code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
# Object-Oriented Python
Object-oriented programming (OOP) is a way of writing programs that represent real-world problem spaces (in terms of objects, functions, classes, attributes, methods, and inheritance). As Allen Downey explains in [__Think Python__](http://www.greenteapress.com/thinkpython/html/thinkpython018.html), in object-oriented programming, we shift away from framing the *function* as the active agent and toward seeing the *object* as the active agent.
In this workshop, we are going to create a class that represents the rational numbers. This tutorial is adapted from content in Anand Chitipothu's [__Python Practice Book__](http://anandology.com/python-practice-book/index.html). It was created by [Rebecca Bilbro](https://github.com/rebeccabilbro/Tutorials/tree/master/OOP)
## Part 1: Classes, methods, modules, and packages.
#### Pair Programming: Partner up with the person sitting next to you
Copy the code below into a file called RatNum.py in your code editor.
It may help to review [built-ins in Python](https://docs.python.org/3.5/library/functions.html) and the [Python data model](https://docs.python.org/3.5/reference/datamodel.html).
```
class RationalNumber:
"""Any number that can be expressed as the quotient or fraction p/q
of two integers, p and q, with the denominator q not equal to zero.
Since q may be equal to 1, every integer is a rational number.
"""
def __init__(self, numerator, denominator=1):
self.n = numerator
self.d = denominator
def __add__(self, other):
# Write a function that allows for the addition of two rational numbers.
# I did this one for you :D
if not isinstance(other, RationalNumber):
other = RationalNumber(other)
n = self.n * other.d + self.d * other.n
d = self.d * other.d
return RationalNumber(n, d)
def __sub__(self, other):
# Write a function that allows for the subtraction of two rational numbers.
pass
def __mul__(self, other):
# Write a function that allows for the multiplication of two rational numbers.
pass
def __truediv__(self, other):
# Write a function that allows for the division of two rational numbers.
pass
def __str__(self):
return "%s/%s" % (self.n, self.d)
__repr__ = __str__
if __name__ == "__main__":
x = RationalNumber(1,2)
y = RationalNumber(3,2)
print ("The first number is {!s}".format(x))
print ("The second number is {!s}".format(y))
print ("Their sum is {!s}".format(x+y))
print ("Their product is {!s}".format(x*y))
print ("Their difference is {!s}".format(x-y))
print ("Their quotient is {!s}".format(x/y))
```
(hint)
|Operation |Method |
|---------------|----------------------------|
|Addition |(a/b) + (c/d) = (ad + bc)/bd|
|Subtraction |(a/b) - (c/d) = (ad - bc)/bd|
|Multiplication |(a/b) x (c/d) = ac/bd |
|Division |(a/b) / (c/d) = ad/bc |
## Modules
Modules are reusable libraries of code. Many libraries come standard with Python. You can import them into a program using the *import* statement. For example:
```
import math
print ("The first few digits of pi are {:f}...".format(math.pi))
```
The math module implements many functions for complex mathematical operations using floating point values, including logarithms, trigonometric operations, and irrational numbers like π.
#### As an exercise, we'll encapsulate your rational numbers script into a module and then import it.
Save the RatNum.py file you've been working in. Open your terminal and navigate whereever you have the file saved.
Type:
python
When you're inside the Python interpreter, enter:
from RatNum import RationalNumber
a = RationalNumber(1,3)
b = RationalNumber(2,3)
print (a*b)
Success! You have just made a module.
## Packages
A package is a directory of modules. For example, we could make a big package by bundling together modules with classes for natural numbers, integers, irrational numbers, and real numbers.
The Python Package Index, or "PyPI", is the official third-party software repository for the Python programming language. It is a comprehensive catalog of all open source Python packages and is maintained by the Python Software Foundation. You can download packages from PyPI with the *pip* command in your terminal.
PyPI packages are uploaded by individual package maintainers. That means you can write and contribute your own Python packages!
#### Now let's turn your module into a package called Mathy.
1. Create a folder called Mathy, and add your RatNum.py file to the folder.
2. Add an empty file to the folder called \_\_init\_\_.py.
3. Create a third file in that folder called MathQuiz.py that imports RationalNumber from RatNum...
4. ...and uses the RationalNumbers class from RatNum. For example:
#MathQuiz.py
from RatNum import RationalNumber
print "Pop quiz! Find the sum, product, difference, and quotient for the following rational numbers:"
x = RationalNumber(1,3)
y = RationalNumber(2,3)
print ("The first number is {!s}".format(x))
print ("The second number is {!s}".format(y))
print ("Their sum is {!s}".format(x+y))
print ("Their product is {!s}".format(x*y))
print ("Their difference is {!s}".format(x-y))
print ("Their quotient is {!s}".format(x/y))
#### In the terminal, navigate to the Mathy folder. When you are inside the folder, type:
python MathQuiz.py
Congrats! You have just made a Python package!
#### Now type:
python RatNum.py
What did you get this time? Is it different from the answer you got for the previous command? Why??
Once you've completed this exercise, move on to Part 2.
## Part 2: Inheritance
Suppose we were to write out another class for another set of numbers, say the integers. What are the rules for addition, subtraction, multiplication, and division? If we can identify shared properties between integers and rational numbers, we could use that information to write a integer class that 'inherits' properties from our rational number class.
#### Let's add an integer class to our RatNum.py file that inherits all the properties of our RationalNumber class.
```
class Integer(RationalNumber):
#What should we add here?
pass
```
#### Now update your \_\_name\_\_ == "\_\_main\_\_" statement at the end of RatNum.py to read:
```
if __name__ == "__main__":
q = Integer(5)
r = Integer(6)
print ("{!s} is an integer expressed as a rational number".format(q))
print ("So is {!s}".format(r))
print ("When you add them you get {!s}".format(q+r))
print ("When you multiply them you get {!s}".format(q*r))
print ("When you subtract them you get {!s}".format(q-r))
print ("When you divide them you get {!s}".format(q/r))
```
Did it work?
Nice job!
|
github_jupyter
|
class RationalNumber:
"""Any number that can be expressed as the quotient or fraction p/q
of two integers, p and q, with the denominator q not equal to zero.
Since q may be equal to 1, every integer is a rational number.
"""
def __init__(self, numerator, denominator=1):
self.n = numerator
self.d = denominator
def __add__(self, other):
# Write a function that allows for the addition of two rational numbers.
# I did this one for you :D
if not isinstance(other, RationalNumber):
other = RationalNumber(other)
n = self.n * other.d + self.d * other.n
d = self.d * other.d
return RationalNumber(n, d)
def __sub__(self, other):
# Write a function that allows for the subtraction of two rational numbers.
pass
def __mul__(self, other):
# Write a function that allows for the multiplication of two rational numbers.
pass
def __truediv__(self, other):
# Write a function that allows for the division of two rational numbers.
pass
def __str__(self):
return "%s/%s" % (self.n, self.d)
__repr__ = __str__
if __name__ == "__main__":
x = RationalNumber(1,2)
y = RationalNumber(3,2)
print ("The first number is {!s}".format(x))
print ("The second number is {!s}".format(y))
print ("Their sum is {!s}".format(x+y))
print ("Their product is {!s}".format(x*y))
print ("Their difference is {!s}".format(x-y))
print ("Their quotient is {!s}".format(x/y))
import math
print ("The first few digits of pi are {:f}...".format(math.pi))
class Integer(RationalNumber):
#What should we add here?
pass
if __name__ == "__main__":
q = Integer(5)
r = Integer(6)
print ("{!s} is an integer expressed as a rational number".format(q))
print ("So is {!s}".format(r))
print ("When you add them you get {!s}".format(q+r))
print ("When you multiply them you get {!s}".format(q*r))
print ("When you subtract them you get {!s}".format(q-r))
print ("When you divide them you get {!s}".format(q/r))
| 0.842475 | 0.917672 |
# Combining Color and Region Selections
Let's combine the mask and color selection to pull only the lane lines out of the image.
Check out the code below. Here we’re doing both the color and region selection steps, requiring that a pixel meet both the mask and color selection requirements to be retained.
```
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import numpy as np
# Read in the image
image = mpimg.imread('img/road_image.jpg')
# Grab the x and y sizes and make two copies of the image
# With one copy we'll extract only the pixels that meet our selection,
# then we'll paint those pixels red in the original image to see our selection
# overlaid on the original.
ysize = image.shape[0]
xsize = image.shape[1]
color_select= np.copy(image)
line_image = np.copy(image)
# Define our color criteria
red_threshold = 200
green_threshold = 200
blue_threshold = 200
rgb_threshold = [red_threshold, green_threshold, blue_threshold]
# Define a triangle region of interest (Note: if you run this code,
# Keep in mind the origin (x=0, y=0) is in the upper left in image processing
# you'll find these are not sensible values!!
# But you'll get a chance to play with them soon in a quiz ;)
left_bottom = [0, 539]
right_bottom = [939, 539]
apex = [450, 320]
fit_left = np.polyfit((left_bottom[0], apex[0]), (left_bottom[1], apex[1]), 1)
fit_right = np.polyfit((right_bottom[0], apex[0]), (right_bottom[1], apex[1]), 1)
fit_bottom = np.polyfit((left_bottom[0], right_bottom[0]), (left_bottom[1], right_bottom[1]), 1)
# Mask pixels below the threshold
color_thresholds = (image[:,:,0] < rgb_threshold[0]) | \
(image[:,:,1] < rgb_threshold[1]) | \
(image[:,:,2] < rgb_threshold[2])
# Find the region inside the lines
XX, YY = np.meshgrid(np.arange(0, xsize), np.arange(0, ysize))
region_thresholds = (YY > (XX*fit_left[0] + fit_left[1])) & \
(YY > (XX*fit_right[0] + fit_right[1])) & \
(YY < (XX*fit_bottom[0] + fit_bottom[1]))
# Mask color selection
color_select[color_thresholds] = [0,0,0]
# Find where image is both colored right and in the region
line_image[~color_thresholds & region_thresholds] = [255,0,0]
# Display our two output images
plt.imshow(color_select)
plt.imshow(line_image)
# uncomment if plot does not display
# plt.show()
# Display the image and show region and color selections
plt.imshow(image)
x = [left_bottom[0], right_bottom[0], apex[0], left_bottom[0]]
y = [left_bottom[1], right_bottom[1], apex[1], left_bottom[1]]
plt.plot(x, y, 'b--', lw=4)
plt.imshow(color_select)
plt.show()
plt.imshow(line_image)
plt.show()
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import numpy as np
# Read in the image
image = mpimg.imread('img/road_image.jpg')
# Grab the x and y sizes and make two copies of the image
# With one copy we'll extract only the pixels that meet our selection,
# then we'll paint those pixels red in the original image to see our selection
# overlaid on the original.
ysize = image.shape[0]
xsize = image.shape[1]
color_select= np.copy(image)
line_image = np.copy(image)
# Define our color criteria
red_threshold = 200
green_threshold = 200
blue_threshold = 200
rgb_threshold = [red_threshold, green_threshold, blue_threshold]
# Define a triangle region of interest (Note: if you run this code,
# Keep in mind the origin (x=0, y=0) is in the upper left in image processing
# you'll find these are not sensible values!!
# But you'll get a chance to play with them soon in a quiz ;)
left_bottom = [0, 539]
right_bottom = [939, 539]
apex = [450, 320]
fit_left = np.polyfit((left_bottom[0], apex[0]), (left_bottom[1], apex[1]), 1)
fit_right = np.polyfit((right_bottom[0], apex[0]), (right_bottom[1], apex[1]), 1)
fit_bottom = np.polyfit((left_bottom[0], right_bottom[0]), (left_bottom[1], right_bottom[1]), 1)
# Mask pixels below the threshold
color_thresholds = (image[:,:,0] < rgb_threshold[0]) | \
(image[:,:,1] < rgb_threshold[1]) | \
(image[:,:,2] < rgb_threshold[2])
# Find the region inside the lines
XX, YY = np.meshgrid(np.arange(0, xsize), np.arange(0, ysize))
region_thresholds = (YY > (XX*fit_left[0] + fit_left[1])) & \
(YY > (XX*fit_right[0] + fit_right[1])) & \
(YY < (XX*fit_bottom[0] + fit_bottom[1]))
# Mask color selection
color_select[color_thresholds] = [0,0,0]
# Find where image is both colored right and in the region
line_image[~color_thresholds & region_thresholds] = [255,0,0]
# Display our two output images
plt.imshow(color_select)
plt.imshow(line_image)
# uncomment if plot does not display
# plt.show()
# Display the image and show region and color selections
plt.imshow(image)
x = [left_bottom[0], right_bottom[0], apex[0], left_bottom[0]]
y = [left_bottom[1], right_bottom[1], apex[1], left_bottom[1]]
plt.plot(x, y, 'b--', lw=4)
plt.imshow(color_select)
plt.show()
plt.imshow(line_image)
plt.show()
| 0.592077 | 0.985855 |
```
import numpy as np
import matplotlib.pyplot as plt
def generate_data():
size = 1000
x = np.linspace(0, 1, size)
y = -10*x + 5
y += 15*np.logical_and(x > 0.75, x < 0.8).astype(float)
return x, y
```
# 1. Inspect the data (0.5 points)
Using `matplotlib`, create a scatter plot of the data returned by `generate_data()`.
What is out of the ordinary about this line?
```
x, y = generate_data()
plt.scatter(x, y)
```
What's odd is that the line segment between x = 0.75 and x = 0.8 is moved upward by 15.
# 2. Implement linear regression (2 points)
Implement a basic linear regression model which is fit to the data from `generate_data` using gradient descent.
Your model should take the form `y = m*x + b`, where `y` is the output, `x` is the input, `m` is a weight parameter, and `b` is a bias parameter.
You must use only `numpy` and derive any derivatives yourself (i.e. no autograd from TensorFlow, MXNet, Pytorch, JAX etc!).
You should use a squared-error loss function.
You are welcome to use any technique you want to decide when to stop training.
Make sure you tune your optimization hyperparameters so that the model converges.
Print out or plot the loss over the course of training.
```
# Initialize the parameters m and b
n = y.shape[0] # n is the number of training example
np.random.seed(12)
b = np.random.randn()
m = np.random.randn()
# Initialize loss list for the convenience of plotting loss curve
losses = []
# Gradient Descent
# first set the learning rate
learning_rate = 0.1
for iter in range(1000):
# calculate the estimate of y
y_estimate = m * x + b
# calculate the error using squared-error loss function
loss = np.sum((y_estimate - y) ** 2) / (2*n)
losses.append(loss)
# take the derivative w.r.t m and b
dm = 1/n * np.sum((y_estimate - y) * x)
db = 1/n * np.sum(y_estimate - y)
# update w and b
m = m - learning_rate * dm
b = b - learning_rate * db
print(losses[998])
print(losses[999])
plt.plot(losses)
plt.xlabel("Iterations")
plt.ylabel("$J(\Theta)$")
plt.title("Values of Loss Function over iterations of Gradient Descent");
```
The losses keep decreasing over each iteration, which means the learning rate
0.1 is appropriate.
The difference of loss between 999th and 1000th iteration is less than $\mathrm{10}^{-3}$. Thus, we can declare convergence.
# 3. Analyze the result (0.5 points)
Print out the values of `w` and `b` found by your model after training and compare them to the ground truth values (which can be found inside the code of the `generate_data` function).
Are they close?
Recreate the scatter plot you generated in question 1 and plot the model as a line on the same plot.
What went wrong?
```
print("The value of estimated w is: " + "\n" + str(m))
print("The value of estimated b is: " + "\n" + str(b))
y_pred = m*x+b
plt.scatter(x, y)
plt.plot(x, y_pred, c="red")
```
The ground truth value for w is -10 and the truth value for the biase term is 5.
The values estimated by my model are -7.518030824377429 for w and
4.508682516078752 for b, which are not close to their ground truth value.
It is because this linear regression model is strongly influenced by the outliers between x=0.75 and x=0.8.
# 4. "Robust" linear regression (0.5 points)
Implement a linear regression model exactly like the one you created in question 2, except using a L1 loss (absolute difference) instead of a squared L2 loss (squared error).
You should be able to copy and paste your code from question 2 and only change a few lines.
Print out or plot the loss over the course of training.
What is different about the loss trajectory compared to the squared-error linear regression?
```
# Initialize the parameters m and b
n = y.shape[0] # n is the number of training example
np.random.seed(12)
b2 = np.random.randn()
m2 = np.random.randn()
# Initialize loss list for the convenience of plotting loss curve
losses2 = []
# Gradient Descent using L1 loss
# first set the learning rate
learning_rate = 0.01
for iter in range(5500):
# calculate the estimate of y
y_estimate = m2 * x + b2
# calculate the error using squared-error loss function
loss = np.sum(np.abs(y_estimate - y)) / n
losses2.append(loss)
# take the derivative w.r.t m and b
dm2 = 1/n * np.sum((y_estimate - y) / np.abs(y_estimate - y) * x)
db2 = 1/n * np.sum((y_estimate - y) / np.abs(y_estimate - y))
# update w and b
m2 = m2 - learning_rate * dm2
b2 = b2 - learning_rate * db2
losses2[5499]
plt.plot(losses2)
plt.xlabel("Iterations")
plt.ylabel("$J(\Theta)$")
plt.title("Values of Loss Function over iterations of Gradient Descent");
```
After a steady decrease, the loss trajectory becomes a horizontal line, which indicates convergence.
The loss trajectory for linear regression with L1 loss is a straight line, while that for squared-error linear regression is a convex curve.
# 5. Analyze the result (0.5 points)
Print out the new values of `w` and `b` found by your model after training.
Are they closer to the true values used in `generate_data`?
Plot the model as a line again.
Why do you think the behavior is different?
```
print("The value of estimated w is: " + "\n" + str(m2))
print("The value of estimated b is: " + "\n" + str(b2))
y_pred2 = m*x+b
plt.scatter(x, y)
plt.plot(x, y_pred2, c="red")
```
Now, with L1 loss function, the estimated value of w is -10.001669242803908, and the estimated value of b is 4.996945831490282. They are much closer to the ground true values compared to the squared-error linear regression. The behavior is different due to the use of a different loss function. The L1 (absolute difference) loss function is more robust to outliers than the L2 (squared error) loss function.
## Acknowledgement
I collaberate with Yicheng Zou to work on this assignment.
Part of the code in this assignment is adapted from Andrew Ng's Machine Learning course on Coursera.
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
def generate_data():
size = 1000
x = np.linspace(0, 1, size)
y = -10*x + 5
y += 15*np.logical_and(x > 0.75, x < 0.8).astype(float)
return x, y
x, y = generate_data()
plt.scatter(x, y)
# Initialize the parameters m and b
n = y.shape[0] # n is the number of training example
np.random.seed(12)
b = np.random.randn()
m = np.random.randn()
# Initialize loss list for the convenience of plotting loss curve
losses = []
# Gradient Descent
# first set the learning rate
learning_rate = 0.1
for iter in range(1000):
# calculate the estimate of y
y_estimate = m * x + b
# calculate the error using squared-error loss function
loss = np.sum((y_estimate - y) ** 2) / (2*n)
losses.append(loss)
# take the derivative w.r.t m and b
dm = 1/n * np.sum((y_estimate - y) * x)
db = 1/n * np.sum(y_estimate - y)
# update w and b
m = m - learning_rate * dm
b = b - learning_rate * db
print(losses[998])
print(losses[999])
plt.plot(losses)
plt.xlabel("Iterations")
plt.ylabel("$J(\Theta)$")
plt.title("Values of Loss Function over iterations of Gradient Descent");
print("The value of estimated w is: " + "\n" + str(m))
print("The value of estimated b is: " + "\n" + str(b))
y_pred = m*x+b
plt.scatter(x, y)
plt.plot(x, y_pred, c="red")
# Initialize the parameters m and b
n = y.shape[0] # n is the number of training example
np.random.seed(12)
b2 = np.random.randn()
m2 = np.random.randn()
# Initialize loss list for the convenience of plotting loss curve
losses2 = []
# Gradient Descent using L1 loss
# first set the learning rate
learning_rate = 0.01
for iter in range(5500):
# calculate the estimate of y
y_estimate = m2 * x + b2
# calculate the error using squared-error loss function
loss = np.sum(np.abs(y_estimate - y)) / n
losses2.append(loss)
# take the derivative w.r.t m and b
dm2 = 1/n * np.sum((y_estimate - y) / np.abs(y_estimate - y) * x)
db2 = 1/n * np.sum((y_estimate - y) / np.abs(y_estimate - y))
# update w and b
m2 = m2 - learning_rate * dm2
b2 = b2 - learning_rate * db2
losses2[5499]
plt.plot(losses2)
plt.xlabel("Iterations")
plt.ylabel("$J(\Theta)$")
plt.title("Values of Loss Function over iterations of Gradient Descent");
print("The value of estimated w is: " + "\n" + str(m2))
print("The value of estimated b is: " + "\n" + str(b2))
y_pred2 = m*x+b
plt.scatter(x, y)
plt.plot(x, y_pred2, c="red")
| 0.818338 | 0.982922 |
## PS2-3 Bayesian Interpretation of Regularization
#### (a)
Proof:
\begin{align*}
\theta_{\mathrm{MAP}} & = \arg \max_\theta p(\theta \ \vert \ x, y) \\
& = \arg \max_\theta \frac{p(y \ \vert \ x, \theta) \ p(x, \theta)}{p(x, y)} \\
& = \arg \max_\theta \frac{p(y \ \vert \ x, \theta) \ p(\theta \ \vert \ x) \ p(x)}{p(x, y)} \\
& = \arg \max_\theta \frac{p(y \ \vert \ x, \theta) \ p(\theta) \ p(x)}{p(x, y)} \\
& = \arg \max_\theta p(y \ \vert \ x, \theta) \ p(\theta)
\end{align*}
#### (b)
Since $p(\theta) \sim \mathcal{N} (0, \eta^2 I)$,
\begin{align*}
\theta_{\mathrm{MAP}} & = \arg \max_\theta p(y \ \vert \ x, \theta) \ p(\theta) \\
& = \arg \min_\theta - \log p(y \ \vert \ x, \theta) - \log p(\theta) \\
& = \arg \min_\theta - \log p(y \ \vert \ x, \theta) - \log \frac{1}{(2 \pi)^{d / 2} \vert \Sigma \vert^{1/2}} \exp \big( -\frac{1}{2} (\theta - \mu)^T \Sigma^{-1} (\theta - \mu) \big) \\
& = \arg \min_\theta - \log p(y \ \vert \ x, \theta) + \frac{1}{2} \theta^T \Sigma^{-1} \theta \\
& = \arg \min_\theta - \log p(y \ \vert \ x, \theta) + \frac{1}{2 \eta^2} \Vert \theta \Vert_2^2 \\
& = \arg \min_\theta - \log p(y \ \vert \ x, \theta) + \lambda \Vert \theta \Vert_2^2
\end{align*}
where $\lambda = 1 / (2 \eta^2)$.
#### (c)
Given $y = \theta^T x + \epsilon$ where $\epsilon \sim \mathcal{N} (0, \sigma^2)$, i.e. $y \ \vert \ x; \ \theta \sim \mathcal{N} (\theta^T x, \sigma^2)$,
\begin{align*}
\theta_{\mathrm{MAP}} & = \arg \min_\theta - \sum_{i = 1}^{m} \log \frac{1}{\sqrt{2 \pi} \sigma} \exp \big( - \frac{(y^{(i)} - \theta^T x^{(i)})^2}{2 \sigma^2} \big) + \lambda \Vert \theta \Vert_2^2 \\
& = \arg \min_\theta \frac{1}{2 \sigma^2} \sum_{i = 1}^{m} (y^{(i)} - \theta^T x^{(i)})^2 + \frac{1}{2 \eta^2} \Vert \theta \Vert_2^2 \\
& = \arg \min_\theta \frac{1}{2 \sigma^2} (\vec{y} - X \theta)^T (\vec{y} - X \theta) + \frac{1}{2 \eta^2} \Vert \theta \Vert_2^2 \\
& = \arg \min_\theta J(\theta)
\end{align*}
By solving
\begin{align*}
\nabla_\theta J(\theta) & = \nabla_\theta \big( \frac{1}{2 \sigma^2} (\vec{y} - X \theta)^T (\vec{y} - X \theta) + \frac{1}{2 \eta^2} \Vert \theta \Vert_2^2 \big) \\
& = \frac{1}{2 \sigma^2} \nabla_\theta (\theta^T X^T X \theta - 2 \vec{y}^T X \theta + \frac{\sigma^2}{\eta^2} \theta^T \theta) \\
& = \frac{1}{\sigma^2} (X^T X \theta - X^T \vec{y} + \frac{\sigma^2}{\eta^2} \theta) \\
& = 0
\end{align*}
we obtain
$$\theta_{\mathrm{MAP}} = (X^T X + \frac{\sigma^2}{\eta^2} I)^{-1} X^T \vec{y}$$
#### (d)
Assume $\theta \in \mathbb{R}^n$. Given $\theta_i \sim \mathcal{L} (0, b)$ and $y = \theta^T x + \epsilon$ where $\epsilon \sim \mathcal{N} (0, \sigma^2)$, we have
\begin{align*}
\theta_{\mathrm{MAP}} & = \arg \min_\theta - \sum_{i = 1}^{m} \log \frac{1}{\sqrt{2 \pi} \sigma} \exp \big( - \frac{(y^{(i)} - \theta^T x^{(i)})^2}{2 \sigma^2} \big)- \sum_{i = 1}^{n} \log \frac{1}{2 b} \exp \big( - \frac{\vert \theta_i - 0 \vert}{b} \big) \\
& = \arg \min_\theta \frac{1}{2 \sigma^2} \sum_{i = 1}^{m} (y^{(i)} - \theta^T x^{(i)})^2 + \sum_{i = 1}^{n} \frac{1}{b} \vert \theta_i \vert \\
& = \arg \min_\theta \frac{1}{2 \sigma^2} \Vert X \theta - \vec{y} \Vert_2^2 + \frac{1}{b} \Vert \theta \Vert_1 \\
& = \arg \min_\theta \Vert X \theta - \vec{y} \Vert_2^2 + \frac{2 \sigma^2}{b} \Vert \theta \Vert_1
\end{align*}
Therefore,
$$J(\theta) = \Vert X \theta - \vec{y} \Vert_2^2 + \frac{2 \sigma^2}{b} \Vert \theta \Vert_1$$
|
github_jupyter
|
## PS2-3 Bayesian Interpretation of Regularization
#### (a)
Proof:
\begin{align*}
\theta_{\mathrm{MAP}} & = \arg \max_\theta p(\theta \ \vert \ x, y) \\
& = \arg \max_\theta \frac{p(y \ \vert \ x, \theta) \ p(x, \theta)}{p(x, y)} \\
& = \arg \max_\theta \frac{p(y \ \vert \ x, \theta) \ p(\theta \ \vert \ x) \ p(x)}{p(x, y)} \\
& = \arg \max_\theta \frac{p(y \ \vert \ x, \theta) \ p(\theta) \ p(x)}{p(x, y)} \\
& = \arg \max_\theta p(y \ \vert \ x, \theta) \ p(\theta)
\end{align*}
#### (b)
Since $p(\theta) \sim \mathcal{N} (0, \eta^2 I)$,
\begin{align*}
\theta_{\mathrm{MAP}} & = \arg \max_\theta p(y \ \vert \ x, \theta) \ p(\theta) \\
& = \arg \min_\theta - \log p(y \ \vert \ x, \theta) - \log p(\theta) \\
& = \arg \min_\theta - \log p(y \ \vert \ x, \theta) - \log \frac{1}{(2 \pi)^{d / 2} \vert \Sigma \vert^{1/2}} \exp \big( -\frac{1}{2} (\theta - \mu)^T \Sigma^{-1} (\theta - \mu) \big) \\
& = \arg \min_\theta - \log p(y \ \vert \ x, \theta) + \frac{1}{2} \theta^T \Sigma^{-1} \theta \\
& = \arg \min_\theta - \log p(y \ \vert \ x, \theta) + \frac{1}{2 \eta^2} \Vert \theta \Vert_2^2 \\
& = \arg \min_\theta - \log p(y \ \vert \ x, \theta) + \lambda \Vert \theta \Vert_2^2
\end{align*}
where $\lambda = 1 / (2 \eta^2)$.
#### (c)
Given $y = \theta^T x + \epsilon$ where $\epsilon \sim \mathcal{N} (0, \sigma^2)$, i.e. $y \ \vert \ x; \ \theta \sim \mathcal{N} (\theta^T x, \sigma^2)$,
\begin{align*}
\theta_{\mathrm{MAP}} & = \arg \min_\theta - \sum_{i = 1}^{m} \log \frac{1}{\sqrt{2 \pi} \sigma} \exp \big( - \frac{(y^{(i)} - \theta^T x^{(i)})^2}{2 \sigma^2} \big) + \lambda \Vert \theta \Vert_2^2 \\
& = \arg \min_\theta \frac{1}{2 \sigma^2} \sum_{i = 1}^{m} (y^{(i)} - \theta^T x^{(i)})^2 + \frac{1}{2 \eta^2} \Vert \theta \Vert_2^2 \\
& = \arg \min_\theta \frac{1}{2 \sigma^2} (\vec{y} - X \theta)^T (\vec{y} - X \theta) + \frac{1}{2 \eta^2} \Vert \theta \Vert_2^2 \\
& = \arg \min_\theta J(\theta)
\end{align*}
By solving
\begin{align*}
\nabla_\theta J(\theta) & = \nabla_\theta \big( \frac{1}{2 \sigma^2} (\vec{y} - X \theta)^T (\vec{y} - X \theta) + \frac{1}{2 \eta^2} \Vert \theta \Vert_2^2 \big) \\
& = \frac{1}{2 \sigma^2} \nabla_\theta (\theta^T X^T X \theta - 2 \vec{y}^T X \theta + \frac{\sigma^2}{\eta^2} \theta^T \theta) \\
& = \frac{1}{\sigma^2} (X^T X \theta - X^T \vec{y} + \frac{\sigma^2}{\eta^2} \theta) \\
& = 0
\end{align*}
we obtain
$$\theta_{\mathrm{MAP}} = (X^T X + \frac{\sigma^2}{\eta^2} I)^{-1} X^T \vec{y}$$
#### (d)
Assume $\theta \in \mathbb{R}^n$. Given $\theta_i \sim \mathcal{L} (0, b)$ and $y = \theta^T x + \epsilon$ where $\epsilon \sim \mathcal{N} (0, \sigma^2)$, we have
\begin{align*}
\theta_{\mathrm{MAP}} & = \arg \min_\theta - \sum_{i = 1}^{m} \log \frac{1}{\sqrt{2 \pi} \sigma} \exp \big( - \frac{(y^{(i)} - \theta^T x^{(i)})^2}{2 \sigma^2} \big)- \sum_{i = 1}^{n} \log \frac{1}{2 b} \exp \big( - \frac{\vert \theta_i - 0 \vert}{b} \big) \\
& = \arg \min_\theta \frac{1}{2 \sigma^2} \sum_{i = 1}^{m} (y^{(i)} - \theta^T x^{(i)})^2 + \sum_{i = 1}^{n} \frac{1}{b} \vert \theta_i \vert \\
& = \arg \min_\theta \frac{1}{2 \sigma^2} \Vert X \theta - \vec{y} \Vert_2^2 + \frac{1}{b} \Vert \theta \Vert_1 \\
& = \arg \min_\theta \Vert X \theta - \vec{y} \Vert_2^2 + \frac{2 \sigma^2}{b} \Vert \theta \Vert_1
\end{align*}
Therefore,
$$J(\theta) = \Vert X \theta - \vec{y} \Vert_2^2 + \frac{2 \sigma^2}{b} \Vert \theta \Vert_1$$
| 0.664649 | 0.995042 |
# Interval based time series classification in sktime
Interval based approaches look at phase dependant intervals of the full series, calculating summary statistics from selected subseries to be used in classification.
Currently 5 univariate interval based approaches are implemented in sktime. Time Series Forest (TSF) \[1\], the Random Interval Spectral Ensemble (RISE) \[2\], Supervised Time Series Forest (STSF) \[3\], the Canonical Interval Forest (CIF) \[4\] and the Diverse Representation Canonical Interval Forest (DrCIF). Both CIF and DrCIF have multivariate capabilities.
In this notebook, we will demonstrate how to use these classifiers on the ItalyPowerDemand and BasicMotions datasets.
#### References:
\[1\] Deng, H., Runger, G., Tuv, E., & Vladimir, M. (2013). A time series forest for classification and feature extraction. Information Sciences, 239, 142-153.
\[2\] Flynn, M., Large, J., & Bagnall, T. (2019). The contract random interval spectral ensemble (c-RISE): the effect of contracting a classifier on accuracy. In International Conference on Hybrid Artificial Intelligence Systems (pp. 381-392). Springer, Cham.
\[3\] Cabello, N., Naghizade, E., Qi, J., & Kulik, L. (2020). Fast and Accurate Time Series Classification Through Supervised Interval Search. In IEEE International Conference on Data Mining.
\[4\] Middlehurst, M., Large, J., & Bagnall, A. (2020). The Canonical Interval Forest (CIF) Classifier for Time Series Classification. arXiv preprint arXiv:2008.09172.
\[5\] Lubba, C. H., Sethi, S. S., Knaute, P., Schultz, S. R., Fulcher, B. D., & Jones, N. S. (2019). catch22: CAnonical Time-series CHaracteristics. Data Mining and Knowledge Discovery, 33(6), 1821-1852.
## 1. Imports
```
from sklearn import metrics
from sktime.classification.interval_based import (
CanonicalIntervalForest,
DrCIF,
RandomIntervalSpectralEnsemble,
SupervisedTimeSeriesForest,
TimeSeriesForestClassifier,
)
from sktime.datasets import load_basic_motions, load_italy_power_demand
```
## 2. Load data
```
X_train, y_train = load_italy_power_demand(split="train", return_X_y=True)
X_test, y_test = load_italy_power_demand(split="test", return_X_y=True)
X_test = X_test[:50]
y_test = y_test[:50]
print(X_train.shape, y_train.shape, X_test.shape, y_test.shape)
X_train_mv, y_train_mv = load_basic_motions(split="train", return_X_y=True)
X_test_mv, y_test_mv = load_basic_motions(split="test", return_X_y=True)
X_train_mv = X_train_mv[:50]
y_train_mv = y_train_mv[:50]
X_test_mv = X_test_mv[:50]
y_test_mv = y_test_mv[:50]
print(X_train_mv.shape, y_train_mv.shape, X_test_mv.shape, y_test_mv.shape)
```
## 3. Time Series Forest (TSF)
TSF is an ensemble of tree classifiers built on the summary statistics of randomly selected intervals.
For each tree sqrt(series_length) intervals are randomly selected.
From each of these intervals the mean, standard deviation and slope is extracted from each time series and concatenated into a feature vector.
These new features are then used to build a tree, which is added to the ensemble.
```
tsf = TimeSeriesForestClassifier(n_estimators=50, random_state=47)
tsf.fit(X_train, y_train)
tsf_preds = tsf.predict(X_test)
print("TSF Accuracy: " + str(metrics.accuracy_score(y_test, tsf_preds)))
```
## 4. Random Interval Spectral Ensemble (RISE)
RISE is a tree based interval ensemble aimed at classifying audio data. Unlike TSF, it uses a single interval for each tree, and it uses spectral features rather than summary statistics.
```
rise = RandomIntervalSpectralEnsemble(n_estimators=50, random_state=47)
rise.fit(X_train, y_train)
rise_preds = rise.predict(X_test)
print("RISE Accuracy: " + str(metrics.accuracy_score(y_test, rise_preds)))
```
## 5. Supervised Time Series Forest (STSF)
STSF makes a number of adjustments from the original TSF algorithm. A supervised method of selecting intervals replaces random selection. Features are extracted from intervals generated from additional representations in periodogram and 1st order differences. Median, min, max and interquartile range are included in the summary statistics extracted.
```
stsf = SupervisedTimeSeriesForest(n_estimators=50, random_state=47)
stsf.fit(X_train, y_train)
stsf_preds = stsf.predict(X_test)
print("STSF Accuracy: " + str(metrics.accuracy_score(y_test, stsf_preds)))
```
## 6. Canonical Interval Forest (CIF)
CIF extends from the TSF algorithm. In addition to the 3 summary statistics used by TSF, CIF makes use of the features from the `Catch22` \[5\] transform.
To increase the diversity of the ensemble, the number of TSF and catch22 attributes is randomly subsampled per tree.
### Univariate
```
cif = CanonicalIntervalForest(n_estimators=50, att_subsample_size=8, random_state=47)
cif.fit(X_train, y_train)
cif_preds = cif.predict(X_test)
print("CIF Accuracy: " + str(metrics.accuracy_score(y_test, cif_preds)))
```
### Multivariate
```
cif_m = CanonicalIntervalForest(n_estimators=50, att_subsample_size=8, random_state=47)
cif_m.fit(X_train_mv, y_train_mv)
cif_m_preds = cif_m.predict(X_test_mv)
print("CIF Accuracy: " + str(metrics.accuracy_score(y_test_mv, cif_m_preds)))
```
## 6. Diverse Representation Canonical Interval Forest (DrCIF)
DrCIF makes use of the periodogram and differences representations used by STSF as well as the addition summary statistics in CIF.
### Univariate
```
drcif = DrCIF(n_estimators=5, att_subsample_size=10, random_state=47)
drcif.fit(X_train, y_train)
drcif_preds = drcif.predict(X_test)
print("DrCIF Accuracy: " + str(metrics.accuracy_score(y_test, drcif_preds)))
```
### Multivariate
```
drcif_m = DrCIF(n_estimators=5, att_subsample_size=10, random_state=47)
drcif_m.fit(X_train_mv, y_train_mv)
drcif_m_preds = drcif_m.predict(X_test_mv)
print("DrCIF Accuracy: " + str(metrics.accuracy_score(y_test_mv, drcif_m_preds)))
```
|
github_jupyter
|
from sklearn import metrics
from sktime.classification.interval_based import (
CanonicalIntervalForest,
DrCIF,
RandomIntervalSpectralEnsemble,
SupervisedTimeSeriesForest,
TimeSeriesForestClassifier,
)
from sktime.datasets import load_basic_motions, load_italy_power_demand
X_train, y_train = load_italy_power_demand(split="train", return_X_y=True)
X_test, y_test = load_italy_power_demand(split="test", return_X_y=True)
X_test = X_test[:50]
y_test = y_test[:50]
print(X_train.shape, y_train.shape, X_test.shape, y_test.shape)
X_train_mv, y_train_mv = load_basic_motions(split="train", return_X_y=True)
X_test_mv, y_test_mv = load_basic_motions(split="test", return_X_y=True)
X_train_mv = X_train_mv[:50]
y_train_mv = y_train_mv[:50]
X_test_mv = X_test_mv[:50]
y_test_mv = y_test_mv[:50]
print(X_train_mv.shape, y_train_mv.shape, X_test_mv.shape, y_test_mv.shape)
tsf = TimeSeriesForestClassifier(n_estimators=50, random_state=47)
tsf.fit(X_train, y_train)
tsf_preds = tsf.predict(X_test)
print("TSF Accuracy: " + str(metrics.accuracy_score(y_test, tsf_preds)))
rise = RandomIntervalSpectralEnsemble(n_estimators=50, random_state=47)
rise.fit(X_train, y_train)
rise_preds = rise.predict(X_test)
print("RISE Accuracy: " + str(metrics.accuracy_score(y_test, rise_preds)))
stsf = SupervisedTimeSeriesForest(n_estimators=50, random_state=47)
stsf.fit(X_train, y_train)
stsf_preds = stsf.predict(X_test)
print("STSF Accuracy: " + str(metrics.accuracy_score(y_test, stsf_preds)))
cif = CanonicalIntervalForest(n_estimators=50, att_subsample_size=8, random_state=47)
cif.fit(X_train, y_train)
cif_preds = cif.predict(X_test)
print("CIF Accuracy: " + str(metrics.accuracy_score(y_test, cif_preds)))
cif_m = CanonicalIntervalForest(n_estimators=50, att_subsample_size=8, random_state=47)
cif_m.fit(X_train_mv, y_train_mv)
cif_m_preds = cif_m.predict(X_test_mv)
print("CIF Accuracy: " + str(metrics.accuracy_score(y_test_mv, cif_m_preds)))
drcif = DrCIF(n_estimators=5, att_subsample_size=10, random_state=47)
drcif.fit(X_train, y_train)
drcif_preds = drcif.predict(X_test)
print("DrCIF Accuracy: " + str(metrics.accuracy_score(y_test, drcif_preds)))
drcif_m = DrCIF(n_estimators=5, att_subsample_size=10, random_state=47)
drcif_m.fit(X_train_mv, y_train_mv)
drcif_m_preds = drcif_m.predict(X_test_mv)
print("DrCIF Accuracy: " + str(metrics.accuracy_score(y_test_mv, drcif_m_preds)))
| 0.619126 | 0.977045 |
# Continuous Control
---
Congratulations for completing the second project of the [Deep Reinforcement Learning Nanodegree](https://www.udacity.com/course/deep-reinforcement-learning-nanodegree--nd893) program! In this notebook, you will learn how to control an agent in a more challenging environment, where the goal is to train a creature with four arms to walk forward. **Note that this exercise is optional!**
### 1. Start the Environment
We begin by importing the necessary packages. If the code cell below returns an error, please revisit the project instructions to double-check that you have installed [Unity ML-Agents](https://github.com/Unity-Technologies/ml-agents/blob/master/docs/Installation.md) and [NumPy](http://www.numpy.org/).
```
from unityagents import UnityEnvironment
import numpy as np
```
Next, we will start the environment! **_Before running the code cell below_**, change the `file_name` parameter to match the location of the Unity environment that you downloaded.
- **Mac**: `"path/to/Crawler.app"`
- **Windows** (x86): `"path/to/Crawler_Windows_x86/Crawler.exe"`
- **Windows** (x86_64): `"path/to/Crawler_Windows_x86_64/Crawler.exe"`
- **Linux** (x86): `"path/to/Crawler_Linux/Crawler.x86"`
- **Linux** (x86_64): `"path/to/Crawler_Linux/Crawler.x86_64"`
- **Linux** (x86, headless): `"path/to/Crawler_Linux_NoVis/Crawler.x86"`
- **Linux** (x86_64, headless): `"path/to/Crawler_Linux_NoVis/Crawler.x86_64"`
For instance, if you are using a Mac, then you downloaded `Crawler.app`. If this file is in the same folder as the notebook, then the line below should appear as follows:
```
env = UnityEnvironment(file_name="Crawler.app")
```
```
env = UnityEnvironment(file_name='../../crawler/Crawler.app')
```
Environments contain **_brains_** which are responsible for deciding the actions of their associated agents. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.
```
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
```
### 2. Examine the State and Action Spaces
Run the code cell below to print some information about the environment.
```
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents
num_agents = len(env_info.agents)
print('Number of agents:', num_agents)
# size of each action
action_size = brain.vector_action_space_size
print('Size of each action:', action_size)
# examine the state space
states = env_info.vector_observations
state_size = states.shape[1]
print('There are {} agents. Each observes a state with length: {}'.format(states.shape[0], state_size))
print('The state for the first agent looks like:', states[0])
```
### 3. Take Random Actions in the Environment
In the next code cell, you will learn how to use the Python API to control the agent and receive feedback from the environment.
Once this cell is executed, you will watch the agent's performance, if it selects an action at random with each time step. A window should pop up that allows you to observe the agent, as it moves through the environment.
Of course, as part of the project, you'll have to change the code so that the agent is able to use its experience to gradually choose better actions when interacting with the environment!
```
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state (for each agent)
scores = np.zeros(num_agents) # initialize the score (for each agent)
while True:
actions = np.random.randn(num_agents, action_size) # select an action (for each agent)
actions = np.clip(actions, -1, 1) # all actions between -1 and 1
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state (for each agent)
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
break
print('Total score (averaged over agents) this episode: {}'.format(np.mean(scores)))
```
When finished, you can close the environment.
```
env.close()
```
### 4. It's Your Turn!
Now it's your turn to train your own agent to solve the environment! When training the environment, set `train_mode=True`, so that the line for resetting the environment looks like the following:
```python
env_info = env.reset(train_mode=True)[brain_name]
```
|
github_jupyter
|
from unityagents import UnityEnvironment
import numpy as np
env = UnityEnvironment(file_name="Crawler.app")
env = UnityEnvironment(file_name='../../crawler/Crawler.app')
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents
num_agents = len(env_info.agents)
print('Number of agents:', num_agents)
# size of each action
action_size = brain.vector_action_space_size
print('Size of each action:', action_size)
# examine the state space
states = env_info.vector_observations
state_size = states.shape[1]
print('There are {} agents. Each observes a state with length: {}'.format(states.shape[0], state_size))
print('The state for the first agent looks like:', states[0])
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state (for each agent)
scores = np.zeros(num_agents) # initialize the score (for each agent)
while True:
actions = np.random.randn(num_agents, action_size) # select an action (for each agent)
actions = np.clip(actions, -1, 1) # all actions between -1 and 1
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state (for each agent)
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
break
print('Total score (averaged over agents) this episode: {}'.format(np.mean(scores)))
env.close()
env_info = env.reset(train_mode=True)[brain_name]
| 0.331444 | 0.980876 |
# Hacking Consulting Project
<div style="text-align: right"><h4>- Based on K-Means Clustering using PySpark MLLib Library</h4></div>
A large technology firm needs your help, they've been hacked! Luckily their forensic engineers have grabbed valuable data about the hacks, including information like session time,locations, wpm typing speed, etc. The forensic engineer relates to you what she has been able to figure out so far, she has been able to grab meta data of each session that the hackers used to connect to their servers. These are the features of the data:
- 'Session_Connection_Time': How long the session lasted in minutes
- 'Bytes Transferred': Number of MB transferred during session
- 'Kali_Trace_Used': Indicates if the hacker was using Kali Linux
- 'Servers_Corrupted': Number of server corrupted during the attack
- 'Pages_Corrupted': Number of pages illegally accessed
- 'Location': Location attack came from (Probably useless because the hackers used VPNs)
- 'WPM_Typing_Speed': Their estimated typing speed based on session logs.
The technology firm has 3 potential hackers that perpetrated the attack. Their certain of the first two hackers but they aren't very sure if the third hacker was involved or not. They have requested your help! Can you help figure out whether or not the third suspect had anything to do with the attacks, or was it just two hackers? It's probably not possible to know for sure, but maybe what you've just learned about Clustering can help!
One last key fact, the forensic engineer knows that the hackers trade off attacks. Meaning they should each have roughly the same amount of attacks. For example if there were 100 total attacks, then in a 2 hacker situation each should have about 50 hacks, in a three hacker situation each would have about 33 hacks. The engineer believes this is the key element to solving this, but doesn't know how to distinguish this unlabeled data into groups of hackers.
#### Import necessary libraries and datasets
```
import findspark
findspark.init('E:\DATA\Apps\hadoop-env\spark-2.4.7-bin-hadoop2.7')
from pyspark.sql import SparkSession
spark = SparkSession.builder.appName('hack_find').getOrCreate()
dataset = spark.read.csv("hack_data.csv",header=True,inferSchema=True)
```
#### Explore the dataset
```
dataset.columns
dataset.head()
dataset.describe().show()
```
#### Transform the data
```
from pyspark.ml.clustering import KMeans
from pyspark.ml.linalg import Vectors
from pyspark.ml.feature import VectorAssembler
feat_cols = ['Session_Connection_Time', 'Bytes Transferred', 'Kali_Trace_Used',
'Servers_Corrupted', 'Pages_Corrupted','WPM_Typing_Speed']
vec_assembler = VectorAssembler(inputCols = feat_cols, outputCol='features')
final_data = vec_assembler.transform(dataset)
final_data.head()
from pyspark.ml.feature import StandardScaler
scaler = StandardScaler(inputCol="features", outputCol="scaledFeatures", withStd=True, withMean=False)
scalerModel = scaler.fit(final_data)
cluster_final_data = scalerModel.transform(final_data)
cluster_final_data.head()
```
#### Start building the model
```
kmeans3 = KMeans(featuresCol='scaledFeatures',k=3)
kmeans2 = KMeans(featuresCol='scaledFeatures',k=2)
model_k3 = kmeans3.fit(cluster_final_data)
model_k2 = kmeans2.fit(cluster_final_data)
model_k3.transform(cluster_final_data).groupBy('prediction').count().show()
model_k2.transform(cluster_final_data).groupBy('prediction').count().show()
```
This confirms that there are two hackers.
```
wssse_k3 = model_k3.computeCost(cluster_final_data)
wssse_k2 = model_k2.computeCost(cluster_final_data)
print("With K=3")
print("Within Set Sum of Squared Errors = " + str(wssse_k3))
print('--'*30)
print("With K=2")
print("Within Set Sum of Squared Errors = " + str(wssse_k2))
```
#### Check for different values of k
```
for k in range(2,9):
kmeans = KMeans(featuresCol='scaledFeatures', k=k)
model = kmeans.fit(cluster_final_data)
wssse = model.computeCost(cluster_final_data)
print("With K={}".format(k))
print("Within Set Sum of Squared Errors = " + str(wssse))
model.transform(cluster_final_data).groupBy('prediction').count().show()
print('--'*30)
```
Hence, as mentioned by the engineers, the attacks were evenly distributed between 2 hackers.
|
github_jupyter
|
import findspark
findspark.init('E:\DATA\Apps\hadoop-env\spark-2.4.7-bin-hadoop2.7')
from pyspark.sql import SparkSession
spark = SparkSession.builder.appName('hack_find').getOrCreate()
dataset = spark.read.csv("hack_data.csv",header=True,inferSchema=True)
dataset.columns
dataset.head()
dataset.describe().show()
from pyspark.ml.clustering import KMeans
from pyspark.ml.linalg import Vectors
from pyspark.ml.feature import VectorAssembler
feat_cols = ['Session_Connection_Time', 'Bytes Transferred', 'Kali_Trace_Used',
'Servers_Corrupted', 'Pages_Corrupted','WPM_Typing_Speed']
vec_assembler = VectorAssembler(inputCols = feat_cols, outputCol='features')
final_data = vec_assembler.transform(dataset)
final_data.head()
from pyspark.ml.feature import StandardScaler
scaler = StandardScaler(inputCol="features", outputCol="scaledFeatures", withStd=True, withMean=False)
scalerModel = scaler.fit(final_data)
cluster_final_data = scalerModel.transform(final_data)
cluster_final_data.head()
kmeans3 = KMeans(featuresCol='scaledFeatures',k=3)
kmeans2 = KMeans(featuresCol='scaledFeatures',k=2)
model_k3 = kmeans3.fit(cluster_final_data)
model_k2 = kmeans2.fit(cluster_final_data)
model_k3.transform(cluster_final_data).groupBy('prediction').count().show()
model_k2.transform(cluster_final_data).groupBy('prediction').count().show()
wssse_k3 = model_k3.computeCost(cluster_final_data)
wssse_k2 = model_k2.computeCost(cluster_final_data)
print("With K=3")
print("Within Set Sum of Squared Errors = " + str(wssse_k3))
print('--'*30)
print("With K=2")
print("Within Set Sum of Squared Errors = " + str(wssse_k2))
for k in range(2,9):
kmeans = KMeans(featuresCol='scaledFeatures', k=k)
model = kmeans.fit(cluster_final_data)
wssse = model.computeCost(cluster_final_data)
print("With K={}".format(k))
print("Within Set Sum of Squared Errors = " + str(wssse))
model.transform(cluster_final_data).groupBy('prediction').count().show()
print('--'*30)
| 0.607896 | 0.926968 |
**Importing the libraries:**
```
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from keras.models import Sequential
from keras.layers import Dense, Dropout
```
**Uploading the dataset**
```
from google.colab import files
uploaded = files.upload()
```
**Loading the dataset and viewing the first few rows:**
```
# Dataset can be downloaded at https://archive.ics.uci.edu/ml/machine-learning-databases/00275/
data = pd.read_csv('hour.csv')
data.head()
```
**Dimensions of the dataset:**
```
data.shape
```
**Extracting the features:**
```
# Feature engineering
ohe_features = ['season', 'weathersit', 'mnth', 'hr', 'weekday']
for feature in ohe_features:
dummies = pd.get_dummies(data[feature],
prefix = feature,
drop_first = False)
data = pd.concat([data, dummies], axis = 1)
drop_features = ['instant', 'dteday', 'season', 'weathersit',
'weekday', 'atemp', 'mnth', 'workingday',
'hr', 'casual', 'registered']
data = data.drop(drop_features, axis = 1)
```
**Normalizing the features:**
```
norm_features = ['cnt', 'temp', 'hum', 'windspeed']
scaled_features = {}
for feature in norm_features:
mean, std = data[feature].mean(), data[feature].std()
scaled_features[feature] = [mean, std]
data.loc[:, feature] = (data[feature] - mean) / std
```
**Splitting the dataset for training, validation and testing:**
```
# Save the final month for testing
test_data = data[-31 * 24:]
data = data[:-31 * 24]
# Extract the target field
target_fields = ['cnt']
features, targets = data.drop(target_fields, axis = 1), data[target_fields]
test_features, test_targets = test_data.drop(target_fields, axis = 1), test_data[target_fields]
# Create a validation set (based on the last )
X_train, y_train = features[: -30 * 24], targets[: -30 * 24]
X_val, y_val = features[-30 * 24: ], targets[-30 * 24:]
```
**Viewing the first few rows of the modified dataset:**
```
data.head()
```
**Defining the model:**
```
model = Sequential()
model.add(Dense(250, input_dim = X_train.shape[1],
activation = 'relu'))
model.add(Dense(150, activation = 'relu'))
model.add(Dense(50, activation = 'relu'))
model.add(Dense(25, activation = 'relu'))
model.add(Dense(1, activation = 'linear'))
# Compile model
model.compile(loss = 'mse',
optimizer = 'sgd',
metrics = ['mse'])
```
**Setting the hyperparameters and training the model:**
```
n_epochs = 1000
batch_size = 1024
history = model.fit(X_train.values, y_train['cnt'],
validation_data = (X_val.values, y_val['cnt']),
batch_size = batch_size,
epochs = n_epochs,
verbose = 0)
```
**Plotting the training and validation losses:**
```
plt.plot(np.arange(len(history.history['loss'])),
history.history['loss'],
label = 'training')
plt.plot(np.arange(len(history.history['val_loss'])),
history.history['val_loss'],
label = 'validation')
plt.title('Overfit on Bike Sharing dataset')
plt.xlabel('epochs')
plt.ylabel('loss')
plt.legend(loc = 0)
plt.show()
# Model overfits on the training data
```
**Printing the minimum loss:**
```
print('Minimum loss: ', min(history.history['val_loss']),
'\nAfter ', np.argmin(history.history['val_loss']), ' epochs')
```
**Adding dropouts to the network architecture to prevent overfitting:**
```
model_drop = Sequential()
model_drop.add(Dense(250, input_dim = X_train.shape[1],
activation = 'relu'))
model_drop.add(Dropout(0.20))
model_drop.add(Dense(150, activation = 'relu'))
model_drop.add(Dropout(0.20))
model_drop.add(Dense(50, activation = 'relu'))
model_drop.add(Dropout(0.20))
model_drop.add(Dense(25, activation = 'relu'))
model_drop.add(Dropout(0.20))
model_drop.add(Dense(1, activation = 'linear'))
# Compile model
model_drop.compile(loss = 'mse',
optimizer = 'sgd',
metrics = ['mse'])
```
**Training the new model:**
```
history_drop = model_drop.fit(X_train.values, y_train['cnt'],
validation_data = (X_val.values, y_val['cnt']),
batch_size = batch_size,
epochs = n_epochs,
verbose = 0)
```
**Plotting the results:**
```
plt.plot(np.arange(len(history_drop.history['loss'])),
history_drop.history['loss'],
label = 'training')
plt.plot(np.arange(len(history_drop.history['val_loss'])),
history_drop.history['val_loss'],
label = 'validation')
plt.title('Using dropout for Bike Sharing dataset')
plt.xlabel('epochs')
plt.ylabel('loss')
plt.legend(loc = 0)
plt.show()
```
**Printing the statistics:**
```
print('Minimum loss:', min(history_drop.history['val_loss']),
'\nAfter ', np.argmin(history_drop.history['val_loss']), ' epochs')
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from keras.models import Sequential
from keras.layers import Dense, Dropout
from google.colab import files
uploaded = files.upload()
# Dataset can be downloaded at https://archive.ics.uci.edu/ml/machine-learning-databases/00275/
data = pd.read_csv('hour.csv')
data.head()
data.shape
# Feature engineering
ohe_features = ['season', 'weathersit', 'mnth', 'hr', 'weekday']
for feature in ohe_features:
dummies = pd.get_dummies(data[feature],
prefix = feature,
drop_first = False)
data = pd.concat([data, dummies], axis = 1)
drop_features = ['instant', 'dteday', 'season', 'weathersit',
'weekday', 'atemp', 'mnth', 'workingday',
'hr', 'casual', 'registered']
data = data.drop(drop_features, axis = 1)
norm_features = ['cnt', 'temp', 'hum', 'windspeed']
scaled_features = {}
for feature in norm_features:
mean, std = data[feature].mean(), data[feature].std()
scaled_features[feature] = [mean, std]
data.loc[:, feature] = (data[feature] - mean) / std
# Save the final month for testing
test_data = data[-31 * 24:]
data = data[:-31 * 24]
# Extract the target field
target_fields = ['cnt']
features, targets = data.drop(target_fields, axis = 1), data[target_fields]
test_features, test_targets = test_data.drop(target_fields, axis = 1), test_data[target_fields]
# Create a validation set (based on the last )
X_train, y_train = features[: -30 * 24], targets[: -30 * 24]
X_val, y_val = features[-30 * 24: ], targets[-30 * 24:]
data.head()
model = Sequential()
model.add(Dense(250, input_dim = X_train.shape[1],
activation = 'relu'))
model.add(Dense(150, activation = 'relu'))
model.add(Dense(50, activation = 'relu'))
model.add(Dense(25, activation = 'relu'))
model.add(Dense(1, activation = 'linear'))
# Compile model
model.compile(loss = 'mse',
optimizer = 'sgd',
metrics = ['mse'])
n_epochs = 1000
batch_size = 1024
history = model.fit(X_train.values, y_train['cnt'],
validation_data = (X_val.values, y_val['cnt']),
batch_size = batch_size,
epochs = n_epochs,
verbose = 0)
plt.plot(np.arange(len(history.history['loss'])),
history.history['loss'],
label = 'training')
plt.plot(np.arange(len(history.history['val_loss'])),
history.history['val_loss'],
label = 'validation')
plt.title('Overfit on Bike Sharing dataset')
plt.xlabel('epochs')
plt.ylabel('loss')
plt.legend(loc = 0)
plt.show()
# Model overfits on the training data
print('Minimum loss: ', min(history.history['val_loss']),
'\nAfter ', np.argmin(history.history['val_loss']), ' epochs')
model_drop = Sequential()
model_drop.add(Dense(250, input_dim = X_train.shape[1],
activation = 'relu'))
model_drop.add(Dropout(0.20))
model_drop.add(Dense(150, activation = 'relu'))
model_drop.add(Dropout(0.20))
model_drop.add(Dense(50, activation = 'relu'))
model_drop.add(Dropout(0.20))
model_drop.add(Dense(25, activation = 'relu'))
model_drop.add(Dropout(0.20))
model_drop.add(Dense(1, activation = 'linear'))
# Compile model
model_drop.compile(loss = 'mse',
optimizer = 'sgd',
metrics = ['mse'])
history_drop = model_drop.fit(X_train.values, y_train['cnt'],
validation_data = (X_val.values, y_val['cnt']),
batch_size = batch_size,
epochs = n_epochs,
verbose = 0)
plt.plot(np.arange(len(history_drop.history['loss'])),
history_drop.history['loss'],
label = 'training')
plt.plot(np.arange(len(history_drop.history['val_loss'])),
history_drop.history['val_loss'],
label = 'validation')
plt.title('Using dropout for Bike Sharing dataset')
plt.xlabel('epochs')
plt.ylabel('loss')
plt.legend(loc = 0)
plt.show()
print('Minimum loss:', min(history_drop.history['val_loss']),
'\nAfter ', np.argmin(history_drop.history['val_loss']), ' epochs')
| 0.635562 | 0.956877 |
# Creating your own dataset from Google Images
*by: Francisco Ingham and Jeremy Howard. Inspired by [Adrian Rosebrock](https://www.pyimagesearch.com/2017/12/04/how-to-create-a-deep-learning-dataset-using-google-images/)*
In this tutorial we will see how to easily create an image dataset through Google Images. **Note**: You will have to repeat these steps for any new category you want to Google (e.g once for dogs and once for cats).
```
from fastai.vision import *
```
## Get a list of URLs
### Search and scroll
Go to [Google Images](http://images.google.com) and search for the images you are interested in. The more specific you are in your Google Search, the better the results and the less manual pruning you will have to do.
Scroll down until you've seen all the images you want to download, or until you see a button that says 'Show more results'. All the images you scrolled past are now available to download. To get more, click on the button, and continue scrolling. The maximum number of images Google Images shows is 700.
It is a good idea to put things you want to exclude into the search query, for instance if you are searching for the Eurasian wolf, "canis lupus lupus", it might be a good idea to exclude other variants:
"canis lupus lupus" -dog -arctos -familiaris -baileyi -occidentalis
You can also limit your results to show only photos by clicking on Tools and selecting Photos from the Type dropdown.
### Download into file
Now you must run some Javascript code in your browser which will save the URLs of all the images you want for you dataset.
In Google Chrome press <kbd>Ctrl</kbd><kbd>Shift</kbd><kbd>j</kbd> on Windows/Linux and <kbd>Cmd</kbd><kbd>Opt</kbd><kbd>j</kbd> on macOS, and a small window the javascript 'Console' will appear. In Firefox press <kbd>Ctrl</kbd><kbd>Shift</kbd><kbd>k</kbd> on Windows/Linux or <kbd>Cmd</kbd><kbd>Opt</kbd><kbd>k</kbd> on macOS. That is where you will paste the JavaScript commands.
You will need to get the urls of each of the images. Before running the following commands, you may want to disable ad blocking extensions (uBlock, AdBlockPlus etc.) in Chrome. Otherwise the window.open() command doesn't work. Then you can run the following commands:
```javascript
urls=Array.from(document.querySelectorAll('.rg_i')).map(el=> el.hasAttribute('data-src')?el.getAttribute('data-src'):el.getAttribute('data-iurl'));
window.open('data:text/csv;charset=utf-8,' + escape(urls.join('\n')));
```
### Create directory and upload urls file into your server
Choose an appropriate name for your labeled images. You can run these steps multiple times to create different labels.
```
folder = 'black'
file = 'urls_black.csv'
folder = 'teddys'
file = 'urls_teddys.csv'
folder = 'grizzly'
file = 'urls_grizzly.csv'
```
You will need to run this cell once per each category.
```
path = Path('data/bears')
dest = path/folder
dest.mkdir(parents=True, exist_ok=True)
path.ls()
```
Finally, upload your urls file. You just need to press 'Upload' in your working directory and select your file, then click 'Upload' for each of the displayed files.

## Download images
Now you will need to download your images from their respective urls.
fast.ai has a function that allows you to do just that. You just have to specify the urls filename as well as the destination folder and this function will download and save all images that can be opened. If they have some problem in being opened, they will not be saved.
Let's download our images! Notice you can choose a maximum number of images to be downloaded. In this case we will not download all the urls.
You will need to run this line once for every category.
```
classes = ['teddys','grizzly','black']
download_images(path/file, dest, max_pics=200)
# If you have problems download, try with `max_workers=0` to see exceptions:
download_images(path/file, dest, max_pics=20, max_workers=0)
```
Then we can remove any images that can't be opened:
```
for c in classes:
print(c)
verify_images(path/c, delete=True, max_size=500)
```
## View data
```
np.random.seed(42)
data = ImageDataBunch.from_folder(path, train=".", valid_pct=0.2,
ds_tfms=get_transforms(), size=224, num_workers=4).normalize(imagenet_stats)
# If you already cleaned your data, run this cell instead of the one before
# np.random.seed(42)
# data = ImageDataBunch.from_csv(path, folder=".", valid_pct=0.2, csv_labels='cleaned.csv',
# ds_tfms=get_transforms(), size=224, num_workers=4).normalize(imagenet_stats)
```
Good! Let's take a look at some of our pictures then.
```
data.classes
data.show_batch(rows=3, figsize=(7,8))
data.classes, data.c, len(data.train_ds), len(data.valid_ds)
```
## Train model
```
learn = cnn_learner(data, models.resnet34, metrics=error_rate)
learn.fit_one_cycle(4)
learn.save('stage-1')
learn.unfreeze()
learn.lr_find()
# If the plot is not showing try to give a start and end learning rate
# learn.lr_find(start_lr=1e-5, end_lr=1e-1)
learn.recorder.plot()
learn.fit_one_cycle(2, max_lr=slice(3e-5,3e-4))
learn.save('stage-2')
```
## Interpretation
```
learn.load('stage-2');
interp = ClassificationInterpretation.from_learner(learn)
interp.plot_confusion_matrix()
```
## Cleaning Up
Some of our top losses aren't due to bad performance by our model. There are images in our data set that shouldn't be.
Using the `ImageCleaner` widget from `fastai.widgets` we can prune our top losses, removing photos that don't belong.
```
from fastai.widgets import *
```
First we need to get the file paths from our top_losses. We can do this with `.from_toplosses`. We then feed the top losses indexes and corresponding dataset to `ImageCleaner`.
Notice that the widget will not delete images directly from disk but it will create a new csv file `cleaned.csv` from where you can create a new ImageDataBunch with the corrected labels to continue training your model.
In order to clean the entire set of images, we need to create a new dataset without the split. The video lecture demostrated the use of the `ds_type` param which no longer has any effect. See [the thread](https://forums.fast.ai/t/duplicate-widget/30975/10) for more details.
```
db = (ImageList.from_folder(path)
.split_none()
.label_from_folder()
.transform(get_transforms(), size=224)
.databunch()
)
# If you already cleaned your data using indexes from `from_toplosses`,
# run this cell instead of the one before to proceed with removing duplicates.
# Otherwise all the results of the previous step would be overwritten by
# the new run of `ImageCleaner`.
# db = (ImageList.from_csv(path, 'cleaned.csv', folder='.')
# .split_none()
# .label_from_df()
# .transform(get_transforms(), size=224)
# .databunch()
# )
```
Then we create a new learner to use our new databunch with all the images.
```
learn_cln = cnn_learner(db, models.resnet34, metrics=error_rate)
learn_cln.load('stage-2');
ds, idxs = DatasetFormatter().from_toplosses(learn_cln)
```
Make sure you're running this notebook in Jupyter Notebook, not Jupyter Lab. That is accessible via [/tree](/tree), not [/lab](/lab). Running the `ImageCleaner` widget in Jupyter Lab is [not currently supported](https://github.com/fastai/fastai/issues/1539).
```
# Don't run this in google colab or any other instances running jupyter lab.
# If you do run this on Jupyter Lab, you need to restart your runtime and
# runtime state including all local variables will be lost.
ImageCleaner(ds, idxs, path)
```
If the code above does not show any GUI(contains images and buttons) rendered by widgets but only text output, that may caused by the configuration problem of ipywidgets. Try the solution in this [link](https://github.com/fastai/fastai/issues/1539#issuecomment-505999861) to solve it.
Flag photos for deletion by clicking 'Delete'. Then click 'Next Batch' to delete flagged photos and keep the rest in that row. `ImageCleaner` will show you a new row of images until there are no more to show. In this case, the widget will show you images until there are none left from `top_losses.ImageCleaner(ds, idxs)`
You can also find duplicates in your dataset and delete them! To do this, you need to run `.from_similars` to get the potential duplicates' ids and then run `ImageCleaner` with `duplicates=True`. The API works in a similar way as with misclassified images: just choose the ones you want to delete and click 'Next Batch' until there are no more images left.
Make sure to recreate the databunch and `learn_cln` from the `cleaned.csv` file. Otherwise the file would be overwritten from scratch, losing all the results from cleaning the data from toplosses.
```
ds, idxs = DatasetFormatter().from_similars(learn_cln)
ImageCleaner(ds, idxs, path, duplicates=True)
```
Remember to recreate your ImageDataBunch from your `cleaned.csv` to include the changes you made in your data!
## Putting your model in production
First thing first, let's export the content of our `Learner` object for production:
```
learn.export()
```
This will create a file named 'export.pkl' in the directory where we were working that contains everything we need to deploy our model (the model, the weights but also some metadata like the classes or the transforms/normalization used).
You probably want to use CPU for inference, except at massive scale (and you almost certainly don't need to train in real-time). If you don't have a GPU that happens automatically. You can test your model on CPU like so:
```
defaults.device = torch.device('cpu')
img = open_image(path/'black'/'00000021.jpg')
img
```
We create our `Learner` in production enviromnent like this, just make sure that `path` contains the file 'export.pkl' from before.
```
learn = load_learner(path)
pred_class,pred_idx,outputs = learn.predict(img)
pred_class.obj
```
So you might create a route something like this ([thanks](https://github.com/simonw/cougar-or-not) to Simon Willison for the structure of this code):
```python
@app.route("/classify-url", methods=["GET"])
async def classify_url(request):
bytes = await get_bytes(request.query_params["url"])
img = open_image(BytesIO(bytes))
_,_,losses = learner.predict(img)
return JSONResponse({
"predictions": sorted(
zip(cat_learner.data.classes, map(float, losses)),
key=lambda p: p[1],
reverse=True
)
})
```
(This example is for the [Starlette](https://www.starlette.io/) web app toolkit.)
## Things that can go wrong
- Most of the time things will train fine with the defaults
- There's not much you really need to tune (despite what you've heard!)
- Most likely are
- Learning rate
- Number of epochs
### Learning rate (LR) too high
```
learn = cnn_learner(data, models.resnet34, metrics=error_rate)
learn.fit_one_cycle(1, max_lr=0.5)
```
### Learning rate (LR) too low
```
learn = cnn_learner(data, models.resnet34, metrics=error_rate)
```
Previously we had this result:
```
Total time: 00:57
epoch train_loss valid_loss error_rate
1 1.030236 0.179226 0.028369 (00:14)
2 0.561508 0.055464 0.014184 (00:13)
3 0.396103 0.053801 0.014184 (00:13)
4 0.316883 0.050197 0.021277 (00:15)
```
```
learn.fit_one_cycle(5, max_lr=1e-5)
learn.recorder.plot_losses()
```
As well as taking a really long time, it's getting too many looks at each image, so may overfit.
### Too few epochs
```
learn = cnn_learner(data, models.resnet34, metrics=error_rate, pretrained=False)
learn.fit_one_cycle(1)
```
### Too many epochs
```
np.random.seed(42)
data = ImageDataBunch.from_folder(path, train=".", valid_pct=0.9, bs=32,
ds_tfms=get_transforms(do_flip=False, max_rotate=0, max_zoom=1, max_lighting=0, max_warp=0
),size=224, num_workers=4).normalize(imagenet_stats)
learn = cnn_learner(data, models.resnet50, metrics=error_rate, ps=0, wd=0)
learn.unfreeze()
learn.fit_one_cycle(40, slice(1e-6,1e-4))
```
|
github_jupyter
|
from fastai.vision import *
urls=Array.from(document.querySelectorAll('.rg_i')).map(el=> el.hasAttribute('data-src')?el.getAttribute('data-src'):el.getAttribute('data-iurl'));
window.open('data:text/csv;charset=utf-8,' + escape(urls.join('\n')));
folder = 'black'
file = 'urls_black.csv'
folder = 'teddys'
file = 'urls_teddys.csv'
folder = 'grizzly'
file = 'urls_grizzly.csv'
path = Path('data/bears')
dest = path/folder
dest.mkdir(parents=True, exist_ok=True)
path.ls()
classes = ['teddys','grizzly','black']
download_images(path/file, dest, max_pics=200)
# If you have problems download, try with `max_workers=0` to see exceptions:
download_images(path/file, dest, max_pics=20, max_workers=0)
for c in classes:
print(c)
verify_images(path/c, delete=True, max_size=500)
np.random.seed(42)
data = ImageDataBunch.from_folder(path, train=".", valid_pct=0.2,
ds_tfms=get_transforms(), size=224, num_workers=4).normalize(imagenet_stats)
# If you already cleaned your data, run this cell instead of the one before
# np.random.seed(42)
# data = ImageDataBunch.from_csv(path, folder=".", valid_pct=0.2, csv_labels='cleaned.csv',
# ds_tfms=get_transforms(), size=224, num_workers=4).normalize(imagenet_stats)
data.classes
data.show_batch(rows=3, figsize=(7,8))
data.classes, data.c, len(data.train_ds), len(data.valid_ds)
learn = cnn_learner(data, models.resnet34, metrics=error_rate)
learn.fit_one_cycle(4)
learn.save('stage-1')
learn.unfreeze()
learn.lr_find()
# If the plot is not showing try to give a start and end learning rate
# learn.lr_find(start_lr=1e-5, end_lr=1e-1)
learn.recorder.plot()
learn.fit_one_cycle(2, max_lr=slice(3e-5,3e-4))
learn.save('stage-2')
learn.load('stage-2');
interp = ClassificationInterpretation.from_learner(learn)
interp.plot_confusion_matrix()
from fastai.widgets import *
db = (ImageList.from_folder(path)
.split_none()
.label_from_folder()
.transform(get_transforms(), size=224)
.databunch()
)
# If you already cleaned your data using indexes from `from_toplosses`,
# run this cell instead of the one before to proceed with removing duplicates.
# Otherwise all the results of the previous step would be overwritten by
# the new run of `ImageCleaner`.
# db = (ImageList.from_csv(path, 'cleaned.csv', folder='.')
# .split_none()
# .label_from_df()
# .transform(get_transforms(), size=224)
# .databunch()
# )
learn_cln = cnn_learner(db, models.resnet34, metrics=error_rate)
learn_cln.load('stage-2');
ds, idxs = DatasetFormatter().from_toplosses(learn_cln)
# Don't run this in google colab or any other instances running jupyter lab.
# If you do run this on Jupyter Lab, you need to restart your runtime and
# runtime state including all local variables will be lost.
ImageCleaner(ds, idxs, path)
ds, idxs = DatasetFormatter().from_similars(learn_cln)
ImageCleaner(ds, idxs, path, duplicates=True)
learn.export()
defaults.device = torch.device('cpu')
img = open_image(path/'black'/'00000021.jpg')
img
learn = load_learner(path)
pred_class,pred_idx,outputs = learn.predict(img)
pred_class.obj
@app.route("/classify-url", methods=["GET"])
async def classify_url(request):
bytes = await get_bytes(request.query_params["url"])
img = open_image(BytesIO(bytes))
_,_,losses = learner.predict(img)
return JSONResponse({
"predictions": sorted(
zip(cat_learner.data.classes, map(float, losses)),
key=lambda p: p[1],
reverse=True
)
})
learn = cnn_learner(data, models.resnet34, metrics=error_rate)
learn.fit_one_cycle(1, max_lr=0.5)
learn = cnn_learner(data, models.resnet34, metrics=error_rate)
Total time: 00:57
epoch train_loss valid_loss error_rate
1 1.030236 0.179226 0.028369 (00:14)
2 0.561508 0.055464 0.014184 (00:13)
3 0.396103 0.053801 0.014184 (00:13)
4 0.316883 0.050197 0.021277 (00:15)
learn.fit_one_cycle(5, max_lr=1e-5)
learn.recorder.plot_losses()
learn = cnn_learner(data, models.resnet34, metrics=error_rate, pretrained=False)
learn.fit_one_cycle(1)
np.random.seed(42)
data = ImageDataBunch.from_folder(path, train=".", valid_pct=0.9, bs=32,
ds_tfms=get_transforms(do_flip=False, max_rotate=0, max_zoom=1, max_lighting=0, max_warp=0
),size=224, num_workers=4).normalize(imagenet_stats)
learn = cnn_learner(data, models.resnet50, metrics=error_rate, ps=0, wd=0)
learn.unfreeze()
learn.fit_one_cycle(40, slice(1e-6,1e-4))
| 0.651798 | 0.921358 |
# CLX Cheat Sheets sample code
(c) 2020 NVIDIA, Blazing SQL
Distributed under Apache License 2.0
```
import cudf
import dask_cudf
import s3fs
from os import path
from clx.analytics.cybert import Cybert
```
---
# CyBERT
---
## Model
```
CLX_S3_BASE_PATH = 'rapidsai-data/cyber/clx'
HF_S3_BASE_PATH = 'models.huggingface.co/bert/raykallen/cybert_apache_parser'
MODEL_DIR = '../models/CyBERT'
DATA_DIR = '../data'
CONFIG_FILENAME = 'config.json'
MODEL_FILENAME = 'pytorch_model.bin'
APACHE_SAMPLE_CSV = 'apache_sample_1k.csv'
if not path.exists(f'{MODEL_DIR}/{MODEL_FILENAME}'):
fs = s3fs.S3FileSystem(anon=True)
fs.get(
f'{HF_S3_BASE_PATH}/{MODEL_FILENAME}'
, f'{MODEL_DIR}/{MODEL_FILENAME}'
)
if not path.exists(f'{MODEL_DIR}/{CONFIG_FILENAME}'):
fs = s3fs.S3FileSystem(anon=True)
fs.get(
f'{HF_S3_BASE_PATH}/{CONFIG_FILENAME}'
, f'{MODEL_DIR}/{CONFIG_FILENAME}'
)
if not path.exists(APACHE_SAMPLE_CSV):
fs = s3fs.S3FileSystem(anon=True)
fs.get(
f'{CLX_S3_BASE_PATH}/{APACHE_SAMPLE_CSV}'
, f'{DATA_DIR}/{APACHE_SAMPLE_CSV}')
```
#### clx.analytics.cybert.Cybert.load_model()
```
cybert = Cybert()
cybert.load_model(
f'{MODEL_DIR}/{MODEL_FILENAME}'
, f'{MODEL_DIR}/{CONFIG_FILENAME}'
)
```
#### clx.analytics.cybert.Cybert.inference()
```
logs_df = cudf.read_csv(f'{DATA_DIR}/{APACHE_SAMPLE_CSV}')
parsed_df, confidence_df = cybert.inference(logs_df["raw"])
parsed_df.head()
confidence_df.head()
```
#### clx.analytics.cybert.Cybert.preprocess()
```
logs_df = cudf.read_csv(f'{DATA_DIR}/{APACHE_SAMPLE_CSV}')
input_ids, attention_masks, meta = cybert.preprocess(logs_df["raw"])
input_ids
attention_masks
meta
```
# DGA Detector
## Model
```
import os
import wget
import time
import cudf
import torch
import shutil
import zipfile
import numpy as np
from datetime import datetime
from sklearn.metrics import accuracy_score, average_precision_score
from clx.analytics.dga_dataset import DGADataset
from clx.analytics.dga_detector import DGADetector
from cuml.preprocessing.model_selection import train_test_split
from clx.utils.data.dataloader import DataLoader
dga = {
"source": "DGA",
"url": "https://data.netlab.360.com/feeds/dga/dga.txt",
"compression": None,
"storage_path": "../data/dga_feed",
}
benign = {
"source": "Benign",
"url": "http://s3.amazonaws.com/alexa-static/top-1m.csv.zip",
"compression": "zip",
"storage_path": "../data/top-1m",
}
def unpack(compression_type, filepath, output_dir):
if compression_type == 'zip':
with zipfile.ZipFile(filepath, 'r') as f:
f.extractall(output_dir)
os.remove(filepath)
def download_file(f):
output_dir = f['storage_path']
filepath = f'{output_dir}/{f["url"].split("/")[-1]}'
if not os.path.exists(filepath):
if not os.path.exists(output_dir):
os.makedirs(output_dir)
print(f'Downloading {f["url"]}...')
filepath = wget.download(f['url'], out=output_dir)
print(f'Unpacking {filepath}')
unpack(f['compression'], filepath, output_dir)
print(f'{f["source"]} data is stored to location {output_dir}')
download_file(dga)
download_file(benign)
def load_input_data(dga, benign):
dga_df = cudf.read_csv(
dga['storage_path'] + '/*'
, names=['generator', 'domain', 'dt_from', 'dt_to']
, usecols=['domain']
, skiprows=18
, delimiter='\t'
)
dga_df['type'] = 0
benign_df = cudf.read_csv(
benign['storage_path'] + '/*'
, names=["line_num","domain"]
, usecols=['domain']
)
benign_df['type'] = 1
input_df = cudf.concat([benign_df, dga_df], ignore_index=True)
return input_df
def create_df(domain_df, type_series):
df = cudf.DataFrame()
df['domain'] = domain_df['domain'].reset_index(drop=True)
df['type'] = type_series.reset_index(drop=True)
return df
def create_dir(dir_path):
print("Verify if directory `%s` already exists." % (dir_path))
if not os.path.exists(dir_path):
print("Directory `%s` does not exist." % (dir_path))
print("Creating directory `%s` to store trained models." % (dir_path))
os.makedirs(dir_path)
def cleanup_cache():
# release memory.
torch.cuda.empty_cache()
input_df = load_input_data(dga, benign)
(
domain_train
, domain_test
, type_train
, type_test
) = train_test_split(input_df, 'type', train_size=0.7)
train_df = domain_train['domain'].reset_index(drop=True)
train_labels = type_train.reset_index(drop=True)
test_df = create_df(domain_test, type_test)
```
#### clx.analytics.dga_detector.DGADetector.init_model()
```
LR = 0.001
N_LAYERS = 3
CHAR_VOCAB = 128
HIDDEN_SIZE = 100
N_DOMAIN_TYPE = 2
dd = DGADetector(lr=LR)
dd.init_model(
n_layers=N_LAYERS
, char_vocab=CHAR_VOCAB
, hidden_size=HIDDEN_SIZE
, n_domain_type=N_DOMAIN_TYPE
)
```
#### clx.analytics.dga_detector.DGADetector.train_model() Yes
```
batch_size = 10000
train_dataset = {'features': train_df, 'labels': train_labels}
test_dataset = DataLoader(DGADataset(test_df), batch_size)
def train_and_eval(dd, train_dataset, test_dataset, epoch, model_dir):
print("Initiating model training")
create_dir(model_dir)
max_accuracy = 0
prev_model_file_path = ""
for i in range(1, epoch + 1):
print("---------")
print("Epoch: %s" % (i))
print("---------")
dd.train_model(train_dataset['features'], train_dataset['labels'])
accuracy = dd.evaluate_model(test_dataset)
now = datetime.now()
output_filepath = (
model_dir
+ "/"
+ "rnn_classifier_{}.pth".format(now.strftime("%Y-%m-%d_%H_%M_%S"))
)
if accuracy > max_accuracy:
dd.save_model(output_filepath)
max_accuracy = accuracy
if prev_model_file_path:
os.remove(prev_model_file_path)
prev_model_file_path = output_filepath
print("Model with highest accuracy (%s) is stored to location %s" % (max_accuracy, prev_model_file_path))
return prev_model_file_path
%%time
epoch = 2
model_dir='../models/DGA_Detector'
model_filepath = train_and_eval(dd, train_dataset, test_dataset, epoch, model_dir)
cleanup_cache()
```
#### clx.analytics.dga_detector.DGADetector.evaluate_model()
```
accuracy = dd.evaluate_model(DataLoader(DGADataset(test_df), 10000))
```
#### clx.analytics.dga_detector.DGADetector.predict()
```
dd = DGADetector()
dd.load_model('../models/DGA_Detector/rnn_classifier_2021-02-22_20_54_32.pth')
pred_results = []
true_results = []
for partition in test_dataset.get_chunks():
pred_results.append(list(dd.predict(partition['domain']).values_host))
true_results.append(list(partition['type'].values_host))
pred_results = np.concatenate(pred_results)
true_results = np.concatenate(true_results)
accuracy_score = accuracy_score(pred_results, true_results)
print('Model accuracy: %s'%(accuracy_score))
cleanup_cache()
```
# Phishing Detector
## Model
```
import cudf;
from cuml.preprocessing.model_selection import train_test_split
from clx.analytics.sequence_classifier import SequenceClassifier
import s3fs;
from os import path
DATA_DIR = '../data/phishing'
CLAIR_TSV = "Phishing_Dataset_Clair_Collection.tsv"
SPAM_TSV = "spam_assassin_spam_200_20021010.tsv"
EASY_HAM_TSV = "spam_assassin_easyham_200_20021010.tsv"
HARD_HAM_TSV = "spam_assassin_hardham_200_20021010.tsv"
ENRON_TSV = "enron_10000.tsv"
S3_BASE_PATH = "rapidsai-data/cyber/clx"
def maybe_download(f, output_dir):
if not path.exists(f'{output_dir}/{f}'):
print(f'Downloading: {f}')
fs = s3fs.S3FileSystem(anon=True)
fs.get(S3_BASE_PATH + "/" + f, f'{output_dir}/{f}')
def read_dataset(f, data_dir):
maybe_download(f, data_dir)
return cudf.read_csv(
f'{data_dir}/{f}'
, delimiter='\t'
, header=None
, names=['label', 'email']
)
dfclair = read_dataset(CLAIR_TSV, DATA_DIR)
dfspam = read_dataset(SPAM_TSV, DATA_DIR)
dfeasyham = read_dataset(EASY_HAM_TSV, DATA_DIR)
dfhardham = read_dataset(HARD_HAM_TSV, DATA_DIR)
dfenron = read_dataset(ENRON_TSV, DATA_DIR)
```
#### clx.analytics.phishing_detector.PhishingDetector.init_model()
```
phish_detect = SequenceClassifier()
phish_detect.init_model(model_or_path='bert-base-uncased')
```
#### clx.analytics.phishing_detector.PhishingDetector.train_model()
```
df_all = cudf.concat([
dfclair
, dfspam
, dfeasyham
, dfhardham
, dfenron
])
(
X_train
, X_test
, y_train
, y_test
) = train_test_split(df_all, 'label', train_size=0.8)
phish_detect.train_model(X_train, y_train, epochs=1)
```
#### clx.analytics.phishing_detector.PhishingDetector.evaluate_model()
```
phish_detect.evaluate_model(X_test['email'], y_test)
```
#### clx.analytics.phishing_detector.PhishingDetector.save_model()
```
phish_detect.save_model('../models/phishing')
```
#### clx.analytics.phishing_detector.PhishingDetector.predict()
```
phish_detect_trained = SequenceClassifier()
phish_detect_trained.init_model(model_or_path='../models/phishing')
phish_detect_trained.predict(X_test['email'])
```
|
github_jupyter
|
import cudf
import dask_cudf
import s3fs
from os import path
from clx.analytics.cybert import Cybert
CLX_S3_BASE_PATH = 'rapidsai-data/cyber/clx'
HF_S3_BASE_PATH = 'models.huggingface.co/bert/raykallen/cybert_apache_parser'
MODEL_DIR = '../models/CyBERT'
DATA_DIR = '../data'
CONFIG_FILENAME = 'config.json'
MODEL_FILENAME = 'pytorch_model.bin'
APACHE_SAMPLE_CSV = 'apache_sample_1k.csv'
if not path.exists(f'{MODEL_DIR}/{MODEL_FILENAME}'):
fs = s3fs.S3FileSystem(anon=True)
fs.get(
f'{HF_S3_BASE_PATH}/{MODEL_FILENAME}'
, f'{MODEL_DIR}/{MODEL_FILENAME}'
)
if not path.exists(f'{MODEL_DIR}/{CONFIG_FILENAME}'):
fs = s3fs.S3FileSystem(anon=True)
fs.get(
f'{HF_S3_BASE_PATH}/{CONFIG_FILENAME}'
, f'{MODEL_DIR}/{CONFIG_FILENAME}'
)
if not path.exists(APACHE_SAMPLE_CSV):
fs = s3fs.S3FileSystem(anon=True)
fs.get(
f'{CLX_S3_BASE_PATH}/{APACHE_SAMPLE_CSV}'
, f'{DATA_DIR}/{APACHE_SAMPLE_CSV}')
cybert = Cybert()
cybert.load_model(
f'{MODEL_DIR}/{MODEL_FILENAME}'
, f'{MODEL_DIR}/{CONFIG_FILENAME}'
)
logs_df = cudf.read_csv(f'{DATA_DIR}/{APACHE_SAMPLE_CSV}')
parsed_df, confidence_df = cybert.inference(logs_df["raw"])
parsed_df.head()
confidence_df.head()
logs_df = cudf.read_csv(f'{DATA_DIR}/{APACHE_SAMPLE_CSV}')
input_ids, attention_masks, meta = cybert.preprocess(logs_df["raw"])
input_ids
attention_masks
meta
import os
import wget
import time
import cudf
import torch
import shutil
import zipfile
import numpy as np
from datetime import datetime
from sklearn.metrics import accuracy_score, average_precision_score
from clx.analytics.dga_dataset import DGADataset
from clx.analytics.dga_detector import DGADetector
from cuml.preprocessing.model_selection import train_test_split
from clx.utils.data.dataloader import DataLoader
dga = {
"source": "DGA",
"url": "https://data.netlab.360.com/feeds/dga/dga.txt",
"compression": None,
"storage_path": "../data/dga_feed",
}
benign = {
"source": "Benign",
"url": "http://s3.amazonaws.com/alexa-static/top-1m.csv.zip",
"compression": "zip",
"storage_path": "../data/top-1m",
}
def unpack(compression_type, filepath, output_dir):
if compression_type == 'zip':
with zipfile.ZipFile(filepath, 'r') as f:
f.extractall(output_dir)
os.remove(filepath)
def download_file(f):
output_dir = f['storage_path']
filepath = f'{output_dir}/{f["url"].split("/")[-1]}'
if not os.path.exists(filepath):
if not os.path.exists(output_dir):
os.makedirs(output_dir)
print(f'Downloading {f["url"]}...')
filepath = wget.download(f['url'], out=output_dir)
print(f'Unpacking {filepath}')
unpack(f['compression'], filepath, output_dir)
print(f'{f["source"]} data is stored to location {output_dir}')
download_file(dga)
download_file(benign)
def load_input_data(dga, benign):
dga_df = cudf.read_csv(
dga['storage_path'] + '/*'
, names=['generator', 'domain', 'dt_from', 'dt_to']
, usecols=['domain']
, skiprows=18
, delimiter='\t'
)
dga_df['type'] = 0
benign_df = cudf.read_csv(
benign['storage_path'] + '/*'
, names=["line_num","domain"]
, usecols=['domain']
)
benign_df['type'] = 1
input_df = cudf.concat([benign_df, dga_df], ignore_index=True)
return input_df
def create_df(domain_df, type_series):
df = cudf.DataFrame()
df['domain'] = domain_df['domain'].reset_index(drop=True)
df['type'] = type_series.reset_index(drop=True)
return df
def create_dir(dir_path):
print("Verify if directory `%s` already exists." % (dir_path))
if not os.path.exists(dir_path):
print("Directory `%s` does not exist." % (dir_path))
print("Creating directory `%s` to store trained models." % (dir_path))
os.makedirs(dir_path)
def cleanup_cache():
# release memory.
torch.cuda.empty_cache()
input_df = load_input_data(dga, benign)
(
domain_train
, domain_test
, type_train
, type_test
) = train_test_split(input_df, 'type', train_size=0.7)
train_df = domain_train['domain'].reset_index(drop=True)
train_labels = type_train.reset_index(drop=True)
test_df = create_df(domain_test, type_test)
LR = 0.001
N_LAYERS = 3
CHAR_VOCAB = 128
HIDDEN_SIZE = 100
N_DOMAIN_TYPE = 2
dd = DGADetector(lr=LR)
dd.init_model(
n_layers=N_LAYERS
, char_vocab=CHAR_VOCAB
, hidden_size=HIDDEN_SIZE
, n_domain_type=N_DOMAIN_TYPE
)
batch_size = 10000
train_dataset = {'features': train_df, 'labels': train_labels}
test_dataset = DataLoader(DGADataset(test_df), batch_size)
def train_and_eval(dd, train_dataset, test_dataset, epoch, model_dir):
print("Initiating model training")
create_dir(model_dir)
max_accuracy = 0
prev_model_file_path = ""
for i in range(1, epoch + 1):
print("---------")
print("Epoch: %s" % (i))
print("---------")
dd.train_model(train_dataset['features'], train_dataset['labels'])
accuracy = dd.evaluate_model(test_dataset)
now = datetime.now()
output_filepath = (
model_dir
+ "/"
+ "rnn_classifier_{}.pth".format(now.strftime("%Y-%m-%d_%H_%M_%S"))
)
if accuracy > max_accuracy:
dd.save_model(output_filepath)
max_accuracy = accuracy
if prev_model_file_path:
os.remove(prev_model_file_path)
prev_model_file_path = output_filepath
print("Model with highest accuracy (%s) is stored to location %s" % (max_accuracy, prev_model_file_path))
return prev_model_file_path
%%time
epoch = 2
model_dir='../models/DGA_Detector'
model_filepath = train_and_eval(dd, train_dataset, test_dataset, epoch, model_dir)
cleanup_cache()
accuracy = dd.evaluate_model(DataLoader(DGADataset(test_df), 10000))
dd = DGADetector()
dd.load_model('../models/DGA_Detector/rnn_classifier_2021-02-22_20_54_32.pth')
pred_results = []
true_results = []
for partition in test_dataset.get_chunks():
pred_results.append(list(dd.predict(partition['domain']).values_host))
true_results.append(list(partition['type'].values_host))
pred_results = np.concatenate(pred_results)
true_results = np.concatenate(true_results)
accuracy_score = accuracy_score(pred_results, true_results)
print('Model accuracy: %s'%(accuracy_score))
cleanup_cache()
import cudf;
from cuml.preprocessing.model_selection import train_test_split
from clx.analytics.sequence_classifier import SequenceClassifier
import s3fs;
from os import path
DATA_DIR = '../data/phishing'
CLAIR_TSV = "Phishing_Dataset_Clair_Collection.tsv"
SPAM_TSV = "spam_assassin_spam_200_20021010.tsv"
EASY_HAM_TSV = "spam_assassin_easyham_200_20021010.tsv"
HARD_HAM_TSV = "spam_assassin_hardham_200_20021010.tsv"
ENRON_TSV = "enron_10000.tsv"
S3_BASE_PATH = "rapidsai-data/cyber/clx"
def maybe_download(f, output_dir):
if not path.exists(f'{output_dir}/{f}'):
print(f'Downloading: {f}')
fs = s3fs.S3FileSystem(anon=True)
fs.get(S3_BASE_PATH + "/" + f, f'{output_dir}/{f}')
def read_dataset(f, data_dir):
maybe_download(f, data_dir)
return cudf.read_csv(
f'{data_dir}/{f}'
, delimiter='\t'
, header=None
, names=['label', 'email']
)
dfclair = read_dataset(CLAIR_TSV, DATA_DIR)
dfspam = read_dataset(SPAM_TSV, DATA_DIR)
dfeasyham = read_dataset(EASY_HAM_TSV, DATA_DIR)
dfhardham = read_dataset(HARD_HAM_TSV, DATA_DIR)
dfenron = read_dataset(ENRON_TSV, DATA_DIR)
phish_detect = SequenceClassifier()
phish_detect.init_model(model_or_path='bert-base-uncased')
df_all = cudf.concat([
dfclair
, dfspam
, dfeasyham
, dfhardham
, dfenron
])
(
X_train
, X_test
, y_train
, y_test
) = train_test_split(df_all, 'label', train_size=0.8)
phish_detect.train_model(X_train, y_train, epochs=1)
phish_detect.evaluate_model(X_test['email'], y_test)
phish_detect.save_model('../models/phishing')
phish_detect_trained = SequenceClassifier()
phish_detect_trained.init_model(model_or_path='../models/phishing')
phish_detect_trained.predict(X_test['email'])
| 0.391755 | 0.517083 |
```
#PyQt5的学习博客
#http://www.cnblogs.com/archisama/p/5442071.html
#!/usr/bin/python3
# -*- coding: utf-8 -*-
"""
PyQt5 教程
在这个例子中, 我们用PyQt5创建了一个简单的窗口。
"""
#面向过程的方式
import sys
from PyQt5.QtWidgets import QApplication, QWidget
if __name__ == '__main__':
app = QApplication(sys.argv) #所有的PyQt5应用必须创建一个应用(Application)对象。
w = QWidget() #Qwidget组件是PyQt5中所有用户界面类的基础类
w.resize(250, 150)
w.move(300, 300)
w.setWindowTitle('Hello PyQt5!')
w.show()
sys.exit(app.exec_())
#-*- coding: utf-8 -*-
"""
使用面向对象的方式,来进行开发
"""
import sys
from PyQt5.QtWidgets import QApplication, QWidget
from PyQt5.QtGui import QIcon
#面向对象的方式
class Example(QWidget):#继承了QWidget
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.setGeometry(300, 300, 300, 220)#设置坐标和大小
self.setWindowTitle('Icon')
self.setWindowIcon(QIcon('F:\\MyTemp\\ICO\\application.ico')) #设置图标
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_()) #退出循环
# -*- coding: utf-8 -*-
"""
设置了图标,添加了一个button
"""
import sys
from PyQt5.QtWidgets import (QWidget, QToolTip, QPushButton, QApplication)
from PyQt5.QtGui import QFont,QIcon
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
QToolTip.setFont(QFont('SansSerif', 10))
self.setToolTip('This is a <b>QWidget</b> widget')#设置TooTip
btn = QPushButton('Button', self)#设置Button
btn.setToolTip('This is a <b>QPushButton</b> widget')#设置Button的TooTip
btn.resize(btn.sizeHint())
btn.move(50, 50)
self.setGeometry(300, 300, 300, 200)
self.setWindowTitle('Tooltips')
self.setWindowIcon(QIcon('F:\\MyTemp\\ICO\\application.ico'))
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
为添加的button关联了一个退出方法
This program creates a quit
button. When we press the button,
the application terminates.
"""
import sys
from PyQt5.QtWidgets import QWidget, QPushButton, QApplication
from PyQt5.QtCore import QCoreApplication
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
#一个应用的组件是分层结构的。在这个分层内,大多数组件都有父类。没有父类的组件是顶级窗口
qbtn = QPushButton('Quit', self)
#在PyQt5中,事件处理系统由信号&槽机制建立。如果我们点击了按钮,信号clicked被发送。
#槽可以是Qt内置的槽或Python 的一个方法调用。
qbtn.clicked.connect(QCoreApplication.instance().quit)
qbtn.resize(qbtn.sizeHint())
qbtn.move(50, 50)
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Quit button')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
MessageBox的使用例子
This program shows a confirmation
message box when we click on the close
button of the application window.
"""
import sys
from PyQt5.QtWidgets import QWidget, QMessageBox, QApplication
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Message box')
self.show()
def closeEvent(self, event):
#如果我们关闭一个QWidget,QCloseEvent类事件将被生成。要修改组件动作我们需要重新实现closeEvent()事件处理方法。
reply = QMessageBox.question(self, 'Message',
"Are you sure to quit?", QMessageBox.Yes |
QMessageBox.No, QMessageBox.No)
if reply == QMessageBox.Yes:
event.accept()
else:
event.ignore()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
将程序框放在屏幕中间
This program centers a window
on the screen.
"""
import sys
from PyQt5.QtWidgets import QWidget, QDesktopWidget, QApplication
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.resize(250, 150)
self.center()
self.setWindowTitle('Center')
self.show()
def center(self):
#我们获得主窗口的一个矩形特定几何图形。这包含了窗口的框架。
qr = self.frameGeometry()
#我们算出相对于显示器的绝对值。并且从这个绝对值中,我们获得了屏幕中心点。
cp = QDesktopWidget().availableGeometry().center()
#移动到中心
qr.moveCenter(cp)
self.move(qr.topLeft())
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
状态栏的使用
This program creates a statusbar.
"""
import sys
from PyQt5.QtWidgets import QMainWindow, QApplication
#QMainWindow类提供了一个应用主窗口。默认创建一个拥有状态栏、工具栏和菜单栏的经典应用窗口骨架。
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.statusBar().showMessage('Ready')#状态栏
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Statusbar')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
添加了菜单栏,以及菜单
This program creates a menubar. The
menubar has one menu with an exit action.
"""
import sys
from PyQt5.QtWidgets import QMainWindow, QAction, qApp, QApplication
from PyQt5.QtGui import QIcon
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
#QAction是一个用于菜单栏、工具栏或自定义快捷键的抽象动作行为。
exitAction = QAction(QIcon('exit.png'), '&Exit', self)
exitAction.setShortcut('Ctrl+Q')
exitAction.setStatusTip('Exit application')
exitAction.triggered.connect(qApp.quit)
self.statusBar()
#menuBar()方法创建了一个菜单栏。我们创建一个file菜单,然后将退出动作添加到file菜单中。
menubar = self.menuBar()
fileMenu = menubar.addMenu('&File')
fileMenu.addAction(exitAction)
self.setGeometry(300, 300, 300, 200)
self.setWindowTitle('Menubar')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
添加了工具栏
This program creates a toolbar.
The toolbar has one action, which
terminates the application, if triggered.
"""
import sys
from PyQt5.QtWidgets import QMainWindow, QAction, qApp, QApplication
from PyQt5.QtGui import QIcon
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
#QAction是一个用于菜单栏、工具栏或自定义快捷键的抽象动作行为。
exitAction = QAction(QIcon('F:\\MyTemp\\ICO\\remove-ticket.ico'), 'Exit', self)
exitAction.setShortcut('Ctrl+Q')
exitAction.triggered.connect(qApp.quit)
#toolbar是工具栏
self.toolbar = self.addToolBar('Exit')
self.toolbar.addAction(exitAction)
self.setGeometry(300, 300, 300, 200)
self.setWindowTitle('Toolbar')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
有状态栏,菜单栏,工具栏以及一个中心组件的传统应用
This program creates a skeleton of
a classic GUI application with a menubar,
toolbar, statusbar, and a central widget.
"""
import sys
from PyQt5.QtWidgets import QMainWindow, QTextEdit, QAction, QApplication
from PyQt5.QtGui import QIcon
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
textEdit = QTextEdit()#文本
self.setCentralWidget(textEdit)
exitAction = QAction(QIcon('F:\\MyTemp\\ICO\\remove-ticket.ico'), 'Exit', self)#QAction
exitAction.setShortcut('Ctrl+Q')
exitAction.setStatusTip('Exit application')
exitAction.triggered.connect(self.close)
self.statusBar()#状态栏
menubar = self.menuBar()#菜单按钮
fileMenu = menubar.addMenu('&File')
fileMenu.addAction(exitAction)
toolbar = self.addToolBar('Exit')#工具栏
toolbar.addAction(exitAction)
self.setGeometry(300, 300, 350, 250)
self.setWindowTitle('Main window')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
绝对的布局方式
This example shows three labels on a window
using absolute positioning.
"""
import sys
from PyQt5.QtWidgets import QWidget, QLabel, QApplication
#绝对布局方式
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
lbl1 = QLabel('Zetcode', self)
lbl1.move(15, 10)#使用move()方法来定位我们的组件
lbl2 = QLabel('tutorials', self)
lbl2.move(35, 40)
lbl3 = QLabel('for programmers', self)
lbl3.move(55, 70)
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Absolute')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
箱式布局
In this example, we position two push
buttons in the bottom-right corner
of the window.
"""
import sys
from PyQt5.QtWidgets import (QWidget, QPushButton,
QHBoxLayout, QVBoxLayout, QApplication)
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
okButton = QPushButton("OK")
cancelButton = QPushButton("Cancel")
hbox = QHBoxLayout()#QHBoxLayout布局类,水平布局类
hbox.addStretch(1)#添加空隙
hbox.addWidget(okButton)
hbox.addWidget(cancelButton)
vbox = QVBoxLayout()#QVBoxLayout布局类,垂直布局类
vbox.addStretch(1)#添加空隙
vbox.addLayout(hbox)
self.setLayout(vbox)
self.setGeometry(300, 300, 300, 150)#设置大小
self.setWindowTitle('Buttons')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
网格布局
In this example, we create a skeleton
of a calculator using a QGridLayout.
"""
import sys
from PyQt5.QtWidgets import (QWidget, QGridLayout,
QPushButton, QApplication)
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
grid = QGridLayout()#创建一个网格布局
self.setLayout(grid)
names = ['Cls', 'Bck', '', 'Close',
'7', '8', '9', '/',
'4', '5', '6', '*',
'1', '2', '3', '-',
'0', '.', '=', '+']
positions = [(i,j) for i in range(5) for j in range(4)]
for position, name in zip(positions, names):
if name == '':
continue
button = QPushButton(name)
grid.addWidget(button, *position)
self.move(300, 150)
self.setWindowTitle('Calculator')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
比较复杂的一个布局应用
In this example, we create a bit
more complicated window layout using
the QGridLayout manager.
"""
import sys
from PyQt5.QtWidgets import (QWidget, QLabel, QLineEdit,
QTextEdit, QGridLayout, QApplication)
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
title = QLabel('Title')#label
author = QLabel('Author')
review = QLabel('Review')
titleEdit = QLineEdit()#行式的输入框
authorEdit = QLineEdit()
reviewEdit = QTextEdit()#文本输入框
grid = QGridLayout()#网格布局
grid.setSpacing(10) #Label和输入框之间的间距
grid.addWidget(title, 1, 0)
grid.addWidget(titleEdit, 1, 1)
grid.addWidget(author, 2, 0)
grid.addWidget(authorEdit, 2, 1)
grid.addWidget(review, 3, 0)
grid.addWidget(reviewEdit, 3, 1, 5, 1)
self.setLayout(grid)
self.setGeometry(300, 300, 350, 300)
self.setWindowTitle('Review')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
所有的GUI应用都是事件驱动的,事件源,事件对象,事件目标是事件模型的三个参与者。
事件源是状态发生改变的对象。它产生了事件。
事件对象(evnet)封装了事件源中的状态变化。
事件目标是想要被通知的对象。事件源对象代表了处理一个事件直到事件目标做出响应的任务。
PyQt5有一个独一无二的信号和槽机制来处理事件。信号和槽用于对象之间的通信。
当指定事件发生,一个事件信号会被发射。槽可以被任何Python脚本调用。当和槽连接的信号被发射时,槽会被调用。
In this example, we connect a signal
of a QSlider to a slot of a QLCDNumber.
"""
import sys
from PyQt5.QtCore import Qt
from PyQt5.QtWidgets import (QWidget, QLCDNumber, QSlider,
QVBoxLayout, QApplication)
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
lcd = QLCDNumber(self)#数字
sld = QSlider(Qt.Horizontal, self)#滑块
vbox = QVBoxLayout()
vbox.addWidget(lcd)
vbox.addWidget(sld)
self.setLayout(vbox)
sld.valueChanged.connect(lcd.display)#此处将滑块条的valueChanged信号和lcd数字显示的display槽连接在一起
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Signal & slot')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
重写事件处理函数
In this example, we reimplement an
event handler.
"""
import sys
from PyQt5.QtCore import Qt
from PyQt5.QtWidgets import QWidget, QApplication
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Event handler')
self.show()
#重写了事件处理函数,问题:在Python之中,如何就是重写了事件处理函数呢?有没有什么标志?
def keyPressEvent(self, e):
if e.key() == Qt.Key_Escape:
self.close()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
#!/usr/bin/python3
# -*- coding: utf-8 -*-
"""
有时需要方便的知道哪一个组件是信号发送者。
因此,PyQt5拥有了sender()方法来解决这个问题。
In this example, we determine the event sender
object.
"""
import sys
from PyQt5.QtWidgets import QMainWindow, QPushButton, QApplication
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
btn1 = QPushButton("Button 1", self)
btn1.move(30, 50)
btn2 = QPushButton("Button 2", self)
btn2.move(150, 50)
btn1.clicked.connect(self.buttonClicked)#此处绑定了处理的方法
btn2.clicked.connect(self.buttonClicked)
self.statusBar()
self.setGeometry(300, 300, 290, 150)
self.setWindowTitle('Event sender')
self.show()
def buttonClicked(self):
sender = self.sender()#事件的发送者
self.statusBar().showMessage(sender.text() + ' was pressed')
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
从QObejct生成的对象可以发送信号。
在下面的例子中我们将会看到怎样去发送自定义的信号。
In this example, we show how to emit a
signal.
"""
import sys
from PyQt5.QtCore import pyqtSignal, QObject
from PyQt5.QtWidgets import QMainWindow, QApplication
class Communicate(QObject):
closeApp = pyqtSignal()
#从QObejct生成的对象可以发送信号
#我们创建一个新的信号叫做closeApp。当触发鼠标点击事件时信号会被发射。
#信号连接到了QMainWindow的close()方法。
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.c = Communicate()
self.c.closeApp.connect(self.close)
self.setGeometry(300, 300, 290, 150)
self.setWindowTitle('Emit signal')
self.show()
#把自定义的closeApp信号连接到QMainWindow的close()槽上。
#感觉是重写了这个方法
def mousePressEvent(self, event):
self.c.closeApp.emit()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
```
|
github_jupyter
|
#PyQt5的学习博客
#http://www.cnblogs.com/archisama/p/5442071.html
#!/usr/bin/python3
# -*- coding: utf-8 -*-
"""
PyQt5 教程
在这个例子中, 我们用PyQt5创建了一个简单的窗口。
"""
#面向过程的方式
import sys
from PyQt5.QtWidgets import QApplication, QWidget
if __name__ == '__main__':
app = QApplication(sys.argv) #所有的PyQt5应用必须创建一个应用(Application)对象。
w = QWidget() #Qwidget组件是PyQt5中所有用户界面类的基础类
w.resize(250, 150)
w.move(300, 300)
w.setWindowTitle('Hello PyQt5!')
w.show()
sys.exit(app.exec_())
#-*- coding: utf-8 -*-
"""
使用面向对象的方式,来进行开发
"""
import sys
from PyQt5.QtWidgets import QApplication, QWidget
from PyQt5.QtGui import QIcon
#面向对象的方式
class Example(QWidget):#继承了QWidget
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.setGeometry(300, 300, 300, 220)#设置坐标和大小
self.setWindowTitle('Icon')
self.setWindowIcon(QIcon('F:\\MyTemp\\ICO\\application.ico')) #设置图标
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_()) #退出循环
# -*- coding: utf-8 -*-
"""
设置了图标,添加了一个button
"""
import sys
from PyQt5.QtWidgets import (QWidget, QToolTip, QPushButton, QApplication)
from PyQt5.QtGui import QFont,QIcon
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
QToolTip.setFont(QFont('SansSerif', 10))
self.setToolTip('This is a <b>QWidget</b> widget')#设置TooTip
btn = QPushButton('Button', self)#设置Button
btn.setToolTip('This is a <b>QPushButton</b> widget')#设置Button的TooTip
btn.resize(btn.sizeHint())
btn.move(50, 50)
self.setGeometry(300, 300, 300, 200)
self.setWindowTitle('Tooltips')
self.setWindowIcon(QIcon('F:\\MyTemp\\ICO\\application.ico'))
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
为添加的button关联了一个退出方法
This program creates a quit
button. When we press the button,
the application terminates.
"""
import sys
from PyQt5.QtWidgets import QWidget, QPushButton, QApplication
from PyQt5.QtCore import QCoreApplication
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
#一个应用的组件是分层结构的。在这个分层内,大多数组件都有父类。没有父类的组件是顶级窗口
qbtn = QPushButton('Quit', self)
#在PyQt5中,事件处理系统由信号&槽机制建立。如果我们点击了按钮,信号clicked被发送。
#槽可以是Qt内置的槽或Python 的一个方法调用。
qbtn.clicked.connect(QCoreApplication.instance().quit)
qbtn.resize(qbtn.sizeHint())
qbtn.move(50, 50)
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Quit button')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
MessageBox的使用例子
This program shows a confirmation
message box when we click on the close
button of the application window.
"""
import sys
from PyQt5.QtWidgets import QWidget, QMessageBox, QApplication
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Message box')
self.show()
def closeEvent(self, event):
#如果我们关闭一个QWidget,QCloseEvent类事件将被生成。要修改组件动作我们需要重新实现closeEvent()事件处理方法。
reply = QMessageBox.question(self, 'Message',
"Are you sure to quit?", QMessageBox.Yes |
QMessageBox.No, QMessageBox.No)
if reply == QMessageBox.Yes:
event.accept()
else:
event.ignore()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
将程序框放在屏幕中间
This program centers a window
on the screen.
"""
import sys
from PyQt5.QtWidgets import QWidget, QDesktopWidget, QApplication
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.resize(250, 150)
self.center()
self.setWindowTitle('Center')
self.show()
def center(self):
#我们获得主窗口的一个矩形特定几何图形。这包含了窗口的框架。
qr = self.frameGeometry()
#我们算出相对于显示器的绝对值。并且从这个绝对值中,我们获得了屏幕中心点。
cp = QDesktopWidget().availableGeometry().center()
#移动到中心
qr.moveCenter(cp)
self.move(qr.topLeft())
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
状态栏的使用
This program creates a statusbar.
"""
import sys
from PyQt5.QtWidgets import QMainWindow, QApplication
#QMainWindow类提供了一个应用主窗口。默认创建一个拥有状态栏、工具栏和菜单栏的经典应用窗口骨架。
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.statusBar().showMessage('Ready')#状态栏
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Statusbar')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
添加了菜单栏,以及菜单
This program creates a menubar. The
menubar has one menu with an exit action.
"""
import sys
from PyQt5.QtWidgets import QMainWindow, QAction, qApp, QApplication
from PyQt5.QtGui import QIcon
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
#QAction是一个用于菜单栏、工具栏或自定义快捷键的抽象动作行为。
exitAction = QAction(QIcon('exit.png'), '&Exit', self)
exitAction.setShortcut('Ctrl+Q')
exitAction.setStatusTip('Exit application')
exitAction.triggered.connect(qApp.quit)
self.statusBar()
#menuBar()方法创建了一个菜单栏。我们创建一个file菜单,然后将退出动作添加到file菜单中。
menubar = self.menuBar()
fileMenu = menubar.addMenu('&File')
fileMenu.addAction(exitAction)
self.setGeometry(300, 300, 300, 200)
self.setWindowTitle('Menubar')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
添加了工具栏
This program creates a toolbar.
The toolbar has one action, which
terminates the application, if triggered.
"""
import sys
from PyQt5.QtWidgets import QMainWindow, QAction, qApp, QApplication
from PyQt5.QtGui import QIcon
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
#QAction是一个用于菜单栏、工具栏或自定义快捷键的抽象动作行为。
exitAction = QAction(QIcon('F:\\MyTemp\\ICO\\remove-ticket.ico'), 'Exit', self)
exitAction.setShortcut('Ctrl+Q')
exitAction.triggered.connect(qApp.quit)
#toolbar是工具栏
self.toolbar = self.addToolBar('Exit')
self.toolbar.addAction(exitAction)
self.setGeometry(300, 300, 300, 200)
self.setWindowTitle('Toolbar')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
有状态栏,菜单栏,工具栏以及一个中心组件的传统应用
This program creates a skeleton of
a classic GUI application with a menubar,
toolbar, statusbar, and a central widget.
"""
import sys
from PyQt5.QtWidgets import QMainWindow, QTextEdit, QAction, QApplication
from PyQt5.QtGui import QIcon
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
textEdit = QTextEdit()#文本
self.setCentralWidget(textEdit)
exitAction = QAction(QIcon('F:\\MyTemp\\ICO\\remove-ticket.ico'), 'Exit', self)#QAction
exitAction.setShortcut('Ctrl+Q')
exitAction.setStatusTip('Exit application')
exitAction.triggered.connect(self.close)
self.statusBar()#状态栏
menubar = self.menuBar()#菜单按钮
fileMenu = menubar.addMenu('&File')
fileMenu.addAction(exitAction)
toolbar = self.addToolBar('Exit')#工具栏
toolbar.addAction(exitAction)
self.setGeometry(300, 300, 350, 250)
self.setWindowTitle('Main window')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
绝对的布局方式
This example shows three labels on a window
using absolute positioning.
"""
import sys
from PyQt5.QtWidgets import QWidget, QLabel, QApplication
#绝对布局方式
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
lbl1 = QLabel('Zetcode', self)
lbl1.move(15, 10)#使用move()方法来定位我们的组件
lbl2 = QLabel('tutorials', self)
lbl2.move(35, 40)
lbl3 = QLabel('for programmers', self)
lbl3.move(55, 70)
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Absolute')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
箱式布局
In this example, we position two push
buttons in the bottom-right corner
of the window.
"""
import sys
from PyQt5.QtWidgets import (QWidget, QPushButton,
QHBoxLayout, QVBoxLayout, QApplication)
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
okButton = QPushButton("OK")
cancelButton = QPushButton("Cancel")
hbox = QHBoxLayout()#QHBoxLayout布局类,水平布局类
hbox.addStretch(1)#添加空隙
hbox.addWidget(okButton)
hbox.addWidget(cancelButton)
vbox = QVBoxLayout()#QVBoxLayout布局类,垂直布局类
vbox.addStretch(1)#添加空隙
vbox.addLayout(hbox)
self.setLayout(vbox)
self.setGeometry(300, 300, 300, 150)#设置大小
self.setWindowTitle('Buttons')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
网格布局
In this example, we create a skeleton
of a calculator using a QGridLayout.
"""
import sys
from PyQt5.QtWidgets import (QWidget, QGridLayout,
QPushButton, QApplication)
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
grid = QGridLayout()#创建一个网格布局
self.setLayout(grid)
names = ['Cls', 'Bck', '', 'Close',
'7', '8', '9', '/',
'4', '5', '6', '*',
'1', '2', '3', '-',
'0', '.', '=', '+']
positions = [(i,j) for i in range(5) for j in range(4)]
for position, name in zip(positions, names):
if name == '':
continue
button = QPushButton(name)
grid.addWidget(button, *position)
self.move(300, 150)
self.setWindowTitle('Calculator')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
比较复杂的一个布局应用
In this example, we create a bit
more complicated window layout using
the QGridLayout manager.
"""
import sys
from PyQt5.QtWidgets import (QWidget, QLabel, QLineEdit,
QTextEdit, QGridLayout, QApplication)
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
title = QLabel('Title')#label
author = QLabel('Author')
review = QLabel('Review')
titleEdit = QLineEdit()#行式的输入框
authorEdit = QLineEdit()
reviewEdit = QTextEdit()#文本输入框
grid = QGridLayout()#网格布局
grid.setSpacing(10) #Label和输入框之间的间距
grid.addWidget(title, 1, 0)
grid.addWidget(titleEdit, 1, 1)
grid.addWidget(author, 2, 0)
grid.addWidget(authorEdit, 2, 1)
grid.addWidget(review, 3, 0)
grid.addWidget(reviewEdit, 3, 1, 5, 1)
self.setLayout(grid)
self.setGeometry(300, 300, 350, 300)
self.setWindowTitle('Review')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
所有的GUI应用都是事件驱动的,事件源,事件对象,事件目标是事件模型的三个参与者。
事件源是状态发生改变的对象。它产生了事件。
事件对象(evnet)封装了事件源中的状态变化。
事件目标是想要被通知的对象。事件源对象代表了处理一个事件直到事件目标做出响应的任务。
PyQt5有一个独一无二的信号和槽机制来处理事件。信号和槽用于对象之间的通信。
当指定事件发生,一个事件信号会被发射。槽可以被任何Python脚本调用。当和槽连接的信号被发射时,槽会被调用。
In this example, we connect a signal
of a QSlider to a slot of a QLCDNumber.
"""
import sys
from PyQt5.QtCore import Qt
from PyQt5.QtWidgets import (QWidget, QLCDNumber, QSlider,
QVBoxLayout, QApplication)
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
lcd = QLCDNumber(self)#数字
sld = QSlider(Qt.Horizontal, self)#滑块
vbox = QVBoxLayout()
vbox.addWidget(lcd)
vbox.addWidget(sld)
self.setLayout(vbox)
sld.valueChanged.connect(lcd.display)#此处将滑块条的valueChanged信号和lcd数字显示的display槽连接在一起
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Signal & slot')
self.show()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
重写事件处理函数
In this example, we reimplement an
event handler.
"""
import sys
from PyQt5.QtCore import Qt
from PyQt5.QtWidgets import QWidget, QApplication
class Example(QWidget):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.setGeometry(300, 300, 250, 150)
self.setWindowTitle('Event handler')
self.show()
#重写了事件处理函数,问题:在Python之中,如何就是重写了事件处理函数呢?有没有什么标志?
def keyPressEvent(self, e):
if e.key() == Qt.Key_Escape:
self.close()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
#!/usr/bin/python3
# -*- coding: utf-8 -*-
"""
有时需要方便的知道哪一个组件是信号发送者。
因此,PyQt5拥有了sender()方法来解决这个问题。
In this example, we determine the event sender
object.
"""
import sys
from PyQt5.QtWidgets import QMainWindow, QPushButton, QApplication
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
btn1 = QPushButton("Button 1", self)
btn1.move(30, 50)
btn2 = QPushButton("Button 2", self)
btn2.move(150, 50)
btn1.clicked.connect(self.buttonClicked)#此处绑定了处理的方法
btn2.clicked.connect(self.buttonClicked)
self.statusBar()
self.setGeometry(300, 300, 290, 150)
self.setWindowTitle('Event sender')
self.show()
def buttonClicked(self):
sender = self.sender()#事件的发送者
self.statusBar().showMessage(sender.text() + ' was pressed')
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
# -*- coding: utf-8 -*-
"""
从QObejct生成的对象可以发送信号。
在下面的例子中我们将会看到怎样去发送自定义的信号。
In this example, we show how to emit a
signal.
"""
import sys
from PyQt5.QtCore import pyqtSignal, QObject
from PyQt5.QtWidgets import QMainWindow, QApplication
class Communicate(QObject):
closeApp = pyqtSignal()
#从QObejct生成的对象可以发送信号
#我们创建一个新的信号叫做closeApp。当触发鼠标点击事件时信号会被发射。
#信号连接到了QMainWindow的close()方法。
class Example(QMainWindow):
def __init__(self):
super().__init__()
self.initUI()
def initUI(self):
self.c = Communicate()
self.c.closeApp.connect(self.close)
self.setGeometry(300, 300, 290, 150)
self.setWindowTitle('Emit signal')
self.show()
#把自定义的closeApp信号连接到QMainWindow的close()槽上。
#感觉是重写了这个方法
def mousePressEvent(self, event):
self.c.closeApp.emit()
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
| 0.221014 | 0.195364 |
# Thermal Emission
The first example we'll look at is that of thermal emission from a galaxy cluster. In this case, the gas in the core of the cluster is "sloshing" in the center, producing spiral-shaped cold fronts. The dataset we want to use for this example is available for download from the [yt Project](http://yt-project.org) at [this link](http://yt-project.org/data/GasSloshing.tar.gz).
First, import our necessary modules:
```
%matplotlib inline
import yt
import pyxsim
import soxs
```
Next, we `load` the dataset with yt. Note that this dataset does not have
species fields in it, so we'll set `default_species_fields="ionized"` to
assume full ionization (as appropriate for galaxy clusters):
```
ds = yt.load("GasSloshing/sloshing_nomag2_hdf5_plt_cnt_0150",
default_species_fields="ionized")
```
Let's use yt to take a slice of density and temperature through the center of the dataset so we can see what we're looking at:
```
slc = yt.SlicePlot(ds, "z", ["density", "temperature"], width=(1.0,"Mpc"))
slc.show()
```
Ok, sloshing gas as advertised. Next, we'll create a sphere object to serve as a source for the photons. Place it at the center of the domain with `"c"`, and use a radius of 500 kpc:
```
sp = ds.sphere("c", (500.,"kpc"))
```
Now, we need to set up a source model. We said we were going to look at the thermal emission from the hot plasma, so to do that we can set up a `ThermalSourceModel`. The first argument specifies which model we want to use. Currently the only option available in pyXSIM is `"apec"`. The next three arguments are the maximum and minimum energies, and the number of bins in the spectrum. We've chosen these numbers so that the spectrum has an energy resolution of about 1 eV.
`ThermalSourceModel` takes a lot of optional arguments, which you can investigate in the docs, but here we'll do something simple and say that the metallicity is a constant $Z = 0.3~Z_\odot$:
```
source_model = pyxsim.ThermalSourceModel("apec", 0.05, 11.0, 1000, Zmet=0.3)
```
We're almost ready to go to generate the photons from this source, but first we should decide what our redshift, collecting area, and exposure time should be. Let's pick big numbers, because remember the point of this first step is to create a Monte-Carlo sample from which to draw smaller sub-samples for mock observations. Note these are all (value, unit) tuples:
```
exp_time = (300., "ks") # exposure time
area = (1000.0, "cm**2") # collecting area
redshift = 0.05
```
So, that's everything--let's create the photons! We use the `make_photons` function for this:
```
n_photons, n_cells = pyxsim.make_photons("sloshing_photons", sp, redshift, area, exp_time, source_model)
```
Ok, that was easy. Now we have a photon list that we can use to create events using the `project_photons` function. Here, we'll just do a simple projection along the z-axis, and center the photons at RA, Dec = (45, 30) degrees. Since we want to be realistic, we'll want to apply foreground galactic absorption using the `"tbabs"` model, assuming a neutral hydrogen column of $N_H = 4 \times 10^{20}~{\rm cm}^{-2}$:
```
n_events = pyxsim.project_photons("sloshing_photons", "sloshing_events", "z", (45.,30.), absorb_model="tbabs", nH=0.04)
```
Now that we have a set of "events" on the sky, we can read them in and write them to a SIMPUT file:
```
events = pyxsim.EventList("sloshing_events.h5")
events.write_to_simput("sloshing", overwrite=True)
```
We can then use this SIMPUT file as an input to the instrument simulator in SOXS. We'll use a small exposure time (100 ks instead of 300 ks), and observe it with the as-launched ACIS-I model:
```
soxs.instrument_simulator("sloshing_simput.fits", "evt.fits", (100.0, "ks"), "chandra_acisi_cy0",
[45., 30.], overwrite=True)
```
We can use the `write_image()` function in SOXS to bin the events into an image and write them to a file, restricting the energies between 0.5 and 2.0 keV:
```
soxs.write_image("evt.fits", "img.fits", emin=0.5, emax=2.0, overwrite=True)
```
Now we can take a quick look at the image:
```
soxs.plot_image("img.fits", stretch='sqrt', cmap='arbre', vmin=0.0, vmax=10.0, width=0.2)
```
|
github_jupyter
|
%matplotlib inline
import yt
import pyxsim
import soxs
ds = yt.load("GasSloshing/sloshing_nomag2_hdf5_plt_cnt_0150",
default_species_fields="ionized")
slc = yt.SlicePlot(ds, "z", ["density", "temperature"], width=(1.0,"Mpc"))
slc.show()
sp = ds.sphere("c", (500.,"kpc"))
source_model = pyxsim.ThermalSourceModel("apec", 0.05, 11.0, 1000, Zmet=0.3)
exp_time = (300., "ks") # exposure time
area = (1000.0, "cm**2") # collecting area
redshift = 0.05
n_photons, n_cells = pyxsim.make_photons("sloshing_photons", sp, redshift, area, exp_time, source_model)
n_events = pyxsim.project_photons("sloshing_photons", "sloshing_events", "z", (45.,30.), absorb_model="tbabs", nH=0.04)
events = pyxsim.EventList("sloshing_events.h5")
events.write_to_simput("sloshing", overwrite=True)
soxs.instrument_simulator("sloshing_simput.fits", "evt.fits", (100.0, "ks"), "chandra_acisi_cy0",
[45., 30.], overwrite=True)
soxs.write_image("evt.fits", "img.fits", emin=0.5, emax=2.0, overwrite=True)
soxs.plot_image("img.fits", stretch='sqrt', cmap='arbre', vmin=0.0, vmax=10.0, width=0.2)
| 0.379493 | 0.99148 |
# Comparison to the literature of Galaxy Builder bulges and bars
```
%load_ext autoreload
%autoreload 2
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from IPython.display import display
import os
from os.path import join
from tqdm import tqdm
import scipy.stats as st
import json
import lib.galaxy_utilities as gu
from gzbuilder_analysis import load_aggregation_results, load_fit_results
import gzbuilder_analysis.parsing as pa
import gzbuilder_analysis.fitting as fg
# %run make_bulge_bar_dataframes.py
def number_with_comp(a):
return sum(i is not None for i in a)
def clean_column_names(df):
df_ = df.copy()
df_.columns = [i.strip().replace(' ', '_') for i in df.columns]
return df_
def get_pbar(gal):
n = gal['t03_bar_a06_bar_debiased'] + gal['t03_bar_a07_no_bar_debiased']
return gal['t03_bar_a06_bar_debiased'] / n
from gzbuilder_analysis import load_aggregation_results
agg_results = load_aggregation_results('output_files/aggregation_results')
# load files contain info relating gzb subject ids to GZ2 bulge / bar results:
bulge_df = pd.read_pickle('lib/bulge_fractions.pkl').dropna()
bar_df = pd.read_pickle('lib/bar_fractions.pkl').dropna()
comparison_df = agg_results.agg(dict(
cls=lambda a: len(a.input_models),
disk=lambda a: a.input_models.apply(lambda a: bool(a['disk'])).sum(),
bulge=lambda a: a.input_models.apply(lambda a: bool(a['bulge'])).sum(),
bar=lambda a: a.input_models.apply(lambda a: bool(a['bar'])).sum(),
)).unstack().T
comparison_df = comparison_df.assign(
disk_frac=comparison_df.disk / comparison_df.cls,
bulge_frac=comparison_df.bulge / comparison_df.cls,
bar_frac=comparison_df.bar / comparison_df.cls,
)
comparison_df = comparison_df.assign(
disk_frac_err=np.sqrt(comparison_df.disk_frac * (1 - comparison_df.disk_frac) / comparison_df.cls),
bulge_frac_err=np.sqrt(comparison_df.bulge_frac * (1 - comparison_df.bulge_frac) / comparison_df.cls),
bar_frac_err=np.sqrt(comparison_df.bar_frac * (1 - comparison_df.bar_frac) / comparison_df.cls),
)
# Let's also incorporate knowledge about the aggregagte model (did we cluster a component)
comparison_df = comparison_df.combine_first(
agg_results.apply(lambda a: a.model).apply(pd.Series).applymap(bool).add_prefix('agg_')
)
# and finaly add in information about GZ2:
comparison_df = comparison_df.assign(
GZ2_no_bulge=bulge_df['GZ2 no bulge'],
GZ2_bar_fraction=bar_df['GZ2 bar fraction'],
).dropna().pipe(clean_column_names)
```
Let's also incorporate knowledge about the aggregagte model (did we cluster a component)
```
comparison_df.head()
comparison_df.query('GZ2_bar_fraction < 0.2').bar_frac.describe()
comparison_df.query('GZ2_bar_fraction > 0.5').bar_frac.describe()
f, ax = plt.subplots(ncols=2, figsize=(17, 8))
plt.sca(ax[0])
plt.errorbar(
1 - comparison_df['GZ2_no_bulge'],
comparison_df['bulge_frac'], yerr=comparison_df['bulge_frac_err'],
fmt='.', c='C1', elinewidth=1, capsize=1
)
plt.xlim(-0.02, 1.02)
plt.ylim(-0.02, 1.02)
plt.gca().add_line(plt.Line2D((-10, 10), (-10, 10), c='k', alpha=0.1))
plt.xlabel('1 - Galaxy Zoo 2 "no bulge" fraction')
plt.ylabel('Fraction of classifications with a bulge in Galaxy Builder')
gz2_no_bulge, gzb_bulge = comparison_df[['GZ2_no_bulge', 'bulge_frac']].dropna().values.T
bar_corr = st.pearsonr(1 - gz2_no_bulge, gzb_bulge)
plt.title('Pearson correlation coefficient {:.3f}, p={:.3e}'.format(*bar_corr));
plt.sca(ax[1])
plt.errorbar(
comparison_df['GZ2_bar_fraction'], comparison_df['bar_frac'],
yerr=comparison_df['bar_frac_err'],
fmt='.', c='C2', elinewidth=1, capsize=1
)
plt.xlim(-0.02, 1.02)
plt.ylim(-0.02, 1.02)
plt.axvline(0.2, c='k', ls=':')
plt.axvline(0.5, c='k', ls=':')
plt.errorbar(
0.1,
**comparison_df.query('GZ2_bar_fraction < 0.2').bar_frac.describe()
.rename(index=dict(mean='y', std='yerr'))[['y', 'yerr']],
zorder=10, fmt='o', capsize=10, color='k', ms=10
)
plt.errorbar(
0.8,
**comparison_df.query('GZ2_bar_fraction > 0.5').bar_frac.describe()
.rename(index=dict(mean='y', std='yerr'))[['y', 'yerr']],
zorder=10, fmt='o', capsize=10, color='k', ms=10
)
plt.text(0.2 - 0.01, 1.01, 'No Bar', horizontalalignment='right', verticalalignment='top')
plt.text(0.5 + 0.01, 1.01, 'Strongly Barred', horizontalalignment='left', verticalalignment='top')
plt.gca().add_line(plt.Line2D((-10, 10), (-10, 10), c='k', alpha=0.1))
plt.xlabel('Galaxy Zoo 2 "has bar" fraction')
plt.ylabel('Fraction of classifications with a bar in Galaxy Builder')
bar_corr = st.pearsonr(*comparison_df[['GZ2_bar_fraction', 'bar_frac']].dropna().values.T)
plt.title('Pearson correlation coefficient {:.3f}, p={:.2e}'.format(*bar_corr));
```
Let's add in some indormation about whether the aggregate model contained this component:
```
f, ax = plt.subplots(ncols=2, figsize=(17, 8))
plt.sca(ax[0])
for i in (False, True):
mask = comparison_df.agg_bulge == i
plt.errorbar(
1 - comparison_df['GZ2_no_bulge'][mask],
comparison_df['bulge_frac'][mask], yerr=comparison_df['bulge_frac_err'][mask],
fmt='o', c=('C2' if i else 'r'), elinewidth=1, capsize=1,
label=('Aggregate has bulge' if i else 'Aggregate does not have bulge')
)
plt.xlim(-0.02, 1.02)
plt.ylim(-0.02, 1.02)
plt.gca().add_line(plt.Line2D((-10, 10), (-10, 10), c='k', alpha=0.1))
plt.xlabel('1 - Galaxy Zoo 2 "no bulge" fraction')
plt.ylabel('Fraction of classifications with a bulge in Galaxy Builder')
gz2_no_bulge, gzb_bulge = comparison_df[['GZ2_no_bulge', 'bulge_frac']].dropna().values.T
bar_corr = st.pearsonr(1 - gz2_no_bulge, gzb_bulge)
plt.title('Pearson correlation coefficient {:.3f}, p={:.3e}'.format(*bar_corr));
plt.sca(ax[1])
for i in (False, True):
mask = comparison_df.agg_bar == i
plt.errorbar(
comparison_df['GZ2_bar_fraction'][mask], comparison_df['bar_frac'][mask],
yerr=comparison_df['bar_frac_err'][mask],
fmt='o', c=('C2' if i else 'r'), ms=5, elinewidth=1, capsize=1,
label=('Aggregate has bar' if i else 'Aggregate does not have bar')
)
plt.xlim(-0.02, 1.02)
plt.ylim(-0.02, 1.02)
plt.axvline(0.2, c='k', ls=':')
plt.axvline(0.5, c='k', ls=':')
plt.errorbar(
0.1,
**comparison_df.query('GZ2_bar_fraction < 0.2').bar_frac.describe()
.rename(index=dict(mean='y', std='yerr'))[['y', 'yerr']],
zorder=10, fmt='o', capsize=10, color='k', ms=10
)
plt.errorbar(
0.8,
**comparison_df.query('GZ2_bar_fraction > 0.5').bar_frac.describe()
.rename(index=dict(mean='y', std='yerr'))[['y', 'yerr']],
zorder=10, fmt='o', capsize=10, color='k', ms=10
)
plt.text(0.2 - 0.01, 1.01, 'No Bar', horizontalalignment='right', verticalalignment='top')
plt.text(0.5 + 0.01, 1.01, 'Strongly Barred', horizontalalignment='left', verticalalignment='top')
plt.legend()
plt.gca().add_line(plt.Line2D((-10, 10), (-10, 10), c='k', alpha=0.1))
plt.xlabel('Galaxy Zoo 2 "has bar" fraction')
plt.ylabel('Fraction of classifications with a bar in Galaxy Builder')
bar_corr = st.pearsonr(*comparison_df[['GZ2_bar_fraction', 'bar_frac']].dropna().values.T)
plt.title('Pearson correlation coefficient {:.3f}, p={:.2e}'.format(*bar_corr));
```
## Relative component fractions by volunteer
Do some volunteers systematically make use of bulges or bars, or is it dependant on the galaxy?
```
%%time
classifications = pd.read_csv('lib/galaxy-builder-classifications.csv', index_col=0)
models = (
classifications.query('workflow_version == 61.107')
.apply(pa.parse_classification, image_size=(512, 512),
axis=1, ignore_scale=True)
.apply(pd.Series)
.assign(subject_ids=classifications['subject_ids'].astype('category'))
)
n_cls_by_usr = (
classifications.query('workflow_version == 61.107')
.user_name
.value_counts()
.sort_values()
)
model_freq = (
models.assign(user_name=classifications.reindex(models.index)['user_name'])
.drop(columns=['spiral', 'subject_ids'])
.groupby('user_name')
.agg(number_with_comp)
.reindex(n_cls_by_usr.index)
.T
/ n_cls_by_usr
).T
model_freq.assign(N_classifications=n_cls_by_usr).tail(10)
```
Restricting to users with more than 30 classifications, what can we see?
```
plt.figure(figsize=(12, 4), dpi=80)
for c in model_freq.columns:
plt.hist(
model_freq[n_cls_by_usr > 20][c].dropna(),
bins='scott', density=True, label=c, alpha=0.4
)
print('Identified {} users with more than 20 classifications'.format(
(n_cls_by_usr > 30).sum()
))
plt.xlabel('Fraction of classifications with component')
plt.ylabel('Density')
plt.legend()
```
Looks like volunteers used discs and bulges almost all the time, with a wide spread in the use of bars (some never, some always).
To be certain of this, we'll calculate the Beta conjugate prior for $N$ classifications with $s$ instances of a component:
$$P(q = x | s, N) = \frac{x^s(1 - x)^{N - s}}{B(s+1,\ N-s+1)}$$
```
from scipy.special import beta
def updated_bn(N, s):
return lambda x: x**(s)*(1 - x)**(N - s) / beta(s + 1, N - s + 1)
x = np.linspace(0, 1, 500)
_f_df = (models.assign(user_name=classifications.reindex(models.index)['user_name'])
.drop(columns=['spiral', 'subject_ids'])
.groupby('user_name')
.agg(number_with_comp)
.reindex(n_cls_by_usr.index)
.assign(n=n_cls_by_usr)
.query('n > 20')
.astype(object)
.apply(
lambda a: pd.Series(np.vectorize(
lambda p: updated_bn(a.n, p)
)(a.iloc[:-1]), index=a.index[:-1]),
axis=1,
)
.applymap(lambda f: f(x))
)
plt.figure(figsize=(8, 3.3), dpi=100)
for i, k in enumerate(('disk', 'bulge', 'bar')):
plt.plot(x, np.mean(_f_df[k]), color=f'C{i}', label=k.capitalize())
plt.fill_between(x, 0, np.mean(_f_df[k]), alpha=0.2, color=f'C{i}')
plt.xlabel(r'$p_{\mathrm{component}}$')
plt.xlim(0, 1)
plt.legend()
plt.tight_layout();
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from IPython.display import display
import os
from os.path import join
from tqdm import tqdm
import scipy.stats as st
import json
import lib.galaxy_utilities as gu
from gzbuilder_analysis import load_aggregation_results, load_fit_results
import gzbuilder_analysis.parsing as pa
import gzbuilder_analysis.fitting as fg
# %run make_bulge_bar_dataframes.py
def number_with_comp(a):
return sum(i is not None for i in a)
def clean_column_names(df):
df_ = df.copy()
df_.columns = [i.strip().replace(' ', '_') for i in df.columns]
return df_
def get_pbar(gal):
n = gal['t03_bar_a06_bar_debiased'] + gal['t03_bar_a07_no_bar_debiased']
return gal['t03_bar_a06_bar_debiased'] / n
from gzbuilder_analysis import load_aggregation_results
agg_results = load_aggregation_results('output_files/aggregation_results')
# load files contain info relating gzb subject ids to GZ2 bulge / bar results:
bulge_df = pd.read_pickle('lib/bulge_fractions.pkl').dropna()
bar_df = pd.read_pickle('lib/bar_fractions.pkl').dropna()
comparison_df = agg_results.agg(dict(
cls=lambda a: len(a.input_models),
disk=lambda a: a.input_models.apply(lambda a: bool(a['disk'])).sum(),
bulge=lambda a: a.input_models.apply(lambda a: bool(a['bulge'])).sum(),
bar=lambda a: a.input_models.apply(lambda a: bool(a['bar'])).sum(),
)).unstack().T
comparison_df = comparison_df.assign(
disk_frac=comparison_df.disk / comparison_df.cls,
bulge_frac=comparison_df.bulge / comparison_df.cls,
bar_frac=comparison_df.bar / comparison_df.cls,
)
comparison_df = comparison_df.assign(
disk_frac_err=np.sqrt(comparison_df.disk_frac * (1 - comparison_df.disk_frac) / comparison_df.cls),
bulge_frac_err=np.sqrt(comparison_df.bulge_frac * (1 - comparison_df.bulge_frac) / comparison_df.cls),
bar_frac_err=np.sqrt(comparison_df.bar_frac * (1 - comparison_df.bar_frac) / comparison_df.cls),
)
# Let's also incorporate knowledge about the aggregagte model (did we cluster a component)
comparison_df = comparison_df.combine_first(
agg_results.apply(lambda a: a.model).apply(pd.Series).applymap(bool).add_prefix('agg_')
)
# and finaly add in information about GZ2:
comparison_df = comparison_df.assign(
GZ2_no_bulge=bulge_df['GZ2 no bulge'],
GZ2_bar_fraction=bar_df['GZ2 bar fraction'],
).dropna().pipe(clean_column_names)
comparison_df.head()
comparison_df.query('GZ2_bar_fraction < 0.2').bar_frac.describe()
comparison_df.query('GZ2_bar_fraction > 0.5').bar_frac.describe()
f, ax = plt.subplots(ncols=2, figsize=(17, 8))
plt.sca(ax[0])
plt.errorbar(
1 - comparison_df['GZ2_no_bulge'],
comparison_df['bulge_frac'], yerr=comparison_df['bulge_frac_err'],
fmt='.', c='C1', elinewidth=1, capsize=1
)
plt.xlim(-0.02, 1.02)
plt.ylim(-0.02, 1.02)
plt.gca().add_line(plt.Line2D((-10, 10), (-10, 10), c='k', alpha=0.1))
plt.xlabel('1 - Galaxy Zoo 2 "no bulge" fraction')
plt.ylabel('Fraction of classifications with a bulge in Galaxy Builder')
gz2_no_bulge, gzb_bulge = comparison_df[['GZ2_no_bulge', 'bulge_frac']].dropna().values.T
bar_corr = st.pearsonr(1 - gz2_no_bulge, gzb_bulge)
plt.title('Pearson correlation coefficient {:.3f}, p={:.3e}'.format(*bar_corr));
plt.sca(ax[1])
plt.errorbar(
comparison_df['GZ2_bar_fraction'], comparison_df['bar_frac'],
yerr=comparison_df['bar_frac_err'],
fmt='.', c='C2', elinewidth=1, capsize=1
)
plt.xlim(-0.02, 1.02)
plt.ylim(-0.02, 1.02)
plt.axvline(0.2, c='k', ls=':')
plt.axvline(0.5, c='k', ls=':')
plt.errorbar(
0.1,
**comparison_df.query('GZ2_bar_fraction < 0.2').bar_frac.describe()
.rename(index=dict(mean='y', std='yerr'))[['y', 'yerr']],
zorder=10, fmt='o', capsize=10, color='k', ms=10
)
plt.errorbar(
0.8,
**comparison_df.query('GZ2_bar_fraction > 0.5').bar_frac.describe()
.rename(index=dict(mean='y', std='yerr'))[['y', 'yerr']],
zorder=10, fmt='o', capsize=10, color='k', ms=10
)
plt.text(0.2 - 0.01, 1.01, 'No Bar', horizontalalignment='right', verticalalignment='top')
plt.text(0.5 + 0.01, 1.01, 'Strongly Barred', horizontalalignment='left', verticalalignment='top')
plt.gca().add_line(plt.Line2D((-10, 10), (-10, 10), c='k', alpha=0.1))
plt.xlabel('Galaxy Zoo 2 "has bar" fraction')
plt.ylabel('Fraction of classifications with a bar in Galaxy Builder')
bar_corr = st.pearsonr(*comparison_df[['GZ2_bar_fraction', 'bar_frac']].dropna().values.T)
plt.title('Pearson correlation coefficient {:.3f}, p={:.2e}'.format(*bar_corr));
f, ax = plt.subplots(ncols=2, figsize=(17, 8))
plt.sca(ax[0])
for i in (False, True):
mask = comparison_df.agg_bulge == i
plt.errorbar(
1 - comparison_df['GZ2_no_bulge'][mask],
comparison_df['bulge_frac'][mask], yerr=comparison_df['bulge_frac_err'][mask],
fmt='o', c=('C2' if i else 'r'), elinewidth=1, capsize=1,
label=('Aggregate has bulge' if i else 'Aggregate does not have bulge')
)
plt.xlim(-0.02, 1.02)
plt.ylim(-0.02, 1.02)
plt.gca().add_line(plt.Line2D((-10, 10), (-10, 10), c='k', alpha=0.1))
plt.xlabel('1 - Galaxy Zoo 2 "no bulge" fraction')
plt.ylabel('Fraction of classifications with a bulge in Galaxy Builder')
gz2_no_bulge, gzb_bulge = comparison_df[['GZ2_no_bulge', 'bulge_frac']].dropna().values.T
bar_corr = st.pearsonr(1 - gz2_no_bulge, gzb_bulge)
plt.title('Pearson correlation coefficient {:.3f}, p={:.3e}'.format(*bar_corr));
plt.sca(ax[1])
for i in (False, True):
mask = comparison_df.agg_bar == i
plt.errorbar(
comparison_df['GZ2_bar_fraction'][mask], comparison_df['bar_frac'][mask],
yerr=comparison_df['bar_frac_err'][mask],
fmt='o', c=('C2' if i else 'r'), ms=5, elinewidth=1, capsize=1,
label=('Aggregate has bar' if i else 'Aggregate does not have bar')
)
plt.xlim(-0.02, 1.02)
plt.ylim(-0.02, 1.02)
plt.axvline(0.2, c='k', ls=':')
plt.axvline(0.5, c='k', ls=':')
plt.errorbar(
0.1,
**comparison_df.query('GZ2_bar_fraction < 0.2').bar_frac.describe()
.rename(index=dict(mean='y', std='yerr'))[['y', 'yerr']],
zorder=10, fmt='o', capsize=10, color='k', ms=10
)
plt.errorbar(
0.8,
**comparison_df.query('GZ2_bar_fraction > 0.5').bar_frac.describe()
.rename(index=dict(mean='y', std='yerr'))[['y', 'yerr']],
zorder=10, fmt='o', capsize=10, color='k', ms=10
)
plt.text(0.2 - 0.01, 1.01, 'No Bar', horizontalalignment='right', verticalalignment='top')
plt.text(0.5 + 0.01, 1.01, 'Strongly Barred', horizontalalignment='left', verticalalignment='top')
plt.legend()
plt.gca().add_line(plt.Line2D((-10, 10), (-10, 10), c='k', alpha=0.1))
plt.xlabel('Galaxy Zoo 2 "has bar" fraction')
plt.ylabel('Fraction of classifications with a bar in Galaxy Builder')
bar_corr = st.pearsonr(*comparison_df[['GZ2_bar_fraction', 'bar_frac']].dropna().values.T)
plt.title('Pearson correlation coefficient {:.3f}, p={:.2e}'.format(*bar_corr));
%%time
classifications = pd.read_csv('lib/galaxy-builder-classifications.csv', index_col=0)
models = (
classifications.query('workflow_version == 61.107')
.apply(pa.parse_classification, image_size=(512, 512),
axis=1, ignore_scale=True)
.apply(pd.Series)
.assign(subject_ids=classifications['subject_ids'].astype('category'))
)
n_cls_by_usr = (
classifications.query('workflow_version == 61.107')
.user_name
.value_counts()
.sort_values()
)
model_freq = (
models.assign(user_name=classifications.reindex(models.index)['user_name'])
.drop(columns=['spiral', 'subject_ids'])
.groupby('user_name')
.agg(number_with_comp)
.reindex(n_cls_by_usr.index)
.T
/ n_cls_by_usr
).T
model_freq.assign(N_classifications=n_cls_by_usr).tail(10)
plt.figure(figsize=(12, 4), dpi=80)
for c in model_freq.columns:
plt.hist(
model_freq[n_cls_by_usr > 20][c].dropna(),
bins='scott', density=True, label=c, alpha=0.4
)
print('Identified {} users with more than 20 classifications'.format(
(n_cls_by_usr > 30).sum()
))
plt.xlabel('Fraction of classifications with component')
plt.ylabel('Density')
plt.legend()
from scipy.special import beta
def updated_bn(N, s):
return lambda x: x**(s)*(1 - x)**(N - s) / beta(s + 1, N - s + 1)
x = np.linspace(0, 1, 500)
_f_df = (models.assign(user_name=classifications.reindex(models.index)['user_name'])
.drop(columns=['spiral', 'subject_ids'])
.groupby('user_name')
.agg(number_with_comp)
.reindex(n_cls_by_usr.index)
.assign(n=n_cls_by_usr)
.query('n > 20')
.astype(object)
.apply(
lambda a: pd.Series(np.vectorize(
lambda p: updated_bn(a.n, p)
)(a.iloc[:-1]), index=a.index[:-1]),
axis=1,
)
.applymap(lambda f: f(x))
)
plt.figure(figsize=(8, 3.3), dpi=100)
for i, k in enumerate(('disk', 'bulge', 'bar')):
plt.plot(x, np.mean(_f_df[k]), color=f'C{i}', label=k.capitalize())
plt.fill_between(x, 0, np.mean(_f_df[k]), alpha=0.2, color=f'C{i}')
plt.xlabel(r'$p_{\mathrm{component}}$')
plt.xlim(0, 1)
plt.legend()
plt.tight_layout();
| 0.584271 | 0.778481 |
```
%logstop
%logstart -ortq ~/.logs/PY_Pythonic.py append
%matplotlib inline
import matplotlib
import seaborn as sns
sns.set()
matplotlib.rcParams['figure.dpi'] = 144
import expectexception
```
# Pythonisms
Much of what we covered in the previous notebook can be fairly generally applicable. Even the Python syntax is quite similar to other languages in the C family. But there are a few things that every language chooses how to do beyond just syntax (although many new languages do take some inspiration from the Python way of doing things). The things we will go over here
* What is Pythonic?
* Float Division
* Python `import` system
* Exceptions
* How to debug Python
Lets start by what we mean by the Python way of doing things.
## `Pythonic`
When learning Python, you will probably browse blogs and other web resources that claim certain things are `Pythonic`. Python has an opinionated way of doing things, mostly captured in the Zen of Python
```
import this
```
`Pythonic` practices are those which the general Python community has agreed are preferable, sometimes this is purely a stylistic consideration and other times it may be related to the way the Python runs.
Making your code `Pythonic` can also be useful when other Python programmers need to interact with it as they will be familiar with the idioms and paradigms you use.
## Imports
In the cells above you might have noticed we used the `import <package>` syntax. This construct allows us to include code from other python files or more generally modules (collections of files) and packages (collection of modules) into the current code we are working with. For the purposes of this course, we have installed all the packages you will need on your machine, but for working with packages, some recommended tools are
- conda
- pip
With installed packages (usually installed with one of those two "package managers"), we can import the package with the `import` command. We can also import only parts of the package. For example, one package we will use in the course is called `pandas`. We can import `pandas`
```
import pandas
pandas
```
We can also import pandas, but call it something else (saves a bit of typing and is conventional for some of the main packages in the Python scientific stack).
```
import pandas as pd
pd
```
Now when we want to use a function or class from pandas, we need to call it with the syntax `pd.function` or `pd.class`. For example, the `DataFrame` object
```
pd.DataFrame
```
Note that this DataFrame does not exist in the main namespace.
```
%%expect_exception NameError
DataFrame
```
We can also just import parts of a package, we can even import them and give them another name!
```
from pandas import DataFrame as dframe
dframe
```
Another thing we can do is to import everything into the main namespace using the syntax
```python
from pandas import *
```
This is highly discouraged because it can cause problems when multiple packages have a function or class with the same name (not uncommon, think about a function like `.info`).
We have covered the basic mechanics of the import system, but what does it allow us to do? Having a sane packaging system allows Python users to package bundles of functionality into modules and packages which can be imported into other bits of codes. If well written, these packages operate mostly like black boxes, where the user understands _what_ the package is doing, but not necessarily _how_ it is performing its functionality.
While it may seem like this is giving up too much control, most of us don't understand exactly how our computer processor works, or even the keyboard, yet we are perfectly comfortable using them to serve their purpose. Packages are similar and when written well can be invaluable tools that allows us incorporate well written tested code that does powerful things into our applications with very little difficulty.
## Standard Library
One useful thing we can do with `import` statements is import packages in the Python standard library. These are packages which are packaged with the interpreter and available on (almost) any Python installation. These packages server a wide variety of purposes, here we have listed just a few along with their description. For the rest, checkout the [documentation](https://docs.python.org/2/library/).
- `collections` - containers
- `re` - regular expressions
- `datetime` - date and time handling
- `heapq` - the heap queue algorithm
- `itertools` - functions for help with iteration
- `functools` - function to assist with functional programming
- `os` - operating system interfaces
- `sys` - system functions
- `pickle` - serialize Python objects
- `gzip` - work with Gzipped files
- `time` - time access
- `argparse` - command line argument handling
- `threading` - threading interface
- `multiprocessing` - process based "threading"
- `subprocess` - subprocess management
- `unittest` - testing tools
- `pdb` - debugger
These packages are optimized, reliable, and available anywhere there is a Python installation, so use them when you can!
## Exceptions
An exception is something that deviates from the norm. In Python its no different, exceptions are when your program deviates from expected behavior. The Python interpreter will attempt to execute any code that it's given and when it can't, it will raise an `Exception`. In our notebooks you will note the `%%expect_exception` magic. This is just a sign that we know there will be an exception in that cell. For example, lets try to add a number to a string.
```
%%expect_exception TypeError
2 + '3'
```
We can see that this raises a `TypeError` because Python doesn't know how to add a string and an integer together (Python will not coerce one of the values into a different type; remember the Zen of Python: 'In the face of ambiguity, refuse the temptation to guess'). Exceptions are often very readable and helpful to debug code, however, we can also write code to handle exceptions when they occur. Lets write a function which adds to things together (basically just another version of the add function) except it will catch the `TypeError` and do some conversion.
```
def add(x, y):
try:
return x + y
except TypeError:
return float(x) + float(y)
```
Now lets run something similar to the previous example
```
add(2, '3')
```
As seen above, the way to handle Exceptions is with the `try` and `except` keywords. The `try` block specifies a bit of code to try to run and the `except` block handles all exceptions that are specifically enumerated. One can also catch all exceptions by doing
```python
try:
func()
except:
handle_exception()
```
But this is not generally a good idea since Python uses Exceptions for all sorts of things (sometimes even exiting programs) and you don't want to catch Exceptions which Python is using for a different purpose. Think of `Exception` handling as handling the small probability things that will happen in your code, not as a tool to anticipate anything.
We have seen exceptions, but what are the alternative? One option, used by other languages is to test ahead of time that conditions necessary to proceed are met. We can rewrite the add function in a different way.
```
def add_2(x,y):
if not isinstance(x, (float, int)):
x = float(x)
if not isinstance(y, (float, int)):
y = float(y)
return x + y
add_2(2, '3')
```
This also works, but its not Pythonic. The Pythonic way of thinking about this is roughly analogous to "its easier to ask for forgiveness than permission". Throwing exceptions actually has other positive benefits, such as the ability to handle errors at higher level code instead of in low level functions.
What we mean by this is if we have a series of functions `f_a,f_b,f_c` and `f_a` calls `f_b` which calls `f_c`, we can choose to handle an exception in `f_c` in any of these functions!
## Python Debugging
We have seen how to handle errors with `Exceptions`, but how do we figure out whats wrong when we have errors that we haven't handled?
Lets look again at our previous example.
```
%%expect_exception TypeError
2 + '3'
```
If we look at the returned text, referred to as a `Traceback`, we can see much useful information. Tracebacks should be read starting from the bottom and working up. In this case the Traceback tells us exactly what happened, we tried to add an `int` and a `str` and there is no way to do this. It even points to the exact line of code where this error occurs.
Lets take a look at a more complicated Traceback. We will create a pandas `DataFrame` with illegal arguments.
```
%%expect_exception ValueError
pd.DataFrame(['one','two','three'],['test'])
```
If we look to the bottom, we can see that this is caused by an improper shape of the arrays we have passed into the `DataFrame` function. We can trace our way back up through the code to see all the functions which were called in order to get to this error. In this case, there were four called, `DataFrame, _init_ndarray, create_block_manager_from_blocks, construction_error`.
Learning how to read Tracebacks and especially to figure out why simple bits of code are failing is an important part to becoming a good Python programmer.
### Exercise
Run the following bits of code in new cells and determine the error, fix the errors in a sensible way.
```python
# Example 1
float([1])
# Example 2
a = []
a[1]
# Example 3
pd.DataFrame(['one','two','three'],['test'])
```
*Copyright © 2019 The Data Incubator. All rights reserved.*
|
github_jupyter
|
%logstop
%logstart -ortq ~/.logs/PY_Pythonic.py append
%matplotlib inline
import matplotlib
import seaborn as sns
sns.set()
matplotlib.rcParams['figure.dpi'] = 144
import expectexception
import this
import pandas
pandas
import pandas as pd
pd
pd.DataFrame
%%expect_exception NameError
DataFrame
from pandas import DataFrame as dframe
dframe
from pandas import *
%%expect_exception TypeError
2 + '3'
def add(x, y):
try:
return x + y
except TypeError:
return float(x) + float(y)
add(2, '3')
try:
func()
except:
handle_exception()
def add_2(x,y):
if not isinstance(x, (float, int)):
x = float(x)
if not isinstance(y, (float, int)):
y = float(y)
return x + y
add_2(2, '3')
%%expect_exception TypeError
2 + '3'
%%expect_exception ValueError
pd.DataFrame(['one','two','three'],['test'])
# Example 1
float([1])
# Example 2
a = []
a[1]
# Example 3
pd.DataFrame(['one','two','three'],['test'])
| 0.437343 | 0.905865 |
```
# Зависимости
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import random
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.compose import ColumnTransformer
from sklearn.tree import DecisionTreeRegressor, DecisionTreeClassifier, plot_tree
from sklearn.metrics import mean_squared_error, f1_score
from sklearn.datasets import load_iris
from sklearn import tree
# Генерируем уникальный seed
my_code = "Маматбеков"
seed_limit = 2 ** 32
my_seed = int.from_bytes(my_code.encode(), "little") % seed_limit
# Читаем данные из файла
example_data = pd.read_csv("../datasets/Fish.csv")
example_data.head()
# Определим размер валидационной и тестовой выборок
val_test_size = round(0.2*len(example_data))
print(val_test_size)
# Создадим обучающую, валидационную и тестовую выборки
random_state = my_seed
train_val, test = train_test_split(example_data, test_size=val_test_size, random_state=random_state)
train, val = train_test_split(train_val, test_size=val_test_size, random_state=random_state)
print(len(train), len(val), len(test))
# Значения в числовых столбцах преобразуем к отрезку [0,1].
# Для настройки скалировщика используем только обучающую выборку.
num_columns = ['Weight', 'Length1', 'Length2', 'Length3', 'Height', 'Width']
ct = ColumnTransformer(transformers=[('numerical', MinMaxScaler(), num_columns)], remainder='passthrough')
ct.fit(train)
# Преобразуем значения, тип данных приводим к DataFrame
sc_train = pd.DataFrame(ct.transform(train))
sc_test = pd.DataFrame(ct.transform(test))
sc_val = pd.DataFrame(ct.transform(val))
# Устанавливаем названия столбцов
column_names = num_columns + ['Species']
sc_train.columns = column_names
sc_test.columns = column_names
sc_val.columns = column_names
sc_train
# Задание №1 - анализ деревьев принятия решений в задаче регрессии
# https://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeRegressor.html
# criterion : {“mse”, “friedman_mse”, “mae”, “poisson”}, default=”mse”
# splitter : {“best”, “random”}, default=”best”
# max_depth : int, default=None
# min_samples_split : int or float, default=2
# min_samples_leaf : int or float, default=1
# Выбираем 4 числовых переменных, три их них будут предикторами, одна - зависимой переменной
n = 4
labels = random.sample(num_columns, n)
y_label = labels[0]
x_labels = labels[1:]
print(x_labels)
print(y_label)
# Отберем необходимые параметры
x_train = sc_train[x_labels]
x_test = sc_test[x_labels]
x_val = sc_val[x_labels]
y_train = sc_train[y_label]
y_test = sc_test[y_label]
y_val = sc_val[y_label]
x_train
# Создайте 4 модели с различными критериями ветвления criterion: 'mse', 'friedman_mse', 'mae', 'poisson'.
# Решите получившуюся задачу регрессии с помощью созданных моделей и сравните их эффективность.
# При необходимости применяйте параметры splitter, max_depth, min_samples_split, min_samples_leaf
# Укажите, какая модель решает задачу лучше других.
r_model1 = DecisionTreeRegressor(criterion='mse', splitter='random', max_depth=2, min_samples_split=4, min_samples_leaf=0.5)
r_model2 = DecisionTreeRegressor(criterion='friedman_mse', splitter='best', max_depth=3, min_samples_split=4, min_samples_leaf=0.5)
r_model3 = DecisionTreeRegressor(criterion='mae')
r_model4 = DecisionTreeRegressor(criterion='poisson', splitter='random', max_depth=1, min_samples_split=2, min_samples_leaf=1)
r_models = []
r_models.append(r_model1)
r_models.append(r_model2)
r_models.append(r_model3)
r_models.append(r_model4)
# Обучаем модели
for model in r_models:
model.fit(x_train, y_train)
# Оценииваем качество работы моделей на валидационной выборке
mses = []
for model in r_models:
val_pred = model.predict(x_val)
mse = mean_squared_error(y_val, val_pred)
mses.append(mse)
print(mse)
# Выбираем лучшую модель
i_min = mses.index(min(mses))
best_r_model = r_models[i_min]
best_r_model.get_params()
# Вычислим ошибку лучшей модели на тестовой выборке.
test_pred = best_r_model.predict(x_test)
mse = mean_squared_error(y_test, test_pred)
print(mse)
# Вывод на экран дерева tree.
# max_depth - максимальная губина отображения, по умолчанию выводится дерево целиком.
plot_tree(best_r_model, max_depth=1)
plt.show()
plot_tree(best_r_model)
plt.show()
# Задание №2 - анализ деревьев принятия решений в задаче классификации
# https://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeClassifier.html
# criterion : {“gini”, “entropy”}, default=”gini”
# splitter : {“best”, “random”}, default=”best”
# max_depth : int, default=None
# min_samples_split : int or float, default=2
# min_samples_leaf : int or float, default=1
# Выбираем 2 числовых переменных, которые будут параметрами элементов набора данных
# Метка класса всегда 'Species'
n = 2
x_labels = random.sample(num_columns, n)
y_label = 'Species'
print(x_labels)
print(y_label)
# Отберем необходимые параметры
x_train = sc_train[x_labels]
x_test = sc_test[x_labels]
x_val = sc_val[x_labels]
y_train = sc_train[y_label]
y_test = sc_test[y_label]
y_val = sc_val[y_label]
x_train
# Создайте 4 модели с различными критериями ветвления criterion : 'gini', 'entropy' и splitter : 'best', 'random'.
# Решите получившуюся задачу классификации с помощью созданных моделей и сравните их эффективность.
# При необходимости применяйте параметры max_depth, min_samples_split, min_samples_leaf
# Укажите, какая модель решает задачу лучше других.
d_model1 = DecisionTreeClassifier(criterion='gini', splitter='best', max_depth=2, min_samples_split=3, min_samples_leaf=1)
d_model2 = DecisionTreeClassifier(criterion='gini', splitter='random', max_depth=1, min_samples_split=4, min_samples_leaf=2)
d_model3 = DecisionTreeClassifier(criterion='entropy', splitter='best', max_depth=1, min_samples_split=2, min_samples_leaf=2)
d_model4 = DecisionTreeClassifier(criterion='entropy', splitter='random', max_depth=2, min_samples_split=5, min_samples_leaf=1)
d_models = []
d_models.append(d_model1)
d_models.append(d_model2)
d_models.append(d_model3)
d_models.append(d_model4)
# Обучаем модели
for model in d_models:
model.fit(x_train, y_train)
# Оценииваем качество работы моделей на валидационной выборке.
f1s = []
for model in d_models:
val_pred = model.predict(x_val)
f1 = f1_score(y_val, val_pred, average='weighted')
f1s.append(f1)
print(f1)
# Выбираем лучшую модель
i_max = f1s.index(max(f1s))
best_d_model = d_models[i_max]
best_d_model.get_params()
# Вычислим ошибку лучшей модели на тестовой выборке.
test_pred = best_d_model.predict(x_test)
f1 = f1_score(y_test, test_pred, average='weighted')
print(f1)
# Вывод на экран дерева tree.
# max_depth - максимальная губина отображения, по умолчанию выводится дерево целиком.
plot_tree(best_d_model, max_depth=1)
plt.show()
plot_tree(best_d_model)
plt.show()
```
|
github_jupyter
|
# Зависимости
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import random
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.compose import ColumnTransformer
from sklearn.tree import DecisionTreeRegressor, DecisionTreeClassifier, plot_tree
from sklearn.metrics import mean_squared_error, f1_score
from sklearn.datasets import load_iris
from sklearn import tree
# Генерируем уникальный seed
my_code = "Маматбеков"
seed_limit = 2 ** 32
my_seed = int.from_bytes(my_code.encode(), "little") % seed_limit
# Читаем данные из файла
example_data = pd.read_csv("../datasets/Fish.csv")
example_data.head()
# Определим размер валидационной и тестовой выборок
val_test_size = round(0.2*len(example_data))
print(val_test_size)
# Создадим обучающую, валидационную и тестовую выборки
random_state = my_seed
train_val, test = train_test_split(example_data, test_size=val_test_size, random_state=random_state)
train, val = train_test_split(train_val, test_size=val_test_size, random_state=random_state)
print(len(train), len(val), len(test))
# Значения в числовых столбцах преобразуем к отрезку [0,1].
# Для настройки скалировщика используем только обучающую выборку.
num_columns = ['Weight', 'Length1', 'Length2', 'Length3', 'Height', 'Width']
ct = ColumnTransformer(transformers=[('numerical', MinMaxScaler(), num_columns)], remainder='passthrough')
ct.fit(train)
# Преобразуем значения, тип данных приводим к DataFrame
sc_train = pd.DataFrame(ct.transform(train))
sc_test = pd.DataFrame(ct.transform(test))
sc_val = pd.DataFrame(ct.transform(val))
# Устанавливаем названия столбцов
column_names = num_columns + ['Species']
sc_train.columns = column_names
sc_test.columns = column_names
sc_val.columns = column_names
sc_train
# Задание №1 - анализ деревьев принятия решений в задаче регрессии
# https://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeRegressor.html
# criterion : {“mse”, “friedman_mse”, “mae”, “poisson”}, default=”mse”
# splitter : {“best”, “random”}, default=”best”
# max_depth : int, default=None
# min_samples_split : int or float, default=2
# min_samples_leaf : int or float, default=1
# Выбираем 4 числовых переменных, три их них будут предикторами, одна - зависимой переменной
n = 4
labels = random.sample(num_columns, n)
y_label = labels[0]
x_labels = labels[1:]
print(x_labels)
print(y_label)
# Отберем необходимые параметры
x_train = sc_train[x_labels]
x_test = sc_test[x_labels]
x_val = sc_val[x_labels]
y_train = sc_train[y_label]
y_test = sc_test[y_label]
y_val = sc_val[y_label]
x_train
# Создайте 4 модели с различными критериями ветвления criterion: 'mse', 'friedman_mse', 'mae', 'poisson'.
# Решите получившуюся задачу регрессии с помощью созданных моделей и сравните их эффективность.
# При необходимости применяйте параметры splitter, max_depth, min_samples_split, min_samples_leaf
# Укажите, какая модель решает задачу лучше других.
r_model1 = DecisionTreeRegressor(criterion='mse', splitter='random', max_depth=2, min_samples_split=4, min_samples_leaf=0.5)
r_model2 = DecisionTreeRegressor(criterion='friedman_mse', splitter='best', max_depth=3, min_samples_split=4, min_samples_leaf=0.5)
r_model3 = DecisionTreeRegressor(criterion='mae')
r_model4 = DecisionTreeRegressor(criterion='poisson', splitter='random', max_depth=1, min_samples_split=2, min_samples_leaf=1)
r_models = []
r_models.append(r_model1)
r_models.append(r_model2)
r_models.append(r_model3)
r_models.append(r_model4)
# Обучаем модели
for model in r_models:
model.fit(x_train, y_train)
# Оценииваем качество работы моделей на валидационной выборке
mses = []
for model in r_models:
val_pred = model.predict(x_val)
mse = mean_squared_error(y_val, val_pred)
mses.append(mse)
print(mse)
# Выбираем лучшую модель
i_min = mses.index(min(mses))
best_r_model = r_models[i_min]
best_r_model.get_params()
# Вычислим ошибку лучшей модели на тестовой выборке.
test_pred = best_r_model.predict(x_test)
mse = mean_squared_error(y_test, test_pred)
print(mse)
# Вывод на экран дерева tree.
# max_depth - максимальная губина отображения, по умолчанию выводится дерево целиком.
plot_tree(best_r_model, max_depth=1)
plt.show()
plot_tree(best_r_model)
plt.show()
# Задание №2 - анализ деревьев принятия решений в задаче классификации
# https://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeClassifier.html
# criterion : {“gini”, “entropy”}, default=”gini”
# splitter : {“best”, “random”}, default=”best”
# max_depth : int, default=None
# min_samples_split : int or float, default=2
# min_samples_leaf : int or float, default=1
# Выбираем 2 числовых переменных, которые будут параметрами элементов набора данных
# Метка класса всегда 'Species'
n = 2
x_labels = random.sample(num_columns, n)
y_label = 'Species'
print(x_labels)
print(y_label)
# Отберем необходимые параметры
x_train = sc_train[x_labels]
x_test = sc_test[x_labels]
x_val = sc_val[x_labels]
y_train = sc_train[y_label]
y_test = sc_test[y_label]
y_val = sc_val[y_label]
x_train
# Создайте 4 модели с различными критериями ветвления criterion : 'gini', 'entropy' и splitter : 'best', 'random'.
# Решите получившуюся задачу классификации с помощью созданных моделей и сравните их эффективность.
# При необходимости применяйте параметры max_depth, min_samples_split, min_samples_leaf
# Укажите, какая модель решает задачу лучше других.
d_model1 = DecisionTreeClassifier(criterion='gini', splitter='best', max_depth=2, min_samples_split=3, min_samples_leaf=1)
d_model2 = DecisionTreeClassifier(criterion='gini', splitter='random', max_depth=1, min_samples_split=4, min_samples_leaf=2)
d_model3 = DecisionTreeClassifier(criterion='entropy', splitter='best', max_depth=1, min_samples_split=2, min_samples_leaf=2)
d_model4 = DecisionTreeClassifier(criterion='entropy', splitter='random', max_depth=2, min_samples_split=5, min_samples_leaf=1)
d_models = []
d_models.append(d_model1)
d_models.append(d_model2)
d_models.append(d_model3)
d_models.append(d_model4)
# Обучаем модели
for model in d_models:
model.fit(x_train, y_train)
# Оценииваем качество работы моделей на валидационной выборке.
f1s = []
for model in d_models:
val_pred = model.predict(x_val)
f1 = f1_score(y_val, val_pred, average='weighted')
f1s.append(f1)
print(f1)
# Выбираем лучшую модель
i_max = f1s.index(max(f1s))
best_d_model = d_models[i_max]
best_d_model.get_params()
# Вычислим ошибку лучшей модели на тестовой выборке.
test_pred = best_d_model.predict(x_test)
f1 = f1_score(y_test, test_pred, average='weighted')
print(f1)
# Вывод на экран дерева tree.
# max_depth - максимальная губина отображения, по умолчанию выводится дерево целиком.
plot_tree(best_d_model, max_depth=1)
plt.show()
plot_tree(best_d_model)
plt.show()
| 0.448426 | 0.609117 |
```
%run homework_modules.ipynb
import torch
from torch.autograd import Variable
import numpy as np
import unittest
class TestLayers(unittest.TestCase):
def test_Linear(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in, n_out = 2, 3, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.Linear(n_in, n_out)
custom_layer = Linear(n_in, n_out)
custom_layer.W = torch_layer.weight.data.numpy()
custom_layer.b = torch_layer.bias.data.numpy()
layer_input = np.random.uniform(-10, 10, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-10, 10, (batch_size, n_out)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
# 3. check layer parameters grad
custom_layer.accGradParameters(layer_input, next_layer_grad)
weight_grad = custom_layer.gradW
bias_grad = custom_layer.gradb
torch_weight_grad = torch_layer.weight.grad.data.numpy()
torch_bias_grad = torch_layer.bias.grad.data.numpy()
self.assertTrue(np.allclose(torch_weight_grad, weight_grad, atol=1e-6))
self.assertTrue(np.allclose(torch_bias_grad, bias_grad, atol=1e-6))
def test_SoftMax(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.Softmax(dim=1)
custom_layer = SoftMax()
layer_input = np.random.uniform(-10, 10, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.random((batch_size, n_in)).astype(np.float32)
next_layer_grad /= next_layer_grad.sum(axis=-1, keepdims=True)
next_layer_grad = next_layer_grad.clip(1e-5,1.)
next_layer_grad = 1. / next_layer_grad
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-5))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-5))
def test_LogSoftMax(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.LogSoftmax(dim=1)
custom_layer = LogSoftMax()
layer_input = np.random.uniform(-10, 10, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.random((batch_size, n_in)).astype(np.float32)
next_layer_grad /= next_layer_grad.sum(axis=-1, keepdims=True)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
def test_BatchNormalization(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 32, 16
for _ in range(100):
# layers initialization
slope = np.random.uniform(0.01, 0.05)
alpha = 0.9
custom_layer = BatchNormalization(alpha)
custom_layer.train()
torch_layer = torch.nn.BatchNorm1d(n_in, eps=custom_layer.EPS, momentum=1.-alpha, affine=False)
custom_layer.moving_mean = torch_layer.running_mean.numpy().copy()
custom_layer.moving_variance = torch_layer.running_var.numpy().copy()
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-5))
# 3. check moving mean
##print(custom_layer.moving_mean, torch_layer.running_mean.numpy())
self.assertTrue(np.allclose(custom_layer.moving_mean, torch_layer.running_mean.numpy()))
# we don't check moving_variance because pytorch uses slightly different formula for it:
# it computes moving average for unbiased variance (i.e var*N/(N-1))
#self.assertTrue(np.allclose(custom_layer.moving_variance, torch_layer.running_var.numpy()))
# 4. check evaluation mode
custom_layer.moving_variance = torch_layer.running_var.numpy().copy()
custom_layer.evaluate()
custom_layer_output = custom_layer.updateOutput(layer_input)
torch_layer.eval()
torch_layer_output_var = torch_layer(layer_input_var)
# for x,y in zip(torch_layer_output_var.data.numpy(), custom_layer_output):
# print(x)
# print(y)
# print()
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
def test_Sequential(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
alpha = 0.9
torch_layer = torch.nn.BatchNorm1d(n_in, eps=BatchNormalization.EPS, momentum=1.-alpha, affine=True)
torch_layer.bias.data = torch.from_numpy(np.random.random(n_in).astype(np.float32))
custom_layer = Sequential()
bn_layer = BatchNormalization(alpha)
bn_layer.moving_mean = torch_layer.running_mean.numpy().copy()
bn_layer.moving_variance = torch_layer.running_var.numpy().copy()
custom_layer.add(bn_layer)
scaling_layer = ChannelwiseScaling(n_in)
scaling_layer.gamma = torch_layer.weight.data.numpy()
scaling_layer.beta = torch_layer.bias.data.numpy()
custom_layer.add(scaling_layer)
custom_layer.train()
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.backward(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-5))
# 3. check layer parameters grad
weight_grad, bias_grad = custom_layer.getGradParameters()[1]
torch_weight_grad = torch_layer.weight.grad.data.numpy()
torch_bias_grad = torch_layer.bias.grad.data.numpy()
self.assertTrue(np.allclose(torch_weight_grad, weight_grad, atol=1e-6))
self.assertTrue(np.allclose(torch_bias_grad, bias_grad, atol=1e-6))
def test_Dropout(self):
np.random.seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
p = np.random.uniform(0.3, 0.7)
layer = Dropout(p)
layer.train()
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
layer_output = layer.updateOutput(layer_input)
self.assertTrue(np.all(np.logical_or(np.isclose(layer_output, 0),
np.isclose(layer_output*(1.-p), layer_input))))
# 2. check layer input grad
layer_grad = layer.updateGradInput(layer_input, next_layer_grad)
self.assertTrue(np.all(np.logical_or(np.isclose(layer_grad, 0),
np.isclose(layer_grad*(1.-p), next_layer_grad))))
# 3. check evaluation mode
layer.evaluate()
layer_output = layer.updateOutput(layer_input)
self.assertTrue(np.allclose(layer_output, layer_input))
# 4. check mask
p = 0.0
layer = Dropout(p)
layer.train()
layer_output = layer.updateOutput(layer_input)
self.assertTrue(np.allclose(layer_output, layer_input))
p = 0.5
layer = Dropout(p)
layer.train()
layer_input = np.random.uniform(5, 10, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(5, 10, (batch_size, n_in)).astype(np.float32)
layer_output = layer.updateOutput(layer_input)
zeroed_elem_mask = np.isclose(layer_output, 0)
layer_grad = layer.updateGradInput(layer_input, next_layer_grad)
self.assertTrue(np.all(zeroed_elem_mask == np.isclose(layer_grad, 0)))
# 5. dropout mask should be generated independently for every input matrix element, not for row/column
batch_size, n_in = 1000, 1
p = 0.8
layer = Dropout(p)
layer.train()
layer_input = np.random.uniform(5, 10, (batch_size, n_in)).astype(np.float32)
layer_output = layer.updateOutput(layer_input)
self.assertTrue(np.sum(np.isclose(layer_output, 0)) != layer_input.size)
layer_input = layer_input.T
layer_output = layer.updateOutput(layer_input)
self.assertTrue(np.sum(np.isclose(layer_output, 0)) != layer_input.size)
def test_LeakyReLU(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
slope = np.random.uniform(0.01, 0.05)
torch_layer = torch.nn.LeakyReLU(slope)
custom_layer = LeakyReLU(slope)
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
def test_ELU(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
alpha = 1.0
torch_layer = torch.nn.ELU(alpha)
custom_layer = ELU(alpha)
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
def test_SoftPlus(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.Softplus()
custom_layer = SoftPlus()
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
def test_ClassNLLCriterionUnstable(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.NLLLoss()
custom_layer = ClassNLLCriterionUnstable()
layer_input = np.random.uniform(0, 1, (batch_size, n_in)).astype(np.float32)
layer_input /= layer_input.sum(axis=-1, keepdims=True)
layer_input = layer_input.clip(custom_layer.EPS, 1. - custom_layer.EPS) # unifies input
target_labels = np.random.choice(n_in, batch_size)
target = np.zeros((batch_size, n_in), np.float32)
target[np.arange(batch_size), target_labels] = 1 # one-hot encoding
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input, target)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(torch.log(layer_input_var),
Variable(torch.from_numpy(target_labels), requires_grad=False))
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, target)
torch_layer_output_var.backward()
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
def test_ClassNLLCriterion(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.NLLLoss()
custom_layer = ClassNLLCriterion()
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
layer_input = torch.nn.LogSoftmax(dim=1)(Variable(torch.from_numpy(layer_input))).data.numpy()
target_labels = np.random.choice(n_in, batch_size)
target = np.zeros((batch_size, n_in), np.float32)
target[np.arange(batch_size), target_labels] = 1 # one-hot encoding
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input, target)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var,
Variable(torch.from_numpy(target_labels), requires_grad=False))
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, target)
torch_layer_output_var.backward()
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
suite = unittest.TestLoader().loadTestsFromTestCase(TestLayers)
unittest.TextTestRunner(verbosity=2).run(suite)
```
|
github_jupyter
|
%run homework_modules.ipynb
import torch
from torch.autograd import Variable
import numpy as np
import unittest
class TestLayers(unittest.TestCase):
def test_Linear(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in, n_out = 2, 3, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.Linear(n_in, n_out)
custom_layer = Linear(n_in, n_out)
custom_layer.W = torch_layer.weight.data.numpy()
custom_layer.b = torch_layer.bias.data.numpy()
layer_input = np.random.uniform(-10, 10, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-10, 10, (batch_size, n_out)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
# 3. check layer parameters grad
custom_layer.accGradParameters(layer_input, next_layer_grad)
weight_grad = custom_layer.gradW
bias_grad = custom_layer.gradb
torch_weight_grad = torch_layer.weight.grad.data.numpy()
torch_bias_grad = torch_layer.bias.grad.data.numpy()
self.assertTrue(np.allclose(torch_weight_grad, weight_grad, atol=1e-6))
self.assertTrue(np.allclose(torch_bias_grad, bias_grad, atol=1e-6))
def test_SoftMax(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.Softmax(dim=1)
custom_layer = SoftMax()
layer_input = np.random.uniform(-10, 10, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.random((batch_size, n_in)).astype(np.float32)
next_layer_grad /= next_layer_grad.sum(axis=-1, keepdims=True)
next_layer_grad = next_layer_grad.clip(1e-5,1.)
next_layer_grad = 1. / next_layer_grad
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-5))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-5))
def test_LogSoftMax(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.LogSoftmax(dim=1)
custom_layer = LogSoftMax()
layer_input = np.random.uniform(-10, 10, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.random((batch_size, n_in)).astype(np.float32)
next_layer_grad /= next_layer_grad.sum(axis=-1, keepdims=True)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
def test_BatchNormalization(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 32, 16
for _ in range(100):
# layers initialization
slope = np.random.uniform(0.01, 0.05)
alpha = 0.9
custom_layer = BatchNormalization(alpha)
custom_layer.train()
torch_layer = torch.nn.BatchNorm1d(n_in, eps=custom_layer.EPS, momentum=1.-alpha, affine=False)
custom_layer.moving_mean = torch_layer.running_mean.numpy().copy()
custom_layer.moving_variance = torch_layer.running_var.numpy().copy()
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-5))
# 3. check moving mean
##print(custom_layer.moving_mean, torch_layer.running_mean.numpy())
self.assertTrue(np.allclose(custom_layer.moving_mean, torch_layer.running_mean.numpy()))
# we don't check moving_variance because pytorch uses slightly different formula for it:
# it computes moving average for unbiased variance (i.e var*N/(N-1))
#self.assertTrue(np.allclose(custom_layer.moving_variance, torch_layer.running_var.numpy()))
# 4. check evaluation mode
custom_layer.moving_variance = torch_layer.running_var.numpy().copy()
custom_layer.evaluate()
custom_layer_output = custom_layer.updateOutput(layer_input)
torch_layer.eval()
torch_layer_output_var = torch_layer(layer_input_var)
# for x,y in zip(torch_layer_output_var.data.numpy(), custom_layer_output):
# print(x)
# print(y)
# print()
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
def test_Sequential(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
alpha = 0.9
torch_layer = torch.nn.BatchNorm1d(n_in, eps=BatchNormalization.EPS, momentum=1.-alpha, affine=True)
torch_layer.bias.data = torch.from_numpy(np.random.random(n_in).astype(np.float32))
custom_layer = Sequential()
bn_layer = BatchNormalization(alpha)
bn_layer.moving_mean = torch_layer.running_mean.numpy().copy()
bn_layer.moving_variance = torch_layer.running_var.numpy().copy()
custom_layer.add(bn_layer)
scaling_layer = ChannelwiseScaling(n_in)
scaling_layer.gamma = torch_layer.weight.data.numpy()
scaling_layer.beta = torch_layer.bias.data.numpy()
custom_layer.add(scaling_layer)
custom_layer.train()
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.backward(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-5))
# 3. check layer parameters grad
weight_grad, bias_grad = custom_layer.getGradParameters()[1]
torch_weight_grad = torch_layer.weight.grad.data.numpy()
torch_bias_grad = torch_layer.bias.grad.data.numpy()
self.assertTrue(np.allclose(torch_weight_grad, weight_grad, atol=1e-6))
self.assertTrue(np.allclose(torch_bias_grad, bias_grad, atol=1e-6))
def test_Dropout(self):
np.random.seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
p = np.random.uniform(0.3, 0.7)
layer = Dropout(p)
layer.train()
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
layer_output = layer.updateOutput(layer_input)
self.assertTrue(np.all(np.logical_or(np.isclose(layer_output, 0),
np.isclose(layer_output*(1.-p), layer_input))))
# 2. check layer input grad
layer_grad = layer.updateGradInput(layer_input, next_layer_grad)
self.assertTrue(np.all(np.logical_or(np.isclose(layer_grad, 0),
np.isclose(layer_grad*(1.-p), next_layer_grad))))
# 3. check evaluation mode
layer.evaluate()
layer_output = layer.updateOutput(layer_input)
self.assertTrue(np.allclose(layer_output, layer_input))
# 4. check mask
p = 0.0
layer = Dropout(p)
layer.train()
layer_output = layer.updateOutput(layer_input)
self.assertTrue(np.allclose(layer_output, layer_input))
p = 0.5
layer = Dropout(p)
layer.train()
layer_input = np.random.uniform(5, 10, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(5, 10, (batch_size, n_in)).astype(np.float32)
layer_output = layer.updateOutput(layer_input)
zeroed_elem_mask = np.isclose(layer_output, 0)
layer_grad = layer.updateGradInput(layer_input, next_layer_grad)
self.assertTrue(np.all(zeroed_elem_mask == np.isclose(layer_grad, 0)))
# 5. dropout mask should be generated independently for every input matrix element, not for row/column
batch_size, n_in = 1000, 1
p = 0.8
layer = Dropout(p)
layer.train()
layer_input = np.random.uniform(5, 10, (batch_size, n_in)).astype(np.float32)
layer_output = layer.updateOutput(layer_input)
self.assertTrue(np.sum(np.isclose(layer_output, 0)) != layer_input.size)
layer_input = layer_input.T
layer_output = layer.updateOutput(layer_input)
self.assertTrue(np.sum(np.isclose(layer_output, 0)) != layer_input.size)
def test_LeakyReLU(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
slope = np.random.uniform(0.01, 0.05)
torch_layer = torch.nn.LeakyReLU(slope)
custom_layer = LeakyReLU(slope)
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
def test_ELU(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
alpha = 1.0
torch_layer = torch.nn.ELU(alpha)
custom_layer = ELU(alpha)
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
def test_SoftPlus(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.Softplus()
custom_layer = SoftPlus()
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var)
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)
torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
def test_ClassNLLCriterionUnstable(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.NLLLoss()
custom_layer = ClassNLLCriterionUnstable()
layer_input = np.random.uniform(0, 1, (batch_size, n_in)).astype(np.float32)
layer_input /= layer_input.sum(axis=-1, keepdims=True)
layer_input = layer_input.clip(custom_layer.EPS, 1. - custom_layer.EPS) # unifies input
target_labels = np.random.choice(n_in, batch_size)
target = np.zeros((batch_size, n_in), np.float32)
target[np.arange(batch_size), target_labels] = 1 # one-hot encoding
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input, target)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(torch.log(layer_input_var),
Variable(torch.from_numpy(target_labels), requires_grad=False))
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, target)
torch_layer_output_var.backward()
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
def test_ClassNLLCriterion(self):
np.random.seed(42)
torch.manual_seed(42)
batch_size, n_in = 2, 4
for _ in range(100):
# layers initialization
torch_layer = torch.nn.NLLLoss()
custom_layer = ClassNLLCriterion()
layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)
layer_input = torch.nn.LogSoftmax(dim=1)(Variable(torch.from_numpy(layer_input))).data.numpy()
target_labels = np.random.choice(n_in, batch_size)
target = np.zeros((batch_size, n_in), np.float32)
target[np.arange(batch_size), target_labels] = 1 # one-hot encoding
# 1. check layer output
custom_layer_output = custom_layer.updateOutput(layer_input, target)
layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)
torch_layer_output_var = torch_layer(layer_input_var,
Variable(torch.from_numpy(target_labels), requires_grad=False))
self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))
# 2. check layer input grad
custom_layer_grad = custom_layer.updateGradInput(layer_input, target)
torch_layer_output_var.backward()
torch_layer_grad_var = layer_input_var.grad
self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))
suite = unittest.TestLoader().loadTestsFromTestCase(TestLayers)
unittest.TextTestRunner(verbosity=2).run(suite)
| 0.748076 | 0.697177 |

[](https://colab.research.google.com/github/JohnSnowLabs/spark-nlp-workshop/blob/master/tutorials/streamlit_notebooks/NER_PT.ipynb)
# **Detect entities in Portuguese text**
## 1. Colab Setup
```
# Install Java
! apt-get update -qq
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
! java -version
# Install pyspark
! pip install --ignore-installed -q pyspark==2.4.4
# Install SparkNLP
! pip install --ignore-installed spark-nlp
```
## 2. Start the Spark session
```
import os
import json
os.environ['JAVA_HOME'] = "/usr/lib/jvm/java-8-openjdk-amd64"
import pandas as pd
import numpy as np
from pyspark.ml import Pipeline
from pyspark.sql import SparkSession
import pyspark.sql.functions as F
from sparknlp.annotator import *
from sparknlp.base import *
import sparknlp
from sparknlp.pretrained import PretrainedPipeline
spark = sparknlp.start()
```
## 3. Select the DL model
```
# If you change the model, re-run all the cells below.
# Applicable models: wikiner_840B_300, wikiner_6B_300, wikiner_6B_100
MODEL_NAME = "wikiner_840B_300"
```
## 4. Some sample examples
```
# Enter examples to be transformed as strings in this list
text_list = [
"""William Henry Gates III (nascido em 28 de outubro de 1955) é um magnata americano de negócios, desenvolvedor de software, investidor e filantropo. Ele é mais conhecido como co-fundador da Microsoft Corporation. Durante sua carreira na Microsoft, Gates ocupou os cargos de presidente, diretor executivo (CEO), presidente e diretor de arquitetura de software, além de ser o maior acionista individual até maio de 2014. Ele é um dos empreendedores e pioneiros mais conhecidos da revolução dos microcomputadores nas décadas de 1970 e 1980. Nascido e criado em Seattle, Washington, Gates co-fundou a Microsoft com o amigo de infância Paul Allen em 1975, em Albuquerque, Novo México; tornou-se a maior empresa de software de computador pessoal do mundo. Gates liderou a empresa como presidente e CEO até deixar o cargo em janeiro de 2000, mas ele permaneceu como presidente e tornou-se arquiteto-chefe de software. No final dos anos 90, Gates foi criticado por suas táticas de negócios, que foram consideradas anticompetitivas. Esta opinião foi confirmada por várias decisões judiciais. Em junho de 2006, Gates anunciou que iria passar para um cargo de meio período na Microsoft e trabalhar em período integral na Fundação Bill & Melinda Gates, a fundação de caridade privada que ele e sua esposa, Melinda Gates, estabeleceram em 2000. [ 9] Ele gradualmente transferiu seus deveres para Ray Ozzie e Craig Mundie. Ele deixou o cargo de presidente da Microsoft em fevereiro de 2014 e assumiu um novo cargo como consultor de tecnologia para apoiar a recém-nomeada CEO Satya Nadella.""",
"""A Mona Lisa é uma pintura a óleo do século XVI, criada por Leonardo. É realizada no Louvre, em Paris."""
]
```
## 5. Define Spark NLP pipeline
```
document_assembler = DocumentAssembler() \
.setInputCol('text') \
.setOutputCol('document')
tokenizer = Tokenizer() \
.setInputCols(['document']) \
.setOutputCol('token')
# The wikiner_840B_300 is trained with glove_840B_300, so the embeddings in the
# pipeline should match. Same applies for the other available models.
if MODEL_NAME == "wikiner_840B_300":
embeddings = WordEmbeddingsModel.pretrained('glove_840B_300', lang='xx') \
.setInputCols(['document', 'token']) \
.setOutputCol('embeddings')
elif MODEL_NAME == "wikiner_6B_300":
embeddings = WordEmbeddingsModel.pretrained('glove_6B_300', lang='xx') \
.setInputCols(['document', 'token']) \
.setOutputCol('embeddings')
elif MODEL_NAME == "wikiner_6B_100":
embeddings = WordEmbeddingsModel.pretrained('glove_100d') \
.setInputCols(['document', 'token']) \
.setOutputCol('embeddings')
ner_model = NerDLModel.pretrained(MODEL_NAME, 'pt') \
.setInputCols(['document', 'token', 'embeddings']) \
.setOutputCol('ner')
ner_converter = NerConverter() \
.setInputCols(['document', 'token', 'ner']) \
.setOutputCol('ner_chunk')
nlp_pipeline = Pipeline(stages=[
document_assembler,
tokenizer,
embeddings,
ner_model,
ner_converter
])
```
## 6. Run the pipeline
```
empty_df = spark.createDataFrame([['']]).toDF('text')
pipeline_model = nlp_pipeline.fit(empty_df)
df = spark.createDataFrame(pd.DataFrame({'text': text_list}))
result = pipeline_model.transform(df)
```
## 7. Visualize results
```
result.select(
F.explode(
F.arrays_zip('ner_chunk.result', 'ner_chunk.metadata')
).alias("cols")
).select(
F.expr("cols['0']").alias('chunk'),
F.expr("cols['1']['entity']").alias('ner_label')
).show(truncate=False)
```
|
github_jupyter
|
# Install Java
! apt-get update -qq
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
! java -version
# Install pyspark
! pip install --ignore-installed -q pyspark==2.4.4
# Install SparkNLP
! pip install --ignore-installed spark-nlp
import os
import json
os.environ['JAVA_HOME'] = "/usr/lib/jvm/java-8-openjdk-amd64"
import pandas as pd
import numpy as np
from pyspark.ml import Pipeline
from pyspark.sql import SparkSession
import pyspark.sql.functions as F
from sparknlp.annotator import *
from sparknlp.base import *
import sparknlp
from sparknlp.pretrained import PretrainedPipeline
spark = sparknlp.start()
# If you change the model, re-run all the cells below.
# Applicable models: wikiner_840B_300, wikiner_6B_300, wikiner_6B_100
MODEL_NAME = "wikiner_840B_300"
# Enter examples to be transformed as strings in this list
text_list = [
"""William Henry Gates III (nascido em 28 de outubro de 1955) é um magnata americano de negócios, desenvolvedor de software, investidor e filantropo. Ele é mais conhecido como co-fundador da Microsoft Corporation. Durante sua carreira na Microsoft, Gates ocupou os cargos de presidente, diretor executivo (CEO), presidente e diretor de arquitetura de software, além de ser o maior acionista individual até maio de 2014. Ele é um dos empreendedores e pioneiros mais conhecidos da revolução dos microcomputadores nas décadas de 1970 e 1980. Nascido e criado em Seattle, Washington, Gates co-fundou a Microsoft com o amigo de infância Paul Allen em 1975, em Albuquerque, Novo México; tornou-se a maior empresa de software de computador pessoal do mundo. Gates liderou a empresa como presidente e CEO até deixar o cargo em janeiro de 2000, mas ele permaneceu como presidente e tornou-se arquiteto-chefe de software. No final dos anos 90, Gates foi criticado por suas táticas de negócios, que foram consideradas anticompetitivas. Esta opinião foi confirmada por várias decisões judiciais. Em junho de 2006, Gates anunciou que iria passar para um cargo de meio período na Microsoft e trabalhar em período integral na Fundação Bill & Melinda Gates, a fundação de caridade privada que ele e sua esposa, Melinda Gates, estabeleceram em 2000. [ 9] Ele gradualmente transferiu seus deveres para Ray Ozzie e Craig Mundie. Ele deixou o cargo de presidente da Microsoft em fevereiro de 2014 e assumiu um novo cargo como consultor de tecnologia para apoiar a recém-nomeada CEO Satya Nadella.""",
"""A Mona Lisa é uma pintura a óleo do século XVI, criada por Leonardo. É realizada no Louvre, em Paris."""
]
document_assembler = DocumentAssembler() \
.setInputCol('text') \
.setOutputCol('document')
tokenizer = Tokenizer() \
.setInputCols(['document']) \
.setOutputCol('token')
# The wikiner_840B_300 is trained with glove_840B_300, so the embeddings in the
# pipeline should match. Same applies for the other available models.
if MODEL_NAME == "wikiner_840B_300":
embeddings = WordEmbeddingsModel.pretrained('glove_840B_300', lang='xx') \
.setInputCols(['document', 'token']) \
.setOutputCol('embeddings')
elif MODEL_NAME == "wikiner_6B_300":
embeddings = WordEmbeddingsModel.pretrained('glove_6B_300', lang='xx') \
.setInputCols(['document', 'token']) \
.setOutputCol('embeddings')
elif MODEL_NAME == "wikiner_6B_100":
embeddings = WordEmbeddingsModel.pretrained('glove_100d') \
.setInputCols(['document', 'token']) \
.setOutputCol('embeddings')
ner_model = NerDLModel.pretrained(MODEL_NAME, 'pt') \
.setInputCols(['document', 'token', 'embeddings']) \
.setOutputCol('ner')
ner_converter = NerConverter() \
.setInputCols(['document', 'token', 'ner']) \
.setOutputCol('ner_chunk')
nlp_pipeline = Pipeline(stages=[
document_assembler,
tokenizer,
embeddings,
ner_model,
ner_converter
])
empty_df = spark.createDataFrame([['']]).toDF('text')
pipeline_model = nlp_pipeline.fit(empty_df)
df = spark.createDataFrame(pd.DataFrame({'text': text_list}))
result = pipeline_model.transform(df)
result.select(
F.explode(
F.arrays_zip('ner_chunk.result', 'ner_chunk.metadata')
).alias("cols")
).select(
F.expr("cols['0']").alias('chunk'),
F.expr("cols['1']['entity']").alias('ner_label')
).show(truncate=False)
| 0.597843 | 0.89974 |
# Mesh basics
In order to solve a model numerically in a region, we have to discretise it. There are two main ways of discretising the space: finite-difference and finite-element discretisation. `discretisedfield` deals only with finite-difference discretisation at the moment. This means that we are dividing our cubic region into smaller "chunks" - small cubes. We refer to the discretised region as a **mesh**:
$$\text{MESH} = \text{REGION} + \text{DISCRETISATION}$$
In this tutorial, we show how to define it, as well as some basic operations we can perform with meshes.
As we showed in previous tutorials, region is always cubic and it is defined by any two diagonally opposite corner points. We are going to use the same region as before, defined by the following two diagonally opposite points
$$\mathbf{p}_{1} = (0, 0, 0)$$
$$\mathbf{p}_{2} = (l_{x}, l_{y}, l_{z})$$
with $l_{x} = 100 \,\text{nm}$, $l_{y} = 50 \,\text{nm}$, and $l_{z} = 20 \,\text{nm}$.
So, let us start by defining the region:
```
import discretisedfield as df
p1 = (0, 0, 0)
p2 = (100e-9, 50e-9, 20e-9)
region = df.Region(p1=p1, p2=p2)
```
The region is now defined. Another missing piece is the discretisation and we need to decide how we are going to discretise the region. In other words, we need to decide into what size "chunks" we are going to discretise our region in. We refer to the "chunk" as the **discretisation cell**. In `discretisedfield`, there are two ways how we can define the discretisation. We can define either:
1. The number of discretisation cells in all 3 ($x$, $y$, and $z$) directions, or
2. The size of a single discretisation cell.
Let us start with the first case. The number of discretisation cells in all three directions can be passed using `n` argument, which is a length-3 tuple:
$$n = (n_{x}, n_{y}, n_{z})$$
For instance, we want to discretise our region in 5 cells in the x-direction, 2 in the y-direction and 1 cell in the z-direction. Therefore, knowing the region as well as the discretisation `n`, we pass them both to `Mesh`:
```
n = (5, 2, 1)
mesh = df.Mesh(region=region, n=n)
```
The mesh is defined. Based on the region dimensions and the number of discretisation cells, we can ask the mesh to give us the size of a single discretisation cell:
```
mesh.cell
```
Knowing this value, we could have defined the mesh passing this value using `cell` argument, and we would have got exactly the same mesh.
```
cell = (20e-9, 25e-9, 20e-9)
mesh = df.Mesh(region=region, cell=cell)
```
If we now ask our new mesh about the number of discretisation cells:
```
mesh.n
```
There is no difference whatsoever how we are going to define the mesh. However, defining the mesh with `cell` can result in an error, if the region cannot be divided into chunks of that size. For instance:
```
try:
mesh = df.Mesh(region=region, cell=(3e-9, 3e-9, 3e-9))
except ValueError:
print('Exception raised.')
```
Let us now have a look at some basic properties we can ask the mesh object for. First of all, region object is a part of the mesh object:
```
mesh.region
```
Therefore, we can perform all the operations on the region we saw previously, but now through the mesh object (`mesh.region`). For instance:
```
mesh.region.pmin # minimum point
mesh.region.edges # edge lenghts
mesh.region.centre # centre point
```
By asking the mesh object directly, we can get the number of discretisation cells in all three directions $n = (n_{x}, n_{y}, n_{z})$:
```
mesh.n
```
and the size of a single discretisation cell:
```
mesh.cell
```
The total number of discretisation cells is:
```
len(mesh)
```
This number is simply $n_{x}n_{y}n_{z}$. We can conclude that the entire region is now divided into 100 small cubes (discretisation cells). Each cell in the mesh has its index and its coordinate. We can get indices of all discretisation cells:
```
# NBVAL_IGNORE_OUTPUT
mesh.indices
```
This gives us a generator object we can use as an iterable in different Pyhton contexts. For instance, we can give it to the `list`.
```
list(mesh.indices)
```
List function now "unpacks" the generator and gives us a list of tuples. Each tuple has three unsigned integers. For instance, we can interpret index `(2, 1, 0)` as an index which belongs to the third cell in the x-direction, second in the y, and the first in the z direction. Please note that indexing in Python starts from 0. Therefore, we say that the "fifth element" has index 4.
Another thing we can associate to every discretisation cell is its coordinate. The coordinate of the cell is the coordinate of its centre point. So, the coordinate of index `(2, 1, 0)` cell is:
```
index = (2, 1, 0)
mesh.index2point(index)
```
It is very often the case we need to iterate through all discretisation cells and use their coordinates. For that, we can use the mesh object itself, which is also an iterable:
```
list(mesh)
```
Since mesh object is an iterator itself, we can use it, for example, in for loops:
```
for point in mesh:
print(point)
```
A function, which is opposite to `index2point`, is `point2index`. This function takes any point in the region and returns the index of a cell it belongs to:
```
point = (41.6e-9, 35.2e-9, 4.71e-9)
mesh.point2index(point)
```
We can also ask the mesh to give us the points along a certain axis:
```
list(mesh.axis_points('x'))
list(mesh.axis_points('y'))
```
We can compare meshes using `==` and `!=` relational operators. Let us define two meshes and compare them to the one we have already defined:
```
mesh_same = df.Mesh(region=region, n=(5, 2, 1))
mesh_different = df.Mesh(region=region, n=(10, 5, 7))
mesh == mesh_same
mesh == mesh_different
mesh != mesh_different
```
Finally, mesh has its representation string:
```
mesh
```
In the representation string, we see `p1`, `p2`, and `n` we discussed earlier, but there are also `bc` and `subregions` we did not and we will look at some more advanced mesh properties in the next tutorials.
|
github_jupyter
|
import discretisedfield as df
p1 = (0, 0, 0)
p2 = (100e-9, 50e-9, 20e-9)
region = df.Region(p1=p1, p2=p2)
n = (5, 2, 1)
mesh = df.Mesh(region=region, n=n)
mesh.cell
cell = (20e-9, 25e-9, 20e-9)
mesh = df.Mesh(region=region, cell=cell)
mesh.n
try:
mesh = df.Mesh(region=region, cell=(3e-9, 3e-9, 3e-9))
except ValueError:
print('Exception raised.')
mesh.region
mesh.region.pmin # minimum point
mesh.region.edges # edge lenghts
mesh.region.centre # centre point
mesh.n
mesh.cell
len(mesh)
# NBVAL_IGNORE_OUTPUT
mesh.indices
list(mesh.indices)
index = (2, 1, 0)
mesh.index2point(index)
list(mesh)
for point in mesh:
print(point)
point = (41.6e-9, 35.2e-9, 4.71e-9)
mesh.point2index(point)
list(mesh.axis_points('x'))
list(mesh.axis_points('y'))
mesh_same = df.Mesh(region=region, n=(5, 2, 1))
mesh_different = df.Mesh(region=region, n=(10, 5, 7))
mesh == mesh_same
mesh == mesh_different
mesh != mesh_different
mesh
| 0.309232 | 0.994264 |
# Introdução
### Pesquisa Nacional por Amostra de Domicílios - PNAD
A Pesquisa Nacional por Amostra de Domicílios - PNAD, de periodicidade anual, foi encerrada em 2016, com a divulgação das informações referentes a 2015. Planejada para produzir resultados para Brasil, Grandes Regiões, Unidades da Federação e nove Regiões Metropolitanas (Belém, Fortaleza, Recife, Salvador, Belo Horizonte, Rio de Janeiro, São Paulo, Curitiba e Porto Alegre), ela pesquisava, de forma permanente, características gerais da população, educação, trabalho, rendimento e habitação, e, com periodicidade variável, outros temas, de acordo com as necessidades de informação para o País, tendo como unidade de investigação o domicílio. A PNAD foi substituída, com metodologia atualizada, pela Pesquisa Nacional por Amostra de Domicílios Contínua - PNAD Contínua, que propicia uma cobertura territorial mais abrangente e disponibiliza informações conjunturais trimestrais sobre a força de trabalho em âmbito nacional.
# Importando as bibliotecas de trabalho
```
import pandas as pd
import numpy as np
from statsmodels.stats.weightstats import ttest_ind, ztest
from scipy.stats import norm
from scipy.stats import t as t_student
import matplotlib.pyplot as plt
import seaborn as sns
from matplotlib.pyplot import figure
sns.set()
%matplotlib inline
# Dataset
dataset = 'pnad_2015.csv'
# Imporatndo o Dataset
df = pd.read_csv(dataset)
df.head()
```
## Análise exploratória e descritiva
### Comparando as colunas de forma geral
```
# Gráfico com a quatidade de homens e mulheres
print('Gráfico com a quatidade de homens e mulheres')
graph_sex = sns.histplot(df['Sexo'].map({0:'Homens', 1:'Mulheres'})).set(title = 'Distribuição de sexos IBGE-PNAD-2015', ylabel = 'Número de pessoas')
plt.show()
graph_cor = sns.histplot(df['Cor'].map({0:'Indígena', 2:'Branca', 4:'Preta', 6:'Amarela', 8:'Parda', 9:'Sem declaração'})).set(title = 'Distribuição de cor IBGE-PNAD-2015', ylabel = 'Número de pessoas')
plt.show()
```
### Número de pessoas por Macrorregião e estados
```
graph_macro_reg = sns.histplot(df['UF'].map({11:'Norte', 12: 'Norte', 13:'Norte', 14:'Norte', 15:'Norte', 16:'Norte', 17:'Norte', 21: 'Nordeste', 22: 'Nordeste', 23: 'Nordeste', 24: 'Nordeste', 25: 'Nordeste', 26: 'Nordeste', 27: 'Nordeste', 28:'Nordeste', 29:'Nordeste', 31:'Sudeste', 32:'Sudeste', 33:'Sudeste', 35:'Sudeste', 41:'Sul', 42:'Sul',43:'Sul', 50:'Sul', 51:'Centro-Oeste', 52:'Centro-Oeste', 53:'Centro-Oeste'})).set(title = 'Distribuição de pessoas por Macrorregião', ylabel = 'Quantidade', xlabel = 'Macrorregiões')
plt.show()
graph_estados = sns.histplot(df['UF'].map({11:'Rondônia', 12: 'Acre', 13:'Amazonas', 14:'Roraima', 15:'Pará', 16:'Amapá', 17:'Tocantins', 21: 'Maranhão', 22: 'Piauí', 23: 'Ceará', 24: 'Rio Grande do Norte', 25: 'Paraíba', 26: 'Pernambuco', 27: 'Alagoas', 28:'Sergipe', 29:'Bahia', 31:'Minas Gerais', 32:'EspíritoSanto', 33:'Rio de Janeiro', 35:'São Paulo', 41:'Paraná', 42:'Santa Catarina',43:'Rio Grande do Sul', 50:'Mato Grosso do Sul', 51:'Mato Grosso', 52:'Goiás', 53:'Distrito Federal'})).set(title = 'Distribuição de pessoas por estado', ylabel = 'Quantidade', xlabel = 'Estados')
plt.xticks(rotation=90)
plt.show()
```
### Distribuição das idades
Analisando a distribuição das idades notamos uma similaridade visual com a distribuição normal, sendo novamente visualmente simétrica.
Calculando sua média, moda e mediana torna possível determinar a simetria da distribuição.
```
graph_idades = sns.histplot(df['Idade'], bins = [0,10,20,30,40,50,60,70,80,90,100], kde = True).set(title = 'Distribuição de idades', ylabel = 'Quantidade')
plt.show()
idade_media_br = df['Idade'].mean()
idade_media_homens = df.query('Sexo == 0')['Idade'].mean()
idade_media_mulheres = df.query('Sexo == 1')['Idade'].mean()
idade_mediana_br = df['Idade'].median()
idade_mediana_homens = df.query('Sexo == 0')['Idade'].median()
idade_mediana_mulheres = df.query('Sexo == 1')['Idade'].median()
idade_moda_br = df['Idade'].mode()
idade_moda_homens = df.query('Sexo == 0')['Idade'].mode()
idade_moda_mulheres = df.query('Sexo == 1')['Idade'].mode()
print(f'''
Idade média da população brasileira: {idade_media_br}
Idade média dos homens brasileiros: {idade_media_homens}
Idade média das mulheres brasileiras: {idade_media_mulheres}
Mediana da idade da população brasileira: {idade_mediana_br}
Mediana da idade dos homens brasileiros: {idade_mediana_homens}
Mediana da idade das mulheres brasileiras: {idade_mediana_mulheres}
Moda da idade da população brasileira: {idade_moda_br[0]}
Moda da idade dos homens brasileiros: {idade_moda_homens[0]}
Moda da idade das mulheres brasileiras: {idade_moda_mulheres[0]}
''')
```
Com esses valores determinados basta comparar qual o maior para determinar a simetria da distribuição:
Idade Média:
$\begin{cases}
\bar{x}_{Br} = 44.07\\
\bar{x}_M = 44.12\\
\bar{x}_H = 44.04
\end{cases}
$
Mediana da Idade:
$\begin{cases}
m_{Br} = 43\\
m_M = 43\\
m_H = 44
\end{cases}
$
Moda da Idade:
$\begin{cases}
m_{Br} = 40\\
m_M = 50\\
m_H = 40
\end{cases}
$
A partir destes resultados notamos que a média e a mediana estão bem próximas, que se aproximam da moda dessa análise. Sendo rigoroso temos que $\bar{x}_{Br} > m_{Br} > m_{Br}$ e, portanto temos uma distribuição ligeiramente assimétria à direita.
No entanto, podemos considerar essa distribuição como se fosse simétrica pois embora seja assimétrica à direita, sua assimetria é pequena. Além de simétrica, essa distribuição se parece bastante com uma distribuição normal. Logo nossa distribuição é parecida com uma distribuição simétrica.
Além disso, podemos ver visualmente, a linha de distribuição muito semelhança à normal, sendo um pouco menos suave, talvez por conta do número de dados.
### Distribuição dos Anos de Estudo
```
graph_anos_de_estudo = sns.histplot(df['Anos de Estudo'], bins = [y for y in range(21)], kde = True).set(title = 'Distribuição de estudo', ylabel = 'Quantidade')
plt.show()
Renda_corte = '50000'
plt.scatter(df.query('Sexo == 0')['Renda'], df.query('Sexo == 0')['Anos de Estudo'])
plt.scatter(df.query('Sexo == 1')['Renda'], df.query('Sexo == 1')['Anos de Estudo'], color = 'red')
plt.legend(['Homens','Mulheres'])
plt.ylabel('Anos de Estudo')
plt.xlabel('Renda')
plt.show()
plt.scatter(df.query('Sexo == 0 and Renda <= ' + Renda_corte)['Renda'], df.query('Sexo == 0 and Renda <= ' + Renda_corte)['Anos de Estudo'])
plt.scatter(df.query('Sexo == 1 and Renda <= ' + Renda_corte)['Renda'], df.query('Sexo == 1 and Renda <= ' + Renda_corte)['Anos de Estudo'], color = 'red')
plt.legend(['Homens','Mulheres'])
plt.ylabel('Anos de Estudo')
plt.xlabel('Renda')
plt.show()
plt.scatter(df.query('Renda <= ' + Renda_corte)['Renda'], df.query('Renda <= ' + Renda_corte)['Anos de Estudo'])
plt.ylabel('Anos de Estudo')
plt.xlabel('Renda')
plt.show()
```
#### Anos de Estudo x Renda Média
### Distribuição da Renda
A renda é uma variável contínua e para visualizar sua distribuição melhor utilizei um plot na forma de histograma para permitir ver a distribuição da renda brasileira geral, masculina e feminina na forma de barras e no boxplot para visualizar a densidade dos dados.
Analisando a distribuição de renda podemos notar visualmente a diferença de renda entre homens e mulheres apresentada pela amostra. Para ter está visão mais nítida basta olhar os gráficos Boxplot das distribuições de renda masculina e feminina.
```
graph_renda = sns.histplot(df['Renda'], bins = [i*1000 for i in range(21)]).set(title = 'Distribuição de Renda', ylabel = 'Quantidade')
plt.show()
sns.boxplot(x = df['Renda']).set(title = 'Renda geral com outliers')
plt.show()
sns.boxplot(x = df['Renda'], showfliers = False).set(title = 'Renda geral sem outliers')
plt.show()
graph_renda = sns.histplot(df.query('Sexo == 0')['Renda'], bins = [i*1000 for i in range(21)]).set(title = 'Distribuição de Renda masculina', ylabel = 'Quantidade')
plt.show()
sns.boxplot(x = df.query('Sexo == 0')['Renda'], showfliers = False).set(title = 'Renda dos homens')
plt.show()
graph_renda = sns.histplot(df.query('Sexo == 1')['Renda'], bins = [i*1000 for i in range(21)]).set(title = 'Distribuição de Renda feminina', ylabel = 'Quantidade')
plt.show()
sns.boxplot(x = df.query('Sexo == 1')['Renda'], showfliers = False).set(title = 'Renda das mulheres')
plt.show()
renda_media_br = df['Renda'].mean()
renda_media_homens = df.query('Sexo == 0')['Renda'].mean()
renda_media_mulheres = df.query('Sexo == 1')['Renda'].mean()
renda_mediana_br = df['Renda'].median()
renda_mediana_homens = df.query('Sexo == 0')['Renda'].median()
renda_mediana_mulheres = df.query('Sexo == 1')['Renda'].median()
renda_moda_br = df['Renda'].mode()
renda_moda_homens = df.query('Sexo == 0')['Renda'].mode()
renda_moda_mulheres = df.query('Sexo == 1')['Renda'].mode()
print(f'''
Renda média da população brasileira: {renda_media_br}
Renda média dos homens brasileiros: {renda_media_homens}
Renda média das mulheres brasileiras: {renda_media_mulheres}
Mediana da renda da população brasileira: {renda_mediana_homens}
Mediana da renda dos homens brasileiros: {renda_mediana_homens}
Mediana da renda das mulheres brasileiras: {renda_mediana_mulheres}
Moda da renda da população brasileira: {renda_moda_br[0]}
Moda da renda dos homens brasileiros: {renda_moda_homens[0]}
Moda da renda das mulheres brasileiras: {renda_moda_mulheres[0]}
''')
```
Notamos que a moda é o salário mínimo, o que infelizmente quer dizer que o valor de salário mais recebido pelas pessoas da amostra é 1 salário mínimo. Além disso, calculamos e visualizamos a diferença das rendas médias entre os homens e as mulheres. A partir desta amostra pode-se pensar que os homens relamente ganham mais do que as mulheres, mas não de maneira amostral, mas sim populacional, sendo algo geral para o Brasil como um todo. Considerando tal afirmação para a população brasileira como um todo. Esse raciocínio será elaborado mais abaixo.
### Analisando por Macrorregião
### Analisando por Cor
```
sns.barplot(data = df, x = df['Cor'].map({0:'Indígena', 2:'Branca', 4:'Preta', 6:'Amarela', 8:'Parda', 9:'Sem declaração'}), y = df['Renda']).set(title = 'Média de Renda por cor')
plt.show()
sns.barplot(data = df, x = df['Cor'].map({0:'Indígena', 2:'Branca', 4:'Preta', 6:'Amarela', 8:'Parda', 9:'Sem declaração'}), y = df['Anos de Estudo']).set(title = 'Média de Anos de Estudo por cor')
plt.show()
sns.barplot(data = df, x = df['Cor'].map({0:'Indígena', 2:'Branca', 4:'Preta', 6:'Amarela', 8:'Parda', 9:'Sem declaração'}), y = df['Idade']).set(title = 'Média de Idade por cor')
plt.show()
sns.barplot(data = df, x = df['Cor'].map({0:'Indígena', 2:'Branca', 4:'Preta', 6:'Amarela', 8:'Parda', 9:'Sem declaração'}), y = df['Altura']).set(title = 'Média de Altura por cor')
plt.show()
```
### Analisando por Sexo
```
sns.barplot(data = df, x = df['Sexo'].map({0:'Homens', 1:'Mulheres'}), y = df['Renda']).set(title = 'Média de Renda por Sexo')
plt.show()
sns.barplot(data = df, x = df['Sexo'].map({0:'Homens', 1:'Mulheres'}), y = df['Anos de Estudo']).set(title = 'Média de Anos de Estudo por sexo')
plt.show()
sns.barplot(data = df, x = df['Sexo'].map({0:'Homens', 1:'Mulheres'}), y = df['Idade']).set(title = 'Média de Idade por sexo')
plt.show()
sns.barplot(data = df, x = df['Sexo'].map({0:'Homens', 1:'Mulheres'}), y = df['Altura']).set(title = 'Média de Altura por sexo')
plt.show()
```
#### Média de Renda por estado
```
fig = sns.barplot(data = df, x = df['UF'].map({11:'Rondônia', 12: 'Acre', 13:'Amazonas', 14:'Roraima', 15:'Pará', 16:'Amapá', 17:'Tocantins', 21: 'Maranhão', 22: 'Piauí', 23: 'Ceará', 24: 'Rio Grande do Norte', 25: 'Paraíba', 26: 'Pernambuco', 27: 'Alagoas', 28:'Sergipe', 29:'Bahia', 31:'Minas Gerais', 32:'EspíritoSanto', 33:'Rio de Janeiro', 35:'São Paulo', 41:'Paraná', 42:'Santa Catarina',43:'Rio Grande do Sul', 50:'Mato Grosso do Sul', 51:'Mato Grosso', 52:'Goiás', 53:'Distrito Federal'}), y = df['Renda']).set(title = 'Média de Renda por estado', xlabel = 'Estados')
plt.xticks(rotation=90)
plt.figure(figsize = (10,12))
plt.savefig('teste123.pdf', dpi = 'figure')
plt.show()
```
#### Média de Anos de Estudo por estado
### Distribuição das Alturas
```
sns.histplot(df['Altura'], bins = [1 + 0.1 * i for i in range(12)], kde = True).set(title = 'Distribuição de altura brasileira', ylabel = 'Número de pessoas')
plt.show()
sns.boxplot(x = df['Altura']).set(title = 'Altura brasileira')
plt.show()
sns.histplot(df.query('Sexo == 0')['Altura'], bins = [1 + 0.1 * i for i in range(12)], kde = True).set(title = 'Distribuição de altura masculina', ylabel = 'Número de pessoas')
plt.show()
sns.boxplot(x = df.query('Sexo == 0')['Altura']).set(title = 'Altura masculina')
plt.show()
sns.histplot(df.query('Sexo == 1')['Altura'], bins = [1 + 0.1 * i for i in range(12)], kde = True).set(title = 'Distribuição de altura feminina', ylabel = 'Número de pessoas')
plt.show()
sns.boxplot(x = df.query('Sexo == 1')['Altura']).set(title = 'Altura feminina')
plt.show()
```
A distribuição da variável das alturas comporta-se como uma normal, como pode ser visto nos histogramas acima.
```
Altura_media_br = df['Altura'].mean()
Altura_media_homens = df.query('Sexo == 0')['Altura'].mean()
Altura_media_mulheres = df.query('Sexo == 1')['Altura'].mean()
Altura_mediana_br = df['Altura'].median()
Altura_mediana_homens = df.query('Sexo == 0')['Altura'].median()
Altura_mediana_mulheres = df.query('Sexo == 1')['Altura'].median()
Altura_moda_br = df['Altura'].mode()
Altura_moda_homens = df.query('Sexo == 0')['Altura'].mode()
Altura_moda_mulheres = df.query('Sexo == 1')['Altura'].mode()
print(f'''
Altura média da população brasileira: {Altura_media_br}
Altura média dos homens brasileiros: {Altura_media_homens}
Altura média das mulheres brasileiras: {Altura_media_mulheres}
Mediana da altura da população brasileira: {Altura_mediana_homens}
Mediana da altura dos homens brasileiros: {Altura_mediana_homens}
Mediana da altura das mulheres brasileiras: {Altura_mediana_mulheres}
Moda da altura da população brasileira: {Altura_moda_br[0]}
Moda da altura dos homens brasileiros: {Altura_moda_homens[0]}
Moda da altura das mulheres brasileiras: {Altura_moda_mulheres[0]}
''')
```
Parece haver algo estranho com relação as alturas das garotas, a média de altura das mulheres está maior que a média masculina, o que sabemos ser errado. Comparando com dados de outras pesquisas do IBGE temos:
> Altura Esperada
> Homens 173.3 cm (5' 8.25'')
> Mulheres 161,1 cm (5' 3.5'')
A partir disto a média amostral brasileira da altura está próxima do seu valor populacional, no entanto, a média amostral da altura feminina está bem distante do seu valor populacional.
## Análise dos dados
```
cores = (list(set(df['Cor'])))
df_adapt = df.copy()
tabela1 = pd.crosstab(df['Renda'] ,df_adapt['Cor'].map({0:'Indígena',2:'Branca',4:'Preta',6:'Amarela',8:'Parda'}))
freq_salario = pd.DataFrame()
freq_salario['Absoluta'] = pd.cut(df['Renda'], bins = [i*1000 for i in range(20)]).value_counts()
# por cor
freq_salario2 = pd.DataFrame()
freq_salario2 = pd.cut(df.query('Cor == 2')['Renda'], bins = [i*1000 for i in range(20)]).value_counts()
rendas = dict()
estudos = dict()
for cor in cores:
estudo_medio = df[df.Cor == int(cor)]['Anos de Estudo'].mean()
rendas[cor] = renda_media
estudos[cor] = estudo_medio
print(estudos)
rendas
freq_salario2
```
## Inferências à população brasileira
## Cálculo de parâmetros
```
n_M, n_H = 500, 500
significancia = 0.01
confianca = 1 - significancia
n = n_M + n_H
amostra_H = df[df.Sexo == 0]['Renda'].sample(n = n_H, random_state = 1)
amostra_M = df[df.Sexo == 1]['Renda'].sample(n = n_M, random_state = 1)
media_H = amostra_H.mean()
media_M = amostra_M.mean()
media_pop_H = df[df.Sexo == 0]['Renda'].mean()
media_pop_M = df[df.Sexo == 1]['Renda'].mean()
desvio_H = amostra_H.std()
desvio_M = amostra_M.std()
```
## Formulando algumas hipóteses para o problema
> Teste Bicaudal
> $H_0$: A Média salarial dos homens é igual a média salarial das mulheres
> $H_1$ As médias são diferentes
$\begin{cases}
H_0: \mu_M = \mu_H \\
H_1: \mu_M \neq \mu_H
\end{cases}
$
> Sendo $H_0$ a hipótese nula e $H_1$ a hipótese alternativa
## Realizando teste da hipótese nula
Testando as hipóteses
> Teste Bicaudal
> 1. Rejeita-se $H_0$ se $z \leq -z{\alpha / 2}$ ou se $z \geq z_{\alpha / 2}$
> 2. Rejeita-se $H_0$ se o valor de $p <\alpha$
```
# Z_alpha para bicaudal
probabilidade = confianca + significancia / 2
z_alpha = norm.ppf(probabilidade)
z_alpha2 = norm.ppf(1 - probabilidade)
z_alpha
# Ztest
graus_de_liberdade = n = 500
# Two-sided -> Bicaudal
z, p = ztest(amostra_H, amostra_M, alternative = 'two-sided')
print (f'z = {z} e p = {p}')
t = t_student.ppf(probabilidade, graus_de_liberdade)
print (f't = {t}')
```
### Testando 1 e 2
```
if z >= z_alpha or z <= z_alpha2: # Teste 1
print(f'A hipótese alternativa está correta com {confianca:.0%} de confiança.')
else:
if p < significancia: # Teste 2
print(f'A hipótese nula está correta com {confianca:.0%} de confiança.')
else:
print(f'A hipótese alternativa está correta com {confianca:.0%} de confiança.')
```
Dessa maneira, nossa hipótese nula aparenta estar correta com uma confiança de 99%. Isso que dizer que as médias salariais muito provavelmente possuem médias diferentes. Dessa forma, resta analisar para que lado tende essa diferença e se possível tentar metrificá-la.
Para isso será preciso analisar de forma unicaudal o problema, sendo que temos duas distribuições unicaudais possíveis, uma com a média das mulheres sendo maiores e outra com a média dos homens sendo maiores. Como:
$\begin{cases}
\mu_H = 2059.212 \\
\mu_M = 1548.274
\end{cases}
$
Sendo assim,
$
\mu_H >= \mu_M \\
$
Dessa maneira irei realizar o teste estatístico considerando as hipóteses a seguir:
$\begin{cases}
H_0: \mu_M =< \mu_H \\
H_1: \mu_H > \mu_M
\end{cases}
$
```
# Z_alpha para unicaudal
probabilidade = confianca
z_alpha = norm.ppf(probabilidade)
z_alpha2 = norm.ppf(1 - probabilidade)
z_alpha, z_alpha2
# Ztest
graus_de_liberdade = n = 500
# Two-sided -> Unicaudal
z, p = ztest(amostra_H, amostra_M, alternative = 'smaller')
print (f'z = {z} e p = {p}')
t = t_student.ppf(probabilidade, graus_de_liberdade)
print (f't = {t}')
```
### Testando 1 e 2
```
if z <= z_alpha: # Teste 1
print(f'A hipótese alternativa está correta com {confianca:.0%} de confiança.')
else:
if p < significancia: # Teste 2
print(f'A hipótese nula está correta com {confianca:.0%} de confiança.')
else:
print(f'A hipótese alternativa está correta com {confianca:.0%} de confiança.')
```
Dessa maneira chegamos a conclusão de que com uma confiança de 99% podemos afirmar que a média salarial dos homens é maior do que a média salarial das mulheres. Ou seja, que a nossa Hipótese Nula era verdadeira.
```
media_H, media_M ,media_pop_H, media_pop_M
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from statsmodels.stats.weightstats import ttest_ind, ztest
from scipy.stats import norm
from scipy.stats import t as t_student
import matplotlib.pyplot as plt
import seaborn as sns
from matplotlib.pyplot import figure
sns.set()
%matplotlib inline
# Dataset
dataset = 'pnad_2015.csv'
# Imporatndo o Dataset
df = pd.read_csv(dataset)
df.head()
# Gráfico com a quatidade de homens e mulheres
print('Gráfico com a quatidade de homens e mulheres')
graph_sex = sns.histplot(df['Sexo'].map({0:'Homens', 1:'Mulheres'})).set(title = 'Distribuição de sexos IBGE-PNAD-2015', ylabel = 'Número de pessoas')
plt.show()
graph_cor = sns.histplot(df['Cor'].map({0:'Indígena', 2:'Branca', 4:'Preta', 6:'Amarela', 8:'Parda', 9:'Sem declaração'})).set(title = 'Distribuição de cor IBGE-PNAD-2015', ylabel = 'Número de pessoas')
plt.show()
graph_macro_reg = sns.histplot(df['UF'].map({11:'Norte', 12: 'Norte', 13:'Norte', 14:'Norte', 15:'Norte', 16:'Norte', 17:'Norte', 21: 'Nordeste', 22: 'Nordeste', 23: 'Nordeste', 24: 'Nordeste', 25: 'Nordeste', 26: 'Nordeste', 27: 'Nordeste', 28:'Nordeste', 29:'Nordeste', 31:'Sudeste', 32:'Sudeste', 33:'Sudeste', 35:'Sudeste', 41:'Sul', 42:'Sul',43:'Sul', 50:'Sul', 51:'Centro-Oeste', 52:'Centro-Oeste', 53:'Centro-Oeste'})).set(title = 'Distribuição de pessoas por Macrorregião', ylabel = 'Quantidade', xlabel = 'Macrorregiões')
plt.show()
graph_estados = sns.histplot(df['UF'].map({11:'Rondônia', 12: 'Acre', 13:'Amazonas', 14:'Roraima', 15:'Pará', 16:'Amapá', 17:'Tocantins', 21: 'Maranhão', 22: 'Piauí', 23: 'Ceará', 24: 'Rio Grande do Norte', 25: 'Paraíba', 26: 'Pernambuco', 27: 'Alagoas', 28:'Sergipe', 29:'Bahia', 31:'Minas Gerais', 32:'EspíritoSanto', 33:'Rio de Janeiro', 35:'São Paulo', 41:'Paraná', 42:'Santa Catarina',43:'Rio Grande do Sul', 50:'Mato Grosso do Sul', 51:'Mato Grosso', 52:'Goiás', 53:'Distrito Federal'})).set(title = 'Distribuição de pessoas por estado', ylabel = 'Quantidade', xlabel = 'Estados')
plt.xticks(rotation=90)
plt.show()
graph_idades = sns.histplot(df['Idade'], bins = [0,10,20,30,40,50,60,70,80,90,100], kde = True).set(title = 'Distribuição de idades', ylabel = 'Quantidade')
plt.show()
idade_media_br = df['Idade'].mean()
idade_media_homens = df.query('Sexo == 0')['Idade'].mean()
idade_media_mulheres = df.query('Sexo == 1')['Idade'].mean()
idade_mediana_br = df['Idade'].median()
idade_mediana_homens = df.query('Sexo == 0')['Idade'].median()
idade_mediana_mulheres = df.query('Sexo == 1')['Idade'].median()
idade_moda_br = df['Idade'].mode()
idade_moda_homens = df.query('Sexo == 0')['Idade'].mode()
idade_moda_mulheres = df.query('Sexo == 1')['Idade'].mode()
print(f'''
Idade média da população brasileira: {idade_media_br}
Idade média dos homens brasileiros: {idade_media_homens}
Idade média das mulheres brasileiras: {idade_media_mulheres}
Mediana da idade da população brasileira: {idade_mediana_br}
Mediana da idade dos homens brasileiros: {idade_mediana_homens}
Mediana da idade das mulheres brasileiras: {idade_mediana_mulheres}
Moda da idade da população brasileira: {idade_moda_br[0]}
Moda da idade dos homens brasileiros: {idade_moda_homens[0]}
Moda da idade das mulheres brasileiras: {idade_moda_mulheres[0]}
''')
graph_anos_de_estudo = sns.histplot(df['Anos de Estudo'], bins = [y for y in range(21)], kde = True).set(title = 'Distribuição de estudo', ylabel = 'Quantidade')
plt.show()
Renda_corte = '50000'
plt.scatter(df.query('Sexo == 0')['Renda'], df.query('Sexo == 0')['Anos de Estudo'])
plt.scatter(df.query('Sexo == 1')['Renda'], df.query('Sexo == 1')['Anos de Estudo'], color = 'red')
plt.legend(['Homens','Mulheres'])
plt.ylabel('Anos de Estudo')
plt.xlabel('Renda')
plt.show()
plt.scatter(df.query('Sexo == 0 and Renda <= ' + Renda_corte)['Renda'], df.query('Sexo == 0 and Renda <= ' + Renda_corte)['Anos de Estudo'])
plt.scatter(df.query('Sexo == 1 and Renda <= ' + Renda_corte)['Renda'], df.query('Sexo == 1 and Renda <= ' + Renda_corte)['Anos de Estudo'], color = 'red')
plt.legend(['Homens','Mulheres'])
plt.ylabel('Anos de Estudo')
plt.xlabel('Renda')
plt.show()
plt.scatter(df.query('Renda <= ' + Renda_corte)['Renda'], df.query('Renda <= ' + Renda_corte)['Anos de Estudo'])
plt.ylabel('Anos de Estudo')
plt.xlabel('Renda')
plt.show()
graph_renda = sns.histplot(df['Renda'], bins = [i*1000 for i in range(21)]).set(title = 'Distribuição de Renda', ylabel = 'Quantidade')
plt.show()
sns.boxplot(x = df['Renda']).set(title = 'Renda geral com outliers')
plt.show()
sns.boxplot(x = df['Renda'], showfliers = False).set(title = 'Renda geral sem outliers')
plt.show()
graph_renda = sns.histplot(df.query('Sexo == 0')['Renda'], bins = [i*1000 for i in range(21)]).set(title = 'Distribuição de Renda masculina', ylabel = 'Quantidade')
plt.show()
sns.boxplot(x = df.query('Sexo == 0')['Renda'], showfliers = False).set(title = 'Renda dos homens')
plt.show()
graph_renda = sns.histplot(df.query('Sexo == 1')['Renda'], bins = [i*1000 for i in range(21)]).set(title = 'Distribuição de Renda feminina', ylabel = 'Quantidade')
plt.show()
sns.boxplot(x = df.query('Sexo == 1')['Renda'], showfliers = False).set(title = 'Renda das mulheres')
plt.show()
renda_media_br = df['Renda'].mean()
renda_media_homens = df.query('Sexo == 0')['Renda'].mean()
renda_media_mulheres = df.query('Sexo == 1')['Renda'].mean()
renda_mediana_br = df['Renda'].median()
renda_mediana_homens = df.query('Sexo == 0')['Renda'].median()
renda_mediana_mulheres = df.query('Sexo == 1')['Renda'].median()
renda_moda_br = df['Renda'].mode()
renda_moda_homens = df.query('Sexo == 0')['Renda'].mode()
renda_moda_mulheres = df.query('Sexo == 1')['Renda'].mode()
print(f'''
Renda média da população brasileira: {renda_media_br}
Renda média dos homens brasileiros: {renda_media_homens}
Renda média das mulheres brasileiras: {renda_media_mulheres}
Mediana da renda da população brasileira: {renda_mediana_homens}
Mediana da renda dos homens brasileiros: {renda_mediana_homens}
Mediana da renda das mulheres brasileiras: {renda_mediana_mulheres}
Moda da renda da população brasileira: {renda_moda_br[0]}
Moda da renda dos homens brasileiros: {renda_moda_homens[0]}
Moda da renda das mulheres brasileiras: {renda_moda_mulheres[0]}
''')
sns.barplot(data = df, x = df['Cor'].map({0:'Indígena', 2:'Branca', 4:'Preta', 6:'Amarela', 8:'Parda', 9:'Sem declaração'}), y = df['Renda']).set(title = 'Média de Renda por cor')
plt.show()
sns.barplot(data = df, x = df['Cor'].map({0:'Indígena', 2:'Branca', 4:'Preta', 6:'Amarela', 8:'Parda', 9:'Sem declaração'}), y = df['Anos de Estudo']).set(title = 'Média de Anos de Estudo por cor')
plt.show()
sns.barplot(data = df, x = df['Cor'].map({0:'Indígena', 2:'Branca', 4:'Preta', 6:'Amarela', 8:'Parda', 9:'Sem declaração'}), y = df['Idade']).set(title = 'Média de Idade por cor')
plt.show()
sns.barplot(data = df, x = df['Cor'].map({0:'Indígena', 2:'Branca', 4:'Preta', 6:'Amarela', 8:'Parda', 9:'Sem declaração'}), y = df['Altura']).set(title = 'Média de Altura por cor')
plt.show()
sns.barplot(data = df, x = df['Sexo'].map({0:'Homens', 1:'Mulheres'}), y = df['Renda']).set(title = 'Média de Renda por Sexo')
plt.show()
sns.barplot(data = df, x = df['Sexo'].map({0:'Homens', 1:'Mulheres'}), y = df['Anos de Estudo']).set(title = 'Média de Anos de Estudo por sexo')
plt.show()
sns.barplot(data = df, x = df['Sexo'].map({0:'Homens', 1:'Mulheres'}), y = df['Idade']).set(title = 'Média de Idade por sexo')
plt.show()
sns.barplot(data = df, x = df['Sexo'].map({0:'Homens', 1:'Mulheres'}), y = df['Altura']).set(title = 'Média de Altura por sexo')
plt.show()
fig = sns.barplot(data = df, x = df['UF'].map({11:'Rondônia', 12: 'Acre', 13:'Amazonas', 14:'Roraima', 15:'Pará', 16:'Amapá', 17:'Tocantins', 21: 'Maranhão', 22: 'Piauí', 23: 'Ceará', 24: 'Rio Grande do Norte', 25: 'Paraíba', 26: 'Pernambuco', 27: 'Alagoas', 28:'Sergipe', 29:'Bahia', 31:'Minas Gerais', 32:'EspíritoSanto', 33:'Rio de Janeiro', 35:'São Paulo', 41:'Paraná', 42:'Santa Catarina',43:'Rio Grande do Sul', 50:'Mato Grosso do Sul', 51:'Mato Grosso', 52:'Goiás', 53:'Distrito Federal'}), y = df['Renda']).set(title = 'Média de Renda por estado', xlabel = 'Estados')
plt.xticks(rotation=90)
plt.figure(figsize = (10,12))
plt.savefig('teste123.pdf', dpi = 'figure')
plt.show()
sns.histplot(df['Altura'], bins = [1 + 0.1 * i for i in range(12)], kde = True).set(title = 'Distribuição de altura brasileira', ylabel = 'Número de pessoas')
plt.show()
sns.boxplot(x = df['Altura']).set(title = 'Altura brasileira')
plt.show()
sns.histplot(df.query('Sexo == 0')['Altura'], bins = [1 + 0.1 * i for i in range(12)], kde = True).set(title = 'Distribuição de altura masculina', ylabel = 'Número de pessoas')
plt.show()
sns.boxplot(x = df.query('Sexo == 0')['Altura']).set(title = 'Altura masculina')
plt.show()
sns.histplot(df.query('Sexo == 1')['Altura'], bins = [1 + 0.1 * i for i in range(12)], kde = True).set(title = 'Distribuição de altura feminina', ylabel = 'Número de pessoas')
plt.show()
sns.boxplot(x = df.query('Sexo == 1')['Altura']).set(title = 'Altura feminina')
plt.show()
Altura_media_br = df['Altura'].mean()
Altura_media_homens = df.query('Sexo == 0')['Altura'].mean()
Altura_media_mulheres = df.query('Sexo == 1')['Altura'].mean()
Altura_mediana_br = df['Altura'].median()
Altura_mediana_homens = df.query('Sexo == 0')['Altura'].median()
Altura_mediana_mulheres = df.query('Sexo == 1')['Altura'].median()
Altura_moda_br = df['Altura'].mode()
Altura_moda_homens = df.query('Sexo == 0')['Altura'].mode()
Altura_moda_mulheres = df.query('Sexo == 1')['Altura'].mode()
print(f'''
Altura média da população brasileira: {Altura_media_br}
Altura média dos homens brasileiros: {Altura_media_homens}
Altura média das mulheres brasileiras: {Altura_media_mulheres}
Mediana da altura da população brasileira: {Altura_mediana_homens}
Mediana da altura dos homens brasileiros: {Altura_mediana_homens}
Mediana da altura das mulheres brasileiras: {Altura_mediana_mulheres}
Moda da altura da população brasileira: {Altura_moda_br[0]}
Moda da altura dos homens brasileiros: {Altura_moda_homens[0]}
Moda da altura das mulheres brasileiras: {Altura_moda_mulheres[0]}
''')
cores = (list(set(df['Cor'])))
df_adapt = df.copy()
tabela1 = pd.crosstab(df['Renda'] ,df_adapt['Cor'].map({0:'Indígena',2:'Branca',4:'Preta',6:'Amarela',8:'Parda'}))
freq_salario = pd.DataFrame()
freq_salario['Absoluta'] = pd.cut(df['Renda'], bins = [i*1000 for i in range(20)]).value_counts()
# por cor
freq_salario2 = pd.DataFrame()
freq_salario2 = pd.cut(df.query('Cor == 2')['Renda'], bins = [i*1000 for i in range(20)]).value_counts()
rendas = dict()
estudos = dict()
for cor in cores:
estudo_medio = df[df.Cor == int(cor)]['Anos de Estudo'].mean()
rendas[cor] = renda_media
estudos[cor] = estudo_medio
print(estudos)
rendas
freq_salario2
n_M, n_H = 500, 500
significancia = 0.01
confianca = 1 - significancia
n = n_M + n_H
amostra_H = df[df.Sexo == 0]['Renda'].sample(n = n_H, random_state = 1)
amostra_M = df[df.Sexo == 1]['Renda'].sample(n = n_M, random_state = 1)
media_H = amostra_H.mean()
media_M = amostra_M.mean()
media_pop_H = df[df.Sexo == 0]['Renda'].mean()
media_pop_M = df[df.Sexo == 1]['Renda'].mean()
desvio_H = amostra_H.std()
desvio_M = amostra_M.std()
# Z_alpha para bicaudal
probabilidade = confianca + significancia / 2
z_alpha = norm.ppf(probabilidade)
z_alpha2 = norm.ppf(1 - probabilidade)
z_alpha
# Ztest
graus_de_liberdade = n = 500
# Two-sided -> Bicaudal
z, p = ztest(amostra_H, amostra_M, alternative = 'two-sided')
print (f'z = {z} e p = {p}')
t = t_student.ppf(probabilidade, graus_de_liberdade)
print (f't = {t}')
if z >= z_alpha or z <= z_alpha2: # Teste 1
print(f'A hipótese alternativa está correta com {confianca:.0%} de confiança.')
else:
if p < significancia: # Teste 2
print(f'A hipótese nula está correta com {confianca:.0%} de confiança.')
else:
print(f'A hipótese alternativa está correta com {confianca:.0%} de confiança.')
# Z_alpha para unicaudal
probabilidade = confianca
z_alpha = norm.ppf(probabilidade)
z_alpha2 = norm.ppf(1 - probabilidade)
z_alpha, z_alpha2
# Ztest
graus_de_liberdade = n = 500
# Two-sided -> Unicaudal
z, p = ztest(amostra_H, amostra_M, alternative = 'smaller')
print (f'z = {z} e p = {p}')
t = t_student.ppf(probabilidade, graus_de_liberdade)
print (f't = {t}')
if z <= z_alpha: # Teste 1
print(f'A hipótese alternativa está correta com {confianca:.0%} de confiança.')
else:
if p < significancia: # Teste 2
print(f'A hipótese nula está correta com {confianca:.0%} de confiança.')
else:
print(f'A hipótese alternativa está correta com {confianca:.0%} de confiança.')
media_H, media_M ,media_pop_H, media_pop_M
| 0.300848 | 0.898589 |
```
%load_ext autoreload
%autoreload 2
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import os, math
import numpy as np, pandas as pd
import matplotlib.pyplot as plt, seaborn as sns
from tqdm import tqdm, tqdm_notebook
from pathlib import Path
pd.set_option('display.max_columns', 1000)
pd.set_option('display.max_rows', 400)
sns.set()
os.chdir('../..')
from src import utils
DATA = Path('data')
RAW = DATA/'raw'
INTERIM = DATA/'interim'
PROCESSED = DATA/'processed'
SUBMISSIONS = DATA/'submissions'
from src.utils import get_weeks, week_num
week_labels = get_weeks(day_from=20160104, num_weeks=121)[91:]
NEURALNET = INTERIM/'neuralnet'
%%time
train = pd.read_feather(NEURALNET/'train_preproc.feather')
val = pd.read_feather(NEURALNET/'val_preproc.feather')
test = pd.read_feather(NEURALNET/'test_preproc.feather')
challenge = pd.read_csv(RAW/'Challenge_20180423.csv', low_memory=False)
# customer = pd.read_csv(RAW/'Customer.csv', low_memory=False)
# isin = pd.read_csv(RAW/'Isin.csv', low_memory=False)
# submission = pd.read_csv(RAW/'sample_submission.csv', low_memory=False)
# trade = pd.read_csv(RAW/'Trade.csv', low_memory=False)
market = pd.read_csv(RAW/'Market.csv', low_memory=False)
macro = pd.read_csv(RAW/'MarketData_Macro.csv', low_memory=False)
market = market[market.DateKey >= week_labels[0]].copy()
market['Week'] = market.DateKey.apply(
lambda x: week_num(week_labels, x))
market.head()
market['Price'] = market.Price - 100
weeks_mean = market.groupby(['IsinIdx', 'Week'], as_index=False) \
['Price', 'Yield', 'ZSpread'].agg('mean')
weeks_std = market.groupby(['IsinIdx', 'Week'], as_index=False) \
['Price', 'Yield', 'ZSpread'].agg({'Price': 'std',
'Yield': 'std',
'ZSpread': 'std'})
n_weeks = weeks_mean.Week.nunique()
price_dict = {}
yield_dict = {}
zspread_dict = {}
df = weeks_mean.drop_duplicates('IsinIdx')
for i in df.IsinIdx:
price_dict[i] = [0] * n_weeks
yield_dict[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
df = challenge.drop_duplicates('IsinIdx')
for i in df.IsinIdx:
price_dict[i] = [0] * n_weeks
yield_dict[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
for i in train.IsinIdx.unique():
price_dict[i] = [0] * n_weeks
yield_dict[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
for i in val.IsinIdx.unique():
price_dict[i] = [0] * n_weeks
yield_dict[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
for i in test.IsinIdx.unique():
price_dict[i] = [0] * n_weeks
yield_dict[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
for i, w, p, y, z in zip(*[weeks_mean[c] for c in \
['IsinIdx', 'Week', 'Price', 'Yield', 'ZSpread']]):
price_dict[i][w] = p
yield_dict[i][w] = y
zspread_dict[i][w] = z
price_dict_std = {}
yield_dict_std = {}
zspread_dict_std = {}
df = weeks_mean.drop_duplicates('IsinIdx')
for i in df.IsinIdx:
price_dict_std[i] = [0] * n_weeks
yield_dict_std[i] = [0] * n_weeks
zspread_dict_std[i] = [0] * n_weeks
df = challenge.drop_duplicates('IsinIdx')
for i in df.IsinIdx:
price_dict_std[i] = [0] * n_weeks
yield_dict_std[i] = [0] * n_weeks
zspread_dict_std[i] = [0] * n_weeks
for i in train.IsinIdx.unique():
price_dict_std[i] = [0] * n_weeks
yield_dict_std[i] = [0] * n_weeks
zspread_dict_std[i] = [0] * n_weeks
for i in val.IsinIdx.unique():
price_dict_std[i] = [0] * n_weeks
yield_dict_std[i] = [0] * n_weeks
zspread_dict_std[i] = [0] * n_weeks
for i in test.IsinIdx.unique():
price_dict_std[i] = [0] * n_weeks
yield_dict_std[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
for i, w, p, y, z in zip(*[weeks_std[c] for c in \
['IsinIdx', 'Week', 'Price', 'Yield', 'ZSpread']]):
price_dict_std[i][w] = p
yield_dict_std[i][w] = y
zspread_dict_std[i][w] = z
```
## Assign
```
from src.structurednet import shift_right
def roll_sequences(prices, yields, zspreads,
prices_std, yields_std, zspreads_std,
i, w, n_weeks):
return [shift_right(prices[i], w, n_weeks),
shift_right(prices_std[i], w, n_weeks),
shift_right(yields[i], w, n_weeks),
shift_right(yields_std[i], w, n_weeks),
shift_right(zspreads[i], w, n_weeks),
shift_right(zspreads_std[i], w, n_weeks),
]
def extract_seqs(df, prices, yields, zspreads,
prices_std, yields_std, zspreads_std, n_weeks):
return np.array([roll_sequences(prices, yields, zspreads,
prices_std, yields_std, zspreads_std,
i, w, n_weeks) \
for i,w in tqdm_notebook(zip(df.IsinIdx,
df.Week), total=len(df))])
%%time
n_weeks = len(week_labels)
train_seqs = extract_seqs(train, price_dict, yield_dict,
zspread_dict,
price_dict_std, yield_dict_std, zspread_dict_std, n_weeks)
%%time
val_seqs = extract_seqs(val, transactions, buysells,
customers, isins, n_weeks)
test_seqs = extract_seqs(test, transactions, buysells,
customers, isins, n_weeks)
%%time
import pickle
with open(NEURALNET/'market_train_seqs.pkl', 'wb') as f:
pickle.dump(train_seqs, f, pickle.HIGHEST_PROTOCOL)
with open(NEURALNET/'market_val_seqs.pkl', 'wb') as f:
pickle.dump(val_seqs, f, pickle.HIGHEST_PROTOCOL)
with open(NEURALNET/'market_test_seqs.pkl', 'wb') as f:
pickle.dump(test_seqs, f, pickle.HIGHEST_PROTOCOL)
```
## Model
```
from torch.utils.data import DataLoader
from torch import optim
import torch.nn as nn
from src.structured_lstm import MultimodalDataset, MultimodalNet, train_model
%%time
import pickle
with open(NEURALNET/'train_seqs.pkl', 'rb') as f:
orig_train_seqs = pickle.load(f)
with open(NEURALNET/'val_seqs.pkl', 'rb') as f:
orig_val_seqs = pickle.load(f)
with open(NEURALNET/'test_seqs.pkl', 'rb') as f:
orig_test_seqs = pickle.load(f)
orig_train_seqs.shape, train_seqs.shape
np.concatenate([orig_train_seqs, train_seqs]).shape
train_seqs = np.concatenate([orig_train_seqs, train_seqs])
val_seqs = np.concatenate([orig_val_seqs, val_seqs])
test_seqs = np.concatenate([orig_test_seqs, test_seqs])
%%time
train_ds = MultimodalDataset(train[cat_cols], train[num_cols],
train_seqs, train[target_col])
val_ds = MultimodalDataset(val[cat_cols], val[num_cols],
val_seqs, val[target_col])
test_ds = MultimodalDataset(test[cat_cols], test[num_cols],
test_seqs, test[target_col])
cat_cols = ['Sector', 'Subsector', 'Region_x', 'Country',
'TickerIdx', 'Seniority', 'Currency', 'ActivityGroup',
'Region_y', 'Activity', 'RiskCaptain', 'Owner',
'IndustrySector', 'IndustrySubgroup', 'MarketIssue', 'CouponType',
'CompositeRatingCat', 'CustomerIdxCat', 'IsinIdxCat', 'BuySellCat']
num_cols = ['ActualMaturityDateKey', 'IssueDateKey', 'IssuedAmount',
'BondDuration', 'BondRemaining', 'BondLife',
'Day', 'CompositeRating', 'BuySellCont',
'DaysSinceBuySell', 'DaysSinceTransaction', 'DaysSinceCustomerActivity',
'DaysSinceBondActivity', 'DaysCountBuySell', 'DaysCountTransaction',
'DaysCountCustomerActivity', 'DaysCountBondActivity', 'SVD_CustomerBias',
'SVD_IsinBuySellBias', 'SVD_Recommend', 'SVD_CustomerFactor00',
'SVD_CustomerFactor01', 'SVD_CustomerFactor02', 'SVD_CustomerFactor03',
'SVD_CustomerFactor04', 'SVD_CustomerFactor05', 'SVD_CustomerFactor06',
'SVD_CustomerFactor07', 'SVD_CustomerFactor08', 'SVD_CustomerFactor09',
'SVD_CustomerFactor10', 'SVD_CustomerFactor11', 'SVD_CustomerFactor12',
'SVD_CustomerFactor13', 'SVD_CustomerFactor14']
id_cols = ['CustomerIdx', 'IsinIdx', 'BuySell']
target_col = 'CustomerInterest'
```
## Model
```
from torch.utils.data import DataLoader
from torch import optim
import torch.nn as nn
from src.structured_lstm import MultimodalDataset, MultimodalNet, train_model
%%time
train_ds = MultimodalDataset(train[cat_cols], train[num_cols],
train_seqs, train[target_col])
val_ds = MultimodalDataset(val[cat_cols], val[num_cols],
val_seqs, val[target_col])
test_ds = MultimodalDataset(test[cat_cols], test[num_cols],
test_seqs, test[target_col])
all_train_ds = torch.utils.data.ConcatDataset([train_ds, val_ds])
%%time
all_train_dl = DataLoader(all_train_ds, batch_size=128, shuffle=True)
test_dl = DataLoader(test_ds, batch_size=128)
USE_CUDA = True
model = MultimodalNet(emb_szs, n_cont=len(num_cols), emb_drop=0.2,
szs=[1000,500], drops=[0.5, 0.5],
rnn_hidden_sz=64, rnn_input_sz=10, rnn_n_layers=2,
rnn_drop=0.5)
if USE_CUDA: model = model.cuda()
optimizer = optim.Adam(model.parameters(), lr=1e-3)
criterion = nn.BCEWithLogitsLoss()
%%time
model, train_losses, _, _ = train_model(
model, all_train_dl, None, optimizer, criterion,
n_epochs=1, USE_CUDA=USE_CUDA, print_every=800)
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import os, math
import numpy as np, pandas as pd
import matplotlib.pyplot as plt, seaborn as sns
from tqdm import tqdm, tqdm_notebook
from pathlib import Path
pd.set_option('display.max_columns', 1000)
pd.set_option('display.max_rows', 400)
sns.set()
os.chdir('../..')
from src import utils
DATA = Path('data')
RAW = DATA/'raw'
INTERIM = DATA/'interim'
PROCESSED = DATA/'processed'
SUBMISSIONS = DATA/'submissions'
from src.utils import get_weeks, week_num
week_labels = get_weeks(day_from=20160104, num_weeks=121)[91:]
NEURALNET = INTERIM/'neuralnet'
%%time
train = pd.read_feather(NEURALNET/'train_preproc.feather')
val = pd.read_feather(NEURALNET/'val_preproc.feather')
test = pd.read_feather(NEURALNET/'test_preproc.feather')
challenge = pd.read_csv(RAW/'Challenge_20180423.csv', low_memory=False)
# customer = pd.read_csv(RAW/'Customer.csv', low_memory=False)
# isin = pd.read_csv(RAW/'Isin.csv', low_memory=False)
# submission = pd.read_csv(RAW/'sample_submission.csv', low_memory=False)
# trade = pd.read_csv(RAW/'Trade.csv', low_memory=False)
market = pd.read_csv(RAW/'Market.csv', low_memory=False)
macro = pd.read_csv(RAW/'MarketData_Macro.csv', low_memory=False)
market = market[market.DateKey >= week_labels[0]].copy()
market['Week'] = market.DateKey.apply(
lambda x: week_num(week_labels, x))
market.head()
market['Price'] = market.Price - 100
weeks_mean = market.groupby(['IsinIdx', 'Week'], as_index=False) \
['Price', 'Yield', 'ZSpread'].agg('mean')
weeks_std = market.groupby(['IsinIdx', 'Week'], as_index=False) \
['Price', 'Yield', 'ZSpread'].agg({'Price': 'std',
'Yield': 'std',
'ZSpread': 'std'})
n_weeks = weeks_mean.Week.nunique()
price_dict = {}
yield_dict = {}
zspread_dict = {}
df = weeks_mean.drop_duplicates('IsinIdx')
for i in df.IsinIdx:
price_dict[i] = [0] * n_weeks
yield_dict[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
df = challenge.drop_duplicates('IsinIdx')
for i in df.IsinIdx:
price_dict[i] = [0] * n_weeks
yield_dict[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
for i in train.IsinIdx.unique():
price_dict[i] = [0] * n_weeks
yield_dict[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
for i in val.IsinIdx.unique():
price_dict[i] = [0] * n_weeks
yield_dict[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
for i in test.IsinIdx.unique():
price_dict[i] = [0] * n_weeks
yield_dict[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
for i, w, p, y, z in zip(*[weeks_mean[c] for c in \
['IsinIdx', 'Week', 'Price', 'Yield', 'ZSpread']]):
price_dict[i][w] = p
yield_dict[i][w] = y
zspread_dict[i][w] = z
price_dict_std = {}
yield_dict_std = {}
zspread_dict_std = {}
df = weeks_mean.drop_duplicates('IsinIdx')
for i in df.IsinIdx:
price_dict_std[i] = [0] * n_weeks
yield_dict_std[i] = [0] * n_weeks
zspread_dict_std[i] = [0] * n_weeks
df = challenge.drop_duplicates('IsinIdx')
for i in df.IsinIdx:
price_dict_std[i] = [0] * n_weeks
yield_dict_std[i] = [0] * n_weeks
zspread_dict_std[i] = [0] * n_weeks
for i in train.IsinIdx.unique():
price_dict_std[i] = [0] * n_weeks
yield_dict_std[i] = [0] * n_weeks
zspread_dict_std[i] = [0] * n_weeks
for i in val.IsinIdx.unique():
price_dict_std[i] = [0] * n_weeks
yield_dict_std[i] = [0] * n_weeks
zspread_dict_std[i] = [0] * n_weeks
for i in test.IsinIdx.unique():
price_dict_std[i] = [0] * n_weeks
yield_dict_std[i] = [0] * n_weeks
zspread_dict[i] = [0] * n_weeks
for i, w, p, y, z in zip(*[weeks_std[c] for c in \
['IsinIdx', 'Week', 'Price', 'Yield', 'ZSpread']]):
price_dict_std[i][w] = p
yield_dict_std[i][w] = y
zspread_dict_std[i][w] = z
from src.structurednet import shift_right
def roll_sequences(prices, yields, zspreads,
prices_std, yields_std, zspreads_std,
i, w, n_weeks):
return [shift_right(prices[i], w, n_weeks),
shift_right(prices_std[i], w, n_weeks),
shift_right(yields[i], w, n_weeks),
shift_right(yields_std[i], w, n_weeks),
shift_right(zspreads[i], w, n_weeks),
shift_right(zspreads_std[i], w, n_weeks),
]
def extract_seqs(df, prices, yields, zspreads,
prices_std, yields_std, zspreads_std, n_weeks):
return np.array([roll_sequences(prices, yields, zspreads,
prices_std, yields_std, zspreads_std,
i, w, n_weeks) \
for i,w in tqdm_notebook(zip(df.IsinIdx,
df.Week), total=len(df))])
%%time
n_weeks = len(week_labels)
train_seqs = extract_seqs(train, price_dict, yield_dict,
zspread_dict,
price_dict_std, yield_dict_std, zspread_dict_std, n_weeks)
%%time
val_seqs = extract_seqs(val, transactions, buysells,
customers, isins, n_weeks)
test_seqs = extract_seqs(test, transactions, buysells,
customers, isins, n_weeks)
%%time
import pickle
with open(NEURALNET/'market_train_seqs.pkl', 'wb') as f:
pickle.dump(train_seqs, f, pickle.HIGHEST_PROTOCOL)
with open(NEURALNET/'market_val_seqs.pkl', 'wb') as f:
pickle.dump(val_seqs, f, pickle.HIGHEST_PROTOCOL)
with open(NEURALNET/'market_test_seqs.pkl', 'wb') as f:
pickle.dump(test_seqs, f, pickle.HIGHEST_PROTOCOL)
from torch.utils.data import DataLoader
from torch import optim
import torch.nn as nn
from src.structured_lstm import MultimodalDataset, MultimodalNet, train_model
%%time
import pickle
with open(NEURALNET/'train_seqs.pkl', 'rb') as f:
orig_train_seqs = pickle.load(f)
with open(NEURALNET/'val_seqs.pkl', 'rb') as f:
orig_val_seqs = pickle.load(f)
with open(NEURALNET/'test_seqs.pkl', 'rb') as f:
orig_test_seqs = pickle.load(f)
orig_train_seqs.shape, train_seqs.shape
np.concatenate([orig_train_seqs, train_seqs]).shape
train_seqs = np.concatenate([orig_train_seqs, train_seqs])
val_seqs = np.concatenate([orig_val_seqs, val_seqs])
test_seqs = np.concatenate([orig_test_seqs, test_seqs])
%%time
train_ds = MultimodalDataset(train[cat_cols], train[num_cols],
train_seqs, train[target_col])
val_ds = MultimodalDataset(val[cat_cols], val[num_cols],
val_seqs, val[target_col])
test_ds = MultimodalDataset(test[cat_cols], test[num_cols],
test_seqs, test[target_col])
cat_cols = ['Sector', 'Subsector', 'Region_x', 'Country',
'TickerIdx', 'Seniority', 'Currency', 'ActivityGroup',
'Region_y', 'Activity', 'RiskCaptain', 'Owner',
'IndustrySector', 'IndustrySubgroup', 'MarketIssue', 'CouponType',
'CompositeRatingCat', 'CustomerIdxCat', 'IsinIdxCat', 'BuySellCat']
num_cols = ['ActualMaturityDateKey', 'IssueDateKey', 'IssuedAmount',
'BondDuration', 'BondRemaining', 'BondLife',
'Day', 'CompositeRating', 'BuySellCont',
'DaysSinceBuySell', 'DaysSinceTransaction', 'DaysSinceCustomerActivity',
'DaysSinceBondActivity', 'DaysCountBuySell', 'DaysCountTransaction',
'DaysCountCustomerActivity', 'DaysCountBondActivity', 'SVD_CustomerBias',
'SVD_IsinBuySellBias', 'SVD_Recommend', 'SVD_CustomerFactor00',
'SVD_CustomerFactor01', 'SVD_CustomerFactor02', 'SVD_CustomerFactor03',
'SVD_CustomerFactor04', 'SVD_CustomerFactor05', 'SVD_CustomerFactor06',
'SVD_CustomerFactor07', 'SVD_CustomerFactor08', 'SVD_CustomerFactor09',
'SVD_CustomerFactor10', 'SVD_CustomerFactor11', 'SVD_CustomerFactor12',
'SVD_CustomerFactor13', 'SVD_CustomerFactor14']
id_cols = ['CustomerIdx', 'IsinIdx', 'BuySell']
target_col = 'CustomerInterest'
from torch.utils.data import DataLoader
from torch import optim
import torch.nn as nn
from src.structured_lstm import MultimodalDataset, MultimodalNet, train_model
%%time
train_ds = MultimodalDataset(train[cat_cols], train[num_cols],
train_seqs, train[target_col])
val_ds = MultimodalDataset(val[cat_cols], val[num_cols],
val_seqs, val[target_col])
test_ds = MultimodalDataset(test[cat_cols], test[num_cols],
test_seqs, test[target_col])
all_train_ds = torch.utils.data.ConcatDataset([train_ds, val_ds])
%%time
all_train_dl = DataLoader(all_train_ds, batch_size=128, shuffle=True)
test_dl = DataLoader(test_ds, batch_size=128)
USE_CUDA = True
model = MultimodalNet(emb_szs, n_cont=len(num_cols), emb_drop=0.2,
szs=[1000,500], drops=[0.5, 0.5],
rnn_hidden_sz=64, rnn_input_sz=10, rnn_n_layers=2,
rnn_drop=0.5)
if USE_CUDA: model = model.cuda()
optimizer = optim.Adam(model.parameters(), lr=1e-3)
criterion = nn.BCEWithLogitsLoss()
%%time
model, train_losses, _, _ = train_model(
model, all_train_dl, None, optimizer, criterion,
n_epochs=1, USE_CUDA=USE_CUDA, print_every=800)
| 0.361277 | 0.388908 |
# Multivariate Logistic Regression Demo
_Source: 🤖[Homemade Machine Learning](https://github.com/trekhleb/homemade-machine-learning) repository_
> ☝Before moving on with this demo you might want to take a look at:
> - 📗[Math behind the Logistic Regression](https://github.com/trekhleb/homemade-machine-learning/tree/master/homemade/logistic_regression)
> - ⚙️[Logistic Regression Source Code](https://github.com/trekhleb/homemade-machine-learning/blob/master/homemade/logistic_regression/logistic_regression.py)
**Logistic regression** is the appropriate regression analysis to conduct when the dependent variable is dichotomous (binary). Like all regression analyses, the logistic regression is a predictive analysis. Logistic regression is used to describe data and to explain the relationship between one dependent binary variable and one or more nominal, ordinal, interval or ratio-level independent variables.
Logistic Regression is used when the dependent variable (target) is categorical.
For example:
- To predict whether an email is spam (`1`) or (`0`).
- Whether online transaction is fraudulent (`1`) or not (`0`).
- Whether the tumor is malignant (`1`) or not (`0`).
> **Demo Project:** In this example we will train clothes classifier that will recognize clothes types (10 categories) from `28x28` pixel images.
```
# To make debugging of logistic_regression module easier we enable imported modules autoreloading feature.
# By doing this you may change the code of logistic_regression library and all these changes will be available here.
%load_ext autoreload
%autoreload 2
# Add project root folder to module loading paths.
import sys
sys.path.append('../..')
```
### Import Dependencies
- [pandas](https://pandas.pydata.org/) - library that we will use for loading and displaying the data in a table
- [numpy](http://www.numpy.org/) - library that we will use for linear algebra operations
- [matplotlib](https://matplotlib.org/) - library that we will use for plotting the data
- [math](https://docs.python.org/3/library/math.html) - math library that we will use to calculate sqaure roots etc.
- [logistic_regression](https://github.com/trekhleb/homemade-machine-learning/blob/master/homemade/logistic_regression/logistic_regression.py) - custom implementation of logistic regression
```
# Import 3rd party dependencies.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import math
# Import custom logistic regression implementation.
from homemade.logistic_regression import LogisticRegression
```
### Load the Data
In this demo we will use a sample of [Fashion MNIST dataset in a CSV format](https://www.kaggle.com/zalando-research/fashionmnist).
Fashion-MNIST is a dataset of Zalando's article images—consisting of a training set. Each example is a 28x28 grayscale image, associated with a label from 10 classes. Zalando intends Fashion-MNIST to serve as a direct drop-in replacement for the original MNIST dataset for benchmarking machine learning algorithms. It shares the same image size and structure of training and testing splits.
Instead of using full dataset with 60000 training examples we will use cut dataset of just 5000 examples that we will also split into training and testing sets.
Each row in the dataset consists of 785 values: the first value is the label (a category from 0 to 9) and the remaining 784 values (28x28 pixels image) are the pixel values (a number from 0 to 255).
Each training and test example is assigned to one of the following labels:
- 0 T-shirt/top
- 1 Trouser
- 2 Pullover
- 3 Dress
- 4 Coat
- 5 Sandal
- 6 Shirt
- 7 Sneaker
- 8 Bag
- 9 Ankle boot
```
# Load the data.
data = pd.read_csv('../../data/fashion-mnist-demo.csv')
# Laets create the mapping between numeric category and category name.
label_map = {
0: 'T-shirt/top',
1: 'Trouser',
2: 'Pullover',
3: 'Dress',
4: 'Coat',
5: 'Sandal',
6: 'Shirt',
7: 'Sneaker',
8: 'Bag',
9: 'Ankle boot',
}
# Print the data table.
data.head(10)
```
### Plot the Data
Let's peek first 25 rows of the dataset and display them as an images to have an example of clothes we will be working with.
```
# How many images to display.
numbers_to_display = 25
# Calculate the number of cells that will hold all the images.
num_cells = math.ceil(math.sqrt(numbers_to_display))
# Make the plot a little bit bigger than default one.
plt.figure(figsize=(10, 10))
# Go through the first images in a training set and plot them.
for plot_index in range(numbers_to_display):
# Extrace image data.
digit = data[plot_index:plot_index + 1].values
digit_label = digit[0][0]
digit_pixels = digit[0][1:]
# Calculate image size (remember that each picture has square proportions).
image_size = int(math.sqrt(digit_pixels.shape[0]))
# Convert image vector into the matrix of pixels.
frame = digit_pixels.reshape((image_size, image_size))
# Plot the image matrix.
plt.subplot(num_cells, num_cells, plot_index + 1)
plt.imshow(frame, cmap='Greys')
plt.title(label_map[digit_label])
plt.tick_params(axis='both', which='both', bottom=False, left=False, labelbottom=False, labelleft=False)
# Plot all subplots.
plt.subplots_adjust(hspace=0.5, wspace=0.5)
plt.show()
```
### Split the Data Into Training and Test Sets
In this step we will split our dataset into _training_ and _testing_ subsets (in proportion 80/20%).
Training data set will be used for training of our model. Testing dataset will be used for validating of the model. All data from testing dataset will be new to model and we may check how accurate are model predictions.
```
# Split data set on training and test sets with proportions 80/20.
# Function sample() returns a random sample of items.
pd_train_data = data.sample(frac=0.8)
pd_test_data = data.drop(pd_train_data.index)
# Convert training and testing data from Pandas to NumPy format.
train_data = pd_train_data.values
test_data = pd_test_data.values
# Extract training/test labels and features.
num_training_examples = 3000
x_train = train_data[:num_training_examples, 1:]
y_train = train_data[:num_training_examples, [0]]
x_test = test_data[:, 1:]
y_test = test_data[:, [0]]
```
### Init and Train Logistic Regression Model
> ☝🏻This is the place where you might want to play with model configuration.
- `polynomial_degree` - this parameter will allow you to add additional polynomial features of certain degree. More features - more curved the line will be.
- `max_iterations` - this is the maximum number of iterations that gradient descent algorithm will use to find the minimum of a cost function. Low numbers may prevent gradient descent from reaching the minimum. High numbers will make the algorithm work longer without improving its accuracy.
- `regularization_param` - parameter that will fight overfitting. The higher the parameter, the simplier is the model will be.
- `polynomial_degree` - the degree of additional polynomial features (`x1^2 * x2, x1^2 * x2^2, ...`). This will allow you to curve the predictions.
- `sinusoid_degree` - the degree of sinusoid parameter multipliers of additional features (`sin(x), sin(2*x), ...`). This will allow you to curve the predictions by adding sinusoidal component to the prediction curve.
- `normalize_data` - boolean flag that indicates whether data normalization is needed or not.
```
# Set up linear regression parameters.
max_iterations = 10000 # Max number of gradient descent iterations.
regularization_param = 25 # Helps to fight model overfitting.
polynomial_degree = 0 # The degree of additional polynomial features.
sinusoid_degree = 0 # The degree of sinusoid parameter multipliers of additional features.
normalize_data = True # Whether we need to normalize data to make it more unifrom or not.
# Init logistic regression instance.
logistic_regression = LogisticRegression(x_train, y_train, polynomial_degree, sinusoid_degree, normalize_data)
# Train logistic regression.
(thetas, costs) = logistic_regression.train(regularization_param, max_iterations)
```
### Print Training Results
Let's see how model parameters (thetas) look like. For each digit class (from 0 to 9) we've just trained a set of 784 parameters (one theta for each image pixel). These parameters represents the importance of every pixel for specific digit recognition.
```
# Print thetas table.
pd.DataFrame(thetas)
```
### Illustrate Hidden Layers Perceptrons
Each perceptron in the hidden layer learned something from the training process. What it learned is represented by input theta parameters for it. Each perceptron in the hidden layer has 28x28 input thetas (one for each input image pizel). Each theta represents how valuable each pixel is for this particuar perceptron. So let's try to plot how valuable each pixel of input image is for each perceptron based on its theta values.
```
# How many images to display.
numbers_to_display = 9
# Calculate the number of cells that will hold all the images.
num_cells = math.ceil(math.sqrt(numbers_to_display))
# Make the plot a little bit bigger than default one.
plt.figure(figsize=(10, 10))
# Go through the thetas and print them.
for plot_index in range(numbers_to_display):
# Extrace thetas data.
digit_pixels = thetas[plot_index][1:]
# Calculate image size (remember that each picture has square proportions).
image_size = int(math.sqrt(digit_pixels.shape[0]))
# Convert image vector into the matrix of pixels.
frame = digit_pixels.reshape((image_size, image_size))
# Plot the thetas matrix.
plt.subplot(num_cells, num_cells, plot_index + 1)
plt.imshow(frame, cmap='Greys')
plt.title(plot_index)
plt.tick_params(axis='both', which='both', bottom=False, left=False, labelbottom=False, labelleft=False)
# Plot all subplots.
plt.subplots_adjust(hspace=0.5, wspace=0.5)
plt.show()
```
### Analyze Gradient Descent Progress
The plot below illustrates how the cost function value changes over each iteration. You should see it decreasing.
In case if cost function value increases it may mean that gradient descent missed the cost function minimum and with each step it goes further away from it.
From this plot you may also get an understanding of how many iterations you need to get an optimal value of the cost function.
```
# Draw gradient descent progress for each label.
labels = logistic_regression.unique_labels
for index, label in enumerate(labels):
plt.plot(range(len(costs[index])), costs[index], label=label_map[labels[index]])
plt.xlabel('Gradient Steps')
plt.ylabel('Cost')
plt.legend()
plt.show()
```
### Calculate Model Training Precision
Calculate how many of training and test examples have been classified correctly. Normally we need test precission to be as high as possible. In case if training precision is high and test precission is low it may mean that our model is overfitted (it works really well with the training data set but it is not good at classifying new unknown data from the test dataset). In this case you may want to play with `regularization_param` parameter to fighth the overfitting.
```
# Make training set predictions.
y_train_predictions = logistic_regression.predict(x_train)
y_test_predictions = logistic_regression.predict(x_test)
# Check what percentage of them are actually correct.
train_precision = np.sum(y_train_predictions == y_train) / y_train.shape[0] * 100
test_precision = np.sum(y_test_predictions == y_test) / y_test.shape[0] * 100
print('Training Precision: {:5.4f}%'.format(train_precision))
print('Test Precision: {:5.4f}%'.format(test_precision))
```
### Plot Test Dataset Predictions
In order to illustrate how our model classifies unknown examples let's plot first 64 predictions for testing dataset. All green clothes on the plot below have been recognized corrctly but all the red clothes have not been recognized correctly by our classifier. On top of each image you may see the clothes class (type) that has been recognized on the image.
```
# How many numbers to display.
numbers_to_display = 64
# Calculate the number of cells that will hold all the numbers.
num_cells = math.ceil(math.sqrt(numbers_to_display))
# Make the plot a little bit bigger than default one.
plt.figure(figsize=(15, 15))
# Go through the first numbers in a test set and plot them.
for plot_index in range(numbers_to_display):
# Extrace digit data.
digit_label = y_test[plot_index, 0]
digit_pixels = x_test[plot_index, :]
# Predicted label.
predicted_label = y_test_predictions[plot_index][0]
# Calculate image size (remember that each picture has square proportions).
image_size = int(math.sqrt(digit_pixels.shape[0]))
# Convert image vector into the matrix of pixels.
frame = digit_pixels.reshape((image_size, image_size))
# Plot the number matrix.
color_map = 'Greens' if predicted_label == digit_label else 'Reds'
plt.subplot(num_cells, num_cells, plot_index + 1)
plt.imshow(frame, cmap=color_map)
plt.title(label_map[predicted_label])
plt.tick_params(axis='both', which='both', bottom=False, left=False, labelbottom=False, labelleft=False)
# Plot all subplots.
plt.subplots_adjust(hspace=0.5, wspace=0.5)
plt.show()
```
|
github_jupyter
|
# To make debugging of logistic_regression module easier we enable imported modules autoreloading feature.
# By doing this you may change the code of logistic_regression library and all these changes will be available here.
%load_ext autoreload
%autoreload 2
# Add project root folder to module loading paths.
import sys
sys.path.append('../..')
# Import 3rd party dependencies.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import math
# Import custom logistic regression implementation.
from homemade.logistic_regression import LogisticRegression
# Load the data.
data = pd.read_csv('../../data/fashion-mnist-demo.csv')
# Laets create the mapping between numeric category and category name.
label_map = {
0: 'T-shirt/top',
1: 'Trouser',
2: 'Pullover',
3: 'Dress',
4: 'Coat',
5: 'Sandal',
6: 'Shirt',
7: 'Sneaker',
8: 'Bag',
9: 'Ankle boot',
}
# Print the data table.
data.head(10)
# How many images to display.
numbers_to_display = 25
# Calculate the number of cells that will hold all the images.
num_cells = math.ceil(math.sqrt(numbers_to_display))
# Make the plot a little bit bigger than default one.
plt.figure(figsize=(10, 10))
# Go through the first images in a training set and plot them.
for plot_index in range(numbers_to_display):
# Extrace image data.
digit = data[plot_index:plot_index + 1].values
digit_label = digit[0][0]
digit_pixels = digit[0][1:]
# Calculate image size (remember that each picture has square proportions).
image_size = int(math.sqrt(digit_pixels.shape[0]))
# Convert image vector into the matrix of pixels.
frame = digit_pixels.reshape((image_size, image_size))
# Plot the image matrix.
plt.subplot(num_cells, num_cells, plot_index + 1)
plt.imshow(frame, cmap='Greys')
plt.title(label_map[digit_label])
plt.tick_params(axis='both', which='both', bottom=False, left=False, labelbottom=False, labelleft=False)
# Plot all subplots.
plt.subplots_adjust(hspace=0.5, wspace=0.5)
plt.show()
# Split data set on training and test sets with proportions 80/20.
# Function sample() returns a random sample of items.
pd_train_data = data.sample(frac=0.8)
pd_test_data = data.drop(pd_train_data.index)
# Convert training and testing data from Pandas to NumPy format.
train_data = pd_train_data.values
test_data = pd_test_data.values
# Extract training/test labels and features.
num_training_examples = 3000
x_train = train_data[:num_training_examples, 1:]
y_train = train_data[:num_training_examples, [0]]
x_test = test_data[:, 1:]
y_test = test_data[:, [0]]
# Set up linear regression parameters.
max_iterations = 10000 # Max number of gradient descent iterations.
regularization_param = 25 # Helps to fight model overfitting.
polynomial_degree = 0 # The degree of additional polynomial features.
sinusoid_degree = 0 # The degree of sinusoid parameter multipliers of additional features.
normalize_data = True # Whether we need to normalize data to make it more unifrom or not.
# Init logistic regression instance.
logistic_regression = LogisticRegression(x_train, y_train, polynomial_degree, sinusoid_degree, normalize_data)
# Train logistic regression.
(thetas, costs) = logistic_regression.train(regularization_param, max_iterations)
# Print thetas table.
pd.DataFrame(thetas)
# How many images to display.
numbers_to_display = 9
# Calculate the number of cells that will hold all the images.
num_cells = math.ceil(math.sqrt(numbers_to_display))
# Make the plot a little bit bigger than default one.
plt.figure(figsize=(10, 10))
# Go through the thetas and print them.
for plot_index in range(numbers_to_display):
# Extrace thetas data.
digit_pixels = thetas[plot_index][1:]
# Calculate image size (remember that each picture has square proportions).
image_size = int(math.sqrt(digit_pixels.shape[0]))
# Convert image vector into the matrix of pixels.
frame = digit_pixels.reshape((image_size, image_size))
# Plot the thetas matrix.
plt.subplot(num_cells, num_cells, plot_index + 1)
plt.imshow(frame, cmap='Greys')
plt.title(plot_index)
plt.tick_params(axis='both', which='both', bottom=False, left=False, labelbottom=False, labelleft=False)
# Plot all subplots.
plt.subplots_adjust(hspace=0.5, wspace=0.5)
plt.show()
# Draw gradient descent progress for each label.
labels = logistic_regression.unique_labels
for index, label in enumerate(labels):
plt.plot(range(len(costs[index])), costs[index], label=label_map[labels[index]])
plt.xlabel('Gradient Steps')
plt.ylabel('Cost')
plt.legend()
plt.show()
# Make training set predictions.
y_train_predictions = logistic_regression.predict(x_train)
y_test_predictions = logistic_regression.predict(x_test)
# Check what percentage of them are actually correct.
train_precision = np.sum(y_train_predictions == y_train) / y_train.shape[0] * 100
test_precision = np.sum(y_test_predictions == y_test) / y_test.shape[0] * 100
print('Training Precision: {:5.4f}%'.format(train_precision))
print('Test Precision: {:5.4f}%'.format(test_precision))
# How many numbers to display.
numbers_to_display = 64
# Calculate the number of cells that will hold all the numbers.
num_cells = math.ceil(math.sqrt(numbers_to_display))
# Make the plot a little bit bigger than default one.
plt.figure(figsize=(15, 15))
# Go through the first numbers in a test set and plot them.
for plot_index in range(numbers_to_display):
# Extrace digit data.
digit_label = y_test[plot_index, 0]
digit_pixels = x_test[plot_index, :]
# Predicted label.
predicted_label = y_test_predictions[plot_index][0]
# Calculate image size (remember that each picture has square proportions).
image_size = int(math.sqrt(digit_pixels.shape[0]))
# Convert image vector into the matrix of pixels.
frame = digit_pixels.reshape((image_size, image_size))
# Plot the number matrix.
color_map = 'Greens' if predicted_label == digit_label else 'Reds'
plt.subplot(num_cells, num_cells, plot_index + 1)
plt.imshow(frame, cmap=color_map)
plt.title(label_map[predicted_label])
plt.tick_params(axis='both', which='both', bottom=False, left=False, labelbottom=False, labelleft=False)
# Plot all subplots.
plt.subplots_adjust(hspace=0.5, wspace=0.5)
plt.show()
| 0.758511 | 0.994066 |
# Fashion-MNIST
```
%matplotlib inline
from matplotlib import pyplot as plt
from mxnet.gluon import data as gdata
from mxnet import ndarray as nd
from mxnet import autograd, nd
import math
import sys
import time
```
### Download the dataset
```
mnist_train = gdata.vision.FashionMNIST(train=True)
mnist_test = gdata.vision.FashionMNIST(train=False)
print(len(mnist_train),len(mnist_test))
```
### Access one experiment
```
feature, label = mnist_train[0]
print(feature.shape, feature.dtype, label, type(label), label.dtype)
```
### Set labels
```
def get_fashion_mnist_labels(labels):
text_labels = ['t-shirt', 'trouser', 'pullover', 'dress', 'coat',
'sandal', 'shirt', 'sneaker', 'bag', 'ankle boot']
return [text_labels[int(i)] for i in labels]
```
### Display some of training data
```
def show_fashion_mnist(images, labels):
fig, axs = plt.subplots(1, len(images),figsize=(12,12))
for f, img, lbl in zip(axs, images, labels):
f.imshow(img.reshape((28,28)).asnumpy())
f.set_title(lbl)
f.axes.get_xaxis().set_visible(False)
f.axes.get_yaxis().set_visible(False)
X, y = mnist_train[0:9]
show_fashion_mnist(X, get_fashion_mnist_labels(y))
```
### Load data in batches
```
batch_size = 256
transformer = gdata.vision.transforms.ToTensor()
if sys.platform.startswith('win'):
num_workers = 0
else:
num_workers = 4
train_iter = gdata.DataLoader(mnist_train.transform_first(transformer),
batch_size, shuffle=True,
num_workers=num_workers)
test_iter = gdata.DataLoader(mnist_test.transform_first(transformer),
batch_size, shuffle=False,
num_workers=num_workers)
```
### Initialize model parameter
```
num_inputs = 784 # we have 28x28 images
num_outputs = 10 # different labels
W = nd.random.normal(scale=0.01, shape=(num_inputs, num_outputs))
b = nd.zeros(num_outputs)
# Attach gradients
W.attach_grad()
b.attach_grad()
```
### Define the model
```
def softmax(X):
X_exp = X.exp()
partition = X_exp.sum(axis=1, keepdims=True)
return X_exp / partition # using broadcast
def net(X):
return softmax(nd.dot(X.reshape((-1, num_inputs)), W) + b)
```
### Define the loss function
```
def cross_entropy(y_hat, y):
return -nd.pick(y_hat, y).log()
```
### Define classification accuracy
```
def accuracy(y_hat, y):
return (y_hat.argmax(axis=1) == y_astype('float32')).mean().asscalar()
def evaluate_accuracy(data_iter, net):
acc_sum, n = 0.0, 0
for X, y in data_iter:
y = y.astype('float32')
acc_sum += (net(X).argmax(axis=1) == y).sum().asscalar()
n += y.size
return acc_sum / n
evaluate_accuracy(test_iter, net)
```
### Training loop
```
# Model training helper
def train(net, train_iter, test_iter, loss, num_epochs, batch_size,
params=None, lr=None, trainer=None):
for epoch in range(num_epochs):
train_l_sum, train_acc_sum, n = 0.0, 0.0, 0
for X, y in train_iter:
with autograd.record():
y_hat = net(X)
l = loss(y_hat, y).sum()
l.backward()
if trainer is None:
for param in params:
param[:] = param - lr * param.grad / batch_size
else:
trainer.step(batch_size)
y = y.astype('float32')
train_l_sum += l.asscalar()
train_acc_sum += (y_hat.argmax(axis=1)==y).sum().asscalar()
n += y.size
test_acc = evaluate_accuracy(test_iter, net)
print('epoch %d, loss %.4f, train acc %.3f, test acc %.3f'
% (epoch+1, train_l_sum / n, train_acc_sum / n, test_acc))
num_epochs, lr = 10, 0.1
train(net, train_iter, test_iter, cross_entropy, num_epochs,
batch_size, [W,b], lr)
```
### Prediction
```
for X, y in test_iter:
break
true_labels = get_fashion_mnist_labels(y.asnumpy())
pred_labels = get_fashion_mnist_labels(net(X).argmax(axis=1).asnumpy())
titles = [truelabel + '\n' + predlabel for truelabel, predlabel in zip(true_labels, pred_labels)]
show_fashion_mnist(X[0:9], titles[0:9])
```
# Using Gluon
### Define the model
```
from mxnet.gluon import nn
net = nn.Sequential()
net.add(nn.Dense(10))
```
### Define classification accuracy
```
def accuracy(y_hat, y):
return (y_hat.argmax(axis=1) == y.astype('float32')).sum().asscalar()
def evaluate_accuracy(net, data_iter, ctx=None):
if not ctx: # Query the first device the first parameter is on.
ctx = list(net.collect_params().values())[0].list_ctx()[0]
metric = [0.0, 0] # num_corrected_examples, num_examples
for X, y in data_iter:
X, y = X.as_in_context(ctx), y.as_in_context(ctx)
metric[0] = metric[0] + accuracy(net(X), y)
metric[1] = metric[1] + y.size
return metric[0]/metric[1]
from mxnet import context
# Helper
def try_gpu(i=0):
"""Return gpu(i) if exists, otherwise return cpu()."""
return context.gpu(i) if context.num_gpus() >= i + 1 else context.cpu()
```
### Training loop
```
from mxnet import gluon, init
def train(net, train_iter, test_iter, num_epochs, lr, ctx=try_gpu()):
net.initialize(force_reinit=True, ctx=ctx, init=init.Xavier())
loss = gluon.loss.SoftmaxCrossEntropyLoss()
trainer = gluon.Trainer(net.collect_params(),
'sgd', {'learning_rate': lr})
for epoch in range(num_epochs):
metric = [0.0,0.0,0] # train_loss, train_acc, num_examples
for i, (X, y) in enumerate(train_iter):
X, y = X.as_in_context(ctx), y.as_in_context(ctx)
with autograd.record():
y_hat = net(X)
l = loss(y_hat, y)
l.backward()
trainer.step(X.shape[0])
# Update metrics
metric[0] = metric[0] + l.sum().asscalar()
metric[1] = metric[1] + accuracy(y_hat, y)
metric[2] = metric[2] + X.shape[0]
train_loss, train_acc = metric[0]/metric[2], metric[1]/metric[2]
test_acc = evaluate_accuracy(net, test_iter)
print('epoch %d, loss %.4f, train acc %.3f, test acc %.3f'
% (epoch+1, train_loss, train_acc, test_acc))
print('loss %.3f, train acc %.3f, test acc %.3f' % (train_loss, train_acc, test_acc))
lr, num_epochs = 0.9, 10
train(net, train_iter, test_iter, num_epochs, lr)
```
### Prediction
```
for X, y in test_iter:
break
true_labels = get_fashion_mnist_labels(y.asnumpy())
pred_labels = get_fashion_mnist_labels(net(X).argmax(axis=1).asnumpy())
titles = [truelabel + '\n' + predlabel for truelabel, predlabel in zip(true_labels, pred_labels)]
show_fashion_mnist(X[0:9], titles[0:9])
```
|
github_jupyter
|
%matplotlib inline
from matplotlib import pyplot as plt
from mxnet.gluon import data as gdata
from mxnet import ndarray as nd
from mxnet import autograd, nd
import math
import sys
import time
mnist_train = gdata.vision.FashionMNIST(train=True)
mnist_test = gdata.vision.FashionMNIST(train=False)
print(len(mnist_train),len(mnist_test))
feature, label = mnist_train[0]
print(feature.shape, feature.dtype, label, type(label), label.dtype)
def get_fashion_mnist_labels(labels):
text_labels = ['t-shirt', 'trouser', 'pullover', 'dress', 'coat',
'sandal', 'shirt', 'sneaker', 'bag', 'ankle boot']
return [text_labels[int(i)] for i in labels]
def show_fashion_mnist(images, labels):
fig, axs = plt.subplots(1, len(images),figsize=(12,12))
for f, img, lbl in zip(axs, images, labels):
f.imshow(img.reshape((28,28)).asnumpy())
f.set_title(lbl)
f.axes.get_xaxis().set_visible(False)
f.axes.get_yaxis().set_visible(False)
X, y = mnist_train[0:9]
show_fashion_mnist(X, get_fashion_mnist_labels(y))
batch_size = 256
transformer = gdata.vision.transforms.ToTensor()
if sys.platform.startswith('win'):
num_workers = 0
else:
num_workers = 4
train_iter = gdata.DataLoader(mnist_train.transform_first(transformer),
batch_size, shuffle=True,
num_workers=num_workers)
test_iter = gdata.DataLoader(mnist_test.transform_first(transformer),
batch_size, shuffle=False,
num_workers=num_workers)
num_inputs = 784 # we have 28x28 images
num_outputs = 10 # different labels
W = nd.random.normal(scale=0.01, shape=(num_inputs, num_outputs))
b = nd.zeros(num_outputs)
# Attach gradients
W.attach_grad()
b.attach_grad()
def softmax(X):
X_exp = X.exp()
partition = X_exp.sum(axis=1, keepdims=True)
return X_exp / partition # using broadcast
def net(X):
return softmax(nd.dot(X.reshape((-1, num_inputs)), W) + b)
def cross_entropy(y_hat, y):
return -nd.pick(y_hat, y).log()
def accuracy(y_hat, y):
return (y_hat.argmax(axis=1) == y_astype('float32')).mean().asscalar()
def evaluate_accuracy(data_iter, net):
acc_sum, n = 0.0, 0
for X, y in data_iter:
y = y.astype('float32')
acc_sum += (net(X).argmax(axis=1) == y).sum().asscalar()
n += y.size
return acc_sum / n
evaluate_accuracy(test_iter, net)
# Model training helper
def train(net, train_iter, test_iter, loss, num_epochs, batch_size,
params=None, lr=None, trainer=None):
for epoch in range(num_epochs):
train_l_sum, train_acc_sum, n = 0.0, 0.0, 0
for X, y in train_iter:
with autograd.record():
y_hat = net(X)
l = loss(y_hat, y).sum()
l.backward()
if trainer is None:
for param in params:
param[:] = param - lr * param.grad / batch_size
else:
trainer.step(batch_size)
y = y.astype('float32')
train_l_sum += l.asscalar()
train_acc_sum += (y_hat.argmax(axis=1)==y).sum().asscalar()
n += y.size
test_acc = evaluate_accuracy(test_iter, net)
print('epoch %d, loss %.4f, train acc %.3f, test acc %.3f'
% (epoch+1, train_l_sum / n, train_acc_sum / n, test_acc))
num_epochs, lr = 10, 0.1
train(net, train_iter, test_iter, cross_entropy, num_epochs,
batch_size, [W,b], lr)
for X, y in test_iter:
break
true_labels = get_fashion_mnist_labels(y.asnumpy())
pred_labels = get_fashion_mnist_labels(net(X).argmax(axis=1).asnumpy())
titles = [truelabel + '\n' + predlabel for truelabel, predlabel in zip(true_labels, pred_labels)]
show_fashion_mnist(X[0:9], titles[0:9])
from mxnet.gluon import nn
net = nn.Sequential()
net.add(nn.Dense(10))
def accuracy(y_hat, y):
return (y_hat.argmax(axis=1) == y.astype('float32')).sum().asscalar()
def evaluate_accuracy(net, data_iter, ctx=None):
if not ctx: # Query the first device the first parameter is on.
ctx = list(net.collect_params().values())[0].list_ctx()[0]
metric = [0.0, 0] # num_corrected_examples, num_examples
for X, y in data_iter:
X, y = X.as_in_context(ctx), y.as_in_context(ctx)
metric[0] = metric[0] + accuracy(net(X), y)
metric[1] = metric[1] + y.size
return metric[0]/metric[1]
from mxnet import context
# Helper
def try_gpu(i=0):
"""Return gpu(i) if exists, otherwise return cpu()."""
return context.gpu(i) if context.num_gpus() >= i + 1 else context.cpu()
from mxnet import gluon, init
def train(net, train_iter, test_iter, num_epochs, lr, ctx=try_gpu()):
net.initialize(force_reinit=True, ctx=ctx, init=init.Xavier())
loss = gluon.loss.SoftmaxCrossEntropyLoss()
trainer = gluon.Trainer(net.collect_params(),
'sgd', {'learning_rate': lr})
for epoch in range(num_epochs):
metric = [0.0,0.0,0] # train_loss, train_acc, num_examples
for i, (X, y) in enumerate(train_iter):
X, y = X.as_in_context(ctx), y.as_in_context(ctx)
with autograd.record():
y_hat = net(X)
l = loss(y_hat, y)
l.backward()
trainer.step(X.shape[0])
# Update metrics
metric[0] = metric[0] + l.sum().asscalar()
metric[1] = metric[1] + accuracy(y_hat, y)
metric[2] = metric[2] + X.shape[0]
train_loss, train_acc = metric[0]/metric[2], metric[1]/metric[2]
test_acc = evaluate_accuracy(net, test_iter)
print('epoch %d, loss %.4f, train acc %.3f, test acc %.3f'
% (epoch+1, train_loss, train_acc, test_acc))
print('loss %.3f, train acc %.3f, test acc %.3f' % (train_loss, train_acc, test_acc))
lr, num_epochs = 0.9, 10
train(net, train_iter, test_iter, num_epochs, lr)
for X, y in test_iter:
break
true_labels = get_fashion_mnist_labels(y.asnumpy())
pred_labels = get_fashion_mnist_labels(net(X).argmax(axis=1).asnumpy())
titles = [truelabel + '\n' + predlabel for truelabel, predlabel in zip(true_labels, pred_labels)]
show_fashion_mnist(X[0:9], titles[0:9])
| 0.436862 | 0.981022 |
# Practice Notebook: Reading and Writing Files
In this exercise, we will test your knowledge of reading and writing files by playing around with some text files.
<br><br>
Let's say we have a text file containing current visitors at a hotel. We'll call it, *guests.txt*. Run the following code to create the file. The file will automatically populate with each initial guest's first name on its own line.
```
guests = open("guests.txt", "w")
initial_guests = ["Bob", "Andrea", "Manuel", "Polly", "Khalid"]
for i in initial_guests:
guests.write(i + "\n")
guests.close()
```
No output is generated for the above code cell. To check the contents of the newly created *guests.txt* file, run the following code.
```
with open("guests.txt") as guests:
for line in guests:
print(line)
```
The output shows that our *guests.txt* file is correctly populated with each initial guest's first name on its own line. Cool!
<br><br>
Now suppose we want to update our file as guests check in and out. Fill in the missing code in the following cell to add guests to the *guests.txt* file as they check in.
```
new_guests = ["Sam", "Danielle", "Jacob"]
with open("guests.txt", "a") as guests:
for i in new_guests:
guests.write(i + "\n")
guests.close()
```
To check whether your code correctly added the new guests to the *guests.txt* file, run the following cell.
```
with open("guests.txt") as guests:
for line in guests:
print(line)
```
The current names in the *guests.txt* file should be: Bob, Andrea, Manuel, Polly, Khalid, Sam, Danielle and Jacob.
<br><br>
Was the *guests.txt* file correctly appended with the new guests? If not, go back and edit your code making sure to fill in the gaps appropriately so that the new guests are correctly added to the *guests.txt* file. Once the new guests are successfully added, you have filled in the missing code correctly. Great!
<br><br>
Now let's remove the guests that have checked out already. There are several ways to do this, however, the method we will choose for this exercise is outlined as follows:
1. Open the file in "read" mode.
2. Iterate over each line in the file and put each guest's name into a Python list.
3. Open the file once again in "write" mode.
4. Add each guest's name in the Python list to the file one by one.
<br>
Ready? Fill in the missing code in the following cell to remove the guests that have checked out already.
```
checked_out=["Andrea", "Manuel", "Khalid"]
temp_list=[]
with open("guests.txt", "r") as guests:
for g in guests:
temp_list.append(g.strip())
with open("guests.txt", "w") as guests:
for name in temp_list:
if name not in checked_out:
guests.write(name + "\n")
```
To check whether your code correctly removed the checked out guests from the *guests.txt* file, run the following cell.
```
with open("guests.txt") as guests:
for line in guests:
print(line)
```
The current names in the *guests.txt* file should be: Bob, Polly, Sam, Danielle and Jacob.
<br><br>
Were the names of the checked out guests correctly removed from the *guests.txt* file? If not, go back and edit your code making sure to fill in the gaps appropriately so that the checked out guests are correctly removed from the *guests.txt* file. Once the checked out guests are successfully removed, you have filled in the missing code correctly. Awesome!
<br><br>
Now let's check whether Bob and Andrea are still checked in. How could we do this? We'll just read through each line in the file to see if their name is in there. Run the following code to check whether Bob and Andrea are still checked in.
```
guests_to_check = ['Bob', 'Andrea']
checked_in = []
with open("guests.txt","r") as guests:
for g in guests:
checked_in.append(g.strip())
for check in guests_to_check:
if check in checked_in:
print("{} is checked in".format(check))
else:
print("{} is not checked in".format(check))
```
We can see that Bob is checked in while Andrea is not. Nice work! You've learned the basics of reading and writing files in Python!
|
github_jupyter
|
guests = open("guests.txt", "w")
initial_guests = ["Bob", "Andrea", "Manuel", "Polly", "Khalid"]
for i in initial_guests:
guests.write(i + "\n")
guests.close()
with open("guests.txt") as guests:
for line in guests:
print(line)
new_guests = ["Sam", "Danielle", "Jacob"]
with open("guests.txt", "a") as guests:
for i in new_guests:
guests.write(i + "\n")
guests.close()
with open("guests.txt") as guests:
for line in guests:
print(line)
checked_out=["Andrea", "Manuel", "Khalid"]
temp_list=[]
with open("guests.txt", "r") as guests:
for g in guests:
temp_list.append(g.strip())
with open("guests.txt", "w") as guests:
for name in temp_list:
if name not in checked_out:
guests.write(name + "\n")
with open("guests.txt") as guests:
for line in guests:
print(line)
guests_to_check = ['Bob', 'Andrea']
checked_in = []
with open("guests.txt","r") as guests:
for g in guests:
checked_in.append(g.strip())
for check in guests_to_check:
if check in checked_in:
print("{} is checked in".format(check))
else:
print("{} is not checked in".format(check))
| 0.063759 | 0.893402 |
```
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import matplotlib as mpl
import pickle
mpl.rcParams['pdf.fonttype'] = 42
mpl.rcParams['ps.fonttype'] = 42
mpl.rcParams['font.family'] = 'Arial'
fig = plt.figure(figsize=(6.4,4.8))
ax1 = plt.axes([0.1,0.1,0.55,0.80])
ax2 = plt.axes([0.75,0.1,0.20,0.80])
ax1.spines['right'].set_visible(False)
ax1.spines['top'].set_visible(False)
ax2.spines['right'].set_visible(False)
ax2.spines['top'].set_visible(False)
x1, x2, x3 = [1,6], [2,7], [3,8]
y1, y2, y3 = [3,4], [1,3], [4,8]
'''
Note for the y1,y2,y3:
deephlapan result can be accessed in ../data/deephlapan_result_cell.csv
IEDB result can be accessed in ../data/ori_test_cells.csv
DeepImmuno-CNN results are the highest score for 10-fold validation, detail can be seen in Supp_fig_3
'''
ax1.bar(x1,y1,color='#E36DF2',width=0.8,label='DeepHLApan')
ax1.bar(x2,y2,color='#04BF7B',width=0.8,label='IEDB')
ax1.bar(x3,y3,color='#F26D6D',width=0.8,label='DeepImmuno-CNN')
ax1.set_xticks([2,7])
ax1.set_ylabel('Number of immunogenic peptides')
ax1.set_xticklabels(['Top20','Top50'])
ax1.legend(frameon=True)
ax1.grid(alpha=0.3,axis='y')
tmp1x = [1,2,3,6,7,8]
tmp1y = [3,1,4,4,3,8]
for i in range(len(tmp1x)):
ax1.text(tmp1x[i]-0.2,tmp1y[i]+0.1,s=tmp1y[i])
ax2.bar([1,2,3],[0.34,0.63,0.83],color=['#E36DF2','#04BF7B','#F26D6D'])
ax2.set_ylim([0,1.05])
ax2.set_ylabel('Sensitivity')
ax2.grid(alpha=0.3,axis='y')
ax2.set_xticks([2])
ax2.set_xticklabels(['hard cutoff'])
tmp2x = [1,2,3]
tmp2y = [0.34,0.63,0.83]
for i in range(len(tmp2x)):
ax2.text(tmp2x[i]-0.3,tmp2y[i]+0.01,s=tmp2y[i])
'''
Again, the data used to plot the bar chart can be accessed in as below
DeepHLApan: ../data/covid_predicted_result.csv
IEDB: ../data/sars_cov_2_result.csv
'''
fig = plt.figure(figsize=(6.4,4.8))
ax1 = plt.axes([0.1,0.1,0.35,0.80])
ax2 = plt.axes([0.55,0.1,0.35,0.80])
ax1.spines['right'].set_visible(False)
ax1.spines['top'].set_visible(False)
ax2.spines['right'].set_visible(False)
ax2.spines['top'].set_visible(False)
ax1.bar([0,5],[0.4,0.14],color='#E36DF2',label='Deephlapan',width=0.8)
ax1.bar([1,6],[0.52,0.38],color='#04BF7B',label='IEDB',width=0.8)
ax1.bar([2,7],[0.68,0.88],color='#F26D6D',label='DeepImmuno-CNN',width=0.8)
ax1.set_xticks([1,6])
ax1.set_xticklabels(['Convalescent','Unexposed'])
ax1.legend(frameon=True)
ax1.grid(True,alpha=0.3,axis='y')
ax1.set_ylabel('Recall')
ax2.bar([10,15],[0.28,0.05],color='#E36DF2',label='Deephlapan',width=0.8)
ax2.bar([11,16],[0.25,0.02],color='#04BF7B',label='IEDB',width=0.8)
ax2.bar([12,17],[0.28,0.11],color='#F26D6D',label='DeepImmuno-CNN',width=0.8)
ax2.set_xticks([11,16])
ax2.set_xticklabels(['Convalescent','Unexposed'])
ax2.set_ylabel('Precision')
ax2.grid(True,alpha=0.3,axis='y')
x1 = [0,1,2,5,6,7]
y1 = [0.4,0.52,0.68,0.14,0.38,0.88]
x2 = [10,11,12,15,16,17]
y2 = [0.28,0.25,0.28,0.05,0.02,0.11]
for i in range(len(x1)):
ax1.text(x1[i]-0.3,y1[i]+0.02,s=y1[i],fontsize=8)
for i in range(len(x2)):
ax2.text(x2[i]-0.35,y2[i]+0.002,s=y2[i],fontsize=8)
```
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import matplotlib as mpl
import pickle
mpl.rcParams['pdf.fonttype'] = 42
mpl.rcParams['ps.fonttype'] = 42
mpl.rcParams['font.family'] = 'Arial'
fig = plt.figure(figsize=(6.4,4.8))
ax1 = plt.axes([0.1,0.1,0.55,0.80])
ax2 = plt.axes([0.75,0.1,0.20,0.80])
ax1.spines['right'].set_visible(False)
ax1.spines['top'].set_visible(False)
ax2.spines['right'].set_visible(False)
ax2.spines['top'].set_visible(False)
x1, x2, x3 = [1,6], [2,7], [3,8]
y1, y2, y3 = [3,4], [1,3], [4,8]
'''
Note for the y1,y2,y3:
deephlapan result can be accessed in ../data/deephlapan_result_cell.csv
IEDB result can be accessed in ../data/ori_test_cells.csv
DeepImmuno-CNN results are the highest score for 10-fold validation, detail can be seen in Supp_fig_3
'''
ax1.bar(x1,y1,color='#E36DF2',width=0.8,label='DeepHLApan')
ax1.bar(x2,y2,color='#04BF7B',width=0.8,label='IEDB')
ax1.bar(x3,y3,color='#F26D6D',width=0.8,label='DeepImmuno-CNN')
ax1.set_xticks([2,7])
ax1.set_ylabel('Number of immunogenic peptides')
ax1.set_xticklabels(['Top20','Top50'])
ax1.legend(frameon=True)
ax1.grid(alpha=0.3,axis='y')
tmp1x = [1,2,3,6,7,8]
tmp1y = [3,1,4,4,3,8]
for i in range(len(tmp1x)):
ax1.text(tmp1x[i]-0.2,tmp1y[i]+0.1,s=tmp1y[i])
ax2.bar([1,2,3],[0.34,0.63,0.83],color=['#E36DF2','#04BF7B','#F26D6D'])
ax2.set_ylim([0,1.05])
ax2.set_ylabel('Sensitivity')
ax2.grid(alpha=0.3,axis='y')
ax2.set_xticks([2])
ax2.set_xticklabels(['hard cutoff'])
tmp2x = [1,2,3]
tmp2y = [0.34,0.63,0.83]
for i in range(len(tmp2x)):
ax2.text(tmp2x[i]-0.3,tmp2y[i]+0.01,s=tmp2y[i])
'''
Again, the data used to plot the bar chart can be accessed in as below
DeepHLApan: ../data/covid_predicted_result.csv
IEDB: ../data/sars_cov_2_result.csv
'''
fig = plt.figure(figsize=(6.4,4.8))
ax1 = plt.axes([0.1,0.1,0.35,0.80])
ax2 = plt.axes([0.55,0.1,0.35,0.80])
ax1.spines['right'].set_visible(False)
ax1.spines['top'].set_visible(False)
ax2.spines['right'].set_visible(False)
ax2.spines['top'].set_visible(False)
ax1.bar([0,5],[0.4,0.14],color='#E36DF2',label='Deephlapan',width=0.8)
ax1.bar([1,6],[0.52,0.38],color='#04BF7B',label='IEDB',width=0.8)
ax1.bar([2,7],[0.68,0.88],color='#F26D6D',label='DeepImmuno-CNN',width=0.8)
ax1.set_xticks([1,6])
ax1.set_xticklabels(['Convalescent','Unexposed'])
ax1.legend(frameon=True)
ax1.grid(True,alpha=0.3,axis='y')
ax1.set_ylabel('Recall')
ax2.bar([10,15],[0.28,0.05],color='#E36DF2',label='Deephlapan',width=0.8)
ax2.bar([11,16],[0.25,0.02],color='#04BF7B',label='IEDB',width=0.8)
ax2.bar([12,17],[0.28,0.11],color='#F26D6D',label='DeepImmuno-CNN',width=0.8)
ax2.set_xticks([11,16])
ax2.set_xticklabels(['Convalescent','Unexposed'])
ax2.set_ylabel('Precision')
ax2.grid(True,alpha=0.3,axis='y')
x1 = [0,1,2,5,6,7]
y1 = [0.4,0.52,0.68,0.14,0.38,0.88]
x2 = [10,11,12,15,16,17]
y2 = [0.28,0.25,0.28,0.05,0.02,0.11]
for i in range(len(x1)):
ax1.text(x1[i]-0.3,y1[i]+0.02,s=y1[i],fontsize=8)
for i in range(len(x2)):
ax2.text(x2[i]-0.35,y2[i]+0.002,s=y2[i],fontsize=8)
| 0.181916 | 0.637257 |
<a href="https://colab.research.google.com/github/Tessellate-Imaging/monk_v1/blob/master/study_roadmaps/1_getting_started_roadmap/1_getting_started_with_monk/1)%20Dog%20Vs%20Cat%20Classifier%20Using%20Mxnet-Gluon%20Backend.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Table of Contents
## [0. Install](#0)
## [1. Importing mxnet-gluoncv backend](#1)
## [2. Creating and Managing experiments](#1)
## [3. Training a Cat Vs Dog image classifier](#2)
## [4. Validating the trained classifier](#3)
## [5. Running inference on test images](#4)
<a id='0'></a>
# Install Monk
- git clone https://github.com/Tessellate-Imaging/monk_v1.git
- cd monk_v1/installation/Linux && pip install -r requirements_cu9.txt
- (Select the requirements file as per OS and CUDA version)
```
!git clone https://github.com/Tessellate-Imaging/monk_v1.git
# Select the requirements file as per OS and CUDA version
!cd monk_v1/installation/Linux && pip install -r requirements_cu9.txt
```
<a id='1'></a>
# Imports
```
# Monk
import os
import sys
sys.path.append("monk_v1/monk/");
#Using mxnet-gluon backend
from gluon_prototype import prototype
```
<a id='2'></a>
# Creating and managing experiments
- Provide project name
- Provide experiment name
- For a specific data create a single project
- Inside each project multiple experiments can be created
- Every experiment can be have diferent hyper-parameters attached to it
```
gtf = prototype(verbose=1);
gtf.Prototype("sample-project-1", "sample-experiment-1");
```
### This creates files and directories as per the following structure
workspace
|
|--------sample-project-1 (Project name can be different)
|
|
|-----sample-experiment-1 (Experiment name can be different)
|
|-----experiment-state.json
|
|-----output
|
|------logs (All training logs and graphs saved here)
|
|------models (all trained models saved here)
<a id='2'></a>
# Training a Cat Vs Dog image classifier
## Quick mode training
- Using Default Function
- dataset_path
- model_name
- num_epochs
## Dataset folder structure
parent_directory
|
|
|------cats
|
|------img1.jpg
|------img2.jpg
|------.... (and so on)
|------dogs
|
|------img1.jpg
|------img2.jpg
|------.... (and so on)
```
gtf.Default(dataset_path="monk_v1/monk/system_check_tests/datasets/dataset_cats_dogs_train",
model_name="resnet18_v1",
num_epochs=5);
#Read the summary generated once you run this cell.
#Start Training
gtf.Train();
#Read the training summary generated once you run the cell and training is completed
```
<a id='4'></a>
# Validating the trained classifier
## Load the experiment in validation mode
- Set flag eval_infer as True
```
gtf = prototype(verbose=1);
gtf.Prototype("sample-project-1", "sample-experiment-1", eval_infer=True);
```
## Load the validation dataset
```
gtf.Dataset_Params(dataset_path="monk_v1/monk/system_check_tests/datasets/dataset_cats_dogs_eval");
gtf.Dataset();
```
## Run validation
```
accuracy, class_based_accuracy = gtf.Evaluate();
```
<a id='5'></a>
# Running inference on test images
## Load the experiment in inference mode
- Set flag eval_infer as True
```
gtf = prototype(verbose=1);
gtf.Prototype("sample-project-1", "sample-experiment-1", eval_infer=True);
```
## Select image and Run inference
```
img_name = "monk_v1/monk/system_check_tests/datasets/dataset_cats_dogs_test/0.jpg";
predictions = gtf.Infer(img_name=img_name);
#Display
from IPython.display import Image
Image(filename=img_name)
img_name = "monk_v1/monk/system_check_tests/datasets/dataset_cats_dogs_test/90.jpg";
predictions = gtf.Infer(img_name=img_name);
#Display
from IPython.display import Image
Image(filename=img_name)
```
|
github_jupyter
|
!git clone https://github.com/Tessellate-Imaging/monk_v1.git
# Select the requirements file as per OS and CUDA version
!cd monk_v1/installation/Linux && pip install -r requirements_cu9.txt
# Monk
import os
import sys
sys.path.append("monk_v1/monk/");
#Using mxnet-gluon backend
from gluon_prototype import prototype
gtf = prototype(verbose=1);
gtf.Prototype("sample-project-1", "sample-experiment-1");
gtf.Default(dataset_path="monk_v1/monk/system_check_tests/datasets/dataset_cats_dogs_train",
model_name="resnet18_v1",
num_epochs=5);
#Read the summary generated once you run this cell.
#Start Training
gtf.Train();
#Read the training summary generated once you run the cell and training is completed
gtf = prototype(verbose=1);
gtf.Prototype("sample-project-1", "sample-experiment-1", eval_infer=True);
gtf.Dataset_Params(dataset_path="monk_v1/monk/system_check_tests/datasets/dataset_cats_dogs_eval");
gtf.Dataset();
accuracy, class_based_accuracy = gtf.Evaluate();
gtf = prototype(verbose=1);
gtf.Prototype("sample-project-1", "sample-experiment-1", eval_infer=True);
img_name = "monk_v1/monk/system_check_tests/datasets/dataset_cats_dogs_test/0.jpg";
predictions = gtf.Infer(img_name=img_name);
#Display
from IPython.display import Image
Image(filename=img_name)
img_name = "monk_v1/monk/system_check_tests/datasets/dataset_cats_dogs_test/90.jpg";
predictions = gtf.Infer(img_name=img_name);
#Display
from IPython.display import Image
Image(filename=img_name)
| 0.343892 | 0.949059 |
```
%matplotlib inline
from matplotlib import style
style.use('fivethirtyeight')
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import datetime as dt
```
# Reflect Tables into SQLAlchemy ORM
```
# Python SQL toolkit and Object Relational Mapper
import sqlalchemy
from matplotlib.pyplot import figure
from sqlalchemy.ext.automap import automap_base
from sqlalchemy.orm import Session
from sqlalchemy import create_engine, func
from sqlalchemy import inspect
engine = create_engine("sqlite:///Resources/hawaii.sqlite")
conn = engine.connect()
# reflect an existing database into a new model
Base = automap_base()
# reflect the tables
Base.prepare(engine, reflect=True)
# We can view all of the classes that automap found
Base.classes.keys()
# Save references to each table
Measurement = Base.classes.measurement
Station = Base.classes.station
# Create our session (link) from Python to the DB
session = Session(engine)
measurement_df = pd.read_sql('select * FROM measurement', conn)
measurement_df.head()
station_df = pd.read_sql('select * FROM station', conn)
station_df.head()
```
# Exploratory Climate Analysis
* Design a query to retrieve the last 12 months of precipitation data and plot the results
* Calculate the date 1 year ago from the last data point in the database
* Perform a query to retrieve the data and precipitation scores
* Save the query results as a Pandas DataFrame and set the index to the date column
* Sort the dataframe by date
* Use Pandas Plotting with Matplotlib to plot the data
```
# Design a query to retrieve the last 12 months of precipitation data and plot the results
##find latest date
mostRctDt = session.query(Measurement.date).order_by(Measurement.date.desc()).first()
print("Most recent date: ",mostRctDt)
# Calculate the date 1 year ago from the last data point in the database
yearAgo = dt.date(2017, 8, 23) - dt.timedelta(days=365)
print("1 year ago: ",yearAgo)
# Perform a query to retrieve the date and precipitation scores
# Save the query results as a Pandas DataFrame and set the index to the date column
# Sort the dataframe by date
precipData = session.query(Measurement.date, Measurement.prcp).filter(Measurement.date).\
filter(Measurement.date >= "2016-08-23").all()
precipData_df = pd.DataFrame(precipData, columns=['date', 'prcp'])
precipData_df.set_index('date', inplace=True)
precipData_df.head()
# Use Pandas Plotting with Matplotlib to plot the data
precipPlot = pd.DataFrame(precipData_df)
fig, ax = plt.subplots(figsize=(22,10))
precipPlot = precipPlot.sort_index(ascending=True)
precipPlot.plot(ax=ax)
plt.xticks(rotation=90)
plt.title("12 Months of Precipitation Data from Most Recent Recorded Date ")
plt.xlabel("Date")
plt.ylabel('Precipitation (in)' )
# Use Pandas to calculate the summary statistics for the precipitation data
precipData_df.describe()
# Design a query to show how many stations are available in this dataset?
stationCount = session.query(Station.id).count()
stationCount
# What are the most active stations? (i.e. what stations have the most rows)?
# List the stations and the counts in descending order.
activeStations = session.query(Measurement.station, func.count(Measurement.station)).\
group_by(Measurement.station).\
order_by(func.count(Measurement.station).desc()).all()
activeStations
# Using the station id from the previous query, calculate the lowest temperature recorded,
# highest temperature recorded, and average temperature of the most active station?
tempMeasure = session.query(Measurement.station,func.min(Measurement.tobs),
func.max(Measurement.tobs),
func.avg(Measurement.tobs)).filter(Measurement.station == 'USC00519281').all()
tempMeasure
# Choose the station with the highest number of temperature observations.
# Query the last 12 months of temperature observation data for this station and plot the results as a histogram
activeStation = session.query(Measurement.tobs, Measurement.station).filter(Measurement.date).\
filter(Measurement.station == 'USC00519281').\
filter(Measurement.date >= "2016-08-23").all()
activeStation
tempData_df = pd.DataFrame(activeStation, columns=['Temperature', 'Station ID'])
tempData_df.set_index('Station ID', inplace=True)
tempData_df
tempData = pd.DataFrame(tempData_df)
tempData
tempData.plot(kind='hist', bins=12, figsize=(10,7))
plt.title("12 Months of Temperature Observations", fontsize='large', fontweight='bold')
plt.xlabel('Temperature (F)', fontsize='large', fontweight='bold')
plt.ylabel('Frequency', fontsize='large', fontweight='bold')
```
|
github_jupyter
|
%matplotlib inline
from matplotlib import style
style.use('fivethirtyeight')
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import datetime as dt
# Python SQL toolkit and Object Relational Mapper
import sqlalchemy
from matplotlib.pyplot import figure
from sqlalchemy.ext.automap import automap_base
from sqlalchemy.orm import Session
from sqlalchemy import create_engine, func
from sqlalchemy import inspect
engine = create_engine("sqlite:///Resources/hawaii.sqlite")
conn = engine.connect()
# reflect an existing database into a new model
Base = automap_base()
# reflect the tables
Base.prepare(engine, reflect=True)
# We can view all of the classes that automap found
Base.classes.keys()
# Save references to each table
Measurement = Base.classes.measurement
Station = Base.classes.station
# Create our session (link) from Python to the DB
session = Session(engine)
measurement_df = pd.read_sql('select * FROM measurement', conn)
measurement_df.head()
station_df = pd.read_sql('select * FROM station', conn)
station_df.head()
# Design a query to retrieve the last 12 months of precipitation data and plot the results
##find latest date
mostRctDt = session.query(Measurement.date).order_by(Measurement.date.desc()).first()
print("Most recent date: ",mostRctDt)
# Calculate the date 1 year ago from the last data point in the database
yearAgo = dt.date(2017, 8, 23) - dt.timedelta(days=365)
print("1 year ago: ",yearAgo)
# Perform a query to retrieve the date and precipitation scores
# Save the query results as a Pandas DataFrame and set the index to the date column
# Sort the dataframe by date
precipData = session.query(Measurement.date, Measurement.prcp).filter(Measurement.date).\
filter(Measurement.date >= "2016-08-23").all()
precipData_df = pd.DataFrame(precipData, columns=['date', 'prcp'])
precipData_df.set_index('date', inplace=True)
precipData_df.head()
# Use Pandas Plotting with Matplotlib to plot the data
precipPlot = pd.DataFrame(precipData_df)
fig, ax = plt.subplots(figsize=(22,10))
precipPlot = precipPlot.sort_index(ascending=True)
precipPlot.plot(ax=ax)
plt.xticks(rotation=90)
plt.title("12 Months of Precipitation Data from Most Recent Recorded Date ")
plt.xlabel("Date")
plt.ylabel('Precipitation (in)' )
# Use Pandas to calculate the summary statistics for the precipitation data
precipData_df.describe()
# Design a query to show how many stations are available in this dataset?
stationCount = session.query(Station.id).count()
stationCount
# What are the most active stations? (i.e. what stations have the most rows)?
# List the stations and the counts in descending order.
activeStations = session.query(Measurement.station, func.count(Measurement.station)).\
group_by(Measurement.station).\
order_by(func.count(Measurement.station).desc()).all()
activeStations
# Using the station id from the previous query, calculate the lowest temperature recorded,
# highest temperature recorded, and average temperature of the most active station?
tempMeasure = session.query(Measurement.station,func.min(Measurement.tobs),
func.max(Measurement.tobs),
func.avg(Measurement.tobs)).filter(Measurement.station == 'USC00519281').all()
tempMeasure
# Choose the station with the highest number of temperature observations.
# Query the last 12 months of temperature observation data for this station and plot the results as a histogram
activeStation = session.query(Measurement.tobs, Measurement.station).filter(Measurement.date).\
filter(Measurement.station == 'USC00519281').\
filter(Measurement.date >= "2016-08-23").all()
activeStation
tempData_df = pd.DataFrame(activeStation, columns=['Temperature', 'Station ID'])
tempData_df.set_index('Station ID', inplace=True)
tempData_df
tempData = pd.DataFrame(tempData_df)
tempData
tempData.plot(kind='hist', bins=12, figsize=(10,7))
plt.title("12 Months of Temperature Observations", fontsize='large', fontweight='bold')
plt.xlabel('Temperature (F)', fontsize='large', fontweight='bold')
plt.ylabel('Frequency', fontsize='large', fontweight='bold')
| 0.770119 | 0.941601 |
### Loading the required libraries
```
import os
import sys
import cv2
import math
import datetime
import numpy as np
import logging as log
import matplotlib.pyplot as plt
from openvino.inference_engine import IENetwork, IECore, IEPlugin
```
### Class for working with the Inference Engine
```
class Network:
'''
Load and store information for working with the Inference Engine,
and any loaded models.
'''
def __init__(self):
self.plugin = None
self.network = None
self.input_blob = None
self.output_blob = None
self.exec_network = None
self.infer_request = None
def load_model(self, model, device="GPU"):
'''
Load the model given IR files.
Defaults to CPU as device for use in the workspace.
Synchronous requests made within.
'''
model_xml = model
model_bin = os.path.splitext(model_xml)[0] + ".bin"
# Initialize the plugin
self.plugin = IECore()
# Read the IR as a IENetwork
self.network = IENetwork(model=model_xml, weights=model_bin)
# Load the IENetwork into the plugin
self.exec_network = self.plugin.load_network(
self.network, device_name=device)
# Get the input layer
self.input_blob = next(iter(self.network.inputs))
self.output_blob = next(iter(self.network.outputs))
return
def get_input_shape(self):
'''
Gets the input shape of the network
'''
return self.network.inputs[self.input_blob].shape
def async_inference(self, image):
'''
Makes an asynchronous inference request, given an input image.
'''
self.exec_network.start_async(
request_id=0, inputs={self.input_blob: image})
return
def wait(self):
'''
Checks the status of the inference request.
'''
status = self.exec_network.requests[0].wait(-1)
return status
def extract_output(self):
'''
Returns a list of the results for the output layer of the network.
'''
return self.exec_network.requests[0].outputs[self.output_blob]
```
### Initialize the Inference Engine
```
plugin = Network()
```
### Load the network model into the IE
```
plugin.load_model("models/ppn-model-2.xml", "HETERO:GPU,CPU")
```
### Check the input shape
```
print(plugin.network.batch_size)
net_input_shape = plugin.get_input_shape()
print(net_input_shape)
```
### Creating input
```
img_size = 200
scale = 0.3
z_size = 7
pattern_change_speed = 0.5
def createInputVec(z, x, y):
r = math.sqrt(((x * scale - (img_size * scale / 2))**2) + (
(y * scale - (img_size * scale / 2))**2))
z_size = len(z)
input = np.random.rand(1, z_size + 3)
for i in range(z_size):
input[0][i] = z[i] * scale
input[0][z_size] = x * scale
input[0][z_size + 1] = y * scale
input[0][z_size + 2] = r
return input
```
### Process output after inference
```
def run_and_plot(fps, seconds):
frames = fps * seconds
file_name = str(datetime.datetime.now()).replace(":", "-").replace(".", "-") + '.avi'
print(file_name, end="")
fourcc = cv2.VideoWriter_fourcc('M', 'J', 'P', 'G')
out = cv2.VideoWriter(file_name, fourcc, fps, (img_size, img_size))
#out = cv2.VideoWriter(file_name, cv2.VideoWriter_fourcc(*'DIVX'), fps, (img_size, img_size))
z = np.random.rand(z_size)
directions = np.ones(z_size)
for frame in range(frames):
reverse_directions = np.where(np.logical_or(z > 100, z < -100), -1, 1)
directions = directions * reverse_directions
change = np.random.rand(z_size) * pattern_change_speed
z = z + change * directions
input_batch = np.zeros((img_size, img_size, z_size + 3))
for i in range(img_size):
for j in range(img_size):
input_batch[i, j] = createInputVec(z, i, j)
input_batch.resize(img_size * img_size, z_size + 3)
# Perform inference on the input
plugin.async_inference(input_batch)
# Get the output of inference
if plugin.wait() == 0:
output_frame = plugin.extract_output()
output_frame = np.resize(output_frame, (img_size, img_size, 3))
output_frame = (output_frame * 255).astype(np.uint8)
out.write(output_frame)
# saving each output_frame as PNG image
#plt.imsave(str(datetime.datetime.now()).replace(":", "-").replace(".", "-") + '.png', output_frame, format="png")
# displaying each output_frame
#imgplot = plt.imshow(output_frame)
#plt.show()
if (frame + 1) % fps == 1:
print("\nSec {:03d}:".format(int(frame / fps) + 1), end=" ")
print("{:3d}".format(frame + 1), end=" ")
out.release()
print("\n")
```
### Perform Inference / Generate patterns
```
run_and_plot(fps = 15, seconds = 10)
```
|
github_jupyter
|
import os
import sys
import cv2
import math
import datetime
import numpy as np
import logging as log
import matplotlib.pyplot as plt
from openvino.inference_engine import IENetwork, IECore, IEPlugin
class Network:
'''
Load and store information for working with the Inference Engine,
and any loaded models.
'''
def __init__(self):
self.plugin = None
self.network = None
self.input_blob = None
self.output_blob = None
self.exec_network = None
self.infer_request = None
def load_model(self, model, device="GPU"):
'''
Load the model given IR files.
Defaults to CPU as device for use in the workspace.
Synchronous requests made within.
'''
model_xml = model
model_bin = os.path.splitext(model_xml)[0] + ".bin"
# Initialize the plugin
self.plugin = IECore()
# Read the IR as a IENetwork
self.network = IENetwork(model=model_xml, weights=model_bin)
# Load the IENetwork into the plugin
self.exec_network = self.plugin.load_network(
self.network, device_name=device)
# Get the input layer
self.input_blob = next(iter(self.network.inputs))
self.output_blob = next(iter(self.network.outputs))
return
def get_input_shape(self):
'''
Gets the input shape of the network
'''
return self.network.inputs[self.input_blob].shape
def async_inference(self, image):
'''
Makes an asynchronous inference request, given an input image.
'''
self.exec_network.start_async(
request_id=0, inputs={self.input_blob: image})
return
def wait(self):
'''
Checks the status of the inference request.
'''
status = self.exec_network.requests[0].wait(-1)
return status
def extract_output(self):
'''
Returns a list of the results for the output layer of the network.
'''
return self.exec_network.requests[0].outputs[self.output_blob]
plugin = Network()
plugin.load_model("models/ppn-model-2.xml", "HETERO:GPU,CPU")
print(plugin.network.batch_size)
net_input_shape = plugin.get_input_shape()
print(net_input_shape)
img_size = 200
scale = 0.3
z_size = 7
pattern_change_speed = 0.5
def createInputVec(z, x, y):
r = math.sqrt(((x * scale - (img_size * scale / 2))**2) + (
(y * scale - (img_size * scale / 2))**2))
z_size = len(z)
input = np.random.rand(1, z_size + 3)
for i in range(z_size):
input[0][i] = z[i] * scale
input[0][z_size] = x * scale
input[0][z_size + 1] = y * scale
input[0][z_size + 2] = r
return input
def run_and_plot(fps, seconds):
frames = fps * seconds
file_name = str(datetime.datetime.now()).replace(":", "-").replace(".", "-") + '.avi'
print(file_name, end="")
fourcc = cv2.VideoWriter_fourcc('M', 'J', 'P', 'G')
out = cv2.VideoWriter(file_name, fourcc, fps, (img_size, img_size))
#out = cv2.VideoWriter(file_name, cv2.VideoWriter_fourcc(*'DIVX'), fps, (img_size, img_size))
z = np.random.rand(z_size)
directions = np.ones(z_size)
for frame in range(frames):
reverse_directions = np.where(np.logical_or(z > 100, z < -100), -1, 1)
directions = directions * reverse_directions
change = np.random.rand(z_size) * pattern_change_speed
z = z + change * directions
input_batch = np.zeros((img_size, img_size, z_size + 3))
for i in range(img_size):
for j in range(img_size):
input_batch[i, j] = createInputVec(z, i, j)
input_batch.resize(img_size * img_size, z_size + 3)
# Perform inference on the input
plugin.async_inference(input_batch)
# Get the output of inference
if plugin.wait() == 0:
output_frame = plugin.extract_output()
output_frame = np.resize(output_frame, (img_size, img_size, 3))
output_frame = (output_frame * 255).astype(np.uint8)
out.write(output_frame)
# saving each output_frame as PNG image
#plt.imsave(str(datetime.datetime.now()).replace(":", "-").replace(".", "-") + '.png', output_frame, format="png")
# displaying each output_frame
#imgplot = plt.imshow(output_frame)
#plt.show()
if (frame + 1) % fps == 1:
print("\nSec {:03d}:".format(int(frame / fps) + 1), end=" ")
print("{:3d}".format(frame + 1), end=" ")
out.release()
print("\n")
run_and_plot(fps = 15, seconds = 10)
| 0.542621 | 0.725126 |
# OCR model for reading Captchas
**Author:** [A_K_Nain](https://twitter.com/A_K_Nain)<br>
**Date created:** 2020/06/14<br>
**Last modified:** 2020/06/26<br>
**Description:** How to implement an OCR model using CNNs, RNNs and CTC loss.
## Introduction
This example demonstrates a simple OCR model built with the Functional API. Apart from
combining CNN and RNN, it also illustrates how you can instantiate a new layer
and use it as an "Endpoint layer" for implementing CTC loss. For a detailed
guide to layer subclassing, please check out
[this page](https://keras.io/guides/making_new_layers_and_models_via_subclassing/)
in the developer guides.
## Setup
```
import os
import numpy as np
import matplotlib.pyplot as plt
from pathlib import Path
from collections import Counter
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
```
## Load the data: [Captcha Images](https://www.kaggle.com/fournierp/captcha-version-2-images)
Let's download the data.
```
!curl -LO https://github.com/AakashKumarNain/CaptchaCracker/raw/master/captcha_images_v2.zip
!unzip -qq captcha_images_v2.zip
```
The dataset contains 1040 captcha files as `png` images. The label for each sample is a string,
the name of the file (minus the file extension).
We will map each character in the string to an integer for training the model. Similary,
we will need to map the predictions of the model back to strings. For this purpose
we will maintain two dictionaries, mapping characters to integers, and integers to characters,
respectively.
```
# Path to the data directory
data_dir = Path("./captcha_images_v2/")
# Get list of all the images
images = sorted(list(map(str, list(data_dir.glob("*.png")))))
labels = [img.split(os.path.sep)[-1].split(".png")[0] for img in images]
characters = set(char for label in labels for char in label)
print("Number of images found: ", len(images))
print("Number of labels found: ", len(labels))
print("Number of unique characters: ", len(characters))
print("Characters present: ", characters)
# Batch size for training and validation
batch_size = 16
# Desired image dimensions
img_width = 200
img_height = 50
# Factor by which the image is going to be downsampled
# by the convolutional blocks. We will be using two
# convolution blocks and each block will have
# a pooling layer which downsample the features by a factor of 2.
# Hence total downsampling factor would be 4.
downsample_factor = 4
# Maximum length of any captcha in the dataset
max_length = max([len(label) for label in labels])
```
## Preprocessing
```
# Mapping characters to integers
char_to_num = layers.StringLookup(
vocabulary=list(characters), mask_token=None
)
# Mapping integers back to original characters
num_to_char = layers.StringLookup(
vocabulary=char_to_num.get_vocabulary(), mask_token=None, invert=True
)
def split_data(images, labels, train_size=0.9, shuffle=True):
# 1. Get the total size of the dataset
size = len(images)
# 2. Make an indices array and shuffle it, if required
indices = np.arange(size)
if shuffle:
np.random.shuffle(indices)
# 3. Get the size of training samples
train_samples = int(size * train_size)
# 4. Split data into training and validation sets
x_train, y_train = images[indices[:train_samples]], labels[indices[:train_samples]]
x_valid, y_valid = images[indices[train_samples:]], labels[indices[train_samples:]]
return x_train, x_valid, y_train, y_valid
# Splitting data into training and validation sets
x_train, x_valid, y_train, y_valid = split_data(np.array(images), np.array(labels))
def encode_single_sample(img_path, label):
# 1. Read image
img = tf.io.read_file(img_path)
# 2. Decode and convert to grayscale
img = tf.io.decode_png(img, channels=1)
# 3. Convert to float32 in [0, 1] range
img = tf.image.convert_image_dtype(img, tf.float32)
# 4. Resize to the desired size
img = tf.image.resize(img, [img_height, img_width])
# 5. Transpose the image because we want the time
# dimension to correspond to the width of the image.
img = tf.transpose(img, perm=[1, 0, 2])
# 6. Map the characters in label to numbers
label = char_to_num(tf.strings.unicode_split(label, input_encoding="UTF-8"))
# 7. Return a dict as our model is expecting two inputs
return {"image": img, "label": label}
```
## Create `Dataset` objects
```
train_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))
train_dataset = (
train_dataset.map(
encode_single_sample, num_parallel_calls=tf.data.AUTOTUNE
)
.batch(batch_size)
.prefetch(buffer_size=tf.data.AUTOTUNE)
)
validation_dataset = tf.data.Dataset.from_tensor_slices((x_valid, y_valid))
validation_dataset = (
validation_dataset.map(
encode_single_sample, num_parallel_calls=tf.data.AUTOTUNE
)
.batch(batch_size)
.prefetch(buffer_size=tf.data.AUTOTUNE)
)
```
## Visualize the data
```
_, ax = plt.subplots(4, 4, figsize=(10, 5))
for batch in train_dataset.take(1):
images = batch["image"]
labels = batch["label"]
for i in range(16):
img = (images[i] * 255).numpy().astype("uint8")
label = tf.strings.reduce_join(num_to_char(labels[i])).numpy().decode("utf-8")
ax[i // 4, i % 4].imshow(img[:, :, 0].T, cmap="gray")
ax[i // 4, i % 4].set_title(label)
ax[i // 4, i % 4].axis("off")
plt.show()
```
## Model
```
class CTCLayer(layers.Layer):
def __init__(self, name=None):
super().__init__(name=name)
self.loss_fn = keras.backend.ctc_batch_cost
def call(self, y_true, y_pred):
# Compute the training-time loss value and add it
# to the layer using `self.add_loss()`.
batch_len = tf.cast(tf.shape(y_true)[0], dtype="int64")
input_length = tf.cast(tf.shape(y_pred)[1], dtype="int64")
label_length = tf.cast(tf.shape(y_true)[1], dtype="int64")
input_length = input_length * tf.ones(shape=(batch_len, 1), dtype="int64")
label_length = label_length * tf.ones(shape=(batch_len, 1), dtype="int64")
loss = self.loss_fn(y_true, y_pred, input_length, label_length)
self.add_loss(loss)
# At test time, just return the computed predictions
return y_pred
def build_model():
# Inputs to the model
input_img = layers.Input(
shape=(img_width, img_height, 1), name="image", dtype="float32"
)
labels = layers.Input(name="label", shape=(None,), dtype="float32")
# First conv block
x = layers.Conv2D(
32,
(3, 3),
activation="relu",
kernel_initializer="he_normal",
padding="same",
name="Conv1",
)(input_img)
x = layers.MaxPooling2D((2, 2), name="pool1")(x)
# Second conv block
x = layers.Conv2D(
64,
(3, 3),
activation="relu",
kernel_initializer="he_normal",
padding="same",
name="Conv2",
)(x)
x = layers.MaxPooling2D((2, 2), name="pool2")(x)
# We have used two max pool with pool size and strides 2.
# Hence, downsampled feature maps are 4x smaller. The number of
# filters in the last layer is 64. Reshape accordingly before
# passing the output to the RNN part of the model
new_shape = ((img_width // 4), (img_height // 4) * 64)
x = layers.Reshape(target_shape=new_shape, name="reshape")(x)
x = layers.Dense(64, activation="relu", name="dense1")(x)
x = layers.Dropout(0.2)(x)
# RNNs
x = layers.Bidirectional(layers.LSTM(128, return_sequences=True, dropout=0.25))(x)
x = layers.Bidirectional(layers.LSTM(64, return_sequences=True, dropout=0.25))(x)
# Output layer
x = layers.Dense(
len(char_to_num.get_vocabulary()) + 1, activation="softmax", name="dense2"
)(x)
# Add CTC layer for calculating CTC loss at each step
output = CTCLayer(name="ctc_loss")(labels, x)
# Define the model
model = keras.models.Model(
inputs=[input_img, labels], outputs=output, name="ocr_model_v1"
)
# Optimizer
opt = keras.optimizers.Adam()
# Compile the model and return
model.compile(optimizer=opt)
return model
# Get the model
model = build_model()
model.summary()
```
## Training
```
epochs = 100
early_stopping_patience = 10
# Add early stopping
early_stopping = keras.callbacks.EarlyStopping(
monitor="val_loss", patience=early_stopping_patience, restore_best_weights=True
)
# Train the model
history = model.fit(
train_dataset,
validation_data=validation_dataset,
epochs=epochs,
callbacks=[early_stopping],
)
```
## Inference
You can use the trained model hosted on [Hugging Face Hub](https://huggingface.co/keras-io/ocr-for-captcha) and try the demo on [Hugging Face Spaces](https://huggingface.co/spaces/keras-io/ocr-for-captcha).
```
# Get the prediction model by extracting layers till the output layer
prediction_model = keras.models.Model(
model.get_layer(name="image").input, model.get_layer(name="dense2").output
)
prediction_model.summary()
# A utility function to decode the output of the network
def decode_batch_predictions(pred):
input_len = np.ones(pred.shape[0]) * pred.shape[1]
# Use greedy search. For complex tasks, you can use beam search
results = keras.backend.ctc_decode(pred, input_length=input_len, greedy=True)[0][0][
:, :max_length
]
# Iterate over the results and get back the text
output_text = []
for res in results:
res = tf.strings.reduce_join(num_to_char(res)).numpy().decode("utf-8")
output_text.append(res)
return output_text
# Let's check results on some validation samples
for batch in validation_dataset.take(1):
batch_images = batch["image"]
batch_labels = batch["label"]
preds = prediction_model.predict(batch_images)
pred_texts = decode_batch_predictions(preds)
orig_texts = []
for label in batch_labels:
label = tf.strings.reduce_join(num_to_char(label)).numpy().decode("utf-8")
orig_texts.append(label)
_, ax = plt.subplots(4, 4, figsize=(15, 5))
for i in range(len(pred_texts)):
img = (batch_images[i, :, :, 0] * 255).numpy().astype(np.uint8)
img = img.T
title = f"Prediction: {pred_texts[i]}"
ax[i // 4, i % 4].imshow(img, cmap="gray")
ax[i // 4, i % 4].set_title(title)
ax[i // 4, i % 4].axis("off")
plt.show()
```
|
github_jupyter
|
import os
import numpy as np
import matplotlib.pyplot as plt
from pathlib import Path
from collections import Counter
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
!curl -LO https://github.com/AakashKumarNain/CaptchaCracker/raw/master/captcha_images_v2.zip
!unzip -qq captcha_images_v2.zip
# Path to the data directory
data_dir = Path("./captcha_images_v2/")
# Get list of all the images
images = sorted(list(map(str, list(data_dir.glob("*.png")))))
labels = [img.split(os.path.sep)[-1].split(".png")[0] for img in images]
characters = set(char for label in labels for char in label)
print("Number of images found: ", len(images))
print("Number of labels found: ", len(labels))
print("Number of unique characters: ", len(characters))
print("Characters present: ", characters)
# Batch size for training and validation
batch_size = 16
# Desired image dimensions
img_width = 200
img_height = 50
# Factor by which the image is going to be downsampled
# by the convolutional blocks. We will be using two
# convolution blocks and each block will have
# a pooling layer which downsample the features by a factor of 2.
# Hence total downsampling factor would be 4.
downsample_factor = 4
# Maximum length of any captcha in the dataset
max_length = max([len(label) for label in labels])
# Mapping characters to integers
char_to_num = layers.StringLookup(
vocabulary=list(characters), mask_token=None
)
# Mapping integers back to original characters
num_to_char = layers.StringLookup(
vocabulary=char_to_num.get_vocabulary(), mask_token=None, invert=True
)
def split_data(images, labels, train_size=0.9, shuffle=True):
# 1. Get the total size of the dataset
size = len(images)
# 2. Make an indices array and shuffle it, if required
indices = np.arange(size)
if shuffle:
np.random.shuffle(indices)
# 3. Get the size of training samples
train_samples = int(size * train_size)
# 4. Split data into training and validation sets
x_train, y_train = images[indices[:train_samples]], labels[indices[:train_samples]]
x_valid, y_valid = images[indices[train_samples:]], labels[indices[train_samples:]]
return x_train, x_valid, y_train, y_valid
# Splitting data into training and validation sets
x_train, x_valid, y_train, y_valid = split_data(np.array(images), np.array(labels))
def encode_single_sample(img_path, label):
# 1. Read image
img = tf.io.read_file(img_path)
# 2. Decode and convert to grayscale
img = tf.io.decode_png(img, channels=1)
# 3. Convert to float32 in [0, 1] range
img = tf.image.convert_image_dtype(img, tf.float32)
# 4. Resize to the desired size
img = tf.image.resize(img, [img_height, img_width])
# 5. Transpose the image because we want the time
# dimension to correspond to the width of the image.
img = tf.transpose(img, perm=[1, 0, 2])
# 6. Map the characters in label to numbers
label = char_to_num(tf.strings.unicode_split(label, input_encoding="UTF-8"))
# 7. Return a dict as our model is expecting two inputs
return {"image": img, "label": label}
train_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))
train_dataset = (
train_dataset.map(
encode_single_sample, num_parallel_calls=tf.data.AUTOTUNE
)
.batch(batch_size)
.prefetch(buffer_size=tf.data.AUTOTUNE)
)
validation_dataset = tf.data.Dataset.from_tensor_slices((x_valid, y_valid))
validation_dataset = (
validation_dataset.map(
encode_single_sample, num_parallel_calls=tf.data.AUTOTUNE
)
.batch(batch_size)
.prefetch(buffer_size=tf.data.AUTOTUNE)
)
_, ax = plt.subplots(4, 4, figsize=(10, 5))
for batch in train_dataset.take(1):
images = batch["image"]
labels = batch["label"]
for i in range(16):
img = (images[i] * 255).numpy().astype("uint8")
label = tf.strings.reduce_join(num_to_char(labels[i])).numpy().decode("utf-8")
ax[i // 4, i % 4].imshow(img[:, :, 0].T, cmap="gray")
ax[i // 4, i % 4].set_title(label)
ax[i // 4, i % 4].axis("off")
plt.show()
class CTCLayer(layers.Layer):
def __init__(self, name=None):
super().__init__(name=name)
self.loss_fn = keras.backend.ctc_batch_cost
def call(self, y_true, y_pred):
# Compute the training-time loss value and add it
# to the layer using `self.add_loss()`.
batch_len = tf.cast(tf.shape(y_true)[0], dtype="int64")
input_length = tf.cast(tf.shape(y_pred)[1], dtype="int64")
label_length = tf.cast(tf.shape(y_true)[1], dtype="int64")
input_length = input_length * tf.ones(shape=(batch_len, 1), dtype="int64")
label_length = label_length * tf.ones(shape=(batch_len, 1), dtype="int64")
loss = self.loss_fn(y_true, y_pred, input_length, label_length)
self.add_loss(loss)
# At test time, just return the computed predictions
return y_pred
def build_model():
# Inputs to the model
input_img = layers.Input(
shape=(img_width, img_height, 1), name="image", dtype="float32"
)
labels = layers.Input(name="label", shape=(None,), dtype="float32")
# First conv block
x = layers.Conv2D(
32,
(3, 3),
activation="relu",
kernel_initializer="he_normal",
padding="same",
name="Conv1",
)(input_img)
x = layers.MaxPooling2D((2, 2), name="pool1")(x)
# Second conv block
x = layers.Conv2D(
64,
(3, 3),
activation="relu",
kernel_initializer="he_normal",
padding="same",
name="Conv2",
)(x)
x = layers.MaxPooling2D((2, 2), name="pool2")(x)
# We have used two max pool with pool size and strides 2.
# Hence, downsampled feature maps are 4x smaller. The number of
# filters in the last layer is 64. Reshape accordingly before
# passing the output to the RNN part of the model
new_shape = ((img_width // 4), (img_height // 4) * 64)
x = layers.Reshape(target_shape=new_shape, name="reshape")(x)
x = layers.Dense(64, activation="relu", name="dense1")(x)
x = layers.Dropout(0.2)(x)
# RNNs
x = layers.Bidirectional(layers.LSTM(128, return_sequences=True, dropout=0.25))(x)
x = layers.Bidirectional(layers.LSTM(64, return_sequences=True, dropout=0.25))(x)
# Output layer
x = layers.Dense(
len(char_to_num.get_vocabulary()) + 1, activation="softmax", name="dense2"
)(x)
# Add CTC layer for calculating CTC loss at each step
output = CTCLayer(name="ctc_loss")(labels, x)
# Define the model
model = keras.models.Model(
inputs=[input_img, labels], outputs=output, name="ocr_model_v1"
)
# Optimizer
opt = keras.optimizers.Adam()
# Compile the model and return
model.compile(optimizer=opt)
return model
# Get the model
model = build_model()
model.summary()
epochs = 100
early_stopping_patience = 10
# Add early stopping
early_stopping = keras.callbacks.EarlyStopping(
monitor="val_loss", patience=early_stopping_patience, restore_best_weights=True
)
# Train the model
history = model.fit(
train_dataset,
validation_data=validation_dataset,
epochs=epochs,
callbacks=[early_stopping],
)
# Get the prediction model by extracting layers till the output layer
prediction_model = keras.models.Model(
model.get_layer(name="image").input, model.get_layer(name="dense2").output
)
prediction_model.summary()
# A utility function to decode the output of the network
def decode_batch_predictions(pred):
input_len = np.ones(pred.shape[0]) * pred.shape[1]
# Use greedy search. For complex tasks, you can use beam search
results = keras.backend.ctc_decode(pred, input_length=input_len, greedy=True)[0][0][
:, :max_length
]
# Iterate over the results and get back the text
output_text = []
for res in results:
res = tf.strings.reduce_join(num_to_char(res)).numpy().decode("utf-8")
output_text.append(res)
return output_text
# Let's check results on some validation samples
for batch in validation_dataset.take(1):
batch_images = batch["image"]
batch_labels = batch["label"]
preds = prediction_model.predict(batch_images)
pred_texts = decode_batch_predictions(preds)
orig_texts = []
for label in batch_labels:
label = tf.strings.reduce_join(num_to_char(label)).numpy().decode("utf-8")
orig_texts.append(label)
_, ax = plt.subplots(4, 4, figsize=(15, 5))
for i in range(len(pred_texts)):
img = (batch_images[i, :, :, 0] * 255).numpy().astype(np.uint8)
img = img.T
title = f"Prediction: {pred_texts[i]}"
ax[i // 4, i % 4].imshow(img, cmap="gray")
ax[i // 4, i % 4].set_title(title)
ax[i // 4, i % 4].axis("off")
plt.show()
| 0.823967 | 0.960915 |
```
import keras
keras.__version__
```
# 5.1 - 합성곱 신경망 소개
이 노트북은 [케라스 창시자에게 배우는 딥러닝](https://tensorflow.blog/케라스-창시자에게-배우는-딥러닝/) 책의 5장 1절의 코드 예제입니다. 책에는 더 많은 내용과 그림이 있습니다. 이 노트북에는 소스 코드에 관련된 설명만 포함합니다. 이 노트북의 설명은 케라스 버전 2.2.2에 맞추어져 있습니다. 케라스 최신 버전이 릴리스되면 노트북을 다시 테스트하기 때문에 설명과 코드의 결과가 조금 다를 수 있습니다.
----
컨브넷의 정의와 컨브넷이 컴퓨터 비전 관련 작업에 잘 맞는 이유에 대해 이론적 배경을 알아보겠습니다. 하지만 먼저 간단한 컨브넷 예제를 둘러 보죠. 2장에서 완전 연결 네트워크로 풀었던(이 방식의 테스트 정확도는 97.8%였습니다) MNIST 숫자 이미지 분류에 컨브넷을 사용해 보겠습니다. 기본적인 컨브넷이더라도 2장의 완전 연결된 모델의 성능을 훨씬 앞지를 것입니다.
다음 코드는 기본적인 컨브넷의 모습입니다. `Conv2D`와 `MaxPooling2D` 층을 쌓아 올렸습니다. 잠시 후에 이들이 무엇인지 배우겠습니다.
컨브넷이 `(image_height, image_width, image_channels)` 크기의 입력 텐서를 사용한다는 점이 중요합니다(배치 차원은 포함하지 않습니다). 이 예제에서는 MNIST 이미지 포맷인 `(28, 28, 1)` 크기의 입력을 처리하도록 컨브넷을 설정해야 합니다. 이 때문에 첫 번째 층의 매개변수로 `input_shape=(28, 28, 1)`을 전달합니다.
```
from keras import layers
from keras import models
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu', input_shape=(28, 28, 1)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
```
지금까지 컨브넷 구조를 출력해 보죠:
```
model.summary()
```
`Conv2D`와 `MaxPooling2D` 층의 출력은 `(height, width, channels)` 크기의 3D 텐서입니다. 높이와 넓이 차원은 네트워크가 깊어질수록 작아지는 경향이 있습니다. 채널의 수는 `Conv2D` 층에 전달된 첫 번째 매개변수에 의해 조절됩니다(32개 또는 64개).
다음 단계에서 마지막 층의 (`(3, 3, 64)` 크기인) 출력 텐서를 완전 연결 네트워크에 주입합니다. 이 네트워크는 이미 익숙하게 보았던 `Dense` 층을 쌓은 분류기입니다. 이 분류기는 1D 벡터를 처리하는데 이전 층의 출력이 3D 텐서입니다. 그래서 먼저 3D 출력을 1D 텐서로 펼쳐야 합니다. 그다음 몇 개의 `Dense` 층을 추가합니다:
```
model.add(layers.Flatten())
model.add(layers.Dense(64, activation='relu'))
model.add(layers.Dense(10, activation='softmax'))
```
10개의 클래스를 분류하기 위해 마지막 층의 출력 크기를 10으로 하고 소프트맥스 활성화 함수를 사용합니다. 이제 전체 네트워크는 다음과 같습니다:
```
model.summary()
```
여기에서 볼 수 있듯이 `(3, 3, 64)` 출력이 `(576,)` 크기의 벡터로 펼쳐진 후 `Dense` 층으로 주입되었습니다.
이제 MNIST 숫자 이미지에 이 컨브넷을 훈련합니다. 2장의 MNIST 예제 코드를 많이 재사용하겠습니다.
```
from keras.datasets import mnist
from keras.utils import to_categorical
(train_images, train_labels), (test_images, test_labels) = mnist.load_data()
train_images = train_images.reshape((60000, 28, 28, 1))
train_images = train_images.astype('float32') / 255
test_images = test_images.reshape((10000, 28, 28, 1))
test_images = test_images.astype('float32') / 255
train_labels = to_categorical(train_labels)
test_labels = to_categorical(test_labels)
model.compile(optimizer='rmsprop',
loss='categorical_crossentropy',
metrics=['accuracy'])
model.fit(train_images, train_labels, epochs=5, batch_size=64)
```
테스트 데이터에서 모델을 평가해 보죠:
```
test_loss, test_acc = model.evaluate(test_images, test_labels)
test_acc
```
2장의 완전 연결 네트워크는 97.8%의 테스트 정확도를 얻은 반면, 기본적인 컨브넷은 99.2%의 테스트 정확도를 얻었습니다. 에러율이 (상대적으로) 64%나 줄었습니다. 나쁘지 않군요!
|
github_jupyter
|
import keras
keras.__version__
from keras import layers
from keras import models
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu', input_shape=(28, 28, 1)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.summary()
model.add(layers.Flatten())
model.add(layers.Dense(64, activation='relu'))
model.add(layers.Dense(10, activation='softmax'))
model.summary()
from keras.datasets import mnist
from keras.utils import to_categorical
(train_images, train_labels), (test_images, test_labels) = mnist.load_data()
train_images = train_images.reshape((60000, 28, 28, 1))
train_images = train_images.astype('float32') / 255
test_images = test_images.reshape((10000, 28, 28, 1))
test_images = test_images.astype('float32') / 255
train_labels = to_categorical(train_labels)
test_labels = to_categorical(test_labels)
model.compile(optimizer='rmsprop',
loss='categorical_crossentropy',
metrics=['accuracy'])
model.fit(train_images, train_labels, epochs=5, batch_size=64)
test_loss, test_acc = model.evaluate(test_images, test_labels)
test_acc
| 0.838614 | 0.92756 |
# Cleanup
After building completing the notebooks you may want to delete the following to prevent any unwanted charges:
* Forecasts
* Predictors
* Datasets
* Dataset Groups
The code snippets below will cover the base use case of creating items in notebooks 1 - 3. You can expand upon this to delete content created in other notebooks.
## Imports and Connectins to AWS
The following lines import all the necessary libraries and then connect you to Amazon Forecast.
```
import sys
import os
import json
import time
import boto3
import pandas as pd
# importing forecast notebook utility from notebooks/common directory
sys.path.insert( 0, os.path.abspath("../../common") )
import util
```
The line below will retrieve your shared variables from the earlier notebooks.
```
%store -r
```
Once again connect to the Forecast APIs via the SDK.
```
session = boto3.Session(region_name=region)
forecast = session.client(service_name='forecast')
forecastquery = session.client(service_name='forecastquery')
```
## Defining the Things to Cleanup
In the previous notebooks you stored several variables at the end of each, now that they have been retrieved above, the cells below will delete the items that were created one at a time until all items that were created have been removed.
```
# Delete the Foreacst:
util.wait_till_delete(lambda: forecast.delete_forecast(ForecastArn=forecast_arn))
# Delete the Predictor:
util.wait_till_delete(lambda: forecast.delete_predictor(PredictorArn=predictor_arn))
# Delete Import
util.wait_till_delete(lambda: forecast.delete_dataset_import_job(DatasetImportJobArn=ds_import_job_arn))
# Delete the Dataset:
util.wait_till_delete(lambda: forecast.delete_dataset(DatasetArn=datasetArn))
# Delete the DatasetGroup:
util.wait_till_delete(lambda: forecast.delete_dataset_group(DatasetGroupArn=datasetGroupArn))
# Delete your file in S3
boto3.Session().resource('s3').Bucket(bucket_name).Object(key).delete()
```
## IAM Policy Cleanup
The very last step in the notebooks is to remove the policies that were attached to a role and then to delete it. No changes should need to be made here, just execute the cell.
```
# IAM policies should also be removed
iam = boto3.client("iam")
iam.detach_role_policy(PolicyArn="arn:aws:iam::aws:policy/AmazonS3FullAccess", RoleName=role_name)
iam.detach_role_policy(PolicyArn="arn:aws:iam::aws:policy/AmazonForecastFullAccess",RoleName=role_name)
iam.delete_role(RoleName=role_name)
```
All that remains to cleanup here is to now go back to the CloudFormation console and delete the stack. You have successfully removed all resources that were created.
|
github_jupyter
|
import sys
import os
import json
import time
import boto3
import pandas as pd
# importing forecast notebook utility from notebooks/common directory
sys.path.insert( 0, os.path.abspath("../../common") )
import util
%store -r
session = boto3.Session(region_name=region)
forecast = session.client(service_name='forecast')
forecastquery = session.client(service_name='forecastquery')
# Delete the Foreacst:
util.wait_till_delete(lambda: forecast.delete_forecast(ForecastArn=forecast_arn))
# Delete the Predictor:
util.wait_till_delete(lambda: forecast.delete_predictor(PredictorArn=predictor_arn))
# Delete Import
util.wait_till_delete(lambda: forecast.delete_dataset_import_job(DatasetImportJobArn=ds_import_job_arn))
# Delete the Dataset:
util.wait_till_delete(lambda: forecast.delete_dataset(DatasetArn=datasetArn))
# Delete the DatasetGroup:
util.wait_till_delete(lambda: forecast.delete_dataset_group(DatasetGroupArn=datasetGroupArn))
# Delete your file in S3
boto3.Session().resource('s3').Bucket(bucket_name).Object(key).delete()
# IAM policies should also be removed
iam = boto3.client("iam")
iam.detach_role_policy(PolicyArn="arn:aws:iam::aws:policy/AmazonS3FullAccess", RoleName=role_name)
iam.detach_role_policy(PolicyArn="arn:aws:iam::aws:policy/AmazonForecastFullAccess",RoleName=role_name)
iam.delete_role(RoleName=role_name)
| 0.291485 | 0.789153 |
```
import numpy as np
from sklearn.cluster import KMeans
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split as tts
from sklearn.linear_model import LogisticRegression, Lasso, LassoCV
from yellowbrick.cluster import *
from yellowbrick.features import FeatureImportances
from yellowbrick.classifier import ROCAUC, DiscriminationThreshold
from yellowbrick.classifier import ClassPredictionError, ConfusionMatrix
from yellowbrick.datasets import load_occupancy, load_energy, load_credit
from yellowbrick.classifier import ClassificationReport, PrecisionRecallCurve
from yellowbrick.regressor import PredictionError, ResidualsPlot, AlphaSelection
```
# Check if fitted on Classifiers
```
X, y = load_occupancy(return_dataset=True).to_numpy()
X_train, X_test, y_train, y_test = tts(X, y, test_size=0.20)
unfitted_model = LogisticRegression(solver='lbfgs')
fitted_model = unfitted_model.fit(X_train, y_train)
oz = ClassPredictionError(fitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ClassPredictionError(unfitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ClassificationReport(fitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ClassificationReport(unfitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ConfusionMatrix(fitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ConfusionMatrix(unfitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = PrecisionRecallCurve(fitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = PrecisionRecallCurve(unfitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ROCAUC(fitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ROCAUC(unfitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = DiscriminationThreshold(fitted_model)
oz.fit(X, y)
oz.show()
oz = DiscriminationThreshold(unfitted_model)
oz.fit(X, y)
oz.show()
```
# Check if fitted on Feature Visualizers*
Just the ones that inherit from `ModelVisualizer`
```
viz = FeatureImportances(fitted_model)
viz.fit(X, y)
viz.show()
viz = FeatureImportances(unfitted_model)
viz.fit(X, y)
viz.show()
# NOTE: Not sure how to deal with Recursive Feature Elimination
```
# Check if fitted on Regressors
```
X, y = load_energy(return_dataset=True).to_numpy()
X_train, X_test, y_train, y_test = tts(X, y, test_size=0.20)
unfitted_nonlinear_model = RandomForestRegressor(n_estimators=10)
fitted_nonlinear_model = unfitted_nonlinear_model.fit(X_train, y_train)
unfitted_linear_model = Lasso()
fitted_linear_model = unfitted_linear_model.fit(X_train, y_train)
oz = PredictionError(unfitted_linear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = PredictionError(fitted_linear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ResidualsPlot(unfitted_linear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ResidualsPlot(fitted_linear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ResidualsPlot(unfitted_nonlinear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ResidualsPlot(fitted_nonlinear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
unfitted_cv_model = LassoCV(alphas=[.01,1,10], cv=3)
fitted_cv_model = unfitted_cv_model.fit(X, y)
oz = AlphaSelection(unfitted_cv_model)
oz.fit(X, y)
oz.show()
oz = AlphaSelection(fitted_cv_model)
oz.fit(X, y)
oz.show()
```
# Check if fitted on Clusterers
```
X, _ = load_credit(return_dataset=True).to_numpy()
unfitted_cluster_model = KMeans(6)
fitted_cluster_model = unfitted_cluster_model.fit(X)
# NOTE: Not sure how to deal with K-Elbow and prefitted models...
# visualizer = KElbowVisualizer(unfitted_cluster_model, k=(4,12))
# visualizer.fit(X)
# visualizer.show()
# visualizer = KElbowVisualizer(fitted_cluster_model, k=(4,12))
# visualizer.fit(X)
# visualizer.show()
# NOTE: Silhouette Scores doesn't have a quick method
visualizer = SilhouetteVisualizer(unfitted_cluster_model)
visualizer.fit(X)
visualizer.show()
visualizer = SilhouetteVisualizer(fitted_cluster_model)
visualizer.fit(X)
visualizer.show()
visualizer = InterclusterDistance(unfitted_cluster_model)
visualizer.fit(X)
visualizer.show()
visualizer = InterclusterDistance(fitted_cluster_model)
visualizer.fit(X)
visualizer.show()
```
# Check if fitted on Model Selection Visualizers
_NOTE: Not sure how to proceed with multi-model visualizers -- is already fitted a real use case here?_
|
github_jupyter
|
import numpy as np
from sklearn.cluster import KMeans
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split as tts
from sklearn.linear_model import LogisticRegression, Lasso, LassoCV
from yellowbrick.cluster import *
from yellowbrick.features import FeatureImportances
from yellowbrick.classifier import ROCAUC, DiscriminationThreshold
from yellowbrick.classifier import ClassPredictionError, ConfusionMatrix
from yellowbrick.datasets import load_occupancy, load_energy, load_credit
from yellowbrick.classifier import ClassificationReport, PrecisionRecallCurve
from yellowbrick.regressor import PredictionError, ResidualsPlot, AlphaSelection
X, y = load_occupancy(return_dataset=True).to_numpy()
X_train, X_test, y_train, y_test = tts(X, y, test_size=0.20)
unfitted_model = LogisticRegression(solver='lbfgs')
fitted_model = unfitted_model.fit(X_train, y_train)
oz = ClassPredictionError(fitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ClassPredictionError(unfitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ClassificationReport(fitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ClassificationReport(unfitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ConfusionMatrix(fitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ConfusionMatrix(unfitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = PrecisionRecallCurve(fitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = PrecisionRecallCurve(unfitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ROCAUC(fitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ROCAUC(unfitted_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = DiscriminationThreshold(fitted_model)
oz.fit(X, y)
oz.show()
oz = DiscriminationThreshold(unfitted_model)
oz.fit(X, y)
oz.show()
viz = FeatureImportances(fitted_model)
viz.fit(X, y)
viz.show()
viz = FeatureImportances(unfitted_model)
viz.fit(X, y)
viz.show()
# NOTE: Not sure how to deal with Recursive Feature Elimination
X, y = load_energy(return_dataset=True).to_numpy()
X_train, X_test, y_train, y_test = tts(X, y, test_size=0.20)
unfitted_nonlinear_model = RandomForestRegressor(n_estimators=10)
fitted_nonlinear_model = unfitted_nonlinear_model.fit(X_train, y_train)
unfitted_linear_model = Lasso()
fitted_linear_model = unfitted_linear_model.fit(X_train, y_train)
oz = PredictionError(unfitted_linear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = PredictionError(fitted_linear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ResidualsPlot(unfitted_linear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ResidualsPlot(fitted_linear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ResidualsPlot(unfitted_nonlinear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
oz = ResidualsPlot(fitted_nonlinear_model)
oz.fit(X_train, y_train)
oz.score(X_test, y_test)
oz.show()
unfitted_cv_model = LassoCV(alphas=[.01,1,10], cv=3)
fitted_cv_model = unfitted_cv_model.fit(X, y)
oz = AlphaSelection(unfitted_cv_model)
oz.fit(X, y)
oz.show()
oz = AlphaSelection(fitted_cv_model)
oz.fit(X, y)
oz.show()
X, _ = load_credit(return_dataset=True).to_numpy()
unfitted_cluster_model = KMeans(6)
fitted_cluster_model = unfitted_cluster_model.fit(X)
# NOTE: Not sure how to deal with K-Elbow and prefitted models...
# visualizer = KElbowVisualizer(unfitted_cluster_model, k=(4,12))
# visualizer.fit(X)
# visualizer.show()
# visualizer = KElbowVisualizer(fitted_cluster_model, k=(4,12))
# visualizer.fit(X)
# visualizer.show()
# NOTE: Silhouette Scores doesn't have a quick method
visualizer = SilhouetteVisualizer(unfitted_cluster_model)
visualizer.fit(X)
visualizer.show()
visualizer = SilhouetteVisualizer(fitted_cluster_model)
visualizer.fit(X)
visualizer.show()
visualizer = InterclusterDistance(unfitted_cluster_model)
visualizer.fit(X)
visualizer.show()
visualizer = InterclusterDistance(fitted_cluster_model)
visualizer.fit(X)
visualizer.show()
| 0.872252 | 0.92164 |
This notebook was prepared by [Donne Martin](https://github.com/donnemartin). Source and license info is on [GitHub](https://github.com/donnemartin/interactive-coding-challenges).
# Solution Notebook
## Problem: Find the single different char between two strings.
* [Constraints](#Constraints)
* [Test Cases](#Test-Cases)
* [Algorithm](#Algorithm)
* [Code](#Code)
* [Unit Test](#Unit-Test)
## Constraints
* Can we assume the strings are ASCII?
* Yes
* Is case important?
* The strings are lower case
* Can we assume the inputs are valid?
* No, check for None
* Otherwise, assume there is only a single different char between the two strings
* Can we assume this fits memory?
* Yes
## Test Cases
* None input -> TypeError
* 'ab', 'aab' -> 'a'
* 'aab', 'ab' -> 'a'
* 'abcd', 'abcde' -> 'e'
* 'aaabbcdd', 'abdbacade' -> 'e'
## Algorithm
### Dictionary
* Keep a dictionary of seen values in s
* Loop through t, decrementing the seen values
* If the char is not there or if the decrement results in a negative value, return the char
* Return the differing char from the dictionary
Complexity:
* Time: O(m+n), where m and n are the lengths of s, t
* Space: O(h), for the dict, where h is the unique chars in s
### XOR
* XOR the two strings, which will isolate the differing char
Complexity:
* Time: O(m+n), where m and n are the lengths of s, t
* Space: O(1)
## Code
```
class Solution(object):
def find_diff(self, str1, str2):
if str1 is None or str2 is None:
raise TypeError('str1 or str2 cannot be None')
seen = {}
for char in str1:
if char in seen:
seen[char] += 1
else:
seen[char] = 1
for char in str2:
try:
seen[char] -= 1
except KeyError:
return char
if seen[char] < 0:
return char
for char, count in seen.items():
return char
def find_diff_xor(self, str1, str2):
if str1 is None or str2 is None:
raise TypeError('str1 or str2 cannot be None')
result = 0
for char in str1:
result ^= ord(char)
for char in str2:
result ^= ord(char)
return chr(result)
```
## Unit Test
```
%%writefile test_str_diff.py
from nose.tools import assert_equal, assert_raises
class TestFindDiff(object):
def test_find_diff(self):
solution = Solution()
assert_raises(TypeError, solution.find_diff, None)
assert_equal(solution.find_diff('ab', 'aab'), 'a')
assert_equal(solution.find_diff('aab', 'ab'), 'a')
assert_equal(solution.find_diff('abcd', 'abcde'), 'e')
assert_equal(solution.find_diff('aaabbcdd', 'abdbacade'), 'e')
assert_equal(solution.find_diff_xor('ab', 'aab'), 'a')
assert_equal(solution.find_diff_xor('aab', 'ab'), 'a')
assert_equal(solution.find_diff_xor('abcd', 'abcde'), 'e')
assert_equal(solution.find_diff_xor('aaabbcdd', 'abdbacade'), 'e')
print('Success: test_find_diff')
def main():
test = TestFindDiff()
test.test_find_diff()
if __name__ == '__main__':
main()
%run -i test_str_diff.py
```
|
github_jupyter
|
class Solution(object):
def find_diff(self, str1, str2):
if str1 is None or str2 is None:
raise TypeError('str1 or str2 cannot be None')
seen = {}
for char in str1:
if char in seen:
seen[char] += 1
else:
seen[char] = 1
for char in str2:
try:
seen[char] -= 1
except KeyError:
return char
if seen[char] < 0:
return char
for char, count in seen.items():
return char
def find_diff_xor(self, str1, str2):
if str1 is None or str2 is None:
raise TypeError('str1 or str2 cannot be None')
result = 0
for char in str1:
result ^= ord(char)
for char in str2:
result ^= ord(char)
return chr(result)
%%writefile test_str_diff.py
from nose.tools import assert_equal, assert_raises
class TestFindDiff(object):
def test_find_diff(self):
solution = Solution()
assert_raises(TypeError, solution.find_diff, None)
assert_equal(solution.find_diff('ab', 'aab'), 'a')
assert_equal(solution.find_diff('aab', 'ab'), 'a')
assert_equal(solution.find_diff('abcd', 'abcde'), 'e')
assert_equal(solution.find_diff('aaabbcdd', 'abdbacade'), 'e')
assert_equal(solution.find_diff_xor('ab', 'aab'), 'a')
assert_equal(solution.find_diff_xor('aab', 'ab'), 'a')
assert_equal(solution.find_diff_xor('abcd', 'abcde'), 'e')
assert_equal(solution.find_diff_xor('aaabbcdd', 'abdbacade'), 'e')
print('Success: test_find_diff')
def main():
test = TestFindDiff()
test.test_find_diff()
if __name__ == '__main__':
main()
%run -i test_str_diff.py
| 0.461017 | 0.941331 |
# Exercise Set 12: Linear regression models.
*Afternoon, August 19, 2019*
In this Exercise Set 12 we will work with linear regression models.
We import our standard stuff. Notice that we are not interested in seeing the convergence warning in scikit-learn so we suppress them for now.
```
import warnings
from sklearn.exceptions import ConvergenceWarning
warnings.filterwarnings(action='ignore', category=ConvergenceWarning)
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
%matplotlib inline
```
## Exercise Section 12.1: Estimating linear models with gradient decent
Normally we use OLS to estimate linear models. In this exercise we replace the OLS-estimator with a new estimator that we code up from scratch. We solve the numerical optimization using the gradient decent algorithm. Using our algorithm we will fit it to some data, and compare our own solution to the standard solution from `sklearn`
> **Ex. 12.1.0**: Import the dataset `tips` from the `seaborn`.
*Hint*: use the `load_dataset` method in seaborn
```
# [Answer to Ex. 12.1.0]
# Load the example tips dataset
tips = sns.load_dataset("tips")
```
> **Ex. 12.1.1**: Convert non-numeric variables to dummy variables for each category (remember to leave one column out for each catagorical variable, so you have a reference). Restructure the data so we get a dataset `y` containing the variable tip, and a dataset `X` containing the
features.
>> *Hint*: You might want to use the `get_dummies` method in pandas, with the `drop_first = True` parameter.
```
# [Answer to Ex. 12.1.1]
tips_num = pd.get_dummies(tips, drop_first=True)
X = tips_num.drop('tip', axis = 1)
y = tips_num['tip']
```
> **Ex. 12.1.2**: Divide the features and target into test and train data. Make the split 50 pct. of each. The split data should be called `X_train`, `X_test`, `y_train`, `y_test`.
>> *Hint*: You may use `train_test_split` in `sklearn.model_selection`.
```
# [Answer to Ex. 12.1.2]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y, test_size=.5)
```
> **Ex. 12.1.3**: Normalize your features by converting to zero mean and one std. deviation.
>> *Hint 1*: Take a look at `StandardScaler` in `sklearn.preprocessing`.
>> *Hint 2*: If in doubt about which distribution to scale, you may read [this post](https://stats.stackexchange.com/questions/174823/how-to-apply-standardization-normalization-to-train-and-testset-if-prediction-i).
```
# [Answer to Ex. 12.1.3]
from sklearn.preprocessing import StandardScaler, PolynomialFeatures
norm_scaler = StandardScaler().fit(X_train)
X_train = norm_scaler.transform(X_train)
X_test = norm_scaler.transform(X_test)
```
> **Ex. 12.1.4**: Make a function called `compute_error` to compute the prediction errors given input target `y_`, input features `X_` and input weights `w_`. You should use matrix multiplication.
>
>> *Hint 1:* You can use the net-input fct. from yesterday.
>>
>> *Hint 2:* If you run the following code,
>> ```python
y__ = np.array([1,1])
X__ = np.array([[1,0],[0,1]])
w__ = np.array([0,1,1])
compute_error(y__, X__, w__)
```
>> then you should get output:
```python
array([0,0])
```
```
# [Answer to Ex. 12.1.4]
def net_input(X_, w_):
''' Computes the matrix product between X and w. Note that
X is assumed not to contain a bias/intercept column.'''
return np.dot(X_, w_[1:]) + w_[0] # We have to add w_[0] separately because this is the constant term. We could also have added a constant term (columns of 1's to X_ and multipliced it to all of w_)
def compute_error(y_, X_, w_):
return y_ - net_input(X_, w_)
```
> **Ex. 12.1.5**: Make a function to update the weights given input target `y_`, input features `X_` and input weights `w_` as well as learning rate, $\eta$, i.e. greek `eta`. You should use matrix multiplication.
```
# [Answer to Ex. 12.1.5]
def update_weight(y_, X_, w_, eta):
error = compute_error(y_, X_, w_)
w_[1:] += eta * (X_.T.dot(error))
w_[0] += eta * (error).sum()
```
> **Ex. 12.1.6**: Use the code below to initialize weights `w` at zero given feature set `X`. Notice how we include an extra weight that includes the bias term. Set the learning rate `eta` to 0.001. Make a loop with 50 iterations where you iteratively apply your weight updating function.
>```python
w = np.zeros(1+X.shape[1])
```
```
# [Answer to Ex. 12.1.6]
w = np.zeros(1+X.shape[1])
error_train, error_test = [], []
for i in range(50):
update_weight(y_train, X_train, w, 10**-3)
```
> **Ex. 12.1.7**: Make a function to compute the mean squared error. Alter the loop so it makes 100 iterations and computes the MSE for test and train after each iteration, plot these in one figure.
>> Hint: You can use the following code to check that your model works:
>>```python
from sklearn.linear_model import LinearRegression
reg = LinearRegression()
reg.fit(X_train, y_train)
assert((w[1:] - reg.coef_).sum() < 0.01)
```
```
# [Answer to Ex. 12.1.7]
def MSE(y_, X_, w_):
error_squared = compute_error(y_, X_, w_)**2
return error_squared.sum() / len(y_)
w = np.zeros(X.shape[1]+1)
MSE_train = [MSE(y_train, X_train, w)]
MSE_test = [MSE(y_test, X_test, w)]
for i in range(100):
update_weight(y_train, X_train, w, 10**-3)
MSE_train.append(MSE(y_train, X_train, w))
MSE_test.append(MSE(y_test, X_test, w))
pd.Series(MSE_train).plot()
pd.Series(MSE_test).plot()
```
The following bonus exercises are for those who have completed all other exercises until now and have a deep motivation for learning more.
> **Ex. 12.1.8 (BONUS)**: Implement your linear regression model as a class.
> ANSWER: A solution is found on p. 320 in Python for Machine Learning.
> **Ex. 12.1.9 (BONUS)**: Is it possible to adjust our linear model to become a Lasso? Is there a simple fix?
> ANSWER: No, we cannot exactly solve for the Lasso with gradient descent. However, we can make an approximate solution which is pretty close and quite intuitive - see good explanation [here](https://stats.stackexchange.com/questions/177800/why-proximal-gradient-descent-instead-of-plain-subgradient-methods-for-lasso).
## Exercise Section 12.2: Houseprices
In this example we will try to predict houseprices using a lot of variable (or features as they are called in Machine Learning). We are going to work with Kaggle's dataset on house prices, see information [here](https://www.kaggle.com/c/house-prices-advanced-regression-techniques). Kaggle is an organization that hosts competitions in building predictive models.
> **Ex. 12.2.0:** Load the california housing data with scikit-learn using the code below. Inspect the data set.
```
# The exercise will be part of assignment 2
```
> **Ex.12.2.1**: Generate interactions between all features to third degree, make sure you **exclude** the bias/intercept term. How many variables are there? Will OLS fail?
> After making interactions rescale the features to have zero mean, unit std. deviation. Should you use the distribution of the training data to rescale the test data?
>> *Hint 1*: Try importing `PolynomialFeatures` from `sklearn.preprocessing`
>> *Hint 2*: If in doubt about which distribution to scale, you may read [this post](https://stats.stackexchange.com/questions/174823/how-to-apply-standardization-normalization-to-train-and-testset-if-prediction-i).
```
# The exercise will be part of assignment 2
```
> **Ex.12.2.2**: Estimate the Lasso model on the train data set, using values of $\lambda$ in the range from $10^{-4}$ to $10^4$. For each $\lambda$ calculate and save the Root Mean Squared Error (RMSE) for the test and train data.
> *Hint*: use `logspace` in numpy to create the range.
```
# The exercise will be part of assignment 2
```
> **Ex.12.2.3**: Make a plot with on the x-axis and the RMSE measures on the y-axis. What happens to RMSE for train and test data as $\lambda$ increases? The x-axis should be log scaled. Which one are we interested in minimizing?
> Bonus: Can you find the lambda that gives the lowest MSE-test score?
```
# The exercise will be part of assignment 2
```
|
github_jupyter
|
import warnings
from sklearn.exceptions import ConvergenceWarning
warnings.filterwarnings(action='ignore', category=ConvergenceWarning)
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
%matplotlib inline
# [Answer to Ex. 12.1.0]
# Load the example tips dataset
tips = sns.load_dataset("tips")
# [Answer to Ex. 12.1.1]
tips_num = pd.get_dummies(tips, drop_first=True)
X = tips_num.drop('tip', axis = 1)
y = tips_num['tip']
# [Answer to Ex. 12.1.2]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y, test_size=.5)
# [Answer to Ex. 12.1.3]
from sklearn.preprocessing import StandardScaler, PolynomialFeatures
norm_scaler = StandardScaler().fit(X_train)
X_train = norm_scaler.transform(X_train)
X_test = norm_scaler.transform(X_test)
y__ = np.array([1,1])
X__ = np.array([[1,0],[0,1]])
w__ = np.array([0,1,1])
compute_error(y__, X__, w__)
> **Ex. 12.1.5**: Make a function to update the weights given input target `y_`, input features `X_` and input weights `w_` as well as learning rate, $\eta$, i.e. greek `eta`. You should use matrix multiplication.
> **Ex. 12.1.6**: Use the code below to initialize weights `w` at zero given feature set `X`. Notice how we include an extra weight that includes the bias term. Set the learning rate `eta` to 0.001. Make a loop with 50 iterations where you iteratively apply your weight updating function.
>```python
w = np.zeros(1+X.shape[1])
# [Answer to Ex. 12.1.6]
w = np.zeros(1+X.shape[1])
error_train, error_test = [], []
for i in range(50):
update_weight(y_train, X_train, w, 10**-3)
from sklearn.linear_model import LinearRegression
reg = LinearRegression()
reg.fit(X_train, y_train)
assert((w[1:] - reg.coef_).sum() < 0.01)
# [Answer to Ex. 12.1.7]
def MSE(y_, X_, w_):
error_squared = compute_error(y_, X_, w_)**2
return error_squared.sum() / len(y_)
w = np.zeros(X.shape[1]+1)
MSE_train = [MSE(y_train, X_train, w)]
MSE_test = [MSE(y_test, X_test, w)]
for i in range(100):
update_weight(y_train, X_train, w, 10**-3)
MSE_train.append(MSE(y_train, X_train, w))
MSE_test.append(MSE(y_test, X_test, w))
pd.Series(MSE_train).plot()
pd.Series(MSE_test).plot()
# The exercise will be part of assignment 2
# The exercise will be part of assignment 2
# The exercise will be part of assignment 2
# The exercise will be part of assignment 2
| 0.682679 | 0.981095 |
# Advanced simulation
In essence, a Model object is able to change the state of the system given a sample and evaluate certain metrics.

Model objects are able to drastically cut simulation time by sorting the samples to minimize perturbations to the system between simulations. This decreases the number of iterations required to solve recycle systems. The following examples show how Model objects can be used.
### Create a model object
**Model objects are used to evaluate metrics around multiple parameters of a system.**
Create a Model object of the lipidcane biorefinery with internal rate of return as a metric:
```
from biosteam.biorefineries import lipidcane as lc
import biosteam as bst
solve_IRR = lc.lipidcane_tea.solve_IRR
metrics = bst.Metric('IRR', solve_IRR),
model = bst.Model(lc.lipidcane_sys, metrics)
```
The Model object begins with no paramters:
```
model
```
Note: Here we defined only one metric, but more metrics are possible.
### Add design parameters
**A design parameter is a Unit attribute that changes design requirements but does not affect mass and energy balances.**
Add number of fermentation reactors as a "design" parameter:
```
R301 = bst.find.unit.R301 # The Fermentation Unit
@model.parameter(element=R301, kind='design', name='Number of reactors')
def set_N_reactors(N):
R301.N = N
```
The decorator returns a Parameter object and adds it to the model:
```
set_N_reactors
```
Calling a Parameter object will update the parameter and results:
```
set_N_reactors(5)
print('Puchase cost at 5 reactors: ' + str(R301.purchase_cost))
set_N_reactors(8)
print('Puchase cost at 8 reactors: ' + str(R301.purchase_cost))
```
### Add cost parameters
**A cost parameter is a Unit attribute that affects cost but does not change design requirements.**
Add the fermentation unit base cost as a "cost" parameter:
```
@model.parameter(element=R301, kind='cost') # Note: name argument not given this time
def set_base_cost(cost):
R301.cost_items['Reactors'].cost = cost
original = R301.cost_items['Reactors'].cost
set_base_cost(10e6)
print('Purchase cost at 10 million USD: ' + str(R301.purchase_cost))
set_base_cost(844e3)
print('Purchase cost at 844,000 USD: ' + str(R301.purchase_cost))
```
If the name was not defined, it defaults to the setter's signature:
```
set_base_cost
```
### Add isolated parameters
**An isolated parameter should not affect Unit objects in any way.**
Add feedstock price as a "isolated" parameter:
```
lipid_cane = lc.lipid_cane # The feedstock stream
@model.parameter(element=lipid_cane, kind='isolated')
def set_feed_price(feedstock_price):
lipid_cane.price = feedstock_price
```
### Add coupled parameters
**A coupled parameter affects mass and energy balances of the system.**
Add lipid fraction as a "coupled" parameter:
```
set_lipid_fraction = model.parameter(lc.set_lipid_fraction, element=lipid_cane, kind='coupled')
set_lipid_fraction(0.10)
print('IRR at 10% lipid: ' + str(solve_IRR()))
set_lipid_fraction(0.05)
print('IRR at 5% lipid: ' + str(solve_IRR()))
```
Add fermentation efficiency as a "coupled" parameter:
```
@model.parameter(element=R301, kind='coupled')
def set_fermentation_efficiency(efficiency):
R301.efficiency = efficiency
```
### Evaluate metric given a sample
**The model can be called to evaluate a sample of parameters.**
All parameters are stored in the model with highly coupled parameters first:
```
model
```
Get all parameters as ordered in the model:
```
model.get_parameters()
```
Evaluate sample:
```
model([0.05, 0.85, 8, 100000, 0.040])
```
### Evaluate metric across samples
Evaluate at give parameter values:
```
import numpy as np
samples = np.array([(0.05, 0.85, 8, 100000, 0.040),
(0.05, 0.90, 7, 100000, 0.040),
(0.09, 0.95, 8, 100000, 0.042)])
model.load_samples(samples)
model.evaluate()
model.table # All evaluations are stored as a pandas DataFrame
```
Note that coupled parameters are on the left most columns, and are ordered from upstream to downstream (e.g. <Stream: Lipid cane> is upstream from <Fermentation: R301>)
### Evaluate multiple metrics
Reset the metrics to include total utility cost:
```
def total_utility_cost():
"""Return utility costs in 10^6 USD/yr"""
return lc.lipidcane_tea.utility_cost / 10**6
# This time use detailed names and units for appearance
model.metrics = (bst.Metric('Internal rate of return', lc.lipidcane_tea.solve_IRR, '%'),
bst.Metric('Utility cost', total_utility_cost, 'USD/yr'))
model
model.evaluate()
model.table
```
### Behind the scenes

Model objects work with the help of Block and Parameter objects that are able to tell the relative importance of parameters through the `element` it affects and the `kind` (how it affects the system). Before a new parameter is made, if its `kind` is "coupled", then the Model object creates a Block object that simulates only the objects affected by the parameter. The Block object, in turn, helps to create a Parameter object by passing its simulation method.
|
github_jupyter
|
from biosteam.biorefineries import lipidcane as lc
import biosteam as bst
solve_IRR = lc.lipidcane_tea.solve_IRR
metrics = bst.Metric('IRR', solve_IRR),
model = bst.Model(lc.lipidcane_sys, metrics)
model
R301 = bst.find.unit.R301 # The Fermentation Unit
@model.parameter(element=R301, kind='design', name='Number of reactors')
def set_N_reactors(N):
R301.N = N
set_N_reactors
set_N_reactors(5)
print('Puchase cost at 5 reactors: ' + str(R301.purchase_cost))
set_N_reactors(8)
print('Puchase cost at 8 reactors: ' + str(R301.purchase_cost))
@model.parameter(element=R301, kind='cost') # Note: name argument not given this time
def set_base_cost(cost):
R301.cost_items['Reactors'].cost = cost
original = R301.cost_items['Reactors'].cost
set_base_cost(10e6)
print('Purchase cost at 10 million USD: ' + str(R301.purchase_cost))
set_base_cost(844e3)
print('Purchase cost at 844,000 USD: ' + str(R301.purchase_cost))
set_base_cost
lipid_cane = lc.lipid_cane # The feedstock stream
@model.parameter(element=lipid_cane, kind='isolated')
def set_feed_price(feedstock_price):
lipid_cane.price = feedstock_price
set_lipid_fraction = model.parameter(lc.set_lipid_fraction, element=lipid_cane, kind='coupled')
set_lipid_fraction(0.10)
print('IRR at 10% lipid: ' + str(solve_IRR()))
set_lipid_fraction(0.05)
print('IRR at 5% lipid: ' + str(solve_IRR()))
@model.parameter(element=R301, kind='coupled')
def set_fermentation_efficiency(efficiency):
R301.efficiency = efficiency
model
model.get_parameters()
model([0.05, 0.85, 8, 100000, 0.040])
import numpy as np
samples = np.array([(0.05, 0.85, 8, 100000, 0.040),
(0.05, 0.90, 7, 100000, 0.040),
(0.09, 0.95, 8, 100000, 0.042)])
model.load_samples(samples)
model.evaluate()
model.table # All evaluations are stored as a pandas DataFrame
def total_utility_cost():
"""Return utility costs in 10^6 USD/yr"""
return lc.lipidcane_tea.utility_cost / 10**6
# This time use detailed names and units for appearance
model.metrics = (bst.Metric('Internal rate of return', lc.lipidcane_tea.solve_IRR, '%'),
bst.Metric('Utility cost', total_utility_cost, 'USD/yr'))
model
model.evaluate()
model.table
| 0.489503 | 0.970521 |
# Exploring the application of quantum circuits in convolutional neural networks
This tutorial will guide you through implementing a hybrid quantum-classical convolutional neural network using Tequila along with other packages such as Tensorflow. We will then train the model on the MNIST dataset, which contains images of handwritten numbers classifed according to the digit they represent. Finally, we will compare the accuracy and loss of models with and without the quantum preprocessing.
Inspriation for this tutorial comes from [Pennylane: Quanvolutional Neural Networks](https://pennylane.ai/qml/demos/tutorial_quanvolution.html). We will similarly follow the method proposed in the reference paper used for this tutorial, [Henderson at al (2020)](https://doi.org/10.1007/s42484-020-00012-y).
## Background
#### Convolutional Neural Nets
An excellent high-level explanation of convolutional neural networks can be found [here](https://www.youtube.com/watch?v=FmpDIaiMIeA). Alternatively, an excellent written explanation can be found [here](http://neuralnetworksanddeeplearning.com/chap6.html) and for more information, the wikipedia article can be found [here](https://en.wikipedia.org/wiki/Convolutional_neural_network).
In summary, a convolutional neural network includes preprocessing layers prior to optimisation layers so that features in the input (which are often images) are extracted and amplified. The result is a model with greater predictive power. This processing also improves classification of images as it extracts features even if they are translocated between images. This means that searching for a particular pixel distribution (for example the shape of a curve or line may be a useful feature when classifying digits) is not dependant on the distribution being in an identical location in each image where it is present. The convolutional process extracts this information even if it is slightly rotated or translocated.
The implementation of the convolutional layer involves a grid for each feature being passed over the entire image. At each location, a score is calculated representing how well the feature and the section of the image match, and this becomes the value of the corresponding pixel in the output image. As a guide, a large score represents a close match, generally meaning that the feature is present at that location of the image, and a low score represents the absence of a match.
#### Our Approach
Our general approach is similar to that used in a conventional convolutional neural network however the initial processing occurs by running the images through a quantum circuit instead of a convolutional filter. Each simulation of a circuit represents one 3x3 filter being applied to one 3x3 region of one image. The construction of the circuit is randomised (see below), however this construction only occurs once per filter such that each region of the image being transformed by the same filter gets run through the same circuit. A single, scalar output is generated from the circuit which is used as the pixel strength of the output image, and the remainder of the neural net uses only classical processing, specifically two further convolutional layers, max pooling and two fully connected layers. This architecture has been chosen to closely mimic the structure used in our reference paper (Henderson et al, 2020), however as they note, "The QNN topology chosen in this work is not fixed by nature ... the QNN framework was designed to give users complete control over the number and order of quanvolutional layers in the architecture. The topology explored in this work was chosen because it was the simplest QNN architecture to use as a baseline for comparison against other purely classical networks. Future work would focus on exploring the impact of more complex architectural variations."
<img src="Quanv_Neural_Net/Our_approach.jpg" width="700" />
#### Quantum Processing
Henderson et al summarise the use of quantum circuits as convolutional layers: "Quanvolutional layers are made up of a group of N quantum filters which operate much like their classical convolutional layer counterparts, producing feature maps by locally transforming input data. The key difference is that quanvolutional filters extract features from input data by transforming spatially local subsections of data using quantum circuits." Our approach to the circuit design is based on the paper and is as follows:
1) The input images are iterated over and each 3x3 region is embedded into the quantum circuit using the threshold function:
$$|\psi \rangle = \begin{cases}
|0\rangle & if & strength\leq 0 \\
|1\rangle & if & strength > 0
\end{cases}$$
As the pixel strengths are normalised to values between -0.5 and 0.5, it is expected that brighter regions of the image will intialise their corresponding qubit in the state $|1\rangle$ and darker regions will intitialise the state $|0\rangle$. Each pixel is represented by one qubit, such that 9 qubits are used in total, and this quantum circuit is reused for each 3x3 square in the filter.
2) We next apply a random circuit to the qubits. To implement this, a random choice from Rx, Ry and Rz gates is applied to a random qubit, and the total number of gates applied in each layer is equal to the number of qubits. With a set probability (which we set to 0.3), a CNOT gate will be applied instead of the rotation to two random qubits. We have chosen to set the parameters of rotation with random numbers between (0,2π) however futher optimisation of the model could be found from using a variational circuit and optimising these parameters.
3) Further layers could be applied of the random gates. To simplify, we only apply one layer.
4) A scalar is outputted from the circuit and used as the corresponding pixel in the output image. We generate this number using the following method. The state vector of the final state of the circuit is simulated and the state corresponding to the most likely output (largest modulus) is selected. We then calculate the number of qubits for this state which are measured as a $|1\rangle$.
5) A total of four filters are applied to each image, and for each filter steps 1-3 are repeated with a different randomised circuit. The output image therefore contains a third dimension with four channels representing the four different outputted values which each filters produced.
<img src="Quanv_Neural_Net/Quantum_circuit.jpg" width="700" />
## Code and Running the Program
The following code cell is used to import the necessary packages and to set parameters.
```
import math
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
import tequila as tq
from operator import itemgetter
from tensorflow import keras
n_filters = 4 # Number of convolutional filters
filter_size = 3 # Size of filter = nxn (here 3x3)
pool_size = 2 # Used for the pooling layer
n_qubits = filter_size ** 2 # Number of qubits
n_layers = 1 # Number of quantum circuit layers
n_train = 1000 # Size of the training dataset
n_test = 200 # Size of the testing dataset
n_epochs = 100 # Number of optimization epochs
SAVE_PATH = "quanvolution/" # Data saving folder
PREPROCESS = False # If False, skip quantum processing and load data from SAVE_PATH
tf.random.set_seed(1) # Seed for TensorFlow random number generator
```
We start by creating the Dataset class. Here, we load the images and labels of handwritten digits from the MNIST dataset. We then reduce the number of images from 60,000 and 10,000 (for the training and testing sets respectively) down to the variables n_train and n_test, normalise the pixel values to within the range (-0.5,0.5) and reshape the images by adding a third dimension. Each image's shape is therefore transformed from (28, 28) to (28, 28, 1) as this is necessary for the convolutional layer.
```
class Dataset:
def __init__(self):
# Loading the full dataset of images from keras
# Shape of self.train_images is (60000, 28, 28), shape of self.train_labels is (60000,)
# For self.test_images and self.test_labels, shapes are (10000, 28, 28) and (10000,)
mnist_dataset = keras.datasets.mnist
(self.train_images, self.train_labels), (self.test_images, self.test_labels) = mnist_dataset.load_data()
# Reduce dataset size to n_train and n_test
# First dimension of shapes are reduced to n_train and n_test
self.train_images = self.train_images[:n_train]
self.train_labels = self.train_labels[:n_train]
self.test_images = self.test_images[:n_test]
self.test_labels = self.test_labels[:n_test]
# Normalize pixel values within -0.5 and +0.5
self.train_images = (self.train_images / 255) - 0.5
self.test_images = (self.test_images / 255) - 0.5
# Add extra dimension for convolution channels
self.train_images = self.train_images[..., tf.newaxis]
self.test_images = self.test_images[..., tf.newaxis]
```
The next code cell contains the class used to generate the quantum circuit. In theory, the circuit could be either structured or random. We form a randomised circuit to match the reference paper (Henderson et al, 2020), however for simplicity, our implementation differs in some ways. We choose to use only use single qubit Rx($\theta$), Ry($\theta$) and Rz($\theta$) gates and the two qubit CNOT gate compared to the choice of single qubit X($\theta$), Y($\theta$), Z($\theta$), U($\theta$), P, T, H and two qubit CNOT, SWAP, SQRTSWAP, or CU gates used in the paper. Furthermore, we chose to assign a two qubit gate to any random qubits with a certain probability (labelled ratio_imprim, set to 0.3) rather than setting a connection probabiltiy between each pair of qubits (this approach follows the Pennylane tutorial). The seed is used for reproducability and its value is set depending on which filter the circuit represents (see QuantumModel below).
The parameters used for the rotation gates have the potential to be optimised using a cost function. For simplicity, and to mirror the paper, here we will use random parameters and we will not include these in the optimisation of the model. This means that the quantum processing only needs to happen once, prior to creating the neural net.
```
class QuantumCircuit:
def __init__(self, seed=None):
# Set random seed for reproducability
if seed: np.random.seed(seed)
# Encode classical information into quantum circuit
# Bit flip gate is applied if the pixel strength > 0
self.circ = tq.QCircuit()
for i in range(n_qubits):
self.circ += tq.gates.X(i, power='input_{}'.format(i))
# Add random layers to the circuit
self.circ += self.random_layers()
def random_layers(self, ratio_imprim=0.3):
# Initialise circuit
circuit = tq.QCircuit()
# Iterate over the number of layers, adding rotational and CNOT gates
# The number of rotational gates added per layer is equal to the number of qubits in the circuit
for i in range(n_layers):
j = 0
while (j < n_qubits):
if np.random.random() > ratio_imprim:
# Applies a random rotation gate to a random qubit with probability (1 - ratio_imprim)
rnd_qubit = np.random.randint(n_qubits)
circuit += np.random.choice(
[tq.gates.Rx(angle='l_{},th_{}'.format(i,j), target=rnd_qubit),
tq.gates.Ry(angle='l_{},th_{}'.format(i,j), target=rnd_qubit),
tq.gates.Rz(angle='l_{},th_{}'.format(i,j), target=rnd_qubit)])
j += 1
else:
# Applies the CNOT gate to 2 random qubits with probability ratio_imprim
if n_qubits > 1:
rnd_qubits = np.random.choice(range(n_qubits), 2, replace=False)
circuit += tq.gates.CNOT(target=rnd_qubits[0], control=rnd_qubits[1])
return circuit
```
As an example to show the circuit used in this program, an instance of a circuit is drawn below. This will differ between calls if you remove the seed variable due to the random nature of forming the circuit.
```
circuit = QuantumCircuit(seed=2)
tq.draw(circuit.circ, backend='qiskit')
```
We next show the QuantumModel class, used to generate the neural network for the images which undergo pre-processing through the quantum convolutional layer. If PREPROCESSING is set to True, each image from the dataset undergoes processing through a number of quantum circuits, determined by n_filters. The embedding used, the structure of the circuit and the method of extracting the output are described in the background of this tutorial.
We use tensorflow to construct the neural net. The implementation we use contains two conventional convolutional layers, each followed by max pooling, and then one fully connected with 1024 nodes before the softmax output layer. We use a Relu activation function for the convolutional and fully connected layers. See the background section of this tutorial for some context on this choice of neural net.
```
class QuantumModel:
def __init__(self, dataset, parameters):
# Initialize dataset and parameters
self.ds = dataset
self.params = parameters
# The images are run through the quantum convolutional layer
self.convolutional_layer()
# The model is initialized
self.model = keras.models.Sequential([
keras.layers.Conv2D(n_filters, filter_size, activation='relu'),
keras.layers.MaxPooling2D(pool_size=pool_size),
keras.layers.Conv2D(n_filters, filter_size, activation='relu'),
keras.layers.MaxPooling2D(pool_size=pool_size),
keras.layers.Flatten(),
keras.layers.Dense(1024, activation="relu"),
keras.layers.Dense(10, activation="softmax")
])
# Compile model using the Adam optimiser
self.model.compile(
optimizer=keras.optimizers.Adam(learning_rate=0.00001),
loss="sparse_categorical_crossentropy",
metrics=["accuracy"]
)
def convolutional_layer(self):
if PREPROCESS == True:
# Initate arrays to store processed images
self.q_train_images = [np.zeros((28-2, 28-2, n_filters)) for _ in range(len(self.ds.train_images))]
self.q_test_images = [np.zeros((28-2, 28-2, n_filters)) for _ in range(len(self.ds.test_images))]
# Loop over the number of filters, applying a different randomised quantum circuit for each
for i in range(n_filters):
print('Filter {}/{}\n'.format(i+1, n_filters))
# Construct circuit
# We set the seed to be i+1 so that the circuits are reproducable but the design differs between filters
# We use i+1 not i to avoid setting the seed as 0 which sometimes produces random behaviour
circuit = QuantumCircuit(seed=i+1)
# Apply the quantum processing to the train_images, analogous to a convolutional layer
print("Quantum pre-processing of train images:")
for j, img in enumerate(self.ds.train_images):
print("{}/{} ".format(j+1, n_train), end="\r")
self.q_train_images[j][...,i] = (self.filter_(img, circuit, self.params[i]))
print('\n')
# Similarly for the test_images
print("Quantum pre-processing of test images:")
for j, img in enumerate(self.ds.test_images):
print("{}/{} ".format(j+1, n_test), end="\r")
self.q_test_images[j][...,i] = (self.filter_(img, circuit, self.params[i]))
print('\n')
# Transform images to numpy array
self.q_train_images = np.asarray(self.q_train_images)
self.q_test_images = np.asarray(self.q_test_images)
# Save pre-processed images
np.save(SAVE_PATH + "q_train_images.npy", self.q_train_images)
np.save(SAVE_PATH + "q_test_images.npy", self.q_test_images)
# Load pre-processed images
self.q_train_images = np.load(SAVE_PATH + "q_train_images.npy")
self.q_test_images = np.load(SAVE_PATH + "q_test_images.npy")
def filter_(self, image, circuit, variables):
# Initialize output image
output = np.zeros((28-2, 28-2))
# Loop over the image co-ordinates (i,j) using a 3x3 square filter
for i in range(28-2):
for j in range(28-2):
# Extract the value of each pixel in the 3x3 filter grid
image_pixels = [
image[i,j,0],
image[i,j+1,0],
image[i,j+2,0],
image[i+1,j,0],
image[i+1,j+1,0],
image[i+1,j+2,0],
image[i+2,j,0],
image[i+2,j+1,0],
image[i+2,j+2,0]
]
# Construct parameters used to embed the pixel strength into the circuit
input_variables = {}
for idx, strength in enumerate(image_pixels):
# If strength > 0, the power of the bit flip gate is 1
# Therefore this qubit starts in state |1>
if strength > 0:
input_variables['input_{}'.format(idx)] = 1
# Otherwise the gate is not applied and the initial state is |0>
else:
input_variables['input_{}'.format(idx)] = 0
# Find the statevector of the circuit and determine the state which is most likely to be measured
wavefunction = tq.simulate(circuit.circ, variables={**variables, **input_variables})
amplitudes = [(k,(abs(wavefunction(k)))) for k in range(2**n_qubits) if wavefunction(k)]
max_idx = max(amplitudes,key=itemgetter(1))[0]
# Count the number of qubits which output '1' in this state
result = len([k for k in str(bin(max_idx))[2::] if k == '1'])
output[i,j] = result
return output
def train(self):
# Train the model on the dataset
self.history = self.model.fit(
self.q_train_images,
self.ds.train_labels,
validation_data=(self.q_test_images, self.ds.test_labels),
batch_size=4,
epochs=n_epochs,
verbose=2
)
```
We also create a ClassicalModel class to run the images through a conventional convolutional neural network. The design of the neural net used here is identical to the QuantumModel class, however the images used are directly from the dataset and therefore have not been processed through the quantum layer. We include this as a control to compare the results from the quantum model.
```
class ClassicalModel:
def __init__(self, dataset):
# Initialize dataset and parameters
self.ds = dataset
# The model is initialized
self.model = keras.models.Sequential([
keras.layers.Conv2D(n_filters, filter_size, activation='relu'),
keras.layers.MaxPooling2D(pool_size=pool_size),
keras.layers.Conv2D(n_filters, filter_size, activation='relu'),
keras.layers.MaxPooling2D(pool_size=pool_size),
keras.layers.Flatten(),
keras.layers.Dense(1024, activation="relu"),
keras.layers.Dense(10, activation="softmax")
])
# Compile model using the Adam optimiser
self.model.compile(
optimizer=keras.optimizers.Adam(learning_rate=0.00005),
loss="sparse_categorical_crossentropy",
metrics=["accuracy"]
)
def train(self):
# Train the model on the dataset
self.history = self.model.fit(
self.ds.train_images,
self.ds.train_labels,
validation_data=(self.ds.test_images, self.ds.test_labels),
batch_size=4,
epochs=n_epochs,
verbose=2
)
```
We are now able to run our program! The following code does this using the quantum_model and classical_model functions. Although the implementations are similar, quantum_model additionally defines the parameters used for the rotational gates in the circuit. We have limited the value of each parameter to the range (0,2π).
Running the program takes some time. Our results are plotted below, so if you would rather not wait, either reduce the numbers in n_train and n_test or skip ahead!
```
def quantum_model():
# Generating parameters, each maps to a random number between 0 and 2*π
# parameters is a list of dictionaries, where each dictionary represents the parameter
# mapping for one filter
parameters = []
for i in range(n_filters):
filter_params = {}
for j in range(n_layers):
for k in range(n_qubits):
filter_params[tq.Variable(name='l_{},th_{}'.format(j,k))] = np.random.uniform(high=2*np.pi)
parameters.append(filter_params)
# Initalise the dataset
ds = Dataset()
# Initialise and train the model
model = QuantumModel(ds, parameters=parameters)
model.train()
# Store the loss and accuracy of the model to return
loss = model.history.history['val_loss']
accuracy = model.history.history['val_accuracy']
return model
def classical_model():
# Initialise the dataset
ds = Dataset()
# Initialise and train the model
model = ClassicalModel(ds)
model.train()
# Store the loss and accuracy of the model to return
loss = model.history.history['val_loss']
accuracy = model.history.history['val_accuracy']
return model
model_q = quantum_model()
model_c = classical_model()
```
## Plotting the Results
The graphs showing the accuracy and loss of our models are included in this text box. These were generated using the function plot, available below. As shown, the results from the quantum processing lead to a model comparable to the classical control in both accuracy and loss. After running for 100 epochs, the quantum model results in a validation set accuracy of 0.9350, compared to the fully classical model which has a validation set accuracy of 0.9150.
<img src="Quanv_Neural_Net/Plots.png" />
```
def plot(model_q, model_c):
plt.style.use("seaborn")
fig, (ax1, ax2) = plt.subplots(2, 1, figsize=(6, 9))
# Plotting the graph for accuracy
ax1.plot(model_q.history.history['val_accuracy'], color="tab:red", label="Quantum")
ax1.plot(model_c.history.history['val_accuracy'], color="tab:green", label="Classical")
ax1.set_ylabel("Accuracy")
ax1.set_ylim([0,1])
ax1.set_xlabel("Epoch")
ax1.legend()
# Plotting the graph for loss
ax2.plot(model_q.history.history['val_loss'], color="tab:red", label="Quantum")
ax2.plot(model_c.history.history['val_loss'], color="tab:green", label="Classical")
ax2.set_ylabel("Loss")
ax2.set_xlabel("Epoch")
ax2.legend()
plt.tight_layout()
plt.show()
plot(model_q, model_c)
```
## Evaluating the Model
Let us now compare the behaviour of the two models. We do this by running the test images through each with the optimised weights and biases and seeing the results of the classification. This process is implemented using the Classification class, shown below.
Overall, our quantum model misclassified images 34, 37, 42, 54, 67, 74, 120, 127, 143, 150, 152, 166, and 185. The classical model misclassified images 8, 16, 21, 23, 54, 60, 61, 67, 74, 93, 113, 125, 134, 160, 168, 178, and 196. This means that in total, the quantum model misclassified 13 images and the classical model misclassified 17 images. Of these, only images 54, 67, and 74 were misclassified by both.
```
from termcolor import colored
class Classification:
def __init__(self, model, test_images):
# Initialising parameters
self.model = model
self.test_images = test_images
self.test_labels = model.ds.test_labels
def classify(self):
# Create predictions on the test set
self.predictions = np.argmax(self.model.model.predict(self.test_images), axis=-1)
# Keep track of the indices of images which were classified correctly and incorrectly
self.correct_indices = np.nonzero(self.predictions == self.test_labels)[0]
self.incorrect_indices = np.nonzero(self.predictions != self.test_labels)[0]
def print_(self):
# Printing the total number of correctly and incorrectly classified images
print(len(self.correct_indices)," classified correctly")
print(len(self.incorrect_indices)," classified incorrectly")
print('\n')
# Printing the classification of each image
for i in range(n_test):
print("Image {}/{}".format(i+1, n_test))
if i in self.correct_indices:
# The image was correctly classified
print('model predicts: {} - true classification: {}'.format(
self.predictions[i], self.test_labels[i]))
else:
# The image was not classified correctly
print(colored('model predicts: {} - true classification: {}'.format(
self.predictions[i], self.test_labels[i]), 'red'))
print('Quantum Model')
q_class = Classification(model_q, model_q.q_test_images)
q_class.classify()
q_class.print_()
print('\n')
print('Classical Model')
c_class = Classification(model_c, model_c.ds.test_images)
c_class.classify()
c_class.print_()
```
Lastly, we can see the effect that the quantum convolutional layer actually has on the images by plotting images after they have been run through the quantum filters, and to do this we use the function visualise, shown below. Included in this text box is a plot showing four images which have been run through our filters. The top row shows images from the original dataset, and each subsequent row shows the result from each of the four filters on that original image. It can be seen that the processing preserves the global shape of the digit while introducing local distortion.
<img src="Quanv_Neural_Net/Filters.png" />
```
def visualise(model):
# Setting n_samples to be the number of images to print
n_samples = 4
fig, axes = plt.subplots(1 + n_filters, n_samples, figsize=(10, 10))
# Iterate over each image
for i in range(n_samples):
# Plot the original image from the dataset
axes[0, 0].set_ylabel("Input")
if i != 0:
axes[0, i].yaxis.set_visible(False)
axes[0, i].imshow(model.ds.train_images[i, :, :, 0], cmap="gray")
# Plot the images generated by each filter
for c in range(n_filters):
axes[c + 1, 0].set_ylabel("Output [ch. {}]".format(c))
if i != 0:
axes[c, i].yaxis.set_visible(False)
axes[c + 1, i].imshow(model.q_train_images[i, :, :, c], cmap="gray")
plt.tight_layout()
plt.show()
visualise(model_q)
```
#### Resources used to make this tutorial:
1. [Pennylane: Quanvolutional Neural Networks](https://pennylane.ai/qml/demos/tutorial_quanvolution.html)
2. Henderson, M., Shakya, S., Pradhan, S. et al. Quanvolutional neural networks: powering image recognition with quantum circuits. Quantum Mach. Intell. 2, 1–9 (2020). https://doi.org/10.1007/s42484-020-00012-y
3. [Keras for Beginners: Implementing a Convolutional Neural Network. Victor Zhou](https://victorzhou.com/blog/keras-cnn-tutorial/).
4. [CNNs, Part 1: An Introduction to Convolutional Neural Networks. Victor Zhou](https://victorzhou.com/blog/intro-to-cnns-part-1/).
5. [How Convolutional Neural Networks work](https://www.youtube.com/watch?v=FmpDIaiMIeA)
6. [Neural Networks and Deep Learning, chapter 6. Michael Nielsen](http://neuralnetworksanddeeplearning.com/chap6.html)
|
github_jupyter
|
import math
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
import tequila as tq
from operator import itemgetter
from tensorflow import keras
n_filters = 4 # Number of convolutional filters
filter_size = 3 # Size of filter = nxn (here 3x3)
pool_size = 2 # Used for the pooling layer
n_qubits = filter_size ** 2 # Number of qubits
n_layers = 1 # Number of quantum circuit layers
n_train = 1000 # Size of the training dataset
n_test = 200 # Size of the testing dataset
n_epochs = 100 # Number of optimization epochs
SAVE_PATH = "quanvolution/" # Data saving folder
PREPROCESS = False # If False, skip quantum processing and load data from SAVE_PATH
tf.random.set_seed(1) # Seed for TensorFlow random number generator
class Dataset:
def __init__(self):
# Loading the full dataset of images from keras
# Shape of self.train_images is (60000, 28, 28), shape of self.train_labels is (60000,)
# For self.test_images and self.test_labels, shapes are (10000, 28, 28) and (10000,)
mnist_dataset = keras.datasets.mnist
(self.train_images, self.train_labels), (self.test_images, self.test_labels) = mnist_dataset.load_data()
# Reduce dataset size to n_train and n_test
# First dimension of shapes are reduced to n_train and n_test
self.train_images = self.train_images[:n_train]
self.train_labels = self.train_labels[:n_train]
self.test_images = self.test_images[:n_test]
self.test_labels = self.test_labels[:n_test]
# Normalize pixel values within -0.5 and +0.5
self.train_images = (self.train_images / 255) - 0.5
self.test_images = (self.test_images / 255) - 0.5
# Add extra dimension for convolution channels
self.train_images = self.train_images[..., tf.newaxis]
self.test_images = self.test_images[..., tf.newaxis]
class QuantumCircuit:
def __init__(self, seed=None):
# Set random seed for reproducability
if seed: np.random.seed(seed)
# Encode classical information into quantum circuit
# Bit flip gate is applied if the pixel strength > 0
self.circ = tq.QCircuit()
for i in range(n_qubits):
self.circ += tq.gates.X(i, power='input_{}'.format(i))
# Add random layers to the circuit
self.circ += self.random_layers()
def random_layers(self, ratio_imprim=0.3):
# Initialise circuit
circuit = tq.QCircuit()
# Iterate over the number of layers, adding rotational and CNOT gates
# The number of rotational gates added per layer is equal to the number of qubits in the circuit
for i in range(n_layers):
j = 0
while (j < n_qubits):
if np.random.random() > ratio_imprim:
# Applies a random rotation gate to a random qubit with probability (1 - ratio_imprim)
rnd_qubit = np.random.randint(n_qubits)
circuit += np.random.choice(
[tq.gates.Rx(angle='l_{},th_{}'.format(i,j), target=rnd_qubit),
tq.gates.Ry(angle='l_{},th_{}'.format(i,j), target=rnd_qubit),
tq.gates.Rz(angle='l_{},th_{}'.format(i,j), target=rnd_qubit)])
j += 1
else:
# Applies the CNOT gate to 2 random qubits with probability ratio_imprim
if n_qubits > 1:
rnd_qubits = np.random.choice(range(n_qubits), 2, replace=False)
circuit += tq.gates.CNOT(target=rnd_qubits[0], control=rnd_qubits[1])
return circuit
circuit = QuantumCircuit(seed=2)
tq.draw(circuit.circ, backend='qiskit')
class QuantumModel:
def __init__(self, dataset, parameters):
# Initialize dataset and parameters
self.ds = dataset
self.params = parameters
# The images are run through the quantum convolutional layer
self.convolutional_layer()
# The model is initialized
self.model = keras.models.Sequential([
keras.layers.Conv2D(n_filters, filter_size, activation='relu'),
keras.layers.MaxPooling2D(pool_size=pool_size),
keras.layers.Conv2D(n_filters, filter_size, activation='relu'),
keras.layers.MaxPooling2D(pool_size=pool_size),
keras.layers.Flatten(),
keras.layers.Dense(1024, activation="relu"),
keras.layers.Dense(10, activation="softmax")
])
# Compile model using the Adam optimiser
self.model.compile(
optimizer=keras.optimizers.Adam(learning_rate=0.00001),
loss="sparse_categorical_crossentropy",
metrics=["accuracy"]
)
def convolutional_layer(self):
if PREPROCESS == True:
# Initate arrays to store processed images
self.q_train_images = [np.zeros((28-2, 28-2, n_filters)) for _ in range(len(self.ds.train_images))]
self.q_test_images = [np.zeros((28-2, 28-2, n_filters)) for _ in range(len(self.ds.test_images))]
# Loop over the number of filters, applying a different randomised quantum circuit for each
for i in range(n_filters):
print('Filter {}/{}\n'.format(i+1, n_filters))
# Construct circuit
# We set the seed to be i+1 so that the circuits are reproducable but the design differs between filters
# We use i+1 not i to avoid setting the seed as 0 which sometimes produces random behaviour
circuit = QuantumCircuit(seed=i+1)
# Apply the quantum processing to the train_images, analogous to a convolutional layer
print("Quantum pre-processing of train images:")
for j, img in enumerate(self.ds.train_images):
print("{}/{} ".format(j+1, n_train), end="\r")
self.q_train_images[j][...,i] = (self.filter_(img, circuit, self.params[i]))
print('\n')
# Similarly for the test_images
print("Quantum pre-processing of test images:")
for j, img in enumerate(self.ds.test_images):
print("{}/{} ".format(j+1, n_test), end="\r")
self.q_test_images[j][...,i] = (self.filter_(img, circuit, self.params[i]))
print('\n')
# Transform images to numpy array
self.q_train_images = np.asarray(self.q_train_images)
self.q_test_images = np.asarray(self.q_test_images)
# Save pre-processed images
np.save(SAVE_PATH + "q_train_images.npy", self.q_train_images)
np.save(SAVE_PATH + "q_test_images.npy", self.q_test_images)
# Load pre-processed images
self.q_train_images = np.load(SAVE_PATH + "q_train_images.npy")
self.q_test_images = np.load(SAVE_PATH + "q_test_images.npy")
def filter_(self, image, circuit, variables):
# Initialize output image
output = np.zeros((28-2, 28-2))
# Loop over the image co-ordinates (i,j) using a 3x3 square filter
for i in range(28-2):
for j in range(28-2):
# Extract the value of each pixel in the 3x3 filter grid
image_pixels = [
image[i,j,0],
image[i,j+1,0],
image[i,j+2,0],
image[i+1,j,0],
image[i+1,j+1,0],
image[i+1,j+2,0],
image[i+2,j,0],
image[i+2,j+1,0],
image[i+2,j+2,0]
]
# Construct parameters used to embed the pixel strength into the circuit
input_variables = {}
for idx, strength in enumerate(image_pixels):
# If strength > 0, the power of the bit flip gate is 1
# Therefore this qubit starts in state |1>
if strength > 0:
input_variables['input_{}'.format(idx)] = 1
# Otherwise the gate is not applied and the initial state is |0>
else:
input_variables['input_{}'.format(idx)] = 0
# Find the statevector of the circuit and determine the state which is most likely to be measured
wavefunction = tq.simulate(circuit.circ, variables={**variables, **input_variables})
amplitudes = [(k,(abs(wavefunction(k)))) for k in range(2**n_qubits) if wavefunction(k)]
max_idx = max(amplitudes,key=itemgetter(1))[0]
# Count the number of qubits which output '1' in this state
result = len([k for k in str(bin(max_idx))[2::] if k == '1'])
output[i,j] = result
return output
def train(self):
# Train the model on the dataset
self.history = self.model.fit(
self.q_train_images,
self.ds.train_labels,
validation_data=(self.q_test_images, self.ds.test_labels),
batch_size=4,
epochs=n_epochs,
verbose=2
)
class ClassicalModel:
def __init__(self, dataset):
# Initialize dataset and parameters
self.ds = dataset
# The model is initialized
self.model = keras.models.Sequential([
keras.layers.Conv2D(n_filters, filter_size, activation='relu'),
keras.layers.MaxPooling2D(pool_size=pool_size),
keras.layers.Conv2D(n_filters, filter_size, activation='relu'),
keras.layers.MaxPooling2D(pool_size=pool_size),
keras.layers.Flatten(),
keras.layers.Dense(1024, activation="relu"),
keras.layers.Dense(10, activation="softmax")
])
# Compile model using the Adam optimiser
self.model.compile(
optimizer=keras.optimizers.Adam(learning_rate=0.00005),
loss="sparse_categorical_crossentropy",
metrics=["accuracy"]
)
def train(self):
# Train the model on the dataset
self.history = self.model.fit(
self.ds.train_images,
self.ds.train_labels,
validation_data=(self.ds.test_images, self.ds.test_labels),
batch_size=4,
epochs=n_epochs,
verbose=2
)
def quantum_model():
# Generating parameters, each maps to a random number between 0 and 2*π
# parameters is a list of dictionaries, where each dictionary represents the parameter
# mapping for one filter
parameters = []
for i in range(n_filters):
filter_params = {}
for j in range(n_layers):
for k in range(n_qubits):
filter_params[tq.Variable(name='l_{},th_{}'.format(j,k))] = np.random.uniform(high=2*np.pi)
parameters.append(filter_params)
# Initalise the dataset
ds = Dataset()
# Initialise and train the model
model = QuantumModel(ds, parameters=parameters)
model.train()
# Store the loss and accuracy of the model to return
loss = model.history.history['val_loss']
accuracy = model.history.history['val_accuracy']
return model
def classical_model():
# Initialise the dataset
ds = Dataset()
# Initialise and train the model
model = ClassicalModel(ds)
model.train()
# Store the loss and accuracy of the model to return
loss = model.history.history['val_loss']
accuracy = model.history.history['val_accuracy']
return model
model_q = quantum_model()
model_c = classical_model()
def plot(model_q, model_c):
plt.style.use("seaborn")
fig, (ax1, ax2) = plt.subplots(2, 1, figsize=(6, 9))
# Plotting the graph for accuracy
ax1.plot(model_q.history.history['val_accuracy'], color="tab:red", label="Quantum")
ax1.plot(model_c.history.history['val_accuracy'], color="tab:green", label="Classical")
ax1.set_ylabel("Accuracy")
ax1.set_ylim([0,1])
ax1.set_xlabel("Epoch")
ax1.legend()
# Plotting the graph for loss
ax2.plot(model_q.history.history['val_loss'], color="tab:red", label="Quantum")
ax2.plot(model_c.history.history['val_loss'], color="tab:green", label="Classical")
ax2.set_ylabel("Loss")
ax2.set_xlabel("Epoch")
ax2.legend()
plt.tight_layout()
plt.show()
plot(model_q, model_c)
from termcolor import colored
class Classification:
def __init__(self, model, test_images):
# Initialising parameters
self.model = model
self.test_images = test_images
self.test_labels = model.ds.test_labels
def classify(self):
# Create predictions on the test set
self.predictions = np.argmax(self.model.model.predict(self.test_images), axis=-1)
# Keep track of the indices of images which were classified correctly and incorrectly
self.correct_indices = np.nonzero(self.predictions == self.test_labels)[0]
self.incorrect_indices = np.nonzero(self.predictions != self.test_labels)[0]
def print_(self):
# Printing the total number of correctly and incorrectly classified images
print(len(self.correct_indices)," classified correctly")
print(len(self.incorrect_indices)," classified incorrectly")
print('\n')
# Printing the classification of each image
for i in range(n_test):
print("Image {}/{}".format(i+1, n_test))
if i in self.correct_indices:
# The image was correctly classified
print('model predicts: {} - true classification: {}'.format(
self.predictions[i], self.test_labels[i]))
else:
# The image was not classified correctly
print(colored('model predicts: {} - true classification: {}'.format(
self.predictions[i], self.test_labels[i]), 'red'))
print('Quantum Model')
q_class = Classification(model_q, model_q.q_test_images)
q_class.classify()
q_class.print_()
print('\n')
print('Classical Model')
c_class = Classification(model_c, model_c.ds.test_images)
c_class.classify()
c_class.print_()
def visualise(model):
# Setting n_samples to be the number of images to print
n_samples = 4
fig, axes = plt.subplots(1 + n_filters, n_samples, figsize=(10, 10))
# Iterate over each image
for i in range(n_samples):
# Plot the original image from the dataset
axes[0, 0].set_ylabel("Input")
if i != 0:
axes[0, i].yaxis.set_visible(False)
axes[0, i].imshow(model.ds.train_images[i, :, :, 0], cmap="gray")
# Plot the images generated by each filter
for c in range(n_filters):
axes[c + 1, 0].set_ylabel("Output [ch. {}]".format(c))
if i != 0:
axes[c, i].yaxis.set_visible(False)
axes[c + 1, i].imshow(model.q_train_images[i, :, :, c], cmap="gray")
plt.tight_layout()
plt.show()
visualise(model_q)
| 0.897594 | 0.996278 |
## Defining the Dataset
In this dataset we will be detecting 3 types of objects: Vehicles, People and animals. The structure of the dataset is as below.
1. A numpy array of all the RGB Images (3x300x400)
2. A numpy array of all the masks (300x400)
3. List of ground truth labels per image
4. List of ground truth bounding box per image. The four numbers are the upper left and lower right coordinates
```
import os
import cv2
import argparse
from PIL import Image
import h5py
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import DataLoader
from torchvision import datasets, transforms
import matplotlib.pyplot as plt
import scipy
%matplotlib inline
# Created the Class for the custom dataset
class CustomDataset(torch.utils.data.Dataset):
def __init__(self, root_img, root_mask, root_npy_labels, root_npy_bboxes, transforms = None):
"""
Inputs:
root_img: The path to the root directory where the image .h5 files are stored
root_mask: The path to the root directory where the mask .h5 files are stored
root_npy_labels: The path to the .npy dataset for labels
root_npy_bboxes: The path to the .npy dataset for the ground truth bounding boxes
transforms: Apply a Pytorch transform to each instance of the image
"""
self.root_img = root_img
self.root_mask = root_mask
self.root_npy_labels = root_npy_labels
self.root_npy_bboxes = root_npy_bboxes
self.transforms = transforms
self.imgs = h5py.File(self.root_img, 'r')
self.mask = h5py.File(self.root_mask, 'r')
self.labels = np.load(self.root_npy_labels, allow_pickle = True)
self.bboxes = np.load(self.root_npy_bboxes, allow_pickle = True)
# To support indexing when an object of the CustomDataset Class is created
def __getitem__(self, index):
# Convert the Masks and the input image into an array
image = np.array(self.imgs['data']).astype('int32')
masks = np.array(self.mask['data']).astype('int32')
# Convert the Mask, image, bounding boxes and labels to a Pytorch Tensor
image = torch.as_tensor(image[index])
masks = torch.as_tensor(masks[index])
bounding_boxes = torch.as_tensor(self.bboxes[index])
labels = torch.as_tensor(self.labels[index])
batch = {}
batch["bounding_boxes"] = bounding_boxes
batch["masks"] = masks
batch["labels"] = labels
if self.transforms is not None:
image, batch = self.transforms(image,batch)
return image, batch
root1 = 'C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_img_comp_zlib.h5'
root2 = 'C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_mask_comp_zlib.h5'
root3_npy = 'C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_labels_comp_zlib.npy'
root4_npy = 'C://Users//shant//Mask_RCNN_Segmentation//dataset/mycocodata_bboxes_comp_zlib.npy'
dataset = CustomDataset(root1, root2, root3_npy, root4_npy)
dataset[12]
root1 = 'C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_img_comp_zlib.h5'
root2 = 'C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_mask_comp_zlib.h5'
img = h5py.File(root1,'r')
# You can Inspect what is inside the dataset by using the command list(x.keys())
imgs = np.array(img['data']).astype('int32')
mask = h5py.File(root2,'r')
torch.as_tensor(imgs[0])
#masks = np.array(mask['data'])
#print(f'Number of images: {imgs.shape} Number of Mask: {masks.shape}')
labels = np.load('C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_labels_comp_zlib.npy', allow_pickle=True)
bounding_box = np.load('C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_bboxes_comp_zlib.npy', allow_pickle = True)
#torch.as_tensor(labels[0])
imgs
```
|
github_jupyter
|
import os
import cv2
import argparse
from PIL import Image
import h5py
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import DataLoader
from torchvision import datasets, transforms
import matplotlib.pyplot as plt
import scipy
%matplotlib inline
# Created the Class for the custom dataset
class CustomDataset(torch.utils.data.Dataset):
def __init__(self, root_img, root_mask, root_npy_labels, root_npy_bboxes, transforms = None):
"""
Inputs:
root_img: The path to the root directory where the image .h5 files are stored
root_mask: The path to the root directory where the mask .h5 files are stored
root_npy_labels: The path to the .npy dataset for labels
root_npy_bboxes: The path to the .npy dataset for the ground truth bounding boxes
transforms: Apply a Pytorch transform to each instance of the image
"""
self.root_img = root_img
self.root_mask = root_mask
self.root_npy_labels = root_npy_labels
self.root_npy_bboxes = root_npy_bboxes
self.transforms = transforms
self.imgs = h5py.File(self.root_img, 'r')
self.mask = h5py.File(self.root_mask, 'r')
self.labels = np.load(self.root_npy_labels, allow_pickle = True)
self.bboxes = np.load(self.root_npy_bboxes, allow_pickle = True)
# To support indexing when an object of the CustomDataset Class is created
def __getitem__(self, index):
# Convert the Masks and the input image into an array
image = np.array(self.imgs['data']).astype('int32')
masks = np.array(self.mask['data']).astype('int32')
# Convert the Mask, image, bounding boxes and labels to a Pytorch Tensor
image = torch.as_tensor(image[index])
masks = torch.as_tensor(masks[index])
bounding_boxes = torch.as_tensor(self.bboxes[index])
labels = torch.as_tensor(self.labels[index])
batch = {}
batch["bounding_boxes"] = bounding_boxes
batch["masks"] = masks
batch["labels"] = labels
if self.transforms is not None:
image, batch = self.transforms(image,batch)
return image, batch
root1 = 'C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_img_comp_zlib.h5'
root2 = 'C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_mask_comp_zlib.h5'
root3_npy = 'C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_labels_comp_zlib.npy'
root4_npy = 'C://Users//shant//Mask_RCNN_Segmentation//dataset/mycocodata_bboxes_comp_zlib.npy'
dataset = CustomDataset(root1, root2, root3_npy, root4_npy)
dataset[12]
root1 = 'C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_img_comp_zlib.h5'
root2 = 'C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_mask_comp_zlib.h5'
img = h5py.File(root1,'r')
# You can Inspect what is inside the dataset by using the command list(x.keys())
imgs = np.array(img['data']).astype('int32')
mask = h5py.File(root2,'r')
torch.as_tensor(imgs[0])
#masks = np.array(mask['data'])
#print(f'Number of images: {imgs.shape} Number of Mask: {masks.shape}')
labels = np.load('C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_labels_comp_zlib.npy', allow_pickle=True)
bounding_box = np.load('C:\\Users\\shant\\Mask_RCNN_Segmentation\\dataset\\mycocodata_bboxes_comp_zlib.npy', allow_pickle = True)
#torch.as_tensor(labels[0])
imgs
| 0.6508 | 0.901531 |
# Incorporating masks into calibrated science images
There are three ways of determining which pixels in a CCD image may need to be
masked (this is in addition to whatever mask or bit fields the observatory at
which you are taking images may provide).
Two of them are the same for all of the science images:
+ Hot pixels unlikely to be properly calibrated by subtracting dark current,
discussed in [Identifying hot pixels](08-01-Identifying-hot-pixels.ipynb).
+ Bad pixels identified by `ccdproc.ccdmask` from flat field images, discussed
in [Creating a mask with `ccdmask`](08-02-Creating-a-mask.ipynb).
The third, identifying cosmic rays, discussed in
[Cosmic ray removal](08-03-Cosmic-ray-removal.ipynb), will by its nature be different for each
science image.
The first two masks could be added to science images at the time the science
images are calibrated, if desired. They are added to the science images here, as
a separate step, because in many situations it is fine to omit masking entirely
and there is no particular advantage to introducing it earlier.
We begin, as usual, with a couple of imports.
```
from pathlib import Path
from astropy import units as u
from astropy.nddata import CCDData
import ccdproc as ccdp
```
## Read masks that are the same for all of the science images
In previous notebooks we constructed a mask based on the dark current and a mask
created by `ccdmask` from a flat image. Displaying the summary of the the
information about the reduced images is a handy way to determine which files are
the masks.
```
ex2_path = Path('example2-reduced')
ifc = ccdp.ImageFileCollection(ex2_path)
ifc.summary['file', 'imagetyp']
```
We read each of those in below, converting the mask to boolean after we read it.
```
mask_ccdmask = CCDData.read(ex2_path / 'mask_from_ccdmask.fits', unit=u.dimensionless_unscaled)
mask_ccdmask.data = mask_ccdmask.data.astype('bool')
mask_hot_pix = CCDData.read(ex2_path / 'mask_from_dark_current.fits', unit=u.dimensionless_unscaled)
mask_hot_pix.data = mask_hot_pix.data.astype('bool')
```
### Combining the masks
We combine the masks using a logical "OR" since we want to mask out pixels that are
bad for any reason.
```
combined_mask = mask_ccdmask.data | mask_hot_pix.data
```
It turns out we are masking roughly 0.056% of the pixels so far.
```
combined_mask.sum()
```
## Detect cosmic rays
Cosmic ray detection was discussed in detail in an
[earlier section](08-03-Cosmic-ray-removal.ipynb). Here we loop over all of the calibrated
science images and:
+ detect cosmic rays in them,
+ combine the cosmic ray mask with the mask that applies to all images,
+ set the mask of the image to the overall mask, and
+ save the image, overwriting the calibrated science image without the mask.
Since the cosmic ray detection takes a while, a status message is displayed
before each image is processed.
```
ifc.files_filtered()
for ccd, file_name in ifc.ccds(imagetyp='light', return_fname=True):
print('Working on file {}'.format(file_name))
new_ccd = ccdp.cosmicray_lacosmic(ccd, readnoise=10, sigclip=8, verbose=True)
overall_mask = new_ccd.mask | combined_mask
# If there was already a mask, keep it.
if ccd.mask is not None:
ccd.mask = ccd.mask | overall_mask
else:
ccd.mask = overall_mask
# Files can be overwritten only with an explicit option
ccd.write(ifc.location / file_name, overwrite=True)
```
|
github_jupyter
|
from pathlib import Path
from astropy import units as u
from astropy.nddata import CCDData
import ccdproc as ccdp
ex2_path = Path('example2-reduced')
ifc = ccdp.ImageFileCollection(ex2_path)
ifc.summary['file', 'imagetyp']
mask_ccdmask = CCDData.read(ex2_path / 'mask_from_ccdmask.fits', unit=u.dimensionless_unscaled)
mask_ccdmask.data = mask_ccdmask.data.astype('bool')
mask_hot_pix = CCDData.read(ex2_path / 'mask_from_dark_current.fits', unit=u.dimensionless_unscaled)
mask_hot_pix.data = mask_hot_pix.data.astype('bool')
combined_mask = mask_ccdmask.data | mask_hot_pix.data
combined_mask.sum()
ifc.files_filtered()
for ccd, file_name in ifc.ccds(imagetyp='light', return_fname=True):
print('Working on file {}'.format(file_name))
new_ccd = ccdp.cosmicray_lacosmic(ccd, readnoise=10, sigclip=8, verbose=True)
overall_mask = new_ccd.mask | combined_mask
# If there was already a mask, keep it.
if ccd.mask is not None:
ccd.mask = ccd.mask | overall_mask
else:
ccd.mask = overall_mask
# Files can be overwritten only with an explicit option
ccd.write(ifc.location / file_name, overwrite=True)
| 0.505127 | 0.986688 |
# Transporter statistics and taxonomic profiles
## Overview
In this notebook some overview statistics of the datasets are computed and taxonomic profiles investigated. The notebook uses data produced by running the [01.process_data](01.process_data.ipynb) notebook.
```
import numpy as np
import pandas as pd
import seaborn as sns
import glob
import os
import matplotlib.pyplot as plt, matplotlib
%matplotlib inline
%config InlineBackend.figure_format = 'svg'
plt.style.use('ggplot')
def make_tax_table(df,name="",rank="superkingdom"):
df_t = df.groupby(rank).sum()
df_tp = df_t.div(df_t.sum())*100
df_tp_mean = df_tp.mean(axis=1)
df_tp_max = df_tp.max(axis=1)
df_tp_min = df_tp.min(axis=1)
df_tp_sd = df_tp.std(axis=1)
table = pd.concat([df_tp_mean,df_tp_max,df_tp_min,df_tp_sd],axis=1)
table.columns = [name+" mean(%)",name+" max(%)",name+" min(%)",name+" std"]
table.rename(index=lambda x: x.split("_")[0], inplace=True)
return table
```
## Load the data
```
transinfo = pd.read_csv("selected_transporters_classified.tab", header=0, sep="\t", index_col=0)
transinfo.head()
```
Read gene abundance values with taxonomic annotations.
```
mg_cov = pd.read_table("data/mg/all_genes.tpm.taxonomy.tsv.gz", header=0, sep="\t", index_col=0)
mt_cov = pd.read_table("data/mt/all_genes.tpm.taxonomy.tsv.gz", header=0, sep="\t", index_col=0)
```
Read orf level transporter data.
```
mg_transcov = pd.read_table("results/mg/all_transporters.tpm.taxonomy.tsv.gz", header=0, sep="\t", index_col=0)
mt_transcov = pd.read_table("results/mt/all_transporters.tpm.taxonomy.tsv.gz", header=0, sep="\t", index_col=0)
mg_select_transcov = pd.read_table("results/mg/select_trans_genes.tpm.tsv", header=0, sep="\t", index_col=0)
mt_select_transcov = pd.read_table("results/mt/select_trans_genes.tpm.tsv", header=0, sep="\t", index_col=0)
```
Read transporter abundances.
```
mg_trans = pd.read_csv("results/mg/all_trans.tpm.tsv", header=0, sep="\t", index_col=0)
mt_trans = pd.read_csv("results/mt/all_trans.tpm.tsv", header=0, sep="\t", index_col=0)
```
## Generate taxonomic overview table
```
mg_tax_table = make_tax_table(mg_cov,name="MG ")
mg_tax_table_cyano = make_tax_table(mg_cov,name="MG ",rank="phylum").loc["Cyanobacteria"]
mg_tax_table = pd.concat([mg_tax_table,pd.DataFrame(mg_tax_table_cyano).T])
mg_tax_table
mt_tax_table = make_tax_table(mt_cov,name="MT ")
mt_tax_table_cyano = make_tax_table(mt_cov,name="MT ",rank="phylum").loc["Cyanobacteria"]
mt_tax_table = pd.concat([mt_tax_table,pd.DataFrame(mt_tax_table_cyano).T])
mt_tax_table
```
Concatenate overview tables.
```
tax_table = pd.concat([mg_tax_table,mt_tax_table],axis=1).round(2)
```
## Generate general overview of transporters
Make table with number of ORFs, ORFs classified as transporters, min, mean and max coverage for transporter ORFs.
```
num_genes = len(mg_cov)
gene_lengths = pd.read_table("data/mg/all_genes.tpm.tsv.gz", usecols=[1])
gene_lengths = np.round(gene_lengths.mean())
def generate_transporter_stats(df):
# Number of transporter genes (genes with sum > 0)
num_trans_genes = len(df.loc[df.groupby(level=0).sum().sum(axis=1)>0])
# Percent of transporter genes
num_trans_genes_p = np.round((num_trans_genes / float(num_genes))*100,2)
# Mean total coverage for transporter genes across the samples
transcov_mean = np.round(((df.groupby(level=0).sum().sum().mean()) / 1e6)*100,2)
# Minimum total coverage for transporter genes across the samples
transcov_min = np.round(((df.groupby(level=0).sum().sum().min()) / 1e6)*100,2)
# Maximum ...
transcov_max = np.round(((df.groupby(level=0).sum().sum().max()) / 1e6)*100,2)
# Standard dev
transcov_std = np.round(((df.groupby(level=0).sum().sum() / 1e6)*100).std(),2)
return num_trans_genes, num_trans_genes_p, transcov_mean, transcov_min, transcov_max, transcov_std
mg_num_trans_genes, mg_num_trans_genes_p, mg_transcov_mean, mg_transcov_min, mg_transcov_max, mg_transcov_std = generate_transporter_stats(mg_transcov)
mt_num_trans_genes, mt_num_trans_genes_p, mt_transcov_mean, mt_transcov_min, mt_transcov_max, mt_transcov_std = generate_transporter_stats(mt_transcov)
```
Create table with transporter statistics for MG and MT datasets
```
stats_df = pd.DataFrame(data={
"Transporter genes": ["{} ({}%)".format(mg_num_trans_genes,mg_num_trans_genes_p),"{} ({}%)".format(mt_num_trans_genes,mt_num_trans_genes_p)],
"Transporter mean": ["{}%".format(mg_transcov_mean),"{}%".format(mt_transcov_mean)],
"Transporter min": ["{}%".format(mg_transcov_min),"{}%".format(mt_transcov_min)],
"Transporter max": ["{}%".format(mg_transcov_max),"{}%".format(mt_transcov_max)],
"Transporter std": ["{}%".format(mg_transcov_std),"{}%".format(mt_transcov_std)]},index=["MG","MT"]).T
stats_df
```
Do the same with the selected transporters.
```
mg_select_num_trans_genes, mg_select_num_trans_genes_p, mg_select_transcov_mean, mg_select_transcov_min, mg_select_transcov_max, mg_select_transcov_std = generate_transporter_stats(mg_select_transcov)
mt_select_num_trans_genes, mt_select_num_trans_genes_p, mt_select_transcov_mean, mt_select_transcov_min, mt_select_transcov_max, mt_select_transcov_std = generate_transporter_stats(mt_select_transcov)
select_stats_df = pd.DataFrame(data={
"Selected transporter genes": ["{} ({}%)".format(mg_select_num_trans_genes,mg_select_num_trans_genes_p),"{} ({}%)".format(mt_select_num_trans_genes,mt_select_num_trans_genes_p)],
"Selected transporter mean": ["{}%".format(mg_select_transcov_mean),"{}%".format(mt_select_transcov_mean)],
"Selected transporter min": ["{}%".format(mg_select_transcov_min),"{}%".format(mt_select_transcov_min)],
"Selected transporter max": ["{}%".format(mg_select_transcov_max),"{}%".format(mt_select_transcov_max)],
"Selected transporter std": ["{}%".format(mg_select_transcov_std),"{}%".format(mt_select_transcov_std)]},index=["mg_select","mt_select"]).T
select_stats_df.to_csv("results/selected_transporter_stats.tab",sep="\t")
select_stats_df
```
## Generate kingdom/phylum level taxonomic plots
```
def get_euk_taxa(taxa, df, rank):
euk_taxa = []
for t in taxa:
k = df.loc[df[rank]==t, "superkingdom"].unique()[0]
if k=="Eukaryota":
euk_taxa.append(t)
return euk_taxa
def set_euk_hatches(ax):
for patch in ax.patches:
t = color2taxmap[patch.properties()['facecolor'][0:-1]]
if t in euk_taxa:
patch.set_hatch("////")
```
Generate profiles for metagenomes.
```
# Get sum of abundances at superkingdom level
mg_k = mg_cov.groupby("superkingdom").sum()
# Normalize to %
mg_kn = mg_k.div(mg_k.sum())*100
mg_kn = mg_kn.loc[["Archaea","Bacteria","Eukaryota","Viruses","Unclassified.sequences","other sequences"]]
mg_kn = mg_kn.loc[mg_kn.sum(axis=1).sort_values(ascending=False).index]
# Swtich Proteobacterial classes to phylum
mg_cov.loc[mg_cov.phylum=="Proteobacteria","phylum"] = mg_cov.loc[mg_cov.phylum=="Proteobacteria","class"]
# Normalize at phylum level
mg_p = mg_cov.groupby("phylum").sum()
mg_pn = mg_p.div(mg_p.sum())*100
_ = mg_pn.mean(axis=1).sort_values(ascending=False)
_.loc[~_.index.str.contains("Unclassified")].head(8)
```
Create the taxonomic overview of the 7 most abundant phyla in the metagenomic dataset. This is **Figure 2** in the paper.
```
select_taxa = ["Verrucomicrobia","Actinobacteria","Alphaproteobacteria","Gammaproteobacteria","Cyanobacteria","Bacteroidetes","Betaproteobacteria"]
from datetime import datetime
newdates = [datetime.strptime(date, "%y%m%d").strftime("%d %B") for date in list(mg_pn.columns)]
# Sort taxa by mean abundance
taxa_order = mg_pn.loc[select_taxa].mean(axis=1).sort_values(ascending=False).index
ax = mg_pn.loc[taxa_order].T.plot(kind="area",stacked=True)
ax.legend(bbox_to_anchor=(1,1))
ax.set_ylabel("% normalized abundance");
xticks = list(range(0,33))
ax.set_xticks(xticks);
ax.set_xticklabels(newdates, rotation=90);
plt.savefig("figures/Figure_2.eps", bbox_inches="tight")
```
Generate profiles for metatranscriptomes.
```
# Get sum of abundances at superkingdom level
mt_k = mt_cov.groupby("superkingdom").sum()
# Normalize to %
mt_kn = mt_k.div(mt_k.sum())*100
mt_kn = mt_kn.loc[["Archaea","Bacteria","Eukaryota","Viruses","Unclassified.sequences","other sequences"]]
mt_kn = mt_kn.loc[mt_kn.sum(axis=1).sort_values(ascending=False).index]
# Swtich Proteobacterial classes to phylum
mt_cov.loc[mt_cov.phylum=="Proteobacteria","phylum"] = mt_cov.loc[mt_cov.phylum=="Proteobacteria","class"]
# Normalize at phylum level
mt_p = mt_cov.groupby("phylum").sum()
mt_pn = mt_p.div(mt_p.sum())*100
```
Get common taxa for both datasets by taking the union of the top 15 most abundant taxa
```
mg_taxa = mg_pn.mean(axis=1).sort_values(ascending=False).head(15).index
mt_taxa = mt_pn.mean(axis=1).sort_values(ascending=False).head(15).index
taxa = set(mg_taxa).union(set(mt_taxa))
```
Single out eukaryotic taxa
```
euk_taxa = get_euk_taxa(taxa, mg_cov, rank="phylum")
```
Sort the taxa by their mean abundance in the mg data
```
taxa_sort = mg_pn.loc[taxa].mean(axis=1).sort_values(ascending=False).index
taxa_colors = dict(zip(taxa_sort,(sns.color_palette("Set1",7)+sns.color_palette("Set2",7)+sns.color_palette("Dark2",5))))
color2taxmap = {}
for t, c in taxa_colors.items():
color2taxmap[c] = t
```
Calculate total number of orders.
```
mg_ordersum = mg_cov.groupby("order").sum()
mg_total_orders = len(mg_ordersum.loc[mg_ordersum.sum(axis=1)>0])
print("{} orders in the entire mg dataset".format(mg_total_orders))
mg_trans_ordersum = mg_select_transcov.groupby("order").sum()
mg_trans_total_orders = len(mg_trans_ordersum.loc[mg_trans_ordersum.sum(axis=1)>0])
print("{} orders in the transporter mg dataset".format(mg_trans_total_orders))
mt_ordersum = mt_cov.groupby("order").sum()
mt_total_orders = len(mt_ordersum.loc[mt_ordersum.sum(axis=1)>0])
print("{} orders in the entire mt dataset".format(mt_total_orders))
mt_trans_ordersum = mt_select_transcov.groupby("order").sum()
mt_trans_total_orders = len(mt_trans_ordersum.loc[mt_trans_ordersum.sum(axis=1)>0])
print("{} orders in the transporter mt dataset".format(mt_trans_total_orders))
```
## Calculate and plot distributions per taxonomic subsets.
Extract ORFs belonging to each subset.
```
cya_orfs = mg_transcov.loc[mg_transcov.phylum=="Cyanobacteria"].index
bac_orfs = mg_transcov.loc[(mg_transcov.phylum!="Cyanobacteria")&(mg_transcov.superkingdom=="Bacteria")].index
euk_orfs = mg_transcov.loc[mg_transcov.superkingdom=="Eukaryota"].index
```
Calculate contribution of taxonomic subsets to the identified transporters.
```
taxgroup_df = pd.DataFrame(columns=["MG","MT"],index=["Bacteria","Cyanobacteria","Eukaryota"])
mg_all_transcov_info = pd.merge(transinfo,mg_transcov,left_index=True,right_on="transporter")
mg_bac_transcov_info = pd.merge(transinfo,mg_transcov.loc[bac_orfs],left_index=True,right_on="transporter")
mg_euk_transcov_info = pd.merge(transinfo,mg_transcov.loc[euk_orfs],left_index=True,right_on="transporter")
mg_cya_transcov_info = pd.merge(transinfo,mg_transcov.loc[cya_orfs],left_index=True,right_on="transporter")
mt_all_transcov_info = pd.merge(transinfo,mt_transcov,left_index=True,right_on="transporter")
mt_bac_transcov_info = pd.merge(transinfo,mt_transcov.loc[bac_orfs],left_index=True,right_on="transporter")
mt_euk_transcov_info = pd.merge(transinfo,mt_transcov.loc[euk_orfs],left_index=True,right_on="transporter")
mt_cya_transcov_info = pd.merge(transinfo,mt_transcov.loc[cya_orfs],left_index=True,right_on="transporter")
mg_cya_part = mg_cya_transcov_info.groupby("transporter").sum().sum().div(mg_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mg_cya_part.min(),mg_cya_part.max(),mg_cya_part.mean()
taxgroup_df.loc["Cyanobacteria","MG"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
mg_euk_part = mg_euk_transcov_info.groupby("transporter").sum().sum().div(mg_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mg_euk_part.min(),mg_euk_part.max(),mg_euk_part.mean()
taxgroup_df.loc["Eukaryota","MG"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
mg_bac_part = mg_bac_transcov_info.groupby("transporter").sum().sum().div(mg_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mg_bac_part.min(),mg_bac_part.max(),mg_bac_part.mean()
taxgroup_df.loc["Bacteria","MG"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
mt_cya_part = mt_cya_transcov_info.groupby("transporter").sum().sum().div(mt_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mt_cya_part.min(),mt_cya_part.max(),mt_cya_part.mean()
taxgroup_df.loc["Cyanobacteria","MT"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
mt_euk_part = mt_euk_transcov_info.groupby("transporter").sum().sum().div(mt_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mt_euk_part.min(),mt_euk_part.max(),mt_euk_part.mean()
taxgroup_df.loc["Eukaryota","MT"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
mt_bac_part = mt_bac_transcov_info.groupby("transporter").sum().sum().div(mt_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mt_bac_part.min(),mt_bac_part.max(),mt_bac_part.mean()
taxgroup_df.loc["Bacteria","MT"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
taxgroup_df
```
### Taxonomic subsets per substrate category
```
def calculate_mean_total_substrate_subset(df,df_sum,subset,var_name="Sample",value_name="%"):
cols = ["fam","transporter","substrate_category","name"]
# Sum to protein family
x = df.groupby(["fam","transporter","substrate_category","name"]).sum().reset_index()
cols.pop(cols.index("fam"))
# Calculate mean of transporters
x.groupby(cols).mean().reset_index()
xt = x.copy()
# Normalize to sum of all transporters
x.iloc[:,4:] = x.iloc[:,4:].div(df_sum)*100
# Sum percent to substrate category
x = x.groupby("substrate_category").sum()
# Melt dataframe and add subset column
x["substrate_category"] = x.index
xm = pd.melt(x,id_vars="substrate_category", var_name="Sample",value_name="%")
xm = xm.assign(Subset=pd.Series(data=subset,index=xm.index))
return xm,xt
# Get contribution of bacterial transporters to total for substrate category
mg_bac_cat_melt,mg_bac_cat = calculate_mean_total_substrate_subset(mg_bac_transcov_info,mg_trans.sum(),"Bacteria")
# Get contribution of eukaryotic transporters to total for substrate category
mg_euk_cat_melt,mg_euk_cat = calculate_mean_total_substrate_subset(mg_euk_transcov_info,mg_trans.sum(),"Eukaryota")
# Get contribution of cyanobacterial transporters to total for substrate category
mg_cya_cat_melt,mg_cya_cat = calculate_mean_total_substrate_subset(mg_cya_transcov_info,mg_trans.sum(),"Cyanobacteria")
# Get contribution of bacterial transporters to total for substrate category
mt_bac_cat_melt,mt_bac_cat = calculate_mean_total_substrate_subset(mt_bac_transcov_info,mt_trans.sum(),"Bacteria")
# Get contribution of eukaryotic transporters to total for substrate category
mt_euk_cat_melt,mt_euk_cat = calculate_mean_total_substrate_subset(mt_euk_transcov_info,mt_trans.sum(),"Eukaryota")
# Get contribution of cyanobacterial transporters to total for substrate category
mt_cya_cat_melt,mt_cya_cat = calculate_mean_total_substrate_subset(mt_cya_transcov_info,mt_trans.sum(),"Cyanobacteria")
# Concatenate dataframes for metagenomes
mg_subsets_cat = pd.concat([pd.concat([mg_bac_cat_melt,mg_euk_cat_melt]),mg_cya_cat_melt])
mg_subsets_cat = mg_subsets_cat.assign(dataset=pd.Series(data="MG",index=mg_subsets_cat.index))
# Concatenate dataframes for metagenomes
mt_subsets_cat = pd.concat([pd.concat([mt_bac_cat_melt,mt_euk_cat_melt]),mt_cya_cat_melt])
mt_subsets_cat = mt_subsets_cat.assign(dataset=pd.Series(data="MT",index=mt_subsets_cat.index))
```
**Concatenate MG and MT**
```
subsets_cat = pd.concat([mg_subsets_cat,mt_subsets_cat])
```
### Plot substrate category distributions
```
cats = transinfo.substrate_category.unique()
# Update Eukaryota subset label
subsets_cat.loc[subsets_cat.Subset=="Eukaryota","Subset"] = ["Picoeukaryota"]*len(subsets_cat.loc[subsets_cat.Subset=="Eukaryota","Subset"])
```
This is **Figure 3** in paper.
```
sns.set(font_scale=0.8)
ax = sns.catplot(kind="bar",data=subsets_cat.loc[subsets_cat.substrate_category.isin(cats)],hue="dataset",
y="substrate_category", x="%", col="Subset",
errwidth=1, height=3, palette="Set1", aspect=1)
ax.set_titles("{col_name}")
ax.set_axis_labels("% of normalized transporter abundance","Substrate category")
plt.savefig("figures/Figure_3.eps", bbox_inches="tight")
_ = mg_transcov.groupby(["fam","transporter"]).sum().reset_index()
_ = _.groupby("transporter").mean()
_ = pd.merge(transinfo, _, left_index=True, right_index=True)
_ = _.loc[_.substrate_category=="Carbohydrate"].groupby("name").sum()
(_.div(_.sum())*100).mean(axis=1).sort_values(ascending=False).head(3).sum()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import seaborn as sns
import glob
import os
import matplotlib.pyplot as plt, matplotlib
%matplotlib inline
%config InlineBackend.figure_format = 'svg'
plt.style.use('ggplot')
def make_tax_table(df,name="",rank="superkingdom"):
df_t = df.groupby(rank).sum()
df_tp = df_t.div(df_t.sum())*100
df_tp_mean = df_tp.mean(axis=1)
df_tp_max = df_tp.max(axis=1)
df_tp_min = df_tp.min(axis=1)
df_tp_sd = df_tp.std(axis=1)
table = pd.concat([df_tp_mean,df_tp_max,df_tp_min,df_tp_sd],axis=1)
table.columns = [name+" mean(%)",name+" max(%)",name+" min(%)",name+" std"]
table.rename(index=lambda x: x.split("_")[0], inplace=True)
return table
transinfo = pd.read_csv("selected_transporters_classified.tab", header=0, sep="\t", index_col=0)
transinfo.head()
mg_cov = pd.read_table("data/mg/all_genes.tpm.taxonomy.tsv.gz", header=0, sep="\t", index_col=0)
mt_cov = pd.read_table("data/mt/all_genes.tpm.taxonomy.tsv.gz", header=0, sep="\t", index_col=0)
mg_transcov = pd.read_table("results/mg/all_transporters.tpm.taxonomy.tsv.gz", header=0, sep="\t", index_col=0)
mt_transcov = pd.read_table("results/mt/all_transporters.tpm.taxonomy.tsv.gz", header=0, sep="\t", index_col=0)
mg_select_transcov = pd.read_table("results/mg/select_trans_genes.tpm.tsv", header=0, sep="\t", index_col=0)
mt_select_transcov = pd.read_table("results/mt/select_trans_genes.tpm.tsv", header=0, sep="\t", index_col=0)
mg_trans = pd.read_csv("results/mg/all_trans.tpm.tsv", header=0, sep="\t", index_col=0)
mt_trans = pd.read_csv("results/mt/all_trans.tpm.tsv", header=0, sep="\t", index_col=0)
mg_tax_table = make_tax_table(mg_cov,name="MG ")
mg_tax_table_cyano = make_tax_table(mg_cov,name="MG ",rank="phylum").loc["Cyanobacteria"]
mg_tax_table = pd.concat([mg_tax_table,pd.DataFrame(mg_tax_table_cyano).T])
mg_tax_table
mt_tax_table = make_tax_table(mt_cov,name="MT ")
mt_tax_table_cyano = make_tax_table(mt_cov,name="MT ",rank="phylum").loc["Cyanobacteria"]
mt_tax_table = pd.concat([mt_tax_table,pd.DataFrame(mt_tax_table_cyano).T])
mt_tax_table
tax_table = pd.concat([mg_tax_table,mt_tax_table],axis=1).round(2)
num_genes = len(mg_cov)
gene_lengths = pd.read_table("data/mg/all_genes.tpm.tsv.gz", usecols=[1])
gene_lengths = np.round(gene_lengths.mean())
def generate_transporter_stats(df):
# Number of transporter genes (genes with sum > 0)
num_trans_genes = len(df.loc[df.groupby(level=0).sum().sum(axis=1)>0])
# Percent of transporter genes
num_trans_genes_p = np.round((num_trans_genes / float(num_genes))*100,2)
# Mean total coverage for transporter genes across the samples
transcov_mean = np.round(((df.groupby(level=0).sum().sum().mean()) / 1e6)*100,2)
# Minimum total coverage for transporter genes across the samples
transcov_min = np.round(((df.groupby(level=0).sum().sum().min()) / 1e6)*100,2)
# Maximum ...
transcov_max = np.round(((df.groupby(level=0).sum().sum().max()) / 1e6)*100,2)
# Standard dev
transcov_std = np.round(((df.groupby(level=0).sum().sum() / 1e6)*100).std(),2)
return num_trans_genes, num_trans_genes_p, transcov_mean, transcov_min, transcov_max, transcov_std
mg_num_trans_genes, mg_num_trans_genes_p, mg_transcov_mean, mg_transcov_min, mg_transcov_max, mg_transcov_std = generate_transporter_stats(mg_transcov)
mt_num_trans_genes, mt_num_trans_genes_p, mt_transcov_mean, mt_transcov_min, mt_transcov_max, mt_transcov_std = generate_transporter_stats(mt_transcov)
stats_df = pd.DataFrame(data={
"Transporter genes": ["{} ({}%)".format(mg_num_trans_genes,mg_num_trans_genes_p),"{} ({}%)".format(mt_num_trans_genes,mt_num_trans_genes_p)],
"Transporter mean": ["{}%".format(mg_transcov_mean),"{}%".format(mt_transcov_mean)],
"Transporter min": ["{}%".format(mg_transcov_min),"{}%".format(mt_transcov_min)],
"Transporter max": ["{}%".format(mg_transcov_max),"{}%".format(mt_transcov_max)],
"Transporter std": ["{}%".format(mg_transcov_std),"{}%".format(mt_transcov_std)]},index=["MG","MT"]).T
stats_df
mg_select_num_trans_genes, mg_select_num_trans_genes_p, mg_select_transcov_mean, mg_select_transcov_min, mg_select_transcov_max, mg_select_transcov_std = generate_transporter_stats(mg_select_transcov)
mt_select_num_trans_genes, mt_select_num_trans_genes_p, mt_select_transcov_mean, mt_select_transcov_min, mt_select_transcov_max, mt_select_transcov_std = generate_transporter_stats(mt_select_transcov)
select_stats_df = pd.DataFrame(data={
"Selected transporter genes": ["{} ({}%)".format(mg_select_num_trans_genes,mg_select_num_trans_genes_p),"{} ({}%)".format(mt_select_num_trans_genes,mt_select_num_trans_genes_p)],
"Selected transporter mean": ["{}%".format(mg_select_transcov_mean),"{}%".format(mt_select_transcov_mean)],
"Selected transporter min": ["{}%".format(mg_select_transcov_min),"{}%".format(mt_select_transcov_min)],
"Selected transporter max": ["{}%".format(mg_select_transcov_max),"{}%".format(mt_select_transcov_max)],
"Selected transporter std": ["{}%".format(mg_select_transcov_std),"{}%".format(mt_select_transcov_std)]},index=["mg_select","mt_select"]).T
select_stats_df.to_csv("results/selected_transporter_stats.tab",sep="\t")
select_stats_df
def get_euk_taxa(taxa, df, rank):
euk_taxa = []
for t in taxa:
k = df.loc[df[rank]==t, "superkingdom"].unique()[0]
if k=="Eukaryota":
euk_taxa.append(t)
return euk_taxa
def set_euk_hatches(ax):
for patch in ax.patches:
t = color2taxmap[patch.properties()['facecolor'][0:-1]]
if t in euk_taxa:
patch.set_hatch("////")
# Get sum of abundances at superkingdom level
mg_k = mg_cov.groupby("superkingdom").sum()
# Normalize to %
mg_kn = mg_k.div(mg_k.sum())*100
mg_kn = mg_kn.loc[["Archaea","Bacteria","Eukaryota","Viruses","Unclassified.sequences","other sequences"]]
mg_kn = mg_kn.loc[mg_kn.sum(axis=1).sort_values(ascending=False).index]
# Swtich Proteobacterial classes to phylum
mg_cov.loc[mg_cov.phylum=="Proteobacteria","phylum"] = mg_cov.loc[mg_cov.phylum=="Proteobacteria","class"]
# Normalize at phylum level
mg_p = mg_cov.groupby("phylum").sum()
mg_pn = mg_p.div(mg_p.sum())*100
_ = mg_pn.mean(axis=1).sort_values(ascending=False)
_.loc[~_.index.str.contains("Unclassified")].head(8)
select_taxa = ["Verrucomicrobia","Actinobacteria","Alphaproteobacteria","Gammaproteobacteria","Cyanobacteria","Bacteroidetes","Betaproteobacteria"]
from datetime import datetime
newdates = [datetime.strptime(date, "%y%m%d").strftime("%d %B") for date in list(mg_pn.columns)]
# Sort taxa by mean abundance
taxa_order = mg_pn.loc[select_taxa].mean(axis=1).sort_values(ascending=False).index
ax = mg_pn.loc[taxa_order].T.plot(kind="area",stacked=True)
ax.legend(bbox_to_anchor=(1,1))
ax.set_ylabel("% normalized abundance");
xticks = list(range(0,33))
ax.set_xticks(xticks);
ax.set_xticklabels(newdates, rotation=90);
plt.savefig("figures/Figure_2.eps", bbox_inches="tight")
# Get sum of abundances at superkingdom level
mt_k = mt_cov.groupby("superkingdom").sum()
# Normalize to %
mt_kn = mt_k.div(mt_k.sum())*100
mt_kn = mt_kn.loc[["Archaea","Bacteria","Eukaryota","Viruses","Unclassified.sequences","other sequences"]]
mt_kn = mt_kn.loc[mt_kn.sum(axis=1).sort_values(ascending=False).index]
# Swtich Proteobacterial classes to phylum
mt_cov.loc[mt_cov.phylum=="Proteobacteria","phylum"] = mt_cov.loc[mt_cov.phylum=="Proteobacteria","class"]
# Normalize at phylum level
mt_p = mt_cov.groupby("phylum").sum()
mt_pn = mt_p.div(mt_p.sum())*100
mg_taxa = mg_pn.mean(axis=1).sort_values(ascending=False).head(15).index
mt_taxa = mt_pn.mean(axis=1).sort_values(ascending=False).head(15).index
taxa = set(mg_taxa).union(set(mt_taxa))
euk_taxa = get_euk_taxa(taxa, mg_cov, rank="phylum")
taxa_sort = mg_pn.loc[taxa].mean(axis=1).sort_values(ascending=False).index
taxa_colors = dict(zip(taxa_sort,(sns.color_palette("Set1",7)+sns.color_palette("Set2",7)+sns.color_palette("Dark2",5))))
color2taxmap = {}
for t, c in taxa_colors.items():
color2taxmap[c] = t
mg_ordersum = mg_cov.groupby("order").sum()
mg_total_orders = len(mg_ordersum.loc[mg_ordersum.sum(axis=1)>0])
print("{} orders in the entire mg dataset".format(mg_total_orders))
mg_trans_ordersum = mg_select_transcov.groupby("order").sum()
mg_trans_total_orders = len(mg_trans_ordersum.loc[mg_trans_ordersum.sum(axis=1)>0])
print("{} orders in the transporter mg dataset".format(mg_trans_total_orders))
mt_ordersum = mt_cov.groupby("order").sum()
mt_total_orders = len(mt_ordersum.loc[mt_ordersum.sum(axis=1)>0])
print("{} orders in the entire mt dataset".format(mt_total_orders))
mt_trans_ordersum = mt_select_transcov.groupby("order").sum()
mt_trans_total_orders = len(mt_trans_ordersum.loc[mt_trans_ordersum.sum(axis=1)>0])
print("{} orders in the transporter mt dataset".format(mt_trans_total_orders))
cya_orfs = mg_transcov.loc[mg_transcov.phylum=="Cyanobacteria"].index
bac_orfs = mg_transcov.loc[(mg_transcov.phylum!="Cyanobacteria")&(mg_transcov.superkingdom=="Bacteria")].index
euk_orfs = mg_transcov.loc[mg_transcov.superkingdom=="Eukaryota"].index
taxgroup_df = pd.DataFrame(columns=["MG","MT"],index=["Bacteria","Cyanobacteria","Eukaryota"])
mg_all_transcov_info = pd.merge(transinfo,mg_transcov,left_index=True,right_on="transporter")
mg_bac_transcov_info = pd.merge(transinfo,mg_transcov.loc[bac_orfs],left_index=True,right_on="transporter")
mg_euk_transcov_info = pd.merge(transinfo,mg_transcov.loc[euk_orfs],left_index=True,right_on="transporter")
mg_cya_transcov_info = pd.merge(transinfo,mg_transcov.loc[cya_orfs],left_index=True,right_on="transporter")
mt_all_transcov_info = pd.merge(transinfo,mt_transcov,left_index=True,right_on="transporter")
mt_bac_transcov_info = pd.merge(transinfo,mt_transcov.loc[bac_orfs],left_index=True,right_on="transporter")
mt_euk_transcov_info = pd.merge(transinfo,mt_transcov.loc[euk_orfs],left_index=True,right_on="transporter")
mt_cya_transcov_info = pd.merge(transinfo,mt_transcov.loc[cya_orfs],left_index=True,right_on="transporter")
mg_cya_part = mg_cya_transcov_info.groupby("transporter").sum().sum().div(mg_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mg_cya_part.min(),mg_cya_part.max(),mg_cya_part.mean()
taxgroup_df.loc["Cyanobacteria","MG"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
mg_euk_part = mg_euk_transcov_info.groupby("transporter").sum().sum().div(mg_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mg_euk_part.min(),mg_euk_part.max(),mg_euk_part.mean()
taxgroup_df.loc["Eukaryota","MG"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
mg_bac_part = mg_bac_transcov_info.groupby("transporter").sum().sum().div(mg_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mg_bac_part.min(),mg_bac_part.max(),mg_bac_part.mean()
taxgroup_df.loc["Bacteria","MG"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
mt_cya_part = mt_cya_transcov_info.groupby("transporter").sum().sum().div(mt_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mt_cya_part.min(),mt_cya_part.max(),mt_cya_part.mean()
taxgroup_df.loc["Cyanobacteria","MT"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
mt_euk_part = mt_euk_transcov_info.groupby("transporter").sum().sum().div(mt_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mt_euk_part.min(),mt_euk_part.max(),mt_euk_part.mean()
taxgroup_df.loc["Eukaryota","MT"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
mt_bac_part = mt_bac_transcov_info.groupby("transporter").sum().sum().div(mt_all_transcov_info.groupby("transporter").sum().sum())*100
mi,ma,me = mt_bac_part.min(),mt_bac_part.max(),mt_bac_part.mean()
taxgroup_df.loc["Bacteria","MT"] = "{}% ({}-{}%)".format(round(me,2),round(mi,2),round(ma,2))
taxgroup_df
def calculate_mean_total_substrate_subset(df,df_sum,subset,var_name="Sample",value_name="%"):
cols = ["fam","transporter","substrate_category","name"]
# Sum to protein family
x = df.groupby(["fam","transporter","substrate_category","name"]).sum().reset_index()
cols.pop(cols.index("fam"))
# Calculate mean of transporters
x.groupby(cols).mean().reset_index()
xt = x.copy()
# Normalize to sum of all transporters
x.iloc[:,4:] = x.iloc[:,4:].div(df_sum)*100
# Sum percent to substrate category
x = x.groupby("substrate_category").sum()
# Melt dataframe and add subset column
x["substrate_category"] = x.index
xm = pd.melt(x,id_vars="substrate_category", var_name="Sample",value_name="%")
xm = xm.assign(Subset=pd.Series(data=subset,index=xm.index))
return xm,xt
# Get contribution of bacterial transporters to total for substrate category
mg_bac_cat_melt,mg_bac_cat = calculate_mean_total_substrate_subset(mg_bac_transcov_info,mg_trans.sum(),"Bacteria")
# Get contribution of eukaryotic transporters to total for substrate category
mg_euk_cat_melt,mg_euk_cat = calculate_mean_total_substrate_subset(mg_euk_transcov_info,mg_trans.sum(),"Eukaryota")
# Get contribution of cyanobacterial transporters to total for substrate category
mg_cya_cat_melt,mg_cya_cat = calculate_mean_total_substrate_subset(mg_cya_transcov_info,mg_trans.sum(),"Cyanobacteria")
# Get contribution of bacterial transporters to total for substrate category
mt_bac_cat_melt,mt_bac_cat = calculate_mean_total_substrate_subset(mt_bac_transcov_info,mt_trans.sum(),"Bacteria")
# Get contribution of eukaryotic transporters to total for substrate category
mt_euk_cat_melt,mt_euk_cat = calculate_mean_total_substrate_subset(mt_euk_transcov_info,mt_trans.sum(),"Eukaryota")
# Get contribution of cyanobacterial transporters to total for substrate category
mt_cya_cat_melt,mt_cya_cat = calculate_mean_total_substrate_subset(mt_cya_transcov_info,mt_trans.sum(),"Cyanobacteria")
# Concatenate dataframes for metagenomes
mg_subsets_cat = pd.concat([pd.concat([mg_bac_cat_melt,mg_euk_cat_melt]),mg_cya_cat_melt])
mg_subsets_cat = mg_subsets_cat.assign(dataset=pd.Series(data="MG",index=mg_subsets_cat.index))
# Concatenate dataframes for metagenomes
mt_subsets_cat = pd.concat([pd.concat([mt_bac_cat_melt,mt_euk_cat_melt]),mt_cya_cat_melt])
mt_subsets_cat = mt_subsets_cat.assign(dataset=pd.Series(data="MT",index=mt_subsets_cat.index))
subsets_cat = pd.concat([mg_subsets_cat,mt_subsets_cat])
cats = transinfo.substrate_category.unique()
# Update Eukaryota subset label
subsets_cat.loc[subsets_cat.Subset=="Eukaryota","Subset"] = ["Picoeukaryota"]*len(subsets_cat.loc[subsets_cat.Subset=="Eukaryota","Subset"])
sns.set(font_scale=0.8)
ax = sns.catplot(kind="bar",data=subsets_cat.loc[subsets_cat.substrate_category.isin(cats)],hue="dataset",
y="substrate_category", x="%", col="Subset",
errwidth=1, height=3, palette="Set1", aspect=1)
ax.set_titles("{col_name}")
ax.set_axis_labels("% of normalized transporter abundance","Substrate category")
plt.savefig("figures/Figure_3.eps", bbox_inches="tight")
_ = mg_transcov.groupby(["fam","transporter"]).sum().reset_index()
_ = _.groupby("transporter").mean()
_ = pd.merge(transinfo, _, left_index=True, right_index=True)
_ = _.loc[_.substrate_category=="Carbohydrate"].groupby("name").sum()
(_.div(_.sum())*100).mean(axis=1).sort_values(ascending=False).head(3).sum()
| 0.291384 | 0.908089 |
# Bayesian Switchpoint Analysis
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/probability/blob/master/tensorflow_probability/examples/jupyter_notebooks/Bayesian_Switchpoint_Analysis.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/probability/blob/master/tensorflow_probability/examples/jupyter_notebooks/Bayesian_Switchpoint_Analysis.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
</table>
This notebook reimplements and extends the Bayesian “Change point analysis” example from the [pymc3 documentation](https://docs.pymc.io/notebooks/getting_started.html#Case-study-2:-Coal-mining-disasters).
## Prerequisites
```
import tensorflow.compat.v2 as tf
tf.enable_v2_behavior()
import tensorflow_probability as tfp
tfd = tfp.distributions
tfb = tfp.bijectors
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (15,8)
%config InlineBackend.figure_format = 'retina'
import numpy as np
import pandas as pd
```
## Dataset
The dataset is from [here](https://pymc-devs.github.io/pymc/tutorial.html#two-types-of-variables). Note, there is another version of this example [floating around](https://docs.pymc.io/notebooks/getting_started.html#Case-study-2:-Coal-mining-disasters), but it has “missing” data – in which case you’d need to impute missing values. (Otherwise your model will not ever leave its initial parameters because the likelihood function will be undefined.)
```
disaster_data = np.array([ 4, 5, 4, 0, 1, 4, 3, 4, 0, 6, 3, 3, 4, 0, 2, 6,
3, 3, 5, 4, 5, 3, 1, 4, 4, 1, 5, 5, 3, 4, 2, 5,
2, 2, 3, 4, 2, 1, 3, 2, 2, 1, 1, 1, 1, 3, 0, 0,
1, 0, 1, 1, 0, 0, 3, 1, 0, 3, 2, 2, 0, 1, 1, 1,
0, 1, 0, 1, 0, 0, 0, 2, 1, 0, 0, 0, 1, 1, 0, 2,
3, 3, 1, 1, 2, 1, 1, 1, 1, 2, 4, 2, 0, 0, 1, 4,
0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1])
years = np.arange(1851, 1962)
plt.plot(years, disaster_data, 'o', markersize=8);
plt.ylabel('Disaster count')
plt.xlabel('Year')
plt.title('Mining disaster data set')
plt.show()
```
## Probabilistic Model
The model assumes a “switch point” (e.g. a year during which safety regulations changed), and Poisson-distributed disaster rate with constant (but potentially different) rates before and after that switch point.
The actual disaster count is fixed (observed); any sample of this model will need to specify both the switchpoint and the “early” and “late” rate of disasters.
Original model from [pymc3 documentation example](https://pymc-devs.github.io/pymc/tutorial.html):
$$
\begin{align*}
(D_t|s,e,l)&\sim \text{Poisson}(r_t), \\
& \,\quad\text{with}\; r_t = \begin{cases}e & \text{if}\; t < s\\l &\text{if}\; t \ge s\end{cases} \\
s&\sim\text{Discrete Uniform}(t_l,\,t_h) \\
e&\sim\text{Exponential}(r_e)\\
l&\sim\text{Exponential}(r_l)
\end{align*}
$$
However, the mean disaster rate $r_t$ has a discontinuity at the switchpoint $s$, which makes it not differentiable. Thus it provides no gradient signal to the Hamiltonian Monte Carlo (HMC) algorithm – but because the $s$ prior is continuous, HMC’s fallback to a random walk is good enough to find the areas of high probability mass in this example.
As a second model, we modify the original model using a [sigmoid “switch”](https://en.wikipedia.org/wiki/Sigmoid_function) between *e* and *l* to make the transition differentiable, and use a continuous uniform distribution for the switchpoint $s$. (One could argue this model is more true to reality, as a “switch” in mean rate would likely be stretched out over multiple years.) The new model is thus:
$$
\begin{align*}
(D_t|s,e,l)&\sim\text{Poisson}(r_t), \\
& \,\quad \text{with}\; r_t = e + \frac{1}{1+\exp(s-t)}(l-e) \\
s&\sim\text{Uniform}(t_l,\,t_h) \\
e&\sim\text{Exponential}(r_e)\\
l&\sim\text{Exponential}(r_l)
\end{align*}
$$
In the absence of more information we assume $r_e = r_l = 1$ as parameters for the priors. We’ll run both models and compare their inference results.
```
def disaster_count_model(disaster_rate_fn):
disaster_count = tfd.JointDistributionNamed(dict(
e=tfd.Exponential(rate=1.),
l=tfd.Exponential(rate=1.),
s=tfd.Uniform(0., high=len(years)),
d_t=lambda s, l, e: tfd.Independent(
tfd.Poisson(rate=disaster_rate_fn(np.arange(len(years)), s, l, e)),
reinterpreted_batch_ndims=1)
))
return disaster_count
def disaster_rate_switch(ys, s, l, e):
return tf.where(ys < s, e, l)
def disaster_rate_sigmoid(ys, s, l, e):
return e + tf.sigmoid(ys - s) * (l - e)
model_switch = disaster_count_model(disaster_rate_switch)
model_sigmoid = disaster_count_model(disaster_rate_sigmoid)
```
The above code defines the model via JointDistributionSequential distributions. The `disaster_rate` functions are called with an array of `[0, ..., len(years)-1]` to produce a vector of `len(years)` random variables – the years before the `switchpoint` are `early_disaster_rate`, the ones after `late_disaster_rate` (modulo the sigmoid transition).
Here is a sanity-check that the target log prob function is sane:
```
def target_log_prob_fn(model, s, e, l):
return model.log_prob(s=s, e=e, l=l, d_t=disaster_data)
models = [model_switch, model_sigmoid]
print([target_log_prob_fn(m, 40., 3., .9).numpy() for m in models]) # Somewhat likely result
print([target_log_prob_fn(m, 60., 1., 5.).numpy() for m in models]) # Rather unlikely result
print([target_log_prob_fn(m, -10., 1., 1.).numpy() for m in models]) # Impossible result
```
## HMC to do Bayesian inference
We define the number of results and burn-in steps required; the code is mostly modeled after [the documentation of tfp.mcmc.HamiltonianMonteCarlo](https://www.tensorflow.org/probability/api_docs/python/tfp/mcmc/HamiltonianMonteCarlo). It uses an adaptive step size (otherwise the outcome is very sensitive to the step size value chosen). We use values of one as the initial state of the chain.
This is not the full story though. If you go back to the model definition above, you’ll note that some of the probability distributions are not well-defined on the whole real number line. Therefore we constrain the space that HMC shall examine by wrapping the HMC kernel with a [TransformedTransitionKernel](https://www.tensorflow.org/probability/api_docs/python/tfp/mcmc/TransformedTransitionKernel) that specifies the forward bijectors to transform the real numbers onto the domain that the probability distribution is defined on (see comments in the code below).
```
num_results = 10000
num_burnin_steps = 3000
@tf.function(autograph=False, experimental_compile=True)
def make_chain(target_log_prob_fn):
kernel = tfp.mcmc.TransformedTransitionKernel(
inner_kernel=tfp.mcmc.HamiltonianMonteCarlo(
target_log_prob_fn=target_log_prob_fn,
step_size=0.05,
num_leapfrog_steps=3),
bijector=[
# The switchpoint is constrained between zero and len(years).
# Hence we supply a bijector that maps the real numbers (in a
# differentiable way) to the interval (0;len(yers))
tfb.Sigmoid(low=0., high=tf.cast(len(years), dtype=tf.float32)),
# Early and late disaster rate: The exponential distribution is
# defined on the positive real numbers
tfb.Softplus(),
tfb.Softplus(),
])
kernel = tfp.mcmc.SimpleStepSizeAdaptation(
inner_kernel=kernel,
num_adaptation_steps=int(0.8*num_burnin_steps))
states = tfp.mcmc.sample_chain(
num_results=num_results,
num_burnin_steps=num_burnin_steps,
current_state=[
# The three latent variables
tf.ones([], name='init_switchpoint'),
tf.ones([], name='init_early_disaster_rate'),
tf.ones([], name='init_late_disaster_rate'),
],
trace_fn=None,
kernel=kernel)
return states
switch_samples = [s.numpy() for s in make_chain(
lambda *args: target_log_prob_fn(model_switch, *args))]
sigmoid_samples = [s.numpy() for s in make_chain(
lambda *args: target_log_prob_fn(model_sigmoid, *args))]
switchpoint, early_disaster_rate, late_disaster_rate = zip(
switch_samples, sigmoid_samples)
```
Run both models in parallel:
## Visualize the result
We visualize the result as histograms of samples of the posterior distribution for the early and late disaster rate, as well as the switchpoint. The histograms are overlaid with a solid line representing the sample median, as well as the 95%ile credible interval bounds as dashed lines.
```
def _desc(v):
return '(median: {}; 95%ile CI: $[{}, {}]$)'.format(
*np.round(np.percentile(v, [50, 2.5, 97.5]), 2))
for t, v in [
('Early disaster rate ($e$) posterior samples', early_disaster_rate),
('Late disaster rate ($l$) posterior samples', late_disaster_rate),
('Switch point ($s$) posterior samples', years[0] + switchpoint),
]:
fig, ax = plt.subplots(nrows=1, ncols=2, sharex=True)
for (m, i) in (('Switch', 0), ('Sigmoid', 1)):
a = ax[i]
a.hist(v[i], bins=50)
a.axvline(x=np.percentile(v[i], 50), color='k')
a.axvline(x=np.percentile(v[i], 2.5), color='k', ls='dashed', alpha=.5)
a.axvline(x=np.percentile(v[i], 97.5), color='k', ls='dashed', alpha=.5)
a.set_title(m + ' model ' + _desc(v[i]))
fig.suptitle(t)
plt.show()
```
|
github_jupyter
|
import tensorflow.compat.v2 as tf
tf.enable_v2_behavior()
import tensorflow_probability as tfp
tfd = tfp.distributions
tfb = tfp.bijectors
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (15,8)
%config InlineBackend.figure_format = 'retina'
import numpy as np
import pandas as pd
disaster_data = np.array([ 4, 5, 4, 0, 1, 4, 3, 4, 0, 6, 3, 3, 4, 0, 2, 6,
3, 3, 5, 4, 5, 3, 1, 4, 4, 1, 5, 5, 3, 4, 2, 5,
2, 2, 3, 4, 2, 1, 3, 2, 2, 1, 1, 1, 1, 3, 0, 0,
1, 0, 1, 1, 0, 0, 3, 1, 0, 3, 2, 2, 0, 1, 1, 1,
0, 1, 0, 1, 0, 0, 0, 2, 1, 0, 0, 0, 1, 1, 0, 2,
3, 3, 1, 1, 2, 1, 1, 1, 1, 2, 4, 2, 0, 0, 1, 4,
0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1])
years = np.arange(1851, 1962)
plt.plot(years, disaster_data, 'o', markersize=8);
plt.ylabel('Disaster count')
plt.xlabel('Year')
plt.title('Mining disaster data set')
plt.show()
def disaster_count_model(disaster_rate_fn):
disaster_count = tfd.JointDistributionNamed(dict(
e=tfd.Exponential(rate=1.),
l=tfd.Exponential(rate=1.),
s=tfd.Uniform(0., high=len(years)),
d_t=lambda s, l, e: tfd.Independent(
tfd.Poisson(rate=disaster_rate_fn(np.arange(len(years)), s, l, e)),
reinterpreted_batch_ndims=1)
))
return disaster_count
def disaster_rate_switch(ys, s, l, e):
return tf.where(ys < s, e, l)
def disaster_rate_sigmoid(ys, s, l, e):
return e + tf.sigmoid(ys - s) * (l - e)
model_switch = disaster_count_model(disaster_rate_switch)
model_sigmoid = disaster_count_model(disaster_rate_sigmoid)
def target_log_prob_fn(model, s, e, l):
return model.log_prob(s=s, e=e, l=l, d_t=disaster_data)
models = [model_switch, model_sigmoid]
print([target_log_prob_fn(m, 40., 3., .9).numpy() for m in models]) # Somewhat likely result
print([target_log_prob_fn(m, 60., 1., 5.).numpy() for m in models]) # Rather unlikely result
print([target_log_prob_fn(m, -10., 1., 1.).numpy() for m in models]) # Impossible result
num_results = 10000
num_burnin_steps = 3000
@tf.function(autograph=False, experimental_compile=True)
def make_chain(target_log_prob_fn):
kernel = tfp.mcmc.TransformedTransitionKernel(
inner_kernel=tfp.mcmc.HamiltonianMonteCarlo(
target_log_prob_fn=target_log_prob_fn,
step_size=0.05,
num_leapfrog_steps=3),
bijector=[
# The switchpoint is constrained between zero and len(years).
# Hence we supply a bijector that maps the real numbers (in a
# differentiable way) to the interval (0;len(yers))
tfb.Sigmoid(low=0., high=tf.cast(len(years), dtype=tf.float32)),
# Early and late disaster rate: The exponential distribution is
# defined on the positive real numbers
tfb.Softplus(),
tfb.Softplus(),
])
kernel = tfp.mcmc.SimpleStepSizeAdaptation(
inner_kernel=kernel,
num_adaptation_steps=int(0.8*num_burnin_steps))
states = tfp.mcmc.sample_chain(
num_results=num_results,
num_burnin_steps=num_burnin_steps,
current_state=[
# The three latent variables
tf.ones([], name='init_switchpoint'),
tf.ones([], name='init_early_disaster_rate'),
tf.ones([], name='init_late_disaster_rate'),
],
trace_fn=None,
kernel=kernel)
return states
switch_samples = [s.numpy() for s in make_chain(
lambda *args: target_log_prob_fn(model_switch, *args))]
sigmoid_samples = [s.numpy() for s in make_chain(
lambda *args: target_log_prob_fn(model_sigmoid, *args))]
switchpoint, early_disaster_rate, late_disaster_rate = zip(
switch_samples, sigmoid_samples)
def _desc(v):
return '(median: {}; 95%ile CI: $[{}, {}]$)'.format(
*np.round(np.percentile(v, [50, 2.5, 97.5]), 2))
for t, v in [
('Early disaster rate ($e$) posterior samples', early_disaster_rate),
('Late disaster rate ($l$) posterior samples', late_disaster_rate),
('Switch point ($s$) posterior samples', years[0] + switchpoint),
]:
fig, ax = plt.subplots(nrows=1, ncols=2, sharex=True)
for (m, i) in (('Switch', 0), ('Sigmoid', 1)):
a = ax[i]
a.hist(v[i], bins=50)
a.axvline(x=np.percentile(v[i], 50), color='k')
a.axvline(x=np.percentile(v[i], 2.5), color='k', ls='dashed', alpha=.5)
a.axvline(x=np.percentile(v[i], 97.5), color='k', ls='dashed', alpha=.5)
a.set_title(m + ' model ' + _desc(v[i]))
fig.suptitle(t)
plt.show()
| 0.613352 | 0.988679 |
## ASSIGNMENT 2
# PHÂN TÍCH DỮ LIỆU TMDB MOVIE
## Dựa vào dataset để phân tích và trả lời 3 câu hỏi sau:
### 1. Khu vực nào có ảnh hưởng nhất tới doanh thu?
### 2. Thể loại phim ảnh hưởng đến doanh thu và điểm trung bình như thế nào?
### 3. Ngày phát hành ảnh hưởng như thế nào đến doanh thu?
```
import numpy as np
import pandas as pd
import json
import matplotlib.pyplot as plt
import warnings
import sklearn
import seaborn as sns
from scipy.stats import f_oneway
from sklearn.preprocessing import PowerTransformer
warnings.filterwarnings('ignore')
warnings.simplefilter('ignore')
df_movies = pd.read_csv("C:/Users/Admin/Desktop/1CBDRobotic/res/week2/tmdb_5000_movies.csv/5000_movies.csv")
df_movies['release_date'] = pd.to_datetime(df_movies['release_date']).apply(lambda x: x.date())
```
### Dataset insight
```
print(df_movies.columns)
print(df_movies.shape)
df_movies.head(5)
print(df_movies.info())
df_movies = df_movies[["genres","production_countries","release_date","revenue","vote_average"]]
print(df_movies.info())
print(df_movies.info())
```
### insight Null values
```
print(df_movies.loc[df_movies['release_date'].isnull()])
def parse_countries(production_countries):
load_countries = json.loads(production_countries)
countries = []
for country in load_countries:
countries.append(country["iso_3166_1"])
return countries
df_movies_revenue_by_countries = df_movies[["production_countries","revenue"]]
df_movies_revenue_by_countries.replace(['[]','',0], np.nan, inplace=True)
df_movies_revenue_by_countries.dropna(inplace=True)
df_movies_revenue_by_countries["countries"] = df_movies["production_countries"].apply(lambda x:parse_countries(x))
df_movies_revenue_by_countries.head(10)
countries_list = (",".join([",".join(x) for x in df_movies_revenue_by_countries["countries"]])).split(",")
countries_list = list(dict.fromkeys(countries_list))
print(len(countries_list))
print(countries_list)
dict_revenue_by_country = {}
for index, row in df_movies_revenue_by_countries.iterrows():
for country in row["countries"]:
if country in dict_revenue_by_country:
dict_revenue_by_country[country].append(row["revenue"])
else:
dict_revenue_by_country[country] = [row["revenue"]]
for key in list(dict_revenue_by_country):
if len(dict_revenue_by_country[key]) < 20:
dict_revenue_by_country.pop(key)
print(len(dict_revenue_by_country))
# print(dict_revenue_by_country)
plt.hist(dict_revenue_by_country.get('US'), 50)
plt.title("Histogram of revenue in US ")
plt.show()
```
### Ta thấy data chưa được đưa về dạng normal distribution.
```
for key in dict_revenue_by_country.keys():
temp = np.array(dict_revenue_by_country.get(key)).reshape(-1,1)
transform_model = PowerTransformer().fit(temp)
plt.hist(transform_distribution(temp),100)
plt.title("Histogram of revenue in " + key)
plt.show()
temp = [(dict_revenue_by_country.get(key)) for key in dict_revenue_by_country.keys()]
anova_test = f_oneway(*temp)
print(anova_test)
```
### => Ta thấy pvalue < 0.05, do đó có thể phủ định H0 và chấp nhận giả thuyết H1: khu vực có ảnh hưởng đến doanh thu
### Ta thấy khu vực ảnh hưởng nhất tới doanh thu sẽ là khu vực có số lượng bộ phim nhiều nhất
```
temp = []
for key in dict_revenue_by_country.keys():
temp.append(len(dict_revenue_by_country.get(key)))
print(temp)
plt.bar(list(dict_revenue_by_country),temp)
```
### => Ta thấy US có số lượng bộ phim vượt trội so với các quốc gia khác, có thể nhận xét: US là nước có ảnh hưởng nhất tới revenue của phim.
## 2. Thể loại phim ảnh hưởng đến doanh thu và điểm trung bình như thế nào?
```
df_revenue_score_genre = df_movies[["revenue","vote_average","genres"]]
df_revenue_score_genre.head(5)
```
### Kiểm định xem thể loại phim có ảnh hưởng đến doanh thu hay không?
```
def parse_genre(genres):
load_genre = json.loads(genres)
genre_names = []
for genre in load_genre:
genre_names.append(genre["name"])
return genre_names
df_revenue_score_genre.replace(['[]','',0], np.nan, inplace=True)
df_revenue_score_genre.dropna(inplace=True)
df_revenue_score_genre["genres"] = df_revenue_score_genre["genres"].apply(lambda x:parse_genre(x))
df_revenue_score_genre.head(3)
genres_list = (",".join([",".join(x) for x in df_revenue_score_genre["genres"]])).split(",")
genres_list = list(dict.fromkeys(genres_list))
print(len(genres_list))
print(genres_list)
dict_revenue_by_genres = {}
for index, row in df_revenue_score_genre.iterrows():
for genres in row["genres"]:
if genres in dict_revenue_by_genres:
dict_revenue_by_genres[genres].append(row["revenue"])
else:
dict_revenue_by_genres[genres] = [row["revenue"]]
temp = []
for key in dict_revenue_by_genres.keys():
temp.append(len(dict_revenue_by_genres.get(key)))
print(temp)
plt.figure(figsize=(25,6))
plt.bar(list(dict_revenue_by_genres),temp, 0.5)
#print(dict_revenue_by_genres)
for key in list(dict_revenue_by_genres):
if len(dict_revenue_by_genres[key]) < np.mean(temp):
dict_revenue_by_genres.pop(key)
temp = []
for key in dict_revenue_by_genres.keys():
temp.append(len(dict_revenue_by_genres.get(key)))
print(temp)
plt.figure(figsize=(12,5))
plt.bar(list(dict_revenue_by_genres),temp, 0.5)
plt.hist(dict_revenue_by_genres.get('Action'), 50)
plt.title("Histogram of revenue in Action")
plt.show()
for key in dict_revenue_by_genres.keys():
temp = np.array(dict_revenue_by_genres.get(key)).reshape(-1,1)
transform_model = PowerTransformer().fit(temp)
plt.hist(transform_distribution(temp), 100)
plt.title("Histogram of revenue in " + key)
plt.show()
temp = [(dict_revenue_by_genres.get(key)) for key in dict_revenue_by_genres.keys()]
anova_test = f_oneway(*temp)
print(anova_test)
```
### Kiểm định xem thể loại phim có ảnh hưởng đến điểm trung bình hay không?
```
dict_score_by_genres = {}
for index, row in df_revenue_score_genre.iterrows():
for genres in row["genres"]:
if genres in dict_score_by_genres:
dict_score_by_genres[genres].append(row["vote_average"])
else:
dict_score_by_genres[genres] = [row["vote_average"]]
temp = []
for key in dict_score_by_genres.keys():
temp.append(len(dict_score_by_genres.get(key)))
for key in list(dict_score_by_genres):
if len(dict_score_by_genres[key]) < np.mean(temp):
dict_score_by_genres.pop(key)
print(temp)
plt.hist(dict_revenue_by_genres.get('Action'), 50)
plt.title("Histogram of score in Action")
plt.show()
for key in dict_score_by_genres.keys():
temp = np.array(dict_score_by_genres.get(key)).reshape(-1,1)
transform_model = PowerTransformer().fit(temp)
plt.hist(transform_distribution(temp), 100)
plt.title("Histogram of score in " + key)
plt.show()
temp = [(dict_score_by_genres.get(key)) for key in dict_score_by_genres.keys()]
anova_test = f_oneway(*temp)
print(anova_test)
```
### => Cả pvalue của genres_and_avenue và pvalue của genres_and_score đều < 0.05
### => ta có thể phủ định H0 và chấp nhận giả thuyết H1: thể loại phim có ảnh hưởng đến doanh thu và điểm trung bình
```
mean_revenue_by_genres =[]
for key in dict_revenue_by_genres.keys():
mean_revenue_by_genres.append(np.mean(dict_revenue_by_genres.get(key)))
print(mean_revenue_by_genres)
plt.bar(list(dict_revenue_by_genres), mean_revenue_by_genres,0.5)
```
### ta thấy adventure là thể loại có doanh thu trung bình cao nhất theo sau là thể loại Action, và trong cùng 1 bộ phim thì cũng thường bao gồm cả 2 thể loại này.
```
mean_score_by_genres =[]
for key in dict_score_by_genres.keys():
mean_score_by_genres.append(np.mean(dict_score_by_genres.get(key)))
print(mean_score_by_genres)
plt.bar(list(dict_score_by_genres), mean_score_by_genres,0.5)
```
### ta thấy Drama là thể loại có điểm số trung bình cao nhất, theo sau là thể loại Crime, những bộ phim chú trọng vào xây dựng cốt truyện và diễn biến tâm lí nhân vật.
## 3. Ngày phát hành ảnh hưởng như thế nào đến doanh thu?
### Kiểm định xem tháng phát hành có ảnh hưởng đến doanh thu hay không
```
df_revenue_date = df_movies[["revenue","release_date"]]
df_revenue_date.replace(['[]','',0], np.nan, inplace=True)
df_revenue_date.dropna(inplace=True)
temp_months = []
temp_years = []
for date in df_revenue_date['release_date']:
temp_months.append(int(date.month))
temp_years.append(int(date.year))
df_revenue_date['months'] = temp_months
df_revenue_date['years'] = temp_years
print(df_revenue_date)
```
### Kiểm tra với Months
```
dict_revenue_by_months = {}
dict_revenue_by_genres = {}
for index, row in df_revenue_date.iterrows():
if row['months'] in dict_revenue_by_months:
dict_revenue_by_months[row['months']].append(row["revenue"])
else:
dict_revenue_by_months[row['months']] = [row["revenue"]]
plt.hist(dict_revenue_by_months.get(1), 50)
plt.title("Histogram of score in month 1")
plt.show()
for key in dict_revenue_by_months.keys():
temp = np.array(dict_revenue_by_months.get(key)).reshape(-1,1)
transform_model = PowerTransformer().fit(temp)
plt.hist(transform_distribution(temp), 100)
plt.title("Histogram of revenue in month " + str(key))
plt.show()
temp = [(dict_revenue_by_months.get(key)) for key in dict_revenue_by_months.keys()]
anova_test = f_oneway(*temp)
print(anova_test)
```
### chấp nhận H1: các tháng có ảnh hưởng đến revenue
```
plt.figure(figsize=(18,6))
test = sns.boxplot(x='months',y='revenue',data=df_revenue_date)
plt.show()
```
### Ta thấy tháng 5, 6 và tháng 11, 12 là những cặp tháng có doanh thu cao hơn so với các tháng lân cận, phim phát hành vào thời điểm này thường có doanh thu cao, có thể là vì tháng 5, 6 là giai đoạn bước vào kì nghỉ hè, còn dịp cuối năm là dịp có nhiều ngày nghỉ lễ ở Châu Âu.
### Kiếm tra với years
```
dict_revenue_by_years = {}
for index, row in df_revenue_date.iterrows():
if row['years'] in dict_revenue_by_years:
dict_revenue_by_years[row['years']].append(row["revenue"])
else:
dict_revenue_by_years[row['years']] = [row["revenue"]]
print(len(list(dict_revenue_by_years)))
for key in list(dict_revenue_by_years):
if len(dict_revenue_by_years[key]) < 100:
dict_revenue_by_years.pop(key)
print(len(list(dict_revenue_by_years)))
plt.hist(dict_revenue_by_years.get(2000), 50)
plt.title("Histogram of score in year 2000")
plt.show()
for key in dict_revenue_by_years.keys():
temp = np.array(dict_revenue_by_years.get(key)).reshape(-1,1)
transform_model = PowerTransformer().fit(temp)
plt.hist(transform_distribution(temp), 100)
plt.title("Histogram of revenue in year " + str(key))
plt.show()
temp = [(dict_revenue_by_years.get(key)) for key in dict_revenue_by_years.keys()]
anova_test = f_oneway(*temp)
print(anova_test)
```
### chấp nhận H1: năm sản xuất có ảnh hưởng đến revenue
```
plt.figure(figsize=(18,6))
test = sns.boxplot(x='years',y='revenue',data=df_revenue_date)
plt.show()
```
#### Kiểm tra correlation
```
a = np.corrcoef(df_revenue_date['years'], df_revenue_date['revenue'])
print(a)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import json
import matplotlib.pyplot as plt
import warnings
import sklearn
import seaborn as sns
from scipy.stats import f_oneway
from sklearn.preprocessing import PowerTransformer
warnings.filterwarnings('ignore')
warnings.simplefilter('ignore')
df_movies = pd.read_csv("C:/Users/Admin/Desktop/1CBDRobotic/res/week2/tmdb_5000_movies.csv/5000_movies.csv")
df_movies['release_date'] = pd.to_datetime(df_movies['release_date']).apply(lambda x: x.date())
print(df_movies.columns)
print(df_movies.shape)
df_movies.head(5)
print(df_movies.info())
df_movies = df_movies[["genres","production_countries","release_date","revenue","vote_average"]]
print(df_movies.info())
print(df_movies.info())
print(df_movies.loc[df_movies['release_date'].isnull()])
def parse_countries(production_countries):
load_countries = json.loads(production_countries)
countries = []
for country in load_countries:
countries.append(country["iso_3166_1"])
return countries
df_movies_revenue_by_countries = df_movies[["production_countries","revenue"]]
df_movies_revenue_by_countries.replace(['[]','',0], np.nan, inplace=True)
df_movies_revenue_by_countries.dropna(inplace=True)
df_movies_revenue_by_countries["countries"] = df_movies["production_countries"].apply(lambda x:parse_countries(x))
df_movies_revenue_by_countries.head(10)
countries_list = (",".join([",".join(x) for x in df_movies_revenue_by_countries["countries"]])).split(",")
countries_list = list(dict.fromkeys(countries_list))
print(len(countries_list))
print(countries_list)
dict_revenue_by_country = {}
for index, row in df_movies_revenue_by_countries.iterrows():
for country in row["countries"]:
if country in dict_revenue_by_country:
dict_revenue_by_country[country].append(row["revenue"])
else:
dict_revenue_by_country[country] = [row["revenue"]]
for key in list(dict_revenue_by_country):
if len(dict_revenue_by_country[key]) < 20:
dict_revenue_by_country.pop(key)
print(len(dict_revenue_by_country))
# print(dict_revenue_by_country)
plt.hist(dict_revenue_by_country.get('US'), 50)
plt.title("Histogram of revenue in US ")
plt.show()
for key in dict_revenue_by_country.keys():
temp = np.array(dict_revenue_by_country.get(key)).reshape(-1,1)
transform_model = PowerTransformer().fit(temp)
plt.hist(transform_distribution(temp),100)
plt.title("Histogram of revenue in " + key)
plt.show()
temp = [(dict_revenue_by_country.get(key)) for key in dict_revenue_by_country.keys()]
anova_test = f_oneway(*temp)
print(anova_test)
temp = []
for key in dict_revenue_by_country.keys():
temp.append(len(dict_revenue_by_country.get(key)))
print(temp)
plt.bar(list(dict_revenue_by_country),temp)
df_revenue_score_genre = df_movies[["revenue","vote_average","genres"]]
df_revenue_score_genre.head(5)
def parse_genre(genres):
load_genre = json.loads(genres)
genre_names = []
for genre in load_genre:
genre_names.append(genre["name"])
return genre_names
df_revenue_score_genre.replace(['[]','',0], np.nan, inplace=True)
df_revenue_score_genre.dropna(inplace=True)
df_revenue_score_genre["genres"] = df_revenue_score_genre["genres"].apply(lambda x:parse_genre(x))
df_revenue_score_genre.head(3)
genres_list = (",".join([",".join(x) for x in df_revenue_score_genre["genres"]])).split(",")
genres_list = list(dict.fromkeys(genres_list))
print(len(genres_list))
print(genres_list)
dict_revenue_by_genres = {}
for index, row in df_revenue_score_genre.iterrows():
for genres in row["genres"]:
if genres in dict_revenue_by_genres:
dict_revenue_by_genres[genres].append(row["revenue"])
else:
dict_revenue_by_genres[genres] = [row["revenue"]]
temp = []
for key in dict_revenue_by_genres.keys():
temp.append(len(dict_revenue_by_genres.get(key)))
print(temp)
plt.figure(figsize=(25,6))
plt.bar(list(dict_revenue_by_genres),temp, 0.5)
#print(dict_revenue_by_genres)
for key in list(dict_revenue_by_genres):
if len(dict_revenue_by_genres[key]) < np.mean(temp):
dict_revenue_by_genres.pop(key)
temp = []
for key in dict_revenue_by_genres.keys():
temp.append(len(dict_revenue_by_genres.get(key)))
print(temp)
plt.figure(figsize=(12,5))
plt.bar(list(dict_revenue_by_genres),temp, 0.5)
plt.hist(dict_revenue_by_genres.get('Action'), 50)
plt.title("Histogram of revenue in Action")
plt.show()
for key in dict_revenue_by_genres.keys():
temp = np.array(dict_revenue_by_genres.get(key)).reshape(-1,1)
transform_model = PowerTransformer().fit(temp)
plt.hist(transform_distribution(temp), 100)
plt.title("Histogram of revenue in " + key)
plt.show()
temp = [(dict_revenue_by_genres.get(key)) for key in dict_revenue_by_genres.keys()]
anova_test = f_oneway(*temp)
print(anova_test)
dict_score_by_genres = {}
for index, row in df_revenue_score_genre.iterrows():
for genres in row["genres"]:
if genres in dict_score_by_genres:
dict_score_by_genres[genres].append(row["vote_average"])
else:
dict_score_by_genres[genres] = [row["vote_average"]]
temp = []
for key in dict_score_by_genres.keys():
temp.append(len(dict_score_by_genres.get(key)))
for key in list(dict_score_by_genres):
if len(dict_score_by_genres[key]) < np.mean(temp):
dict_score_by_genres.pop(key)
print(temp)
plt.hist(dict_revenue_by_genres.get('Action'), 50)
plt.title("Histogram of score in Action")
plt.show()
for key in dict_score_by_genres.keys():
temp = np.array(dict_score_by_genres.get(key)).reshape(-1,1)
transform_model = PowerTransformer().fit(temp)
plt.hist(transform_distribution(temp), 100)
plt.title("Histogram of score in " + key)
plt.show()
temp = [(dict_score_by_genres.get(key)) for key in dict_score_by_genres.keys()]
anova_test = f_oneway(*temp)
print(anova_test)
mean_revenue_by_genres =[]
for key in dict_revenue_by_genres.keys():
mean_revenue_by_genres.append(np.mean(dict_revenue_by_genres.get(key)))
print(mean_revenue_by_genres)
plt.bar(list(dict_revenue_by_genres), mean_revenue_by_genres,0.5)
mean_score_by_genres =[]
for key in dict_score_by_genres.keys():
mean_score_by_genres.append(np.mean(dict_score_by_genres.get(key)))
print(mean_score_by_genres)
plt.bar(list(dict_score_by_genres), mean_score_by_genres,0.5)
df_revenue_date = df_movies[["revenue","release_date"]]
df_revenue_date.replace(['[]','',0], np.nan, inplace=True)
df_revenue_date.dropna(inplace=True)
temp_months = []
temp_years = []
for date in df_revenue_date['release_date']:
temp_months.append(int(date.month))
temp_years.append(int(date.year))
df_revenue_date['months'] = temp_months
df_revenue_date['years'] = temp_years
print(df_revenue_date)
dict_revenue_by_months = {}
dict_revenue_by_genres = {}
for index, row in df_revenue_date.iterrows():
if row['months'] in dict_revenue_by_months:
dict_revenue_by_months[row['months']].append(row["revenue"])
else:
dict_revenue_by_months[row['months']] = [row["revenue"]]
plt.hist(dict_revenue_by_months.get(1), 50)
plt.title("Histogram of score in month 1")
plt.show()
for key in dict_revenue_by_months.keys():
temp = np.array(dict_revenue_by_months.get(key)).reshape(-1,1)
transform_model = PowerTransformer().fit(temp)
plt.hist(transform_distribution(temp), 100)
plt.title("Histogram of revenue in month " + str(key))
plt.show()
temp = [(dict_revenue_by_months.get(key)) for key in dict_revenue_by_months.keys()]
anova_test = f_oneway(*temp)
print(anova_test)
plt.figure(figsize=(18,6))
test = sns.boxplot(x='months',y='revenue',data=df_revenue_date)
plt.show()
dict_revenue_by_years = {}
for index, row in df_revenue_date.iterrows():
if row['years'] in dict_revenue_by_years:
dict_revenue_by_years[row['years']].append(row["revenue"])
else:
dict_revenue_by_years[row['years']] = [row["revenue"]]
print(len(list(dict_revenue_by_years)))
for key in list(dict_revenue_by_years):
if len(dict_revenue_by_years[key]) < 100:
dict_revenue_by_years.pop(key)
print(len(list(dict_revenue_by_years)))
plt.hist(dict_revenue_by_years.get(2000), 50)
plt.title("Histogram of score in year 2000")
plt.show()
for key in dict_revenue_by_years.keys():
temp = np.array(dict_revenue_by_years.get(key)).reshape(-1,1)
transform_model = PowerTransformer().fit(temp)
plt.hist(transform_distribution(temp), 100)
plt.title("Histogram of revenue in year " + str(key))
plt.show()
temp = [(dict_revenue_by_years.get(key)) for key in dict_revenue_by_years.keys()]
anova_test = f_oneway(*temp)
print(anova_test)
plt.figure(figsize=(18,6))
test = sns.boxplot(x='years',y='revenue',data=df_revenue_date)
plt.show()
a = np.corrcoef(df_revenue_date['years'], df_revenue_date['revenue'])
print(a)
| 0.073759 | 0.63641 |
```
import nltk
from nltk.corpus import stopwords
from nltk.cluster.util import cosine_distance
import numpy as np
import networkx as nx
def read_article(file_name):
file = open("H:\Machine Learning\Summery.txt", "r")
filedata = file.readlines()
article = filedata[0].split(". ")
sentences = []
for sentence in article:
print(sentence)
sentences.append(sentence.replace("[^a-zA-Z]", " ").split(" "))
sentences.pop()
return sentences
def sentence_similarity(sent1, sent2, stopwords=None):
if stopwords is None:
stopwords = []
sent1 = [w.lower() for w in sent1]
sent2 = [w.lower() for w in sent2]
all_words = list(set(sent1 + sent2))
vector1 = [0] * len(all_words)
vector2 = [0] * len(all_words)
# build the vector for the first sentence
for w in sent1:
if w in stopwords:
continue
vector1[all_words.index(w)] += 1
# build the vector for the second sentence
for w in sent2:
if w in stopwords:
continue
vector2[all_words.index(w)] += 1
return 1 - cosine_distance(vector1, vector2)
def build_similarity_matrix(sentences, stop_words):
# Create an empty similarity matrix
similarity_matrix = np.zeros((len(sentences), len(sentences)))
for idx1 in range(len(sentences)):
for idx2 in range(len(sentences)):
if idx1 == idx2: #ignore if both are same sentences
continue
similarity_matrix[idx1][idx2] = sentence_similarity(sentences[idx1], sentences[idx2], stop_words)
return similarity_matrix
def generate_summary(file_name, top_n=5):
stop_words = stopwords.words('english')
summarize_text = []
# Step 1 - Read text anc split it
sentences = read_article(file_name)
# Step 2 - Generate Similary Martix across sentences
sentence_similarity_martix = build_similarity_matrix(sentences, stop_words)
# Step 3 - Rank sentences in similarity martix
sentence_similarity_graph = nx.from_numpy_array(sentence_similarity_martix)
scores = nx.pagerank(sentence_similarity_graph)
# Step 4 - Sort the rank and pick top sentences
ranked_sentence = sorted(((scores[i],s) for i,s in enumerate(sentences)), reverse=True)
print("Indexes of top ranked_sentence order are ", ranked_sentence)
for i in range(top_n):
summarize_text.append(" ".join(ranked_sentence[i][1]))
# Step 5 - Offcourse, output the summarize texr
print("Summarize Text: \n", ". ".join(summarize_text))
# let's begin
generate_summary( "msft.txt", 2)
```
|
github_jupyter
|
import nltk
from nltk.corpus import stopwords
from nltk.cluster.util import cosine_distance
import numpy as np
import networkx as nx
def read_article(file_name):
file = open("H:\Machine Learning\Summery.txt", "r")
filedata = file.readlines()
article = filedata[0].split(". ")
sentences = []
for sentence in article:
print(sentence)
sentences.append(sentence.replace("[^a-zA-Z]", " ").split(" "))
sentences.pop()
return sentences
def sentence_similarity(sent1, sent2, stopwords=None):
if stopwords is None:
stopwords = []
sent1 = [w.lower() for w in sent1]
sent2 = [w.lower() for w in sent2]
all_words = list(set(sent1 + sent2))
vector1 = [0] * len(all_words)
vector2 = [0] * len(all_words)
# build the vector for the first sentence
for w in sent1:
if w in stopwords:
continue
vector1[all_words.index(w)] += 1
# build the vector for the second sentence
for w in sent2:
if w in stopwords:
continue
vector2[all_words.index(w)] += 1
return 1 - cosine_distance(vector1, vector2)
def build_similarity_matrix(sentences, stop_words):
# Create an empty similarity matrix
similarity_matrix = np.zeros((len(sentences), len(sentences)))
for idx1 in range(len(sentences)):
for idx2 in range(len(sentences)):
if idx1 == idx2: #ignore if both are same sentences
continue
similarity_matrix[idx1][idx2] = sentence_similarity(sentences[idx1], sentences[idx2], stop_words)
return similarity_matrix
def generate_summary(file_name, top_n=5):
stop_words = stopwords.words('english')
summarize_text = []
# Step 1 - Read text anc split it
sentences = read_article(file_name)
# Step 2 - Generate Similary Martix across sentences
sentence_similarity_martix = build_similarity_matrix(sentences, stop_words)
# Step 3 - Rank sentences in similarity martix
sentence_similarity_graph = nx.from_numpy_array(sentence_similarity_martix)
scores = nx.pagerank(sentence_similarity_graph)
# Step 4 - Sort the rank and pick top sentences
ranked_sentence = sorted(((scores[i],s) for i,s in enumerate(sentences)), reverse=True)
print("Indexes of top ranked_sentence order are ", ranked_sentence)
for i in range(top_n):
summarize_text.append(" ".join(ranked_sentence[i][1]))
# Step 5 - Offcourse, output the summarize texr
print("Summarize Text: \n", ". ".join(summarize_text))
# let's begin
generate_summary( "msft.txt", 2)
| 0.329715 | 0.385751 |
```
import numpy as np
from sklearn.preprocessing import MinMaxScaler
from sklearn.utils import shuffle
import datetime
import csv
import matplotlib
import matplotlib.pyplot as plt
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_error
from keras.models import load_model
%matplotlib inline
from google.colab import drive
drive.mount('/content/drive')
def read_csv(file_path):
data = list()
with open(file_path, 'r') as file:
reader = csv.reader(file)
next(reader)
for raw in reader:
data.append([float(i) for i in raw])
return data
def split_dataset(data, per):
total_num = len(data)
test_num = total_num * per // 100
train_data = data[0:(total_num - test_num)]
test_data = data[(total_num - test_num):]
return train_data, test_data
ph = [1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 12]
ph_mse_data = [[] for _ in range(len(ph))]
ph_networks = [['/content/drive/My Drive/mlp_model.h5', "/content/drive/My Drive/simple_rnn_model.h5", "/content/drive/My Drive/lstm_model.h5"],
['/content/drive/My Drive/mlp_model2.h5', "/content/drive/My Drive/simple_rnn_model2.h5", "/content/drive/My Drive/lstm_model2.h5"],
['/content/drive/My Drive/mlp_model3.h5', "/content/drive/My Drive/simple_rnn_model3.h5", "/content/drive/My Drive/lstm_model3.h5"],
['/content/drive/My Drive/mlp_model4.h5', "/content/drive/My Drive/simple_rnn_model4.h5", "/content/drive/My Drive/lstm_model4.h5"],
['/content/drive/My Drive/mlp_model6.h5', "/content/drive/My Drive/simple_rnn_model6.h5", "/content/drive/My Drive/lstm_model6.h5"],
['/content/drive/My Drive/mlp_model7.h5', "/content/drive/My Drive/simple_rnn_model7.h5", "/content/drive/My Drive/lstm_model7.h5"],
['/content/drive/My Drive/mlp_model8.h5', "/content/drive/My Drive/simple_rnn_model8.h5", "/content/drive/My Drive/lstm_model8.h5"],
['/content/drive/My Drive/mlp_model9.h5', "/content/drive/My Drive/simple_rnn_model9.h5", "/content/drive/My Drive/lstm_model9.h5"],
['/content/drive/My Drive/mlp_model10.h5', "/content/drive/My Drive/simple_rnn_model10.h5", "/content/drive/My Drive/lstm_model10.h5"],
['/content/drive/My Drive/mlp_model11.h5', "/content/drive/My Drive/simple_rnn_model11.h5", "/content/drive/My Drive/lstm_model11.h5"],
['/content/drive/My Drive/mlp_model12.h5', "/content/drive/My Drive/simple_rnn_model12.h5", "/content/drive/My Drive/lstm_model12.h5"]]
print(ph)
print(ph[3])
#ph1_networks = ['Paper\\Networks\\mlp_model.h5', "Paper\\Networks\\simple_rnn_model.h5", "Paper\\Networks\\lstm_model.h5", "Paper\\Networks\\gru_model.h5"]
#ph5_networks = ['Paper\\Networks\\mlp_model5.h5', "Paper\\Networks\\simple_rnn_model5.h5", "Paper\\Networks\\lstm_model5.h5"]
#ph9_networks = ['Paper\\Networks\\mlp_model9.h5', "Paper\\Networks\\simple_rnn_model9.h5", "Paper\\Networks\\lstm_model9.h5"]
def prepare_data(duration, ph):
input_data = list()
output_data = list()
for j in range(29):
file_path = f"/content/drive/My Drive/ColabNotebooks/paperwithvedadi/data(10subjects)/filtered/filtered_{j+1}.csv"
data = read_csv(file_path)
#num = len (data) - duration - ((1*i)+1)
num = len (data) - duration - ph
for k in range(num):
input_data.append([row for row in data[k:k+duration]])
#output_data.append(data[j+duration+(1*i)])
output_data.append(data[k+duration+(ph-1)])
return np.array(input_data), np.array(output_data)
for j in range(len(ph)):
input_data, output_data = prepare_data(200//4, ph[j])
input_data = np.squeeze(input_data)
train_data, test_data = split_dataset(input_data, 15)
train_label, test_label = split_dataset(output_data, 15)
n_timesteps = train_data.shape[1]
n_features = train_data.shape[2]
test_data = test_data.reshape(test_data.shape[0], n_timesteps * n_features)
NewScale = MinMaxScaler(feature_range=(0,1), copy=True)
test_data = NewScale.fit_transform(test_data)
test_label = NewScale.fit_transform(test_label)
test_data = test_data.reshape(test_data.shape[0], n_timesteps, n_features)
for i in range(len(ph_networks[j])):
#test_data, test_label = shuffle(test_data, test_label, random_state=1)
if i == 0:
#train_data = train_data.reshape(train_data.shape[0], n_timesteps * n_features)
test_data = test_data.reshape(test_data.shape[0], n_timesteps * n_features)
else:
#train_data = train_data.reshape(train_data.shape[0], n_timesteps, n_features)
test_data = test_data.reshape(test_data.shape[0], n_timesteps, n_features)
model = load_model(ph_networks[j][i])
prediction = model.predict(test_data)
print("ph = " , j, " network = ", ph_networks[j][i]," ", np.shape(prediction) )
mse_data = ((test_label[:, 8] - prediction[:, 8])**2)
ph_mse_data[j].append(mse_data)
def set_box_color(bp, color):
plt.setp(bp['boxes'], color=color)
plt.setp(bp['whiskers'], color=color)
plt.setp(bp['caps'], color=color)
plt.setp(bp['medians'], color=color)
fig, ax = plt.subplots(figsize=(30, 7))
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.set_ylabel('Squared Error')
ax.grid(color='grey', axis='y', linestyle='-', linewidth=0.25, alpha=0.5)
#ticks = ['Fully Connected', 'Simple Recurrent', 'LSTM', 'GRU']
ticks = ['Fully Connected', 'Simple Recurrent', 'LSTM']
bp1 = ax.boxplot(ph_mse_data[0], positions=np.array(range(len(ph_mse_data[0])))*6.0-2.5, sym='', widths=0.4, showfliers=True)
bp2 = ax.boxplot(ph_mse_data[1], positions=np.array(range(len(ph_mse_data[1])))*6.0-2.0, sym='', widths=0.4, showfliers=True)
bp3 = ax.boxplot(ph_mse_data[2], positions=np.array(range(len(ph_mse_data[2])))*6.0-1.5, sym='', widths=0.4, showfliers=True)
bp4 = ax.boxplot(ph_mse_data[3], positions=np.array(range(len(ph_mse_data[3])))*6.0-1.0, sym='', widths=0.4, showfliers=True)
bp6 = ax.boxplot(ph_mse_data[4], positions=np.array(range(len(ph_mse_data[4])))*6.0-0.5, sym='', widths=0.4, showfliers=True)
bp7 = ax.boxplot(ph_mse_data[5], positions=np.array(range(len(ph_mse_data[5])))*6.0, sym='', widths=0.4, showfliers=True)
bp8 = ax.boxplot(ph_mse_data[6], positions=np.array(range(len(ph_mse_data[6])))*6.0+0.5, sym='', widths=0.4, showfliers=True)
bp9 = ax.boxplot(ph_mse_data[7], positions=np.array(range(len(ph_mse_data[7])))*6.0+1.0, sym='', widths=0.4, showfliers=True)
bp10 = ax.boxplot(ph_mse_data[8], positions=np.array(range(len(ph_mse_data[8])))*6.0+1.5, sym='', widths=0.4, showfliers=True)
bp11 = ax.boxplot(ph_mse_data[9], positions=np.array(range(len(ph_mse_data[9])))*6.0+2.0, sym='', widths=0.4, showfliers=True)
bp12 = ax.boxplot(ph_mse_data[10], positions=np.array(range(len(ph_mse_data[10])))*6.0+2.5, sym='', widths=0.4, showfliers=True)
plt.xticks(range(0, len(ticks) * 2, 2), ticks)
set_box_color(bp1, '#D7191C') # colors are from http://colorbrewer2.org/
set_box_color(bp2, '#2C7BB6')
set_box_color(bp3, '#feb24c')
set_box_color(bp4, '#c51b8a') # colors are from http://colorbrewer2.org/
set_box_color(bp6, '#fa9fb5')
set_box_color(bp7, '#2b8cbe')
set_box_color(bp8, '#fc9272') # colors are from http://colorbrewer2.org/
set_box_color(bp9, '#31a354')
set_box_color(bp10, '#a1d99b')
set_box_color(bp11, '#c994c7') # colors are from http://colorbrewer2.org/
set_box_color(bp12, '#636363')
plt.plot([], c='#D7191C', label='Prediction Horizon = 1')
plt.plot([], c='#2C7BB6', label='Prediction Horizon = 2')
plt.plot([], c='#feb24c', label='Prediction Horizon = 3')
plt.plot([], c='#c51b8a', label='Prediction Horizon = 4')
plt.plot([], c='#fa9fb5', label='Prediction Horizon = 6')
plt.plot([], c='#2b8cbe', label='Prediction Horizon = 7')
plt.plot([], c='#fc9272', label='Prediction Horizon = 8')
plt.plot([], c='#31a354', label='Prediction Horizon = 9')
plt.plot([], c='#a1d99b', label='Prediction Horizon = 10')
plt.plot([], c='#c994c7', label='Prediction Horizon = 11')
plt.plot([], c='#636363', label='Prediction Horizon = 12')
#plt.legend()
plt.savefig(f'/content/drive/My Drive/ColabNotebooks/paperwithvedadi/SquaredErrorDistribution.png', dpi = 900, bbox_inches='tight')
plt.show
train_data = train_data.reshape(train_data.shape[0], n_timesteps * n_features)
test_data = test_data.reshape(test_data.shape[0], n_timesteps * n_features)
train_data = train_data.reshape(train_data.shape[0], n_timesteps , n_features)
test_data = test_data.reshape(test_data.shape[0], n_timesteps , n_features)
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_error
from keras.models import load_model
model = load_model("Paper\\Networks\\lstm_model9.h5")
#test_data, test_label = shuffle(test_data, test_label, random_state=1)
start = datetime.datetime.now()
prediction = model.predict(test_data)
end = datetime.datetime.now()
test_time = end - start
test_loss = model.evaluate(test_data, test_label)
mse = mean_squared_error(test_label[:, 8], prediction[:, 8])
r2 = r2_score(test_label[:, 8], prediction[:, 8])
print("test time is : ",test_time)
print('Test - Loss:', test_loss)
print('Test - MSE :', mse)
print('Test - R2 Score :', r2)
import matplotlib as mpl
from pylab import cm
#mpl.rcParams['font.family'] = 'Times New Roman'
plt.rcParams['font.size'] = 16
plt.rcParams['axes.linewidth'] = 2
colors = cm.get_cmap('rainbow', 2)
fig = plt.figure(figsize = [10,5])
ax = fig.add_axes([0, 0, 1, 1])
ax.spines['right'].set_visible(False)
ax.spines['top'].set_visible(False)
time = np.arange(0, 15, 15/300)
ax.plot(time, test_label[10000:13000:10, 8], label='Actual', linewidth=2, c='r', linestyle='-')
ax.plot(time, prediction[10000:13000:10, 8], label='Prediction', linewidth=2, c='b', linestyle='--')
plt.legend(frameon=False)
ax.set_xlabel('Time (s)', labelpad=10)
ax.set_ylabel('Normalized Force', labelpad=10)
ax.set_title("LSTM Network")
#ax.set_title("Fully Connected Network")
#ax.set_title("Simple Recurrent Network")
ax.legend(bbox_to_anchor=(1, 1), loc=0, frameon=False, fontsize=14)
plt.savefig('Paper\\PerformanceImages\\lstm_prediction9.png', dpi = 900, bbox_inches='tight')
```
|
github_jupyter
|
import numpy as np
from sklearn.preprocessing import MinMaxScaler
from sklearn.utils import shuffle
import datetime
import csv
import matplotlib
import matplotlib.pyplot as plt
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_error
from keras.models import load_model
%matplotlib inline
from google.colab import drive
drive.mount('/content/drive')
def read_csv(file_path):
data = list()
with open(file_path, 'r') as file:
reader = csv.reader(file)
next(reader)
for raw in reader:
data.append([float(i) for i in raw])
return data
def split_dataset(data, per):
total_num = len(data)
test_num = total_num * per // 100
train_data = data[0:(total_num - test_num)]
test_data = data[(total_num - test_num):]
return train_data, test_data
ph = [1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 12]
ph_mse_data = [[] for _ in range(len(ph))]
ph_networks = [['/content/drive/My Drive/mlp_model.h5', "/content/drive/My Drive/simple_rnn_model.h5", "/content/drive/My Drive/lstm_model.h5"],
['/content/drive/My Drive/mlp_model2.h5', "/content/drive/My Drive/simple_rnn_model2.h5", "/content/drive/My Drive/lstm_model2.h5"],
['/content/drive/My Drive/mlp_model3.h5', "/content/drive/My Drive/simple_rnn_model3.h5", "/content/drive/My Drive/lstm_model3.h5"],
['/content/drive/My Drive/mlp_model4.h5', "/content/drive/My Drive/simple_rnn_model4.h5", "/content/drive/My Drive/lstm_model4.h5"],
['/content/drive/My Drive/mlp_model6.h5', "/content/drive/My Drive/simple_rnn_model6.h5", "/content/drive/My Drive/lstm_model6.h5"],
['/content/drive/My Drive/mlp_model7.h5', "/content/drive/My Drive/simple_rnn_model7.h5", "/content/drive/My Drive/lstm_model7.h5"],
['/content/drive/My Drive/mlp_model8.h5', "/content/drive/My Drive/simple_rnn_model8.h5", "/content/drive/My Drive/lstm_model8.h5"],
['/content/drive/My Drive/mlp_model9.h5', "/content/drive/My Drive/simple_rnn_model9.h5", "/content/drive/My Drive/lstm_model9.h5"],
['/content/drive/My Drive/mlp_model10.h5', "/content/drive/My Drive/simple_rnn_model10.h5", "/content/drive/My Drive/lstm_model10.h5"],
['/content/drive/My Drive/mlp_model11.h5', "/content/drive/My Drive/simple_rnn_model11.h5", "/content/drive/My Drive/lstm_model11.h5"],
['/content/drive/My Drive/mlp_model12.h5', "/content/drive/My Drive/simple_rnn_model12.h5", "/content/drive/My Drive/lstm_model12.h5"]]
print(ph)
print(ph[3])
#ph1_networks = ['Paper\\Networks\\mlp_model.h5', "Paper\\Networks\\simple_rnn_model.h5", "Paper\\Networks\\lstm_model.h5", "Paper\\Networks\\gru_model.h5"]
#ph5_networks = ['Paper\\Networks\\mlp_model5.h5', "Paper\\Networks\\simple_rnn_model5.h5", "Paper\\Networks\\lstm_model5.h5"]
#ph9_networks = ['Paper\\Networks\\mlp_model9.h5', "Paper\\Networks\\simple_rnn_model9.h5", "Paper\\Networks\\lstm_model9.h5"]
def prepare_data(duration, ph):
input_data = list()
output_data = list()
for j in range(29):
file_path = f"/content/drive/My Drive/ColabNotebooks/paperwithvedadi/data(10subjects)/filtered/filtered_{j+1}.csv"
data = read_csv(file_path)
#num = len (data) - duration - ((1*i)+1)
num = len (data) - duration - ph
for k in range(num):
input_data.append([row for row in data[k:k+duration]])
#output_data.append(data[j+duration+(1*i)])
output_data.append(data[k+duration+(ph-1)])
return np.array(input_data), np.array(output_data)
for j in range(len(ph)):
input_data, output_data = prepare_data(200//4, ph[j])
input_data = np.squeeze(input_data)
train_data, test_data = split_dataset(input_data, 15)
train_label, test_label = split_dataset(output_data, 15)
n_timesteps = train_data.shape[1]
n_features = train_data.shape[2]
test_data = test_data.reshape(test_data.shape[0], n_timesteps * n_features)
NewScale = MinMaxScaler(feature_range=(0,1), copy=True)
test_data = NewScale.fit_transform(test_data)
test_label = NewScale.fit_transform(test_label)
test_data = test_data.reshape(test_data.shape[0], n_timesteps, n_features)
for i in range(len(ph_networks[j])):
#test_data, test_label = shuffle(test_data, test_label, random_state=1)
if i == 0:
#train_data = train_data.reshape(train_data.shape[0], n_timesteps * n_features)
test_data = test_data.reshape(test_data.shape[0], n_timesteps * n_features)
else:
#train_data = train_data.reshape(train_data.shape[0], n_timesteps, n_features)
test_data = test_data.reshape(test_data.shape[0], n_timesteps, n_features)
model = load_model(ph_networks[j][i])
prediction = model.predict(test_data)
print("ph = " , j, " network = ", ph_networks[j][i]," ", np.shape(prediction) )
mse_data = ((test_label[:, 8] - prediction[:, 8])**2)
ph_mse_data[j].append(mse_data)
def set_box_color(bp, color):
plt.setp(bp['boxes'], color=color)
plt.setp(bp['whiskers'], color=color)
plt.setp(bp['caps'], color=color)
plt.setp(bp['medians'], color=color)
fig, ax = plt.subplots(figsize=(30, 7))
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.set_ylabel('Squared Error')
ax.grid(color='grey', axis='y', linestyle='-', linewidth=0.25, alpha=0.5)
#ticks = ['Fully Connected', 'Simple Recurrent', 'LSTM', 'GRU']
ticks = ['Fully Connected', 'Simple Recurrent', 'LSTM']
bp1 = ax.boxplot(ph_mse_data[0], positions=np.array(range(len(ph_mse_data[0])))*6.0-2.5, sym='', widths=0.4, showfliers=True)
bp2 = ax.boxplot(ph_mse_data[1], positions=np.array(range(len(ph_mse_data[1])))*6.0-2.0, sym='', widths=0.4, showfliers=True)
bp3 = ax.boxplot(ph_mse_data[2], positions=np.array(range(len(ph_mse_data[2])))*6.0-1.5, sym='', widths=0.4, showfliers=True)
bp4 = ax.boxplot(ph_mse_data[3], positions=np.array(range(len(ph_mse_data[3])))*6.0-1.0, sym='', widths=0.4, showfliers=True)
bp6 = ax.boxplot(ph_mse_data[4], positions=np.array(range(len(ph_mse_data[4])))*6.0-0.5, sym='', widths=0.4, showfliers=True)
bp7 = ax.boxplot(ph_mse_data[5], positions=np.array(range(len(ph_mse_data[5])))*6.0, sym='', widths=0.4, showfliers=True)
bp8 = ax.boxplot(ph_mse_data[6], positions=np.array(range(len(ph_mse_data[6])))*6.0+0.5, sym='', widths=0.4, showfliers=True)
bp9 = ax.boxplot(ph_mse_data[7], positions=np.array(range(len(ph_mse_data[7])))*6.0+1.0, sym='', widths=0.4, showfliers=True)
bp10 = ax.boxplot(ph_mse_data[8], positions=np.array(range(len(ph_mse_data[8])))*6.0+1.5, sym='', widths=0.4, showfliers=True)
bp11 = ax.boxplot(ph_mse_data[9], positions=np.array(range(len(ph_mse_data[9])))*6.0+2.0, sym='', widths=0.4, showfliers=True)
bp12 = ax.boxplot(ph_mse_data[10], positions=np.array(range(len(ph_mse_data[10])))*6.0+2.5, sym='', widths=0.4, showfliers=True)
plt.xticks(range(0, len(ticks) * 2, 2), ticks)
set_box_color(bp1, '#D7191C') # colors are from http://colorbrewer2.org/
set_box_color(bp2, '#2C7BB6')
set_box_color(bp3, '#feb24c')
set_box_color(bp4, '#c51b8a') # colors are from http://colorbrewer2.org/
set_box_color(bp6, '#fa9fb5')
set_box_color(bp7, '#2b8cbe')
set_box_color(bp8, '#fc9272') # colors are from http://colorbrewer2.org/
set_box_color(bp9, '#31a354')
set_box_color(bp10, '#a1d99b')
set_box_color(bp11, '#c994c7') # colors are from http://colorbrewer2.org/
set_box_color(bp12, '#636363')
plt.plot([], c='#D7191C', label='Prediction Horizon = 1')
plt.plot([], c='#2C7BB6', label='Prediction Horizon = 2')
plt.plot([], c='#feb24c', label='Prediction Horizon = 3')
plt.plot([], c='#c51b8a', label='Prediction Horizon = 4')
plt.plot([], c='#fa9fb5', label='Prediction Horizon = 6')
plt.plot([], c='#2b8cbe', label='Prediction Horizon = 7')
plt.plot([], c='#fc9272', label='Prediction Horizon = 8')
plt.plot([], c='#31a354', label='Prediction Horizon = 9')
plt.plot([], c='#a1d99b', label='Prediction Horizon = 10')
plt.plot([], c='#c994c7', label='Prediction Horizon = 11')
plt.plot([], c='#636363', label='Prediction Horizon = 12')
#plt.legend()
plt.savefig(f'/content/drive/My Drive/ColabNotebooks/paperwithvedadi/SquaredErrorDistribution.png', dpi = 900, bbox_inches='tight')
plt.show
train_data = train_data.reshape(train_data.shape[0], n_timesteps * n_features)
test_data = test_data.reshape(test_data.shape[0], n_timesteps * n_features)
train_data = train_data.reshape(train_data.shape[0], n_timesteps , n_features)
test_data = test_data.reshape(test_data.shape[0], n_timesteps , n_features)
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_error
from keras.models import load_model
model = load_model("Paper\\Networks\\lstm_model9.h5")
#test_data, test_label = shuffle(test_data, test_label, random_state=1)
start = datetime.datetime.now()
prediction = model.predict(test_data)
end = datetime.datetime.now()
test_time = end - start
test_loss = model.evaluate(test_data, test_label)
mse = mean_squared_error(test_label[:, 8], prediction[:, 8])
r2 = r2_score(test_label[:, 8], prediction[:, 8])
print("test time is : ",test_time)
print('Test - Loss:', test_loss)
print('Test - MSE :', mse)
print('Test - R2 Score :', r2)
import matplotlib as mpl
from pylab import cm
#mpl.rcParams['font.family'] = 'Times New Roman'
plt.rcParams['font.size'] = 16
plt.rcParams['axes.linewidth'] = 2
colors = cm.get_cmap('rainbow', 2)
fig = plt.figure(figsize = [10,5])
ax = fig.add_axes([0, 0, 1, 1])
ax.spines['right'].set_visible(False)
ax.spines['top'].set_visible(False)
time = np.arange(0, 15, 15/300)
ax.plot(time, test_label[10000:13000:10, 8], label='Actual', linewidth=2, c='r', linestyle='-')
ax.plot(time, prediction[10000:13000:10, 8], label='Prediction', linewidth=2, c='b', linestyle='--')
plt.legend(frameon=False)
ax.set_xlabel('Time (s)', labelpad=10)
ax.set_ylabel('Normalized Force', labelpad=10)
ax.set_title("LSTM Network")
#ax.set_title("Fully Connected Network")
#ax.set_title("Simple Recurrent Network")
ax.legend(bbox_to_anchor=(1, 1), loc=0, frameon=False, fontsize=14)
plt.savefig('Paper\\PerformanceImages\\lstm_prediction9.png', dpi = 900, bbox_inches='tight')
| 0.342572 | 0.202778 |
## Imports
```
import pandas as pd
import networkx as nx
import numpy as np
import matplotlib.pyplot as plt
```
## Loading the data
```
people_df = pd.read_csv("./data/data_people_dump.zip", usecols=[1, 2, 3, 4])
people_df.drop_duplicates(subset='uuid', inplace=True)
people_df.set_index('uuid', inplace=True)
df = pd.read_csv('./data/data_survey_dump.zip')
df = df[(df.selected != 0) & (~df.uuid.isna())]
df['not_selected'] = np.where(df.selected != df.option_a, df.option_a, df.option_b)
```
## Defining the functions to create and analyze the graphs
```
def pref_graph_from_df(df_user, default_edge_params=None):
default_edge_params = default_edge_params if default_edge_params is not None else {}
g = nx.DiGraph()
for record in df_user.itertuples():
g.add_edge(record.selected, record.not_selected, **default_edge_params)
return g
def find_n_cycles(graph, max_n_cycles=100):
cyles_iterator = nx.simple_cycles(graph)
cycles = []
for __ in range(max_n_cycles):
try:
cycle = next(cyles_iterator)
cycles.append(cycle)
except StopIteration:
break
return cycles
def check_possible_inconsistencies(df_user):
g = pref_graph_from_df(df_user)
try:
nx.find_cycle(g, orientation='ignore')
return True
except nx.exception.NetworkXNoCycle:
return False
def find_inconsistencies(df_user, max_n_inconsistencies=100):
g = pref_graph_from_df(df_user)
inconsistencies = find_n_cycles(g, max_n_cycles=max_n_inconsistencies)
return pd.Series([inconsistencies])
def draw_preferences_graph(df_user, max_cycles=100):
default_edge_parameters = {'edge_line_color': 'black', 'edge_line_width': 0.3, 'weight': 1}
g = pref_graph_from_df(df_user, default_edge_parameters)
cycles = find_n_cycles(g, max_cycles)
for cycle in cycles:
cycle_shifted = cycle[1:] + cycle[:1]
paired_cycle_nodes = zip(cycle, cycle_shifted)
for start_node, end_node in paired_cycle_nodes:
cycle_edge_parameters = {'edge_line_color': 'red', 'edge_line_width': 1.5, 'weight': 0.001}
g.add_edge(start_node, end_node, **cycle_edge_parameters)
edge_line_colors = list(nx.get_edge_attributes(g, 'edge_line_color').values())
edge_line_widths = list(nx.get_edge_attributes(g, 'edge_line_width').values())
node_labels = dict(zip(g.nodes, g.nodes))
plt.figure(figsize=(20, 10))
pos = nx.spring_layout(g, weight='weight')
nx.draw_networkx_edges(g, pos, edge_color=edge_line_colors, width=edge_line_widths, arrowsize=17)
nx.draw_networkx_nodes(g, pos, node_size=600, alpha=0.5)
nx.draw_networkx_labels(g, pos, node_labels, font_weight='bold', font_size=12)
uuid = df_user.uuid.iloc[0]
plt.title(f"Preferences of {uuid} (#Inconsistencies = {len(cycles)})")
plt.show()
def count_unique_options(df_user):
all_options = pd.concat([df_user.selected, df_user.not_selected])
unique_options = all_options.unique()
return len(unique_options)
sample_uuids = df.uuid.drop_duplicates()#.sample()
sample_df = df[df.uuid.isin(sample_uuids)].copy()
gr_data = pd.DataFrame(index=sample_df.uuid.drop_duplicates())
gr_data['n_questions'] = sample_df.groupby('uuid').size()
gr_data['n_unique_options'] = sample_df.groupby('uuid').apply(count_unique_options)
gr_data['inconsistencies'] = sample_df.groupby('uuid').apply(find_inconsistencies)
gr_data['has_possible_inconsistencies'] = sample_df.groupby('uuid').apply(check_possible_inconsistencies)
gr_data['n_nonunique_options'] = gr_data.n_questions*2
gr_data['n_inconsistencies'] = gr_data.inconsistencies.apply(len)
gr_data['has_inconsistencies'] = gr_data.n_inconsistencies > 0
gr_data['options_density'] = 1 - (gr_data.n_unique_options/gr_data.n_nonunique_options)
```
## Random example
```
sample_user = df.uuid.drop_duplicates().sample().iloc[0]
user_df = df[df.uuid == sample_user]
draw_preferences_graph(user_df)
```
## Percentage of users that show intransitive preferences
```
percentage_intransitive = 100*(gr_data.has_inconsistencies.sum() / gr_data.has_possible_inconsistencies.sum())
print(f"Percentage of users who can show intransitive preferences and actualy show one or more {percentage_intransitive:.4}")
```
|
github_jupyter
|
import pandas as pd
import networkx as nx
import numpy as np
import matplotlib.pyplot as plt
people_df = pd.read_csv("./data/data_people_dump.zip", usecols=[1, 2, 3, 4])
people_df.drop_duplicates(subset='uuid', inplace=True)
people_df.set_index('uuid', inplace=True)
df = pd.read_csv('./data/data_survey_dump.zip')
df = df[(df.selected != 0) & (~df.uuid.isna())]
df['not_selected'] = np.where(df.selected != df.option_a, df.option_a, df.option_b)
def pref_graph_from_df(df_user, default_edge_params=None):
default_edge_params = default_edge_params if default_edge_params is not None else {}
g = nx.DiGraph()
for record in df_user.itertuples():
g.add_edge(record.selected, record.not_selected, **default_edge_params)
return g
def find_n_cycles(graph, max_n_cycles=100):
cyles_iterator = nx.simple_cycles(graph)
cycles = []
for __ in range(max_n_cycles):
try:
cycle = next(cyles_iterator)
cycles.append(cycle)
except StopIteration:
break
return cycles
def check_possible_inconsistencies(df_user):
g = pref_graph_from_df(df_user)
try:
nx.find_cycle(g, orientation='ignore')
return True
except nx.exception.NetworkXNoCycle:
return False
def find_inconsistencies(df_user, max_n_inconsistencies=100):
g = pref_graph_from_df(df_user)
inconsistencies = find_n_cycles(g, max_n_cycles=max_n_inconsistencies)
return pd.Series([inconsistencies])
def draw_preferences_graph(df_user, max_cycles=100):
default_edge_parameters = {'edge_line_color': 'black', 'edge_line_width': 0.3, 'weight': 1}
g = pref_graph_from_df(df_user, default_edge_parameters)
cycles = find_n_cycles(g, max_cycles)
for cycle in cycles:
cycle_shifted = cycle[1:] + cycle[:1]
paired_cycle_nodes = zip(cycle, cycle_shifted)
for start_node, end_node in paired_cycle_nodes:
cycle_edge_parameters = {'edge_line_color': 'red', 'edge_line_width': 1.5, 'weight': 0.001}
g.add_edge(start_node, end_node, **cycle_edge_parameters)
edge_line_colors = list(nx.get_edge_attributes(g, 'edge_line_color').values())
edge_line_widths = list(nx.get_edge_attributes(g, 'edge_line_width').values())
node_labels = dict(zip(g.nodes, g.nodes))
plt.figure(figsize=(20, 10))
pos = nx.spring_layout(g, weight='weight')
nx.draw_networkx_edges(g, pos, edge_color=edge_line_colors, width=edge_line_widths, arrowsize=17)
nx.draw_networkx_nodes(g, pos, node_size=600, alpha=0.5)
nx.draw_networkx_labels(g, pos, node_labels, font_weight='bold', font_size=12)
uuid = df_user.uuid.iloc[0]
plt.title(f"Preferences of {uuid} (#Inconsistencies = {len(cycles)})")
plt.show()
def count_unique_options(df_user):
all_options = pd.concat([df_user.selected, df_user.not_selected])
unique_options = all_options.unique()
return len(unique_options)
sample_uuids = df.uuid.drop_duplicates()#.sample()
sample_df = df[df.uuid.isin(sample_uuids)].copy()
gr_data = pd.DataFrame(index=sample_df.uuid.drop_duplicates())
gr_data['n_questions'] = sample_df.groupby('uuid').size()
gr_data['n_unique_options'] = sample_df.groupby('uuid').apply(count_unique_options)
gr_data['inconsistencies'] = sample_df.groupby('uuid').apply(find_inconsistencies)
gr_data['has_possible_inconsistencies'] = sample_df.groupby('uuid').apply(check_possible_inconsistencies)
gr_data['n_nonunique_options'] = gr_data.n_questions*2
gr_data['n_inconsistencies'] = gr_data.inconsistencies.apply(len)
gr_data['has_inconsistencies'] = gr_data.n_inconsistencies > 0
gr_data['options_density'] = 1 - (gr_data.n_unique_options/gr_data.n_nonunique_options)
sample_user = df.uuid.drop_duplicates().sample().iloc[0]
user_df = df[df.uuid == sample_user]
draw_preferences_graph(user_df)
percentage_intransitive = 100*(gr_data.has_inconsistencies.sum() / gr_data.has_possible_inconsistencies.sum())
print(f"Percentage of users who can show intransitive preferences and actualy show one or more {percentage_intransitive:.4}")
| 0.367043 | 0.685167 |
<!--NOTEBOOK_HEADER-->
*This notebook contains material from [PyRosetta](https://RosettaCommons.github.io/PyRosetta.notebooks);
content is available [on Github](https://github.com/RosettaCommons/PyRosetta.notebooks.git).*
<!--NAVIGATION-->
< [Modeling Membrane Proteins](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/15.00-Modeling-Membrane-Proteins.ipynb) | [Contents](toc.ipynb) | [Index](index.ipynb) | [Predicting the ∆∆G of single point mutations](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/15.02-Membrane-Protein-ddG-of-mutation.ipynb) ><p><a href="https://colab.research.google.com/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/15.01-Accounting-for-the-lipid-bilayer.ipynb"><img align="left" src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab" title="Open in Google Colaboratory"></a>
# Setting up a membrane protein in the bilayer
Keywords: membrane, bilayer, AddMembraneMover, OCTOPUS
## Getting Started: Setting up the protein in the lipid bilayer
To start modeling membrane proteins, we must place the protein in the lipid bilayer. This begs an important question: how should the protein be oriented? The orientation of a protein in the bilayer is driven by a number of biophysical factors, such as burying nonpolar side chains in the hydrophobic membrane. For RosettaMP, there are three ways to choose the initial orientation. The choice is up to you, and often depends on how much information you have about your protein beforehand.
```
# Notebook setup
import sys
if 'google.colab' in sys.modules:
!pip install pyrosettacolabsetup
import pyrosettacolabsetup
pyrosettacolabsetup.setup()
print ("Notebook is set for PyRosetta use in Colab. Have fun!")
from pyrosetta import *
pyrosetta.init()
```
Make sure you are in the right directory for accessing the `.pdb` files:
`cd google_drive/My\ Drive/student-notebooks/`
```
#cd google_drive/My\ Drive/student-notebooks/
```
### Option 1: Download a pre-transformed PDB from the OPM database
```
from pyrosetta.toolbox import cleanATOM
cleanATOM("inputs/1afo.pdb")
pose = pose_from_pdb("inputs/1afo.clean.pdb")
```
Then, initialize RosettaMP using AddMembraneMover. In this option, the orientation is known and you can estimate the transmembrane spans from the orientation. Therefore, we tell RosettaMP to estimate the spanning topology from structure:
```
from pyrosetta.rosetta.protocols.membrane import *
addmem = AddMembraneMover("from_structure")
addmem.apply(pose)
```
### Option 2: Estimate the transmembrane spans and use this information to choose an orientation
In this option, you will need to figure out what the transmembrane spans are. For this, you can used a sequence-based server such as OCTOPUS (http://octopus.cbr.su.se ). You will need to find the sequence of 1AFO on the PDB, copy/paste the sequence of one of the chains into OCTOPUS, and then save the output as a text file. Then, you will need to convert the output from OCTOPUS to the Rosetta format using the `octopus2memb` script.
Next, initialize RosettaMP with AddMembraneMover. Here, instead of specifying “from_structure”, you will specify the path to your spanning topology file:
```
from pyrosetta.rosetta.protocols.membrane import *
if not os.getenv("DEBUG"):
addmem = AddMembraneMover("inputs/1afo.span")
addmem.apply(pose)
```
## Key Concepts for the membrane representation
1. AddMembraneMover adds an additional residue to the protein called the Membrane residue. It is not a physical residue, but it contains information about the membrane. Note that AddMembraneMover attaches the MEM residue to the protein in Rosetta’s representation, but it does not physically exist as a residue. This is a special kind of connection called a “jump edge” whereas connections between the actual residues like are called “peptide edges” (more on that in the fold tree section).
2. The spanning information is stored in a SpanningTopology object
Let’s check some information about our current pose:
print(pose.conformation())
print(pose.conformation().membrane_info())
pose.conformation() shows information about all residues in the pose, fold_tree() shows information about the Edges of the FoldTree, and membrane_info() shows information about the membrane residue.
```
if not os.getenv("DEBUG"):
### BEGIN SOLUTION
print(pose.conformation())
print(pose.conformation().membrane_info())
### END SOLUTION
```
**Questions:**
How many residues compose 1AFO? ___
Which residue is the Membrane residue? ___
How many transmembrane spans does membrane_info() say there are?
## Fold Tree
Understanding the fold tree is necessary to use movers that move parts of the protein with respect to other parts of the protein. For example, TiltMover requires a jump number and tilts the section after the jump number by a specified amount. SpinAroundPartnerMover spins one partner with respect to another, which also requires a jump number. We will explain the terminology shortly! Enter this code in the Python command line:
`print(pose.conformation().fold_tree())`
```
if not os.getenv("DEBUG"):
### BEGIN SOLUTION
print(pose.conformation().fold_tree())
### END SOLUTION
```
1AFO is a relatively simple protein with 2 chains, however PyMOL shows 3 chains. Next to the “1AFO_AB.pdb” line in PyMOL, click “label” and then “chains”. Select Chain C, then select “label” and then “residue name”. What is the only residue in Chain C, and therefore what does the third chain represent? Does it make sense that Chain C is the membrane representation and not physically part of the protein?
This information is shown in the fold tree data above, where we see one jump edge between residues 1 and 41, and a second jump edge for the membrane representation connecting MEM “residue” 81 to residue 1. Jump edges have a positive final number which increments for each jump. The edges with a negative final number indicate a peptide edge. **Jump edges represent parts of the protein that are not physically connected to each other, and peptide edges represent parts that are physically connected.**
Edge 1 40 -1 means that the edge connects residue 1 to residue 40, and it’s a physical connection. Therefore what does this Edge represent? **It represents Chain A.**
Edge 1 41 1 means that there is a *physical separation* between residues 1 and 41. Therefore what does this Edge represent? **It represents the separation between Chain A and Chain B.**
For a more in-depth review of fold trees, look at Rosetta documentation (https://www.rosettacommons.org/demos/latest/tutorials/fold_tree/fold_tree).
The key takeaway is that if we wanted to tilt one part of the protein with respect to another part of the protein, it doesn’t make sense to give TiltMover jump number 2, which is the membrane jump. It does make sense to give TiltMover jump number 1, because then we’re asking TiltMover to tilt Chain B with respect to Chain A.
<!--NAVIGATION-->
< [Modeling Membrane Proteins](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/15.00-Modeling-Membrane-Proteins.ipynb) | [Contents](toc.ipynb) | [Index](index.ipynb) | [Predicting the ∆∆G of single point mutations](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/15.02-Membrane-Protein-ddG-of-mutation.ipynb) ><p><a href="https://colab.research.google.com/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/15.01-Accounting-for-the-lipid-bilayer.ipynb"><img align="left" src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab" title="Open in Google Colaboratory"></a>
|
github_jupyter
|
# Notebook setup
import sys
if 'google.colab' in sys.modules:
!pip install pyrosettacolabsetup
import pyrosettacolabsetup
pyrosettacolabsetup.setup()
print ("Notebook is set for PyRosetta use in Colab. Have fun!")
from pyrosetta import *
pyrosetta.init()
#cd google_drive/My\ Drive/student-notebooks/
from pyrosetta.toolbox import cleanATOM
cleanATOM("inputs/1afo.pdb")
pose = pose_from_pdb("inputs/1afo.clean.pdb")
from pyrosetta.rosetta.protocols.membrane import *
addmem = AddMembraneMover("from_structure")
addmem.apply(pose)
from pyrosetta.rosetta.protocols.membrane import *
if not os.getenv("DEBUG"):
addmem = AddMembraneMover("inputs/1afo.span")
addmem.apply(pose)
if not os.getenv("DEBUG"):
### BEGIN SOLUTION
print(pose.conformation())
print(pose.conformation().membrane_info())
### END SOLUTION
if not os.getenv("DEBUG"):
### BEGIN SOLUTION
print(pose.conformation().fold_tree())
### END SOLUTION
| 0.206454 | 0.958924 |
# How do I add members to one my _projects_?
### Overview
Here we focus on _adding a member_ to one of your projects. Importantly, you **must** be the admin of this project.
### Prerequisites
1. You need to be a member (or owner) of _at least one_ project.
2. You need your _authentication token_ and the API needs to know about it. See <a href="Setup_API_environment.ipynb">**Setup_API_environment.ipynb**</a> for details.
3. You understand how to <a href="projects_listAll.ipynb" target="_blank">list</a> projects you are a member of (we will just use that call directly here).
## Imports
We import the _Api_ class from the official sevenbridges-python bindings below.
```
import sevenbridges as sbg
```
## Initialize the object
The `Api` object needs to know your **auth\_token** and the correct path. Here we assume you are using the credentials file in your home directory. For other options see <a href="Setup_API_environment.ipynb">Setup_API_environment.ipynb</a>
```
# [USER INPUT] specify credentials file profile {cgc, sbg, default}
prof = 'default'
config_file = sbg.Config(profile=prof)
api = sbg.Api(config=config_file)
```
## List all your projects
We start by listing all of your projects, then get more information on the first one.
```
# [USER INPUT] Set project name here to add members to:
# Note that you can have multiple apps or projects with the same name. It is best practice to reference entities by ID.
project_name = 'MAL'
# check if this project already exists. LIST all projects and check for name match
my_project = api.projects.query(name=project_name)
if not my_project: # exploit fact that empty list is False
print('Target project ({}) not found, please check spelling'.format(project_name))
raise KeyboardInterrupt
else:
my_project = my_project[0]
my_project = api.projects.get(id=my_project.id)
print('You have selected project {}.'.format(my_project.name))
if hasattr(my_project, 'description'):
# Need to check if description has been entered, GUI created project have default text,
# but it is not in the metadata.
print('Project description: {} \n'.format(my_project.description))
```
## Add members
In the list **user\_names** below, add some actual user names. Then run the script and they will be added to your project.
```
user_names =['',
'']
# here we are assigning all users in the list the same permissions, this could also be a list
user_permissions = {'write': True,
'read': True,
'copy': True,
'execute': False,
'admin': False}
for name in user_names:
my_project.add_member(user=name, permissions=user_permissions)
```
## Additional Information
Detailed documentation of this particular REST architectural style request is available [here](http://docs.sevenbridges.com/docs/add-a-member-to-a-project)
|
github_jupyter
|
import sevenbridges as sbg
# [USER INPUT] specify credentials file profile {cgc, sbg, default}
prof = 'default'
config_file = sbg.Config(profile=prof)
api = sbg.Api(config=config_file)
# [USER INPUT] Set project name here to add members to:
# Note that you can have multiple apps or projects with the same name. It is best practice to reference entities by ID.
project_name = 'MAL'
# check if this project already exists. LIST all projects and check for name match
my_project = api.projects.query(name=project_name)
if not my_project: # exploit fact that empty list is False
print('Target project ({}) not found, please check spelling'.format(project_name))
raise KeyboardInterrupt
else:
my_project = my_project[0]
my_project = api.projects.get(id=my_project.id)
print('You have selected project {}.'.format(my_project.name))
if hasattr(my_project, 'description'):
# Need to check if description has been entered, GUI created project have default text,
# but it is not in the metadata.
print('Project description: {} \n'.format(my_project.description))
user_names =['',
'']
# here we are assigning all users in the list the same permissions, this could also be a list
user_permissions = {'write': True,
'read': True,
'copy': True,
'execute': False,
'admin': False}
for name in user_names:
my_project.add_member(user=name, permissions=user_permissions)
| 0.165054 | 0.724749 |
# Changes in religious affiliation and attendance
Analysis based on data from the [CIRP Freshman Survey](https://heri.ucla.edu/cirp-freshman-survey/)
Copyright Allen Downey
[MIT License](https://en.wikipedia.org/wiki/MIT_License)
```
%matplotlib inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from utils import decorate, savefig
import statsmodels.formula.api as smf
import warnings
warnings.filterwarnings('error')
```
Read the data. Note: I transcribed these data manually from published documents, so data entry errors are possible.
```
df = pd.read_csv('freshman_survey.csv', skiprows=2, index_col='year')
df[df.columns] /= 10
df.head()
df.tail()
```
Compute time variables for regression analysis, centered on 1966 (which makes the estimated intercept more interpretable).
```
df['time'] = df.index - 1966
df['time2'] = df.time**2
```
The following functions fits a regression model and uses a permutation method to estimate uncertainty due to random sampling.
```
def make_error_model(df, y, formula, n=100):
"""Makes a model that captures sample error and residual error.
df: DataFrame
y: Series
formula: string representation of the regression model
n: number of simulations to run
returns: (fittedvalues, sample_error, total_error)
"""
# make the best fit
df['y'] = y
results = smf.ols(formula, data=df).fit()
fittedvalues = results.fittedvalues
resid = results.resid
# permute residuals and generate hypothetical fits
fits = []
for i in range(n):
df['y'] = fittedvalues + np.random.permutation(results.resid)
fake_results = smf.ols(formula, data=df).fit()
fits.append(fake_results.fittedvalues)
# compute the variance of the fits
fits = np.array(fits)
sample_var = fits.var(axis=0)
# add sample_var and the variance of the residuals
total_var = sample_var + resid.var()
# standard errors are square roots of the variances
return fittedvalues, np.sqrt(sample_var), np.sqrt(total_var)
```
Plot a region showing a confidence interval.
```
def fill_between(fittedvalues, stderr, **options):
"""Fills in the 95% confidence interval.
fittedvalues: series
stderr: standard error
"""
low = fittedvalues - 2 * stderr
high = fittedvalues + 2 * stderr
plt.fill_between(fittedvalues.index, low, high, **options)
```
Plot a line of best fit, a region showing the confidence interval of the estimate and the predictive interval.
```
def plot_model(df, y, formula, **options):
"""Run a model and plot the results.
df: DataFrame
y: Series of actual data
formula: Patsy string for the regression model
options: dictional of options used to plot the data
"""
fittedvalues, sample_error, total_error = make_error_model(
df, y, formula)
fill_between(fittedvalues, total_error, color='0.9')
fill_between(fittedvalues, sample_error, color='0.8')
fittedvalues.plot(color='0.7', label='_nolegend')
y.plot(**options)
```
Plot the fraction of respondents with no religious preference along with a quadratic model.
```
y = df['noneall']
y1 = y.loc[1966:2014]
y1
y2 = y.loc[2015:]
y2
```
Put all figures on the same x-axis for easier comparison.
```
xlim = [1965, 2022]
formula = 'y ~ time + time2'
plot_model(df, y, formula, color='C0', alpha=0.7, label='None')
y2.plot(color='C1', label='Atheist,Agnostic,None')
decorate(title='No religious preference',
xlabel='Year of survey',
ylabel='Percent',
xlim=xlim,
ylim=[0, 38])
savefig('heri.1')
```
Fitting a quadratic model to percentages is a bit nonsensical, since percentages can't exceed 1. It would probably be better to work in terms of log-odds, particularly if we are interested in forecasting what might happen after we cross the 50% line. But for now the simple model is fine.
```
ps = df.noneall / 100
odds = ps / (1-ps)
log_odds = np.log(odds)
log_odds
plot_model(df, log_odds, formula, color='C0', label='None')
decorate(xlabel='Year of survey', xlim=xlim, ylabel='Log odds')
```
Plot the fraction of students reporting attendance at religious services, along with a quadratic model.
```
attend = df['attendedall'].copy()
# I'm discarding the data point from 1966,
# which seems unreasonably low
attend[1966] = np.nan
plot_model(df, attend, formula,
color='C2', alpha=0.7,
label='_nolegend')
decorate(title='Attendance at religious services',
xlabel='Year of survey',
ylabel='Percent',
xlim=xlim,
ylim=[60, 100])
savefig('heri.3')
```
Plot the gender gap along with a quadratic model.
```
diff = df.nonemen - df.nonewomen
diff = diff.loc[1973:]
plot_model(df, diff, formula,
color='C4', alpha=1, label='_nolegend')
decorate(title='Gender gap',
xlabel='Year of survey',
ylabel='Difference (percentage points)',
xlim=xlim)
savefig('heri.2')
```
To see whether the gender gap is still increasing, we can fit a quadatic model to the most recent data.
```
diff = df.nonemen - df.nonewomen
diff = diff.loc[1986:]
plot_model(df, diff, formula, color='C4', label='Gender gap')
decorate(xlabel='Year of survey',
ylabel='Difference (percentage points)')
```
A linear model for the most recent data suggests that the gap might not be growing.
```
diff = df.nonemen - df.nonewomen
diff = diff.loc[1986:]
plot_model(df, diff, 'y ~ time', color='C4', label='Gender gap')
decorate(xlabel='Year of survey',
ylabel='Difference (percentage points)')
```
|
github_jupyter
|
%matplotlib inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from utils import decorate, savefig
import statsmodels.formula.api as smf
import warnings
warnings.filterwarnings('error')
df = pd.read_csv('freshman_survey.csv', skiprows=2, index_col='year')
df[df.columns] /= 10
df.head()
df.tail()
df['time'] = df.index - 1966
df['time2'] = df.time**2
def make_error_model(df, y, formula, n=100):
"""Makes a model that captures sample error and residual error.
df: DataFrame
y: Series
formula: string representation of the regression model
n: number of simulations to run
returns: (fittedvalues, sample_error, total_error)
"""
# make the best fit
df['y'] = y
results = smf.ols(formula, data=df).fit()
fittedvalues = results.fittedvalues
resid = results.resid
# permute residuals and generate hypothetical fits
fits = []
for i in range(n):
df['y'] = fittedvalues + np.random.permutation(results.resid)
fake_results = smf.ols(formula, data=df).fit()
fits.append(fake_results.fittedvalues)
# compute the variance of the fits
fits = np.array(fits)
sample_var = fits.var(axis=0)
# add sample_var and the variance of the residuals
total_var = sample_var + resid.var()
# standard errors are square roots of the variances
return fittedvalues, np.sqrt(sample_var), np.sqrt(total_var)
def fill_between(fittedvalues, stderr, **options):
"""Fills in the 95% confidence interval.
fittedvalues: series
stderr: standard error
"""
low = fittedvalues - 2 * stderr
high = fittedvalues + 2 * stderr
plt.fill_between(fittedvalues.index, low, high, **options)
def plot_model(df, y, formula, **options):
"""Run a model and plot the results.
df: DataFrame
y: Series of actual data
formula: Patsy string for the regression model
options: dictional of options used to plot the data
"""
fittedvalues, sample_error, total_error = make_error_model(
df, y, formula)
fill_between(fittedvalues, total_error, color='0.9')
fill_between(fittedvalues, sample_error, color='0.8')
fittedvalues.plot(color='0.7', label='_nolegend')
y.plot(**options)
y = df['noneall']
y1 = y.loc[1966:2014]
y1
y2 = y.loc[2015:]
y2
xlim = [1965, 2022]
formula = 'y ~ time + time2'
plot_model(df, y, formula, color='C0', alpha=0.7, label='None')
y2.plot(color='C1', label='Atheist,Agnostic,None')
decorate(title='No religious preference',
xlabel='Year of survey',
ylabel='Percent',
xlim=xlim,
ylim=[0, 38])
savefig('heri.1')
ps = df.noneall / 100
odds = ps / (1-ps)
log_odds = np.log(odds)
log_odds
plot_model(df, log_odds, formula, color='C0', label='None')
decorate(xlabel='Year of survey', xlim=xlim, ylabel='Log odds')
attend = df['attendedall'].copy()
# I'm discarding the data point from 1966,
# which seems unreasonably low
attend[1966] = np.nan
plot_model(df, attend, formula,
color='C2', alpha=0.7,
label='_nolegend')
decorate(title='Attendance at religious services',
xlabel='Year of survey',
ylabel='Percent',
xlim=xlim,
ylim=[60, 100])
savefig('heri.3')
diff = df.nonemen - df.nonewomen
diff = diff.loc[1973:]
plot_model(df, diff, formula,
color='C4', alpha=1, label='_nolegend')
decorate(title='Gender gap',
xlabel='Year of survey',
ylabel='Difference (percentage points)',
xlim=xlim)
savefig('heri.2')
diff = df.nonemen - df.nonewomen
diff = diff.loc[1986:]
plot_model(df, diff, formula, color='C4', label='Gender gap')
decorate(xlabel='Year of survey',
ylabel='Difference (percentage points)')
diff = df.nonemen - df.nonewomen
diff = diff.loc[1986:]
plot_model(df, diff, 'y ~ time', color='C4', label='Gender gap')
decorate(xlabel='Year of survey',
ylabel='Difference (percentage points)')
| 0.722233 | 0.986258 |
```
# Imports
import numpy as np
import matplotlib.pyplot as plt
from keras.datasets import boston_housing
from keras import (models, layers)
# Extract training and test samples
(train_data, train_targets), (test_data, test_targets) = boston_housing.load_data()
# Normalize data
mean = train_data.mean(axis=0)
train_data -= mean
std = train_data.std(axis=0)
train_data /= std
# Normalize test with train mean/std
test_data -= mean
test_data /= std
# Define model
def build_model():
model = models.Sequential()
model.add(layers.Dense(64, activation="relu",
input_shape=(train_data.shape[1],)
))
model.add(layers.Dense(64, activation="relu"))
model.add(layers.Dense(1))
model.compile(optimizer="rmsprop", loss="mse", metrics=["mae"])
return model
# K-fold validation
k = 4
num_val_samples = len(train_data) // k
num_epochs = 500
all_mae_histories = []
for i in range(k):
print("Processing fold {}".format(i))
val_data = train_data[i * num_val_samples : (i + 1) * num_val_samples]
val_targets = train_targets[i * num_val_samples : (i + 1) * num_val_samples]
partial_train_data = np.concatenate(
[train_data[:i * num_val_samples],
train_data[(i + 1) * num_val_samples:]],
axis=0)
partial_train_targets = np.concatenate(
[train_targets[:i * num_val_samples],
train_targets[(i + 1) * num_val_samples:]],
axis=0)
model = build_model()
history = model.fit(partial_train_data, partial_train_targets,
epochs=num_epochs, batch_size=1, verbose=0
)
mae_history = history.history["mae"]
all_mae_histories.append(mae_history)
# Build average history for epochs
average_mae_history = [
np.mean([x[i] for x in all_mae_histories]) for i in range(num_epochs)
]
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_mae_history = smooth_curve(average_mae_history[10:])
# Plot
plt.plot(range(1, len(smooth_mae_history) + 1) , smooth_mae_history)
plt.xlabel("Epochs")
plt.ylabel("Validation MAE")
plt.show()
# Train final model
model = build_model()
model.fit(train_data, train_targets,
epochs=80, batch_size=16, verbose=0)
test_mse_score, test_mae_score = model.evaluate(test_data, test_targets)
```
|
github_jupyter
|
# Imports
import numpy as np
import matplotlib.pyplot as plt
from keras.datasets import boston_housing
from keras import (models, layers)
# Extract training and test samples
(train_data, train_targets), (test_data, test_targets) = boston_housing.load_data()
# Normalize data
mean = train_data.mean(axis=0)
train_data -= mean
std = train_data.std(axis=0)
train_data /= std
# Normalize test with train mean/std
test_data -= mean
test_data /= std
# Define model
def build_model():
model = models.Sequential()
model.add(layers.Dense(64, activation="relu",
input_shape=(train_data.shape[1],)
))
model.add(layers.Dense(64, activation="relu"))
model.add(layers.Dense(1))
model.compile(optimizer="rmsprop", loss="mse", metrics=["mae"])
return model
# K-fold validation
k = 4
num_val_samples = len(train_data) // k
num_epochs = 500
all_mae_histories = []
for i in range(k):
print("Processing fold {}".format(i))
val_data = train_data[i * num_val_samples : (i + 1) * num_val_samples]
val_targets = train_targets[i * num_val_samples : (i + 1) * num_val_samples]
partial_train_data = np.concatenate(
[train_data[:i * num_val_samples],
train_data[(i + 1) * num_val_samples:]],
axis=0)
partial_train_targets = np.concatenate(
[train_targets[:i * num_val_samples],
train_targets[(i + 1) * num_val_samples:]],
axis=0)
model = build_model()
history = model.fit(partial_train_data, partial_train_targets,
epochs=num_epochs, batch_size=1, verbose=0
)
mae_history = history.history["mae"]
all_mae_histories.append(mae_history)
# Build average history for epochs
average_mae_history = [
np.mean([x[i] for x in all_mae_histories]) for i in range(num_epochs)
]
def smooth_curve(points, factor=0.9):
smoothed_points = []
for point in points:
if smoothed_points:
previous = smoothed_points[-1]
smoothed_points.append(previous * factor + point * (1 - factor))
else:
smoothed_points.append(point)
return smoothed_points
smooth_mae_history = smooth_curve(average_mae_history[10:])
# Plot
plt.plot(range(1, len(smooth_mae_history) + 1) , smooth_mae_history)
plt.xlabel("Epochs")
plt.ylabel("Validation MAE")
plt.show()
# Train final model
model = build_model()
model.fit(train_data, train_targets,
epochs=80, batch_size=16, verbose=0)
test_mse_score, test_mae_score = model.evaluate(test_data, test_targets)
| 0.781747 | 0.736306 |
## 1. User Input
```
%load_ext autoreload
%autoreload 2
api_key = "API_KEY"
rh_user = "user"
rh_pass = "pass"
demo_run = True
```
## 2. Downloading Robinhood orders
It will prompt MFA (if you have it enabled). Run again if you want refreshed Robinhood data.
```
import pandas as pd
import numpy as np
from pyrh import Robinhood
if not demo_run:
client = Robinhood(username=rh_user, password=rh_pass)
client.login()
else:
client = None
from backend.robinhood_data import RobinhoodData
rh = RobinhoodData('data/', client)
if demo_run:
demo_orders = rh.demo_orders()
demo_dividends= rh.demo_dividends()
else:
demo_orders = None
demo_dividends= None
dividends, orders, open_positions, closed_positions = rh.download(demo_orders, demo_dividends)
```
## 3. Download stock prices and market index
Rerun if you want fresh market data
```
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
from backend.market_data import download_save_market_data
market = download_save_market_data(
api_key=api_key,
symbols=orders.symbol.unique(),
start_date=orders.date.min(),
end_date=pd.Timestamp("today", tz='UTC'))
```
## 4. Portfolio Models
```
from backend.portfolio_models import PortfolioModels
import empyrical as emp
# main calculations section
ptf = PortfolioModels('data')
summary = ptf.portfolio_summary()
stocks = ptf.stocks_risk()
df_corr, df_cov = ptf.stocks_correlation()
ptf_stats = ptf.portfolio_stats()
markowitz = ptf.markowitz_portfolios()
investment, dividends = ptf.portfolio_returns()
```
## 5. Results
### 5.1 Portfolio summary
```
summary
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
import matplotlib.dates as mdates
sns.set_style("whitegrid")
MY_DPI = 450
f, ax1 = plt.subplots(figsize=(9, 4), dpi=MY_DPI)
ax2 = ax1.twinx()
ax1.plot(investment, linewidth=1, color='#67a9cf')
ax1.axhline(y=0, color='#ca0020', linestyle='-', linewidth=0.5)
ax1.set_ylabel("Portfolio returns")
ax2.plot(dividends, linewidth=0.5, color='#ef8a62')
ax2.set_ylabel("Dividends")
# format y-axis
ax1.get_yaxis().set_major_formatter(
ticker.FuncFormatter(lambda x, p: '${:.0f}'.format(x)))
ax2.get_yaxis().set_major_formatter(
ticker.FuncFormatter(lambda x, p: '${:.0f}'.format(x)))
# format dates and grids
date_fmt = mdates.DateFormatter('%b-%Y')
ax1.xaxis.set_major_formatter(date_fmt)
ax1.grid(False, axis='both', linestyle='-', linewidth=0.5, color="#deebf7")
ax1.grid(b=None, axis='y')
ax2.grid(False, axis='both', linestyle='-', linewidth=0.5, color="#deebf7")
ax2.grid(b=None, axis='y')
plt.show()
```
### 5.2 Portfolio Stats
```
ptf_stats
```
### 5.3 Stocks performance
```
stocks
```
### 5.4 Correlations
```
import seaborn as sns
fig, ax = plt.subplots(figsize=(10,10))
sns.heatmap(ax=ax, data=df_corr, center=0, annot=True)
```
### 5.5 Markowitz
```
for l in markowitz:
print(l['name'])
print(l['weights'])
print('\n')
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
api_key = "API_KEY"
rh_user = "user"
rh_pass = "pass"
demo_run = True
import pandas as pd
import numpy as np
from pyrh import Robinhood
if not demo_run:
client = Robinhood(username=rh_user, password=rh_pass)
client.login()
else:
client = None
from backend.robinhood_data import RobinhoodData
rh = RobinhoodData('data/', client)
if demo_run:
demo_orders = rh.demo_orders()
demo_dividends= rh.demo_dividends()
else:
demo_orders = None
demo_dividends= None
dividends, orders, open_positions, closed_positions = rh.download(demo_orders, demo_dividends)
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
from backend.market_data import download_save_market_data
market = download_save_market_data(
api_key=api_key,
symbols=orders.symbol.unique(),
start_date=orders.date.min(),
end_date=pd.Timestamp("today", tz='UTC'))
from backend.portfolio_models import PortfolioModels
import empyrical as emp
# main calculations section
ptf = PortfolioModels('data')
summary = ptf.portfolio_summary()
stocks = ptf.stocks_risk()
df_corr, df_cov = ptf.stocks_correlation()
ptf_stats = ptf.portfolio_stats()
markowitz = ptf.markowitz_portfolios()
investment, dividends = ptf.portfolio_returns()
summary
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
import matplotlib.dates as mdates
sns.set_style("whitegrid")
MY_DPI = 450
f, ax1 = plt.subplots(figsize=(9, 4), dpi=MY_DPI)
ax2 = ax1.twinx()
ax1.plot(investment, linewidth=1, color='#67a9cf')
ax1.axhline(y=0, color='#ca0020', linestyle='-', linewidth=0.5)
ax1.set_ylabel("Portfolio returns")
ax2.plot(dividends, linewidth=0.5, color='#ef8a62')
ax2.set_ylabel("Dividends")
# format y-axis
ax1.get_yaxis().set_major_formatter(
ticker.FuncFormatter(lambda x, p: '${:.0f}'.format(x)))
ax2.get_yaxis().set_major_formatter(
ticker.FuncFormatter(lambda x, p: '${:.0f}'.format(x)))
# format dates and grids
date_fmt = mdates.DateFormatter('%b-%Y')
ax1.xaxis.set_major_formatter(date_fmt)
ax1.grid(False, axis='both', linestyle='-', linewidth=0.5, color="#deebf7")
ax1.grid(b=None, axis='y')
ax2.grid(False, axis='both', linestyle='-', linewidth=0.5, color="#deebf7")
ax2.grid(b=None, axis='y')
plt.show()
ptf_stats
stocks
import seaborn as sns
fig, ax = plt.subplots(figsize=(10,10))
sns.heatmap(ax=ax, data=df_corr, center=0, annot=True)
for l in markowitz:
print(l['name'])
print(l['weights'])
print('\n')
| 0.406862 | 0.593756 |
<a href="https://colab.research.google.com/github/chefdarek/DS-Unit-2-Classification-1/blob/master/DS_Sprint_Challenge_7_Classification_1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
_Lambda School Data Science, Unit 2_
# Classification 1 Sprint Challenge: Predict Steph Curry's shots 🏀
For your Sprint Challenge, you'll use a dataset with all Steph Curry's NBA field goal attempts. (Regular season and playoff games, from October 28, 2009, through June 5, 2019.)
You'll use information about the shot and the game to predict whether the shot was made. This is hard to predict! Try for an accuracy score in the high 50's or low 60's. The dataset was collected with the [nba_api](https://github.com/swar/nba_api) Python library.
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
from sklearn.preprocessing import StandardScaler
pd.set_option('display.max_rows', 500)
pd.set_option('display.max_columns', 500)
pd.set_option('display.width', 1000)
%matplotlib inline
!pip install category_encoders
import category_encoders as ce
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import QuantileTransformer
from sklearn.preprocessing import RobustScaler
from sklearn.impute import SimpleImputer
from sklearn.linear_model import LogisticRegression
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
from sklearn.pipeline import make_pipeline
import pandas as pd
url = 'https://drive.google.com/uc?export=download&id=1fL7KPyxgGYfQDsuJoBWHIWwCAf-HTFpX'
df = pd.read_csv(url, parse_dates=['game_date']).set_index('game_date')
assert df.shape == (13958, 19)
df.dtypes
df.index
df.tail()
df.isna().sum()
conts = df.select_dtypes('number')
conts.describe()
from yellowbrick.features import Rank2D
X = conts
y = df.shot_made_flag
visualizer = Rank2D(algorithm="pearson")
visualizer.fit_transform(X,y)
visualizer.poof()
cats = df.select_dtypes('object')
cats.describe()
df.describe(exclude='number').T.sort_values(by='unique')
df = df.drop(['game_id', 'game_event_id','player_name',], axis=1)
```
Baseline Predictions
```
y_train = df['shot_made_flag']
y_train.value_counts(normalize=True)
majority_class = y_train.mode()[0]
y_pred = [majority_class] * len(y_train)
#autopredict on AS
from sklearn.metrics import accuracy_score
accuracy_score(y_train, y_pred)
#predicting 47%
df.shot_made_flag.mean()
```
Test Train Validate Split
```
df_train = df['2009-10-28':'2017-9-28']
y_train = df_train['shot_made_flag']
df_train = df_train.drop('shot_made_flag', axis=1).copy()
print(df_train.shape)
df_train.head()
df_train.info()
df_val = df['2017-9-29':'2018-9-28'].copy()
y_val = df_val['shot_made_flag']
df_val = df_val.drop('shot_made_flag', axis=1)
print(df_val.shape)
df_val.info()
df_test = df['2018-9-1':]
y_test = df_test['shot_made_flag']
df_test = df_test.drop('shot_made_flag', axis=1).copy()
print(df_test.shape)
df_test.info()
catcode = [
'action_type','shot_zone_basic',
'shot_zone_area','shot_zone_range',
'htm','vtm',
]
numeric_features = df_train.select_dtypes('number').columns.tolist()
features = catcode + numeric_features
X_train_subset = df_train[features]
X_val_subset = df_val[features]
X_test = df_test[features]
```
Random Forest
```
Rf = RandomForestClassifier(n_estimators=800, n_jobs=-1)
pipeline = make_pipeline(
ce.OneHotEncoder(use_cat_names=True),
QuantileTransformer(),
IterativeImputer(),
Rf
)
# Fit on train, score on val, predict on test
pipeline.fit(X_train_subset, y_train)
print('Train Accuracy', pipeline.score(X_train_subset, y_train))
print('Validation Accuracy', pipeline.score(X_val_subset, y_val))
y_pred = pipeline.predict(X_test)
# Get feature importances
encoder = pipeline.named_steps['onehotencoder']
rf = pipeline.named_steps['randomforestclassifier']
feature_names = encoder.transform(X_train_subset).columns
importances = pd.Series(Rf.feature_importances_, feature_names)
#feature importances
n = 20
plt.figure(figsize=(10,n/2))
plt.title(f'Top {n} features')
importances.sort_values()[-n:].plot.barh(color='red');
from sklearn.metrics import confusion_matrix
confusion_matrix(y_val, y_pred[:1168])
pipeline.named_steps['randomforestclassifier'].classes_
from sklearn.utils.multiclass import unique_labels
unique_labels(y_val)
def plot_confusion_matrix(y_true, y_pred):
labels = unique_labels(y_true)
columns = [f'Predicted {label}' for label in labels]
index = [f'Actual {label}' for label in labels]
table = pd.DataFrame(confusion_matrix(y_true, y_pred),
columns=columns, index=index)
return sns.heatmap(table, annot=True, fmt='d', cmap='viridis')
plot_confusion_matrix(y_val, y_pred[:1168]);
from sklearn.metrics import classification_report
print(classification_report(y_val, y_pred[:1168]))
```
Logistic Regression
```
Lr = LogisticRegression(solver='lbfgs', multi_class='auto', max_iter=1000)
pipeline = make_pipeline(
ce.OneHotEncoder(use_cat_names=True),
QuantileTransformer(),
IterativeImputer(),
Lr
)
# Fit on train, score on val, predict on test
pipeline.fit(X_train_subset, y_train)
print('Train Accuracy', pipeline.score(X_train_subset, y_train))
print('Validation Accuracy', pipeline.score(X_val_subset, y_val))
y_pred2 = pipeline.predict(X_test)
```
This Sprint Challenge has two parts. To demonstrate mastery on each part, do all the required, numbered instructions. To earn a score of "3" for the part, also do the stretch goals.
## Part 1. Prepare to model
### Required
1. **Do train/validate/test split.** Use the 2009-10 season through 2016-17 season to train, the 2017-18 season to validate, and the 2018-19 season to test. NBA seasons begin in October and end in June. You'll know you've split the data correctly when your train set has 11081 observations, your validation set has 1168 observations, and your test set has 1709 observations.
2. **Begin with baselines for classification.** Your target to predict is `shot_made_flag`. What is the baseline accuracy for the validation set, if you guessed the majority class for every prediction?
3. **Use Ordinal Encoding _or_ One-Hot Encoding,** for the categorical features you select.
4. **Train a Random Forest _or_ Logistic Regression** with the features you select.
### Stretch goals
Engineer at least 4 of these 5 features:
- **Homecourt Advantage**: Is the home team (`htm`) the Golden State Warriors (`GSW`) ?
- **Opponent**: Who is the other team playing the Golden State Warriors?
- **Seconds remaining in the period**: Combine minutes remaining with seconds remaining, to get the total number of seconds remaining in the period.
- **Seconds remaining in the game**: Combine period, and seconds remaining in the period, to get the total number of seconds remaining in the game. A basketball game has 4 periods, each 12 minutes long.
- **Made previous shot**: Was Steph Curry's previous shot successful?
## Part 2. Evaluate models
### Required
1. Get your model's **validation accuracy.** (Multiple times if you try multiple iterations.)
2. Get your model's **test accuracy.** (One time, at the end.)
3. Get and plot your Random Forest's **feature importances** _or_ your Logistic Regression's **coefficients.**
4. Imagine this is the confusion matrix for a binary classification model. **Calculate accuracy, precision, and recall for this confusion matrix:**
<table>
<tr>
<td colspan="2" rowspan="2"></td>
<td colspan="2">Predicted</td>
</tr>
<tr>
<td>Negative</td>
<td>Positive</td>
</tr>
<tr>
<td rowspan="2">Actual</td>
<td>Negative</td>
<td style="border: solid">85</td>
<td style="border: solid">58</td>
</tr>
<tr>
<td>Positive</td>
<td style="border: solid">8</td>
<td style="border: solid"> 36</td>
</tr>
</table>
### Stretch goals
- Calculate F1 score for the provided, imaginary confusion matrix.
- Plot a real confusion matrix for your basketball model, with row and column labels.
- Print the classification report for your model.
```
#Accuracy on matrix
correct_predictions = 85 +36
total_predictions = 85 + 36 + 8 + 58
correct_predictions / total_predictions
#Recall of Positive Matrix
precision_Pos = 36
total_predictions_pos = 8+58+36
precision_Pos/total_predictions_pos
#Precision of matrix
actual_pos = 48
correct_pos = 36
correct_pos/actual_pos
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
from sklearn.preprocessing import StandardScaler
pd.set_option('display.max_rows', 500)
pd.set_option('display.max_columns', 500)
pd.set_option('display.width', 1000)
%matplotlib inline
!pip install category_encoders
import category_encoders as ce
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import QuantileTransformer
from sklearn.preprocessing import RobustScaler
from sklearn.impute import SimpleImputer
from sklearn.linear_model import LogisticRegression
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
from sklearn.pipeline import make_pipeline
import pandas as pd
url = 'https://drive.google.com/uc?export=download&id=1fL7KPyxgGYfQDsuJoBWHIWwCAf-HTFpX'
df = pd.read_csv(url, parse_dates=['game_date']).set_index('game_date')
assert df.shape == (13958, 19)
df.dtypes
df.index
df.tail()
df.isna().sum()
conts = df.select_dtypes('number')
conts.describe()
from yellowbrick.features import Rank2D
X = conts
y = df.shot_made_flag
visualizer = Rank2D(algorithm="pearson")
visualizer.fit_transform(X,y)
visualizer.poof()
cats = df.select_dtypes('object')
cats.describe()
df.describe(exclude='number').T.sort_values(by='unique')
df = df.drop(['game_id', 'game_event_id','player_name',], axis=1)
y_train = df['shot_made_flag']
y_train.value_counts(normalize=True)
majority_class = y_train.mode()[0]
y_pred = [majority_class] * len(y_train)
#autopredict on AS
from sklearn.metrics import accuracy_score
accuracy_score(y_train, y_pred)
#predicting 47%
df.shot_made_flag.mean()
df_train = df['2009-10-28':'2017-9-28']
y_train = df_train['shot_made_flag']
df_train = df_train.drop('shot_made_flag', axis=1).copy()
print(df_train.shape)
df_train.head()
df_train.info()
df_val = df['2017-9-29':'2018-9-28'].copy()
y_val = df_val['shot_made_flag']
df_val = df_val.drop('shot_made_flag', axis=1)
print(df_val.shape)
df_val.info()
df_test = df['2018-9-1':]
y_test = df_test['shot_made_flag']
df_test = df_test.drop('shot_made_flag', axis=1).copy()
print(df_test.shape)
df_test.info()
catcode = [
'action_type','shot_zone_basic',
'shot_zone_area','shot_zone_range',
'htm','vtm',
]
numeric_features = df_train.select_dtypes('number').columns.tolist()
features = catcode + numeric_features
X_train_subset = df_train[features]
X_val_subset = df_val[features]
X_test = df_test[features]
Rf = RandomForestClassifier(n_estimators=800, n_jobs=-1)
pipeline = make_pipeline(
ce.OneHotEncoder(use_cat_names=True),
QuantileTransformer(),
IterativeImputer(),
Rf
)
# Fit on train, score on val, predict on test
pipeline.fit(X_train_subset, y_train)
print('Train Accuracy', pipeline.score(X_train_subset, y_train))
print('Validation Accuracy', pipeline.score(X_val_subset, y_val))
y_pred = pipeline.predict(X_test)
# Get feature importances
encoder = pipeline.named_steps['onehotencoder']
rf = pipeline.named_steps['randomforestclassifier']
feature_names = encoder.transform(X_train_subset).columns
importances = pd.Series(Rf.feature_importances_, feature_names)
#feature importances
n = 20
plt.figure(figsize=(10,n/2))
plt.title(f'Top {n} features')
importances.sort_values()[-n:].plot.barh(color='red');
from sklearn.metrics import confusion_matrix
confusion_matrix(y_val, y_pred[:1168])
pipeline.named_steps['randomforestclassifier'].classes_
from sklearn.utils.multiclass import unique_labels
unique_labels(y_val)
def plot_confusion_matrix(y_true, y_pred):
labels = unique_labels(y_true)
columns = [f'Predicted {label}' for label in labels]
index = [f'Actual {label}' for label in labels]
table = pd.DataFrame(confusion_matrix(y_true, y_pred),
columns=columns, index=index)
return sns.heatmap(table, annot=True, fmt='d', cmap='viridis')
plot_confusion_matrix(y_val, y_pred[:1168]);
from sklearn.metrics import classification_report
print(classification_report(y_val, y_pred[:1168]))
Lr = LogisticRegression(solver='lbfgs', multi_class='auto', max_iter=1000)
pipeline = make_pipeline(
ce.OneHotEncoder(use_cat_names=True),
QuantileTransformer(),
IterativeImputer(),
Lr
)
# Fit on train, score on val, predict on test
pipeline.fit(X_train_subset, y_train)
print('Train Accuracy', pipeline.score(X_train_subset, y_train))
print('Validation Accuracy', pipeline.score(X_val_subset, y_val))
y_pred2 = pipeline.predict(X_test)
#Accuracy on matrix
correct_predictions = 85 +36
total_predictions = 85 + 36 + 8 + 58
correct_predictions / total_predictions
#Recall of Positive Matrix
precision_Pos = 36
total_predictions_pos = 8+58+36
precision_Pos/total_predictions_pos
#Precision of matrix
actual_pos = 48
correct_pos = 36
correct_pos/actual_pos
| 0.631594 | 0.909103 |
```
# 查看当前挂载的数据集目录, 该目录下的变更重启环境后会自动还原
# View dataset directory.
# This directory will be recovered automatically after resetting environment.
!ls /home/aistudio/data
# 查看工作区文件, 该目录下的变更将会持久保存. 请及时清理不必要的文件, 避免加载过慢.
# View personal work directory.
# All changes under this directory will be kept even after reset.
# Please clean unnecessary files in time to speed up environment loading.
!ls /home/aistudio/work
# 如果需要进行持久化安装, 需要使用持久化路径, 如下方代码示例:
# If a persistence installation is required,
# you need to use the persistence path as the following:
!mkdir /home/aistudio/external-libraries
!pip install beautifulsoup4 -t /home/aistudio/external-libraries
!pip install paddleseg
# 同时添加如下代码, 这样每次环境(kernel)启动的时候只要运行下方代码即可:
# Also add the following code,
# so that every time the environment (kernel) starts,
# just run the following code:
import sys
sys.path.append('/home/aistudio/external-libraries')
import paddle
from paddle.io import Dataset
from paddle.vision.transforms import Compose, Transpose, ColorJitter, vflip, Grayscale, hflip, Normalize
from PIL import Image
import numpy as np
import random
import os
# 读取数据集
class Dataloader(Dataset):
def __init__(self, model=''):
super(Dataloader).__init__()
self.image_floader = ''
if model == 'train':
self.file = 'work/train.txt'
else:
self.file = 'work/test.txt'
self.jpg_list, self.label_list= self.read_list()
def read_list(self):
data_list = []
jpg_list = []
label_list = []
with open(self.file) as lines:
for line in lines:
jpg_path = os.path.join("img_train", line.split(',')[0])
label_path = os.path.join("lab_train", line.split(',')[1].replace('\n', ''))
data_list.append((jpg_path, label_path))
random.shuffle(data_list)
for k in data_list:
jpg_list.append(k[0])
label_list.append(k[1])
return jpg_list, label_list
def _load_img(self, jpg_path, label_path):
jpg = np.array(Image.open(jpg_path))
jpg = 1 / (1 + np.exp(-((jpg - 127.5) / 127.5)))
label = Image.open(label_path)
return Compose([Transpose()])(jpg), Compose([Grayscale(), Transpose()])(label)
def __getitem__(self, idx):
train_image, label_image= self._load_img(self.jpg_list[idx], self.label_list[idx])
train_image = np.array(train_image, dtype='float32')
label_image = np.array(label_image, dtype='int64')
label_image[label_image>4]=4
zero_image=np.zeros(shape=label_image.shape)
zero_image[label_image!=4]=1
data=2-np.sum(zero_image)/256/256
label_image=np.concatenate([label_image,data*zero_image])
return train_image, label_image
def __len__(self):
return len(self.label_list)
# 计算损失函数
class LOSS_CROSS_IOU(paddle.nn.Layer):
def __init__(self, weights, num_class):
super(LOSS_CROSS_IOU, self).__init__()
self.weights_list = weights
self.num_class = num_class
def forward(self, input, label):
if(len(input)>1):
input_1 = paddle.transpose(input[1], [0, 2, 3, 1])
input = paddle.transpose(input[0], [0, 2, 3, 1])
label_1 = paddle.cast(paddle.transpose(paddle.unsqueeze(label[:,0,:,:],axis=1), [0, 2, 3, 1]),dtype="int64")
iou_loss = paddle.abs(paddle.mean(paddle.nn.functional.dice_loss(input,label_1)))
cross_loss = paddle.mean(paddle.nn.functional.softmax_with_cross_entropy(logits=input, label=label_1))+paddle.mean(paddle.nn.functional.softmax_with_cross_entropy(logits=input_1, label=label_1))
else:
input = paddle.transpose(input[0], [0, 2, 3, 1])
label_1 = paddle.cast(paddle.transpose(paddle.unsqueeze(label[:,0,:,:],axis=1), [0, 2, 3, 1]),dtype="int64")
iou_loss = paddle.abs(paddle.mean(paddle.nn.functional.dice_loss(input,label_1)))
cross_loss = paddle.mean(paddle.nn.functional.softmax_with_cross_entropy(logits=input, label=label_1))
return paddle.add(iou_loss, cross_loss)
def train():
from paddleseg.models.backbones.hrnet import HRNet_W48
from paddleseg.models.ocrnet import OCRNet
from paddleseg.models.backbones import ResNet101_vd
from paddleseg.models.deeplab import DeepLabV3P
NET=OCRNet(5,HRNet_W48(pretrained='https://bj.bcebos.com/paddleseg/dygraph/hrnet_w48_ssld.tar.gz'),(0,))
# BackBone=ResNet101_vd()
# NET = DeepLabV3P(5,backbone=BackBone,pretrained="https://bj.bcebos.com/paddleseg/dygraph/resnet101_vd_ssld.tar.gz")
epoch=20
batch_size = 48
step = 0
step2 = 0
PrintNum = 20
modelDir = "Model/"
# layer_state_dict = paddle.load("Model/3/model.pdparams")
# NET.set_state_dict(layer_state_dict)
ans_list=[]
NET.train()
# 训练测试数据集
train_dataset = Dataloader(model='train')
val_dataset = Dataloader(model='test')
train_loader = paddle.io.DataLoader(train_dataset, batch_size=batch_size, drop_last=True, shuffle=True)
val_loader = paddle.io.DataLoader(val_dataset, batch_size=batch_size, drop_last=True, shuffle=True)
# 设置学习率,设置优化器
scheduler = paddle.optimizer.lr.StepDecay(learning_rate=0.01, step_size=2, gamma=0.8, verbose=True)
sgd = paddle.optimizer.SGD(learning_rate=scheduler, parameters=NET.parameters())
loss_fn = LOSS_CROSS_IOU(0, 5)
for i in range(epoch):
losses = []
# 按Batch循环
for batch_id, data in enumerate(train_loader()):
step += 1
x_data = data[0] # 训练数据
y_data = data[1]
predicts = NET(x_data)
loss = loss_fn(predicts, y_data)
losses.append(loss.numpy())
if batch_id % PrintNum == 0:
print('AFTER ', i + 1, ' epochs', batch_id + 1, ' batch iou:', sum(losses) / len(losses))
loss.backward()
sgd.step()
# 梯度清零
sgd.clear_grad()
scheduler.step()
print('epoch iou:', sum(losses) / len(losses))
NET.eval()
with paddle.no_grad():
aiou = []
for batch_id, data in enumerate(val_loader()):
step2 += 1
x_data = data[0] # 训练数据
y_data = data[1]
predicts = NET(x_data)
loss = loss_fn(predicts, y_data)
aiou.append(loss.numpy())
if batch_id % PrintNum == 0:
print('test biou ', sum(aiou) / len(aiou))
print('test biou all', sum(aiou) / len(aiou))
ans_list.append(sum(aiou) / len(aiou))
NET.train()
if ans_list[-1] == min(ans_list):
model_path = modelDir + str(i)
paddle.save(NET.state_dict(), os.path.join(model_path, 'model.pdparams'))
paddle.save(sgd.state_dict(), os.path.join(model_path, 'model.pdopt'))
else:
print(ans_list[-1], min(ans_list), 'no save')
return 0
if __name__ == '__main__':
train()
```
请点击[此处](https://ai.baidu.com/docs#/AIStudio_Project_Notebook/a38e5576)查看本环境基本用法. <br>
Please click [here ](https://ai.baidu.com/docs#/AIStudio_Project_Notebook/a38e5576) for more detailed instructions.
```
import paddle
from paddle.io import Dataset
from paddle.vision.transforms import Compose, Transpose, ColorJitter, vflip, Grayscale, hflip, Normalize
from PIL import Image
import numpy as np
from tqdm import tqdm
import os
from paddleseg.models.backbones.hrnet import HRNet_W48
from paddleseg.models.ocrnet import OCRNet
def predict(model_path="Model/5/model.pdparams",pic_dir="img_testA",save_dir="ans"):
files=os.listdir(pic_dir)
model = OCRNet(5, HRNet_W48(pretrained=None), (0,))
layer_state_dict = paddle.load(model_path)
model.set_state_dict(layer_state_dict)
model.eval()
for i in tqdm(files):
path=os.path.join(pic_dir,i)
data=np.array(Image.open(path))
data = 1 / (1 + np.exp(-((data - 127.5) / 127.5)))
data=np.reshape(data,[1,256,256,3])
pic_data = paddle.transpose(paddle.cast(paddle.to_tensor(data), dtype='float32'), [0, 3, 1, 2])
predicts = model(pic_data)
pic = paddle.transpose(predicts[0], [0, 2, 3, 1])
ans = paddle.argmax(pic, axis=-1)
ans=ans.numpy()
ans=np.reshape(ans,[256,256])
im=Image.fromarray(ans.astype("uint8"))
im.save(os.path.join(save_dir,i.replace("jpg","png")))
return
def change(pic_dir="img_testA"):
files=os.listdir(pic_dir)
for i in tqdm(files):
path=os.path.join(pic_dir,i)
data=np.array(Image.open(path))
data[data>3]=1
im=Image.fromarray(data.astype("uint8"))
im.save(path)
if __name__ == '__main__':
# change()
# predict()
```
|
github_jupyter
|
# 查看当前挂载的数据集目录, 该目录下的变更重启环境后会自动还原
# View dataset directory.
# This directory will be recovered automatically after resetting environment.
!ls /home/aistudio/data
# 查看工作区文件, 该目录下的变更将会持久保存. 请及时清理不必要的文件, 避免加载过慢.
# View personal work directory.
# All changes under this directory will be kept even after reset.
# Please clean unnecessary files in time to speed up environment loading.
!ls /home/aistudio/work
# 如果需要进行持久化安装, 需要使用持久化路径, 如下方代码示例:
# If a persistence installation is required,
# you need to use the persistence path as the following:
!mkdir /home/aistudio/external-libraries
!pip install beautifulsoup4 -t /home/aistudio/external-libraries
!pip install paddleseg
# 同时添加如下代码, 这样每次环境(kernel)启动的时候只要运行下方代码即可:
# Also add the following code,
# so that every time the environment (kernel) starts,
# just run the following code:
import sys
sys.path.append('/home/aistudio/external-libraries')
import paddle
from paddle.io import Dataset
from paddle.vision.transforms import Compose, Transpose, ColorJitter, vflip, Grayscale, hflip, Normalize
from PIL import Image
import numpy as np
import random
import os
# 读取数据集
class Dataloader(Dataset):
def __init__(self, model=''):
super(Dataloader).__init__()
self.image_floader = ''
if model == 'train':
self.file = 'work/train.txt'
else:
self.file = 'work/test.txt'
self.jpg_list, self.label_list= self.read_list()
def read_list(self):
data_list = []
jpg_list = []
label_list = []
with open(self.file) as lines:
for line in lines:
jpg_path = os.path.join("img_train", line.split(',')[0])
label_path = os.path.join("lab_train", line.split(',')[1].replace('\n', ''))
data_list.append((jpg_path, label_path))
random.shuffle(data_list)
for k in data_list:
jpg_list.append(k[0])
label_list.append(k[1])
return jpg_list, label_list
def _load_img(self, jpg_path, label_path):
jpg = np.array(Image.open(jpg_path))
jpg = 1 / (1 + np.exp(-((jpg - 127.5) / 127.5)))
label = Image.open(label_path)
return Compose([Transpose()])(jpg), Compose([Grayscale(), Transpose()])(label)
def __getitem__(self, idx):
train_image, label_image= self._load_img(self.jpg_list[idx], self.label_list[idx])
train_image = np.array(train_image, dtype='float32')
label_image = np.array(label_image, dtype='int64')
label_image[label_image>4]=4
zero_image=np.zeros(shape=label_image.shape)
zero_image[label_image!=4]=1
data=2-np.sum(zero_image)/256/256
label_image=np.concatenate([label_image,data*zero_image])
return train_image, label_image
def __len__(self):
return len(self.label_list)
# 计算损失函数
class LOSS_CROSS_IOU(paddle.nn.Layer):
def __init__(self, weights, num_class):
super(LOSS_CROSS_IOU, self).__init__()
self.weights_list = weights
self.num_class = num_class
def forward(self, input, label):
if(len(input)>1):
input_1 = paddle.transpose(input[1], [0, 2, 3, 1])
input = paddle.transpose(input[0], [0, 2, 3, 1])
label_1 = paddle.cast(paddle.transpose(paddle.unsqueeze(label[:,0,:,:],axis=1), [0, 2, 3, 1]),dtype="int64")
iou_loss = paddle.abs(paddle.mean(paddle.nn.functional.dice_loss(input,label_1)))
cross_loss = paddle.mean(paddle.nn.functional.softmax_with_cross_entropy(logits=input, label=label_1))+paddle.mean(paddle.nn.functional.softmax_with_cross_entropy(logits=input_1, label=label_1))
else:
input = paddle.transpose(input[0], [0, 2, 3, 1])
label_1 = paddle.cast(paddle.transpose(paddle.unsqueeze(label[:,0,:,:],axis=1), [0, 2, 3, 1]),dtype="int64")
iou_loss = paddle.abs(paddle.mean(paddle.nn.functional.dice_loss(input,label_1)))
cross_loss = paddle.mean(paddle.nn.functional.softmax_with_cross_entropy(logits=input, label=label_1))
return paddle.add(iou_loss, cross_loss)
def train():
from paddleseg.models.backbones.hrnet import HRNet_W48
from paddleseg.models.ocrnet import OCRNet
from paddleseg.models.backbones import ResNet101_vd
from paddleseg.models.deeplab import DeepLabV3P
NET=OCRNet(5,HRNet_W48(pretrained='https://bj.bcebos.com/paddleseg/dygraph/hrnet_w48_ssld.tar.gz'),(0,))
# BackBone=ResNet101_vd()
# NET = DeepLabV3P(5,backbone=BackBone,pretrained="https://bj.bcebos.com/paddleseg/dygraph/resnet101_vd_ssld.tar.gz")
epoch=20
batch_size = 48
step = 0
step2 = 0
PrintNum = 20
modelDir = "Model/"
# layer_state_dict = paddle.load("Model/3/model.pdparams")
# NET.set_state_dict(layer_state_dict)
ans_list=[]
NET.train()
# 训练测试数据集
train_dataset = Dataloader(model='train')
val_dataset = Dataloader(model='test')
train_loader = paddle.io.DataLoader(train_dataset, batch_size=batch_size, drop_last=True, shuffle=True)
val_loader = paddle.io.DataLoader(val_dataset, batch_size=batch_size, drop_last=True, shuffle=True)
# 设置学习率,设置优化器
scheduler = paddle.optimizer.lr.StepDecay(learning_rate=0.01, step_size=2, gamma=0.8, verbose=True)
sgd = paddle.optimizer.SGD(learning_rate=scheduler, parameters=NET.parameters())
loss_fn = LOSS_CROSS_IOU(0, 5)
for i in range(epoch):
losses = []
# 按Batch循环
for batch_id, data in enumerate(train_loader()):
step += 1
x_data = data[0] # 训练数据
y_data = data[1]
predicts = NET(x_data)
loss = loss_fn(predicts, y_data)
losses.append(loss.numpy())
if batch_id % PrintNum == 0:
print('AFTER ', i + 1, ' epochs', batch_id + 1, ' batch iou:', sum(losses) / len(losses))
loss.backward()
sgd.step()
# 梯度清零
sgd.clear_grad()
scheduler.step()
print('epoch iou:', sum(losses) / len(losses))
NET.eval()
with paddle.no_grad():
aiou = []
for batch_id, data in enumerate(val_loader()):
step2 += 1
x_data = data[0] # 训练数据
y_data = data[1]
predicts = NET(x_data)
loss = loss_fn(predicts, y_data)
aiou.append(loss.numpy())
if batch_id % PrintNum == 0:
print('test biou ', sum(aiou) / len(aiou))
print('test biou all', sum(aiou) / len(aiou))
ans_list.append(sum(aiou) / len(aiou))
NET.train()
if ans_list[-1] == min(ans_list):
model_path = modelDir + str(i)
paddle.save(NET.state_dict(), os.path.join(model_path, 'model.pdparams'))
paddle.save(sgd.state_dict(), os.path.join(model_path, 'model.pdopt'))
else:
print(ans_list[-1], min(ans_list), 'no save')
return 0
if __name__ == '__main__':
train()
import paddle
from paddle.io import Dataset
from paddle.vision.transforms import Compose, Transpose, ColorJitter, vflip, Grayscale, hflip, Normalize
from PIL import Image
import numpy as np
from tqdm import tqdm
import os
from paddleseg.models.backbones.hrnet import HRNet_W48
from paddleseg.models.ocrnet import OCRNet
def predict(model_path="Model/5/model.pdparams",pic_dir="img_testA",save_dir="ans"):
files=os.listdir(pic_dir)
model = OCRNet(5, HRNet_W48(pretrained=None), (0,))
layer_state_dict = paddle.load(model_path)
model.set_state_dict(layer_state_dict)
model.eval()
for i in tqdm(files):
path=os.path.join(pic_dir,i)
data=np.array(Image.open(path))
data = 1 / (1 + np.exp(-((data - 127.5) / 127.5)))
data=np.reshape(data,[1,256,256,3])
pic_data = paddle.transpose(paddle.cast(paddle.to_tensor(data), dtype='float32'), [0, 3, 1, 2])
predicts = model(pic_data)
pic = paddle.transpose(predicts[0], [0, 2, 3, 1])
ans = paddle.argmax(pic, axis=-1)
ans=ans.numpy()
ans=np.reshape(ans,[256,256])
im=Image.fromarray(ans.astype("uint8"))
im.save(os.path.join(save_dir,i.replace("jpg","png")))
return
def change(pic_dir="img_testA"):
files=os.listdir(pic_dir)
for i in tqdm(files):
path=os.path.join(pic_dir,i)
data=np.array(Image.open(path))
data[data>3]=1
im=Image.fromarray(data.astype("uint8"))
im.save(path)
if __name__ == '__main__':
# change()
# predict()
| 0.357792 | 0.330188 |
Lambda School Data Science, Unit 2: Predictive Modeling
# Kaggle Challenge, Module 4
## Assignment
- [ ] If you haven't yet, [review requirements for your portfolio project](https://lambdaschool.github.io/ds/unit2), then submit your dataset.
- [ ] Plot a confusion matrix for your Tanzania Waterpumps model.
- [ ] Continue to participate in our Kaggle challenge. Every student should have made at least one submission that scores at least 60% accuracy (above the majority class baseline).
- [ ] Submit your final predictions to our Kaggle competition. Optionally, go to **My Submissions**, and _"you may select up to 1 submission to be used to count towards your final leaderboard score."_
- [ ] Commit your notebook to your fork of the GitHub repo.
- [ ] Read [Maximizing Scarce Maintenance Resources with Data: Applying predictive modeling, precision at k, and clustering to optimize impact](https://towardsdatascience.com/maximizing-scarce-maintenance-resources-with-data-8f3491133050), by Lambda DS3 student Michael Brady. His blog post extends the Tanzania Waterpumps scenario, far beyond what's in the lecture notebook.
## Stretch Goals
### Reading
- [Attacking discrimination with smarter machine learning](https://research.google.com/bigpicture/attacking-discrimination-in-ml/), by Google Research, with interactive visualizations. _"A threshold classifier essentially makes a yes/no decision, putting things in one category or another. We look at how these classifiers work, ways they can potentially be unfair, and how you might turn an unfair classifier into a fairer one. As an illustrative example, we focus on loan granting scenarios where a bank may grant or deny a loan based on a single, automatically computed number such as a credit score."_
- [Notebook about how to calculate expected value from a confusion matrix by treating it as a cost-benefit matrix](https://github.com/podopie/DAT18NYC/blob/master/classes/13-expected_value_cost_benefit_analysis.ipynb)
- [Simple guide to confusion matrix terminology](https://www.dataschool.io/simple-guide-to-confusion-matrix-terminology/) by Kevin Markham, with video
- [Visualizing Machine Learning Thresholds to Make Better Business Decisions](https://blog.insightdatascience.com/visualizing-machine-learning-thresholds-to-make-better-business-decisions-4ab07f823415)
### Doing
- [ ] Share visualizations in our Slack channel!
- [ ] RandomizedSearchCV / GridSearchCV, for model selection. (See module 3 assignment notebook)
- [ ] More Categorical Encoding. (See module 2 assignment notebook)
- [ ] Stacking Ensemble. (See below)
### Stacking Ensemble
Here's some code you can use to "stack" multiple submissions, which is another form of ensembling:
```python
import pandas as pd
# Filenames of your submissions you want to ensemble
files = ['submission-01.csv', 'submission-02.csv', 'submission-03.csv']
target = 'status_group'
submissions = (pd.read_csv(file)[[target]] for file in files)
ensemble = pd.concat(submissions, axis='columns')
majority_vote = ensemble.mode(axis='columns')[0]
sample_submission = pd.read_csv('sample_submission.csv')
submission = sample_submission.copy()
submission[target] = majority_vote
submission.to_csv('my-ultimate-ensemble-submission.csv', index=False)
```
```
%%capture
import sys
# If you're on Colab:
if 'google.colab' in sys.modules:
DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Kaggle-Challenge/master/data/'
!pip install category_encoders==2.*
# If you're working locally:
else:
DATA_PATH = '../data/'
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
# Merge train_features.csv & train_labels.csv
train = pd.merge(pd.read_csv(DATA_PATH+'waterpumps/train_features.csv'),
pd.read_csv(DATA_PATH+'waterpumps/train_labels.csv'))
# Read test_features.csv & sample_submission.csv
test = pd.read_csv(DATA_PATH+'waterpumps/test_features.csv')
sample_submission = pd.read_csv(DATA_PATH+'waterpumps/sample_submission.csv')
# (This is from a previous version of the assignment notebook)
target = 'status_group'
train, val = train_test_split(train, test_size=len(test),
stratify=train[target], random_state=42)
# Copying my earlier code
def remove_zeroes(X):
X = X.copy()
X['latitude'] = X['latitude'].replace(-2e-08, 0)
zeroes = ['gps_height', 'longitude', 'latitude', 'population', 'construction_year']
for col in zeroes:
X[col] = X[col].replace(0, np.nan)
return X
def datetime_features(X):
X = X.copy()
X['date_recorded'] = pd.to_datetime(X['date_recorded'], infer_datetime_format=True)
X['year_recorded'] = X['date_recorded'].dt.year
X['construction_year'] = X['construction_year'].fillna(np.around(np.mean(X['construction_year']), decimals=0))
X['time_to_inspection'] = X['year_recorded'] - X['construction_year']
return X
def drop_redundant(X):
X = X.copy()
redundant_cols = ['recorded_by', 'payment_type', 'region_code', 'date_recorded', 'id']
for col in redundant_cols:
X = X.drop(col, axis=1)
return X
def wrangle(X):
X = X.copy()
X = remove_zeroes(X)
X = datetime_features(X)
X = drop_redundant(X)
return X
X_train = wrangle(train).drop(target, axis=1)
y_train = train[target]
X_val = wrangle(val).drop(target, axis=1)
y_val = val[target]
X_test = wrangle(test)
import category_encoders as ce
from sklearn.impute import SimpleImputer
from sklearn.ensemble import RandomForestClassifier
from sklearn.pipeline import make_pipeline, Pipeline
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='mean'),
RandomForestClassifier(max_depth=20, max_features=0.7, n_estimators=200, random_state=99)
)
pipeline.fit(X_train, y_train)
print(pipeline.score(X_val, y_val))
from sklearn.model_selection import GridSearchCV
pipeline = Pipeline([
('encoder', ce.BinaryEncoder()),
('imputer', SimpleImputer()),
('classifier', RandomForestClassifier())
])
param_grid = {
'encoder': [ce.BinaryEncoder(), ce.OrdinalEncoder()],
'imputer__strategy': ['mean', 'median', 'most_frequent'],
'classifier__n_estimators': [200],
'classifier__max_depth': [20],
'classifier__max_features': [0.7]
}
grid = GridSearchCV(pipeline, param_grid=param_grid, scoring='accuracy', cv=5, n_jobs=-1)
grid.fit(X_train, y_train);
print('Best hyperparameters', grid.best_params_)
print('Accuracy', grid.best_score_)
pipeline = make_pipeline(
ce.BinaryEncoder(cols=None, drop_invariant=False, handle_missing='value', handle_unknown='value',
mapping=None, return_df=True, verbose=0),
SimpleImputer(strategy='most_frequent'),
RandomForestClassifier(max_depth=20, max_features=0.7, n_estimators=200, random_state=99)
)
pipeline.fit(X_train, y_train)
print(pipeline.score(X_val, y_val))
y_pred = pd.DataFrame(pipeline.predict(X_test), columns=['status_group'])
submission1 = pd.concat([test['id'], y_pred], axis=1)
submission1.to_csv('water-submission-14.csv', index=None, header=True)
pipeline = make_pipeline(
ce.BinaryEncoder(cols=None, drop_invariant=False, handle_missing='value', handle_unknown='value',
mapping=None, return_df=True, verbose=0),
SimpleImputer(strategy='median'),
RandomForestClassifier(n_estimators=200, random_state=99)
)
pipeline.fit(X_train, y_train)
print(pipeline.score(X_val, y_val))
y_pred = pd.DataFrame(pipeline.predict(X_test), columns=['status_group'])
submission2 = pd.concat([test['id'], y_pred], axis=1)
submission2.to_csv('water-submission-15.csv', index=None, header=True)
from sklearn.metrics import confusion_matrix
confusion_matrix(y_val, y_pred)
%matplotlib inline
from sklearn.utils.multiclass import unique_labels
import seaborn as sns
labels = unique_labels(y_val)
columns = [f'Predicted {label}' for label in labels]
index = [f'Actual {label}' for label in labels]
table = pd.DataFrame(confusion_matrix(y_val, y_pred), columns=columns, index=index)
sns.heatmap(table, annot=True, fmt='d', cmap='viridis');
files = ['water-submission-13.csv', 'water-submission-14.csv', 'water-submission-15.csv']
target = 'status_group'
submissions = (pd.read_csv(file)[[target]] for file in files)
ensemble = pd.concat(submissions, axis='columns')
majority_vote = ensemble.mode(axis='columns')[0]
submission = sample_submission.copy()
submission[target] = majority_vote
submission.to_csv('water-submission-16.csv', index=False)
```
|
github_jupyter
|
import pandas as pd
# Filenames of your submissions you want to ensemble
files = ['submission-01.csv', 'submission-02.csv', 'submission-03.csv']
target = 'status_group'
submissions = (pd.read_csv(file)[[target]] for file in files)
ensemble = pd.concat(submissions, axis='columns')
majority_vote = ensemble.mode(axis='columns')[0]
sample_submission = pd.read_csv('sample_submission.csv')
submission = sample_submission.copy()
submission[target] = majority_vote
submission.to_csv('my-ultimate-ensemble-submission.csv', index=False)
%%capture
import sys
# If you're on Colab:
if 'google.colab' in sys.modules:
DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Kaggle-Challenge/master/data/'
!pip install category_encoders==2.*
# If you're working locally:
else:
DATA_PATH = '../data/'
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
# Merge train_features.csv & train_labels.csv
train = pd.merge(pd.read_csv(DATA_PATH+'waterpumps/train_features.csv'),
pd.read_csv(DATA_PATH+'waterpumps/train_labels.csv'))
# Read test_features.csv & sample_submission.csv
test = pd.read_csv(DATA_PATH+'waterpumps/test_features.csv')
sample_submission = pd.read_csv(DATA_PATH+'waterpumps/sample_submission.csv')
# (This is from a previous version of the assignment notebook)
target = 'status_group'
train, val = train_test_split(train, test_size=len(test),
stratify=train[target], random_state=42)
# Copying my earlier code
def remove_zeroes(X):
X = X.copy()
X['latitude'] = X['latitude'].replace(-2e-08, 0)
zeroes = ['gps_height', 'longitude', 'latitude', 'population', 'construction_year']
for col in zeroes:
X[col] = X[col].replace(0, np.nan)
return X
def datetime_features(X):
X = X.copy()
X['date_recorded'] = pd.to_datetime(X['date_recorded'], infer_datetime_format=True)
X['year_recorded'] = X['date_recorded'].dt.year
X['construction_year'] = X['construction_year'].fillna(np.around(np.mean(X['construction_year']), decimals=0))
X['time_to_inspection'] = X['year_recorded'] - X['construction_year']
return X
def drop_redundant(X):
X = X.copy()
redundant_cols = ['recorded_by', 'payment_type', 'region_code', 'date_recorded', 'id']
for col in redundant_cols:
X = X.drop(col, axis=1)
return X
def wrangle(X):
X = X.copy()
X = remove_zeroes(X)
X = datetime_features(X)
X = drop_redundant(X)
return X
X_train = wrangle(train).drop(target, axis=1)
y_train = train[target]
X_val = wrangle(val).drop(target, axis=1)
y_val = val[target]
X_test = wrangle(test)
import category_encoders as ce
from sklearn.impute import SimpleImputer
from sklearn.ensemble import RandomForestClassifier
from sklearn.pipeline import make_pipeline, Pipeline
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='mean'),
RandomForestClassifier(max_depth=20, max_features=0.7, n_estimators=200, random_state=99)
)
pipeline.fit(X_train, y_train)
print(pipeline.score(X_val, y_val))
from sklearn.model_selection import GridSearchCV
pipeline = Pipeline([
('encoder', ce.BinaryEncoder()),
('imputer', SimpleImputer()),
('classifier', RandomForestClassifier())
])
param_grid = {
'encoder': [ce.BinaryEncoder(), ce.OrdinalEncoder()],
'imputer__strategy': ['mean', 'median', 'most_frequent'],
'classifier__n_estimators': [200],
'classifier__max_depth': [20],
'classifier__max_features': [0.7]
}
grid = GridSearchCV(pipeline, param_grid=param_grid, scoring='accuracy', cv=5, n_jobs=-1)
grid.fit(X_train, y_train);
print('Best hyperparameters', grid.best_params_)
print('Accuracy', grid.best_score_)
pipeline = make_pipeline(
ce.BinaryEncoder(cols=None, drop_invariant=False, handle_missing='value', handle_unknown='value',
mapping=None, return_df=True, verbose=0),
SimpleImputer(strategy='most_frequent'),
RandomForestClassifier(max_depth=20, max_features=0.7, n_estimators=200, random_state=99)
)
pipeline.fit(X_train, y_train)
print(pipeline.score(X_val, y_val))
y_pred = pd.DataFrame(pipeline.predict(X_test), columns=['status_group'])
submission1 = pd.concat([test['id'], y_pred], axis=1)
submission1.to_csv('water-submission-14.csv', index=None, header=True)
pipeline = make_pipeline(
ce.BinaryEncoder(cols=None, drop_invariant=False, handle_missing='value', handle_unknown='value',
mapping=None, return_df=True, verbose=0),
SimpleImputer(strategy='median'),
RandomForestClassifier(n_estimators=200, random_state=99)
)
pipeline.fit(X_train, y_train)
print(pipeline.score(X_val, y_val))
y_pred = pd.DataFrame(pipeline.predict(X_test), columns=['status_group'])
submission2 = pd.concat([test['id'], y_pred], axis=1)
submission2.to_csv('water-submission-15.csv', index=None, header=True)
from sklearn.metrics import confusion_matrix
confusion_matrix(y_val, y_pred)
%matplotlib inline
from sklearn.utils.multiclass import unique_labels
import seaborn as sns
labels = unique_labels(y_val)
columns = [f'Predicted {label}' for label in labels]
index = [f'Actual {label}' for label in labels]
table = pd.DataFrame(confusion_matrix(y_val, y_pred), columns=columns, index=index)
sns.heatmap(table, annot=True, fmt='d', cmap='viridis');
files = ['water-submission-13.csv', 'water-submission-14.csv', 'water-submission-15.csv']
target = 'status_group'
submissions = (pd.read_csv(file)[[target]] for file in files)
ensemble = pd.concat(submissions, axis='columns')
majority_vote = ensemble.mode(axis='columns')[0]
submission = sample_submission.copy()
submission[target] = majority_vote
submission.to_csv('water-submission-16.csv', index=False)
| 0.411229 | 0.910346 |
<a href="https://colab.research.google.com/github/kwbt-kzk/github-slideshow/blob/main/Untitled2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
# Problem 1
import os
import geopandas as gpd
import matplotlib.pyplot as plt
from shapely.geometry import Polygon
longitudes = [29.99671173095703, 31.58196258544922, 27.738052368164062, 26.50013542175293, 26.652359008789062, 25.921663284301758, 22.90027618408203, 23.257217407226562,
23.335693359375, 22.87444305419922, 23.08465003967285, 22.565473556518555, 21.452774047851562, 21.66388702392578, 21.065969467163086, 21.67659568786621,
21.496871948242188, 22.339998245239258, 22.288192749023438, 24.539581298828125, 25.444232940673828, 25.303749084472656, 24.669166564941406, 24.689163208007812,
24.174999237060547, 23.68471908569336, 24.000761032104492, 23.57332992553711, 23.76513671875, 23.430830001831055, 23.6597900390625, 20.580928802490234, 21.320831298828125,
22.398330688476562, 23.97638702392578, 24.934917449951172, 25.7611083984375, 25.95930290222168, 26.476804733276367, 27.91069221496582, 29.1027774810791, 29.29846954345703,
28.4355525970459, 28.817358016967773, 28.459857940673828, 30.028610229492188, 29.075136184692383, 30.13492774963379, 29.818885803222656, 29.640830993652344, 30.57735824584961,
29.99671173095703]
latitudes = [63.748023986816406, 62.90789794921875, 60.511383056640625, 60.44499588012695, 60.646385192871094, 60.243743896484375, 59.806800842285156, 59.91944122314453,
60.02395248413086, 60.14555358886719, 60.3452033996582, 60.211936950683594, 60.56249237060547, 61.54027557373047, 62.59798049926758, 63.02013397216797,
63.20353698730469, 63.27652359008789, 63.525691986083984, 64.79915618896484, 64.9533920288086, 65.51513671875, 65.65470886230469, 65.89610290527344, 65.79151916503906,
66.26332092285156, 66.80228424072266, 67.1570053100586, 67.4168701171875, 67.47978210449219, 67.94589233398438, 69.060302734375, 69.32611083984375, 68.71110534667969,
68.83248901367188, 68.580810546875, 68.98916625976562, 69.68568420410156, 69.9363784790039, 70.08860778808594, 69.70597076416016, 69.48533630371094, 68.90263366699219,
68.84700012207031, 68.53485107421875, 67.69471740722656, 66.90360260009766, 65.70887756347656, 65.6533203125, 64.92096710205078, 64.22373962402344, 63.748023986816406]
coordpairs=None
coordpairs=[]
for poi in range(len(longitudes)):
coordpairs.append([longitudes[poi],latitudes[poi]])
poly = None
poly=Polygon(coordpairs)
print(coordpairs[0])
print(poly.geom_type)
geo=gpd.GeoDataFrame()
geo['geometry'] = None
geo.at['geometry'] = poly
print(geo.head())
print(len(geo))
geo.plot()
fp = 'polygon.shp'
geo.to_file(fp)
assert os.path.isfile(fp), "Output file does not exits."
# Problem 2
def func1():
return len(coordpairs)
def func2():
return poly.geom_type
def func3():
return geo
def func4():
return geo
# Problem 2
import pandas as pd
import geopandas as gpd
from shapely.geometry import Point
data = None
data = pd.read_csv('data/some_posts.csv')
Points = lambda row:Point(row['lat'],row['lon'])
data['geometry'] = data.apply(Points, axis=1)
print("Number of rows:", len(data))
print(data['geometry'].head())
import geopandas as gpd
from pyproj import CRS
geo=None
geo = gpd.GeoDataFrame(data, geometry='geometry',crs=CRS.from_epsg(4326).to_wkt())
print("Number of rows:", len(geo))
print(geo.head())
fp = 'Kruger_posts.shp'
geo.to_file(fp)
import os
assert os.path.isfile(fp), "output shapefile does not exist"
geo.plot()
def func5():
return data
def func6():
return geo
```
|
github_jupyter
|
# Problem 1
import os
import geopandas as gpd
import matplotlib.pyplot as plt
from shapely.geometry import Polygon
longitudes = [29.99671173095703, 31.58196258544922, 27.738052368164062, 26.50013542175293, 26.652359008789062, 25.921663284301758, 22.90027618408203, 23.257217407226562,
23.335693359375, 22.87444305419922, 23.08465003967285, 22.565473556518555, 21.452774047851562, 21.66388702392578, 21.065969467163086, 21.67659568786621,
21.496871948242188, 22.339998245239258, 22.288192749023438, 24.539581298828125, 25.444232940673828, 25.303749084472656, 24.669166564941406, 24.689163208007812,
24.174999237060547, 23.68471908569336, 24.000761032104492, 23.57332992553711, 23.76513671875, 23.430830001831055, 23.6597900390625, 20.580928802490234, 21.320831298828125,
22.398330688476562, 23.97638702392578, 24.934917449951172, 25.7611083984375, 25.95930290222168, 26.476804733276367, 27.91069221496582, 29.1027774810791, 29.29846954345703,
28.4355525970459, 28.817358016967773, 28.459857940673828, 30.028610229492188, 29.075136184692383, 30.13492774963379, 29.818885803222656, 29.640830993652344, 30.57735824584961,
29.99671173095703]
latitudes = [63.748023986816406, 62.90789794921875, 60.511383056640625, 60.44499588012695, 60.646385192871094, 60.243743896484375, 59.806800842285156, 59.91944122314453,
60.02395248413086, 60.14555358886719, 60.3452033996582, 60.211936950683594, 60.56249237060547, 61.54027557373047, 62.59798049926758, 63.02013397216797,
63.20353698730469, 63.27652359008789, 63.525691986083984, 64.79915618896484, 64.9533920288086, 65.51513671875, 65.65470886230469, 65.89610290527344, 65.79151916503906,
66.26332092285156, 66.80228424072266, 67.1570053100586, 67.4168701171875, 67.47978210449219, 67.94589233398438, 69.060302734375, 69.32611083984375, 68.71110534667969,
68.83248901367188, 68.580810546875, 68.98916625976562, 69.68568420410156, 69.9363784790039, 70.08860778808594, 69.70597076416016, 69.48533630371094, 68.90263366699219,
68.84700012207031, 68.53485107421875, 67.69471740722656, 66.90360260009766, 65.70887756347656, 65.6533203125, 64.92096710205078, 64.22373962402344, 63.748023986816406]
coordpairs=None
coordpairs=[]
for poi in range(len(longitudes)):
coordpairs.append([longitudes[poi],latitudes[poi]])
poly = None
poly=Polygon(coordpairs)
print(coordpairs[0])
print(poly.geom_type)
geo=gpd.GeoDataFrame()
geo['geometry'] = None
geo.at['geometry'] = poly
print(geo.head())
print(len(geo))
geo.plot()
fp = 'polygon.shp'
geo.to_file(fp)
assert os.path.isfile(fp), "Output file does not exits."
# Problem 2
def func1():
return len(coordpairs)
def func2():
return poly.geom_type
def func3():
return geo
def func4():
return geo
# Problem 2
import pandas as pd
import geopandas as gpd
from shapely.geometry import Point
data = None
data = pd.read_csv('data/some_posts.csv')
Points = lambda row:Point(row['lat'],row['lon'])
data['geometry'] = data.apply(Points, axis=1)
print("Number of rows:", len(data))
print(data['geometry'].head())
import geopandas as gpd
from pyproj import CRS
geo=None
geo = gpd.GeoDataFrame(data, geometry='geometry',crs=CRS.from_epsg(4326).to_wkt())
print("Number of rows:", len(geo))
print(geo.head())
fp = 'Kruger_posts.shp'
geo.to_file(fp)
import os
assert os.path.isfile(fp), "output shapefile does not exist"
geo.plot()
def func5():
return data
def func6():
return geo
| 0.198608 | 0.838548 |
```
import pandas as pd
from bs4 import BeautifulSoup as bs
from pprint import pprint
import pymongo
import requests
from splinter import Browser
```
## NASA Mars News
* Scrape the NASA Mars News Site and collect the latest News Title and Paragraph Text. Assign the text to variables that you can reference later.
```
executable_path = {'executable_path': '/Users/ekaster/Documents/DATABOOTCAMP-MATERIAL/chromedriver.exe'}
browser = Browser('chrome', **executable_path, headless=False)
mars_news_url = 'https://mars.nasa.gov/news/'
browser.visit(mars_news_url)
response = requests.get(mars_news_url)
news_soup = bs(response.text, 'html.parser')
news_title = news_soup.find("div", class_="content_title").text
news_p = news_soup.find("div", class_="rollover_description_inner").text
print(news_title)
print(news_p)
```
## JPL Mars Space Images - Featured Image
* Visit the url for JPL Featured Space Image here.
* Use splinter to navigate the site and find the image url for the current Featured Mars Image and assign the url string to a variable called featured_image_url.
* Make sure to find the image url to the full size .jpg image.
* Make sure to save a complete url string for this image.
```
executable_path = {'executable_path' : '/Users/ekaster/Documents/DATABOOTCAMP-MATERIAL/chromedriver.exe'}
browser = Browser('chrome', **executable_path, headless=False)
mars_img_url = "https://www.jpl.nasa.gov/spaceimages/?search=&category=Mars"
browser.visit(mars_img_url)
browser.click_link_by_partial_text('FULL IMAGE')
browser.click_link_by_partial_text('more info')
browser_soup = bs(browser.html, 'html.parser')
get_img_url = browser_soup.find('img', class_='main_image')
img_src_url = get_img_url.get('src')
featured_image_url = "https://www.jpl.nasa.gov" + img_src_url
print(featured_image_url)
```
## Mars Weather
* Visit the Mars Weather twitter account here and scrape the latest Mars weather tweet from the page. Save the tweet text for the weather report as a variable called mars_weather.
* Note: Be sure you are not signed in to twitter, or scraping may become more difficult.
* Note: Twitter frequently changes how information is presented on their website. If you are having difficulty getting the correct html tag data, consider researching Regular Expression Patterns and how they can be used in combination with the .find() method.
```
executable_path = {'executable_path' : '/Users/ekaster/Documents/DATABOOTCAMP-MATERIAL/chromedriver.exe'}
browser = Browser('chrome', **executable_path, headless=False)
mars_twitter_url = 'https://twitter.com/marswxreport?lang=en'
browser.visit(mars_twitter_url)
twitter_soup = bs(browser.html, 'html.parser')
print(twitter_soup.prettify())
latest_tweets = twitter_soup.find('div', class_='css-901oao r-hkyrab r-1qd0xha r-a023e6 r-16dba41 r-ad9z0x r-bcqeeo r-bnwqim r-qvutc0').text
print(latest_tweets)
```
## Mars Facts
* Visit the Mars Facts webpage here and use Pandas to scrape the table containing facts about the planet including Diameter, Mass, etc.
* Use Pandas to convert the data to a HTML table string.
```
request_mars_space_facts = requests.get("https://space-facts.com/mars/")
mars_space_table_read = pd.read_html(request_mars_space_facts.text)
mars_space_table_read
mars_df = mars_space_table_read[0]
mars_df.columns = ['Description','Value']
mars_df.set_index(['Description'], inplace=True)
mars_df
mars_data_html = mars_df.to_html()
mars_data_html
mars_data_html.replace('\n', '')
```
## Mars Hemispheres
* Visit the USGS Astrogeology site here to obtain high resolution images for each of Mar's hemispheres.
* You will need to click each of the links to the hemispheres in order to find the image url to the full resolution image.
* Save both the image url string for the full resolution hemisphere image, and the Hemisphere title containing the hemisphere name. Use a Python dictionary to store the data using the keys img_url and title.
* Append the dictionary with the image url string and the hemisphere title to a list. This list will contain one dictionary for each hemisphere.
```
executable_path = {'executable_path': '/Users/ekaster/Documents/DATABOOTCAMP-MATERIAL/chromedriver.exe'}
browser = Browser('chrome', **executable_path, headless=False)
mars_hemi_url = 'https://astrogeology.usgs.gov/search/results?q=hemisphere+enhanced&k1=target&v1=Mars'
browser.visit(mars_hemi_url)
mars_hemi_html = browser.html
mars_hemi_soup = bs(mars_hemi_html, 'html.parser')
items = mars_hemi_soup.find_all('div', class_='item')
mars_hemi_img_url = []
mars_hemi_main_url = 'https://astrogeology.usgs.gov'
for i in items:
title = i.find('h3').text
partial_img_url = i.find('a', class_='itemLink product-item')['href']
browser.visit(mars_hemi_main_url + partial_img_url)
partial_img_html = browser.html
soup = bs( partial_img_html, 'html.parser')
img_url = mars_hemi_main_url + soup.find('img', class_='wide-image')['src']
mars_hemi_img_url.append({"title" : title, "img_url" : img_url})
mars_hemi_img_url
```
|
github_jupyter
|
import pandas as pd
from bs4 import BeautifulSoup as bs
from pprint import pprint
import pymongo
import requests
from splinter import Browser
executable_path = {'executable_path': '/Users/ekaster/Documents/DATABOOTCAMP-MATERIAL/chromedriver.exe'}
browser = Browser('chrome', **executable_path, headless=False)
mars_news_url = 'https://mars.nasa.gov/news/'
browser.visit(mars_news_url)
response = requests.get(mars_news_url)
news_soup = bs(response.text, 'html.parser')
news_title = news_soup.find("div", class_="content_title").text
news_p = news_soup.find("div", class_="rollover_description_inner").text
print(news_title)
print(news_p)
executable_path = {'executable_path' : '/Users/ekaster/Documents/DATABOOTCAMP-MATERIAL/chromedriver.exe'}
browser = Browser('chrome', **executable_path, headless=False)
mars_img_url = "https://www.jpl.nasa.gov/spaceimages/?search=&category=Mars"
browser.visit(mars_img_url)
browser.click_link_by_partial_text('FULL IMAGE')
browser.click_link_by_partial_text('more info')
browser_soup = bs(browser.html, 'html.parser')
get_img_url = browser_soup.find('img', class_='main_image')
img_src_url = get_img_url.get('src')
featured_image_url = "https://www.jpl.nasa.gov" + img_src_url
print(featured_image_url)
executable_path = {'executable_path' : '/Users/ekaster/Documents/DATABOOTCAMP-MATERIAL/chromedriver.exe'}
browser = Browser('chrome', **executable_path, headless=False)
mars_twitter_url = 'https://twitter.com/marswxreport?lang=en'
browser.visit(mars_twitter_url)
twitter_soup = bs(browser.html, 'html.parser')
print(twitter_soup.prettify())
latest_tweets = twitter_soup.find('div', class_='css-901oao r-hkyrab r-1qd0xha r-a023e6 r-16dba41 r-ad9z0x r-bcqeeo r-bnwqim r-qvutc0').text
print(latest_tweets)
request_mars_space_facts = requests.get("https://space-facts.com/mars/")
mars_space_table_read = pd.read_html(request_mars_space_facts.text)
mars_space_table_read
mars_df = mars_space_table_read[0]
mars_df.columns = ['Description','Value']
mars_df.set_index(['Description'], inplace=True)
mars_df
mars_data_html = mars_df.to_html()
mars_data_html
mars_data_html.replace('\n', '')
executable_path = {'executable_path': '/Users/ekaster/Documents/DATABOOTCAMP-MATERIAL/chromedriver.exe'}
browser = Browser('chrome', **executable_path, headless=False)
mars_hemi_url = 'https://astrogeology.usgs.gov/search/results?q=hemisphere+enhanced&k1=target&v1=Mars'
browser.visit(mars_hemi_url)
mars_hemi_html = browser.html
mars_hemi_soup = bs(mars_hemi_html, 'html.parser')
items = mars_hemi_soup.find_all('div', class_='item')
mars_hemi_img_url = []
mars_hemi_main_url = 'https://astrogeology.usgs.gov'
for i in items:
title = i.find('h3').text
partial_img_url = i.find('a', class_='itemLink product-item')['href']
browser.visit(mars_hemi_main_url + partial_img_url)
partial_img_html = browser.html
soup = bs( partial_img_html, 'html.parser')
img_url = mars_hemi_main_url + soup.find('img', class_='wide-image')['src']
mars_hemi_img_url.append({"title" : title, "img_url" : img_url})
mars_hemi_img_url
| 0.18396 | 0.608536 |
```
# Speed Test -- Trajectory Plotting
import numpy as np
import pandas as pd
from geopandas import GeoDataFrame, read_file
from shapely.geometry import Point, LineString, Polygon
from datetime import datetime, timedelta
import matplotlib.pyplot as plt
import movingpandas as mpd
from holoviews import opts, dim
gdf = read_file('../data/geolife_small.gpkg')
runtimes={}
%%time
t0 = datetime.now()
gdf.plot()
runtime = datetime.now()-t0
runtimes['GeoDataFrame.plot'] = runtime
print(runtime)
traj_collection = mpd.TrajectoryCollection(gdf, 'trajectory_id', t='t')
%%time
t0 = datetime.now()
traj_collection.plot()
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.plot'] = runtime
print(runtime)
%%time
t0 = datetime.now()
traj_collection.hvplot(line_width=7)
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.hvplot'] = runtime
print(runtime)
%%time
t0 = datetime.now()
traj_collection.hvplot(line_width=7, frame_width=300, frame_height=300)
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.hvplot (smaller)'] = runtime
print(runtime)
%%time
generalized_traj = mpd.DouglasPeuckerGeneralizer(traj_collection).generalize(tolerance=0.01)
t0 = datetime.now()
generalized_traj.hvplot(line_width=7)
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.hvplot (generalized)'] = runtime
print(runtime)
%%time
t0 = datetime.now()
gdf.hvplot(geo=True, tiles='OSM')
runtime = datetime.now()-t0
runtimes['GeoDataFrame.hvplot'] = runtime
print(runtime)
%%time
line_gdf = traj_collection.to_line_gdf()
t0 = datetime.now()
line_gdf.hvplot(geo=True, tiles='OSM', line_width=7)
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.to_line_gdf.hvplot'] = runtime
print(runtime)
%%time
line_gdf = traj_collection.to_line_gdf()
t0 = datetime.now()
line_gdf.hvplot()
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.to_line_gdf.hvplot (no basemap)'] = runtime
print(runtime)
%%time
traj_gdf = traj_collection.to_traj_gdf()
t0 = datetime.now()
traj_gdf.hvplot(geo=True, tiles='OSM', line_width=7)
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.to_traj_gdf.hvplot'] = runtime
print(runtime)
for key, value in sorted(runtimes.items()):
print(f'{key}: {value}')
result = pd.DataFrame.from_dict(runtimes, orient='index', columns=['runtime'])
result['seconds'] = result.runtime.dt.total_seconds()
result
result.sort_values('seconds').hvplot.barh(y='seconds', title='Runtimes in seconds')
import geopandas
print(f'GeoPandas {geopandas.__version__}')
import geoviews
print(f'Geoviews {geoviews.__version__}')
import cartopy
print(f'Cartopy {cartopy.__version__}')
```
|
github_jupyter
|
# Speed Test -- Trajectory Plotting
import numpy as np
import pandas as pd
from geopandas import GeoDataFrame, read_file
from shapely.geometry import Point, LineString, Polygon
from datetime import datetime, timedelta
import matplotlib.pyplot as plt
import movingpandas as mpd
from holoviews import opts, dim
gdf = read_file('../data/geolife_small.gpkg')
runtimes={}
%%time
t0 = datetime.now()
gdf.plot()
runtime = datetime.now()-t0
runtimes['GeoDataFrame.plot'] = runtime
print(runtime)
traj_collection = mpd.TrajectoryCollection(gdf, 'trajectory_id', t='t')
%%time
t0 = datetime.now()
traj_collection.plot()
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.plot'] = runtime
print(runtime)
%%time
t0 = datetime.now()
traj_collection.hvplot(line_width=7)
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.hvplot'] = runtime
print(runtime)
%%time
t0 = datetime.now()
traj_collection.hvplot(line_width=7, frame_width=300, frame_height=300)
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.hvplot (smaller)'] = runtime
print(runtime)
%%time
generalized_traj = mpd.DouglasPeuckerGeneralizer(traj_collection).generalize(tolerance=0.01)
t0 = datetime.now()
generalized_traj.hvplot(line_width=7)
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.hvplot (generalized)'] = runtime
print(runtime)
%%time
t0 = datetime.now()
gdf.hvplot(geo=True, tiles='OSM')
runtime = datetime.now()-t0
runtimes['GeoDataFrame.hvplot'] = runtime
print(runtime)
%%time
line_gdf = traj_collection.to_line_gdf()
t0 = datetime.now()
line_gdf.hvplot(geo=True, tiles='OSM', line_width=7)
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.to_line_gdf.hvplot'] = runtime
print(runtime)
%%time
line_gdf = traj_collection.to_line_gdf()
t0 = datetime.now()
line_gdf.hvplot()
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.to_line_gdf.hvplot (no basemap)'] = runtime
print(runtime)
%%time
traj_gdf = traj_collection.to_traj_gdf()
t0 = datetime.now()
traj_gdf.hvplot(geo=True, tiles='OSM', line_width=7)
runtime = datetime.now()-t0
runtimes['TrajectoryCollection.to_traj_gdf.hvplot'] = runtime
print(runtime)
for key, value in sorted(runtimes.items()):
print(f'{key}: {value}')
result = pd.DataFrame.from_dict(runtimes, orient='index', columns=['runtime'])
result['seconds'] = result.runtime.dt.total_seconds()
result
result.sort_values('seconds').hvplot.barh(y='seconds', title='Runtimes in seconds')
import geopandas
print(f'GeoPandas {geopandas.__version__}')
import geoviews
print(f'Geoviews {geoviews.__version__}')
import cartopy
print(f'Cartopy {cartopy.__version__}')
| 0.418222 | 0.424472 |
# Order statistics
This notebook introduces order statistics numerically.
# Setup
```
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
plt.rcParams.update({"text.usetex": True, 'font.size':14, 'font.family':'serif'})
np.random.seed(1337)
R = 1000000
```
# Theory: Order statistics
Let $(X_1,...,X_N)$ be a sample of $N$ draws. The notation $(X_{(1)}, ..., X_{(N)})$ denotes the *sorted* vector of the $N$ draws. That is, $X_{(1)}$ is the minimum, $X_{(n)}$ is the maximums, and $X_{(k)}$ is the $k$'th smallest, which we call the $k$'th order statistic.
In auction theory, it becomes useful to think of the distribution of the $k$'th order statistic. Just like the sample average, $ \bar{X} \equiv N^{-1} \sum_{i=1}^N X_i$, the $k$'th order statistic is just a function of our $n$ stochastic variables. Since any function of a stochastic variable is itself a stochastic variable, the $k$'th order statistic has a distribution, which will have a mean, a std.dev., etc.
## Normally distributed variables with $N=5$
```
N = 5 # number of samples each time (e.g. number of bidders in our auction)
u = np.random.normal(0,1,(N,R))
U = pd.DataFrame(u)
fig, ax = plt.subplots()
U.max(0).hist(bins=100, ax=ax, label='Max', alpha=0.9, density=True);
U.mean(0).hist(bins=100, ax=ax, label='Mean',alpha=0.6, density=True);
ax.legend(loc='best');
sns.despine(); ax.grid(False);
ax.set_xlabel('$x$'); ax.set_ylabel(f'Density ($N={N}, R={R:,}$)');
plt.savefig('img/normal_maxmean.pdf');
```
Plotting the unsorted data
```
fig, ax = plt.subplots()
for k in range(u.shape[0]):
ax.hist(u[k, :], density=True, alpha=0.6, label=f'${k+1}$', bins=100);
ax.legend(loc='best', title="$k$'th player"); sns.despine();
ax.grid(False); ax.set_xlabel('$x$'); ax.set_ylabel('Density');
plt.savefig('img/normal_hist_unsorted.pdf');
```
Now let's instead sort the dataset
```
u_sort = np.sort(u, 0)
```
... and show what it looks like separately for each *order* position ($k$).
```
u_sort = np.sort(u, 0)
fig, ax = plt.subplots()
for k in range(u_sort.shape[0]):
ax.hist(u_sort[k, :], density=True, alpha=0.6, label=f'${k+1}$', bins=100);
ax.legend(loc='best', title="$k$'th order"); sns.despine();
ax.set_xlabel('$x$'); ax.set_ylabel('Density');
for k in range(u_sort.shape[0]):
ax.axvline(u_sort[k, :].mean(), color='gray', linestyle=':');
plt.savefig('img/normal_hist_sorted_meanlines.pdf');
```
## Varying $N$
Next, we can consider what the *average* of the $k$'th order statistic looks like depending on the sample size, $N$, for $k = n-1,n$ (i.e. the largest and 2nd largest values).
```
NN = np.array([2,3,4,5,6])
kk = np.array([-1, -2])
k_labs = ['N', 'N-1']
yy = np.empty((len(kk), len(NN)))
for iN,N in enumerate(NN):
u = np.random.normal(0,1,(N,R))
u = np.sort(u, 0)
for ik,k in enumerate(kk):
yy[ik, iN] = u[k, :].mean()
```
And let's plot this.
```
fig, ax = plt.subplots();
for ik, k in enumerate(kk):
ax.plot(NN, yy[ik, :], '-o', label=f'$k={k_labs[ik]}$');
ax.legend(loc='best'); ax.set_xlabel('$N$ (number of draws from $N(0,1)$)');
ax.set_ylabel(f'Average (over $R={R:,}$ draws)'); sns.despine();
plt.savefig('img/normal_largest_and_2nd_largest.pdf')
```
# Uniform distribution
Next, we do it all again, but this time for the uniform distribution
```
# take R draws of independent values
N = 5
u = np.random.uniform(0,1,(N,R))
fig, ax = plt.subplots()
for k in range(u.shape[0]):
ax.hist(u[k, :], density=True, alpha=0.6, label=f'${k+1}$', bins=100);
ax.legend(loc='lower center', title="$k$'th player"); sns.despine();
ax.grid(False); ax.set_xlabel('$x$'); ax.set_ylabel('Density');
plt.savefig('img/uniform_hist_unsorted.pdf');
u_sort = np.sort(u, 0)
fig, ax = plt.subplots()
for r in range(u_sort.shape[0]):
ax.hist(u_sort[r, :], density=True, alpha=0.6, label=f'${r+1}$', bins=100);
ax.legend(loc='best', title="$k$'th order"); sns.despine();
ax.set_xlabel('$v$'); ax.set_ylabel('Density');
```
## Varying $N$
```
NN = np.array([2,3,4,5,6])
kk = np.array([-1, -2])
k_labs = ['N', 'N-1']
yy = np.empty((len(kk), len(NN)))
for iN,N in enumerate(NN):
u = np.random.uniform(0,1,(N,R))
u = np.sort(u, 0)
for ik,k in enumerate(kk):
yy[ik, iN] = u[k, :].mean()
fig, ax = plt.subplots();
for ik, k in enumerate(kk):
ax.plot(NN, yy[ik, :], '-o', label=f'$k={k_labs[ik]}$');
ax.legend(loc='best'); ax.set_xlabel('$N$ (number of draws from $U(0,1)$)');
ax.set_ylabel(f'Average (over $R={R:,}$ draws)'); sns.despine();
plt.savefig('img/uniform_largest_and_2nd_largest.pdf')
```
## Verifying with analytic formulas
We know that if $X_i \sim U(0,1)$, then $X_{(k)} \sim \mathcal{B}(k, n+1-k)$. That is, when $X_i$ are uniformly distributed, the order statistics are beta distributed. This implies that the expected values are simply:
$$ \mathbb{E}\left( X_{(k)} ) \right) = \frac{k}{n+1}, \quad k = 1, ..., n. $$
(and just to be clear, our notation is that $X_{(1)}$ is the minimum and $X_{(n)}$ is the maximum.)
**First:** We comapre the expected value formula to the average of a large number of simulations.
```
N=6
u = np.random.uniform(0,1,(N,R))
u = np.sort(u, 0)
for k in range(1,N+1):
# compute
mean_analytic = k/(N+1)
mean_simulated = u[k-1,:].mean() # "k-1" to get from base 1 to base 0
# print
print(f'--- k={k} ---')
print(f'{"Simulated":<15} = {mean_simulated: 10.5f}')
print(f'{"Formula":<15} = {mean_analytic: 10.5f}')
```
**Second:** we compare the ***actual distributions***. It turns out, that if $X_i \sim \mathcal{U}(0,1)$ and we draw $N$ samples, then
$$X_{(k)} \sim \mathcal{B}(k,N+1-k).$$
That is, the order statistics from the uniform distribution are beta distributed!
```
k = N-1 # pick which order statistic to look at
b = np.random.beta(k,N+1-k,(R,))
fig, ax = plt.subplots();
ax.hist(u[k-1, :], label='Simulated', density=True, alpha=0.6, bins=100);
ax.hist(b, label=f'Beta({k}, {N+1-k})', density=True, alpha=0.5, bins=100);
ax.legend(loc='best'); sns.despine(); ax.set_xlabel('$X$'); ax.set_ylabel('Density');
plt.savefig('img/beta_vs_simulation.pdf');
```
# Truncated distributions
```
u = np.random.normal(0,1,(R,))
v = np.random.normal(0,1,(R,))
trunc = 1.0
v = v[v <= trunc] # deleting truncated rows
fig, ax = plt.subplots();
ax.hist(u, density=True, alpha=0.8, bins=100, label='Normal')
ax.hist(v, density=True, alpha=0.5, bins=100, label='Truncated')
ax.legend(loc='best'); ax.set_ylabel('Density'); ax.axvline(trunc, color='gray', linestyle=':');
sns.despine();
plt.savefig('img/truncated.pdf');
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
plt.rcParams.update({"text.usetex": True, 'font.size':14, 'font.family':'serif'})
np.random.seed(1337)
R = 1000000
N = 5 # number of samples each time (e.g. number of bidders in our auction)
u = np.random.normal(0,1,(N,R))
U = pd.DataFrame(u)
fig, ax = plt.subplots()
U.max(0).hist(bins=100, ax=ax, label='Max', alpha=0.9, density=True);
U.mean(0).hist(bins=100, ax=ax, label='Mean',alpha=0.6, density=True);
ax.legend(loc='best');
sns.despine(); ax.grid(False);
ax.set_xlabel('$x$'); ax.set_ylabel(f'Density ($N={N}, R={R:,}$)');
plt.savefig('img/normal_maxmean.pdf');
fig, ax = plt.subplots()
for k in range(u.shape[0]):
ax.hist(u[k, :], density=True, alpha=0.6, label=f'${k+1}$', bins=100);
ax.legend(loc='best', title="$k$'th player"); sns.despine();
ax.grid(False); ax.set_xlabel('$x$'); ax.set_ylabel('Density');
plt.savefig('img/normal_hist_unsorted.pdf');
u_sort = np.sort(u, 0)
u_sort = np.sort(u, 0)
fig, ax = plt.subplots()
for k in range(u_sort.shape[0]):
ax.hist(u_sort[k, :], density=True, alpha=0.6, label=f'${k+1}$', bins=100);
ax.legend(loc='best', title="$k$'th order"); sns.despine();
ax.set_xlabel('$x$'); ax.set_ylabel('Density');
for k in range(u_sort.shape[0]):
ax.axvline(u_sort[k, :].mean(), color='gray', linestyle=':');
plt.savefig('img/normal_hist_sorted_meanlines.pdf');
NN = np.array([2,3,4,5,6])
kk = np.array([-1, -2])
k_labs = ['N', 'N-1']
yy = np.empty((len(kk), len(NN)))
for iN,N in enumerate(NN):
u = np.random.normal(0,1,(N,R))
u = np.sort(u, 0)
for ik,k in enumerate(kk):
yy[ik, iN] = u[k, :].mean()
fig, ax = plt.subplots();
for ik, k in enumerate(kk):
ax.plot(NN, yy[ik, :], '-o', label=f'$k={k_labs[ik]}$');
ax.legend(loc='best'); ax.set_xlabel('$N$ (number of draws from $N(0,1)$)');
ax.set_ylabel(f'Average (over $R={R:,}$ draws)'); sns.despine();
plt.savefig('img/normal_largest_and_2nd_largest.pdf')
# take R draws of independent values
N = 5
u = np.random.uniform(0,1,(N,R))
fig, ax = plt.subplots()
for k in range(u.shape[0]):
ax.hist(u[k, :], density=True, alpha=0.6, label=f'${k+1}$', bins=100);
ax.legend(loc='lower center', title="$k$'th player"); sns.despine();
ax.grid(False); ax.set_xlabel('$x$'); ax.set_ylabel('Density');
plt.savefig('img/uniform_hist_unsorted.pdf');
u_sort = np.sort(u, 0)
fig, ax = plt.subplots()
for r in range(u_sort.shape[0]):
ax.hist(u_sort[r, :], density=True, alpha=0.6, label=f'${r+1}$', bins=100);
ax.legend(loc='best', title="$k$'th order"); sns.despine();
ax.set_xlabel('$v$'); ax.set_ylabel('Density');
NN = np.array([2,3,4,5,6])
kk = np.array([-1, -2])
k_labs = ['N', 'N-1']
yy = np.empty((len(kk), len(NN)))
for iN,N in enumerate(NN):
u = np.random.uniform(0,1,(N,R))
u = np.sort(u, 0)
for ik,k in enumerate(kk):
yy[ik, iN] = u[k, :].mean()
fig, ax = plt.subplots();
for ik, k in enumerate(kk):
ax.plot(NN, yy[ik, :], '-o', label=f'$k={k_labs[ik]}$');
ax.legend(loc='best'); ax.set_xlabel('$N$ (number of draws from $U(0,1)$)');
ax.set_ylabel(f'Average (over $R={R:,}$ draws)'); sns.despine();
plt.savefig('img/uniform_largest_and_2nd_largest.pdf')
N=6
u = np.random.uniform(0,1,(N,R))
u = np.sort(u, 0)
for k in range(1,N+1):
# compute
mean_analytic = k/(N+1)
mean_simulated = u[k-1,:].mean() # "k-1" to get from base 1 to base 0
# print
print(f'--- k={k} ---')
print(f'{"Simulated":<15} = {mean_simulated: 10.5f}')
print(f'{"Formula":<15} = {mean_analytic: 10.5f}')
k = N-1 # pick which order statistic to look at
b = np.random.beta(k,N+1-k,(R,))
fig, ax = plt.subplots();
ax.hist(u[k-1, :], label='Simulated', density=True, alpha=0.6, bins=100);
ax.hist(b, label=f'Beta({k}, {N+1-k})', density=True, alpha=0.5, bins=100);
ax.legend(loc='best'); sns.despine(); ax.set_xlabel('$X$'); ax.set_ylabel('Density');
plt.savefig('img/beta_vs_simulation.pdf');
u = np.random.normal(0,1,(R,))
v = np.random.normal(0,1,(R,))
trunc = 1.0
v = v[v <= trunc] # deleting truncated rows
fig, ax = plt.subplots();
ax.hist(u, density=True, alpha=0.8, bins=100, label='Normal')
ax.hist(v, density=True, alpha=0.5, bins=100, label='Truncated')
ax.legend(loc='best'); ax.set_ylabel('Density'); ax.axvline(trunc, color='gray', linestyle=':');
sns.despine();
plt.savefig('img/truncated.pdf');
| 0.338624 | 0.916484 |
```
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from sklearn import preprocessing
import seaborn as sns
from scipy.stats import norm
from scipy import stats
from scipy.stats import skew
train_df = pd.read_csv('../data/orignal/train.csv', index_col = 0)
test_df = pd.read_csv('../data/orignal/test.csv', index_col = 0)
combine_df = pd.concat([train_df, test_df])
# 相关性检测
#correlation matrix
corrmat = train_df.corr()
f, ax = plt.subplots(figsize=(12, 9))
sns.heatmap(corrmat, vmax=.8, square=True)
def fixSkew(feature_df, name):
skewed_feat = skew(feature_df[name])
if skewed_feat > 0.75:
print('fix')
return np.log1p(feature_df[name])
else:
print('notfix')
return feature_df[name]
plt.show()
#saleprice correlation matrix
k = 10 #number of variables for heatmap
cols = corrmat.nlargest(k, 'SalePrice')['SalePrice'].index
cm = np.corrcoef(train_df[cols].values.T)
sns.set(font_scale=0.75)
hm = sns.heatmap(cm, cbar=True, annot=True, square=True, fmt='.2f', annot_kws={'size': 10}, yticklabels=cols.values, xticklabels=cols.values)
plt.show()
```
### MSSubClass
涉及销售的寓所类型
```
combine_df[combine_df['MSSubClass'].isnull()]
```
### MSZoning
售卖的地产区域类型
```
combine_df['MSZoning'] = combine_df['MSZoning'].fillna('RL')
le = preprocessing.LabelEncoder()
le.fit(combine_df['MSZoning'])
combine_df['MSZoning'] = le.transform(combine_df['MSZoning'])
```
### LotFrontage
距离最近的街道的直线距离
填充中位数
数值标准化
```
lot_frontage_df = combine_df['LotFrontage'].fillna(combine_df['LotFrontage'].median())
lot_frontage_df = pd.DataFrame(preprocessing.scale(lot_frontage_df.values), np.array(range(1, 2920)), columns=['LotFrontage'])
lot_frontage_df.index.name = 'Id'
```
### LotArea
房产占地面积
数值标准化
```
lot_area_df = pd.DataFrame(preprocessing.scale(combine_df['LotArea']), np.array(range(1, 2920)), columns=['LotArea'])
lot_area_df.index.name = 'Id'
```
### Street
取值不平衡 丢弃该特征
```
combine_df['Street'].value_counts()
```
### Alley
```
combine_df['Alley_Access'] = combine_df['Alley'].apply(lambda x : 0 if pd.isnull(x) else 1)
combine_df['Alley'] = combine_df['Alley'].fillna('NoAccess')
combine_df['Alley'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['Alley'])
combine_df['Alley'] = le.transform(combine_df['Alley'])
```
### LotShape
住宅的房型
```
combine_df['LotShape'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['LotShape'])
combine_df['LotShape'] = le.transform(combine_df['LotShape'])
```
### LandContour
住宅的地面是否平坦
```
combine_df['LandContour'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['LandContour'])
combine_df['LandContour'] = le.transform(combine_df['LandContour'])
```
### Utilities
配套设施
[不平衡] 丢弃
```
combine_df['Utilities'].value_counts()
```
### LotConfig
住宅的地理类型
```
combine_df['LotConfig'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['LotConfig'])
combine_df['LotConfig'] = le.transform(combine_df['LotConfig'])
```
### LandSlope
住宅的倾斜度
```
combine_df['LandSlope'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['LandSlope'])
combine_df['LandSlope'] = le.transform(combine_df['LandSlope'])
```
### Neighborhood
在AME城中的物理位置
```
combine_df['Neighborhood'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['Neighborhood'])
combine_df['Neighborhood'] = le.transform(combine_df['Neighborhood'])
```
### Condition1
附近的情况
```
le = preprocessing.LabelEncoder()
le.fit(combine_df['Condition1'])
combine_df['Condition1'] = le.transform(combine_df['Condition1'])
combine_df['Condition1'].value_counts()
```
### Condition2
附近的情况
```
le = preprocessing.LabelEncoder()
le.fit(combine_df['Condition2'])
combine_df['Condition2'] = le.transform(combine_df['Condition2'])
combine_df['Condition2'].value_counts()
```
### BldgType
住宅类型
```
le = preprocessing.LabelEncoder()
le.fit(combine_df['BldgType'])
combine_df['BldgType'] = le.transform(combine_df['BldgType'])
combine_df['BldgType'].value_counts()
```
### HouseStyle
住宅风格
```
le = preprocessing.LabelEncoder()
le.fit(combine_df['HouseStyle'])
combine_df['HouseStyle'] = le.transform(combine_df['HouseStyle'])
combine_df['HouseStyle'].value_counts()
```
### OverallQual
装修覆盖率及装修完成度
```
sns.distplot(np.log(combine_df['OverallQual']), fit=norm);
fig = plt.figure()
plt.show()
combine_df['OverallQual']
overall_qual_df = pd.DataFrame(preprocessing.scale(fixSkew(combine_df, 'OverallQual')), np.array(range(1, 2920)), columns=['OverallQual'])
overall_qual_df.index.name = 'Id'
```
### OverallCond
住宅的整体状况
```
overall_cond_df = pd.DataFrame(preprocessing.scale(combine_df['OverallCond'].values), np.array(range(1, 2920)), columns=['OverallCond'])
overall_cond_df.index.name = 'Id'
```
### YearBuilt
原始施工日期
计算原始施工日到目前(2016年)总共多少年
```
combine_df['YearBuilt'] = combine_df['YearBuilt'].apply(lambda x : 2016 - x)
year_built_df = pd.DataFrame(preprocessing.scale(fixSkew(combine_df, 'YearBuilt')), np.array(range(1, 2920)), columns=['YearBuilt'])
```
### YearRemodAdd
改造时间年份
计算原始施工日到目前(2016年)总共多少年
```
year_remodadd_df = pd.DataFrame(2016 - combine_df['YearRemodAdd'])
```
### RoofStyle
屋顶类型
```
le = preprocessing.LabelEncoder()
le.fit(combine_df['RoofStyle'])
combine_df['RoofStyle'] = le.transform(combine_df['RoofStyle'])
combine_df['RoofStyle'].value_counts()
```
### RoofMatl
屋顶材料
```
le = preprocessing.LabelEncoder()
le.fit(combine_df['RoofMatl'])
combine_df['RoofMatl'] = le.transform(combine_df['RoofMatl'])
combine_df['RoofMatl'].value_counts()
```
### Exterior1st
房子的外观
```
combine_df[combine_df['Exterior1st'].isnull()]
combine_df['Exterior1st'] = combine_df['Exterior1st'].fillna('VinylSd')
le = preprocessing.LabelEncoder()
le.fit(combine_df['Exterior1st'])
combine_df['Exterior1st'] = le.transform(combine_df['Exterior1st'])
combine_df['Exterior1st'].value_counts()
```
### Exterior2nd
房子的外观
```
combine_df[combine_df['Exterior2nd'].isnull()]
combine_df['Exterior2nd'] = combine_df['Exterior2nd'].fillna('VinylSd')
le = preprocessing.LabelEncoder()
le.fit(combine_df['Exterior2nd'])
combine_df['Exterior2nd'] = le.transform(combine_df['Exterior2nd'])
combine_df['Exterior2nd'].value_counts()
```
### MasVnrType
表层砌体类型
```
combine_df['MasVnrType'] = combine_df['MasVnrType'].fillna('None')
le = preprocessing.LabelEncoder()
le.fit(combine_df['MasVnrType'])
combine_df['MasVnrType'] = le.transform(combine_df['MasVnrType'])
combine_df['MasVnrType'].value_counts()
```
### MasVnrArea
表层砌面面积
```
combine_df['MasVnrArea'].median()
combine_df['MasVnrArea'] = combine_df['MasVnrArea'].fillna(combine_df['MasVnrArea'].median())
mas_vnr_area_df = pd.DataFrame(preprocessing.scale(combine_df['MasVnrArea']), np.array(range(1, 2920)), columns=['MasVnrArea'])
mas_vnr_area_df.index.name = 'Id'
```
### ExterQual
外观材料质量
```
combine_df['ExterQual'].isnull().any()
le = preprocessing.LabelEncoder()
le.fit(combine_df['ExterQual'])
combine_df['ExterQual'] = le.transform(combine_df['ExterQual'])
combine_df['ExterQual'].value_counts()
```
### ExterCond
外部材料现状
```
combine_df['ExterCond'].isnull().any()
le = preprocessing.LabelEncoder()
le.fit(combine_df['ExterCond'])
combine_df['ExterCond'] = le.transform(combine_df['ExterCond'])
combine_df['ExterCond'].value_counts()
```
### Foundation
地基类型
```
combine_df['Foundation'].isnull().any()
le = preprocessing.LabelEncoder()
le.fit(combine_df['Foundation'])
combine_df['Foundation'] = le.transform(combine_df['Foundation'])
combine_df['Foundation'].value_counts()
```
### Bsmt
是否有地下室
```
combine_df['Has_Bsmt'] = combine_df['BsmtQual'].apply(lambda x : 0 if pd.isnull(x) else 1)
```
### BsmtQual
地下室高度
```
combine_df['BsmtQual'] = combine_df['BsmtQual'].fillna('No_Bsmt')
le = preprocessing.LabelEncoder()
le.fit(combine_df['BsmtQual'])
combine_df['BsmtQual'] = le.transform(combine_df['BsmtQual'])
```
### BsmtCond
地下室的环境条件
```
combine_df['BsmtCond'] = combine_df['BsmtCond'].fillna('No_Bsmt')
le = preprocessing.LabelEncoder()
le.fit(combine_df['BsmtCond'])
combine_df['BsmtCond'] = le.transform(combine_df['BsmtCond'])
```
### BsmtExposure
光照条件
```
combine_df['BsmtExposure'] = combine_df['BsmtExposure'].fillna('No_Bsmt')
le = preprocessing.LabelEncoder()
le.fit(combine_df['BsmtExposure'])
combine_df['BsmtExposure'] = le.transform(combine_df['BsmtExposure'])
```
### BsmtFinType1
地下室装修完成度
```
combine_df['BsmtFinType1'] = combine_df['BsmtFinType1'].fillna('No_Bsmt')
le = preprocessing.LabelEncoder()
le.fit(combine_df['BsmtFinType1'])
combine_df['BsmtFinType1'] = le.transform(combine_df['BsmtFinType1'])
```
### BsmtFinSF1
Type1完成的面积
```
combine_df['BsmtFinSF1'] = combine_df['BsmtFinSF1'].fillna(0)
bsmt_fin_SF1_df = pd.DataFrame(preprocessing.scale(combine_df['BsmtFinSF1']), np.array(range(1, 2920)), columns=['BsmtFinSF1'])
bsmt_fin_SF1_df.index.name = 'Id'
```
### BsmtFinType2
地下室装修完成度
```
combine_df['BsmtFinType2'] = combine_df['BsmtFinType2'].fillna('No_Bsmt')
le = preprocessing.LabelEncoder()
le.fit(combine_df['BsmtFinType2'])
combine_df['BsmtFinType2'] = le.transform(combine_df['BsmtFinType2'])
```
### BsmtFinSF2
Type2完成的面积
```
combine_df['BsmtFinSF2'] = combine_df['BsmtFinSF2'].fillna(0)
bsmt_fin_SF2_df = pd.DataFrame(preprocessing.scale(combine_df['BsmtFinSF2']), np.array(range(1, 2920)), columns=['BsmtFinSF2'])
bsmt_fin_SF2_df.index.name = 'Id'
```
### BsmtUnfSF
未完成的地下室面积
```
combine_df[combine_df['BsmtUnfSF'].isnull()]
combine_df.ix[2121, 'BsmtUnfSF'] = 0
bsmt_unf_sf_df = pd.DataFrame(preprocessing.scale(combine_df['BsmtUnfSF']), np.array(range(1, 2920)), columns=['BsmtUnfSF'])
bsmt_unf_sf_df.index.name = 'Id'
```
### TotalBsmtSF
地下室总面积
```
combine_df[combine_df['TotalBsmtSF'].isnull()]
combine_df.ix[2121, 'TotalBsmtSF'] = 0
total_bsmt_sf_df = pd.DataFrame(preprocessing.scale(fixSkew(combine_df, 'TotalBsmtSF')), np.array(range(1, 2920)), columns=['TotalBsmtSF'])
total_bsmt_sf_df.index.name = 'Id'
combine_df['TotalBsmtSF'].describe()
```
### Heating
供暖类型
```
combine_df[combine_df['Heating'].isnull()]
le = preprocessing.LabelEncoder()
le.fit(combine_df['Heating'])
combine_df['Heating'] = le.transform(combine_df['Heating'])
```
### HeatingQC
供暖效果
```
combine_df[combine_df['HeatingQC'].isnull()]
le = preprocessing.LabelEncoder()
le.fit(combine_df['HeatingQC'])
combine_df['HeatingQC'] = le.transform(combine_df['HeatingQC'])
```
### CentralAir
中央空调
```
combine_df[combine_df['CentralAir'].isnull()]
le = preprocessing.LabelEncoder()
le.fit(combine_df['CentralAir'])
combine_df['CentralAir'] = le.transform(combine_df['CentralAir'])
```
### Electrical
电力系统
```
combine_df[combine_df['Electrical'].isnull()]
combine_df['Electrical'].value_counts()
combine_df.ix[1380, 'Electrical'] = 'SBrkr'
le = preprocessing.LabelEncoder()
le.fit(combine_df['Electrical'])
combine_df['Electrical'] = le.transform(combine_df['Electrical'])
```
### 1stFlrSF
一楼面积
```
combine_df[combine_df['1stFlrSF'].isnull()]
fst_flr_sf_df = pd.DataFrame(preprocessing.scale(combine_df['1stFlrSF']), np.array(range(1, 2920)), columns=['1stFlrSF'])
fst_flr_sf_df.index.name = 'Id'
```
### 2ndFlrSF
二楼面积
```
combine_df[combine_df['2ndFlrSF'].isnull()]
snd_flr_sf_df = pd.DataFrame(preprocessing.scale(combine_df['2ndFlrSF']), np.array(range(1, 2920)), columns=['2ndFlrSF'])
snd_flr_sf_df.index.name = 'Id'
```
### LowQualFinSF
低质量完成的面积
```
combine_df[combine_df['LowQualFinSF'].isnull()]
low_qual_fin_sf_df = pd.DataFrame(preprocessing.scale(combine_df['LowQualFinSF']), np.array(range(1, 2920)), columns=['LowQualFinSF'])
low_qual_fin_sf_df.index.name = 'Id'
```
### GrLivArea
地面以上居住面积
```
combine_df[combine_df['GrLivArea'].isnull()]
gr_liv_area_df = pd.DataFrame(preprocessing.scale(fixSkew(combine_df, 'GrLivArea')), np.array(range(1, 2920)), columns=['GrLivArea'])
gr_liv_area_df.index.name = 'Id'
```
### BsmtFullBath
地下室全浴室
```
combine_df[combine_df['BsmtFullBath'].isnull()]
combine_df['BsmtFullBath'].value_counts()
combine_df.ix[2121, 'Has_Bsmt']
combine_df.ix[2189, 'Has_Bsmt']
combine_df['BsmtFullBath'] = combine_df['BsmtFullBath'].fillna(0).astype(np.int)
```
### BsmtHalfBath
底下室半浴室
```
combine_df[combine_df['BsmtHalfBath'].isnull()]
combine_df['BsmtHalfBath'].value_counts()
combine_df['BsmtHalfBath'] = combine_df['BsmtHalfBath'].fillna(0).astype(np.int)
```
### FullBath
地上全浴室个数
```
combine_df[combine_df['FullBath'].isnull()]
combine_df['FullBath'].value_counts()
```
### HalfBath
地上半浴室个数
```
combine_df[combine_df['HalfBath'].isnull()]
combine_df['HalfBath'].value_counts()
```
### BedroomAbvGr
地上卧室
```
combine_df[combine_df['BedroomAbvGr'].isnull()]
combine_df['BedroomAbvGr'].value_counts()
```
### KitchenAbvGr
地上厨房
```
combine_df[combine_df['KitchenAbvGr'].isnull()]
combine_df['KitchenAbvGr'].value_counts()
```
### KitchenQual
厨房质量
```
combine_df[combine_df['KitchenQual'].isnull()]
combine_df['KitchenQual'].value_counts()
combine_df.ix[1556, 'KitchenQual'] = 'TA'
le = preprocessing.LabelEncoder()
le.fit(combine_df['KitchenQual'])
combine_df['KitchenQual'] = le.transform(combine_df['KitchenQual'])
```
### TotRmsAbvGrd
地上的房间总数量
```
combine_df[combine_df['TotRmsAbvGrd'].isnull()]
combine_df['TotRmsAbvGrd'].value_counts()
```
### Functional
家庭功能
```
combine_df[combine_df['Functional'].isnull()]
combine_df['Functional'].value_counts()
combine_df.ix[2217, 'Functional'] = 'Typ'
combine_df.ix[2474, 'Functional'] = 'Typ'
le = preprocessing.LabelEncoder()
le.fit(combine_df['Functional'])
combine_df['Functional'] = le.transform(combine_df['Functional'])
```
### Fireplaces
壁炉数量
```
combine_df[combine_df['Fireplaces'].isnull()]
combine_df['Fireplaces'].value_counts()
```
### HasFireplace
是否有壁炉
```
combine_df['Has_Fireplace'] = combine_df['FireplaceQu'].apply(lambda x : 0 if pd.isnull(x) else 1)
```
### FireplaceQu
壁炉质量
```
combine_df['FireplaceQu'] = combine_df['FireplaceQu'].fillna('No_Fp')
le = preprocessing.LabelEncoder()
le.fit(combine_df['FireplaceQu'])
combine_df['FireplaceQu'] = le.transform(combine_df['FireplaceQu'])
```
### Has_GarageType
是否有车库、
```
combine_df['Has_Garage'] = combine_df['GarageType'].apply(lambda x : 0 if pd.isnull(x) else 1)
combine_df.ix[2127, 'Has_Garage'] = 0
combine_df.ix[2577, 'Has_Garage'] = 0
```
### GarageType
车库所在位置
```
type_df = combine_df[combine_df['GarageType'].isnull()]
combine_df['GarageType'] = combine_df['GarageType'].fillna('No_GT')
combine_df.ix[2127, 'GarageType'] = 'No_GT'
combine_df.ix[2577, 'GarageType'] = 'No_GT'
le = preprocessing.LabelEncoder()
le.fit(combine_df['GarageType'])
combine_df['GarageType'] = le.transform(combine_df['GarageType'])
```
### GarageYrBlt
车库建造年份
```
yt_df = combine_df[combine_df['GarageYrBlt'].isnull()]
set(yt_df.index) - set(type_df.index)
combine_df['GarageYrBlt'] = combine_df['GarageYrBlt'].fillna(2016)
year_garage_df = 2016 - combine_df['GarageYrBlt']
```
### GarageCars
车库能停几辆车
```
combine_df[combine_df['GarageCars'].isnull()]
combine_df['GarageCars'].median()
combine_df.ix[2577, 'GarageCars'] = 0
garage_cars_df = pd.DataFrame(preprocessing.scale(fixSkew(combine_df, 'GarageCars')), np.array(range(1, 2920)), columns=['GarageCars'])
garage_cars_df.index.name = 'Id'
```
### GarageArea
车库面积
```
combine_df[combine_df['GarageArea'].isnull()]
combine_df.ix[2577, 'GarageArea'] = 0
garage_area_df = pd.DataFrame(preprocessing.scale(combine_df['GarageArea']), np.array(range(1, 2920)), columns=['GarageArea'])
garage_area_df.index.name = 'Id'
```
### GarageQual
车库质量
```
combine_df[combine_df['GarageQual'].isnull() & (combine_df['Has_Garage'] == 1)]
combine_df['GarageQual'] = combine_df['GarageQual'].fillna('No_GT')
le = preprocessing.LabelEncoder()
le.fit(combine_df['GarageQual'])
combine_df['GarageQual'] = le.transform(combine_df['GarageQual'])
```
### GarageCond
车库条件
```
combine_df[combine_df['GarageCond'].isnull() & (combine_df['Has_Garage'] == 1)]
combine_df['GarageCond'] = combine_df['GarageQual'].fillna('No_GT')
le = preprocessing.LabelEncoder()
le.fit(combine_df['GarageCond'])
combine_df['GarageCond'] = le.transform(combine_df['GarageCond'])
```
### PavedDrive
汽车开的道路情况
```
combine_df[combine_df['PavedDrive'].isnull()]
le = preprocessing.LabelEncoder()
le.fit(combine_df['PavedDrive'])
combine_df['PavedDrive'] = le.transform(combine_df['PavedDrive'])
```
### WoodDeckSF
木甲板面积平方英尺
```
combine_df[combine_df['WoodDeckSF'].isnull()]
wood_deck_df = pd.DataFrame(preprocessing.scale(combine_df['WoodDeckSF']), np.array(range(1, 2920)), columns=['WoodDeckSF'])
wood_deck_df.index.name = 'Id'
```
### OpenPorchSF
开放玄关面积平方英尺
```
combine_df[combine_df['OpenPorchSF'].isnull()]
open_porch_sf_df = pd.DataFrame(preprocessing.scale(combine_df['OpenPorchSF']), np.array(range(1, 2920)), columns=['OpenPorchSF'])
open_porch_sf_df.index.name = 'Id'
```
### EnclosedPorch
封闭走廊地区平方英尺
```
combine_df[combine_df['EnclosedPorch'].isnull()]
enclose_porch_df = pd.DataFrame(preprocessing.scale(combine_df['EnclosedPorch']), np.array(range(1, 2920)), columns=['EnclosedPorch'])
enclose_porch_df.index.name = 'Id'
```
### 3SsnPorch
三面玄关面积平方英尺
```
combine_df[combine_df['3SsnPorch'].isnull()]
three_ssn_porch_df = pd.DataFrame(preprocessing.scale(combine_df['3SsnPorch']), np.array(range(1, 2920)), columns=['3SsnPorch'])
three_ssn_porch_df.index.name = 'Id'
```
### ScreenPorch
窗口玄关面积平方英尺
```
combine_df[combine_df['ScreenPorch'].isnull()]
screen_porch_df = pd.DataFrame(preprocessing.scale(combine_df['ScreenPorch']), np.array(range(1, 2920)), columns=['ScreenPorch'])
screen_porch_df.index.name = 'Id'
```
### Has_Pool
是否有游泳池
```
combine_df['Has_Pool'] = combine_df['PoolArea'].apply(lambda x:0 if x == 0 else 1 )
```
### PoolArea
游泳池面积
```
combine_df[combine_df['PoolArea'].isnull()]
pool_area_df = pd.DataFrame(preprocessing.scale(combine_df['PoolArea']), np.array(range(1, 2920)), columns=['PoolArea'])
pool_area_df.index.name = 'Id'
```
### PoolQC
游泳池质量
```
combine_df[combine_df['PoolQC'].isnull()]
combine_df['PoolQC'] = combine_df['PoolQC'].fillna('No_Pool')
le = preprocessing.LabelEncoder()
le.fit(combine_df['PoolQC'])
combine_df['PoolQC'] = le.transform(combine_df['PoolQC'])
```
### Fence
栅栏质量
```
combine_df[combine_df['Fence'].isnull()]
combine_df['Fence'] = combine_df['Fence'].fillna('No_Fence')
le = preprocessing.LabelEncoder()
le.fit(combine_df['Fence'])
combine_df['Fence'] = le.transform(combine_df['Fence'])
```
### MoSold
销售的月份
```
combine_df[combine_df['MoSold'].isnull()]
combine_df['MoSold'].value_counts()
```
### YrSold
销售的年份
```
combine_df[combine_df['YrSold'].isnull()]
combine_df['YrSold'].value_counts()
```
### SaleType
销售的类型
```
combine_df[combine_df['SaleType'].isnull()]
combine_df['SaleType'].value_counts()
combine_df.ix[2490, 'SaleType'] = 'WD'
le = preprocessing.LabelEncoder()
le.fit(combine_df['SaleType'])
combine_df['SaleType'] = le.transform(combine_df['SaleType'])
```
### SaleCondition
销售条件
```
combine_df[combine_df['SaleCondition'].isnull()]
le = preprocessing.LabelEncoder()
le.fit(combine_df['SaleCondition'])
combine_df['SaleCondition'] = le.transform(combine_df['SaleCondition'])
### MiscFeature
#另外一些特征
combine_df['MiscFeature'].value_counts()
```
#### 排除的特征
【Street】:不平衡
【Utilities】:不平衡
【Condition2】:不平衡
```
sns.distplot(garage_cars_df, fit=norm)
plt.show()
```
### 特征合并
合并所有特征
分离训练集和测试集
```
#单变量相关性低
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['Heating']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['Alley_Access']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['Alley']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['Has_Pool']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pool_area_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['BldgType']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtCond']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['GarageCond']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, low_qual_fin_sf_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtHalfBath']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['ExterQual']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['Has_Fireplace']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['KitchenAbvGr']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, enclose_porch_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, three_ssn_porch_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, screen_porch_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['MoSold']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['YrSold']), left_index=True, right_index=True)
#*******************************************************************************************************************************
#多变量相关性较强的特征
# X_df = pd.merge(X_df, garage_area_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['TotRmsAbvGrd']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, fst_flr_sf_df, left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(gr_liv_area_df, overall_qual_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, bsmt_fin_SF1_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['GarageQual']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Electrical']), left_index=True, right_index=True)
X_df = pd.merge(X_df, total_bsmt_sf_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['LotShape']), left_index=True, right_index=True)
X_df = pd.merge(X_df, lot_area_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, lot_frontage_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['LandContour']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['LotConfig']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Neighborhood']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Condition1']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Condition2']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['HouseStyle']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, overall_cond_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, year_built_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, year_remodadd_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['RoofStyle']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['RoofMatl']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Exterior1st']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Exterior2nd']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['MasVnrType']), left_index=True, right_index=True)
X_df = pd.merge(X_df, mas_vnr_area_df, left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['ExterCond']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Foundation']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Has_Bsmt']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtQual']), left_index=True, right_index=True)
#*******************************************************************************************************************************
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtExposure']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtFinType1']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtFinType2']), left_index=True, right_index=True)
X_df = pd.merge(X_df, bsmt_fin_SF2_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, bsmt_unf_sf_df, left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['HeatingQC']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['CentralAir']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, snd_flr_sf_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtFullBath']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['FullBath']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['HalfBath']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BedroomAbvGr']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['KitchenQual']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Functional']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['FireplaceQu']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Has_Garage']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['GarageType']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['GarageYrBlt']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, garage_cars_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, wood_deck_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, open_porch_sf_df, left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['PoolQC']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Fence']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['SaleType']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['SaleCondition']), left_index=True, right_index=True)
X_train_df = X_df.loc[1:1460]
X_test_df = X_df.loc[1461:2919]
#norm_y = preprocessing.scale(train_df['SalePrice'])
y_train_df = np.log1p(train_df['SalePrice'])
sns.distplot(y_train_df, fit=norm);
fig = plt.figure()
res = stats.probplot(y_train_df, plot=plt)
plt.show()
X_train_df.to_csv('../data/offline/X_train.csv', header = True, index=True)
X_test_df.to_csv('../data/offline/X_test.csv', header = True, index=True)
y_train_df.to_csv('../data/offline/y_train.csv', header = True, index=True)
len(X_test_df)
y_train_df.describe()
y_train_df
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from sklearn import preprocessing
import seaborn as sns
from scipy.stats import norm
from scipy import stats
from scipy.stats import skew
train_df = pd.read_csv('../data/orignal/train.csv', index_col = 0)
test_df = pd.read_csv('../data/orignal/test.csv', index_col = 0)
combine_df = pd.concat([train_df, test_df])
# 相关性检测
#correlation matrix
corrmat = train_df.corr()
f, ax = plt.subplots(figsize=(12, 9))
sns.heatmap(corrmat, vmax=.8, square=True)
def fixSkew(feature_df, name):
skewed_feat = skew(feature_df[name])
if skewed_feat > 0.75:
print('fix')
return np.log1p(feature_df[name])
else:
print('notfix')
return feature_df[name]
plt.show()
#saleprice correlation matrix
k = 10 #number of variables for heatmap
cols = corrmat.nlargest(k, 'SalePrice')['SalePrice'].index
cm = np.corrcoef(train_df[cols].values.T)
sns.set(font_scale=0.75)
hm = sns.heatmap(cm, cbar=True, annot=True, square=True, fmt='.2f', annot_kws={'size': 10}, yticklabels=cols.values, xticklabels=cols.values)
plt.show()
combine_df[combine_df['MSSubClass'].isnull()]
combine_df['MSZoning'] = combine_df['MSZoning'].fillna('RL')
le = preprocessing.LabelEncoder()
le.fit(combine_df['MSZoning'])
combine_df['MSZoning'] = le.transform(combine_df['MSZoning'])
lot_frontage_df = combine_df['LotFrontage'].fillna(combine_df['LotFrontage'].median())
lot_frontage_df = pd.DataFrame(preprocessing.scale(lot_frontage_df.values), np.array(range(1, 2920)), columns=['LotFrontage'])
lot_frontage_df.index.name = 'Id'
lot_area_df = pd.DataFrame(preprocessing.scale(combine_df['LotArea']), np.array(range(1, 2920)), columns=['LotArea'])
lot_area_df.index.name = 'Id'
combine_df['Street'].value_counts()
combine_df['Alley_Access'] = combine_df['Alley'].apply(lambda x : 0 if pd.isnull(x) else 1)
combine_df['Alley'] = combine_df['Alley'].fillna('NoAccess')
combine_df['Alley'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['Alley'])
combine_df['Alley'] = le.transform(combine_df['Alley'])
combine_df['LotShape'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['LotShape'])
combine_df['LotShape'] = le.transform(combine_df['LotShape'])
combine_df['LandContour'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['LandContour'])
combine_df['LandContour'] = le.transform(combine_df['LandContour'])
combine_df['Utilities'].value_counts()
combine_df['LotConfig'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['LotConfig'])
combine_df['LotConfig'] = le.transform(combine_df['LotConfig'])
combine_df['LandSlope'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['LandSlope'])
combine_df['LandSlope'] = le.transform(combine_df['LandSlope'])
combine_df['Neighborhood'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['Neighborhood'])
combine_df['Neighborhood'] = le.transform(combine_df['Neighborhood'])
le = preprocessing.LabelEncoder()
le.fit(combine_df['Condition1'])
combine_df['Condition1'] = le.transform(combine_df['Condition1'])
combine_df['Condition1'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['Condition2'])
combine_df['Condition2'] = le.transform(combine_df['Condition2'])
combine_df['Condition2'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['BldgType'])
combine_df['BldgType'] = le.transform(combine_df['BldgType'])
combine_df['BldgType'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['HouseStyle'])
combine_df['HouseStyle'] = le.transform(combine_df['HouseStyle'])
combine_df['HouseStyle'].value_counts()
sns.distplot(np.log(combine_df['OverallQual']), fit=norm);
fig = plt.figure()
plt.show()
combine_df['OverallQual']
overall_qual_df = pd.DataFrame(preprocessing.scale(fixSkew(combine_df, 'OverallQual')), np.array(range(1, 2920)), columns=['OverallQual'])
overall_qual_df.index.name = 'Id'
overall_cond_df = pd.DataFrame(preprocessing.scale(combine_df['OverallCond'].values), np.array(range(1, 2920)), columns=['OverallCond'])
overall_cond_df.index.name = 'Id'
combine_df['YearBuilt'] = combine_df['YearBuilt'].apply(lambda x : 2016 - x)
year_built_df = pd.DataFrame(preprocessing.scale(fixSkew(combine_df, 'YearBuilt')), np.array(range(1, 2920)), columns=['YearBuilt'])
year_remodadd_df = pd.DataFrame(2016 - combine_df['YearRemodAdd'])
le = preprocessing.LabelEncoder()
le.fit(combine_df['RoofStyle'])
combine_df['RoofStyle'] = le.transform(combine_df['RoofStyle'])
combine_df['RoofStyle'].value_counts()
le = preprocessing.LabelEncoder()
le.fit(combine_df['RoofMatl'])
combine_df['RoofMatl'] = le.transform(combine_df['RoofMatl'])
combine_df['RoofMatl'].value_counts()
combine_df[combine_df['Exterior1st'].isnull()]
combine_df['Exterior1st'] = combine_df['Exterior1st'].fillna('VinylSd')
le = preprocessing.LabelEncoder()
le.fit(combine_df['Exterior1st'])
combine_df['Exterior1st'] = le.transform(combine_df['Exterior1st'])
combine_df['Exterior1st'].value_counts()
combine_df[combine_df['Exterior2nd'].isnull()]
combine_df['Exterior2nd'] = combine_df['Exterior2nd'].fillna('VinylSd')
le = preprocessing.LabelEncoder()
le.fit(combine_df['Exterior2nd'])
combine_df['Exterior2nd'] = le.transform(combine_df['Exterior2nd'])
combine_df['Exterior2nd'].value_counts()
combine_df['MasVnrType'] = combine_df['MasVnrType'].fillna('None')
le = preprocessing.LabelEncoder()
le.fit(combine_df['MasVnrType'])
combine_df['MasVnrType'] = le.transform(combine_df['MasVnrType'])
combine_df['MasVnrType'].value_counts()
combine_df['MasVnrArea'].median()
combine_df['MasVnrArea'] = combine_df['MasVnrArea'].fillna(combine_df['MasVnrArea'].median())
mas_vnr_area_df = pd.DataFrame(preprocessing.scale(combine_df['MasVnrArea']), np.array(range(1, 2920)), columns=['MasVnrArea'])
mas_vnr_area_df.index.name = 'Id'
combine_df['ExterQual'].isnull().any()
le = preprocessing.LabelEncoder()
le.fit(combine_df['ExterQual'])
combine_df['ExterQual'] = le.transform(combine_df['ExterQual'])
combine_df['ExterQual'].value_counts()
combine_df['ExterCond'].isnull().any()
le = preprocessing.LabelEncoder()
le.fit(combine_df['ExterCond'])
combine_df['ExterCond'] = le.transform(combine_df['ExterCond'])
combine_df['ExterCond'].value_counts()
combine_df['Foundation'].isnull().any()
le = preprocessing.LabelEncoder()
le.fit(combine_df['Foundation'])
combine_df['Foundation'] = le.transform(combine_df['Foundation'])
combine_df['Foundation'].value_counts()
combine_df['Has_Bsmt'] = combine_df['BsmtQual'].apply(lambda x : 0 if pd.isnull(x) else 1)
combine_df['BsmtQual'] = combine_df['BsmtQual'].fillna('No_Bsmt')
le = preprocessing.LabelEncoder()
le.fit(combine_df['BsmtQual'])
combine_df['BsmtQual'] = le.transform(combine_df['BsmtQual'])
combine_df['BsmtCond'] = combine_df['BsmtCond'].fillna('No_Bsmt')
le = preprocessing.LabelEncoder()
le.fit(combine_df['BsmtCond'])
combine_df['BsmtCond'] = le.transform(combine_df['BsmtCond'])
combine_df['BsmtExposure'] = combine_df['BsmtExposure'].fillna('No_Bsmt')
le = preprocessing.LabelEncoder()
le.fit(combine_df['BsmtExposure'])
combine_df['BsmtExposure'] = le.transform(combine_df['BsmtExposure'])
combine_df['BsmtFinType1'] = combine_df['BsmtFinType1'].fillna('No_Bsmt')
le = preprocessing.LabelEncoder()
le.fit(combine_df['BsmtFinType1'])
combine_df['BsmtFinType1'] = le.transform(combine_df['BsmtFinType1'])
combine_df['BsmtFinSF1'] = combine_df['BsmtFinSF1'].fillna(0)
bsmt_fin_SF1_df = pd.DataFrame(preprocessing.scale(combine_df['BsmtFinSF1']), np.array(range(1, 2920)), columns=['BsmtFinSF1'])
bsmt_fin_SF1_df.index.name = 'Id'
combine_df['BsmtFinType2'] = combine_df['BsmtFinType2'].fillna('No_Bsmt')
le = preprocessing.LabelEncoder()
le.fit(combine_df['BsmtFinType2'])
combine_df['BsmtFinType2'] = le.transform(combine_df['BsmtFinType2'])
combine_df['BsmtFinSF2'] = combine_df['BsmtFinSF2'].fillna(0)
bsmt_fin_SF2_df = pd.DataFrame(preprocessing.scale(combine_df['BsmtFinSF2']), np.array(range(1, 2920)), columns=['BsmtFinSF2'])
bsmt_fin_SF2_df.index.name = 'Id'
combine_df[combine_df['BsmtUnfSF'].isnull()]
combine_df.ix[2121, 'BsmtUnfSF'] = 0
bsmt_unf_sf_df = pd.DataFrame(preprocessing.scale(combine_df['BsmtUnfSF']), np.array(range(1, 2920)), columns=['BsmtUnfSF'])
bsmt_unf_sf_df.index.name = 'Id'
combine_df[combine_df['TotalBsmtSF'].isnull()]
combine_df.ix[2121, 'TotalBsmtSF'] = 0
total_bsmt_sf_df = pd.DataFrame(preprocessing.scale(fixSkew(combine_df, 'TotalBsmtSF')), np.array(range(1, 2920)), columns=['TotalBsmtSF'])
total_bsmt_sf_df.index.name = 'Id'
combine_df['TotalBsmtSF'].describe()
combine_df[combine_df['Heating'].isnull()]
le = preprocessing.LabelEncoder()
le.fit(combine_df['Heating'])
combine_df['Heating'] = le.transform(combine_df['Heating'])
combine_df[combine_df['HeatingQC'].isnull()]
le = preprocessing.LabelEncoder()
le.fit(combine_df['HeatingQC'])
combine_df['HeatingQC'] = le.transform(combine_df['HeatingQC'])
combine_df[combine_df['CentralAir'].isnull()]
le = preprocessing.LabelEncoder()
le.fit(combine_df['CentralAir'])
combine_df['CentralAir'] = le.transform(combine_df['CentralAir'])
combine_df[combine_df['Electrical'].isnull()]
combine_df['Electrical'].value_counts()
combine_df.ix[1380, 'Electrical'] = 'SBrkr'
le = preprocessing.LabelEncoder()
le.fit(combine_df['Electrical'])
combine_df['Electrical'] = le.transform(combine_df['Electrical'])
combine_df[combine_df['1stFlrSF'].isnull()]
fst_flr_sf_df = pd.DataFrame(preprocessing.scale(combine_df['1stFlrSF']), np.array(range(1, 2920)), columns=['1stFlrSF'])
fst_flr_sf_df.index.name = 'Id'
combine_df[combine_df['2ndFlrSF'].isnull()]
snd_flr_sf_df = pd.DataFrame(preprocessing.scale(combine_df['2ndFlrSF']), np.array(range(1, 2920)), columns=['2ndFlrSF'])
snd_flr_sf_df.index.name = 'Id'
combine_df[combine_df['LowQualFinSF'].isnull()]
low_qual_fin_sf_df = pd.DataFrame(preprocessing.scale(combine_df['LowQualFinSF']), np.array(range(1, 2920)), columns=['LowQualFinSF'])
low_qual_fin_sf_df.index.name = 'Id'
combine_df[combine_df['GrLivArea'].isnull()]
gr_liv_area_df = pd.DataFrame(preprocessing.scale(fixSkew(combine_df, 'GrLivArea')), np.array(range(1, 2920)), columns=['GrLivArea'])
gr_liv_area_df.index.name = 'Id'
combine_df[combine_df['BsmtFullBath'].isnull()]
combine_df['BsmtFullBath'].value_counts()
combine_df.ix[2121, 'Has_Bsmt']
combine_df.ix[2189, 'Has_Bsmt']
combine_df['BsmtFullBath'] = combine_df['BsmtFullBath'].fillna(0).astype(np.int)
combine_df[combine_df['BsmtHalfBath'].isnull()]
combine_df['BsmtHalfBath'].value_counts()
combine_df['BsmtHalfBath'] = combine_df['BsmtHalfBath'].fillna(0).astype(np.int)
combine_df[combine_df['FullBath'].isnull()]
combine_df['FullBath'].value_counts()
combine_df[combine_df['HalfBath'].isnull()]
combine_df['HalfBath'].value_counts()
combine_df[combine_df['BedroomAbvGr'].isnull()]
combine_df['BedroomAbvGr'].value_counts()
combine_df[combine_df['KitchenAbvGr'].isnull()]
combine_df['KitchenAbvGr'].value_counts()
combine_df[combine_df['KitchenQual'].isnull()]
combine_df['KitchenQual'].value_counts()
combine_df.ix[1556, 'KitchenQual'] = 'TA'
le = preprocessing.LabelEncoder()
le.fit(combine_df['KitchenQual'])
combine_df['KitchenQual'] = le.transform(combine_df['KitchenQual'])
combine_df[combine_df['TotRmsAbvGrd'].isnull()]
combine_df['TotRmsAbvGrd'].value_counts()
combine_df[combine_df['Functional'].isnull()]
combine_df['Functional'].value_counts()
combine_df.ix[2217, 'Functional'] = 'Typ'
combine_df.ix[2474, 'Functional'] = 'Typ'
le = preprocessing.LabelEncoder()
le.fit(combine_df['Functional'])
combine_df['Functional'] = le.transform(combine_df['Functional'])
combine_df[combine_df['Fireplaces'].isnull()]
combine_df['Fireplaces'].value_counts()
combine_df['Has_Fireplace'] = combine_df['FireplaceQu'].apply(lambda x : 0 if pd.isnull(x) else 1)
combine_df['FireplaceQu'] = combine_df['FireplaceQu'].fillna('No_Fp')
le = preprocessing.LabelEncoder()
le.fit(combine_df['FireplaceQu'])
combine_df['FireplaceQu'] = le.transform(combine_df['FireplaceQu'])
combine_df['Has_Garage'] = combine_df['GarageType'].apply(lambda x : 0 if pd.isnull(x) else 1)
combine_df.ix[2127, 'Has_Garage'] = 0
combine_df.ix[2577, 'Has_Garage'] = 0
type_df = combine_df[combine_df['GarageType'].isnull()]
combine_df['GarageType'] = combine_df['GarageType'].fillna('No_GT')
combine_df.ix[2127, 'GarageType'] = 'No_GT'
combine_df.ix[2577, 'GarageType'] = 'No_GT'
le = preprocessing.LabelEncoder()
le.fit(combine_df['GarageType'])
combine_df['GarageType'] = le.transform(combine_df['GarageType'])
yt_df = combine_df[combine_df['GarageYrBlt'].isnull()]
set(yt_df.index) - set(type_df.index)
combine_df['GarageYrBlt'] = combine_df['GarageYrBlt'].fillna(2016)
year_garage_df = 2016 - combine_df['GarageYrBlt']
combine_df[combine_df['GarageCars'].isnull()]
combine_df['GarageCars'].median()
combine_df.ix[2577, 'GarageCars'] = 0
garage_cars_df = pd.DataFrame(preprocessing.scale(fixSkew(combine_df, 'GarageCars')), np.array(range(1, 2920)), columns=['GarageCars'])
garage_cars_df.index.name = 'Id'
combine_df[combine_df['GarageArea'].isnull()]
combine_df.ix[2577, 'GarageArea'] = 0
garage_area_df = pd.DataFrame(preprocessing.scale(combine_df['GarageArea']), np.array(range(1, 2920)), columns=['GarageArea'])
garage_area_df.index.name = 'Id'
combine_df[combine_df['GarageQual'].isnull() & (combine_df['Has_Garage'] == 1)]
combine_df['GarageQual'] = combine_df['GarageQual'].fillna('No_GT')
le = preprocessing.LabelEncoder()
le.fit(combine_df['GarageQual'])
combine_df['GarageQual'] = le.transform(combine_df['GarageQual'])
combine_df[combine_df['GarageCond'].isnull() & (combine_df['Has_Garage'] == 1)]
combine_df['GarageCond'] = combine_df['GarageQual'].fillna('No_GT')
le = preprocessing.LabelEncoder()
le.fit(combine_df['GarageCond'])
combine_df['GarageCond'] = le.transform(combine_df['GarageCond'])
combine_df[combine_df['PavedDrive'].isnull()]
le = preprocessing.LabelEncoder()
le.fit(combine_df['PavedDrive'])
combine_df['PavedDrive'] = le.transform(combine_df['PavedDrive'])
combine_df[combine_df['WoodDeckSF'].isnull()]
wood_deck_df = pd.DataFrame(preprocessing.scale(combine_df['WoodDeckSF']), np.array(range(1, 2920)), columns=['WoodDeckSF'])
wood_deck_df.index.name = 'Id'
combine_df[combine_df['OpenPorchSF'].isnull()]
open_porch_sf_df = pd.DataFrame(preprocessing.scale(combine_df['OpenPorchSF']), np.array(range(1, 2920)), columns=['OpenPorchSF'])
open_porch_sf_df.index.name = 'Id'
combine_df[combine_df['EnclosedPorch'].isnull()]
enclose_porch_df = pd.DataFrame(preprocessing.scale(combine_df['EnclosedPorch']), np.array(range(1, 2920)), columns=['EnclosedPorch'])
enclose_porch_df.index.name = 'Id'
combine_df[combine_df['3SsnPorch'].isnull()]
three_ssn_porch_df = pd.DataFrame(preprocessing.scale(combine_df['3SsnPorch']), np.array(range(1, 2920)), columns=['3SsnPorch'])
three_ssn_porch_df.index.name = 'Id'
combine_df[combine_df['ScreenPorch'].isnull()]
screen_porch_df = pd.DataFrame(preprocessing.scale(combine_df['ScreenPorch']), np.array(range(1, 2920)), columns=['ScreenPorch'])
screen_porch_df.index.name = 'Id'
combine_df['Has_Pool'] = combine_df['PoolArea'].apply(lambda x:0 if x == 0 else 1 )
combine_df[combine_df['PoolArea'].isnull()]
pool_area_df = pd.DataFrame(preprocessing.scale(combine_df['PoolArea']), np.array(range(1, 2920)), columns=['PoolArea'])
pool_area_df.index.name = 'Id'
combine_df[combine_df['PoolQC'].isnull()]
combine_df['PoolQC'] = combine_df['PoolQC'].fillna('No_Pool')
le = preprocessing.LabelEncoder()
le.fit(combine_df['PoolQC'])
combine_df['PoolQC'] = le.transform(combine_df['PoolQC'])
combine_df[combine_df['Fence'].isnull()]
combine_df['Fence'] = combine_df['Fence'].fillna('No_Fence')
le = preprocessing.LabelEncoder()
le.fit(combine_df['Fence'])
combine_df['Fence'] = le.transform(combine_df['Fence'])
combine_df[combine_df['MoSold'].isnull()]
combine_df['MoSold'].value_counts()
combine_df[combine_df['YrSold'].isnull()]
combine_df['YrSold'].value_counts()
combine_df[combine_df['SaleType'].isnull()]
combine_df['SaleType'].value_counts()
combine_df.ix[2490, 'SaleType'] = 'WD'
le = preprocessing.LabelEncoder()
le.fit(combine_df['SaleType'])
combine_df['SaleType'] = le.transform(combine_df['SaleType'])
combine_df[combine_df['SaleCondition'].isnull()]
le = preprocessing.LabelEncoder()
le.fit(combine_df['SaleCondition'])
combine_df['SaleCondition'] = le.transform(combine_df['SaleCondition'])
### MiscFeature
#另外一些特征
combine_df['MiscFeature'].value_counts()
sns.distplot(garage_cars_df, fit=norm)
plt.show()
#单变量相关性低
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['Heating']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['Alley_Access']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['Alley']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['Has_Pool']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pool_area_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['BldgType']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtCond']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['GarageCond']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, low_qual_fin_sf_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtHalfBath']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['ExterQual']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['Has_Fireplace']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['KitchenAbvGr']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, enclose_porch_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, three_ssn_porch_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, screen_porch_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['MoSold']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['YrSold']), left_index=True, right_index=True)
#*******************************************************************************************************************************
#多变量相关性较强的特征
# X_df = pd.merge(X_df, garage_area_df, left_index=True, right_index=True)
# X_df = pd.merge(X_df, pd.DataFrame(combine_df['TotRmsAbvGrd']), left_index=True, right_index=True)
# X_df = pd.merge(X_df, fst_flr_sf_df, left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(gr_liv_area_df, overall_qual_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, bsmt_fin_SF1_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['GarageQual']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Electrical']), left_index=True, right_index=True)
X_df = pd.merge(X_df, total_bsmt_sf_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['LotShape']), left_index=True, right_index=True)
X_df = pd.merge(X_df, lot_area_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, lot_frontage_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['LandContour']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['LotConfig']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Neighborhood']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Condition1']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Condition2']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['HouseStyle']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, overall_cond_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, year_built_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, year_remodadd_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['RoofStyle']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['RoofMatl']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Exterior1st']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Exterior2nd']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['MasVnrType']), left_index=True, right_index=True)
X_df = pd.merge(X_df, mas_vnr_area_df, left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['ExterCond']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Foundation']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Has_Bsmt']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtQual']), left_index=True, right_index=True)
#*******************************************************************************************************************************
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtExposure']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtFinType1']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtFinType2']), left_index=True, right_index=True)
X_df = pd.merge(X_df, bsmt_fin_SF2_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, bsmt_unf_sf_df, left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['HeatingQC']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['CentralAir']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, snd_flr_sf_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BsmtFullBath']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['FullBath']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['HalfBath']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['BedroomAbvGr']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['KitchenQual']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Functional']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['FireplaceQu']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Has_Garage']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['GarageType']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['GarageYrBlt']), left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, garage_cars_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, wood_deck_df, left_index=True, right_index=True)
X_df = pd.merge(X_df, open_porch_sf_df, left_index=True, right_index=True)
#*******************************************************************************************************************************
X_df = pd.merge(X_df, pd.DataFrame(combine_df['PoolQC']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['Fence']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['SaleType']), left_index=True, right_index=True)
X_df = pd.merge(X_df, pd.DataFrame(combine_df['SaleCondition']), left_index=True, right_index=True)
X_train_df = X_df.loc[1:1460]
X_test_df = X_df.loc[1461:2919]
#norm_y = preprocessing.scale(train_df['SalePrice'])
y_train_df = np.log1p(train_df['SalePrice'])
sns.distplot(y_train_df, fit=norm);
fig = plt.figure()
res = stats.probplot(y_train_df, plot=plt)
plt.show()
X_train_df.to_csv('../data/offline/X_train.csv', header = True, index=True)
X_test_df.to_csv('../data/offline/X_test.csv', header = True, index=True)
y_train_df.to_csv('../data/offline/y_train.csv', header = True, index=True)
len(X_test_df)
y_train_df.describe()
y_train_df
| 0.314156 | 0.767951 |
```
import numpy as np
a=np.array([[1,2,3,4]])
np.broadcast(a,(3,2))
x=np.array([1,2,3])
x
x.shape
y=np.array([[1],[2],[3]])
y.shape
y
z=np.array([[[2],[4],[5]],[[6],[7],[9]]])
z
z.shape
np.broadcast_arrays(x,y,z)
y=np.expand_dims(x,axis=0)
y
y.shape
z=np.expand_dims(x,axis=1)
z
z.shape
np.newaxis is None
x=np.array([[[0],[1],[2]]])
x
x.shape
y=np.squeeze(x)
y
y.shape
x
np.squeeze(x,axis=0).shape
np.asfarray([2,3],dtype='float') ##floating array
np.asfarray([2,3],dtype='int8')
a=np.array([[1,2],[3,4]])
a
b=np.array([[5,6]])
b.shape
np.concatenate((a,b),axis=0)
np.concatenate((a,b.T),axis=1)
a=np.ma.arange(3)
a
a[1]=np.ma.masked
a
b=np.arange(2,5)
b
np.ma.concatenate([a,b]) ##b is also converted into mask when add with masked array
```
10+2
```
10+10
_
[np.random.randn(3,4) for _ in range(10)]
[np.random.randn(3,4) for _ in range(10)]
np.stack(arrays,axis=0).shape
np.stack(arrays,axis=1).shape
a=([[1,2,3]])
a
b=([2,3,6])
b
np.column_stack((a,b))
np.hstack((a,b))
import numpy as np
p=np.arange(1,5,2)
p
np.diagflat([[1,2], [3,4]])
M = ri(1,100,25).reshape(5,5) # Matrix of random interegrs
print("\n5x5 Matrix of random integers\n",'-'*50,"\n",M)
print("\nHere is the sorted matrix along each row\n",'-'*50,"\n",np.sort(M, kind='merge
sort')) # Default axis =1
print("\nHere is the sorted matrix along each column\n",'-'*50,"\n",np.sort(M, axis=0,
M = ri(1,100,25).reshape(5,5) # Matrix of random interegrs
print("\n5x5 Matrix of random integers\n",'-'*50,"\n",M)
print("\nHere is the sorted matrix along each row\n",'-'*50,"\n",np.sort(M, kind='merge
sort')) # Default axis =1
print("\nHere is the sorted matrix along each column\n",'-'*50,"\n",np.sort(M, axis=0,
###friday
l1=[1,2,3]
l2=[4,5,6]
l3=[7,8,9]
```
#
```
l1
##friday if we give a sqre bracket then it vl consider indexes
import numpy as np
x=np.arange(12.0)
x
np.split(x,[3,4])
y=np.array([11,22,33,44,55,66,77,88,99])
y
np.split(y,[3,4,8,10])
np.array_split(y,8)
x=np.arange(16.0).reshape(2,2,4)
x
np.dsplit(x,2)
np.dsplit(x,np.array([3,4]))
x
np.dsplit(x,np.array([2,2]))
np.dsplit(x,np.array([3,2])) ###here first 3 takes based on
x
np.dsplit(x,np.array([1,3])) ###here takes last two and 3 takess first 3
np.dsplit(x,np.array([2,2])) ###here takes last two and 3 takess first 3
#### np.array it gives the range in bw 2 nums,suppose (2,5).here the range is 2,3,4 so it creates in middle rest of the arrays created above and belo of this
x
##practice
from numpy.random import randint as ri
import numpy as np
mat=np.array(ri(10,100,15)).reshape(3,5)
```
print('random int from 10 to 100\n',mat)
```
print('random int from 10 to 100\n:',mat)
a=ri(1,100,30)
a
print("shape of a\n:",a.shape)
a.reshape(2,5,3)
j=a>25
j
```
|
github_jupyter
|
import numpy as np
a=np.array([[1,2,3,4]])
np.broadcast(a,(3,2))
x=np.array([1,2,3])
x
x.shape
y=np.array([[1],[2],[3]])
y.shape
y
z=np.array([[[2],[4],[5]],[[6],[7],[9]]])
z
z.shape
np.broadcast_arrays(x,y,z)
y=np.expand_dims(x,axis=0)
y
y.shape
z=np.expand_dims(x,axis=1)
z
z.shape
np.newaxis is None
x=np.array([[[0],[1],[2]]])
x
x.shape
y=np.squeeze(x)
y
y.shape
x
np.squeeze(x,axis=0).shape
np.asfarray([2,3],dtype='float') ##floating array
np.asfarray([2,3],dtype='int8')
a=np.array([[1,2],[3,4]])
a
b=np.array([[5,6]])
b.shape
np.concatenate((a,b),axis=0)
np.concatenate((a,b.T),axis=1)
a=np.ma.arange(3)
a
a[1]=np.ma.masked
a
b=np.arange(2,5)
b
np.ma.concatenate([a,b]) ##b is also converted into mask when add with masked array
10+10
_
[np.random.randn(3,4) for _ in range(10)]
[np.random.randn(3,4) for _ in range(10)]
np.stack(arrays,axis=0).shape
np.stack(arrays,axis=1).shape
a=([[1,2,3]])
a
b=([2,3,6])
b
np.column_stack((a,b))
np.hstack((a,b))
import numpy as np
p=np.arange(1,5,2)
p
np.diagflat([[1,2], [3,4]])
M = ri(1,100,25).reshape(5,5) # Matrix of random interegrs
print("\n5x5 Matrix of random integers\n",'-'*50,"\n",M)
print("\nHere is the sorted matrix along each row\n",'-'*50,"\n",np.sort(M, kind='merge
sort')) # Default axis =1
print("\nHere is the sorted matrix along each column\n",'-'*50,"\n",np.sort(M, axis=0,
M = ri(1,100,25).reshape(5,5) # Matrix of random interegrs
print("\n5x5 Matrix of random integers\n",'-'*50,"\n",M)
print("\nHere is the sorted matrix along each row\n",'-'*50,"\n",np.sort(M, kind='merge
sort')) # Default axis =1
print("\nHere is the sorted matrix along each column\n",'-'*50,"\n",np.sort(M, axis=0,
###friday
l1=[1,2,3]
l2=[4,5,6]
l3=[7,8,9]
l1
##friday if we give a sqre bracket then it vl consider indexes
import numpy as np
x=np.arange(12.0)
x
np.split(x,[3,4])
y=np.array([11,22,33,44,55,66,77,88,99])
y
np.split(y,[3,4,8,10])
np.array_split(y,8)
x=np.arange(16.0).reshape(2,2,4)
x
np.dsplit(x,2)
np.dsplit(x,np.array([3,4]))
x
np.dsplit(x,np.array([2,2]))
np.dsplit(x,np.array([3,2])) ###here first 3 takes based on
x
np.dsplit(x,np.array([1,3])) ###here takes last two and 3 takess first 3
np.dsplit(x,np.array([2,2])) ###here takes last two and 3 takess first 3
#### np.array it gives the range in bw 2 nums,suppose (2,5).here the range is 2,3,4 so it creates in middle rest of the arrays created above and belo of this
x
##practice
from numpy.random import randint as ri
import numpy as np
mat=np.array(ri(10,100,15)).reshape(3,5)
print('random int from 10 to 100\n:',mat)
a=ri(1,100,30)
a
print("shape of a\n:",a.shape)
a.reshape(2,5,3)
j=a>25
j
| 0.311008 | 0.77343 |
In this notebook, I look through the data from 2020 and construct a model for each team who got to play.
Set up the libraries and connect to the database.
```
import numpy
import pandas
from sqlalchemy import create_engine
import matplotlib.pyplot as plt
%matplotlib inline
import pymc3 as pm
engine = create_engine('postgresql://cheese:cheesepass4279@localhost:5432/cheesecake')
```
In the first query, pull each match, with each row representing each team. Then, process the data to have columns represent breakdown attributes.
```
query = """
select alliance.key, alliance.color, alliance_teams.team_key, match.match_number, alliance.score,
match.score_breakdown->'red' as breakdown_red,
match.score_breakdown->'blue' as breakdown_blue,
alliance_teams.position
from match
inner join alliance on alliance.match_key = match.key
inner join alliance_teams on alliance_teams.alliance_id = alliance.key
where comp_level = 'qm' and alliance.key like '2020%%'
"""
with engine.connect() as conn, conn.begin():
data = pandas.read_sql(query, conn)
data.loc[data.color == 'red', 'breakdown'] = data.loc[data.color == 'red', 'breakdown_red']
data.loc[data.color == 'blue', 'breakdown'] = data.loc[data.color == 'blue', 'breakdown_blue']
data = data.drop(['breakdown_red', 'breakdown_blue'], axis=1)
df = pandas.concat([
data.drop(['breakdown'], axis=1),
data['breakdown'].apply(pandas.Series)
], axis=1)
df
```
Translate team keys to numbers for the model, then run the model.
```
id2team = dict(enumerate(data['team_key'].unique()))
team2id = dict(zip(id2team.values(), id2team.keys()))
tms1 = data['team_key'][0::3].apply(lambda x: team2id.get(x)).values
tms2 = data['team_key'][1::3].apply(lambda x: team2id.get(x)).values
tms3 = data['team_key'][2::3].apply(lambda x: team2id.get(x)).values
with pm.Model() as model:
auto_score = pm.Gamma("auto_score", alpha=1.5, beta=0.1, shape=len(id2team))
tele_score = pm.Gamma("tele_score", alpha=1.5, beta=0.1, shape=len(id2team))
theta_auto = (auto_score[tms1] + auto_score[tms2] + auto_score[tms3])
theta_tele = (tele_score[tms1] + tele_score[tms2] + tele_score[tms3])
points = pm.Poisson('autoCellPoints', mu=theta_auto, observed=df['autoCellPoints'][1::3].values)
telepoints = pm.Poisson('teleopCellPoints', mu=theta_tele, observed=df['teleopCellPoints'][1::3].values)
trace = pm.sample(1000)
```
Construct a dataframe for each scoring section.
```
post = pandas.DataFrame({
'auto': numpy.median(trace['auto_score'], axis=0),
'tele': numpy.median(trace['tele_score'], axis=0)
}, index=[id2team[i] for i in range(trace['auto_score'].shape[1])])
for i in range(0, 3):
df.loc[df.position == i, 'initLine'] = df.loc[df.position == i, 'initLineRobot{}'.format(i + 1)]
df.loc[:, 'initLine'] = (df['initLine'] == 'Exited') * 5
df.loc[:, ['key', 'team_key', 'initLine']]
for i in range(0, 3):
df.loc[df.position == i, 'endgame'] = df.loc[df.position == i, 'endgameRobot{}'.format(i + 1)]
val_map = {'Hang': 25, 'Park': 5, 'None': 0}
df.loc[:, 'endgame'] = df['endgame'].replace(val_map)
post['initLine'] = df.groupby('team_key')['initLine'].mean()
post['endgame'] = df.groupby('team_key')['endgame'].mean()
climb_pts = ((((df['position'] == 0) & (df['endgameRobot1'] == "Hang")) |
((df['position'] == 1) & (df['endgameRobot2'] == "Hang")) |
((df['position'] == 2) & (df['endgameRobot3'] == "Hang"))) &
(df['endgameRungIsLevel'] == 'IsLevel')) * 15
df['balance_points'] = (climb_pts / df['tba_numRobotsHanging']).replace(numpy.inf, 0).fillna(0)
post['endgame_balance'] = df.groupby('team_key')['balance_points'].mean()
post
post.sum(axis=1).quantile([.1, .25, .5, .75, .9])
post.sum(axis=1).hist(bins=20, range=(0,100), density=True)
post['auto'].hist(bins=30, range=(0,30))
post['tele'].hist(bins=25, range=(0,50))
post['initLine'].hist(bins=5, range=(0,5))
post['endgame'].hist(bins=15, range=(0,30))
post['endgame_balance'].hist(bins=15, range=(0,15))
post['tele'].median()
```
## OPR
```
df.groupby('key')
data = df[df.key.str.startswith('2020ncwak')]
data
teams.sort()
matrix = []
scores = []
for i, (x, y) in enumerate(data.groupby('key')):
li = []
for team in teams:
li.append(team in list(y['team_key']))
matrix.append(li)
scores.append(y['autoCellPoints'].unique()[0])
ma = numpy.matrix(matrix) * 1
scores = numpy.array(scores)
opr = numpy.linalg.solve(
numpy.transpose(ma).dot(ma),
numpy.transpose(ma).dot(numpy.transpose(numpy.matrix(scores)))
)
for i, r in enumerate(opr):
print(teams[i], r)
teams = data['team_key'].unique()
teams.sort()
matrix = []
scores = []
for i, (x, y) in enumerate(data.groupby('key')):
li = []
for team in teams:
li.append(team in list(y['team_key']))
matrix.append(li)
scores.append(y['autoPoints'].unique()[0])
ma = numpy.matrix(matrix) * 1
scores = numpy.array(scores)
opr = numpy.linalg.solve(
numpy.transpose(ma).dot(ma),
numpy.transpose(ma).dot(numpy.transpose(numpy.matrix(scores)))
)
for i, r in enumerate(opr):
print(teams[i], r)
```
## Team Component Scores
```
post[post.index.isin(teams)]
post.sum(axis=1).sort_values(ascending=False)[0:10]
post[post.index == 'frc973']
post.sum(axis=1).median()
(post['endgame'] + post['endgame_balance']).sort_values(ascending=False)[0:10]
post.corr()
post.rank(ascending=False)[post.index=='frc973']
post.rank(ascending=False)[post.index=='frc1533']
post[post.index.isin(teams)]
post[post.index.isin(teams)].sum(axis=1).sort_values()
post[post.index.isin(teams)].sum(axis=1).hist(bins=5, range=(0,50))
```
### Models for success rates
Construct priors.
```
df.groupby('team_key')['initLine'].value_counts().unstack().fillna(0)
import scipy
rates = df.groupby('team_key')['initLine'].sum() / df.groupby('team_key')['initLine'].count() / 5
rates.hist()
alpha, beta, lim, scale = scipy.stats.beta.fit(rates)
x = numpy.arange(0, 1, 0.01)
y = scipy.stats.beta.pdf(x, alpha, beta)
plt.plot(x,y)
rates = (
df[df['endgame'] == 25].groupby('team_key')['endgame'].count() / df.groupby('team_key')['endgame'].count()
).fillna(0)
rates.hist()
alpha, beta, lim, scale = scipy.stats.beta.fit(rates)
x = numpy.arange(0, 1, 0.01)
y = scipy.stats.beta.pdf(x, alpha, beta, scale=scale)
plt.plot(x,y)
alpha, beta
success = df.groupby('team_key')['endgame'].value_counts().unstack().fillna(0)[25]
failure = df.groupby('team_key')['endgame'].count() - df.groupby('team_key')['endgame'].value_counts().unstack().fillna(0)[25]
success
failure
x = numpy.arange(0, 1, 0.01)
y = scipy.stats.beta.pdf(x, alpha + 1, beta + 9)
plt.plot(x,y)
y = scipy.stats.beta.pdf(x, alpha + 7, beta + 5)
plt.plot(x,y)
y2 = scipy.stats.beta.pdf(x, alpha + 8, beta + 2)
plt.plot(x,y2)
y2 = scipy.stats.beta.pdf(x, alpha + 11, beta + 0)
plt.plot(x,y2)
x = numpy.arange(0, 1, 0.01)
y = scipy.stats.beta.cdf(x, alpha + 1, beta + 9)
plt.plot(x,y)
y = scipy.stats.beta.cdf(x, alpha + 7, beta + 5)
plt.plot(x,y)
y2 = scipy.stats.beta.cdf(x, alpha + 8, beta + 2)
plt.plot(x,y2)
y2 = scipy.stats.beta.cdf(x, alpha + 11, beta + 0)
plt.plot(x,y2)
numpy.sum(scipy.stats.beta.rvs(
alpha + 11, beta + 0, size=1000
) > scipy.stats.beta.rvs(
alpha + 7, beta + 5, size=1000
))/1000
numpy.sum(scipy.stats.beta.rvs(
alpha + 11, beta + 0, size=1000
) > 0.75)/1000
```
|
github_jupyter
|
import numpy
import pandas
from sqlalchemy import create_engine
import matplotlib.pyplot as plt
%matplotlib inline
import pymc3 as pm
engine = create_engine('postgresql://cheese:cheesepass4279@localhost:5432/cheesecake')
query = """
select alliance.key, alliance.color, alliance_teams.team_key, match.match_number, alliance.score,
match.score_breakdown->'red' as breakdown_red,
match.score_breakdown->'blue' as breakdown_blue,
alliance_teams.position
from match
inner join alliance on alliance.match_key = match.key
inner join alliance_teams on alliance_teams.alliance_id = alliance.key
where comp_level = 'qm' and alliance.key like '2020%%'
"""
with engine.connect() as conn, conn.begin():
data = pandas.read_sql(query, conn)
data.loc[data.color == 'red', 'breakdown'] = data.loc[data.color == 'red', 'breakdown_red']
data.loc[data.color == 'blue', 'breakdown'] = data.loc[data.color == 'blue', 'breakdown_blue']
data = data.drop(['breakdown_red', 'breakdown_blue'], axis=1)
df = pandas.concat([
data.drop(['breakdown'], axis=1),
data['breakdown'].apply(pandas.Series)
], axis=1)
df
id2team = dict(enumerate(data['team_key'].unique()))
team2id = dict(zip(id2team.values(), id2team.keys()))
tms1 = data['team_key'][0::3].apply(lambda x: team2id.get(x)).values
tms2 = data['team_key'][1::3].apply(lambda x: team2id.get(x)).values
tms3 = data['team_key'][2::3].apply(lambda x: team2id.get(x)).values
with pm.Model() as model:
auto_score = pm.Gamma("auto_score", alpha=1.5, beta=0.1, shape=len(id2team))
tele_score = pm.Gamma("tele_score", alpha=1.5, beta=0.1, shape=len(id2team))
theta_auto = (auto_score[tms1] + auto_score[tms2] + auto_score[tms3])
theta_tele = (tele_score[tms1] + tele_score[tms2] + tele_score[tms3])
points = pm.Poisson('autoCellPoints', mu=theta_auto, observed=df['autoCellPoints'][1::3].values)
telepoints = pm.Poisson('teleopCellPoints', mu=theta_tele, observed=df['teleopCellPoints'][1::3].values)
trace = pm.sample(1000)
post = pandas.DataFrame({
'auto': numpy.median(trace['auto_score'], axis=0),
'tele': numpy.median(trace['tele_score'], axis=0)
}, index=[id2team[i] for i in range(trace['auto_score'].shape[1])])
for i in range(0, 3):
df.loc[df.position == i, 'initLine'] = df.loc[df.position == i, 'initLineRobot{}'.format(i + 1)]
df.loc[:, 'initLine'] = (df['initLine'] == 'Exited') * 5
df.loc[:, ['key', 'team_key', 'initLine']]
for i in range(0, 3):
df.loc[df.position == i, 'endgame'] = df.loc[df.position == i, 'endgameRobot{}'.format(i + 1)]
val_map = {'Hang': 25, 'Park': 5, 'None': 0}
df.loc[:, 'endgame'] = df['endgame'].replace(val_map)
post['initLine'] = df.groupby('team_key')['initLine'].mean()
post['endgame'] = df.groupby('team_key')['endgame'].mean()
climb_pts = ((((df['position'] == 0) & (df['endgameRobot1'] == "Hang")) |
((df['position'] == 1) & (df['endgameRobot2'] == "Hang")) |
((df['position'] == 2) & (df['endgameRobot3'] == "Hang"))) &
(df['endgameRungIsLevel'] == 'IsLevel')) * 15
df['balance_points'] = (climb_pts / df['tba_numRobotsHanging']).replace(numpy.inf, 0).fillna(0)
post['endgame_balance'] = df.groupby('team_key')['balance_points'].mean()
post
post.sum(axis=1).quantile([.1, .25, .5, .75, .9])
post.sum(axis=1).hist(bins=20, range=(0,100), density=True)
post['auto'].hist(bins=30, range=(0,30))
post['tele'].hist(bins=25, range=(0,50))
post['initLine'].hist(bins=5, range=(0,5))
post['endgame'].hist(bins=15, range=(0,30))
post['endgame_balance'].hist(bins=15, range=(0,15))
post['tele'].median()
df.groupby('key')
data = df[df.key.str.startswith('2020ncwak')]
data
teams.sort()
matrix = []
scores = []
for i, (x, y) in enumerate(data.groupby('key')):
li = []
for team in teams:
li.append(team in list(y['team_key']))
matrix.append(li)
scores.append(y['autoCellPoints'].unique()[0])
ma = numpy.matrix(matrix) * 1
scores = numpy.array(scores)
opr = numpy.linalg.solve(
numpy.transpose(ma).dot(ma),
numpy.transpose(ma).dot(numpy.transpose(numpy.matrix(scores)))
)
for i, r in enumerate(opr):
print(teams[i], r)
teams = data['team_key'].unique()
teams.sort()
matrix = []
scores = []
for i, (x, y) in enumerate(data.groupby('key')):
li = []
for team in teams:
li.append(team in list(y['team_key']))
matrix.append(li)
scores.append(y['autoPoints'].unique()[0])
ma = numpy.matrix(matrix) * 1
scores = numpy.array(scores)
opr = numpy.linalg.solve(
numpy.transpose(ma).dot(ma),
numpy.transpose(ma).dot(numpy.transpose(numpy.matrix(scores)))
)
for i, r in enumerate(opr):
print(teams[i], r)
post[post.index.isin(teams)]
post.sum(axis=1).sort_values(ascending=False)[0:10]
post[post.index == 'frc973']
post.sum(axis=1).median()
(post['endgame'] + post['endgame_balance']).sort_values(ascending=False)[0:10]
post.corr()
post.rank(ascending=False)[post.index=='frc973']
post.rank(ascending=False)[post.index=='frc1533']
post[post.index.isin(teams)]
post[post.index.isin(teams)].sum(axis=1).sort_values()
post[post.index.isin(teams)].sum(axis=1).hist(bins=5, range=(0,50))
df.groupby('team_key')['initLine'].value_counts().unstack().fillna(0)
import scipy
rates = df.groupby('team_key')['initLine'].sum() / df.groupby('team_key')['initLine'].count() / 5
rates.hist()
alpha, beta, lim, scale = scipy.stats.beta.fit(rates)
x = numpy.arange(0, 1, 0.01)
y = scipy.stats.beta.pdf(x, alpha, beta)
plt.plot(x,y)
rates = (
df[df['endgame'] == 25].groupby('team_key')['endgame'].count() / df.groupby('team_key')['endgame'].count()
).fillna(0)
rates.hist()
alpha, beta, lim, scale = scipy.stats.beta.fit(rates)
x = numpy.arange(0, 1, 0.01)
y = scipy.stats.beta.pdf(x, alpha, beta, scale=scale)
plt.plot(x,y)
alpha, beta
success = df.groupby('team_key')['endgame'].value_counts().unstack().fillna(0)[25]
failure = df.groupby('team_key')['endgame'].count() - df.groupby('team_key')['endgame'].value_counts().unstack().fillna(0)[25]
success
failure
x = numpy.arange(0, 1, 0.01)
y = scipy.stats.beta.pdf(x, alpha + 1, beta + 9)
plt.plot(x,y)
y = scipy.stats.beta.pdf(x, alpha + 7, beta + 5)
plt.plot(x,y)
y2 = scipy.stats.beta.pdf(x, alpha + 8, beta + 2)
plt.plot(x,y2)
y2 = scipy.stats.beta.pdf(x, alpha + 11, beta + 0)
plt.plot(x,y2)
x = numpy.arange(0, 1, 0.01)
y = scipy.stats.beta.cdf(x, alpha + 1, beta + 9)
plt.plot(x,y)
y = scipy.stats.beta.cdf(x, alpha + 7, beta + 5)
plt.plot(x,y)
y2 = scipy.stats.beta.cdf(x, alpha + 8, beta + 2)
plt.plot(x,y2)
y2 = scipy.stats.beta.cdf(x, alpha + 11, beta + 0)
plt.plot(x,y2)
numpy.sum(scipy.stats.beta.rvs(
alpha + 11, beta + 0, size=1000
) > scipy.stats.beta.rvs(
alpha + 7, beta + 5, size=1000
))/1000
numpy.sum(scipy.stats.beta.rvs(
alpha + 11, beta + 0, size=1000
) > 0.75)/1000
| 0.180576 | 0.893495 |
```
import tensorflow as tf
print(tf.__version__)
# additional imports
import numpy as np
import matplotlib.pyplot as plt
from tensorflow.keras.layers import Input, Conv2D, Dense, Flatten, Dropout, GlobalMaxPooling2D, MaxPooling2D, BatchNormalization
from tensorflow.keras.models import Model
# Load in the data
cifar10 = tf.keras.datasets.cifar10
(x_train, y_train), (x_test, y_test) = cifar10.load_data()
x_train, x_test = x_train / 255.0, x_test / 255.0
y_train, y_test = y_train.flatten(), y_test.flatten()
print("x_train.shape:", x_train.shape)
print("y_train.shape", y_train.shape)
# number of classes
K = len(set(y_train))
print("number of classes:", K)
# Build the model using the functional API
i = Input(shape=x_train[0].shape)
# x = Conv2D(32, (3, 3), strides=2, activation='relu')(i)
# x = Conv2D(64, (3, 3), strides=2, activation='relu')(x)
# x = Conv2D(128, (3, 3), strides=2, activation='relu')(x)
x = Conv2D(32, (3, 3), activation='relu', padding='same')(i)
x = BatchNormalization()(x)
x = Conv2D(32, (3, 3), activation='relu', padding='same')(x)
x = BatchNormalization()(x)
x = MaxPooling2D((2, 2))(x)
# x = Dropout(0.2)(x)
x = Conv2D(64, (3, 3), activation='relu', padding='same')(x)
x = BatchNormalization()(x)
x = Conv2D(64, (3, 3), activation='relu', padding='same')(x)
x = BatchNormalization()(x)
x = MaxPooling2D((2, 2))(x)
# x = Dropout(0.2)(x)
x = Conv2D(128, (3, 3), activation='relu', padding='same')(x)
x = BatchNormalization()(x)
x = Conv2D(128, (3, 3), activation='relu', padding='same')(x)
x = BatchNormalization()(x)
x = MaxPooling2D((2, 2))(x)
# x = Dropout(0.2)(x)
# x = GlobalMaxPooling2D()(x)
x = Flatten()(x)
x = Dropout(0.2)(x)
x = Dense(1024, activation='relu')(x)
x = Dropout(0.2)(x)
x = Dense(K, activation='softmax')(x)
model = Model(i, x)
# Compile
# Note: make sure you are using the GPU for this!
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# Fit
r = model.fit(x_train, y_train, validation_data=(x_test, y_test), epochs=50)
# Fit with data augmentation
# Note: if you run this AFTER calling the previous model.fit(), it will CONTINUE training where it left off
batch_size = 32
data_generator = tf.keras.preprocessing.image.ImageDataGenerator(width_shift_range=0.1, height_shift_range=0.1, horizontal_flip=True)
train_generator = data_generator.flow(x_train, y_train, batch_size)
steps_per_epoch = x_train.shape[0] // batch_size
r = model.fit(train_generator, validation_data=(x_test, y_test), steps_per_epoch=steps_per_epoch, epochs=50)
# Plot loss per iteration
import matplotlib.pyplot as plt
plt.plot(r.history['loss'], label='loss')
plt.plot(r.history['val_loss'], label='val_loss')
plt.legend()
# Plot accuracy per iteration
plt.plot(r.history['accuracy'], label='acc')
plt.plot(r.history['val_accuracy'], label='val_acc')
plt.legend()
# Plot confusion matrix
from sklearn.metrics import confusion_matrix
import itertools
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
plt.show()
p_test = model.predict(x_test).argmax(axis=1)
cm = confusion_matrix(y_test, p_test)
plot_confusion_matrix(cm, list(range(10)))
# label mapping
labels = '''airplane
automobile
bird
cat
deer
dog
frog
horse
ship
truck'''.split()
# Show some misclassified examples
misclassified_idx = np.where(p_test != y_test)[0]
i = np.random.choice(misclassified_idx)
plt.imshow(x_test[i], cmap='gray')
plt.title("True label: %s Predicted: %s" % (labels[y_test[i]], labels[p_test[i]]));
# Now that the model is so large, it's useful to summarize it
model.summary()
```
|
github_jupyter
|
import tensorflow as tf
print(tf.__version__)
# additional imports
import numpy as np
import matplotlib.pyplot as plt
from tensorflow.keras.layers import Input, Conv2D, Dense, Flatten, Dropout, GlobalMaxPooling2D, MaxPooling2D, BatchNormalization
from tensorflow.keras.models import Model
# Load in the data
cifar10 = tf.keras.datasets.cifar10
(x_train, y_train), (x_test, y_test) = cifar10.load_data()
x_train, x_test = x_train / 255.0, x_test / 255.0
y_train, y_test = y_train.flatten(), y_test.flatten()
print("x_train.shape:", x_train.shape)
print("y_train.shape", y_train.shape)
# number of classes
K = len(set(y_train))
print("number of classes:", K)
# Build the model using the functional API
i = Input(shape=x_train[0].shape)
# x = Conv2D(32, (3, 3), strides=2, activation='relu')(i)
# x = Conv2D(64, (3, 3), strides=2, activation='relu')(x)
# x = Conv2D(128, (3, 3), strides=2, activation='relu')(x)
x = Conv2D(32, (3, 3), activation='relu', padding='same')(i)
x = BatchNormalization()(x)
x = Conv2D(32, (3, 3), activation='relu', padding='same')(x)
x = BatchNormalization()(x)
x = MaxPooling2D((2, 2))(x)
# x = Dropout(0.2)(x)
x = Conv2D(64, (3, 3), activation='relu', padding='same')(x)
x = BatchNormalization()(x)
x = Conv2D(64, (3, 3), activation='relu', padding='same')(x)
x = BatchNormalization()(x)
x = MaxPooling2D((2, 2))(x)
# x = Dropout(0.2)(x)
x = Conv2D(128, (3, 3), activation='relu', padding='same')(x)
x = BatchNormalization()(x)
x = Conv2D(128, (3, 3), activation='relu', padding='same')(x)
x = BatchNormalization()(x)
x = MaxPooling2D((2, 2))(x)
# x = Dropout(0.2)(x)
# x = GlobalMaxPooling2D()(x)
x = Flatten()(x)
x = Dropout(0.2)(x)
x = Dense(1024, activation='relu')(x)
x = Dropout(0.2)(x)
x = Dense(K, activation='softmax')(x)
model = Model(i, x)
# Compile
# Note: make sure you are using the GPU for this!
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# Fit
r = model.fit(x_train, y_train, validation_data=(x_test, y_test), epochs=50)
# Fit with data augmentation
# Note: if you run this AFTER calling the previous model.fit(), it will CONTINUE training where it left off
batch_size = 32
data_generator = tf.keras.preprocessing.image.ImageDataGenerator(width_shift_range=0.1, height_shift_range=0.1, horizontal_flip=True)
train_generator = data_generator.flow(x_train, y_train, batch_size)
steps_per_epoch = x_train.shape[0] // batch_size
r = model.fit(train_generator, validation_data=(x_test, y_test), steps_per_epoch=steps_per_epoch, epochs=50)
# Plot loss per iteration
import matplotlib.pyplot as plt
plt.plot(r.history['loss'], label='loss')
plt.plot(r.history['val_loss'], label='val_loss')
plt.legend()
# Plot accuracy per iteration
plt.plot(r.history['accuracy'], label='acc')
plt.plot(r.history['val_accuracy'], label='val_acc')
plt.legend()
# Plot confusion matrix
from sklearn.metrics import confusion_matrix
import itertools
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
plt.show()
p_test = model.predict(x_test).argmax(axis=1)
cm = confusion_matrix(y_test, p_test)
plot_confusion_matrix(cm, list(range(10)))
# label mapping
labels = '''airplane
automobile
bird
cat
deer
dog
frog
horse
ship
truck'''.split()
# Show some misclassified examples
misclassified_idx = np.where(p_test != y_test)[0]
i = np.random.choice(misclassified_idx)
plt.imshow(x_test[i], cmap='gray')
plt.title("True label: %s Predicted: %s" % (labels[y_test[i]], labels[p_test[i]]));
# Now that the model is so large, it's useful to summarize it
model.summary()
| 0.940946 | 0.812161 |
```
from helpers.utilities import *
%run helpers/notebook_setup.ipynb
indexed_by_target_path = 'data/clean/protein/indexed_by_target.csv'
patients_variance_at_one_path ='data/clean/protein/z_log_10-patients_variance_at_one.csv'
zz_log_path = 'data/clean/protein/zz_log_10.csv'
log_matrix_path = 'data/clean/protein/log_10.csv'
clinical_path = 'data/clean/protein/clinical_data_ordered_to_match_proteins_matrix.csv'
raw_protein_matrix = read_csv(indexed_by_target_path, index_col=0)
log_matrix = read_csv(log_matrix_path, index_col=0)
zz_log_matrix = read_csv(zz_log_path, index_col=0)
patients_variance_at_one = read_csv(patients_variance_at_one_path, index_col=0)
clinical = read_csv(clinical_path, index_col=0)
by_condition = clinical['Meningitis']
conditions = sorted(set(by_condition))
matrices = {
'raw': raw_protein_matrix,
'log': log_matrix,
'z-score patients then z-score proteins': zz_log_matrix,
'z-score patients': patients_variance_at_one
}
```
## Patient-to-patient variance of each of the proteins, grouped by their condition:
```
proteins_variance = []
for condition in conditions:
for transform, matrix in matrices.items():
for protein, variance in matrix.T[by_condition == condition].T.var(axis=1).items():
proteins_variance.append(
{
'condition': condition,
'transform': transform,
'variance': variance,
'protein': protein,
'variance_ratio': variance / matrix.loc[protein].var()
}
)
proteins_variance = DataFrame(proteins_variance)
proteins_variance.head()
%%R -i proteins_variance -w 800
(
ggplot(proteins_variance, aes(x=condition, y=variance, fill=transform))
+ geom_boxplot()
+ scale_y_log10()
+ theme(legend.position='bottom')
)
```
If we assume that the variance of proteins between patients in the same group should be lower than the variance between proteins among all samples, we can get a meaningful comparison of the proposed transformations:
```
%%R -i proteins_variance -w 800
(
ggplot(proteins_variance, aes(x=condition, y=variance_ratio, fill=transform))
+ geom_boxplot()
+ scale_y_log10()
+ theme(legend.position='bottom')
)
```
- It is not obvious if the comparison of raw values to log transformed values gives us a clear picture (because the division by log transformed value and by raw value is not the same thing), but
- it seems that the benefit of log transform is the greatest in the TB group (worth remembering)
- the same goes for fixing the variance of each patient at one (benefit visible in the TB group)
- and for the double z-score transform. While in the absolute terms the in-group variance for this procedure increased (see the plot above), when we compared it to the global variance, this method is as good as single z-score - which is contrary to my consternations
## Protein-to-protein variance of each of the patients, grouped by their condition:
This time I compare how variable is each sample (patient) against the group that they belong to. This one has no strong biological intuition, but one could say that very similar patients should have comparable variances. This is of course an oversimplification and generally not true in many situations.
```
patients_variance = []
for condition in conditions:
for transform, matrix in matrices.items():
for patient, variance in matrix.T[by_condition == condition].T.var(axis=0).items():
patients_variance.append(
{
'condition': condition,
'transform': transform,
'variance': variance,
'patient': patient,
'variance_ratio': variance / matrix.T[by_condition == condition].T.var().mean(),
}
)
patients_variance = DataFrame(patients_variance)
patients_variance.head()
%%R -i patients_variance -w 800
(
ggplot(patients_variance, aes(x=condition, y=variance, fill=transform))
+ geom_boxplot()
+ scale_y_log10()
+ theme(legend.position='bottom')
)
%%R -i patients_variance -w 800
(
ggplot(patients_variance, aes(x=condition, y=variance_ratio, fill=transform))
+ geom_boxplot()
+ theme(legend.position='bottom')
)
# closer to one = variances more similar = the desired output
```
- the single z-score transform has unit variance on patients by definition thus is not worth additional discussion
- the double z-score performs (on average) considerably worse than other methods (as expected - this is the cost of the trade-off of having fixed variance in proteins rather than patients)
### Mean abundance of proteins in each of the patients, compared against the mean of the group
In non-transformed data this is influenced by:
- the disease
- the technical variation
If we compare the patients proteins abundance to the mean abundance of the disease group, we expect to get comparable results.
A good transformation would reduce the technical variation thus reducing the variance in such comparison.
```
patients_mean_protein_abundance = []
for condition in conditions:
for transform, matrix in matrices.items():
mean_condition = matrix.T[by_condition == condition].T.mean(axis=0).mean()
# print(condition, transform, mean_condition)
for patient, mean in matrix.T[by_condition == condition].T.mean(axis=0).items():
patients_mean_protein_abundance.append(
{
'condition': condition,
'transform': transform,
'mean': mean,
'patient': patient,
'mean_ratio': mean / mean_condition,
}
)
patients_mean_protein_abundance = DataFrame(patients_mean_protein_abundance)
patients_mean_protein_abundance.head()
%%R -i patients_mean_protein_abundance -w 800
(
ggplot(patients_mean_protein_abundance, aes(x=condition, y=mean, fill=transform))
+ geom_boxplot()
+ scale_y_log10()
+ theme(legend.position='bottom')
)
%%R -i patients_mean_protein_abundance -w 800
(
ggplot(patients_mean_protein_abundance, aes(x=condition, y=mean_ratio, fill=transform))
+ geom_boxplot()
+ theme(legend.position='bottom')
)
```
- single z-score (patients variance = 1, mean = 0) performed worse. It should not. This may be due to instability of the computations (we are dividing a number which is essentially a 0 by another number which is a mean of multiple "almost" zeros, e.g. -3.598654e-16 / 6.011499383558319e-17 (see below)
- the results for viral/double z-score are concerning. This seems to the most important finding in this notebook.
```
patients_mean_protein_abundance[patients_mean_protein_abundance['transform']=='z-score patients'].head()
```
### Why double z-score transform increases the difference between means in the viral group?
The data must not conform to our expectations. There is nothing unstable numerically:
```
double = patients_mean_protein_abundance[patients_mean_protein_abundance['transform']=='z-score patients then z-score proteins']
double.head()
double[double.condition == 'Viral'].head()
```
How is the Viral group different?
- fewer samples
- potentially different diseases?
Hypothesis 1: there are to few samples
TODO: test by simulation (permutations test)
|
github_jupyter
|
from helpers.utilities import *
%run helpers/notebook_setup.ipynb
indexed_by_target_path = 'data/clean/protein/indexed_by_target.csv'
patients_variance_at_one_path ='data/clean/protein/z_log_10-patients_variance_at_one.csv'
zz_log_path = 'data/clean/protein/zz_log_10.csv'
log_matrix_path = 'data/clean/protein/log_10.csv'
clinical_path = 'data/clean/protein/clinical_data_ordered_to_match_proteins_matrix.csv'
raw_protein_matrix = read_csv(indexed_by_target_path, index_col=0)
log_matrix = read_csv(log_matrix_path, index_col=0)
zz_log_matrix = read_csv(zz_log_path, index_col=0)
patients_variance_at_one = read_csv(patients_variance_at_one_path, index_col=0)
clinical = read_csv(clinical_path, index_col=0)
by_condition = clinical['Meningitis']
conditions = sorted(set(by_condition))
matrices = {
'raw': raw_protein_matrix,
'log': log_matrix,
'z-score patients then z-score proteins': zz_log_matrix,
'z-score patients': patients_variance_at_one
}
proteins_variance = []
for condition in conditions:
for transform, matrix in matrices.items():
for protein, variance in matrix.T[by_condition == condition].T.var(axis=1).items():
proteins_variance.append(
{
'condition': condition,
'transform': transform,
'variance': variance,
'protein': protein,
'variance_ratio': variance / matrix.loc[protein].var()
}
)
proteins_variance = DataFrame(proteins_variance)
proteins_variance.head()
%%R -i proteins_variance -w 800
(
ggplot(proteins_variance, aes(x=condition, y=variance, fill=transform))
+ geom_boxplot()
+ scale_y_log10()
+ theme(legend.position='bottom')
)
%%R -i proteins_variance -w 800
(
ggplot(proteins_variance, aes(x=condition, y=variance_ratio, fill=transform))
+ geom_boxplot()
+ scale_y_log10()
+ theme(legend.position='bottom')
)
patients_variance = []
for condition in conditions:
for transform, matrix in matrices.items():
for patient, variance in matrix.T[by_condition == condition].T.var(axis=0).items():
patients_variance.append(
{
'condition': condition,
'transform': transform,
'variance': variance,
'patient': patient,
'variance_ratio': variance / matrix.T[by_condition == condition].T.var().mean(),
}
)
patients_variance = DataFrame(patients_variance)
patients_variance.head()
%%R -i patients_variance -w 800
(
ggplot(patients_variance, aes(x=condition, y=variance, fill=transform))
+ geom_boxplot()
+ scale_y_log10()
+ theme(legend.position='bottom')
)
%%R -i patients_variance -w 800
(
ggplot(patients_variance, aes(x=condition, y=variance_ratio, fill=transform))
+ geom_boxplot()
+ theme(legend.position='bottom')
)
# closer to one = variances more similar = the desired output
patients_mean_protein_abundance = []
for condition in conditions:
for transform, matrix in matrices.items():
mean_condition = matrix.T[by_condition == condition].T.mean(axis=0).mean()
# print(condition, transform, mean_condition)
for patient, mean in matrix.T[by_condition == condition].T.mean(axis=0).items():
patients_mean_protein_abundance.append(
{
'condition': condition,
'transform': transform,
'mean': mean,
'patient': patient,
'mean_ratio': mean / mean_condition,
}
)
patients_mean_protein_abundance = DataFrame(patients_mean_protein_abundance)
patients_mean_protein_abundance.head()
%%R -i patients_mean_protein_abundance -w 800
(
ggplot(patients_mean_protein_abundance, aes(x=condition, y=mean, fill=transform))
+ geom_boxplot()
+ scale_y_log10()
+ theme(legend.position='bottom')
)
%%R -i patients_mean_protein_abundance -w 800
(
ggplot(patients_mean_protein_abundance, aes(x=condition, y=mean_ratio, fill=transform))
+ geom_boxplot()
+ theme(legend.position='bottom')
)
patients_mean_protein_abundance[patients_mean_protein_abundance['transform']=='z-score patients'].head()
double = patients_mean_protein_abundance[patients_mean_protein_abundance['transform']=='z-score patients then z-score proteins']
double.head()
double[double.condition == 'Viral'].head()
| 0.380759 | 0.754734 |
```
import chseg
import numpy as np
import tensorflow as tf
from sklearn.metrics import classification_report
params = {
'seq_len': 50,
'batch_size': 128,
'n_class': 4,
'hidden_dim': 128,
'clip_norm': 5.0,
'text_iter_step': 10,
'lr': {'start': 5e-3, 'end': 5e-4},
'n_epoch': 1,
'display_step': 50,
}
def to_test_seq(*args):
return [np.reshape(x[:(len(x)-len(x)%params['seq_len'])],
[-1,params['seq_len']]) for x in args]
def iter_seq(x):
return np.array([x[i: i+params['seq_len']] for i in range(
0, len(x)-params['seq_len'], params['text_iter_step'])])
def to_train_seq(*args):
return [iter_seq(x) for x in args]
def pipeline_train(X, y, sess):
dataset = tf.data.Dataset.from_tensor_slices((X, y))
dataset = dataset.shuffle(len(X)).batch(params['batch_size'])
iterator = dataset.make_initializable_iterator()
X_ph = tf.placeholder(tf.int32, [None, params['seq_len']])
y_ph = tf.placeholder(tf.int32, [None, params['seq_len']])
init_dict = {X_ph: X, y_ph: y}
sess.run(iterator.initializer, init_dict)
return iterator, init_dict
def pipeline_test(X, sess):
dataset = tf.data.Dataset.from_tensor_slices(X)
dataset = dataset.batch(params['batch_size'])
iterator = dataset.make_initializable_iterator()
X_ph = tf.placeholder(tf.int32, [None, params['seq_len']])
init_dict = {X_ph: X}
sess.run(iterator.initializer, init_dict)
return iterator, init_dict
x_train, y_train, x_test, y_test, params['vocab_size'], word2idx, idx2word = chseg.load_data()
X_train, Y_train = to_train_seq(x_train, y_train)
X_test, Y_test = to_test_seq(x_test, y_test)
sess = tf.Session()
params['lr']['steps'] = len(X_train) // params['batch_size']
iter_train, init_dict_train = pipeline_train(X_train, Y_train, sess)
iter_test, init_dict_test = pipeline_test(X_test, sess)
def rnn_cell():
return tf.nn.rnn_cell.GRUCell(params['hidden_dim'],
kernel_initializer=tf.orthogonal_initializer())
def clip_grads(loss):
variables = tf.trainable_variables()
grads = tf.gradients(loss, variables)
clipped_grads, _ = tf.clip_by_global_norm(grads, params['clip_norm'])
return zip(clipped_grads, variables)
def forward(x, reuse, is_training):
with tf.variable_scope('model', reuse=reuse):
x = tf.contrib.layers.embed_sequence(x, params['vocab_size'], params['hidden_dim'])
x = tf.layers.dropout(x, 0.1, training=is_training)
bi_outputs, _ = tf.nn.bidirectional_dynamic_rnn(
rnn_cell(), rnn_cell(), x, dtype=tf.float32)
x = tf.concat(bi_outputs, -1)
logits = tf.layers.dense(x, params['n_class'])
return logits
ops = {}
X_train_batch, y_train_batch = iter_train.get_next()
X_test_batch = iter_test.get_next()
logits_tr = forward(X_train_batch, reuse=False, is_training=True)
logits_te = forward(X_test_batch, reuse=True, is_training=False)
log_likelihood, trans_params = tf.contrib.crf.crf_log_likelihood(
logits_tr, y_train_batch, tf.count_nonzero(X_train_batch, 1))
ops['loss'] = tf.reduce_mean(-log_likelihood)
ops['global_step'] = tf.Variable(0, trainable=False)
ops['lr'] = tf.train.exponential_decay(
params['lr']['start'], ops['global_step'], params['lr']['steps'],
params['lr']['end']/params['lr']['start'])
ops['train'] = tf.train.AdamOptimizer(ops['lr']).apply_gradients(
clip_grads(ops['loss']), global_step=ops['global_step'])
ops['crf_decode'] = tf.contrib.crf.crf_decode(
logits_te, trans_params, tf.count_nonzero(X_test_batch, 1))[0]
sess.run(tf.global_variables_initializer())
for epoch in range(1, params['n_epoch']+1):
while True:
try:
sess.run(ops['train'])
except tf.errors.OutOfRangeError:
break
else:
step = sess.run(ops['global_step'])
if step % params['display_step'] == 0 or step == 1:
loss, lr = sess.run([ops['loss'], ops['lr']])
print("Epoch %d | Step %d | Loss %.3f | LR: %.4f" % (epoch, step, loss, lr))
Y_pred = []
while True:
try:
Y_pred.append(sess.run(ops['crf_decode']))
except tf.errors.OutOfRangeError:
break
Y_pred = np.concatenate(Y_pred)
if epoch != params['n_epoch']:
sess.run(iter_train.initializer, init_dict_train)
sess.run(iter_test.initializer, init_dict_test)
print(classification_report(Y_test.ravel(), Y_pred.ravel(), target_names=['B','M','E','S']))
sample = '我来到大学读书,希望学到知识'
x = np.atleast_2d([word2idx[w] for w in sample] + [0]*(params['seq_len']-len(sample)))
ph = tf.placeholder(tf.int32, [None, params['seq_len']])
logits = forward(ph, reuse=True, is_training=False)
inference = tf.contrib.crf.crf_decode(logits, trans_params, tf.count_nonzero(ph, 1))[0]
x = sess.run(inference, {ph: x})[0][:len(sample)]
res = ''
for i, l in enumerate(x):
c = sample[i]
if l == 2 or l == 3:
c += ' '
res += c
print(res)
```
|
github_jupyter
|
import chseg
import numpy as np
import tensorflow as tf
from sklearn.metrics import classification_report
params = {
'seq_len': 50,
'batch_size': 128,
'n_class': 4,
'hidden_dim': 128,
'clip_norm': 5.0,
'text_iter_step': 10,
'lr': {'start': 5e-3, 'end': 5e-4},
'n_epoch': 1,
'display_step': 50,
}
def to_test_seq(*args):
return [np.reshape(x[:(len(x)-len(x)%params['seq_len'])],
[-1,params['seq_len']]) for x in args]
def iter_seq(x):
return np.array([x[i: i+params['seq_len']] for i in range(
0, len(x)-params['seq_len'], params['text_iter_step'])])
def to_train_seq(*args):
return [iter_seq(x) for x in args]
def pipeline_train(X, y, sess):
dataset = tf.data.Dataset.from_tensor_slices((X, y))
dataset = dataset.shuffle(len(X)).batch(params['batch_size'])
iterator = dataset.make_initializable_iterator()
X_ph = tf.placeholder(tf.int32, [None, params['seq_len']])
y_ph = tf.placeholder(tf.int32, [None, params['seq_len']])
init_dict = {X_ph: X, y_ph: y}
sess.run(iterator.initializer, init_dict)
return iterator, init_dict
def pipeline_test(X, sess):
dataset = tf.data.Dataset.from_tensor_slices(X)
dataset = dataset.batch(params['batch_size'])
iterator = dataset.make_initializable_iterator()
X_ph = tf.placeholder(tf.int32, [None, params['seq_len']])
init_dict = {X_ph: X}
sess.run(iterator.initializer, init_dict)
return iterator, init_dict
x_train, y_train, x_test, y_test, params['vocab_size'], word2idx, idx2word = chseg.load_data()
X_train, Y_train = to_train_seq(x_train, y_train)
X_test, Y_test = to_test_seq(x_test, y_test)
sess = tf.Session()
params['lr']['steps'] = len(X_train) // params['batch_size']
iter_train, init_dict_train = pipeline_train(X_train, Y_train, sess)
iter_test, init_dict_test = pipeline_test(X_test, sess)
def rnn_cell():
return tf.nn.rnn_cell.GRUCell(params['hidden_dim'],
kernel_initializer=tf.orthogonal_initializer())
def clip_grads(loss):
variables = tf.trainable_variables()
grads = tf.gradients(loss, variables)
clipped_grads, _ = tf.clip_by_global_norm(grads, params['clip_norm'])
return zip(clipped_grads, variables)
def forward(x, reuse, is_training):
with tf.variable_scope('model', reuse=reuse):
x = tf.contrib.layers.embed_sequence(x, params['vocab_size'], params['hidden_dim'])
x = tf.layers.dropout(x, 0.1, training=is_training)
bi_outputs, _ = tf.nn.bidirectional_dynamic_rnn(
rnn_cell(), rnn_cell(), x, dtype=tf.float32)
x = tf.concat(bi_outputs, -1)
logits = tf.layers.dense(x, params['n_class'])
return logits
ops = {}
X_train_batch, y_train_batch = iter_train.get_next()
X_test_batch = iter_test.get_next()
logits_tr = forward(X_train_batch, reuse=False, is_training=True)
logits_te = forward(X_test_batch, reuse=True, is_training=False)
log_likelihood, trans_params = tf.contrib.crf.crf_log_likelihood(
logits_tr, y_train_batch, tf.count_nonzero(X_train_batch, 1))
ops['loss'] = tf.reduce_mean(-log_likelihood)
ops['global_step'] = tf.Variable(0, trainable=False)
ops['lr'] = tf.train.exponential_decay(
params['lr']['start'], ops['global_step'], params['lr']['steps'],
params['lr']['end']/params['lr']['start'])
ops['train'] = tf.train.AdamOptimizer(ops['lr']).apply_gradients(
clip_grads(ops['loss']), global_step=ops['global_step'])
ops['crf_decode'] = tf.contrib.crf.crf_decode(
logits_te, trans_params, tf.count_nonzero(X_test_batch, 1))[0]
sess.run(tf.global_variables_initializer())
for epoch in range(1, params['n_epoch']+1):
while True:
try:
sess.run(ops['train'])
except tf.errors.OutOfRangeError:
break
else:
step = sess.run(ops['global_step'])
if step % params['display_step'] == 0 or step == 1:
loss, lr = sess.run([ops['loss'], ops['lr']])
print("Epoch %d | Step %d | Loss %.3f | LR: %.4f" % (epoch, step, loss, lr))
Y_pred = []
while True:
try:
Y_pred.append(sess.run(ops['crf_decode']))
except tf.errors.OutOfRangeError:
break
Y_pred = np.concatenate(Y_pred)
if epoch != params['n_epoch']:
sess.run(iter_train.initializer, init_dict_train)
sess.run(iter_test.initializer, init_dict_test)
print(classification_report(Y_test.ravel(), Y_pred.ravel(), target_names=['B','M','E','S']))
sample = '我来到大学读书,希望学到知识'
x = np.atleast_2d([word2idx[w] for w in sample] + [0]*(params['seq_len']-len(sample)))
ph = tf.placeholder(tf.int32, [None, params['seq_len']])
logits = forward(ph, reuse=True, is_training=False)
inference = tf.contrib.crf.crf_decode(logits, trans_params, tf.count_nonzero(ph, 1))[0]
x = sess.run(inference, {ph: x})[0][:len(sample)]
res = ''
for i, l in enumerate(x):
c = sample[i]
if l == 2 or l == 3:
c += ' '
res += c
print(res)
| 0.657209 | 0.417182 |
```
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow import keras
import seaborn as sns
from os.path import join
plt.style.use(["seaborn", "thesis"])
plt.rc("figure", figsize=(8,4))
figure_save_path = "/home/jo/Repos/MastersThesis/Application/figures/"
```
# Data
```
from SCFInitialGuess.utilities.dataset import ScreenedData
target = "P"
r_max = 10 # 10 angstrom
data = ScreenedData(r_max)
data.include(data_path = "../../dataset/MethanT/", postfix = "MethanT", target=target)
data.include(data_path = "../../dataset/MethanT2/", postfix = "MethanT2", target=target)
data.include(data_path = "../../dataset/MethanT3/", postfix = "MethanT3", target=target)
data.include(data_path = "../../dataset/MethanT4/", postfix = "MethanT4", target=target)
data.include(data_path = "../../dataset/EthanT/", postfix = "EthanT", target=target)
data.include(data_path = "../../dataset/EthanT2/", postfix = "EthanT2", target=target)
data.include(data_path = "../../dataset/EthanT3/", postfix = "EthanT3", target=target)
data.include(data_path = "../../dataset/EthanT4/", postfix = "EthanT4", target=target)
data.include(data_path = "../../dataset/EthanT5/", postfix = "EthanT5", target=target)
data.include(data_path = "../../dataset/EthanT6/", postfix = "EthanT6", target=target)
data.include(data_path = "../../dataset/EthenT/", postfix = "EthenT", target=target)
data.include(data_path = "../../dataset/EthenT2/", postfix = "EthenT2", target=target)
data.include(data_path = "../../dataset/EthenT3/", postfix = "EthenT3", target=target)
data.include(data_path = "../../dataset/EthenT4/", postfix = "EthenT4", target=target)
data.include(data_path = "../../dataset/EthenT5/", postfix = "EthenT5", target=target)
data.include(data_path = "../../dataset/EthenT6/", postfix = "EthenT6", target=target)
data.include(data_path = "../../dataset/EthinT/", postfix = "EthinT", target=target)
data.include(data_path = "../../dataset/EthinT2/", postfix = "EthinT2", target=target)
data.include(data_path = "../../dataset/EthinT3/", postfix = "EthinT3", target=target)
#data.include(data_path = "../../dataset/QM9/", postfix = "QM9-300")
```
# Analysize
```
len(data.molecules[0]), len(data.molecules[1]), len(data.molecules[2])
np.sum([len(data.molecules[0]), len(data.molecules[1]), len(data.molecules[2])])
counter = {}
for mol in (data.molecules[0] + data.molecules[1] + data.molecules[2]):
for atom in mol.species:
counter[atom] = counter.get(atom, 0) + 1
print(counter)
```
# Distances
```
def distances(mol):
r = []
for i, geom_i in enumerate(mol.geometry):
for j, geom_j in enumerate(mol.geometry):
# avoid duplicates
if i < j:
continue
# only count C-H distances
if set([geom_i[0], geom_j[0]]) == set(["H", "C"]):
r.append(
np.sqrt(np.sum((np.array(geom_i[1]) - np.array(geom_j[1]))**2))
)
return r
def distances_batch(moles):
r = []
for mol in moles:
r += distances(mol)
return r
r_train = distances_batch(data.molecules[0])
r_validation = distances_batch(data.molecules[1])
r_test = distances_batch(data.molecules[2])
np.sum(np.array(r_test) > 15)
```
## Histogram
```
n_bins = 30
#offset = np.min(E)
hist_train, edges = np.histogram(r_train, bins=n_bins, density=True)
hist_validation, _ = np.histogram(r_validation, bins=edges, density=True)
hist_test, _ = np.histogram(r_test, bins=edges, density=True)
centers = (edges[:-1] + edges[1:]) / 2
width = np.mean(np.diff(centers)) * 0.23
plt.bar(centers - width, hist_train, width=width, label="Train")
plt.bar(centers, hist_validation, width=width, label="Validation")
plt.bar(centers + width, hist_test, width=width, label="Test")
plt.ylabel("Relative Frequency / 1")
plt.xlabel("C-H distance / $\AA$")
plt.tight_layout()
plt.legend()
plt.savefig(figure_save_path + "CHDistanceDistributionCarbs.pdf")
plt.show()
```
# Energies
```
from SCFInitialGuess.utilities.analysis import measure_hf_energy
E_train = np.array(measure_hf_energy(data.T[0], data.molecules[0]))
E_validation = np.array(measure_hf_energy(data.T[1], data.molecules[1]))
E_test = np.array(measure_hf_energy(data.T[2], data.molecules[2]))
n_bins = 50
hist_train, edges = np.histogram(E_train, bins=n_bins, density=True)
hist_validation, _ = np.histogram(E_validation, bins=edges, density=True)
hist_test, _ = np.histogram(E_test, bins=edges, density=True)
centers = (edges[:-1] + edges[1:]) / 2
width = np.mean(np.diff(centers)) * 0.3
plt.bar(centers - width, hist_train, width=width)
plt.bar(centers , hist_validation, width=width)
plt.bar(centers, hist_test, width=width)
plt.ylabel("Relative Frequency / 1")
plt.xlabel("HF Energy / Hartree")
plt.tight_layout()
plt.savefig(figure_save_path + "EnergyDistributionDataset.pdf")
plt.show()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow import keras
import seaborn as sns
from os.path import join
plt.style.use(["seaborn", "thesis"])
plt.rc("figure", figsize=(8,4))
figure_save_path = "/home/jo/Repos/MastersThesis/Application/figures/"
from SCFInitialGuess.utilities.dataset import ScreenedData
target = "P"
r_max = 10 # 10 angstrom
data = ScreenedData(r_max)
data.include(data_path = "../../dataset/MethanT/", postfix = "MethanT", target=target)
data.include(data_path = "../../dataset/MethanT2/", postfix = "MethanT2", target=target)
data.include(data_path = "../../dataset/MethanT3/", postfix = "MethanT3", target=target)
data.include(data_path = "../../dataset/MethanT4/", postfix = "MethanT4", target=target)
data.include(data_path = "../../dataset/EthanT/", postfix = "EthanT", target=target)
data.include(data_path = "../../dataset/EthanT2/", postfix = "EthanT2", target=target)
data.include(data_path = "../../dataset/EthanT3/", postfix = "EthanT3", target=target)
data.include(data_path = "../../dataset/EthanT4/", postfix = "EthanT4", target=target)
data.include(data_path = "../../dataset/EthanT5/", postfix = "EthanT5", target=target)
data.include(data_path = "../../dataset/EthanT6/", postfix = "EthanT6", target=target)
data.include(data_path = "../../dataset/EthenT/", postfix = "EthenT", target=target)
data.include(data_path = "../../dataset/EthenT2/", postfix = "EthenT2", target=target)
data.include(data_path = "../../dataset/EthenT3/", postfix = "EthenT3", target=target)
data.include(data_path = "../../dataset/EthenT4/", postfix = "EthenT4", target=target)
data.include(data_path = "../../dataset/EthenT5/", postfix = "EthenT5", target=target)
data.include(data_path = "../../dataset/EthenT6/", postfix = "EthenT6", target=target)
data.include(data_path = "../../dataset/EthinT/", postfix = "EthinT", target=target)
data.include(data_path = "../../dataset/EthinT2/", postfix = "EthinT2", target=target)
data.include(data_path = "../../dataset/EthinT3/", postfix = "EthinT3", target=target)
#data.include(data_path = "../../dataset/QM9/", postfix = "QM9-300")
len(data.molecules[0]), len(data.molecules[1]), len(data.molecules[2])
np.sum([len(data.molecules[0]), len(data.molecules[1]), len(data.molecules[2])])
counter = {}
for mol in (data.molecules[0] + data.molecules[1] + data.molecules[2]):
for atom in mol.species:
counter[atom] = counter.get(atom, 0) + 1
print(counter)
def distances(mol):
r = []
for i, geom_i in enumerate(mol.geometry):
for j, geom_j in enumerate(mol.geometry):
# avoid duplicates
if i < j:
continue
# only count C-H distances
if set([geom_i[0], geom_j[0]]) == set(["H", "C"]):
r.append(
np.sqrt(np.sum((np.array(geom_i[1]) - np.array(geom_j[1]))**2))
)
return r
def distances_batch(moles):
r = []
for mol in moles:
r += distances(mol)
return r
r_train = distances_batch(data.molecules[0])
r_validation = distances_batch(data.molecules[1])
r_test = distances_batch(data.molecules[2])
np.sum(np.array(r_test) > 15)
n_bins = 30
#offset = np.min(E)
hist_train, edges = np.histogram(r_train, bins=n_bins, density=True)
hist_validation, _ = np.histogram(r_validation, bins=edges, density=True)
hist_test, _ = np.histogram(r_test, bins=edges, density=True)
centers = (edges[:-1] + edges[1:]) / 2
width = np.mean(np.diff(centers)) * 0.23
plt.bar(centers - width, hist_train, width=width, label="Train")
plt.bar(centers, hist_validation, width=width, label="Validation")
plt.bar(centers + width, hist_test, width=width, label="Test")
plt.ylabel("Relative Frequency / 1")
plt.xlabel("C-H distance / $\AA$")
plt.tight_layout()
plt.legend()
plt.savefig(figure_save_path + "CHDistanceDistributionCarbs.pdf")
plt.show()
from SCFInitialGuess.utilities.analysis import measure_hf_energy
E_train = np.array(measure_hf_energy(data.T[0], data.molecules[0]))
E_validation = np.array(measure_hf_energy(data.T[1], data.molecules[1]))
E_test = np.array(measure_hf_energy(data.T[2], data.molecules[2]))
n_bins = 50
hist_train, edges = np.histogram(E_train, bins=n_bins, density=True)
hist_validation, _ = np.histogram(E_validation, bins=edges, density=True)
hist_test, _ = np.histogram(E_test, bins=edges, density=True)
centers = (edges[:-1] + edges[1:]) / 2
width = np.mean(np.diff(centers)) * 0.3
plt.bar(centers - width, hist_train, width=width)
plt.bar(centers , hist_validation, width=width)
plt.bar(centers, hist_test, width=width)
plt.ylabel("Relative Frequency / 1")
plt.xlabel("HF Energy / Hartree")
plt.tight_layout()
plt.savefig(figure_save_path + "EnergyDistributionDataset.pdf")
plt.show()
| 0.434941 | 0.654571 |
<a href="https://colab.research.google.com/github/amanjain252002/Stock-Price-Prediction/blob/main/Deep_Learning_Model.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import math
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tensorflow as tf
from sklearn.metrics import mean_squared_error, mean_absolute_error
import warnings
warnings.filterwarnings('ignore')
StkData = pd.read_csv('Data/FB_16_21.csv')
```
### Data Proprocessing
```
def Dataset(Data, Date):
Train_Data = Data['Adj Close'][Data['Date'] < Date].to_numpy()
Data_Train = []
Data_Train_X = []
Data_Train_Y = []
for i in range(0, len(Train_Data), 5):
try:
Data_Train.append(Train_Data[i : i + 5])
except:
pass
if len(Data_Train[-1]) < 5:
Data_Train.pop(-1)
Data_Train_X = Data_Train[0 : -1]
Data_Train_X = np.array(Data_Train_X)
Data_Train_X = Data_Train_X.reshape((-1, 5, 1))
Data_Train_Y = Data_Train[1 : len(Data_Train)]
Data_Train_Y = np.array(Data_Train_Y)
Data_Train_Y = Data_Train_Y.reshape((-1, 5, 1))
Test_Data = Data['Adj Close'][Data['Date'] >= Date].to_numpy()
Data_Test = []
Data_Test_X = []
Data_Test_Y = []
for i in range(0, len(Test_Data), 5):
try:
Data_Test.append(Test_Data[i : i + 5])
except:
pass
if len(Data_Test[-1]) < 5:
Data_Test.pop(-1)
Data_Test_X = Data_Test[0 : -1]
Data_Test_X = np.array(Data_Test_X)
Data_Test_X = Data_Test_X.reshape((-1, 5, 1))
Data_Test_Y = Data_Test[1 : len(Data_Test)]
Data_Test_Y = np.array(Data_Test_Y)
Data_Test_Y = Data_Test_Y.reshape((-1, 5, 1))
return Data_Train_X, Data_Train_Y, Data_Test_X, Data_Test_Y
```
### Model
```
def Model():
model = tf.keras.models.Sequential([
tf.keras.layers.LSTM(200, input_shape = (5, 1), activation = tf.nn.leaky_relu, return_sequences = True),
tf.keras.layers.LSTM(200, activation = tf.nn.leaky_relu),
tf.keras.layers.Dense(200, activation = tf.nn.leaky_relu),
tf.keras.layers.Dense(100, activation = tf.nn.leaky_relu),
tf.keras.layers.Dense(50, activation = tf.nn.leaky_relu),
tf.keras.layers.Dense(5, activation = tf.nn.leaky_relu)
])
return model
model = Model()
tf.keras.utils.plot_model(model, show_shapes=True)
model.summary()
```
### Custom Learning Rate
```
def scheduler(epoch):
if epoch <= 150:
lrate = (10 ** -5) * (epoch / 150)
elif epoch <= 400:
initial_lrate = (10 ** -5)
k = 0.01
lrate = initial_lrate * math.exp(-k * (epoch - 150))
else:
lrate = (10 ** -6)
return lrate
epochs = [i for i in range(1, 1001, 1)]
lrate = [scheduler(i) for i in range(1, 1001, 1)]
plt.plot(epochs, lrate)
callback = tf.keras.callbacks.LearningRateScheduler(scheduler)
```
#Apple
```
StkData.head()
StkData.info()
# Change Dtype of Date column
StkData["Date"] = pd.to_datetime(StkData["Date"])
```
###Split the Data into Training and Test set
```
StkData_Date = '2021-04-08'
StkData_Train_X, StkData_Train_Y, StkData_Test_X, StkData_Test_Y = Dataset(StkData, StkData_Date)
```
### Model Fitting
```
StkData_Model = Model()
StkData_Model.compile(optimizer = tf.keras.optimizers.Adam(), loss = 'mse', metrics = [tf.keras.metrics.RootMeanSquaredError()])
StkData_hist = StkData_Model.fit(StkData_Train_X, StkData_Train_Y, epochs = 1000, validation_data = (StkData_Test_X, StkData_Test_Y), callbacks=[callback])
history_dict = StkData_hist.history
loss = history_dict["loss"]
root_mean_squared_error = history_dict["root_mean_squared_error"]
val_loss = history_dict["val_loss"]
val_root_mean_squared_error = history_dict["val_root_mean_squared_error"]
epochs = range(1, len(loss) + 1)
fig, (ax1, ax2) = plt.subplots(1, 2)
fig.set_figheight(5)
fig.set_figwidth(15)
ax1.plot(epochs, loss, label = 'Training Loss')
ax1.plot(epochs, val_loss, label = 'Validation Loss')
ax1.set(xlabel = "Epochs", ylabel = "Loss")
ax1.legend()
ax2.plot(epochs, root_mean_squared_error, label = "Training Root Mean Squared Error")
ax2.plot(epochs, val_root_mean_squared_error, label = "Validation Root Mean Squared Error")
ax2.set(xlabel = "Epochs", ylabel = "Loss")
ax2.legend()
```
### Predicting the closing stock price
```
StkData_prediction = StkData_Model.predict(StkData_Test_X)
plt.figure(figsize=(20, 5))
plt.plot(StkData['Date'][StkData['Date'] < '2021-07-09'], StkData['Adj Close'][StkData['Date'] < '2021-07-09'], label = 'Training')
plt.plot(StkData['Date'][StkData['Date'] >= '2021-04-19'], StkData['Adj Close'][StkData['Date'] >= '2021-04-19'], label = 'Testing')
plt.plot(StkData['Date'][StkData['Date'] >= '2021-04-19'], StkData_prediction.reshape(-1), label = 'Predictions')
plt.xlabel('Time')
plt.ylabel('Closing Price')
plt.legend(loc = 'best')
plt.show()
rmse = math.sqrt(mean_squared_error(StkData_Test_Y.reshape(-1, 5), StkData_prediction))
mape = np.mean(np.abs(StkData_prediction - StkData_Test_Y.reshape(-1, 5))/np.abs(StkData_Test_Y.reshape(-1, 5)))
print(f'RMSE: {rmse}')
print(f'MAPE: {mape}')
```
|
github_jupyter
|
import math
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tensorflow as tf
from sklearn.metrics import mean_squared_error, mean_absolute_error
import warnings
warnings.filterwarnings('ignore')
StkData = pd.read_csv('Data/FB_16_21.csv')
def Dataset(Data, Date):
Train_Data = Data['Adj Close'][Data['Date'] < Date].to_numpy()
Data_Train = []
Data_Train_X = []
Data_Train_Y = []
for i in range(0, len(Train_Data), 5):
try:
Data_Train.append(Train_Data[i : i + 5])
except:
pass
if len(Data_Train[-1]) < 5:
Data_Train.pop(-1)
Data_Train_X = Data_Train[0 : -1]
Data_Train_X = np.array(Data_Train_X)
Data_Train_X = Data_Train_X.reshape((-1, 5, 1))
Data_Train_Y = Data_Train[1 : len(Data_Train)]
Data_Train_Y = np.array(Data_Train_Y)
Data_Train_Y = Data_Train_Y.reshape((-1, 5, 1))
Test_Data = Data['Adj Close'][Data['Date'] >= Date].to_numpy()
Data_Test = []
Data_Test_X = []
Data_Test_Y = []
for i in range(0, len(Test_Data), 5):
try:
Data_Test.append(Test_Data[i : i + 5])
except:
pass
if len(Data_Test[-1]) < 5:
Data_Test.pop(-1)
Data_Test_X = Data_Test[0 : -1]
Data_Test_X = np.array(Data_Test_X)
Data_Test_X = Data_Test_X.reshape((-1, 5, 1))
Data_Test_Y = Data_Test[1 : len(Data_Test)]
Data_Test_Y = np.array(Data_Test_Y)
Data_Test_Y = Data_Test_Y.reshape((-1, 5, 1))
return Data_Train_X, Data_Train_Y, Data_Test_X, Data_Test_Y
def Model():
model = tf.keras.models.Sequential([
tf.keras.layers.LSTM(200, input_shape = (5, 1), activation = tf.nn.leaky_relu, return_sequences = True),
tf.keras.layers.LSTM(200, activation = tf.nn.leaky_relu),
tf.keras.layers.Dense(200, activation = tf.nn.leaky_relu),
tf.keras.layers.Dense(100, activation = tf.nn.leaky_relu),
tf.keras.layers.Dense(50, activation = tf.nn.leaky_relu),
tf.keras.layers.Dense(5, activation = tf.nn.leaky_relu)
])
return model
model = Model()
tf.keras.utils.plot_model(model, show_shapes=True)
model.summary()
def scheduler(epoch):
if epoch <= 150:
lrate = (10 ** -5) * (epoch / 150)
elif epoch <= 400:
initial_lrate = (10 ** -5)
k = 0.01
lrate = initial_lrate * math.exp(-k * (epoch - 150))
else:
lrate = (10 ** -6)
return lrate
epochs = [i for i in range(1, 1001, 1)]
lrate = [scheduler(i) for i in range(1, 1001, 1)]
plt.plot(epochs, lrate)
callback = tf.keras.callbacks.LearningRateScheduler(scheduler)
StkData.head()
StkData.info()
# Change Dtype of Date column
StkData["Date"] = pd.to_datetime(StkData["Date"])
StkData_Date = '2021-04-08'
StkData_Train_X, StkData_Train_Y, StkData_Test_X, StkData_Test_Y = Dataset(StkData, StkData_Date)
StkData_Model = Model()
StkData_Model.compile(optimizer = tf.keras.optimizers.Adam(), loss = 'mse', metrics = [tf.keras.metrics.RootMeanSquaredError()])
StkData_hist = StkData_Model.fit(StkData_Train_X, StkData_Train_Y, epochs = 1000, validation_data = (StkData_Test_X, StkData_Test_Y), callbacks=[callback])
history_dict = StkData_hist.history
loss = history_dict["loss"]
root_mean_squared_error = history_dict["root_mean_squared_error"]
val_loss = history_dict["val_loss"]
val_root_mean_squared_error = history_dict["val_root_mean_squared_error"]
epochs = range(1, len(loss) + 1)
fig, (ax1, ax2) = plt.subplots(1, 2)
fig.set_figheight(5)
fig.set_figwidth(15)
ax1.plot(epochs, loss, label = 'Training Loss')
ax1.plot(epochs, val_loss, label = 'Validation Loss')
ax1.set(xlabel = "Epochs", ylabel = "Loss")
ax1.legend()
ax2.plot(epochs, root_mean_squared_error, label = "Training Root Mean Squared Error")
ax2.plot(epochs, val_root_mean_squared_error, label = "Validation Root Mean Squared Error")
ax2.set(xlabel = "Epochs", ylabel = "Loss")
ax2.legend()
StkData_prediction = StkData_Model.predict(StkData_Test_X)
plt.figure(figsize=(20, 5))
plt.plot(StkData['Date'][StkData['Date'] < '2021-07-09'], StkData['Adj Close'][StkData['Date'] < '2021-07-09'], label = 'Training')
plt.plot(StkData['Date'][StkData['Date'] >= '2021-04-19'], StkData['Adj Close'][StkData['Date'] >= '2021-04-19'], label = 'Testing')
plt.plot(StkData['Date'][StkData['Date'] >= '2021-04-19'], StkData_prediction.reshape(-1), label = 'Predictions')
plt.xlabel('Time')
plt.ylabel('Closing Price')
plt.legend(loc = 'best')
plt.show()
rmse = math.sqrt(mean_squared_error(StkData_Test_Y.reshape(-1, 5), StkData_prediction))
mape = np.mean(np.abs(StkData_prediction - StkData_Test_Y.reshape(-1, 5))/np.abs(StkData_Test_Y.reshape(-1, 5)))
print(f'RMSE: {rmse}')
print(f'MAPE: {mape}')
| 0.510985 | 0.929568 |
## Expectation Reflection + Least Absolute Deviations
In the following, we demonstrate how to apply Least Absolute Deviations (LAD) for classification task such as medical diagnosis.
We import the necessary packages to the Jupyter notebook:
```
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split,KFold
from sklearn.utils import shuffle
from sklearn.metrics import confusion_matrix,accuracy_score,precision_score,\
recall_score,roc_curve,auc
import expectation_reflection as ER
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import MinMaxScaler
from function import split_train_test,make_data_balance
np.random.seed(1)
```
First of all, the processed data are imported.
```
data_list = np.loadtxt('data_list_30sets.txt',dtype='str')
#data_list = ['29parkinson','30paradox2','31renal','32patientcare','33svr','34newt','35pcos']
print(data_list)
def read_data(data_id):
data_name = data_list[data_id]
print('data_name:',data_name)
Xy = np.loadtxt('../classification_data/%s/data_processed_knn5.dat'%data_name)
X = Xy[:,:-1]
#y = Xy[:,-1]
# 2020.07.15: convert y from {-1,+1} to {0,1}:
y = (Xy[:,-1]+1)/2.
#print(np.unique(y,return_counts=True))
X,y = make_data_balance(X,y)
print(np.unique(y,return_counts=True))
X, y = shuffle(X, y, random_state=1)
X_train,X_test,y_train,y_test = train_test_split(X,y,test_size=0.5,random_state = 1)
sc = MinMaxScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
return X_train,X_test,y_train,y_test
def measure_performance(X_train,X_test,y_train,y_test):
n = X_train.shape[1]
l2 = [0.0001,0.001,0.01,0.1,1.,10.,100.]
#l2 = [0.0001,0.001,0.01,0.1,1.,10.]
nl2 = len(l2)
# cross validation
kf = 4
kfold = KFold(n_splits=kf,shuffle=False)
h01 = np.zeros(kf)
w1 = np.zeros((kf,n))
cost1 = np.zeros(kf)
h0 = np.zeros(nl2)
w = np.zeros((nl2,n))
cost = np.zeros(nl2)
for il2 in range(len(l2)):
for i,(train_index,val_index) in enumerate(kfold.split(y_train)):
X_train1, X_val = X_train[train_index], X_train[val_index]
y_train1, y_val = y_train[train_index], y_train[val_index]
#h01[i],w1[i,:] = ER.fit(X_train1,y_train1,niter_max=100,l2=l2[il2])
h01[i],w1[i,:] = ER.fit_LAD(X_train1,y_train1,niter_max=100,l2=l2[il2])
y_val_pred,p_val_pred = ER.predict(X_val,h01[i],w1[i])
cost1[i] = ((p_val_pred - y_val)**2).mean()
h0[il2] = h01.mean(axis=0)
w[il2,:] = w1.mean(axis=0)
cost[il2] = cost1.mean()
# optimal value of l2:
il2_opt = np.argmin(cost)
print('optimal l2:',l2[il2_opt])
# performance:
y_test_pred,p_test_pred = ER.predict(X_test,h0[il2_opt],w[il2_opt,:])
fp,tp,thresholds = roc_curve(y_test, p_test_pred, drop_intermediate=False)
roc_auc = auc(fp,tp)
#print('AUC:', roc_auc)
acc = accuracy_score(y_test,y_test_pred)
#print('Accuracy:', acc)
precision = precision_score(y_test,y_test_pred)
#print('Precision:',precision)
recall = recall_score(y_test,y_test_pred)
#print('Recall:',recall)
f1_score = 2*precision*recall/(precision+recall)
return acc,roc_auc,precision,recall,f1_score
n_data = len(data_list)
roc_auc = np.zeros(n_data) ; acc = np.zeros(n_data)
precision = np.zeros(n_data) ; recall = np.zeros(n_data)
f1_score = np.zeros(n_data)
for data_id in range(n_data):
X_train,X_test,y_train,y_test = read_data(data_id)
acc[data_id],roc_auc[data_id],precision[data_id],recall[data_id],f1_score[data_id] =\
measure_performance(X_train,X_test,y_train,y_test)
print(data_id,acc[data_id],roc_auc[data_id],precision[data_id],recall[data_id],f1_score[data_id])
print('acc_mean:',acc.mean())
print('roc_mean:',roc_auc.mean())
print('precision:',precision.mean())
print('recall:',recall.mean())
print('f1_score:',f1_score.mean())
np.savetxt('result_knn5_ER_LAD.dat',(roc_auc,acc,precision,recall,f1_score),fmt='%f')
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split,KFold
from sklearn.utils import shuffle
from sklearn.metrics import confusion_matrix,accuracy_score,precision_score,\
recall_score,roc_curve,auc
import expectation_reflection as ER
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import MinMaxScaler
from function import split_train_test,make_data_balance
np.random.seed(1)
data_list = np.loadtxt('data_list_30sets.txt',dtype='str')
#data_list = ['29parkinson','30paradox2','31renal','32patientcare','33svr','34newt','35pcos']
print(data_list)
def read_data(data_id):
data_name = data_list[data_id]
print('data_name:',data_name)
Xy = np.loadtxt('../classification_data/%s/data_processed_knn5.dat'%data_name)
X = Xy[:,:-1]
#y = Xy[:,-1]
# 2020.07.15: convert y from {-1,+1} to {0,1}:
y = (Xy[:,-1]+1)/2.
#print(np.unique(y,return_counts=True))
X,y = make_data_balance(X,y)
print(np.unique(y,return_counts=True))
X, y = shuffle(X, y, random_state=1)
X_train,X_test,y_train,y_test = train_test_split(X,y,test_size=0.5,random_state = 1)
sc = MinMaxScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
return X_train,X_test,y_train,y_test
def measure_performance(X_train,X_test,y_train,y_test):
n = X_train.shape[1]
l2 = [0.0001,0.001,0.01,0.1,1.,10.,100.]
#l2 = [0.0001,0.001,0.01,0.1,1.,10.]
nl2 = len(l2)
# cross validation
kf = 4
kfold = KFold(n_splits=kf,shuffle=False)
h01 = np.zeros(kf)
w1 = np.zeros((kf,n))
cost1 = np.zeros(kf)
h0 = np.zeros(nl2)
w = np.zeros((nl2,n))
cost = np.zeros(nl2)
for il2 in range(len(l2)):
for i,(train_index,val_index) in enumerate(kfold.split(y_train)):
X_train1, X_val = X_train[train_index], X_train[val_index]
y_train1, y_val = y_train[train_index], y_train[val_index]
#h01[i],w1[i,:] = ER.fit(X_train1,y_train1,niter_max=100,l2=l2[il2])
h01[i],w1[i,:] = ER.fit_LAD(X_train1,y_train1,niter_max=100,l2=l2[il2])
y_val_pred,p_val_pred = ER.predict(X_val,h01[i],w1[i])
cost1[i] = ((p_val_pred - y_val)**2).mean()
h0[il2] = h01.mean(axis=0)
w[il2,:] = w1.mean(axis=0)
cost[il2] = cost1.mean()
# optimal value of l2:
il2_opt = np.argmin(cost)
print('optimal l2:',l2[il2_opt])
# performance:
y_test_pred,p_test_pred = ER.predict(X_test,h0[il2_opt],w[il2_opt,:])
fp,tp,thresholds = roc_curve(y_test, p_test_pred, drop_intermediate=False)
roc_auc = auc(fp,tp)
#print('AUC:', roc_auc)
acc = accuracy_score(y_test,y_test_pred)
#print('Accuracy:', acc)
precision = precision_score(y_test,y_test_pred)
#print('Precision:',precision)
recall = recall_score(y_test,y_test_pred)
#print('Recall:',recall)
f1_score = 2*precision*recall/(precision+recall)
return acc,roc_auc,precision,recall,f1_score
n_data = len(data_list)
roc_auc = np.zeros(n_data) ; acc = np.zeros(n_data)
precision = np.zeros(n_data) ; recall = np.zeros(n_data)
f1_score = np.zeros(n_data)
for data_id in range(n_data):
X_train,X_test,y_train,y_test = read_data(data_id)
acc[data_id],roc_auc[data_id],precision[data_id],recall[data_id],f1_score[data_id] =\
measure_performance(X_train,X_test,y_train,y_test)
print(data_id,acc[data_id],roc_auc[data_id],precision[data_id],recall[data_id],f1_score[data_id])
print('acc_mean:',acc.mean())
print('roc_mean:',roc_auc.mean())
print('precision:',precision.mean())
print('recall:',recall.mean())
print('f1_score:',f1_score.mean())
np.savetxt('result_knn5_ER_LAD.dat',(roc_auc,acc,precision,recall,f1_score),fmt='%f')
| 0.291485 | 0.831417 |
<a href="https://colab.research.google.com/github/wakamezake/Notebooks/blob/master/Deeplearning_tutorial_for_cell_image.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
<iframe src="//www.slideshare.net/slideshow/embed_code/key/wWQoShy9DDyj2y" width="595" height="485" frameborder="0" marginwidth="0" marginheight="0" scrolling="no" style="border:1px solid #CCC; border-width:1px; margin-bottom:5px; max-width: 100%;" allowfullscreen> </iframe> <div style="margin-bottom:5px"> <strong> <a href="//www.slideshare.net/DaisukeTakao/ss-234001580" title="チュートリアル:細胞画像を使った初めてのディープラーニング" target="_blank">チュートリアル:細胞画像を使った初めてのディープラーニング</a> </strong> from <strong><a href="https://www.slideshare.net/DaisukeTakao" target="_blank">DaisukeTakao</a></strong> </div>
```
!wget https://zenodo.org/record/3825945/files/G2.zip
!wget https://zenodo.org/record/3825945/files/notG2.zip
!unzip -qq G2.zip
!unzip -qq notG2.zip
!mkdir dataset
!mv G2/ dataset
!mv notG2 dataset
import numpy as np
import matplotlib.pyplot as plt
from PIL import Image
from tqdm import tqdm
import copy
import time
import torch.optim as optim
from torch.optim import lr_scheduler
import torch.nn as nn
from torch.autograd import Variable
import torch
import torchvision
from torchvision import datasets, models, transforms
dataset_root_path = './dataset'
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
transformers = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
set_keys = ['train', 'val']
batch_size = 32
image_dataset = datasets.ImageFolder(dataset_root_path, transform=transformers)
train_size = int(0.8 * len(image_dataset))
validation_size = len(image_dataset) - train_size
data_train, data_validation = torch.utils.data.random_split(image_dataset,
[train_size, validation_size])
dataloaders = {x: torch.utils.data.DataLoader(d, batch_size=32,
shuffle=True, num_workers=4)
for x, d in zip(set_keys, [data_train, data_validation])}
dataset_sizes = {"train":train_size, "val":validation_size}
class_names = image_dataset.classes
image_dataset.class_to_idx
def imshow(inp, title=None):
"""Imshow for Tensor."""
inp = inp.numpy().transpose((1, 2, 0))
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
inp = std * inp + mean
inp = np.clip(inp, 0, 1)
plt.imshow(inp)
if title is not None:
plt.title(title)
inputs, classes = next(iter(dataloaders['train']))
out = torchvision.utils.make_grid(inputs)
imshow(out, title=[class_names[x] for x in classes])
def train_model(model, criterion, optimizer, scheduler, num_epochs=25):
since = time.time()
best_model_wts = copy.deepcopy(model.state_dict())
best_acc = 0.0
for epoch in range(num_epochs):
print('Epoch {}/{}'.format(epoch, num_epochs - 1))
print('-' * 10)
# Each epoch has a training and validation phase
for phase in ['train', 'val']:
if phase == 'train':
model.train() # Set model to training mode
else:
model.eval() # Set model to evaluate mode
running_loss = 0.0
running_corrects = 0
# Iterate over data.
for inputs, labels in dataloaders[phase]:
inputs = inputs.to(device)
labels = labels.to(device)
# zero the parameter gradients
optimizer.zero_grad()
# forward
# track history if only in train
with torch.set_grad_enabled(phase == 'train'):
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
# backward + optimize only if in training phase
if phase == 'train':
loss.backward()
optimizer.step()
# statistics
running_loss += loss.item() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
if phase == 'train':
scheduler.step()
epoch_loss = running_loss / dataset_sizes[phase]
epoch_acc = running_corrects.double() / dataset_sizes[phase]
print('{} Loss: {:.4f} Acc: {:.4f}'.format(
phase, epoch_loss, epoch_acc))
# deep copy the model
if phase == 'val' and epoch_acc > best_acc:
best_acc = epoch_acc
best_model_wts = copy.deepcopy(model.state_dict())
print()
time_elapsed = time.time() - since
print('Training complete in {:.0f}m {:.0f}s'.format(
time_elapsed // 60, time_elapsed % 60))
print('Best val Acc: {:4f}'.format(best_acc))
# load best model weights
model.load_state_dict(best_model_wts)
return model
model_ft = models.resnet18(pretrained=True)
num_ftrs = model_ft.fc.in_features
# Here the size of each output sample is set to 2.
# Alternatively, it can be generalized to nn.Linear(num_ftrs, len(class_names)).
model_ft.fc = nn.Linear(num_ftrs, 2)
model_ft = model_ft.to(device)
criterion = nn.CrossEntropyLoss()
# Observe that all parameters are being optimized
optimizer_ft = optim.SGD(model_ft.parameters(), lr=0.001, momentum=0.9)
# Decay LR by a factor of 0.1 every 7 epochs
exp_lr_scheduler = lr_scheduler.StepLR(optimizer_ft, step_size=7, gamma=0.1)
model_ft = train_model(model_ft, criterion, optimizer_ft, exp_lr_scheduler, num_epochs=25)
torch.__version__
def visualize_model(model, num_images=6):
was_training = model.training
model.eval()
images_so_far = 0
fig = plt.figure()
with torch.no_grad():
for i, (inputs, labels) in enumerate(dataloaders['val']):
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
for j in range(inputs.size()[0]):
images_so_far += 1
ax = plt.subplot(num_images//2, 2, images_so_far)
ax.axis('off')
ax.set_title('predicted: {}'.format(class_names[preds[j]]))
imshow(inputs.cpu().data[j])
if images_so_far == num_images:
model.train(mode=was_training)
return
model.train(mode=was_training)
visualize_model(model_ft)
```
|
github_jupyter
|
!wget https://zenodo.org/record/3825945/files/G2.zip
!wget https://zenodo.org/record/3825945/files/notG2.zip
!unzip -qq G2.zip
!unzip -qq notG2.zip
!mkdir dataset
!mv G2/ dataset
!mv notG2 dataset
import numpy as np
import matplotlib.pyplot as plt
from PIL import Image
from tqdm import tqdm
import copy
import time
import torch.optim as optim
from torch.optim import lr_scheduler
import torch.nn as nn
from torch.autograd import Variable
import torch
import torchvision
from torchvision import datasets, models, transforms
dataset_root_path = './dataset'
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
transformers = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
set_keys = ['train', 'val']
batch_size = 32
image_dataset = datasets.ImageFolder(dataset_root_path, transform=transformers)
train_size = int(0.8 * len(image_dataset))
validation_size = len(image_dataset) - train_size
data_train, data_validation = torch.utils.data.random_split(image_dataset,
[train_size, validation_size])
dataloaders = {x: torch.utils.data.DataLoader(d, batch_size=32,
shuffle=True, num_workers=4)
for x, d in zip(set_keys, [data_train, data_validation])}
dataset_sizes = {"train":train_size, "val":validation_size}
class_names = image_dataset.classes
image_dataset.class_to_idx
def imshow(inp, title=None):
"""Imshow for Tensor."""
inp = inp.numpy().transpose((1, 2, 0))
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
inp = std * inp + mean
inp = np.clip(inp, 0, 1)
plt.imshow(inp)
if title is not None:
plt.title(title)
inputs, classes = next(iter(dataloaders['train']))
out = torchvision.utils.make_grid(inputs)
imshow(out, title=[class_names[x] for x in classes])
def train_model(model, criterion, optimizer, scheduler, num_epochs=25):
since = time.time()
best_model_wts = copy.deepcopy(model.state_dict())
best_acc = 0.0
for epoch in range(num_epochs):
print('Epoch {}/{}'.format(epoch, num_epochs - 1))
print('-' * 10)
# Each epoch has a training and validation phase
for phase in ['train', 'val']:
if phase == 'train':
model.train() # Set model to training mode
else:
model.eval() # Set model to evaluate mode
running_loss = 0.0
running_corrects = 0
# Iterate over data.
for inputs, labels in dataloaders[phase]:
inputs = inputs.to(device)
labels = labels.to(device)
# zero the parameter gradients
optimizer.zero_grad()
# forward
# track history if only in train
with torch.set_grad_enabled(phase == 'train'):
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
# backward + optimize only if in training phase
if phase == 'train':
loss.backward()
optimizer.step()
# statistics
running_loss += loss.item() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
if phase == 'train':
scheduler.step()
epoch_loss = running_loss / dataset_sizes[phase]
epoch_acc = running_corrects.double() / dataset_sizes[phase]
print('{} Loss: {:.4f} Acc: {:.4f}'.format(
phase, epoch_loss, epoch_acc))
# deep copy the model
if phase == 'val' and epoch_acc > best_acc:
best_acc = epoch_acc
best_model_wts = copy.deepcopy(model.state_dict())
print()
time_elapsed = time.time() - since
print('Training complete in {:.0f}m {:.0f}s'.format(
time_elapsed // 60, time_elapsed % 60))
print('Best val Acc: {:4f}'.format(best_acc))
# load best model weights
model.load_state_dict(best_model_wts)
return model
model_ft = models.resnet18(pretrained=True)
num_ftrs = model_ft.fc.in_features
# Here the size of each output sample is set to 2.
# Alternatively, it can be generalized to nn.Linear(num_ftrs, len(class_names)).
model_ft.fc = nn.Linear(num_ftrs, 2)
model_ft = model_ft.to(device)
criterion = nn.CrossEntropyLoss()
# Observe that all parameters are being optimized
optimizer_ft = optim.SGD(model_ft.parameters(), lr=0.001, momentum=0.9)
# Decay LR by a factor of 0.1 every 7 epochs
exp_lr_scheduler = lr_scheduler.StepLR(optimizer_ft, step_size=7, gamma=0.1)
model_ft = train_model(model_ft, criterion, optimizer_ft, exp_lr_scheduler, num_epochs=25)
torch.__version__
def visualize_model(model, num_images=6):
was_training = model.training
model.eval()
images_so_far = 0
fig = plt.figure()
with torch.no_grad():
for i, (inputs, labels) in enumerate(dataloaders['val']):
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
for j in range(inputs.size()[0]):
images_so_far += 1
ax = plt.subplot(num_images//2, 2, images_so_far)
ax.axis('off')
ax.set_title('predicted: {}'.format(class_names[preds[j]]))
imshow(inputs.cpu().data[j])
if images_so_far == num_images:
model.train(mode=was_training)
return
model.train(mode=was_training)
visualize_model(model_ft)
| 0.859192 | 0.917525 |
```
# Зависимости
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import random
import os
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler, OneHotEncoder
from sklearn.compose import ColumnTransformer
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import f1_score
import tensorflow as tf
from sklearn.neighbors import KNeighborsClassifier
# Инициализируем все известные генераторы случаынйх чисел / Setting all known random seeds
my_code = "Johnson"
seed_limit = 2 ** 32
my_seed = int.from_bytes(my_code.encode(), "little") % seed_limit
os.environ['PYTHONHASHSEED']=str(my_seed)
random.seed(my_seed)
np.random.seed(my_seed)
tf.compat.v1.set_random_seed(my_seed)
session_conf = tf.compat.v1.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
sess = tf.compat.v1.Session(graph=tf.compat.v1.get_default_graph(), config=session_conf)
tf.compat.v1.keras.backend.set_session(sess)
# Читаем данные из файла
train_data = pd.read_csv("datasets/iris_train.csv")
train_data.head()
# Определим размер валидационной выборки
val_size = round(0.2*len(train_data))
print(val_size)
# Создадим обучающую и валидационную выборки
random_state = my_seed
train, val = train_test_split(train_data, test_size=val_size, random_state=random_state)
print(len(train), len(val))
# Значения в числовых столбцах преобразуем к отрезку [0,1].
# Для настройки скалировщика используем только обучающую выборку.
num_columns = ['sepal_length', 'sepal_width', 'petal_length', 'petal_width']
ord_columns = ['species']
ct = ColumnTransformer(transformers=[('numerical', MinMaxScaler(), num_columns)], remainder='passthrough')
ct.fit(train)
# Преобразуем значения, тип данных приводим к DataFrame
sc_train = pd.DataFrame(ct.transform(train))
sc_val = pd.DataFrame(ct.transform(val))
# Устанавливаем названия столбцов
column_names = num_columns + ord_columns
sc_train.columns = column_names
sc_val.columns = column_names
sc_train
# Отберем необходимые параметры
x_train = sc_train[num_columns]
x_val = sc_val[num_columns]
y_train = (sc_train[ord_columns].values).flatten()
y_val = (sc_val[ord_columns].values).flatten()
# Создадим простую модель логистической регрессии
model = KNeighborsClassifier(n_neighbors=18)
# Обучим модель
model.fit(x_train, y_train)
# Проверим работу обученной нейронной сети на валидационной выборке
pred_val = model.predict(x_val)
f1 = f1_score(y_val, pred_val, average='weighted')
print(f1)
test = pd.read_csv("datasets/iris_test.csv")
test['species'] = ''
test.head()
sc_test = pd.DataFrame(ct.transform(test))
sc_test.columns = column_names
x_test = sc_test[num_columns]
test['species'] = model.predict(x_test)
test.head()
test.to_csv('Soloviev.csv', index=False)
```
|
github_jupyter
|
# Зависимости
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import random
import os
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler, OneHotEncoder
from sklearn.compose import ColumnTransformer
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import f1_score
import tensorflow as tf
from sklearn.neighbors import KNeighborsClassifier
# Инициализируем все известные генераторы случаынйх чисел / Setting all known random seeds
my_code = "Johnson"
seed_limit = 2 ** 32
my_seed = int.from_bytes(my_code.encode(), "little") % seed_limit
os.environ['PYTHONHASHSEED']=str(my_seed)
random.seed(my_seed)
np.random.seed(my_seed)
tf.compat.v1.set_random_seed(my_seed)
session_conf = tf.compat.v1.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
sess = tf.compat.v1.Session(graph=tf.compat.v1.get_default_graph(), config=session_conf)
tf.compat.v1.keras.backend.set_session(sess)
# Читаем данные из файла
train_data = pd.read_csv("datasets/iris_train.csv")
train_data.head()
# Определим размер валидационной выборки
val_size = round(0.2*len(train_data))
print(val_size)
# Создадим обучающую и валидационную выборки
random_state = my_seed
train, val = train_test_split(train_data, test_size=val_size, random_state=random_state)
print(len(train), len(val))
# Значения в числовых столбцах преобразуем к отрезку [0,1].
# Для настройки скалировщика используем только обучающую выборку.
num_columns = ['sepal_length', 'sepal_width', 'petal_length', 'petal_width']
ord_columns = ['species']
ct = ColumnTransformer(transformers=[('numerical', MinMaxScaler(), num_columns)], remainder='passthrough')
ct.fit(train)
# Преобразуем значения, тип данных приводим к DataFrame
sc_train = pd.DataFrame(ct.transform(train))
sc_val = pd.DataFrame(ct.transform(val))
# Устанавливаем названия столбцов
column_names = num_columns + ord_columns
sc_train.columns = column_names
sc_val.columns = column_names
sc_train
# Отберем необходимые параметры
x_train = sc_train[num_columns]
x_val = sc_val[num_columns]
y_train = (sc_train[ord_columns].values).flatten()
y_val = (sc_val[ord_columns].values).flatten()
# Создадим простую модель логистической регрессии
model = KNeighborsClassifier(n_neighbors=18)
# Обучим модель
model.fit(x_train, y_train)
# Проверим работу обученной нейронной сети на валидационной выборке
pred_val = model.predict(x_val)
f1 = f1_score(y_val, pred_val, average='weighted')
print(f1)
test = pd.read_csv("datasets/iris_test.csv")
test['species'] = ''
test.head()
sc_test = pd.DataFrame(ct.transform(test))
sc_test.columns = column_names
x_test = sc_test[num_columns]
test['species'] = model.predict(x_test)
test.head()
test.to_csv('Soloviev.csv', index=False)
| 0.31321 | 0.547706 |
```
import pandas as pd
import numpy as np
import datetime
import pandas_datareader.data as web
from datetime import datetime,timedelta
import ffn
import scipy
import math
import operator
import os
class summarystats:
def __init__(self,region,datapath,outputpath):
self.region=region
self.datapath=datapath
self.outputpath=outputpath
def calcMMIndex(self,df,colname,idxname):
df.loc[df.index[0],idxname]= 1
prev_dt= df.index[0]
for dt in df.index[1:]:
caldays= (dt- prev_dt).days
df.loc[dt,idxname]= df.loc[prev_dt,idxname]*(1+df.loc[prev_dt,colname]/360*caldays/100)
prev_dt=dt
df.drop(columns=colname,inplace=True)
return df
def getMMIndex(self):
if (self.region=='US'):
yld=web.DataReader('DGS1MO', 'fred',start='2000-01-01').dropna()## download 1-Month Treasury Constant Maturity Rate from FRB St louis
yld.rename_axis(index={'DATE':'Date'},inplace=True)
idx=self.calcMMIndex(yld.copy(),'DGS1MO','1MTBillIndex')
if(self.region=='EUR'):
yld= pd.read_csv(self.datapath+'\\1MEuribor.csv',skiprows=5,header=None).rename(columns={1:'Euribor'})
yld['Date']= yld[0].apply(lambda x: pd.to_datetime(datetime.strptime(x,'%Y%b')))
yld=yld.drop(columns=0).set_index('Date')
idx= self.calcMMIndex(yld.copy(),'Euribor','1MEuriborIndex')
return idx
def rollingreturns(self,all_idxs,windows=[36,60]):
mnth_end_rets= all_idxs.asfreq('M',method='ffill').pct_change()[1:]
df= pd.DataFrame(columns=all_idxs.columns)
rolling= {}
for window in windows:
rolling[window]={}
for k in ['Returns','Risk','Returns-Risk']:
rolling[window][k]= pd.DataFrame(columns=all_idxs.columns)
for i in range(window,len(mnth_end_rets)+1):
idx= mnth_end_rets.index[i-1]
rolling[window]['Returns'].loc[idx,:]=scipy.stats.gmean(1+mnth_end_rets.iloc[i-window:i,:])**12-1
rolling[window]['Risk'].loc[idx,:]= mnth_end_rets.iloc[i-window:i,:].std()*np.sqrt(12)
rolling[window]['Returns-Risk'].loc[idx,:]= rolling[window]['Returns'].loc[idx,:]/rolling[window]['Risk'].loc[idx,:]
for k in ['Returns','Risk','Returns-Risk']:
df.loc['Average '+str(window)+ 'months rolling returns',:]= np.round(100*rolling[window]['Returns'].mean(),2)
df.loc['Average '+str(window)+ 'months rolling risk',:]= np.round(rolling[window]['Risk'].mean()*100,2)
df.loc['Average '+str(window)+ 'months rolling return/risk',:]= np.round(rolling[window]['Returns-Risk'].mean().astype(float),2)
return df,rolling
def PerformanceSummaryWrapper(self,indexlevels,benchmark=True,simulationname=''):
indexnames=indexlevels.columns
benchmarkname = indexnames[0]
enddate=max(indexlevels.index)
indexlevels= indexlevels.fillna(method='ffill').dropna()
stats = ffn.core.GroupStats(indexlevels)
Perf = stats.stats.loc[{'start','end','cagr','monthly_mean',
'monthly_vol','max_drawdown','monthly_skew','monthly_kurt','calmar'},
indexlevels.columns]
RiskSummary = stats.stats.loc[{'start','end','monthly_vol','max_drawdown','monthly_skew','monthly_kurt','calmar'},
indexlevels.columns]
RiskSummary.loc['start'] = [startdt.strftime('%Y-%m-%d') for startdt in RiskSummary.loc['start']]
RiskSummary.loc['end'] = [enddt.strftime('%Y-%m-%d') for enddt in RiskSummary.loc['end']]
drawdownseries = ffn.core.to_drawdown_series(indexlevels)
RiskSummary.loc['Max Drawdown Period'] = [max(drawdownseries[(drawdownseries[column]==0)&
(drawdownseries[column].index<min(drawdownseries[drawdownseries[column]==
min(drawdownseries[column])].index))].index).strftime('%Y-%m-%d') + ' to '+
max(drawdownseries[drawdownseries[column]==min(drawdownseries[column])].index).strftime('%Y-%m-%d')
for column in indexlevels.columns]
RiskSummary.loc['Max Downstreak Years (Absolute)'] = [max([x - drawdownseries[drawdownseries[column]==0].index[i - 1]
for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)][1:]).days/365.0 for column in indexlevels.columns]
RiskSummary.loc['Max Downstreak Period (Absolute)'] = [max(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[[np.argmax([x - drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[i - 1] for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)])-1]]).strftime('%Y-%m-%d')+' to '+
max(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[[np.argmax([x - drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[i - 1] for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)])]]).strftime('%Y-%m-%d') for column in indexlevels.columns]
rfr=pd.DataFrame()
if (self.region=='US'):
rfr = ffn.core.to_monthly(self.getMMIndex()).to_returns()[1:]
elif(self.region=='EUR'):
rfr= self.getMMIndex().to_returns()[1:]
rfr.rename(columns={rfr.columns[0]:'Rtn'},inplace=True)
rfr['Rtn'] = 1 + rfr['Rtn']
# Calculate the geometric mean of risk-free rates from start-date to end-date
Perf.loc['RFR'] = [scipy.stats.gmean(rfr['Rtn'][(rfr.index>start) & (rfr.index<=end)]) for (start,end) in zip(Perf.loc['start'], Perf.loc['end'])]
Perf.loc['RFR'] = Perf.loc['RFR']**12 -1
Perf.loc['Sharpe-Ratio'] = (Perf.loc['cagr'] - Perf.loc['RFR']) / Perf.loc['monthly_vol']
Perf.loc['start'] = [startdt.strftime('%Y-%m-%d') for startdt in Perf.loc['start']]
Perf.loc['end'] = [enddt.strftime('%Y-%m-%d') for enddt in Perf.loc['end']]
Perf.loc['Return/Risk'] = Perf.loc['cagr'] / Perf.loc['monthly_vol']
# round and multiply a few columns by 100
Perf.loc[['cagr','monthly_mean','monthly_vol','max_drawdown'],:]= np.round(100*Perf.loc[['cagr','monthly_mean','monthly_vol','max_drawdown'],:].astype('float'),2)
if benchmark:
strategyreturns = ffn.core.to_monthly(indexlevels).to_returns()
benchmarkreturns = ffn.core.to_monthly(indexlevels[[benchmarkname]]).to_returns()
excessreturns = strategyreturns - np.tile(benchmarkreturns,len(indexnames))
gmreturns=strategyreturns+1
relativeperformancelevels = (indexlevels.loc[:,indexlevels.columns[1:]] /np.transpose(np.tile(indexlevels.loc[:,benchmarkname],(len(indexnames)-1,1)))).rebase()
drawdownseries =ffn.core.to_drawdown_series(relativeperformancelevels)
RiskSummary.loc['Max Downstreak Years (Relative)'] = [0]+[max([x - drawdownseries[drawdownseries[column]==0].index[i - 1]
for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)][1:]).days/365.0 for column in indexlevels.columns[1:]]
RiskSummary.loc['Max Downstreak Period (Relative)'] = ['']+[max(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[[np.argmax([x - drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[i - 1] for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)])-1]]).strftime('%Y-%m-%d')+' to '+
max(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[[np.argmax([x - drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[i - 1] for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)])]]).strftime('%Y-%m-%d') for column in indexlevels.columns[1:]]
RiskSummary.loc['Downside Risk (%)']=np.round([math.sqrt(np.mean(np.square(np.minimum((strategyreturns[column] - np.mean(strategyreturns[column])),np.zeros(len((strategyreturns[column] - np.mean(strategyreturns[column]))))))))*100*math.sqrt(12) for column in strategyreturns.columns],2)
Perf.loc['Active Return (%)'] = Perf.loc['cagr'] - np.tile(Perf.loc['cagr',[benchmarkname]],len(indexnames))
Perf.loc['Tracking Error (%)']= (excessreturns.std()*np.sqrt(12)*100).values
Perf.loc['Tracking Error (%)',benchmarkname] = np.NaN
Perf.loc['Information Ratio'] = Perf.loc['Active Return (%)'] /Perf.loc['Tracking Error (%)']
RiskSummary.loc['Correlation'] = strategyreturns.corr()[benchmarkname]
RiskSummary.loc['Beta'] = strategyreturns.cov()[benchmarkname] /np.tile(strategyreturns.var()[benchmarkname],len(indexnames))
Perf.loc[['Active Return (%)','Tracking Error (%)','Information Ratio'],:]= np.round(Perf.loc[['Active Return (%)','Tracking Error (%)','Information Ratio'],:].astype('float'),2)
RiskSummary.loc['Monthly Batting Average (%)']= np.round([x*100 for x in list(map(operator.truediv, [len(excessreturns[excessreturns[column]>0]) for column in excessreturns.columns], [len(excessreturns[column])-1 for column in excessreturns.columns]))],2)
RiskSummary.loc['Upside Capture Ratio']= np.round([(scipy.stats.mstats.gmean(gmreturns[column] [gmreturns[benchmarkname]>1])-1)/(scipy.stats.mstats.gmean(gmreturns[benchmarkname] [gmreturns[benchmarkname]>1])-1) for column in gmreturns.columns],4)
RiskSummary.loc['Downside Capture Ratio']= np.round([(scipy.stats.mstats.gmean(gmreturns[column][gmreturns[benchmarkname]<1])-1)/(scipy.stats.mstats.gmean(gmreturns[benchmarkname] [gmreturns[benchmarkname]<1])-1) for column in gmreturns.columns],4)
RiskSummary.loc[['monthly_skew','monthly_kurt','Beta','calmar','Correlation','Max Downstreak Years (Absolute)',
'Max Downstreak Years (Relative)'],:]= np.round(RiskSummary.loc[['monthly_skew','monthly_kurt','Beta',
'Correlation','Max Downstreak Years (Absolute)','Max Downstreak Years (Relative)'],:].astype('float'),2)
RiskSummary.loc[['max_drawdown','monthly_vol'],:]= np.round(100*RiskSummary.loc[['max_drawdown','monthly_vol'],:].astype('float'),2)
RiskSummary = RiskSummary.loc[['start','end','monthly_vol','Downside Risk (%)','max_drawdown','calmar','Max Drawdown Period','Max Downstreak Years (Absolute)','Max Downstreak Period (Absolute)','Max Downstreak Years (Relative)',
'Max Downstreak Period (Relative)','Monthly Batting Average (%)','Upside Capture Ratio','Downside Capture Ratio','monthly_skew',\
'monthly_kurt','Correlation','Beta'],:]
RiskSummary.rename(index={'max_drawdown':'Maximum Drawdown (%)',\
'monthly_vol':'Risk (%)','monthly_skew':'Skewness',\
'monthly_kurt':'Kurtosis','calmar':'Calmar Ratio'},inplace=True)
else:
strategyreturns = ffn.core.to_monthly(indexlevels).to_returns()
RiskSummary.loc['Downside Risk (%)']=np.round([math.sqrt(np.mean(np.square(np.minimum((strategyreturns[column] - np.mean(strategyreturns[column])),np.zeros(len((strategyreturns[column] - np.mean(strategyreturns[column]))))))))*100*math.sqrt(12) for column in strategyreturns.columns],2)
RiskSummary.loc[['monthly_skew','monthly_kurt','calmar'],:]= np.round(RiskSummary.loc[['monthly_skew','monthly_kurt','calmar'],:].astype('float'),2)
RiskSummary.loc[['max_drawdown','monthly_vol'],:]= np.round(100*RiskSummary.loc[['max_drawdown','monthly_vol'],:].astype('float'),2)
RiskSummary = RiskSummary.loc[['start','end','monthly_vol','Downside Risk (%)','max_drawdown',\
'Max Drawdown Period','calmar','Max Downstreak Years (Absolute)',\
'Max Downstreak Period (Absolute)','monthly_skew','monthly_kurt'],:]
RiskSummary.rename(index={'max_drawdown':'Maximum Drawdown (%)',\
'monthly_vol':'Risk (%)','monthly_skew':'Skewness',\
'monthly_kurt':'Kurtosis','calmar':'Calmar Ratio'},inplace=True)
AdditionalPerf = Perf.loc[{'start','end'}]
horizons = ['three_month','six_month','ytd','one_year','three_year','five_year','ten_year']
commonhorizon = set(horizons) & set(stats.stats.index)
commonhorizon = [ch for ch in horizons if ch in commonhorizon]
horizonreturns = stats.stats.loc[commonhorizon,
indexlevels.columns]*100
AdditionalPerf=AdditionalPerf.append(np.round(horizonreturns.astype('float'),2))
calendaryearreturns = np.round(indexlevels.to_monthly().pct_change(periods=12)*100,2)
calendaryearreturns = calendaryearreturns[calendaryearreturns.index.month==12].dropna()
calendaryearreturns.index = calendaryearreturns.index.year
AdditionalPerf = AdditionalPerf.append(calendaryearreturns)
Perf.loc['Downside Risk (%)']=RiskSummary.loc['Downside Risk (%)']
Perf.loc['Sortino-Ratio']= (Perf.loc['cagr'] - Perf.loc['RFR']) / Perf.loc['Downside Risk (%)']
Perf.loc['Return/Max Drawdown']=Perf.loc['cagr']/np.abs(Perf.loc['max_drawdown'])
Perf.loc[['Return/Risk','Sharpe-Ratio','Sortino-Ratio','monthly_skew','monthly_kurt','calmar','Return/Max Drawdown'],:]= np.round(Perf.loc[['Return/Risk','Sharpe-Ratio',\
'Sortino-Ratio','monthly_skew','monthly_kurt','calmar','Return/Max Drawdown'],:].astype('float'),2)
Perf.loc[['Sortino-Ratio'],:]= np.round(Perf.loc[['Sortino-Ratio'],:].astype('float'),2)
Perf = Perf.loc[['start','end','cagr','monthly_mean','monthly_vol','Downside Risk (%)','Return/Risk', 'monthly_skew',\
'monthly_kurt','Sharpe-Ratio','Sortino-Ratio',\
'max_drawdown','calmar','Return/Max Drawdown'],:]
Perf.rename(index={'max_drawdown':'Maximum Drawdown (%)',\
'monthly_vol':'Risk (%)','cagr':'Annualized Compunded Return/CAGR(%)',\
'monthly_mean':'Annualized Arthimetic mean(%)','calmar':'Calmar Ratio',\
'monthly_skew':'Skewness',\
'monthly_kurt':'Kurtosis'},inplace=True)
# RiskSummary.index = [indexsummarylabels.get(indexname,indexname) for indexname in RiskSummary.index]
simulname= self.region+'-Simulation-'+datetime.now().strftime('%Y%m%d-%H%M')+simulationname
# os.mkdir(self.outputpath+'//results//'+simulname)
# newpath=self.outputpath+'//results//'+simulname+'//'
writer= pd.ExcelWriter(self.outputpath+simulname+'.xlsx')
Perf.to_excel(writer,'PerformanceSummary')
# Perf.to_csv(newpath+'PerformanceSummary.csv')
RiskSummary.to_excel(writer,'RiskSummary')
# RiskSummary.to_csv(newpath+'RiskSummary.csv')
AdditionalPerf.to_excel(writer,'Horizon Returns')
# AdditionalPerf.to_csv(newpath+'Horizon Returns.csv')
strategyreturns.to_excel(writer,'Strategy Returns')
# strategyreturns.to_csv(self.outputpath+'strategyreturns.csv')
strategyreturns.corr().to_excel(writer,'Correlation')
dfroll,rolling= self.rollingreturns(indexlevels)
dfroll.to_excel(writer,'Average Rolling Stats')
for i in rolling.keys():
for j in rolling[i].keys():
rolling[i][j].to_excel(writer, 'rolling '+str(i)+'M '+str(j))
writer.close()
# strategyreturns.corr().to_csv(self.outputpath+'Correlation.csv')
return Perf
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import datetime
import pandas_datareader.data as web
from datetime import datetime,timedelta
import ffn
import scipy
import math
import operator
import os
class summarystats:
def __init__(self,region,datapath,outputpath):
self.region=region
self.datapath=datapath
self.outputpath=outputpath
def calcMMIndex(self,df,colname,idxname):
df.loc[df.index[0],idxname]= 1
prev_dt= df.index[0]
for dt in df.index[1:]:
caldays= (dt- prev_dt).days
df.loc[dt,idxname]= df.loc[prev_dt,idxname]*(1+df.loc[prev_dt,colname]/360*caldays/100)
prev_dt=dt
df.drop(columns=colname,inplace=True)
return df
def getMMIndex(self):
if (self.region=='US'):
yld=web.DataReader('DGS1MO', 'fred',start='2000-01-01').dropna()## download 1-Month Treasury Constant Maturity Rate from FRB St louis
yld.rename_axis(index={'DATE':'Date'},inplace=True)
idx=self.calcMMIndex(yld.copy(),'DGS1MO','1MTBillIndex')
if(self.region=='EUR'):
yld= pd.read_csv(self.datapath+'\\1MEuribor.csv',skiprows=5,header=None).rename(columns={1:'Euribor'})
yld['Date']= yld[0].apply(lambda x: pd.to_datetime(datetime.strptime(x,'%Y%b')))
yld=yld.drop(columns=0).set_index('Date')
idx= self.calcMMIndex(yld.copy(),'Euribor','1MEuriborIndex')
return idx
def rollingreturns(self,all_idxs,windows=[36,60]):
mnth_end_rets= all_idxs.asfreq('M',method='ffill').pct_change()[1:]
df= pd.DataFrame(columns=all_idxs.columns)
rolling= {}
for window in windows:
rolling[window]={}
for k in ['Returns','Risk','Returns-Risk']:
rolling[window][k]= pd.DataFrame(columns=all_idxs.columns)
for i in range(window,len(mnth_end_rets)+1):
idx= mnth_end_rets.index[i-1]
rolling[window]['Returns'].loc[idx,:]=scipy.stats.gmean(1+mnth_end_rets.iloc[i-window:i,:])**12-1
rolling[window]['Risk'].loc[idx,:]= mnth_end_rets.iloc[i-window:i,:].std()*np.sqrt(12)
rolling[window]['Returns-Risk'].loc[idx,:]= rolling[window]['Returns'].loc[idx,:]/rolling[window]['Risk'].loc[idx,:]
for k in ['Returns','Risk','Returns-Risk']:
df.loc['Average '+str(window)+ 'months rolling returns',:]= np.round(100*rolling[window]['Returns'].mean(),2)
df.loc['Average '+str(window)+ 'months rolling risk',:]= np.round(rolling[window]['Risk'].mean()*100,2)
df.loc['Average '+str(window)+ 'months rolling return/risk',:]= np.round(rolling[window]['Returns-Risk'].mean().astype(float),2)
return df,rolling
def PerformanceSummaryWrapper(self,indexlevels,benchmark=True,simulationname=''):
indexnames=indexlevels.columns
benchmarkname = indexnames[0]
enddate=max(indexlevels.index)
indexlevels= indexlevels.fillna(method='ffill').dropna()
stats = ffn.core.GroupStats(indexlevels)
Perf = stats.stats.loc[{'start','end','cagr','monthly_mean',
'monthly_vol','max_drawdown','monthly_skew','monthly_kurt','calmar'},
indexlevels.columns]
RiskSummary = stats.stats.loc[{'start','end','monthly_vol','max_drawdown','monthly_skew','monthly_kurt','calmar'},
indexlevels.columns]
RiskSummary.loc['start'] = [startdt.strftime('%Y-%m-%d') for startdt in RiskSummary.loc['start']]
RiskSummary.loc['end'] = [enddt.strftime('%Y-%m-%d') for enddt in RiskSummary.loc['end']]
drawdownseries = ffn.core.to_drawdown_series(indexlevels)
RiskSummary.loc['Max Drawdown Period'] = [max(drawdownseries[(drawdownseries[column]==0)&
(drawdownseries[column].index<min(drawdownseries[drawdownseries[column]==
min(drawdownseries[column])].index))].index).strftime('%Y-%m-%d') + ' to '+
max(drawdownseries[drawdownseries[column]==min(drawdownseries[column])].index).strftime('%Y-%m-%d')
for column in indexlevels.columns]
RiskSummary.loc['Max Downstreak Years (Absolute)'] = [max([x - drawdownseries[drawdownseries[column]==0].index[i - 1]
for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)][1:]).days/365.0 for column in indexlevels.columns]
RiskSummary.loc['Max Downstreak Period (Absolute)'] = [max(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[[np.argmax([x - drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[i - 1] for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)])-1]]).strftime('%Y-%m-%d')+' to '+
max(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[[np.argmax([x - drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[i - 1] for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)])]]).strftime('%Y-%m-%d') for column in indexlevels.columns]
rfr=pd.DataFrame()
if (self.region=='US'):
rfr = ffn.core.to_monthly(self.getMMIndex()).to_returns()[1:]
elif(self.region=='EUR'):
rfr= self.getMMIndex().to_returns()[1:]
rfr.rename(columns={rfr.columns[0]:'Rtn'},inplace=True)
rfr['Rtn'] = 1 + rfr['Rtn']
# Calculate the geometric mean of risk-free rates from start-date to end-date
Perf.loc['RFR'] = [scipy.stats.gmean(rfr['Rtn'][(rfr.index>start) & (rfr.index<=end)]) for (start,end) in zip(Perf.loc['start'], Perf.loc['end'])]
Perf.loc['RFR'] = Perf.loc['RFR']**12 -1
Perf.loc['Sharpe-Ratio'] = (Perf.loc['cagr'] - Perf.loc['RFR']) / Perf.loc['monthly_vol']
Perf.loc['start'] = [startdt.strftime('%Y-%m-%d') for startdt in Perf.loc['start']]
Perf.loc['end'] = [enddt.strftime('%Y-%m-%d') for enddt in Perf.loc['end']]
Perf.loc['Return/Risk'] = Perf.loc['cagr'] / Perf.loc['monthly_vol']
# round and multiply a few columns by 100
Perf.loc[['cagr','monthly_mean','monthly_vol','max_drawdown'],:]= np.round(100*Perf.loc[['cagr','monthly_mean','monthly_vol','max_drawdown'],:].astype('float'),2)
if benchmark:
strategyreturns = ffn.core.to_monthly(indexlevels).to_returns()
benchmarkreturns = ffn.core.to_monthly(indexlevels[[benchmarkname]]).to_returns()
excessreturns = strategyreturns - np.tile(benchmarkreturns,len(indexnames))
gmreturns=strategyreturns+1
relativeperformancelevels = (indexlevels.loc[:,indexlevels.columns[1:]] /np.transpose(np.tile(indexlevels.loc[:,benchmarkname],(len(indexnames)-1,1)))).rebase()
drawdownseries =ffn.core.to_drawdown_series(relativeperformancelevels)
RiskSummary.loc['Max Downstreak Years (Relative)'] = [0]+[max([x - drawdownseries[drawdownseries[column]==0].index[i - 1]
for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)][1:]).days/365.0 for column in indexlevels.columns[1:]]
RiskSummary.loc['Max Downstreak Period (Relative)'] = ['']+[max(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[[np.argmax([x - drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[i - 1] for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)])-1]]).strftime('%Y-%m-%d')+' to '+
max(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[[np.argmax([x - drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
[i - 1] for i, x in enumerate(drawdownseries[drawdownseries[column]==0].index.append(pd.DatetimeIndex([enddate]))
)])]]).strftime('%Y-%m-%d') for column in indexlevels.columns[1:]]
RiskSummary.loc['Downside Risk (%)']=np.round([math.sqrt(np.mean(np.square(np.minimum((strategyreturns[column] - np.mean(strategyreturns[column])),np.zeros(len((strategyreturns[column] - np.mean(strategyreturns[column]))))))))*100*math.sqrt(12) for column in strategyreturns.columns],2)
Perf.loc['Active Return (%)'] = Perf.loc['cagr'] - np.tile(Perf.loc['cagr',[benchmarkname]],len(indexnames))
Perf.loc['Tracking Error (%)']= (excessreturns.std()*np.sqrt(12)*100).values
Perf.loc['Tracking Error (%)',benchmarkname] = np.NaN
Perf.loc['Information Ratio'] = Perf.loc['Active Return (%)'] /Perf.loc['Tracking Error (%)']
RiskSummary.loc['Correlation'] = strategyreturns.corr()[benchmarkname]
RiskSummary.loc['Beta'] = strategyreturns.cov()[benchmarkname] /np.tile(strategyreturns.var()[benchmarkname],len(indexnames))
Perf.loc[['Active Return (%)','Tracking Error (%)','Information Ratio'],:]= np.round(Perf.loc[['Active Return (%)','Tracking Error (%)','Information Ratio'],:].astype('float'),2)
RiskSummary.loc['Monthly Batting Average (%)']= np.round([x*100 for x in list(map(operator.truediv, [len(excessreturns[excessreturns[column]>0]) for column in excessreturns.columns], [len(excessreturns[column])-1 for column in excessreturns.columns]))],2)
RiskSummary.loc['Upside Capture Ratio']= np.round([(scipy.stats.mstats.gmean(gmreturns[column] [gmreturns[benchmarkname]>1])-1)/(scipy.stats.mstats.gmean(gmreturns[benchmarkname] [gmreturns[benchmarkname]>1])-1) for column in gmreturns.columns],4)
RiskSummary.loc['Downside Capture Ratio']= np.round([(scipy.stats.mstats.gmean(gmreturns[column][gmreturns[benchmarkname]<1])-1)/(scipy.stats.mstats.gmean(gmreturns[benchmarkname] [gmreturns[benchmarkname]<1])-1) for column in gmreturns.columns],4)
RiskSummary.loc[['monthly_skew','monthly_kurt','Beta','calmar','Correlation','Max Downstreak Years (Absolute)',
'Max Downstreak Years (Relative)'],:]= np.round(RiskSummary.loc[['monthly_skew','monthly_kurt','Beta',
'Correlation','Max Downstreak Years (Absolute)','Max Downstreak Years (Relative)'],:].astype('float'),2)
RiskSummary.loc[['max_drawdown','monthly_vol'],:]= np.round(100*RiskSummary.loc[['max_drawdown','monthly_vol'],:].astype('float'),2)
RiskSummary = RiskSummary.loc[['start','end','monthly_vol','Downside Risk (%)','max_drawdown','calmar','Max Drawdown Period','Max Downstreak Years (Absolute)','Max Downstreak Period (Absolute)','Max Downstreak Years (Relative)',
'Max Downstreak Period (Relative)','Monthly Batting Average (%)','Upside Capture Ratio','Downside Capture Ratio','monthly_skew',\
'monthly_kurt','Correlation','Beta'],:]
RiskSummary.rename(index={'max_drawdown':'Maximum Drawdown (%)',\
'monthly_vol':'Risk (%)','monthly_skew':'Skewness',\
'monthly_kurt':'Kurtosis','calmar':'Calmar Ratio'},inplace=True)
else:
strategyreturns = ffn.core.to_monthly(indexlevels).to_returns()
RiskSummary.loc['Downside Risk (%)']=np.round([math.sqrt(np.mean(np.square(np.minimum((strategyreturns[column] - np.mean(strategyreturns[column])),np.zeros(len((strategyreturns[column] - np.mean(strategyreturns[column]))))))))*100*math.sqrt(12) for column in strategyreturns.columns],2)
RiskSummary.loc[['monthly_skew','monthly_kurt','calmar'],:]= np.round(RiskSummary.loc[['monthly_skew','monthly_kurt','calmar'],:].astype('float'),2)
RiskSummary.loc[['max_drawdown','monthly_vol'],:]= np.round(100*RiskSummary.loc[['max_drawdown','monthly_vol'],:].astype('float'),2)
RiskSummary = RiskSummary.loc[['start','end','monthly_vol','Downside Risk (%)','max_drawdown',\
'Max Drawdown Period','calmar','Max Downstreak Years (Absolute)',\
'Max Downstreak Period (Absolute)','monthly_skew','monthly_kurt'],:]
RiskSummary.rename(index={'max_drawdown':'Maximum Drawdown (%)',\
'monthly_vol':'Risk (%)','monthly_skew':'Skewness',\
'monthly_kurt':'Kurtosis','calmar':'Calmar Ratio'},inplace=True)
AdditionalPerf = Perf.loc[{'start','end'}]
horizons = ['three_month','six_month','ytd','one_year','three_year','five_year','ten_year']
commonhorizon = set(horizons) & set(stats.stats.index)
commonhorizon = [ch for ch in horizons if ch in commonhorizon]
horizonreturns = stats.stats.loc[commonhorizon,
indexlevels.columns]*100
AdditionalPerf=AdditionalPerf.append(np.round(horizonreturns.astype('float'),2))
calendaryearreturns = np.round(indexlevels.to_monthly().pct_change(periods=12)*100,2)
calendaryearreturns = calendaryearreturns[calendaryearreturns.index.month==12].dropna()
calendaryearreturns.index = calendaryearreturns.index.year
AdditionalPerf = AdditionalPerf.append(calendaryearreturns)
Perf.loc['Downside Risk (%)']=RiskSummary.loc['Downside Risk (%)']
Perf.loc['Sortino-Ratio']= (Perf.loc['cagr'] - Perf.loc['RFR']) / Perf.loc['Downside Risk (%)']
Perf.loc['Return/Max Drawdown']=Perf.loc['cagr']/np.abs(Perf.loc['max_drawdown'])
Perf.loc[['Return/Risk','Sharpe-Ratio','Sortino-Ratio','monthly_skew','monthly_kurt','calmar','Return/Max Drawdown'],:]= np.round(Perf.loc[['Return/Risk','Sharpe-Ratio',\
'Sortino-Ratio','monthly_skew','monthly_kurt','calmar','Return/Max Drawdown'],:].astype('float'),2)
Perf.loc[['Sortino-Ratio'],:]= np.round(Perf.loc[['Sortino-Ratio'],:].astype('float'),2)
Perf = Perf.loc[['start','end','cagr','monthly_mean','monthly_vol','Downside Risk (%)','Return/Risk', 'monthly_skew',\
'monthly_kurt','Sharpe-Ratio','Sortino-Ratio',\
'max_drawdown','calmar','Return/Max Drawdown'],:]
Perf.rename(index={'max_drawdown':'Maximum Drawdown (%)',\
'monthly_vol':'Risk (%)','cagr':'Annualized Compunded Return/CAGR(%)',\
'monthly_mean':'Annualized Arthimetic mean(%)','calmar':'Calmar Ratio',\
'monthly_skew':'Skewness',\
'monthly_kurt':'Kurtosis'},inplace=True)
# RiskSummary.index = [indexsummarylabels.get(indexname,indexname) for indexname in RiskSummary.index]
simulname= self.region+'-Simulation-'+datetime.now().strftime('%Y%m%d-%H%M')+simulationname
# os.mkdir(self.outputpath+'//results//'+simulname)
# newpath=self.outputpath+'//results//'+simulname+'//'
writer= pd.ExcelWriter(self.outputpath+simulname+'.xlsx')
Perf.to_excel(writer,'PerformanceSummary')
# Perf.to_csv(newpath+'PerformanceSummary.csv')
RiskSummary.to_excel(writer,'RiskSummary')
# RiskSummary.to_csv(newpath+'RiskSummary.csv')
AdditionalPerf.to_excel(writer,'Horizon Returns')
# AdditionalPerf.to_csv(newpath+'Horizon Returns.csv')
strategyreturns.to_excel(writer,'Strategy Returns')
# strategyreturns.to_csv(self.outputpath+'strategyreturns.csv')
strategyreturns.corr().to_excel(writer,'Correlation')
dfroll,rolling= self.rollingreturns(indexlevels)
dfroll.to_excel(writer,'Average Rolling Stats')
for i in rolling.keys():
for j in rolling[i].keys():
rolling[i][j].to_excel(writer, 'rolling '+str(i)+'M '+str(j))
writer.close()
# strategyreturns.corr().to_csv(self.outputpath+'Correlation.csv')
return Perf
| 0.283583 | 0.274239 |
### DataFrame Indexing and Loading
```
import pandas as pd
purchase_1=pd.Series({'Name':'chris',
'item_purschased':'Dog food',
'cost':22.50})
purchase_2=pd.Series({'Name':'Keyvn',
'item_purschased':'kitty litter',
'cost':2.50})
purchase_3=pd.Series({'Name':'Vinod',
'item_purschased':'bird seed',
'cost':5.0})
df=pd.DataFrame([purchase_1,purchase_2,purchase_3],index=['store1','store1','store2'])
df
costs=df['cost']
costs
```
### applying Broadcasting to df
```
costs+=2
costs
df
# this command will not work in window, it will only work in linux or MacOS
!cat olympics.csv
df0=pd.read_csv('olympics.csv')
df0.head()
```
### Making 1st row as Column labels and 1st row as df label
```
df0=pd.read_csv('olympics.csv', index_col=0, skiprows=1)
df0.head()
```
### settting the pandas name using the pandas' column name property
```
df0.columns
for col in df0.columns:
if col[:2] == '01':
print(col[:2])
df0.rename(columns={col:'Gold'+col[4:]}, inplace=True)
if col[:2] == '02':
df0.rename(columns={col:'Silver'+col[4:]}, inplace=True)
print(col[:2],'Silver')
if col[:2] == '03':
df0.rename(columns={col:'Bronze'+col[4:]}, inplace=True)
print(col[:2],'Bronze')
if col[:2] == '№ ':
df0.rename(columns={col:'Gold'+col[4:]}, inplace=True)
df0.head()
```
# Querying a Data Frame
### generating Mask :
countries who won gold Medal
```
df0['Gold']>0
```
### Masking the original Data Frame :
#### to filter countries with who won gold medal
```
only_gold=df0.where(df0['Gold']>0)
only_gold.head()
only_gold['Gold'].count()
df0['Gold'].count()
```
### Droping the NAN values
```
only_gold=only_gold.dropna()
only_gold.head()
```
### Method 2: shortcut method pandas developer allows taking boolean as index value
using condition in index , this method also removes NaN values automatically
```
# vountries whow won gold in olympics
only_gold2=df0[df0['Gold']>0]
only_gold2.head()
```
## To find out countries who won gold either is summer or in winter
#### the out put of two Boolean mask compared with bitwise operator (& |) is another boolean mask
this we can chain together a bunch of AND & , OR | statements in order to create a more complex query and the result is single boolean mask
```
df0[(df0['Gold']>0) | (df0['Gold.1']>0)]
```
#### now to count the number of Rows each country who has gold in either summer or winter we use len( )
```
len(df0[(df0['Gold']>0) | (df0['Gold.1']>0)])
```
### Any country which had won gold in winter olympics but not in summer olympics
```
df0[(df0['Gold.1']>0) & (df0['Gold']==0)]
```
# Video 07: Indexing a Data Frame
1 ) index is a Row level Label, both Series and DataFrame contain Index
2 ) Row correspond to axis zero
3) Index could Either be inferred . i.e when we create a new Series without an index, in which case we get numeric values for index.
4) Index can be set explicitly like when we use Dictionary object to create the Series or when we load the data from CSV file and specify the header
5) Another option to an index is using a Set Index function , this function takes a list of column and promotes those coulmns as set of index.
6) Set index is distructive process. It does't keep the current index
7) of you want to keep the current index, you need to manully create a new column and copy into it values from the Index attribute
```
df0.head()
```
## 1. lets say we dont want to Index a DataFrame by Countries but by Gold Medal Won at Summer games
#### firstly we have to preseve the country names into a New column
```
df0['Country']=df0.index
df0
```
#### Now we set the Gold Medal Summer column as our Index
```
df0=df0.set_index('Gold')
df0.head()
```
Since we are creating a new index from an existing column it apppears that an new empty row has been added.
these empty value are none or NaN in case of Numeric Datatype.
and jupyter notebook has provided a way of accommodating the Name of index column
## 2. Create a Default Numbered Index
```
df0=df0.reset_index()
df0.head()
```
## 3. Multi Level Indexing or Hirerrical Indexing
This similar to composite keys in Relational database system
- To create a multi level index we simply call set index and give it a list column in promoting to an index
Pandas will search through in order, finding the distinct data and forming composite indices.
A good example is also found when dealing with geograpical Data, which is sorted by regions or demographics.
### 3a Importing USA population Census Data Breakdown at county level
```
df1=pd.read_csv('census.csv')
df1.head()
df1['SUMLEV'].unique()
```
#### lets keep only the county data and filter all the Rows that are summary at state level
```
df2=df1[df1['SUMLEV']==50]
df2
```
### 3b.Lets Reduce the Data to Total Population and Total number of Birth
```
list_column=df2.columns
list_column
column_to_keep=['STNAME', 'CTYNAME',
'BIRTHS2010', 'BIRTHS2011', 'BIRTHS2012', 'BIRTHS2013', 'BIRTHS2014','BIRTHS2015',
'POPESTIMATE2010', 'POPESTIMATE2011', 'POPESTIMATE2012', 'POPESTIMATE2013','POPESTIMATE2014', 'POPESTIMATE2015']
df3=df2[column_to_keep]
df3.head()
df3=df3.set_index(['STNAME','CTYNAME'])
df3
```
## 3c How can we Query this Data? i.e what is the population of Washtenaw County?
loc attribute can take multiple arguments and it could query both Row and columns
```
df3.loc['Michigan','Washtenaw County']
```
## 4. How can we filter two Counties?
```
df3.loc[[('Michigan','Washtenaw County'),('Michigan','Wayne')]]
```
# Video #08 Handling Missing Values
```
df=pd.read_csv('log.csv')
df.head()
df=df.fillna(99)
df.head()
```
### We can also use F_fill or B_fill (forward fill or Backward fill)
### Before filling any value using f_fill or B_fill ; we will index our Data by timestamp and then sort it by time stamp
```
df=pd.read_csv('log.csv')
df=df.set_index('time')
df.head()
df.sort_index()
```
### applying ffill on sorted values
```
df.ffill()
```
### here index isn't Unique , two user can use same index(two user can use the system at same time, this is comon in parallel processing)
```
df=df.reset_index()
df.head()
# df=df.reset_index()
df=df.set_index(['time','user'])
df.sort_index()
df
```
### fill missing values
```
df.fillna(99)
```
# Statistical function will ignore NaN or missing values
|
github_jupyter
|
import pandas as pd
purchase_1=pd.Series({'Name':'chris',
'item_purschased':'Dog food',
'cost':22.50})
purchase_2=pd.Series({'Name':'Keyvn',
'item_purschased':'kitty litter',
'cost':2.50})
purchase_3=pd.Series({'Name':'Vinod',
'item_purschased':'bird seed',
'cost':5.0})
df=pd.DataFrame([purchase_1,purchase_2,purchase_3],index=['store1','store1','store2'])
df
costs=df['cost']
costs
costs+=2
costs
df
# this command will not work in window, it will only work in linux or MacOS
!cat olympics.csv
df0=pd.read_csv('olympics.csv')
df0.head()
df0=pd.read_csv('olympics.csv', index_col=0, skiprows=1)
df0.head()
df0.columns
for col in df0.columns:
if col[:2] == '01':
print(col[:2])
df0.rename(columns={col:'Gold'+col[4:]}, inplace=True)
if col[:2] == '02':
df0.rename(columns={col:'Silver'+col[4:]}, inplace=True)
print(col[:2],'Silver')
if col[:2] == '03':
df0.rename(columns={col:'Bronze'+col[4:]}, inplace=True)
print(col[:2],'Bronze')
if col[:2] == '№ ':
df0.rename(columns={col:'Gold'+col[4:]}, inplace=True)
df0.head()
df0['Gold']>0
only_gold=df0.where(df0['Gold']>0)
only_gold.head()
only_gold['Gold'].count()
df0['Gold'].count()
only_gold=only_gold.dropna()
only_gold.head()
# vountries whow won gold in olympics
only_gold2=df0[df0['Gold']>0]
only_gold2.head()
df0[(df0['Gold']>0) | (df0['Gold.1']>0)]
len(df0[(df0['Gold']>0) | (df0['Gold.1']>0)])
df0[(df0['Gold.1']>0) & (df0['Gold']==0)]
df0.head()
df0['Country']=df0.index
df0
df0=df0.set_index('Gold')
df0.head()
df0=df0.reset_index()
df0.head()
df1=pd.read_csv('census.csv')
df1.head()
df1['SUMLEV'].unique()
df2=df1[df1['SUMLEV']==50]
df2
list_column=df2.columns
list_column
column_to_keep=['STNAME', 'CTYNAME',
'BIRTHS2010', 'BIRTHS2011', 'BIRTHS2012', 'BIRTHS2013', 'BIRTHS2014','BIRTHS2015',
'POPESTIMATE2010', 'POPESTIMATE2011', 'POPESTIMATE2012', 'POPESTIMATE2013','POPESTIMATE2014', 'POPESTIMATE2015']
df3=df2[column_to_keep]
df3.head()
df3=df3.set_index(['STNAME','CTYNAME'])
df3
df3.loc['Michigan','Washtenaw County']
df3.loc[[('Michigan','Washtenaw County'),('Michigan','Wayne')]]
df=pd.read_csv('log.csv')
df.head()
df=df.fillna(99)
df.head()
df=pd.read_csv('log.csv')
df=df.set_index('time')
df.head()
df.sort_index()
df.ffill()
df=df.reset_index()
df.head()
# df=df.reset_index()
df=df.set_index(['time','user'])
df.sort_index()
df
df.fillna(99)
| 0.094469 | 0.830422 |
# Decision Trees
> Chapter 6
- permalink: /06_decision_trees
_This notebook contains all the sample code and solutions to the exercises in chapter 6._
# Setup
First, let's import a few common modules, ensure MatplotLib plots figures inline and prepare a function to save the figures. We also check that Python 3.5 or later is installed (although Python 2.x may work, it is deprecated so we strongly recommend you use Python 3 instead), as well as Scikit-Learn ≥0.20.
```
#collapse-show
# Python ≥3.5 is required
import sys
assert sys.version_info >= (3, 5)
# Scikit-Learn ≥0.20 is required
import sklearn
assert sklearn.__version__ >= "0.20"
# Common imports
import numpy as np
import os
# to make this notebook's output stable across runs
np.random.seed(42)
# To plot pretty figures
%matplotlib inline
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rc('axes', labelsize=14)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
# Where to save the figures
PROJECT_ROOT_DIR = "."
CHAPTER_ID = "decision_trees"
IMAGES_PATH = os.path.join(PROJECT_ROOT_DIR, "images", CHAPTER_ID)
os.makedirs(IMAGES_PATH, exist_ok=True)
def save_fig(fig_id, tight_layout=True, fig_extension="png", resolution=300):
path = os.path.join(IMAGES_PATH, fig_id + "." + fig_extension)
print("Saving figure", fig_id)
if tight_layout:
plt.tight_layout()
plt.savefig(path, format=fig_extension, dpi=resolution)
```
# Training and visualizing
```
#collapse-show
from sklearn.datasets import load_iris
from sklearn.tree import DecisionTreeClassifier
iris = load_iris()
X = iris.data[:, 2:] # petal length and width
y = iris.target
tree_clf = DecisionTreeClassifier(max_depth=2, random_state=42)
tree_clf.fit(X, y)
from graphviz import Source
from sklearn.tree import export_graphviz
export_graphviz(
tree_clf,
out_file=os.path.join(IMAGES_PATH, "iris_tree.dot"),
feature_names=iris.feature_names[2:],
class_names=iris.target_names,
rounded=True,
filled=True
)
Source.from_file(os.path.join(IMAGES_PATH, "iris_tree.dot"))
#collapse-show
from matplotlib.colors import ListedColormap
def plot_decision_boundary(clf, X, y, axes=[0, 7.5, 0, 3], iris=True, legend=False, plot_training=True):
x1s = np.linspace(axes[0], axes[1], 100)
x2s = np.linspace(axes[2], axes[3], 100)
x1, x2 = np.meshgrid(x1s, x2s)
X_new = np.c_[x1.ravel(), x2.ravel()]
y_pred = clf.predict(X_new).reshape(x1.shape)
custom_cmap = ListedColormap(['#fafab0','#9898ff','#a0faa0'])
plt.contourf(x1, x2, y_pred, alpha=0.3, cmap=custom_cmap)
if not iris:
custom_cmap2 = ListedColormap(['#7d7d58','#4c4c7f','#507d50'])
plt.contour(x1, x2, y_pred, cmap=custom_cmap2, alpha=0.8)
if plot_training:
plt.plot(X[:, 0][y==0], X[:, 1][y==0], "yo", label="Iris setosa")
plt.plot(X[:, 0][y==1], X[:, 1][y==1], "bs", label="Iris versicolor")
plt.plot(X[:, 0][y==2], X[:, 1][y==2], "g^", label="Iris virginica")
plt.axis(axes)
if iris:
plt.xlabel("Petal length", fontsize=14)
plt.ylabel("Petal width", fontsize=14)
else:
plt.xlabel(r"$x_1$", fontsize=18)
plt.ylabel(r"$x_2$", fontsize=18, rotation=0)
if legend:
plt.legend(loc="lower right", fontsize=14)
plt.figure(figsize=(8, 4))
plot_decision_boundary(tree_clf, X, y)
plt.plot([2.45, 2.45], [0, 3], "k-", linewidth=2)
plt.plot([2.45, 7.5], [1.75, 1.75], "k--", linewidth=2)
plt.plot([4.95, 4.95], [0, 1.75], "k:", linewidth=2)
plt.plot([4.85, 4.85], [1.75, 3], "k:", linewidth=2)
plt.text(1.40, 1.0, "Depth=0", fontsize=15)
plt.text(3.2, 1.80, "Depth=1", fontsize=13)
plt.text(4.05, 0.5, "(Depth=2)", fontsize=11)
save_fig("decision_tree_decision_boundaries_plot")
plt.show()
```
# Predicting classes and class probabilities
```
tree_clf.predict_proba([[5, 1.5]])
tree_clf.predict([[5, 1.5]])
```
# Sensitivity to training set details
```
X[(X[:, 1]==X[:, 1][y==1].max()) & (y==1)] # widest Iris versicolor flower
not_widest_versicolor = (X[:, 1]!=1.8) | (y==2)
X_tweaked = X[not_widest_versicolor]
y_tweaked = y[not_widest_versicolor]
tree_clf_tweaked = DecisionTreeClassifier(max_depth=2, random_state=40)
tree_clf_tweaked.fit(X_tweaked, y_tweaked)
#collapse-show
plt.figure(figsize=(8, 4))
plot_decision_boundary(tree_clf_tweaked, X_tweaked, y_tweaked, legend=False)
plt.plot([0, 7.5], [0.8, 0.8], "k-", linewidth=2)
plt.plot([0, 7.5], [1.75, 1.75], "k--", linewidth=2)
plt.text(1.0, 0.9, "Depth=0", fontsize=15)
plt.text(1.0, 1.80, "Depth=1", fontsize=13)
save_fig("decision_tree_instability_plot")
plt.show()
#collapse-show
from sklearn.datasets import make_moons
Xm, ym = make_moons(n_samples=100, noise=0.25, random_state=53)
deep_tree_clf1 = DecisionTreeClassifier(random_state=42)
deep_tree_clf2 = DecisionTreeClassifier(min_samples_leaf=4, random_state=42)
deep_tree_clf1.fit(Xm, ym)
deep_tree_clf2.fit(Xm, ym)
fig, axes = plt.subplots(ncols=2, figsize=(10, 4), sharey=True)
plt.sca(axes[0])
plot_decision_boundary(deep_tree_clf1, Xm, ym, axes=[-1.5, 2.4, -1, 1.5], iris=False)
plt.title("No restrictions", fontsize=16)
plt.sca(axes[1])
plot_decision_boundary(deep_tree_clf2, Xm, ym, axes=[-1.5, 2.4, -1, 1.5], iris=False)
plt.title("min_samples_leaf = {}".format(deep_tree_clf2.min_samples_leaf), fontsize=14)
plt.ylabel("")
save_fig("min_samples_leaf_plot")
plt.show()
#collapse-show
angle = np.pi / 180 * 20
rotation_matrix = np.array([[np.cos(angle), -np.sin(angle)], [np.sin(angle), np.cos(angle)]])
Xr = X.dot(rotation_matrix)
tree_clf_r = DecisionTreeClassifier(random_state=42)
tree_clf_r.fit(Xr, y)
plt.figure(figsize=(8, 3))
plot_decision_boundary(tree_clf_r, Xr, y, axes=[0.5, 7.5, -1.0, 1], iris=False)
plt.show()
#collapse-show
np.random.seed(6)
Xs = np.random.rand(100, 2) - 0.5
ys = (Xs[:, 0] > 0).astype(np.float32) * 2
angle = np.pi / 4
rotation_matrix = np.array([[np.cos(angle), -np.sin(angle)], [np.sin(angle), np.cos(angle)]])
Xsr = Xs.dot(rotation_matrix)
tree_clf_s = DecisionTreeClassifier(random_state=42)
tree_clf_s.fit(Xs, ys)
tree_clf_sr = DecisionTreeClassifier(random_state=42)
tree_clf_sr.fit(Xsr, ys)
fig, axes = plt.subplots(ncols=2, figsize=(10, 4), sharey=True)
plt.sca(axes[0])
plot_decision_boundary(tree_clf_s, Xs, ys, axes=[-0.7, 0.7, -0.7, 0.7], iris=False)
plt.sca(axes[1])
plot_decision_boundary(tree_clf_sr, Xsr, ys, axes=[-0.7, 0.7, -0.7, 0.7], iris=False)
plt.ylabel("")
save_fig("sensitivity_to_rotation_plot")
plt.show()
```
# Regression trees
```
# Quadratic training set + noise
np.random.seed(42)
m = 200
X = np.random.rand(m, 1)
y = 4 * (X - 0.5) ** 2
y = y + np.random.randn(m, 1) / 10
from sklearn.tree import DecisionTreeRegressor
tree_reg = DecisionTreeRegressor(max_depth=2, random_state=42)
tree_reg.fit(X, y)
#collapse-show
from sklearn.tree import DecisionTreeRegressor
tree_reg1 = DecisionTreeRegressor(random_state=42, max_depth=2)
tree_reg2 = DecisionTreeRegressor(random_state=42, max_depth=3)
tree_reg1.fit(X, y)
tree_reg2.fit(X, y)
def plot_regression_predictions(tree_reg, X, y, axes=[0, 1, -0.2, 1], ylabel="$y$"):
x1 = np.linspace(axes[0], axes[1], 500).reshape(-1, 1)
y_pred = tree_reg.predict(x1)
plt.axis(axes)
plt.xlabel("$x_1$", fontsize=18)
if ylabel:
plt.ylabel(ylabel, fontsize=18, rotation=0)
plt.plot(X, y, "b.")
plt.plot(x1, y_pred, "r.-", linewidth=2, label=r"$\hat{y}$")
fig, axes = plt.subplots(ncols=2, figsize=(10, 4), sharey=True)
plt.sca(axes[0])
plot_regression_predictions(tree_reg1, X, y)
for split, style in ((0.1973, "k-"), (0.0917, "k--"), (0.7718, "k--")):
plt.plot([split, split], [-0.2, 1], style, linewidth=2)
plt.text(0.21, 0.65, "Depth=0", fontsize=15)
plt.text(0.01, 0.2, "Depth=1", fontsize=13)
plt.text(0.65, 0.8, "Depth=1", fontsize=13)
plt.legend(loc="upper center", fontsize=18)
plt.title("max_depth=2", fontsize=14)
plt.sca(axes[1])
plot_regression_predictions(tree_reg2, X, y, ylabel=None)
for split, style in ((0.1973, "k-"), (0.0917, "k--"), (0.7718, "k--")):
plt.plot([split, split], [-0.2, 1], style, linewidth=2)
for split in (0.0458, 0.1298, 0.2873, 0.9040):
plt.plot([split, split], [-0.2, 1], "k:", linewidth=1)
plt.text(0.3, 0.5, "Depth=2", fontsize=13)
plt.title("max_depth=3", fontsize=14)
save_fig("tree_regression_plot")
plt.show()
export_graphviz(
tree_reg1,
out_file=os.path.join(IMAGES_PATH, "regression_tree.dot"),
feature_names=["x1"],
rounded=True,
filled=True
)
Source.from_file(os.path.join(IMAGES_PATH, "regression_tree.dot"))
#collapse-show
tree_reg1 = DecisionTreeRegressor(random_state=42)
tree_reg2 = DecisionTreeRegressor(random_state=42, min_samples_leaf=10)
tree_reg1.fit(X, y)
tree_reg2.fit(X, y)
x1 = np.linspace(0, 1, 500).reshape(-1, 1)
y_pred1 = tree_reg1.predict(x1)
y_pred2 = tree_reg2.predict(x1)
fig, axes = plt.subplots(ncols=2, figsize=(10, 4), sharey=True)
plt.sca(axes[0])
plt.plot(X, y, "b.")
plt.plot(x1, y_pred1, "r.-", linewidth=2, label=r"$\hat{y}$")
plt.axis([0, 1, -0.2, 1.1])
plt.xlabel("$x_1$", fontsize=18)
plt.ylabel("$y$", fontsize=18, rotation=0)
plt.legend(loc="upper center", fontsize=18)
plt.title("No restrictions", fontsize=14)
plt.sca(axes[1])
plt.plot(X, y, "b.")
plt.plot(x1, y_pred2, "r.-", linewidth=2, label=r"$\hat{y}$")
plt.axis([0, 1, -0.2, 1.1])
plt.xlabel("$x_1$", fontsize=18)
plt.title("min_samples_leaf={}".format(tree_reg2.min_samples_leaf), fontsize=14)
save_fig("tree_regression_regularization_plot")
plt.show()
```
# Exercise solutions
## 1. to 6.
See appendix A.
## 7.
_Exercise: train and fine-tune a Decision Tree for the moons dataset._
a. Generate a moons dataset using `make_moons(n_samples=10000, noise=0.4)`.
Adding `random_state=42` to make this notebook's output constant:
```
from sklearn.datasets import make_moons
X, y = make_moons(n_samples=10000, noise=0.4, random_state=42)
```
b. Split it into a training set and a test set using `train_test_split()`.
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
```
c. Use grid search with cross-validation (with the help of the `GridSearchCV` class) to find good hyperparameter values for a `DecisionTreeClassifier`. Hint: try various values for `max_leaf_nodes`.
```
from sklearn.model_selection import GridSearchCV
params = {'max_leaf_nodes': list(range(2, 100)), 'min_samples_split': [2, 3, 4]}
grid_search_cv = GridSearchCV(DecisionTreeClassifier(random_state=42), params, verbose=1, cv=3)
grid_search_cv.fit(X_train, y_train)
grid_search_cv.best_estimator_
```
d. Train it on the full training set using these hyperparameters, and measure your model's performance on the test set. You should get roughly 85% to 87% accuracy.
By default, `GridSearchCV` trains the best model found on the whole training set (you can change this by setting `refit=False`), so we don't need to do it again. We can simply evaluate the model's accuracy:
```
from sklearn.metrics import accuracy_score
y_pred = grid_search_cv.predict(X_test)
accuracy_score(y_test, y_pred)
```
## 8.
_Exercise: Grow a forest._
a. Continuing the previous exercise, generate 1,000 subsets of the training set, each containing 100 instances selected randomly. Hint: you can use Scikit-Learn's `ShuffleSplit` class for this.
```
from sklearn.model_selection import ShuffleSplit
n_trees = 1000
n_instances = 100
mini_sets = []
rs = ShuffleSplit(n_splits=n_trees, test_size=len(X_train) - n_instances, random_state=42)
for mini_train_index, mini_test_index in rs.split(X_train):
X_mini_train = X_train[mini_train_index]
y_mini_train = y_train[mini_train_index]
mini_sets.append((X_mini_train, y_mini_train))
```
b. Train one Decision Tree on each subset, using the best hyperparameter values found above. Evaluate these 1,000 Decision Trees on the test set. Since they were trained on smaller sets, these Decision Trees will likely perform worse than the first Decision Tree, achieving only about 80% accuracy.
```
from sklearn.base import clone
forest = [clone(grid_search_cv.best_estimator_) for _ in range(n_trees)]
accuracy_scores = []
for tree, (X_mini_train, y_mini_train) in zip(forest, mini_sets):
tree.fit(X_mini_train, y_mini_train)
y_pred = tree.predict(X_test)
accuracy_scores.append(accuracy_score(y_test, y_pred))
np.mean(accuracy_scores)
```
c. Now comes the magic. For each test set instance, generate the predictions of the 1,000 Decision Trees, and keep only the most frequent prediction (you can use SciPy's `mode()` function for this). This gives you _majority-vote predictions_ over the test set.
```
Y_pred = np.empty([n_trees, len(X_test)], dtype=np.uint8)
for tree_index, tree in enumerate(forest):
Y_pred[tree_index] = tree.predict(X_test)
from scipy.stats import mode
y_pred_majority_votes, n_votes = mode(Y_pred, axis=0)
```
d. Evaluate these predictions on the test set: you should obtain a slightly higher accuracy than your first model (about 0.5 to 1.5% higher). Congratulations, you have trained a Random Forest classifier!
```
accuracy_score(y_test, y_pred_majority_votes.reshape([-1]))
```
|
github_jupyter
|
#collapse-show
# Python ≥3.5 is required
import sys
assert sys.version_info >= (3, 5)
# Scikit-Learn ≥0.20 is required
import sklearn
assert sklearn.__version__ >= "0.20"
# Common imports
import numpy as np
import os
# to make this notebook's output stable across runs
np.random.seed(42)
# To plot pretty figures
%matplotlib inline
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rc('axes', labelsize=14)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
# Where to save the figures
PROJECT_ROOT_DIR = "."
CHAPTER_ID = "decision_trees"
IMAGES_PATH = os.path.join(PROJECT_ROOT_DIR, "images", CHAPTER_ID)
os.makedirs(IMAGES_PATH, exist_ok=True)
def save_fig(fig_id, tight_layout=True, fig_extension="png", resolution=300):
path = os.path.join(IMAGES_PATH, fig_id + "." + fig_extension)
print("Saving figure", fig_id)
if tight_layout:
plt.tight_layout()
plt.savefig(path, format=fig_extension, dpi=resolution)
#collapse-show
from sklearn.datasets import load_iris
from sklearn.tree import DecisionTreeClassifier
iris = load_iris()
X = iris.data[:, 2:] # petal length and width
y = iris.target
tree_clf = DecisionTreeClassifier(max_depth=2, random_state=42)
tree_clf.fit(X, y)
from graphviz import Source
from sklearn.tree import export_graphviz
export_graphviz(
tree_clf,
out_file=os.path.join(IMAGES_PATH, "iris_tree.dot"),
feature_names=iris.feature_names[2:],
class_names=iris.target_names,
rounded=True,
filled=True
)
Source.from_file(os.path.join(IMAGES_PATH, "iris_tree.dot"))
#collapse-show
from matplotlib.colors import ListedColormap
def plot_decision_boundary(clf, X, y, axes=[0, 7.5, 0, 3], iris=True, legend=False, plot_training=True):
x1s = np.linspace(axes[0], axes[1], 100)
x2s = np.linspace(axes[2], axes[3], 100)
x1, x2 = np.meshgrid(x1s, x2s)
X_new = np.c_[x1.ravel(), x2.ravel()]
y_pred = clf.predict(X_new).reshape(x1.shape)
custom_cmap = ListedColormap(['#fafab0','#9898ff','#a0faa0'])
plt.contourf(x1, x2, y_pred, alpha=0.3, cmap=custom_cmap)
if not iris:
custom_cmap2 = ListedColormap(['#7d7d58','#4c4c7f','#507d50'])
plt.contour(x1, x2, y_pred, cmap=custom_cmap2, alpha=0.8)
if plot_training:
plt.plot(X[:, 0][y==0], X[:, 1][y==0], "yo", label="Iris setosa")
plt.plot(X[:, 0][y==1], X[:, 1][y==1], "bs", label="Iris versicolor")
plt.plot(X[:, 0][y==2], X[:, 1][y==2], "g^", label="Iris virginica")
plt.axis(axes)
if iris:
plt.xlabel("Petal length", fontsize=14)
plt.ylabel("Petal width", fontsize=14)
else:
plt.xlabel(r"$x_1$", fontsize=18)
plt.ylabel(r"$x_2$", fontsize=18, rotation=0)
if legend:
plt.legend(loc="lower right", fontsize=14)
plt.figure(figsize=(8, 4))
plot_decision_boundary(tree_clf, X, y)
plt.plot([2.45, 2.45], [0, 3], "k-", linewidth=2)
plt.plot([2.45, 7.5], [1.75, 1.75], "k--", linewidth=2)
plt.plot([4.95, 4.95], [0, 1.75], "k:", linewidth=2)
plt.plot([4.85, 4.85], [1.75, 3], "k:", linewidth=2)
plt.text(1.40, 1.0, "Depth=0", fontsize=15)
plt.text(3.2, 1.80, "Depth=1", fontsize=13)
plt.text(4.05, 0.5, "(Depth=2)", fontsize=11)
save_fig("decision_tree_decision_boundaries_plot")
plt.show()
tree_clf.predict_proba([[5, 1.5]])
tree_clf.predict([[5, 1.5]])
X[(X[:, 1]==X[:, 1][y==1].max()) & (y==1)] # widest Iris versicolor flower
not_widest_versicolor = (X[:, 1]!=1.8) | (y==2)
X_tweaked = X[not_widest_versicolor]
y_tweaked = y[not_widest_versicolor]
tree_clf_tweaked = DecisionTreeClassifier(max_depth=2, random_state=40)
tree_clf_tweaked.fit(X_tweaked, y_tweaked)
#collapse-show
plt.figure(figsize=(8, 4))
plot_decision_boundary(tree_clf_tweaked, X_tweaked, y_tweaked, legend=False)
plt.plot([0, 7.5], [0.8, 0.8], "k-", linewidth=2)
plt.plot([0, 7.5], [1.75, 1.75], "k--", linewidth=2)
plt.text(1.0, 0.9, "Depth=0", fontsize=15)
plt.text(1.0, 1.80, "Depth=1", fontsize=13)
save_fig("decision_tree_instability_plot")
plt.show()
#collapse-show
from sklearn.datasets import make_moons
Xm, ym = make_moons(n_samples=100, noise=0.25, random_state=53)
deep_tree_clf1 = DecisionTreeClassifier(random_state=42)
deep_tree_clf2 = DecisionTreeClassifier(min_samples_leaf=4, random_state=42)
deep_tree_clf1.fit(Xm, ym)
deep_tree_clf2.fit(Xm, ym)
fig, axes = plt.subplots(ncols=2, figsize=(10, 4), sharey=True)
plt.sca(axes[0])
plot_decision_boundary(deep_tree_clf1, Xm, ym, axes=[-1.5, 2.4, -1, 1.5], iris=False)
plt.title("No restrictions", fontsize=16)
plt.sca(axes[1])
plot_decision_boundary(deep_tree_clf2, Xm, ym, axes=[-1.5, 2.4, -1, 1.5], iris=False)
plt.title("min_samples_leaf = {}".format(deep_tree_clf2.min_samples_leaf), fontsize=14)
plt.ylabel("")
save_fig("min_samples_leaf_plot")
plt.show()
#collapse-show
angle = np.pi / 180 * 20
rotation_matrix = np.array([[np.cos(angle), -np.sin(angle)], [np.sin(angle), np.cos(angle)]])
Xr = X.dot(rotation_matrix)
tree_clf_r = DecisionTreeClassifier(random_state=42)
tree_clf_r.fit(Xr, y)
plt.figure(figsize=(8, 3))
plot_decision_boundary(tree_clf_r, Xr, y, axes=[0.5, 7.5, -1.0, 1], iris=False)
plt.show()
#collapse-show
np.random.seed(6)
Xs = np.random.rand(100, 2) - 0.5
ys = (Xs[:, 0] > 0).astype(np.float32) * 2
angle = np.pi / 4
rotation_matrix = np.array([[np.cos(angle), -np.sin(angle)], [np.sin(angle), np.cos(angle)]])
Xsr = Xs.dot(rotation_matrix)
tree_clf_s = DecisionTreeClassifier(random_state=42)
tree_clf_s.fit(Xs, ys)
tree_clf_sr = DecisionTreeClassifier(random_state=42)
tree_clf_sr.fit(Xsr, ys)
fig, axes = plt.subplots(ncols=2, figsize=(10, 4), sharey=True)
plt.sca(axes[0])
plot_decision_boundary(tree_clf_s, Xs, ys, axes=[-0.7, 0.7, -0.7, 0.7], iris=False)
plt.sca(axes[1])
plot_decision_boundary(tree_clf_sr, Xsr, ys, axes=[-0.7, 0.7, -0.7, 0.7], iris=False)
plt.ylabel("")
save_fig("sensitivity_to_rotation_plot")
plt.show()
# Quadratic training set + noise
np.random.seed(42)
m = 200
X = np.random.rand(m, 1)
y = 4 * (X - 0.5) ** 2
y = y + np.random.randn(m, 1) / 10
from sklearn.tree import DecisionTreeRegressor
tree_reg = DecisionTreeRegressor(max_depth=2, random_state=42)
tree_reg.fit(X, y)
#collapse-show
from sklearn.tree import DecisionTreeRegressor
tree_reg1 = DecisionTreeRegressor(random_state=42, max_depth=2)
tree_reg2 = DecisionTreeRegressor(random_state=42, max_depth=3)
tree_reg1.fit(X, y)
tree_reg2.fit(X, y)
def plot_regression_predictions(tree_reg, X, y, axes=[0, 1, -0.2, 1], ylabel="$y$"):
x1 = np.linspace(axes[0], axes[1], 500).reshape(-1, 1)
y_pred = tree_reg.predict(x1)
plt.axis(axes)
plt.xlabel("$x_1$", fontsize=18)
if ylabel:
plt.ylabel(ylabel, fontsize=18, rotation=0)
plt.plot(X, y, "b.")
plt.plot(x1, y_pred, "r.-", linewidth=2, label=r"$\hat{y}$")
fig, axes = plt.subplots(ncols=2, figsize=(10, 4), sharey=True)
plt.sca(axes[0])
plot_regression_predictions(tree_reg1, X, y)
for split, style in ((0.1973, "k-"), (0.0917, "k--"), (0.7718, "k--")):
plt.plot([split, split], [-0.2, 1], style, linewidth=2)
plt.text(0.21, 0.65, "Depth=0", fontsize=15)
plt.text(0.01, 0.2, "Depth=1", fontsize=13)
plt.text(0.65, 0.8, "Depth=1", fontsize=13)
plt.legend(loc="upper center", fontsize=18)
plt.title("max_depth=2", fontsize=14)
plt.sca(axes[1])
plot_regression_predictions(tree_reg2, X, y, ylabel=None)
for split, style in ((0.1973, "k-"), (0.0917, "k--"), (0.7718, "k--")):
plt.plot([split, split], [-0.2, 1], style, linewidth=2)
for split in (0.0458, 0.1298, 0.2873, 0.9040):
plt.plot([split, split], [-0.2, 1], "k:", linewidth=1)
plt.text(0.3, 0.5, "Depth=2", fontsize=13)
plt.title("max_depth=3", fontsize=14)
save_fig("tree_regression_plot")
plt.show()
export_graphviz(
tree_reg1,
out_file=os.path.join(IMAGES_PATH, "regression_tree.dot"),
feature_names=["x1"],
rounded=True,
filled=True
)
Source.from_file(os.path.join(IMAGES_PATH, "regression_tree.dot"))
#collapse-show
tree_reg1 = DecisionTreeRegressor(random_state=42)
tree_reg2 = DecisionTreeRegressor(random_state=42, min_samples_leaf=10)
tree_reg1.fit(X, y)
tree_reg2.fit(X, y)
x1 = np.linspace(0, 1, 500).reshape(-1, 1)
y_pred1 = tree_reg1.predict(x1)
y_pred2 = tree_reg2.predict(x1)
fig, axes = plt.subplots(ncols=2, figsize=(10, 4), sharey=True)
plt.sca(axes[0])
plt.plot(X, y, "b.")
plt.plot(x1, y_pred1, "r.-", linewidth=2, label=r"$\hat{y}$")
plt.axis([0, 1, -0.2, 1.1])
plt.xlabel("$x_1$", fontsize=18)
plt.ylabel("$y$", fontsize=18, rotation=0)
plt.legend(loc="upper center", fontsize=18)
plt.title("No restrictions", fontsize=14)
plt.sca(axes[1])
plt.plot(X, y, "b.")
plt.plot(x1, y_pred2, "r.-", linewidth=2, label=r"$\hat{y}$")
plt.axis([0, 1, -0.2, 1.1])
plt.xlabel("$x_1$", fontsize=18)
plt.title("min_samples_leaf={}".format(tree_reg2.min_samples_leaf), fontsize=14)
save_fig("tree_regression_regularization_plot")
plt.show()
from sklearn.datasets import make_moons
X, y = make_moons(n_samples=10000, noise=0.4, random_state=42)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
from sklearn.model_selection import GridSearchCV
params = {'max_leaf_nodes': list(range(2, 100)), 'min_samples_split': [2, 3, 4]}
grid_search_cv = GridSearchCV(DecisionTreeClassifier(random_state=42), params, verbose=1, cv=3)
grid_search_cv.fit(X_train, y_train)
grid_search_cv.best_estimator_
from sklearn.metrics import accuracy_score
y_pred = grid_search_cv.predict(X_test)
accuracy_score(y_test, y_pred)
from sklearn.model_selection import ShuffleSplit
n_trees = 1000
n_instances = 100
mini_sets = []
rs = ShuffleSplit(n_splits=n_trees, test_size=len(X_train) - n_instances, random_state=42)
for mini_train_index, mini_test_index in rs.split(X_train):
X_mini_train = X_train[mini_train_index]
y_mini_train = y_train[mini_train_index]
mini_sets.append((X_mini_train, y_mini_train))
from sklearn.base import clone
forest = [clone(grid_search_cv.best_estimator_) for _ in range(n_trees)]
accuracy_scores = []
for tree, (X_mini_train, y_mini_train) in zip(forest, mini_sets):
tree.fit(X_mini_train, y_mini_train)
y_pred = tree.predict(X_test)
accuracy_scores.append(accuracy_score(y_test, y_pred))
np.mean(accuracy_scores)
Y_pred = np.empty([n_trees, len(X_test)], dtype=np.uint8)
for tree_index, tree in enumerate(forest):
Y_pred[tree_index] = tree.predict(X_test)
from scipy.stats import mode
y_pred_majority_votes, n_votes = mode(Y_pred, axis=0)
accuracy_score(y_test, y_pred_majority_votes.reshape([-1]))
| 0.458349 | 0.937954 |
```
%load_ext autoreload
%autoreload 2
import os
import sys
# This is done so that the notebook can see files
# placed in a different folder
module_path = os.path.abspath(os.path.join('..'))
if module_path not in sys.path:
sys.path.append(module_path)
import matplotlib.image as mpimg
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import osmnx as ox
import functions as F
```
## Data collection
### Voting data
Data for the 2020 local elections, for each locality.
```
import requests
import time
import json
coduri = ['ab', 'ar', 'ag', 'bc', 'bh', 'bn', 'bt', 'br', 'bv', 'bz', 'cl', 'cs', 'cj', 'ct', 'cv', 'db',
'dj', 'gl', 'gr', 'gj', 'hr', 'hd', 'il', 'is', 'if', 'mm', 'mh', 'ms', 'nt', 'ot',
'ph', 'sj', 'sm', 'sb', 'sv', 'tr', 'tm', 'tl', 'vl', 'vs', 'vn', 'b']
res_list = []
for cod in coduri:
url = f'https://prezenta.roaep.ro/locale27092020/data/json/sicpv/pv/pv_{cod}_final.json?_={time.time_ns() // 1_000_000}'
r = requests.get(url)
print(f'fetched {cod}')
json_data = json.loads(r.text)
results = json_data['stages']['FINAL']['scopes']['UAT']['categories']['P']['table']
for k in results:
try:
top_candidate = F.get_top_candidate(results[k])
res_list.append(top_candidate)
except ValueError:
continue
vote_data = pd.DataFrame(res_list)
# vote_data = vote_data.append(pd.DataFrame.from_dict({'partid':['MIX'], 'localitate':['MUNICIPIUL BUCURESTI'], 'siruta':[179132], 'judet':['bucuresti']}))
# vote_data['localitate'] = vote_data['localitate'].apply(lambda x: F.normalize(x, strip_city=False))
# vote_data['judet'] = vote_data['judet'].apply(lambda x: F.normalize(x, True))
# vote_data.to_csv("vote_data_2020_primar.csv", sep=",", header=True, index=False)
vote_data = pd.read_csv("vote_data_2020_primar.csv")
```
### Vaccination data
Updated with 5th of August data
```
from tika import parser
import re
raw = parser.from_file('AV-pe-uat-5.10.2021.pdf')
p = re.compile('(.*)(\s[A-Z]{1}\s)(.*)(\s\d{1,2}\.\d{1,2})')
records = []
counter = 0
for line in raw['content'].split('\n'):
if len(line) > 10:
entry_dict = {}
match = p.match(line)
if match is None:
continue
localitate = match.group(3).strip()
# print(localitate)
if not counter % 100:
print(counter)
counter += 1
tip_uat = match.group(2).strip()
judet = match.group(1).strip()
if tip_uat == 'M':
# Strip 'MUNICIPIUL' from big city names.
localitate = localitate[11:]
if tip_uat == 'O':
# Strip 'MUNICIPIUL' from big city names.
localitate = localitate[5:]
if judet == "MUNICIPIUL BUCUREŞTI":
localitate = "BUCURESTI"
# try:
# loc_gdf = ox.graph_from_place(f'{localitate}, {judet}, Romania', which_result=0)
# except IndexError:
# print("No gdf for " + localitate)
# continue
# lat, long = F.get_middle_coords(loc_gdf)
entry_dict['localitate'] = localitate
entry_dict['judet'] = judet
entry_dict['tip_uat'] = tip_uat
entry_dict['procent_vacc'] = match.group(4).strip()
# entry_dict['lat'] = lat
# entry_dict['long'] = long
records.append(entry_dict)
vacc_data = pd.DataFrame.from_records(records)
vacc_data['judet'] = vacc_data['judet'].apply(lambda x: F.normalize(x, True))
vacc_data['localitate'] = vacc_data['localitate'].apply(lambda x: F.normalize(x, False))
# vacc_data['judet'] = vacc_data['judet'].apply(lambda x: F.normalize(x, True))
# vacc_data = vacc_data[:3181]
# vacc_data = vacc_data.astype({'procent_vacc': 'float64'})
# vacc_data.to_csv("vacc_data_5_aug_2021_cu_judet.csv", sep=",", header=True, index=False)
vacc_data = pd.read_csv('vacc_data_5_oct_2021_cu_judet.csv')
```
### Vaccination locations
```
locations = pd.read_csv('centre_covid.csv')
vacc_data['min_dist'] = 100000
vacc_data['nearest_count'] = 0
vacc_data['nearest_std'] = 0
for i in range(vacc_data.shape[0]):
min_dist, nc, nsd = F.nearest_centers(vacc_data.iloc[i]['lat'],
vacc_data.iloc[i]['long'], locations)
vacc_data.at[i, 'min_dist'] = min_dist
vacc_data.at[i, 'nearest_count'] = nc
vacc_data.at[i, 'nearest_std'] = nsd
```
#### Use Google cloud API to get location elevations
```
step = 100
start_idxs = [i for i in range(0, vacc_data.shape[0], step)] + [vacc_data.shape[0]]
all_elevations = []
for i in range(len(start_idxs)-1):
print(f'{i} {start_idxs[i]} {start_idxs[i+1]}')
response = get_elevations(vacc_data, start_idxs[i], start_idxs[i+1], '[API key here]')
all_elevations += elevations_from_response(response)
```
### Age data
```
age_data = pd.read_csv('sR_Tab_31.csv')
age_data = F.add_county(age_data)
age_data = F.filter_rows(age_data)
age_data = age_data.rename(columns={'uat':'localitate'})
age_data['localitate'] = age_data['localitate'].apply(lambda x: F.normalize(x, strip_city=False))
age_data['judet'] = age_data['judet'].apply(lambda x: F.normalize(x, True))
age_data['over65_ratio'] = (age_data['65_69'] + age_data['70_74'] + age_data['75_79'] +
age_data['80_84'] + age_data['peste_85']) / age_data['total']
age_data['over60_ratio'] = (age_data['60_64'] + age_data['65_69'] + age_data['70_74'] + age_data['75_79'] +
age_data['80_84'] + age_data['peste_85']) / age_data['total']
age_data['under40_ratio'] = (age_data['sub_5'] + age_data['5_9'] + age_data['10_14'] +
age_data['15_19'] + age_data['20_24'] + age_data['25_29'] +
age_data['30_34'] + age_data['35_39']) / age_data['total']
```
### Education data
```
ed_data = pd.read_csv('sR_TAB_161.csv')
ed_data = F.add_county(ed_data)
ed_data = F.filter_rows(ed_data)
ed_data = ed_data.rename(columns={'uat':'localitate'})
ed_data['localitate'] = ed_data['localitate'].apply(lambda x: F.normalize(x, strip_city=False))
ed_data['judet'] = ed_data['judet'].apply(lambda x: F.normalize(x, True))
ed_data['tertiary_ratio'] = ed_data['total_high'] / ed_data['total_peste10']
ed_data['illiterate_ratio'] = (ed_data['no_school'] + ed_data['illiterate']) / ed_data['total_peste10']
```
### Religion data
```
rel_data = pd.read_csv('sR_TAB_13.csv')
rel_data['sex'] = 'Ambele sexe'
rel_data = rel_data.fillna(0)
rel_data = F.add_county(rel_data, step=1, start=0)
rel_data = F.filter_rows(rel_data)
rel_data = rel_data.rename(columns={'uat':'localitate'})
rel_data['localitate'] = rel_data['localitate'].apply(lambda x: F.normalize(x, strip_city=False))
rel_data['judet'] = rel_data['judet'].apply(lambda x: F.normalize(x, True))
rel_data['orthodox_ratio'] = (rel_data['ortodoxa'] + rel_data['crestina_rit_vechi'] +
rel_data['ortodoxa_sarba'] + rel_data['armeana']) / rel_data['total']
rel_data['catholic_ratio'] = (rel_data['romano_catolica'] + rel_data['greco_catolica']) / rel_data['total']
rel_data['protestant_ratio'] = (rel_data['reformata'] + rel_data['lutherana'] + rel_data['unitariana'] +
rel_data['evanghelica'] + rel_data['evanghelica_augustana']) / rel_data['total']
rel_data['neoprot_ratio'] = (rel_data['penticostala'] + rel_data['baptista'] +
rel_data['crestina_dupa_evanghelie'] +
rel_data['adventista_z7'] + rel_data['martorii_lui_iehova']) / rel_data['total']
rel_data['other_ratio'] = (rel_data['musulmana'] + rel_data['mozaica'] +
rel_data['alta_religie']) / rel_data['total']
rel_data['irreligious_ratio'] = (rel_data['fara_religie'] + rel_data['atei']) / rel_data['total']
rel_data['na_ratio'] = rel_data['na'] / rel_data['total']
```
### Joining data
```
ed_data = ed_data.set_index(['localitate', 'judet'])
age_data = age_data.set_index(['localitate', 'judet'])
vote_data = vote_data.set_index(['localitate', 'judet'])
vacc_data = vacc_data.set_index(['localitate', 'judet'])
rel_data = rel_data.set_index(['localitate', 'judet'])
age_ed = ed_data[['illiterate_ratio', 'tertiary_ratio']].join(age_data[['over65_ratio', 'under40_ratio', 'total']])
age_ed_rel_gr = age_ed.join(rel_data[['orthodox_ratio','catholic_ratio','protestant_ratio',
'neoprot_ratio','other_ratio','irreligious_ratio','na_ratio']])
age_ed_vote_gr = age_ed_rel_gr.join(vote_data)
age_ed_vote_gr = age_ed_vote_gr.reset_index()
age_ed_vote_gr['localitate'] = age_ed_vote_gr['localitate'].apply(lambda x: F.normalize(x, strip_city=True))
age_ed_vote_gr = age_ed_vote_gr.set_index(['localitate', 'judet'])
age_ed_vote_vacc_gr = age_ed_vote_gr.join(vacc_data)
age_ed_vote_vacc_gr = age_ed_vote_vacc_gr.drop(age_ed_vote_vacc_gr[age_ed_vote_vacc_gr['procent_vacc'].isnull()].index)
age_ed_vote_vacc_gr = age_ed_vote_vacc_gr.reset_index()
age_ed_vote_vacc_gr.to_csv('vacc_vote_age_ed_rel_data.csv', sep=",", header=True, index=False)
```
### Covid rates
```
covid_data = pd.read_csv('covid_rate_14oct21_over3.csv')
covid_data['judet'] = covid_data['judet'].apply(lambda x: F.normalize(x, True))
covid_data['localitate'] = covid_data['localitate'].apply(lambda x: F.normalize(x, False))
covid_data.set_index(['localitate', 'judet'], inplace=True)
age_ed_vote_vacc_gr.set_index(['localitate', 'judet'], inplace=True)
covid_data = covid_data.join(age_ed_vote_vacc_gr, how='inner')
covid_data.reset_index(inplace=True)
covid_data.to_csv('all_with_covid_rate_14oct21.csv', sep=",", header=True, index=False)
covid_data[covid_data['incidenta'] >= 15][['populatie', 'cazuri', 'incidenta', 'procent_vacc']]
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
import os
import sys
# This is done so that the notebook can see files
# placed in a different folder
module_path = os.path.abspath(os.path.join('..'))
if module_path not in sys.path:
sys.path.append(module_path)
import matplotlib.image as mpimg
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import osmnx as ox
import functions as F
import requests
import time
import json
coduri = ['ab', 'ar', 'ag', 'bc', 'bh', 'bn', 'bt', 'br', 'bv', 'bz', 'cl', 'cs', 'cj', 'ct', 'cv', 'db',
'dj', 'gl', 'gr', 'gj', 'hr', 'hd', 'il', 'is', 'if', 'mm', 'mh', 'ms', 'nt', 'ot',
'ph', 'sj', 'sm', 'sb', 'sv', 'tr', 'tm', 'tl', 'vl', 'vs', 'vn', 'b']
res_list = []
for cod in coduri:
url = f'https://prezenta.roaep.ro/locale27092020/data/json/sicpv/pv/pv_{cod}_final.json?_={time.time_ns() // 1_000_000}'
r = requests.get(url)
print(f'fetched {cod}')
json_data = json.loads(r.text)
results = json_data['stages']['FINAL']['scopes']['UAT']['categories']['P']['table']
for k in results:
try:
top_candidate = F.get_top_candidate(results[k])
res_list.append(top_candidate)
except ValueError:
continue
vote_data = pd.DataFrame(res_list)
# vote_data = vote_data.append(pd.DataFrame.from_dict({'partid':['MIX'], 'localitate':['MUNICIPIUL BUCURESTI'], 'siruta':[179132], 'judet':['bucuresti']}))
# vote_data['localitate'] = vote_data['localitate'].apply(lambda x: F.normalize(x, strip_city=False))
# vote_data['judet'] = vote_data['judet'].apply(lambda x: F.normalize(x, True))
# vote_data.to_csv("vote_data_2020_primar.csv", sep=",", header=True, index=False)
vote_data = pd.read_csv("vote_data_2020_primar.csv")
from tika import parser
import re
raw = parser.from_file('AV-pe-uat-5.10.2021.pdf')
p = re.compile('(.*)(\s[A-Z]{1}\s)(.*)(\s\d{1,2}\.\d{1,2})')
records = []
counter = 0
for line in raw['content'].split('\n'):
if len(line) > 10:
entry_dict = {}
match = p.match(line)
if match is None:
continue
localitate = match.group(3).strip()
# print(localitate)
if not counter % 100:
print(counter)
counter += 1
tip_uat = match.group(2).strip()
judet = match.group(1).strip()
if tip_uat == 'M':
# Strip 'MUNICIPIUL' from big city names.
localitate = localitate[11:]
if tip_uat == 'O':
# Strip 'MUNICIPIUL' from big city names.
localitate = localitate[5:]
if judet == "MUNICIPIUL BUCUREŞTI":
localitate = "BUCURESTI"
# try:
# loc_gdf = ox.graph_from_place(f'{localitate}, {judet}, Romania', which_result=0)
# except IndexError:
# print("No gdf for " + localitate)
# continue
# lat, long = F.get_middle_coords(loc_gdf)
entry_dict['localitate'] = localitate
entry_dict['judet'] = judet
entry_dict['tip_uat'] = tip_uat
entry_dict['procent_vacc'] = match.group(4).strip()
# entry_dict['lat'] = lat
# entry_dict['long'] = long
records.append(entry_dict)
vacc_data = pd.DataFrame.from_records(records)
vacc_data['judet'] = vacc_data['judet'].apply(lambda x: F.normalize(x, True))
vacc_data['localitate'] = vacc_data['localitate'].apply(lambda x: F.normalize(x, False))
# vacc_data['judet'] = vacc_data['judet'].apply(lambda x: F.normalize(x, True))
# vacc_data = vacc_data[:3181]
# vacc_data = vacc_data.astype({'procent_vacc': 'float64'})
# vacc_data.to_csv("vacc_data_5_aug_2021_cu_judet.csv", sep=",", header=True, index=False)
vacc_data = pd.read_csv('vacc_data_5_oct_2021_cu_judet.csv')
locations = pd.read_csv('centre_covid.csv')
vacc_data['min_dist'] = 100000
vacc_data['nearest_count'] = 0
vacc_data['nearest_std'] = 0
for i in range(vacc_data.shape[0]):
min_dist, nc, nsd = F.nearest_centers(vacc_data.iloc[i]['lat'],
vacc_data.iloc[i]['long'], locations)
vacc_data.at[i, 'min_dist'] = min_dist
vacc_data.at[i, 'nearest_count'] = nc
vacc_data.at[i, 'nearest_std'] = nsd
step = 100
start_idxs = [i for i in range(0, vacc_data.shape[0], step)] + [vacc_data.shape[0]]
all_elevations = []
for i in range(len(start_idxs)-1):
print(f'{i} {start_idxs[i]} {start_idxs[i+1]}')
response = get_elevations(vacc_data, start_idxs[i], start_idxs[i+1], '[API key here]')
all_elevations += elevations_from_response(response)
age_data = pd.read_csv('sR_Tab_31.csv')
age_data = F.add_county(age_data)
age_data = F.filter_rows(age_data)
age_data = age_data.rename(columns={'uat':'localitate'})
age_data['localitate'] = age_data['localitate'].apply(lambda x: F.normalize(x, strip_city=False))
age_data['judet'] = age_data['judet'].apply(lambda x: F.normalize(x, True))
age_data['over65_ratio'] = (age_data['65_69'] + age_data['70_74'] + age_data['75_79'] +
age_data['80_84'] + age_data['peste_85']) / age_data['total']
age_data['over60_ratio'] = (age_data['60_64'] + age_data['65_69'] + age_data['70_74'] + age_data['75_79'] +
age_data['80_84'] + age_data['peste_85']) / age_data['total']
age_data['under40_ratio'] = (age_data['sub_5'] + age_data['5_9'] + age_data['10_14'] +
age_data['15_19'] + age_data['20_24'] + age_data['25_29'] +
age_data['30_34'] + age_data['35_39']) / age_data['total']
ed_data = pd.read_csv('sR_TAB_161.csv')
ed_data = F.add_county(ed_data)
ed_data = F.filter_rows(ed_data)
ed_data = ed_data.rename(columns={'uat':'localitate'})
ed_data['localitate'] = ed_data['localitate'].apply(lambda x: F.normalize(x, strip_city=False))
ed_data['judet'] = ed_data['judet'].apply(lambda x: F.normalize(x, True))
ed_data['tertiary_ratio'] = ed_data['total_high'] / ed_data['total_peste10']
ed_data['illiterate_ratio'] = (ed_data['no_school'] + ed_data['illiterate']) / ed_data['total_peste10']
rel_data = pd.read_csv('sR_TAB_13.csv')
rel_data['sex'] = 'Ambele sexe'
rel_data = rel_data.fillna(0)
rel_data = F.add_county(rel_data, step=1, start=0)
rel_data = F.filter_rows(rel_data)
rel_data = rel_data.rename(columns={'uat':'localitate'})
rel_data['localitate'] = rel_data['localitate'].apply(lambda x: F.normalize(x, strip_city=False))
rel_data['judet'] = rel_data['judet'].apply(lambda x: F.normalize(x, True))
rel_data['orthodox_ratio'] = (rel_data['ortodoxa'] + rel_data['crestina_rit_vechi'] +
rel_data['ortodoxa_sarba'] + rel_data['armeana']) / rel_data['total']
rel_data['catholic_ratio'] = (rel_data['romano_catolica'] + rel_data['greco_catolica']) / rel_data['total']
rel_data['protestant_ratio'] = (rel_data['reformata'] + rel_data['lutherana'] + rel_data['unitariana'] +
rel_data['evanghelica'] + rel_data['evanghelica_augustana']) / rel_data['total']
rel_data['neoprot_ratio'] = (rel_data['penticostala'] + rel_data['baptista'] +
rel_data['crestina_dupa_evanghelie'] +
rel_data['adventista_z7'] + rel_data['martorii_lui_iehova']) / rel_data['total']
rel_data['other_ratio'] = (rel_data['musulmana'] + rel_data['mozaica'] +
rel_data['alta_religie']) / rel_data['total']
rel_data['irreligious_ratio'] = (rel_data['fara_religie'] + rel_data['atei']) / rel_data['total']
rel_data['na_ratio'] = rel_data['na'] / rel_data['total']
ed_data = ed_data.set_index(['localitate', 'judet'])
age_data = age_data.set_index(['localitate', 'judet'])
vote_data = vote_data.set_index(['localitate', 'judet'])
vacc_data = vacc_data.set_index(['localitate', 'judet'])
rel_data = rel_data.set_index(['localitate', 'judet'])
age_ed = ed_data[['illiterate_ratio', 'tertiary_ratio']].join(age_data[['over65_ratio', 'under40_ratio', 'total']])
age_ed_rel_gr = age_ed.join(rel_data[['orthodox_ratio','catholic_ratio','protestant_ratio',
'neoprot_ratio','other_ratio','irreligious_ratio','na_ratio']])
age_ed_vote_gr = age_ed_rel_gr.join(vote_data)
age_ed_vote_gr = age_ed_vote_gr.reset_index()
age_ed_vote_gr['localitate'] = age_ed_vote_gr['localitate'].apply(lambda x: F.normalize(x, strip_city=True))
age_ed_vote_gr = age_ed_vote_gr.set_index(['localitate', 'judet'])
age_ed_vote_vacc_gr = age_ed_vote_gr.join(vacc_data)
age_ed_vote_vacc_gr = age_ed_vote_vacc_gr.drop(age_ed_vote_vacc_gr[age_ed_vote_vacc_gr['procent_vacc'].isnull()].index)
age_ed_vote_vacc_gr = age_ed_vote_vacc_gr.reset_index()
age_ed_vote_vacc_gr.to_csv('vacc_vote_age_ed_rel_data.csv', sep=",", header=True, index=False)
covid_data = pd.read_csv('covid_rate_14oct21_over3.csv')
covid_data['judet'] = covid_data['judet'].apply(lambda x: F.normalize(x, True))
covid_data['localitate'] = covid_data['localitate'].apply(lambda x: F.normalize(x, False))
covid_data.set_index(['localitate', 'judet'], inplace=True)
age_ed_vote_vacc_gr.set_index(['localitate', 'judet'], inplace=True)
covid_data = covid_data.join(age_ed_vote_vacc_gr, how='inner')
covid_data.reset_index(inplace=True)
covid_data.to_csv('all_with_covid_rate_14oct21.csv', sep=",", header=True, index=False)
covid_data[covid_data['incidenta'] >= 15][['populatie', 'cazuri', 'incidenta', 'procent_vacc']]
| 0.056724 | 0.562837 |
```
### load the rpy2 extension
%load_ext rpy2.ipython
```
## WordCloud from a text file
```
%%R -w 400 -h 400 -u px # instead of px, you can also choose 'in', 'cm', or 'mm'
library(tm)
library(wordcloud)
library(SnowballC)
contents <- readLines('d:/temp/wordcloud/contention.txt')
doc.vec <- VectorSource(contents)
doc.corpus <- Corpus(doc.vec)
#xkcd.df <- read.csv(file.path(path, datafiles))
#xkcd.corpus <- Corpus(DataframeSource(data.frame(xkcd.df[, 3])))
cleaned <- tm_map(doc.corpus,stripWhitespace)
cleaned <- tm_map(cleaned, content_transformer(tolower))
cleaned <- tm_map(cleaned,removeWords,stopwords("english"))
#cleaned <- tm_map(cleaned,stemDocument)
cleaned <- tm_map(cleaned,removeNumbers)
cleaned <- tm_map(cleaned,removePunctuation)
cleaned <- tm_map(cleaned,removeWords, "customer")
cleaned <- tm_map(cleaned,removeWords, "cust")
cleaned <- tm_map(cleaned,removeWords, "client")
cleaned <- tm_map(cleaned,removeWords, "replace")
cleaned <- tm_map(cleaned,removeWords, "replaced")
cleaned <- tm_map(cleaned,removeWords, "repl")
cleaned <- tm_map(cleaned,removeWords, "states")
cleaned <- tm_map(cleaned,removeWords, "perform")
cleaned <- tm_map(cleaned,removeWords, "performed")
cleaned <- tm_map(cleaned,removeWords, "checked")
cleaned <- tm_map(cleaned,removeWords, "found")
cleaned <- tm_map(cleaned,removeWords, "advise")
cleaned <- tm_map(cleaned,removeWords, "inspect")
cleaned <- tm_map(cleaned,removeWords, "inspected")
cleaned <- tm_map(cleaned,removeWords, "tech")
cleaned <- tm_map(cleaned,removeWords, "technician")
cleaned <- tm_map(cleaned,removeWords, "new")
cleaned <- tm_map(cleaned,removeWords, "test")
cleaned <- tm_map(cleaned,removeWords, "please")
wordcloud(cleaned, max.words=100, colors=brewer.pal(7,"Dark2"),random.order=FALSE, scale=c(5,0.5))
```
## WordCloud from an Excel file using pandas
```
import pandas as pd
contention = pd.read_excel(r'\\hdcnas02\AQ_MarketQuality\DensoOBD\WarrantySummaries\35830 - Sunroof Switch\12G_Civic_35830.xlsx','Claims')
%R -i contention
%%R -w 400 -h 400 -u px # instead of px, you can also choose 'in', 'cm', or 'mm'
df <- as.data.frame(contention)
library(tm)
library(wordcloud)
library(SnowballC)
doc.corpus <- Corpus(VectorSource(contention$CUSTOMER_CONTENTION_TEXT))
cleaned <- tm_map(doc.corpus,stripWhitespace)
cleaned <- tm_map(cleaned, content_transformer(tolower))
cleaned <- tm_map(cleaned,removeWords,stopwords("english"))
#cleaned <- tm_map(cleaned,stemDocument)
cleaned <- tm_map(cleaned,removeNumbers)
cleaned <- tm_map(cleaned,removePunctuation)
# Remove "worthless" words
cleaned <- tm_map(cleaned,removeWords, "customer")
cleaned <- tm_map(cleaned,removeWords, "cust")
cleaned <- tm_map(cleaned,removeWords, "client")
cleaned <- tm_map(cleaned,removeWords, "replace")
cleaned <- tm_map(cleaned,removeWords, "replaced")
cleaned <- tm_map(cleaned,removeWords, "repl")
cleaned <- tm_map(cleaned,removeWords, "states")
cleaned <- tm_map(cleaned,removeWords, "perform")
cleaned <- tm_map(cleaned,removeWords, "performed")
cleaned <- tm_map(cleaned,removeWords, "checked")
cleaned <- tm_map(cleaned,removeWords, "found")
cleaned <- tm_map(cleaned,removeWords, "advise")
cleaned <- tm_map(cleaned,removeWords, "inspect")
cleaned <- tm_map(cleaned,removeWords, "inspected")
cleaned <- tm_map(cleaned,removeWords, "tech")
cleaned <- tm_map(cleaned,removeWords, "technician")
cleaned <- tm_map(cleaned,removeWords, "new")
cleaned <- tm_map(cleaned,removeWords, "test")
cleaned <- tm_map(cleaned,removeWords, "please")
wordcloud(cleaned, max.words=50, colors=brewer.pal(6,"Dark2"),random.order=FALSE, scale=c(5,0.5))
```
|
github_jupyter
|
### load the rpy2 extension
%load_ext rpy2.ipython
%%R -w 400 -h 400 -u px # instead of px, you can also choose 'in', 'cm', or 'mm'
library(tm)
library(wordcloud)
library(SnowballC)
contents <- readLines('d:/temp/wordcloud/contention.txt')
doc.vec <- VectorSource(contents)
doc.corpus <- Corpus(doc.vec)
#xkcd.df <- read.csv(file.path(path, datafiles))
#xkcd.corpus <- Corpus(DataframeSource(data.frame(xkcd.df[, 3])))
cleaned <- tm_map(doc.corpus,stripWhitespace)
cleaned <- tm_map(cleaned, content_transformer(tolower))
cleaned <- tm_map(cleaned,removeWords,stopwords("english"))
#cleaned <- tm_map(cleaned,stemDocument)
cleaned <- tm_map(cleaned,removeNumbers)
cleaned <- tm_map(cleaned,removePunctuation)
cleaned <- tm_map(cleaned,removeWords, "customer")
cleaned <- tm_map(cleaned,removeWords, "cust")
cleaned <- tm_map(cleaned,removeWords, "client")
cleaned <- tm_map(cleaned,removeWords, "replace")
cleaned <- tm_map(cleaned,removeWords, "replaced")
cleaned <- tm_map(cleaned,removeWords, "repl")
cleaned <- tm_map(cleaned,removeWords, "states")
cleaned <- tm_map(cleaned,removeWords, "perform")
cleaned <- tm_map(cleaned,removeWords, "performed")
cleaned <- tm_map(cleaned,removeWords, "checked")
cleaned <- tm_map(cleaned,removeWords, "found")
cleaned <- tm_map(cleaned,removeWords, "advise")
cleaned <- tm_map(cleaned,removeWords, "inspect")
cleaned <- tm_map(cleaned,removeWords, "inspected")
cleaned <- tm_map(cleaned,removeWords, "tech")
cleaned <- tm_map(cleaned,removeWords, "technician")
cleaned <- tm_map(cleaned,removeWords, "new")
cleaned <- tm_map(cleaned,removeWords, "test")
cleaned <- tm_map(cleaned,removeWords, "please")
wordcloud(cleaned, max.words=100, colors=brewer.pal(7,"Dark2"),random.order=FALSE, scale=c(5,0.5))
import pandas as pd
contention = pd.read_excel(r'\\hdcnas02\AQ_MarketQuality\DensoOBD\WarrantySummaries\35830 - Sunroof Switch\12G_Civic_35830.xlsx','Claims')
%R -i contention
%%R -w 400 -h 400 -u px # instead of px, you can also choose 'in', 'cm', or 'mm'
df <- as.data.frame(contention)
library(tm)
library(wordcloud)
library(SnowballC)
doc.corpus <- Corpus(VectorSource(contention$CUSTOMER_CONTENTION_TEXT))
cleaned <- tm_map(doc.corpus,stripWhitespace)
cleaned <- tm_map(cleaned, content_transformer(tolower))
cleaned <- tm_map(cleaned,removeWords,stopwords("english"))
#cleaned <- tm_map(cleaned,stemDocument)
cleaned <- tm_map(cleaned,removeNumbers)
cleaned <- tm_map(cleaned,removePunctuation)
# Remove "worthless" words
cleaned <- tm_map(cleaned,removeWords, "customer")
cleaned <- tm_map(cleaned,removeWords, "cust")
cleaned <- tm_map(cleaned,removeWords, "client")
cleaned <- tm_map(cleaned,removeWords, "replace")
cleaned <- tm_map(cleaned,removeWords, "replaced")
cleaned <- tm_map(cleaned,removeWords, "repl")
cleaned <- tm_map(cleaned,removeWords, "states")
cleaned <- tm_map(cleaned,removeWords, "perform")
cleaned <- tm_map(cleaned,removeWords, "performed")
cleaned <- tm_map(cleaned,removeWords, "checked")
cleaned <- tm_map(cleaned,removeWords, "found")
cleaned <- tm_map(cleaned,removeWords, "advise")
cleaned <- tm_map(cleaned,removeWords, "inspect")
cleaned <- tm_map(cleaned,removeWords, "inspected")
cleaned <- tm_map(cleaned,removeWords, "tech")
cleaned <- tm_map(cleaned,removeWords, "technician")
cleaned <- tm_map(cleaned,removeWords, "new")
cleaned <- tm_map(cleaned,removeWords, "test")
cleaned <- tm_map(cleaned,removeWords, "please")
wordcloud(cleaned, max.words=50, colors=brewer.pal(6,"Dark2"),random.order=FALSE, scale=c(5,0.5))
| 0.205456 | 0.74911 |
### Stats
Jay Urbain, PhD
7/25/2018
Topics:
Sampling
Central Tendencies
Deviations
Correlaton
Data Visualization
References:
https://matplotlib.org/users/index.html
Data Science from Scratch,, Joel Grus, 2015.
Python Data Science Handbook, Jake VanderPlas, 2017.
```
from collections import Counter
import math
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
import pandas as pd
num_friends = [100,49,41,40,25,21,21,19,19,18,18,16,15,15,15,15,14,14,13,13,13,13,12,12,11,10,10,10,10,10,10,10,10,10,10,10,10,10,10,10,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,8,8,8,8,8,8,8,8,8,8,8,8,8,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1]
plt.hist(num_friends, bins=10)
plt.axis([0,101,0,25])
plt.title("Histogram of Friend Counts")
plt.xlabel("# of friends")
plt.ylabel("# of people")
num_friends_np = np.array(num_friends)
num_friends_pd = pd.DataFrame(num_friends)
print('num_points', len(num_friends))
print('num_points', len(num_friends_np))
print('num_points', num_friends_np.size)
print('num_points', num_friends_np.shape[0])
print('num_points', len(num_friends_pd))
print('num_points', num_friends_pd.size) # careful: number elements in df
print('num_points', num_friends_pd.shape[0])
print('largest_value', max(num_friends))
print('largest_value', max(num_friends_np))
print('largest_value', np.max(num_friends_np))
print('largest_value', num_friends_np.max())
print('smallest_value', min(num_friends))
print('smallest_value', min(num_friends_np))
print('smallest_value', np.min(num_friends_np))
print('smallest_value', num_friends_np.min())
sorted_values = sorted(num_friends)
print('sorted_values', sorted_values)
smallest_value = sorted_values[0]
print('smallest_value', smallest_value)
second_smallest_value = sorted_values[1]
print('second_smallest_value', second_smallest_value)
second_largest_value = sorted_values[-2]
print('second_largest_value', second_largest_value)
max(num_friends)
```
#### Characterizing Distributions
```
import matplotlib.pyplot as plt
import numpy as np
import math
import scipy.stats
mu = 0
variance = 1
sigma = math.sqrt(variance)
x = np.linspace(mu - 3*sigma, mu + 5*sigma, 100)
y = scipy.stats.norm.pdf(x, mu, sigma)
median = np.median(y)
plt.plot(x, scipy.stats.norm.pdf(x, mu, sigma))
# place a text box in upper left in axes coords
textstr = '$\mu=%.2f$\n$\mathrm{median}=%.2f$\n$\sigma=%.2f$'%(mu, median, sigma)
plt.text(0.0001, 0.85, textstr, transform=ax.transAxes, fontsize=10,
verticalalignment='top', bbox=props)
plt.title("Normal distribution, right skew")
plt.text(0.0001, 0.85, textstr, transform=ax.transAxes, fontsize=10,
verticalalignment='top', bbox=props)
import matplotlib.pyplot as plt
import numpy as np
import math
import scipy.stats
mu = 0
variance = 1
sigma = math.sqrt(variance)
x = np.linspace(mu - 3*sigma, mu + 3*sigma, 100)
y = scipy.stats.norm.pdf(x, mu, sigma)
median = np.median(y)
plt.plot(x, scipy.stats.norm.pdf(x, mu, sigma))
# place a text box in upper left in axes coords
textstr = '$\mu=%.2f$\n$\mathrm{median}=%.2f$\n$\sigma=%.2f$'%(mu, median, sigma)
plt.text(0.0001, 0.85, textstr, transform=ax.transAxes, fontsize=10,
verticalalignment='top', bbox=props)
import matplotlib.pyplot as plt
mu, sigma = 0, 0.1 # mean and standard deviation
s = np.random.normal(mu, sigma, 1000)
count, bins, ignored = plt.hist(s, 30, normed=True)
plt.plot(bins, 1/(sigma * np.sqrt(2 * np.pi)) *
np.exp( - (bins - mu)**2 / (2 * sigma**2) ),
linewidth=2, color='r')
import numpy as np
import matplotlib.pyplot as plt
mu = 0
variance = 1
sigma = math.sqrt(variance)
x1 = np.linspace(mu - 3*sigma, mu + 3*sigma, 100)
y1 = scipy.stats.norm.pdf(x1, mu, sigma)
fig, ax = plt.subplots()
ax.plot(x1, y1, color='black', label='No skew', alpha=1.0)
ax.axvline(x1.mean(), color='blue', linewidth=1, alpha=0.5)
ax.axvline(np.median(x1), color='green', linewidth=1, alpha=0.5)
plt.legend(('Distribution', 'Mean', 'Std'),
loc='upper right', shadow=True)
# plt.legend('mean: {:0.2f}'.format(x1.mean()),
# 'median: {:0.2f}'.format(np.median(x1)),
# loc='upper right', shadow=True)
textstr = '$\mu=%.2f$\n$\mathrm{median}=%.2f$\n$\sigma=%.2f$'%(x1.mean(), np.median(x1), sigma)
plt.text(0.1, 0.95, textstr, transform=ax.transAxes, fontsize=10,
verticalalignment='top', bbox=props)
ax.margins(0.05)
import numpy as np
import matplotlib.pyplot as plt
def norm_dist_plit(mu, variance, x, title):
sigma = math.sqrt(variance)
x1 = np.linspace(mu - 3*sigma, mu + 3*sigma, 100)
y1 = scipy.stats.norm.pdf(x1, mu, sigma)
fig, ax = plt.subplots()
ax.plot(x1, y1, color='black', label='No skew', alpha=1.0)
ax.axvline(x1.mean(), color='blue', linewidth=1, alpha=0.5)
ax.axvline(np.median(x1), color='green', linewidth=1, alpha=0.5)
plt.legend(('Distribution', 'Mean', 'Std'),
loc='upper right', shadow=True)
textstr = '$\mu=%.2f$\n$\mathrm{median}=%.2f$\n$\sigma=%.2f$'%(x1.mean(), np.median(x1), sigma)
plt.text(0.1, 0.95, textstr, transform=ax.transAxes, fontsize=10,
verticalalignment='top', bbox=props)
plt.title(title)
ax.margins(0.05)
x = np.linspace(mu - 3*sigma, mu + 3*sigma, 100)
norm_dist_plit(mu, variance, x, "Normal dist, no skew")
x = np.linspace(mu - 3*sigma, mu + 5*sigma, 100)
norm_dist_plit(mu, variance, x, "Normal dist, right skew")
%matplotlib inline
import matplotlib.pyplot as plt
plt.style.use('seaborn-white')
from __future__ import print_function
"""
Edward Tufte uses this example from Anscombe to show 4 datasets of x
and y that have the same mean, standard deviation, and regression
line, but which are qualitatively different.
matplotlib fun for a rainy day
"""
import matplotlib.pyplot as plt
import numpy as np
x = np.array([10, 8, 13, 9, 11, 14, 6, 4, 12, 7, 5])
y1 = np.array([8.04, 6.95, 7.58, 8.81, 8.33, 9.96, 7.24, 4.26, 10.84, 4.82, 5.68])
y2 = np.array([9.14, 8.14, 8.74, 8.77, 9.26, 8.10, 6.13, 3.10, 9.13, 7.26, 4.74])
y3 = np.array([7.46, 6.77, 12.74, 7.11, 7.81, 8.84, 6.08, 5.39, 8.15, 6.42, 5.73])
x4 = np.array([8, 8, 8, 8, 8, 8, 8, 19, 8, 8, 8])
y4 = np.array([6.58, 5.76, 7.71, 8.84, 8.47, 7.04, 5.25, 12.50, 5.56, 7.91, 6.89])
def fit(x):
return 3 + 0.5 * x
xfit = np.array([np.min(x), np.max(x)])
plt.subplot(221)
plt.plot(x, y1, 'ks', xfit, fit(xfit), 'r-', lw=2)
plt.axis([2, 20, 2, 14])
plt.setp(plt.gca(), xticklabels=[], yticks=(4, 8, 12), xticks=(0, 10, 20))
plt.text(3, 12, 'I', fontsize=20)
plt.subplot(222)
plt.plot(x, y2, 'ks', xfit, fit(xfit), 'r-', lw=2)
plt.axis([2, 20, 2, 14])
plt.setp(plt.gca(), xticks=(0, 10, 20), xticklabels=[],
yticks=(4, 8, 12), yticklabels=[], )
plt.text(3, 12, 'II', fontsize=20)
plt.subplot(223)
plt.plot(x, y3, 'ks', xfit, fit(xfit), 'r-', lw=2)
plt.axis([2, 20, 2, 14])
plt.text(3, 12, 'III', fontsize=20)
plt.setp(plt.gca(), yticks=(4, 8, 12), xticks=(0, 10, 20))
plt.subplot(224)
xfit = np.array([np.min(x4), np.max(x4)])
plt.plot(x4, y4, 'ks', xfit, fit(xfit), 'r-', lw=2)
plt.axis([2, 20, 2, 14])
plt.setp(plt.gca(), yticklabels=[], yticks=(4, 8, 12), xticks=(0, 10, 20))
plt.text(3, 12, 'IV', fontsize=20)
# verify the stats
pairs = (x, y1), (x, y2), (x, y3), (x4, y4)
for x, y in pairs:
print('mean=%1.2f, std=%1.2f, r=%1.2f' % (np.mean(y), np.std(y),
np.corrcoef(x, y)[0][1]))
plt.show()
```
|
github_jupyter
|
from collections import Counter
import math
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
import pandas as pd
num_friends = [100,49,41,40,25,21,21,19,19,18,18,16,15,15,15,15,14,14,13,13,13,13,12,12,11,10,10,10,10,10,10,10,10,10,10,10,10,10,10,10,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,8,8,8,8,8,8,8,8,8,8,8,8,8,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1]
plt.hist(num_friends, bins=10)
plt.axis([0,101,0,25])
plt.title("Histogram of Friend Counts")
plt.xlabel("# of friends")
plt.ylabel("# of people")
num_friends_np = np.array(num_friends)
num_friends_pd = pd.DataFrame(num_friends)
print('num_points', len(num_friends))
print('num_points', len(num_friends_np))
print('num_points', num_friends_np.size)
print('num_points', num_friends_np.shape[0])
print('num_points', len(num_friends_pd))
print('num_points', num_friends_pd.size) # careful: number elements in df
print('num_points', num_friends_pd.shape[0])
print('largest_value', max(num_friends))
print('largest_value', max(num_friends_np))
print('largest_value', np.max(num_friends_np))
print('largest_value', num_friends_np.max())
print('smallest_value', min(num_friends))
print('smallest_value', min(num_friends_np))
print('smallest_value', np.min(num_friends_np))
print('smallest_value', num_friends_np.min())
sorted_values = sorted(num_friends)
print('sorted_values', sorted_values)
smallest_value = sorted_values[0]
print('smallest_value', smallest_value)
second_smallest_value = sorted_values[1]
print('second_smallest_value', second_smallest_value)
second_largest_value = sorted_values[-2]
print('second_largest_value', second_largest_value)
max(num_friends)
import matplotlib.pyplot as plt
import numpy as np
import math
import scipy.stats
mu = 0
variance = 1
sigma = math.sqrt(variance)
x = np.linspace(mu - 3*sigma, mu + 5*sigma, 100)
y = scipy.stats.norm.pdf(x, mu, sigma)
median = np.median(y)
plt.plot(x, scipy.stats.norm.pdf(x, mu, sigma))
# place a text box in upper left in axes coords
textstr = '$\mu=%.2f$\n$\mathrm{median}=%.2f$\n$\sigma=%.2f$'%(mu, median, sigma)
plt.text(0.0001, 0.85, textstr, transform=ax.transAxes, fontsize=10,
verticalalignment='top', bbox=props)
plt.title("Normal distribution, right skew")
plt.text(0.0001, 0.85, textstr, transform=ax.transAxes, fontsize=10,
verticalalignment='top', bbox=props)
import matplotlib.pyplot as plt
import numpy as np
import math
import scipy.stats
mu = 0
variance = 1
sigma = math.sqrt(variance)
x = np.linspace(mu - 3*sigma, mu + 3*sigma, 100)
y = scipy.stats.norm.pdf(x, mu, sigma)
median = np.median(y)
plt.plot(x, scipy.stats.norm.pdf(x, mu, sigma))
# place a text box in upper left in axes coords
textstr = '$\mu=%.2f$\n$\mathrm{median}=%.2f$\n$\sigma=%.2f$'%(mu, median, sigma)
plt.text(0.0001, 0.85, textstr, transform=ax.transAxes, fontsize=10,
verticalalignment='top', bbox=props)
import matplotlib.pyplot as plt
mu, sigma = 0, 0.1 # mean and standard deviation
s = np.random.normal(mu, sigma, 1000)
count, bins, ignored = plt.hist(s, 30, normed=True)
plt.plot(bins, 1/(sigma * np.sqrt(2 * np.pi)) *
np.exp( - (bins - mu)**2 / (2 * sigma**2) ),
linewidth=2, color='r')
import numpy as np
import matplotlib.pyplot as plt
mu = 0
variance = 1
sigma = math.sqrt(variance)
x1 = np.linspace(mu - 3*sigma, mu + 3*sigma, 100)
y1 = scipy.stats.norm.pdf(x1, mu, sigma)
fig, ax = plt.subplots()
ax.plot(x1, y1, color='black', label='No skew', alpha=1.0)
ax.axvline(x1.mean(), color='blue', linewidth=1, alpha=0.5)
ax.axvline(np.median(x1), color='green', linewidth=1, alpha=0.5)
plt.legend(('Distribution', 'Mean', 'Std'),
loc='upper right', shadow=True)
# plt.legend('mean: {:0.2f}'.format(x1.mean()),
# 'median: {:0.2f}'.format(np.median(x1)),
# loc='upper right', shadow=True)
textstr = '$\mu=%.2f$\n$\mathrm{median}=%.2f$\n$\sigma=%.2f$'%(x1.mean(), np.median(x1), sigma)
plt.text(0.1, 0.95, textstr, transform=ax.transAxes, fontsize=10,
verticalalignment='top', bbox=props)
ax.margins(0.05)
import numpy as np
import matplotlib.pyplot as plt
def norm_dist_plit(mu, variance, x, title):
sigma = math.sqrt(variance)
x1 = np.linspace(mu - 3*sigma, mu + 3*sigma, 100)
y1 = scipy.stats.norm.pdf(x1, mu, sigma)
fig, ax = plt.subplots()
ax.plot(x1, y1, color='black', label='No skew', alpha=1.0)
ax.axvline(x1.mean(), color='blue', linewidth=1, alpha=0.5)
ax.axvline(np.median(x1), color='green', linewidth=1, alpha=0.5)
plt.legend(('Distribution', 'Mean', 'Std'),
loc='upper right', shadow=True)
textstr = '$\mu=%.2f$\n$\mathrm{median}=%.2f$\n$\sigma=%.2f$'%(x1.mean(), np.median(x1), sigma)
plt.text(0.1, 0.95, textstr, transform=ax.transAxes, fontsize=10,
verticalalignment='top', bbox=props)
plt.title(title)
ax.margins(0.05)
x = np.linspace(mu - 3*sigma, mu + 3*sigma, 100)
norm_dist_plit(mu, variance, x, "Normal dist, no skew")
x = np.linspace(mu - 3*sigma, mu + 5*sigma, 100)
norm_dist_plit(mu, variance, x, "Normal dist, right skew")
%matplotlib inline
import matplotlib.pyplot as plt
plt.style.use('seaborn-white')
from __future__ import print_function
"""
Edward Tufte uses this example from Anscombe to show 4 datasets of x
and y that have the same mean, standard deviation, and regression
line, but which are qualitatively different.
matplotlib fun for a rainy day
"""
import matplotlib.pyplot as plt
import numpy as np
x = np.array([10, 8, 13, 9, 11, 14, 6, 4, 12, 7, 5])
y1 = np.array([8.04, 6.95, 7.58, 8.81, 8.33, 9.96, 7.24, 4.26, 10.84, 4.82, 5.68])
y2 = np.array([9.14, 8.14, 8.74, 8.77, 9.26, 8.10, 6.13, 3.10, 9.13, 7.26, 4.74])
y3 = np.array([7.46, 6.77, 12.74, 7.11, 7.81, 8.84, 6.08, 5.39, 8.15, 6.42, 5.73])
x4 = np.array([8, 8, 8, 8, 8, 8, 8, 19, 8, 8, 8])
y4 = np.array([6.58, 5.76, 7.71, 8.84, 8.47, 7.04, 5.25, 12.50, 5.56, 7.91, 6.89])
def fit(x):
return 3 + 0.5 * x
xfit = np.array([np.min(x), np.max(x)])
plt.subplot(221)
plt.plot(x, y1, 'ks', xfit, fit(xfit), 'r-', lw=2)
plt.axis([2, 20, 2, 14])
plt.setp(plt.gca(), xticklabels=[], yticks=(4, 8, 12), xticks=(0, 10, 20))
plt.text(3, 12, 'I', fontsize=20)
plt.subplot(222)
plt.plot(x, y2, 'ks', xfit, fit(xfit), 'r-', lw=2)
plt.axis([2, 20, 2, 14])
plt.setp(plt.gca(), xticks=(0, 10, 20), xticklabels=[],
yticks=(4, 8, 12), yticklabels=[], )
plt.text(3, 12, 'II', fontsize=20)
plt.subplot(223)
plt.plot(x, y3, 'ks', xfit, fit(xfit), 'r-', lw=2)
plt.axis([2, 20, 2, 14])
plt.text(3, 12, 'III', fontsize=20)
plt.setp(plt.gca(), yticks=(4, 8, 12), xticks=(0, 10, 20))
plt.subplot(224)
xfit = np.array([np.min(x4), np.max(x4)])
plt.plot(x4, y4, 'ks', xfit, fit(xfit), 'r-', lw=2)
plt.axis([2, 20, 2, 14])
plt.setp(plt.gca(), yticklabels=[], yticks=(4, 8, 12), xticks=(0, 10, 20))
plt.text(3, 12, 'IV', fontsize=20)
# verify the stats
pairs = (x, y1), (x, y2), (x, y3), (x4, y4)
for x, y in pairs:
print('mean=%1.2f, std=%1.2f, r=%1.2f' % (np.mean(y), np.std(y),
np.corrcoef(x, y)[0][1]))
plt.show()
| 0.552781 | 0.751101 |
# Sklearn
## sklearn.tree
документация: http://scikit-learn.org/stable/modules/classes.html#module-sklearn.tree
примеры: http://scikit-learn.org/stable/modules/classes.html#module-sklearn.tree
```
from matplotlib.colors import ListedColormap
from sklearn import cross_validation, datasets, metrics, tree
import numpy as np
%pylab inline
```
### Генерация данных
```
classification_problem = datasets.make_classification(n_features = 2, n_informative = 2,
n_classes = 3, n_redundant=0,
n_clusters_per_class=1, random_state=3)
colors = ListedColormap(['red', 'blue', 'yellow'])
light_colors = ListedColormap(['lightcoral', 'lightblue', 'lightyellow'])
pylab.figure(figsize=(8,6))
pylab.scatter(map(lambda x: x[0], classification_problem[0]), map(lambda x: x[1], classification_problem[0]),
c=classification_problem[1], cmap=colors, s=100)
train_data, test_data, train_labels, test_labels = cross_validation.train_test_split(classification_problem[0],
classification_problem[1],
test_size = 0.3,
random_state = 1)
```
### Модель DecisionTreeClassifier
```
clf = tree.DecisionTreeClassifier(random_state=1)
clf.fit(train_data, train_labels)
predictions = clf.predict(test_data)
metrics.accuracy_score(test_labels, predictions)
predictions
```
### Разделяющая поверхность
```
def get_meshgrid(data, step=.05, border=.5,):
x_min, x_max = data[:, 0].min() - border, data[:, 0].max() + border
y_min, y_max = data[:, 1].min() - border, data[:, 1].max() + border
return np.meshgrid(np.arange(x_min, x_max, step), np.arange(y_min, y_max, step))
def plot_decision_surface(estimator, train_data, train_labels, test_data, test_labels,
colors = colors, light_colors = light_colors):
#fit model
estimator.fit(train_data, train_labels)
#set figure size
pyplot.figure(figsize = (16, 6))
#plot decision surface on the train data
pyplot.subplot(1,2,1)
xx, yy = get_meshgrid(train_data)
mesh_predictions = np.array(estimator.predict(np.c_[xx.ravel(), yy.ravel()])).reshape(xx.shape)
pyplot.pcolormesh(xx, yy, mesh_predictions, cmap = light_colors)
pyplot.scatter(train_data[:, 0], train_data[:, 1], c = train_labels, s = 100, cmap = colors)
pyplot.title('Train data, accuracy={:.2f}'.format(metrics.accuracy_score(train_labels, estimator.predict(train_data))))
#plot decision surface on the test data
pyplot.subplot(1,2,2)
pyplot.pcolormesh(xx, yy, mesh_predictions, cmap = light_colors)
pyplot.scatter(test_data[:, 0], test_data[:, 1], c = test_labels, s = 100, cmap = colors)
pyplot.title('Test data, accuracy={:.2f}'.format(metrics.accuracy_score(test_labels, estimator.predict(test_data))))
estimator = tree.DecisionTreeClassifier(random_state = 1, max_depth = 1)
plot_decision_surface(estimator, train_data, train_labels, test_data, test_labels)
plot_decision_surface(tree.DecisionTreeClassifier(random_state = 1, max_depth = 2),
train_data, train_labels, test_data, test_labels)
plot_decision_surface(tree.DecisionTreeClassifier(random_state = 1, max_depth = 3),
train_data, train_labels, test_data, test_labels)
plot_decision_surface(tree.DecisionTreeClassifier(random_state = 1),
train_data, train_labels, test_data, test_labels)
plot_decision_surface(tree.DecisionTreeClassifier(random_state = 1, min_samples_leaf = 3),
train_data, train_labels, test_data, test_labels)
```
|
github_jupyter
|
from matplotlib.colors import ListedColormap
from sklearn import cross_validation, datasets, metrics, tree
import numpy as np
%pylab inline
classification_problem = datasets.make_classification(n_features = 2, n_informative = 2,
n_classes = 3, n_redundant=0,
n_clusters_per_class=1, random_state=3)
colors = ListedColormap(['red', 'blue', 'yellow'])
light_colors = ListedColormap(['lightcoral', 'lightblue', 'lightyellow'])
pylab.figure(figsize=(8,6))
pylab.scatter(map(lambda x: x[0], classification_problem[0]), map(lambda x: x[1], classification_problem[0]),
c=classification_problem[1], cmap=colors, s=100)
train_data, test_data, train_labels, test_labels = cross_validation.train_test_split(classification_problem[0],
classification_problem[1],
test_size = 0.3,
random_state = 1)
clf = tree.DecisionTreeClassifier(random_state=1)
clf.fit(train_data, train_labels)
predictions = clf.predict(test_data)
metrics.accuracy_score(test_labels, predictions)
predictions
def get_meshgrid(data, step=.05, border=.5,):
x_min, x_max = data[:, 0].min() - border, data[:, 0].max() + border
y_min, y_max = data[:, 1].min() - border, data[:, 1].max() + border
return np.meshgrid(np.arange(x_min, x_max, step), np.arange(y_min, y_max, step))
def plot_decision_surface(estimator, train_data, train_labels, test_data, test_labels,
colors = colors, light_colors = light_colors):
#fit model
estimator.fit(train_data, train_labels)
#set figure size
pyplot.figure(figsize = (16, 6))
#plot decision surface on the train data
pyplot.subplot(1,2,1)
xx, yy = get_meshgrid(train_data)
mesh_predictions = np.array(estimator.predict(np.c_[xx.ravel(), yy.ravel()])).reshape(xx.shape)
pyplot.pcolormesh(xx, yy, mesh_predictions, cmap = light_colors)
pyplot.scatter(train_data[:, 0], train_data[:, 1], c = train_labels, s = 100, cmap = colors)
pyplot.title('Train data, accuracy={:.2f}'.format(metrics.accuracy_score(train_labels, estimator.predict(train_data))))
#plot decision surface on the test data
pyplot.subplot(1,2,2)
pyplot.pcolormesh(xx, yy, mesh_predictions, cmap = light_colors)
pyplot.scatter(test_data[:, 0], test_data[:, 1], c = test_labels, s = 100, cmap = colors)
pyplot.title('Test data, accuracy={:.2f}'.format(metrics.accuracy_score(test_labels, estimator.predict(test_data))))
estimator = tree.DecisionTreeClassifier(random_state = 1, max_depth = 1)
plot_decision_surface(estimator, train_data, train_labels, test_data, test_labels)
plot_decision_surface(tree.DecisionTreeClassifier(random_state = 1, max_depth = 2),
train_data, train_labels, test_data, test_labels)
plot_decision_surface(tree.DecisionTreeClassifier(random_state = 1, max_depth = 3),
train_data, train_labels, test_data, test_labels)
plot_decision_surface(tree.DecisionTreeClassifier(random_state = 1),
train_data, train_labels, test_data, test_labels)
plot_decision_surface(tree.DecisionTreeClassifier(random_state = 1, min_samples_leaf = 3),
train_data, train_labels, test_data, test_labels)
| 0.847983 | 0.932207 |
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#Dependencies" data-toc-modified-id="Dependencies-1">Dependencies</a></span></li><li><span><a href="#Paths" data-toc-modified-id="Paths-2">Paths</a></span></li><li><span><a href="#Main" data-toc-modified-id="Main-3">Main</a></span><ul class="toc-item"><li><span><a href="#Cleanup-according-to-CRC-review" data-toc-modified-id="Cleanup-according-to-CRC-review-3.1">Cleanup according to CRC review</a></span></li><li><span><a href="#Modify-disease-types-based-on-my-path-reviews" data-toc-modified-id="Modify-disease-types-based-on-my-path-reviews-3.2">Modify disease types based on my path reviews</a></span></li><li><span><a href="#Group-Ganglioglioma-with-pedLGG" data-toc-modified-id="Group-Ganglioglioma-with-pedLGG-3.3">Group Ganglioglioma with pedLGG</a></span></li><li><span><a href="#Group-DNET-with-pedLGG" data-toc-modified-id="Group-DNET-with-pedLGG-3.4">Group DNET with pedLGG</a></span></li><li><span><a href="#Group-SEGA-(from-Others-group)-with-pedLGG" data-toc-modified-id="Group-SEGA-(from-Others-group)-with-pedLGG-3.5">Group SEGA (from Others group) with pedLGG</a></span></li><li><span><a href="#Group-PA-(ICGC-data)-with-pedLGG" data-toc-modified-id="Group-PA-(ICGC-data)-with-pedLGG-3.6">Group PA (ICGC data) with pedLGG</a></span></li><li><span><a href="#Remove-met-NBL-cases-from-CBTTC-dataset:-7316-224-and-7316-3311" data-toc-modified-id="Remove-met-NBL-cases-from-CBTTC-dataset:-7316-224-and-7316-3311-3.7">Remove met NBL cases from CBTTC dataset: 7316-224 and 7316-3311</a></span></li><li><span><a href="#Bucket-DIPGs-into-pedHGG" data-toc-modified-id="Bucket-DIPGs-into-pedHGG-3.8">Bucket DIPGs into pedHGG</a></span></li><li><span><a href="#Reannotate-Choroid-plexus-as-CP" data-toc-modified-id="Reannotate-Choroid-plexus-as-CP-3.9">Reannotate Choroid plexus as CP</a></span></li><li><span><a href="#Reannotate-Ewings-as-ES" data-toc-modified-id="Reannotate-Ewings-as-ES-3.10">Reannotate Ewings as ES</a></span></li><li><span><a href="#Reannotate-two-embryonal-tumours-as-ETMR" data-toc-modified-id="Reannotate-two-embryonal-tumours-as-ETMR-3.11">Reannotate two embryonal tumours as ETMR</a></span></li><li><span><a href="#Cleanup-Others-group-by-path-reports-and-remove-remaining-samples" data-toc-modified-id="Cleanup-Others-group-by-path-reports-and-remove-remaining-samples-3.12">Cleanup Others group by path reports and remove remaining samples</a></span></li><li><span><a href="#Remove-PNET-group" data-toc-modified-id="Remove-PNET-group-3.13">Remove PNET group</a></span></li><li><span><a href="#Check-if-all-ATRT-cases-have-SMARCB1-mutation" data-toc-modified-id="Check-if-all-ATRT-cases-have-SMARCB1-mutation-3.14">Check if all ATRT cases have SMARCB1 mutation</a></span></li><li><span><a href="#Modify-cohort-abbreviations" data-toc-modified-id="Modify-cohort-abbreviations-3.15">Modify cohort abbreviations</a></span></li></ul></li></ul></div>
# Dependencies
# Paths
```
manifestpath <- "/Users/anabbi/OneDrive - UHN/Documents/IPD2/Manifests/"
datapath <- "/Users/anabbi/OneDrive - UHN/Documents/IPD2/Data/"
plotpath <- "/Users/anabbi/OneDrive - UHN/Documents/IPD2/Plots/"
```
# Main
```
load(file = paste0(datapath,"ESTIMATE/estimate_manifest_primary.RData"))
estimate_manifest_primary_clean <- estimate_manifest_primary
```
## Cleanup according to CRC review
Checked with CRC, see excel file CBTTC Dataset_pathrepchecked_CRCreviewed in Manifests
```
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-740"] <- "EP WHO Grade III"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-740"] <- "EP"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-952"] <- "Composite DNET and GGM"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-715"] <- "GBM"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-715"] <- "pedHGG"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-1855"] <- "Burkitts Lymphoma"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-2153"] <- "Hemangiopericytoma"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-3817"] <- "GBM"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-3817"] <- "pedHGG"
```
## Modify disease types based on my path reviews
Bucket DNET, GG, PA, PXA and otherLGG into pedLGG
See excel sheet CBTTC Dataset_pathrepchecked_primary
```
pathchecked <- read.csv(file = paste0(manifestpath, "CBTTC Dataset_pathrepchecked_primary.csv"),
header = T, na.strings = "", stringsAsFactors = F)
pathchecked$Cohort[pathchecked$CBTTC.Event.ID == "7316-740"] <- "EP"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-373"] <- "Ganglioglioma Grade III"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-2980"] <- "Malignant glioma with features of PXA"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-3922"] <- "Anaplastic PA"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-373" |
estimate_manifest_primary_clean$sample_id == "7316-2980" |
estimate_manifest_primary_clean$sample_id == "7316-3922"] <- "pedHGG"
pxas <- pathchecked$CBTTC.Event.ID[grepl("PXA",pathchecked$Final.Dx.on.path.report)]
pxas <- pxas[pxas != "7316-2980"]
pas <- pathchecked$CBTTC.Event.ID[grepl("PA",pathchecked$Final.Dx.on.path.report)|
grepl("ilomyxoid",pathchecked$Final.Dx.on.path.report)]
pas <- pas[pas != "7316-3922"]
otherlggs <- pathchecked$CBTTC.Event.ID[pathchecked$Cohort == "LGG" &
!pathchecked$CBTTC.Event.ID %in% pxas &
!pathchecked$CBTTC.Event.ID %in% pas]
otherlggs <- otherlggs[otherlggs != "7316-3922"]
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id %in% pxas] <- "PXA"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id %in% pas] <- "PA"
```
If path report was available for otherlggs, add disease type, if not, keep as is: Low grade glioma/astrocytoma WHO grade I/II
```
myotherlggdx <- pathchecked[pathchecked$CBTTC.Event.ID %in% otherlggs,]
myotherlggdx <- myotherlggdx[myotherlggdx$Final.Dx.on.path.report != "na",]
for(i in 1:nrow(myotherlggdx)){
estimate_manifest_primary_clean$disease_type[match(myotherlggdx$CBTTC.Event.ID[i],
estimate_manifest_primary_clean$sample_id)] <- myotherlggdx$Final.Dx.on.path.report[i]
}
```
## Group Ganglioglioma with pedLGG
```
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Ganglioglioma"] <- "pedLGG"
```
## Group DNET with pedLGG
```
estimate_manifest_primary_clean$cohort[grepl("DNET",estimate_manifest_primary_clean$disease_type)] <- "pedLGG"
```
## Group SEGA (from Others group) with pedLGG
```
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Glial-neuronal tumor NOS"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Subependymal Giant Cell Astrocytoma (SEGA)"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Papillary Glioneuronal"] <- "pedLGG"
```
## Group PA (ICGC data) with pedLGG
```
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "PA"] <- "pedLGG"
```
## Remove met NBL cases from CBTTC dataset: 7316-224 and 7316-3311
```
estimate_manifest_primary_clean <- estimate_manifest_primary_clean[estimate_manifest_primary_clean$sample_id != "7316-3311" &
estimate_manifest_primary_clean$sample_id != "7316-224",]
```
## Bucket DIPGs into pedHGG
```
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Brainstem glioma- Diffuse intrinsic pontine glioma"] <- "pedHGG"
```
## Reannotate Choroid plexus as CP
```
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Choroid plexus carcinoma"] <- "CP"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Choroid plexus papilloma"] <- "CP"
```
## Reannotate Ewings as ES
```
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Ewings Sarcoma"] <- "ES"
```
## Reannotate two embryonal tumours as ETMR
```
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "ET"] <- "ETMR"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Medulloepithelioma"] <- "ETMR"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Embryonal Tumor with Multilayer Rosettes, ROS (WHO Grade IV)"] <- "ETMR"
```
## Cleanup Others group by path reports and remove remaining samples
```
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-1845"] <- "astrocytoma fibrillary type with intrinsic vascular malformation with necrosis"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-1845"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-133"] <- "pedLGG"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-157"] <- "Atypical DNET/GG/Cortical dysplasia"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-157"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-2495"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-2513"] <- "pedLGG"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-71"] <- "Gliomatosis cerebri with extensive anaplasia"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-71"] <- "pedHGG"
estimate_manifest_primary_clean <- estimate_manifest_primary_clean[estimate_manifest_primary_clean$cohort != "Other",]
```
## Remove PNET group
```
estimate_manifest_primary_clean <- estimate_manifest_primary_clean[estimate_manifest_primary_clean$cohort != "PNET",]
```
## Check if all ATRT cases have SMARCB1 mutation
```
ATRTsample_ids <- estimate_manifest_primary_clean$sample_id[estimate_manifest_primary_clean$cohort == "ATRT" &
estimate_manifest_primary_clean$group == "CBTTC"]
```
I checked all path reports, see excel sheet CBTTC Dataset_Arash Nabbi 12.04.18_pathrepchecked. The majority of ATRT samples either had loss of INI1 expression assessed by IHC or had genetic test (eg SNParray) confirming the deletion of SMARCB1 locus. Exceptions are the following:
7316-479
7316-1073: No path report
7316-2090
7316-3937
I will remove them...
```
estimate_manifest_primary_clean <- estimate_manifest_primary_clean[estimate_manifest_primary_clean$sample_id != "7316-479" &
estimate_manifest_primary_clean$sample_id != "7316-1073" &
estimate_manifest_primary_clean$sample_id != "7316-2090" &
estimate_manifest_primary_clean$sample_id != "7316-3937",]
```
## Modify cohort abbreviations
```
table(estimate_manifest_primary_clean$cohort)
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "MN"] <- "MNG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "SCHN"] <- "SCHW"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "EP"] <- "EPN"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "ES"] <- "EWS"
table(estimate_manifest_primary_clean$cohort)
```
Remove EWS and TT/GN. They are less than 10% of the entire cohort.
```
estimate_manifest_primary_clean <-
estimate_manifest_primary_clean[!estimate_manifest_primary_clean$cohort %in% c("EWS", "TT/GN"),]
table(estimate_manifest_primary_clean$group)
dim(estimate_manifest_primary_clean)
table(estimate_manifest_primary_clean$group)
```
Replace CBTTC with CBTN and DKFZ with ICGC
```
estimate_manifest_primary_clean$group[estimate_manifest_primary_clean$group == "DKFZ"] <- "ICGC"
estimate_manifest_primary_clean$group[estimate_manifest_primary_clean$group == "CBTTC"] <- "CBTN"
save(estimate_manifest_primary_clean,
file = paste0(datapath,"ESTIMATE/estimate_manifest_primary_clean.RData"))
```
|
github_jupyter
|
manifestpath <- "/Users/anabbi/OneDrive - UHN/Documents/IPD2/Manifests/"
datapath <- "/Users/anabbi/OneDrive - UHN/Documents/IPD2/Data/"
plotpath <- "/Users/anabbi/OneDrive - UHN/Documents/IPD2/Plots/"
load(file = paste0(datapath,"ESTIMATE/estimate_manifest_primary.RData"))
estimate_manifest_primary_clean <- estimate_manifest_primary
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-740"] <- "EP WHO Grade III"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-740"] <- "EP"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-952"] <- "Composite DNET and GGM"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-715"] <- "GBM"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-715"] <- "pedHGG"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-1855"] <- "Burkitts Lymphoma"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-2153"] <- "Hemangiopericytoma"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-3817"] <- "GBM"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-3817"] <- "pedHGG"
pathchecked <- read.csv(file = paste0(manifestpath, "CBTTC Dataset_pathrepchecked_primary.csv"),
header = T, na.strings = "", stringsAsFactors = F)
pathchecked$Cohort[pathchecked$CBTTC.Event.ID == "7316-740"] <- "EP"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-373"] <- "Ganglioglioma Grade III"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-2980"] <- "Malignant glioma with features of PXA"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-3922"] <- "Anaplastic PA"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-373" |
estimate_manifest_primary_clean$sample_id == "7316-2980" |
estimate_manifest_primary_clean$sample_id == "7316-3922"] <- "pedHGG"
pxas <- pathchecked$CBTTC.Event.ID[grepl("PXA",pathchecked$Final.Dx.on.path.report)]
pxas <- pxas[pxas != "7316-2980"]
pas <- pathchecked$CBTTC.Event.ID[grepl("PA",pathchecked$Final.Dx.on.path.report)|
grepl("ilomyxoid",pathchecked$Final.Dx.on.path.report)]
pas <- pas[pas != "7316-3922"]
otherlggs <- pathchecked$CBTTC.Event.ID[pathchecked$Cohort == "LGG" &
!pathchecked$CBTTC.Event.ID %in% pxas &
!pathchecked$CBTTC.Event.ID %in% pas]
otherlggs <- otherlggs[otherlggs != "7316-3922"]
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id %in% pxas] <- "PXA"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id %in% pas] <- "PA"
myotherlggdx <- pathchecked[pathchecked$CBTTC.Event.ID %in% otherlggs,]
myotherlggdx <- myotherlggdx[myotherlggdx$Final.Dx.on.path.report != "na",]
for(i in 1:nrow(myotherlggdx)){
estimate_manifest_primary_clean$disease_type[match(myotherlggdx$CBTTC.Event.ID[i],
estimate_manifest_primary_clean$sample_id)] <- myotherlggdx$Final.Dx.on.path.report[i]
}
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Ganglioglioma"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[grepl("DNET",estimate_manifest_primary_clean$disease_type)] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Glial-neuronal tumor NOS"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Subependymal Giant Cell Astrocytoma (SEGA)"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Papillary Glioneuronal"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "PA"] <- "pedLGG"
estimate_manifest_primary_clean <- estimate_manifest_primary_clean[estimate_manifest_primary_clean$sample_id != "7316-3311" &
estimate_manifest_primary_clean$sample_id != "7316-224",]
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Brainstem glioma- Diffuse intrinsic pontine glioma"] <- "pedHGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Choroid plexus carcinoma"] <- "CP"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Choroid plexus papilloma"] <- "CP"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Ewings Sarcoma"] <- "ES"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "ET"] <- "ETMR"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Medulloepithelioma"] <- "ETMR"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$disease_type == "Embryonal Tumor with Multilayer Rosettes, ROS (WHO Grade IV)"] <- "ETMR"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-1845"] <- "astrocytoma fibrillary type with intrinsic vascular malformation with necrosis"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-1845"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-133"] <- "pedLGG"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-157"] <- "Atypical DNET/GG/Cortical dysplasia"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-157"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-2495"] <- "pedLGG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-2513"] <- "pedLGG"
estimate_manifest_primary_clean$disease_type[estimate_manifest_primary_clean$sample_id == "7316-71"] <- "Gliomatosis cerebri with extensive anaplasia"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$sample_id == "7316-71"] <- "pedHGG"
estimate_manifest_primary_clean <- estimate_manifest_primary_clean[estimate_manifest_primary_clean$cohort != "Other",]
estimate_manifest_primary_clean <- estimate_manifest_primary_clean[estimate_manifest_primary_clean$cohort != "PNET",]
ATRTsample_ids <- estimate_manifest_primary_clean$sample_id[estimate_manifest_primary_clean$cohort == "ATRT" &
estimate_manifest_primary_clean$group == "CBTTC"]
estimate_manifest_primary_clean <- estimate_manifest_primary_clean[estimate_manifest_primary_clean$sample_id != "7316-479" &
estimate_manifest_primary_clean$sample_id != "7316-1073" &
estimate_manifest_primary_clean$sample_id != "7316-2090" &
estimate_manifest_primary_clean$sample_id != "7316-3937",]
table(estimate_manifest_primary_clean$cohort)
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "MN"] <- "MNG"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "SCHN"] <- "SCHW"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "EP"] <- "EPN"
estimate_manifest_primary_clean$cohort[estimate_manifest_primary_clean$cohort == "ES"] <- "EWS"
table(estimate_manifest_primary_clean$cohort)
estimate_manifest_primary_clean <-
estimate_manifest_primary_clean[!estimate_manifest_primary_clean$cohort %in% c("EWS", "TT/GN"),]
table(estimate_manifest_primary_clean$group)
dim(estimate_manifest_primary_clean)
table(estimate_manifest_primary_clean$group)
estimate_manifest_primary_clean$group[estimate_manifest_primary_clean$group == "DKFZ"] <- "ICGC"
estimate_manifest_primary_clean$group[estimate_manifest_primary_clean$group == "CBTTC"] <- "CBTN"
save(estimate_manifest_primary_clean,
file = paste0(datapath,"ESTIMATE/estimate_manifest_primary_clean.RData"))
| 0.261425 | 0.781018 |
<a href="https://colab.research.google.com/github/Educat8n/Reinforcement-Learning-for-Game-Playing-and-More/blob/main/Module1/MultiArmedBandits_Example.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
# Import modules
%matplotlib notebook
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.animation import FuncAnimation
import gym
from tqdm import tqdm
import time
from gym import spaces
from gym.utils import seeding
class ArmedBandits(gym.Env):
"""
Multi-Armed Bandit Environment implemented using gym interface.
"""
def __init__(self, mean, stddev):
super(ArmedBandits, self).__init__()
assert len(mean.shape) == 2
assert len(stddev.shape) == 2
# Define action and state space
self.num_bandits = mean.shape[1]
self.num_experiments = mean.shape[0]
self.action_space = spaces.Discrete(self.num_bandits)
# The state Multi-armed bandits problem is static
self.state_space = spaces.Discrete(1)
self.mean = mean
self.stddev = stddev
def step(self, action):
assert (action < self.num_bandits).all()
# Assign reward from the action assigned reward distribution
sampled_means = self.mean[np.arange(self.num_experiments),action]
sampled_stddevs = self.stddev[np.arange(self.num_experiments),action]
reward = np.random.normal(sampled_means, sampled_stddevs, (self.num_experiments,))
# Return a constant state of 0
state, done, info = 0, False, dict()
return state, reward, done, info
def _seed(self, seed=None):
self.np_random, seed = seeding.np.random(seed)
return [seed]
# The mean and standard deviation for a four-armed bandit.
mean = np.array([[5, 1, 0, -5]])
stdev = np.array([[1, 0.1, 0.5, 0.1]])
# Create the environment
env = ArmedBandits(mean, stdev)
for i in range(4):
action = np.array([i])
_, reward, _, _ = env.step(action)
print("Bandit:", i, " gave a reward of:", reward[0])
def argMax(q_table):
"""
Takes in the Q-table(n*k) and returns the index of the
item with the highest value for each row (action).
In case of tie, breaks it randomly.
"""
noise = 1e-6*np.random.random(q_table.shape)
mask = q_table == q_table.max(axis=1)[:, None]
return np.argmax(noise*mask,axis=1)
class GreedyAgent:
def __init__(self, reward_estimates):
"""
The agent guesses the reward it will receive from the environment and
update it incrementally as the based on the interaction with the environment.
"""
assert len(reward_estimates.shape) == 2
self.num_bandits = reward_estimates.shape[1]
self.num_experiments = reward_estimates.shape[0]
self.reward_estimates = reward_estimates.astype(np.float64)
self.action_count = np.zeros(reward_estimates.shape)
def get_action(self):
# Greedy agent takes the action with maximum reward estimate
action = argMax(self.reward_estimates)
# Keep a counter of the action
self.action_count[np.arange(self.num_experiments), action] += 1
return action
def update_estimates(self, reward, action):
"""
Using the rewards obtained from the previuos interaction
update the future estimates incrementally
"""
n = self.action_count[np.arange(self.num_experiments), action]
# Find the difference between the received rewards and estimated estimates
error = reward - self.reward_estimates[np.arange(self.num_experiments), action]
# Update the reward difference incrementally
self.reward_estimates[np.arange(self.num_experiments), action] += (1/n)*error
# Initialize the multi-armed bandit environment
num_steps = 500
num_experiments = 2
num_bandits = 8
mean = np.random.normal(size=(num_experiments, num_bandits))
stdev = np.ones((num_experiments, num_bandits))
env = ArmedBandits(mean, stdev)
# Initialize the agent with zero reward estimates
agent = GreedyAgent(np.zeros((num_experiments,num_bandits)))
# Let us plot the performance as the agent interacts
fig, axs = plt.subplots(1, num_experiments, figsize=(10, 4))
x_pos = np.arange(num_bandits)
def init():
for i in range(num_experiments):
initialize(i)
def initialize(i):
ax = axs[i]
ax.clear()
ax.set_ylim(-4, 4)
ax.set_xlim(-0.5, num_bandits-.5)
ax.set_xlabel('Actions', fontsize=14)
ax.set_ylabel('Value', fontsize=14)
ax.set_title(label='Estimated Values vs. Real values', fontsize=15)
ax.plot(x_pos, env.mean[i], marker='D', linestyle='', alpha=0.8, color='r', label='Real Values')
ax.axhline(0, color='black', lw=1)
# Implement a step, which involves the agent acting upon the
# environment and learning from the received reward.
def step(g):
action = agent.get_action()
_, reward, _, _ = env.step(action)
agent.update_estimates(reward, action)
for i in range(num_experiments):
initialize(i)
ax = axs[i]
# Plot the estimated values from the agent compared to the real values
estimates = agent.reward_estimates[i]
values = ax.bar(x_pos, estimates, align='center', color='blue', alpha=0.5)
anim = FuncAnimation(fig, func=step, frames=np.arange(num_steps), init_func=init, interval=10, repeat=False)
from IPython.display import HTML
HTML(anim.to_html5_video())
```
|
github_jupyter
|
# Import modules
%matplotlib notebook
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.animation import FuncAnimation
import gym
from tqdm import tqdm
import time
from gym import spaces
from gym.utils import seeding
class ArmedBandits(gym.Env):
"""
Multi-Armed Bandit Environment implemented using gym interface.
"""
def __init__(self, mean, stddev):
super(ArmedBandits, self).__init__()
assert len(mean.shape) == 2
assert len(stddev.shape) == 2
# Define action and state space
self.num_bandits = mean.shape[1]
self.num_experiments = mean.shape[0]
self.action_space = spaces.Discrete(self.num_bandits)
# The state Multi-armed bandits problem is static
self.state_space = spaces.Discrete(1)
self.mean = mean
self.stddev = stddev
def step(self, action):
assert (action < self.num_bandits).all()
# Assign reward from the action assigned reward distribution
sampled_means = self.mean[np.arange(self.num_experiments),action]
sampled_stddevs = self.stddev[np.arange(self.num_experiments),action]
reward = np.random.normal(sampled_means, sampled_stddevs, (self.num_experiments,))
# Return a constant state of 0
state, done, info = 0, False, dict()
return state, reward, done, info
def _seed(self, seed=None):
self.np_random, seed = seeding.np.random(seed)
return [seed]
# The mean and standard deviation for a four-armed bandit.
mean = np.array([[5, 1, 0, -5]])
stdev = np.array([[1, 0.1, 0.5, 0.1]])
# Create the environment
env = ArmedBandits(mean, stdev)
for i in range(4):
action = np.array([i])
_, reward, _, _ = env.step(action)
print("Bandit:", i, " gave a reward of:", reward[0])
def argMax(q_table):
"""
Takes in the Q-table(n*k) and returns the index of the
item with the highest value for each row (action).
In case of tie, breaks it randomly.
"""
noise = 1e-6*np.random.random(q_table.shape)
mask = q_table == q_table.max(axis=1)[:, None]
return np.argmax(noise*mask,axis=1)
class GreedyAgent:
def __init__(self, reward_estimates):
"""
The agent guesses the reward it will receive from the environment and
update it incrementally as the based on the interaction with the environment.
"""
assert len(reward_estimates.shape) == 2
self.num_bandits = reward_estimates.shape[1]
self.num_experiments = reward_estimates.shape[0]
self.reward_estimates = reward_estimates.astype(np.float64)
self.action_count = np.zeros(reward_estimates.shape)
def get_action(self):
# Greedy agent takes the action with maximum reward estimate
action = argMax(self.reward_estimates)
# Keep a counter of the action
self.action_count[np.arange(self.num_experiments), action] += 1
return action
def update_estimates(self, reward, action):
"""
Using the rewards obtained from the previuos interaction
update the future estimates incrementally
"""
n = self.action_count[np.arange(self.num_experiments), action]
# Find the difference between the received rewards and estimated estimates
error = reward - self.reward_estimates[np.arange(self.num_experiments), action]
# Update the reward difference incrementally
self.reward_estimates[np.arange(self.num_experiments), action] += (1/n)*error
# Initialize the multi-armed bandit environment
num_steps = 500
num_experiments = 2
num_bandits = 8
mean = np.random.normal(size=(num_experiments, num_bandits))
stdev = np.ones((num_experiments, num_bandits))
env = ArmedBandits(mean, stdev)
# Initialize the agent with zero reward estimates
agent = GreedyAgent(np.zeros((num_experiments,num_bandits)))
# Let us plot the performance as the agent interacts
fig, axs = plt.subplots(1, num_experiments, figsize=(10, 4))
x_pos = np.arange(num_bandits)
def init():
for i in range(num_experiments):
initialize(i)
def initialize(i):
ax = axs[i]
ax.clear()
ax.set_ylim(-4, 4)
ax.set_xlim(-0.5, num_bandits-.5)
ax.set_xlabel('Actions', fontsize=14)
ax.set_ylabel('Value', fontsize=14)
ax.set_title(label='Estimated Values vs. Real values', fontsize=15)
ax.plot(x_pos, env.mean[i], marker='D', linestyle='', alpha=0.8, color='r', label='Real Values')
ax.axhline(0, color='black', lw=1)
# Implement a step, which involves the agent acting upon the
# environment and learning from the received reward.
def step(g):
action = agent.get_action()
_, reward, _, _ = env.step(action)
agent.update_estimates(reward, action)
for i in range(num_experiments):
initialize(i)
ax = axs[i]
# Plot the estimated values from the agent compared to the real values
estimates = agent.reward_estimates[i]
values = ax.bar(x_pos, estimates, align='center', color='blue', alpha=0.5)
anim = FuncAnimation(fig, func=step, frames=np.arange(num_steps), init_func=init, interval=10, repeat=False)
from IPython.display import HTML
HTML(anim.to_html5_video())
| 0.844056 | 0.955486 |
# Google Earth Engine Python API Methods Case Study
## Identifying Vegetation Change in Tolima Department, Columbia
### Doug's Testing
## Introduction
This workflow implements a vegetation change detection analysis with the [Google Earth Engine](https://earthengine.google.com/) (GEE) Python application programming interface (API). The workflow applies these methods to a study area in the Tolima Department, Columbia, during the 2017 Semester A growing season, from peak green (June) to post-harvest (September).
## Environment Setup
This workflow uses [Python 3.8](https://www.python.org/downloads/release/python-380/) and requires the following packages:
* `ee`
* `geemap`
* `vegetation_change`
The `ee` and `geemap` packages are available from [Conda-Forge](https://conda-forge.org/). The `vegetation_change` package (from `vegetation_change.py`) provides custom functions that implement GEE functionality for the benefit of this analysis. The Conda environment provided with this analysis (contained in `environment.yml`) includes all packages needed from Conda-Forge. The custom script exists in the same folder as this Jupyter Notebook.
```
# Import packages
import ee
import geemap as gm
import vegetation_change as vc
```
The workflow [authenticates](https://developers.google.com/earth-engine/python_install-conda#get_credentials) to GEE with an active account and then initializes the GEE library (if the authentication succeeds).
```
# Initialze GEE Python API; authenticate if necessary
try:
ee.Initialize()
except Exception as error:
ee.Authenticate()
ee.Initialize()
```
The workflow defines the user name and public [GEE Assets](https://developers.google.com/earth-engine/asset_manager) folder for accessing the study area features within and exporting the results of the analysis to GEE Assets. The import of the study area features step must use these variables, unless a user has exported them to another location prior to the analysis. The export of the results may omit this path and use a path specific user who executes the workflow.
```
# Define output folder (GEE username + folder location)
gee_username = "calekochenour"
gee_asset_folder = "vegetation-change"
```
## Data Preparation
The workflow imports the study area boundary and study area canals, each as individual `ee.FeatureCollection` object.
```
# Create FeatureCollections for study area
study_area_boundary = ee.FeatureCollection(
"users/calekochenour/vegetation-change/drtt_study_area_boundary")
study_area_canals = ee.FeatureCollection(
"users/calekochenour/vegetation-change/drtt_study_area_canals")
```
The workflow loads two [Landsat 8 Surface Reflectance Tier 1](https://developers.google.com/earth-engine/datasets/catalog/LANDSAT_LC08_C01_T1_SR) images, June 2, 2017 (peak green) and September 6, 2017 (post-harvest), as `ee.Image` objects and clips each image to the study area boundary. The workflow then creates an `ee.ImageCollection` object that contains the two images.
```
# Load and clip imagery for 2017 Semester A growing season
peak_green = ee.Image(
'LANDSAT/LC08/C01/T1_SR/LC08_008057_20170602').clip(study_area_boundary)
post_harvest = ee.Image(
'LANDSAT/LC08/C01/T1_SR/LC08_008057_20170906').clip(study_area_boundary)
# Create ImageCollection for Peak Green and Post-Harvest
collection = ee.ImageCollection([peak_green, post_harvest])
```
## Data Processing
The workflow uses a custom function from the `vegetation_change.py` script, `ndvi_diff_landsat8()` (lines 140-189), to:
* Mask each image in the `ee.ImageCollection` for clouds and cloud shadows;
* Compute and add the [normalized difference vegetation index](https://www.usgs.gov/land-resources/nli/landsat/landsat-normalized-difference-vegetation-index?qt-science_support_page_related_con=0#qt-science_support_page_related_con) (NDVI) band to each image in the `ee.ImageCollection`;
* Convert the `ee.ImageCollection` to an `ee.List` object;
* Subtract the peak green NDVI band from the post-harvest NDVI band; and,
* Return the NDVI difference (post-harvest NDVI - peak green NDVI).
The `vegetation_change.py` script defines the helper functions for `ndvi_diff_landsat8()` in the following locations:
* Mask image
* Function: `mask_landsat8_sr()`
* Location: Lines 9-43
* Compute NDVI band
* Function: `add_ndvi()`
* Location: Lines 46-70
* Convert to list
* Function: `image_collection_to_list()`
* Location: Lines 73-95
* Subtract NDVI bands
* Function: `subtract_ndvi_bands()`
* Location: Lines 98-135
```
# Compute NDVI difference raster for Peak Green to Post-Harvest
ndvi_diff = vc.ndvi_diff_landsat8(collection, 1, 0)
```
The workflow defines two threshold ranges for NDVI change, -2.0 to -0.5 for the primary change (largest change) and -0.5 to -0.35 for secondary change (second largest change). The workflow refined the threshold ranges based on the specific imagery used in this study area, during the time period analyzed. The workflow computed the NDVI change by subtracting the pre-change image (peak green, June) from the post-change image (post-harvest, September). NDVI change values less than 0 indicate change from green vegetation to no vegetation. NDVI change values greater than 0 indicate change from no vegetation to green vegetation.
```
# Define NDVI thresholds for classification;
# indices 0/1 identify min/max for primary class;
# indices 2/3 identify min/max for secondary class
ndvi_change_thresholds = [-2.0, -0.5, -0.5, -0.35]
```
The workflow uses a custom function from the `vegetation_change.py` script, `segment_snic()` (lines 192-270), to:
* Segment the NDVI difference image;
* Classify the image based on the defined NDVI thresholds;
* Extract classified features (primary and secondary NDVI difference ranges); and,
* Return the extracted features as `ee.Image` objects stored in a Python dictionary.
```
# Segment, classify, and extract features
change_features = vc.segment_snic(
ndvi_diff, study_area_boundary, ndvi_change_thresholds)
```
The workflow uses a custom function from the `vegetation_change.py` script, `raster_to_vector()` (lines 273-311), to:
* Convert the classified extracted features (in `ee.Image` format) to `ee.FeatureCollection` objects; and,
* Return the vectorized versions of the extracted features.
```
# Convert rasters to vectors
change_primary_vector = vc.raster_to_vector(
change_features.get('primary'), study_area_boundary)
change_secondary_vector = vc.raster_to_vector(
change_features.get('secondary'), study_area_boundary)
```
## Data Export
The workflow defines output folder locations within a valid GEE Assets folder and uses a custom function from the `vegetation_change.py` script, `export_vector()` (lines 314-385), to export the extracted (vectorized) features to the defined GEE Assets folder.
A user must change the `gee_username` variable within this Jupyter Notebook (cell 9, line 2) to the user name used to authenticate to GEE at the beginning of this workflow in order for the export to succeed. The user must also change the `gee_asset_folder` within this Jupyter Notebook (cell 9, line 3) to a valid GEE Assets folder within the user account. A user may (optional) change the name of the output files, `vegetation_change_primary` and `vegetation_change_secondary` (cell 26, lines 2 and 3 in this Jupyter Notebook).
The workflow implements a check for the existence of the file path and output file names prior to exporting and skips the export if a file with the specified file name already exists.
```
# Define output GEE Asset names
change_primary_asset_name = f'users/{gee_username}/{gee_asset_folder}/vegetation_change_primary'
change_secondary_asset_name = f'users/{gee_username}/{gee_asset_folder}/vegetation_change_secondary'
# Check if GEE Asset already exists prior to export; primary change
if (change_primary_asset := ee.FeatureCollection(change_primary_asset_name)):
# Skip export
print(
f"GEE Asset ID '{change_primary_asset_name}' already exists. Skipping export...")
else:
# Export vectors to GEE Asset
change_primary_export = vc.export_vector(
vector=change_primary_vector,
description='Primary Change',
output_name=change_primary_asset_name,
output_method='asset')
# Check if GEE Asset already exists prior to export; secondary change
if (change_secondary_asset := ee.FeatureCollection(change_secondary_asset_name)):
# Skip export
print(
f"GEE Asset ID '{change_secondary_asset_name}' already exists. Skipping export...")
else:
# Export vectors to GEE Asset
change_secondary_export = vc.export_vector(
vector=change_secondary_vector,
description='Secondary Change',
output_name=change_secondary_asset_name,
output_method='asset')
```
## Data Visualization
The workflow uses a function from the `geemap` package, `Map()`, to create an interactive map that displays layers in the analysis.
The workflow uses the a function from the `geemap` package, `addLayer()`, to add all layers from the analysis to the interactive map, to include:
* Study area vector files;
* Pre and post-change imagery (reg/green/blue and color-infrared);
* NDVI difference image;
* Classified clusters (rasters);
* Classified clusters (vectors) from the raster to vector conversion; and,
* Classified clusters (vectors) imported from the GEE Assets, which the workflow exported during the Data Export step.
The `vegetation_change.py` script defines the display of each layer with visualization parameters, some with single colors (vector layers, classified/extracted features), and others with pre-defined visualization parameters, to include imagery (red/gree/blue and color-infrared) and NDVI difference (continuous and discrete color ramps). The `vegetation_change.py` script defines the visualization parameters in the following locations:
* Reg/Green/Blue
* Variable: `vis_params_rgb`
* Location: Lines 390-394
* Color Infrared
* Variable: `vis_params_cir`
* Location: Lines 397-401
* NDVI
* Variable: `vis_params_ndvi`
* Location: Lines 404-408
* NDVI Difference (Continuous)
* Variable: `vis_params_ndvi_diff`
* Location: Lines 411-415
* NDVI Difference (Discrete)
* Variable: `vis_params_ndvi_diff_sld`
* Location: Lines 418-428
```
# Create map for visualization
vegetation_change_map = gm.Map()
vegetation_change_map.setOptions('SATELLITE')
# Center map to study area
vegetation_change_map.setCenter(-75.0978, 3.7722, 12)
# Add pre-change and post-change images to map, RGB and CIR
vegetation_change_map.addLayer(
peak_green,
vc.vis_params_rgb,
'Landsat 8 - RGB - 2017 - Semester A - Peak Green - Pre-Change')
vegetation_change_map.addLayer(
post_harvest,
vc.vis_params_rgb,
'Landsat 8 - RGB - 2017 - Semester A - Post Harvest - Post-Change')
vegetation_change_map.addLayer(
peak_green,
vc.vis_params_cir,
'Landsat 8 - CIR - 2017 - Semester A - Peak Green - Pre-Change')
vegetation_change_map.addLayer(
post_harvest,
vc.vis_params_cir,
'Landsat 8 - CIR - 2017 - Semester A - Post Harvest - Post-Change')
# Add NDVI difference to map, continuous and discrete
vegetation_change_map.addLayer(
ndvi_diff,
vc.vis_params_ndvi_diff,
"NDVI Difference - Continuous - 2017 - Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
vegetation_change_map.addLayer(
ndvi_diff.sldStyle(vc.vis_params_ndvi_diff_sld),
{},
'NDVI Difference - Discrete - 2017 - Semester A - Peak Green to Post-Harvest - Pre to Post-Change')
# Add classified/extracted rasters (primary and secondary)
vegetation_change_map.addLayer(
change_features.get('primary'),
{'palette': ['green']},
"Classified Clusters - Raster - Primary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
vegetation_change_map.addLayer(
change_features.get('secondary'),
{'palette': ['lightgreen']},
"Classified Clusters - Raster - Secondary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
# Add classified/extracted vectors (from internal workflow)
vegetation_change_map.addLayer(
change_primary_vector,
{'color': 'green'},
"Classified Clusters - Vector - Primary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
vegetation_change_map.addLayer(
change_secondary_vector,
{'color': 'lightgreen'},
"Classified Clusters - Vector - Secondary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
# Add classified/extracted vectors (from GEE Asset export)
vegetation_change_map.addLayer(
change_primary_asset,
{'color': 'green'},
"Classified Clusters - GEE Asset - Primary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
vegetation_change_map.addLayer(
change_secondary_asset,
{'color': 'lightgreen'},
"Classified Clusters - GEE Asset - Secondary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
# Add study area boundary and canals to map
empty = ee.Image().byte()
study_area_boundary_vis = empty.paint(
featureCollection=study_area_boundary, color=1, width=3)
study_area_canals_vis = empty.paint(
featureCollection=study_area_canals, color=1, width=3)
vegetation_change_map.addLayer(
study_area_boundary_vis,
{'palette': 'FF0000'},
'Study Area - Boundary')
vegetation_change_map.addLayer(
study_area_canals_vis,
{'palette': 'blue'},
'Study Area - Canals')
```
The workflow displays the interative map that contains all layers used in the analysis, to include:
* Study area vector files;
* Pre and post-change imagery (reg/green/blue and color-infrared);
* NDVI difference image;
* Classified clusters (rasters);
* Classified clusters (vectors) from the raster to vector conversion; and,
* Classified clusters (vectors) imported from the GEE Assets, which the workflow exported during the Data Export step.
```
# Display map
vegetation_change_map
```
|
github_jupyter
|
# Import packages
import ee
import geemap as gm
import vegetation_change as vc
# Initialze GEE Python API; authenticate if necessary
try:
ee.Initialize()
except Exception as error:
ee.Authenticate()
ee.Initialize()
# Define output folder (GEE username + folder location)
gee_username = "calekochenour"
gee_asset_folder = "vegetation-change"
# Create FeatureCollections for study area
study_area_boundary = ee.FeatureCollection(
"users/calekochenour/vegetation-change/drtt_study_area_boundary")
study_area_canals = ee.FeatureCollection(
"users/calekochenour/vegetation-change/drtt_study_area_canals")
# Load and clip imagery for 2017 Semester A growing season
peak_green = ee.Image(
'LANDSAT/LC08/C01/T1_SR/LC08_008057_20170602').clip(study_area_boundary)
post_harvest = ee.Image(
'LANDSAT/LC08/C01/T1_SR/LC08_008057_20170906').clip(study_area_boundary)
# Create ImageCollection for Peak Green and Post-Harvest
collection = ee.ImageCollection([peak_green, post_harvest])
# Compute NDVI difference raster for Peak Green to Post-Harvest
ndvi_diff = vc.ndvi_diff_landsat8(collection, 1, 0)
# Define NDVI thresholds for classification;
# indices 0/1 identify min/max for primary class;
# indices 2/3 identify min/max for secondary class
ndvi_change_thresholds = [-2.0, -0.5, -0.5, -0.35]
# Segment, classify, and extract features
change_features = vc.segment_snic(
ndvi_diff, study_area_boundary, ndvi_change_thresholds)
# Convert rasters to vectors
change_primary_vector = vc.raster_to_vector(
change_features.get('primary'), study_area_boundary)
change_secondary_vector = vc.raster_to_vector(
change_features.get('secondary'), study_area_boundary)
# Define output GEE Asset names
change_primary_asset_name = f'users/{gee_username}/{gee_asset_folder}/vegetation_change_primary'
change_secondary_asset_name = f'users/{gee_username}/{gee_asset_folder}/vegetation_change_secondary'
# Check if GEE Asset already exists prior to export; primary change
if (change_primary_asset := ee.FeatureCollection(change_primary_asset_name)):
# Skip export
print(
f"GEE Asset ID '{change_primary_asset_name}' already exists. Skipping export...")
else:
# Export vectors to GEE Asset
change_primary_export = vc.export_vector(
vector=change_primary_vector,
description='Primary Change',
output_name=change_primary_asset_name,
output_method='asset')
# Check if GEE Asset already exists prior to export; secondary change
if (change_secondary_asset := ee.FeatureCollection(change_secondary_asset_name)):
# Skip export
print(
f"GEE Asset ID '{change_secondary_asset_name}' already exists. Skipping export...")
else:
# Export vectors to GEE Asset
change_secondary_export = vc.export_vector(
vector=change_secondary_vector,
description='Secondary Change',
output_name=change_secondary_asset_name,
output_method='asset')
# Create map for visualization
vegetation_change_map = gm.Map()
vegetation_change_map.setOptions('SATELLITE')
# Center map to study area
vegetation_change_map.setCenter(-75.0978, 3.7722, 12)
# Add pre-change and post-change images to map, RGB and CIR
vegetation_change_map.addLayer(
peak_green,
vc.vis_params_rgb,
'Landsat 8 - RGB - 2017 - Semester A - Peak Green - Pre-Change')
vegetation_change_map.addLayer(
post_harvest,
vc.vis_params_rgb,
'Landsat 8 - RGB - 2017 - Semester A - Post Harvest - Post-Change')
vegetation_change_map.addLayer(
peak_green,
vc.vis_params_cir,
'Landsat 8 - CIR - 2017 - Semester A - Peak Green - Pre-Change')
vegetation_change_map.addLayer(
post_harvest,
vc.vis_params_cir,
'Landsat 8 - CIR - 2017 - Semester A - Post Harvest - Post-Change')
# Add NDVI difference to map, continuous and discrete
vegetation_change_map.addLayer(
ndvi_diff,
vc.vis_params_ndvi_diff,
"NDVI Difference - Continuous - 2017 - Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
vegetation_change_map.addLayer(
ndvi_diff.sldStyle(vc.vis_params_ndvi_diff_sld),
{},
'NDVI Difference - Discrete - 2017 - Semester A - Peak Green to Post-Harvest - Pre to Post-Change')
# Add classified/extracted rasters (primary and secondary)
vegetation_change_map.addLayer(
change_features.get('primary'),
{'palette': ['green']},
"Classified Clusters - Raster - Primary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
vegetation_change_map.addLayer(
change_features.get('secondary'),
{'palette': ['lightgreen']},
"Classified Clusters - Raster - Secondary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
# Add classified/extracted vectors (from internal workflow)
vegetation_change_map.addLayer(
change_primary_vector,
{'color': 'green'},
"Classified Clusters - Vector - Primary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
vegetation_change_map.addLayer(
change_secondary_vector,
{'color': 'lightgreen'},
"Classified Clusters - Vector - Secondary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
# Add classified/extracted vectors (from GEE Asset export)
vegetation_change_map.addLayer(
change_primary_asset,
{'color': 'green'},
"Classified Clusters - GEE Asset - Primary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
vegetation_change_map.addLayer(
change_secondary_asset,
{'color': 'lightgreen'},
"Classified Clusters - GEE Asset - Secondary Change - 2017 Semester A - Peak Green to Post-Harvest - Pre to Post-Change")
# Add study area boundary and canals to map
empty = ee.Image().byte()
study_area_boundary_vis = empty.paint(
featureCollection=study_area_boundary, color=1, width=3)
study_area_canals_vis = empty.paint(
featureCollection=study_area_canals, color=1, width=3)
vegetation_change_map.addLayer(
study_area_boundary_vis,
{'palette': 'FF0000'},
'Study Area - Boundary')
vegetation_change_map.addLayer(
study_area_canals_vis,
{'palette': 'blue'},
'Study Area - Canals')
# Display map
vegetation_change_map
| 0.722135 | 0.976758 |
# Step 13: Create knowledge-graph-from-topic-model

|**[Overview](#Overview)** |**[Prior-steps](#Prior-steps)**|**[How-to-use](#How-to-use)**|**[Next-steps](#Next-steps)**|**[Postscript](#Postscript)**|**[Acknowledgements](#Acknowledgments)|
# Overview
Step 12 has identified two topics, each characterised by certain words. Now we use the knowledge graph to understand the business domain covered by the document library we are looking at. In consultation with the portfolio stakeholders, and domain experts, we can modify the knowledge-graph to reflect a sensible way of handling the business domain.
The immediate output is either:
- a knowledge graph, which can handle multiple views or facets reflecting different stakeholder understanding
or
- tree data structure, cut down from the knowledge graph, that reflects one convergent, dominating knowledge structure suitable for organising portfolio services in this business domain.
# Installation
At this point you will need Neo4j (or you can do this in YEd or Gephi).
# Prior-steps
Step 5 which provides records for the whole library
Step 12 which creates a topic model.
# How-to-use
## Open Neo4j
# How-to-use
## Open Neo4j
```
#hide
#Use either Neo4j Desktop, or create a Neo4j sandbox.
#Change security settings for APOC. Where you do this is different between v 3.5 and 4
apoc.import.file.enabled=true
apoc.export.file.enabled=true
#See https://neo4j.com/docs/labs/apoc/current/import/graphml
```
# Start with Topic 0: Supply chain and security
The prime words are
0 security
1 official
2 purchaser
3 chain
4 suppliers
5 supply
6 supplier
7 commissioning
8 dutyholder’s
9 resilience
## Topic 1: Decommissioning & Safety
The prime topic words
0 waste
1 decommissioning
2 radioactive
3 licensee
4 alarp
5 psa
6 change
7 project
8 licensee’s
## Sketch obvious relationships
Each topic was sketched on paper in a way that made sense at the time, making obvious links between the prime topic words
## Create two graphs in Neo4j from the sketches
One way of entry is to use the [arrows tool](xx).
Otherwise:
1. add in nodes with CREATE (m {id: 'title'}) etc
2. add in relations with MATCH (m {id:'Decommission'}),(n {id:'ALARP'}) CREATE (m)-[r:RELATED]->(n) etc.
3. add in a generic label for all with : 'MATCH (n) SET n:topics RETURN n.name, labels(n) AS labels
4. Select 'topics' label and adjust to show id in the visualisation
5. After checking its all there.
Stand back and look for insight. At this stage, we will assume it looks okay.
CALL apoc.export.graphml.all("Topic-graph.graphml", {})
Take the graph out from the folder area, and copy it into interim results
|
github_jupyter
|
#hide
#Use either Neo4j Desktop, or create a Neo4j sandbox.
#Change security settings for APOC. Where you do this is different between v 3.5 and 4
apoc.import.file.enabled=true
apoc.export.file.enabled=true
#See https://neo4j.com/docs/labs/apoc/current/import/graphml
| 0.349755 | 0.932392 |
<table class="ee-notebook-buttons" align="left">
<td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/Array/linear_regression.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
<td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Array/linear_regression.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
<td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Array/linear_regression.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
</table>
## Install Earth Engine API and geemap
Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://geemap.org). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
```
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('Installing geemap ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
import ee
import geemap
```
## Create an interactive map
The default basemap is `Google Maps`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/basemaps.py) can be added using the `Map.add_basemap()` function.
```
Map = geemap.Map(center=[40,-100], zoom=4)
Map
```
## Add Earth Engine Python script
```
# Add Earth Engine dataset
# Simple regression of year versus NDVI.
# Define the start date and position to get images covering Montezuma Castle,
# Arizona, from 2000-2010.
start = '2000-01-01'
end = '2010-01-01'
lng = -111.83533
lat = 34.57499
region = ee.Geometry.Point(lng, lat)
# Filter to Landsat 7 images in the given time and place, filter to a regular
# time of year to avoid seasonal affects, and for each image create the bands
# we will regress on:
# 1. A 1, so the resulting array has a column of ones to capture the offset.
# 2. Fractional year past 2000-01-01.
# 3. NDVI.
def addBand(image):
date = ee.Date(image.get('system:time_start'))
yearOffset = date.difference(ee.Date(start), 'year')
ndvi = image.normalizedDifference(['B4', 'B3'])
return ee.Image(1).addBands(yearOffset).addBands(ndvi).toDouble()
images = ee.ImageCollection('LANDSAT/LE07/C01/T1') \
.filterDate(start, end) \
.filter(ee.Filter.dayOfYear(160, 240)) \
.filterBounds(region) \
.map(addBand)
# date = ee.Date(image.get('system:time_start'))
# yearOffset = date.difference(ee.Date(start), 'year')
# ndvi = image.normalizedDifference(['B4', 'B3'])
# return ee.Image(1).addBands(yearOffset).addBands(ndvi).toDouble()
# })
# Convert to an array. Give the axes names for more readable code.
array = images.toArray()
imageAxis = 0
bandAxis = 1
# Slice off the year and ndvi, and solve for the coefficients.
x = array.arraySlice(bandAxis, 0, 2)
y = array.arraySlice(bandAxis, 2)
fit = x.matrixSolve(y)
# Get the coefficient for the year, effectively the slope of the long-term
# NDVI trend.
slope = fit.arrayGet([1, 0])
Map.setCenter(lng, lat, 12)
Map.addLayer(slope, {'min': -0.03, 'max': 0.03}, 'Slope')
```
## Display Earth Engine data layers
```
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
```
|
github_jupyter
|
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('Installing geemap ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
import ee
import geemap
Map = geemap.Map(center=[40,-100], zoom=4)
Map
# Add Earth Engine dataset
# Simple regression of year versus NDVI.
# Define the start date and position to get images covering Montezuma Castle,
# Arizona, from 2000-2010.
start = '2000-01-01'
end = '2010-01-01'
lng = -111.83533
lat = 34.57499
region = ee.Geometry.Point(lng, lat)
# Filter to Landsat 7 images in the given time and place, filter to a regular
# time of year to avoid seasonal affects, and for each image create the bands
# we will regress on:
# 1. A 1, so the resulting array has a column of ones to capture the offset.
# 2. Fractional year past 2000-01-01.
# 3. NDVI.
def addBand(image):
date = ee.Date(image.get('system:time_start'))
yearOffset = date.difference(ee.Date(start), 'year')
ndvi = image.normalizedDifference(['B4', 'B3'])
return ee.Image(1).addBands(yearOffset).addBands(ndvi).toDouble()
images = ee.ImageCollection('LANDSAT/LE07/C01/T1') \
.filterDate(start, end) \
.filter(ee.Filter.dayOfYear(160, 240)) \
.filterBounds(region) \
.map(addBand)
# date = ee.Date(image.get('system:time_start'))
# yearOffset = date.difference(ee.Date(start), 'year')
# ndvi = image.normalizedDifference(['B4', 'B3'])
# return ee.Image(1).addBands(yearOffset).addBands(ndvi).toDouble()
# })
# Convert to an array. Give the axes names for more readable code.
array = images.toArray()
imageAxis = 0
bandAxis = 1
# Slice off the year and ndvi, and solve for the coefficients.
x = array.arraySlice(bandAxis, 0, 2)
y = array.arraySlice(bandAxis, 2)
fit = x.matrixSolve(y)
# Get the coefficient for the year, effectively the slope of the long-term
# NDVI trend.
slope = fit.arrayGet([1, 0])
Map.setCenter(lng, lat, 12)
Map.addLayer(slope, {'min': -0.03, 'max': 0.03}, 'Slope')
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
| 0.593845 | 0.978031 |
# 6. Advanced Visualisation tools

Hello and welcome to session 6 of the Visual Analytics with Python and Power BI, hosted by the DataKirk. Once again, this session is about visualisation tools available in Python, with the module Matplotlib.
If you're working on the server at https://jupyterhub.thedatakirk.org.uk/ then all the relevant libraries (Matplotlib, Pandas etc) should already be installed and ready to use. However, if you're running the code on your own computer (which we do advise at this point as it will accelerate your learning!) then you'll need to make sure that you have installed them. This can be done by opening up a command prompt and typing
```
pip install matplotlib pandas
```
To check whether you have these libraries installed, run the cell below.
```
import pandas as pd
import matplotlib.pyplot as plt
```
As long as you get no errors, you are good to go! Once the libraries are imported, it's also a good idea to run the line in the cell below:
```
%matplotlib notebook
```
The will make all Matplotlib plots interactive so you can pan, zoom and move around within the figure. Once you have run this cell you should see these options at the bottom of every plot:

If you don't see them when you begin plotting, try going back and running the cell above again.
Here are some useful links for the session:
1. List of matplotlib colours: https://matplotlib.org/3.1.0/gallery/color/named_colors.html
2. Google colour picker: https://www.google.com/search?q=color+picker
Like last time, this notebook is more or less a blank canvas for you to experiment with different visualisations. Here, you will find the code to load in various different pandas dataframes. It is you decision how you would like to proceed from there. Some tips:
1. Always start each new visualisation with `plt.figure()`
2. To plot a single column against the dataframe index, try `plt.plot(df.index, df['column name])`.
3. Two columns can be scattered against each other via `plt.scatter(df['col 1'], df['col 2'])`
# 1. Coronavirus data UK
The first dataset contains information about the spread of coronavirus in Scotland, England, Wales and Northern Ireland.
Potential analysis suggestions:
1. To what extent are caseloads correlated amongst the four nations?
2. What aspects are the same and different about the first and second wave?
3. How are the different nations comparing in the fight against covid?
```
data_sco = pd.read_csv('../5. Examples of Visual Analytics in Python/data/covid/Corona_Scot.csv', index_col=0, parse_dates=True)
data_wal = pd.read_csv('../5. Examples of Visual Analytics in Python/data/covid/Corona_Wales.csv', index_col=0, parse_dates=True)
data_eng = pd.read_csv('../5. Examples of Visual Analytics in Python/data/covid/Corona_Eng.csv', index_col=0, parse_dates=True)
data_nir = pd.read_csv('../5. Examples of Visual Analytics in Python/data/covid/Corona_NI.csv', index_col=0, parse_dates=True)
```
# 2. Stock price data
The second dataset contains stock price information since 2012 for the largest 20 companies listed in the United Kingdom:
Potential analysis suggestions:
1. How has coronavirus affected the stock market in the UK?
2. How correlated are stocks in the UK?
3. Which stocks are the biggest winners and the biggest losers in the last 8 years?
```
price_data = pd.read_csv('../5. Examples of Visual Analytics in Python/data/stocks/FTSE_stock_prices.csv', index_col='Date', parse_dates=True)
company_info = pd.read_csv('../5. Examples of Visual Analytics in Python/data/stocks/companies.csv')
```
# 3. Income, Inequality and Environment
The third dataset contains annual data for GDP, inequality and carbon emissions for 192 countries around the world.
Potential analysis suggestions:
1. What is the relation between GDP and carbon emissions?
2. What trends in time can you identify?
3. Is there a relation between carbon emissions and inequality?
Helpful hint:
Sometimes is can be helpful to set the scale of an axis to logarithmic - this can be done by calling
```python
plt.xscale('log')
```
or
```python
plt.yscale('log')
```
```
population = pd.read_csv('../5. Examples of Visual Analytics in Python/data/national/population.csv', index_col=0, parse_dates=True)
co2_per_cap = pd.read_csv('../5. Examples of Visual Analytics in Python/data/national/co2_emissions_tonnes_per_person.csv', index_col=0, parse_dates=True)
gdp_per_cap = pd.read_csv('../5. Examples of Visual Analytics in Python/data/national/gdppercapita_us_inflation_adjusted.csv', index_col=0, parse_dates=True)
inequality_metric = pd.read_csv('../5. Examples of Visual Analytics in Python/data/national/gini.csv', index_col=0, parse_dates=True)
```
## UK Geographical Data
The final dataset contains the elevation profile of the UK and the coordinates of around 50 of the most populated cities.
Potential analysis:
1. Can you use `imshow` to view the elevation profile?
2. Where are the population centres of the UK?
3. Can you scatter the cities over the elevation profile?
```
cities = pd.read_csv('data/UK_cities.csv', index_col=0)
elevation = pd.read_csv('data/UK_elevation.csv', index_col=0)
```
|
github_jupyter
|
pip install matplotlib pandas
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib notebook
data_sco = pd.read_csv('../5. Examples of Visual Analytics in Python/data/covid/Corona_Scot.csv', index_col=0, parse_dates=True)
data_wal = pd.read_csv('../5. Examples of Visual Analytics in Python/data/covid/Corona_Wales.csv', index_col=0, parse_dates=True)
data_eng = pd.read_csv('../5. Examples of Visual Analytics in Python/data/covid/Corona_Eng.csv', index_col=0, parse_dates=True)
data_nir = pd.read_csv('../5. Examples of Visual Analytics in Python/data/covid/Corona_NI.csv', index_col=0, parse_dates=True)
price_data = pd.read_csv('../5. Examples of Visual Analytics in Python/data/stocks/FTSE_stock_prices.csv', index_col='Date', parse_dates=True)
company_info = pd.read_csv('../5. Examples of Visual Analytics in Python/data/stocks/companies.csv')
plt.xscale('log')
plt.yscale('log')
population = pd.read_csv('../5. Examples of Visual Analytics in Python/data/national/population.csv', index_col=0, parse_dates=True)
co2_per_cap = pd.read_csv('../5. Examples of Visual Analytics in Python/data/national/co2_emissions_tonnes_per_person.csv', index_col=0, parse_dates=True)
gdp_per_cap = pd.read_csv('../5. Examples of Visual Analytics in Python/data/national/gdppercapita_us_inflation_adjusted.csv', index_col=0, parse_dates=True)
inequality_metric = pd.read_csv('../5. Examples of Visual Analytics in Python/data/national/gini.csv', index_col=0, parse_dates=True)
cities = pd.read_csv('data/UK_cities.csv', index_col=0)
elevation = pd.read_csv('data/UK_elevation.csv', index_col=0)
| 0.379608 | 0.985286 |
```
import sys
print(sys.version)
"""
Created on Jun 17 2020
@author: Neven Caplar
@contact: ncaplar@princeton.edu
1. Name and place the data in DATA_FOLDER. For example, on my system I have them at
/Users/nevencaplar/Documents/PFS/ReducedData/
2. (OPTIONAL)Next cell contains some extensions that I use that make life much easier when using jupyter notebook
Without them this notebook becomes reallllly huge and hard to deal with
These can be downloaded from https://github.com/ipython-contrib/jupyter_contrib_nbextensions
"""
%%javascript
try {
require(['base/js/utils'], function (utils) {
utils.load_extension('code_prettify/code_prettify');
utils.load_extension('collapsible_headings/main');
utils.load_extension('codefolding/edit');
utils.load_extension('codefolding/main');
utils.load_extension('execute_time/ExecuteTime');
utils.load_extension('toc2/main');
});
}
catch (err) {
console.log('toc2 load error:', err);
}
# make notebook nice and wide to fill the entire screen
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
DATA_FOLDER='/Users/nevencaplar/Documents/PFS/ReducedData/'
import numpy as np
#matplotlib
import matplotlib
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
matplotlib.rcParams.update({'font.size': 18})
%config InlineBackend.rc = {}
%matplotlib inline
%config IPython.matplotlib.backend = "retina"
```
# Init
```
# Extract data from /tigress/ncaplar/Data/Data_for_Brent_June_2020
# Specify dataset and arc
# dataset is one of [1,2,3,4,5]
# dataset = 0; not avaliable
# dataset = 1; F/3.2 stop, February 2019 data
# dataset = 2; F/2.8 stop, May 2019 data
# dataset = 3; F/2.5 stop, June 2019 data
# dataset =4,5; F=2.8 stop, taken in July 2019, reduced in August
# arc can be HgAr, Ne or Kr for dataset [2,4,5]
# arc can be HgAr or Ne for dataset [1,3]
# specify defocus
# one of the followign values
# defocus=['-4.0','-3.5','-3.0','-2.5','-2','-1.5','-1','-0.5','0','0.5','1','1.5','2','2.5','3.0','3.5','4']
arc='HgAr'
dataset=2
defocus_value='-3.0'
if dataset==1:
STAMPS_FOLDER=DATA_FOLDER+"Data_Feb_5/Stamps_cleaned/"
if dataset==2:
STAMPS_FOLDER=DATA_FOLDER+"Data_May_28/Stamps_cleaned/"
if dataset==3:
STAMPS_FOLDER=DATA_FOLDER+"Data_Jun_25/Stamps_cleaned/"
if dataset==4 or dataset==5:
STAMPS_FOLDER=DATA_FOLDER+"Data_Aug_14/Stamps_cleaned/"
defocus=['-4.0','-3.5','-3.0','-2.5','-2','-1.5','-1','-0.5','0','0.5','1','1.5','2','2.5','3.0','3.5','4']
if dataset==1:
# F/3.2 stop
if arc is not None:
if arc=="HgAr":
single_number_focus=11748
final_Arc=np.load(DATA_FOLDER+'Data_Feb_5/Dataframes/finalHgAr_Feb2019.pkl',allow_pickle=True)
elif arc=="Ne":
single_number_focus=11748+607
final_Arc=np.load(DATA_FOLDER+'Data_Feb_5/Dataframes/finalNe_Feb2019.pkl',allow_pickle=True)
if dataset==2:
# F/2.8 stop
if arc is not None:
if arc=="HgAr":
single_number_focus=17017+54
final_Arc=np.load(DATA_FOLDER+'Data_May_28/Dataframes/finalHgAr_Feb2020',allow_pickle=True)
if arc=="Ne":
single_number_focus=16292
final_Arc=np.load(DATA_FOLDER+'Data_May_28/Dataframes/finalNe_Feb2020',allow_pickle=True)
if arc=="Kr":
single_number_focus=17310+54
final_Arc=np.load(DATA_FOLDER+'Data_May_28/Dataframes/finalKr_Feb2020',allow_pickle=True)
if dataset==3:
# F/2.5 stop
if arc is not None:
if arc=="HgAr":
single_number_focus=19238+54
final_Arc=np.load(DATA_FOLDER+'Data_May_28/Dataframes/finalHgAr_May2019.pkl',allow_pickle=True)
elif arc=="Ne":
single_number_focus=19472
final_Arc=np.load(DATA_FOLDER+'Data_May_28/Dataframes/finalNe_May2019.pkl',allow_pickle=True)
if dataset==4 or dataset==5:
# F/2.8 stop
if arc is not None:
if arc=="HgAr":
single_number_focus=21346+54
final_Arc=np.load(DATA_FOLDER+'Data_Aug_14/Dataframes/finalHgAr_Feb2020',allow_pickle=True)
if arc=="Ne":
single_number_focus=21550+54
final_Arc=np.load(DATA_FOLDER+'Data_Aug_14/Dataframes/finalNe_Feb2020',allow_pickle=True)
if str(arc)=="Kr":
single_number_focus=21754+54
final_Arc=np.load(DATA_FOLDER+'Data_Aug_14/Dataframes/finalKr_Feb2020',allow_pickle=True)
defocus_modification=(defocus.index(defocus_value)-9)*6
obs=single_number_focus+defocus_modification
```
# Show data
```
# which spot do you wish to look at
single_number=48
# information about the spot
final_Arc.loc[single_number]
# load and show the stamps
sci_image =np.load(STAMPS_FOLDER+'sci'+str(obs)+str(single_number)+str(arc)+'_Stacked.npy')
mask_image =np.load(STAMPS_FOLDER+'mask'+str(obs)+str(single_number)+str(arc)+'_Stacked.npy')
var_image =np.load(STAMPS_FOLDER+'var'+str(obs)+str(single_number)+str(arc)+'_Stacked.npy')
plt.figure(figsize=(30,10))
plt.subplot(131)
plt.imshow(sci_image)
plt.subplot(132)
plt.imshow(mask_image)
plt.subplot(133)
plt.imshow(var_image)
```
|
github_jupyter
|
import sys
print(sys.version)
"""
Created on Jun 17 2020
@author: Neven Caplar
@contact: ncaplar@princeton.edu
1. Name and place the data in DATA_FOLDER. For example, on my system I have them at
/Users/nevencaplar/Documents/PFS/ReducedData/
2. (OPTIONAL)Next cell contains some extensions that I use that make life much easier when using jupyter notebook
Without them this notebook becomes reallllly huge and hard to deal with
These can be downloaded from https://github.com/ipython-contrib/jupyter_contrib_nbextensions
"""
%%javascript
try {
require(['base/js/utils'], function (utils) {
utils.load_extension('code_prettify/code_prettify');
utils.load_extension('collapsible_headings/main');
utils.load_extension('codefolding/edit');
utils.load_extension('codefolding/main');
utils.load_extension('execute_time/ExecuteTime');
utils.load_extension('toc2/main');
});
}
catch (err) {
console.log('toc2 load error:', err);
}
# make notebook nice and wide to fill the entire screen
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
DATA_FOLDER='/Users/nevencaplar/Documents/PFS/ReducedData/'
import numpy as np
#matplotlib
import matplotlib
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
matplotlib.rcParams.update({'font.size': 18})
%config InlineBackend.rc = {}
%matplotlib inline
%config IPython.matplotlib.backend = "retina"
# Extract data from /tigress/ncaplar/Data/Data_for_Brent_June_2020
# Specify dataset and arc
# dataset is one of [1,2,3,4,5]
# dataset = 0; not avaliable
# dataset = 1; F/3.2 stop, February 2019 data
# dataset = 2; F/2.8 stop, May 2019 data
# dataset = 3; F/2.5 stop, June 2019 data
# dataset =4,5; F=2.8 stop, taken in July 2019, reduced in August
# arc can be HgAr, Ne or Kr for dataset [2,4,5]
# arc can be HgAr or Ne for dataset [1,3]
# specify defocus
# one of the followign values
# defocus=['-4.0','-3.5','-3.0','-2.5','-2','-1.5','-1','-0.5','0','0.5','1','1.5','2','2.5','3.0','3.5','4']
arc='HgAr'
dataset=2
defocus_value='-3.0'
if dataset==1:
STAMPS_FOLDER=DATA_FOLDER+"Data_Feb_5/Stamps_cleaned/"
if dataset==2:
STAMPS_FOLDER=DATA_FOLDER+"Data_May_28/Stamps_cleaned/"
if dataset==3:
STAMPS_FOLDER=DATA_FOLDER+"Data_Jun_25/Stamps_cleaned/"
if dataset==4 or dataset==5:
STAMPS_FOLDER=DATA_FOLDER+"Data_Aug_14/Stamps_cleaned/"
defocus=['-4.0','-3.5','-3.0','-2.5','-2','-1.5','-1','-0.5','0','0.5','1','1.5','2','2.5','3.0','3.5','4']
if dataset==1:
# F/3.2 stop
if arc is not None:
if arc=="HgAr":
single_number_focus=11748
final_Arc=np.load(DATA_FOLDER+'Data_Feb_5/Dataframes/finalHgAr_Feb2019.pkl',allow_pickle=True)
elif arc=="Ne":
single_number_focus=11748+607
final_Arc=np.load(DATA_FOLDER+'Data_Feb_5/Dataframes/finalNe_Feb2019.pkl',allow_pickle=True)
if dataset==2:
# F/2.8 stop
if arc is not None:
if arc=="HgAr":
single_number_focus=17017+54
final_Arc=np.load(DATA_FOLDER+'Data_May_28/Dataframes/finalHgAr_Feb2020',allow_pickle=True)
if arc=="Ne":
single_number_focus=16292
final_Arc=np.load(DATA_FOLDER+'Data_May_28/Dataframes/finalNe_Feb2020',allow_pickle=True)
if arc=="Kr":
single_number_focus=17310+54
final_Arc=np.load(DATA_FOLDER+'Data_May_28/Dataframes/finalKr_Feb2020',allow_pickle=True)
if dataset==3:
# F/2.5 stop
if arc is not None:
if arc=="HgAr":
single_number_focus=19238+54
final_Arc=np.load(DATA_FOLDER+'Data_May_28/Dataframes/finalHgAr_May2019.pkl',allow_pickle=True)
elif arc=="Ne":
single_number_focus=19472
final_Arc=np.load(DATA_FOLDER+'Data_May_28/Dataframes/finalNe_May2019.pkl',allow_pickle=True)
if dataset==4 or dataset==5:
# F/2.8 stop
if arc is not None:
if arc=="HgAr":
single_number_focus=21346+54
final_Arc=np.load(DATA_FOLDER+'Data_Aug_14/Dataframes/finalHgAr_Feb2020',allow_pickle=True)
if arc=="Ne":
single_number_focus=21550+54
final_Arc=np.load(DATA_FOLDER+'Data_Aug_14/Dataframes/finalNe_Feb2020',allow_pickle=True)
if str(arc)=="Kr":
single_number_focus=21754+54
final_Arc=np.load(DATA_FOLDER+'Data_Aug_14/Dataframes/finalKr_Feb2020',allow_pickle=True)
defocus_modification=(defocus.index(defocus_value)-9)*6
obs=single_number_focus+defocus_modification
# which spot do you wish to look at
single_number=48
# information about the spot
final_Arc.loc[single_number]
# load and show the stamps
sci_image =np.load(STAMPS_FOLDER+'sci'+str(obs)+str(single_number)+str(arc)+'_Stacked.npy')
mask_image =np.load(STAMPS_FOLDER+'mask'+str(obs)+str(single_number)+str(arc)+'_Stacked.npy')
var_image =np.load(STAMPS_FOLDER+'var'+str(obs)+str(single_number)+str(arc)+'_Stacked.npy')
plt.figure(figsize=(30,10))
plt.subplot(131)
plt.imshow(sci_image)
plt.subplot(132)
plt.imshow(mask_image)
plt.subplot(133)
plt.imshow(var_image)
| 0.204978 | 0.459561 |
<a href="https://colab.research.google.com/github/JiaminJIAN/20MA573/blob/master/src/Finite_Difference_Method.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Finite Defference Method
## (1) Abstract
### 1. Goal:
The goal of this course is:
* Learn various types of the first order derivative approximation: FFD, BFD, CFD operators
* Understand convergence rate of operators
* learn python functions
### 2. Problem
Let $f(x) = \sin x$. Plot, with $h = .5$
- its explicit first order derivative $f'$,
- FFD $\delta_h f$,
- BFD $\delta_{-h}f$,
- and CFD $\delta_{\pm h}f$
### 3. Anal
Given a smooth function $f: \mathbb R \mapsto \mathbb R$, its derivative is
$$f'(x) = \lim_{h\to 0} \frac{f(x+h) - f(x)}{h}.$$
This means, if $h$ is small enough, then
$$f'(x) \simeq \frac{f(x+h) - f(x)}{h} := \delta_h f.$$
We call $\delta_h$ by Finite Difference (FD) operator. In particular,
- If $h>0$, then $\delta_h$ is Forward Finite Difference (FFD);
- If $h<0$, then $\delta_h$ is Backward Finite Difference (BFD);
- The average of FFD and BFD is Central Finite Difference (CFD), denoted by
$$\delta_{\pm h} f (x) := \frac 1 2 (\delta_h f (x) + \delta_{-h} f(x)) \simeq f'(x).$$
### 4. Definition(FFD, BFD and CFD)
The definition of **FFD** is as follow:
$$\delta_{h} f(x) = \frac{f(x+h) - f(x)}{h}, \quad h > 0;$$
and for **BFD**
$$\delta_{-h} f(x) = \frac{f(x-h) - f(x)}{-h} = \frac{f(x) - f(x-h)}{h}, \quad h > 0;$$
and then for **CFD**
$$\delta_{\pm h} f (x) = \frac{f(x+h) - f(x-h)}{2h}, \quad h>0.$$
### 5. Definition(Convergence):
Suppose there exists a sequence of number $X_{h}$ s.t.
$$\lim_{h \to 0} X_{h} = a, $$
then we say $X_h$ is convergence to a.
If
$$|X_{h} - a| < K h^{\alpha}$$
for some $K >0$, then we say $X_{h} \to a$ with order $\alpha$.
### 6. Proposition
- Both FFD and BFD has convergence order $1$; i.e.
$$|\delta_h f(x) - f'(x)| = O(h).$$
- CFD has convergence order $2$.
$$|\delta_{\pm h} f(x) - f'(x)| = O(h^2).$$
### 7. Exercise
Prove the above proposition.
**Proof:**
By the Taylor expansion, we have
$$f(x+h) = f(x) + f'(x)h + \frac{1}{2} f''(x) h^{2} + O(h^{3}),$$
and then
$$\delta_{h} (x) = f'(x) + \frac{1}{2} f''(x) h + O(h^{2}).$$
Since $f \in C^{2}$, the term $f''(x)$ is bounded. We have
$$|\delta_h f(x) - f'(x)| = |\frac{1}{2} f''(x) + O(h)|h \leq K h,$$
so FFD has convergence order 1. Similarly we know that BFD has convergence order 1. Using the $-h$ to subsititute the $h$, we have
$$\delta_{-h} (x) = f'(x) - \frac{1}{2} f''(x) h + O(h^{2}),$$
and
$$\delta_{\pm h} f (x) = f'(x) + O(h^{2}).$$
Then we have
$$|\delta_{\pm h} f(x) - f'(x)|= O(h^{2}).$$
## (2) Code for finite differentiation method
We shall import all needed packages first.
```
import numpy as np
import matplotlib.pyplot as plt
```
Math operators ffd, bfd, cfd will be defined here as python functions.
```
def ffd(f, x, h):
return (f(x+h) - f(x))/h
def bfd(f, x, h):
return (f(x) - f(x-h))/h
def cfd(f, x, h):
return (f(x+h) - f(x-h))/h/2
```
Next, for the original function $f(x) = \sin x$,
we shall plot its exact derivative
$$f'(x) = \cos x, $$
then, with $h = .5$, plot
- ffd $\delta_h f$,
- bfd $\delta_{-h}f$,
- and cfd $\delta_{\pm}f$
From the graph, it is obvious that cfd is the closest one to original $f'$.
```
h = .5 #step size
x_co = np.linspace(0, 2*np.pi, 100)
plt.plot(x_co, np.cos(x_co), label = 'cosine')
plt.plot(x_co, ffd(np.sin, x_co, h), label = 'FFD')
plt.plot(x_co, bfd(np.sin, x_co, h), label = 'BFD')
plt.plot(x_co, cfd(np.sin, x_co, h), label = 'CFD')
plt.legend()
```
## (3) Demonstrate the convergence rate
### 1. Problem
Let $f(x) = \sin x$. We shall demonstrate its FFD convergence rate being $1$.
### 2. Anal
Given a smooth function $f: \mathbb R \mapsto \mathbb R$, recall that FFD is defined by
$$f'(x) \simeq \frac{f(x+h) - f(x)}{h} := \delta_h f.$$
Moreover, FFD has convergence order $1$; i.e.
$$|\delta_h f(x) - f'(x)| = O(h) \simeq K h.$$
A usual approach to demonstrate the convergence rate $1$ is as follows. Let's denote the aboslute error term (the left hand side of the above equation) as
$\epsilon(h)$ and its convergence rate is $\alpha$, then the error term behaves as
$$\epsilon(h) \simeq K h^\alpha.$$
To demonstrate its convergence rate being $1$, we want to visualize $\alpha =1$. To proceed, we could compute $\epsilon(h)$ for the values
$$h \in \{2^{-n}: n = 5, 6, \ldots, 10\}.$$
Write
$$\epsilon_n = \epsilon(2^{-n}) \simeq K 2^{-n\alpha}.$$
Take $log_2$ both sides, we have
$$\log_2 \epsilon_n \simeq \log_2 K - \alpha \cdot n.$$
We can plot a $n$ vs $\ln \epsilon_n$ as $n$ varies from small number to a big number.
If the convergence analysis is correct, the plot shall show a line with slope $\alpha$.
### 3. Example:
Verify FFD convergence rate with at $\pi/3$ with $h = 2^{-n}$, where $n$ ranges from 5 to 10.
```
import numpy as np
import matplotlib.pyplot as plt
```
finite difference operators
```
def ffd(f, x, h):
return (f(x+h) - f(x))/h
def bfd(f, x, h):
return (f(x) - f(x-h))/h
def cfd(f, x, h):
return (f(x+h) - f(x-h))/h/2
x_target = np.pi/3 #target point to be examined
y_target = np.cos(x_target) #exact derivative value at the target point
nn = np.arange(5, 11)
hh = 1/np.power(2, nn) #step sizes to be taken
err = ffd(np.sin, x_target, hh) - y_target #errors corresponding to each step size
yy = np.log2(np.abs(err))
plt.plot(nn, yy)
import scipy.stats as ss
out = ss.linregress(nn,yy)
print('the convergence order is ' + str(-out[0]))
```
So, from the above code, we can see the FFD converdence rate is 1. For the CFD, we can do same thing as before.
```
err2 = cfd(np.sin, x_target, hh) - y_target #errors corresponding to each step size
yy2 = np.log2(np.abs(err2))
plt.plot(nn, yy2)
out2 = ss.linregress(nn,yy2)
print('the convergence order is ' + str(-out2[0]))
```
So we can see the CFD converdence rate is 2.
## (4) The second order derivative approximation by finite difference method
### 1. Abstract
- Goal:
- Learn the second order derivative approximation: second order central finite difference
- Understand convergence rate
### 2. Problem
Let $f(x) = \sin x$. Plot $f''$ and $\delta_{-h} \delta_h f$ with $h = .5$
### 3. Anal
One of the commonly used FD for $f''$ is the following:
$$f''(x) = \frac{d}{dx} f'(x) \simeq \delta_h f'(x) \simeq \delta_h \delta_{-h} f(x).$$
If we write it explicitly, then
$$f''(x) \simeq \frac{f(x+h) - 2 f(x) + f(x-h)}{h^2}.$$
__Prop__
The central finite difference for the second order has convergence order $2$.
__Proof__
For the second order central finite different, we have
$$\delta_{h} \delta_{-h} f(x) = \frac{f(x+h) - 2 f(x) + f(x-h)}{h^2}.$$
Recall the Taylor series expansion for $f(x + h)$ and $f(x-h)$ at $x$, when$f \in C^{4} (\mathbb{R})$ and $f^{(4)}(x) \neq 0$, we have
$$f(x+h) = f(x) + f'(x) h + \frac{1}{2} f''(x) h^{2} + \frac{1}{3!} f^{(3)}(x) h^{3} + O(h^{4}), $$
and
$$f(x-h) = f(x) - f'(x) h + \frac{1}{2} f''(x) h^{2} - \frac{1}{3!} f^{(3)}(x) h^{3} + O(h^{4}), $$
so we have
$$f(x+h) + f(x-h) -2f(x)= f''(x) h^{2} +O(h^{4}).$$
So by the definition of second order central finite different, we have
$$\delta_{h} \delta_{-h} f(x) = f''(x) + O(h^{2}),$$
and then
$$|\delta_{h} \delta_{-h} f(x) - f''(x)| = O(h^{2}).$$
By the definition of convergence order, we know that the central finite difference for the second order has convergence order $2$.
**A Commen**
Recall the general form of Taylor expansion, we have
$$f(x+h) = \sum_{k = 0}^{+ \infty} \frac{f^{(k)}(x)}{k!} h^{k},$$
and
$$f(x-h) = \sum_{k = 0}^{+ \infty} \frac{f^{(k)}(x)}{k!} (-h)^{k},$$
so we can get
$$f(x+h) + f(x-h) = \sum_{k = 0}^{+ \infty} \frac{f^{(k)}(x)}{k!} \Big(h^{k} + (-h)^{k} \Big) = 2 \sum_{n = 0}^{+ \infty} \frac{f^{(2n)}(x)}{(2n)!} h^{2n}.$$
Then we have
$$\delta_{h} \delta_{-h} f(x) - f''(x) = 2 \sum_{n = 2}^{+ \infty} \frac{f^{(2n)}(x)}{(2n)!} h^{2n-2}.$$
When $f^{(4)}(x) \neq 0$, we know that the central finite difference for the second order has convergence order 2. But when $f^{(4)} (x) = 0$ and there exists $m > 2$ and $f^{(2m)}(x) \neq 0$, the convergence rate will be more higher.
And consider a speical case $f(x) = sin(x)$, we know that
$$f^{(n)} (x) = sin(\frac{n \pi}{2} + x).$$
For $x = \pi$ and $k \in \mathbb{N}$, we have
$$f^{(2k)} (x) = sin(k \pi + x) = 0,$$
so we know that
$$\delta_{h} \delta_{-h} f(x) - f''(x) = 0.$$
### 4. Code
```
import numpy as np
import matplotlib.pyplot as plt
from pylab import plt
plt.style.use('seaborn')
%matplotlib inline
def sfd(f, x, h):
return (f(x+h) + f(x-h) - 2 * f(x)) / (h**2)
h = .5 #step size
x_co = np.linspace(0, 2*np.pi, 100)
plt.plot(x_co, - np.sin(x_co), label = '$-sin(x)$');
plt.plot(x_co, sfd(np.sin, x_co, h), label = 'sfd');
plt.legend();
```
plot log-log chart for the demonstration of convergence rate, find convergence order using linear regression.
```
x_target = np.pi/3 #target point to be examined
y_target = -np.sin(x_target) #exact derivative value at the target point
nn = np.arange(5, 11)
hh = 1/np.power(2, nn) #step sizes to be taken
err = sfd(np.sin, x_target, hh) - y_target #errors corresponding to each step size
yy = np.log2(np.abs(err))
plt.plot(nn, yy)
import scipy.stats as ss
out = ss.linregress(nn,yy)
print('the convergence order is ' + str(-out[0]))
```
We can change the point 𝑥 from 𝑥=𝜋/3 to 𝑥=𝜋 , then we can see that:
```
x_target = np.pi #target point to be examined
y_target = -np.sin(x_target) #exact derivative value at the target point
nn = np.arange(5, 11)
hh = 1/np.power(2, nn) #step sizes to be taken
err = sfd(np.sin, x_target, hh) - y_target #errors corresponding to each step size
yy = np.log2(np.abs(err))
plt.plot(nn, yy)
import scipy.stats as ss
out = ss.linregress(nn,yy)
print('the convergence order is ' + str(-out[0]))
```
The convergence order is a negative number, it is unreasonable. Such that in this phenomenon, we can use this method to measure the speed of convergence. One interpretation is when n is big enough, the bias between the estimator and target value is very small. In this condition the accuracy may not be improved.
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
def ffd(f, x, h):
return (f(x+h) - f(x))/h
def bfd(f, x, h):
return (f(x) - f(x-h))/h
def cfd(f, x, h):
return (f(x+h) - f(x-h))/h/2
h = .5 #step size
x_co = np.linspace(0, 2*np.pi, 100)
plt.plot(x_co, np.cos(x_co), label = 'cosine')
plt.plot(x_co, ffd(np.sin, x_co, h), label = 'FFD')
plt.plot(x_co, bfd(np.sin, x_co, h), label = 'BFD')
plt.plot(x_co, cfd(np.sin, x_co, h), label = 'CFD')
plt.legend()
import numpy as np
import matplotlib.pyplot as plt
def ffd(f, x, h):
return (f(x+h) - f(x))/h
def bfd(f, x, h):
return (f(x) - f(x-h))/h
def cfd(f, x, h):
return (f(x+h) - f(x-h))/h/2
x_target = np.pi/3 #target point to be examined
y_target = np.cos(x_target) #exact derivative value at the target point
nn = np.arange(5, 11)
hh = 1/np.power(2, nn) #step sizes to be taken
err = ffd(np.sin, x_target, hh) - y_target #errors corresponding to each step size
yy = np.log2(np.abs(err))
plt.plot(nn, yy)
import scipy.stats as ss
out = ss.linregress(nn,yy)
print('the convergence order is ' + str(-out[0]))
err2 = cfd(np.sin, x_target, hh) - y_target #errors corresponding to each step size
yy2 = np.log2(np.abs(err2))
plt.plot(nn, yy2)
out2 = ss.linregress(nn,yy2)
print('the convergence order is ' + str(-out2[0]))
import numpy as np
import matplotlib.pyplot as plt
from pylab import plt
plt.style.use('seaborn')
%matplotlib inline
def sfd(f, x, h):
return (f(x+h) + f(x-h) - 2 * f(x)) / (h**2)
h = .5 #step size
x_co = np.linspace(0, 2*np.pi, 100)
plt.plot(x_co, - np.sin(x_co), label = '$-sin(x)$');
plt.plot(x_co, sfd(np.sin, x_co, h), label = 'sfd');
plt.legend();
x_target = np.pi/3 #target point to be examined
y_target = -np.sin(x_target) #exact derivative value at the target point
nn = np.arange(5, 11)
hh = 1/np.power(2, nn) #step sizes to be taken
err = sfd(np.sin, x_target, hh) - y_target #errors corresponding to each step size
yy = np.log2(np.abs(err))
plt.plot(nn, yy)
import scipy.stats as ss
out = ss.linregress(nn,yy)
print('the convergence order is ' + str(-out[0]))
x_target = np.pi #target point to be examined
y_target = -np.sin(x_target) #exact derivative value at the target point
nn = np.arange(5, 11)
hh = 1/np.power(2, nn) #step sizes to be taken
err = sfd(np.sin, x_target, hh) - y_target #errors corresponding to each step size
yy = np.log2(np.abs(err))
plt.plot(nn, yy)
import scipy.stats as ss
out = ss.linregress(nn,yy)
print('the convergence order is ' + str(-out[0]))
| 0.481454 | 0.990821 |
```
import os
os.environ['CUDA_VISIBLE_DEVICES'] = ''
from malaya_speech.train.model import hubert, ctc
from malaya_speech.train.model.conformer.model import Model as ConformerModel
import malaya_speech
import tensorflow as tf
import numpy as np
import json
from glob import glob
import string
unique_vocab = [''] + list(
string.ascii_lowercase + string.digits
) + [' ']
len(unique_vocab)
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/language-model/bahasa-manglish-combined/model.trie.klm
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/ctc-decoder/ctc_decoders-1.0-cp36-cp36m-linux_x86_64.whl
# !pip3 install ctc_decoders-1.0-cp36-cp36m-linux_x86_64.whl
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/malay-test.tar.gz
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/singlish-test.tar.gz
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/mandarin-test.tar.gz
# !tar -zxf malay-test.tar.gz
# !tar -zxf singlish-test.tar.gz
# !tar -zxf mandarin-test.tar.gz
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/malay-test.json
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/singlish-test.json
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/mandarin-test.json
from glob import glob
malay = sorted(glob('malay-test/*.wav'), key = lambda x: int(x.split('/')[1].replace('.wav', '')))
singlish = sorted(glob('singlish-test/*.wav'), key = lambda x: int(x.split('/')[1].replace('.wav', '')))
mandarin = sorted(glob('mandarin-test/*.wav'), key = lambda x: int(x.split('/')[1].replace('.wav', '')))
len(malay), len(singlish), len(mandarin)
with open('malay-test.json') as fopen:
malay_label = json.load(fopen)
with open('singlish-test.json') as fopen:
singlish_label = json.load(fopen)
with open('mandarin-test.json') as fopen:
mandarin_label = json.load(fopen)
len(malay_label), len(singlish_label), len(mandarin_label)
from sklearn.utils import shuffle
audio = malay + singlish + mandarin
labels = malay_label + singlish_label + mandarin_label
audio, labels = shuffle(audio, labels)
test_set = list(zip(audio, labels))
test_set[:10]
from ctc_decoders import Scorer
from ctc_decoders import ctc_beam_search_decoder
n_mels = 80
sr = 16000
maxlen = 18
minlen_text = 1
def mp3_to_wav(file, sr = sr):
audio = AudioSegment.from_file(file)
audio = audio.set_frame_rate(sr).set_channels(1)
sample = np.array(audio.get_array_of_samples())
return malaya_speech.astype.int_to_float(sample), sr
def generate():
audios, cleaned_texts = audio, labels
for i in range(len(audios)):
try:
if audios[i].endswith('.mp3'):
wav_data, _ = mp3_to_wav(audios[i])
else:
wav_data, _ = malaya_speech.load(audios[i], sr = sr)
t = [unique_vocab.index(c) for c in cleaned_texts[i]]
yield {
'waveforms': wav_data,
'waveforms_length': [len(wav_data)],
'targets': t,
'targets_length': [len(t)],
}
except Exception as e:
print(e)
def get_dataset(
batch_size = 3,
shuffle_size = 20,
thread_count = 24,
maxlen_feature = 1800,
):
def get():
dataset = tf.data.Dataset.from_generator(
generate,
{
'waveforms': tf.float32,
'waveforms_length': tf.int32,
'targets': tf.int32,
'targets_length': tf.int32,
},
output_shapes = {
'waveforms': tf.TensorShape([None]),
'waveforms_length': tf.TensorShape([None]),
'targets': tf.TensorShape([None]),
'targets_length': tf.TensorShape([None]),
},
)
dataset = dataset.prefetch(tf.contrib.data.AUTOTUNE)
dataset = dataset.padded_batch(
batch_size,
padded_shapes = {
'waveforms': tf.TensorShape([None]),
'waveforms_length': tf.TensorShape([None]),
'targets': tf.TensorShape([None]),
'targets_length': tf.TensorShape([None]),
},
padding_values = {
'waveforms': tf.constant(0, dtype = tf.float32),
'waveforms_length': tf.constant(0, dtype = tf.int32),
'targets': tf.constant(0, dtype = tf.int32),
'targets_length': tf.constant(0, dtype = tf.int32),
},
)
return dataset
return get
dev_dataset = get_dataset()()
features = dev_dataset.make_one_shot_iterator().get_next()
features
training = True
class Encoder:
def __init__(self, config):
self.config = config
self.encoder = ConformerModel(**self.config)
def __call__(self, x, input_mask, training = True):
return self.encoder(x, training = training)
config_conformer = malaya_speech.config.conformer_large_encoder_config
config_conformer['subsampling']['type'] = 'none'
config_conformer['dropout'] = 0.0
encoder = Encoder(config_conformer)
cfg = hubert.HuBERTConfig(
extractor_mode='layer_norm',
dropout=0.0,
attention_dropout=0.0,
encoder_layerdrop=0.0,
dropout_input=0.0,
dropout_features=0.0,
final_dim=768,
)
model = hubert.Model(cfg, encoder, ['pad', 'eos', 'unk'] + [str(i) for i in range(100)])
X = features['waveforms']
X_len = features['waveforms_length'][:, 0]
r = model(X, padding_mask = X_len, features_only = True, mask = False)
logits = tf.layers.dense(r['x'], len(unique_vocab) + 1)
log_probs = tf.nn.log_softmax(logits)
seq_lens = tf.reduce_sum(
tf.cast(tf.logical_not(r['padding_mask']), tf.int32), axis = 1
)
logits = tf.transpose(logits, [1, 0, 2])
logits = tf.identity(logits, name = 'logits')
seq_lens = tf.identity(seq_lens, name = 'seq_lens')
# decoded = tf.nn.ctc_beam_search_decoder(
# logits,
# seq_lens,
# beam_width = beam_size,
# top_paths = 1,
# merge_repeated = True)[0][0]
# decoded._indices, decoded._values
logits, seq_lens, log_probs
decoded = tf.nn.ctc_beam_search_decoder(logits, seq_lens, beam_width=10, top_paths=1, merge_repeated=True)
preds = tf.sparse.to_dense(tf.to_int32(decoded[0][0]))
preds = tf.identity(preds, 'preds')
log_probs = tf.identity(log_probs, 'log_probs')
preds, log_probs
sess = tf.Session()
sess.run(tf.global_variables_initializer())
var_list = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES)
saver = tf.train.Saver(var_list = var_list)
saver.restore(sess, 'hubert-conformer-large-3mixed-ctc/model.ckpt-1800000')
import six
import string
from typing import List
def decode(ids, lookup: List[str] = None):
"""
Decode integer representation to string based on ascii table or lookup variable.
Parameters
-----------
ids: List[int]
lookup: List[str], optional (default=None)
list of unique strings.
Returns
--------
result: str
"""
decoded_ids = []
int2byte = six.int2byte
for id_ in ids:
if lookup:
decoded_ids.append(lookup[id_])
else:
decoded_ids.append(
int2byte(id_ - NUM_RESERVED_TOKENS).decode('utf-8')
)
return ''.join(decoded_ids)
# %%time
# kenlm_model = kenlm.Model('model.trie.klm')
# decoder = build_ctcdecoder(
# unique_vocab + ['_'],
# kenlm_model,
# alpha=0.1,
# beta=3.0,
# ctc_token_idx=len(unique_vocab)
# )
%%time
from pyctcdecode import build_ctcdecoder
import kenlm
kenlm_model = kenlm.Model('model.trie.klm')
decoder = build_ctcdecoder(
unique_vocab + ['_'],
kenlm_model,
alpha=0.2,
beta=1.0,
ctc_token_idx=len(unique_vocab)
)
scorer = Scorer(0.5, 1.0, 'model.trie.klm', unique_vocab)
logits_t = tf.nn.softmax(tf.transpose(logits, [1, 0, 2]))
# r = sess.run([preds, logits_t, seq_lens, features['targets']])
# out = decoder2.decode_beams(r[1][1,:r[2][1]],
# prune_history=True)
# text, lm_state, timesteps, logit_score, lm_score = out[0]
# text, ctc_beam_search_decoder(r[1][1,:r[2][1]], unique_vocab, 20, ext_scoring_func = scorer)[0][1]
# out = decoder.decode_beams(np.pad(r[1][:,0], [[0,0], [1,0]], constant_values = -13.0),
# prune_history=True)
# text, lm_state, timesteps, logit_score, lm_score = out[0]
# text
# out = decoder2.decode_beams(np.pad(r[1][:,0], [[0,0], [1,0]], constant_values = -13.0),
# prune_history=True)
# text, lm_state, timesteps, logit_score, lm_score = out[0]
# text
# decode(r[0][1], unique_vocab), decode(r[-1][1], unique_vocab)
# %%time
# ctc_beam_search_decoder(r[1][0,:r[2][0]], unique_vocab, 20, ext_scoring_func = scorer)[0][1]
# %%time
# ctc_beam_search_decoder(r[1][1,:r[2][1]], unique_vocab, 20, ext_scoring_func = scorer)[0]
from malaya_speech.utils import metrics, char
wer, cer, wer_lm, cer_lm = [], [], [], []
wer_lm2, cer_lm2 = [], []
index = 0
while True:
try:
r = sess.run([preds, logits_t, seq_lens, features['targets']])
for no, row in enumerate(r[0]):
d = decode(row, lookup = unique_vocab).replace('<PAD>', '')
t = decode(r[-1][no], lookup = unique_vocab).replace('<PAD>', '')
wer.append(malaya_speech.metrics.calculate_wer(t, d))
cer.append(malaya_speech.metrics.calculate_cer(t, d))
d_lm = ctc_beam_search_decoder(r[1][no,:r[2][no]],
unique_vocab, 20, ext_scoring_func = scorer)[0][1]
wer_lm.append(malaya_speech.metrics.calculate_wer(t, d_lm))
cer_lm.append(malaya_speech.metrics.calculate_cer(t, d_lm))
out = decoder.decode_beams(r[1][no,:r[2][no]], prune_history=True)
d_lm2, lm_state, timesteps, logit_score, lm_score = out[0]
wer_lm2.append(malaya_speech.metrics.calculate_wer(t, d_lm2))
cer_lm2.append(malaya_speech.metrics.calculate_cer(t, d_lm2))
index += 1
except Exception as e:
break
np.mean(wer), np.mean(cer), np.mean(wer_lm), np.mean(cer_lm), np.mean(wer_lm2), np.mean(cer_lm2)
```
|
github_jupyter
|
import os
os.environ['CUDA_VISIBLE_DEVICES'] = ''
from malaya_speech.train.model import hubert, ctc
from malaya_speech.train.model.conformer.model import Model as ConformerModel
import malaya_speech
import tensorflow as tf
import numpy as np
import json
from glob import glob
import string
unique_vocab = [''] + list(
string.ascii_lowercase + string.digits
) + [' ']
len(unique_vocab)
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/language-model/bahasa-manglish-combined/model.trie.klm
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/ctc-decoder/ctc_decoders-1.0-cp36-cp36m-linux_x86_64.whl
# !pip3 install ctc_decoders-1.0-cp36-cp36m-linux_x86_64.whl
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/malay-test.tar.gz
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/singlish-test.tar.gz
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/mandarin-test.tar.gz
# !tar -zxf malay-test.tar.gz
# !tar -zxf singlish-test.tar.gz
# !tar -zxf mandarin-test.tar.gz
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/malay-test.json
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/singlish-test.json
# !wget https://f000.backblazeb2.com/file/malaya-speech-model/asr-dataset/mandarin-test.json
from glob import glob
malay = sorted(glob('malay-test/*.wav'), key = lambda x: int(x.split('/')[1].replace('.wav', '')))
singlish = sorted(glob('singlish-test/*.wav'), key = lambda x: int(x.split('/')[1].replace('.wav', '')))
mandarin = sorted(glob('mandarin-test/*.wav'), key = lambda x: int(x.split('/')[1].replace('.wav', '')))
len(malay), len(singlish), len(mandarin)
with open('malay-test.json') as fopen:
malay_label = json.load(fopen)
with open('singlish-test.json') as fopen:
singlish_label = json.load(fopen)
with open('mandarin-test.json') as fopen:
mandarin_label = json.load(fopen)
len(malay_label), len(singlish_label), len(mandarin_label)
from sklearn.utils import shuffle
audio = malay + singlish + mandarin
labels = malay_label + singlish_label + mandarin_label
audio, labels = shuffle(audio, labels)
test_set = list(zip(audio, labels))
test_set[:10]
from ctc_decoders import Scorer
from ctc_decoders import ctc_beam_search_decoder
n_mels = 80
sr = 16000
maxlen = 18
minlen_text = 1
def mp3_to_wav(file, sr = sr):
audio = AudioSegment.from_file(file)
audio = audio.set_frame_rate(sr).set_channels(1)
sample = np.array(audio.get_array_of_samples())
return malaya_speech.astype.int_to_float(sample), sr
def generate():
audios, cleaned_texts = audio, labels
for i in range(len(audios)):
try:
if audios[i].endswith('.mp3'):
wav_data, _ = mp3_to_wav(audios[i])
else:
wav_data, _ = malaya_speech.load(audios[i], sr = sr)
t = [unique_vocab.index(c) for c in cleaned_texts[i]]
yield {
'waveforms': wav_data,
'waveforms_length': [len(wav_data)],
'targets': t,
'targets_length': [len(t)],
}
except Exception as e:
print(e)
def get_dataset(
batch_size = 3,
shuffle_size = 20,
thread_count = 24,
maxlen_feature = 1800,
):
def get():
dataset = tf.data.Dataset.from_generator(
generate,
{
'waveforms': tf.float32,
'waveforms_length': tf.int32,
'targets': tf.int32,
'targets_length': tf.int32,
},
output_shapes = {
'waveforms': tf.TensorShape([None]),
'waveforms_length': tf.TensorShape([None]),
'targets': tf.TensorShape([None]),
'targets_length': tf.TensorShape([None]),
},
)
dataset = dataset.prefetch(tf.contrib.data.AUTOTUNE)
dataset = dataset.padded_batch(
batch_size,
padded_shapes = {
'waveforms': tf.TensorShape([None]),
'waveforms_length': tf.TensorShape([None]),
'targets': tf.TensorShape([None]),
'targets_length': tf.TensorShape([None]),
},
padding_values = {
'waveforms': tf.constant(0, dtype = tf.float32),
'waveforms_length': tf.constant(0, dtype = tf.int32),
'targets': tf.constant(0, dtype = tf.int32),
'targets_length': tf.constant(0, dtype = tf.int32),
},
)
return dataset
return get
dev_dataset = get_dataset()()
features = dev_dataset.make_one_shot_iterator().get_next()
features
training = True
class Encoder:
def __init__(self, config):
self.config = config
self.encoder = ConformerModel(**self.config)
def __call__(self, x, input_mask, training = True):
return self.encoder(x, training = training)
config_conformer = malaya_speech.config.conformer_large_encoder_config
config_conformer['subsampling']['type'] = 'none'
config_conformer['dropout'] = 0.0
encoder = Encoder(config_conformer)
cfg = hubert.HuBERTConfig(
extractor_mode='layer_norm',
dropout=0.0,
attention_dropout=0.0,
encoder_layerdrop=0.0,
dropout_input=0.0,
dropout_features=0.0,
final_dim=768,
)
model = hubert.Model(cfg, encoder, ['pad', 'eos', 'unk'] + [str(i) for i in range(100)])
X = features['waveforms']
X_len = features['waveforms_length'][:, 0]
r = model(X, padding_mask = X_len, features_only = True, mask = False)
logits = tf.layers.dense(r['x'], len(unique_vocab) + 1)
log_probs = tf.nn.log_softmax(logits)
seq_lens = tf.reduce_sum(
tf.cast(tf.logical_not(r['padding_mask']), tf.int32), axis = 1
)
logits = tf.transpose(logits, [1, 0, 2])
logits = tf.identity(logits, name = 'logits')
seq_lens = tf.identity(seq_lens, name = 'seq_lens')
# decoded = tf.nn.ctc_beam_search_decoder(
# logits,
# seq_lens,
# beam_width = beam_size,
# top_paths = 1,
# merge_repeated = True)[0][0]
# decoded._indices, decoded._values
logits, seq_lens, log_probs
decoded = tf.nn.ctc_beam_search_decoder(logits, seq_lens, beam_width=10, top_paths=1, merge_repeated=True)
preds = tf.sparse.to_dense(tf.to_int32(decoded[0][0]))
preds = tf.identity(preds, 'preds')
log_probs = tf.identity(log_probs, 'log_probs')
preds, log_probs
sess = tf.Session()
sess.run(tf.global_variables_initializer())
var_list = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES)
saver = tf.train.Saver(var_list = var_list)
saver.restore(sess, 'hubert-conformer-large-3mixed-ctc/model.ckpt-1800000')
import six
import string
from typing import List
def decode(ids, lookup: List[str] = None):
"""
Decode integer representation to string based on ascii table or lookup variable.
Parameters
-----------
ids: List[int]
lookup: List[str], optional (default=None)
list of unique strings.
Returns
--------
result: str
"""
decoded_ids = []
int2byte = six.int2byte
for id_ in ids:
if lookup:
decoded_ids.append(lookup[id_])
else:
decoded_ids.append(
int2byte(id_ - NUM_RESERVED_TOKENS).decode('utf-8')
)
return ''.join(decoded_ids)
# %%time
# kenlm_model = kenlm.Model('model.trie.klm')
# decoder = build_ctcdecoder(
# unique_vocab + ['_'],
# kenlm_model,
# alpha=0.1,
# beta=3.0,
# ctc_token_idx=len(unique_vocab)
# )
%%time
from pyctcdecode import build_ctcdecoder
import kenlm
kenlm_model = kenlm.Model('model.trie.klm')
decoder = build_ctcdecoder(
unique_vocab + ['_'],
kenlm_model,
alpha=0.2,
beta=1.0,
ctc_token_idx=len(unique_vocab)
)
scorer = Scorer(0.5, 1.0, 'model.trie.klm', unique_vocab)
logits_t = tf.nn.softmax(tf.transpose(logits, [1, 0, 2]))
# r = sess.run([preds, logits_t, seq_lens, features['targets']])
# out = decoder2.decode_beams(r[1][1,:r[2][1]],
# prune_history=True)
# text, lm_state, timesteps, logit_score, lm_score = out[0]
# text, ctc_beam_search_decoder(r[1][1,:r[2][1]], unique_vocab, 20, ext_scoring_func = scorer)[0][1]
# out = decoder.decode_beams(np.pad(r[1][:,0], [[0,0], [1,0]], constant_values = -13.0),
# prune_history=True)
# text, lm_state, timesteps, logit_score, lm_score = out[0]
# text
# out = decoder2.decode_beams(np.pad(r[1][:,0], [[0,0], [1,0]], constant_values = -13.0),
# prune_history=True)
# text, lm_state, timesteps, logit_score, lm_score = out[0]
# text
# decode(r[0][1], unique_vocab), decode(r[-1][1], unique_vocab)
# %%time
# ctc_beam_search_decoder(r[1][0,:r[2][0]], unique_vocab, 20, ext_scoring_func = scorer)[0][1]
# %%time
# ctc_beam_search_decoder(r[1][1,:r[2][1]], unique_vocab, 20, ext_scoring_func = scorer)[0]
from malaya_speech.utils import metrics, char
wer, cer, wer_lm, cer_lm = [], [], [], []
wer_lm2, cer_lm2 = [], []
index = 0
while True:
try:
r = sess.run([preds, logits_t, seq_lens, features['targets']])
for no, row in enumerate(r[0]):
d = decode(row, lookup = unique_vocab).replace('<PAD>', '')
t = decode(r[-1][no], lookup = unique_vocab).replace('<PAD>', '')
wer.append(malaya_speech.metrics.calculate_wer(t, d))
cer.append(malaya_speech.metrics.calculate_cer(t, d))
d_lm = ctc_beam_search_decoder(r[1][no,:r[2][no]],
unique_vocab, 20, ext_scoring_func = scorer)[0][1]
wer_lm.append(malaya_speech.metrics.calculate_wer(t, d_lm))
cer_lm.append(malaya_speech.metrics.calculate_cer(t, d_lm))
out = decoder.decode_beams(r[1][no,:r[2][no]], prune_history=True)
d_lm2, lm_state, timesteps, logit_score, lm_score = out[0]
wer_lm2.append(malaya_speech.metrics.calculate_wer(t, d_lm2))
cer_lm2.append(malaya_speech.metrics.calculate_cer(t, d_lm2))
index += 1
except Exception as e:
break
np.mean(wer), np.mean(cer), np.mean(wer_lm), np.mean(cer_lm), np.mean(wer_lm2), np.mean(cer_lm2)
| 0.540439 | 0.19787 |
```
from sklearn.datasets import make_classification
import xgboost as xgb
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, roc_curve, auc, confusion_matrix, classification_report
from sklearn import preprocessing
import sklearn
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import shap
import lime
from examine_explanation import examine_interpretation
from examine_explanation import examine_local_fidelity
from examine_explanation import get_lipschitz
from examine_explanation import get_lipschitz
```
# Fidelity
### Create synthetic dataset
```
n_features = 4
X, y = make_classification(n_samples=1000, n_informative=2, n_features=n_features, n_redundant=2)
X = preprocessing.normalize(X)
X=pd.DataFrame(data=X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
```
### XGB Model
```
X_train.columns = ['0','1','2','3']
X_test.columns = ['0','1','2','3']
xgb_model = xgb.XGBClassifier()
xgb_model.fit(X_train, y_train)
#xgb_model.get_booster().feature_names = X_train.columns
xgb_preds = xgb_model.predict(X_test)
print(accuracy_score(y_test, xgb_preds))
print(classification_report(y_test, xgb_preds))
```
## Random forest
```
rf = sklearn.ensemble.RandomForestClassifier(n_estimators=50)
rf.fit(X_train, y_train)
sklearn.metrics.accuracy_score(y_test, rf.predict(X_test))
```
### Get interpretation importances
```
import eli5
from eli5.sklearn import PermutationImportance
from sklearn.externals.six import StringIO
from IPython.display import Image
from sklearn.tree import export_graphviz
import pydotplus
from pdpbox import pdp, get_dataset, info_plots
import shap
import lime
shap.initjs()
```
# Preturbation based on permutation importances
```
perm = PermutationImportance(xgb_model, random_state=1).fit(X_test, y_test)
perm_importances = perm.feature_importances_
feature_names = [str(i) for i in range(n_features)]
eli5.show_weights(perm, feature_names=feature_names)
preturbed_perm_accuraties = examine_interpretation(xgb_model, X_test, y_test, perm_importances, epsilon=4, resolution=50, proportionality_mode=0)
```
# Perturbation based on local importances
### SHAP
```
examine_local_fidelity(xgb_model, X_test, y_test, epsilon=3)
```
### LIME
```
examine_local_fidelity(rf, X_test, y_test, epsilon=3,framework='lime' )
```
# Preturbation based on shapley values
```
xgb_explainer = shap.TreeExplainer(xgb_model)
xgb_shap_values = xgb_explainer.shap_values(X_test)
shap_imps = []
transposed_shap = [*zip(*xgb_shap_values)]
for idx, col in enumerate(transposed_shap):
shap_imps.append(np.mean(list(map(lambda x: abs(x), col))))
abs_importances = list(map(abs, shap_imps))
total_importance = (sum(abs_importances))
importance_shares = list(map(lambda x: x/total_importance, abs_importances))
max_importance = max(shap_imps)
reversed_importances = list(map(lambda x: max_importance - x, abs_importances))
total_reversed_importance = (sum(reversed_importances))
inverse_importance_shares = list(map(lambda x: x/total_reversed_importance, reversed_importances))
shap.summary_plot(xgb_shap_values, X_test, plot_type="bar", color='red')
for i in range(len(importance_shares)):
d=np.linspace(inverse_importance_shares[i], importance_shares[i],100)
plt.plot(d)
print
plt.legend(['Feature 0', 'Feature 1', 'Feature 2', 'Feature 3'])
plt.xlabel('Percentile of perturbation range', fontsize=13)
plt.ylabel('Share of feature to be perturbed', fontsize=13)
```
## Mode 0
```
preturbed_shap_accuracies = examine_interpretation(xgb_model, X_test, y_test, shap_imps, epsilon=4, resolution=50)
```
## Mode 1
```
preturbed_shap_accuracies = examine_interpretation(xgb_model, X_test, y_test, shap_imps, epsilon=2, resolution=50, proportionality_mode=1)
```
# Test for dataset without noise features
```
newx, newy = make_classification(n_samples=1000, n_informative=2, n_features=2, n_redundant=0)
newx = preprocessing.normalize(newx)
newx=pd.DataFrame(data=newx)
sns.scatterplot(x=newx[0],y=newx[1],hue=newy)
X_train2, X_test2, y_train2, y_test2 = train_test_split(newx, newy, test_size=0.33, random_state=42)
xgb_model2 = xgb.XGBClassifier()
xgb_model2.fit(X_train2, y_train2)
xgb_preds2 = xgb_model2.predict(X_test2)
print(accuracy_score(y_test2, xgb_preds2))
print(classification_report(y_test2, xgb_preds2))
```
## Permutation importances
```
perm2 = PermutationImportance(xgb_model2, random_state=1).fit(X_test2, y_test2)
perm_importances2 = perm2.feature_importances_
eli5.show_weights(perm2)
```
## Mode 0
```
preturbed_perm_accuraties2 = examine_interpretation(xgb_model2, X_test2, y_test2, perm_importances2, epsilon=2, resolution=50, proportionality_mode=0)
```
# Mode 1
```
preturbed_perm_accuraties2 = examine_interpretation(xgb_model2, X_test2, y_test2, perm_importances2, epsilon=2, resolution=50, proportionality_mode=1)
shap_imps2 = []
transposed_shap2 = [*zip(*xgb_shap_values2)]
for idx, col in enumerate(transposed_shap2):
shap_imps2.append(np.mean(list(map(lambda x: abs(x), col))))
shap.summary_plot(xgb_shap_values2, X_test2, plot_type="bar", color='red')
```
## Mode 0
```
preturbed_shap_accuracies2 = examine_interpretation(xgb_model2, X_test2, y_test2, shap_imps2, epsilon=10, resolution=50)
```
## Mode 1
```
preturbed_shap_accuracies2 = examine_interpretation(xgb_model2, X_test2, y_test2, shap_imps2, epsilon=10, resolution=50, proportionality_mode=1, count_per_step=50)
lime_lips = get_lipschitz(rf, X_test, epsilon=3, framework='lime')
shap_lips = get_lipschitz(rf, X_test.iloc[:12], epsilon=3, sample_num=5)
lip_df = pd.DataFrame({'lime':lime_lips, 'shap':shap_lips})
sns.boxplot(x="variable", y="value", data=pd.melt(lip_df))
m1_forest = sklearn.ensemble.RandomForestClassifier(n_estimators=50)
m1_forest.fit(X_train, y_train)
#sklearn.metrics.accuracy_score(y_test, rf.predict(X_test))
m2_xgb = xgb.XGBClassifier()
m2_xgb.fit(X_train, y_train)
cvals = check_consistency([m1_forest, m2_xgb], X_test, y_test, sample_num = 10)
```
|
github_jupyter
|
from sklearn.datasets import make_classification
import xgboost as xgb
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, roc_curve, auc, confusion_matrix, classification_report
from sklearn import preprocessing
import sklearn
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import shap
import lime
from examine_explanation import examine_interpretation
from examine_explanation import examine_local_fidelity
from examine_explanation import get_lipschitz
from examine_explanation import get_lipschitz
n_features = 4
X, y = make_classification(n_samples=1000, n_informative=2, n_features=n_features, n_redundant=2)
X = preprocessing.normalize(X)
X=pd.DataFrame(data=X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
X_train.columns = ['0','1','2','3']
X_test.columns = ['0','1','2','3']
xgb_model = xgb.XGBClassifier()
xgb_model.fit(X_train, y_train)
#xgb_model.get_booster().feature_names = X_train.columns
xgb_preds = xgb_model.predict(X_test)
print(accuracy_score(y_test, xgb_preds))
print(classification_report(y_test, xgb_preds))
rf = sklearn.ensemble.RandomForestClassifier(n_estimators=50)
rf.fit(X_train, y_train)
sklearn.metrics.accuracy_score(y_test, rf.predict(X_test))
import eli5
from eli5.sklearn import PermutationImportance
from sklearn.externals.six import StringIO
from IPython.display import Image
from sklearn.tree import export_graphviz
import pydotplus
from pdpbox import pdp, get_dataset, info_plots
import shap
import lime
shap.initjs()
perm = PermutationImportance(xgb_model, random_state=1).fit(X_test, y_test)
perm_importances = perm.feature_importances_
feature_names = [str(i) for i in range(n_features)]
eli5.show_weights(perm, feature_names=feature_names)
preturbed_perm_accuraties = examine_interpretation(xgb_model, X_test, y_test, perm_importances, epsilon=4, resolution=50, proportionality_mode=0)
examine_local_fidelity(xgb_model, X_test, y_test, epsilon=3)
examine_local_fidelity(rf, X_test, y_test, epsilon=3,framework='lime' )
xgb_explainer = shap.TreeExplainer(xgb_model)
xgb_shap_values = xgb_explainer.shap_values(X_test)
shap_imps = []
transposed_shap = [*zip(*xgb_shap_values)]
for idx, col in enumerate(transposed_shap):
shap_imps.append(np.mean(list(map(lambda x: abs(x), col))))
abs_importances = list(map(abs, shap_imps))
total_importance = (sum(abs_importances))
importance_shares = list(map(lambda x: x/total_importance, abs_importances))
max_importance = max(shap_imps)
reversed_importances = list(map(lambda x: max_importance - x, abs_importances))
total_reversed_importance = (sum(reversed_importances))
inverse_importance_shares = list(map(lambda x: x/total_reversed_importance, reversed_importances))
shap.summary_plot(xgb_shap_values, X_test, plot_type="bar", color='red')
for i in range(len(importance_shares)):
d=np.linspace(inverse_importance_shares[i], importance_shares[i],100)
plt.plot(d)
print
plt.legend(['Feature 0', 'Feature 1', 'Feature 2', 'Feature 3'])
plt.xlabel('Percentile of perturbation range', fontsize=13)
plt.ylabel('Share of feature to be perturbed', fontsize=13)
preturbed_shap_accuracies = examine_interpretation(xgb_model, X_test, y_test, shap_imps, epsilon=4, resolution=50)
preturbed_shap_accuracies = examine_interpretation(xgb_model, X_test, y_test, shap_imps, epsilon=2, resolution=50, proportionality_mode=1)
newx, newy = make_classification(n_samples=1000, n_informative=2, n_features=2, n_redundant=0)
newx = preprocessing.normalize(newx)
newx=pd.DataFrame(data=newx)
sns.scatterplot(x=newx[0],y=newx[1],hue=newy)
X_train2, X_test2, y_train2, y_test2 = train_test_split(newx, newy, test_size=0.33, random_state=42)
xgb_model2 = xgb.XGBClassifier()
xgb_model2.fit(X_train2, y_train2)
xgb_preds2 = xgb_model2.predict(X_test2)
print(accuracy_score(y_test2, xgb_preds2))
print(classification_report(y_test2, xgb_preds2))
perm2 = PermutationImportance(xgb_model2, random_state=1).fit(X_test2, y_test2)
perm_importances2 = perm2.feature_importances_
eli5.show_weights(perm2)
preturbed_perm_accuraties2 = examine_interpretation(xgb_model2, X_test2, y_test2, perm_importances2, epsilon=2, resolution=50, proportionality_mode=0)
preturbed_perm_accuraties2 = examine_interpretation(xgb_model2, X_test2, y_test2, perm_importances2, epsilon=2, resolution=50, proportionality_mode=1)
shap_imps2 = []
transposed_shap2 = [*zip(*xgb_shap_values2)]
for idx, col in enumerate(transposed_shap2):
shap_imps2.append(np.mean(list(map(lambda x: abs(x), col))))
shap.summary_plot(xgb_shap_values2, X_test2, plot_type="bar", color='red')
preturbed_shap_accuracies2 = examine_interpretation(xgb_model2, X_test2, y_test2, shap_imps2, epsilon=10, resolution=50)
preturbed_shap_accuracies2 = examine_interpretation(xgb_model2, X_test2, y_test2, shap_imps2, epsilon=10, resolution=50, proportionality_mode=1, count_per_step=50)
lime_lips = get_lipschitz(rf, X_test, epsilon=3, framework='lime')
shap_lips = get_lipschitz(rf, X_test.iloc[:12], epsilon=3, sample_num=5)
lip_df = pd.DataFrame({'lime':lime_lips, 'shap':shap_lips})
sns.boxplot(x="variable", y="value", data=pd.melt(lip_df))
m1_forest = sklearn.ensemble.RandomForestClassifier(n_estimators=50)
m1_forest.fit(X_train, y_train)
#sklearn.metrics.accuracy_score(y_test, rf.predict(X_test))
m2_xgb = xgb.XGBClassifier()
m2_xgb.fit(X_train, y_train)
cvals = check_consistency([m1_forest, m2_xgb], X_test, y_test, sample_num = 10)
| 0.697094 | 0.906115 |
```
from pynvml import *
nvmlInit()
vram = nvmlDeviceGetMemoryInfo(nvmlDeviceGetHandleByIndex(1)).free/1024.**2
print('GPU1 Memory: %dMB' % vram)
if vram < 8000:
raise Exception('GPU Memory too low')
nvmlShutdown()
import os
import cv2
import h5py
import numpy as np
import matplotlib.pyplot as plt
from IPython.display import *
from collections import Counter
import seaborn as sns
from tqdm import tqdm
import pandas as pd
import re
import time
import random
from keras.models import *
import keras.backend as K
from make_parallel import make_parallel
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
characters = u'0123456789()+-*/=君不见黄河之水天上来奔流到海复回烟锁池塘柳深圳铁板烧; '
n_len = 45
rnn_length = 110
n, width, height, n_class, channels = 100000, 900, 81, len(characters), 3
def decode(out):
return ''.join([characters[x] for x in out if x < n_class-1 and x > -1])
def disp3(index):
s = decode(out[index])
plt.figure(figsize=(16, 4))
plt.imshow(X[index].transpose(1, 0, 2))
plt.title('pred:%s'%s)
def disp2(img):
cv2.imwrite('a.png', img)
return Image('a.png')
def disp(img, txt=None, first=False):
global index
if first:
index = 1
plt.figure(figsize=(16, 9))
else:
index += 1
plt.subplot(4, 1, index)
if len(img.shape) == 2:
plt.imshow(img, cmap='gray')
else:
plt.imshow(img[:,:,::-1])
if txt:
plt.title(txt)
```
# 读取测试集
```
X = np.zeros((n, width, height, channels), dtype=np.uint8)
for i in tqdm(range(n)):
img = cv2.imread('crop_split2_test/%d.png'%i).transpose(1, 0, 2)
a, b, _ = img.shape
X[i, :a, :b] = img
```
# 预测
```
z = '0.997754'
base_model = load_model('model_346_split2_3_%s.h5' % z)
base_model2 = make_parallel(base_model, 4)
y_pred = base_model2.predict(X, batch_size=500, verbose=1)
out = K.get_value(K.ctc_decode(y_pred[:,2:], input_length=np.ones(y_pred.shape[0])*rnn_length)[0][0])[:, :n_len]
ss = map(decode, out)
vals = []
errs = []
errsid = []
for i in tqdm(range(100000)):
val = ''
try:
a = ss[i].split(';')
s = a[-1]
for x in a[:-1]:
x, c = x.split('=')
s = s.replace(x, c+'.0')
val = '%.2f' % eval(s)
except:
# disp3(i)
errs.append(ss[i])
errsid.append(i)
ss[i] = ''
vals.append(val)
with open('result_%s.txt' % z, 'w') as f:
f.write('\n'.join(map(' '.join, list(zip(ss, vals)))).encode('utf-8'))
print len(errs)
print 1-len(errs)/100000.
z = '0.997559'
base_model = load_model('model_346_split2_3_%s.h5' % z)
base_model2 = make_parallel(base_model, 4)
y_pred = base_model2.predict(X, batch_size=500, verbose=1)
out = K.get_value(K.ctc_decode(y_pred[:,2:], input_length=np.ones(y_pred.shape[0])*rnn_length)[0][0])[:, :n_len]
ss = map(decode, out)
vals = []
errs = []
errsid = []
for i in tqdm(range(100000)):
val = ''
try:
a = ss[i].split(';')
s = a[-1]
for x in a[:-1]:
x, c = x.split('=')
s = s.replace(x, c+'.0')
val = '%.2f' % eval(s)
except:
# disp3(i)
errs.append(ss[i])
errsid.append(i)
ss[i] = ''
vals.append(val)
with open('result_%s_%d.txt' % (z, len(errs)), 'w') as f:
f.write('\n'.join(map(' '.join, list(zip(ss, vals)))).encode('utf-8'))
print len(errs)
print 1-len(errs)/100000.
```
|
github_jupyter
|
from pynvml import *
nvmlInit()
vram = nvmlDeviceGetMemoryInfo(nvmlDeviceGetHandleByIndex(1)).free/1024.**2
print('GPU1 Memory: %dMB' % vram)
if vram < 8000:
raise Exception('GPU Memory too low')
nvmlShutdown()
import os
import cv2
import h5py
import numpy as np
import matplotlib.pyplot as plt
from IPython.display import *
from collections import Counter
import seaborn as sns
from tqdm import tqdm
import pandas as pd
import re
import time
import random
from keras.models import *
import keras.backend as K
from make_parallel import make_parallel
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
characters = u'0123456789()+-*/=君不见黄河之水天上来奔流到海复回烟锁池塘柳深圳铁板烧; '
n_len = 45
rnn_length = 110
n, width, height, n_class, channels = 100000, 900, 81, len(characters), 3
def decode(out):
return ''.join([characters[x] for x in out if x < n_class-1 and x > -1])
def disp3(index):
s = decode(out[index])
plt.figure(figsize=(16, 4))
plt.imshow(X[index].transpose(1, 0, 2))
plt.title('pred:%s'%s)
def disp2(img):
cv2.imwrite('a.png', img)
return Image('a.png')
def disp(img, txt=None, first=False):
global index
if first:
index = 1
plt.figure(figsize=(16, 9))
else:
index += 1
plt.subplot(4, 1, index)
if len(img.shape) == 2:
plt.imshow(img, cmap='gray')
else:
plt.imshow(img[:,:,::-1])
if txt:
plt.title(txt)
X = np.zeros((n, width, height, channels), dtype=np.uint8)
for i in tqdm(range(n)):
img = cv2.imread('crop_split2_test/%d.png'%i).transpose(1, 0, 2)
a, b, _ = img.shape
X[i, :a, :b] = img
z = '0.997754'
base_model = load_model('model_346_split2_3_%s.h5' % z)
base_model2 = make_parallel(base_model, 4)
y_pred = base_model2.predict(X, batch_size=500, verbose=1)
out = K.get_value(K.ctc_decode(y_pred[:,2:], input_length=np.ones(y_pred.shape[0])*rnn_length)[0][0])[:, :n_len]
ss = map(decode, out)
vals = []
errs = []
errsid = []
for i in tqdm(range(100000)):
val = ''
try:
a = ss[i].split(';')
s = a[-1]
for x in a[:-1]:
x, c = x.split('=')
s = s.replace(x, c+'.0')
val = '%.2f' % eval(s)
except:
# disp3(i)
errs.append(ss[i])
errsid.append(i)
ss[i] = ''
vals.append(val)
with open('result_%s.txt' % z, 'w') as f:
f.write('\n'.join(map(' '.join, list(zip(ss, vals)))).encode('utf-8'))
print len(errs)
print 1-len(errs)/100000.
z = '0.997559'
base_model = load_model('model_346_split2_3_%s.h5' % z)
base_model2 = make_parallel(base_model, 4)
y_pred = base_model2.predict(X, batch_size=500, verbose=1)
out = K.get_value(K.ctc_decode(y_pred[:,2:], input_length=np.ones(y_pred.shape[0])*rnn_length)[0][0])[:, :n_len]
ss = map(decode, out)
vals = []
errs = []
errsid = []
for i in tqdm(range(100000)):
val = ''
try:
a = ss[i].split(';')
s = a[-1]
for x in a[:-1]:
x, c = x.split('=')
s = s.replace(x, c+'.0')
val = '%.2f' % eval(s)
except:
# disp3(i)
errs.append(ss[i])
errsid.append(i)
ss[i] = ''
vals.append(val)
with open('result_%s_%d.txt' % (z, len(errs)), 'w') as f:
f.write('\n'.join(map(' '.join, list(zip(ss, vals)))).encode('utf-8'))
print len(errs)
print 1-len(errs)/100000.
| 0.175326 | 0.435601 |
# Svet Superjunakov Stan Leeja
## Projektna naloga pri predmetu Programiranje 1
## Avtor: Nejc Zajc
V tej datoteki analiziram zbrane in urejene podatke glede ustvarjenih likov Stana Leeja pri Marvel stripih.
## UVOD
Pri projektni nalogi analize podatkov sem si ogledal like, ki jih je v svoji bogati karieri ustvaril oziroma pomagal ustvariti leta 2018 preminuli **Stan Lee**. Bil je eden izmed največjih ustvarjalcev stripov na svetu, like pa je ustvarjal pod znamko *Marvel Comics*.
Ob vseh dogodkih, ki so obkrožali Marvel v lanskem letu, sem se odločil na superjunake njihovega velikega vesolja pogledati še iz malo drugačne smeri.
Iz spletne baze podatkov o [Marvelovih likih](https://marvel.fandom.com/wiki/Marvel_Database) sem pobral podatke o likih Stan Leeja, kot sem to opisal v *README.md*, v tej datoteki pa zbrane podatke raziščem.
## 0. Priprava
Najprej naložim pakete in shranim podatke v spremenljivke. Prav tako nastavim nekaj osnovnih nastavitev za uporabljanje *Jupyter Notebooka*.
```
# naložim paketa, s katerimi obdelujem podatke
import pandas as pd
import numpy as np
# zaradi preglednosti v tabelah izpisujem le 20 vrstic
pd.options.display.max_rows = 20
# izberem stil grafov
%matplotlib notebook
# zaradi zaporednih ukazov, pandas knjižnica včasih navlkjub željenemu delovanju opozarja na kakšne stvari, da se temu
# v končnem izdelku izognem izključim ta opozorila
import warnings
warnings.simplefilter(action='ignore')
# naložim še podatke, s katerimi delam, ker imam vse podatke urejene po "id"-ju, ki je enak končnici spletne strani,
# na kateri se nahaja lik, vrstic v tabelah ni potrebno dodatno številčiti
podatki = pd.read_csv('podatki/podatki.csv', index_col = "id")
avtorji = pd.read_csv('podatki/avtorji.csv', index_col = "id")
moci = pd.read_csv('podatki/moci.csv', index_col = "id")
tabele = pd.read_csv('podatki/tabele.csv', index_col = "id")
```
Za začetek si ogledam koliko in katere podatke imam.
```
podatki.groupby("tip").count()
```
Vse podatke sem zbral le za osebe, zato se bom v drugem delu analize posvetil le njim.
Podatke glede stripov, ko se je lik pojavil prvič in podatke o avtorjih (shranjeni v *avtorji*) pa sem zbral za vse vnose. Na začetku se bom tako posvetil splošni analizi.
## 1. Prve pojavitve likov
V **prvem delu** naloge si ogledam, kaj lahko ugotovim glede stripov v katerih je Stan Lee prvič omenil like.
## 1.1. Najbolj aktivna leta
Stanley Martin Lieber je živel med leti 1922 in 2018. Najprej si pogledam, v katerih letih je svetu predstavil največ likov.
```
podatki.groupby("leto").size().plot()
```
Vidim, da je v šestdesetih letih napisal daleč največ novih likov.
Ko poiščem pod katerim naslovom je bilo skupno predstavljeno največ novih likov, vidim da so bili to *Fantastični štirje*. Zato ne preseneča rezultat da so ti stripi izhajali ravno v šestdesetih letih.
```
podatki.groupby("naslov").size().sort_values(ascending = False).head(1) # = "Fantastic Four"
podatki[podatki.naslov == "Fantastic Four"].groupby("leto").size()
```
V omenjenih šestdesetih letih je popularnost Stan Leeja eksplodirala, prav tako je pomagal k vzponu Marvela, ki so ga poimenovali kar **Marvel revolution**.
## 1.2. Izdaje v seriji & meseci
V tem delu preverim smiselno idejo, da se novi liki najpogosteje pojavijo v prvih izdajah serij.
```
st_zbranih_izdaj = podatki.groupby("izdaja").size().sum() # 2236 - za toliko likov sem zbral izdajo
v_prvih = podatki[podatki.izdaja == 1].shape[0] # 233 - število likov v prvih delih serij
v_prvih / st_zbranih_izdaj
```
Nepresenetljivo se je največ predstavitev likov zgodilo v prvih delih serij stripov. To pa se vseeno ni zgodilo tako pogosto, kot bi pričakoval. Nov lik se je prvič pojavil v prvem delu namreč le v dobrih 10 % primerov.
Zanima me še, katera je bila najvišja izdaja, v kateri je bil predstavljen lik.
```
podatki[podatki.izdaja < 1900].groupby("izdaja").size().tail(20) # opazim nekaj primerov izdaj nad 500!
podatki[(podatki.izdaja > 500) & (podatki.izdaja < 1900)]
```
Ko iz izbora odstranim strip z naslovom *Spider-Man Newspaper Strips*, ki je številčil izdaje po letih, ko je izšel, sem vseeno presenečen nad rezultati. Stripa *Amazing Spider-Man* in *Thor* sta celo po svoji 500. izdaji še vedno predstavljala nove like.
Ogledam si še kako na število novih likov vpliva del leta.
```
pogledam_po_mesecih = podatki[(podatki.mesec != "Spring") & (podatki.mesec != "Summer") &
(podatki.mesec != "Fall") & (podatki.mesec != "Winter")]
meseci = ["January", "February", "March", "April", "May", "June",
"July", "August", "September", "October", "November", "December"]
pogledam_po_mesecih['mesec'] = pd.Categorical(pogledam_po_mesecih['mesec'], categories=meseci, ordered=True)
pogledam_po_mesecih.groupby("mesec").size().to_frame("st_likov").plot.bar()
```
Opazim višek v drugi polovici leta, natančneje v začetku šolskega in študijskega leta.
## 2. Vesolja & sodelavci
V *Marvel Comics Multiverse* (vesolju Marvelovih stripov) glavno Zemljo imenujejo **Earth-616**. Posledično se večino dogajanja dogaja tam, prav tako se tam pojavlja največ likov.
Kaj pa ostale Zemlje? Ali je Stan Lee s katerim izmed sodelavcev sodeloval večinoma *na drugi Zemlji*?
Ker podatkov za vesolja nisem zbiral za like, ki niso osebe, se sedaj omejim samo na te.
```
osebe = podatki[podatki.tip == "Character"]
avtorji.groupby("avtor").size().sort_values(ascending = False)
osebe.groupby("vesolje").size().sort_values(ascending = False).head(10)
```
Najprej sem se prepričal o predpostavkah, da je glavna Zemlja res najpogostejša in da je Stan Lee sodeloval največkrat s Jackom Kirbyjem.
Zdaj raziščem še sodelovanja glede na svetove.
```
skupni = pd.merge(osebe, avtorji, left_on="id", right_on="id") # združim tabeli
po_avtorjih = avtorji.groupby("avtor").size().to_frame("st_sodelovanj") # spremenim v tabelo
po_avtorjih["Earth-616"] = skupni[skupni.vesolje == "Earth-616"].groupby("avtor").size() # dodam koliko likov iz glavne Zemlje
po_avtorjih["delez"] = po_avtorjih["Earth-616"] / po_avtorjih.st_sodelovanj # ter še stolpec, z deležem le-teh
po_avtorjih.sort_values("delez").dropna() # za prikaz izpustim tiste, ki niso nikoli sodelovali z likom iz glavne Zemlje
skupni[skupni.avtor == "John Romita Jr."]
skupni[skupni.avtor == "Larry Lieber"].groupby("vesolje").size()
```
Tako sem našel sodelavnca *John Romita Jr.*-ja, ki je skoraj izključno sodeloval s Stan Leejem na projektih, ki se niso dogajali na glavni Zemlji. Ko pa si le te natančneje ogledam, se izkaže, da je bil skupni projekt (na ostalih Zemljah) le en, a je ta prispeval veliko novih oseb. Te osebe so znani liki, a ker se strip dogaja na tuji Zemlji, sta jih tako skupaj predstavila svetu.
Drugi izstopajoči je *Larry Lieber* (Stanov brat), ki pa je navlkjub manjhnemu deležu, pri več kot polovici skupnih projektov ustvaril lik iz glavne Zemlje.
*Opomba*: Pri prikazu deležev izpustim vrstice, ki nimajo podatkov za glavno Zemljo, do česar lahko pride tudi v primeru, ko za skupni lik ni podatka o vesolju (ne le v primeru, ko noben izmed skupnih likov ni iz glavne Zemlje).
## 3. Osebe
Sedaj se še malo bolj podrobno spustim v podatke zbrane za osebe.
## 3.1. Status razmerja
Za osebe sem poleg ostalih zbiral tudi podatke o stanju njihovj razmerij.
```
osebe.groupby("razmerje").size()
```
Opazim, da je največ oseb samskih. Takšnih je celo več kot polovica, kar ovrže mojo drugo hipotezo.
```
osebe.groupby("razmerje").size().Single / osebe.groupby("razmerje").size().sum()
```
Poleg tistih v razmerju pa jih je kar nekaj tudi takšnih, ki so bili v razmerju, a so to zvezo izgubili - torej *Divorced*, *Separated* in *Widowed*.
Malo raziskovanja me pripelje do naslednje zanimive povezave.
Ločim osebe, za katere imam podatek o razmerju, na tiste, katerih naziv je enak njihovemu pravemu imenu in tiste, za katere to ne drži.
```
isto = osebe[osebe.naziv == osebe.pravo_ime].count().razmerje # ti imajo naziv enak imenu - teh je 334
spremenjeno = osebe[osebe.naziv != osebe.pravo_ime].count().razmerje # naziv teh je drugačen - teh je 425
a = osebe[osebe.naziv == osebe.pravo_ime].groupby("razmerje").size() # le po istih pogledam njihova razmerja
a
b = osebe[osebe.naziv != osebe.pravo_ime].groupby("razmerje").size() # enako storim za spremenjene
delez_isti = (a.Divorced + a.Separated + a.Widowed) / isto # izračunam delež tistih,
delez_spremenjeno = (b.Divorced + b.Separated + b.Widowed) / spremenjeno # ki so izgubili zvezo za obe skupini
(delez_isti, delez_spremenjeno)
```
Osebe, ki si za naziv izberejo ime različno od njihovega pravega, so torej kar precej manj uspešne v ohranjanju razmerij.
Druga možnost je, da si osebe po končanem razmerju izberejo naziv, ki je različen od njihovega pravega imena.
Kateri razlagi verjamemo, je odvisno od lastne presoje.
## 3.2. Najmočnejše osebe
V tem delu si pogledam **različne moči**, ki jih imajo zbrane osebe.
```
moci.groupby("moc").size().sort_values()
moci_po_osebah = moci.groupby("id").size().to_frame("st_moci")
moci_po_osebah.sort_values("st_moci")
```
Kot pričakovano, so najpogostejše moči nadčloveška fizična moč, hitrost, odpornost in podobne.
Izmed oseb je glede na različne moči najmočnejša *Jean Grey*, ki je *Omega Level Mutant*. Skupaj z *Adamom Warlockom* ima precej več moči kot ostali, kar kaže na njuno premoč glede raznolikosti moči.
Za osebe sem zbiral tudi **tabele ocen** njihove moči po področjih. Vsaka tabela vsebuje oceno od 1 (najšibkejše) do 7 (najmočnejše) za področja: *INT* - inteligenca, *STR* - moč, *SPD* - hitrost, *DUR* - vzdržljivost, *ENP* - sproščanje energije in *FGT* - spretnost pretepanja.
Tabeli dodam stolpec vsote vseh 6 ocen, in tako dobim še eno lestvico oseb po njihovi moči.
```
tabele["SUM"] = tabele.INT + tabele.STR + tabele.SPD + tabele.DUR + tabele.ENP + tabele.FGT
tabele.sort_values("SUM")
tabele.mean().sort_values()
```
Lestvica pri vrhu ni podobna prejšnji, opazim pa da tu ni oseb, ki bi močno izstopale.
Popolnega rezultata (42 točk) ni, če pa gledam vsote brez enega izmed stolpcev, dobim ko izpustim spretnost pretepanja kar 7 polnih vsot - tu najdem kozmična bitja z močjo izjemnih razsežnosti, ki ne potrebujejo navadnega pretepanja. Edini drugi primer vsote 35 točk po petih stoplcih se zgodi ob izpuščeni inteligenci pri *Super Adaptoidu* - super-robotu, ki kopira druge osebe ter njihove moči.
Da mnogi junaki niso odvisni le od pretepanja kaže tudi najmanjša povprečna vrednost pri tej lastnosti.
```
tabele.loc["Jean_Grey_(Earth-616)"]
moci.loc["Dormammu_(Earth-616)"]
```
Oba "najmočnejša" na lestvicah sta iz drugega vidika sicer nadpovprečna, a ne povsem pri vrhu.
Zato poskusim ugotoviti, koliko imata lestvici sploh skupnega.
```
zdruzeni = pd.merge(moci_po_osebah, tabele[["SUM"]], left_on = "id", right_on = "id")
# naredim tabelo z dvema stolpcema obeh lestvic
zdruzeni.sort_values("SUM").plot().axes.get_xaxis().set_ticks([])# narisem graf, na x-osi so osebe
```
V grafu sem po velikosti uredil vsoto točk v tabeli. Glede tega ali število različnih moči narašča je težko veliko reči. Zato združim podatke in pogledam ali po manjših skupinah povprečno število moči narašča.
Osebam v spremenljivki *zdruzeni* pripišem vrednost vsote v tabeli zaokrožene na 4 (za natančnejšo zaokrožitev, pred računanjem celega dela prištejem 1). Pri tako ustvarjenih razredih si ponovno ogledam graf števila različnih moči.
```
zdruzeni["na_stiri"] = 4 * ((zdruzeni.SUM + 1) // 4)
na_stiri = zdruzeni.groupby("na_stiri")
na_stiri.mean()[["st_moci"]].plot.bar()
```
Zdaj se lepo vidi, da sta lestvici povezani, saj povprečno število moči očitno narašča z zaokroženo vsoto v tabeli.
## 3.3. Avtorji & število moči
Zanima me še, ali je kateri izmed sodelavcev ustvarjal nad- ali podpovprečno močne osebe. Tega vprašanja se lotim s primerjanjem povprečnega števila moči glede na avtorja, omejim pa se le na takšne, ki so s Stan Leejem sodelovali najpogosteje.
```
moci_po_a = pd.merge(avtorji, moci_po_osebah, left_on = "id", right_on = "id").groupby("avtor").mean().sort_values("st_moci")
# zdruzim tabelo avtorjev in moči po osebah, in tako dobim tabelo oseb z dodanim stolpcem "st_moci",
# to nato uredim po skupinah po avtorju ter izračunam povprečno število moči
pogosti = avtorji.groupby("avtor").size().sort_values().tail(10).to_frame("zadetki")
# naredim stolpec 10 najbolj pogostih sodelovanj Stan Leeja
pd.merge(moci_po_a, pogosti, right_on = "avtor", left_on = "avtor").sort_values("zadetki")
```
Ugotovim torej, da je med pogostimi sodelavci Stan Leeja najmočnejše osebe ustvarjal *Steve Ditko*. Izrazito šibke pa so bile osebe *Dicka Ayersa*.
## 4. Marvel's Avengers
V zadnjem delu naloge, si ogledam kako močni so najbolj slavni junaki iz filmov zadnjih let.
Pogledam si 6 članov *Maščevalcev* iz filma [The Avengers](https://www.imdb.com/title/tt0848228/).
Pogledam njihove zadetke in izberem verzije iz glavne Zemlje.
Tabeli dodam ocene iz tabele in pogledam ugotovitve.
```
podatki[podatki.naziv == "Spider-Man"]
podatki[podatki.naziv == "Iron Man"]
podatki[podatki.naziv == "Hulk"]
podatki[podatki.naziv == "Black Widow"]
podatki[podatki.naziv == "Hawkeye"]
podatki[podatki.naziv == "Thor"]
avengers = podatki.loc[["Thor_Odinson_(Earth-616)", "Clinton_Barton_(Earth-616)", "Natalia_Romanova_(Earth-616)",
"Bruce_Banner_(Earth-616)", "Anthony_Stark_(Earth-616)", "Peter_Parker_(Earth-616)"]]
pd.merge(avengers, tabele, left_on = "id", right_on = "id")
```
Glede na ocene iz tabele sta v šibkejšem delu ekipe *Black Widow* in *Hawkeye*, saj sta "le navadna" človeka. Prav tako se med šibkejšimi znajde *Spider-Man*.
Pričakovano sta najpametnejša člana zasedbe znastvenika *Tony Stark* in *Bruce Banner*, najmočnejša pa *Thor* in *Hulk*.
## ZAKLJUČEK
Tekom analize podatkov sem uspešno potrdil vse 3 izmed 4 delovnih hipotez. Dotaknil pa sem se tudi nekaterih drugih področij in povezav med zbranimi podatki.
Možna dodatka z zbranim podatkom, ki bi omogočila še bolj zanimivo analizo bi bila lahko:
- število prodanih stripov posameznih serij, s čimer bi lahko ugotovil ali so med bralci popularnejši močnejši liki ali ne,
- oznaka junak / zlobnež za vsako osebo, ki bi omogočila vpogled v razporeditev moči v posameznih bitkah.
Avtor: Nejc Zajc, študijsko leto: 2019/2020
|
github_jupyter
|
# naložim paketa, s katerimi obdelujem podatke
import pandas as pd
import numpy as np
# zaradi preglednosti v tabelah izpisujem le 20 vrstic
pd.options.display.max_rows = 20
# izberem stil grafov
%matplotlib notebook
# zaradi zaporednih ukazov, pandas knjižnica včasih navlkjub željenemu delovanju opozarja na kakšne stvari, da se temu
# v končnem izdelku izognem izključim ta opozorila
import warnings
warnings.simplefilter(action='ignore')
# naložim še podatke, s katerimi delam, ker imam vse podatke urejene po "id"-ju, ki je enak končnici spletne strani,
# na kateri se nahaja lik, vrstic v tabelah ni potrebno dodatno številčiti
podatki = pd.read_csv('podatki/podatki.csv', index_col = "id")
avtorji = pd.read_csv('podatki/avtorji.csv', index_col = "id")
moci = pd.read_csv('podatki/moci.csv', index_col = "id")
tabele = pd.read_csv('podatki/tabele.csv', index_col = "id")
podatki.groupby("tip").count()
podatki.groupby("leto").size().plot()
podatki.groupby("naslov").size().sort_values(ascending = False).head(1) # = "Fantastic Four"
podatki[podatki.naslov == "Fantastic Four"].groupby("leto").size()
st_zbranih_izdaj = podatki.groupby("izdaja").size().sum() # 2236 - za toliko likov sem zbral izdajo
v_prvih = podatki[podatki.izdaja == 1].shape[0] # 233 - število likov v prvih delih serij
v_prvih / st_zbranih_izdaj
podatki[podatki.izdaja < 1900].groupby("izdaja").size().tail(20) # opazim nekaj primerov izdaj nad 500!
podatki[(podatki.izdaja > 500) & (podatki.izdaja < 1900)]
pogledam_po_mesecih = podatki[(podatki.mesec != "Spring") & (podatki.mesec != "Summer") &
(podatki.mesec != "Fall") & (podatki.mesec != "Winter")]
meseci = ["January", "February", "March", "April", "May", "June",
"July", "August", "September", "October", "November", "December"]
pogledam_po_mesecih['mesec'] = pd.Categorical(pogledam_po_mesecih['mesec'], categories=meseci, ordered=True)
pogledam_po_mesecih.groupby("mesec").size().to_frame("st_likov").plot.bar()
osebe = podatki[podatki.tip == "Character"]
avtorji.groupby("avtor").size().sort_values(ascending = False)
osebe.groupby("vesolje").size().sort_values(ascending = False).head(10)
skupni = pd.merge(osebe, avtorji, left_on="id", right_on="id") # združim tabeli
po_avtorjih = avtorji.groupby("avtor").size().to_frame("st_sodelovanj") # spremenim v tabelo
po_avtorjih["Earth-616"] = skupni[skupni.vesolje == "Earth-616"].groupby("avtor").size() # dodam koliko likov iz glavne Zemlje
po_avtorjih["delez"] = po_avtorjih["Earth-616"] / po_avtorjih.st_sodelovanj # ter še stolpec, z deležem le-teh
po_avtorjih.sort_values("delez").dropna() # za prikaz izpustim tiste, ki niso nikoli sodelovali z likom iz glavne Zemlje
skupni[skupni.avtor == "John Romita Jr."]
skupni[skupni.avtor == "Larry Lieber"].groupby("vesolje").size()
osebe.groupby("razmerje").size()
osebe.groupby("razmerje").size().Single / osebe.groupby("razmerje").size().sum()
isto = osebe[osebe.naziv == osebe.pravo_ime].count().razmerje # ti imajo naziv enak imenu - teh je 334
spremenjeno = osebe[osebe.naziv != osebe.pravo_ime].count().razmerje # naziv teh je drugačen - teh je 425
a = osebe[osebe.naziv == osebe.pravo_ime].groupby("razmerje").size() # le po istih pogledam njihova razmerja
a
b = osebe[osebe.naziv != osebe.pravo_ime].groupby("razmerje").size() # enako storim za spremenjene
delez_isti = (a.Divorced + a.Separated + a.Widowed) / isto # izračunam delež tistih,
delez_spremenjeno = (b.Divorced + b.Separated + b.Widowed) / spremenjeno # ki so izgubili zvezo za obe skupini
(delez_isti, delez_spremenjeno)
moci.groupby("moc").size().sort_values()
moci_po_osebah = moci.groupby("id").size().to_frame("st_moci")
moci_po_osebah.sort_values("st_moci")
tabele["SUM"] = tabele.INT + tabele.STR + tabele.SPD + tabele.DUR + tabele.ENP + tabele.FGT
tabele.sort_values("SUM")
tabele.mean().sort_values()
tabele.loc["Jean_Grey_(Earth-616)"]
moci.loc["Dormammu_(Earth-616)"]
zdruzeni = pd.merge(moci_po_osebah, tabele[["SUM"]], left_on = "id", right_on = "id")
# naredim tabelo z dvema stolpcema obeh lestvic
zdruzeni.sort_values("SUM").plot().axes.get_xaxis().set_ticks([])# narisem graf, na x-osi so osebe
zdruzeni["na_stiri"] = 4 * ((zdruzeni.SUM + 1) // 4)
na_stiri = zdruzeni.groupby("na_stiri")
na_stiri.mean()[["st_moci"]].plot.bar()
moci_po_a = pd.merge(avtorji, moci_po_osebah, left_on = "id", right_on = "id").groupby("avtor").mean().sort_values("st_moci")
# zdruzim tabelo avtorjev in moči po osebah, in tako dobim tabelo oseb z dodanim stolpcem "st_moci",
# to nato uredim po skupinah po avtorju ter izračunam povprečno število moči
pogosti = avtorji.groupby("avtor").size().sort_values().tail(10).to_frame("zadetki")
# naredim stolpec 10 najbolj pogostih sodelovanj Stan Leeja
pd.merge(moci_po_a, pogosti, right_on = "avtor", left_on = "avtor").sort_values("zadetki")
podatki[podatki.naziv == "Spider-Man"]
podatki[podatki.naziv == "Iron Man"]
podatki[podatki.naziv == "Hulk"]
podatki[podatki.naziv == "Black Widow"]
podatki[podatki.naziv == "Hawkeye"]
podatki[podatki.naziv == "Thor"]
avengers = podatki.loc[["Thor_Odinson_(Earth-616)", "Clinton_Barton_(Earth-616)", "Natalia_Romanova_(Earth-616)",
"Bruce_Banner_(Earth-616)", "Anthony_Stark_(Earth-616)", "Peter_Parker_(Earth-616)"]]
pd.merge(avengers, tabele, left_on = "id", right_on = "id")
| 0.221014 | 0.80651 |
# Presentación y objetivo
El objeto del presente proyecto es el de desarrollar una Red Neuronal Recurrente (RNN) que sea capaz de clasificar con éxito tweets relacionados con el COVID-19. Para este fin, se va a llevar a cabo un análisis de sentimiento de los textos extraídos directamente de Twitter aplicando técnicas de Procesamiento de Lenguaje Natural (NLP).

El conjunto de datos incluye tweets ya etiquetados, por lo que se van a aplicar métodos de aprendizaje supervisados. En esencia, se trata de un problema de clasificación multiclase.
# Carga de librerías necesarias
```
# Cargando librerías para EDA y modelado
import numpy as np
import pandas as pd
import os
import re
import warnings
warnings.filterwarnings('ignore')
import tensorflow as tf
from tensorflow.keras import layers
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import LSTM, Embedding, Bidirectional, Dense, Dropout
from tensorflow.keras.losses import CategoricalCrossentropy
from tensorflow.keras.optimizers import Adam
from sklearn.model_selection import train_test_split
# Para codificaciones one-hot
from tensorflow.keras.utils import to_categorical
# Para tratamiento y limpieza de texto
from tensorflow.keras import preprocessing
from tensorflow.keras.layers.experimental.preprocessing import TextVectorization
# Visualización
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
# Evaluación
from sklearn.metrics import classification_report, accuracy_score
```
# Cargando los datos
Se va a trabajar con dos archivos de partida:
- "Corona_NLP_train.csv", que serán aquellos datos que utilizaremos para entrenar el modelo.
- "Corona_NLP_test.csv", que será el dataset a partir del cual validaremos las clasificaciones obtenidas.
En primer lugar, definimos la cabecera:
```
col_names = [
'User',
'Name',
'Location',
'Date',
'Tweet Content',
'Sentiment'
]
df_train = pd.read_csv('Corona_NLP_train.csv', header = 0, names = col_names, encoding = 'latin1')
df_train.head()
```
El dataset contiene únicamente 6 parámetros, donde **UserName** y **ScreenName** se han representado como códigos numéricos a fin de preservar la privacidad de los usuarios.
Procedemos de forma análoga con los datos de test:
```
df_test = pd.read_csv('Corona_NLP_test.csv', header=0, names = col_names, encoding='latin1')
df_test.head()
```
# Análisis Exploratorio (EDA)
Uno de los pasos preliminares más importantes a la hora de realizar cualquier modelo consiste en llevar a cabo la exploración de los datos con objeto de obtener y comprender toda la información que éstos contienen. Además, esta etapa permite determinar si se presentan datos vacíos o faltantes y si éstos son relevantes para la Red Neuronal.
```
df_train.shape
```
El dataframe de entrenamiento cuenta con 41.157 filas y 6 columnas.
```
df_train.info()
```
Donde 8590 tweets no cuentan con la Ubicación (Location) establecida.
```
df_test.shape
```
El dataframe de test cuenta con 3.798 filas y 6 columnas.
```
df_test.info()
```
Nuevamente, se presentan 834 tweets sin localización.
Para el proyecto, los parámetros relevantes van a ser el contenido de los tweets (texto) así como el sentimiento vinculado a los mismos, por lo que no es necesario complementar la información faltante o suprimir las filas con estos parámetros nulos.
Los posibles estados emocionales asociados a los tweets, que se corresponden con las etiquetas del modelo supervisado, son:
```
for sentiment in df_train['Sentiment'].unique():
print(sentiment)
num_classes = len(df_train['Sentiment'].unique())
print(f"El número total de sentimientos asociados a los textos es de {num_classes}")
```
Para comprobar el número total de tweets ligados a cada estado emocional:
```
print(df_train.Sentiment.value_counts())
```
Comprobamos esta distribución con el siguiente diagrama:
```
plt.figure(figsize=(7, 7))
colors = ['#1b4f72', '#2874a6', '#3498db', '#85c1e9', '#d6eaf8']
df_train['Sentiment'].value_counts().plot(kind='pie', colors = colors, autopct="%0.1f %%")
plt.pie([1,0,0,0], radius = 0.5, colors = 'w')
plt.show()
```
## Hashtags
En este apartado, comprobamos cuáles son los hashtags más frecuentes en los datos de entrenamiento:
```
def find_hash(text):
line = re.findall(r'(?<=#)\w+',text)
return " ".join(line)
df_train['hash'] = df_train['Tweet Content'].apply(lambda x: find_hash(x))
temp = df_train['hash'].value_counts()[:][1:11]
temp = temp.to_frame().reset_index().rename(columns = {'index':'Hashtag','hash':'count'})
plt.figure(figsize =(15, 5))
sns.set_style("darkgrid")
sns.barplot(x = "Hashtag", y = "count", data = temp, palette = 'CMRmap_r')
plt.show()
```
## Menciones
Las menciones más frecuentes se muestran en la siguiente gráfica:
```
def mentions(text):
line = re.findall(r'(?<=@)\w+',text)
return " ".join(line)
df_train['mentions'] = df_train['Tweet Content'].apply(lambda x: mentions(x))
temp = df_train['mentions'].value_counts()[:][1:11]
temp = temp.to_frame().reset_index().rename(columns = {'index':'Mentions','mentions':'count'})
plt.figure(figsize =(15, 5))
sns.barplot(x = "Mentions",y = "count", data = temp, palette = 'RdYlBu_r')
plt.show()
```
# Limpieza de los datos (preprocesamiento)
Se realiza una primera limpieza de los datos:
```
def clean(text):
# eliminando urls
text = re.sub(r'http\S+', '', text)
# eliminando menciones
text = re.sub(r'@\w+','',text)
# eliminando hastags
text = re.sub(r'#\w+', '', text)
# eliminando caracteres extraños
text = re.sub(r'\n', '', text)
text = re.sub(r'\r', '', text)
# eliminando etiquetas
text = re.sub('r<.*?>','', text)
return text
df_train['Tweet Content'] = df_train['Tweet Content'].apply(lambda x: clean(x))
```
Para posteriormente realizar la descomposición de los datos de df_train en entrenamiento y validación:
```
X_train, X_val, y_train, y_val = train_test_split(df_train['Tweet Content'], df_train['Sentiment'], shuffle=True, test_size=0.2)
X_train = np.array(X_train)
X_train
```
Seguidamente definimos el tamaño del vocabulario a partir del cual se compondrán los tokens en vectores así como la forma en que se preprocesarán los textos (todo en minúsculas y sin signos de puntuación, eliminando espacios en blanco):
```
# Tamaño del vocabulario (un índice por token)
vocab_size = 10000
encoder = TextVectorization(
max_tokens = vocab_size,
standardize = "lower_and_strip_punctuation",
split = "whitespace",
ngrams=None,
output_mode = "int",
output_sequence_length=None,
pad_to_max_tokens=False,
vocabulary=None
)
```
Aplicamos el método *adapt* para ajustar el estado de la capa de preprocesamiento al conjunto de datos. Esto hará que el modelo cree un índice de cadenas a números enteros.
***Nota: únicamente se han de emplear los datos de entrenamiento al llamar al método adapt (usar el conjunto de prueba podría filtrar información).***
```
encoder.adapt(X_train)
```
El método .adapt establece el vocabulario. Después del relleno y los tokens desconocidos, se ordenan por frecuencia resultando el siguiente vocabulario:
```
vocab = (encoder.get_vocabulary())
```
Para el cual mostramos las primeras 200 palabras:
```
print(vocab[:200])
vocab = np.array(vocab)
count = 0
vocab = np.array(encoder.get_vocabulary())
vocab
for palabra in vocab:
count += 1
print("El número total de palabras de que se compone el vocabulario es:",count)
```
Vamos a comprobar cómo quedaría el texto codificado para un tweet aleatorio:
```
sample_tweet = X_train[250]
sample_tweet
# Vamos a ver cómo queda este ejemplo codificado
codified_sample = encoder(sample_tweet).numpy()
codified_sample
```
Como se puede apreciar, se ha descompuesto el tweet del ejemplo en un vector con los índices de los tokens en el vocabulario definido.
# Creando el modelo
Transformación one-hot de las variables categóricas:
```
l = {"Neutral" : 0, "Positive" : 1, "Extremely Positive" : 2, "Negative" : 3, "Extremely Negative" : 4}
y_train = y_train.map(l)
y_val = y_val.map(l)
y_train
y_train = to_categorical(y_train, num_classes = 5)
y_val = to_categorical(y_val, num_classes = 5)
y_train
```
Diseñamos las capas de que se compone el modelo:
```
model = Sequential()
model.add(encoder)
model.add(Embedding(input_dim = len(encoder.get_vocabulary()),
output_dim = 32,
# Usamos masking para añadir padding y de este
# modo poder procesar secuencias de distintos tamaños
mask_zero = True))
model.add(Bidirectional(LSTM(32)))
model.add(Dense(32, activation = "relu"))
model.add(Dropout(0.6))
model.add(Dense(5, activation = 'softmax'))
```
Lo compilamos:
```
model.compile(loss = 'categorical_crossentropy',
optimizer = "adam",
metrics = ["accuracy"])
```
Definimos los callbacks:
```
call_back = tf.keras.callbacks.EarlyStopping(patience = 3, restore_best_weights = True)
```
Para finalmente instanciarlo y entrenarlo:
```
%%time
history = model.fit(X_train, y_train,
epochs = 15,
callbacks = [call_back],
validation_data = (X_val, y_val))
sns.set_style("whitegrid")
fig = plt.figure(figsize=(15, 5))
# Plot accuracy
ax = fig.add_subplot(121)
ax.plot(range(7), history.history['accuracy'])
ax.plot(range(7), history.history['val_accuracy'])
ax.legend(['training_acc', 'validation_acc'])
ax.set_title('Accuracy')
# Plot loss
ax2 = fig.add_subplot(122)
ax2.plot(range(7), history.history['loss'])
ax2.plot(range(7), history.history['val_loss'])
ax2.legend(['training_loss', 'validation_loss'])
ax2.set_title('Loss')
plt.show()
```
Se observa como se llega rápidamente al sobreajuste en el momento en que la pérdida de entrenamiento sigue reduciéndose pero en cambio la de validación aumenta. Este hecho se produce a partir de la segunda epoch y se mantiene tantas epochs como patience hayamos definido en el early-stopping.
```
train_lstm_results = model.evaluate(X_train, y_train, verbose=0, batch_size=64)
test_lstm_results = model.evaluate(X_val, y_val, verbose=0, batch_size=64)
print(f'Precisión datos entrenamiento: {train_lstm_results[1]*100:0.2f}')
print(f'Precisión datos validación: {test_lstm_results[1]*100:0.2f}')
```
Con esta evaluación comprobamos que con el modelo alcanzamos una precisión cercana al 89% para los datos de entrenamiento y ligeramente superior al 75% en los de validación.
# Realizando las predicciones
Preprocesamos los datos de test en primer lugar:
```
df_test['Tweet Content'] = df_test['Tweet Content'].apply(lambda x: clean(x))
X_test = df_test['Tweet Content']
y_test = df_test['Sentiment']
y_test = y_test.map(l)
y_test = to_categorical(y_test, num_classes = 5)
y_test
```
Siendo las predicciones:
```
pred = model.predict_classes(np.array(X_test))
pred
pred_cat = to_categorical(pred, num_classes = 5)
pred_cat
```
La precisión sobre los datos de test llega casi al 73%.
```
accuracy_score(y_test, pred_cat)
print(classification_report(y_test, pred_cat))
```
Gracias al classification_report, comprobamos la precisión conseguida para cada una de las etiquetas. Se observa que los textos extremadamente positivos obtienen las mayores precisiones mientras que aquellos cuyo contenido es positivo o negativo son los que menos aciertos obtienen en su clasificación.
|
github_jupyter
|
# Cargando librerías para EDA y modelado
import numpy as np
import pandas as pd
import os
import re
import warnings
warnings.filterwarnings('ignore')
import tensorflow as tf
from tensorflow.keras import layers
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import LSTM, Embedding, Bidirectional, Dense, Dropout
from tensorflow.keras.losses import CategoricalCrossentropy
from tensorflow.keras.optimizers import Adam
from sklearn.model_selection import train_test_split
# Para codificaciones one-hot
from tensorflow.keras.utils import to_categorical
# Para tratamiento y limpieza de texto
from tensorflow.keras import preprocessing
from tensorflow.keras.layers.experimental.preprocessing import TextVectorization
# Visualización
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
# Evaluación
from sklearn.metrics import classification_report, accuracy_score
col_names = [
'User',
'Name',
'Location',
'Date',
'Tweet Content',
'Sentiment'
]
df_train = pd.read_csv('Corona_NLP_train.csv', header = 0, names = col_names, encoding = 'latin1')
df_train.head()
df_test = pd.read_csv('Corona_NLP_test.csv', header=0, names = col_names, encoding='latin1')
df_test.head()
df_train.shape
df_train.info()
df_test.shape
df_test.info()
for sentiment in df_train['Sentiment'].unique():
print(sentiment)
num_classes = len(df_train['Sentiment'].unique())
print(f"El número total de sentimientos asociados a los textos es de {num_classes}")
print(df_train.Sentiment.value_counts())
plt.figure(figsize=(7, 7))
colors = ['#1b4f72', '#2874a6', '#3498db', '#85c1e9', '#d6eaf8']
df_train['Sentiment'].value_counts().plot(kind='pie', colors = colors, autopct="%0.1f %%")
plt.pie([1,0,0,0], radius = 0.5, colors = 'w')
plt.show()
def find_hash(text):
line = re.findall(r'(?<=#)\w+',text)
return " ".join(line)
df_train['hash'] = df_train['Tweet Content'].apply(lambda x: find_hash(x))
temp = df_train['hash'].value_counts()[:][1:11]
temp = temp.to_frame().reset_index().rename(columns = {'index':'Hashtag','hash':'count'})
plt.figure(figsize =(15, 5))
sns.set_style("darkgrid")
sns.barplot(x = "Hashtag", y = "count", data = temp, palette = 'CMRmap_r')
plt.show()
def mentions(text):
line = re.findall(r'(?<=@)\w+',text)
return " ".join(line)
df_train['mentions'] = df_train['Tweet Content'].apply(lambda x: mentions(x))
temp = df_train['mentions'].value_counts()[:][1:11]
temp = temp.to_frame().reset_index().rename(columns = {'index':'Mentions','mentions':'count'})
plt.figure(figsize =(15, 5))
sns.barplot(x = "Mentions",y = "count", data = temp, palette = 'RdYlBu_r')
plt.show()
def clean(text):
# eliminando urls
text = re.sub(r'http\S+', '', text)
# eliminando menciones
text = re.sub(r'@\w+','',text)
# eliminando hastags
text = re.sub(r'#\w+', '', text)
# eliminando caracteres extraños
text = re.sub(r'\n', '', text)
text = re.sub(r'\r', '', text)
# eliminando etiquetas
text = re.sub('r<.*?>','', text)
return text
df_train['Tweet Content'] = df_train['Tweet Content'].apply(lambda x: clean(x))
X_train, X_val, y_train, y_val = train_test_split(df_train['Tweet Content'], df_train['Sentiment'], shuffle=True, test_size=0.2)
X_train = np.array(X_train)
X_train
# Tamaño del vocabulario (un índice por token)
vocab_size = 10000
encoder = TextVectorization(
max_tokens = vocab_size,
standardize = "lower_and_strip_punctuation",
split = "whitespace",
ngrams=None,
output_mode = "int",
output_sequence_length=None,
pad_to_max_tokens=False,
vocabulary=None
)
encoder.adapt(X_train)
vocab = (encoder.get_vocabulary())
print(vocab[:200])
vocab = np.array(vocab)
count = 0
vocab = np.array(encoder.get_vocabulary())
vocab
for palabra in vocab:
count += 1
print("El número total de palabras de que se compone el vocabulario es:",count)
sample_tweet = X_train[250]
sample_tweet
# Vamos a ver cómo queda este ejemplo codificado
codified_sample = encoder(sample_tweet).numpy()
codified_sample
l = {"Neutral" : 0, "Positive" : 1, "Extremely Positive" : 2, "Negative" : 3, "Extremely Negative" : 4}
y_train = y_train.map(l)
y_val = y_val.map(l)
y_train
y_train = to_categorical(y_train, num_classes = 5)
y_val = to_categorical(y_val, num_classes = 5)
y_train
model = Sequential()
model.add(encoder)
model.add(Embedding(input_dim = len(encoder.get_vocabulary()),
output_dim = 32,
# Usamos masking para añadir padding y de este
# modo poder procesar secuencias de distintos tamaños
mask_zero = True))
model.add(Bidirectional(LSTM(32)))
model.add(Dense(32, activation = "relu"))
model.add(Dropout(0.6))
model.add(Dense(5, activation = 'softmax'))
model.compile(loss = 'categorical_crossentropy',
optimizer = "adam",
metrics = ["accuracy"])
call_back = tf.keras.callbacks.EarlyStopping(patience = 3, restore_best_weights = True)
%%time
history = model.fit(X_train, y_train,
epochs = 15,
callbacks = [call_back],
validation_data = (X_val, y_val))
sns.set_style("whitegrid")
fig = plt.figure(figsize=(15, 5))
# Plot accuracy
ax = fig.add_subplot(121)
ax.plot(range(7), history.history['accuracy'])
ax.plot(range(7), history.history['val_accuracy'])
ax.legend(['training_acc', 'validation_acc'])
ax.set_title('Accuracy')
# Plot loss
ax2 = fig.add_subplot(122)
ax2.plot(range(7), history.history['loss'])
ax2.plot(range(7), history.history['val_loss'])
ax2.legend(['training_loss', 'validation_loss'])
ax2.set_title('Loss')
plt.show()
train_lstm_results = model.evaluate(X_train, y_train, verbose=0, batch_size=64)
test_lstm_results = model.evaluate(X_val, y_val, verbose=0, batch_size=64)
print(f'Precisión datos entrenamiento: {train_lstm_results[1]*100:0.2f}')
print(f'Precisión datos validación: {test_lstm_results[1]*100:0.2f}')
df_test['Tweet Content'] = df_test['Tweet Content'].apply(lambda x: clean(x))
X_test = df_test['Tweet Content']
y_test = df_test['Sentiment']
y_test = y_test.map(l)
y_test = to_categorical(y_test, num_classes = 5)
y_test
pred = model.predict_classes(np.array(X_test))
pred
pred_cat = to_categorical(pred, num_classes = 5)
pred_cat
accuracy_score(y_test, pred_cat)
print(classification_report(y_test, pred_cat))
| 0.590661 | 0.919751 |
# Whole-body manipulation
The objective of this exercise is to reach multiple targets while keeping balance in the Talos humanoid robot.
<img src="https://robots.ieee.org/robots/talos/Photos/SD/talos-photo2-full.jpg" alt="drawing" width="250"/>
This exercise focuses on a multi-contact optimal control problem of the form:
\begin{equation}\nonumber
\begin{aligned}
\min_{\mathbf{x}_s,\mathbf{u}_s}
&\hspace{-2.em}
& & \hspace{-0.75em}l_N(\mathbf{x}_{N})+\sum_{k=0}^{N-1} \int_{t_k}^{t_k+\Delta t_k}\hspace{-2.em} l_k(\mathbf{x}_k,\mathbf{u}_k)dt \hspace{-8.em}&\\
& \hspace{-1em}\textrm{s.t.}
& & \mathbf{q}_{k+1} = \mathbf{q}_k \oplus \int_{t_k}^{t_k+\Delta t_k}\hspace{-2.em}\mathbf{v}_{k+1}\,dt, &\textrm{(integrator)}\\
& & & \mathbf{v}_{k+1} = \mathbf{v}_k + \int_{t_k}^{t_k+\Delta t_k}\hspace{-2.em}\mathbf{\dot{v}}_k\,dt, &\\
& & & \hspace{-1em}\left[\begin{matrix}\mathbf{\dot{v}}_k \\ -\boldsymbol{\lambda}_k\end{matrix}\right] =
\left[\begin{matrix}\mathbf{M} & \mathbf{J}^{\top}_c \\ {\mathbf{J}_{c}} & \mathbf{0} \end{matrix}\right]^{-1}
\left[\begin{matrix}\boldsymbol{\tau}_b \\ -\mathbf{a}_0 \\\end{matrix}\right], &\textrm{(contact dynamics)}\\
& & & \mathbf{R}\boldsymbol{\lambda}_{\mathcal{C}(k)} \leq \mathbf{\mathbf{r}}, &\textrm{(friction-cone)}\\
& & & \mathbf{\bar{x}} \leq \mathbf{x}_k \leq \mathbf{\underline{x}}, &\textrm{(state bounds)}
\end{aligned}
\end{equation}}
where $l_i(\mathbf{x}_i, \mathbf{u}_i) = w_{hand}\|\log{(\mathbf{p}_{\mathcal{G}(k)}(\mathbf{q}_k)^{-1} \mathbf{^oM}_{\mathbf{f}_{\mathcal{G}(k)}})}\| + w_{xreg}\|\mathbf{x} - \mathbf{x}_0\|_{Q} + w_{ureg}\|\mathbf{u}\|_{R}$. Note that (1) the first term is the hand placement cost and (2) the terminal cost does not include the control regularization term.
Below there is a basic example that defines the above problem for reaching one target. Later, you will have to build the problem on top of it.
Without no more preamble, let's first declare the robot model and the foot and hand names!
```
import crocoddyl
import example_robot_data
import numpy as np
import pinocchio as pin
# Load robot
robot = example_robot_data.load('talos')
rmodel = robot.model
q0 = rmodel.referenceConfigurations["half_sitting"]
x0 = np.concatenate([q0, np.zeros(rmodel.nv)])
# Declaring the foot and hand names
rf_name = "right_sole_link"
lf_name = "left_sole_link"
lh_name = "gripper_left_joint"
# Getting the frame ids
rf_id = rmodel.getFrameId(rf_name)
lf_id = rmodel.getFrameId(lf_name)
lh_id = rmodel.getFrameId(lh_name)
# Define the robot's state and actuation
state = crocoddyl.StateMultibody(rmodel)
actuation = crocoddyl.ActuationModelFloatingBase(state)
```
With the following function, we could build a differential action model giving a desired hand target.
The function builds a double-support contact phase and defines a hand-placement task. The cost function also includes:
- state and control regularization terms
- state limits penalization
- friction cone penalization
```
def createActionModel(target):
# Creating a double-support contact (feet support)
contacts = crocoddyl.ContactModelMultiple(state, actuation.nu)
lf_contact = crocoddyl.ContactModel6D(state, lf_id, pin.SE3.Identity(), actuation.nu, np.array([0, 0]))
rf_contact = crocoddyl.ContactModel6D(state, rf_id, pin.SE3.Identity(), actuation.nu, np.array([0, 0]))
contacts.addContact("lf_contact", lf_contact)
contacts.addContact("rf_contact", rf_contact)
# Define the cost sum (cost manager)
costs = crocoddyl.CostModelSum(state, actuation.nu)
# Adding the hand-placement cost
w_hand = np.array([1] * 3 + [0.0001] * 3)
lh_Mref = pin.SE3(np.eye(3), target)
activation_hand = crocoddyl.ActivationModelWeightedQuad(w_hand**2)
lh_cost = crocoddyl.CostModelFramePlacement(state, activation_hand, lh_id, lh_Mref, actuation.nu)
costs.addCost("lh_goal", lh_cost, 1e2)
# Adding state and control regularization terms
w_x = np.array([0] * 3 + [10.] * 3 + [0.01] * (state.nv - 6) + [10] * state.nv)
activation_xreg = crocoddyl.ActivationModelWeightedQuad(w_x**2)
x_reg_cost = crocoddyl.CostModelState(state, activation_xreg, x0, actuation.nu)
u_reg_cost = crocoddyl.CostModelControl(state, actuation.nu)
costs.addCost("xReg", x_reg_cost, 1e-3)
costs.addCost("uReg", u_reg_cost, 1e-4)
# Adding the state limits penalization
x_lb = np.concatenate([state.lb[1:state.nv + 1], state.lb[-state.nv:]])
x_ub = np.concatenate([state.ub[1:state.nv + 1], state.ub[-state.nv:]])
activation_xbounds = crocoddyl.ActivationModelQuadraticBarrier(crocoddyl.ActivationBounds(x_lb, x_ub))
x_bounds = crocoddyl.CostModelState(state, activation_xbounds, 0 * x0, actuation.nu)
costs.addCost("xBounds", x_bounds, 1.)
# Adding the friction cone penalization
nsurf, mu = np.array([0, 0, 1]), 0.7
cone = crocoddyl.FrictionCone(nsurf, mu, 4, False)
activation_friction = crocoddyl.ActivationModelQuadraticBarrier(crocoddyl.ActivationBounds(cone.lb, cone.ub))
lf_friction = crocoddyl.CostModelContactFrictionCone(state, activation_friction, lf_id, cone, actuation.nu)
rf_friction = crocoddyl.CostModelContactFrictionCone(state, activation_friction, rf_id, cone, actuation.nu)
costs.addCost("lf_friction", lf_friction, 1e1)
costs.addCost("rf_friction", rf_friction, 1e1)
# Creating the action model
dmodel = crocoddyl.DifferentialActionModelContactFwdDynamics(state, actuation, contacts, costs)
return dmodel
```
And to easily build a sequence of tasks, we have the following function
```
def createSequence(dmodels, DT, N):
return [[crocoddyl.IntegratedActionModelEuler(m, DT)] * N +
[crocoddyl.IntegratedActionModelEuler(m, 0.)] for m in dmodels]
```
Finally, the following function allows us to display the motions and desired targets:
```
import meshcat.geometry as g
import meshcat.transformations as tf
def createDisplay(targets):
display = crocoddyl.MeshcatDisplay(robot, 4, 4, False)
for i, target in enumerate(targets):
display.robot.viewer["target_" + str(i)].set_object(g.Sphere(0.05))
Href = np.array([[1., 0., 0., target[0]],
[0., 1., 0., target[1]],
[0., 0., 1., target[2]],
[0., 0., 0., 1.]])
display.robot.viewer["target_" + str(i)].set_transform(np.array([[1., 0., 0., target[0]],
[0., 1., 0., target[1]],
[0., 0., 1., target[2]],
[0., 0., 0., 1.]]))
return display
```
Now, we create an optimal control problem to reach a single target
```
DT, N = 5e-2, 20
target = np.array([0.4, 0, 1.2])
# Creating a running model for the target
dmodel = createActionModel(target)
seqs = createSequence([dmodel], DT, N)
# Defining the problem and the solver
problem = crocoddyl.ShootingProblem(x0, sum(seqs, [])[:-1], seqs[-1][-1])
fddp = crocoddyl.SolverFDDP(problem)
# Creating display
display = createDisplay([target])
# Adding callbacks to inspect the evolution of the solver (logs are printed in the terminal)
fddp.setCallbacks([crocoddyl.CallbackVerbose(), crocoddyl.CallbackDisplay(display)])
# Embedded in this cell
display.robot.viewer.jupyter_cell()
```
Let's solve this problem!
```
print("Problem solved:", fddp.solve())
print("Number of iterations:", fddp.iter)
print("Total cost:", fddp.cost)
print("Gradient norm:", fddp.stoppingCriteria())
```
You could display again the final solution
```
display.rate = -1
display.freq = 1
display.displayFromSolver(fddp)
```
## Modifying the example
Let's build an optimal control problem to reach 4 targets as described below:
```
targets = []
targets += [np.array([0.4, 0.1, 1.2])]
targets += [np.array([0.6, 0.1, 1.2])]
targets += [np.array([0.6, -0.1, 1.2])]
targets += [np.array([0.4, -0.1, 1.2])]
```
Now let's display the targets in Meshcat. Do not forget to embed again the display into the jupyter cell
```
display = createDisplay(targets)
# Embedded in this cell
display.robot.viewer.jupyter_cell()
```
After checking that everything is alright, it's time to build the sequence!
Do not forget to create the problem as well :)
And we solve it as before
```
# Create the FDDP solver
fddp = crocoddyl.SolverFDDP(problem)
fddp.setCallbacks([crocoddyl.CallbackVerbose(), crocoddyl.CallbackDisplay(display)])
# Solves the problem
print("Problem solved:", fddp.solve())
print("Number of iterations:", fddp.iter)
print("Total cost:", fddp.cost)
print("Gradient norm:", fddp.stoppingCriteria())
```
Do not miss the change to display the motion at the right display speed!
```
display.rate = -1
display.freq = 1
display.displayFromSolver(fddp)
```
## Same targets with right hand
You've learned how to reach 4 targets with the left hand, congratulations!
To keep playing within this problem, you should create a new createActionModel to achieve the same task for the right hand.
```
def createActionModel(target):
# now god is with you xD
# time to show you up!
```
And here you need to create the problem and solve.
Do not forget to display the results
|
github_jupyter
|
import crocoddyl
import example_robot_data
import numpy as np
import pinocchio as pin
# Load robot
robot = example_robot_data.load('talos')
rmodel = robot.model
q0 = rmodel.referenceConfigurations["half_sitting"]
x0 = np.concatenate([q0, np.zeros(rmodel.nv)])
# Declaring the foot and hand names
rf_name = "right_sole_link"
lf_name = "left_sole_link"
lh_name = "gripper_left_joint"
# Getting the frame ids
rf_id = rmodel.getFrameId(rf_name)
lf_id = rmodel.getFrameId(lf_name)
lh_id = rmodel.getFrameId(lh_name)
# Define the robot's state and actuation
state = crocoddyl.StateMultibody(rmodel)
actuation = crocoddyl.ActuationModelFloatingBase(state)
def createActionModel(target):
# Creating a double-support contact (feet support)
contacts = crocoddyl.ContactModelMultiple(state, actuation.nu)
lf_contact = crocoddyl.ContactModel6D(state, lf_id, pin.SE3.Identity(), actuation.nu, np.array([0, 0]))
rf_contact = crocoddyl.ContactModel6D(state, rf_id, pin.SE3.Identity(), actuation.nu, np.array([0, 0]))
contacts.addContact("lf_contact", lf_contact)
contacts.addContact("rf_contact", rf_contact)
# Define the cost sum (cost manager)
costs = crocoddyl.CostModelSum(state, actuation.nu)
# Adding the hand-placement cost
w_hand = np.array([1] * 3 + [0.0001] * 3)
lh_Mref = pin.SE3(np.eye(3), target)
activation_hand = crocoddyl.ActivationModelWeightedQuad(w_hand**2)
lh_cost = crocoddyl.CostModelFramePlacement(state, activation_hand, lh_id, lh_Mref, actuation.nu)
costs.addCost("lh_goal", lh_cost, 1e2)
# Adding state and control regularization terms
w_x = np.array([0] * 3 + [10.] * 3 + [0.01] * (state.nv - 6) + [10] * state.nv)
activation_xreg = crocoddyl.ActivationModelWeightedQuad(w_x**2)
x_reg_cost = crocoddyl.CostModelState(state, activation_xreg, x0, actuation.nu)
u_reg_cost = crocoddyl.CostModelControl(state, actuation.nu)
costs.addCost("xReg", x_reg_cost, 1e-3)
costs.addCost("uReg", u_reg_cost, 1e-4)
# Adding the state limits penalization
x_lb = np.concatenate([state.lb[1:state.nv + 1], state.lb[-state.nv:]])
x_ub = np.concatenate([state.ub[1:state.nv + 1], state.ub[-state.nv:]])
activation_xbounds = crocoddyl.ActivationModelQuadraticBarrier(crocoddyl.ActivationBounds(x_lb, x_ub))
x_bounds = crocoddyl.CostModelState(state, activation_xbounds, 0 * x0, actuation.nu)
costs.addCost("xBounds", x_bounds, 1.)
# Adding the friction cone penalization
nsurf, mu = np.array([0, 0, 1]), 0.7
cone = crocoddyl.FrictionCone(nsurf, mu, 4, False)
activation_friction = crocoddyl.ActivationModelQuadraticBarrier(crocoddyl.ActivationBounds(cone.lb, cone.ub))
lf_friction = crocoddyl.CostModelContactFrictionCone(state, activation_friction, lf_id, cone, actuation.nu)
rf_friction = crocoddyl.CostModelContactFrictionCone(state, activation_friction, rf_id, cone, actuation.nu)
costs.addCost("lf_friction", lf_friction, 1e1)
costs.addCost("rf_friction", rf_friction, 1e1)
# Creating the action model
dmodel = crocoddyl.DifferentialActionModelContactFwdDynamics(state, actuation, contacts, costs)
return dmodel
def createSequence(dmodels, DT, N):
return [[crocoddyl.IntegratedActionModelEuler(m, DT)] * N +
[crocoddyl.IntegratedActionModelEuler(m, 0.)] for m in dmodels]
import meshcat.geometry as g
import meshcat.transformations as tf
def createDisplay(targets):
display = crocoddyl.MeshcatDisplay(robot, 4, 4, False)
for i, target in enumerate(targets):
display.robot.viewer["target_" + str(i)].set_object(g.Sphere(0.05))
Href = np.array([[1., 0., 0., target[0]],
[0., 1., 0., target[1]],
[0., 0., 1., target[2]],
[0., 0., 0., 1.]])
display.robot.viewer["target_" + str(i)].set_transform(np.array([[1., 0., 0., target[0]],
[0., 1., 0., target[1]],
[0., 0., 1., target[2]],
[0., 0., 0., 1.]]))
return display
DT, N = 5e-2, 20
target = np.array([0.4, 0, 1.2])
# Creating a running model for the target
dmodel = createActionModel(target)
seqs = createSequence([dmodel], DT, N)
# Defining the problem and the solver
problem = crocoddyl.ShootingProblem(x0, sum(seqs, [])[:-1], seqs[-1][-1])
fddp = crocoddyl.SolverFDDP(problem)
# Creating display
display = createDisplay([target])
# Adding callbacks to inspect the evolution of the solver (logs are printed in the terminal)
fddp.setCallbacks([crocoddyl.CallbackVerbose(), crocoddyl.CallbackDisplay(display)])
# Embedded in this cell
display.robot.viewer.jupyter_cell()
print("Problem solved:", fddp.solve())
print("Number of iterations:", fddp.iter)
print("Total cost:", fddp.cost)
print("Gradient norm:", fddp.stoppingCriteria())
display.rate = -1
display.freq = 1
display.displayFromSolver(fddp)
targets = []
targets += [np.array([0.4, 0.1, 1.2])]
targets += [np.array([0.6, 0.1, 1.2])]
targets += [np.array([0.6, -0.1, 1.2])]
targets += [np.array([0.4, -0.1, 1.2])]
display = createDisplay(targets)
# Embedded in this cell
display.robot.viewer.jupyter_cell()
# Create the FDDP solver
fddp = crocoddyl.SolverFDDP(problem)
fddp.setCallbacks([crocoddyl.CallbackVerbose(), crocoddyl.CallbackDisplay(display)])
# Solves the problem
print("Problem solved:", fddp.solve())
print("Number of iterations:", fddp.iter)
print("Total cost:", fddp.cost)
print("Gradient norm:", fddp.stoppingCriteria())
display.rate = -1
display.freq = 1
display.displayFromSolver(fddp)
def createActionModel(target):
# now god is with you xD
# time to show you up!
| 0.776877 | 0.959573 |
Para entrar no modo apresentação, execute a seguinte célula e pressione `-`
```
%reload_ext slide
```
<img src="images/intro.png" alt="João Felipe Pimentel. Universidade Federal Fluminense" width="800"/>
<span class="notebook-slide-start"/>
Para manter a **qualidade de software** é necessário **monitorar** e **extrair métricas**.
<img src="images/extractmetrics.svg" alt="Software sendo analizado" width="100"/>
Durante a evolução do software, não só funcionalidades vão sendo adicionadas e removidas, mas o processo de desenvolvimento também muda. Às vezes novas métricas são adicionadas. Às vezes métricas antigas deixam de fazer sentido.
<img src="images/evolution.svg" alt="Evolução do software" width="auto"/>
Em algumas situações, o processo de monitoramento precisa ser **exploratório** para que se obtenham informações relevantes para o momento.
<img src="images/complexity.svg" alt="Software sendo analizado" width="auto"/>
Para facilitar explorações e melhorias contínuas do processo, é interessante que as análises sejam feitas de forma **interativa**, ou seja, com a possibilidade de fazer análises explorativas e integrar análises prontas ao processo.
Para obter dados para análises, precisamos **minerar** repositórios de software sob demanda.
<img src="images/miningv0.svg" alt="Mineração e Análise Interativa de Software" width="auto"/>
Este minicurso está dividido em 2 partes:
- Interatividade
- Mineração
<img src="images/mining.svg" alt="Mineração e Análise Interativa de Software" width="auto"/>
## Por que Jupyter?
Ferramenta que permite combinar código, texto, visualização, e widgets interativos.
O código fica organizado em células que podem ser executadas e re-executadas em qualquer ordem e de acordo com o desejo do usuário.
Em uma análise explorativa, é possivel manter resultados parciais, evitando esforço computacional.
Extensões ao Python facilitam algumas tarefas.
## Interatividade
Será apresentado o Jupyter com as modificações ao Python proporcionadas pelo IPython, tais como **bang expressions**, **line magics** e **cell magics**.
Para visualização, usaremos a biblioteca **matplotlib** e e estenderemos a visualização rica do Jupyter para formar grafos com programa **GraphViz**.
Por fim, widgets interativos do **ipywidgets** serão apresentados.
## Mineração
Para mineração, criaremos um servidor de proxy em **Flask**, usaremos a biblioteca **requests** para fazer requisições web, extrairemos informações de repositórios usando comandos do **git** e usaremos a biblioteca **Pygit2** para auxiliar a extração dessas informações.
As requisições web serão feitas com 3 objetivos:
- Obter uma página HTML e usar a biblioteca **BeautifulSoup** para extrair informações dela.
- Acessar a API v3 do GitHub, que utiliza REST
- Acessar a API v4 do GitHub, que utiliza GraphQL
## Minicurso
O minicurso está disponível no GitHub:
> https://github.com/JoaoFelipe/minicurso-mineracao-interativa
URL curta:
> https://cutit.org/MIGHUB
Ao longo do minicurso, passarei exercícios. A melhor forma de acompanhar sem perder tempo instalando dependências é pelo GitPod
> https://gitpod.io/#https://github.com/JoaoFelipe/minicurso-mineracao-interativa
URL curta:
> https://cutit.org/MIGPOD
## Gitpod
> https://cutit.org/MIGPOD
Para entrar no GitPod, basta autorizar a conexão com uma conta do GitHub.
Ao iniciar o ambiente, digite `"echo $jupynb"` no terminal e copie e cole o resultado também no terminal:
```
jupyter notebook --NotebookApp.allow_origin=\'$(gp url 8888)\' --ip='*' --NotebookApp.token='' --NotebookApp.password=''
```
Isso iniciará o Jupyter com toda a apresentação. Estamos no arquivo [1.Introducao.ipynb](1.Introducao.ipynb)
## Agenda
Vou começar apresentando o **Jupyter** com algumas funcionalidades para interatividade, apresentadas no Notebook [2.Jupyter.ipynb](2.Jupyter.ipynb) e [3.IPython.ipynb](3.IPython.ipynb).
Em seguida vou falar de **mineração de repositórios**, apresentando os notebooks [4.Proxy.ipynb](4.Proxy.ipynb), [5.Crawling.ipynb](5.Crawling.ipynb), [6.API.v3.ipynb](6.API.v3.ipynb), [7.API.v4.ipynb](7.API.v4.ipynb), [8.Git.ipynb](8.Git.ipynb), [9.Pygit2.ipynb](9.Pygit2.ipynb).
Por fim, vou voltar para a parte de interatividade para falar sobre formas de **estender** o Jupyter e **ipywidgets**, apresentando os notebooks [10.Visualizacao.Rica.ipynb](10.Visualizacao.Rica.ipynb) e [11.Widgets.ipynb](11.Widgets.ipynb).
Programação do dia:
- **09:00 - 10:00: Minicurso**
- 10:00 - 10:30: Coffee Break
- **10:30 - 12:00: Minicurso**
- 12:00 - 13:30: Almoço
- **13:30 - 15:45: Minicurso**
## Um pedido de desculpas
Descrição do minicurso
> O mini-curso tem o objetivo de apresentar mineração interativa de repositórios com o objetivo de melhoria continua de processos. O mini-curso abordará 4 tópicos: interatividade, coleta de dados, análise e visualização. Para interatividade, será apresentada a ferramenta Jupyter Notebook, indicando como ela pode ser usada para tarefas exploratórias e para a construção de dashboards. Para a coleta de dados, será usada a API do GitHub para obter issues de um repositório e a biblioteca PyGit2 para navegar no histórico. Para análise dos dados, será usada a biblioteca pandas. Por fim, para a visualização dos dados, será usada a biblioteca Matplotlib. O mini-curso será guiado por tarefas tais como observar a densidade de defeitos do projeto com o passar do tempo, descobrir quem são os desenvolvedores que mais contibuíram com o projeto no decorrer do tempo, ~~medir a cobertura de testes ao longo do tempo~~, etc.
O projeto que vou usar de exemplo ao longo da apresentação (`gems-uff/sapos`) mudou bastante ao longo do tempo e preparar um ambiente para medir **cobertura de testes** ao longo do tempo se mostrou mais complicado do que eu gostaria. Por conta disso, essa operação foi substituída por uma mais simples: medir a quantidade de linhas ao longo do tempo.
Continua: [2.Jupyter.ipynb](2.Jupyter.ipynb)
|
github_jupyter
|
%reload_ext slide
jupyter notebook --NotebookApp.allow_origin=\'$(gp url 8888)\' --ip='*' --NotebookApp.token='' --NotebookApp.password=''
| 0.241311 | 0.93115 |
STAT 453: Deep Learning (Spring 2020)
Instructor: Sebastian Raschka (sraschka@wisc.edu)
- Course website: http://pages.stat.wisc.edu/~sraschka/teaching/stat453-ss2020/
- GitHub repository: https://github.com/rasbt/stat453-deep-learning-ss20
# RNN with LSTM
Demo of a simple RNN for sentiment classification (here: a binary classification problem with two labels, positive and negative) using LSTM (Long Short Term Memory) cells.
```
%load_ext watermark
%watermark -a 'Sebastian Raschka' -v -p torch
import torch
import torch.nn.functional as F
from torchtext import data
from torchtext import datasets
import time
import random
torch.backends.cudnn.deterministic = True
```
## General Settings
```
RANDOM_SEED = 123
torch.manual_seed(RANDOM_SEED)
VOCABULARY_SIZE = 20000
LEARNING_RATE = 1e-4
BATCH_SIZE = 128
NUM_EPOCHS = 15
DEVICE = torch.device('cuda:1' if torch.cuda.is_available() else 'cpu')
EMBEDDING_DIM = 128
HIDDEN_DIM = 256
OUTPUT_DIM = 1
```
## Dataset
Load the IMDB Movie Review dataset:
```
TEXT = data.Field(tokenize='spacy',
include_lengths=True) # necessary for packed_padded_sequence
LABEL = data.LabelField(dtype=torch.float)
train_data, test_data = datasets.IMDB.splits(TEXT, LABEL)
train_data, valid_data = train_data.split(random_state=random.seed(RANDOM_SEED),
split_ratio=0.8)
print(f'Num Train: {len(train_data)}')
print(f'Num Valid: {len(valid_data)}')
print(f'Num Test: {len(test_data)}')
```
Build the vocabulary based on the top "VOCABULARY_SIZE" words:
```
TEXT.build_vocab(train_data, max_size=VOCABULARY_SIZE)
LABEL.build_vocab(train_data)
print(f'Vocabulary size: {len(TEXT.vocab)}')
print(f'Number of classes: {len(LABEL.vocab)}')
```
The TEXT.vocab dictionary will contain the word counts and indices. The reason why the number of words is VOCABULARY_SIZE + 2 is that it contains to special tokens for padding and unknown words: `<unk>` and `<pad>`.
Make dataset iterators:
```
train_loader, valid_loader, test_loader = data.BucketIterator.splits(
(train_data, valid_data, test_data),
batch_size=BATCH_SIZE,
sort_within_batch=True, # necessary for packed_padded_sequence
device=DEVICE)
```
Testing the iterators (note that the number of rows depends on the longest document in the respective batch):
```
print('Train')
for batch in train_loader:
print(f'Text matrix size: {batch.text[0].size()}')
print(f'Target vector size: {batch.label.size()}')
break
print('\nValid:')
for batch in valid_loader:
print(f'Text matrix size: {batch.text[0].size()}')
print(f'Target vector size: {batch.label.size()}')
break
print('\nTest:')
for batch in test_loader:
print(f'Text matrix size: {batch.text[0].size()}')
print(f'Target vector size: {batch.label.size()}')
break
```
## Model
```
import torch.nn as nn
class RNN(nn.Module):
def __init__(self, input_dim, embedding_dim, hidden_dim, output_dim):
super().__init__()
self.embedding = nn.Embedding(input_dim, embedding_dim)
self.rnn = nn.LSTM(embedding_dim, hidden_dim)
self.fc = nn.Linear(hidden_dim, output_dim)
def forward(self, text, text_length):
#[sentence len, batch size] => [sentence len, batch size, embedding size]
embedded = self.embedding(text)
packed = torch.nn.utils.rnn.pack_padded_sequence(embedded, text_length)
#[sentence len, batch size, embedding size] =>
# output: [sentence len, batch size, hidden size]
# hidden: [1, batch size, hidden size]
packed_output, (hidden, cell) = self.rnn(packed)
return self.fc(hidden.squeeze(0)).view(-1)
INPUT_DIM = len(TEXT.vocab)
torch.manual_seed(RANDOM_SEED)
model = RNN(INPUT_DIM, EMBEDDING_DIM, HIDDEN_DIM, OUTPUT_DIM)
model = model.to(DEVICE)
optimizer = torch.optim.Adam(model.parameters(), lr=LEARNING_RATE)
```
## Training
```
def compute_binary_accuracy(model, data_loader, device):
model.eval()
correct_pred, num_examples = 0, 0
with torch.no_grad():
for batch_idx, batch_data in enumerate(data_loader):
text, text_lengths = batch_data.text
logits = model(text, text_lengths)
predicted_labels = (torch.sigmoid(logits) > 0.5).long()
num_examples += batch_data.label.size(0)
correct_pred += (predicted_labels == batch_data.label.long()).sum()
return correct_pred.float()/num_examples * 100
start_time = time.time()
for epoch in range(NUM_EPOCHS):
model.train()
for batch_idx, batch_data in enumerate(train_loader):
text, text_lengths = batch_data.text
### FORWARD AND BACK PROP
logits = model(text, text_lengths)
cost = F.binary_cross_entropy_with_logits(logits, batch_data.label)
optimizer.zero_grad()
cost.backward()
### UPDATE MODEL PARAMETERS
optimizer.step()
### LOGGING
if not batch_idx % 50:
print (f'Epoch: {epoch+1:03d}/{NUM_EPOCHS:03d} | '
f'Batch {batch_idx:03d}/{len(train_loader):03d} | '
f'Cost: {cost:.4f}')
with torch.set_grad_enabled(False):
print(f'training accuracy: '
f'{compute_binary_accuracy(model, train_loader, DEVICE):.2f}%'
f'\nvalid accuracy: '
f'{compute_binary_accuracy(model, valid_loader, DEVICE):.2f}%')
print(f'Time elapsed: {(time.time() - start_time)/60:.2f} min')
print(f'Total Training Time: {(time.time() - start_time)/60:.2f} min')
print(f'Test accuracy: {compute_binary_accuracy(model, test_loader, DEVICE):.2f}%')
import spacy
nlp = spacy.load('en')
def predict_sentiment(model, sentence):
# based on:
# https://github.com/bentrevett/pytorch-sentiment-analysis/blob/
# master/2%20-%20Upgraded%20Sentiment%20Analysis.ipynb
model.eval()
tokenized = [tok.text for tok in nlp.tokenizer(sentence)]
indexed = [TEXT.vocab.stoi[t] for t in tokenized]
length = [len(indexed)]
tensor = torch.LongTensor(indexed).to(DEVICE)
tensor = tensor.unsqueeze(1)
length_tensor = torch.LongTensor(length)
prediction = torch.sigmoid(model(tensor, length_tensor))
return prediction.item()
print('Probability positive:')
predict_sentiment(model, "I really love this movie. This movie is so great!")
```
|
github_jupyter
|
%load_ext watermark
%watermark -a 'Sebastian Raschka' -v -p torch
import torch
import torch.nn.functional as F
from torchtext import data
from torchtext import datasets
import time
import random
torch.backends.cudnn.deterministic = True
RANDOM_SEED = 123
torch.manual_seed(RANDOM_SEED)
VOCABULARY_SIZE = 20000
LEARNING_RATE = 1e-4
BATCH_SIZE = 128
NUM_EPOCHS = 15
DEVICE = torch.device('cuda:1' if torch.cuda.is_available() else 'cpu')
EMBEDDING_DIM = 128
HIDDEN_DIM = 256
OUTPUT_DIM = 1
TEXT = data.Field(tokenize='spacy',
include_lengths=True) # necessary for packed_padded_sequence
LABEL = data.LabelField(dtype=torch.float)
train_data, test_data = datasets.IMDB.splits(TEXT, LABEL)
train_data, valid_data = train_data.split(random_state=random.seed(RANDOM_SEED),
split_ratio=0.8)
print(f'Num Train: {len(train_data)}')
print(f'Num Valid: {len(valid_data)}')
print(f'Num Test: {len(test_data)}')
TEXT.build_vocab(train_data, max_size=VOCABULARY_SIZE)
LABEL.build_vocab(train_data)
print(f'Vocabulary size: {len(TEXT.vocab)}')
print(f'Number of classes: {len(LABEL.vocab)}')
train_loader, valid_loader, test_loader = data.BucketIterator.splits(
(train_data, valid_data, test_data),
batch_size=BATCH_SIZE,
sort_within_batch=True, # necessary for packed_padded_sequence
device=DEVICE)
print('Train')
for batch in train_loader:
print(f'Text matrix size: {batch.text[0].size()}')
print(f'Target vector size: {batch.label.size()}')
break
print('\nValid:')
for batch in valid_loader:
print(f'Text matrix size: {batch.text[0].size()}')
print(f'Target vector size: {batch.label.size()}')
break
print('\nTest:')
for batch in test_loader:
print(f'Text matrix size: {batch.text[0].size()}')
print(f'Target vector size: {batch.label.size()}')
break
import torch.nn as nn
class RNN(nn.Module):
def __init__(self, input_dim, embedding_dim, hidden_dim, output_dim):
super().__init__()
self.embedding = nn.Embedding(input_dim, embedding_dim)
self.rnn = nn.LSTM(embedding_dim, hidden_dim)
self.fc = nn.Linear(hidden_dim, output_dim)
def forward(self, text, text_length):
#[sentence len, batch size] => [sentence len, batch size, embedding size]
embedded = self.embedding(text)
packed = torch.nn.utils.rnn.pack_padded_sequence(embedded, text_length)
#[sentence len, batch size, embedding size] =>
# output: [sentence len, batch size, hidden size]
# hidden: [1, batch size, hidden size]
packed_output, (hidden, cell) = self.rnn(packed)
return self.fc(hidden.squeeze(0)).view(-1)
INPUT_DIM = len(TEXT.vocab)
torch.manual_seed(RANDOM_SEED)
model = RNN(INPUT_DIM, EMBEDDING_DIM, HIDDEN_DIM, OUTPUT_DIM)
model = model.to(DEVICE)
optimizer = torch.optim.Adam(model.parameters(), lr=LEARNING_RATE)
def compute_binary_accuracy(model, data_loader, device):
model.eval()
correct_pred, num_examples = 0, 0
with torch.no_grad():
for batch_idx, batch_data in enumerate(data_loader):
text, text_lengths = batch_data.text
logits = model(text, text_lengths)
predicted_labels = (torch.sigmoid(logits) > 0.5).long()
num_examples += batch_data.label.size(0)
correct_pred += (predicted_labels == batch_data.label.long()).sum()
return correct_pred.float()/num_examples * 100
start_time = time.time()
for epoch in range(NUM_EPOCHS):
model.train()
for batch_idx, batch_data in enumerate(train_loader):
text, text_lengths = batch_data.text
### FORWARD AND BACK PROP
logits = model(text, text_lengths)
cost = F.binary_cross_entropy_with_logits(logits, batch_data.label)
optimizer.zero_grad()
cost.backward()
### UPDATE MODEL PARAMETERS
optimizer.step()
### LOGGING
if not batch_idx % 50:
print (f'Epoch: {epoch+1:03d}/{NUM_EPOCHS:03d} | '
f'Batch {batch_idx:03d}/{len(train_loader):03d} | '
f'Cost: {cost:.4f}')
with torch.set_grad_enabled(False):
print(f'training accuracy: '
f'{compute_binary_accuracy(model, train_loader, DEVICE):.2f}%'
f'\nvalid accuracy: '
f'{compute_binary_accuracy(model, valid_loader, DEVICE):.2f}%')
print(f'Time elapsed: {(time.time() - start_time)/60:.2f} min')
print(f'Total Training Time: {(time.time() - start_time)/60:.2f} min')
print(f'Test accuracy: {compute_binary_accuracy(model, test_loader, DEVICE):.2f}%')
import spacy
nlp = spacy.load('en')
def predict_sentiment(model, sentence):
# based on:
# https://github.com/bentrevett/pytorch-sentiment-analysis/blob/
# master/2%20-%20Upgraded%20Sentiment%20Analysis.ipynb
model.eval()
tokenized = [tok.text for tok in nlp.tokenizer(sentence)]
indexed = [TEXT.vocab.stoi[t] for t in tokenized]
length = [len(indexed)]
tensor = torch.LongTensor(indexed).to(DEVICE)
tensor = tensor.unsqueeze(1)
length_tensor = torch.LongTensor(length)
prediction = torch.sigmoid(model(tensor, length_tensor))
return prediction.item()
print('Probability positive:')
predict_sentiment(model, "I really love this movie. This movie is so great!")
| 0.874573 | 0.923108 |
```
print('Hello world')
from IPython.display import Image
from IPython.core.display import HTML
```
# Tervetuloa opintojaksolle Johdanto datatieteeseen
Tiedot opintojakson suorittamisesta löytyy <a href="https://infotuni.github.io/joda2022/">GitHub:sta</a>.
Luennoijana toimii <a href="https://www.tuni.fi/fi/jukka-huhtamaki">Jukka Huhtamäki</a> ([@jnkka](https://twitter.com/jnkka)).
Opetusassistenttina toimivat Erjon Skenderi, Pihla Toivanen ja Saeid Heshmatisafa.
Luentomuistion valmisteli [Arho Suominen](https://www.tuni.fi/fi/ajankohtaista/kun-teknologia-muuttuu-yrityksen-taytyy-loytaa-keinot-sopeutua-muutokseen).
## Odotukset keväälle - mitä ihmettä on Johdanto datatieteeseen!
"Data Scientist" kääntyy suomeksi yleensä muotoon datatieteilijä. Mitä muita vaihtoehtoja voisi olla? Tietojen tutkija? Mitä tarkoitetaan datatieteellä ja mitä odotuksia opiskelijoilla on opintojaksolle?
<img src="https://upload.wikimedia.org/wikipedia/commons/7/7f/Data_scientist_Venn_diagram.png" alt="Datatieteilijän osaamisprofiili" width="600" />
Datatiede rakentuu neljän laajan kokonaisuuden varaan:
* liiketoimintaosaaminen,
* ohjelmointi- ja tietokantaosaaminen,
* tilastollinen analyysi ja
* datalähtöinen viestintä ja visualisointi.
Opiskelijoilta toivotaan perusosaamista näiltä aloilta. Opintojaksolla on tavoitteena syventyä näihin aiheisiin datatieteen näkökulmasta sekä esitellä opiskelijoille riittävät tiedot datatiedeosaamiseen kuuluvien taitojen hankkimiseen Tampereen yliopiston opetustarjonnasta.
Vaihtoehtoinen nimi opintojaksolle voisi olla Tietojohtamisen datatiede. Johdannosta huolimatta tällä opintojaksolla myös toteutetaan datatiedeprosesseja käytännössä!
## Suorittaminen
Suorittamisohjeet löytyvät <a href="https://infotuni.github.io/joda2022/suorittaminen/">opintojakson kotisivulta</a>.
## Harjoitukset ja harjoitustyöt
Ohjeet harjoitustyöhön suorittamiseksi löytyvät <a href="https://infotuni.github.io/joda2022/harjoitustyo/">opintojakson kotisivulta</a>.
# Mitä on datatiede?
## Määritelmä
Tietojen tutkijan tai datatieteilijän (data scientist) rooli organisaatiossa on moninainen. Työtä on kuvattu monitieteiseksi, yhdistäen ainakin tietoteknistä, matemaattista ja liiketoiminnallista osaamista. Harvard Business Review nosti ammattinimikkeen laajempaan tietoisuuteen vuonna 2012 artikkelillaan [Data Scientist: The Sexiest Job of the 21st Century](https://hbr.org/2012/10/data-scientist-the-sexiest-job-of-the-21st-century):
<blockquote>
"...he started to see possibilities. He began forming theories, testing hunches, and finding patterns that allowed him to predict whose networks a given profile would land in. He could imagine that new features capitalizing on the heuristics he was developing might provide value to users."
</blockquote>
Alunperin datatieteestä käytettiin termiä [datalogy](https://dl.acm.org/citation.cfm?id=366510). Mielenkiintoista luettavaa on esimerkiksi artikkeli [Datalogy — The copenhagen tradition of computer science](https://link.springer.com/article/10.1007/BF01941128) datalogy-termin kehittäjä Naurin luomasta Kööpenhaminalaisesta tietotekniikan koulutuksen traditiosta.
Datatiedettä voi tarkastella useilla eri painotuksilla. Se voidaan nähdä markkinointiterminä [tilastotieteelle](http://www2.isye.gatech.edu/~jeffwu/presentations/datascience.pdf). On kuitenkin selvää, että tämä hyvin kapea käsitys ei kuvaa datatiedettä riittävällä tavalla, vaan tilastotiede on nähtävä yhtenä osana [datatieteilijän osaamista](https://arxiv.org/ftp/arxiv/papers/1410/1410.3127.pdf). Simon Lindgren ([2021](https://www.wiley.com/en-us/Data+Theory%3A+Interpretive+Sociology+and+Computational+Methods-p-9781509539277)) ehdottaa että datatiede voidaan myös tulkita erääksi tavaksi tehdä etnografista tutkimusta (ks. [Tikka et al., 2021](https://journal.fi/mediaviestinta/article/view/109871)).
Datatieteessä keskeistä on kyky muokata laajoja aineistoja sekä hyödyntää ohjelmointia eri tavoin. Esimerkkinä voidaan pitää siirtymää tilasto-ohjelmista kuten [R](https://fi.wikipedia.org/wiki/R_(ohjelmointikieli)) kohti täysiverisiä ohjelmointikieliä kuten [Python](https://fi.wikipedia.org/wiki/Python_(ohjelmointikieli)). Sekä R että Python ovat käytännössä ohjelmointikieliä, mutta R erikoistuu nimenomaisesti tilastolliseen laskentaa ja grafiikan tuottamiseen. Mikä on siis muutos, joka on tapahtunut kun Python kasvattaa suosiotaan osin R:n kustannuksella?
## Malleja ja käsitekarttoja aiheeseen
[CRISP-DM -malli](https://en.wikipedia.org/wiki/Cross-industry_standard_process_for_data_mining) esittelee avoimen standardin prosessikuvauksen datatieteen prosessista:
<img src="https://upload.wikimedia.org/wikipedia/commons/b/b9/CRISP-DM_Process_Diagram.png" alt="CRISP-DM" width="400"/>
[Houston Analyticsin CRISP-DM-sovellus](https://www.houston-analytics.com/project-methodology) laajentaa tarkastelukulman organisaatioon kokonaisuudessaan.
Microsofting tiimidatatiedemalli kuvaa hyvin käytännön datatiedeprojekteja:
<img src="https://docs.microsoft.com/en-us/azure/architecture/data-science-process/media/overview/tdsp-lifecycle2.png" />
Prosessimallin kautta pystyy myös ymmärtämään mitä edellytetään [hyvältä datieteilijältä](https://www.schoolofdatascience.amsterdam/news/skills-need-become-modern-data-scientist/). [Datatieteen metrokarttaan](http://nirvacana.com/thoughts/2013/07/08/becoming-a-data-scientist/) kokoaa ja kytkee yhteen aihepiirin menetelmiä ja teknologioita.
On aivan olennaista pohtia, onko realistista että yksi henkilö hallitsee näin laajan kokonaisuuden ja minkälaisia painotuksia oheisen metrokartan sisällä voi tehdä niin että osaaminen on edelleen relevanttia. Mikä on datatieteen tavoite ja onko se parhaimmillaan [erikois- vai yleisosaajien toteuttamaa](https://hbr.org/2019/03/why-data-science-teams-need-generalists-not-specialists)?
# Datatieteen edellytykset
## Etiikka!
Data ja sen pohjalta rakennettavat sovellukset ovat mitä suurinta vallankäyttöä nykyaikaisessa yhteiskunnannassa ja siksi [eettiset kysymykset](https://medium.com/big-data-at-berkeley/things-you-need-to-know-before-you-become-a-data-scientist-a-beginners-guide-to-data-ethics-8f9aa21af742) on syytä nostaa esiin heti datatiedeopintojen alussa.
## Data
Ihan ensiksi kannattaa kysyä että mitä se data on? [Piileekö datassa totuus](https://twitter.com/jnkka/status/1434783168201216000)?
Globaalisti käytössämme on **ennennäkemätön määrä dataa**. Arvioimme että vuoteen 2025 mennessä käytössä on [163 Zetabittiä](https://www.forbes.com/sites/andrewcave/2017/04/13/what-will-we-do-when-the-worlds-data-hits-163-zettabytes-in-2025/) dataa, tai kuvattuna toisella tavalla, luomme joka minuutti [käsittämättömän määrän dataa](https://www.domo.com/learn/data-never-sleeps-5?aid=ogsm072517_1&sf100871281=1). Onko realistista että edes ymmärrämme onko tämä määrä dataa hyödyllistä tai mitä tällä datalla voidaan saada aikaan?
Data on avainasemassa tekoälyn toisessa aallossa, jossa keskiössä on nimenomaan **tilastollinen oppiminen**. Nykyinen tekoälyyn liittyvä toiminta fokusoituu nimenomaan koneoppimiseen ja erityisesti syviin neuroverkkoihin. Tämä ei ole ihme, sillä viime vuosien merkittävimmät menestystarinat perustuvat juuri näihin teknologioihin. Käytettävissä olevat valtavat datamäärät, hyvät kehitystyökalut ja vuosittain kasvava laskentateho vauhdittavat kehitystä.
Koneoppimisen voittokulusta etenkin ihmisiin ja organisoitumiseen liittyvässä datatieteessä myös **kartoittavalla ja kuvailevalla analytiikalla** on keskeinen rooli.
Valitaan ensimmäinen dataesimerkki [Hans Roslingin innoittamana](https://www.ted.com/talks/hans_rosling_the_best_stats_you_ve_ever_seen).
```
# pip install gapminder
import pandas as pd
from gapminder import gapminder
gapminder.head(10)
# pip install plotly_express
import plotly_express as px
px.scatter(px.data.gapminder(), x="gdpPercap", y="lifeExp", animation_frame="year", animation_group="country",
size="pop", color="continent", hover_name="country",
log_x = True,
size_max=45, range_x=[100,100000], range_y=[25,90])
```
Dataa on myös julkisesti saatavilla enemmän kuin koskaan ennnen. Hyvänä esimerkkinä on vaikka [Kaggle](www.kaggle.com), joka antaa mahdollisuuden ladata itselle mielenkiintoisia aineistoja eri tarkoituksiin.
```
df = pd.read_csv("Mall_Customers.csv")
df.head()
df.tail()
df['Gender'] = df['Genre']
import numpy as np
df.pivot_table(df, index=["Gender"], aggfunc=np.mean)
```
**Keskustelua ongelmista, reunaehdoista ja rajoitteista.**
## Laskentateho
Laskentatehokasvu on selkeästi yksi merkittävin mekanismi data tieteen kehittymiseen. Kaikkihan tavalla tai toisella liittyy Mooren lakiin, eli kykyymme suorittaa laskutoimituksia.

Laskentatehon kasvun lisäksi tekniset ratkaisut skaalata yksittäisen koneen tai klusterin laskentatehoa ovat kehittyneet merkittävästi. Nämä tekevät jopa yksittäisestä koneesta huomattavan tehokkaan työyksikön
```
from IPython.core.display import HTML
HTML('<iframe width="560" height="315" src="https://www.youtube.com/embed/tQBovBvSDvA?start=1808" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>')
```
## Analyysiympäristö
Erityyppiset laskentaympäristöt voidaan karkeasti jakaa kuuteen. Vaihtoehdot kasvavat henkilökohtaisesta koneesta aina pilviratkaisuihin tai laskentaklustereihin.
```
HTML('<iframe width="560" height="315" src="https://www.youtube.com/embed/4paAY2kseCE" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>')
```
## Työvälineet
Käytettävissä olevien työvälineiden määrä on kasvanut huimasti. Aikaisemmin käytössä oli lähinnä tilastolaskentaympäristöt kuten [R](https://www.r-project.org/), joita korvaamaan/lisäämään on nyt tullut Python-pohjaiset ympäristöt. Tämän sisällä keskeisiä työkaluja ovat esimerkiksi [Pandas](https://pandas.pydata.org/), [Scikit-learn](https://scikit-learn.org/stable/) ja visualisointityövälineet kuten [Holoviews](http://holoviews.org/).
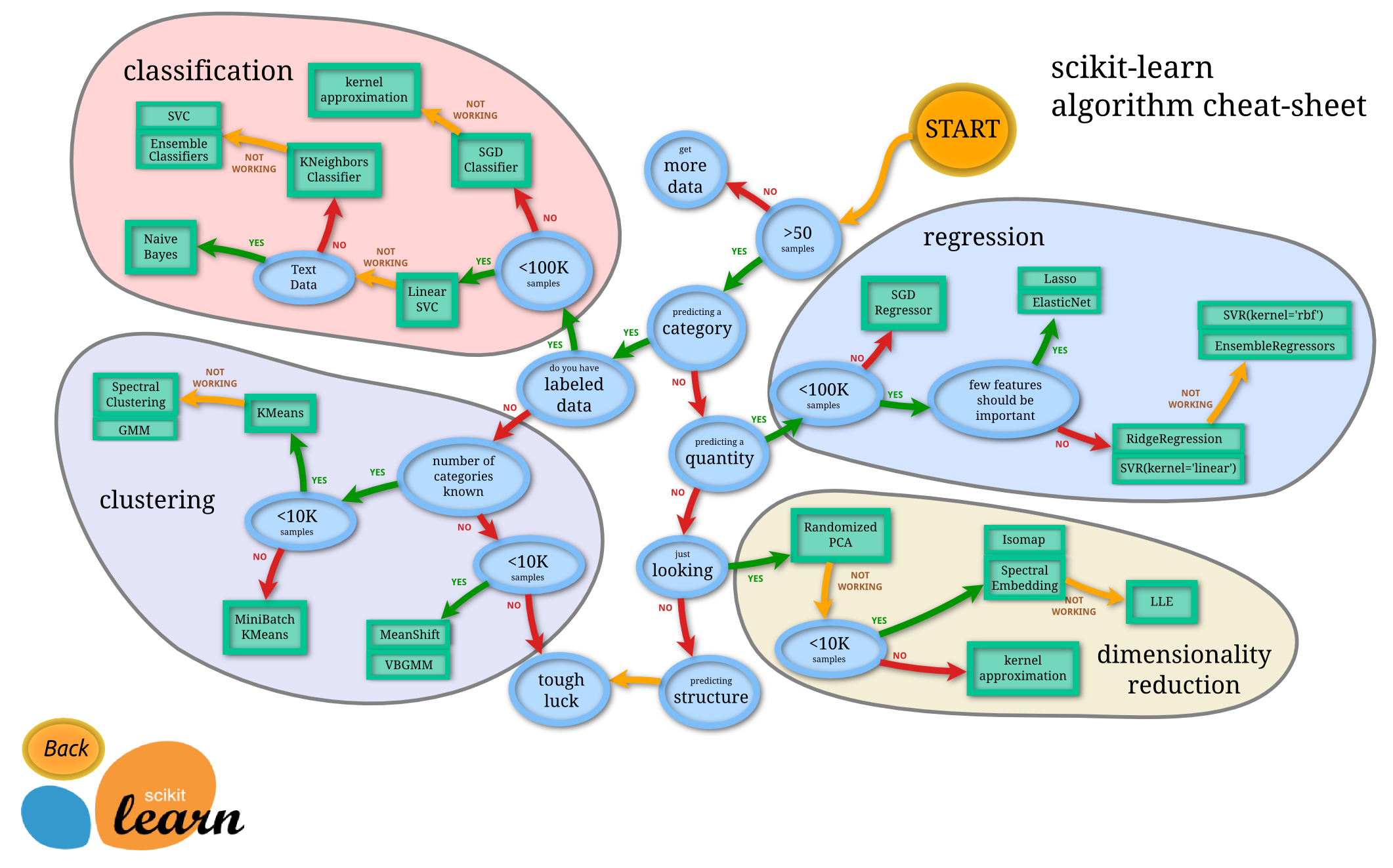
```
HTML('<iframe width="560" height="315" src="https://www.youtube.com/embed/k27MJJLJNT4?start=1808" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>')
```
Tämä luentomateriaali esitetään Jupyter Notebook -muodossa. Voit ottaa käyttöön oman työkirjapohjaisen laskentaympäristön useilla eri tavoilla:
* [CSC Notebooks](https://www.csc.fi/web/blog/post/-/blogs/notebooks-enemman-aikaa-opetuksen-ytimelle)
* [JupyterLab](https://jupyterlab.readthedocs.io/en/stable/getting_started/installation.html)
* [Google Colaboratory](https://colab.research.google.com/)
* [Anaconda](https://anaconda.org/)
## Lopuksi
Dataa on paljon, laskentatehoa on tarjolla pilvessä loputtomasti ja välineitä riittää. Tietojohtamisen näkökulmasta tarkastellen datatiedettä ei kuitenkaan voi erottaa ihmisestä eikä organisaatiosta. Miten datasta tuotetaan **informaatiota, tietämystä ja lopulta viisautta**?
Data on lopulta ihmisen toiminnan tuotosta ja usein tietoisesti ja tarkoituksella tuotettua ([boyd & Crawford, 2012](https://doi.org/10.1080/1369118X.2012.678878), [Pink ja muut, 2018](https://doi.org/10.1177/2053951717753228)). Lindgren esittelee kirjassaan [Data Theory: Interpretive Sociology and Computational Methods](https://www.wiley.com/en-us/Data+Theory%3A+Interpretive+Sociology+and+Computational+Methods-p-9781509539277) kiinnostavan provokaation: datatiede ja etnografia tarvitsevat toisiaan.
|
github_jupyter
|
print('Hello world')
from IPython.display import Image
from IPython.core.display import HTML
# pip install gapminder
import pandas as pd
from gapminder import gapminder
gapminder.head(10)
# pip install plotly_express
import plotly_express as px
px.scatter(px.data.gapminder(), x="gdpPercap", y="lifeExp", animation_frame="year", animation_group="country",
size="pop", color="continent", hover_name="country",
log_x = True,
size_max=45, range_x=[100,100000], range_y=[25,90])
df = pd.read_csv("Mall_Customers.csv")
df.head()
df.tail()
df['Gender'] = df['Genre']
import numpy as np
df.pivot_table(df, index=["Gender"], aggfunc=np.mean)
from IPython.core.display import HTML
HTML('<iframe width="560" height="315" src="https://www.youtube.com/embed/tQBovBvSDvA?start=1808" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>')
HTML('<iframe width="560" height="315" src="https://www.youtube.com/embed/4paAY2kseCE" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>')
HTML('<iframe width="560" height="315" src="https://www.youtube.com/embed/k27MJJLJNT4?start=1808" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>')
| 0.487795 | 0.729062 |
<table class="ee-notebook-buttons" align="left">
<td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/FeatureCollection/filtering_feature_collection.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
<td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/FeatureCollection/filtering_feature_collection.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
<td><a target="_blank" href="https://mybinder.org/v2/gh/giswqs/earthengine-py-notebooks/master?filepath=FeatureCollection/filtering_feature_collection.ipynb"><img width=58px src="https://mybinder.org/static/images/logo_social.png" />Run in binder</a></td>
<td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/FeatureCollection/filtering_feature_collection.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
</table>
## Install Earth Engine API
Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geehydro](https://github.com/giswqs/geehydro). The **geehydro** Python package builds on the [folium](https://github.com/python-visualization/folium) package and implements several methods for displaying Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, `Map.centerObject()`, and `Map.setOptions()`.
The magic command `%%capture` can be used to hide output from a specific cell.
```
# %%capture
# !pip install earthengine-api
# !pip install geehydro
```
Import libraries
```
import ee
import folium
import geehydro
```
Authenticate and initialize Earth Engine API. You only need to authenticate the Earth Engine API once. Uncomment the line `ee.Authenticate()`
if you are running this notebook for this first time or if you are getting an authentication error.
```
# ee.Authenticate()
ee.Initialize()
```
## Create an interactive map
This step creates an interactive map using [folium](https://github.com/python-visualization/folium). The default basemap is the OpenStreetMap. Additional basemaps can be added using the `Map.setOptions()` function.
The optional basemaps can be `ROADMAP`, `SATELLITE`, `HYBRID`, `TERRAIN`, or `ESRI`.
```
Map = folium.Map(location=[40, -100], zoom_start=4)
Map.setOptions('HYBRID')
```
## Add Earth Engine Python script
```
def cal_area(feature):
num = ee.Number.parse(feature.get('areasqkm'))
return feature.set('areasqkm', num)
# Load watersheds from a data table.
sheds = ee.FeatureCollection('USGS/WBD/2017/HUC06') \
# .map(cal_area)
# Define a region roughly covering the continental US.
continentalUS = ee.Geometry.Rectangle(-127.18, 19.39, -62.75, 51.29)
# Filter the table geographically: only watersheds in the continental US.
filtered = sheds.filterBounds(continentalUS) \
.map(cal_area)
# Check the number of watersheds after filtering for location.
print('Count after filter:', filtered.size().getInfo())
# Filter to get only larger continental US watersheds.
largeSheds = filtered.filter(ee.Filter.gt('areasqkm', 25000))
# Check the number of watersheds after filtering for size and location.
print('Count after filtering by size:', largeSheds.size().getInfo())
```
## Display Earth Engine data layers
```
Map.setControlVisibility(layerControl=True, fullscreenControl=True, latLngPopup=True)
Map
```
|
github_jupyter
|
# %%capture
# !pip install earthengine-api
# !pip install geehydro
import ee
import folium
import geehydro
# ee.Authenticate()
ee.Initialize()
Map = folium.Map(location=[40, -100], zoom_start=4)
Map.setOptions('HYBRID')
def cal_area(feature):
num = ee.Number.parse(feature.get('areasqkm'))
return feature.set('areasqkm', num)
# Load watersheds from a data table.
sheds = ee.FeatureCollection('USGS/WBD/2017/HUC06') \
# .map(cal_area)
# Define a region roughly covering the continental US.
continentalUS = ee.Geometry.Rectangle(-127.18, 19.39, -62.75, 51.29)
# Filter the table geographically: only watersheds in the continental US.
filtered = sheds.filterBounds(continentalUS) \
.map(cal_area)
# Check the number of watersheds after filtering for location.
print('Count after filter:', filtered.size().getInfo())
# Filter to get only larger continental US watersheds.
largeSheds = filtered.filter(ee.Filter.gt('areasqkm', 25000))
# Check the number of watersheds after filtering for size and location.
print('Count after filtering by size:', largeSheds.size().getInfo())
Map.setControlVisibility(layerControl=True, fullscreenControl=True, latLngPopup=True)
Map
| 0.716417 | 0.960694 |
<img src="images/usm.jpg" width="480" height="240" align="left"/>
# MAT281 - Laboratorio N°02
## Objetivos de la clase
* Reforzar los conceptos básicos de numpy.
## Contenidos
* [Problema 01](#p1)
* [Problema 02](#p2)
* [Problema 03](#p3)
<a id='p1'></a>
## Problema 01
Una **media móvil simple** (SMA) es el promedio de los últimos $k$ datos anteriores, es decir, sea $a_1$,$a_2$,...,$a_n$ un arreglo $n$-dimensional, entonces la SMA se define por:
$$sma(k) =\dfrac{1}{k}(a_{n}+a_{n-1}+...+a_{n-(k-1)}) = \dfrac{1}{k}\sum_{i=0}^{k-1}a_{n-i} $$
Por otro lado podemos definir el SMA con una venta móvil de $n$ si el resultado nos retorna la el promedio ponderado avanzando de la siguiente forma:
* $a = [1,2,3,4,5]$, la SMA con una ventana de $n=2$ sería:
* sma(2): [mean(1,2),mean(2,3),mean(3,4)] = [1.5, 2.5, 3.5, 4.5]
* sma(3): [mean(1,2,3),mean(2,3,4),mean(3,4,5)] = [2.,3.,4.]
Implemente una función llamada `sma` cuyo input sea un arreglo unidimensional $a$ y un entero $n$, y cuyo ouput retorne el valor de la media móvil simple sobre el arreglo de la siguiente forma:
* **Ejemplo**: *sma([5,3,8,10,2,1,5,1,0,2], 2)* = $[4. , 5.5, 9. , 6. , 1.5, 3. , 3. , 0.5, 1. ]$
En este caso, se esta calculando el SMA para un arreglo con una ventana de $n=2$.
**Hint**: utilice la función `numpy.cumsum`
```
import numpy as np
def sma(arreglo,n)->np.array:
"""
sma(arreglo,n)
Calcula la media móvil simple de n datos anteriores del arreglo
Parameters
----------
arreglo: array
Arreglo con valores númericos
Returns
-------
output : array
Arreglo con las medias móviles simples
Examples
--------
>>> sma([1,2,3,4,5],3)
[2.0, 3.0, 4.0]
>>>sma([5,3,8,10,2,1,5,1,0,2], 2)
[4.0, 5.5, 9.0, 6.0, 1.5, 3.0, 3.0, 0.5, 1.0]
"""
i=0 # Contador
dev=[] # Arreglo a devolver
sumas=np.cumsum(arreglo) # arreglo con todas las sumas acumuladas del arreglo
promedio=0 # variable para guardar el promedio de n variables del arreglo
sup=0 # suma de n valores
inf=0 # suma de n-i valores, esta sera la cola inferior de la sumatoria
while i<=len(arreglo)-n: #itero la cantidad especificada
if i==0: #caso en que no deba eleminar una cola inferior de la suma
sup=sumas[n-1]
inf=0
promedio=(sup-inf)/n
dev.append(promedio)
i+=1
if i>0: # resto de casos donde si debo eliminar una cola de la suma total de elementos
sup=sumas[i+n-1]
inf=sumas[i-1]
promedio = (sup-inf)/n
dev.append(promedio)
i+=1
return dev
```
<a id='p2'></a>
## Problema 02
La función **strides($a,n,p$)**, corresponde a transformar un arreglo unidimensional $a$ en una matriz de $n$ columnas, en el cual las filas se van construyendo desfasando la posición del arreglo en $p$ pasos hacia adelante.
* Para el arreglo unidimensional $a$ = [ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10], la función strides($a,4,2$), corresponde a crear una matriz de $4$ columnas, cuyos desfaces hacia adelante se hacen de dos en dos.
El resultado tendría que ser algo así:$$\begin{pmatrix}
1& 2 &3 &4 \\
3& 4&5&6 \\
5& 6 &7 &8 \\
7& 8 &9 &10 \\
\end{pmatrix}$$
Implemente una función llamada `strides(a,4,2)` cuyo input sea un arreglo unidimensional y retorne la matriz de $4$ columnas, cuyos desfaces hacia adelante se hacen de dos en dos.
* **Ejemplo**: *strides($a$,4,2)* =$\begin{pmatrix}
1& 2 &3 &4 \\
3& 4&5&6 \\
5& 6 &7 &8 \\
7& 8 &9 &10 \\
\end{pmatrix}$
```
import numpy as np
arr=[1,2,3,4,5,6,7,8,9,10]
def strides(a,n,p)->np.array:
"""
strides(a,n,p)
Transforma un arreglo en una matriz de n columnas,
en la cual las filas se van construyendo desfasando
la posición del arreglo en 𝑝 pasos hacia adelante.
Parameters
-----------
a : array
Arreglo que se desea transformar
n : int
Cantidad de columnas de la matriz
p : int
Desface para las filas
Returns
--------
output : np.array((n,n))
Examples
--------
>>>mat=strides(arr,4,2)
[[ 1. 2. 3. 4.]
[ 3. 4. 5. 6.]
[ 5. 6. 7. 8.]
[ 7. 8. 9. 10.]]
>>>mat=strides(arr,4,1)
[[1. 2. 3. 4.]
[2. 3. 4. 5.]
[3. 4. 5. 6.]
[4. 5. 6. 7.]]
"""
i=0
aj=np.zeros((n,n))
while i<n:
j=i*p
aj[i]=a[j:j+n]
i+=1
return aj
mat=strides(arr,4,2)
print(mat)
mat=strides(arr,4,1)
print(mat)
import numpy as np
arr=[1,2,3,4,5,6,7,8,9,10]
aj=np.zeros((4,4))
print(aj)
aj[1]=[1,2,3,4]
print(aj)
```
<a id='p3'></a>
## Problema 03
Un **cuadrado mágico** es una matriz de tamaño $n \times n$ de números enteros positivos tal que
la suma de los números por columnas, filas y diagonales principales sea la misma. Usualmente, los números empleados para rellenar las casillas son consecutivos, de 1 a $n^2$, siendo $n$ el número de columnas y filas del cuadrado mágico.
Si los números son consecutivos de 1 a $n^2$, la suma de los números por columnas, filas y diagonales principales
es igual a : $$M_{n} = \dfrac{n(n^2+1)}{2}$$
Por ejemplo,
* $A= \begin{pmatrix}
4& 9 &2 \\
3& 5&7 \\
8& 1 &6
\end{pmatrix}$,
es un cuadrado mágico.
* $B= \begin{pmatrix}
4& 2 &9 \\
3& 5&7 \\
8& 1 &6
\end{pmatrix}$, no es un cuadrado mágico.
Implemente una función llamada `es_cudrado_magico` cuyo input sea una matriz cuadrada de tamaño $n$ con números consecutivos de $1$ a $n^2$ y cuyo ouput retorne *True* si es un cuadrado mágico o 'False', en caso contrario
* **Ejemplo**: *es_cudrado_magico($A$)* = True, *es_cudrado_magico($B$)* = False
**Hint**: Cree una función que valide la mariz es cuadrada y que sus números son consecutivos del 1 a $n^2$.
```
import numpy as np
A=np.zeros((3,3))
B=np.zeros((3,3))
A[0]=[4,9,2]
A[1]=[3,5,7]
A[2]=[8,1,6]
B[0]=[4,2,9]
B[1]=[3,5,7]
B[2]=[8,1,6]
def cuadrada(a)->bool:
"""
cuadrada(a)
Determina si una matriz es cuadrada
Parameters
----------
a : np.array
Matriz de numpy
Returns
--------
output : bool
Valor de verdad de que la matriz entregada sea cuadrada
"""
if a.shape[0]!=a.shape[1]: #determina si la matriz es cuadrada
return False # Retorna falso si no lo es
return True # Retorna verdadero si, sí lo es
def ordenada(a,n):
"""
ordenada(a,n)
Determina si la matriz tiene todos los elementos del 1 al n^2, donde n es
el tamaño de la matriz
Parameters
----------
a : np.array
matriz de numpy
n : int
Tamaño de la matriz cuadrada
Returns
--------
output : bool
Valor de verdad de que la matriz contenga los elementos deseados
"""
i=0 #contador
arr=list(range(1,n**2+1)) #arreglo ordenado de los elementos que la matriz debe tener
ordenada=np.sort(a,None) # arreglo ordenado de todos los elementos de la matriz entregada
while i<n^2: #recorro los arreglos elemento a elemento
if arr[i]==ordenada[i]: #compruebo que sean iguales
i+=1
else:return False # si en al menos un elemento no son iguales se retorna False
return True #si los elementos son igual se retorna True
def magia(a,n):
"""
magia(a,n)
Se determina si las filas y diagonales principales
de la matriz dada suman la constante magica correspondiente a su tamaño
Parameters
----------
a : np.array
matriz dada candidata a magica
n : int
Tamaño de la matriz dada
Returns
--------
output : bool
Valor de verdad de que la matriz sea magica
"""
magico=n*((n**2)+1)/2 # calculo la constante correspondiente al tamaño de la matriz
for i in range(n): #contador para recorrer la matriz fila a fila, y colimna a colmna
if np.sum(a,1)[i]!=magico: #determina si las filas cumplen la condicion
return False
if np.sum(a.T,1)[i]!=magico: #determina si las columnas cumples las condiciones
return False
if a.trace()!=magico: #determina si la diagonal principal cumple la condicion
return False
if a[::-1].trace()!=magico: #determina si la diagonal secundaria cumple la condicion
return False
return True
def es_cuadrado_magico(a)->bool:
"""
cuadrado_magico(a)
Determina si la matriz dada es magica y retorna un valor de verdad
Parameters
----------
a : np.array
matriz candidata
Returns
--------
output : bool
Valor de verdad de que la matriz dada sea magica
Examples
---------
>>>print(es_cuadrado_magico(A))
True
>>>print(es_cuadrado_magico(B))
False
"""
flag=True
flag=cuadrada(a)
if flag==False:
return False
largo=a.shape[0]
flag=ordenada(a,largo)
if flag==False:
return False
flag=magia(a,largo)
if flag==False:
return False
return flag
```
|
github_jupyter
|
import numpy as np
def sma(arreglo,n)->np.array:
"""
sma(arreglo,n)
Calcula la media móvil simple de n datos anteriores del arreglo
Parameters
----------
arreglo: array
Arreglo con valores númericos
Returns
-------
output : array
Arreglo con las medias móviles simples
Examples
--------
>>> sma([1,2,3,4,5],3)
[2.0, 3.0, 4.0]
>>>sma([5,3,8,10,2,1,5,1,0,2], 2)
[4.0, 5.5, 9.0, 6.0, 1.5, 3.0, 3.0, 0.5, 1.0]
"""
i=0 # Contador
dev=[] # Arreglo a devolver
sumas=np.cumsum(arreglo) # arreglo con todas las sumas acumuladas del arreglo
promedio=0 # variable para guardar el promedio de n variables del arreglo
sup=0 # suma de n valores
inf=0 # suma de n-i valores, esta sera la cola inferior de la sumatoria
while i<=len(arreglo)-n: #itero la cantidad especificada
if i==0: #caso en que no deba eleminar una cola inferior de la suma
sup=sumas[n-1]
inf=0
promedio=(sup-inf)/n
dev.append(promedio)
i+=1
if i>0: # resto de casos donde si debo eliminar una cola de la suma total de elementos
sup=sumas[i+n-1]
inf=sumas[i-1]
promedio = (sup-inf)/n
dev.append(promedio)
i+=1
return dev
import numpy as np
arr=[1,2,3,4,5,6,7,8,9,10]
def strides(a,n,p)->np.array:
"""
strides(a,n,p)
Transforma un arreglo en una matriz de n columnas,
en la cual las filas se van construyendo desfasando
la posición del arreglo en 𝑝 pasos hacia adelante.
Parameters
-----------
a : array
Arreglo que se desea transformar
n : int
Cantidad de columnas de la matriz
p : int
Desface para las filas
Returns
--------
output : np.array((n,n))
Examples
--------
>>>mat=strides(arr,4,2)
[[ 1. 2. 3. 4.]
[ 3. 4. 5. 6.]
[ 5. 6. 7. 8.]
[ 7. 8. 9. 10.]]
>>>mat=strides(arr,4,1)
[[1. 2. 3. 4.]
[2. 3. 4. 5.]
[3. 4. 5. 6.]
[4. 5. 6. 7.]]
"""
i=0
aj=np.zeros((n,n))
while i<n:
j=i*p
aj[i]=a[j:j+n]
i+=1
return aj
mat=strides(arr,4,2)
print(mat)
mat=strides(arr,4,1)
print(mat)
import numpy as np
arr=[1,2,3,4,5,6,7,8,9,10]
aj=np.zeros((4,4))
print(aj)
aj[1]=[1,2,3,4]
print(aj)
import numpy as np
A=np.zeros((3,3))
B=np.zeros((3,3))
A[0]=[4,9,2]
A[1]=[3,5,7]
A[2]=[8,1,6]
B[0]=[4,2,9]
B[1]=[3,5,7]
B[2]=[8,1,6]
def cuadrada(a)->bool:
"""
cuadrada(a)
Determina si una matriz es cuadrada
Parameters
----------
a : np.array
Matriz de numpy
Returns
--------
output : bool
Valor de verdad de que la matriz entregada sea cuadrada
"""
if a.shape[0]!=a.shape[1]: #determina si la matriz es cuadrada
return False # Retorna falso si no lo es
return True # Retorna verdadero si, sí lo es
def ordenada(a,n):
"""
ordenada(a,n)
Determina si la matriz tiene todos los elementos del 1 al n^2, donde n es
el tamaño de la matriz
Parameters
----------
a : np.array
matriz de numpy
n : int
Tamaño de la matriz cuadrada
Returns
--------
output : bool
Valor de verdad de que la matriz contenga los elementos deseados
"""
i=0 #contador
arr=list(range(1,n**2+1)) #arreglo ordenado de los elementos que la matriz debe tener
ordenada=np.sort(a,None) # arreglo ordenado de todos los elementos de la matriz entregada
while i<n^2: #recorro los arreglos elemento a elemento
if arr[i]==ordenada[i]: #compruebo que sean iguales
i+=1
else:return False # si en al menos un elemento no son iguales se retorna False
return True #si los elementos son igual se retorna True
def magia(a,n):
"""
magia(a,n)
Se determina si las filas y diagonales principales
de la matriz dada suman la constante magica correspondiente a su tamaño
Parameters
----------
a : np.array
matriz dada candidata a magica
n : int
Tamaño de la matriz dada
Returns
--------
output : bool
Valor de verdad de que la matriz sea magica
"""
magico=n*((n**2)+1)/2 # calculo la constante correspondiente al tamaño de la matriz
for i in range(n): #contador para recorrer la matriz fila a fila, y colimna a colmna
if np.sum(a,1)[i]!=magico: #determina si las filas cumplen la condicion
return False
if np.sum(a.T,1)[i]!=magico: #determina si las columnas cumples las condiciones
return False
if a.trace()!=magico: #determina si la diagonal principal cumple la condicion
return False
if a[::-1].trace()!=magico: #determina si la diagonal secundaria cumple la condicion
return False
return True
def es_cuadrado_magico(a)->bool:
"""
cuadrado_magico(a)
Determina si la matriz dada es magica y retorna un valor de verdad
Parameters
----------
a : np.array
matriz candidata
Returns
--------
output : bool
Valor de verdad de que la matriz dada sea magica
Examples
---------
>>>print(es_cuadrado_magico(A))
True
>>>print(es_cuadrado_magico(B))
False
"""
flag=True
flag=cuadrada(a)
if flag==False:
return False
largo=a.shape[0]
flag=ordenada(a,largo)
if flag==False:
return False
flag=magia(a,largo)
if flag==False:
return False
return flag
| 0.51879 | 0.934395 |
<!--BOOK_INFORMATION-->
<img align="left" style="padding-right:10px;" src="fig/cover-small.jpg">
*This notebook contains an excerpt from the [Whirlwind Tour of Python](http://www.oreilly.com/programming/free/a-whirlwind-tour-of-python.csp) by Jake VanderPlas; the content is available [on GitHub](https://github.com/jakevdp/WhirlwindTourOfPython).*
*The text and code are released under the [CC0](https://github.com/jakevdp/WhirlwindTourOfPython/blob/master/LICENSE) license; see also the companion project, the [Python Data Science Handbook](https://github.com/jakevdp/PythonDataScienceHandbook).*
<!--NAVIGATION-->
< [Control Flow](07-Control-Flow-Statements.ipynb) | [Contents](Index.ipynb) | [Errors and Exceptions](09-Errors-and-Exceptions.ipynb) >
# Defining and Using Functions
So far, our scripts have been simple, single-use code blocks.
One way to organize our Python code and to make it more readable and reusable is to factor-out useful pieces into reusable *functions*.
Here we'll cover two ways of creating functions: the ``def`` statement, useful for any type of function, and the ``lambda`` statement, useful for creating short anonymous functions.
## Using Functions
Functions are groups of code that have a name, and can be called using parentheses.
We've seen functions before. For example, ``print`` in Python 3 is a function:
```
print('abc')
```
Here ``print`` is the function name, and ``'abc'`` is the function's *argument*.
In addition to arguments, there are *keyword arguments* that are specified by name.
One available keyword argument for the ``print()`` function (in Python 3) is ``sep``, which tells what character or characters should be used to separate multiple items:
```
print(1, 2, 3)
print(1, 2, 3, sep='--')
```
When non-keyword arguments are used together with keyword arguments, the keyword arguments must come at the end.
## Defining Functions
Functions become even more useful when we begin to define our own, organizing functionality to be used in multiple places.
In Python, functions are defined with the ``def`` statement.
```
def add_1(x):
return x + 1
add_1(10)
```
We can even create a docstring for our functions:
```
def add_1(x):
"""Adds 1 to the input.
Args:
x (int/float): A numberical value
Returns:
x + 1
"""
return x + 1
help(add_1)
```
For example, we can encapsulate a version of our Fibonacci sequence code from the previous section as follows:
```
def fibonacci(N):
"""Calculates the first N Fibonacci numbers, with initial condition a, b = 0, 1.
Args:
N (int): Number of Fibonacci numbers to be returned
Returns:
L (list): A list of Fibonacci numbers
"""
L = []
a, b = 0, 1
while len(L) < N:
a, b = b, a + b
L.append(a)
return L
help(fibonacci)
```
Now we have a function named ``fibonacci`` which takes a single argument ``N``, does something with this argument, and ``return``s a value; in this case, a list of the first ``N`` Fibonacci numbers:
```
fibonacci(10)
```
If you're familiar with strongly-typed languages like ``C``, you'll immediately notice that there is no type information associated with the function inputs or outputs.
Python functions can return any Python object, simple or compound, which means constructs that may be difficult in other languages are straightforward in Python.
For example, multiple return values are simply put in a tuple, which is indicated by commas:
```
def real_imag_conj(val):
"""Returns real, imaginary, and complex conjugate of a complex number.
Args:
val (complex): A complex number
Returns:
val.real, val.imag, val.conjugate()
"""
return val.real, val.imag, val.conjugate()
r, i, c = real_imag_conj(3 + 4j)
print(r, i, c)
```
## Default Argument Values
Often when defining a function, there are certain values that we want the function to use *most* of the time, but we'd also like to give the user some flexibility.
In this case, we can use *default values* for arguments.
Consider the ``fibonacci`` function from before.
What if we would like the user to be able to play with the starting values?
We could do that as follows:
```
def fibonacci(N, a=0, b=1):
L = []
while len(L) < N:
a, b = b, a + b
L.append(a)
return L
```
With a single argument, the result of the function call is identical to before:
```
fibonacci(10)
```
But now we can use the function to explore new things, such as the effect of new starting values:
```
fibonacci(10, 0, 2)
```
The values can also be specified by name if desired, in which case the order of the named values does not matter:
```
fibonacci(10, b=3, a=1)
```
## ``*args`` and ``**kwargs``: Flexible Arguments
Sometimes you might wish to write a function in which you don't initially know how many arguments the user will pass.
In this case, you can use the special form ``*args`` and ``**kwargs`` to catch all arguments that are passed.
Here is an example:
```
def catch_all(*args, **kwargs):
print("args =", args)
print("kwargs = ", kwargs)
catch_all(1, 2, 3, a=4, b=5)
catch_all('a', keyword=2)
```
Here it is not the names ``args`` and ``kwargs`` that are important, but the ``*`` characters preceding them.
``args`` and ``kwargs`` are just the variable names often used by convention, short for "arguments" and "keyword arguments".
The operative difference is the asterisk characters: a single ``*`` before a variable means "expand this as a sequence", while a double ``**`` before a variable means "expand this as a dictionary".
In fact, this syntax can be used not only with the function definition, but with the function call as well!
```
inputs = (1, 2, 3)
keywords = {'pi': 3.14}
catch_all(*inputs, **keywords)
```
## Anonymous (``lambda``) Functions
Earlier we quickly covered the most common way of defining functions, the ``def`` statement.
You'll likely come across another way of defining short, one-off functions with the ``lambda`` statement.
It looks something like this:
```
add = lambda x, y: x + y
add(1, 2)
```
This lambda function is roughly equivalent to
```
def add(x, y):
return x + y
```
So why would you ever want to use such a thing?
Primarily, it comes down to the fact that *everything is an object* in Python, even functions themselves!
That means that functions can be passed as arguments to functions.
As an example of this, suppose we have some data stored in a list of dictionaries:
```
data = [{'first':'Guido', 'last':'Van Rossum', 'YOB':1956},
{'first':'Grace', 'last':'Hopper', 'YOB':1906},
{'first':'Alan', 'last':'Turing', 'YOB':1912}]
```
Now suppose we want to sort this data.
Python has a ``sorted`` function that does this:
```
sorted([2,4,3,5,1,6])
```
But dictionaries are not orderable: we need a way to tell the function *how* to sort our data.
We can do this by specifying the ``key`` function, a function which given an item returns the sorting key for that item:
```
# sort alphabetically by first name
sorted(data, key=lambda item: item['first'])
# sort by year of birth
sorted(data, key=lambda item: item['YOB'])
```
While these key functions could certainly be created by the normal, ``def`` syntax, the ``lambda`` syntax is convenient for such short one-off functions like these.
<!--NAVIGATION-->
< [Control Flow](07-Control-Flow-Statements.ipynb) | [Contents](Index.ipynb) | [Errors and Exceptions](09-Errors-and-Exceptions.ipynb) >
|
github_jupyter
|
print('abc')
print(1, 2, 3)
print(1, 2, 3, sep='--')
def add_1(x):
return x + 1
add_1(10)
def add_1(x):
"""Adds 1 to the input.
Args:
x (int/float): A numberical value
Returns:
x + 1
"""
return x + 1
help(add_1)
def fibonacci(N):
"""Calculates the first N Fibonacci numbers, with initial condition a, b = 0, 1.
Args:
N (int): Number of Fibonacci numbers to be returned
Returns:
L (list): A list of Fibonacci numbers
"""
L = []
a, b = 0, 1
while len(L) < N:
a, b = b, a + b
L.append(a)
return L
help(fibonacci)
fibonacci(10)
def real_imag_conj(val):
"""Returns real, imaginary, and complex conjugate of a complex number.
Args:
val (complex): A complex number
Returns:
val.real, val.imag, val.conjugate()
"""
return val.real, val.imag, val.conjugate()
r, i, c = real_imag_conj(3 + 4j)
print(r, i, c)
def fibonacci(N, a=0, b=1):
L = []
while len(L) < N:
a, b = b, a + b
L.append(a)
return L
fibonacci(10)
fibonacci(10, 0, 2)
fibonacci(10, b=3, a=1)
def catch_all(*args, **kwargs):
print("args =", args)
print("kwargs = ", kwargs)
catch_all(1, 2, 3, a=4, b=5)
catch_all('a', keyword=2)
inputs = (1, 2, 3)
keywords = {'pi': 3.14}
catch_all(*inputs, **keywords)
add = lambda x, y: x + y
add(1, 2)
def add(x, y):
return x + y
data = [{'first':'Guido', 'last':'Van Rossum', 'YOB':1956},
{'first':'Grace', 'last':'Hopper', 'YOB':1906},
{'first':'Alan', 'last':'Turing', 'YOB':1912}]
sorted([2,4,3,5,1,6])
# sort alphabetically by first name
sorted(data, key=lambda item: item['first'])
# sort by year of birth
sorted(data, key=lambda item: item['YOB'])
| 0.666822 | 0.989652 |

[](https://colab.research.google.com/github/JohnSnowLabs/nlu/blob/master/examples/colab/Training/multi_lingual/multi_class_text_classification/NLU_training_multi_lingual_multi_class_text_classifier_demo.ipynb)
# Training a Deep Learning Classifier with NLU
## ClassifierDL (Multi-class Text Classification)
With the [ClassifierDL model](https://nlp.johnsnowlabs.com/docs/en/annotators#classifierdl-multi-class-text-classification) from Spark NLP you can achieve State Of the Art results on any multi class text classification problem
This notebook showcases the following features :
- How to train the deep learning classifier
- How to store a pipeline to disk
- How to load the pipeline from disk (Enables NLU offline mode)
You can achieve these results or even better on this dataset with training data :

<br>
You can achieve these results or even better on this dataset with test data :
<br>

# 1. Install Java 8 and NLU
```
import os
! apt-get update -qq > /dev/null
# Install java
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["PATH"] = os.environ["JAVA_HOME"] + "/bin:" + os.environ["PATH"]
! pip install nlu > /dev/null pyspark==2.4.7
import nlu
```
# 2. Download news classification dataset
```
! wget http://ckl-it.de/wp-content/uploads/2021/02/news_category_test_multi_lingual.csv
import pandas as pd
test_path = '/content/news_category_test_multi_lingual.csv'
train_df = pd.read_csv(test_path)
from sklearn.model_selection import train_test_split
train_df, test_df = train_test_split(train_df, test_size=0.2)
train_df
```
# 3. Train Deep Learning Classifier using nlu.load('train.classifier')
By default, the Universal Sentence Encoder Embeddings (USE) are beeing downloaded to provide embeddings for the classifier. You can use any of the 50+ other sentence Emeddings in NLU tough!
You dataset label column should be named 'y' and the feature column with text data should be named 'text'
```
trainable_pipe = nlu.load('xx.embed_sentence.labse train.classifier')
# We need to train longer and user smaller LR for NON-USE based sentence embeddings usually
# We could tune the hyperparameters further with hyperparameter tuning methods like gridsearch
# Also longer training gives more accuracy
trainable_pipe['classifier_dl'].setMaxEpochs(60)
trainable_pipe['classifier_dl'].setLr(0.005)
fitted_pipe = trainable_pipe.fit(train_df.iloc[:1500])
# predict with the trainable pipeline on dataset and get predictions
preds = fitted_pipe.predict(train_df.iloc[:1500],output_level='document')
#sentence detector that is part of the pipe generates sone NaNs. lets drop them first
preds.dropna(inplace=True)
from sklearn.metrics import classification_report
print(classification_report(preds['y'], preds['category']))
preds
```
# 3.1 evaluate on Test Data
```
preds = fitted_pipe.predict(test_df,output_level='document')
#sentence detector that is part of the pipe generates sone NaNs. lets drop them first
preds.dropna(inplace=True)
print(classification_report(preds['y'], preds['category']))
```
# 4. Test Model with 20 languages!
```
train_df = pd.read_csv("news_category_test_multi_lingual.csv")
preds = fitted_pipe.predict(train_df[["test_sentences","y"]].iloc[:100],output_level='document')
#sentence detector that is part of the pipe generates sone NaNs. lets drop them first
preds.dropna(inplace=True)
print(classification_report(preds['y'], preds['category']))
preds
```
# The Model understands Englsih

```
fitted_pipe.predict("There have been a great increase in businesses over the last decade ")
fitted_pipe.predict("Science has advanced rapidly over the last century ")
```
# The Model understands German

```
# German for: 'Businesses are the best way of making profit'
fitted_pipe.predict("Unternehmen sind der beste Weg, um Gewinn zu erzielen")
# German for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Die Wissenschaft hat im letzten Jahrhundert rasante Fortschritte gemacht ")
```
# The Model understands Chinese

```
# Chinese for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("在过去的十年中,业务有了很大的增长 ")
# Chinese for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("在上个世纪,科学发展迅速 ")
```
# Model understands Afrikaans

```
# Afrikaans for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("Daar het die afgelope dekade 'n groot toename in besighede plaasgevind ")
# Afrikaans for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Die wetenskap het die afgelope eeu vinnig gevorder ")
```
# The model understands Vietnamese

```
# Vietnamese for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("Đã có sự gia tăng đáng kể trong các doanh nghiệp trong thập kỷ qua ")
# Vietnamese for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Khoa học đã phát triển nhanh chóng trong thế kỷ qua ")
```
# The model understands Japanese

```
# Japanese for: 'Businesses are the best way of making profit'
fitted_pipe.predict("ビジネスは利益を上げるための最良の方法です")
# Japanese for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("科学は前世紀にわたって急速に進歩しました ")
```
# The model understands Zulu

```
# Zulu for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("Kube nokwanda okukhulu emabhizinisini kule minyaka eyishumi edlule ")
# Zulu for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Isayensi ithuthuke ngokushesha ngekhulu leminyaka elidlule ")
```
# The Model understands Turkish

```
# Turkish for: 'Businesses are the best way of making profit'
fitted_pipe.predict("İşletmeler kar elde etmenin en iyi yoludur ")
# Turkish for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Bilim, geçen yüzyılda hızla ilerledi ")
```
# The Model understands Hebrew

```
# Hebrew for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("חלה עלייה גדולה בעסקים בעשור האחרון ")
# Hebrew for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("המדע התקדם במהירות במהלך המאה האחרונה ")
```
# The Model understands Telugu

```
# Telugu for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("గత దశాబ్దంలో వ్యాపారాలలో గొప్ప పెరుగుదల ఉంది ")
# Telugu for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("గత శతాబ్దంలో సైన్స్ వేగంగా అభివృద్ధి చెందింది ")
```
# Model understands Russian

```
# Russian for: 'Businesses are the best way of making profit'
fitted_pipe.predict("Бизнес - лучший способ получения прибыли")
# Russian for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Наука стремительно развивалась за последнее столетие ")
```
# Model understands Urdu

```
# Urdu for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("پچھلے ایک دہائی کے دوران کاروباروں میں زبردست اضافہ ہوا ہے ")
# Urdu for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("سائنس گذشتہ صدی کے دوران تیزی سے ترقی کرچکی ہے ")
```
# Model understands Hindi

```
# hindi for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("पिछले दशक में व्यवसायों में बहुत वृद्धि हुई है ")
# hindi for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("विज्ञान पिछली सदी में तेजी से आगे बढ़ा है ")
```
# The model understands Tartar

```
# Tartar for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("Соңгы ун елда бизнеста зур үсеш булды ")
# Tartar for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Соңгы гасырда фән тиз үсә ")
```
# The Model understands French

```
# French for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("Il y a eu une forte augmentation des entreprises au cours de la dernière décennie ")
# French for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("La science a progressé rapidement au cours du siècle dernier ")
```
# The Model understands Thai

```
# Thai for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("มีธุรกิจเพิ่มขึ้นอย่างมากในช่วงทศวรรษที่ผ่านมา ")
# Thai for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("วิทยาศาสตร์ก้าวหน้าอย่างรวดเร็วในช่วงศตวรรษที่ผ่านมา ")
```
# The Model understands Khmer

```
# Khmer for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("មានការរីកចម្រើនយ៉ាងខ្លាំងនៅក្នុងអាជីវកម្មក្នុងរយៈពេលមួយទសវត្សចុងក្រោយនេះ ")
# Khmer for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("វិទ្យាសាស្ត្របានជឿនលឿនយ៉ាងលឿនក្នុងរយៈពេលមួយសតវត្សចុងក្រោយនេះ ")
```
# The Model understands Yiddish

```
# Yiddish for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("די לעצטע יאָרצענדלינג איז געווען אַ גרויס פאַרגרעסערן אין געשעפטן ")
# Yiddish for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("וויסנשאַפֿט איז ראַפּאַדלי אַוואַנסירטע איבער די לעצטע יאָרהונדערט ")
```
# The Model understands Kygrgyz

```
# Kygrgyz for: 'Businesses are the best way of making profit'
fitted_pipe.predict("Бизнес - бул киреше табуунун эң мыкты жолу ")
# Kygrgyz for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Илим акыркы кылымда тездик менен өнүккөн ")
```
# The Model understands Tamil

```
# Tamil for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("கடந்த தசாப்தத்தில் வணிகங்களில் பெரும் அதிகரிப்பு ஏற்பட்டுள்ளது ")
# Tamil for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("கடந்த நூற்றாண்டில் அறிவியல் வேகமாக முன்னேறியுள்ளது ")
```
# 5. Lets save the model
```
stored_model_path = './models/classifier_dl_trained'
fitted_pipe.save(stored_model_path)
```
# 6. Lets load the model from HDD.
This makes Offlien NLU usage possible!
You need to call nlu.load(path=path_to_the_pipe) to load a model/pipeline from disk.
```
stored_model_path = './models/classifier_dl_trained'
hdd_pipe = nlu.load(path=stored_model_path)
preds = hdd_pipe.predict('Tesla plans to invest 10M into the ML sector')
preds
hdd_pipe.print_info()
```
|
github_jupyter
|
import os
! apt-get update -qq > /dev/null
# Install java
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["PATH"] = os.environ["JAVA_HOME"] + "/bin:" + os.environ["PATH"]
! pip install nlu > /dev/null pyspark==2.4.7
import nlu
! wget http://ckl-it.de/wp-content/uploads/2021/02/news_category_test_multi_lingual.csv
import pandas as pd
test_path = '/content/news_category_test_multi_lingual.csv'
train_df = pd.read_csv(test_path)
from sklearn.model_selection import train_test_split
train_df, test_df = train_test_split(train_df, test_size=0.2)
train_df
trainable_pipe = nlu.load('xx.embed_sentence.labse train.classifier')
# We need to train longer and user smaller LR for NON-USE based sentence embeddings usually
# We could tune the hyperparameters further with hyperparameter tuning methods like gridsearch
# Also longer training gives more accuracy
trainable_pipe['classifier_dl'].setMaxEpochs(60)
trainable_pipe['classifier_dl'].setLr(0.005)
fitted_pipe = trainable_pipe.fit(train_df.iloc[:1500])
# predict with the trainable pipeline on dataset and get predictions
preds = fitted_pipe.predict(train_df.iloc[:1500],output_level='document')
#sentence detector that is part of the pipe generates sone NaNs. lets drop them first
preds.dropna(inplace=True)
from sklearn.metrics import classification_report
print(classification_report(preds['y'], preds['category']))
preds
preds = fitted_pipe.predict(test_df,output_level='document')
#sentence detector that is part of the pipe generates sone NaNs. lets drop them first
preds.dropna(inplace=True)
print(classification_report(preds['y'], preds['category']))
train_df = pd.read_csv("news_category_test_multi_lingual.csv")
preds = fitted_pipe.predict(train_df[["test_sentences","y"]].iloc[:100],output_level='document')
#sentence detector that is part of the pipe generates sone NaNs. lets drop them first
preds.dropna(inplace=True)
print(classification_report(preds['y'], preds['category']))
preds
fitted_pipe.predict("There have been a great increase in businesses over the last decade ")
fitted_pipe.predict("Science has advanced rapidly over the last century ")
# German for: 'Businesses are the best way of making profit'
fitted_pipe.predict("Unternehmen sind der beste Weg, um Gewinn zu erzielen")
# German for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Die Wissenschaft hat im letzten Jahrhundert rasante Fortschritte gemacht ")
# Chinese for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("在过去的十年中,业务有了很大的增长 ")
# Chinese for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("在上个世纪,科学发展迅速 ")
# Afrikaans for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("Daar het die afgelope dekade 'n groot toename in besighede plaasgevind ")
# Afrikaans for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Die wetenskap het die afgelope eeu vinnig gevorder ")
# Vietnamese for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("Đã có sự gia tăng đáng kể trong các doanh nghiệp trong thập kỷ qua ")
# Vietnamese for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Khoa học đã phát triển nhanh chóng trong thế kỷ qua ")
# Japanese for: 'Businesses are the best way of making profit'
fitted_pipe.predict("ビジネスは利益を上げるための最良の方法です")
# Japanese for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("科学は前世紀にわたって急速に進歩しました ")
# Zulu for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("Kube nokwanda okukhulu emabhizinisini kule minyaka eyishumi edlule ")
# Zulu for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Isayensi ithuthuke ngokushesha ngekhulu leminyaka elidlule ")
# Turkish for: 'Businesses are the best way of making profit'
fitted_pipe.predict("İşletmeler kar elde etmenin en iyi yoludur ")
# Turkish for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Bilim, geçen yüzyılda hızla ilerledi ")
# Hebrew for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("חלה עלייה גדולה בעסקים בעשור האחרון ")
# Hebrew for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("המדע התקדם במהירות במהלך המאה האחרונה ")
# Telugu for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("గత దశాబ్దంలో వ్యాపారాలలో గొప్ప పెరుగుదల ఉంది ")
# Telugu for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("గత శతాబ్దంలో సైన్స్ వేగంగా అభివృద్ధి చెందింది ")
# Russian for: 'Businesses are the best way of making profit'
fitted_pipe.predict("Бизнес - лучший способ получения прибыли")
# Russian for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Наука стремительно развивалась за последнее столетие ")
# Urdu for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("پچھلے ایک دہائی کے دوران کاروباروں میں زبردست اضافہ ہوا ہے ")
# Urdu for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("سائنس گذشتہ صدی کے دوران تیزی سے ترقی کرچکی ہے ")
# hindi for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("पिछले दशक में व्यवसायों में बहुत वृद्धि हुई है ")
# hindi for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("विज्ञान पिछली सदी में तेजी से आगे बढ़ा है ")
# Tartar for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("Соңгы ун елда бизнеста зур үсеш булды ")
# Tartar for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Соңгы гасырда фән тиз үсә ")
# French for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("Il y a eu une forte augmentation des entreprises au cours de la dernière décennie ")
# French for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("La science a progressé rapidement au cours du siècle dernier ")
# Thai for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("มีธุรกิจเพิ่มขึ้นอย่างมากในช่วงทศวรรษที่ผ่านมา ")
# Thai for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("วิทยาศาสตร์ก้าวหน้าอย่างรวดเร็วในช่วงศตวรรษที่ผ่านมา ")
# Khmer for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("មានការរីកចម្រើនយ៉ាងខ្លាំងនៅក្នុងអាជីវកម្មក្នុងរយៈពេលមួយទសវត្សចុងក្រោយនេះ ")
# Khmer for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("វិទ្យាសាស្ត្របានជឿនលឿនយ៉ាងលឿនក្នុងរយៈពេលមួយសតវត្សចុងក្រោយនេះ ")
# Yiddish for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("די לעצטע יאָרצענדלינג איז געווען אַ גרויס פאַרגרעסערן אין געשעפטן ")
# Yiddish for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("וויסנשאַפֿט איז ראַפּאַדלי אַוואַנסירטע איבער די לעצטע יאָרהונדערט ")
# Kygrgyz for: 'Businesses are the best way of making profit'
fitted_pipe.predict("Бизнес - бул киреше табуунун эң мыкты жолу ")
# Kygrgyz for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("Илим акыркы кылымда тездик менен өнүккөн ")
# Tamil for: 'There have been a great increase in businesses over the last decade'
fitted_pipe.predict("கடந்த தசாப்தத்தில் வணிகங்களில் பெரும் அதிகரிப்பு ஏற்பட்டுள்ளது ")
# Tamil for: 'Science has advanced rapidly over the last century'
fitted_pipe.predict("கடந்த நூற்றாண்டில் அறிவியல் வேகமாக முன்னேறியுள்ளது ")
stored_model_path = './models/classifier_dl_trained'
fitted_pipe.save(stored_model_path)
stored_model_path = './models/classifier_dl_trained'
hdd_pipe = nlu.load(path=stored_model_path)
preds = hdd_pipe.predict('Tesla plans to invest 10M into the ML sector')
preds
hdd_pipe.print_info()
| 0.397704 | 0.776792 |
# Testing rotation period optimisation.
The purpose of this project is to attempt to optimise the measurement of stellar rotation periods from TESS data. This will involve the following stages:
1) Calculate single & double Lomb-Scargle periodograms, ACFs and PDMs of Kepler light curves with and without measured rotation periods (McQuillan et al. 2014).
2) Calculate features/statistics of these: heights and positions of tallest peaks, etc.
3) Train a random forest classifier to classify rotators, non-rotators, and non-periodic rotators.
4) Train a random forest regressor to measure a rotation period from these features.
5) Repeat with Kepler light curves cut into 27 days sectors.
6) Build a training set from TESS CVZ stars.
7) Repeat using this as a training set.
```
%matplotlib inline
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import starrotate as sr
import kepler_data as kd
import sigma_clip as sc
import calc_statistics as cs
```
First let's just try one star. Load the McQuillan tables.
```
mc1 = pd.read_csv("../data/Table_1_Periodic.txt")
mc2 = pd.read_csv("../data/Table_2_Non_Periodic.txt")
mc1.head()
```
Load the first light curve.
```
i = 1
kplr_path = "/Users/rangus/.kplr/data/lightcurves"
path_to_light_curve = os.path.join(kplr_path, str(int(mc1.iloc[i].kepid)).zfill(9))
x, y, yerr = cs.load_and_process(path_to_light_curve)
rm = sr.RotationModel(x, y, yerr)
```
Calculate LS periodogram and ACF.
```
highest_peak_period = rm.LS_rotation()
highest_peak_acf = rm.ACF_rotation(interval=0.02043365)
```
Calculate some statistics. Ideas:
Lomb-Scargle:
1st 3 Peak positions, 1st 3 Peak heights, RMS, MAD
ACF
1st 3 Peak positions, 1st 3 Peak heights, Highest 3 Peak positions, Highest 3 Peak heights
Light curve:
Rvar, Rvar_10day, Rvar_20day, Rvar_50_day
Get statistics.
```
ls_h, ls_p, acf_h1, acf_p1, acf_h2, acf_p2, acf_pgram_h, acf_pgram_p, ls_mad, ls_rms, \
acf_mad, acf_rms, Rvar, acf_freqs, acf_pgram = cs.get_statistics(
y, 1./rm.freq, rm.power, rm.lags, rm.acf)
fig = plt.figure(figsize=(10, 10))
ax1 = fig.add_subplot(311)
ax1.plot(1./rm.freq, rm.power)
ax1.set_xlim(0, np.min([10*highest_peak_period, max(1./rm.freq)]))
ax1.set_ylabel("$\mathrm{Power}$")
ax1.axvline(ls_p[0], color="C1", ls="--")
ax1.axvline(ls_p[1], color="C1", ls="--")
ax1.axvline(ls_p[2], color="C1", ls="--")
ax2 = fig.add_subplot(312, sharex=ax1)
ax2.plot(rm.lags, rm.acf)
ax2.set_ylabel("$\mathrm{ACF}$")
ax2.axvline(acf_p2[0], color="C1", ls="--")
ax2.axvline(acf_p2[1], color="C1", ls="--")
ax2.axvline(acf_p2[2], color="C1", ls="--")
ax3 = fig.add_subplot(313, sharex=ax1)
ax3.plot(1./acf_freqs, acf_pgram)
ax3.axvline(acf_pgram_p[0], color="C1", ls="--")
ax3.axvline(acf_pgram_p[1], color="C1", ls="--")
ax3.axvline(acf_pgram_p[2], color="C1", ls="--")
ax3.set_ylabel("$\mathrm{Power}$")
ax3.set_xlabel("$\mathrm{Time~[Days]}$")
plt.subplots_adjust(hspace=0, right=0.83)
plt.setp(ax1.get_xticklabels(), visible=False);
fig = plt.figure(figsize=(10, 10))
ax1 = fig.add_subplot(311)
ax1.plot(x, y, "k.", ms=1, alpha=.5)
ax1.set_ylabel("$\mathrm{Flux}$")
ax1.set_xlabel("$\mathrm{Time~[Days]}$")
ax2 = fig.add_subplot(312)
ax2.plot(x, y, "k.", ms=1)
ax2.set_xlim(x[0], x[0]+100)
ax2.set_ylabel("$\mathrm{Flux}$")
ax2.set_xlabel("$\mathrm{Time~[Days]}$")
ax3 = fig.add_subplot(313)
phase = (x % ls_p[0])/ls_p[0]
m = x < 50*ls_p[0]
ax3.plot(phase[m], y[m], "k.", ms=2, alpha=.5)
ax3.set_ylabel("$\mathrm{Flux}$")
ax3.set_xlabel("$\mathrm{Phase}$")
plt.tight_layout()
print(ls_p[0], acf_p2[0], acf_pgram_p[0], mc1.iloc[i].Prot)
```
|
github_jupyter
|
%matplotlib inline
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import starrotate as sr
import kepler_data as kd
import sigma_clip as sc
import calc_statistics as cs
mc1 = pd.read_csv("../data/Table_1_Periodic.txt")
mc2 = pd.read_csv("../data/Table_2_Non_Periodic.txt")
mc1.head()
i = 1
kplr_path = "/Users/rangus/.kplr/data/lightcurves"
path_to_light_curve = os.path.join(kplr_path, str(int(mc1.iloc[i].kepid)).zfill(9))
x, y, yerr = cs.load_and_process(path_to_light_curve)
rm = sr.RotationModel(x, y, yerr)
highest_peak_period = rm.LS_rotation()
highest_peak_acf = rm.ACF_rotation(interval=0.02043365)
ls_h, ls_p, acf_h1, acf_p1, acf_h2, acf_p2, acf_pgram_h, acf_pgram_p, ls_mad, ls_rms, \
acf_mad, acf_rms, Rvar, acf_freqs, acf_pgram = cs.get_statistics(
y, 1./rm.freq, rm.power, rm.lags, rm.acf)
fig = plt.figure(figsize=(10, 10))
ax1 = fig.add_subplot(311)
ax1.plot(1./rm.freq, rm.power)
ax1.set_xlim(0, np.min([10*highest_peak_period, max(1./rm.freq)]))
ax1.set_ylabel("$\mathrm{Power}$")
ax1.axvline(ls_p[0], color="C1", ls="--")
ax1.axvline(ls_p[1], color="C1", ls="--")
ax1.axvline(ls_p[2], color="C1", ls="--")
ax2 = fig.add_subplot(312, sharex=ax1)
ax2.plot(rm.lags, rm.acf)
ax2.set_ylabel("$\mathrm{ACF}$")
ax2.axvline(acf_p2[0], color="C1", ls="--")
ax2.axvline(acf_p2[1], color="C1", ls="--")
ax2.axvline(acf_p2[2], color="C1", ls="--")
ax3 = fig.add_subplot(313, sharex=ax1)
ax3.plot(1./acf_freqs, acf_pgram)
ax3.axvline(acf_pgram_p[0], color="C1", ls="--")
ax3.axvline(acf_pgram_p[1], color="C1", ls="--")
ax3.axvline(acf_pgram_p[2], color="C1", ls="--")
ax3.set_ylabel("$\mathrm{Power}$")
ax3.set_xlabel("$\mathrm{Time~[Days]}$")
plt.subplots_adjust(hspace=0, right=0.83)
plt.setp(ax1.get_xticklabels(), visible=False);
fig = plt.figure(figsize=(10, 10))
ax1 = fig.add_subplot(311)
ax1.plot(x, y, "k.", ms=1, alpha=.5)
ax1.set_ylabel("$\mathrm{Flux}$")
ax1.set_xlabel("$\mathrm{Time~[Days]}$")
ax2 = fig.add_subplot(312)
ax2.plot(x, y, "k.", ms=1)
ax2.set_xlim(x[0], x[0]+100)
ax2.set_ylabel("$\mathrm{Flux}$")
ax2.set_xlabel("$\mathrm{Time~[Days]}$")
ax3 = fig.add_subplot(313)
phase = (x % ls_p[0])/ls_p[0]
m = x < 50*ls_p[0]
ax3.plot(phase[m], y[m], "k.", ms=2, alpha=.5)
ax3.set_ylabel("$\mathrm{Flux}$")
ax3.set_xlabel("$\mathrm{Phase}$")
plt.tight_layout()
print(ls_p[0], acf_p2[0], acf_pgram_p[0], mc1.iloc[i].Prot)
| 0.245356 | 0.976625 |
```
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from statsmodels.graphics.regressionplots import influence_plot
import statsmodels.formula.api as smf
import numpy as np
a=pd.read_csv('ToyotaCorolla.csv')
a
a.columns
a1=a.drop(['Id', 'Model','Mfg_Month', 'Mfg_Year','Fuel_Type','Met_Color','Color','Automatic', 'Cylinders','Mfr_Guarantee',
'BOVAG_Guarantee', 'Guarantee_Period', 'ABS', 'Airbag_1', 'Airbag_2',
'Airco', 'Automatic_airco', 'Boardcomputer', 'CD_Player',
'Central_Lock', 'Powered_Windows', 'Power_Steering', 'Radio',
'Mistlamps', 'Sport_Model', 'Backseat_Divider', 'Metallic_Rim',
'Radio_cassette', 'Tow_Bar'],axis=1)
a1
a1.corr()
a2=a1.rename({'Price':'pr','Age_08_04':'age','Doors':'dr','Gears':'gr','Quarterly_Tax':'qt','Weight':'wt'},axis=1)
a2
a2.corr()
a2.columns
model=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a2).fit()
(model.rsquared,model.rsquared_adj)
```
# iteration 1
```
model_influence=model.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a2)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a3=a2.drop([80],axis=0)
a4=a3.reset_index()
a5=a4.drop(['index'],axis=1)
model1=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a5).fit()
(model1.rsquared,model1.rsquared_adj)
```
# iteration 2
```
model_influence=model1.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a5)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a6=a5.drop([220],axis=0)
a7=a6.reset_index()
a8=a7.drop(['index'],axis=1)
model2=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a8).fit()
(model2.rsquared,model2.rsquared_adj)
```
# iteration 3
```
from statsmodels.graphics.regressionplots import influence_plot
influence_plot(model2)
plt.show()
a9=a8.drop([958,599,989],axis=0)
a10=a9.reset_index()
a11=a10.drop(['index'],axis=1)
model3=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a11).fit()
(model3.rsquared,model3.rsquared_adj)
```
# iteration 4
```
model_influence=model3.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a11)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a12=a11.drop([651],axis=0)
a13=a12.reset_index()
a14=a13.drop(['index'],axis=1)
model4=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a14).fit()
(model4.rsquared,model4.rsquared_adj)
```
# iteration 5
```
influence_plot(model4)
plt.show()
a15=a14.drop([108],axis=0)
a16=a15.reset_index()
a17=a16.drop(['index'],axis=1)
model5=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a17).fit()
(model5.rsquared,model5.rsquared_adj)
```
# iteration 6
```
model_influence=model4.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a14)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a18=a14.drop([190],axis=0)
a19=a18.reset_index()
a20=a19.drop(['index'],axis=1)
model6=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a20).fit()
(model6.rsquared,model6.rsquared_adj)
```
# iteration 8
```
model_influence=model6.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a20)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a21=a20.drop([1051],axis=0)
a22=a21.reset_index()
a23=a22.drop(['index'],axis=1)
model7=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a23).fit()
(model7.rsquared,model7.rsquared_adj)
```
# iteration 7
```
model_influence=model7.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a23)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a24=a23.drop([190],axis=0)
a25=a24.reset_index()
a26=a25.drop(['index'],axis=1)
model8=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a26).fit()
(model8.rsquared,model8.rsquared_adj)
```
# iteration 9
```
a27=a26**(1/2)
a27.head()
model9=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a27).fit()
model9.rsquared,model9.rsquared_adj
# here squareroot transformation is not working ,hence we are again going to drop another entries from the dataset.
```
# iteration 10
```
model_influence=model8.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a26)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a28=a27.drop([398],axis=0)
a29=a28.reset_index()
a30=a29.drop(['index'],axis=1)
model10=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a30).fit()
(model10.rsquared,model10.rsquared_adj)
# here we can see that corresponding models accuracy is started decreasing over here.
df={'models':['basic model','model1','model2','model3','model4','model5','model6','model7','model8','model10'],'rsquared values':[model.rsquared,model1.rsquared,model2.rsquared,model3.rsquared,model4.rsquared,model5.rsquared,model6.rsquared,model7.rsquared,model8.rsquared,model10.rsquared]}
b2=pd.DataFrame(df)
b2
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from statsmodels.graphics.regressionplots import influence_plot
import statsmodels.formula.api as smf
import numpy as np
a=pd.read_csv('ToyotaCorolla.csv')
a
a.columns
a1=a.drop(['Id', 'Model','Mfg_Month', 'Mfg_Year','Fuel_Type','Met_Color','Color','Automatic', 'Cylinders','Mfr_Guarantee',
'BOVAG_Guarantee', 'Guarantee_Period', 'ABS', 'Airbag_1', 'Airbag_2',
'Airco', 'Automatic_airco', 'Boardcomputer', 'CD_Player',
'Central_Lock', 'Powered_Windows', 'Power_Steering', 'Radio',
'Mistlamps', 'Sport_Model', 'Backseat_Divider', 'Metallic_Rim',
'Radio_cassette', 'Tow_Bar'],axis=1)
a1
a1.corr()
a2=a1.rename({'Price':'pr','Age_08_04':'age','Doors':'dr','Gears':'gr','Quarterly_Tax':'qt','Weight':'wt'},axis=1)
a2
a2.corr()
a2.columns
model=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a2).fit()
(model.rsquared,model.rsquared_adj)
model_influence=model.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a2)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a3=a2.drop([80],axis=0)
a4=a3.reset_index()
a5=a4.drop(['index'],axis=1)
model1=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a5).fit()
(model1.rsquared,model1.rsquared_adj)
model_influence=model1.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a5)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a6=a5.drop([220],axis=0)
a7=a6.reset_index()
a8=a7.drop(['index'],axis=1)
model2=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a8).fit()
(model2.rsquared,model2.rsquared_adj)
from statsmodels.graphics.regressionplots import influence_plot
influence_plot(model2)
plt.show()
a9=a8.drop([958,599,989],axis=0)
a10=a9.reset_index()
a11=a10.drop(['index'],axis=1)
model3=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a11).fit()
(model3.rsquared,model3.rsquared_adj)
model_influence=model3.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a11)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a12=a11.drop([651],axis=0)
a13=a12.reset_index()
a14=a13.drop(['index'],axis=1)
model4=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a14).fit()
(model4.rsquared,model4.rsquared_adj)
influence_plot(model4)
plt.show()
a15=a14.drop([108],axis=0)
a16=a15.reset_index()
a17=a16.drop(['index'],axis=1)
model5=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a17).fit()
(model5.rsquared,model5.rsquared_adj)
model_influence=model4.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a14)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a18=a14.drop([190],axis=0)
a19=a18.reset_index()
a20=a19.drop(['index'],axis=1)
model6=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a20).fit()
(model6.rsquared,model6.rsquared_adj)
model_influence=model6.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a20)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a21=a20.drop([1051],axis=0)
a22=a21.reset_index()
a23=a22.drop(['index'],axis=1)
model7=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a23).fit()
(model7.rsquared,model7.rsquared_adj)
model_influence=model7.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a23)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a24=a23.drop([190],axis=0)
a25=a24.reset_index()
a26=a25.drop(['index'],axis=1)
model8=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a26).fit()
(model8.rsquared,model8.rsquared_adj)
a27=a26**(1/2)
a27.head()
model9=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a27).fit()
model9.rsquared,model9.rsquared_adj
# here squareroot transformation is not working ,hence we are again going to drop another entries from the dataset.
model_influence=model8.get_influence()
(c_V,_)=model_influence.cooks_distance
fig=plt.subplots(figsize=(20,7))
plt.stem(np.arange(len(a26)),np.round(c_V,3))
plt.xlabel('row index')
plt.ylabel('cooks distance')
(np.argmax(c_V),np.max(c_V))
a28=a27.drop([398],axis=0)
a29=a28.reset_index()
a30=a29.drop(['index'],axis=1)
model10=smf.ols('pr~age+KM+HP+cc+dr+gr+qt+wt',data=a30).fit()
(model10.rsquared,model10.rsquared_adj)
# here we can see that corresponding models accuracy is started decreasing over here.
df={'models':['basic model','model1','model2','model3','model4','model5','model6','model7','model8','model10'],'rsquared values':[model.rsquared,model1.rsquared,model2.rsquared,model3.rsquared,model4.rsquared,model5.rsquared,model6.rsquared,model7.rsquared,model8.rsquared,model10.rsquared]}
b2=pd.DataFrame(df)
b2
| 0.378919 | 0.698407 |
# Introduction to data
Statistics is the study of how best to collect, analyze and draw conclusions from data following a general process of investigation :
1. Identify a question or problem
2. Collect relevant data on topic
3. Analyze the data
4. Form a conslusion
Statistics should answer 3 primary questions :
* how best we collect data ?
* how should it be analyzed ?
* What can we infer from the analysis ?
## 1.1 Case Study : using stents to prevent strokes
**Generating a table of observations for trreatment and control group of a stent efficiency study**
```
import pandas as pd
import numpy as np
pd.options.display.max_rows = 20 # control the number of rows printed
```
Creating 2 distinct groups respecting the amount of observations given in the book :
```
#Creating the treatment group
treatment = pd.DataFrame(columns=['group','thirty_days','365days'])
treatment['group'] = ['treatment']*224
treatment['thirty_days'] = (['no event']*191) + (['stroke']*33)
treatment['365days'] = (['no event']*179) + (['stroke'] * 45)
treatment
#creating the control group
control = pd.DataFrame(columns=['group','thirty_days','365days'])
control['group'] = ['control']*227
control['thirty_days'] = (['no event']*214)+(['stroke']*13)
control['365days'] = (['no event'] * 199)+(['stroke']*28)
control
```
Merge the 2 Datraframes to produce the same table as the example :
```
frames = [treatment,control]
patients = pd.concat(frames,ignore_index=True)
patients
```
Shuffling The rows
```
patients = patients.sample(frac=1).reset_index(drop=True)
patients
```
** Summary Statistics : ** summarizing a large amount of data with a single number like
* The proportion of people who had a stroke in the treatment group : 45/224 = 20%
helps getting a first insight information.
<br/>
Still caution is important, many parameters should be taken into account and hastly inference could be misleading.
## 1.2 Data Basics
Each **row** of the matrice represents a **case**. And each column a **variable**
Types of variables
```
from IPython.display import Image
Image(filename="data_types.png")
```
## 1.3 Overview of data collection principles
**Population :** the target of a statistical analysis.
** Sample : ** being too expensive to collect an exhaustivity of data in a population, we tend take a small fraction of the population. This can be done by raffles, even thou there is always a risk to take biased data.
**Anecdotal evidence : ** data collected in a haphazard fashion, that may only represents extraordinary cases.
It's important to avoid commn traps when collecting and interpreting data :
* **non-reponse : ** a big non response rate makes it hard to get a clear answer
* **convenience sample : ** elements easily found in gathering data (close neighborhood for instance) generates a big bias
** Explanatory and response variables : ** if we suspect a data **X** to affect another **Y**. Then we can call X the **explanatory variable** and Y the **response variable**.
<br/>
Ex : poverty(explanatory) affecting federal spending(response) in a county.
## 1.4 Observational Studies and sampling strategies
** Prospective study : ** indentifies individuals and collects information as events unfold
** Retrospective studies : ** collect data after events have taken place
** Sampling methods **:
* **Simple Random sampling : ** Simple raffle like paper in a bucket
* **Stratified sampling : ** divide the population in stratas of identical attributes
* **Cluster sample : **
* **Multistage Sampe : **
## 1.5 Experiments :
Principles of experiment design :
* **Controlling :** control differences in an experiment (control group)
* **Randomization :** randomizing pateints into treatment group
* **Replication :** the more cases researchers observe, the more accurately they can estimate the effect
* **Blocking :** grouping individuals into specific blocks
```
from IPython.display import Image
Image(filename="blocking.png")
```
## 1.6 Examining Numerical Data
using the **email50** dataset :
```
#importing plotting libs
import matplotlib.pyplot as plt
%matplotlib inline
e50 = pd.read_csv('Dataset/email50.csv')
e50.head()
```
### 1.6.1 Scatterplot
Scatterplot offers a case-by-case view of data for 2 numerical variables
```
e50.plot.scatter(figsize=(12,6),x='num_char',y='line_breaks')
plt.xlabel("Number of characters (in thousands)",fontsize=16)
plt.ylabel('Number of Lines',fontsize=16)
```
## The mean
Sometimes called the average, is a common way to to measure the center of the distribution.
Mean of the number of characters in a mail :
```
e50['num_char'].mean()
```
* The sample **mean** is usualy refered to as $\bar{X}$
**rule : **
<br/>
$\bar{X}$ = $\frac{1}{n}$ ($\displaystyle\sum_{i=1}^{n} x_i$) = $\frac{x_1+x_2+\dots+x_n}{n}$
* The population mean is refered to as $\mu$
### 1.6.2 Histograms
Histograms provide a view of the **data density**
```
e50['num_char'].plot.hist()
```
we can notice that the data trail off to the right. in this case we talk about **right skew**. The opposite is true for **left skew**.
**Mode** : is represented by a prominent peakin the distribution. in this example we see that the mode is the first bar. However we can find such graph with 2 to more modes. In this case we talk about **Bimodal** to **Mutlimodal** distributions.
## 1.6.2 Variance and Standard Deviation
The **standard deviation** describes how far away the typical observation is from the mean. It is a mean to calculate the dispersion of data. The lower the standard deviation, the closer it is from the mean.
<br/><br/>
We call the distance of an observation from its mean its **deviation**
<br/><br/>
The notation for the sample standard deviation is **s**.
<br/>
The notation for the population standard deviation is **$\delta$**
**Formula for the Sample standard deviation : **
<br/>
<br/>
$\delta$ = $\sqrt\frac{1}{N-1}\displaystyle\sum_{i=1}^{N}(x_i - \bar{x})^2$
Although we mostly use the standard deviation, the **Variance** represents the squared differences from the mean and is obtained by squaring the standard deviation.
<br/>
$s = \sqrt Variance$
### 1.6.3 Box plots, quartiles and median
A **box plot** summarizes a dataset using 5 statistics :
* Minimum
* First Quartile Q1
* Median
* Third Quartile Q3
* Maximum
```
#importing seaborn to use the boxplot
import seaborn as sns
```
box plot of the **num_char** variable
```
sns.boxplot(data=e50,y='num_char')
```
- The dark line represents the **median**. The median is what we called a **robust estimates** that designate the data right in the middle out of a set of ordered data.
- The full length of the box is the **IQR** (Interquartile range) that covers the 25th percentile Q1 till the 75th percentile Q3. The larger the IQR and standard deviation, the more variable is the data.
```
from IPython.display import Image
Image(filename="boxplot.png")
```
- the **whiskers** capture the data outside the box. Their reach however is never allowed to trespass 1.5 times the IQR. everything over these bars is considered as **outlier**
- an **outlier** is a data that looks extreme relative to the rest of the data
Median and IQR are called **robust estimates** because extreme observations have little effect on their values. The mean and standard deviation are much more affected by extravagant data.
### 1.6.4 Transforming data
**Transforming** data makes them easier to model when they are strongly skewed. It is a rescaling of the data using a function. Data becomes therefore less skewed and outliers less extreme.
* we work with the Major Baseball League Salaray Dataset (mlb.csv)
**Annex 2 : ** retrieve the data in a csv format from the text file mlb.txt (mlb_Salary.py)
```
mlb = pd.read_csv('Dataset/mlb.csv')
mlb.head()
```
Showing the distribution of the salaries from 2010 without transformation
```
mlb['salary'].plot.hist()
```
Data distribution after a log transform ()
```
np.log(mlb['salary']).plot.hist()
```
* example with the email50 datas
```
e50.plot.scatter(x='num_char',y='line_breaks')
plt.title("Scatter plot of line breaks against number of characters raw")
x = np.log(e50['num_char'],dtype='float64')
y = np.log(e50['line_breaks'], dtype='float64')
plt.scatter(x,y)
plt.title("Scatter plot of line breaks against number of characters after log transform")
```
## 1.7 Categorical data
### 1.7.1 Contingency and frequency table
```
e50.shape
e50.head()
```
- A table that summarizes data for 2 categorical varaibles is valled a **contingency table**
```
#calling a function to count instead of using lambda function for further use
def counting(x):
return x.count()
contingency = e50.pivot_table(index='spam',columns='number', values='num_char', aggfunc=[counting])
cont_plot = contingency.copy()
contingency
#Counting the total of all the observations
contingency.loc[:,('counting','total')] = contingency.sum(axis=1)
contingency
```
- A table for a single variable is called a **frequency table**.
(Using the 'email50' example to cut down the rows from 3921 to 50)
```
frequency = e50.pivot_table(columns='number', values='num_char', aggfunc=[counting])
frequency
#Counting the total of all the observations
frequency.loc[:,('counting','total')] = frequency.loc[:,('counting','big')] + frequency.loc[:,('counting','none')] + frequency.loc[:,('counting','small')]
frequency
```
### 1.7.2 Row and column proportions
- **Column Proportion** makes the corresponding the contingency table to their proportions
```
#The Contingency Table
contingency
#It's Column proportion
contingency.loc[:,('counting')].div(contingency.loc[:,('counting','total')],axis=0)#divide all the other columns by one column
contingency.info()
```
### 1.7.3 Segmented bar plot
```
# saved cont_plot on In [26]
cont_plot.T.plot.bar(stacked=True)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
pd.options.display.max_rows = 20 # control the number of rows printed
#Creating the treatment group
treatment = pd.DataFrame(columns=['group','thirty_days','365days'])
treatment['group'] = ['treatment']*224
treatment['thirty_days'] = (['no event']*191) + (['stroke']*33)
treatment['365days'] = (['no event']*179) + (['stroke'] * 45)
treatment
#creating the control group
control = pd.DataFrame(columns=['group','thirty_days','365days'])
control['group'] = ['control']*227
control['thirty_days'] = (['no event']*214)+(['stroke']*13)
control['365days'] = (['no event'] * 199)+(['stroke']*28)
control
frames = [treatment,control]
patients = pd.concat(frames,ignore_index=True)
patients
patients = patients.sample(frac=1).reset_index(drop=True)
patients
from IPython.display import Image
Image(filename="data_types.png")
from IPython.display import Image
Image(filename="blocking.png")
#importing plotting libs
import matplotlib.pyplot as plt
%matplotlib inline
e50 = pd.read_csv('Dataset/email50.csv')
e50.head()
e50.plot.scatter(figsize=(12,6),x='num_char',y='line_breaks')
plt.xlabel("Number of characters (in thousands)",fontsize=16)
plt.ylabel('Number of Lines',fontsize=16)
e50['num_char'].mean()
e50['num_char'].plot.hist()
#importing seaborn to use the boxplot
import seaborn as sns
sns.boxplot(data=e50,y='num_char')
from IPython.display import Image
Image(filename="boxplot.png")
mlb = pd.read_csv('Dataset/mlb.csv')
mlb.head()
mlb['salary'].plot.hist()
np.log(mlb['salary']).plot.hist()
e50.plot.scatter(x='num_char',y='line_breaks')
plt.title("Scatter plot of line breaks against number of characters raw")
x = np.log(e50['num_char'],dtype='float64')
y = np.log(e50['line_breaks'], dtype='float64')
plt.scatter(x,y)
plt.title("Scatter plot of line breaks against number of characters after log transform")
e50.shape
e50.head()
#calling a function to count instead of using lambda function for further use
def counting(x):
return x.count()
contingency = e50.pivot_table(index='spam',columns='number', values='num_char', aggfunc=[counting])
cont_plot = contingency.copy()
contingency
#Counting the total of all the observations
contingency.loc[:,('counting','total')] = contingency.sum(axis=1)
contingency
frequency = e50.pivot_table(columns='number', values='num_char', aggfunc=[counting])
frequency
#Counting the total of all the observations
frequency.loc[:,('counting','total')] = frequency.loc[:,('counting','big')] + frequency.loc[:,('counting','none')] + frequency.loc[:,('counting','small')]
frequency
#The Contingency Table
contingency
#It's Column proportion
contingency.loc[:,('counting')].div(contingency.loc[:,('counting','total')],axis=0)#divide all the other columns by one column
contingency.info()
# saved cont_plot on In [26]
cont_plot.T.plot.bar(stacked=True)
| 0.528777 | 0.989722 |
<img width="10%" alt="Naas" src="https://landen.imgix.net/jtci2pxwjczr/assets/5ice39g4.png?w=160"/>
# WorldBank - Most populated countries
<a href="https://app.naas.ai/user-redirect/naas/downloader?url=https://raw.githubusercontent.com/jupyter-naas/awesome-notebooks/master/WorldBank/WorldBank_Most_populated_countries.ipynb" target="_parent"><img src="https://naasai-public.s3.eu-west-3.amazonaws.com/open_in_naas.svg"/></a>
**Tags:** #worldbank #opendata #snippet #plotly #matplotlib
**Author:** [Jeremy Ravenel](https://www.linkedin.com/in/ACoAAAJHE7sB5OxuKHuzguZ9L6lfDHqw--cdnJg/)
**Notebook d'exemple pour classer les pays les plus peuplés**
**Sources:**
OECD -> Organisation for economic co-operation and Development
## Input
### Import library
```
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import requests
import io
import numpy as np
import plotly.graph_objects as go
import plotly.express as px
from pydrive.auth import GoogleAuth
from pydrive.drive import GoogleDrive
from google.colab import auth
from oauth2client.client import GoogleCredentials
from pandas import DataFrame
import plotly.graph_objects as go
```
## Model
### Lets search the file frome gdrive
```
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
downloaded = drive.CreateFile({'id':"1FjX4NTIq1z3zS9vCdAdpddtj9mKa0wIW"}) # replace the id with id of file you want to access
downloaded.GetContentFile('POP_PROJ_20042020112713800.csv')
```
### Stock the data in a variable
```
data = pd.read_csv("POP_PROJ_20042020112713800.csv", usecols=["Country", "Time", "Value"])
data.rename(columns = {'Country':'COUNTRY', 'Time':'TIME',
'Value':'VALUE'}, inplace = True)
data
```
### Fonction
```
firstOccur = []
secondOccur = []
firstYear = 2000
secondYear = 2030
def tambouille_first(number1):
first = []
for index, row in data.iterrows():
if(row["TIME"] == number1):
first.append(row)
first = DataFrame(first)
first = first.sort_values(by ="VALUE",ascending=True)
first = first.tail(10)
return first
def tambouille_second(number2):
second = []
for index, row in data.iterrows():
if(row["TIME"] == number2):
second.append(row)
second = DataFrame(second)
second =second.sort_values(by ="VALUE",ascending=True)
second = second.tail(10)
return second
firstOccur = tambouille_first(firstYear)
secondOccur = tambouille_second(secondYear)
firstOccur
```
## Output
### Display plot
```
fig = go.Figure(data=[
go.Bar(name=str(firstYear), y=firstOccur["COUNTRY"], x=firstOccur["VALUE"],orientation='h'),
go.Bar(name=str(secondYear), y=secondOccur["COUNTRY"], x=secondOccur["VALUE"],orientation='h'),
])
fig.update_layout(title_text="TOP 10 des pays les plus peuplés en 2000 avec prévision 2030", annotations=[
dict(
x=1,
y=-0.15,
showarrow=False,
text="Source : OECD -> 2019",
xref="paper",
yref="paper"
)])
fig.show()
```
**Tutorial video (in french)**
https://drive.google.com/file/d/14QhRJTWxlV6HyHmrLuSGsJ6NuFrV2GCZ/view
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import requests
import io
import numpy as np
import plotly.graph_objects as go
import plotly.express as px
from pydrive.auth import GoogleAuth
from pydrive.drive import GoogleDrive
from google.colab import auth
from oauth2client.client import GoogleCredentials
from pandas import DataFrame
import plotly.graph_objects as go
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
downloaded = drive.CreateFile({'id':"1FjX4NTIq1z3zS9vCdAdpddtj9mKa0wIW"}) # replace the id with id of file you want to access
downloaded.GetContentFile('POP_PROJ_20042020112713800.csv')
data = pd.read_csv("POP_PROJ_20042020112713800.csv", usecols=["Country", "Time", "Value"])
data.rename(columns = {'Country':'COUNTRY', 'Time':'TIME',
'Value':'VALUE'}, inplace = True)
data
firstOccur = []
secondOccur = []
firstYear = 2000
secondYear = 2030
def tambouille_first(number1):
first = []
for index, row in data.iterrows():
if(row["TIME"] == number1):
first.append(row)
first = DataFrame(first)
first = first.sort_values(by ="VALUE",ascending=True)
first = first.tail(10)
return first
def tambouille_second(number2):
second = []
for index, row in data.iterrows():
if(row["TIME"] == number2):
second.append(row)
second = DataFrame(second)
second =second.sort_values(by ="VALUE",ascending=True)
second = second.tail(10)
return second
firstOccur = tambouille_first(firstYear)
secondOccur = tambouille_second(secondYear)
firstOccur
fig = go.Figure(data=[
go.Bar(name=str(firstYear), y=firstOccur["COUNTRY"], x=firstOccur["VALUE"],orientation='h'),
go.Bar(name=str(secondYear), y=secondOccur["COUNTRY"], x=secondOccur["VALUE"],orientation='h'),
])
fig.update_layout(title_text="TOP 10 des pays les plus peuplés en 2000 avec prévision 2030", annotations=[
dict(
x=1,
y=-0.15,
showarrow=False,
text="Source : OECD -> 2019",
xref="paper",
yref="paper"
)])
fig.show()
| 0.345105 | 0.85183 |
<!--NOTEBOOK_HEADER-->
*This notebook contains material from [PyRosetta](https://RosettaCommons.github.io/PyRosetta.notebooks);
content is available [on Github](https://github.com/RosettaCommons/PyRosetta.notebooks.git).*
<!--NAVIGATION-->
< [Getting spatial features from a Pose](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/02.04-Getting-Spatial-Features-from-Pose.ipynb) | [Contents](toc.ipynb) | [Index](index.ipynb) | [Visualization with the `PyMOLMover`](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/02.06-Visualization-and-PyMOL-Mover.ipynb) ><p><a href="https://colab.research.google.com/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/02.05-Protein-Geometry.ipynb"><img align="left" src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab" title="Open in Google Colaboratory"></a>
# Protein Geometry
Keywords: pose_from_sequence(), bond_angle(), set_phi(), set_psi(), xyz()
```
# Notebook setup
import sys
if 'google.colab' in sys.modules:
!pip install pyrosettacolabsetup
import pyrosettacolabsetup
pyrosettacolabsetup.mount_pyrosetta_install()
print ("Notebook is set for PyRosetta use in Colab. Have fun!")
from pyrosetta import *
from pyrosetta.teaching import *
init()
```
**From previous section:**
Make sure you are in the directory with the pdb files:
`cd google_drive/My\ Drive/student-notebooks/`
```
pose = pose_from_pdb("inputs/5tj3.pdb")
resid = pose.pdb_info().pdb2pose('A', 28)
res_28 = pose.residue(resid)
N28 = AtomID(res_28.atom_index("N"), resid)
CA28 = AtomID(res_28.atom_index("CA"), resid)
C28 = AtomID(res_28.atom_index("C"), resid)
```
## Rosetta Database Files
Let's take a look at Rosetta's ideal values for this amino acid's bond lengths and see how these values compare. First find Pyrosetta's database directory on your computer (hint: it should have shown up when you ran `init()` at the beginning of this Jupyter notebook.) Here's an example:
```
from IPython.display import Image
Image('./Media/init-path.png',width='700')
```
Head to the subdirectory `chemical/residue_type_sets/fa_standard/` to find the residue you're looking at. Let's look at valine, which can be found in the `l-caa` folder, since it is a standard amino acid. The `ICOOR_INTERNAL` lines will provide torsion angles, bond angles, and bond lengths between subsequent atoms in this residue. From this you should be able to deduce Rosetta's ideal $N$-$C_\alpha$ and $C_\alpha$-$C$ bond lengths.
These ideal values would for instance be used if we generated a new pose from an amino acid sequence. In fact, let's try that here:
```
one_res_seq = "V"
pose_one_res = pose_from_sequence(one_res_seq)
print(pose_one_res.sequence())
N_xyz = pose_one_res.residue(1).xyz("N")
CA_xyz = pose_one_res.residue(1).xyz("CA")
C_xyz = pose_one_res.residue(1).xyz("C")
print((CA_xyz - N_xyz).norm())
print((CA_xyz - C_xyz).norm())
```
Now lets figure out how to get angles in the protein. If the `Conformation` class has the angle we're looking for, we can use the AtomID objects we've already created:
```
angle = pose.conformation().bond_angle(N28, CA28, C28)
print(angle)
```
Notice that `.bond_angle()` gives us the angle in radians. We can compute the above angle in degrees:
```
import math
angle*180/math.pi
```
Note how this compares to the expected angle based on a tetrahedral geometry for the $C_\alpha$ carbon.
### Exercise 5: Calculating psi angle
Try to calculate this angle using the xyz atom positions for N, CA, and C of residue A:28 in the protein. You can use the `Vector` function `v3 = v1.dot(v2)` along with `v1.norm()`. The vector angle between two vectors BA and BC is $\cos^{-1}(\frac{BA \cdot BC}{|BA| |BC|})$.
## Manipulating Protein Geometry
We can also alter the geometry of the protein, with particular interest in manipulating the protein backbone and $\chi$ dihedrals.
### Exercise 6: Changing phi/psi angles
Perform each of the following manipulations, and give the coordinates of the CB atom of Pose residue 2 afterward.
- Set the $\phi$ of residue 2 to -60
- Set the $\psi$ of residue 2 to -43
```
# three alanines
tripeptide = pose_from_sequence("AAA")
orig_phi = tripeptide.phi(2)
orig_psi = tripeptide.psi(2)
print("original phi:", orig_phi)
print("original psi:", orig_psi)
# print the xyz coordinates of the CB atom of residue 2 here BEFORE setting
### BEGIN SOLUTION
print("xyz coordinates:", tripeptide.residue(2).xyz("CB"))
### END SOLUTION
# set the phi and psi here
### BEGIN SOLUTION
tripeptide.set_phi(2, -60)
tripeptide.set_psi(2, -43)
print("new phi:", tripeptide.phi(2))
print("new psi:", tripeptide.psi(2))
### END SOLUTION
# print the xyz coordinates of the CB atom of residue 2 here AFTER setting
### BEGIN SOLUTION
print("xyz coordinates:", tripeptide.residue(2).xyz("CB"))
### END SOLUTION
# did changing the phi and psi angle change the xyz coordinates of the CB atom of alanine 2?
```
By printing the pose (see below command), we can see that the whole protein is in a single chain from residue 1 to 524 (or 519, depending on if the pose was cleaned).
The `FOLD_TREE` controls how changes to residue geometry propagate through the protein (left to right in the FoldTree chain.) We will go over the FoldTree in another lecture, but based on how you think perturbing the backbone of a protein structure affects the overall protein conformation, consider this question: If you changed a torsion angle for residue 5, would the Cartesian coordinaes for residue 7 change? What about the coordinates for residue 3?
Try looking at the pose in PyMOL before and after you set the backbone $\phi$ and $\psi$ for a chosen residue.
```
print(pose)
```
<!--NAVIGATION-->
< [Getting spatial features from a Pose](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/02.04-Getting-Spatial-Features-from-Pose.ipynb) | [Contents](toc.ipynb) | [Index](index.ipynb) | [Visualization with the `PyMOLMover`](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/02.06-Visualization-and-PyMOL-Mover.ipynb) ><p><a href="https://colab.research.google.com/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/02.05-Protein-Geometry.ipynb"><img align="left" src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab" title="Open in Google Colaboratory"></a>
|
github_jupyter
|
# Notebook setup
import sys
if 'google.colab' in sys.modules:
!pip install pyrosettacolabsetup
import pyrosettacolabsetup
pyrosettacolabsetup.mount_pyrosetta_install()
print ("Notebook is set for PyRosetta use in Colab. Have fun!")
from pyrosetta import *
from pyrosetta.teaching import *
init()
pose = pose_from_pdb("inputs/5tj3.pdb")
resid = pose.pdb_info().pdb2pose('A', 28)
res_28 = pose.residue(resid)
N28 = AtomID(res_28.atom_index("N"), resid)
CA28 = AtomID(res_28.atom_index("CA"), resid)
C28 = AtomID(res_28.atom_index("C"), resid)
from IPython.display import Image
Image('./Media/init-path.png',width='700')
one_res_seq = "V"
pose_one_res = pose_from_sequence(one_res_seq)
print(pose_one_res.sequence())
N_xyz = pose_one_res.residue(1).xyz("N")
CA_xyz = pose_one_res.residue(1).xyz("CA")
C_xyz = pose_one_res.residue(1).xyz("C")
print((CA_xyz - N_xyz).norm())
print((CA_xyz - C_xyz).norm())
angle = pose.conformation().bond_angle(N28, CA28, C28)
print(angle)
import math
angle*180/math.pi
# three alanines
tripeptide = pose_from_sequence("AAA")
orig_phi = tripeptide.phi(2)
orig_psi = tripeptide.psi(2)
print("original phi:", orig_phi)
print("original psi:", orig_psi)
# print the xyz coordinates of the CB atom of residue 2 here BEFORE setting
### BEGIN SOLUTION
print("xyz coordinates:", tripeptide.residue(2).xyz("CB"))
### END SOLUTION
# set the phi and psi here
### BEGIN SOLUTION
tripeptide.set_phi(2, -60)
tripeptide.set_psi(2, -43)
print("new phi:", tripeptide.phi(2))
print("new psi:", tripeptide.psi(2))
### END SOLUTION
# print the xyz coordinates of the CB atom of residue 2 here AFTER setting
### BEGIN SOLUTION
print("xyz coordinates:", tripeptide.residue(2).xyz("CB"))
### END SOLUTION
# did changing the phi and psi angle change the xyz coordinates of the CB atom of alanine 2?
print(pose)
| 0.305283 | 0.954732 |
[](https://www.pythonista.io)
# *RESTful APIs*.
## La arquitectura *RESTful* o *REST*.
*REST* es el acrónimo de "Transferencia Representacional de Estado", la cual es una propuesta de arquitectura de servcios web basada en los métodos definidos para *HTTP*. A estos servicios también se les conoce como *RESTFul*.
La arquitectura *REST* fue propuesta por primera vez en [la tesis doctoral](https://www.ics.uci.edu/~fielding/pubs/dissertation/top.htm) de Roy Fieding en el año 2000.
*REST* es una arquitectura que resulta flexible y simple en comparación con otras propuestas y en vista de que no restringe el uso de algún formato de datos ni tampoco exige el apego a esquema predefinidos, puede ser ampliamante extendida.
En este capítulo utilizaremos *JSON* como el formato por defecto para la comunicación de mensajes de datos.
## Las interfaces de programación de aplicaciones (*API*).
Una *API* permite definir las reglas y el modo en los que un usuario puede interactuar con un sistema mediante la construcción de expresiones con instrucciones y esquemas de datos específicos.
Una *API* web es aquella que permite enviar instrucciones por medio del uso de *URLs* (*end points*) y los datos que se envían a ésta.
## Objetivos de este capítulo.
Se creará una *API* que realizará operaciones de altas, bajas y cambios o *CRUD* (el acrónimo de "crear, leer, actualizar y eliminar" en inglés) en una base de datos rudimentaria.
* Cada una de las operaciones serán definidas mapeando un método *HTTP* a una función que realice las operaciones utilizando los datos enviados en la petición.
* Los *endpoints* corresponden a una *URL* compuesta por una ruta fija desde ```/api/``` añadiendo un número que correspondería al campo ```"cuenta"``` de un registro en la base de datos.
* El resto de la información será enviada en formato *JSON* con los campos obligatorios:
* ```"nombre"```
* ```"primer_apellido"```
* ```"carrera"```
* ```"semestre"```
* ```"promedio"```
* ```"al_corriente"```
* El campo ```"segundo_apellido"``` es opcional y en caso de no ser enviado será sustituido por una cadena de caracteres vacía en la base de datos.
* Los campos deben de cumplir ciertas reglas y apegarse a la estructura descrita. De lo contrario, la operación no se realizaría.
## Importación de módulos y datos.
### El paquete ```data```.
El paquete data corresponde al directorio local [```data/```](data/), el cual contiene al *script* [```data/__init__.py```](data/__init__.py) con el siguiente código.
``` python
#! /usr/bin/python3
# La ruta en la que se encuentra la base de datos.
ruta = 'data/alumnos.py'
# Define los campos que conforman la estructura de un mensaje completo.
orden = ('nombre', 'primer_apellido', 'segundo_apellido', 'carrera','semestre', 'promedio', 'al_corriente')
# Indica el tipo de dato de cada campo en un registro de la base de datos, y si este es obligatorio (True).
campos ={'cuenta': (int, True), 'nombre': (str, True), 'primer_apellido': (str, True), 'segundo_apellido': (str, False), 'carrera': (str, True), 'semestre': (int, True), 'promedio': (float, True), 'al_corriente': (bool, True)}
# Listado de las cadenas de caracteres que deben aceptarse en el campo "Carreras".
carreras = ("Sistemas", "Derecho", "Actuaría", "Arquitectura", "Administración")
```
```
from flask import Flask, jsonify, request, abort
from json import loads
from data import ruta, campos, orden, carreras
ruta
campos
orden
carreras
```
## Definición de funciones.
### Funciones de gestión de la base de datos.
En este caso la base de datos no es otra cosa más que un archivo de texto que representa a un objeto de tipo ```list``` de *Python*.
La base de datos puede ser consultada en [data/alumnos.py](data/alumnos.py).
### Función de carga de datos.
```
def carga_base(ruta):
'''Función que carga la representación de un objeto de Python
localizada en un script de Python.'''
with open(ruta, 'tr') as base:
return eval(base.read())
```
### Función de escritura de datos.
```
def escribe_base(lista, ruta):
'''Función que excribe la representación de un objeto de Python
localizada en un script de Python.'''
with open(ruta, 'wt') as base:
base.write(str(lista))
```
### Función de búsqueda en la base de datos.
* Busca dentro del campo ```'cuenta'``` de cada elemento de ```base``` al número entero correspondiente al parámetro ```cuenta```.
* En caso de encontrar una coincidencia, regresa al objeto correspondiente.
* En caso de no encontrar coincidencia regresa ```False```.
```
def busca_base(cuenta, base):
'''Función que busaca un valor dado como cuenta en una lista
de objetos tipo dict que contenga el campo "cuenta".'''
for alumno in base:
try:
if alumno['cuenta'] == int(cuenta):
return alumno
except:
return False
return False
```
## Funciones de validación de datos.
### Función que valida el tipo de dato.
```
def es_tipo(dato, tipo):
'''Función que valida el tipo de dato.'''
if tipo == str:
return True
else:
try:
return tipo(dato) is dato
except:
return False
```
### Función que valida las reglas de los datos.
* Los campos ```'nombre"``` y ```'primer_apellido'``` no deben de ser una cadena de caracteres vacía.
* El campo ```semestre``` debe de ser un entero mayor a ```1```.
* La cadena de caracteres del campo ```'carrera'``` debe de estar dentro de las cadenas listadas en ```datos.carrera```.
* El campo ```promedio``` debe de ser un número entre ```0``` y ```10```.
```
def reglas(dato, campo):
'''Función que valida las reglas de datos.'''
if campo == "carrera" and dato not in carreras:
return False
elif campo == "semestre" and dato < 1:
return False
elif campo == "promedio" and (dato < 0 or dato > 10):
return False
elif (campo in ("nombre", "primer_apellido") and (dato == "")):
return False
else:
return True
```
### Función de validación de tipos y reglas.
```
def valida(dato, campo):
'''Función que valida tipo y reglas.'''
return es_tipo(dato, campos[campo][0]) and reglas(dato, campo)
```
### Función que valida que el mensaje contiene todos los campos obligatorios.
```
def recurso_completo(base, ruta, cuenta, peticion):
'''Función que valida la estructura de datos.'''
try:
candidato = {'cuenta': int(cuenta)}
peticion = loads(peticion)
if (set(peticion)).issubset(set(orden)):
for campo in orden:
if not campos[campo][1] and campo not in peticion:
candidato[campo] = ''
elif valida(peticion[campo], campo):
candidato[campo] = peticion[campo]
else:
abort(400)
else:
abort(400)
except:
abort(400)
base.append(candidato)
escribe_base(base, ruta)
return jsonify(candidato)
```
## Código del servidor.
* El servidor correrá en http://localhost:5000/api/. Si se accede a la raíz, se desplegará un listado de todos los alumnos en formato *JSON*.
* El servidor soporta los métodos:
* **GET**: para obtener la información de un alumno por su número de cuenta.
* **POST**: para crear un registro nuevo.
* **PUT**: para sustituir por completo un registro existente.
* **PATCH**: para modificar ciertos datos de un registro existente.
* **DELETE**: para eliminar un registro existente.
```
app = Flask(__name__)
@app.route('/api/', methods=['GET'])
def index():
with open(ruta, 'tr') as base:
return jsonify(eval(base.read()))
@app.route('/api/<cuenta>', methods=['GET', 'POST', 'PUT', 'PATCH', 'DELETE'])
def api(cuenta):
if request.method == 'GET':
base = carga_base(ruta)
alumno = busca_base(cuenta, base)
if alumno:
return jsonify(alumno)
else:
abort(404)
if request.method == 'DELETE':
base = carga_base(ruta)
alumno = busca_base(cuenta, base)
if alumno:
base.remove(alumno)
escribe_base(base, ruta)
return jsonify(alumno)
else:
abort(404)
if request.method == 'POST':
base = carga_base(ruta)
alumno = busca_base(cuenta, base)
if alumno:
abort(409)
else:
return recurso_completo(base, ruta, cuenta, request.data)
if request.method == 'PUT':
base = carga_base(ruta)
alumno = busca_base(cuenta, base)
if not alumno:
abort(404)
else:
base.remove(alumno)
return recurso_completo(base, ruta, cuenta, request.data)
if request.method == 'PATCH':
base = carga_base(ruta)
alumno = busca_base(cuenta, base)
if not alumno:
abort(404)
else:
indice = base.index(alumno)
try:
peticion = loads(request.data)
if (set(peticion)).issubset(set(orden)):
for campo in peticion:
dato = peticion[campo]
if valida(dato, campo):
alumno[campo] = dato
else:
abort(400)
else:
abort(400)
except:
abort(400)
base[indice] = alumno
escribe_base(base, ruta)
return jsonify(alumno)
app.run('0.0.0.0')
```
### Notas:
* **No reinicie o detenga el kernel de la notebook hasta que los clientes que accedan a esta aplicación hayan terminado sus sesiones.**
* Debido a que el código de la celda de arriba levanta el servidor de Flask, ésta se ejecutará indefinidamente y desplegará los mensajes de respuesta a las peticiones de los clientes que se conecten.
<p style="text-align: center"><a rel="license" href="http://creativecommons.org/licenses/by/4.0/"><img alt="Licencia Creative Commons" style="border-width:0" src="https://i.creativecommons.org/l/by/4.0/80x15.png" /></a><br />Esta obra está bajo una <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Licencia Creative Commons Atribución 4.0 Internacional</a>.</p>
<p style="text-align: center">© José Luis Chiquete Valdivieso. 2021.</p>
|
github_jupyter
|
* ```"carrera"```
* ```"semestre"```
* ```"promedio"```
* ```"al_corriente"```
* El campo ```"segundo_apellido"``` es opcional y en caso de no ser enviado será sustituido por una cadena de caracteres vacía en la base de datos.
* Los campos deben de cumplir ciertas reglas y apegarse a la estructura descrita. De lo contrario, la operación no se realizaría.
## Importación de módulos y datos.
### El paquete ```data```.
El paquete data corresponde al directorio local [```data/```](data/), el cual contiene al *script* [```data/__init__.py```](data/__init__.py) con el siguiente código.
## Definición de funciones.
### Funciones de gestión de la base de datos.
En este caso la base de datos no es otra cosa más que un archivo de texto que representa a un objeto de tipo ```list``` de *Python*.
La base de datos puede ser consultada en [data/alumnos.py](data/alumnos.py).
### Función de carga de datos.
### Función de escritura de datos.
### Función de búsqueda en la base de datos.
* Busca dentro del campo ```'cuenta'``` de cada elemento de ```base``` al número entero correspondiente al parámetro ```cuenta```.
* En caso de encontrar una coincidencia, regresa al objeto correspondiente.
* En caso de no encontrar coincidencia regresa ```False```.
## Funciones de validación de datos.
### Función que valida el tipo de dato.
### Función que valida las reglas de los datos.
* Los campos ```'nombre"``` y ```'primer_apellido'``` no deben de ser una cadena de caracteres vacía.
* El campo ```semestre``` debe de ser un entero mayor a ```1```.
* La cadena de caracteres del campo ```'carrera'``` debe de estar dentro de las cadenas listadas en ```datos.carrera```.
* El campo ```promedio``` debe de ser un número entre ```0``` y ```10```.
### Función de validación de tipos y reglas.
### Función que valida que el mensaje contiene todos los campos obligatorios.
## Código del servidor.
* El servidor correrá en http://localhost:5000/api/. Si se accede a la raíz, se desplegará un listado de todos los alumnos en formato *JSON*.
* El servidor soporta los métodos:
* **GET**: para obtener la información de un alumno por su número de cuenta.
* **POST**: para crear un registro nuevo.
* **PUT**: para sustituir por completo un registro existente.
* **PATCH**: para modificar ciertos datos de un registro existente.
* **DELETE**: para eliminar un registro existente.
| 0.603231 | 0.934515 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.