computation was implemented by using Keras Neural Network to examine the given heart disease<br> dataset and provide effective model to predict the presence of heart disease.
The main objective is to investigate various techniques for data preprocessing and imputation of missing values in the dataset.
Also, trying different neural network architectures, batch sizes and number of epochs for training data is to find the best possible model for prediction.The results are then compared<br> using performance metrics such as accuracy and loss function to see whether which components affect the efficiency of the outcomes.
# Model 1: Imputing the missing values with Zero and preprocessing data using StandardScaler
## Data
The dataset used for computing the model was collected from V.A. Medical Center, Long Beach and Cleveland Clinic Foundation by the investigator<br> named Robert Detrano, M.D., Ph.D.
```
%pylab inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
```
### Read and prepare the dataset
```
cols = ['age', 'sex', 'cp', 'trestbps', 'chol', 'fbs', 'restecg', 'thalach', 'exang', 'oldpeak', 'slope', 'ca', 'thal', 'num']
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/heart-disease/processed.cleveland.data', index_col=None, header=None, names=cols, na_values='?')
df
```
There are 303 number of instances in this dataset. Each instance has 14 attributed variables as following :
1. age : age in years
2. sex : sex (1 = male; 0 = female)
3. cp : chest pain type
-- Value 1: typical angina
-- Value 2: atypical angina
-- Value 3: non-anginal pain
-- Value 4: asymptomatic
4. trestbps : resting blood pressure (in mm Hg on admission to the hospital)
5. chol : serum cholestoral in mg/dl
6. fbs : (fasting blood sugar > 120 mg/dl) (1 = true; 0 = false)
7. restecg : resting electrocardiographic results
-- Value 0: normal
-- Value 1: having ST-T wave abnormality (T wave inversions and/or ST elevation or depression of > 0.05 mV)
-- Value 2: showing probable or definite left ventricular hypertrophy by Estes' criteria
8. thalach : maximum heart rate achieved
9. exang : exercise induced angina (1 = yes; 0 = no)
10. oldpeak : ST depression induced by exercise relative to rest
11. slope : the slope of the peak exercise ST segment
-- Value 1: upsloping
-- Value 2: flat
-- Value 3: downsloping
12. ca : number of major vessels (0-3) colored by flourosopy
13. thal : 3 = normal; 6 = fixed defect; 7 = reversable defect
14. num : diagnosis of heart disease (angiographic disease status)
-- Value 0: < 50% diameter narrowing
-- Value 1: > 50% diameter narrowing
(in any major vessel: attributes 59 through 68 are vessels)
<br>The descriptive statistics of this dataset is shown below:
```
df.describe()
```
#### Check missing values in the dataset
```
df[df.isna().any(axis=1)]
```
The dataset contains some missing values in the following variables: ca and thal.
Those values are imputated by using various techniques such as filling with zero, mean, median or random values before training in the neural network.
#### Fill the missing value with zero
```
df = df.fillna(0)
df
```
#### Separate Data and Label
```
data = df.iloc[:, 0:13]
labels = df.iloc[:, 13]
```
The first 13 variables are used as data for heart disease analysis while the last attribute is the labeled value.
#### Treat any value higher than one as one
```
labels = labels.replace(2,1)
labels = labels.replace(3,1)
labels = labels.replace(4,1)
```
In the original dataset, predicted variables have values range from 0 to 4 representing a healthy condition of heart. The value 0 indicates healthy heart while the value 4 indicates severely unhealthy heart. For this document, 0 to 4 class labels were changed to a binary classification. The predicted class can be <br> either 0 or 1, meaning the heart is either Healthy(0) or Unhealthy(1). Therefore, the value that is greater than 0 is decided to be labeled as class 1.
### Data Preprocessing
Before building the model, the raw variables are changed into a representation that is more suitable for computating in neural network.<br> Techniques such as StandardScaler and Normalization are implemented in preprocessing phase.
#### Convert Dataframe to Numpy array
```
data = data.to_numpy()
labels = labels.to_numpy()
```
#### Preprocessing data using StandardScaler
```
scaler = preprocessing.StandardScaler().fit(data)
data_scaler = scaler.transform(data)
```
## Models and Training
### Split data into Training set, Validation set and Testing set
```
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
```
The data is split using the ratio of (60:20:20). First, the data is divided into training set and testing set with a ratio of (80:20).<br>Then the training set is split into train set and validation set with a ratio of (75:25).
Total : 303 data --- Training set : 181 data , Validation set : 61 data , Testing set : 61 data <br>
### Construct the Model Architecture
```
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
```
We used the sequential model for this case with 1 input layer, 1 hidden layer with 13 neurons and rectified linear unit (ReLU) activation function mathematically defined as y = max(0,x) , and 1 output layer with 1 neuron and sigmoid activation function mathematically defined as y = 1/1+e^-x.
Then we compile the model with adaptive moment estimation (Adam) optimizer which is the combination of RMSprop and Momentum, binary cross-entropy loss function which is the entropy of classification and output the accuracy of the model.
### Build the Model
We train the model using epoch equal to 200 and batch sizes equal to 1 to find the best epoch for our model.
```
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
```
## Results
### Plot the training and validation Loss and Accuracy Graph
```
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Cut off the first few data points
```
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Smoothen the previous curve for clarity
```
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.grid()
plt.show()
```
From the graph, it shows that the validation accuracy reaches the highest point of approximately 0.88 at around 33 epochs, so we try to rebuild the model by changing the epoch size to the optimal one.
### Rebuild the Model
```
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=35, batch_size=1)
```
### Test the Model
```
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
```
# Model 2: Imputing the missing values with Zero and preprocessing data using Normalization
## Data
### Data preprocessing
#### Preprocessing data using Normalization
```
minmax_scaler = preprocessing.MinMaxScaler()
data = minmax_scaler.fit_transform(data)
```
## Models and Training
### Split data into Training set, Validation set and Testing set
```
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
```
### Construct the Model Architecture
```
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
```
### Build the Model
We train the model using epoch equal to 200 and batch sizes equal to 1 to find the best epoch for our model.
```
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
```
## Results
### Plot the training and validation Loss and Accuracy Graph
```
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Cut off the first few data points
```
# Cut off the first few data points
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Smoothen the previous curve for clarity
```
# Smoothen the previous curve for clarity
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.show()
```
From the graph, it shows that the validation accuracy reaches the highest point of approximately 0.91 at around 60 epochs, so we try to rebuild the model by changing the epoch size to the optimal one.
### Rebuild the Model
```
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=60, batch_size=1)
```
### Test the Model
```
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
```
# Model 3: Imputing the missing values with Mean and Preprocessing data using StandardScaler
## Data
### Read in the dataset from .csv file
```
cols = ['age', 'sex', 'cp', 'trestbps', 'chol', 'fbs', 'restecg', 'thalach', 'exang', 'oldpeak', 'slope', 'ca', 'thal', 'num']
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/heart-disease/processed.cleveland.data', index_col=None, header=None, names=cols, na_values='?')
```
#### Fill the missing value with mean
```
df = df.fillna(df.mean())
df
```
#### Separate Data and Label
```
data = df.iloc[:, 0:13]
labels = df.iloc[:, 13]
```
#### Treat any value higher than one as one
```
labels = labels.replace(2,1)
labels = labels.replace(3,1)
labels = labels.replace(4,1)
```
### Data Preprocessing
#### Convert Dataframe to Numpy Array
```
data = data.to_numpy()
labels = labels.to_numpy()
```
#### Preprocessing data using StandardScaler
```
scaler = preprocessing.StandardScaler().fit(data)
data_scaler = scaler.transform(data)
```
## Models and Training
### Split data into Training set, Validation set and Testing set
```
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
```
### Construct the Model Architecture
```
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
```
### Build the Model
We train the model using epoch equal to 200 and batch sizes equal to 1 to find the best epoch for our model.
```
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
```
## Results
### Plot the training and validation Loss and Accuracy Graph
```
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Cut off the first few data points
```
# Cut off the first few data points
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Smoothen the previous curve for clarity
```
# Smoothen the previous curve for clarity
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.show()
```
From the graph, it shows that the validation accuracy reaches the highest point of approximately 0.88 at around 160 epochs, so we try to rebuild the model by changing the epoch size to the optimal one.
### Rebuild the Model
```
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=160, batch_size=1)
```
### Test the Model
```
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
```
# Model 4: Imputing the missing values with Mean and preprocessing data using Normalization
## Data
### Data preprocessing
#### Preprocessing data using Normalization
```
minmax_scaler = preprocessing.MinMaxScaler()
data = minmax_scaler.fit_transform(data)
```
## Models and Training
### Split data into Training set, Validation set and Testing set
```
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
```
### Construct the Model Architecture
```
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
```
### Build the Model
We train the model using epoch equal to 200 and batch sizes equal to 1 to find the best epoch for our model.
```
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
```
## Results
### Plot the training and validation Loss and Accuracy Graph
```
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Cut off the first few data points
```
# Cut off the first few data points
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Smoothen the previous curve for clarity
```
# Smoothen the previous curve for clarity
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.show()
```
From the graph, it shows that the validation accuracy reaches the highest point of approximately 0.88 at around 85 epochs, so we try to rebuild the model by changing the epoch size to the optimal one.
### Rebuild the Model
```
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=85, batch_size=1)
```
### Test the Model
```
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
```
# Model 5: Imputing the missing values with Median and preprocessing data using StandardScaler
## Data
### Read in the dataset from .csv file
```
cols = ['age', 'sex', 'cp', 'trestbps', 'chol', 'fbs', 'restecg', 'thalach', 'exang', 'oldpeak', 'slope', 'ca', 'thal', 'num']
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/heart-disease/processed.cleveland.data', index_col=None, header=None, names=cols, na_values='?')
```
#### Fill the missing value with mean
```
df = df.fillna(df.median())
df
```
#### Separate Data and Label
```
data = df.iloc[:, 0:13]
labels = df.iloc[:, 13]
```
#### Treat any value higher than one as one
```
labels = labels.replace(2,1)
labels = labels.replace(3,1)
labels = labels.replace(4,1)
```
### Data Preprocessing
#### Convert Dataframe to Numpy Array
```
data = data.to_numpy()
labels = labels.to_numpy()
```
#### Preprocessing data using StandardScaler
```
scaler = preprocessing.StandardScaler().fit(data)
data_scaler = scaler.transform(data)
```
## Models and Training
### Split data into Training set, Validation set and Testing set
```
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
```
### Construct the Model Architecture
```
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
```
### Build the Model
We train the model using epoch equal to 200 and batch sizes equal to 1 to find the best epoch for our model.
```
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
```
## Results
### Plot the training and validation Loss and Accuracy Graph
```
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Cut off the first few data points
```
# Cut off the first few data points
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Smoothen the previous curve for clarity
```
# Smoothen the previous curve for clarity
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.show()
```
From the graph, it shows that the validation accuracy reaches the highest point of approximately 0.91 at around 60 epochs, so we try to rebuild the model by changing the epoch size to the optimal one.
### Rebuild the Model
```
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=60, batch_size=1)
```
### Test the Model
```
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
```
# Model 6: Imputing the missing values with Median and preprocessing data using Normalization
## Data
### Data preprocessing
#### Preprocessing data using Normalization
```
minmax_scaler = preprocessing.MinMaxScaler()
data = minmax_scaler.fit_transform(data)
```
## Models and Training
### Split data into Training set, Validation set and Testing set
```
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
```
### Construct the Model Architecture
```
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
```
### Build the Model
We train the model using epoch equal to 200 and batch sizes equal to 1 to find the best epoch for our model.
```
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
```
## Results
### Plot the training and validation Loss and Accuracy Graph
```
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Cut off the first few data points
```
# Cut off the first few data points
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
```
### Smoothen the previous curve for clarity
```
# Smoothen the previous curve for clarity
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.show()
```
From the graph, it shows that the validation accuracy reaches the highest point of approximately 0.9 at around 95 epochs, so we try to rebuild the model by changing the epoch size to the optimal one.
### Rebuild the Model
```
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=95, batch_size=1)
```
### Test the Model
```
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
```
# Finalized Result and Discussion
In this case 1, we compare each model base on the accuracy score and loss values that each model gets from test set data. It outcomes with six results from different six models which are as following :
Accuracy
<table style="width:80%">
<tr>
<th></th>
<th>StandardScaler</th>
<th>Normalization</th>
</tr>
<tr>
<td>Nan=0</td>
<td>0.8197</td>
<td>0.7869</td>
</tr>
<tr>
<td>Nan=Mean</td>
<td>0.8033</td>
<td>0.8361</td>
</tr>
<tr>
<td>Nan=Median </td>
<td>0.7869</td>
<td>0.8361</td>
</tr>
</table>
Loss
<table style="width:80%">
<tr>
<th></th>
<th>StandardScaler</th>
<th>Normalization</th>
</tr>
<tr>
<td>Nan=0</td>
<td>0.5637</td>
<td>0.6105 </td>
</tr>
<tr>
<td>Nan=Mean</td>
<td>0.7471</td>
<td>0.5744</td>
</tr>
<tr>
<td>Nan=Median </td>
<td>0.7104 </td>
<td>0.6305</td>
</tr>
</table>
# Conclusion
In conclusion, we have investigated several techniques for data preprocessing and imputating missing values in the dataset to find the effective model for predicting the presence of heart disease. The result comes out that the best techniques used to predict the model from this dataset are "Imputating missing values with mean and preprocessing data using normalization", which gives the accuracy of 0.8361 and loss value of 0.5744.
In constructing neural network model architectures, we tested various values of batch sizes and number of epochs for training data. We tried on a batch size of 1, 5, 10 and 20 respectively and found that increasing batch size doesn't make this model more effective since the accuracy doesn't increase. Therefore, we decided to use the batch size of 1 to predict the model.
Also, we changed the number of epochs according to the outcome from the 'Validation accuracy vs. Epoch graph' of the result after training each model. Thus, we noticed what the smallest number of epoches is that gives highest accuracy value.
Moreover, we designed the model architecture with 1 input layer, 1 hidden layer with 13 neurons and rectified linear unit (ReLU) activation function, and 1 output layer with 1 neuron and sigmoid activation function. Since there are 13 input variables used for training the model, so the least there should be 13 neurons in the hidden layer. Upon testing model, we tried increasing number of neurons and layers, but the outcome did not give better result than the previous model.
Therefore, we built the model with 13 neurons in 1 hidden layer to simplify the model.
To finalize, our model is accurate but since the number of samples used for training the model is a small set, only 303 samples, the prediction model is not stable every time the program is rerun. The accuracy might be increase or decrease slightly compared with the previous results. Therefore, to improve the accuracy of this model, more group of samples should be used to train the model.
|
github_jupyter
|
%pylab inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
cols = ['age', 'sex', 'cp', 'trestbps', 'chol', 'fbs', 'restecg', 'thalach', 'exang', 'oldpeak', 'slope', 'ca', 'thal', 'num']
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/heart-disease/processed.cleveland.data', index_col=None, header=None, names=cols, na_values='?')
df
df.describe()
df[df.isna().any(axis=1)]
df = df.fillna(0)
df
data = df.iloc[:, 0:13]
labels = df.iloc[:, 13]
labels = labels.replace(2,1)
labels = labels.replace(3,1)
labels = labels.replace(4,1)
data = data.to_numpy()
labels = labels.to_numpy()
scaler = preprocessing.StandardScaler().fit(data)
data_scaler = scaler.transform(data)
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.grid()
plt.show()
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=35, batch_size=1)
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
minmax_scaler = preprocessing.MinMaxScaler()
data = minmax_scaler.fit_transform(data)
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
# Cut off the first few data points
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
# Smoothen the previous curve for clarity
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.show()
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=60, batch_size=1)
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
cols = ['age', 'sex', 'cp', 'trestbps', 'chol', 'fbs', 'restecg', 'thalach', 'exang', 'oldpeak', 'slope', 'ca', 'thal', 'num']
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/heart-disease/processed.cleveland.data', index_col=None, header=None, names=cols, na_values='?')
df = df.fillna(df.mean())
df
data = df.iloc[:, 0:13]
labels = df.iloc[:, 13]
labels = labels.replace(2,1)
labels = labels.replace(3,1)
labels = labels.replace(4,1)
data = data.to_numpy()
labels = labels.to_numpy()
scaler = preprocessing.StandardScaler().fit(data)
data_scaler = scaler.transform(data)
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
# Cut off the first few data points
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
# Smoothen the previous curve for clarity
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.show()
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=160, batch_size=1)
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
minmax_scaler = preprocessing.MinMaxScaler()
data = minmax_scaler.fit_transform(data)
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
# Cut off the first few data points
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
# Smoothen the previous curve for clarity
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.show()
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=85, batch_size=1)
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
cols = ['age', 'sex', 'cp', 'trestbps', 'chol', 'fbs', 'restecg', 'thalach', 'exang', 'oldpeak', 'slope', 'ca', 'thal', 'num']
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/heart-disease/processed.cleveland.data', index_col=None, header=None, names=cols, na_values='?')
df = df.fillna(df.median())
df
data = df.iloc[:, 0:13]
labels = df.iloc[:, 13]
labels = labels.replace(2,1)
labels = labels.replace(3,1)
labels = labels.replace(4,1)
data = data.to_numpy()
labels = labels.to_numpy()
scaler = preprocessing.StandardScaler().fit(data)
data_scaler = scaler.transform(data)
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
# Cut off the first few data points
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
# Smoothen the previous curve for clarity
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.show()
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=60, batch_size=1)
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
minmax_scaler = preprocessing.MinMaxScaler()
data = minmax_scaler.fit_transform(data)
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, random_state=112, test_size=0.20)
data_train_partial, data_val, labels_train_partial, labels_val = train_test_split(data_train, labels_train, random_state=112, test_size=0.25)
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
history = model.fit(data_train_partial, labels_train_partial, epochs=200, batch_size=1, validation_data=(data_val, labels_val))
print(history.history.keys())
loss = history.history['loss']
val_loss = history.history['val_loss']
accuracy = history.history['accuracy']
val_accuracy = history.history['val_accuracy']
time = range(1,len(loss)+1)
plt.plot(time, loss, 'b-')
plt.plot(time, val_loss, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
plt.plot(time, accuracy, 'b-')
plt.plot(time, val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
# Cut off the first few data points
short_val_accuracy = val_accuracy[10:]
plt.plot(range(1,len(short_val_accuracy)+1), short_val_accuracy, 'r-')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.show()
# Smoothen the previous curve for clarity
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_val_accuracy = smooth_curve(val_accuracy[10:])
plt.plot(range(1, len(smooth_val_accuracy) + 1), smooth_val_accuracy)
plt.xlabel('Epochs')
plt.ylabel('Validation Accuracy')
plt.show()
# Build a fresh model ...
model = Sequential()
model.add(Dense(13, activation='relu', input_dim=13))
model.add(Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# ... and train it for a shorter time for less overfitting
# (now with all the training data, including the samples previously used for validation)
history = model.fit(data_train, labels_train, epochs=95, batch_size=1)
# Test the model with new unseen data
test_mse_score, test_accuracy_score = model.evaluate(data_test, labels_test)
# Taking a look at test data predictions versus true targets
results = model.predict(data_test)
for i in range(len(results)):
print(labels_test[i], results[i])
| 0.699562 | 0.932638 |
<table style="background-color: white; text-align: center"><tr>
<td style="background-color: white"> <img src="UL_logo.png" alt="Drawing" style="width: 100px;"/> </td>
<td style="background-color: white"> <img src="FCUL_logo.png" alt="Drawing" style="width: 200px;"/> </td>
</tr></table>
<p style="text-align: center; font-size: 20px;">Faculdade de Ciências da Universidade de Lisboa</p>
<br>
<br>
<br>
<p style="text-align: center; font-size: 18px; font-style: italic;">Projecto fase I</p>
<br>
<br>
<br>
<br>
<br>
<br>
<p style="text-align: center; font-size: 16px; font-style: italic;">Fundamentos da Web-semântica</p>
<p style="text-align: center; font-size: 16px; font-style: italic;">Docente: André Falcão</p>
<br>
<br>
<br>
<br>
<br>
<br>
<br>
<p style="text-align: right; font-size: 14px;">João Pinto Nº 55018</p>
<p style="text-align: right; font-size: 14px;">Ricardo Meireles Nº 55841</p>
<p style="text-align: center; font-size: 12px; font-weight: bold; font-style: italic;">Ano Lectivo 2020-2021</p>
-------------------------------------------------------------------------
### Caracterização do dataset
Conforme escolhido pelos alunos, e aprovado pelo docente desta unidade curricular, este dataset da NFL é constituído por três partes sendo estas:
<ul>
<li>Estatísticas Base – Os dados base do jogador, como certos identificadores e propriedades do mesmo;</li>
<li>Estatísticas de Carreira – <em>Kickoff</em> – Estatísticas referentes aos pontapés de saída sobre cada jogador em específico na sua carreira;</li>
<li>Estatísticas de Carreira – <em>Passing</em> - Estatísticas referentes a passes efetuados por cada jogador em específico na sua carreira.</li>
</ul>
Referente aos dados base de cada jogador, podemos observar os seguintes campos:
<ul>
<li>Player Id – A identificação do jogador perante o dataset. É composto pelo nome e por um número atribuído ao jogador. Este será o campo que irá permitir a ligação de jogadores referentemente às suas estatísticas de carreira;</li>
<li>Name – O nome do jogador em causa;</li>
<li>Age – A idade do jogador em causa;</li>
<li>Birth Place – O local de nascimento do jogador;</li>
<li>Birthday – A data em que o jogador em causa nasceu;</li>
<li>High School – A escola secundária que o jogador frequentou;</li>
<li>High School Location – A localização de a escola secundária que o jogador frequentou;</li>
<li>College – A faculdade que o jogador em causa frequentou;</li>
<li>Number – O número do jogador em campo;</li>
<li>Current Status – O seu estado corrente, denominado por dois valores: ativo (Active) ou reformado (Retired);</li>
<li>Current Team – A equipa corrente do jogador. Representada como vazia para jogadores já retirados;</li>
<li>Experience – A experiência do jogador baseada em épocas;</li>
<li>Height (inches) – A altura do jogador em polegadas <em>(inches)</em>;</li>
<li>Weight (lbs) – O peso do jogador em libras <em>(lbs)</em>;</li>
<li>Years Played – O período, em intervalo de anos, em que o jogador jogou.</li>
</ul>
Respeitante às estatísticas de pontapé de saída de cada jogador, podemos observar os seguintes campos:
<ul>
<li>Player Id – A identificação do jogador perante o dataset. É composto pelo nome e por um número atribuído ao jogador – Permite a ligação aos seus dados base;</li>
<li>Name – O nome do jogador em causa;</li>
<li>Position – A posição do jogador na equipa;</li>
<li>Year – O ano a que estas estatísticas se aplicam;</li>
<li>Team – A equipa do jogador para o qual estas estatísticas se aplicam;</li>
<li>Games Played – A quantidade de jogos em que o jogador jogou;</li>
<li>Kickoffs – A quantidade de pontapés de saída realizados por o jogador em causa;</li>
<li>Kickoff Yards – Somatório da distância medida em jardas <em>(yards)</em> de até aonde chegou a bola;</li>
<li>Out of Bounds Kickoffs – A quantidade de pontapés de saída que resultaram na bola em sair fora dos limites do campo;</li>
<li>Yards per Kickoff – Média de jardas alcançadas por <em>kickoff</em> referente ao jogador;</li>
<li>Touchbacks – A quantidade de <em>touchbacks</em> efetuados;</li>
<li>Touchback Percentage – A percentagem de touchbacks efetuados pelo jogador na totalidade;</li>
<li>Kickoffs Returned – A quantidade de pontapés de saída de equipas adversárias que foram retornados pelo jogador;</li>
<li>Average Returned Yards – A média de jardas retornadas nos <em>kickoffs</em> efetuados pelo jogador;</li>
<li>Kickoffs Resulting in TDs – A quantidade de pontapés de saída resultantes em <em>Touchdowns</em>;</li>
<li>On Sides Kicks – A quantidade de on side kicks efetuados pelo jogador. Um on side kick trata-se de um pontapé de saída direcionado às linhas laterais do campo;</li>
<li>On Sides Kicks Returned – A quantidade de on side kicks da equipa adversária retornados pelo jogador.</li>
</ul>
Alusivo às estatísticas de passes efetuados, podemos observar os seguintes dados:
<ul>
<li>Player Id – A identificação do jogador perante o dataset. É composto pelo nome e por um número atribuído ao jogador – Permite a ligação aos seus dados base;</li>
<li>Name – O nome do jogador em causa;</li>
<li>Position – A posição do jogador na equipa;</li>
<li>Year – O ano a que estas estatísticas se aplicam;</li>
<li>Team – A equipa do jogador para o qual estas estatísticas se aplicam;</li>
<li>Games Played – A quantidade de jogos em que o jogador jogou;</li>
<li>Passes Attempted – A quantidade de passes tentados pelo jogador;</li>
<li>Passes Completed – A quantidade de passes completados pelo jogador;</li>
<li>Completion Percentage – A percentagem de passes completados pelo jogador;</li>
<li>Pass Attempts Per Game – O número de passes tentados pelo jogador;</li>
<li>Passing Yards – O somatório de jardas resultantes de passes do jogador;</li>
<li>Passing Yards Per Attempt – A média de jardas de passe por tentativa;</li>
<li>Passing Yards Per Game – A média de jardas de passe por jogo;</li>
<li>TD Passes – A quantidade de passes resultantes em uma Touchdown;</li>
<li>Percentage of TDs per Attempts – A percentagem de Touchdowns por tentativa;</li>
<li>Ints – O número de interceções efetuadas pelo jogador;</li>
<li>Int Rate – A taxa de interceções realizadas pelo jogador por passe;</li>
<li>Longest Pass – A maior distância alcançável pelo jogador num passe;</li>
<li>Passes Longer than 20 Yards – A quantidade de passes cuja distância ultrapassa as vinte jardas <em>(yards)</em>;</li>
<li>Passes Longer than 40 Yards - A quantidade de passes cuja distância ultrapassa as quarenta jardas <em>(yards)</em>;</li>
<li>Sacks – A quantidade de sacks efetuados pelo jogador;</li>
<li>Sacked Yards Lost - A quantidade de jardas <em>Sacked</em> perdidas</li>
<li>Passer Rating – Mede a eficiência do jogador enquanto <em>passer</em>.</li>
</ul>
### 1 - Load de Libraries, criação de Namespaces, read de .csv's e inicialização de Triple Store como grafo
```
import csv
import rdflib
from rdflib import Graph, Literal, Namespace, URIRef, BNode
from rdflib.namespace import DCTERMS, RDF, RDFS, XSD
from urllib.parse import quote
# Criação de namespaces
NFL = Namespace('http://www.nfl.com/')
pff = Namespace('http://www.pff.com/')
statid=1
# Import de dados
dados_base = csv.DictReader(open("Basic_Stats_Sample.csv"))
dados_passing = csv.DictReader(open("Career_Stats_Passing_Sample.csv"))
dados_kickoff = csv.DictReader(open("Career_Stats_Kickoff_Sample.csv"))
# Inicialização de Triple Store como grafo
dados = Graph()
```
### 2 - Associação de campos a variáveis e geração de GUID's para os dados base
```
for row in dados_base:
# Categorização de dados conforme a sua referencialidade como URI
# Categorização de dados únicos no seu contexto como Literals
id =row['Player Id']
player = URIRef(NFL+id)
current_team = URIRef(NFL+quote(row['Current Team']))
name = URIRef(NFL+quote(row['Name']))
position = URIRef(NFL+quote(row['Position']))
player_number = Literal(row['Number'],datatype=XSD.int)
current_status = URIRef(NFL+quote(row['Current Status']))
experience = Literal(row['Experience'])
birth_place = URIRef(NFL+quote(row['Birth Place']))
college = URIRef(NFL+quote(row['College']))
high_school = URIRef(NFL+quote(row['High School']))
high_school_location = URIRef(NFL+quote(row['High School Location']))
height = Literal(row['Height (inches)'],datatype=XSD.int)
weight = Literal(row['Weight (lbs)'],datatype=XSD.int)
years_played = Literal(row['Years Played'])
age = Literal(row['Age'],datatype=XSD.int)
# Adicionar o triplo, identificado o tipo de objecto conforme o que se trata
dados.add((player, RDF.type, NFL.player))
dados.add((player, RDFS.label, Literal(id, lang="en")))
dados.add((player, NFL.name, name ))
dados.add((player, NFL.current_team, current_team))
dados.add((player, NFL.position, position))
dados.add((player, NFL.player_number, player_number))
dados.add((player, NFL.current_status, current_status))
dados.add((player, NFL.experience, experience))
dados.add((player, NFL.birth_place, birth_place))
dados.add((player, NFL.college, college))
dados.add((player, NFL.high_school, NFL.high_school))
dados.add((player, NFL.high_school_location, high_school_location))
dados.add((player, NFL.height, height))
dados.add((player, NFL.weight, weight))
dados.add((player, NFL.years_played, years_played))
```
#### 2.1 - Associação de campos a variáveis e geração de GUID's para os dados de passes
```
for row in dados_passing:
# Categorização de dados conforme a sua referencialidade como URI
# Categorização de dados únicos no seu contexto como Literals
stat = URIRef(pff+str(statid))
statid= statid+1
player = URIRef(NFL+id)
name = URIRef(NFL+quote(row['Name']))
team = URIRef(NFL+quote(row['Team']))
games_played = Literal(row['Games Played'])
passes_attempted = Literal(row['Passes Attempted'])
passes_completed = Literal(row['Passes Completed'])
completion_percentage = Literal(row['Completion Percentage'])
pass_att_per_game = Literal(row['Pass Attempts Per Game'])
passing_yards = Literal(row['Passing Yards'])
year = Literal(row['Year'])
passing_yards_att = Literal(row['Passing Yards Per Attempt'])
passing_yards_game = Literal(row['Passing Yards Per Game'])
td_passes = Literal(row['TD Passes'])
percentage_tds_per_attemp = Literal(row['Percentage of TDs per Attempts'])
interceptions = Literal(row['Ints'])
interceptions_rate = Literal(row['Ints'])
longest_pass = Literal(row['Longest Pass'])
passes_longer_20yards = Literal(row['Passes Longer than 20 Yards'])
passes_longer_40yards = Literal(row['Passes Longer than 40 Yards'])
sacks = Literal(row['Sacks'])
sacked_yards_lost = Literal(row['Sacked Yards Lost'])
passer_rating = Literal(row['Passer Rating'])
# Adicionar o triplo, identificado o tipo de objecto conforme o que se trata
dados.add((stat, RDF.type, pff.stat))
dados.add((stat, NFL.name, name))
dados.add((stat, NFL.player, player))
dados.add((stat,pff.passing_yards,passing_yards))
dados.add((stat,NFL.team,team))
dados.add((stat,pff.year,year))
dados.add((stat,pff.games_played,games_played))
dados.add((stat,pff.passes_attempted,passes_attempted))
dados.add((stat,pff.passes_completed,passes_completed))
dados.add((stat,pff.completion_percentage,completion_percentage))
dados.add((stat,pff.pass_att_per_game,pass_att_per_game))
dados.add((stat,pff.passing_yards,passing_yards))
dados.add((stat,pff.passing_yards_att,passing_yards_att))
dados.add((stat,pff.passing_yards_game,passing_yards_game))
dados.add((stat,pff.td_passes,td_passes))
dados.add((stat,pff.percentage_tds_per_attemp,percentage_tds_per_attemp))
dados.add((stat,pff.interceptions,interceptions))
dados.add((stat,pff.interceptions_rate,interceptions_rate))
dados.add((stat,pff.longest_pass,longest_pass))
dados.add((stat,pff.passes_longer_20yards,passes_longer_20yards))
dados.add((stat,pff.passes_longer_40yards,passes_longer_40yards))
dados.add((stat,pff.sacks,sacks))
dados.add((stat,pff.sacked_yards_lost,sacked_yards_lost))
dados.add((stat,pff.passer_rating,passer_rating))
```
#### 2.2 - Associação de campos a variáveis e geração de GUID's para os dados de kickoff
```
for row in dados_kickoff:
# Categorização de dados conforme a sua referencialidade como URI
# Categorização de dados únicos no seu contexto como Literals
stat_kickoff = URIRef(pff+str(statid))
statid= statid+1
player = URIRef(NFL+id)
name = URIRef(NFL+quote(row['Name']))
team = URIRef(NFL+quote(row['Team']))
position = URIRef(NFL+quote(row['Position']))
year = Literal(row['Year'])
team = URIRef(NFL+quote(row['Team']))
games_played = Literal(row['Games Played'])
kickoffs = Literal(row['Kickoffs'])
kickoff_yards = Literal(row['Kickoff Yards'])
oob_kickoffs = Literal(row['Out of Bounds Kickoffs'])
yards_per_kickoff = Literal(row['Yards Per Kickoff'])
touchbacks = Literal(row['Touchbacks'])
touchbacks_percentage = Literal(row['Touchback Percentage'])
kickoffs_returned = Literal(row['Kickoffs Returned'])
avg_returned_yards = Literal(row['Average Returned Yards'])
kickoff_resulting_in_TDs = Literal(row['Kickoffs Resulting in TDs'])
on_sides_kicks = Literal(row['On Sides Kicks'])
on_sides_kicks_returned = Literal(row['On Sides Kicks Returned'])
# Adicionar o triplo, identificado o tipo de objecto conforme o que se trata
dados.add((stat_kickoff, RDF.type, pff.stat))
dados.add((stat_kickoff, NFL.player, player))
dados.add((stat_kickoff, NFL.name, name))
dados.add((stat_kickoff, NFL.position,position))
dados.add((stat_kickoff, pff.year, year))
dados.add((stat_kickoff,pff.position,position))
dados.add((stat_kickoff,NFL.team,team))
dados.add((stat_kickoff,pff.games_played,games_played))
dados.add((stat_kickoff,pff.kickoffs,kickoffs))
dados.add((stat_kickoff,pff.kickoff_yards,kickoff_yards))
dados.add((stat_kickoff,pff.oob_kickoffs,oob_kickoffs))
dados.add((stat_kickoff,pff.yards_per_kickoff,yards_per_kickoff))
dados.add((stat_kickoff,pff.touchbacks,touchbacks))
dados.add((stat_kickoff,pff.touchbacks_percentage,touchbacks_percentage))
dados.add((stat_kickoff,pff.kickoffs_returned,kickoffs_returned))
dados.add((stat_kickoff,pff.avg_returned_yards,avg_returned_yards))
dados.add((stat_kickoff,pff.kickoff_resulting_in_TDs,kickoff_resulting_in_TDs))
dados.add((stat_kickoff,pff.on_sides_kicks,on_sides_kicks))
dados.add((stat_kickoff,pff.on_sides_kicks_returned,on_sides_kicks_returned))
```
### 3 - Bind de dados à Triple Store
```
dados.bind("NFL",NFL)
dados.bind("pff", pff)
dados.bind("Pro football Focus - Kickoff", stat_kickoff)
```
### 4 - Print de dados em formato Turtle
```
graph_data = (dados.serialize(format="turtle").decode("utf-8"))
print(graph_data)
```
#### 4.1 - Save de dados como nfl_data.ttl para reutilização
```
graph_data=dados.serialize(format='turtle')
fil=open("nfl_data.ttl", "wb")
fil.write(graph_data)
fil.close()
```
## 5 - Queries
```
# Query 1 -> Nome de todos os jogadores cuja posiçao é Quarterback e a sua equipa
qres = dados.query(
"""SELECT ?name ?equipa
WHERE {
?p a NFL:player .
?p NFL:name ?name .
?p NFL:current_team ?equipa .
?p NFL:position NFL:QB .
}LIMIT 50""")
for row in qres:
print("Name: %s Team: %s" % row)
# Query 2 -> Nome de todos os jogadores que tenham jogado exactamente 3 epocas
qres = dados.query(
"""SELECT ?name
WHERE {
?p a NFL:player .
?p NFL:name ?name .
?p NFL:experience '3 Seasons' .
}LIMIT 50""")
for row in qres:
print("Name: %s" % row)
# Query 3 -> Nome e peso de todos os jogadores com peso superior a 200
qres = dados.query(
"""SELECT ?name ?weight
WHERE {
?p a NFL:player .
?p NFL:name ?name .
?p NFL:weight ?weight .
FILTER (?weight > 200)
}LIMIT 50""")
for row in qres:
print("Name: %s Weight %s" % row)
# Query 4 -> Nome de todos os jogadores que jogaram em 1950 com mais de 10 "sacks"
qres = dados.query(
"""SELECT ?name
WHERE {
?a a NFL:player .
?a NFL:name ?name .
?b pff:year '1950' .
?b pff:sacks ?sacks
filter (?sacks > 10)
}LIMIT 50""")
for row in qres:
print("Name: %s " % row)
# Query 5 -> Todos os jogadores com nome, jogos jogados e passes completados
qres = dados.query(
"""SELECT ?name ?gp ?pa
WHERE {
?p a NFL:player .
?p NFL:name ?name .
?a pff:games_played ?gp .
?a pff:passes_attempted ?pa .
}LIMIT 50""")
for row in qres:
print("Name: %s Games Played: %s Passes Attempted: %s" % row)
# Query 6 -> Nome e equipa de todos os jogadores que tenham jogado pelo menos 16 jogos em 2008 e feito mais de 70 kickoffs.
qres = dados.query(
"""SELECT ?name ?team
WHERE {
?a a NFL:player .
?a NFL:name ?name .
?b NFL:team ?team .
?b pff:year '2008' .
?b pff:games_played '16' .
?b pff:kickoffs ?kickoffs .
filter (?kickoffs > 70)
}LIMIT 50""")
for row in qres:
print("Name: %s Team: %s " % row)
```
|
github_jupyter
|
import csv
import rdflib
from rdflib import Graph, Literal, Namespace, URIRef, BNode
from rdflib.namespace import DCTERMS, RDF, RDFS, XSD
from urllib.parse import quote
# Criação de namespaces
NFL = Namespace('http://www.nfl.com/')
pff = Namespace('http://www.pff.com/')
statid=1
# Import de dados
dados_base = csv.DictReader(open("Basic_Stats_Sample.csv"))
dados_passing = csv.DictReader(open("Career_Stats_Passing_Sample.csv"))
dados_kickoff = csv.DictReader(open("Career_Stats_Kickoff_Sample.csv"))
# Inicialização de Triple Store como grafo
dados = Graph()
for row in dados_base:
# Categorização de dados conforme a sua referencialidade como URI
# Categorização de dados únicos no seu contexto como Literals
id =row['Player Id']
player = URIRef(NFL+id)
current_team = URIRef(NFL+quote(row['Current Team']))
name = URIRef(NFL+quote(row['Name']))
position = URIRef(NFL+quote(row['Position']))
player_number = Literal(row['Number'],datatype=XSD.int)
current_status = URIRef(NFL+quote(row['Current Status']))
experience = Literal(row['Experience'])
birth_place = URIRef(NFL+quote(row['Birth Place']))
college = URIRef(NFL+quote(row['College']))
high_school = URIRef(NFL+quote(row['High School']))
high_school_location = URIRef(NFL+quote(row['High School Location']))
height = Literal(row['Height (inches)'],datatype=XSD.int)
weight = Literal(row['Weight (lbs)'],datatype=XSD.int)
years_played = Literal(row['Years Played'])
age = Literal(row['Age'],datatype=XSD.int)
# Adicionar o triplo, identificado o tipo de objecto conforme o que se trata
dados.add((player, RDF.type, NFL.player))
dados.add((player, RDFS.label, Literal(id, lang="en")))
dados.add((player, NFL.name, name ))
dados.add((player, NFL.current_team, current_team))
dados.add((player, NFL.position, position))
dados.add((player, NFL.player_number, player_number))
dados.add((player, NFL.current_status, current_status))
dados.add((player, NFL.experience, experience))
dados.add((player, NFL.birth_place, birth_place))
dados.add((player, NFL.college, college))
dados.add((player, NFL.high_school, NFL.high_school))
dados.add((player, NFL.high_school_location, high_school_location))
dados.add((player, NFL.height, height))
dados.add((player, NFL.weight, weight))
dados.add((player, NFL.years_played, years_played))
for row in dados_passing:
# Categorização de dados conforme a sua referencialidade como URI
# Categorização de dados únicos no seu contexto como Literals
stat = URIRef(pff+str(statid))
statid= statid+1
player = URIRef(NFL+id)
name = URIRef(NFL+quote(row['Name']))
team = URIRef(NFL+quote(row['Team']))
games_played = Literal(row['Games Played'])
passes_attempted = Literal(row['Passes Attempted'])
passes_completed = Literal(row['Passes Completed'])
completion_percentage = Literal(row['Completion Percentage'])
pass_att_per_game = Literal(row['Pass Attempts Per Game'])
passing_yards = Literal(row['Passing Yards'])
year = Literal(row['Year'])
passing_yards_att = Literal(row['Passing Yards Per Attempt'])
passing_yards_game = Literal(row['Passing Yards Per Game'])
td_passes = Literal(row['TD Passes'])
percentage_tds_per_attemp = Literal(row['Percentage of TDs per Attempts'])
interceptions = Literal(row['Ints'])
interceptions_rate = Literal(row['Ints'])
longest_pass = Literal(row['Longest Pass'])
passes_longer_20yards = Literal(row['Passes Longer than 20 Yards'])
passes_longer_40yards = Literal(row['Passes Longer than 40 Yards'])
sacks = Literal(row['Sacks'])
sacked_yards_lost = Literal(row['Sacked Yards Lost'])
passer_rating = Literal(row['Passer Rating'])
# Adicionar o triplo, identificado o tipo de objecto conforme o que se trata
dados.add((stat, RDF.type, pff.stat))
dados.add((stat, NFL.name, name))
dados.add((stat, NFL.player, player))
dados.add((stat,pff.passing_yards,passing_yards))
dados.add((stat,NFL.team,team))
dados.add((stat,pff.year,year))
dados.add((stat,pff.games_played,games_played))
dados.add((stat,pff.passes_attempted,passes_attempted))
dados.add((stat,pff.passes_completed,passes_completed))
dados.add((stat,pff.completion_percentage,completion_percentage))
dados.add((stat,pff.pass_att_per_game,pass_att_per_game))
dados.add((stat,pff.passing_yards,passing_yards))
dados.add((stat,pff.passing_yards_att,passing_yards_att))
dados.add((stat,pff.passing_yards_game,passing_yards_game))
dados.add((stat,pff.td_passes,td_passes))
dados.add((stat,pff.percentage_tds_per_attemp,percentage_tds_per_attemp))
dados.add((stat,pff.interceptions,interceptions))
dados.add((stat,pff.interceptions_rate,interceptions_rate))
dados.add((stat,pff.longest_pass,longest_pass))
dados.add((stat,pff.passes_longer_20yards,passes_longer_20yards))
dados.add((stat,pff.passes_longer_40yards,passes_longer_40yards))
dados.add((stat,pff.sacks,sacks))
dados.add((stat,pff.sacked_yards_lost,sacked_yards_lost))
dados.add((stat,pff.passer_rating,passer_rating))
for row in dados_kickoff:
# Categorização de dados conforme a sua referencialidade como URI
# Categorização de dados únicos no seu contexto como Literals
stat_kickoff = URIRef(pff+str(statid))
statid= statid+1
player = URIRef(NFL+id)
name = URIRef(NFL+quote(row['Name']))
team = URIRef(NFL+quote(row['Team']))
position = URIRef(NFL+quote(row['Position']))
year = Literal(row['Year'])
team = URIRef(NFL+quote(row['Team']))
games_played = Literal(row['Games Played'])
kickoffs = Literal(row['Kickoffs'])
kickoff_yards = Literal(row['Kickoff Yards'])
oob_kickoffs = Literal(row['Out of Bounds Kickoffs'])
yards_per_kickoff = Literal(row['Yards Per Kickoff'])
touchbacks = Literal(row['Touchbacks'])
touchbacks_percentage = Literal(row['Touchback Percentage'])
kickoffs_returned = Literal(row['Kickoffs Returned'])
avg_returned_yards = Literal(row['Average Returned Yards'])
kickoff_resulting_in_TDs = Literal(row['Kickoffs Resulting in TDs'])
on_sides_kicks = Literal(row['On Sides Kicks'])
on_sides_kicks_returned = Literal(row['On Sides Kicks Returned'])
# Adicionar o triplo, identificado o tipo de objecto conforme o que se trata
dados.add((stat_kickoff, RDF.type, pff.stat))
dados.add((stat_kickoff, NFL.player, player))
dados.add((stat_kickoff, NFL.name, name))
dados.add((stat_kickoff, NFL.position,position))
dados.add((stat_kickoff, pff.year, year))
dados.add((stat_kickoff,pff.position,position))
dados.add((stat_kickoff,NFL.team,team))
dados.add((stat_kickoff,pff.games_played,games_played))
dados.add((stat_kickoff,pff.kickoffs,kickoffs))
dados.add((stat_kickoff,pff.kickoff_yards,kickoff_yards))
dados.add((stat_kickoff,pff.oob_kickoffs,oob_kickoffs))
dados.add((stat_kickoff,pff.yards_per_kickoff,yards_per_kickoff))
dados.add((stat_kickoff,pff.touchbacks,touchbacks))
dados.add((stat_kickoff,pff.touchbacks_percentage,touchbacks_percentage))
dados.add((stat_kickoff,pff.kickoffs_returned,kickoffs_returned))
dados.add((stat_kickoff,pff.avg_returned_yards,avg_returned_yards))
dados.add((stat_kickoff,pff.kickoff_resulting_in_TDs,kickoff_resulting_in_TDs))
dados.add((stat_kickoff,pff.on_sides_kicks,on_sides_kicks))
dados.add((stat_kickoff,pff.on_sides_kicks_returned,on_sides_kicks_returned))
dados.bind("NFL",NFL)
dados.bind("pff", pff)
dados.bind("Pro football Focus - Kickoff", stat_kickoff)
graph_data = (dados.serialize(format="turtle").decode("utf-8"))
print(graph_data)
graph_data=dados.serialize(format='turtle')
fil=open("nfl_data.ttl", "wb")
fil.write(graph_data)
fil.close()
# Query 1 -> Nome de todos os jogadores cuja posiçao é Quarterback e a sua equipa
qres = dados.query(
"""SELECT ?name ?equipa
WHERE {
?p a NFL:player .
?p NFL:name ?name .
?p NFL:current_team ?equipa .
?p NFL:position NFL:QB .
}LIMIT 50""")
for row in qres:
print("Name: %s Team: %s" % row)
# Query 2 -> Nome de todos os jogadores que tenham jogado exactamente 3 epocas
qres = dados.query(
"""SELECT ?name
WHERE {
?p a NFL:player .
?p NFL:name ?name .
?p NFL:experience '3 Seasons' .
}LIMIT 50""")
for row in qres:
print("Name: %s" % row)
# Query 3 -> Nome e peso de todos os jogadores com peso superior a 200
qres = dados.query(
"""SELECT ?name ?weight
WHERE {
?p a NFL:player .
?p NFL:name ?name .
?p NFL:weight ?weight .
FILTER (?weight > 200)
}LIMIT 50""")
for row in qres:
print("Name: %s Weight %s" % row)
# Query 4 -> Nome de todos os jogadores que jogaram em 1950 com mais de 10 "sacks"
qres = dados.query(
"""SELECT ?name
WHERE {
?a a NFL:player .
?a NFL:name ?name .
?b pff:year '1950' .
?b pff:sacks ?sacks
filter (?sacks > 10)
}LIMIT 50""")
for row in qres:
print("Name: %s " % row)
# Query 5 -> Todos os jogadores com nome, jogos jogados e passes completados
qres = dados.query(
"""SELECT ?name ?gp ?pa
WHERE {
?p a NFL:player .
?p NFL:name ?name .
?a pff:games_played ?gp .
?a pff:passes_attempted ?pa .
}LIMIT 50""")
for row in qres:
print("Name: %s Games Played: %s Passes Attempted: %s" % row)
# Query 6 -> Nome e equipa de todos os jogadores que tenham jogado pelo menos 16 jogos em 2008 e feito mais de 70 kickoffs.
qres = dados.query(
"""SELECT ?name ?team
WHERE {
?a a NFL:player .
?a NFL:name ?name .
?b NFL:team ?team .
?b pff:year '2008' .
?b pff:games_played '16' .
?b pff:kickoffs ?kickoffs .
filter (?kickoffs > 70)
}LIMIT 50""")
for row in qres:
print("Name: %s Team: %s " % row)
| 0.293708 | 0.89616 |
# Generative Adversarial Networks (GANs)
So far in CS231N, all the applications of neural networks that we have explored have been **discriminative models** that take an input and are trained to produce a labeled output. This has ranged from straightforward classification of image categories to sentence generation (which was still phrased as a classification problem, our labels were in vocabulary space and we’d learned a recurrence to capture multi-word labels). In this notebook, we will expand our repetoire, and build **generative models** using neural networks. Specifically, we will learn how to build models which generate novel images that resemble a set of training images.
### What is a GAN?
In 2014, [Goodfellow et al.](https://arxiv.org/abs/1406.2661) presented a method for training generative models called Generative Adversarial Networks (GANs for short). In a GAN, we build two different neural networks. Our first network is a traditional classification network, called the **discriminator**. We will train the discriminator to take images, and classify them as being real (belonging to the training set) or fake (not present in the training set). Our other network, called the **generator**, will take random noise as input and transform it using a neural network to produce images. The goal of the generator is to fool the discriminator into thinking the images it produced are real.
We can think of this back and forth process of the generator ($G$) trying to fool the discriminator ($D$), and the discriminator trying to correctly classify real vs. fake as a minimax game:
$$\underset{G}{\text{minimize}}\; \underset{D}{\text{maximize}}\; \mathbb{E}_{x \sim p_\text{data}}\left[\log D(x)\right] + \mathbb{E}_{z \sim p(z)}\left[\log \left(1-D(G(z))\right)\right]$$
where $z \sim p(z)$ are the random noise samples, $G(z)$ are the generated images using the neural network generator $G$, and $D$ is the output of the discriminator, specifying the probability of an input being real. In [Goodfellow et al.](https://arxiv.org/abs/1406.2661), they analyze this minimax game and show how it relates to minimizing the Jensen-Shannon divergence between the training data distribution and the generated samples from $G$.
To optimize this minimax game, we will aternate between taking gradient *descent* steps on the objective for $G$, and gradient *ascent* steps on the objective for $D$:
1. update the **generator** ($G$) to minimize the probability of the __discriminator making the correct choice__.
2. update the **discriminator** ($D$) to maximize the probability of the __discriminator making the correct choice__.
While these updates are useful for analysis, they do not perform well in practice. Instead, we will use a different objective when we update the generator: maximize the probability of the **discriminator making the incorrect choice**. This small change helps to allevaiate problems with the generator gradient vanishing when the discriminator is confident. This is the standard update used in most GAN papers, and was used in the original paper from [Goodfellow et al.](https://arxiv.org/abs/1406.2661).
In this assignment, we will alternate the following updates:
1. Update the generator ($G$) to maximize the probability of the discriminator making the incorrect choice on generated data:
$$\underset{G}{\text{maximize}}\; \mathbb{E}_{z \sim p(z)}\left[\log D(G(z))\right]$$
2. Update the discriminator ($D$), to maximize the probability of the discriminator making the correct choice on real and generated data:
$$\underset{D}{\text{maximize}}\; \mathbb{E}_{x \sim p_\text{data}}\left[\log D(x)\right] + \mathbb{E}_{z \sim p(z)}\left[\log \left(1-D(G(z))\right)\right]$$
### What else is there?
Since 2014, GANs have exploded into a huge research area, with massive [workshops](https://sites.google.com/site/nips2016adversarial/), and [hundreds of new papers](https://github.com/hindupuravinash/the-gan-zoo). Compared to other approaches for generative models, they often produce the highest quality samples but are some of the most difficult and finicky models to train (see [this github repo](https://github.com/soumith/ganhacks) that contains a set of 17 hacks that are useful for getting models working). Improving the stabiilty and robustness of GAN training is an open research question, with new papers coming out every day! For a more recent tutorial on GANs, see [here](https://arxiv.org/abs/1701.00160). There is also some even more recent exciting work that changes the objective function to Wasserstein distance and yields much more stable results across model architectures: [WGAN](https://arxiv.org/abs/1701.07875), [WGAN-GP](https://arxiv.org/abs/1704.00028).
GANs are not the only way to train a generative model! For other approaches to generative modeling check out the [deep generative model chapter](http://www.deeplearningbook.org/contents/generative_models.html) of the Deep Learning [book](http://www.deeplearningbook.org). Another popular way of training neural networks as generative models is Variational Autoencoders (co-discovered [here](https://arxiv.org/abs/1312.6114) and [here](https://arxiv.org/abs/1401.4082)). Variatonal autoencoders combine neural networks with variationl inference to train deep generative models. These models tend to be far more stable and easier to train but currently don't produce samples that are as pretty as GANs.
Here's an example of what your outputs from the 3 different models you're going to train should look like... note that GANs are sometimes finicky, so your outputs might not look exactly like this... this is just meant to be a *rough* guideline of the kind of quality you can expect:

## Setup
```
!pip install http://download.pytorch.org/whl/torch-0.2.0.post3-cp36-cp36m-macosx_10_7_x86_64.whl
!pip install torchvision
import torch
import torch.nn as nn
from torch.nn import init
from torch.autograd import Variable
import torchvision
import torchvision.transforms as T
import torch.optim as optim
from torch.utils.data import DataLoader
from torch.utils.data import sampler
import torchvision.datasets as dset
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
%matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
def show_images(images):
images = np.reshape(images, [images.shape[0], -1]) # images reshape to (batch_size, D)
sqrtn = int(np.ceil(np.sqrt(images.shape[0])))
sqrtimg = int(np.ceil(np.sqrt(images.shape[1])))
fig = plt.figure(figsize=(sqrtn, sqrtn))
gs = gridspec.GridSpec(sqrtn, sqrtn)
gs.update(wspace=0.05, hspace=0.05)
for i, img in enumerate(images):
ax = plt.subplot(gs[i])
plt.axis('off')
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.set_aspect('equal')
plt.imshow(img.reshape([sqrtimg,sqrtimg]))
return
def preprocess_img(x):
return 2 * x - 1.0
def deprocess_img(x):
return (x + 1.0) / 2.0
def rel_error(x,y):
return np.max(np.abs(x - y) / (np.maximum(1e-8, np.abs(x) + np.abs(y))))
def count_params(model):
"""Count the number of parameters in the current TensorFlow graph """
param_count = np.sum([np.prod(p.size()) for p in model.parameters()])
return param_count
answers = np.load('gan-checks-tf.npz')
```
## Dataset
GANs are notoriously finicky with hyperparameters, and also require many training epochs. In order to make this assignment approachable without a GPU, we will be working on the MNIST dataset, which is 60,000 training and 10,000 test images. Each picture contains a centered image of white digit on black background (0 through 9). This was one of the first datasets used to train convolutional neural networks and it is fairly easy -- a standard CNN model can easily exceed 99% accuracy.
To simplify our code here, we will use the PyTorch MNIST wrapper, which downloads and loads the MNIST dataset. See the [documentation](https://github.com/pytorch/vision/blob/master/torchvision/datasets/mnist.py) for more information about the interface. The default parameters will take 5,000 of the training examples and place them into a validation dataset. The data will be saved into a folder called `MNIST_data`.
```
class ChunkSampler(sampler.Sampler):
"""Samples elements sequentially from some offset.
Arguments:
num_samples: # of desired datapoints
start: offset where we should start selecting from
"""
def __init__(self, num_samples, start=0):
self.num_samples = num_samples
self.start = start
def __iter__(self):
return iter(range(self.start, self.start + self.num_samples))
def __len__(self):
return self.num_samples
NUM_TRAIN = 50000
NUM_VAL = 5000
NOISE_DIM = 96
batch_size = 128
mnist_train = dset.MNIST('./cs231n/datasets/MNIST_data', train=True, download=True,
transform=T.ToTensor())
loader_train = DataLoader(mnist_train, batch_size=batch_size,
sampler=ChunkSampler(NUM_TRAIN, 0))
mnist_val = dset.MNIST('./cs231n/datasets/MNIST_data', train=True, download=True,
transform=T.ToTensor())
loader_val = DataLoader(mnist_val, batch_size=batch_size,
sampler=ChunkSampler(NUM_VAL, NUM_TRAIN))
imgs = loader_train.__iter__().next()[0].view(batch_size, 784).numpy().squeeze()
show_images(imgs)
```
## Random Noise
Generate uniform noise from -1 to 1 with shape `[batch_size, dim]`.
Hint: use `torch.rand`.
```
def sample_noise(batch_size, dim):
"""
Generate a PyTorch Tensor of uniform random noise.
Input:
- batch_size: Integer giving the batch size of noise to generate.
- dim: Integer giving the dimension of noise to generate.
Output:
- A PyTorch Tensor of shape (batch_size, dim) containing uniform
random noise in the range (-1, 1).
"""
return torch.Tensor(batch_size, dim).uniform_(-1, 1)
```
Make sure noise is the correct shape and type:
```
def test_sample_noise():
batch_size = 3
dim = 4
torch.manual_seed(231)
z = sample_noise(batch_size, dim)
np_z = z.cpu().numpy()
assert np_z.shape == (batch_size, dim)
assert torch.is_tensor(z)
assert np.all(np_z >= -1.0) and np.all(np_z <= 1.0)
assert np.any(np_z < 0.0) and np.any(np_z > 0.0)
print('All tests passed!')
test_sample_noise()
```
## Flatten
Recall our Flatten operation from previous notebooks... this time we also provide an Unflatten, which you might want to use when implementing the convolutional generator. We also provide a weight initializer (and call it for you) that uses Xavier initialization instead of PyTorch's uniform default.
```
class Flatten(nn.Module):
def forward(self, x):
N, C, H, W = x.size() # read in N, C, H, W
return x.view(N, -1) # "flatten" the C * H * W values into a single vector per image
class Unflatten(nn.Module):
"""
An Unflatten module receives an input of shape (N, C*H*W) and reshapes it
to produce an output of shape (N, C, H, W).
"""
def __init__(self, N=-1, C=128, H=7, W=7):
super(Unflatten, self).__init__()
self.N = N
self.C = C
self.H = H
self.W = W
def forward(self, x):
return x.view(self.N, self.C, self.H, self.W)
def initialize_weights(m):
if isinstance(m, nn.Linear) or isinstance(m, nn.ConvTranspose2d):
init.xavier_uniform(m.weight.data)
```
## CPU / GPU
By default all code will run on CPU. GPUs are not needed for this assignment, but will help you to train your models faster. If you do want to run the code on a GPU, then change the `dtype` variable in the following cell.
```
dtype = torch.FloatTensor
# dtype = torch.cuda.FloatTensor ## UNCOMMENT THIS LINE IF YOU'RE ON A GPU!
```
# Discriminator
Our first step is to build a discriminator. Fill in the architecture as part of the `nn.Sequential` constructor in the function below. All fully connected layers should include bias terms. The architecture is:
* Fully connected layer from size 784 to 256
* LeakyReLU with alpha 0.01
* Fully connected layer from 256 to 256
* LeakyReLU with alpha 0.01
* Fully connected layer from 256 to 1
Recall that the Leaky ReLU nonlinearity computes $f(x) = \max(\alpha x, x)$ for some fixed constant $\alpha$; for the LeakyReLU nonlinearities in the architecture above we set $\alpha=0.01$.
The output of the discriminator should have shape `[batch_size, 1]`, and contain real numbers corresponding to the scores that each of the `batch_size` inputs is a real image.
```
def discriminator():
"""
Build and return a PyTorch model implementing the architecture above.
"""
model = nn.Sequential(
Flatten(),
nn.Linear(784, 256),
nn.LeakyReLU(0.01, inplace=True),
nn.Linear(256, 256),
nn.LeakyReLU(0.01, inplace=True),
nn.Linear(256, 1)
)
return model
```
Test to make sure the number of parameters in the discriminator is correct:
```
def test_discriminator(true_count=267009):
model = discriminator()
cur_count = count_params(model)
if cur_count != true_count:
print('Incorrect number of parameters in discriminator. Check your achitecture.')
else:
print('Correct number of parameters in discriminator.')
test_discriminator()
```
# Generator
Now to build the generator network:
* Fully connected layer from noise_dim to 1024
* ReLU
* Fully connected layer with size 1024
* ReLU
* Fully connected layer with size 784
* TanH
* To clip the image to be [-1,1]
```
from itertools import product
for v in [((4 + a)*1024 + (1024 + b)*1024 + (1024 + c)*784, a, b, c) for a, b, c in product([0, 1], repeat=3)]:
print(v)
def generator(noise_dim=NOISE_DIM):
"""
Build and return a PyTorch model implementing the architecture above.
"""
model = nn.Sequential(
nn.Linear(noise_dim, 1024, bias=True),
nn.ReLU(inplace=True),
nn.Linear(1024, 1024, bias=True),
nn.ReLU(inplace=True),
nn.Linear(1024, 784, bias=True),
nn.Tanh()
)
return model
```
Test to make sure the number of parameters in the generator is correct:
```
def test_generator(true_count=1858320):
model = generator(4)
cur_count = count_params(model)
if cur_count != true_count:
print('Incorrect number of parameters in generator. Check your achitecture.')
else:
print('Correct number of parameters in generator.')
test_generator()
```
# GAN Loss
Compute the generator and discriminator loss.
Note that these are negated from the equations presented earlier as we will be *minimizing* these losses.
**HINTS**: You should use the `bce_loss` function defined below to compute the binary cross entropy loss which is needed to compute the log probability of the true label given the logits output from the discriminator. Given a score $s\in\mathbb{R}$ and a label $y\in\{0, 1\}$, the binary cross entropy loss is
$$ bce(s, y) = y * \log(s) + (1 - y) * \log(1 - s) $$
A naive implementation of this formula can be numerically unstable, so we have provided a numerically stable implementation for you below.
You will also need to compute labels corresponding to real or fake and use the logit arguments to determine their size. Make sure you cast these labels to the correct data type using the global `dtype` variable, for example:
`true_labels = Variable(torch.ones(size)).type(dtype)`
Instead of computing the expectation, we will be averaging over elements of the minibatch, so make sure to combine the loss by averaging instead of summing.
```
def bce_loss(input, target):
"""
Numerically stable version of the binary cross-entropy loss function.
As per https://github.com/pytorch/pytorch/issues/751
See the TensorFlow docs for a derivation of this formula:
https://www.tensorflow.org/api_docs/python/tf/nn/sigmoid_cross_entropy_with_logits
Inputs:
- input: PyTorch Variable of shape (N, ) giving scores.
- target: PyTorch Variable of shape (N,) containing 0 and 1 giving targets.
Returns:
- A PyTorch Variable containing the mean BCE loss over the minibatch of input data.
"""
neg_abs = - input.abs()
loss = input.clamp(min=0) - input * target + (1 + neg_abs.exp()).log()
return loss.mean()
```
The generator loss is: $$\ell_G = -\mathbb{E}_{z \sim p(z)}\left[\log D(G(z))\right]$$
```
def generator_loss(logits_fake):
"""
Computes the generator loss described above.
Inputs:
- logits_fake: PyTorch Variable of shape (N,) giving scores for the fake data.
Returns:
- loss: PyTorch Variable containing the (scalar) loss for the generator.
"""
return bce_loss(logits_fake, Variable(torch.ones(logits_fake.size())).type(dtype))
```
The discriminator loss is:
$$ \ell_D = -\mathbb{E}_{x \sim p_\text{data}}\left[\log D(x)\right] - \mathbb{E}_{z \sim p(z)}\left[\log \left(1-D(G(z))\right)\right]$$
```
def discriminator_loss(logits_real, logits_fake):
"""
Computes the discriminator loss described above.
Inputs:
- logits_real: PyTorch Variable of shape (N,) giving scores for the real data.
- logits_fake: PyTorch Variable of shape (N,) giving scores for the fake data.
Returns:
- loss: PyTorch Variable containing (scalar) the loss for the discriminator.
"""
loss_real = bce_loss(logits_real, Variable(torch.ones(logits_real.size())).type(dtype))
loss_fake = bce_loss(logits_fake, Variable(torch.zeros(logits_fake.size())).type(dtype))
return loss_real + loss_fake
```
Test your generator and discriminator loss. You should see errors < 1e-7.
```
def test_discriminator_loss(logits_real, logits_fake, d_loss_true):
d_loss = discriminator_loss(Variable(torch.Tensor(logits_real)).type(dtype),
Variable(torch.Tensor(logits_fake)).type(dtype)).data.cpu().numpy()
print("Maximum error in d_loss: %g"%rel_error(d_loss_true, d_loss))
test_discriminator_loss(answers['logits_real'], answers['logits_fake'],
answers['d_loss_true'])
def test_generator_loss(logits_fake, g_loss_true):
g_loss = generator_loss(Variable(torch.Tensor(logits_fake)).type(dtype)).data.cpu().numpy()
print("Maximum error in g_loss: %g"%rel_error(g_loss_true, g_loss))
test_generator_loss(answers['logits_fake'], answers['g_loss_true'])
```
# Optimizing our loss
Make a function that returns an `optim.Adam` optimizer for the given model with a 1e-3 learning rate, beta1=0.5, beta2=0.999. You'll use this to construct optimizers for the generators and discriminators for the rest of the notebook.
```
def get_optimizer(model):
"""
Construct and return an Adam optimizer for the model with learning rate 1e-3,
beta1=0.5, and beta2=0.999.
Input:
- model: A PyTorch model that we want to optimize.
Returns:
- An Adam optimizer for the model with the desired hyperparameters.
"""
optimizer = optim.Adam(model.parameters(), lr=1e-3, betas=(0.5, 0.999))
return optimizer
```
# Training a GAN!
We provide you the main training loop... you won't need to change this function, but we encourage you to read through and understand it.
```
def run_a_gan(D, G, D_solver, G_solver, discriminator_loss, generator_loss, show_every=250,
batch_size=128, noise_size=96, num_epochs=10):
"""
Train a GAN!
Inputs:
- D, G: PyTorch models for the discriminator and generator
- D_solver, G_solver: torch.optim Optimizers to use for training the
discriminator and generator.
- discriminator_loss, generator_loss: Functions to use for computing the generator and
discriminator loss, respectively.
- show_every: Show samples after every show_every iterations.
- batch_size: Batch size to use for training.
- noise_size: Dimension of the noise to use as input to the generator.
- num_epochs: Number of epochs over the training dataset to use for training.
"""
iter_count = 0
for epoch in range(num_epochs):
for x, _ in loader_train:
if len(x) != batch_size:
continue
D_solver.zero_grad()
real_data = Variable(x).type(dtype)
logits_real = D(2* (real_data - 0.5)).type(dtype)
g_fake_seed = Variable(sample_noise(batch_size, noise_size)).type(dtype)
fake_images = G(g_fake_seed).detach()
logits_fake = D(fake_images.view(batch_size, 1, 28, 28))
d_total_error = discriminator_loss(logits_real, logits_fake)
d_total_error.backward()
D_solver.step()
G_solver.zero_grad()
g_fake_seed = Variable(sample_noise(batch_size, noise_size)).type(dtype)
fake_images = G(g_fake_seed)
gen_logits_fake = D(fake_images.view(batch_size, 1, 28, 28))
g_error = generator_loss(gen_logits_fake)
g_error.backward()
G_solver.step()
if (iter_count % show_every == 0):
print('Iter: {}, D: {:.4}, G:{:.4}'.format(iter_count,d_total_error.data[0],g_error.data[0]))
imgs_numpy = fake_images.data.cpu().numpy()
show_images(imgs_numpy[0:16])
plt.show()
print()
iter_count += 1
```
Well that wasn't so hard, was it? In the iterations in the low 100s you should see black backgrounds, fuzzy shapes as you approach iteration 1000, and decent shapes, about half of which will be sharp and clearly recognizable as we pass 3000.
```
# Make the discriminator
D = discriminator().type(dtype)
# Make the generator
G = generator().type(dtype)
# Use the function you wrote earlier to get optimizers for the Discriminator and the Generator
D_solver = get_optimizer(D)
G_solver = get_optimizer(G)
# Run it!
run_a_gan(D, G, D_solver, G_solver, discriminator_loss, generator_loss)
```
# Least Squares GAN
We'll now look at [Least Squares GAN](https://arxiv.org/abs/1611.04076), a newer, more stable alernative to the original GAN loss function. For this part, all we have to do is change the loss function and retrain the model. We'll implement equation (9) in the paper, with the generator loss:
$$\ell_G = \frac{1}{2}\mathbb{E}_{z \sim p(z)}\left[\left(D(G(z))-1\right)^2\right]$$
and the discriminator loss:
$$ \ell_D = \frac{1}{2}\mathbb{E}_{x \sim p_\text{data}}\left[\left(D(x)-1\right)^2\right] + \frac{1}{2}\mathbb{E}_{z \sim p(z)}\left[ \left(D(G(z))\right)^2\right]$$
**HINTS**: Instead of computing the expectation, we will be averaging over elements of the minibatch, so make sure to combine the loss by averaging instead of summing. When plugging in for $D(x)$ and $D(G(z))$ use the direct output from the discriminator (`scores_real` and `scores_fake`).
```
def ls_discriminator_loss(scores_real, scores_fake):
"""
Compute the Least-Squares GAN loss for the discriminator.
Inputs:
- scores_real: PyTorch Variable of shape (N,) giving scores for the real data.
- scores_fake: PyTorch Variable of shape (N,) giving scores for the fake data.
Outputs:
- loss: A PyTorch Variable containing the loss.
"""
loss_real = torch.mean(torch.pow(scores_real - 1.0, 2.0))
loss_fake = torch.mean(torch.pow(scores_fake, 2.0))
return 0.5 * (loss_real + loss_fake)
def ls_generator_loss(scores_fake):
"""
Computes the Least-Squares GAN loss for the generator.
Inputs:
- scores_fake: PyTorch Variable of shape (N,) giving scores for the fake data.
Outputs:
- loss: A PyTorch Variable containing the loss.
"""
return torch.mean(torch.pow(scores_fake - 1, 2.0)) / 2
```
Before running a GAN with our new loss function, let's check it:
```
def test_lsgan_loss(score_real, score_fake, d_loss_true, g_loss_true):
d_loss = ls_discriminator_loss(torch.FloatTensor(score_real), torch.FloatTensor(score_fake))
g_loss = ls_generator_loss(torch.FloatTensor(score_fake))
print("Maximum error in d_loss: %g"%rel_error(d_loss_true, d_loss))
print("Maximum error in g_loss: %g"%rel_error(g_loss_true, g_loss))
test_lsgan_loss(answers['logits_real'], answers['logits_fake'],
answers['d_loss_lsgan_true'], answers['g_loss_lsgan_true'])
D_LS = discriminator().type(dtype)
G_LS = generator().type(dtype)
D_LS_solver = get_optimizer(D_LS)
G_LS_solver = get_optimizer(G_LS)
run_a_gan(D_LS, G_LS, D_LS_solver, G_LS_solver, ls_discriminator_loss, ls_generator_loss)
```
# INLINE QUESTION 1
Describe how the visual quality of the samples changes over the course of training. Do you notice anything about the distribution of the samples? How do the results change across different training runs?
Some digits visually look better.
# Deeply Convolutional GANs
In the first part of the notebook, we implemented an almost direct copy of the original GAN network from Ian Goodfellow. However, this network architecture allows no real spatial reasoning. It is unable to reason about things like "sharp edges" in general because it lacks any convolutional layers. Thus, in this section, we will implement some of the ideas from [DCGAN](https://arxiv.org/abs/1511.06434), where we use convolutional networks
#### Discriminator
We will use a discriminator inspired by the TensorFlow MNIST classification tutorial, which is able to get above 99% accuracy on the MNIST dataset fairly quickly.
* Reshape into image tensor (Use Unflatten!)
* 32 Filters, 5x5, Stride 1, Leaky ReLU(alpha=0.01)
* Max Pool 2x2, Stride 2
* 64 Filters, 5x5, Stride 1, Leaky ReLU(alpha=0.01)
* Max Pool 2x2, Stride 2
* Flatten
* Fully Connected size 4 x 4 x 64, Leaky ReLU(alpha=0.01)
* Fully Connected size 1
```
def out_dim(sz, filter_size, padding, stride):
"""
Computes the size of dimension after convolution.
Input:
- sz: Original size of dimension
- filter_size: Filter size applied in convolution
- padding: Applied to the original dimension
- stride: Between the two applications of convolution
Returns a tuple of:
- out: The size of the dimension after the convolution is computed
"""
return 1 + int((sz + 2 * padding - filter_size) / stride)
def build_dc_classifier():
"""
Build and return a PyTorch model for the DCGAN discriminator implementing
the architecture above.
"""
from functools import reduce
conv_relu_pool_sizes = [
lambda N: out_dim(N, 5, 0, 1),
lambda N: out_dim(N, 2, 0, 2)
] * 2
n_to_Flatten = reduce(lambda value, f: f(value), conv_relu_pool_sizes, 28)
return nn.Sequential(
Unflatten(batch_size, 1, 28, 28),
nn.Conv2d(1, 32, 5, 1),
nn.LeakyReLU(0.01, inplace=True),
nn.MaxPool2d(2, 2),
nn.Conv2d(32, 64, 5, 1),
nn.LeakyReLU(0.01, inplace=True),
nn.MaxPool2d(2, 2),
Flatten(),
nn.Linear(n_to_Flatten**2*64, 4*4*64),
nn.LeakyReLU(0.01, inplace=True),
nn.Linear(4*4*64, 1)
)
data = Variable(loader_train.__iter__().next()[0]).type(dtype)
b = build_dc_classifier().type(dtype)
out = b(data)
```
Check the number of parameters in your classifier as a sanity check:
```
def test_dc_classifer(true_count=1102721):
model = build_dc_classifier()
cur_count = count_params(model)
if cur_count != true_count:
print('Incorrect number of parameters in generator. Check your achitecture.')
else:
print('Correct number of parameters in generator.')
test_dc_classifer()
```
#### Generator
For the generator, we will copy the architecture exactly from the [InfoGAN paper](https://arxiv.org/pdf/1606.03657.pdf). See Appendix C.1 MNIST. See the documentation for [torch.nn.ConvTranspose2d](http://pytorch.org/docs/master/nn.html#convtranspose2d). We are always "training" in GAN mode.
* Fully connected of size 1024, ReLU
* BatchNorm
* Fully connected of size 7 x 7 x 128, ReLU
* BatchNorm
* Reshape into Image Tensor
* 64 conv2d^T filters of 4x4, stride 2, 'same' padding, ReLU
* BatchNorm
* 1 conv2d^T filter of 4x4, stride 2, 'same' padding, TanH
* Should have a 28x28x1 image, reshape back into 784 vector
```
def build_dc_generator(noise_dim=NOISE_DIM):
"""
Build and return a PyTorch model implementing the DCGAN generator using
the architecture described above.
"""
return nn.Sequential(
nn.Linear(noise_dim, 1024),
nn.ReLU(inplace=True),
nn.BatchNorm1d(1024),
nn.Linear(1024, 7*7*128),
nn.ReLU(inplace=True),
nn.BatchNorm1d(7*7*128),
Unflatten(batch_size, 128, 7, 7),
nn.ConvTranspose2d(128, 64, 4, 2, 1),
nn.ReLU(inplace=True),
nn.BatchNorm2d(64),
nn.ConvTranspose2d(64, 1, 4, 2, 1),
nn.Tanh(),
Flatten()
)
test_g_gan = build_dc_generator().type(dtype)
test_g_gan.apply(initialize_weights)
fake_seed = Variable(torch.randn(batch_size, NOISE_DIM)).type(dtype)
fake_images = test_g_gan.forward(fake_seed)
fake_images.size()
```
Check the number of parameters in your generator as a sanity check:
```
def test_dc_generator(true_count=6580801):
model = build_dc_generator(4)
cur_count = count_params(model)
if cur_count != true_count:
print('Incorrect number of parameters in generator. Check your achitecture.')
else:
print('Correct number of parameters in generator.')
test_dc_generator()
D_DC = build_dc_classifier().type(dtype)
D_DC.apply(initialize_weights)
G_DC = build_dc_generator().type(dtype)
G_DC.apply(initialize_weights)
D_DC_solver = get_optimizer(D_DC)
G_DC_solver = get_optimizer(G_DC)
run_a_gan(D_DC, G_DC, D_DC_solver, G_DC_solver, discriminator_loss, generator_loss, num_epochs=5)
```
# INLINE QUESTION 2
What differences do you see between the DCGAN results and the original GAN results?
Quality is very nice. Digits are sharp and could be visually associated with numbers. No extra noise is present on those images.
## Extra Credit
** Be sure you don't destroy your results above, but feel free to copy+paste code to get results below **
* For a small amount of extra credit, you can implement additional new GAN loss functions below, provided they converge. See AFI, BiGAN, Softmax GAN, Conditional GAN, InfoGAN, etc.
* Likewise for an improved architecture or using a convolutional GAN (or even implement a VAE)
* For a bigger chunk of extra credit, load the CIFAR10 data (see last assignment) and train a compelling generative model on CIFAR-10
* Something new/cool.
#### Describe what you did here
** TBD **
|
github_jupyter
|
!pip install http://download.pytorch.org/whl/torch-0.2.0.post3-cp36-cp36m-macosx_10_7_x86_64.whl
!pip install torchvision
import torch
import torch.nn as nn
from torch.nn import init
from torch.autograd import Variable
import torchvision
import torchvision.transforms as T
import torch.optim as optim
from torch.utils.data import DataLoader
from torch.utils.data import sampler
import torchvision.datasets as dset
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
%matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
def show_images(images):
images = np.reshape(images, [images.shape[0], -1]) # images reshape to (batch_size, D)
sqrtn = int(np.ceil(np.sqrt(images.shape[0])))
sqrtimg = int(np.ceil(np.sqrt(images.shape[1])))
fig = plt.figure(figsize=(sqrtn, sqrtn))
gs = gridspec.GridSpec(sqrtn, sqrtn)
gs.update(wspace=0.05, hspace=0.05)
for i, img in enumerate(images):
ax = plt.subplot(gs[i])
plt.axis('off')
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.set_aspect('equal')
plt.imshow(img.reshape([sqrtimg,sqrtimg]))
return
def preprocess_img(x):
return 2 * x - 1.0
def deprocess_img(x):
return (x + 1.0) / 2.0
def rel_error(x,y):
return np.max(np.abs(x - y) / (np.maximum(1e-8, np.abs(x) + np.abs(y))))
def count_params(model):
"""Count the number of parameters in the current TensorFlow graph """
param_count = np.sum([np.prod(p.size()) for p in model.parameters()])
return param_count
answers = np.load('gan-checks-tf.npz')
class ChunkSampler(sampler.Sampler):
"""Samples elements sequentially from some offset.
Arguments:
num_samples: # of desired datapoints
start: offset where we should start selecting from
"""
def __init__(self, num_samples, start=0):
self.num_samples = num_samples
self.start = start
def __iter__(self):
return iter(range(self.start, self.start + self.num_samples))
def __len__(self):
return self.num_samples
NUM_TRAIN = 50000
NUM_VAL = 5000
NOISE_DIM = 96
batch_size = 128
mnist_train = dset.MNIST('./cs231n/datasets/MNIST_data', train=True, download=True,
transform=T.ToTensor())
loader_train = DataLoader(mnist_train, batch_size=batch_size,
sampler=ChunkSampler(NUM_TRAIN, 0))
mnist_val = dset.MNIST('./cs231n/datasets/MNIST_data', train=True, download=True,
transform=T.ToTensor())
loader_val = DataLoader(mnist_val, batch_size=batch_size,
sampler=ChunkSampler(NUM_VAL, NUM_TRAIN))
imgs = loader_train.__iter__().next()[0].view(batch_size, 784).numpy().squeeze()
show_images(imgs)
def sample_noise(batch_size, dim):
"""
Generate a PyTorch Tensor of uniform random noise.
Input:
- batch_size: Integer giving the batch size of noise to generate.
- dim: Integer giving the dimension of noise to generate.
Output:
- A PyTorch Tensor of shape (batch_size, dim) containing uniform
random noise in the range (-1, 1).
"""
return torch.Tensor(batch_size, dim).uniform_(-1, 1)
def test_sample_noise():
batch_size = 3
dim = 4
torch.manual_seed(231)
z = sample_noise(batch_size, dim)
np_z = z.cpu().numpy()
assert np_z.shape == (batch_size, dim)
assert torch.is_tensor(z)
assert np.all(np_z >= -1.0) and np.all(np_z <= 1.0)
assert np.any(np_z < 0.0) and np.any(np_z > 0.0)
print('All tests passed!')
test_sample_noise()
class Flatten(nn.Module):
def forward(self, x):
N, C, H, W = x.size() # read in N, C, H, W
return x.view(N, -1) # "flatten" the C * H * W values into a single vector per image
class Unflatten(nn.Module):
"""
An Unflatten module receives an input of shape (N, C*H*W) and reshapes it
to produce an output of shape (N, C, H, W).
"""
def __init__(self, N=-1, C=128, H=7, W=7):
super(Unflatten, self).__init__()
self.N = N
self.C = C
self.H = H
self.W = W
def forward(self, x):
return x.view(self.N, self.C, self.H, self.W)
def initialize_weights(m):
if isinstance(m, nn.Linear) or isinstance(m, nn.ConvTranspose2d):
init.xavier_uniform(m.weight.data)
dtype = torch.FloatTensor
# dtype = torch.cuda.FloatTensor ## UNCOMMENT THIS LINE IF YOU'RE ON A GPU!
def discriminator():
"""
Build and return a PyTorch model implementing the architecture above.
"""
model = nn.Sequential(
Flatten(),
nn.Linear(784, 256),
nn.LeakyReLU(0.01, inplace=True),
nn.Linear(256, 256),
nn.LeakyReLU(0.01, inplace=True),
nn.Linear(256, 1)
)
return model
def test_discriminator(true_count=267009):
model = discriminator()
cur_count = count_params(model)
if cur_count != true_count:
print('Incorrect number of parameters in discriminator. Check your achitecture.')
else:
print('Correct number of parameters in discriminator.')
test_discriminator()
from itertools import product
for v in [((4 + a)*1024 + (1024 + b)*1024 + (1024 + c)*784, a, b, c) for a, b, c in product([0, 1], repeat=3)]:
print(v)
def generator(noise_dim=NOISE_DIM):
"""
Build and return a PyTorch model implementing the architecture above.
"""
model = nn.Sequential(
nn.Linear(noise_dim, 1024, bias=True),
nn.ReLU(inplace=True),
nn.Linear(1024, 1024, bias=True),
nn.ReLU(inplace=True),
nn.Linear(1024, 784, bias=True),
nn.Tanh()
)
return model
def test_generator(true_count=1858320):
model = generator(4)
cur_count = count_params(model)
if cur_count != true_count:
print('Incorrect number of parameters in generator. Check your achitecture.')
else:
print('Correct number of parameters in generator.')
test_generator()
def bce_loss(input, target):
"""
Numerically stable version of the binary cross-entropy loss function.
As per https://github.com/pytorch/pytorch/issues/751
See the TensorFlow docs for a derivation of this formula:
https://www.tensorflow.org/api_docs/python/tf/nn/sigmoid_cross_entropy_with_logits
Inputs:
- input: PyTorch Variable of shape (N, ) giving scores.
- target: PyTorch Variable of shape (N,) containing 0 and 1 giving targets.
Returns:
- A PyTorch Variable containing the mean BCE loss over the minibatch of input data.
"""
neg_abs = - input.abs()
loss = input.clamp(min=0) - input * target + (1 + neg_abs.exp()).log()
return loss.mean()
def generator_loss(logits_fake):
"""
Computes the generator loss described above.
Inputs:
- logits_fake: PyTorch Variable of shape (N,) giving scores for the fake data.
Returns:
- loss: PyTorch Variable containing the (scalar) loss for the generator.
"""
return bce_loss(logits_fake, Variable(torch.ones(logits_fake.size())).type(dtype))
def discriminator_loss(logits_real, logits_fake):
"""
Computes the discriminator loss described above.
Inputs:
- logits_real: PyTorch Variable of shape (N,) giving scores for the real data.
- logits_fake: PyTorch Variable of shape (N,) giving scores for the fake data.
Returns:
- loss: PyTorch Variable containing (scalar) the loss for the discriminator.
"""
loss_real = bce_loss(logits_real, Variable(torch.ones(logits_real.size())).type(dtype))
loss_fake = bce_loss(logits_fake, Variable(torch.zeros(logits_fake.size())).type(dtype))
return loss_real + loss_fake
def test_discriminator_loss(logits_real, logits_fake, d_loss_true):
d_loss = discriminator_loss(Variable(torch.Tensor(logits_real)).type(dtype),
Variable(torch.Tensor(logits_fake)).type(dtype)).data.cpu().numpy()
print("Maximum error in d_loss: %g"%rel_error(d_loss_true, d_loss))
test_discriminator_loss(answers['logits_real'], answers['logits_fake'],
answers['d_loss_true'])
def test_generator_loss(logits_fake, g_loss_true):
g_loss = generator_loss(Variable(torch.Tensor(logits_fake)).type(dtype)).data.cpu().numpy()
print("Maximum error in g_loss: %g"%rel_error(g_loss_true, g_loss))
test_generator_loss(answers['logits_fake'], answers['g_loss_true'])
def get_optimizer(model):
"""
Construct and return an Adam optimizer for the model with learning rate 1e-3,
beta1=0.5, and beta2=0.999.
Input:
- model: A PyTorch model that we want to optimize.
Returns:
- An Adam optimizer for the model with the desired hyperparameters.
"""
optimizer = optim.Adam(model.parameters(), lr=1e-3, betas=(0.5, 0.999))
return optimizer
def run_a_gan(D, G, D_solver, G_solver, discriminator_loss, generator_loss, show_every=250,
batch_size=128, noise_size=96, num_epochs=10):
"""
Train a GAN!
Inputs:
- D, G: PyTorch models for the discriminator and generator
- D_solver, G_solver: torch.optim Optimizers to use for training the
discriminator and generator.
- discriminator_loss, generator_loss: Functions to use for computing the generator and
discriminator loss, respectively.
- show_every: Show samples after every show_every iterations.
- batch_size: Batch size to use for training.
- noise_size: Dimension of the noise to use as input to the generator.
- num_epochs: Number of epochs over the training dataset to use for training.
"""
iter_count = 0
for epoch in range(num_epochs):
for x, _ in loader_train:
if len(x) != batch_size:
continue
D_solver.zero_grad()
real_data = Variable(x).type(dtype)
logits_real = D(2* (real_data - 0.5)).type(dtype)
g_fake_seed = Variable(sample_noise(batch_size, noise_size)).type(dtype)
fake_images = G(g_fake_seed).detach()
logits_fake = D(fake_images.view(batch_size, 1, 28, 28))
d_total_error = discriminator_loss(logits_real, logits_fake)
d_total_error.backward()
D_solver.step()
G_solver.zero_grad()
g_fake_seed = Variable(sample_noise(batch_size, noise_size)).type(dtype)
fake_images = G(g_fake_seed)
gen_logits_fake = D(fake_images.view(batch_size, 1, 28, 28))
g_error = generator_loss(gen_logits_fake)
g_error.backward()
G_solver.step()
if (iter_count % show_every == 0):
print('Iter: {}, D: {:.4}, G:{:.4}'.format(iter_count,d_total_error.data[0],g_error.data[0]))
imgs_numpy = fake_images.data.cpu().numpy()
show_images(imgs_numpy[0:16])
plt.show()
print()
iter_count += 1
# Make the discriminator
D = discriminator().type(dtype)
# Make the generator
G = generator().type(dtype)
# Use the function you wrote earlier to get optimizers for the Discriminator and the Generator
D_solver = get_optimizer(D)
G_solver = get_optimizer(G)
# Run it!
run_a_gan(D, G, D_solver, G_solver, discriminator_loss, generator_loss)
def ls_discriminator_loss(scores_real, scores_fake):
"""
Compute the Least-Squares GAN loss for the discriminator.
Inputs:
- scores_real: PyTorch Variable of shape (N,) giving scores for the real data.
- scores_fake: PyTorch Variable of shape (N,) giving scores for the fake data.
Outputs:
- loss: A PyTorch Variable containing the loss.
"""
loss_real = torch.mean(torch.pow(scores_real - 1.0, 2.0))
loss_fake = torch.mean(torch.pow(scores_fake, 2.0))
return 0.5 * (loss_real + loss_fake)
def ls_generator_loss(scores_fake):
"""
Computes the Least-Squares GAN loss for the generator.
Inputs:
- scores_fake: PyTorch Variable of shape (N,) giving scores for the fake data.
Outputs:
- loss: A PyTorch Variable containing the loss.
"""
return torch.mean(torch.pow(scores_fake - 1, 2.0)) / 2
def test_lsgan_loss(score_real, score_fake, d_loss_true, g_loss_true):
d_loss = ls_discriminator_loss(torch.FloatTensor(score_real), torch.FloatTensor(score_fake))
g_loss = ls_generator_loss(torch.FloatTensor(score_fake))
print("Maximum error in d_loss: %g"%rel_error(d_loss_true, d_loss))
print("Maximum error in g_loss: %g"%rel_error(g_loss_true, g_loss))
test_lsgan_loss(answers['logits_real'], answers['logits_fake'],
answers['d_loss_lsgan_true'], answers['g_loss_lsgan_true'])
D_LS = discriminator().type(dtype)
G_LS = generator().type(dtype)
D_LS_solver = get_optimizer(D_LS)
G_LS_solver = get_optimizer(G_LS)
run_a_gan(D_LS, G_LS, D_LS_solver, G_LS_solver, ls_discriminator_loss, ls_generator_loss)
def out_dim(sz, filter_size, padding, stride):
"""
Computes the size of dimension after convolution.
Input:
- sz: Original size of dimension
- filter_size: Filter size applied in convolution
- padding: Applied to the original dimension
- stride: Between the two applications of convolution
Returns a tuple of:
- out: The size of the dimension after the convolution is computed
"""
return 1 + int((sz + 2 * padding - filter_size) / stride)
def build_dc_classifier():
"""
Build and return a PyTorch model for the DCGAN discriminator implementing
the architecture above.
"""
from functools import reduce
conv_relu_pool_sizes = [
lambda N: out_dim(N, 5, 0, 1),
lambda N: out_dim(N, 2, 0, 2)
] * 2
n_to_Flatten = reduce(lambda value, f: f(value), conv_relu_pool_sizes, 28)
return nn.Sequential(
Unflatten(batch_size, 1, 28, 28),
nn.Conv2d(1, 32, 5, 1),
nn.LeakyReLU(0.01, inplace=True),
nn.MaxPool2d(2, 2),
nn.Conv2d(32, 64, 5, 1),
nn.LeakyReLU(0.01, inplace=True),
nn.MaxPool2d(2, 2),
Flatten(),
nn.Linear(n_to_Flatten**2*64, 4*4*64),
nn.LeakyReLU(0.01, inplace=True),
nn.Linear(4*4*64, 1)
)
data = Variable(loader_train.__iter__().next()[0]).type(dtype)
b = build_dc_classifier().type(dtype)
out = b(data)
def test_dc_classifer(true_count=1102721):
model = build_dc_classifier()
cur_count = count_params(model)
if cur_count != true_count:
print('Incorrect number of parameters in generator. Check your achitecture.')
else:
print('Correct number of parameters in generator.')
test_dc_classifer()
def build_dc_generator(noise_dim=NOISE_DIM):
"""
Build and return a PyTorch model implementing the DCGAN generator using
the architecture described above.
"""
return nn.Sequential(
nn.Linear(noise_dim, 1024),
nn.ReLU(inplace=True),
nn.BatchNorm1d(1024),
nn.Linear(1024, 7*7*128),
nn.ReLU(inplace=True),
nn.BatchNorm1d(7*7*128),
Unflatten(batch_size, 128, 7, 7),
nn.ConvTranspose2d(128, 64, 4, 2, 1),
nn.ReLU(inplace=True),
nn.BatchNorm2d(64),
nn.ConvTranspose2d(64, 1, 4, 2, 1),
nn.Tanh(),
Flatten()
)
test_g_gan = build_dc_generator().type(dtype)
test_g_gan.apply(initialize_weights)
fake_seed = Variable(torch.randn(batch_size, NOISE_DIM)).type(dtype)
fake_images = test_g_gan.forward(fake_seed)
fake_images.size()
def test_dc_generator(true_count=6580801):
model = build_dc_generator(4)
cur_count = count_params(model)
if cur_count != true_count:
print('Incorrect number of parameters in generator. Check your achitecture.')
else:
print('Correct number of parameters in generator.')
test_dc_generator()
D_DC = build_dc_classifier().type(dtype)
D_DC.apply(initialize_weights)
G_DC = build_dc_generator().type(dtype)
G_DC.apply(initialize_weights)
D_DC_solver = get_optimizer(D_DC)
G_DC_solver = get_optimizer(G_DC)
run_a_gan(D_DC, G_DC, D_DC_solver, G_DC_solver, discriminator_loss, generator_loss, num_epochs=5)
| 0.923147 | 0.990569 |
<a href="https://colab.research.google.com/github/microprediction/m6/blob/main/notebook_examples/auto_completion_example.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
!pip install --upgrade git+https://github.com/microprediction/winning.git
!pip install --upgrade scipy
!pip install array-to-latex
```
### Fill table of 5-way rank probabilities from "best stock" probabilities
```
from winning.lattice import skew_normal_density
from winning.lattice_plot import densitiesPlot
from pprint import pprint
```
We infer 5-way rank probabilities from the probability that a stock will perform the best.
This method assumes that you believe stocks have the same return distribution (one month hence) up to a translation. Of course this isn't the only assumption that might be applied. See the other notebooks.
```
a = 1.0 # Skew parameter
unit = 0.1 # Lattice step size
density = skew_normal_density(L=100, unit=unit, scale=1.0, loc=0, a=a)
assets = ['Asset '+str(i) for i in range(1,6)]
p = [ 0.25, 0.15, 0.2, 0.22, 0.18 ] # Probability of being lowest return stock
```
Infer relative location parameters, called "abilities" here
```
from winning.lattice_calibration import state_price_implied_ability
abilities = state_price_implied_ability(density=density, prices=p, unit=unit)
print(abilities)
```
Plot the offset densities
```
from winning.lattice import densities_from_offsets
densities = densities_from_offsets( density=density, offsets = [abl/unit for abl in abilities])
densitiesPlot(densities=densities,unit=0.1,legend=assets)
from winning.lattice_copula import gaussian_copula_five
import time
st = time.time()
rank_probs = gaussian_copula_five(densities, rho=0.0)
print({'elapsed':time.time()-st})
```
We show the 5-way rank probabilties and check that the probability of a stock being the worst is roughly the same as what we assumed at the outset, at least with rho=0
```
import pandas as pd
index = ['Rank '+str(i) for i in range(1,6)]
df = pd.DataFrame(columns=assets,data=rank_probs, index=index).transpose()
df['p']=p
df['Rank 1 - p'] = df['Rank 1']-df['p']
df
```
Next we'll assume 25% correlation between the assets (or technically, between auxiliary random N(0,1) variables ... not the assets themselves ... see the paper)
```
rank_probs = gaussian_copula_five(densities, rho=0.25)
df = pd.DataFrame(columns=assets,data=rank_probs, index=index).transpose()
df['p']=p
df['Rank 1 - p'] = df['Rank 1']-df['p']
df
```
That's pretty much the end of the show, although I need a few things below for writeup and comparisons elsewhere.
You'll notice that this time the probability of the stock being the worst deviates a little more, but is still extremely close. In general the rank probabilities will be largely insensitive to the choice of correlation parameter rho, because this is something of an Urn game and the common factor increases or decreases their absolute probabilities by roughly the same amount.
```
import array_to_latex as a2l
import numpy as np
a2l.to_ltx(np.asarray(rank_probs), frmt = '{:6.3f}', arraytype = 'array')
a2l.to_ltx(df, frmt = '{:6.3f}')
df_not = df.copy()
for i in [2,3,4,5]:
df_not['Rank '+str(i)]=['?']*5
del df_not['p']
del df_not['Rank 1 - p']
df_not
a2l.to_ltx(df_not)
```
Now adding some more output by request...
```
from winning.lattice import symmetric_lattice, implied_L
L = implied_L(densities[0])
symmetric_lattice(L=L,unit=0.1)
pprint(densities)
```
|
github_jupyter
|
!pip install --upgrade git+https://github.com/microprediction/winning.git
!pip install --upgrade scipy
!pip install array-to-latex
from winning.lattice import skew_normal_density
from winning.lattice_plot import densitiesPlot
from pprint import pprint
a = 1.0 # Skew parameter
unit = 0.1 # Lattice step size
density = skew_normal_density(L=100, unit=unit, scale=1.0, loc=0, a=a)
assets = ['Asset '+str(i) for i in range(1,6)]
p = [ 0.25, 0.15, 0.2, 0.22, 0.18 ] # Probability of being lowest return stock
from winning.lattice_calibration import state_price_implied_ability
abilities = state_price_implied_ability(density=density, prices=p, unit=unit)
print(abilities)
from winning.lattice import densities_from_offsets
densities = densities_from_offsets( density=density, offsets = [abl/unit for abl in abilities])
densitiesPlot(densities=densities,unit=0.1,legend=assets)
from winning.lattice_copula import gaussian_copula_five
import time
st = time.time()
rank_probs = gaussian_copula_five(densities, rho=0.0)
print({'elapsed':time.time()-st})
import pandas as pd
index = ['Rank '+str(i) for i in range(1,6)]
df = pd.DataFrame(columns=assets,data=rank_probs, index=index).transpose()
df['p']=p
df['Rank 1 - p'] = df['Rank 1']-df['p']
df
rank_probs = gaussian_copula_five(densities, rho=0.25)
df = pd.DataFrame(columns=assets,data=rank_probs, index=index).transpose()
df['p']=p
df['Rank 1 - p'] = df['Rank 1']-df['p']
df
import array_to_latex as a2l
import numpy as np
a2l.to_ltx(np.asarray(rank_probs), frmt = '{:6.3f}', arraytype = 'array')
a2l.to_ltx(df, frmt = '{:6.3f}')
df_not = df.copy()
for i in [2,3,4,5]:
df_not['Rank '+str(i)]=['?']*5
del df_not['p']
del df_not['Rank 1 - p']
df_not
a2l.to_ltx(df_not)
from winning.lattice import symmetric_lattice, implied_L
L = implied_L(densities[0])
symmetric_lattice(L=L,unit=0.1)
pprint(densities)
| 0.474875 | 0.974362 |
# Stochastic Differential Equation: Ion channel noise in Hodgkin-Huxley neurons
In the following, we estimate parameters of the stochastic differential equation model of ion channel noise in Hodgkin-Huxley neurons presented in:
> Goldwyn, Joshua H., Nikita S. Imennov, Michael Famulare, and Eric Shea-Brown. “Stochastic Differential Equation Models for Ion Channel Noise in Hodgkin-Huxley Neurons.” Physical Review E 83, no. 4 (2011): 041908. doi:10.1103/PhysRevE.83.041908.
The code was implemented in Fortran 95 and made available in ModelDB: [ModelDB](https://senselab.med.yale.edu/ModelDB/showmodel.cshtml?model=128502).
(The code is not included in pyABC and neither developed nor maintained by the pyABC developers.)
## Download and compilation of the Fortran model
We start by downloading the code from ModelDB. For this, the ``requests`` package is needed.
```
import requests
URL = ("https://senselab.med.yale.edu/modeldb/"
"eavBinDown.cshtml?o=128502&a=23&mime=application/zip")
req = requests.request("GET", URL)
```
The zip file to which ``URL`` points is stored in memory.
The code is then extracted into a temporary directory and compiled
using ``make HH_run`` provided as part of the download from ModelDB.
The Fortran compiler ``gfortran`` is required for compilation.
```
import os
from zipfile import ZipFile
import subprocess
import tempfile
from io import BytesIO
tempdir = tempfile.mkdtemp()
archive = ZipFile(BytesIO(req.content))
archive.extractall(tempdir)
ret = subprocess.run(
["make", "HH_run"],
cwd=os.path.join(tempdir, "ModelDBFolder"))
EXEC = os.path.join(tempdir, "ModelDBFolder", "HH_run")
print(f"The executable location is {EXEC}")
```
The variable ``EXEC`` points to the executable.
A simulate function is defined which uses the ``subprocess.run`` function to execute the external binary.
The external binary writes to stdout. The output is captured and stored in a pandas dataframe.
This dataframe is returned by the ``simulate`` function.
```
import pandas as pd
import numpy as np
def simulate(model=2, membrane_dim=10, time_steps=1e4,
time_step_size=0.01, isi=100, dc=20, noise=0,
sine_amplitude=0, sine_frequency=0,
voltage_clamp=0, data_to_print=1, rng_seed=None):
"""
Simulate the SDE Ion Channel Model defined
in an external Fortran simulator.
Returns: pandas.DataFrame
Index: t, Time
Columns: V, Na, K
V: Voltage
Na, K: Proportion of open channels
"""
if rng_seed is None:
rng_seed = np.random.randint(2**32-2) + 1
membrane_area = membrane_dim**2
re = subprocess.run(
[EXEC, str(model),
# the binary cannot very long floats
f"{membrane_area:.5f}", str(time_steps),
str(time_step_size), str(isi), f"{dc:.5f}",
str(noise), str(sine_amplitude), str(sine_frequency),
str(voltage_clamp), str(data_to_print),
str(rng_seed)],
stdout=subprocess.PIPE)
df = pd.read_table(BytesIO(re.stdout),
delim_whitespace=True,
header=None, index_col=0,
names=["t", "V", "Na", "K"])
return df
```
## Generating the observed data
We run a simulations and plot the fraction of open "K" channels and open "Na" channels:
```
import matplotlib.pyplot as plt
%matplotlib inline
gt = {"dc": 20, "membrane_dim": 10}
fig, axes = plt.subplots(nrows=2, sharex=True)
fig.set_size_inches((12,8))
for _ in range(10):
observation = simulate(**gt)
observation.plot(y="K", color="C1", ax=axes[0]);
observation.plot(y="Na", color="C0", ax=axes[1]);
for ax in axes:
ax.legend().set_visible(False)
axes[0].set_title("K")
axes[0].set_ylabel("K")
axes[1].set_title("Na")
axes[1].set_ylabel("Na");
```
We observe how the channels open and close and also that the individual trajectores differ from realization to realization, even though we simulate for the exact same parameter set. We take the last simulation as observed data.
## Defining distance and prior
We'll now demonstrate how to use pyABC to estimate parameters of the model.
Here, we'll focus on the ``dc`` and the ``membrane_dim`` parameters.
The ``dc`` parameter describes the input current, the ``membrane_dim`` is the square root of the membrane area.
We choose uniform priors:
```
from pyabc import Distribution, RV, ABCSMC
dcmin, dcmax = 2, 30
memmin, memmax = 1, 12
prior = Distribution(
dc=RV("uniform", dcmin, dcmax - dcmin),
membrane_dim=RV("uniform", memmin, memmax - memmin))
```
The distance function is defined as $L_2$ norm between the fractions of open "K" channels.
```
def distance(x, y):
diff = x["data"]["K"] - y["data"]["K"]
dist = np.sqrt(np.sum(diff**2))
return dist
```
We also define a small ``simulate_pyabc`` wrapper function, which wraps the ``simulate`` function.
This is needed to comply with the interface expected by ``pyABC``.
```
def simulate_pyabc(parameter):
res = simulate(**parameter)
return {"data": res}
```
## Performing parameter inference with pyABC
We are now ready to start pyABC.
As usually, we first initialize the ABCSMC object,
then pass the observed data and the database location in which to store
the logged parameters and summary statistics,
and finally run the inference until the maximum number of allowed populations
``max_nr_populations`` or the final acceptance threshold ``minimum_epsilon`` is reached.
```
abc = ABCSMC(simulate_pyabc, prior, distance,
population_size=35)
abc_id = abc.new("sqlite:///"
+ os.path.join(tempdir, "test.db"),
{"data": observation})
history = abc.run(max_nr_populations=10, minimum_epsilon=6)
```
## Visualization of the estimated parameters
We plot the posterior distribution after a few generations together with the parameters
generating the observed data (the dotted line and the orange dot).
```
from pyabc.visualization import plot_kde_matrix
dfw = history.get_distribution(m=0)
grid = plot_kde_matrix(*dfw,
limits={"dc": (dcmin, dcmax),
"membrane_dim": (memmin, memmax)})
grid.map_diag(lambda x, **kwargs: plt.gca().axvline(
gt[x.name], color="k", linestyle="dotted"))
grid.map_lower(lambda x, y, **kwargs: plt.gca().scatter(
[gt[x.name]], [gt[y.name]], color="orange"))
plt.gcf().set_size_inches(8, 8)
```
The ``dc`` parameter is very well detected.
(Don't get confused by the y-axis. It applies to the scatterplot, not to the marginal distribution.)
The distribution of ``membrane_dim`` is broader.
(Note that even the exact posterior is not necessarily peaked at the ground truth parameters).
## Evaluation of the fit
We compare four types of data:
1. samples from the prior distribution,
2. samples from the posterior distribution,
3. the data generated by the accepted parameters, and
4. the observation.
```
from pyabc.transition import MultivariateNormalTransition
fig, axes = plt.subplots(nrows=3, sharex=True)
fig.set_size_inches(8, 12)
n = 5 # Number of samples to plot from each category
# Plot samples from the prior
alpha = .5
for _ in range(n):
prior_sample = simulate(**prior.rvs())
prior_sample.plot(y="K", ax=axes[0],
color="C1", alpha=alpha)
# Fit a posterior KDE and plot samples form it
posterior = MultivariateNormalTransition()
posterior.fit(*history.get_distribution(m=0))
for _ in range(n):
posterior_sample = simulate(**posterior.rvs())
posterior_sample.plot(y="K", ax=axes[1],
color="C0", alpha=alpha)
# Plot the stored summary statistics
sum_stats = history.get_weighted_sum_stats_for_model(m=0, t=history.max_t)
for stored in sum_stats[1][:n]:
stored["data"].plot(y="K", ax=axes[2],
color="C2", alpha=alpha)
# Plot the observation
for ax in axes:
observation.plot(y="K", ax=ax, color="k", linewidth=1.5)
ax.legend().set_visible(False)
ax.set_ylabel("K");
# Add a legend with pseudo artists to first plot
axes[0].legend([plt.plot([0], color="C1")[0],
plt.plot([0], color="C0")[0],
plt.plot([0], color="C2")[0],
plt.plot([0], color="k")[0]],
["Prior", "Posterior",
"Stored, accepted", "Observation"],
bbox_to_anchor=(.5, 1),
loc="lower center",
ncol=4);
```
We observe that the samples from the prior exhibit the largest variation and do not resemble the observation well.
The samples from the posterior are much closer to the observed data.
Even a little bit closer are the samples generated by the accepted parameters.
This has at least two reasons: First, the posterior KDE-fit smoothes the particle populations. Second, the sample generated by a parameter that was accepted is biased towards being more similar to the observed data as compared to a random sample from that parameter.
|
github_jupyter
|
import requests
URL = ("https://senselab.med.yale.edu/modeldb/"
"eavBinDown.cshtml?o=128502&a=23&mime=application/zip")
req = requests.request("GET", URL)
import os
from zipfile import ZipFile
import subprocess
import tempfile
from io import BytesIO
tempdir = tempfile.mkdtemp()
archive = ZipFile(BytesIO(req.content))
archive.extractall(tempdir)
ret = subprocess.run(
["make", "HH_run"],
cwd=os.path.join(tempdir, "ModelDBFolder"))
EXEC = os.path.join(tempdir, "ModelDBFolder", "HH_run")
print(f"The executable location is {EXEC}")
import pandas as pd
import numpy as np
def simulate(model=2, membrane_dim=10, time_steps=1e4,
time_step_size=0.01, isi=100, dc=20, noise=0,
sine_amplitude=0, sine_frequency=0,
voltage_clamp=0, data_to_print=1, rng_seed=None):
"""
Simulate the SDE Ion Channel Model defined
in an external Fortran simulator.
Returns: pandas.DataFrame
Index: t, Time
Columns: V, Na, K
V: Voltage
Na, K: Proportion of open channels
"""
if rng_seed is None:
rng_seed = np.random.randint(2**32-2) + 1
membrane_area = membrane_dim**2
re = subprocess.run(
[EXEC, str(model),
# the binary cannot very long floats
f"{membrane_area:.5f}", str(time_steps),
str(time_step_size), str(isi), f"{dc:.5f}",
str(noise), str(sine_amplitude), str(sine_frequency),
str(voltage_clamp), str(data_to_print),
str(rng_seed)],
stdout=subprocess.PIPE)
df = pd.read_table(BytesIO(re.stdout),
delim_whitespace=True,
header=None, index_col=0,
names=["t", "V", "Na", "K"])
return df
import matplotlib.pyplot as plt
%matplotlib inline
gt = {"dc": 20, "membrane_dim": 10}
fig, axes = plt.subplots(nrows=2, sharex=True)
fig.set_size_inches((12,8))
for _ in range(10):
observation = simulate(**gt)
observation.plot(y="K", color="C1", ax=axes[0]);
observation.plot(y="Na", color="C0", ax=axes[1]);
for ax in axes:
ax.legend().set_visible(False)
axes[0].set_title("K")
axes[0].set_ylabel("K")
axes[1].set_title("Na")
axes[1].set_ylabel("Na");
from pyabc import Distribution, RV, ABCSMC
dcmin, dcmax = 2, 30
memmin, memmax = 1, 12
prior = Distribution(
dc=RV("uniform", dcmin, dcmax - dcmin),
membrane_dim=RV("uniform", memmin, memmax - memmin))
def distance(x, y):
diff = x["data"]["K"] - y["data"]["K"]
dist = np.sqrt(np.sum(diff**2))
return dist
def simulate_pyabc(parameter):
res = simulate(**parameter)
return {"data": res}
abc = ABCSMC(simulate_pyabc, prior, distance,
population_size=35)
abc_id = abc.new("sqlite:///"
+ os.path.join(tempdir, "test.db"),
{"data": observation})
history = abc.run(max_nr_populations=10, minimum_epsilon=6)
from pyabc.visualization import plot_kde_matrix
dfw = history.get_distribution(m=0)
grid = plot_kde_matrix(*dfw,
limits={"dc": (dcmin, dcmax),
"membrane_dim": (memmin, memmax)})
grid.map_diag(lambda x, **kwargs: plt.gca().axvline(
gt[x.name], color="k", linestyle="dotted"))
grid.map_lower(lambda x, y, **kwargs: plt.gca().scatter(
[gt[x.name]], [gt[y.name]], color="orange"))
plt.gcf().set_size_inches(8, 8)
from pyabc.transition import MultivariateNormalTransition
fig, axes = plt.subplots(nrows=3, sharex=True)
fig.set_size_inches(8, 12)
n = 5 # Number of samples to plot from each category
# Plot samples from the prior
alpha = .5
for _ in range(n):
prior_sample = simulate(**prior.rvs())
prior_sample.plot(y="K", ax=axes[0],
color="C1", alpha=alpha)
# Fit a posterior KDE and plot samples form it
posterior = MultivariateNormalTransition()
posterior.fit(*history.get_distribution(m=0))
for _ in range(n):
posterior_sample = simulate(**posterior.rvs())
posterior_sample.plot(y="K", ax=axes[1],
color="C0", alpha=alpha)
# Plot the stored summary statistics
sum_stats = history.get_weighted_sum_stats_for_model(m=0, t=history.max_t)
for stored in sum_stats[1][:n]:
stored["data"].plot(y="K", ax=axes[2],
color="C2", alpha=alpha)
# Plot the observation
for ax in axes:
observation.plot(y="K", ax=ax, color="k", linewidth=1.5)
ax.legend().set_visible(False)
ax.set_ylabel("K");
# Add a legend with pseudo artists to first plot
axes[0].legend([plt.plot([0], color="C1")[0],
plt.plot([0], color="C0")[0],
plt.plot([0], color="C2")[0],
plt.plot([0], color="k")[0]],
["Prior", "Posterior",
"Stored, accepted", "Observation"],
bbox_to_anchor=(.5, 1),
loc="lower center",
ncol=4);
| 0.67694 | 0.944944 |
```
!git clone https://github.com/mueedurrehman/distiller.git
%cd distiller
!pip3 install -e .
from google.colab import drive
drive.mount('/content/drive')
import torch
import torchvision
import torchvision.transforms as transforms
import torchvision.models as models
from torchsummary import summary
import matplotlib.pyplot as plt
import numpy as np
import torch.nn as nn
import torch.nn.functional as F
import time
import math
import yaml
import distiller
from collections import OrderedDict
import os
import pandas as pd
!cp "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Baseline Models/baseResnet56" .
# !cp ../../../drive/MyDrive/CS6787\ -\ Final\ Project/Google\ Colab\ Scripts/Quantization\ Models/Resnet56\ Quantization\ Stats/resnet56_quant_stats.yaml .
```
Asymmetric U
```
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/4bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/6bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/8bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/10bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/12bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/14bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/16bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
```
Asymetric S
```
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/4bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/6bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/8bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/10bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/12bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/14bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/16bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
```
Symetric
```
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/4bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/6bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/8bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/10bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/12bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/14bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/16bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
```
|
github_jupyter
|
!git clone https://github.com/mueedurrehman/distiller.git
%cd distiller
!pip3 install -e .
from google.colab import drive
drive.mount('/content/drive')
import torch
import torchvision
import torchvision.transforms as transforms
import torchvision.models as models
from torchsummary import summary
import matplotlib.pyplot as plt
import numpy as np
import torch.nn as nn
import torch.nn.functional as F
import time
import math
import yaml
import distiller
from collections import OrderedDict
import os
import pandas as pd
!cp "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Baseline Models/baseResnet56" .
# !cp ../../../drive/MyDrive/CS6787\ -\ Final\ Project/Google\ Colab\ Scripts/Quantization\ Models/Resnet56\ Quantization\ Stats/resnet56_quant_stats.yaml .
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/4bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/6bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/8bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/10bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/12bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/14bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/16bitAsymmetricUPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/4bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/6bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/8bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/10bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/12bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/14bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/16bitAsymmetricSPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/4bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/6bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/8bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/10bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/12bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/14bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
!python3 "/content/distiller/examples/classifier_compression/compress_classifier.py" --arch resnet56_cifar -p 10 ./data --resume="baseResnet56" --out-dir "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults" --evaluate --quantize-eval --qe-config-file "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingSchedules/None/16bitSymmetricPost.yaml" -o "/content/drive/MyDrive/Colab Notebooks/6787 Notebooks/Google Colab Scripts/Schedules/Quantization Schedules/PostTrainingResults"
| 0.415966 | 0.264622 |
# Slice sampling
## 1. introduction(why do we need slice sampling?)
MH algorithm and Gibbs sampling are fundation of MCMC algorithm. These two algorithm are widely used for inference problems, but they are belong to **random walk** sampling algorithms and they are very sensitive to the step size of proposal distribution.
**Random walk** means that the next state is predicted based on current state, and this process is essentially random walk. What's the limitation of random walk algorithm:
1. overshoot and oscillate at pintches
2. very sensitive to the step size: large step size leads to lots of rejected samples, and small step size will take a lot of time to travel the whole "distribution". Soemtimes, to specifies the step size is hard because the detail balance puts a constraint preventing any proposal width scale parameters from being set based on past iterations of the current chain.
3. In very high dimension, MH algorithm is very inefficient due to random walk.
4. MH cannot travel a long distance between isolate local minimum.
5. large correlation.
## 2. What's the splice sampling?
- Pick an initial point x0x0 from our posterior
- Draw y0y0 from U(0, f(x0x0))
- Repeat for N samples
- Select the interval (e.g. stepping out, etc)
- Sample xixi from that interval (e.g. shrinkage)
- Draw yiyi from U(0, f(xixi))
### 2.1 The univariate case
1. Initially, randomly selecting $x^{(k)}$ when $k=0$.
2. Draw $y^{(k)}$ from $U(0, f(x^{(k)}))$.
3. Find an interval $I=(L, R)$ around $x^{k}$ corresponding to $S=x,s.t. f(x)>y^{(k)}$
4. Draw $x^{(k+1)}$ from $U(I)$.
5. Repeat step (2).
Note, $S$ is the perfect region and $I$ is the interval we choose to corrsepond to this.
```
import numpy as np
import scipy as sp
import matplotlib.pyplot as plt
import seaborn as sns
from scipy.stats import norm
%matplotlib inline
mu1 = 3; mu2=10; sigma1=1; sigma2=2; l1=.0; l2=1.0;
normal = 1./np.sqrt(2*np.pi*sigma2**2)
fun = lambda x: l1*norm.pdf(x, mu1, sigma1) + l2*norm.pdf(x, mu2, sigma2)
invfunR = lambda y: np.sqrt(-2*sigma2**2*np.log(y/normal))+mu2
invfunL = lambda y: -np.sqrt(-2*sigma2**2*np.log(y/normal))+mu2
x = np.linspace(0,20, 100)
plt.figure(figsize=[20,12])
plt.subplot(3,2,1)
plt.plot(x, fun(x), 'b')
np.random.seed(17)
x0=np.random.uniform(low=5, high=15, size=1)
plt.plot( [x0 ,x0], [0, 0.2], 'r-.')
plt.title('Step 1: Initialize')
plt.annotate( '$x^{0}$', [x0-0.2,.01], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.subplot(3,2,2)
plt.plot(x, fun(x), 'b')
plt.annotate( '$x^{0}$', [x0-0.2,.01], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.annotate( '$f(x^0)$', [x0+0.2, fun(x0)], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.title('Step 2: Draw $y^{0}$')
y0=np.random.uniform(low=0, high=fun(x0), size=1)
plt.plot( [x0,x0], [y0, y0], 'bs')
plt.annotate( '$y^{0}$', [x0,y0], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.subplot(3,2,3)
plt.plot(x, fun(x), 'b')
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.plot( [x0,x0], [y0, y0], 'bs')
plt.plot( [invfunL(y0), invfunR(y0)] , [y0, y0], 'r-.')
#plt.plot( [2,4] , [y0, y0], 'r-.')
plt.title('Step 3: Find interval I')
plt.subplot(3,2,4)
plt.plot(x, fun(x), 'b')
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
x1=np.random.uniform(low=8.5, high=11.5, size=1)
plt.plot( [x1,x1], [y0, y0], 'bs')
plt.plot( [invfunL(y0), invfunR(y0)] , [y0, y0], 'r-.')
#plt.plot( [2,4] , [y0, y0], 'r-.')
plt.title('Step 4: Sample $x^{1}$ from interval I')
plt.annotate( '$x^{1}$', [x1-0.7,y0], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.subplot(3,2,5)
plt.plot(x, fun(x), 'b')
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.plot( [x1 ,x1], [0, fun(x1)], 'g-.')
y1=np.random.uniform(low=0, high=fun(x1), size=1)
plt.plot( [x1,x1], [y0, y0], 'bs')
plt.plot( [x1,x1], [y1, y1], 'gs')
plt.plot( [invfunL(y0), invfunR(y0)] , [y0, y0], 'r-.')
#plt.plot( [2,4] , [y0, y0], 'r-.')
plt.title('Step 5: Draw $y^1$')
#plt.annotate( '$x^{1}$', [9.5,y0], xytext=None, xycoords='data',
# textcoords='data', arrowprops=None)
```
### 2.2 The problem of the multimodal case
```
mu1=3; mu2=10; sigma1=1; sigma2=2; l1=.30; l2=.70;
normal = 1./np.sqrt(2*np.pi*sigma2**2)
fun=lambda x: l1*norm.pdf(x, mu1, sigma1)+l2*norm.pdf(x, mu2, sigma2)
x = np.linspace(0,20, 100)
plt.figure(figsize=[20,12])
plt.subplot(2,2,1)
plt.plot(x, fun(x), 'b')
np.random.seed(16)
x0=np.random.uniform(low=0, high=20, size=1)
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.title('Step 1: Initialize')
plt.annotate( '$x^{0}$', [x0+0.1,.001], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.subplot(2,2,2)
plt.plot(x, fun(x), 'b')
plt.annotate( '$x^{0}$', [x0,.001], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.annotate( '$f(x^0)$', [x0,fun(x0)], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.title('Step 2: Draw $y^{0}$')
y0=np.random.uniform(low=0, high=fun(x0), size=1)
plt.plot( [x0,x0], [y0, y0], 'bs')
plt.annotate( '$y^{0}$', [10.5,.15], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.subplot(2,2,3)
plt.plot(x, fun(x), 'b')
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.plot( [x0,x0], [y0, y0], 'bs')
plt.plot( [5.7,14.2] , [y0, y0], 'r-.')
plt.plot( [1.3,5.1] , [y0, y0], 'r-.')
plt.title('Step 3: Find interval I')
plt.subplot(2,2,4)
plt.plot(x, fun(x), 'b')
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.plot( [9,9], [y0, y0], 'bs')
plt.plot( [5.7,14.2] , [y0, y0], 'r-.')
plt.plot( [1.3,5.1] , [y0, y0], 'r-.')
plt.title('Step 4: Sample $x^{1}$ from interval I')
plt.annotate( '$x^{1}$', [9.5,y0], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
```
## 3 Stepping out algorithm
"step out" is procedure to determine the proposed interval.
The idea behind stepping out is that you expand your interval by fixed widths ww until your endpoints are outside of S. The full algorithm is as follows:
- Set w = width of your interval expansions
- Draw u, v ~ Unif(0,1)
- set $L = x(0)−wu$,$R=L+w$ (so $x^{(0)}$ lies in $[L, R]$ )
- while $y < f(L)$ (here’s where we extend left interval)
- L = L - w
- while $y < f(R)$ ( here’s where we extend the right interval)
- R = R + w
The final interval will be larger than S. We will later see how we accept/reject to ensure our samples are from within S.
```
import time
def plot_bounds(k, kmax, L, R, y, ts=1):
if k <= kmax:
plt.plot( [L,R], [y, y], 'r', marker=".", linestyle='None')
plt.annotate("{}".format(k), xy=(L,y))
plt.annotate("{}".format(k), xy=(R,y))
#time.sleep(ts)
def plot_yline(k, kmax, xprev, y, f, ts
=1):
if k <= kmax:
plt.plot( [x_prev ,x_prev], [0, f(x_prev)], 'r-', lw=1)
plt.plot( [x_prev,x_prev], [y, y], 'rd', alpha=0.2)
plt.annotate("{}".format(k), xy=(x_prev,y))
#time.sleep(ts)
def plot_prop(k, kmax, x_prop, y, accept=None, ts=1):
if accept=="y":
symbol="ro"
alpha=0.3
elif accept=="n":
symbol="ko"
alpha=0.1
else:
symbol="bs"
alpha=0.5
if k <= kmax:
plt.plot( [x_prop, x_prop], [y, y], symbol, alpha=alpha)
plt.annotate("{}".format(k), xy=(x_prop,y))
#time.sleep(ts)
w=1.0
x = np.linspace(0,20, 100)
L=0; R=0;
x_prev = np.random.uniform(low=0, high=17)
fig = plt.figure()
iters=10000
trace=[]
kmax=1
plt.plot(x, fun(x),'g')
for k in range(iters):
y_samp = np.random.uniform(low=0, high=fun(x_prev))
plot_yline(k, 5, x_prev, y_samp, fun)
# widen left
U = np.random.rand()
L=x_prev-U*w
R=x_prev+w*(1.0-U)
while fun(L)>y_samp:
L = L-w
plot_bounds(k, kmax, L, R, y_samp)
while fun(R)>y_samp:
R = R+w
plot_bounds(k, kmax, L, R, y_samp)
#now propose new x on L,R
x_prop= np.random.uniform(low=L, high=R)
if k <= kmax:
print("L,R, xprop", L, R, x_prop)
#now accept if kosher, ie if function at new x is higher than the y we sampled at prev x
#plot_prop(k, kmax, x_prop, y_samp, accept=None)
accept=False
while accept==False:
if y_samp < fun(x_prop):
x_prev = x_prop
trace.append(x_prop)
plot_prop(k, kmax, x_prop, y_samp, accept='y')
accept = True
else: #propose again: in real slice we would shrink
plot_prop(k, kmax, x_prop, y_samp, accept='n')
x_prop= np.random.uniform(low=L, high=R)
plt.show()
plt.hist(trace, bins=50, alpha=0.3, normed=True);
#sns.kdeplot(xsmp)
plt.xlim( [0,20])
plt.plot(x, fun(x))
```
## 4. Shinkage algorithm
Shinkage algorithm is a methods to sample from the univariate window.
The idea behind the shrinkage procedure is that you sample from your interval, if the sample’s not in SS, make that point the new endpoint of your interval decreasing the length of your interval. Otherwise keep the sample and go back to the interval selection problem.
The procedure of Shinkage algorithm:
- Start with interval $I=(L,R)$.
- Current sample is $x^{(k)}$ and $y^{(k)}$.
- repeat until loop exits
- sample $x^{(*)}$ uniformly from $[L, R]$.
- if $y^{(k)}< f(x^{(*)})$:
- accept $x^{(*)}$ and end loop
- else
- if $x^{(*)} < x^{(k)}$, $L=x^{(*)}$.
- if $x^{(*)} > x^{(k)}$, $R=x^{(*)}$
```
w=1.0
x = np.linspace(0,20, 100)
L=0; R=0;
x_prev = np.random.uniform(low=0, high=17)
fig = plt.figure()
iters=10000
trace=[]
kmax=1
plt.plot(x, fun(x),'g')
for k in range(iters):
y_samp = np.random.uniform(low=0, high=fun(x_prev))
plot_yline(k, 5, x_prev, y_samp, fun)
# widen left
U = np.random.rand()
L=x_prev-U*w
R=x_prev+w*(1.0-U)
while fun(L)>y_samp:
L = L-w
plot_bounds(k, kmax, L, R, y_samp)
while fun(R)>y_samp:
R = R+w
plot_bounds(k, kmax, L, R, y_samp)
#now propose new x on L,R
while 1:
x_prop= np.random.uniform(low=L, high=R)
if k <= kmax:
print("L,R, xprop", L, R, x_prop)
if y_samp < fun(x_prop):
x_prev = x_prop
trace.append(x_prop)
plot_prop(k, kmax, x_prop, y_samp, accept='y')
break
elif x_prop > x_prev:
R = x_prop
elif x_prop < x_prev:
L = x_prop
plt.hist(trace, bins=100, alpha=0.3, normed=True);
#sns.kdeplot(xsmp)
plt.xlim( [0,20])
plt.plot(x, fun(x))
```
## 5. Why does slice sampling work?(detail balance)
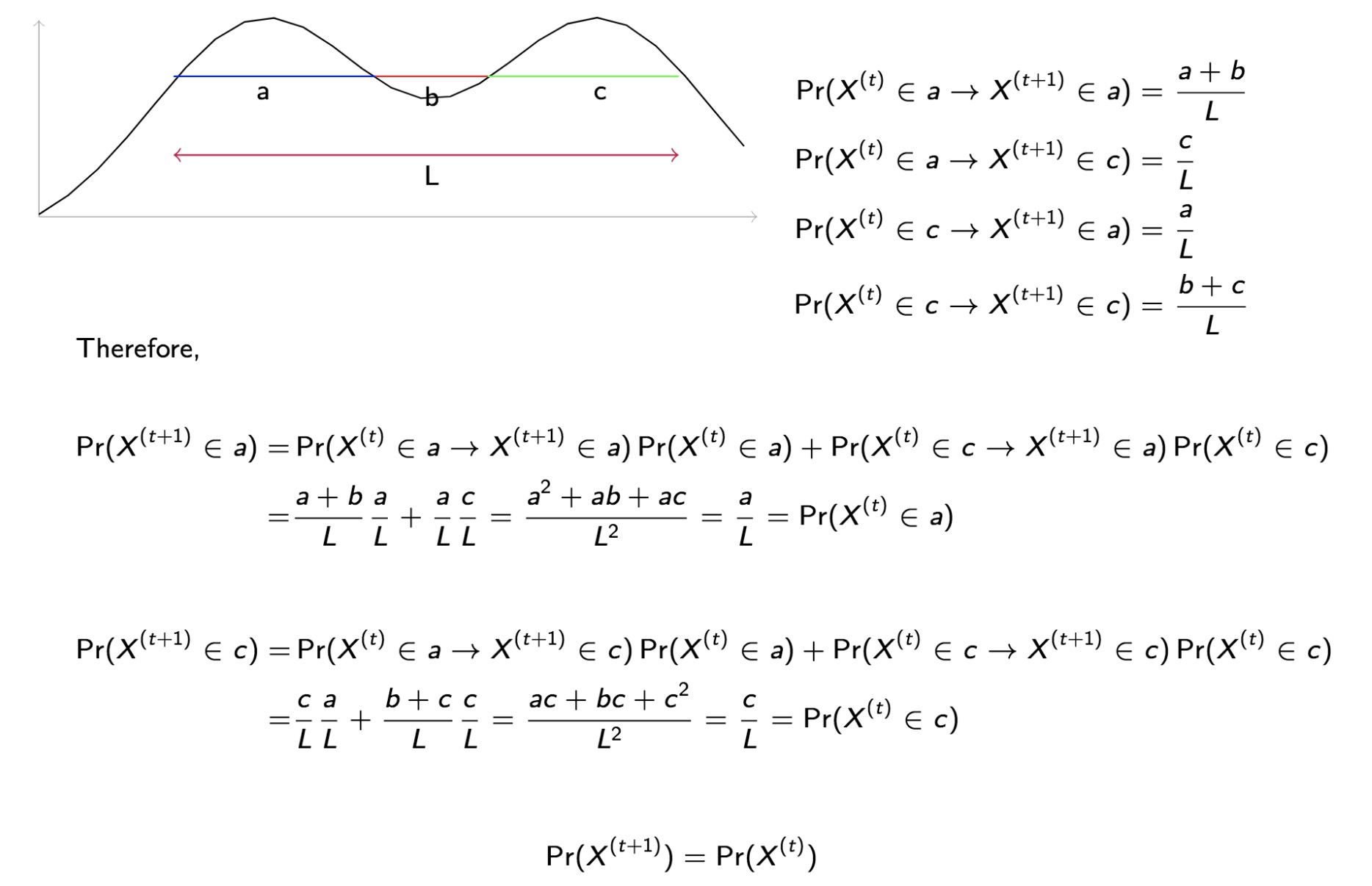
# Reference
```
1.
```
|
github_jupyter
|
import numpy as np
import scipy as sp
import matplotlib.pyplot as plt
import seaborn as sns
from scipy.stats import norm
%matplotlib inline
mu1 = 3; mu2=10; sigma1=1; sigma2=2; l1=.0; l2=1.0;
normal = 1./np.sqrt(2*np.pi*sigma2**2)
fun = lambda x: l1*norm.pdf(x, mu1, sigma1) + l2*norm.pdf(x, mu2, sigma2)
invfunR = lambda y: np.sqrt(-2*sigma2**2*np.log(y/normal))+mu2
invfunL = lambda y: -np.sqrt(-2*sigma2**2*np.log(y/normal))+mu2
x = np.linspace(0,20, 100)
plt.figure(figsize=[20,12])
plt.subplot(3,2,1)
plt.plot(x, fun(x), 'b')
np.random.seed(17)
x0=np.random.uniform(low=5, high=15, size=1)
plt.plot( [x0 ,x0], [0, 0.2], 'r-.')
plt.title('Step 1: Initialize')
plt.annotate( '$x^{0}$', [x0-0.2,.01], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.subplot(3,2,2)
plt.plot(x, fun(x), 'b')
plt.annotate( '$x^{0}$', [x0-0.2,.01], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.annotate( '$f(x^0)$', [x0+0.2, fun(x0)], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.title('Step 2: Draw $y^{0}$')
y0=np.random.uniform(low=0, high=fun(x0), size=1)
plt.plot( [x0,x0], [y0, y0], 'bs')
plt.annotate( '$y^{0}$', [x0,y0], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.subplot(3,2,3)
plt.plot(x, fun(x), 'b')
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.plot( [x0,x0], [y0, y0], 'bs')
plt.plot( [invfunL(y0), invfunR(y0)] , [y0, y0], 'r-.')
#plt.plot( [2,4] , [y0, y0], 'r-.')
plt.title('Step 3: Find interval I')
plt.subplot(3,2,4)
plt.plot(x, fun(x), 'b')
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
x1=np.random.uniform(low=8.5, high=11.5, size=1)
plt.plot( [x1,x1], [y0, y0], 'bs')
plt.plot( [invfunL(y0), invfunR(y0)] , [y0, y0], 'r-.')
#plt.plot( [2,4] , [y0, y0], 'r-.')
plt.title('Step 4: Sample $x^{1}$ from interval I')
plt.annotate( '$x^{1}$', [x1-0.7,y0], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.subplot(3,2,5)
plt.plot(x, fun(x), 'b')
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.plot( [x1 ,x1], [0, fun(x1)], 'g-.')
y1=np.random.uniform(low=0, high=fun(x1), size=1)
plt.plot( [x1,x1], [y0, y0], 'bs')
plt.plot( [x1,x1], [y1, y1], 'gs')
plt.plot( [invfunL(y0), invfunR(y0)] , [y0, y0], 'r-.')
#plt.plot( [2,4] , [y0, y0], 'r-.')
plt.title('Step 5: Draw $y^1$')
#plt.annotate( '$x^{1}$', [9.5,y0], xytext=None, xycoords='data',
# textcoords='data', arrowprops=None)
mu1=3; mu2=10; sigma1=1; sigma2=2; l1=.30; l2=.70;
normal = 1./np.sqrt(2*np.pi*sigma2**2)
fun=lambda x: l1*norm.pdf(x, mu1, sigma1)+l2*norm.pdf(x, mu2, sigma2)
x = np.linspace(0,20, 100)
plt.figure(figsize=[20,12])
plt.subplot(2,2,1)
plt.plot(x, fun(x), 'b')
np.random.seed(16)
x0=np.random.uniform(low=0, high=20, size=1)
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.title('Step 1: Initialize')
plt.annotate( '$x^{0}$', [x0+0.1,.001], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.subplot(2,2,2)
plt.plot(x, fun(x), 'b')
plt.annotate( '$x^{0}$', [x0,.001], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.annotate( '$f(x^0)$', [x0,fun(x0)], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.title('Step 2: Draw $y^{0}$')
y0=np.random.uniform(low=0, high=fun(x0), size=1)
plt.plot( [x0,x0], [y0, y0], 'bs')
plt.annotate( '$y^{0}$', [10.5,.15], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
plt.subplot(2,2,3)
plt.plot(x, fun(x), 'b')
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.plot( [x0,x0], [y0, y0], 'bs')
plt.plot( [5.7,14.2] , [y0, y0], 'r-.')
plt.plot( [1.3,5.1] , [y0, y0], 'r-.')
plt.title('Step 3: Find interval I')
plt.subplot(2,2,4)
plt.plot(x, fun(x), 'b')
plt.plot( [x0 ,x0], [0, fun(x0)], 'r-.')
plt.plot( [9,9], [y0, y0], 'bs')
plt.plot( [5.7,14.2] , [y0, y0], 'r-.')
plt.plot( [1.3,5.1] , [y0, y0], 'r-.')
plt.title('Step 4: Sample $x^{1}$ from interval I')
plt.annotate( '$x^{1}$', [9.5,y0], xytext=None, xycoords='data',
textcoords='data', arrowprops=None)
import time
def plot_bounds(k, kmax, L, R, y, ts=1):
if k <= kmax:
plt.plot( [L,R], [y, y], 'r', marker=".", linestyle='None')
plt.annotate("{}".format(k), xy=(L,y))
plt.annotate("{}".format(k), xy=(R,y))
#time.sleep(ts)
def plot_yline(k, kmax, xprev, y, f, ts
=1):
if k <= kmax:
plt.plot( [x_prev ,x_prev], [0, f(x_prev)], 'r-', lw=1)
plt.plot( [x_prev,x_prev], [y, y], 'rd', alpha=0.2)
plt.annotate("{}".format(k), xy=(x_prev,y))
#time.sleep(ts)
def plot_prop(k, kmax, x_prop, y, accept=None, ts=1):
if accept=="y":
symbol="ro"
alpha=0.3
elif accept=="n":
symbol="ko"
alpha=0.1
else:
symbol="bs"
alpha=0.5
if k <= kmax:
plt.plot( [x_prop, x_prop], [y, y], symbol, alpha=alpha)
plt.annotate("{}".format(k), xy=(x_prop,y))
#time.sleep(ts)
w=1.0
x = np.linspace(0,20, 100)
L=0; R=0;
x_prev = np.random.uniform(low=0, high=17)
fig = plt.figure()
iters=10000
trace=[]
kmax=1
plt.plot(x, fun(x),'g')
for k in range(iters):
y_samp = np.random.uniform(low=0, high=fun(x_prev))
plot_yline(k, 5, x_prev, y_samp, fun)
# widen left
U = np.random.rand()
L=x_prev-U*w
R=x_prev+w*(1.0-U)
while fun(L)>y_samp:
L = L-w
plot_bounds(k, kmax, L, R, y_samp)
while fun(R)>y_samp:
R = R+w
plot_bounds(k, kmax, L, R, y_samp)
#now propose new x on L,R
x_prop= np.random.uniform(low=L, high=R)
if k <= kmax:
print("L,R, xprop", L, R, x_prop)
#now accept if kosher, ie if function at new x is higher than the y we sampled at prev x
#plot_prop(k, kmax, x_prop, y_samp, accept=None)
accept=False
while accept==False:
if y_samp < fun(x_prop):
x_prev = x_prop
trace.append(x_prop)
plot_prop(k, kmax, x_prop, y_samp, accept='y')
accept = True
else: #propose again: in real slice we would shrink
plot_prop(k, kmax, x_prop, y_samp, accept='n')
x_prop= np.random.uniform(low=L, high=R)
plt.show()
plt.hist(trace, bins=50, alpha=0.3, normed=True);
#sns.kdeplot(xsmp)
plt.xlim( [0,20])
plt.plot(x, fun(x))
w=1.0
x = np.linspace(0,20, 100)
L=0; R=0;
x_prev = np.random.uniform(low=0, high=17)
fig = plt.figure()
iters=10000
trace=[]
kmax=1
plt.plot(x, fun(x),'g')
for k in range(iters):
y_samp = np.random.uniform(low=0, high=fun(x_prev))
plot_yline(k, 5, x_prev, y_samp, fun)
# widen left
U = np.random.rand()
L=x_prev-U*w
R=x_prev+w*(1.0-U)
while fun(L)>y_samp:
L = L-w
plot_bounds(k, kmax, L, R, y_samp)
while fun(R)>y_samp:
R = R+w
plot_bounds(k, kmax, L, R, y_samp)
#now propose new x on L,R
while 1:
x_prop= np.random.uniform(low=L, high=R)
if k <= kmax:
print("L,R, xprop", L, R, x_prop)
if y_samp < fun(x_prop):
x_prev = x_prop
trace.append(x_prop)
plot_prop(k, kmax, x_prop, y_samp, accept='y')
break
elif x_prop > x_prev:
R = x_prop
elif x_prop < x_prev:
L = x_prop
plt.hist(trace, bins=100, alpha=0.3, normed=True);
#sns.kdeplot(xsmp)
plt.xlim( [0,20])
plt.plot(x, fun(x))
1.
| 0.396185 | 0.987911 |
```
def load_dataset():
data = pd.read_csv('GoogleStocks.csv')
data['date'] = data['date'].str.replace('/','-')
data=data[1:]
return data
def split_dataset(data):
train, test= train_test_split(data, test_size=0.2, shuffle=False)
return train,test
def initialise_prices(train_data):
open_price = np.array(train_data['open']).astype(np.float)
close_price = np.array(train_data['close']).astype(np.float)
high_price = np.array(train_data['high']).astype(np.float)
low_price = np.array(train_data['low']).astype(np.float)
volume = np.array(data['volume']).astype(np.float)
frac_change = (close_price-open_price)/open_price
frac_high = (high_price - open_price)/open_price
frac_low = (open_price - low_price)/open_price
return frac_low, frac_high, frac_change, volume
def extract_features(data):
open_price = np.array(data['open']).astype(np.float)
close_price = np.array(data['close']).astype(np.float)
high_price = np.array(data['high']).astype(np.float)
low_price = np.array(data['low']).astype(np.float)
volume = np.array(data['volume']).astype(np.float)
frac_change = (close_price - open_price) / open_price
frac_high = (high_price - open_price) / open_price
frac_low = (open_price - low_price) / open_price
return np.column_stack((frac_change, frac_high, frac_low))
def compute_all_possible_outcomes():
frac_change_range = np.linspace(-0.1, 0.1, n_steps_frac_change)
frac_high_range = np.linspace(0, 0.1, n_steps_frac_high)
frac_low_range = np.linspace(0, 0.1, n_steps_frac_low)
return np.array(list(itertools.product(frac_change_range, frac_high_range, frac_low_range)))
def get_most_probable_outcome(day_index):
previous_data_start_index = max(0, day_index - n_latency_days)
previous_data_end_index = max(0, day_index - 1)
previous_data = test_data.iloc[previous_data_end_index: previous_data_start_index]
previous_data_features =extract_features(previous_data)
outcome_score = []
for possible_outcome in possible_outcomes:
total_data = np.row_stack((previous_data_features, possible_outcome))
outcome_score.append(hmm.score(total_data))
most_probable_outcome = possible_outcomes[np.argmax(outcome_score)]
return most_probable_outcome
def predict_open_price(day_index):
close_price = test_data.iloc[day_index]['close']
close_price = float(close_price)
predicted_frac_change, _, _ = get_most_probable_outcome(day_index)
return close_price/(1 + predicted_frac_change)
def predict_open_prices_for_days(days, no_hidden, timesteps):
predicted_open_prices = []
for day_index in range(days):
predicted_open_prices.append(predict_open_price(day_index))
test_data_x = test_data[0: days]
days = np.array(test_data_x['date'], dtype="datetime64[ms]")
actual_open_prices = test_data_x['open']
fig = plt.figure(figsize=(16,6))
axes = fig.add_subplot(111)
axes.plot(days, actual_open_prices, 'b-', label="actual")
axes.plot(days, predicted_open_prices, 'r-', label="predicted")
fig.autofmt_xdate()
plt.legend()
title = "Stock Prediction Using HMM (Hidden States : "+str(no_hidden)+" Time Steps : "+str(timesteps)+" )"
plt.title(title)
plt.show()
data=load_dataset()
train_data,test_data=split_dataset(data)
frac_low, frac_high, frac_change, volume=initialise_prices(train_data)
data_new=np.column_stack((frac_change, frac_high, frac_low))
n_steps_frac_change=20
n_steps_frac_high=20
n_steps_frac_low=20
for i in [4, 8, 12]:
for j in [20, 50, 75]:
hmm = GaussianHMM(n_components=i)
hmm.fit(data_new)
n_latency_days=j
possible_outcomes=compute_all_possible_outcomes()
predict_open_prices_for_days(100, i, j)
```
We can see from above that RNN has worked better in comparision of the two.
HMMs make the Markovian assumption — that is, you assume that the current state depends only on the previous state. That can be true or not depending on the cases which was not here so RNN performed better on Stock Prediction.
|
github_jupyter
|
def load_dataset():
data = pd.read_csv('GoogleStocks.csv')
data['date'] = data['date'].str.replace('/','-')
data=data[1:]
return data
def split_dataset(data):
train, test= train_test_split(data, test_size=0.2, shuffle=False)
return train,test
def initialise_prices(train_data):
open_price = np.array(train_data['open']).astype(np.float)
close_price = np.array(train_data['close']).astype(np.float)
high_price = np.array(train_data['high']).astype(np.float)
low_price = np.array(train_data['low']).astype(np.float)
volume = np.array(data['volume']).astype(np.float)
frac_change = (close_price-open_price)/open_price
frac_high = (high_price - open_price)/open_price
frac_low = (open_price - low_price)/open_price
return frac_low, frac_high, frac_change, volume
def extract_features(data):
open_price = np.array(data['open']).astype(np.float)
close_price = np.array(data['close']).astype(np.float)
high_price = np.array(data['high']).astype(np.float)
low_price = np.array(data['low']).astype(np.float)
volume = np.array(data['volume']).astype(np.float)
frac_change = (close_price - open_price) / open_price
frac_high = (high_price - open_price) / open_price
frac_low = (open_price - low_price) / open_price
return np.column_stack((frac_change, frac_high, frac_low))
def compute_all_possible_outcomes():
frac_change_range = np.linspace(-0.1, 0.1, n_steps_frac_change)
frac_high_range = np.linspace(0, 0.1, n_steps_frac_high)
frac_low_range = np.linspace(0, 0.1, n_steps_frac_low)
return np.array(list(itertools.product(frac_change_range, frac_high_range, frac_low_range)))
def get_most_probable_outcome(day_index):
previous_data_start_index = max(0, day_index - n_latency_days)
previous_data_end_index = max(0, day_index - 1)
previous_data = test_data.iloc[previous_data_end_index: previous_data_start_index]
previous_data_features =extract_features(previous_data)
outcome_score = []
for possible_outcome in possible_outcomes:
total_data = np.row_stack((previous_data_features, possible_outcome))
outcome_score.append(hmm.score(total_data))
most_probable_outcome = possible_outcomes[np.argmax(outcome_score)]
return most_probable_outcome
def predict_open_price(day_index):
close_price = test_data.iloc[day_index]['close']
close_price = float(close_price)
predicted_frac_change, _, _ = get_most_probable_outcome(day_index)
return close_price/(1 + predicted_frac_change)
def predict_open_prices_for_days(days, no_hidden, timesteps):
predicted_open_prices = []
for day_index in range(days):
predicted_open_prices.append(predict_open_price(day_index))
test_data_x = test_data[0: days]
days = np.array(test_data_x['date'], dtype="datetime64[ms]")
actual_open_prices = test_data_x['open']
fig = plt.figure(figsize=(16,6))
axes = fig.add_subplot(111)
axes.plot(days, actual_open_prices, 'b-', label="actual")
axes.plot(days, predicted_open_prices, 'r-', label="predicted")
fig.autofmt_xdate()
plt.legend()
title = "Stock Prediction Using HMM (Hidden States : "+str(no_hidden)+" Time Steps : "+str(timesteps)+" )"
plt.title(title)
plt.show()
data=load_dataset()
train_data,test_data=split_dataset(data)
frac_low, frac_high, frac_change, volume=initialise_prices(train_data)
data_new=np.column_stack((frac_change, frac_high, frac_low))
n_steps_frac_change=20
n_steps_frac_high=20
n_steps_frac_low=20
for i in [4, 8, 12]:
for j in [20, 50, 75]:
hmm = GaussianHMM(n_components=i)
hmm.fit(data_new)
n_latency_days=j
possible_outcomes=compute_all_possible_outcomes()
predict_open_prices_for_days(100, i, j)
| 0.378919 | 0.720467 |
## Linear Regression Assumptions
Linear relationship between target and features
No outliers
No high-leverage points
Homoscedasticity of error terms
Uncorrelated error terms
Independent features
### 1 Linear Relationship Between Target & Features
```
import numpy as np
np.random.seed(20)
x = np.arange(20)
y = [x*2 + np.random.rand(1)*4 for x in range(20)]
x_reshape = x.reshape(-1,1)
from sklearn.linear_model import LinearRegression
linear = LinearRegression()
linear.fit(x_reshape, y)
import seaborn as sns # To get graphs
import matplotlib.pyplot as plt # To get graphs
plt.figure(figsize=(10,10))
plt.scatter(x_reshape, y)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y, color ="r")
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('linear Relationship with Gaussian Noise')
class Stats:
def __init__(self, X, y, model):
self.data = X
self.target = y
self.model = model
## degrees of freedom population dep. variable variance
self._dft = X.shape[0] - 1
## degrees of freedom population error variance
self._dfe = X.shape[0] - X.shape[1] - 1
def sse(self):
'''returns sum of squared errors (model vs actual)'''
squared_errors = (self.target - self.model.predict(self.data)) ** 2
return np.sum(squared_errors)
def sst(self):
'''returns total sum of squared errors (actual vs avg(actual))'''
avg_y = np.mean(self.target)
squared_errors = (self.target - avg_y) ** 2
return np.sum(squared_errors)
def r_squared(self):
'''returns calculated value of r^2'''
return 1 - self.sse()/self.sst()
def adj_r_squared(self):
'''returns calculated value of adjusted r^2'''
return 1 - (self.sse()/self._dfe) / (self.sst()/self._dft)
def pretty_print_stats(stats_obj):
'''returns report of statistics for a given model object'''
items = ( ('sse:', stats_obj.sse()), ('sst:', stats_obj.sst()),
('r^2:', stats_obj.r_squared()), ('adj_r^2:', stats_obj.adj_r_squared()) )
for item in items:
print('{0:8} {1:.4f}'.format(item[0], item[1]))
s1 = Stats(x_reshape, y, linear)
pretty_print_stats(s1)
y_nonlinear = [x**3 + np.random.rand(1)*10 for x in range(20)]
nonlinear = LinearRegression()
nonlinear.fit(x_reshape, y_nonlinear)
```
### Potential Problem: Data w/Nonlinear Pattern
```
y_nonlinear = [x**3 + np.random.rand(1)*10 for x in range(20)]
nonlinear = LinearRegression()
nonlinear.fit(x_reshape, y_nonlinear)
import seaborn as sns # To get graphs
import matplotlib.pyplot as plt # To get graphs
plt.figure(figsize=(10,10))
plt.scatter(x_reshape, y_nonlinear)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y, color ="r")
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('Nonlinear Relationship')
s2 = Stats(x_reshape, y_nonlinear, nonlinear)
pretty_print_stats(s2)
```
No surprise, we see a substantial increases in both SSE and SST as well as substantial ***decreases in R^2 and adjusted R^2.***
## Considerations
We can check to see if our model is capturing the underlying pattern effectively. Specifically, let’s generate side-by-side ***Residual Plots for the linear case and the nonlinear case.***
```
import matplotlib.pyplot as plt
#Linear Grid
fig, axes = plt.subplots(1, 2, sharex=False, sharey=False)
fig.suptitle('[Residual Plots]')
fig.set_size_inches(12,5)
axes[0].plot(linear.predict(x_reshape), y-linear.predict(x_reshape), 'bo')
axes[0].axhline(y=0, color='k')
axes[0].grid()
axes[0].set_title('Linear')
axes[0].set_xlabel('predicted values')
axes[0].set_ylabel('residuals')
#Non-Linear Grid
axes[1].plot(nonlinear.predict(x_reshape), y_nonlinear-nonlinear.predict(x_reshape), 'ro')
axes[1].axhline(y=0, color='k')
axes[1].grid()
axes[1].set_title('Non-Linear')
axes[1].set_xlabel('predicted values')
```
**The nonlinear pattern** is overwhelmingly obvious in the residual plots. You may be wondering why we bothered plotting at all since we saw the nonlinear trend when plotting the observed data. That works well for low dimensional cases that are easy to visualize but how will you know if you have more than 2-3 features? The residual plot is a powerful tool in that case and something you should leverage often.
```
import seaborn as sns
residuals_linear = y - linear.predict(x_reshape)
residuals_nlinear = y_nonlinear - nonlinear.predict(x_reshape)
sns.distplot(residuals_linear);
plt.title('Linear')
```
The histogram of the linear model on linear data looks **approximately Normal (aka Gaussian)** while the second histogram shows a skew. But is there a more quantitative method to test for Normality? Absolutely. SciPy has a normaltest method. Let’s see it in action.
```
from scipy.stats import normaltest
normaltest(residuals_linear)
```
**The null hypothesis** is that the residual distribution is Normally distributed. Since the p-value > 0.05, we cannot reject the null. In other words, we can confidently say the residuals are Normally distributed.
```
normaltest(residuals_nlinear)
```
## Takeaway
**The linear data** exhibits a fair amount of randomness centered around 0 in the residual plot indicating our model has captured nearly all the discernable pattern. On the other hand, the nonlinear data shows a clear nonlinear trend. In other words, using the nonlinear data as-is with our linear model will result in a poor model fit.
## Possible Solutions to Nonlinear Data
1)Consider transforming the features
2)Consider applying a different algorithm
Say we have a single feature x. Assuming we see a nonlinear pattern in the data, we can transform x such that linear regression can pickup the pattern. For example, perhaps there’s a quadratic relationship between x and y. We can model that simply by including x^2 in our data. The x^2 feature now gets its own parameter in the model. This process of modeling transformed features with polynomial terms is called **polynomial regression**. Let’s see it in action.
## Polynomial Regression
```
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import PolynomialFeatures
poly = Pipeline([('poly', PolynomialFeatures(degree=3)),
('linear', LinearRegression(fit_intercept=False))])
poly.fit(x_reshape, y_nonlinear)
plt.figure(figsize=(10,10))
plt.scatter(x_reshape, y_nonlinear)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y_nonlinear, color ="r")
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('Nonlinear Relationship')
```
### 2 No Outliers
```
np.random.seed(20)
x = np.arange(20)
y = [x*2 + np.random.rand(1)*4 for x in range(20)]
y_outlier = y.copy()
y_outlier[8] = np.array([38]) ## insert outlier
# sklearn expects 2D array so have to reshape x
x_reshape = x.reshape(-1,1)
# fit model w/standard data
linear_nooutlier = LinearRegression()
linear_nooutlier.fit(x_reshape, y);
# fit model w/outlier data
linear_outlier = LinearRegression()
linear_outlier.fit(x_reshape, y_outlier);
```
We’ll do the customary reshaping of our 1D x array and fit two models: one with the outlier and one without. Then we’ll investigate the impact on the various stats.
```
plt.figure(figsize=(10,10))
#plt.scatter(x_reshape, y_nonlinear)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y_outlier, color ="b",marker='.', alpha=0.7, linestyle='-')
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('Linear Relationship Outlier')
#With Outliers
s3 = Stats(x_reshape, y_outlier, linear)
pretty_print_stats(s3)
#Without Outliers
pretty_print_stats(s1)
```
### Possible Solutions
1)Investigate the outlier(s). Do NOT assume these cases are just bad data. Some outliers are true examples while others are data entry errors. You need to know which it is before proceeding.
2)Consider imputing or removing outliers.
## 3 No High-Leverage Points
### Generate Dummy Data
```
np.random.seed(20)
x = np.arange(20)
y_linear_leverage = [x*2 + np.random.rand(1)*4 for x in range(20)]
y_linear_leverage[18] = np.array([55]) ## high-leverage point
y_linear_leverage[19] = np.array([58]) ## high-leverage point
x_reshape = x.reshape(-1,1)
```
### Fitting into a Model
```
linear_leverage = LinearRegression()
linear_leverage.fit(x_reshape, y_linear_leverage)
plt.figure(figsize=(10,10))
#plt.scatter(x_reshape, y_nonlinear)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y, label="Without Leverage",marker='.', alpha=0.7, linestyle='dotted')
plt.plot(x_reshape, y_linear_leverage, label="With Leverage" ,marker='.', alpha=0.7, linestyle='-')
plt.legend(loc="upper left")
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('Linear Relationship with High Leverage Points')
import numpy as np
import matplotlib.pyplot as plt
x = np.linspace(0, 20, 1000)
y1 = np.sin(x)
y2 = np.cos(x)
plt.plot(x, y1, "-b", label="sine")
plt.plot(x, y2, "-r", label="cosine")
plt.legend(loc="upper left")
plt.ylim(-1.5, 2.0)
plt.show()
#Without No Leverage
pretty_print_stats(s1)
#With the Leverage
s4 = Stats(x_reshape, y_linear_leverage, linear)
pretty_print_stats(s4)
```
## Normalized Test
```
normaltest(y_outlier-linear_leverage.predict(x_reshape))
```
Fails! The residuals are not **Normally distributed**, statistically speaking that is. This is a key assumption of linear regression and we have violated it.
## Possible Solutions
Explore the data to understand why these data points exist. Are they true data points or mistakes of some kind?
1)Consider imputing or removing them if outliers, but only if you have good reason to do so!
2)Consider a more robust loss function (e.g. Huber).
3)Consider a more robust algorithm (e.g. RANSAC).
```
import matplotlib.pyplot as plt
#Linear Grid
fig, axes = plt.subplots(1, 2, sharex=False, sharey=False)
fig.suptitle('[Residual Plots]')
fig.set_size_inches(12,5)
axes[0].plot(linear.predict(y), y-linear.predict(y_outlier), 'bo')
axes[0].axhline(y=0, color='k')
axes[0].grid()
axes[0].set_title('No Leverage Points')
axes[0].set_xlabel('predicted values')
axes[0].set_ylabel('residuals')
#Non-Linear Grid
axes[1].plot(linear.predict(y), y-linear.predict(y_linear_leverage), 'ro')
axes[1].axhline(y=0, color='k')
axes[1].grid()
axes[1].set_title('Leverage Points Presents')
axes[1].set_xlabel('predicted values')
```
### 4 Homoscedasticity of Error Terms
**Homescedasticity** means the errors exhibit constant variance.
This is a key assumption of linear regression. Heteroscedasticity, on the other hand, is what happens when errors show some sort of growth. The tell tale sign you have heteroscedasticity is a fan-like shape in your residual plot. Let’s take a look
```
#Generate Data
np.random.seed(20)
x = np.arange(20)
y_homo = [x*2 + np.random.rand(1) for x in range(20)] ## homoscedastic error
y_hetero = [x*2 + np.random.rand(1)*2*x for x in range(20)] ## heteroscedastic error
x_reshape = x.reshape(-1,1)
```
#### Fit Model
```
linear_homo = LinearRegression()
linear_homo.fit(x_reshape, y_homo)
linear_hetero = LinearRegression()
linear_hetero.fit(x_reshape, y_hetero)
plt.figure(figsize=(10,10))
#plt.scatter(x_reshape, y_nonlinear)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y_homo, label="Actual",marker='.', alpha=0.7, linestyle='dotted')
plt.plot(x_reshape, y_hetero, label="With Homescedasticity" ,marker='.', alpha=0.7, linestyle='-')
plt.legend(loc="upper left")
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('Homescedasticity of Errors')
```
### Normal Test
```
# homoscedastic data
normaltest(y_homo-linear_homo.predict(x_reshape))
# heteroscedastic data
normaltest(y_hetero-linear_hetero.predict(x_reshape))
```
There’s no reason to ***reject the null that both residual distributions are Normally distributed.***
### Possible Solution
**Consider log transforming the target values**
```
y_hetero_log = np.log10(np.array(y_hetero) + 1e1)
x_reshape_log = np.log10(np.array(x_reshape) + 1e1)
linear_hetero_log = LinearRegression()
linear_hetero_log.fit(x_reshape, y_hetero_log)
linear_hetero_log_log = LinearRegression()
linear_hetero_log_log.fit(x_reshape_log, y_hetero_log)
#Homoscedastic grid
fig, axes = plt.subplots(1, 2, sharex=False, sharey=False)
fig.suptitle('[Residual Plots]')
fig.set_size_inches(12,5)
axes[0].plot(linear.predict(y), y-linear.predict(y_homo), 'bo')
axes[0].axhline(y=0, color='k')
axes[0].grid()
axes[0].set_title('Homoscedastic')
axes[0].set_xlabel('predicted values')
axes[0].set_ylabel('residuals')
#Heteroscedastic
axes[1].plot(linear.predict(y), y-linear.predict(y_hetero_log), 'ro')
axes[1].axhline(y=0, color='k')
axes[1].grid()
axes[1].set_title('Leverage Points Presents')
axes[1].set_xlabel('predicted values')
normaltest(y_hetero_log - linear_hetero_log.predict(x_reshape))
```
The plot on the right shows we addressed **heteroscedasticity** but there’a a fair amount of correlation amongst the errors. That brings us to our next assumption.
### 5 Uncorrelated Error Terms
```
#Generate Dummy Data
np.random.seed(20)
x = np.arange(20)
y_uncorr = [2*x + np.random.rand(1) for x in range(20)]
y_corr = np.sin(x)
x_reshape = x.reshape(-1,1)
linear_uncorr = LinearRegression()
linear_uncorr.fit(x_reshape, y_uncorr)
linear_corr = LinearRegression()
linear_corr.fit(x_reshape, y_corr)
```
### Residual Plot
```
#Homoscedastic grid
fig, axes = plt.subplots(1, 2, sharex=False, sharey=False)
fig.suptitle('[Residual Plots]')
fig.set_size_inches(12,5)
axes[0].plot(linear.predict(x_reshape), y-linear.predict(y_uncorr), 'bo')
axes[0].axhline(y=0, color='k')
axes[0].grid()
axes[0].set_title('Y_Uncorrelation')
axes[0].set_xlabel('predicted values')
axes[0].set_ylabel('residuals')
axes[1].plot(linear.predict(y), y-linear.predict(y_hetero_log), 'ro')
axes[1].axhline(y=0, color='k')
axes[1].grid()
axes[1].set_title('Leverage Points Presents')
axes[1].set_xlabel('predicted values')
```
## Possible Solution
1) Forget linear regression. Use time series modeling instead.
We’ll discuss time series modeling in detail in another post. For now, just know correlated errors is a problem for linear regression because linear regression expects records to be i.i.d.
### 6 Independent features
**Independent features** means no feature is an any way derived from the other features. For example, imagine a simple dataset with three features. The first two features are in no way related. However, the third is simply the sum of the first two features. That means this ficitonal dataset has one linearly dependent feature. That’s a problem for linear regression. Let’s take a look.
```
np.random.seed(39)
x1 = np.arange(20) * 2
x2 = np.random.randint(low=0, high=50, size=20)
x_idp = np.vstack((x1,x2))
ynew = np.add( np.sum(x_idp, axis=0), np.random.randn(20)*5 ) ## y = x1 + x2 + noise
```
### Example
```
from mpl_toolkits.mplot3d import Axes3D
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams['legend.fontsize'] = 10
fig = plt.figure()
ax = fig.gca(projection='3d')
theta = np.linspace(-4 * np.pi, 4 * np.pi, 100)
z = np.linspace(-2, 2, 100)
r = z**2 + 1
x = r * np.sin(theta)
y1 = r * np.cos(theta)
ax.plot(x, y1, z, label='parametric curve')
ax.legend()
plt.show()
ynew.shape
fig = plt.figure()
ax = fig.gca(projection='3d')
ax.plot(x1, x2, ynew, label='parametric curve',linestyle='dotted')
ax.legend()
plt.show()
import pandas as pd
dp_df = pd.DataFrame([x1,x2,(x1+x2)]).T
```
## Fiiting Models
```
lr_idp = LinearRegression()
lr_idp.fit(x_idp.T, ynew)
lr_dp = LinearRegression()
lr_dp.fit(dp_df, ynew)
```
### linearly independent features
sse: 361.5308
sst: 6898.6751
r^2: 0.9476
adj_r^2: 0.9414
### linearly dependent features
1) sse: 361.5308
2) sst: 6898.6751
3) r^2: 0.9476
4) adj_r^2: 0.9378
**We see no difference in SSE, SST, or R^2**.
As we learned in the previous post about metrics, adjusted R^2 is telling us that the additional feature in the linearly dependent feature set adds no new information, which is why we see a decrease in that value. Be careful because linear regression assumes independent features, and looking at simple metrics like SSE, SST, and R^2 alone won’t tip you off that your features are correlated.
|
github_jupyter
|
import numpy as np
np.random.seed(20)
x = np.arange(20)
y = [x*2 + np.random.rand(1)*4 for x in range(20)]
x_reshape = x.reshape(-1,1)
from sklearn.linear_model import LinearRegression
linear = LinearRegression()
linear.fit(x_reshape, y)
import seaborn as sns # To get graphs
import matplotlib.pyplot as plt # To get graphs
plt.figure(figsize=(10,10))
plt.scatter(x_reshape, y)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y, color ="r")
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('linear Relationship with Gaussian Noise')
class Stats:
def __init__(self, X, y, model):
self.data = X
self.target = y
self.model = model
## degrees of freedom population dep. variable variance
self._dft = X.shape[0] - 1
## degrees of freedom population error variance
self._dfe = X.shape[0] - X.shape[1] - 1
def sse(self):
'''returns sum of squared errors (model vs actual)'''
squared_errors = (self.target - self.model.predict(self.data)) ** 2
return np.sum(squared_errors)
def sst(self):
'''returns total sum of squared errors (actual vs avg(actual))'''
avg_y = np.mean(self.target)
squared_errors = (self.target - avg_y) ** 2
return np.sum(squared_errors)
def r_squared(self):
'''returns calculated value of r^2'''
return 1 - self.sse()/self.sst()
def adj_r_squared(self):
'''returns calculated value of adjusted r^2'''
return 1 - (self.sse()/self._dfe) / (self.sst()/self._dft)
def pretty_print_stats(stats_obj):
'''returns report of statistics for a given model object'''
items = ( ('sse:', stats_obj.sse()), ('sst:', stats_obj.sst()),
('r^2:', stats_obj.r_squared()), ('adj_r^2:', stats_obj.adj_r_squared()) )
for item in items:
print('{0:8} {1:.4f}'.format(item[0], item[1]))
s1 = Stats(x_reshape, y, linear)
pretty_print_stats(s1)
y_nonlinear = [x**3 + np.random.rand(1)*10 for x in range(20)]
nonlinear = LinearRegression()
nonlinear.fit(x_reshape, y_nonlinear)
y_nonlinear = [x**3 + np.random.rand(1)*10 for x in range(20)]
nonlinear = LinearRegression()
nonlinear.fit(x_reshape, y_nonlinear)
import seaborn as sns # To get graphs
import matplotlib.pyplot as plt # To get graphs
plt.figure(figsize=(10,10))
plt.scatter(x_reshape, y_nonlinear)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y, color ="r")
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('Nonlinear Relationship')
s2 = Stats(x_reshape, y_nonlinear, nonlinear)
pretty_print_stats(s2)
import matplotlib.pyplot as plt
#Linear Grid
fig, axes = plt.subplots(1, 2, sharex=False, sharey=False)
fig.suptitle('[Residual Plots]')
fig.set_size_inches(12,5)
axes[0].plot(linear.predict(x_reshape), y-linear.predict(x_reshape), 'bo')
axes[0].axhline(y=0, color='k')
axes[0].grid()
axes[0].set_title('Linear')
axes[0].set_xlabel('predicted values')
axes[0].set_ylabel('residuals')
#Non-Linear Grid
axes[1].plot(nonlinear.predict(x_reshape), y_nonlinear-nonlinear.predict(x_reshape), 'ro')
axes[1].axhline(y=0, color='k')
axes[1].grid()
axes[1].set_title('Non-Linear')
axes[1].set_xlabel('predicted values')
import seaborn as sns
residuals_linear = y - linear.predict(x_reshape)
residuals_nlinear = y_nonlinear - nonlinear.predict(x_reshape)
sns.distplot(residuals_linear);
plt.title('Linear')
from scipy.stats import normaltest
normaltest(residuals_linear)
normaltest(residuals_nlinear)
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import PolynomialFeatures
poly = Pipeline([('poly', PolynomialFeatures(degree=3)),
('linear', LinearRegression(fit_intercept=False))])
poly.fit(x_reshape, y_nonlinear)
plt.figure(figsize=(10,10))
plt.scatter(x_reshape, y_nonlinear)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y_nonlinear, color ="r")
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('Nonlinear Relationship')
np.random.seed(20)
x = np.arange(20)
y = [x*2 + np.random.rand(1)*4 for x in range(20)]
y_outlier = y.copy()
y_outlier[8] = np.array([38]) ## insert outlier
# sklearn expects 2D array so have to reshape x
x_reshape = x.reshape(-1,1)
# fit model w/standard data
linear_nooutlier = LinearRegression()
linear_nooutlier.fit(x_reshape, y);
# fit model w/outlier data
linear_outlier = LinearRegression()
linear_outlier.fit(x_reshape, y_outlier);
plt.figure(figsize=(10,10))
#plt.scatter(x_reshape, y_nonlinear)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y_outlier, color ="b",marker='.', alpha=0.7, linestyle='-')
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('Linear Relationship Outlier')
#With Outliers
s3 = Stats(x_reshape, y_outlier, linear)
pretty_print_stats(s3)
#Without Outliers
pretty_print_stats(s1)
np.random.seed(20)
x = np.arange(20)
y_linear_leverage = [x*2 + np.random.rand(1)*4 for x in range(20)]
y_linear_leverage[18] = np.array([55]) ## high-leverage point
y_linear_leverage[19] = np.array([58]) ## high-leverage point
x_reshape = x.reshape(-1,1)
linear_leverage = LinearRegression()
linear_leverage.fit(x_reshape, y_linear_leverage)
plt.figure(figsize=(10,10))
#plt.scatter(x_reshape, y_nonlinear)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y, label="Without Leverage",marker='.', alpha=0.7, linestyle='dotted')
plt.plot(x_reshape, y_linear_leverage, label="With Leverage" ,marker='.', alpha=0.7, linestyle='-')
plt.legend(loc="upper left")
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('Linear Relationship with High Leverage Points')
import numpy as np
import matplotlib.pyplot as plt
x = np.linspace(0, 20, 1000)
y1 = np.sin(x)
y2 = np.cos(x)
plt.plot(x, y1, "-b", label="sine")
plt.plot(x, y2, "-r", label="cosine")
plt.legend(loc="upper left")
plt.ylim(-1.5, 2.0)
plt.show()
#Without No Leverage
pretty_print_stats(s1)
#With the Leverage
s4 = Stats(x_reshape, y_linear_leverage, linear)
pretty_print_stats(s4)
normaltest(y_outlier-linear_leverage.predict(x_reshape))
import matplotlib.pyplot as plt
#Linear Grid
fig, axes = plt.subplots(1, 2, sharex=False, sharey=False)
fig.suptitle('[Residual Plots]')
fig.set_size_inches(12,5)
axes[0].plot(linear.predict(y), y-linear.predict(y_outlier), 'bo')
axes[0].axhline(y=0, color='k')
axes[0].grid()
axes[0].set_title('No Leverage Points')
axes[0].set_xlabel('predicted values')
axes[0].set_ylabel('residuals')
#Non-Linear Grid
axes[1].plot(linear.predict(y), y-linear.predict(y_linear_leverage), 'ro')
axes[1].axhline(y=0, color='k')
axes[1].grid()
axes[1].set_title('Leverage Points Presents')
axes[1].set_xlabel('predicted values')
#Generate Data
np.random.seed(20)
x = np.arange(20)
y_homo = [x*2 + np.random.rand(1) for x in range(20)] ## homoscedastic error
y_hetero = [x*2 + np.random.rand(1)*2*x for x in range(20)] ## heteroscedastic error
x_reshape = x.reshape(-1,1)
linear_homo = LinearRegression()
linear_homo.fit(x_reshape, y_homo)
linear_hetero = LinearRegression()
linear_hetero.fit(x_reshape, y_hetero)
plt.figure(figsize=(10,10))
#plt.scatter(x_reshape, y_nonlinear)
myline = np.linspace(0, 10, 100)
plt.plot(x_reshape, y_homo, label="Actual",marker='.', alpha=0.7, linestyle='dotted')
plt.plot(x_reshape, y_hetero, label="With Homescedasticity" ,marker='.', alpha=0.7, linestyle='-')
plt.legend(loc="upper left")
plt.xlabel('Feature Value')
plt.ylabel('Target Value')
plt.title('Homescedasticity of Errors')
# homoscedastic data
normaltest(y_homo-linear_homo.predict(x_reshape))
# heteroscedastic data
normaltest(y_hetero-linear_hetero.predict(x_reshape))
y_hetero_log = np.log10(np.array(y_hetero) + 1e1)
x_reshape_log = np.log10(np.array(x_reshape) + 1e1)
linear_hetero_log = LinearRegression()
linear_hetero_log.fit(x_reshape, y_hetero_log)
linear_hetero_log_log = LinearRegression()
linear_hetero_log_log.fit(x_reshape_log, y_hetero_log)
#Homoscedastic grid
fig, axes = plt.subplots(1, 2, sharex=False, sharey=False)
fig.suptitle('[Residual Plots]')
fig.set_size_inches(12,5)
axes[0].plot(linear.predict(y), y-linear.predict(y_homo), 'bo')
axes[0].axhline(y=0, color='k')
axes[0].grid()
axes[0].set_title('Homoscedastic')
axes[0].set_xlabel('predicted values')
axes[0].set_ylabel('residuals')
#Heteroscedastic
axes[1].plot(linear.predict(y), y-linear.predict(y_hetero_log), 'ro')
axes[1].axhline(y=0, color='k')
axes[1].grid()
axes[1].set_title('Leverage Points Presents')
axes[1].set_xlabel('predicted values')
normaltest(y_hetero_log - linear_hetero_log.predict(x_reshape))
#Generate Dummy Data
np.random.seed(20)
x = np.arange(20)
y_uncorr = [2*x + np.random.rand(1) for x in range(20)]
y_corr = np.sin(x)
x_reshape = x.reshape(-1,1)
linear_uncorr = LinearRegression()
linear_uncorr.fit(x_reshape, y_uncorr)
linear_corr = LinearRegression()
linear_corr.fit(x_reshape, y_corr)
#Homoscedastic grid
fig, axes = plt.subplots(1, 2, sharex=False, sharey=False)
fig.suptitle('[Residual Plots]')
fig.set_size_inches(12,5)
axes[0].plot(linear.predict(x_reshape), y-linear.predict(y_uncorr), 'bo')
axes[0].axhline(y=0, color='k')
axes[0].grid()
axes[0].set_title('Y_Uncorrelation')
axes[0].set_xlabel('predicted values')
axes[0].set_ylabel('residuals')
axes[1].plot(linear.predict(y), y-linear.predict(y_hetero_log), 'ro')
axes[1].axhline(y=0, color='k')
axes[1].grid()
axes[1].set_title('Leverage Points Presents')
axes[1].set_xlabel('predicted values')
np.random.seed(39)
x1 = np.arange(20) * 2
x2 = np.random.randint(low=0, high=50, size=20)
x_idp = np.vstack((x1,x2))
ynew = np.add( np.sum(x_idp, axis=0), np.random.randn(20)*5 ) ## y = x1 + x2 + noise
from mpl_toolkits.mplot3d import Axes3D
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams['legend.fontsize'] = 10
fig = plt.figure()
ax = fig.gca(projection='3d')
theta = np.linspace(-4 * np.pi, 4 * np.pi, 100)
z = np.linspace(-2, 2, 100)
r = z**2 + 1
x = r * np.sin(theta)
y1 = r * np.cos(theta)
ax.plot(x, y1, z, label='parametric curve')
ax.legend()
plt.show()
ynew.shape
fig = plt.figure()
ax = fig.gca(projection='3d')
ax.plot(x1, x2, ynew, label='parametric curve',linestyle='dotted')
ax.legend()
plt.show()
import pandas as pd
dp_df = pd.DataFrame([x1,x2,(x1+x2)]).T
lr_idp = LinearRegression()
lr_idp.fit(x_idp.T, ynew)
lr_dp = LinearRegression()
lr_dp.fit(dp_df, ynew)
| 0.833121 | 0.943764 |
### Latter Classification Neural Network
Trying to build the best CNN architecture for the images we have regarding to this paper : https://arxiv.org/pdf/1606.02228.pdf
## Library Selections
In order to use state of the art toolset for this research paper we installed CUDA 10 , Pytorch 1.0, Python 3.7 and openCV 3.4 on ubuntu 16.04 with NVDIA 1080 GPU. https://arxiv.org/pdf/1606.02228.pdf
```
# Import required libraries for this section
import numpy as np
import cv2
import torch
import torchvision
import torchvision.datasets as dset
import torchvision.transforms as transforms
from torch.utils.data.sampler import SubsetRandomSampler
import torch.optim as optim
from torch import nn
# Use GPU if it's available
from collections import OrderedDict
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
num_workers = 0
# how many samples per batch to load
batch_size = 20
# percentage of data set to use as test
validation_size = 0.5
test_validation_size = 0.4
transform = transforms.Compose([ transforms.CenterCrop(1000), transforms.RandomResizedCrop(224),
transforms.RandomHorizontalFlip(), transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])])
data_set = dset.ImageFolder(root="data",transform=transform)
dataloader = torch.utils.data.DataLoader(data_set, batch_size=4,shuffle=True,num_workers=2)
# obtain training indices that will be used for test
num_data = len(data_set)
indices = list(range(num_data))
np.random.shuffle(indices)
split = int(np.floor(test_validation_size * num_data))
train_idx, test_idx = indices[split:], indices[:split]
num_train_data = len(test_idx)
split_validation = int(np.floor(validation_size * num_train_data))
test_idx, validation_idx = test_idx[split_validation:], test_idx[:split_validation]
# define samplers for obtaining training and validation batches
train_sampler = SubsetRandomSampler(train_idx)
validation_sampler = SubsetRandomSampler(validation_idx)
test_sampler = SubsetRandomSampler(test_idx)
# prepare data loaders
train_loader = torch.utils.data.DataLoader(data_set, batch_size=batch_size,
sampler = train_sampler, num_workers=num_workers)
validation_loader = torch.utils.data.DataLoader(data_set, batch_size=batch_size,
sampler = test_sampler, num_workers=num_workers)
test_loader = torch.utils.data.DataLoader(data_set, batch_size=batch_size,
sampler = test_sampler, num_workers=num_workers)
classes = ('ebrus','suminagashis')
from torch.autograd import Variable
import torch.nn.functional as F
class MultilayerCNN(torch.nn.Module):
#Our batch shape for input x is (3, 224, 224)
#RELU with batchnorm.
def __init__(self):
super(MultilayerCNN, self).__init__()
self.layer1 = nn.Sequential(
nn.Conv2d(3, 128, kernel_size=5, stride=1, padding=2),
nn.BatchNorm2d(128),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2))
self.layer2 = nn.Sequential(
nn.Conv2d(128, 64, kernel_size=5, stride=1, padding=2),
nn.BatchNorm2d(64),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2))
self.layer3 = nn.Sequential(
nn.Conv2d(64, 32, kernel_size=5, stride=1, padding=2),
nn.BatchNorm2d(32),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2))
self.layer4 = nn.Sequential(
nn.Conv2d(32, 16, kernel_size=5, stride=1, padding=2),
nn.BatchNorm2d(16),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2))
self.layer5 = nn.Sequential(
nn.Conv2d(16, 8, kernel_size=5, stride=1, padding=2),
nn.BatchNorm2d(8),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2))
self.fc = nn.Linear(8 * 7 *7, 2)
def forward(self, x):
x = self.layer1(x)
x = self.layer2(x)
x = self.layer3(x)
x = self.layer4(x)
x = self.layer5(x)
x = x.view(-1, 8 * 7 *7)
x = self.fc(x)
return x
#apply linear learning rate decay
def adjust_lr(init_lr, optimizer, epoch, n_epochs):
lr = init_lr * (1 - (epoch // n_epochs))
for param_group in optimizer.param_groups:
param_group['lr'] = lr
return lr
def createLossAndOptimizer(net, learning_rate=0.001):
#Loss function
loss = torch.nn.CrossEntropyLoss()
#Optimizer
optimizer = optim.Adam(net.parameters(), lr=learning_rate)
return(loss, optimizer)
import time
def trainNet(net, batch_size, n_epochs, learning_rate):
#Print all of the hyperparameters of the training iteration:
print("===== HYPERPARAMETERS =====")
print("batch_size=", batch_size)
print("epochs=", n_epochs)
print("learning_rate=", learning_rate)
print("=" * 30)
#Get training data
n_batches = len(train_loader)
#Time for printing
training_start_time = time.time()
#Loop for n_epochs
for epoch in range(n_epochs):
running_loss = 0.0
print_every = n_batches // 10
start_time = time.time()
total_train_loss = 0
#Create our loss and optimizer functions
loss, optimizer = createLossAndOptimizer(net, learning_rate)
learning_rate = adjust_lr(learning_rate, optimizer, epoch, n_epochs)
for i, data in enumerate(train_loader, 0):
#Get inputs
inputs, labels = data
#Wrap them in a Variable object
inputs, labels = Variable(inputs), Variable(labels)
#Set the parameter gradients to zero
optimizer.zero_grad()
#Forward pass, backward pass, optimize
outputs = net(inputs)
loss_size = loss(outputs, labels)
loss_size.backward()
optimizer.step()
#Print statistics
running_loss += loss_size.data
total_train_loss += loss_size.data
#Print every 10th batch of an epoch
if (i + 1) % (print_every + 1) == 0:
print("Epoch {}, {:d}% \t train_loss: {:.2f} took: {:.2f}s".format(
epoch+1, int(100 * (i+1) / n_batches), running_loss / print_every, time.time() - start_time))
#Reset running loss and time
running_loss = 0.0
start_time = time.time()
#At the end of the epoch, do a pass on the validation set
total_val_loss = 0
for inputs, labels in validation_loader:
#Wrap tensors in Variables
inputs, labels = Variable(inputs), Variable(labels)
#Forward pass
val_outputs = net(inputs)
val_loss_size = loss(val_outputs, labels)
total_val_loss += val_loss_size.item()
total_test_loss = 0
accuracy = 0
net.eval()
with torch.no_grad():
for inputs, labels in test_loader:
#Wrap tensors in Variables
inputs, labels = Variable(inputs), Variable(labels)
#Forward pass
test_outputs = net(inputs)
test_loss_size = loss(test_outputs, labels)
total_test_loss += test_loss_size.item()
ps = torch.exp(test_outputs)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
net.train()
print("Training loss = {:.2f}".format(total_train_loss / len(train_loader)))
print("Validation loss = {:.2f}".format(total_val_loss / len(validation_loader)))
print("Test loss = {:.2f}".format(total_test_loss / len(test_loader)))
print("Test Accuracy = {:.2f}".format(accuracy / len(test_loader)))
print("Training finished, took {:.2f}s".format(time.time() - training_start_time))
CNN = MultilayerCNN()
trainNet(CNN, batch_size=128, n_epochs=5, learning_rate=0.001)
torch.save(CNN.state_dict(), 'Multilayer_CNN_128_CenterCrop.pth')
trainNet(CNN, batch_size=256, n_epochs=5, learning_rate=0.001)
torch.save(CNN.state_dict(), 'Multilayer_CNN_256_CenterCrop.pth')
```
|
github_jupyter
|
# Import required libraries for this section
import numpy as np
import cv2
import torch
import torchvision
import torchvision.datasets as dset
import torchvision.transforms as transforms
from torch.utils.data.sampler import SubsetRandomSampler
import torch.optim as optim
from torch import nn
# Use GPU if it's available
from collections import OrderedDict
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
num_workers = 0
# how many samples per batch to load
batch_size = 20
# percentage of data set to use as test
validation_size = 0.5
test_validation_size = 0.4
transform = transforms.Compose([ transforms.CenterCrop(1000), transforms.RandomResizedCrop(224),
transforms.RandomHorizontalFlip(), transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])])
data_set = dset.ImageFolder(root="data",transform=transform)
dataloader = torch.utils.data.DataLoader(data_set, batch_size=4,shuffle=True,num_workers=2)
# obtain training indices that will be used for test
num_data = len(data_set)
indices = list(range(num_data))
np.random.shuffle(indices)
split = int(np.floor(test_validation_size * num_data))
train_idx, test_idx = indices[split:], indices[:split]
num_train_data = len(test_idx)
split_validation = int(np.floor(validation_size * num_train_data))
test_idx, validation_idx = test_idx[split_validation:], test_idx[:split_validation]
# define samplers for obtaining training and validation batches
train_sampler = SubsetRandomSampler(train_idx)
validation_sampler = SubsetRandomSampler(validation_idx)
test_sampler = SubsetRandomSampler(test_idx)
# prepare data loaders
train_loader = torch.utils.data.DataLoader(data_set, batch_size=batch_size,
sampler = train_sampler, num_workers=num_workers)
validation_loader = torch.utils.data.DataLoader(data_set, batch_size=batch_size,
sampler = test_sampler, num_workers=num_workers)
test_loader = torch.utils.data.DataLoader(data_set, batch_size=batch_size,
sampler = test_sampler, num_workers=num_workers)
classes = ('ebrus','suminagashis')
from torch.autograd import Variable
import torch.nn.functional as F
class MultilayerCNN(torch.nn.Module):
#Our batch shape for input x is (3, 224, 224)
#RELU with batchnorm.
def __init__(self):
super(MultilayerCNN, self).__init__()
self.layer1 = nn.Sequential(
nn.Conv2d(3, 128, kernel_size=5, stride=1, padding=2),
nn.BatchNorm2d(128),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2))
self.layer2 = nn.Sequential(
nn.Conv2d(128, 64, kernel_size=5, stride=1, padding=2),
nn.BatchNorm2d(64),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2))
self.layer3 = nn.Sequential(
nn.Conv2d(64, 32, kernel_size=5, stride=1, padding=2),
nn.BatchNorm2d(32),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2))
self.layer4 = nn.Sequential(
nn.Conv2d(32, 16, kernel_size=5, stride=1, padding=2),
nn.BatchNorm2d(16),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2))
self.layer5 = nn.Sequential(
nn.Conv2d(16, 8, kernel_size=5, stride=1, padding=2),
nn.BatchNorm2d(8),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2))
self.fc = nn.Linear(8 * 7 *7, 2)
def forward(self, x):
x = self.layer1(x)
x = self.layer2(x)
x = self.layer3(x)
x = self.layer4(x)
x = self.layer5(x)
x = x.view(-1, 8 * 7 *7)
x = self.fc(x)
return x
#apply linear learning rate decay
def adjust_lr(init_lr, optimizer, epoch, n_epochs):
lr = init_lr * (1 - (epoch // n_epochs))
for param_group in optimizer.param_groups:
param_group['lr'] = lr
return lr
def createLossAndOptimizer(net, learning_rate=0.001):
#Loss function
loss = torch.nn.CrossEntropyLoss()
#Optimizer
optimizer = optim.Adam(net.parameters(), lr=learning_rate)
return(loss, optimizer)
import time
def trainNet(net, batch_size, n_epochs, learning_rate):
#Print all of the hyperparameters of the training iteration:
print("===== HYPERPARAMETERS =====")
print("batch_size=", batch_size)
print("epochs=", n_epochs)
print("learning_rate=", learning_rate)
print("=" * 30)
#Get training data
n_batches = len(train_loader)
#Time for printing
training_start_time = time.time()
#Loop for n_epochs
for epoch in range(n_epochs):
running_loss = 0.0
print_every = n_batches // 10
start_time = time.time()
total_train_loss = 0
#Create our loss and optimizer functions
loss, optimizer = createLossAndOptimizer(net, learning_rate)
learning_rate = adjust_lr(learning_rate, optimizer, epoch, n_epochs)
for i, data in enumerate(train_loader, 0):
#Get inputs
inputs, labels = data
#Wrap them in a Variable object
inputs, labels = Variable(inputs), Variable(labels)
#Set the parameter gradients to zero
optimizer.zero_grad()
#Forward pass, backward pass, optimize
outputs = net(inputs)
loss_size = loss(outputs, labels)
loss_size.backward()
optimizer.step()
#Print statistics
running_loss += loss_size.data
total_train_loss += loss_size.data
#Print every 10th batch of an epoch
if (i + 1) % (print_every + 1) == 0:
print("Epoch {}, {:d}% \t train_loss: {:.2f} took: {:.2f}s".format(
epoch+1, int(100 * (i+1) / n_batches), running_loss / print_every, time.time() - start_time))
#Reset running loss and time
running_loss = 0.0
start_time = time.time()
#At the end of the epoch, do a pass on the validation set
total_val_loss = 0
for inputs, labels in validation_loader:
#Wrap tensors in Variables
inputs, labels = Variable(inputs), Variable(labels)
#Forward pass
val_outputs = net(inputs)
val_loss_size = loss(val_outputs, labels)
total_val_loss += val_loss_size.item()
total_test_loss = 0
accuracy = 0
net.eval()
with torch.no_grad():
for inputs, labels in test_loader:
#Wrap tensors in Variables
inputs, labels = Variable(inputs), Variable(labels)
#Forward pass
test_outputs = net(inputs)
test_loss_size = loss(test_outputs, labels)
total_test_loss += test_loss_size.item()
ps = torch.exp(test_outputs)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
net.train()
print("Training loss = {:.2f}".format(total_train_loss / len(train_loader)))
print("Validation loss = {:.2f}".format(total_val_loss / len(validation_loader)))
print("Test loss = {:.2f}".format(total_test_loss / len(test_loader)))
print("Test Accuracy = {:.2f}".format(accuracy / len(test_loader)))
print("Training finished, took {:.2f}s".format(time.time() - training_start_time))
CNN = MultilayerCNN()
trainNet(CNN, batch_size=128, n_epochs=5, learning_rate=0.001)
torch.save(CNN.state_dict(), 'Multilayer_CNN_128_CenterCrop.pth')
trainNet(CNN, batch_size=256, n_epochs=5, learning_rate=0.001)
torch.save(CNN.state_dict(), 'Multilayer_CNN_256_CenterCrop.pth')
| 0.923156 | 0.922726 |
```
import math
import torch
import gpytorch
from matplotlib import pyplot as plt
from torch import nn, optim
from torch.autograd import Variable
from gpytorch.kernels import RBFKernel, GridInterpolationKernel
from gpytorch.means import ConstantMean
from gpytorch.likelihoods import GaussianLikelihood
from gpytorch.random_variables import GaussianRandomVariable
%matplotlib inline
train_x = Variable(torch.linspace(0, 1, 1000))
train_y = Variable(torch.sin(train_x.data * (4 * math.pi)) + torch.randn(train_x.size()) * 0.2)
train_x = train_x.cuda()
train_y = train_y.cuda()
class KissGPModel(gpytorch.GPModel):
def __init__(self):
super(KissGPModel, self).__init__(GaussianLikelihood(log_noise_bounds=(-5, 5)))
self.mean_module = ConstantMean(constant_bounds=(-1, 1))
covar_module = RBFKernel(log_lengthscale_bounds=(-3, 5))
self.grid_covar_module = GridInterpolationKernel(covar_module)
self.initialize_interpolation_grid(500, grid_bounds=[(0, 1)])
def forward(self, x):
mean_x = self.mean_module(x)
covar_x = self.grid_covar_module(x)
return GaussianRandomVariable(mean_x, covar_x)
model = KissGPModel().cuda()
model.condition(train_x, train_y)
# Optimize the model
def train():
model.train()
optimizer = optim.Adam(model.parameters(), lr=0.1)
optimizer.n_iter = 0
for i in range(30):
optimizer.zero_grad()
output = model(train_x)
loss = -model.marginal_log_likelihood(output, train_y) * 1. / len(train_x)
loss.backward()
optimizer.n_iter += 1
print('Iter %d/30 - Loss: %.3f' % (i + 1, loss.data[0]))
optimizer.step()
_ = model.eval()
%time train()
def plot_model_and_predictions(model, plot_train_data=True):
f, observed_ax = plt.subplots(1, 1, figsize=(4, 3))
test_x = Variable(torch.linspace(0, 1, 51)).cuda()
observed_pred = model(test_x)
def ax_plot(ax, rand_var, title):
lower, upper = rand_var.confidence_region()
if plot_train_data:
ax.plot(train_x.data.cpu().numpy(), train_y.data.cpu().numpy(), 'k*')
ax.plot(test_x.data.cpu().numpy(), rand_var.mean().data.cpu().numpy(), 'b')
ax.fill_between(test_x.data.cpu().numpy(), lower.data.cpu().numpy(), upper.data.cpu().numpy(), alpha=0.5)
ax.set_ylim([-3, 3])
ax.legend(['Observed Data', 'Mean', 'Confidence'])
ax.set_title(title)
ax_plot(observed_ax, observed_pred, 'Observed Values (Likelihood)')
return f
f = plot_model_and_predictions(model, plot_train_data=False)
```
|
github_jupyter
|
import math
import torch
import gpytorch
from matplotlib import pyplot as plt
from torch import nn, optim
from torch.autograd import Variable
from gpytorch.kernels import RBFKernel, GridInterpolationKernel
from gpytorch.means import ConstantMean
from gpytorch.likelihoods import GaussianLikelihood
from gpytorch.random_variables import GaussianRandomVariable
%matplotlib inline
train_x = Variable(torch.linspace(0, 1, 1000))
train_y = Variable(torch.sin(train_x.data * (4 * math.pi)) + torch.randn(train_x.size()) * 0.2)
train_x = train_x.cuda()
train_y = train_y.cuda()
class KissGPModel(gpytorch.GPModel):
def __init__(self):
super(KissGPModel, self).__init__(GaussianLikelihood(log_noise_bounds=(-5, 5)))
self.mean_module = ConstantMean(constant_bounds=(-1, 1))
covar_module = RBFKernel(log_lengthscale_bounds=(-3, 5))
self.grid_covar_module = GridInterpolationKernel(covar_module)
self.initialize_interpolation_grid(500, grid_bounds=[(0, 1)])
def forward(self, x):
mean_x = self.mean_module(x)
covar_x = self.grid_covar_module(x)
return GaussianRandomVariable(mean_x, covar_x)
model = KissGPModel().cuda()
model.condition(train_x, train_y)
# Optimize the model
def train():
model.train()
optimizer = optim.Adam(model.parameters(), lr=0.1)
optimizer.n_iter = 0
for i in range(30):
optimizer.zero_grad()
output = model(train_x)
loss = -model.marginal_log_likelihood(output, train_y) * 1. / len(train_x)
loss.backward()
optimizer.n_iter += 1
print('Iter %d/30 - Loss: %.3f' % (i + 1, loss.data[0]))
optimizer.step()
_ = model.eval()
%time train()
def plot_model_and_predictions(model, plot_train_data=True):
f, observed_ax = plt.subplots(1, 1, figsize=(4, 3))
test_x = Variable(torch.linspace(0, 1, 51)).cuda()
observed_pred = model(test_x)
def ax_plot(ax, rand_var, title):
lower, upper = rand_var.confidence_region()
if plot_train_data:
ax.plot(train_x.data.cpu().numpy(), train_y.data.cpu().numpy(), 'k*')
ax.plot(test_x.data.cpu().numpy(), rand_var.mean().data.cpu().numpy(), 'b')
ax.fill_between(test_x.data.cpu().numpy(), lower.data.cpu().numpy(), upper.data.cpu().numpy(), alpha=0.5)
ax.set_ylim([-3, 3])
ax.legend(['Observed Data', 'Mean', 'Confidence'])
ax.set_title(title)
ax_plot(observed_ax, observed_pred, 'Observed Values (Likelihood)')
return f
f = plot_model_and_predictions(model, plot_train_data=False)
| 0.903871 | 0.732065 |
```
import torch
import numpy as np
from detectron2.utils.visualizer import Visualizer
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
from matplotlib.patches import Ellipse
import matplotlib.cm as cm
dataset = torch.load("proposals_offsetted_top_1000.pt")
'''
res = dataset['data'][160]
print(res.keys())
image_id = res["image_id"]
instances = res["instances"]
proposals_offsetted = res["proposals_offsetted"]
print(image_id)
print(res['proposals_offsetted'].pred_boxes.tensor.size())
print(res['instances'].pred_boxes.tensor.size())
'''
def scatter(X, ax=None, top=None, color=None,
draw_cov=True, draw_samples=True,
format='proposals', linestyle=None, **kwargs):
'''
plot scatter points or scatter rectangles
dim X = nb of points per image x dim of space
'''
ax = ax or plt.gca()
ax.set_xticks([])
ax.set_yticks([])
N = top if ((top is not None) and top <= X.size(0)) else X.size(0)
R = rect_for_plots(X[:N, :], format=format)
if 'mu' in kwargs:
mu_ini, cov, labels, avg_scores = kwargs['mu'], kwargs['cov'], kwargs['labels'], kwargs['avg_scores']
mu = rect_for_plots(mu_ini, format=format)
ulabels = np.sort(np.unique(labels.cpu().numpy()))[1:]
ulabels = ulabels[np.argsort(params['avg_score'])]
colors = cm.rainbow(np.linspace(0, 1, len(ulabels)))
#Plot l=0 background
if draw_samples:
R_temp = R[labels==0]
for i in range(R_temp.size(0)):
ax.add_patch(
Rectangle((R_temp[i, 0].cpu().data.numpy(), R_temp[i, 1].cpu().data.numpy()),
R_temp[i, 2].cpu().data.numpy(), R_temp[i, 3].cpu().data.numpy(),
fill=False, color='gray', linewidth=1.5, alpha=0.1, linestyle='-'
)
)
#Plot l>1 object clusters
for l, c in zip(ulabels, colors):
R_temp = R[labels==l]
if draw_samples:
#Draw cluster l
for i in range(R_temp.size(0)):
ax.add_patch(
Rectangle((R_temp[i, 0].cpu().data.numpy(), R_temp[i, 1].cpu().data.numpy()),
R_temp[i, 2].cpu().data.numpy(), R_temp[i, 3].cpu().data.numpy(),
fill=False, color=c, linewidth=1.5, alpha=0.4, linestyle='-'
)
)
#Draw means
ax.add_patch(
Rectangle((mu[l - 1, 0].cpu().data.numpy(), mu[l - 1, 1].cpu().data.numpy()),
mu[l - 1, 2].cpu().data.numpy(), mu[l - 1, 3].cpu().data.numpy(),
fill=False, color=c, linewidth=2, alpha=0.8
)
)
if draw_cov:
#Draw variance
draw_ellipse(mu_ini[l - 1,0:2].cpu().detach().numpy(),
torch.diag(cov[l -1 , 0:2]).cpu().detach().numpy(),
ax=ax, fc=c
)
draw_ellipse(mu_ini[l - 1,2:4].cpu().detach().numpy(),
torch.diag(cov[l - 1, 2:4]).cpu().detach().numpy(),
ax=ax, fc=c
)
else:
if color==None:
reds = cm.get_cmap('Reds')
colors = np.flip(reds(np.linspace(0, 1, R.size(0))),0)
else:
colors=[color for i in range(R.size(0))]
linestyle = '-' if linestyle is None else linestyle
for i in reversed(range(R.size(0))):
ax.add_patch(
Rectangle((R[i, 0].cpu().data.numpy(), R[i, 1].cpu().data.numpy()),
R[i, 2].cpu().data.numpy(), R[i, 3].cpu().data.numpy(),
fill=False, color=colors[i], linewidth=1.5, alpha=0.5,
linestyle=linestyle
)
)
ax.axis('equal')
return None
def rect_for_plots(rects, format='proposals'):
'''
input: N x 4 in format (coordinates upper left) x (coordinates bottom right)
'''
if format == 'proposals':
R = torch.zeros_like(rects)
R[:, 0]=rects[:, 0]
R[:, 1]=rects[:, 3]
R[:, 2]=rects[:, 2]-rects[:, 0]
R[:, 3]=rects[:, 1] - rects[:,3]
elif format == 'instances':
R = torch.zeros_like(rects)
R[:, 0] = rects[:,0] - 0.5*rects[:,2]
R[:, 1] = rects[:, 1] - 0.5*rects[:, 3]
R[:, 2] = rects[:, 2]
R[:, 3] = rects[:, 3]
else:
X = torch.hstack((rects[:, 0].unsqueeze(1), rects[:, 2].unsqueeze(1)))
Y = torch.hstack((rects[:, 1].unsqueeze(1), rects[:, 3].unsqueeze(1)))
x_min, _ = torch.min(X, 1)
y_min, _ = torch.min(Y, 1)
x_max, _ = torch.max(X, 1)
y_max, _ = torch.max(Y, 1)
w = x_max - x_min
h = y_max - y_min
R = torch.hstack(
(x_min.unsqueeze(1),
y_min.unsqueeze(1),
w.unsqueeze(1),
h.unsqueeze(1))
)
return R
def draw_ellipse(pos, cov, ax=None, **kwargs):
if type(pos) != np.ndarray:
pos = to_numpy(poxs)
if type(cov) != np.ndarray:
cov = to_numpy(cov)
ax = ax or plt.gca()
U, s, Vt = np.linalg.svd(cov)
angle = np.degrees(np.arctan2(U[1,0], U[0,0]))
width, height = 2 * np.sqrt(s)
for nsig in range(1, 6):
ax.add_patch(Ellipse(pos, nsig*width, nsig*height, angle,
alpha=0.5/nsig, **kwargs))
'''
fig=plt.figure(figsize=(12,9))
ax = plt.gca()
scatter(res['proposals_offsetted'].pred_boxes.tensor, ax = ax, format='proposals', top=500)
scatter(res['instances'].pred_boxes.tensor, ax=ax, format='proposals',color='blue')
'''
from detectron2.structures import pairwise_iou
#The lower the nms_thr, the more the number of samples per cluster
def naive_estimation(res, nms_thr=0.5):
#1- Compute pair-wise IoUs
iou = pairwise_iou(res['instances'].pred_boxes,
res['proposals_offsetted'].pred_boxes
)
#2- Estimate the mixture parameters sequentially
params={'mu':[], 'cov':[], 'pi':[], 'avg_score':[]}
processed=0
proposals_tensor = res['proposals_offsetted'].pred_boxes.tensor.clone()
scores = res['proposals_offsetted'].scores.clone()
mask = torch.ones(proposals_tensor.size(0))
labels = torch.zeros_like(mask).long()
for i in range(len(res['instances'].pred_boxes)):
print('Number of remaining proposals: {}'.format(mask.sum()))
cond = ((iou[i, :] >= nms_thr)*mask).bool()
labels[cond]= i + 1
current_cluster=proposals_tensor[cond]
current_scores=scores[cond]
#Compute mixture parameters
N = current_cluster.size(0)
X = current_cluster
W = current_scores.unsqueeze(1).repeat(1, current_cluster.size(1))
mu = (X*W).sum(0)/W.sum(0)
cov = (1/N)*(((X - mu.repeat(X.size(0),1))**2)*W).sum(0)
pi = N
params['mu'].append(mu)
params['cov'].append(cov)
params['pi'].append(pi)
params['avg_score'].append(current_scores.mean().item())
#Suppress non-maximum boxes
mask[iou[i, :] >= nms_thr] = 0
processed += N
print('{} boxes in cluster {} (mean score: {})'.format(N, i, current_scores.mean()))
params['pi']=[params['pi'][i]/processed for i in range(len(params['pi']))]
print('{} boxes (/{}) used to estimate the mixture parameters'.format(processed, len(res['proposals_offsetted'].pred_boxes)))
return proposals_tensor, labels, params
'''
proposals_tensor, labels, params = naive_estimation(res)
'''
from PIL import Image
def open_img_from_path(img_path: str):
return np.array(Image.open(img_path))
def get_img_from_id(image_id, directory='/u/mernoult/ood-object-detection/datasets/coco/val2017/'):
file_name = directory
for _ in range(12 - len(str(image_id))):
file_name += str(0)
file_name += str(image_id) + '.jpg'
print(file_name)
image = open_img_from_path(file_name)
return image
'''
fig=plt.figure(figsize=(12,9))
image = get_img_from_id(image_id)
plt.imshow(image)
ax = plt.gca()
print(params['mu'])
scatter(proposals_tensor, ax=ax,
mu = torch.vstack(params['mu']),
cov = torch.vstack(params['cov']),
labels=labels,
avg_scores=params['avg_score'],
draw_cov=True,
draw_samples=False
)
scatter(res['instances'].pred_boxes.tensor, ax=ax,
format='proposals',
color='black',
linestyle='--')
'''
#Whole pipeline
#1-Pick an image
res = dataset['data'][175]
#2- Perform naive bbox density estimation
proposals_tensor, labels, params = naive_estimation(res)
#3- Plot
fig=plt.figure(figsize=(12,9))
image = get_img_from_id(res['image_id'])
plt.imshow(image)
ax = plt.gca()
print(params['mu'])
scatter(proposals_tensor, ax=ax,
mu = torch.vstack(params['mu']),
cov = torch.vstack(params['cov']),
labels=labels,
avg_scores=params['avg_score'],
draw_cov=True,
draw_samples=True
)
scatter(res['instances'].pred_boxes.tensor, ax=ax,
format='proposals',
color='black',
linestyle='--')
```
|
github_jupyter
|
import torch
import numpy as np
from detectron2.utils.visualizer import Visualizer
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
from matplotlib.patches import Ellipse
import matplotlib.cm as cm
dataset = torch.load("proposals_offsetted_top_1000.pt")
'''
res = dataset['data'][160]
print(res.keys())
image_id = res["image_id"]
instances = res["instances"]
proposals_offsetted = res["proposals_offsetted"]
print(image_id)
print(res['proposals_offsetted'].pred_boxes.tensor.size())
print(res['instances'].pred_boxes.tensor.size())
'''
def scatter(X, ax=None, top=None, color=None,
draw_cov=True, draw_samples=True,
format='proposals', linestyle=None, **kwargs):
'''
plot scatter points or scatter rectangles
dim X = nb of points per image x dim of space
'''
ax = ax or plt.gca()
ax.set_xticks([])
ax.set_yticks([])
N = top if ((top is not None) and top <= X.size(0)) else X.size(0)
R = rect_for_plots(X[:N, :], format=format)
if 'mu' in kwargs:
mu_ini, cov, labels, avg_scores = kwargs['mu'], kwargs['cov'], kwargs['labels'], kwargs['avg_scores']
mu = rect_for_plots(mu_ini, format=format)
ulabels = np.sort(np.unique(labels.cpu().numpy()))[1:]
ulabels = ulabels[np.argsort(params['avg_score'])]
colors = cm.rainbow(np.linspace(0, 1, len(ulabels)))
#Plot l=0 background
if draw_samples:
R_temp = R[labels==0]
for i in range(R_temp.size(0)):
ax.add_patch(
Rectangle((R_temp[i, 0].cpu().data.numpy(), R_temp[i, 1].cpu().data.numpy()),
R_temp[i, 2].cpu().data.numpy(), R_temp[i, 3].cpu().data.numpy(),
fill=False, color='gray', linewidth=1.5, alpha=0.1, linestyle='-'
)
)
#Plot l>1 object clusters
for l, c in zip(ulabels, colors):
R_temp = R[labels==l]
if draw_samples:
#Draw cluster l
for i in range(R_temp.size(0)):
ax.add_patch(
Rectangle((R_temp[i, 0].cpu().data.numpy(), R_temp[i, 1].cpu().data.numpy()),
R_temp[i, 2].cpu().data.numpy(), R_temp[i, 3].cpu().data.numpy(),
fill=False, color=c, linewidth=1.5, alpha=0.4, linestyle='-'
)
)
#Draw means
ax.add_patch(
Rectangle((mu[l - 1, 0].cpu().data.numpy(), mu[l - 1, 1].cpu().data.numpy()),
mu[l - 1, 2].cpu().data.numpy(), mu[l - 1, 3].cpu().data.numpy(),
fill=False, color=c, linewidth=2, alpha=0.8
)
)
if draw_cov:
#Draw variance
draw_ellipse(mu_ini[l - 1,0:2].cpu().detach().numpy(),
torch.diag(cov[l -1 , 0:2]).cpu().detach().numpy(),
ax=ax, fc=c
)
draw_ellipse(mu_ini[l - 1,2:4].cpu().detach().numpy(),
torch.diag(cov[l - 1, 2:4]).cpu().detach().numpy(),
ax=ax, fc=c
)
else:
if color==None:
reds = cm.get_cmap('Reds')
colors = np.flip(reds(np.linspace(0, 1, R.size(0))),0)
else:
colors=[color for i in range(R.size(0))]
linestyle = '-' if linestyle is None else linestyle
for i in reversed(range(R.size(0))):
ax.add_patch(
Rectangle((R[i, 0].cpu().data.numpy(), R[i, 1].cpu().data.numpy()),
R[i, 2].cpu().data.numpy(), R[i, 3].cpu().data.numpy(),
fill=False, color=colors[i], linewidth=1.5, alpha=0.5,
linestyle=linestyle
)
)
ax.axis('equal')
return None
def rect_for_plots(rects, format='proposals'):
'''
input: N x 4 in format (coordinates upper left) x (coordinates bottom right)
'''
if format == 'proposals':
R = torch.zeros_like(rects)
R[:, 0]=rects[:, 0]
R[:, 1]=rects[:, 3]
R[:, 2]=rects[:, 2]-rects[:, 0]
R[:, 3]=rects[:, 1] - rects[:,3]
elif format == 'instances':
R = torch.zeros_like(rects)
R[:, 0] = rects[:,0] - 0.5*rects[:,2]
R[:, 1] = rects[:, 1] - 0.5*rects[:, 3]
R[:, 2] = rects[:, 2]
R[:, 3] = rects[:, 3]
else:
X = torch.hstack((rects[:, 0].unsqueeze(1), rects[:, 2].unsqueeze(1)))
Y = torch.hstack((rects[:, 1].unsqueeze(1), rects[:, 3].unsqueeze(1)))
x_min, _ = torch.min(X, 1)
y_min, _ = torch.min(Y, 1)
x_max, _ = torch.max(X, 1)
y_max, _ = torch.max(Y, 1)
w = x_max - x_min
h = y_max - y_min
R = torch.hstack(
(x_min.unsqueeze(1),
y_min.unsqueeze(1),
w.unsqueeze(1),
h.unsqueeze(1))
)
return R
def draw_ellipse(pos, cov, ax=None, **kwargs):
if type(pos) != np.ndarray:
pos = to_numpy(poxs)
if type(cov) != np.ndarray:
cov = to_numpy(cov)
ax = ax or plt.gca()
U, s, Vt = np.linalg.svd(cov)
angle = np.degrees(np.arctan2(U[1,0], U[0,0]))
width, height = 2 * np.sqrt(s)
for nsig in range(1, 6):
ax.add_patch(Ellipse(pos, nsig*width, nsig*height, angle,
alpha=0.5/nsig, **kwargs))
'''
fig=plt.figure(figsize=(12,9))
ax = plt.gca()
scatter(res['proposals_offsetted'].pred_boxes.tensor, ax = ax, format='proposals', top=500)
scatter(res['instances'].pred_boxes.tensor, ax=ax, format='proposals',color='blue')
'''
from detectron2.structures import pairwise_iou
#The lower the nms_thr, the more the number of samples per cluster
def naive_estimation(res, nms_thr=0.5):
#1- Compute pair-wise IoUs
iou = pairwise_iou(res['instances'].pred_boxes,
res['proposals_offsetted'].pred_boxes
)
#2- Estimate the mixture parameters sequentially
params={'mu':[], 'cov':[], 'pi':[], 'avg_score':[]}
processed=0
proposals_tensor = res['proposals_offsetted'].pred_boxes.tensor.clone()
scores = res['proposals_offsetted'].scores.clone()
mask = torch.ones(proposals_tensor.size(0))
labels = torch.zeros_like(mask).long()
for i in range(len(res['instances'].pred_boxes)):
print('Number of remaining proposals: {}'.format(mask.sum()))
cond = ((iou[i, :] >= nms_thr)*mask).bool()
labels[cond]= i + 1
current_cluster=proposals_tensor[cond]
current_scores=scores[cond]
#Compute mixture parameters
N = current_cluster.size(0)
X = current_cluster
W = current_scores.unsqueeze(1).repeat(1, current_cluster.size(1))
mu = (X*W).sum(0)/W.sum(0)
cov = (1/N)*(((X - mu.repeat(X.size(0),1))**2)*W).sum(0)
pi = N
params['mu'].append(mu)
params['cov'].append(cov)
params['pi'].append(pi)
params['avg_score'].append(current_scores.mean().item())
#Suppress non-maximum boxes
mask[iou[i, :] >= nms_thr] = 0
processed += N
print('{} boxes in cluster {} (mean score: {})'.format(N, i, current_scores.mean()))
params['pi']=[params['pi'][i]/processed for i in range(len(params['pi']))]
print('{} boxes (/{}) used to estimate the mixture parameters'.format(processed, len(res['proposals_offsetted'].pred_boxes)))
return proposals_tensor, labels, params
'''
proposals_tensor, labels, params = naive_estimation(res)
'''
from PIL import Image
def open_img_from_path(img_path: str):
return np.array(Image.open(img_path))
def get_img_from_id(image_id, directory='/u/mernoult/ood-object-detection/datasets/coco/val2017/'):
file_name = directory
for _ in range(12 - len(str(image_id))):
file_name += str(0)
file_name += str(image_id) + '.jpg'
print(file_name)
image = open_img_from_path(file_name)
return image
'''
fig=plt.figure(figsize=(12,9))
image = get_img_from_id(image_id)
plt.imshow(image)
ax = plt.gca()
print(params['mu'])
scatter(proposals_tensor, ax=ax,
mu = torch.vstack(params['mu']),
cov = torch.vstack(params['cov']),
labels=labels,
avg_scores=params['avg_score'],
draw_cov=True,
draw_samples=False
)
scatter(res['instances'].pred_boxes.tensor, ax=ax,
format='proposals',
color='black',
linestyle='--')
'''
#Whole pipeline
#1-Pick an image
res = dataset['data'][175]
#2- Perform naive bbox density estimation
proposals_tensor, labels, params = naive_estimation(res)
#3- Plot
fig=plt.figure(figsize=(12,9))
image = get_img_from_id(res['image_id'])
plt.imshow(image)
ax = plt.gca()
print(params['mu'])
scatter(proposals_tensor, ax=ax,
mu = torch.vstack(params['mu']),
cov = torch.vstack(params['cov']),
labels=labels,
avg_scores=params['avg_score'],
draw_cov=True,
draw_samples=True
)
scatter(res['instances'].pred_boxes.tensor, ax=ax,
format='proposals',
color='black',
linestyle='--')
| 0.446977 | 0.585397 |
# Notes for Think Stats by Allen B. Downey
```
from typing import List
import numpy as np
import pandas as pd
from scipy import stats
import matplotlib.pyplot as plt
import sklearn
% matplotlib inline
```
## Chapter 01
### Glossary
- anecdotal evidence - is an evidence based on personal experience rather than based on well-designed and scrupulous study.
- cross-sectional study - is a study that colllects data about a population at a particular point in time.
- longitudinal study - is a study that follow the same group repeatedly and collects the data over time.
## Chapter 02
#### Mean - central tendency
$$ \overline{x} = \frac{1}{n} \sum_i x_i \ $$
```
sample = [1, 3, 5, 6]
np.mean(sample)
pd.DataFrame(sample).mean()
```
#### Variance
$$ S^2 = \frac{1}{n} \sum_i (x_i - \overline{x})^2 $$
```
np.var(sample)
# Warning! Pandas variance by default is normalized by N-1!
# That can be changed by using ddof(delta degrees of freedom) = 0
pd.DataFrame(sample).var(ddof = 0)
```
#### Standard Deviation
$$ \sigma = \sqrt{S^{2}} $$
```
np.std(sample)
# Warning! Pandas std is calculated with variance by N-1!
# That can be changed by using ddof(delta degrees of freedom) = 0
pd.DataFrame(sample).std(ddof = 0)
```
#### Effect size - Cohen'd
Having groups **G1** and **G2**, with number of elements given as **N1** and **N2**, the effect size is given as:
$$ Cohen'd = \frac{\overline{G1} - \overline{G2}}{\sqrt{(\sigma (G1) \cdot (N1-1) + \sigma (G2) \cdot (N2-1)) / ((N1-1) + (N2-1))}} $$
```
def effect_size(g1: pd.DataFrame, g2: pd.DataFrame) -> float:
diff = g1.mean() - g2.mean()
var_g1, var_g2 = g1.var(ddof=1), g2.var(ddof=1)
n1, n2 = len(g1), len(g2)
pooled_var = (var_g1 * (n1 - 1) + var_g2 * (n2 - 1)) / ((n1 - 1) + (n2 - 1))
cohen_d = diff / np.sqrt(pooled_var)
return cohen_d
```
It is calculated with delta degree of freedom = 1!
```
effect_size(pd.DataFrame([1, 2, 3, 4]), pd.DataFrame([3, 3, 1, 2]))
```
## Chapter 03
#### Probability Mass Function
Probability mass function maps each value to its probability.
Probability of a group always adds to one.
```
s = pd.Series([1, 2, 3, 4, 2])
def pmf(series: pd.Series) -> pd.Series:
return series.value_counts().sort_index() / series.count()
pmf(s)
```
#### DataFrame Indexing
```
array = np.random.randn(4, 2)
array
df = pd.DataFrame(array)
df
columns = ['A', 'B']
df = pd.DataFrame(data=array,
columns=columns)
df
index = ['a', 'b', 'c', 'd']
df = pd.DataFrame(data=array,
columns=columns,
index=index)
df
df['A']
df.loc['a']
df.iloc[0]
indices = ['a', 'c']
df.loc[indices]
df['a':'c']
df[0:2]
df[:2]
df['A'].loc['a']
```
## Chapter 04
#### Percentile Rank
Percentile rank is a metric that presents how big is the subset of the data that the values in the subset
are equal or below any given value.
```
# Data
a = [1, 2, 3, 3, 4, 5]
stats.percentileofscore(a=a, score=2.5)
stats.percentileofscore(a=a, score=2)
stats.percentileofscore(a=a, score=3)
```
#### Percentile
Percentile is the opposite operation to percentile rank - it maps a percentile rank to a value.
```
np.percentile(a=a, q=50)
np.percentile(a=a, q=70)
# Pandas uses quantiles with different interpolation methods.
pd.DataFrame(data=a).quantile(q=0.5)
```
#### Cumulative Distribution Function
Cumulative Distribution Function is the function that maps from a value to its percentile rank.
```
series = pd.Series(np.random.randn(25))
series.head()
cdf = series.value_counts().sort_index().cumsum()
cdf.plot()
```
It can be plotted using .hist()
```
series.hist(cumulative=True)
```
#### Interquartile Range
Interquartile range is the difference between the 7th and 25th percentiles.
It is used as a measure of the spread of a distribution.
## Chapter 05
#### Exponential Distribution
$$ f(x, \lambda ) = \left\{\begin{matrix}
\lambda e^{- \lambda x} & x \geq 0
\\
0 & x < 0
\end{matrix}\right. $$
Variable $ \lambda $ defines the shape of the distribution.
The exponential distribution is used analyzing a series of events and measure times between them.
If the events are equally likely to occur at any time, the distribution of inverarrival times tends to look like an exponential distribution.
```
r1 = np.random.exponential(scale=1.0, size=100000)
r2 = np.random.exponential(scale=0.5, size=100000)
plt.hist(r1, bins = 200)
plt.xlim((0, 10))
plt.show()
plt.hist(r2, bins = 200)
plt.xlim((0, 10))
plt.show()
```
#### Normal (Gaussian) Distribution
$$ f(x | \mu, \sigma ^{2} ) = \frac{1}{\sqrt{2\pi\sigma ^{2}}} e^{- \frac{(x - \mu )^{2}}{2 \sigma^{2}}} $$
The Gaussian distribution is described by two variables:
- mean $\mu$
- standard deviation $\sigma$
If $\mu=0$ and $\sigma=1$, the distribution is called standard normal distribution.
The Gaussian distribution approximates a lot of natural pheonomena.
It describes the variability in the data, where the forces behind them is additive.
Physical processes are expected to be the sum of many independant processes and often the have distributions nearly the normal distribution.
```
g1 = np.random.normal(loc=0.0, scale=1.0, size=100000)
g2 = np.random.normal(loc=0.0, scale=3.0, size=100000)
plt.hist(g1, bins = 200)
plt.xlim((-10, 10))
plt.show()
plt.hist(g2, bins = 200)
plt.xlim((-10, 10))
plt.show()
```
#### Lognormal Distribution
$$ f(x | \mu, \sigma ^{2} ) = \frac{1}{\sigma x \sqrt{2\pi}} e^{- \frac{(ln(x) - \mu )^{2}}{2 \sigma^{2}}} $$
where:
- $\mu$ is mean of the corresponding Gaussian distribution
- $\sigma$ is standard deviation of the corresponding Gaussian distribution
The lognormal distribution is similar to the Gaussian distribution.
The difference is that it is assumed that the processes behind the outcome are multiplicative, instead of additive as in the Gaussian distribution.
```
l1 = np.random.lognormal(mean=0.0, sigma=0.5, size=100000)
l2= np.random.lognormal(mean=0.0, sigma=1, size=100000)
plt.hist(l1, bins = 1000)
plt.xlim((0, 10))
plt.ylim((0, 4500))
plt.show()
plt.hist(l2, bins = 1000)
plt.xlim((0, 10))
plt.ylim((0, 4500))
plt.show()
```
#### Pareto Distribution
The Pareto distribution originated from the economics as description of wealth ion the society.
If is often described using so called "Matthew principle": "rich get reacher, poor get poorer".
The probability density for the Pareto distribution is given as:
$$p(x) = \frac{am^a}{x^{a+1}}$$
where:
- a is the shape
- m is the scale
Pareto distribution given like that can be obtained:
```python
p = (np.random.pareto(a, size=1000) + 1) * m
```
```
p1 = (np.random.pareto(1, size=1000) + 1) * 1
p2 = (np.random.pareto(2, size=1000) + 1) * 1
plt.hist(p1, bins=100)
plt.ylim((0, 1000))
plt.show()
plt.hist(p2, bins = 100)
plt.ylim((0, 1000))
plt.show()
```
#### Weibull Distribution
The Weibull Distribution is given as:
$$ f(x ; \lambda, a) = \left\{\begin{matrix}
\frac{a}{\lambda}(\frac{x}{\lambda})^{a-1}e^{-(\frac{x}{\lambda})^{a}}
& x \geq 0\\
0 & x < 0
\end{matrix}\right. $$
where:
- a is the shape
- $\lambda$ is the scale
If the quantity X is a "time-to-failure", the Weibull distribution gives a distribution for which the failure rate is proportional to a power of time.
The shape parameter, a, is that power plus one, and so this parameter can be interpreted directly as follows:
1) ** a < 1 ** - indicates that the failure rate decreases over time (Lindy effect). This happens if there is significant "infant mortality", or defective items failing early and the failure rate decreasing over time as the defective items are weeded out of the population. In the context of the diffusion of innovations, this means negative word of mouth: the hazard function is a monotonically decreasing function of the proportion of adopters.
2) ** a = 1 ** - indicates that the failure rate is constant over time. This might suggest random external events are causing mortality, or failure. The Weibull distribution reduces to an exponential distribution.
3) ** a > 1 ** - indicates that the failure rate increases with time. This happens if there is an "aging" process, or parts that are more likely to fail as time goes on. In the context of the diffusion of innovations, this means positive word of mouth: the hazard function is a monotonically increasing function of the proportion of adopters.
```
w1 = np.random.weibull(a=0.8, size=1000000)
w2 = np.random.weibull(a=1, size=1000000)
w3 = np.random.weibull(a=5, size=1000000)
w4 = np.random.weibull(a=10, size=1000000)
plt.hist(w1, bins = 200)
plt.xlim((-1, 15))
plt.ylim((0, 200000))
plt.show()
plt.hist(w2, bins = 200)
plt.xlim((-1, 15))
plt.ylim((0, 200000))
plt.show()
```
Different scale below:
```
plt.hist(w3, bins = 200)
plt.xlim((-1, 5))
plt.ylim((0, 25000))
plt.show()
plt.hist(w4, bins = 200)
plt.xlim((-1, 5))
plt.ylim((0, 25000))
plt.show()
```
## Chapter 06
#### Moments
$k^{th}$ central moment is given as:
$$ m_{k} = \frac{1}{n}\sum_{i}^{ }(x_{i} - \bar{x})^{k} $$
Second order momentu, when $k = 2 $, is the variance.
If the measured value is in f.e. $cm$, the first moment is also in $cm$, but the second is in $cm^{2}$, the third in $cm^{3}$, the forth $cm^{4}$, and so on.
#### Skewness
Skewness is a property that describes the shape of a distribution.
- If the distribution is focused around its central tendency, it is unskewed.
- If the values focues on the left of the central tendency, it is described as "left skewed".
- If the values focues on the right of the central tendency, it is called "right skewed".
#### Pearson's median skewness coefficient
Pearson's median skewness coefficient is a measure of skewness baed on the difference between the sample mean and median:
$$ g_{p}=3 \frac{(\bar x - \tilde{x})}{\sigma} $$
where:
- $ \tilde{x} $ is the median
- $ \sigma $ is the standard deviation
#### Robustness of a statistic
A statistic is robust if the outliers have relatively small impact on the value of the statistic.
## Chapter 07
#### Correlation
A correlation is a statistic intended to quantify the strength of the relationship between two variables.
Few challenges present themselves during such analysis:
- usually the variables have different units
- usually the variables come from different distributions
There are two common approaches trying to solve the challenges:
- Transforming each value to a standard score (example: Pearson product-moment correlation coefficient)
- Transforming each value to a rank (example: Spearman rank correlation coefficient)
#### Covariance
Covariance is a measurement of the tendency of two variables to vary together.
It is given as:
$$ Cov(X, Y) = \frac{1}{n-1}\sum (x_{i} - \bar x)(y_{i} - \bar y) $$
where:
- X and Y are two series of the same lengths
```
Z = np.array([[0, 2], [1, 1], [2, 0]]).T
Z
np.cov(Z, ddof=1)
```
#### Pearson's Correlation
The Pearson's correlation is computed by dividing the deviations by the standard deviations:
$$p = \frac{Conv(X, Y)}{\sigma_{X} \sigma_{Y}}$$
Pearson's correlations ia always between -1 and +1.
If the value $p$ is positive, the correlated values change is similar manner, when one is high, the other one tends to be high as well, when one is low, the other one tends to be low.
If the value $p$ is positive, the correlated values change is similar manner, when one is high, the other one tends to be high as well, when one is low, the other one tends to be low.
The magnitude of the correlation, $p$, describes the strength of the correlation when 1 is the perfect, positive correlation.
Pearson's correlation works
#### Spearman's Rank
Spearman's rank is more robust than the Pearson's correlations. It mitgates the effect of outliers and skewed distributions.
If the relationship is nonlinear, the Pearson'c correlation tends to underestimate the strength of the relationship.
```
stats.spearmanr([1, 2, 3, 4, 5], [5, 6, 7, 8, 7])
```
## Chapter 08
#### Mean Squared Error
Mean squared error is a way to measure a quality of an estimator.
It is important to mention that it is very sensitive to outliers and large values.
$$ MSE = \frac{1}{n}\sum (Y_{i}-\hat{Y_{i}})^{2} $$
```
from sklearn.metrics import mean_squared_error
y_true = [3, -0.5, 2, 7]
y_pred = [2.5, 0.0, 2, 8]
mean_squared_error(y_true, y_pred)
```
## Chapter 09
#### T-test
A t-test is an analysis framework used to determine the difference between two sample means from two normally distributed populations with unknown variances.
#### Chi-Squared Test
Definition:
https://www.chegg.com/homework-help/definitions/chi-square-test-14
Example of chi-squared test methodology.
https://www.spss-tutorials.com/chi-square-independence-test/
#### Holm–Bonferroni method
Holm-Bonferroni method is used to counteract the problem of multiple comparisons.
#### Errors
In hypothesis testing, there are two types of error one can make:
- false positive - assuming that something is significant, when, in reality, it is not.
- false negative - assuming that something is not significant when it is.
## Chapter 10
#### Least Squares Fit
Least square fit is given as:
$$ y = ax + b $$
where:
- a - slope
- b - inter
It is a good approach to estimate an unknown value or correlation between values if the relation is linear.
#### Coefficient of determination
Another way to measure goodness of fit is the coefficient of determination, known as $R^2$ and called R-squared:
There is a relationship between the Pearson's coefficient of correlation:
$$ R^2 = p^2 $$
Thus, if Pearson's correlation is 0.5 or -0.5, then the R-squared is 0.25.
## Chapter 11
#### Logistic Regression
Linear regression can be generalized to handle various kind of dependent variables.
#### Types of variables
Endogenous variables are dependent variables, they are kind of variables one would like to predict.
Exogenous variables are explanatory variables, which are variables used to predict or explain dependent variables.
## Chapter 12
#### Vocabulary
- trend = a smooth function that captures persistent changes
- seasonality = periodic variation (possibly daily, weekly, monghtly, yearly cycles)
- noise = random variations around a longterm trend
#### Moving average
One way to simply measure seasonality is moving average.
It is computed by calculating mean over a certain window and move the window, usually by the smallest period.
```
trend = pd.Series([1, 2, 3, 4, 5, 6, 5, 4, 3, 2, 1])
noise = pd.Series(np.random.random(11))
season = trend + noise
season.name = 'season'
two_day_window = season.rolling(window=2).mean()
two_day_window.name = 'rolling mean'
two_day_window
plt.figure(figsize=(7, 7))
plt.plot(season)
plt.plot(two_day_window)
plt.legend()
```
#### Exponentially-weighted moving average (EWMA)
Another approach is to calculate weighted average where the most recent values has the highest weight and the weights from previous values drop off exponentially.
The span parameter roughly corresponds to the window size of a moving average. It controls how fast the weights drop off, so it determines the number of points that make a non-negligible contribution to each average.
```
ewma2 = season.ewm(span=2).mean()
ewma2.name = 'ewma2'
ewma3 = season.ewm(span=3).mean()
ewma3.name = 'ewma3'
ewma5 = season.ewm(span=5).mean()
ewma5.name = 'ewma5'
plt.figure(figsize=(7, 7))
plt.plot(season)
plt.plot(ewma2)
plt.plot(ewma3)
plt.plot(ewma5)
plt.legend()
```
#### Types of errors in series prediction
- sampling error = the prediction is based on estimated parameters, which depend on random variation in the sample. If we run the experiment again, we expect estimates to vary
- random variation = unexpected random event / variation
- modeling error = inadequate, over engineered or simply wrong models
## Chapter 13
#### Survival Curve
Survival times are data that measure follow-up time from a defined starting point to the occurrence of a given event.
Usually, the underlying distribution or rarely normal, thus standard statistical techniques cannot be applied.
The survival function is gives as:
$$ S(t) = 1 - CDF(t) $$
where:
- $CDF(t)$ is the probability of a lifetime less than or equal to $t$.
#### Hazard function
$$h(t)=\frac{S(t) - S(t+1))}{S(t)}$$
Numerator of the hazard function $h(t)$ is the fraction of lifetimes that end at t.
#### Kaplan-Meier Estimation
The Kaplan-Meier estimate is also called as “product limit estimate”. It involves computing of probabilities of occurrence of event at a certain point of time. We multiply these successive probabilities by any earlier computed probabilities to get the final estimate. The survival probability at any particular time is calculated by the formula given below:
$$ S_{t} = \frac{N_{las} - N_{d}}{N_{las} } $$
where:
- $N_{las}$ - Number of subjects living at the start
- $N_{d}$ - Number of subjects died
|
github_jupyter
|
from typing import List
import numpy as np
import pandas as pd
from scipy import stats
import matplotlib.pyplot as plt
import sklearn
% matplotlib inline
sample = [1, 3, 5, 6]
np.mean(sample)
pd.DataFrame(sample).mean()
np.var(sample)
# Warning! Pandas variance by default is normalized by N-1!
# That can be changed by using ddof(delta degrees of freedom) = 0
pd.DataFrame(sample).var(ddof = 0)
np.std(sample)
# Warning! Pandas std is calculated with variance by N-1!
# That can be changed by using ddof(delta degrees of freedom) = 0
pd.DataFrame(sample).std(ddof = 0)
def effect_size(g1: pd.DataFrame, g2: pd.DataFrame) -> float:
diff = g1.mean() - g2.mean()
var_g1, var_g2 = g1.var(ddof=1), g2.var(ddof=1)
n1, n2 = len(g1), len(g2)
pooled_var = (var_g1 * (n1 - 1) + var_g2 * (n2 - 1)) / ((n1 - 1) + (n2 - 1))
cohen_d = diff / np.sqrt(pooled_var)
return cohen_d
effect_size(pd.DataFrame([1, 2, 3, 4]), pd.DataFrame([3, 3, 1, 2]))
s = pd.Series([1, 2, 3, 4, 2])
def pmf(series: pd.Series) -> pd.Series:
return series.value_counts().sort_index() / series.count()
pmf(s)
array = np.random.randn(4, 2)
array
df = pd.DataFrame(array)
df
columns = ['A', 'B']
df = pd.DataFrame(data=array,
columns=columns)
df
index = ['a', 'b', 'c', 'd']
df = pd.DataFrame(data=array,
columns=columns,
index=index)
df
df['A']
df.loc['a']
df.iloc[0]
indices = ['a', 'c']
df.loc[indices]
df['a':'c']
df[0:2]
df[:2]
df['A'].loc['a']
# Data
a = [1, 2, 3, 3, 4, 5]
stats.percentileofscore(a=a, score=2.5)
stats.percentileofscore(a=a, score=2)
stats.percentileofscore(a=a, score=3)
np.percentile(a=a, q=50)
np.percentile(a=a, q=70)
# Pandas uses quantiles with different interpolation methods.
pd.DataFrame(data=a).quantile(q=0.5)
series = pd.Series(np.random.randn(25))
series.head()
cdf = series.value_counts().sort_index().cumsum()
cdf.plot()
series.hist(cumulative=True)
r1 = np.random.exponential(scale=1.0, size=100000)
r2 = np.random.exponential(scale=0.5, size=100000)
plt.hist(r1, bins = 200)
plt.xlim((0, 10))
plt.show()
plt.hist(r2, bins = 200)
plt.xlim((0, 10))
plt.show()
g1 = np.random.normal(loc=0.0, scale=1.0, size=100000)
g2 = np.random.normal(loc=0.0, scale=3.0, size=100000)
plt.hist(g1, bins = 200)
plt.xlim((-10, 10))
plt.show()
plt.hist(g2, bins = 200)
plt.xlim((-10, 10))
plt.show()
l1 = np.random.lognormal(mean=0.0, sigma=0.5, size=100000)
l2= np.random.lognormal(mean=0.0, sigma=1, size=100000)
plt.hist(l1, bins = 1000)
plt.xlim((0, 10))
plt.ylim((0, 4500))
plt.show()
plt.hist(l2, bins = 1000)
plt.xlim((0, 10))
plt.ylim((0, 4500))
plt.show()
p = (np.random.pareto(a, size=1000) + 1) * m
p1 = (np.random.pareto(1, size=1000) + 1) * 1
p2 = (np.random.pareto(2, size=1000) + 1) * 1
plt.hist(p1, bins=100)
plt.ylim((0, 1000))
plt.show()
plt.hist(p2, bins = 100)
plt.ylim((0, 1000))
plt.show()
w1 = np.random.weibull(a=0.8, size=1000000)
w2 = np.random.weibull(a=1, size=1000000)
w3 = np.random.weibull(a=5, size=1000000)
w4 = np.random.weibull(a=10, size=1000000)
plt.hist(w1, bins = 200)
plt.xlim((-1, 15))
plt.ylim((0, 200000))
plt.show()
plt.hist(w2, bins = 200)
plt.xlim((-1, 15))
plt.ylim((0, 200000))
plt.show()
plt.hist(w3, bins = 200)
plt.xlim((-1, 5))
plt.ylim((0, 25000))
plt.show()
plt.hist(w4, bins = 200)
plt.xlim((-1, 5))
plt.ylim((0, 25000))
plt.show()
Z = np.array([[0, 2], [1, 1], [2, 0]]).T
Z
np.cov(Z, ddof=1)
stats.spearmanr([1, 2, 3, 4, 5], [5, 6, 7, 8, 7])
from sklearn.metrics import mean_squared_error
y_true = [3, -0.5, 2, 7]
y_pred = [2.5, 0.0, 2, 8]
mean_squared_error(y_true, y_pred)
trend = pd.Series([1, 2, 3, 4, 5, 6, 5, 4, 3, 2, 1])
noise = pd.Series(np.random.random(11))
season = trend + noise
season.name = 'season'
two_day_window = season.rolling(window=2).mean()
two_day_window.name = 'rolling mean'
two_day_window
plt.figure(figsize=(7, 7))
plt.plot(season)
plt.plot(two_day_window)
plt.legend()
ewma2 = season.ewm(span=2).mean()
ewma2.name = 'ewma2'
ewma3 = season.ewm(span=3).mean()
ewma3.name = 'ewma3'
ewma5 = season.ewm(span=5).mean()
ewma5.name = 'ewma5'
plt.figure(figsize=(7, 7))
plt.plot(season)
plt.plot(ewma2)
plt.plot(ewma3)
plt.plot(ewma5)
plt.legend()
| 0.59972 | 0.991067 |
```
import pandas as pd
names = pd.read_csv('data/names.csv')
names.head()
new_data_list = ['Aria', 1]
names.loc[4] = new_data_list
names
names.loc['five'] = ['Zach', 3]
names
names.loc[len(names)] = {'Name':'Zayd',
'Age':2}
names
names.loc[len(names)] = {'Name':'Dean',
'Age':32}
names
names = pd.read_csv('data/names.csv')
names.append({'Name':'Aria',
'Age':1}, ignore_index = True)
names.index = ['Canada', 'Canada','USA','USA']
names
s = pd.Series({'Name':'Zach',
'Age':3},
name = len(names))
s
names.append(s)
s1 = pd.Series({'Name': 'Zach',
'Age': 3},
name = len(names))
s2 = pd.Series({'Name': 'Zayd',
'Age': 2},
name = 'USA')
names.append([s1,s2])
bbal_16 = pd.read_csv('data/baseball16.csv')
bbal_16.head()
data_dict = bbal_16.iloc[0].to_dict()
data_dict
import numpy as np
new_data_dict = {k: '' if isinstance(v, str) else np.nan for k,v in data_dict.items()}
new_data_dict
for k,v in data_dict.items():
print(f' k is {k} en v is {v}')
random_data = []
for i in range(1000):
d = dict()
for k, v in data_dict.items():
if isinstance(v, str):
d[k] = np.random.choice(list('abcde'))
else:
d[k] = np.random.randint(10)
random_data.append(pd.Series(d, name = i + len(bbal_16)))
random_data[0].head()
%%timeit
bbal_16_copy = bbal_16.copy()
for row in random_data:
bbal_16_copy = bbal_16_copy.append(row)
%%timeit
bbal_16_copy = bbal_16.copy()
bbal_16_copy = bbal_16_copy.append(random_data)
stocks_2016 = pd.read_csv('data/stocks_2016.csv',
index_col = 'Symbol')
stocks_2016
stocks_2017 = pd.read_csv('data/stocks_2017.csv',
index_col = 'Symbol')
stocks_2017
s_list = [ stocks_2016, stocks_2017]
pd.concat(s_list)
pd.concat(s_list, keys = ['2016', '2017'],
names = ['Year', 'Symbol'])
pd.concat(s_list,
keys = ['2016', '2017'],
axis = 'columns',
sort = True,
names = ['Year', None])
pd.concat(s_list,
join = 'inner',
axis = 'columns',
names = 'Year',
sort = True)
stocks_2016.append(stocks_2017)
trump_url = 'https://en.wikipedia.org/wiki/List_of_generation_VIII_Pok%C3%A9mon'
df_list = pd.read_html(trump_url)
len(df_list)
df_list[3]
df_list = pd.read_html(trump_url,
match = 'Yungoos',
header = 0)
df_list
df_list[0].columns
df_list2 = df_list[0]
df_list2.columns = ['Pokémon species', 'Pokémon']
df_list2.head()
from IPython.display import display_html
years = 2016,2017,2018
stock_tables = [pd.read_csv('data/stocks_{}.csv'.format(year),
index_col = 'Symbol') for year in years]
type(stock_tables)
stock_tables = []
for year in years:
print(year)
stock_tables.append(pd.read_csv('data/stocks_{}.csv'.format(year), index_col = 'Symbol'))
stock_tables[2]
stocks_2018
def display_frames(frames, num_spaces = 0):
t_style = '<table style = "display: inline;"'
tables_html = [df.to_html().replace('<table', t_style) for df in frames]
space = ' ' * num_spaces
# print(tables_html)
display_html(space.join(tables_html), raw = True)
display_frames(stock_tables, 30)
pd.concat(stock_tables, keys = [2016, 2017, 2018])
pd.concat(dict(zip(years, stock_tables)), axis = 'columns')
stocks_2016.join(stocks_2017,
lsuffix = '_2016',
rsuffix = '_2017',
how = 'outer')
other = [stock_tables[1].add_suffix('_2017'),
stock_tables[2].add_suffix('_2018')]
# stocks_2017
stock_tables[2]
stocks_2016.add_suffix('_2016').join(other,
how = 'outer')
stock_join = stocks_2016.add_suffix('_2016').join(other,
how = 'outer')
stock_concat = pd.concat(dict(zip(years, stock_tables)),
axis = 'columns',
sort = True)
stock_concat
level_1 = stock_concat.columns.get_level_values(1)
level_1
level_0 = stock_concat.columns.get_level_values(0).astype(str)
level_0
stock_concat.columns = level_1 + '_' + level_0
stock_concat
stock_join
stock_join.equals(stock_concat)
names = ['prices', 'transactions']
food_tables = [pd.read_csv('data/food_{}.csv'.format(name)) for name in names]
food_tables
food_tables[0]
food_tables[1]
food_prices, food_transactions = food_tables
display_frames(food_tables, 30)
food_transactions.merge(food_prices,
on = ['item', 'store'])
food_prices.query('Date == 2017')
food_transactions.merge(food_prices.query('Date == 2017'),
how = 'left')
food_prices.query('Date == 2017').set_index(['item', 'store'])
food_prices_join = food_prices.query('Date == 2017')\
.set_index(['item', 'store'])
food_prices_join
food_transactions.join(food_prices_join,
on = ['item', 'store'])
import glob
df_list = []
for filename in glob.glob('*/gas prices/*.csv'):
# print(filename)
df_list.append(pd.read_csv(filename,
index_col = 'Week',
parse_dates = ['Week']))
gas = pd.concat(df_list, axis = 'columns')
gas.head()
from sqlalchemy import create_engine
engine = create_engine('sqlite:///data/chinook.db')
tracks = pd.read_sql_table('tracks', engine)
tracks.head()
genres = pd.read_sql_table('genres', engine)
genres.head()
genre_track = genres.merge(tracks[['GenreId', 'Milliseconds']],
on = 'GenreId',
how = 'left')
genre_track.head()
genre_time = genre_track.groupby('Name')['Milliseconds'].mean()
genre_time
pd.to_timedelta(genre_time, unit = 'ms').sort_values().dt.floor('s')
cust = pd.read_sql_table('customers',
engine,
columns = ['CustomerId', 'FirstName', 'LastName'])
cust.head()
invoice = pd.read_sql_table('invoices',
engine,
columns = ['InvoiceId', 'CustomerId'])
invoice.head()
ii = pd.read_sql_table('invoice_items',
engine,
columns = ['InvoiceId', 'UnitPrice', 'Quantity'])
ii.head()
cust_inv = cust.merge(invoice, on='CustomerId')
cust_inv.head(9)
cust_inv = cust.merge(invoice, on='CustomerId')\
.merge(ii, on = 'InvoiceId')
cust_inv.head(8)
cust_inv['Total'] = cust_inv['Quantity'] * cust_inv['UnitPrice']
cols = ['CustomerId', 'FirstName', 'LastName']
# cust_inv.assign(Total = total)
cust_inv.head(8)
cust_inv.groupby(cols)['Total'].sum()\
.sort_values(ascending = False)\
.head()
sql_string1 = '''
select
Name,
time(avg(Milliseconds) / 1000, 'unixepoch') as avg_time
from (
select
g.Name,
t.Milliseconds
from
genres as g
join
tracks as t
on
g.genreid == t.genreid
)
group by
Name
order by
avg_time
'''
pd.read_sql_query(sql_string1, engine)
>>> sql_string2 = '''
select
c.customerid,
c.FirstName,
c.LastName,
sum(ii.quantity * ii.unitprice) as Total
from
customers as c
join
invoices as i
on c.customerid = i.customerid
join
invoice_items as ii
on i.invoiceid = ii.invoiceid
group by
c.customerid, c.FirstName, c.LastName
order by
Total desc
'''
>>> pd.read_sql_query(sql_string2, engine)
import pandas as pd
crime_sort = pd.read_hdf('data/crime.h5', 'crime') \
.set_index('REPORTED_DATE') \
.sort_index()
crime_sort.head()
crime_sort.resample('W')
weekly_crimes = crime_sort.resample('W').size()
weekly_crimes.head()
len(crime_sort.loc[:'2012-1-8'])
len(crime_sort.loc['2012-1-9':'2012-1-15'])
crime_sort.resample('W-THU').size().head()
crime_sort.loc[:'2012-1-5'].size
weekly_crimes_gby = crime_sort.groupby(pd.Grouper(freq = 'W'))\
.size()
weekly_crimes_gby.head()
dir(crime_sort.resample('W'))
crime = pd.read_hdf('data/crime.h5', 'crime')
crime.head()
weekly_crimes2 = crime.resample('W',
on = 'REPORTED_DATE').size()
weekly_crimes2.head()
weekly_crimes2.equals(weekly_crimes)
weekly_crimes_gby2 = crime.groupby(pd.Grouper(key = 'REPORTED_DATE',
freq = 'W')).size()
weekly_crimes_gby2.head()
weekly_crimes_gby2.equals(weekly_crimes_gby)
weekly_crimes.plot(figsize = (16,4),
title = 'All Denver Crimes');
crime_sort = pd.read_hdf('data/crime.h5',
'crime')\
.set_index('REPORTED_DATE')\
.sort_index()
crime_sort.head()
crime_quarterly = crime_sort.resample('Q')['IS_CRIME',
'IS_TRAFFIC'].sum()
crime_quarterly.head()
crime_sort.resample('QS')['IS_CRIME','IS_TRAFFIC'].sum()\
.head()
crime_sort.loc['2012-04-01':'2012-06-30',['IS_CRIME',
'IS_TRAFFIC']]\
.sum()
crime_quarterly2 = crime_sort.groupby(pd.Grouper(freq = 'Q'))\
['IS_CRIME','IS_TRAFFIC'].sum()
crime_quarterly2.head()
crime_quarterly2.equals(crime_quarterly)
plot_kwargs = dict(figsize = (16,4),
color = ['black', 'lightgrey'],
title = 'Denver Crimes and Traffic Accidents')
crime_quarterly.plot(**plot_kwargs);
crime_begin = crime_quarterly.iloc[0]
crime_begin
crime_quarterly.div(crime_begin)\
.sub(1)\
.round(2)\
.plot(**plot_kwargs);
```
|
github_jupyter
|
import pandas as pd
names = pd.read_csv('data/names.csv')
names.head()
new_data_list = ['Aria', 1]
names.loc[4] = new_data_list
names
names.loc['five'] = ['Zach', 3]
names
names.loc[len(names)] = {'Name':'Zayd',
'Age':2}
names
names.loc[len(names)] = {'Name':'Dean',
'Age':32}
names
names = pd.read_csv('data/names.csv')
names.append({'Name':'Aria',
'Age':1}, ignore_index = True)
names.index = ['Canada', 'Canada','USA','USA']
names
s = pd.Series({'Name':'Zach',
'Age':3},
name = len(names))
s
names.append(s)
s1 = pd.Series({'Name': 'Zach',
'Age': 3},
name = len(names))
s2 = pd.Series({'Name': 'Zayd',
'Age': 2},
name = 'USA')
names.append([s1,s2])
bbal_16 = pd.read_csv('data/baseball16.csv')
bbal_16.head()
data_dict = bbal_16.iloc[0].to_dict()
data_dict
import numpy as np
new_data_dict = {k: '' if isinstance(v, str) else np.nan for k,v in data_dict.items()}
new_data_dict
for k,v in data_dict.items():
print(f' k is {k} en v is {v}')
random_data = []
for i in range(1000):
d = dict()
for k, v in data_dict.items():
if isinstance(v, str):
d[k] = np.random.choice(list('abcde'))
else:
d[k] = np.random.randint(10)
random_data.append(pd.Series(d, name = i + len(bbal_16)))
random_data[0].head()
%%timeit
bbal_16_copy = bbal_16.copy()
for row in random_data:
bbal_16_copy = bbal_16_copy.append(row)
%%timeit
bbal_16_copy = bbal_16.copy()
bbal_16_copy = bbal_16_copy.append(random_data)
stocks_2016 = pd.read_csv('data/stocks_2016.csv',
index_col = 'Symbol')
stocks_2016
stocks_2017 = pd.read_csv('data/stocks_2017.csv',
index_col = 'Symbol')
stocks_2017
s_list = [ stocks_2016, stocks_2017]
pd.concat(s_list)
pd.concat(s_list, keys = ['2016', '2017'],
names = ['Year', 'Symbol'])
pd.concat(s_list,
keys = ['2016', '2017'],
axis = 'columns',
sort = True,
names = ['Year', None])
pd.concat(s_list,
join = 'inner',
axis = 'columns',
names = 'Year',
sort = True)
stocks_2016.append(stocks_2017)
trump_url = 'https://en.wikipedia.org/wiki/List_of_generation_VIII_Pok%C3%A9mon'
df_list = pd.read_html(trump_url)
len(df_list)
df_list[3]
df_list = pd.read_html(trump_url,
match = 'Yungoos',
header = 0)
df_list
df_list[0].columns
df_list2 = df_list[0]
df_list2.columns = ['Pokémon species', 'Pokémon']
df_list2.head()
from IPython.display import display_html
years = 2016,2017,2018
stock_tables = [pd.read_csv('data/stocks_{}.csv'.format(year),
index_col = 'Symbol') for year in years]
type(stock_tables)
stock_tables = []
for year in years:
print(year)
stock_tables.append(pd.read_csv('data/stocks_{}.csv'.format(year), index_col = 'Symbol'))
stock_tables[2]
stocks_2018
def display_frames(frames, num_spaces = 0):
t_style = '<table style = "display: inline;"'
tables_html = [df.to_html().replace('<table', t_style) for df in frames]
space = ' ' * num_spaces
# print(tables_html)
display_html(space.join(tables_html), raw = True)
display_frames(stock_tables, 30)
pd.concat(stock_tables, keys = [2016, 2017, 2018])
pd.concat(dict(zip(years, stock_tables)), axis = 'columns')
stocks_2016.join(stocks_2017,
lsuffix = '_2016',
rsuffix = '_2017',
how = 'outer')
other = [stock_tables[1].add_suffix('_2017'),
stock_tables[2].add_suffix('_2018')]
# stocks_2017
stock_tables[2]
stocks_2016.add_suffix('_2016').join(other,
how = 'outer')
stock_join = stocks_2016.add_suffix('_2016').join(other,
how = 'outer')
stock_concat = pd.concat(dict(zip(years, stock_tables)),
axis = 'columns',
sort = True)
stock_concat
level_1 = stock_concat.columns.get_level_values(1)
level_1
level_0 = stock_concat.columns.get_level_values(0).astype(str)
level_0
stock_concat.columns = level_1 + '_' + level_0
stock_concat
stock_join
stock_join.equals(stock_concat)
names = ['prices', 'transactions']
food_tables = [pd.read_csv('data/food_{}.csv'.format(name)) for name in names]
food_tables
food_tables[0]
food_tables[1]
food_prices, food_transactions = food_tables
display_frames(food_tables, 30)
food_transactions.merge(food_prices,
on = ['item', 'store'])
food_prices.query('Date == 2017')
food_transactions.merge(food_prices.query('Date == 2017'),
how = 'left')
food_prices.query('Date == 2017').set_index(['item', 'store'])
food_prices_join = food_prices.query('Date == 2017')\
.set_index(['item', 'store'])
food_prices_join
food_transactions.join(food_prices_join,
on = ['item', 'store'])
import glob
df_list = []
for filename in glob.glob('*/gas prices/*.csv'):
# print(filename)
df_list.append(pd.read_csv(filename,
index_col = 'Week',
parse_dates = ['Week']))
gas = pd.concat(df_list, axis = 'columns')
gas.head()
from sqlalchemy import create_engine
engine = create_engine('sqlite:///data/chinook.db')
tracks = pd.read_sql_table('tracks', engine)
tracks.head()
genres = pd.read_sql_table('genres', engine)
genres.head()
genre_track = genres.merge(tracks[['GenreId', 'Milliseconds']],
on = 'GenreId',
how = 'left')
genre_track.head()
genre_time = genre_track.groupby('Name')['Milliseconds'].mean()
genre_time
pd.to_timedelta(genre_time, unit = 'ms').sort_values().dt.floor('s')
cust = pd.read_sql_table('customers',
engine,
columns = ['CustomerId', 'FirstName', 'LastName'])
cust.head()
invoice = pd.read_sql_table('invoices',
engine,
columns = ['InvoiceId', 'CustomerId'])
invoice.head()
ii = pd.read_sql_table('invoice_items',
engine,
columns = ['InvoiceId', 'UnitPrice', 'Quantity'])
ii.head()
cust_inv = cust.merge(invoice, on='CustomerId')
cust_inv.head(9)
cust_inv = cust.merge(invoice, on='CustomerId')\
.merge(ii, on = 'InvoiceId')
cust_inv.head(8)
cust_inv['Total'] = cust_inv['Quantity'] * cust_inv['UnitPrice']
cols = ['CustomerId', 'FirstName', 'LastName']
# cust_inv.assign(Total = total)
cust_inv.head(8)
cust_inv.groupby(cols)['Total'].sum()\
.sort_values(ascending = False)\
.head()
sql_string1 = '''
select
Name,
time(avg(Milliseconds) / 1000, 'unixepoch') as avg_time
from (
select
g.Name,
t.Milliseconds
from
genres as g
join
tracks as t
on
g.genreid == t.genreid
)
group by
Name
order by
avg_time
'''
pd.read_sql_query(sql_string1, engine)
>>> sql_string2 = '''
select
c.customerid,
c.FirstName,
c.LastName,
sum(ii.quantity * ii.unitprice) as Total
from
customers as c
join
invoices as i
on c.customerid = i.customerid
join
invoice_items as ii
on i.invoiceid = ii.invoiceid
group by
c.customerid, c.FirstName, c.LastName
order by
Total desc
'''
>>> pd.read_sql_query(sql_string2, engine)
import pandas as pd
crime_sort = pd.read_hdf('data/crime.h5', 'crime') \
.set_index('REPORTED_DATE') \
.sort_index()
crime_sort.head()
crime_sort.resample('W')
weekly_crimes = crime_sort.resample('W').size()
weekly_crimes.head()
len(crime_sort.loc[:'2012-1-8'])
len(crime_sort.loc['2012-1-9':'2012-1-15'])
crime_sort.resample('W-THU').size().head()
crime_sort.loc[:'2012-1-5'].size
weekly_crimes_gby = crime_sort.groupby(pd.Grouper(freq = 'W'))\
.size()
weekly_crimes_gby.head()
dir(crime_sort.resample('W'))
crime = pd.read_hdf('data/crime.h5', 'crime')
crime.head()
weekly_crimes2 = crime.resample('W',
on = 'REPORTED_DATE').size()
weekly_crimes2.head()
weekly_crimes2.equals(weekly_crimes)
weekly_crimes_gby2 = crime.groupby(pd.Grouper(key = 'REPORTED_DATE',
freq = 'W')).size()
weekly_crimes_gby2.head()
weekly_crimes_gby2.equals(weekly_crimes_gby)
weekly_crimes.plot(figsize = (16,4),
title = 'All Denver Crimes');
crime_sort = pd.read_hdf('data/crime.h5',
'crime')\
.set_index('REPORTED_DATE')\
.sort_index()
crime_sort.head()
crime_quarterly = crime_sort.resample('Q')['IS_CRIME',
'IS_TRAFFIC'].sum()
crime_quarterly.head()
crime_sort.resample('QS')['IS_CRIME','IS_TRAFFIC'].sum()\
.head()
crime_sort.loc['2012-04-01':'2012-06-30',['IS_CRIME',
'IS_TRAFFIC']]\
.sum()
crime_quarterly2 = crime_sort.groupby(pd.Grouper(freq = 'Q'))\
['IS_CRIME','IS_TRAFFIC'].sum()
crime_quarterly2.head()
crime_quarterly2.equals(crime_quarterly)
plot_kwargs = dict(figsize = (16,4),
color = ['black', 'lightgrey'],
title = 'Denver Crimes and Traffic Accidents')
crime_quarterly.plot(**plot_kwargs);
crime_begin = crime_quarterly.iloc[0]
crime_begin
crime_quarterly.div(crime_begin)\
.sub(1)\
.round(2)\
.plot(**plot_kwargs);
| 0.151467 | 0.230389 |
### About:
4/10/2021 Program calculate the correlation coefficient:
(1) between internet speeds
(2) between states
(3) between states & internet speeds??? -> Self Assigned
#### NOTE: Make sure to unzip the xlsx files
@author: Minh Nguyen @AIA
@credit: Qasim, Andrei @AIA
```
import numpy as np
from numpy.random import randn
from numpy.random import seed
from numpy import array_split
import pandas as pd
from sklearn import metrics as mt
from sklearn import model_selection as md
from matplotlib import pyplot as plt
import seaborn as sns
import sklearn.datasets as ds
import random
```
### Correlation Calculation
```
def correlation_cal(df):
# Instead of just dropping the missing values, we will fill in N/A values
if df.isna().values.any():
while True:
missing_type = input("Please enter the type of missing value replacement: mean, medium, mode, or drop from the row")
missing_type = missing_type.lower()
if(missing_type in ['mean', 'median', 'mode', 'drop']):
if(missing_type == 'mean'):
df.fillna(df.mean(), inplace=True)
elif(missing_type == 'median'):
df.fillna(df.median(), inplace=True)
elif(missing_type == 'mode'):
df.fillna(df.mode(), inplace=True)
else:
df.dropna()
break
else:
print("Please input the option from the list")
# Calculate input data correlation
while True:
corr_type = input("Please enter type of correlation: Pearson, Spearman, or Kendall: ")
corr_type = corr_type.lower()
if corr_type in ['pearson', 'spearman', 'kendall']:
break
else:
print("Please try again")
# Plot correlation matrix
corrMatrix = df.corr(method=corr_type)
_, ax = plt.subplots(figsize=(12, 10))
sns.heatmap(corrMatrix, ax = ax, cmap="gray", linewidths = 0.1) # cmap can also be "YlGnBu"
```
# (1) Correlation Calculation Between Internet Speeds
#### Preprocess Data
```
!ls
def parse_data_bis(file):
""" Function load csv files into csv pandas by internet speed
Function parse data by internet speed for all 50 states + nation (row 0)
"""
dataset = pd.read_excel(file, skiprows=[0,1,2,3], usecols=[2,3,4,5,6,7,8,9,10])
return dataset
df_bis = parse_data_bis("preprocess_data.xlsx")
df_bis.head()
```
### Run 1: Correlation Pearson - Mean
```
# Run Correlation Pearson
correlation_cal(df_bis)
```
### Run 2: Correlation Spearman - Mean
```
# Run Correlation Spearman
correlation_cal(df_bis)
```
### Run 3: Correlation Kendall - Mean
```
# Run Correlation Kendall
correlation_cal(df_bis)
```
### Analysis - Self Assigned
1/ What are the two significant correlation coefficient for Pearson, Spearman, Kendall?
2/ What are other analysis we can do with these graphs?
3/ Any other metric besides Pearson, Spearman, Kendall?
# (2) Correlation Calculation Between States
```
def parse_data_bs(file):
""" Function load csv files into csv pandas by internet speed
Function parse data by states and nation for all 10 types of internet speeds
"""
dataset = pd.read_excel(file, skiprows=[0,1,2,3,4], usecols=[0,2,3,4,5,6,7,8,9,10], drop=True)
dataset.set_index("Nationwide", inplace=True)
dataset = dataset.T
return dataset
df_bs = parse_data_bs("preprocess_data.xlsx")
df_bs.head()
```
### Run 1: Correlation Pearson - Mean
```
# Run Correlation Pearson
correlation_cal(df_bs)
```
### Run 2: Correlation Spearman - Mean
```
# Run Correlation Spearman
correlation_cal(df_bs)
```
### Run 3: Correlation Kendall - Mean
```
# Run Correlation Kendall
correlation_cal(df_bs)
```
### Analysis - Self Assigned
1/ What are the two significant correlation coefficient for Pearson, Spearman, Kendall?
2/ What are other analysis we can do with these graphs?
3/ Any other metric besides Pearson, Spearman, Kendall?
|
github_jupyter
|
import numpy as np
from numpy.random import randn
from numpy.random import seed
from numpy import array_split
import pandas as pd
from sklearn import metrics as mt
from sklearn import model_selection as md
from matplotlib import pyplot as plt
import seaborn as sns
import sklearn.datasets as ds
import random
def correlation_cal(df):
# Instead of just dropping the missing values, we will fill in N/A values
if df.isna().values.any():
while True:
missing_type = input("Please enter the type of missing value replacement: mean, medium, mode, or drop from the row")
missing_type = missing_type.lower()
if(missing_type in ['mean', 'median', 'mode', 'drop']):
if(missing_type == 'mean'):
df.fillna(df.mean(), inplace=True)
elif(missing_type == 'median'):
df.fillna(df.median(), inplace=True)
elif(missing_type == 'mode'):
df.fillna(df.mode(), inplace=True)
else:
df.dropna()
break
else:
print("Please input the option from the list")
# Calculate input data correlation
while True:
corr_type = input("Please enter type of correlation: Pearson, Spearman, or Kendall: ")
corr_type = corr_type.lower()
if corr_type in ['pearson', 'spearman', 'kendall']:
break
else:
print("Please try again")
# Plot correlation matrix
corrMatrix = df.corr(method=corr_type)
_, ax = plt.subplots(figsize=(12, 10))
sns.heatmap(corrMatrix, ax = ax, cmap="gray", linewidths = 0.1) # cmap can also be "YlGnBu"
!ls
def parse_data_bis(file):
""" Function load csv files into csv pandas by internet speed
Function parse data by internet speed for all 50 states + nation (row 0)
"""
dataset = pd.read_excel(file, skiprows=[0,1,2,3], usecols=[2,3,4,5,6,7,8,9,10])
return dataset
df_bis = parse_data_bis("preprocess_data.xlsx")
df_bis.head()
# Run Correlation Pearson
correlation_cal(df_bis)
# Run Correlation Spearman
correlation_cal(df_bis)
# Run Correlation Kendall
correlation_cal(df_bis)
def parse_data_bs(file):
""" Function load csv files into csv pandas by internet speed
Function parse data by states and nation for all 10 types of internet speeds
"""
dataset = pd.read_excel(file, skiprows=[0,1,2,3,4], usecols=[0,2,3,4,5,6,7,8,9,10], drop=True)
dataset.set_index("Nationwide", inplace=True)
dataset = dataset.T
return dataset
df_bs = parse_data_bs("preprocess_data.xlsx")
df_bs.head()
# Run Correlation Pearson
correlation_cal(df_bs)
# Run Correlation Spearman
correlation_cal(df_bs)
# Run Correlation Kendall
correlation_cal(df_bs)
| 0.476336 | 0.908212 |
# Download Publicly Available Neutrophil Dataset
**Gregory Way, 2018**
Here, I download [GSE103706](https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103706) which is associated with [Rincon et al. 2018](https://doi.org/10.1186/s12864-018-4957-6).
This dataset includes two acute myeloid leukemia (AML) cell lines; PLB-985 and HL-60.
There are 14 samples total in this dataset.
The cell lines are exposed to two treatments - DMSO and DMSO+Nutridoma.
The treatments are demonstrated to induce neutrophil differentiation in these cell lines.
We hypothesized that our constructed feature identified through our interpret compression approach would have higher activation patterns in the cell lines with induced neutrophil differentiation.
```
import os
import csv
import pandas as pd
from sklearn import preprocessing
from scripts.utils import download_geo
base_url = 'ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE103nnn/GSE103706/suppl/'
name = 'GSE103706_Merged_data_FPKM_and_normalized.xlsx'
directory = 'download'
download_geo(base_url, name, directory)
path = 'download/GSE103706_Merged_data_FPKM_and_normalized.xlsx'
! sha256sum $path
```
## Process the Data
```
# Load Data
geo_df = pd.read_excel(path, index_col=0, skiprows=1)
geo_df = geo_df[geo_df.symbol != 'N\A']
print(geo_df.shape)
geo_df.head(2)
```
## Update Gene Names
```
# Load curated gene names from versioned resource
commit = '721204091a96e55de6dcad165d6d8265e67e2a48'
url = 'https://raw.githubusercontent.com/cognoma/genes/{}/data/genes.tsv'.format(commit)
gene_df = pd.read_table(url)
# Only consider protein-coding genes
gene_df = (
gene_df.query("gene_type == 'protein-coding'")
)
symbol_to_entrez = dict(zip(gene_df.symbol,
gene_df.entrez_gene_id))
# Add alternative symbols to entrez mapping dictionary
gene_df = gene_df.dropna(axis='rows', subset=['synonyms'])
gene_df.synonyms = gene_df.synonyms.str.split('|')
all_syn = (
gene_df.apply(lambda x: pd.Series(x.synonyms), axis=1)
.stack()
.reset_index(level=1, drop=True)
)
# Name the synonym series and join with rest of genes
all_syn.name = 'all_synonyms'
gene_with_syn_df = gene_df.join(all_syn)
# Remove rows that have redundant symbols in all_synonyms
gene_with_syn_df = (
gene_with_syn_df
# Drop synonyms that are duplicated - can't be sure of mapping
.drop_duplicates(['all_synonyms'], keep=False)
# Drop rows in which the symbol appears in the list of synonyms
.query('symbol not in all_synonyms')
)
# Create a synonym to entrez mapping and add to dictionary
synonym_to_entrez = dict(zip(gene_with_syn_df.all_synonyms,
gene_with_syn_df.entrez_gene_id))
symbol_to_entrez.update(synonym_to_entrez)
# Load gene updater
url = 'https://raw.githubusercontent.com/cognoma/genes/{}/data/updater.tsv'.format(commit)
updater_df = pd.read_table(url)
old_to_new_entrez = dict(zip(updater_df.old_entrez_gene_id,
updater_df.new_entrez_gene_id))
# Update the symbol column to entrez_gene_id
geo_map = geo_df.symbol.replace(symbol_to_entrez)
geo_map = geo_map.replace(old_to_new_entrez)
geo_df.index = geo_map
geo_df.index.name = 'entrez_gene_id'
geo_df = geo_df.drop(['ens_gene_id', 'ncbi_gene_id', 'gene_short', 'symbol'], axis='columns')
geo_df = geo_df.loc[geo_df.index.isin(symbol_to_entrez.values()), :]
geo_df.head()
```
## Scale Data and Output to File
```
# Scale RNAseq data using zero-one normalization
geo_scaled_zeroone_df = preprocessing.MinMaxScaler().fit_transform(geo_df.transpose())
geo_scaled_zeroone_df = (
pd.DataFrame(geo_scaled_zeroone_df,
columns=geo_df.index,
index=geo_df.columns)
.sort_index(axis='columns')
.sort_index(axis='rows')
)
geo_scaled_zeroone_df.columns = geo_scaled_zeroone_df.columns.astype(str)
geo_scaled_zeroone_df = geo_scaled_zeroone_df.loc[:, ~geo_scaled_zeroone_df.columns.duplicated(keep='first')]
os.makedirs('data', exist_ok=True)
file = os.path.join('data', 'GSE103706_processed_matrix.tsv.gz')
geo_scaled_zeroone_df.to_csv(file, sep='\t', compression='gzip')
geo_scaled_zeroone_df.head()
```
|
github_jupyter
|
import os
import csv
import pandas as pd
from sklearn import preprocessing
from scripts.utils import download_geo
base_url = 'ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE103nnn/GSE103706/suppl/'
name = 'GSE103706_Merged_data_FPKM_and_normalized.xlsx'
directory = 'download'
download_geo(base_url, name, directory)
path = 'download/GSE103706_Merged_data_FPKM_and_normalized.xlsx'
! sha256sum $path
# Load Data
geo_df = pd.read_excel(path, index_col=0, skiprows=1)
geo_df = geo_df[geo_df.symbol != 'N\A']
print(geo_df.shape)
geo_df.head(2)
# Load curated gene names from versioned resource
commit = '721204091a96e55de6dcad165d6d8265e67e2a48'
url = 'https://raw.githubusercontent.com/cognoma/genes/{}/data/genes.tsv'.format(commit)
gene_df = pd.read_table(url)
# Only consider protein-coding genes
gene_df = (
gene_df.query("gene_type == 'protein-coding'")
)
symbol_to_entrez = dict(zip(gene_df.symbol,
gene_df.entrez_gene_id))
# Add alternative symbols to entrez mapping dictionary
gene_df = gene_df.dropna(axis='rows', subset=['synonyms'])
gene_df.synonyms = gene_df.synonyms.str.split('|')
all_syn = (
gene_df.apply(lambda x: pd.Series(x.synonyms), axis=1)
.stack()
.reset_index(level=1, drop=True)
)
# Name the synonym series and join with rest of genes
all_syn.name = 'all_synonyms'
gene_with_syn_df = gene_df.join(all_syn)
# Remove rows that have redundant symbols in all_synonyms
gene_with_syn_df = (
gene_with_syn_df
# Drop synonyms that are duplicated - can't be sure of mapping
.drop_duplicates(['all_synonyms'], keep=False)
# Drop rows in which the symbol appears in the list of synonyms
.query('symbol not in all_synonyms')
)
# Create a synonym to entrez mapping and add to dictionary
synonym_to_entrez = dict(zip(gene_with_syn_df.all_synonyms,
gene_with_syn_df.entrez_gene_id))
symbol_to_entrez.update(synonym_to_entrez)
# Load gene updater
url = 'https://raw.githubusercontent.com/cognoma/genes/{}/data/updater.tsv'.format(commit)
updater_df = pd.read_table(url)
old_to_new_entrez = dict(zip(updater_df.old_entrez_gene_id,
updater_df.new_entrez_gene_id))
# Update the symbol column to entrez_gene_id
geo_map = geo_df.symbol.replace(symbol_to_entrez)
geo_map = geo_map.replace(old_to_new_entrez)
geo_df.index = geo_map
geo_df.index.name = 'entrez_gene_id'
geo_df = geo_df.drop(['ens_gene_id', 'ncbi_gene_id', 'gene_short', 'symbol'], axis='columns')
geo_df = geo_df.loc[geo_df.index.isin(symbol_to_entrez.values()), :]
geo_df.head()
# Scale RNAseq data using zero-one normalization
geo_scaled_zeroone_df = preprocessing.MinMaxScaler().fit_transform(geo_df.transpose())
geo_scaled_zeroone_df = (
pd.DataFrame(geo_scaled_zeroone_df,
columns=geo_df.index,
index=geo_df.columns)
.sort_index(axis='columns')
.sort_index(axis='rows')
)
geo_scaled_zeroone_df.columns = geo_scaled_zeroone_df.columns.astype(str)
geo_scaled_zeroone_df = geo_scaled_zeroone_df.loc[:, ~geo_scaled_zeroone_df.columns.duplicated(keep='first')]
os.makedirs('data', exist_ok=True)
file = os.path.join('data', 'GSE103706_processed_matrix.tsv.gz')
geo_scaled_zeroone_df.to_csv(file, sep='\t', compression='gzip')
geo_scaled_zeroone_df.head()
| 0.550849 | 0.920469 |
#DEAP - Enabling Nimbler Evolutions
by François-Michel De Rainville, Félix-Antoine Fortin, Marc-André Gardner, Marc Parizeau, and Christian Gagné
<img src="files/images/deap_highlights.png" width="400px" style="float:right;margin-left:50px;"/>
DEAP is a Distributed Evolutionary Algorithm (EA) framework written in Python and designed to help researchers developing custom evolutionary algorithms. Its design philosophy promotes explicit algorithms and transparent data structures, in contrast with most other evolutionary computation softwares that tend to encapsulate standardized algorithms using the black-box approach. This philosophy sets it apart as a rapid prototyping framework for testing of new ideas in EA research.
##Introduction
The [DEAP framework](#deap-jmlr) is designed over the three following founding principles:
1. Data structures are key to evolutionary computation. They must facilitate the implementation of algorithms and be easy to customize.
2. Operator selection and algorithm parameters have strong influences on evolutions, while often being problem dependent. Users should be able to parametrize every aspect of the algorithms with minimal complexity.
3. EAs are usually embarrassingly parallel. Therefore, mechanisms that implement distribution paradigms should be trivial to use.
With the help of its sister project [SCOOP](#scoop) and the power of the Python programming language, DEAP implements these three principles in a simple and elegant design.
###Data Structures
A very important part of the success for designing any algorithm - if not the most important - is choosing the appropriate data structures. Freedom in type creation is fundamental in the process of designing evolutionary algorithms that solve real world problems. DEAP's *creator* module allows users to:
- create classes with a single line of code (inheritance);
- add attributes (composition);
- group classes in a single module (sandboxing).
In the following listing, we create a minimizing fitness.
```
from deap import base, creator
creator.create("FitnessMin", base.Fitness, weights=(-1.0,))
```
The `create` function expects at least two arguments; the name of the class to be created and the base class it inherits from. The next arguments are used as class attributes.
Thus, the class just created is a `FitnessMin` inheriting from the base class `Fitness` and having a `weights` attribute set to the one element tuple `(-1.0,)`, indicating minimization of a single objective. A multi-objective fitness would be created using a multi-element tuple.
Next, we define with the same mechanism an `Individual` class inheriting from a `list` and composed with a `fitness` attribute.
```
creator.create("Individual", list, fitness=creator.FitnessMin)
```
When an `Individual` is instantiated, its `fitness` is initialized with an instance of the previously defined `FitnessMin` class. This is illustrated in the following example,
```
ind = creator.Individual([1,0,1,0,1])
ind.fitness.values = (sum(ind),)
```
where an individual is created from a list of binary values and the value of its fitness is set to the sum of its elements.
In DEAP, the fitness value is always multi-objective with the single objective case being a tuple of one element. This tuple is instantiated by the comma following the sum operation.
###Operators
Operator selection is another crucial part of evolutionary algorithms. It must be straightforward and its parametrization intuitive. DEAP's *Toolbox* enables users to:
- create aliases for operators;
- register operators' parameters;
- interchange operators efficiently;
- regroup all operators in a single structure.
The next example presents the construction of a toolbox and how operators and their parameters are registered.
```
from deap import tools
toolbox = base.Toolbox()
toolbox.register("mate", tools.cxOnePoint)
toolbox.register("mutate", tools.mutGaussian, mu=0.0, std=1.0)
```
The `register` function expects at least two arguments; the alias of the function and the function itself. The next arguments are passed to the function when called, similarly to the `partial` function from the standard *functools* module.
Thus, the first operator is a one point crossover registered under the alias `mate`. The second operator, a gaussian mutation, is registered with its parameters under the generic name `mutate`. Both operators are available from the *tools* module along with many more instruments to support evolution presented at the end of this paper.
During subsequent experiments, replacing the one point crossover by a two point crossover is as easy as substituting the third line of the previous listing by the following one.
```
toolbox.register("mate", tools.cxTwoPoint)
```
Wherever the generic function `mate` is used, the new two point crossover will be used.
###Parallelization
DEAP is parallel ready. The idea is to use a mapping operation that applies a function to every item of a sequence, for instance to evaluate the individuals fitness. By default, every toolbox is registered with the standard `map` function of Python. For algorithms to evaluate individuals in parallel, one only needs to replace this alias by a parallel map such as the one provided by [SCOOP](#scoop), a library capable of distributing tasks over a cluster of computers.
```
from scoop import futures
toolbox.register("map", futures.map)
```
DEAP is also compatible with the *multiprocessing* standard module, if the user only cares to run on a single multicore machine.
```
import multiprocessing
pool = multiprocessing.Pool()
toolbox.register("map", pool.map)
```
With these powerful tools, DEAP allows scientists and researchers with little programming knowledge to easily implement distributed and parallel EAs.
##Preaching by Example
The best introduction to evolutionary computation with DEAP is to present simple, yet compelling examples. The following sections set forth how algorithms are easy to implement while keeping a strong grip on how they behave. The first section introduces a classical genetic algorithm and exposes different levels of explicitness. The second section presents how genetic programming is implemented in DEAP and the versatility of the GP module. The final example demonstrate how easy it is to implement a generic distributed island model with SCOOP.
###A Simple Genetic Algorithm
A commonly used example in evolutionary computation is the OneMax problem which consists in maximizing the number of ones in a binary solution. The more ones an individual contains, the higher its fitness value is. Using a genetic algorithm to find such an individual is relatively straightforward. Applying crossovers and mutations on a population of randomly generated binary individuals and selecting the fittest ones at each generation usually lead quickly to a perfect (all ones) solution. A problem of this simplicity should be solved with a very simple program.
```
import random
from deap import algorithms, base, creator, tools
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
creator.create("Individual", list, fitness=creator.FitnessMax)
def evalOneMax(individual):
return (sum(individual),)
toolbox = base.Toolbox()
toolbox.register("attr_bool", random.randint, 0, 1)
toolbox.register("individual", tools.initRepeat, creator.Individual, toolbox.attr_bool, n=100)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("evaluate", evalOneMax)
toolbox.register("mate", tools.cxTwoPoint)
toolbox.register("mutate", tools.mutFlipBit, indpb=0.05)
toolbox.register("select", tools.selTournament, tournsize=3)
if __name__ == "__main__":
pop = toolbox.population(n=300)
algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2, ngen=40, verbose=False)
print(tools.selBest(pop, k=1))
```
The preceeding code presents all that is needed to solve the OneMax problem with DEAP. The first two lines import the necessary modules. Next, on lines 3 and 4, two types are created; a maximizing fitness (note the positive weights), and a list individual composed with an instance of this maximizing fitness. Then, on lines 5 and 6, the evaluation function is defined. It counts the number of ones in a binary list by summing its elements (note again the one element returned tuple corresponding to a single objective fitness). Subsequently, a `Toolbox` is instantiated in which the necessary operators are registered. The first operator, on line 8, produces binary values, in this case integers in [0, 1]$, using the standard *random* module. The alias `individual`, on line 9, is assigned to the helper function `initRepeat`, which takes a container as the first argument, a function that generates content as the second argument and the number of repetitions as the last argument.
Thus, calling the individual function instantiates an `Individual` of `n=100` bits by calling repeatedly the registered `attr_bool` function. The same repetition initializer is used on the next line to produce a population as a list of individuals. The missing number of repetition `n` will be given later in the program. Subsequently, on lines 11 to 14, the evaluation, crossover, mutation and selection operators are registered with all of their parameters.
The main program starts at line 16. First, a population of `n=300` individuals is instantiated. Then, the algorithm, provided with the population and the toolbox, is run for `ngen=40` generations with `cxpb=0.5` probability of mating and `mutpb=0.2` probability of mutating an individual for each generation. Finally, on the last line, the best individual of the resulting population is selected and displayed on screen.
###Controlling Everything
When developing, researching or using EAs, pre-implemented canned algorithms seldom do everything that is needed. Usually, developers/researchers/users have to dig into the framework to tune, add or replace a part of the original algorithm. DEAP breaks with the traditional black-box approach on that precise point; it encourages users to rapidly build their own algorithms. With the different tools provided by DEAP, it is possible to design a nimble algorithm that tackles most problems at hand.
```
import random
from deap import algorithms, base, creator, tools
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
creator.create("Individual", list, fitness=creator.FitnessMax)
def evalOneMax(individual):
return (sum(individual),)
toolbox = base.Toolbox()
toolbox.register("attr_bool", random.randint, 0, 1)
toolbox.register("individual", tools.initRepeat, creator.Individual, toolbox.attr_bool, n=100)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("evaluate", evalOneMax)
toolbox.register("mate", tools.cxTwoPoint)
toolbox.register("mutate", tools.mutFlipBit, indpb=0.05)
toolbox.register("select", tools.selTournament, tournsize=3)
if __name__ == "__main__":
pop = toolbox.population(n=300)
ngen, cxpb, mutpb = 40, 0.5, 0.2
fitnesses = toolbox.map(toolbox.evaluate, pop)
for ind, fit in zip(pop, fitnesses):
ind.fitness.values = fit
for g in range(ngen):
pop = toolbox.select(pop, k=len(pop))
pop = algorithms.varAnd(pop, toolbox, cxpb, mutpb)
invalids = [ind for ind in pop if not ind.fitness.valid]
fitnesses = toolbox.map(toolbox.evaluate, invalids)
for ind, fit in zip(invalids, fitnesses):
ind.fitness.values = fit
print(tools.selBest(pop, k=1))
```
Starting from the previous OneMax solution, a first decomposition of the algorithm replaces the canned `eaSimple` function by a generational loop. Again, this example is exhaustive but still very simple. On the first 3 lines, the evaluation is applied to every individual in the population by the `map` function contained in every toolbox. Next, a loop over both the population and the evaluated fitnesses sets each individual's fitness value. Thereafter, the generational loop begins. It starts by selecting `k` individuals from the population. Then, the selected individuals are varied by crossover **and** mutation by the `varAnd` function. A second variation scheme `varOr` can also be used, where the individuals are produced by crossover **or** mutation. Once modified, the individuals are evaluated for the next iteration. Only freshly produced individuals have to be evaluated; they are filtered by their fitness validity. This version of the program provides the possibility to change the stopping criterion and add components to the evolution.
```
import random, math
from deap import algorithms, base, creator, tools
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
creator.create("Individual", list, fitness=creator.FitnessMax)
def evalOneMax(individual):
return (sum(individual),)
toolbox = base.Toolbox()
toolbox.register("attr_bool", random.randint, 0, 1)
toolbox.register("individual", tools.initRepeat, creator.Individual, toolbox.attr_bool, n=100)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("evaluate", evalOneMax)
toolbox.register("mate", tools.cxTwoPoint)
toolbox.register("mutate", tools.mutFlipBit, indpb=0.05)
toolbox.register("select", tools.selTournament, tournsize=3)
if __name__ == "__main__":
pop = toolbox.population(n=300)
ngen, cxpb, mutpb = 40, 0.5, 0.2
fitnesses = toolbox.map(toolbox.evaluate, pop)
for ind, fit in zip(pop, fitnesses):
ind.fitness.values = fit
for g in range(ngen):
pop = toolbox.select(pop, k=len(pop))
pop = [toolbox.clone(ind) for ind in pop]
for child1, child2 in zip(pop[::2], pop[1::2]):
if random.random() < cxpb:
toolbox.mate(child1, child2)
del child1.fitness.values, child2.fitness.values
for mutant in pop:
if random.random() < mutpb:
toolbox.mutate(mutant)
del mutant.fitness.values
invalids = [ind for ind in pop if not ind.fitness.valid]
fitnesses = toolbox.map(toolbox.evaluate, invalids)
for ind, fit in zip(invalids, fitnesses):
ind.fitness.values = fit
print(tools.selBest(pop, k=1))
```
An even greater level of detail can be obtained by substituting the `varAnd` function by its full content, presented in last code example. This listing starts with the duplication of the population by the `clone` tool available in every toolbox. Then, the crossover is applied to a portion of consecutive individuals . Each modified individual sees its fitness invalidated by the deletion of its value. Finally, a percentage of the population is mutated and their fitness values is also deleted. This variant of the algorithm provides control over the application order and the number of operators, among many other aspects.
The explicitness in which algorithm are written with DEAP clarifies the experiments. This eliminates any ambiguity on the different aspects of the algorithm that could, when overlooked, jeopardize the reproducibility and interpretation of results.
###Genetic Programming
DEAP also includes every component necessary to design genetic programming algorithms with the same ease as for genetic algorithms. For example, the most commonly used tree individual can be created as following.
```
import operator, random
from deap import algorithms, base, creator, tools, gp
creator.create("FitnessMin", base.Fitness, weights=(-1.0,))
creator.create("Individual", gp.PrimitiveTree, fitness=creator.FitnessMin)
```
The primitive tree is provided in the *gp* module since it is one of the few data types the Python standard library does not provide. The primitives and terminals that will populate the trees are regrouped in a primitive set. The following listing presents a primitive set instantiation with basic operators provided by the standard library *operator* module. The `arity` of a primitive is its number of operands.
```
pset = gp.PrimitiveSet(name="MAIN", arity=1)
pset.addPrimitive(operator.add, arity=2)
pset.addPrimitive(operator.sub, arity=2)
pset.addPrimitive(operator.mul, arity=2)
pset.addPrimitive(operator.neg, arity=1)
pset.renameArguments(ARG0="x")
```
Functions that initialize individuals and populations are registered in a toolbox just as in the preceding genetic algorithm example. DEAP implements the three initialization methods proposed by [Koza](#koza) to generate trees: full, grow, and half-and-half.
```
toolbox = base.Toolbox()
toolbox.register("expr", gp.genFull, pset=pset, min_=1, max_=3)
toolbox.register("individual", tools.initIterate, creator.Individual, toolbox.expr)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
```
We may now introduce an example of a symbolic regression evaluation function. First, the `gp.compile` function transforms the primitive tree into its executable form, a Python function, using a primitive set `pset` given as the evaluation function's third argument. Then, the rest is simple maths: we compute the root mean squared error between the individual's program and the target x^4+x^3+x^2+x on a set of points, the evaluation function's second arguent.
```
def evaluateRegression(individual, points, pset):
func = gp.compile(expr=individual, pset=pset)
sqerrors = ((func(x) - x**4 - x**3 - x**2 - x)**2 for x in points)
return math.sqrt(sum(sqerrors) / len(points)),
```
Next, the evaluation function and the variation operators are registered similarly to the onemax example, while the other operators remain the same.
```
toolbox.register("evaluate", evaluateRegression, points=[x/10. for x in range(-10, 10)],
pset=pset)
toolbox.register("mate", gp.cxOnePoint)
toolbox.register("expr_mut", gp.genFull, min_=0, max_=2)
toolbox.register("mutate", gp.mutUniform, expr=toolbox.expr_mut, pset=pset)
toolbox.register("select", tools.selTournament, tournsize=3)
if __name__ == "__main__":
pop = toolbox.population(n=300)
algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2, ngen=40, verbose=False)
bests = tools.selBest(pop, k=1)
print(bests[0])
print(bests[0].fitness)
```
Furthermore, using external libraries such as [NetworkX](#networkx) and [PyGraphviz](#pygraphviz), the best primitive trees can be visualized as follow.
```
import matplotlib.pyplot as plt
import networkx
nodes, edges, labels = gp.graph(bests[0])
graph = networkx.Graph()
graph.add_nodes_from(nodes)
graph.add_edges_from(edges)
pos = networkx.graphviz_layout(graph, prog="dot")
plt.figure(figsize=(7,7))
networkx.draw_networkx_nodes(graph, pos, node_size=900, node_color="w")
networkx.draw_networkx_edges(graph, pos)
networkx.draw_networkx_labels(graph, pos, labels)
plt.axis("off")
plt.show()
```
The primitives are not limited to standard library operators, any function or instance method can be added to a primitive set. Terminals can be any type of objects and even functions without argument. The next example takes advantage of this flexibility and reduces the runtime of the previous example by vectorizing the evaluation using [Numpy](#numpy), a library of high-level mathematical functions operating on multi-dimensional arrays.
```
import numpy, random
from deap import algorithms, base, creator, tools, gp
creator.create("FitnessMin", base.Fitness, weights=(-1.0,))
creator.create("Tree", gp.PrimitiveTree, fitness=creator.FitnessMin)
pset = gp.PrimitiveSet(name="MAIN", arity=1)
pset.addPrimitive(numpy.add, arity=2)
pset.addPrimitive(numpy.subtract, arity=2)
pset.addPrimitive(numpy.multiply, arity=2)
pset.addPrimitive(numpy.negative, arity=1)
def evaluateRegression(individual, points, pset):
func = gp.compile(expr=individual, pset=pset)
sqerrors = (func(points)-(points**4 + points**3 + points**2 + points))**2
return (numpy.sqrt(numpy.sum(sqerrors) / len(points)),)
toolbox = base.Toolbox()
toolbox.register("expr", gp.genFull, pset=pset, min_=1, max_=3)
toolbox.register("individual", tools.initIterate, creator.Tree, toolbox.expr)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("evaluate", evaluateRegression, points=numpy.linspace(-1, 1, 1000), pset=pset)
toolbox.register("mate", gp.cxOnePoint)
toolbox.register("expr_mut", gp.genFull, min_=0, max_=2)
toolbox.register("mutate", gp.mutUniform, expr=toolbox.expr_mut, pset=pset)
toolbox.register("select", tools.selTournament, tournsize=3)
if __name__ == "__main__":
pop = toolbox.population(n=300)
algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2, ngen=40, verbose=False)
print(tools.selBest(pop, k=1)[0])
```
The idea is to evolve a program whose argument is a vector instead of a scalar. Most of the code remains identical; only minor modifications are required. First, we replace the operators in the primitive set by Numpy operators that work on vectors. Then, we remove the loop from the evaluation function, since it is implicit in the operators. Finally, we replace the `sum` and `sqrt` functions by their faster Numpy equivalent and our regression problem is now vectorized. The execution is thereby significantly improved as the scalar example runs in around 3 seconds to optimize the regression on 20 points, while the vectorial runtime is identical but for a regression on 1000 points. By modifying only 6 lines of code, not only are we able to vectorize our problem, but the runtime is reduced by a **factor 50**!
In addition to the wide support of function and object types, DEAP's *gp* module also supports automatically defined functions (ADF), strongly typed genetic programming (STGP), and object-oriented genetic programming (OOGP), for which examples are provided in the library documentation.
##Distributed Island Model
*This section is skipped because SCOOP cannot run inside a notebook*
##Evolution Support
DEAP comes with several supporting tools that can be easily integrated into any algorithm. This section presents some of them in the context of the OneMax example.
The first tool, *Statistics*, computes statistics on arbitrary attributes of designated objects, usually the fitness individuals. The attribute is specified by a key function at the statistics object instantiation before starting the algorithm.
```
stats = tools.Statistics(key=operator.attrgetter("fitness.values"))
```
This is followed by the registration of the statistical functions as for a toolbox.
```
stats.register("max", numpy.max)
stats.register("mean", numpy.mean)
stats.register("min", numpy.min)
```
Ultimately, at every generation, a statistical record of the population is compiled using the registered functions.
```
record = stats.compile(pop)
print(record)
```
The statistics compilation produced a dictionary containing the statistical keywords and their respective value. These last lines, added after the evaluation part, will produce a screen log of the evolution statistics.
For posterity and better readability, statistics can also be logged in a \textit{Logbook}, which is simply a list of recorded dictionaries that can be printed with an elegant layout. For example, the following lines create a new logbook, then record the previously computed statistics and print it to screen.
```
logbook = tools.Logbook()
logbook.record(gen=0, nevals=300, fitness=record)
print(logbook)
```
The next tool, named *Hall of Fame*, preserves the best individuals that appeared during an evolution. At every generation, it scans the population and saves the individuals in a separate archive that does not interact with the population. If the best solution disappears during the evolution, it will still be available in the hall of fame. The hall of fame can be provided as an argument to the algorithms as follows:
```
halloffame = tools.HallOfFame(maxsize=10)
pop = toolbox.population(n=300)
pop, logbook = algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2,
ngen=40, halloffame=halloffame, verbose=False)
```
Moreover, the hall of fame can be updated manually right after the population is evaluated with the following line of code.
```
halloffame.update(pop)
```
The hall of fame proposes a list interface where the individuals are sorted in descending order of fitness. Thus, the fittest solution can be retrieved by accessing the list's first element.
```
best = halloffame[0]
```
A Pareto dominance version of the hall of fame is also available. The *Pareto Front* maintains an archive of non-dominated individuals along the evolution. Its interface is the same than the standard hall of fame.
Another tool, called the *History*, tracks the genealogy of the individuals in a population. By wrapping the variation operators, the history saves the parents of each individual. This feature is added to the variation operators of the toolbox with the following lines.
```
history = tools.History()
toolbox.decorate("mate", history.decorator)
toolbox.decorate("mutate", history.decorator)
```
It is therefore possible to determine the genesis of individuals. The next code presents the genealogy of the best individual in the OneMax example for the last 5 generations of the evolution. The graph is produced by the NetworkX library and the following listing.
```
halloffame = tools.HallOfFame(maxsize=1)
pop = toolbox.population(n=300)
pop, logbook = algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2,
ngen=40, halloffame=halloffame, verbose=False)
h = history.getGenealogy(halloffame[0], max_depth=5)
graph = networkx.DiGraph(h)
graph = graph.reverse() # Make the grah top-down
colors = [toolbox.evaluate(history.genealogy_history[i])[0] for i in graph]
pos = networkx.graphviz_layout(graph, prog="dot")
networkx.draw(graph, pos, node_color=colors)
cb = plt.colorbar()
cb.set_label("Error")
plt.show()
```
The last presented tool is a checkpointing facility. Rather than a DEAP object, checkpointing is ensured by the powerful pickle module available in the standard library. Pickle is a module that can serialize almost any Python object. Checkpointing only requires selecting objects that shall be preserved and the write frequency. This is exactly what is done in the following lines that can be added at the end of the generational loop
```
import pickle
freq = 5
g = 0
if g % freq == 0:
cp = dict(population=pop, generation=g, rndstate=random.getstate())
pickle.dump(cp, open("checkpoint.pkl", "w"), 2)
```
These last lines write into a file the population, the generation number, and the random number generator state so that this information can be used latter to restart an evolution from this exact point in time.
Reloading the data is as simple as reading the pickled dictionary and accessing its attributes.
```
cp = pickle.load(open("checkpoint.pkl", "r"))
pop = cp["population"]
g = cp["generation"]
random.setstate(cp["rndstate"])
```
This simple mechanism provides fault tolerance to any sort of evolutionary algorithms implemented with DEAP. This happens to be critical when exploiting large computational resources where chances of failure grow quickly with the number of computing nodes. Even in very stable execution environments, checkpoints can significantly reduce the amount of time spent on experimentation by allowing evolutions to restart and continue beyond the original stopping criteria.
##Conclusion
DEAP proposes an agile framework to easily prototype and execute explicit evolutionary algorithms. Its creator module is instrumental for building custom transparent data structures for the problem at hand. Its toolbox gathers all necessary operators and their arguments in a single handy structure. Its design provides straightforward distributed execution with multiple distribution libraries. The presented examples only cover a small part of DEAP's capabilities that include evolution strategies (including CMA-ES), multi-objective optimization (NSGA-II and SPEA-II), co-evolution, particle swarm optimization, as well as many benchmarks (continuous, binary, regression, and moving peaks), and examples (more than 40).
After more than 4 years of development, DEAP version 1.0 has been released in February 2014.
DEAP is an open source software, licensed under LGPL, developed primarily at the Computer Vision and Systems Laboratory of Université Laval, Québec, Canada. DEAP is compatible with Python 2 and 3. It has a single dependency on Numpy for computing statistics and running CMA-ES. Try it out and become nimbler too: <http://deap.gel.ulaval.ca>.
##References
- <a id="deap-jmlr">[1]</a> F.-A. Fortin, F.-M. De Rainville, M.-A. Gardner, M. Parizeau, and C. Gagné.
DEAP: Evolutionary Algorithms Made Easy.
*Journal of Machine Learning Research*,
13:2171--2175, 2012.
- <a id="deap-gecco">[2]</a> F.-M. De Rainville, F.-A. Fortin, M.-A. Gardner, M. Parizeau, and C. Gagné.
DEAP: A Python Framework for Evolutionary Algorithms.
*In Companion Proceedings of the Genetic and Evolutionary Computation Conference*,
pages 85--92, 2012.
- <a id="scoop">[3]</a> Y. Hold-Geoffroy, O. Gagnon, and M. Parizeau.
SCOOP: Scalable COncurrent Operations in Python.
<http://www.pyscoop.org/>
- <a id="koza">[4]</a> J. R. Koza.
Genetic Programming - On the Programming of Computers by Means of Natural Selection.
MIT Press, 1992.
- <a id="networkx">[5]</a> A. A. Hagberg, D. A. Schult, and P. J. Swart.
Exploring network structure, dynamics, and function using NetworkX.
*In Proceedings of the Python in Science Conference*,
pages 11-15, 2008.
<http://networkx.github.io>
- <a id="pygraphviz">[6]</a> A. A. Hagberg, D. A. Schult, and M. Renieris.
PyGraphviz a Python interface to the Graphviz graph layout and visualization package.
<http://networkx.lanl.gov/pygraphviz>
- <a id="numpy">[7]</a> E. Jones and T. Oliphant and P. Peterson and others.
SciPy: Open source scientific tools for Python.
<http://www.scipy.org>
|
github_jupyter
|
from deap import base, creator
creator.create("FitnessMin", base.Fitness, weights=(-1.0,))
creator.create("Individual", list, fitness=creator.FitnessMin)
ind = creator.Individual([1,0,1,0,1])
ind.fitness.values = (sum(ind),)
from deap import tools
toolbox = base.Toolbox()
toolbox.register("mate", tools.cxOnePoint)
toolbox.register("mutate", tools.mutGaussian, mu=0.0, std=1.0)
toolbox.register("mate", tools.cxTwoPoint)
from scoop import futures
toolbox.register("map", futures.map)
import multiprocessing
pool = multiprocessing.Pool()
toolbox.register("map", pool.map)
import random
from deap import algorithms, base, creator, tools
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
creator.create("Individual", list, fitness=creator.FitnessMax)
def evalOneMax(individual):
return (sum(individual),)
toolbox = base.Toolbox()
toolbox.register("attr_bool", random.randint, 0, 1)
toolbox.register("individual", tools.initRepeat, creator.Individual, toolbox.attr_bool, n=100)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("evaluate", evalOneMax)
toolbox.register("mate", tools.cxTwoPoint)
toolbox.register("mutate", tools.mutFlipBit, indpb=0.05)
toolbox.register("select", tools.selTournament, tournsize=3)
if __name__ == "__main__":
pop = toolbox.population(n=300)
algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2, ngen=40, verbose=False)
print(tools.selBest(pop, k=1))
import random
from deap import algorithms, base, creator, tools
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
creator.create("Individual", list, fitness=creator.FitnessMax)
def evalOneMax(individual):
return (sum(individual),)
toolbox = base.Toolbox()
toolbox.register("attr_bool", random.randint, 0, 1)
toolbox.register("individual", tools.initRepeat, creator.Individual, toolbox.attr_bool, n=100)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("evaluate", evalOneMax)
toolbox.register("mate", tools.cxTwoPoint)
toolbox.register("mutate", tools.mutFlipBit, indpb=0.05)
toolbox.register("select", tools.selTournament, tournsize=3)
if __name__ == "__main__":
pop = toolbox.population(n=300)
ngen, cxpb, mutpb = 40, 0.5, 0.2
fitnesses = toolbox.map(toolbox.evaluate, pop)
for ind, fit in zip(pop, fitnesses):
ind.fitness.values = fit
for g in range(ngen):
pop = toolbox.select(pop, k=len(pop))
pop = algorithms.varAnd(pop, toolbox, cxpb, mutpb)
invalids = [ind for ind in pop if not ind.fitness.valid]
fitnesses = toolbox.map(toolbox.evaluate, invalids)
for ind, fit in zip(invalids, fitnesses):
ind.fitness.values = fit
print(tools.selBest(pop, k=1))
import random, math
from deap import algorithms, base, creator, tools
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
creator.create("Individual", list, fitness=creator.FitnessMax)
def evalOneMax(individual):
return (sum(individual),)
toolbox = base.Toolbox()
toolbox.register("attr_bool", random.randint, 0, 1)
toolbox.register("individual", tools.initRepeat, creator.Individual, toolbox.attr_bool, n=100)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("evaluate", evalOneMax)
toolbox.register("mate", tools.cxTwoPoint)
toolbox.register("mutate", tools.mutFlipBit, indpb=0.05)
toolbox.register("select", tools.selTournament, tournsize=3)
if __name__ == "__main__":
pop = toolbox.population(n=300)
ngen, cxpb, mutpb = 40, 0.5, 0.2
fitnesses = toolbox.map(toolbox.evaluate, pop)
for ind, fit in zip(pop, fitnesses):
ind.fitness.values = fit
for g in range(ngen):
pop = toolbox.select(pop, k=len(pop))
pop = [toolbox.clone(ind) for ind in pop]
for child1, child2 in zip(pop[::2], pop[1::2]):
if random.random() < cxpb:
toolbox.mate(child1, child2)
del child1.fitness.values, child2.fitness.values
for mutant in pop:
if random.random() < mutpb:
toolbox.mutate(mutant)
del mutant.fitness.values
invalids = [ind for ind in pop if not ind.fitness.valid]
fitnesses = toolbox.map(toolbox.evaluate, invalids)
for ind, fit in zip(invalids, fitnesses):
ind.fitness.values = fit
print(tools.selBest(pop, k=1))
import operator, random
from deap import algorithms, base, creator, tools, gp
creator.create("FitnessMin", base.Fitness, weights=(-1.0,))
creator.create("Individual", gp.PrimitiveTree, fitness=creator.FitnessMin)
pset = gp.PrimitiveSet(name="MAIN", arity=1)
pset.addPrimitive(operator.add, arity=2)
pset.addPrimitive(operator.sub, arity=2)
pset.addPrimitive(operator.mul, arity=2)
pset.addPrimitive(operator.neg, arity=1)
pset.renameArguments(ARG0="x")
toolbox = base.Toolbox()
toolbox.register("expr", gp.genFull, pset=pset, min_=1, max_=3)
toolbox.register("individual", tools.initIterate, creator.Individual, toolbox.expr)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
def evaluateRegression(individual, points, pset):
func = gp.compile(expr=individual, pset=pset)
sqerrors = ((func(x) - x**4 - x**3 - x**2 - x)**2 for x in points)
return math.sqrt(sum(sqerrors) / len(points)),
toolbox.register("evaluate", evaluateRegression, points=[x/10. for x in range(-10, 10)],
pset=pset)
toolbox.register("mate", gp.cxOnePoint)
toolbox.register("expr_mut", gp.genFull, min_=0, max_=2)
toolbox.register("mutate", gp.mutUniform, expr=toolbox.expr_mut, pset=pset)
toolbox.register("select", tools.selTournament, tournsize=3)
if __name__ == "__main__":
pop = toolbox.population(n=300)
algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2, ngen=40, verbose=False)
bests = tools.selBest(pop, k=1)
print(bests[0])
print(bests[0].fitness)
import matplotlib.pyplot as plt
import networkx
nodes, edges, labels = gp.graph(bests[0])
graph = networkx.Graph()
graph.add_nodes_from(nodes)
graph.add_edges_from(edges)
pos = networkx.graphviz_layout(graph, prog="dot")
plt.figure(figsize=(7,7))
networkx.draw_networkx_nodes(graph, pos, node_size=900, node_color="w")
networkx.draw_networkx_edges(graph, pos)
networkx.draw_networkx_labels(graph, pos, labels)
plt.axis("off")
plt.show()
import numpy, random
from deap import algorithms, base, creator, tools, gp
creator.create("FitnessMin", base.Fitness, weights=(-1.0,))
creator.create("Tree", gp.PrimitiveTree, fitness=creator.FitnessMin)
pset = gp.PrimitiveSet(name="MAIN", arity=1)
pset.addPrimitive(numpy.add, arity=2)
pset.addPrimitive(numpy.subtract, arity=2)
pset.addPrimitive(numpy.multiply, arity=2)
pset.addPrimitive(numpy.negative, arity=1)
def evaluateRegression(individual, points, pset):
func = gp.compile(expr=individual, pset=pset)
sqerrors = (func(points)-(points**4 + points**3 + points**2 + points))**2
return (numpy.sqrt(numpy.sum(sqerrors) / len(points)),)
toolbox = base.Toolbox()
toolbox.register("expr", gp.genFull, pset=pset, min_=1, max_=3)
toolbox.register("individual", tools.initIterate, creator.Tree, toolbox.expr)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("evaluate", evaluateRegression, points=numpy.linspace(-1, 1, 1000), pset=pset)
toolbox.register("mate", gp.cxOnePoint)
toolbox.register("expr_mut", gp.genFull, min_=0, max_=2)
toolbox.register("mutate", gp.mutUniform, expr=toolbox.expr_mut, pset=pset)
toolbox.register("select", tools.selTournament, tournsize=3)
if __name__ == "__main__":
pop = toolbox.population(n=300)
algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2, ngen=40, verbose=False)
print(tools.selBest(pop, k=1)[0])
stats = tools.Statistics(key=operator.attrgetter("fitness.values"))
stats.register("max", numpy.max)
stats.register("mean", numpy.mean)
stats.register("min", numpy.min)
record = stats.compile(pop)
print(record)
logbook = tools.Logbook()
logbook.record(gen=0, nevals=300, fitness=record)
print(logbook)
halloffame = tools.HallOfFame(maxsize=10)
pop = toolbox.population(n=300)
pop, logbook = algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2,
ngen=40, halloffame=halloffame, verbose=False)
halloffame.update(pop)
best = halloffame[0]
history = tools.History()
toolbox.decorate("mate", history.decorator)
toolbox.decorate("mutate", history.decorator)
halloffame = tools.HallOfFame(maxsize=1)
pop = toolbox.population(n=300)
pop, logbook = algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2,
ngen=40, halloffame=halloffame, verbose=False)
h = history.getGenealogy(halloffame[0], max_depth=5)
graph = networkx.DiGraph(h)
graph = graph.reverse() # Make the grah top-down
colors = [toolbox.evaluate(history.genealogy_history[i])[0] for i in graph]
pos = networkx.graphviz_layout(graph, prog="dot")
networkx.draw(graph, pos, node_color=colors)
cb = plt.colorbar()
cb.set_label("Error")
plt.show()
import pickle
freq = 5
g = 0
if g % freq == 0:
cp = dict(population=pop, generation=g, rndstate=random.getstate())
pickle.dump(cp, open("checkpoint.pkl", "w"), 2)
cp = pickle.load(open("checkpoint.pkl", "r"))
pop = cp["population"]
g = cp["generation"]
random.setstate(cp["rndstate"])
| 0.556882 | 0.984855 |
# Natural language inference: task and datasets
```
__author__ = "Christopher Potts"
__version__ = "CS224u, Stanford, Spring 2022"
```
## Contents
1. [Overview](#Overview)
1. [Our version of the task](#Our-version-of-the-task)
1. [Primary resources](#Primary-resources)
1. [Set-up](#Set-up)
1. [SNLI](#SNLI)
1. [SNLI properties](#SNLI-properties)
1. [Working with SNLI](#Working-with-SNLI)
1. [MultiNLI](#MultiNLI)
1. [MultiNLI properties](#MultiNLI-properties)
1. [Working with MultiNLI](#Working-with-MultiNLI)
1. [Annotated MultiNLI subsets](#Annotated-MultiNLI-subsets)
1. [Adversarial NLI](#Adversarial-NLI)
1. [Adversarial NLI properties](#Adversarial-NLI-properties)
1. [Working with Adversarial NLI](#Working-with-Adversarial-NLI)
1. [Other NLI datasets](#Other-NLI-datasets)
## Overview
Natural Language Inference (NLI) is the task of predicting the logical relationships between words, phrases, sentences, (paragraphs, documents, ...). Such relationships are crucial for all kinds of reasoning in natural language: arguing, debating, problem solving, summarization, and so forth.
[Dagan et al. (2006)](https://u.cs.biu.ac.il/~nlp/RTE1/Proceedings/dagan_et_al.pdf), one of the foundational papers on NLI (also called Recognizing Textual Entailment; RTE), make a case for the generality of this task in NLU:
> It seems that major inferences, as needed by multiple applications, can indeed be cast in terms of textual entailment. For example, __a QA system__ has to identify texts that entail a hypothesized answer. [...] Similarly, for certain __Information Retrieval__ queries the combination of semantic concepts and relations denoted by the query should be entailed from relevant retrieved documents. [...] In __multi-document summarization__ a redundant sentence, to be omitted from the summary, should be entailed from other sentences in the summary. And in __MT evaluation__ a correct translation should be semantically equivalent to the gold standard translation, and thus both translations should entail each other. Consequently, we hypothesize that textual entailment recognition is a suitable generic task for evaluating and comparing applied semantic inference models. Eventually, such efforts can promote the development of entailment recognition "engines" which may provide useful generic modules across applications.
## Our version of the task
Our NLI data will look like this:
| Premise | Relation | Hypothesis |
|:--------|:---------------:|:------------|
| turtle | contradiction | linguist |
| A turtled danced | entails | A turtle moved |
| Every reptile danced | entails | Every turtle moved |
| Some turtles walk | contradicts | No turtles move |
| James Byron Dean refused to move without blue jeans | entails | James Dean didn't dance without pants |
In the [word-entailment bakeoff](hw_wordentail.ipynb), we study a special case of this where the premise and hypothesis are single words. This notebook begins to introduce the problem of NLI more fully.
## Primary resources
We're going to focus on three NLI corpora:
* [The Stanford Natural Language Inference corpus (SNLI)](https://nlp.stanford.edu/projects/snli/)
* [The Multi-Genre NLI Corpus (MultiNLI)](https://www.nyu.edu/projects/bowman/multinli/)
* [The Adversarial NLI Corpus (ANLI)](https://github.com/facebookresearch/anli)
The first was collected by a group at Stanford, led by [Sam Bowman](https://www.nyu.edu/projects/bowman/), and the second was collected by a group at NYU, also led by [Sam Bowman](https://www.nyu.edu/projects/bowman/). Both have the same format and were crowdsourced using the same basic methods. However, SNLI is entirely focused on image captions, whereas MultiNLI includes a greater range of contexts.
The third corpus was collected by a group at Facebook AI and UNC Chapel Hill. The team's goal was to address the fact that datasets like SNLI and MultiNLI seem to be artificially easy – models trained on them can often surpass stated human performance levels but still fail on examples that are simple and intuitive for people. The dataset is "Adversarial" because the annotators were asked to try to construct examples that fooled strong models but still passed muster with other human readers.
This notebook presents tools for working with these corpora. The [second notebook in the unit](nli_02_models.ipynb) concerns models of NLI.
## Set-up
* As usual, you need to be fully set up to work with [the CS224u repository](https://github.com/cgpotts/cs224u/).
* If you haven't already, download [the course data](http://web.stanford.edu/class/cs224u/data/data.tgz), unpack it, and place it in the directory containing the course repository – the same directory as this notebook. (If you want to put it somewhere else, change `DATA_HOME` below.)
```
import nli
import os
import pandas as pd
import random
from datasets import load_dataset
DATA_HOME = os.path.join("data", "nlidata")
ANNOTATIONS_HOME = os.path.join(DATA_HOME, "multinli_1.0_annotations")
```
## SNLI
### SNLI properties
For SNLI (and MultiNLI), MTurk annotators were presented with premise sentences and asked to produce new sentences that entailed, contradicted, or were neutral with respect to the premise. A subset of the examples were then validated by an additional four MTurk annotators.
* All the premises are captions from the [Flickr30K corpus](http://shannon.cs.illinois.edu/DenotationGraph/).
* Some of the sentences rather depressingly reflect stereotypes ([Rudinger et al. 2017](https://www.aclweb.org/anthology/W17-1609)).
* 550,152 train examples; 10K dev; 10K test
* Mean length in tokens:
* Premise: 14.1
* Hypothesis: 8.3
* Clause-types
* Premise S-rooted: 74%
* Hypothesis S-rooted: 88.9%
* Vocab size: 37,026
* 56,951 examples validated by four additional annotators
* 58.3% examples with unanimous gold label
* 91.2% of gold labels match the author's label
* 0.70 overall Fleiss kappa
* Top scores currently around 90%.
### Working with SNLI
```
snli = load_dataset("snli")
```
The dataset has three splits:
```
snli.keys()
```
The class `nli.NLIReader` is used by all the readers discussed here.
Because the datasets are so large, it is often useful to be able to randomly sample from them. This is supported with the keyword argument `samp_percentage`. For example, the following samples approximately 10% of the examples from the SNLI training set:
```
nli.NLIReader(snli['train'], samp_percentage=0.10, random_state=42)
```
The precise number of examples will vary somewhat because of the way the sampling is done. (Here, we choose efficiency over precision in the number of cases we return; see the implementation for details.)
All of the readers have a `read` method that yields `NLIExample` example instances. For SNLI, these have the following attributes:
* __label__: `str`
* __premise__: `str`
* __hypothesis__: `str`
Note: the original SNLI distribution includes a number of other valuable fields, including identifiers for the original caption in the [Flickr 30k corpus](http://shannon.cs.illinois.edu/DenotationGraph/), parses for the examples, and annotation distributions for the validation set. Perhaps someone could update [the dataset on Hugging Face](https://huggingface.co/datasets/snli) to provide access to this information!
The following creates the label distribution for the training data:
```
snli_labels = pd.Series(
[ex.label for ex in nli.NLIReader(
snli['train'], filter_unlabeled=False).read()])
snli_labels.value_counts()
```
Use `filter_unlabeled=True` (the default) to silently drop the examples for which `gold_label` is `-`.
Let's look at a specific example in some detail:
```
snli_iterator = iter(nli.NLIReader(snli['train']).read())
snli_ex = next(snli_iterator)
print(snli_ex)
```
## MultiNLI
### MultiNLI properties
* Train premises drawn from five genres:
1. Fiction: works from 1912–2010 spanning many genres
1. Government: reports, letters, speeches, etc., from government websites
1. The _Slate_ website
1. Telephone: the Switchboard corpus
1. Travel: Berlitz travel guides
* Additional genres just for dev and test (the __mismatched__ condition):
1. The 9/11 report
1. Face-to-face: The Charlotte Narrative and Conversation Collection
1. Fundraising letters
1. Non-fiction from Oxford University Press
1. _Verbatim_ articles about linguistics
* 392,702 train examples; 20K dev; 20K test
* 19,647 examples validated by four additional annotators
* 58.2% examples with unanimous gold label
* 92.6% of gold labels match the author's label
* Test-set labels available as a Kaggle competition.
* Top matched scores currently around 0.81.
* Top mismatched scores currently around 0.83.
### Working with MultiNLI
```
mnli = load_dataset("multi_nli")
```
For MultiNLI, we have the following splits:
* `train`
* `validation_matched`
* `validation_mismatched`
```
mnli.keys()
```
The MultiNLI test sets are available on Kaggle ([matched version](https://www.kaggle.com/c/multinli-matched-open-evaluation) and [mismatched version](https://www.kaggle.com/c/multinli-mismatched-open-evaluation)).
The interface to these is the same as for the SNLI readers:
```
nli.NLIReader(mnli['train'], samp_percentage=0.10, random_state=42)
```
The `NLIExample` instances for MultiNLI have nearly all the attributes that SNLI is supposed to have!
* __promptID__: `str`
* __label__: `str`
* __pairID__: `str`
* __premise__: `str`
* __premise_binary_parse__: `nltk.tree.Tree`
* __premise_parse__: `nltk.tree.Tree`
* __hypothesis__: `str`
* __hypothesis_binary_parse__: `nltk.tree.Tree`
* __hypothesis_parse__: `nltk.tree.Tree`
The only field that is unfortunately missing is __annotator_labels__, which gives all five labels chosen by annotators for the two dev splits. Perhaps someone could [create a PR to bring these fields back in](https://huggingface.co/datasets/multi_nli)!
The full label distribution for the train split:
```
multinli_labels = pd.Series(
[ex.label for ex in nli.NLIReader(
mnli['validation_mismatched'], filter_unlabeled=False).read()])
multinli_labels.value_counts()
```
No examples in the MultiNLI train set lack a gold label. The original corpus distribution does contain some unlabeled examples in its dev-sets, but those seem to have been removed in the Hugging Face distribution. As a result, the value of the `filter_unlabeled` parameter has no effect for `mnli`.
Let's look at a specific example:
```
mnli_iterator = iter(nli.NLIReader(mnli['train']).read())
mnli_ex = next(mnli_iterator)
```
As you can see, there are three versions of the premise and hypothesis sentences:
1. Regular string representations of the data
2. Unlabeled binary parses
3. Labeled parses
```
mnli_ex.premise
```
The binary parses lack node labels; so that we can use `nltk.tree.Tree` with them, the label `X` is added to all of them:
```
mnli_ex.premise_binary_parse
```
Here's the full parse tree with syntactic categories:
```
mnli_ex.premise_parse
```
The leaves of either tree are tokenized versions of them:
```
mnli_ex.premise_parse.leaves()
```
### Annotated MultiNLI subsets
MultiNLI includes additional annotations for a subset of the dev examples. The goal is to help people understand how well their models are doing on crucial NLI-related linguistic phenomena.
```
matched_ann_filename = os.path.join(
ANNOTATIONS_HOME,
"multinli_1.0_matched_annotations.txt")
mismatched_ann_filename = os.path.join(
ANNOTATIONS_HOME,
"multinli_1.0_mismatched_annotations.txt")
def view_random_example(annotations, random_state=42):
random.seed(random_state)
ann_ex = random.choice(list(annotations.items()))
pairid, ann_ex = ann_ex
ex = ann_ex['example']
print("pairID: {}".format(pairid))
print(ann_ex['annotations'])
print(ex.premise)
print(ex.label)
print(ex.hypothesis)
matched_ann = nli.read_annotated_subset(
matched_ann_filename,
mnli['validation_matched'])
view_random_example(matched_ann, random_state=23)
```
## Adversarial NLI
### Adversarial NLI properties
The ANLI dataset was created in response to evidence that datasets like SNLI and MultiNLI are artificially easy for modern machine learning models to solve. The team sought to tackle this weakness head-on, by designing a crowdsourcing task in which annotators were explicitly trying to confuse state-of-the-art models. In broad outline, the task worked like this:
1. The crowdworker is presented with a premise (context) text and asked to construct a hypothesis sentence that entails, contradicts, or is neutral with respect to that premise. (The actual wording is more informal, along the lines of the SNLI/MultiNLI task).
1. The crowdworker submits a hypothesis text.
1. The premise/hypothesis pair is fed to a trained model that makes a prediction about the correct NLI label.
1. If the model's prediction is correct, then the crowdworker loops back to step 2 to try again. If the model's prediction is incorrect, then the example is validated by different crowdworkers.
The dataset consists of three rounds, each involving a different model and a different set of sources for the premise texts:
| Round | Model | Training data | Context sources |
|:------:|:------------|:---------------------------|:-----------------|
| 1 | [BERT-large](https://www.aclweb.org/anthology/N19-1423/) | SNLI + MultiNLI | Wikipedia |
| 2 | [ROBERTa](https://arxiv.org/abs/1907.11692) | SNLI + MultiNLI + [NLI-FEVER](https://github.com/easonnie/combine-FEVER-NSMN/blob/master/other_resources/nli_fever.md) + Round 1 | Wikipedia |
| 3 | [ROBERTa](https://arxiv.org/abs/1907.11692) | SNLI + MultiNLI + [NLI-FEVER](https://github.com/easonnie/combine-FEVER-NSMN/blob/master/other_resources/nli_fever.md) + Round 2 | Various |
Each round has train/dev/test splits. The sizes of these splits and their label distributions are calculated just below.
The [project README](https://github.com/facebookresearch/anli/blob/master/README.md) seeks to establish some rules for how the rounds can be used for training and evaluation.
### Working with Adversarial NLI
```
anli = load_dataset("anli")
```
For ANLI, we have a lot of options. Because it is distributed in three rounds, and the rounds can be used independently or pooled:
```
anli.keys()
```
Here is the fully pooled train setting:
```
anli_pooled_reader = nli.NLIReader(
anli['train_r1'], anli['train_r2'], anli['train_r3'],
filter_unlabeled=False)
anli_pooled_labels = pd.Series([ex.label for ex in anli_pooled_reader.read()])
anli_pooled_labels.value_counts()
for rounds in ((1,), (2,), (3,), (1,2,3)):
splits = [anli['train_r{}'.format(i)] for i in rounds]
count = len(list(nli.NLIReader(*splits).read()))
print("R{0:}: {1:,}".format(rounds, count))
```
The above figures correspond to those in Table 2 of the paper.
Here is a summary of what `NLIExample` instances offer for this corpus:
* __uid__: a unique identifier; akin to `pairID` in SNLI/MultiNLI
* __premise__: the premise; corresponds to `sentence1` in SNLI/MultiNLI
* __hypothesis__: the hypothesis; corresponds to `sentence2` in SNLI/MultiNLI
* __label__: the gold label; corresponds to `gold_label` in SNLI/MultiNLI
* __reason__: a crowdworker's free-text hypothesis about why the model made an incorrect prediction for the current __context__/__hypothesis__ pair
The ANLI distribution contains additional fields that are unfortunately left out of the Hugging Face distribution:
* __model_label__: the label predicted by the model used in the current round
* __emturk__: for dev (and test), this is `True` if the annotator contributed only dev (test) exmples, else `False`; in turn, it is `False` for all train examples.
* __genre__: the source for the __context__ text
* __tag__: information about the round and train/dev/test classification
As with the other datasets, it would be a wonderful service to the field to [improve the interface](https://huggingface.co/datasets/anli)!
```
anli_ex = next(iter(nli.NLIReader(anli['dev_r3']).read()))
anli_ex
```
## Other NLI datasets
There are a lot of NLI datasets available from the Hugging Face `datasets` library. [Here's a link to the full index](https://huggingface.co/datasets?task_ids=task_ids:natural-language-inference), and here are some highlights:
* [The SemEval 2014 semantic relatedness shared task](http://alt.qcri.org/semeval2014/task1/) used an NLI dataset called [Sentences Involving Compositional Knowledge (SICK)](http://alt.qcri.org/semeval2014/task1/index.php?id=data-and-tools). HF: https://huggingface.co/datasets/sick
* [XNLI](https://github.com/facebookresearch/XNLI) is a multilingual collection of test sets derived from MultiNLI. HF: https://huggingface.co/datasets/xnli
* [Turkish NLI](https://github.com/boun-tabi/NLI-TR) provides human-validated machine-translated versions of SNLI and MultiNLI. These seem not to show up in the above index, but they can be accessed via:
```
snli_tr_dataset = load_dataset('nli_tr', 'snli_tr')
multinli_tr_dataset = load_dataset('nli_tr', 'multinli_tr')
```
And here are some tips on additional NLI datasets that aren't yet on Hugging Face:
* [The FraCaS textual inference test suite](http://www-nlp.stanford.edu/~wcmac/downloads/) is a smaller, hand-built dataset that is great for evaluating a model's ability to handle complex logical patterns.
* [SemEval 2013](https://www.cs.york.ac.uk/semeval-2013/) had a wide range of interesting data sets for NLI and related tasks.
* [The SemEval 2014 semantic relatedness shared task](http://alt.qcri.org/semeval2014/task1/) used an NLI dataset called [Sentences Involving Compositional Knowledge (SICK)](http://alt.qcri.org/semeval2014/task1/index.php?id=data-and-tools).
* [MedNLI](https://physionet.org/physiotools/mimic-code/mednli/) is specialized to the medical domain, using data derived from [MIMIC III](https://mimic.physionet.org).
* [Diverse Natural Language Inference Collection (DNC)](http://decomp.io/projects/diverse-natural-language-inference/) transforms existing annotations from other tasks into NLI problems for a diverse range of reasoning challenges.
* [SciTail](http://data.allenai.org/scitail/) is an NLI dataset derived from multiple-choice science exam questions and Web text.
* [NLI Style FEVER](https://github.com/easonnie/combine-FEVER-NSMN/blob/master/other_resources/nli_fever.md) is a version of [the FEVER dataset](http://fever.ai) put into a standard NLI format. It was used by the Adversarial NLI team to train models for their annotation round 2.
* [OCNLI: Original Chinese Natural Language Inference](https://github.com/CLUEbenchmark/OCNLI): A large, high-quality MNLI-style NLI dataset for Mandarin.
|
github_jupyter
|
__author__ = "Christopher Potts"
__version__ = "CS224u, Stanford, Spring 2022"
import nli
import os
import pandas as pd
import random
from datasets import load_dataset
DATA_HOME = os.path.join("data", "nlidata")
ANNOTATIONS_HOME = os.path.join(DATA_HOME, "multinli_1.0_annotations")
snli = load_dataset("snli")
snli.keys()
nli.NLIReader(snli['train'], samp_percentage=0.10, random_state=42)
snli_labels = pd.Series(
[ex.label for ex in nli.NLIReader(
snli['train'], filter_unlabeled=False).read()])
snli_labels.value_counts()
snli_iterator = iter(nli.NLIReader(snli['train']).read())
snli_ex = next(snli_iterator)
print(snli_ex)
mnli = load_dataset("multi_nli")
mnli.keys()
nli.NLIReader(mnli['train'], samp_percentage=0.10, random_state=42)
multinli_labels = pd.Series(
[ex.label for ex in nli.NLIReader(
mnli['validation_mismatched'], filter_unlabeled=False).read()])
multinli_labels.value_counts()
mnli_iterator = iter(nli.NLIReader(mnli['train']).read())
mnli_ex = next(mnli_iterator)
mnli_ex.premise
mnli_ex.premise_binary_parse
mnli_ex.premise_parse
mnli_ex.premise_parse.leaves()
matched_ann_filename = os.path.join(
ANNOTATIONS_HOME,
"multinli_1.0_matched_annotations.txt")
mismatched_ann_filename = os.path.join(
ANNOTATIONS_HOME,
"multinli_1.0_mismatched_annotations.txt")
def view_random_example(annotations, random_state=42):
random.seed(random_state)
ann_ex = random.choice(list(annotations.items()))
pairid, ann_ex = ann_ex
ex = ann_ex['example']
print("pairID: {}".format(pairid))
print(ann_ex['annotations'])
print(ex.premise)
print(ex.label)
print(ex.hypothesis)
matched_ann = nli.read_annotated_subset(
matched_ann_filename,
mnli['validation_matched'])
view_random_example(matched_ann, random_state=23)
anli = load_dataset("anli")
anli.keys()
anli_pooled_reader = nli.NLIReader(
anli['train_r1'], anli['train_r2'], anli['train_r3'],
filter_unlabeled=False)
anli_pooled_labels = pd.Series([ex.label for ex in anli_pooled_reader.read()])
anli_pooled_labels.value_counts()
for rounds in ((1,), (2,), (3,), (1,2,3)):
splits = [anli['train_r{}'.format(i)] for i in rounds]
count = len(list(nli.NLIReader(*splits).read()))
print("R{0:}: {1:,}".format(rounds, count))
anli_ex = next(iter(nli.NLIReader(anli['dev_r3']).read()))
anli_ex
| 0.275714 | 0.985746 |
# Triangle Counting
In this notebook, we will count the numner of trianges in our test dataset. The NetworkX and cuGraph processes will be interleaved so that each step can be compared.
Notebook Credits
* Original Authors: Bradley Rees
* Created: 08/01/2019
* Last Edit: 08/16/2020
RAPIDS Versions: 0.13
Test Hardware
* GV100 32G, CUDA 10.2
## Introduction
Triangle Counting, as the name implies, finds the number of triangles in a graph. Triangles are important in computing the clustering Coefficient and can be used for clustering.
To compute the Pagerank scores for a graph in cuGraph we use:<br>
**cugraph.triangles(G)**
* __G__: cugraph.Graph object
Compute the triangle (number of cycles of length three) count of the input graph.
Parameters
----------
G : cugraph.graph
cuGraph graph descriptor, should contain the connectivity information,
(edge weights are not used in this algorithm)
Returns
-------
count : int64
A 64 bit integer whose value gives the number of triangles in the
graph.
__Reference__:
O. Green, P. Yalamanchili, L.M. Munguia,
“Fast Triangle Counting on GPU”
Irregular Applications: Architectures and Algorithms (IA3), 2014
#### Some notes about vertex IDs...
* The current version of cuGraph requires that vertex IDs be representable as 32-bit integers, meaning graphs currently can contain at most 2^32 unique vertex IDs. However, this limitation is being actively addressed and a version of cuGraph that accommodates more than 2^32 vertices will be available in the near future.
* cuGraph will automatically renumber graphs to an internal format consisting of a contiguous series of integers starting from 0, and convert back to the original IDs when returning data to the caller. If the vertex IDs of the data are already a contiguous series of integers starting from 0, the auto-renumbering step can be skipped for faster graph creation times.
* To skip auto-renumbering, set the `renumber` boolean arg to `False` when calling the appropriate graph creation API (eg. `G.from_cudf_edgelist(gdf_r, source='src', destination='dst', renumber=False)`).
* For more advanced renumbering support, see the examples in `structure/renumber.ipynb` and `structure/renumber-2.ipynb`
### Test Data
We will be using the Zachary Karate club dataset
*W. W. Zachary, An information flow model for conflict and fission in small groups, Journal of
Anthropological Research 33, 452-473 (1977).*

### Prep
```
# Import needed libraries
import cugraph
import cudf
from collections import OrderedDict
# NetworkX libraries
import networkx as nx
from scipy.io import mmread
```
### Some Prep
```
# Define the path to the test data
datafile='../data/karate-data.csv'
```
---
# NetworkX
```
# Read the data, this also created a NetworkX Graph
file = open(datafile, 'rb')
Gnx = nx.read_edgelist(file)
nx_count = nx.triangles(Gnx)
# NetworkX does not give a single count, but list how many triangles each vertex is associated with
nx_count
# To get the number of triangles, we would need to loop through the array and add up each count
count = 0
for key, value in nx_count.items():
count = count + value
count
```
Let's seet how that compares to cuGraph
----
# cuGraph
### Read in the data - GPU
cuGraph depends on cuDF for data loading and the initial Dataframe creation
The data file contains an edge list, which represents the connection of a vertex to another. The `source` to `destination` pairs is in what is known as Coordinate Format (COO). In this test case, the data is just two columns. However a third, `weight`, column is also possible
```
# Test file
gdf = cudf.read_csv(datafile, delimiter='\t', names=['src', 'dst'], dtype=['int32', 'int32'] )
```
### Create a Graph
```
# create a Graph using the source (src) and destination (dst) vertex pairs from the Dataframe
G = cugraph.Graph()
G.from_cudf_edgelist(gdf, source='src', destination='dst')
```
### Call the Triangle Counting
```
# Call cugraph.pagerank to get the pagerank scores
cu_count = cugraph.triangles(G)
cu_count
```
_It was that easy!_
----
___
Copyright (c) 2019-2020, NVIDIA CORPORATION.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
___
|
github_jupyter
|
# Import needed libraries
import cugraph
import cudf
from collections import OrderedDict
# NetworkX libraries
import networkx as nx
from scipy.io import mmread
# Define the path to the test data
datafile='../data/karate-data.csv'
# Read the data, this also created a NetworkX Graph
file = open(datafile, 'rb')
Gnx = nx.read_edgelist(file)
nx_count = nx.triangles(Gnx)
# NetworkX does not give a single count, but list how many triangles each vertex is associated with
nx_count
# To get the number of triangles, we would need to loop through the array and add up each count
count = 0
for key, value in nx_count.items():
count = count + value
count
# Test file
gdf = cudf.read_csv(datafile, delimiter='\t', names=['src', 'dst'], dtype=['int32', 'int32'] )
# create a Graph using the source (src) and destination (dst) vertex pairs from the Dataframe
G = cugraph.Graph()
G.from_cudf_edgelist(gdf, source='src', destination='dst')
# Call cugraph.pagerank to get the pagerank scores
cu_count = cugraph.triangles(G)
cu_count
| 0.412057 | 0.991676 |
___
<a href='http://www.pieriandata.com'> <img src='../Pierian_Data_Logo.png' /></a>
___
# Matplotlib Figure Object
Import the `matplotlib.pyplot` module under the name `plt` (the tidy way):
```
# COMMON MISTAKE!
# DON'T FORGET THE .PYPLOT part
import matplotlib.pyplot as plt
```
**NOTE: For users running .py scripts in an IDE like PyCharm or Sublime Text Editor. You will not see the plots in a notebook, instead if you are using another editor, you'll use: *plt.show()* at the end of all your plotting commands to have the figure pop up in another window.**
___
### Matplotlib Object Oriented Method
Now that we've seen the basics, let's break it all down with a more formal introduction of Matplotlib's Object Oriented API. This means we will instantiate figure objects and then call methods or attributes from that object.
### The Data
```
import numpy as np
a = np.linspace(0,10,11)
b = a ** 4
a
b
x = np.arange(0,10)
y = 2 * x
x
y
```
## Creating a Figure
The main idea in using the more formal Object Oriented method is to create figure objects and then just call methods or attributes off of that object. This approach is nicer when dealing with a canvas that has multiple plots on it.
```
# Creates blank canvas
fig = plt.figure()
```
**NOTE: ALL THE COMMANDS NEED TO GO IN THE SAME CELL!**
To begin we create a figure instance. Then we can add axes to that figure:
```
# Create Figure (empty canvas)
fig = plt.figure()
# Add set of axes to figure
axes = fig.add_axes([0, 0, 1, 1]) # left, bottom, width, height (range 0 to 1)
# Plot on that set of axes
axes.plot(x, y)
plt.show()
# Create Figure (empty canvas)
fig = plt.figure()
# Add set of axes to figure
axes = fig.add_axes([0, 0, 1, 1]) # left, bottom, width, height (range 0 to 1)
# Plot on that set of axes
axes.plot(a, b)
plt.show()
```
### Adding another set of axes to the Figure
So far we've only seen one set of axes on this figure object, but we can keep adding new axes on to it at any location and size we want. We can then plot on that new set of axes.
```
type(fig)
```
Code is a little more complicated, but the advantage is that we now have full control of where the plot axes are placed, and we can easily add more than one axis to the figure. Note how we're plotting a,b twice here
```
# Creates blank canvas
fig = plt.figure()
axes1 = fig.add_axes([0, 0, 1, 1]) # Large figure
axes2 = fig.add_axes([0.2, 0.2, 0.5, 0.5]) # Smaller figure
# Larger Figure Axes 1
axes1.plot(a, b)
# Use set_ to add to the axes figure
axes1.set_xlabel('X Label')
axes1.set_ylabel('Y Label')
axes1.set_title('Big Figure')
# Insert Figure Axes 2
axes2.plot(a,b)
axes2.set_title('Small Figure');
```
Let's move the small figure and edit its parameters.
```
# Creates blank canvas
fig = plt.figure()
axes1 = fig.add_axes([0, 0, 1, 1]) # Large figure
axes2 = fig.add_axes([0.2, 0.5, 0.25, 0.25]) # Smaller figure
# Larger Figure Axes 1
axes1.plot(a, b)
# Use set_ to add to the axes figure
axes1.set_xlabel('X Label')
axes1.set_ylabel('Y Label')
axes1.set_title('Big Figure')
# Insert Figure Axes 2
axes2.plot(a,b)
axes2.set_xlim(8,10)
axes2.set_ylim(4000,10000)
axes2.set_xlabel('X')
axes2.set_ylabel('Y')
axes2.set_title('Zoomed In');
```
You can add as many axes on to the same figure as you want, even outside of the main figure if the length and width correspond to this.
```
# Creates blank canvas
fig = plt.figure()
axes1 = fig.add_axes([0, 0, 1, 1]) # Full figure
axes2 = fig.add_axes([0.2, 0.5, 0.25, 0.25]) # Smaller figure
axes3 = fig.add_axes([1, 1, 0.25, 0.25]) # Starts at top right corner!
# Larger Figure Axes 1
axes1.plot(a, b)
# Use set_ to add to the axes figure
axes1.set_xlabel('X Label')
axes1.set_ylabel('Y Label')
axes1.set_title('Big Figure')
# Insert Figure Axes 2
axes2.plot(a,b)
axes2.set_xlim(8,10)
axes2.set_ylim(4000,10000)
axes2.set_xlabel('X')
axes2.set_ylabel('Y')
axes2.set_title('Zoomed In');
# Insert Figure Axes 3
axes3.plot(a,b)
```
### Figure Parameters
```
# Creates blank canvas
fig = plt.figure(figsize=(12,8),dpi=100)
axes1 = fig.add_axes([0, 0, 1, 1])
axes1.plot(a,b)
```
## Exporting a Figure
```
fig = plt.figure()
axes1 = fig.add_axes([0, 0, 1, 1])
axes1.plot(a,b)
axes1.set_xlabel('X')
# bbox_inches ='tight' automatically makes sure the bounding box is correct
fig.savefig('figure.png',bbox_inches='tight')
```
---
```
# Creates blank canvas
fig = plt.figure(figsize=(12,8))
axes1 = fig.add_axes([0, 0, 1, 1]) # Full figure
axes2 = fig.add_axes([1, 1, 0.25, 0.25]) # Starts at top right corner!
# Larger Figure Axes 1
axes1.plot(x,y)
# Insert Figure Axes 2
axes2.plot(x,y)
fig.savefig('test.png',bbox_inches='tight')
```
----
----
|
github_jupyter
|
# COMMON MISTAKE!
# DON'T FORGET THE .PYPLOT part
import matplotlib.pyplot as plt
import numpy as np
a = np.linspace(0,10,11)
b = a ** 4
a
b
x = np.arange(0,10)
y = 2 * x
x
y
# Creates blank canvas
fig = plt.figure()
# Create Figure (empty canvas)
fig = plt.figure()
# Add set of axes to figure
axes = fig.add_axes([0, 0, 1, 1]) # left, bottom, width, height (range 0 to 1)
# Plot on that set of axes
axes.plot(x, y)
plt.show()
# Create Figure (empty canvas)
fig = plt.figure()
# Add set of axes to figure
axes = fig.add_axes([0, 0, 1, 1]) # left, bottom, width, height (range 0 to 1)
# Plot on that set of axes
axes.plot(a, b)
plt.show()
type(fig)
# Creates blank canvas
fig = plt.figure()
axes1 = fig.add_axes([0, 0, 1, 1]) # Large figure
axes2 = fig.add_axes([0.2, 0.2, 0.5, 0.5]) # Smaller figure
# Larger Figure Axes 1
axes1.plot(a, b)
# Use set_ to add to the axes figure
axes1.set_xlabel('X Label')
axes1.set_ylabel('Y Label')
axes1.set_title('Big Figure')
# Insert Figure Axes 2
axes2.plot(a,b)
axes2.set_title('Small Figure');
# Creates blank canvas
fig = plt.figure()
axes1 = fig.add_axes([0, 0, 1, 1]) # Large figure
axes2 = fig.add_axes([0.2, 0.5, 0.25, 0.25]) # Smaller figure
# Larger Figure Axes 1
axes1.plot(a, b)
# Use set_ to add to the axes figure
axes1.set_xlabel('X Label')
axes1.set_ylabel('Y Label')
axes1.set_title('Big Figure')
# Insert Figure Axes 2
axes2.plot(a,b)
axes2.set_xlim(8,10)
axes2.set_ylim(4000,10000)
axes2.set_xlabel('X')
axes2.set_ylabel('Y')
axes2.set_title('Zoomed In');
# Creates blank canvas
fig = plt.figure()
axes1 = fig.add_axes([0, 0, 1, 1]) # Full figure
axes2 = fig.add_axes([0.2, 0.5, 0.25, 0.25]) # Smaller figure
axes3 = fig.add_axes([1, 1, 0.25, 0.25]) # Starts at top right corner!
# Larger Figure Axes 1
axes1.plot(a, b)
# Use set_ to add to the axes figure
axes1.set_xlabel('X Label')
axes1.set_ylabel('Y Label')
axes1.set_title('Big Figure')
# Insert Figure Axes 2
axes2.plot(a,b)
axes2.set_xlim(8,10)
axes2.set_ylim(4000,10000)
axes2.set_xlabel('X')
axes2.set_ylabel('Y')
axes2.set_title('Zoomed In');
# Insert Figure Axes 3
axes3.plot(a,b)
# Creates blank canvas
fig = plt.figure(figsize=(12,8),dpi=100)
axes1 = fig.add_axes([0, 0, 1, 1])
axes1.plot(a,b)
fig = plt.figure()
axes1 = fig.add_axes([0, 0, 1, 1])
axes1.plot(a,b)
axes1.set_xlabel('X')
# bbox_inches ='tight' automatically makes sure the bounding box is correct
fig.savefig('figure.png',bbox_inches='tight')
# Creates blank canvas
fig = plt.figure(figsize=(12,8))
axes1 = fig.add_axes([0, 0, 1, 1]) # Full figure
axes2 = fig.add_axes([1, 1, 0.25, 0.25]) # Starts at top right corner!
# Larger Figure Axes 1
axes1.plot(x,y)
# Insert Figure Axes 2
axes2.plot(x,y)
fig.savefig('test.png',bbox_inches='tight')
| 0.737442 | 0.983754 |
# Transfer Learning
In this notebook, you'll learn how to use pre-trained networks to solved challenging problems in computer vision. Specifically, you'll use networks trained on [ImageNet](http://www.image-net.org/) [available from torchvision](http://pytorch.org/docs/0.3.0/torchvision/models.html).
ImageNet is a massive dataset with over 1 million labeled images in 1000 categories. It's used to train deep neural networks using an architecture called convolutional layers. I'm not going to get into the details of convolutional networks here, but if you want to learn more about them, please [watch this](https://www.youtube.com/watch?v=2-Ol7ZB0MmU).
Once trained, these models work astonishingly well as feature detectors for images they weren't trained on. Using a pre-trained network on images not in the training set is called transfer learning. Here we'll use transfer learning to train a network that can classify our cat and dog photos with near perfect accuracy.
With `torchvision.models` you can download these pre-trained networks and use them in your applications. We'll include `models` in our imports now.
```
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
import torch
from torch import nn
from torch import optim
import torch.nn.functional as F
from torchvision import datasets, transforms, models
```
Most of the pretrained models require the input to be 224x224 images. Also, we'll need to match the normalization used when the models were trained. Each color channel was normalized separately, the means are `[0.485, 0.456, 0.406]` and the standard deviations are `[0.229, 0.224, 0.225]`.
```
data_dir = 'Cat_Dog_data'
# TODO: Define transforms for the training data and testing data
# Pass transforms in here, then run the next cell to see how the transforms look
train_data = datasets.ImageFolder(data_dir + '/train', transform=train_transforms)
test_data = datasets.ImageFolder(data_dir + '/test', transform=test_transforms)
trainloader = torch.utils.data.DataLoader(train_data, batch_size=64, shuffle=True)
testloader = torch.utils.data.DataLoader(test_data, batch_size=32)
```
We can load in a model such as [DenseNet](http://pytorch.org/docs/0.3.0/torchvision/models.html#id5). Let's print out the model architecture so we can see what's going on.
```
model = models.densenet121(pretrained=True)
model
```
This model is built out of two main parts, the features and the classifier. The features part is a stack of convolutional layers and overall works as a feature detector that can be fed into a classifier. The classifier part is a single fully-connected layer `(classifier): Linear(in_features=1024, out_features=1000)`. This layer was trained on the ImageNet dataset, so it won't work for our specific problem. That means we need to replace the classifier, but the features will work perfectly on their own. In general, I think about pre-trained networks as amazingly good feature detectors that can be used as the input for simple feed-forward classifiers.
```
# Freeze parameters so we don't backprop through them
for param in model.parameters():
param.requires_grad = False
from collections import OrderedDict
classifier = nn.Sequential(OrderedDict([
('fc1', nn.Linear(1024, 500)),
('relu', nn.ReLU()),
('fc2', nn.Linear(500, 2)),
('output', nn.LogSoftmax(dim=1))
]))
model.classifier = classifier
```
With our model built, we need to train the classifier. However, now we're using a **really deep** neural network. If you try to train this on a CPU like normal, it will take a long, long time. Instead, we're going to use the GPU to do the calculations. The linear algebra computations are done in parallel on the GPU leading to 100x increased training speeds. It's also possible to train on multiple GPUs, further decreasing training time.
PyTorch, along with pretty much every other deep learning framework, uses [CUDA](https://developer.nvidia.com/cuda-zone) to efficiently compute the forward and backwards passes on the GPU. In PyTorch, you move your model parameters and other tensors to the GPU memory using `model.to('cuda')`. You can move them back from the GPU with `model.to('cpu')` which you'll commonly do when you need to operate on the network output outside of PyTorch. As a demonstration of the increased speed, I'll compare how long it takes to perform a forward and backward pass with and without a GPU.
```
import time
for device in ['cpu', 'cuda']:
criterion = nn.NLLLoss()
# Only train the classifier parameters, feature parameters are frozen
optimizer = optim.Adam(model.classifier.parameters(), lr=0.001)
model.to(device)
for ii, (inputs, labels) in enumerate(trainloader):
# Move input and label tensors to the GPU
inputs, labels = inputs.to(device), labels.to(device)
start = time.time()
outputs = model.forward(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
if ii==3:
break
print(f"Device = {device}; Time per batch: {(time.time() - start)/3:.3f} seconds")
```
You can write device agnostic code which will automatically use CUDA if it's enabled like so:
```python
# at beginning of the script
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
...
# then whenever you get a new Tensor or Module
# this won't copy if they are already on the desired device
input = data.to(device)
model = MyModule(...).to(device)
```
From here, I'll let you finish training the model. The process is the same as before except now your model is much more powerful. You should get better than 95% accuracy easily.
>**Exercise:** Train a pretrained models to classify the cat and dog images. Continue with the DenseNet model, or try ResNet, it's also a good model to try out first. Make sure you are only training the classifier and the parameters for the features part are frozen.
```
# TODO: Train a model with a pre-trained network
```
|
github_jupyter
|
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
import torch
from torch import nn
from torch import optim
import torch.nn.functional as F
from torchvision import datasets, transforms, models
data_dir = 'Cat_Dog_data'
# TODO: Define transforms for the training data and testing data
# Pass transforms in here, then run the next cell to see how the transforms look
train_data = datasets.ImageFolder(data_dir + '/train', transform=train_transforms)
test_data = datasets.ImageFolder(data_dir + '/test', transform=test_transforms)
trainloader = torch.utils.data.DataLoader(train_data, batch_size=64, shuffle=True)
testloader = torch.utils.data.DataLoader(test_data, batch_size=32)
model = models.densenet121(pretrained=True)
model
# Freeze parameters so we don't backprop through them
for param in model.parameters():
param.requires_grad = False
from collections import OrderedDict
classifier = nn.Sequential(OrderedDict([
('fc1', nn.Linear(1024, 500)),
('relu', nn.ReLU()),
('fc2', nn.Linear(500, 2)),
('output', nn.LogSoftmax(dim=1))
]))
model.classifier = classifier
import time
for device in ['cpu', 'cuda']:
criterion = nn.NLLLoss()
# Only train the classifier parameters, feature parameters are frozen
optimizer = optim.Adam(model.classifier.parameters(), lr=0.001)
model.to(device)
for ii, (inputs, labels) in enumerate(trainloader):
# Move input and label tensors to the GPU
inputs, labels = inputs.to(device), labels.to(device)
start = time.time()
outputs = model.forward(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
if ii==3:
break
print(f"Device = {device}; Time per batch: {(time.time() - start)/3:.3f} seconds")
# at beginning of the script
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
...
# then whenever you get a new Tensor or Module
# this won't copy if they are already on the desired device
input = data.to(device)
model = MyModule(...).to(device)
# TODO: Train a model with a pre-trained network
| 0.489992 | 0.989171 |
# Generative Adversarial Network
In this notebook, we'll be building a generative adversarial network (GAN) trained on the MNIST dataset. From this, we'll be able to generate new handwritten digits!
GANs were [first reported on](https://arxiv.org/abs/1406.2661) in 2014 from Ian Goodfellow and others in Yoshua Bengio's lab. Since then, GANs have exploded in popularity. Here are a few examples to check out:
* [Pix2Pix](https://affinelayer.com/pixsrv/)
* [CycleGAN](https://github.com/junyanz/CycleGAN)
* [A whole list](https://github.com/wiseodd/generative-models)
The idea behind GANs is that you have two networks, a generator $G$ and a discriminator $D$, competing against each other. The generator makes fake data to pass to the discriminator. The discriminator also sees real data and predicts if the data it's received is real or fake. The generator is trained to fool the discriminator, it wants to output data that looks _as close as possible_ to real data. And the discriminator is trained to figure out which data is real and which is fake. What ends up happening is that the generator learns to make data that is indistiguishable from real data to the discriminator.

The general structure of a GAN is shown in the diagram above, using MNIST images as data. The latent sample is a random vector the generator uses to contruct it's fake images. As the generator learns through training, it figures out how to map these random vectors to recognizable images that can fool the discriminator.
The output of the discriminator is a sigmoid function, where 0 indicates a fake image and 1 indicates an real image. If you're interested only in generating new images, you can throw out the discriminator after training. Now, let's see how we build this thing in TensorFlow.
```
%matplotlib inline
import pickle as pkl
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets('MNIST_data')
```
## Model Inputs
First we need to create the inputs for our graph. We need two inputs, one for the discriminator and one for the generator. Here we'll call the discriminator input `inputs_real` and the generator input `inputs_z`. We'll assign them the appropriate sizes for each of the networks.
>**Exercise:** Finish the `model_inputs` function below. Create the placeholders for `inputs_real` and `inputs_z` using the input sizes `real_dim` and `z_dim` respectively.
```
def model_inputs(real_dim, z_dim):
inputs_real = tf.placeholder(tf.float32, shape=[None, real_dim], name="inputs_real")
inputs_z = tf.placeholder(tf.float32, shape=[None, z_dim], name="inputs_z")
return inputs_real, inputs_z
```
## Generator network

Here we'll build the generator network. To make this network a universal function approximator, we'll need at least one hidden layer. We should use a leaky ReLU to allow gradients to flow backwards through the layer unimpeded. A leaky ReLU is like a normal ReLU, except that there is a small non-zero output for negative input values.
#### Variable Scope
Here we need to use `tf.variable_scope` for two reasons. Firstly, we're going to make sure all the variable names start with `generator`. Similarly, we'll prepend `discriminator` to the discriminator variables. This will help out later when we're training the separate networks.
We could just use `tf.name_scope` to set the names, but we also want to reuse these networks with different inputs. For the generator, we're going to train it, but also _sample from it_ as we're training and after training. The discriminator will need to share variables between the fake and real input images. So, we can use the `reuse` keyword for `tf.variable_scope` to tell TensorFlow to reuse the variables instead of creating new ones if we build the graph again.
To use `tf.variable_scope`, you use a `with` statement:
```python
with tf.variable_scope('scope_name', reuse=False):
# code here
```
Here's more from [the TensorFlow documentation](https://www.tensorflow.org/programmers_guide/variable_scope#the_problem) to get another look at using `tf.variable_scope`.
#### Leaky ReLU
TensorFlow doesn't provide an operation for leaky ReLUs, so we'll need to make one . For this you can just take the outputs from a linear fully connected layer and pass them to `tf.maximum`. Typically, a parameter `alpha` sets the magnitude of the output for negative values. So, the output for negative input (`x`) values is `alpha*x`, and the output for positive `x` is `x`:
$$
f(x) = max(\alpha * x, x)
$$
#### Tanh Output
The generator has been found to perform the best with $tanh$ for the generator output. This means that we'll have to rescale the MNIST images to be between -1 and 1, instead of 0 and 1.
>**Exercise:** Implement the generator network in the function below. You'll need to return the tanh output. Make sure to wrap your code in a variable scope, with 'generator' as the scope name, and pass the `reuse` keyword argument from the function to `tf.variable_scope`.
```
def generator(z, out_dim, n_units=128, reuse=False, alpha=0.01):
''' Build the generator network.
Arguments
---------
z : Input tensor for the generator
out_dim : Shape of the generator output
n_units : Number of units in hidden layer
reuse : Reuse the variables with tf.variable_scope
alpha : leak parameter for leaky ReLU
Returns
-------
out, logits:
'''
with tf.variable_scope('generator', reuse=reuse): # finish this
# Hidden layer
h1 = tf.layers.dense(z, n_units, activation=None)
# Leaky ReLU
h1 = tf.maximum(alpha * h1, h1)
# Logits and tanh output
logits = tf.layers.dense(h1, out_dim, activation=None)
out = tf.tanh(logits)
return out
```
## Discriminator
The discriminator network is almost exactly the same as the generator network, except that we're using a sigmoid output layer.
>**Exercise:** Implement the discriminator network in the function below. Same as above, you'll need to return both the logits and the sigmoid output. Make sure to wrap your code in a variable scope, with 'discriminator' as the scope name, and pass the `reuse` keyword argument from the function arguments to `tf.variable_scope`.
```
def discriminator(x, n_units=128, reuse=False, alpha=0.01):
''' Build the discriminator network.
Arguments
---------
x : Input tensor for the discriminator
n_units: Number of units in hidden layer
reuse : Reuse the variables with tf.variable_scope
alpha : leak parameter for leaky ReLU
Returns
-------
out, logits:
'''
with tf.variable_scope('discriminator', reuse=reuse): # finish this
# Hidden layer
h1 = tf.layers.dense(x, n_units, activation=None)
# Leaky ReLU
h1 = tf.maximum(alpha * h1, h1)
logits = tf.layers.dense(h1, 1, activation=None)
out = tf.sigmoid(logits)
return out, logits
```
## Hyperparameters
```
# Size of input image to discriminator
input_size = 784 # 28x28 MNIST images flattened
# Size of latent vector to generator
z_size = 100
# Sizes of hidden layers in generator and discriminator
g_hidden_size = 128
d_hidden_size = 128
# Leak factor for leaky ReLU
alpha = 0.01
# Label smoothing
smooth = 0.1
```
## Build network
Now we're building the network from the functions defined above.
First is to get our inputs, `input_real, input_z` from `model_inputs` using the sizes of the input and z.
Then, we'll create the generator, `generator(input_z, input_size)`. This builds the generator with the appropriate input and output sizes.
Then the discriminators. We'll build two of them, one for real data and one for fake data. Since we want the weights to be the same for both real and fake data, we need to reuse the variables. For the fake data, we're getting it from the generator as `g_model`. So the real data discriminator is `discriminator(input_real)` while the fake discriminator is `discriminator(g_model, reuse=True)`.
>**Exercise:** Build the network from the functions you defined earlier.
```
tf.reset_default_graph()
# Create our input placeholders
input_real, input_z = model_inputs(input_size, z_size)
# Generator network here
g_model = generator(input_z, input_size)
# g_model is the generator output
# Disriminator network here
d_model_real, d_logits_real = discriminator(input_real)
d_model_fake, d_logits_fake = discriminator(g_model, reuse=True)
```
## Discriminator and Generator Losses
Now we need to calculate the losses, which is a little tricky. For the discriminator, the total loss is the sum of the losses for real and fake images, `d_loss = d_loss_real + d_loss_fake`. The losses will by sigmoid cross-entropies, which we can get with `tf.nn.sigmoid_cross_entropy_with_logits`. We'll also wrap that in `tf.reduce_mean` to get the mean for all the images in the batch. So the losses will look something like
```python
tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(logits=logits, labels=labels))
```
For the real image logits, we'll use `d_logits_real` which we got from the discriminator in the cell above. For the labels, we want them to be all ones, since these are all real images. To help the discriminator generalize better, the labels are reduced a bit from 1.0 to 0.9, for example, using the parameter `smooth`. This is known as label smoothing, typically used with classifiers to improve performance. In TensorFlow, it looks something like `labels = tf.ones_like(tensor) * (1 - smooth)`
The discriminator loss for the fake data is similar. The logits are `d_logits_fake`, which we got from passing the generator output to the discriminator. These fake logits are used with labels of all zeros. Remember that we want the discriminator to output 1 for real images and 0 for fake images, so we need to set up the losses to reflect that.
Finally, the generator losses are using `d_logits_fake`, the fake image logits. But, now the labels are all ones. The generator is trying to fool the discriminator, so it wants to discriminator to output ones for fake images.
>**Exercise:** Calculate the losses for the discriminator and the generator. There are two discriminator losses, one for real images and one for fake images. For the real image loss, use the real logits and (smoothed) labels of ones. For the fake image loss, use the fake logits with labels of all zeros. The total discriminator loss is the sum of those two losses. Finally, the generator loss again uses the fake logits from the discriminator, but this time the labels are all ones because the generator wants to fool the discriminator.
```
# Calculate losses
d_loss_real = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_real,
labels=(tf.ones_like(d_logits_real) * (1 - smooth))))
d_loss_fake = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_fake,
labels=(tf.zeros_like(d_logits_real) * (1 - smooth))))
d_loss = d_loss_real + d_loss_fake
g_loss = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_fake,
labels=tf.ones_like(d_logits_fake)))
```
## Optimizers
We want to update the generator and discriminator variables separately. So we need to get the variables for each part and build optimizers for the two parts. To get all the trainable variables, we use `tf.trainable_variables()`. This creates a list of all the variables we've defined in our graph.
For the generator optimizer, we only want to generator variables. Our past selves were nice and used a variable scope to start all of our generator variable names with `generator`. So, we just need to iterate through the list from `tf.trainable_variables()` and keep variables that start with `generator`. Each variable object has an attribute `name` which holds the name of the variable as a string (`var.name == 'weights_0'` for instance).
We can do something similar with the discriminator. All the variables in the discriminator start with `discriminator`.
Then, in the optimizer we pass the variable lists to the `var_list` keyword argument of the `minimize` method. This tells the optimizer to only update the listed variables. Something like `tf.train.AdamOptimizer().minimize(loss, var_list=var_list)` will only train the variables in `var_list`.
>**Exercise: ** Below, implement the optimizers for the generator and discriminator. First you'll need to get a list of trainable variables, then split that list into two lists, one for the generator variables and another for the discriminator variables. Finally, using `AdamOptimizer`, create an optimizer for each network that update the network variables separately.
```
# Optimizers
learning_rate = 0.002
# Get the trainable_variables, split into G and D parts
t_vars = tf.trainable_variables()
g_vars = [var for var in t_vars if var.name.startswith('generator')]
d_vars = [var for var in t_vars if var.name.startswith('discriminator')]
d_train_opt = tf.train.AdamOptimizer(learning_rate).minimize(d_loss, var_list=d_vars)
g_train_opt = tf.train.AdamOptimizer(learning_rate).minimize(g_loss, var_list=g_vars)
```
## Training
```
batch_size = 100
epochs = 100
samples = []
losses = []
saver = tf.train.Saver(var_list = g_vars)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for e in range(epochs):
for ii in range(mnist.train.num_examples//batch_size):
batch = mnist.train.next_batch(batch_size)
# Get images, reshape and rescale to pass to D
batch_images = batch[0].reshape((batch_size, 784))
batch_images = batch_images*2 - 1
# Sample random noise for G
batch_z = np.random.uniform(-1, 1, size=(batch_size, z_size))
# Run optimizers
_ = sess.run(d_train_opt, feed_dict={input_real: batch_images, input_z: batch_z})
_ = sess.run(g_train_opt, feed_dict={input_z: batch_z})
# At the end of each epoch, get the losses and print them out
train_loss_d = sess.run(d_loss, {input_z: batch_z, input_real: batch_images})
train_loss_g = g_loss.eval({input_z: batch_z})
print("Epoch {}/{}...".format(e+1, epochs),
"Discriminator Loss: {:.4f}...".format(train_loss_d),
"Generator Loss: {:.4f}".format(train_loss_g))
# Save losses to view after training
losses.append((train_loss_d, train_loss_g))
# Sample from generator as we're training for viewing afterwards
sample_z = np.random.uniform(-1, 1, size=(16, z_size))
gen_samples = sess.run(
generator(input_z, input_size, reuse=True),
feed_dict={input_z: sample_z})
samples.append(gen_samples)
saver.save(sess, './checkpoints/generator.ckpt')
# Save training generator samples
with open('train_samples.pkl', 'wb') as f:
pkl.dump(samples, f)
```
## Training loss
Here we'll check out the training losses for the generator and discriminator.
```
%matplotlib inline
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
losses = np.array(losses)
plt.plot(losses.T[0], label='Discriminator')
plt.plot(losses.T[1], label='Generator')
plt.title("Training Losses")
plt.legend()
```
## Generator samples from training
Here we can view samples of images from the generator. First we'll look at images taken while training.
```
def view_samples(epoch, samples):
fig, axes = plt.subplots(figsize=(7,7), nrows=4, ncols=4, sharey=True, sharex=True)
for ax, img in zip(axes.flatten(), samples[epoch]):
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
im = ax.imshow(img.reshape((28,28)), cmap='Greys_r')
return fig, axes
# Load samples from generator taken while training
with open('train_samples.pkl', 'rb') as f:
samples = pkl.load(f)
```
These are samples from the final training epoch. You can see the generator is able to reproduce numbers like 5, 7, 3, 0, 9. Since this is just a sample, it isn't representative of the full range of images this generator can make.
```
_ = view_samples(-1, samples)
```
Below I'm showing the generated images as the network was training, every 10 epochs. With bonus optical illusion!
```
rows, cols = 10, 6
fig, axes = plt.subplots(figsize=(7,12), nrows=rows, ncols=cols, sharex=True, sharey=True)
for sample, ax_row in zip(samples[::int(len(samples)/rows)], axes):
for img, ax in zip(sample[::int(len(sample)/cols)], ax_row):
ax.imshow(img.reshape((28,28)), cmap='Greys_r')
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
```
It starts out as all noise. Then it learns to make only the center white and the rest black. You can start to see some number like structures appear out of the noise. Looks like 1, 9, and 8 show up first. Then, it learns 5 and 3.
## Sampling from the generator
We can also get completely new images from the generator by using the checkpoint we saved after training. We just need to pass in a new latent vector $z$ and we'll get new samples!
```
saver = tf.train.Saver(var_list=g_vars)
with tf.Session() as sess:
saver.restore(sess, tf.train.latest_checkpoint('checkpoints'))
sample_z = np.random.uniform(-1, 1, size=(16, z_size))
gen_samples = sess.run(
generator(input_z, input_size, reuse=True),
feed_dict={input_z: sample_z})
view_samples(0, [gen_samples])
```
|
github_jupyter
|
%matplotlib inline
import pickle as pkl
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets('MNIST_data')
def model_inputs(real_dim, z_dim):
inputs_real = tf.placeholder(tf.float32, shape=[None, real_dim], name="inputs_real")
inputs_z = tf.placeholder(tf.float32, shape=[None, z_dim], name="inputs_z")
return inputs_real, inputs_z
with tf.variable_scope('scope_name', reuse=False):
# code here
def generator(z, out_dim, n_units=128, reuse=False, alpha=0.01):
''' Build the generator network.
Arguments
---------
z : Input tensor for the generator
out_dim : Shape of the generator output
n_units : Number of units in hidden layer
reuse : Reuse the variables with tf.variable_scope
alpha : leak parameter for leaky ReLU
Returns
-------
out, logits:
'''
with tf.variable_scope('generator', reuse=reuse): # finish this
# Hidden layer
h1 = tf.layers.dense(z, n_units, activation=None)
# Leaky ReLU
h1 = tf.maximum(alpha * h1, h1)
# Logits and tanh output
logits = tf.layers.dense(h1, out_dim, activation=None)
out = tf.tanh(logits)
return out
def discriminator(x, n_units=128, reuse=False, alpha=0.01):
''' Build the discriminator network.
Arguments
---------
x : Input tensor for the discriminator
n_units: Number of units in hidden layer
reuse : Reuse the variables with tf.variable_scope
alpha : leak parameter for leaky ReLU
Returns
-------
out, logits:
'''
with tf.variable_scope('discriminator', reuse=reuse): # finish this
# Hidden layer
h1 = tf.layers.dense(x, n_units, activation=None)
# Leaky ReLU
h1 = tf.maximum(alpha * h1, h1)
logits = tf.layers.dense(h1, 1, activation=None)
out = tf.sigmoid(logits)
return out, logits
# Size of input image to discriminator
input_size = 784 # 28x28 MNIST images flattened
# Size of latent vector to generator
z_size = 100
# Sizes of hidden layers in generator and discriminator
g_hidden_size = 128
d_hidden_size = 128
# Leak factor for leaky ReLU
alpha = 0.01
# Label smoothing
smooth = 0.1
tf.reset_default_graph()
# Create our input placeholders
input_real, input_z = model_inputs(input_size, z_size)
# Generator network here
g_model = generator(input_z, input_size)
# g_model is the generator output
# Disriminator network here
d_model_real, d_logits_real = discriminator(input_real)
d_model_fake, d_logits_fake = discriminator(g_model, reuse=True)
tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(logits=logits, labels=labels))
# Calculate losses
d_loss_real = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_real,
labels=(tf.ones_like(d_logits_real) * (1 - smooth))))
d_loss_fake = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_fake,
labels=(tf.zeros_like(d_logits_real) * (1 - smooth))))
d_loss = d_loss_real + d_loss_fake
g_loss = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_fake,
labels=tf.ones_like(d_logits_fake)))
# Optimizers
learning_rate = 0.002
# Get the trainable_variables, split into G and D parts
t_vars = tf.trainable_variables()
g_vars = [var for var in t_vars if var.name.startswith('generator')]
d_vars = [var for var in t_vars if var.name.startswith('discriminator')]
d_train_opt = tf.train.AdamOptimizer(learning_rate).minimize(d_loss, var_list=d_vars)
g_train_opt = tf.train.AdamOptimizer(learning_rate).minimize(g_loss, var_list=g_vars)
batch_size = 100
epochs = 100
samples = []
losses = []
saver = tf.train.Saver(var_list = g_vars)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for e in range(epochs):
for ii in range(mnist.train.num_examples//batch_size):
batch = mnist.train.next_batch(batch_size)
# Get images, reshape and rescale to pass to D
batch_images = batch[0].reshape((batch_size, 784))
batch_images = batch_images*2 - 1
# Sample random noise for G
batch_z = np.random.uniform(-1, 1, size=(batch_size, z_size))
# Run optimizers
_ = sess.run(d_train_opt, feed_dict={input_real: batch_images, input_z: batch_z})
_ = sess.run(g_train_opt, feed_dict={input_z: batch_z})
# At the end of each epoch, get the losses and print them out
train_loss_d = sess.run(d_loss, {input_z: batch_z, input_real: batch_images})
train_loss_g = g_loss.eval({input_z: batch_z})
print("Epoch {}/{}...".format(e+1, epochs),
"Discriminator Loss: {:.4f}...".format(train_loss_d),
"Generator Loss: {:.4f}".format(train_loss_g))
# Save losses to view after training
losses.append((train_loss_d, train_loss_g))
# Sample from generator as we're training for viewing afterwards
sample_z = np.random.uniform(-1, 1, size=(16, z_size))
gen_samples = sess.run(
generator(input_z, input_size, reuse=True),
feed_dict={input_z: sample_z})
samples.append(gen_samples)
saver.save(sess, './checkpoints/generator.ckpt')
# Save training generator samples
with open('train_samples.pkl', 'wb') as f:
pkl.dump(samples, f)
%matplotlib inline
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
losses = np.array(losses)
plt.plot(losses.T[0], label='Discriminator')
plt.plot(losses.T[1], label='Generator')
plt.title("Training Losses")
plt.legend()
def view_samples(epoch, samples):
fig, axes = plt.subplots(figsize=(7,7), nrows=4, ncols=4, sharey=True, sharex=True)
for ax, img in zip(axes.flatten(), samples[epoch]):
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
im = ax.imshow(img.reshape((28,28)), cmap='Greys_r')
return fig, axes
# Load samples from generator taken while training
with open('train_samples.pkl', 'rb') as f:
samples = pkl.load(f)
_ = view_samples(-1, samples)
rows, cols = 10, 6
fig, axes = plt.subplots(figsize=(7,12), nrows=rows, ncols=cols, sharex=True, sharey=True)
for sample, ax_row in zip(samples[::int(len(samples)/rows)], axes):
for img, ax in zip(sample[::int(len(sample)/cols)], ax_row):
ax.imshow(img.reshape((28,28)), cmap='Greys_r')
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
saver = tf.train.Saver(var_list=g_vars)
with tf.Session() as sess:
saver.restore(sess, tf.train.latest_checkpoint('checkpoints'))
sample_z = np.random.uniform(-1, 1, size=(16, z_size))
gen_samples = sess.run(
generator(input_z, input_size, reuse=True),
feed_dict={input_z: sample_z})
view_samples(0, [gen_samples])
| 0.833562 | 0.991891 |
## Como usar o pacote `basedosdados` em Python para análise de dados públicos
### 0. Explore nosso catálogo de bases!
Através do [nosso site](https://basedosdados.org/) é possível acessar informações sobre +930 bases de dados públicas nacionais e internacionais de diversas organizações e temas.
Ao buscar por algum tema ou base, você pode descobrir quais dados já tratamos e disponibilizamos no nosso _datalake_ público pelo símbolo <img src="attachment:7ff8f3e3-85d3-4b64-ae55-67a9598b732c.png" width="64">
Neste webinar iremos utilizar o [pacote da `basedosdados` (Python)](https://github.com/basedosdados/mais) para mostrar como acessar as bases disponíveis na <img src="attachment:7ff8f3e3-85d3-4b64-ae55-67a9598b732c.png" width="64">
- Outros recursos: [Tutorial escrito "Base de Dados Python 101"](https://dev.to/basedosdados/base-dos-dados-python-101-44lc)
### 1. Instale o pacote
```
# Todos os pacotes usados neste exemplo estão listados no requirements.txt, você pode instalar com:
# ! pip install -r requirements.txt
# Ou somente instalar nosso pacote, caso já tenha os demais listados:
# ! pip install basedosdados
import basedosdados as bd
```
### 2. Explore os dados disponíveis na BD+
Uma vez importado o pacote, vamos explorar os módulos de requisição de dados do nosso *datalake* público. Você pode ler em detalhes sobre todas as funcionalidades do pacote na nossa documentação: https://basedosdados.github.io/mais/reference_api_py/
* Listar datasets disponíveis:
```
# ?bd.list_datasets
bd.list_datasets(with_description=True)
```
* Listar tabelas disponíveis num determinado dataset:
```
# ?bd.list_dataset_tables
bd.list_dataset_tables(dataset_id="br_inpe_prodes", with_description=True)
```
* Obter metadados das colunas de ums tabela:
```
# ?bd.get_table_columns
bd.get_table_columns(
dataset_id="br_inpe_prodes",
table_id="desmatamento_municipios"
)
```
#### Configurando sua conta no Google Cloud
As funções que necessitam fazer consultas diretas aos dados no *datalake* necessitam de que você crie um projeto para autenticação no Google Cloud. O passo a passo de como fazer isso vai aparecerr direto no erro na sua tela, mas você também pode ver na nossa [documentação](https://basedosdados.github.io/mais/access_data_local/#criando-um-projeto-no-google-cloud).
_Não é necessário passar nenhuma informação de pagamento, você só precisa criar um projeto que será cobrado caso ultrapasse sua cota - são 1TB de processamento por mês, nem a gente consegue ultrapassar esse limite!_
```
# Vamos obter um erro neste comando: não é passado um billing_project_id!
# Na mensagem de erro vão aparecer as instruções de como criar um projeto no GCloud
# e achar seu project_id para ser usado nas consultas aos dados
bd.read_table(dataset_id="br_inpe_prodes", table_id="desmatamento_municipios", limit=100)
```
Uma vez configurado o projeto no Google Cloud, podemos rodar a consulta passando o seu `project_id` no parâmetro `billing_project_id`
```
bd.read_table(
dataset_id="br_inpe_prodes",
table_id="desmatamento_municipios",
limit=10,
billing_project_id="gcp_escoladedados")
```
* Calcular tamanho da tabela:
```
bd.get_table_size(
dataset_id="br_inpe_prodes",
table_id="desmatamento_municipios",
billing_project_id="gcp_escoladedados"
)
```
* Carregar a tabela no ambiente como um `Dataframe`:
```
df_prodes = bd.read_table(
dataset_id="br_inpe_prodes",
table_id="desmatamento_municipios",
billing_project_id="gcp_escoladedados")
df_prodes.info()
df_prodes.head()
```
* Consertar tipos das colunas:
```
cols_float = ["area", "desmatado", "incremento", "floresta", "nuvem", "nao_observado", "nao_floresta", "hidrografia"]
df_prodes[cols_float] = df_prodes[cols_float].astype(float)
```
* Verificar abrangência temporal:
```
df_prodes["ano"].unique()
```
* **Questão: Quais foram os municípios com maior incremento área desmatada em 2018-2019?**
```
(df_prodes[df_prodes["ano"] == 2019]
.sort_values(by="incremento", ascending=False)
.head(10))
```
**Mas afinal, quais são esses municípios?**
### Explorando a tabela de Diretórios Brasileiros
Reunimos numa base única construída pela Base dos Dados os códigos institucionais e informações de entidades brasileiras, como municípios. Você pode saber mais sobre a base aqui: https://basedosdados.org/dataset/br-bd-diretorios-brasil
_Por que construímos essa base? Para resolver os seguintes problemas:_
- Não existe um identificador único para municípios entre instituições brasileiras.
- IDs e nomes mudam tem typos entre anos e entres instituições.
- Municípios são criados ao longo do tempo, ganhando IDs novos.
```
df_diretorios = bd.read_table(
dataset_id="br_bd_diretorios_brasil",
table_id="municipio",
billing_project_id="gcp_escoladedados")
df_diretorios.info()
df_diretorios.head()
df_prodes = df_prodes.merge(df_diretorios[["id_municipio", "municipio", "sigla_uf"]], on="id_municipio", how="left")
df_prodes
```
* Criar coluna com o nome completo `Município/UF`:
```
df_prodes["municipio_uf"] = df_prodes["municipio"] + "/" + df_prodes["sigla_uf"]
```
* Listar municípios com maior incremento em área desmatada em 2018-2019:
```
(df_prodes[df_prodes["ano"] == 2019]
.sort_values(by="incremento", ascending=False)
.head(10))[["municipio_uf", "incremento"]]
```
### Analisando dados históricos de desmatamento
Agora com os dados prontos, vamos explorar algumas visualizações!
#### Qual foi o total de área desmatada ao longo dos anos?
```
area_acumulada = df_prodes.groupby("ano")[["desmatado"]].sum().round(2)
area_acumulada.columns = ["Área total desmatada (km2)"]
area_acumulada
import plotly.express as px
px.bar(area_acumulada,
y="Área total desmatada (km2)",
text="Área total desmatada (km2)",
title="Aumento de área total desmatada (km2) na Amazônia Legal de 2000 a 2019")
```
#### Quais os municípios com maior área desmatada até 2019?
```
ranking_municipios_2019 = (
df_prodes[df_prodes["ano"] == 2019]
.sort_values(by="desmatado", ascending=False)
.head(15))[["municipio_uf", "desmatado"]]
ranking_municipios_2019.columns = ["Município", "Área desmatada (km2)"]
ranking_municipios_2019
px.bar(ranking_municipios_2019.sort_values(by="Área desmatada (km2)"),
x="Área desmatada (km2)",
y="Município",
text="Área desmatada (km2)",
title="Municípios com maior área desmatada (km2) na Amazônia Legal até 2019",
orientation="h",
height=600)
```
#### Qual a relação entre a produção pecuária e a área dematada nos municípios?
Que tal cruzar os dados de desmatamento com a quantidade de rebanho bonivo? Vamos puxar os dados sobre os rebanhos da [Pesquisa Pecuária Municipal (PPM)](https://basedosdados.org/dataset/br-ibge-ppm) disnponíveis também na Base dos Dados para alimentar nossa análise.
```
# Como os dados são desde 1974 e queremos somente 2019, ao invés de carregar a base inteira iremos passar
# uma query em SQL para filtrar os dados antes de carregar no nosso ambiente
query = """
SELECT * FROM `basedosdados.br_ibge_ppm.efetivo_rebanhos`
WHERE ano = 2019;
"""
df_ppm = bd.read_sql(query, billing_project_id="gcp_escoladedados")
df_ppm.info()
df_ppm.head()
df_ppm["tipo_rebanho"].unique()
```
* Filtrar rebanhos bovinos:
```
df_ppm = df_ppm[df_ppm["tipo_rebanho"] == "Bovino"]
df_ppm = df_ppm.merge(df_prodes[["ano", "id_municipio", "municipio_uf", "desmatado"]], on=["id_municipio", "ano"], how="inner")
df_ppm
import numpy as np
fig = px.scatter(
df_ppm.sort_values(by="quantidade_animais", ascending=False, ignore_index=True).loc[1:],
x="quantidade_animais",
y="desmatado",
title="Área desmatada por quantidade de rebanho bovino nos municípios em 2019",
trendline="ols"
)
# Outro jeito de redefinir titulos dos eixos...
fig.update_layout(dict(xaxis=dict(title="Quantidade de rebanho bovino"),
yaxis=dict(title="Área dematada (km2)")))
fig.show()
results = px.get_trendline_results(fig)
results.px_fit_results.iloc[0].summary()
```
|
github_jupyter
|
# Todos os pacotes usados neste exemplo estão listados no requirements.txt, você pode instalar com:
# ! pip install -r requirements.txt
# Ou somente instalar nosso pacote, caso já tenha os demais listados:
# ! pip install basedosdados
import basedosdados as bd
# ?bd.list_datasets
bd.list_datasets(with_description=True)
# ?bd.list_dataset_tables
bd.list_dataset_tables(dataset_id="br_inpe_prodes", with_description=True)
# ?bd.get_table_columns
bd.get_table_columns(
dataset_id="br_inpe_prodes",
table_id="desmatamento_municipios"
)
# Vamos obter um erro neste comando: não é passado um billing_project_id!
# Na mensagem de erro vão aparecer as instruções de como criar um projeto no GCloud
# e achar seu project_id para ser usado nas consultas aos dados
bd.read_table(dataset_id="br_inpe_prodes", table_id="desmatamento_municipios", limit=100)
bd.read_table(
dataset_id="br_inpe_prodes",
table_id="desmatamento_municipios",
limit=10,
billing_project_id="gcp_escoladedados")
bd.get_table_size(
dataset_id="br_inpe_prodes",
table_id="desmatamento_municipios",
billing_project_id="gcp_escoladedados"
)
df_prodes = bd.read_table(
dataset_id="br_inpe_prodes",
table_id="desmatamento_municipios",
billing_project_id="gcp_escoladedados")
df_prodes.info()
df_prodes.head()
cols_float = ["area", "desmatado", "incremento", "floresta", "nuvem", "nao_observado", "nao_floresta", "hidrografia"]
df_prodes[cols_float] = df_prodes[cols_float].astype(float)
df_prodes["ano"].unique()
(df_prodes[df_prodes["ano"] == 2019]
.sort_values(by="incremento", ascending=False)
.head(10))
df_diretorios = bd.read_table(
dataset_id="br_bd_diretorios_brasil",
table_id="municipio",
billing_project_id="gcp_escoladedados")
df_diretorios.info()
df_diretorios.head()
df_prodes = df_prodes.merge(df_diretorios[["id_municipio", "municipio", "sigla_uf"]], on="id_municipio", how="left")
df_prodes
df_prodes["municipio_uf"] = df_prodes["municipio"] + "/" + df_prodes["sigla_uf"]
(df_prodes[df_prodes["ano"] == 2019]
.sort_values(by="incremento", ascending=False)
.head(10))[["municipio_uf", "incremento"]]
area_acumulada = df_prodes.groupby("ano")[["desmatado"]].sum().round(2)
area_acumulada.columns = ["Área total desmatada (km2)"]
area_acumulada
import plotly.express as px
px.bar(area_acumulada,
y="Área total desmatada (km2)",
text="Área total desmatada (km2)",
title="Aumento de área total desmatada (km2) na Amazônia Legal de 2000 a 2019")
ranking_municipios_2019 = (
df_prodes[df_prodes["ano"] == 2019]
.sort_values(by="desmatado", ascending=False)
.head(15))[["municipio_uf", "desmatado"]]
ranking_municipios_2019.columns = ["Município", "Área desmatada (km2)"]
ranking_municipios_2019
px.bar(ranking_municipios_2019.sort_values(by="Área desmatada (km2)"),
x="Área desmatada (km2)",
y="Município",
text="Área desmatada (km2)",
title="Municípios com maior área desmatada (km2) na Amazônia Legal até 2019",
orientation="h",
height=600)
# Como os dados são desde 1974 e queremos somente 2019, ao invés de carregar a base inteira iremos passar
# uma query em SQL para filtrar os dados antes de carregar no nosso ambiente
query = """
SELECT * FROM `basedosdados.br_ibge_ppm.efetivo_rebanhos`
WHERE ano = 2019;
"""
df_ppm = bd.read_sql(query, billing_project_id="gcp_escoladedados")
df_ppm.info()
df_ppm.head()
df_ppm["tipo_rebanho"].unique()
df_ppm = df_ppm[df_ppm["tipo_rebanho"] == "Bovino"]
df_ppm = df_ppm.merge(df_prodes[["ano", "id_municipio", "municipio_uf", "desmatado"]], on=["id_municipio", "ano"], how="inner")
df_ppm
import numpy as np
fig = px.scatter(
df_ppm.sort_values(by="quantidade_animais", ascending=False, ignore_index=True).loc[1:],
x="quantidade_animais",
y="desmatado",
title="Área desmatada por quantidade de rebanho bovino nos municípios em 2019",
trendline="ols"
)
# Outro jeito de redefinir titulos dos eixos...
fig.update_layout(dict(xaxis=dict(title="Quantidade de rebanho bovino"),
yaxis=dict(title="Área dematada (km2)")))
fig.show()
results = px.get_trendline_results(fig)
results.px_fit_results.iloc[0].summary()
| 0.267887 | 0.946498 |
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
a = np.ones((2,6)) * 2
b = np.ones((6, 3)) * 16
h = np.dot(a, b)
print(h.shape)
print(h)
print(h[:, 0])
dataset = pd.read_csv("ex1data1.txt")
dataset
X = dataset.iloc[:,0:1].values #Ev metrekarelerinin olduğu matrix (bu datada feet kullanılmış metre yerine) boyutu Ev SayısıX1
Y = dataset.iloc[:,1:].values # Ev fiyatları ( Gerçek değeri / 1000 şeklinde yer alıyor veride) boyutu Ev sayısıX1
theta = np.ones((2,1)) # Hipotezde kullanılacak olan teta 0 ve teta 1 i vektör halinde yazıyoruz.
X_plot = dataset.iloc[:,0:1].values #plotlamak için kullanacağımız x
X = np.append(np.ones((X.shape[0],1)).astype(int),values = X,axis=1) #theta 0 için Xin başına 1lerden oluşan
#bir adet sütun vektörü eklemeliyiz. Boyut EvSayısıx2 oldu.
m = X.shape[0] # Ev sayısı
J_history = [] #Cost fonksiyonun nasıl değiştiğini gözlemlemek için
alpha = 0.01#Learning rate dediğimiz türevi küçültmek için kullanılan katsayı istediğiniz gibi oynayıp gözlemleyebilirsiniz.
plt.figure()#Veri kümemizi inceleyelim x eksenimiz Evin metrekaresi Y eksenimiz dolar/1000 cinsinden evlerin fiyatları.
plt.scatter(X_plot,Y,c="r")
num_iters = 400 #Gradient descenti kaç kere uygulayacağınız Yine bununla istediğiniz kadar oynayıp gözlem yapabilirsiniz.
def computeCost(J_history,m,X,Y,theta):#Costu hesaplayan fonksiyonumuz.
preds = np.matmul(X,theta) # X EvSayısıx2 lik bir matrixti theta ise 2X1lik bir matrix(vektör de diyebilirsiniz)
#Matrix çarpımının tanımını izlerseniz mXn lik bir matrixi nXt lik bir matrixle çarptığınızda size mXt lik bir matrix döner.
#Bu matrix Çarpımının sonucuda EvSayısıx1 olur.Aslında bu matrix çarpımıyla yaptığımız şey tüm evler için hipotezin verdiği
#değeri bulmak.Yani theta0 + theta1.Metrekare nin bize verdiği sonucu bulmuş oluyoruz.
diffrence =np.square( preds - Y) #Formüldeki kare kısmı için direk matrixin element-wise karesini alabiliyoruz np.square ile.
J = (1/(2*m)) * np.sum(diffrence)#Yine formülden gelen tüm evler için yapılan tahminlerin gerçek fiyatlardan farklarının toplamı
#nın ev sayısı toplamına bölümü bize average costu verecektir.
J_history.append(J)
return J
def computeGradients(theta,X,Y,m):#Gradientleri compute ettiğimiz fonksiyon
preds = np.matmul(X,theta)#Yine yukarıdaki gibi matrix multiplication ile bilgisayarın yaptığı tahminlerin matrixini çıkartıyoruz.
diffrence = 1/m *(preds -Y)#Yine bilgisayarın yaptığı tahminden gerçek tahminleri çıkarıyoruz.
grads = np.matmul(np.transpose(X),diffrence)#Türev formülünü uyguluyoruz.Burda yaptığımız şey fonksiyonXiçinin türevi
#2x96 olması için X in transposeunu alıyoruz.96x2 lik bir matrixi 2x96lık yapıyor diffrence = 96x1 vector mxnXnxt mxt
#sonuç 2x1lik bir gradeint değeri
return grads
def train(num_iters,alpha,X,Y,theta,J_history,m):#Gradient descenti uyguladığımız thetayı bulduğumuz fonksiyon
for i in range(num_iters):
grads = computeGradients(theta,X,Y,m)
theta = theta - alpha *grads
computeCost(J_history,m,X,Y,theta)#Costların logunu tutmak için
if(i % 50 == 0): #Her 50 iterationda bir Line'ın nasıl değiştiğini gözlemleyelim.
plt.clf()
plt.figure()
plt.scatter(X_plot,Y,c= "b")
preds = np.matmul(X,theta)
plt.plot(X_plot[:,0],preds,c="r")
plt.pause(0.000001)
plt.clf()
plt.figure()
plt.plot([i for i in range(num_iters)],J_history)#Gradient descentin Costumuzu
plt.xlabel("Num_iters")
plt.ylabel("Cost")
return theta
J_history = []
theta = train(800,alpha,X,Y,theta,J_history,m)
plt.clf()
plt.figure()
plt.scatter(X_plot,Y,c= "b")
preds = np.matmul(X,theta)
plt.plot(X_plot[:,0],preds,c="r")
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
a = np.ones((2,6)) * 2
b = np.ones((6, 3)) * 16
h = np.dot(a, b)
print(h.shape)
print(h)
print(h[:, 0])
dataset = pd.read_csv("ex1data1.txt")
dataset
X = dataset.iloc[:,0:1].values #Ev metrekarelerinin olduğu matrix (bu datada feet kullanılmış metre yerine) boyutu Ev SayısıX1
Y = dataset.iloc[:,1:].values # Ev fiyatları ( Gerçek değeri / 1000 şeklinde yer alıyor veride) boyutu Ev sayısıX1
theta = np.ones((2,1)) # Hipotezde kullanılacak olan teta 0 ve teta 1 i vektör halinde yazıyoruz.
X_plot = dataset.iloc[:,0:1].values #plotlamak için kullanacağımız x
X = np.append(np.ones((X.shape[0],1)).astype(int),values = X,axis=1) #theta 0 için Xin başına 1lerden oluşan
#bir adet sütun vektörü eklemeliyiz. Boyut EvSayısıx2 oldu.
m = X.shape[0] # Ev sayısı
J_history = [] #Cost fonksiyonun nasıl değiştiğini gözlemlemek için
alpha = 0.01#Learning rate dediğimiz türevi küçültmek için kullanılan katsayı istediğiniz gibi oynayıp gözlemleyebilirsiniz.
plt.figure()#Veri kümemizi inceleyelim x eksenimiz Evin metrekaresi Y eksenimiz dolar/1000 cinsinden evlerin fiyatları.
plt.scatter(X_plot,Y,c="r")
num_iters = 400 #Gradient descenti kaç kere uygulayacağınız Yine bununla istediğiniz kadar oynayıp gözlem yapabilirsiniz.
def computeCost(J_history,m,X,Y,theta):#Costu hesaplayan fonksiyonumuz.
preds = np.matmul(X,theta) # X EvSayısıx2 lik bir matrixti theta ise 2X1lik bir matrix(vektör de diyebilirsiniz)
#Matrix çarpımının tanımını izlerseniz mXn lik bir matrixi nXt lik bir matrixle çarptığınızda size mXt lik bir matrix döner.
#Bu matrix Çarpımının sonucuda EvSayısıx1 olur.Aslında bu matrix çarpımıyla yaptığımız şey tüm evler için hipotezin verdiği
#değeri bulmak.Yani theta0 + theta1.Metrekare nin bize verdiği sonucu bulmuş oluyoruz.
diffrence =np.square( preds - Y) #Formüldeki kare kısmı için direk matrixin element-wise karesini alabiliyoruz np.square ile.
J = (1/(2*m)) * np.sum(diffrence)#Yine formülden gelen tüm evler için yapılan tahminlerin gerçek fiyatlardan farklarının toplamı
#nın ev sayısı toplamına bölümü bize average costu verecektir.
J_history.append(J)
return J
def computeGradients(theta,X,Y,m):#Gradientleri compute ettiğimiz fonksiyon
preds = np.matmul(X,theta)#Yine yukarıdaki gibi matrix multiplication ile bilgisayarın yaptığı tahminlerin matrixini çıkartıyoruz.
diffrence = 1/m *(preds -Y)#Yine bilgisayarın yaptığı tahminden gerçek tahminleri çıkarıyoruz.
grads = np.matmul(np.transpose(X),diffrence)#Türev formülünü uyguluyoruz.Burda yaptığımız şey fonksiyonXiçinin türevi
#2x96 olması için X in transposeunu alıyoruz.96x2 lik bir matrixi 2x96lık yapıyor diffrence = 96x1 vector mxnXnxt mxt
#sonuç 2x1lik bir gradeint değeri
return grads
def train(num_iters,alpha,X,Y,theta,J_history,m):#Gradient descenti uyguladığımız thetayı bulduğumuz fonksiyon
for i in range(num_iters):
grads = computeGradients(theta,X,Y,m)
theta = theta - alpha *grads
computeCost(J_history,m,X,Y,theta)#Costların logunu tutmak için
if(i % 50 == 0): #Her 50 iterationda bir Line'ın nasıl değiştiğini gözlemleyelim.
plt.clf()
plt.figure()
plt.scatter(X_plot,Y,c= "b")
preds = np.matmul(X,theta)
plt.plot(X_plot[:,0],preds,c="r")
plt.pause(0.000001)
plt.clf()
plt.figure()
plt.plot([i for i in range(num_iters)],J_history)#Gradient descentin Costumuzu
plt.xlabel("Num_iters")
plt.ylabel("Cost")
return theta
J_history = []
theta = train(800,alpha,X,Y,theta,J_history,m)
plt.clf()
plt.figure()
plt.scatter(X_plot,Y,c= "b")
preds = np.matmul(X,theta)
plt.plot(X_plot[:,0],preds,c="r")
plt.show()
| 0.32178 | 0.760695 |
# Improving Numerical Integration Methods
The solution and estimation of Dynamic Discrete Choice Models (DDCM) is often constrained by computational feasibility. For instance, Keane and Wolpin (1997) and subsequent work that estimates DDCM's of post-graduation career dynamics abstract from many important determinants of earnings and mobility dynamics. Examples include the isolation from match heterogeneity, permanent skill shocks, and absence of multidimensional skill structures.
Keane and Wolpin (1994, 1997) split their occupational classes into white- and blue-collar occupations. Nevertheless, empirical evidence suggests that skill requirements vary substantially within blue- and white-collar occupations. Arguably any aggregation of occupational classes should be able to account for meaningful skill differences. Acemoglu and Autor (2011) suggest four aggregate groups that are explicitly derived from the task content of classified three digit occupations in the US data.[<sup>1</sup>](#fn1)
Adding elements alike enlarges the computational burden of solving a DDCM enormously, and as already Keane and Wolpin (1994) noted, "[...] for problems of the size we would like to consider a [...] simulation strategy is not computationally practicable". A bottleneck in solving and estimating DDCM's is the solution of the expected value function, the so-called $EMax(\cdot)$. Adding new features to the model increases the required number of function evaluations, which are the costly operation in numerical integration. Results from applied mathematics suggest methods that are more efficient and thus enable a performance increase. For the same number of function evaluations (and hence computational cost) quasi-Monte Carlo methods achieve a significantly higher accuracy.
With **respy** it is possible to employ quasi-Monte Carlo methods to solve and estimate DDCM's. This notebook contains a tutorial on how to specify the numerical integration option in **respy**. For expositional purposes we will rely on the Keane and Wolpin (1994) model. The subsequent sections:
- Will explain how to choose a Monte Carlo method in **respy**.
- Will describe how to choose the number of iterations.
- Provide a simulation study: How does the integration method affects the solution of the model?
The following [slides](https://github.com/HumanCapitalAnalysis/student-project-rafael_suchy/blob/master/slides/numerical_integration_ddcm.pdf) provide a gentle introduction into the Keane and Wolpin (1994) model and highlight its main components. Additionally the reason and basic intuition for the usage of Monte Carlo and quasi-Monte Carlo methods are outlined.
```
%matplotlib inline
import pandas as pd
import respy as rp
import numpy as np
import chaospy as cp
import matplotlib.pyplot as plt
from matplotlib import ticker
from _numerical_integration import *
```
## Numerical Integration in respy
The current functionality of **respy** entails two main methods for the numerical approximation of the $EMax(\cdot)$:
- Monte Carlo Simulation: Chooses points randomly in the domain
- Quasi Monte Carlo Simulation: Chooses points from one of the two low-discrepancy sequences
- Sobol
- Halton
A very short introduction about the nature of low-discrepancy sequences is provided in the following [notebook](https://github.com/HumanCapitalAnalysis/student-project-rafael_suchy/blob/master/notebooks/98_low_discrepancy_sequences_application_integration.ipynb).
Now it is finally time to get our hands on the implementation in **respy**. The main method affected is ``create_base_draws`` within ``shared.py``, which as the name suggests creates a set of draws from the standard normal distribution. The draws are either drawn randomly (Monte Carlo Simulation) or from low-discrepancy sequences (Quasi-Monte Carlo Simulation). Either of the methods is used to calculate the $EMax(\cdot)$ in the solution *and* the choice probabilities in the maximum likelihood estimation.
## How to choose the Monte Carlo Method in respy
As mentioned, we will use the Keane and Wolpin (1994) model within this tutorial. First, we retrieve the parametrization. For details, you are welcomed to consult the previous tutorials of this readme.
```
params, options = rp.get_example_model("kw_94_one", with_data=False)
```
The set of *options* helps to define the underlying model and characterizes the solution methods as well as some functional forms. Inspecting the content we recognize the relevant options `solution_draws` and `monte_carlo_sequence`.
```
options
```
The option `solution_draws` specifies how many points (iteration) are simulated (performed) in order to solve the model. Ceteris paribus, choosing more points leads to a higher precision but also to higher computational costs. The option `monte_carlo_sequence` determines the method.[<sup>2</sup>](#fn2) As we will see, the implementation of quasi-Monte Carlo methods enables to break the trade-off: It is possible to increase the accuracy for a given set of points, switching from Monte Carlo methods to quasi-Monte Carlo methods.
It is possible to choose `monte_carlo_sequence` from the set of $\{$"random", "halton", "sobol" $\}$. In the following we will use the Sobol sequence with 200 draws to solve the model (`rp.solve`), by changing the option parameters as follows.
```
options["monte_carlo_sequence"], options["solution_draws"] = ["sobol", 200]
state_space = rp.solve(params, options)
```
To verify, we can shortly check whether the options were changed correctly.
```
options["monte_carlo_sequence"], options["solution_draws"]
```
Up to this point, all necessary information to change the integration option is mentioned. The eager reader can start to experiment with its own model (or a different Keane and Wolpin specification). In the next section we will provide an example and simulate the Keane and Wolpin (1994) model with different options. We will elaborate on how the employed Monte Carlo methods affects the model.
---
## Simulation study: How does the integration method affects the solution of the model?
We will simulate the model with either of the methods: "random", "halton", "sobol" and vary the amount of points from `POINTS_MIN` to `POINTS_MAX` by increments of (`POINTS_MAX` - `POINTS_MIN`)/`NUM_EVALS` that are used in the solution of the model. A reference value is calculated by using a very large amount of points ``POINTS_TRUTH``. Note, solving the whole model with ``POINTS_TRUTH`` repeatedly would not be possible at all.
```
# For a real application you might change the points as commented
POINTS_TRUTH = 1_000 # 1_000_000
POINTS_MIN = 2 # 100
POINTS_MAX = 4 # 10_000
NUM_EVALS = 10 # 100
POINTS_GRID = np.logspace(
POINTS_MIN,
POINTS_MAX,
NUM_EVALS,
dtype = int
)
METHODS = ["random", "halton", "sobol"]
```
First, we will compute a reference value for the $EMax(\cdot)$ values by using a large amount of points with the "sobol"-sequence. We extract the "true" $EMax(\cdot)$ values into the object `emax_true`.
<center>
<b>Caution:</b> Execution may take some time.
</center>
```
options["monte_carlo_sequence"], options["solution_draws"] = ["sobol", POINTS_TRUTH]
state_space = rp.solve(params, options)
emax_true = state_space.emax_value_functions
```
Second, we will loop over the methods and the number of points to calculate the $EMax(\cdot)$ for each specification of the model.
<center>
<b>Caution:</b> Execution may take even some more time. <br>
Alternatively, you can load the results from the pre-simulated pickle-file.
</center>
```
df_emax_values = pd.DataFrame(index=POINTS_GRID, columns=METHODS)
for method in METHODS:
options["monte_carlo_sequence"] = method
print("Current Iteration:", method.capitalize(), ".")
for points in POINTS_GRID:
options["solution_draws"] = points
state_space = rp.solve(params, options)
df_emax_values.loc[points][method] = state_space.emax_value_functions
```
To prevent doing the calculation over and over again, we will save our results in pickle files.
```
indices = range(
df_emax_values.loc[POINTS_GRID[1],
METHODS[0]].shape[0]
)
int_points = POINTS_GRID
index = list()
for point in int_points:
for idx in indices:
index.append((point, idx))
```
<center>
<b>Caution:</b> Execution may take even some more time. <br>
Alternatively, you can load the results from the pre-simulated pickle-file.
</center>
```
index = pd.MultiIndex.from_tuples(
index,
names=('integration_points', 'state_index')
)
df_emax_values_store = \
pd.DataFrame(
columns=[*METHODS], index=index)
for points in df_emax_values.index:
for method in METHODS:
df_emax_values_store[method].loc[points, :] = df_emax_values.loc[points, method]
```
For exposition of the convergence results, we will look at one particular approximation. For example at iteration number 14.
```
which_integrand = 14
get_single_integrand(df_emax_values_store, which_integrand, 1, METHODS)
```
We see that the quasi-Monte Carlo methods are stable after around 500 iterations (points) whereas the random sequence oscillates even for a large number of points. To get a more elaborate picture, we can zoom-in into the figure and inspect the behavior for a low amount of points. It gets evident that already after 200 points the `halton` and `sobol` options yield stable results.
A more theoterically founded method (vs. pure eyeballing) to evaluate the performance of our numerical integration is the calculation of the Root Mean Squared Error (RMSE). For this reason we first define some evaluation criteria.
```
def rmse(x1, x2, scaling_param = 1, axis = 0):
x1 = np.asanyarray(x1)
x2 = np.asanyarray(x2)
scaling_param = np.asanyarray(scaling_param)
return np.sqrt(
np.mean(((x1 - x2) ** 2)/scaling_param,
axis = axis)
)
df_rmse_comparison = pd.DataFrame(index = POINTS_GRID, columns = METHODS)
df_rel_rmse_comparison = pd.DataFrame(index = POINTS_GRID, columns = METHODS)
for points in POINTS_GRID:
for method in METHODS:
df_rmse_comparison.loc[points][method] = rmse(df_emax_values.loc[points][method], emax_true, 1, 0)
df_rel_rmse_comparison.loc[points][method] = rmse(df_emax_values.loc[points][method], emax_true, df_emax_values.loc[points][method], 0)
```
## How to choose the number of iterations?
Already in 1975 Hoaglin and Andrews set up recommendations how to report results and procedures when performing simulation studies. Hauck and Anderson (1984) surveyed 1,198 papers in five major statistical journals where 216 provided results that were based on simulation. Only 9% of papers that used a simulation study justified the choice of the number of iterations. This is contrary to the recommendations by Hoaglin and Andrews.[<sup>3</sup>](#fn3)
Harwell (2018) provide an update of the survey. The authors survey studies in six statistics journals between 1985 to 2012 and report that the use of simulation studies had doubled since 1984. An improvement of reporting standards was not found: Less than 18 of 371 simulation studies used an experimental design that could be identified. Additionally, 99.9% of the studies relied solely on a visual analysis of the results. In this section we will provide some ideas how to substantiate the number of iterations within (quasi-)Monte Carlo simulations.
### Number of iterations for Monte Carlo Analysis
The variance expression of Monte Carlo methods can guide the calculation of the number of iterations. Suppose we simulate $N$ samples (iterations). The variance is approximately given by
$$
\dfrac{\sigma^2(f)}{N}.
$$
A-priori, the value of $\sigma^2(f)$ is unknown but can be estimated. Similar to our calculation of the `emax_true` we can simulate a sample with `POINTS_TRUTH` number of iterations. Out of this sample we can estimate $\sigma^2(f)$. Specifying a desired precision $\rho$ we can immediately calculate the necessary number of iterations[<sup>4</sup>](#fn4) as
$$
\sqrt{M} \approx \dfrac{\sigma}{\rho}.
$$
A different, less scientific approach acknowledges the diversity of problems than can be tackled with Monte Carlo simulations. A fixed number of iterations may serve as a first guideline and substantiated reasoning that is followed by additional refinement. Quantifying extremely small probabilities needs a different amount of iterations than quantifying a mean. Bukaçi et al. (2016) propose a method that is based solely on convergence plots. For the graphical evaluation it is necessary to calculate the value of interest (for example the value of the $EMax(\cdot)$) repeatedly using different number of iterations. Following Bukaçi et al. (2016) the number of iterations at which the Monte Carlo estimate becomes stable can be used in the simulation study.
### Number of iterations for quasi-Monte Carlo Analysis
To reiterate, with the same computational effort (same number of function evaluations) the quasi-Monte Carlo methods achieve a higher accuracy than the MC method. Hence, having a number of iterations for the Monte Carlo method, we certainly know that employing a quasi-Monte Carlo procedure will perform better. Due to the completely deterministic nature of quasi-Monte Carlo methods (integration) we obtain deterministic, and hence guaranteed error bounds. Theoretically, it is always possible to determine an integration rule that yields a prescribed level of precision.
Formally we can use the Koksma-Hlawka inequality to determine an error bound resulting from quasi-Monte Carlo methods.[<sup>5</sup>](#fn5) If the random variable (integrand) $f$ has bounded variance $\mathbb{V}(f)$ on $\bar{I}^d$, then for any sample of nodes $x_1, \dots, x_N \in \bar{I}^d$ we have
$$
\left| \dfrac{1}{N} \sum_{i=1}^N f(x_i) - \int_{\bar{I}^d} f(u) \mathrm{d}u ~\right| \leq \mathbb{V}(f) D_N^*(x_1, \dots, x_N),
$$
where $D_N^*$ denotes the star discrepancy[<sup>6</sup>](#fn6) of the nodes.
If we are able to calculate all expressions in the Koksma-Hlawka inequality the required number of iterations for a pre-specified precisions follows immediately. The world could be so easy if the derivation of the Koksma-Hlawka inequality would not point towards a drawback: it does not apply to functions that have (simple) discontinuities. To circumvent this issue Brandolini et al. (2013) derive a Koksma-Hlawka-type inequality which applies to piecewise smooth functions. Nevertheless, oftentimes it is not feasible to calculate the expressions involved in the Koksma-Hlawka inequality (at least not without difficulties).
However, the Koksma-Hlawka inequality offers an additional insight: point sets with a small discrepancy guarantee small errors when using quasi-Monte Carlo methods for numerical integration. Number generators based on integer arithmetic module two, like the Sobol-sequence, provide additional equidistribution properties *whenever* the number of iterations $N$ is a power of two, $N = 2^n- 1$. Specifically, our [notebook](https://github.com/HumanCapitalAnalysis/student-project-rafael_suchy/blob/master/notebooks/98_low_discrepancy_sequences_application_integration.ipynb) demonstrates that those sequences are only equidistant, and hence have the minimal star discrepancy, if we have $N = 2^n – 1$ points.[<sup>7</sup>](#fn7) Consequently, employing iterations that satisfy this condition we can improve our procedure without incurring additional costs. This fact came to our mind when elaborating on the concept of Frolov points (Kacwin et al. (2018)).
### Convergence Plots
Another very useful assessment criterion are convergence plots (on log-log scale).
```
indices = range(df_emax_values.loc[POINTS_GRID[1], 'sobol'].shape[0])
int_points = POINTS_GRID
index_rmse = list()
for label in ['absolute', 'relative']:
for point in int_points:
index_rmse.append((label, point))
index_rmse = pd.MultiIndex.from_tuples(index_rmse, names=('measure', 'integration_points'))
df_rmse_total = \
pd.DataFrame(
columns=METHODS, index=index_rmse)
df_rmse_total.loc['relative', :] = df_rel_rmse_comparison.values
df_rmse_total.loc['absolute', :] = df_rmse_comparison.values
```
To compare our achieved convergence rates with theoretical results, we define reference comparison rates. For the choice of the functional form, please refer to "Appendix: Notes on (quasi-)Monte Carlo approximation".
```
rates_comparison = [
[(0.1 * np.log(points)**4)/(points) for points in POINTS_GRID-1],
[(0.05 * np.log(points)**4)/(points)**(1/2) for points in POINTS_GRID-1],
[(6 * np.log(points)**4)/(points)**(3/2) for points in POINTS_GRID-1]
]
```
An inspection of the second figure directly reveals the cost of improving our precision. If we want to decrease our relative RMSE to 10$^{-2}$ then we need to increase our points from 10$^2$ to 10$^3$. Furthermore we see that the options `sobol` and `halton` perform much better than employing a random sequence. In fact for our problem quasi-Monte Carlo methods reach nearly theoretically predicted convergence rates.
```
get_rmse_rate(df_rmse_total, rates_comparison, METHODS)
```
## Evaluation of Economic Relevance
Up to now we have shown that the choice of `monte_carlo_sequence` matters for the RMSE, the precision of our value function calculation, and the number of involved iterations. The next natural question is to ask whether computational decisions translate into economically relevant differences. For example, does the policy-conclusion of a schooling subsidy changes when we employ a different integration method leaving all else equal?
We tackle this question within the Keane and Wolpin (1994) model. First, we fetch the model and load the accompanying data set. We simulate 4000 agents and iterate over the options `monte_carlo_sequence` while comparing the status quo with a tuition subsidy of 500 monetary units. If the reader is interested to simulate other tuition subsidies, the possible sets are included as comments.
```
params, options, _ = rp.get_example_model("kw_94_one")
options["simulation_agents"] = 4_000 # Get the amount of agents we are simulating
options["solution_draws"] = 400 # Manually set the number of points to be drawn
# Initialize simulation
simulate = rp.get_simulate_func(params, options)
# Full set of models: models = np.repeat(["one", "two", "three"], 3)
MODELS = np.repeat(["one"], 2)
# Full set of tuition subsidies: [0, 500, 0, 1_000, 0, 2_000]
TUITION_SUBSIDIES = [0, 500]
index_row = pd.MultiIndex.from_product(
[METHODS, POINTS_GRID],
names = ["Method", "Points"]
)
df_avg_yos = pd.DataFrame(
index = index_row,
columns = TUITION_SUBSIDIES
) # Contains average years of schooling per individual
df_diff_avg_yos = pd.DataFrame(
index = index_row,
columns = TUITION_SUBSIDIES
) # Contains difference to "true" years of schooling per individual
```
<center>
<b>Caution:</b> Execution may take even some more time. <br>
Alternatively, you can load the results from the pre-simulated pickle-file.
</center>
```
subsidy_emax_truth = dict()
for s in TUITION_SUBSIDIES:
print("Runtime check tuition subsidy:", s, u'\u2713')
params.loc[("nonpec_edu", "at_least_twelve_exp_edu"), "value"] += s # Increase the subsidy parameter
options["monte_carlo_sequence"], options["solution_draws"] = ["random", POINTS_TRUTH] # Set options for evaluation of "truth"
simulate = rp.get_simulate_func(params, options) # Build the simulation function
subsidy_emax_truth[s] = simulate(params).groupby("Identifier")['Experience_Edu'].max().mean()
for method in METHODS:
options["monte_carlo_sequence"] = method
print("Runtime check method:", method, u'\u2713')
for points in POINTS_GRID:
options["solution_draws"] = points
simulate = rp.get_simulate_func(params, options)
df_avg_yos[s].loc[method, points] = simulate(params).groupby("Identifier")['Experience_Edu'].max().mean()
df_diff_avg_yos[s].loc[method, points] = df_avg_yos[s].loc[method, points] - subsidy_emax_truth[s]
params.loc[("nonpec_edu", "at_least_twelve_exp_edu"), "value"] -= s # Decrease the subsidy to initial value
get_policy_prediction(df_diff_avg_yos, *TUITION_SUBSIDIES, METHODS)
```
This figure reveals that the change in average years of schooling as a result of the 500 MU tuition subsidy is sensitive to the employed methods and choice of iterations. While `sobol` and `halton` provide reliable and stable results after 200 iterations this does not hold true for the `random` option. Using between 10 and 1000 iterations 8 $\%$ of the policy effect are driven by the choice of the Monte Carlo method.
## Remarks
<span id="fn1"><sup>1</sup>
Over the last few decades the empirical literature established a relatively weak economic performance of occupations that require workers to solve routine cognitive or routine manual tasks. Dimensions of occupational skill requirements, skill prices, and employment shares evolved substantially in the last years. As the authors suggest, distinction between
$~~~~~$ a) abstract, non-routine tasks (e.g. managerial)
$~~~~~$ b) routine cognitive tasks (e.g. administrative and sales),
$~~~~~$ c) routine manual tasks (e.g. production),
$~~~~~$ d) non-routine manual tasks (e.g. service occupations).
may be more appropriate
</span>
<span id="fn2"><sup>2</sup>
Please do not get confused. Although the option name is called `monte_carlo_sequence` it does indeed include the Quasi-Monte Carlo methods. The naming convenation was chosen this way, because in **respy** we regard Monte Carlo methods as the aggregate group.
</span>
<span id="fn3"><sup>3</sup>
A review of papers within Economics would probably indicate a similar state.
</span>
<span id="fn4"><sup>4</sup>
See [Niederreiter (1992)](https://epubs.siam.org/doi/book/10.1137/1.9781611970081) for a thorough treatment including methods for variance reduction.
</span>
<span id="fn5"><sup>5</sup>
A derivation can be found at pp.19f in [Niederreiter (1992)](https://epubs.siam.org/doi/book/10.1137/1.9781611970081).
</span>
<span id="fn6"><sup>6</sup>
See [Niederreiter (1992)](https://epubs.siam.org/doi/book/10.1137/1.9781611970081) p.14 for the definition.
</span>
<span id="fn7"><sup>7</sup>
If 0 is explicitly included in the construction of the sequence then the term “$-1$” can be dropped.
</span>
## Appendix: Notes on (quasi-)Monte Carlo approximation
This section will serve as a short introduction into the idea and notation of (quasi-)Monte Carlo integration, especially why the function $f$ can be treated as integrand and simultaneously as random variable. To discuss an approximate calculation of the integral $\int_{\mathbb{D}} f(u) \mathrm{d}u$ we will assume that the integration domain $\mathbb{D} \subseteq \mathbb{R}^d$ satisfies $0 < \lambda_d(\mathbb{D}) < \infty$. We denote the d-dimensional Lebesgue measure with $\lambda_d$.
By defining the probability meaure $\mathrm{d}\mu = \dfrac{\mathrm{du}}{\lambda_d(\mathbb{D})}$ we turn the domain $\mathbb{D}$ into a probability space. In this notation we can rewrite the integral as
$$
\int_{\mathbb{D}} f(u) \mathrm{d}u = \lambda_d(\mathbb{D}) f \mathrm{d}\mu = \lambda_d(\mathbb{D}) \mathbb{E}[f],
$$
where $\mathbb{E}[f]$ denotes the expected value of the random variable $f$. The Monte Carlo estimate for the expected value $\mathbb{E}[f]$ is obtained by taking $N$ independent $\lambda$-distributed random samples $s_1, \dots, s_N$ from the domain $A$ of the random variable $f$ and let
$$
\mathbb{E}[f] \approx \dfrac{1}{N} \sum_{i=1}^N f(s_i).
$$
A derivation (as provided in Niederreiter) shows that we can formalize the Monte Carlo estimate as follows
$$
\int_{\mathbb{D}} f(u) \mathrm{d}u \approx \dfrac{1}{N} \sum_{i=1}^N f(y_i),
$$
The nodes $y_i, \dots, y_N$ are $N$ independent random samples from a uniform distribution on $\bar{I}^d$, where $\bar{I}^d$ denotes the closed d-dimensional unit cube. We have a probabilistic error bound of $O(N^{-1/2})$.
The quasi-Monte Carlo approximation looks formally like the MC estimate but is used with deterministic nodes $x_1, \dots, x_N \in \bar{I}^d$. We take $\mathbb{D}$ as a subset of $\bar{I}^d$ and choose deterministic points $x_1, \dots, x_N \in \mathbb{D}$. The QMC approximation is given by
$$
\int_\mathbb{D} f(u) \mathrm{d}u \approx \dfrac{1}{N} \sum_{i=1}^N f(x_i).
$$
We get a deterministic error bound of $O(N^{-1}(log(N))^{d-1})$, where $d$ denotes the number of (effective) dimensions as
### Theoretical error bounds as benchmark
The theoretical results on convergence (error bounds) are particularly useful when we want to determine whether our integration problem is suited for (quasi-)Monte Carlo methods. Or vice versa: whether (quasi-)Monte Carlo methods are appropriate to solve our integration problem. As our convergence plots indicate, the rate of the employed quasi-Monte Carlo methods `"halton"` and `"sobol"` nearly attain their theoretical counterpart. Hence, we can conlude that our integration problem is indeed suited to be solved by quasi-Monte Carlo methods.
## References
> Acemoglu, D. and D. Autor (2011). [Skills, Tasks and Technologies: Implications for Employment and Earnings](https://economics.mit.edu/files/7006). *Handbook of Labor Economics*, Chapter 12, Vol. 4b.
> Bellman, R. E. (1957). ["Dynamic Programming"](https://press.princeton.edu/titles/9234.html). Princeton University Press, Princeton, NJ.
> Bukaçi, E. et al. (2016). [Number of iterations needed in Monte Carlo Simulation using reliability analysis for tunnel supports](https://www.ijera.com/papers/Vol6_issue6/Part%20-%203/J0606036064.pdf). *International Journal of Engineering Research and Applications*, 6(6): pp.60-64.
> Brandolini, L., Colzani, L., Gigante, G., and Travaglini, G. (2013). [On the Koksma–Hlawka inequality](https://www.sciencedirect.com/science/article/pii/S0885064X12000854#b000005). *Journal of Complexity*, 29(2): 158-172.
> Caflisch, R.E. (1998). [Monte Carlo and quasi-Monte Carlo methods](http://dsec.pku.edu.cn/~tieli/notes/numer_anal/MCQMC_Caflisch.pdf). *Acta Numerica*, 1-49.
> Caflisch, R.E., Morokoff, W., and Owen, A. (1997). [Valuation of Mortgage backed securities using Brownian bridges to reduce effective dimension](http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.36.3160) *Journal of Computational Finance*, 1: 27-46.
> Davis, P. J. and Rabinowitz, P. (1984). ["Methods of Numerical Integration"](https://store.doverpublications.com/0486453391.html). Dover Publications, Mineola, NY.
> Gerstner, T. and Griebel, M. (1998). ["Numerical Integration using Sparse Grids"](https://rdcu.be/bPFpc). *Numerical Algorithms* 18: 209–232.
> Halton, J. (1964). ["Algorithm 247: Radical-inverse Quasi-Random Point Sequence"](https://dl.acm.org/citation.cfm?id=365104). *Communications of the ACM,* 7: 701-701.
> Hauck, W.H. and Anderson, S. (1984). [A Survey Regarding the Reporting of Simulation Studies](https://amstat.tandfonline.com/doi/abs/10.1080/00031305.1984.10483206#.XjP298hKiUk). *The American Statistician*, 38(3): 214–216.
> Hoaglin, D.C. and Andrews, D.F. (1975). [The Reporting of Computation-Based Results in Statistics](https://www.tandfonline.com/doi/citedby/10.1080/00031305.1975.10477393?scroll=top&needAccess=true). *The American Statistician*, 29(3): 122–126.
> Harwell, M., Nidhi, K., and Peralta-Torres, Y. (2018). [A Survey of Reporting Practices of Computer Simulation Studies in Statistical Research](https://www.tandfonline.com/doi/full/10.1080/00031305.2017.1342692?scroll=top&needAccess=true). *The American Statistician*, 72(4): 321-327.
> Kacwin, C., Oettershagen, J., Ullrich, M., and Ullrich, T. (2018). [Numerical performance of optimized Frolov lattices in tensor product reproducing kernel Sobolev spaces.](https://ins.uni-bonn.de/media/public/content-frolov-frolov/FrolovNumerics.pdf?pk=6) ArXiv e-prints, 2018.
> Keane, M.P. and Wolpin, K.I. (1994). [The Solution and Estimation of Discrete Choice Dynamic Programming Models by Simulation and Interpolation: Monte Carlo Evidence](https://www.jstor.org/stable/2109768?seq=1). *The Review of Economics and Statistics*, 76(4): 648-672.
> Keane, M.P. and Wolpin, K.I. (1997). [The Career Decisions of Young Men](https://doi.org/10.1086/262080). *Journal of Political Economy*, 105(3): 473-522.
> Lemieux, C. (2009). ["Monte Carlo and Quasi-Monte Carlo Sampling"](https://www.springer.com/gp/book/9780387781648). Springer, Boston, MA.
> L’Ecuyer, P. (2017). ["Randomized Quasi-Monte Carlo: An Introduction for Practitioners"](https://hal.inria.fr/hal-01561550/document). 12th International Conference on Monte Carlo and Quasi-Monte Carlo Methods in Scientific Computing (MCQMC 2016), August 2016, Stanford, CA.
> L’Ecuyer, P. and Lemieux, C. (2005). ["Recent Advances in Randomized Quasi-Monte Carlo Methods"](https://link.springer.com/chapter/10.1007%2F0-306-48102-2_20). In: Dror M., L’Ecuyer P., Szidarovszky F. (eds) Modeling Uncertainty. *International Series in Operations Research & Management Science,* Vol. 46, Springer, Boston, MA.
> Niederreiter, H. (1992). [Random Number Generation and Quasi-Monte Carlo Methods](https://epubs.siam.org/doi/book/10.1137/1.9781611970081). *CBMS-NSF Regional Conference Series in Applied Mathematics*.
> Puterman, M. L. (1994). ["Markov Decision Processes: Discrete Stochastic Dynamic Programming (1st ed.)"](https://dl.acm.org/citation.cfm?id=528623). John Wiley & Sons, Inc., New York, NY, USA.
> Skrainka, B. S. and K. L. Judd. (2011). ["High Performance Quadrature Rules: How Numerical Integration Affects a Popular Model of Product Differentiation"](https://www.ucl.ac.uk/~uctpbss/EconSite/Research_files/HighPerformanceQuad.pdf). Unpublished.
> Sobol, I. M. (1967). ["Distribution of Points in a Cube and Approximate Evaluation of Integrals"](http://mi.mathnet.ru/eng/zvmmf7334). *Zh. Vych. Mat. Mat. Fiz.,* 7: 784–802 (in Russian); *U.S.S.R Comput. Maths. Math. Phys.7*: 86–112.
> Wang, X. and Fang, K.T. (2003). [The effective dimension and quasi-Monte Carlo integration](https://www.sciencedirect.com/science/article/pii/S0885064X03000037#BIB3). *Journal of Complexity*, 19(2): 101-124.
> Wolpin, K. I. (2013). [The Limits of Inference without Theory](https://mitpress.mit.edu/books/limits-inference-without-theory). MIT Press, Cambridge, MA.
|
github_jupyter
|
%matplotlib inline
import pandas as pd
import respy as rp
import numpy as np
import chaospy as cp
import matplotlib.pyplot as plt
from matplotlib import ticker
from _numerical_integration import *
params, options = rp.get_example_model("kw_94_one", with_data=False)
options
options["monte_carlo_sequence"], options["solution_draws"] = ["sobol", 200]
state_space = rp.solve(params, options)
options["monte_carlo_sequence"], options["solution_draws"]
# For a real application you might change the points as commented
POINTS_TRUTH = 1_000 # 1_000_000
POINTS_MIN = 2 # 100
POINTS_MAX = 4 # 10_000
NUM_EVALS = 10 # 100
POINTS_GRID = np.logspace(
POINTS_MIN,
POINTS_MAX,
NUM_EVALS,
dtype = int
)
METHODS = ["random", "halton", "sobol"]
options["monte_carlo_sequence"], options["solution_draws"] = ["sobol", POINTS_TRUTH]
state_space = rp.solve(params, options)
emax_true = state_space.emax_value_functions
df_emax_values = pd.DataFrame(index=POINTS_GRID, columns=METHODS)
for method in METHODS:
options["monte_carlo_sequence"] = method
print("Current Iteration:", method.capitalize(), ".")
for points in POINTS_GRID:
options["solution_draws"] = points
state_space = rp.solve(params, options)
df_emax_values.loc[points][method] = state_space.emax_value_functions
indices = range(
df_emax_values.loc[POINTS_GRID[1],
METHODS[0]].shape[0]
)
int_points = POINTS_GRID
index = list()
for point in int_points:
for idx in indices:
index.append((point, idx))
index = pd.MultiIndex.from_tuples(
index,
names=('integration_points', 'state_index')
)
df_emax_values_store = \
pd.DataFrame(
columns=[*METHODS], index=index)
for points in df_emax_values.index:
for method in METHODS:
df_emax_values_store[method].loc[points, :] = df_emax_values.loc[points, method]
which_integrand = 14
get_single_integrand(df_emax_values_store, which_integrand, 1, METHODS)
def rmse(x1, x2, scaling_param = 1, axis = 0):
x1 = np.asanyarray(x1)
x2 = np.asanyarray(x2)
scaling_param = np.asanyarray(scaling_param)
return np.sqrt(
np.mean(((x1 - x2) ** 2)/scaling_param,
axis = axis)
)
df_rmse_comparison = pd.DataFrame(index = POINTS_GRID, columns = METHODS)
df_rel_rmse_comparison = pd.DataFrame(index = POINTS_GRID, columns = METHODS)
for points in POINTS_GRID:
for method in METHODS:
df_rmse_comparison.loc[points][method] = rmse(df_emax_values.loc[points][method], emax_true, 1, 0)
df_rel_rmse_comparison.loc[points][method] = rmse(df_emax_values.loc[points][method], emax_true, df_emax_values.loc[points][method], 0)
indices = range(df_emax_values.loc[POINTS_GRID[1], 'sobol'].shape[0])
int_points = POINTS_GRID
index_rmse = list()
for label in ['absolute', 'relative']:
for point in int_points:
index_rmse.append((label, point))
index_rmse = pd.MultiIndex.from_tuples(index_rmse, names=('measure', 'integration_points'))
df_rmse_total = \
pd.DataFrame(
columns=METHODS, index=index_rmse)
df_rmse_total.loc['relative', :] = df_rel_rmse_comparison.values
df_rmse_total.loc['absolute', :] = df_rmse_comparison.values
rates_comparison = [
[(0.1 * np.log(points)**4)/(points) for points in POINTS_GRID-1],
[(0.05 * np.log(points)**4)/(points)**(1/2) for points in POINTS_GRID-1],
[(6 * np.log(points)**4)/(points)**(3/2) for points in POINTS_GRID-1]
]
get_rmse_rate(df_rmse_total, rates_comparison, METHODS)
params, options, _ = rp.get_example_model("kw_94_one")
options["simulation_agents"] = 4_000 # Get the amount of agents we are simulating
options["solution_draws"] = 400 # Manually set the number of points to be drawn
# Initialize simulation
simulate = rp.get_simulate_func(params, options)
# Full set of models: models = np.repeat(["one", "two", "three"], 3)
MODELS = np.repeat(["one"], 2)
# Full set of tuition subsidies: [0, 500, 0, 1_000, 0, 2_000]
TUITION_SUBSIDIES = [0, 500]
index_row = pd.MultiIndex.from_product(
[METHODS, POINTS_GRID],
names = ["Method", "Points"]
)
df_avg_yos = pd.DataFrame(
index = index_row,
columns = TUITION_SUBSIDIES
) # Contains average years of schooling per individual
df_diff_avg_yos = pd.DataFrame(
index = index_row,
columns = TUITION_SUBSIDIES
) # Contains difference to "true" years of schooling per individual
subsidy_emax_truth = dict()
for s in TUITION_SUBSIDIES:
print("Runtime check tuition subsidy:", s, u'\u2713')
params.loc[("nonpec_edu", "at_least_twelve_exp_edu"), "value"] += s # Increase the subsidy parameter
options["monte_carlo_sequence"], options["solution_draws"] = ["random", POINTS_TRUTH] # Set options for evaluation of "truth"
simulate = rp.get_simulate_func(params, options) # Build the simulation function
subsidy_emax_truth[s] = simulate(params).groupby("Identifier")['Experience_Edu'].max().mean()
for method in METHODS:
options["monte_carlo_sequence"] = method
print("Runtime check method:", method, u'\u2713')
for points in POINTS_GRID:
options["solution_draws"] = points
simulate = rp.get_simulate_func(params, options)
df_avg_yos[s].loc[method, points] = simulate(params).groupby("Identifier")['Experience_Edu'].max().mean()
df_diff_avg_yos[s].loc[method, points] = df_avg_yos[s].loc[method, points] - subsidy_emax_truth[s]
params.loc[("nonpec_edu", "at_least_twelve_exp_edu"), "value"] -= s # Decrease the subsidy to initial value
get_policy_prediction(df_diff_avg_yos, *TUITION_SUBSIDIES, METHODS)
| 0.386763 | 0.993116 |
Copyright (c) Microsoft Corporation. All rights reserved.
Licensed under the MIT License.

# Azure Machine Learning Pipeline with AzureBatchStep
This notebook is used to demonstrate the use of AzureBatchStep in Azure Machine Learning Pipeline.
An AzureBatchStep will submit a job to an AzureBatch Compute to run a simple windows executable.
## Azure Machine Learning and Pipeline SDK-specific Imports
```
import azureml.core
from azureml.core import Workspace, Experiment
from azureml.core.compute import ComputeTarget, BatchCompute
from azureml.core.datastore import Datastore
from azureml.data.data_reference import DataReference
from azureml.exceptions import ComputeTargetException
from azureml.pipeline.core import Pipeline, PipelineData
from azureml.pipeline.steps import AzureBatchStep
import os
from os import path
from tempfile import mkdtemp
# Check core SDK version number
print("SDK version:", azureml.core.VERSION)
```
## Initialize Workspace
Initialize a workspace object from persisted configuration. Make sure the config file is present at .\config.json
If you are using an Azure Machine Learning Notebook VM, you are all set. Otherwise, If you don't have a config.json file, please go through the [configuration Notebook](https://aka.ms/pl-config) located [here](https://github.com/Azure/MachineLearningNotebooks).
This sets you up with a working config file that has information on your workspace, subscription id, etc.
```
ws = Workspace.from_config()
print('Workspace Name: ' + ws.name,
'Azure Region: ' + ws.location,
'Subscription Id: ' + ws.subscription_id,
'Resource Group: ' + ws.resource_group, sep = '\n')
```
## Attach Batch Compute to Workspace
To submit jobs to Azure Batch service, you must attach your Azure Batch account to the workspace.
```
batch_compute_name = 'mybatchcompute' # Name to associate with new compute in workspace
# Batch account details needed to attach as compute to workspace
batch_account_name = "<batch_account_name>" # Name of the Batch account
batch_resource_group = "<batch_resource_group>" # Name of the resource group which contains this account
try:
# check if already attached
batch_compute = BatchCompute(ws, batch_compute_name)
except ComputeTargetException:
print('Attaching Batch compute...')
provisioning_config = BatchCompute.attach_configuration(resource_group=batch_resource_group,
account_name=batch_account_name)
batch_compute = ComputeTarget.attach(ws, batch_compute_name, provisioning_config)
batch_compute.wait_for_completion()
print("Provisioning state:{}".format(batch_compute.provisioning_state))
print("Provisioning errors:{}".format(batch_compute.provisioning_errors))
print("Using Batch compute:{}".format(batch_compute.cluster_resource_id))
```
## Setup Datastore
Setting up the Blob storage associated with the workspace.
The following call retrieves the Azure Blob Store associated with your workspace.
Note that workspaceblobstore is **the name of this store and CANNOT BE CHANGED and must be used as is**.
If you want to register another Datastore, please follow the instructions from here:
https://docs.microsoft.com/en-us/azure/machine-learning/service/how-to-access-data#register-a-datastore
```
datastore = Datastore(ws, "workspaceblobstore")
print('Datastore details:')
print('Datastore Account Name: ' + datastore.account_name)
print('Datastore Workspace Name: ' + datastore.workspace.name)
print('Datastore Container Name: ' + datastore.container_name)
```
## Setup Input and Output
For this example we will upload a file in the provided Datastore. These are some helper methods to achieve that.
```
def create_local_file(content, file_name):
# create a file in a local temporary directory
temp_dir = mkdtemp()
with open(path.join(temp_dir, file_name), 'w') as f:
f.write(content)
return temp_dir
def upload_file_to_datastore(datastore, file_name, content):
src_dir = create_local_file(content=content, file_name=file_name)
datastore.upload(src_dir=src_dir, overwrite=True, show_progress=True)
```
Here we associate the input DataReference with an existing file in the provided Datastore. Feel free to upload the file of your choice manually or use the *upload_file_to_datastore* method.
```
file_name="input.txt"
upload_file_to_datastore(datastore=datastore,
file_name=file_name,
content="this is the content of the file")
testdata = DataReference(datastore=datastore,
path_on_datastore=file_name,
data_reference_name="input")
outputdata = PipelineData(name="output", datastore=datastore)
```
## Setup AzureBatch Job Binaries
AzureBatch can run a task within the job and here we put a simple .cmd file to be executed. Feel free to put any binaries in the folder, or modify the .cmd file as needed, they will be uploaded once we create the AzureBatch Step.
```
binaries_folder = "azurebatch/job_binaries"
if not os.path.isdir(binaries_folder):
os.mkdir(binaries_folder)
file_name="azurebatch.cmd"
with open(path.join(binaries_folder, file_name), 'w') as f:
f.write("copy \"%1\" \"%2\"")
```
## Create an AzureBatchStep
AzureBatchStep is used to submit a job to the attached Azure Batch compute.
- **name:** Name of the step
- **pool_id:** Name of the pool, it can be an existing pool, or one that will be created when the job is submitted
- **inputs:** List of inputs that will be processed by the job
- **outputs:** List of outputs the job will create
- **executable:** The executable that will run as part of the job
- **arguments:** Arguments for the executable. They can be plain string format, inputs, outputs or parameters
- **compute_target:** The compute target where the job will run.
- **source_directory:** The local directory with binaries to be executed by the job
Optional parameters:
- **create_pool:** Boolean flag to indicate whether create the pool before running the jobs
- **delete_batch_job_after_finish:** Boolean flag to indicate whether to delete the job from Batch account after it's finished
- **delete_batch_pool_after_finish:** Boolean flag to indicate whether to delete the pool after the job finishes
- **is_positive_exit_code_failure:** Boolean flag to indicate if the job fails if the task exists with a positive code
- **vm_image_urn:** If create_pool is true and VM uses VirtualMachineConfiguration.
Value format: 'urn:publisher:offer:sku'.
Example: urn:MicrosoftWindowsServer:WindowsServer:2012-R2-Datacenter
For more details:
https://docs.microsoft.com/en-us/azure/virtual-machines/windows/cli-ps-findimage#table-of-commonly-used-windows-images and
https://docs.microsoft.com/en-us/azure/virtual-machines/linux/cli-ps-findimage#find-specific-images
- **run_task_as_admin:** Boolean flag to indicate if the task should run with Admin privileges
- **target_compute_nodes:** Assumes create_pool is true, indicates how many compute nodes will be added to the pool
- **source_directory:** Local folder that contains the module binaries, executable, assemblies etc.
- **executable:** Name of the command/executable that will be executed as part of the job
- **arguments:** Arguments for the command/executable
- **inputs:** List of input port bindings
- **outputs:** List of output port bindings
- **vm_size:** If create_pool is true, indicating Virtual machine size of the compute nodes
- **compute_target:** BatchCompute compute
- **allow_reuse:** Whether the module should reuse previous results when run with the same settings/inputs
- **version:** A version tag to denote a change in functionality for the module
```
step = AzureBatchStep(
name="Azure Batch Job",
pool_id="MyPoolName", # Replace this with the pool name of your choice
inputs=[testdata],
outputs=[outputdata],
executable="azurebatch.cmd",
arguments=[testdata, outputdata],
compute_target=batch_compute,
source_directory=binaries_folder,
)
```
## Build and Submit the Pipeline
```
pipeline = Pipeline(workspace=ws, steps=[step])
pipeline_run = Experiment(ws, 'azurebatch_experiment').submit(pipeline)
```
## Visualize the Running Pipeline
```
from azureml.widgets import RunDetails
RunDetails(pipeline_run).show()
```
|
github_jupyter
|
import azureml.core
from azureml.core import Workspace, Experiment
from azureml.core.compute import ComputeTarget, BatchCompute
from azureml.core.datastore import Datastore
from azureml.data.data_reference import DataReference
from azureml.exceptions import ComputeTargetException
from azureml.pipeline.core import Pipeline, PipelineData
from azureml.pipeline.steps import AzureBatchStep
import os
from os import path
from tempfile import mkdtemp
# Check core SDK version number
print("SDK version:", azureml.core.VERSION)
ws = Workspace.from_config()
print('Workspace Name: ' + ws.name,
'Azure Region: ' + ws.location,
'Subscription Id: ' + ws.subscription_id,
'Resource Group: ' + ws.resource_group, sep = '\n')
batch_compute_name = 'mybatchcompute' # Name to associate with new compute in workspace
# Batch account details needed to attach as compute to workspace
batch_account_name = "<batch_account_name>" # Name of the Batch account
batch_resource_group = "<batch_resource_group>" # Name of the resource group which contains this account
try:
# check if already attached
batch_compute = BatchCompute(ws, batch_compute_name)
except ComputeTargetException:
print('Attaching Batch compute...')
provisioning_config = BatchCompute.attach_configuration(resource_group=batch_resource_group,
account_name=batch_account_name)
batch_compute = ComputeTarget.attach(ws, batch_compute_name, provisioning_config)
batch_compute.wait_for_completion()
print("Provisioning state:{}".format(batch_compute.provisioning_state))
print("Provisioning errors:{}".format(batch_compute.provisioning_errors))
print("Using Batch compute:{}".format(batch_compute.cluster_resource_id))
datastore = Datastore(ws, "workspaceblobstore")
print('Datastore details:')
print('Datastore Account Name: ' + datastore.account_name)
print('Datastore Workspace Name: ' + datastore.workspace.name)
print('Datastore Container Name: ' + datastore.container_name)
def create_local_file(content, file_name):
# create a file in a local temporary directory
temp_dir = mkdtemp()
with open(path.join(temp_dir, file_name), 'w') as f:
f.write(content)
return temp_dir
def upload_file_to_datastore(datastore, file_name, content):
src_dir = create_local_file(content=content, file_name=file_name)
datastore.upload(src_dir=src_dir, overwrite=True, show_progress=True)
file_name="input.txt"
upload_file_to_datastore(datastore=datastore,
file_name=file_name,
content="this is the content of the file")
testdata = DataReference(datastore=datastore,
path_on_datastore=file_name,
data_reference_name="input")
outputdata = PipelineData(name="output", datastore=datastore)
binaries_folder = "azurebatch/job_binaries"
if not os.path.isdir(binaries_folder):
os.mkdir(binaries_folder)
file_name="azurebatch.cmd"
with open(path.join(binaries_folder, file_name), 'w') as f:
f.write("copy \"%1\" \"%2\"")
step = AzureBatchStep(
name="Azure Batch Job",
pool_id="MyPoolName", # Replace this with the pool name of your choice
inputs=[testdata],
outputs=[outputdata],
executable="azurebatch.cmd",
arguments=[testdata, outputdata],
compute_target=batch_compute,
source_directory=binaries_folder,
)
pipeline = Pipeline(workspace=ws, steps=[step])
pipeline_run = Experiment(ws, 'azurebatch_experiment').submit(pipeline)
from azureml.widgets import RunDetails
RunDetails(pipeline_run).show()
| 0.347426 | 0.87982 |
***
***
# 使用Graphlab实现推荐系统的隐语义模型
> ## (Latent Factor Model, LFM)
***
***
```
import graphlab
graphlab.canvas.set_target("ipynb")
rating_sf = graphlab.SFrame('ratings')
users = graphlab.SFrame('users')
items = graphlab.SFrame('items')
rating_sf.show()
dir(graphlab.recommender)
(train, test) = graphlab.recommender.util.random_split_by_user(rating_sf, 'user_id', 'movie_id')
from graphlab import item_similarity_recommender
itemcf = item_similarity_recommender.create(
train[train['rating'] > 4], 'user_id', 'movie_id')
pop = graphlab.popularity_recommender.create(
train[train['rating'] > 4], 'user_id', 'movie_id')
m = graphlab.recommender.create(
train, 'user_id', 'movie_id', 'rating')
m
m['coefficients']
graphlab.recommender.util.compare_models(test[test['rating'] > 4],
[pop, itemcf, m],
user_sample=0.2,
metric='precision_recall')
```
### Optimizing for ranking
```
m_rank = graphlab.recommender.ranking_factorization_recommender.create(
train, 'user_id', 'movie_id', 'rating',
unobserved_rating_value=3)
results = graphlab.recommender.util.compare_models(
test[test['rating'] > 4],
[pop, itemcf, m, m_rank],
user_sample=0.2,
metric='precision_recall')
results[3]['precision_recall_overall']
```
### Experimenting with side features
```
user_sf = graphlab.SFrame('users')
item_sf = graphlab.SFrame('items')
m_user = graphlab.recommender.create(train, 'user_id', 'movie_id', 'rating',
user_data=user_sf)
m_item = graphlab.recommender.create(train, 'user_id', 'movie_id', 'rating',
item_data=item_sf)
m_both = graphlab.recommender.create(train, 'user_id', 'movie_id', 'rating',
user_data=user_sf, item_data=item_sf)
m_both
results = graphlab.recommender.util.compare_models(test, [m, m_user, m_item, m_both], user_sample=0.2)
[results[i]['rmse_overall'] for i in range(len(results))]
results[0]['rmse_by_item'].show()
graphlab.recommender.util.compare_models(test[test['rating'] > 4],
[m_rank, m_both],
user_sample=0.2,
metric='precision_recall')
```
### Factorization machines
```
fm = graphlab.recommender.create(train.head(10000), 'user_id', 'movie_id', 'rating',
method='factorization_model',
item_data=item_sf,
sgd_step_size=0.09,
max_iterations=10)
```
|
github_jupyter
|
import graphlab
graphlab.canvas.set_target("ipynb")
rating_sf = graphlab.SFrame('ratings')
users = graphlab.SFrame('users')
items = graphlab.SFrame('items')
rating_sf.show()
dir(graphlab.recommender)
(train, test) = graphlab.recommender.util.random_split_by_user(rating_sf, 'user_id', 'movie_id')
from graphlab import item_similarity_recommender
itemcf = item_similarity_recommender.create(
train[train['rating'] > 4], 'user_id', 'movie_id')
pop = graphlab.popularity_recommender.create(
train[train['rating'] > 4], 'user_id', 'movie_id')
m = graphlab.recommender.create(
train, 'user_id', 'movie_id', 'rating')
m
m['coefficients']
graphlab.recommender.util.compare_models(test[test['rating'] > 4],
[pop, itemcf, m],
user_sample=0.2,
metric='precision_recall')
m_rank = graphlab.recommender.ranking_factorization_recommender.create(
train, 'user_id', 'movie_id', 'rating',
unobserved_rating_value=3)
results = graphlab.recommender.util.compare_models(
test[test['rating'] > 4],
[pop, itemcf, m, m_rank],
user_sample=0.2,
metric='precision_recall')
results[3]['precision_recall_overall']
user_sf = graphlab.SFrame('users')
item_sf = graphlab.SFrame('items')
m_user = graphlab.recommender.create(train, 'user_id', 'movie_id', 'rating',
user_data=user_sf)
m_item = graphlab.recommender.create(train, 'user_id', 'movie_id', 'rating',
item_data=item_sf)
m_both = graphlab.recommender.create(train, 'user_id', 'movie_id', 'rating',
user_data=user_sf, item_data=item_sf)
m_both
results = graphlab.recommender.util.compare_models(test, [m, m_user, m_item, m_both], user_sample=0.2)
[results[i]['rmse_overall'] for i in range(len(results))]
results[0]['rmse_by_item'].show()
graphlab.recommender.util.compare_models(test[test['rating'] > 4],
[m_rank, m_both],
user_sample=0.2,
metric='precision_recall')
fm = graphlab.recommender.create(train.head(10000), 'user_id', 'movie_id', 'rating',
method='factorization_model',
item_data=item_sf,
sgd_step_size=0.09,
max_iterations=10)
| 0.403449 | 0.630728 |
## Space Project (Part 1) - Our Planets!
This notebook will generate an image of the planets and the distance between each other based on real data<br>
It makes use of simple webscraping, pandas and image processing with Pillow
<br>Source: https://nssdc.gsfc.nasa.gov/planetary/factsheet/
### Imports
```
import pandas as pd
import matplotlib.pyplot as plt
import glob
from PIL import Image
from PIL import ImageFont
from PIL import ImageDraw
from IPython.display import Image as ImageDisplay
```
#### 1. Load Into Dataframe and Set Index
```
#Load in dataframe and set index
df = pd.read_html('https://nssdc.gsfc.nasa.gov/planetary/factsheet/', header = 0)[0]
df = df.set_index('Unnamed: 0')
df.drop(df.tail(1).index,inplace=True)
df.index.name = None
#Processing
df2 = df
df2['MOON'] = df['MOON'].str.replace('*', '', regex=True)
df2 = df.transpose()
df2['Distance from Sun (106 km)'] = df2['Distance from Sun (106 km)'].astype(float)
df2['Diameter (km)'] = df2['Diameter (km)'].astype(float)
#Scales a planet to 1000 times it's size for viewability
df2['Diameter (106 km) times 10^3'] = df2['Diameter (km)'] / 1000000 * 1000
#Show dataframe
df2
#Specify image width and heigth
width = int(df2['Distance from Sun (106 km)'].max()) + 100
heigth = int(df2['Diameter (106 km) times 10^3'].max() * 4)
```
#### 2. Create Image of Our Planets
```
#Specify the location of the planet images
planets = glob.glob('PIC\*.png')
#Create canvas
our_planets = Image.new("RGB", (width, heigth), "black")
#Add planets on canvas
for planet in planets:
with open(planet, 'rb') as file:
#Define variables
planetname = planet.replace('PIC\\','').replace('.png','')
distance_from_sun = df2.at[planetname.upper(), 'Distance from Sun (106 km)']
size_proportion = df2.at[planetname.upper(), 'Diameter (106 km) times 10^3']
if planetname == 'saturn' or planetname == 'uranus':
size_proportion *= 1.35 #Dealing with the ring's in the PNG
number_of_moons = df2.at[planetname.upper(), 'Number of Moons']
mean_temperature = df2.at[planetname.upper(), 'Mean Temperature (C)']
#Image processing, the image has a padding before and after the last planet of 50 pixels
img = Image.open(file)
img.thumbnail((int(size_proportion), int(size_proportion)))
left_padding = 50 + int((distance_from_sun))
top_padding = int((heigth - img.width) / 2)
resized_img = img
#Text
font = ImageFont.truetype("futur.ttf", 10)
draw = ImageDraw.Draw(our_planets)
if int(number_of_moons) > 1:
draw.text((left_padding + (size_proportion / 2), top_padding + size_proportion + 8), f'{planetname}\n{mean_temperature} c\n{number_of_moons} moons', (255, 255, 255), font=font, anchor='ma', align='center')
elif int(number_of_moons) == 1:
draw.text((left_padding + (size_proportion / 2), top_padding + size_proportion + 8), f'{planetname}\n{mean_temperature} c\n{number_of_moons} moon', (255, 255, 255), font=font, anchor='ma', align='center')
else:
draw.text((left_padding + (size_proportion / 2), top_padding + size_proportion + 8), f'{planetname}\n{mean_temperature} c', (255, 255, 255), font=font, anchor='ma', align='center')
#Description
if planetname == 'earth':
font_title = ImageFont.truetype("futur.ttf", 8)
draw.text((width/2, int(heigth/100*96)), f'Planets are displayed thousand times their actual size to scale relative to each other\nsource: nasa', (255, 255, 255), anchor='ma', align='center', font=font_title)
#Put planet on its rightful spot in its rightful size!
our_planets.paste(resized_img, (left_padding, top_padding))
our_planets.save('our_planets.png')
```
#### 3. Result
```
ImageDisplay(filename='our_planets.png')
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import glob
from PIL import Image
from PIL import ImageFont
from PIL import ImageDraw
from IPython.display import Image as ImageDisplay
#Load in dataframe and set index
df = pd.read_html('https://nssdc.gsfc.nasa.gov/planetary/factsheet/', header = 0)[0]
df = df.set_index('Unnamed: 0')
df.drop(df.tail(1).index,inplace=True)
df.index.name = None
#Processing
df2 = df
df2['MOON'] = df['MOON'].str.replace('*', '', regex=True)
df2 = df.transpose()
df2['Distance from Sun (106 km)'] = df2['Distance from Sun (106 km)'].astype(float)
df2['Diameter (km)'] = df2['Diameter (km)'].astype(float)
#Scales a planet to 1000 times it's size for viewability
df2['Diameter (106 km) times 10^3'] = df2['Diameter (km)'] / 1000000 * 1000
#Show dataframe
df2
#Specify image width and heigth
width = int(df2['Distance from Sun (106 km)'].max()) + 100
heigth = int(df2['Diameter (106 km) times 10^3'].max() * 4)
#Specify the location of the planet images
planets = glob.glob('PIC\*.png')
#Create canvas
our_planets = Image.new("RGB", (width, heigth), "black")
#Add planets on canvas
for planet in planets:
with open(planet, 'rb') as file:
#Define variables
planetname = planet.replace('PIC\\','').replace('.png','')
distance_from_sun = df2.at[planetname.upper(), 'Distance from Sun (106 km)']
size_proportion = df2.at[planetname.upper(), 'Diameter (106 km) times 10^3']
if planetname == 'saturn' or planetname == 'uranus':
size_proportion *= 1.35 #Dealing with the ring's in the PNG
number_of_moons = df2.at[planetname.upper(), 'Number of Moons']
mean_temperature = df2.at[planetname.upper(), 'Mean Temperature (C)']
#Image processing, the image has a padding before and after the last planet of 50 pixels
img = Image.open(file)
img.thumbnail((int(size_proportion), int(size_proportion)))
left_padding = 50 + int((distance_from_sun))
top_padding = int((heigth - img.width) / 2)
resized_img = img
#Text
font = ImageFont.truetype("futur.ttf", 10)
draw = ImageDraw.Draw(our_planets)
if int(number_of_moons) > 1:
draw.text((left_padding + (size_proportion / 2), top_padding + size_proportion + 8), f'{planetname}\n{mean_temperature} c\n{number_of_moons} moons', (255, 255, 255), font=font, anchor='ma', align='center')
elif int(number_of_moons) == 1:
draw.text((left_padding + (size_proportion / 2), top_padding + size_proportion + 8), f'{planetname}\n{mean_temperature} c\n{number_of_moons} moon', (255, 255, 255), font=font, anchor='ma', align='center')
else:
draw.text((left_padding + (size_proportion / 2), top_padding + size_proportion + 8), f'{planetname}\n{mean_temperature} c', (255, 255, 255), font=font, anchor='ma', align='center')
#Description
if planetname == 'earth':
font_title = ImageFont.truetype("futur.ttf", 8)
draw.text((width/2, int(heigth/100*96)), f'Planets are displayed thousand times their actual size to scale relative to each other\nsource: nasa', (255, 255, 255), anchor='ma', align='center', font=font_title)
#Put planet on its rightful spot in its rightful size!
our_planets.paste(resized_img, (left_padding, top_padding))
our_planets.save('our_planets.png')
ImageDisplay(filename='our_planets.png')
| 0.549641 | 0.934753 |
# Boolean Indexing
## Recipes
* [Calculating boolean statistics](#Calculating-boolean-statistics)
* [Constructing multiple boolean conditions](#Constructing-multiple-boolean-conditions)
* [Filtering with boolean indexing](#Filtering-with-boolean-indexing)
* [Replicating boolean indexing with index selection](#Replicating-boolean-indexing-with-index-selection)
* [Selecting with unique and sorted indexes](#Selecting-with-unique-and-sorted-indexes)
* [Gaining perspective on stock prices](#Gaining-perspective-on-stock-prices)
* [Translating SQL WHERE clauses](#Translating-SQL-WHERE-clauses)
* [Determining the normality of stock market returns](#Determining-the-normality-of-stock-market-returns)
* [Improving readability of boolean indexing with the query method](#Improving-readability-of-boolean-indexing-with-the-query-method)
* [Preserving Series with the where method](#Preserving-Series-with-the-where-method)
* [Masking DataFrame rows](#Masking-DataFrame-rows)
* [Selecting with booleans, integer location, and labels](#Selecting-with-booleans,-integer-location-and-labels)
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
```
# Introduction
Filtering data from a dataset is one of the most common and basic operations. There are numerous ways to filter (or subset) data in pandas with boolean indexing. Boolean indexing (also known as boolean selection) can be a confusing term, but for the purposes of pandas, it refers to selecting rows by providing a boolean value (True or False) for each row. These boolean values are usually stored in a Series or NumPy ndarray and are usually created by applying a boolean condition to one or more columns in a DataFrame. We begin by creating boolean Series and calculating statistics on them and then move on to creating more complex conditionals before using boolean indexing in a wide variety of ways to filter data.
# Calculating boolean statistics
```
# Read in the movie dataset, set the index to the movie title, and inspect the first few rows:
movie = pd.read_csv('data/movie.csv', index_col='movie_title')
movie.head()
# Determine whether the duration of each movie is longer than two hours by using the greater than comparison operator with the duration Series::
movie_2_hours = movie['duration'] > 120
movie_2_hours.head(10)
# We can now use this Series to determine the number of movies that are longer than two hours:
movie_2_hours.sum()
# To find the percentage of movies in the dataset longer than two hours, use the mean method:
movie_2_hours.mean()
movie['duration'].dropna().gt(120).mean()
movie_2_hours.describe()
```
# Constructing multiple boolean conditions
In Python, boolean expressions use the built-in logical operators and, or, and not. These keywords do not work with boolean indexing in pandas and are respectively replaced with &, |, and ~. Additionally, each expression must be wrapped in parentheses or an error will be raised.
```
# Load in the movie dataset and set the index as the title:
movie = pd.read_csv('data/movie.csv', index_col='movie_title')
# Create a variable to hold each set of criteria independently as a boolean Series:
criteria1 = movie.imdb_score > 8
criteria2 = movie.content_rating == 'PG-13'
criteria3 = (movie.title_year < 2000) | (movie.title_year >= 2010)
criteria2.head()
# Combine all the criteria together into a single boolean Series:
criteria_final = criteria1 & criteria2 & criteria3
criteria_final.head()
```
# Filtering with boolean indexing
Boolean selection for Series and DataFrame objects is virtually identical. Both work by passing a Series of booleans indexed identically to the object being filtered to the indexing operator.
```
# Read in the movie dataset, set the index to the movie_title, and create the first set of criteria:
movie = pd.read_csv('data/movie.csv', index_col='movie_title')
crit_a1 = movie.imdb_score > 8
crit_a2 = movie.content_rating == 'PG-13'
crit_a3 = (movie.title_year < 2000) | (movie.title_year > 2009)
final_crit_a = crit_a1 & crit_a2 & crit_a3
# Create criteria for the second set of movies:
crit_b1 = movie.imdb_score < 5
crit_b2 = movie.content_rating == 'R'
crit_b3 = (movie.title_year >= 2000) & (movie.title_year <= 2010)
final_crit_b = crit_b1 & crit_b2 & crit_b3
```
Combine the two sets of criteria using the pandas or operator. This yields a boolean Series of all movies that are members of either set:
```
final_crit_all = final_crit_a | final_crit_b
final_crit_all.head()
# Once you have your boolean Series, you simply pass it to the indexing operator to filter the data:
movie[final_crit_all].head()
```
We have successfully filtered the data and all the columns of the DataFrame. We can't easily perform a manual check to determine whether the filter worked correctly. Let's filter both rows and columns with the .loc indexer:
```
cols = ['imdb_score', 'content_rating', 'title_year']
movie_filtered = movie.loc[final_crit_all, cols]
movie_filtered.head(10)
```
# Replicating boolean indexing with index selection
It is possible to replicate specific cases of boolean selection by taking advantage of the index. Selection through the index is more intuitive and makes for greater readability.
```
# Read in the college dataset and use boolean indexing to select all institutions from the state of Texas (TX):
college = pd.read_csv('data/college.csv')
college[college['STABBR'] == 'TX'].head()
# To replicate this using index selection, we need to move the STABBR column into the index. We can then use label-based selection with the .loc indexer:
college2 = college.set_index('STABBR')
college2.loc['TX'].head()
# Let's compare the speed of both methods:
%timeit college[college['STABBR'] == 'TX']
%timeit college2.loc['TX']
# Boolean indexing takes three times as long as index selection. As setting the index does not come for free, let's time that operation as well:
%timeit college2 = college.set_index('STABBR')
```
# Selecting with unique and sorted indexes
Index selection performance drastically improves when the index is unique or sorted. The prior recipe used an unsorted index that contained duplicates, which makes for relatively slow selection
```
# Read in the college dataset, create a separate DataFrame with STABBR as the index, and check whether the index is sorted:
college = pd.read_csv('data/college.csv')
college2 = college.set_index('STABBR')
college2.index.is_monotonic
# Sort the index from college2 and store it as another object:
college3 = college2.sort_index()
college3.index.is_monotonic
# Time the selection of the state of Texas (TX) from all three DataFrames:
%timeit college[college['STABBR'] == 'TX']
%timeit college2.loc['TX']
%timeit college3.loc['TX']
# The sorted index performs nearly an order of magnitude faster than boolean selection. Let's now turn towards unique indexes. For this, we use the institution name as the index:
college_unique = college.set_index('INSTNM')
college_unique.index.is_unique
# Let's select Stanford University with boolean indexing:
college[college['INSTNM'] == 'Stanford University']
# Let's select Stanford University with index selection:
college_unique.loc['Stanford University']
# They both produce the same data, just with different objects. Let's time each approach:
%timeit college[college['INSTNM'] == 'Stanford University']
%timeit college_unique.loc['Stanford University']
```
# Gaining perspective on stock prices
Investors who have purchased long stock positions would obviously like to sell stocks at or near their all-time highs. This, of course, is very difficult to do in practice, especially if a stock price has only spent a small portion of its history above a certain threshold. We can use boolean indexing to find all points in time that a stock has spent above or below a certain value. This exercise may help us gain perspective as to what a common range for some stock to be trading within.
```
# Read in the Schlumberger stock data, put the Date column into the index, and convert it to a DatetimeIndex:
slb = pd.read_csv('data/slb_stock.csv', index_col='Date', parse_dates=['Date'])
slb.head()
# Select the closing price as a Series and use the describe method to return summary statistics as a Series:
slb_close = slb['Close']
slb_summary = slb_close.describe(percentiles=[.1, .9])
slb_summary
# Using boolean selection, select all closing prices in the upper or lower tenth percentile:
upper_10 = slb_summary.loc['90%']
lower_10 = slb_summary.loc['10%']
criteria = (slb_close < lower_10) | (slb_close > upper_10)
slb_top_bottom_10 = slb_close[criteria]
```
Plot the resulting filtered Series in light gray on top of all closing prices in black.Use the matplotlib library to draw horizontal lines at the tenth and ninetieth percentiles:
```
slb_close.plot(color='black', figsize=(12,6))
slb_top_bottom_10.plot(marker='o', style=' ', ms=4, color='blue')
xmin = criteria.index[0]
xmax = criteria.index[-1]
plt.hlines(y=[lower_10, upper_10], xmin=xmin, xmax=xmax,color='black')
```
# Determining the normality of stock market returns
In elementary statistics textbooks, the normal distribution is heavily relied upon to describe many different populations of data. Although many random processes do appear to look like normal distributions most of the time, real-life tends to be more complex. Stock market returns are a prime example of a distribution that can look fairly normal but in actuality be quite far off.
```
# Load Amazon stock data and set the date as the index:
amzn = pd.read_csv('data/amzn_stock.csv', index_col='Date', parse_dates=['Date'])
amzn.head()
# Create a Series by selecting only the closing price and then using the pct_change method to get the daily rate of return:
amzn_daily_return = amzn.Close.pct_change()
amzn_daily_return.head()
# Drop the missing value and plot a histogram of the returns to visually inspect the distribution:
amzn_daily_return = amzn_daily_return.dropna()
amzn_daily_return.hist(bins=20)
mean = amzn_daily_return.mean()
std = amzn_daily_return.std()
abs_z_score = amzn_daily_return.sub(mean).abs().div(std)
pcts = [abs_z_score.lt(i).mean() for i in range(1,4)]
print('{:.3f} fall within 1 standard deviation. '
'{:.3f} within 2 and {:.3f} within 3'.format(*pcts))
def test_return_normality(stock_data):
close = stock_data['Close']
daily_return = close.pct_change().dropna()
daily_return.hist(bins=20)
mean = daily_return.mean()
std = daily_return.std()
abs_z_score = abs(daily_return - mean) / std
pcts = [abs_z_score.lt(i).mean() for i in range(1,4)]
print('{:.3f} fall within 1 standard deviation. '
'{:.3f} within 2 and {:.3f} within 3'.format(*pcts))
slb = pd.read_csv('data/slb_stock.csv',
index_col='Date', parse_dates=['Date'])
test_return_normality(slb)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# Read in the movie dataset, set the index to the movie title, and inspect the first few rows:
movie = pd.read_csv('data/movie.csv', index_col='movie_title')
movie.head()
# Determine whether the duration of each movie is longer than two hours by using the greater than comparison operator with the duration Series::
movie_2_hours = movie['duration'] > 120
movie_2_hours.head(10)
# We can now use this Series to determine the number of movies that are longer than two hours:
movie_2_hours.sum()
# To find the percentage of movies in the dataset longer than two hours, use the mean method:
movie_2_hours.mean()
movie['duration'].dropna().gt(120).mean()
movie_2_hours.describe()
# Load in the movie dataset and set the index as the title:
movie = pd.read_csv('data/movie.csv', index_col='movie_title')
# Create a variable to hold each set of criteria independently as a boolean Series:
criteria1 = movie.imdb_score > 8
criteria2 = movie.content_rating == 'PG-13'
criteria3 = (movie.title_year < 2000) | (movie.title_year >= 2010)
criteria2.head()
# Combine all the criteria together into a single boolean Series:
criteria_final = criteria1 & criteria2 & criteria3
criteria_final.head()
# Read in the movie dataset, set the index to the movie_title, and create the first set of criteria:
movie = pd.read_csv('data/movie.csv', index_col='movie_title')
crit_a1 = movie.imdb_score > 8
crit_a2 = movie.content_rating == 'PG-13'
crit_a3 = (movie.title_year < 2000) | (movie.title_year > 2009)
final_crit_a = crit_a1 & crit_a2 & crit_a3
# Create criteria for the second set of movies:
crit_b1 = movie.imdb_score < 5
crit_b2 = movie.content_rating == 'R'
crit_b3 = (movie.title_year >= 2000) & (movie.title_year <= 2010)
final_crit_b = crit_b1 & crit_b2 & crit_b3
final_crit_all = final_crit_a | final_crit_b
final_crit_all.head()
# Once you have your boolean Series, you simply pass it to the indexing operator to filter the data:
movie[final_crit_all].head()
cols = ['imdb_score', 'content_rating', 'title_year']
movie_filtered = movie.loc[final_crit_all, cols]
movie_filtered.head(10)
# Read in the college dataset and use boolean indexing to select all institutions from the state of Texas (TX):
college = pd.read_csv('data/college.csv')
college[college['STABBR'] == 'TX'].head()
# To replicate this using index selection, we need to move the STABBR column into the index. We can then use label-based selection with the .loc indexer:
college2 = college.set_index('STABBR')
college2.loc['TX'].head()
# Let's compare the speed of both methods:
%timeit college[college['STABBR'] == 'TX']
%timeit college2.loc['TX']
# Boolean indexing takes three times as long as index selection. As setting the index does not come for free, let's time that operation as well:
%timeit college2 = college.set_index('STABBR')
# Read in the college dataset, create a separate DataFrame with STABBR as the index, and check whether the index is sorted:
college = pd.read_csv('data/college.csv')
college2 = college.set_index('STABBR')
college2.index.is_monotonic
# Sort the index from college2 and store it as another object:
college3 = college2.sort_index()
college3.index.is_monotonic
# Time the selection of the state of Texas (TX) from all three DataFrames:
%timeit college[college['STABBR'] == 'TX']
%timeit college2.loc['TX']
%timeit college3.loc['TX']
# The sorted index performs nearly an order of magnitude faster than boolean selection. Let's now turn towards unique indexes. For this, we use the institution name as the index:
college_unique = college.set_index('INSTNM')
college_unique.index.is_unique
# Let's select Stanford University with boolean indexing:
college[college['INSTNM'] == 'Stanford University']
# Let's select Stanford University with index selection:
college_unique.loc['Stanford University']
# They both produce the same data, just with different objects. Let's time each approach:
%timeit college[college['INSTNM'] == 'Stanford University']
%timeit college_unique.loc['Stanford University']
# Read in the Schlumberger stock data, put the Date column into the index, and convert it to a DatetimeIndex:
slb = pd.read_csv('data/slb_stock.csv', index_col='Date', parse_dates=['Date'])
slb.head()
# Select the closing price as a Series and use the describe method to return summary statistics as a Series:
slb_close = slb['Close']
slb_summary = slb_close.describe(percentiles=[.1, .9])
slb_summary
# Using boolean selection, select all closing prices in the upper or lower tenth percentile:
upper_10 = slb_summary.loc['90%']
lower_10 = slb_summary.loc['10%']
criteria = (slb_close < lower_10) | (slb_close > upper_10)
slb_top_bottom_10 = slb_close[criteria]
slb_close.plot(color='black', figsize=(12,6))
slb_top_bottom_10.plot(marker='o', style=' ', ms=4, color='blue')
xmin = criteria.index[0]
xmax = criteria.index[-1]
plt.hlines(y=[lower_10, upper_10], xmin=xmin, xmax=xmax,color='black')
# Load Amazon stock data and set the date as the index:
amzn = pd.read_csv('data/amzn_stock.csv', index_col='Date', parse_dates=['Date'])
amzn.head()
# Create a Series by selecting only the closing price and then using the pct_change method to get the daily rate of return:
amzn_daily_return = amzn.Close.pct_change()
amzn_daily_return.head()
# Drop the missing value and plot a histogram of the returns to visually inspect the distribution:
amzn_daily_return = amzn_daily_return.dropna()
amzn_daily_return.hist(bins=20)
mean = amzn_daily_return.mean()
std = amzn_daily_return.std()
abs_z_score = amzn_daily_return.sub(mean).abs().div(std)
pcts = [abs_z_score.lt(i).mean() for i in range(1,4)]
print('{:.3f} fall within 1 standard deviation. '
'{:.3f} within 2 and {:.3f} within 3'.format(*pcts))
def test_return_normality(stock_data):
close = stock_data['Close']
daily_return = close.pct_change().dropna()
daily_return.hist(bins=20)
mean = daily_return.mean()
std = daily_return.std()
abs_z_score = abs(daily_return - mean) / std
pcts = [abs_z_score.lt(i).mean() for i in range(1,4)]
print('{:.3f} fall within 1 standard deviation. '
'{:.3f} within 2 and {:.3f} within 3'.format(*pcts))
slb = pd.read_csv('data/slb_stock.csv',
index_col='Date', parse_dates=['Date'])
test_return_normality(slb)
| 0.79649 | 0.973919 |
```
import h5py
import numpy as np
import argparse as ap
from pathlib import Path
import os
from tqdm import tqdm
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(font_scale=1.9, rc={'text.usetex': True})
sns.set_style('whitegrid')
datasets = ['red_sphere_final', 'blue_sphere_final', 'red_rectangle_final']
titles = ['Same color and shape', 'Different color', 'Different shape']
p_denn_l1 = Path('log/train:red_sphere_repulsive:green_sphere_l:1_b:0.1')
p_denn_l10 = Path('log/train:red_sphere_repulsive:green_sphere_l:10_b:0.1')
p_deep_ensemble = Path('log/new_ensemble_train:red_sphere')
# ===============================================================================
# showing the plots
# ===============================================================================
fig, axes = plt.subplots(1, 3, figsize=(10, 4), squeeze=False, sharey=True)
for i, (dataset, title) in enumerate(zip(datasets, titles)):
ax = axes[0, i]
ax.set_ylim([0, 6e3])
with h5py.File(p_deep_ensemble / '{}.h5'.format(dataset), 'r') as cf:
baseline_test_std = cf.get('std')[:]
with h5py.File(p_denn_l1 / '{}.h5'.format(dataset), 'r') as f:
test_std1 = f.get('std')[:]
with h5py.File(p_denn_l10 / '{}.h5'.format(dataset), 'r') as f:
test_std2 = f.get('std')[:]
# Preparing data for plots
# --> We take the mean since the differences between the actions do not seem to be significative
baseline_test = baseline_test_std.mean(1)
method_test1 = test_std1.mean(1)
method_test2 = test_std2.mean(1)
# display ticks
#ax.set_xticks([0, 0.5])
plt.setp(ax.get_xticklabels(), visible=True)
plt.setp(ax.get_xticklabels()[0], ha='left')
plt.setp(ax.get_xticklabels()[-1], ha='right')
if i == 0:
plt.setp(ax.get_yticklabels()[0], va='bottom')
plt.setp(ax.get_yticklabels()[-1], va='top')
#bins = np.histogram(np.hstack((test_std[:,0], eval_std[:,0], baseline_test_std[:,0], baseline_eval_std[:,0])), bins=40)[1] # defining the bins
bins = np.arange(0, 0.5, 0.01) # defining the bins
# Proposed method
sns.distplot(method_test1, hist=True, kde=False, bins=bins,
kde_kws={'shade': True, 'linewidth': 2},
hist_kws={"histtype": "step", 'log': False, 'linewidth': 3, 'linestyle':('solid'), 'alpha':.5},
label=r'DENN, $\lambda=1$', ax=ax, color='blue')
# Proposed method
sns.distplot(method_test2, hist=True, kde=False, bins=bins,
kde_kws={'shade': True, 'linewidth': 2},
hist_kws={"histtype": "step", 'log': False, 'linewidth': 3, 'linestyle':('dashed'), 'alpha':.5},
label=r'DENN, $\lambda=10$', ax=ax, color='black')
# Baseline method
sns.distplot(baseline_test, hist=True, kde=False, bins=bins,
kde_kws={'shade': True, 'linewidth': 2},
hist_kws={"histtype": "step", 'log': False, 'linewidth': 3, 'linestyle':('solid'), 'alpha':.5},
label='Deep ensemble', ax=ax, color='orangered')
ax.set_xlabel('Avg. std over actions')
if i == 0:
ax.set_ylabel('Count')
else:
ax.set_ylabel('')
ax.set_xlim(0.0, 0.5)
if i == 0:
ax.legend(loc=(.1, .45), prop={'size':18})
ax.set_title(title)
plt.tight_layout()
plt.subplots_adjust(wspace = .1)
plt.ticklabel_format(style='sci', axis='y', scilimits=(0,0))
t = axes[0,0].yaxis.get_offset_text()
t.set_x(-.25)
plt.show()
# figname = 'reacher-predictive-std.pdf'
# pfig = Path('img') / figname
# plt.savefig(pfig)
```
|
github_jupyter
|
import h5py
import numpy as np
import argparse as ap
from pathlib import Path
import os
from tqdm import tqdm
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(font_scale=1.9, rc={'text.usetex': True})
sns.set_style('whitegrid')
datasets = ['red_sphere_final', 'blue_sphere_final', 'red_rectangle_final']
titles = ['Same color and shape', 'Different color', 'Different shape']
p_denn_l1 = Path('log/train:red_sphere_repulsive:green_sphere_l:1_b:0.1')
p_denn_l10 = Path('log/train:red_sphere_repulsive:green_sphere_l:10_b:0.1')
p_deep_ensemble = Path('log/new_ensemble_train:red_sphere')
# ===============================================================================
# showing the plots
# ===============================================================================
fig, axes = plt.subplots(1, 3, figsize=(10, 4), squeeze=False, sharey=True)
for i, (dataset, title) in enumerate(zip(datasets, titles)):
ax = axes[0, i]
ax.set_ylim([0, 6e3])
with h5py.File(p_deep_ensemble / '{}.h5'.format(dataset), 'r') as cf:
baseline_test_std = cf.get('std')[:]
with h5py.File(p_denn_l1 / '{}.h5'.format(dataset), 'r') as f:
test_std1 = f.get('std')[:]
with h5py.File(p_denn_l10 / '{}.h5'.format(dataset), 'r') as f:
test_std2 = f.get('std')[:]
# Preparing data for plots
# --> We take the mean since the differences between the actions do not seem to be significative
baseline_test = baseline_test_std.mean(1)
method_test1 = test_std1.mean(1)
method_test2 = test_std2.mean(1)
# display ticks
#ax.set_xticks([0, 0.5])
plt.setp(ax.get_xticklabels(), visible=True)
plt.setp(ax.get_xticklabels()[0], ha='left')
plt.setp(ax.get_xticklabels()[-1], ha='right')
if i == 0:
plt.setp(ax.get_yticklabels()[0], va='bottom')
plt.setp(ax.get_yticklabels()[-1], va='top')
#bins = np.histogram(np.hstack((test_std[:,0], eval_std[:,0], baseline_test_std[:,0], baseline_eval_std[:,0])), bins=40)[1] # defining the bins
bins = np.arange(0, 0.5, 0.01) # defining the bins
# Proposed method
sns.distplot(method_test1, hist=True, kde=False, bins=bins,
kde_kws={'shade': True, 'linewidth': 2},
hist_kws={"histtype": "step", 'log': False, 'linewidth': 3, 'linestyle':('solid'), 'alpha':.5},
label=r'DENN, $\lambda=1$', ax=ax, color='blue')
# Proposed method
sns.distplot(method_test2, hist=True, kde=False, bins=bins,
kde_kws={'shade': True, 'linewidth': 2},
hist_kws={"histtype": "step", 'log': False, 'linewidth': 3, 'linestyle':('dashed'), 'alpha':.5},
label=r'DENN, $\lambda=10$', ax=ax, color='black')
# Baseline method
sns.distplot(baseline_test, hist=True, kde=False, bins=bins,
kde_kws={'shade': True, 'linewidth': 2},
hist_kws={"histtype": "step", 'log': False, 'linewidth': 3, 'linestyle':('solid'), 'alpha':.5},
label='Deep ensemble', ax=ax, color='orangered')
ax.set_xlabel('Avg. std over actions')
if i == 0:
ax.set_ylabel('Count')
else:
ax.set_ylabel('')
ax.set_xlim(0.0, 0.5)
if i == 0:
ax.legend(loc=(.1, .45), prop={'size':18})
ax.set_title(title)
plt.tight_layout()
plt.subplots_adjust(wspace = .1)
plt.ticklabel_format(style='sci', axis='y', scilimits=(0,0))
t = axes[0,0].yaxis.get_offset_text()
t.set_x(-.25)
plt.show()
# figname = 'reacher-predictive-std.pdf'
# pfig = Path('img') / figname
# plt.savefig(pfig)
| 0.471467 | 0.54583 |
# [Dictionaries](https://docs.python.org/3/library/stdtypes.html#dict)
Collections of `key`-`value` pairs.
Dictionaries can be created by placing a comma-separated list of key: value pairs within braces,
for example: {'jack': 4098, 'sjoerd': 4127} or {4098: 'jack', 4127: 'sjoerd'},
or by the dict constructor.
```
my_dict1 = {'jack': 4098, 'sjoerd': 4127}
my_dict2 = dict({4098: 'jack', 4127: 'sjoerd'})
print(my_dict1)
print(my_dict2)
my_empty_dict = {} # alternative: my_empty_dict = dict()
print('dict: {}, type: {}'.format(my_empty_dict, type(my_empty_dict)))
```
## Initialization
```
dict1 = {'value1': 1.6, 'value2': 10, 'name': 'John Doe'}
dict2 = dict(value1=1.6, value2=10, name='John Doe')
print(dict1)
print(dict2)
print('equal: {}'.format(dict1 == dict2))
print('length: {}'.format(len(dict1)))
```
## Creating dictionary from lists
```
keys = ['Ten', 'Twenty', 'Thirty']
values = [10, 20, 30]
sampleDict = dict(zip(keys, values))
print(sampleDict)
```
## `dict.keys(), dict.values(), dict.items()`
```
print('keys: {}'.format(dict1.keys())) # list of all keys
print('values: {}'.format(dict1.values())) # list of all values
print('items: {}'.format(dict1.items())) # list of all key,value pairs
```
## Accessing and setting values
```
my_dict = {}
my_dict['key1'] = 'value1'
my_dict['key2'] = 99
my_dict['key1'] = 'new value' # overriding existing value
print(my_dict)
print('value of key1: {}'.format(my_dict['key1']))
```
Accessing a nonexistent key will raise `KeyError` (see [`dict.get()`](#dict_get) for workaround):
```
# print(my_dict['nope'])
```
## Deleting
```
my_dict = {'key1': 'value1', 'key2': 99, 'keyX': 'valueX'}
del my_dict['keyX']
print(my_dict)
# Usually better to make sure that the key exists (see also pop() and popitem())
key_to_delete = 'my_key'
if key_to_delete in my_dict:
del my_dict[key_to_delete]
else:
print('{key} is not in {dictionary}'.format(key=key_to_delete, dictionary=my_dict))
```
## Dictionaries are mutable
```
my_dict = {'ham': 'good', 'carrot': 'semi good'}
my_other_dict = my_dict
my_other_dict['carrot'] = 'super tasty'
my_other_dict['sausage'] = 'best ever'
print('my_dict: {}\nother: {}'.format(my_dict, my_other_dict))
print('equal: {}'.format(my_dict == my_other_dict))
```
Create a new `dict` if you want to have a copy:
```
my_dict = {'ham': 'good', 'carrot': 'semi good'}
my_other_dict = dict(my_dict)
my_other_dict['beer'] = 'decent'
print('my_dict: {}\nother: {}'.format(my_dict, my_other_dict))
print('equal: {}'.format(my_dict == my_other_dict))
```
<a id='dict_get'></a>
## `dict.get()`
Returns `None` if `key` is not in `dict`. However, you can also specify `default` return value which will be returned if `key` is not present in the `dict`.
```
my_dict = {'a': 1, 'b': 2, 'c': 3}
d = my_dict.get('d')
print('d: {}'.format(d))
d = my_dict.get('d', 'my default value')
print('d: {}'.format(d))
```
## `dict.pop()`
```
my_dict = dict(food='ham', drink='beer', sport='football')
print('dict before pops: {}'.format(my_dict))
food = my_dict.pop('food')
print('food: {}'.format(food))
print('dict after popping food: {}'.format(my_dict))
food_again = my_dict.pop('food', 'default value for food')
print('food again: {}'.format(food_again))
print('dict after popping food again: {}'.format(my_dict))
```
## `dict.setdefault()`
Returns the `value` of `key` defined as first parameter. If the `key` is not present in the dict, adds `key` with default value (second parameter).
```
my_dict = {'a': 1, 'b': 2, 'c': 3}
a = my_dict.setdefault('a', 'my default value')
d = my_dict.setdefault('d', 'my default value')
print('a: {}\nd: {}\nmy_dict: {}'.format(a, d, my_dict))
```
## `dict.update()`
Merge two `dict`s
```
dict1 = {'a': 1, 'b': 2}
dict2 = {'c': 3}
dict1.update(dict2)
print(dict1)
# If they have same keys:
dict1.update({'c': 4})
print(dict1)
```
Another way to merge two dictionaries
```
dict1 = {'Ten': 10, 'Twenty': 20, 'Thirty': 30}
dict2 = {'Thirty': 30, 'Fourty': 40, 'Fifty': 50}
dict3 = {**dict1, **dict2}
print(dict3)
```
## Looping through `dict`s
```
my_dict = {'hacker': True, 'age': 72, 'name': 'John Doe'}
# Looping through all values in my_dict
for val in my_dict:
print(val)
# Looping through all key,value pairs in my_dict
for key, val in my_dict.items():
print('{}={}'.format(key, val))
```
## The keys of a `dict` have to be immutable
Thus you can not use e.g. a `list` or a `dict` as key because they are mutable types
:
```
# bad_dict = {['my_list'], 'value'} # Raises TypeError
```
Values can be mutable
```
good_dict = {'my key': ['Python', 'is', 'still', 'cool']}
print(good_dict)
```
|
github_jupyter
|
my_dict1 = {'jack': 4098, 'sjoerd': 4127}
my_dict2 = dict({4098: 'jack', 4127: 'sjoerd'})
print(my_dict1)
print(my_dict2)
my_empty_dict = {} # alternative: my_empty_dict = dict()
print('dict: {}, type: {}'.format(my_empty_dict, type(my_empty_dict)))
dict1 = {'value1': 1.6, 'value2': 10, 'name': 'John Doe'}
dict2 = dict(value1=1.6, value2=10, name='John Doe')
print(dict1)
print(dict2)
print('equal: {}'.format(dict1 == dict2))
print('length: {}'.format(len(dict1)))
keys = ['Ten', 'Twenty', 'Thirty']
values = [10, 20, 30]
sampleDict = dict(zip(keys, values))
print(sampleDict)
print('keys: {}'.format(dict1.keys())) # list of all keys
print('values: {}'.format(dict1.values())) # list of all values
print('items: {}'.format(dict1.items())) # list of all key,value pairs
my_dict = {}
my_dict['key1'] = 'value1'
my_dict['key2'] = 99
my_dict['key1'] = 'new value' # overriding existing value
print(my_dict)
print('value of key1: {}'.format(my_dict['key1']))
# print(my_dict['nope'])
my_dict = {'key1': 'value1', 'key2': 99, 'keyX': 'valueX'}
del my_dict['keyX']
print(my_dict)
# Usually better to make sure that the key exists (see also pop() and popitem())
key_to_delete = 'my_key'
if key_to_delete in my_dict:
del my_dict[key_to_delete]
else:
print('{key} is not in {dictionary}'.format(key=key_to_delete, dictionary=my_dict))
my_dict = {'ham': 'good', 'carrot': 'semi good'}
my_other_dict = my_dict
my_other_dict['carrot'] = 'super tasty'
my_other_dict['sausage'] = 'best ever'
print('my_dict: {}\nother: {}'.format(my_dict, my_other_dict))
print('equal: {}'.format(my_dict == my_other_dict))
my_dict = {'ham': 'good', 'carrot': 'semi good'}
my_other_dict = dict(my_dict)
my_other_dict['beer'] = 'decent'
print('my_dict: {}\nother: {}'.format(my_dict, my_other_dict))
print('equal: {}'.format(my_dict == my_other_dict))
my_dict = {'a': 1, 'b': 2, 'c': 3}
d = my_dict.get('d')
print('d: {}'.format(d))
d = my_dict.get('d', 'my default value')
print('d: {}'.format(d))
my_dict = dict(food='ham', drink='beer', sport='football')
print('dict before pops: {}'.format(my_dict))
food = my_dict.pop('food')
print('food: {}'.format(food))
print('dict after popping food: {}'.format(my_dict))
food_again = my_dict.pop('food', 'default value for food')
print('food again: {}'.format(food_again))
print('dict after popping food again: {}'.format(my_dict))
my_dict = {'a': 1, 'b': 2, 'c': 3}
a = my_dict.setdefault('a', 'my default value')
d = my_dict.setdefault('d', 'my default value')
print('a: {}\nd: {}\nmy_dict: {}'.format(a, d, my_dict))
dict1 = {'a': 1, 'b': 2}
dict2 = {'c': 3}
dict1.update(dict2)
print(dict1)
# If they have same keys:
dict1.update({'c': 4})
print(dict1)
dict1 = {'Ten': 10, 'Twenty': 20, 'Thirty': 30}
dict2 = {'Thirty': 30, 'Fourty': 40, 'Fifty': 50}
dict3 = {**dict1, **dict2}
print(dict3)
my_dict = {'hacker': True, 'age': 72, 'name': 'John Doe'}
# Looping through all values in my_dict
for val in my_dict:
print(val)
# Looping through all key,value pairs in my_dict
for key, val in my_dict.items():
print('{}={}'.format(key, val))
# bad_dict = {['my_list'], 'value'} # Raises TypeError
good_dict = {'my key': ['Python', 'is', 'still', 'cool']}
print(good_dict)
| 0.15084 | 0.875468 |
# Generate Anomalies (S | SV | SD | S+SV | S+SD | SV+SD | S+SV+SD)
```
# Basic libs
import os
import glob
import random
import numpy as np
import pandas as pd
from pathlib import Path
from datetime import datetime
# Plot libs
import seaborn as sns
import matplotlib.pyplot as plt
# Display 999 rows of Dataframe
pd.options.display.max_rows = 999
# The csv station ("santa_elena" or "hawkesworth_bridge")
STATION = "santa_elena"
# The start of the test set
TEST_SET_INIT = datetime(2020, 12, 1).date()
# If it is equal to 1, than will be create a csv with anomalies in the entire dataset
# Otherwise, just for test set (after dez/01/2020)
ALL_DATA = 0
```
## Pre-Processing
```
# Combines the csv creating a DataFrame with all data
def read_data(path):
df_all = None
# Show all csv ending with _wtlv1 in all dirs inside this path
for f in Path(path).rglob('*_wtlv1*.csv'):
print(f)
# Ignores other spreadsheets that are not STATION
if not STATION in str(f):
continue
# Get the filename, without the path
station = os.path.basename(f).split('_wtlv1')[0]
# datetime and update_at are combined as one date column
_df = pd.read_csv(f, parse_dates=["datetime", "updated_at"])
_df['station'] = station
if df_all is None:
df_all = _df
else:
df_all = pd.concat([df_all, _df], ignore_index=True)
# Remove updated_at and variable_id columns
df_all.drop(columns=['updated_at', 'variable_id'], inplace=True)
# Remove duplicates rows (by datetime)
df_all.drop_duplicates(subset=["datetime"], keep="last", inplace=True)
# Sort the dataframe and remove the index
df_all.set_index("datetime", inplace=True)
df_all.sort_index(inplace=True)
df_all.reset_index(inplace=True)
return(df_all)
```
## Artificial Anomalies
```
class Anomaly:
# --------------------------------------------------------------------------------
raw_data = None
measured = None
# --------------------------------------------------------------------------------
# Get the measured data to generate anomalies and plot
def init(self, raw_data, day):
# Gets a copy of raw data
self.raw_data = raw_data.copy()
# Select the test part of the data
columns = ['measured']
sample_slice = raw_data[raw_data.datetime.dt.date >= day].copy()
self.measured = sample_slice[columns]["measured"].tolist()
# --------------------------------------------------------------------------------
# Generate one spike anomaly in data
def generate_spike(self, possible_indexes, lower, upper, size):
# Possible indexes
num_examples = len(self.measured)
# Gets a random index
index = random.randint(1, num_examples-2)
# Search an index where there is no anomaly in that and neighbors indexes
while(np.any(possible_indexes[index-1:index+size+1] == True)):
index = random.randint(1, num_examples-2)
# Define a signal (+ ou -)
signal = 1 if(random.random() > 0.5) else -1
# Defines the measure of the anomaly that will be summed with the real measure
magnitude = random.uniform(lower, upper)
anomaly_value = self.measured[index] + (signal * magnitude)
return(anomaly_value, index)
# --------------------------------------------------------------------------------
# Generate one stationary value anomaly in data
def generate_stationary_value(self, possible_indexes, size):
# Possible indexes
num_examples = len(self.measured)
# Gets a random index
index = random.randint(1, num_examples-size)
# Search an index where there is no anomaly in that and neighbors indexes
while(np.any(possible_indexes[index-1:index+size+1] == True)):
index = random.randint(1, num_examples-size)
anomaly_value = self.measured[index-1]
return(anomaly_value, index)
# --------------------------------------------------------------------------------
# Generate one sensor displacement anomaly in data
def generate_sensor_displacement(self, possible_indexes, size):
# Possible indexes
num_examples = len(self.measured)
# Gets a random index
index = random.randint(1, num_examples-size)
# Search an index where there is no anomaly in that and neighbors indexes
while(np.any(possible_indexes[index-1:index+size+1] == True)):
index = random.randint(1, num_examples-size)
displacement = random.uniform(0.2, 1)
anomaly_values = []
indexes = []
# Define a signal (+ ou -)
signal = 1 if(random.random() > 0.5) else -1
# Store the values and indexes in lists
for i in range(index, index+size):
anomaly_values.append(self.measured[i] + (signal * displacement))
indexes.append(i)
return(anomaly_values, indexes)
# --------------------------------------------------------------------------------
# Generate "percent"% of anomalies in data.
## Possible anomaly types: s | sv | sd | s+sv | s+sd | sv+sd | s+sv+sd
def generate_anomalies(self, percent, lower, upper, anomaly_types):
SPIKE = 0
STAT_VALUES = 1
SENSOR_DISP = 2
# Returns the anomalies merged with the mesuared values
anomalies = self.measured.copy()
num_examples = len(self.measured)
# Define an array to control where anomalies will be placed (without sequential anomalies)
possible_indexes = np.full((num_examples,), False)
added_anomalies = 0
# Define a list with the anomalies informed by "anomaly_types"
if(anomaly_types == "s+sv+sd"):
choices = [SPIKE, STAT_VALUES, SENSOR_DISP]
elif(anomaly_types == "s+sv"):
choices = [SPIKE, STAT_VALUES]
elif(anomaly_types == "s+sd"):
choices = [SPIKE, SENSOR_DISP]
elif(anomaly_types == "sv+sd"):
choices = [STAT_VALUES, SENSOR_DISP]
elif(anomaly_types == "sd"):
choices = [SENSOR_DISP]
elif(anomaly_types == "sv"):
choices = [STAT_VALUES]
else:
choices = [SPIKE]
# Inserts the "percent of anomalies"
while(added_anomalies < int(num_examples * percent)):
# Choose, randomly, the anomaly type
anomaly_type = random.choice(choices)
# If it's Spike
if(anomaly_type == SPIKE):
size = 1
# Generates the anomaly
anomaly_value, index = self.generate_spike(possible_indexes, lower, upper, size)
# Inserts the anomaly
anomalies[index] = anomaly_value
anomalies[index:index+size] = [anomaly_value] * size
# Update the control variables (no overlap)
possible_indexes[index:index+size] = [True] * size
added_anomalies += size
# If it's Stationary Values
elif(anomaly_type == STAT_VALUES):
# Size of the anomaly
size = random.randint(2, 10)
# Generates the anomaly
anomaly_value, index = self.generate_stationary_value(possible_indexes, size)
# Insert the anomaly
anomalies[index] = anomaly_value
anomalies[index:index+size] = [anomaly_value] * size
# Update the control variables (no overlap)
possible_indexes[index:index+size] = [True] * size
added_anomalies += size
# If it's Sensor Displacement
else:
# Size of the anomaly
size = random.randint(2, 10)
anomaly_values, indexes = self.generate_sensor_displacement(possible_indexes, size)
# Insert the anomaly
for i in range(len(indexes)):
anomalies[indexes[i]] = anomaly_values[i]
# Update the control variables (no overlap)
possible_indexes[indexes[i]-1:indexes[i]+1] = True
added_anomalies += size
return(anomalies)
# --------------------------------------------------------------------------------
# Plots a range of artificial anomalies
def plot_anomaly(self, anomalies, anomaly_type, date, start, end):
# Size of the figure
sns.set(rc={'figure.figsize':(11, 10)})
# Format x axis
x_axis = date[start:end].strftime("%d %b %H:%M").tolist()
# Define the plot
d1 = pd.DataFrame({"Measured" : np.asarray(self.measured)[start:end]})
d2 = pd.DataFrame({"Anomaly" : np.asarray(anomalies)[start:end]})
fig, ax = plt.subplots()
ax.plot(x_axis, d2, label="Anomaly", marker="s")
ax.plot(x_axis, d1, label="Measured", marker="s")
ax.legend()
ax.set_ylabel("Measured")
ax.set_xlabel("Time")
plt.xticks(rotation=70)
# Define the title with the type of the anomaly
if(anomaly_type == "sv"):
plt.title("Stationary Values (SV) Anomalies")
elif(anomaly_type == "sd"):
plt.title("Sensor Displacement (SD) Anomalies")
else:
plt.title("Spike (S) Anomalies")
plt.show()
# --------------------------------------------------------------------------------
# Function that creates a dataframe to group the anomalies with the other informations
def generate_anomaly_dataframe(self, anomalies, anomaly_type, day):
# Separate the test part (all the training part was used in the regression)
columns = ['datetime', 'measured', 'station_id']
sample_slice = self.raw_data[raw_data.datetime.dt.date >= day].copy()
sample = sample_slice[columns]
# Adds the anomalies columns
sample["anomaly_value"] = anomalies
sample["anomaly_name"] = np.where(sample["anomaly_value"] != sample["measured"], anomaly_type, "-")
return(sample)
# --------------------------------------------------------------------------------
# Function the save the dataframe with the informations and anomalies into a csv
def generate_anomaly_csv_sample(self, sample, anomaly_type, path):
sample.to_csv("{}/artificial-{}-anomaly.csv".format(path, anomaly_type), index=False)
# --------------------------------------------------------------------------------
# ------------------------------------------------------------------------------------
```
# Main
```
# Read the data
raw_data = read_data("../../stations")
# Generate Anomalies in all data
if(ALL_DATA == 1):
date = str(raw_data["datetime"].min()).split(" ")[0].split("-")
year = int(date[0])
month = int(date[1])
day = int(date[2])
date = datetime(year, month, day).date()
output_path = "./csvs/all_data"
# Or, generate anomalies only on the test set
else:
date = TEST_SET_INIT
output_path = "./csvs/test_set"
```
# Generating Anomalies
## Anomalies Class
```
# Create the Anomaly class
anomaly_class = Anomaly()
anomaly_class.init(raw_data, date)
# Params to control the number and interval of anomalies
percent_of_anomalies = 0.25
lower = 0.3
upper = 1.0
start = 0
end = 30
# Get the x_axis (dates)
x_axis = pd.DataFrame({"datetime": pd.to_datetime(raw_data["datetime"], format="%Y-%m-%dT%H:%M:%SZ", errors='coerce')})
x_axis.set_index("datetime", inplace=True)
x_axis = x_axis[x_axis.index.date >= date]
```
## 1) Generate Spike Anomalies
```
# Generate a list with spike anomalies
anomaly_type = "s"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs spike)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
```
## 2) Generate Stationary Values Anomalies
```
# Generate a list with stationary values anomalies
anomaly_type = "sv"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs stationary values)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
```
## 3) Generate Sensor Displacement Anomalies
```
# Generate a list with sensor displacement anomalies
anomaly_type = "sd"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs sensor displacement)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
```
## 4) Generate S + SV Anomalies
```
# Generate a list with s+sv anomalies
anomaly_type = "s+sv"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs s+sv)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
```
## 5) Generating S + SD Anomalies
```
# Generate a list with s+sd anomalies
anomaly_type = "s+sd"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs s+sd)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
```
## 6) Generating SV + SD Anomalies
```
# Generate a list with sv+sd anomalies
anomaly_type = "sv+sd"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs sv+sd)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
```
## 7) Generating S + SV + SD Anomalies
```
# Generate a list with s+sv+sd anomalies
anomaly_type = "s+sv+sd"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs s+sv+sd)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
```
|
github_jupyter
|
# Basic libs
import os
import glob
import random
import numpy as np
import pandas as pd
from pathlib import Path
from datetime import datetime
# Plot libs
import seaborn as sns
import matplotlib.pyplot as plt
# Display 999 rows of Dataframe
pd.options.display.max_rows = 999
# The csv station ("santa_elena" or "hawkesworth_bridge")
STATION = "santa_elena"
# The start of the test set
TEST_SET_INIT = datetime(2020, 12, 1).date()
# If it is equal to 1, than will be create a csv with anomalies in the entire dataset
# Otherwise, just for test set (after dez/01/2020)
ALL_DATA = 0
# Combines the csv creating a DataFrame with all data
def read_data(path):
df_all = None
# Show all csv ending with _wtlv1 in all dirs inside this path
for f in Path(path).rglob('*_wtlv1*.csv'):
print(f)
# Ignores other spreadsheets that are not STATION
if not STATION in str(f):
continue
# Get the filename, without the path
station = os.path.basename(f).split('_wtlv1')[0]
# datetime and update_at are combined as one date column
_df = pd.read_csv(f, parse_dates=["datetime", "updated_at"])
_df['station'] = station
if df_all is None:
df_all = _df
else:
df_all = pd.concat([df_all, _df], ignore_index=True)
# Remove updated_at and variable_id columns
df_all.drop(columns=['updated_at', 'variable_id'], inplace=True)
# Remove duplicates rows (by datetime)
df_all.drop_duplicates(subset=["datetime"], keep="last", inplace=True)
# Sort the dataframe and remove the index
df_all.set_index("datetime", inplace=True)
df_all.sort_index(inplace=True)
df_all.reset_index(inplace=True)
return(df_all)
class Anomaly:
# --------------------------------------------------------------------------------
raw_data = None
measured = None
# --------------------------------------------------------------------------------
# Get the measured data to generate anomalies and plot
def init(self, raw_data, day):
# Gets a copy of raw data
self.raw_data = raw_data.copy()
# Select the test part of the data
columns = ['measured']
sample_slice = raw_data[raw_data.datetime.dt.date >= day].copy()
self.measured = sample_slice[columns]["measured"].tolist()
# --------------------------------------------------------------------------------
# Generate one spike anomaly in data
def generate_spike(self, possible_indexes, lower, upper, size):
# Possible indexes
num_examples = len(self.measured)
# Gets a random index
index = random.randint(1, num_examples-2)
# Search an index where there is no anomaly in that and neighbors indexes
while(np.any(possible_indexes[index-1:index+size+1] == True)):
index = random.randint(1, num_examples-2)
# Define a signal (+ ou -)
signal = 1 if(random.random() > 0.5) else -1
# Defines the measure of the anomaly that will be summed with the real measure
magnitude = random.uniform(lower, upper)
anomaly_value = self.measured[index] + (signal * magnitude)
return(anomaly_value, index)
# --------------------------------------------------------------------------------
# Generate one stationary value anomaly in data
def generate_stationary_value(self, possible_indexes, size):
# Possible indexes
num_examples = len(self.measured)
# Gets a random index
index = random.randint(1, num_examples-size)
# Search an index where there is no anomaly in that and neighbors indexes
while(np.any(possible_indexes[index-1:index+size+1] == True)):
index = random.randint(1, num_examples-size)
anomaly_value = self.measured[index-1]
return(anomaly_value, index)
# --------------------------------------------------------------------------------
# Generate one sensor displacement anomaly in data
def generate_sensor_displacement(self, possible_indexes, size):
# Possible indexes
num_examples = len(self.measured)
# Gets a random index
index = random.randint(1, num_examples-size)
# Search an index where there is no anomaly in that and neighbors indexes
while(np.any(possible_indexes[index-1:index+size+1] == True)):
index = random.randint(1, num_examples-size)
displacement = random.uniform(0.2, 1)
anomaly_values = []
indexes = []
# Define a signal (+ ou -)
signal = 1 if(random.random() > 0.5) else -1
# Store the values and indexes in lists
for i in range(index, index+size):
anomaly_values.append(self.measured[i] + (signal * displacement))
indexes.append(i)
return(anomaly_values, indexes)
# --------------------------------------------------------------------------------
# Generate "percent"% of anomalies in data.
## Possible anomaly types: s | sv | sd | s+sv | s+sd | sv+sd | s+sv+sd
def generate_anomalies(self, percent, lower, upper, anomaly_types):
SPIKE = 0
STAT_VALUES = 1
SENSOR_DISP = 2
# Returns the anomalies merged with the mesuared values
anomalies = self.measured.copy()
num_examples = len(self.measured)
# Define an array to control where anomalies will be placed (without sequential anomalies)
possible_indexes = np.full((num_examples,), False)
added_anomalies = 0
# Define a list with the anomalies informed by "anomaly_types"
if(anomaly_types == "s+sv+sd"):
choices = [SPIKE, STAT_VALUES, SENSOR_DISP]
elif(anomaly_types == "s+sv"):
choices = [SPIKE, STAT_VALUES]
elif(anomaly_types == "s+sd"):
choices = [SPIKE, SENSOR_DISP]
elif(anomaly_types == "sv+sd"):
choices = [STAT_VALUES, SENSOR_DISP]
elif(anomaly_types == "sd"):
choices = [SENSOR_DISP]
elif(anomaly_types == "sv"):
choices = [STAT_VALUES]
else:
choices = [SPIKE]
# Inserts the "percent of anomalies"
while(added_anomalies < int(num_examples * percent)):
# Choose, randomly, the anomaly type
anomaly_type = random.choice(choices)
# If it's Spike
if(anomaly_type == SPIKE):
size = 1
# Generates the anomaly
anomaly_value, index = self.generate_spike(possible_indexes, lower, upper, size)
# Inserts the anomaly
anomalies[index] = anomaly_value
anomalies[index:index+size] = [anomaly_value] * size
# Update the control variables (no overlap)
possible_indexes[index:index+size] = [True] * size
added_anomalies += size
# If it's Stationary Values
elif(anomaly_type == STAT_VALUES):
# Size of the anomaly
size = random.randint(2, 10)
# Generates the anomaly
anomaly_value, index = self.generate_stationary_value(possible_indexes, size)
# Insert the anomaly
anomalies[index] = anomaly_value
anomalies[index:index+size] = [anomaly_value] * size
# Update the control variables (no overlap)
possible_indexes[index:index+size] = [True] * size
added_anomalies += size
# If it's Sensor Displacement
else:
# Size of the anomaly
size = random.randint(2, 10)
anomaly_values, indexes = self.generate_sensor_displacement(possible_indexes, size)
# Insert the anomaly
for i in range(len(indexes)):
anomalies[indexes[i]] = anomaly_values[i]
# Update the control variables (no overlap)
possible_indexes[indexes[i]-1:indexes[i]+1] = True
added_anomalies += size
return(anomalies)
# --------------------------------------------------------------------------------
# Plots a range of artificial anomalies
def plot_anomaly(self, anomalies, anomaly_type, date, start, end):
# Size of the figure
sns.set(rc={'figure.figsize':(11, 10)})
# Format x axis
x_axis = date[start:end].strftime("%d %b %H:%M").tolist()
# Define the plot
d1 = pd.DataFrame({"Measured" : np.asarray(self.measured)[start:end]})
d2 = pd.DataFrame({"Anomaly" : np.asarray(anomalies)[start:end]})
fig, ax = plt.subplots()
ax.plot(x_axis, d2, label="Anomaly", marker="s")
ax.plot(x_axis, d1, label="Measured", marker="s")
ax.legend()
ax.set_ylabel("Measured")
ax.set_xlabel("Time")
plt.xticks(rotation=70)
# Define the title with the type of the anomaly
if(anomaly_type == "sv"):
plt.title("Stationary Values (SV) Anomalies")
elif(anomaly_type == "sd"):
plt.title("Sensor Displacement (SD) Anomalies")
else:
plt.title("Spike (S) Anomalies")
plt.show()
# --------------------------------------------------------------------------------
# Function that creates a dataframe to group the anomalies with the other informations
def generate_anomaly_dataframe(self, anomalies, anomaly_type, day):
# Separate the test part (all the training part was used in the regression)
columns = ['datetime', 'measured', 'station_id']
sample_slice = self.raw_data[raw_data.datetime.dt.date >= day].copy()
sample = sample_slice[columns]
# Adds the anomalies columns
sample["anomaly_value"] = anomalies
sample["anomaly_name"] = np.where(sample["anomaly_value"] != sample["measured"], anomaly_type, "-")
return(sample)
# --------------------------------------------------------------------------------
# Function the save the dataframe with the informations and anomalies into a csv
def generate_anomaly_csv_sample(self, sample, anomaly_type, path):
sample.to_csv("{}/artificial-{}-anomaly.csv".format(path, anomaly_type), index=False)
# --------------------------------------------------------------------------------
# ------------------------------------------------------------------------------------
# Read the data
raw_data = read_data("../../stations")
# Generate Anomalies in all data
if(ALL_DATA == 1):
date = str(raw_data["datetime"].min()).split(" ")[0].split("-")
year = int(date[0])
month = int(date[1])
day = int(date[2])
date = datetime(year, month, day).date()
output_path = "./csvs/all_data"
# Or, generate anomalies only on the test set
else:
date = TEST_SET_INIT
output_path = "./csvs/test_set"
# Create the Anomaly class
anomaly_class = Anomaly()
anomaly_class.init(raw_data, date)
# Params to control the number and interval of anomalies
percent_of_anomalies = 0.25
lower = 0.3
upper = 1.0
start = 0
end = 30
# Get the x_axis (dates)
x_axis = pd.DataFrame({"datetime": pd.to_datetime(raw_data["datetime"], format="%Y-%m-%dT%H:%M:%SZ", errors='coerce')})
x_axis.set_index("datetime", inplace=True)
x_axis = x_axis[x_axis.index.date >= date]
# Generate a list with spike anomalies
anomaly_type = "s"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs spike)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
# Generate a list with stationary values anomalies
anomaly_type = "sv"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs stationary values)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
# Generate a list with sensor displacement anomalies
anomaly_type = "sd"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs sensor displacement)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
# Generate a list with s+sv anomalies
anomaly_type = "s+sv"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs s+sv)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
# Generate a list with s+sd anomalies
anomaly_type = "s+sd"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs s+sd)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
# Generate a list with sv+sd anomalies
anomaly_type = "sv+sd"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs sv+sd)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
# Generate a list with s+sv+sd anomalies
anomaly_type = "s+sv+sd"
anomalies = anomaly_class.generate_anomalies(percent_of_anomalies, lower, upper, anomaly_type)
# Plot it (measured vs s+sv+sd)
anomaly_class.plot_anomaly(anomalies, anomaly_type, x_axis.index, start, end)
# Save into a csv file
sample = anomaly_class.generate_anomaly_dataframe(anomalies, anomaly_type, date)
anomaly_class.generate_anomaly_csv_sample(sample, anomaly_type, output_path)
| 0.548674 | 0.856992 |
```
import pandas as pd
import pandas.io.sql as sqlio
import psycopg2
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(font_scale=1.5, style="whitegrid")
from matplotlib.ticker import (MultipleLocator, FormatStrFormatter,AutoMinorLocator)
from matplotlib.dates import DateFormatter
import matplotlib.dates as mdates
import numpy as np
from connector import getConnection
```
### Interest income distribution
```
conn = getConnection()
sql = """WITH interest_income_query AS(SELECT i.sale_date_key, SUM(i.interest) AS interest_income
FROM installment i
GROUP BY i.sale_date_key)
SELECT iq_2018.month, ROUND(iq_2018.ii::numeric,2) as "interest income 2018",
ROUND(iq_2019.ii::numeric,2) as "interest income 2019"
FROM
(SELECT d.month, SUM(q.interest_income) AS ii
FROM public."date" d, interest_income_query q
WHERE d.date_key = q.sale_date_key AND d.year = 2018
group by d.month) AS iq_2018,
(SELECT d.month, SUM(q.interest_income) AS ii
FROM public."date" d, interest_income_query q
WHERE d.date_key = q.sale_date_key AND d.year = 2019
GROUP BY d.month) AS iq_2019
WHERE iq_2018.month = iq_2019.month
ORDER BY iq_2019.month """
df = sqlio.read_sql_query(sql, conn)
df.head()
conn.close()
df.plot(x="month", y=["interest income 2018", "interest income 2019"], kind="bar",figsize=(15,10), rot = 0, colormap= 'Dark2_r')
conn = getConnection()
sql = """WITH interest_income_query AS(SELECT i.sale_date_key, SUM(i.interest) AS interest_income
FROM installment i
GROUP BY i.sale_date_key)
SELECT 'interest' as income ,ROUND(iq_2018.ii::numeric,2) as "2018",
ROUND(iq_2019.ii::numeric,2) as "2019"
FROM
(SELECT d.year, SUM(q.interest_income) AS ii
FROM public."date" d, interest_income_query q
WHERE d.date_key = q.sale_date_key AND d.year = 2018
group by d.year) AS iq_2018,
(SELECT d.year, SUM(q.interest_income) AS ii
FROM public."date" d, interest_income_query q
WHERE d.date_key = q.sale_date_key AND d.year = 2019
GROUP BY d.year) AS iq_2019
"""
df = sqlio.read_sql_query(sql, conn)
df = df.set_index('income')
conn.close()
df.head()
df.plot(y=["2018", "2019"], kind="bar",figsize=(15,10), rot = 0, colormap='Dark2_r')
```
### Best selling month
```
conn = getConnection()
sql = """SELECT d.month, SUM(s.loan_amount) as granted ,
RANK() OVER ( ORDER BY SUM(s.loan_amount) DESC ) best
FROM sale s, public."date" d
WHERE s.sale_date_key = d.date_key AND d.year = 2018
GROUP BY d.month; """
df = sqlio.read_sql_query(sql, conn)
df = df.set_index('month')
conn.close()
df.head()
df.plot.bar(y=['granted'],figsize=(15,10), rot = 0)
```
### Best 7 days period for sales
```
conn = getConnection()
sql = """SELECT iq.day, total_granted, week_total_sale_avg
FROM
(SELECT q1.date AS date, q1.month ,q1.day ,
SUM(q1.loan_amount) AS total_granted,AVG(SUM(q1.loan_amount))
OVER ( ORDER BY q1.date_key ROWS BETWEEN 3 PRECEDING and 3 FOLLOWING ) AS week_total_sale_avg
FROM
(
SELECT d.*, COALESCE(s.loan_amount ,0) AS loan_amount
FROM
public."date" d LEFT OUTER JOIN sale s
ON d.date_key = s.sale_date_key
WHERE d.year = 2018 AND
d.month in (2,3,4)
)AS q1
GROUP BY q1.date_key,q1.date, q1.month , q1.day
) AS iq
WHERE iq.month = 3 """
df = sqlio.read_sql_query(sql, conn)
df = df.set_index('day')
df = df['week_total_sale_avg']
conn.close()
df.head()
df.plot.line(y=['week_total_sale_avg'],figsize=(15,10), rot = 0)
conn = getConnection()
sql = """with unpaid_query as(
select d.year, d.month, sum(amount) unpaid
from installment i
left join payment p on i.installment_key = p.installment_key
join customer c on sale_customer_key = c.customer_key
join npl n on i.sale_customer_key = n.sale_customer_key
join "date" d on i.due_date_key = d.date_key
where payment_date_key is null
and i.due_date_key < 731 -- 31-12-2019
group by d.year, d.month)
SELECT up_2019.month, round(up_2018.unpaid,2) as "unpaid 2018",
round(up_2019.unpaid,2) as "unpaid 2019"
FROM
(SELECT q.month, unpaid
FROM unpaid_query q
WHERE q.year = 2019) up_2019
left join
(SELECT q.month, unpaid
FROM unpaid_query q
WHERE q.year = 2018) up_2018
ON up_2019.month = up_2018.month
ORDER BY up_2019.month """
df = sqlio.read_sql_query(sql, conn)
df = df.set_index('month')
conn.close()
df.head()
#df.plot(y=["2018", "2019"], kind="bar",figsize=(15,10), rot = 0)
df.plot.bar(figsize=(15,10), rot = 0,colormap='Dark2')
```
### Monthly status
```
conn = getConnection()
sql = """
WITH states AS (select month_end_Date, count(*) as num,
(case when npl.min_date_npl is null then 'BONIS'
when (month_end.date_key -npl.min_date_npl) > 365 then 'DEFAULT'
when (month_end.date_key -npl.min_date_npl) > 180 then 'UTP'
when (month_end.date_key -npl.min_date_npl) > 90 then 'PASTDUE' else 'BONIS' end) status
from sale s left outer join npl on s.customer_key = npl.sale_customer_key,
(select max(date_key) as date_key, max(date) as month_end_Date
from "date"
group by year, MONTH
HAVING max(date) < current_date
ORDER BY YEAR, MONTH ) month_end
WHERE s.sale_date_key < month_end.date_key
GROUP BY month_end_date, status
ORDER BY month_end_date, status )
SELECT s_all.month_end_date, COALESCE(s_BONIS.num,0) AS BONIS, COALESCE(s_PASTDUE.num,0) AS PASTDUE ,
COALESCE(s_UTP.num,0) as UTP, COALESCE(s_DEFAULT.num,0) as DEFAULT
FROM states s_all
LEFT OUTER JOIN states s_BONIS ON s_all.month_end_Date = s_BONIS.month_end_Date AND s_BONIS.status = 'BONIS'
LEFT OUTER JOIN states s_PASTDUE ON s_all.month_end_Date = s_PASTDUE.month_end_Date AND s_PASTDUE.status = 'PASTDUE'
LEFT OUTER JOIN states s_UTP ON s_all.month_end_Date = s_UTP.month_end_Date AND s_UTP.status = 'UTP'
LEFT OUTER JOIN states s_DEFAULT ON s_all.month_end_Date = s_DEFAULT.month_end_Date AND s_DEFAULT.status = 'DEFAULT'
"""
df = sqlio.read_sql_query(sql, conn)
df = df.set_index('month_end_date')
color=(0.2, 0.4, 0.6, 0.6)
normalizedDataFrame = df.divide(df.sum(axis=1), axis=0)
conn.close()
normalizedDataFrame.head()
#df.plot(y=["2018", "2019"], kind="bar",figsize=(15,10), rot = 0)
#df.plot.bar(figsize=(15,10), rot = 0)
normalizedDataFrame.plot.area(stacked=True,figsize=(15,10),ylim = [0.9,1], colormap='cividis');
```
|
github_jupyter
|
import pandas as pd
import pandas.io.sql as sqlio
import psycopg2
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(font_scale=1.5, style="whitegrid")
from matplotlib.ticker import (MultipleLocator, FormatStrFormatter,AutoMinorLocator)
from matplotlib.dates import DateFormatter
import matplotlib.dates as mdates
import numpy as np
from connector import getConnection
conn = getConnection()
sql = """WITH interest_income_query AS(SELECT i.sale_date_key, SUM(i.interest) AS interest_income
FROM installment i
GROUP BY i.sale_date_key)
SELECT iq_2018.month, ROUND(iq_2018.ii::numeric,2) as "interest income 2018",
ROUND(iq_2019.ii::numeric,2) as "interest income 2019"
FROM
(SELECT d.month, SUM(q.interest_income) AS ii
FROM public."date" d, interest_income_query q
WHERE d.date_key = q.sale_date_key AND d.year = 2018
group by d.month) AS iq_2018,
(SELECT d.month, SUM(q.interest_income) AS ii
FROM public."date" d, interest_income_query q
WHERE d.date_key = q.sale_date_key AND d.year = 2019
GROUP BY d.month) AS iq_2019
WHERE iq_2018.month = iq_2019.month
ORDER BY iq_2019.month """
df = sqlio.read_sql_query(sql, conn)
df.head()
conn.close()
df.plot(x="month", y=["interest income 2018", "interest income 2019"], kind="bar",figsize=(15,10), rot = 0, colormap= 'Dark2_r')
conn = getConnection()
sql = """WITH interest_income_query AS(SELECT i.sale_date_key, SUM(i.interest) AS interest_income
FROM installment i
GROUP BY i.sale_date_key)
SELECT 'interest' as income ,ROUND(iq_2018.ii::numeric,2) as "2018",
ROUND(iq_2019.ii::numeric,2) as "2019"
FROM
(SELECT d.year, SUM(q.interest_income) AS ii
FROM public."date" d, interest_income_query q
WHERE d.date_key = q.sale_date_key AND d.year = 2018
group by d.year) AS iq_2018,
(SELECT d.year, SUM(q.interest_income) AS ii
FROM public."date" d, interest_income_query q
WHERE d.date_key = q.sale_date_key AND d.year = 2019
GROUP BY d.year) AS iq_2019
"""
df = sqlio.read_sql_query(sql, conn)
df = df.set_index('income')
conn.close()
df.head()
df.plot(y=["2018", "2019"], kind="bar",figsize=(15,10), rot = 0, colormap='Dark2_r')
conn = getConnection()
sql = """SELECT d.month, SUM(s.loan_amount) as granted ,
RANK() OVER ( ORDER BY SUM(s.loan_amount) DESC ) best
FROM sale s, public."date" d
WHERE s.sale_date_key = d.date_key AND d.year = 2018
GROUP BY d.month; """
df = sqlio.read_sql_query(sql, conn)
df = df.set_index('month')
conn.close()
df.head()
df.plot.bar(y=['granted'],figsize=(15,10), rot = 0)
conn = getConnection()
sql = """SELECT iq.day, total_granted, week_total_sale_avg
FROM
(SELECT q1.date AS date, q1.month ,q1.day ,
SUM(q1.loan_amount) AS total_granted,AVG(SUM(q1.loan_amount))
OVER ( ORDER BY q1.date_key ROWS BETWEEN 3 PRECEDING and 3 FOLLOWING ) AS week_total_sale_avg
FROM
(
SELECT d.*, COALESCE(s.loan_amount ,0) AS loan_amount
FROM
public."date" d LEFT OUTER JOIN sale s
ON d.date_key = s.sale_date_key
WHERE d.year = 2018 AND
d.month in (2,3,4)
)AS q1
GROUP BY q1.date_key,q1.date, q1.month , q1.day
) AS iq
WHERE iq.month = 3 """
df = sqlio.read_sql_query(sql, conn)
df = df.set_index('day')
df = df['week_total_sale_avg']
conn.close()
df.head()
df.plot.line(y=['week_total_sale_avg'],figsize=(15,10), rot = 0)
conn = getConnection()
sql = """with unpaid_query as(
select d.year, d.month, sum(amount) unpaid
from installment i
left join payment p on i.installment_key = p.installment_key
join customer c on sale_customer_key = c.customer_key
join npl n on i.sale_customer_key = n.sale_customer_key
join "date" d on i.due_date_key = d.date_key
where payment_date_key is null
and i.due_date_key < 731 -- 31-12-2019
group by d.year, d.month)
SELECT up_2019.month, round(up_2018.unpaid,2) as "unpaid 2018",
round(up_2019.unpaid,2) as "unpaid 2019"
FROM
(SELECT q.month, unpaid
FROM unpaid_query q
WHERE q.year = 2019) up_2019
left join
(SELECT q.month, unpaid
FROM unpaid_query q
WHERE q.year = 2018) up_2018
ON up_2019.month = up_2018.month
ORDER BY up_2019.month """
df = sqlio.read_sql_query(sql, conn)
df = df.set_index('month')
conn.close()
df.head()
#df.plot(y=["2018", "2019"], kind="bar",figsize=(15,10), rot = 0)
df.plot.bar(figsize=(15,10), rot = 0,colormap='Dark2')
conn = getConnection()
sql = """
WITH states AS (select month_end_Date, count(*) as num,
(case when npl.min_date_npl is null then 'BONIS'
when (month_end.date_key -npl.min_date_npl) > 365 then 'DEFAULT'
when (month_end.date_key -npl.min_date_npl) > 180 then 'UTP'
when (month_end.date_key -npl.min_date_npl) > 90 then 'PASTDUE' else 'BONIS' end) status
from sale s left outer join npl on s.customer_key = npl.sale_customer_key,
(select max(date_key) as date_key, max(date) as month_end_Date
from "date"
group by year, MONTH
HAVING max(date) < current_date
ORDER BY YEAR, MONTH ) month_end
WHERE s.sale_date_key < month_end.date_key
GROUP BY month_end_date, status
ORDER BY month_end_date, status )
SELECT s_all.month_end_date, COALESCE(s_BONIS.num,0) AS BONIS, COALESCE(s_PASTDUE.num,0) AS PASTDUE ,
COALESCE(s_UTP.num,0) as UTP, COALESCE(s_DEFAULT.num,0) as DEFAULT
FROM states s_all
LEFT OUTER JOIN states s_BONIS ON s_all.month_end_Date = s_BONIS.month_end_Date AND s_BONIS.status = 'BONIS'
LEFT OUTER JOIN states s_PASTDUE ON s_all.month_end_Date = s_PASTDUE.month_end_Date AND s_PASTDUE.status = 'PASTDUE'
LEFT OUTER JOIN states s_UTP ON s_all.month_end_Date = s_UTP.month_end_Date AND s_UTP.status = 'UTP'
LEFT OUTER JOIN states s_DEFAULT ON s_all.month_end_Date = s_DEFAULT.month_end_Date AND s_DEFAULT.status = 'DEFAULT'
"""
df = sqlio.read_sql_query(sql, conn)
df = df.set_index('month_end_date')
color=(0.2, 0.4, 0.6, 0.6)
normalizedDataFrame = df.divide(df.sum(axis=1), axis=0)
conn.close()
normalizedDataFrame.head()
#df.plot(y=["2018", "2019"], kind="bar",figsize=(15,10), rot = 0)
#df.plot.bar(figsize=(15,10), rot = 0)
normalizedDataFrame.plot.area(stacked=True,figsize=(15,10),ylim = [0.9,1], colormap='cividis');
| 0.212395 | 0.589894 |
```
import sys
from typing import Dict
import matplotlib
import numpy as np
import xarray as xr
climate_indices_home_path = "/home/james/git/climate_indices"
if climate_indices_home_path not in sys.path:
sys.path.insert(climate_indices_home_path)
from climate_indices import compute, indices, utils
%matplotlib inline
netcdf_precip = '/data/datasets/nclimgrid/nclimgrid_lowres_prcp.nc'
netcdf_gamma = '/data/datasets/nclimgrid/nclimgrid_lowres_gamma.nc'
```
Open the precipitation dataset as an xarray Dataset object.
```
ds_prcp = xr.open_dataset(netcdf_precip)
list(ds_prcp.dims)
```
Get the precipitation data and reshape the array to have the time dimension as the inner-most axis:
```
da_prcp = ds_prcp['prcp'].transpose('lat', 'lon', 'time')
initial_year = int(da_prcp['time'][0].dt.year)
calibration_year_initial = 1900
calibration_year_final = 2000
period_times = 12
total_lats = da_prcp.shape[0]
total_lons = da_prcp.shape[1]
fitting_shape = (total_lats, total_lons, period_times)
scales = [1] # , 2, 3, 6, 9, 12, 24]
periodicity = compute.Periodicity.monthly
```
Define a function that can be used to compute the gamma fitting parameters for a particular month scale:
```
def compute_gammas(
da_precip: xr.DataArray,
gamma_coords: Dict,
scale: int,
calibration_year_initial,
calibration_year_final,
periodicity: compute.Periodicity,
) -> (xr.DataArray, xr.DataArray):
initial_year = int(da_precip['time'][0].dt.year)
if periodicity == compute.Periodicity.monthly:
period_times = 12
elif periodicity == compute.Periodicity.daily:
period_times = 366
total_lats = da_precip.shape[0]
total_lons = da_precip.shape[1]
fitting_shape = (total_lats, total_lons, period_times)
alphas = np.full(shape=fitting_shape, fill_value=np.NaN)
betas = np.full(shape=fitting_shape, fill_value=np.NaN)
# loop over the grid cells and compute the gamma parameters for each
for lat_index in range(total_lats):
for lon_index in range(total_lons):
# get the precipitation values for the lat/lon grid cell
values = da_precip[lat_index, lon_index]
# skip over this grid cell if all NaN values
if (np.ma.is_masked(values) and values.mask.all()) or np.all(np.isnan(values)):
continue
# convolve to scale
scaled_values = \
compute.scale_values(
values,
scale=scale,
periodicity=periodicity,
)
# compute the fitting parameters on the scaled data
alphas[lat_index, lon_index], betas[lat_index, lon_index] = \
compute.gamma_parameters(
scaled_values,
data_start_year=initial_year,
calibration_start_year=calibration_year_initial,
calibration_end_year=calibration_year_final,
periodicity=periodicity,
)
alpha_attrs = {
'description': 'shape parameter of the gamma distribution (also referred to as the concentration) ' + \
f'computed from the {scale}-month scaled precipitation values',
}
da_alpha = xr.DataArray(
data=alphas,
coords=gamma_coords,
dims=tuple(gamma_coords.keys()),
name=f"alpha_{str(scale).zfill(2)}",
attrs=alpha_attrs,
)
beta_attrs = {
'description': '1 / scale of the distribution (also referred to as the rate) ' + \
f'computed from the {scale}-month scaled precipitation values',
}
da_beta = xr.DataArray(
data=betas,
coords=gamma_coords,
dims=tuple(gamma_coords.keys()),
name=f"beta_{str(scale).zfill(2)}",
attrs=beta_attrs,
)
return da_alpha, da_beta
```
Define a function that can be used to compute the SPI for a particular month scale:
```
def compute_spi_gamma(
da_precip: xr.DataArray,
da_alpha: xr.DataArray,
da_beta: xr.DataArray,
scale: int,
periodicity: compute.Periodicity,
) -> xr.DataArray:
initial_year = int(da_precip['time'][0].dt.year)
total_lats = da_precip.shape[0]
total_lons = da_precip.shape[1]
spi = np.full(shape=da_precip.shape, fill_value=np.NaN)
for lat_index in range(total_lats):
for lon_index in range(total_lons):
# get the values for the lat/lon grid cell
values = da_precip[lat_index, lon_index]
# skip over this grid cell if all NaN values
if (np.ma.is_masked(values) and values.mask.all()) or np.all(np.isnan(values)):
continue
gamma_parameters = {
"alphas": da_alpha[lat_index, lon_index],
"betas": da_beta[lat_index, lon_index],
}
# compute the SPI
spi[lat_index, lon_index] = \
indices.spi(
values,
scale=scale,
distribution=indices.Distribution.gamma,
data_start_year=initial_year,
calibration_year_initial=calibration_year_initial,
calibration_year_final=calibration_year_final,
periodicity=compute.Periodicity.monthly,
fitting_params=gamma_parameters,
)
# build a DataArray for this scale's SPI
da_spi = xr.DataArray(
data=spi,
coords=da_precip.coords,
dims=da_precip.dims,
name=f"spi_gamma_{str(scale).zfill(2)}",
)
da_spi.attrs = {
'description': f'SPI ({scale}-{periodicity} gamma) computed from monthly precipitation ' + \
f'data for the period {da_precip.time[0]} through {da_precip.time[-1]} using a ' + \
f'calibration period from {calibration_year_initial} through {calibration_year_final}',
'valid_min': -3.09,
'valid_max': 3.09,
'long_name': f'{scale}-{periodicity} SPI(gamma)',
'calibration_year_initial': calibration_year_initial,
'calibration_year_final': calibration_year_final,
}
return da_spi
```
Copy the attributes from the precipitation dataset that will be applicable to the corresponding gamma fitting parameters and SPI datasets:
```
attrs_to_copy = [
'Conventions',
'ncei_template_version',
'naming_authority',
'standard_name_vocabulary',
'institution',
'geospatial_lat_min',
'geospatial_lat_max',
'geospatial_lon_min',
'geospatial_lon_max',
'geospatial_lat_units',
'geospatial_lon_units',
]
global_attrs = {key: value for (key, value) in ds_prcp.attrs.items() if key in attrs_to_copy}
```
Compute the gamma fitting parameters for all scales and add these into a Dataset that we'll write to NetCDF:
```
%%time
if periodicity == compute.Periodicity.monthly:
period_times = 12
time_coord_name = "month"
elif periodicity == compute.Periodicity.daily:
period_times = 366
time_coord_name = "day"
gamma_coords = {"lat": ds_prcp.lat, "lon": ds_prcp.lon, time_coord_name: range(period_times)}
ds_gamma = xr.Dataset(
coords=gamma_coords,
attrs=global_attrs,
)
for scale in scales:
var_name_alpha = f"alpha_{str(scale).zfill(2)}"
var_name_beta = f"beta_{str(scale).zfill(2)}"
da_alpha, da_beta = compute_gammas(
da_prcp,
gamma_coords,
scale,
calibration_year_initial,
calibration_year_final,
periodicity,
)
ds_gamma[var_name_alpha] = da_alpha
ds_gamma[var_name_beta] = da_beta
ds_gamma.to_netcdf(netcdf_gamma)
ds_gamma
```
Compute the SPI using the pre-computed gamma fitting parameters for all scales and add these into a SPI(gamma) Dataset that we'll write to NetCDF:
```
%%time
ds_spi = xr.Dataset(
coords=ds_prcp.coords,
attrs=global_attrs,
)
for scale in scales:
var_name_alpha = f"alpha_{str(scale).zfill(2)}"
var_name_beta = f"beta_{str(scale).zfill(2)}"
da_spi = compute_spi_gamma(
da_prcp,
ds_gamma[var_name_alpha],
ds_gamma[var_name_alpha],
scale,
periodicity,
)
ds_spi[f"spi_gamma_{str(scale).zfill(2)}"] = da_spi
netcdf_spi = '/home/james/data/nclimgrid/nclimgrid_lowres_spi_gamma.nc'
ds_spi.to_netcdf(netcdf_spi)
ds_spi
```
Plot a time step to validate that the SPI values look reasonable:
```
ds_spi[f"spi_gamma_{str(scale).zfill(2)}"].isel(time=500).plot()
```
Define a function that computes SPI without using pre-computed fitting parameters.
```
def compute_spi_gamma_without_fittings(
da_precip: xr.DataArray,
scale: int,
periodicity: compute.Periodicity,
) -> xr.DataArray:
initial_year = int(da_precip['time'][0].dt.year)
total_lats = da_precip.shape[0]
total_lons = da_precip.shape[1]
spi = np.full(shape=da_precip.shape, fill_value=np.NaN)
for lat_index in range(total_lats):
for lon_index in range(total_lons):
# get the values for the lat/lon grid cell
values = da_precip[lat_index, lon_index]
# skip over this grid cell if all NaN values
if (np.ma.is_masked(values) and values.mask.all()) or np.all(np.isnan(values)):
continue
# compute the SPI
spi[lat_index, lon_index] = \
indices.spi(
values,
scale=scale,
distribution=indices.Distribution.gamma,
data_start_year=initial_year,
calibration_year_initial=calibration_year_initial,
calibration_year_final=calibration_year_final,
periodicity=compute.Periodicity.monthly,
)
# build a DataArray for this scale's SPI
da_spi = xr.DataArray(
data=spi,
coords=da_precip.coords,
dims=da_precip.dims,
name=f"spi_gamma_{str(scale).zfill(2)}",
)
da_spi.attrs = {
'description': f'SPI ({scale}-{periodicity} gamma) computed from monthly precipitation ' + \
f'data for the period {da_precip.time[0]} through {da_precip.time[-1]} using a ' + \
f'calibration period from {calibration_year_initial} through {calibration_year_final}',
'valid_min': -3.09,
'valid_max': 3.09,
'long_name': f'{scale}-{periodicity} SPI(gamma)',
'calibration_year_initial': calibration_year_initial,
'calibration_year_final': calibration_year_final,
}
return da_spi
```
Compute SPI without pre-computed fitting parameters to see if there's a significant time difference.
```
%%time
ds_spi_no_fittings = xr.Dataset(
coords=ds_prcp.coords,
attrs=global_attrs,
)
for scale in scales:
da_spi = compute_spi_gamma_without_fittings(
da_prcp,
scale,
periodicity,
)
ds_spi_no_fittings[f"spi_gamma_{str(scale).zfill(2)}"] = da_spi
netcdf_spi = '/home/james/data/nclimgrid/nclimgrid_lowres_spi_gamma_no_fittings.nc'
ds_spi_no_fittings.to_netcdf(netcdf_spi)
```
|
github_jupyter
|
import sys
from typing import Dict
import matplotlib
import numpy as np
import xarray as xr
climate_indices_home_path = "/home/james/git/climate_indices"
if climate_indices_home_path not in sys.path:
sys.path.insert(climate_indices_home_path)
from climate_indices import compute, indices, utils
%matplotlib inline
netcdf_precip = '/data/datasets/nclimgrid/nclimgrid_lowres_prcp.nc'
netcdf_gamma = '/data/datasets/nclimgrid/nclimgrid_lowres_gamma.nc'
ds_prcp = xr.open_dataset(netcdf_precip)
list(ds_prcp.dims)
da_prcp = ds_prcp['prcp'].transpose('lat', 'lon', 'time')
initial_year = int(da_prcp['time'][0].dt.year)
calibration_year_initial = 1900
calibration_year_final = 2000
period_times = 12
total_lats = da_prcp.shape[0]
total_lons = da_prcp.shape[1]
fitting_shape = (total_lats, total_lons, period_times)
scales = [1] # , 2, 3, 6, 9, 12, 24]
periodicity = compute.Periodicity.monthly
def compute_gammas(
da_precip: xr.DataArray,
gamma_coords: Dict,
scale: int,
calibration_year_initial,
calibration_year_final,
periodicity: compute.Periodicity,
) -> (xr.DataArray, xr.DataArray):
initial_year = int(da_precip['time'][0].dt.year)
if periodicity == compute.Periodicity.monthly:
period_times = 12
elif periodicity == compute.Periodicity.daily:
period_times = 366
total_lats = da_precip.shape[0]
total_lons = da_precip.shape[1]
fitting_shape = (total_lats, total_lons, period_times)
alphas = np.full(shape=fitting_shape, fill_value=np.NaN)
betas = np.full(shape=fitting_shape, fill_value=np.NaN)
# loop over the grid cells and compute the gamma parameters for each
for lat_index in range(total_lats):
for lon_index in range(total_lons):
# get the precipitation values for the lat/lon grid cell
values = da_precip[lat_index, lon_index]
# skip over this grid cell if all NaN values
if (np.ma.is_masked(values) and values.mask.all()) or np.all(np.isnan(values)):
continue
# convolve to scale
scaled_values = \
compute.scale_values(
values,
scale=scale,
periodicity=periodicity,
)
# compute the fitting parameters on the scaled data
alphas[lat_index, lon_index], betas[lat_index, lon_index] = \
compute.gamma_parameters(
scaled_values,
data_start_year=initial_year,
calibration_start_year=calibration_year_initial,
calibration_end_year=calibration_year_final,
periodicity=periodicity,
)
alpha_attrs = {
'description': 'shape parameter of the gamma distribution (also referred to as the concentration) ' + \
f'computed from the {scale}-month scaled precipitation values',
}
da_alpha = xr.DataArray(
data=alphas,
coords=gamma_coords,
dims=tuple(gamma_coords.keys()),
name=f"alpha_{str(scale).zfill(2)}",
attrs=alpha_attrs,
)
beta_attrs = {
'description': '1 / scale of the distribution (also referred to as the rate) ' + \
f'computed from the {scale}-month scaled precipitation values',
}
da_beta = xr.DataArray(
data=betas,
coords=gamma_coords,
dims=tuple(gamma_coords.keys()),
name=f"beta_{str(scale).zfill(2)}",
attrs=beta_attrs,
)
return da_alpha, da_beta
def compute_spi_gamma(
da_precip: xr.DataArray,
da_alpha: xr.DataArray,
da_beta: xr.DataArray,
scale: int,
periodicity: compute.Periodicity,
) -> xr.DataArray:
initial_year = int(da_precip['time'][0].dt.year)
total_lats = da_precip.shape[0]
total_lons = da_precip.shape[1]
spi = np.full(shape=da_precip.shape, fill_value=np.NaN)
for lat_index in range(total_lats):
for lon_index in range(total_lons):
# get the values for the lat/lon grid cell
values = da_precip[lat_index, lon_index]
# skip over this grid cell if all NaN values
if (np.ma.is_masked(values) and values.mask.all()) or np.all(np.isnan(values)):
continue
gamma_parameters = {
"alphas": da_alpha[lat_index, lon_index],
"betas": da_beta[lat_index, lon_index],
}
# compute the SPI
spi[lat_index, lon_index] = \
indices.spi(
values,
scale=scale,
distribution=indices.Distribution.gamma,
data_start_year=initial_year,
calibration_year_initial=calibration_year_initial,
calibration_year_final=calibration_year_final,
periodicity=compute.Periodicity.monthly,
fitting_params=gamma_parameters,
)
# build a DataArray for this scale's SPI
da_spi = xr.DataArray(
data=spi,
coords=da_precip.coords,
dims=da_precip.dims,
name=f"spi_gamma_{str(scale).zfill(2)}",
)
da_spi.attrs = {
'description': f'SPI ({scale}-{periodicity} gamma) computed from monthly precipitation ' + \
f'data for the period {da_precip.time[0]} through {da_precip.time[-1]} using a ' + \
f'calibration period from {calibration_year_initial} through {calibration_year_final}',
'valid_min': -3.09,
'valid_max': 3.09,
'long_name': f'{scale}-{periodicity} SPI(gamma)',
'calibration_year_initial': calibration_year_initial,
'calibration_year_final': calibration_year_final,
}
return da_spi
attrs_to_copy = [
'Conventions',
'ncei_template_version',
'naming_authority',
'standard_name_vocabulary',
'institution',
'geospatial_lat_min',
'geospatial_lat_max',
'geospatial_lon_min',
'geospatial_lon_max',
'geospatial_lat_units',
'geospatial_lon_units',
]
global_attrs = {key: value for (key, value) in ds_prcp.attrs.items() if key in attrs_to_copy}
%%time
if periodicity == compute.Periodicity.monthly:
period_times = 12
time_coord_name = "month"
elif periodicity == compute.Periodicity.daily:
period_times = 366
time_coord_name = "day"
gamma_coords = {"lat": ds_prcp.lat, "lon": ds_prcp.lon, time_coord_name: range(period_times)}
ds_gamma = xr.Dataset(
coords=gamma_coords,
attrs=global_attrs,
)
for scale in scales:
var_name_alpha = f"alpha_{str(scale).zfill(2)}"
var_name_beta = f"beta_{str(scale).zfill(2)}"
da_alpha, da_beta = compute_gammas(
da_prcp,
gamma_coords,
scale,
calibration_year_initial,
calibration_year_final,
periodicity,
)
ds_gamma[var_name_alpha] = da_alpha
ds_gamma[var_name_beta] = da_beta
ds_gamma.to_netcdf(netcdf_gamma)
ds_gamma
%%time
ds_spi = xr.Dataset(
coords=ds_prcp.coords,
attrs=global_attrs,
)
for scale in scales:
var_name_alpha = f"alpha_{str(scale).zfill(2)}"
var_name_beta = f"beta_{str(scale).zfill(2)}"
da_spi = compute_spi_gamma(
da_prcp,
ds_gamma[var_name_alpha],
ds_gamma[var_name_alpha],
scale,
periodicity,
)
ds_spi[f"spi_gamma_{str(scale).zfill(2)}"] = da_spi
netcdf_spi = '/home/james/data/nclimgrid/nclimgrid_lowres_spi_gamma.nc'
ds_spi.to_netcdf(netcdf_spi)
ds_spi
ds_spi[f"spi_gamma_{str(scale).zfill(2)}"].isel(time=500).plot()
def compute_spi_gamma_without_fittings(
da_precip: xr.DataArray,
scale: int,
periodicity: compute.Periodicity,
) -> xr.DataArray:
initial_year = int(da_precip['time'][0].dt.year)
total_lats = da_precip.shape[0]
total_lons = da_precip.shape[1]
spi = np.full(shape=da_precip.shape, fill_value=np.NaN)
for lat_index in range(total_lats):
for lon_index in range(total_lons):
# get the values for the lat/lon grid cell
values = da_precip[lat_index, lon_index]
# skip over this grid cell if all NaN values
if (np.ma.is_masked(values) and values.mask.all()) or np.all(np.isnan(values)):
continue
# compute the SPI
spi[lat_index, lon_index] = \
indices.spi(
values,
scale=scale,
distribution=indices.Distribution.gamma,
data_start_year=initial_year,
calibration_year_initial=calibration_year_initial,
calibration_year_final=calibration_year_final,
periodicity=compute.Periodicity.monthly,
)
# build a DataArray for this scale's SPI
da_spi = xr.DataArray(
data=spi,
coords=da_precip.coords,
dims=da_precip.dims,
name=f"spi_gamma_{str(scale).zfill(2)}",
)
da_spi.attrs = {
'description': f'SPI ({scale}-{periodicity} gamma) computed from monthly precipitation ' + \
f'data for the period {da_precip.time[0]} through {da_precip.time[-1]} using a ' + \
f'calibration period from {calibration_year_initial} through {calibration_year_final}',
'valid_min': -3.09,
'valid_max': 3.09,
'long_name': f'{scale}-{periodicity} SPI(gamma)',
'calibration_year_initial': calibration_year_initial,
'calibration_year_final': calibration_year_final,
}
return da_spi
%%time
ds_spi_no_fittings = xr.Dataset(
coords=ds_prcp.coords,
attrs=global_attrs,
)
for scale in scales:
da_spi = compute_spi_gamma_without_fittings(
da_prcp,
scale,
periodicity,
)
ds_spi_no_fittings[f"spi_gamma_{str(scale).zfill(2)}"] = da_spi
netcdf_spi = '/home/james/data/nclimgrid/nclimgrid_lowres_spi_gamma_no_fittings.nc'
ds_spi_no_fittings.to_netcdf(netcdf_spi)
| 0.563858 | 0.778186 |
# predictions
---
this notebook explores the output of the SSIM net and how its predictions compare to the actual SSIM data. we will also explore some of the filters that the network has learned and see what they look like.
```
import numpy as np
import pandas as pd
import scipy.signal as sig
import matplotlib.pyplot as plt
import iqa_tools as iqa
import matplotlib.gridspec as gridspec
import tensorflow as tf
image_dim, result_dim = 96, 86
input_layer, output_layer = 4, 1
input_layer, first_layer, second_layer, third_layer, fourth_layer, output_layer = 4, 100, 50, 25, 10, 1
filter_dim, filter_dim2 = 11, 1
```
read in the data files
```
# data input
data_path = 'https://raw.githubusercontent.com/michaelneuder/image_quality_analysis/master/data/sample_data/'
# train data --- 500 images, 96x96 pixels
orig_500 = pd.read_csv('{}orig_500.txt'.format(data_path), header=None, delim_whitespace = True)
recon_500 = pd.read_csv('{}recon_500.txt'.format(data_path), header=None, delim_whitespace = True)
# test data --- 140 images, 96x96 pixels
orig_140 = pd.read_csv('{}orig_140.txt'.format(data_path), header=None, delim_whitespace = True)
recon_140 = pd.read_csv('{}recon_140.txt'.format(data_path), header=None, delim_whitespace = True)
# train target --- 500 images, 86x86 pixels (dimension reduction due no zero padding being used)
ssim_500 = pd.read_csv('{}ssim_500_nogauss.csv'.format(data_path), header=None)
ssim_140 = pd.read_csv('{}ssim_140_nogauss.csv'.format(data_path), header=None)
# getting 4 input channels for train and test --- (orig, recon, orig squared, recon squared)
original_images_train = orig_500.values
original_images_train_sq = orig_500.values**2
reconstructed_images_train = recon_500.values
reconstructed_images_train_sq = recon_500.values**2
original_images_test = orig_140.values
original_images_test_sq = orig_140.values**2
reconstructed_images_test = recon_140.values
reconstructed_images_test_sq = recon_140.values**2
# stack inputs
training_input = np.dstack((original_images_train, reconstructed_images_train, original_images_train_sq, reconstructed_images_train_sq))
testing_input = np.dstack((original_images_test, reconstructed_images_test, original_images_test_sq, reconstructed_images_test_sq))
# normalize inputs
training_input_normalized, testing_input_normalized = iqa.normalize_input(training_input, testing_input)
# target values
training_target = ssim_500.values
testing_target = ssim_140.values
# get size of training and testing set
train_size = original_images_train.shape[0]
test_size = original_images_test.shape[0]
# reshaping features to (num images, 96x96, 4 channels)
train_features = np.reshape(training_input_normalized, [train_size,image_dim,image_dim,input_layer])
test_features = np.reshape(testing_input_normalized, [test_size,image_dim,image_dim,input_layer])
# reshaping target to --- (num images, 86x86, 1)
train_target = np.reshape(training_target, [train_size, result_dim, result_dim, output_layer])
test_target = np.reshape(testing_target, [test_size, result_dim, result_dim, output_layer])
plt.figure(figsize = (12,12))
gs1 = gridspec.GridSpec(3, 3)
gs1.update(wspace=0, hspace=0.03)
for i in range(3):
x = np.random.randint(500)
ax1, ax2, ax3 = plt.subplot(gs1[3*i]), plt.subplot(gs1[3*i+1]), plt.subplot(gs1[3*i+2])
for ax in [ax1, ax2, ax3]:
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
if i == 0:
ax1.set_title('original', size=20)
ax2.set_title('reconstructed', size=20)
ax3.set_title('ssim', size=20)
ax1.imshow(train_features[x,:,:,0], cmap='gray')
ax2.imshow(train_features[x,:,:,1], cmap='gray')
ax3.imshow(train_target[x,:,:,0], cmap='plasma')
plt.show()
```
ok now that we have all the data in place we need to load the current weights learned by the network.
```
weights = {}; biases = {};
for entry in ['weights1', 'weights2', 'weights3', 'weights4', 'weights_out']:
temp = np.loadtxt('weights/{}.txt'.format(entry))
weights[entry] = temp
for entry in ['bias1', 'bias2', 'bias3', 'bias4', 'bias_out']:
temp = np.loadtxt('weights/{}.txt'.format(entry))
biases[entry] = temp
weights['weights1'] = weights['weights1'].reshape((filter_dim,filter_dim,input_layer,first_layer))
weights['weights2'] = weights['weights2'].reshape((filter_dim2,filter_dim2,first_layer,second_layer))
weights['weights3'] = weights['weights3'].reshape((filter_dim2,filter_dim2,second_layer,third_layer))
weights['weights4'] = weights['weights4'].reshape((filter_dim2,filter_dim2,third_layer,fourth_layer))
weights['weights_out'] = weights['weights_out'].reshape((filter_dim2,filter_dim2,fourth_layer+third_layer+second_layer+first_layer,output_layer))
biases['bias1'] = biases['bias1'].reshape((first_layer))
biases['bias2'] = biases['bias2'].reshape((second_layer))
biases['bias3'] = biases['bias3'].reshape((third_layer))
biases['bias4'] = biases['bias4'].reshape((fourth_layer))
biases['bias_out'] = biases['bias_out'].reshape((output_layer))
for weight in weights:
print(weights[weight].shape)
for bias in biases:
print(biases[bias].shape)
```
ok now we have all the filters loaded in lets look at a couple for kicks.
```
plt.figure(figsize = (9,9))
gs1 = gridspec.GridSpec(3, 3)
gs1.update(wspace=0, hspace=0.03)
for i in range(3):
ax1, ax2, ax3 = plt.subplot(gs1[3*i]), plt.subplot(gs1[3*i+1]), plt.subplot(gs1[3*i+2])
for ax in [ax1, ax2, ax3]:
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
if i == 0:
ax1.set_title('original', size=20)
ax2.set_title('reconstructed', size=20)
ax3.set_title('orig sq', size=20)
x = np.random.randint(100)
ax1.imshow(weights['weights1'][:,:,0,x], cmap='gray')
ax2.imshow(weights['weights1'][:,:,1,x], cmap='gray')
ax3.imshow(weights['weights1'][:,:,2,x], cmap='gray')
plt.show()
```
cool looking i guess. now lets convolve some images.
```
def convolve_inner_layers(x, W, b):
'''
inner layers of network --- tanh activation
'''
y = tf.nn.conv2d(x, W, strides = [1,1,1,1], padding='VALID')
y = tf.nn.bias_add(y, b)
return tf.nn.tanh(y)
def convolve_ouput_layer(x, W, b):
'''
output layer of network --- linear activation
'''
y = tf.nn.conv2d(x, W, strides = [1,1,1,1], padding='VALID')
y = tf.nn.bias_add(y, b)
return y
def conv_net(x, W, b):
'''
entire conv net. each layer feed to following layer as well as output layer
'''
conv1 = convolve_inner_layers(x, W['weights1'], b['bias1'])
conv2 = convolve_inner_layers(conv1, W['weights2'], b['bias2'])
conv3 = convolve_inner_layers(conv2, W['weights3'], b['bias3'])
conv4 = convolve_inner_layers(conv3, W['weights4'], b['bias4'])
output_feed = tf.concat([conv1, conv2, conv3, conv4],3)
output = convolve_ouput_layer(output_feed, W['weights_out'], b['bias_out'])
return output
test_features.shape
sess = tf.Session()
test_im = np.reshape(train_features[:3,:,:,:], (3,96,96,4))
prediction = sess.run(conv_net(tf.cast(test_im, 'float32'), weights, biases))
test_im.shape
prediction.shape
plt.figure(figsize = (16,12))
gs1 = gridspec.GridSpec(3, 4)
gs1.update(wspace=0, hspace=0.03)
for i in range(3):
ax1, ax2, ax3, ax4 = plt.subplot(gs1[4*i]), plt.subplot(gs1[4*i+1]), plt.subplot(gs1[4*i+2]), plt.subplot(gs1[4*i+3])
for ax in [ax1, ax2, ax3, ax4]:
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
if i == 0:
ax1.set_title('original', size=20)
ax2.set_title('reconstructed', size=20)
ax3.set_title('ssim', size=20)
ax4.set_title('ssim net prediction', size=20)
ax1.imshow(train_features[i,:,:,0], cmap='gray')
ax2.imshow(train_features[i,:,:,1], cmap='gray')
ax3.imshow(train_target[i,:,:,0], cmap='plasma')
ax4.imshow(prediction[i,:,:,0], cmap='plasma')
plt.savefig('prediction_demo1.png')
plt.show()
np.square(np.mean(train_target[:3,:,:,0] - prediction[:,:,:,0])) / np.var(train_target)*100
np.var(train_target)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import scipy.signal as sig
import matplotlib.pyplot as plt
import iqa_tools as iqa
import matplotlib.gridspec as gridspec
import tensorflow as tf
image_dim, result_dim = 96, 86
input_layer, output_layer = 4, 1
input_layer, first_layer, second_layer, third_layer, fourth_layer, output_layer = 4, 100, 50, 25, 10, 1
filter_dim, filter_dim2 = 11, 1
# data input
data_path = 'https://raw.githubusercontent.com/michaelneuder/image_quality_analysis/master/data/sample_data/'
# train data --- 500 images, 96x96 pixels
orig_500 = pd.read_csv('{}orig_500.txt'.format(data_path), header=None, delim_whitespace = True)
recon_500 = pd.read_csv('{}recon_500.txt'.format(data_path), header=None, delim_whitespace = True)
# test data --- 140 images, 96x96 pixels
orig_140 = pd.read_csv('{}orig_140.txt'.format(data_path), header=None, delim_whitespace = True)
recon_140 = pd.read_csv('{}recon_140.txt'.format(data_path), header=None, delim_whitespace = True)
# train target --- 500 images, 86x86 pixels (dimension reduction due no zero padding being used)
ssim_500 = pd.read_csv('{}ssim_500_nogauss.csv'.format(data_path), header=None)
ssim_140 = pd.read_csv('{}ssim_140_nogauss.csv'.format(data_path), header=None)
# getting 4 input channels for train and test --- (orig, recon, orig squared, recon squared)
original_images_train = orig_500.values
original_images_train_sq = orig_500.values**2
reconstructed_images_train = recon_500.values
reconstructed_images_train_sq = recon_500.values**2
original_images_test = orig_140.values
original_images_test_sq = orig_140.values**2
reconstructed_images_test = recon_140.values
reconstructed_images_test_sq = recon_140.values**2
# stack inputs
training_input = np.dstack((original_images_train, reconstructed_images_train, original_images_train_sq, reconstructed_images_train_sq))
testing_input = np.dstack((original_images_test, reconstructed_images_test, original_images_test_sq, reconstructed_images_test_sq))
# normalize inputs
training_input_normalized, testing_input_normalized = iqa.normalize_input(training_input, testing_input)
# target values
training_target = ssim_500.values
testing_target = ssim_140.values
# get size of training and testing set
train_size = original_images_train.shape[0]
test_size = original_images_test.shape[0]
# reshaping features to (num images, 96x96, 4 channels)
train_features = np.reshape(training_input_normalized, [train_size,image_dim,image_dim,input_layer])
test_features = np.reshape(testing_input_normalized, [test_size,image_dim,image_dim,input_layer])
# reshaping target to --- (num images, 86x86, 1)
train_target = np.reshape(training_target, [train_size, result_dim, result_dim, output_layer])
test_target = np.reshape(testing_target, [test_size, result_dim, result_dim, output_layer])
plt.figure(figsize = (12,12))
gs1 = gridspec.GridSpec(3, 3)
gs1.update(wspace=0, hspace=0.03)
for i in range(3):
x = np.random.randint(500)
ax1, ax2, ax3 = plt.subplot(gs1[3*i]), plt.subplot(gs1[3*i+1]), plt.subplot(gs1[3*i+2])
for ax in [ax1, ax2, ax3]:
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
if i == 0:
ax1.set_title('original', size=20)
ax2.set_title('reconstructed', size=20)
ax3.set_title('ssim', size=20)
ax1.imshow(train_features[x,:,:,0], cmap='gray')
ax2.imshow(train_features[x,:,:,1], cmap='gray')
ax3.imshow(train_target[x,:,:,0], cmap='plasma')
plt.show()
weights = {}; biases = {};
for entry in ['weights1', 'weights2', 'weights3', 'weights4', 'weights_out']:
temp = np.loadtxt('weights/{}.txt'.format(entry))
weights[entry] = temp
for entry in ['bias1', 'bias2', 'bias3', 'bias4', 'bias_out']:
temp = np.loadtxt('weights/{}.txt'.format(entry))
biases[entry] = temp
weights['weights1'] = weights['weights1'].reshape((filter_dim,filter_dim,input_layer,first_layer))
weights['weights2'] = weights['weights2'].reshape((filter_dim2,filter_dim2,first_layer,second_layer))
weights['weights3'] = weights['weights3'].reshape((filter_dim2,filter_dim2,second_layer,third_layer))
weights['weights4'] = weights['weights4'].reshape((filter_dim2,filter_dim2,third_layer,fourth_layer))
weights['weights_out'] = weights['weights_out'].reshape((filter_dim2,filter_dim2,fourth_layer+third_layer+second_layer+first_layer,output_layer))
biases['bias1'] = biases['bias1'].reshape((first_layer))
biases['bias2'] = biases['bias2'].reshape((second_layer))
biases['bias3'] = biases['bias3'].reshape((third_layer))
biases['bias4'] = biases['bias4'].reshape((fourth_layer))
biases['bias_out'] = biases['bias_out'].reshape((output_layer))
for weight in weights:
print(weights[weight].shape)
for bias in biases:
print(biases[bias].shape)
plt.figure(figsize = (9,9))
gs1 = gridspec.GridSpec(3, 3)
gs1.update(wspace=0, hspace=0.03)
for i in range(3):
ax1, ax2, ax3 = plt.subplot(gs1[3*i]), plt.subplot(gs1[3*i+1]), plt.subplot(gs1[3*i+2])
for ax in [ax1, ax2, ax3]:
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
if i == 0:
ax1.set_title('original', size=20)
ax2.set_title('reconstructed', size=20)
ax3.set_title('orig sq', size=20)
x = np.random.randint(100)
ax1.imshow(weights['weights1'][:,:,0,x], cmap='gray')
ax2.imshow(weights['weights1'][:,:,1,x], cmap='gray')
ax3.imshow(weights['weights1'][:,:,2,x], cmap='gray')
plt.show()
def convolve_inner_layers(x, W, b):
'''
inner layers of network --- tanh activation
'''
y = tf.nn.conv2d(x, W, strides = [1,1,1,1], padding='VALID')
y = tf.nn.bias_add(y, b)
return tf.nn.tanh(y)
def convolve_ouput_layer(x, W, b):
'''
output layer of network --- linear activation
'''
y = tf.nn.conv2d(x, W, strides = [1,1,1,1], padding='VALID')
y = tf.nn.bias_add(y, b)
return y
def conv_net(x, W, b):
'''
entire conv net. each layer feed to following layer as well as output layer
'''
conv1 = convolve_inner_layers(x, W['weights1'], b['bias1'])
conv2 = convolve_inner_layers(conv1, W['weights2'], b['bias2'])
conv3 = convolve_inner_layers(conv2, W['weights3'], b['bias3'])
conv4 = convolve_inner_layers(conv3, W['weights4'], b['bias4'])
output_feed = tf.concat([conv1, conv2, conv3, conv4],3)
output = convolve_ouput_layer(output_feed, W['weights_out'], b['bias_out'])
return output
test_features.shape
sess = tf.Session()
test_im = np.reshape(train_features[:3,:,:,:], (3,96,96,4))
prediction = sess.run(conv_net(tf.cast(test_im, 'float32'), weights, biases))
test_im.shape
prediction.shape
plt.figure(figsize = (16,12))
gs1 = gridspec.GridSpec(3, 4)
gs1.update(wspace=0, hspace=0.03)
for i in range(3):
ax1, ax2, ax3, ax4 = plt.subplot(gs1[4*i]), plt.subplot(gs1[4*i+1]), plt.subplot(gs1[4*i+2]), plt.subplot(gs1[4*i+3])
for ax in [ax1, ax2, ax3, ax4]:
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
if i == 0:
ax1.set_title('original', size=20)
ax2.set_title('reconstructed', size=20)
ax3.set_title('ssim', size=20)
ax4.set_title('ssim net prediction', size=20)
ax1.imshow(train_features[i,:,:,0], cmap='gray')
ax2.imshow(train_features[i,:,:,1], cmap='gray')
ax3.imshow(train_target[i,:,:,0], cmap='plasma')
ax4.imshow(prediction[i,:,:,0], cmap='plasma')
plt.savefig('prediction_demo1.png')
plt.show()
np.square(np.mean(train_target[:3,:,:,0] - prediction[:,:,:,0])) / np.var(train_target)*100
np.var(train_target)
| 0.464659 | 0.951818 |
```
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
import matplotlib.pyplot as plt
import PIL
from PIL import Image
import cv2
import torch
import torchvision
from torchvision.models.detection import FasterRCNN
from torchvision.models.detection.faster_rcnn import FastRCNNPredictor
from torchvision import transforms
import torch.utils.data
from torch.utils.data import Dataset, DataLoader
import random
import albumentations
from albumentations.pytorch.transforms import ToTensorV2
def get_instance_objectdetection_model(num_classes,path_weight):
# load an instance segmentation model pre-trained on COCO
create_model = torchvision.models.detection.fasterrcnn_resnet50_fpn(pretrained=False,pretrained_backbone=False)
# get the number of input features for the classifier
in_features = create_model.roi_heads.box_predictor.cls_score.in_features
# replace the pre-trained head with a new one
create_model.roi_heads.box_predictor = FastRCNNPredictor(in_features, num_classes)
create_model.load_state_dict(torch.load(path_weight,map_location=torch.device('cpu')))
return create_model
#os.listdir("/kaggle/input/gwd-thirditeration-weights/customtrained_fasterrcnn_resnet50_fpn_augementation_28052020_1340.pth")
#path_weight = "/kaggle/input/global_wheat_detection/fasterrcnn_resnet50_fpn_best.pth"
#path_weight = "/kaggle/input/gwd-augmentation-training-weights-ver01/customtrained_fasterrcnn_resnet50_fpn_augementation (1).pth"
path_weight = "/kaggle/input/gwd-thirditeration-weights/customtrained_fasterrcnn_resnet50_fpn_augementation_28052020_1340.pth"
num_classes = 2
# Why 2 classes - background and wheat-heads
trained_model = get_instance_objectdetection_model(num_classes,path_weight)
def get_test_transform():
return albumentations.Compose([
# A.Resize(512, 512),
ToTensorV2(p=1.0)
])
class GlobalWheatDetectionTestDataset(torch.utils.data.Dataset):
# first lets start with __init__ and initialize any objects
def __init__(self,input_df,input_dir,transforms=None):
self.df=input_df
self.list_images = list(self.df['image_id'].unique())
self.image_dir=input_dir
self.transforms = transforms
# next lets define __getitem__
# very important to note what it returns for EACH image:
# I. image - a PIL image of size (H,W) for ResNet50 FPN image should be scaled
# II. image_id
def __getitem__(self,idx):
# II. image_id
img_id = self.list_images[idx]
# I. Input image
# Specifications: A.RGB format B. scaled (0,1) C. size (H,W) D. PIL format
img = cv2.imread(self.image_dir+"/"+img_id+".jpg")
img_RGB = cv2.cvtColor(img,cv2.COLOR_BGR2RGB)
img_scaled = img_RGB/255.0
img_final = img_scaled
ret_image = {}
ret_image['image']=img_final
if self.transforms is not None:
res = self.transforms(**ret_image)
img_final = res['image']
#img_final = torch.tensor(img_final, dtype=torch.float32)
return img_final, img_id
# next lets define __len__
def __len__(self):
return len(self.df['image_id'].unique())
def collate_fn(batch):
return tuple(zip(*batch))
df_test=pd.read_csv("/kaggle/input/global-wheat-detection/sample_submission.csv")
test_dir="/kaggle/input/global-wheat-detection/test/"
df_test.head()
test_dataset = GlobalWheatDetectionTestDataset(df_test,test_dir,get_test_transform())
test_dataloader = DataLoader(test_dataset, batch_size=8,shuffle=False, num_workers=1,collate_fn=collate_fn)
device = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
print(device)
detection_threshold = 0.45
def format_prediction_string(boxes, scores): ## Define the formate for storing prediction results
pred_strings = []
for j in zip(scores, boxes):
pred_strings.append("{0:.4f} {1} {2} {3} {4}".format(j[0], j[1][0], j[1][1], j[1][2], j[1][3]))
return " ".join(pred_strings)
images, img_ids = next(iter(test_dataloader))
## Lets make the prediction
results=[]
trained_model.to(device)
trained_model.eval()
images = []
outputs =[]
for images_, image_ids in test_dataloader:
images = list(image.to(device,dtype=torch.float) for image in images_)
outputs = trained_model(images)
for i, image in enumerate(images):
boxes = outputs[i]['boxes'].data.cpu().numpy() ##Formate of the output's box is [Xmin,Ymin,Xmax,Ymax]
scores = outputs[i]['scores'].data.cpu().numpy()
boxes = boxes[scores >= detection_threshold].astype(np.int32) #Compare the score of output with the threshold and
scores = scores[scores >= detection_threshold] #slelect only those boxes whose score is greater
# than threshold value
image_id = image_ids[i]
boxes[:, 2] = boxes[:, 2] - boxes[:, 0]
boxes[:, 3] = boxes[:, 3] - boxes[:, 1] #Convert the box formate to [Xmin,Ymin,W,H]
result = { #Store the image id and boxes and scores in result dict.
'image_id': image_id,
'PredictionString': format_prediction_string(boxes, scores)
}
results.append(result) #Append the result dict to Results list
test_df = pd.DataFrame(results, columns=['image_id', 'PredictionString'])
test_df.head()
test_df.to_csv('submission.csv', index=False)
```
|
github_jupyter
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
import matplotlib.pyplot as plt
import PIL
from PIL import Image
import cv2
import torch
import torchvision
from torchvision.models.detection import FasterRCNN
from torchvision.models.detection.faster_rcnn import FastRCNNPredictor
from torchvision import transforms
import torch.utils.data
from torch.utils.data import Dataset, DataLoader
import random
import albumentations
from albumentations.pytorch.transforms import ToTensorV2
def get_instance_objectdetection_model(num_classes,path_weight):
# load an instance segmentation model pre-trained on COCO
create_model = torchvision.models.detection.fasterrcnn_resnet50_fpn(pretrained=False,pretrained_backbone=False)
# get the number of input features for the classifier
in_features = create_model.roi_heads.box_predictor.cls_score.in_features
# replace the pre-trained head with a new one
create_model.roi_heads.box_predictor = FastRCNNPredictor(in_features, num_classes)
create_model.load_state_dict(torch.load(path_weight,map_location=torch.device('cpu')))
return create_model
#os.listdir("/kaggle/input/gwd-thirditeration-weights/customtrained_fasterrcnn_resnet50_fpn_augementation_28052020_1340.pth")
#path_weight = "/kaggle/input/global_wheat_detection/fasterrcnn_resnet50_fpn_best.pth"
#path_weight = "/kaggle/input/gwd-augmentation-training-weights-ver01/customtrained_fasterrcnn_resnet50_fpn_augementation (1).pth"
path_weight = "/kaggle/input/gwd-thirditeration-weights/customtrained_fasterrcnn_resnet50_fpn_augementation_28052020_1340.pth"
num_classes = 2
# Why 2 classes - background and wheat-heads
trained_model = get_instance_objectdetection_model(num_classes,path_weight)
def get_test_transform():
return albumentations.Compose([
# A.Resize(512, 512),
ToTensorV2(p=1.0)
])
class GlobalWheatDetectionTestDataset(torch.utils.data.Dataset):
# first lets start with __init__ and initialize any objects
def __init__(self,input_df,input_dir,transforms=None):
self.df=input_df
self.list_images = list(self.df['image_id'].unique())
self.image_dir=input_dir
self.transforms = transforms
# next lets define __getitem__
# very important to note what it returns for EACH image:
# I. image - a PIL image of size (H,W) for ResNet50 FPN image should be scaled
# II. image_id
def __getitem__(self,idx):
# II. image_id
img_id = self.list_images[idx]
# I. Input image
# Specifications: A.RGB format B. scaled (0,1) C. size (H,W) D. PIL format
img = cv2.imread(self.image_dir+"/"+img_id+".jpg")
img_RGB = cv2.cvtColor(img,cv2.COLOR_BGR2RGB)
img_scaled = img_RGB/255.0
img_final = img_scaled
ret_image = {}
ret_image['image']=img_final
if self.transforms is not None:
res = self.transforms(**ret_image)
img_final = res['image']
#img_final = torch.tensor(img_final, dtype=torch.float32)
return img_final, img_id
# next lets define __len__
def __len__(self):
return len(self.df['image_id'].unique())
def collate_fn(batch):
return tuple(zip(*batch))
df_test=pd.read_csv("/kaggle/input/global-wheat-detection/sample_submission.csv")
test_dir="/kaggle/input/global-wheat-detection/test/"
df_test.head()
test_dataset = GlobalWheatDetectionTestDataset(df_test,test_dir,get_test_transform())
test_dataloader = DataLoader(test_dataset, batch_size=8,shuffle=False, num_workers=1,collate_fn=collate_fn)
device = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
print(device)
detection_threshold = 0.45
def format_prediction_string(boxes, scores): ## Define the formate for storing prediction results
pred_strings = []
for j in zip(scores, boxes):
pred_strings.append("{0:.4f} {1} {2} {3} {4}".format(j[0], j[1][0], j[1][1], j[1][2], j[1][3]))
return " ".join(pred_strings)
images, img_ids = next(iter(test_dataloader))
## Lets make the prediction
results=[]
trained_model.to(device)
trained_model.eval()
images = []
outputs =[]
for images_, image_ids in test_dataloader:
images = list(image.to(device,dtype=torch.float) for image in images_)
outputs = trained_model(images)
for i, image in enumerate(images):
boxes = outputs[i]['boxes'].data.cpu().numpy() ##Formate of the output's box is [Xmin,Ymin,Xmax,Ymax]
scores = outputs[i]['scores'].data.cpu().numpy()
boxes = boxes[scores >= detection_threshold].astype(np.int32) #Compare the score of output with the threshold and
scores = scores[scores >= detection_threshold] #slelect only those boxes whose score is greater
# than threshold value
image_id = image_ids[i]
boxes[:, 2] = boxes[:, 2] - boxes[:, 0]
boxes[:, 3] = boxes[:, 3] - boxes[:, 1] #Convert the box formate to [Xmin,Ymin,W,H]
result = { #Store the image id and boxes and scores in result dict.
'image_id': image_id,
'PredictionString': format_prediction_string(boxes, scores)
}
results.append(result) #Append the result dict to Results list
test_df = pd.DataFrame(results, columns=['image_id', 'PredictionString'])
test_df.head()
test_df.to_csv('submission.csv', index=False)
| 0.684791 | 0.42316 |
# MapReduce using SPARK
```
%pylab inline
import pandas as pd
import seaborn as sns
pd.set_option('display.width', 500)
pd.set_option('display.max_columns', 100)
```
# Table of Contents
* [SPARK](#SPARK)
* Installing Spark locally
* [Spark Context](#Spark-Context)
* [Create A RDD](#Create-A-RDD)
* [Call `collect` on an RDD: Lazy Spark](#Call-collect-on-an-RDD:-Lazy-Spark)
* [Operations on RDDs](#Operations-on-RDDs)
* [Word Examples](#Word-Examples)
* [Key Value Pairs](#Key-Value-Pairs)
* [word count 1](#word-count-1)
* [word count 2: `reduceByKey()`](#word-count-2:--reduceByKey%28%29)
* [Nested Syntax](#Nested-Syntax)
* [Using Cache](#Using-Cache)
* [Fun with words](#Fun-with-words)
* [DataFrames](#DataFrames)
* [Machine Learning](#Machine-Learning)
With shameless stealing of some code and text from:
- https://github.com/tdhopper/rta-pyspark-presentation/blob/master/slides.ipynb
- Databricks and Berkeley Spark MOOC: https://www.edx.org/course/introduction-big-data-apache-spark-uc-berkeleyx-cs100-1x
which you should go check out.
## Installing Spark locally
**Step 1: Install Apache Spark**
For example, for Mac users using Homebrew:
```
$ brew install apache-spark
```
**Step 2: Install the Java SDK version 1.8 or above for your platform (not just the JRE runtime)**
Make sure you can access commands such as `java` on your command line.
**Step 3: Install the latest findspark package using pip**
```
➜ ~ pip install findspark
Collecting findspark
Downloading findspark-0.0.5-py2.py3-none-any.whl
Installing collected packages: findspark
Successfully installed findspark-0.0.5
```
# Spark Context
You can also use it directly from the notebook interface on the mac if you installed `apache-spark` using `brew` and also installed `findspark` above.
```
import findspark
findspark.init('c:\spark')
import pyspark
sc = pyspark.SparkContext()
sc
sc.parallelize([1, 2, 3, 4, 5, 6, 7, 8, 9, 10]).map(lambda x: x**2).sum()
```
### Create A RDD
```
wordsList = ['cat', 'elephant', 'rat', 'rat', 'cat']
wordsRDD = sc.parallelize(wordsList, 4)
# Print out the type of wordsRDD
print type(wordsRDD)
```
### Call `collect` on an RDD: Lazy Spark
Spark is lazy. Until you `collect`, nothing is actually run.
>Instead, they just remember the transformations applied to some base dataset (e.g. a file). The transformations are only computed when an action requires a result to be returned to the driver program.
```
wordsRDD.collect()
```
### Operations on RDDs
From the Spark Programming Guide:
>RDDs support two types of operations: transformations, which create a new dataset from an existing one, and actions, which return a value to the driver program after running a computation on the dataset. For example, map is a transformation that passes each dataset element through a function and returns a new RDD representing the results. On the other hand, reduce is an action that aggregates all the elements of the RDD using some function and returns the final result to the driver program (although there is also a parallel reduceByKey that returns a distributed dataset).
### Word Examples
```
def makePlural(word):
return word + 's'
print makePlural('cat')
```
Transform one RDD into another.
```
pluralRDD = wordsRDD.map(makePlural)
print pluralRDD.first()
print pluralRDD.take(2)
pluralRDD.take(1)
pluralRDD.collect()
```
### Key Value Pairs
```
wordPairs = wordsRDD.map(lambda w: (w, 1))
print wordPairs.collect()
```
### WORD COUNT!
This little exercise shows how to use mapreduce to calculate the counts of individual words in a list.
```
wordsList = ['cat', 'elephant', 'rat', 'rat', 'cat']
wordsRDD = sc.parallelize(wordsList, 4)
wordCountsCollected = (wordsRDD
.map(lambda w: (w, 1))
.reduceByKey(lambda x,y: x+y)
.collect())
print wordCountsCollected
```
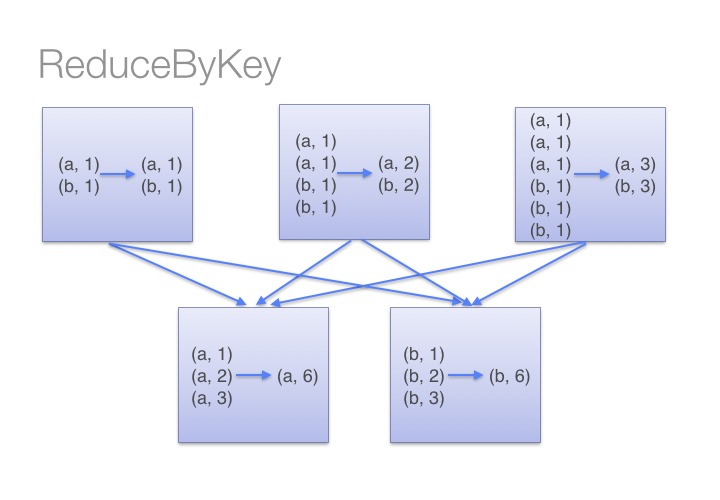
```
print (wordsRDD
.map(lambda w: (w, 1))
.reduceByKey(lambda x,y: x+y)).toDebugString()
```
### Using Cache
```
wordsList = ['cat', 'elephant', 'rat', 'rat', 'cat']
wordsRDD = sc.parallelize(wordsList, 4)
print wordsRDD
wordsRDD.count()
```
Normally, every operation is run from the start. This may be inefficient in many cases. So when appropriate, we may want to cache the result the first time an operation is run on an RDD.
```
#this is rerun from the start
wordsRDD.count()
#default storage level (MEMORY_ONLY)
wordsRDD.cache()#nothing done this is still lazy
#parallelize is rerun and cached because we told it to cache
wordsRDD.count()
#this `sc.parallelize` is not rerun in this case
wordsRDD.count()
```
Where is this useful: it is when you have branching parts or loops, so that you dont do things again and again. Spark, being "lazy" will rerun the chain again. So `cache` or `persist` serves as a checkpoint, breaking the RDD chain or the *lineage*.
```
birdsList=['heron','owl']
animList=wordsList+birdsList
animaldict={}
for e in wordsList:
animaldict[e]='mammal'
for e in birdsList:
animaldict[e]='bird'
animaldict
animsrdd = sc.parallelize(animList, 4)
animsrdd.cache()
#below runs the whole chain but causes cache to be populated
mammalcount=animsrdd.filter(lambda w: animaldict[w]=='mammal').count()
#now only the filter is carried out
birdcount=animsrdd.filter(lambda w: animaldict[w]=='bird').count()
print mammalcount, birdcount
```
### Exercises: Fun with MapReduce
Read http://spark.apache.org/docs/latest/programming-guide.html for some useful background and then try out the following exercises
The file `./sparklect/english.stop.txt` contains a list of English stopwords, while the file `./sparklect/shakes/juliuscaesar.txt` contains the entire text of Shakespeare's 'Julius Caesar'.
* Load all of the stopwords into a Python list
* Load the text of Julius Caesar into an RDD using the `sparkcontext.textFile()` method. Call it `juliusrdd`.
```
# your turn
stopw = pd.read_csv('./sparklect/english.stop.txt')
sc.stop()
sc = pyspark.SparkContext()
juliusrdd = sc.textFile('./sparklect/shakes/juliuscaesar.txt')
```
How many words does Julius Caesar have? *Hint: use `flatMap()`*.
```
# your turn
counts = juliusrdd.flatMap(lambda l: l.split(' ')).map(lambda w: (w,1)).reduceByKey(lambda a,b: a+b).collect()
```
Now print the first 20 words of Julius Caesar as a Python list.
```
# your turn
for i in range(0,19):
print counts[i]
```
Now print the first 20 words of Julius Caesar, **after removing all the stopwords**. *Hint: use `filter()`*.
```
# your turn
def notStopWord(w):
l=[]
l.append(w.encode('ascii','replace'))
if (w == u'') | (w == u'a'): #removing newline; why 'a' is not in the list of stop words?
return False
return ~any(stopw.isin(l))
counts = (juliusrdd
.flatMap(lambda l: l.lower().split(' '))
.map(lambda w: w.strip(".").strip(",").strip('"').strip("'"))
.filter(lambda w: notStopWord(w))
.map(lambda w: (w,1))
.reduceByKey(lambda a,b: a+b)
.collect())
for i in range(0,19):
print counts[i]
```
Now, use the word counting MapReduce code you've seen before. Count the number of times each word occurs and print the top 20 results as a list of tuples of the form `(word, count)`. *Hint: use `takeOrdered()` instead of `take()`*
```
# your turn
counts = (juliusrdd
.flatMap(lambda l: l.lower().split(' '))
.map(lambda w: w.strip(".").strip(",").strip('"').strip("'"))
.filter(lambda w: notStopWord(w))
.map(lambda w: (w,1))
.reduceByKey(lambda a,b: a+b))
top20 = counts.takeOrdered(20, key = lambda x: -x[1])
```
Plot a bar graph. For each of the top 20 words on the X axis, represent the count on the Y axis.
```
# your turn
import matplotlib.pyplot as plt
y, x = zip(*top20)
ax = sns.barplot(x, y)
ax.set(xlabel='Counts', ylabel='Word')
ax.set_title('Most common words in Shakespeare\'s Julius Caesar')
plt.show()
```
### Using partitions for parallelization
In order to make your code more efficient, you want to use all of the available processing power, even on a single laptop. If your machine has multiple cores, you can tune the number of partitions to use all of them! From http://www.stat.berkeley.edu/scf/paciorek-spark-2014.html:
>You want each partition to be able to fit in the memory availalbe on a node, and if you have multi-core nodes, you want that as many partitions as there are cores be able to fit in memory.
>For load-balancing you'll want at least as many partitions as total computational cores in your cluster and probably rather more partitions. The Spark documentation suggests 2-4 partitions (which they also seem to call slices) per CPU. Often there are 100-10,000 partitions. Another rule of thumb is that tasks should take at least 100 ms. If less than that, you may want to repartition to have fewer partitions.
```
shakesrdd=sc.textFile("./sparklect/shakes/*.txt", minPartitions=4)
shakesrdd.take(10)
```
Now calculate the top 20 words in all of the files that you just read.
```
# your turn
counts = (shakesrdd
.flatMap(lambda l: l.lower().split(' '))
.map(lambda w: w.strip(".").strip(",").strip('"').strip("'"))
.filter(lambda w: notStopWord(w))
.map(lambda w: (w,1))
.reduceByKey(lambda a,b: a+b))
top20 = counts.takeOrdered(20, key = lambda x: -x[1])
print top20
```
## Optional topic 1: DataFrames
Pandas and Spark dataframes can be easily converted to each other, making it easier to work with different data formats. This section shows some examples of each.
Convert Spark DataFrame to Pandas
`pandas_df = spark_df.toPandas()`
Create a Spark DataFrame from Pandas
`spark_df = context.createDataFrame(pandas_df)`
Must fit in memory.
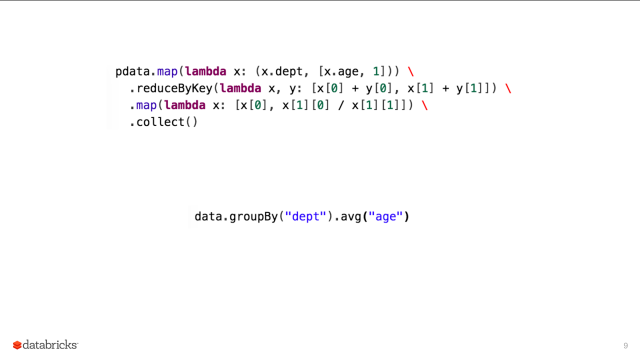
VERY IMPORTANT: DataFrames in Spark are like RDD in the sense that they’re an immutable data structure.
```
df=pd.read_csv("https://dl.dropboxusercontent.com/u/75194/stats/data/01_heights_weights_genders.csv")
df.head()
```
Convert this pandas dataframe to a Spark dataframe
```
from pyspark.sql import SQLContext
sqlsc=SQLContext(sc)
sparkdf = sqlsc.createDataFrame(df)
sparkdf
sparkdf.show(5)
type(sparkdf.Gender)
temp = sparkdf.map(lambda r: r.Gender)
print type(temp)
temp.take(10)
```
## Optional topic 2: Machine Learning using Spark
While we don't go in-depth into machine learning using spark here, this sample code will help you get started.
```
from pyspark.mllib.classification import LogisticRegressionWithLBFGS
from pyspark.mllib.regression import LabeledPoint
```
Now create a data set from the Spark dataframe
```
data=sparkdf.map(lambda row: LabeledPoint(row.Gender=='Male',[row.Height, row.Weight]))
data.take(5)
data2=sparkdf.map(lambda row: LabeledPoint(row[0]=='Male',row[1:]))
data2.take(1)[0].label, data2.take(1)[0].features
```
Split the data set into training and test sets
```
train, test = data.randomSplit([0.7,0.3])
train.cache()
test.cache()
type(train)
```
Train the logistic regression model using MLIB
```
model = LogisticRegressionWithLBFGS.train(train)
model.weights
```
Run it on the test data
```
results = test.map(lambda lp: (lp.label, float(model.predict(lp.features))))
print results.take(10)
type(results)
```
Measure accuracy and other metrics
```
test_accuracy=results.filter(lambda (a,p): a==p).count()/float(results.count())
test_accuracy
from pyspark.mllib.evaluation import BinaryClassificationMetrics
metrics = BinaryClassificationMetrics(results)
print type(metrics)
metrics.areaUnderROC
type(model)
#!rm -rf mylogistic.model
#model.save(sc, "mylogistic.model")
```
The pipeline API automates a lot of this stuff, allowing us to work directly on dataframes. It is not all supported in Python, as yet.
Also see:
- http://jordicasanellas.weebly.com/data-science-blog/machine-learning-with-spark
- http://spark.apache.org/docs/latest/mllib-guide.html
- http://www.techpoweredmath.com/spark-dataframes-mllib-tutorial/
- http://spark.apache.org/docs/latest/api/python/
- http://spark.apache.org/docs/latest/programming-guide.html
`rdd.saveAsTextFile()` saves an RDD as a string.
```
sc.stop()
```
|
github_jupyter
|
%pylab inline
import pandas as pd
import seaborn as sns
pd.set_option('display.width', 500)
pd.set_option('display.max_columns', 100)
$ brew install apache-spark
➜ ~ pip install findspark
Collecting findspark
Downloading findspark-0.0.5-py2.py3-none-any.whl
Installing collected packages: findspark
Successfully installed findspark-0.0.5
import findspark
findspark.init('c:\spark')
import pyspark
sc = pyspark.SparkContext()
sc
sc.parallelize([1, 2, 3, 4, 5, 6, 7, 8, 9, 10]).map(lambda x: x**2).sum()
wordsList = ['cat', 'elephant', 'rat', 'rat', 'cat']
wordsRDD = sc.parallelize(wordsList, 4)
# Print out the type of wordsRDD
print type(wordsRDD)
wordsRDD.collect()
def makePlural(word):
return word + 's'
print makePlural('cat')
pluralRDD = wordsRDD.map(makePlural)
print pluralRDD.first()
print pluralRDD.take(2)
pluralRDD.take(1)
pluralRDD.collect()
wordPairs = wordsRDD.map(lambda w: (w, 1))
print wordPairs.collect()
wordsList = ['cat', 'elephant', 'rat', 'rat', 'cat']
wordsRDD = sc.parallelize(wordsList, 4)
wordCountsCollected = (wordsRDD
.map(lambda w: (w, 1))
.reduceByKey(lambda x,y: x+y)
.collect())
print wordCountsCollected
print (wordsRDD
.map(lambda w: (w, 1))
.reduceByKey(lambda x,y: x+y)).toDebugString()
wordsList = ['cat', 'elephant', 'rat', 'rat', 'cat']
wordsRDD = sc.parallelize(wordsList, 4)
print wordsRDD
wordsRDD.count()
#this is rerun from the start
wordsRDD.count()
#default storage level (MEMORY_ONLY)
wordsRDD.cache()#nothing done this is still lazy
#parallelize is rerun and cached because we told it to cache
wordsRDD.count()
#this `sc.parallelize` is not rerun in this case
wordsRDD.count()
birdsList=['heron','owl']
animList=wordsList+birdsList
animaldict={}
for e in wordsList:
animaldict[e]='mammal'
for e in birdsList:
animaldict[e]='bird'
animaldict
animsrdd = sc.parallelize(animList, 4)
animsrdd.cache()
#below runs the whole chain but causes cache to be populated
mammalcount=animsrdd.filter(lambda w: animaldict[w]=='mammal').count()
#now only the filter is carried out
birdcount=animsrdd.filter(lambda w: animaldict[w]=='bird').count()
print mammalcount, birdcount
# your turn
stopw = pd.read_csv('./sparklect/english.stop.txt')
sc.stop()
sc = pyspark.SparkContext()
juliusrdd = sc.textFile('./sparklect/shakes/juliuscaesar.txt')
# your turn
counts = juliusrdd.flatMap(lambda l: l.split(' ')).map(lambda w: (w,1)).reduceByKey(lambda a,b: a+b).collect()
# your turn
for i in range(0,19):
print counts[i]
# your turn
def notStopWord(w):
l=[]
l.append(w.encode('ascii','replace'))
if (w == u'') | (w == u'a'): #removing newline; why 'a' is not in the list of stop words?
return False
return ~any(stopw.isin(l))
counts = (juliusrdd
.flatMap(lambda l: l.lower().split(' '))
.map(lambda w: w.strip(".").strip(",").strip('"').strip("'"))
.filter(lambda w: notStopWord(w))
.map(lambda w: (w,1))
.reduceByKey(lambda a,b: a+b)
.collect())
for i in range(0,19):
print counts[i]
# your turn
counts = (juliusrdd
.flatMap(lambda l: l.lower().split(' '))
.map(lambda w: w.strip(".").strip(",").strip('"').strip("'"))
.filter(lambda w: notStopWord(w))
.map(lambda w: (w,1))
.reduceByKey(lambda a,b: a+b))
top20 = counts.takeOrdered(20, key = lambda x: -x[1])
# your turn
import matplotlib.pyplot as plt
y, x = zip(*top20)
ax = sns.barplot(x, y)
ax.set(xlabel='Counts', ylabel='Word')
ax.set_title('Most common words in Shakespeare\'s Julius Caesar')
plt.show()
shakesrdd=sc.textFile("./sparklect/shakes/*.txt", minPartitions=4)
shakesrdd.take(10)
# your turn
counts = (shakesrdd
.flatMap(lambda l: l.lower().split(' '))
.map(lambda w: w.strip(".").strip(",").strip('"').strip("'"))
.filter(lambda w: notStopWord(w))
.map(lambda w: (w,1))
.reduceByKey(lambda a,b: a+b))
top20 = counts.takeOrdered(20, key = lambda x: -x[1])
print top20
df=pd.read_csv("https://dl.dropboxusercontent.com/u/75194/stats/data/01_heights_weights_genders.csv")
df.head()
from pyspark.sql import SQLContext
sqlsc=SQLContext(sc)
sparkdf = sqlsc.createDataFrame(df)
sparkdf
sparkdf.show(5)
type(sparkdf.Gender)
temp = sparkdf.map(lambda r: r.Gender)
print type(temp)
temp.take(10)
from pyspark.mllib.classification import LogisticRegressionWithLBFGS
from pyspark.mllib.regression import LabeledPoint
data=sparkdf.map(lambda row: LabeledPoint(row.Gender=='Male',[row.Height, row.Weight]))
data.take(5)
data2=sparkdf.map(lambda row: LabeledPoint(row[0]=='Male',row[1:]))
data2.take(1)[0].label, data2.take(1)[0].features
train, test = data.randomSplit([0.7,0.3])
train.cache()
test.cache()
type(train)
model = LogisticRegressionWithLBFGS.train(train)
model.weights
results = test.map(lambda lp: (lp.label, float(model.predict(lp.features))))
print results.take(10)
type(results)
test_accuracy=results.filter(lambda (a,p): a==p).count()/float(results.count())
test_accuracy
from pyspark.mllib.evaluation import BinaryClassificationMetrics
metrics = BinaryClassificationMetrics(results)
print type(metrics)
metrics.areaUnderROC
type(model)
#!rm -rf mylogistic.model
#model.save(sc, "mylogistic.model")
sc.stop()
| 0.352536 | 0.920504 |
# Tutorial
OceanSpy builds on software packages developed by the Pangeo community, in particular [xarray](http://xarray.pydata.org/en/stable/), [dask](https://dask.org/), and [xgcm](https://xgcm.readthedocs.io/en/stable/). It is preferable to have some familiarity with these packages to get the most out of OceanSpy.
This tutorial will take you through the main features of OceanSpy.
If you are using SciServer, make sure that you are using the Oceanography kernel. The current kernel is displayed in the top-right corner of the notebook. You can change kernel by clicking on `Kernel`>>`Change Kernel`.
To get started, import the oceanspy package:
```
import oceanspy as ospy
```
If you get an error that says `No module named 'oceanspy'`, it means that you are not using the Oceanography kernel. Click on `Kernel`>>`Change Kernel`, then select `Oceanography`.
## Dask Client
As explained [here](https://examples.dask.org/xarray.html#Start-Dask-Client-for-Dashboard), starting a dask client is optional, but useful for optimization purposes because it provides a [dashboard](http://distributed.dask.org/en/latest/web.html) to monitor the computation.
On your local computer, you can access the dask dashboard just by clicking on the link displayed by the client.
The dashboard link is currently not enabled on SciServer. Follow these instructions to visualize the dashboard on SciServer:
1. Switch to JupyterLab if you haven't done so yet (click on `Switch To JupyterLab`).
2. Copy the link of the notebook and paste to a new tab, then substitute whatever is after the last slash with 'dask'.
Here is an example:
* **Notebook**: `https://apps.sciserver.org/dockervm40/b029009b-6b4d-11e9-8a88-5254001d4703/lab?`
* **Dashboard**: `https://apps.sciserver.org/dockervm40/b029009b-6b4d-11e9-8a88-5254001d4703/dask`
The link to the dashboard will be created after you execute the code below.
```
from dask.distributed import Client
client = Client()
client
```
The client configuration can be changed and optimized for the computations needed. The main arguments to change to optimize performance are `n_workers`, `threads_per_worker`, and `memory_limit`.
## OceanDataset
An `xarray.Dataset` (or `ds`) is the only object required to initialize an `oceanspy.OceanDataset` object (or `od`).
An `od` is a collection of objects used by OceanSpy, and it can be initialized using the following command:
```python
od = ospy.OceanDataset(ds)
```
See [Import datasets](Tutorial.ipynb#Import-datasets) for step-by-step instructions on how to import your own dataset.
### Opening
Several datasets are available on SciServer (see [SciServer access](sciserver.rst#sciserver-access)).
Use `open_oceandataset.from_catalog()` to open one of these datasets (see [Dastasets](datasets.rst) for a list of available datasets).
Otherwise, you can run this notebook on any computer by downloading the get_started data and using `open_oceandataset.from_zarr()`.
Set `SciServer = True` to run this notebook on SciServer, otherwise set `SciServer = False`.
```
SciServer = False # True: SciServer - False: any computer
if SciServer:
od = ospy.open_oceandataset.from_catalog("get_started")
else:
import os
if not os.path.isdir("oceanspy_get_started"):
# Download get_started
import subprocess
print("Downloading and uncompressing get_started data...")
print("...it might take a couple of minutes.")
commands = [
"wget -v -O oceanspy_get_started.tar.gz -L "
"https://livejohnshopkins-my.sharepoint.com/"
":u:/g/personal/malmans2_jh_edu/"
"EXjiMbANEHBZhy62oUDjzT4BtoJSW2W0tYtS2qO8_SM5mQ?"
"download=1",
"tar xvzf oceanspy_get_started.tar.gz",
"rm -f oceanspy_get_started.tar.gz",
]
subprocess.call("&&".join(commands), shell=True)
od = ospy.open_oceandataset.from_zarr("oceanspy_get_started")
print()
print(od)
```
### Set functions
All attributes are stored as global attributes (strings) in the `Dataset` object, and decoded by OceanSpy.
Because of this, do not change attributes directly, but use OceanSpy's [Set methods](api.rst#set).
For example:
```
od = od.set_name("oceandataset #1", overwrite=True)
od = od.set_description("This is my first oceandataset", overwrite=True)
print(od)
```
The advantage of storing all the attributes in the ``Dataset`` object is that checkpoints can be created at any time (e.g., storing the Dataset in NetCDF format), and an ``OceanDataset`` object can be easily reconstructed on any computer. Thus, OceanSpy can be used at different stages of the post-processing.
### Modify Dataset objects
Most OceanSpy functions modify or add variables to `od.dataset`.
However, `od.dataset` is just a mirror object constructed from `od._ds`.
If aliases have been set (this is necessary if your dataset uses variable names that differ from the OceanSpy reference names) `od._ds` and `od.dataset` differ from each other.
If you want to modify the ``Dataset`` object without using OceanSpy, you can easily extract it from `od` and change it using `xarray`. Then, you can re-initialize `od` if you want to use OceanSpy again.
Here is an example:
```
# Extract ds
ds = od.dataset
# Compute mean temperature
ds["meanTemp"] = ds["Temp"].mean("time")
# Re-initialize the OceanDataset
od = ospy.OceanDataset(ds)
print(od, "\n" * 3, od.dataset["meanTemp"])
```
Note: Make sure that the global attributes of the Dataset do not get lost, so you will not have to re-set the attributes of the ``OceanDataset``.
Here is an example:
```
import xarray as xr
# Extract ds
ds = od.dataset
# Compute mean salinity
ds = xr.merge([ds, ds["S"].mean("time").rename("meanS")])
# Global attributes have been dropped
print("Global attributes:", ds.attrs, "\n" * 2)
# Re-set global attributes
ds.attrs = od.dataset.attrs
# Re-initialize the OceanDataset
od = ospy.OceanDataset(ds)
print(od, "\n" * 3, od.dataset["meanS"])
```
## Subsampling
There are several functions that subsample the oceandataset in different ways. For example, it is possible to extract mooring sections, conduct ship surveys, or extract particle properties (see [Subsampling](api.rst#subsampling)).
Most OceanSpy [Computing](api.rst#computing) functions make use of the [xgcm](https://xgcm.readthedocs.io/en/latest/grids.html#axes-and-positions) functionality to have multiple axes (e.g., `X` and `Xp1`) along a single physical dimension (e.g., longitude). Because we will still want to be able to perform calculations on the reduced data set, the default behavior in OceanSpy is to retain all axes.
The following commands extract subsets of the data and show this behavior:
```
# Plot the original domain
%matplotlib inline
ax = od.plot.horizontal_section(varName="Depth")
title = ax.set_title(od.name)
```
The following commands cut out and plot a small region from the original `oceandataset`.
```
od_cut = od.subsample.cutout(
XRange=[-21, -13.5], YRange=[69.6, 71.4], ZRange=0, timeRange="2007-09-01"
)
od_cut = od_cut.set_name("cutout", overwrite=False)
# Alternatively, this syntax can be used:
# od_cut = ospy.subsample.cutout(od, ...)
# Plot the cutout domain
ax = od_cut.plot.horizontal_section(varName="Depth")
title = ax.set_title(od_cut.name)
```
The size of the dataset has been reduced, but all axes in the horizontal (`X`, `Xp1`, `Y`, `Yp1`), vertical (`Z`, `Zp1`, `Zu`, `Zl`), and time dimensions (`time`, `time_midp`) have been retained, so that `od_cut` is still compatible with OceanSpy:
```
print("\nOriginal: {} Gigabytes".format(od.dataset.nbytes * 1.0e-9))
print(dict(od.dataset.sizes))
print("\nCutout: {} Megabytes".format(od_cut.dataset.nbytes * 1.0e-6))
print(dict(od_cut.dataset.sizes))
```
Sometimes it could be desirable to change this default behavior, and several additional arguments are available for this purpose (see [Subsampling](api.rst#subsampling)). For example, it is possible to reduce the vertical dimension to a single location:
```
# Extract sea surface, and drop Z-axis.
od_drop = od.subsample.cutout(ZRange=0, dropAxes=True)
print("\nOriginal oceandataset:")
print(dict(od.dataset.sizes))
print(od.grid)
print("\nNew oceandataset:")
print(dict(od_drop.dataset.sizes))
print(od_drop.grid)
```
Now, the vertical dimension is no longer part of the ``xgcm.Grid`` object, and all coordinates along the vertical dimensions have size 1.
## Computing
The compute module contains functions that create new variables (see [Computing](api.rst#computing)). Most OceanSpy functions use lazy evaluation, which means that the actual computation is not done until values are needed (e.g., when plotting).
There are two different types of compute functions:
* Fixed-name: Functions that do not require an input. The name of new variables is fixed.
* Smart-name: Functions that do require an input (e.g., vector calculus). The name of new variables is based on input names.
### Fixed-name
We compute the kinetic energy as an example.
This syntax returns a dataset containing the new variable:
```
ds_KE = ospy.compute.kinetic_energy(od_drop)
print(ds_KE)
```
while this syntax adds the new variable to `od_drop`:
```
od_KE = od_drop.compute.kinetic_energy()
print(od_KE.dataset)
```
Kinetic energy has been lazily evaluated so far.
We can trigger the computation by plotting the mean kinetic energy.
```
od_KE.plot.horizontal_section(varName="KE", meanAxes="time")
```
Note that OceanSpy always computes weighted means rather than regular averages!
### Smart-name
We now compute gradients as an example.
As seen above, `od.compute.gradient(...)` returns a dataset, while `od = ospy.compute.gradient(od, ...)` adds new variables to the oceandataset.
The following cell computes temperature gradients along all dimensions:
```
ds = ospy.compute.gradient(od, varNameList="Temp")
print(ds.data_vars)
```
while the following code computes the temperature, salinity, and density gradients along the time dimension only.
Note that `Sigma0` needs to be computed.
```
ds = ospy.compute.gradient(
od, varNameList=["Temp", "S", "Sigma0"], axesList=["time"]
)
print(ds.data_vars)
```
Here is an overview of the smart-name functions:
```
print("\nGRADIENT")
print(ospy.compute.gradient(od, "Temp").data_vars)
print("\nDIVERGENCE")
print(ospy.compute.divergence(od, iName="U", jName="V", kName="W").data_vars)
print("\nCURL")
print(ospy.compute.curl(od, iName="U", jName="V", kName="W").data_vars)
print("\nLAPLACIAN")
print(ospy.compute.laplacian(od, varNameList="Temp").data_vars)
print("\nWEIGHTED MEAN")
print(ospy.compute.weighted_mean(od, varNameList="Temp").data_vars)
print("\nINTEGRAL")
print(ospy.compute.integral(od, varNameList="Temp").data_vars)
```
All new variables have been lazily evaluated so far. The following cell triggers the evaluation of the weighted mean temperature and salinity along all dimensions:
```
ds = ospy.compute.weighted_mean(
od, varNameList=["Temp", "S"], storeWeights=False
)
for var in ds.data_vars:
print(
"{} = {} {}".format(
var, ds[var].values, ds[var].attrs.pop("units", "")
)
)
```
## Plotting
Some of the plot functions have been used above. See [Plotting](api.rst#plotting) for a list of available functions.
For example, horizontal sections are projected using the attribute `projection` of `od`.
Here we plot the mean sea surface temperature and the isobaths using different projections:
```
ax = od.plot.horizontal_section(
varName="Temp",
contourName="Depth",
meanAxes="time",
center=False,
cmap="Spectral_r",
robust=True,
cutout_kwargs={"ZRange": 0, "dropAxes": True},
)
```
We can change the projection by using one the set functions of the [Set methods](api.rst#set) of the `oceandataset`.
```
# Change projection
od_NPS = od.set_projection("NorthPolarStereo")
ax = od_NPS.plot.horizontal_section(
varName="Temp",
contourName="Depth",
meanAxes=True,
center=False,
cmap="Spectral_r",
robust=True,
cutout_kwargs={"ZRange": 0, "dropAxes": True},
)
```
## Animating
See [Animating](api.rst#animating) for a list of available functions. Plotting and animating functions have identical syntax. For example, just replace `od.plot.horizontal_section` with `od.aimate.horizontal_section` to create an animation of Sea Surface Temperature:
```
anim = od.animate.horizontal_section(
varName="Temp",
contourName="Depth",
center=False,
cmap="Spectral_r",
robust=True,
cutout_kwargs={"ZRange": 0, "dropAxes": True},
display=True,
)
```
## SciServer workflow
The SciServer Interactive mode runs on a Virtual Machine with 16 cores shared between multiple users.
Use it for notebooks that don't require heavy computations, or to test and design your notebooks.
Use the SciServer Jobs mode instead to fully exploit the computational power of SciServer.
For larger jobs, you have exclusive access to 32 logical CPU cores and 240GiB of memory. See [SciServer access](sciserver.rst#sciserver-access) for more details.
## Import datasets
The following step-by-step instructions show how to import any Ocean General Circulation Model data set:
1. Open the dataset using `xarray`. For example,
```python
import xarray as xr
ds = open_mfdataset(paths)
```
2. Create an ``OceanDataset``.
```python
import oceanspy as ospy
od = ospy.OceanDataset(ds)
```
3. Use [Set methods](api.rst#set) to connect the dataset with a `xgcm.Grid`, create aliases, set parameters, ...
For example, setting aliases is necessary if your dataset uses variable names that differ from the OceanSpy reference names.
See below for a list of OceanSpy reference names and parameters. In addition, any variable computed by OceanSpy (e.g., `Sigma0`) can be aliased.
4. Use [Import methods](api.rst#import) if your dataset is not compatible with OceanSpy (e.g., remove NaNs from coordinate variables):
All commands above can be triggered using `ospy.open_dataset.from_catalog` and a configuration file (e.g., see [SciServer catalogs](https://github.com/hainegroup/oceanspy/tree/master/sciserver_catalogs)).
Here we print the OceanSpy parameters that can be set.
```
# Print parameters
print("\n{:>15}: {}\n".format("PARAMETER NAME", "DESCRIPTION"))
for par, desc in sorted(ospy.PARAMETERS_DESCRIPTION.items()):
print("{:>15}: {}".format(par, desc))
```
While here we print the reference names of the variables used by OceanSpy.
```
# Print reference names
if SciServer:
od = ospy.open_oceandataset.from_catalog("get_started")
else:
import os
if not os.path.isdir("oceanspy_get_started"):
# Download get_started
import subprocess
print("Downloading and uncompressing get_started data...")
print("...it might take a couple of minutes.")
commands = [
"wget -v -O oceanspy_get_started.tar.gz -L "
"https://jh.box.com/shared/static/"
"pw83oja1gp6mbf8j34ff0qrxp08kf64q.gz",
"tar xvzf oceanspy_get_started.tar.gz",
"rm -f oceanspy_get_started.tar.gz",
]
subprocess.call("&&".join(commands), shell=True)
od = ospy.open_oceandataset.from_zarr("oceanspy_get_started")
table = {
var: od.dataset[var].attrs.pop(
"description", od.dataset[var].attrs.pop("long_name", None)
)
for var in od.dataset.variables
}
print("\n{:>15}: {}\n".format("REFERENCE NAME", "DESCRIPTION"))
for name, desc in sorted(table.items()):
print("{:>15}: {}".format(name, desc))
```
|
github_jupyter
|
import oceanspy as ospy
from dask.distributed import Client
client = Client()
client
od = ospy.OceanDataset(ds)
SciServer = False # True: SciServer - False: any computer
if SciServer:
od = ospy.open_oceandataset.from_catalog("get_started")
else:
import os
if not os.path.isdir("oceanspy_get_started"):
# Download get_started
import subprocess
print("Downloading and uncompressing get_started data...")
print("...it might take a couple of minutes.")
commands = [
"wget -v -O oceanspy_get_started.tar.gz -L "
"https://livejohnshopkins-my.sharepoint.com/"
":u:/g/personal/malmans2_jh_edu/"
"EXjiMbANEHBZhy62oUDjzT4BtoJSW2W0tYtS2qO8_SM5mQ?"
"download=1",
"tar xvzf oceanspy_get_started.tar.gz",
"rm -f oceanspy_get_started.tar.gz",
]
subprocess.call("&&".join(commands), shell=True)
od = ospy.open_oceandataset.from_zarr("oceanspy_get_started")
print()
print(od)
od = od.set_name("oceandataset #1", overwrite=True)
od = od.set_description("This is my first oceandataset", overwrite=True)
print(od)
# Extract ds
ds = od.dataset
# Compute mean temperature
ds["meanTemp"] = ds["Temp"].mean("time")
# Re-initialize the OceanDataset
od = ospy.OceanDataset(ds)
print(od, "\n" * 3, od.dataset["meanTemp"])
import xarray as xr
# Extract ds
ds = od.dataset
# Compute mean salinity
ds = xr.merge([ds, ds["S"].mean("time").rename("meanS")])
# Global attributes have been dropped
print("Global attributes:", ds.attrs, "\n" * 2)
# Re-set global attributes
ds.attrs = od.dataset.attrs
# Re-initialize the OceanDataset
od = ospy.OceanDataset(ds)
print(od, "\n" * 3, od.dataset["meanS"])
# Plot the original domain
%matplotlib inline
ax = od.plot.horizontal_section(varName="Depth")
title = ax.set_title(od.name)
od_cut = od.subsample.cutout(
XRange=[-21, -13.5], YRange=[69.6, 71.4], ZRange=0, timeRange="2007-09-01"
)
od_cut = od_cut.set_name("cutout", overwrite=False)
# Alternatively, this syntax can be used:
# od_cut = ospy.subsample.cutout(od, ...)
# Plot the cutout domain
ax = od_cut.plot.horizontal_section(varName="Depth")
title = ax.set_title(od_cut.name)
print("\nOriginal: {} Gigabytes".format(od.dataset.nbytes * 1.0e-9))
print(dict(od.dataset.sizes))
print("\nCutout: {} Megabytes".format(od_cut.dataset.nbytes * 1.0e-6))
print(dict(od_cut.dataset.sizes))
# Extract sea surface, and drop Z-axis.
od_drop = od.subsample.cutout(ZRange=0, dropAxes=True)
print("\nOriginal oceandataset:")
print(dict(od.dataset.sizes))
print(od.grid)
print("\nNew oceandataset:")
print(dict(od_drop.dataset.sizes))
print(od_drop.grid)
ds_KE = ospy.compute.kinetic_energy(od_drop)
print(ds_KE)
od_KE = od_drop.compute.kinetic_energy()
print(od_KE.dataset)
od_KE.plot.horizontal_section(varName="KE", meanAxes="time")
ds = ospy.compute.gradient(od, varNameList="Temp")
print(ds.data_vars)
ds = ospy.compute.gradient(
od, varNameList=["Temp", "S", "Sigma0"], axesList=["time"]
)
print(ds.data_vars)
print("\nGRADIENT")
print(ospy.compute.gradient(od, "Temp").data_vars)
print("\nDIVERGENCE")
print(ospy.compute.divergence(od, iName="U", jName="V", kName="W").data_vars)
print("\nCURL")
print(ospy.compute.curl(od, iName="U", jName="V", kName="W").data_vars)
print("\nLAPLACIAN")
print(ospy.compute.laplacian(od, varNameList="Temp").data_vars)
print("\nWEIGHTED MEAN")
print(ospy.compute.weighted_mean(od, varNameList="Temp").data_vars)
print("\nINTEGRAL")
print(ospy.compute.integral(od, varNameList="Temp").data_vars)
ds = ospy.compute.weighted_mean(
od, varNameList=["Temp", "S"], storeWeights=False
)
for var in ds.data_vars:
print(
"{} = {} {}".format(
var, ds[var].values, ds[var].attrs.pop("units", "")
)
)
ax = od.plot.horizontal_section(
varName="Temp",
contourName="Depth",
meanAxes="time",
center=False,
cmap="Spectral_r",
robust=True,
cutout_kwargs={"ZRange": 0, "dropAxes": True},
)
# Change projection
od_NPS = od.set_projection("NorthPolarStereo")
ax = od_NPS.plot.horizontal_section(
varName="Temp",
contourName="Depth",
meanAxes=True,
center=False,
cmap="Spectral_r",
robust=True,
cutout_kwargs={"ZRange": 0, "dropAxes": True},
)
anim = od.animate.horizontal_section(
varName="Temp",
contourName="Depth",
center=False,
cmap="Spectral_r",
robust=True,
cutout_kwargs={"ZRange": 0, "dropAxes": True},
display=True,
)
import xarray as xr
ds = open_mfdataset(paths)
import oceanspy as ospy
od = ospy.OceanDataset(ds)
# Print parameters
print("\n{:>15}: {}\n".format("PARAMETER NAME", "DESCRIPTION"))
for par, desc in sorted(ospy.PARAMETERS_DESCRIPTION.items()):
print("{:>15}: {}".format(par, desc))
# Print reference names
if SciServer:
od = ospy.open_oceandataset.from_catalog("get_started")
else:
import os
if not os.path.isdir("oceanspy_get_started"):
# Download get_started
import subprocess
print("Downloading and uncompressing get_started data...")
print("...it might take a couple of minutes.")
commands = [
"wget -v -O oceanspy_get_started.tar.gz -L "
"https://jh.box.com/shared/static/"
"pw83oja1gp6mbf8j34ff0qrxp08kf64q.gz",
"tar xvzf oceanspy_get_started.tar.gz",
"rm -f oceanspy_get_started.tar.gz",
]
subprocess.call("&&".join(commands), shell=True)
od = ospy.open_oceandataset.from_zarr("oceanspy_get_started")
table = {
var: od.dataset[var].attrs.pop(
"description", od.dataset[var].attrs.pop("long_name", None)
)
for var in od.dataset.variables
}
print("\n{:>15}: {}\n".format("REFERENCE NAME", "DESCRIPTION"))
for name, desc in sorted(table.items()):
print("{:>15}: {}".format(name, desc))
| 0.490236 | 0.97567 |
```
# default_exp lookup
#export
import requests
import tempfile
import os
import re
from io import BytesIO
import shutil
import json
#export
import tightai
from tightai import api_key
from tightai.exceptions import RequestError
from tightai.handlers import credentials
from tightai.conf import API_ENDPOINT, CLI_ENDPOINT, USER_HOME
#hide
test = False
if test:
CLI_ENDPOINT = "http://cli.desalsa.io:8000"
CLI_ENDPOINT
#export
class Lookup:
api = CLI_ENDPOINT
@classmethod
def get_request_path(self, path, next_url=None):
if path.startswith("http"):
request_path = path
else:
if not path.startswith("/"):
path = '/' + path
request_path = "{api}{path}".format(api=self.api, path=path)
if next_url is not None:
request_path = next_url
return request_path
@classmethod
def get_http_headers(self):
api_key = None
try:
api_key = tightai.api_key
except:
pass
if api_key != None:
return {'Authorization': f"Bearer {api_key}"}
token = credentials.get_encoded_token()
authorization_header = f"Token {token}"
return {'Authorization': authorization_header}
@classmethod
def perform_request(self, method, endpoint_path, data=None, next_url=None):
headers = self.get_http_headers()
path = self.get_request_path(endpoint_path)
return requests.request(method, path, data=data, headers=headers)
@classmethod
def http_get(self, endpoint_path, next_url=None, **kwargs):
return self.perform_request("get", endpoint_path, data=kwargs, next_url=next_url)
@classmethod
def http_put(self, endpoint_path, data={}, **kwargs):
return self.perform_request("put", endpoint_path, data=data)
@classmethod
def http_delete(self, endpoint_path, data={}, **kwargs):
return self.perform_request("delete", endpoint_path, data=data)
@classmethod
def http_post(self, endpoint_path, data={}, **kwargs):
return self.perform_request("post", endpoint_path, data=data)
@classmethod
def handle_invalid_lookup(self, response, expected_status_code=200):
r = response
if not r.status_code == expected_status_code:
try:
msg = r.json()
except:
msg = r.text
if isinstance(msg, dict):
if "detail" in msg:
msg = msg['detail']
else:
final_msg = []
for k, v in msg.items():
sub_v = v
if isinstance(v, list):
sub_v = ", ".join(v)
_msg = f"{k}: {sub_v}"
final_msg.append(_msg)
msg = final_msg
if isinstance(msg, list):
msg = ", ".join(msg)
raise RequestError(f"{msg}", status=r.status_code)
#hide
projects = Lookup().http_get("/projects")
print(projects)
projects.json()
```
|
github_jupyter
|
# default_exp lookup
#export
import requests
import tempfile
import os
import re
from io import BytesIO
import shutil
import json
#export
import tightai
from tightai import api_key
from tightai.exceptions import RequestError
from tightai.handlers import credentials
from tightai.conf import API_ENDPOINT, CLI_ENDPOINT, USER_HOME
#hide
test = False
if test:
CLI_ENDPOINT = "http://cli.desalsa.io:8000"
CLI_ENDPOINT
#export
class Lookup:
api = CLI_ENDPOINT
@classmethod
def get_request_path(self, path, next_url=None):
if path.startswith("http"):
request_path = path
else:
if not path.startswith("/"):
path = '/' + path
request_path = "{api}{path}".format(api=self.api, path=path)
if next_url is not None:
request_path = next_url
return request_path
@classmethod
def get_http_headers(self):
api_key = None
try:
api_key = tightai.api_key
except:
pass
if api_key != None:
return {'Authorization': f"Bearer {api_key}"}
token = credentials.get_encoded_token()
authorization_header = f"Token {token}"
return {'Authorization': authorization_header}
@classmethod
def perform_request(self, method, endpoint_path, data=None, next_url=None):
headers = self.get_http_headers()
path = self.get_request_path(endpoint_path)
return requests.request(method, path, data=data, headers=headers)
@classmethod
def http_get(self, endpoint_path, next_url=None, **kwargs):
return self.perform_request("get", endpoint_path, data=kwargs, next_url=next_url)
@classmethod
def http_put(self, endpoint_path, data={}, **kwargs):
return self.perform_request("put", endpoint_path, data=data)
@classmethod
def http_delete(self, endpoint_path, data={}, **kwargs):
return self.perform_request("delete", endpoint_path, data=data)
@classmethod
def http_post(self, endpoint_path, data={}, **kwargs):
return self.perform_request("post", endpoint_path, data=data)
@classmethod
def handle_invalid_lookup(self, response, expected_status_code=200):
r = response
if not r.status_code == expected_status_code:
try:
msg = r.json()
except:
msg = r.text
if isinstance(msg, dict):
if "detail" in msg:
msg = msg['detail']
else:
final_msg = []
for k, v in msg.items():
sub_v = v
if isinstance(v, list):
sub_v = ", ".join(v)
_msg = f"{k}: {sub_v}"
final_msg.append(_msg)
msg = final_msg
if isinstance(msg, list):
msg = ", ".join(msg)
raise RequestError(f"{msg}", status=r.status_code)
#hide
projects = Lookup().http_get("/projects")
print(projects)
projects.json()
| 0.326379 | 0.124798 |
<a href="https://colab.research.google.com/github/chamikasudusinghe/nocml/blob/master/fft_r6_i1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Module Imports for Data Fetiching and Visualization
```
import time
import pandas as pd
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
```
Module Imports for Data Processing
```
from sklearn import preprocessing
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
import pickle
```
Importing Dataset from GitHub
Train Data
```
df1 = pd.read_csv('https://raw.githubusercontent.com/chamikasudusinghe/nocml/master/dos%20results%20ver%204/router-dataset/r6/2-fft-malicious-n-0-15-m-1-r6.csv?token=AKVFSOGWII4PQBWODYO56AC63I2IK')
df9 = pd.read_csv('https://raw.githubusercontent.com/chamikasudusinghe/nocml/master/dos%20results%20ver%204/router-dataset/r6/2-fft-normal-n-0-15-r6.csv?token=AKVFSOB5PQW5BFUJQL6FMNK63I2I6')
df = df1.append(df9, ignore_index=True,sort=False)
df = df.sort_values('timestamp')
df.to_csv('fft-r1-train.csv',index=False)
df = pd.read_csv('fft-r1-train.csv')
df
df.shape
```
Test Data
```
df13 = pd.read_csv('https://raw.githubusercontent.com/chamikasudusinghe/nocml/master/dos%20results%20ver%204/router-dataset/r6/2-fft-malicious-n-0-15-m-11-r6.csv?token=AKVFSODRMP7WFOTNGA43XEC63I2IO')
df14 = pd.read_csv('https://raw.githubusercontent.com/chamikasudusinghe/nocml/master/dos%20results%20ver%204/router-dataset/r6/2-fft-malicious-n-0-15-m-12-r6.csv?token=AKVFSOGFXPLRBVFF2CSTSU263I2RE')
df15 = pd.read_csv('https://raw.githubusercontent.com/chamikasudusinghe/nocml/master/dos%20results%20ver%204/router-dataset/r6/2-fft-malicious-n-0-15-m-7-r6.csv?token=AKVFSOCFZYWDIHTDXDL47DC63I2RG')
print(df13.shape)
print(df14.shape)
print(df15.shape)
```
Processing
```
df.isnull().sum()
df = df.drop(columns=['timestamp','src_ni','src_router','dst_ni','dst_router'])
df.corr()
plt.figure(figsize=(25,25))
sns.heatmap(df.corr(), annot = True)
plt.show()
def find_correlation(data, threshold=0.9):
corr_mat = data.corr()
corr_mat.loc[:, :] = np.tril(corr_mat, k=-1)
already_in = set()
result = []
for col in corr_mat:
perfect_corr = corr_mat[col][abs(corr_mat[col])> threshold].index.tolist()
if perfect_corr and col not in already_in:
already_in.update(set(perfect_corr))
perfect_corr.append(col)
result.append(perfect_corr)
select_nested = [f[1:] for f in result]
select_flat = [i for j in select_nested for i in j]
return select_flat
columns_to_drop = find_correlation(df.drop(columns=['target']))
columns_to_drop
df = df.drop(columns=['inport','cache_coherence_type','flit_id','flit_type','vnet','current_hop','hop_percentage','port_index','cache_coherence_vnet_index','vnet_vc_cc_index'])
plt.figure(figsize=(11,11))
sns.heatmap(df.corr(), annot = True)
plt.show()
plt.figure(figsize=(11,11))
sns.heatmap(df.corr())
plt.show()
```
Processing Dataset for Training
```
train_X = df.drop(columns=['target'])
train_Y = df['target']
#standardization
x = train_X.values
min_max_scaler = preprocessing.MinMaxScaler()
columns = train_X.columns
x_scaled = min_max_scaler.fit_transform(x)
train_X = pd.DataFrame(x_scaled)
train_X.columns = columns
train_X
train_X[train_X.duplicated()].shape
test_X = df13.drop(columns=['target','timestamp','src_ni','src_router','dst_ni','dst_router','inport','cache_coherence_type','flit_id','flit_type','vnet','current_hop','hop_percentage','port_index','cache_coherence_vnet_index','vnet_vc_cc_index'])
test_Y = df13['target']
x = test_X.values
min_max_scaler = preprocessing.MinMaxScaler()
columns = test_X.columns
x_scaled = min_max_scaler.fit_transform(x)
test_X = pd.DataFrame(x_scaled)
test_X.columns = columns
print(test_X[test_X.duplicated()].shape)
test_X
test_X1 = df14.drop(columns=['target','timestamp','src_ni','src_router','dst_ni','dst_router','inport','cache_coherence_type','flit_id','flit_type','vnet','current_hop','hop_percentage','port_index','cache_coherence_vnet_index','vnet_vc_cc_index'])
test_Y1 = df14['target']
x = test_X1.values
min_max_scaler = preprocessing.MinMaxScaler()
columns = test_X1.columns
x_scaled = min_max_scaler.fit_transform(x)
test_X1 = pd.DataFrame(x_scaled)
test_X1.columns = columns
print(test_X1[test_X1.duplicated()].shape)
test_X2 = df15.drop(columns=['target','timestamp','src_ni','src_router','dst_ni','dst_router','inport','cache_coherence_type','flit_id','flit_type','vnet','current_hop','hop_percentage','port_index','cache_coherence_vnet_index','vnet_vc_cc_index'])
test_Y2 = df15['target']
x = test_X2.values
min_max_scaler = preprocessing.MinMaxScaler()
columns = test_X2.columns
x_scaled = min_max_scaler.fit_transform(x)
test_X2 = pd.DataFrame(x_scaled)
test_X2.columns = columns
print(test_X2[test_X2.duplicated()].shape)
```
#### Machine Learning Models
Module Imports for Data Processing and Report Generation in Machine Learning Models
```
from sklearn.model_selection import train_test_split
import statsmodels.api as sm
from sklearn import metrics
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
from sklearn.metrics import roc_curve
from sklearn.metrics import roc_auc_score
from sklearn.metrics import accuracy_score
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import cross_val_score
```
Labels
1. 0 - malicious
2. 1 - good
```
train_Y = df['target']
train_Y.value_counts()
```
Training and Validation Splitting of the Dataset
```
seed = 5
np.random.seed(seed)
X_train, X_test, y_train, y_test = train_test_split(train_X, train_Y, test_size=0.33, random_state=seed, shuffle=True)
```
Feature Selection
```
#SelectKBest for feature selection
bf = SelectKBest(score_func=chi2, k='all')
fit = bf.fit(X_train,y_train)
dfscores = pd.DataFrame(fit.scores_)
dfcolumns = pd.DataFrame(columns)
featureScores = pd.concat([dfcolumns,dfscores],axis=1)
featureScores.columns = ['Specs','Score']
print(featureScores.nlargest(10,'Score'))
featureScores.plot(kind='barh')
```
Decision Tree Classifier
```
#decisiontreee
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import GridSearchCV
dt = DecisionTreeClassifier(max_depth=20,max_features=10,random_state = 42)
dt.fit(X_train,y_train)
pickle.dump(dt, open("dt-r1.pickle.dat", 'wb'))
y_pred_dt= dt.predict(X_test)
dt_score_train = dt.score(X_train,y_train)
print("Train Prediction Score",dt_score_train*100)
dt_score_test = accuracy_score(y_test,y_pred_dt)
print("Test Prediction Score",dt_score_test*100)
y_pred_dt_test= dt.predict(test_X)
dt_score_test = accuracy_score(test_Y,y_pred_dt_test)
print("Test Prediction Score",dt_score_test*100)
y_pred_dt_test= dt.predict(test_X1)
dt_score_test = accuracy_score(test_Y1,y_pred_dt_test)
print("Test Prediction Score",dt_score_test*100)
y_pred_dt_test= dt.predict(test_X2)
dt_score_test = accuracy_score(test_Y2,y_pred_dt_test)
print("Test Prediction Score",dt_score_test*100)
feat_importances = pd.Series(dt.feature_importances_, index=columns)
feat_importances.plot(kind='barh')
cm = confusion_matrix(y_test, y_pred_dt)
class_label = ["Anomalous", "Normal"]
df_cm = pd.DataFrame(cm, index=class_label,columns=class_label)
sns.heatmap(df_cm, annot=True, fmt='d')
plt.title("Confusion Matrix")
plt.xlabel("Predicted Label")
plt.ylabel("True Label")
plt.show()
print(classification_report(y_test,y_pred_dt))
dt_roc_auc = roc_auc_score(y_test, y_pred_dt)
fpr, tpr, thresholds = roc_curve(y_test, dt.predict_proba(X_test)[:,1])
plt.figure()
plt.plot(fpr, tpr, label='DTree (area = %0.2f)' % dt_roc_auc)
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic')
plt.legend(loc="lower right")
plt.savefig('DT_ROC')
plt.show()
```
XGB Classifier
```
from xgboost import XGBClassifier
from xgboost import plot_importance
xgbc = XGBClassifier(max_depth=20,min_child_weight=1,n_estimators=500,random_state=42,learning_rate=0.2)
xgbc.fit(X_train,y_train)
pickle.dump(xgbc, open("xgbc-r6l-i1.pickle.dat", 'wb'))
y_pred_xgbc= xgbc.predict(X_test)
xgbc_score_train = xgbc.score(X_train,y_train)
print("Train Prediction Score",xgbc_score_train*100)
xgbc_score_test = accuracy_score(y_test,y_pred_xgbc)
print("Test Prediction Score",xgbc_score_test*100)
y_pred_xgbc_test= xgbc.predict(test_X)
xgbc_score_test = accuracy_score(test_Y,y_pred_xgbc_test)
print("Test Prediction Score",xgbc_score_test*100)
y_pred_xgbc_test= xgbc.predict(test_X1)
xgbc_score_test = accuracy_score(test_Y1,y_pred_xgbc_test)
print("Test Prediction Score",xgbc_score_test*100)
y_pred_xgbc_test= xgbc.predict(test_X2)
xgbc_score_test = accuracy_score(test_Y2,y_pred_xgbc_test)
print("Test Prediction Score",xgbc_score_test*100)
plot_importance(xgbc)
plt.show()
cm = confusion_matrix(y_test, y_pred_xgbc)
class_label = ["Anomalous", "Normal"]
df_cm = pd.DataFrame(cm, index=class_label,columns=class_label)
sns.heatmap(df_cm, annot=True, fmt='d')
plt.title("Confusion Matrix")
plt.xlabel("Predicted Label")
plt.ylabel("True Label")
plt.show()
print(classification_report(y_test,y_pred_xgbc))
xgb_roc_auc = roc_auc_score(y_test, y_pred_xgbc)
fpr, tpr, thresholds = roc_curve(y_test, xgbc.predict_proba(X_test)[:,1])
plt.figure()
plt.plot(fpr, tpr, label='XGBoost (area = %0.2f)' % xgb_roc_auc)
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic')
plt.legend(loc="lower right")
plt.savefig('XGB_ROC')
plt.show()
```
|
github_jupyter
|
import time
import pandas as pd
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import preprocessing
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
import pickle
df1 = pd.read_csv('https://raw.githubusercontent.com/chamikasudusinghe/nocml/master/dos%20results%20ver%204/router-dataset/r6/2-fft-malicious-n-0-15-m-1-r6.csv?token=AKVFSOGWII4PQBWODYO56AC63I2IK')
df9 = pd.read_csv('https://raw.githubusercontent.com/chamikasudusinghe/nocml/master/dos%20results%20ver%204/router-dataset/r6/2-fft-normal-n-0-15-r6.csv?token=AKVFSOB5PQW5BFUJQL6FMNK63I2I6')
df = df1.append(df9, ignore_index=True,sort=False)
df = df.sort_values('timestamp')
df.to_csv('fft-r1-train.csv',index=False)
df = pd.read_csv('fft-r1-train.csv')
df
df.shape
df13 = pd.read_csv('https://raw.githubusercontent.com/chamikasudusinghe/nocml/master/dos%20results%20ver%204/router-dataset/r6/2-fft-malicious-n-0-15-m-11-r6.csv?token=AKVFSODRMP7WFOTNGA43XEC63I2IO')
df14 = pd.read_csv('https://raw.githubusercontent.com/chamikasudusinghe/nocml/master/dos%20results%20ver%204/router-dataset/r6/2-fft-malicious-n-0-15-m-12-r6.csv?token=AKVFSOGFXPLRBVFF2CSTSU263I2RE')
df15 = pd.read_csv('https://raw.githubusercontent.com/chamikasudusinghe/nocml/master/dos%20results%20ver%204/router-dataset/r6/2-fft-malicious-n-0-15-m-7-r6.csv?token=AKVFSOCFZYWDIHTDXDL47DC63I2RG')
print(df13.shape)
print(df14.shape)
print(df15.shape)
df.isnull().sum()
df = df.drop(columns=['timestamp','src_ni','src_router','dst_ni','dst_router'])
df.corr()
plt.figure(figsize=(25,25))
sns.heatmap(df.corr(), annot = True)
plt.show()
def find_correlation(data, threshold=0.9):
corr_mat = data.corr()
corr_mat.loc[:, :] = np.tril(corr_mat, k=-1)
already_in = set()
result = []
for col in corr_mat:
perfect_corr = corr_mat[col][abs(corr_mat[col])> threshold].index.tolist()
if perfect_corr and col not in already_in:
already_in.update(set(perfect_corr))
perfect_corr.append(col)
result.append(perfect_corr)
select_nested = [f[1:] for f in result]
select_flat = [i for j in select_nested for i in j]
return select_flat
columns_to_drop = find_correlation(df.drop(columns=['target']))
columns_to_drop
df = df.drop(columns=['inport','cache_coherence_type','flit_id','flit_type','vnet','current_hop','hop_percentage','port_index','cache_coherence_vnet_index','vnet_vc_cc_index'])
plt.figure(figsize=(11,11))
sns.heatmap(df.corr(), annot = True)
plt.show()
plt.figure(figsize=(11,11))
sns.heatmap(df.corr())
plt.show()
train_X = df.drop(columns=['target'])
train_Y = df['target']
#standardization
x = train_X.values
min_max_scaler = preprocessing.MinMaxScaler()
columns = train_X.columns
x_scaled = min_max_scaler.fit_transform(x)
train_X = pd.DataFrame(x_scaled)
train_X.columns = columns
train_X
train_X[train_X.duplicated()].shape
test_X = df13.drop(columns=['target','timestamp','src_ni','src_router','dst_ni','dst_router','inport','cache_coherence_type','flit_id','flit_type','vnet','current_hop','hop_percentage','port_index','cache_coherence_vnet_index','vnet_vc_cc_index'])
test_Y = df13['target']
x = test_X.values
min_max_scaler = preprocessing.MinMaxScaler()
columns = test_X.columns
x_scaled = min_max_scaler.fit_transform(x)
test_X = pd.DataFrame(x_scaled)
test_X.columns = columns
print(test_X[test_X.duplicated()].shape)
test_X
test_X1 = df14.drop(columns=['target','timestamp','src_ni','src_router','dst_ni','dst_router','inport','cache_coherence_type','flit_id','flit_type','vnet','current_hop','hop_percentage','port_index','cache_coherence_vnet_index','vnet_vc_cc_index'])
test_Y1 = df14['target']
x = test_X1.values
min_max_scaler = preprocessing.MinMaxScaler()
columns = test_X1.columns
x_scaled = min_max_scaler.fit_transform(x)
test_X1 = pd.DataFrame(x_scaled)
test_X1.columns = columns
print(test_X1[test_X1.duplicated()].shape)
test_X2 = df15.drop(columns=['target','timestamp','src_ni','src_router','dst_ni','dst_router','inport','cache_coherence_type','flit_id','flit_type','vnet','current_hop','hop_percentage','port_index','cache_coherence_vnet_index','vnet_vc_cc_index'])
test_Y2 = df15['target']
x = test_X2.values
min_max_scaler = preprocessing.MinMaxScaler()
columns = test_X2.columns
x_scaled = min_max_scaler.fit_transform(x)
test_X2 = pd.DataFrame(x_scaled)
test_X2.columns = columns
print(test_X2[test_X2.duplicated()].shape)
from sklearn.model_selection import train_test_split
import statsmodels.api as sm
from sklearn import metrics
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
from sklearn.metrics import roc_curve
from sklearn.metrics import roc_auc_score
from sklearn.metrics import accuracy_score
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import cross_val_score
train_Y = df['target']
train_Y.value_counts()
seed = 5
np.random.seed(seed)
X_train, X_test, y_train, y_test = train_test_split(train_X, train_Y, test_size=0.33, random_state=seed, shuffle=True)
#SelectKBest for feature selection
bf = SelectKBest(score_func=chi2, k='all')
fit = bf.fit(X_train,y_train)
dfscores = pd.DataFrame(fit.scores_)
dfcolumns = pd.DataFrame(columns)
featureScores = pd.concat([dfcolumns,dfscores],axis=1)
featureScores.columns = ['Specs','Score']
print(featureScores.nlargest(10,'Score'))
featureScores.plot(kind='barh')
#decisiontreee
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import GridSearchCV
dt = DecisionTreeClassifier(max_depth=20,max_features=10,random_state = 42)
dt.fit(X_train,y_train)
pickle.dump(dt, open("dt-r1.pickle.dat", 'wb'))
y_pred_dt= dt.predict(X_test)
dt_score_train = dt.score(X_train,y_train)
print("Train Prediction Score",dt_score_train*100)
dt_score_test = accuracy_score(y_test,y_pred_dt)
print("Test Prediction Score",dt_score_test*100)
y_pred_dt_test= dt.predict(test_X)
dt_score_test = accuracy_score(test_Y,y_pred_dt_test)
print("Test Prediction Score",dt_score_test*100)
y_pred_dt_test= dt.predict(test_X1)
dt_score_test = accuracy_score(test_Y1,y_pred_dt_test)
print("Test Prediction Score",dt_score_test*100)
y_pred_dt_test= dt.predict(test_X2)
dt_score_test = accuracy_score(test_Y2,y_pred_dt_test)
print("Test Prediction Score",dt_score_test*100)
feat_importances = pd.Series(dt.feature_importances_, index=columns)
feat_importances.plot(kind='barh')
cm = confusion_matrix(y_test, y_pred_dt)
class_label = ["Anomalous", "Normal"]
df_cm = pd.DataFrame(cm, index=class_label,columns=class_label)
sns.heatmap(df_cm, annot=True, fmt='d')
plt.title("Confusion Matrix")
plt.xlabel("Predicted Label")
plt.ylabel("True Label")
plt.show()
print(classification_report(y_test,y_pred_dt))
dt_roc_auc = roc_auc_score(y_test, y_pred_dt)
fpr, tpr, thresholds = roc_curve(y_test, dt.predict_proba(X_test)[:,1])
plt.figure()
plt.plot(fpr, tpr, label='DTree (area = %0.2f)' % dt_roc_auc)
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic')
plt.legend(loc="lower right")
plt.savefig('DT_ROC')
plt.show()
from xgboost import XGBClassifier
from xgboost import plot_importance
xgbc = XGBClassifier(max_depth=20,min_child_weight=1,n_estimators=500,random_state=42,learning_rate=0.2)
xgbc.fit(X_train,y_train)
pickle.dump(xgbc, open("xgbc-r6l-i1.pickle.dat", 'wb'))
y_pred_xgbc= xgbc.predict(X_test)
xgbc_score_train = xgbc.score(X_train,y_train)
print("Train Prediction Score",xgbc_score_train*100)
xgbc_score_test = accuracy_score(y_test,y_pred_xgbc)
print("Test Prediction Score",xgbc_score_test*100)
y_pred_xgbc_test= xgbc.predict(test_X)
xgbc_score_test = accuracy_score(test_Y,y_pred_xgbc_test)
print("Test Prediction Score",xgbc_score_test*100)
y_pred_xgbc_test= xgbc.predict(test_X1)
xgbc_score_test = accuracy_score(test_Y1,y_pred_xgbc_test)
print("Test Prediction Score",xgbc_score_test*100)
y_pred_xgbc_test= xgbc.predict(test_X2)
xgbc_score_test = accuracy_score(test_Y2,y_pred_xgbc_test)
print("Test Prediction Score",xgbc_score_test*100)
plot_importance(xgbc)
plt.show()
cm = confusion_matrix(y_test, y_pred_xgbc)
class_label = ["Anomalous", "Normal"]
df_cm = pd.DataFrame(cm, index=class_label,columns=class_label)
sns.heatmap(df_cm, annot=True, fmt='d')
plt.title("Confusion Matrix")
plt.xlabel("Predicted Label")
plt.ylabel("True Label")
plt.show()
print(classification_report(y_test,y_pred_xgbc))
xgb_roc_auc = roc_auc_score(y_test, y_pred_xgbc)
fpr, tpr, thresholds = roc_curve(y_test, xgbc.predict_proba(X_test)[:,1])
plt.figure()
plt.plot(fpr, tpr, label='XGBoost (area = %0.2f)' % xgb_roc_auc)
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic')
plt.legend(loc="lower right")
plt.savefig('XGB_ROC')
plt.show()
| 0.401453 | 0.874935 |
# Run MLFlow Model in Seldon Core
This notebook shows how you can easily train a model using [MLFlow](https://mlflow.org/) and serve requests within Seldon Core on Kubernetes.
Dependencies
* ```pip install seldon-core```
* ```pip install mlflow```
## Train Example MlFlow Model
```
!git clone https://github.com/mlflow/mlflow
!python mlflow/examples/sklearn_elasticnet_wine/train.py
```
## Test Inference Locally
```
!pygmentize MyMlflowModel.py
!s2i build -E environment_rest . seldonio/seldon-core-s2i-python3:0.10 mlflow_model:0.1
!docker run --name "mlflow_model" -d --rm -p 5000:5000 mlflow_model:0.1
!curl -H "Content-Type: application/x-www-form-urlencoded" -g 0.0.0.0:5000/predict -d 'json={"data":{"names":["alcohol", "chlorides", "citric acid", "density", "fixed acidity", "free sulfur dioxide", "pH", "residual sugar", "sulphates", "total sulfur dioxide", "volatile acidity"],"ndarray":[[12.8, 0.029, 0.48, 0.98, 6.2, 29, 3.33, 1.2, 0.39, 75, 0.66]]}}'
!curl -H "Content-Type: application/x-www-form-urlencoded" -g 0.0.0.0:5000/predict -d 'json={"data":{"ndarray":[[12.8, 0.029, 0.48, 0.98, 6.2, 29, 3.33, 1.2, 0.39, 75, 0.66]]}}'
!docker rm mlflow_model --force
```
## Test Inference on Minikube
**Due to a [minikube/s2i issue](https://github.com/SeldonIO/seldon-core/issues/253) you will need [s2i >= 1.1.13](https://github.com/openshift/source-to-image/releases/tag/v1.1.13)**
```
!minikube start --memory 4096
!kubectl create clusterrolebinding kube-system-cluster-admin --clusterrole=cluster-admin --serviceaccount=kube-system:default
!helm init
!kubectl rollout status deploy/tiller-deploy -n kube-system
!helm install ../../../helm-charts/seldon-core-operator --name seldon-core --set usageMetrics.enabled=true --namespace seldon-system
!kubectl rollout status statefulset.apps/seldon-operator-controller-manager -n seldon-system
```
## Setup Ingress
Please note: There are reported gRPC issues with ambassador (see https://github.com/SeldonIO/seldon-core/issues/473).
```
!helm install stable/ambassador --name ambassador --set crds.keep=false
!kubectl rollout status deployment.apps/ambassador
!eval $(minikube docker-env) && s2i build -E environment_rest . seldonio/seldon-core-s2i-python3:0.10 mlflow_model:0.1
!kubectl create -f deployment.json
!kubectl rollout status deployment/mlflow-dep-mlflow-pred-d580056
!seldon-core-api-tester contract.json `minikube ip` `kubectl get svc ambassador -o jsonpath='{.spec.ports[0].nodePort}'` \
mlflow-example --namespace seldon -p
!minikube delete
```
|
github_jupyter
|
* ```pip install mlflow```
## Train Example MlFlow Model
## Test Inference Locally
## Test Inference on Minikube
**Due to a [minikube/s2i issue](https://github.com/SeldonIO/seldon-core/issues/253) you will need [s2i >= 1.1.13](https://github.com/openshift/source-to-image/releases/tag/v1.1.13)**
## Setup Ingress
Please note: There are reported gRPC issues with ambassador (see https://github.com/SeldonIO/seldon-core/issues/473).
| 0.639849 | 0.902995 |
```
import xgboost as xgb
from sklearn.metrics import mean_squared_error
import pandas as pd
import numpy as np
from pathlib import Path
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from sklearn.model_selection import train_test_split, RandomizedSearchCV, PredefinedSplit
from sklearn.feature_extraction import DictVectorizer
from scipy.stats import uniform, randint
from matplotlib import pyplot as plt
from helpers import *
# path to project directory
path = Path('./')
# read in training dataset
train_df = pd.read_csv(path/'data/train_v4.csv', index_col=0, dtype={'season':str})
```
## XGBoost model
XGboost is a ensemble tree-based predictive algorithm that performs well across a range of applications. Applying it to a time series problem, where metrics from recent time periods can be predicitve, requires us to add in window features (e.g. points scored last gameweek). These are created using the player_lag_features function from 00_fpl_features.
```
# add a bunch of player lag features
lag_train_df, team_lag_vars = team_lag_features(train_df, ['total_points'], ['all', 1, 2, 3, 4, 5, 10])
lag_train_df, player_lag_vars = player_lag_features(lag_train_df, ['total_points'], ['all', 1, 2, 3, 4, 5, 10])
```
We have introduced a number of lag (window) features for each player's points per game, their team's points per game and the opposition team's points per game over the previous 1, 2, 3, 4, 5, 10 and all gameweeks.
Next we can set the validation point and length as well as the categorical and continuous features we'll be using to predict the dependent variable, total points for each game. These are used in the create_lag_train function to get an our training set (including appropriate lag values in the validation set).
The gameweeks and seasons are ordered, so we want to have them as ordered categories with the correct order (2016-17 is before 2017-18 etc.).
```
# set validaton point/length and categorical/continuous variables
valid_season = '1920'
valid_gw = 20
valid_len = 6
# cat_vars = ['season', 'position', 'team', 'opponent_team', 'was_home']
cat_vars = ['season', 'position', 'was_home']
cont_vars = ['gw', 'minutes']
dep_var = ['total_points']
cat_vars = cat_vars + ['minutes_last_all',
'minutes_last_1',
'minutes_last_2',
'minutes_last_3',
'minutes_last_4',
'minutes_last_5',
'minutes_last_10']
# we want to set gw and season as ordered categorical variables
# need lists with ordered categories
ordered_gws = list(range(1,39))
ordered_seasons = ['1617', '1718', '1819', '1920']
# set as categories with correct order
lag_train_df['gw'] = lag_train_df['gw'].astype('category')
lag_train_df['season'] = lag_train_df['season'].astype('category')
lag_train_df['gw'].cat.set_categories(ordered_gws, ordered=True, inplace=True)
lag_train_df['season'].cat.set_categories(ordered_seasons, ordered=True, inplace=True)
# create dataset with adjusted post-validation lag numbers
lag_train_df, train_idx, valid_idx = create_lag_train(lag_train_df,
cat_vars, cont_vars,
player_lag_vars, team_lag_vars, dep_var,
valid_season, valid_gw, valid_len)
# take a look at the dataframe
lag_train_df.info()
```
We can now build the input (X) and dependent (y) variable datasets. This includes encoding the categorical features so that each level is represented in it's own column (e.g. postition_1, position_2, etc.).
```
# split out dependent variable
X, y = lag_train_df[cat_vars + cont_vars + player_lag_vars + team_lag_vars].copy(), lag_train_df[dep_var].copy()
# since position is categorical, it should be a string
X['position'] = X['position'].apply(str)
# need to transform season
enc = LabelEncoder()
X['season'] = enc.fit_transform(X['season'])
X_dict = X.to_dict("records")
# Create the DictVectorizer object: dv
dv = DictVectorizer(sparse=False, separator='_')
# Apply dv on df: df_encoded
X_encoded = dv.fit_transform(X_dict)
X_df = pd.DataFrame(X_encoded, columns=dv.feature_names_)
```
You can see the resulting dataset with the categorical features split out into levels. This can now be used in the XGBoost API.
```
X_df.info()
X[player_lag_vars + team_lag_vars].info()
```
We can start by instatiating an XGBRegressor (since the dependent variable is continuous) and do a single train with arbitrary parameters. We split out the validation set and use it after training to create predictions and calculate the RMSE versus actuals.
```
# split out training and validation sets
X_train = X_df.iloc[train_idx]
y_train = y.iloc[train_idx]
X_test = X_df.iloc[valid_idx]
y_test = y.iloc[valid_idx]
# instatiate and train XGB Regressor
# print result
xg_reg = xgb.XGBRegressor(gamma=0.05, learning_rate=0.08, max_depth=5, n_estimators=75, subsample=0.7)
xg_reg.fit(X_train, y_train)
preds = xg_reg.predict(X_test)
print("RMSE: %f" % (r_mse(preds, y_test['total_points'])))
max(preds)
# thinking about whether we should include 0 minute rows in training and/or validation
```
This is a clear improvement on the baseline approach, but perhaps it can be improved by doing a parameter search.
To do this we will first define the grid of parameters to be searched.
```
xgb.plot_importance(xg_reg, max_num_features=15)
plt.show()
# parameter search space
params = {#"colsample_bytree": uniform(0.7, 0.3),
"gamma": uniform(0, 0.5),
"learning_rate": uniform(0.003, 0.3), # default 0.1
"max_depth": randint(2, 6), # default 3
"n_estimators": randint(25, 200), # default 100
"subsample": uniform(0.6, 0.4)}
```
In this case we will pass both train and validation parts of the dataset, along with a series telling the XGBRegressor object which rows to use for training, and which for validation.
```
X_train = X_df
y_train = y
test_fold = np.repeat([-1, 0], [valid_idx[0], valid_idx[-1] - valid_idx[0] + 1])
ps = PredefinedSplit(test_fold)
```
We can then again instatiate the XGBRegressor object, but this time pass it to a randomised search validation object, along with the parameter grid, validation splits, and number of iterations we want to run.
We then fit this to the training data - 25 random parameter selections will be made and the best parameters for the validation set can be found (may take a few minutes to run).
```
# Instantiate the regressor: gbm
gbm = xgb.XGBRegressor(objective="reg:squarederror")
# Perform random search: grid_mse
randomized_mse = RandomizedSearchCV(estimator=gbm,
param_distributions=params,
scoring="neg_mean_squared_error",
n_iter=25,
cv=ps,
verbose=1)
# Fit randomized_mse to the data
randomized_mse.fit(X_train, y_train)
# Print the best parameters and lowest RMSE
print("Best parameters found: ", randomized_mse.best_params_)
print("Lowest RMSE found: ", np.sqrt(np.abs(randomized_mse.best_score_)))
```
A slight improvement on the above.
```
# Create the parameter grid: gbm_param_grid
gbm_param_grid = {
'n_estimators': [50, 100, 200],
'max_depth': [2, 3, 5],
#'colsample_bytree': [0.1, 0.5, 0.8, 1],
'learning_rate': [0.1]
}
```
|
github_jupyter
|
import xgboost as xgb
from sklearn.metrics import mean_squared_error
import pandas as pd
import numpy as np
from pathlib import Path
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from sklearn.model_selection import train_test_split, RandomizedSearchCV, PredefinedSplit
from sklearn.feature_extraction import DictVectorizer
from scipy.stats import uniform, randint
from matplotlib import pyplot as plt
from helpers import *
# path to project directory
path = Path('./')
# read in training dataset
train_df = pd.read_csv(path/'data/train_v4.csv', index_col=0, dtype={'season':str})
# add a bunch of player lag features
lag_train_df, team_lag_vars = team_lag_features(train_df, ['total_points'], ['all', 1, 2, 3, 4, 5, 10])
lag_train_df, player_lag_vars = player_lag_features(lag_train_df, ['total_points'], ['all', 1, 2, 3, 4, 5, 10])
# set validaton point/length and categorical/continuous variables
valid_season = '1920'
valid_gw = 20
valid_len = 6
# cat_vars = ['season', 'position', 'team', 'opponent_team', 'was_home']
cat_vars = ['season', 'position', 'was_home']
cont_vars = ['gw', 'minutes']
dep_var = ['total_points']
cat_vars = cat_vars + ['minutes_last_all',
'minutes_last_1',
'minutes_last_2',
'minutes_last_3',
'minutes_last_4',
'minutes_last_5',
'minutes_last_10']
# we want to set gw and season as ordered categorical variables
# need lists with ordered categories
ordered_gws = list(range(1,39))
ordered_seasons = ['1617', '1718', '1819', '1920']
# set as categories with correct order
lag_train_df['gw'] = lag_train_df['gw'].astype('category')
lag_train_df['season'] = lag_train_df['season'].astype('category')
lag_train_df['gw'].cat.set_categories(ordered_gws, ordered=True, inplace=True)
lag_train_df['season'].cat.set_categories(ordered_seasons, ordered=True, inplace=True)
# create dataset with adjusted post-validation lag numbers
lag_train_df, train_idx, valid_idx = create_lag_train(lag_train_df,
cat_vars, cont_vars,
player_lag_vars, team_lag_vars, dep_var,
valid_season, valid_gw, valid_len)
# take a look at the dataframe
lag_train_df.info()
# split out dependent variable
X, y = lag_train_df[cat_vars + cont_vars + player_lag_vars + team_lag_vars].copy(), lag_train_df[dep_var].copy()
# since position is categorical, it should be a string
X['position'] = X['position'].apply(str)
# need to transform season
enc = LabelEncoder()
X['season'] = enc.fit_transform(X['season'])
X_dict = X.to_dict("records")
# Create the DictVectorizer object: dv
dv = DictVectorizer(sparse=False, separator='_')
# Apply dv on df: df_encoded
X_encoded = dv.fit_transform(X_dict)
X_df = pd.DataFrame(X_encoded, columns=dv.feature_names_)
X_df.info()
X[player_lag_vars + team_lag_vars].info()
# split out training and validation sets
X_train = X_df.iloc[train_idx]
y_train = y.iloc[train_idx]
X_test = X_df.iloc[valid_idx]
y_test = y.iloc[valid_idx]
# instatiate and train XGB Regressor
# print result
xg_reg = xgb.XGBRegressor(gamma=0.05, learning_rate=0.08, max_depth=5, n_estimators=75, subsample=0.7)
xg_reg.fit(X_train, y_train)
preds = xg_reg.predict(X_test)
print("RMSE: %f" % (r_mse(preds, y_test['total_points'])))
max(preds)
# thinking about whether we should include 0 minute rows in training and/or validation
xgb.plot_importance(xg_reg, max_num_features=15)
plt.show()
# parameter search space
params = {#"colsample_bytree": uniform(0.7, 0.3),
"gamma": uniform(0, 0.5),
"learning_rate": uniform(0.003, 0.3), # default 0.1
"max_depth": randint(2, 6), # default 3
"n_estimators": randint(25, 200), # default 100
"subsample": uniform(0.6, 0.4)}
X_train = X_df
y_train = y
test_fold = np.repeat([-1, 0], [valid_idx[0], valid_idx[-1] - valid_idx[0] + 1])
ps = PredefinedSplit(test_fold)
# Instantiate the regressor: gbm
gbm = xgb.XGBRegressor(objective="reg:squarederror")
# Perform random search: grid_mse
randomized_mse = RandomizedSearchCV(estimator=gbm,
param_distributions=params,
scoring="neg_mean_squared_error",
n_iter=25,
cv=ps,
verbose=1)
# Fit randomized_mse to the data
randomized_mse.fit(X_train, y_train)
# Print the best parameters and lowest RMSE
print("Best parameters found: ", randomized_mse.best_params_)
print("Lowest RMSE found: ", np.sqrt(np.abs(randomized_mse.best_score_)))
# Create the parameter grid: gbm_param_grid
gbm_param_grid = {
'n_estimators': [50, 100, 200],
'max_depth': [2, 3, 5],
#'colsample_bytree': [0.1, 0.5, 0.8, 1],
'learning_rate': [0.1]
}
| 0.496338 | 0.820793 |
```
%pylab inline
from constant import *
Vgrid = np.load("Value03.npy")
num = 10000
# calculate the stationary distribution
S_distribution = jnp.ones(nS)/nS
for _ in range(100):
S_distribution = jnp.matmul(S_distribution, Ps)
#P(0,1)
P01 = jnp.dot(Pe[:,0],S_distribution)
#P(1,0)
P10 = jnp.dot(Pe[:,1],S_distribution)
jnp.array([[1-P01, P01],[P10, 1-P10]])
E_distribution = jnp.ones(2)/2
for _ in range(100):
E_distribution = jnp.matmul(E_distribution, jnp.array([[1-P01, P01],[P10, 1-P10]]))
'''
x = [w,n,m,s,e,o]
x = [5,0,0,0,0,0]
'''
from jax import random
def simulation(key):
initE = random.choice(a = nE, p=E_distribution, key = key)
initS = random.choice(a = nS, p=S_distribution, key = key)
x = [5, 0, 0, initS, initE, 0]
path = []
move = []
for t in range(T_min, T_max):
_, key = random.split(key)
if t == T_max-1:
_,a = V(t,Vgrid[:,:,:,:,:,:,t],x)
else:
_,a = V(t,Vgrid[:,:,:,:,:,:,t+1],x)
xp = transition(t,a.reshape((1,-1)),x)
p = xp[:,-1]
x_next = xp[:,:-1]
path.append(x)
move.append(a)
x = x_next[random.choice(a = nS*nE, p=p, key = key)]
path.append(x)
return jnp.array(path), jnp.array(move)
%%time
# simulation part
keys = vmap(random.PRNGKey)(jnp.arange(num))
Paths, Moves = vmap(simulation)(keys)
# x = [w,n,m,s,e,o]
# x = [0,1,2,3,4,5]
ws = Paths[:,:,0].T
ns = Paths[:,:,1].T
ms = Paths[:,:,2].T
ss = Paths[:,:,3].T
es = Paths[:,:,4].T
os = Paths[:,:,5].T
cs = Moves[:,:,0].T
bs = Moves[:,:,1].T
ks = Moves[:,:,2].T
hs = Moves[:,:,3].T
actions = Moves[:,:,4].T
plt.figure(figsize = [16,8])
plt.title("The mean values of simulation")
plt.plot(range(20, T_max + 21),jnp.mean(ws + H*pt*os - ms,axis = 1), label = "wealth + home equity")
plt.plot(range(20, T_max + 21),jnp.mean(ws,axis = 1), label = "wealth")
plt.plot(range(20, T_max + 20),jnp.mean(cs,axis = 1), label = "consumption")
plt.plot(range(20, T_max + 20),jnp.mean(bs,axis = 1), label = "bond")
plt.plot(range(20, T_max + 20),jnp.mean(ks,axis = 1), label = "stock")
plt.legend()
plt.title("housing consumption")
plt.plot(range(20, T_max + 20),(hs).mean(axis = 1), label = "housing")
plt.title("house owner percentage in the population")
plt.plot(range(20, T_max + 21),(os).mean(axis = 1), label = "owning")
# agent number, x = [w,n,m,s,e,o]
agentNum = 13
plt.figure(figsize = [16,8])
plt.plot(range(20, T_max + 21),ws[:,agentNum], label = "wealth")
plt.plot(range(20, T_max + 21),ns[:,agentNum], label = "401k")
plt.plot(range(20, T_max + 21),ms[:,agentNum], label = "mortgage")
plt.plot(range(20, T_max + 20),cs[:,agentNum], label = "consumption")
plt.plot(range(20, T_max + 20),bs[:,agentNum], label = "bond")
plt.plot(range(20, T_max + 20),ks[:,agentNum], label = "stock")
plt.plot(range(20, T_max + 21),os[:,agentNum]*100, label = "ownership", color = "k")
plt.legend()
```
|
github_jupyter
|
%pylab inline
from constant import *
Vgrid = np.load("Value03.npy")
num = 10000
# calculate the stationary distribution
S_distribution = jnp.ones(nS)/nS
for _ in range(100):
S_distribution = jnp.matmul(S_distribution, Ps)
#P(0,1)
P01 = jnp.dot(Pe[:,0],S_distribution)
#P(1,0)
P10 = jnp.dot(Pe[:,1],S_distribution)
jnp.array([[1-P01, P01],[P10, 1-P10]])
E_distribution = jnp.ones(2)/2
for _ in range(100):
E_distribution = jnp.matmul(E_distribution, jnp.array([[1-P01, P01],[P10, 1-P10]]))
'''
x = [w,n,m,s,e,o]
x = [5,0,0,0,0,0]
'''
from jax import random
def simulation(key):
initE = random.choice(a = nE, p=E_distribution, key = key)
initS = random.choice(a = nS, p=S_distribution, key = key)
x = [5, 0, 0, initS, initE, 0]
path = []
move = []
for t in range(T_min, T_max):
_, key = random.split(key)
if t == T_max-1:
_,a = V(t,Vgrid[:,:,:,:,:,:,t],x)
else:
_,a = V(t,Vgrid[:,:,:,:,:,:,t+1],x)
xp = transition(t,a.reshape((1,-1)),x)
p = xp[:,-1]
x_next = xp[:,:-1]
path.append(x)
move.append(a)
x = x_next[random.choice(a = nS*nE, p=p, key = key)]
path.append(x)
return jnp.array(path), jnp.array(move)
%%time
# simulation part
keys = vmap(random.PRNGKey)(jnp.arange(num))
Paths, Moves = vmap(simulation)(keys)
# x = [w,n,m,s,e,o]
# x = [0,1,2,3,4,5]
ws = Paths[:,:,0].T
ns = Paths[:,:,1].T
ms = Paths[:,:,2].T
ss = Paths[:,:,3].T
es = Paths[:,:,4].T
os = Paths[:,:,5].T
cs = Moves[:,:,0].T
bs = Moves[:,:,1].T
ks = Moves[:,:,2].T
hs = Moves[:,:,3].T
actions = Moves[:,:,4].T
plt.figure(figsize = [16,8])
plt.title("The mean values of simulation")
plt.plot(range(20, T_max + 21),jnp.mean(ws + H*pt*os - ms,axis = 1), label = "wealth + home equity")
plt.plot(range(20, T_max + 21),jnp.mean(ws,axis = 1), label = "wealth")
plt.plot(range(20, T_max + 20),jnp.mean(cs,axis = 1), label = "consumption")
plt.plot(range(20, T_max + 20),jnp.mean(bs,axis = 1), label = "bond")
plt.plot(range(20, T_max + 20),jnp.mean(ks,axis = 1), label = "stock")
plt.legend()
plt.title("housing consumption")
plt.plot(range(20, T_max + 20),(hs).mean(axis = 1), label = "housing")
plt.title("house owner percentage in the population")
plt.plot(range(20, T_max + 21),(os).mean(axis = 1), label = "owning")
# agent number, x = [w,n,m,s,e,o]
agentNum = 13
plt.figure(figsize = [16,8])
plt.plot(range(20, T_max + 21),ws[:,agentNum], label = "wealth")
plt.plot(range(20, T_max + 21),ns[:,agentNum], label = "401k")
plt.plot(range(20, T_max + 21),ms[:,agentNum], label = "mortgage")
plt.plot(range(20, T_max + 20),cs[:,agentNum], label = "consumption")
plt.plot(range(20, T_max + 20),bs[:,agentNum], label = "bond")
plt.plot(range(20, T_max + 20),ks[:,agentNum], label = "stock")
plt.plot(range(20, T_max + 21),os[:,agentNum]*100, label = "ownership", color = "k")
plt.legend()
| 0.469277 | 0.48121 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.