code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
```
from pymatgen.core import Structure
from pymatgen.analysis.magnetism.analyzer import MagneticStructureEnumerator, CollinearMagneticStructureAnalyzer
from pymatgen.io.vasp.inputs import Poscar, Incar, Kpoints
from pymatgen.io.vasp.sets import MPRelaxSet, MPStaticSet, MPSOCSet
from shutil import copy
import os
from pymatgen.io.vasp.outputs import Vasprun
import numpy as np
import warnings
from time import sleep
warnings.filterwarnings('ignore')
def create_job_script(out_path, job_id=None):
"""
Args:
out_path (str) - folder where job script will be created.
job_id (str) - preferable name of your job in squeue,
and also the name of the folder for your vasp files.
As a result in the folder with name 'job_id' will be created job script
"""
if not job_id:
job_id = os.path.basename(out_path)
job_script_text = f"""#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks=8
#SBATCH --time=06:00:00
#SBATCH --job-name={job_id}
#SBATCH --output=log
#SBATCH --error=err
#SBATCH -p lenovo
module load mpi/impi-5.0.3 intel/mkl-11.2.3 vasp/vasp-5.4.4
mpirun vasp_std"""
if not os.path.exists(out_path):
os.mkdir(out_path)
with open(f'{out_path}/jobscript.sh', 'w') as job:
job.writelines(job_script_text)
def afm_atom_creator(in_data: list, custom_atom='Po') -> list:
"""
Args:
in_data (list) - list of rows from POSCAR type file.
Add one type of "Fake" atom into the POSCAR structure.
This allows it to be treated as an atoms with spin up and down respectively,
thus estimate number of positive and negative contributions into the total energy.
"""
out_data = in_data.copy()
out_data[5] = 'Po ' + out_data[5]
return out_data
def up_down_spin_counter(in_data: list) -> list:
spin_down = 0
spin_up = 0
no_spin = 0
for row in in_data[8:]:
if 'spin=-' in row:
spin_down += 1
elif 'spin=' in row:
spin_up += 1
else:
no_spin += 1
return [spin_up, spin_down, no_spin]
def spin_row_replacer(in_data: list) -> list:
out_data = in_data.copy()
out_data[6] = ' '.join(str(i) for i in up_down_spin_counter(in_data)) + '\n'
return out_data
def siman_POSCAR_writer(in_path: str, out_path: str) -> None:
"""
Args:
in_path (str) - path to the POSCAR type file which needs to be made
readable for siman
out_path (str) - path where refactored version of this file will be
written
"""
with open(in_path) as in_f:
in_data = in_f.readlines()
out_data = spin_row_replacer(afm_atom_creator(in_data))
with open(out_path, 'w+') as out_f:
out_f.writelines(out_data)
def get_siman_inputs(input_path: str):
out_path = os.path.join(input_path, 'siman_inputs')
if not os.path.exists(out_path):
os.mkdir(out_path)
vasp_inputs_path = os.path.join(input_path, 'vasp_inputs')
afm_foldrs = [os.path.join(vasp_inputs_path, i)
for i in [i for i in os.listdir(vasp_inputs_path) if 'afm' in i]]
for folder in afm_foldrs:
tmp_out_path = os.path.join(out_path, 'POSCAR_' + folder.split("/")[-1])
siman_POSCAR_writer(in_path=os.path.join(folder, 'POSCAR'), out_path=tmp_out_path)
def submit_all_jobs(input_folder: str) -> None:
vasp_inputs_path = os.path.join(input_folder, 'vasp_inputs')
initial_path = os.getcwd()
for folder_name in os.listdir(vasp_inputs_path):
os.chdir(initial_path)
tmp_path = os.path.join(vasp_inputs_path, folder_name)
os.chdir(tmp_path)
os.system('sbatch jobscript.sh')
os.chdir(initial_path)
LDAUJ_dict = {'Co': 0, 'Cr': 0, 'Fe': 0, 'Mn': 0, 'Mo': 0, 'Ni': 0, 'V': 0, 'W': 0,
'Nb': 0, 'Sc': 0, 'Ru': 0, 'Rh': 0, 'Pd': 0, 'Cu': 0, 'Y': 0, 'Os': 0, 'Ti': 0, 'Zr': 0, 'Re': 0, 'Hf': 0, 'Pt': 0, 'La': 0}
LDAUU_dict = {'Co': 3.32, 'Cr': 3.7, 'Fe': 5.3, 'Mn': 3.9, 'Mo': 4.38, 'Ni': 6.2, 'V': 3.25, 'W': 6.2,
'Nb': 1.45, 'Sc': 4.18, 'Ru': 4.29, 'Rh': 4.17, 'Pd': 2.96, 'Cu': 7.71, 'Y': 3.23, 'Os': 2.47, 'Ti': 5.89, 'Zr': 5.55,
'Re': 1.28, 'Hf': 4.77, 'Pt': 2.95, 'La': 5.3}
LDAUL_dict = {'Co': 2, 'Cr': 2, 'Fe': 2, 'Mn': 2, 'Mo': 2, 'Ni': 2, 'V': 2, 'W': 2,
'Nb': 2, 'Sc': 2, 'Ru': 2, 'Rh': 2, 'Pd': 2, 'Cu': 2, 'Y': 2, 'Os': 2, 'Ti': 2, 'Zr': 2, 'Re': 2, 'Hf': 2, 'Pt': 2, 'La': 2}
relx_dict = {'ISMEAR': 0, 'SIGMA': 0.01, 'ISIF': 4, 'EDIFF': 1E-4, 'POTIM': 0.3,
'EDIFFG': -0.01, 'SYMPREC': 1E-8, 'NCORE': 4, 'LCHARG': False, 'ICHARG': 2,
'LDAU': True, 'LDAUJ': LDAUJ_dict, 'LDAUL': LDAUL_dict, 'LDAUU': LDAUU_dict, 'LWAVE': False,
'LDAUPRINT': 1, 'LDAUTYPE': 2, 'LASPH': True, 'LMAXMIX': 4}
stat_dict = {'ISMEAR': -5, 'EDIFF': 1E-6, 'SYMPREC': 1E-8, 'NCORE': 4, 'ICHARG': 2,
'LDAU': True, 'LDAUJ': LDAUJ_dict, 'LDAUL': LDAUL_dict, 'LDAUU': LDAUU_dict, 'NELM': 120, 'LVHAR': False,
'LDAUPRINT': 1, 'LDAUTYPE': 2, 'LASPH': True, 'LMAXMIX': 4, 'LWAVE': False, 'LVTOT': False}
def write_static_set(structure, vasp_static_path: str, static_dict: dict) -> None:
"""
Args:
structure (pymatgen.core.structure.Structure)
vasp_static_path (str) - path to the folder for static VASP run
static_dict (dict) - dictionary with VASP INCAR keywords
Write the following files into specified folder:
INCAR_stat
jobscript.sh
"""
if not os.path.exists(vasp_static_path):
os.mkdir(vasp_static_path)
static_set = MPStaticSet(structure,
user_incar_settings=stat_dict,
reciprocal_density=300,
force_gamma=True)
static_set.incar.write_file(os.path.join(vasp_static_path, 'INCAR_stat'))
create_job_script(vasp_static_path)
def write_relax_set(structure, vasp_relax_path: str, relax_dict: dict) -> None:
"""
Args:
structure (pymatgen.core.structure.Structure)
vasp_static_path (str) - path to the folder for static VASP run
static_dict (dict) - dictionary with VASP INCAR keywords
Write the following files into specified folder:
INCAR
POSCAR
POTCAR
KPOINTS
jobscript.sh
"""
if not os.path.exists(vasp_relax_path):
os.mkdir(vasp_relax_path)
relax_set = MPRelaxSet(structure=structure,
user_incar_settings=relx_dict,
user_kpoints_settings={'reciprocal_density': 300},
force_gamma=True)
relax_set.get_vasp_input().write_input(vasp_relax_path)
create_job_script(vasp_relax_path)
def get_VASP_inputs(input_path: str, relx_dict: dict, static_dict: dict) -> None:
init_structure = Structure.from_file(os.path.join(input_path, 'POSCAR'))
enum_struct_list = MagneticStructureEnumerator(init_structure,
transformation_kwargs={'symm_prec': 0.1,
'enum_precision_parameter': 0.00001},
strategies=('ferromagnetic', 'antiferromagnetic'))
if not os.path.exists(os.path.join(input_path, 'vasp_inputs')):
os.mkdir(os.path.join(input_path, 'vasp_inputs'))
for i, magnetic_structure in enumerate(enum_struct_list.ordered_structures):
magnetic_type = enum_struct_list.ordered_structure_origins[i]
str_id = magnetic_type + str(i)
vasp_out_path = os.path.join(input_path, 'vasp_inputs', str_id)
if not os.path.exists(vasp_out_path):
os.mkdir(vasp_out_path)
write_relax_set(structure=magnetic_structure,
vasp_relax_path=vasp_out_path,
relax_dict=relx_dict)
write_static_set(structure=magnetic_structure,
vasp_static_path=vasp_out_path,
static_dict=stat_dict)
def static_changer(vasprun_path: str):
"""
1. Replace INCAR relax for INCAR_stat
2. Replace POSCAR with relaxed CONTCAR
Namely prepare the folder for more accurate run of VASP
for total energy estimation.
"""
base_path = '/'.join(vasprun_path.split('/')[:-1])
inc_path = os.path.join(base_path, 'INCAR')
inc_stat_path = os.path.join(base_path, 'INCAR_stat')
inc_relax_path = os.path.join(base_path, 'INCAR_relax')
contcar_path = os.path.join(base_path, 'CONTCAR')
poscar_path = os.path.join(base_path, 'POSCAR')
log_relax = os.path.join(base_path, 'log_relax')
log = os.path.join(base_path, 'log')
out_relax = os.path.join(base_path, 'OUTCAR_relax')
out = os.path.join(base_path, 'OUTCAR')
copy(inc_path, inc_relax_path) # INCAR -> INCAR_relax
copy(inc_stat_path, inc_path) # INCAR_stat -> INCAR
copy(contcar_path, poscar_path) # CONTCAR -> POSCAR
copy(log, log_relax) # log -> log_relax
copy(out, out_relax) # OUTCAR -> OUTCAR_relax
def vasprun_checker(input_path):
vasp_inputs_path = os.path.join(input_path, 'vasp_inputs')
vasprun_pathes = sorted([os.path.join(vasp_inputs_path, i, 'vasprun.xml')
for i in os.listdir(vasp_inputs_path)])
tmp_vasprun = vasprun_pathes.copy()
while 1:
print(len(vasprun_pathes))
for i, vasprun_path in enumerate(vasprun_pathes):
print(i + 1, end=' ')
if os.path.exists(vasprun_path):
try:
vasprun = Vasprun(vasprun_path, parse_dos=False,
parse_eigen=False, exception_on_bad_xml=False)
if vasprun.converged and vasprun.converged_ionic and vasprun.converged_electronic:
print(f'Converged! {vasprun_path}')
tmp_vasprun.remove(vasprun_path)
else:
print(f'Not converged! {vasprun_path}')
tmp_vasprun.remove(vasprun_path)
except Exception:
print('Still running')
else:
print(f'{vasprun_path} not written yet!')
vasprun_pathes = tmp_vasprun.copy()
print('\n')
sleep(20)
if not vasprun_pathes:
print('All done!')
break
def file_builder(input_path: str):
assert os.path.exists(input_path), f'Input path: {input_path} does not exist!'
assert os.path.exists(os.path.join(input_path, 'POSCAR')
), f'Please specify POSCAR file in you input folder: {input_path}'
get_VASP_inputs(input_path=input_path,
relx_dict=relx_dict,
static_dict=stat_dict)
get_siman_inputs(input_path)
copy(os.path.join(input_path, 'POSCAR'), os.path.join(
input_path, 'siman_inputs', 'POSCAR_fm0'))
# submit_all_jobs(input_path)
# vasprun_checker(input_path)
in_path = '../data/test/'
all_pathes= [os.path.join(in_path, i) for i in os.listdir(in_path)]
bad_structutres = []
for i in all_pathes:
try:
file_builder(i)
except:
bad_structutres += [i]
```
|
github_jupyter
|
from pymatgen.core import Structure
from pymatgen.analysis.magnetism.analyzer import MagneticStructureEnumerator, CollinearMagneticStructureAnalyzer
from pymatgen.io.vasp.inputs import Poscar, Incar, Kpoints
from pymatgen.io.vasp.sets import MPRelaxSet, MPStaticSet, MPSOCSet
from shutil import copy
import os
from pymatgen.io.vasp.outputs import Vasprun
import numpy as np
import warnings
from time import sleep
warnings.filterwarnings('ignore')
def create_job_script(out_path, job_id=None):
"""
Args:
out_path (str) - folder where job script will be created.
job_id (str) - preferable name of your job in squeue,
and also the name of the folder for your vasp files.
As a result in the folder with name 'job_id' will be created job script
"""
if not job_id:
job_id = os.path.basename(out_path)
job_script_text = f"""#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks=8
#SBATCH --time=06:00:00
#SBATCH --job-name={job_id}
#SBATCH --output=log
#SBATCH --error=err
#SBATCH -p lenovo
module load mpi/impi-5.0.3 intel/mkl-11.2.3 vasp/vasp-5.4.4
mpirun vasp_std"""
if not os.path.exists(out_path):
os.mkdir(out_path)
with open(f'{out_path}/jobscript.sh', 'w') as job:
job.writelines(job_script_text)
def afm_atom_creator(in_data: list, custom_atom='Po') -> list:
"""
Args:
in_data (list) - list of rows from POSCAR type file.
Add one type of "Fake" atom into the POSCAR structure.
This allows it to be treated as an atoms with spin up and down respectively,
thus estimate number of positive and negative contributions into the total energy.
"""
out_data = in_data.copy()
out_data[5] = 'Po ' + out_data[5]
return out_data
def up_down_spin_counter(in_data: list) -> list:
spin_down = 0
spin_up = 0
no_spin = 0
for row in in_data[8:]:
if 'spin=-' in row:
spin_down += 1
elif 'spin=' in row:
spin_up += 1
else:
no_spin += 1
return [spin_up, spin_down, no_spin]
def spin_row_replacer(in_data: list) -> list:
out_data = in_data.copy()
out_data[6] = ' '.join(str(i) for i in up_down_spin_counter(in_data)) + '\n'
return out_data
def siman_POSCAR_writer(in_path: str, out_path: str) -> None:
"""
Args:
in_path (str) - path to the POSCAR type file which needs to be made
readable for siman
out_path (str) - path where refactored version of this file will be
written
"""
with open(in_path) as in_f:
in_data = in_f.readlines()
out_data = spin_row_replacer(afm_atom_creator(in_data))
with open(out_path, 'w+') as out_f:
out_f.writelines(out_data)
def get_siman_inputs(input_path: str):
out_path = os.path.join(input_path, 'siman_inputs')
if not os.path.exists(out_path):
os.mkdir(out_path)
vasp_inputs_path = os.path.join(input_path, 'vasp_inputs')
afm_foldrs = [os.path.join(vasp_inputs_path, i)
for i in [i for i in os.listdir(vasp_inputs_path) if 'afm' in i]]
for folder in afm_foldrs:
tmp_out_path = os.path.join(out_path, 'POSCAR_' + folder.split("/")[-1])
siman_POSCAR_writer(in_path=os.path.join(folder, 'POSCAR'), out_path=tmp_out_path)
def submit_all_jobs(input_folder: str) -> None:
vasp_inputs_path = os.path.join(input_folder, 'vasp_inputs')
initial_path = os.getcwd()
for folder_name in os.listdir(vasp_inputs_path):
os.chdir(initial_path)
tmp_path = os.path.join(vasp_inputs_path, folder_name)
os.chdir(tmp_path)
os.system('sbatch jobscript.sh')
os.chdir(initial_path)
LDAUJ_dict = {'Co': 0, 'Cr': 0, 'Fe': 0, 'Mn': 0, 'Mo': 0, 'Ni': 0, 'V': 0, 'W': 0,
'Nb': 0, 'Sc': 0, 'Ru': 0, 'Rh': 0, 'Pd': 0, 'Cu': 0, 'Y': 0, 'Os': 0, 'Ti': 0, 'Zr': 0, 'Re': 0, 'Hf': 0, 'Pt': 0, 'La': 0}
LDAUU_dict = {'Co': 3.32, 'Cr': 3.7, 'Fe': 5.3, 'Mn': 3.9, 'Mo': 4.38, 'Ni': 6.2, 'V': 3.25, 'W': 6.2,
'Nb': 1.45, 'Sc': 4.18, 'Ru': 4.29, 'Rh': 4.17, 'Pd': 2.96, 'Cu': 7.71, 'Y': 3.23, 'Os': 2.47, 'Ti': 5.89, 'Zr': 5.55,
'Re': 1.28, 'Hf': 4.77, 'Pt': 2.95, 'La': 5.3}
LDAUL_dict = {'Co': 2, 'Cr': 2, 'Fe': 2, 'Mn': 2, 'Mo': 2, 'Ni': 2, 'V': 2, 'W': 2,
'Nb': 2, 'Sc': 2, 'Ru': 2, 'Rh': 2, 'Pd': 2, 'Cu': 2, 'Y': 2, 'Os': 2, 'Ti': 2, 'Zr': 2, 'Re': 2, 'Hf': 2, 'Pt': 2, 'La': 2}
relx_dict = {'ISMEAR': 0, 'SIGMA': 0.01, 'ISIF': 4, 'EDIFF': 1E-4, 'POTIM': 0.3,
'EDIFFG': -0.01, 'SYMPREC': 1E-8, 'NCORE': 4, 'LCHARG': False, 'ICHARG': 2,
'LDAU': True, 'LDAUJ': LDAUJ_dict, 'LDAUL': LDAUL_dict, 'LDAUU': LDAUU_dict, 'LWAVE': False,
'LDAUPRINT': 1, 'LDAUTYPE': 2, 'LASPH': True, 'LMAXMIX': 4}
stat_dict = {'ISMEAR': -5, 'EDIFF': 1E-6, 'SYMPREC': 1E-8, 'NCORE': 4, 'ICHARG': 2,
'LDAU': True, 'LDAUJ': LDAUJ_dict, 'LDAUL': LDAUL_dict, 'LDAUU': LDAUU_dict, 'NELM': 120, 'LVHAR': False,
'LDAUPRINT': 1, 'LDAUTYPE': 2, 'LASPH': True, 'LMAXMIX': 4, 'LWAVE': False, 'LVTOT': False}
def write_static_set(structure, vasp_static_path: str, static_dict: dict) -> None:
"""
Args:
structure (pymatgen.core.structure.Structure)
vasp_static_path (str) - path to the folder for static VASP run
static_dict (dict) - dictionary with VASP INCAR keywords
Write the following files into specified folder:
INCAR_stat
jobscript.sh
"""
if not os.path.exists(vasp_static_path):
os.mkdir(vasp_static_path)
static_set = MPStaticSet(structure,
user_incar_settings=stat_dict,
reciprocal_density=300,
force_gamma=True)
static_set.incar.write_file(os.path.join(vasp_static_path, 'INCAR_stat'))
create_job_script(vasp_static_path)
def write_relax_set(structure, vasp_relax_path: str, relax_dict: dict) -> None:
"""
Args:
structure (pymatgen.core.structure.Structure)
vasp_static_path (str) - path to the folder for static VASP run
static_dict (dict) - dictionary with VASP INCAR keywords
Write the following files into specified folder:
INCAR
POSCAR
POTCAR
KPOINTS
jobscript.sh
"""
if not os.path.exists(vasp_relax_path):
os.mkdir(vasp_relax_path)
relax_set = MPRelaxSet(structure=structure,
user_incar_settings=relx_dict,
user_kpoints_settings={'reciprocal_density': 300},
force_gamma=True)
relax_set.get_vasp_input().write_input(vasp_relax_path)
create_job_script(vasp_relax_path)
def get_VASP_inputs(input_path: str, relx_dict: dict, static_dict: dict) -> None:
init_structure = Structure.from_file(os.path.join(input_path, 'POSCAR'))
enum_struct_list = MagneticStructureEnumerator(init_structure,
transformation_kwargs={'symm_prec': 0.1,
'enum_precision_parameter': 0.00001},
strategies=('ferromagnetic', 'antiferromagnetic'))
if not os.path.exists(os.path.join(input_path, 'vasp_inputs')):
os.mkdir(os.path.join(input_path, 'vasp_inputs'))
for i, magnetic_structure in enumerate(enum_struct_list.ordered_structures):
magnetic_type = enum_struct_list.ordered_structure_origins[i]
str_id = magnetic_type + str(i)
vasp_out_path = os.path.join(input_path, 'vasp_inputs', str_id)
if not os.path.exists(vasp_out_path):
os.mkdir(vasp_out_path)
write_relax_set(structure=magnetic_structure,
vasp_relax_path=vasp_out_path,
relax_dict=relx_dict)
write_static_set(structure=magnetic_structure,
vasp_static_path=vasp_out_path,
static_dict=stat_dict)
def static_changer(vasprun_path: str):
"""
1. Replace INCAR relax for INCAR_stat
2. Replace POSCAR with relaxed CONTCAR
Namely prepare the folder for more accurate run of VASP
for total energy estimation.
"""
base_path = '/'.join(vasprun_path.split('/')[:-1])
inc_path = os.path.join(base_path, 'INCAR')
inc_stat_path = os.path.join(base_path, 'INCAR_stat')
inc_relax_path = os.path.join(base_path, 'INCAR_relax')
contcar_path = os.path.join(base_path, 'CONTCAR')
poscar_path = os.path.join(base_path, 'POSCAR')
log_relax = os.path.join(base_path, 'log_relax')
log = os.path.join(base_path, 'log')
out_relax = os.path.join(base_path, 'OUTCAR_relax')
out = os.path.join(base_path, 'OUTCAR')
copy(inc_path, inc_relax_path) # INCAR -> INCAR_relax
copy(inc_stat_path, inc_path) # INCAR_stat -> INCAR
copy(contcar_path, poscar_path) # CONTCAR -> POSCAR
copy(log, log_relax) # log -> log_relax
copy(out, out_relax) # OUTCAR -> OUTCAR_relax
def vasprun_checker(input_path):
vasp_inputs_path = os.path.join(input_path, 'vasp_inputs')
vasprun_pathes = sorted([os.path.join(vasp_inputs_path, i, 'vasprun.xml')
for i in os.listdir(vasp_inputs_path)])
tmp_vasprun = vasprun_pathes.copy()
while 1:
print(len(vasprun_pathes))
for i, vasprun_path in enumerate(vasprun_pathes):
print(i + 1, end=' ')
if os.path.exists(vasprun_path):
try:
vasprun = Vasprun(vasprun_path, parse_dos=False,
parse_eigen=False, exception_on_bad_xml=False)
if vasprun.converged and vasprun.converged_ionic and vasprun.converged_electronic:
print(f'Converged! {vasprun_path}')
tmp_vasprun.remove(vasprun_path)
else:
print(f'Not converged! {vasprun_path}')
tmp_vasprun.remove(vasprun_path)
except Exception:
print('Still running')
else:
print(f'{vasprun_path} not written yet!')
vasprun_pathes = tmp_vasprun.copy()
print('\n')
sleep(20)
if not vasprun_pathes:
print('All done!')
break
def file_builder(input_path: str):
assert os.path.exists(input_path), f'Input path: {input_path} does not exist!'
assert os.path.exists(os.path.join(input_path, 'POSCAR')
), f'Please specify POSCAR file in you input folder: {input_path}'
get_VASP_inputs(input_path=input_path,
relx_dict=relx_dict,
static_dict=stat_dict)
get_siman_inputs(input_path)
copy(os.path.join(input_path, 'POSCAR'), os.path.join(
input_path, 'siman_inputs', 'POSCAR_fm0'))
# submit_all_jobs(input_path)
# vasprun_checker(input_path)
in_path = '../data/test/'
all_pathes= [os.path.join(in_path, i) for i in os.listdir(in_path)]
bad_structutres = []
for i in all_pathes:
try:
file_builder(i)
except:
bad_structutres += [i]
| 0.466846 | 0.308099 |
## Read File containing times
```
import re
def convert_time(str_time):
# Expect time with format 'Minutes:seconds'
m, s = str_time.split(':')
return float(m)*60+float(s)
def read_log_file(log_file):
with open(log_file) as time_file:
line = time_file.readline()
list_logged_info = ["Command being timed", "User time (seconds)", "System time (seconds)", "Percent of CPU this job got", "Elapsed (wall clock) time (h:mm:ss or m:ss)", "Average shared text size (kbytes)", "Average unshared data size (kbytes)", "Average stack size (kbytes)", "Average total size (kbytes)", "Maximum resident set size (kbytes)", "Average resident set size (kbytes)", "Major (requiring I/O) page faults", "Minor (reclaiming a frame) page faults", "Voluntary context switches", "Involuntary context switches", "Swaps", "File system inputs", "File system outputs", "Socket messages sent", "Socket messages received", "Signals delivered", "Page size (bytes)", "Exit status"]
all_targets = []
number_of_lines = 0
nb_info = len(list_logged_info)
previous_info = nb_info - 1 # Initialize with previous info being the last element of list_logged_info
skipped_lines = []
while line:
line = line.strip()
#print line
try:
key, info = line.split(": ")
# Did we find the expected info line (i.e. are we on the next list_logged_info line)
expected_key = list_logged_info[(previous_info+1)%nb_info]
if key != expected_key:
raise Exception('Found key: "%s". Expecting: "%s". Previous key: "%s\nCommand line: %s\nline #: %d"'
%(key, expected_key, list_logged_info[previous_info], line, number_of_lines))
previous_info = (previous_info + 1)%nb_info
# Save info
if key == 'Command being timed':
cmd = info
elif key == 'Percent of CPU this job got':
percent_cpu = float(info.split('%')[0])
elif key == 'Elapsed (wall clock) time (h:mm:ss or m:ss)':
# Safe check
if cmd is None:
raise Exception('cmd is None. Line: %s' %line)
if percent_cpu is None:
raise Exception('percent_cpu is None. Line: %s' %line)
all_targets.append((cmd, convert_time(info)*percent_cpu/100.0))
# Reset `cmd` and `percent_cpu` just in case
cmd = None
percent_cpu = None
# Move to next line
line = time_file.readline()
number_of_lines += 1
except ValueError as e:
print "Skipping a line that has multiple ':' character"
skipped_lines.append(line)
# Find next command that corresponds to first key
found_first_key = False
while not found_first_key:
line = time_file.readline()
number_of_lines += 1
line = line.strip()
split_line = line.split(": ")
if split_line[0] == list_logged_info[0]:
found_first_key = True
print("Number of lines: %d" % number_of_lines)
print("Number of skipped lines: %s" % len(skipped_lines))
print("Number of targets: %s" % len(all_targets))
return all_targets, skipped_lines
def TargetTime(all_targets,total_make_time, name, pattern, debug=False):
sum_time = 0.0
nb_targets = 0
for cmd, time in all_targets[:-2]: # Skip last target that is total time
if pattern.match(cmd):
sum_time += time
nb_targets +=1
elif debug:
print cmd
print("Sum time %s targets (%d): %f (%f%%)" % (name, nb_targets, sum_time, sum_time/total_make_time*100.0))
def AnalyzeCompilationTimes(log_file_name):
print("File name: %s" % log_file_name)
all_targets, skipped_lines = read_log_file(log_file_name)
total_make_time = all_targets[-1][1]
print("Total time (%s): %f" % (all_targets[-1][0], total_make_time))
sum_time = 0.0
for cmd, time in all_targets[:-2]: # Skip last target that is total time
sum_time += time
print("Sum time all targets: %f" % sum_time)
# Find list commands used:
set_commands = set()
for cmd, time in all_targets[:-2]:
cmd_split = cmd.split(" ")
set_commands.add(cmd_split[0])
print("All commands: %s" % str(set_commands))
TargetTime(all_targets, total_make_time, 'swig', re.compile('.*swig.*bin.*swig.*python.*'))
TargetTime(all_targets, total_make_time,'igenerator', re.compile('.*igenerator.py.*'))
TargetTime(all_targets, total_make_time,'C++ Tests', re.compile('.*test.*Test\.cxx.*'))
TargetTime(all_targets, total_make_time,'Link static libs', re.compile('.*/usr/bin/ar.*'))
TargetTime(all_targets, total_make_time,'Compilation', re.compile('.*/usr/bin/c.*'))
TargetTime(all_targets, total_make_time,'Compilation C', re.compile('.*/usr/bin/cc.*'))
TargetTime(all_targets, total_make_time,'Compilation C++', re.compile('.*/usr/bin/c\+\+.*'))
TargetTime(all_targets, total_make_time,'CMake', re.compile('.*/usr/local/bin/cmake.*'))
TargetTime(all_targets, total_make_time,'Ranlib', re.compile('.*/usr/bin/ranlib.*'))
TargetTime(all_targets, total_make_time,'castxml', re.compile('.*castxml/bin/castxml.*'))
TargetTime(all_targets, total_make_time,'make', re.compile('.*make.*'))
TargetTime(all_targets, total_make_time,'env', re.compile('.*env.*'))
TargetTime(all_targets, total_make_time,'H5make_libsettings', re.compile('.*H5make_libsettings.*'))
TargetTime(all_targets, total_make_time,'python', re.compile('.*bin/python.*'))
TargetTime(all_targets, total_make_time,'H5detect', re.compile('.*H5detect.*'))
TargetTime(all_targets, total_make_time,'itkmkg3states', re.compile('.*itkmkg3states.*'))
TargetTime(all_targets, total_make_time,'Wrapping', re.compile('.*Wrapping.*'))
TargetTime(all_targets, total_make_time,'HeaderTest', re.compile('.*HeaderTest.*'))
AnalyzeCompilationTimes("log_time_verbose_2digits")
# This was run with `CTest` so total time includes configuration and testing time on top of compilation time.
AnalyzeCompilationTimes("log_time_testdrivers_not_compiled")
print('Speed up disabling TestDrivers compilation: %f' %(41875.057500-30539.744700) )
```
|
github_jupyter
|
import re
def convert_time(str_time):
# Expect time with format 'Minutes:seconds'
m, s = str_time.split(':')
return float(m)*60+float(s)
def read_log_file(log_file):
with open(log_file) as time_file:
line = time_file.readline()
list_logged_info = ["Command being timed", "User time (seconds)", "System time (seconds)", "Percent of CPU this job got", "Elapsed (wall clock) time (h:mm:ss or m:ss)", "Average shared text size (kbytes)", "Average unshared data size (kbytes)", "Average stack size (kbytes)", "Average total size (kbytes)", "Maximum resident set size (kbytes)", "Average resident set size (kbytes)", "Major (requiring I/O) page faults", "Minor (reclaiming a frame) page faults", "Voluntary context switches", "Involuntary context switches", "Swaps", "File system inputs", "File system outputs", "Socket messages sent", "Socket messages received", "Signals delivered", "Page size (bytes)", "Exit status"]
all_targets = []
number_of_lines = 0
nb_info = len(list_logged_info)
previous_info = nb_info - 1 # Initialize with previous info being the last element of list_logged_info
skipped_lines = []
while line:
line = line.strip()
#print line
try:
key, info = line.split(": ")
# Did we find the expected info line (i.e. are we on the next list_logged_info line)
expected_key = list_logged_info[(previous_info+1)%nb_info]
if key != expected_key:
raise Exception('Found key: "%s". Expecting: "%s". Previous key: "%s\nCommand line: %s\nline #: %d"'
%(key, expected_key, list_logged_info[previous_info], line, number_of_lines))
previous_info = (previous_info + 1)%nb_info
# Save info
if key == 'Command being timed':
cmd = info
elif key == 'Percent of CPU this job got':
percent_cpu = float(info.split('%')[0])
elif key == 'Elapsed (wall clock) time (h:mm:ss or m:ss)':
# Safe check
if cmd is None:
raise Exception('cmd is None. Line: %s' %line)
if percent_cpu is None:
raise Exception('percent_cpu is None. Line: %s' %line)
all_targets.append((cmd, convert_time(info)*percent_cpu/100.0))
# Reset `cmd` and `percent_cpu` just in case
cmd = None
percent_cpu = None
# Move to next line
line = time_file.readline()
number_of_lines += 1
except ValueError as e:
print "Skipping a line that has multiple ':' character"
skipped_lines.append(line)
# Find next command that corresponds to first key
found_first_key = False
while not found_first_key:
line = time_file.readline()
number_of_lines += 1
line = line.strip()
split_line = line.split(": ")
if split_line[0] == list_logged_info[0]:
found_first_key = True
print("Number of lines: %d" % number_of_lines)
print("Number of skipped lines: %s" % len(skipped_lines))
print("Number of targets: %s" % len(all_targets))
return all_targets, skipped_lines
def TargetTime(all_targets,total_make_time, name, pattern, debug=False):
sum_time = 0.0
nb_targets = 0
for cmd, time in all_targets[:-2]: # Skip last target that is total time
if pattern.match(cmd):
sum_time += time
nb_targets +=1
elif debug:
print cmd
print("Sum time %s targets (%d): %f (%f%%)" % (name, nb_targets, sum_time, sum_time/total_make_time*100.0))
def AnalyzeCompilationTimes(log_file_name):
print("File name: %s" % log_file_name)
all_targets, skipped_lines = read_log_file(log_file_name)
total_make_time = all_targets[-1][1]
print("Total time (%s): %f" % (all_targets[-1][0], total_make_time))
sum_time = 0.0
for cmd, time in all_targets[:-2]: # Skip last target that is total time
sum_time += time
print("Sum time all targets: %f" % sum_time)
# Find list commands used:
set_commands = set()
for cmd, time in all_targets[:-2]:
cmd_split = cmd.split(" ")
set_commands.add(cmd_split[0])
print("All commands: %s" % str(set_commands))
TargetTime(all_targets, total_make_time, 'swig', re.compile('.*swig.*bin.*swig.*python.*'))
TargetTime(all_targets, total_make_time,'igenerator', re.compile('.*igenerator.py.*'))
TargetTime(all_targets, total_make_time,'C++ Tests', re.compile('.*test.*Test\.cxx.*'))
TargetTime(all_targets, total_make_time,'Link static libs', re.compile('.*/usr/bin/ar.*'))
TargetTime(all_targets, total_make_time,'Compilation', re.compile('.*/usr/bin/c.*'))
TargetTime(all_targets, total_make_time,'Compilation C', re.compile('.*/usr/bin/cc.*'))
TargetTime(all_targets, total_make_time,'Compilation C++', re.compile('.*/usr/bin/c\+\+.*'))
TargetTime(all_targets, total_make_time,'CMake', re.compile('.*/usr/local/bin/cmake.*'))
TargetTime(all_targets, total_make_time,'Ranlib', re.compile('.*/usr/bin/ranlib.*'))
TargetTime(all_targets, total_make_time,'castxml', re.compile('.*castxml/bin/castxml.*'))
TargetTime(all_targets, total_make_time,'make', re.compile('.*make.*'))
TargetTime(all_targets, total_make_time,'env', re.compile('.*env.*'))
TargetTime(all_targets, total_make_time,'H5make_libsettings', re.compile('.*H5make_libsettings.*'))
TargetTime(all_targets, total_make_time,'python', re.compile('.*bin/python.*'))
TargetTime(all_targets, total_make_time,'H5detect', re.compile('.*H5detect.*'))
TargetTime(all_targets, total_make_time,'itkmkg3states', re.compile('.*itkmkg3states.*'))
TargetTime(all_targets, total_make_time,'Wrapping', re.compile('.*Wrapping.*'))
TargetTime(all_targets, total_make_time,'HeaderTest', re.compile('.*HeaderTest.*'))
AnalyzeCompilationTimes("log_time_verbose_2digits")
# This was run with `CTest` so total time includes configuration and testing time on top of compilation time.
AnalyzeCompilationTimes("log_time_testdrivers_not_compiled")
print('Speed up disabling TestDrivers compilation: %f' %(41875.057500-30539.744700) )
| 0.390825 | 0.469703 |
```
import numpy as np
import matplotlib.pylab as plt
import numpy.random as rd
from sklearn.datasets import fetch_openml # MNIST data
from sklearn.model_selection import train_test_split
#os.environ["CUDA_VISIBLE_DEVICES"] = '0'
#sess = tf.Session(config=tf.ConfigProto(device_count={'GPU': 0}))
from keras.models import Sequential
from keras.models import Model
from keras.layers import Dense
from keras.layers import Flatten
from keras.layers import Conv2D
from keras.layers import MaxPooling2D
from keras.layers import UpSampling2D
from keras.layers import Input
from keras.layers import concatenate
from keras.layers import Cropping2D
```
## A1 - U-Net
```
# padding "valid" -> no padding
# INPUT
visible = Input(shape=(572,572,1))
# DOWNCONVOLUTION
conv1 = (Conv2D(filters = 64, input_shape=(572,572,1), kernel_size=3, padding="valid", strides=(1,1)))(inputdata)
conv2 =(Conv2D(filters = 64, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv1)
pool1 = (MaxPooling2D(pool_size=(2,2)))(conv2)
conv3 = (Conv2D(filters = 128, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(pool1)
conv4 = (Conv2D(filters = 128, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv3)
pool2 = (MaxPooling2D(pool_size=(2,2)))(conv4)
conv5 = (Conv2D(filters = 256, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(pool2)
conv6 = (Conv2D(filters = 256, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv5)
pool3 = (MaxPooling2D(pool_size=(2,2)))(conv6)
conv7 = (Conv2D(filters = 512, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(pool3)
conv8 = (Conv2D(filters = 512, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv7)
pool4 = (MaxPooling2D(pool_size=(2,2)))(conv8)
conv9 = (Conv2D(filters = 1024, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(pool4)
conv10 = (Conv2D(filters = 1024, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv9)
# UPCONVOLUTION
upsamp1 = (UpSampling2D(size = 2))(conv10)
conv8_cr = (Cropping2D(cropping= (4,4) ))(conv8) # 64-56 = 8, symm. crop by 4
concat1 = concatenate([conv8_cr, upsamp1])
conv11 = (Conv2D(filters = 512, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(concat1)
conv12 = (Conv2D(filters = 512, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv11)
upsamp2 = (UpSampling2D(size = 2))(conv12)
conv6_cr = (Cropping2D(cropping= (16,16) ))(conv6) # 136-104 = 32, symm. crop by 16
concat2 = concatenate([conv6_cr, upsamp2])
conv13 = (Conv2D(filters = 256, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(concat2)
conv14 = (Conv2D(filters = 256, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv13)
upsamp3 = (UpSampling2D(size = 2))(conv14)
conv4_cr = (Cropping2D(cropping= (40,40) ))(conv4) # 280-200 = 80, symm. crop by 40
concat3 = concatenate([conv4_cr, upsamp3])
conv15 = (Conv2D(filters = 128, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(concat3)
conv16 = (Conv2D(filters = 128, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv15)
upsamp4 =(UpSampling2D(size = 2))(conv16)
conv2_cr = (Cropping2D(cropping= (88,88) ))(conv2) # 568-392 = 176, symm. crop by 88
concat4 = concatenate([conv2_cr, upsamp4])
conv17 = (Conv2D(filters = 64, kernel_size=3, padding="valid", strides=(1,1)))(concat4)
conv18 = (Conv2D(filters = 64, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv17)
output = (Conv2D(filters = 2, kernel_size=1, padding="valid", strides=(1,1), activation='relu'))(conv18)
# COMPILE
model = Model(inputs=inputdata, outputs = output)
model.summary()
```
## A2 - AlexNet
```
model = Sequential()
# l1
model.add(Conv2D(filters = 96, kernel_size=11, input_shape = (227, 227, 3), padding="valid", strides=(4,4), activation='relu', data_format="channels_last"))
model.add(MaxPooling2D(pool_size = (3,3), strides = (2,2), padding = "valid"))
# l2
model.add(Conv2D(filters = 256, kernel_size=5, padding="same", strides=(1,1), activation='relu'))
model.add(MaxPooling2D(pool_size = (3,3), strides = (2,2), padding = "valid"))
# l3
model.add(Conv2D(filters = 384, kernel_size=3, padding="same", strides=(1,1), activation='relu'))
# l4
model.add(Conv2D(filters = 384, kernel_size=3, padding="same", strides=(1,1), activation='relu'))
# l5
model.add(Conv2D(filters = 256, kernel_size=3, padding="same", strides=(1,1), activation='relu'))
model.add(MaxPooling2D(pool_size = (3,3), strides = (2,2), padding = "valid"))
# dense
model.add(Flatten())
model.add(Dense(4096, activation="relu"))
model.add(Dense(4069, activation="relu"))
model.add(Dense(1000, activation="softmax"))
# compile
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
model.summary()
```
## A3 - PCA
$d$ is the first principal component, the eigenvector corresponding to the largest eigenvalue of $X^T X$.
```
from sklearn.decomposition import PCA
X = []
for n in range(100):
x = np.random.normal(loc = 0, scale = 5)
y = np.random.normal(loc = 0, scale = 1)
X.append([x,y])
plt.scatter([el[0] for el in X], [el[1] for el in X])
pca = PCA(n_components=2)
pca.fit(X)
PCA(copy=True, iterated_power='auto', n_components=2, random_state=None,
svd_solver='auto', tol=0.0, whiten=False)
print("Explained by 1st component: %g %%" % round(100*pca.explained_variance_ratio_[0],2))
print("Explained by 2nd component: %g %%" % round(100*pca.explained_variance_ratio_[1],2))
#print(pca.singular_values_)
```
For a change in variance by a factor of 5, it can be observed that the first component of a PCA explains about 95% of the overall variance and is therefore the relevant "feature" in this data set.
|
github_jupyter
|
import numpy as np
import matplotlib.pylab as plt
import numpy.random as rd
from sklearn.datasets import fetch_openml # MNIST data
from sklearn.model_selection import train_test_split
#os.environ["CUDA_VISIBLE_DEVICES"] = '0'
#sess = tf.Session(config=tf.ConfigProto(device_count={'GPU': 0}))
from keras.models import Sequential
from keras.models import Model
from keras.layers import Dense
from keras.layers import Flatten
from keras.layers import Conv2D
from keras.layers import MaxPooling2D
from keras.layers import UpSampling2D
from keras.layers import Input
from keras.layers import concatenate
from keras.layers import Cropping2D
# padding "valid" -> no padding
# INPUT
visible = Input(shape=(572,572,1))
# DOWNCONVOLUTION
conv1 = (Conv2D(filters = 64, input_shape=(572,572,1), kernel_size=3, padding="valid", strides=(1,1)))(inputdata)
conv2 =(Conv2D(filters = 64, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv1)
pool1 = (MaxPooling2D(pool_size=(2,2)))(conv2)
conv3 = (Conv2D(filters = 128, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(pool1)
conv4 = (Conv2D(filters = 128, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv3)
pool2 = (MaxPooling2D(pool_size=(2,2)))(conv4)
conv5 = (Conv2D(filters = 256, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(pool2)
conv6 = (Conv2D(filters = 256, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv5)
pool3 = (MaxPooling2D(pool_size=(2,2)))(conv6)
conv7 = (Conv2D(filters = 512, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(pool3)
conv8 = (Conv2D(filters = 512, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv7)
pool4 = (MaxPooling2D(pool_size=(2,2)))(conv8)
conv9 = (Conv2D(filters = 1024, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(pool4)
conv10 = (Conv2D(filters = 1024, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv9)
# UPCONVOLUTION
upsamp1 = (UpSampling2D(size = 2))(conv10)
conv8_cr = (Cropping2D(cropping= (4,4) ))(conv8) # 64-56 = 8, symm. crop by 4
concat1 = concatenate([conv8_cr, upsamp1])
conv11 = (Conv2D(filters = 512, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(concat1)
conv12 = (Conv2D(filters = 512, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv11)
upsamp2 = (UpSampling2D(size = 2))(conv12)
conv6_cr = (Cropping2D(cropping= (16,16) ))(conv6) # 136-104 = 32, symm. crop by 16
concat2 = concatenate([conv6_cr, upsamp2])
conv13 = (Conv2D(filters = 256, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(concat2)
conv14 = (Conv2D(filters = 256, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv13)
upsamp3 = (UpSampling2D(size = 2))(conv14)
conv4_cr = (Cropping2D(cropping= (40,40) ))(conv4) # 280-200 = 80, symm. crop by 40
concat3 = concatenate([conv4_cr, upsamp3])
conv15 = (Conv2D(filters = 128, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(concat3)
conv16 = (Conv2D(filters = 128, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv15)
upsamp4 =(UpSampling2D(size = 2))(conv16)
conv2_cr = (Cropping2D(cropping= (88,88) ))(conv2) # 568-392 = 176, symm. crop by 88
concat4 = concatenate([conv2_cr, upsamp4])
conv17 = (Conv2D(filters = 64, kernel_size=3, padding="valid", strides=(1,1)))(concat4)
conv18 = (Conv2D(filters = 64, kernel_size=3, padding="valid", strides=(1,1), activation='relu'))(conv17)
output = (Conv2D(filters = 2, kernel_size=1, padding="valid", strides=(1,1), activation='relu'))(conv18)
# COMPILE
model = Model(inputs=inputdata, outputs = output)
model.summary()
model = Sequential()
# l1
model.add(Conv2D(filters = 96, kernel_size=11, input_shape = (227, 227, 3), padding="valid", strides=(4,4), activation='relu', data_format="channels_last"))
model.add(MaxPooling2D(pool_size = (3,3), strides = (2,2), padding = "valid"))
# l2
model.add(Conv2D(filters = 256, kernel_size=5, padding="same", strides=(1,1), activation='relu'))
model.add(MaxPooling2D(pool_size = (3,3), strides = (2,2), padding = "valid"))
# l3
model.add(Conv2D(filters = 384, kernel_size=3, padding="same", strides=(1,1), activation='relu'))
# l4
model.add(Conv2D(filters = 384, kernel_size=3, padding="same", strides=(1,1), activation='relu'))
# l5
model.add(Conv2D(filters = 256, kernel_size=3, padding="same", strides=(1,1), activation='relu'))
model.add(MaxPooling2D(pool_size = (3,3), strides = (2,2), padding = "valid"))
# dense
model.add(Flatten())
model.add(Dense(4096, activation="relu"))
model.add(Dense(4069, activation="relu"))
model.add(Dense(1000, activation="softmax"))
# compile
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
model.summary()
from sklearn.decomposition import PCA
X = []
for n in range(100):
x = np.random.normal(loc = 0, scale = 5)
y = np.random.normal(loc = 0, scale = 1)
X.append([x,y])
plt.scatter([el[0] for el in X], [el[1] for el in X])
pca = PCA(n_components=2)
pca.fit(X)
PCA(copy=True, iterated_power='auto', n_components=2, random_state=None,
svd_solver='auto', tol=0.0, whiten=False)
print("Explained by 1st component: %g %%" % round(100*pca.explained_variance_ratio_[0],2))
print("Explained by 2nd component: %g %%" % round(100*pca.explained_variance_ratio_[1],2))
#print(pca.singular_values_)
| 0.721351 | 0.877844 |
# Simple RNN Practice

## 1. Settings
### 1) Import Required Libraries
```
import torch
import torch.nn as nn
import torch.optim as optim
from torch.autograd import Variable
import numpy as np
```
### 2) Hyperparameter & Data
```
# Preprocessing string data
# alphabet(0-25), others(26~32), start(33), end(34) -> 35 chars
n_hidden = 50
lr = 0.01
epochs = 2000
string = "hello pytorch.how long can a rnn cell remember?"# show us your limit!"
chars = "abcdefghijklmnopqrstuvwxyz ?!.,:;01"
char_list = [i for i in chars]
char_len = len(char_list)
n_letters = len(char_list)
```
### 3) String to One-hot
```
# String to onehot vector
# a -> [1 0 0 ... 0 0]
def string_to_onehot(string):
start = np.zeros(shape=len(char_list) ,dtype=int)
end = np.zeros(shape=len(char_list) ,dtype=int)
start[-2] = 1
end[-1] = 1
for i in string:
idx = char_list.index(i)
zero = np.zeros(shape=char_len ,dtype=int)
zero[idx]=1
start = np.vstack([start,zero])
output = np.vstack([start,end])
return output
```
### 4) One-hot to Character
```
# Onehot vector to word
# [1 0 0 ... 0 0] -> a
def onehot_to_word(onehot_1):
onehot = torch.Tensor.numpy(onehot_1)
return char_list[onehot.argmax()]
```
## 2. RNN class
```
# RNN with 1 hidden layer
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
super(RNN, self).__init__()
self.input_size = input_size
self.hidden_size = hidden_size
self.output_size = output_size
self.i2h = nn.Linear(input_size, hidden_size)
self.h2h = nn.Linear(hidden_size, hidden_size)
self.i2o = nn.Linear(hidden_size, output_size)
self.act_fn = nn.Tanh()
def forward(self, input, hidden):
hidden = self.act_fn(self.i2h(input)+self.h2h(hidden))
output = self.i2o(hidden)
return output, hidden
def init_hidden(self):
return Variable(torch.zeros(1, self.hidden_size))
rnn = RNN(n_letters, n_hidden, n_letters)
```
## 3. Loss function & Optimizer
```
loss_func = nn.MSELoss()
optimizer = torch.optim.Adam(rnn.parameters(), lr=lr)
```
## 4. Train
```
one_hot = torch.from_numpy(string_to_onehot(string)).type_as(torch.FloatTensor())
for i in range(epochs):
rnn.zero_grad()
total_loss = 0
hidden = rnn.init_hidden()
for j in range(one_hot.size()[0]-1):
input = Variable(one_hot[j:j+1,:])
output, hidden = rnn.forward(input, hidden)
target = Variable(one_hot[j+1])
loss = loss_func(output.view(-1),target.view(-1))
total_loss += loss
input = output
total_loss.backward()
optimizer.step()
if i % 10 == 0:
print(total_loss)
```
## 5. Test
```
hidden = rnn.init_hidden()
input = Variable(one_hot[0:1,:])
for i in range(len(string)):
output, hidden = rnn.forward(input, hidden)
print(onehot_to_word(output.data),end="")
input = output
```
|
github_jupyter
|
import torch
import torch.nn as nn
import torch.optim as optim
from torch.autograd import Variable
import numpy as np
# Preprocessing string data
# alphabet(0-25), others(26~32), start(33), end(34) -> 35 chars
n_hidden = 50
lr = 0.01
epochs = 2000
string = "hello pytorch.how long can a rnn cell remember?"# show us your limit!"
chars = "abcdefghijklmnopqrstuvwxyz ?!.,:;01"
char_list = [i for i in chars]
char_len = len(char_list)
n_letters = len(char_list)
# String to onehot vector
# a -> [1 0 0 ... 0 0]
def string_to_onehot(string):
start = np.zeros(shape=len(char_list) ,dtype=int)
end = np.zeros(shape=len(char_list) ,dtype=int)
start[-2] = 1
end[-1] = 1
for i in string:
idx = char_list.index(i)
zero = np.zeros(shape=char_len ,dtype=int)
zero[idx]=1
start = np.vstack([start,zero])
output = np.vstack([start,end])
return output
# Onehot vector to word
# [1 0 0 ... 0 0] -> a
def onehot_to_word(onehot_1):
onehot = torch.Tensor.numpy(onehot_1)
return char_list[onehot.argmax()]
# RNN with 1 hidden layer
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
super(RNN, self).__init__()
self.input_size = input_size
self.hidden_size = hidden_size
self.output_size = output_size
self.i2h = nn.Linear(input_size, hidden_size)
self.h2h = nn.Linear(hidden_size, hidden_size)
self.i2o = nn.Linear(hidden_size, output_size)
self.act_fn = nn.Tanh()
def forward(self, input, hidden):
hidden = self.act_fn(self.i2h(input)+self.h2h(hidden))
output = self.i2o(hidden)
return output, hidden
def init_hidden(self):
return Variable(torch.zeros(1, self.hidden_size))
rnn = RNN(n_letters, n_hidden, n_letters)
loss_func = nn.MSELoss()
optimizer = torch.optim.Adam(rnn.parameters(), lr=lr)
one_hot = torch.from_numpy(string_to_onehot(string)).type_as(torch.FloatTensor())
for i in range(epochs):
rnn.zero_grad()
total_loss = 0
hidden = rnn.init_hidden()
for j in range(one_hot.size()[0]-1):
input = Variable(one_hot[j:j+1,:])
output, hidden = rnn.forward(input, hidden)
target = Variable(one_hot[j+1])
loss = loss_func(output.view(-1),target.view(-1))
total_loss += loss
input = output
total_loss.backward()
optimizer.step()
if i % 10 == 0:
print(total_loss)
hidden = rnn.init_hidden()
input = Variable(one_hot[0:1,:])
for i in range(len(string)):
output, hidden = rnn.forward(input, hidden)
print(onehot_to_word(output.data),end="")
input = output
| 0.860398 | 0.921039 |
```
import os
os.environ['CUDA_VISIBLE_DEVICES'] = ''
from malaya_speech.train.model import hubert, ctc
from malaya_speech.train.model.conformer.model import Model as ConformerModel
import malaya_speech
import tensorflow as tf
import numpy as np
import json
from glob import glob
with open('malaya-speech-sst-vocab.json') as fopen:
unique_vocab = json.load(fopen) + ['{', '}', '[']
subwords = malaya_speech.subword.load('transducer.subword')
X = tf.compat.v1.placeholder(tf.float32, [None, None], name = 'X_placeholder')
X_len = tf.compat.v1.placeholder(tf.int32, [None], name = 'X_len_placeholder')
training = True
class Encoder:
def __init__(self, config):
self.config = config
self.encoder = ConformerModel(**self.config)
def __call__(self, x, input_mask, training = True):
return self.encoder(x, training = training)
config_conformer = malaya_speech.config.conformer_base_encoder_config
config_conformer['subsampling']['type'] = 'none'
config_conformer['dropout'] = 0.0
encoder = Encoder(config_conformer)
cfg = hubert.HuBERTConfig(
extractor_mode='layer_norm',
dropout=0.0,
attention_dropout=0.0,
encoder_layerdrop=0.0,
dropout_input=0.0,
dropout_features=0.0,
final_dim=256,
)
model = hubert.Model(cfg, encoder, ['pad', 'eos', 'unk'] + [str(i) for i in range(100)])
r = model(X, padding_mask = X_len, features_only = True, mask = False)
logits = tf.layers.dense(r['x'], subwords.vocab_size + 1)
seq_lens = tf.reduce_sum(
tf.cast(tf.logical_not(r['padding_mask']), tf.int32), axis = 1
)
logits = tf.transpose(logits, [1, 0, 2])
logits = tf.identity(logits, name = 'logits')
seq_lens = tf.identity(seq_lens, name = 'seq_lens')
sess = tf.Session()
sess.run(tf.global_variables_initializer())
var_list = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES)
saver = tf.train.Saver(var_list = var_list)
saver.restore(sess, 'hubert-conformer-base-ctc/model.ckpt-810000')
saver = tf.train.Saver()
saver.save(sess, 'output-hubert-conformer-base-ctc/model.ckpt')
strings = ','.join(
[
n.name
for n in tf.get_default_graph().as_graph_def().node
if ('Variable' in n.op
or 'gather' in n.op.lower()
or 'placeholder' in n.name
or 'logits' in n.name
or 'seq_lens' in n.name)
and 'adam' not in n.name
and 'global_step' not in n.name
and 'Assign' not in n.name
and 'ReadVariableOp' not in n.name
and 'Gather' not in n.name
]
)
strings.split(',')
def freeze_graph(model_dir, output_node_names):
if not tf.gfile.Exists(model_dir):
raise AssertionError(
"Export directory doesn't exists. Please specify an export "
'directory: %s' % model_dir
)
checkpoint = tf.train.get_checkpoint_state(model_dir)
input_checkpoint = checkpoint.model_checkpoint_path
absolute_model_dir = '/'.join(input_checkpoint.split('/')[:-1])
output_graph = absolute_model_dir + '/frozen_model.pb'
clear_devices = True
with tf.Session(graph = tf.Graph()) as sess:
saver = tf.train.import_meta_graph(
input_checkpoint + '.meta', clear_devices = clear_devices
)
saver.restore(sess, input_checkpoint)
output_graph_def = tf.graph_util.convert_variables_to_constants(
sess,
tf.get_default_graph().as_graph_def(),
output_node_names.split(','),
)
with tf.gfile.GFile(output_graph, 'wb') as f:
f.write(output_graph_def.SerializeToString())
print('%d ops in the final graph.' % len(output_graph_def.node))
freeze_graph('output-hubert-conformer-base-ctc', strings)
def load_graph(frozen_graph_filename):
with tf.gfile.GFile(frozen_graph_filename, 'rb') as f:
graph_def = tf.GraphDef()
graph_def.ParseFromString(f.read())
with tf.Graph().as_default() as graph:
tf.import_graph_def(graph_def)
return graph
files = [
'speech/record/savewav_2020-11-26_22-36-06_294832.wav',
'speech/record/savewav_2020-11-26_22-40-56_929661.wav',
'speech/record/675.wav',
'speech/record/664.wav',
'speech/example-speaker/husein-zolkepli.wav',
'speech/example-speaker/mas-aisyah.wav',
'speech/example-speaker/khalil-nooh.wav',
'speech/example-speaker/shafiqah-idayu.wav',
'speech/khutbah/wadi-annuar.wav',
]
ys = [malaya_speech.load(f)[0] for f in files]
padded, lens = malaya_speech.padding.sequence_1d(ys, return_len = True)
g = load_graph('output-hubert-conformer-base-ctc/frozen_model.pb')
input_nodes = [
'X_placeholder',
'X_len_placeholder',
]
output_nodes = [
'logits',
'seq_lens',
]
inputs = {n: g.get_tensor_by_name(f'import/{n}:0') for n in input_nodes}
outputs = {n: g.get_tensor_by_name(f'import/{n}:0') for n in output_nodes}
test_sess = tf.Session(graph = g)
r = test_sess.run(outputs['logits'], feed_dict = {inputs['X_placeholder']: padded,
inputs['X_len_placeholder']: lens})
from tensorflow.tools.graph_transforms import TransformGraph
transforms = ['add_default_attributes',
'remove_nodes(op=Identity, op=CheckNumerics, op=Dropout)',
'fold_batch_norms',
'fold_old_batch_norms',
'quantize_weights(fallback_min=-10, fallback_max=10)',
'strip_unused_nodes',
'sort_by_execution_order']
pb = 'output-hubert-conformer-base-ctc/frozen_model.pb'
input_graph_def = tf.GraphDef()
with tf.gfile.FastGFile(pb, 'rb') as f:
input_graph_def.ParseFromString(f.read())
transformed_graph_def = TransformGraph(input_graph_def,
input_nodes,
output_nodes, transforms)
with tf.gfile.GFile(f'{pb}.quantized', 'wb') as f:
f.write(transformed_graph_def.SerializeToString())
g = load_graph('output-hubert-conformer-base-ctc/frozen_model.pb.quantized')
!tar -czvf output-hubert-conformer-base-ctc.tar.gz output-hubert-conformer-base-ctc
b2_application_key_id = os.environ['b2_application_key_id']
b2_application_key = os.environ['b2_application_key']
from b2sdk.v1 import *
info = InMemoryAccountInfo()
b2_api = B2Api(info)
application_key_id = b2_application_key_id
application_key = b2_application_key
b2_api.authorize_account("production", application_key_id, application_key)
file_info = {'how': 'good-file'}
b2_bucket = b2_api.get_bucket_by_name('malaya-speech-model')
key = 'output-hubert-conformer-base-ctc.tar.gz'
outPutname = "pretrained/output-hubert-conformer-base-ctc-subwords.tar.gz"
b2_bucket.upload_local_file(
local_file=key,
file_name=outPutname,
file_infos=file_info,
)
file = 'output-hubert-conformer-base-ctc/frozen_model.pb'
outPutname = 'speech-to-text-ctc/hubert-conformer-subword/model.pb'
b2_bucket.upload_local_file(
local_file=file,
file_name=outPutname,
file_infos=file_info,
)
file = 'output-hubert-conformer-base-ctc/frozen_model.pb.quantized'
outPutname = 'speech-to-text-ctc/hubert-conformer-subword-quantized/model.pb'
b2_bucket.upload_local_file(
local_file=file,
file_name=outPutname,
file_infos=file_info,
)
!rm -rf output-hubert-conformer-base-ctc output-hubert-conformer-base-ctc.tar.gz
```
|
github_jupyter
|
import os
os.environ['CUDA_VISIBLE_DEVICES'] = ''
from malaya_speech.train.model import hubert, ctc
from malaya_speech.train.model.conformer.model import Model as ConformerModel
import malaya_speech
import tensorflow as tf
import numpy as np
import json
from glob import glob
with open('malaya-speech-sst-vocab.json') as fopen:
unique_vocab = json.load(fopen) + ['{', '}', '[']
subwords = malaya_speech.subword.load('transducer.subword')
X = tf.compat.v1.placeholder(tf.float32, [None, None], name = 'X_placeholder')
X_len = tf.compat.v1.placeholder(tf.int32, [None], name = 'X_len_placeholder')
training = True
class Encoder:
def __init__(self, config):
self.config = config
self.encoder = ConformerModel(**self.config)
def __call__(self, x, input_mask, training = True):
return self.encoder(x, training = training)
config_conformer = malaya_speech.config.conformer_base_encoder_config
config_conformer['subsampling']['type'] = 'none'
config_conformer['dropout'] = 0.0
encoder = Encoder(config_conformer)
cfg = hubert.HuBERTConfig(
extractor_mode='layer_norm',
dropout=0.0,
attention_dropout=0.0,
encoder_layerdrop=0.0,
dropout_input=0.0,
dropout_features=0.0,
final_dim=256,
)
model = hubert.Model(cfg, encoder, ['pad', 'eos', 'unk'] + [str(i) for i in range(100)])
r = model(X, padding_mask = X_len, features_only = True, mask = False)
logits = tf.layers.dense(r['x'], subwords.vocab_size + 1)
seq_lens = tf.reduce_sum(
tf.cast(tf.logical_not(r['padding_mask']), tf.int32), axis = 1
)
logits = tf.transpose(logits, [1, 0, 2])
logits = tf.identity(logits, name = 'logits')
seq_lens = tf.identity(seq_lens, name = 'seq_lens')
sess = tf.Session()
sess.run(tf.global_variables_initializer())
var_list = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES)
saver = tf.train.Saver(var_list = var_list)
saver.restore(sess, 'hubert-conformer-base-ctc/model.ckpt-810000')
saver = tf.train.Saver()
saver.save(sess, 'output-hubert-conformer-base-ctc/model.ckpt')
strings = ','.join(
[
n.name
for n in tf.get_default_graph().as_graph_def().node
if ('Variable' in n.op
or 'gather' in n.op.lower()
or 'placeholder' in n.name
or 'logits' in n.name
or 'seq_lens' in n.name)
and 'adam' not in n.name
and 'global_step' not in n.name
and 'Assign' not in n.name
and 'ReadVariableOp' not in n.name
and 'Gather' not in n.name
]
)
strings.split(',')
def freeze_graph(model_dir, output_node_names):
if not tf.gfile.Exists(model_dir):
raise AssertionError(
"Export directory doesn't exists. Please specify an export "
'directory: %s' % model_dir
)
checkpoint = tf.train.get_checkpoint_state(model_dir)
input_checkpoint = checkpoint.model_checkpoint_path
absolute_model_dir = '/'.join(input_checkpoint.split('/')[:-1])
output_graph = absolute_model_dir + '/frozen_model.pb'
clear_devices = True
with tf.Session(graph = tf.Graph()) as sess:
saver = tf.train.import_meta_graph(
input_checkpoint + '.meta', clear_devices = clear_devices
)
saver.restore(sess, input_checkpoint)
output_graph_def = tf.graph_util.convert_variables_to_constants(
sess,
tf.get_default_graph().as_graph_def(),
output_node_names.split(','),
)
with tf.gfile.GFile(output_graph, 'wb') as f:
f.write(output_graph_def.SerializeToString())
print('%d ops in the final graph.' % len(output_graph_def.node))
freeze_graph('output-hubert-conformer-base-ctc', strings)
def load_graph(frozen_graph_filename):
with tf.gfile.GFile(frozen_graph_filename, 'rb') as f:
graph_def = tf.GraphDef()
graph_def.ParseFromString(f.read())
with tf.Graph().as_default() as graph:
tf.import_graph_def(graph_def)
return graph
files = [
'speech/record/savewav_2020-11-26_22-36-06_294832.wav',
'speech/record/savewav_2020-11-26_22-40-56_929661.wav',
'speech/record/675.wav',
'speech/record/664.wav',
'speech/example-speaker/husein-zolkepli.wav',
'speech/example-speaker/mas-aisyah.wav',
'speech/example-speaker/khalil-nooh.wav',
'speech/example-speaker/shafiqah-idayu.wav',
'speech/khutbah/wadi-annuar.wav',
]
ys = [malaya_speech.load(f)[0] for f in files]
padded, lens = malaya_speech.padding.sequence_1d(ys, return_len = True)
g = load_graph('output-hubert-conformer-base-ctc/frozen_model.pb')
input_nodes = [
'X_placeholder',
'X_len_placeholder',
]
output_nodes = [
'logits',
'seq_lens',
]
inputs = {n: g.get_tensor_by_name(f'import/{n}:0') for n in input_nodes}
outputs = {n: g.get_tensor_by_name(f'import/{n}:0') for n in output_nodes}
test_sess = tf.Session(graph = g)
r = test_sess.run(outputs['logits'], feed_dict = {inputs['X_placeholder']: padded,
inputs['X_len_placeholder']: lens})
from tensorflow.tools.graph_transforms import TransformGraph
transforms = ['add_default_attributes',
'remove_nodes(op=Identity, op=CheckNumerics, op=Dropout)',
'fold_batch_norms',
'fold_old_batch_norms',
'quantize_weights(fallback_min=-10, fallback_max=10)',
'strip_unused_nodes',
'sort_by_execution_order']
pb = 'output-hubert-conformer-base-ctc/frozen_model.pb'
input_graph_def = tf.GraphDef()
with tf.gfile.FastGFile(pb, 'rb') as f:
input_graph_def.ParseFromString(f.read())
transformed_graph_def = TransformGraph(input_graph_def,
input_nodes,
output_nodes, transforms)
with tf.gfile.GFile(f'{pb}.quantized', 'wb') as f:
f.write(transformed_graph_def.SerializeToString())
g = load_graph('output-hubert-conformer-base-ctc/frozen_model.pb.quantized')
!tar -czvf output-hubert-conformer-base-ctc.tar.gz output-hubert-conformer-base-ctc
b2_application_key_id = os.environ['b2_application_key_id']
b2_application_key = os.environ['b2_application_key']
from b2sdk.v1 import *
info = InMemoryAccountInfo()
b2_api = B2Api(info)
application_key_id = b2_application_key_id
application_key = b2_application_key
b2_api.authorize_account("production", application_key_id, application_key)
file_info = {'how': 'good-file'}
b2_bucket = b2_api.get_bucket_by_name('malaya-speech-model')
key = 'output-hubert-conformer-base-ctc.tar.gz'
outPutname = "pretrained/output-hubert-conformer-base-ctc-subwords.tar.gz"
b2_bucket.upload_local_file(
local_file=key,
file_name=outPutname,
file_infos=file_info,
)
file = 'output-hubert-conformer-base-ctc/frozen_model.pb'
outPutname = 'speech-to-text-ctc/hubert-conformer-subword/model.pb'
b2_bucket.upload_local_file(
local_file=file,
file_name=outPutname,
file_infos=file_info,
)
file = 'output-hubert-conformer-base-ctc/frozen_model.pb.quantized'
outPutname = 'speech-to-text-ctc/hubert-conformer-subword-quantized/model.pb'
b2_bucket.upload_local_file(
local_file=file,
file_name=outPutname,
file_infos=file_info,
)
!rm -rf output-hubert-conformer-base-ctc output-hubert-conformer-base-ctc.tar.gz
| 0.475605 | 0.151718 |
A typical digital image is made by stacking Red Blue and Green pixel arrays of intensities ranging from 0 to 255.
<img src="RGB.png">
A grayscale image does not contain color but only shades of gray. The pixel intensity in a grayscale image varies from black (0 intensity) to white (255 full intensity) to make it what we usually call as a Black & White image.
Digits dataset is a grayscale image dataset of handwritten digit having 1797 8×8 images.
```
#importing the dataset
```
sklearn.datasets module makes it quick to import digits data by importing load_digits class from it. The shape of the digit data is (1797, 64). 8×8 pixels are flattened to create a vector of length 64 for every image.
```
#taking a sample image to view
#Remember image is in the form of numpy array.
```
Now, using PCA, let’s reduce the image dimensions from 64 to just 2 so that we can visualize the dataset using a Scatterplot.
```
# Import required modules
```
We can also pass a float value less than 1 instead of an integer number. i.e. PCA(0.90) this means the algorithm will find the principal components which explain 90% of the variance in data.
Another cool application of PCA is in Image compression. Let’s have a look at how can we achieve this with python.
```
# Importing required libraries
import cv2
#! pip install opencv-python
# Loading the image
img = cv2.imread('my_doggo_sample.jpeg') # you can use any that you want
plt.imshow(img)
# Splitting the image in R,G,B arrays.
#it will split the original image into Blue, Green and Red arrays.
img.shape
```
OpenCV will split into Blue, Green, and Red channels instead of Red, Blue, and Green. Be very careful of the sequence here.
```
# Initialize PCA with first 20 principal components
# Applying to the red channel and then inverse transform to transformed array
# Applying to the green channel and then inverse transform to transformed array
# Applying to the blue channel and then inverse transform to transformed array
img_not_compressed =
#viewing the not compressed image
plt.imshow(img_not_compressed)
```
In the process of reconstructing the original dimensions from the reduced dimensions, some information is lost as we keep only selected principal components, 20 in this case.
```
img_compressed =
```
Stacking the inverted arrays using dstack function. Here it is important to specify the datatype of our arrays, as most images are of 8 bit. Each pixel is represented by one 8-bit byte.
```
#viewing the compressed image
plt.imshow(img_compressed)
```
The output above is what we get when considering just 20 Principal components.
If we increase the number of Principal components the output image will get clear.
1) Now check with how many Principal Components your eyes can't see the difference with the original!
2) The dog should not be so blue, fix it!
|
github_jupyter
|
#importing the dataset
#taking a sample image to view
#Remember image is in the form of numpy array.
# Import required modules
# Importing required libraries
import cv2
#! pip install opencv-python
# Loading the image
img = cv2.imread('my_doggo_sample.jpeg') # you can use any that you want
plt.imshow(img)
# Splitting the image in R,G,B arrays.
#it will split the original image into Blue, Green and Red arrays.
img.shape
# Initialize PCA with first 20 principal components
# Applying to the red channel and then inverse transform to transformed array
# Applying to the green channel and then inverse transform to transformed array
# Applying to the blue channel and then inverse transform to transformed array
img_not_compressed =
#viewing the not compressed image
plt.imshow(img_not_compressed)
img_compressed =
#viewing the compressed image
plt.imshow(img_compressed)
| 0.466846 | 0.974653 |
# pylearn2 tutorial: Convolutional network
by [Ian Goodfellow](http://www-etud.iro.umontreal.ca/~goodfeli)
## Introduction
This ipython notebook will teach you the basics of how convolutional networks work, and show you how to use multilayer perceptrons in pylearn2.
To do this, we will go over several concepts:
Part 1: What pylearn2 is doing for you in this example
- Review of multilayer perceptrons, and how convolutional networks are similar
- Convolution and the equivariance property
- Pooling and the invariance property
- A note on using convolution in research papers
Part 2: How to use pylearn2 to train a convolutional network
- pylearn2 Spaces
- MNIST classification example
Note that this won't explain in detail how the individual classes are implemented. The classes
follow pretty good naming conventions and have pretty good docstrings, but if you have trouble
understanding them, write to me and I might add a part 3 explaining how some of the parts work
under the hood.
Please write to pylearn-dev@googlegroups.com if you encounter any problem with this tutorial.
## Requirements
Before running this notebook, you must have installed pylearn2.
Follow the [download and installation instructions](http://deeplearning.net/software/pylearn2/#download-and-installation)
if you have not yet done so.
This tutorial also assumes you already know about multilayer perceptrons, and know how to train and evaluate a multilayer perceptron in pylearn2. If not, work through multilayer_perceptron.ipynb before starting this tutorial.
It's also strongly recommend that you run this notebook with THEANO_FLAGS="device=gpu". This is a processing intensive example and the GPU will make it run a lot faster, if you have one available. Execute the next cell to verify that you are using the GPU.
```
import theano
print theano.config.device
```
## Part 1: What pylearn2 is doing for you in this example
In this part, we won't get into any specifics of pylearn2 yet. We'll just discuss what a convolutional network is. If you already know about convolutional networks, feel free to skip to part 2.
### Review of multilayer perceptrons, and how convolutional networks are similar
In multilayer_perceptron.ipynb, we saw how the multilayer perceptron (MLP) is a versatile model that can do many things. In this series of tutorials, we think of it as a classification model that learns to map an input vector $x$ to a probability distribution $p(y\mid x)$ where $y$ is a categorical value with $k$ different values. Using a dataset $\mathcal{D}$ of $(x, y)$, we can train any such probabilistic model by maximizing the log likelihood,
$$ \sum_{x,y \in \mathcal{D} } \log P(y \mid x). $$
The multilayer perceptron defines $P(y \mid x)$ to be the composition of several simpler functions. Each function being composed can be thought of as another "layer" or "stage" of processing.
A convolutional network is nothing but a multilayer perceptron where some layers take a very special form, which we will call "convolutional layers". These layers are specially designed for processing inputs where the indices of the elements have some topological significance.
For example, if we represent a grayscale image as an array $I$ with the array indices corresponding to physical locations in the image, then we know that the element $I_{i,j}$ represents something that is spatially close to the element $I_{i+1,j}$. This is in contrast to a vector representation of an image. If $I$ is a vector, then $I_i$ might not be very close at all to $I_{i+1}$, depending on whether the image was converted to vector form in row-major or column major format and depending on whether $i$ is close to the end of a row or column.
Other kinds of data with topological in the indices include time series data, where some series $S$ can be indexed by a time variable $t$. We know that $S_t$ and $S_{t+1}$ come from close together in time. We can also think of the (row, column, time) indices of video data as providing topological information.
Suppose $T$ is a function that can translate (move) an input in the space defined by its indices by some amount $x$.
In other words,
$T(S,x)_i = S_j$ where $j=i-x$ (a MathJax or ipython bug seems to prevent me from putting $i-x$ in a subscript).
Convolutional layers are an example of a function $f$ designed with the property $f(T(S,x)) \approx f(S)$ for small x.
This means if a neural network can recognize a handwritten digit in one position, it can recognize it when it is slightly shifted to a nearby position. Being able to recognize shifted versions of previously seen inputs greatly improves the generalization performance of convolutional networks.
## Convolution and the equivariance property
TODO
## Pooling and the invariance property
TODO
## A note on using convolution in research papers
TODO
#Part 2: How to use pylearn2 to train an MLP
Now that we've described the theory of what we're going to do, it's time to do it! This part describes
how to use pylearn2 to run the algorithms described above.
As in the MLP tutorial, we will use the convolutional net to do optical character recognition on the MNIST dataset.
## pylearn2 Spaces
In many places in pylearn2, we would like to be able to process several different kinds of data. In previous tutorials, we've just talked about data that could be preprocessed into a vector representation. Our algorithms all worked on vector spaces. However, it's often useful to format data in other ways. The pylearn2 Space object is used to specify the format for data. The VectorSpace class represents the typical vector formatted data we've used so far. The only thing it needs to encode about the data is its dimensionality, i.e., how many elements the vector has. In this tutorial we will start to explicitly represent images as having 2D structure, so we need to use the Conv2DSpace. The Conv2DSpace object describes how to represent a collection of images as a 4-tensor.
One thing the Conv2DSpace object needs to describe is the shape of the space--how big is the image in terms of rows and columns of pixels? Also, the image may have multiple channels. In this example, we use a grayscale input image, so the input only has one channel. Color images require three channels to store the red, green, and blue pixels at each location. We can also think of the output of each convolution layer as living in a Conv2DSpace, where each kernel outputs a different channel. Finally, the Conv2DSpace specifies what each axis of the 4-tensor means. The default is for the first axis to index over different examples, the second axis to index over channels, and the last two to index over rows and columns, respectively. This is the format that theano's 2D convolution code uses, but other libraries exist that use other formats and we often need to convert between them.
## MNIST classification example
Setting up a convolutional network in pylearn2 is essentially the same as setting up any other MLP. In the YAML experiment description below, there are really just two things to take note of.
First, rather than using "nvis" to specify the input that the MLP will take, we use a parameter called "input_space". "nvis" is actually shorthand; if you pass an integer n to nvis, it will set input_space to VectorSpace(n). Now that we are using a convolutional network, we need the input to be formatted as a collection of images so that the convolution operator will have a 2D space to work on.
Second, we make a few layers of the network be "ConvRectifiedLinear" layers. Putting some convolutional layers in the network makes those layers invariant to small translations, so the job of the remaining layers is much easier.
We don't need to do anything special to make the Softmax layer on top work with these convolutional layers. The MLP class will tell the Softmax class that its input is now coming from a Conv2DSpace. The Softmax layer will then use the Conv2DSpace's convert method to convert the 2D output from the convolutional layer into a batch of vector-valued examples.
The model and training is defined in conv.yaml file. Here we load it and set some of it's hyper-parameters.
```
!wget https://raw.githubusercontent.com/lisa-lab/pylearn2/master/pylearn2/scripts/tutorials/convolutional_network/conv.yaml
train = open('conv.yaml', 'r').read()
train_params = {'train_stop': 50000,
'valid_stop': 60000,
'test_stop': 10000,
'batch_size': 100,
'output_channels_h2': 64,
'output_channels_h3': 64,
'max_epochs': 500,
'save_path': '.'}
train = train % (train_params)
print train
```
Now, we use pylearn2's yaml_parse.load to construct the Train object, and run its main loop. The same thing could be accomplished by running pylearn2's train.py script on a file containing the yaml string.
Execute the next cell to train the model. This will take several minutes and possible as much as a few hours depending on how fast your computer is.
Make sure the dataset is present:
```
%%bash
mkdir -p /disk/scratch/neuroglycerin/mnist/
cd /disk/scratch/neuroglycerin/mnist/
wget http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz
wget http://yann.lecun.com/exdb/mnist/train-labels-idx1-ubyte.gz
wget http://yann.lecun.com/exdb/mnist/t10k-images-idx3-ubyte.gz
wget http://yann.lecun.com/exdb/mnist/t10k-labels-idx1-ubyte.gz
gzip -d *
cd -
```
And make sure the pylearn2 environment variable for it is set:
```
%env PYLEARN2_DATA_PATH=/disk/scratch/neuroglycerin
!echo $PYLEARN2_DATA_PATH
%pdb
from pylearn2.config import yaml_parse
train = yaml_parse.load(train)
train.main_loop()
```
__Giving up on that yaml file__. Trying the `mnist.yaml` that is supplied with the repo:
```
%pdb
%run /afs/inf.ed.ac.uk/user/s08/s0805516/repos/pylearn2/pylearn2/scripts/train.py --time-budget 600 ~/repos/pylearn2/pylearn2/scripts/papers/maxout/mnist.yaml
!du -h /afs/inf.ed.ac.uk/user/s08/s0805516/repos/pylearn2/pylearn2/scripts/papers/maxout/mnist_best.pkl
!rm /afs/inf.ed.ac.uk/user/s08/s0805516/repos/pylearn2/pylearn2/scripts/papers/maxout/mnist_best.pkl
```
Compiling the theano functions used to run the network will take a long time for this example. This is because the number of theano variables and ops used to specify the computation is relatively large. There is no single theano op for doing max pooling with overlapping pooling windows, so pylearn2 builds a large expression graph using indexing operations to accomplish the max pooling.
After the model is trained, we can use the print_monitor script to print the last monitoring entry of a saved model. By running it on "convolutional_network_best.pkl", we can see the performance of the model at the point where it did the best on the validation set.
```
!print_monitor.py convolutional_network_best.pkl | grep test_y_misclass
```
The test set error has dropped to 0.74%! This is a big improvement over the standard MLP.
We can also look at the convolution kernels learned by the first layer, to see that the network is looking for shifted versions of small pieces of penstrokes.
```
!show_weights.py convolutional_network_best.pkl
```
## Further reading
You can find more information on convolutional networks from the following sources:
[LISA lab's Deep Learning Tutorials: Convolutional Neural Networks (LeNet)](http://deeplearning.net/tutorial/lenet.html)
This is by no means a complete list.
|
github_jupyter
|
import theano
print theano.config.device
!wget https://raw.githubusercontent.com/lisa-lab/pylearn2/master/pylearn2/scripts/tutorials/convolutional_network/conv.yaml
train = open('conv.yaml', 'r').read()
train_params = {'train_stop': 50000,
'valid_stop': 60000,
'test_stop': 10000,
'batch_size': 100,
'output_channels_h2': 64,
'output_channels_h3': 64,
'max_epochs': 500,
'save_path': '.'}
train = train % (train_params)
print train
%%bash
mkdir -p /disk/scratch/neuroglycerin/mnist/
cd /disk/scratch/neuroglycerin/mnist/
wget http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz
wget http://yann.lecun.com/exdb/mnist/train-labels-idx1-ubyte.gz
wget http://yann.lecun.com/exdb/mnist/t10k-images-idx3-ubyte.gz
wget http://yann.lecun.com/exdb/mnist/t10k-labels-idx1-ubyte.gz
gzip -d *
cd -
%env PYLEARN2_DATA_PATH=/disk/scratch/neuroglycerin
!echo $PYLEARN2_DATA_PATH
%pdb
from pylearn2.config import yaml_parse
train = yaml_parse.load(train)
train.main_loop()
%pdb
%run /afs/inf.ed.ac.uk/user/s08/s0805516/repos/pylearn2/pylearn2/scripts/train.py --time-budget 600 ~/repos/pylearn2/pylearn2/scripts/papers/maxout/mnist.yaml
!du -h /afs/inf.ed.ac.uk/user/s08/s0805516/repos/pylearn2/pylearn2/scripts/papers/maxout/mnist_best.pkl
!rm /afs/inf.ed.ac.uk/user/s08/s0805516/repos/pylearn2/pylearn2/scripts/papers/maxout/mnist_best.pkl
!print_monitor.py convolutional_network_best.pkl | grep test_y_misclass
!show_weights.py convolutional_network_best.pkl
| 0.30819 | 0.99341 |
# Reconstruction Sandbox
This notebook is a test-bed for regularization and reconstruction methods
```
%matplotlib notebook
%load_ext autoreload
%autoreload 2
# Load motiondeblur module and Dataset class
import libwallerlab.projects.motiondeblur as md
from libwallerlab.utilities.io import Dataset, isDataset
# Platform imports
import os, glob
from os.path import expanduser
# Debugging imports
import llops as yp
import matplotlib.pyplot as plt
import numpy as np
yp.config.setDefaultBackend('arrayfire')
yp.config.setDefaultDatatype('float32')
```
## Define Output Path
```
output_path = os.path.expanduser('~/datasets/motiondeblur/beads_output')
if not os.path.exists(output_path):
os.mkdir(output_path)
```
## Load Data
```
!open /Users/zfphil/Dropbox/02-04-19-MotionDeblur-beads2/beads2_line_45ms_strobe_raster_1_motion_deblur_2019_02_04_16_45_48/
# 4.9um beads, 45ms brightness
# dataset_full_path = '/Users/zfphil/Dropbox/02-04-19-MotionDeblur-v1/beads_line_45ms_coded_raster_100_motion_deblur_2019_02_04_16_20_26/'
# 4.9um beads2, 45ms brightness
dataset_full_path = '/Users/zfphil/Dropbox/02-04-19-MotionDeblur-beads2/beads2_line_45ms_coded_raster_100_motion_deblur_2019_02_04_16_45_36'
# dataset_full_path = '/Users/zfphil/Dropbox/02-04-19-MotionDeblur-beads2/beads2_line_45ms_constant_raster_1_motion_deblur_2019_02_04_16_45_23/'
# dataset_full_path = '/Users/zfphil/Dropbox/02-04-19-MotionDeblur-beads2/beads2_line_45ms_strobe_raster_1_motion_deblur_2019_02_04_16_45_48/'
# dataset_full_path = '/Users/zfphil/Dropbox/02-04-19-MotionDeblur-beads2/beads2_line_45ms_stopandstare_stop_and_stare_2019_02_04_16_44_59/'
# Create dataset object (loads metadata)
dataset = Dataset(dataset_full_path, use_median_filter=False, subtract_mean_dark_current=False, force_type='motion_deblur')
```
## Perform Registration and Normalization
```
force = True
# Perform registration
dataset.motiondeblur.register(force=force)
# Perform normalization
dataset.motiondeblur.normalize(force=force)
# Perform kernel shape
dataset.metadata.calibration['blur_vector'] = {'scale': {'axis': 1, 'factor': 1}}
```
## Solve For Single Segment
```
dataset.frame_mask = [10,11,12,13,14]
# Create recon object
recon = md.recon.Reconstruction(dataset, alpha_blend_distance=1000, normalize=False, use_psf=False, estimate_background_poly=True)
# Perform reconstruction
recon.reconstruct(iteration_count=100, step_size=1, frame_number=4, mode='global', reg_types={})
# Save result
recon.save(output_path, filename=recon.dataset.metadata.file_header + '_no_median', formats=['npz'], save_raw=False, downsample=4)
plt.figure()
plt.imshow(recon.object_recovered)
plt.clim(400, 4000)
recon.show()
plt.clim(0,300)
```
## Calculate DNF
```
x = dataset.motiondeblur.blur_vectors()[0][0][0]
# Normalize
x = x / yp.scalar(yp.sum(x))
# Take fourier transform intensity
x_fft = yp.Ft(x)
sigma_x = yp.abs(x_fft) ** 2
# Calculate DNF
np.sqrt(1 / len(x) * np.sum(1 / sigma_x))
```
## Loop Over Segments
```
for segment_index in dataset.motiondeblur.position_segment_indicies_full:
# Set segment index
dataset.motiondeblur.position_segment_indicies = [segment_index]
# Create recon object
recon = md.recon.Reconstruction(dataset, alpha_blend_distance=1000, pad_mode='mean', )
# Perform reconstruction
# recon.reconstruct(iteration_count=-1, step_size=1, frame_number=4, mode='global', reg_types={'l2': 1e-4})
recon.reconstruct(iteration_count=100, step_size=1, frame_number=4, mode='global', reg_types={'l2': 1e-4})
# Save result
recon.save(output_path, filename=recon.dataset.metadata.file_header, formats=['png', 'npz'], save_raw=True, downsample=4)
# Try with L1 Sparsity
```
|
github_jupyter
|
%matplotlib notebook
%load_ext autoreload
%autoreload 2
# Load motiondeblur module and Dataset class
import libwallerlab.projects.motiondeblur as md
from libwallerlab.utilities.io import Dataset, isDataset
# Platform imports
import os, glob
from os.path import expanduser
# Debugging imports
import llops as yp
import matplotlib.pyplot as plt
import numpy as np
yp.config.setDefaultBackend('arrayfire')
yp.config.setDefaultDatatype('float32')
output_path = os.path.expanduser('~/datasets/motiondeblur/beads_output')
if not os.path.exists(output_path):
os.mkdir(output_path)
!open /Users/zfphil/Dropbox/02-04-19-MotionDeblur-beads2/beads2_line_45ms_strobe_raster_1_motion_deblur_2019_02_04_16_45_48/
# 4.9um beads, 45ms brightness
# dataset_full_path = '/Users/zfphil/Dropbox/02-04-19-MotionDeblur-v1/beads_line_45ms_coded_raster_100_motion_deblur_2019_02_04_16_20_26/'
# 4.9um beads2, 45ms brightness
dataset_full_path = '/Users/zfphil/Dropbox/02-04-19-MotionDeblur-beads2/beads2_line_45ms_coded_raster_100_motion_deblur_2019_02_04_16_45_36'
# dataset_full_path = '/Users/zfphil/Dropbox/02-04-19-MotionDeblur-beads2/beads2_line_45ms_constant_raster_1_motion_deblur_2019_02_04_16_45_23/'
# dataset_full_path = '/Users/zfphil/Dropbox/02-04-19-MotionDeblur-beads2/beads2_line_45ms_strobe_raster_1_motion_deblur_2019_02_04_16_45_48/'
# dataset_full_path = '/Users/zfphil/Dropbox/02-04-19-MotionDeblur-beads2/beads2_line_45ms_stopandstare_stop_and_stare_2019_02_04_16_44_59/'
# Create dataset object (loads metadata)
dataset = Dataset(dataset_full_path, use_median_filter=False, subtract_mean_dark_current=False, force_type='motion_deblur')
force = True
# Perform registration
dataset.motiondeblur.register(force=force)
# Perform normalization
dataset.motiondeblur.normalize(force=force)
# Perform kernel shape
dataset.metadata.calibration['blur_vector'] = {'scale': {'axis': 1, 'factor': 1}}
dataset.frame_mask = [10,11,12,13,14]
# Create recon object
recon = md.recon.Reconstruction(dataset, alpha_blend_distance=1000, normalize=False, use_psf=False, estimate_background_poly=True)
# Perform reconstruction
recon.reconstruct(iteration_count=100, step_size=1, frame_number=4, mode='global', reg_types={})
# Save result
recon.save(output_path, filename=recon.dataset.metadata.file_header + '_no_median', formats=['npz'], save_raw=False, downsample=4)
plt.figure()
plt.imshow(recon.object_recovered)
plt.clim(400, 4000)
recon.show()
plt.clim(0,300)
x = dataset.motiondeblur.blur_vectors()[0][0][0]
# Normalize
x = x / yp.scalar(yp.sum(x))
# Take fourier transform intensity
x_fft = yp.Ft(x)
sigma_x = yp.abs(x_fft) ** 2
# Calculate DNF
np.sqrt(1 / len(x) * np.sum(1 / sigma_x))
for segment_index in dataset.motiondeblur.position_segment_indicies_full:
# Set segment index
dataset.motiondeblur.position_segment_indicies = [segment_index]
# Create recon object
recon = md.recon.Reconstruction(dataset, alpha_blend_distance=1000, pad_mode='mean', )
# Perform reconstruction
# recon.reconstruct(iteration_count=-1, step_size=1, frame_number=4, mode='global', reg_types={'l2': 1e-4})
recon.reconstruct(iteration_count=100, step_size=1, frame_number=4, mode='global', reg_types={'l2': 1e-4})
# Save result
recon.save(output_path, filename=recon.dataset.metadata.file_header, formats=['png', 'npz'], save_raw=True, downsample=4)
# Try with L1 Sparsity
| 0.551332 | 0.777258 |
# NumPy Basics: Arrays and Vectorized Computation
```
import numpy as np
np.random.seed(12345)
import matplotlib.pyplot as plt
plt.rc('figure', figsize=(10, 6))
np.set_printoptions(precision=4, suppress=True)
import numpy as np
my_arr = np.arange(1000000)
my_list = list(range(1000000))
%time for _ in range(10): my_arr2 = my_arr * 2
%time for _ in range(10): my_list2 = [x * 2 for x in my_list]
```
## The NumPy ndarray: A Multidimensional Array Object
```
import numpy as np
# Generate some random data
data = np.random.randn(2, 3)
data
data * 10
data + data
data.shape
data.dtype
```
### Creating ndarrays
```
data1 = [6, 7.5, 8, 0, 1]
arr1 = np.array(data1)
arr1
data2 = [[1, 2, 3, 4], [5, 6, 7, 8]]
arr2 = np.array(data2)
arr2
arr2.ndim
arr2.shape
arr1.dtype
arr2.dtype
np.zeros(10)
np.zeros((3, 6))
np.empty((2, 3, 2))
np.arange(15)
```
### Data Types for ndarrays
```
arr1 = np.array([1, 2, 3], dtype=np.float64)
arr2 = np.array([1, 2, 3], dtype=np.int32)
arr1.dtype
arr2.dtype
arr = np.array([1, 2, 3, 4, 5])
arr.dtype
float_arr = arr.astype(np.float64)
float_arr.dtype
arr = np.array([3.7, -1.2, -2.6, 0.5, 12.9, 10.1])
arr
arr.astype(np.int32)
numeric_strings = np.array(['1.25', '-9.6', '42'], dtype=np.string_)
numeric_strings.astype(float)
int_array = np.arange(10)
calibers = np.array([.22, .270, .357, .380, .44, .50], dtype=np.float64)
int_array.astype(calibers.dtype)
empty_uint32 = np.empty(8, dtype='u4')
empty_uint32
```
### Arithmetic with NumPy Arrays
```
arr = np.array([[1., 2., 3.], [4., 5., 6.]])
arr
arr * arr
arr - arr
1 / arr
arr ** 0.5
arr2 = np.array([[0., 4., 1.], [7., 2., 12.]])
arr2
arr2 > arr
```
### Basic Indexing and Slicing
```
arr = np.arange(10)
arr
arr[5]
arr[5:8]
arr[5:8] = 12
arr
arr_slice = arr[5:8]
arr_slice
arr_slice[1] = 12345
arr
arr_slice[:] = 64
arr
arr2d = np.array([[1, 2, 3], [4, 5, 6], [7, 8, 9]])
arr2d[2]
arr2d[0][2]
arr2d[0, 2]
arr3d = np.array([[[1, 2, 3], [4, 5, 6]], [[7, 8, 9], [10, 11, 12]]])
arr3d
arr3d[0]
old_values = arr3d[0].copy()
arr3d[0] = 42
arr3d
arr3d[0] = old_values
arr3d
arr3d[1, 0]
x = arr3d[1]
x
x[0]
```
#### Indexing with slices
```
arr
arr[1:6]
arr2d
arr2d[:2]
arr2d[:2, 1:]
arr2d[1, :2]
arr2d[:2, 2]
arr2d[:, :1]
arr2d[:2, 1:] = 0
arr2d
```
### Boolean Indexing
```
names = np.array(['Bob', 'Joe', 'Will', 'Bob', 'Will', 'Joe', 'Joe'])
data = np.random.randn(7, 4)
names
data
names == 'Bob'
data[names == 'Bob']
data[names == 'Bob', 2:]
data[names == 'Bob', 3]
names != 'Bob'
data[~(names == 'Bob')]
cond = names == 'Bob'
data[~cond]
mask = (names == 'Bob') | (names == 'Will')
mask
data[mask]
data[data < 0] = 0
data
data[names != 'Joe'] = 7
data
```
### Fancy Indexing
```
arr = np.empty((8, 4))
for i in range(8):
arr[i] = i
arr
arr[[4, 3, 0, 6]]
arr[[-3, -5, -7]]
arr = np.arange(32).reshape((8, 4))
arr
arr[[1, 5, 7, 2], [0, 3, 1, 2]]
arr[[1, 5, 7, 2]][:, [0, 3, 1, 2]]
```
### Transposing Arrays and Swapping Axes
```
arr = np.arange(15).reshape((3, 5))
arr
arr.T
arr = np.random.randn(6, 3)
arr
np.dot(arr.T, arr)
arr = np.arange(16).reshape((2, 2, 4))
arr
arr.transpose((1, 0, 2))
arr
arr.swapaxes(1, 2)
```
## Universal Functions: Fast Element-Wise Array Functions
```
arr = np.arange(10)
arr
np.sqrt(arr)
np.exp(arr)
x = np.random.randn(8)
y = np.random.randn(8)
x
y
np.maximum(x, y)
arr = np.random.randn(7) * 5
arr
remainder, whole_part = np.modf(arr)
remainder
whole_part
arr
np.sqrt(arr)
np.sqrt(arr, arr)
arr
```
## Array-Oriented Programming with Arrays
```
points = np.arange(-5, 5, 0.01) # 1000 equally spaced points
xs, ys = np.meshgrid(points, points)
ys
z = np.sqrt(xs ** 2 + ys ** 2)
z
import matplotlib.pyplot as plt
plt.imshow(z, cmap=plt.cm.gray); plt.colorbar()
plt.title("Image plot of $\sqrt{x^2 + y^2}$ for a grid of values")
plt.draw()
plt.close('all')
```
### Expressing Conditional Logic as Array Operations
```
xarr = np.array([1.1, 1.2, 1.3, 1.4, 1.5])
yarr = np.array([2.1, 2.2, 2.3, 2.4, 2.5])
cond = np.array([True, False, True, True, False])
result = [(x if c else y)
for x, y, c in zip(xarr, yarr, cond)]
result
result = np.where(cond, xarr, yarr)
result
arr = np.random.randn(4, 4)
arr
arr > 0
np.where(arr > 0, 2, -2)
np.where(arr > 0, 2, arr) # set only positive values to 2
```
### Mathematical and Statistical Methods
```
arr = np.random.randn(5, 4)
arr
arr.mean()
np.mean(arr)
arr.sum()
arr.mean(axis=1)
arr.sum(axis=0)
arr = np.array([0, 1, 2, 3, 4, 5, 6, 7])
arr.cumsum()
arr = np.array([[0, 1, 2], [3, 4, 5], [6, 7, 8]])
arr
arr.cumsum(axis=0)
arr.cumprod(axis=1)
```
### Methods for Boolean Arrays
```
arr = np.random.randn(100)
(arr > 0).sum() # Number of positive values
bools = np.array([False, False, True, False])
bools.any()
bools.all()
```
### Sorting
```
arr = np.random.randn(6)
arr
arr.sort()
arr
arr = np.random.randn(5, 3)
arr
arr.sort(1)
arr
large_arr = np.random.randn(1000)
large_arr.sort()
large_arr[int(0.05 * len(large_arr))] # 5% quantile
```
### Unique and Other Set Logic
```
names = np.array(['Bob', 'Joe', 'Will', 'Bob', 'Will', 'Joe', 'Joe'])
np.unique(names)
ints = np.array([3, 3, 3, 2, 2, 1, 1, 4, 4])
np.unique(ints)
sorted(set(names))
values = np.array([6, 0, 0, 3, 2, 5, 6])
np.in1d(values, [2, 3, 6])
```
## File Input and Output with Arrays
```
arr = np.arange(10)
np.save('some_array', arr)
np.load('some_array.npy')
np.savez('array_archive.npz', a=arr, b=arr)
arch = np.load('array_archive.npz')
arch['b']
np.savez_compressed('arrays_compressed.npz', a=arr, b=arr)
!rm some_array.npy
!rm array_archive.npz
!rm arrays_compressed.npz
```
## Linear Algebra
```
x = np.array([[1., 2., 3.], [4., 5., 6.]])
y = np.array([[6., 23.], [-1, 7], [8, 9]])
x
y
x.dot(y)
np.dot(x, y)
np.dot(x, np.ones(3))
x @ np.ones(3)
from numpy.linalg import inv, qr
X = np.random.randn(5, 5)
mat = X.T.dot(X)
inv(mat)
mat.dot(inv(mat))
q, r = qr(mat)
r
```
## Pseudorandom Number Generation
```
samples = np.random.normal(size=(4, 4))
samples
from random import normalvariate
N = 1000000
%timeit samples = [normalvariate(0, 1) for _ in range(N)]
%timeit np.random.normal(size=N)
np.random.seed(1234)
rng = np.random.RandomState(1234)
rng.randn(10)
```
## Example: Random Walks
```
import random
position = 0
walk = [position]
steps = 1000
for i in range(steps):
step = 1 if random.randint(0, 1) else -1
position += step
walk.append(position)
plt.figure()
plt.plot(walk[:100])
np.random.seed(12345)
nsteps = 1000
draws = np.random.randint(0, 2, size=nsteps)
steps = np.where(draws > 0, 1, -1)
walk = steps.cumsum()
walk.min()
walk.max()
(np.abs(walk) >= 10).argmax()
```
### Simulating Many Random Walks at Once
```
nwalks = 5000
nsteps = 1000
draws = np.random.randint(0, 2, size=(nwalks, nsteps)) # 0 or 1
steps = np.where(draws > 0, 1, -1)
walks = steps.cumsum(1)
walks
walks.max()
walks.min()
hits30 = (np.abs(walks) >= 30).any(1)
hits30
hits30.sum() # Number that hit 30 or -30
crossing_times = (np.abs(walks[hits30]) >= 30).argmax(1)
crossing_times.mean()
steps = np.random.normal(loc=0, scale=0.25,
size=(nwalks, nsteps))
```
## Conclusion
|
github_jupyter
|
import numpy as np
np.random.seed(12345)
import matplotlib.pyplot as plt
plt.rc('figure', figsize=(10, 6))
np.set_printoptions(precision=4, suppress=True)
import numpy as np
my_arr = np.arange(1000000)
my_list = list(range(1000000))
%time for _ in range(10): my_arr2 = my_arr * 2
%time for _ in range(10): my_list2 = [x * 2 for x in my_list]
import numpy as np
# Generate some random data
data = np.random.randn(2, 3)
data
data * 10
data + data
data.shape
data.dtype
data1 = [6, 7.5, 8, 0, 1]
arr1 = np.array(data1)
arr1
data2 = [[1, 2, 3, 4], [5, 6, 7, 8]]
arr2 = np.array(data2)
arr2
arr2.ndim
arr2.shape
arr1.dtype
arr2.dtype
np.zeros(10)
np.zeros((3, 6))
np.empty((2, 3, 2))
np.arange(15)
arr1 = np.array([1, 2, 3], dtype=np.float64)
arr2 = np.array([1, 2, 3], dtype=np.int32)
arr1.dtype
arr2.dtype
arr = np.array([1, 2, 3, 4, 5])
arr.dtype
float_arr = arr.astype(np.float64)
float_arr.dtype
arr = np.array([3.7, -1.2, -2.6, 0.5, 12.9, 10.1])
arr
arr.astype(np.int32)
numeric_strings = np.array(['1.25', '-9.6', '42'], dtype=np.string_)
numeric_strings.astype(float)
int_array = np.arange(10)
calibers = np.array([.22, .270, .357, .380, .44, .50], dtype=np.float64)
int_array.astype(calibers.dtype)
empty_uint32 = np.empty(8, dtype='u4')
empty_uint32
arr = np.array([[1., 2., 3.], [4., 5., 6.]])
arr
arr * arr
arr - arr
1 / arr
arr ** 0.5
arr2 = np.array([[0., 4., 1.], [7., 2., 12.]])
arr2
arr2 > arr
arr = np.arange(10)
arr
arr[5]
arr[5:8]
arr[5:8] = 12
arr
arr_slice = arr[5:8]
arr_slice
arr_slice[1] = 12345
arr
arr_slice[:] = 64
arr
arr2d = np.array([[1, 2, 3], [4, 5, 6], [7, 8, 9]])
arr2d[2]
arr2d[0][2]
arr2d[0, 2]
arr3d = np.array([[[1, 2, 3], [4, 5, 6]], [[7, 8, 9], [10, 11, 12]]])
arr3d
arr3d[0]
old_values = arr3d[0].copy()
arr3d[0] = 42
arr3d
arr3d[0] = old_values
arr3d
arr3d[1, 0]
x = arr3d[1]
x
x[0]
arr
arr[1:6]
arr2d
arr2d[:2]
arr2d[:2, 1:]
arr2d[1, :2]
arr2d[:2, 2]
arr2d[:, :1]
arr2d[:2, 1:] = 0
arr2d
names = np.array(['Bob', 'Joe', 'Will', 'Bob', 'Will', 'Joe', 'Joe'])
data = np.random.randn(7, 4)
names
data
names == 'Bob'
data[names == 'Bob']
data[names == 'Bob', 2:]
data[names == 'Bob', 3]
names != 'Bob'
data[~(names == 'Bob')]
cond = names == 'Bob'
data[~cond]
mask = (names == 'Bob') | (names == 'Will')
mask
data[mask]
data[data < 0] = 0
data
data[names != 'Joe'] = 7
data
arr = np.empty((8, 4))
for i in range(8):
arr[i] = i
arr
arr[[4, 3, 0, 6]]
arr[[-3, -5, -7]]
arr = np.arange(32).reshape((8, 4))
arr
arr[[1, 5, 7, 2], [0, 3, 1, 2]]
arr[[1, 5, 7, 2]][:, [0, 3, 1, 2]]
arr = np.arange(15).reshape((3, 5))
arr
arr.T
arr = np.random.randn(6, 3)
arr
np.dot(arr.T, arr)
arr = np.arange(16).reshape((2, 2, 4))
arr
arr.transpose((1, 0, 2))
arr
arr.swapaxes(1, 2)
arr = np.arange(10)
arr
np.sqrt(arr)
np.exp(arr)
x = np.random.randn(8)
y = np.random.randn(8)
x
y
np.maximum(x, y)
arr = np.random.randn(7) * 5
arr
remainder, whole_part = np.modf(arr)
remainder
whole_part
arr
np.sqrt(arr)
np.sqrt(arr, arr)
arr
points = np.arange(-5, 5, 0.01) # 1000 equally spaced points
xs, ys = np.meshgrid(points, points)
ys
z = np.sqrt(xs ** 2 + ys ** 2)
z
import matplotlib.pyplot as plt
plt.imshow(z, cmap=plt.cm.gray); plt.colorbar()
plt.title("Image plot of $\sqrt{x^2 + y^2}$ for a grid of values")
plt.draw()
plt.close('all')
xarr = np.array([1.1, 1.2, 1.3, 1.4, 1.5])
yarr = np.array([2.1, 2.2, 2.3, 2.4, 2.5])
cond = np.array([True, False, True, True, False])
result = [(x if c else y)
for x, y, c in zip(xarr, yarr, cond)]
result
result = np.where(cond, xarr, yarr)
result
arr = np.random.randn(4, 4)
arr
arr > 0
np.where(arr > 0, 2, -2)
np.where(arr > 0, 2, arr) # set only positive values to 2
arr = np.random.randn(5, 4)
arr
arr.mean()
np.mean(arr)
arr.sum()
arr.mean(axis=1)
arr.sum(axis=0)
arr = np.array([0, 1, 2, 3, 4, 5, 6, 7])
arr.cumsum()
arr = np.array([[0, 1, 2], [3, 4, 5], [6, 7, 8]])
arr
arr.cumsum(axis=0)
arr.cumprod(axis=1)
arr = np.random.randn(100)
(arr > 0).sum() # Number of positive values
bools = np.array([False, False, True, False])
bools.any()
bools.all()
arr = np.random.randn(6)
arr
arr.sort()
arr
arr = np.random.randn(5, 3)
arr
arr.sort(1)
arr
large_arr = np.random.randn(1000)
large_arr.sort()
large_arr[int(0.05 * len(large_arr))] # 5% quantile
names = np.array(['Bob', 'Joe', 'Will', 'Bob', 'Will', 'Joe', 'Joe'])
np.unique(names)
ints = np.array([3, 3, 3, 2, 2, 1, 1, 4, 4])
np.unique(ints)
sorted(set(names))
values = np.array([6, 0, 0, 3, 2, 5, 6])
np.in1d(values, [2, 3, 6])
arr = np.arange(10)
np.save('some_array', arr)
np.load('some_array.npy')
np.savez('array_archive.npz', a=arr, b=arr)
arch = np.load('array_archive.npz')
arch['b']
np.savez_compressed('arrays_compressed.npz', a=arr, b=arr)
!rm some_array.npy
!rm array_archive.npz
!rm arrays_compressed.npz
x = np.array([[1., 2., 3.], [4., 5., 6.]])
y = np.array([[6., 23.], [-1, 7], [8, 9]])
x
y
x.dot(y)
np.dot(x, y)
np.dot(x, np.ones(3))
x @ np.ones(3)
from numpy.linalg import inv, qr
X = np.random.randn(5, 5)
mat = X.T.dot(X)
inv(mat)
mat.dot(inv(mat))
q, r = qr(mat)
r
samples = np.random.normal(size=(4, 4))
samples
from random import normalvariate
N = 1000000
%timeit samples = [normalvariate(0, 1) for _ in range(N)]
%timeit np.random.normal(size=N)
np.random.seed(1234)
rng = np.random.RandomState(1234)
rng.randn(10)
import random
position = 0
walk = [position]
steps = 1000
for i in range(steps):
step = 1 if random.randint(0, 1) else -1
position += step
walk.append(position)
plt.figure()
plt.plot(walk[:100])
np.random.seed(12345)
nsteps = 1000
draws = np.random.randint(0, 2, size=nsteps)
steps = np.where(draws > 0, 1, -1)
walk = steps.cumsum()
walk.min()
walk.max()
(np.abs(walk) >= 10).argmax()
nwalks = 5000
nsteps = 1000
draws = np.random.randint(0, 2, size=(nwalks, nsteps)) # 0 or 1
steps = np.where(draws > 0, 1, -1)
walks = steps.cumsum(1)
walks
walks.max()
walks.min()
hits30 = (np.abs(walks) >= 30).any(1)
hits30
hits30.sum() # Number that hit 30 or -30
crossing_times = (np.abs(walks[hits30]) >= 30).argmax(1)
crossing_times.mean()
steps = np.random.normal(loc=0, scale=0.25,
size=(nwalks, nsteps))
| 0.294012 | 0.960175 |
# Chicago Crime Prediction Pipeline
An example notebook that demonstrates how to:
* Download data from BigQuery
* Create a Kubeflow pipeline
* Include Google Cloud AI Platform components to train and deploy the model in the pipeline
* Submit a job for execution
The model forecasts how many crimes are expected to be reported the next day, based on how many were reported over the previous `n` days.
## Imports
```
%%capture
# Install the SDK (Uncomment the code if the SDK is not installed before)
!python3 -m pip install 'kfp>=0.1.31' --quiet
!python3 -m pip install pandas --upgrade -q
import json
import kfp
import kfp.components as comp
import kfp.dsl as dsl
import kfp.gcp as gcp
import pandas as pd
import time
```
## Pipeline
### Constants
```
# Required Parameters
project_id = '<ADD GCP PROJECT HERE>'
output = 'gs://<ADD STORAGE LOCATION HERE>' # No ending slash
# Optional Parameters
REGION = 'us-central1'
RUNTIME_VERSION = '1.13'
PACKAGE_URIS=json.dumps(['gs://chicago-crime/chicago_crime_trainer-0.0.tar.gz'])
TRAINER_OUTPUT_GCS_PATH = output + '/train/output/' + str(int(time.time())) + '/'
DATA_GCS_PATH = output + '/reports.csv'
PYTHON_MODULE = 'trainer.task'
PIPELINE_NAME = 'Chicago Crime Prediction'
PIPELINE_FILENAME_PREFIX = 'chicago'
PIPELINE_DESCRIPTION = ''
MODEL_NAME = 'chicago_pipeline_model' + str(int(time.time()))
MODEL_VERSION = 'chicago_pipeline_model_v1' + str(int(time.time()))
```
### Download data
Define a download function that uses the BigQuery component
```
bigquery_query_op = comp.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/4e7e6e866c1256e641b0c3effc55438e6e4b30f6/components/gcp/bigquery/query/component.yaml')
QUERY = """
SELECT count(*) as count, TIMESTAMP_TRUNC(date, DAY) as day
FROM `bigquery-public-data.chicago_crime.crime`
GROUP BY day
ORDER BY day
"""
def download(project_id, data_gcs_path):
return bigquery_query_op(
query=QUERY,
project_id=project_id,
output_gcs_path=data_gcs_path
).apply(
gcp.use_gcp_secret('user-gcp-sa')
)
```
### Train the model
Run training code that will pre-process the data and then submit a training job to the AI Platform.
```
mlengine_train_op = comp.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/4e7e6e866c1256e641b0c3effc55438e6e4b30f6/components/gcp/ml_engine/train/component.yaml')
def train(project_id,
trainer_args,
package_uris,
trainer_output_gcs_path,
gcs_working_dir,
region,
python_module,
runtime_version):
return mlengine_train_op(
project_id=project_id,
python_module=python_module,
package_uris=package_uris,
region=region,
args=trainer_args,
job_dir=trainer_output_gcs_path,
runtime_version=runtime_version
).apply(gcp.use_gcp_secret('user-gcp-sa'))
```
### Deploy model
Deploy the model with the ID given from the training step
```
mlengine_deploy_op = comp.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/4e7e6e866c1256e641b0c3effc55438e6e4b30f6/components/gcp/ml_engine/deploy/component.yaml')
def deploy(
project_id,
model_uri,
model_id,
model_version,
runtime_version):
return mlengine_deploy_op(
model_uri=model_uri,
project_id=project_id,
model_id=model_id,
version_id=model_version,
runtime_version=runtime_version,
replace_existing_version=True,
set_default=True).apply(gcp.use_gcp_secret('user-gcp-sa'))
```
### Define pipeline
```
@dsl.pipeline(
name=PIPELINE_NAME,
description=PIPELINE_DESCRIPTION
)
def pipeline(
data_gcs_path=DATA_GCS_PATH,
gcs_working_dir=output,
project_id=project_id,
python_module=PYTHON_MODULE,
region=REGION,
runtime_version=RUNTIME_VERSION,
package_uris=PACKAGE_URIS,
trainer_output_gcs_path=TRAINER_OUTPUT_GCS_PATH,
):
download_task = download(project_id,
data_gcs_path)
train_task = train(project_id,
json.dumps(
['--data-file-url',
'%s' % download_task.outputs['output_gcs_path'],
'--job-dir',
output]
),
package_uris,
trainer_output_gcs_path,
gcs_working_dir,
region,
python_module,
runtime_version)
deploy_task = deploy(project_id,
train_task.outputs['job_dir'],
MODEL_NAME,
MODEL_VERSION,
runtime_version)
return True
# Reference for invocation later
pipeline_func = pipeline
```
### Submit the pipeline for execution
```
pipeline = kfp.Client().create_run_from_pipeline_func(pipeline, arguments={})
```
### Wait for the pipeline to finish
```
pipeline.wait_for_run_completion(timeout=1800)
```
### Clean models
```
!gcloud ml-engine versions delete $MODEL_VERSION --model $MODEL_NAME
!gcloud ml-engine models delete $MODEL_NAME
```
|
github_jupyter
|
%%capture
# Install the SDK (Uncomment the code if the SDK is not installed before)
!python3 -m pip install 'kfp>=0.1.31' --quiet
!python3 -m pip install pandas --upgrade -q
import json
import kfp
import kfp.components as comp
import kfp.dsl as dsl
import kfp.gcp as gcp
import pandas as pd
import time
# Required Parameters
project_id = '<ADD GCP PROJECT HERE>'
output = 'gs://<ADD STORAGE LOCATION HERE>' # No ending slash
# Optional Parameters
REGION = 'us-central1'
RUNTIME_VERSION = '1.13'
PACKAGE_URIS=json.dumps(['gs://chicago-crime/chicago_crime_trainer-0.0.tar.gz'])
TRAINER_OUTPUT_GCS_PATH = output + '/train/output/' + str(int(time.time())) + '/'
DATA_GCS_PATH = output + '/reports.csv'
PYTHON_MODULE = 'trainer.task'
PIPELINE_NAME = 'Chicago Crime Prediction'
PIPELINE_FILENAME_PREFIX = 'chicago'
PIPELINE_DESCRIPTION = ''
MODEL_NAME = 'chicago_pipeline_model' + str(int(time.time()))
MODEL_VERSION = 'chicago_pipeline_model_v1' + str(int(time.time()))
bigquery_query_op = comp.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/4e7e6e866c1256e641b0c3effc55438e6e4b30f6/components/gcp/bigquery/query/component.yaml')
QUERY = """
SELECT count(*) as count, TIMESTAMP_TRUNC(date, DAY) as day
FROM `bigquery-public-data.chicago_crime.crime`
GROUP BY day
ORDER BY day
"""
def download(project_id, data_gcs_path):
return bigquery_query_op(
query=QUERY,
project_id=project_id,
output_gcs_path=data_gcs_path
).apply(
gcp.use_gcp_secret('user-gcp-sa')
)
mlengine_train_op = comp.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/4e7e6e866c1256e641b0c3effc55438e6e4b30f6/components/gcp/ml_engine/train/component.yaml')
def train(project_id,
trainer_args,
package_uris,
trainer_output_gcs_path,
gcs_working_dir,
region,
python_module,
runtime_version):
return mlengine_train_op(
project_id=project_id,
python_module=python_module,
package_uris=package_uris,
region=region,
args=trainer_args,
job_dir=trainer_output_gcs_path,
runtime_version=runtime_version
).apply(gcp.use_gcp_secret('user-gcp-sa'))
mlengine_deploy_op = comp.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/4e7e6e866c1256e641b0c3effc55438e6e4b30f6/components/gcp/ml_engine/deploy/component.yaml')
def deploy(
project_id,
model_uri,
model_id,
model_version,
runtime_version):
return mlengine_deploy_op(
model_uri=model_uri,
project_id=project_id,
model_id=model_id,
version_id=model_version,
runtime_version=runtime_version,
replace_existing_version=True,
set_default=True).apply(gcp.use_gcp_secret('user-gcp-sa'))
@dsl.pipeline(
name=PIPELINE_NAME,
description=PIPELINE_DESCRIPTION
)
def pipeline(
data_gcs_path=DATA_GCS_PATH,
gcs_working_dir=output,
project_id=project_id,
python_module=PYTHON_MODULE,
region=REGION,
runtime_version=RUNTIME_VERSION,
package_uris=PACKAGE_URIS,
trainer_output_gcs_path=TRAINER_OUTPUT_GCS_PATH,
):
download_task = download(project_id,
data_gcs_path)
train_task = train(project_id,
json.dumps(
['--data-file-url',
'%s' % download_task.outputs['output_gcs_path'],
'--job-dir',
output]
),
package_uris,
trainer_output_gcs_path,
gcs_working_dir,
region,
python_module,
runtime_version)
deploy_task = deploy(project_id,
train_task.outputs['job_dir'],
MODEL_NAME,
MODEL_VERSION,
runtime_version)
return True
# Reference for invocation later
pipeline_func = pipeline
pipeline = kfp.Client().create_run_from_pipeline_func(pipeline, arguments={})
pipeline.wait_for_run_completion(timeout=1800)
!gcloud ml-engine versions delete $MODEL_VERSION --model $MODEL_NAME
!gcloud ml-engine models delete $MODEL_NAME
| 0.483892 | 0.826081 |
```
%load_ext autoreload
%autoreload 2
%matplotlib inline
from pathlib2 import Path
from IPython.core.debugger import set_trace
from fastai import datasets
import pickle, gzip, math, torch, matplotlib as mpl
import matplotlib.pyplot as plt
from torch import tensor
torch.cuda.set_device(0)
```
### Matrix multiplication
#### with elementwise operations
```
a = tensor([[1, 2, 3], [4, 5, 6], [7, 8, 9]]).to('cuda')
print(a.device, a.type())
b = torch.randint(high=5, size=(3,3)).to('cuda')
def matmul(a,b):
ar,ac = a.shape
br,bc = b.shape
assert ar==bc
c = torch.zeros(ar,bc).to('cuda')
for i in range(ar):
for j in range(bc):
c[i,j] = (a[i,:]*b[:,j]).sum(dim=0)
return c
%timeit -n 10 _=matmul(a,b)
```
#### with broadcasting
Broadcasting only makes sense if one is the inputs needs it, so we will add another case 'c'. We will still test the performance on the square matrices
```
c = tensor([1, 2, 3]).to('cuda')
def matmul_br(a,b):
ar,ac = a.shape
br,bc = b.shape
assert ac==br
c = torch.zeros(ar, bc).to('cuda')
for i in range(ar):
c[i] = (a[i,:].unsqueeze(-1) * b).sum(dim=0)
return c
%timeit -n 10 _=matmul_br(a,c[:,None])
%timeit -n 10 _=matmul_br(a,b)
```
After re-running this notebook several times, I found that the square matrices are always faster and has less std
```
assert (matmul(a,b) == matmul_br(a,b)).all()
```
We will always check above condition for all variants of matmul_* with matmul being the base case
#### with einsum
Cuda only supports floating point so we will push einsum to cuda
```
def matmul_es(a,b): return torch.einsum('ik,kj->ij', a, b).to('cuda')
a = a.float(); b = b.float(); c = c.float()
%timeit -n 10 _=matmul_es(a,b)
```
__We did push the einsum to gpu but it still does the compute on CPU, WHY?__
```
%timeit -n 10 _=matmul_es(a,c[:,None])
```
__Why is this on GPU? The CPU cache issue is with a and b only!__
```
assert (matmul(a,b) == matmul_es(a,b)).all()
```
#### with pytorch
```
%timeit -n 10 _=a.matmul(b)
%timeit -n 10 _=a.matmul(c)
assert (a.matmul(b) == matmul(a,b)).all()
assert (a.matmul(b) == matmul_es(a,b)).all()
%timeit -n 10 _=a@b
%timeit -n 10 _=a@c
assert (a@b == matmul_es(a,b)).all()
```
It is interesting to observe that matmul_br still shows the trend with square matrices multiplying faster, we cannot compare for matmul_es because of inconsistent runs between GPU and CPU
But pytorch's implementation is faster for non square matrices, is it because it is greedy with its memory footprint and access for calculations?
__TODO: Need to understand this in details__
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
%matplotlib inline
from pathlib2 import Path
from IPython.core.debugger import set_trace
from fastai import datasets
import pickle, gzip, math, torch, matplotlib as mpl
import matplotlib.pyplot as plt
from torch import tensor
torch.cuda.set_device(0)
a = tensor([[1, 2, 3], [4, 5, 6], [7, 8, 9]]).to('cuda')
print(a.device, a.type())
b = torch.randint(high=5, size=(3,3)).to('cuda')
def matmul(a,b):
ar,ac = a.shape
br,bc = b.shape
assert ar==bc
c = torch.zeros(ar,bc).to('cuda')
for i in range(ar):
for j in range(bc):
c[i,j] = (a[i,:]*b[:,j]).sum(dim=0)
return c
%timeit -n 10 _=matmul(a,b)
c = tensor([1, 2, 3]).to('cuda')
def matmul_br(a,b):
ar,ac = a.shape
br,bc = b.shape
assert ac==br
c = torch.zeros(ar, bc).to('cuda')
for i in range(ar):
c[i] = (a[i,:].unsqueeze(-1) * b).sum(dim=0)
return c
%timeit -n 10 _=matmul_br(a,c[:,None])
%timeit -n 10 _=matmul_br(a,b)
assert (matmul(a,b) == matmul_br(a,b)).all()
def matmul_es(a,b): return torch.einsum('ik,kj->ij', a, b).to('cuda')
a = a.float(); b = b.float(); c = c.float()
%timeit -n 10 _=matmul_es(a,b)
%timeit -n 10 _=matmul_es(a,c[:,None])
assert (matmul(a,b) == matmul_es(a,b)).all()
%timeit -n 10 _=a.matmul(b)
%timeit -n 10 _=a.matmul(c)
assert (a.matmul(b) == matmul(a,b)).all()
assert (a.matmul(b) == matmul_es(a,b)).all()
%timeit -n 10 _=a@b
%timeit -n 10 _=a@c
assert (a@b == matmul_es(a,b)).all()
| 0.599368 | 0.882276 |
# Radial Beam Sampling
```
!pip install --upgrade pip
!TMPDIR=/var/tmp pip install tensorflow-gpu==2.8.0
!TMPDIR=/var/tmp pip install tensorflow_datasets
!TMPDIR=/var/tmp pip install tensorflow_addons
!TMPDIR=/var/tmp pip install matplotlib
!TMPDIR=/var/tmp pip install numpy --upgrade
import os
os.environ["CUDA_VISIBLE_DEVICES"] = str(0)
import math
import copy
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
import tensorflow as tf
import tensorflow_datasets as tfds
import tensorflow_addons as tfa
```
### Load the dataset
```
dataset_name = 'coil100'
dataset = tfds.load(dataset_name, split='train', shuffle_files=True)
```
### Specify Beam Paramters
```
# beam length (D)
target_size = 128 // 2
# number of beams (|B|)
beam_set_size = 32
# zero padding (delta)
margin_padding = math.ceil(target_size * (math.sqrt(2) - 1))
# image size (W,H)
img_size = int(dataset.element_spec['image'].shape[0])
# new image size (W+delta, H+delta)
w_dim = img_size + margin_padding
h_dim = img_size + margin_padding
# center (start of beams) (c)
center = (w_dim // 2, h_dim // 2)
```
### Compute Endpoints of Beams
By iterating along all four edges and collect endpoints.
```
vecs_per_quarter = (beam_set_size + 4) // 4
w_atom = math.floor(w_dim / (vecs_per_quarter - 1))
h_atom = math.floor(h_dim / (vecs_per_quarter - 1))
# upper border (left to right)
endpoints = [(i * w_atom, 0) for i in range(vecs_per_quarter - 1)] + [(w_dim - 1, 0)]
# right border (top to bottom)
endpoints += [(w_dim - 1, i * h_atom) for i in range(1, vecs_per_quarter - 1)]
# lower border (right to left)
endpoints += [(i * w_atom, h_dim - 1) for i in range(vecs_per_quarter - 1, 0, -1)]
# left border (bottom to top)
endpoints += [(0, i * h_atom) for i in range(vecs_per_quarter - 1, 0, -1)]
endpoints = np.array(endpoints)
```
### Compute resulting angles/directions of beams
```
# atan2
def angle_between(p1, p2, degree=False, gpu=False):
if gpu:
ang1 = tf.math.atan2(*p1[::-1])
ang2 = tf.math.atan2(*p2[::-1])
else:
ang1 = np.arctan2(*p1[::-1])
ang2 = np.arctan2(*p2[::-1])
angle = (ang1 - ang2) % (2 * np.pi)
if degree:
return np.rad2deg(angle)
return angle
angles = [0., ]
# compute vector pointing from center to endpoint
# by translating the image to the center as the new coordinate origin
_endpoints = copy.deepcopy(endpoints)
for i in range(len(_endpoints)):
_endpoints[i][0] -= center[0]
_endpoints[i][1] = center[1] - _endpoints[i][1]
# compute the angle between two beams
for i in range(len(_endpoints) - 1):
relative_angle = angle_between(_endpoints[i], _endpoints[i+1], degree=True)
angles.append(angles[-1] + relative_angle)
```
### Bresenham to estimate pixel positions
```
def bresenham(start, end, length=None):
# Setup initial conditions
x1, y1 = start
x2, y2 = end
dx = x2 - x1
dy = y2 - y1
# Determine how steep the line is
is_steep = abs(dy) > abs(dx)
# Rotate line
if is_steep:
x1, y1 = y1, x1
x2, y2 = y2, x2
# Swap start and end points if necessary and store swap state
swapped = False
if x1 > x2:
x1, x2 = x2, x1
y1, y2 = y2, y1
swapped = True
# Recalculate differentials
dx = x2 - x1
dy = y2 - y1
# Calculate error
error = int(dx / 2.0)
ystep = 1 if y1 < y2 else -1
# Iterate over bounding box generating points between start and end
y = y1
points, left, right = [], [], []
for x in range(x1, x2 + 1):
coord = [y, x] if is_steep else [x, y]
points.append(coord)
proxy_a = [coord[0] - 1, coord[1]] if is_steep else [coord[0], coord[1] - 1]
proxy_b = [coord[0] + 1, coord[1]] if is_steep else [coord[0], coord[1] + 1]
if swapped or is_steep:
left.append(proxy_b)
right.append(proxy_a)
else:
left.append(proxy_a)
right.append(proxy_b)
error -= abs(dy)
if error < 0:
y += ystep
error += dx
# Reverse the list if the coordinates were swapped
if swapped:
points.reverse()
left.reverse()
right.reverse()
if length is not None:
return np.array(left)[:length], np.array(points)[:length], np.array(right)[:length]
return np.array(left), np.array(points), np.array(right)
lines = [bresenham(center, endpoint, length=target_size) for endpoint in endpoints]
# sanity check and validate
lines = tf.clip_by_value(tf.cast(lines, tf.int32), 0, w_dim - 1)
```
### sample radial beams from images in dataset
```
# add padding
dataset = dataset.map(lambda x: {'image': tf.pad(x['image'], [
[tf.math.maximum(0, (h_dim - x['image'].shape[1]) // 2),
tf.math.maximum(0, (h_dim - x['image'].shape[1]) // 2)],
[tf.math.maximum(0, (w_dim - x['image'].shape[0]) // 2),
tf.math.maximum(0, (w_dim - x['image'].shape[0]) // 2)],
[0, 0]], "CONSTANT")})
```
### Add arbitrary rotated images
```
def rotate_image(image, angles: list, angle=None):
if angle is not None:
return {'rotated': tfa.image.rotate(image, angle, interpolation='bilinear'),
'angle': angle}
# Dataset does not execute eagerly, so the python randint is executed once to create the
# graph and the returned value is used.
rnd_idx = int(tf.random.uniform([], minval=0, maxval=len(angles)))
# 360 - angle since `tfa.image.rotate` will rotate counter clockwise
# but the angle matrix in the model is build for clockwise rotations
angle = (angles[rnd_idx] / 180.) * math.pi
# angle = 2 * math.pi - angle
return {'rotated': tfa.image.rotate(image, angle, interpolation='bilinear'),
'angle': tf.one_hot(rnd_idx, len(angles))}
# rotate images
angle = tf.random.uniform(shape=[dataset.cardinality()], minval=0, maxval=2*math.pi)
dataset = tf.data.Dataset.zip((dataset, tf.data.Dataset.from_tensor_slices({'angle': angle})))
dataset = dataset.map(lambda x, y: {**x, **rotate_image(x['image'], angles, angle=y['angle'])})
# map vector evaluations
dataset = dataset.map(lambda x: {**x, 'beam': tf.gather_nd(x['image'], lines),
'beam_rot': tf.gather_nd(x['rotated'], lines)})
n_beams, _, n_pixels, n_channels = dataset.element_spec['beam'].shape
```
### Pick one image out of the dataset and plot it
```
img_id = 51
for d, data in enumerate(dataset):
image_vanilla = tf.cast(data['image'], tf.float32) / 255.
image_rotated = tf.cast(data['rotated'], tf.float32) / 255.
if d == img_id:
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].imshow(image_vanilla)
ax[1].imshow(image_rotated)
plt.show()
break
```
### Plot the beams overlay
```
def img_color_beams(ax, image, lines, colors):
image = np.array(image)
for line, color in zip(lines, colors[:, :3] / 255):
image[line[1, :, 0], line[1, :, 1]] = color
image[line[0, :, 0], line[0, :, 1]] = color
image[line[2, :, 0], line[2, :, 1]] = color
ax.imshow(image)
ax.set_xlabel('')
ax.set_ylabel('')
return ax
# unique color codes for each beam
color_codes = np.array([matplotlib.cm.get_cmap('Spectral')(i, bytes=True)
for i in np.linspace(0, 1, n_beams)])
# circle color codes by rotation angle
colors = np.array([np.roll(color_codes, i, axis=0) for i in range(len(angles))])
# plot X0 and Xbeta with color beams as a sanity check
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0] = img_color_beams(ax[0], image_vanilla, lines, colors[0])
ax[1] = img_color_beams(ax[1], image_rotated, lines, colors[1])
plt.show()
```
### Store the dataset
```
splits = [0.8, 0.2, 0.0]
n_train = int(splits[0] * float(dataset.cardinality()))
n_val = int(splits[1] * float(dataset.cardinality()))
n_test = int(splits[2] * float(dataset.cardinality()))
train_dataset = dataset.take(n_train)
val_dataset = dataset.skip(n_train).take(n_val)
test_dataset = dataset.skip(n_train).skip(n_val).take(n_train)
tf.data.experimental.save(train_dataset, 'data/{}_train'.format(dataset_name))
tf.data.experimental.save(val_dataset, 'data/{}_val'.format(dataset_name))
tf.data.experimental.save(test_dataset, 'data/{}_test'.format(dataset_name))
```
|
github_jupyter
|
!pip install --upgrade pip
!TMPDIR=/var/tmp pip install tensorflow-gpu==2.8.0
!TMPDIR=/var/tmp pip install tensorflow_datasets
!TMPDIR=/var/tmp pip install tensorflow_addons
!TMPDIR=/var/tmp pip install matplotlib
!TMPDIR=/var/tmp pip install numpy --upgrade
import os
os.environ["CUDA_VISIBLE_DEVICES"] = str(0)
import math
import copy
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
import tensorflow as tf
import tensorflow_datasets as tfds
import tensorflow_addons as tfa
dataset_name = 'coil100'
dataset = tfds.load(dataset_name, split='train', shuffle_files=True)
# beam length (D)
target_size = 128 // 2
# number of beams (|B|)
beam_set_size = 32
# zero padding (delta)
margin_padding = math.ceil(target_size * (math.sqrt(2) - 1))
# image size (W,H)
img_size = int(dataset.element_spec['image'].shape[0])
# new image size (W+delta, H+delta)
w_dim = img_size + margin_padding
h_dim = img_size + margin_padding
# center (start of beams) (c)
center = (w_dim // 2, h_dim // 2)
vecs_per_quarter = (beam_set_size + 4) // 4
w_atom = math.floor(w_dim / (vecs_per_quarter - 1))
h_atom = math.floor(h_dim / (vecs_per_quarter - 1))
# upper border (left to right)
endpoints = [(i * w_atom, 0) for i in range(vecs_per_quarter - 1)] + [(w_dim - 1, 0)]
# right border (top to bottom)
endpoints += [(w_dim - 1, i * h_atom) for i in range(1, vecs_per_quarter - 1)]
# lower border (right to left)
endpoints += [(i * w_atom, h_dim - 1) for i in range(vecs_per_quarter - 1, 0, -1)]
# left border (bottom to top)
endpoints += [(0, i * h_atom) for i in range(vecs_per_quarter - 1, 0, -1)]
endpoints = np.array(endpoints)
# atan2
def angle_between(p1, p2, degree=False, gpu=False):
if gpu:
ang1 = tf.math.atan2(*p1[::-1])
ang2 = tf.math.atan2(*p2[::-1])
else:
ang1 = np.arctan2(*p1[::-1])
ang2 = np.arctan2(*p2[::-1])
angle = (ang1 - ang2) % (2 * np.pi)
if degree:
return np.rad2deg(angle)
return angle
angles = [0., ]
# compute vector pointing from center to endpoint
# by translating the image to the center as the new coordinate origin
_endpoints = copy.deepcopy(endpoints)
for i in range(len(_endpoints)):
_endpoints[i][0] -= center[0]
_endpoints[i][1] = center[1] - _endpoints[i][1]
# compute the angle between two beams
for i in range(len(_endpoints) - 1):
relative_angle = angle_between(_endpoints[i], _endpoints[i+1], degree=True)
angles.append(angles[-1] + relative_angle)
def bresenham(start, end, length=None):
# Setup initial conditions
x1, y1 = start
x2, y2 = end
dx = x2 - x1
dy = y2 - y1
# Determine how steep the line is
is_steep = abs(dy) > abs(dx)
# Rotate line
if is_steep:
x1, y1 = y1, x1
x2, y2 = y2, x2
# Swap start and end points if necessary and store swap state
swapped = False
if x1 > x2:
x1, x2 = x2, x1
y1, y2 = y2, y1
swapped = True
# Recalculate differentials
dx = x2 - x1
dy = y2 - y1
# Calculate error
error = int(dx / 2.0)
ystep = 1 if y1 < y2 else -1
# Iterate over bounding box generating points between start and end
y = y1
points, left, right = [], [], []
for x in range(x1, x2 + 1):
coord = [y, x] if is_steep else [x, y]
points.append(coord)
proxy_a = [coord[0] - 1, coord[1]] if is_steep else [coord[0], coord[1] - 1]
proxy_b = [coord[0] + 1, coord[1]] if is_steep else [coord[0], coord[1] + 1]
if swapped or is_steep:
left.append(proxy_b)
right.append(proxy_a)
else:
left.append(proxy_a)
right.append(proxy_b)
error -= abs(dy)
if error < 0:
y += ystep
error += dx
# Reverse the list if the coordinates were swapped
if swapped:
points.reverse()
left.reverse()
right.reverse()
if length is not None:
return np.array(left)[:length], np.array(points)[:length], np.array(right)[:length]
return np.array(left), np.array(points), np.array(right)
lines = [bresenham(center, endpoint, length=target_size) for endpoint in endpoints]
# sanity check and validate
lines = tf.clip_by_value(tf.cast(lines, tf.int32), 0, w_dim - 1)
# add padding
dataset = dataset.map(lambda x: {'image': tf.pad(x['image'], [
[tf.math.maximum(0, (h_dim - x['image'].shape[1]) // 2),
tf.math.maximum(0, (h_dim - x['image'].shape[1]) // 2)],
[tf.math.maximum(0, (w_dim - x['image'].shape[0]) // 2),
tf.math.maximum(0, (w_dim - x['image'].shape[0]) // 2)],
[0, 0]], "CONSTANT")})
def rotate_image(image, angles: list, angle=None):
if angle is not None:
return {'rotated': tfa.image.rotate(image, angle, interpolation='bilinear'),
'angle': angle}
# Dataset does not execute eagerly, so the python randint is executed once to create the
# graph and the returned value is used.
rnd_idx = int(tf.random.uniform([], minval=0, maxval=len(angles)))
# 360 - angle since `tfa.image.rotate` will rotate counter clockwise
# but the angle matrix in the model is build for clockwise rotations
angle = (angles[rnd_idx] / 180.) * math.pi
# angle = 2 * math.pi - angle
return {'rotated': tfa.image.rotate(image, angle, interpolation='bilinear'),
'angle': tf.one_hot(rnd_idx, len(angles))}
# rotate images
angle = tf.random.uniform(shape=[dataset.cardinality()], minval=0, maxval=2*math.pi)
dataset = tf.data.Dataset.zip((dataset, tf.data.Dataset.from_tensor_slices({'angle': angle})))
dataset = dataset.map(lambda x, y: {**x, **rotate_image(x['image'], angles, angle=y['angle'])})
# map vector evaluations
dataset = dataset.map(lambda x: {**x, 'beam': tf.gather_nd(x['image'], lines),
'beam_rot': tf.gather_nd(x['rotated'], lines)})
n_beams, _, n_pixels, n_channels = dataset.element_spec['beam'].shape
img_id = 51
for d, data in enumerate(dataset):
image_vanilla = tf.cast(data['image'], tf.float32) / 255.
image_rotated = tf.cast(data['rotated'], tf.float32) / 255.
if d == img_id:
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].imshow(image_vanilla)
ax[1].imshow(image_rotated)
plt.show()
break
def img_color_beams(ax, image, lines, colors):
image = np.array(image)
for line, color in zip(lines, colors[:, :3] / 255):
image[line[1, :, 0], line[1, :, 1]] = color
image[line[0, :, 0], line[0, :, 1]] = color
image[line[2, :, 0], line[2, :, 1]] = color
ax.imshow(image)
ax.set_xlabel('')
ax.set_ylabel('')
return ax
# unique color codes for each beam
color_codes = np.array([matplotlib.cm.get_cmap('Spectral')(i, bytes=True)
for i in np.linspace(0, 1, n_beams)])
# circle color codes by rotation angle
colors = np.array([np.roll(color_codes, i, axis=0) for i in range(len(angles))])
# plot X0 and Xbeta with color beams as a sanity check
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0] = img_color_beams(ax[0], image_vanilla, lines, colors[0])
ax[1] = img_color_beams(ax[1], image_rotated, lines, colors[1])
plt.show()
splits = [0.8, 0.2, 0.0]
n_train = int(splits[0] * float(dataset.cardinality()))
n_val = int(splits[1] * float(dataset.cardinality()))
n_test = int(splits[2] * float(dataset.cardinality()))
train_dataset = dataset.take(n_train)
val_dataset = dataset.skip(n_train).take(n_val)
test_dataset = dataset.skip(n_train).skip(n_val).take(n_train)
tf.data.experimental.save(train_dataset, 'data/{}_train'.format(dataset_name))
tf.data.experimental.save(val_dataset, 'data/{}_val'.format(dataset_name))
tf.data.experimental.save(test_dataset, 'data/{}_test'.format(dataset_name))
| 0.601359 | 0.878887 |
# Stand-Alone Self-Attention in Vision Models Summary
> Introducing Self-Attention as a stand-alone layer in Convolutional Architectures by Prajit Ramachandran,Niki Parmar,Ashish Vaswani,Jonathon Shlens,Anselm Levskaya,Irwan Bello
- toc: true
- badges: false
- comments: true
- categories: [jupyter]
- image: images/SATCONV.png
[Paper Link](https://arxiv.org/pdf/1906.05909.pdf)
## What did the authors want to achieve ?
Combat the deficits of Conolutional Architectures, (local connectivity, failing to reason globally) by introducing Attention to as a stand-alone layer. The authors prove that this can be both more accurate and more efficient at the same time. Architectures that are attention only, and a mixed version of convolutional and attention architectures are introduced and compared to the vanilla convolutional implementations.
## Methods
<br/><br/>
### Conv Block deficits
Capturing long range interactions is challenging for convolutions as they do not scale well with large receptives. Since attention has been used to tackle long range dependencies in sequence modeling, since architectures like SE Nets model attention on a chanel wise basis successfully. However in these cases attention was only an add-on to a traditional architecture style. In this paper the authors propose to use attention mechanisms as stand alone layers.
### Self-Attention
Attention was originally introduced in order to allow for summarization from variable length source sentences. Attention focuses on the important parts of the input and thereby can serve as a primary representation learning mechanism and fully replace recurrence. The word self means that it just considers a single context (query,keys and values are extracted from the same image). The breakthrough in this paper is the use of self-attention layers instead of convolutional layers.
In this work, already existing mechanisms are used which are not optimized for the image domain. Therefore it is permutation equivariant and has limited expression capability for vision tasks.
<br/><br/>

<br/><br/>
The process is as follows :
1) given a pixel $x_{i,j} \in R^{d_{in}}$ in positions $ab \in N_{k}(i,j)$ a local kernel with kernel size $k$ is extracted. $x_{i,j}$ is the middle of the kernel, which is called *memory block*. Prior work only performed global attention, which can only be done with a downsized sample as it is very compute expensive.
<br/><br/>
2) Single headed attention is computed :
<br/><br/>
$y_{i,j} = \sum_{ab \in N_{k}(i,j)} softmax_{a,b}(q_{ij}^{T}k_{ab})v_{ab}$
<br/><br/>
where the ${queries}$ $q_{ij} = W_Q x_{ij}$
<br/><br/>
${keys}$ $k_{ab} = W_K x_{ab}$
<br/><br/>
and ${values}$ $v_{ab} = W_V x_{ab}$
<br/><br/>
are linear transformations of the pixel in position and it's neighbors in the kernel.
<br/><br/>
$\texttt{softmax}_{a b}$ is a softmax, which is applied to all logits computed in the neighborhood of $ij$.
$W_Q, W_K, W_V \in \mathbb{R}^{d_{out} \times d_{in}}$ are learned transforms.
<br/><br/>
Local self-attention is similar to convolution in the way that it aggregates spatial information in the neighborhoods, multi attention heads are used to learn unique representations of the input. This is done by partitioning pixel features into $N$ groups and then computing single-headed attention on each one seperately with the transforms $W_Q, W_K, W_V \in \mathbb{R}^{d_{out} \times d_{in}}$. The outputs of the heads are then concatenated.
<br/><br/>
2D relative pose embeddings,relative attention is used :
<br/><br/>
1) relative attention computes relative distances of the pixel to each one in the neighborhood : row $(a-i)$ and column offset $(b-j)$
2) row and column offset are associated with an embedding and concatenated into a vector $r_{a-i,b-j}$
3) Spatial-relative attention is then defined as :
<br/><br/>
$y_{ij} = \sum_{a,b\in N_{k}(i, j)} softmax_{ab}(q_{ij}^{T}k_{ab}+q_{ij}^{T}r_{a-i,b-j})v_{ab}$
<br/><br/>
The logit measuring similarity between the query and an element results from the content of the element and the relative distance of the element from the query. By inlcuding this spatial information, self-attention also has translation equivariance, just like conv layers. Unlike conv layers, self-attentions parameter count is independent of its spatial extent.
The compute cost also grows slower :
For example, if $d_{in} = d_{out} = 128$, a convolution layer with $k = 3$ has the same computational cost as an attention layer with $k = 19$.
Using this as their basis, a fully attentional architecture is created in two steps:
#### Replacing Spatial Convolutions
A spatial conv is defined to have spatial extent k > 1, which also includes 1x1 convolutions. These can be viewed as fully connected layers.
Here the authors want to replace conv blocks in a straightforward way, specificially focusing on ResNet. Therefore the 3x3 convolution in Path B is swapped with a self-attention layer as defined above. All the other blocks are not changed, this might be supobtimal but promises potential improvements using architecture search.
#### Replacing the Convolutional Stem (intial layers of the CNN)
This part focuses on replacing the inital layers, as they are the most compute expensive due to the large input size of the image. In the OG ResNet the input is a 7x7 kernel with stride 2, followed by 3x3 max pooling with stride 2. At the beginning RGB pixels are individually uninformative and information is heavily spatially correlated through low level features such as edges. Edge detectors are difficult to learn for self-attention due to spatial correlation, convs learn these easily through distance based weight parameterization. The authors inject spatially-varying linear transforms into the pointwise 1x1 softmax convolution.
$\tilde{v}_{a b} = \left(\sum_m p(a, b, m) W_V^{m}\right) x_{a b}$
The results is similar to convolutions, weights are learned based on a local neighborhood basis. So in total the stem consists of spatially aware value features, followed by max-pooling. A more detailed explanation of this can be found in the appendix of the paper (page 14/15).
## Results
Implementation details for both classification and object detection are in the appendix.
### ImageNet
The multi head self-attention layer uses a spatial width of k=7 and 8 attention heads. The position-aware attention stem as described above is used.The stem performs self-attention within each 4×4 block of the original image, followed by batch normalization and a 4×4 max pool operation. Results are below :

### Coco Object Detection
Here Retina Net is used with a classification backbone, followed by an FPN, the network has 2 detection heads. Results are in the table below :

We can see that using attention based backbone we can achieve results on par with a conv backbone, but with 22% less parameters. This can be extended by additionaly making the FPN and the detection heads attention-based and thereby reducing paraneter count by 34% and more importantly FLOPS by 39%.
### Where is stand-alone attention most useful ?

Results are schown in the tables above.
#### Stem
The basic results for the stem is that, convolutions perform very well here, as described above self-attention can not easily learn edges due to the high spatial correlation which is captured very well by conv layers though.
#### Full Net
The authors basically state what has been described above, conv layers capture low level features very well, while attention is able to model global relations effectively. Therefore an optimal architecture should contain both attention and convolutional layers.
### Which attention features are important ?
- Effect of spatial extent of self-attention (Table 4) :
The value of spatial extent k should generally be larger (for example k=11), the exact optimal setting depends on hyperaparameter choices.
- Importance of positional information (Table 5 + 6)
3 types of encoding were used : no positional encoding, sinusodial encoing and absolute pixel position. Relativ encoding performs 2% better than absolute one. Removing content-content interaction only descreases accuracy by 0.5%. Therefore the positional encoding seems to be very important and can be a strong focus of their future research.
- Importance of spatially-aware attention stem (Table 7)
Using stand-alone attention in the stem with spatially-aware values, it outperforms vanilla stand-alone attention by 1.4%, while having similar FLOPS. Using a spatial convolution to the values instead of spatially-aware point-wise transformations (see above), leads to more FLOPS and slightly worse results. A future goal of the authors is to unify attention used across the stem and main
|
github_jupyter
|
# Stand-Alone Self-Attention in Vision Models Summary
> Introducing Self-Attention as a stand-alone layer in Convolutional Architectures by Prajit Ramachandran,Niki Parmar,Ashish Vaswani,Jonathon Shlens,Anselm Levskaya,Irwan Bello
- toc: true
- badges: false
- comments: true
- categories: [jupyter]
- image: images/SATCONV.png
[Paper Link](https://arxiv.org/pdf/1906.05909.pdf)
## What did the authors want to achieve ?
Combat the deficits of Conolutional Architectures, (local connectivity, failing to reason globally) by introducing Attention to as a stand-alone layer. The authors prove that this can be both more accurate and more efficient at the same time. Architectures that are attention only, and a mixed version of convolutional and attention architectures are introduced and compared to the vanilla convolutional implementations.
## Methods
<br/><br/>
### Conv Block deficits
Capturing long range interactions is challenging for convolutions as they do not scale well with large receptives. Since attention has been used to tackle long range dependencies in sequence modeling, since architectures like SE Nets model attention on a chanel wise basis successfully. However in these cases attention was only an add-on to a traditional architecture style. In this paper the authors propose to use attention mechanisms as stand alone layers.
### Self-Attention
Attention was originally introduced in order to allow for summarization from variable length source sentences. Attention focuses on the important parts of the input and thereby can serve as a primary representation learning mechanism and fully replace recurrence. The word self means that it just considers a single context (query,keys and values are extracted from the same image). The breakthrough in this paper is the use of self-attention layers instead of convolutional layers.
In this work, already existing mechanisms are used which are not optimized for the image domain. Therefore it is permutation equivariant and has limited expression capability for vision tasks.
<br/><br/>

<br/><br/>
The process is as follows :
1) given a pixel $x_{i,j} \in R^{d_{in}}$ in positions $ab \in N_{k}(i,j)$ a local kernel with kernel size $k$ is extracted. $x_{i,j}$ is the middle of the kernel, which is called *memory block*. Prior work only performed global attention, which can only be done with a downsized sample as it is very compute expensive.
<br/><br/>
2) Single headed attention is computed :
<br/><br/>
$y_{i,j} = \sum_{ab \in N_{k}(i,j)} softmax_{a,b}(q_{ij}^{T}k_{ab})v_{ab}$
<br/><br/>
where the ${queries}$ $q_{ij} = W_Q x_{ij}$
<br/><br/>
${keys}$ $k_{ab} = W_K x_{ab}$
<br/><br/>
and ${values}$ $v_{ab} = W_V x_{ab}$
<br/><br/>
are linear transformations of the pixel in position and it's neighbors in the kernel.
<br/><br/>
$\texttt{softmax}_{a b}$ is a softmax, which is applied to all logits computed in the neighborhood of $ij$.
$W_Q, W_K, W_V \in \mathbb{R}^{d_{out} \times d_{in}}$ are learned transforms.
<br/><br/>
Local self-attention is similar to convolution in the way that it aggregates spatial information in the neighborhoods, multi attention heads are used to learn unique representations of the input. This is done by partitioning pixel features into $N$ groups and then computing single-headed attention on each one seperately with the transforms $W_Q, W_K, W_V \in \mathbb{R}^{d_{out} \times d_{in}}$. The outputs of the heads are then concatenated.
<br/><br/>
2D relative pose embeddings,relative attention is used :
<br/><br/>
1) relative attention computes relative distances of the pixel to each one in the neighborhood : row $(a-i)$ and column offset $(b-j)$
2) row and column offset are associated with an embedding and concatenated into a vector $r_{a-i,b-j}$
3) Spatial-relative attention is then defined as :
<br/><br/>
$y_{ij} = \sum_{a,b\in N_{k}(i, j)} softmax_{ab}(q_{ij}^{T}k_{ab}+q_{ij}^{T}r_{a-i,b-j})v_{ab}$
<br/><br/>
The logit measuring similarity between the query and an element results from the content of the element and the relative distance of the element from the query. By inlcuding this spatial information, self-attention also has translation equivariance, just like conv layers. Unlike conv layers, self-attentions parameter count is independent of its spatial extent.
The compute cost also grows slower :
For example, if $d_{in} = d_{out} = 128$, a convolution layer with $k = 3$ has the same computational cost as an attention layer with $k = 19$.
Using this as their basis, a fully attentional architecture is created in two steps:
#### Replacing Spatial Convolutions
A spatial conv is defined to have spatial extent k > 1, which also includes 1x1 convolutions. These can be viewed as fully connected layers.
Here the authors want to replace conv blocks in a straightforward way, specificially focusing on ResNet. Therefore the 3x3 convolution in Path B is swapped with a self-attention layer as defined above. All the other blocks are not changed, this might be supobtimal but promises potential improvements using architecture search.
#### Replacing the Convolutional Stem (intial layers of the CNN)
This part focuses on replacing the inital layers, as they are the most compute expensive due to the large input size of the image. In the OG ResNet the input is a 7x7 kernel with stride 2, followed by 3x3 max pooling with stride 2. At the beginning RGB pixels are individually uninformative and information is heavily spatially correlated through low level features such as edges. Edge detectors are difficult to learn for self-attention due to spatial correlation, convs learn these easily through distance based weight parameterization. The authors inject spatially-varying linear transforms into the pointwise 1x1 softmax convolution.
$\tilde{v}_{a b} = \left(\sum_m p(a, b, m) W_V^{m}\right) x_{a b}$
The results is similar to convolutions, weights are learned based on a local neighborhood basis. So in total the stem consists of spatially aware value features, followed by max-pooling. A more detailed explanation of this can be found in the appendix of the paper (page 14/15).
## Results
Implementation details for both classification and object detection are in the appendix.
### ImageNet
The multi head self-attention layer uses a spatial width of k=7 and 8 attention heads. The position-aware attention stem as described above is used.The stem performs self-attention within each 4×4 block of the original image, followed by batch normalization and a 4×4 max pool operation. Results are below :

### Coco Object Detection
Here Retina Net is used with a classification backbone, followed by an FPN, the network has 2 detection heads. Results are in the table below :

We can see that using attention based backbone we can achieve results on par with a conv backbone, but with 22% less parameters. This can be extended by additionaly making the FPN and the detection heads attention-based and thereby reducing paraneter count by 34% and more importantly FLOPS by 39%.
### Where is stand-alone attention most useful ?

Results are schown in the tables above.
#### Stem
The basic results for the stem is that, convolutions perform very well here, as described above self-attention can not easily learn edges due to the high spatial correlation which is captured very well by conv layers though.
#### Full Net
The authors basically state what has been described above, conv layers capture low level features very well, while attention is able to model global relations effectively. Therefore an optimal architecture should contain both attention and convolutional layers.
### Which attention features are important ?
- Effect of spatial extent of self-attention (Table 4) :
The value of spatial extent k should generally be larger (for example k=11), the exact optimal setting depends on hyperaparameter choices.
- Importance of positional information (Table 5 + 6)
3 types of encoding were used : no positional encoding, sinusodial encoing and absolute pixel position. Relativ encoding performs 2% better than absolute one. Removing content-content interaction only descreases accuracy by 0.5%. Therefore the positional encoding seems to be very important and can be a strong focus of their future research.
- Importance of spatially-aware attention stem (Table 7)
Using stand-alone attention in the stem with spatially-aware values, it outperforms vanilla stand-alone attention by 1.4%, while having similar FLOPS. Using a spatial convolution to the values instead of spatially-aware point-wise transformations (see above), leads to more FLOPS and slightly worse results. A future goal of the authors is to unify attention used across the stem and main
| 0.935095 | 0.842798 |
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib notebook
```
# Principal Component Analysis
This notebook will demonstrate performing principal component analysis using eigenvectors of the covariance matrix as well as SVD.
First, let's set up a mock example. Let's say that we have 3 sensors that observe the location of a ball attached to a spring. We are going to use PCA to determine the underlying dynamics of this system.
```
x_a = np.random.multivariate_normal([-1, 1], [[0.9, 0.5], [0.05, 0.05]], 100)
x_b = np.random.multivariate_normal([-1, 1], [[-0.9, 0.5], [-0.1, 0]], 100)
x_c = np.random.multivariate_normal([-1, 1], [[0, 1], [0.03, 0]], 100)
fig = plt.figure()
ax = fig.add_subplot(111)
ax.scatter(x_a[:, 0], x_a[:, 1], c='b')
ax.scatter(x_b[:, 0], x_b[:, 1], c='r')
ax.scatter(x_c[:, 0], x_c[:, 1], c='g')
```
# PCA via Eigenvectors of the Covariance Matrix
With PCA, we are attempting to find a change-of-basis matrix $P$ such that our original data $X$ is transformed into a new representation $Y$. We want the new representation $Y$ to reduce the redunancy among the features. That is, find a $P$ such that $S_{Y} = \frac{1}{n-1}YY^{T}$ will be diagonalized.
We start by constructing a covariance matrix between the features that we have observed: $S_{X} = \frac{1}{n-1}XX^{T}$.
So, how do we find this? We start by noting that
\begin{align*}
S_{Y} &= \frac{1}{n-1}YY^{T}\\
&= \frac{1}{n-1}(PX)(PX)^{T}\\
&= \frac{1}{n-1}PXX^{T}P^{T}\\
&= \frac{1}{n-1}P(XX^{T})P^{T}\\
&= \frac{1}{n-1}PAP^{T}
\end{align*}
Here, $A = XX^{T}$ is symmetric. We also learned about a useful factorization of symmetric matrices: $A = EDE^{T}$, where $D$ is a diagonal matrix representing the eigenvalues of $A$ and $E$ represents the corresponding eigenvectors.
Getting back to our original $P$. We want $P$ such that each row is an eigenvector of $A$. This is simply $E^{T}$ from the factorization above! Thus, $P = E^{T}$. We can now substitute $A = P^{T}DP$ into $S_{Y} = \frac{1}{n-1}PAP^{T}$:
\begin{align*}
S_{Y} &= \frac{1}{n-1}PAP^{T}\\
&= \frac{1}{n-1}P(P^{T}DP)P^{T}\\
&= \frac{1}{n-1}(PP^{T})D(PP^{T})\\
&= \frac{1}{n-1}(PP^{-1})D(PP^{-1})\\
&= \frac{1}{n-1}D\\
\end{align*}
We are left with $P$, the principal components of $X$, and $S_{Y}$, the variance of $X$ along the rows of $P$.
```
# Construct the matrix X out of our 3 sensors
X = np.vstack([x_b.T, x_a.T, x_c.T])
num_features, num_samples = X.shape
print(num_features, num_samples)
# Subtract the mean from each dimension
X_mean = np.mean(X, 1)
X = X - X_mean[:, np.newaxis]
# Calculate the covariance
X_cov = 1 / (num_samples - 1) * X @ X.T
fig = plt.figure()
ax = fig.add_subplot(111)
ax.matshow(X_cov)
# Compute the eigendecomposition
D, P = np.linalg.eig(X_cov)
```
# Dimensionality Reduction
Clearly we do not need 6 dimensions to describe the underlying dynamics of the given system. The eigenvectors corresponding the largest eigenvalues capture the largest variance of our dataset.
We can select only a few of these eigenvectors and project our data to a lower dimension. In this case, we will transform each 6D feature vector back to 2D using the 2 eigenvectors that best capture the variance of the data.
```
# Sort eigenvalues and eigenvectors in decreasing order
sort_idx = D.argsort()[::-1]
D = D[sort_idx]
P = P[:,sort_idx]
n_comps = 2 # number of eigenvectors to use
W = P[:n_comps]
```
# Visualizing the projected space
Now that we've computed the principal components of our original data. We can see what they look like when projected to the new space.
This will qualitatively provide the answer to the question of what is the most salient feature in our data?
```
# Project the original samples using the principal components corresponding to the highest variance.
X_proj = W @ X
fig = plt.figure()
ax = fig.add_subplot(111)
ax.scatter(X_proj[0, :], X_proj[1, :], c='b') # when using 2 components
# ax.scatter(X_proj[0, :], np.zeros(X_proj.shape[1]), c='b') # when using 1 component
ax.set_xlim([-3, 3])
ax.set_ylim([-3, 3])
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
%matplotlib notebook
x_a = np.random.multivariate_normal([-1, 1], [[0.9, 0.5], [0.05, 0.05]], 100)
x_b = np.random.multivariate_normal([-1, 1], [[-0.9, 0.5], [-0.1, 0]], 100)
x_c = np.random.multivariate_normal([-1, 1], [[0, 1], [0.03, 0]], 100)
fig = plt.figure()
ax = fig.add_subplot(111)
ax.scatter(x_a[:, 0], x_a[:, 1], c='b')
ax.scatter(x_b[:, 0], x_b[:, 1], c='r')
ax.scatter(x_c[:, 0], x_c[:, 1], c='g')
# Construct the matrix X out of our 3 sensors
X = np.vstack([x_b.T, x_a.T, x_c.T])
num_features, num_samples = X.shape
print(num_features, num_samples)
# Subtract the mean from each dimension
X_mean = np.mean(X, 1)
X = X - X_mean[:, np.newaxis]
# Calculate the covariance
X_cov = 1 / (num_samples - 1) * X @ X.T
fig = plt.figure()
ax = fig.add_subplot(111)
ax.matshow(X_cov)
# Compute the eigendecomposition
D, P = np.linalg.eig(X_cov)
# Sort eigenvalues and eigenvectors in decreasing order
sort_idx = D.argsort()[::-1]
D = D[sort_idx]
P = P[:,sort_idx]
n_comps = 2 # number of eigenvectors to use
W = P[:n_comps]
# Project the original samples using the principal components corresponding to the highest variance.
X_proj = W @ X
fig = plt.figure()
ax = fig.add_subplot(111)
ax.scatter(X_proj[0, :], X_proj[1, :], c='b') # when using 2 components
# ax.scatter(X_proj[0, :], np.zeros(X_proj.shape[1]), c='b') # when using 1 component
ax.set_xlim([-3, 3])
ax.set_ylim([-3, 3])
| 0.805211 | 0.988799 |
```
import pandas as pd
import numpy as np
from lightgbm import LGBMRegressor
from xgboost import XGBRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import GridSearchCV
from sklearn.dummy import DummyRegressor
from scipy.stats import zscore, uniform
from sklearn.preprocessing import LabelEncoder, MinMaxScaler
from math import sqrt
import altair as alt
alt.renderers.enable('notebook')
alt.data_transformers.enable('json')
from sklearn.model_selection import train_test_split
# read data
airbnb_ny = pd.read_csv('../data/raw_data.csv')
# drop features
airbnb_ny.drop(['id','name', 'host_id', 'host_name','last_review'], axis=1, inplace=True)
# fill nas in reviews per month
airbnb_ny = airbnb_ny.fillna({'reviews_per_month':0})
# split to X and Y
X = airbnb_ny.drop(['price'], axis=1)
y = airbnb_ny.price
# split to test and train
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state = 1)
# combine X and y for test and train respectively
full_train = pd.concat((X_train, y_train), axis= 1)
full_test = pd.concat((X_test, y_test), axis= 1)
full_train.head()
full_train.info()
full_train.shape
X_train = full_train.drop(['price'], axis=1)
y_train = full_train['price']
X_test = full_test.drop(['price'], axis=1)
y_test = full_test['price']
categorical_features = [
'neighbourhood_group',
'neighbourhood',
'room_type'
]
for feature in categorical_features:
le = LabelEncoder()
le.fit(X_train[feature])
X_train[feature] = le.transform(X_train[feature])
X_test[feature] = le.transform(X_test[feature])
X_train.head()
y_train
null_model = DummyRegressor(strategy='median')
null_model.fit(X_train, y_train)
```
The MSE of the null model is:
```
mean_absolute_error(y_test, null_model.predict(X_test))
random_forest_tuning_parameters = {
'max_depth': [10, 50],
'min_samples_split': [5, 20],
'n_estimators': [600, 1500],
'criterion': ['mse'],
'random_state': [0]
}
rf = GridSearchCV(
estimator=RandomForestRegressor(),
param_grid=random_forest_tuning_parameters,
cv=4,
verbose=2,
n_jobs=-1,
scoring='neg_mean_absolute_error'
)
rf.fit(X_train, y_train)
def print_model_info(grid_model, model_name):
print(model_name + ' info:')
print('')
print('MAE:')
print(mean_absolute_error(y_test, grid_model.best_estimator_.predict(X_test)))
print('')
print('Best params: ')
print(grid_model.best_params_)
feature_importantance_series = pd.Series(grid_model.best_estimator_.feature_importances_)
feature_importantance_series.index = X_test.columns
print('')
print('Feature importance:')
print(feature_importantance_series.sort_values(ascending=False))
print_model_info(rf, 'Random Forest Regressor')
light_regressor_tuning_parameters = {
'min_data_in_leaf': [100, 300, 500, 1000, 1500],
'num_leaves': [15, 30, 40, 50, 60],
'max_depth': [15, 30, 45],
'random_state': [0]
}
light_reg = GridSearchCV(
estimator=LGBMRegressor(),
param_grid=light_regressor_tuning_parameters,
cv=4,
verbose=2,
n_jobs=-1,
scoring='neg_mean_absolute_error'
)
light_reg.fit(X_train, y_train, eval_metric='l1')
print_model_info(light_reg, 'LightGBM Regressor')
xgb_regressor_tuning_parameters = {
'max_depth': [5, 7, 10],
'colsample_bytree': [0.6, 0.7, 0.8],
'n_estimators': [500, 1000],
'random_state': [0]
}
xgb_reg = GridSearchCV(
estimator=XGBRegressor(),
param_grid=xgb_regressor_tuning_parameters,
cv=4,
verbose=2,
n_jobs=-1,
scoring='neg_mean_absolute_error'
)
xgb_reg.fit(X_train, y_train, eval_metric='mae')
print_model_info(xgb_reg, 'XGBoost Regressor')
test_average_ensemble_prediction = np.average([
rf.best_estimator_.predict(X_test),
light_reg.best_estimator_.predict(X_test),
xgb_reg.best_estimator_.predict(X_test)
], axis=0)
mean_absolute_error(y_test, test_average_ensemble_prediction)
ensemble_residual_df = pd.DataFrame({
'true_price': y_test,
'average_ensemble_residual': y_test - test_average_ensemble_prediction
})
residual_chart = alt.Chart(ensemble_residual_df).mark_circle(size=30, opacity=0.4).encode(
x=alt.X('true_price', title='Price'),
y=alt.Y('average_ensemble_residual', title='Average ensembling residual')
).properties(
width=350,
height=500
).properties(
title='Average Ensembling Residuals on Test Data'
)
residual_dist_chart = alt.Chart(ensemble_residual_df).mark_bar().encode(
x=alt.X(
'average_ensemble_residual',
title='Average ensembling residual',
bin=alt.Bin(extent=[-1000, 1000], step=50)
),
y='count()'
).properties(
width=350,
height=500
).properties(
title='Ensembling Residual Distribution'
)
model_result_charts = (residual_chart | residual_dist_chart).configure_axis(
labelFontSize=15,
titleFontSize=15
)
with alt.data_transformers.enable('default'):
model_result_charts.save("../results/plots/model_result_charts.png")
feature_important_df = pd.DataFrame({
'Random Forest': rf.best_estimator_.feature_importances_,
'XGBoost': xgb_reg.best_estimator_.feature_importances_,
'LightGBM': light_reg.best_estimator_.feature_importances_/sum(light_reg.best_estimator_.feature_importances_)
})
feature_important_df.index = X_test.columns
feature_important_df
train_average_ensemble_prediction = np.average([
rf.best_estimator_.predict(X_train),
light_reg.best_estimator_.predict(X_train),
xgb_reg.best_estimator_.predict(X_train)
], axis=0)
mean_absolute_error_df = pd.DataFrame({
'train_mean_absolute_error': [
mean_absolute_error(y_train, null_model.predict(X_train)),
mean_absolute_error(y_train, rf.predict(X_train)),
mean_absolute_error(y_train, xgb_reg.predict(X_train)),
mean_absolute_error(y_train, light_reg.predict(X_train)),
mean_absolute_error(y_train, train_average_ensemble_prediction),
],
'test_mean_absolute_error': [
mean_absolute_error(y_test, null_model.predict(X_test)),
mean_absolute_error(y_test, rf.predict(X_test)),
mean_absolute_error(y_test, xgb_reg.predict(X_test)),
mean_absolute_error(y_test, light_reg.predict(X_test)),
mean_absolute_error(y_test, test_average_ensemble_prediction),
]
})
mean_absolute_error_df.index = [
'Median Null Model',
'Random Forest',
'XGBoost',
'LightGBM',
'Average Ensembling'
]
mean_absolute_error_df
mean_absolute_error_df.to_csv("../results/tables/mean_absolute_error_table.csv")
feature_important_df.to_csv('../results/tables/feature_importance_table.csv')
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from lightgbm import LGBMRegressor
from xgboost import XGBRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import GridSearchCV
from sklearn.dummy import DummyRegressor
from scipy.stats import zscore, uniform
from sklearn.preprocessing import LabelEncoder, MinMaxScaler
from math import sqrt
import altair as alt
alt.renderers.enable('notebook')
alt.data_transformers.enable('json')
from sklearn.model_selection import train_test_split
# read data
airbnb_ny = pd.read_csv('../data/raw_data.csv')
# drop features
airbnb_ny.drop(['id','name', 'host_id', 'host_name','last_review'], axis=1, inplace=True)
# fill nas in reviews per month
airbnb_ny = airbnb_ny.fillna({'reviews_per_month':0})
# split to X and Y
X = airbnb_ny.drop(['price'], axis=1)
y = airbnb_ny.price
# split to test and train
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state = 1)
# combine X and y for test and train respectively
full_train = pd.concat((X_train, y_train), axis= 1)
full_test = pd.concat((X_test, y_test), axis= 1)
full_train.head()
full_train.info()
full_train.shape
X_train = full_train.drop(['price'], axis=1)
y_train = full_train['price']
X_test = full_test.drop(['price'], axis=1)
y_test = full_test['price']
categorical_features = [
'neighbourhood_group',
'neighbourhood',
'room_type'
]
for feature in categorical_features:
le = LabelEncoder()
le.fit(X_train[feature])
X_train[feature] = le.transform(X_train[feature])
X_test[feature] = le.transform(X_test[feature])
X_train.head()
y_train
null_model = DummyRegressor(strategy='median')
null_model.fit(X_train, y_train)
mean_absolute_error(y_test, null_model.predict(X_test))
random_forest_tuning_parameters = {
'max_depth': [10, 50],
'min_samples_split': [5, 20],
'n_estimators': [600, 1500],
'criterion': ['mse'],
'random_state': [0]
}
rf = GridSearchCV(
estimator=RandomForestRegressor(),
param_grid=random_forest_tuning_parameters,
cv=4,
verbose=2,
n_jobs=-1,
scoring='neg_mean_absolute_error'
)
rf.fit(X_train, y_train)
def print_model_info(grid_model, model_name):
print(model_name + ' info:')
print('')
print('MAE:')
print(mean_absolute_error(y_test, grid_model.best_estimator_.predict(X_test)))
print('')
print('Best params: ')
print(grid_model.best_params_)
feature_importantance_series = pd.Series(grid_model.best_estimator_.feature_importances_)
feature_importantance_series.index = X_test.columns
print('')
print('Feature importance:')
print(feature_importantance_series.sort_values(ascending=False))
print_model_info(rf, 'Random Forest Regressor')
light_regressor_tuning_parameters = {
'min_data_in_leaf': [100, 300, 500, 1000, 1500],
'num_leaves': [15, 30, 40, 50, 60],
'max_depth': [15, 30, 45],
'random_state': [0]
}
light_reg = GridSearchCV(
estimator=LGBMRegressor(),
param_grid=light_regressor_tuning_parameters,
cv=4,
verbose=2,
n_jobs=-1,
scoring='neg_mean_absolute_error'
)
light_reg.fit(X_train, y_train, eval_metric='l1')
print_model_info(light_reg, 'LightGBM Regressor')
xgb_regressor_tuning_parameters = {
'max_depth': [5, 7, 10],
'colsample_bytree': [0.6, 0.7, 0.8],
'n_estimators': [500, 1000],
'random_state': [0]
}
xgb_reg = GridSearchCV(
estimator=XGBRegressor(),
param_grid=xgb_regressor_tuning_parameters,
cv=4,
verbose=2,
n_jobs=-1,
scoring='neg_mean_absolute_error'
)
xgb_reg.fit(X_train, y_train, eval_metric='mae')
print_model_info(xgb_reg, 'XGBoost Regressor')
test_average_ensemble_prediction = np.average([
rf.best_estimator_.predict(X_test),
light_reg.best_estimator_.predict(X_test),
xgb_reg.best_estimator_.predict(X_test)
], axis=0)
mean_absolute_error(y_test, test_average_ensemble_prediction)
ensemble_residual_df = pd.DataFrame({
'true_price': y_test,
'average_ensemble_residual': y_test - test_average_ensemble_prediction
})
residual_chart = alt.Chart(ensemble_residual_df).mark_circle(size=30, opacity=0.4).encode(
x=alt.X('true_price', title='Price'),
y=alt.Y('average_ensemble_residual', title='Average ensembling residual')
).properties(
width=350,
height=500
).properties(
title='Average Ensembling Residuals on Test Data'
)
residual_dist_chart = alt.Chart(ensemble_residual_df).mark_bar().encode(
x=alt.X(
'average_ensemble_residual',
title='Average ensembling residual',
bin=alt.Bin(extent=[-1000, 1000], step=50)
),
y='count()'
).properties(
width=350,
height=500
).properties(
title='Ensembling Residual Distribution'
)
model_result_charts = (residual_chart | residual_dist_chart).configure_axis(
labelFontSize=15,
titleFontSize=15
)
with alt.data_transformers.enable('default'):
model_result_charts.save("../results/plots/model_result_charts.png")
feature_important_df = pd.DataFrame({
'Random Forest': rf.best_estimator_.feature_importances_,
'XGBoost': xgb_reg.best_estimator_.feature_importances_,
'LightGBM': light_reg.best_estimator_.feature_importances_/sum(light_reg.best_estimator_.feature_importances_)
})
feature_important_df.index = X_test.columns
feature_important_df
train_average_ensemble_prediction = np.average([
rf.best_estimator_.predict(X_train),
light_reg.best_estimator_.predict(X_train),
xgb_reg.best_estimator_.predict(X_train)
], axis=0)
mean_absolute_error_df = pd.DataFrame({
'train_mean_absolute_error': [
mean_absolute_error(y_train, null_model.predict(X_train)),
mean_absolute_error(y_train, rf.predict(X_train)),
mean_absolute_error(y_train, xgb_reg.predict(X_train)),
mean_absolute_error(y_train, light_reg.predict(X_train)),
mean_absolute_error(y_train, train_average_ensemble_prediction),
],
'test_mean_absolute_error': [
mean_absolute_error(y_test, null_model.predict(X_test)),
mean_absolute_error(y_test, rf.predict(X_test)),
mean_absolute_error(y_test, xgb_reg.predict(X_test)),
mean_absolute_error(y_test, light_reg.predict(X_test)),
mean_absolute_error(y_test, test_average_ensemble_prediction),
]
})
mean_absolute_error_df.index = [
'Median Null Model',
'Random Forest',
'XGBoost',
'LightGBM',
'Average Ensembling'
]
mean_absolute_error_df
mean_absolute_error_df.to_csv("../results/tables/mean_absolute_error_table.csv")
feature_important_df.to_csv('../results/tables/feature_importance_table.csv')
| 0.517083 | 0.604662 |
### PCA and t-SNE using scikit-learn
### Edgar Acuna
### Dataset: Diabetes and bees_2p
#### March 2020
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
from matplotlib.ticker import NullFormatter
from sklearn import manifold
from time import time
```
### T-SNE for Diabetes
```
#Leyendo el conjunto de datos pima-diabetes
url= "http://academic.uprm.edu/eacuna/diabetes.dat"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
data = pd.read_table(url, names=names,header=None)
data.head()
y=data['class']
X=data.iloc[:,0:8]
```
### t-SNE visualization
```
n_components = 2
(fig, subplots) = plt.subplots(1, 6, figsize=(15, 8),squeeze=False)
perplexities = [5, 20, 30, 40,50,60]
red = y == 1
green = y == 2
for i, perplexity in enumerate(perplexities):
ax = subplots[0][i]
t0 = time()
tsne = manifold.TSNE(n_components=n_components, init='random',
random_state=0, perplexity=perplexity)
Y = tsne.fit_transform(X)
t1 = time()
print("circles, perplexity=%d in %.2g sec" % (perplexity, t1 - t0))
ax.set_title("Perplexity=%d" % perplexity)
ax.scatter(Y[red, 0], Y[red, 1], c="r")
ax.scatter(Y[green, 0], Y[green, 1], c="g")
ax.xaxis.set_major_formatter(NullFormatter())
ax.yaxis.set_major_formatter(NullFormatter())
ax.axis('tight')
plt.show()
```
### t-SNE for bees_2p
```
bees=pd.read_csv("https://academic.uprm.edu/eacuna/bees_2p.csv")
X1=bees.iloc[:,3:23]
#dropping two outliers bees 153 y 369
X2=X1.drop([152,368])
X2.describe()
from sklearn.cluster import KMeans
from sklearn.preprocessing import StandardScaler
from sklearn.metrics.pairwise import pairwise_distances
from sklearn.neighbors import DistanceMetric
from scipy.spatial import distance
kmeans = KMeans(n_clusters=2, random_state=0).fit(X2)
clustlabels=kmeans.labels_
print(clustlabels)
y=clustlabels
n_components = 2
(fig, subplots) = plt.subplots(1, 6, figsize=(15, 8),squeeze=False)
perplexities = [5, 10, 15, 20, 30, 40]
red = y == 0
green = y == 1
for i, perplexity in enumerate(perplexities):
ax = subplots[0][i]
t0 = time()
tsne = manifold.TSNE(n_components=n_components, init='random',
random_state=0, perplexity=perplexity)
Y = tsne.fit_transform(X2)
t1 = time()
print("bees, perplexity=%d in %.2g sec" % (perplexity, t1 - t0))
ax.set_title("Perplexity=%d" % perplexity)
ax.scatter(Y[red, 0], Y[red, 1], c="r")
ax.scatter(Y[green, 0], Y[green, 1], c="g")
ax.xaxis.set_major_formatter(NullFormatter())
ax.yaxis.set_major_formatter(NullFormatter())
ax.axis('tight')
plt.show()
```
### t-SNE for bees_2p after PCA
```
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
pca = PCA(n_components=9)
X2 = StandardScaler().fit_transform(X2)
principalComponents = pca.fit_transform(X2)
pcaDF=pd.DataFrame(data = principalComponents, columns = ['PC1', 'PC2','PC3','PC4','PC5','PC6','PC7','PC8','PC9'])
print(pca.explained_variance_)
print(pca.explained_variance_ratio_)
print(pca.explained_variance_ratio_.cumsum())
y=clustlabels
n_components = 2
(fig, subplots) = plt.subplots(1, 6, figsize=(15, 8),squeeze=False)
perplexities = [5, 10, 15, 20, 50, 60]
red = y == 0
green = y == 1
for i, perplexity in enumerate(perplexities):
ax = subplots[0][i]
t0 = time()
tsne = manifold.TSNE(n_components=n_components, init='random',
random_state=0, perplexity=perplexity)
Y = tsne.fit_transform(pcaDF)
t1 = time()
print("bees, perplexity=%d in %.2g sec" % (perplexity, t1 - t0))
ax.set_title("Perplexity=%d" % perplexity)
ax.scatter(Y[red, 0], Y[red, 1], c="r")
ax.scatter(Y[green, 0], Y[green, 1], c="g")
ax.xaxis.set_major_formatter(NullFormatter())
ax.yaxis.set_major_formatter(NullFormatter())
ax.axis('tight')
plt.show()
```
### PCA for bess_2p
```
from matplotlib.colors import ListedColormap
from itertools import cycle, islice
colors = np.array(list(islice(cycle(['#377eb8', '#ff7f00', '#4daf4a',
'#f781bf', '#a65628', '#984ea3',
'#999999', '#e41a1c', '#dede00']),
int(max(clustlabels) + 1))))
plt.scatter(pcaDF['PC1'], pcaDF['PC2'],color=colors[clustlabels])
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
from matplotlib.ticker import NullFormatter
from sklearn import manifold
from time import time
#Leyendo el conjunto de datos pima-diabetes
url= "http://academic.uprm.edu/eacuna/diabetes.dat"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
data = pd.read_table(url, names=names,header=None)
data.head()
y=data['class']
X=data.iloc[:,0:8]
n_components = 2
(fig, subplots) = plt.subplots(1, 6, figsize=(15, 8),squeeze=False)
perplexities = [5, 20, 30, 40,50,60]
red = y == 1
green = y == 2
for i, perplexity in enumerate(perplexities):
ax = subplots[0][i]
t0 = time()
tsne = manifold.TSNE(n_components=n_components, init='random',
random_state=0, perplexity=perplexity)
Y = tsne.fit_transform(X)
t1 = time()
print("circles, perplexity=%d in %.2g sec" % (perplexity, t1 - t0))
ax.set_title("Perplexity=%d" % perplexity)
ax.scatter(Y[red, 0], Y[red, 1], c="r")
ax.scatter(Y[green, 0], Y[green, 1], c="g")
ax.xaxis.set_major_formatter(NullFormatter())
ax.yaxis.set_major_formatter(NullFormatter())
ax.axis('tight')
plt.show()
bees=pd.read_csv("https://academic.uprm.edu/eacuna/bees_2p.csv")
X1=bees.iloc[:,3:23]
#dropping two outliers bees 153 y 369
X2=X1.drop([152,368])
X2.describe()
from sklearn.cluster import KMeans
from sklearn.preprocessing import StandardScaler
from sklearn.metrics.pairwise import pairwise_distances
from sklearn.neighbors import DistanceMetric
from scipy.spatial import distance
kmeans = KMeans(n_clusters=2, random_state=0).fit(X2)
clustlabels=kmeans.labels_
print(clustlabels)
y=clustlabels
n_components = 2
(fig, subplots) = plt.subplots(1, 6, figsize=(15, 8),squeeze=False)
perplexities = [5, 10, 15, 20, 30, 40]
red = y == 0
green = y == 1
for i, perplexity in enumerate(perplexities):
ax = subplots[0][i]
t0 = time()
tsne = manifold.TSNE(n_components=n_components, init='random',
random_state=0, perplexity=perplexity)
Y = tsne.fit_transform(X2)
t1 = time()
print("bees, perplexity=%d in %.2g sec" % (perplexity, t1 - t0))
ax.set_title("Perplexity=%d" % perplexity)
ax.scatter(Y[red, 0], Y[red, 1], c="r")
ax.scatter(Y[green, 0], Y[green, 1], c="g")
ax.xaxis.set_major_formatter(NullFormatter())
ax.yaxis.set_major_formatter(NullFormatter())
ax.axis('tight')
plt.show()
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
pca = PCA(n_components=9)
X2 = StandardScaler().fit_transform(X2)
principalComponents = pca.fit_transform(X2)
pcaDF=pd.DataFrame(data = principalComponents, columns = ['PC1', 'PC2','PC3','PC4','PC5','PC6','PC7','PC8','PC9'])
print(pca.explained_variance_)
print(pca.explained_variance_ratio_)
print(pca.explained_variance_ratio_.cumsum())
y=clustlabels
n_components = 2
(fig, subplots) = plt.subplots(1, 6, figsize=(15, 8),squeeze=False)
perplexities = [5, 10, 15, 20, 50, 60]
red = y == 0
green = y == 1
for i, perplexity in enumerate(perplexities):
ax = subplots[0][i]
t0 = time()
tsne = manifold.TSNE(n_components=n_components, init='random',
random_state=0, perplexity=perplexity)
Y = tsne.fit_transform(pcaDF)
t1 = time()
print("bees, perplexity=%d in %.2g sec" % (perplexity, t1 - t0))
ax.set_title("Perplexity=%d" % perplexity)
ax.scatter(Y[red, 0], Y[red, 1], c="r")
ax.scatter(Y[green, 0], Y[green, 1], c="g")
ax.xaxis.set_major_formatter(NullFormatter())
ax.yaxis.set_major_formatter(NullFormatter())
ax.axis('tight')
plt.show()
from matplotlib.colors import ListedColormap
from itertools import cycle, islice
colors = np.array(list(islice(cycle(['#377eb8', '#ff7f00', '#4daf4a',
'#f781bf', '#a65628', '#984ea3',
'#999999', '#e41a1c', '#dede00']),
int(max(clustlabels) + 1))))
plt.scatter(pcaDF['PC1'], pcaDF['PC2'],color=colors[clustlabels])
plt.show()
| 0.468547 | 0.901097 |
## Dates and Times
A special type of categorical variable are those that instead of taking traditional labels, like color (blue, red), or city (London, Manchester), take dates and / or time as values. For example, date of birth ('29-08-1987', '12-01-2012'), or date of application ('2016-Dec', '2013-March').
Datetime variables can contain dates only, time only, or date and time.
We don't usually work with a datetime variable in their raw format because:
- Date variables contain a huge number of different categories
- We can extract much more information from datetime variables by preprocessing them correctly
In addition, often, date variables will contain dates that were not present in the dataset used to train the machine learning model. In fact, date variables will usually contain dates placed in the future, respect to the dates in the training dataset. Therefore, the machine learning model will not know what to do with them, because it never saw them while being trained.
**I will cover different was of pre-processing / engineering datetime variables in the section "Engineering Datetime Variables". later in this course**
=============================================================================
## In this demo: Peer to peer lending (Finance)
In this demo, we will use a toy data set which simulates data from a peer-o-peer finance company to inspect discrete and continuous numerical variables.
- You should have downloaded the **Datasets** together with the Jupyter notebooks in **Section 1**.
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# let's load the dataset
# Variable definitions:
#-------------------------
# disbursed amount: loan amount lent to the borrower
# market: risk band in which borrowers are placed
# loan purpose: intended use of the loan
# date_issued: date the loan was issued
# date_last_payment: date of last payment towards repyaing the loan
data = pd.read_csv('../loan.csv')
data.head()
# pandas assigns type 'object' when reading dates
# and considers them strings.
# Let's have a look
data[['date_issued', 'date_last_payment']].dtypes
```
Both date_issued and date_last_payment are casted as objects. Therefore, pandas will treat them as strings or categorical variables.
In order to instruct pandas to treat them as dates, we need to re-cast them into datetime format. See below.
```
# now let's parse the dates, currently coded as strings, into datetime format
# this will allow us to make some analysis afterwards
data['date_issued_dt'] = pd.to_datetime(data['date_issued'])
data['date_last_payment_dt'] = pd.to_datetime(data['date_last_payment'])
data[['date_issued', 'date_issued_dt', 'date_last_payment', 'date_last_payment_dt']].head()
# let's extract the month and the year from the variable date
# to make nicer plots
# more on this in section 12 of the course
data['month'] = data['date_issued_dt'].dt.month
data['year'] = data['date_issued_dt'].dt.year
# let's see how much money Lending Club has disbursed
# (i.e., lent) over the years to the different risk
# markets (grade variable)
fig = data.groupby(['year','month', 'market'])['disbursed_amount'].sum().unstack().plot(
figsize=(14, 8), linewidth=2)
fig.set_title('Disbursed amount in time')
fig.set_ylabel('Disbursed Amount')
```
This toy finance company seems to have increased the amount of money lent from 2012 onwards. The tendency indicates that they continue to grow. In addition, we can see that their major business comes from lending money to C and B grades.
'A' grades are the lower risk borrowers, borrowers that most likely will be able to repay their loans, as they are typically in a better financial situation. Borrowers within this grade are charged lower interest rates.
D and E grades represent the riskier borrowers. Usually borrowers in somewhat tighter financial situations, or for whom there is not sufficient financial history to make a reliable credit assessment. They are typically charged higher rates, as the business, and therefore the investors, take a higher risk when lending them money.
**That is all for this demonstration. I hope you enjoyed the notebook, and see you in the next one.**
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# let's load the dataset
# Variable definitions:
#-------------------------
# disbursed amount: loan amount lent to the borrower
# market: risk band in which borrowers are placed
# loan purpose: intended use of the loan
# date_issued: date the loan was issued
# date_last_payment: date of last payment towards repyaing the loan
data = pd.read_csv('../loan.csv')
data.head()
# pandas assigns type 'object' when reading dates
# and considers them strings.
# Let's have a look
data[['date_issued', 'date_last_payment']].dtypes
# now let's parse the dates, currently coded as strings, into datetime format
# this will allow us to make some analysis afterwards
data['date_issued_dt'] = pd.to_datetime(data['date_issued'])
data['date_last_payment_dt'] = pd.to_datetime(data['date_last_payment'])
data[['date_issued', 'date_issued_dt', 'date_last_payment', 'date_last_payment_dt']].head()
# let's extract the month and the year from the variable date
# to make nicer plots
# more on this in section 12 of the course
data['month'] = data['date_issued_dt'].dt.month
data['year'] = data['date_issued_dt'].dt.year
# let's see how much money Lending Club has disbursed
# (i.e., lent) over the years to the different risk
# markets (grade variable)
fig = data.groupby(['year','month', 'market'])['disbursed_amount'].sum().unstack().plot(
figsize=(14, 8), linewidth=2)
fig.set_title('Disbursed amount in time')
fig.set_ylabel('Disbursed Amount')
| 0.284377 | 0.940408 |
# COVID-19
This notebook analyzes the growth of the COVID-19 pandemy. It relies on the data provided by Johns Hopkins CSSE at https://github.com/CSSEGISandData/COVID-19 . The main question is: how will the number of infected people change over time. We will use a very simple approach, that should not be used for serious predictions of spreads of deseases, but which is well supported in PySpark. For a better mathematical model, please read https://de.wikipedia.org/wiki/SIR-Modell . Unfortunately there is no support in PySpark for estimating model parameters within a more meaningful model.
So this notebook is mainly about getting some basic insights into machine learning with PySpark.
# 0. Spark Context & Imports
```
import matplotlib.pyplot as plt
import pyspark.sql.functions as f
from pyspark.sql import SparkSession
if not 'spark' in locals():
spark = (
SparkSession.builder.master("local[*]")
.config("spark.driver.memory", "64G")
.getOrCreate()
)
spark
%matplotlib inline
```
# 1. Load Data
The original data is available at https://github.com/CSSEGISandData/COVID-19 provided by Johns Hopkins CSSE. There are several different representations of the data, we will peek into different versions and then select the most appropriate to work with.
```
basedir = 's3://dimajix-training/data/covid-19'
# basedir = '/dimajix/data/COVID-19/csse_covid_19_data'
```
## 1.1 Load Time Series
The repository already contains time series data. This is nice to look at, but specifically for PySpark maybe a little bit hard to work with. Each line in the file contains a full time series of the number of positive tested persons. This means that the number of columns change with every update.
```
series = spark.read.option("header", True).csv(
basedir + "/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv"
)
series.toPandas()
```
## 1.2 Load Daily Reports
The repository also contains more detailed files containing the daily reports of the total number of positively tested persons. Within those files, every line represents exactly one region and one time. Therefore the schema stays stable with every update, only new records are appended. But there are some small technical challenges that we need to take.
### Date parser helper
First small challenge: All records contain a date, some of them a datetime. But the format has changed several times. In order to handle the different cases, we provide a small PySpark UDF (User Defined Function) that is capable of parsing all formats and which returns the extracted date.
```
import datetime
from pyspark.sql.types import *
@f.udf(DateType())
def parse_date(date):
if "/" in date:
date = date.split(" ")[0]
(m, d, y) = date.split("/")
y = int(y)
m = int(m)
d = int(d)
if y < 2000:
y += 2000
else:
date = date[0:10]
(y, m, d) = date.split("-")
y = int(y)
m = int(m)
d = int(d)
return datetime.date(year=y, month=m, day=d)
# print(parse_date("2020-03-01"))
# print(parse_date("1/22/2020"))
# print(parse_date("2020-03-01T23:45:50"))
```
### Read in data, old schema
Next challenge is that the schema did change, namely between 2020-03-21 and 2020-03-22. The column names have changed, new columns have been added and so on. Therefore we cannot read in all files within a single `spark.read.csv`, but we need to split them up into two separate batches with different schemas.
```
# Last date to read
today = datetime.date(2020, 4, 7) # datetime.date.today()
# First date to read
start_date = datetime.date(2020, 1, 22)
# First date with new schema
schema_switch_date = datetime.date(2020, 3, 22)
```
The first bunch of files is stored as CSV and has the following columns:
* `Province_State`
* `Country_Region`
* `Last_Update` date of the last update
* `Confirmed` the number of confirmed cases
* `Deaths` the number of confirmed cases, which have died
* `Recovered` the number of recovered cases
* `Latitude` and `Longitude` geo coordinates of the province
The metrics (confirmed, deaths and recovered) are always totals, they already contain all cases from the past.
```
daily_reports_dir = basedir + "/csse_covid_19_daily_reports"
# Define old schema for first batch of files
schema_1 = StructType(
[
StructField("Province_State", StringType()),
StructField("Country_Region", StringType()),
StructField("Last_Update", StringType()),
StructField("Confirmed", LongType()),
StructField("Deaths", LongType()),
StructField("Recovered", LongType()),
StructField("Latitude", DoubleType()),
StructField("Longitude", DoubleType()),
]
)
# Generate all dates with old schema
schema_1_dates = [
start_date + datetime.timedelta(days=d)
for d in range(0, (schema_switch_date - start_date).days)
]
# Generate file names with old schema
schema_1_files = [
daily_reports_dir + "/" + d.strftime("%m-%d-%Y") + ".csv" for d in schema_1_dates
]
# Read in all files with old schema
cases_1 = spark.read.schema(schema_1).option("header", True).csv(schema_1_files)
# Peek inside
cases_1.toPandas()
```
### Read in data, new schema
Now we perform exactly the same logical step, we read in all files with the new schema. The second bunch of files is stored as CSV and has the following columns:
* `FIPS` country code
* `Admin2` administrative name below province (i.e. counties)
* `Province_State`
* `Country_Region`
* `Last_Update` date of the last update
* `Latitude` and `Longitude` geo coordinates of the province
* `Confirmed` the number of confirmed cases
* `Deaths` the number of confirmed cases, which have died
* `Recovered` the number of recovered cases
* `Active` the number of currently active cases
* `Combined_Key` a combination of `Admin2`, `Province_State` and `Country_Region`
The metrics (confirmed, deaths and recovered) are always totals, they already contain all cases from the past.
```
from pyspark.sql.types import *
daily_reports_dir = basedir + "/csse_covid_19_daily_reports"
# New schema
schema_2 = StructType(
[
StructField("FIPS", StringType()),
StructField("Admin2", StringType()),
StructField("Province_State", StringType()),
StructField("Country_Region", StringType()),
StructField("Last_Update", StringType()),
StructField("Latitude", DoubleType()),
StructField("Longitude", DoubleType()),
StructField("Confirmed", LongType()),
StructField("Deaths", LongType()),
StructField("Recovered", LongType()),
StructField("Active", LongType()),
StructField("Combined_Key", StringType()),
]
)
# Generate all dates with new schema
schema_2_dates = [
schema_switch_date + datetime.timedelta(days=d)
for d in range(0, (today - schema_switch_date).days)
]
# Generate file names with new schema
schema_2_files = [
daily_reports_dir + "/" + d.strftime("%m-%d-%Y") + ".csv" for d in schema_2_dates
]
# Read in all CSV files with new schema
cases_2 = spark.read.schema(schema_2).option("header", True).csv(schema_2_files)
cases_2.toPandas()
```
### Unify Records
Now we union both data sets `cases_1` and `cases_2` into a bigger data set with a common schema. The target schema should contain the following columns:
* `Country_Region`
* `Province_State`
* `Admin2`
* `Last_Update`
* `Confirmed`
* `Deaths`
* `Recovered`
In case a specific column is not present in onw of the two input DataFrames, simply provide a NULL value (`None` in Python) instead.
```
all_cases = cases_1.select(
f.col("Country_Region"),
f.col("Province_State"),
f.lit(None).cast(StringType()).alias("Admin2"),
f.col("Last_Update"),
f.col("Confirmed"),
f.col("Deaths"),
f.col("Recovered"),
).union(
cases_2.select(
f.col("Country_Region"),
f.col("Province_State"),
f.col("Admin2"),
f.col("Last_Update"),
f.col("Confirmed"),
f.col("Deaths"),
f.col("Recovered"),
)
)
all_cases.toPandas()
```
## 1.3 Aggragate
The records can contain multiple updates per day. But we only want to have the latest update per administrative region on each day. Therefore we perform a simple grouped aggregation and simply pick the maximum of all metrics of interest (`Confirmed`, `Deaths`, `Recovered`). This means we require a grouped aggregation with the grouping keys `Last_Update`, `Country_Region`, `Province_State` and `Admin2`.
```
all_cases_eod = all_cases.groupBy(
parse_date(f.col("Last_Update")).alias("Last_Update"),
f.col("Country_Region"),
f.col("Province_State"),
f.col("Admin2"),
).agg(
f.max(f.col("Confirmed")).alias("Confirmed"),
f.max(f.col("Deaths")).alias("Deaths"),
f.max(f.col("Recovered")).alias("Recovered"),
)
all_cases_eod.show()
```
## 1.4 Sanity Checks
Since we have now a nice data set containing all records, lets peek inside and let us perform some sanity checks if the numbers are correct.
```
all_cases_eod.where(f.col("Country_Region") == f.lit("US")).orderBy(
f.col("Confirmed").desc()
).show(truncate=False)
```
### Count cases in US
Let us count the cases in the US for a specific date, maybe compare it to some resource on the web. This can be done by summing up all confirmed case where `Country_Region` equals to `US` and where `Last_Update` equals some date of your choice (for example `2020-04-05`).
```
all_cases_eod.where(f.col("Country_Region") == f.lit("US")).where(
f.col("Last_Update") == f.lit("2020-04-05")
).select(f.sum(f.col("Confirmed"))).toPandas()
```
### Count cases in Germany
Let us now sum up the confirmed cases for Germany.
```
all_cases_eod.where(f.col("Country_Region") == f.lit("Germany")).where(
f.col("Last_Update") == f.lit("2020-04-06")
).select(f.sum(f.col("Confirmed"))).toPandas()
```
# 2. Inspect & Visualize
Now that we have a meaningful dataset, let us create some visualizations.
## 2.1 Additional Preparations
Before doing deeper analyzis, we still need to perform some simple preparations in order to make the resuls more meaningful.
### Cases pre country and day
We are not interested in the specific numbers of different provinces or states within a single country. The problem with data per province is that they may contain too few cases for following any theoretical law or for forming any meaningful probability distribution. Therefore we sum up all cases per country per day.
```
all_country_cases = all_cases_eod.groupBy("Country_Region", "Last_Update").agg(
f.sum(f.col("Confirmed")).alias("Confirmed"),
f.sum(f.col("Deaths")).alias("Deaths"),
f.sum(f.col("Recovered")).alias("Recovered"),
)
```
### Calculate age in days
Before continuing, we will add one extra column, namely the day of the epedemy for every country. The desease started on different dates in different countries (start being defined as the date of the first record in the data set). To be able to compare the development of the desease between different countries, it is advisable to add a country specific `day` column, which simply counts the days since the first infection in the particular country.
```
from pyspark.sql.window import Window
all_countries_age_cases = all_country_cases.withColumn(
"First_Update",
f.min(f.col("Last_Update")).over(
Window.partitionBy("Country_Region").orderBy("Last_Update")
),
).withColumn("day", f.datediff(f.col("Last_Update"), f.col("First_Update")))
all_countries_age_cases.show()
```
## 2.2 Pick single country
For the whole analysis, we focus on a single country. I decided to pick Germany, but you can also pick a different country.
The selection can easily be done by filtering using the column `Country_Region` to contain the desired country (for example `Germany`).
```
country_cases = all_countries_age_cases.where(f.col("Country_Region") == "Germany")
# Show first 10 days of data in the correct order
country_cases.orderBy(f.col("day")).show(10)
```
## 2.3 Plot
Let us make a simple plot which shows the number of cases over time. This can be achieved by using the matplotlib function `plt.plot`, which takes two arguments: the data on the horizontal axis (x-axis) and the data on the vertical axis (y-axis). One can also specify the size of the plot by using the function `plt.figure` as below.
```
df = country_cases.toPandas()
# Set size of the figure
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
# Make an image usig plt.plot
plt.plot(df["day"], df["Confirmed"])
```
### Plot on logarithmic scale
The spread of a epedemy follows an exponential pattern (specifically at the beginning), this can also be seen from the plot above. Therefore it is a good idea to change the scale from a linear scale to a logarithmic scale. With the logarithmic scale, you can spot the relativ rate of increase, which is the slope of the curve. Changing the scale can easily be done with the function `plt.yscale('log')`.
```
df = country_cases.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.yscale('log')
plt.plot(df["day"], df["Confirmed"], color='blue', lw=2)
```
### Plot of daily increases
In this case, we are also interested in the number of new cases on every day. This means, that we need to subtract the current number of confirmed cases from the last number of confirmed cases. This is a good example where so called *windowed aggregation* can help us in PySpark. Normally all rows of a DataFrame are processed independently, but for this question (the difference of the number of confirmed cases between two days), we would need to access the rows from two different days. That can be done with window functions.
```
daily_increase = country_cases.withColumn(
"Confirmed_Increase",
f.col("Confirmed")
- f.last(f.col("Confirmed")).over(
Window.partitionBy("Country_Region").orderBy("day").rowsBetween(-100, -1)
),
)
daily_increase.show(10)
```
Now we have an additional column "Confirmed_Increase", which we can now plot. A continuous line plot doesn't make so much sense, since the metric is very discrete by its nature. Therefore we opt for a bar chart instead by using the method `plt.bar` instead of `plt.plot`.
```
df = daily_increase.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.bar(df["day"], df["Confirmed_Increase"], color='blue', lw=2)
```
# 3. Machine Learning
Now we want to use some methods of machine learning in order to predict the further development of the desease within a single country.
```
# Import relevant packages
from pyspark.ml import *
from pyspark.ml.evaluation import *
from pyspark.ml.feature import *
from pyspark.ml.regression import *
```
## 3.1 Split Data
The very first step in every machine learning project is to split up the whole data into two data sets, the *training data* and the *validation data*. The basic idea is that in order to validate our model, we need some data that was not used during training. That is what the *validation data* will be used for. If we did not exclude the data from the training, we could only infer information about how good the model fits to our data - but we would have no information about how good the model copes with new data. And the second aspect is crucial in prediction applications, where the model will be used on new data points which have never been seen before.
There are different approaches how to do that, but not all approaches work in every scenario. In our use case we are looking at a time series, therefore we need to split the data at a specific date - we need to hide out some information for the training phase. For time series it is important not to perform a random sampling, since this would imply information creep from the future. I.e. if we exclude day 40 and include day 50 in our training data set, day 50 obviously has some information on day 40, which normally would not be available.
```
all_days = country_cases.select("day").orderBy(f.col("day")).distinct().collect()
all_days = [row[0] for row in all_days]
num_days = len(all_days)
cut_day = all_days[int(0.7 * num_days)]
print("cut_day = " + str(cut_day))
# We might want to skip some days where there was no real growth
first_day = 28
# Select training records from first_day until cut_day (inclusive)
training_records = country_cases.where(
(f.col("day") <= cut_day) & (f.col("day") >= first_day)
).cache()
# Select validation records from cut_day (exclusive)
validation_records = country_cases.where(f.col("day") > cut_day).cache()
```
## 3.2 Simple Regression
The most simple approach is to use a very basic linear regression. We skip this super simple approach, since we already know that our data has some exponential ingredients. Therefore we already use a so called *generalized linear model* (GLM), which transforms our values into a logarithmic space before performing a linear regression. Here we already know that this won't work out nicely, since the plots above already indicate a curved shape over time - something a trivial linear model cannot catch. We will take care of that in a later step.
### PySpark ML Pipelines
Spark ML encourages to use so called *pipelines*, which combine a list of transformation blocks. For the very first very simple example, we need two building blocks:
* `VectorAssembler` is required to collect all features into a single column of the special type `Vector`. Most machine learning algorithms require the independant variables (the predictor variables) to packaged together into a single column of type `Vector`. This can be easily done by using the `VectorAssembler`-
* `GeneralizedLinearRegression` provides the regression algorithm a a building block. It needs to be configured with the indepedant variable (the features column), the dependant variable (the label column) and the prediction column where the predictions should be stored in.
```
pipeline = Pipeline(
stages=[
VectorAssembler(inputCols=["day"], outputCol="features"),
GeneralizedLinearRegression(
family="gaussian",
link="log",
featuresCol="features",
labelCol="Confirmed",
predictionCol="Predict",
),
]
)
```
### Fit model
Once we have specified all building blocks in the pipeline, we can *fit* the whole pipeline to obtain a *pipeline model*. The fitting operation either applies a transformation (like the `VectorAssembler`) or recursively fits any embedded estimator (like the `GeneralizedLinearRegression`).
```
model = pipeline.fit(training_records)
```
### Perform prediction
One we have obtained the model, it can be used as a transformer again in order to produce predictions. For plotting a graph, we will apply the model not only to the validation set, but to the whole data set. This can be done with the `model.transform` method applied to the `country_cases` DataFrame.
```
pred = model.transform(country_cases)
```
### Visualize
Finally we want to visualize the real values and the predictions.
```
df = pred.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.yscale('log')
plt.plot(df["day"], df["Confirmed"], color='blue', lw=2)
plt.plot(df["day"], df["Predict"], color='red', lw=2)
plt.vlines(cut_day, ymin=0, ymax=400000)
```
### Measure Perfrmance
The picture already doesn't make much hope that the model generalizes well to new data. But in order to compare different models, we should also quantify the fit. We use a built in metric called *root mean squared error* provided by the class `RegressionEvaluator`. One an instance of the class is created, you can evaluate the predictions by using the `evaluate` function.
Since we are only interested in the ability to generalize, we use the `validation_records` DataFrame for measuring the qulity of fit.
```
evaluator = RegressionEvaluator(
predictionCol='Predict', labelCol='Confirmed', metricName='rmse'
)
pred = model.transform(validation_records)
evaluator.evaluate(pred)
```
## 3.3 Improve Model
The first shot is not very satisfactory, specifically if looking at the logarithmic plot. The data seems to describe a curve (which is good), we could try to fit a polynom of order 2. This means that we will use (day^2, day, const) as features. This *polynomial expansion* of the original feature `day` can be generated by the `PolynomialExpansion` feature transformer.
This means that we will slightly extend our pipeline as follows.
```
pipeline = Pipeline(
stages=[
VectorAssembler(inputCols=["day"], outputCol="day_vec"),
PolynomialExpansion(inputCol="day_vec", outputCol="features", degree=2),
GeneralizedLinearRegression(
family="gaussian",
link="log",
featuresCol="features",
labelCol="Confirmed",
predictionCol="Predict",
),
]
)
```
### Fit and predict
Now we will again fit the pipeline to retrieve a model and immediately apply the model to all cases in order to get the data for another plot.
```
model = pipeline.fit(training_records)
pred = model.transform(country_cases)
```
### Visualize
The next visualization looks better, especially the critical part of the graph is estimated much better. Note that we did not use data before day 28, since there was no real growth before that day.
Note that our predicted values are above the measured values. This can mean multiple things:
* *Pessimist*: Our model does not perform as good as desired
* *Optimist*: Actions taken by politics change the real model parameters in a favorable way, such that the real number of infections do not grow any more as predicted
```
df = pred.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.yscale('log')
plt.plot(df["day"], df["Confirmed"], color='blue', lw=2)
plt.plot(df["day"], df["Predict"], color='red', lw=2)
plt.vlines(cut_day, ymin=0, ymax=400000)
```
Although the image looks quite promising on the logarithmic scale, let us have a look at the linear scale. We will notice that we overpredict the number of cases by a factor of two and our prediction will look even worse for the future.
```
df = pred.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.plot(df["day"], df["Confirmed"], color='blue', lw=2)
plt.plot(df["day"], df["Predict"], color='red', lw=2)
plt.vlines(cut_day, ymin=0, ymax=300000)
```
### Measure performance
Again we will use the `RegressionEvaluator` as before to quantify the prediction error. The error should be much lower now.
```
evaluator = RegressionEvaluator(
predictionCol='Predict', labelCol='Confirmed', metricName='rmse'
)
pred = model.transform(validation_records)
evaluator.evaluate(pred)
```
## 3.5 Change training/validation split
If we change the split of training to validation, things look much better. Of course this might be already expected, since we predict less data, but even the non-logarithmic plot looks really good
```
# Use 80% for training
cut_day = all_days[int(0.8 * num_days)]
print("cut_day = " + str(cut_day))
training_records_80 = country_cases.where(
(f.col("day") <= cut_day) & (f.col("day") >= first_day)
).cache()
validation_records_80 = country_cases.where(f.col("day") > cut_day).cache()
model = pipeline.fit(training_records_80)
pred = model.transform(country_cases)
df = pred.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.plot(df["day"], df["Confirmed"], color='blue', lw=2)
plt.plot(df["day"], df["Predict"], color='red', lw=2)
plt.vlines(cut_day, ymin=0, ymax=120000)
```
This result should let you look optimistic into the future, as it may indicate that the underlying process really has changed between day 50 and 60 and that the infection really slows down.
### Measure performance
Again we will use the `RegressionEvaluator` as before to quantify the prediction error. The error should be much lower now.
```
evaluator = RegressionEvaluator(
predictionCol='Predict', labelCol='Confirmed', metricName='rmse'
)
pred = model.transform(validation_records)
evaluator.evaluate(pred)
```
# 4. Final Note
As already mentioned in the beginning, the whole approach is somewhat questionable. We are throwing a very generic machinery at a very specific problem which has a very specific structure. Therefore other approaches involving more meaningful models like https://de.wikipedia.org/wiki/SIR-Modell could give better prediction results. But those models require a completely different numerical approach for fitting the model to the data. We used the tool at hand (in this case PySpark) to generate a model, which does only make very mild (and possibly wrong) assumptions about the development process of the desease. Nevertheless such approaches might also give good results, since on the other hand specific mathematical models also rely on very specific assumptions and simplifications, which may also not be justified.
|
github_jupyter
|
import matplotlib.pyplot as plt
import pyspark.sql.functions as f
from pyspark.sql import SparkSession
if not 'spark' in locals():
spark = (
SparkSession.builder.master("local[*]")
.config("spark.driver.memory", "64G")
.getOrCreate()
)
spark
%matplotlib inline
basedir = 's3://dimajix-training/data/covid-19'
# basedir = '/dimajix/data/COVID-19/csse_covid_19_data'
series = spark.read.option("header", True).csv(
basedir + "/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv"
)
series.toPandas()
import datetime
from pyspark.sql.types import *
@f.udf(DateType())
def parse_date(date):
if "/" in date:
date = date.split(" ")[0]
(m, d, y) = date.split("/")
y = int(y)
m = int(m)
d = int(d)
if y < 2000:
y += 2000
else:
date = date[0:10]
(y, m, d) = date.split("-")
y = int(y)
m = int(m)
d = int(d)
return datetime.date(year=y, month=m, day=d)
# print(parse_date("2020-03-01"))
# print(parse_date("1/22/2020"))
# print(parse_date("2020-03-01T23:45:50"))
# Last date to read
today = datetime.date(2020, 4, 7) # datetime.date.today()
# First date to read
start_date = datetime.date(2020, 1, 22)
# First date with new schema
schema_switch_date = datetime.date(2020, 3, 22)
daily_reports_dir = basedir + "/csse_covid_19_daily_reports"
# Define old schema for first batch of files
schema_1 = StructType(
[
StructField("Province_State", StringType()),
StructField("Country_Region", StringType()),
StructField("Last_Update", StringType()),
StructField("Confirmed", LongType()),
StructField("Deaths", LongType()),
StructField("Recovered", LongType()),
StructField("Latitude", DoubleType()),
StructField("Longitude", DoubleType()),
]
)
# Generate all dates with old schema
schema_1_dates = [
start_date + datetime.timedelta(days=d)
for d in range(0, (schema_switch_date - start_date).days)
]
# Generate file names with old schema
schema_1_files = [
daily_reports_dir + "/" + d.strftime("%m-%d-%Y") + ".csv" for d in schema_1_dates
]
# Read in all files with old schema
cases_1 = spark.read.schema(schema_1).option("header", True).csv(schema_1_files)
# Peek inside
cases_1.toPandas()
from pyspark.sql.types import *
daily_reports_dir = basedir + "/csse_covid_19_daily_reports"
# New schema
schema_2 = StructType(
[
StructField("FIPS", StringType()),
StructField("Admin2", StringType()),
StructField("Province_State", StringType()),
StructField("Country_Region", StringType()),
StructField("Last_Update", StringType()),
StructField("Latitude", DoubleType()),
StructField("Longitude", DoubleType()),
StructField("Confirmed", LongType()),
StructField("Deaths", LongType()),
StructField("Recovered", LongType()),
StructField("Active", LongType()),
StructField("Combined_Key", StringType()),
]
)
# Generate all dates with new schema
schema_2_dates = [
schema_switch_date + datetime.timedelta(days=d)
for d in range(0, (today - schema_switch_date).days)
]
# Generate file names with new schema
schema_2_files = [
daily_reports_dir + "/" + d.strftime("%m-%d-%Y") + ".csv" for d in schema_2_dates
]
# Read in all CSV files with new schema
cases_2 = spark.read.schema(schema_2).option("header", True).csv(schema_2_files)
cases_2.toPandas()
all_cases = cases_1.select(
f.col("Country_Region"),
f.col("Province_State"),
f.lit(None).cast(StringType()).alias("Admin2"),
f.col("Last_Update"),
f.col("Confirmed"),
f.col("Deaths"),
f.col("Recovered"),
).union(
cases_2.select(
f.col("Country_Region"),
f.col("Province_State"),
f.col("Admin2"),
f.col("Last_Update"),
f.col("Confirmed"),
f.col("Deaths"),
f.col("Recovered"),
)
)
all_cases.toPandas()
all_cases_eod = all_cases.groupBy(
parse_date(f.col("Last_Update")).alias("Last_Update"),
f.col("Country_Region"),
f.col("Province_State"),
f.col("Admin2"),
).agg(
f.max(f.col("Confirmed")).alias("Confirmed"),
f.max(f.col("Deaths")).alias("Deaths"),
f.max(f.col("Recovered")).alias("Recovered"),
)
all_cases_eod.show()
all_cases_eod.where(f.col("Country_Region") == f.lit("US")).orderBy(
f.col("Confirmed").desc()
).show(truncate=False)
all_cases_eod.where(f.col("Country_Region") == f.lit("US")).where(
f.col("Last_Update") == f.lit("2020-04-05")
).select(f.sum(f.col("Confirmed"))).toPandas()
all_cases_eod.where(f.col("Country_Region") == f.lit("Germany")).where(
f.col("Last_Update") == f.lit("2020-04-06")
).select(f.sum(f.col("Confirmed"))).toPandas()
all_country_cases = all_cases_eod.groupBy("Country_Region", "Last_Update").agg(
f.sum(f.col("Confirmed")).alias("Confirmed"),
f.sum(f.col("Deaths")).alias("Deaths"),
f.sum(f.col("Recovered")).alias("Recovered"),
)
from pyspark.sql.window import Window
all_countries_age_cases = all_country_cases.withColumn(
"First_Update",
f.min(f.col("Last_Update")).over(
Window.partitionBy("Country_Region").orderBy("Last_Update")
),
).withColumn("day", f.datediff(f.col("Last_Update"), f.col("First_Update")))
all_countries_age_cases.show()
country_cases = all_countries_age_cases.where(f.col("Country_Region") == "Germany")
# Show first 10 days of data in the correct order
country_cases.orderBy(f.col("day")).show(10)
df = country_cases.toPandas()
# Set size of the figure
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
# Make an image usig plt.plot
plt.plot(df["day"], df["Confirmed"])
df = country_cases.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.yscale('log')
plt.plot(df["day"], df["Confirmed"], color='blue', lw=2)
daily_increase = country_cases.withColumn(
"Confirmed_Increase",
f.col("Confirmed")
- f.last(f.col("Confirmed")).over(
Window.partitionBy("Country_Region").orderBy("day").rowsBetween(-100, -1)
),
)
daily_increase.show(10)
df = daily_increase.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.bar(df["day"], df["Confirmed_Increase"], color='blue', lw=2)
# Import relevant packages
from pyspark.ml import *
from pyspark.ml.evaluation import *
from pyspark.ml.feature import *
from pyspark.ml.regression import *
all_days = country_cases.select("day").orderBy(f.col("day")).distinct().collect()
all_days = [row[0] for row in all_days]
num_days = len(all_days)
cut_day = all_days[int(0.7 * num_days)]
print("cut_day = " + str(cut_day))
# We might want to skip some days where there was no real growth
first_day = 28
# Select training records from first_day until cut_day (inclusive)
training_records = country_cases.where(
(f.col("day") <= cut_day) & (f.col("day") >= first_day)
).cache()
# Select validation records from cut_day (exclusive)
validation_records = country_cases.where(f.col("day") > cut_day).cache()
pipeline = Pipeline(
stages=[
VectorAssembler(inputCols=["day"], outputCol="features"),
GeneralizedLinearRegression(
family="gaussian",
link="log",
featuresCol="features",
labelCol="Confirmed",
predictionCol="Predict",
),
]
)
model = pipeline.fit(training_records)
pred = model.transform(country_cases)
df = pred.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.yscale('log')
plt.plot(df["day"], df["Confirmed"], color='blue', lw=2)
plt.plot(df["day"], df["Predict"], color='red', lw=2)
plt.vlines(cut_day, ymin=0, ymax=400000)
evaluator = RegressionEvaluator(
predictionCol='Predict', labelCol='Confirmed', metricName='rmse'
)
pred = model.transform(validation_records)
evaluator.evaluate(pred)
pipeline = Pipeline(
stages=[
VectorAssembler(inputCols=["day"], outputCol="day_vec"),
PolynomialExpansion(inputCol="day_vec", outputCol="features", degree=2),
GeneralizedLinearRegression(
family="gaussian",
link="log",
featuresCol="features",
labelCol="Confirmed",
predictionCol="Predict",
),
]
)
model = pipeline.fit(training_records)
pred = model.transform(country_cases)
df = pred.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.yscale('log')
plt.plot(df["day"], df["Confirmed"], color='blue', lw=2)
plt.plot(df["day"], df["Predict"], color='red', lw=2)
plt.vlines(cut_day, ymin=0, ymax=400000)
df = pred.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.plot(df["day"], df["Confirmed"], color='blue', lw=2)
plt.plot(df["day"], df["Predict"], color='red', lw=2)
plt.vlines(cut_day, ymin=0, ymax=300000)
evaluator = RegressionEvaluator(
predictionCol='Predict', labelCol='Confirmed', metricName='rmse'
)
pred = model.transform(validation_records)
evaluator.evaluate(pred)
# Use 80% for training
cut_day = all_days[int(0.8 * num_days)]
print("cut_day = " + str(cut_day))
training_records_80 = country_cases.where(
(f.col("day") <= cut_day) & (f.col("day") >= first_day)
).cache()
validation_records_80 = country_cases.where(f.col("day") > cut_day).cache()
model = pipeline.fit(training_records_80)
pred = model.transform(country_cases)
df = pred.toPandas()
plt.figure(figsize=(16, 6), dpi=80, facecolor='w', edgecolor='k')
plt.plot(df["day"], df["Confirmed"], color='blue', lw=2)
plt.plot(df["day"], df["Predict"], color='red', lw=2)
plt.vlines(cut_day, ymin=0, ymax=120000)
evaluator = RegressionEvaluator(
predictionCol='Predict', labelCol='Confirmed', metricName='rmse'
)
pred = model.transform(validation_records)
evaluator.evaluate(pred)
| 0.345989 | 0.973418 |
```
import csv
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
import pandas as pd
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from sklearn.model_selection import train_test_split
from tensorflow.keras.utils import to_categorical
from keras.callbacks import ReduceLROnPlateau
def get_data_train(filename):
with open(filename) as training_file:
file = csv.reader(training_file, delimiter = ",")
images = []
labels = []
ignore = 1
for row in file:
if ignore == 1:
ignore = 0
continue
labels.append(row[0])
images.append(np.array_split(row[1:],28))
return np.array(images).astype("int32"), np.array(labels).astype("int32")
def get_data_test(filename):
with open(filename) as training_file:
file = csv.reader(training_file, delimiter = ",")
images = []
ignore = 1
for row in file:
if ignore == 1:
ignore = 0
continue
images.append(np.array_split(row,28))
return np.array(images).astype("int32")
train_path = 'data/train.csv'
test_path = 'data/test.csv'
train_images, train_labels = get_data_train(train_path)
test_images = get_data_test(test_path)
seed = 1
x_train, x_test, y_train, y_test = train_test_split(train_images, train_labels, test_size=0.2, random_state=seed)
print(train_images.shape)
print(test_images.shape)
print(x_train.shape)
print(x_test.shape)
print(y_train.shape)
print(y_test.shape)
x_train = np.expand_dims(x_train, axis=3)
x_test = np.expand_dims(x_test, axis=3)
train_datagen = ImageDataGenerator(rescale = 1./255,
rotation_range=15,
width_shift_range=0.1,
height_shift_range=0.1,
shear_range=0.1,
zoom_range=0.1,
horizontal_flip=False,
fill_mode='nearest'
)
validation_datagen = ImageDataGenerator(rescale = 1./255)
print(x_train.shape)
print(x_test.shape)
model = tf.keras.models.Sequential([
tf.keras.layers.Conv2D(64, (5,5), activation=tf.nn.relu,padding='Same',input_shape=(28, 28, 1)),
#tf.keras.layers.BatchNormalization(),
#tf.keras.layers.Conv2D(64, (5,5), activation=tf.nn.relu,padding='Same'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(128, (3,3), activation=tf.nn.relu,padding = 'Same'),
#tf.keras.layers.BatchNormalization(),
#tf.keras.layers.Conv2D(128, (3,3), activation=tf.nn.relu,padding = 'Same'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(128,activation=tf.nn.relu),
#tf.keras.layers.Dropout(0.2),
tf.keras.layers.Dense(10, activation = tf.nn.softmax)
])
y_train_cat = to_categorical(y_train)
y_test_cat = to_categorical(y_test)
training_generator = train_datagen.flow(x_train, y_train_cat, batch_size=64)
validation_generator = validation_datagen.flow(x_test, y_test_cat, batch_size=64)
model.compile(loss = 'categorical_crossentropy', optimizer= tf.keras.optimizers.Adam(), metrics=['acc'])
learning_rate_reduction = ReduceLROnPlateau(monitor='val_acc',
patience=2,
verbose=1,
factor=0.5,
min_lr=0.000003)
history = model.fit(training_generator,
epochs = 35,
verbose = 1,
validation_data = validation_generator,
callbacks=[learning_rate_reduction])
%matplotlib inline
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'r', label='Training accuracy')
plt.plot(epochs, val_acc, 'b', label='Validation accuracy')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'r', label='Training Loss')
plt.plot(epochs, val_loss, 'b', label='Validation Loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
#final model
final_model = tf.keras.models.Sequential([
tf.keras.layers.Conv2D(64, (5,5), activation=tf.nn.relu,padding='Same',input_shape=(28, 28, 1)),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(128, (3,3), activation=tf.nn.relu,padding = 'Same'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(128,activation=tf.nn.relu),
tf.keras.layers.Dense(10, activation = tf.nn.softmax)
])
final_model.compile(loss = 'categorical_crossentropy', optimizer= tf.keras.optimizers.Adam(), metrics=['acc'])
final_model.summary()
train_labels_cat = to_categorical(train_labels)
train_images = np.expand_dims(train_images, axis=3)
learning_rate_reduction_final = ReduceLROnPlateau(monitor='acc',
patience=2,
verbose=1,
factor=0.5,
min_lr=0.000003)
final_model.fit(train_datagen.flow(train_images, train_labels_cat, batch_size=64),
epochs = 25,
verbose = 1,
callbacks=[learning_rate_reduction_final])
test_datagen = ImageDataGenerator(rescale = 1./255)
test_images = np.expand_dims(test_images, axis=3)
results = final_model.predict(test_datagen.flow(test_images))
results = np.argmax(results,axis = 1)
results = pd.Series(results,name="Label")
submission = pd.concat([pd.Series(range(1,28001),name = "ImageId"),results],axis = 1)
submission.to_csv("submission.csv",index=False)
get_ipython().run_cell_magic('javascript', '', '<!-- Save the notebook -->\nIPython.notebook.save_checkpoint();')
```
|
github_jupyter
|
import csv
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
import pandas as pd
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from sklearn.model_selection import train_test_split
from tensorflow.keras.utils import to_categorical
from keras.callbacks import ReduceLROnPlateau
def get_data_train(filename):
with open(filename) as training_file:
file = csv.reader(training_file, delimiter = ",")
images = []
labels = []
ignore = 1
for row in file:
if ignore == 1:
ignore = 0
continue
labels.append(row[0])
images.append(np.array_split(row[1:],28))
return np.array(images).astype("int32"), np.array(labels).astype("int32")
def get_data_test(filename):
with open(filename) as training_file:
file = csv.reader(training_file, delimiter = ",")
images = []
ignore = 1
for row in file:
if ignore == 1:
ignore = 0
continue
images.append(np.array_split(row,28))
return np.array(images).astype("int32")
train_path = 'data/train.csv'
test_path = 'data/test.csv'
train_images, train_labels = get_data_train(train_path)
test_images = get_data_test(test_path)
seed = 1
x_train, x_test, y_train, y_test = train_test_split(train_images, train_labels, test_size=0.2, random_state=seed)
print(train_images.shape)
print(test_images.shape)
print(x_train.shape)
print(x_test.shape)
print(y_train.shape)
print(y_test.shape)
x_train = np.expand_dims(x_train, axis=3)
x_test = np.expand_dims(x_test, axis=3)
train_datagen = ImageDataGenerator(rescale = 1./255,
rotation_range=15,
width_shift_range=0.1,
height_shift_range=0.1,
shear_range=0.1,
zoom_range=0.1,
horizontal_flip=False,
fill_mode='nearest'
)
validation_datagen = ImageDataGenerator(rescale = 1./255)
print(x_train.shape)
print(x_test.shape)
model = tf.keras.models.Sequential([
tf.keras.layers.Conv2D(64, (5,5), activation=tf.nn.relu,padding='Same',input_shape=(28, 28, 1)),
#tf.keras.layers.BatchNormalization(),
#tf.keras.layers.Conv2D(64, (5,5), activation=tf.nn.relu,padding='Same'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(128, (3,3), activation=tf.nn.relu,padding = 'Same'),
#tf.keras.layers.BatchNormalization(),
#tf.keras.layers.Conv2D(128, (3,3), activation=tf.nn.relu,padding = 'Same'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(128,activation=tf.nn.relu),
#tf.keras.layers.Dropout(0.2),
tf.keras.layers.Dense(10, activation = tf.nn.softmax)
])
y_train_cat = to_categorical(y_train)
y_test_cat = to_categorical(y_test)
training_generator = train_datagen.flow(x_train, y_train_cat, batch_size=64)
validation_generator = validation_datagen.flow(x_test, y_test_cat, batch_size=64)
model.compile(loss = 'categorical_crossentropy', optimizer= tf.keras.optimizers.Adam(), metrics=['acc'])
learning_rate_reduction = ReduceLROnPlateau(monitor='val_acc',
patience=2,
verbose=1,
factor=0.5,
min_lr=0.000003)
history = model.fit(training_generator,
epochs = 35,
verbose = 1,
validation_data = validation_generator,
callbacks=[learning_rate_reduction])
%matplotlib inline
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'r', label='Training accuracy')
plt.plot(epochs, val_acc, 'b', label='Validation accuracy')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'r', label='Training Loss')
plt.plot(epochs, val_loss, 'b', label='Validation Loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
#final model
final_model = tf.keras.models.Sequential([
tf.keras.layers.Conv2D(64, (5,5), activation=tf.nn.relu,padding='Same',input_shape=(28, 28, 1)),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(128, (3,3), activation=tf.nn.relu,padding = 'Same'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(128,activation=tf.nn.relu),
tf.keras.layers.Dense(10, activation = tf.nn.softmax)
])
final_model.compile(loss = 'categorical_crossentropy', optimizer= tf.keras.optimizers.Adam(), metrics=['acc'])
final_model.summary()
train_labels_cat = to_categorical(train_labels)
train_images = np.expand_dims(train_images, axis=3)
learning_rate_reduction_final = ReduceLROnPlateau(monitor='acc',
patience=2,
verbose=1,
factor=0.5,
min_lr=0.000003)
final_model.fit(train_datagen.flow(train_images, train_labels_cat, batch_size=64),
epochs = 25,
verbose = 1,
callbacks=[learning_rate_reduction_final])
test_datagen = ImageDataGenerator(rescale = 1./255)
test_images = np.expand_dims(test_images, axis=3)
results = final_model.predict(test_datagen.flow(test_images))
results = np.argmax(results,axis = 1)
results = pd.Series(results,name="Label")
submission = pd.concat([pd.Series(range(1,28001),name = "ImageId"),results],axis = 1)
submission.to_csv("submission.csv",index=False)
get_ipython().run_cell_magic('javascript', '', '<!-- Save the notebook -->\nIPython.notebook.save_checkpoint();')
| 0.697506 | 0.641387 |
Notebook criado para aplicação dos conhecimentos sobre previsões de séries temporais utilizando os modelos ARIMA, SARIMA e SARIMAX.
Considero esse tema como um dos mais difíceis de machine learning, por isso esse notebook estara em constante atualização conforme eu for melhorando meus conhecimentos.
Objetivo: Criar um modelo para realizar previsões de demanda dos passageiros em empresas aéreas.
```
!pip install pmdarima
# todas as importações que serão necessárias durante a criação do modelo
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import warnings
from statsmodels.tools.eval_measures import rmse, mse
from statsmodels.tsa.statespace.sarimax import SARIMAX
from statsmodels.tsa.seasonal import seasonal_decompose
from statsmodels.graphics.tsaplots import plot_acf,plot_pacf
from statsmodels.tsa.statespace.tools import diff
from pmdarima import auto_arima
from statsmodels.tsa.stattools import adfuller
from statsmodels.tsa.arima_model import ARMA,ARIMA,ARMAResults, ARIMAResults
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline
#utilização do colab para importar os arquivos
from google.colab import files
files.upload()
df = pd.read_csv("airline_passengers.csv", parse_dates=True, index_col=0)
df.index.freq="MS"
df.head()
df.isnull().sum()
df.describe()
```
Passado a primeira parte da análise dos dados, definição do índice como a data, podemos visualizar o dataset durante o tempo. Vemos que:
1 - Existe uma tendência de alta
2 - Existe sazonalidade em períodos
```
plt.figure(figsize=(12,5))
df['Thousands of Passengers'].plot(legend=True)
plt.title("Passengers in airlines over time")
plt.ylabel("Passengers")
plt.xlabel("Time")
plot_acf(df['Thousands of Passengers'], lags=40);
plot_pacf(df['Thousands of Passengers'], lags=30);
seasonal = seasonal_decompose(df['Thousands of Passengers'], model='multiplicative');
```
Podemos então dividir o primeiro gráfico entre a tendência, a sazonalidade e o residuo
A tendência é em que forma que o gráfico está se aprensentando? Em alta? Em queda?
Sazonalidade é os períodos que se repetem, por exemplo, no inverno as pessoas costumam viajar mais do que no verão, e isso se repete todo ano
Residuo é tudo aquilo que não pode ser explicado pela tendência e pela sazonalidade
```
seasonal.plot();
```
Vemos claramente nesse gráfico que existe uma sazonalidade , tendo uma alta no verão e uma baixa de demanda no inverno
```
plt.figure(figsize=(12,5))
seasonal.seasonal.plot();
plt.title("Seasonal")
plt.xlabel('Date')
plt.figure(figsize=(12,5))
seasonal.trend.plot()
plt.title("Tendência")
plt.xlabel('Date')
plt.figure(figsize=(12,5))
seasonal.resid.plot()
plt.title("Residual")
plt.xlabel('Date')
```
A seguir devemos então verificar se a série é estacionária ou não, para isso é utilizaremos o teste do adfuller
O seguinte teste retornará uma tupla de estatísticas do teste do ADF, como Estatística do teste Valor-P; Número de defasagens usadas; Número de observações usadas para a regressão do ADF e um dicionário de Valores críticos.
Resumindo, se o valor-p encontrado pelo teste for < 0,05, a série é estacionária, ja se o valor for acima de 0,05 a série não é estacionária e por esse motivo devemos então normalizala.
Utilizei uma função para simplificar a visualização dos valores, e o valor p encontrado foi superior a 0,05, ou seja, a série não é estacionaria.
```
# função criada para uma melhor visualização dos resultados do adfuller test
def adf_test(series,title=''):
"""
Pass in a time series and an optional title, returns an ADF report
"""
print(f'Augmented Dickey-Fuller Test: {title}')
result = adfuller(series.dropna(),autolag='AIC') # .dropna() handles differenced data
labels = ['ADF test statistic','p-value','# lags used','# observations']
out = pd.Series(result[0:4],index=labels)
for key,val in result[4].items():
out[f'critical value ({key})']=val
print(out.to_string()) # .to_string() removes the line "dtype: float64"
if result[1] <= 0.05:
print("Strong evidence against the null hypothesis")
print("Reject the null hypothesis")
print("Data has no unit root and is stationary")
else:
print("Weak evidence against the null hypothesis")
print("Fail to reject the null hypothesis")
print("Data has a unit root and is non-stationary")
adf_test(df['Thousands of Passengers'])
```
Como podemos transformar uma série não estacionária, em uma série estacionária? Podemos aplicar simplesmente a "differencing":
```
df1 = df.copy()
plt.figure(figsize=(12,5))
df1['d1'] = diff(df1['Thousands of Passengers'],k_diff=1)
df1['d1'].plot();
plt.title("Stacionary timeseries")
plt.ylabel("Date")
```
MODELO SARIMA
De uma forma resumida, o ARIMA model é composto por:
AR(p) = Autoregression model, prevemos utilizando a combinação de valores passados da próprima variável. Gera modelos lineares. Representado pela letra P
MA(d) = é o modelo de média móvel.
ARMA(p,q) = A junção dos dois acima - representado pela letra Q
ARIMA(p,q,d) = O mesmo processo que ocorre para o ARMA + aplicação da diferenciação para tornar a série estacionária.
Temos então o SARIMAX (termo genérico), além dos parâmetros (p,q,d) aceita também o (P,D,Q)m, descrevendo os componentes sazonais. P,D e Q representa a regressão sazonal, diferenciação, e média movel, m representa o número de pontos para cada ciclo.
O X representa a variável exógena, como não utilizaremos ela, não entrarei em maiores detalhes.
Para a definição dos melhores parâmetros existe a forma manual e a forma automática, onde a função definirá os melhores parâmetros para nós...
Função: Auto_arima, devo informar ainda o meu dataset, definir os pontos de start do "p" e do "q", definir a sazonalidade e o período de sazonalidade, no caso são de 12 meses.
O objetivo é achar os melhores parâmetros em base no valor AIC, que deve ser o menor possível, com a menor complexidade
```
sarima = auto_arima(df['Thousands of Passengers'],start_p=0, start_q=0,
seasonal=True,trace=True, m=12)
sarima
```
Temos então com o modelo ARIMA(0,1,1) que com a seasonal order (2,1,1,12) se transforma em SARIMA
```
sarima.summary()
train = df.iloc[:132]
test = df.iloc[132:]
start = len(train)
end = len(train) + len(test) - 1
model = SARIMAX(train['Thousands of Passengers'], order=(0, 1, 1),seasonal_order=(2, 1, 1, 12)).fit()
model.summary()
```
Após a divisão de dataset de treino e dataset de test, treinamos nosso modelo e então podemos verificar como nosso modelo performou
```
predictions = model.predict(start,end,typ="levels").rename("SARIMAX(0, 1, 1)x(2, 1, 1, 12)")
plt.figure(figsize=(12,5))
test['Thousands of Passengers'].plot(label="Test set",legend=True)
predictions.plot(legend=True)
train['Thousands of Passengers'].plot(label = "Train set",legend=True)
plt.title("Forcast")
plt.ylabel('Passengers')
plt.xlabel("Date")
```
Dando um zoom na nossa previsão, podemos ver que o modelo fez boas previsões, e chegou próximo ao real. As métricas utilizadas para verificar a performance do nosso modelo será o Mean Absolute Error e Root Mean Squared Error
```
test['Thousands of Passengers'].plot()
predictions.plot()
mse(test['Thousands of Passengers'],predictions)
rmse(test['Thousands of Passengers'],predictions) #valor muito bom comparado com a média dos passageiros no mesmo período
start = len(df['Thousands of Passengers'])
end = len(df['Thousands of Passengers']) + 24
```
Após definirmos o modelo, podemos então realizar o treinamento no dataset completo, e realizar previsões para o futuro... no caso defini que irei fazer previsões para os próximos 24 meses
```
model = SARIMAX(df['Thousands of Passengers'], order=(0, 1, 1),seasonal_order=(2, 1, 1, 12)).fit()
predictions = model.predict(start,end,typ="levels").rename("SARIMAX(0, 1, 1)x(2, 1, 1, 12) - Final")
plt.figure(figsize=(12,5))
df['Thousands of Passengers'].plot(label="Train set", legend=True)
predictions.plot(label = "predictions", legend=True)
plt.ylabel("Passengers")
plt.xlabel("Time (years)")
plt.title("Forcast Passengers")
```
|
github_jupyter
|
!pip install pmdarima
# todas as importações que serão necessárias durante a criação do modelo
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import warnings
from statsmodels.tools.eval_measures import rmse, mse
from statsmodels.tsa.statespace.sarimax import SARIMAX
from statsmodels.tsa.seasonal import seasonal_decompose
from statsmodels.graphics.tsaplots import plot_acf,plot_pacf
from statsmodels.tsa.statespace.tools import diff
from pmdarima import auto_arima
from statsmodels.tsa.stattools import adfuller
from statsmodels.tsa.arima_model import ARMA,ARIMA,ARMAResults, ARIMAResults
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline
#utilização do colab para importar os arquivos
from google.colab import files
files.upload()
df = pd.read_csv("airline_passengers.csv", parse_dates=True, index_col=0)
df.index.freq="MS"
df.head()
df.isnull().sum()
df.describe()
plt.figure(figsize=(12,5))
df['Thousands of Passengers'].plot(legend=True)
plt.title("Passengers in airlines over time")
plt.ylabel("Passengers")
plt.xlabel("Time")
plot_acf(df['Thousands of Passengers'], lags=40);
plot_pacf(df['Thousands of Passengers'], lags=30);
seasonal = seasonal_decompose(df['Thousands of Passengers'], model='multiplicative');
seasonal.plot();
plt.figure(figsize=(12,5))
seasonal.seasonal.plot();
plt.title("Seasonal")
plt.xlabel('Date')
plt.figure(figsize=(12,5))
seasonal.trend.plot()
plt.title("Tendência")
plt.xlabel('Date')
plt.figure(figsize=(12,5))
seasonal.resid.plot()
plt.title("Residual")
plt.xlabel('Date')
# função criada para uma melhor visualização dos resultados do adfuller test
def adf_test(series,title=''):
"""
Pass in a time series and an optional title, returns an ADF report
"""
print(f'Augmented Dickey-Fuller Test: {title}')
result = adfuller(series.dropna(),autolag='AIC') # .dropna() handles differenced data
labels = ['ADF test statistic','p-value','# lags used','# observations']
out = pd.Series(result[0:4],index=labels)
for key,val in result[4].items():
out[f'critical value ({key})']=val
print(out.to_string()) # .to_string() removes the line "dtype: float64"
if result[1] <= 0.05:
print("Strong evidence against the null hypothesis")
print("Reject the null hypothesis")
print("Data has no unit root and is stationary")
else:
print("Weak evidence against the null hypothesis")
print("Fail to reject the null hypothesis")
print("Data has a unit root and is non-stationary")
adf_test(df['Thousands of Passengers'])
df1 = df.copy()
plt.figure(figsize=(12,5))
df1['d1'] = diff(df1['Thousands of Passengers'],k_diff=1)
df1['d1'].plot();
plt.title("Stacionary timeseries")
plt.ylabel("Date")
sarima = auto_arima(df['Thousands of Passengers'],start_p=0, start_q=0,
seasonal=True,trace=True, m=12)
sarima
sarima.summary()
train = df.iloc[:132]
test = df.iloc[132:]
start = len(train)
end = len(train) + len(test) - 1
model = SARIMAX(train['Thousands of Passengers'], order=(0, 1, 1),seasonal_order=(2, 1, 1, 12)).fit()
model.summary()
predictions = model.predict(start,end,typ="levels").rename("SARIMAX(0, 1, 1)x(2, 1, 1, 12)")
plt.figure(figsize=(12,5))
test['Thousands of Passengers'].plot(label="Test set",legend=True)
predictions.plot(legend=True)
train['Thousands of Passengers'].plot(label = "Train set",legend=True)
plt.title("Forcast")
plt.ylabel('Passengers')
plt.xlabel("Date")
test['Thousands of Passengers'].plot()
predictions.plot()
mse(test['Thousands of Passengers'],predictions)
rmse(test['Thousands of Passengers'],predictions) #valor muito bom comparado com a média dos passageiros no mesmo período
start = len(df['Thousands of Passengers'])
end = len(df['Thousands of Passengers']) + 24
model = SARIMAX(df['Thousands of Passengers'], order=(0, 1, 1),seasonal_order=(2, 1, 1, 12)).fit()
predictions = model.predict(start,end,typ="levels").rename("SARIMAX(0, 1, 1)x(2, 1, 1, 12) - Final")
plt.figure(figsize=(12,5))
df['Thousands of Passengers'].plot(label="Train set", legend=True)
predictions.plot(label = "predictions", legend=True)
plt.ylabel("Passengers")
plt.xlabel("Time (years)")
plt.title("Forcast Passengers")
| 0.543833 | 0.949153 |
# Intern name: Anmol Pant
## TASK 3: To Explore Unsupervised Machine Learning
###### From the given ‘Iris’ dataset, predict the optimum number of clusters and represent it visually.
DataSet: https://drive.google.com/file/d/11Iq7YvbWZbt8VXjfm06brx66b10YiwK-/view?usp=sharing
```
# Importing the Required libraries
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
from sklearn import datasets
# Reading the data
iris_df = pd.read_csv("Iris.csv", index_col = 0)
print("Let's see a part of the whole dataset - \n")
iris_df.head() # See the first 5 rows
print ("The info about the datset is as follows - \n")
iris_df.info()
```
As we can see there are no null values present. We can use the dataset as it is.
Let's plot a pair plot to visualise all the attributes's dependency on each other in one go.
```
# Plotting the pair plot
sns.pairplot(iris_df, hue = 'Species')
# Defining 'X'
X = iris_df.iloc[:, [0, 1, 2, 3]].values
# Finding the optimum number of clusters for k-means classification
from sklearn.cluster import KMeans
wcss = []
for i in range(1, 11):
kmeans = KMeans(n_clusters = i, init = 'k-means++',
max_iter = 300, n_init = 10, random_state = 0)
kmeans.fit(X)
wcss.append(kmeans.inertia_)
# Plotting the results onto a line graph,
# Allowing us to observe 'The elbow'
plt.plot(range(1, 11), wcss)
plt.title('The elbow method')
plt.xlabel('Number of clusters')
plt.ylabel('WCSS') # Within cluster sum of squares
plt.show()
sns.set(rc={'figure.figsize':(7,5)})
```
It can be clearly see why it is called 'The elbow method' from the above graph, the optimum clusters is where the elbow occurs. This is when the within cluster sum of squares (WCSS) doesn't decrease significantly with every iteration.
From this we choose the number of clusters as **3**.
```
# Applying kmeans to the dataset / Creating the kmeans classifier
kmeans = KMeans(n_clusters = 3, init = 'k-means++',
max_iter = 300, n_init = 10, random_state = 0)
y_kmeans = kmeans.fit_predict(X)
y_kmeans
# Visualising the clusters - On the first two columns
plt.scatter(X[y_kmeans == 0, 0], X[y_kmeans == 0, 1],
s = 100, c = 'blue', label = 'Iris-setosa')
plt.scatter(X[y_kmeans == 1, 0], X[y_kmeans == 1, 1],
s = 100, c = 'orange', label = 'Iris-versicolour')
plt.scatter(X[y_kmeans == 2, 0], X[y_kmeans == 2, 1],
s = 100, c = 'green', label = 'Iris-virginica')
# Plotting the centroids of the clusters
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:,1],
s = 100, c = 'red', label = 'Centroids')
plt.legend()
sns.set(rc={'figure.figsize':(16,8)})
```
|
github_jupyter
|
# Importing the Required libraries
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
from sklearn import datasets
# Reading the data
iris_df = pd.read_csv("Iris.csv", index_col = 0)
print("Let's see a part of the whole dataset - \n")
iris_df.head() # See the first 5 rows
print ("The info about the datset is as follows - \n")
iris_df.info()
# Plotting the pair plot
sns.pairplot(iris_df, hue = 'Species')
# Defining 'X'
X = iris_df.iloc[:, [0, 1, 2, 3]].values
# Finding the optimum number of clusters for k-means classification
from sklearn.cluster import KMeans
wcss = []
for i in range(1, 11):
kmeans = KMeans(n_clusters = i, init = 'k-means++',
max_iter = 300, n_init = 10, random_state = 0)
kmeans.fit(X)
wcss.append(kmeans.inertia_)
# Plotting the results onto a line graph,
# Allowing us to observe 'The elbow'
plt.plot(range(1, 11), wcss)
plt.title('The elbow method')
plt.xlabel('Number of clusters')
plt.ylabel('WCSS') # Within cluster sum of squares
plt.show()
sns.set(rc={'figure.figsize':(7,5)})
# Applying kmeans to the dataset / Creating the kmeans classifier
kmeans = KMeans(n_clusters = 3, init = 'k-means++',
max_iter = 300, n_init = 10, random_state = 0)
y_kmeans = kmeans.fit_predict(X)
y_kmeans
# Visualising the clusters - On the first two columns
plt.scatter(X[y_kmeans == 0, 0], X[y_kmeans == 0, 1],
s = 100, c = 'blue', label = 'Iris-setosa')
plt.scatter(X[y_kmeans == 1, 0], X[y_kmeans == 1, 1],
s = 100, c = 'orange', label = 'Iris-versicolour')
plt.scatter(X[y_kmeans == 2, 0], X[y_kmeans == 2, 1],
s = 100, c = 'green', label = 'Iris-virginica')
# Plotting the centroids of the clusters
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:,1],
s = 100, c = 'red', label = 'Centroids')
plt.legend()
sns.set(rc={'figure.figsize':(16,8)})
| 0.776284 | 0.945751 |
<img src="../Images/Level1Beginner.png" alt="Beginner" width="128" height="128" align="right">
## Archivos en Python
### File Handler o Manejador de Archivo
En Python un **file object** es una variable que referencia a un objeto que representa a un archivo, es por medio de ese objeto que se pueden realizar operaciones con el archivo.
#### Abrir un archvo
Mediante la función **open(...)** se asigna el **file object** a una variable; el argumento obligatorio es el nombre del archivo incluida la ruta de acceso al mismo si fuese necesario.
Un segundo argumento opcional permite determinar el modo de acceso al archivo y el tratamiento del archivo binario o texto; por defecto el modo de acceso es "**r**" - **Read** y el tratamiento es "**t**" - **Text**.
Cuando la operación de apertura del archivo no se puede realizar se lanza una excepción.
En el tratamiento de archivos se deben considerar muchas situaciones excepcionales, el código siempre debe contemplar el tratamiento de las excepciones.
Enlace a la lista [excepciones específicas](https://docs.python.org/es/3/library/exceptions.html#concrete-exceptions)
```
# Abrir un archivo con valores por defecto
try :
file = open("rockandrollyfiebre.txt", mode='rt')
print(file)
except Exception as e:
print("Error de Entrada/Salida:", e)
else :
print("Cerrando el archivo")
file.close()
```
---
### Tipos de archivos
En Python se consideran dos tipos de archivos, los que tienen líneas de texto y los que tienen secuencias de bytes.
Una línea de texto es una secuencia de caracteres que finaliza con un caracter especial conocideo como **salto de linea** que se representa mediante **'\\n'**
Una secuencia de bytes corresponde a algún tipo de convención, o acuerdo que indica el significado de cada uno de esos bytes; en otras palabras para trabajar con secuencias de bytes es necesario conocer la estructura o formato en que esa secuencia de bytes fue construida.
---
### Lectura de archivos de texto
El manejador de archivo (file handle) permite acceder secuencialmente a todas las lineas de texto contenidas en el archivo.
```
# Lectura de cada una de las lineas de un archivo de text
try :
file = open("rockandrollyfiebre.txt", mode='rt')
for line in file :
print(line)
except Exception as e:
print("Error de Entrada/Salida:", e)
else :
#print("Cerrando el archivo")
file.close()
```
Es posible leer todo el contenido del archivo.
```
# Lectura de todo el contenido de un archivo de texto
try :
file = open("rockandrollyfiebre.txt", mode='rt', encoding='UTF8')
text = file.read()
print(text)
except Exception as e:
print("Error de Entrada/Salida:", e)
else :
#print("Cerrando el archivo")
file.close()
```
Es posible leer todas las lineas del archivo.
```
# Lectura de todas las lineas de un archivo de texto
try :
file = open("rockandrollyfiebre.txt", mode='rt', encoding='UTF8')
lines = file.readlines()
print(lines)
except Exception as e:
print("Error de Entrada/Salida:", e)
else :
#print("Cerrando el archivo")
file.close()
```
---
### Escritura de archivos de texto
Para **escribir** en un archivo de texto es necesario abrirlo en alguna de estas modalidades:
**a** agrega texto al final del archivo, el archivo debe existir
**w** trunca el archivo y permite agregar texto, crea el archivo si no existe
**x** crea el archivo y permite agregar texto, falla si el archivo ya existe
```
# Escritura de una archivo
try :
file = open("pepe.txt", mode='wt', encoding='UTF8')
file.write("Primera linea en el archivo\n")
file.write("\n\nFin.")
except Exception as e:
print("Error de Entrada/Salida:", e)
else :
#print("Cerrando el archivo")
file.close()
```
<img src="../Images/Level2Intermediate.png" alt="Intermediate" width="128" height="128" align="right">
### Directorios
Los sistemas de almacenamiento de archivos suelen organizarse en forma de **carpetas** o **directorios**, en Python se cuenta con el paquete **os** que facilita el tratamiento de estas estructuras.
```
import os
```
Mediante **os.listdir(...)** es posible obtener una lista de todos los archivos y directorios en un directorio en particular.
```
# Lista de archivos y directorios en el directorio actual
print(os.listdir("."))
# Lista de archivos y directorios en el directorio anterior al actual
print(os.listdir(".."))
# Lista de archivos y directorios en dos directorios anteriores al actual
print(os.listdir("../.."))
```
En el paquete **os** se encuentra un subpaquete **path** con funciones útiles para manipular la información de un directorio.
```
def listfiles(filepath=".") :
u""" Returns a list of only files in filepath """
return [filename for filename in os.listdir(filepath) if os.path.isfile(os.path.join(filepath, filename))]
def listdirectories(filepath=".") :
u""" Returns a list of only directories in filepath """
return [filename for filename in os.listdir(filepath) if not os.path.isfile(os.path.join(filepath, filename))]
print(listfiles(".."))
print(listdirectories(".."))
```
|
github_jupyter
|
# Abrir un archivo con valores por defecto
try :
file = open("rockandrollyfiebre.txt", mode='rt')
print(file)
except Exception as e:
print("Error de Entrada/Salida:", e)
else :
print("Cerrando el archivo")
file.close()
# Lectura de cada una de las lineas de un archivo de text
try :
file = open("rockandrollyfiebre.txt", mode='rt')
for line in file :
print(line)
except Exception as e:
print("Error de Entrada/Salida:", e)
else :
#print("Cerrando el archivo")
file.close()
# Lectura de todo el contenido de un archivo de texto
try :
file = open("rockandrollyfiebre.txt", mode='rt', encoding='UTF8')
text = file.read()
print(text)
except Exception as e:
print("Error de Entrada/Salida:", e)
else :
#print("Cerrando el archivo")
file.close()
# Lectura de todas las lineas de un archivo de texto
try :
file = open("rockandrollyfiebre.txt", mode='rt', encoding='UTF8')
lines = file.readlines()
print(lines)
except Exception as e:
print("Error de Entrada/Salida:", e)
else :
#print("Cerrando el archivo")
file.close()
# Escritura de una archivo
try :
file = open("pepe.txt", mode='wt', encoding='UTF8')
file.write("Primera linea en el archivo\n")
file.write("\n\nFin.")
except Exception as e:
print("Error de Entrada/Salida:", e)
else :
#print("Cerrando el archivo")
file.close()
import os
# Lista de archivos y directorios en el directorio actual
print(os.listdir("."))
# Lista de archivos y directorios en el directorio anterior al actual
print(os.listdir(".."))
# Lista de archivos y directorios en dos directorios anteriores al actual
print(os.listdir("../.."))
def listfiles(filepath=".") :
u""" Returns a list of only files in filepath """
return [filename for filename in os.listdir(filepath) if os.path.isfile(os.path.join(filepath, filename))]
def listdirectories(filepath=".") :
u""" Returns a list of only directories in filepath """
return [filename for filename in os.listdir(filepath) if not os.path.isfile(os.path.join(filepath, filename))]
print(listfiles(".."))
print(listdirectories(".."))
| 0.104569 | 0.909385 |
# OPeNDAP Access
```
#https://ghrc.nsstc.nasa.gov/opendap/globalir/data/2020/0525/globir.20146.0000
from netCDF4 import Dataset
import xarray as xr
import dask
import os
import requests
#Allows us to visualize the dask progress for parallel operations
from dask.diagnostics import ProgressBar
ProgressBar().register()
import urllib
from urllib import request
from http.cookiejar import CookieJar
import json
from urllib import request, parse
from http.cookiejar import CookieJar
import getpass
import netrc
def get_token( url: str,client_id: str, user_ip: str,endpoint: str) -> str:
try:
username, _, password = netrc.netrc().authenticators(endpoint)
token: str = ''
xml: str = """<?xml version='1.0' encoding='utf-8'?>
<token><username>{}</username><password>{}</password><client_id>{}</client_id>
<user_ip_address>{}</user_ip_address></token>""".format(username, password, client_id, user_ip)
headers: Dict = {'Content-Type': 'application/xml','Accept': 'application/json'}
resp = requests.post(url, headers=headers, data=xml)
response_content: Dict = json.loads(resp.content)
token = response_content['token']['id']
except:
print("Error getting the token - check user name and password")
return token
def setup_earthdata_login_auth(endpoint):
"""
Set up the request library so that it authenticates against the given Earthdata Login
endpoint and is able to track cookies between requests. This looks in the .netrc file
first and if no credentials are found, it prompts for them.
Valid endpoints include:
urs.earthdata.nasa.gov - Earthdata Login production
"""
try:
username, _, password = netrc.netrc().authenticators(endpoint)
except (FileNotFoundError, TypeError):
# FileNotFound = There's no .netrc file
# TypeError = The endpoint isn't in the netrc file, causing the above to try unpacking None
print('Please provide your Earthdata Login credentials to allow data access')
print('Your credentials will only be passed to %s and will not be exposed in Jupyter' % (endpoint))
username = input('Username:')
password = getpass.getpass()
manager = request.HTTPPasswordMgrWithDefaultRealm()
manager.add_password(None, endpoint, username, password)
auth = request.HTTPBasicAuthHandler(manager)
jar = CookieJar()
processor = request.HTTPCookieProcessor(jar)
opener = request.build_opener(auth, processor)
request.install_opener(opener)
edl="urs.earthdata.nasa.gov"
cmr="cmr.earthdata.nasa.gov"
setup_earthdata_login_auth(edl)
token_url="https://"+cmr+"/legacy-services/rest/tokens"
token=get_token(token_url,'podaac-subscriber', "127.0.0.1",edl)
#CMR Link to use
#https://cmr.earthdata.nasa.gov/search/granules.umm_json?collection_concept_id=C1625128926-GHRC_CLOUD&temporal=2019-01-01T10:00:00Z,2019-12-31T23:59:59Z
r = requests.get('https://cmr.earthdata.nasa.gov/search/granules.umm_json?collection_concept_id=C1996881146-POCLOUD&temporal=2019-01-01T10:00:00Z,2019-02-01T00:00:00Z&pageSize=365&token='+ token)
response_body = r.json()
od_files = []
for itm in response_body['items']:
for urls in itm['umm']['RelatedUrls']:
if 'OPeNDAP' in urls['Description']:
od_files.append(urls['URL'])
od_files
len(od_files)
for f in od_files:
print (" opening " + f)
data_url = f'{f}.dap.nc4'
# The notation below is [start index, step, end index]
# lat[ /lat= 0..17998] start index. = -90
# lon[ /lon= 0..35999] start index. = -180
# time[ /time= 0..0]
required_variables = {'analysed_sst[0:1:0][000:1:9000][000:1:9000]',
'analysis_error[0:1:0][000:1:9000][000:1:9000]',
'lat[000:1:9000]',
'lon[000:1:9000]',
'time[0:1:0]'}
#upper latitude, left longitude, lower latitude, right longitude
basename = os.path.basename(data_url)
request_params = {'dap4.ce': ';'.join(required_variables)}
#identity encoding to work around an issue with server side response compression (??)
response = requests.get(data_url, params=request_params, headers={'Accept-Encoding': 'identity'})
if response.ok:
with open(basename, 'wb') as file_handler:
file_handler.write(response.content)
else:
print(f'Request failed: {response.text}')
import xarray as xr
cloud_data = xr.open_mfdataset('*.dap.nc4', engine='h5netcdf')
cloud_data
#Histogram
cloud_data['analysed_sst'].plot()
# Choose one time segment, plot the data
cloud_data['analysed_sst'].isel(time=4).plot()
#Plot a single point over time
cloud_data['analysed_sst'].isel(lat=7000, lon=7000).plot()
```
|
github_jupyter
|
#https://ghrc.nsstc.nasa.gov/opendap/globalir/data/2020/0525/globir.20146.0000
from netCDF4 import Dataset
import xarray as xr
import dask
import os
import requests
#Allows us to visualize the dask progress for parallel operations
from dask.diagnostics import ProgressBar
ProgressBar().register()
import urllib
from urllib import request
from http.cookiejar import CookieJar
import json
from urllib import request, parse
from http.cookiejar import CookieJar
import getpass
import netrc
def get_token( url: str,client_id: str, user_ip: str,endpoint: str) -> str:
try:
username, _, password = netrc.netrc().authenticators(endpoint)
token: str = ''
xml: str = """<?xml version='1.0' encoding='utf-8'?>
<token><username>{}</username><password>{}</password><client_id>{}</client_id>
<user_ip_address>{}</user_ip_address></token>""".format(username, password, client_id, user_ip)
headers: Dict = {'Content-Type': 'application/xml','Accept': 'application/json'}
resp = requests.post(url, headers=headers, data=xml)
response_content: Dict = json.loads(resp.content)
token = response_content['token']['id']
except:
print("Error getting the token - check user name and password")
return token
def setup_earthdata_login_auth(endpoint):
"""
Set up the request library so that it authenticates against the given Earthdata Login
endpoint and is able to track cookies between requests. This looks in the .netrc file
first and if no credentials are found, it prompts for them.
Valid endpoints include:
urs.earthdata.nasa.gov - Earthdata Login production
"""
try:
username, _, password = netrc.netrc().authenticators(endpoint)
except (FileNotFoundError, TypeError):
# FileNotFound = There's no .netrc file
# TypeError = The endpoint isn't in the netrc file, causing the above to try unpacking None
print('Please provide your Earthdata Login credentials to allow data access')
print('Your credentials will only be passed to %s and will not be exposed in Jupyter' % (endpoint))
username = input('Username:')
password = getpass.getpass()
manager = request.HTTPPasswordMgrWithDefaultRealm()
manager.add_password(None, endpoint, username, password)
auth = request.HTTPBasicAuthHandler(manager)
jar = CookieJar()
processor = request.HTTPCookieProcessor(jar)
opener = request.build_opener(auth, processor)
request.install_opener(opener)
edl="urs.earthdata.nasa.gov"
cmr="cmr.earthdata.nasa.gov"
setup_earthdata_login_auth(edl)
token_url="https://"+cmr+"/legacy-services/rest/tokens"
token=get_token(token_url,'podaac-subscriber', "127.0.0.1",edl)
#CMR Link to use
#https://cmr.earthdata.nasa.gov/search/granules.umm_json?collection_concept_id=C1625128926-GHRC_CLOUD&temporal=2019-01-01T10:00:00Z,2019-12-31T23:59:59Z
r = requests.get('https://cmr.earthdata.nasa.gov/search/granules.umm_json?collection_concept_id=C1996881146-POCLOUD&temporal=2019-01-01T10:00:00Z,2019-02-01T00:00:00Z&pageSize=365&token='+ token)
response_body = r.json()
od_files = []
for itm in response_body['items']:
for urls in itm['umm']['RelatedUrls']:
if 'OPeNDAP' in urls['Description']:
od_files.append(urls['URL'])
od_files
len(od_files)
for f in od_files:
print (" opening " + f)
data_url = f'{f}.dap.nc4'
# The notation below is [start index, step, end index]
# lat[ /lat= 0..17998] start index. = -90
# lon[ /lon= 0..35999] start index. = -180
# time[ /time= 0..0]
required_variables = {'analysed_sst[0:1:0][000:1:9000][000:1:9000]',
'analysis_error[0:1:0][000:1:9000][000:1:9000]',
'lat[000:1:9000]',
'lon[000:1:9000]',
'time[0:1:0]'}
#upper latitude, left longitude, lower latitude, right longitude
basename = os.path.basename(data_url)
request_params = {'dap4.ce': ';'.join(required_variables)}
#identity encoding to work around an issue with server side response compression (??)
response = requests.get(data_url, params=request_params, headers={'Accept-Encoding': 'identity'})
if response.ok:
with open(basename, 'wb') as file_handler:
file_handler.write(response.content)
else:
print(f'Request failed: {response.text}')
import xarray as xr
cloud_data = xr.open_mfdataset('*.dap.nc4', engine='h5netcdf')
cloud_data
#Histogram
cloud_data['analysed_sst'].plot()
# Choose one time segment, plot the data
cloud_data['analysed_sst'].isel(time=4).plot()
#Plot a single point over time
cloud_data['analysed_sst'].isel(lat=7000, lon=7000).plot()
| 0.361954 | 0.327413 |
### Import Libraries
```
# pandas to handle csv
import pandas as pd
# numpy to handle array
import numpy as np
# standard scaler to normalize the dataset
from sklearn.preprocessing import StandardScaler
# seaborn, scikit plot & matplotlib for vizualisation
import seaborn as sns
import matplotlib.pyplot as plt
import scikitplot as skplt
# train test splitter and grid search cross validation
from sklearn.model_selection import train_test_split, GridSearchCV
# xgboost classifier
# https://xgboost.readthedocs.io/en/latest/python/python_api.html
from xgboost import XGBClassifier
# random forest classifier
# https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html#sklearn.ensemble.RandomForestClassifier
from sklearn.ensemble import RandomForestClassifier
# metrics functions
from sklearn.metrics import classification_report, accuracy_score, plot_confusion_matrix
```
### Import Dataset
```
# read the dataset
dataset = pd.read_csv('Attribute_list_all_features_37.csv')
# drop unwanted columns
dataset.drop(['Rider_name', 'Date', 'Race_Name', 'Rank', 'RaceStrength', 'RiderStrength', 'Year',
'last_60_10','last_60_5','last_60_3','last_120_10','last_12_5','last_120_3','last_180_10',
'last_180_5','last_180_3','last_280_10','last_280_5','last_280_3'], axis=1, inplace=True)
# remove rows with blank cells
dataset.dropna(axis=1, inplace=True)
# display first 5 rows of the dataset
dataset.head()
```
### Feature Engineering
```
# convert categorical features
dataset = pd.get_dummies(dataset,columns=['Class','SprinterType'])
# change succes to integers
dataset.loc[(dataset.Succes == 'Succesfull'), 'Succes'] = 1
dataset.loc[(dataset.Succes == 'Non-succesfull'), 'Succes'] = 0
dataset.loc[(dataset.Succes == 'Failure'), 'Succes'] = 0
# set the datatype of succes columns to numeric
dataset['Succes'] = pd.to_numeric(dataset['Succes'])
# create feature dataset
feature_dataset = dataset[['Normalized_power', 'Duration', 'Stage', 'max10secpower', 'FTP',
'Kilogram', 'CP', 'Wprime', 'power_mean', 'TSS', 'Count',
'CumSum', 'Count10', 'Count30', 'Count60', 'CumSumAbsolute',
'SSDAboveCP', 'SSD', 'Var', 'Skew', 'Kurtosis', 'WpCount',
'WpDischarge', 'L1', 'L2', 'L3', 'L4', 'L5', 'L6', 'L7', 'entire_CP',
'entire_10', 'entire_5', 'entire_3', 'entire_2', 'Class_.1', 'Class_HC',
'Class_WT', 'SprinterType_Medium', 'SprinterType_Not',
'SprinterType_Sprinter']].copy()
# create target dataset
target = dataset[['Succes']].copy()
```
### Feature Selection
```
# create a correlation heatmap to understand the correlation of features to target
# any high correlation features can be removed to see if any accuracy improvement happens to the models
f, ax = plt.subplots(figsize=(12, 12))
corr = dataset.corr()
sns.heatmap(corr, mask=np.zeros_like(corr, dtype=np.bool), cmap=sns.diverging_palette(220, 10, as_cmap=True),
square=True, ax=ax)
plt.title('Correlation HeatMap')
```
Target(Succes) doesn't have any high correlation with features present in the data. Hence proceeding with scaling and modeling
### Feature Scaling
```
# create feature array/dataframe
# Normalize the dataset so that the algorithm can be trained easily
X = pd.DataFrame(StandardScaler().fit_transform(feature_dataset))
X.columns = feature_dataset.columns
# create target array
y = target.values.ravel()
```
### Train Test Split
```
# reserve 33% of data for testing
# random state is set inorder to regenrate the same result in every run
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
```
### XGBoost Classifier
#### Grid Search and Cross Validation
```
# a function to define the gridsearch inorder to get the best fit model with optimized parameters
# function return the best fit optimized model
# https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GridSearchCV.html
def gridsearchfn(X_train_data, X_test_data, y_train_data, y_test_data,
model, param_grid, cv=10, scoring_fit='accuracy'):
gs = GridSearchCV(
estimator=model,
param_grid=param_grid,
cv=cv,
n_jobs=-1,
scoring=scoring_fit,
verbose=2
)
fitted_model = gs.fit(X_train_data, y_train_data)
return fitted_model
# define xgboost classifier with random state as model.
model = XGBClassifier(random_state=42)
# parameter grid is defined inorder to search through and get the best fit parameters
param_grid = {
'max_depth': [5,10,15,20,25,30,35,40,45,50],
'eta': [0.001, 0.01, 0.1, 1],
'gamma': [0, 0.1 , 1, 2, 5]
}
# invoke the gridsearch function to get the best fit model and parameters
model = gridsearchfn(X_train, X_test, y_train, y_test, model, param_grid, cv=3)
# print the best parameters
print('Best Parameters : ', model.best_params_)
```
#### Model Finalization
```
# Best Parameters : {'eta': 0.01, 'gamma': 0, 'max_depth': 10}
# define the model with best parameters from grid search cross validation
xgbclf = XGBClassifier(eta= 0.01, gamma= 0, max_depth= 10, random_state=0)
xgbclf.fit(X_train, y_train)
```
#### Train Data Metrics
```
# plot the confusion matrix
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.plot_confusion_matrix.html
plt.rcParams["figure.figsize"] = (10,10)
plot_confusion_matrix(xgbclf, X_train, y_train)
plt.title('Train Data Confusion Matrix - XGBoost')
# generate perfomance report with f1 score, precision, accuracy and recall parameters
y_pred = xgbclf.predict(X_train)
print ('Train Data Perfomance Details')
print (classification_report(y_train, y_pred))
```
#### Test Data Metrics
```
# plot the confusion matrix
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.plot_confusion_matrix.html
plt.rcParams["figure.figsize"] = (10,10)
plot_confusion_matrix(xgbclf, X_test, y_test)
plt.title('Test Data Confusion Matrix - XGBoost')
# generate perfomance report with f1 score, precision, accuracy and recall parameters
print ('Test Data Perfomance Details')
y_pred = xgbclf.predict(X_test)
xgbclf_acc_score = accuracy_score(y_test, y_pred)
print (classification_report(y_test, y_pred))
```
#### Test Data ROC Curve
```
# plot roc curve for test data
# https://libraries.io/pypi/scikit-plot
y_true = y_test
y_probas = xgbclf.predict_proba(X_test)
plt.rcParams["figure.figsize"] = (10,10)
skplt.metrics.plot_roc(y_true, y_probas)
plt.title('ROC Curves - XGBoost')
plt.show()
```
#### Feature Importance - XGBoost
```
# extract the feature importance data from the model
feat_imp_df = pd.DataFrame(xgbclf.get_booster().get_score( importance_type='gain'), index=[0]).T
feat_imp_df.columns = ['gain']
feat_imp_df.sort_values(by='gain', inplace=True)
# create a hroizontal bar plot to vizualise the feature importance
plt.rcParams["figure.figsize"] = (20,10)
feat_imp_df.plot.barh()
plt.title('Feature Importance - XGBoost')
```
### Random Forest Classifier
#### Grid Search and Cross Validation
```
# define xgboost classifier with random state as 42 as model.
model = RandomForestClassifier(random_state=42)
# parameter grid is defined inorder to search through and get the best fit parameters
param_grid = {
'n_estimators': [10,20,40,60,120,240,480,960]
}
# invoke the gridsearch function to get the best fit model and parameters
model = gridsearchfn(X_train, X_test, y_train, y_test, model, param_grid, cv=5)
print('Best Parameters : ', model.best_params_)
```
#### Model Finalization
```
# Best Parameters : {'n_estimators': 60}
rndfclf = XGBClassifier(n_estimators= 60, random_state=42)
rndfclf.fit(X_train, y_train)
```
#### Train Data Metrics
```
# plot the confusion matrix
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.plot_confusion_matrix.html
plt.rcParams["figure.figsize"] = (10,10)
plot_confusion_matrix(rndfclf, X_train, y_train)
plt.title('Train Data Confusion Matrix - Random Forest')
# generate perfomance report with f1 score, precision, accuracy and recall parameters
y_pred = rndfclf.predict(X_train)
print ('Train Data Perfomance Details')
print (classification_report(y_train, y_pred))
```
#### Test Data Metrics
```
# plot the confusion matrix
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.plot_confusion_matrix.html
plt.rcParams["figure.figsize"] = (10,10)
plot_confusion_matrix(rndfclf, X_test, y_test)
plt.title('Test Data Confusion Matrix - Random Forest')
# generate perfomance report with f1 score, precision, accuracy and recall parameters
print ('Test Data Perfomance Details')
y_pred = rndfclf.predict(X_test)
rndf_acc_score = accuracy_score(y_test, y_pred)
print (classification_report(y_test, y_pred))
```
#### Test Data ROC Curve
```
# plot roc curve for test data
# https://libraries.io/pypi/scikit-plot
y_true = y_test
y_probas = rndfclf.predict_proba(X_test)
plt.rcParams["figure.figsize"] = (10,10)
skplt.metrics.plot_roc(y_true, y_probas)
plt.title('ROC Curves - Random Forest')
plt.show()
```
#### Feature Importance - Random Forest
```
# extract the feature importance data from the model
feat_imp_df = pd.DataFrame(rndfclf.get_booster().get_score( importance_type='gain'), index=[0]).T
feat_imp_df.columns = ['gain']
feat_imp_df.sort_values(by='gain', inplace=True)
# create a hroizontal bar plot to vizualise the feature importance
plt.rcParams["figure.figsize"] = (20,10)
feat_imp_df.plot.barh()
plt.title('Feature Importance - Random Forest')
```
### Highest Feature Importance Factor (FTP) and Success
```
# generate a grouped data inorder to establish a relation between FTP and occurace of success
graph_df = dataset[['FTP', 'Succes']].copy()
graph_df['count'] = 1
data_df = graph_df.groupby(['FTP', 'Succes']).sum()
data_df.to_csv('FTP and Success.csv')
plt.rcParams["figure.figsize"] = (20,10)
data_df.plot.bar()
plt.title('FTP and Success')
plt.ylabel('Occurance')
```
Failure tends to happen alot with FTP 442, 438, 410, 400. Success tends to happen a lot with FTP 450, 434.
### Accuracy Comparison
```
# generate a accuracy comparison plot for the model defined above and two benchmark scenarios (all zeroes and all ones)
y_pred = np.zeros(y_test.shape)
all_zero_acc = accuracy_score(y_test, y_pred)
y_pred = np.ones(y_test.shape)
all_ones_acc = accuracy_score(y_test, y_pred)
arr_dict = {'All Zero': all_zero_acc,
'All Ones': all_ones_acc,
'XGBoost': xgbclf_acc_score,
'Random Forest': rndf_acc_score}
acc_cmp_df = pd.DataFrame(arr_dict, index=[0]).T
acc_cmp_df.columns = ['Accuracy Score']
plt.rcParams["figure.figsize"] = (10,8)
acc_cmp_df.plot.bar()
plt.title('Accuracy Comparison')
print(all_zero_acc,all_ones_acc,xgbclf_acc_score,rndf_acc_score)
```
### Conclusion
XGBoost was found to be 63% accurate on test data.
Random Forest was found to be 66% accurate on test data. Hence Random Forest can be suggested as an optimal model.
|
github_jupyter
|
# pandas to handle csv
import pandas as pd
# numpy to handle array
import numpy as np
# standard scaler to normalize the dataset
from sklearn.preprocessing import StandardScaler
# seaborn, scikit plot & matplotlib for vizualisation
import seaborn as sns
import matplotlib.pyplot as plt
import scikitplot as skplt
# train test splitter and grid search cross validation
from sklearn.model_selection import train_test_split, GridSearchCV
# xgboost classifier
# https://xgboost.readthedocs.io/en/latest/python/python_api.html
from xgboost import XGBClassifier
# random forest classifier
# https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html#sklearn.ensemble.RandomForestClassifier
from sklearn.ensemble import RandomForestClassifier
# metrics functions
from sklearn.metrics import classification_report, accuracy_score, plot_confusion_matrix
# read the dataset
dataset = pd.read_csv('Attribute_list_all_features_37.csv')
# drop unwanted columns
dataset.drop(['Rider_name', 'Date', 'Race_Name', 'Rank', 'RaceStrength', 'RiderStrength', 'Year',
'last_60_10','last_60_5','last_60_3','last_120_10','last_12_5','last_120_3','last_180_10',
'last_180_5','last_180_3','last_280_10','last_280_5','last_280_3'], axis=1, inplace=True)
# remove rows with blank cells
dataset.dropna(axis=1, inplace=True)
# display first 5 rows of the dataset
dataset.head()
# convert categorical features
dataset = pd.get_dummies(dataset,columns=['Class','SprinterType'])
# change succes to integers
dataset.loc[(dataset.Succes == 'Succesfull'), 'Succes'] = 1
dataset.loc[(dataset.Succes == 'Non-succesfull'), 'Succes'] = 0
dataset.loc[(dataset.Succes == 'Failure'), 'Succes'] = 0
# set the datatype of succes columns to numeric
dataset['Succes'] = pd.to_numeric(dataset['Succes'])
# create feature dataset
feature_dataset = dataset[['Normalized_power', 'Duration', 'Stage', 'max10secpower', 'FTP',
'Kilogram', 'CP', 'Wprime', 'power_mean', 'TSS', 'Count',
'CumSum', 'Count10', 'Count30', 'Count60', 'CumSumAbsolute',
'SSDAboveCP', 'SSD', 'Var', 'Skew', 'Kurtosis', 'WpCount',
'WpDischarge', 'L1', 'L2', 'L3', 'L4', 'L5', 'L6', 'L7', 'entire_CP',
'entire_10', 'entire_5', 'entire_3', 'entire_2', 'Class_.1', 'Class_HC',
'Class_WT', 'SprinterType_Medium', 'SprinterType_Not',
'SprinterType_Sprinter']].copy()
# create target dataset
target = dataset[['Succes']].copy()
# create a correlation heatmap to understand the correlation of features to target
# any high correlation features can be removed to see if any accuracy improvement happens to the models
f, ax = plt.subplots(figsize=(12, 12))
corr = dataset.corr()
sns.heatmap(corr, mask=np.zeros_like(corr, dtype=np.bool), cmap=sns.diverging_palette(220, 10, as_cmap=True),
square=True, ax=ax)
plt.title('Correlation HeatMap')
# create feature array/dataframe
# Normalize the dataset so that the algorithm can be trained easily
X = pd.DataFrame(StandardScaler().fit_transform(feature_dataset))
X.columns = feature_dataset.columns
# create target array
y = target.values.ravel()
# reserve 33% of data for testing
# random state is set inorder to regenrate the same result in every run
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
# a function to define the gridsearch inorder to get the best fit model with optimized parameters
# function return the best fit optimized model
# https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GridSearchCV.html
def gridsearchfn(X_train_data, X_test_data, y_train_data, y_test_data,
model, param_grid, cv=10, scoring_fit='accuracy'):
gs = GridSearchCV(
estimator=model,
param_grid=param_grid,
cv=cv,
n_jobs=-1,
scoring=scoring_fit,
verbose=2
)
fitted_model = gs.fit(X_train_data, y_train_data)
return fitted_model
# define xgboost classifier with random state as model.
model = XGBClassifier(random_state=42)
# parameter grid is defined inorder to search through and get the best fit parameters
param_grid = {
'max_depth': [5,10,15,20,25,30,35,40,45,50],
'eta': [0.001, 0.01, 0.1, 1],
'gamma': [0, 0.1 , 1, 2, 5]
}
# invoke the gridsearch function to get the best fit model and parameters
model = gridsearchfn(X_train, X_test, y_train, y_test, model, param_grid, cv=3)
# print the best parameters
print('Best Parameters : ', model.best_params_)
# Best Parameters : {'eta': 0.01, 'gamma': 0, 'max_depth': 10}
# define the model with best parameters from grid search cross validation
xgbclf = XGBClassifier(eta= 0.01, gamma= 0, max_depth= 10, random_state=0)
xgbclf.fit(X_train, y_train)
# plot the confusion matrix
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.plot_confusion_matrix.html
plt.rcParams["figure.figsize"] = (10,10)
plot_confusion_matrix(xgbclf, X_train, y_train)
plt.title('Train Data Confusion Matrix - XGBoost')
# generate perfomance report with f1 score, precision, accuracy and recall parameters
y_pred = xgbclf.predict(X_train)
print ('Train Data Perfomance Details')
print (classification_report(y_train, y_pred))
# plot the confusion matrix
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.plot_confusion_matrix.html
plt.rcParams["figure.figsize"] = (10,10)
plot_confusion_matrix(xgbclf, X_test, y_test)
plt.title('Test Data Confusion Matrix - XGBoost')
# generate perfomance report with f1 score, precision, accuracy and recall parameters
print ('Test Data Perfomance Details')
y_pred = xgbclf.predict(X_test)
xgbclf_acc_score = accuracy_score(y_test, y_pred)
print (classification_report(y_test, y_pred))
# plot roc curve for test data
# https://libraries.io/pypi/scikit-plot
y_true = y_test
y_probas = xgbclf.predict_proba(X_test)
plt.rcParams["figure.figsize"] = (10,10)
skplt.metrics.plot_roc(y_true, y_probas)
plt.title('ROC Curves - XGBoost')
plt.show()
# extract the feature importance data from the model
feat_imp_df = pd.DataFrame(xgbclf.get_booster().get_score( importance_type='gain'), index=[0]).T
feat_imp_df.columns = ['gain']
feat_imp_df.sort_values(by='gain', inplace=True)
# create a hroizontal bar plot to vizualise the feature importance
plt.rcParams["figure.figsize"] = (20,10)
feat_imp_df.plot.barh()
plt.title('Feature Importance - XGBoost')
# define xgboost classifier with random state as 42 as model.
model = RandomForestClassifier(random_state=42)
# parameter grid is defined inorder to search through and get the best fit parameters
param_grid = {
'n_estimators': [10,20,40,60,120,240,480,960]
}
# invoke the gridsearch function to get the best fit model and parameters
model = gridsearchfn(X_train, X_test, y_train, y_test, model, param_grid, cv=5)
print('Best Parameters : ', model.best_params_)
# Best Parameters : {'n_estimators': 60}
rndfclf = XGBClassifier(n_estimators= 60, random_state=42)
rndfclf.fit(X_train, y_train)
# plot the confusion matrix
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.plot_confusion_matrix.html
plt.rcParams["figure.figsize"] = (10,10)
plot_confusion_matrix(rndfclf, X_train, y_train)
plt.title('Train Data Confusion Matrix - Random Forest')
# generate perfomance report with f1 score, precision, accuracy and recall parameters
y_pred = rndfclf.predict(X_train)
print ('Train Data Perfomance Details')
print (classification_report(y_train, y_pred))
# plot the confusion matrix
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.plot_confusion_matrix.html
plt.rcParams["figure.figsize"] = (10,10)
plot_confusion_matrix(rndfclf, X_test, y_test)
plt.title('Test Data Confusion Matrix - Random Forest')
# generate perfomance report with f1 score, precision, accuracy and recall parameters
print ('Test Data Perfomance Details')
y_pred = rndfclf.predict(X_test)
rndf_acc_score = accuracy_score(y_test, y_pred)
print (classification_report(y_test, y_pred))
# plot roc curve for test data
# https://libraries.io/pypi/scikit-plot
y_true = y_test
y_probas = rndfclf.predict_proba(X_test)
plt.rcParams["figure.figsize"] = (10,10)
skplt.metrics.plot_roc(y_true, y_probas)
plt.title('ROC Curves - Random Forest')
plt.show()
# extract the feature importance data from the model
feat_imp_df = pd.DataFrame(rndfclf.get_booster().get_score( importance_type='gain'), index=[0]).T
feat_imp_df.columns = ['gain']
feat_imp_df.sort_values(by='gain', inplace=True)
# create a hroizontal bar plot to vizualise the feature importance
plt.rcParams["figure.figsize"] = (20,10)
feat_imp_df.plot.barh()
plt.title('Feature Importance - Random Forest')
# generate a grouped data inorder to establish a relation between FTP and occurace of success
graph_df = dataset[['FTP', 'Succes']].copy()
graph_df['count'] = 1
data_df = graph_df.groupby(['FTP', 'Succes']).sum()
data_df.to_csv('FTP and Success.csv')
plt.rcParams["figure.figsize"] = (20,10)
data_df.plot.bar()
plt.title('FTP and Success')
plt.ylabel('Occurance')
# generate a accuracy comparison plot for the model defined above and two benchmark scenarios (all zeroes and all ones)
y_pred = np.zeros(y_test.shape)
all_zero_acc = accuracy_score(y_test, y_pred)
y_pred = np.ones(y_test.shape)
all_ones_acc = accuracy_score(y_test, y_pred)
arr_dict = {'All Zero': all_zero_acc,
'All Ones': all_ones_acc,
'XGBoost': xgbclf_acc_score,
'Random Forest': rndf_acc_score}
acc_cmp_df = pd.DataFrame(arr_dict, index=[0]).T
acc_cmp_df.columns = ['Accuracy Score']
plt.rcParams["figure.figsize"] = (10,8)
acc_cmp_df.plot.bar()
plt.title('Accuracy Comparison')
print(all_zero_acc,all_ones_acc,xgbclf_acc_score,rndf_acc_score)
| 0.775009 | 0.892187 |
# Classification of dummy data using various classifers
### Import necessary libraries
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
```
### Read Data
```
train1 = pd.read_csv("train.csv")
train1.shape
train1.head()
```
### Check for empty cells
```
train1.isnull().sum()
```
### Plotting the Correlation Heat Map for visualization
```
%matplotlib inline
plt.figure(figsize = (10,10))
sns.heatmap(train1.corr(),annot=True , linewidths=.5)
```
### Checking more information about the data
```
train1.describe()
train1.corr() #correlation in the dataset
val1 = train1.drop(['Victory Status','Id','Number of tanks'],axis=1)
#drop unnecessary columns
```
### Import MinMaxScaler() from sklearn library for normalising the data
```
from sklearn.preprocessing import MinMaxScaler
mnmx = MinMaxScaler()
```
### Classified Column
```
val2 = train1['Victory Status']
```
### Columns with the training parameters
```
df_numcols = pd.DataFrame(mnmx.fit_transform(val1.iloc[:]),
columns = val1.iloc[:].columns.tolist())
print(df_numcols.describe())
```
### Import necessary components and split the dataset into train and test
```
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, precision_score
X_train, X_test, y_train, y_test = train_test_split(df_numcols,
val2,test_size=0.49,
random_state=40)
#df_numcols is the training prameters while val2 is the output for each
```
### Using Logistic Regression for classification
```
from sklearn.linear_model import LogisticRegression
LR = LogisticRegression()
LR.fit(X_train,y_train)
predictions= LR.predict(X_test)
print('Accuracy score: ', format(accuracy_score(y_test, predictions)))
print('Precision score: ', format(precision_score(y_test, predictions)))
```
### Using Naive Bayes for classification
```
from sklearn.naive_bayes import MultinomialNB
NB = MultinomialNB()
NB.fit(X_train,y_train)
predictions= NB.predict(X_test)
print('Accuracy score: ', format(accuracy_score(y_test, predictions)))
print('Precision score: ', format(precision_score(y_test, predictions)))
```
### Using SVM Classifier
```
from sklearn import svm
SVC_model = svm.SVC()
SVC_model.fit(X_train, y_train)
predictions = SVC_model.predict(X_test)
print('Accuracy score: ', format(accuracy_score(y_test, predictions)))
print('Precision score: ', format(precision_score(y_test, predictions)))
```
### Using Random Forest for classification
```
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(n_estimators = 1000) #n_estimators = number of trees in the forest
clf.fit(X_train,y_train)
predictions= clf.predict(X_test)
print('Accuracy score: ', format(accuracy_score(y_test, predictions)))
print('Precision score: ', format(precision_score(y_test, predictions)))
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
train1 = pd.read_csv("train.csv")
train1.shape
train1.head()
train1.isnull().sum()
%matplotlib inline
plt.figure(figsize = (10,10))
sns.heatmap(train1.corr(),annot=True , linewidths=.5)
train1.describe()
train1.corr() #correlation in the dataset
val1 = train1.drop(['Victory Status','Id','Number of tanks'],axis=1)
#drop unnecessary columns
from sklearn.preprocessing import MinMaxScaler
mnmx = MinMaxScaler()
val2 = train1['Victory Status']
df_numcols = pd.DataFrame(mnmx.fit_transform(val1.iloc[:]),
columns = val1.iloc[:].columns.tolist())
print(df_numcols.describe())
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, precision_score
X_train, X_test, y_train, y_test = train_test_split(df_numcols,
val2,test_size=0.49,
random_state=40)
#df_numcols is the training prameters while val2 is the output for each
from sklearn.linear_model import LogisticRegression
LR = LogisticRegression()
LR.fit(X_train,y_train)
predictions= LR.predict(X_test)
print('Accuracy score: ', format(accuracy_score(y_test, predictions)))
print('Precision score: ', format(precision_score(y_test, predictions)))
from sklearn.naive_bayes import MultinomialNB
NB = MultinomialNB()
NB.fit(X_train,y_train)
predictions= NB.predict(X_test)
print('Accuracy score: ', format(accuracy_score(y_test, predictions)))
print('Precision score: ', format(precision_score(y_test, predictions)))
from sklearn import svm
SVC_model = svm.SVC()
SVC_model.fit(X_train, y_train)
predictions = SVC_model.predict(X_test)
print('Accuracy score: ', format(accuracy_score(y_test, predictions)))
print('Precision score: ', format(precision_score(y_test, predictions)))
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(n_estimators = 1000) #n_estimators = number of trees in the forest
clf.fit(X_train,y_train)
predictions= clf.predict(X_test)
print('Accuracy score: ', format(accuracy_score(y_test, predictions)))
print('Precision score: ', format(precision_score(y_test, predictions)))
| 0.586286 | 0.939192 |
# 2D Advection-Diffusion equation
in this notebook we provide a simple example of the DeepMoD algorithm and apply it on the 2D advection-diffusion equation.
```
# General imports
import numpy as np
import torch
# DeepMoD functions
from deepymod import DeepMoD
from deepymod.model.func_approx import NN
from deepymod.model.library import Library2D_third
from deepymod.model.constraint import LeastSquares
from deepymod.model.sparse_estimators import Threshold,PDEFIND
from deepymod.training import train
from deepymod.training.sparsity_scheduler import TrainTestPeriodic
from scipy.io import loadmat
# Settings for reproducibility
np.random.seed(1)
torch.manual_seed(1)
if torch.cuda.is_available():
device = 'cuda'
else:
device = 'cpu'
```
## Prepare the data
Next, we prepare the dataset.
```
data = loadmat('Diffusion_2D_space41.mat')
data = np.real(data['Expression1']).reshape((41,41,41,4))[:,:,:,3]
x_dim, y_dim, t_dim = data.shape
time_range = [4,6,8,10,12,14]
for i in time_range:
# Downsample data and prepare data without noise:
down_data= np.take(np.take(np.take(data,np.arange(0,x_dim,2),axis=0),np.arange(0,y_dim,2),axis=1),np.arange(0,t_dim,i),axis=2)
print("Dowmsampled shape:",down_data.shape, "Total number of data points:", np.product(down_data.shape))
index = len(np.arange(0,t_dim,i))
width, width_2, steps = down_data.shape
x_arr, y_arr, t_arr = np.linspace(0,1,width), np.linspace(0,1,width_2), np.linspace(0,1,steps)
x_grid, y_grid, t_grid = np.meshgrid(x_arr, y_arr, t_arr, indexing='ij')
X, y = np.transpose((t_grid.flatten(), x_grid.flatten(), y_grid.flatten())), np.float32(down_data.reshape((down_data.size, 1)))
# Add noise
noise_level = 0.0
y_noisy = y + noise_level * np.std(y) * np.random.randn(y.size, 1)
# Randomize data
idx = np.random.permutation(y.shape[0])
X_train = torch.tensor(X[idx, :], dtype=torch.float32, requires_grad=True).to(device)
y_train = torch.tensor(y_noisy[idx, :], dtype=torch.float32).to(device)
# Configure DeepMoD
network = NN(3, [40, 40, 40, 40], 1)
library = Library2D_third(poly_order=0)
estimator = Threshold(0.05)
sparsity_scheduler = TrainTestPeriodic(periodicity=50, patience=200, delta=1e-5)
constraint = LeastSquares()
model = DeepMoD(network, library, estimator, constraint).to(device)
optimizer = torch.optim.Adam(model.parameters(), betas=(0.99, 0.99), amsgrad=True, lr=2e-3)
logdir='final_runs/no_noise_x21/'+str(index)+'/'
train(model, X_train, y_train, optimizer,sparsity_scheduler, log_dir=logdir, split=0.8, max_iterations=50000, delta=1e-6, patience=200)
```
|
github_jupyter
|
# General imports
import numpy as np
import torch
# DeepMoD functions
from deepymod import DeepMoD
from deepymod.model.func_approx import NN
from deepymod.model.library import Library2D_third
from deepymod.model.constraint import LeastSquares
from deepymod.model.sparse_estimators import Threshold,PDEFIND
from deepymod.training import train
from deepymod.training.sparsity_scheduler import TrainTestPeriodic
from scipy.io import loadmat
# Settings for reproducibility
np.random.seed(1)
torch.manual_seed(1)
if torch.cuda.is_available():
device = 'cuda'
else:
device = 'cpu'
data = loadmat('Diffusion_2D_space41.mat')
data = np.real(data['Expression1']).reshape((41,41,41,4))[:,:,:,3]
x_dim, y_dim, t_dim = data.shape
time_range = [4,6,8,10,12,14]
for i in time_range:
# Downsample data and prepare data without noise:
down_data= np.take(np.take(np.take(data,np.arange(0,x_dim,2),axis=0),np.arange(0,y_dim,2),axis=1),np.arange(0,t_dim,i),axis=2)
print("Dowmsampled shape:",down_data.shape, "Total number of data points:", np.product(down_data.shape))
index = len(np.arange(0,t_dim,i))
width, width_2, steps = down_data.shape
x_arr, y_arr, t_arr = np.linspace(0,1,width), np.linspace(0,1,width_2), np.linspace(0,1,steps)
x_grid, y_grid, t_grid = np.meshgrid(x_arr, y_arr, t_arr, indexing='ij')
X, y = np.transpose((t_grid.flatten(), x_grid.flatten(), y_grid.flatten())), np.float32(down_data.reshape((down_data.size, 1)))
# Add noise
noise_level = 0.0
y_noisy = y + noise_level * np.std(y) * np.random.randn(y.size, 1)
# Randomize data
idx = np.random.permutation(y.shape[0])
X_train = torch.tensor(X[idx, :], dtype=torch.float32, requires_grad=True).to(device)
y_train = torch.tensor(y_noisy[idx, :], dtype=torch.float32).to(device)
# Configure DeepMoD
network = NN(3, [40, 40, 40, 40], 1)
library = Library2D_third(poly_order=0)
estimator = Threshold(0.05)
sparsity_scheduler = TrainTestPeriodic(periodicity=50, patience=200, delta=1e-5)
constraint = LeastSquares()
model = DeepMoD(network, library, estimator, constraint).to(device)
optimizer = torch.optim.Adam(model.parameters(), betas=(0.99, 0.99), amsgrad=True, lr=2e-3)
logdir='final_runs/no_noise_x21/'+str(index)+'/'
train(model, X_train, y_train, optimizer,sparsity_scheduler, log_dir=logdir, split=0.8, max_iterations=50000, delta=1e-6, patience=200)
| 0.602763 | 0.969295 |
# Lesson 3 Class Exercises: Pandas Part 1
With these class exercises we learn a few new things. When new knowledge is introduced you'll see the icon shown on the right:
<span style="float:right; margin-left:10px; clear:both;"></span>
## Reminder
The first checkin-in of the project is due next Tueday. After today, you should have everything you need to know to accomplish that first part.
## Get Started
Import the Numpy and Pandas packages
```
import numpy as np
import pandas as pd
```
## Exercise 1: Import Iris Data
Import the Iris dataset made available to you in the last class period for the Numpy part2 exercises. Save it to a variable naemd `iris`. Print the first 5 rows and the dimensions to ensure it was read in properly.
```
iris = pd.read_csv('data/iris.csv')
print(iris.head())
iris.shape
```
Notice how much easier this was to import compared to the Numpy `genfromtxt`. We did not have to skip the headers, we did not have to specify the data type and we can have mixed data types in the same matrix.
## Exercise 2: Import Legislators Data
For portions of this notebook we will use a public dataset that contains all of the current legistators of the United States Congress. This dataset can be found [here](https://github.com/unitedstates/congress-legislators).
Import the data directly from this URL: https://theunitedstates.io/congress-legislators/legislators-current.csv
Save the data in a variable named `legistators`. Print the first 5 lines, and the dimensions.
```
legislators = pd.read_csv("https://theunitedstates.io/congress-legislators/legislators-current.csv")
print(legislators.head())
legistators.shape
```
## Exercise 3: Explore the Data
### Task 1
Print the column names of the legistators dataframe and explore the type of data in the data frame.
```
legislators.columns
```
### Task 2
Show the datatypes of all of the columns in the legislator data. Do all of the data types seem appropriate for the data?
```
legislators.dtypes
```
Show all of the datayptes in the iris dataframe
```
iris.dtypes
```
### Task 3
It's always important to know where the missing values are in your data. Are there any missing values in the legislators dataframe? How many per column?
Hint: we didn't learn how to find missing values in the lesson, but we can use the `isna()` function.
```
legislators.isna().sum()
```
How about in the iris dataframe?
```
iris.isna().sum()
```
### Task 4
It is also important to know if you have any duplicatd rows. If you are performing statistcal analyses and you have duplicated entries they can affect the results. So, let's find out. Are there any duplicated rows in the legislators dataframe? Print then number of duplicates. If there are duplicates print the rows. What function could we used to find out if we have duplicated rows?
```
legislators.duplicated().sum()
```
Do we have duplicated rows in the iris dataset? Print the number of duplicates? If there are duplicates print the rows.
```
iris.duplicated().sum()
iris[iris.duplicated()]
```
If there are duplicated rows should we remove them or keep them?
### Task 5
It is important to also check that the range of values in our data matches expectations. For example, if we expect to have four species in our iris data, we should check that we see four species. How many political parties should we expect in the legislators data? If all we saw were a single part perhaps the data is incomplete.... Let's check. You can find out how many unique values there are per column using the `nunique` function. Try it for both the legislators and the iris data set.
```
legislators.nunique()
iris.nunique()
```
What do you think? Do we see what we might expect? Are there fields where this type of check doesn't matter? In what fields might this type of exploration matter?
Check to see if you have all of the values expected for a given field. Pick a column you know should have a set number of values and print all of the unique values in that column. Do so for both the legislator and iris datasets.
```
print(legislators['gender'].unique())
print(iris['species'].unique())
```
## Exercise 5: Describe the data
For both the legislators and the iris data, get descriptive statistics for each numeric field.
```
iris.describe()
legislators.describe()
```
## Exercise 6: Row Index Labels
For the legislator dataframe, let's change the row labels from numerical indexes to something more recognizable. Take a look at the columns of data, is there anything you might want to substitue as a row label? Pick one and set the index lables. Then print the top 5 rows to see if the index labels are present.
```
legislators.index = legislators['full_name']
print(legislators.head(5))
```
## Exercise 7: Indexing & Sampling
Randomly select 15 Republicans or Democrats (your choice) from the senate.
```
legislators[(~legislators['senate_class'].isna()) & (legislators['party'] == "Republican")].sample(15)
```
## Exercise 8: Dates
<span style="float:right; margin-left:10px; clear:both;"></span>
Let's learn something not covered in the Pandas 1 lesson regarding dates. We have the birthdates for each legislator, but they are in a String format. Let's convert it to a datetime object. We can do this using the `pd.to_datetime` function. Take a look at the online documentation to see how to use this function. Convert the `legislators['birthday']` column to a `datetime` object. Confirm that the column is now a datetime object.
```
legislators['birthday'] = pd.to_datetime(legislators['birthday'])
legislators['birthday'].head()
```
Now that we have the birthdays in a `datetime` object, how can we calculate their age? Hint: we can use the `pd.Timestamp.now()` function to get a datetime object for this moment. Let's subtract the current time from their birthdays. Print the top 5 results.
```
(pd.Timestamp.now() - legislators['birthday']).head()
```
Notice that the result of subtracting two `datetime` objects is a `timedelta` object. It contains the difference between two time values. The value we calculated therefore gives us the number of days old. However, we want the number of years.
To get the number of years we can divide the number of days old by the number of days in a year (i.e. 365). However, we need to extract out the days from the `datetime` object. To get this, the Pandas Series object has an accessor for extracting components of `datetime` objects and `timedelta` objects. It's named `dt` and it works for both. You can learn more about the attributes of this accessor at the [datetime objects page](https://pandas.pydata.org/pandas-docs/stable/reference/series.html#datetime-properties) and the [timedelta objects page](https://pandas.pydata.org/pandas-docs/stable/reference/series.html#timedelta-properties) by clicking. Take a moment to look over that documentation.
How would then extract the days in order to divide by 365 to get the years? Once you've figurd it out. Do so, convert the years to an integer and add the resulting series back into the legislator dataframe as a new column named `age`. Hint: use the [astype](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.Series.astype.html) function of Pandas to convert the type.
```
age = ((pd.Timestamp.now() - legislators['birthday']).dt.days / 365).astype('int')
legislators['age'] = age
```
Next, find the youngest, oldest and average age of all legislators
```
legislators.describe()
```
Who are the oldest and youngest legislators?
```
legislators[(legislators['age'] == 86) | (legislators['age']==30)]
```
## Exercise 9: Indexing with loc and iloc
Reindex the legislators dataframe using the state, and find all legislators from your home state using the `loc` accessor.
```
legislators.index = legislators['state']
legislators.loc['SC']
```
Use the loc command to find all legislators from South Carolina and North Carolina
```
legislators.loc[['SC', 'NC']]
```
Use the loc command to retrieve all legislators from California, Oregon and Washington and only get their full name, state, party and age
```
legislators.loc[['CA', 'OR', 'WA'], ['full_name', 'state', 'party', 'age']]
```
## Exercise 10: Economics Data Example
### Task 1: Explore the data
Import the data from the [Lectures in Quantiatives Economics](https://github.com/QuantEcon/lecture-source-py) regarding minimum wages in countries round the world in US Dollars. You can view the data [here](https://github.com/QuantEcon/lecture-source-py/blob/master/source/_static/lecture_specific/pandas_panel/realwage.csv) and you can access the data file here: https://raw.githubusercontent.com/QuantEcon/lecture-source-py/master/source/_static/lecture_specific/pandas_panel/realwage.csv. Then perform the following
Import and print the first 5 lines of data to explore what is there.
```
minwages = pd.read_csv('https://raw.githubusercontent.com/QuantEcon/lecture-source-py/master/source/_static/lecture_specific/pandas_panel/realwage.csv')
minwages.head()
```
Find the shape of the data.
```
minwages.shape
```
List the column names.
```
minwages.columns
```
Identify the data types. Do they match what you would expect?
```
minwages.dtypes
```
Identify columns with missing values.
```
minwages.isna().sum()
```
Identify if there are duplicated entires.
```
minwages.duplicated().sum()
```
How many unique values per row are there. Do these look reasonable for the data type and what you know about what is stored in the column?
```
minwages.nunique()
```
### Task 2: Explore More
Retrieve descriptive statistics for the data.
```
minwages.describe()
```
Identify all of the countries listed in the data.
```
minwages['Country'].unique()
```
Convert the time column to a datetime object.
```
minwages['Time'] = pd.to_datetime(minwages['Time'])
```
Identify the time points that were used for data collection. How many years of data collection were there? What time of year were the data collected?
```
minwages['Time'].unique()
```
Because we only have one data point collected per year per country, simplify this by adding a new column with just the year. Print the first 5 rows to confirm the column was added.
```
minwages['Year'] = minwages['Time'].dt.year
minwages.head()
```
There are two pay periods. Retrieve them in a list of just the two strings
```
minwages['Pay period'].unique()
```
### Task 3: Clean the data
We have no duplicates in this data so we do not need to consider removing those, but we do have missing values in the `value` column. Lets remove those. Check the dimensions afterwards to make sure they rows with missing values are gone.
```
minwages.dropna(inplace=True)
minwages.shape
```
### Task 4: Indexing
Use boolean indexing to retrieve the rows of annual salary in United States
```
minwages[(minwages['Country'] == "United States") & (minwages['Pay period'] == 'Annual')]
```
Do we have enough data to calculate descriptive statistics for annual salary in the United States in 2016?
```
minwages[(minwages['Country'] == "United States") & (minwages['Pay period'] == 'Annual') & (minwages['Year'] == 2016)]
```
Use loc to calculate descriptive statistics for the hourly salary in the United States and then again separately for Ireland. Hint: you will have to set row indexes.
```
minwages.index = minwages['Country']
minwages[minwages['Pay period'] == 'Hourly'].loc['United States'].describe()
minwages[minwages['Pay period'] == 'Hourly'].loc['Ireland'].describe()
```
Now do the same for Annual salary
```
minwages[minwages['Pay period'] == 'Annual'].loc['Ireland'].describe()
minwages[minwages['Pay period'] == 'Annual'].loc['United States'].describe()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
iris = pd.read_csv('data/iris.csv')
print(iris.head())
iris.shape
legislators = pd.read_csv("https://theunitedstates.io/congress-legislators/legislators-current.csv")
print(legislators.head())
legistators.shape
legislators.columns
legislators.dtypes
iris.dtypes
legislators.isna().sum()
iris.isna().sum()
legislators.duplicated().sum()
iris.duplicated().sum()
iris[iris.duplicated()]
legislators.nunique()
iris.nunique()
print(legislators['gender'].unique())
print(iris['species'].unique())
iris.describe()
legislators.describe()
legislators.index = legislators['full_name']
print(legislators.head(5))
legislators[(~legislators['senate_class'].isna()) & (legislators['party'] == "Republican")].sample(15)
legislators['birthday'] = pd.to_datetime(legislators['birthday'])
legislators['birthday'].head()
(pd.Timestamp.now() - legislators['birthday']).head()
age = ((pd.Timestamp.now() - legislators['birthday']).dt.days / 365).astype('int')
legislators['age'] = age
legislators.describe()
legislators[(legislators['age'] == 86) | (legislators['age']==30)]
legislators.index = legislators['state']
legislators.loc['SC']
legislators.loc[['SC', 'NC']]
legislators.loc[['CA', 'OR', 'WA'], ['full_name', 'state', 'party', 'age']]
minwages = pd.read_csv('https://raw.githubusercontent.com/QuantEcon/lecture-source-py/master/source/_static/lecture_specific/pandas_panel/realwage.csv')
minwages.head()
minwages.shape
minwages.columns
minwages.dtypes
minwages.isna().sum()
minwages.duplicated().sum()
minwages.nunique()
minwages.describe()
minwages['Country'].unique()
minwages['Time'] = pd.to_datetime(minwages['Time'])
minwages['Time'].unique()
minwages['Year'] = minwages['Time'].dt.year
minwages.head()
minwages['Pay period'].unique()
minwages.dropna(inplace=True)
minwages.shape
minwages[(minwages['Country'] == "United States") & (minwages['Pay period'] == 'Annual')]
minwages[(minwages['Country'] == "United States") & (minwages['Pay period'] == 'Annual') & (minwages['Year'] == 2016)]
minwages.index = minwages['Country']
minwages[minwages['Pay period'] == 'Hourly'].loc['United States'].describe()
minwages[minwages['Pay period'] == 'Hourly'].loc['Ireland'].describe()
minwages[minwages['Pay period'] == 'Annual'].loc['Ireland'].describe()
minwages[minwages['Pay period'] == 'Annual'].loc['United States'].describe()
| 0.263694 | 0.99083 |
# Лабораторная работа 1 Условные операторы и циклы в языке Python
### Цель работы:
приобретение навыков программирования разветвляющихся алгоритмов и
алгоритмов циклической структуры. Освоить операторы языка Python версии 3.x if , while ,
for , break и continue , позволяющих реализовывать разветвляющиеся алгоритмы и
алгоритмы циклической структуры.
### Практическая часть:
### Индивидуальные задания:
**Задание 1.** Дано целое число такое, что |C|<9. Вывести это число в словесной форме, учитывая его знак.
```
C= int(input("введите число:"))
if C>-9 and C<9:
if(C==-8):
print("Минус восемь")
if(C==-7):
print("Минус семь")
if(C==-6):
print("Минус шесть")
if(C==-5):
print("Минус пять")
if(C==-4):
print("Минус четыри")
if(C==-3):
print("Минус три")
if(C==-2):
print("Минус два")
if(C==-1):
print("Минус один")
if(C==0):
print("Ноль")
if(C==8):
print("Восемь")
if(C==7):
print("Семь")
if(C==6):
print("Шесть")
if(C==5):
print("Пять")
if(C==4):
print("Четыри")
if(C==3):
print("Три")
if(C==2):
print("Два")
if(C==1):
print("Один")
else:
print("Вы ввели число вне предела |C|<9")
```
**Задание 2.** Решить квадратное неравенство `ax^2 + bx + c > 0` `(a!=0)`, где `a,b` и `c`- действительные числа.
```
import math
a= float(input("введите число a: "))
if(a==0):
print("Ошибка. Вы ввели число a равное нулю")
else:
b= float(input("введите число b: "))
c= float(input("введите число c: "))
print("Решение")
D = b ** 2 - 4 * a * c
print("Дискриминант D = %.2f" % D)
if D > 0:
x1 = (-b + math.sqrt(D)) / (2 * a)
x2 = (-b - math.sqrt(D)) / (2 * a)
print("x1 = %.2f \nx2 = %.2f" % (x1, x2))
if(x1>x2):
print("Ответ: x>%.2f, x<%.2f" % (x1, x2))
if(x1<x2):
print("Ответ: x>%.2f, x<%.2f" % (x2, x1))
elif D == 0:
x = -b / (2 * a)
print("x = %.2f" % x)
print("Ответ: x>%.2f" % x)
else:
print("Ответ: корней нет")
```
**Задание 3.** Напечатать таблицу соответствия между весом в фунтах и весом в кг для значений от 1 до ***а***
фунтов с шагом 1 фунт, если 1 фунт = 400 г.
```
i= int(input("Введите число a (предельное значение фунтов): "))
f=1
beg=1
kg=0.4
print("\nТаблица соответствия фунтов и килограмм:")
while f<=i:
print("%.2f Фунт=%.2f Кг"% (f, kg))
f+=1
kg*=2
```
**Задание повышенной сложности** произвести вычисления значения специальной функции по ее разложению в ряд с точностью \\(\epsilon=10^{-10}\\), аргумент функции вводится с клавиатуры. $$ J_n(x) = (\frac{x}{2})^{n} \displaystyle\sum_{\kappa=0}^{\infty} \frac{{(x^2/4)}^\kappa}{k!(\kappa+n)!} $$
```
import mpmath
n= int(input("введите число n:"))
x= float(input("введите число x: "))
J=mpmath.besselj(n, x, derivative=0)
print("J(x)=",J)
```
|
github_jupyter
|
C= int(input("введите число:"))
if C>-9 and C<9:
if(C==-8):
print("Минус восемь")
if(C==-7):
print("Минус семь")
if(C==-6):
print("Минус шесть")
if(C==-5):
print("Минус пять")
if(C==-4):
print("Минус четыри")
if(C==-3):
print("Минус три")
if(C==-2):
print("Минус два")
if(C==-1):
print("Минус один")
if(C==0):
print("Ноль")
if(C==8):
print("Восемь")
if(C==7):
print("Семь")
if(C==6):
print("Шесть")
if(C==5):
print("Пять")
if(C==4):
print("Четыри")
if(C==3):
print("Три")
if(C==2):
print("Два")
if(C==1):
print("Один")
else:
print("Вы ввели число вне предела |C|<9")
import math
a= float(input("введите число a: "))
if(a==0):
print("Ошибка. Вы ввели число a равное нулю")
else:
b= float(input("введите число b: "))
c= float(input("введите число c: "))
print("Решение")
D = b ** 2 - 4 * a * c
print("Дискриминант D = %.2f" % D)
if D > 0:
x1 = (-b + math.sqrt(D)) / (2 * a)
x2 = (-b - math.sqrt(D)) / (2 * a)
print("x1 = %.2f \nx2 = %.2f" % (x1, x2))
if(x1>x2):
print("Ответ: x>%.2f, x<%.2f" % (x1, x2))
if(x1<x2):
print("Ответ: x>%.2f, x<%.2f" % (x2, x1))
elif D == 0:
x = -b / (2 * a)
print("x = %.2f" % x)
print("Ответ: x>%.2f" % x)
else:
print("Ответ: корней нет")
i= int(input("Введите число a (предельное значение фунтов): "))
f=1
beg=1
kg=0.4
print("\nТаблица соответствия фунтов и килограмм:")
while f<=i:
print("%.2f Фунт=%.2f Кг"% (f, kg))
f+=1
kg*=2
import mpmath
n= int(input("введите число n:"))
x= float(input("введите число x: "))
J=mpmath.besselj(n, x, derivative=0)
print("J(x)=",J)
| 0.071142 | 0.910386 |
# Introduction
> This course introduces students to basic microeconmetric methods. The objective is to learn how to make and evaluate causal claims. By the end of the course, students should to able to apply each of the methods discussed and critically evaluate research based on them.
I just want to discuss some basic features of the course. We discuss the core references, the tooling for the course, student projects, and illustrate the basics of the potential outcomes model and causal graphs.
#### Causal questions
What is the causal effect of ...
* neighborhood of residence on educational performance, deviance, and youth development
* school vouchers on learning?
* of charter schools on learning?
* worker training on earnings?
* ...
What causal question brought you here?
### Core reference Test
The whole course is built on the following textbook:
* Winship, C., & Morgan, S. L. (2007). [*Counterfactuals and causal inference: Methods and principles for social research*](https://www.amazon.com/Counterfactuals-Causal-Inference-Principles-Analytical/dp/1107694167/ref=dp_ob_title_bk). Cambridge, England: Cambridge University Press.
This is a rather non-standard textbook in economics. However, I very much enjoy working with it as it provides a coherent conceptual framework for a host of different methods for causal analysis. It then clearly delineates the special cases that allow the application of particular methods. We will follow their lead and structure our thinking around the **counterfactual approach to causal analysis** and its two key ingredients **potential outcome model** and **directed graphs**.
It also is one of the few textbooks that includes extensive simulation studies to convey the economic assumptions required to apply certain estimation strategies.
It is not very technical at all, so will also need to draw on more conventional resources to fill this gap.
* Wooldridge, J. M. (2001). [*Econometric analysis of cross section and panel data*](https://mitpress.mit.edu/books/econometric-analysis-cross-section-and-panel-data). Cambridge, MA: The MIT Press.
* Angrist, J. D., & Pischke, J. (2009). [*Mostly harmless econometrics: An empiricists companion*](https://www.amazon.com/Mostly-Harmless-Econometrics-Empiricists-Companion/dp/0691120358/ref=sr_1_1?keywords=mostly+harmless+econometrics&qid=1553511192&s=gateway&sr=8-1). Princeton, NJ: Princeton University Press.
* Frölich, M., and Sperlich, S. (2019). [*Impact evaluation: Treatment effects and causal analysis*](https://www.cambridge.org/core/books/impact-evaluation/F07A859F06FF131D78DA7FC81939A6DC). Cambridge, England: Cambridge University Press.
Focusing on the conceptual framework as much as we do in the class has its cost. We might not get to discuss all the approaches you might be particularly interested in. However, my goal is that all of you can draw on this framework later on to think about your econometric problem in a structured way. This then enables you to choose the right approach for the analysis and study it in more detail on your own.
<img src="material/fig-dunning-kruger.png" width="500">
Combining this counterfactual approach to causal analysis with sufficient domain-expertise will allow you to leave the valley of despair.
### Lectures
We follow the general structure of Winship & Morgan (2007).
* Counterfactuals, potential outcomes and causal graphs
* Estimating causal effects by conditioning on observables
* regression, matching, ...
* Estimating causal effects by other means
* instrumental variables, mechanism-based estimation, regression discontinuity design, ...
### Tooling
We will use open-source software and some of the tools building on it extensively throughout the course.
* [Course website](https://ose-data-science.readthedocs.io/en/latest/)
* [GitHub](https://github.com)
* [Zulip](https://zulip.com)
* [Python](https://python.org)
* [SciPy](https://www.scipy.org/index.html) and [statsmodels](https://www.statsmodels.org/stable/index.html)
* [Jupyterlan](https://jupyterlab.readthedocs.io/en/stable/)
* [GitHub Actions](https://github.com/features/actions)
We will briefly discuss each of these components over the next week. By then end of the term, you hopefully have a good sense on how we combine all of them to produce sound empirical research. Transparency and reproducibility are a the absolute minimum of sound data science and all then can be very achieved using the kind of tools of our class.
Compared to other classes on the topic, we will do quite some programming in class. I think I have a good reason to do so. From my own experience in learning and teaching the material, there is nothing better to understand the potential and limitations of the approaches we discuss than to implemented them in a simulation setup where we have full control of the underlying data generating process.
To cite Richard Feynman: What I cannot create, I cannot understand.
However, it is often problematic that students have a very, very heterogeneous background regarding their prior programming experience and some feel intimidated by the need to not only learn the material we discuss in class but also catch up on the programming. To mitigate this valid concern, we started several accompanying initiatives that will get you up to speed such as additional workshop, help desks, etc. Make sure to join our Q&A channels in Zulip and attend the our [Computing Primer](https://github.com/OpenSourceEconomics/ose-course-primer).
### Problem sets
Thanks to [Mila Kiseleva](https://github.com/milakis), [Tim Mensinger](https://github.com/timmens/), and [Sebastian Gsell](https://github.com/segsell) we now have four problem sets available on our website.
* Potential outcome model
* Matching
* Regression-discontinuity design
* Generalized Roy model
Just as the whole course, they do not only require you to further digest the material in the course but also require you to do some programming. They are available on our course website and we will discuss them in due course.
### Projects
Applying methods from data science and understanding their potential and limitations is only possible when bringing them to bear on one's one research project. So we will work on student projects during the course. More details are available [here](https://ose-data-science.readthedocs.io/en/latest/projects/index.html).
### Data sources
Throughout the course, we will use several data sets that commonly serve as teaching examples. We collected them from several textbooks and are available in a central place in our online repository [here](https://github.com/OpenSourceEconomics/ose-course-data-science/tree/master/datasets).
### Potential outcome model
The potential outcome model serves us several purposes:
* help stipulate assumptions
* evaluate alternative data analysis techniques
* think carefully about process of causal exposure
#### Basic setup
There are three simple variables:
* $D$, treatment
* $Y_1$, outcome in the treatment state
* $Y_0$, outcome in the no-treatment state
#### Examples
* economics of education
* health economics
* industrial organization
* $...$
#### Exploration
We will use our first dataset to illustrate the basic problems of causal analysis. We will use the original data from the article below:
* LaLonde, R. J. (1986). [Evaluating the econometric evaluations of training programs with experimental data](https://www.jstor.org/stable/1806062). *The American Economic Review*, 76(4), 604-620.
He summarizes the basic setup as follows:
> The National Supported Work Demonstration (NSW) was temporary employment program desinged to help disadvantaged workers lacking basic job skills move into the labor market by giving them work experience and counseling in sheltered environment. Unlike other federally sponsored employment programs, the NSW program assigned qualified applications randomly. Those assigned to the treatment group received all the benefits of the NSW program, while those assigned to the control group were left to fend for themselves.
What is the *effect* of the program?
We will have a quick look at a subset of the data to illustrate the **fundamental problem of evaluation**, i.e. we only observe one of the potential outcomes depending on the treatment status but never both.
```
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import numpy as np
# We collected a host of data from two other influential textbooks.
df = pd.read_csv("../../datasets/processed/dehejia_waba/nsw_lalonde.csv")
df.index.set_names("Individual", inplace=True)
df.describe()
# It is important to check for missing values first.
for column in df.columns:
assert not df[column].isna().any()
```
Note that this lecture, just as all other lectures, is available on [](https://mybinder.org/v2/gh/HumanCapitalAnalysis/microeconometrics/master?filepath=lectures%2F01_introduction%2Flecture.ipynb) so you can easily continue working on it and take your exploration to another direction.
There are numerous discrete variables in this dataset describing the individual's background. How does their distribution look like?
```
columns_background = [
"treat",
"age",
"education",
"black",
"hispanic",
"married",
"nodegree",
]
for column in columns_background:
sns.countplot(x=df[column], color="#1f77b4")
plt.show()
```
How about the continous earnings variable?
```
columns_outcome = ["re75", "re78"]
for column in columns_outcome:
earnings = df[column]
# We drop all earnings at zero.
earnings = earnings.loc[earnings > 0]
ax = sns.histplot(earnings)
ax.set_xlim([0, None])
plt.show()
```
We work under the assumption that the data is generated by an experiment. Let's make sure by checking the distribution of the background variables by treatment status.
```
info = ["count", "mean", "std"]
for column in columns_background:
print("\n\n", column.capitalize())
print(df.groupby("treat")[column].describe()[info])
```
What is the data that corresponds to $(Y, Y_1, Y_0, D)$?
```
# We first create True / False
is_treated = df["treat"] == 1
df["Y"] = df["re78"]
df["Y_0"] = df.loc[~is_treated, "re78"]
df["Y_1"] = df.loc[is_treated, "re78"]
df["D"] = np.nan
df.loc[~is_treated, "D"] = 0
df.loc[is_treated, "D"] = 1
df[["Y", "Y_1", "Y_0", "D"]].sample(10)
```
Let us get a basic impression on how the distribution of earnings looks like by treatment status.
```
df.groupby("D")["re78"].describe()
ax = sns.histplot(df.loc[~is_treated, "Y"], label="untreated", color = "red")
ax = sns.histplot(df.loc[df.treat == 1].Y , label="treated")
ax.set_xlim(0, None)
ax.legend()
```
We are now ready to reproduce one of the key findings from this article. What is the difference in earnings in 1978 between those that did participate in the program and those that did not?
```
stat = df.loc[is_treated, "Y"].mean() - df.loc[~is_treated, "Y"].mean()
f"{stat:.2f}"
```
Earnings are \$886.30 higher among those that participate in the treatment compared to those that do not. Can we say even more?
**References**
Here are some further references for the potential outcome model.
* Heckman, J. J., and Vytlacil, E. J. (2007a). `Econometric evaluation of social programs, part I: Causal effects, structural models and econometric policy evaluation <https://www.sciencedirect.com/science/article/pii/S1573441207060709>`_. In J. J. Heckman, and E. E. Leamer (Eds.), *Handbook of Econometrics* (Vol. 6B, pp. 4779–4874). Amsterdam, Netherlands: Elsevier Science.
* Imbens G. W., and Rubin D. B. (2015). `Causal inference for statistics, social, and biomedical sciences: An introduction <https://www.cambridge.org/core/books/causal-inference-for-statistics-social-and-biomedical-sciences/71126BE90C58F1A431FE9B2DD07938AB>`_. Cambridge, England: Cambridge University Press.
* Rosenbaum, P. R. (2017). `Observation and experiment: An introduction to causal inference <https://www.hup.harvard.edu/catalog.php?isbn=9780674975576>`_. Cambridge, MA: Harvard University Press.
### Causal graphs
One unique feature of our core textbook is the heavy use of causal graphs to investigate and assess the validity of different estimation strategies. There are three general strategies to estimate causal effects and their applicability depends on the exact structure of the causal graph.
* condition on variables, i.e. matching and regression-based estimation
* exogenous variation, i.e. instrumental variables estimation
* establish an exhaustive and isolated mechanism, i.e. structural estimation
Here are some examples of what to expect.
<img src="material/fig-causal-graph-1.png" width=500>
<img src="material/fig-causal-graph-2.png" width=500>
<img src="material/fig-causal-graph-3.png" width=500>
The key message for now:
* There is often more than one way to estimate a causal effect with differing demands about knowledge and observability
Pearl (2009) is the seminal reference on the use of graphs to represent general causal representations.
**References**
* Pearl, J. (2014). `Causality <https://www.cambridge.org/core/books/causality/B0046844FAE10CBF274D4ACBDAEB5F5B>`_. Cambridge, England: Cambridge University Press.
* Pearl, J., and Mackenzie, D. (2018). `The book of why: The new science of cause and effect <https://www.basicbooks.com/titles/judea-pearl/the-book-of-why/9780465097609/>`_. New York, NY: Basic Books.
* Pearl J., Glymour M., and Jewell N. P. (2016). `Causal inference in statistics: A primer <https://www.wiley.com/en-us/Causal+Inference+in+Statistics%3A+A+Primer-p-9781119186847>`_. Chichester, UK: Wiley.
### Resources
* **LaLonde, R. J. (1986)**. [Evaluating the econometric evaluations of training programs with experimental data](https://www.jstor.org/stable/1806062). *The American Economic Review*, 76(4), 604-620.
|
github_jupyter
|
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import numpy as np
# We collected a host of data from two other influential textbooks.
df = pd.read_csv("../../datasets/processed/dehejia_waba/nsw_lalonde.csv")
df.index.set_names("Individual", inplace=True)
df.describe()
# It is important to check for missing values first.
for column in df.columns:
assert not df[column].isna().any()
columns_background = [
"treat",
"age",
"education",
"black",
"hispanic",
"married",
"nodegree",
]
for column in columns_background:
sns.countplot(x=df[column], color="#1f77b4")
plt.show()
columns_outcome = ["re75", "re78"]
for column in columns_outcome:
earnings = df[column]
# We drop all earnings at zero.
earnings = earnings.loc[earnings > 0]
ax = sns.histplot(earnings)
ax.set_xlim([0, None])
plt.show()
info = ["count", "mean", "std"]
for column in columns_background:
print("\n\n", column.capitalize())
print(df.groupby("treat")[column].describe()[info])
# We first create True / False
is_treated = df["treat"] == 1
df["Y"] = df["re78"]
df["Y_0"] = df.loc[~is_treated, "re78"]
df["Y_1"] = df.loc[is_treated, "re78"]
df["D"] = np.nan
df.loc[~is_treated, "D"] = 0
df.loc[is_treated, "D"] = 1
df[["Y", "Y_1", "Y_0", "D"]].sample(10)
df.groupby("D")["re78"].describe()
ax = sns.histplot(df.loc[~is_treated, "Y"], label="untreated", color = "red")
ax = sns.histplot(df.loc[df.treat == 1].Y , label="treated")
ax.set_xlim(0, None)
ax.legend()
stat = df.loc[is_treated, "Y"].mean() - df.loc[~is_treated, "Y"].mean()
f"{stat:.2f}"
| 0.436382 | 0.988492 |
```
# Dependencies and Setup
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
#File to Load
us_Pollution = "Datasets/AQI_data.csv"
us_GDP_Industry = "Datasets/GDP_and_Personal_Income.csv"
#us_GDP_State = "Datasets/GDP_by_State.csv"
#Read the dataframes
us_pollution_df = pd.read_csv(us_Pollution)
us_GDP_Industry_df = pd.read_csv(us_GDP_Industry)
#Display us_Pollution DataFrame
us_pollution_df.head()
#Find the Average pollution by pollutants for the US
us_pollution_df_Overall = us_pollution_df.groupby(["Year"]).mean()
#us_pollution_df_Overall
#Drop Columns not needed for analysis
us_pollution_df_Overall = us_pollution_df_Overall.loc[:,['NO2 AQI','O3 AQI','SO2 AQI','CO AQI']]
us_pollution_df_Overall
#Display the us_Pollution DataFrame
us_GDP_Industry_df.head(3)
#Merge Both DataFrames
merge_table = pd.merge(us_pollution_df_Overall, us_GDP_Industry_df, on="Year", how="outer")
merge_table
#Generate Visuals: GDP Trends by Industry
x_axis = [2000,2001,2002,2003,2004,2005,2006,2007,2008,2009,2010,2011,2012,2013,2014,2015,2016]
fig, ax1 = plt.subplots()
Total_GDP = ax1.plot(x_axis,merge_table['Total Industry'],marker="o",
color="red", linewidth=1, label="Total",ls='--')
#Second axes that shares the same x-axis
ax2 = ax1.twinx()
Mining = ax2.plot(x_axis,merge_table['Mining, quarrying, and oil and gas extraction'],marker="^",
color="blue", linewidth=1, label="Mining",ls='--')
Manufacturing = ax2.plot(x_axis,merge_table['Manufacturing'],marker="s",
color="green", linewidth=1, label="Manufacturing",ls='--')
Transportation_Warehousing = ax2.plot(x_axis,merge_table['Transportation and warehousing'],marker="d",
color="black", linewidth=1, label="Transportation/Warehousing",ls='--')
GDP = Total_GDP+Mining+Manufacturing+Transportation_Warehousing
labs=[l.get_label() for l in GDP]
ax1.legend(GDP, labs, loc='center left', bbox_to_anchor=(1.2, 0.5) )
ax1.set_ylabel('Total GDP')
ax2.set_ylabel('Specific Industries')
plt.title('GDP Trends by Industry (In Trillions)')
plt.xlabel('Years')
plt.grid()
#Save the Figure
plt.savefig("Images/GDP by Industry.png")
#Generate Visuals: N02 AQI Pollutant vs Total Industry GDP
x_axis = [2000,2001,2002,2003,2004,2005,2006,2007,2008,2009,2010,2011,2012,2013,2014,2015,2016]
fig, ax1 = plt.subplots()
color = 'tab:red'
ax1.set_xlabel('Years')
ax1.set_ylabel('Total GDP (in Trillions)', color=color)
ax1.plot(x_axis, merge_table['Total Industry'], color=color)
ax1.tick_params(axis='y', labelcolor=color)
#Second axes that shares the same x-axis
ax2 = ax1.twinx()
color = 'tab:blue'
ax2.set_ylabel('Nitrogen dioxide (NO2)', color=color)
ax2.plot(x_axis, merge_table['NO2 AQI'], color=color)
ax2.tick_params(axis='y', labelcolor=color)
#Display graph
fig.tight_layout()
plt.show()
#Save the Figure
fig.savefig("Images/GDPvsNO2AQI.png")
#Generate Visuals: O3 AQI Pollutant vs Total Industry GDP
x_axis = [2000,2001,2002,2003,2004,2005,2006,2007,2008,2009,2010,2011,2012,2013,2014,2015,2016]
fig, ax1 = plt.subplots()
color = 'tab:red'
ax1.set_xlabel('Years')
ax1.set_ylabel('Total GDP (in Trillions)', color=color)
ax1.plot(x_axis, merge_table['Total Industry'], color=color)
ax1.tick_params(axis='y', labelcolor=color)
#Second axes that shares the same x-axis
ax2 = ax1.twinx()
color = 'tab:blue'
ax2.set_ylabel('Ozone (O3)', color=color)
ax2.plot(x_axis, merge_table['O3 AQI'], color=color)
ax2.tick_params(axis='y', labelcolor=color)
#Display Graph
fig.tight_layout()
plt.show()
#Save the Figure
fig.savefig("Images/GDPvs03AQI.png")
#Generate Visuals: O3 AQI Pollutant vs Total Industry GDP
x_axis = [2000,2001,2002,2003,2004,2005,2006,2007,2008,2009,2010,2011,2012,2013,2014,2015,2016]
fig, ax1 = plt.subplots()
color = 'tab:red'
ax1.set_xlabel('Years')
ax1.set_ylabel('Total GDP (in Trillions)', color=color)
ax1.plot(x_axis, merge_table['Total Industry'], color=color)
ax1.tick_params(axis='y', labelcolor=color)
#Second axes that shares the same x-axis
ax2 = ax1.twinx()
color = 'tab:blue'
ax2.set_ylabel('Sulphur dioxide (SO2)', color=color)
ax2.plot(x_axis, merge_table['SO2 AQI'], color=color)
ax2.tick_params(axis='y', labelcolor=color)
#Display Graph
fig.tight_layout()
plt.show()
#Save the Figure
fig.savefig("Images/GDPvsS02AQI.png")
#Generate Visuals: O3 AQI Pollutant vs Total Industry GDP
x_axis = [2000,2001,2002,2003,2004,2005,2006,2007,2008,2009,2010,2011,2012,2013,2014,2015,2016]
fig, ax1 = plt.subplots()
color = 'tab:red'
ax1.set_xlabel('Years')
ax1.set_ylabel('Total GDP (in Trillions)', color=color)
ax1.plot(x_axis, merge_table['Total Industry'], color=color)
ax1.tick_params(axis='y', labelcolor=color)
#Second axes that shares the same x-axis
ax2 = ax1.twinx()
color = 'tab:blue'
ax2.set_ylabel('Carbon monoxide (CO)', color=color)
ax2.plot(x_axis, merge_table['CO AQI'], color=color)
ax2.tick_params(axis='y', labelcolor=color)
#Display Graph
fig.tight_layout()
plt.show()
#Save the Figure
fig.savefig("Images/GDPvsC0AQI.png")
```
|
github_jupyter
|
# Dependencies and Setup
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
#File to Load
us_Pollution = "Datasets/AQI_data.csv"
us_GDP_Industry = "Datasets/GDP_and_Personal_Income.csv"
#us_GDP_State = "Datasets/GDP_by_State.csv"
#Read the dataframes
us_pollution_df = pd.read_csv(us_Pollution)
us_GDP_Industry_df = pd.read_csv(us_GDP_Industry)
#Display us_Pollution DataFrame
us_pollution_df.head()
#Find the Average pollution by pollutants for the US
us_pollution_df_Overall = us_pollution_df.groupby(["Year"]).mean()
#us_pollution_df_Overall
#Drop Columns not needed for analysis
us_pollution_df_Overall = us_pollution_df_Overall.loc[:,['NO2 AQI','O3 AQI','SO2 AQI','CO AQI']]
us_pollution_df_Overall
#Display the us_Pollution DataFrame
us_GDP_Industry_df.head(3)
#Merge Both DataFrames
merge_table = pd.merge(us_pollution_df_Overall, us_GDP_Industry_df, on="Year", how="outer")
merge_table
#Generate Visuals: GDP Trends by Industry
x_axis = [2000,2001,2002,2003,2004,2005,2006,2007,2008,2009,2010,2011,2012,2013,2014,2015,2016]
fig, ax1 = plt.subplots()
Total_GDP = ax1.plot(x_axis,merge_table['Total Industry'],marker="o",
color="red", linewidth=1, label="Total",ls='--')
#Second axes that shares the same x-axis
ax2 = ax1.twinx()
Mining = ax2.plot(x_axis,merge_table['Mining, quarrying, and oil and gas extraction'],marker="^",
color="blue", linewidth=1, label="Mining",ls='--')
Manufacturing = ax2.plot(x_axis,merge_table['Manufacturing'],marker="s",
color="green", linewidth=1, label="Manufacturing",ls='--')
Transportation_Warehousing = ax2.plot(x_axis,merge_table['Transportation and warehousing'],marker="d",
color="black", linewidth=1, label="Transportation/Warehousing",ls='--')
GDP = Total_GDP+Mining+Manufacturing+Transportation_Warehousing
labs=[l.get_label() for l in GDP]
ax1.legend(GDP, labs, loc='center left', bbox_to_anchor=(1.2, 0.5) )
ax1.set_ylabel('Total GDP')
ax2.set_ylabel('Specific Industries')
plt.title('GDP Trends by Industry (In Trillions)')
plt.xlabel('Years')
plt.grid()
#Save the Figure
plt.savefig("Images/GDP by Industry.png")
#Generate Visuals: N02 AQI Pollutant vs Total Industry GDP
x_axis = [2000,2001,2002,2003,2004,2005,2006,2007,2008,2009,2010,2011,2012,2013,2014,2015,2016]
fig, ax1 = plt.subplots()
color = 'tab:red'
ax1.set_xlabel('Years')
ax1.set_ylabel('Total GDP (in Trillions)', color=color)
ax1.plot(x_axis, merge_table['Total Industry'], color=color)
ax1.tick_params(axis='y', labelcolor=color)
#Second axes that shares the same x-axis
ax2 = ax1.twinx()
color = 'tab:blue'
ax2.set_ylabel('Nitrogen dioxide (NO2)', color=color)
ax2.plot(x_axis, merge_table['NO2 AQI'], color=color)
ax2.tick_params(axis='y', labelcolor=color)
#Display graph
fig.tight_layout()
plt.show()
#Save the Figure
fig.savefig("Images/GDPvsNO2AQI.png")
#Generate Visuals: O3 AQI Pollutant vs Total Industry GDP
x_axis = [2000,2001,2002,2003,2004,2005,2006,2007,2008,2009,2010,2011,2012,2013,2014,2015,2016]
fig, ax1 = plt.subplots()
color = 'tab:red'
ax1.set_xlabel('Years')
ax1.set_ylabel('Total GDP (in Trillions)', color=color)
ax1.plot(x_axis, merge_table['Total Industry'], color=color)
ax1.tick_params(axis='y', labelcolor=color)
#Second axes that shares the same x-axis
ax2 = ax1.twinx()
color = 'tab:blue'
ax2.set_ylabel('Ozone (O3)', color=color)
ax2.plot(x_axis, merge_table['O3 AQI'], color=color)
ax2.tick_params(axis='y', labelcolor=color)
#Display Graph
fig.tight_layout()
plt.show()
#Save the Figure
fig.savefig("Images/GDPvs03AQI.png")
#Generate Visuals: O3 AQI Pollutant vs Total Industry GDP
x_axis = [2000,2001,2002,2003,2004,2005,2006,2007,2008,2009,2010,2011,2012,2013,2014,2015,2016]
fig, ax1 = plt.subplots()
color = 'tab:red'
ax1.set_xlabel('Years')
ax1.set_ylabel('Total GDP (in Trillions)', color=color)
ax1.plot(x_axis, merge_table['Total Industry'], color=color)
ax1.tick_params(axis='y', labelcolor=color)
#Second axes that shares the same x-axis
ax2 = ax1.twinx()
color = 'tab:blue'
ax2.set_ylabel('Sulphur dioxide (SO2)', color=color)
ax2.plot(x_axis, merge_table['SO2 AQI'], color=color)
ax2.tick_params(axis='y', labelcolor=color)
#Display Graph
fig.tight_layout()
plt.show()
#Save the Figure
fig.savefig("Images/GDPvsS02AQI.png")
#Generate Visuals: O3 AQI Pollutant vs Total Industry GDP
x_axis = [2000,2001,2002,2003,2004,2005,2006,2007,2008,2009,2010,2011,2012,2013,2014,2015,2016]
fig, ax1 = plt.subplots()
color = 'tab:red'
ax1.set_xlabel('Years')
ax1.set_ylabel('Total GDP (in Trillions)', color=color)
ax1.plot(x_axis, merge_table['Total Industry'], color=color)
ax1.tick_params(axis='y', labelcolor=color)
#Second axes that shares the same x-axis
ax2 = ax1.twinx()
color = 'tab:blue'
ax2.set_ylabel('Carbon monoxide (CO)', color=color)
ax2.plot(x_axis, merge_table['CO AQI'], color=color)
ax2.tick_params(axis='y', labelcolor=color)
#Display Graph
fig.tight_layout()
plt.show()
#Save the Figure
fig.savefig("Images/GDPvsC0AQI.png")
| 0.503174 | 0.530905 |
```
from helpers.svg_wrapper import SVGImg
import helpers.plotting as pt
pt.import_lato_font_in_notebook()
```
# Statistics for everyone
How to use statistics to make better decisions
# Intuitive decision making can lead you astray
- B. F. Skinner [showed](https://www.youtube.com/watch?v=TtfQlkGwE2U) that rewards reinforce behaviours.
- Hence, animals can be [trained](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1473025/) to do [many tasks](https://www.theatlantic.com/technology/archive/2013/06/skinner-marketing-were-the-rats-and-facebook-likes-are-the-reward/276613/)
- Randomly rewarding [pigeons](https://www.all-about-psychology.com/support-files/superstition-in-the-pigeon.pdf) [reinforced](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2615824/) apparently random behaviours ("[superstitions](https://www.psychologicalscience.org/observer/the-many-lives-of-superstition)").
```
SVGImg('images/pigeon.svg', width='40%', output_dir='slides')
```
# Reasoning and discussing isn't always enough
- Participants in the TV show [Trick or Treat (S2/E6)](https://en.wikipedia.org/wiki/Trick_or_Treat_%28TV_series%29) had to find out how to score points.
- They developed complex ideas on how using various available objects scored points
```
SVGImg('images/experiment.svg', width='70%', output_dir='slides')
```
# Humans detect patterns where there are none
In the show, points increased every time a fish swam across an aquarium
```
SVGImg('images/fish.svg', width='50%', output_dir='slides')
```
"We got to a hundred points, so we obviously did it right." - A participant.
# Focusing on spurious signals can be costly
- Participants missed actual hints that would have allowed them<br> to win more money more easily
- See [part 1](https://www.youtube.com/watch?v=IDi2NlsA4nI) and [part 2](https://www.youtube.com/watch?v=yzXSSPp4Epg)
```
SVGImg('images/sign.svg', width='70%', output_dir='slides')
```
# Even quantitative data can mislead<br> without proper analysis
```
SVGImg('images/spurious_correlations.svg', output_dir='slides')
```
See also: [Spurious correlations](http://www.tylervigen.com/spurious-correlations), [Why not to use two axes](https://blog.datawrapper.de/dualaxis)
# Algorithms can make similar mistakes
A common problem in machine learning is [over- or underfitting](https://en.wikipedia.org/wiki/Overfitting) variations in training data.<br> The resulting models don't generalise well to future observations.
# Statistics can help to tell the signal from the noise
### For example:
- In 2016, many were certain that Clinton would win the US election.
- On Nov. 4, four days before the election, [a statistics website reported](https://fivethirtyeight.com/features/trump-is-just-a-normal-polling-error-behind-clinton):<br> "Trump Is Just A Normal Polling Error Behind Clinton"
# You can get better at statistics!
- Follow this course to improve your decision making skills
- Suitable for general audiences
Next section: [An introductory experiment](1_introductory_card_experiment.slides.html)
|
github_jupyter
|
from helpers.svg_wrapper import SVGImg
import helpers.plotting as pt
pt.import_lato_font_in_notebook()
SVGImg('images/pigeon.svg', width='40%', output_dir='slides')
SVGImg('images/experiment.svg', width='70%', output_dir='slides')
SVGImg('images/fish.svg', width='50%', output_dir='slides')
SVGImg('images/sign.svg', width='70%', output_dir='slides')
SVGImg('images/spurious_correlations.svg', output_dir='slides')
| 0.362969 | 0.922203 |
# Body position recognition using FastAI
> Recognising body parts during golf swing, step 1
- toc: true
- badges: true
- categories: [jupyter]
- image: images/chart-preview.png

## Introduction
This is a first step in a project to analyse golf swings.
In this first step I try to identify different parts of the body and golf equipment during the golf swing. This step is of limited success for overall analysis but the steps used are useful for the lessons learnt.
This work uses deep learning to identify locations (vectors) on images and fitting by regression.
In this step I will use a dataSet found at <a href='https://github.com/wmcnally/golfdb'> Git Hub GolfSwing</a> and the paper of the work https://arxiv.org/abs/1903.06528.
What this dataset/paper does is split the golf swing into a number of sequences based on the position of the body and golf club, e.g. start, golf club parallel to ground, striking the ball etc. We will call these the golf positions. These positions are shown below.
<iframe src="https://giphy.com/embed/YiSd21YzHLbtbXxShr" width="480" height="270" frameBorder="0" class="giphy-embed" allowFullScreen></iframe><p><a href="https://giphy.com/gifs/YiSd21YzHLbtbXxShr">via GIPHY</a></p>

The dataset includes a series of videos that have been characterised based on the different swing sequences.
### Steps in this page
1. Download the video dataset and the details of the frames of the different positions
1. Create images at the different positions from the videos
1. Classify points on the images and a file for each image of these
1. Upload data to GitHub and download on notebook for analysis
1. Use deep learning to identify the positions on the images
## Use the video analysis dataset to create images of golf swings
First I cloned the directory (https://github.com/wmcnally/golfdb) onto my local PC. I then need to identify which videos to use- I want the ones behind the golfer and preferably of lower frame rate.
Below are the names of the videos I selected
```
import numpy as np
import os
useVids=[1,3,5,7,13,24,43,46,48,71,77,81,83,89,93,242,681,1060]
np.shape(useVids)
```
I now want to find the frames in each video that represent the selected positions.
These exist in a '.pkl' file. So we open the file and then select the videos (rows) we want to use.
```
import pandas as pd
import pickle
file_path= cda + "\data\\golfDB.pkl"
data = pickle.load(open(file_path,"rb"))
aa=[]
i=0
for ii in useVids:
if i==0:
aa=data[ii==data.id]
else:
aa=aa.append(data[ii==data.id])
i=i+1
aa.reset_index(drop=True,inplace=True)
aa.tail()
```

In the DataFrame (aa) the details we want are just the 'events' so we know what frames to save as images from the videos
First we create a function that takes a video location and details of the frames (or the selected golf positions) and then creates a new folder containing images of those frames.
This uses the library `cv2` and a secondary check to normalise the positions if it is different from that given (this was useful in earlier versions but later ones the frame number matched that given by the aa dataFrame).
The function works by finding a frame rate then stepping through the video by adding the time per frame after each step. If the frame is at a position given by the input (from aa) it is saved as an image.
```
def createImages(fila,pos):
'''
Given a video file location (fila) it will save as images to a folder
Given positions in video (pos) these images from the video are saved
pos is created based on positions of swings
'''
import cv2
import numpy as np
import os
# create a video capture object
cap = cv2.VideoCapture(fila)
# get details of the video clip
duration = cap.get(cv2.CAP_PROP_POS_MSEC)
frame_count = cap.get(cv2.CAP_PROP_FRAME_COUNT)
fps = cap.get(cv2.CAP_PROP_FPS)
duration_seconds = frame_count / fps
print('duration is ',duration,'. frame_count is ',frame_count,'. fps is ',fps,'. duration sec is',duration_seconds)
#alter pos based on frame count
posb4=pos
pos=(pos/(np.max(pos)/frame_count))
pos=np.array([int(nn) for nn in pos])
pos=pos[1:-2]#ignore first value and last two
# create a folder if it doesn't exist
folder = fila.split('\\')[-1].split('.')[0]
folder = '_images'+folder
print(folder)
try:
os.mkdir(folder)
except:
pass
vidcap = cap
# this function creates an image from part of a video and
# saves as a JPG file
def getFrame(sec,go):
vidcap.set(cv2.CAP_PROP_POS_MSEC,sec)
hasFrames,image = vidcap.read()
if hasFrames and go:
cv2.imwrite(os.path.join(folder,"frame{:d}.jpg".format(count)), image) # save frame as JPG file
return hasFrames
# goes through the video clip and steps through based on frame rate
sec = 0
frameRate = 1000/fps
count=1
go=0
success = True
while success:
count = count + 1
sec = sec + frameRate
#only saves images if at positions in pos
if count in pos:
go=1
else:
go=0
success = getFrame(sec,go)
print("{} images are extacted in {}.".format(count,folder))
```
And below I call the script for the videos I selected
```
import cv2
fila = cda + '\\data\\videos_160\\'
for ii,aai in enumerate(aa.id):
fold = fila + str(aai)+'.mp4'
pos=aa.iloc[ii,7]
pos=pos-pos[0]
if ii>1:
cII(fold,pos)
cap = createImages.VideoCapture(fold)
```
So now we have a series of folders for each video with images given by the selected positions
## Manually classify points on the images
To be able to perform analysis on the images they first need to be labelled.
To do this I decided to take the **manual** approach and classify the images myself. I decided to choose the following regions in each image:
- The ball
- The end of the golf club (clubhead)
- The back wrist
- the back elbow
- the top of the head
This is done using the follwing function
```
def imDo(im):
fig=plt.figure(figsize=(20, 15))
plt.imshow(im)
def tellme(s):
print(s)
plt.title(s, fontsize=16)
tellme('You will define golf swing, click to begin')
plt.waitforbuttonpress()
while True:
pts = []
while len(pts) < 5:
tellme('Select golf ball-golf club- wrist- elbow- head with mouse')
pts = np.asarray(plt.ginput(5, timeout=-1))
if len(pts) < 5:
tellme('Too few points, starting over')
time.sleep(1) # Wait a second
ph = plt.plot(pts[:, 0], pts[:, 1], marker='x',markersize=20,markeredgewidth=3)
tellme('Happy? Key click for yes, mouse click for no')
if plt.waitforbuttonpress():
break
plt.close(fig)
return pts
```
Before we can call this function we want to make sure the image appears as a new window
Also some imports
```
import fastbook
from fastbook import *
from fastai.vision.all import *
import matplotlib
cda = os.getcwd()
matplotlib.use('TKAgg')
```
Now for each image file created, the script below runs `imDo` which plots the image then asks the user to select 5 points on the image for classification.
these points are then save as txt file with the same name as the image file to be used later in modeling

```
foldOuta=cda+'//_dataSel//'
lsa = os.listdir(foldOuta)
lsa
ptsALL=[]
for ii,folds in enumerate(lsa):
if ii>0:
print(folds)
img_files = get_image_files(foldOuta+folds)
for fils in img_files:
im = PILImage.create(fils)
pts=imDo(im)
ptsALL.append(pts)
fnom=str(fils).split('\\')[-1].split('.')[0]
np.savetxt(foldOuta+folds+'\\'+fnom+'.txt',pts)
```
## Upload data for use in modeling
Fastai has a function called `untar_data` that prepares images in a .tgz folder ready to use for analysis.
A tgz file can be made by a Python script, but all the ones I tried produced an error, so instead I used
To create a tar file see https://opensource.com/article/17/7/how-unzip-targz-file
Open up a terminal go to the folder that contains the folder wanting to compress and then tar with the command line
<code>tar --create --verbose --file GC.tgz GolfComb</code>
I have then uploaded it to GitHub. Go to the file on Github open it and right click on 'view raw' and select copy link.

## Model the data
The rest needs to be done with a GPU. I have done this with https://colab.research.google.com/ (free time is limited but details not published) and the code tab for a notebook on https://www.kaggle.com/ (36 h per month for free)
First import the fastai stuff
```
!pip install -Uqq fastbook
import fastbook
fastbook.setup_book()
from fastbook import *
from fastai.vision.all import *
import os
import re
import numpy
```
untar the data and set the path
```
url='https://github.com/ThomasHSimm/GolfSwingTSimm/blob/main/_dataSel/GC.tgz?raw=true'
path = untar_data(url)
Path.BASE_PATH = path
```
Have a look at the data
```
(path/'Test').ls()
```

A function to classify the points on the image
Loads the text file for each image and returns a TensorPoint object of points on the image
```
def get_pointa_img(fileo):
txtfile = str(fileo)[0:-4] + '.txt'
# print(txtfile)
pts=np.loadtxt(txtfile)
pp=pts[-1,:]
# print(pp)
return TensorPoint(pp)
```
Create a DataBlock
The DataBlock is the foundation of the model. It needs to know
- the location of the images,
- the label for the images (points on images in this case)
- separation of data into test and validation sets (done automatically if not specified)
- the type of data used `blocks=(ImageBlock, PointBlock)`
- any resizing of images
- any transforms (Data Augmentation)
```
item_tfms = [Resize(448, method='squish')]
biwi = DataBlock(
blocks=(ImageBlock, PointBlock),
get_items=get_image_files,
item_tfms=item_tfms,
get_y=get_pointa_img,
batch_tfms=[*aug_transforms(size=224, min_scale=0.75),
Normalize.from_stats(*imagenet_stats)])
```
Now create a DataLoaders object which has the path of the data and the batch size (here 30)
Batch size is important to specify to avoid memory issues
```
dls = biwi.dataloaders(path,30)
```
Now create the learner
Pass it the dataLoaders, we're doing transfer learning from resnet50 (imageNet trained model), what metrics we'll use for loss, and the range in y values we want
```
learn = cnn_learner(dls, resnet50, metrics=partial(accuracy_multi, thresh=0.02),y_range=(-1,1))
```
Have a look at the data. Can see the transforms
```
dls.show_batch(max_n=8, figsize=(12,12))
```
Find the best learning rate
```
learn.lr_find()
```

To fit the model we have a few options:
- `learn.fit(10,lr=4e-3)`
- `learn.fit_one_cycle()`
- `learn.fine_tune(10, base_lr=1e-3, freeze_epochs=7)`
- `learn.fine_tune(15, lr)`
FastAI adds an extra 2 layers on the end of neural network these can then be fitted using `fine_tune`. It is recommended to do a few fits frozen before unfreezing. This is normally the best option for transfer learning.
But the other ones can be used. In general fit can be more unstable and lead to bigger losses, but can be useful if fine_tune is not bringing losses down.
https://forums.fast.ai/t/fine-tune-vs-fit-one-cycle/66029/6
fit_one_cycle = New Model
fine_tuning = with Transfer Learning?
I’d say yes but with a very strong but, only because it’s easy to fall into a trap that way. fine_tuning is geared towards transfer learning specifically, but you can also just do fit_one_cycle as well! (Or flat_cos).
For beginners it’s a great starting fit function (and advanced too), but also don’t forget that you can then build on what that function is doing. For instance, I wonder how modifying/adapting that function for Ranger/flat_cos would need to change!
```
learn.fine_tune(10, base_lr=1e-3, freeze_epochs=7)
```

```
learn.lr_find()
```

Some more fitting, reducing the learning rate after steps
```
learn.fit(20,lr=1e-4)
```

Some more fitting
Mixing fit with fine_tune and reducing learning rate seems to work best for reducing loss
Loss here is:
train_loss valid_loss
0.054042 0.008305
## Results
Look at the results, pretty good for ~10 mins of 81 images of learning although doesn't always get the top of the head.
```
learn.show_results()
```

```
#save the model
learn.export(fname='headTry1.pkl')
```
However, when this is generalised to other points, such as hands and clubhead, that are less static the results are poor.
Presumably a combination of the low resolution of the images making it difficult to identify features and the lack of images.

Increasing the res of the images/videos improves the classification considerably.
But still not quite there, probably needs more labelling
train_loss valid_loss
0.030079 0.031188

|
github_jupyter
|
import numpy as np
import os
useVids=[1,3,5,7,13,24,43,46,48,71,77,81,83,89,93,242,681,1060]
np.shape(useVids)
import pandas as pd
import pickle
file_path= cda + "\data\\golfDB.pkl"
data = pickle.load(open(file_path,"rb"))
aa=[]
i=0
for ii in useVids:
if i==0:
aa=data[ii==data.id]
else:
aa=aa.append(data[ii==data.id])
i=i+1
aa.reset_index(drop=True,inplace=True)
aa.tail()
def createImages(fila,pos):
'''
Given a video file location (fila) it will save as images to a folder
Given positions in video (pos) these images from the video are saved
pos is created based on positions of swings
'''
import cv2
import numpy as np
import os
# create a video capture object
cap = cv2.VideoCapture(fila)
# get details of the video clip
duration = cap.get(cv2.CAP_PROP_POS_MSEC)
frame_count = cap.get(cv2.CAP_PROP_FRAME_COUNT)
fps = cap.get(cv2.CAP_PROP_FPS)
duration_seconds = frame_count / fps
print('duration is ',duration,'. frame_count is ',frame_count,'. fps is ',fps,'. duration sec is',duration_seconds)
#alter pos based on frame count
posb4=pos
pos=(pos/(np.max(pos)/frame_count))
pos=np.array([int(nn) for nn in pos])
pos=pos[1:-2]#ignore first value and last two
# create a folder if it doesn't exist
folder = fila.split('\\')[-1].split('.')[0]
folder = '_images'+folder
print(folder)
try:
os.mkdir(folder)
except:
pass
vidcap = cap
# this function creates an image from part of a video and
# saves as a JPG file
def getFrame(sec,go):
vidcap.set(cv2.CAP_PROP_POS_MSEC,sec)
hasFrames,image = vidcap.read()
if hasFrames and go:
cv2.imwrite(os.path.join(folder,"frame{:d}.jpg".format(count)), image) # save frame as JPG file
return hasFrames
# goes through the video clip and steps through based on frame rate
sec = 0
frameRate = 1000/fps
count=1
go=0
success = True
while success:
count = count + 1
sec = sec + frameRate
#only saves images if at positions in pos
if count in pos:
go=1
else:
go=0
success = getFrame(sec,go)
print("{} images are extacted in {}.".format(count,folder))
import cv2
fila = cda + '\\data\\videos_160\\'
for ii,aai in enumerate(aa.id):
fold = fila + str(aai)+'.mp4'
pos=aa.iloc[ii,7]
pos=pos-pos[0]
if ii>1:
cII(fold,pos)
cap = createImages.VideoCapture(fold)
def imDo(im):
fig=plt.figure(figsize=(20, 15))
plt.imshow(im)
def tellme(s):
print(s)
plt.title(s, fontsize=16)
tellme('You will define golf swing, click to begin')
plt.waitforbuttonpress()
while True:
pts = []
while len(pts) < 5:
tellme('Select golf ball-golf club- wrist- elbow- head with mouse')
pts = np.asarray(plt.ginput(5, timeout=-1))
if len(pts) < 5:
tellme('Too few points, starting over')
time.sleep(1) # Wait a second
ph = plt.plot(pts[:, 0], pts[:, 1], marker='x',markersize=20,markeredgewidth=3)
tellme('Happy? Key click for yes, mouse click for no')
if plt.waitforbuttonpress():
break
plt.close(fig)
return pts
import fastbook
from fastbook import *
from fastai.vision.all import *
import matplotlib
cda = os.getcwd()
matplotlib.use('TKAgg')
foldOuta=cda+'//_dataSel//'
lsa = os.listdir(foldOuta)
lsa
ptsALL=[]
for ii,folds in enumerate(lsa):
if ii>0:
print(folds)
img_files = get_image_files(foldOuta+folds)
for fils in img_files:
im = PILImage.create(fils)
pts=imDo(im)
ptsALL.append(pts)
fnom=str(fils).split('\\')[-1].split('.')[0]
np.savetxt(foldOuta+folds+'\\'+fnom+'.txt',pts)
!pip install -Uqq fastbook
import fastbook
fastbook.setup_book()
from fastbook import *
from fastai.vision.all import *
import os
import re
import numpy
url='https://github.com/ThomasHSimm/GolfSwingTSimm/blob/main/_dataSel/GC.tgz?raw=true'
path = untar_data(url)
Path.BASE_PATH = path
(path/'Test').ls()
def get_pointa_img(fileo):
txtfile = str(fileo)[0:-4] + '.txt'
# print(txtfile)
pts=np.loadtxt(txtfile)
pp=pts[-1,:]
# print(pp)
return TensorPoint(pp)
item_tfms = [Resize(448, method='squish')]
biwi = DataBlock(
blocks=(ImageBlock, PointBlock),
get_items=get_image_files,
item_tfms=item_tfms,
get_y=get_pointa_img,
batch_tfms=[*aug_transforms(size=224, min_scale=0.75),
Normalize.from_stats(*imagenet_stats)])
dls = biwi.dataloaders(path,30)
learn = cnn_learner(dls, resnet50, metrics=partial(accuracy_multi, thresh=0.02),y_range=(-1,1))
dls.show_batch(max_n=8, figsize=(12,12))
learn.lr_find()
learn.fine_tune(10, base_lr=1e-3, freeze_epochs=7)
learn.lr_find()
learn.fit(20,lr=1e-4)
learn.show_results()
#save the model
learn.export(fname='headTry1.pkl')
| 0.277375 | 0.937954 |
```
import tensorflow as tf
tf.test.gpu_device_name()
import numpy as np
from tensorflow import keras
from tensorflow.keras import layers
import cv2
def get_model():
# Create a simple model.
inputs = keras.Input(shape=(32,))
outputs = keras.layers.Dense(1)(inputs)
model = keras.Model(inputs, outputs)
model.compile(optimizer="adam", loss="mean_squared_error")
return model
##model = get_model()
##model = keras.Sequential([keras.Input((32,)), keras.layers.Dense(1)])
model = keras.models.Sequential()
model.add(keras.layers.InputLayer(
input_shape=(128, 128, 3)
))
model.add(
keras.layers.Conv2D(
filters=32,
kernel_size=(5,5),
strides = (1,1),
padding='same',
activation='relu',
name='Conv_1'))
model.add(
keras.layers.MaxPool2D(
pool_size = (2,2),
name = 'Pool_1'))#Image_size: 32*64*64(32 filters,image_size 64*64)
model.add(
keras.layers.Conv2D(
filters = 64,
kernel_size = (5,5),
strides = (1,1),
padding = 'same',
activation = 'relu',
name = 'Conv_2'))
model.add(
keras.layers.MaxPool2D(
pool_size = (2,2),
name = 'Pool_2'))#Image_size: 64*32*32(64 filters,image_size 32*32)
model.add(
keras.layers.Conv2D(
filters = 128,
kernel_size = (5,5),
strides = (1,1),
padding = 'same',
activation = 'relu',
name = 'Conv_3'))
model.add(
keras.layers.MaxPool2D(
pool_size = (2,2),
name = 'Pool_3'))#Image_size: 128*16*16(128 filters,image_size 16*16)
model.add(
keras.layers.Conv2D(
filters = 256,
kernel_size = (5,5),
strides = (1,1),
padding = 'same',
activation = 'relu',
name = 'Conv_4'))
model.add(
keras.layers.MaxPool2D(
pool_size = (2,2),
name = 'Pool_4'))#Image_size: 256*8*8(256 filters,image_size 8*8)
model.add(keras.layers.Flatten())
model.add(keras.layers.Dense(units=1024, activation='relu',name = 'fc_1'))
model.add(keras.layers.Dropout(rate=0.2))
model.add(keras.layers.Dense(units=512, activation='relu',name = 'fc_2'))
model.add(keras.layers.Dense(units=10,activation='softmax',name = 'fc_3'))
model.save('/tmp/model')
model.built = True
model.load_weights("/content/Train_weights_1.h5");
```
Now we have the same model, ready to test on our own input.
```
img_cv = cv2.imread("/content/img_6.jpg")
img_cv_r = cv2.resize(img_cv,(128,128)) # testing purposes only
# function that takes path to the image- and above lines
# c0- safe driving
# c1- texting
# c2- talking on phone
# c3- operating center console
# c4- drinking
# c5- reaching behind
# c6- hair/makeup
# c7- talking to passenger
def outputLabel (predict):
if (predict == 1 | predict == 3) :
return 1
if ((predict == 2 | predict == 4)) :
return 2
return (predict - 2)
img_cv = cv2.imread("/content/img_6.jpg")
img_cv_r = cv2.resize(img_cv,(128,128))
img_cv_predict = np.reshape(img_cv_r,[1,128,128,3]) # 128 by 128 dimension, 3 because 3 channel rgb for color
arr_predict = model.predict(img_cv_predict,batch_size = 1)
print(arr_predict)
print(np.argmax(arr_predict))
#int label = outputLabel(np.argmax(arr_predict))
#print(label)
# write a function to group together the labels, put the final class in json file to merge
```
|
github_jupyter
|
import tensorflow as tf
tf.test.gpu_device_name()
import numpy as np
from tensorflow import keras
from tensorflow.keras import layers
import cv2
def get_model():
# Create a simple model.
inputs = keras.Input(shape=(32,))
outputs = keras.layers.Dense(1)(inputs)
model = keras.Model(inputs, outputs)
model.compile(optimizer="adam", loss="mean_squared_error")
return model
##model = get_model()
##model = keras.Sequential([keras.Input((32,)), keras.layers.Dense(1)])
model = keras.models.Sequential()
model.add(keras.layers.InputLayer(
input_shape=(128, 128, 3)
))
model.add(
keras.layers.Conv2D(
filters=32,
kernel_size=(5,5),
strides = (1,1),
padding='same',
activation='relu',
name='Conv_1'))
model.add(
keras.layers.MaxPool2D(
pool_size = (2,2),
name = 'Pool_1'))#Image_size: 32*64*64(32 filters,image_size 64*64)
model.add(
keras.layers.Conv2D(
filters = 64,
kernel_size = (5,5),
strides = (1,1),
padding = 'same',
activation = 'relu',
name = 'Conv_2'))
model.add(
keras.layers.MaxPool2D(
pool_size = (2,2),
name = 'Pool_2'))#Image_size: 64*32*32(64 filters,image_size 32*32)
model.add(
keras.layers.Conv2D(
filters = 128,
kernel_size = (5,5),
strides = (1,1),
padding = 'same',
activation = 'relu',
name = 'Conv_3'))
model.add(
keras.layers.MaxPool2D(
pool_size = (2,2),
name = 'Pool_3'))#Image_size: 128*16*16(128 filters,image_size 16*16)
model.add(
keras.layers.Conv2D(
filters = 256,
kernel_size = (5,5),
strides = (1,1),
padding = 'same',
activation = 'relu',
name = 'Conv_4'))
model.add(
keras.layers.MaxPool2D(
pool_size = (2,2),
name = 'Pool_4'))#Image_size: 256*8*8(256 filters,image_size 8*8)
model.add(keras.layers.Flatten())
model.add(keras.layers.Dense(units=1024, activation='relu',name = 'fc_1'))
model.add(keras.layers.Dropout(rate=0.2))
model.add(keras.layers.Dense(units=512, activation='relu',name = 'fc_2'))
model.add(keras.layers.Dense(units=10,activation='softmax',name = 'fc_3'))
model.save('/tmp/model')
model.built = True
model.load_weights("/content/Train_weights_1.h5");
img_cv = cv2.imread("/content/img_6.jpg")
img_cv_r = cv2.resize(img_cv,(128,128)) # testing purposes only
# function that takes path to the image- and above lines
# c0- safe driving
# c1- texting
# c2- talking on phone
# c3- operating center console
# c4- drinking
# c5- reaching behind
# c6- hair/makeup
# c7- talking to passenger
def outputLabel (predict):
if (predict == 1 | predict == 3) :
return 1
if ((predict == 2 | predict == 4)) :
return 2
return (predict - 2)
img_cv = cv2.imread("/content/img_6.jpg")
img_cv_r = cv2.resize(img_cv,(128,128))
img_cv_predict = np.reshape(img_cv_r,[1,128,128,3]) # 128 by 128 dimension, 3 because 3 channel rgb for color
arr_predict = model.predict(img_cv_predict,batch_size = 1)
print(arr_predict)
print(np.argmax(arr_predict))
#int label = outputLabel(np.argmax(arr_predict))
#print(label)
# write a function to group together the labels, put the final class in json file to merge
| 0.659076 | 0.530358 |
# Lab session 3: Global Optimization with Gaussian Processes
Gaussian Process Summer School, 16th September 2015.
Written by Javier Gonzalez and Zhenwen Dai.
The goal of this lab session is to illustrate the concepts seen during the tutorial in Gaussian processes for Global optimization. We will focus on two aspects of Bayesian Optimization (BO): (1) the choice of the model (2) the choice of the acquisition function.
The technical material associated to the methods used in this lab can be found in the slides of the tutorial.
## 1. Getting started
In addition to GPy, this lab uses GPyOpt (http://sheffieldml.github.io/GPy/), a satellite module of GPy useful to solve global optimization problems. Please be sure that it is correctly installed before starting. The easiest way is using pip. In Ubuntu machines you can do:
```
pip install gpyopt
```
Some of the options of GPyOpt depend on other external packages: DIRECT, cma, pyDOE. Please be sure that this are installed if you want to use all the options. With everything installed, you are ready to start.
Now, just as in the previous lab, specify to include plots in the notebook and to import relevant libraries.
```
%pylab inline
import GPy
import GPyOpt
import numpy as np
from numpy.random import seed
seed(12345)
```
### Remembering the basics
Before starting with the lab, remember that (BO) is an heuristic for global optimization of black-box functions. Let $f: {\mathcal X} \to R$ be a 'well behaved' continuous function defined on a compact subset ${\mathcal X} \subseteq R^d$. Our goal is to solve the global optimization problem of finding
$$ x_{M} = \arg \min_{x \in {\mathcal X}} f(x). $$
We assume that $f$ is a *black-box* from which only perturbed evaluations of the type $y_i = f(x_i) + \epsilon_i$, with $\epsilon_i \sim\mathcal{N}(0,\sigma^2)$, are available. The goal is to find $x_M$ by minimizing the number of evaluations of $f$. To do this, we need to determine two crucial bits:
1. A **Gaussian process** that will capture the our beliefs on $f$.
2. An **acquisition function** that based on the model will be useful to determine where to collect new evaluations of f.
Remember that every time a new data point is collected the model is updated and the acquisition function optimized again.
### Running example
We start with a one-dimensional example. Consider here the Forrester function
$$f(x) =(6x-2)^2 \sin(12x-4),$$ defined on the interval $[0, 1]$.
The minimum of this function is located at $x_{min}=0.78$. We assume that the evaluations of $f$ to are perturbed by zero-mean Gaussian noise with standard deviation 0.25. The Forrester function is part of the benchmark of functions of GPyOpt. To create the true function, the perturbed version and the boundaries of the problem you need to run the following cell.
```
f_true = GPyOpt.fmodels.experiments1d.forrester() # true function object
f_sim = GPyOpt.fmodels.experiments1d.forrester(sd=.25) # noisy version
bounds = f_true.bounds # problem constrains (implemented by default)
f_objective = f_sim.f # objective function
```
To plot the true $f$, simply write:
```
f_true.plot(bounds)
```
f_objective contains the function that we are going to optimize. You can define your own objective but it should be able to map any numpy array of dimension $n\times d$ (inputs) to a numpy array of dimension $n\times 1$ (outputs). For instance:
```
n = 8
x = np.random.rand(n).reshape(n,1)
x
f_objective(x)
```
The bounds of the problem shoould be defined as a tuple containing the upper and lower limits of the box in which the optimization will be performed. In our example:
```
bounds
```
To use BO to solve this problem, we need to create a GPyOpt object in which we need to specify the following elements:
* The function to optimize.
* The box constrains of the problem.
* The model, that is fixed by default to be a GP with a SE kernel.
* The acquisition function (and its parameters).
We create an SE kernel as we do in GPy
```
k = GPy.kern.RBF(1)
```
And now we have all the elements to start optimizing $f$. We create the optimization problem instance. Note that you don't need to specify the evaluation budget of. This is because at this stage we are not running the optimization, we are just initializing the different elements of the BO algorithm.
```
# Creation of the object that we will use to run BO.
seed(1234)
myBopt = GPyOpt.methods.BayesianOptimization(f = f_objective, # function to optimize
bounds = bounds, # box-constrains of the problem
kernel = k, # kernel of the GP
acquisition='EI') # acquisition = Expected improvement
?GPyOpt.methods.BayesianOptimization(
```
At this point you can access to a number of elements in myBopt, including the GP model and the current dataset (initialized at 3 random locations by default).
```
myBopt.X
myBopt.Y
myBopt.model
```
The noise variance is automatically bounded to avoid numerical problems. In case of having a problem where the evaluations of $f$ are exact you only need to include 'exact_feval=True' when creating the BO object as above. Now, to run the optimization for certain number of iterations you only need to write:
```
# Run the optimization (may take a few senconds)
max_iter = 15 # evaluation budget
myBopt.run_optimization(max_iter) # run optimization
```
And that's it! You should have receive a message describing if the method converged (two equal x's are selected in consecutive steps of the optimization) or if the maximum number of iterations was reached. In one dimensional examples, you can visualize the model and the acquisition function (normalized between 0 and 1) as follows.
```
myBopt.plot_acquisition()
```
You can only make the previous plot if the dimension of the problem is 1 or 2. However, you can always how the optimization evolved by running:
```
myBopt.plot_convergence()
```
The first plot shows the distance between the last two collected observations at each iteration. This plot is useful to evaluate the convergence of the method. The second plot shows the best found value at each iteration. It is useful to compare different methods. The fastest the curve decreases the best is the method. The last one shows the predicted sdev. at the next location. This plot is useful to study the exploration/exploitation that is carried out at each step of the optimization. See how the method converged after 10 iterations. You can also print the updated GP
```
myBopt.model
```
### Exercise 1
Use Bayesian optimization to find the minimum of the function $f(x)= x^2 + 10 \sin(x)$ in the interval [-10, 10].
1.1 Define the bounds of the problem, the function and check that it admits a numpy array of observations as input.
```
# Answer to 1.1 here:
```
1.2 Create a GPyOpt object for global optimization using a Mattern52 kernel and adding a gitter of $0.1$ to the expected improvement acquisition (Hint: when creating the object use the option acquisition_par = 0.1).
```
# Answer to 1.2 here:
```
1.3 For stability reasons, constrain the noise of the model to be 10e-4.
```
# Answer to 1.3 here:
```
1.4 Run the optimization for 10 iterations. Make and comment the convergence plots. Has the method converged?
```
# Answer to 1.4 here:
```
## 2. Acquisition functions
In this section we are going to have a look to different acquisition functions. In GPyOpt you can use the expected improvement ('EI') the maximum probability of improvement ('MPI') and the lower confidence bound. When using GPyOpt you can simply specify the acquisition that you want at the moment of creating the BO object. However, you can also load these acquisitions as separate objects.
```
from GPyOpt.core.acquisition import AcquisitionEI, AcquisitionLCB, AcquisitionMPI
```
To access these acquisitions 'externally' we create a GP model using the objective function in Section 1 evaluated in 10 locations.
```
seed(1234)
n = 10
X = np.random.rand(n).reshape(n,1)
Y = f_objective(X)
m = GPy.models.GPRegression(X,Y)
m.optimize()
m.plot([0,1])
```
Now we create thee objects, one for each acquisition. The gitter parameter, to balance exploration and exploitation, need to be specified.
```
acq_EI = AcquisitionEI(acquisition_par = 0)
acq_LCB = AcquisitionLCB(acquisition_par = 2)
acq_MPI = AcquisitionMPI(acquisition_par = 0)
```
These objects are empty, until we initialize them with the above computed model.
```
acq_EI.set_model(m)
acq_LCB.set_model(m)
acq_MPI.set_model(m)
```
The objects acq_EI, acq_LCB, acq_MPI contain the acquisition functions and their gradients. By running the following piece of code you can visualize the three acquisitions. Note that we add a negative sign before each acquisition. This is because within GPyOpt these functions are minimized (instead of maximized) using gradient optimizers (like BFGS) to select new locations. In this plot, however, the larger is the value of the acquisition, the better is the point.
```
# Plot the three acquisition functions (factor 0.1 added in in the LCB for visualization)
X_grid = np.linspace(0,1,200)[:, None]
plt.figure(figsize=(10,6))
plt.title('Acquisition functions',size=25)
plt.plot(X_grid, - 0.1*acq_LCB.acquisition_function(X_grid),label='LCB')
plt.plot(X_grid, -acq_EI.acquisition_function(X_grid),label='EI')
plt.plot(X_grid, -acq_MPI.acquisition_function(X_grid),label='MPI')
plt.xlabel('x',size=15)
plt.ylabel('a(x)',size=15)
legend()
```
### Exercise 2
2.1 According to the previous plot, what areas in the domain are worth exoloring and why? How can we interpret the previous plot in terms of the exploration/exploitation trade off of each one of the three acquisitions?
```
# Answer to 2.1 here:
```
2.2 Now make a plot comparing the shape of the LCB acquisition (of GP-UCB in the literature) with values different values of parameters. Use the values $[0,0.1,0.25,0.5,1,2,5]$. How does the decision about where to collect the sample change when we increase the value of the parameter?
```
# Answer to 2.2 here:
```
### Exercise 3
Consider the sixhumpcamel function defined as
$$f(x_1,x_2) = \left(4-2.1x_1^2 + \frac{x_1^4}{3} \right)x_1^2 + x_1x_2 + (-4 +4x_2^2)x_2^2,$$
in $[-2,2]\times [-1,1]$. This function has two global minima, at $(0.0898,-0.7126)$ and $(-0.0898,0.7126)$. This function is also implemented in GPyOpt so, to load and visualize it simply run.
```
GPyOpt.fmodels.experiments2d.sixhumpcamel().plot()
f_shc = GPyOpt.fmodels.experiments2d.sixhumpcamel(sd = 0.1).f # simulated version with some noise
bounds_shc = GPyOpt.fmodels.experiments2d.sixhumpcamel().bounds
```
3.1 Create three objects to optimize this function using the the 'EI' (with parameter equal to zero), the LCB (with parameter equal to 2) and the MPI (with parameter equal to zero). Use the same initial data in the three cases (Hint: use the options 'X' and 'Y' when creating the BO object).
```
# Answer to 3.1
```
3.2 In the three cases run the optimization for 30 iterations
```
# Answer to 3.2 here:
```
3.3 Now make a plot comparing the three methods. The x axis should contain the number of iterations and y axis the best found value (Hint: use .Y_best to extract from the BO objects the best current value at each iteration). Which acquisition is has the best performance in this example?
```
# Answer to 3.2 here:
```
3.4 Compare the models and the acquisition functions in the three cases (after the 30 iterations). What do you observe?
```
# Answer to 3.4 here:
```
## 3. A Practical Example of Tuning the Parameters for SVR
After learning some theory about Bayesian Optimization, let's have a look at how to use GPyOpt to tune the hyper-parameters of a practical algorithm. Here shows how to tune the hyper-parameters for Support Vector Regression (SVR) with the toy dataset that we have seen from Lab 1: the Olympic marathon dataset.
We split the original dataset into the training data (first 20 datapoints) and testing data (last 7 datapoints). The performance of SVR is evaluated in terms of Rooted Mean Squared Error (RMSE) on the testing data.
```
# Let's load the dataset
GPy.util.datasets.authorize_download = lambda x: True # prevents requesting authorization for download.
data = GPy.util.datasets.olympic_marathon_men()
X = data['X']
Y = data['Y']
X_train = X[:20]
Y_train = Y[:20,0]
X_test = X[20:]
Y_test = Y[20:,0]
```
Let's first see the results with the default kernel parameters.
```
from sklearn import svm
svr = svm.SVR()
svr.fit(X_train,Y_train)
Y_train_pred = svr.predict(X_train)
Y_test_pred = svr.predict(X_test)
print "The default parameters obtained: C="+str(svr.C)+", epilson="+str(svr.epsilon)+", gamma="+str(svr.gamma)
```
We compute the RMSE on the testing data and plot the prediction. With the default parameters, SVR does not give an OK fit to the training data but completely miss out the testing data well.
```
plot(X_train,Y_train_pred,'b',label='pred-train')
plot(X_test,Y_test_pred,'g',label='pred-test')
plot(X_train,Y_train,'rx',label='ground truth')
plot(X_test,Y_test,'rx')
legend(loc='best')
print "RMSE = "+str(np.sqrt(np.square(Y_test_pred-Y_test).mean()))
```
Now let's try **Bayesian Optimization**. We first write a wrap function for fitting with SVR. The objective is the RMSE from cross-validation. We optimize the parameters in *log* space.
```
nfold = 3
def fit_svr_val(x):
x = np.atleast_2d(np.exp(x))
fs = np.zeros((x.shape[0],1))
for i in range(x.shape[0]):
fs[i] = 0
for n in range(nfold):
idx = np.array(range(X_train.shape[0]))
idx_valid = np.logical_and(idx>=X_train.shape[0]/nfold*n, idx<X_train.shape[0]/nfold*(n+1))
idx_train = np.logical_not(idx_valid)
svr = svm.SVR(C=x[i,0], epsilon=x[i,1],gamma=x[i,2])
svr.fit(X_train[idx_train],Y_train[idx_train])
fs[i] += np.sqrt(np.square(svr.predict(X_train[idx_valid])-Y_train[idx_valid]).mean())
fs[i] *= 1./nfold
return fs
```
We set the search interval of $C$ to be roughly $[0,1000]$ and the search interval of $\epsilon$ and $\gamma$ to be roughtly $[1\times 10^{-5},0.1]$.
```
bounds = [(0.,7.),(-12.,-2.),(-12.,-2.)]
```
We, then, create the GPyOpt object and run the optimization procedure. It might take a while.
```
opt = GPyOpt.methods.BayesianOptimization(f = fit_svr_val, # function to optimize
bounds = bounds, # box-constrains of the problem
acquisition='LCB', # LCB acquisition
acquisition_par = 0.1) # acquisition = Expected improvement
# it may take a few seconds
opt.run_optimization(max_iter=50)
opt.plot_convergence()
```
Let's show the best parameters found. They differ significantly from the default parameters.
```
x_best = np.exp(opt.X[np.argmin(opt.Y)])
print "The best parameters obtained: C="+str(x_best[0])+", epilson="+str(x_best[1])+", gamma="+str(x_best[2])
svr = svm.SVR(C=x_best[0], epsilon=x_best[1],gamma=x_best[2])
svr.fit(X_train,Y_train)
Y_train_pred = svr.predict(X_train)
Y_test_pred = svr.predict(X_test)
```
We can see SVR does a reasonable fit to the data. The result could be further improved by increasing the *max_iter*.
```
plot(X_train,Y_train_pred,'b',label='pred-train')
plot(X_test,Y_test_pred,'g',label='pred-test')
plot(X_train,Y_train,'rx',label='ground truth')
plot(X_test,Y_test,'rx')
legend(loc='best')
print "RMSE = "+str(np.sqrt(np.square(Y_test_pred-Y_test).mean()))
```
### Exercise 4
Why we do not directly use the RMSE of the whole training dataset as the objective? Why bother with cross-validation?
|
github_jupyter
|
pip install gpyopt
%pylab inline
import GPy
import GPyOpt
import numpy as np
from numpy.random import seed
seed(12345)
f_true = GPyOpt.fmodels.experiments1d.forrester() # true function object
f_sim = GPyOpt.fmodels.experiments1d.forrester(sd=.25) # noisy version
bounds = f_true.bounds # problem constrains (implemented by default)
f_objective = f_sim.f # objective function
f_true.plot(bounds)
n = 8
x = np.random.rand(n).reshape(n,1)
x
f_objective(x)
bounds
k = GPy.kern.RBF(1)
# Creation of the object that we will use to run BO.
seed(1234)
myBopt = GPyOpt.methods.BayesianOptimization(f = f_objective, # function to optimize
bounds = bounds, # box-constrains of the problem
kernel = k, # kernel of the GP
acquisition='EI') # acquisition = Expected improvement
?GPyOpt.methods.BayesianOptimization(
myBopt.X
myBopt.Y
myBopt.model
# Run the optimization (may take a few senconds)
max_iter = 15 # evaluation budget
myBopt.run_optimization(max_iter) # run optimization
myBopt.plot_acquisition()
myBopt.plot_convergence()
myBopt.model
# Answer to 1.1 here:
# Answer to 1.2 here:
# Answer to 1.3 here:
# Answer to 1.4 here:
from GPyOpt.core.acquisition import AcquisitionEI, AcquisitionLCB, AcquisitionMPI
seed(1234)
n = 10
X = np.random.rand(n).reshape(n,1)
Y = f_objective(X)
m = GPy.models.GPRegression(X,Y)
m.optimize()
m.plot([0,1])
acq_EI = AcquisitionEI(acquisition_par = 0)
acq_LCB = AcquisitionLCB(acquisition_par = 2)
acq_MPI = AcquisitionMPI(acquisition_par = 0)
acq_EI.set_model(m)
acq_LCB.set_model(m)
acq_MPI.set_model(m)
# Plot the three acquisition functions (factor 0.1 added in in the LCB for visualization)
X_grid = np.linspace(0,1,200)[:, None]
plt.figure(figsize=(10,6))
plt.title('Acquisition functions',size=25)
plt.plot(X_grid, - 0.1*acq_LCB.acquisition_function(X_grid),label='LCB')
plt.plot(X_grid, -acq_EI.acquisition_function(X_grid),label='EI')
plt.plot(X_grid, -acq_MPI.acquisition_function(X_grid),label='MPI')
plt.xlabel('x',size=15)
plt.ylabel('a(x)',size=15)
legend()
# Answer to 2.1 here:
# Answer to 2.2 here:
GPyOpt.fmodels.experiments2d.sixhumpcamel().plot()
f_shc = GPyOpt.fmodels.experiments2d.sixhumpcamel(sd = 0.1).f # simulated version with some noise
bounds_shc = GPyOpt.fmodels.experiments2d.sixhumpcamel().bounds
# Answer to 3.1
# Answer to 3.2 here:
# Answer to 3.2 here:
# Answer to 3.4 here:
# Let's load the dataset
GPy.util.datasets.authorize_download = lambda x: True # prevents requesting authorization for download.
data = GPy.util.datasets.olympic_marathon_men()
X = data['X']
Y = data['Y']
X_train = X[:20]
Y_train = Y[:20,0]
X_test = X[20:]
Y_test = Y[20:,0]
from sklearn import svm
svr = svm.SVR()
svr.fit(X_train,Y_train)
Y_train_pred = svr.predict(X_train)
Y_test_pred = svr.predict(X_test)
print "The default parameters obtained: C="+str(svr.C)+", epilson="+str(svr.epsilon)+", gamma="+str(svr.gamma)
plot(X_train,Y_train_pred,'b',label='pred-train')
plot(X_test,Y_test_pred,'g',label='pred-test')
plot(X_train,Y_train,'rx',label='ground truth')
plot(X_test,Y_test,'rx')
legend(loc='best')
print "RMSE = "+str(np.sqrt(np.square(Y_test_pred-Y_test).mean()))
nfold = 3
def fit_svr_val(x):
x = np.atleast_2d(np.exp(x))
fs = np.zeros((x.shape[0],1))
for i in range(x.shape[0]):
fs[i] = 0
for n in range(nfold):
idx = np.array(range(X_train.shape[0]))
idx_valid = np.logical_and(idx>=X_train.shape[0]/nfold*n, idx<X_train.shape[0]/nfold*(n+1))
idx_train = np.logical_not(idx_valid)
svr = svm.SVR(C=x[i,0], epsilon=x[i,1],gamma=x[i,2])
svr.fit(X_train[idx_train],Y_train[idx_train])
fs[i] += np.sqrt(np.square(svr.predict(X_train[idx_valid])-Y_train[idx_valid]).mean())
fs[i] *= 1./nfold
return fs
bounds = [(0.,7.),(-12.,-2.),(-12.,-2.)]
opt = GPyOpt.methods.BayesianOptimization(f = fit_svr_val, # function to optimize
bounds = bounds, # box-constrains of the problem
acquisition='LCB', # LCB acquisition
acquisition_par = 0.1) # acquisition = Expected improvement
# it may take a few seconds
opt.run_optimization(max_iter=50)
opt.plot_convergence()
x_best = np.exp(opt.X[np.argmin(opt.Y)])
print "The best parameters obtained: C="+str(x_best[0])+", epilson="+str(x_best[1])+", gamma="+str(x_best[2])
svr = svm.SVR(C=x_best[0], epsilon=x_best[1],gamma=x_best[2])
svr.fit(X_train,Y_train)
Y_train_pred = svr.predict(X_train)
Y_test_pred = svr.predict(X_test)
plot(X_train,Y_train_pred,'b',label='pred-train')
plot(X_test,Y_test_pred,'g',label='pred-test')
plot(X_train,Y_train,'rx',label='ground truth')
plot(X_test,Y_test,'rx')
legend(loc='best')
print "RMSE = "+str(np.sqrt(np.square(Y_test_pred-Y_test).mean()))
| 0.684475 | 0.990803 |
# Investigating Stop and Frisk
**GOALS**:
- Access datasets as external files
- Create `DataFrame` from file
- Select rows and columns from the `DataFrame`
- Filter values in `DataFrame` based on logical conditions
- Split, Apply, Combine using `groupby` method
- Use *histograms, boxplots, barplots,* and *countplots* to investigate distributions of quantitative variables
- Use `.corr()` to explore relationships between variables and visualize with `heatmap`
- Use *scatterplots* to examine relationships between quantitative variables
## The Stop and Frisk Data
To start, we want to create a `DataFrame` from our `.csv` file we downloaded from the NYPD shared data.

```
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import seaborn as sns
#/ this sets up variables for this data set. variables make it easier to interpret a dataset.
df = pd.read_csv('data/sqf-2017.csv')
#this imports the dataset!
df.head()
#this step organizes the content within the dataset, by displaying the first five rows.
df.info()
# this defines the info contained within the dataset
df.OBSERVED_DURATION_MINUTES.hist()
#this creates a bar graph for the information regarding the total amount of minutes observed for each stop and frisk.
df.OBSERVED_DURATION_MINUTES.mean()
# this calculates the mean for the duration of minutes per stop and frisk. In this case, the mean is 28.9 (or 29), minutes per stop.
df.OBSERVED_DURATION_MINUTES.describe()
# this describes the total number of minutes spent for stop and frisk, which is what the first bar graph described. it also explains the average number of minutes for 25%, 50%, and 75% of the stop and frisk data.
df.SUSPECT_RACE_DESCRIPTION.value_counts()
# these are the values counts for each ethnic group represented in the stop and frisk dataset.
df.head()
# once again, the . head () feature compresses the information in to display first five rows of the dataset
df.groupby('STOP_LOCATION_BORO_NAME')['FRISKED_FLAG'].count()
#this is the amount of stops per borough
df.groupby('STOP_LOCATION_BORO_NAME')['FRISKED_FLAG'].count().plot(kind = 'bar', color = 'blue', title = 'Count of Frisks by Borough', figsize = (10, 5))
# this is a bar graph representing the number of frisks per borough and the amount of frisks that were flagged.
df.groupby('STOP_LOCATION_BORO_NAME')[['SEARCHED_FLAG','FRISKED_FLAG']].count().plot(kind = 'bar', color = ['blue', 'red'], title = 'Count of Frisks by Borough', figsize = (10, 5))
# this is a bar graph that represents the search flags in conjunction with the frisked flags
five_boroughs = df[(df['STOP_LOCATION_BORO_NAME'] == 'MANHATTAN') | (df['STOP_LOCATION_BORO_NAME'] == 'BRONX') | (df['STOP_LOCATION_BORO_NAME'] == 'BROOKLYN') | (df['STOP_LOCATION_BORO_NAME'] == 'STATEN IS') | (df['STOP_LOCATION_BORO_NAME'] == 'QUEENS')]
#the variable, five_boroughs is defined by the following function
five_boroughs.STOP_LOCATION_BORO_NAME.value_counts()
# this accounts for the number of stops per borough within the five boroughs of New York City
five_boroughs.groupby('STOP_LOCATION_BORO_NAME')['FRISKED_FLAG'].count().plot(kind = 'bar', color = 'blue', title = 'Count of Frisks by Borough', figsize = (10, 5))
# this is a bar graph presenting the number of stops per borough within new york city.
df['OBSERVED_DURATION_MINUTES'].describe()
# this provides further information on the observed duration per each stop and frisk
df.shape
df[df['OBSERVED_DURATION_MINUTES'] <= 120].shape
11629 - 11521
108/11629
# cells 25 - 28 detail an equation for the observed duration per stop and frisk
trimmed_time = df[df['OBSERVED_DURATION_MINUTES'] <= 120]
# this is creating a variable to interpret the math problem in steps 25 - 28
trimmed_time['OBSERVED_DURATION_MINUTES'].hist()
# this is a bar graph representing the equation from cells 25 - 28
trimmed_time['OBSERVED_DURATION_MINUTES'].plot(kind = 'box')
# this is creating a box plot from the same information in the above cell
df['SUSPECT_RACE_DESCRIPTION'].value_counts()
# these are the value counts for suspect stops based on ethnicity
df.groupby('SUSPECT_RACE_DESCRIPTION')['STOP_LOCATION_BORO_NAME'].count()
df.groupby('STOP_LOCATION_BORO_NAME')['SUSPECT_RACE_DESCRIPTION'].count()
df.groupby('STOP_LOCATION_BORO_NAME')['SUSPECT_RACE_DESCRIPTION'].count().plot(kind = 'bar', color = 'blue', title = 'Race Descriptions By Borough', figsize = (10, 5))
# the bar graph represents the information in the previous cells, by examining the suspect race description stopped per borough
df['STOP_DURATION_MINUTES'].describe()
# this represents the amount of time each stop took not to be confused with the previous section, in which the observed duration was determined.
df.shape
df[df['STOP_DURATION_MINUTES'] <= 120].shape
11629 - 11537
92/11629
trimmed_time = df[df['STOP_DURATION_MINUTES'] <= 120]
# a variable is used to define the information contained within the stop duration minutes data
trimmed_time['STOP_DURATION_MINUTES'].hist()
# a histogram describing the above information, stop duration times.
df['SUSPECT_SEX'].describe()
# a description of the suspect sex per stop and frisk. this signifies that the majority of individuals stopped were male.
df['SUSPECT_SEX'].value_counts
# these are the value counts for both males and females per stop and frisk.
gender = df[(df['SUSPECT_SEX'] == 'FEMALE') | (df['SUSPECT_SEX'] == 'MALE')]
# creating this value allows for the information to be organized by sex. rather than interpret a large list of columns and rows, the data is narrowed down to males and females
gender.SUSPECT_SEX.value_counts()
# this step compares the amount of males to the amount of females who were stopped. Males make up the majority of individuals stopped.
df.groupby('SUSPECT_SEX')['SUSPECT_REPORTED_AGE'].count()
# this was meant to compare data from the suspects' age to their sex
age = df[(df['SUSPECT_SEX'] == 'MALE')
| (df['SUSPECT_REPORTED_AGE'] == '18')
| (df['SUSPECT_REPORTED_AGE'] == '19')
| (df['SUSPECT_REPORTED_AGE'] == '20')
| (df['SUSPECT_REPORTED_AGE'] == '21')
| (df['SUSPECT_REPORTED_AGE'] == '22')
| (df['SUSPECT_REPORTED_AGE'] == '23')]
# this is a variable meant to scaled down the sex and reported age of suspects
df['PHYSICAL_FORCE_CEW_FLAG'].describe()
# this accounts for the physical force used per stop and frisk
df['PHYSICAL_FORCE_DRAW_POINT_FIREARM_FLAG'].describe()
# this accounts for the number of times in which a firearm was used during a stop and frisk encounter
df['PHYSICAL_FORCE_HANDCUFF_SUSPECT_FLAG'].describe()
# this describes when physical force was used when handcuffing suspects
df['SUSPECT_ARREST_OFFENSE'].describe()
# this accounts for the arrest offenses of suspects
df.groupby('SUSPECT_ARREST_OFFENSE')['SUSPECT_RACE_DESCRIPTION'].count()
# this defines the variety and frequency of suspect arrest offenses
```
New Lab!
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import seaborn as sns
#/ this sets up variables for this data set. variables make it easier to interpret a dataset.
df = pd.read_csv('data/sqf-2017.csv')
#this imports the dataset!
df.head()
#this step organizes the content within the dataset, by displaying the first five rows.
df.info()
# this defines the info contained within the dataset
df.OBSERVED_DURATION_MINUTES.hist()
#this creates a bar graph for the information regarding the total amount of minutes observed for each stop and frisk.
df.OBSERVED_DURATION_MINUTES.mean()
# this calculates the mean for the duration of minutes per stop and frisk. In this case, the mean is 28.9 (or 29), minutes per stop.
df.OBSERVED_DURATION_MINUTES.describe()
# this describes the total number of minutes spent for stop and frisk, which is what the first bar graph described. it also explains the average number of minutes for 25%, 50%, and 75% of the stop and frisk data.
df.SUSPECT_RACE_DESCRIPTION.value_counts()
# these are the values counts for each ethnic group represented in the stop and frisk dataset.
df.head()
# once again, the . head () feature compresses the information in to display first five rows of the dataset
df.groupby('STOP_LOCATION_BORO_NAME')['FRISKED_FLAG'].count()
#this is the amount of stops per borough
df.groupby('STOP_LOCATION_BORO_NAME')['FRISKED_FLAG'].count().plot(kind = 'bar', color = 'blue', title = 'Count of Frisks by Borough', figsize = (10, 5))
# this is a bar graph representing the number of frisks per borough and the amount of frisks that were flagged.
df.groupby('STOP_LOCATION_BORO_NAME')[['SEARCHED_FLAG','FRISKED_FLAG']].count().plot(kind = 'bar', color = ['blue', 'red'], title = 'Count of Frisks by Borough', figsize = (10, 5))
# this is a bar graph that represents the search flags in conjunction with the frisked flags
five_boroughs = df[(df['STOP_LOCATION_BORO_NAME'] == 'MANHATTAN') | (df['STOP_LOCATION_BORO_NAME'] == 'BRONX') | (df['STOP_LOCATION_BORO_NAME'] == 'BROOKLYN') | (df['STOP_LOCATION_BORO_NAME'] == 'STATEN IS') | (df['STOP_LOCATION_BORO_NAME'] == 'QUEENS')]
#the variable, five_boroughs is defined by the following function
five_boroughs.STOP_LOCATION_BORO_NAME.value_counts()
# this accounts for the number of stops per borough within the five boroughs of New York City
five_boroughs.groupby('STOP_LOCATION_BORO_NAME')['FRISKED_FLAG'].count().plot(kind = 'bar', color = 'blue', title = 'Count of Frisks by Borough', figsize = (10, 5))
# this is a bar graph presenting the number of stops per borough within new york city.
df['OBSERVED_DURATION_MINUTES'].describe()
# this provides further information on the observed duration per each stop and frisk
df.shape
df[df['OBSERVED_DURATION_MINUTES'] <= 120].shape
11629 - 11521
108/11629
# cells 25 - 28 detail an equation for the observed duration per stop and frisk
trimmed_time = df[df['OBSERVED_DURATION_MINUTES'] <= 120]
# this is creating a variable to interpret the math problem in steps 25 - 28
trimmed_time['OBSERVED_DURATION_MINUTES'].hist()
# this is a bar graph representing the equation from cells 25 - 28
trimmed_time['OBSERVED_DURATION_MINUTES'].plot(kind = 'box')
# this is creating a box plot from the same information in the above cell
df['SUSPECT_RACE_DESCRIPTION'].value_counts()
# these are the value counts for suspect stops based on ethnicity
df.groupby('SUSPECT_RACE_DESCRIPTION')['STOP_LOCATION_BORO_NAME'].count()
df.groupby('STOP_LOCATION_BORO_NAME')['SUSPECT_RACE_DESCRIPTION'].count()
df.groupby('STOP_LOCATION_BORO_NAME')['SUSPECT_RACE_DESCRIPTION'].count().plot(kind = 'bar', color = 'blue', title = 'Race Descriptions By Borough', figsize = (10, 5))
# the bar graph represents the information in the previous cells, by examining the suspect race description stopped per borough
df['STOP_DURATION_MINUTES'].describe()
# this represents the amount of time each stop took not to be confused with the previous section, in which the observed duration was determined.
df.shape
df[df['STOP_DURATION_MINUTES'] <= 120].shape
11629 - 11537
92/11629
trimmed_time = df[df['STOP_DURATION_MINUTES'] <= 120]
# a variable is used to define the information contained within the stop duration minutes data
trimmed_time['STOP_DURATION_MINUTES'].hist()
# a histogram describing the above information, stop duration times.
df['SUSPECT_SEX'].describe()
# a description of the suspect sex per stop and frisk. this signifies that the majority of individuals stopped were male.
df['SUSPECT_SEX'].value_counts
# these are the value counts for both males and females per stop and frisk.
gender = df[(df['SUSPECT_SEX'] == 'FEMALE') | (df['SUSPECT_SEX'] == 'MALE')]
# creating this value allows for the information to be organized by sex. rather than interpret a large list of columns and rows, the data is narrowed down to males and females
gender.SUSPECT_SEX.value_counts()
# this step compares the amount of males to the amount of females who were stopped. Males make up the majority of individuals stopped.
df.groupby('SUSPECT_SEX')['SUSPECT_REPORTED_AGE'].count()
# this was meant to compare data from the suspects' age to their sex
age = df[(df['SUSPECT_SEX'] == 'MALE')
| (df['SUSPECT_REPORTED_AGE'] == '18')
| (df['SUSPECT_REPORTED_AGE'] == '19')
| (df['SUSPECT_REPORTED_AGE'] == '20')
| (df['SUSPECT_REPORTED_AGE'] == '21')
| (df['SUSPECT_REPORTED_AGE'] == '22')
| (df['SUSPECT_REPORTED_AGE'] == '23')]
# this is a variable meant to scaled down the sex and reported age of suspects
df['PHYSICAL_FORCE_CEW_FLAG'].describe()
# this accounts for the physical force used per stop and frisk
df['PHYSICAL_FORCE_DRAW_POINT_FIREARM_FLAG'].describe()
# this accounts for the number of times in which a firearm was used during a stop and frisk encounter
df['PHYSICAL_FORCE_HANDCUFF_SUSPECT_FLAG'].describe()
# this describes when physical force was used when handcuffing suspects
df['SUSPECT_ARREST_OFFENSE'].describe()
# this accounts for the arrest offenses of suspects
df.groupby('SUSPECT_ARREST_OFFENSE')['SUSPECT_RACE_DESCRIPTION'].count()
# this defines the variety and frequency of suspect arrest offenses
| 0.528777 | 0.914634 |
# Complex Fourier Transform
## Complex numbers
Although complex numbers are fundamentally disconnected from our reality, they can be used to solve science and engineering problems in two ways:
1. As parameters from a real world problem than can be substituted into a complex form.
2. As complex numbers that can be mathematically equivalent to the physical problem.
This second approach leads to the complex Fourier Transform, a more sophisticated version of the real Fourier Transform.
## Review of Real DFT
We defined the real version of the Discrete Fourier Transform according to the equations:
$$\mathbf{Re}X[k] = \sum^{N-1}_{n=0}x[n]\cos{(2\pi kn/N)}$$
$$\mathbf{Im}X[k] = -\sum^{N-1}_{n=0}x[n]\sin{(2\pi kn/N)}$$
where $0\leq k \leq N/2$
By introducing the normalization factor $2/N$, which comes from $Re\bar{X}[k]$ and $Im\bar{X}[k]$, we can write:
$$\mathbf{Re}X[k] = \frac{2}{N}\sum^{N-1}_{n=0}x[n]\cos{(2\pi kn/N)}$$
$$\mathbf{Im}X[k] = -\frac{2}{N}\sum^{N-1}_{n=0}x[n]\sin{(2\pi kn/N)}$$
The amplitudes of the cosine waves are contained in $Re X[k]$, while the amplitudes of the sine waves are contained in $ImX[k]$. These equations operate by correlating the respective cosine or sine wave with the time domain signal. In spite of using the names: real part and imaginary part, there are no complex numbers in these equations.
Even though the real DFT uses only real numbers, substitution allows the frequency domain to be represented using complex numbers. As suggested by the names of the arrays. In other words, we place a $j$ with each value in the imaginary part, and add the result to the real part. However, do not make the mistake of thinking that this is the **"complex DFT"**. This is nothing more than the real DFT with complex substitution.
While the real DFT is adequate for many applications in science and engineering, it is mathematically awkward in three respects:
1. Only takes advantage of complex numbers through the use of substitution, therefore complex numbers don't have a meaning here.
2. Poor handling of the negative frequency portion of the spectrum.
3. $Re X[0]$ and $Re X[N/2]$ need special handling.
## Euler's Refresher
We can use Euler's formula to express the relationship between the trigonometric functions and the complex exponential function as:
$$e^{jx}=\cos{(x)}+j\sin{(x)}$$
Using this formula, we can express sine and cosines as follows:
$$e^{-jx}=\cos{(-x)}+j\sin{(-x)}$$
Since cosine is an even and sine an odd function we can get:
$$e^{-jx}=\cos{(x)}-j\sin{(x)}$$
If we add $e^{jx}$ and $e^{-jx}$ we can get an expression for cosine as:
$$\cos(x) = \frac{e^{jx}+e^{-jx}}{2}$$
If we subtract $e^{jx}$ and $e^{-jx}$ we can get an expression for sine as:
$$\sin(x) = \frac{e^{jx}-e^{-jx}}{2j}$$
Rewriting for $x=\omega t$
$$\cos(\omega t) =\frac{1}{2} e^{j\omega t}+\frac{1}{2} e^{-j\omega t}$$
$$\sin(\omega t) =\frac{1}{2j}e^{j\omega t}-\frac{1}{2j}e^{-j\omega t}$$
With Euler's formula we see that the sum of exponential contains a positive frequency $\omega$ and a negative frequency $-\omega$.
# Complex DFT
The Complex Discrete Fourier Transform is defined as:
$$X[k] = \frac{1}{N}\sum\limits^{N-1}_{n=0}{x[n]e^{-j\frac{2\pi k n}{N}}} $$
Where $X[k]$ has $N-1$ points.
By using Euler's formula we can get a rectangular form for the Complex DFT:
$$X[k] = \frac{1}{N}\sum\limits^{N-1}_{n=0}{x[n]\left[\cos{\left(\frac{2\pi k n}{N}\right)} -j\sin{\left(\frac{2\pi k n}{N}\right)} \right]} $$
### Differences between Real DFT and Complex DFT
1. Real DFT converts a real time domain signal, $x[n]$ into two real frequency domain signals $Re X[k]$ and $Im X[k]$. In Complex DFT, $x[n]$ and $X[k]$ are arrays of complex numbers.
2. Real DFT uses only positive frequencies (k goes from 0 to N/2). Complex DFT uses positive and negative frequencies (k goes from 0 to N-1, positive frequencies go from 0 to N/2 and negative from N/2 to N-1).
3. Real DFT adds $j$ to the sine wave allowing the frequency spectrum to be represented by complex numbers. To convert back to sine and cosine waves we drop the $j$ and sum terms. This is mathematically incorrect!
4. Scaling factors of two is not needed in Complex DFT, since this is dealt by the positive and negative frequency nature of the transformation.
5. Complex DFT doesn't require special handling of $Re X[0]$ and $Re X[N/2]$.
```
import sys
sys.path.insert(0, '../../')
import numpy as np
import matplotlib.pyplot as plt
from Common import common_plots
from Common import statistics
cplots = common_plots.Plot()
file = {'x':'Signals/InputSignal_f32_1kHz_15kHz.dat'}
x = np.loadtxt(file['x'])
N,M = x.shape
x = x.reshape(N*M, 1)
cplots.plot_single(x.T, style='line')
plt.xlabel('samples')
plt.ylabel('amplitude');
```
### Create a FourierComplex Class
In this part you will create a class called `FourierComplex` which has the methods described in the implementation. The method `complex_dft` uses the equation described before to implement the Complex Fourier Transform. You have to take special care of your numpy arrays because they will hold complex values.
```
class FourierComplex():
def __init__(self, signal, domain='fraction', **kwargs):
"""
Function that calculates the Complex DFT of an input signal.
Parameters:
signal (numpy array): Array of numbers representing the signal to transform.
domain (string): String value that selects between frequency domain's
independent variable.
'samples' returns number of samples between 0 to N/2
'fraction' returns a fraction of the sampling rate between 0 to 0.5
'natural' returns the natural frequency between 0 and pi.
'analog' returns analog frequency between 0 and fsamp/2
kwargs: - fsamp (float): value representing the sampling frequency.
(Only used for 'analog' style).
Attributes:
signal (numpy array): orignal signal.
dft (complex numpy array): complex Fourier Transform of input signal.
rex (numpy array): real DFT part of input signal.
imx (numpy array): imaginary DFT part of input signal.
domain (numpy array): Frequency domain's independent variable.
"""
self.signal = None
self.dft = None
self.rex = None
self.imx = None
self.domain = None
return
def complex_dft(self):
"""
Function that calculates the Complex DFT of an input signal.
Returns:
complex numpy array: complex DFT of input signal of type imaginary.
"""
return None
def real_dft(self):
"""
Function that calculates the real part of the Complex DFT of
an input signal.
Returns:
numpy array: real part of the Complex DFT of input signal.
"""
return None
def imag_dft(self):
"""
Function that calculates the imaginary part of the Complex DFT of
an input signal.
Returns:
numpy array: imaginary part of the Complex DFT of input signal.
"""
return None
def frequency_domain(self, style='fraction', **kwargs):
"""
Function that calculates the frequency domain independent variable.
Parameters:
obtain the frequency domain.
style (string): String value that selects between frequency domain's
independent variable.
'samples' returns number of samples between 0 to N/2
'fraction' returns a fraction of the sampling rate between 0 to 0.5
'natural' returns the natural frequency between 0 and pi.
'analog' returns analog frequency between 0 and fsamp/2
fsamp (float): Float value representing the sampling frequency.
(Only used for 'analog' style).
Returns:
numpy array: Returns frequency domain's independent variable.
"""
N = self.dft.shape[0]
t = np.arange(N)
if(style=='fraction'):
return t/(N-1)
elif(style=='natural'):
return np.pi*(t/(N-1))
elif(style=='analog'):
return kwargs['fsamp']*t/(N-1)
elif(style=='samples'):
return t
else:
return t
```
### Test your FourierComplex Class
You can test your implementation and compare it with SciPy, if there is any mismatch try to correct your code.
```
from scipy.fftpack import fft
#SciPy Calculations
y =fft(x.flatten())
N = y.shape[0]
rey = (np.real(y)).reshape(-1,1)/N
imy = (np.imag(y)).reshape(-1,1)/N
#Our Calculation
X = FourierComplex(x, domain='fraction')
plt.suptitle("Comparison between Scipy and Our Implementation", fontsize=14)
plt.subplot(1,2,1)
plt.plot(X.domain, X.rex, label='Our Implementation')
plt.plot(X.domain, rey, label='SciPy Implementation')
plt.xlabel('Fraction Domain')
plt.ylabel('Amplitude')
plt.legend()
plt.grid('on');
plt.subplot(1,2,2)
plt.plot(X.domain, X.imx, label='Our Implementation')
plt.plot(X.domain, imy, label='SciPy Implementation')
plt.xlabel('Fraction Domain')
plt.ylabel('Amplitude')
plt.legend()
plt.grid('on');
```
## Complex IDFT
The Complex Inverse Discrete Fourier Transform is defined as:
$$x[n] = \sum\limits^{N-1}_{k=0}{X[k]e^{j\frac{2\pi k n}{N}}} $$
Where $x[n]$ has $N-1$ points.
By using Euler's formula we can get a rectangular form for the Complex IDFT:
$$x[n] = \sum\limits^{N-1}_{k=0}{\left(Re X[k]+j ImX[k] \right)e^{j\frac{2\pi k n}{N}}} $$
$$ = \sum\limits^{N-1}_{k=0}{Re X[k] e^{j\frac{2\pi k n}{N}}} + \sum\limits^{N-1}_{k=0}{j Im X[k] e^{j\frac{2\pi k n}{N}}} $$
with:
$$e^{j\frac{2\pi k n}{N}} = \left[\cos{\left(\frac{2\pi k n}{N}\right)} +j\sin{\left(\frac{2\pi k n}{N}\right)} \right]$$
therefore:
$$x[n] = \sum\limits^{N-1}_{k=0}{Re X[k] \left[\cos{\left(\frac{2\pi k n}{N}\right)} +j\sin{\left(\frac{2\pi k n}{N}\right)} \right]} + \sum\limits^{N-1}_{k=0}{Im X[k] \left[-\sin{\left(\frac{2\pi k n}{N}\right)} +j\cos{\left(\frac{2\pi k n}{N}\right)} \right]} $$
In words, each value in the real part of the frequency domain contributes a real cosine wave and an imaginary sine wave to the time domain. Likewise, each value in the imaginary part of the frequency domain contributes a real sine wave and an imaginary cosine wave. The time domain is found by adding all these real and imaginary sinusoids. The important concept is that each value in the frequency domain produces both a real sinusoid and an imaginary sinusoid in the time domain.
### Create a ComplexFourierTransform Class
Now you will implement a class called `ComplexFourierTransform` which extends your previous class `FourierComplex` and inherits all of its attributes. You can search about the `super` function for this.
```
class ComplexFourierTransform():
def __init__(self, signal, domain='fraction', **kwargs):
"""
Function that calculates the Complex DFT and IDFT of an input signal.
Parameters:
Same parameters as FourierComplex class.
Attributes:
Ihnerits same attributes as FourierComplex class.
idft (complex numpy array): complex IDFT of the signal
"""
self.idft = None
return
def complex_idft(self):
"""
Function that calculates the Complex IDFT of an input signal.
Returns:
complex numpy array: complex IDFT of input signal of type imaginary.
"""
return None
```
### Test your ComplexFourierTransform Class
You can test your implementation and compare it with the original signal, if there is any mismatch try to correct your code. Try to understand both the real and imaginary signals that the Complex IDFT generates.
```
#Our Calculation
X = ComplexFourierTransform(x, domain='fraction')
plt.suptitle("Complex IDFT", fontsize=14)
plt.subplot(2,1,1)
plt.plot(x, label='Original Signal')
plt.plot(np.real(X.idft), label='Complex IDT -Real Part')
plt.xlabel('Sample')
plt.ylabel('Amplitude')
plt.legend()
plt.grid('on');
plt.subplot(2,1,2)
plt.plot(np.imag(X.idft), label='Complex IDT -Imaginary Part')
plt.xlabel('Sample')
plt.ylabel('Amplitude')
plt.legend()
plt.grid('on');
```
Find the mean and variance of the real and imaginary IDFT signal using the `Statistics` class developed before.
```
stat = None
print('Mean of the real IDFT signal = {:.3f}'.format(stat.mean(np.real(X.idft))))
print('Mean of the imaginary IDFT signal = {:.3f}'.format(stat.mean(np.imag(X.idft))))
print('\nVariance of the real IDFT signal = {:.3f}'.format(stat.variance(np.real(X.idft))))
print('Variance of the imaginary IDFT signal = {:.3f}'.format(stat.variance(np.imag(X.idft))))
```
You can see that our signal can be though as "pure" real signal.
As a final exercise, save your `ComplexFourierTransform` class in the `Common` folder as `complex_fourier_transform.py`
|
github_jupyter
|
import sys
sys.path.insert(0, '../../')
import numpy as np
import matplotlib.pyplot as plt
from Common import common_plots
from Common import statistics
cplots = common_plots.Plot()
file = {'x':'Signals/InputSignal_f32_1kHz_15kHz.dat'}
x = np.loadtxt(file['x'])
N,M = x.shape
x = x.reshape(N*M, 1)
cplots.plot_single(x.T, style='line')
plt.xlabel('samples')
plt.ylabel('amplitude');
class FourierComplex():
def __init__(self, signal, domain='fraction', **kwargs):
"""
Function that calculates the Complex DFT of an input signal.
Parameters:
signal (numpy array): Array of numbers representing the signal to transform.
domain (string): String value that selects between frequency domain's
independent variable.
'samples' returns number of samples between 0 to N/2
'fraction' returns a fraction of the sampling rate between 0 to 0.5
'natural' returns the natural frequency between 0 and pi.
'analog' returns analog frequency between 0 and fsamp/2
kwargs: - fsamp (float): value representing the sampling frequency.
(Only used for 'analog' style).
Attributes:
signal (numpy array): orignal signal.
dft (complex numpy array): complex Fourier Transform of input signal.
rex (numpy array): real DFT part of input signal.
imx (numpy array): imaginary DFT part of input signal.
domain (numpy array): Frequency domain's independent variable.
"""
self.signal = None
self.dft = None
self.rex = None
self.imx = None
self.domain = None
return
def complex_dft(self):
"""
Function that calculates the Complex DFT of an input signal.
Returns:
complex numpy array: complex DFT of input signal of type imaginary.
"""
return None
def real_dft(self):
"""
Function that calculates the real part of the Complex DFT of
an input signal.
Returns:
numpy array: real part of the Complex DFT of input signal.
"""
return None
def imag_dft(self):
"""
Function that calculates the imaginary part of the Complex DFT of
an input signal.
Returns:
numpy array: imaginary part of the Complex DFT of input signal.
"""
return None
def frequency_domain(self, style='fraction', **kwargs):
"""
Function that calculates the frequency domain independent variable.
Parameters:
obtain the frequency domain.
style (string): String value that selects between frequency domain's
independent variable.
'samples' returns number of samples between 0 to N/2
'fraction' returns a fraction of the sampling rate between 0 to 0.5
'natural' returns the natural frequency between 0 and pi.
'analog' returns analog frequency between 0 and fsamp/2
fsamp (float): Float value representing the sampling frequency.
(Only used for 'analog' style).
Returns:
numpy array: Returns frequency domain's independent variable.
"""
N = self.dft.shape[0]
t = np.arange(N)
if(style=='fraction'):
return t/(N-1)
elif(style=='natural'):
return np.pi*(t/(N-1))
elif(style=='analog'):
return kwargs['fsamp']*t/(N-1)
elif(style=='samples'):
return t
else:
return t
from scipy.fftpack import fft
#SciPy Calculations
y =fft(x.flatten())
N = y.shape[0]
rey = (np.real(y)).reshape(-1,1)/N
imy = (np.imag(y)).reshape(-1,1)/N
#Our Calculation
X = FourierComplex(x, domain='fraction')
plt.suptitle("Comparison between Scipy and Our Implementation", fontsize=14)
plt.subplot(1,2,1)
plt.plot(X.domain, X.rex, label='Our Implementation')
plt.plot(X.domain, rey, label='SciPy Implementation')
plt.xlabel('Fraction Domain')
plt.ylabel('Amplitude')
plt.legend()
plt.grid('on');
plt.subplot(1,2,2)
plt.plot(X.domain, X.imx, label='Our Implementation')
plt.plot(X.domain, imy, label='SciPy Implementation')
plt.xlabel('Fraction Domain')
plt.ylabel('Amplitude')
plt.legend()
plt.grid('on');
class ComplexFourierTransform():
def __init__(self, signal, domain='fraction', **kwargs):
"""
Function that calculates the Complex DFT and IDFT of an input signal.
Parameters:
Same parameters as FourierComplex class.
Attributes:
Ihnerits same attributes as FourierComplex class.
idft (complex numpy array): complex IDFT of the signal
"""
self.idft = None
return
def complex_idft(self):
"""
Function that calculates the Complex IDFT of an input signal.
Returns:
complex numpy array: complex IDFT of input signal of type imaginary.
"""
return None
#Our Calculation
X = ComplexFourierTransform(x, domain='fraction')
plt.suptitle("Complex IDFT", fontsize=14)
plt.subplot(2,1,1)
plt.plot(x, label='Original Signal')
plt.plot(np.real(X.idft), label='Complex IDT -Real Part')
plt.xlabel('Sample')
plt.ylabel('Amplitude')
plt.legend()
plt.grid('on');
plt.subplot(2,1,2)
plt.plot(np.imag(X.idft), label='Complex IDT -Imaginary Part')
plt.xlabel('Sample')
plt.ylabel('Amplitude')
plt.legend()
plt.grid('on');
stat = None
print('Mean of the real IDFT signal = {:.3f}'.format(stat.mean(np.real(X.idft))))
print('Mean of the imaginary IDFT signal = {:.3f}'.format(stat.mean(np.imag(X.idft))))
print('\nVariance of the real IDFT signal = {:.3f}'.format(stat.variance(np.real(X.idft))))
print('Variance of the imaginary IDFT signal = {:.3f}'.format(stat.variance(np.imag(X.idft))))
| 0.831861 | 0.990025 |
## Visualize the Bosch Small Traffic Lights Dataset
The Bosch small traffic lights dataset can be downloaded from here: https://hci.iwr.uni-heidelberg.de/node/6132
The dataset used in this notebook is the RGB dataset
### 1. Import necessary modules
```
import os, yaml
import glob
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from pandas.io.json import json_normalize
import tensorflow as tf
import matplotlib.pyplot as plt
%matplotlib inline
```
### 2. Load the dataset
```
#Define path to the dataset and annotation filenames
DATA_FOLDER = os.path.join('.', 'data', 'bosch')
TRAIN_DATA_FOLDER = os.path.join(DATA_FOLDER, 'dataset_train_rgb')
TEST_DATA_FOLDER = os.path.join(DATA_FOLDER, 'dataset_test_rgb')
TRAIN_IMAGE_FOLDER = os.path.join(TRAIN_DATA_FOLDER, 'rgb', 'train')
TEST_IMAGE_FOLDER = os.path.join(TEST_DATA_FOLDER, 'rgb', 'test')
TRAIN_ANNOTATIONS_FILE = os.path.join(TRAIN_DATA_FOLDER, 'train.yaml')
TEST_ANNOTATIONS_FILE = os.path.join(TEST_DATA_FOLDER, 'test.yaml')
#Read in all the image files
train_image_files = glob.glob(os.path.join(TRAIN_IMAGE_FOLDER,'**','*.png'), recursive=True)
test_image_files = glob.glob(os.path.join(TEST_IMAGE_FOLDER,'*.png'), recursive=True)
#Read in all the annotations
train_annotations = yaml.load(open(TRAIN_ANNOTATIONS_FILE, 'rb').read())
test_annotations = yaml.load(open(TEST_ANNOTATIONS_FILE, 'rb').read())
assert(len(train_image_files) == len(train_annotations)), "Number of training annotations does not match training images!"
assert(len(test_image_files) == len(test_annotations)), "Number of test annotations does not match test images!"
```
### 3. Explore the data
```
#Summarize the data
n_train_samples = len(train_annotations)
n_test_samples = len(test_annotations)
sample_train = train_annotations[10]
sample_test = test_annotations[10]
print("Number of training examples: {:d}".format(n_train_samples))
print("Number of test examples: {:d}\n".format(n_test_samples))
print('The annotation files are a {} of {} with the following keys: \n{}\n'
.format(type(train_annotations).__name__,
type(sample_train).__name__,
sample_train.keys()))
print('The boxes key has values that are a {} of {} with keys: \n{}\n'
.format(type(sample_train['boxes']).__name__,
type(sample_train['boxes'][0]).__name__,
sample_train['boxes'][0].keys()))
print('The path key in the training dataset has the following format: \n{}\n'.format(sample_train['path']))
print('The path key in the test dataset has the following format: \n{}\n'.format(sample_test['path']))
#Load the data into dataframes to get the unique labels and instances of each label
train_df = pd.io.json.json_normalize(train_annotations)
test_df = pd.io.json.json_normalize(test_annotations)
trainIdx = train_df.set_index(['path']).boxes.apply(pd.Series).stack().index
testIdx = test_df.set_index(['path']).boxes.apply(pd.Series).stack().index
train_df = pd.DataFrame(train_df.set_index(['path'])
.boxes.apply(pd.Series).stack().values.tolist(),index=trainIdx).reset_index().drop('level_1',1)
test_df = pd.DataFrame(test_df.set_index(['path'])
.boxes.apply(pd.Series).stack().values.tolist(),index=testIdx).reset_index().drop('level_1',1)
print('The training annotations have the following class distributions :\n{}\n'.format(train_df.label.value_counts()))
print('The test annotations have the following class distribution:\n{}\n'.format(test_df.label.value_counts()))
plt.figure(figsize = (22,8))
plt.subplot(1,2,1)
pd.value_counts(test_df['label']).plot(kind='barh', color=['g', 'r', 'k', 'y'])
plt.title('Test annotations class distribution')
plt.subplot(1,2,2)
pd.value_counts(train_df['label']).plot(kind='barh')
plt.title('Test annotations class distribution')
plt.show()
train_df.groupby(['occluded', 'label'])['label'].count().unstack('occluded').plot(kind='barh', stacked=True, figsize=(10,5) )
plt.title('Training annotation class distribution')
test_df.groupby(['occluded', 'label'])['label'].count().unstack('occluded').plot(kind='barh', stacked=True, figsize=(10,5))
plt.title('Test annotation class distribution')
plt.show()
```
|
github_jupyter
|
import os, yaml
import glob
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from pandas.io.json import json_normalize
import tensorflow as tf
import matplotlib.pyplot as plt
%matplotlib inline
#Define path to the dataset and annotation filenames
DATA_FOLDER = os.path.join('.', 'data', 'bosch')
TRAIN_DATA_FOLDER = os.path.join(DATA_FOLDER, 'dataset_train_rgb')
TEST_DATA_FOLDER = os.path.join(DATA_FOLDER, 'dataset_test_rgb')
TRAIN_IMAGE_FOLDER = os.path.join(TRAIN_DATA_FOLDER, 'rgb', 'train')
TEST_IMAGE_FOLDER = os.path.join(TEST_DATA_FOLDER, 'rgb', 'test')
TRAIN_ANNOTATIONS_FILE = os.path.join(TRAIN_DATA_FOLDER, 'train.yaml')
TEST_ANNOTATIONS_FILE = os.path.join(TEST_DATA_FOLDER, 'test.yaml')
#Read in all the image files
train_image_files = glob.glob(os.path.join(TRAIN_IMAGE_FOLDER,'**','*.png'), recursive=True)
test_image_files = glob.glob(os.path.join(TEST_IMAGE_FOLDER,'*.png'), recursive=True)
#Read in all the annotations
train_annotations = yaml.load(open(TRAIN_ANNOTATIONS_FILE, 'rb').read())
test_annotations = yaml.load(open(TEST_ANNOTATIONS_FILE, 'rb').read())
assert(len(train_image_files) == len(train_annotations)), "Number of training annotations does not match training images!"
assert(len(test_image_files) == len(test_annotations)), "Number of test annotations does not match test images!"
#Summarize the data
n_train_samples = len(train_annotations)
n_test_samples = len(test_annotations)
sample_train = train_annotations[10]
sample_test = test_annotations[10]
print("Number of training examples: {:d}".format(n_train_samples))
print("Number of test examples: {:d}\n".format(n_test_samples))
print('The annotation files are a {} of {} with the following keys: \n{}\n'
.format(type(train_annotations).__name__,
type(sample_train).__name__,
sample_train.keys()))
print('The boxes key has values that are a {} of {} with keys: \n{}\n'
.format(type(sample_train['boxes']).__name__,
type(sample_train['boxes'][0]).__name__,
sample_train['boxes'][0].keys()))
print('The path key in the training dataset has the following format: \n{}\n'.format(sample_train['path']))
print('The path key in the test dataset has the following format: \n{}\n'.format(sample_test['path']))
#Load the data into dataframes to get the unique labels and instances of each label
train_df = pd.io.json.json_normalize(train_annotations)
test_df = pd.io.json.json_normalize(test_annotations)
trainIdx = train_df.set_index(['path']).boxes.apply(pd.Series).stack().index
testIdx = test_df.set_index(['path']).boxes.apply(pd.Series).stack().index
train_df = pd.DataFrame(train_df.set_index(['path'])
.boxes.apply(pd.Series).stack().values.tolist(),index=trainIdx).reset_index().drop('level_1',1)
test_df = pd.DataFrame(test_df.set_index(['path'])
.boxes.apply(pd.Series).stack().values.tolist(),index=testIdx).reset_index().drop('level_1',1)
print('The training annotations have the following class distributions :\n{}\n'.format(train_df.label.value_counts()))
print('The test annotations have the following class distribution:\n{}\n'.format(test_df.label.value_counts()))
plt.figure(figsize = (22,8))
plt.subplot(1,2,1)
pd.value_counts(test_df['label']).plot(kind='barh', color=['g', 'r', 'k', 'y'])
plt.title('Test annotations class distribution')
plt.subplot(1,2,2)
pd.value_counts(train_df['label']).plot(kind='barh')
plt.title('Test annotations class distribution')
plt.show()
train_df.groupby(['occluded', 'label'])['label'].count().unstack('occluded').plot(kind='barh', stacked=True, figsize=(10,5) )
plt.title('Training annotation class distribution')
test_df.groupby(['occluded', 'label'])['label'].count().unstack('occluded').plot(kind='barh', stacked=True, figsize=(10,5))
plt.title('Test annotation class distribution')
plt.show()
| 0.557123 | 0.881207 |
# HowTo - 3 - Get offset stars and make finding charts
# 0) Load test data
### We begin again by loading the modules and the test data
```
import pandas as pd
from qso_toolbox import catalog_tools as ct, image_tools as it
pd.options.mode.chained_assignment = None
df_hdf = pd.read_hdf('./data/stripe82_milliquas_190210.hdf5',key='data')
# We use the first 10 entries of the test set we used for HowTo-1-DownloadingImageCutouts
test_set = df_hdf.query('340 < mq_ra < 350 and -1.26 < mq_dec < 0')[:10]
test_set
```
# 1) Get offset stars
### The main routines to get offset stars are found in catalog_tools. The routines use the astroquery package (get_astroquery_offset), the NOAO datalab python package (get_offset_stars_datalab) and the MAST website for PanSTARRS (get_offset_stars_ps1). Due to the way that different surveys scripted region queries, this seemed to be the most comfortable way of coding it up at this time. In the future one would want to consolidate these into one function.
### For each object in the supplied dataframe the function calls a region query around the object. The closest 'n' objects (fulfilling all specified criteria) are retrieved and stored in a new dataframe. After each object the new dataframe is stored in 'temp_offset_df.csv' in case one region query fails. In the end all functions return a dataframe with a maximum of 'n' offset starts per object.
### The dataframe includes the original target name, RA and Dec, the offset name (including its suffix), offset Ra and Dec, one magnitude for the offset, the separation (arcseconds), position angle, Ra and Dec offset (arcseconds).
### The offset stars in the dataframe will be given alphabetical suffices (e.g. '_A',' _B', etc.). Currently retrieval of a maximum of n=5 offset stars is possible.
### All offset star routines use essentially three different functions.
### a) query_region_"astroquery, ps1, datalab"()
Returns the catalog data of sources within a given radius of a defined
position.
### b) get_"astroquery, ps1, datalab"_offset()
Return the n nearest offset stars specified by the quality criteria
around a given target.
### c) get_offset_stars_"astroquery, ps1, datalab"()
Get offset stars for all targets in the input DataFrame.
### While one would usually use function c) to retrieve offset stars, functions a) and b) can also be used indepdently from c).
# 1.1) Astroquery offsets
### Currently options to get offset stars from 2MASS, NOMAD and VHSDR6 using the IRSA, Vizier and VSA astroquery services. It is trivial to add more surveys for these three services.
### These surveys are summarized in the internal variable astroquery_dict. Please have a look into the dictionary as it also sets, which magnitude will be retrieved.
astroquery_dict = {
'tmass': {'service': 'irsa', 'catalog': 'fp_psc',
'ra': 'ra', 'dec': 'dec', 'mag_name':
'TMASS_J', 'mag': 'j_m', 'distance':
'dist', 'data_release': None},
'nomad': {'service': 'vizier', 'catalog': 'NOMAD',
'ra': 'RAJ2000', 'dec': 'DECJ2000',
'mag_name': 'R', 'mag': 'Rmag', 'distance':
'distance', 'data_release': None},
'vhsdr6': {'service': 'vsa', 'catalog': 'VHS',
'ra': 'ra', 'dec': 'dec',
'data_release': 'VHSDR6', 'mag_name': 'VHS_J',
'mag': 'jAperMag3', 'distance': 'distance'}
}
## Example:
### We will retrieve offset stars for the test_set from VHS DR6. It will automatically retrieve the J-band magnitude ('jAperMag3') along with the position. The additional quality query ensures that we do not retrieve the target itself, limits the objects to a certain J-band magnitude range and set a simple quality flag for the J-band.
```
# Quality query needs to be written in pandas dataframe query syntax. Knowledge about the
# catalog columns returned from the query_region_astroquery() function is very useful.
quality_query = 'distance > 3/60. and 10 < jAperMag3 < 20 and jppErrBits==0'
offset_df = ct.get_offset_stars_astroquery(test_set, 'mq_name', 'mq_ra',
'mq_dec', 90, catalog='vhsdr6',
quality_query=quality_query, n=3, verbosity=2)
```
### Let's display (a few rows from) the offset dataframe:
```
offset_df[:5]
```
# 1.2) Datalab offsets
### The datalab version offers access to a different range of surveys than astroquery. At this point only access to DES DR1 data is included here. Similar to the astroquery routines a simple dictionary sets this limitation an can be easily expanded. The current dictionary reads:
datalab_offset_dict = {'des_dr1.main': {'ra': 'ra', 'dec': 'dec',
'mag': 'mag_auto_z',
'mag_name': 'mag_auto_z'}}
### Main difference to the astroquery offset routines: The 'quality_query' keyword argument is replaced by 'where', which is now in ADQL (Astronomical Data Query Language) syntax.
## Example:
### We will again provide a quick example.
```
# The where keyword argument will be added to the ADQL query to the NOAO datalab.
where = 'mag_auto_z > 15 AND mag_auto_z < 19'
offset_df = ct.get_offset_stars_datalab(test_set, 'mq_name', 'mq_ra',
'mq_dec', 90, survey='des_dr1', table='main',
n=3, where=where, verbosity=2)
```
### Let's display (a few rows from) the offset dataframe:
```
offset_df[:5]
```
# 1.3) PanSTARRS offsets
### The PanSTARRS version of the offset query uses the MAST website to query the archive using a url and retrieving the provided data table.
### The 'qualiy_query' keyword argument in pandas query syntax then applies quality criteria to the retrieved data table.
### WARNING: As the response from the MAST PanSTARRS archive is slow the routine will take a while for large target catalogs.
## Example:
```
quality_query = '10 < zMeanPSFMag < 19'
offset_df = ct.get_offset_stars_ps1(test_set, 'mq_name', 'mq_ra',
'mq_dec', 90, data_release='dr2',
catalog='mean', quality_query=quality_query, verbosity=2)
```
### Let's display (a few rows from) the offset dataframe:
```
offset_df[:5]
```
# 2) Get Finding Charts
### The qso_toolbox image_tools package also provides routines for finding chart generation. However, before finding charts can be made the images need to be downloaded and, if necessary, offset stars need to be found.
### The following example downloads the images and retrives offset starts for our test set:
```
# Download finding chart images
surveys = ['vhsdr6']
bands = ['J']
fovs = [300]
ct.get_photometry(test_set, 'mq_ra', 'mq_dec', surveys, bands, './cutouts/', fovs, verbosity=0)
# Get offset stars
quality_query = 'distance > 3/60. and 10 < jAperMag3 < 20 and jppErrBits==0'
offset_df = ct.get_offset_stars_astroquery(test_set, 'mq_name', 'mq_ra',
'mq_dec', 90, catalog='vhsdr6',
quality_query=quality_query, n=3, verbosity=0)
```
### Now we are ready to produce finding charts. The routine that you probably want to use is image_tools.make_finding_charts(). It generates finding charts for the entire target list (pandas dataframe) and saves them either in png or pdf format.
### Before we demonstrate this routine, we will show how you generate only one finding chart in the following example:
```
# We first reduce the offsets in the catalog to the ones for this particular target
offset_target = offset_df.query('target_name=="{}"'.format('SDSS J224007.89-011522.1'))
print(offset_target)
# Make one finding chart
ra = test_set.loc[test_set.index[1],'mq_ra']
dec = test_set.loc[test_set.index[1],'mq_dec']
print(ra, dec)
it.make_finding_chart(ra, dec, 'vhsdr6', 'J', 2, 240,
'./cutouts/',
offset_df=offset_target,
offset_ra_column_name='offset_ra',
offset_dec_column_name='offset_dec',
# offset_mag_column_name='jAperMag3',
# offset_id_column_name='offset_shortname',
slit_width=1.0, slit_length=20,
position_angle=0, verbosity=2)
```
### Generating finding charts for your entire target catalog is comparatively easy. You just call the function below supplied with your target catalog and the offset star catalog.
```
# Make finding charts
it.make_finding_charts(test_set[:2], 'mq_ra', 'mq_dec',
'mq_name', 'vhsdr6', 'J',
2.5, 120, './cutouts',
offset_table=offset_df,
offset_focus=True,
offset_ra_column_name='offset_ra',
offset_dec_column_name='offset_dec',
pos_angle_column_name=None,
offset_mag_column_name='jAperMag3',
offset_id_column_name='offset_shortname',
label_position='topleft',
slit_width=1.0, slit_length=50,
format='png', verbosity=0,
auto_download=True)
```
### In the end make sure that your finding charts reflect the depth, sensitivity and field of view of your acquisition camera. So you will likely need to tweak the code here and there.
### In addition, make sure to check all finding charts! If you take a look at the once we generated you will notice that a few extended sources are selected as offset stars. This should not be the case, so make sure to include the necessary quality criteria when you retrieve offset stars.
|
github_jupyter
|
import pandas as pd
from qso_toolbox import catalog_tools as ct, image_tools as it
pd.options.mode.chained_assignment = None
df_hdf = pd.read_hdf('./data/stripe82_milliquas_190210.hdf5',key='data')
# We use the first 10 entries of the test set we used for HowTo-1-DownloadingImageCutouts
test_set = df_hdf.query('340 < mq_ra < 350 and -1.26 < mq_dec < 0')[:10]
test_set
# Quality query needs to be written in pandas dataframe query syntax. Knowledge about the
# catalog columns returned from the query_region_astroquery() function is very useful.
quality_query = 'distance > 3/60. and 10 < jAperMag3 < 20 and jppErrBits==0'
offset_df = ct.get_offset_stars_astroquery(test_set, 'mq_name', 'mq_ra',
'mq_dec', 90, catalog='vhsdr6',
quality_query=quality_query, n=3, verbosity=2)
offset_df[:5]
# The where keyword argument will be added to the ADQL query to the NOAO datalab.
where = 'mag_auto_z > 15 AND mag_auto_z < 19'
offset_df = ct.get_offset_stars_datalab(test_set, 'mq_name', 'mq_ra',
'mq_dec', 90, survey='des_dr1', table='main',
n=3, where=where, verbosity=2)
offset_df[:5]
quality_query = '10 < zMeanPSFMag < 19'
offset_df = ct.get_offset_stars_ps1(test_set, 'mq_name', 'mq_ra',
'mq_dec', 90, data_release='dr2',
catalog='mean', quality_query=quality_query, verbosity=2)
offset_df[:5]
# Download finding chart images
surveys = ['vhsdr6']
bands = ['J']
fovs = [300]
ct.get_photometry(test_set, 'mq_ra', 'mq_dec', surveys, bands, './cutouts/', fovs, verbosity=0)
# Get offset stars
quality_query = 'distance > 3/60. and 10 < jAperMag3 < 20 and jppErrBits==0'
offset_df = ct.get_offset_stars_astroquery(test_set, 'mq_name', 'mq_ra',
'mq_dec', 90, catalog='vhsdr6',
quality_query=quality_query, n=3, verbosity=0)
# We first reduce the offsets in the catalog to the ones for this particular target
offset_target = offset_df.query('target_name=="{}"'.format('SDSS J224007.89-011522.1'))
print(offset_target)
# Make one finding chart
ra = test_set.loc[test_set.index[1],'mq_ra']
dec = test_set.loc[test_set.index[1],'mq_dec']
print(ra, dec)
it.make_finding_chart(ra, dec, 'vhsdr6', 'J', 2, 240,
'./cutouts/',
offset_df=offset_target,
offset_ra_column_name='offset_ra',
offset_dec_column_name='offset_dec',
# offset_mag_column_name='jAperMag3',
# offset_id_column_name='offset_shortname',
slit_width=1.0, slit_length=20,
position_angle=0, verbosity=2)
# Make finding charts
it.make_finding_charts(test_set[:2], 'mq_ra', 'mq_dec',
'mq_name', 'vhsdr6', 'J',
2.5, 120, './cutouts',
offset_table=offset_df,
offset_focus=True,
offset_ra_column_name='offset_ra',
offset_dec_column_name='offset_dec',
pos_angle_column_name=None,
offset_mag_column_name='jAperMag3',
offset_id_column_name='offset_shortname',
label_position='topleft',
slit_width=1.0, slit_length=50,
format='png', verbosity=0,
auto_download=True)
| 0.50415 | 0.98537 |
# Now You Code In Class: Getting Directions!
Let's continue on our zomato journey, mashing it up with the Bing Maps API so that we can get driving instructions to the restaurant.
Bing Maps Route API is here: https://docs.microsoft.com/en-us/bingmaps/rest-services/routes/calculate-a-route
The zomato functions are the the `zomato.ipynb` file. We can import one noteook into another using the `import_ipynb` python module.
```
import requests
import import_ipynb
import zomato as zomato
# testing
zomato_key = 'Zomato-Key'
zomato.example(zomato_key)
```
## Step 1a: Learning The Bing Route API
FIRST: Sign up and get an API Key: https://docs.microsoft.com/en-us/bingmaps/getting-started/bing-maps-dev-center-help/getting-a-bing-maps-key
NEXT: Read the Bing Maps Route API is here: https://docs.microsoft.com/en-us/bingmaps/rest-services/routes/calculate-a-route
WHAT IS REQUIRED TO EXECUTE THE API (INPUTS):
PROMPT 1
WHAT DOES THE API GIVE US BACK (OUTPUTS):
PROMPT 2
Algorithm (Steps in Program):
Algorithms to call API's are the same, so the concerns are:
- What do we need to call the API?
- What do I need from the response?
## Step 1b: Code the API: Proof of Concept
```
# PROMPT 3 Demonstrate you can call the API
start = 'Manley Field House, Syracuse, NY 13244'
finish = 'Carrier Dome, Syracuse, NY 13204'
bing_key = 'Bing-Key'
# PROMPT 4 What do I need from the response?
```
## Step 1.c: Refactor into a function `getDirections()`
Now the we understand the API, let's refactor the code into a useable function
FUNCTION INPUTS:
PROMPT 5
FUNCTION OUTPUTS:
PROMPT 6
ALGORITHM:
PROMPT 7
TEST (How do we know it's right?)
PROMPT 8
```
# PROMPT 9 - write function
```
## Step 1.d: Test the function
```
# PROMPT 10
bing_key = 'bing-Key'
start = 'Manley Field House, Syracuse, NY 13244'
finish = 'Carrier Dome, Syracuse, NY 13204'
```
## Step 2: Putting it all together
PROMPTS:
11. Input your home address, then city where you want to eat.
12. Use Zomato to find the cityid
13. Use Zomato to get the trending restaurants
14. Build a list of trending restaurants
15. Start Interact - upon selection of a restaurant
16. find restaurant ID
17. get restaurant detail
18. show restaurant details name, cuisine, hours, address, phone
19. find driving directions from home address to restaurant
```
from IPython.display import display, HTML
from ipywidgets import widgets, interact
import requests
display(HTML("<h1>Fudge's Food Finder</h1>"))
bing_key = 'Bing-Key'
zomato_key = 'Zomato-Key'
# PROMPTS
# run this code to turn in your work!
from coursetools.submission import Submission
Submission().submit_now()
```
|
github_jupyter
|
import requests
import import_ipynb
import zomato as zomato
# testing
zomato_key = 'Zomato-Key'
zomato.example(zomato_key)
# PROMPT 3 Demonstrate you can call the API
start = 'Manley Field House, Syracuse, NY 13244'
finish = 'Carrier Dome, Syracuse, NY 13204'
bing_key = 'Bing-Key'
# PROMPT 4 What do I need from the response?
# PROMPT 9 - write function
# PROMPT 10
bing_key = 'bing-Key'
start = 'Manley Field House, Syracuse, NY 13244'
finish = 'Carrier Dome, Syracuse, NY 13204'
from IPython.display import display, HTML
from ipywidgets import widgets, interact
import requests
display(HTML("<h1>Fudge's Food Finder</h1>"))
bing_key = 'Bing-Key'
zomato_key = 'Zomato-Key'
# PROMPTS
# run this code to turn in your work!
from coursetools.submission import Submission
Submission().submit_now()
| 0.273866 | 0.892796 |
# MULTI-LAYER PERCEPTRON
**Import MINST Data**
```
# Import MINST data
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("MNIST_data/", one_hot=True)
import tensorflow as tf
```
# Setting parameters for layers
We need to setup model parameters that will be used to make model more accurate and tuning purpose
```
# Parameters
learning_rate = 0.001
training_epochs = 15
batch_size = 100
display_step = 1
# Network Parameters
n_hidden_1 = 256 # 1st layer number of features
n_hidden_2 = 256 # 2nd layer number of features
n_input = 784 # MNIST data input (img shape: 28*28)
n_classes = 10 # MNIST total classes (0-9 digits)
# tf Graph input
x = tf.placeholder("float", [None, n_input])
y = tf.placeholder("float", [None, n_classes])
```
# Create multilayer perceptron function
Here we have one hidden layer i.e. Rectified linear unit and output layer with linear activation
```
# Create model
def multilayer_perceptron(x, weights, biases):
# Hidden layer with RELU activation
layer_1 = tf.add(tf.matmul(x, weights['h1']), biases['b1'])
#rectified linear unit
layer_1 = tf.nn.relu(layer_1)
# Hidden layer with RELU activation
layer_2 = tf.add(tf.matmul(layer_1, weights['h2']), biases['b2'])
layer_2 = tf.nn.relu(layer_2)
# Output layer with linear activation
out_layer = tf.matmul(layer_2, weights['out']) + biases['out']
return out_layer
```
# Layers weight and bias
```
# Store layers weight & bias
weights = {
'h1': tf.Variable(tf.random_normal([n_input, n_hidden_1])),
'h2': tf.Variable(tf.random_normal([n_hidden_1, n_hidden_2])),
'out': tf.Variable(tf.random_normal([n_hidden_2, n_classes]))
}
biases = {
'b1': tf.Variable(tf.random_normal([n_hidden_1])),
'b2': tf.Variable(tf.random_normal([n_hidden_2])),
'out': tf.Variable(tf.random_normal([n_classes]))
}
# Construct model
pred = multilayer_perceptron(x, weights, biases)
# Define loss and optimizer
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))
optimizer = tf.train.AdamOptimizer(learning_rate=learning_rate).minimize(cost)
# Initializing the variables
init = tf.global_variables_initializer()
```
# Training and testing of model with data
```
# Launch the graph
with tf.Session() as sess:
sess.run(init)
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
batch_x, batch_y = mnist.train.next_batch(batch_size)
# Run optimization op (backprop) and cost op (to get loss value)
_, c = sess.run([optimizer, cost], feed_dict={x: batch_x,
y: batch_y})
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if epoch % display_step == 0:
print("Epoch:", '%04d' % (epoch + 1), \
"cost=", "{:.9f}".format(avg_cost))
print("Optimization Finished!")
# Test model
correct_prediction = tf.equal(tf.argmax(pred, 1), tf.argmax(y, 1))
# Calculate accuracy
accuracy = tf.reduce_mean(tf.cast(correct_prediction, "float"))
print("Accuracy:", accuracy.eval({x: mnist.test.images, y: mnist.test.labels}))
```
|
github_jupyter
|
# Import MINST data
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("MNIST_data/", one_hot=True)
import tensorflow as tf
# Parameters
learning_rate = 0.001
training_epochs = 15
batch_size = 100
display_step = 1
# Network Parameters
n_hidden_1 = 256 # 1st layer number of features
n_hidden_2 = 256 # 2nd layer number of features
n_input = 784 # MNIST data input (img shape: 28*28)
n_classes = 10 # MNIST total classes (0-9 digits)
# tf Graph input
x = tf.placeholder("float", [None, n_input])
y = tf.placeholder("float", [None, n_classes])
# Create model
def multilayer_perceptron(x, weights, biases):
# Hidden layer with RELU activation
layer_1 = tf.add(tf.matmul(x, weights['h1']), biases['b1'])
#rectified linear unit
layer_1 = tf.nn.relu(layer_1)
# Hidden layer with RELU activation
layer_2 = tf.add(tf.matmul(layer_1, weights['h2']), biases['b2'])
layer_2 = tf.nn.relu(layer_2)
# Output layer with linear activation
out_layer = tf.matmul(layer_2, weights['out']) + biases['out']
return out_layer
# Store layers weight & bias
weights = {
'h1': tf.Variable(tf.random_normal([n_input, n_hidden_1])),
'h2': tf.Variable(tf.random_normal([n_hidden_1, n_hidden_2])),
'out': tf.Variable(tf.random_normal([n_hidden_2, n_classes]))
}
biases = {
'b1': tf.Variable(tf.random_normal([n_hidden_1])),
'b2': tf.Variable(tf.random_normal([n_hidden_2])),
'out': tf.Variable(tf.random_normal([n_classes]))
}
# Construct model
pred = multilayer_perceptron(x, weights, biases)
# Define loss and optimizer
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))
optimizer = tf.train.AdamOptimizer(learning_rate=learning_rate).minimize(cost)
# Initializing the variables
init = tf.global_variables_initializer()
# Launch the graph
with tf.Session() as sess:
sess.run(init)
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
batch_x, batch_y = mnist.train.next_batch(batch_size)
# Run optimization op (backprop) and cost op (to get loss value)
_, c = sess.run([optimizer, cost], feed_dict={x: batch_x,
y: batch_y})
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if epoch % display_step == 0:
print("Epoch:", '%04d' % (epoch + 1), \
"cost=", "{:.9f}".format(avg_cost))
print("Optimization Finished!")
# Test model
correct_prediction = tf.equal(tf.argmax(pred, 1), tf.argmax(y, 1))
# Calculate accuracy
accuracy = tf.reduce_mean(tf.cast(correct_prediction, "float"))
print("Accuracy:", accuracy.eval({x: mnist.test.images, y: mnist.test.labels}))
| 0.918105 | 0.96175 |
Copyright (c) Microsoft Corporation. All rights reserved.
Licensed under the MIT License.

# Automated Machine Learning
**BikeShare Demand Forecasting**
## Contents
1. [Introduction](#Introduction)
1. [Setup](#Setup)
1. [Compute](#Compute)
1. [Data](#Data)
1. [Train](#Train)
1. [Featurization](#Featurization)
1. [Evaluate](#Evaluate)
## Introduction
This notebook demonstrates demand forecasting for a bike-sharing service using AutoML.
AutoML highlights here include built-in holiday featurization, accessing engineered feature names, and working with the `forecast` function. Please also look at the additional forecasting notebooks, which document lagging, rolling windows, forecast quantiles, other ways to use the forecast function, and forecaster deployment.
Make sure you have executed the [configuration notebook](../../../configuration.ipynb) before running this notebook.
Notebook synopsis:
1. Creating an Experiment in an existing Workspace
2. Configuration and local run of AutoML for a time-series model with lag and holiday features
3. Viewing the engineered names for featurized data and featurization summary for all raw features
4. Evaluating the fitted model using a rolling test
## Setup
```
import azureml.core
import pandas as pd
import numpy as np
import logging
from azureml.core import Workspace, Experiment, Dataset
from azureml.train.automl import AutoMLConfig
from datetime import datetime
```
This sample notebook may use features that are not available in previous versions of the Azure ML SDK.
```
print("This notebook was created using version 1.16.0 of the Azure ML SDK")
print("You are currently using version", azureml.core.VERSION, "of the Azure ML SDK")
```
As part of the setup you have already created a <b>Workspace</b>. To run AutoML, you also need to create an <b>Experiment</b>. An Experiment corresponds to a prediction problem you are trying to solve, while a Run corresponds to a specific approach to the problem.
```
ws = Workspace.from_config()
# choose a name for the run history container in the workspace
experiment_name = 'automl-bikeshareforecasting'
experiment = Experiment(ws, experiment_name)
output = {}
output['Subscription ID'] = ws.subscription_id
output['Workspace'] = ws.name
output['SKU'] = ws.sku
output['Resource Group'] = ws.resource_group
output['Location'] = ws.location
output['Run History Name'] = experiment_name
pd.set_option('display.max_colwidth', -1)
outputDf = pd.DataFrame(data = output, index = [''])
outputDf.T
```
## Compute
You will need to create a [compute target](https://docs.microsoft.com/en-us/azure/machine-learning/service/how-to-set-up-training-targets#amlcompute) for your AutoML run. In this tutorial, you create AmlCompute as your training compute resource.
#### Creation of AmlCompute takes approximately 5 minutes.
If the AmlCompute with that name is already in your workspace this code will skip the creation process.
As with other Azure services, there are limits on certain resources (e.g. AmlCompute) associated with the Azure Machine Learning service. Please read this article on the default limits and how to request more quota.
```
from azureml.core.compute import ComputeTarget, AmlCompute
from azureml.core.compute_target import ComputeTargetException
# Choose a name for your cluster.
amlcompute_cluster_name = "bike-cluster"
# Verify that cluster does not exist already
try:
compute_target = ComputeTarget(workspace=ws, name=amlcompute_cluster_name)
print('Found existing cluster, use it.')
except ComputeTargetException:
compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_D2_V2',
max_nodes=4)
compute_target = ComputeTarget.create(ws, amlcompute_cluster_name, compute_config)
compute_target.wait_for_completion(show_output=True)
```
## Data
The [Machine Learning service workspace](https://docs.microsoft.com/en-us/azure/machine-learning/service/concept-workspace) is paired with the storage account, which contains the default data store. We will use it to upload the bike share data and create [tabular dataset](https://docs.microsoft.com/en-us/python/api/azureml-core/azureml.data.tabulardataset?view=azure-ml-py) for training. A tabular dataset defines a series of lazily-evaluated, immutable operations to load data from the data source into tabular representation.
```
datastore = ws.get_default_datastore()
datastore.upload_files(files = ['./bike-no.csv'], target_path = 'dataset/', overwrite = True,show_progress = True)
```
Let's set up what we know about the dataset.
**Target column** is what we want to forecast.
**Time column** is the time axis along which to predict.
```
target_column_name = 'cnt'
time_column_name = 'date'
dataset = Dataset.Tabular.from_delimited_files(path = [(datastore, 'dataset/bike-no.csv')]).with_timestamp_columns(fine_grain_timestamp=time_column_name)
dataset.take(5).to_pandas_dataframe().reset_index(drop=True)
```
### Split the data
The first split we make is into train and test sets. Note we are splitting on time. Data before 9/1 will be used for training, and data after and including 9/1 will be used for testing.
```
# select data that occurs before a specified date
train = dataset.time_before(datetime(2012, 8, 31), include_boundary=True)
train.to_pandas_dataframe().tail(5).reset_index(drop=True)
test = dataset.time_after(datetime(2012, 9, 1), include_boundary=True)
test.to_pandas_dataframe().head(5).reset_index(drop=True)
```
## Forecasting Parameters
To define forecasting parameters for your experiment training, you can leverage the ForecastingParameters class. The table below details the forecasting parameter we will be passing into our experiment.
|Property|Description|
|-|-|
|**time_column_name**|The name of your time column.|
|**forecast_horizon**|The forecast horizon is how many periods forward you would like to forecast. This integer horizon is in units of the timeseries frequency (e.g. daily, weekly).|
|**country_or_region_for_holidays**|The country/region used to generate holiday features. These should be ISO 3166 two-letter country/region codes (i.e. 'US', 'GB').|
|**target_lags**|The target_lags specifies how far back we will construct the lags of the target variable.|
|**drop_column_names**|Name(s) of columns to drop prior to modeling|
## Train
Instantiate a AutoMLConfig object. This defines the settings and data used to run the experiment.
|Property|Description|
|-|-|
|**task**|forecasting|
|**primary_metric**|This is the metric that you want to optimize.<br> Forecasting supports the following primary metrics <br><i>spearman_correlation</i><br><i>normalized_root_mean_squared_error</i><br><i>r2_score</i><br><i>normalized_mean_absolute_error</i>
|**blocked_models**|Models in blocked_models won't be used by AutoML. All supported models can be found at [here](https://docs.microsoft.com/en-us/python/api/azureml-train-automl-client/azureml.train.automl.constants.supportedmodels.forecasting?view=azure-ml-py).|
|**experiment_timeout_hours**|Experimentation timeout in hours.|
|**training_data**|Input dataset, containing both features and label column.|
|**label_column_name**|The name of the label column.|
|**compute_target**|The remote compute for training.|
|**n_cross_validations**|Number of cross validation splits.|
|**enable_early_stopping**|If early stopping is on, training will stop when the primary metric is no longer improving.|
|**forecasting_parameters**|A class that holds all the forecasting related parameters.|
This notebook uses the blocked_models parameter to exclude some models that take a longer time to train on this dataset. You can choose to remove models from the blocked_models list but you may need to increase the experiment_timeout_hours parameter value to get results.
### Setting forecaster maximum horizon
The forecast horizon is the number of periods into the future that the model should predict. Here, we set the horizon to 14 periods (i.e. 14 days). Notice that this is much shorter than the number of days in the test set; we will need to use a rolling test to evaluate the performance on the whole test set. For more discussion of forecast horizons and guiding principles for setting them, please see the [energy demand notebook](https://github.com/Azure/MachineLearningNotebooks/tree/master/how-to-use-azureml/automated-machine-learning/forecasting-energy-demand).
```
forecast_horizon = 14
```
### Config AutoML
```
from azureml.automl.core.forecasting_parameters import ForecastingParameters
forecasting_parameters = ForecastingParameters(
time_column_name=time_column_name,
forecast_horizon=forecast_horizon,
country_or_region_for_holidays='US', # set country_or_region will trigger holiday featurizer
target_lags='auto', # use heuristic based lag setting
drop_column_names=['casual', 'registered'] # these columns are a breakdown of the total and therefore a leak
)
automl_config = AutoMLConfig(task='forecasting',
primary_metric='normalized_root_mean_squared_error',
blocked_models = ['ExtremeRandomTrees'],
experiment_timeout_hours=0.3,
training_data=train,
label_column_name=target_column_name,
compute_target=compute_target,
enable_early_stopping=True,
n_cross_validations=3,
max_concurrent_iterations=4,
max_cores_per_iteration=-1,
verbosity=logging.INFO,
forecasting_parameters=forecasting_parameters)
```
We will now run the experiment, you can go to Azure ML portal to view the run details.
```
remote_run = experiment.submit(automl_config, show_output=False)
remote_run
remote_run.wait_for_completion()
```
### Retrieve the Best Model
Below we select the best model from all the training iterations using get_output method.
```
best_run, fitted_model = remote_run.get_output()
fitted_model.steps
```
## Featurization
You can access the engineered feature names generated in time-series featurization. Note that a number of named holiday periods are represented. We recommend that you have at least one year of data when using this feature to ensure that all yearly holidays are captured in the training featurization.
```
fitted_model.named_steps['timeseriestransformer'].get_engineered_feature_names()
```
### View the featurization summary
You can also see what featurization steps were performed on different raw features in the user data. For each raw feature in the user data, the following information is displayed:
- Raw feature name
- Number of engineered features formed out of this raw feature
- Type detected
- If feature was dropped
- List of feature transformations for the raw feature
```
# Get the featurization summary as a list of JSON
featurization_summary = fitted_model.named_steps['timeseriestransformer'].get_featurization_summary()
# View the featurization summary as a pandas dataframe
pd.DataFrame.from_records(featurization_summary)
```
## Evaluate
We now use the best fitted model from the AutoML Run to make forecasts for the test set. We will do batch scoring on the test dataset which should have the same schema as training dataset.
The scoring will run on a remote compute. In this example, it will reuse the training compute.
```
test_experiment = Experiment(ws, experiment_name + "_test")
```
### Retrieving forecasts from the model
To run the forecast on the remote compute we will use a helper script: forecasting_script. This script contains the utility methods which will be used by the remote estimator. We copy the script to the project folder to upload it to remote compute.
```
import os
import shutil
script_folder = os.path.join(os.getcwd(), 'forecast')
os.makedirs(script_folder, exist_ok=True)
shutil.copy('forecasting_script.py', script_folder)
```
For brevity, we have created a function called run_forecast that submits the test data to the best model determined during the training run and retrieves forecasts. The test set is longer than the forecast horizon specified at train time, so the forecasting script uses a so-called rolling evaluation to generate predictions over the whole test set. A rolling evaluation iterates the forecaster over the test set, using the actuals in the test set to make lag features as needed.
```
from run_forecast import run_rolling_forecast
remote_run = run_rolling_forecast(test_experiment, compute_target, best_run, test, target_column_name)
remote_run
remote_run.wait_for_completion(show_output=False)
```
### Download the prediction result for metrics calcuation
The test data with predictions are saved in artifact outputs/predictions.csv. You can download it and calculation some error metrics for the forecasts and vizualize the predictions vs. the actuals.
```
remote_run.download_file('outputs/predictions.csv', 'predictions.csv')
df_all = pd.read_csv('predictions.csv')
from azureml.automl.core.shared import constants
from azureml.automl.runtime.shared.score import scoring
from sklearn.metrics import mean_absolute_error, mean_squared_error
from matplotlib import pyplot as plt
# use automl metrics module
scores = scoring.score_regression(
y_test=df_all[target_column_name],
y_pred=df_all['predicted'],
metrics=list(constants.Metric.SCALAR_REGRESSION_SET))
print("[Test data scores]\n")
for key, value in scores.items():
print('{}: {:.3f}'.format(key, value))
# Plot outputs
%matplotlib inline
test_pred = plt.scatter(df_all[target_column_name], df_all['predicted'], color='b')
test_test = plt.scatter(df_all[target_column_name], df_all[target_column_name], color='g')
plt.legend((test_pred, test_test), ('prediction', 'truth'), loc='upper left', fontsize=8)
plt.show()
```
Since we did a rolling evaluation on the test set, we can analyze the predictions by their forecast horizon relative to the rolling origin. The model was initially trained at a forecast horizon of 14, so each prediction from the model is associated with a horizon value from 1 to 14. The horizon values are in a column named, "horizon_origin," in the prediction set. For example, we can calculate some of the error metrics grouped by the horizon:
```
from metrics_helper import MAPE, APE
df_all.groupby('horizon_origin').apply(
lambda df: pd.Series({'MAPE': MAPE(df[target_column_name], df['predicted']),
'RMSE': np.sqrt(mean_squared_error(df[target_column_name], df['predicted'])),
'MAE': mean_absolute_error(df[target_column_name], df['predicted'])}))
```
To drill down more, we can look at the distributions of APE (absolute percentage error) by horizon. From the chart, it is clear that the overall MAPE is being skewed by one particular point where the actual value is of small absolute value.
```
df_all_APE = df_all.assign(APE=APE(df_all[target_column_name], df_all['predicted']))
APEs = [df_all_APE[df_all['horizon_origin'] == h].APE.values for h in range(1, forecast_horizon + 1)]
%matplotlib inline
plt.boxplot(APEs)
plt.yscale('log')
plt.xlabel('horizon')
plt.ylabel('APE (%)')
plt.title('Absolute Percentage Errors by Forecast Horizon')
plt.show()
```
|
github_jupyter
|
import azureml.core
import pandas as pd
import numpy as np
import logging
from azureml.core import Workspace, Experiment, Dataset
from azureml.train.automl import AutoMLConfig
from datetime import datetime
print("This notebook was created using version 1.16.0 of the Azure ML SDK")
print("You are currently using version", azureml.core.VERSION, "of the Azure ML SDK")
ws = Workspace.from_config()
# choose a name for the run history container in the workspace
experiment_name = 'automl-bikeshareforecasting'
experiment = Experiment(ws, experiment_name)
output = {}
output['Subscription ID'] = ws.subscription_id
output['Workspace'] = ws.name
output['SKU'] = ws.sku
output['Resource Group'] = ws.resource_group
output['Location'] = ws.location
output['Run History Name'] = experiment_name
pd.set_option('display.max_colwidth', -1)
outputDf = pd.DataFrame(data = output, index = [''])
outputDf.T
from azureml.core.compute import ComputeTarget, AmlCompute
from azureml.core.compute_target import ComputeTargetException
# Choose a name for your cluster.
amlcompute_cluster_name = "bike-cluster"
# Verify that cluster does not exist already
try:
compute_target = ComputeTarget(workspace=ws, name=amlcompute_cluster_name)
print('Found existing cluster, use it.')
except ComputeTargetException:
compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_D2_V2',
max_nodes=4)
compute_target = ComputeTarget.create(ws, amlcompute_cluster_name, compute_config)
compute_target.wait_for_completion(show_output=True)
datastore = ws.get_default_datastore()
datastore.upload_files(files = ['./bike-no.csv'], target_path = 'dataset/', overwrite = True,show_progress = True)
target_column_name = 'cnt'
time_column_name = 'date'
dataset = Dataset.Tabular.from_delimited_files(path = [(datastore, 'dataset/bike-no.csv')]).with_timestamp_columns(fine_grain_timestamp=time_column_name)
dataset.take(5).to_pandas_dataframe().reset_index(drop=True)
# select data that occurs before a specified date
train = dataset.time_before(datetime(2012, 8, 31), include_boundary=True)
train.to_pandas_dataframe().tail(5).reset_index(drop=True)
test = dataset.time_after(datetime(2012, 9, 1), include_boundary=True)
test.to_pandas_dataframe().head(5).reset_index(drop=True)
forecast_horizon = 14
from azureml.automl.core.forecasting_parameters import ForecastingParameters
forecasting_parameters = ForecastingParameters(
time_column_name=time_column_name,
forecast_horizon=forecast_horizon,
country_or_region_for_holidays='US', # set country_or_region will trigger holiday featurizer
target_lags='auto', # use heuristic based lag setting
drop_column_names=['casual', 'registered'] # these columns are a breakdown of the total and therefore a leak
)
automl_config = AutoMLConfig(task='forecasting',
primary_metric='normalized_root_mean_squared_error',
blocked_models = ['ExtremeRandomTrees'],
experiment_timeout_hours=0.3,
training_data=train,
label_column_name=target_column_name,
compute_target=compute_target,
enable_early_stopping=True,
n_cross_validations=3,
max_concurrent_iterations=4,
max_cores_per_iteration=-1,
verbosity=logging.INFO,
forecasting_parameters=forecasting_parameters)
remote_run = experiment.submit(automl_config, show_output=False)
remote_run
remote_run.wait_for_completion()
best_run, fitted_model = remote_run.get_output()
fitted_model.steps
fitted_model.named_steps['timeseriestransformer'].get_engineered_feature_names()
# Get the featurization summary as a list of JSON
featurization_summary = fitted_model.named_steps['timeseriestransformer'].get_featurization_summary()
# View the featurization summary as a pandas dataframe
pd.DataFrame.from_records(featurization_summary)
test_experiment = Experiment(ws, experiment_name + "_test")
import os
import shutil
script_folder = os.path.join(os.getcwd(), 'forecast')
os.makedirs(script_folder, exist_ok=True)
shutil.copy('forecasting_script.py', script_folder)
from run_forecast import run_rolling_forecast
remote_run = run_rolling_forecast(test_experiment, compute_target, best_run, test, target_column_name)
remote_run
remote_run.wait_for_completion(show_output=False)
remote_run.download_file('outputs/predictions.csv', 'predictions.csv')
df_all = pd.read_csv('predictions.csv')
from azureml.automl.core.shared import constants
from azureml.automl.runtime.shared.score import scoring
from sklearn.metrics import mean_absolute_error, mean_squared_error
from matplotlib import pyplot as plt
# use automl metrics module
scores = scoring.score_regression(
y_test=df_all[target_column_name],
y_pred=df_all['predicted'],
metrics=list(constants.Metric.SCALAR_REGRESSION_SET))
print("[Test data scores]\n")
for key, value in scores.items():
print('{}: {:.3f}'.format(key, value))
# Plot outputs
%matplotlib inline
test_pred = plt.scatter(df_all[target_column_name], df_all['predicted'], color='b')
test_test = plt.scatter(df_all[target_column_name], df_all[target_column_name], color='g')
plt.legend((test_pred, test_test), ('prediction', 'truth'), loc='upper left', fontsize=8)
plt.show()
from metrics_helper import MAPE, APE
df_all.groupby('horizon_origin').apply(
lambda df: pd.Series({'MAPE': MAPE(df[target_column_name], df['predicted']),
'RMSE': np.sqrt(mean_squared_error(df[target_column_name], df['predicted'])),
'MAE': mean_absolute_error(df[target_column_name], df['predicted'])}))
df_all_APE = df_all.assign(APE=APE(df_all[target_column_name], df_all['predicted']))
APEs = [df_all_APE[df_all['horizon_origin'] == h].APE.values for h in range(1, forecast_horizon + 1)]
%matplotlib inline
plt.boxplot(APEs)
plt.yscale('log')
plt.xlabel('horizon')
plt.ylabel('APE (%)')
plt.title('Absolute Percentage Errors by Forecast Horizon')
plt.show()
| 0.657978 | 0.956877 |
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from ESRNN import ESRNN
plt.style.use('ggplot')
pd.options.display.max_rows = 999
np.set_printoptions(threshold=np.inf)
```
## Useful funtions
```
# Plot
def plot_prediction(y, y_hat):
n_y = len(y)
n_yhat = len(y_hat)
ds_y = np.array(range(n_y))
ds_yhat = np.array(range(n_y, n_y+n_yhat))
plt.plot(ds_y, y, label = 'y')
plt.plot(ds_yhat, y_hat, label='y_hat')
plt.legend(loc='upper left')
plt.show()
def ffill_missing_dates_particular_serie(serie, min_date, max_date, freq):
date_range = pd.date_range(start=min_date, end=max_date, freq=freq)
unique_id = serie['unique_id'].unique()
df_balanced = pd.DataFrame({'ds':date_range, 'key':[1]*len(date_range), 'unique_id': unique_id[0]})
# Check balance
check_balance = df_balanced.groupby(['unique_id']).size().reset_index(name='count')
assert len(set(check_balance['count'].values)) <= 1
df_balanced = df_balanced.merge(serie, how="left", on=['unique_id', 'ds'])
df_balanced['y'] = df_balanced['y'].fillna(method='ffill')
df_balanced['x'] = df_balanced['x'].fillna(method='ffill')
return df_balanced
def ffill_missing_dates_per_serie(df, freq, fixed_max_date=None):
"""Receives a DataFrame with a date column and forward fills the missing gaps in dates, not filling dates before
the first appearance of a unique key
Parameters
----------
df: DataFrame
Input DataFrame
key: str or list
Name(s) of the column(s) which make a unique time series
date_col: str
Name of the column that contains the time column
freq: str
Pandas time frequency standard strings, like "W-THU" or "D" or "M"
numeric_to_fill: str or list
Name(s) of the columns with numeric values to fill "fill_value" with
"""
if fixed_max_date is None:
df_max_min_dates = df[['unique_id', 'ds']].groupby('unique_id').agg(['min', 'max']).reset_index()
else:
df_max_min_dates = df[['unique_id', 'ds']].groupby('unique_id').agg(['min']).reset_index()
df_max_min_dates['max'] = fixed_max_date
df_max_min_dates.columns = df_max_min_dates.columns.droplevel()
df_max_min_dates.columns = ['unique_id', 'min_date', 'max_date']
df_list = []
for index, row in df_max_min_dates.iterrows():
df_id = df[df['unique_id'] == row['unique_id']]
df_id = ffill_missing_dates_particular_serie(df_id, row['min_date'], row['max_date'], freq)
df_list.append(df_id)
df_dates = pd.concat(df_list).reset_index(drop=True).drop('key', axis=1)[['unique_id', 'ds', 'y','x']]
return df_dates
```
## Data cleaning
### Train
```
# Original stock data
data = pd.read_csv('data/train.csv')
data['Date'] = data['Year'].astype(str)+'-'+data['Date'].astype(str)
data['Date'] = pd.to_datetime(data['Date'])
data = data[['Company','Year','Date','Close']]
data.head()
# Clean data (model assumes this name columns)
data['unique_id'] = data['Company']+"_"+data['Year'].astype(str)
data = data.rename(columns={'Date':'ds', 'Close':'y'})
data['x'] = data['Year'].astype(str)
data.head()
#Series must be complete in the frequency
data = ffill_missing_dates_per_serie(data,'D')
X_train = data[['unique_id','ds','x']]
y_train = data[['unique_id','ds','y']]
```
### Test
```
data_test = pd.read_csv('data/test.csv')
data_test['Date'] = data_test['Year'].astype(str)+'-'+data_test['Date'].astype(str)
data_test['Date'] = pd.to_datetime(data_test['Date'])
data_test = data_test[['Company','Year','Date','Close']]
data_test.head()
# Clean data (model assumes this name columns)
data_test['unique_id'] = data_test['Company']+"_"+data_test['Year'].astype(str)
data_test = data_test.rename(columns={'Date':'ds', 'Close':'y'})
data_test['x'] = data_test['Year'].astype(str)
X_test = data_test[['unique_id','ds']]
X_test.head()
```
## Model train example to view initial vs trained predictions
Note: to reduce wigglines of prediction train with more epochs and/or increase level_variability_penalty hyperpar
```
# Model with no train to see initial prediction
esrnn = ESRNN(max_epochs=0, batch_size=8, learning_rate=1e-3,
seasonality=30, input_size=30, output_size=60)
esrnn.fit(X_train, y_train, random_seed=1)
y_hat = esrnn.predict(y_train[['unique_id']])
uniques = X_train['unique_id'].unique()
plot_id = 0
y_test_plot = y_train.loc[y_train['unique_id']==uniques[plot_id]]
y_hat_test_plot = y_hat.loc[y_hat['unique_id']==uniques[plot_id]]
plot_prediction(y_test_plot['y'], y_hat_test_plot['y_hat'])
# Train model
esrnn = ESRNN(max_epochs=50, batch_size=8, learning_rate=1e-3,
seasonality=30, input_size=30, output_size=60)
esrnn.fit(X_train, y_train, random_seed=1)
y_hat = esrnn.predict(y_train[['unique_id']])
plot_id = 0
y_test_plot = y_train.loc[y_train['unique_id']==uniques[plot_id]]
y_hat_test_plot = y_hat.loc[y_hat['unique_id']==uniques[plot_id]]
plot_prediction(y_test_plot['y'], y_hat_test_plot['y_hat'])
```
## Predictions in stock test data
```
y_hat = esrnn.predict(X_test)
y_hat
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from ESRNN import ESRNN
plt.style.use('ggplot')
pd.options.display.max_rows = 999
np.set_printoptions(threshold=np.inf)
# Plot
def plot_prediction(y, y_hat):
n_y = len(y)
n_yhat = len(y_hat)
ds_y = np.array(range(n_y))
ds_yhat = np.array(range(n_y, n_y+n_yhat))
plt.plot(ds_y, y, label = 'y')
plt.plot(ds_yhat, y_hat, label='y_hat')
plt.legend(loc='upper left')
plt.show()
def ffill_missing_dates_particular_serie(serie, min_date, max_date, freq):
date_range = pd.date_range(start=min_date, end=max_date, freq=freq)
unique_id = serie['unique_id'].unique()
df_balanced = pd.DataFrame({'ds':date_range, 'key':[1]*len(date_range), 'unique_id': unique_id[0]})
# Check balance
check_balance = df_balanced.groupby(['unique_id']).size().reset_index(name='count')
assert len(set(check_balance['count'].values)) <= 1
df_balanced = df_balanced.merge(serie, how="left", on=['unique_id', 'ds'])
df_balanced['y'] = df_balanced['y'].fillna(method='ffill')
df_balanced['x'] = df_balanced['x'].fillna(method='ffill')
return df_balanced
def ffill_missing_dates_per_serie(df, freq, fixed_max_date=None):
"""Receives a DataFrame with a date column and forward fills the missing gaps in dates, not filling dates before
the first appearance of a unique key
Parameters
----------
df: DataFrame
Input DataFrame
key: str or list
Name(s) of the column(s) which make a unique time series
date_col: str
Name of the column that contains the time column
freq: str
Pandas time frequency standard strings, like "W-THU" or "D" or "M"
numeric_to_fill: str or list
Name(s) of the columns with numeric values to fill "fill_value" with
"""
if fixed_max_date is None:
df_max_min_dates = df[['unique_id', 'ds']].groupby('unique_id').agg(['min', 'max']).reset_index()
else:
df_max_min_dates = df[['unique_id', 'ds']].groupby('unique_id').agg(['min']).reset_index()
df_max_min_dates['max'] = fixed_max_date
df_max_min_dates.columns = df_max_min_dates.columns.droplevel()
df_max_min_dates.columns = ['unique_id', 'min_date', 'max_date']
df_list = []
for index, row in df_max_min_dates.iterrows():
df_id = df[df['unique_id'] == row['unique_id']]
df_id = ffill_missing_dates_particular_serie(df_id, row['min_date'], row['max_date'], freq)
df_list.append(df_id)
df_dates = pd.concat(df_list).reset_index(drop=True).drop('key', axis=1)[['unique_id', 'ds', 'y','x']]
return df_dates
# Original stock data
data = pd.read_csv('data/train.csv')
data['Date'] = data['Year'].astype(str)+'-'+data['Date'].astype(str)
data['Date'] = pd.to_datetime(data['Date'])
data = data[['Company','Year','Date','Close']]
data.head()
# Clean data (model assumes this name columns)
data['unique_id'] = data['Company']+"_"+data['Year'].astype(str)
data = data.rename(columns={'Date':'ds', 'Close':'y'})
data['x'] = data['Year'].astype(str)
data.head()
#Series must be complete in the frequency
data = ffill_missing_dates_per_serie(data,'D')
X_train = data[['unique_id','ds','x']]
y_train = data[['unique_id','ds','y']]
data_test = pd.read_csv('data/test.csv')
data_test['Date'] = data_test['Year'].astype(str)+'-'+data_test['Date'].astype(str)
data_test['Date'] = pd.to_datetime(data_test['Date'])
data_test = data_test[['Company','Year','Date','Close']]
data_test.head()
# Clean data (model assumes this name columns)
data_test['unique_id'] = data_test['Company']+"_"+data_test['Year'].astype(str)
data_test = data_test.rename(columns={'Date':'ds', 'Close':'y'})
data_test['x'] = data_test['Year'].astype(str)
X_test = data_test[['unique_id','ds']]
X_test.head()
# Model with no train to see initial prediction
esrnn = ESRNN(max_epochs=0, batch_size=8, learning_rate=1e-3,
seasonality=30, input_size=30, output_size=60)
esrnn.fit(X_train, y_train, random_seed=1)
y_hat = esrnn.predict(y_train[['unique_id']])
uniques = X_train['unique_id'].unique()
plot_id = 0
y_test_plot = y_train.loc[y_train['unique_id']==uniques[plot_id]]
y_hat_test_plot = y_hat.loc[y_hat['unique_id']==uniques[plot_id]]
plot_prediction(y_test_plot['y'], y_hat_test_plot['y_hat'])
# Train model
esrnn = ESRNN(max_epochs=50, batch_size=8, learning_rate=1e-3,
seasonality=30, input_size=30, output_size=60)
esrnn.fit(X_train, y_train, random_seed=1)
y_hat = esrnn.predict(y_train[['unique_id']])
plot_id = 0
y_test_plot = y_train.loc[y_train['unique_id']==uniques[plot_id]]
y_hat_test_plot = y_hat.loc[y_hat['unique_id']==uniques[plot_id]]
plot_prediction(y_test_plot['y'], y_hat_test_plot['y_hat'])
y_hat = esrnn.predict(X_test)
y_hat
| 0.635336 | 0.746093 |
```
''' Data collection for characterizing Quick Current Sensor
Use keithley 177/179 with USB adapter to collect data.
Use DC load to step current
'''
import os
import serial
import time
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.linear_model import HuberRegressor
from sklearn.preprocessing import StandardScaler
current_dmm_port = '/dev/ttyUSB0'
voltage_dmm_port = '/dev/ttyUSB1'
current_dmm_range = 1E-4 # how many decimal places to shift
voltage_dmm_range = 1E-3
serBaud = 115200
serTimeout = 2
# Open both ports
currentDmm = serial.Serial(port=current_dmm_port, baudrate=serBaud, timeout=serTimeout)
voltageDmm = serial.Serial(port=voltage_dmm_port, baudrate=serBaud, timeout=serTimeout)
# Voltage to Current Transfer Funciton for 177 with 0.1ohm shunt resistor
Vx = np.array([0.0103, 0.0510, 0.1019, 0.1528, 0.2037, 0.2547, 0.3057, 0.3567]) # volts
Cy = np.array([0.101, 0.501, 1.001, 1.502, 2.002, 2.503, 3.004, 3.505]) # amps
z = np.polyfit(Vx, Cy, 1)
VtoI = np.poly1d(z)
sample_duration = 110 #seconds
delay_between_samples = 1 #seconds
print_as_sampling = False
nSamples = int(sample_duration / delay_between_samples)
print("Colleciton Duration: {} seconds".format(sample_duration))
print("Number Of Samples: {}\n".format(nSamples))
print("Collecting Data...")
# Open ports if they are closed
if (currentDmm.is_open == False):
currentDmm.open()
if (voltageDmm.is_open == False):
voltageDmm.open()
time.sleep(1)
current_list = []
voltage_list = []
for n in range(0, nSamples):
currentDmm.reset_input_buffer()
cval = currentDmm.readline().decode('ascii')
try:
cint = int(cval.lstrip('\x00').rstrip('\r\n'))
except:
cint = 0
current_float = VtoI(cint * current_dmm_range)
voltageDmm.reset_input_buffer()
vval = voltageDmm.readline().decode('ascii')
try:
vint = int(vval.lstrip('\x00').rstrip('\r\n'))
except:
vint = 0
voltage_float = vint * voltage_dmm_range
if (print_as_sampling):
print("Current: {:.3f}, Voltage: {:.3f}".format(current_float, voltage_float))
current_list.append(current_float)
voltage_list.append(voltage_float)
time.sleep(delay_between_samples)
currentDmm.close()
voltageDmm.close()
# Get a transfer funciton
# https://stackoverflow.com/questions/61143998/numpy-best-fit-line-with-outliers
# make np array for data wrangling
x = np.array(current_list)
y = np.array(voltage_list)
# standardize
x_scaler, y_scaler = StandardScaler(), StandardScaler()
x_train = x_scaler.fit_transform(x[..., None])
y_train = y_scaler.fit_transform(y[..., None])
# fit model
model = HuberRegressor(epsilon=1)
model.fit(x_train, y_train.ravel())
# do some predictions
test_x = np.array([0, 12])
predictions = y_scaler.inverse_transform(model.predict(x_scaler.transform(test_x[..., None])))
# use polyfit to get transfer funciton
z = np.polyfit(test_x, predictions, 1)
print('vout = {:.3f} * current + {:.3f}'.format(z[0], z[1]))
print('{:.1f}V/A'.format(z[0]))
# plot
fig, ax = plt.subplots(figsize=(16,9), dpi=80)
ax.plot(x, y, '*')
ax.plot(test_x, predictions, '--')
ax.set_xlabel('Current [A]')
ax.set_ylabel('Output Voltage [V]')
plt.show()
```
|
github_jupyter
|
''' Data collection for characterizing Quick Current Sensor
Use keithley 177/179 with USB adapter to collect data.
Use DC load to step current
'''
import os
import serial
import time
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.linear_model import HuberRegressor
from sklearn.preprocessing import StandardScaler
current_dmm_port = '/dev/ttyUSB0'
voltage_dmm_port = '/dev/ttyUSB1'
current_dmm_range = 1E-4 # how many decimal places to shift
voltage_dmm_range = 1E-3
serBaud = 115200
serTimeout = 2
# Open both ports
currentDmm = serial.Serial(port=current_dmm_port, baudrate=serBaud, timeout=serTimeout)
voltageDmm = serial.Serial(port=voltage_dmm_port, baudrate=serBaud, timeout=serTimeout)
# Voltage to Current Transfer Funciton for 177 with 0.1ohm shunt resistor
Vx = np.array([0.0103, 0.0510, 0.1019, 0.1528, 0.2037, 0.2547, 0.3057, 0.3567]) # volts
Cy = np.array([0.101, 0.501, 1.001, 1.502, 2.002, 2.503, 3.004, 3.505]) # amps
z = np.polyfit(Vx, Cy, 1)
VtoI = np.poly1d(z)
sample_duration = 110 #seconds
delay_between_samples = 1 #seconds
print_as_sampling = False
nSamples = int(sample_duration / delay_between_samples)
print("Colleciton Duration: {} seconds".format(sample_duration))
print("Number Of Samples: {}\n".format(nSamples))
print("Collecting Data...")
# Open ports if they are closed
if (currentDmm.is_open == False):
currentDmm.open()
if (voltageDmm.is_open == False):
voltageDmm.open()
time.sleep(1)
current_list = []
voltage_list = []
for n in range(0, nSamples):
currentDmm.reset_input_buffer()
cval = currentDmm.readline().decode('ascii')
try:
cint = int(cval.lstrip('\x00').rstrip('\r\n'))
except:
cint = 0
current_float = VtoI(cint * current_dmm_range)
voltageDmm.reset_input_buffer()
vval = voltageDmm.readline().decode('ascii')
try:
vint = int(vval.lstrip('\x00').rstrip('\r\n'))
except:
vint = 0
voltage_float = vint * voltage_dmm_range
if (print_as_sampling):
print("Current: {:.3f}, Voltage: {:.3f}".format(current_float, voltage_float))
current_list.append(current_float)
voltage_list.append(voltage_float)
time.sleep(delay_between_samples)
currentDmm.close()
voltageDmm.close()
# Get a transfer funciton
# https://stackoverflow.com/questions/61143998/numpy-best-fit-line-with-outliers
# make np array for data wrangling
x = np.array(current_list)
y = np.array(voltage_list)
# standardize
x_scaler, y_scaler = StandardScaler(), StandardScaler()
x_train = x_scaler.fit_transform(x[..., None])
y_train = y_scaler.fit_transform(y[..., None])
# fit model
model = HuberRegressor(epsilon=1)
model.fit(x_train, y_train.ravel())
# do some predictions
test_x = np.array([0, 12])
predictions = y_scaler.inverse_transform(model.predict(x_scaler.transform(test_x[..., None])))
# use polyfit to get transfer funciton
z = np.polyfit(test_x, predictions, 1)
print('vout = {:.3f} * current + {:.3f}'.format(z[0], z[1]))
print('{:.1f}V/A'.format(z[0]))
# plot
fig, ax = plt.subplots(figsize=(16,9), dpi=80)
ax.plot(x, y, '*')
ax.plot(test_x, predictions, '--')
ax.set_xlabel('Current [A]')
ax.set_ylabel('Output Voltage [V]')
plt.show()
| 0.659953 | 0.495972 |
## Probability, Sets and Visualization
First of all i will introduce the concept of set, is important the notion of sets in probability, A set can define a group of possible events that can occur.
Defined the event as uppercase $ X $ and the outcomes of the event as $ x $ so mathematically we define $ x \in X $, with this idea we can define set $ A = \{ 1, 2, 3, 5 \}$ with 4 outcomes,so if we choose a number from 1 to 5 they will have a chance of $4/5$ to be part of the set $ A $, as $ P(A) = 4/5$ here $ P(A) $ defines the event of getting a element from 1 to 5.
## Interception of events
Given the set $ A = \{ 1, 2, 3, 5 \}$ and $ B = \{ 2, 3, 4 \} $ we can define the interception as $ A \cap B = \{2, 3\} $ here all elements of set $ A $ also belongs to $ B $.
The probability of element of interception given 2 events is defined by $ P( A \cap B ) = P(A) . P(B) $,
basically im saying that event A and event B occurred.
knowing that $P(B) = 3/5$ is second event and appling this formula we can discover $ P( A \cap B ) = 3/5 . 4/5 = 12/25 = 0.48 $.
That is how we can implement and visualize in python:
```
# First of all install "pip install matplotlib-venn" in your jupyter-notebook environment
import matplotlib.pyplot as plt
from matplotlib_venn import venn2, venn3
n_e = 5 # Number of possible elements.
A = set(['1' , '2', '3', '5'])
B = set(['2', '3' , '4'])
p_A = len(A) / n_e # Probability of getting a element in set A.
p_B = len(B) / n_e # Probability of getting a element in set B.
Interception_A_B = A & B
p_Interception_A_B = p_A * p_B
v = venn2([A, B])
v.get_label_by_id('10').set_text(p_A)
v.get_label_by_id('01').set_text(p_B)
v.get_label_by_id('11').set_text(p_Interception_A_B)
plt.show()
print("Interception = {" + ' , '.join(Interception_A_B) + "}")
print("Probability of getting two events being a interception = %.2f" % p_Interception_A_B)
```
## Union of events
Given the set $ A = \{ 1, 2, 3, 5 \}$ and $ B = \{ 2, 3, 4 \} $ we can define the union as $ A \cup B = \{1, 2, 3, 4, 5\} $ here the union contains the outcomes of set $ A $ and $ B $.
The probability of element of union is defined by $ P( A \cup B ) = P(A) + P(B) - P( A \cap B ) $.
That is how we can implement and visualize in python:
```
UNION_A_B = A | B
p_UNION_A_B = p_A + p_B - p_Interception_A_B
v = venn3([A, B, UNION_A_B] , ('A', 'B', 'UNION'))
v.get_label_by_id('111').set_text(format(p_UNION_A_B, '.2f'))
v.get_label_by_id('101').set_text(p_A)
v.get_label_by_id('011').set_text(p_B)
v.get_label_by_id('001').set_text(' ')
plt.show()
print("Union = {" + ' , '.join(UNION_A_B) + "}")
print("Probability of two events be a union = %.2f" % p_UNION_A_B)
```
So the chance of getting a element in union given 2 events is $92\%$, why? In this case i saying that event A or event B occurred in order ${\{A, B\}}$ or ${\{B, A\}}$ and never ${\{A, B'\}}$ and ${\{B, A'\}}$ if i get $4$ two times they will not be part of the union of the probability!
## Complements
Given $ A = \{ 1, 2, 3 , 5 \} $ the complement $ A' $ is every element that $ x \notin A $ and $ x \in A'$ like $ x = 4$ considering that value 4 exists in universal set $ U $ (which set contains all possible values)
Here $ P(A') = 1 - P(A) $
```
print("Has '4' not in A ? " + str('4' not in A))
print("Probability of not occurring event A: %.2f" % (1 - p_A))
```
## Dependence of events
The notation $ P( A | B)$ define the dependence of events, so they are not independent anymore occurring only in order ${\{A, B\}}$ and never in ${\{B, A\}}$
The $ P( A | B) $ is defined as $ P( A | B) = \frac{P( A \cap B)}{P(B)} $
```
p_a_depends_b = p_Interception_A_B / p_B
print(p_a_depends_b) # In this case they are statistically independent resulting in P(A)
```
# Bayes' theorem
The bayes theorem is defined by the correlation $ P( A | B) = \frac{P( B | A) . P(A)}{P(B)} $
So we conclude that.
```
p_b_depends_a = p_Interception_A_B / p_A
print(p_a_depends_b == ((p_b_depends_a * p_A) / p_B) )
```
You can see more about in next jbook of Bayes theorem and Bayesian statistics and relation between posterior, likehood, prior, evidence.
|
github_jupyter
|
# First of all install "pip install matplotlib-venn" in your jupyter-notebook environment
import matplotlib.pyplot as plt
from matplotlib_venn import venn2, venn3
n_e = 5 # Number of possible elements.
A = set(['1' , '2', '3', '5'])
B = set(['2', '3' , '4'])
p_A = len(A) / n_e # Probability of getting a element in set A.
p_B = len(B) / n_e # Probability of getting a element in set B.
Interception_A_B = A & B
p_Interception_A_B = p_A * p_B
v = venn2([A, B])
v.get_label_by_id('10').set_text(p_A)
v.get_label_by_id('01').set_text(p_B)
v.get_label_by_id('11').set_text(p_Interception_A_B)
plt.show()
print("Interception = {" + ' , '.join(Interception_A_B) + "}")
print("Probability of getting two events being a interception = %.2f" % p_Interception_A_B)
UNION_A_B = A | B
p_UNION_A_B = p_A + p_B - p_Interception_A_B
v = venn3([A, B, UNION_A_B] , ('A', 'B', 'UNION'))
v.get_label_by_id('111').set_text(format(p_UNION_A_B, '.2f'))
v.get_label_by_id('101').set_text(p_A)
v.get_label_by_id('011').set_text(p_B)
v.get_label_by_id('001').set_text(' ')
plt.show()
print("Union = {" + ' , '.join(UNION_A_B) + "}")
print("Probability of two events be a union = %.2f" % p_UNION_A_B)
print("Has '4' not in A ? " + str('4' not in A))
print("Probability of not occurring event A: %.2f" % (1 - p_A))
p_a_depends_b = p_Interception_A_B / p_B
print(p_a_depends_b) # In this case they are statistically independent resulting in P(A)
p_b_depends_a = p_Interception_A_B / p_A
print(p_a_depends_b == ((p_b_depends_a * p_A) / p_B) )
| 0.356111 | 0.973139 |
# Introduction to Programming for Everyone
## Python 3
### Who am I?
- Name: __Hemma__ (名) __Philamore__ (姓)
- Country: __England__
- Lab: __Matsuno mechatronics lab__, Kyoto University, Katsura Campus
- Research: __Bio-inspired, energy-autonomous robots__
- Contact: __philamore.hemma.5s@kyoto-u.ac.jp__
<img src="img/row_bot.jpg" alt="Drawing" style="width: 300px;"/>
### Teaching Assistant
Ask him anything.
<br>He speaks excellent Japanese!
### Who are you?
Please write your personal information on the sheet:
- Name
- Email address (that you will use to submit homework)
- Major
- Year of study
Today is an introductory seminar.
- Course content
- How to access the course material.
- Software.
- Assessment.
- Homework and class necessary class preparation.
- Software Installation
## Why study programming?
- Increased use of computing in everyday life.
- A tool you can use for the other subject you study.
- A growing sector of the jobs market.
- Coding in jobs, not traditionally related to computing.
- It's fun!
## Why study Python?
- Free and open source
- Easy to learn
- "High level"
- Community
- Increasingly used in industry
## Why study Python?
- Fast development/progression time.
<img src="img/optimizing-what.png" width="600">
## Course Goal
To develop:
- A good standalone programming toolkit.
- Skills to improve the quality of your work in other subjects.
- A fundamental base from which to start developing further as a programmer.
## Course Entry Level
Beginner, no prior programming knowlegde.
## Course Theme
### Computer Game Development.
- mathematics
- data handling
- physical interfaces (keyboard, mouse...)
- image manipulation and simulation
- computer architecture
- __.... something for everyone.__
## Computers
Please use your personal computers for this course.
<br>You will need to install two things.
- Anaconda
- git
<br>
Lab PCs may also be used.
- __Course content__
- How to access the course material
- Software
- Assessment
- Homework and class necessary class preparation
- Software Installation
## Course Content
- Basic Operators and Control Flow
- Data types, data structures and Imported Packages
- Importing, Drawing and Animating Images
- Physical User Interfaces
- Functions
- Text, Sound and Multi-Player Games
- Outputs and Data Analysis
- *Coursework Assigment : Build your own game*.
- Introduction to Object Oriented Programming
- Introduction to Network Programming
- Error Handling and Exceptions
- Testing
- Introduction to Version Control
## Version Control and Testing
- Code management skills.
- Essential techniques used by professional programmers.
- Storing, documenting, and sharing your code with others.
- Improve the quality of your work.
<table><tr><td>
<img src='img/ginea_pigs1.jpg' style="width: 500px;"> </td><td>
<img src='img/ginea_pigs2.jpg' style="width: 500px;"> </td><td>
<img src='img/ginea_pigs3.jpg' style="width: 500px;"> </td></tr>
</table>
- Course content
- __How to access the course material__
- Software
- Assessment
- Homework and class necessary class preparation
- Software Installation
## How To Access the Course Material
The course notes can be downloaded from:
<br>https://github.com/hphilamore/ILAS_Python_for_everyone
## How To Access the Course Material
You will use the command line to download the course notes.
This will allow yout to:
1. Edit the course notes in class
1. Download updated notes, solutions to exercises, corrections *without* overwriting the notes you already have.
## Introduction to the Command Line.
Let's open a terminal...
On a Windows PC:
- press "win key" + "R"
- type: __cmd__
- press enter
On a Mac:
- open Applications >> Utilities >> Terminal
A terminal will launch.
Using the terminal, you will navigate to where you want to store the course notes.
Type:
>`dir`
..and press enter.
You will see all the folders in your personal user area.
We can move down the filesystem of the computer by typing:
>`cd`
followed by the name of the folder we want to move to.
>`cd Documents`
Type:
>`dir`
again to view the contents of your Documents folder.
The folder must be:
- on the same branch
- one step from our current location
<img src="img/directory_tree.gif" alt="Drawing" style="width: 400px;"/>
Type:
>`dir`
again to view the contents of the folder that you have moved into.
To move back up by one step, type:
>`cd ..`
Try this now.
<img src="img/directory_tree.gif" alt="Drawing" style="width: 400px;"/>
#### If using a university computer.
The *command prompt* will say something like:
C:¥Users¥Username:
The C tells us that we are on the C drive of the computer.
Switch to the M drive where the user (you!) can save files.
<img src="img/KUterminal.png" alt="Drawing" style="width: 500px;"/>
In the terminal type:
>`M:`
...and press enter.
You should see the command prompt change.
<img src="img/KUterminalMdrive.png" alt="Drawing" style="width: 700px;"/>
## How to Access the Course Material : <br> "Cloning" the Textbook Using Git
Go to the Github site we opened earlier.
We are going to download the course notes by *cloning*.
If you do not have git installed yet, do this step later.
Click the button "Clone or download" and copy the link by presssing Ctrl , C
<img src="img/clone-URL.png" alt="Drawing" style="width: 500px;"/>
In the terminal type `git clone`.
After the word `clone` __leave a space__ and then paste the URL that you just copied:
> `git clone` https://github.com/hphilamore/ILAS_Python_for_everyone
`Clone` copies all the files from the repository at the URL you have entered.
In the terminal type:
> `dir`
A folder called "ILAS_python_for_everyone" should have appeared.
Go into the folder and view the content by typing:
>`cd ILAS_pyhon`
><br>`dir`
- Course content
- How to access the course material
- __Software__
- Assessment
- Homework and class necessary class preparation
- Software Installation
## Software : Python Environments
There are different *environments* in which we can write and run python code.
Each environment has different advantages and is suitable for different work types.
## Software : Python Environments - Jupyter Notebook
You can view the course notes via a *web browser* or using *Jupyter notebook*.
To launch Jupyter notebook.
In the terminal type:
> `jupyter notebook`
and press enter.
__OR__
Choose Jupyter notebook from your computer's start menu e.g.:
> Start >> Programs (すべてのプログラム) >> Programming >> Anaconda3 >> Jupyter notebook
<img src="img/jupyter_notebook.png" alt="Drawing" style="width: 300px;"/>
You can type code in the boxes.
You can run the code by pressing:
>"Shift" + "Enter"
This is an easy way to try out code next to printed instructions and images.
Cells are executed in the order that the user runs them.
By convention, Jupyter notebooks are __expected__ to be run from top to bottom.
If you skip a cell or run the cells in a different order you may get unexpected behaviour.
For example, in the next cell enter:
```python
x = 10
```
then re-run the cell above containing:
```python
y = x - 2
print(y)
```
and you should see the value of y change.
The original value of `x` has been replaced by the new definition `x = 10`.
<img src="img/jupyter_change_value.png" alt="Drawing" style="width: 500px;"/>
Now run the cell containing:
```python
x = 2 + 3
```
then the cell containing:
```python
y = x - 2
print(y)
```
and you will see the value of `y` change back to it's original value.
<img src="img/jupyter_value_change2.png" alt="Drawing" style="width: 500px;"/>
To run all the cells sequentially from top to bottom click:
Cell >> Run All
This menu also contains other options for running cells.
## Software : Python Environments - Python interpreter
We can start the interpreter by typing
>``python3``
in the terminal.
The command prompt changes to:
>``>>>``
This is useful for doing small calculations. But the standard python interpreter is not very convenient for longer pieces of code.
<!-- <img src="files/images/python-screenshot.jpg" width="600"> -->
<img src="img/python-screenshot.jpg" width="600">
## Software : Python Environments - Spyder
Spyder is an integrated development environment (IDE).
An IDE is a program used to write and edit code.
To launch spyder
In the terminal type:
> `spyder`
and press enter.
__OR__
Choose Spyder from your computer's start menu e.g.:
> Start >> Programs (すべてのプログラム) >> Programming >> Anaconda3 >> Spyder
### Writing and running code in the Spyder IDE
You can write code by tying in the box on the left (editor).
You can run the code you have typed by clicking the green triangle in the menu bar at the top of the window.
<img src="img/run_spyder.png" alt="Drawing" style="width: 300px;"/>
The output should appear in the box on the bottom right (console)
The box in the top, right can be used to diplay the file system or stored variables (much ilke in MATLAB).
### Writing code in the Spyder IDE and and running code in the Spyder IDE
### Save your code
Click:
File >> Save as
Save your file with `.py` extension.
### Running your program without using Spyder.
Spyder and other IDEs provide a convenient way to write, run and save code.
However, once you have saved your program you can run it without the need for another program.
In the terminal, use `cd` to navigate to within the folder where you just saved your file.
Type:
> `python3`
...followed by a space, then the name of the program that you want to run.
For example, to run a file ``my-program.py``:
> `python my-program.py`
<img src="img/python-screenshot.jpg" width="600">
### Creating Python files without using Spyder.
In fact, you don't even need an IDE to create a Python file.
You can write Python code in a normal text editor (Notepad (Windows), Wordpad (Windows), TextEdit (Mac), Text Editor (Ubuntu Linux).
Provided you add the file extension .py when saving the file, you can run the file exactly the same way as we just did from the command line.
We will use:
- __Jupyter notebook__ for short coding practise exercises.
- __Spyder__ for longer, more complete programs.
We will go through how to explain:
- Git
- Jupyter notebook
- Spyder
at the end of today's class.
- Course content
- How to access the course material
- Software
- __Assessment__
- Homework and class necessary class preparation
- Software Installation
## Assessment
- Game development project
- 50% final grade
- in pairs
- week 8
<br>
- Exam
- 50% final grade
- individual
- open book
- week 15
- Course content
- How to access the course material
- Software
- Assessment
- __Homework and class necessary class preparation__
- Software Installation
### Class Preparation
Lesson structure (learning by doing):
- Doing examples together
- Trying exercises by yourself
I will be asking you to *independently* read the notes ahead of / during class to prepare and complete exercises.
### Homework ("Test-Yourself" exercises)
There are "Test-Yourself" exercises for you to complete at the end of the class notes for each class.
Work not completed in class should be completed for homework and submitted by email.
### Review Excericises
There are also Review Exercises at the end of the notes for each class.
Review exercises are not assessed but are good preparation for the exam.
I strongly advise you to complete them.
- Course content
- How to access the course material
- Software
- Assessment
- Homework and class necessary class preparation
- __Software Installation__
Before next class you *must* have the following software installed on the computer that you are going to use to take the course.
- Git
- Anaconda (Jupyter notebook and Spyder)
<br>
Before next class you *must* have downloaded (cloned) the class notes.
### Are there any questions?
## Software Installation
### Instructions if using personal computer
__Anaconda__ (which includes Jupyter notebook) can be downloaded from: <br>https://www.anaconda.com/download/
><br>Windows, Mac and Linux versions are available
><br>Python 3.6 version and Python 2.7 version are available.
><br>Choose Python 3.6 version
<br>
__Git__ can be installed using the instructions found here:
https://gist.github.com/derhuerst/1b15ff4652a867391f03
><br>Windows, Mac and Linux versions are available
><br>For Windows choose the appropriate file for your operating system.
><br>Git-2.16.3-32-bit.exe for 32 bit operating system.
><br>Git-2.16.3-64-bit.exe for 64 bit operating system.
### Instructions if using Kyoto University computer
__Anaconda__ is already installed.
You need install __Git__ in your local user area.
The following instructions tell you how to do this.
__IMPORTANT NOTE__
If you are going to use your personal computer to complete work in/outside of the seminars, you DO NOT need to complete this step.
Download the Git program from here:
https://github.com/git-for-windows/git/releases/tag/v2.14.1.windows.1
The version you need is:
PortableGit-2.14.1-32-bit.7z.exe
When prompted, choose to __run the file__ 実行(R).
<img src="img/GitHubInstallRun.png" alt="Drawing" style="width: 200px;"/>
When prompted, change the location to save the file to:
M:¥Documents¥PortableGit
<img src="img/GitLocation.png" alt="Drawing" style="width: 200px;"/>
Press OK
The download may take some time.
Once the download has completed...
To open the terminal:
- press "win key" + "R"
- type: __cmd__
- press enter
In the terminal type:
>`M:`
...and press enter, to switch to the M drive.
You should see the command prompt change.
<img src="img/KUterminalMdrive.png" alt="Drawing" style="width: 700px;"/>
To navigate to documents type:
>`cd Documents`
cd stands for "change directory".
You should now see a folder called PortableGit in the contents list of __Documents__ folder.
Type:
>cd PortableGit
to move into your PortableGit folder.
To check git has installed type:
>`git-bash.exe`
A new terminal window will open. In this window type:
>`git --version`
If Git has installed, the version of the program will be dipolayed. You should see something like this:
<img src="img/git-version.gif" alt="Drawing" style="width: 500px;"/>
Close the window.
The final thing we need to do is to tell the computer where to look for the Git program.
Move one step up from the Git folder. In the original terminal window, type:
> `cd ..`
Now enter the following in the terminal:
> PATH=M:¥Documents¥PortableGit¥bin;%PATH%
(you may need to have your keyboard set to JP to achieve this)
<img src="img/windows_change_lang.png" alt="Drawing" style="width: 400px;"/>
You can type this or __copy and paste__ it from the README section on the github page we looked at earlier.
<img src="img/readme_.png" alt="Drawing" style="width: 500px;"/>
__Whenever to use Git on a Kyoto University computer outside of the computer lab (Room North wing 21, Academic Center Bldg., Yoshida-South Campus), you must first opena terminal and type the line of code above to tell the computer where to look for the Git program.__
The program Git has its own terminal commands.
Each one starts with the word `git`
You can check git is working by typing:
>`git status`
You should see something like this:
<img src="img/git-version.gif" alt="Drawing" style="width: 500px;"/>
|
github_jupyter
|
x = 10
y = x - 2
print(y)
x = 2 + 3
y = x - 2
print(y)
| 0.170646 | 0.603056 |
# Vector-space models: retrofitting
```
__author__ = "Christopher Potts"
__version__ = "CS224u, Stanford, Spring 2021"
```
## Contents
1. [Overview](#Overview)
1. [Set-up](#Set-up)
1. [The retrofitting model](#The-retrofitting-model)
1. [Examples](#Examples)
1. [Only node 0 has outgoing edges](#Only-node-0-has-outgoing-edges)
1. [All nodes connected to all others](#All-nodes-connected-to-all-others)
1. [As before, but now 2 has no outgoing edges](#As-before,-but-now-2-has-no-outgoing-edges)
1. [All nodes connected to all others, but $\alpha = 0$](#All-nodes-connected-to-all-others,-but-$\alpha-=-0$)
1. [WordNet](#WordNet)
1. [Background on WordNet](#Background-on-WordNet)
1. [WordNet and VSMs](#WordNet-and-VSMs)
1. [Reproducing the WordNet synonym graph experiment](#Reproducing-the-WordNet-synonym-graph-experiment)
1. [Other retrofitting models and ideas](#Other-retrofitting-models-and-ideas)
## Overview
Thus far, all of the information in our word vectors has come solely from co-occurrences patterns in text. This information is often very easy to obtain – though one does need a __lot__ of text – and it is striking how rich the resulting representations can be.
Nonetheless, it seems clear that there is important information that we will miss this way – relationships that just aren't encoded at all in co-occurrences or that get distorted by such patterns.
For example, it is probably straightforward to learn representations that will support the inference that all puppies are dogs (_puppy_ entails _dog_), but it might be difficult to learn that _dog_ entails _mammal_ because of the unusual way that very broad taxonomic terms like _mammal_ are used in text.
The question then arises: how can we bring structured information – labels – into our representations? If we can do that, then we might get the best of both worlds: the ease of using co-occurrence data and the refinement that comes from using labeled data.
In this notebook, we look at one powerful method for doing this: the __retrofitting__ model of [Faruqui et al. 2016](http://www.aclweb.org/anthology/N15-1184). In this model, one learns (or just downloads) distributed representations for nodes in a knowledge graph and then updates those representations to bring connected nodes closer to each other.
This is an incredibly fertile idea; the final section of the notebook reviews some recent extensions, and new ones are likely appearing all the time.
## Set-up
```
from collections import defaultdict
from nltk.corpus import wordnet as wn
import numpy as np
import os
import pandas as pd
import retrofitting
from retrofitting import Retrofitter
import utils
data_home = 'data'
```
__Note__: To make full use of this notebook, you will need the NLTK data distribution – or, at the very least, its WordNet files. Anaconda comes with NLTK but not with its data distribution. To install that, open a Python interpreter and run
```import nltk; nltk.download()```
If you decide to download the data to a different directory than the default, then you'll have to set `NLTK_DATA` in your shell profile. (If that doesn't make sense to you, then we recommend choosing the default download directory!)
## The retrofitting model
For an __an existing VSM__ $\widehat{Q}$ of dimension $m \times n$, and a set of __edges__ $E$ (pairs of indices into rows in $\widehat{Q}$), the retrofitting objective is to obtain a new VSM $Q$ (also dimension $m \times n$) according to the following objective:
$$\sum_{i=1}^{m} \left[
\alpha_{i}\|q_{i} - \widehat{q}_{i}\|_{2}^{2}
+
\sum_{j : (i,j) \in E}\beta_{ij}\|q_{i} - q_{j}\|_{2}^{2}
\right]$$
The left term encodes a pressure to stay like the original vector. The right term encodes a pressure to be more like one's neighbors. In minimizing this objective, we should be able to strike a balance between old and new, VSM and graph.
Definitions:
1. $\|u - v\|_{2}^{2}$ gives the __squared euclidean distance__ from $u$ to $v$.
1. $\alpha$ and $\beta$ are weights we set by hand, controlling the relative strength of the two pressures. In the paper, they use $\alpha=1$ and $\beta = \frac{1}{\{j : (i, j) \in E\}}$.
## Examples
To get a feel for what's happening, it's helpful to visualize the changes that occur in small, easily understood VSMs and graphs. The function `retrofitting.plot_retro_path` helps with this.
```
Q_hat = pd.DataFrame(
[[0.0, 0.0],
[0.0, 0.5],
[0.5, 0.0]],
columns=['x', 'y'])
Q_hat
```
### Only node 0 has outgoing edges
```
edges_0 = {0: {1, 2}, 1: set(), 2: set()}
_ = retrofitting.plot_retro_path(Q_hat, edges_0)
```
### All nodes connected to all others
```
edges_all = {0: {1, 2}, 1: {0, 2}, 2: {0, 1}}
_ = retrofitting.plot_retro_path(Q_hat, edges_all)
```
### As before, but now 2 has no outgoing edges
```
edges_isolated = {0: {1, 2}, 1: {0, 2}, 2: set()}
_ = retrofitting.plot_retro_path(Q_hat, edges_isolated)
```
### All nodes connected to all others, but $\alpha = 0$
```
_ = retrofitting.plot_retro_path(
Q_hat, edges_all,
retrofitter=Retrofitter(alpha=lambda x: 0))
```
## WordNet
Faruqui et al. conduct experiments on three knowledge graphs: [WordNet](https://wordnet.princeton.edu), [FrameNet](https://framenet.icsi.berkeley.edu/fndrupal/), and the [Penn Paraphrase Database (PPDB)](http://paraphrase.org/). [The repository for their paper](https://github.com/mfaruqui/retrofitting) includes the graphs that they derived for their experiments.
Here, we'll reproduce just one of the two WordNet experiments they report, in which the graph is formed based on synonymy.
### Background on WordNet
WordNet is an incredible, hand-built lexical resource capturing a wealth of information about English words and their inter-relationships. ([Here is a collection of WordNets in other languages.](http://globalwordnet.org)) For a detailed overview using NLTK, see [this tutorial](http://compprag.christopherpotts.net/wordnet.html).
The core concepts:
* A __lemma__ is something like our usual notion of __word__. Lemmas are highly sense-disambiguated. For instance, there are six lemmas that are consistent with the string `crane`: the bird, the machine, the poets, ...
* A __synset__ is a collection of lemmas that are synonymous in the WordNet sense (which is WordNet-specific; words with intuitively different meanings might still be grouped together into synsets.).
WordNet is a graph of relations between lemmas and between synsets, capturing things like hypernymy, antonymy, and many others. For the most part, the relations are defined between nouns; the graph is sparser for other areas of the lexicon.
```
lems = wn.lemmas('crane', pos=None)
for lem in lems:
ss = lem.synset()
print("="*70)
print("Lemma name: {}".format(lem.name()))
print("Lemma Synset: {}".format(ss))
print("Synset definition: {}".format(ss.definition()))
```
### WordNet and VSMs
A central challenge of working with WordNet is that one doesn't usually encounter lemmas or synsets in the wild. One probably gets just strings, or maybe strings with part-of-speech tags. Mapping these objects to lemmas is incredibly difficult.
For our experiments with VSMs, we simply collapse together all the senses that a given string can have. This is expedient, of course. It might also be a good choice linguistically: senses are flexible and thus hard to individuate, and we might hope that our vectors can model multiple senses at the same time.
(That said, there is excellent work on creating sense-vectors; see [Reisinger and Mooney 2010](http://www.aclweb.org/anthology/N10-1013); [Huang et al 2012](http://www.aclweb.org/anthology/P12-1092).)
The following code uses the NLTK WordNet API to create the edge dictionary we need for using the `Retrofitter` class:
```
def get_wordnet_edges():
edges = defaultdict(set)
for ss in wn.all_synsets():
lem_names = {lem.name() for lem in ss.lemmas()}
for lem in lem_names:
edges[lem] |= lem_names
return edges
wn_edges = get_wordnet_edges()
```
### Reproducing the WordNet synonym graph experiment
For our VSM, let's use the 300d file included in this distribution from the GloVe team, as it is close to or identical to the one used in the paper:
http://nlp.stanford.edu/data/glove.6B.zip
If you download this archive, place it in `vsmdata`, and unpack it, then the following will load the file into a dictionary for you:
```
glove_dict = utils.glove2dict(
os.path.join(data_home, 'glove.6B', 'glove.6B.300d.txt'))
```
This is the initial embedding space $\widehat{Q}$:
```
X_glove = pd.DataFrame(glove_dict).T
X_glove.T.shape
```
Now we just need to replace all of the strings in `edges` with indices into `X_glove`:
```
def convert_edges_to_indices(edges, Q):
lookup = dict(zip(Q.index, range(Q.shape[0])))
index_edges = defaultdict(set)
for start, finish_nodes in edges.items():
s = lookup.get(start)
if s:
f = {lookup[n] for n in finish_nodes if n in lookup}
if f:
index_edges[s] = f
return index_edges
wn_index_edges = convert_edges_to_indices(wn_edges, X_glove)
```
And now we can retrofit:
```
wn_retro = Retrofitter(verbose=True)
X_retro = wn_retro.fit(X_glove, wn_index_edges)
```
You can now evaluate `X_retro` using the homework/bake-off notebook [hw_wordrelatedness.ipynb](hw_wordrelatedness.ipynb)!
```
# Optionally write `X_retro` to disk for use elsewhere:
#
X_retro.to_csv(
os.path.join(data_home, 'glove6B300d-retrofit-wn.csv.gz'),
compression='gzip')
```
## Other retrofitting models and ideas
* The retrofitting idea is very close to __graph embedding__, in which one learns distributed representations of nodes based on their position in the graph. See [Hamilton et al. 2017](https://arxiv.org/pdf/1709.05584.pdf) for an overview of these methods. There are numerous parallels with the material we've reviewed here.
* If you think of the input VSM as a "warm start" for graph embedding algorithms, then you're essentially retrofitting. This connection opens up a number of new opportunities to go beyond the similarity-based semantics that underlies Faruqui et al.'s model. See [Lengerich et al. 2017](https://arxiv.org/pdf/1708.00112.pdf), section 3.2, for more on these connections.
* [Mrkšić et al. 2016](https://www.aclweb.org/anthology/N16-1018) address the limitation of Faruqui et al's model that it assumes connected nodes in the graph are similar. In a graph with complex, varied edge semantics, this is likely to be false. They address the case of antonymy in particular.
* [Lengerich et al. 2017](https://arxiv.org/pdf/1708.00112.pdf) present a __functional retrofitting__ framework in which the edge meanings are explicitly modeled, and they evaluate instantiations of the framework with linear and neural edge penalty functions. (The Faruqui et al. model emerges as a specific instantiation of this framework.)
|
github_jupyter
|
__author__ = "Christopher Potts"
__version__ = "CS224u, Stanford, Spring 2021"
from collections import defaultdict
from nltk.corpus import wordnet as wn
import numpy as np
import os
import pandas as pd
import retrofitting
from retrofitting import Retrofitter
import utils
data_home = 'data'
If you decide to download the data to a different directory than the default, then you'll have to set `NLTK_DATA` in your shell profile. (If that doesn't make sense to you, then we recommend choosing the default download directory!)
## The retrofitting model
For an __an existing VSM__ $\widehat{Q}$ of dimension $m \times n$, and a set of __edges__ $E$ (pairs of indices into rows in $\widehat{Q}$), the retrofitting objective is to obtain a new VSM $Q$ (also dimension $m \times n$) according to the following objective:
$$\sum_{i=1}^{m} \left[
\alpha_{i}\|q_{i} - \widehat{q}_{i}\|_{2}^{2}
+
\sum_{j : (i,j) \in E}\beta_{ij}\|q_{i} - q_{j}\|_{2}^{2}
\right]$$
The left term encodes a pressure to stay like the original vector. The right term encodes a pressure to be more like one's neighbors. In minimizing this objective, we should be able to strike a balance between old and new, VSM and graph.
Definitions:
1. $\|u - v\|_{2}^{2}$ gives the __squared euclidean distance__ from $u$ to $v$.
1. $\alpha$ and $\beta$ are weights we set by hand, controlling the relative strength of the two pressures. In the paper, they use $\alpha=1$ and $\beta = \frac{1}{\{j : (i, j) \in E\}}$.
## Examples
To get a feel for what's happening, it's helpful to visualize the changes that occur in small, easily understood VSMs and graphs. The function `retrofitting.plot_retro_path` helps with this.
### Only node 0 has outgoing edges
### All nodes connected to all others
### As before, but now 2 has no outgoing edges
### All nodes connected to all others, but $\alpha = 0$
## WordNet
Faruqui et al. conduct experiments on three knowledge graphs: [WordNet](https://wordnet.princeton.edu), [FrameNet](https://framenet.icsi.berkeley.edu/fndrupal/), and the [Penn Paraphrase Database (PPDB)](http://paraphrase.org/). [The repository for their paper](https://github.com/mfaruqui/retrofitting) includes the graphs that they derived for their experiments.
Here, we'll reproduce just one of the two WordNet experiments they report, in which the graph is formed based on synonymy.
### Background on WordNet
WordNet is an incredible, hand-built lexical resource capturing a wealth of information about English words and their inter-relationships. ([Here is a collection of WordNets in other languages.](http://globalwordnet.org)) For a detailed overview using NLTK, see [this tutorial](http://compprag.christopherpotts.net/wordnet.html).
The core concepts:
* A __lemma__ is something like our usual notion of __word__. Lemmas are highly sense-disambiguated. For instance, there are six lemmas that are consistent with the string `crane`: the bird, the machine, the poets, ...
* A __synset__ is a collection of lemmas that are synonymous in the WordNet sense (which is WordNet-specific; words with intuitively different meanings might still be grouped together into synsets.).
WordNet is a graph of relations between lemmas and between synsets, capturing things like hypernymy, antonymy, and many others. For the most part, the relations are defined between nouns; the graph is sparser for other areas of the lexicon.
### WordNet and VSMs
A central challenge of working with WordNet is that one doesn't usually encounter lemmas or synsets in the wild. One probably gets just strings, or maybe strings with part-of-speech tags. Mapping these objects to lemmas is incredibly difficult.
For our experiments with VSMs, we simply collapse together all the senses that a given string can have. This is expedient, of course. It might also be a good choice linguistically: senses are flexible and thus hard to individuate, and we might hope that our vectors can model multiple senses at the same time.
(That said, there is excellent work on creating sense-vectors; see [Reisinger and Mooney 2010](http://www.aclweb.org/anthology/N10-1013); [Huang et al 2012](http://www.aclweb.org/anthology/P12-1092).)
The following code uses the NLTK WordNet API to create the edge dictionary we need for using the `Retrofitter` class:
### Reproducing the WordNet synonym graph experiment
For our VSM, let's use the 300d file included in this distribution from the GloVe team, as it is close to or identical to the one used in the paper:
http://nlp.stanford.edu/data/glove.6B.zip
If you download this archive, place it in `vsmdata`, and unpack it, then the following will load the file into a dictionary for you:
This is the initial embedding space $\widehat{Q}$:
Now we just need to replace all of the strings in `edges` with indices into `X_glove`:
And now we can retrofit:
You can now evaluate `X_retro` using the homework/bake-off notebook [hw_wordrelatedness.ipynb](hw_wordrelatedness.ipynb)!
| 0.810216 | 0.952086 |
# Text Generation
Here I'm using [Shakespeare plays](https://www.kaggle.com/kingburrito666/shakespeare-plays) by [LiamLarsen](https://www.kaggle.com/kingburrito666) to create a deep learning model which will be able to `generate text` using some input text.
**While doing all of this we will go through:**
- `Preprocessing` text data
- Building multilayer `Bidirectional RNN` model
- Saving `wording embeddings` learned by the learning algorithm
```
import io
import re
import json
import string
import unicodedata
from random import randint
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from nltk.corpus import stopwords
from wordcloud import STOPWORDS, WordCloud
import tensorflow as tf
from tensorflow.keras import Sequential
from tensorflow.keras.metrics import AUC
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.callbacks import Callback, ReduceLROnPlateau
from tensorflow.keras.layers import LSTM, Bidirectional, Dense, Embedding
!pip install contractions
!pip install tensorflow-addons
from tensorflow_addons.metrics import F1Score
from contractions import contractions_dict
for key, value in list(contractions_dict.items())[:10]:
print(f'{key} == {value}')
```
## ✈️ Getting data
Actual word of text file is `4583798`
```
def load_data(size_to_read=100_000):
filename = '/kaggle/input/shakespeare-plays/alllines.txt'
with open(filename, 'r') as f:
data = f.read(size_to_read)
return data
data = load_data(size_to_read=500_000)
data[:100]
```
## 🍭 Data preparation
Splitting our text data into `corpus` which in turn will be our `training samples`.
```
corpus = data.lower().split('\n')
print(corpus[:10])
```

```
def expand_contractions(text, contraction_map=contractions_dict):
# Using regex for getting all contracted words
contractions_keys = '|'.join(contraction_map.keys())
contractions_pattern = re.compile(f'({contractions_keys})', flags=re.DOTALL)
def expand_match(contraction):
# Getting entire matched sub-string
match = contraction.group(0)
expanded_contraction = contraction_map.get(match)
if not expand_contractions:
print(match)
return match
return expanded_contraction
expanded_text = contractions_pattern.sub(expand_match, text)
expanded_text = re.sub("'", "", expanded_text)
return expanded_text
expand_contractions("y'all can't expand contractions i'd think")
for idx, sentence in enumerate(corpus):
corpus[idx] = expand_contractions(sentence)
corpus[:5]
# Remove puncuation from word
def rm_punc_from_word(word):
clean_alphabet_list = [
alphabet for alphabet in word if alphabet not in string.punctuation
]
return ''.join(clean_alphabet_list)
print(rm_punc_from_word('#cool!'))
# Remove puncuation from text
def rm_punc_from_text(text):
clean_word_list = [rm_punc_from_word(word) for word in text]
return ''.join(clean_word_list)
print(rm_punc_from_text("Frankly, my dear, I don't give a damn"))
# Remove numbers from text
def rm_number_from_text(text):
text = re.sub('[0-9]+', '', text)
return ' '.join(text.split()) # to rm `extra` white space
print(rm_number_from_text('You are 100times more sexier than me'))
print(rm_number_from_text('If you taught yes then you are 10 times more delusional than me'))
# Remove stopwords from text
def rm_stopwords_from_text(text):
_stopwords = stopwords.words('english')
text = text.split()
word_list = [word for word in text if word not in _stopwords]
return ' '.join(word_list)
rm_stopwords_from_text("Love means never having to say you're sorry")
# Cleaning text
def clean_text(text):
text = text.lower()
text = rm_punc_from_text(text)
text = rm_number_from_text(text)
text = rm_stopwords_from_text(text)
# there are hyphen(–) in many titles, so replacing it with empty str
# this hyphen(–) is different from normal hyphen(-)
text = re.sub('–', '', text)
text = ' '.join(text.split()) # removing `extra` white spaces
# Removing unnecessary characters from text
text = re.sub("(\\t)", ' ', str(text)).lower()
text = re.sub("(\\r)", ' ', str(text)).lower()
text = re.sub("(\\n)", ' ', str(text)).lower()
# remove accented chars ('Sómě Áccěntěd těxt' => 'Some Accented text')
text = unicodedata.normalize('NFKD', text).encode('ascii', 'ignore').decode(
'utf-8', 'ignore'
)
text = re.sub("(__+)", ' ', str(text)).lower()
text = re.sub("(--+)", ' ', str(text)).lower()
text = re.sub("(~~+)", ' ', str(text)).lower()
text = re.sub("(\+\++)", ' ', str(text)).lower()
text = re.sub("(\.\.+)", ' ', str(text)).lower()
text = re.sub(r"[<>()|&©ø\[\]\'\",;?~*!]", ' ', str(text)).lower()
text = re.sub("(mailto:)", ' ', str(text)).lower()
text = re.sub(r"(\\x9\d)", ' ', str(text)).lower()
text = re.sub("([iI][nN][cC]\d+)", 'INC_NUM', str(text)).lower()
text = re.sub("([cC][mM]\d+)|([cC][hH][gG]\d+)", 'CM_NUM',
str(text)).lower()
text = re.sub("(\.\s+)", ' ', str(text)).lower()
text = re.sub("(\-\s+)", ' ', str(text)).lower()
text = re.sub("(\:\s+)", ' ', str(text)).lower()
text = re.sub("(\s+.\s+)", ' ', str(text)).lower()
try:
url = re.search(r'((https*:\/*)([^\/\s]+))(.[^\s]+)', str(text))
repl_url = url.group(3)
text = re.sub(r'((https*:\/*)([^\/\s]+))(.[^\s]+)', repl_url, str(text))
except:
pass
text = re.sub("(\s+)", ' ', str(text)).lower()
text = re.sub("(\s+.\s+)", ' ', str(text)).lower()
return text
clean_text("Mrs. Robinson, you're trying to seduce me, aren't you?")
for idx, sentence in enumerate(corpus):
corpus[idx] = clean_text(sentence)
corpus[:10]
```
Removing sentences whose length is `1`.
```
print(f'Corpus size before: {len(corpus)}')
corpus = [sentence for sentence in corpus if len(sentence.split(' ')) > 1]
print(f'Corpus size now: {len(corpus)}')
corpus[:10]
```
**🕶 Data visulaization**

```
# To customize colours of wordcloud texts
def wc_blue_color_func(word, font_size, position, orientation, random_state=None, **kwargs):
return "hsl(214, 67%%, %d%%)" % randint(60, 100)
# stopwords for wordcloud
def get_wc_stopwords():
wc_stopwords = set(STOPWORDS)
# Adding words to stopwords
# these words showed up while plotting wordcloud for text
wc_stopwords.add('s')
wc_stopwords.add('one')
wc_stopwords.add('using')
wc_stopwords.add('example')
wc_stopwords.add('work')
wc_stopwords.add('use')
wc_stopwords.add('make')
return wc_stopwords
# plot wordcloud
def plot_wordcloud(text, color_func):
wc_stopwords = get_wc_stopwords()
wc = WordCloud(
stopwords=wc_stopwords, width=1200, height=400, random_state=0
).generate(text)
f, axs = plt.subplots(figsize=(20, 10))
with sns.axes_style("ticks"):
sns.despine(offset=10, trim=True)
plt.imshow(
wc.recolor(color_func=color_func, random_state=0),
interpolation="bilinear"
)
plt.xlabel('Title WordCloud')
plot_wordcloud(' '.join(corpus), wc_blue_color_func)
```
**Preparing data for neural network**
```
print(f"Vocab size: {len(set(' '.join(corpus).split(' ')))}")
oov_token = '<UNK>'
tokenizer = Tokenizer(oov_token=oov_token)
tokenizer.fit_on_texts(corpus)
word_index = tokenizer.word_index
total_words = len(word_index) + 1
'''
Adding 1 to total_words to avoid below error
IndexError Traceback (most recent call last)
<ipython-input-136-16f89b53d516> in <module>
----> 1 y = tf.keras.utils.to_categorical(labels, num_classes=total_words)
2 print(y[1])
/opt/conda/lib/python3.7/site-packages/tensorflow/python/keras/utils/np_utils.py in to_categorical(y, num_classes, dtype)
76 n = y.shape[0]
77 categorical = np.zeros((n, num_classes), dtype=dtype)
---> 78 categorical[np.arange(n), y] = 1
79 output_shape = input_shape + (num_classes,)
80 categorical = np.reshape(categorical, output_shape)
IndexError: index 3049 is out of bounds for axis 1 with size 3049
This is because of reserving padding (i.e. index zero).
Stackoverflow post for more into: https://stackoverflow.com/questions/53525994/how-to-find-num-words-or-vocabulary-size-of-keras-tokenizer-when-one-is-not-as
'''
print(total_words)
# Converting the text to sequence using the tokenizer
def get_input_sequences(corpus, tokenizer):
input_sequences = []
for line in corpus:
tokens_list = tokenizer.texts_to_sequences([line])[0]
for i in range(1, len(tokens_list)):
n_gram_sequence = tokens_list[:i + 1]
input_sequences.append(n_gram_sequence)
return input_sequences
input_sequences = get_input_sequences(corpus, tokenizer)
print(input_sequences[:5])
# getting the max len of among all sequences
max_sequence_len = max([len(x) for x in input_sequences])
print(max_sequence_len)
# padding the input sequence
padded_input_sequences = np.array(pad_sequences(input_sequences, maxlen=max_sequence_len, padding='pre'))
print(padded_input_sequences[1])
```
Padding from starting since we are going to `predict the last word`.
```
# shuffling the data
np.random.shuffle(padded_input_sequences)
def map_sequence_to_text(x, y, index_word):
text = ''
for index in x:
text += index_word[index] + ' '
text += f'[{index_word[y]}]'
return text
# Removing the input and output texts
x = padded_input_sequences[:, :-1]
labels = padded_input_sequences[:, -1]
print(x[17])
print(labels[17])
def map_sequence_to_text(x, y, index_word):
text = ''
for index in x:
if index == 0:
# index 0 == padded char
continue
text += index_word[index] + ' '
text += f'[{index_word[y]}]'
return text
map_sequence_to_text(x[17], labels[17], tokenizer.index_word)
y = tf.keras.utils.to_categorical(labels, num_classes=total_words)
print(y[1])
```
## 👩🔬 Modelling
```
lstm_units = 512
embedding_dim = 512
optimizer = tf.keras.optimizers.Adam(learning_rate=0.001)
loss = 'categorical_crossentropy'
num_epochs = 30
metrics = [
'accuracy',
AUC(curve='ROC', multi_label=True, name='auc_roc'),
F1Score(num_classes=total_words, average='weighted')
]
class CustomCallback(Callback):
def on_epoch_start(self, epoch, logs=None):
print()
def on_epoch_end(self, epoch, logs=None):
loss = logs['loss']
accuracy = logs['accuracy']
f1_score = logs['f1_score']
auc_roc = logs['auc_roc']
info = {
'loss': round(loss, 5),
'accuracy': round(accuracy, 4),
'auc_roc': round(auc_roc, 4),
'f1_score': round(f1_score, 4),
}
print(f'\n{json.dumps(info, indent=2)}')
callbacks = [
ReduceLROnPlateau(monitor='val_loss', factor=0.1, patience=2, min_lr=0.000001, verbose=1),
CustomCallback()
]
def build_model(
total_words,
max_sequence_len,
lstm_units=lstm_units,
embedding_dim=embedding_dim,
loss=loss,
optimizer=optimizer,
metrics=metrics
):
model = Sequential([
Embedding(total_words, embedding_dim, input_length=max_sequence_len - 1, trainable=True),
Bidirectional(LSTM(lstm_units)),
Dense(total_words, activation='softmax')
])
model.compile(loss=loss, optimizer=optimizer, metrics=metrics)
return model
model = build_model(total_words, max_sequence_len, lstm_units, embedding_dim)
model.summary()
history = model.fit(x, y, epochs=num_epochs, callbacks=callbacks, verbose=1)
```
**Plotting model's performance**
```
# Accuracy
plt.plot(history.history['accuracy'][1:], label='train acc')
plt.xlabel('Epoch')
plt.ylabel('Accuracy')
plt.legend(loc='lower right')
# Loss
plt.plot(history.history['loss'][1:], label='train loss')
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.legend(loc='lower right')
# Saving embedding vectors and words to visualize embeddings using `Tensorflow Projector`.
# Getting weights of our embedding layer
embedding_layer = model.layers[0]
embedding_layer_weigths = embedding_layer.get_weights()[0]
print(embedding_layer_weigths.shape)
# Reversing the `word_index`
word_index = tokenizer.word_index
reverse_word_index = {value: key for key, value in word_index.items()}
# Writing vectors and their meta data which when entered to Tensorflow Project,
# it will display our Word Embedding
out_vectors = io.open('vecs.tsv', 'w', encoding='utf-8')
out_metadata = io.open('meta.tsv', 'w', encoding='utf-8')
# Skipping over the first word in vocabulary which is '<OOV>' (if set oov_token parameter set then)
for word_num in range(1, total_words):
words = reverse_word_index[word_num]
embeddings = embedding_layer_weigths[word_num]
out_metadata.write(words + '\n')
out_vectors.write('\t'.join([str(x) for x in embeddings]) + '\n')
out_vectors.close()
out_metadata.close()
```
The first value of the above output is the `vocab_size(total_words)` and second value is the `embedding_dim`.
## 🔮 Predictions
```
def predict_next(model, text, tokenizer, max_sequence_len, num_of_words=10):
# predict next num_of_words for text
for _ in range(num_of_words):
input_sequences = tokenizer.texts_to_sequences([text])[0]
padded_input_sequences = pad_sequences(
[input_sequences], maxlen=max_sequence_len - 1, padding='pre'
)
predicted = model.predict_classes(padded_input_sequences, verbose=0)
output_word = ''
for word, index in tokenizer.word_index.items():
if index == predicted:
output_word = word
break
text += ' ' + output_word
return text
seed_text = 'The sky is'
print(predict_next(model, seed_text, tokenizer, max_sequence_len, num_of_words=4))
seed_text = 'Everything is fair in love and'
print(predict_next(model, seed_text, tokenizer, max_sequence_len, num_of_words=10))
seed_text = 'My life'
print(predict_next(model, seed_text, tokenizer, max_sequence_len, num_of_words=15))
seed_text = 'You are a type of guy that'
print(predict_next(model, seed_text, tokenizer, max_sequence_len, num_of_words=20))
seed_text = 'FROM off a hill whose concave womb reworded A plaintful'
print(predict_next(model, seed_text, tokenizer, max_sequence_len, num_of_words=5))
```
## 🎁 Saving the model
```
model.save('model')
```
---
If this kernel helped you then don't forget to 👍 `like` and give your 🎙 `feedback`.

---
|
github_jupyter
|
import io
import re
import json
import string
import unicodedata
from random import randint
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from nltk.corpus import stopwords
from wordcloud import STOPWORDS, WordCloud
import tensorflow as tf
from tensorflow.keras import Sequential
from tensorflow.keras.metrics import AUC
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.callbacks import Callback, ReduceLROnPlateau
from tensorflow.keras.layers import LSTM, Bidirectional, Dense, Embedding
!pip install contractions
!pip install tensorflow-addons
from tensorflow_addons.metrics import F1Score
from contractions import contractions_dict
for key, value in list(contractions_dict.items())[:10]:
print(f'{key} == {value}')
def load_data(size_to_read=100_000):
filename = '/kaggle/input/shakespeare-plays/alllines.txt'
with open(filename, 'r') as f:
data = f.read(size_to_read)
return data
data = load_data(size_to_read=500_000)
data[:100]
corpus = data.lower().split('\n')
print(corpus[:10])
def expand_contractions(text, contraction_map=contractions_dict):
# Using regex for getting all contracted words
contractions_keys = '|'.join(contraction_map.keys())
contractions_pattern = re.compile(f'({contractions_keys})', flags=re.DOTALL)
def expand_match(contraction):
# Getting entire matched sub-string
match = contraction.group(0)
expanded_contraction = contraction_map.get(match)
if not expand_contractions:
print(match)
return match
return expanded_contraction
expanded_text = contractions_pattern.sub(expand_match, text)
expanded_text = re.sub("'", "", expanded_text)
return expanded_text
expand_contractions("y'all can't expand contractions i'd think")
for idx, sentence in enumerate(corpus):
corpus[idx] = expand_contractions(sentence)
corpus[:5]
# Remove puncuation from word
def rm_punc_from_word(word):
clean_alphabet_list = [
alphabet for alphabet in word if alphabet not in string.punctuation
]
return ''.join(clean_alphabet_list)
print(rm_punc_from_word('#cool!'))
# Remove puncuation from text
def rm_punc_from_text(text):
clean_word_list = [rm_punc_from_word(word) for word in text]
return ''.join(clean_word_list)
print(rm_punc_from_text("Frankly, my dear, I don't give a damn"))
# Remove numbers from text
def rm_number_from_text(text):
text = re.sub('[0-9]+', '', text)
return ' '.join(text.split()) # to rm `extra` white space
print(rm_number_from_text('You are 100times more sexier than me'))
print(rm_number_from_text('If you taught yes then you are 10 times more delusional than me'))
# Remove stopwords from text
def rm_stopwords_from_text(text):
_stopwords = stopwords.words('english')
text = text.split()
word_list = [word for word in text if word not in _stopwords]
return ' '.join(word_list)
rm_stopwords_from_text("Love means never having to say you're sorry")
# Cleaning text
def clean_text(text):
text = text.lower()
text = rm_punc_from_text(text)
text = rm_number_from_text(text)
text = rm_stopwords_from_text(text)
# there are hyphen(–) in many titles, so replacing it with empty str
# this hyphen(–) is different from normal hyphen(-)
text = re.sub('–', '', text)
text = ' '.join(text.split()) # removing `extra` white spaces
# Removing unnecessary characters from text
text = re.sub("(\\t)", ' ', str(text)).lower()
text = re.sub("(\\r)", ' ', str(text)).lower()
text = re.sub("(\\n)", ' ', str(text)).lower()
# remove accented chars ('Sómě Áccěntěd těxt' => 'Some Accented text')
text = unicodedata.normalize('NFKD', text).encode('ascii', 'ignore').decode(
'utf-8', 'ignore'
)
text = re.sub("(__+)", ' ', str(text)).lower()
text = re.sub("(--+)", ' ', str(text)).lower()
text = re.sub("(~~+)", ' ', str(text)).lower()
text = re.sub("(\+\++)", ' ', str(text)).lower()
text = re.sub("(\.\.+)", ' ', str(text)).lower()
text = re.sub(r"[<>()|&©ø\[\]\'\",;?~*!]", ' ', str(text)).lower()
text = re.sub("(mailto:)", ' ', str(text)).lower()
text = re.sub(r"(\\x9\d)", ' ', str(text)).lower()
text = re.sub("([iI][nN][cC]\d+)", 'INC_NUM', str(text)).lower()
text = re.sub("([cC][mM]\d+)|([cC][hH][gG]\d+)", 'CM_NUM',
str(text)).lower()
text = re.sub("(\.\s+)", ' ', str(text)).lower()
text = re.sub("(\-\s+)", ' ', str(text)).lower()
text = re.sub("(\:\s+)", ' ', str(text)).lower()
text = re.sub("(\s+.\s+)", ' ', str(text)).lower()
try:
url = re.search(r'((https*:\/*)([^\/\s]+))(.[^\s]+)', str(text))
repl_url = url.group(3)
text = re.sub(r'((https*:\/*)([^\/\s]+))(.[^\s]+)', repl_url, str(text))
except:
pass
text = re.sub("(\s+)", ' ', str(text)).lower()
text = re.sub("(\s+.\s+)", ' ', str(text)).lower()
return text
clean_text("Mrs. Robinson, you're trying to seduce me, aren't you?")
for idx, sentence in enumerate(corpus):
corpus[idx] = clean_text(sentence)
corpus[:10]
print(f'Corpus size before: {len(corpus)}')
corpus = [sentence for sentence in corpus if len(sentence.split(' ')) > 1]
print(f'Corpus size now: {len(corpus)}')
corpus[:10]
# To customize colours of wordcloud texts
def wc_blue_color_func(word, font_size, position, orientation, random_state=None, **kwargs):
return "hsl(214, 67%%, %d%%)" % randint(60, 100)
# stopwords for wordcloud
def get_wc_stopwords():
wc_stopwords = set(STOPWORDS)
# Adding words to stopwords
# these words showed up while plotting wordcloud for text
wc_stopwords.add('s')
wc_stopwords.add('one')
wc_stopwords.add('using')
wc_stopwords.add('example')
wc_stopwords.add('work')
wc_stopwords.add('use')
wc_stopwords.add('make')
return wc_stopwords
# plot wordcloud
def plot_wordcloud(text, color_func):
wc_stopwords = get_wc_stopwords()
wc = WordCloud(
stopwords=wc_stopwords, width=1200, height=400, random_state=0
).generate(text)
f, axs = plt.subplots(figsize=(20, 10))
with sns.axes_style("ticks"):
sns.despine(offset=10, trim=True)
plt.imshow(
wc.recolor(color_func=color_func, random_state=0),
interpolation="bilinear"
)
plt.xlabel('Title WordCloud')
plot_wordcloud(' '.join(corpus), wc_blue_color_func)
print(f"Vocab size: {len(set(' '.join(corpus).split(' ')))}")
oov_token = '<UNK>'
tokenizer = Tokenizer(oov_token=oov_token)
tokenizer.fit_on_texts(corpus)
word_index = tokenizer.word_index
total_words = len(word_index) + 1
'''
Adding 1 to total_words to avoid below error
IndexError Traceback (most recent call last)
<ipython-input-136-16f89b53d516> in <module>
----> 1 y = tf.keras.utils.to_categorical(labels, num_classes=total_words)
2 print(y[1])
/opt/conda/lib/python3.7/site-packages/tensorflow/python/keras/utils/np_utils.py in to_categorical(y, num_classes, dtype)
76 n = y.shape[0]
77 categorical = np.zeros((n, num_classes), dtype=dtype)
---> 78 categorical[np.arange(n), y] = 1
79 output_shape = input_shape + (num_classes,)
80 categorical = np.reshape(categorical, output_shape)
IndexError: index 3049 is out of bounds for axis 1 with size 3049
This is because of reserving padding (i.e. index zero).
Stackoverflow post for more into: https://stackoverflow.com/questions/53525994/how-to-find-num-words-or-vocabulary-size-of-keras-tokenizer-when-one-is-not-as
'''
print(total_words)
# Converting the text to sequence using the tokenizer
def get_input_sequences(corpus, tokenizer):
input_sequences = []
for line in corpus:
tokens_list = tokenizer.texts_to_sequences([line])[0]
for i in range(1, len(tokens_list)):
n_gram_sequence = tokens_list[:i + 1]
input_sequences.append(n_gram_sequence)
return input_sequences
input_sequences = get_input_sequences(corpus, tokenizer)
print(input_sequences[:5])
# getting the max len of among all sequences
max_sequence_len = max([len(x) for x in input_sequences])
print(max_sequence_len)
# padding the input sequence
padded_input_sequences = np.array(pad_sequences(input_sequences, maxlen=max_sequence_len, padding='pre'))
print(padded_input_sequences[1])
# shuffling the data
np.random.shuffle(padded_input_sequences)
def map_sequence_to_text(x, y, index_word):
text = ''
for index in x:
text += index_word[index] + ' '
text += f'[{index_word[y]}]'
return text
# Removing the input and output texts
x = padded_input_sequences[:, :-1]
labels = padded_input_sequences[:, -1]
print(x[17])
print(labels[17])
def map_sequence_to_text(x, y, index_word):
text = ''
for index in x:
if index == 0:
# index 0 == padded char
continue
text += index_word[index] + ' '
text += f'[{index_word[y]}]'
return text
map_sequence_to_text(x[17], labels[17], tokenizer.index_word)
y = tf.keras.utils.to_categorical(labels, num_classes=total_words)
print(y[1])
lstm_units = 512
embedding_dim = 512
optimizer = tf.keras.optimizers.Adam(learning_rate=0.001)
loss = 'categorical_crossentropy'
num_epochs = 30
metrics = [
'accuracy',
AUC(curve='ROC', multi_label=True, name='auc_roc'),
F1Score(num_classes=total_words, average='weighted')
]
class CustomCallback(Callback):
def on_epoch_start(self, epoch, logs=None):
print()
def on_epoch_end(self, epoch, logs=None):
loss = logs['loss']
accuracy = logs['accuracy']
f1_score = logs['f1_score']
auc_roc = logs['auc_roc']
info = {
'loss': round(loss, 5),
'accuracy': round(accuracy, 4),
'auc_roc': round(auc_roc, 4),
'f1_score': round(f1_score, 4),
}
print(f'\n{json.dumps(info, indent=2)}')
callbacks = [
ReduceLROnPlateau(monitor='val_loss', factor=0.1, patience=2, min_lr=0.000001, verbose=1),
CustomCallback()
]
def build_model(
total_words,
max_sequence_len,
lstm_units=lstm_units,
embedding_dim=embedding_dim,
loss=loss,
optimizer=optimizer,
metrics=metrics
):
model = Sequential([
Embedding(total_words, embedding_dim, input_length=max_sequence_len - 1, trainable=True),
Bidirectional(LSTM(lstm_units)),
Dense(total_words, activation='softmax')
])
model.compile(loss=loss, optimizer=optimizer, metrics=metrics)
return model
model = build_model(total_words, max_sequence_len, lstm_units, embedding_dim)
model.summary()
history = model.fit(x, y, epochs=num_epochs, callbacks=callbacks, verbose=1)
# Accuracy
plt.plot(history.history['accuracy'][1:], label='train acc')
plt.xlabel('Epoch')
plt.ylabel('Accuracy')
plt.legend(loc='lower right')
# Loss
plt.plot(history.history['loss'][1:], label='train loss')
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.legend(loc='lower right')
# Saving embedding vectors and words to visualize embeddings using `Tensorflow Projector`.
# Getting weights of our embedding layer
embedding_layer = model.layers[0]
embedding_layer_weigths = embedding_layer.get_weights()[0]
print(embedding_layer_weigths.shape)
# Reversing the `word_index`
word_index = tokenizer.word_index
reverse_word_index = {value: key for key, value in word_index.items()}
# Writing vectors and their meta data which when entered to Tensorflow Project,
# it will display our Word Embedding
out_vectors = io.open('vecs.tsv', 'w', encoding='utf-8')
out_metadata = io.open('meta.tsv', 'w', encoding='utf-8')
# Skipping over the first word in vocabulary which is '<OOV>' (if set oov_token parameter set then)
for word_num in range(1, total_words):
words = reverse_word_index[word_num]
embeddings = embedding_layer_weigths[word_num]
out_metadata.write(words + '\n')
out_vectors.write('\t'.join([str(x) for x in embeddings]) + '\n')
out_vectors.close()
out_metadata.close()
def predict_next(model, text, tokenizer, max_sequence_len, num_of_words=10):
# predict next num_of_words for text
for _ in range(num_of_words):
input_sequences = tokenizer.texts_to_sequences([text])[0]
padded_input_sequences = pad_sequences(
[input_sequences], maxlen=max_sequence_len - 1, padding='pre'
)
predicted = model.predict_classes(padded_input_sequences, verbose=0)
output_word = ''
for word, index in tokenizer.word_index.items():
if index == predicted:
output_word = word
break
text += ' ' + output_word
return text
seed_text = 'The sky is'
print(predict_next(model, seed_text, tokenizer, max_sequence_len, num_of_words=4))
seed_text = 'Everything is fair in love and'
print(predict_next(model, seed_text, tokenizer, max_sequence_len, num_of_words=10))
seed_text = 'My life'
print(predict_next(model, seed_text, tokenizer, max_sequence_len, num_of_words=15))
seed_text = 'You are a type of guy that'
print(predict_next(model, seed_text, tokenizer, max_sequence_len, num_of_words=20))
seed_text = 'FROM off a hill whose concave womb reworded A plaintful'
print(predict_next(model, seed_text, tokenizer, max_sequence_len, num_of_words=5))
model.save('model')
| 0.462473 | 0.734976 |
# TAT-C: Collect Observations Web Request Example
Dr. Paul Grogan, I. Josue Tapia-Tamayo, Isaac Feldman
Collective Design (CoDe) Lab
Stevens Institute of Technology, School of Systems and Enterprises
This example demonstrates how to use HTTP requests to a TAT-C server to model observations of single point by a single satellite using an orbit derived from an existing Two Line Element (TLE) set.
## Dependencies
### Standard Python Libraries
This example is compatible with python 3.8.10 and makes use of the `geopandas`, `datetime`, and `requests` libraries.
```
from datetime import datetime, timedelta, timezone
import geopandas
import requests
```
## Establish Mission Window
First we define the mission window using the `datetime`, `timedelta`, and `timezone` objects from the python `datetime` library
```
# Set the start date to be January, 1, 2021 at noon (12 pm) UTC
start = datetime(year=2021, month=1, day=1, hour=12, minute=0, second=0, tzinfo=timezone.utc)
# Set the end date to be 30 days after the start date
end = start + timedelta(days=30)
```
## Make the HTTP Request
Next we make the HTTP request to our server running TATC
### Authentication
Before we can make the HTTP request for observation gathering, we have to authenticate with the server and receive an authentication token
```
# The server URL
url = 'https://localhost:8000'
# Post login information to the server
r = requests.post(url + '/auth/login', data={'username':'admin@example.com', 'password':'admin'})
# Save the authentication token from the response
token = r.json()['access_token']
print('Token:', token)
# Create a header using the authentication token
headers = {'Content-Type': 'application/json',
'accept': 'application/json', 'Authorization':'Bearer ' + token}
print('\n'+'Headers:', headers)
```
### Post the Task Request
Here we use the `requests` library and TAT-C CollectObservationsRequest schemas format, found at <h>https://tatc.code-lab.org/docs#/</h>, to submit the task request to the server.
```
# Post the task request
r1 = requests.post(url + '/collect/observations', json={
'points': [ # List of oints on the ground to be observed
{
'id': 0, # Point with ID: 0
'latitude': 40.74259, # Point is located at latitude 40.74259 degrees
'longitude': -74.02686 # Point is located at longitude -7402686 degrees
}
],
'satellite': { # The satellite of interest
'type': "satellite", # The type of satellite representation
'name': "NOAA 1", # A name for the satellite
'orbit': { # The satellites orbit
'type': 'tle', # The type of information they orbit is to be derived from
'tle': [
'1 04793U 70106A 22044.41526573 -.00000035 00000+0 54783-4 0 9991', # The first line of the orbit TLE
'2 04793 101.5750 111.2777 0031771 203.0437 167.8944 12.54003052342042' # The second line of the orbit TLE
]
},
'instruments': [ # List of instruments on the satellite
{
'name':'NOAA 1 Instrument', # Name of the instrument
'field_of_regard': 180 # Instrument field of regard
}
]
},
'instrument': 0, # List index of the instrument to be used for observations
'start': start.isoformat(), # Start of the mission window
'end': end.isoformat(), # End of the mission Window
'omit_solar': True
}, headers=headers)
# Save the task ID
print('r1 content:', r1.content)
print(r1)
```
### Retreiving the result
```
# Save the task ID
print('r1 content:', r1.content)
print(r1)
task_id = '/' + r1.json()["task_id"]
# A boolean to represent readiness of the result
ready = False
# Until the result is ready
while not ready:
# Get the status of the task
r2 = requests.get(url + '/collect/observations' + task_id + '/status', headers=headers)
# Check if the task is ready
if r2.json()['ready'] == True and r2.json()['successful'] == True:
ready = True
# Report if the task is ready
print(r2.json())
# Delay next loop for 0.1 seconds to prevent overloading the system with requests
time.sleep(0.1)
# Get the result
r3 = requests.get(url + task + task_id + '/results', headers=headers)
# Save the result to a GeoDataFrame
results_gdf = gpd.GeoDataFrame(r3.json())
```
|
github_jupyter
|
from datetime import datetime, timedelta, timezone
import geopandas
import requests
# Set the start date to be January, 1, 2021 at noon (12 pm) UTC
start = datetime(year=2021, month=1, day=1, hour=12, minute=0, second=0, tzinfo=timezone.utc)
# Set the end date to be 30 days after the start date
end = start + timedelta(days=30)
# The server URL
url = 'https://localhost:8000'
# Post login information to the server
r = requests.post(url + '/auth/login', data={'username':'admin@example.com', 'password':'admin'})
# Save the authentication token from the response
token = r.json()['access_token']
print('Token:', token)
# Create a header using the authentication token
headers = {'Content-Type': 'application/json',
'accept': 'application/json', 'Authorization':'Bearer ' + token}
print('\n'+'Headers:', headers)
# Post the task request
r1 = requests.post(url + '/collect/observations', json={
'points': [ # List of oints on the ground to be observed
{
'id': 0, # Point with ID: 0
'latitude': 40.74259, # Point is located at latitude 40.74259 degrees
'longitude': -74.02686 # Point is located at longitude -7402686 degrees
}
],
'satellite': { # The satellite of interest
'type': "satellite", # The type of satellite representation
'name': "NOAA 1", # A name for the satellite
'orbit': { # The satellites orbit
'type': 'tle', # The type of information they orbit is to be derived from
'tle': [
'1 04793U 70106A 22044.41526573 -.00000035 00000+0 54783-4 0 9991', # The first line of the orbit TLE
'2 04793 101.5750 111.2777 0031771 203.0437 167.8944 12.54003052342042' # The second line of the orbit TLE
]
},
'instruments': [ # List of instruments on the satellite
{
'name':'NOAA 1 Instrument', # Name of the instrument
'field_of_regard': 180 # Instrument field of regard
}
]
},
'instrument': 0, # List index of the instrument to be used for observations
'start': start.isoformat(), # Start of the mission window
'end': end.isoformat(), # End of the mission Window
'omit_solar': True
}, headers=headers)
# Save the task ID
print('r1 content:', r1.content)
print(r1)
# Save the task ID
print('r1 content:', r1.content)
print(r1)
task_id = '/' + r1.json()["task_id"]
# A boolean to represent readiness of the result
ready = False
# Until the result is ready
while not ready:
# Get the status of the task
r2 = requests.get(url + '/collect/observations' + task_id + '/status', headers=headers)
# Check if the task is ready
if r2.json()['ready'] == True and r2.json()['successful'] == True:
ready = True
# Report if the task is ready
print(r2.json())
# Delay next loop for 0.1 seconds to prevent overloading the system with requests
time.sleep(0.1)
# Get the result
r3 = requests.get(url + task + task_id + '/results', headers=headers)
# Save the result to a GeoDataFrame
results_gdf = gpd.GeoDataFrame(r3.json())
| 0.471467 | 0.908212 |
# Credit Card Fraud Detection
Throughout the financial sector, machine learning algorithms are being developed to detect fraudulent transactions. In this project, that is exactly what we are going to be doing as well.
Using a dataset of of nearly 28,500 credit card transactions and multiple unsupervised anomaly detection algorithms, we are going to identify transactions with a high probability of being credit card fraud.
Dataset is from Kaggle or UCI.
Furthermore, using metrics suchs as precision, recall, and F1-scores, we will investigate why the classification accuracy for these algorithms can be misleading.
In addition, we will explore the use of data visualization techniques common in data science, such as parameter histograms and correlation matrices, to gain a better understanding of the underlying distribution of data in our data set.
Let's get started!
### Importing the library
```
import pandas as pd
import numpy as np
import seaborn as sns
```
### Importing the dataset
```
data = pd.read_csv("F:\\PROJECT\\7. Credit Card Fraud Detection\\Dataset\\creditcard.csv")
data.head()
```
### Visualising the dataset
```
# Plot histograms of each parameter
import matplotlib.pyplot as plt
data.hist(figsize = (20, 20))
plt.show()
```
### Checking the correlation between variable
```
# Strong correlation between the Class, Time, Amount, V2, V3, .........
import seaborn as sns
plt.figure(figsize=(30,20))
sns.heatmap(data.corr(), annot=True)
```
### Applying the feature scaling to the dataset
```
from sklearn.preprocessing import StandardScaler
data['normalizedAmount'] = StandardScaler().fit_transform(data['Amount'].values.reshape(-1,1))
data = data.drop(['Amount'], axis = 1)
data = data.drop(['Time'], axis = 1)
data.head()
X = data.iloc[:, data.columns != 'Class']
X.head()
Y = data.iloc[:, data.columns == 'Class']
Y.head()
```
### Spliting the dataset into trainset and test set
```
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size = 0.3, random_state = 0)
X_train = np.array(X_train)
X_test = np.array(X_test)
Y_train = np.array(Y_train)
Y_test = np.array(Y_test)
```
#### Applying the Decision tree Model
```
from sklearn.tree import DecisionTreeClassifier
classifier = DecisionTreeClassifier()
classifier.fit(X_train, Y_train)
```
#### Predicting the Result to the test set
```
Y_pred = classifier.predict(X_test)
Y_pred
classifier.score(X_test, Y_test)
```
#### Analyse the result using Confusion matrix
```
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(Y_test, Y_pred)
sns.heatmap(cm, annot = True)
```
#### Analyse the model Report
```
from sklearn.metrics import classification_report
print(classification_report(Y_test, Y_pred))
```
#### We get the accuracy of 99.9262% to Model by using the DecisionTree Classifier
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
data = pd.read_csv("F:\\PROJECT\\7. Credit Card Fraud Detection\\Dataset\\creditcard.csv")
data.head()
# Plot histograms of each parameter
import matplotlib.pyplot as plt
data.hist(figsize = (20, 20))
plt.show()
# Strong correlation between the Class, Time, Amount, V2, V3, .........
import seaborn as sns
plt.figure(figsize=(30,20))
sns.heatmap(data.corr(), annot=True)
from sklearn.preprocessing import StandardScaler
data['normalizedAmount'] = StandardScaler().fit_transform(data['Amount'].values.reshape(-1,1))
data = data.drop(['Amount'], axis = 1)
data = data.drop(['Time'], axis = 1)
data.head()
X = data.iloc[:, data.columns != 'Class']
X.head()
Y = data.iloc[:, data.columns == 'Class']
Y.head()
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size = 0.3, random_state = 0)
X_train = np.array(X_train)
X_test = np.array(X_test)
Y_train = np.array(Y_train)
Y_test = np.array(Y_test)
from sklearn.tree import DecisionTreeClassifier
classifier = DecisionTreeClassifier()
classifier.fit(X_train, Y_train)
Y_pred = classifier.predict(X_test)
Y_pred
classifier.score(X_test, Y_test)
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(Y_test, Y_pred)
sns.heatmap(cm, annot = True)
from sklearn.metrics import classification_report
print(classification_report(Y_test, Y_pred))
| 0.707607 | 0.989182 |
```
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
dataset=pd.read_csv("TestDataset.csv")
print(dataset.shape)
data_size=len(dataset)
print (data_size)
import re
import nltk
from urllib.parse import urlparse
#nltk.download('stopwords')
from nltk.corpus import stopwords
from nltk.stem.porter import PorterStemmer
corpus=[]
for i in range(0,data_size):
tweet=dataset['text'][i]
tweet = re.sub('[^a-zA-Z]', ' ', dataset['text'][i])
tweet=tweet.lower()
tweet=tweet.split()
ps=PorterStemmer()
tweet=[ps.stem(word) for word in tweet if word not in set(stopwords.words('english')) and len(word)>2]
tweet=' '.join(tweet)
corpus.append(tweet)
words=[]
for i in corpus:
for word in i:
words.append(i)
print (len(words))
print ((corpus[0]))
import collections
counter=collections.Counter(words)
common=counter.most_common(50)
common_words=[]
for i in range(0,50):
wo=common[i][0].split()
for w in wo:
common_words.append(w)
print (len(common_words))
common_words=list(set(common_words))
print (len(common_words))
print (common_words)
imp_words=['medic','need','give','relief','fund','food','donat','aid','water','meal','send','offer','financ','blood']
common_words=[i for i in common_words if i not in imp_words]
print (len(common_words))
dataset=pd.read_csv("LabelledTestDataset.csv")
print(dataset.shape)
data_size=len(dataset)
print (data_size)
need={}
availability={}
for i in range(0,data_size):
if dataset['class'][i]==1:
need[dataset['id'][i]]=str(dataset['text'][i])
elif dataset['class'][i]==2:
availability[dataset['id'][i]]=str(dataset['text'][i])
need_size=len(need)
availability_size=len(availability)
print (need_size)
print (availability_size)
import re
from nltk.corpus import stopwords
from nltk.stem.porter import PorterStemmer
from textblob import TextBlob
import enchant
from collections import defaultdict
d=enchant.Dict("en_US")
need_keys=list(need.keys())
availability_keys=list(availability.keys())
for i in need_keys:
need[i] = re.sub('[^a-zA-Z]', ' ', need[i])
need[i]=need[i].lower()
for i in availability_keys:
availability[i] = re.sub('[^a-zA-Z]', ' ', availability[i])
availability[i]=availability[i].lower()
for i in need_keys:
blob=TextBlob(need[i])
need[i]=([n for n,t in blob.tags if t == 'NN' and d.check(str(n))==True])
ps=PorterStemmer()
need[i]=[ps.stem(word) for word in need[i] if word not in set(stopwords.words('english')) and len(word)>2]
need[i]=[w for w in need[i] if w not in common_words]
#print(need[i])
#need[i]=need[i].split()
#print(need[i])
for i in availability_keys:
blob=TextBlob(availability[i])
availability[i]=([n for n,t in blob.tags if t=="NN" and d.check(str(n))==True])
ps=PorterStemmer()
availability[i]=[ps.stem(word) for word in availability[i] if word not in set(stopwords.words('english')) and len(word)>2]
#availability[i]=availability[i].split()
#print (availability[i])
match=defaultdict(list)
for i in need_keys:
for j in availability_keys:
for item in need[i]:
#print(need[i])
#print (item)
flag=False
for av in availability[j]:
#print (av)
if(str(item)==str(av)):
match[i].append(j)
flag=True
break
if(flag==True):
break
for i in need_keys:
print (len(match[i]))
print (match[need_keys[0]])
print (need[need_keys[0]])
print (need_keys[0])
output=[]
for i in need_keys:
output.append(match[i])
print (output[0:10])
import csv
with open("outputPhase2.txt","w+") as f:
#writer=csv.writer(f,delimiter=',',doublequote=False,escapechar='')
for i in need_keys:
out=str(i)+":"
for j in range(0,5):
if(len(match[i])>=j+1):
out+=str(match[i][j])+","
if len(match[i])!=0:
f.write(str(out[0:-1])+"\n")
f.close()
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
dataset=pd.read_csv("TestDataset.csv")
print(dataset.shape)
data_size=len(dataset)
print (data_size)
import re
import nltk
from urllib.parse import urlparse
#nltk.download('stopwords')
from nltk.corpus import stopwords
from nltk.stem.porter import PorterStemmer
corpus=[]
for i in range(0,data_size):
tweet=dataset['text'][i]
tweet = re.sub('[^a-zA-Z]', ' ', dataset['text'][i])
tweet=tweet.lower()
tweet=tweet.split()
ps=PorterStemmer()
tweet=[ps.stem(word) for word in tweet if word not in set(stopwords.words('english')) and len(word)>2]
tweet=' '.join(tweet)
corpus.append(tweet)
words=[]
for i in corpus:
for word in i:
words.append(i)
print (len(words))
print ((corpus[0]))
import collections
counter=collections.Counter(words)
common=counter.most_common(50)
common_words=[]
for i in range(0,50):
wo=common[i][0].split()
for w in wo:
common_words.append(w)
print (len(common_words))
common_words=list(set(common_words))
print (len(common_words))
print (common_words)
imp_words=['medic','need','give','relief','fund','food','donat','aid','water','meal','send','offer','financ','blood']
common_words=[i for i in common_words if i not in imp_words]
print (len(common_words))
dataset=pd.read_csv("LabelledTestDataset.csv")
print(dataset.shape)
data_size=len(dataset)
print (data_size)
need={}
availability={}
for i in range(0,data_size):
if dataset['class'][i]==1:
need[dataset['id'][i]]=str(dataset['text'][i])
elif dataset['class'][i]==2:
availability[dataset['id'][i]]=str(dataset['text'][i])
need_size=len(need)
availability_size=len(availability)
print (need_size)
print (availability_size)
import re
from nltk.corpus import stopwords
from nltk.stem.porter import PorterStemmer
from textblob import TextBlob
import enchant
from collections import defaultdict
d=enchant.Dict("en_US")
need_keys=list(need.keys())
availability_keys=list(availability.keys())
for i in need_keys:
need[i] = re.sub('[^a-zA-Z]', ' ', need[i])
need[i]=need[i].lower()
for i in availability_keys:
availability[i] = re.sub('[^a-zA-Z]', ' ', availability[i])
availability[i]=availability[i].lower()
for i in need_keys:
blob=TextBlob(need[i])
need[i]=([n for n,t in blob.tags if t == 'NN' and d.check(str(n))==True])
ps=PorterStemmer()
need[i]=[ps.stem(word) for word in need[i] if word not in set(stopwords.words('english')) and len(word)>2]
need[i]=[w for w in need[i] if w not in common_words]
#print(need[i])
#need[i]=need[i].split()
#print(need[i])
for i in availability_keys:
blob=TextBlob(availability[i])
availability[i]=([n for n,t in blob.tags if t=="NN" and d.check(str(n))==True])
ps=PorterStemmer()
availability[i]=[ps.stem(word) for word in availability[i] if word not in set(stopwords.words('english')) and len(word)>2]
#availability[i]=availability[i].split()
#print (availability[i])
match=defaultdict(list)
for i in need_keys:
for j in availability_keys:
for item in need[i]:
#print(need[i])
#print (item)
flag=False
for av in availability[j]:
#print (av)
if(str(item)==str(av)):
match[i].append(j)
flag=True
break
if(flag==True):
break
for i in need_keys:
print (len(match[i]))
print (match[need_keys[0]])
print (need[need_keys[0]])
print (need_keys[0])
output=[]
for i in need_keys:
output.append(match[i])
print (output[0:10])
import csv
with open("outputPhase2.txt","w+") as f:
#writer=csv.writer(f,delimiter=',',doublequote=False,escapechar='')
for i in need_keys:
out=str(i)+":"
for j in range(0,5):
if(len(match[i])>=j+1):
out+=str(match[i][j])+","
if len(match[i])!=0:
f.write(str(out[0:-1])+"\n")
f.close()
| 0.034278 | 0.173183 |
## Geopandas and a Map of the Middle World:
On the way to wondering how to create a map of the middle kingdom for Islamic history I also got thinking about creating a personalized Barcelona map. Turns out there are compete geojson files for Barca which are in the included folder.
This [website](https://docs.astraea.earth/hc/en-us/articles/360043919911-Read-a-GeoJSON-File-into-a-GeoPandas-DataFrame) offers some guidance for reading such files into a geopandas data frame. Let's see if it works.
[Hurricane Tracks:](https://blog.matthewgove.com/2021/06/11/python-geopandas-easily-create-stunning-maps-without-a-gis-program/) This might be helpful in layering patches for the extent of the middle world.
[Geopandas Reference:](https://geopandas.org/en/stable/index.html)
```
import geopandas as gpd
import geoplot as gp
import matplotlib.pyplot as plt
from matplotlib.colors import colorConverter
from IPython.display import display, Image
from shapely.geometry import Point
```
### Global Geopandas Data Sources
The datasets given below are broadly available for general use. Seems potentially useful for drawing low res maps of the world.
```
gpd.datasets.available
```
### Barcelona Geojson Data Sets
The bcn-geodata contains a multitude of geographic data sets in geojson format. The 2020 data was imported from [this github](https://github.com/martgnz/bcn-geodata).
A range of other data is available on the [Barcelona municipal open data site](https://opendata-ajuntament.barcelona.cat/en/) which seems like an AWESOME resource. This is the [mapping article](https://diegoquintanav.github.io/folium-barcelona-EN.html) that got me turned on to the data. Need to explore more for sure.
Next two cells read some data in from the database and then print out the .crs (coordinate reference systems) for the dataframe and the type of dataframe to be sure everything is on track.
[Geopandas Projection Info:](https://geopandas.org/en/stable/docs/user_guide/projections.html) There were some issues with the projections at various points in the analysis so itn was suggested to project the data into a Mercator projection as per the linked documentation. The Mercator projection doesn't use lat/long so the geometry values are much larger for determining offsets. Actual units are meters!! Any lat/long information will need to be projected to [EPSG:3395](https://epsg.io/3395) for consistency.
```
path_to_earth = gpd.datasets.get_path("naturalearth_lowres")
path_to_cities = gpd.datasets.get_path("naturalearth_cities")
earthDF = gpd.read_file(path_to_earth)
citiesDF = gpd.read_file(path_to_cities)
# remove antarctica and reproject to Mercator
earthDF = earthDF[(earthDF.name != "Antarctica") & (earthDF.name != "Fr. S. Antarctic Lands")]
earthDF = earthDF.to_crs("EPSG:3395")
citiesDF = citiesDF.to_crs("EPSG:3395")
# create extra columns of needed info
# centroid is a method so centroid data is not in data base
earthDF['centroid_col'] = earthDF.centroid
# Need a label location for cities that can
# be manipulated relative to location for city.
# Some places I need to move label relative to city
citiesDF['label_location'] = citiesDF['geometry']
earthDF.centroid_col
citiesDF.columns
earthDF.columns
```
### ..and then there's plotting!
So much to figure out here and comment. Much learning happening - yikes!
To control the density of the facecolor differently than the density of the edgecolor a stackexchange thread suggested using colorConverter to define a face color WITH the alpha already packaged in. Note that I needed to import the colorConverter library to do this.
Realized that the plot is built up in layers so the order in which I display the layers affects which color is laid on top of the other. Thoughtful choices seem important here.
**Questions:**
* How do I identify the names of the specific regions. The names don't appear to be in the dataframe -- opening up the geojson file indicated that there
```
# continents to help sort through naming conventions
#europe = earthDF[earthDF['continent'] == "Europe"]
#asia = earthDF[earthDF['continent'] == "Asia"]
# countries of interest
# Southern Europe
portugal = earthDF[earthDF.name == 'Portugal']
spain = earthDF[earthDF.name == 'Spain']
france = earthDF[earthDF.name == 'France']
italy = earthDF[earthDF.name == 'Italy']
greece = earthDF[earthDF.name == 'Greece']
slovenia = earthDF[earthDF.name == 'Slovenia']
croatia = earthDF[earthDF.name == 'Croatia']
bosnia = earthDF[earthDF.name == 'Bosnia and Herz.']
montenegro = earthDF[earthDF.name == 'Montenegro']
albania = earthDF[earthDF.name == 'Albania']
austria = earthDF[earthDF.name == 'Austria']
serbia = earthDF[earthDF.name == 'Serbia']
macedonia = earthDF[earthDF.name == 'Macedonia']
kosovo = earthDF[earthDF.name == 'Kosovo']
bulgaria = earthDF[earthDF.name == 'Bulgaria']
balkans = croatia.append(bosnia).append(montenegro).append(albania).append(serbia).append(kosovo).append(macedonia)
# France is an multipolygon country. This set of steps breaks up and retains
# only those parts I want.
exploded = france.explode()
exploded.reset_index(drop = True, inplace= True)
exploded.drop(0, inplace = True)
france = exploded
france['centroid_col'] = exploded.centroid
france
# North Africa
libya = earthDF[earthDF.name == 'Libya']
egypt = earthDF[earthDF.name == 'Egypt']
algeria = earthDF[earthDF.name == 'Algeria']
tunisia = earthDF[earthDF.name == 'Tunisia']
morocco = earthDF[earthDF.name == 'Morocco']
# eastern middle world
india = earthDF[earthDF.name == 'India']
pakistan = earthDF[earthDF.name == 'Pakistan']
afghanistan = earthDF[earthDF.name == 'Afghanistan']
iran = earthDF[earthDF.name == 'Iran']
iraq = earthDF[earthDF.name == 'Iraq']
# Arabian Peninsula
saudiArabia = earthDF[earthDF.name == 'Saudi Arabia']
yemen = earthDF[earthDF.name == 'Yemen']
oman = earthDF[earthDF.name == 'Oman']
kuwait = earthDF[earthDF.name == 'Kuwait']
uae = earthDF[earthDF.name == 'United Arab Emirates']
# the Levant
israel = earthDF[earthDF.name == 'Israel']
lebanon = earthDF[earthDF.name == 'Lebanon']
syria = earthDF[earthDF.name == 'Syria']
jordan = earthDF[earthDF.name == 'Jordan']
# append to form regions
northAfrica = libya.append(algeria).append(egypt).append(tunisia).append(morocco)
southEurope = portugal.append(spain).append(france).append(italy).append(greece).append(slovenia)
southEurope = southEurope.append(balkans).append(austria).append(serbia).append(slovenia).append(bulgaria)
persiaIndia = india.append(pakistan).append(afghanistan).append(iran).append(iraq)
arabia = saudiArabia.append(kuwait).append(oman).append(uae).append(yemen)
levant = israel.append(lebanon).append(jordan).append(syria)
# tidy up indices which are from original file
northAfrica.reset_index(drop = True, inplace= True)
southEurope.reset_index(drop = True, inplace= True)
persiaIndia.reset_index(drop = True, inplace= True)
arabia.reset_index(drop = True, inplace= True)
levant.reset_index(drop = True, inplace= True)
northAfrica
citiesDF.label_location.x
southEurope
# scaleable offset for labels
labeloffset = .5
# cities of interest
# North Africa
cairo = citiesDF[citiesDF.name == 'Cairo']
tunis = citiesDF[citiesDF.name == 'Tunis']
tripoli = citiesDF[citiesDF.name == 'Tripoli']
algiers = citiesDF[citiesDF.name == 'Algiers']
#casablanca = citiesDF[citiesDF.name == 'Casablanca'] #apparently no Casablanca
rabat = citiesDF[citiesDF.name == 'Rabat']
# Southern Europe
paris = citiesDF[citiesDF.name == 'Paris']
lisbon = citiesDF[citiesDF.name == 'Lisbon']
madrid = citiesDF[citiesDF.name == 'Madrid']
rome = citiesDF[citiesDF.name == 'Rome']
athens = citiesDF[citiesDF.name == 'Athens']
# Eastern Middle World
kabul = citiesDF[citiesDF.name == 'Kabul']
tehran = citiesDF[citiesDF.name == 'Tehran']
baghdad = citiesDF[citiesDF.name == 'Baghdad']
islamabad = citiesDF[citiesDF.name == 'Islamabad']
newDelhi = citiesDF[citiesDF.name == 'New Delhi']
# Arabian Peninsula
riyadh = citiesDF[citiesDF.name == 'Riyadh']
sanaa = citiesDF[citiesDF.name == 'Sanaa']
muscat = citiesDF[citiesDF.name == 'Muscat']
abuDhabi = citiesDF[citiesDF.name == 'Abu Dhabi']
mecca = citiesDF[citiesDF.name == 'Mecca']
# move labels as needed
# Levant
jerusalem = citiesDF[citiesDF.name == 'Jerusalem']
beirut = citiesDF[citiesDF.name == 'Beirut']
damascus = citiesDF[citiesDF.name == 'Damascus']
amman = citiesDF[citiesDF.name == 'Amman']
# Group cities
citiesNA = cairo.append(tunis).append(tripoli).append(algiers).append(rabat)
citiesSE = paris.append(lisbon).append(madrid).append(rome).append(athens)
citiesMWE = kabul.append(tehran).append(baghdad).append(islamabad).append(newDelhi)
citiesAP = riyadh.append(sanaa).append(abuDhabi).append(mecca).append(muscat)
citiesLV = jerusalem.append(beirut).append(damascus).append(amman)
citiesNA.reset_index(drop = True, inplace= True)
citiesSE.reset_index(drop = True, inplace= True)
citiesMWE.reset_index(drop = True, inplace= True)
citiesAP.reset_index(drop = True, inplace= True)
citiesLV.reset_index(drop = True, inplace= True)
citiesSE
citiesAP.at[2, 'name']
citiesSE
```
### Name and City for a single country
Typically I pull the name from the name column and the centroid is the 'center of mass' of the polygon that represents the country
```
cities.geometry.x
fig, ax = plt.subplots(figsize=(24,16))
plotLabelOffset = 150000.
fc1 = colorConverter.to_rgba('green', alpha=0.2)
northAfrica.plot(ax=ax, cmap = 'viridis',edgecolor='k', linewidth=0.3, alpha = 0.2)
southEurope.plot(ax=ax, cmap = 'viridis',edgecolor='k', linewidth=0.3, alpha = 0.2)
citiesNA.plot(ax=ax, facecolor = 'r',edgecolor='r', linewidth=0.2)
citiesSE.plot(ax=ax, facecolor = 'r',edgecolor='r', linewidth=0.2)
for count,name in enumerate(northAfrica.name):
ax.annotate(text = name, xy = (northAfrica.centroid_col.x[count], northAfrica.centroid_col.y[count]), ha='center', fontsize=14)
for countC,nameC in enumerate(citiesNA.name):
ax.annotate(text = nameC, xy = (citiesNA.geometry.x[countC], citiesNA.geometry.y[countC] - plotLabelOffset), ha='center', fontsize=14)
for count,name in enumerate(southEurope.name):
ax.annotate(text = name, xy = (southEurope.centroid_col.x[count], southEurope.centroid_col.y[count] + plotLabelOffset/5.), ha='center', fontsize=14)
for countC,nameC in enumerate(citiesSE.name):
ax.annotate(text = nameC, xy = (citiesSE.geometry.x[countC], citiesSE.geometry.y[countC] - plotLabelOffset), ha='center', fontsize=14)
plt.title("Middle World (West)", fontsize=40, color='grey')
#plt.xlim(2.125, 2.150)
#plt.ylim(41.375, 41.400)
#plt.axis('equal')
plt.show()
```
### Eastern Half of Middle World
```
fig2, ax2 = plt.subplots(figsize=(24,16))
plotLabelOffsetB = 75000.
fc1 = colorConverter.to_rgba('green', alpha=0.2)
persiaIndia.plot(ax=ax2, cmap = 'viridis',edgecolor='k', linewidth=0.3, alpha = 0.2)
arabia.plot(ax=ax2, cmap = 'viridis',edgecolor='k', linewidth=0.3, alpha = 0.2)
levant.plot(ax=ax2, cmap = 'viridis',edgecolor='k', linewidth=0.3, alpha = 0.2)
citiesMWE.plot(ax=ax2, facecolor = 'r',edgecolor='r', linewidth=0.2)
citiesAP.plot(ax=ax2, facecolor = 'r',edgecolor='r', linewidth=0.2)
muscat.plot(ax=ax2, facecolor = 'r',edgecolor='r', linewidth=0.2)
citiesLV.plot(ax=ax2, facecolor = 'r',edgecolor='r', linewidth=0.2)
for count,name in enumerate(persiaIndia.name):
ax2.annotate(text = name, xy = (persiaIndia.centroid_col.x[count], persiaIndia.centroid_col.y[count]-plotLabelOffsetB), ha='center', fontsize=14)
for count,name in enumerate(arabia.name):
ax2.annotate(text = name, xy = (arabia.centroid_col.x[count], arabia.centroid_col.y[count] - plotLabelOffsetB), ha='center', fontsize=14)
for count,name in enumerate(levant.name):
ax2.annotate(text = name, xy = (levant.centroid_col.x[count], levant.centroid_col.y[count] - plotLabelOffsetB), ha='center', fontsize=14)
for countC,nameC in enumerate(citiesMWE.name):
ax2.annotate(text = nameC, xy = (citiesMWE.geometry.x[countC], citiesMWE.geometry.y[countC]+plotLabelOffsetB), ha='center', fontsize=14)
for countC,nameC in enumerate(citiesAP.name):
ax2.annotate(text = nameC, xy = (citiesAP.geometry.x[countC], citiesAP.geometry.y[countC]+plotLabelOffsetB), ha='center', fontsize=14)
for countC,nameC in enumerate(citiesLV.name):
ax2.annotate(text = nameC, xy = (citiesLV.geometry.x[countC], citiesLV.geometry.y[countC]+plotLabelOffsetB), ha='center', fontsize=14)
plt.title("Middle World (East)", fontsize=40, color='grey')
#plt.xlim(2.125, 2.150)
#plt.ylim(41.375, 41.400)
#plt.axis('equal')
plt.show()
```
|
github_jupyter
|
import geopandas as gpd
import geoplot as gp
import matplotlib.pyplot as plt
from matplotlib.colors import colorConverter
from IPython.display import display, Image
from shapely.geometry import Point
gpd.datasets.available
path_to_earth = gpd.datasets.get_path("naturalearth_lowres")
path_to_cities = gpd.datasets.get_path("naturalearth_cities")
earthDF = gpd.read_file(path_to_earth)
citiesDF = gpd.read_file(path_to_cities)
# remove antarctica and reproject to Mercator
earthDF = earthDF[(earthDF.name != "Antarctica") & (earthDF.name != "Fr. S. Antarctic Lands")]
earthDF = earthDF.to_crs("EPSG:3395")
citiesDF = citiesDF.to_crs("EPSG:3395")
# create extra columns of needed info
# centroid is a method so centroid data is not in data base
earthDF['centroid_col'] = earthDF.centroid
# Need a label location for cities that can
# be manipulated relative to location for city.
# Some places I need to move label relative to city
citiesDF['label_location'] = citiesDF['geometry']
earthDF.centroid_col
citiesDF.columns
earthDF.columns
# continents to help sort through naming conventions
#europe = earthDF[earthDF['continent'] == "Europe"]
#asia = earthDF[earthDF['continent'] == "Asia"]
# countries of interest
# Southern Europe
portugal = earthDF[earthDF.name == 'Portugal']
spain = earthDF[earthDF.name == 'Spain']
france = earthDF[earthDF.name == 'France']
italy = earthDF[earthDF.name == 'Italy']
greece = earthDF[earthDF.name == 'Greece']
slovenia = earthDF[earthDF.name == 'Slovenia']
croatia = earthDF[earthDF.name == 'Croatia']
bosnia = earthDF[earthDF.name == 'Bosnia and Herz.']
montenegro = earthDF[earthDF.name == 'Montenegro']
albania = earthDF[earthDF.name == 'Albania']
austria = earthDF[earthDF.name == 'Austria']
serbia = earthDF[earthDF.name == 'Serbia']
macedonia = earthDF[earthDF.name == 'Macedonia']
kosovo = earthDF[earthDF.name == 'Kosovo']
bulgaria = earthDF[earthDF.name == 'Bulgaria']
balkans = croatia.append(bosnia).append(montenegro).append(albania).append(serbia).append(kosovo).append(macedonia)
# France is an multipolygon country. This set of steps breaks up and retains
# only those parts I want.
exploded = france.explode()
exploded.reset_index(drop = True, inplace= True)
exploded.drop(0, inplace = True)
france = exploded
france['centroid_col'] = exploded.centroid
france
# North Africa
libya = earthDF[earthDF.name == 'Libya']
egypt = earthDF[earthDF.name == 'Egypt']
algeria = earthDF[earthDF.name == 'Algeria']
tunisia = earthDF[earthDF.name == 'Tunisia']
morocco = earthDF[earthDF.name == 'Morocco']
# eastern middle world
india = earthDF[earthDF.name == 'India']
pakistan = earthDF[earthDF.name == 'Pakistan']
afghanistan = earthDF[earthDF.name == 'Afghanistan']
iran = earthDF[earthDF.name == 'Iran']
iraq = earthDF[earthDF.name == 'Iraq']
# Arabian Peninsula
saudiArabia = earthDF[earthDF.name == 'Saudi Arabia']
yemen = earthDF[earthDF.name == 'Yemen']
oman = earthDF[earthDF.name == 'Oman']
kuwait = earthDF[earthDF.name == 'Kuwait']
uae = earthDF[earthDF.name == 'United Arab Emirates']
# the Levant
israel = earthDF[earthDF.name == 'Israel']
lebanon = earthDF[earthDF.name == 'Lebanon']
syria = earthDF[earthDF.name == 'Syria']
jordan = earthDF[earthDF.name == 'Jordan']
# append to form regions
northAfrica = libya.append(algeria).append(egypt).append(tunisia).append(morocco)
southEurope = portugal.append(spain).append(france).append(italy).append(greece).append(slovenia)
southEurope = southEurope.append(balkans).append(austria).append(serbia).append(slovenia).append(bulgaria)
persiaIndia = india.append(pakistan).append(afghanistan).append(iran).append(iraq)
arabia = saudiArabia.append(kuwait).append(oman).append(uae).append(yemen)
levant = israel.append(lebanon).append(jordan).append(syria)
# tidy up indices which are from original file
northAfrica.reset_index(drop = True, inplace= True)
southEurope.reset_index(drop = True, inplace= True)
persiaIndia.reset_index(drop = True, inplace= True)
arabia.reset_index(drop = True, inplace= True)
levant.reset_index(drop = True, inplace= True)
northAfrica
citiesDF.label_location.x
southEurope
# scaleable offset for labels
labeloffset = .5
# cities of interest
# North Africa
cairo = citiesDF[citiesDF.name == 'Cairo']
tunis = citiesDF[citiesDF.name == 'Tunis']
tripoli = citiesDF[citiesDF.name == 'Tripoli']
algiers = citiesDF[citiesDF.name == 'Algiers']
#casablanca = citiesDF[citiesDF.name == 'Casablanca'] #apparently no Casablanca
rabat = citiesDF[citiesDF.name == 'Rabat']
# Southern Europe
paris = citiesDF[citiesDF.name == 'Paris']
lisbon = citiesDF[citiesDF.name == 'Lisbon']
madrid = citiesDF[citiesDF.name == 'Madrid']
rome = citiesDF[citiesDF.name == 'Rome']
athens = citiesDF[citiesDF.name == 'Athens']
# Eastern Middle World
kabul = citiesDF[citiesDF.name == 'Kabul']
tehran = citiesDF[citiesDF.name == 'Tehran']
baghdad = citiesDF[citiesDF.name == 'Baghdad']
islamabad = citiesDF[citiesDF.name == 'Islamabad']
newDelhi = citiesDF[citiesDF.name == 'New Delhi']
# Arabian Peninsula
riyadh = citiesDF[citiesDF.name == 'Riyadh']
sanaa = citiesDF[citiesDF.name == 'Sanaa']
muscat = citiesDF[citiesDF.name == 'Muscat']
abuDhabi = citiesDF[citiesDF.name == 'Abu Dhabi']
mecca = citiesDF[citiesDF.name == 'Mecca']
# move labels as needed
# Levant
jerusalem = citiesDF[citiesDF.name == 'Jerusalem']
beirut = citiesDF[citiesDF.name == 'Beirut']
damascus = citiesDF[citiesDF.name == 'Damascus']
amman = citiesDF[citiesDF.name == 'Amman']
# Group cities
citiesNA = cairo.append(tunis).append(tripoli).append(algiers).append(rabat)
citiesSE = paris.append(lisbon).append(madrid).append(rome).append(athens)
citiesMWE = kabul.append(tehran).append(baghdad).append(islamabad).append(newDelhi)
citiesAP = riyadh.append(sanaa).append(abuDhabi).append(mecca).append(muscat)
citiesLV = jerusalem.append(beirut).append(damascus).append(amman)
citiesNA.reset_index(drop = True, inplace= True)
citiesSE.reset_index(drop = True, inplace= True)
citiesMWE.reset_index(drop = True, inplace= True)
citiesAP.reset_index(drop = True, inplace= True)
citiesLV.reset_index(drop = True, inplace= True)
citiesSE
citiesAP.at[2, 'name']
citiesSE
cities.geometry.x
fig, ax = plt.subplots(figsize=(24,16))
plotLabelOffset = 150000.
fc1 = colorConverter.to_rgba('green', alpha=0.2)
northAfrica.plot(ax=ax, cmap = 'viridis',edgecolor='k', linewidth=0.3, alpha = 0.2)
southEurope.plot(ax=ax, cmap = 'viridis',edgecolor='k', linewidth=0.3, alpha = 0.2)
citiesNA.plot(ax=ax, facecolor = 'r',edgecolor='r', linewidth=0.2)
citiesSE.plot(ax=ax, facecolor = 'r',edgecolor='r', linewidth=0.2)
for count,name in enumerate(northAfrica.name):
ax.annotate(text = name, xy = (northAfrica.centroid_col.x[count], northAfrica.centroid_col.y[count]), ha='center', fontsize=14)
for countC,nameC in enumerate(citiesNA.name):
ax.annotate(text = nameC, xy = (citiesNA.geometry.x[countC], citiesNA.geometry.y[countC] - plotLabelOffset), ha='center', fontsize=14)
for count,name in enumerate(southEurope.name):
ax.annotate(text = name, xy = (southEurope.centroid_col.x[count], southEurope.centroid_col.y[count] + plotLabelOffset/5.), ha='center', fontsize=14)
for countC,nameC in enumerate(citiesSE.name):
ax.annotate(text = nameC, xy = (citiesSE.geometry.x[countC], citiesSE.geometry.y[countC] - plotLabelOffset), ha='center', fontsize=14)
plt.title("Middle World (West)", fontsize=40, color='grey')
#plt.xlim(2.125, 2.150)
#plt.ylim(41.375, 41.400)
#plt.axis('equal')
plt.show()
fig2, ax2 = plt.subplots(figsize=(24,16))
plotLabelOffsetB = 75000.
fc1 = colorConverter.to_rgba('green', alpha=0.2)
persiaIndia.plot(ax=ax2, cmap = 'viridis',edgecolor='k', linewidth=0.3, alpha = 0.2)
arabia.plot(ax=ax2, cmap = 'viridis',edgecolor='k', linewidth=0.3, alpha = 0.2)
levant.plot(ax=ax2, cmap = 'viridis',edgecolor='k', linewidth=0.3, alpha = 0.2)
citiesMWE.plot(ax=ax2, facecolor = 'r',edgecolor='r', linewidth=0.2)
citiesAP.plot(ax=ax2, facecolor = 'r',edgecolor='r', linewidth=0.2)
muscat.plot(ax=ax2, facecolor = 'r',edgecolor='r', linewidth=0.2)
citiesLV.plot(ax=ax2, facecolor = 'r',edgecolor='r', linewidth=0.2)
for count,name in enumerate(persiaIndia.name):
ax2.annotate(text = name, xy = (persiaIndia.centroid_col.x[count], persiaIndia.centroid_col.y[count]-plotLabelOffsetB), ha='center', fontsize=14)
for count,name in enumerate(arabia.name):
ax2.annotate(text = name, xy = (arabia.centroid_col.x[count], arabia.centroid_col.y[count] - plotLabelOffsetB), ha='center', fontsize=14)
for count,name in enumerate(levant.name):
ax2.annotate(text = name, xy = (levant.centroid_col.x[count], levant.centroid_col.y[count] - plotLabelOffsetB), ha='center', fontsize=14)
for countC,nameC in enumerate(citiesMWE.name):
ax2.annotate(text = nameC, xy = (citiesMWE.geometry.x[countC], citiesMWE.geometry.y[countC]+plotLabelOffsetB), ha='center', fontsize=14)
for countC,nameC in enumerate(citiesAP.name):
ax2.annotate(text = nameC, xy = (citiesAP.geometry.x[countC], citiesAP.geometry.y[countC]+plotLabelOffsetB), ha='center', fontsize=14)
for countC,nameC in enumerate(citiesLV.name):
ax2.annotate(text = nameC, xy = (citiesLV.geometry.x[countC], citiesLV.geometry.y[countC]+plotLabelOffsetB), ha='center', fontsize=14)
plt.title("Middle World (East)", fontsize=40, color='grey')
#plt.xlim(2.125, 2.150)
#plt.ylim(41.375, 41.400)
#plt.axis('equal')
plt.show()
| 0.245899 | 0.980747 |
# Tools for verifying that every record is correctly processed and saved
## Initialize database files
Manually resets the master.db file which results are saved into
```
%cd twitteranalysis
from DataTools.SqliteTools import initialize_master_db, delete_master_db, initialize_word_map_db
import environment
delete_master_db()
initialize_master_db()
initialize_word_map_db(environment.TWEET_DB_NO_STOP)
test = '%s/user-test.db' % environment.LOG_FOLDER_PATH
initialize_word_map_db(test)
```
## Run the user parser
```
%cd twitteranalysis
%run -i Executables/process_user_descriptions_into_words2.py
```
## Manually clear server queue
This should not normally be needed.
```
%cd twitteranalysis
from Servers.ClientSide import Client
c = Client()
# Each of the listening request handlers needs its queue flushed
c.send_flush_command()
#j = c.send_shutdown_command()
j
# add_indexes(environment.TWEET_DB_MASTER)
```
# Check integrity of saved data
```
%cd twitteranalysis
import environment
import sqlite3
from DataTools import SqliteDataTools as DT
actualUsers = 1328927
numberProcessed = 4352
numberEmpty = 332
expectedUsers = numberProcessed - numberEmpty
print('should have %s' % expectedUsers)
DT.count_rows(environment.TWEET_DB_NO_STOP)
%%time
count_words(environment.USER_DB_NO_STOP)
# environment.USER_DB_NO_STOP
# 11.4 without index
# 6.31 with index
DT.count_rows(environment.TWEET_DB_MASTER)
DT.count_tweets(environment.TWEET_DB_MASTER)
num_users = DT.count_users()
missing = expectedUsers - num_users
pct_problem = missing / numberProcessed
expected_missing = round(actualUsers * pct_problem)
print("%s users were not saved; this is %s pct of the total processed" %( missing, pct_problem))
print("Projecting %s problem cases" % expected_missing)
```
NB:
- numberUsers processed = 4352
- users w empty descriptions = 332
- users non-english = 129
- expected users: 4020 (less empty)
5/15 6.06
- master only
- 3984 unique ids based on master
- 67500 rows in master
- 36 missing
- 10993 projected problems
5/15 1.36
- lock added to sqlite writer
- 4085 unique ids based on files
- 3950 unique ids based on master
- 67002 rows in master and based on files
5/15 11.12
- 4059 unique ids based on files
- 3923 unique ids based on master
- 66507 rows in master and based on files
5/15 10.39
- 3653 unique ids based on files
- 3529 unique ids based on master
- 60060 rows in master and based on files
5/14 9.00
- 4014 unique ids based on files
- 2510 unique ids based on master
- 41973 rows in master and based on files
Before changed to class method w separate call to flush
- 3591 Unique user ids
- 47952 rows in master.db
After
- 4014 Unique user ids
- 67940 rows in master.db
### Figure out which users are missing
```
%cd twitteranalysis
import sqlite3
import environment
import DataTools.Cursors
cursor = DataTools.Cursors.WindowedUserCursor( language='en' )
cursor.next()
def find_missing_users():
"""Finds users which were not saved to master.db"""
missing = []
try:
conn = sqlite3.connect(environment.MASTER_DB)
curs = conn.cursor() # Connect a cursor
while True:
user = cursor.next()
q = "select * from word_map_deux where user_id = %s" % user.userID
r1 = curs.execute(q)
r = r1.fetchone()
if r is None:
missing.append(user.userID)
except StopIteration:
curs.close()
conn.close()
return missing
missing = find_missing_users()
print(len(missing))
import pandas as pd
from DataTools.DataConnections import MySqlConnection, DAO
conn = MySqlConnection(environment.CREDENTIAL_FILE)
conn._make_engine()
def get_description_for_id(userId):
"""Loads the description from master.db
Returns a tuple (userId, description)
"""
q = 'select description from users where userID = %s' % userId
v = pd.read_sql_query(q, conn.engine).iloc[0].values[0]
return (userId, v)
# figure out which users were not missing due to an empty profile
descripts = []
# get the descriptions for each user
for userId in missing:
descripts.append(get_description_for_id(userId))
# determine which are substantive problems
substantive = [x for x in descripts if x[1] != '']
substantive_ids = [x[0] for x in descripts if x[1] != '']
print("%s users had non-empty profiles but were not saved. These are 'substantive errors'" % len(substantive))
substantive
names=['timestamp', 'userid', 'note']
# when a user was enqued for processing on client
proc = pd.read_csv(environment.PROCESSING_ENQUE_LOG_FILE, header=None, names=names)
# when a user was enqued for saving on the client
enq = pd.read_csv(environment.CLIENT_ENQUE_LOG_FILE, header=None, names=names)
# when a user was sent to the server
sent = pd.read_csv(environment.CLIENT_SEND_LOG_FILE, header=None, names=names)
# when the server received each request
srv = pd.read_csv(environment.SERVER_RECEIVE_LOG_FILE, header=None, names=names)
print("%s users processed; %s users received by server" % (len(proc), len(set(srv.userid.tolist()))))
def h(frame, userId):
try:
return frame[frame.userid == userId].index[0]
except:
return None
def get_indexes(userId):
d = {'id': userId}
d['processed'] = h(proc, userId)
d['clientEnque'] = h(enq, userId)
d['sent'] = h(sent, userId)
d['received'] = h(srv, userId)
return d
get_indexes(1956700424)
proc[proc.userid == 1956700424].index[0]
sent[sent.userid == 1956700424].index
```
# figure out where in the process the substantive errors happened
## substantive errors which were enqued in processing
```
p = proc[proc.userid.isin(substantive_ids)]
# sub
s = srv[srv.userid.isin(substantive_ids)]
snt = sent[sent.userid.isin(substantive_ids)]
ceq = enq[enq.userid.isin(substantive_ids)]
processed_ids = set(p.userid.tolist())
server_received_ids = set(s.userid.tolist())
sent_ids = set(snt.userid.tolist())
client_enq_ids = set(ceq.userid.tolist())
print("The following concerns the flow of users with substative errors through the system")
print("%s were enqueued for processing" % (len(processed_ids)))
print("%s were enqueued on the client to be sent to the server" % len(client_enq_ids))
print("%s were sent to the server" % len(sent_ids))
print("%s were received by the server" % len(server_received_ids))
```
### Processed but not enqueued for saving
```
proc_not_enq = [p for p in processed_ids if p not in client_enq_ids]
proc_not_enq
[get_description_for_id(id) for id in proc_not_enq]
```
### Enqueued on client but not sent
```
not_sent = [p for p in client_enq_ids if p not in sent_ids ]
not_sent
[get_description_for_id(id) for id in not_sent]
d = pd.DataFrame([get_indexes(id) for id in not_sent])
d.set_index('id', inplace=True)
d.sort_values('processed')
```
### Are these the same users each time?
```
prev_run = [1956700424.0, 1965229962.0,
1943096588.0,
2150423437.0,
2163358988.0,
1943901734.0,
2163604778.0,
1946121392.0,
1958085936.0,
2147790896.0,
2167298995.0,
2148304566.0,
2151409467.0,
2177120316.0,
1966904126.0,
1977458240.0,
1978158529.0,
2168963268.0,
1967229895.0,
1952156365.0,
1974223567.0,
1961129809.0,
1947484375.0,
2157188568.0,
1942653919.0,
2187999841.0,
2153422184.0,
2153945834.0,
2148022776.0,
1971054716.0]
[x for x in not_sent if x in prev_run]
processed_and_received_sub_errors = processed_ids.intersection(server_received_ids)
len(processed_and_received_sub_errors)
# these were processed by client and received by server
# but were not recorded
j = [x for x in substantive if x[0] in processed_and_received_sub_errors]
j
```
EXCLUSIVE
- 30 seconds
- no server side queue induced errors
IMMEDIATE
- 30 seconds
- no server side queue induced errors
DEFERRED
- 30 seconds
- no server side queue induced errors
Default (bare BEGIN)
- 29 seconds
- 60 server side errors
Autocommit
- Long
- 258 server side errors
```
len(sent_ids)
client_enq_ids
def c(row):
return p.i[row.index + 1].timestamp - row.timestamp
j = p.apply(lambda x: c(x))
p
```
uids = []
rows = []
print("Unique user ids; rows")
for db in otherDbNames:
dbPath = '%s/%s' % (environment.DB_FOLDER, db)
conn = sqlite3.connect( dbPath ) # Connect to the main database
curs = conn.cursor() # Connect a cursor
r1 = conn.execute("select count( distinct user_id) from word_map_deux")
v = r1.fetchone()
uids.append(v[0])
r2 = conn.execute("select count( word) from word_map_deux")
v2 = r2.fetchone()
rows.append(v2[0])
print("%s : %s; %s " % (db, v[0], v2[0]))
conn.close()
|
github_jupyter
|
%cd twitteranalysis
from DataTools.SqliteTools import initialize_master_db, delete_master_db, initialize_word_map_db
import environment
delete_master_db()
initialize_master_db()
initialize_word_map_db(environment.TWEET_DB_NO_STOP)
test = '%s/user-test.db' % environment.LOG_FOLDER_PATH
initialize_word_map_db(test)
%cd twitteranalysis
%run -i Executables/process_user_descriptions_into_words2.py
%cd twitteranalysis
from Servers.ClientSide import Client
c = Client()
# Each of the listening request handlers needs its queue flushed
c.send_flush_command()
#j = c.send_shutdown_command()
j
# add_indexes(environment.TWEET_DB_MASTER)
%cd twitteranalysis
import environment
import sqlite3
from DataTools import SqliteDataTools as DT
actualUsers = 1328927
numberProcessed = 4352
numberEmpty = 332
expectedUsers = numberProcessed - numberEmpty
print('should have %s' % expectedUsers)
DT.count_rows(environment.TWEET_DB_NO_STOP)
%%time
count_words(environment.USER_DB_NO_STOP)
# environment.USER_DB_NO_STOP
# 11.4 without index
# 6.31 with index
DT.count_rows(environment.TWEET_DB_MASTER)
DT.count_tweets(environment.TWEET_DB_MASTER)
num_users = DT.count_users()
missing = expectedUsers - num_users
pct_problem = missing / numberProcessed
expected_missing = round(actualUsers * pct_problem)
print("%s users were not saved; this is %s pct of the total processed" %( missing, pct_problem))
print("Projecting %s problem cases" % expected_missing)
%cd twitteranalysis
import sqlite3
import environment
import DataTools.Cursors
cursor = DataTools.Cursors.WindowedUserCursor( language='en' )
cursor.next()
def find_missing_users():
"""Finds users which were not saved to master.db"""
missing = []
try:
conn = sqlite3.connect(environment.MASTER_DB)
curs = conn.cursor() # Connect a cursor
while True:
user = cursor.next()
q = "select * from word_map_deux where user_id = %s" % user.userID
r1 = curs.execute(q)
r = r1.fetchone()
if r is None:
missing.append(user.userID)
except StopIteration:
curs.close()
conn.close()
return missing
missing = find_missing_users()
print(len(missing))
import pandas as pd
from DataTools.DataConnections import MySqlConnection, DAO
conn = MySqlConnection(environment.CREDENTIAL_FILE)
conn._make_engine()
def get_description_for_id(userId):
"""Loads the description from master.db
Returns a tuple (userId, description)
"""
q = 'select description from users where userID = %s' % userId
v = pd.read_sql_query(q, conn.engine).iloc[0].values[0]
return (userId, v)
# figure out which users were not missing due to an empty profile
descripts = []
# get the descriptions for each user
for userId in missing:
descripts.append(get_description_for_id(userId))
# determine which are substantive problems
substantive = [x for x in descripts if x[1] != '']
substantive_ids = [x[0] for x in descripts if x[1] != '']
print("%s users had non-empty profiles but were not saved. These are 'substantive errors'" % len(substantive))
substantive
names=['timestamp', 'userid', 'note']
# when a user was enqued for processing on client
proc = pd.read_csv(environment.PROCESSING_ENQUE_LOG_FILE, header=None, names=names)
# when a user was enqued for saving on the client
enq = pd.read_csv(environment.CLIENT_ENQUE_LOG_FILE, header=None, names=names)
# when a user was sent to the server
sent = pd.read_csv(environment.CLIENT_SEND_LOG_FILE, header=None, names=names)
# when the server received each request
srv = pd.read_csv(environment.SERVER_RECEIVE_LOG_FILE, header=None, names=names)
print("%s users processed; %s users received by server" % (len(proc), len(set(srv.userid.tolist()))))
def h(frame, userId):
try:
return frame[frame.userid == userId].index[0]
except:
return None
def get_indexes(userId):
d = {'id': userId}
d['processed'] = h(proc, userId)
d['clientEnque'] = h(enq, userId)
d['sent'] = h(sent, userId)
d['received'] = h(srv, userId)
return d
get_indexes(1956700424)
proc[proc.userid == 1956700424].index[0]
sent[sent.userid == 1956700424].index
p = proc[proc.userid.isin(substantive_ids)]
# sub
s = srv[srv.userid.isin(substantive_ids)]
snt = sent[sent.userid.isin(substantive_ids)]
ceq = enq[enq.userid.isin(substantive_ids)]
processed_ids = set(p.userid.tolist())
server_received_ids = set(s.userid.tolist())
sent_ids = set(snt.userid.tolist())
client_enq_ids = set(ceq.userid.tolist())
print("The following concerns the flow of users with substative errors through the system")
print("%s were enqueued for processing" % (len(processed_ids)))
print("%s were enqueued on the client to be sent to the server" % len(client_enq_ids))
print("%s were sent to the server" % len(sent_ids))
print("%s were received by the server" % len(server_received_ids))
proc_not_enq = [p for p in processed_ids if p not in client_enq_ids]
proc_not_enq
[get_description_for_id(id) for id in proc_not_enq]
not_sent = [p for p in client_enq_ids if p not in sent_ids ]
not_sent
[get_description_for_id(id) for id in not_sent]
d = pd.DataFrame([get_indexes(id) for id in not_sent])
d.set_index('id', inplace=True)
d.sort_values('processed')
prev_run = [1956700424.0, 1965229962.0,
1943096588.0,
2150423437.0,
2163358988.0,
1943901734.0,
2163604778.0,
1946121392.0,
1958085936.0,
2147790896.0,
2167298995.0,
2148304566.0,
2151409467.0,
2177120316.0,
1966904126.0,
1977458240.0,
1978158529.0,
2168963268.0,
1967229895.0,
1952156365.0,
1974223567.0,
1961129809.0,
1947484375.0,
2157188568.0,
1942653919.0,
2187999841.0,
2153422184.0,
2153945834.0,
2148022776.0,
1971054716.0]
[x for x in not_sent if x in prev_run]
processed_and_received_sub_errors = processed_ids.intersection(server_received_ids)
len(processed_and_received_sub_errors)
# these were processed by client and received by server
# but were not recorded
j = [x for x in substantive if x[0] in processed_and_received_sub_errors]
j
len(sent_ids)
client_enq_ids
def c(row):
return p.i[row.index + 1].timestamp - row.timestamp
j = p.apply(lambda x: c(x))
p
| 0.364438 | 0.717798 |
# リアルタイム推論サービスを作成する
予測モデルのトレーニング後、クライアントが新しいデータから予測を取得するために使用できるリアルタイム サービスとしてモデルをデプロイできます。
## ワークスペースに接続する
作業を開始するには、ワークスペースに接続します。
> **注**: Azure サブスクリプションでまだ認証済みのセッションを確立していない場合は、リンクをクリックして認証コードを入力し、Azure にサインインして認証するよう指示されます。
```
import azureml.core
from azureml.core import Workspace
# 保存された構成ファイルからワークスペースを読み込む
ws = Workspace.from_config()
print('Ready to use Azure ML {} to work with {}'.format(azureml.core.VERSION, ws.name))
```
## モデルをトレーニングして登録する
それでは、モデルをトレーニングして登録しましょう。
```
from azureml.core import Experiment
from azureml.core import Model
import pandas as pd
import numpy as np
import joblib
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
# ワークスペースで Azure 実験を作成する
experiment = Experiment(workspace=ws, name="mslearn-train-diabetes")
run = experiment.start_logging()
print("Starting experiment:", experiment.name)
# 糖尿病データセットを読み込む
print("Loading Data...")
diabetes = pd.read_csv('data/diabetes.csv')
# 特徴とラベルを分離する
X, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values
# データをトレーニング セットとテスト セットに分割する
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)
# デシジョン ツリー モデルをトレーニングする
print('Training a decision tree model')
model = DecisionTreeClassifier().fit(X_train, y_train)
# 精度を計算する
y_hat = model.predict(X_test)
acc = np.average(y_hat == y_test)
print('Accuracy:', acc)
run.log('Accuracy', np.float(acc))
# AUC を計算する
y_scores = model.predict_proba(X_test)
auc = roc_auc_score(y_test,y_scores[:,1])
print('AUC: ' + str(auc))
run.log('AUC', np.float(auc))
# トレーニング済みモデルを保存する
model_file = 'diabetes_model.pkl'
joblib.dump(value=model, filename=model_file)
run.upload_file(name = 'outputs/' + model_file, path_or_stream = './' + model_file)
# 実行を完了する
run.complete()
# モデルを登録する
run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',
tags={'Training context':'Inline Training'},
properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})
print('Model trained and registered.')
```
## モデルを Web サービスとして公開する
糖尿病の可能性に基づいて患者を分類する機械学習モデルをトレーニングし、登録しました。このモデルは、糖尿病の臨床検査を受ける必要があるとリスクがあると考えられる患者のみが必要な医師の手術などの運用環境で使用できます。このシナリオをサポートするには、モデルを Web サービスとしてデプロイします。
まず、ワークスペースに登録したモデルを決定しましょう。
```
from azureml.core import Model
for model in Model.list(ws):
print(model.name, 'version:', model.version)
for tag_name in model.tags:
tag = model.tags[tag_name]
print ('\t',tag_name, ':', tag)
for prop_name in model.properties:
prop = model.properties[prop_name]
print ('\t',prop_name, ':', prop)
print('\n')
```
それでは、デプロイしたいモデルを取得しましょう。既定では、モデル名を指定すると、最新バージョンが返されます。
```
model = ws.models['diabetes_model']
print(model.name, 'version', model.version)
```
このモデルをホストする Web サービスを作成しますが、これにはコードと構成ファイルが必要です。そのため、それらのフォルダーを作成してみましょう。
```
import os
folder_name = 'diabetes_service'
# Web サービス ファイル用フォルダーを作成する
experiment_folder = './' + folder_name
os.makedirs(experiment_folder, exist_ok=True)
print(folder_name, 'folder created.')
# スクリプトと環境ファイルをスコアリングするためのパスを設定する
script_file = os.path.join(experiment_folder,"score_diabetes.py")
env_file = os.path.join(experiment_folder,"diabetes_env.yml")
```
モデルをデプロイする Web サービスでは、入力データを読み込み、ワークスペースからモデルを取得し、予測を生成して返すために、Python コードが必要になります。このコードは、Web サービスにデプロイされる*エントリ スクリプト* (頻繁に*スコアリング スクリプト*と呼ばれます) に保存します。
```
%%writefile $script_file
import json
import joblib
import numpy as np
from azureml.core.model import Model
# サービスの読み込み時に呼び出される
def init():
global model
# Get the path to the deployed model file and load it
model_path = Model.get_model_path('diabetes_model')
model = joblib.load(model_path)
# 要求の受信時に呼び出される
def run(raw_data):
# Get the input data as a numpy array
data = np.array(json.loads(raw_data)['data'])
# Get a prediction from the model
predictions = model.predict(data)
# Get the corresponding classname for each prediction (0 or 1)
classnames = ['not-diabetic', 'diabetic']
predicted_classes = []
for prediction in predictions:
predicted_classes.append(classnames[prediction])
# Return the predictions as JSON
return json.dumps(predicted_classes)
```
Web サービスはコンテナーでホストされ、コンテナーは初期化されるときに必要な Python 依存関係をインストールする必要があります。この場合、スコアリング コードには **scikit-learn** と **azureml-defaults** パッケージが必要なので、コンテナー ホストに環境にインストールするよう指示する .yml ファイルを作成します。
```
%%writefile $env_file
name: inference_env
dependencies:
- python=3.6.2
- scikit-learn
- pip
- pip:
- azureml-defaults
```
これでデプロイする準備ができました。コンテナーに **diabetes-service**.という名前のサービスをデプロイします。デプロイ プロセスには、次のステップが含まれます。
1. モデルの読み込みと使用に必要なスコアリング ファイルと環境ファイルを含む推論構成を定義します。
2. サービスをホストする実行環境を定義するデプロイメント構成を定義します。この場合、Azure Container Instances。
3. モデルを Web サービスとしてデプロイする
4. デプロイされたサービスの状態を確認します。
> **詳細情報**: モデル デプロイ、ターゲット実行環境のオプションの詳細については、[ドキュメント](https://docs.microsoft.com/azure/machine-learning/how-to-deploy-and-where)を参照してください。
デプロイは、最初にコンテナー イメージを作成するプロセスを実行し、そのイメージに基づいて Web サービスを作成するプロセスを実行するため、時間がかかります。デプロイが正常に完了すると、**正常**な状態が表示されます。
```
from azureml.core.webservice import AciWebservice
from azureml.core.model import InferenceConfig
# スコアリング環境を構成する
inference_config = InferenceConfig(runtime= "python",
entry_script=script_file,
conda_file=env_file)
deployment_config = AciWebservice.deploy_configuration(cpu_cores = 1, memory_gb = 1)
service_name = "diabetes-service"
service = Model.deploy(ws, service_name, [model], inference_config, deployment_config)
service.wait_for_deployment(True)
print(service.state)
```
うまくいけば、デプロイが成功し、**正常**な状態を確認できます。確認できない場合は、次のコードを使用して、トラブルシューティングに役立つサービス ログを取得できます。
```
print(service.get_logs())
# 変更を行って再デプロイする必要がある場合は、次のコードを使用して異常なサービスを削除することが必要となる可能性があります。
#service.delete()
```
[Azure Machine Learning Studio](https://ml.azure.com) でワークスペースを確認し、ワークスペースにデプロイされたサービスを示す**エンドポイント**ページを表示します。
次のコードを実行して、ワークスペース内の Web サービスの名前を取得することもできます。
```
for webservice_name in ws.webservices:
print(webservice_name)
```
## Web サービスを使用する
サービスをデプロイしたら、クライアント アプリケーションからサービスを使用できます。
```
import json
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22]]
print ('Patient: {}'.format(x_new[0]))
# JSON ドキュメントでシリアル化可能なリストに配列を変換する
input_json = json.dumps({"data": x_new})
# 入力データを渡して Web サービスを呼び出す (Web サービスはバイナリ形式のデータも受け入れます)
predictions = service.run(input_data = input_json)
# 予測されたクラスを取得する - それは最初の (そして唯一の) クラスになります。
predicted_classes = json.loads(predictions)
print(predicted_classes[0])
```
また、複数の患者の観察をサービスに送信し、それぞれの予測を取得することもできます。
```
import json
# 今回の入力は、2 つの特徴配列のひとつです。
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22],
[0,148,58,11,179,39.19207553,0.160829008,45]]
# JSON ドキュメント内のシリアル化可能なリストに配列を変換する
input_json = json.dumps({"data": x_new})
# Web サービスを呼び出して入力データを渡す
predictions = service.run(input_data = input_json)
# 予測されたクラスを取得する
predicted_classes = json.loads(predictions)
for i in range(len(x_new)):
print ("Patient {}".format(x_new[i]), predicted_classes[i] )
```
上記のコードでは、Azure Machine Learning SDK を使用してコンテナー化された Web サービスに接続し、それを使用して糖尿病分類モデルから予測を生成しています。運用環境では、Azure Machine Learning SDK を使用せず、単に Web サービスに HTTP 要求を行うビジネス アプリケーションによってモデルが使用される可能性があります。
これらのアプリケーションが要求を送信する必要がある URL を決定しましょう。
```
endpoint = service.scoring_uri
print(endpoint)
```
エンドポイント URI がわかったので、アプリケーションは HTTP 要求を行い、患者データを JSON 形式で送信し、予測されたクラスを受け取ることができます。
```
import requests
import json
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22],
[0,148,58,11,179,39.19207553,0.160829008,45]]
# JSON ドキュメントでシリアル化可能なリストに配列を変換する
input_json = json.dumps({"data": x_new})
# コンテンツ タイプを設定する
headers = { 'Content-Type':'application/json' }
predictions = requests.post(endpoint, input_json, headers = headers)
predicted_classes = json.loads(predictions.json())
for i in range(len(x_new)):
print ("Patient {}".format(x_new[i]), predicted_classes[i] )
```
認証を必要としない Azure Container Instances (ACI) サービスとして Web サービスをデプロイしました。これは開発とテストには適していますが、運用環境では Azure Kubernetes Service (AKS) クラスターへのデプロイとトークンベースの認証の有効化を検討する必要があります。これには、**Authorization** ヘッダーを含める REST 要求が必要です。
## サービスを削除する
サービスが不要になった場合は、不要な料金が発生しないように削除する必要があります。
```
service.delete()
print ('Service deleted.')
```
モデルをサービスとして公開する方法の詳細については、[ドキュメント](https://docs.microsoft.com/azure/machine-learning/how-to-deploy-and-where)を参照してください。
|
github_jupyter
|
import azureml.core
from azureml.core import Workspace
# 保存された構成ファイルからワークスペースを読み込む
ws = Workspace.from_config()
print('Ready to use Azure ML {} to work with {}'.format(azureml.core.VERSION, ws.name))
from azureml.core import Experiment
from azureml.core import Model
import pandas as pd
import numpy as np
import joblib
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
# ワークスペースで Azure 実験を作成する
experiment = Experiment(workspace=ws, name="mslearn-train-diabetes")
run = experiment.start_logging()
print("Starting experiment:", experiment.name)
# 糖尿病データセットを読み込む
print("Loading Data...")
diabetes = pd.read_csv('data/diabetes.csv')
# 特徴とラベルを分離する
X, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values
# データをトレーニング セットとテスト セットに分割する
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)
# デシジョン ツリー モデルをトレーニングする
print('Training a decision tree model')
model = DecisionTreeClassifier().fit(X_train, y_train)
# 精度を計算する
y_hat = model.predict(X_test)
acc = np.average(y_hat == y_test)
print('Accuracy:', acc)
run.log('Accuracy', np.float(acc))
# AUC を計算する
y_scores = model.predict_proba(X_test)
auc = roc_auc_score(y_test,y_scores[:,1])
print('AUC: ' + str(auc))
run.log('AUC', np.float(auc))
# トレーニング済みモデルを保存する
model_file = 'diabetes_model.pkl'
joblib.dump(value=model, filename=model_file)
run.upload_file(name = 'outputs/' + model_file, path_or_stream = './' + model_file)
# 実行を完了する
run.complete()
# モデルを登録する
run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',
tags={'Training context':'Inline Training'},
properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})
print('Model trained and registered.')
from azureml.core import Model
for model in Model.list(ws):
print(model.name, 'version:', model.version)
for tag_name in model.tags:
tag = model.tags[tag_name]
print ('\t',tag_name, ':', tag)
for prop_name in model.properties:
prop = model.properties[prop_name]
print ('\t',prop_name, ':', prop)
print('\n')
model = ws.models['diabetes_model']
print(model.name, 'version', model.version)
import os
folder_name = 'diabetes_service'
# Web サービス ファイル用フォルダーを作成する
experiment_folder = './' + folder_name
os.makedirs(experiment_folder, exist_ok=True)
print(folder_name, 'folder created.')
# スクリプトと環境ファイルをスコアリングするためのパスを設定する
script_file = os.path.join(experiment_folder,"score_diabetes.py")
env_file = os.path.join(experiment_folder,"diabetes_env.yml")
%%writefile $script_file
import json
import joblib
import numpy as np
from azureml.core.model import Model
# サービスの読み込み時に呼び出される
def init():
global model
# Get the path to the deployed model file and load it
model_path = Model.get_model_path('diabetes_model')
model = joblib.load(model_path)
# 要求の受信時に呼び出される
def run(raw_data):
# Get the input data as a numpy array
data = np.array(json.loads(raw_data)['data'])
# Get a prediction from the model
predictions = model.predict(data)
# Get the corresponding classname for each prediction (0 or 1)
classnames = ['not-diabetic', 'diabetic']
predicted_classes = []
for prediction in predictions:
predicted_classes.append(classnames[prediction])
# Return the predictions as JSON
return json.dumps(predicted_classes)
%%writefile $env_file
name: inference_env
dependencies:
- python=3.6.2
- scikit-learn
- pip
- pip:
- azureml-defaults
from azureml.core.webservice import AciWebservice
from azureml.core.model import InferenceConfig
# スコアリング環境を構成する
inference_config = InferenceConfig(runtime= "python",
entry_script=script_file,
conda_file=env_file)
deployment_config = AciWebservice.deploy_configuration(cpu_cores = 1, memory_gb = 1)
service_name = "diabetes-service"
service = Model.deploy(ws, service_name, [model], inference_config, deployment_config)
service.wait_for_deployment(True)
print(service.state)
print(service.get_logs())
# 変更を行って再デプロイする必要がある場合は、次のコードを使用して異常なサービスを削除することが必要となる可能性があります。
#service.delete()
for webservice_name in ws.webservices:
print(webservice_name)
import json
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22]]
print ('Patient: {}'.format(x_new[0]))
# JSON ドキュメントでシリアル化可能なリストに配列を変換する
input_json = json.dumps({"data": x_new})
# 入力データを渡して Web サービスを呼び出す (Web サービスはバイナリ形式のデータも受け入れます)
predictions = service.run(input_data = input_json)
# 予測されたクラスを取得する - それは最初の (そして唯一の) クラスになります。
predicted_classes = json.loads(predictions)
print(predicted_classes[0])
import json
# 今回の入力は、2 つの特徴配列のひとつです。
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22],
[0,148,58,11,179,39.19207553,0.160829008,45]]
# JSON ドキュメント内のシリアル化可能なリストに配列を変換する
input_json = json.dumps({"data": x_new})
# Web サービスを呼び出して入力データを渡す
predictions = service.run(input_data = input_json)
# 予測されたクラスを取得する
predicted_classes = json.loads(predictions)
for i in range(len(x_new)):
print ("Patient {}".format(x_new[i]), predicted_classes[i] )
endpoint = service.scoring_uri
print(endpoint)
import requests
import json
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22],
[0,148,58,11,179,39.19207553,0.160829008,45]]
# JSON ドキュメントでシリアル化可能なリストに配列を変換する
input_json = json.dumps({"data": x_new})
# コンテンツ タイプを設定する
headers = { 'Content-Type':'application/json' }
predictions = requests.post(endpoint, input_json, headers = headers)
predicted_classes = json.loads(predictions.json())
for i in range(len(x_new)):
print ("Patient {}".format(x_new[i]), predicted_classes[i] )
service.delete()
print ('Service deleted.')
| 0.435902 | 0.897291 |
# Lecture \#2: Setting up your Development Environment
Here is what I intend to cover today:
* Basics of Python
* What is Interactive Python (IPython)?
* What are Jupyter Notebooks?
* What are version control systems, and what is Git?
* What is GitHub?
* How do I share code through a version control system like Git?
At the end of this process, I would like for each of you to be able to create an Jupyter Notebook locally on your computer, and then be able to allow anyone else in the course (if pushing it to our course's repository) or in the world (if pushing it to a public repository) to see it.
This very same file we have on the screen now will make that journey.
## Before you begin
Things you'll need to do ahead of time:
1. Create an account on [github.com](http://github.com)
2. Install the [Anaconda Python distribution](https://www.anaconda.com/distribution/)
3. Install git on your computer, which you can get [here](http://git-scm.com/)
In addition to the references posted on the slide deck, here are some references that will be **very** helpful to ensure you understand what we are doing and how it all works:
1. Git References
* What it is and what is it used for?
* [Official Documentation](https://git-scm.com/documentation), especially the first three videos on this page.
* [Official Git Tutorial](https://git-scm.com/docs/gittutorial), if you are already familiar with the command line interface to some other version control software and just need to get started.
* How does it work?
* [Visual, interactive explanation](https://onlywei.github.io/explain-git-with-d3/): this is really valuable if you want to wrap your head around the basic of what's happening under the hood.
* [Git from the inside out](https://codewords.recurse.com/issues/two/git-from-the-inside-out): an in-depth discussion of how things really work under the hood.
2. Python References
* Why Python and how is it useful for for Scientific Computing?
* First, a quick intro to [Python for Scientific Computing](http://nbviewer.ipython.org/github/jrjohansson/scientific-python-lectures/blob/master/Lecture-0-Scientific-Computing-with-Python.ipynb)
* Other cool sources:
* [SciPy lecture notes](http://www.scipy-lectures.org/): heavy focus on Python fundamentals.
* [Quantitative Economics with Python](http://lectures.quantecon.org/): the name says it all.
* Online Courses:
* [Introduction to Scientifi Python](https://web.stanford.edu/class/cme193/) (Stanford)
* [Practical Data Science](http://datasciencecourse.org/) (CMU)
* [Computational Statistics in Python](http://people.duke.edu/~ccc14/sta-663/index.html) (Duke)
* How to use Python?
* Here's [a tutorial](http://nbviewer.ipython.org/github/jrjohansson/scientific-python-lectures/blob/master/Lecture-1-Introduction-to-Python-Programming.ipynb).
* Two libraries that we are going to be making extensive use of are numpy and matplotlib. The same person who wrote the tutorial above has tutorials for [numpy](http://nbviewer.ipython.org/github/jrjohansson/scientific-python-lectures/blob/master/Lecture-2-Numpy.ipynb) and [matplotlib](http://nbviewer.ipython.org/github/jrjohansson/scientific-python-lectures/blob/master/Lecture-4-Matplotlib.ipynb)
OK. Let's get you started.
## Cloning the course's git repository
This file you are currently viewing is part of the course's git repository, which you can find here:
[https://github.com/INFERLab/S22-12-770](https://github.com/INFERLab/S22-12-770)
Since it is in a private repository, you need to be invited as a contributor to be able to clone that repository into your computer and edit each file locally. So make sure you've shared your Github username with me before proceeding. Once you are a contributor, you could either clone it using the command line interface to git, or a graphical user interface (whichever you installed on your computer if you chose to install git). From the command line, for instance, you would issue this command to clone it:
```
git clone http://github.com/marioberges/S21-12-752.git
```
Make sure that you can clone the repository in your computer by issuing that command.
If you are successful, you will be able to see a new folder called `S22-12-770` inside the folder where you issued the command. A copy of this Jupyter Notebook file should be in there as well, and you can view it by opening an Jupyter Notebook Server as follows:
```
jupyter notebook
```
Just make sure you issue this last command on the corresponding folder.
# Creating and using your own repositories
The steps we followed above were for cloning the course's official repository. However, you will want to repeat these steps for any other repository you may be interested in working with, especially the ones that you end up creating under your Github account. Thus, let's practice importing one of your repositories.
Follow these steps:
1. Head over to github.com and log in using your credentials.
2. Create a new repository and name it whatever you like.
3. At the end of the process you will be given a checkout string. Copy that.
4. Use the checkout string to replace the one we used earlier that looked like this:
```
git clone http://github.com/yourusername/yourrepository.git
```
5. Try issuing that command on your computer (obviously, replacing `yourusername` and `yourrepository` with the right information)
6. If all goes well, you'll have your (empty) repository available for use in your computer.
Now it's time for you to practice some of your recently learned git skills.
Create a new Jupyter notebook, making sure to place it inside the folder of the repository you just cloned.
Add a couple of Python commands to it, or some comments, and save it.
Now go back to the terminal and add, commit and push the changes to your repository:
```
git add yourfile.ipynb
git commit -m "Made my first commit"
git push origin master
```
If this worked, you should be able to see the file added to your repository by simply pointing your browser to:
`http://github.com/yourusername/yourrepository`
## Doing away with the terminal
Because Jupyter can be used to issue commands to a shell, directly, you can avoid having to switch to a terminal screen if you want to. This means we could have performed all of the above git manipulation directly from this notebook. The trick is to create a *Code* cell (the default type of cells) in the Jupyter notebook and then issuing the commands preceded by a `!` sign, as follows:
```
!git status
```
Try running the above cell and see what you get.
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
np.arange
```
|
github_jupyter
|
git clone http://github.com/marioberges/S21-12-752.git
jupyter notebook
git clone http://github.com/yourusername/yourrepository.git
```
5. Try issuing that command on your computer (obviously, replacing `yourusername` and `yourrepository` with the right information)
6. If all goes well, you'll have your (empty) repository available for use in your computer.
Now it's time for you to practice some of your recently learned git skills.
Create a new Jupyter notebook, making sure to place it inside the folder of the repository you just cloned.
Add a couple of Python commands to it, or some comments, and save it.
Now go back to the terminal and add, commit and push the changes to your repository:
If this worked, you should be able to see the file added to your repository by simply pointing your browser to:
`http://github.com/yourusername/yourrepository`
## Doing away with the terminal
Because Jupyter can be used to issue commands to a shell, directly, you can avoid having to switch to a terminal screen if you want to. This means we could have performed all of the above git manipulation directly from this notebook. The trick is to create a *Code* cell (the default type of cells) in the Jupyter notebook and then issuing the commands preceded by a `!` sign, as follows:
Try running the above cell and see what you get.
| 0.637031 | 0.941223 |
```
%reload_ext autoreload
%autoreload 2
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
from loguru import logger
datapath = "data/data.csv"
```
# Load Data
> **Note**: It seems pandas couldn't read a large chunk of data. It's only able to load 6K data points.
I am sure I could have solved this, but the solution is just a crude way to approach the given problem for tagging a description. So, I let it be. (Plus, it was race against time)
### Important
If we're going to train a BERT-based Language Model, we will only use a subset of the data where
the length of the description < 512 (since BERT can only process 512 input tokens at a time).
> **NOTE**:
- This can be solved by treating classification of single data point as chunks where we could break the long text.
- Or, we can use other robust models like [Longformer](https://github.com/allenai/longformer) that can process
at max 4096 input characters.
```
def parse_kws(kw_str, level=2):
res = kw_str.split(",")
res = map(lambda kw: [_.strip().lower() for _ in kw.split(">")], res)
res = map(lambda x: x[level if level<len(x) else len(x)-1], res)
return list(set(res))
def load_data(path, level=0):
logger.info(f"Loading data from {path}. [KW Level={level}]")
df = pd.read_csv(path)
df["desc"] = df["desc"].apply(str.strip)
df["labels"] = df["keywords"].apply(lambda x: parse_kws(x, level))
df["textlen"] = df["desc"].apply(len)
return df
DATA = load_data(datapath, level=1)
# Like I said, we're only able to load 6K data points which can be fixed in future!
DATA.shape
DATA.head()
```
# Analysis
```
from collections import Counter, defaultdict
from loguru import logger
def analyze_labels(df):
df = df.copy()
labels = [l for ls in df["labels"] for l in ls]
uniques = set(labels)
logger.info(f"{len(uniques)} unique labels")
analyze_labels(DATA)
```
### Text Length Analysis
```
_data = DATA.copy()
_data = _data[_data["textlen"]>0]
logger.debug(_data.shape)
# BERT can only process 512 sequence length at once
# So, what % of text satisfy that pre-condition?
len(_data[_data["textlen"] <= 512]) / len(_data), len(_data[_data["textlen"] <= 1024]) / len(_data)
plt.figure(figsize=(20, 15))
sns.histplot(data=_data, x="textlen", bins=100).set(xlim=(0, 3000))
```
## MultiLabelEncoder Analysis
Since, the problem is a multi-label clasification (if we treat the problem as a classification problem), we need to see if we can encode the labels accordingly!
```
from sklearn.preprocessing import MultiLabelBinarizer
DATA_TO_USE = DATA.copy()
DATA_TO_USE = DATA_TO_USE[DATA_TO_USE["textlen"]<=500]
DATA_TO_USE.shape
DATA_TO_USE.head()
# Note: Now our level 1 keywords are <29 (N=22) classes because we have filtered the data
analyze_labels(DATA_TO_USE)
LE = MultiLabelBinarizer()
LABELS_ENCODED = LE.fit_transform(DATA_TO_USE["labels"])
# 22 classes for level=1 keywords
LABELS_ENCODED.shape
LABELS_ENCODED[:10]
LE.classes_
# Test if inverse works
# Note: encoded lables should be 1s and 0s
LE.inverse_transform(LABELS_ENCODED[:10])
DATA_TO_USE["labels_encoded"] = list(LABELS_ENCODED)
DATA_TO_USE.head()
```
|
github_jupyter
|
%reload_ext autoreload
%autoreload 2
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
from loguru import logger
datapath = "data/data.csv"
def parse_kws(kw_str, level=2):
res = kw_str.split(",")
res = map(lambda kw: [_.strip().lower() for _ in kw.split(">")], res)
res = map(lambda x: x[level if level<len(x) else len(x)-1], res)
return list(set(res))
def load_data(path, level=0):
logger.info(f"Loading data from {path}. [KW Level={level}]")
df = pd.read_csv(path)
df["desc"] = df["desc"].apply(str.strip)
df["labels"] = df["keywords"].apply(lambda x: parse_kws(x, level))
df["textlen"] = df["desc"].apply(len)
return df
DATA = load_data(datapath, level=1)
# Like I said, we're only able to load 6K data points which can be fixed in future!
DATA.shape
DATA.head()
from collections import Counter, defaultdict
from loguru import logger
def analyze_labels(df):
df = df.copy()
labels = [l for ls in df["labels"] for l in ls]
uniques = set(labels)
logger.info(f"{len(uniques)} unique labels")
analyze_labels(DATA)
_data = DATA.copy()
_data = _data[_data["textlen"]>0]
logger.debug(_data.shape)
# BERT can only process 512 sequence length at once
# So, what % of text satisfy that pre-condition?
len(_data[_data["textlen"] <= 512]) / len(_data), len(_data[_data["textlen"] <= 1024]) / len(_data)
plt.figure(figsize=(20, 15))
sns.histplot(data=_data, x="textlen", bins=100).set(xlim=(0, 3000))
from sklearn.preprocessing import MultiLabelBinarizer
DATA_TO_USE = DATA.copy()
DATA_TO_USE = DATA_TO_USE[DATA_TO_USE["textlen"]<=500]
DATA_TO_USE.shape
DATA_TO_USE.head()
# Note: Now our level 1 keywords are <29 (N=22) classes because we have filtered the data
analyze_labels(DATA_TO_USE)
LE = MultiLabelBinarizer()
LABELS_ENCODED = LE.fit_transform(DATA_TO_USE["labels"])
# 22 classes for level=1 keywords
LABELS_ENCODED.shape
LABELS_ENCODED[:10]
LE.classes_
# Test if inverse works
# Note: encoded lables should be 1s and 0s
LE.inverse_transform(LABELS_ENCODED[:10])
DATA_TO_USE["labels_encoded"] = list(LABELS_ENCODED)
DATA_TO_USE.head()
| 0.341802 | 0.854763 |
# Name
Data preparation using Spark on YARN with Cloud Dataproc
# Label
Cloud Dataproc, GCP, Cloud Storage, Spark, Kubeflow, pipelines, components, YARN
# Summary
A Kubeflow Pipeline component to prepare data by submitting a Spark job on YARN to Cloud Dataproc.
# Details
## Intended use
Use the component to run an Apache Spark job as one preprocessing step in a Kubeflow Pipeline.
## Runtime arguments
Argument | Description | Optional | Data type | Accepted values | Default |
:--- | :---------- | :--- | :------- | :------| :------|
project_id | The ID of the Google Cloud Platform (GCP) project that the cluster belongs to.|No | GCPProjectID | | |
region | The Cloud Dataproc region to handle the request. | No | GCPRegion | | |
cluster_name | The name of the cluster to run the job. | No | String | | |
main_jar_file_uri | The Hadoop Compatible Filesystem (HCFS) URI of the JAR file that contains the main class. | No | GCSPath | | |
main_class | The name of the driver's main class. The JAR file that contains the class must be either in the default CLASSPATH or specified in `spark_job.jarFileUris`.| No | | | |
args | The arguments to pass to the driver. Do not include arguments, such as --conf, that can be set as job properties, since a collision may occur that causes an incorrect job submission.| Yes | | | |
spark_job | The payload of a [SparkJob](https://cloud.google.com/dataproc/docs/reference/rest/v1/SparkJob).| Yes | | | |
job | The payload of a [Dataproc job](https://cloud.google.com/dataproc/docs/reference/rest/v1/projects.regions.jobs). | Yes | | | |
wait_interval | The number of seconds to wait between polling the operation. | Yes | | | 30 |
## Output
Name | Description | Type
:--- | :---------- | :---
job_id | The ID of the created job. | String
## Cautions & requirements
To use the component, you must:
* Set up a GCP project by following this [guide](https://cloud.google.com/dataproc/docs/guides/setup-project).
* [Create a new cluster](https://cloud.google.com/dataproc/docs/guides/create-cluster).
* Run the component under a secret [Kubeflow user service account](https://www.kubeflow.org/docs/started/getting-started-gke/#gcp-service-accounts) in a Kubeflow cluster. For example:
```
component_op(...).apply(gcp.use_gcp_secret('user-gcp-sa'))
```
* Grant the Kubeflow user service account the role `roles/dataproc.editor` on the project.
## Detailed description
This component creates a Spark job from [Dataproc submit job REST API](https://cloud.google.com/dataproc/docs/reference/rest/v1/projects.regions.jobs/submit).
Follow these steps to use the component in a pipeline:
1. Install the Kubeflow Pipeline SDK:
```
%%capture --no-stderr
KFP_PACKAGE = 'https://storage.googleapis.com/ml-pipeline/release/0.1.14/kfp.tar.gz'
!pip3 install $KFP_PACKAGE --upgrade
```
2. Load the component using KFP SDK
```
import kfp.components as comp
dataproc_submit_spark_job_op = comp.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/74d8e592174ae90175f66c3c00ba76a835cfba6d/components/gcp/dataproc/submit_spark_job/component.yaml')
help(dataproc_submit_spark_job_op)
```
### Sample
Note: The following sample code works in an IPython notebook or directly in Python code.
#### Set up a Dataproc cluster
[Create a new Dataproc cluster](https://cloud.google.com/dataproc/docs/guides/create-cluster) (or reuse an existing one) before running the sample code.
#### Prepare a Spark job
Upload your Spark JAR file to a Cloud Storage bucket. In the sample, we use a JAR file that is preinstalled in the main cluster: `file:///usr/lib/spark/examples/jars/spark-examples.jar`.
Here is the [source code of the sample](https://github.com/apache/spark/blob/master/examples/src/main/java/org/apache/spark/examples/JavaSparkPi.java).
To package a self-contained Spark application, follow these [instructions](https://spark.apache.org/docs/latest/quick-start.html#self-contained-applications).
#### Set sample parameters
```
PROJECT_ID = '<Please put your project ID here>'
CLUSTER_NAME = '<Please put your existing cluster name here>'
REGION = 'us-central1'
SPARK_FILE_URI = 'file:///usr/lib/spark/examples/jars/spark-examples.jar'
MAIN_CLASS = 'org.apache.spark.examples.SparkPi'
ARGS = ['1000']
EXPERIMENT_NAME = 'Dataproc - Submit Spark Job'
```
#### Example pipeline that uses the component
```
import kfp.dsl as dsl
import kfp.gcp as gcp
import json
@dsl.pipeline(
name='Dataproc submit Spark job pipeline',
description='Dataproc submit Spark job pipeline'
)
def dataproc_submit_spark_job_pipeline(
project_id = PROJECT_ID,
region = REGION,
cluster_name = CLUSTER_NAME,
main_jar_file_uri = '',
main_class = MAIN_CLASS,
args = json.dumps(ARGS),
spark_job=json.dumps({ 'jarFileUris': [ SPARK_FILE_URI ] }),
job='{}',
wait_interval='30'
):
dataproc_submit_spark_job_op(
project_id=project_id,
region=region,
cluster_name=cluster_name,
main_jar_file_uri=main_jar_file_uri,
main_class=main_class,
args=args,
spark_job=spark_job,
job=job,
wait_interval=wait_interval).apply(gcp.use_gcp_secret('user-gcp-sa'))
```
#### Compile the pipeline
```
pipeline_func = dataproc_submit_spark_job_pipeline
pipeline_filename = pipeline_func.__name__ + '.zip'
import kfp.compiler as compiler
compiler.Compiler().compile(pipeline_func, pipeline_filename)
```
#### Submit the pipeline for execution
```
#Specify pipeline argument values
arguments = {}
#Get or create an experiment and submit a pipeline run
import kfp
client = kfp.Client()
experiment = client.create_experiment(EXPERIMENT_NAME)
#Submit a pipeline run
run_name = pipeline_func.__name__ + ' run'
run_result = client.run_pipeline(experiment.id, run_name, pipeline_filename, arguments)
```
## References
* [Component Python code](https://github.com/kubeflow/pipelines/blob/master/component_sdk/python/kfp_component/google/dataproc/_submit_spark_job.py)
* [Component Docker file](https://github.com/kubeflow/pipelines/blob/master/components/gcp/container/Dockerfile)
* [Sample notebook](https://github.com/kubeflow/pipelines/blob/master/components/gcp/dataproc/submit_spark_job/sample.ipynb)
* [Dataproc SparkJob](https://cloud.google.com/dataproc/docs/reference/rest/v1/SparkJob)
## License
By deploying or using this software you agree to comply with the [AI Hub Terms of Service](https://aihub.cloud.google.com/u/0/aihub-tos) and the [Google APIs Terms of Service](https://developers.google.com/terms/). To the extent of a direct conflict of terms, the AI Hub Terms of Service will control.
|
github_jupyter
|
component_op(...).apply(gcp.use_gcp_secret('user-gcp-sa'))
```
* Grant the Kubeflow user service account the role `roles/dataproc.editor` on the project.
## Detailed description
This component creates a Spark job from [Dataproc submit job REST API](https://cloud.google.com/dataproc/docs/reference/rest/v1/projects.regions.jobs/submit).
Follow these steps to use the component in a pipeline:
1. Install the Kubeflow Pipeline SDK:
2. Load the component using KFP SDK
### Sample
Note: The following sample code works in an IPython notebook or directly in Python code.
#### Set up a Dataproc cluster
[Create a new Dataproc cluster](https://cloud.google.com/dataproc/docs/guides/create-cluster) (or reuse an existing one) before running the sample code.
#### Prepare a Spark job
Upload your Spark JAR file to a Cloud Storage bucket. In the sample, we use a JAR file that is preinstalled in the main cluster: `file:///usr/lib/spark/examples/jars/spark-examples.jar`.
Here is the [source code of the sample](https://github.com/apache/spark/blob/master/examples/src/main/java/org/apache/spark/examples/JavaSparkPi.java).
To package a self-contained Spark application, follow these [instructions](https://spark.apache.org/docs/latest/quick-start.html#self-contained-applications).
#### Set sample parameters
#### Example pipeline that uses the component
#### Compile the pipeline
#### Submit the pipeline for execution
| 0.811153 | 0.941493 |
# Overview: Generating Model Grids
This notebook provides an example of how to generate a custom grid of models within `brutus`, for us as part of the `fitting` module. More detailed information can be found in the documentation and in other notebooks. Files needed to run various parts of the code can also be found online.
```
import brutus
from brutus import seds
```
New grids can be generated using the `SEDmaker` class, which requires an input **neural network (NN) file** to generate photometry and **equivalent evolutionary point (EEP) track file** to generate the stellar models. These can also take in particular filters from the `filters` module. If no filters are passed, photometry is by default generated over all available bands.
```
# initialize the SED maker for the MIST v1.2 models
mist = seds.SEDmaker(nnfile='../data/DATAFILES/nn_c3k.h5',
mistfile='../data/DATAFILES/MIST_1.2_EEPtrk.h5')
```
Generating the grid is as simple as running the `make_grid` function. This can generate a grid over initial mass (`mini`), initial metallicity (`feh`), EEP (`eep`), alpha-element abundance variation (`afe`), and secondary mass fraction (`smf`). Note that while defaults are specified, the function is designed with the expectation that users will pass at least some additional arguments. For additional information on the available options, please see the release paper and the documentation.
```
# build the SED grid
mist.make_grid(smf_grid=np.array([0.]), # no binaries
afe_grid=np.array([0.])) # no afe
```
The output grid is saved internally, with the grid labels (`grid_label`), output stellar parameters (`grid_param`), and spectral energy distributions (SEDs) (`grid_sed`) saved. Since not all models on the grid are well-defined (i.e. there are no models for evolved low-mass stars since they are distinctly unphysical), there is also a selection array (`grid_sel`) created.
For compatibility with the `load_models` function in the `utils` module, the models have to saved to disk in hdf5 format with the internal structure shown below.
```
# dump results to disk
import h5py
grid_vers = 'v9' # version number of MIST grid
with h5py.File("../data/grid_mist_{}.h5".format(grid_vers), "w") as out:
# selection array
sel = mist.grid_sel
# labels used to generate the grid
labels = out.create_dataset("labels", data=mist.grid_label[sel])
# parameters generated interpolating over the MIST isochrones
pars = out.create_dataset("parameters", data=mist.grid_param[sel])
# SEDS generated using the NN from the stellar parameters
seds = out.create_dataset("mag_coeffs", data=mist.grid_sed[sel])
```
And we're done! We can now use this stellar model grid in other parts of the code.
|
github_jupyter
|
import brutus
from brutus import seds
# initialize the SED maker for the MIST v1.2 models
mist = seds.SEDmaker(nnfile='../data/DATAFILES/nn_c3k.h5',
mistfile='../data/DATAFILES/MIST_1.2_EEPtrk.h5')
# build the SED grid
mist.make_grid(smf_grid=np.array([0.]), # no binaries
afe_grid=np.array([0.])) # no afe
# dump results to disk
import h5py
grid_vers = 'v9' # version number of MIST grid
with h5py.File("../data/grid_mist_{}.h5".format(grid_vers), "w") as out:
# selection array
sel = mist.grid_sel
# labels used to generate the grid
labels = out.create_dataset("labels", data=mist.grid_label[sel])
# parameters generated interpolating over the MIST isochrones
pars = out.create_dataset("parameters", data=mist.grid_param[sel])
# SEDS generated using the NN from the stellar parameters
seds = out.create_dataset("mag_coeffs", data=mist.grid_sed[sel])
| 0.380068 | 0.980636 |
# Speaker-Identification
```
# Importing the libraries
import os
import pandas as pd
import re
import librosa
from datasets import Dataset
```
# Loading the dataset
```
# Defining the root directory
data_directory = "../Badaga_Corpus-v.0.1.0/"
tagged_file = "Badaga-v0.1.0.xlsx"
# loading the dataset
tagged_file_path = os.path.join(data_directory, tagged_file)
# loading the transcription file
data_frame = pd.read_excel(tagged_file_path)
# droping the missing values
data_frame.dropna(inplace=True)
# loading the audio files
data_frame["audio_file_name"] = data_frame["audio_file_name"].apply(lambda x: os.path.join(data_directory, "clips", x))
# splitting the data into train and test using split_index from transcription file
train_df = data_frame[data_frame["split_label"]!="test"]
test_df = data_frame[data_frame["split_label"]=="test"]
# printing the data
data_frame
# converting variables such as "audio_file_name" and "user_id" as list and renaming them as "path" and "label"
train_df["path"] = list(train_df["audio_file_name"])
train_df["label"] = list(train_df["user_id"])
test_df["path"] = list(test_df["audio_file_name"])
test_df["label"] = list(test_df["user_id"])
# creating a new datasets using the above list for both training and testing set
train_df = train_df[["path", "label"]]
test_df = test_df[["path", "label"]]
# printing the shape of train and test
train_df.shape, test_df.shape
# printing the data
train_df
# dropping the index for the newly created dataset
train_df = train_df.reset_index(drop=True)
test_df = test_df.reset_index(drop=True)
# saving it as csv files for both training and testing
train_df.to_csv("files/speaker_train.csv", sep="\t", encoding="utf-8", index=False)
test_df.to_csv("filesspeaker_test.csv", sep="\t", encoding="utf-8", index=False)
# We need to specify the input and output column
input_column = "path"
output_column = "label"
# Loading the created dataset using datasets
from datasets import load_dataset, load_metric
data_files = {
"train": "files/speaker_train.csv",
"validation": "files/speaker_test.csv",
}
dataset = load_dataset("csv", data_files=data_files, delimiter="\t", )
train_dataset = dataset["train"]
eval_dataset = dataset["validation"]
print(train_dataset)
print(eval_dataset)
# we need to distinguish the unique labels
label_list = train_dataset.unique(output_column)
label_list.sort() # Let's sort it for determinism
num_labels = len(label_list)
print(f"A classification problem with {num_labels} classes: {label_list}")
```
# Feature Extraction
```
# loading the feature extractor and processor from the transformers
from transformers import AutoConfig, Wav2Vec2Processor, Wav2Vec2FeatureExtractor
```
# Loading the Wav2Vec Model
```
# loading the model
model_name_or_path = "facebook/wav2vec2-large-xlsr-53"
pooling_mode = "mean"
# config
config = AutoConfig.from_pretrained(
model_name_or_path,
num_labels=num_labels,
label2id={label: i for i, label in enumerate(label_list)},
id2label={i: label for i, label in enumerate(label_list)},
finetuning_task="wav2vec2_clf",
)
setattr(config, 'pooling_mode', pooling_mode)
# feature extraction
feature_extractor = Wav2Vec2FeatureExtractor.from_pretrained(model_name_or_path,)
target_sampling_rate = feature_extractor.sampling_rate
print(f"The target sampling rate: {target_sampling_rate}")
# loading the audio data using librosa
import librosa
def speech_file_to_array_fn(path):
a, s = librosa.load(path, sr=16000)
# speech_array, sampling_rate = torchaudio.load(path)
# resampler = torchaudio.transforms.Resample(sampling_rate, target_sampling_rate)
# speech = resampler(speech_array).squeeze().numpy()
return a
def label_to_id(label, label_list):
if len(label_list) > 0:
return label_list.index(label) if label in label_list else -1
return label
# function for pre-processing
def preprocess_function(examples):
speech_list = [speech_file_to_array_fn(path) for path in examples[input_column]]
target_list = [label_to_id(label, label_list) for label in examples[output_column]]
result = feature_extractor(speech_list, sampling_rate=target_sampling_rate)
result["labels"] = list(target_list)
return result
# using map function to map the pre-processed files to the train adn test sets
import torchaudio
train_dataset = train_dataset.map(
preprocess_function,
batch_size=100,
batched=True,
num_proc=4
)
eval_dataset = eval_dataset.map(
preprocess_function,
batch_size=100,
batched=True,
num_proc=4
)
# defining the dataclass for speech classifier
from dataclasses import dataclass
from typing import Optional, Tuple
import torch
from transformers.file_utils import ModelOutput
@dataclass
class SpeechClassifierOutput(ModelOutput):
loss: Optional[torch.FloatTensor] = None
logits: torch.FloatTensor = None
hidden_states: Optional[Tuple[torch.FloatTensor]] = None
attentions: Optional[Tuple[torch.FloatTensor]] = None
```
# Fine-Tuning Wav2Vec Pretrained Model for Speaker-Identification
```
# defining the classifier class
import torch
import torch.nn as nn
from torch.nn import BCEWithLogitsLoss, CrossEntropyLoss, MSELoss
from transformers.models.wav2vec2.modeling_wav2vec2 import (
Wav2Vec2PreTrainedModel,
Wav2Vec2Model
)
class Wav2Vec2ClassificationHead(nn.Module):
"""Head for wav2vec classification task."""
def __init__(self, config):
super().__init__()
self.dense = nn.Linear(config.hidden_size, config.hidden_size)
self.dropout = nn.Dropout(config.final_dropout)
self.out_proj = nn.Linear(config.hidden_size, config.num_labels)
def forward(self, features, **kwargs):
x = features
x = self.dropout(x)
x = self.dense(x)
x = torch.tanh(x)
x = self.dropout(x)
x = self.out_proj(x)
return x
class Wav2Vec2ForSpeechClassification(Wav2Vec2PreTrainedModel):
def __init__(self, config):
super().__init__(config)
self.num_labels = config.num_labels
self.pooling_mode = config.pooling_mode
self.config = config
self.wav2vec2 = Wav2Vec2Model(config)
self.classifier = Wav2Vec2ClassificationHead(config)
self.init_weights()
def freeze_feature_extractor(self):
self.wav2vec2.feature_extractor._freeze_parameters()
def merged_strategy(
self,
hidden_states,
mode="mean"
):
if mode == "mean":
outputs = torch.mean(hidden_states, dim=1)
elif mode == "sum":
outputs = torch.sum(hidden_states, dim=1)
elif mode == "max":
outputs = torch.max(hidden_states, dim=1)[0]
else:
raise Exception(
"The pooling method hasn't been defined! Your pooling mode must be one of these ['mean', 'sum', 'max']")
return outputs
def forward(
self,
input_values,
attention_mask=None,
output_attentions=None,
output_hidden_states=None,
return_dict=None,
labels=None,
):
return_dict = return_dict if return_dict is not None else self.config.use_return_dict
outputs = self.wav2vec2(
input_values,
attention_mask=attention_mask,
output_attentions=output_attentions,
output_hidden_states=output_hidden_states,
return_dict=return_dict,
)
hidden_states = outputs[0]
hidden_states = self.merged_strategy(hidden_states, mode=self.pooling_mode)
logits = self.classifier(hidden_states)
loss = None
if labels is not None:
if self.config.problem_type is None:
if self.num_labels == 1:
self.config.problem_type = "regression"
elif self.num_labels > 1 and (labels.dtype == torch.long or labels.dtype == torch.int):
self.config.problem_type = "single_label_classification"
else:
self.config.problem_type = "multi_label_classification"
if self.config.problem_type == "regression":
loss_fct = MSELoss()
loss = loss_fct(logits.view(-1, self.num_labels), labels)
elif self.config.problem_type == "single_label_classification":
loss_fct = CrossEntropyLoss()
loss = loss_fct(logits.view(-1, self.num_labels), labels.view(-1))
elif self.config.problem_type == "multi_label_classification":
loss_fct = BCEWithLogitsLoss()
loss = loss_fct(logits, labels)
if not return_dict:
output = (logits,) + outputs[2:]
return ((loss,) + output) if loss is not None else output
return SpeechClassifierOutput(
loss=loss,
logits=logits,
hidden_states=outputs.hidden_states,
attentions=outputs.attentions,
)
# defining function for datacollator and padding
from dataclasses import dataclass
from typing import Dict, List, Optional, Union
import torch
import transformers
from transformers import Wav2Vec2Processor, Wav2Vec2FeatureExtractor
@dataclass
class DataCollatorCTCWithPadding:
"""
Data collator that will dynamically pad the inputs received.
Args:
feature_extractor (:class:`~transformers.Wav2Vec2FeatureExtractor`)
The feature_extractor used for proccessing the data.
padding (:obj:`bool`, :obj:`str` or :class:`~transformers.tokenization_utils_base.PaddingStrategy`, `optional`, defaults to :obj:`True`):
Select a strategy to pad the returned sequences (according to the model's padding side and padding index)
among:
* :obj:`True` or :obj:`'longest'`: Pad to the longest sequence in the batch (or no padding if only a single
sequence if provided).
* :obj:`'max_length'`: Pad to a maximum length specified with the argument :obj:`max_length` or to the
maximum acceptable input length for the model if that argument is not provided.
* :obj:`False` or :obj:`'do_not_pad'` (default): No padding (i.e., can output a batch with sequences of
different lengths).
max_length (:obj:`int`, `optional`):
Maximum length of the ``input_values`` of the returned list and optionally padding length (see above).
max_length_labels (:obj:`int`, `optional`):
Maximum length of the ``labels`` returned list and optionally padding length (see above).
pad_to_multiple_of (:obj:`int`, `optional`):
If set will pad the sequence to a multiple of the provided value.
This is especially useful to enable the use of Tensor Cores on NVIDIA hardware with compute capability >=
7.5 (Volta).
"""
feature_extractor: Wav2Vec2FeatureExtractor
padding: Union[bool, str] = True
max_length: Optional[int] = None
max_length_labels: Optional[int] = None
pad_to_multiple_of: Optional[int] = None
pad_to_multiple_of_labels: Optional[int] = None
def __call__(self, features: List[Dict[str, Union[List[int], torch.Tensor]]]) -> Dict[str, torch.Tensor]:
input_features = [{"input_values": feature["input_values"]} for feature in features]
label_features = [feature["labels"] for feature in features]
d_type = torch.long if isinstance(label_features[0], int) else torch.float
batch = self.feature_extractor.pad(
input_features,
padding=self.padding,
max_length=self.max_length,
pad_to_multiple_of=self.pad_to_multiple_of,
return_tensors="pt",
)
batch["labels"] = torch.tensor(label_features, dtype=d_type)
return batch
# setting only for classification (regression is set)
is_regression = False
# function for computing metrics for evaluation
import numpy as np
from transformers import EvalPrediction
def compute_metrics(p: EvalPrediction):
preds = p.predictions[0] if isinstance(p.predictions, tuple) else p.predictions
preds = np.squeeze(preds) if is_regression else np.argmax(preds, axis=1)
if is_regression:
return {"mse": ((preds - p.label_ids) ** 2).mean().item()}
else:
return {"accuracy": (preds == p.label_ids).astype(np.float32).mean().item()}
# classification
model = Wav2Vec2ForSpeechClassification.from_pretrained(
model_name_or_path,
config=config,
)
model.freeze_feature_extractor()
# setting up the arguments for training
from transformers import TrainingArguments
training_args = TrainingArguments(
output_dir="wav2vec2-rbg-badaga-speaker",
per_device_train_batch_size=4,
per_device_eval_batch_size=4,
gradient_accumulation_steps=2,
evaluation_strategy="steps",
num_train_epochs=1.0,
fp16=True,
save_steps=10,
eval_steps=10,
logging_steps=10,
learning_rate=1e-4,
save_total_limit=2,
)
# setting up the trainer function
from typing import Any, Dict, Union
import torch
from packaging import version
from torch import nn
from transformers import (
Trainer,
is_apex_available,
)
if is_apex_available():
from apex import amp
if version.parse(torch.__version__) >= version.parse("1.6"):
_is_native_amp_available = True
from torch.cuda.amp import autocast
class CTCTrainer(Trainer):
def training_step(self, model: nn.Module, inputs: Dict[str, Union[torch.Tensor, Any]]) -> torch.Tensor:
"""
Perform a training step on a batch of inputs.
Subclass and override to inject custom behavior.
Args:
model (:obj:`nn.Module`):
The model to train.
inputs (:obj:`Dict[str, Union[torch.Tensor, Any]]`):
The inputs and targets of the model.
The dictionary will be unpacked before being fed to the model. Most models expect the targets under the
argument :obj:`labels`. Check your model's documentation for all accepted arguments.
Return:
:obj:`torch.Tensor`: The tensor with training loss on this batch.
"""
model.train()
inputs = self._prepare_inputs(inputs)
if self.use_amp:
with autocast():
loss = self.compute_loss(model, inputs)
else:
loss = self.compute_loss(model, inputs)
if self.args.gradient_accumulation_steps > 1:
loss = loss / self.args.gradient_accumulation_steps
if self.use_amp:
self.scaler.scale(loss).backward()
elif self.use_apex:
with amp.scale_loss(loss, self.optimizer) as scaled_loss:
scaled_loss.backward()
elif self.deepspeed:
self.deepspeed.backward(loss)
else:
loss.backward()
return loss.detach()
# calling the data collator with padding
data_collator = DataCollatorCTCWithPadding(feature_extractor=feature_extractor, padding=True)
## setting up CTCT trainer
trainer = CTCTrainer(
model=model,
data_collator=data_collator,
args=training_args,
compute_metrics=compute_metrics,
train_dataset=train_dataset,
eval_dataset=eval_dataset,
tokenizer=feature_extractor,
)
# training
trainer.train()
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"Device: {device}")
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model_name_or_path = "wav2vec2-rbg-badaga-speaker/"
config = AutoConfig.from_pretrained(model_name_or_path)
feature_extractor = Wav2Vec2FeatureExtractor.from_pretrained(model_name_or_path)
sampling_rate = feature_extractor.sampling_rate
model = Wav2Vec2ForSpeechClassification.from_pretrained(model_name_or_path).to(device)
```
|
github_jupyter
|
# Importing the libraries
import os
import pandas as pd
import re
import librosa
from datasets import Dataset
# Defining the root directory
data_directory = "../Badaga_Corpus-v.0.1.0/"
tagged_file = "Badaga-v0.1.0.xlsx"
# loading the dataset
tagged_file_path = os.path.join(data_directory, tagged_file)
# loading the transcription file
data_frame = pd.read_excel(tagged_file_path)
# droping the missing values
data_frame.dropna(inplace=True)
# loading the audio files
data_frame["audio_file_name"] = data_frame["audio_file_name"].apply(lambda x: os.path.join(data_directory, "clips", x))
# splitting the data into train and test using split_index from transcription file
train_df = data_frame[data_frame["split_label"]!="test"]
test_df = data_frame[data_frame["split_label"]=="test"]
# printing the data
data_frame
# converting variables such as "audio_file_name" and "user_id" as list and renaming them as "path" and "label"
train_df["path"] = list(train_df["audio_file_name"])
train_df["label"] = list(train_df["user_id"])
test_df["path"] = list(test_df["audio_file_name"])
test_df["label"] = list(test_df["user_id"])
# creating a new datasets using the above list for both training and testing set
train_df = train_df[["path", "label"]]
test_df = test_df[["path", "label"]]
# printing the shape of train and test
train_df.shape, test_df.shape
# printing the data
train_df
# dropping the index for the newly created dataset
train_df = train_df.reset_index(drop=True)
test_df = test_df.reset_index(drop=True)
# saving it as csv files for both training and testing
train_df.to_csv("files/speaker_train.csv", sep="\t", encoding="utf-8", index=False)
test_df.to_csv("filesspeaker_test.csv", sep="\t", encoding="utf-8", index=False)
# We need to specify the input and output column
input_column = "path"
output_column = "label"
# Loading the created dataset using datasets
from datasets import load_dataset, load_metric
data_files = {
"train": "files/speaker_train.csv",
"validation": "files/speaker_test.csv",
}
dataset = load_dataset("csv", data_files=data_files, delimiter="\t", )
train_dataset = dataset["train"]
eval_dataset = dataset["validation"]
print(train_dataset)
print(eval_dataset)
# we need to distinguish the unique labels
label_list = train_dataset.unique(output_column)
label_list.sort() # Let's sort it for determinism
num_labels = len(label_list)
print(f"A classification problem with {num_labels} classes: {label_list}")
# loading the feature extractor and processor from the transformers
from transformers import AutoConfig, Wav2Vec2Processor, Wav2Vec2FeatureExtractor
# loading the model
model_name_or_path = "facebook/wav2vec2-large-xlsr-53"
pooling_mode = "mean"
# config
config = AutoConfig.from_pretrained(
model_name_or_path,
num_labels=num_labels,
label2id={label: i for i, label in enumerate(label_list)},
id2label={i: label for i, label in enumerate(label_list)},
finetuning_task="wav2vec2_clf",
)
setattr(config, 'pooling_mode', pooling_mode)
# feature extraction
feature_extractor = Wav2Vec2FeatureExtractor.from_pretrained(model_name_or_path,)
target_sampling_rate = feature_extractor.sampling_rate
print(f"The target sampling rate: {target_sampling_rate}")
# loading the audio data using librosa
import librosa
def speech_file_to_array_fn(path):
a, s = librosa.load(path, sr=16000)
# speech_array, sampling_rate = torchaudio.load(path)
# resampler = torchaudio.transforms.Resample(sampling_rate, target_sampling_rate)
# speech = resampler(speech_array).squeeze().numpy()
return a
def label_to_id(label, label_list):
if len(label_list) > 0:
return label_list.index(label) if label in label_list else -1
return label
# function for pre-processing
def preprocess_function(examples):
speech_list = [speech_file_to_array_fn(path) for path in examples[input_column]]
target_list = [label_to_id(label, label_list) for label in examples[output_column]]
result = feature_extractor(speech_list, sampling_rate=target_sampling_rate)
result["labels"] = list(target_list)
return result
# using map function to map the pre-processed files to the train adn test sets
import torchaudio
train_dataset = train_dataset.map(
preprocess_function,
batch_size=100,
batched=True,
num_proc=4
)
eval_dataset = eval_dataset.map(
preprocess_function,
batch_size=100,
batched=True,
num_proc=4
)
# defining the dataclass for speech classifier
from dataclasses import dataclass
from typing import Optional, Tuple
import torch
from transformers.file_utils import ModelOutput
@dataclass
class SpeechClassifierOutput(ModelOutput):
loss: Optional[torch.FloatTensor] = None
logits: torch.FloatTensor = None
hidden_states: Optional[Tuple[torch.FloatTensor]] = None
attentions: Optional[Tuple[torch.FloatTensor]] = None
# defining the classifier class
import torch
import torch.nn as nn
from torch.nn import BCEWithLogitsLoss, CrossEntropyLoss, MSELoss
from transformers.models.wav2vec2.modeling_wav2vec2 import (
Wav2Vec2PreTrainedModel,
Wav2Vec2Model
)
class Wav2Vec2ClassificationHead(nn.Module):
"""Head for wav2vec classification task."""
def __init__(self, config):
super().__init__()
self.dense = nn.Linear(config.hidden_size, config.hidden_size)
self.dropout = nn.Dropout(config.final_dropout)
self.out_proj = nn.Linear(config.hidden_size, config.num_labels)
def forward(self, features, **kwargs):
x = features
x = self.dropout(x)
x = self.dense(x)
x = torch.tanh(x)
x = self.dropout(x)
x = self.out_proj(x)
return x
class Wav2Vec2ForSpeechClassification(Wav2Vec2PreTrainedModel):
def __init__(self, config):
super().__init__(config)
self.num_labels = config.num_labels
self.pooling_mode = config.pooling_mode
self.config = config
self.wav2vec2 = Wav2Vec2Model(config)
self.classifier = Wav2Vec2ClassificationHead(config)
self.init_weights()
def freeze_feature_extractor(self):
self.wav2vec2.feature_extractor._freeze_parameters()
def merged_strategy(
self,
hidden_states,
mode="mean"
):
if mode == "mean":
outputs = torch.mean(hidden_states, dim=1)
elif mode == "sum":
outputs = torch.sum(hidden_states, dim=1)
elif mode == "max":
outputs = torch.max(hidden_states, dim=1)[0]
else:
raise Exception(
"The pooling method hasn't been defined! Your pooling mode must be one of these ['mean', 'sum', 'max']")
return outputs
def forward(
self,
input_values,
attention_mask=None,
output_attentions=None,
output_hidden_states=None,
return_dict=None,
labels=None,
):
return_dict = return_dict if return_dict is not None else self.config.use_return_dict
outputs = self.wav2vec2(
input_values,
attention_mask=attention_mask,
output_attentions=output_attentions,
output_hidden_states=output_hidden_states,
return_dict=return_dict,
)
hidden_states = outputs[0]
hidden_states = self.merged_strategy(hidden_states, mode=self.pooling_mode)
logits = self.classifier(hidden_states)
loss = None
if labels is not None:
if self.config.problem_type is None:
if self.num_labels == 1:
self.config.problem_type = "regression"
elif self.num_labels > 1 and (labels.dtype == torch.long or labels.dtype == torch.int):
self.config.problem_type = "single_label_classification"
else:
self.config.problem_type = "multi_label_classification"
if self.config.problem_type == "regression":
loss_fct = MSELoss()
loss = loss_fct(logits.view(-1, self.num_labels), labels)
elif self.config.problem_type == "single_label_classification":
loss_fct = CrossEntropyLoss()
loss = loss_fct(logits.view(-1, self.num_labels), labels.view(-1))
elif self.config.problem_type == "multi_label_classification":
loss_fct = BCEWithLogitsLoss()
loss = loss_fct(logits, labels)
if not return_dict:
output = (logits,) + outputs[2:]
return ((loss,) + output) if loss is not None else output
return SpeechClassifierOutput(
loss=loss,
logits=logits,
hidden_states=outputs.hidden_states,
attentions=outputs.attentions,
)
# defining function for datacollator and padding
from dataclasses import dataclass
from typing import Dict, List, Optional, Union
import torch
import transformers
from transformers import Wav2Vec2Processor, Wav2Vec2FeatureExtractor
@dataclass
class DataCollatorCTCWithPadding:
"""
Data collator that will dynamically pad the inputs received.
Args:
feature_extractor (:class:`~transformers.Wav2Vec2FeatureExtractor`)
The feature_extractor used for proccessing the data.
padding (:obj:`bool`, :obj:`str` or :class:`~transformers.tokenization_utils_base.PaddingStrategy`, `optional`, defaults to :obj:`True`):
Select a strategy to pad the returned sequences (according to the model's padding side and padding index)
among:
* :obj:`True` or :obj:`'longest'`: Pad to the longest sequence in the batch (or no padding if only a single
sequence if provided).
* :obj:`'max_length'`: Pad to a maximum length specified with the argument :obj:`max_length` or to the
maximum acceptable input length for the model if that argument is not provided.
* :obj:`False` or :obj:`'do_not_pad'` (default): No padding (i.e., can output a batch with sequences of
different lengths).
max_length (:obj:`int`, `optional`):
Maximum length of the ``input_values`` of the returned list and optionally padding length (see above).
max_length_labels (:obj:`int`, `optional`):
Maximum length of the ``labels`` returned list and optionally padding length (see above).
pad_to_multiple_of (:obj:`int`, `optional`):
If set will pad the sequence to a multiple of the provided value.
This is especially useful to enable the use of Tensor Cores on NVIDIA hardware with compute capability >=
7.5 (Volta).
"""
feature_extractor: Wav2Vec2FeatureExtractor
padding: Union[bool, str] = True
max_length: Optional[int] = None
max_length_labels: Optional[int] = None
pad_to_multiple_of: Optional[int] = None
pad_to_multiple_of_labels: Optional[int] = None
def __call__(self, features: List[Dict[str, Union[List[int], torch.Tensor]]]) -> Dict[str, torch.Tensor]:
input_features = [{"input_values": feature["input_values"]} for feature in features]
label_features = [feature["labels"] for feature in features]
d_type = torch.long if isinstance(label_features[0], int) else torch.float
batch = self.feature_extractor.pad(
input_features,
padding=self.padding,
max_length=self.max_length,
pad_to_multiple_of=self.pad_to_multiple_of,
return_tensors="pt",
)
batch["labels"] = torch.tensor(label_features, dtype=d_type)
return batch
# setting only for classification (regression is set)
is_regression = False
# function for computing metrics for evaluation
import numpy as np
from transformers import EvalPrediction
def compute_metrics(p: EvalPrediction):
preds = p.predictions[0] if isinstance(p.predictions, tuple) else p.predictions
preds = np.squeeze(preds) if is_regression else np.argmax(preds, axis=1)
if is_regression:
return {"mse": ((preds - p.label_ids) ** 2).mean().item()}
else:
return {"accuracy": (preds == p.label_ids).astype(np.float32).mean().item()}
# classification
model = Wav2Vec2ForSpeechClassification.from_pretrained(
model_name_or_path,
config=config,
)
model.freeze_feature_extractor()
# setting up the arguments for training
from transformers import TrainingArguments
training_args = TrainingArguments(
output_dir="wav2vec2-rbg-badaga-speaker",
per_device_train_batch_size=4,
per_device_eval_batch_size=4,
gradient_accumulation_steps=2,
evaluation_strategy="steps",
num_train_epochs=1.0,
fp16=True,
save_steps=10,
eval_steps=10,
logging_steps=10,
learning_rate=1e-4,
save_total_limit=2,
)
# setting up the trainer function
from typing import Any, Dict, Union
import torch
from packaging import version
from torch import nn
from transformers import (
Trainer,
is_apex_available,
)
if is_apex_available():
from apex import amp
if version.parse(torch.__version__) >= version.parse("1.6"):
_is_native_amp_available = True
from torch.cuda.amp import autocast
class CTCTrainer(Trainer):
def training_step(self, model: nn.Module, inputs: Dict[str, Union[torch.Tensor, Any]]) -> torch.Tensor:
"""
Perform a training step on a batch of inputs.
Subclass and override to inject custom behavior.
Args:
model (:obj:`nn.Module`):
The model to train.
inputs (:obj:`Dict[str, Union[torch.Tensor, Any]]`):
The inputs and targets of the model.
The dictionary will be unpacked before being fed to the model. Most models expect the targets under the
argument :obj:`labels`. Check your model's documentation for all accepted arguments.
Return:
:obj:`torch.Tensor`: The tensor with training loss on this batch.
"""
model.train()
inputs = self._prepare_inputs(inputs)
if self.use_amp:
with autocast():
loss = self.compute_loss(model, inputs)
else:
loss = self.compute_loss(model, inputs)
if self.args.gradient_accumulation_steps > 1:
loss = loss / self.args.gradient_accumulation_steps
if self.use_amp:
self.scaler.scale(loss).backward()
elif self.use_apex:
with amp.scale_loss(loss, self.optimizer) as scaled_loss:
scaled_loss.backward()
elif self.deepspeed:
self.deepspeed.backward(loss)
else:
loss.backward()
return loss.detach()
# calling the data collator with padding
data_collator = DataCollatorCTCWithPadding(feature_extractor=feature_extractor, padding=True)
## setting up CTCT trainer
trainer = CTCTrainer(
model=model,
data_collator=data_collator,
args=training_args,
compute_metrics=compute_metrics,
train_dataset=train_dataset,
eval_dataset=eval_dataset,
tokenizer=feature_extractor,
)
# training
trainer.train()
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"Device: {device}")
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model_name_or_path = "wav2vec2-rbg-badaga-speaker/"
config = AutoConfig.from_pretrained(model_name_or_path)
feature_extractor = Wav2Vec2FeatureExtractor.from_pretrained(model_name_or_path)
sampling_rate = feature_extractor.sampling_rate
model = Wav2Vec2ForSpeechClassification.from_pretrained(model_name_or_path).to(device)
| 0.758153 | 0.827096 |
## EXAMPLE - 3
**Tasks :- Answerability detection**
**Tasks Description**
``answerability`` :- This is modeled as a sentence pair classification task where the first sentence is a query and second sentence is a context passage. The objective of this task is to determine whether the query can be answered from the context passage or not.
**Conversational Utility** :- This can be a useful component for building a question-answering/ machine comprehension based system. In such cases, it becomes very important to determine whether the given query can be answered with given context passage or not before extracting/abstracting an answer from it. Performing question-answering for a query which is not answerable from the context, could lead to incorrect answer extraction.
**Data** :- In this example, we are using the <a href="https://msmarco.blob.core.windows.net/msmarcoranking/triples.train.small.tar.gz">MSMARCO triples</a> data which is having sentence pairs and labels.
The data contains triplets where the first entry is the query, second one is the context passage from which the query can be answered (positive passage) , while the third entry is a context passage from which the query cannot be answered (negative passage).
Data is transformed into sentence pair classification format, with query-positive context pair labeled as 1 (answerable) and query-negative context pair labeled as 0 (non-answerable)
The data can be downloaded using the following ``wget`` command and extracted using ``tar`` command. The data is fairly large to download (7.4GB).
```
!wget https://msmarco.blob.core.windows.net/msmarcoranking/triples.train.small.tar.gz -P msmarco_data
!tar -xvzf msmarco_data/triples.train.small.tar.gz -C msmarco_data/
!rm msmarco_data/triples.train.small.tar.gz
```
# Step - 1: Transforming data
The data is present in *JSONL* format where each object contains a sample having the two sentences as ``sentence1`` and ``sentence2``. We consider ``gold_label`` field as the label which can have value: entailment, contradiction or neutral.
We already provide a sample transformation function ``msmarco_answerability_detection_to_tsv`` to convert this data to required tsv format. Data is transformed into sentence pair classification format, with query-positive context pair labeled as 1 (answerable) and query-negative context pair labeled as 0 (non-answerable)
Running data transformations will save the required train, dev and test tsv data files under ``data`` directory in root of library. For more details on the data transformation process, refer to <a href="https://multi-task-nlp.readthedocs.io/en/latest/data_transformations.html">data transformations</a> in documentation.
The transformation file should have the following details which is already created ``transform_file_snli.yml``.
```
transform1:
transform_func: msmarco_answerability_detection_to_tsv
transform_params:
data_frac : 0.02
read_file_names:
- triples.train.small.tsv
read_dir : msmarco_data
save_dir: ../../data
```
Following command can be used to run the data transformation for the tasks.
# Step -2 Data Preparation
For more details on the data preparation process, refer to <a href="https://multi-task-nlp.readthedocs.io/en/latest/training.html#running-data-preparation">data preparation</a> in documentation.
Defining tasks file for training single model for entailment task. The file is already created at ``tasks_file_answerability.yml``
```
answerability:
model_type: BERT
config_name: bert-base-uncased
dropout_prob: 0.2
class_num: 2
metrics:
- classification_accuracy
loss_type: CrossEntropyLoss
task_type: SentencePairClassification
file_names:
- msmarco_answerability_train.tsv
- msmarco_answerability_dev.tsv
- msmarco_answerability_test.tsv
```
```
!python ../../data_preparation.py \
--task_file 'tasks_file_answerability.yml' \
--data_dir '../../data' \
--max_seq_len 324
```
# Step - 3 Running train
Following command will start the training for the tasks. The log file reporting the loss, metrics and the tensorboard logs will be present in a time-stamped directory.
For knowing more details about the train process, refer to <a href= "https://multi-task-nlp.readthedocs.io/en/latest/training.html#running-train">running training</a> in documentation.
```
!python ../../train.py \
--data_dir '../../data/bert-base-uncased_prepared_data' \
--task_file 'tasks_file_answerability.yml' \
--out_dir 'msmarco_answerability_bert_base' \
--epochs 3 \
--train_batch_size 8 \
--eval_batch_size 16 \
--grad_accumulation_steps 2 \
--log_per_updates 250 \
--max_seq_len 324 \
--save_per_updates 16000 \
--eval_while_train \
--test_while_train \
--silent
```
# Step - 4 Infering
You can import and use the ``inferPipeline`` to get predictions for the required tasks.
The trained model and maximum sequence length to be used needs to be specified.
For knowing more details about infering, refer to <a href="https://multi-task-nlp.readthedocs.io/en/latest/infering.html">infer pipeline</a> in documentation.
```
import sys
sys.path.insert(1, '../../')
from infer_pipeline import inferPipeline
```
|
github_jupyter
|
!wget https://msmarco.blob.core.windows.net/msmarcoranking/triples.train.small.tar.gz -P msmarco_data
!tar -xvzf msmarco_data/triples.train.small.tar.gz -C msmarco_data/
!rm msmarco_data/triples.train.small.tar.gz
transform1:
transform_func: msmarco_answerability_detection_to_tsv
transform_params:
data_frac : 0.02
read_file_names:
- triples.train.small.tsv
read_dir : msmarco_data
save_dir: ../../data
```
Following command can be used to run the data transformation for the tasks.
# Step -2 Data Preparation
For more details on the data preparation process, refer to <a href="https://multi-task-nlp.readthedocs.io/en/latest/training.html#running-data-preparation">data preparation</a> in documentation.
Defining tasks file for training single model for entailment task. The file is already created at ``tasks_file_answerability.yml``
# Step - 3 Running train
Following command will start the training for the tasks. The log file reporting the loss, metrics and the tensorboard logs will be present in a time-stamped directory.
For knowing more details about the train process, refer to <a href= "https://multi-task-nlp.readthedocs.io/en/latest/training.html#running-train">running training</a> in documentation.
# Step - 4 Infering
You can import and use the ``inferPipeline`` to get predictions for the required tasks.
The trained model and maximum sequence length to be used needs to be specified.
For knowing more details about infering, refer to <a href="https://multi-task-nlp.readthedocs.io/en/latest/infering.html">infer pipeline</a> in documentation.
| 0.768081 | 0.954605 |
<a href="https://colab.research.google.com/github/nayanemaia/Projects_MachineLearning/blob/main/Medium_RandomForestRegressor.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
#**Machine learning: Tudo o que você precisa saber**
Vamos para a parte prática, porque nem só de teoria vive o homem, não é mesmo???
Para exemplificar o nosso estudo vamos usar um Dataset de notas de alunos de uma escola do Reino Unido, disponível no Kagle (https://www.kaggle.com/dipam7/student-grade-prediction).

#### Abaixo vamos ensinar como:
1. Abrir o dataset e explorar esses dados, verificar anormalidades
2. Transformar dados categóricos em númericos para fins de regressão
3. Seleção de variáveis e como fazer isso
5. Achar os melhores parâmetros e calibrar o modelo
6. Evitar que ocorra Overfitting ou Underfitting
7. Testar o modelo e avaliar
#### Quem que vos escreve né?
Aqui quem fala é a Nayane Maia, sou engenheira agrônoma e trabalho com Data Science desde 2018, quando fiz uma aulinha despretenciosa de Python sem nem saber mexer no Excel direito. Atualmente trabalho como cientista de dados na área da pesquisa em Agronomia e mudanças climáticas, mas sempre me arriscando em coisas aleatórias para aprender mais. E aqui estou ajudando o Matheus Cardoso a escrever o seu primeiro artigo de DS para a comunidade e para o Neuron-DS&AI.... Big responsibility!
Vocês podem me achar no GitHub (https://github.com/nayanemaia) e LinkedIn (https://www.linkedin.com/in/nayane-maia/).
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# Base de dados: https://www.kaggle.com/dipam7/student-grade-prediction
df = pd.read_csv('student-mat.csv')
df.head()
```
Para iniciar, vamos fazer uma análise exploratória desses dados e verificar tendências. Primeiramente vamos abrir uma informação do dataset, percebemos que existem muitos dados categóricos (objects). Se a gente quisesse realizar uma classificação, seria uma belezinha, pois os nossos dados podem ser agrupados por classificação e clusterização. Mas como queremos fazer a estimação das notas, que são dados numéricos e inteiros (int64), precisamos transformar esses dados em codificação númerica, como veremos a seguir.
```
df.info()
```
Ao realizar o describe dos nossos dados percebemos que não tem nenhum dado faltante, portanto, podemos seguir fazendo a análise exploratória dos dados.
```
# estatística descritiva
df.describe()
# distribuição das notas finais (G1)
plt.figure(figsize=(8, 4))
sns.countplot(df['G1'])
# reason to choose this school
sns.boxplot(x="reason", y='G1', data=df)
# Nota G1 x idade
plt.figure(figsize=(8, 4))
sns.boxplot(x="age", y='G1', data=df)
# Nota G1 x faltas
plt.figure(figsize=(10, 5))
sns.boxplot(x="absences", y='G1', data=df)
# Matriz de Correlação
corr = df.corr()
plt.figure(figsize=(14, 6))
mask = np.triu(np.ones_like(corr, dtype=bool))
sns.heatmap(corr, annot=True, mask=mask)
```
Como falei anteriormente, precisamos codificiar os dados categoricos em numéricos. Existem diversas formas de fazer isso, que você pode encontrar aqui nesse artigo algumas formas (https://contactsunny.medium.com/label-encoder-vs-one-hot-encoder-in-machine-learning-3fc273365621). Nesse projeto vamos usar o LabelEncoder, como:
```
# Codificação dos atributos categóricos
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
#seleciona os atributos categóricos
categoricas = df.select_dtypes(include='object').columns
# aplica o label encoder
df[categoricas] = df[categoricas].apply(lambda col: le.fit_transform(col))
df[categoricas].head()
```
Vamos abrir o dataset completo atualizado e codificado:
```
df.head()
```
Queremos estimar a nota dos alunos do primeiro período letivo (G1), para isso, precisamos definir quem vai ser as nossas variáveis dependentes e independentes, como abaixo:
```
X = df.iloc[:,0:30]
y = df['G1']
```
Como podemos ver, temos 30 features como variáveis independentes (X). Esse número grandes de variáveis pode deixar o treinamento do modelo muito lento, ou pode ter muitas variáveis com multicolinearidade, que afetam o desempenho do nosso modelo, além disso, pode deixar o nosso modelo com Underfitting (https://didatica.tech/underfitting-e-overfitting/). Para evitar isso, vamos usar um método para seleção de variáveis, para saber um pouco mais sobre alguns métodos, você pode encontrar aqui https://medium.com/neurondsai/comparando-m%C3%A9todos-de-sele%C3%A7%C3%A3o-de-vari%C3%A1veis-ef6ffe4f501. No nosso caso, vamos usar um método super simples, que é o método MDI (Mean Decrease Impurity) usando o Random Forest.
### Usando o método de seleção de variáveis pelo método de RandomForestClassifier
```
def mean_decrease_impurity(model, feature_names):
# Feature importance based on in-sample (IS) mean impurity reduction
feature_imp_df = {i: tree.feature_importances_ for i, tree in enumerate(model.estimators_)}
feature_imp_df = pd.DataFrame.from_dict(feature_imp_df, orient='index')
feature_imp_df.columns = feature_names
# Make sure that features with zero importance are not averaged, since the only reason for a 0 is that the feature
# was not randomly chosen. Replace those values with np.nan
feature_imp_df = feature_imp_df.replace(0, np.nan) # Because max_features = 1
importance = pd.concat({'mean': feature_imp_df.mean(),
'std': feature_imp_df.std() * feature_imp_df.shape[0] ** -0.5}, axis=1)
importance /= importance['mean'].sum()
return importance
import matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams['figure.figsize'] = (16, 6)
def plot_feature_importance(importance_df, oob_score, oos_score):
# Plot mean imp bars with std
plt.figure(figsize=(10, importance_df.shape[0] / 5))
importance_df.sort_values('mean', ascending=True, inplace=True)
importance_df['mean'].plot(kind='barh', color='b', alpha=0.25, xerr=importance_df['std'], error_kw={'ecolor': 'r'})
plt.title('Feature importance. OOB Score:{}; OOS score:{}'.format(round(oob_score, 4), round(oos_score, 4)))
plt.show()
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, BaggingClassifier
from sklearn.model_selection import cross_val_score
from sklearn.metrics import accuracy_score, make_scorer
#setup accuracy as score metric
scorer = make_scorer(accuracy_score)
#fit classifier
clf = RandomForestClassifier(max_features=1, oob_score=True)
fit = clf.fit(X, y)
#get importances dataframe
mdi_importances = mean_decrease_impurity(fit, X.columns)
#get oob & oos scores
oob_score = fit.oob_score_
oos_score = cross_val_score(clf, X, y, cv=3, scoring=scorer).mean()
plot_feature_importance(mdi_importances, oob_score=oob_score, oos_score=oos_score)
```
Para escolher as melhores variáveis não nos baseamos apenas pelas saídas geradas pelo RF, pois analisando o gráfico de correlação, percebemos que as variáveis selecionadas tinha uma baixa correlação, então mesclamos com as variáveis que melhor poderiam explicar o resultado final. Pois o principal objetivo do Machine Learning é exatamente esse, usar soluções que sejam palpáveis e explicavéis. Pensando sempre do ponto de vista do negócio. Qual solução eu quero trazer e de qual forma posso ajudar o meu cliente?
# Testando o modelo de Machine Learning
Importante lembrar que o número de variáveis selecionadas depende muito do ponto de vista do negócio também, do que adianta eu selecionar 10 variáveis , mas que não fazem sentido nenhum para o negócio ou responder as minhas dúvidas. No entanto, o contrário também existe, pode ser que apenas 4 ou 5 variáveis não seja o suficiente para explicar a sua solução. Então nessa fase, pesa muito as noções de negócio e seu entendimento de mundo como cientista de dados da área a qual você é especialista.
```
# inserindo apenas as variáveis que achamos importantes
X = X [['Medu','absences','Fedu','failures']]
y = y
```
Para o modelo não identificar e decorar tendências nos dados, vamos dividir o dataset em treinamento e teste. Em que 70% dos dados vão para treinamentos e 30% dos dados para teste, como:
```
# separando treinamento e teste
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_score
from sklearn.metrics import r2_score
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_squared_error
from math import sqrt
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=1)
print(X_train.shape); print(X_test.shape)
```
## Testando o algoritmo Random Forest Regressor
Chegou a hora de treinar o seu modelo de Machine Learning e não deixar a peruca cair!

> Para calibrar o nosso modelo vamos usar o GridSearch, dessa forma não perdemos tempo inserindo diversos parâmetros até achar um resultado razoável. Automaticamente ele ja seleciona os melhores parâmetros usando o crossvalidation (CV), deixando nosso modelo mais robusto, sem risco dele decorar o dataset e causar um Overfitting. Para saber mais sobre o CV veja aqui (https://medium.com/data-hackers/crossvalidation-de-maneira-did%C3%A1tica-79c9b080a6ec).
```
from sklearn.model_selection import cross_val_score, GridSearchCV
from sklearn.ensemble import RandomForestRegressor
model_params = {
'max_depth' : [2,3,4,5,6,10],
'n_estimators': [10,30,50,100,150]}
# create random forest model
rf_model = RandomForestRegressor(random_state=1)
clf = GridSearchCV(rf_model, model_params, cv=5)
model = clf.fit(X_train, y_train)
print('Train R^2 Treinamento: %.3f'%clf.best_estimator_.score(X_train, y_train))
print('Test R^2 Teste: %.3f'%clf.best_estimator_.score(X_test, y_test))
print('Best Parameters : ',model.best_params_)
```
Após o GridSearch fazer a seleção das melhores variáveis dividindo o treinamento em 5 partes (CV=5), tudo isso para ele não decorar os dados. Você deve ta se perguntando, mas para quê eu separei 70% dos meus dados para treinamento e agora preciso fazer isso de novo?! Como falei antes, mesmo você dividindo o dataset, o algoritmo pode aprender muito bem nessa fase de treinamento (em outras palavras, ele vai decorar os seus dados!) e ter um péssimo desempenho no teste final. Fazendo isso, os riscos são menores dele gravar as tendências.
Agora que ja temos os nossos *Best Parameters*, vamos agora inserir os melhores resultados no nosso modelo de regressão e avaliar o desempenho dele:
```
from sklearn.ensemble import RandomForestRegressor
regressor = RandomForestRegressor(n_estimators=150,max_depth= 2, random_state=1)
regressor.fit(X_train, y_train)
y_pred = regressor.predict(X_test)
# Avaliação do desempenho do modelo
print('R2 do teste:', r2_score(y_test, y_pred))
print('RMSE:' , sqrt(mean_squared_error(y_test, y_pred)))
print("MAPE=",np.mean(np.abs((y_test - y_pred) / y_test))* 100 )
```
Logo abaixo podemos ver a porcentagem de erro para cada repetição dos dados (Target), podemos ver que teve nota de um aluno que teve uma diferença de 41%:
```
# - Test prediction
performance = pd.DataFrame(y_pred, columns=['Prediction'])
# - Target data
y_test = y_test.reset_index(drop=True)
performance['Target'] = y_test
# - The difference in %
performance['Difference (%)']= np.absolute((performance['Target']
- performance['Prediction'])/
performance['Target']*100)
performance.head()
```
### Vamos avaliar as variáveis que foram escolhidas para estimar as notas dos alunos do primeiro período G1:
*Failures* é o número de reprovações de classe anteriores
*Absences* é o número de faltas durante o período letivo
*Fedu* é o nível de educação do pai.
*Medu* é o nível de educação da mãe.
Em que: 0 - nenhuma instrução, 1 - primary education (4th grade), 2 - 5th to 9th grade, 3 - secondary education, 4 - graduation/pos graduation
```
import seaborn as sns
import matplotlib.pyplot as plt
#definindo o tamanho das figuras
fig = plt.figure(figsize=(10, 10))
fig.add_subplot(221)
sns.violinplot(data=df, x="Fedu", y="G1",
split=True, inner="quart", linewidth=1,)
sns.despine(left=True)
fig.add_subplot(222)
sns.boxplot(x="Medu", y='G1', data=df)
sns.despine(left=True)
fig.add_subplot(223)
sns.boxplot(x='absences', y='G1', data=df)
sns.despine(left=True)
fig.add_subplot(224)
p2 = sns.barplot(x = 'G1', y = 'failures', data = df)
sns.despine(left=True)
```
### Resultados e Soluções
Analisando as informações das figuras anteriores, podemos perceber que o nível de instrução do pai e da mãe pode ter relação sim com as notas dos alunos, pois os mesmos podem ajudar ou não ajudar os filhos com as tarefas escolares. Da mesma forma que o número de faltas e a reprovação de classes anteriores também pode ajudar identificar as notas ruins.
Podemos perceber que a precisão do modelo realmente foi ruim, pois os dados estimados (vermelho) das notas dos alunos passaram bem longe dos dados reais (azul):
```
import seaborn as sns
sns.distplot(y_pred, hist = False, color = 'r', label = 'Predicted Values')
sns.distplot(y_test, hist = False, color = 'b', label = 'Observado Values')
plt.title('Observado vs Estimado', fontsize = 16)
plt.xlabel('Values', fontsize = 12)
plt.ylabel('Frequency', fontsize = 12)
plt.legend(loc = 'upper left', fontsize = 13)
```
---
# Questionamentos
## 
O que poderia ser feito nesse modelo para aumentar a precisão??
Talvez inserir outras features?
Outro método de seleção de variáveis?
Transformar e padronizar os dados de entrada?
Fazer teste de multicolinearidade e eliminar variáveis tendenciosas?
Fica aí esses questionamentos para vocês resolverem! #peace
No kagle (https://www.kaggle.com/dipam7/student-grade-prediction/code), percebi que a maior precisão encontrada para esse Dataset usando o Random Forest foi de R2=0.86. No entanto, quando fui analisar as features de entrada que eles usaram, uma das melhores variáveis selecionadas foram as notas do bimestre dois e notas finais (G2, G3). Decidimos não inserir essas features como variáveis independentes porque achamos que seria muito tendencioso, ja que foram as variáveis com a maior correlação de pearson.
No kagle mesmo eles dizem que: "o atributo alvo G3 tem forte correlação com os atributos G2 e G1. Isso ocorre porque G3 é a nota do último ano (emitida no 3º período), enquanto G1 e G2 correspondem às notas do 1º e 2º períodos. É mais difícil prever G3 sem G2 e G1, mas essa previsão é muito mais útil (consulte a fonte do artigo para obter mais detalhes)."
Vamos imaginar que você seja contratado como cientista de dados para estimar as notas dos alunos de uma escola, e se acontecer de uma escola não ter outras turmas para estimar as notas ou outros períodos letivos? E se todas as variáveis disponíveis forem ausência, sexo, saúde, razões, etc?? Para evitar que nosso modelo fosse tendencioso por causa das notas das outras turmas, colocamos todas as variáveis, exceto as notas dos outros períodos avaliativos e notas finas do bimestre, talvez por isso a precisão foi tão baixa do nosso modelo.
Mas temos que lembrar que nem sempre podemos olhar apenas a precisão do modelo, e lembrar que o nosso erro de notas foi baixo, variando de 2.8 para mais ou para menos (RMSE), o que representa um erro médio de aproximadamente 24% (MAPE). Portanto, o ideal aqui seria testar outros modelos para ver se a acurácia e a precisão vão melhorar.

|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# Base de dados: https://www.kaggle.com/dipam7/student-grade-prediction
df = pd.read_csv('student-mat.csv')
df.head()
df.info()
# estatística descritiva
df.describe()
# distribuição das notas finais (G1)
plt.figure(figsize=(8, 4))
sns.countplot(df['G1'])
# reason to choose this school
sns.boxplot(x="reason", y='G1', data=df)
# Nota G1 x idade
plt.figure(figsize=(8, 4))
sns.boxplot(x="age", y='G1', data=df)
# Nota G1 x faltas
plt.figure(figsize=(10, 5))
sns.boxplot(x="absences", y='G1', data=df)
# Matriz de Correlação
corr = df.corr()
plt.figure(figsize=(14, 6))
mask = np.triu(np.ones_like(corr, dtype=bool))
sns.heatmap(corr, annot=True, mask=mask)
# Codificação dos atributos categóricos
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
#seleciona os atributos categóricos
categoricas = df.select_dtypes(include='object').columns
# aplica o label encoder
df[categoricas] = df[categoricas].apply(lambda col: le.fit_transform(col))
df[categoricas].head()
df.head()
X = df.iloc[:,0:30]
y = df['G1']
def mean_decrease_impurity(model, feature_names):
# Feature importance based on in-sample (IS) mean impurity reduction
feature_imp_df = {i: tree.feature_importances_ for i, tree in enumerate(model.estimators_)}
feature_imp_df = pd.DataFrame.from_dict(feature_imp_df, orient='index')
feature_imp_df.columns = feature_names
# Make sure that features with zero importance are not averaged, since the only reason for a 0 is that the feature
# was not randomly chosen. Replace those values with np.nan
feature_imp_df = feature_imp_df.replace(0, np.nan) # Because max_features = 1
importance = pd.concat({'mean': feature_imp_df.mean(),
'std': feature_imp_df.std() * feature_imp_df.shape[0] ** -0.5}, axis=1)
importance /= importance['mean'].sum()
return importance
import matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams['figure.figsize'] = (16, 6)
def plot_feature_importance(importance_df, oob_score, oos_score):
# Plot mean imp bars with std
plt.figure(figsize=(10, importance_df.shape[0] / 5))
importance_df.sort_values('mean', ascending=True, inplace=True)
importance_df['mean'].plot(kind='barh', color='b', alpha=0.25, xerr=importance_df['std'], error_kw={'ecolor': 'r'})
plt.title('Feature importance. OOB Score:{}; OOS score:{}'.format(round(oob_score, 4), round(oos_score, 4)))
plt.show()
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, BaggingClassifier
from sklearn.model_selection import cross_val_score
from sklearn.metrics import accuracy_score, make_scorer
#setup accuracy as score metric
scorer = make_scorer(accuracy_score)
#fit classifier
clf = RandomForestClassifier(max_features=1, oob_score=True)
fit = clf.fit(X, y)
#get importances dataframe
mdi_importances = mean_decrease_impurity(fit, X.columns)
#get oob & oos scores
oob_score = fit.oob_score_
oos_score = cross_val_score(clf, X, y, cv=3, scoring=scorer).mean()
plot_feature_importance(mdi_importances, oob_score=oob_score, oos_score=oos_score)
# inserindo apenas as variáveis que achamos importantes
X = X [['Medu','absences','Fedu','failures']]
y = y
# separando treinamento e teste
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_score
from sklearn.metrics import r2_score
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_squared_error
from math import sqrt
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=1)
print(X_train.shape); print(X_test.shape)
from sklearn.model_selection import cross_val_score, GridSearchCV
from sklearn.ensemble import RandomForestRegressor
model_params = {
'max_depth' : [2,3,4,5,6,10],
'n_estimators': [10,30,50,100,150]}
# create random forest model
rf_model = RandomForestRegressor(random_state=1)
clf = GridSearchCV(rf_model, model_params, cv=5)
model = clf.fit(X_train, y_train)
print('Train R^2 Treinamento: %.3f'%clf.best_estimator_.score(X_train, y_train))
print('Test R^2 Teste: %.3f'%clf.best_estimator_.score(X_test, y_test))
print('Best Parameters : ',model.best_params_)
from sklearn.ensemble import RandomForestRegressor
regressor = RandomForestRegressor(n_estimators=150,max_depth= 2, random_state=1)
regressor.fit(X_train, y_train)
y_pred = regressor.predict(X_test)
# Avaliação do desempenho do modelo
print('R2 do teste:', r2_score(y_test, y_pred))
print('RMSE:' , sqrt(mean_squared_error(y_test, y_pred)))
print("MAPE=",np.mean(np.abs((y_test - y_pred) / y_test))* 100 )
# - Test prediction
performance = pd.DataFrame(y_pred, columns=['Prediction'])
# - Target data
y_test = y_test.reset_index(drop=True)
performance['Target'] = y_test
# - The difference in %
performance['Difference (%)']= np.absolute((performance['Target']
- performance['Prediction'])/
performance['Target']*100)
performance.head()
import seaborn as sns
import matplotlib.pyplot as plt
#definindo o tamanho das figuras
fig = plt.figure(figsize=(10, 10))
fig.add_subplot(221)
sns.violinplot(data=df, x="Fedu", y="G1",
split=True, inner="quart", linewidth=1,)
sns.despine(left=True)
fig.add_subplot(222)
sns.boxplot(x="Medu", y='G1', data=df)
sns.despine(left=True)
fig.add_subplot(223)
sns.boxplot(x='absences', y='G1', data=df)
sns.despine(left=True)
fig.add_subplot(224)
p2 = sns.barplot(x = 'G1', y = 'failures', data = df)
sns.despine(left=True)
import seaborn as sns
sns.distplot(y_pred, hist = False, color = 'r', label = 'Predicted Values')
sns.distplot(y_test, hist = False, color = 'b', label = 'Observado Values')
plt.title('Observado vs Estimado', fontsize = 16)
plt.xlabel('Values', fontsize = 12)
plt.ylabel('Frequency', fontsize = 12)
plt.legend(loc = 'upper left', fontsize = 13)
| 0.667906 | 0.970465 |
<link rel="stylesheet" href="../../styles/theme_style.css">
<!--link rel="stylesheet" href="../../styles/header_style.css"-->
<link rel="stylesheet" href="https://cdnjs.cloudflare.com/ajax/libs/font-awesome/4.7.0/css/font-awesome.min.css">
<table width="100%">
<tr>
<td id="image_td" width="15%" class="header_image_color_4"><div id="image_img" class="header_image_4"></div></td>
<td class="header_text"> Generation of a time axis (conversion of samples into seconds) </td>
</tr>
</table>
<div id="flex-container">
<div id="diff_level" class="flex-item">
<strong>Difficulty Level:</strong> <span class="fa fa-star checked"></span>
<span class="fa fa-star checked"></span>
<span class="fa fa-star"></span>
<span class="fa fa-star"></span>
<span class="fa fa-star"></span>
</div>
<div id="tag" class="flex-item-tag">
<span id="tag_list">
<table id="tag_list_table">
<tr>
<td class="shield_left">Tags</td>
<td class="shield_right" id="tags">pre-process☁time☁conversion</td>
</tr>
</table>
</span>
<!-- [OR] Visit https://img.shields.io in order to create a tag badge-->
</div>
</div>
All electrophysiological signals, collected by PLUX acquisition systems, are, in its essence, time series.
Raw data contained in the generated .txt, .h5 and .edf files consists in samples and each sample value is in a raw value with 8 or 16 bits that needs to be converted to a physical unit by the respective transfer function.
PLUX has examples of conversion rules for each sensor (in separate .pdf files), which may be accessed at <a href="http://biosignalsplux.com/en/learn/documentation">"Documentation>>Sensors" section <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></a> of <strong><span class="color2">biosignalsplux</span></strong> website.
<img src="../../images/pre-process/generation_of_time_axis/sensors_section.gif">
Although each file returned by <strong><span class="color2">OpenSignals</span></strong> contains a sequence number linked to each sample, giving a notion of "time order" and that can be used as x axis, working with real time units is, in many occasions, more intuitive.
So, in the present **<span class="color5">Jupyter Notebook</span>** is described how to associate a time axis to an acquired signal, taking into consideration the number of acquired samples and the respective sampling rate.
<hr>
<p class="steps">1 - Importation of the needed packages </p>
```
# Package dedicated to download files remotely
from wget import download
# Package used for loading data from the input text file and for generation of a time axis
from numpy import loadtxt, linspace
# Package used for loading data from the input h5 file
import h5py
# biosignalsnotebooks own package.
import biosignalsnotebooks as bsnb
```
<p class="steps"> A - Text Files</p>
<p class="steps">A1 - Load of support data inside .txt file (described in a <span class="color5">Jupyter Notebook</span> entitled <a href="../Load/open_txt.ipynb"><strong> "Load acquired data from .txt file" <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a>) </p>
```
# Download of the text file followed by content loading.
txt_file_url = "https://drive.google.com/uc?export=download&id=1m7E7PnKLfcd4HtOASH6vRmyBbCmIEkLf"
txt_file = download(txt_file_url, out="download_file_name.txt")
txt_file = open(txt_file, "r")
# [Internal code for overwrite file if already exists]
import os
import shutil
txt_file.close()
if os.path.exists("download_file_name.txt"):
shutil.move(txt_file.name,"download_file_name.txt")
txt_file = "download_file_name.txt"
txt_file = open(txt_file, "r")
```
<p class="steps">A2 - Load of acquisition samples (in this case from the third column of the text file - list entry 2)</p>
```
txt_signal = loadtxt(txt_file)[:, 2]
```
<p class="steps">A3 - Determination of the number of acquired samples</p>
```
# Number of acquired samples
nbr_samples_txt = len(txt_signal)
from sty import fg, rs
print(fg(98,195,238) + "\033[1mNumber of samples (.txt file):\033[0m" + fg.rs + " " + str(nbr_samples_txt))
```
<p class="steps"> B - H5 Files</p>
<p class="steps">B1 - Load of support data inside .h5 file (described in the <span class="color5">Jupyter Notebook</span> entitled <a href="../Load/open_h5.ipynb"><strong> "Load acquired data from .h5 file"<img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a>) </p>
```
# Download of the .h5 file followed by content loading.
h5_file_url = "https://drive.google.com/uc?export=download&id=1UgOKuOMvHTm3LlQ_e7b6R_qZL5cdL4Rv"
h5_file = download(h5_file_url, out="download_file_name.h5")
h5_object = h5py.File(h5_file)
# [Internal code for overwrite file if already exists]
import os
import shutil
h5_object.close()
if os.path.exists("download_file_name.h5"):
shutil.move(h5_file,"download_file_name.h5")
h5_file = "download_file_name.h5"
h5_object = h5py.File(h5_file)
```
<p class="steps">B2 - Load of acquisition samples inside .h5 file</p>
```
# Device mac-address.
mac_address = list(h5_object.keys())[0]
# Access to signal data acquired by the device identified by "mac_address" in "channel_1".
h5_signal = list(h5_object.get(mac_address).get("raw").get("channel_1"))
```
<p class="steps">B3 - Determination of the number of acquired samples</p>
```
# Number of acquired samples
nbr_samples_h5 = len(h5_signal)
print(fg(232,77,14) + "\033[1mNumber of samples (.h5 file):\033[0m" + fg.rs + " " + str(nbr_samples_h5))
```
As it can be seen, the number of samples is equal for both file types.
```
print(fg(98,195,238) + "\033[1mNumber of samples (.txt file):\033[0m" + fg.rs + " " + str(nbr_samples_txt))
print(fg(232,77,14) + "\033[1mNumber of samples (.h5 file):\033[0m" + fg.rs + " " + str(nbr_samples_h5))
```
So, we can simplify and reduce the number of variables:
```
nbr_samples = nbr_samples_txt
```
Like described in the Notebook intro, for generating a time-axis it is needed the <strong><span class="color4">number of acquired samples</span></strong> and the <strong><span class="color7">sampling rate</span></strong>.
Currently the only unknown parameter is the <strong><span class="color7">sampling rate</span></strong>, which can be easily accessed for .txt and .h5 files as described in <a href="../Load/signal_loading_preparatory_steps.ipynb" target="_blank">"Signal Loading - Working with File Header"<img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></a>.
For our acquisition the sampling rate is:
```
sampling_rate = 1000 # Hz
```
<p class="steps">AB4 - Determination of acquisition time in seconds</p>
```
# Conversion between sample number and seconds
acq_time = nbr_samples / sampling_rate
print ("Acquisition Time: " + str(acq_time) + " s")
```
<p class="steps">AB5 - Creation of the time axis (between 0 and 417.15 seconds) through <span class="color4">linspace</span> function</p>
```
time_axis = linspace(0, acq_time, nbr_samples)
print ("Time-Axis: \n" + str(time_axis))
```
<p class="steps">AB6 - Plot of the acquired signal (first 10 seconds) with the generated time-axis</p>
```
bsnb.plot(time_axis[:10*sampling_rate], txt_signal[:10*sampling_rate])
```
*This procedure can be automatically done by **generate_time** function in **conversion** module of **<span class="color2">biosignalsnotebooks</span>** package*
```
time_axis_auto = bsnb.generate_time(h5_file_url)
from numpy import array
print ("Time-Axis returned by generateTime function:")
print (array(time_axis_auto))
```
Time is a really important "dimension" in our daily lives and particularly on signal processing analysis. Without a time "anchor" like <strong><span class="color7">sampling rate</span></strong> it is very difficult to link the acquired digital data with real events.
Concepts like "temporal duration" or "time rate" become meaningless, being more difficult to take adequate conclusions.
However, as can be seen, a researcher in possession of the data to process and a single parameter (sampling rate) can easily generate a time-axis, following the demonstrated procedure.
<strong><span class="color7">We hope that you have enjoyed this guide. </span><span class="color2">biosignalsnotebooks</span><span class="color4"> is an environment in continuous expansion, so don't stop your journey and learn more with the remaining <a href="../MainFiles/biosignalsnotebooks.ipynb">Notebooks <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></a></span></strong> !
<span class="color6">**Auxiliary Code Segment (should not be replicated by the user)**</span>
```
from biosignalsnotebooks.__notebook_support__ import css_style_apply
css_style_apply()
%%html
<script>
// AUTORUN ALL CELLS ON NOTEBOOK-LOAD!
require(
['base/js/namespace', 'jquery'],
function(jupyter, $) {
$(jupyter.events).on("kernel_ready.Kernel", function () {
console.log("Auto-running all cells-below...");
jupyter.actions.call('jupyter-notebook:run-all-cells-below');
jupyter.actions.call('jupyter-notebook:save-notebook');
});
}
);
</script>
```
|
github_jupyter
|
# Package dedicated to download files remotely
from wget import download
# Package used for loading data from the input text file and for generation of a time axis
from numpy import loadtxt, linspace
# Package used for loading data from the input h5 file
import h5py
# biosignalsnotebooks own package.
import biosignalsnotebooks as bsnb
# Download of the text file followed by content loading.
txt_file_url = "https://drive.google.com/uc?export=download&id=1m7E7PnKLfcd4HtOASH6vRmyBbCmIEkLf"
txt_file = download(txt_file_url, out="download_file_name.txt")
txt_file = open(txt_file, "r")
# [Internal code for overwrite file if already exists]
import os
import shutil
txt_file.close()
if os.path.exists("download_file_name.txt"):
shutil.move(txt_file.name,"download_file_name.txt")
txt_file = "download_file_name.txt"
txt_file = open(txt_file, "r")
txt_signal = loadtxt(txt_file)[:, 2]
# Number of acquired samples
nbr_samples_txt = len(txt_signal)
from sty import fg, rs
print(fg(98,195,238) + "\033[1mNumber of samples (.txt file):\033[0m" + fg.rs + " " + str(nbr_samples_txt))
# Download of the .h5 file followed by content loading.
h5_file_url = "https://drive.google.com/uc?export=download&id=1UgOKuOMvHTm3LlQ_e7b6R_qZL5cdL4Rv"
h5_file = download(h5_file_url, out="download_file_name.h5")
h5_object = h5py.File(h5_file)
# [Internal code for overwrite file if already exists]
import os
import shutil
h5_object.close()
if os.path.exists("download_file_name.h5"):
shutil.move(h5_file,"download_file_name.h5")
h5_file = "download_file_name.h5"
h5_object = h5py.File(h5_file)
# Device mac-address.
mac_address = list(h5_object.keys())[0]
# Access to signal data acquired by the device identified by "mac_address" in "channel_1".
h5_signal = list(h5_object.get(mac_address).get("raw").get("channel_1"))
# Number of acquired samples
nbr_samples_h5 = len(h5_signal)
print(fg(232,77,14) + "\033[1mNumber of samples (.h5 file):\033[0m" + fg.rs + " " + str(nbr_samples_h5))
print(fg(98,195,238) + "\033[1mNumber of samples (.txt file):\033[0m" + fg.rs + " " + str(nbr_samples_txt))
print(fg(232,77,14) + "\033[1mNumber of samples (.h5 file):\033[0m" + fg.rs + " " + str(nbr_samples_h5))
nbr_samples = nbr_samples_txt
sampling_rate = 1000 # Hz
# Conversion between sample number and seconds
acq_time = nbr_samples / sampling_rate
print ("Acquisition Time: " + str(acq_time) + " s")
time_axis = linspace(0, acq_time, nbr_samples)
print ("Time-Axis: \n" + str(time_axis))
bsnb.plot(time_axis[:10*sampling_rate], txt_signal[:10*sampling_rate])
time_axis_auto = bsnb.generate_time(h5_file_url)
from numpy import array
print ("Time-Axis returned by generateTime function:")
print (array(time_axis_auto))
from biosignalsnotebooks.__notebook_support__ import css_style_apply
css_style_apply()
%%html
<script>
// AUTORUN ALL CELLS ON NOTEBOOK-LOAD!
require(
['base/js/namespace', 'jquery'],
function(jupyter, $) {
$(jupyter.events).on("kernel_ready.Kernel", function () {
console.log("Auto-running all cells-below...");
jupyter.actions.call('jupyter-notebook:run-all-cells-below');
jupyter.actions.call('jupyter-notebook:save-notebook');
});
}
);
</script>
| 0.426083 | 0.59752 |
# Determine the matrix dimension for a given number of matrix elements
```
import sys
sys.path.append('..')
import sparsity_pattern as spat
import math
import scipy.sparse
import numpy as np
from typing import List
def print_matrix(pat: List[List[int]], n_rows, n_cols):
# convert to matrix
mat = scipy.sparse.lil_matrix((n_rows, n_cols), dtype=np.int64)
idx_rows, idx_cols = np.array(pat)[:, 0], np.array(pat)[:, 1]
mat[idx_rows, idx_cols] = 1
# print
print("Sparsity Pattern:")
print(pat)
print("Matrix:")
print(mat.todense())
```
# `'circle'` pattern
The matrix dimension `n` and the number of `offsets` determine the number of matrix elements for a quadratic sparse matrix with 'circle' pattern.
If the number of matrix elements `n_max_params` is given, the `get_matrix_dims_circle` function searches for hyperparameter combinations (n, offsets) that result in circle patterns with exactly or slightly less than `n_max_params` matrix elements.
### How to use `get_matrix_dims_circle`
The function `get_matrix_dims_circle` returns a list of dict.
```
n_max_params = 50
hyperparams = spat.utils.get_matrix_dims_circle(n_max_params)
hyperparams
```
Each dict is a feasible combination of a matrix dimension and offsets for the 'circle' pattern.
```
pat = spat.get("circle", **hyperparams[3])
n_dim = hyperparams[3]['n']
print_matrix(pat, n_dim, n_dim)
```
### How does it work?
Assume we want to squeeze up to `n_max_params` weights into a quadratic sparse matrix with 'circle' pattern.
```
n_max_params = 50
```
The square root `int(sqr(n_max_params))` is the first smallest matrix dimension `n_min_dim` that can hold most of the desired `n_max_params` of weights. However, we need to subtract `-1` because the diagonals are assumed to be 0. `n_min_dim` is also the the maximum number of offsets we can squeeze into such a matrix.
```
n_min_dim = int(math.sqrt(n_max_params)) - 1
```
We loop over `n_offsets = [1, 2, .. n_min_dim]`.
The matrix dimension `n_dim` for the desired number of weights `n_max_params` and number of offsets `n_offsets` is `int(n_max_params / n_offsets)`.
```
for n_offsets in range(1, n_min_dim+1):
n_dim = n_max_params // n_offsets
result = {"n_dim": n_dim, "offsets": list(range(1, n_offsets + 1))}
# add more information
if True:
pat = spat.get("circle", n_dim, range(1, n_offsets + 1))
n_act_params = len(pat)
result = {**result, "n_act_params": n_act_params, "ratio_squeezed": n_act_params / n_max_params}
print(result)
```
|
github_jupyter
|
import sys
sys.path.append('..')
import sparsity_pattern as spat
import math
import scipy.sparse
import numpy as np
from typing import List
def print_matrix(pat: List[List[int]], n_rows, n_cols):
# convert to matrix
mat = scipy.sparse.lil_matrix((n_rows, n_cols), dtype=np.int64)
idx_rows, idx_cols = np.array(pat)[:, 0], np.array(pat)[:, 1]
mat[idx_rows, idx_cols] = 1
# print
print("Sparsity Pattern:")
print(pat)
print("Matrix:")
print(mat.todense())
n_max_params = 50
hyperparams = spat.utils.get_matrix_dims_circle(n_max_params)
hyperparams
pat = spat.get("circle", **hyperparams[3])
n_dim = hyperparams[3]['n']
print_matrix(pat, n_dim, n_dim)
n_max_params = 50
n_min_dim = int(math.sqrt(n_max_params)) - 1
for n_offsets in range(1, n_min_dim+1):
n_dim = n_max_params // n_offsets
result = {"n_dim": n_dim, "offsets": list(range(1, n_offsets + 1))}
# add more information
if True:
pat = spat.get("circle", n_dim, range(1, n_offsets + 1))
n_act_params = len(pat)
result = {**result, "n_act_params": n_act_params, "ratio_squeezed": n_act_params / n_max_params}
print(result)
| 0.191933 | 0.954351 |
```
%load_ext autoreload
%autoreload 2
%aimport utils_1_1
import pandas as pd
import numpy as np
import altair as alt
from altair_saver import save
import datetime
import dateutil.parser
from os.path import join
from constants_1_1 import SITE_FILE_TYPES
from utils_1_1 import (
get_site_file_paths,
get_site_file_info,
get_site_ids,
get_visualization_subtitle,
get_country_color_map,
)
from theme import apply_theme
from web import for_website
alt.data_transformers.disable_max_rows(); # Allow using rows more than 5000
data_release='2021-04-29'
consistent_date = {
'2020-Mar-Apr': "'20 Mar - '20 Apr",
'2020-May-Jun': "'20 May - '20 Jun",
'2020-Jul-Aug': "'20 Jul - '20 Aug",
'2020-Sep-Oct': "'20 Sep - '20 Oct",
'2020-Nov-2021-Jan': "'20 Nov - '21 Jan"
}
date = ['Mar - Apr', 'May - Jun', 'Jul - Aug', 'Sep - Oct', 'Since Nov']
date = ["'20 Mar - '20 Apr", "'20 May - '20 Jun", "'20 Jul - '20 Aug", "'20 Sep - '20 Oct", "'20 Nov - '21 Jan"]
sites = ['META', 'APHP', 'FRBDX', 'ICSM', 'NWU', 'BIDMC', 'MGB', 'UCLA', 'UMICH', 'UPENN', 'UPITT', 'VA1', 'VA2', 'VA3', 'VA4', 'VA5']
site_colors = ['black', '#D45E00', '#0072B2', '#CB7AA7', '#E79F00', '#029F73', '#DBD03C', '#57B4E9', '#57B4E9', '#57B4E9', '#57B4E9', '#57B4E9']
df = pd.read_csv(join("..", "data", "Phase2.1SurvivalRSummariesPublic", "ToShare", "table.score.toShare.csv"))
print(df.head())
# Rename columns
df = df.drop(columns=["Unnamed: 0"])
df = df.rename(columns={
'siteid': 'site',
'calendar_month': 'month'
})
# More readable values
df.site = df.site.apply(lambda x: x.upper())
print(df.site.unique().tolist())
print(df.month.unique().tolist())
# Drop "combine" sites
df = df[df.site != "COMBINE"]
# df = pd.melt(df, id_vars=['siteid'], value_vars=date, var_name='date', value_name='value')
df.month = df.month.apply(lambda x: consistent_date[x])
# Add a reference (META)
# df['reference'] = df.date.apply(lambda x: df[(df.date == x) & (df.siteid == 'META')].value.sum())
df.head()
def risk(_d, metric='pos'):
d = _d.copy()
"""
DATA PREPROCESSING...
"""
d.loc[d.site == 'combine', 'site'] = 'All Sites'
d.cat = d.cat.apply(lambda x: {
'L':'Low Risk',
'M': 'Medium Risk',
'H': 'High Risk',
'H/M': 'High/Medium',
'L/M': 'Low/Medium'
}[x])
"""
PLOT!
"""
y_title = '% of Patients in Each Category' if metric == 'pos' else '% of Event in Each Category'
colors = ['#7BADD1', '#427BB5', '#14366E'] if metric == 'pos' else ['#A8DED1', '#3FA86F', '#005A24'] if metric == 'ppv' else ['red', 'salmon']
colorDomain = ['High/Medium', 'Low/Medium'] if metric == 'rr' else ['Low Risk', 'Medium Risk', 'High Risk']
width = 300
size = 50
y_scale = alt.Scale(domain=[0, 1]) if metric == 'pos' or metric=='ppv' else alt.Scale()
bar = alt.Chart(
d
).transform_calculate(
order="{'Low Risk':0, 'Medium Risk': 1, 'High Risk': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title='Month', scale=alt.Scale(domain=['Mar-Apr', 'May-Jun', 'Jul-Aug', 'Sep-Oct', 'Since Nov'])),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Category', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width
)
if metric == 'pos':
bar = bar.mark_bar(
size=size, stroke='black'
)
else:
bar = bar.mark_line(
size=3, point=True, opacity=0.8
)
d['visibility'] = d['value'] > 0.08
text = alt.Chart(
d
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).mark_text(size=16, dx=0, dy=5, color='white', baseline='top', fontWeight=500).encode(
x=alt.X('month:N'),
y=alt.Y('value:Q', stack='zero'),
detail='cat:N',
text=alt.Text('value:Q', format='.0%'),
order="order:O",
opacity=alt.Opacity('visibility:N', scale=alt.Scale(domain=[True, False], range=[1, 0]))
)
# .transform_filter(
# (f'datum.value > 0.10')
# )
if metric == 'pos':
bar = (bar + text)
bar = bar.facet(
column=alt.Column('site:N', header=alt.Header(title=None)),
)
"""
COMBINE
"""
res = bar.properties(
title={
"text": [
f"Distribution of Risk Scores" if metric == 'pos' else f"Event Rate of Risk Scores"
],
"dx": 80,
"subtitle": [
# lab, #.title(),
get_visualization_subtitle(data_release=data_release, with_num_sites=False)
],
"subtitleColor": "gray",
}
)
return res
d = df.copy()
width = 160
height = 180
height2 = 140
size = 28
point=alt.OverlayMarkDef(filled=False, fill='white', strokeWidth=2)
"""
DATA PREPROCESSING...
"""
d.loc[d.site == 'combine', 'site'] = 'All Sites'
d.cat = d.cat.apply(lambda x: {
'L':'Low',
'M': 'Medium',
'H': 'High',
'H/M': 'H/M',
'L/M': 'L/M'
}[x])
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%% TOP %%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='pos'
y_title = '% of Patients in Each Category'
y_scale = alt.Scale(domain=[0, 1])
colors = ['#7BADD1', '#427BB5', '#14366E']
# colorDomain = ['Low Risk', 'Medium Risk', 'High Risk']
colorDomain = ['Low', 'Medium', 'High']
# colorDomain = ['L', 'M', 'H']
bar = alt.Chart(
d
).transform_calculate(
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Risk Level', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height
)
bar = bar.mark_bar(
size=size, stroke='black'
)
d['visibility'] = d['value'] > 0.08
text = alt.Chart(
d
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).mark_text(size=14, dx=0, dy=5, color='white', baseline='top', fontWeight=500).encode(
x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55, domain=False), sort=date),
y=alt.Y('value:Q', stack='zero'),
detail='cat:N',
text=alt.Text('value:Q', format='.0%'),
order="order:O",
opacity=alt.Opacity('visibility:N', scale=alt.Scale(domain=[True, False], range=[1, 0]), legend=None)
)
if metric == 'pos':
bar = (bar + text)
bar = bar.facet(
column=alt.Column('site:N', header=alt.Header(title=None), sort=sites),
spacing=20
)
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%% Bottom %%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='ppv'
y_title = '% of Event'
colors = ['#A8DED1', '#3FA86F', '#005A24']
colors = ['#00A87E', '#00634B', 'black']
y_scale = alt.Scale(domain=[0, 1])
line = alt.Chart(
d
).transform_calculate(
# order="{'Low Risk':0, 'Medium Risk': 1, 'High Risk': 2}[datum.variable]"
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
# x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
x=alt.X("month:N", title='Month', scale=alt.Scale(domain=date), axis=alt.Axis(grid=False, ticks=False, labels=False, domain=False, title=None, labelAngle=-55)),
y=alt.Y("value:Q", title=y_title, scale=y_scale),
color=alt.Color("cat:N", title='Risk Level', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height2
)
line = line.mark_line(
size=3, point=point, opacity=0.8
).facet(
column=alt.Column('site:N', header=alt.Header(title=None), sort=sites),
)
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%% Bottom 2 %%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='rr'
y_title = 'Ratio Between Risk Levels'
# colors = ['red', 'salmon']
colors = ['#D45E00', '#351800']
# colorDomain = ['High/Medium', 'Low/Medium']
colorDomain = ['L/M', 'H/M']
y_scale = alt.Scale(domain=[0, 10], clamp=True)
line2 = alt.Chart(
d
).transform_calculate(
# order="{'High/Medium':0, 'Low/Medium': 1}[datum.variable]"
order="{'H/M':0, 'L/M': 1}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
# x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
x=alt.X("month:N", title=None, scale=alt.Scale(domain=date), axis=alt.Axis(grid=False, domain=False, labelAngle=-55)),
# x=alt.X("month:N", title='Month', scale=alt.Scale(domain=date), axis=alt.Axis(grid=True, ticks=False, labels=False, domain=False, title=None)),
y=alt.Y("value:Q", title=y_title, scale=y_scale),
color=alt.Color("cat:N", title='Risk Ratio', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height2
)
line2 = line2.mark_line(
size=3, point=point, opacity=0.8
).facet(
column=alt.Column('site:N', header=alt.Header(title=None, labels=False), sort=sites),
)
# Just to show color legend and left axis
# line_not_first = line
# line_not_first = line.encode(
# x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
# y=alt.Y("value:Q", title=None, axis=alt.Axis(format='.0%', ticks=False, labels=False, domain=False), scale=y_scale),
# color=alt.Color("cat:N", title='Risk Level', scale=alt.Scale(domain=colorDomain, range=colors), legend=None),
# order="order:O"
# )
# line2_first = line2.encode(
# x=alt.X("month:N", title=None, sort=date),
# y=alt.Y("value:Q", title=None, axis=alt.Axis(format='.0%', ticks=False, labels=False, domain=False), scale=y_scale),
# color=alt.Color("cat:N", title='Risk Ratio (Right Y Axis)', scale=alt.Scale(domain=colorDomain, range=colors)),
# order="order:O"
# )
# line2_last = line2.encode(
# x=alt.X("month:N", title=None, sort=date),
# y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
# color=alt.Color("cat:N", title='Risk Ratio', scale=alt.Scale(domain=colorDomain, range=colors), legend=None),
# order="order:O"
# )
# line = alt.concat(*(
# alt.layer(line, line2_first, title={
# "text": site,
# "fontSize": 16,
# "dx": 130}).transform_filter(alt.datum.site == site).resolve_scale(y='independent', color='independent') if site == 'META' else
# alt.layer(line_not_first, line2_last, title={
# "text": site,
# "fontSize": 16,
# "dx": 85}).transform_filter(alt.datum.site == site).resolve_scale(y='independent', color='independent') if site == sites[-1] else
# alt.layer(line_not_first, line2, title={
# "text": site,
# "fontSize": 16,
# "dx": 85}).transform_filter(alt.datum.site == site).resolve_scale(y='independent', color='independent')
# for site in sites
# ), spacing=3).resolve_scale(color='shared')
print(d.site.unique())
"""
COMBINE
"""
top = bar.properties(
title={
"text": [
f"Distribution Of Parients By Risk Level"
],
"fontWeight": "normal",
"dx": 170,
"subtitle": [
get_visualization_subtitle(data_release=data_release, with_num_sites=False)
],
"subtitleColor": "gray",
}
)
bot = line.properties(
title={
"text": [
f"Event Rate Of Risk Scores"
],
"dx": 150,
# "subtitle": [
# get_visualization_subtitle(data_release=data_release, with_num_sites=False)
# ],
"subtitleColor": "gray",
}
)
# top.display()
# bot.display()
# line2.display()
res = alt.vconcat(
top,
bot,
line2,
spacing=10
).resolve_scale(y='independent', color='independent')
"""
STYLE
"""
res = apply_theme(
res,
axis_y_title_font_size=16,
title_anchor='start',
legend_orient='left',
header_label_orient='top',
# legend_title_orient='left',
axis_label_font_size=14,
header_label_font_size=16,
point_size=90
)
res
d = df.copy()
d = d[d.site == 'META']
width = 200
height = 200
size = 30
point=alt.OverlayMarkDef(filled=False, fill='white', strokeWidth=2)
"""
DATA PREPROCESSING...
"""
d.loc[d.site == 'combine', 'site'] = 'All Sites'
d.cat = d.cat.apply(lambda x: {
'L':'Low',
'M': 'Medium',
'H': 'High',
'H/M': 'H/M',
'L/M': 'L/M'
}[x])
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%% TOP %%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='pos'
y_title = '% of Patients in Each Category'
y_scale = alt.Scale(domain=[0, 1])
colors = ['#7BADD1', '#427BB5', '#14366E']
# colorDomain = ['Low Risk', 'Medium Risk', 'High Risk']
colorDomain = ['Low', 'Medium', 'High']
# colorDomain = ['L', 'M', 'H']
bar = alt.Chart(
d
).transform_calculate(
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Risk Level', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height
)
bar = bar.mark_bar(
size=size, stroke='black'
)
d['visibility'] = d['value'] > 0.08
text = alt.Chart(
d
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).mark_text(size=16, dx=0, dy=5, color='white', baseline='top', fontWeight=500).encode(
x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
y=alt.Y('value:Q', stack='zero'),
detail='cat:N',
text=alt.Text('value:Q', format='.0%'),
order="order:O",
opacity=alt.Opacity('visibility:N', scale=alt.Scale(domain=[True, False], range=[1, 0]), legend=None)
)
if metric == 'pos':
bar = (bar + text)
# bar = bar.facet(
# column=alt.Column('site:N', header=alt.Header(title=None), sort=sites),
# spacing=20
# )
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%% Bottom %%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='ppv'
y_title = '% of Event'
colors = ['#A8DED1', '#3FA86F', '#005A24']
colors = ['#00A87E', '#00634B', 'black']
y_scale = alt.Scale(domain=[0, 1])
line = alt.Chart(
d
).transform_calculate(
# order="{'Low Risk':0, 'Medium Risk': 1, 'High Risk': 2}[datum.variable]"
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
# x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
x=alt.X("month:N", title=None, scale=alt.Scale(domain=date), axis=alt.Axis(labelAngle=-55)),
y=alt.Y("value:Q", title=y_title, scale=y_scale),
color=alt.Color("cat:N", title='Risk Level', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height
)
line = line.mark_line(
size=3, point=point, opacity=0.8
)
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%% Bottom 2 %%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='rr'
y_title = 'Ratio Between Risk Levels'
# colors = ['red', 'salmon']
colors = ['#D45E00', '#351800']
# colorDomain = ['High/Medium', 'Low/Medium']
colorDomain = ['L/M', 'H/M']
y_scale = alt.Scale(domain=[0, 10], clamp=True)
line2 = alt.Chart(
d
).transform_calculate(
# order="{'High/Medium':0, 'Low/Medium': 1}[datum.variable]"
order="{'H/M':0, 'L/M': 1}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
# x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
x=alt.X("month:N", title=None, scale=alt.Scale(domain=date), axis=alt.Axis(grid=False,labelAngle=-55)),
# x=alt.X("month:N", title='Month', scale=alt.Scale(domain=date), axis=alt.Axis(grid=True, ticks=False, labels=False, domain=False, title=None)),
y=alt.Y("value:Q", title=y_title, scale=y_scale),
color=alt.Color("cat:N", title='Risk Ratio', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height
)
line2 = line2.mark_line(
size=3, point=point, opacity=0.8
)
print(d.site.unique())
"""
COMBINE
"""
res = alt.hconcat(
bar,
line,
line2,
spacing=30
).resolve_scale(y='independent', color='independent')
res = res.properties(
title={
"text": [
f"Meta-Analysis Of Risk Scores"
],
"dx": 70,
# "subtitle": [
# get_visualization_subtitle(data_release=data_release, with_num_sites=False)
# ],
"subtitleColor": "gray",
}
)
"""
STYLE
"""
res = apply_theme(
res,
axis_y_title_font_size=16,
title_anchor='start',
legend_orient='top',
# legend_title_orient='left',
axis_label_font_size=14,
header_label_font_size=16,
point_size=90,
)
res
```
# Deprecated
```
pos = risk(df, metric='pos')
ppv = risk(df, metric='ppv')
res = alt.vconcat(pos, ppv, spacing=30).resolve_scale(color='independent', x='independent')
res = apply_theme(
res,
axis_y_title_font_size=16,
title_anchor='start',
legend_orient='right',
header_label_font_size=16
)
res.display()
d = df.copy()
width = 280
height = 200
height2 = 140
size = 50
"""
DATA PREPROCESSING...
"""
d.loc[d.site == 'combine', 'site'] = 'All Sites'
d.cat = d.cat.apply(lambda x: {
'L':'Low',
'M': 'Medium',
'H': 'High',
'H/M': 'H/M',
'L/M': 'L/M'
}[x])
# d.cat = d.cat.apply(lambda x: {
# 'L':'Low Risk',
# 'M': 'Medium Risk',
# 'H': 'High Risk',
# 'H/M': 'High/Medium',
# 'L/M': 'Low/Medium'
# }[x])
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%% TOP %%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='pos'
y_title = '% of Patients in Each Category'
y_scale = alt.Scale(domain=[0, 1])
colors = ['#7BADD1', '#427BB5', '#14366E']
# colorDomain = ['Low Risk', 'Medium Risk', 'High Risk']
colorDomain = ['Low', 'Medium', 'High']
# colorDomain = ['L', 'M', 'H']
bar = alt.Chart(
d
).transform_calculate(
# order="{'Low Risk':0, 'Medium Risk': 1, 'High Risk': 2}[datum.variable]"
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title='Month', scale=alt.Scale(domain=['Mar-Apr', 'May-Jun', 'Jul-Aug', 'Sep-Oct', 'Since Nov']), axis=alt.Axis(grid=True)),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Risk', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height
)
bar = bar.mark_bar(
size=size, stroke='black'
)
d['visibility'] = d['value'] > 0.08
text = alt.Chart(
d
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).mark_text(size=16, dx=0, dy=5, color='white', baseline='top', fontWeight=500).encode(
x=alt.X('month:N'),
y=alt.Y('value:Q', stack='zero'),
detail='cat:N',
text=alt.Text('value:Q', format='.0%'),
order="order:O",
opacity=alt.Opacity('visibility:N', scale=alt.Scale(domain=[True, False], range=[1, 0]), legend=None)
)
if metric == 'pos':
bar = (bar + text)
bar = bar.facet(
column=alt.Column('site:N', header=alt.Header(title=None)),
)
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%% Bottom %%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='ppv'
y_title = '% of Risk Event'
colors = ['#A8DED1', '#3FA86F', '#005A24']
colors = ['#00A87E', '#00634B', 'black']
y_scale = alt.Scale(domain=[0, 0.6])
line = alt.Chart(
d
).transform_calculate(
# order="{'Low Risk':0, 'Medium Risk': 1, 'High Risk': 2}[datum.variable]"
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title='Month', scale=alt.Scale(domain=['Mar-Apr', 'May-Jun', 'Jul-Aug', 'Sep-Oct', 'Since Nov']), axis=alt.Axis(grid=True, ticks=False, labels=False, domain=False, title=None)),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Risk', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height2
)
line = line.mark_line(
size=3, point=True, opacity=0.8
).facet(
column=alt.Column('site:N', header=alt.Header(title=None)),
)
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%% Bottom 2 %%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='rr'
y_title = 'Ratio Between Risks'
colors = ['#D45E00', '#351800']
# colorDomain = ['High/Medium', 'Low/Medium']
colorDomain = ['L/M', 'H/M']
y_scale = alt.Scale(domain=[0, 4.2])
line2 = alt.Chart(
d
).transform_calculate(
# order="{'High/Medium':0, 'Low/Medium': 1}[datum.variable]"
order="{'H/M':0, 'L/M': 1}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title='Month', scale=alt.Scale(domain=['Mar-Apr', 'May-Jun', 'Jul-Aug', 'Sep-Oct', 'Since Nov']), axis=alt.Axis(grid=True)),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Risk Ratio', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O",
# shape="site:N"
).properties(
width=width,
height=height2
)
line2 = line2.mark_line(
size=3, opacity=0.8, point=True
).facet(
column=alt.Column('site:N', header=alt.Header(title=None, labels=False)),
)
"""
COMBINE
"""
top = bar.properties(
title={
"text": [
f"Distribution of Risk Scores"
],
"dx": 180,
"subtitle": [
get_visualization_subtitle(data_release=data_release, with_num_sites=False)
],
"subtitleColor": "gray",
}
)
line = line.properties(
title={
"text": [
f"Event Rate of Risk Scores"
],
"dx": 180,
"subtitle": [
get_visualization_subtitle(data_release=data_release, with_num_sites=False)
],
"subtitleColor": "gray",
}
)
# line2 = line2.properties(
# title={
# "text": [
# f"Risk Ratio"
# ],
# "dx": 180,
# "subtitle": [
# get_visualization_subtitle(data_release=data_release, with_num_sites=False)
# ],
# "subtitleColor": "gray",
# }
# )
res = alt.vconcat(top, line, line2, spacing=10).resolve_scale(color='independent')
"""
STYLE
"""
res = apply_theme(
res,
axis_y_title_font_size=14,
axis_title_font_size=14,
axis_label_font_size=12,
title_anchor='start',
legend_orient='left',
header_label_font_size=16
)
res
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
%aimport utils_1_1
import pandas as pd
import numpy as np
import altair as alt
from altair_saver import save
import datetime
import dateutil.parser
from os.path import join
from constants_1_1 import SITE_FILE_TYPES
from utils_1_1 import (
get_site_file_paths,
get_site_file_info,
get_site_ids,
get_visualization_subtitle,
get_country_color_map,
)
from theme import apply_theme
from web import for_website
alt.data_transformers.disable_max_rows(); # Allow using rows more than 5000
data_release='2021-04-29'
consistent_date = {
'2020-Mar-Apr': "'20 Mar - '20 Apr",
'2020-May-Jun': "'20 May - '20 Jun",
'2020-Jul-Aug': "'20 Jul - '20 Aug",
'2020-Sep-Oct': "'20 Sep - '20 Oct",
'2020-Nov-2021-Jan': "'20 Nov - '21 Jan"
}
date = ['Mar - Apr', 'May - Jun', 'Jul - Aug', 'Sep - Oct', 'Since Nov']
date = ["'20 Mar - '20 Apr", "'20 May - '20 Jun", "'20 Jul - '20 Aug", "'20 Sep - '20 Oct", "'20 Nov - '21 Jan"]
sites = ['META', 'APHP', 'FRBDX', 'ICSM', 'NWU', 'BIDMC', 'MGB', 'UCLA', 'UMICH', 'UPENN', 'UPITT', 'VA1', 'VA2', 'VA3', 'VA4', 'VA5']
site_colors = ['black', '#D45E00', '#0072B2', '#CB7AA7', '#E79F00', '#029F73', '#DBD03C', '#57B4E9', '#57B4E9', '#57B4E9', '#57B4E9', '#57B4E9']
df = pd.read_csv(join("..", "data", "Phase2.1SurvivalRSummariesPublic", "ToShare", "table.score.toShare.csv"))
print(df.head())
# Rename columns
df = df.drop(columns=["Unnamed: 0"])
df = df.rename(columns={
'siteid': 'site',
'calendar_month': 'month'
})
# More readable values
df.site = df.site.apply(lambda x: x.upper())
print(df.site.unique().tolist())
print(df.month.unique().tolist())
# Drop "combine" sites
df = df[df.site != "COMBINE"]
# df = pd.melt(df, id_vars=['siteid'], value_vars=date, var_name='date', value_name='value')
df.month = df.month.apply(lambda x: consistent_date[x])
# Add a reference (META)
# df['reference'] = df.date.apply(lambda x: df[(df.date == x) & (df.siteid == 'META')].value.sum())
df.head()
def risk(_d, metric='pos'):
d = _d.copy()
"""
DATA PREPROCESSING...
"""
d.loc[d.site == 'combine', 'site'] = 'All Sites'
d.cat = d.cat.apply(lambda x: {
'L':'Low Risk',
'M': 'Medium Risk',
'H': 'High Risk',
'H/M': 'High/Medium',
'L/M': 'Low/Medium'
}[x])
"""
PLOT!
"""
y_title = '% of Patients in Each Category' if metric == 'pos' else '% of Event in Each Category'
colors = ['#7BADD1', '#427BB5', '#14366E'] if metric == 'pos' else ['#A8DED1', '#3FA86F', '#005A24'] if metric == 'ppv' else ['red', 'salmon']
colorDomain = ['High/Medium', 'Low/Medium'] if metric == 'rr' else ['Low Risk', 'Medium Risk', 'High Risk']
width = 300
size = 50
y_scale = alt.Scale(domain=[0, 1]) if metric == 'pos' or metric=='ppv' else alt.Scale()
bar = alt.Chart(
d
).transform_calculate(
order="{'Low Risk':0, 'Medium Risk': 1, 'High Risk': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title='Month', scale=alt.Scale(domain=['Mar-Apr', 'May-Jun', 'Jul-Aug', 'Sep-Oct', 'Since Nov'])),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Category', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width
)
if metric == 'pos':
bar = bar.mark_bar(
size=size, stroke='black'
)
else:
bar = bar.mark_line(
size=3, point=True, opacity=0.8
)
d['visibility'] = d['value'] > 0.08
text = alt.Chart(
d
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).mark_text(size=16, dx=0, dy=5, color='white', baseline='top', fontWeight=500).encode(
x=alt.X('month:N'),
y=alt.Y('value:Q', stack='zero'),
detail='cat:N',
text=alt.Text('value:Q', format='.0%'),
order="order:O",
opacity=alt.Opacity('visibility:N', scale=alt.Scale(domain=[True, False], range=[1, 0]))
)
# .transform_filter(
# (f'datum.value > 0.10')
# )
if metric == 'pos':
bar = (bar + text)
bar = bar.facet(
column=alt.Column('site:N', header=alt.Header(title=None)),
)
"""
COMBINE
"""
res = bar.properties(
title={
"text": [
f"Distribution of Risk Scores" if metric == 'pos' else f"Event Rate of Risk Scores"
],
"dx": 80,
"subtitle": [
# lab, #.title(),
get_visualization_subtitle(data_release=data_release, with_num_sites=False)
],
"subtitleColor": "gray",
}
)
return res
d = df.copy()
width = 160
height = 180
height2 = 140
size = 28
point=alt.OverlayMarkDef(filled=False, fill='white', strokeWidth=2)
"""
DATA PREPROCESSING...
"""
d.loc[d.site == 'combine', 'site'] = 'All Sites'
d.cat = d.cat.apply(lambda x: {
'L':'Low',
'M': 'Medium',
'H': 'High',
'H/M': 'H/M',
'L/M': 'L/M'
}[x])
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%% TOP %%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='pos'
y_title = '% of Patients in Each Category'
y_scale = alt.Scale(domain=[0, 1])
colors = ['#7BADD1', '#427BB5', '#14366E']
# colorDomain = ['Low Risk', 'Medium Risk', 'High Risk']
colorDomain = ['Low', 'Medium', 'High']
# colorDomain = ['L', 'M', 'H']
bar = alt.Chart(
d
).transform_calculate(
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Risk Level', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height
)
bar = bar.mark_bar(
size=size, stroke='black'
)
d['visibility'] = d['value'] > 0.08
text = alt.Chart(
d
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).mark_text(size=14, dx=0, dy=5, color='white', baseline='top', fontWeight=500).encode(
x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55, domain=False), sort=date),
y=alt.Y('value:Q', stack='zero'),
detail='cat:N',
text=alt.Text('value:Q', format='.0%'),
order="order:O",
opacity=alt.Opacity('visibility:N', scale=alt.Scale(domain=[True, False], range=[1, 0]), legend=None)
)
if metric == 'pos':
bar = (bar + text)
bar = bar.facet(
column=alt.Column('site:N', header=alt.Header(title=None), sort=sites),
spacing=20
)
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%% Bottom %%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='ppv'
y_title = '% of Event'
colors = ['#A8DED1', '#3FA86F', '#005A24']
colors = ['#00A87E', '#00634B', 'black']
y_scale = alt.Scale(domain=[0, 1])
line = alt.Chart(
d
).transform_calculate(
# order="{'Low Risk':0, 'Medium Risk': 1, 'High Risk': 2}[datum.variable]"
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
# x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
x=alt.X("month:N", title='Month', scale=alt.Scale(domain=date), axis=alt.Axis(grid=False, ticks=False, labels=False, domain=False, title=None, labelAngle=-55)),
y=alt.Y("value:Q", title=y_title, scale=y_scale),
color=alt.Color("cat:N", title='Risk Level', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height2
)
line = line.mark_line(
size=3, point=point, opacity=0.8
).facet(
column=alt.Column('site:N', header=alt.Header(title=None), sort=sites),
)
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%% Bottom 2 %%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='rr'
y_title = 'Ratio Between Risk Levels'
# colors = ['red', 'salmon']
colors = ['#D45E00', '#351800']
# colorDomain = ['High/Medium', 'Low/Medium']
colorDomain = ['L/M', 'H/M']
y_scale = alt.Scale(domain=[0, 10], clamp=True)
line2 = alt.Chart(
d
).transform_calculate(
# order="{'High/Medium':0, 'Low/Medium': 1}[datum.variable]"
order="{'H/M':0, 'L/M': 1}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
# x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
x=alt.X("month:N", title=None, scale=alt.Scale(domain=date), axis=alt.Axis(grid=False, domain=False, labelAngle=-55)),
# x=alt.X("month:N", title='Month', scale=alt.Scale(domain=date), axis=alt.Axis(grid=True, ticks=False, labels=False, domain=False, title=None)),
y=alt.Y("value:Q", title=y_title, scale=y_scale),
color=alt.Color("cat:N", title='Risk Ratio', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height2
)
line2 = line2.mark_line(
size=3, point=point, opacity=0.8
).facet(
column=alt.Column('site:N', header=alt.Header(title=None, labels=False), sort=sites),
)
# Just to show color legend and left axis
# line_not_first = line
# line_not_first = line.encode(
# x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
# y=alt.Y("value:Q", title=None, axis=alt.Axis(format='.0%', ticks=False, labels=False, domain=False), scale=y_scale),
# color=alt.Color("cat:N", title='Risk Level', scale=alt.Scale(domain=colorDomain, range=colors), legend=None),
# order="order:O"
# )
# line2_first = line2.encode(
# x=alt.X("month:N", title=None, sort=date),
# y=alt.Y("value:Q", title=None, axis=alt.Axis(format='.0%', ticks=False, labels=False, domain=False), scale=y_scale),
# color=alt.Color("cat:N", title='Risk Ratio (Right Y Axis)', scale=alt.Scale(domain=colorDomain, range=colors)),
# order="order:O"
# )
# line2_last = line2.encode(
# x=alt.X("month:N", title=None, sort=date),
# y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
# color=alt.Color("cat:N", title='Risk Ratio', scale=alt.Scale(domain=colorDomain, range=colors), legend=None),
# order="order:O"
# )
# line = alt.concat(*(
# alt.layer(line, line2_first, title={
# "text": site,
# "fontSize": 16,
# "dx": 130}).transform_filter(alt.datum.site == site).resolve_scale(y='independent', color='independent') if site == 'META' else
# alt.layer(line_not_first, line2_last, title={
# "text": site,
# "fontSize": 16,
# "dx": 85}).transform_filter(alt.datum.site == site).resolve_scale(y='independent', color='independent') if site == sites[-1] else
# alt.layer(line_not_first, line2, title={
# "text": site,
# "fontSize": 16,
# "dx": 85}).transform_filter(alt.datum.site == site).resolve_scale(y='independent', color='independent')
# for site in sites
# ), spacing=3).resolve_scale(color='shared')
print(d.site.unique())
"""
COMBINE
"""
top = bar.properties(
title={
"text": [
f"Distribution Of Parients By Risk Level"
],
"fontWeight": "normal",
"dx": 170,
"subtitle": [
get_visualization_subtitle(data_release=data_release, with_num_sites=False)
],
"subtitleColor": "gray",
}
)
bot = line.properties(
title={
"text": [
f"Event Rate Of Risk Scores"
],
"dx": 150,
# "subtitle": [
# get_visualization_subtitle(data_release=data_release, with_num_sites=False)
# ],
"subtitleColor": "gray",
}
)
# top.display()
# bot.display()
# line2.display()
res = alt.vconcat(
top,
bot,
line2,
spacing=10
).resolve_scale(y='independent', color='independent')
"""
STYLE
"""
res = apply_theme(
res,
axis_y_title_font_size=16,
title_anchor='start',
legend_orient='left',
header_label_orient='top',
# legend_title_orient='left',
axis_label_font_size=14,
header_label_font_size=16,
point_size=90
)
res
d = df.copy()
d = d[d.site == 'META']
width = 200
height = 200
size = 30
point=alt.OverlayMarkDef(filled=False, fill='white', strokeWidth=2)
"""
DATA PREPROCESSING...
"""
d.loc[d.site == 'combine', 'site'] = 'All Sites'
d.cat = d.cat.apply(lambda x: {
'L':'Low',
'M': 'Medium',
'H': 'High',
'H/M': 'H/M',
'L/M': 'L/M'
}[x])
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%% TOP %%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='pos'
y_title = '% of Patients in Each Category'
y_scale = alt.Scale(domain=[0, 1])
colors = ['#7BADD1', '#427BB5', '#14366E']
# colorDomain = ['Low Risk', 'Medium Risk', 'High Risk']
colorDomain = ['Low', 'Medium', 'High']
# colorDomain = ['L', 'M', 'H']
bar = alt.Chart(
d
).transform_calculate(
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Risk Level', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height
)
bar = bar.mark_bar(
size=size, stroke='black'
)
d['visibility'] = d['value'] > 0.08
text = alt.Chart(
d
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).mark_text(size=16, dx=0, dy=5, color='white', baseline='top', fontWeight=500).encode(
x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
y=alt.Y('value:Q', stack='zero'),
detail='cat:N',
text=alt.Text('value:Q', format='.0%'),
order="order:O",
opacity=alt.Opacity('visibility:N', scale=alt.Scale(domain=[True, False], range=[1, 0]), legend=None)
)
if metric == 'pos':
bar = (bar + text)
# bar = bar.facet(
# column=alt.Column('site:N', header=alt.Header(title=None), sort=sites),
# spacing=20
# )
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%% Bottom %%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='ppv'
y_title = '% of Event'
colors = ['#A8DED1', '#3FA86F', '#005A24']
colors = ['#00A87E', '#00634B', 'black']
y_scale = alt.Scale(domain=[0, 1])
line = alt.Chart(
d
).transform_calculate(
# order="{'Low Risk':0, 'Medium Risk': 1, 'High Risk': 2}[datum.variable]"
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
# x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
x=alt.X("month:N", title=None, scale=alt.Scale(domain=date), axis=alt.Axis(labelAngle=-55)),
y=alt.Y("value:Q", title=y_title, scale=y_scale),
color=alt.Color("cat:N", title='Risk Level', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height
)
line = line.mark_line(
size=3, point=point, opacity=0.8
)
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%% Bottom 2 %%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='rr'
y_title = 'Ratio Between Risk Levels'
# colors = ['red', 'salmon']
colors = ['#D45E00', '#351800']
# colorDomain = ['High/Medium', 'Low/Medium']
colorDomain = ['L/M', 'H/M']
y_scale = alt.Scale(domain=[0, 10], clamp=True)
line2 = alt.Chart(
d
).transform_calculate(
# order="{'High/Medium':0, 'Low/Medium': 1}[datum.variable]"
order="{'H/M':0, 'L/M': 1}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
# x=alt.X("month:N", title=None, axis=alt.Axis(labelAngle=-55), sort=date),
x=alt.X("month:N", title=None, scale=alt.Scale(domain=date), axis=alt.Axis(grid=False,labelAngle=-55)),
# x=alt.X("month:N", title='Month', scale=alt.Scale(domain=date), axis=alt.Axis(grid=True, ticks=False, labels=False, domain=False, title=None)),
y=alt.Y("value:Q", title=y_title, scale=y_scale),
color=alt.Color("cat:N", title='Risk Ratio', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height
)
line2 = line2.mark_line(
size=3, point=point, opacity=0.8
)
print(d.site.unique())
"""
COMBINE
"""
res = alt.hconcat(
bar,
line,
line2,
spacing=30
).resolve_scale(y='independent', color='independent')
res = res.properties(
title={
"text": [
f"Meta-Analysis Of Risk Scores"
],
"dx": 70,
# "subtitle": [
# get_visualization_subtitle(data_release=data_release, with_num_sites=False)
# ],
"subtitleColor": "gray",
}
)
"""
STYLE
"""
res = apply_theme(
res,
axis_y_title_font_size=16,
title_anchor='start',
legend_orient='top',
# legend_title_orient='left',
axis_label_font_size=14,
header_label_font_size=16,
point_size=90,
)
res
pos = risk(df, metric='pos')
ppv = risk(df, metric='ppv')
res = alt.vconcat(pos, ppv, spacing=30).resolve_scale(color='independent', x='independent')
res = apply_theme(
res,
axis_y_title_font_size=16,
title_anchor='start',
legend_orient='right',
header_label_font_size=16
)
res.display()
d = df.copy()
width = 280
height = 200
height2 = 140
size = 50
"""
DATA PREPROCESSING...
"""
d.loc[d.site == 'combine', 'site'] = 'All Sites'
d.cat = d.cat.apply(lambda x: {
'L':'Low',
'M': 'Medium',
'H': 'High',
'H/M': 'H/M',
'L/M': 'L/M'
}[x])
# d.cat = d.cat.apply(lambda x: {
# 'L':'Low Risk',
# 'M': 'Medium Risk',
# 'H': 'High Risk',
# 'H/M': 'High/Medium',
# 'L/M': 'Low/Medium'
# }[x])
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%% TOP %%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='pos'
y_title = '% of Patients in Each Category'
y_scale = alt.Scale(domain=[0, 1])
colors = ['#7BADD1', '#427BB5', '#14366E']
# colorDomain = ['Low Risk', 'Medium Risk', 'High Risk']
colorDomain = ['Low', 'Medium', 'High']
# colorDomain = ['L', 'M', 'H']
bar = alt.Chart(
d
).transform_calculate(
# order="{'Low Risk':0, 'Medium Risk': 1, 'High Risk': 2}[datum.variable]"
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title='Month', scale=alt.Scale(domain=['Mar-Apr', 'May-Jun', 'Jul-Aug', 'Sep-Oct', 'Since Nov']), axis=alt.Axis(grid=True)),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Risk', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height
)
bar = bar.mark_bar(
size=size, stroke='black'
)
d['visibility'] = d['value'] > 0.08
text = alt.Chart(
d
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).mark_text(size=16, dx=0, dy=5, color='white', baseline='top', fontWeight=500).encode(
x=alt.X('month:N'),
y=alt.Y('value:Q', stack='zero'),
detail='cat:N',
text=alt.Text('value:Q', format='.0%'),
order="order:O",
opacity=alt.Opacity('visibility:N', scale=alt.Scale(domain=[True, False], range=[1, 0]), legend=None)
)
if metric == 'pos':
bar = (bar + text)
bar = bar.facet(
column=alt.Column('site:N', header=alt.Header(title=None)),
)
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%% Bottom %%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='ppv'
y_title = '% of Risk Event'
colors = ['#A8DED1', '#3FA86F', '#005A24']
colors = ['#00A87E', '#00634B', 'black']
y_scale = alt.Scale(domain=[0, 0.6])
line = alt.Chart(
d
).transform_calculate(
# order="{'Low Risk':0, 'Medium Risk': 1, 'High Risk': 2}[datum.variable]"
order="{'L':0, 'M': 1, 'H': 2}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title='Month', scale=alt.Scale(domain=['Mar-Apr', 'May-Jun', 'Jul-Aug', 'Sep-Oct', 'Since Nov']), axis=alt.Axis(grid=True, ticks=False, labels=False, domain=False, title=None)),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Risk', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O"
).properties(
width=width,
height=height2
)
line = line.mark_line(
size=3, point=True, opacity=0.8
).facet(
column=alt.Column('site:N', header=alt.Header(title=None)),
)
"""
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%% Bottom 2 %%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
"""
metric='rr'
y_title = 'Ratio Between Risks'
colors = ['#D45E00', '#351800']
# colorDomain = ['High/Medium', 'Low/Medium']
colorDomain = ['L/M', 'H/M']
y_scale = alt.Scale(domain=[0, 4.2])
line2 = alt.Chart(
d
).transform_calculate(
# order="{'High/Medium':0, 'Low/Medium': 1}[datum.variable]"
order="{'H/M':0, 'L/M': 1}[datum.variable]"
).transform_filter(
{'field': 'metric', 'oneOf': [metric]}
).encode(
x=alt.X("month:N", title='Month', scale=alt.Scale(domain=['Mar-Apr', 'May-Jun', 'Jul-Aug', 'Sep-Oct', 'Since Nov']), axis=alt.Axis(grid=True)),
y=alt.Y("value:Q", title=y_title, axis=alt.Axis(format='.0%'), scale=y_scale),
color=alt.Color("cat:N", title='Risk Ratio', scale=alt.Scale(domain=colorDomain, range=colors)),
order="order:O",
# shape="site:N"
).properties(
width=width,
height=height2
)
line2 = line2.mark_line(
size=3, opacity=0.8, point=True
).facet(
column=alt.Column('site:N', header=alt.Header(title=None, labels=False)),
)
"""
COMBINE
"""
top = bar.properties(
title={
"text": [
f"Distribution of Risk Scores"
],
"dx": 180,
"subtitle": [
get_visualization_subtitle(data_release=data_release, with_num_sites=False)
],
"subtitleColor": "gray",
}
)
line = line.properties(
title={
"text": [
f"Event Rate of Risk Scores"
],
"dx": 180,
"subtitle": [
get_visualization_subtitle(data_release=data_release, with_num_sites=False)
],
"subtitleColor": "gray",
}
)
# line2 = line2.properties(
# title={
# "text": [
# f"Risk Ratio"
# ],
# "dx": 180,
# "subtitle": [
# get_visualization_subtitle(data_release=data_release, with_num_sites=False)
# ],
# "subtitleColor": "gray",
# }
# )
res = alt.vconcat(top, line, line2, spacing=10).resolve_scale(color='independent')
"""
STYLE
"""
res = apply_theme(
res,
axis_y_title_font_size=14,
axis_title_font_size=14,
axis_label_font_size=12,
title_anchor='start',
legend_orient='left',
header_label_font_size=16
)
res
| 0.38341 | 0.349616 |
## 3. 데이터 탐색적 분석 - EDA
`$ pip install plotly_express`
#### 3.2 데이터 읽기
```
import pandas as pd
titanic_url = 'https://raw.githubusercontent.com/PinkWink/ML_tutorial/master/dataset/titanic.xls'
titanic = pd.read_excel(titanic_url)
titanic.head()
```
#### 3.3 생존 상황
```
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
f, ax = plt.subplots(1, 2, figsize=(18, 8))
titanic['survived'].value_counts().plot.pie(explode=[0, 0.01],
autopct='%1.1f%%', ax=ax[0], shadow=True)
ax[0].set_title('Pie plot - Survived')
ax[0].set_ylabel('')
sns.countplot('survived', data=titanic, ax=ax[1])
ax[1].set_title('Count plot - Survived')
plt.show()
```
#### 3.5 성별에 따른 생존 상황은?
```
f, ax = plt.subplots(1, 2, figsize=(18, 8))
sns.countplot('sex', data=titanic, ax=ax[0])
ax[0].set_title('Count of Passengers of SEx')
ax[0].set_ylabel('')
sns.countplot('sex', hue='survived', data=titanic, ax=ax[1])
ax[1].set_title('Sex:Survived and Unsurvived')
plt.show()
```
#### 3.7 경제력 대비 생존률
```
pd.crosstab(titanic['pclass'], titanic['survived'], margins=True)
```
#### 3.8 선실 등급별 성별 상황
```
import seaborn as sns
grid = sns.FacetGrid(titanic, row='pclass', col='sex', height=4, aspect=2)
grid.map(plt.hist, 'age', alpha=.8, bins=20)
grid.add_legend();
##3등실에는 특히 남성승객이 많았다~
```
#### 3.10 나이별 승객 현황을 알아보자
```
import plotly.express as px
fig = px.histogram(titanic, x="age")
fig.show()
##아이들과 20~30대가 많았다
```
#### 3.12 등실별 생존률을 연령별로 관찰해보자
```
grid = sns.FacetGrid(titanic, col='survived', row='pclass', height=4, aspect=2)
grid.map(plt.hist, 'age', alpha=.5, bins=20)
grid.add_legend();
##선실등급이 높으면 생존률이 확실히 높은듯 하다
```
#### 3.14 나이를 5단계로 정리하기
```
titanic['age_cat'] = pd.cut(titanic['age'], bins=[0,7,15,30,60,100],
include_lowest=True,
labels=['baby', 'teen', 'young', 'adult', 'old'])
titanic.head()
```
#### 3.15 나이,성별,등급별 생존자 수를 한번에 파악할 수 있을까?
```
plt.figure(figsize=(12,4))
plt.subplot(131)
sns.barplot('pclass', 'survived', data=titanic)
plt.subplot(132)
sns.barplot('age_cat', 'survived', data=titanic)
plt.subplot(133)
sns.barplot('sex', 'survived', data=titanic)
plt.subplots_adjust(top=1, bottom=0.1, left=0.1, right=1, hspace=0.5, wspace=0.5)
```
#### 3.17 남/여 나이별 생존 상황을 보다 더 들여다 보자
```
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(14,6))
women = titanic[titanic['sex']=='female']
men = titanic[titanic['sex']=='male']
ax = sns.distplot(women[women['survived']==1]['age'], bins=20,
label = 'survived', ax = axes[0], kde=False)
ax = sns.distplot(women[women['survived']==0]['age'], bins=40,
label = 'not_survived', ax = axes[0], kde=False)
ax.legend(); ax.set_title('Female')
ax = sns.distplot(men[men['survived']==1]['age'], bins=18,
label = 'survived', ax = axes[1], kde=False)
ax = sns.distplot(men[men['survived']==0]['age'], bins=40,
label = 'not_survived', ax = axes[1], kde=False)
ax.legend(); ax = ax.set_title('Male')
```
#### 3.19 탑승객의 이름에서 신분을 알 수 있다
```
for idx, dataset in titanic.iterrows():
print(dataset['name'])
```
#### 3.20 정규식을 이용해서 문장 사이의 신분에 대한 정보를 얻을 수 있다
```
import re
for idx, dataset in titanic.iterrows():
tmp = dataset['name']
print(idx)
print(re.search('\,\s\w+(\s\w+)?\.', tmp).group())
# .group() : 글자를 반환한다
```
#### 3.21 살짝 응용하면 -> 사회적 신분만 얻을 수 있다
```
import re
title = []
for idx, dataset in titanic.iterrows():
title.append(re.search('\,\s\w+(\s\w+)?\.', dataset['name']).group()[2:-1])
titanic['title'] = title
titanic.head()
```
#### 3.23 성별별로 본 귀족
```
pd.crosstab(titanic['title'], titanic['sex'])
```
#### 3.24 사회적 신분을 조금더 정리하자
```
titanic['title'] = titanic['title'].replace('Mlle', "Miss")
titanic['title'] = titanic['title'].replace('Ms', "Miss")
titanic['title'] = titanic['title'].replace('Mme', "Mrs")
Rare_f = ['Dona', 'Dr', 'Lady', 'the Countess']
Rare_m = ['Capt', 'Col', 'Don', 'Major', 'Rev', 'Sir', 'Jonkheer', 'Master']
for each in Rare_f:
titanic['title'] = titanic['title'].replace(each, 'Rare_f')
for each in Rare_m:
titanic['title'] = titanic['title'].replace(each, 'Rare_m')
titanic['title'].unique()
```
#### 3.25 이 결과는 또 어떻게 해석할 수 있을까
```
titanic[['title', 'survived']].groupby(['title'], as_index=False).mean()
```
## 4. 머신러닝을 이용한 생존자 예측
#### 4.1 간단히 구조확인
```
titanic.info()
```
#### 4.2 머신러닝을 위해 해당컬럼을 숫자로 변경해야겠다
```
titanic['sex'].unique()
```
#### 4.3 Label Encode를 사용하면 편하다
```
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
le.fit(titanic['sex'])
titanic['gender'] = le.transform(titanic['sex'])
titanic.head()
```
#### 4.4 결측치는 어쩔 수 없이 포기하자
```
titanic = titanic[titanic['age'].notnull()]
titanic = titanic[titanic['fare'].notnull()]
titanic.info()
```
#### 4.5 상관관계
```
correlation_matrix = titanic.corr().round(1)
sns.heatmap(data=correlation_matrix, annot=True, cmap='bwr')
```
#### 4.6 먼저 특성을 선택하고, 데이터를 나누자
```
from sklearn.model_selection import train_test_split
x = titanic[['pclass', 'age', 'sibsp', 'parch', 'fare', 'gender']] # 특성 선정
y = titanic['survived']
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, random_state=13)
```
#### 4.7 일단 DecisionTree
```
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score
dt = DecisionTreeClassifier(max_depth=4, random_state=13)
dt.fit(x_train, y_train)
pred = dt.predict(x_test)
print(accuracy_score(y_test, pred))
```
## 5. 디카프리오의 생존율은?
#### 5.2 디카프리오의 생존률
```
import numpy as np
dicaprio = np.array([[3, 18, 0, 0, 5, 1]])
print('Decaprio : ', dt.predict_proba(dicaprio)[0,1])
```
#### 5.3 윈슬릿의 생존률
```
winslet = np.array([[1, 16, 1, 1, 100, 0]])
print('Winslet : ', dt.predict_proba(winslet)[0,1])
```
|
github_jupyter
|
import pandas as pd
titanic_url = 'https://raw.githubusercontent.com/PinkWink/ML_tutorial/master/dataset/titanic.xls'
titanic = pd.read_excel(titanic_url)
titanic.head()
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
f, ax = plt.subplots(1, 2, figsize=(18, 8))
titanic['survived'].value_counts().plot.pie(explode=[0, 0.01],
autopct='%1.1f%%', ax=ax[0], shadow=True)
ax[0].set_title('Pie plot - Survived')
ax[0].set_ylabel('')
sns.countplot('survived', data=titanic, ax=ax[1])
ax[1].set_title('Count plot - Survived')
plt.show()
f, ax = plt.subplots(1, 2, figsize=(18, 8))
sns.countplot('sex', data=titanic, ax=ax[0])
ax[0].set_title('Count of Passengers of SEx')
ax[0].set_ylabel('')
sns.countplot('sex', hue='survived', data=titanic, ax=ax[1])
ax[1].set_title('Sex:Survived and Unsurvived')
plt.show()
pd.crosstab(titanic['pclass'], titanic['survived'], margins=True)
import seaborn as sns
grid = sns.FacetGrid(titanic, row='pclass', col='sex', height=4, aspect=2)
grid.map(plt.hist, 'age', alpha=.8, bins=20)
grid.add_legend();
##3등실에는 특히 남성승객이 많았다~
import plotly.express as px
fig = px.histogram(titanic, x="age")
fig.show()
##아이들과 20~30대가 많았다
grid = sns.FacetGrid(titanic, col='survived', row='pclass', height=4, aspect=2)
grid.map(plt.hist, 'age', alpha=.5, bins=20)
grid.add_legend();
##선실등급이 높으면 생존률이 확실히 높은듯 하다
titanic['age_cat'] = pd.cut(titanic['age'], bins=[0,7,15,30,60,100],
include_lowest=True,
labels=['baby', 'teen', 'young', 'adult', 'old'])
titanic.head()
plt.figure(figsize=(12,4))
plt.subplot(131)
sns.barplot('pclass', 'survived', data=titanic)
plt.subplot(132)
sns.barplot('age_cat', 'survived', data=titanic)
plt.subplot(133)
sns.barplot('sex', 'survived', data=titanic)
plt.subplots_adjust(top=1, bottom=0.1, left=0.1, right=1, hspace=0.5, wspace=0.5)
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(14,6))
women = titanic[titanic['sex']=='female']
men = titanic[titanic['sex']=='male']
ax = sns.distplot(women[women['survived']==1]['age'], bins=20,
label = 'survived', ax = axes[0], kde=False)
ax = sns.distplot(women[women['survived']==0]['age'], bins=40,
label = 'not_survived', ax = axes[0], kde=False)
ax.legend(); ax.set_title('Female')
ax = sns.distplot(men[men['survived']==1]['age'], bins=18,
label = 'survived', ax = axes[1], kde=False)
ax = sns.distplot(men[men['survived']==0]['age'], bins=40,
label = 'not_survived', ax = axes[1], kde=False)
ax.legend(); ax = ax.set_title('Male')
for idx, dataset in titanic.iterrows():
print(dataset['name'])
import re
for idx, dataset in titanic.iterrows():
tmp = dataset['name']
print(idx)
print(re.search('\,\s\w+(\s\w+)?\.', tmp).group())
# .group() : 글자를 반환한다
import re
title = []
for idx, dataset in titanic.iterrows():
title.append(re.search('\,\s\w+(\s\w+)?\.', dataset['name']).group()[2:-1])
titanic['title'] = title
titanic.head()
pd.crosstab(titanic['title'], titanic['sex'])
titanic['title'] = titanic['title'].replace('Mlle', "Miss")
titanic['title'] = titanic['title'].replace('Ms', "Miss")
titanic['title'] = titanic['title'].replace('Mme', "Mrs")
Rare_f = ['Dona', 'Dr', 'Lady', 'the Countess']
Rare_m = ['Capt', 'Col', 'Don', 'Major', 'Rev', 'Sir', 'Jonkheer', 'Master']
for each in Rare_f:
titanic['title'] = titanic['title'].replace(each, 'Rare_f')
for each in Rare_m:
titanic['title'] = titanic['title'].replace(each, 'Rare_m')
titanic['title'].unique()
titanic[['title', 'survived']].groupby(['title'], as_index=False).mean()
titanic.info()
titanic['sex'].unique()
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
le.fit(titanic['sex'])
titanic['gender'] = le.transform(titanic['sex'])
titanic.head()
titanic = titanic[titanic['age'].notnull()]
titanic = titanic[titanic['fare'].notnull()]
titanic.info()
correlation_matrix = titanic.corr().round(1)
sns.heatmap(data=correlation_matrix, annot=True, cmap='bwr')
from sklearn.model_selection import train_test_split
x = titanic[['pclass', 'age', 'sibsp', 'parch', 'fare', 'gender']] # 특성 선정
y = titanic['survived']
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, random_state=13)
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score
dt = DecisionTreeClassifier(max_depth=4, random_state=13)
dt.fit(x_train, y_train)
pred = dt.predict(x_test)
print(accuracy_score(y_test, pred))
import numpy as np
dicaprio = np.array([[3, 18, 0, 0, 5, 1]])
print('Decaprio : ', dt.predict_proba(dicaprio)[0,1])
winslet = np.array([[1, 16, 1, 1, 100, 0]])
print('Winslet : ', dt.predict_proba(winslet)[0,1])
| 0.355887 | 0.952794 |
<a href="https://colab.research.google.com/github/RileyWClarke/MLTSA_RClarke/blob/master/HW3/rclarke_HW3.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# MLTSA - Riley Clarke - HW3
```
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
%matplotlib inline
plt.style.use("https://raw.githubusercontent.com/fedhere/MLTSA_FBianco/master/fbb.mplstyle")
```
## Installing/importing FB Prophet
```
!pip install fbprophet
import fbprophet
from fbprophet import Prophet
from fbprophet import diagnostics
```
## Reading in Uber data from the following repo:
https://github.com/fivethirtyeight/uber-tlc-foil-response.
```
#Builds a list of pandas dataframes from the github file urls:
a = []
for mon in ['apr', 'may', 'jun', 'jul', 'aug', 'sep']:
df = pd.read_csv('https://raw.githubusercontent.com/fivethirtyeight/uber-tlc-foil-response/master/uber-trip-data/uber-raw-data-'
+ mon + '14.csv')
a.append(df)
#concatenates the list into a single df
uberrides = pd.concat(a)
uberrides.head()
uberrides.shape
```
### DATA PREPARATION 1: Converting time column to datetime type and creating new df with number of rides per day
```
uberrides["time"] = pd.to_datetime(uberrides['Date/Time'])
uberrides.head()
uberrides.iloc[0]["time"].date
uberbyday = uberrides.groupby(uberrides["time"].dt.date).count()
# Returns pandas df with the number of rows in uberrides for each distinct date
uberbyday.head()
uberbyday = uberbyday.drop(columns=['Lat','Lon','Base','time'])
uberbyday.head()
```
FB Prophet needs us to rename our columns as 'y' for the endogenous variable (# of rides) and 'ds' for the exogenous variable (calendar date).
```
uberbyday['ds'] = uberbyday.index
uberbyday.rename(columns={'Date/Time':'y'}, inplace=True)
uberbyday.head()
#Plotting the initial dataset
uberbyday.plot(x="ds", y="y");
```
Figure 1: Uber trips per day in NYC from 04-2014 to 10-2014. The data appears to display an increasing trend, with a possible point-of-change at the end of August. There also seems to be a quasi-periodicity with a frequency of 4/month, suggesting number of rides tend to spike on the weekends.
## DATA PREPARATION 2: Standardization Boogaloo
Standarization will make the data more tractable and also convert the data type to floating points, which will aid regression later.
```
uberbyday['y'] = (uberbyday['y'] - uberbyday['y'].mean()) / uberbyday['y'].std()
uberbyday.plot(x="ds", y="y");
```
Figure 2: Same dataset as Figure 1, but with the data standarized to mean zero and standard deviation 1.
## DATA ANALYSIS
### MODEL 1: trends and seasonality
```
simplem = Prophet() #Invoking the Prophet model object
simplem.fit(uberbyday) #Fitting the model on the data
# Create a dataframe of dates one month past the end of the dataset
d=[]
for i in range(1,32):
if i <= 9:
d.append('2014-10-0'+str(i))
if i > 9:
d.append('2014-10-'+str(i))
future = pd.DataFrame(data = {'ds': d})
# and stitch it onto the end of the dataset
past_and_future = pd.concat([uberbyday, future])
#use the fitted model to create a forecast of the dataset one month into the future
forecast = simplem.predict(past_and_future)
#Peek at the last 5 rows of the forecast:
forecast.iloc[-5:]
```
As a check, we can see that the forecast dataframe extends to Oct 31st, one month past the original dataframe.
```
forecast['ds'].iloc[-31]
#Increase in trend value over month of October
forecast['trend'].iloc[-1] - forecast['trend'].iloc[-31]
```
From Oct 1st to Oct 31st, the forecasted trend increased by about 0.4 standard deviations
```
#Plotting original data + forecast
fig1 = simplem.plot(forecast)
plt.plot(uberbyday.y, '--')
plt.xticks(rotation=90);
```
Figure 3: Data from Figures 1 & 2 (scatterplot/dashed blue line) superimposed with forecasted data (solid blue line with shaded uncertainties). The forecast clearly shows the upward trend continuing into October, with an additional ~0.4$\sigma$ growth.
```
fig2 = simplem.plot_components(forecast)
plt.xticks(fontsize=14)
plt.show()
```
Figure 4: The upper plot shows the total forecasted trend, which more clearly illustrates the ~0.4$\sigma$ increase. The lower plot shows the weekly trend of the forecast, showing that rides actually tend to spike on Thursday/Friday, not over the weekend like I predicted eariler!
(Also, not sure how to fix the upper xticks since the plot_components method does not return a matplotlib subplots object)
## Model Validation
Crossvalidation and Chi-Square GOF Estimation
```
#Check number of model params
simplem.params
```
This model has 5 parameters: $\beta$, $\delta$, k, m, and $\sigma_{obs}$
```
def rchi2(y, ytrue, nparams):
'''
Returns the reduced Chi-Square goodness of fit metric for a given number of model parameters
===============
y: y-values produced by model
ytrue: y-values of real dataset
nparams: number of model parameters
returns:
reduced chi-square GOF (float)
'''
return np.sum( ((y-ytrue)**2) ) / (len(y) - nparams) #Note uncertainties are set to 1
#Computing the cross-validaton
cvresult_simplem = diagnostics.cross_validation(simplem, '30 days')
cvresult_simplem
rchi2(cvresult_simplem['yhat'], cvresult_simplem['y'], 5)
```
# DATA ACQUISITION 2: WWO Weather Data
Collecting NYC weather data for the relevant timeframe from the www.worldweatheronline.com API
```
!pip install wwo_hist
from wwo_hist import retrieve_hist_data
from google.colab import drive
drive.mount("/content/gdrive")
cd gdrive/My\ Drive
!pwd
frequency = 24 #frequency in hours (1/24 hours = 1/day)
start_date = '2014-04-01'
end_date = '2014-09-30'
api_key = pd.read_csv("my_apis.csv", header=None, index_col=0).loc["wwo"].values[0]
location_list = ['New+york,ny']
hist_weather_data = retrieve_hist_data(api_key,
location_list,
start_date,
end_date,
frequency,
location_label = False,
export_csv = True)
weather = pd.read_csv("New+york,ny.csv")
weather['date_time'] = pd.to_datetime(weather['date_time'])#Convert weather timestamps to pandas datetime type
weather.head()
weather.rename(columns = {'date_time':'ds'}, inplace=True) #Rename datetime column to ds to merge with uberbyday df
weather.head()
#Convert uberbyday['ds'] to datetime because it wasn't so before?
uberbyday['ds'] = pd.to_datetime(uberbyday['ds'])
uberbyday['ds']
#Merge the dataframes:
uberweather = uberbyday.merge(weather, on='ds')
uberweather
fig, ax = plt.subplots(3, figsize = (15,10))
uberweather.plot(y="y", ax=ax[0])
uberweather.plot(y="tempC", ax=ax[1])
uberweather.plot(y="precipMM", ax=ax[2])
plt.xlabel('Date')
ax[0].set_ylabel('Rides/day (standardized)', fontsize=12)
ax[1].set_ylabel('Temperature (Celsius)', fontsize=12)
ax[2].set_ylabel('Precipiation (mm)', fontsize=12)
ax[0].set_xticks([])
ax[1].set_xticks([])
plt.xticks(range(0,len(uberweather),20), uberweather['ds'].dt.date,rotation=90, fontsize=12);
```
Figure 5: The top plot shows our original series (standarized rides per day) with days since 2014-04-01 on the x-axis. The middle plot shows average daily temperature in degrees Celsius over that time period, and the bottoms shows daily precipitation in millimeters of rainwater over the time period. We might naively expect days with high precipitation and/or lower average temperatures to produce increases in rides/day. We'll see if adding these features as regressors improves the GOF of our model.
## MODEL 2: Regressors
Simple model used earlier, but with temperature and precipitation features added as regressors.
```
regrm = Prophet()
regrm.add_regressor('tempC')
regrm.add_regressor('precipMM');
regrm.fit(uberweather)
cvresult_regrm = diagnostics.cross_validation(regrm, '30 days')
nowcast = regrm.predict(uberweather)
regrm.plot(nowcast)
plt.ylabel('# of Rides (standardized)')
plt.xlabel('Date')
plt.show()
```
Figure 4: Standardized rides/day with temperature and precipitation regressors added to our model. Comparing to Figure 3 (the simple model w/o regressors) we can see that weather has a pronounced effect on the model, causing some short-timescale changes while preserving the overall trend.
```
regrm.params
```
Again, our model has the same 5 parameters
```
rchi2(cvresult_regrm['yhat'], cvresult_regrm['y'], 5)
```
Hmm our chi-square increased after adding the regressors...
# MODEL 3 add holidays
Holidays naturally affect rides per day, so we can implement US holidays using the add_country_holidays() Prophet method.
```
regholm = Prophet()
regholm.add_regressor('tempC')
regholm.add_regressor('precipMM');
regholm.add_country_holidays(country_name='US')
regholm.fit(uberweather)
herecast = regholm.predict(uberweather)
regholm.plot(herecast)
plt.ylabel('# of Rides (standardized)')
plt.xlabel('Date')
plt.show()
```
Figure 6: Rides per day model with temperature and precipitation added as regressors, as well as US holidays added. Note the major dip in ridership the end of August (Labor Day?) and the beginning of July (Independence Day?)
```
cvresult_regholm = diagnostics.cross_validation(regholm, '30 days')
regholm.params
rchi2(cvresult_regholm['yhat'], cvresult_regholm['y'], 5)
```
Ok, so the chi-square decreased when adding holidays, suggesting holidays indeed have a pronounced effect on Uber ridership
# MODEL 4: Markov-Chain Monte Carlo optimization
FB Prophet comes with a built-in MCMC optimization functionality, so we'll build a fourth model with a 5000-sample MCMC
```
regmcmc = Prophet(mcmc_samples=5000)
regmcmc.add_regressor('tempC')
regmcmc.add_regressor('precipMM');
regmcmc.add_country_holidays(country_name='US')
regmcmc.fit(uberweather)
mcmccast = regmcmc.predict(uberweather)
regmcmc.plot(mcmccast)
plt.ylabel('# of Rides (standardized)')
plt.xlabel('Date')
plt.show()
```
Figure 7: Rides per day model with temperature/precipitation regressors, holidays, and MCMC optimization. Bit tough to see by eye if the MCMC changed the forecast by much compared to previous models, but we can check the chi-square result.
```
cvresult_regmcmc = diagnostics.cross_validation(regmcmc, '30 days')
regmcmc.params
rchi2(cvresult_regmcmc['y'], cvresult_regmcmc['yhat'], 5)
```
So the chi-square increased a bit compared to the regression+holidays model. I would have expected the chi square to decrease, but without knowing the details of Prophet's MCMC implementation, it's difficult to pin down why that didn't happen.
### Summary of Model Performance:
Model | $\chi^2$ |
-------|-------------------|
simplem|0.47610571629587883|
regrm |0.5490993626794071 |
regholm|0.4639629930684204 |
regmcmc|0.49364441086582517|
|
github_jupyter
|
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
%matplotlib inline
plt.style.use("https://raw.githubusercontent.com/fedhere/MLTSA_FBianco/master/fbb.mplstyle")
!pip install fbprophet
import fbprophet
from fbprophet import Prophet
from fbprophet import diagnostics
#Builds a list of pandas dataframes from the github file urls:
a = []
for mon in ['apr', 'may', 'jun', 'jul', 'aug', 'sep']:
df = pd.read_csv('https://raw.githubusercontent.com/fivethirtyeight/uber-tlc-foil-response/master/uber-trip-data/uber-raw-data-'
+ mon + '14.csv')
a.append(df)
#concatenates the list into a single df
uberrides = pd.concat(a)
uberrides.head()
uberrides.shape
uberrides["time"] = pd.to_datetime(uberrides['Date/Time'])
uberrides.head()
uberrides.iloc[0]["time"].date
uberbyday = uberrides.groupby(uberrides["time"].dt.date).count()
# Returns pandas df with the number of rows in uberrides for each distinct date
uberbyday.head()
uberbyday = uberbyday.drop(columns=['Lat','Lon','Base','time'])
uberbyday.head()
uberbyday['ds'] = uberbyday.index
uberbyday.rename(columns={'Date/Time':'y'}, inplace=True)
uberbyday.head()
#Plotting the initial dataset
uberbyday.plot(x="ds", y="y");
uberbyday['y'] = (uberbyday['y'] - uberbyday['y'].mean()) / uberbyday['y'].std()
uberbyday.plot(x="ds", y="y");
simplem = Prophet() #Invoking the Prophet model object
simplem.fit(uberbyday) #Fitting the model on the data
# Create a dataframe of dates one month past the end of the dataset
d=[]
for i in range(1,32):
if i <= 9:
d.append('2014-10-0'+str(i))
if i > 9:
d.append('2014-10-'+str(i))
future = pd.DataFrame(data = {'ds': d})
# and stitch it onto the end of the dataset
past_and_future = pd.concat([uberbyday, future])
#use the fitted model to create a forecast of the dataset one month into the future
forecast = simplem.predict(past_and_future)
#Peek at the last 5 rows of the forecast:
forecast.iloc[-5:]
forecast['ds'].iloc[-31]
#Increase in trend value over month of October
forecast['trend'].iloc[-1] - forecast['trend'].iloc[-31]
#Plotting original data + forecast
fig1 = simplem.plot(forecast)
plt.plot(uberbyday.y, '--')
plt.xticks(rotation=90);
fig2 = simplem.plot_components(forecast)
plt.xticks(fontsize=14)
plt.show()
#Check number of model params
simplem.params
def rchi2(y, ytrue, nparams):
'''
Returns the reduced Chi-Square goodness of fit metric for a given number of model parameters
===============
y: y-values produced by model
ytrue: y-values of real dataset
nparams: number of model parameters
returns:
reduced chi-square GOF (float)
'''
return np.sum( ((y-ytrue)**2) ) / (len(y) - nparams) #Note uncertainties are set to 1
#Computing the cross-validaton
cvresult_simplem = diagnostics.cross_validation(simplem, '30 days')
cvresult_simplem
rchi2(cvresult_simplem['yhat'], cvresult_simplem['y'], 5)
!pip install wwo_hist
from wwo_hist import retrieve_hist_data
from google.colab import drive
drive.mount("/content/gdrive")
cd gdrive/My\ Drive
!pwd
frequency = 24 #frequency in hours (1/24 hours = 1/day)
start_date = '2014-04-01'
end_date = '2014-09-30'
api_key = pd.read_csv("my_apis.csv", header=None, index_col=0).loc["wwo"].values[0]
location_list = ['New+york,ny']
hist_weather_data = retrieve_hist_data(api_key,
location_list,
start_date,
end_date,
frequency,
location_label = False,
export_csv = True)
weather = pd.read_csv("New+york,ny.csv")
weather['date_time'] = pd.to_datetime(weather['date_time'])#Convert weather timestamps to pandas datetime type
weather.head()
weather.rename(columns = {'date_time':'ds'}, inplace=True) #Rename datetime column to ds to merge with uberbyday df
weather.head()
#Convert uberbyday['ds'] to datetime because it wasn't so before?
uberbyday['ds'] = pd.to_datetime(uberbyday['ds'])
uberbyday['ds']
#Merge the dataframes:
uberweather = uberbyday.merge(weather, on='ds')
uberweather
fig, ax = plt.subplots(3, figsize = (15,10))
uberweather.plot(y="y", ax=ax[0])
uberweather.plot(y="tempC", ax=ax[1])
uberweather.plot(y="precipMM", ax=ax[2])
plt.xlabel('Date')
ax[0].set_ylabel('Rides/day (standardized)', fontsize=12)
ax[1].set_ylabel('Temperature (Celsius)', fontsize=12)
ax[2].set_ylabel('Precipiation (mm)', fontsize=12)
ax[0].set_xticks([])
ax[1].set_xticks([])
plt.xticks(range(0,len(uberweather),20), uberweather['ds'].dt.date,rotation=90, fontsize=12);
regrm = Prophet()
regrm.add_regressor('tempC')
regrm.add_regressor('precipMM');
regrm.fit(uberweather)
cvresult_regrm = diagnostics.cross_validation(regrm, '30 days')
nowcast = regrm.predict(uberweather)
regrm.plot(nowcast)
plt.ylabel('# of Rides (standardized)')
plt.xlabel('Date')
plt.show()
regrm.params
rchi2(cvresult_regrm['yhat'], cvresult_regrm['y'], 5)
regholm = Prophet()
regholm.add_regressor('tempC')
regholm.add_regressor('precipMM');
regholm.add_country_holidays(country_name='US')
regholm.fit(uberweather)
herecast = regholm.predict(uberweather)
regholm.plot(herecast)
plt.ylabel('# of Rides (standardized)')
plt.xlabel('Date')
plt.show()
cvresult_regholm = diagnostics.cross_validation(regholm, '30 days')
regholm.params
rchi2(cvresult_regholm['yhat'], cvresult_regholm['y'], 5)
regmcmc = Prophet(mcmc_samples=5000)
regmcmc.add_regressor('tempC')
regmcmc.add_regressor('precipMM');
regmcmc.add_country_holidays(country_name='US')
regmcmc.fit(uberweather)
mcmccast = regmcmc.predict(uberweather)
regmcmc.plot(mcmccast)
plt.ylabel('# of Rides (standardized)')
plt.xlabel('Date')
plt.show()
cvresult_regmcmc = diagnostics.cross_validation(regmcmc, '30 days')
regmcmc.params
rchi2(cvresult_regmcmc['y'], cvresult_regmcmc['yhat'], 5)
| 0.565299 | 0.969527 |
# 1A.1 - D'une structure de données à l'autre
Ce notebook s'amuse à passer d'une structure de données à une autre, d'une liste à une dictionnaire, d'une liste de liste à un dictionnaire, avec toujours les mêmes données. list, dict, tuple.
```
from jyquickhelper import add_notebook_menu
add_notebook_menu()
```
## histogramme et dictionnaire
### liste à dictionnaire
Un histogramme est le moyen le plus simple de calculer la distribution d'une variable, de compter la fréquence des éléments d'une liste.
```
ens = ["a", "b", "gh", "er", "b", "gh"]
hist = {}
for e in ens:
hist[e] = hist.get(e, 0) + 1
hist
```
La méthode [get](https://docs.python.org/3/library/stdtypes.html?highlight=get#dict.get) comme beaucoup de fonctions implémente un besoin fréquent. Elle regarde si une clé appartient au dictionnaire, retourne la valeur associée ou une valeur par défault dans le cas contraire. Sans utiliser cette méthode, le code précédent devient :
```
ens = ["a", "b", "gh", "er", "b", "gh"]
hist = {}
for e in ens:
if e in hist:
hist[e] += 1
else:
hist[e] = 1
hist
```
### dictionnaire à liste
A priori l'histogramme représente la même information que la liste initiale `ens`. Il doit exister un moyen de recontruire la liste initiale.
```
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
ens = []
for k, v in hist.items():
for i in range(v):
ens.append(k)
ens
```
La liste initiale est retrouvée excepté l'ordre qui est différent. Les éléments identiques sont côte à côte. La méthode [items](https://docs.python.org/3/library/stdtypes.html?highlight=get#dict.items) retourne des couples `(clé, valeur)` ou plutôt une vue, c'est-à-dire une façon de parcourir un ensemble.
```
hist.items()
```
Pour vérifier que la méthode [items](https://docs.python.org/3/library/stdtypes.html?highlight=get#dict.items) ne retourne pas un ensemble mais une façon de parcours un ensemble, on regarde sa taille avec la fonction [getsizeof](https://docs.python.org/3/library/sys.html?highlight=getsizeof#sys.getsizeof) :
```
import sys
vue = hist.items()
sys.getsizeof(ens), sys.getsizeof(hist), sys.getsizeof(vue)
```
Et pour un dictionnaire plus grand, la taille du dictionnaire.
```
d = {i:i for i in range(1000)}
sys.getsizeof(d), sys.getsizeof(d.items())
```
On peut ne pas utiliser la méthode [items](https://docs.python.org/3/library/stdtypes.html?highlight=get#dict.items) :
```
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
ens = []
for k in hist:
v = hist[k]
for i in range(v):
ens.append(k)
ens
```
### dictionnaire et deux listes
Cette fois-ci, on met les clés d'un côté et les valeurs de l'autre.
```
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
cles = [k for k in hist]
vals = [hist[k] for k in hist]
cles, vals
```
On peut écrire aussi ce programme
```
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
cles = list(hist.keys())
vals = list(hist.values())
cles, vals
```
Toutefois, cette écriture n'est pas recommandée car il est possible que l'expression ``for k in hist`` ou ``list(hist.keys())`` parcourent les clés d'un dictionnaire de deux façons différentes si le dictionnaire est modifié entre temps. Mais on ne s'en pas toujours compte car cela dépend de l'implémentation des méthodes associées à la classe [dict](https://docs.python.org/3.5/library/stdtypes.html?highlight=dict#dict) (voir [cpython](https://github.com/python/cpython/tree/master/Python)). C'est pourquoi on préfère ne parcourir qu'une seule fois le dictionnaire tout en créant les deux listes.
```
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
cles = []
vals = []
for k, v in hist.items():
cles.append(k)
vals.append(v)
cles, vals
```
### deux listes et dictionnaires
On effectue l'opération inverse.
```
cles, vals = ['a', 'gh', 'er', 'b'], [1, 2, 1, 2]
hist = {a:b for a, b in zip(cles, vals)}
hist
```
Et si on ne veut pas utiliser la fonction [zip](https://docs.python.org/3/library/functions.html#zip) :
```
cles, vals = ['a', 'gh', 'er', 'b'], [1, 2, 1, 2]
hist = {}
for i in range(len(cles)):
hist[cles[i]] = vals[i]
hist
```
### zip reverse
La fonction [zip](https://docs.python.org/3/library/functions.html#zip) permet de parcourir deux listes en parallèles. Cela permet de raccourcir le code pour créer un dictionnaire à partir de clés et de valeurs séparés. Ca paraît bien plus long que de créer les listes des clés et des valeurs. Et pourtant le code suivant peut être considérablement raccourci :
```
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
cles = []
vals = []
for k, v in hist.items():
cles.append(k)
vals.append(v)
cles, vals
```
Cela devient :
```
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
cles, vals = zip(*hist.items())
cles, vals
```
Petite différence, `cles`, `vals` sont sous forme de [tuple](https://docs.python.org/3.5/library/stdtypes.html?highlight=tuple#tuple) mais cela reste très élégant.
## matrices et dictionnaires
### liste de listes et dictionnaires
Une liste de listes est la représentation la plus naturelle. Essayons de la transformer sous forme de dictionnaire. On utilise la fonction [enumerate](https://docs.python.org/3/library/functions.html#enumerate).
```
mat = [[1, 2],
[3, 4]]
dv = {}
for i, row in enumerate(mat):
for j, x in enumerate(row):
dv[i,j] = x
dv
```
### dictionnaires et liste de listes
On effectue l'opération inverse. Nous n'avons pas perdu d'information, nous devrions retrouver la liste de listes originale.
```
dx = {(0, 0): 1, (0, 1): 2, (1, 0): 3, (1, 1): 4}
max_i = max(k[0] for k in dx) + 1
max_j = max(k[1] for k in dx) + 1
mat = [[0] * max_j for i in range(max_i)]
for k, v in dv.items():
mat[k[0]][k[1]] = v
mat
```
La différence principale entre un dictionnaire ``d`` et une liste ``l`` est que l'instruction ``d[k]`` ajoute un élément d'indice ``k`` (quel que soit ``k``) alors que l'instruction ``l[k]``) suppose que l'élément d'indice ``k`` existe dans la liste. C'est pour cela qu'on commence à calculer les indices maximaux largeur, longueur.
### matrice sparse
On utilise cette répresentation surtout lorsque pour des matrices sparses : la majorité des coefficients sont nuls. Dans ce cas, le dictionnaire final ne contient que les coefficients non nuls.
```
mat = [[1, 0, 0],
[0, 4, 0]]
dv = {}
for i, row in enumerate(mat):
for j, x in enumerate(row):
if x != 0:
dv[i,j] = x
dv
```
Si on ne conserve pas les dimensions de la matrice originale, on perd un peu d'information dans un cas précis : si la matrice se termine par une colonne ou une ligne de zéros.
```
dx = {(0, 0): 1, (1, 1): 4}
max_i = max(k[0] for k in dx) + 1
max_j = max(k[1] for k in dx) + 1
mat = [[0] * max_j for i in range(max_i)]
for k, v in dv.items():
mat[k[0]][k[1]] = v
mat
```
## matrices et tableaux
### 2 dimensions logiques, 1 dimension en mémoire
On préfère représenter une matrice par un seul vecteur même si logiquement elle en contient car cela prend moins de place en mémoire. Dans ce cas, on met les lignes bout à bout.
```
mat = [[1, 0, 0],
[0, 4, 0],
[1, 2, 3]]
arr = []
for i, row in enumerate(mat):
for j, x in enumerate(row):
arr.append(x)
arr
```
D'un côté, nous avons 4 listes avec `mat` et une seule avec `arr`. Vérifions les tailles :
```
import sys
sys.getsizeof(mat), sys.getsizeof(arr)
```
Etrange ! Mais pour comprendre, il faut lire la documentation de la fonction [getsizeof](https://docs.python.org/3/library/sys.html?highlight=getsizeof#sys.getsizeof) qui ne compte pas la somme des objets référencés par celui dont on mesure la taille. Autrement dit, dans le cas d'une liste de listes, la fonction ne mesure que la taille de la première liste. Pour corriger le tir, on utilise la fonction suggérée par la documentation de Python.
```
from ensae_teaching_cs.helpers.size_helper import total_size
total_size(mat), total_size(arr)
```
On peut aussi utiliser le module [pympler](https://pythonhosted.org/Pympler/) et la fonction [asizeof](https://pythonhosted.org/Pympler/asizeof.html#asizeof).
```
from pympler.asizeof import asizeof
asizeof(mat), asizeof(arr)
```
Cela prend énormément de place pour 9 *float* (soit 9x8 octets) mais Python stocke beaucoup plus d'informations qu'un langage compilé type C++. Ca tombe bien, c'est que fait le module [numpy](http://www.numpy.org/).
```
from numpy import array
amat = array(mat)
aarr = array(arr)
asizeof(amat), asizeof(aarr)
```
Et si on augmente le nombre de réels pour faire disparaître les coûts fixes :
```
n = 100000
li = list(float(x) for x in range(n))
ar = array(li)
asizeof(li) /n, asizeof(ar) / n
```
Python prend 4 fois plus de place que numpy.
### du tableau à la liste de listes
A moins que la matrice soit carrée, il faut conserver une des dimensions du tableau original, le nombre de lignes par exemple.
```
arr = [1, 0, 0, 0, 4, 0, 1, 2, 3]
nb_lin = 3
nb_col = len(arr) // nb_lin
mat = []
pos = 0
for i in range(nb_lin):
row = []
for j in range(nb_col):
row.append(arr[pos])
pos += 1
mat.append(row)
mat
```
On peut aussi faire comme ceci :
```
arr = [1, 0, 0, 0, 4, 0, 1, 2, 3]
nb_lin = 3
nb_col = len(arr) // nb_lin
mat = [[0] * nb_col for i in range(nb_lin)]
for pos, x in enumerate(arr):
i = pos // nb_lin
j = pos % nb_lin
mat[i][j] = x
mat
```
|
github_jupyter
|
from jyquickhelper import add_notebook_menu
add_notebook_menu()
ens = ["a", "b", "gh", "er", "b", "gh"]
hist = {}
for e in ens:
hist[e] = hist.get(e, 0) + 1
hist
ens = ["a", "b", "gh", "er", "b", "gh"]
hist = {}
for e in ens:
if e in hist:
hist[e] += 1
else:
hist[e] = 1
hist
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
ens = []
for k, v in hist.items():
for i in range(v):
ens.append(k)
ens
hist.items()
import sys
vue = hist.items()
sys.getsizeof(ens), sys.getsizeof(hist), sys.getsizeof(vue)
d = {i:i for i in range(1000)}
sys.getsizeof(d), sys.getsizeof(d.items())
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
ens = []
for k in hist:
v = hist[k]
for i in range(v):
ens.append(k)
ens
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
cles = [k for k in hist]
vals = [hist[k] for k in hist]
cles, vals
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
cles = list(hist.keys())
vals = list(hist.values())
cles, vals
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
cles = []
vals = []
for k, v in hist.items():
cles.append(k)
vals.append(v)
cles, vals
cles, vals = ['a', 'gh', 'er', 'b'], [1, 2, 1, 2]
hist = {a:b for a, b in zip(cles, vals)}
hist
cles, vals = ['a', 'gh', 'er', 'b'], [1, 2, 1, 2]
hist = {}
for i in range(len(cles)):
hist[cles[i]] = vals[i]
hist
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
cles = []
vals = []
for k, v in hist.items():
cles.append(k)
vals.append(v)
cles, vals
hist = {'a': 1, 'b': 2, 'er': 1, 'gh': 2}
cles, vals = zip(*hist.items())
cles, vals
mat = [[1, 2],
[3, 4]]
dv = {}
for i, row in enumerate(mat):
for j, x in enumerate(row):
dv[i,j] = x
dv
dx = {(0, 0): 1, (0, 1): 2, (1, 0): 3, (1, 1): 4}
max_i = max(k[0] for k in dx) + 1
max_j = max(k[1] for k in dx) + 1
mat = [[0] * max_j for i in range(max_i)]
for k, v in dv.items():
mat[k[0]][k[1]] = v
mat
mat = [[1, 0, 0],
[0, 4, 0]]
dv = {}
for i, row in enumerate(mat):
for j, x in enumerate(row):
if x != 0:
dv[i,j] = x
dv
dx = {(0, 0): 1, (1, 1): 4}
max_i = max(k[0] for k in dx) + 1
max_j = max(k[1] for k in dx) + 1
mat = [[0] * max_j for i in range(max_i)]
for k, v in dv.items():
mat[k[0]][k[1]] = v
mat
mat = [[1, 0, 0],
[0, 4, 0],
[1, 2, 3]]
arr = []
for i, row in enumerate(mat):
for j, x in enumerate(row):
arr.append(x)
arr
import sys
sys.getsizeof(mat), sys.getsizeof(arr)
from ensae_teaching_cs.helpers.size_helper import total_size
total_size(mat), total_size(arr)
from pympler.asizeof import asizeof
asizeof(mat), asizeof(arr)
from numpy import array
amat = array(mat)
aarr = array(arr)
asizeof(amat), asizeof(aarr)
n = 100000
li = list(float(x) for x in range(n))
ar = array(li)
asizeof(li) /n, asizeof(ar) / n
arr = [1, 0, 0, 0, 4, 0, 1, 2, 3]
nb_lin = 3
nb_col = len(arr) // nb_lin
mat = []
pos = 0
for i in range(nb_lin):
row = []
for j in range(nb_col):
row.append(arr[pos])
pos += 1
mat.append(row)
mat
arr = [1, 0, 0, 0, 4, 0, 1, 2, 3]
nb_lin = 3
nb_col = len(arr) // nb_lin
mat = [[0] * nb_col for i in range(nb_lin)]
for pos, x in enumerate(arr):
i = pos // nb_lin
j = pos % nb_lin
mat[i][j] = x
mat
| 0.083473 | 0.953794 |
```
import dagstermill
context = dagstermill.get_context()
eastbound_delays = 'eastbound_delays'
westbound_delays = 'westbound_delays'
db_url = context.resources.db_info.url
import math
import os
import geopandas as geo
import matplotlib.pyplot as plt
import pandas as pd
import sqlalchemy as sa
from dagster.utils import mkdir_p
engine = sa.create_engine(db_url)
from matplotlib.backends.backend_pdf import PdfPages
plots_path = os.path.join(os.getcwd(), 'plots')
mkdir_p(plots_path)
pdf_path = os.path.join(plots_path, 'delays_by_geography.pdf')
pp = PdfPages(pdf_path)
westbound_delays = pd.read_sql('''
select * from {westbound_delays}
'''.format(westbound_delays=westbound_delays), engine)
eastbound_delays = pd.read_sql('''
select * from {eastbound_delays}
'''.format(eastbound_delays=eastbound_delays), engine)
states = geo.read_file('https://d2ad6b4ur7yvpq.cloudfront.net/naturalearth-3.3.0/ne_110m_admin_1_states_provinces_shp.geojson')
continental_us = states[
states['iso_3166_2'].map(lambda x: 'HI' not in x) &
states['iso_3166_2'].map(lambda x: 'AK' not in x)
]
eastbound_delays
ax = continental_us.plot(
alpha=0.1,
edgecolor='black',
)
# These "should" be great circles, but to keep the demo dependencies lightweight we aren't using basemap
for i in eastbound_delays.index:
if eastbound_delays['avg_arrival_delay'][i] > 1:
plt.plot(
(eastbound_delays['origin_longitude'][i], eastbound_delays['dest_longitude'][i]),
(eastbound_delays['origin_latitude'][i], eastbound_delays['dest_latitude'][i]),
'k-',
linewidth=math.sqrt(eastbound_delays['avg_arrival_delay'][i]),
alpha=min(math.sqrt(eastbound_delays['avg_arrival_delay'][i]) * .1, 1)
)
plt.title('Average Delays on Eastbound Routes', fontsize=8)
pp.savefig()
westbound_delays
ax = continental_us.plot(
alpha=0.1,
edgecolor='black',
)
# These "should" be great circles, but to keep the demo dependencies lightweight we aren't using basemap
for i in westbound_delays.index:
if westbound_delays['avg_arrival_delay'][i] > 1:
plt.plot(
(westbound_delays['origin_longitude'][i], westbound_delays['dest_longitude'][i]),
(westbound_delays['origin_latitude'][i], westbound_delays['dest_latitude'][i]),
'k-',
linewidth=math.sqrt(westbound_delays['avg_arrival_delay'][i]),
alpha=min(math.sqrt(westbound_delays['avg_arrival_delay'][i]) * .1, 1)
)
plt.title('Average Delays on Westbound Routes', fontsize=8)
pp.savefig()
pp.close()
from dagster import LocalFileHandle
dagstermill.yield_result(LocalFileHandle(pdf_path))
```
|
github_jupyter
|
import dagstermill
context = dagstermill.get_context()
eastbound_delays = 'eastbound_delays'
westbound_delays = 'westbound_delays'
db_url = context.resources.db_info.url
import math
import os
import geopandas as geo
import matplotlib.pyplot as plt
import pandas as pd
import sqlalchemy as sa
from dagster.utils import mkdir_p
engine = sa.create_engine(db_url)
from matplotlib.backends.backend_pdf import PdfPages
plots_path = os.path.join(os.getcwd(), 'plots')
mkdir_p(plots_path)
pdf_path = os.path.join(plots_path, 'delays_by_geography.pdf')
pp = PdfPages(pdf_path)
westbound_delays = pd.read_sql('''
select * from {westbound_delays}
'''.format(westbound_delays=westbound_delays), engine)
eastbound_delays = pd.read_sql('''
select * from {eastbound_delays}
'''.format(eastbound_delays=eastbound_delays), engine)
states = geo.read_file('https://d2ad6b4ur7yvpq.cloudfront.net/naturalearth-3.3.0/ne_110m_admin_1_states_provinces_shp.geojson')
continental_us = states[
states['iso_3166_2'].map(lambda x: 'HI' not in x) &
states['iso_3166_2'].map(lambda x: 'AK' not in x)
]
eastbound_delays
ax = continental_us.plot(
alpha=0.1,
edgecolor='black',
)
# These "should" be great circles, but to keep the demo dependencies lightweight we aren't using basemap
for i in eastbound_delays.index:
if eastbound_delays['avg_arrival_delay'][i] > 1:
plt.plot(
(eastbound_delays['origin_longitude'][i], eastbound_delays['dest_longitude'][i]),
(eastbound_delays['origin_latitude'][i], eastbound_delays['dest_latitude'][i]),
'k-',
linewidth=math.sqrt(eastbound_delays['avg_arrival_delay'][i]),
alpha=min(math.sqrt(eastbound_delays['avg_arrival_delay'][i]) * .1, 1)
)
plt.title('Average Delays on Eastbound Routes', fontsize=8)
pp.savefig()
westbound_delays
ax = continental_us.plot(
alpha=0.1,
edgecolor='black',
)
# These "should" be great circles, but to keep the demo dependencies lightweight we aren't using basemap
for i in westbound_delays.index:
if westbound_delays['avg_arrival_delay'][i] > 1:
plt.plot(
(westbound_delays['origin_longitude'][i], westbound_delays['dest_longitude'][i]),
(westbound_delays['origin_latitude'][i], westbound_delays['dest_latitude'][i]),
'k-',
linewidth=math.sqrt(westbound_delays['avg_arrival_delay'][i]),
alpha=min(math.sqrt(westbound_delays['avg_arrival_delay'][i]) * .1, 1)
)
plt.title('Average Delays on Westbound Routes', fontsize=8)
pp.savefig()
pp.close()
from dagster import LocalFileHandle
dagstermill.yield_result(LocalFileHandle(pdf_path))
| 0.322419 | 0.261873 |
<!-- ---
title: Collective Communication with Ignite
weight: 1
date: 2021-10-5
downloads: true
sidebar: true
tags:
- idist
- all_gather
- all_reduce
- broadcast
- barrier
--- -->
# Collective Communication with Ignite
In this tutorial, we will see how to use advanced distributed functions like `all_reduce()`, `all_gather()`, `broadcast()` and `barrier()`. We will discuss unique use cases for all of them and represent them visually.
<!--more-->
## Required Dependencies
```
!pip install pytorch-ignite
```
## Imports
```
import torch
import ignite.distributed as idist
```
## All Reduce
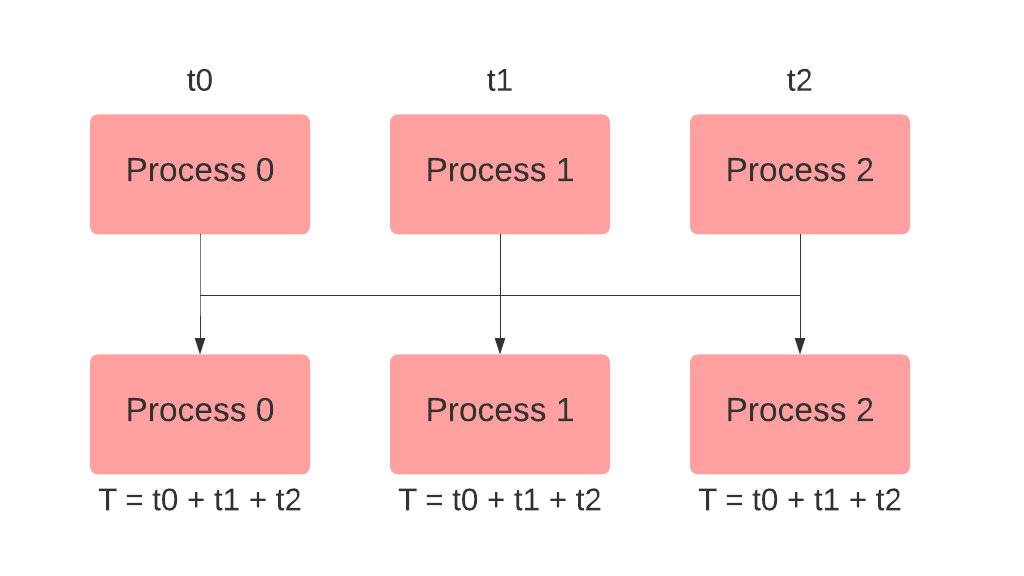
The [`all_reduce()`](https://pytorch.org/ignite/distributed.html#ignite.distributed.utils.all_reduce) method is used to collect specified tensors from each process and make them available on every node then perform a specified operation (sum, product, min, max, etc) on them. Let's spawn 3 processes with ranks 0, 1 and 2 and define a `tensor` on all of them. If we performed `all_reduce` with the operation SUM on `tensor` then `tensor` on all ranks will be gathered, added and stored in `tensor` as shown below:
```
def all_reduce_example(local_rank):
tensor = torch.arange(2, dtype=torch.int64) + 1 + 2 * local_rank
print(f"Rank {local_rank}, Initial value: {tensor}")
idist.all_reduce(tensor, op="SUM")
print(f"Rank {local_rank}, After performing all_reduce: {tensor}")
```
We can use [idist.spawn](https://pytorch.org/ignite/distributed.html#ignite.distributed.utils.spawn) to spawn 3 processes (`nproc_per_node`) and execute the above function.
```
idist.spawn(backend="gloo", fn=all_reduce_example, args=(), nproc_per_node=3)
```
Now let's assume a more real world scenario - You need to find the average of all the gradients available on different processes.
> First, we get the number of GPUs available, with the get_world_size method. Then, for every model parameter, we do the following:
>
> 1. Gather the gradients on each process
> 2. Apply the sum operation on the gradients
> 3. Divide by the world size to average them
>
> Finally, we can go on to update the model parameters using the averaged gradients!
>
> -- <cite>[Distributed Deep Learning 101: Introduction](https://towardsdatascience.com/distributed-deep-learning-101-introduction-ebfc1bcd59d9)</cite>
You can get the number of GPUs (processes) available using another helper method [`idist.get_world_size()`](https://pytorch.org/ignite/distributed.html#ignite.distributed.utils.get_world_size) and then use `all_reduce()` to collect the gradients and apply the SUM operation.
```
def average_gradients(model):
num_processes = idist.get_world_size()
for param in model.parameters():
idist.all_reduce(param.grad.data, op="SUM")
param.grad.data = param.grad.data / num_processes
```
## All Gather
The [`all_gather()`](https://pytorch.org/ignite/distributed.html#ignite.distributed.utils.all_gather) method is used when you just want to collect a tensor, number or string across all participating processes. As a basic example, suppose you have to collect all the different values stored in `num` on all ranks. You can achieve this by using `all_gather` as below:
```
def all_gather_example(local_rank):
num = 2.0 * idist.get_rank()
print(f"Rank {local_rank}, Initial value: {num}")
all_nums = idist.all_gather(num)
print(f"Rank {local_rank}, After performing all_gather: {all_nums}")
idist.spawn(backend="gloo", fn=all_gather_example, args=(), nproc_per_node=3)
```
Now let's assume you need to gather the predicted values which are distributed across all the processes on the main process so you could store them to a file. Here is how you can do it:
```
def write_preds_to_file(predictions, filename):
prediction_tensor = torch.tensor(predictions)
prediction_tensor = idist.all_gather(prediction_tensor)
if idist.get_rank() == 0:
torch.save(prediction_tensor, filename)
```
**Note:** In the above example, only the main process required the gathered values and not all the processes. This can be also be done via the `gather()` method, however one of the backends [`nccl` does not support `gather()`](https://pytorch.org/docs/stable/distributed.html) hence we had to use `all_gather()`.
## Broadcast
The [`broadcast()`](https://pytorch.org/ignite/distributed.html#ignite.distributed.utils.broadcast) method copies a tensor, float or string from a source process to all the other processes. For example, you need to send a message from rank 0 to all other ranks. You can do this by creating the actual message on rank 0 and a placeholder on all other ranks, then broadcast the message mentioning a source rank. You can also use `safe_mode=True` in case the placeholder is not defined on all ranks.
```
def broadcast_example(local_rank):
message = f"hello from rank {idist.get_rank()}"
print(f"Rank {local_rank}, Initial value: {message}")
message = idist.broadcast(message, src=0)
print(f"Rank {local_rank}, After performing broadcast: {message}")
idist.spawn(backend="gloo", fn=broadcast_example, args=(), nproc_per_node=3)
```
For a real world use case, let's assume you need to gather the predicted and actual values from all the processes on rank 0 for computing a metric and avoiding a memory error. You can do do this by first using `all_gather()`, then computing the metric and finally using `broadcast()` to share the result with all processes. `src` below refers to the rank of the source process.
```
def compute_metric(prediction_tensor, target_tensor):
prediction_tensor = idist.all_gather(prediction_tensor)
target_tensor = idist.all_gather(target_tensor)
result = 0.0
if idist.get_rank() == 0:
result = compute_fn(prediction_tensor, target_tensor)
result = idist.broadcast(result, src=0)
return result
```
## Barrier
The [`barrier()`](https://pytorch.org/ignite/distributed.html#ignite.distributed.utils.barrier) method helps synchronize all processes. For example - while downloading data during training, we have to make sure only the main process (`rank = 0`) downloads the datasets to prevent the sub processes (`rank > 0`) from downloading the same file to the same path at the same time. This way all sub processes get a copy of this already downloaded dataset. This is where we can utilize `barrier()` to make the sub processes wait until the main process downloads the datasets. Once that is done, all the subprocesses instantiate the datasets, while the main process waits. Finally, all the processes are synced up.
```
def get_datasets(config):
if idist.get_local_rank() > 0:
idist.barrier()
train_dataset, test_dataset = get_train_test_datasets(config["data_path"])
if idist.get_local_rank() == 0:
idist.barrier()
return train_dataset, test_dataset
```
|
github_jupyter
|
!pip install pytorch-ignite
import torch
import ignite.distributed as idist
def all_reduce_example(local_rank):
tensor = torch.arange(2, dtype=torch.int64) + 1 + 2 * local_rank
print(f"Rank {local_rank}, Initial value: {tensor}")
idist.all_reduce(tensor, op="SUM")
print(f"Rank {local_rank}, After performing all_reduce: {tensor}")
idist.spawn(backend="gloo", fn=all_reduce_example, args=(), nproc_per_node=3)
def average_gradients(model):
num_processes = idist.get_world_size()
for param in model.parameters():
idist.all_reduce(param.grad.data, op="SUM")
param.grad.data = param.grad.data / num_processes
def all_gather_example(local_rank):
num = 2.0 * idist.get_rank()
print(f"Rank {local_rank}, Initial value: {num}")
all_nums = idist.all_gather(num)
print(f"Rank {local_rank}, After performing all_gather: {all_nums}")
idist.spawn(backend="gloo", fn=all_gather_example, args=(), nproc_per_node=3)
def write_preds_to_file(predictions, filename):
prediction_tensor = torch.tensor(predictions)
prediction_tensor = idist.all_gather(prediction_tensor)
if idist.get_rank() == 0:
torch.save(prediction_tensor, filename)
def broadcast_example(local_rank):
message = f"hello from rank {idist.get_rank()}"
print(f"Rank {local_rank}, Initial value: {message}")
message = idist.broadcast(message, src=0)
print(f"Rank {local_rank}, After performing broadcast: {message}")
idist.spawn(backend="gloo", fn=broadcast_example, args=(), nproc_per_node=3)
def compute_metric(prediction_tensor, target_tensor):
prediction_tensor = idist.all_gather(prediction_tensor)
target_tensor = idist.all_gather(target_tensor)
result = 0.0
if idist.get_rank() == 0:
result = compute_fn(prediction_tensor, target_tensor)
result = idist.broadcast(result, src=0)
return result
def get_datasets(config):
if idist.get_local_rank() > 0:
idist.barrier()
train_dataset, test_dataset = get_train_test_datasets(config["data_path"])
if idist.get_local_rank() == 0:
idist.barrier()
return train_dataset, test_dataset
| 0.541651 | 0.945248 |
## Learning OpenCV 4 Computer Vision with Python 3 - Pg 93
https://github.com/PacktPublishing/Learning-OpenCV-4-Computer-Vision-with-Python-Third-Edition/blob/master/chapter04/disparity.py
```
import cv2
import numpy as np
# Setting Parameters
minDisparity = 16 # Minimum possible disparity value. Normally, it is zero but sometimes rectification algorithms can shift images, so this parameter needs to be adjusted accordingly.
numDisparities = 192-minDisparity # Maximum disparity minus minimum disparity. The value is always greater than zero. In the current implementation, this parameter must be divisible by 16.
blockSize = 5 # Matched block size. It must be an odd number >=1 . Normally, it should be somewhere in the 3..11 range
'''
uniquenessRatio:
Margin in percentage by which the best (minimum) computed cost function value should "win" the second best value to
consider the found match correct.
Normally, a value within the 5-15 range is good enough.
'''
uniquenessRatio = 1 #
speckleWindowSize = 3 # Maximum size of smooth disparity regions to consider their noise speckles and invalidate. Set it to 0 to disable speckle filtering. Otherwise, set it somewhere in the 50-200 range.
speckleRange = 3 # Maximum disparity variation within each connected component. If you do speckle filtering, set the parameter to a positive value, it will be implicitly multiplied by 16. Normally, 1 or 2 is good enough.
disp12MaxDiff = 200
'''
P1, P2
The second parameter controlling the disparity smoothness. The larger the values are, the smoother the disparity is.
P1 is the penalty on the disparity change by plus or minus 1 between neighbor pixels.
P2 is the penalty on the disparity change by more than 1 between neighbor pixels.
The algorithm requires P2 > P1 .
See stereo_match.cpp sample where some reasonably good P1 and P2 values are shown
(like 8*number_of_image_channels*blockSize*blockSize and 32*number_of_image_channels*blockSize*blockSize , respectively).
'''
P1 = 8 * 3 * blockSize ** 2
P2 = 32 * 3 * blockSize ** 2 # P2 must be greater than P1
# MODE (unused) - Set it to StereoSGBM::MODE_HH to run the full-scale two-pass dynamic programming algorithm. It will consume O(W*H*numDisparities) bytes, which is large for 640x480 stereo and huge for HD-size pictures. By default, it is set to false .
# Semi-Global Block Matching
stereo = cv2.StereoSGBM_create(
minDisparity=minDisparity,
numDisparities=numDisparities,
blockSize=blockSize,
uniquenessRatio=uniquenessRatio,
speckleRange=speckleRange,
speckleWindowSize=speckleWindowSize,
disp12MaxDiff=disp12MaxDiff,
P1=P1,
P2=P2
)
imgL = cv2.imread('1lsm.png')
imgR = cv2.imread('1rsm.png')
def update(sliderValue = 0):
'''
This is updating the Trackbar values
'''
stereo.setBlockSize(
cv2.getTrackbarPos('blockSize', 'Disparity'))
stereo.setUniquenessRatio(
cv2.getTrackbarPos('uniquenessRatio', 'Disparity'))
stereo.setSpeckleWindowSize(
cv2.getTrackbarPos('speckleWindowSize', 'Disparity'))
stereo.setSpeckleRange(
cv2.getTrackbarPos('speckleRange', 'Disparity'))
stereo.setDisp12MaxDiff(
cv2.getTrackbarPos('disp12MaxDiff', 'Disparity'))
disparity = stereo.compute(
imgL, imgR).astype(np.float32) / 16.0
cv2.imshow('Left', imgL)
cv2.imshow('Right', imgR)
cv2.imshow('Disparity', (disparity - minDisparity) / numDisparities)
# Create Window and Trackbars
cv2.namedWindow('Disparity')
cv2.createTrackbar('blockSize', 'Disparity', blockSize, 21, update) # Keep an eye on this, must be odd number > 1
cv2.createTrackbar('uniquenessRatio', 'Disparity', uniquenessRatio, 50, update)
cv2.createTrackbar('speckleWindowSize', 'Disparity', speckleWindowSize, 200, update)
cv2.createTrackbar('speckleRange', 'Disparity', speckleRange, 50, update)
cv2.createTrackbar('disp12MaxDiff', 'Disparity', disp12MaxDiff, 250, update)
# Initialize the disparity map. Show the disparity map and images.
update()
# Wait for the user to press any key.
# Meanwhile, update() will be called anytime the user moves a slider.
cv2.waitKey()
cv2.destroyAllWindows()
```
https://vision.middlebury.edu/stereo/data/
|
github_jupyter
|
import cv2
import numpy as np
# Setting Parameters
minDisparity = 16 # Minimum possible disparity value. Normally, it is zero but sometimes rectification algorithms can shift images, so this parameter needs to be adjusted accordingly.
numDisparities = 192-minDisparity # Maximum disparity minus minimum disparity. The value is always greater than zero. In the current implementation, this parameter must be divisible by 16.
blockSize = 5 # Matched block size. It must be an odd number >=1 . Normally, it should be somewhere in the 3..11 range
'''
uniquenessRatio:
Margin in percentage by which the best (minimum) computed cost function value should "win" the second best value to
consider the found match correct.
Normally, a value within the 5-15 range is good enough.
'''
uniquenessRatio = 1 #
speckleWindowSize = 3 # Maximum size of smooth disparity regions to consider their noise speckles and invalidate. Set it to 0 to disable speckle filtering. Otherwise, set it somewhere in the 50-200 range.
speckleRange = 3 # Maximum disparity variation within each connected component. If you do speckle filtering, set the parameter to a positive value, it will be implicitly multiplied by 16. Normally, 1 or 2 is good enough.
disp12MaxDiff = 200
'''
P1, P2
The second parameter controlling the disparity smoothness. The larger the values are, the smoother the disparity is.
P1 is the penalty on the disparity change by plus or minus 1 between neighbor pixels.
P2 is the penalty on the disparity change by more than 1 between neighbor pixels.
The algorithm requires P2 > P1 .
See stereo_match.cpp sample where some reasonably good P1 and P2 values are shown
(like 8*number_of_image_channels*blockSize*blockSize and 32*number_of_image_channels*blockSize*blockSize , respectively).
'''
P1 = 8 * 3 * blockSize ** 2
P2 = 32 * 3 * blockSize ** 2 # P2 must be greater than P1
# MODE (unused) - Set it to StereoSGBM::MODE_HH to run the full-scale two-pass dynamic programming algorithm. It will consume O(W*H*numDisparities) bytes, which is large for 640x480 stereo and huge for HD-size pictures. By default, it is set to false .
# Semi-Global Block Matching
stereo = cv2.StereoSGBM_create(
minDisparity=minDisparity,
numDisparities=numDisparities,
blockSize=blockSize,
uniquenessRatio=uniquenessRatio,
speckleRange=speckleRange,
speckleWindowSize=speckleWindowSize,
disp12MaxDiff=disp12MaxDiff,
P1=P1,
P2=P2
)
imgL = cv2.imread('1lsm.png')
imgR = cv2.imread('1rsm.png')
def update(sliderValue = 0):
'''
This is updating the Trackbar values
'''
stereo.setBlockSize(
cv2.getTrackbarPos('blockSize', 'Disparity'))
stereo.setUniquenessRatio(
cv2.getTrackbarPos('uniquenessRatio', 'Disparity'))
stereo.setSpeckleWindowSize(
cv2.getTrackbarPos('speckleWindowSize', 'Disparity'))
stereo.setSpeckleRange(
cv2.getTrackbarPos('speckleRange', 'Disparity'))
stereo.setDisp12MaxDiff(
cv2.getTrackbarPos('disp12MaxDiff', 'Disparity'))
disparity = stereo.compute(
imgL, imgR).astype(np.float32) / 16.0
cv2.imshow('Left', imgL)
cv2.imshow('Right', imgR)
cv2.imshow('Disparity', (disparity - minDisparity) / numDisparities)
# Create Window and Trackbars
cv2.namedWindow('Disparity')
cv2.createTrackbar('blockSize', 'Disparity', blockSize, 21, update) # Keep an eye on this, must be odd number > 1
cv2.createTrackbar('uniquenessRatio', 'Disparity', uniquenessRatio, 50, update)
cv2.createTrackbar('speckleWindowSize', 'Disparity', speckleWindowSize, 200, update)
cv2.createTrackbar('speckleRange', 'Disparity', speckleRange, 50, update)
cv2.createTrackbar('disp12MaxDiff', 'Disparity', disp12MaxDiff, 250, update)
# Initialize the disparity map. Show the disparity map and images.
update()
# Wait for the user to press any key.
# Meanwhile, update() will be called anytime the user moves a slider.
cv2.waitKey()
cv2.destroyAllWindows()
| 0.627837 | 0.873754 |
# 7.7 Transformerの学習・推論、判定根拠の可視化を実装
- 本ファイルでは、ここまでで作成したTransformerモデルとIMDbのDataLoaderを使用してクラス分類を学習させます。テストデータで推論をし、さらに判断根拠となるAttentionを可視化します
# 7.7 学習目標
1. Transformerの学習を実装できるようになる
2. Transformerの判定時のAttention可視化を実装できるようになる
# 事前準備
- フォルダ「utils」内の関数やクラスを使用します
```
# パッケージのimport
import numpy as np
import random
import torch
import torch.nn as nn
import torch.optim as optim
import torchtext
# 乱数のシードを設定
torch.manual_seed(1234)
np.random.seed(1234)
random.seed(1234)
```
# DatasetとDataLoaderを作成
```
from utils.dataloader import get_IMDb_DataLoaders_and_TEXT
# 読み込み
train_dl, val_dl, test_dl, TEXT = get_IMDb_DataLoaders_and_TEXT(
max_length=256, batch_size=64)
# 辞書オブジェクトにまとめる
dataloaders_dict = {"train": train_dl, "val": val_dl}
```
# ネットワークモデルの作成
```
from utils.transformer import TransformerClassification
# モデル構築
net = TransformerClassification(
text_embedding_vectors=TEXT.vocab.vectors, d_model=300, max_seq_len=256, output_dim=2)
# ネットワークの初期化を定義
def weights_init(m):
classname = m.__class__.__name__
if classname.find('Linear') != -1:
# Liner層の初期化
nn.init.kaiming_normal_(m.weight)
if m.bias is not None:
nn.init.constant_(m.bias, 0.0)
# 訓練モードに設定
net.train()
# TransformerBlockモジュールを初期化実行
net.net3_1.apply(weights_init)
net.net3_2.apply(weights_init)
print('ネットワーク設定完了')
```
# 損失関数と最適化手法を定義
```
# 損失関数の設定
criterion = nn.CrossEntropyLoss()
# nn.LogSoftmax()を計算してからnn.NLLLoss(negative log likelihood loss)を計算
# 最適化手法の設定
learning_rate = 2e-5
optimizer = optim.Adam(net.parameters(), lr=learning_rate)
```
# 学習・検証を実施
```
# モデルを学習させる関数を作成
def train_model(net, dataloaders_dict, criterion, optimizer, num_epochs):
# GPUが使えるかを確認
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
print("使用デバイス:", device)
print('-----start-------')
# ネットワークをGPUへ
net.to(device)
# ネットワークがある程度固定であれば、高速化させる
torch.backends.cudnn.benchmark = True
# epochのループ
for epoch in range(num_epochs):
# epochごとの訓練と検証のループ
for phase in ['train', 'val']:
if phase == 'train':
net.train() # モデルを訓練モードに
else:
net.eval() # モデルを検証モードに
epoch_loss = 0.0 # epochの損失和
epoch_corrects = 0 # epochの正解数
# データローダーからミニバッチを取り出すループ
for batch in (dataloaders_dict[phase]):
# batchはTextとLableの辞書オブジェクト
# GPUが使えるならGPUにデータを送る
inputs = batch.Text[0].to(device) # 文章
labels = batch.Label.to(device) # ラベル
# optimizerを初期化
optimizer.zero_grad()
# 順伝搬(forward)計算
with torch.set_grad_enabled(phase == 'train'):
# mask作成
input_pad = 1 # 単語のIDにおいて、'<pad>': 1 なので
input_mask = (inputs != input_pad)
# Transformerに入力
outputs, _, _ = net(inputs, input_mask)
loss = criterion(outputs, labels) # 損失を計算
_, preds = torch.max(outputs, 1) # ラベルを予測
# 訓練時はバックプロパゲーション
if phase == 'train':
loss.backward()
optimizer.step()
# 結果の計算
epoch_loss += loss.item() * inputs.size(0) # lossの合計を更新
# 正解数の合計を更新
epoch_corrects += torch.sum(preds == labels.data)
# epochごとのlossと正解率
epoch_loss = epoch_loss / len(dataloaders_dict[phase].dataset)
epoch_acc = epoch_corrects.double(
) / len(dataloaders_dict[phase].dataset)
print('Epoch {}/{} | {:^5} | Loss: {:.4f} Acc: {:.4f}'.format(epoch+1, num_epochs,
phase, epoch_loss, epoch_acc))
return net
# 学習・検証を実行する 15分ほどかかります
num_epochs = 10
net_trained = train_model(net, dataloaders_dict,
criterion, optimizer, num_epochs=num_epochs)
```
# テストデータでの正解率を求める
```
# device
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
net_trained.eval() # モデルを検証モードに
net_trained.to(device)
epoch_corrects = 0 # epochの正解数
for batch in (test_dl): # testデータのDataLoader
# batchはTextとLableの辞書オブジェクト
# GPUが使えるならGPUにデータを送る
inputs = batch.Text[0].to(device) # 文章
labels = batch.Label.to(device) # ラベル
# 順伝搬(forward)計算
with torch.set_grad_enabled(False):
# mask作成
input_pad = 1 # 単語のIDにおいて、'<pad>': 1 なので
input_mask = (inputs != input_pad)
# Transformerに入力
outputs, _, _ = net_trained(inputs, input_mask)
_, preds = torch.max(outputs, 1) # ラベルを予測
# 結果の計算
# 正解数の合計を更新
epoch_corrects += torch.sum(preds == labels.data)
# 正解率
epoch_acc = epoch_corrects.double() / len(test_dl.dataset)
print('テストデータ{}個での正解率:{:.4f}'.format(len(test_dl.dataset),epoch_acc))
```
# Attentionの可視化で判定根拠を探る
```
# HTMLを作成する関数を実装
def highlight(word, attn):
"Attentionの値が大きいと文字の背景が濃い赤になるhtmlを出力させる関数"
html_color = '#%02X%02X%02X' % (
255, int(255*(1 - attn)), int(255*(1 - attn)))
return '<span style="background-color: {}"> {}</span>'.format(html_color, word)
def mk_html(index, batch, preds, normlized_weights_1, normlized_weights_2, TEXT):
"HTMLデータを作成する"
# indexの結果を抽出
sentence = batch.Text[0][index] # 文章
label = batch.Label[index] # ラベル
pred = preds[index] # 予測
# indexのAttentionを抽出と規格化
attens1 = normlized_weights_1[index, 0, :] # 0番目の<cls>のAttention
attens1 /= attens1.max()
attens2 = normlized_weights_2[index, 0, :] # 0番目の<cls>のAttention
attens2 /= attens2.max()
# ラベルと予測結果を文字に置き換え
if label == 0:
label_str = "Negative"
else:
label_str = "Positive"
if pred == 0:
pred_str = "Negative"
else:
pred_str = "Positive"
# 表示用のHTMLを作成する
html = '正解ラベル:{}<br>推論ラベル:{}<br><br>'.format(label_str, pred_str)
# 1段目のAttention
html += '[TransformerBlockの1段目のAttentionを可視化]<br>'
for word, attn in zip(sentence, attens1):
html += highlight(TEXT.vocab.itos[word], attn)
html += "<br><br>"
# 2段目のAttention
html += '[TransformerBlockの2段目のAttentionを可視化]<br>'
for word, attn in zip(sentence, attens2):
html += highlight(TEXT.vocab.itos[word], attn)
html += "<br><br>"
return html
from IPython.display import HTML
# Transformerで処理
# ミニバッチの用意
batch = next(iter(test_dl))
# GPUが使えるならGPUにデータを送る
inputs = batch.Text[0].to(device) # 文章
labels = batch.Label.to(device) # ラベル
# mask作成
input_pad = 1 # 単語のIDにおいて、'<pad>': 1 なので
input_mask = (inputs != input_pad)
# Transformerに入力
outputs, normlized_weights_1, normlized_weights_2 = net_trained(
inputs, input_mask)
_, preds = torch.max(outputs, 1) # ラベルを予測
index = 3 # 出力させたいデータ
html_output = mk_html(index, batch, preds, normlized_weights_1,
normlized_weights_2, TEXT) # HTML作成
HTML(html_output) # HTML形式で出力
index = 61 # 出力させたいデータ
html_output = mk_html(index, batch, preds, normlized_weights_1,
normlized_weights_2, TEXT) # HTML作成
HTML(html_output) # HTML形式で出力
```
|
github_jupyter
|
# パッケージのimport
import numpy as np
import random
import torch
import torch.nn as nn
import torch.optim as optim
import torchtext
# 乱数のシードを設定
torch.manual_seed(1234)
np.random.seed(1234)
random.seed(1234)
from utils.dataloader import get_IMDb_DataLoaders_and_TEXT
# 読み込み
train_dl, val_dl, test_dl, TEXT = get_IMDb_DataLoaders_and_TEXT(
max_length=256, batch_size=64)
# 辞書オブジェクトにまとめる
dataloaders_dict = {"train": train_dl, "val": val_dl}
from utils.transformer import TransformerClassification
# モデル構築
net = TransformerClassification(
text_embedding_vectors=TEXT.vocab.vectors, d_model=300, max_seq_len=256, output_dim=2)
# ネットワークの初期化を定義
def weights_init(m):
classname = m.__class__.__name__
if classname.find('Linear') != -1:
# Liner層の初期化
nn.init.kaiming_normal_(m.weight)
if m.bias is not None:
nn.init.constant_(m.bias, 0.0)
# 訓練モードに設定
net.train()
# TransformerBlockモジュールを初期化実行
net.net3_1.apply(weights_init)
net.net3_2.apply(weights_init)
print('ネットワーク設定完了')
# 損失関数の設定
criterion = nn.CrossEntropyLoss()
# nn.LogSoftmax()を計算してからnn.NLLLoss(negative log likelihood loss)を計算
# 最適化手法の設定
learning_rate = 2e-5
optimizer = optim.Adam(net.parameters(), lr=learning_rate)
# モデルを学習させる関数を作成
def train_model(net, dataloaders_dict, criterion, optimizer, num_epochs):
# GPUが使えるかを確認
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
print("使用デバイス:", device)
print('-----start-------')
# ネットワークをGPUへ
net.to(device)
# ネットワークがある程度固定であれば、高速化させる
torch.backends.cudnn.benchmark = True
# epochのループ
for epoch in range(num_epochs):
# epochごとの訓練と検証のループ
for phase in ['train', 'val']:
if phase == 'train':
net.train() # モデルを訓練モードに
else:
net.eval() # モデルを検証モードに
epoch_loss = 0.0 # epochの損失和
epoch_corrects = 0 # epochの正解数
# データローダーからミニバッチを取り出すループ
for batch in (dataloaders_dict[phase]):
# batchはTextとLableの辞書オブジェクト
# GPUが使えるならGPUにデータを送る
inputs = batch.Text[0].to(device) # 文章
labels = batch.Label.to(device) # ラベル
# optimizerを初期化
optimizer.zero_grad()
# 順伝搬(forward)計算
with torch.set_grad_enabled(phase == 'train'):
# mask作成
input_pad = 1 # 単語のIDにおいて、'<pad>': 1 なので
input_mask = (inputs != input_pad)
# Transformerに入力
outputs, _, _ = net(inputs, input_mask)
loss = criterion(outputs, labels) # 損失を計算
_, preds = torch.max(outputs, 1) # ラベルを予測
# 訓練時はバックプロパゲーション
if phase == 'train':
loss.backward()
optimizer.step()
# 結果の計算
epoch_loss += loss.item() * inputs.size(0) # lossの合計を更新
# 正解数の合計を更新
epoch_corrects += torch.sum(preds == labels.data)
# epochごとのlossと正解率
epoch_loss = epoch_loss / len(dataloaders_dict[phase].dataset)
epoch_acc = epoch_corrects.double(
) / len(dataloaders_dict[phase].dataset)
print('Epoch {}/{} | {:^5} | Loss: {:.4f} Acc: {:.4f}'.format(epoch+1, num_epochs,
phase, epoch_loss, epoch_acc))
return net
# 学習・検証を実行する 15分ほどかかります
num_epochs = 10
net_trained = train_model(net, dataloaders_dict,
criterion, optimizer, num_epochs=num_epochs)
# device
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
net_trained.eval() # モデルを検証モードに
net_trained.to(device)
epoch_corrects = 0 # epochの正解数
for batch in (test_dl): # testデータのDataLoader
# batchはTextとLableの辞書オブジェクト
# GPUが使えるならGPUにデータを送る
inputs = batch.Text[0].to(device) # 文章
labels = batch.Label.to(device) # ラベル
# 順伝搬(forward)計算
with torch.set_grad_enabled(False):
# mask作成
input_pad = 1 # 単語のIDにおいて、'<pad>': 1 なので
input_mask = (inputs != input_pad)
# Transformerに入力
outputs, _, _ = net_trained(inputs, input_mask)
_, preds = torch.max(outputs, 1) # ラベルを予測
# 結果の計算
# 正解数の合計を更新
epoch_corrects += torch.sum(preds == labels.data)
# 正解率
epoch_acc = epoch_corrects.double() / len(test_dl.dataset)
print('テストデータ{}個での正解率:{:.4f}'.format(len(test_dl.dataset),epoch_acc))
# HTMLを作成する関数を実装
def highlight(word, attn):
"Attentionの値が大きいと文字の背景が濃い赤になるhtmlを出力させる関数"
html_color = '#%02X%02X%02X' % (
255, int(255*(1 - attn)), int(255*(1 - attn)))
return '<span style="background-color: {}"> {}</span>'.format(html_color, word)
def mk_html(index, batch, preds, normlized_weights_1, normlized_weights_2, TEXT):
"HTMLデータを作成する"
# indexの結果を抽出
sentence = batch.Text[0][index] # 文章
label = batch.Label[index] # ラベル
pred = preds[index] # 予測
# indexのAttentionを抽出と規格化
attens1 = normlized_weights_1[index, 0, :] # 0番目の<cls>のAttention
attens1 /= attens1.max()
attens2 = normlized_weights_2[index, 0, :] # 0番目の<cls>のAttention
attens2 /= attens2.max()
# ラベルと予測結果を文字に置き換え
if label == 0:
label_str = "Negative"
else:
label_str = "Positive"
if pred == 0:
pred_str = "Negative"
else:
pred_str = "Positive"
# 表示用のHTMLを作成する
html = '正解ラベル:{}<br>推論ラベル:{}<br><br>'.format(label_str, pred_str)
# 1段目のAttention
html += '[TransformerBlockの1段目のAttentionを可視化]<br>'
for word, attn in zip(sentence, attens1):
html += highlight(TEXT.vocab.itos[word], attn)
html += "<br><br>"
# 2段目のAttention
html += '[TransformerBlockの2段目のAttentionを可視化]<br>'
for word, attn in zip(sentence, attens2):
html += highlight(TEXT.vocab.itos[word], attn)
html += "<br><br>"
return html
from IPython.display import HTML
# Transformerで処理
# ミニバッチの用意
batch = next(iter(test_dl))
# GPUが使えるならGPUにデータを送る
inputs = batch.Text[0].to(device) # 文章
labels = batch.Label.to(device) # ラベル
# mask作成
input_pad = 1 # 単語のIDにおいて、'<pad>': 1 なので
input_mask = (inputs != input_pad)
# Transformerに入力
outputs, normlized_weights_1, normlized_weights_2 = net_trained(
inputs, input_mask)
_, preds = torch.max(outputs, 1) # ラベルを予測
index = 3 # 出力させたいデータ
html_output = mk_html(index, batch, preds, normlized_weights_1,
normlized_weights_2, TEXT) # HTML作成
HTML(html_output) # HTML形式で出力
index = 61 # 出力させたいデータ
html_output = mk_html(index, batch, preds, normlized_weights_1,
normlized_weights_2, TEXT) # HTML作成
HTML(html_output) # HTML形式で出力
| 0.534855 | 0.879458 |
So far we have used function arguments in a basic way; this is the way that is familiar from mathematics:
```
# Load the Numpy package, and rename to "np"
import numpy as np
np.cos(0)
```
Here is another Numpy function, from the `random` sub-package of the Numpy library. We get to the sub-packages with the dot `.` - so to get to the `random` sub-package, we use `np.random`. Then, to get to the functions in this sub-package, we use the dot again, like this:
```
np.random.randint(0, 2)
```
Remember, this is a random integer from 0 up to, but *not including* 2, so it is a random integer that can either be 0 or 1.
Now let us look at the help for the `np.random.randint` function. As usual, we do this by appending `?` to the function name, and pressing Enter in the notebook.
```
# To see the help for np.random.randint, remove the # at the beginning
# of the next line, and execute this cell.
# np.random.randint?
```
We find that the function can accept up to four arguments. We have passed two.
The first sets the argument called `low` to be 0, and the second sets the
argument called `high` to be 2.
To take another example, in this case we are asking for a random number
starting at 1 up to, but not including 11. This gives us a random integer from
1 through 10. `low` is 1 and `high` is 11.
```
# Random integer from 1 through 10.
np.random.randint(1, 11)
```
If we pass three arguments, we also set the `size` argument. This tells the function how many random numbers to return. The following asks for an array of four random integers from 1 through 20:
```
# Four random integers from 1 through 20.
np.random.randint(1, 21, 4)
```
Notice that this is an *array*.
Now look again at the help. Notice that the help gives each argument a *name*
--- `low`, `high`, `size`. We can also use these names when we set these
arguments. For example, the cell below does exactly the same thing as the cell
above.
```
# Four random integers from 1 through 20, using keyword arguments.
np.random.randint(low=1, high=21, size=4)
```
When we call the function using the arguments with their names like this, the named arguments are called *keyword* arguments.
Passing the arguments like this, using keywords, can be very useful, to make it
clearer what each argument means. For example, it's a common pattern to call a function with one or a few keyword arguments, like this:
```
# Four random integers from 1 through 20.
np.random.randint(1, 21, size=4)
```
Writing the call like the cell gives exactly the same result as the cell below,
but the cell above can be easier to follow, because the person reading the code
does not have to guess what the 4 means --- they can see that it means the size
of the output array.
```
# Four random integers from 1 through 20 - but no keyword argument.
np.random.randint(1, 21, size=4)
```
To take another example, we have already seen the function `round`. Inspect
the help for `round` with `round?` and Enter in a notebook cell.
`round` takes up to two arguments. If we pass one argument, it is just the value that `round` will round to the nearest integer:
```
round(3.1415)
```
If we pass two arguments, the first argument is the value we will round, and the second is the number of digits to round to, like this:
```
round(3.1415, 2)
```
As you saw in the help, the second argument has the name `ndigits`, so we can also write:
```
round(3.1415, ndigits=2)
```
As before, this makes the code a little bit easier to read and understand,
because it is immediately clear from the name `ndigits` that the 2 means the
number of digits to round to.
|
github_jupyter
|
# Load the Numpy package, and rename to "np"
import numpy as np
np.cos(0)
np.random.randint(0, 2)
# To see the help for np.random.randint, remove the # at the beginning
# of the next line, and execute this cell.
# np.random.randint?
# Random integer from 1 through 10.
np.random.randint(1, 11)
# Four random integers from 1 through 20.
np.random.randint(1, 21, 4)
# Four random integers from 1 through 20, using keyword arguments.
np.random.randint(low=1, high=21, size=4)
# Four random integers from 1 through 20.
np.random.randint(1, 21, size=4)
# Four random integers from 1 through 20 - but no keyword argument.
np.random.randint(1, 21, size=4)
round(3.1415)
round(3.1415, 2)
round(3.1415, ndigits=2)
| 0.61555 | 0.989347 |
```
%matplotlib inline
import cv2
import numpy as np
import os
import sys
import time
from matplotlib import pyplot as plt
from IPython import display
#load jumper template
jumper_template = cv2.imread('jumper.png')
template_h,template_w = jumper_template.shape[0:2]
screen_x, screen_y = 1080,1920
screen_x_eff, screen_y_eff = 1125,1958
jumper_foot_offset = 20
holdDt = 1.392
tap_x, tap_y = 600,1000
background_xy = (10,980)
header_y,foot_y = 500,1200
img = cv2.imread('scrshot - Copy.png')
background_color = img[background_xy[1],background_xy[0],:]
res = cv2.matchTemplate(img,jumper_template,cv2.TM_SQDIFF_NORMED) #find jumper template matching
min_val, max_val, min_loc, max_loc = cv2.minMaxLoc(res)
top_left = min_loc
bottom_right = (top_left[0] + template_w, top_left[1] + template_h)
jumper_xy = (top_left[0]+int(template_w*0.5), top_left[1]+template_h-jumper_foot_offset) #jumper base location
mirrored_xy = (screen_x_eff-jumper_xy[0],screen_y_eff-jumper_xy[1]) #mirror the jumper base location to get the target base location
img[top_left[1]:bottom_right[1]+1,top_left[0]:bottom_right[0]+1,:]=background_color
img = cv2.circle(img,jumper_xy, 10, (0,0,255), 2) # highlight where the jumper center is found
#img = cv2.GaussianBlur(img,(3,3),0)
canny = cv2.Canny(img, 1,10)
canny[0:header_y,:] = 0
canny[foot_y:,:] = 0
y_top = np.nonzero([max(row) for row in canny])[0][0]
x_top_left = np.min(np.nonzero(canny[y_top]))
x_top_right = np.max(np.nonzero(canny[y_top]))
x_top = int(np.mean(np.nonzero(canny[y_top])))
topcorner_xy = (x_top,y_top);
if (x_top_right-x_top_left>7): #then this is a circle disk
y_bot_startSearch = y_top+80
y_bot = np.nonzero(canny[y_bot_startSearch:,x_top])[0][0]+y_bot_startSearch
cv2.line(img,(x_top,y_top),(x_top,y_bot),255,2)
target_xy = (x_top,int(y_top+y_bot)/2)
else: #this is a box
#search left boundary
target_color = img[y_top+1,x_top];
y_current = y_top;
x_current = x_top;
while True:
y_current += 1;
x_search_range = range(x_current-5,x_current+1);
color_diff = np.linalg.norm(img[y_current,x_search_range,:]-target_color,axis=1)
if np.min(color_diff)<30:
x_current = x_search_range[np.argmin(color_diff)];
target_color = img[y_current,x_current];
else: #found corner
leftcorner_xy = (x_current,y_current-1);
break;
#search right boundary
target_color = img[y_top+1,x_top];
y_current = y_top;
x_current = x_top;
while True:
y_current += 1;
x_search_range = range(x_current+5,x_current,-1);
color_diff = np.linalg.norm(img[y_current,x_search_range,:]-target_color,axis=1)
if np.min(color_diff)<30:
x_current = x_search_range[np.argmin(color_diff)];
target_color = img[y_current,x_current];
else: #found corner
rightcorner_xy = (x_current,y_current-1);
break;
img = cv2.line(img,topcorner_xy,leftcorner_xy, (0,255,0), 2) # highlight where the jumper center is found
img = cv2.line(img,topcorner_xy,rightcorner_xy, (0,255,0), 2) # highlight where the jumper center is found
target_xy = mirrored_xy;
fig=plt.figure(figsize=(18, 16))
plt.subplot(111)
plt.imshow(img,cmap='Greys')
plt.xticks([]), plt.yticks([])
plt.show()
```
|
github_jupyter
|
%matplotlib inline
import cv2
import numpy as np
import os
import sys
import time
from matplotlib import pyplot as plt
from IPython import display
#load jumper template
jumper_template = cv2.imread('jumper.png')
template_h,template_w = jumper_template.shape[0:2]
screen_x, screen_y = 1080,1920
screen_x_eff, screen_y_eff = 1125,1958
jumper_foot_offset = 20
holdDt = 1.392
tap_x, tap_y = 600,1000
background_xy = (10,980)
header_y,foot_y = 500,1200
img = cv2.imread('scrshot - Copy.png')
background_color = img[background_xy[1],background_xy[0],:]
res = cv2.matchTemplate(img,jumper_template,cv2.TM_SQDIFF_NORMED) #find jumper template matching
min_val, max_val, min_loc, max_loc = cv2.minMaxLoc(res)
top_left = min_loc
bottom_right = (top_left[0] + template_w, top_left[1] + template_h)
jumper_xy = (top_left[0]+int(template_w*0.5), top_left[1]+template_h-jumper_foot_offset) #jumper base location
mirrored_xy = (screen_x_eff-jumper_xy[0],screen_y_eff-jumper_xy[1]) #mirror the jumper base location to get the target base location
img[top_left[1]:bottom_right[1]+1,top_left[0]:bottom_right[0]+1,:]=background_color
img = cv2.circle(img,jumper_xy, 10, (0,0,255), 2) # highlight where the jumper center is found
#img = cv2.GaussianBlur(img,(3,3),0)
canny = cv2.Canny(img, 1,10)
canny[0:header_y,:] = 0
canny[foot_y:,:] = 0
y_top = np.nonzero([max(row) for row in canny])[0][0]
x_top_left = np.min(np.nonzero(canny[y_top]))
x_top_right = np.max(np.nonzero(canny[y_top]))
x_top = int(np.mean(np.nonzero(canny[y_top])))
topcorner_xy = (x_top,y_top);
if (x_top_right-x_top_left>7): #then this is a circle disk
y_bot_startSearch = y_top+80
y_bot = np.nonzero(canny[y_bot_startSearch:,x_top])[0][0]+y_bot_startSearch
cv2.line(img,(x_top,y_top),(x_top,y_bot),255,2)
target_xy = (x_top,int(y_top+y_bot)/2)
else: #this is a box
#search left boundary
target_color = img[y_top+1,x_top];
y_current = y_top;
x_current = x_top;
while True:
y_current += 1;
x_search_range = range(x_current-5,x_current+1);
color_diff = np.linalg.norm(img[y_current,x_search_range,:]-target_color,axis=1)
if np.min(color_diff)<30:
x_current = x_search_range[np.argmin(color_diff)];
target_color = img[y_current,x_current];
else: #found corner
leftcorner_xy = (x_current,y_current-1);
break;
#search right boundary
target_color = img[y_top+1,x_top];
y_current = y_top;
x_current = x_top;
while True:
y_current += 1;
x_search_range = range(x_current+5,x_current,-1);
color_diff = np.linalg.norm(img[y_current,x_search_range,:]-target_color,axis=1)
if np.min(color_diff)<30:
x_current = x_search_range[np.argmin(color_diff)];
target_color = img[y_current,x_current];
else: #found corner
rightcorner_xy = (x_current,y_current-1);
break;
img = cv2.line(img,topcorner_xy,leftcorner_xy, (0,255,0), 2) # highlight where the jumper center is found
img = cv2.line(img,topcorner_xy,rightcorner_xy, (0,255,0), 2) # highlight where the jumper center is found
target_xy = mirrored_xy;
fig=plt.figure(figsize=(18, 16))
plt.subplot(111)
plt.imshow(img,cmap='Greys')
plt.xticks([]), plt.yticks([])
plt.show()
| 0.269806 | 0.215433 |
# 更多字符串和特殊方法
- 前面我们已经学了类,在Python中还有一些特殊的方法起着非常重要的作用,这里会介绍一些特殊的方法和运算符的重载,以及使用特殊方法设计类
## str 类
- 一个str对象是不可变的,也就是说,一旦创建了这个字符串,那么它的内容在认为不改变的情况下是不会变的
- s1 = str()#字符串
- s2 = str('welcome to Python')
```
a = ''
print(a)
type(a)
a = 100
b = str(a)
b + 'liyifeng'
```
## 创建两个对象,分别观察两者id
- id为Python内存地址
```
a = 100
b = 1000
print(id(a),id(b))
a = '100'
b = '1000'
print(id(a),id(b))
```
## 处理字符串的函数
- len
- max
- min
- 字符串一切是按照ASCII码值进行比较
```
length = len('liyifeng')
length
c = 'mijianan'
d = 'liyifeng'
max(c,d)
```
## 下角标运算符 []
- 一个字符串是一个字符序列,可以通过索引进行访问
- 观察字符串是否是一个可迭代序列 \__iter__
```
print(c)
c[-5]
print(c)
c[31]#越界
a = 'mijiayuan'
i=0
while i<len(a):
print(a[i])
i += 1
A = '李易峰要开演唱会啦'
print(len(A))
A[0:6]
A[0:9:2]
A[2:5:1]
A[::-1]
```
## 切片 [start: end]
- start 默认值为0
- end 默认值为-1 前闭后开
1.和 for loop 一样,如果步长为正,那么strat<end
2.如果步长为负,那么strat>end
3.如果想要整体翻转(左右),[::-1]
```
def Search(index):
path = 'C:/Users/Administrator/Desktop/Python/Photo/mail.txt'
with open(file=path,mode='r') as f:
for i in range(1000):
line = f.readline()
new_line = line[0:index+1]
if new_line[-1] == '@':
print(new_line[:-1])
Search(6)
```
## 链接运算符 + 和复制运算符 *
- \+ 链接多个字符串,同时''.join()也是
- \* 复制多个字符串
```
a ='mijianan'
b ='liyifeng'
a+b
a * 10
'*'.join('liyifeng')
```
## in 和 not in 运算符
- in :判断某个字符是否在字符串内
- not in :判断某个字符是否不在字符串内
- 返回的是布尔值
```
'J' in 'Joker'
```
## 比较字符串
- ==, !=, >=, <=, >, <
- 依照ASCII码值进行比较
```
'a'<'b'
```
## 测试字符串

- 注意:
> - isalnum() 中是不能包含空格,否则会返回False
```
len(a)
a = '1256'
a.isdigit()
def Panduan():
a=input('请输入密码:')
if len(a)>8 and a.supper() == true and a.lower() == false and a.digit() == true:
print('true')
else:
print('false')
Panduan()
password = input('请输入密码:')
A1=0#代表大写
A2=0#代表小写
A3=0#代表数字
if len(password) <= 8:
print('密码必须大于8位')
for i in password:
if i.isupper():
N1 +=1
```
## 搜索子串

```
def Search(index):
path = 'C:/Users/Administrator/Desktop/Python/Photo/mail.txt'
with open(file=path,mode='r') as f:
for i in range(1000):
line = f.readline()
if line.find('258109664') != -1:
print(line)
Search(9)
```
## 转换字符串

## 删除字符串

```
a = 'MI JIA NAN'
print(a)
a.strip('JIA')
```
## 格式化字符串

## EP:
- 1

- 2
随机参数100个数字,将www.baidu.com/?page=进行拼接
## Python高级使用方法 -- 字符串
- 我们经常使用的方法实际上就是调用Python的运算重载

# Homework
- 1

```
def SSN():
anquan=987-25-123#固定一个社会安全号码
haoma=eval(input('请输入一个社会安全号码>>'))
if anquan == haoma:#if条件,如果输入的号码等于固定的安全号码
print('Valid SSN')#输出Valid SSN
else: #如果输入的号码不等于安全号码
print('Invalid SSN')#输出Invalid SSN
SSN()
SSN()
```
- 2

```
def jiance():
a=input('请输入一个字符串a>>')#输入第一个字符串
b=input('请输入一个字符串b>>')#输入第二个字符串
if a.find(b)==-1:#检测
print('b不是a的子串')#输出
else:
print('b是a的是子串')
jiance()
```
- 3

```
password = input('请输入密码:>>')
N1 = 0 #大写
N2 = 0 #小写
N3 = 0 #数字
for i in password:
if i.isupper():#查找大写字母
N1 +=1#计数
if i.islower():#查找小写字母
N2 +=1#计数
if i.isdigit():#查找数字
N3 +=1#计数
if len(password)>=8 and N1+N2>0 and N3>=2:#如果密码长度不低于8个且包含字母且至少包含两个数字
print('valid password')#输出valid password
elif len(password)<8 or N1+N2<=0 or N3<2:#如果密码长度小于8个或者没有包含字母或者数字少于两个
print('invalid password')#输出invalid password
```
- 4

```
def countletters(*args):
A = eval(input('请输入:'))#输入字符串
xiaoxie = 0
daxie = 0
for i in A:
ASCII = ord(i)#ASCII转化
if 65<=ASCII<=90:#查找大写
daxie += 1
elif 97<=ASCII<=122:#查找小写
xiaoxie += 1
return daxie+xiaoxie
countletters()
```
- 5

```
def getnumber():
haoma =input('请输入号码:')
low = 'a','b','c','d','e','f','g','h','i','j','k','l','m','n','o','p','q','r','s','t','u','v','w','x','y','z'#小写
upper = 'A','B','C','D','E','F','G','H','I','J','K','L','M','N','O','P','Q','R','S','T','U','V','W','X','Y','Z'#大写
for i in haoma:
if i in low:#如果字符串存在于小写
ASCII1 = ord(i)#转化ASCII
print(ASCII1)
elif i in upper:#如果字符串存在于大写
ASCII2 = ord(i)
print(ASCII2)
elif i not in low:#如果字符串不存在于小写
print(i)
elif i not in upper:#如果字符串不存在于大写
print(i)
getnumber()
```
- 6

```
def reverse():
s= input('请输入:>>')
return s[::-1]#从后向前
reverse()
```
- 7

- 8

- 9

```
class Rectangle:
def __init__(self):
print('我初始化啦!')
def bianchang(self):
self.width=eval(input('输入长'))#输入长
self.height=eval(input('输入宽'))#输入宽
def getArea(self):
self.S=self.width*self.height#长方形面积公式:长x宽
print('面积是:',self.S)
def getPerimeter(self):
self.F=2*(self.width+self.height)#长方形周长:(长+宽)x2
print('周长是:',self.F)
mjn=Rectangle()
mjn.bianchang()
mjn.getArea()
mjn.getPerimeter()
```
|
github_jupyter
|
a = ''
print(a)
type(a)
a = 100
b = str(a)
b + 'liyifeng'
a = 100
b = 1000
print(id(a),id(b))
a = '100'
b = '1000'
print(id(a),id(b))
length = len('liyifeng')
length
c = 'mijianan'
d = 'liyifeng'
max(c,d)
print(c)
c[-5]
print(c)
c[31]#越界
a = 'mijiayuan'
i=0
while i<len(a):
print(a[i])
i += 1
A = '李易峰要开演唱会啦'
print(len(A))
A[0:6]
A[0:9:2]
A[2:5:1]
A[::-1]
def Search(index):
path = 'C:/Users/Administrator/Desktop/Python/Photo/mail.txt'
with open(file=path,mode='r') as f:
for i in range(1000):
line = f.readline()
new_line = line[0:index+1]
if new_line[-1] == '@':
print(new_line[:-1])
Search(6)
a ='mijianan'
b ='liyifeng'
a+b
a * 10
'*'.join('liyifeng')
'J' in 'Joker'
'a'<'b'
len(a)
a = '1256'
a.isdigit()
def Panduan():
a=input('请输入密码:')
if len(a)>8 and a.supper() == true and a.lower() == false and a.digit() == true:
print('true')
else:
print('false')
Panduan()
password = input('请输入密码:')
A1=0#代表大写
A2=0#代表小写
A3=0#代表数字
if len(password) <= 8:
print('密码必须大于8位')
for i in password:
if i.isupper():
N1 +=1
def Search(index):
path = 'C:/Users/Administrator/Desktop/Python/Photo/mail.txt'
with open(file=path,mode='r') as f:
for i in range(1000):
line = f.readline()
if line.find('258109664') != -1:
print(line)
Search(9)
a = 'MI JIA NAN'
print(a)
a.strip('JIA')
def SSN():
anquan=987-25-123#固定一个社会安全号码
haoma=eval(input('请输入一个社会安全号码>>'))
if anquan == haoma:#if条件,如果输入的号码等于固定的安全号码
print('Valid SSN')#输出Valid SSN
else: #如果输入的号码不等于安全号码
print('Invalid SSN')#输出Invalid SSN
SSN()
SSN()
def jiance():
a=input('请输入一个字符串a>>')#输入第一个字符串
b=input('请输入一个字符串b>>')#输入第二个字符串
if a.find(b)==-1:#检测
print('b不是a的子串')#输出
else:
print('b是a的是子串')
jiance()
password = input('请输入密码:>>')
N1 = 0 #大写
N2 = 0 #小写
N3 = 0 #数字
for i in password:
if i.isupper():#查找大写字母
N1 +=1#计数
if i.islower():#查找小写字母
N2 +=1#计数
if i.isdigit():#查找数字
N3 +=1#计数
if len(password)>=8 and N1+N2>0 and N3>=2:#如果密码长度不低于8个且包含字母且至少包含两个数字
print('valid password')#输出valid password
elif len(password)<8 or N1+N2<=0 or N3<2:#如果密码长度小于8个或者没有包含字母或者数字少于两个
print('invalid password')#输出invalid password
def countletters(*args):
A = eval(input('请输入:'))#输入字符串
xiaoxie = 0
daxie = 0
for i in A:
ASCII = ord(i)#ASCII转化
if 65<=ASCII<=90:#查找大写
daxie += 1
elif 97<=ASCII<=122:#查找小写
xiaoxie += 1
return daxie+xiaoxie
countletters()
def getnumber():
haoma =input('请输入号码:')
low = 'a','b','c','d','e','f','g','h','i','j','k','l','m','n','o','p','q','r','s','t','u','v','w','x','y','z'#小写
upper = 'A','B','C','D','E','F','G','H','I','J','K','L','M','N','O','P','Q','R','S','T','U','V','W','X','Y','Z'#大写
for i in haoma:
if i in low:#如果字符串存在于小写
ASCII1 = ord(i)#转化ASCII
print(ASCII1)
elif i in upper:#如果字符串存在于大写
ASCII2 = ord(i)
print(ASCII2)
elif i not in low:#如果字符串不存在于小写
print(i)
elif i not in upper:#如果字符串不存在于大写
print(i)
getnumber()
def reverse():
s= input('请输入:>>')
return s[::-1]#从后向前
reverse()
class Rectangle:
def __init__(self):
print('我初始化啦!')
def bianchang(self):
self.width=eval(input('输入长'))#输入长
self.height=eval(input('输入宽'))#输入宽
def getArea(self):
self.S=self.width*self.height#长方形面积公式:长x宽
print('面积是:',self.S)
def getPerimeter(self):
self.F=2*(self.width+self.height)#长方形周长:(长+宽)x2
print('周长是:',self.F)
mjn=Rectangle()
mjn.bianchang()
mjn.getArea()
mjn.getPerimeter()
| 0.10307 | 0.830078 |
# Beyond Predictive Models- *What if* my rejected loan application got accepted?

In the recent years we have seen an acute rise in the adoption of Machine Learning by businesses to make *Data-Driven* decisions. One of the several industries making immense use of it is the Finance Industry wherein machine learning is increasingly being used to solve problems in *Portfolio Management*, *Algorithmic Trading*, *Fraud Detection* etc. Here we focus on one particular application, i.e. to gain a better understanding of **Loan Defaults** so as to minimize them. Using past data, a straightforward application of machine learning would be to design a predictive model which can accurately predict the probability of default. This model would then be used in tandem with the loan officers instincts to decide as to whether the loan application should be approved or not.
But is that score generated by a black box machine learning classifier enough for the loan officer to make his decision? In addition, what if the applicant who has just been denied the loan asks for an explanation. Will the loan officer be able to explain the decision made by the black box? or simply what the applicant should do differently in order to get an approval? In extreme cases, a highly accurate model might also be biased towards a particular gender, race which would ultimately hurt the banks reputation for being unethical.
In this case study we consider a large and rich dataset from the U.S. Small Business Administration (SBA). Our task is to create a ML Classifier which can accurately predict the probability of default and can assist the loan officer in answering:-
***As a representative of the bank, should I grant this loan to a particular small business (Company 'X')? Why or why not? If not, then what should 'X' do differently in order to secure an approval?***
This notebook is divided into 2 parts, in ***Part-I***, we focus on feature engineering based on domain knowledge and design accurate predictive models.
In ***Part-II***, we shift our focus towards explaining individual predictions made by the black box classifier designed in *Part-I* through *diverse* counterfactual explantions which lie in the *vicinity* of the original instance being evaluated. An vital aspect of our explanations would also involve probing the model to dig out the latent biases(if any) embedded deep inside the model.
## Part-I: Designing Predictive Models
### a) Data Cleaning
A raw version of the dataset ('SBANational.csv') associated with this notebook can be downloaded [here](https://www.tandfonline.com/doi/suppl/10.1080/10691898.2018.1434342?scroll=top)
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# Read dataset and store date in the correct format
dataset = pd.read_csv('SBAnational.csv')
print('The shape of the dataset is: ',dataset.shape)
from datetime import datetime
date_list = list(dataset['ApprovalDate'])
date_list_updated = list()
for date_str in date_list:
d = datetime.strptime(date_str,'%d-%b-%y')
date_list_updated.append(d)
dataset['ApprovalDate']=date_list_updated
dataset.sample(10)
```
### b) Feature Engineering
There are a number of variables that consistently emerge as indicators of risk that could explain the variation of loan default rates. Seven variables that are discussed below include *Location (State), Industry, Gross Disbursement, New versus Established Business, Loans Backed by Real Estate, Economic Recession, and SBA’s Guaranteed Portion of Approved Loan*.
Based on domain knowledge and the work done by [M.Li et al. 2017](https://amstat.tandfonline.com/doi/full/10.1080/10691898.2018.1434342), we create the following new features:-
#### 1) Default (*Response Variable*)
Response variable *'Default'* attains the value 1 if *MIS_Status='CHGOFF'*, and 0 otherwise.
```
dataset['Default']=0
dataset.loc[dataset['MIS_Status']=='CHGOFF','Default']=1 #Default if MIS_Status = CHGOFF
#dataset.sample(10)
```
#### 2) Was the Loan active during the Great Recession?
Creating a new feature ***Recession*** which denotes whether the loan was active during the recession period (b/w 1/12/2007 to 31/06/2009). A dummy variable ***“Recession”*** where ***“Recession”*** is 1 if the loans were active in between December 2007 to June 2009, and equals 0 for all other times.
```
dataset['Recession']=0
dataset.loc[dataset['ApprovalDate']>='2007-12-01','Recession']=1
dataset.loc[dataset['ApprovalDate']>='2009-06-30','Recession']=0
#dataset.sample(10)
```
#### 3) Is the loan backed by real estate?
Creating a new feature ***Backed_by_real_estate*** which denotes whether the loan was backed by real estate or not. Loans that are backed by real estate generally have terms greater than 20 years, so our newly created feature takes the value as 1, if the term is greater than 20 years and 0 otherwise.
```
dataset['Backed_by_Real_Estate']=0
dataset.loc[dataset['Term']>=240,'Backed_by_Real_Estate']=1
#dataset.sample(10)
```
#### 4) Which industry category did the small business seeking loan belong to?
Creating a new feature ***Industry_code*** which tells us which category the given company belongs to. By default, the industry code is present in the first two characters of NAICS.
The industry codes are as follows:-

Table taken from: [M.Li et al. 2017](https://amstat.tandfonline.com/doi/full/10.1080/10691898.2018.1434342)
```
# Extract Industry Code from NAICS to create a new Industry Feature
dataset['Industry']=dataset['NAICS'].astype('str').str[0:2]
```
#### 5) How much *portion* of the loan amount is guaranteed by SBA?
Another risk indicator ***Portion*** was engineered which represents the percentage of the loan that is guaranteed by SBA. This is one of the variables generated by calculating the ratio of the
amount of the loan SBA has been guaranteed and the gross amount approved by the bank. A *guaranteed loan* is a type of loan in which a third party agrees to pay if the borrower should default and is used by borrowers with poor credit or little in the way of financial resources; it enables financially unattractive candidates to qualify for a loan and assures that the lender won't lose money.
$Portion = \frac{SBA_{Approved}}{Gross_{Approved}} $
```
dataset['SBA_Appv'] = dataset['SBA_Appv'].str.replace(',', '').str.replace('$', '').str.replace('.', '').astype(int)
dataset['GrAppv'] = dataset['GrAppv'].str.replace(',', '').str.replace('$', '').str.replace('.', '').astype(int)
dataset['Portion']=dataset['SBA_Appv']/dataset['GrAppv']
```
We create a plot to check the distribution of Default v/s Non-Default Cases over the years. There seems to be a high default rate from 2006 till 2008.
```
import seaborn as sns
plt.figure(figsize=(15,8))
dataset_sorted = dataset.sort_values(by='ApprovalDate')
ax = sns.countplot(x='ApprovalFY',hue='Default',data=dataset_sorted)
plt.xticks(rotation=60)
plt.show()
```
#### 6) What is the total amount disbursed?
Gross disbursement (represented as ***“DisbursementGross”*** in the dataset) is another risk indicator identified as a key variable to consider. The rationale behind selecting ***“DisbursementGross”*** is that the larger the loan size, the more likely the underlying business will be established and expanding (i.e., purchasing assets that have some resale value), thereby increasing the likelihood of paying off the loan.
#### 7 & 8.
Finally we claim that the Location of the company represented by ***State*** and whether its a newly established company or not might also serve as potential risk indicator. It may be argued that established businesses already
have a proven track record of success and are requesting a loan to expand on what they already do successfully. Whereas, new businesses sometimes do not anticipate the obstacles they may face and may be unable to successfully. So a feature ***NewExist*** was engineered, which attains the value = 1 if the business is less than 2 years old and 0 if the business is more than 2 years old.
```
dataset_shortlisted = dataset_sorted[['State','Industry','DisbursementGross','NewExist','Backed_by_Real_Estate',
'Recession','Portion', 'Default']]
dataset_shortlisted = dataset_shortlisted.dropna()
```
This is how our final dataset looks like. Using these risk indicators we will now focus on designing Classifiers.
```
dataset_shortlisted['DisbursementGross'] = dataset_shortlisted['DisbursementGross'].str.replace(',', '').str.replace('$', '').str.replace('.', '').astype(int)
dataset_shortlisted['NewExist']=dataset_shortlisted['NewExist'].astype(int)
dataset_shortlisted.sample(10)
# Check counts of New v/s Established Companies
dataset_shortlisted['NewExist'].value_counts()
# NewExist contains some values as 0 which does not carry any significance, so we remove them
dataset_shortlisted=dataset_shortlisted.loc[(dataset_shortlisted.NewExist == 1) | (dataset_shortlisted.NewExist == 2),:]
```
One Hot Encode all the categorical columns present in the dataset
```
dataset_encoded= pd.get_dummies(dataset_shortlisted,columns=['Industry','State'])
dataset_encoded.shape
```
As expected.There is a clear class imbalance present in the dataset!
```
dataset_encoded['Default'].value_counts()
```
## Treating Data for Imbalanced Classification
As seen above, the distribution of *Default* v/s *Not-Default* classes in our dataset is highly skewed. If we go on to train models on this dataset, then even the best performing models would give us misleading results. So it might happen that even though the overall accuracy would be high(falsely) but the model would not be working well in idenifying default cases(Low Specificity and Low Kappa Scores).
There are several methods which can be used to treat imbalanced class problems but since we have access to a good amount of instances for each class, therefore we simply downsample instances from the majority class (**Non-Default**) to match the minority class (**Default**)
```
dataset_bal_0 = dataset_encoded[dataset_encoded['Default'].isin(["0"])].sample(157480,random_state=26)
dataset_bal_1 = dataset_encoded[dataset_encoded['Default'].isin(["1"])]
dataset_balanced = pd.concat([dataset_bal_0,dataset_bal_1])#.sample(26000,random_state=26)
```
# Artificial Neural Network
We tried out several linear and non-linear Machine Learning Classifiers and the best performing linear classifier (Logistic Reg.) gave us a balanced accuracy of 50% whereas the non-linear classifier(XGBoost) gave us a balanced accuracy of 70%.
Keeping the [Universal approximation theorem](http://cognitivemedium.com/magic_paper/assets/Hornik.pdf) in mind we construct a single layer ANN Classifier which will be used as an input to *DiCE* for generating counterfactuals. After experimenting with different choices of hyperparameters, the architechture described below gave us the best validation score on several metrics.
```
# supress deprecation warnings from TF
import tensorflow as tf
from tensorflow import keras
tf.compat.v1.logging.set_verbosity(tf.compat.v1.logging.ERROR)
tf.random.set_seed(26)
# Adjusting for class imbalance by downsampling
dataset_bal_0 = dataset_shortlisted[dataset_shortlisted['Default'].isin(["0"])].sample(157480,random_state=26)
dataset_bal_1 = dataset_shortlisted[dataset_shortlisted['Default'].isin(["1"])]
dataset_balanced_dice = pd.concat([dataset_bal_0,dataset_bal_1])
import dice_ml
sess = tf.compat.v1.InteractiveSession()
d = dice_ml.Data(dataframe=dataset_balanced_dice, continuous_features=['DisbursementGross', 'NewExist',
'Backed_by_Real_Estate', 'Recession', 'Portion'], outcome_name='Default')
train, _ = d.split_data(d.normalize_data(d.one_hot_encoded_data))
X_train = train.loc[:, train.columns != 'Default']
y_train = train.loc[:, train.columns == 'Default']
X_train.head()
from sklearn.metrics import roc_auc_score
METRICS = [
#keras.metrics.TruePositives(name='tp'),
#keras.metrics.FalsePositives(name='fp'),
#keras.metrics.TrueNegatives(name='tn'),
#keras.metrics.FalseNegatives(name='fn'),
keras.metrics.BinaryAccuracy(name='accuracy'),
keras.metrics.Precision(name='precision'),
keras.metrics.Recall(name='recall'),
keras.metrics.AUC(name='auc'),
]
ann_model = keras.Sequential()
ann_model.add(keras.layers.Dense(200, input_shape=(X_train.shape[1],),
kernel_regularizer=keras.regularizers.l2(0.001),
activation=tf.nn.relu))
ann_model.add(keras.layers.Dense(1, activation=tf.nn.sigmoid))
ann_model.compile(loss='binary_crossentropy',
optimizer=tf.keras.optimizers.Adam(0.001),
metrics=METRICS)#=['accuracy',auroc])
ann_model.fit(X_train, y_train, validation_split=0.20, epochs=26, verbose=1)
# the training will take some time for 26 epochs.
# you can wait or set verbose=1 or 0 to see(not see) the progress of training.
```
## Enter *DiCE*
## Part- II: Counterfactual Explanations for individual instances using the trained model
Now that we have trained a black box classifier to identify default cases, we aim to generate explanations which can assist the loan officer in making an informed decision and at the same time provide actionable insights so that even if the applicant is denied a loan at present, they can secure it in the future after encorporating the suggested changes.
Given a trained classifier and the instance needing explanation, [*DiCE*](https://arxiv.org/abs/1905.07697) focuses on generating a set of counterfactual explanations by adressing a ***diversity-proximity*** tradeoff.
In addition to facilitate actionability, *DiCE* is flexible enough to support user-provided inputs based on domain knowledge, such as custom weights for individual features (A higher feature weight means that the feature is harder to change than others) and restrictions on the perturbation of certain features that are difficult to modify in the real world.
```
# provide the trained ML model to DiCE's model object
backend = 'TF'+tf.__version__[0]
m = dice_ml.Model(model=ann_model, backend=backend)
```
## Generate diverse counterfactuals
Based on the data object *d* and the model object *m*, we can now instantiate the DiCE class for generating explanations.
```
# initiate DiCE
exp = dice_ml.Dice(d, m)
```
## Would a loan be granted to a Real Estate business based in New York during COVID'19?
Let us consider a newly established small business based in *New York* that advertizes itself to be the best in helping fellow new yorkers and newcomers in finding the right apartment to rent (despite the abnormally high costs of real estate in New York!). It goes to JP Morgan & Chase and puts forward their case before a loan officer requesting for a loan of USD 125,000. To make their application strong, they have a guranteed payback of USD 112,500 (*Portion=90%*) which will be paid by a third party in case of default. However, at this stage they don't have any backing in the form of real estate which can be used by the bank in case of default. Another important point to note is that the US Job market is going through recession right now due to COVID'19.
We now provide this instance (in the form of a 7-d vector) whose decision will first be evaluated by our ANN Classifier and then explained by *DICE*.
```
# query instance in the form of a dictionary; keys: feature name, values: feature value
query_instance = {'State': 'NY', 'Industry': '53', 'DisbursementGross': 125000, 'NewExist': 1, 'Backed_by_Real_Estate': 0, 'Recession': 1, 'Portion': 0.90}
print("Counterfactual sample: {}".format(query_instance))
```
## Try-1. Naive use of DiCE
We start by generating counterfactuals using default parameter values so as to know what all needs to be tweaked.
The results obtained suggest that we might need to restrict change in some features that are difficult to change in the real world. Also continous features might need a scaling factor to prevent them from attaining abnormally high values in the generated counterfactuals.
```
# generate counterfactuals
dice_exp = exp.generate_counterfactuals(query_instance, total_CFs=4, desired_class="opposite")
# visualize the resutls by highlighting only the changes
dice_exp.visualize_as_dataframe(show_only_changes=True)
```
## Try-2. Slightly better use of DiCE
We notice 2 major problems in the counterfactuals being generated in Try-1:-
1) **Model biased towards certain State's and Industry type's**- Given that *New York* witnesses comparably high default percentages (with *California* being the highest) and that the *Real-Estate* Industry also has the highest default percentage, there seems to be some bias towards these two categories. This is confirmed when the generated counterfactuals suggest to change *State* and *Industry-Type* respectively.
Removing *State* & *Industry-type* from the *features_to_change* list can solve this problem.
2) **Abnormally high values for *GrossDisbursement***- It may be the case that some features are harder to change than others(In our case *DisbursementGross* seem to attain erratic values). DiCE allows input of relative difficulty in changing a feature through specifying feature weights. A higher feature weight means that the feature is harder to change than others. For instance, one way is to use the mean absolute deviation from the median as a measure of relative difficulty of changing a continuous feature. Let's see what their values are by computing them below:
```
# get MAD
mads = d.get_mads(normalized=True)
# create feature weights
feature_weights = {}
for feature in mads:
feature_weights[feature] = round(1/mads[feature], 2)
print(feature_weights)
# Assigning custom weight for Gross Disbursement because counterfactuals being generated are having higher values of this feature.
feature_weights = {'DisbursementGross': 13000, 'Portion': 1} # Setting weight of DisbursementGross to 130.1 still gave bad results, so we went for an even higher value
dice_exp = exp.generate_counterfactuals(query_instance, total_CFs=4, desired_class="opposite",feature_weights=feature_weights ,
features_to_vary=['DisbursementGross','NewExist','Backed_by_Real_Estate','Recession','Portion'])
# visualize the results by highlighting only the changes
dice_exp.visualize_as_dataframe(show_only_changes=True)
```
## Try 3.
Most of the counterfactuals being generated in Try-2 (despite assigning custom weights) still seem to suggest scenarios wherein the *DisbursementGross* is too high. More importantly they recommend that the customer should wait for the COVID'19 recession to get over and get some security in terms of Real Estate Backing to support their application.
If waiting for the recession to get over is not an option, then the only realistic option for them to secure the loan is if they increase the gross disbursement to USD 813,923 and get some real estate backing to support their application.
Now what if the business is certain that it cannot increase the gross disbursement to USD 813,923? because that would imply that they have to a secure a bigger amount as guaranteed payback as well.
In our final try, we explore if its possible to get the loan without any changes in Gross Disbursement. To do so, we remove *DisbursementGross* from the *features_to_vary* list and run DiCE once again.
```
# assigning custom weight for Gross Disbursement because counterfactuals being generated are having higher values of this feature.
#feature_weights = {'DisbursementGross': 13000, 'Portion': 1}
dice_exp = exp.generate_counterfactuals(query_instance, total_CFs=4, desired_class="opposite",feature_weights=feature_weights ,
features_to_vary=['NewExist','Backed_by_Real_Estate','Recession','Portion'])
# visualize the results by highlighting only the changes
dice_exp.visualize_as_dataframe(show_only_changes=True)
```
#### **Voila! As it turns out, quite luckily there is a feasible counterfactual scenario wherein the business would just have to get a real estate backing and they can even get away with a guaranteed loan portion of 80% (given that they were already willing to guarantee 90% of the loan portion)**
So now the loan officer can safely approve their loan application once they have made these suggested changes!
A caveat to note is that currently the counterfactual explanations being generated by *DiCE* suffer from lack of causal knowledge about the features being modified. As it turns out features do not exist in a vaccum, they come from a data generating process which often constrains their independent modification.
In our case, the causal process might take the form of the graphical model mentioned below. This would constrain the way in which risk indicators affecting loan defaults are perturbed. Future versions of DiCE would incorporate a post hoc filtering procedure based on the constraints specified by the causal graphical model given as input by the user.

|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# Read dataset and store date in the correct format
dataset = pd.read_csv('SBAnational.csv')
print('The shape of the dataset is: ',dataset.shape)
from datetime import datetime
date_list = list(dataset['ApprovalDate'])
date_list_updated = list()
for date_str in date_list:
d = datetime.strptime(date_str,'%d-%b-%y')
date_list_updated.append(d)
dataset['ApprovalDate']=date_list_updated
dataset.sample(10)
dataset['Default']=0
dataset.loc[dataset['MIS_Status']=='CHGOFF','Default']=1 #Default if MIS_Status = CHGOFF
#dataset.sample(10)
dataset['Recession']=0
dataset.loc[dataset['ApprovalDate']>='2007-12-01','Recession']=1
dataset.loc[dataset['ApprovalDate']>='2009-06-30','Recession']=0
#dataset.sample(10)
dataset['Backed_by_Real_Estate']=0
dataset.loc[dataset['Term']>=240,'Backed_by_Real_Estate']=1
#dataset.sample(10)
# Extract Industry Code from NAICS to create a new Industry Feature
dataset['Industry']=dataset['NAICS'].astype('str').str[0:2]
dataset['SBA_Appv'] = dataset['SBA_Appv'].str.replace(',', '').str.replace('$', '').str.replace('.', '').astype(int)
dataset['GrAppv'] = dataset['GrAppv'].str.replace(',', '').str.replace('$', '').str.replace('.', '').astype(int)
dataset['Portion']=dataset['SBA_Appv']/dataset['GrAppv']
import seaborn as sns
plt.figure(figsize=(15,8))
dataset_sorted = dataset.sort_values(by='ApprovalDate')
ax = sns.countplot(x='ApprovalFY',hue='Default',data=dataset_sorted)
plt.xticks(rotation=60)
plt.show()
dataset_shortlisted = dataset_sorted[['State','Industry','DisbursementGross','NewExist','Backed_by_Real_Estate',
'Recession','Portion', 'Default']]
dataset_shortlisted = dataset_shortlisted.dropna()
dataset_shortlisted['DisbursementGross'] = dataset_shortlisted['DisbursementGross'].str.replace(',', '').str.replace('$', '').str.replace('.', '').astype(int)
dataset_shortlisted['NewExist']=dataset_shortlisted['NewExist'].astype(int)
dataset_shortlisted.sample(10)
# Check counts of New v/s Established Companies
dataset_shortlisted['NewExist'].value_counts()
# NewExist contains some values as 0 which does not carry any significance, so we remove them
dataset_shortlisted=dataset_shortlisted.loc[(dataset_shortlisted.NewExist == 1) | (dataset_shortlisted.NewExist == 2),:]
dataset_encoded= pd.get_dummies(dataset_shortlisted,columns=['Industry','State'])
dataset_encoded.shape
dataset_encoded['Default'].value_counts()
dataset_bal_0 = dataset_encoded[dataset_encoded['Default'].isin(["0"])].sample(157480,random_state=26)
dataset_bal_1 = dataset_encoded[dataset_encoded['Default'].isin(["1"])]
dataset_balanced = pd.concat([dataset_bal_0,dataset_bal_1])#.sample(26000,random_state=26)
# supress deprecation warnings from TF
import tensorflow as tf
from tensorflow import keras
tf.compat.v1.logging.set_verbosity(tf.compat.v1.logging.ERROR)
tf.random.set_seed(26)
# Adjusting for class imbalance by downsampling
dataset_bal_0 = dataset_shortlisted[dataset_shortlisted['Default'].isin(["0"])].sample(157480,random_state=26)
dataset_bal_1 = dataset_shortlisted[dataset_shortlisted['Default'].isin(["1"])]
dataset_balanced_dice = pd.concat([dataset_bal_0,dataset_bal_1])
import dice_ml
sess = tf.compat.v1.InteractiveSession()
d = dice_ml.Data(dataframe=dataset_balanced_dice, continuous_features=['DisbursementGross', 'NewExist',
'Backed_by_Real_Estate', 'Recession', 'Portion'], outcome_name='Default')
train, _ = d.split_data(d.normalize_data(d.one_hot_encoded_data))
X_train = train.loc[:, train.columns != 'Default']
y_train = train.loc[:, train.columns == 'Default']
X_train.head()
from sklearn.metrics import roc_auc_score
METRICS = [
#keras.metrics.TruePositives(name='tp'),
#keras.metrics.FalsePositives(name='fp'),
#keras.metrics.TrueNegatives(name='tn'),
#keras.metrics.FalseNegatives(name='fn'),
keras.metrics.BinaryAccuracy(name='accuracy'),
keras.metrics.Precision(name='precision'),
keras.metrics.Recall(name='recall'),
keras.metrics.AUC(name='auc'),
]
ann_model = keras.Sequential()
ann_model.add(keras.layers.Dense(200, input_shape=(X_train.shape[1],),
kernel_regularizer=keras.regularizers.l2(0.001),
activation=tf.nn.relu))
ann_model.add(keras.layers.Dense(1, activation=tf.nn.sigmoid))
ann_model.compile(loss='binary_crossentropy',
optimizer=tf.keras.optimizers.Adam(0.001),
metrics=METRICS)#=['accuracy',auroc])
ann_model.fit(X_train, y_train, validation_split=0.20, epochs=26, verbose=1)
# the training will take some time for 26 epochs.
# you can wait or set verbose=1 or 0 to see(not see) the progress of training.
# provide the trained ML model to DiCE's model object
backend = 'TF'+tf.__version__[0]
m = dice_ml.Model(model=ann_model, backend=backend)
# initiate DiCE
exp = dice_ml.Dice(d, m)
# query instance in the form of a dictionary; keys: feature name, values: feature value
query_instance = {'State': 'NY', 'Industry': '53', 'DisbursementGross': 125000, 'NewExist': 1, 'Backed_by_Real_Estate': 0, 'Recession': 1, 'Portion': 0.90}
print("Counterfactual sample: {}".format(query_instance))
# generate counterfactuals
dice_exp = exp.generate_counterfactuals(query_instance, total_CFs=4, desired_class="opposite")
# visualize the resutls by highlighting only the changes
dice_exp.visualize_as_dataframe(show_only_changes=True)
# get MAD
mads = d.get_mads(normalized=True)
# create feature weights
feature_weights = {}
for feature in mads:
feature_weights[feature] = round(1/mads[feature], 2)
print(feature_weights)
# Assigning custom weight for Gross Disbursement because counterfactuals being generated are having higher values of this feature.
feature_weights = {'DisbursementGross': 13000, 'Portion': 1} # Setting weight of DisbursementGross to 130.1 still gave bad results, so we went for an even higher value
dice_exp = exp.generate_counterfactuals(query_instance, total_CFs=4, desired_class="opposite",feature_weights=feature_weights ,
features_to_vary=['DisbursementGross','NewExist','Backed_by_Real_Estate','Recession','Portion'])
# visualize the results by highlighting only the changes
dice_exp.visualize_as_dataframe(show_only_changes=True)
# assigning custom weight for Gross Disbursement because counterfactuals being generated are having higher values of this feature.
#feature_weights = {'DisbursementGross': 13000, 'Portion': 1}
dice_exp = exp.generate_counterfactuals(query_instance, total_CFs=4, desired_class="opposite",feature_weights=feature_weights ,
features_to_vary=['NewExist','Backed_by_Real_Estate','Recession','Portion'])
# visualize the results by highlighting only the changes
dice_exp.visualize_as_dataframe(show_only_changes=True)
| 0.442637 | 0.965641 |
---
## Universidad de Costa Rica
### Escuela de Ingeniería Eléctrica
#### IE0405 - Modelos Probabilísticos de Señales y Sistemas
Segundo semestre del 2020
---
* Estudiante: **Nombre completo**
* Carné: **B12345**
* Grupo: **1**
# `P2` - *La demanda energética de electricidad en Costa Rica*
> Esta actividad reúne las herramientas de programación y la definición, propiedades y funciones de la variable aleatoria continua para analizar los registros de la demanda energética del Sistema Eléctrico Nacional (SEN) durante el año 2019, para determinar un modelo probabilístico de mejor ajuste basado en pruebas de bondad.
---
* Elaboración de nota teórica y demostración: **Jeaustin Sirias Chacón**, como parte de IE0499 - Proyecto Eléctrico: *Estudio y simulación de aplicaciones de la teoría de probabilidad en la ingeniería eléctrica*.
* Revisión: **Fabián Abarca Calderón**
---
## 1. - Introducción: la variable aleatoria se aprende para no olvidarse
El concepto de la variable aleatoria es quizás uno de los conceptos más relevantes en la construcción de modelos probabilísticos, de modo que resulta imprescindible tener una noción clara.
La variable aleatoria podría tener un dominio que no es un conjunto numérico sino un espacio abstracto que contiene "sucesos", u ocurrencias de un experimento aleatorio. A este dominio se le llama *espacio muestral*. ¿Qué ocurre entonces con el ámbito? Aunque no es posible asociar un espacio abstracto directamente con valores numéricos, sí se puede relacionar numéricamente todos sus resultados posibles. Pero, ¿qué ocurre cuando este espacio muestral tiene infinitas probabilidades? Es acá cuando surge la **variable aleatoria continua**. Supóngase el siguiente contraejemplo:
> Es usual observar tres tipos de medios de transporte en la ciudad: automóviles, motocicletas y bicicletas. El día de hoy me propuse contar 150 vehículos de forma aleatoria en la carretera principal de San Pedro de Montes de Oca mientras iba por el pan de la tarde para mi madre. Cuando volví a casa tabulé mis resultados y los representé de la siguiente manera según el tipo de vehículo:
| Dominio $S$ | Variable aleatoria $X$ |
|-----------------------|------------------------|
| <img src="https://i.pinimg.com/564x/2c/c6/2e/2cc62efc5998bc3bfdec089acf1e12c4.jpg" width="30"></img> | $x_1$ |
| <img src="https://cdn.onlinewebfonts.com/svg/img_323762.png" width="20"></img> | $x_2$ |
| <img src="https://cdn.onlinewebfonts.com/svg/img_22991.png" width="20"></img> | $x_3$ |
> Luego de contabilizar la frecuencia de automóviles, bicicletas y motocicletas observadas durante el experimento me enteré de que el espacio muestral estaba limitado a solamente tres posibilidades, y aunque si mañana repitiese el ejercicio y la frecuencia de los automóviles vistos posiblemente variará, solo tendré la oportunidad de contar autos, motos y bicis...
El caso anterior representa un variable aleatoria **discreta**, puesto que puede obtenerse un número contable de ocurrencias $x_1$, $x_2$ y $x_3$, sin embargo, ¿qué ocurrirá si ahora desea repertirse el experimento anterior pero para conocer el peso en kilogramos de cada vehículo observado en la carretera?, ¿será posible que al menos dos vehículos compartan exactamente el mismo peso?, ¿estará el espacio muestral $S$ limitado a un número de magnitudes de peso en kg? Efectivamente, no. Si de forma ideal se colocara una váscula en cada uno de los vehículos podría apreciarse que existirán valores parecidos, pero no iguales; por ejemplo, dos autos que pesen 1340,5683 kg y 1340,7324 kg, respectivamente, entonces existe una cantidad no mensurable de probabiblidades en el espacio muestral $S$. En general se dirá que la probabilidad de encontrar un valor *puntual* (de precisión infinita) en una variable aleatoria continua es cero. Por ejemplo:
$$\displaystyle \frac{1500.\overline{0} \text{ kg}}{\text{Infinitos pesos entre 10 kg y 4000 kg}} \approx 0$$
---
## 2. - Pruebas de bondad de ajuste de los modelos
Hasta el momento en el curso hemos encontrado los parámetros de mejor ajuste entre un conjunto de datos (una "muestra") y un modelo probabilístico particular, quizá elegido *arbitrariamente* o por un desarrollado sentido de la intuición, del tipo: "hmm, esa distribución me parece exponencial". Sin embargo, está claro que algunos modelos hacen una mejor descripción de los datos que otros, y no siempre se puede encontrar "a puro ojo".
¿Cómo se evalúa entonces esta "bondad de ajuste" (*goodness of fit*) de cada modelo, de forma tal que se puedan comparar con una sola métrica todas las distribuciones analizadas y tomar una decisión? Existe alrededor de una docena de pruebas, pero aquí usaremos dos de las más comunes:
* [La prueba de Kolmogorov–Smirnov](https://es.wikipedia.org/wiki/Prueba_de_Kolmogorov-Smirnov), o *KS test*.
* [La prueba chi-cuadrado de Pearson](https://en.wikipedia.org/wiki/Chi-squared_test), o $\chi^2$.
La explicación de cada una de estas pruebas se sale del objetivo de esta etapa del curso, por eso se mencionan aquí nada más.
#### Algunas distribuciones a utilizar
> ¿Qué puede esperarse de la demanda energética nacional si fuese una variable aleatoria?, ¿sería esta última, discreta o continua?, ¿podría aproximarse su distribución anual, mensual, diaria u horaria hacia un modelo de densidad probabilístico?
Al igual que en el ejemplo del peso en los vehículos, el espacio muestral de la demanda de energía es infinito para cualquier intervalo de valores $[a, b]$. Podría ocurrir que a las **00:00** de hoy la demanda registrada sea **909.8934 MW** mientras que mañana a la misma hora será **909.2232 MW** y al siguiente, **909.873666641 MW**; es decir, el experimento de medir la demanda en ese período tiene un sinnúmero de posibilidades, de modo que es una variable aleatoria *continua*.
Las funciones continuas de de probabilidad son muy variadas, las hay de todas formas. Algunas de ellas describen sistemas habituales y serán aquí utilizadas:
* Distribución normal
* Distribución de Rayleigh
* Distribución de Burr tipo XII
* Distribución gamma
* Distribución beta
* Distribución alfa
**Nota**: Algunas librerías de programación para encontrar el mejor ajuste hacen pruebas con *una gran cantidad* de distribuciones disponibles (más de 80), sin hacer ninguna presuposición. Nosotros, sin embargo, usaremos estas nada más, asumiendo que tienen "formas similares".
#### ¿Qué hace a una distribución mejor que otra al ajustar una población?
En términos relativos, depende en gran medida del sistema o proceso que se estudia. Como se expuso anteriormente hay una enorme familia de funciones de probabilidad. Habrá una de ellas que describa un conjunto de datos mejor que las demás. A esto se le denomina **bondad de ajuste** y se basa en evaluar discrepancias, residuos y/o frecuencias de dos o más distribuciones, generalmente con la intención de conocer si las muestras provienen de una misma distribución, si las muestras observadas siguien una distribución en particular o bien para evaluar qué tanto se ajusta un modelo probabilístico construido a partir de datos observados.
En su mayoría se parte de una hipótesis nula $H_{O}$ que supone la siguiente premisa:
> Los datos observados y los predichos son iguales hasta que no se pruebe lo contrario.
Aparte de $\chi^2$ y *KS test* mencionados antes, se hace uso de índices de error como la [raíz del error cuadrático medio](https://es.wikipedia.org/wiki/Ra%C3%ADz_del_error_cuadr%C3%A1tico_medio) (RMSE) o el [error cuadrático medio](https://es.wikipedia.org/wiki/Error_cuadr%C3%A1tico_medio#:~:text=En%20el%20an%C3%A1lisis%20de%20regresi%C3%B3n,n%C3%BAmero%20de%20grados%20de%20libertad.) (SSE) para contrastar las muestras de una población.
---
## 3. - Contexto: el *Sistema Eléctrico Nacional* (SEN) de Costa Rica
El [Centro Nacional de Control de Energía](https://apps.grupoice.com/CenceWeb/CenceMain.jsf) (CENCE) es el ente estatal encargado de registrar, manipular y analizar el sistema eléctrico nacional de Costa Rica en los ámbitos de generación, distribución y demanda de la energía eléctrica en el país. La matriz energética nacional está administrada por siete empresas distribuidoras, a saber:
* **Instituto Costarricense de Electricidad** (ICE)
* **Compañía Nacional de Fuerza y Luz** (CNFL)
* **Junta Administrativa del Servicio Eléctrico Municipal de Cartago** (JASEC)
* **Coopeguanacaste R.L.**
* **Coopelesca R.L.**
* **Coopesantos R.L.**
* **Empresa de Servicios Públicos de Heredia** (ESPH)
<img align='center' src='https://i.imgur.com/pPc9mIA.png' width ="650"/>
El servicio y el costo de las tarifas eléctricas ofrecidas por cada una de las empresas depende de la ubicación, el sector que lo solicita (residencial, industrial, comercial...) y las disposiciones de la [Autoridad Reguladora de los Servicios Públicos](https://aresep.go.cr/electricidad) (ARESEP). A nivel nacional se hallan establecidos tres períodos por concepto de actividad de consumo energético durante el día:
* **Período de punta**: Entre las **10:01** y las **12:30** horas, y entre las **17:31** y las **20:00** horas para un total de cinco horas diarias.
* **Período de valle**: Se comprende entre las **06:01** y las **10:00** horas, y entre las **12:31** y las **17:30** para total de nueve horas diarias.
* **Período nocturno**: Desde las **20:01** hasta las **06:00** del próximo día, para un total de 10 horas.
La demanda energética a nivel nacional es registrada en intervalos de 15 minutos durante todo el año. Existen temporadas o situaciones cuando la demanda es particularmente mayor por temas sociales y/o económicos. Por ejemplo, las fiestas de fin de año se caracterizan por celebrar la **Navidad** y el **Año Nuevo**: las casas, las vías públicas y los parques se iluminan con luces festivas al menos durante todo el mes de diciembre y poco antes. Asimismo, aumenta el uso de los hornos eléctricos en las familias para elaborar recetas propias de la fecha.
Otro caso es la actual [emergencia nacional por el COVID-19](https://www.facebook.com/watch/?v=862104867616321), la cual ha repercutido considerablemente en todas las actividades habituales.
### 3.1. - Aplicación: construyendo un modelo probabilístico basado en demanda energética
Para la siguiente actividad, existe una base de datos que contiene la demanda energética nacional del año 2019 por hora, como se muestra a continuación:
<img align='center' src='https://i.imgur.com/2PwdGF0.png' width ="700"/>
Dicha "población" es una variable aleatoria continua. Es deseable hallar un modelo probabilístico que se ajuste lo mejor posible a lo observado de acuerdo con las pruebas de bondad de ajuste mencionadas anteriormente. Por ejemplo, se quiere analizar el comportamiendo de la demanda a las **18:00** horas durante todos los días en estudio. El módulo [`stats`](https://docs.scipy.org/doc/scipy/reference/stats.html) de SciPy es útil para ejemplificar la presente aplicación de forma programada. La estrategia a implementar se elaborará bajo los siguientes pasos:
1. Acondicionar la base de datos para obtener las muestras a la hora de interés.
2. Ajustar varios modelos probabilísticos a evaluar sobre la muestra observada.
3. Determinar el mejor modelo probabilístico mediante las pruebas de bondad de ajuste **chi-cuadrado** ($\chi^2$), **Kolmogorov-Smirnov** (*KS test*) y el índice de error **RMSE**.
4. Determinar los cuatro primeros momentos centrales para el mejor modelo.
5. Visualizar los resultados obtenidos.
Para lograr los puntos anteriores se emplean entonces las siguientes librerías:
```python
import numpy as np # para manipular datos
import matplotlib.pyplot as plt # para visualizar resultados
import pandas as pd # para acondicionar la base de datos
from scipy import stats # la música de la fiesta
from datetime import datetime # funciones de conversión de fechas y horas
```
### 3.2. - Lectura y acondicionamiento de los datos
Es una buena práctica de programación desarrollar código empleando funciones, puesto que permite la generalización del proceso. Por ejemplo, para este caso es útil elaborar una función que no solamente acondicione la demanda a las **18:00**, sino a cualquier hora. Dicho de este modo entonces la hora debe ser un parámetro de ajuste variable en los argumentos.
#### 3.2.1. - Sobre el formato JSON
[JSON](https://es.wikipedia.org/wiki/JSON) (extensión `.json`) es un formato de texto de alto nivel, muy utilizado en el intercambio de información por su alta legibilidad y fácil manejo de la sintaxis. La librería de manipulación de datos, [Pandas](https://pandas.pydata.org/pandas-docs/stable/index.html), ofrece un método especialmente adecuado para leer y manipular dicho formato. Para esta ocasión la base de datos importada se encuentra escrita en JSON para familiarizar su uso.
Los datos a analizar lucen de la siguiente manera:
```json
{
"data":[
{
"fechaHora": "2019-01-01 00:00:00",
"MW": 958.05,
"MW_P": 1
},
{
"fechaHora": "2019-01-01 01:00:00",
"MW": 917.04,
"MW_P": 2
},
{
"fechaHora": "2019-01-01 02:00:00",
"MW": 856.19,
"MW_P": 3
},
{
"fechaHora": "2019-01-01 03:00:00",
"MW": 803.04,
"MW_P": 4
},
(...miles de datos más...)
]
}
```
Y pueden interpretarse como una tabla donde `"fechaHora"`, `"MW"` y `"MW_P"` son los encabezados de cada columna, es decir:
| `"fechaHora"` | `"MW"` | `"MW_P"` |
|-----------------------|--------|----------|
| "2019-01-01 00:00:00" | 958.05 | 1 |
| "2019-01-01 01:00:00" | 917.04 | 2 |
| "2019-01-01 02:00:00" | 856.19 | 3 |
| "2019-01-01 03:00:00" | 803.04 | 4 |
| ... | ... | ... |
##### Formato ISO de la fecha y hora
El formato `'YYYY-MM-DD hh:mm:ss'` es conocido como **formato ISO**, según el estándar ISO 8601.
### 3.3. - Funciones desarrolladas
Para la resolución de este proyecto se presentan dos funciones y una función auxiliar:
* `extraer_datos(archivo_json, hora)`: Importa la base de datos completa y devuelve los datos de potencia a la hora indicada en un *array* de valores.
* `evaluar_modelos(datos, distribuciones, divisiones, hora)`: Evalúa la bondad de ajuste de los datos con los modelos utilizados y grafica cada modelo.
* `horas_asignadas(digitos)`: Elige una hora A en periodo punta y una hora B de los otros periodos, con los dígitos del carné como *seed*.
```
import pandas as pd
import numpy as np
from datetime import datetime
def extraer_datos(archivo_json, hora):
'''Importa la base de datos completa y devuelve los
datos de potencia a la hora indicada en un
array de valores.
'''
# Cargar el "DataFrame"
df = pd.read_json(archivo_json)
# Convertir en un array de NumPy
datos = np.array(df)
# Crear vector con los valores demanda en una hora
demanda = []
# Extraer la demanda en la hora seleccionada
for i in range(len(datos)):
instante = datetime.fromisoformat(datos[i][0]['fechaHora'])
if instante.hour == hora:
demanda.append(datos[i][0]['MW'])
return demanda
```
Observar que, en la función anterior, la variable `datos` tiene la siguiente forma:
```python
[[{'fechaHora': '2019-01-01 00:00:00', 'MW': 958.05, 'MW_P': 1}]
[{'fechaHora': '2019-01-01 01:00:00', 'MW': 917.04, 'MW_P': 2}]
[{'fechaHora': '2019-01-01 02:00:00', 'MW': 856.19, 'MW_P': 3}]
...
[{'fechaHora': '2019-09-12 22:00:00', 'MW': 1184.73, 'MW_P': 1174.2}]
[{'fechaHora': '2019-09-12 23:00:00', 'MW': 1044.81, 'MW_P': 1064.9}]
[{'fechaHora': '2019-09-13 00:00:00', 'MW': 975.18, 'MW_P': 995}]]
```
que muestra un conjunto de diccionarios. Por tanto, la instrucción
```python
datos[i][0]['fechaHora']
```
accesa el `i`-ésimo elemento, `[0]` representa el diccionario mismo (el único elemento que hay) y `['fechaHora']` devuelve el *valor* asociado con la *llave* `'fechaHora'`. Por ejemplo:
```python
>>> datos[1][0]['fechaHora']
'2019-01-01 01:00:00'
>>> datos[2][0]['MW']
856.19
```
### 3.4. - Parámetros de mejor ajuste
La siguiente función determina cuáles son los parámetros de mejor ajuste para ciertas distribuciones elegidas, utilizando `scipy.stats`.
```
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
from datetime import datetime
def evaluar_modelos(datos, distribuciones, divisiones, hora):
'''Evalúa la bondad de ajuste de los datos con los
modelos utilizados y grafica cada modelo.
'''
# Distribución de frecuencia relativa
ocurrencias_exp, limites = np.histogram(datos, bins=divisiones)
# Eliminar los ceros de la frecuencia relativa
for i in range(divisiones):
if ocurrencias_exp[i] == 0:
ocurrencias_exp[i] = 1
# Encontrar el valor central de las divisiones
bins_centrados = (limites + np.roll(limites, -1))[:-1] / 2.0
escala = len(datos) * (max(datos) - min(datos)) / len(bins_centrados)
# Crear subfiguras para visualización (1 x 2)
fig, ax = plt.subplots(1, 2, figsize=(15, 5))
# Información de la figura 1
ax[0].set_title('Ajuste de las distribuciones')
ax[0].set_ylabel('Frecuencia')
ax[0].set_xlabel('Potencia [MW]')
# Información de la figura 3
ax[1].set_title('Distribución con mejor criterio de bondad de ajuste')
ax[1].set_ylabel('Frecuencia')
ax[1].set_xlabel('Potencia [MW]')
# Visualizar datos
ax[0].hist(datos, bins=divisiones, histtype='bar', color='palevioletred', rwidth=0.8)
ax[1].hist(datos, bins=divisiones, histtype='bar', color='b')
# Condiciones iniciales de las pruebas de ajuste
rmse_min = np.inf # el mayor índice de error
p_max = 0 # el mejor p en chisqr test (0 es el "peor")
kspmax = 0 # el mejor p en KStest (0 es el "peor")
np.seterr(all='ignore') # ignorar errores con números de punto flotante
# Evaluar las distribuciones, extraer parámetros y visualizar
for distribucion in distribuciones:
# Extraer de scipy.stats la distribución ("get attribute")
dist = getattr(stats, distribucion)
# Parámetros de mejor ajuste para la distribución
param = dist.fit(datos)
# Evaluar la PDF en el valor central de las divisiones
pdf = dist.pdf(bins_centrados, *param)
# Convertir frecuencia relativa en ocurrencias (número absoluto)
ocurrencias_teo = [int(round(i)) for i in escala*pdf]
# Soporte para la gráfica
d = np.arange(min(datos)*0.96, max(datos)*1.04, 1)
# Graficar en ax[1]
pdf_plot = dist.pdf(d, *param)
ax[0].plot(d, escala*pdf_plot, lw=3.5, label='{}'.format(distribucion))
# Prueba de bondad de ajuste por chi-cuadrado
coef_chi, p = stats.chisquare(f_obs=ocurrencias_teo, f_exp=ocurrencias_exp)
if p > p_max: # si el p actual es mayor
p_max = p # designarlo como el máximo
dist_chi = distribucion # elegir la distribución como la de mejor ajuste
mod_chi = dist, param, pdf
# Bondad de ajuste por RMSE (Root-Mean-Square Error)
diferencia = (ocurrencias_teo - ocurrencias_exp)**2
rmse = np.sqrt(np.mean(diferencia))
if rmse < rmse_min:
rmse_min = rmse
dist_rmse = distribucion
mod_rmse = dist, param, pdf
# Bondad de ajuste por Kolgomorov - Smirnov
D, ksp = stats.kstest(datos, distribucion, args=param)
if ksp > kspmax:
kspmax = ksp
dist_ks = distribucion
# Decidir el mejor modelo
if dist_chi == dist_rmse or dist_chi == dist_ks:
params = mod_chi[1]
mejor_ajuste = dist_chi
ax[1].hist(datos, bins=divisiones, color='cornflowerblue', label='Distribución observada')
ax[1].bar(bins_centrados, mod_chi[2] * escala, width=6, color='r', label='Mejor ajuste: {}'.format(dist_chi))
m, v, s, k = mod_chi[0].stats(*params, moments='mvsk')
elif dist_rmse == dist_ks:
params = mod_rmse[1]
mejor_ajuste = dist_rmse
ax[1].hist(datos, bins = divisiones, color='cornflowerblue', label='Distribución observada')
ax[1].bar(bins_centrados, mod_rmse[2] * escala, width=6, color='r', label='Mejor ajuste: {}'.format(dist_rmse))
m, v, s, k = mod_rmse[0].stats(*params, moments='mvsk')
# Imprimir resumen y resultados
print('-------\nResumen\n-------')
print('Cantidad de muestras:', len(datos), 'días a las', hora, 'horas')
print('Máximo:', max(datos), 'MW')
print('Mínimo:', min(datos), 'MW')
print('Tipo: Demanda energética horaria')
print('------\nAjuste\n------')
print('Menor error RMS es:', dist_rmse)
print('Mejor bondad de ajuste en la prueba de chi-cuadrado es:', dist_chi)
print('Mejor bondad de ajuste en la prueba de Kolmogorov–Smirnov es:', dist_ks)
print('Distribución elegida:', mejor_ajuste)
print('--------\nMomentos\n--------')
print('Media:', m, '\nVarianza:', v, '\nDesviación estándar:', np.sqrt(v), '\nCoeficiente simetría:', s, '\nKurtosis:', k)
print('--------\nGráficas\n--------')
ax[0].legend()
ax[1].legend()
plt.show()
```
### 3.5. - Evaluando los datos
Llegado a este punto, ahora solo se requiere llamar las dos funciones desarrolladas y elegir **la base de datos**, **las distribuciones** (de la galería de distribuciones continuas disponibles en el módulo [`stats`](https://docs.scipy.org/doc/scipy/reference/stats.html#continuous-distributions)) y **la hora** a la que desea evaluarse.
```
# Hora, en el intervalo [0, 23] (tipo int)
hora = 18
# Distribuciones a evaluar
distribuciones = ['norm', 'rayleigh', 'burr12', 'alpha', 'gamma', 'beta']
# Llamar a las funciones
demandas = extraer_datos('demanda_2019.json', hora)
evaluar_modelos(demandas, distribuciones, 25, hora)
```
---
## 4. - Asignaciones del proyecto
### 4.1. - Comparación de consumo de potencia para distintas horas del día
La curva de consumo de potencia diaria del SEN muestra cambios importantes durante el día, así que es esperable encontrar comportamientos distintos en la distribución de probabilidad para cada hora.
* (40%) Encuentre **la distribución de mejor ajuste y sus parámetros** para las dos horas asignadas.
Puede encontrar las horas asignadas con la función `horas_asignadas(digitos)`, donde `digitos` son los dígitos numéricos de su carné (por ejemplo: para B12345 `digitos = 12345`)
```
import random
def horas_asignadas(digitos):
'''Elige una hora A en periodo punta
y una hora B de los otros periodos,
con los dígitos del carné como "seed"
'''
random.seed(digitos)
punta = [11, 12, 18, 19, 20]
valle = [7, 8, 9, 10, 13, 14, 15, 16, 17]
nocturno = [21, 22, 23, 0, 1, 2, 3, 4, 5, 6]
otro = valle + nocturno
HA = punta[random.randrange(0, len(punta))]
HB = otro[random.randrange(0, len(otro))]
horas = 'Hora A = {}, hora B = {}'.format(HA, HB)
return horas
horas_asignadas(12345)
# 4.1. - Comparación de consumo de potencia
```
### 4.2. - Obtención de los momentos de los modelos de distribución por hora
Resuma estos hallazgos en una tabla con los cuatro momentos más importantes (y la desviación estándar) para cada modelo de cada hora analizada.
* (30%) Complete la tabla de resultados de los momentos, haciendo los cálculos respectivos con Python o con sus modelos (mostrando las ecuaciones respectivas).
```
# 4.2. - Obtención de los momentos de los modelos
```
#### Expresiones analíticas parametrizadas de los momentos
<!-- Ejemplo para la distribución beta -->
| Momento | Expresión analítica parametrizada de la distribución |
|-------------|------------------------------------------------------|
| Media | $\displaystyle E[X] = \frac{\alpha}{\alpha+\beta}\!$ |
| Varianza | $\displaystyle \operatorname{var}[X] = \frac{\alpha\beta}{(\alpha+\beta)^2 (\alpha+\beta+1)}\!$ |
| Inclinación | $\displaystyle S_X = \frac{2\,(\beta-\alpha)\sqrt{\alpha+\beta+1}}{(\alpha+\beta+2)\sqrt{\alpha\beta}}$ |
| Kurtosis | $\displaystyle \kappa_X = \frac{6[(\alpha - \beta)^2 (\alpha +\beta + 1) - \alpha \beta (\alpha + \beta + 2)]}{\alpha \beta (\alpha + \beta + 2) (\alpha + \beta + 3)}$ |
#### Valores obtenidos para el modelo y los datos de la muestra
Análisis para las horas A = XX:XX y B = YY:YY.
| Momento | Fuente | A = 7:00 am | B = 7:00 pm |
|-----------------|----------|-------------|-------------|
| **Media** | *Modelo* | mmm | mmm |
| **Media** | *Datos* | mmm | mmm |
| **Varianza** | *Modelo* | vvv | vvv |
| **Varianza** | *Datos* | vvv | vvv |
| **Desviación** | *Modelo* | sdsd | sdsd |
| **Desviación** | *Datos* | sdsd | sdsd |
| **Inclinación** | *Modelo* | sss | sss |
| **Inclinación** | *Datos* | sss | sss |
| **Kurtosis** | *Modelo* | kkk | kkk |
| **Kurtosis** | *Datos* | kkk | kkk |
**Nota**: utilizar cuatro decimales.
### 4.3. - Análisis de los datos obtenidos
De la comparación de las horas estudiadas,
* (30%) Explique las posibles razones de las diferencias observadas, desde una interpretación estadística.
<!-- Inicie aquí la explicación. Puede incluir imágenes, tablas, fragmentos de código o lo que considere necesario. -->
### Análisis
<!-- Utilice las mejores prácticas de edición de formato de Markdown: https://www.markdownguide.org/basic-syntax/ -->
Aquí va el análisis y las ecuaciones y las tablas y las figuras...
$$
x_{1,2} = \frac{-b \pm \sqrt{b^2 - 4ac}}{2a}
$$
Y luego también.
#### Análisis de la media
#### Análisis de la varianza y desviación estándar
#### Análisis de la inclinación
#### Análisis de la kurtosis
---
### Universidad de Costa Rica
#### Facultad de Ingeniería
##### Escuela de Ingeniería Eléctrica
---
|
github_jupyter
|
import numpy as np # para manipular datos
import matplotlib.pyplot as plt # para visualizar resultados
import pandas as pd # para acondicionar la base de datos
from scipy import stats # la música de la fiesta
from datetime import datetime # funciones de conversión de fechas y horas
{
"data":[
{
"fechaHora": "2019-01-01 00:00:00",
"MW": 958.05,
"MW_P": 1
},
{
"fechaHora": "2019-01-01 01:00:00",
"MW": 917.04,
"MW_P": 2
},
{
"fechaHora": "2019-01-01 02:00:00",
"MW": 856.19,
"MW_P": 3
},
{
"fechaHora": "2019-01-01 03:00:00",
"MW": 803.04,
"MW_P": 4
},
(...miles de datos más...)
]
}
import pandas as pd
import numpy as np
from datetime import datetime
def extraer_datos(archivo_json, hora):
'''Importa la base de datos completa y devuelve los
datos de potencia a la hora indicada en un
array de valores.
'''
# Cargar el "DataFrame"
df = pd.read_json(archivo_json)
# Convertir en un array de NumPy
datos = np.array(df)
# Crear vector con los valores demanda en una hora
demanda = []
# Extraer la demanda en la hora seleccionada
for i in range(len(datos)):
instante = datetime.fromisoformat(datos[i][0]['fechaHora'])
if instante.hour == hora:
demanda.append(datos[i][0]['MW'])
return demanda
[[{'fechaHora': '2019-01-01 00:00:00', 'MW': 958.05, 'MW_P': 1}]
[{'fechaHora': '2019-01-01 01:00:00', 'MW': 917.04, 'MW_P': 2}]
[{'fechaHora': '2019-01-01 02:00:00', 'MW': 856.19, 'MW_P': 3}]
...
[{'fechaHora': '2019-09-12 22:00:00', 'MW': 1184.73, 'MW_P': 1174.2}]
[{'fechaHora': '2019-09-12 23:00:00', 'MW': 1044.81, 'MW_P': 1064.9}]
[{'fechaHora': '2019-09-13 00:00:00', 'MW': 975.18, 'MW_P': 995}]]
datos[i][0]['fechaHora']
>>> datos[1][0]['fechaHora']
'2019-01-01 01:00:00'
>>> datos[2][0]['MW']
856.19
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
from datetime import datetime
def evaluar_modelos(datos, distribuciones, divisiones, hora):
'''Evalúa la bondad de ajuste de los datos con los
modelos utilizados y grafica cada modelo.
'''
# Distribución de frecuencia relativa
ocurrencias_exp, limites = np.histogram(datos, bins=divisiones)
# Eliminar los ceros de la frecuencia relativa
for i in range(divisiones):
if ocurrencias_exp[i] == 0:
ocurrencias_exp[i] = 1
# Encontrar el valor central de las divisiones
bins_centrados = (limites + np.roll(limites, -1))[:-1] / 2.0
escala = len(datos) * (max(datos) - min(datos)) / len(bins_centrados)
# Crear subfiguras para visualización (1 x 2)
fig, ax = plt.subplots(1, 2, figsize=(15, 5))
# Información de la figura 1
ax[0].set_title('Ajuste de las distribuciones')
ax[0].set_ylabel('Frecuencia')
ax[0].set_xlabel('Potencia [MW]')
# Información de la figura 3
ax[1].set_title('Distribución con mejor criterio de bondad de ajuste')
ax[1].set_ylabel('Frecuencia')
ax[1].set_xlabel('Potencia [MW]')
# Visualizar datos
ax[0].hist(datos, bins=divisiones, histtype='bar', color='palevioletred', rwidth=0.8)
ax[1].hist(datos, bins=divisiones, histtype='bar', color='b')
# Condiciones iniciales de las pruebas de ajuste
rmse_min = np.inf # el mayor índice de error
p_max = 0 # el mejor p en chisqr test (0 es el "peor")
kspmax = 0 # el mejor p en KStest (0 es el "peor")
np.seterr(all='ignore') # ignorar errores con números de punto flotante
# Evaluar las distribuciones, extraer parámetros y visualizar
for distribucion in distribuciones:
# Extraer de scipy.stats la distribución ("get attribute")
dist = getattr(stats, distribucion)
# Parámetros de mejor ajuste para la distribución
param = dist.fit(datos)
# Evaluar la PDF en el valor central de las divisiones
pdf = dist.pdf(bins_centrados, *param)
# Convertir frecuencia relativa en ocurrencias (número absoluto)
ocurrencias_teo = [int(round(i)) for i in escala*pdf]
# Soporte para la gráfica
d = np.arange(min(datos)*0.96, max(datos)*1.04, 1)
# Graficar en ax[1]
pdf_plot = dist.pdf(d, *param)
ax[0].plot(d, escala*pdf_plot, lw=3.5, label='{}'.format(distribucion))
# Prueba de bondad de ajuste por chi-cuadrado
coef_chi, p = stats.chisquare(f_obs=ocurrencias_teo, f_exp=ocurrencias_exp)
if p > p_max: # si el p actual es mayor
p_max = p # designarlo como el máximo
dist_chi = distribucion # elegir la distribución como la de mejor ajuste
mod_chi = dist, param, pdf
# Bondad de ajuste por RMSE (Root-Mean-Square Error)
diferencia = (ocurrencias_teo - ocurrencias_exp)**2
rmse = np.sqrt(np.mean(diferencia))
if rmse < rmse_min:
rmse_min = rmse
dist_rmse = distribucion
mod_rmse = dist, param, pdf
# Bondad de ajuste por Kolgomorov - Smirnov
D, ksp = stats.kstest(datos, distribucion, args=param)
if ksp > kspmax:
kspmax = ksp
dist_ks = distribucion
# Decidir el mejor modelo
if dist_chi == dist_rmse or dist_chi == dist_ks:
params = mod_chi[1]
mejor_ajuste = dist_chi
ax[1].hist(datos, bins=divisiones, color='cornflowerblue', label='Distribución observada')
ax[1].bar(bins_centrados, mod_chi[2] * escala, width=6, color='r', label='Mejor ajuste: {}'.format(dist_chi))
m, v, s, k = mod_chi[0].stats(*params, moments='mvsk')
elif dist_rmse == dist_ks:
params = mod_rmse[1]
mejor_ajuste = dist_rmse
ax[1].hist(datos, bins = divisiones, color='cornflowerblue', label='Distribución observada')
ax[1].bar(bins_centrados, mod_rmse[2] * escala, width=6, color='r', label='Mejor ajuste: {}'.format(dist_rmse))
m, v, s, k = mod_rmse[0].stats(*params, moments='mvsk')
# Imprimir resumen y resultados
print('-------\nResumen\n-------')
print('Cantidad de muestras:', len(datos), 'días a las', hora, 'horas')
print('Máximo:', max(datos), 'MW')
print('Mínimo:', min(datos), 'MW')
print('Tipo: Demanda energética horaria')
print('------\nAjuste\n------')
print('Menor error RMS es:', dist_rmse)
print('Mejor bondad de ajuste en la prueba de chi-cuadrado es:', dist_chi)
print('Mejor bondad de ajuste en la prueba de Kolmogorov–Smirnov es:', dist_ks)
print('Distribución elegida:', mejor_ajuste)
print('--------\nMomentos\n--------')
print('Media:', m, '\nVarianza:', v, '\nDesviación estándar:', np.sqrt(v), '\nCoeficiente simetría:', s, '\nKurtosis:', k)
print('--------\nGráficas\n--------')
ax[0].legend()
ax[1].legend()
plt.show()
# Hora, en el intervalo [0, 23] (tipo int)
hora = 18
# Distribuciones a evaluar
distribuciones = ['norm', 'rayleigh', 'burr12', 'alpha', 'gamma', 'beta']
# Llamar a las funciones
demandas = extraer_datos('demanda_2019.json', hora)
evaluar_modelos(demandas, distribuciones, 25, hora)
import random
def horas_asignadas(digitos):
'''Elige una hora A en periodo punta
y una hora B de los otros periodos,
con los dígitos del carné como "seed"
'''
random.seed(digitos)
punta = [11, 12, 18, 19, 20]
valle = [7, 8, 9, 10, 13, 14, 15, 16, 17]
nocturno = [21, 22, 23, 0, 1, 2, 3, 4, 5, 6]
otro = valle + nocturno
HA = punta[random.randrange(0, len(punta))]
HB = otro[random.randrange(0, len(otro))]
horas = 'Hora A = {}, hora B = {}'.format(HA, HB)
return horas
horas_asignadas(12345)
# 4.1. - Comparación de consumo de potencia
# 4.2. - Obtención de los momentos de los modelos
| 0.338296 | 0.938237 |
# XGBoost (template)
July 2018
Rupert T
rup...@.....com
Derived from work at DataKind Summer Datadive 2018
* Non-functional! Template for future work only *
Summary:
.....
Model 1: Regression
Model 2: Classification
```
%matplotlib inline
import matplotlib
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os
# data import
df = pd.read_csv('xxx.csv')
```
### Pre-process
```
df.shape
df.head()
```
### Exploratory analysis
```
fig, ax = plt.subplots(figsize=(10,8))
pairings_filtered.boxplot(column=['tut_per_day'], by='Tutor Age Bracket', ax=ax)
plt.suptitle('')
plt.title('Pairing performance: Tutorials per day')
plt.ylim((0,0.2))
```
### Feature engineering
### Correlations between features and target
```
from scipy.stats import pearsonr
feature_names = model_data.loc[:,feature_col_names].columns.tolist()
correlations = {}
for f in feature_names:
data_temp = model_data[[f,target]].dropna()
x1 = data_temp[f].values
x2 = data_temp[target].values
key = f + ' vs ' + target
correlations[key] = pearsonr(x1,x2)[0]
data_correlations = pd.DataFrame(correlations, index=['Value']).T
data_correlations.loc[data_correlations['Value'].abs().sort_values(ascending=False).index]
# Look at raw data
sorted = model_data.sort_values(target, ascending=True)#.values
idx = np.arange(sorted.shape[0])
n_cols = model_data.shape[1]
fig = plt.figure(figsize=(15,14))
for j in range(n_cols):
plt.subplot(n_cols,1,j+1)
plt.plot(idx,sorted.loc[:,model_col_names[j]])
plt.title('Features vs target')
plt.ylabel(model_col_names[j])
# plt.subplot(n_cols,1,6)
# plt.plot(idx,sorted[:,5],'r')
# plt.ylabel(target)
# plt.ylim((0,0.2))
plt.show()
```
### Model 1: XGBoost regression
...
```
# preprocess
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
feature_col_names = ['School Year', 'School', 'student_subject', 'Allocated Level', 'Organisation Name', 'Occupation', 'TAP priority status', 'City', 'Business partner', 'tutor_qualification_level', 'Tutor Age', 'allocated_subject_prefered', 'pairing_subject_prefered', 'pairing_subject_first_degree']
# feature_col_names = ['Tutor Age', 'allocated_subject_prefered', 'pairing_subject_prefered', 'pairing_subject_first_degree', 'tutor_qualification_level']
target = 'Total Happened Tutorials'
model_col_names = feature_col_names.copy()
model_col_names.append(target)
model_data = pairings_filtered.loc[:, model_col_names]
filtered_model_data = model_data #.loc[model_data[target]<0.2, :]
# Force to string
force_str_cols = ['School Year', 'School', 'student_subject', 'Allocated Level', 'Organisation Name', 'Occupation', 'TAP priority status', 'City', 'Business partner', 'tutor_qualification_level']
for col_name in force_str_cols:
if col_name in filtered_model_data:
filtered_model_data[col_name] = filtered_model_data[col_name].astype(str)
# Split into features and labels - all values even nan
X = filtered_model_data.loc[:,feature_col_names].as_matrix()
y = filtered_model_data.loc[:,[target]].as_matrix()
X.shape, y.shape
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import OneHotEncoder
encode_cols = 10 # first 6 cols
# encode string input values as integers
encoded_x = None
for i in range(0, encode_cols):
label_encoder = LabelEncoder()
feature = label_encoder.fit_transform(X[:,i])
feature = feature.reshape(X.shape[0], 1)
onehot_encoder = OneHotEncoder(sparse=False)
feature = onehot_encoder.fit_transform(feature)
print(feature.shape)
if encoded_x is None:
encoded_x = feature
else:
encoded_x = np.concatenate((encoded_x, feature), axis=1)
print("enoded X shape: : ", encoded_x.shape)
all_X = np.concatenate((encoded_x, X[:,encode_cols:]), axis=1)
print("all X shape: : ", all_X.shape)
# # encode string class values as integers
# label_encoder = LabelEncoder()
# label_encoder = label_encoder.fit(Y)
# label_encoded_y = label_encoder.transform(Y)
# Split into test and train and validation
X_train, X_test, y_train, y_test = train_test_split(encoded_x, y, test_size=.3, random_state=42)
#X_train, X_test, y_train_with, y_test_with_id = train_test_split(X_train, y_train_with_id, test_size=.2, random_state=42)
# y_train = y_train_with_id['tut_per_day']#.as_matrix()
# y_test = y_test_with_id['tut_per_day']#.as_matrix()
# y_valid = y_valid_with_id['tut_per_day']#.as_matrix()
(X_train.shape, X_test.shape), (y_train.shape, y_test.shape)
import xgboost
xgb = xgboost.XGBRegressor(n_estimators=500, learning_rate=0.1, gamma=0, subsample=0.75,
colsample_bytree=1, max_depth=15)
xgb.fit(X_train,y_train)
from sklearn.metrics import explained_variance_score
predictions = xgb.predict(X_test)
print(explained_variance_score(predictions,y_test))
fig = plt.figure(figsize=(8,8))
#plt.subplot(5,1,1)
plt.plot(y_test,predictions,'k.')
plt.plot([0,70],[0,70], 'r--')
plt.title('XGBoost performance: Truth vs prediction')
plt.ylabel('Prediction')
plt.xlabel('Truth')
plt.xlim((0,70))
plt.ylim((0,70))
feature_scores = xgb.feature_importances_
feature_scores_dict = {feature_name: feature_scores[idx] for idx, feature_name in enumerate(feature_col_names)}
feature_scores_dict
# Feature importance
importance_df = pd.DataFrame.from_dict(feature_scores_dict,orient='index')
importance_df.columns = ['fscore']
importance_df = importance_df.sort_values(by = 'fscore', ascending=True)
importance_df.ix[-40:,:].plot(kind='barh', legend=False, figsize=(6, 10))
plt.title('XGBoost Feature Importance')
plt.xlabel('relative importance')
#plt.gcf().savefig('feature_importance_xgb.png', facecolor='w', transparent=False)
```
### Model 2: XGBoost classifier
...
```
feature_col_names = ['School Year', 'School', 'student_subject', 'Allocated Level', 'Organisation Name', 'Occupation', 'TAP priority status', 'City', 'Business partner', 'tutor_qualification_level', 'Tutor Age', 'allocated_subject_prefered', 'pairing_subject_prefered', 'pairing_subject_first_degree']
# feature_col_names = ['Tutor Age', 'allocated_subject_prefered', 'pairing_subject_prefered', 'pairing_subject_first_degree', 'tutor_qualification_level']
target = 'success_tut'
model_col_names = feature_col_names.copy()
model_col_names.append(target)
model_data = pairings_filtered.loc[:, model_col_names]
filtered_model_data = model_data #.loc[model_data[target]<0.2, :]
# Force to string
force_str_cols = ['School Year', 'School', 'student_subject', 'Allocated Level', 'Organisation Name', 'Occupation', 'TAP priority status', 'City', 'Business partner', 'tutor_qualification_level']
for col_name in force_str_cols:
if col_name in filtered_model_data:
filtered_model_data[col_name] = filtered_model_data[col_name].astype(str)
# Rebalance classes
balanced_model_data = pd.concat([filtered_model_data,
filtered_model_data.loc[filtered_model_data['success_tut'],:],
filtered_model_data.loc[filtered_model_data['success_tut'],:]], axis=0).sample(frac=1)
balanced_model_data.shape
# Split into features and labels - all values even nan
X = balanced_model_data.loc[:,feature_col_names].as_matrix()
y = balanced_model_data.loc[:,[target]].as_matrix()
X.shape, y.shape
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import OneHotEncoder
encode_cols = 10 # first 6 cols
# encode string input values as integers
encoded_x = None
for i in range(0, encode_cols):
label_encoder = LabelEncoder()
feature = label_encoder.fit_transform(X[:,i])
feature = feature.reshape(X.shape[0], 1)
onehot_encoder = OneHotEncoder(sparse=False)
feature = onehot_encoder.fit_transform(feature)
print(feature.shape)
if encoded_x is None:
encoded_x = feature
else:
encoded_x = np.concatenate((encoded_x, feature), axis=1)
print("enoded X shape: : ", encoded_x.shape)
all_X = np.concatenate((encoded_x, X[:,encode_cols:]), axis=1)
print("all X shape: : ", all_X.shape)
# # encode string class values as integers
# label_encoder = LabelEncoder()
# label_encoder = label_encoder.fit(Y)
# label_encoded_y = label_encoder.transform(Y)
# Split into test and train and validation
X_train, X_test, y_train, y_test = train_test_split(encoded_x, y, test_size=.3, random_state=42)
#X_train, X_test, y_train_with, y_test_with_id = train_test_split(X_train, y_train_with_id, test_size=.2, random_state=42)
y_train = y_train.astype(np.int32)
y_test = y_test.astype(np.int32)
(X_train.shape, X_test.shape), (y_train.shape, y_test.shape)
import xgboost
xgb = xgboost.XGBClassifier(n_estimators=200, learning_rate=0.5, gamma=0, subsample=0.75,
colsample_bytree=1, max_depth=10)
xgb.fit(X_train,y_train)
pred = xgb.predict(X_test)
# Confusion matrix
from sklearn.metrics import confusion_matrix
import itertools
import numpy as np
import matplotlib.pyplot as plt
def plot_confusion_matrix2(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
num_formatter = '%.1f%%'
multiplier = 100
else:
print('Confusion matrix')
num_formatter = '%d'
multiplier = 1
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
#plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
print(cm)
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, num_formatter % (multiplier * cm[i, j]),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
# Plot confusion matrix
from sklearn.metrics import confusion_matrix
figure = plt.figure(figsize=(8, 8))
pred_bool = pred #(pred > 0.5) * 1
cnf_matrix = confusion_matrix(y_test, pred_bool)
class_names = ['0','1']
np.set_printoptions(precision=2)
ax = plt.subplot(122)
plot_confusion_matrix2(cnf_matrix, classes=class_names,
title='Confusion matrix', normalize=False)
#plt.tight_layout()
plt.show()
#figure.savefig('images4/xgboost_3-way_confusion matrix_all_sensors.png', facecolor='w', transparent=False)
# Plot confusion matrix - with %
from sklearn.metrics import confusion_matrix
figure = plt.figure(figsize=(8, 8))
pred_bool = pred #(pred > 0.5) * 1
cnf_matrix = confusion_matrix(y_test, pred_bool)
class_names = ['Pair not successful','Pair successful']
np.set_printoptions(precision=2)
ax = plt.subplot(122)
plot_confusion_matrix2(cnf_matrix, classes=class_names,
title='Confusion matrix', normalize=True)
#plt.tight_layout()
plt.show()
#figure.savefig('images4/xgboost_3-way_confusion matrix_all_sensors.png', facecolor='w', transparent=False)
```
|
github_jupyter
|
%matplotlib inline
import matplotlib
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os
# data import
df = pd.read_csv('xxx.csv')
df.shape
df.head()
fig, ax = plt.subplots(figsize=(10,8))
pairings_filtered.boxplot(column=['tut_per_day'], by='Tutor Age Bracket', ax=ax)
plt.suptitle('')
plt.title('Pairing performance: Tutorials per day')
plt.ylim((0,0.2))
from scipy.stats import pearsonr
feature_names = model_data.loc[:,feature_col_names].columns.tolist()
correlations = {}
for f in feature_names:
data_temp = model_data[[f,target]].dropna()
x1 = data_temp[f].values
x2 = data_temp[target].values
key = f + ' vs ' + target
correlations[key] = pearsonr(x1,x2)[0]
data_correlations = pd.DataFrame(correlations, index=['Value']).T
data_correlations.loc[data_correlations['Value'].abs().sort_values(ascending=False).index]
# Look at raw data
sorted = model_data.sort_values(target, ascending=True)#.values
idx = np.arange(sorted.shape[0])
n_cols = model_data.shape[1]
fig = plt.figure(figsize=(15,14))
for j in range(n_cols):
plt.subplot(n_cols,1,j+1)
plt.plot(idx,sorted.loc[:,model_col_names[j]])
plt.title('Features vs target')
plt.ylabel(model_col_names[j])
# plt.subplot(n_cols,1,6)
# plt.plot(idx,sorted[:,5],'r')
# plt.ylabel(target)
# plt.ylim((0,0.2))
plt.show()
# preprocess
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
feature_col_names = ['School Year', 'School', 'student_subject', 'Allocated Level', 'Organisation Name', 'Occupation', 'TAP priority status', 'City', 'Business partner', 'tutor_qualification_level', 'Tutor Age', 'allocated_subject_prefered', 'pairing_subject_prefered', 'pairing_subject_first_degree']
# feature_col_names = ['Tutor Age', 'allocated_subject_prefered', 'pairing_subject_prefered', 'pairing_subject_first_degree', 'tutor_qualification_level']
target = 'Total Happened Tutorials'
model_col_names = feature_col_names.copy()
model_col_names.append(target)
model_data = pairings_filtered.loc[:, model_col_names]
filtered_model_data = model_data #.loc[model_data[target]<0.2, :]
# Force to string
force_str_cols = ['School Year', 'School', 'student_subject', 'Allocated Level', 'Organisation Name', 'Occupation', 'TAP priority status', 'City', 'Business partner', 'tutor_qualification_level']
for col_name in force_str_cols:
if col_name in filtered_model_data:
filtered_model_data[col_name] = filtered_model_data[col_name].astype(str)
# Split into features and labels - all values even nan
X = filtered_model_data.loc[:,feature_col_names].as_matrix()
y = filtered_model_data.loc[:,[target]].as_matrix()
X.shape, y.shape
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import OneHotEncoder
encode_cols = 10 # first 6 cols
# encode string input values as integers
encoded_x = None
for i in range(0, encode_cols):
label_encoder = LabelEncoder()
feature = label_encoder.fit_transform(X[:,i])
feature = feature.reshape(X.shape[0], 1)
onehot_encoder = OneHotEncoder(sparse=False)
feature = onehot_encoder.fit_transform(feature)
print(feature.shape)
if encoded_x is None:
encoded_x = feature
else:
encoded_x = np.concatenate((encoded_x, feature), axis=1)
print("enoded X shape: : ", encoded_x.shape)
all_X = np.concatenate((encoded_x, X[:,encode_cols:]), axis=1)
print("all X shape: : ", all_X.shape)
# # encode string class values as integers
# label_encoder = LabelEncoder()
# label_encoder = label_encoder.fit(Y)
# label_encoded_y = label_encoder.transform(Y)
# Split into test and train and validation
X_train, X_test, y_train, y_test = train_test_split(encoded_x, y, test_size=.3, random_state=42)
#X_train, X_test, y_train_with, y_test_with_id = train_test_split(X_train, y_train_with_id, test_size=.2, random_state=42)
# y_train = y_train_with_id['tut_per_day']#.as_matrix()
# y_test = y_test_with_id['tut_per_day']#.as_matrix()
# y_valid = y_valid_with_id['tut_per_day']#.as_matrix()
(X_train.shape, X_test.shape), (y_train.shape, y_test.shape)
import xgboost
xgb = xgboost.XGBRegressor(n_estimators=500, learning_rate=0.1, gamma=0, subsample=0.75,
colsample_bytree=1, max_depth=15)
xgb.fit(X_train,y_train)
from sklearn.metrics import explained_variance_score
predictions = xgb.predict(X_test)
print(explained_variance_score(predictions,y_test))
fig = plt.figure(figsize=(8,8))
#plt.subplot(5,1,1)
plt.plot(y_test,predictions,'k.')
plt.plot([0,70],[0,70], 'r--')
plt.title('XGBoost performance: Truth vs prediction')
plt.ylabel('Prediction')
plt.xlabel('Truth')
plt.xlim((0,70))
plt.ylim((0,70))
feature_scores = xgb.feature_importances_
feature_scores_dict = {feature_name: feature_scores[idx] for idx, feature_name in enumerate(feature_col_names)}
feature_scores_dict
# Feature importance
importance_df = pd.DataFrame.from_dict(feature_scores_dict,orient='index')
importance_df.columns = ['fscore']
importance_df = importance_df.sort_values(by = 'fscore', ascending=True)
importance_df.ix[-40:,:].plot(kind='barh', legend=False, figsize=(6, 10))
plt.title('XGBoost Feature Importance')
plt.xlabel('relative importance')
#plt.gcf().savefig('feature_importance_xgb.png', facecolor='w', transparent=False)
feature_col_names = ['School Year', 'School', 'student_subject', 'Allocated Level', 'Organisation Name', 'Occupation', 'TAP priority status', 'City', 'Business partner', 'tutor_qualification_level', 'Tutor Age', 'allocated_subject_prefered', 'pairing_subject_prefered', 'pairing_subject_first_degree']
# feature_col_names = ['Tutor Age', 'allocated_subject_prefered', 'pairing_subject_prefered', 'pairing_subject_first_degree', 'tutor_qualification_level']
target = 'success_tut'
model_col_names = feature_col_names.copy()
model_col_names.append(target)
model_data = pairings_filtered.loc[:, model_col_names]
filtered_model_data = model_data #.loc[model_data[target]<0.2, :]
# Force to string
force_str_cols = ['School Year', 'School', 'student_subject', 'Allocated Level', 'Organisation Name', 'Occupation', 'TAP priority status', 'City', 'Business partner', 'tutor_qualification_level']
for col_name in force_str_cols:
if col_name in filtered_model_data:
filtered_model_data[col_name] = filtered_model_data[col_name].astype(str)
# Rebalance classes
balanced_model_data = pd.concat([filtered_model_data,
filtered_model_data.loc[filtered_model_data['success_tut'],:],
filtered_model_data.loc[filtered_model_data['success_tut'],:]], axis=0).sample(frac=1)
balanced_model_data.shape
# Split into features and labels - all values even nan
X = balanced_model_data.loc[:,feature_col_names].as_matrix()
y = balanced_model_data.loc[:,[target]].as_matrix()
X.shape, y.shape
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import OneHotEncoder
encode_cols = 10 # first 6 cols
# encode string input values as integers
encoded_x = None
for i in range(0, encode_cols):
label_encoder = LabelEncoder()
feature = label_encoder.fit_transform(X[:,i])
feature = feature.reshape(X.shape[0], 1)
onehot_encoder = OneHotEncoder(sparse=False)
feature = onehot_encoder.fit_transform(feature)
print(feature.shape)
if encoded_x is None:
encoded_x = feature
else:
encoded_x = np.concatenate((encoded_x, feature), axis=1)
print("enoded X shape: : ", encoded_x.shape)
all_X = np.concatenate((encoded_x, X[:,encode_cols:]), axis=1)
print("all X shape: : ", all_X.shape)
# # encode string class values as integers
# label_encoder = LabelEncoder()
# label_encoder = label_encoder.fit(Y)
# label_encoded_y = label_encoder.transform(Y)
# Split into test and train and validation
X_train, X_test, y_train, y_test = train_test_split(encoded_x, y, test_size=.3, random_state=42)
#X_train, X_test, y_train_with, y_test_with_id = train_test_split(X_train, y_train_with_id, test_size=.2, random_state=42)
y_train = y_train.astype(np.int32)
y_test = y_test.astype(np.int32)
(X_train.shape, X_test.shape), (y_train.shape, y_test.shape)
import xgboost
xgb = xgboost.XGBClassifier(n_estimators=200, learning_rate=0.5, gamma=0, subsample=0.75,
colsample_bytree=1, max_depth=10)
xgb.fit(X_train,y_train)
pred = xgb.predict(X_test)
# Confusion matrix
from sklearn.metrics import confusion_matrix
import itertools
import numpy as np
import matplotlib.pyplot as plt
def plot_confusion_matrix2(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
num_formatter = '%.1f%%'
multiplier = 100
else:
print('Confusion matrix')
num_formatter = '%d'
multiplier = 1
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
#plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
print(cm)
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, num_formatter % (multiplier * cm[i, j]),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
# Plot confusion matrix
from sklearn.metrics import confusion_matrix
figure = plt.figure(figsize=(8, 8))
pred_bool = pred #(pred > 0.5) * 1
cnf_matrix = confusion_matrix(y_test, pred_bool)
class_names = ['0','1']
np.set_printoptions(precision=2)
ax = plt.subplot(122)
plot_confusion_matrix2(cnf_matrix, classes=class_names,
title='Confusion matrix', normalize=False)
#plt.tight_layout()
plt.show()
#figure.savefig('images4/xgboost_3-way_confusion matrix_all_sensors.png', facecolor='w', transparent=False)
# Plot confusion matrix - with %
from sklearn.metrics import confusion_matrix
figure = plt.figure(figsize=(8, 8))
pred_bool = pred #(pred > 0.5) * 1
cnf_matrix = confusion_matrix(y_test, pred_bool)
class_names = ['Pair not successful','Pair successful']
np.set_printoptions(precision=2)
ax = plt.subplot(122)
plot_confusion_matrix2(cnf_matrix, classes=class_names,
title='Confusion matrix', normalize=True)
#plt.tight_layout()
plt.show()
#figure.savefig('images4/xgboost_3-way_confusion matrix_all_sensors.png', facecolor='w', transparent=False)
| 0.469277 | 0.880746 |
## Functions
### Definition
We use `def` to define a function, and `return` to pass back a value:
```
def double(x):
return x*2
print(double(5), double([5]), double('five'))
```
### Default Parameters
We can specify default values for parameters:
```
def jeeves(name = "Sir"):
return "Very good, {}".format(name)
jeeves()
jeeves('James')
```
If you have some parameters with defaults, and some without, those with defaults **must** go later.
If you have multiple default arguments, you can specify neither, one or both:
```
def jeeves(greeting="Very good", name="Sir"):
return "{}, {}".format(greeting, name)
jeeves()
jeeves("Hello")
jeeves(name = "James")
jeeves(greeting="Suits you")
jeeves("Hello", "Sailor")
```
### Side effects
Functions can do things to change their **mutable** arguments,
so `return` is optional.
This is pretty awful style, in general, functions should normally be side-effect free.
Here is a contrived example of a function that makes plausible use of a side-effect
```
def double_inplace(vec):
vec[:] = [element*2 for element in vec]
z = list(range(4))
double_inplace(z)
print(z)
letters = ['a', 'b', 'c', 'd', 'e', 'f', 'g']
letters[:] = []
```
In this example, we're using `[:]` to access into the same list, and write it's data.
vec = [element*2 for element in vec]
would just move a local label, not change the input.
But I'd usually just write this as a function which **returned** the output:
```
def double(vec):
return [element*2 for element in vec]
```
Let's remind ourselves of the behaviour for modifying lists in-place using `[:]` with a simple array:
```
x = 5
x = 7
x = ['a','b','c']
y = x
x
x[:] = ["Hooray!", "Yippee"]
y
```
### Early Return
Return without arguments can be used to exit early from a function
Here's a slightly more plausibly useful function-with-side-effects to extend a list with a specified padding datum.
```
def extend(to, vec, pad):
if len(vec) >= to:
return # Exit early, list is already long enough.
vec[:] = vec + [pad]*(to-len(vec))
x = list(range(3))
extend(6, x, 'a')
print(x)
z = range(9)
extend(6, z, 'a')
print(z)
```
### Unpacking arguments
If a vector is supplied to a function with a '*', its elements
are used to fill each of a function's arguments.
```
def arrow(before, after):
return str(before) + " -> " + str(after)
arrow(1, 3)
x = [1,-1]
arrow(*x)
```
This can be quite powerful:
```
charges = {"neutron": 0, "proton": 1, "electron": -1}
for particle in charges.items():
print(arrow(*particle))
```
### Sequence Arguments
Similiarly, if a `*` is used in the **definition** of a function, multiple
arguments are absorbed into a list **inside** the function:
```
def doubler(*sequence):
return [x*2 for x in sequence]
doubler(1,2,3)
doubler(5, 2, "Wow!")
```
### Keyword Arguments
If two asterisks are used, named arguments are supplied inside the function as a dictionary:
```
def arrowify(**args):
for key, value in args.items():
print(key + " -> " + value)
arrowify(neutron="n", proton="p", electron="e")
```
These different approaches can be mixed:
```
def somefunc(a, b, *args, **kwargs):
print("A:", a)
print("B:", b)
print("args:", args)
print("keyword args", kwargs)
somefunc(1, 2, 3, 4, 5, fish="Haddock")
```
|
github_jupyter
|
def double(x):
return x*2
print(double(5), double([5]), double('five'))
def jeeves(name = "Sir"):
return "Very good, {}".format(name)
jeeves()
jeeves('James')
def jeeves(greeting="Very good", name="Sir"):
return "{}, {}".format(greeting, name)
jeeves()
jeeves("Hello")
jeeves(name = "James")
jeeves(greeting="Suits you")
jeeves("Hello", "Sailor")
def double_inplace(vec):
vec[:] = [element*2 for element in vec]
z = list(range(4))
double_inplace(z)
print(z)
letters = ['a', 'b', 'c', 'd', 'e', 'f', 'g']
letters[:] = []
def double(vec):
return [element*2 for element in vec]
x = 5
x = 7
x = ['a','b','c']
y = x
x
x[:] = ["Hooray!", "Yippee"]
y
def extend(to, vec, pad):
if len(vec) >= to:
return # Exit early, list is already long enough.
vec[:] = vec + [pad]*(to-len(vec))
x = list(range(3))
extend(6, x, 'a')
print(x)
z = range(9)
extend(6, z, 'a')
print(z)
def arrow(before, after):
return str(before) + " -> " + str(after)
arrow(1, 3)
x = [1,-1]
arrow(*x)
charges = {"neutron": 0, "proton": 1, "electron": -1}
for particle in charges.items():
print(arrow(*particle))
def doubler(*sequence):
return [x*2 for x in sequence]
doubler(1,2,3)
doubler(5, 2, "Wow!")
def arrowify(**args):
for key, value in args.items():
print(key + " -> " + value)
arrowify(neutron="n", proton="p", electron="e")
def somefunc(a, b, *args, **kwargs):
print("A:", a)
print("B:", b)
print("args:", args)
print("keyword args", kwargs)
somefunc(1, 2, 3, 4, 5, fish="Haddock")
| 0.33372 | 0.976175 |
# Importing Brevitas networks into FINN
In this notebook we'll go through an example of how to import a Brevitas-trained QNN into FINN. The steps will be as follows:
1. Load up the trained PyTorch model
2. Call Brevitas FINN-ONNX export and visualize with Netron
3. Import into FINN and call cleanup transformations
We'll use the following showSrc function to print the source code for function calls in the Jupyter notebook:
```
import onnx
import inspect
def showSrc(what):
print("".join(inspect.getsourcelines(what)[0]))
```
## 1. Load up the trained PyTorch model
The FINN Docker image comes with several [example Brevitas networks](https://github.com/maltanar/brevitas_cnv_lfc), and we'll use the LFC-w1a1 model as the example network here. This is a binarized fully connected network trained on the MNIST dataset. Let's start by looking at what the PyTorch network definition looks like:
```
from models.LFC import LFC
showSrc(LFC)
```
We can see that the network topology is constructed using a few helper functions that generate the quantized linear layers and quantized activations. The bitwidth of the layers is actually parametrized in the constructor, so let's instantiate a 1-bit weights and activations version of this network. We also have pretrained weights for this network, which we will load into the model.
```
import torch
trained_lfc_w1a1_checkpoint = "/workspace/brevitas_cnv_lfc/pretrained_models/LFC_1W1A/checkpoints/best.tar"
lfc = LFC(weight_bit_width=1, act_bit_width=1, in_bit_width=1).eval()
checkpoint = torch.load(trained_lfc_w1a1_checkpoint, map_location="cpu")
lfc.load_state_dict(checkpoint["state_dict"])
lfc
```
We have now instantiated our trained PyTorch network. Let's try to run an example MNIST image through the network using PyTorch.
```
import matplotlib.pyplot as plt
from pkgutil import get_data
import onnx
import onnx.numpy_helper as nph
raw_i = get_data("finn", "data/onnx/mnist-conv/test_data_set_0/input_0.pb")
input_tensor = onnx.load_tensor_from_string(raw_i)
input_tensor_npy = nph.to_array(input_tensor)
input_tensor_pyt = torch.from_numpy(input_tensor_npy).float()
imgplot = plt.imshow(input_tensor_npy.reshape(28,28), cmap='gray')
from torch.nn.functional import softmax
# do forward pass in PyTorch/Brevitas
produced = lfc.forward(input_tensor_pyt).detach()
probabilities = softmax(produced, dim=-1).flatten()
probabilities
import numpy as np
objects = [str(x) for x in range(10)]
y_pos = np.arange(len(objects))
plt.bar(y_pos, probabilities, align='center', alpha=0.5)
plt.xticks(y_pos, objects)
plt.ylabel('Predicted Probability')
plt.title('LFC-w1a1 Predictions for Image')
plt.show()
```
## 2. Call Brevitas FINN-ONNX export and visualize with Netron
Brevitas comes with built-in FINN-ONNX export functionality. This is similar to the regular ONNX export capabilities of PyTorch, with a few differences:
1. The weight quantization logic is not exported as part of the graph; rather, the quantized weights themselves are exported.
2. Special quantization annotations are used to preserve the low-bit quantization information. ONNX (at the time of writing) supports 8-bit quantization as the minimum bitwidth, whereas FINN-ONNX quantization annotations can go down to binary/bipolar quantization.
3. Low-bit quantized activation functions are exported as MultiThreshold operators.
It's actually quite straightforward to export ONNX from our Brevitas model as follows:
```
import brevitas.onnx as bo
export_onnx_path = "/tmp/LFCW1A1.onnx"
input_shape = (1, 1, 28, 28)
bo.export_finn_onnx(lfc, input_shape, export_onnx_path)
```
Let's examine what the exported ONNX model looks like. For this, we will use the Netron visualizer:
```
import netron
netron.start(export_onnx_path, port=8081, host="0.0.0.0")
%%html
<iframe src="http://0.0.0.0:8081/" style="position: relative; width: 100%;" height="400"></iframe>
```
When running this notebook in the FINN Docker container, you should be able to see an interactive visualization of the imported network above, and click on individual nodes to inspect their parameters. If you look at any of the MatMul nodes, you should be able to see that the weights are all {-1, +1} values, and the activations are Sign functions.
## 3. Import into FINN and call cleanup transformations
We will now import this ONNX model into FINN using the ModelWrapper, and examine some of the graph attributes from Python.
```
from finn.core.modelwrapper import ModelWrapper
model = ModelWrapper(export_onnx_path)
model.graph.node[9]
```
The ModelWrapper exposes a range of other useful functions as well. For instance, by convention the second input of the MatMul node will be a pre-initialized weight tensor, which we can view using the following:
```
model.get_initializer(model.graph.node[9].input[1])
```
We can also examine the quantization annotations and shapes of various tensors using the convenience functions provided by ModelWrapper.
```
model.get_tensor_datatype(model.graph.node[9].input[1])
model.get_tensor_shape(model.graph.node[9].input[1])
```
If we want to operate further on this model in FINN, it is a good idea to execute certain "cleanup" transformations on this graph. Here, we will run shape inference and constant folding on this graph, and visualize the resulting graph in Netron again.
```
from finn.transformation.fold_constants import FoldConstants
from finn.transformation.infer_shapes import InferShapes
model = model.transform(InferShapes())
model = model.transform(FoldConstants())
export_onnx_path_transformed = "/tmp/LFCW1A1-clean.onnx"
model.save(export_onnx_path_transformed)
netron.start(export_onnx_path_transformed, port=8081, host="0.0.0.0")
%%html
<iframe src="http://0.0.0.0:8081/" style="position: relative; width: 100%;" height="400"></iframe>
```
We can see that the resulting graph has become smaller and simpler. Specifically, the input reshaping is now a single Reshape node instead of the Shape -> Gather -> Unsqueeze -> Concat -> Reshape sequence. We can now use the internal ONNX execution capabilities of FINN to ensure that we still get the same output from this model as we did with PyTorch.
```
import finn.core.onnx_exec as oxe
input_dict = {"0": nph.to_array(input_tensor)}
output_dict = oxe.execute_onnx(model, input_dict)
produced_finn = output_dict[list(output_dict.keys())[0]]
produced_finn
np.isclose(produced, produced_finn).all()
```
We have succesfully verified that the transformed and cleaned-up FINN graph still produces the same output, and can now use this model for further processing in FINN.
|
github_jupyter
|
import onnx
import inspect
def showSrc(what):
print("".join(inspect.getsourcelines(what)[0]))
from models.LFC import LFC
showSrc(LFC)
import torch
trained_lfc_w1a1_checkpoint = "/workspace/brevitas_cnv_lfc/pretrained_models/LFC_1W1A/checkpoints/best.tar"
lfc = LFC(weight_bit_width=1, act_bit_width=1, in_bit_width=1).eval()
checkpoint = torch.load(trained_lfc_w1a1_checkpoint, map_location="cpu")
lfc.load_state_dict(checkpoint["state_dict"])
lfc
import matplotlib.pyplot as plt
from pkgutil import get_data
import onnx
import onnx.numpy_helper as nph
raw_i = get_data("finn", "data/onnx/mnist-conv/test_data_set_0/input_0.pb")
input_tensor = onnx.load_tensor_from_string(raw_i)
input_tensor_npy = nph.to_array(input_tensor)
input_tensor_pyt = torch.from_numpy(input_tensor_npy).float()
imgplot = plt.imshow(input_tensor_npy.reshape(28,28), cmap='gray')
from torch.nn.functional import softmax
# do forward pass in PyTorch/Brevitas
produced = lfc.forward(input_tensor_pyt).detach()
probabilities = softmax(produced, dim=-1).flatten()
probabilities
import numpy as np
objects = [str(x) for x in range(10)]
y_pos = np.arange(len(objects))
plt.bar(y_pos, probabilities, align='center', alpha=0.5)
plt.xticks(y_pos, objects)
plt.ylabel('Predicted Probability')
plt.title('LFC-w1a1 Predictions for Image')
plt.show()
import brevitas.onnx as bo
export_onnx_path = "/tmp/LFCW1A1.onnx"
input_shape = (1, 1, 28, 28)
bo.export_finn_onnx(lfc, input_shape, export_onnx_path)
import netron
netron.start(export_onnx_path, port=8081, host="0.0.0.0")
%%html
<iframe src="http://0.0.0.0:8081/" style="position: relative; width: 100%;" height="400"></iframe>
from finn.core.modelwrapper import ModelWrapper
model = ModelWrapper(export_onnx_path)
model.graph.node[9]
model.get_initializer(model.graph.node[9].input[1])
model.get_tensor_datatype(model.graph.node[9].input[1])
model.get_tensor_shape(model.graph.node[9].input[1])
from finn.transformation.fold_constants import FoldConstants
from finn.transformation.infer_shapes import InferShapes
model = model.transform(InferShapes())
model = model.transform(FoldConstants())
export_onnx_path_transformed = "/tmp/LFCW1A1-clean.onnx"
model.save(export_onnx_path_transformed)
netron.start(export_onnx_path_transformed, port=8081, host="0.0.0.0")
%%html
<iframe src="http://0.0.0.0:8081/" style="position: relative; width: 100%;" height="400"></iframe>
import finn.core.onnx_exec as oxe
input_dict = {"0": nph.to_array(input_tensor)}
output_dict = oxe.execute_onnx(model, input_dict)
produced_finn = output_dict[list(output_dict.keys())[0]]
produced_finn
np.isclose(produced, produced_finn).all()
| 0.47025 | 0.982389 |
# This colab notebook must be run on a **P100** GPU instance otherwise it will crash. Use the Cell-1 to ensure that it has a **P100** GPU instance
Cell-1: Ensure the required gpu instance (P100)
```
#no.of sockets i.e available slots for physical processors
!lscpu | grep 'Socket(s):'
#no.of cores each processor is having
!lscpu | grep 'Core(s) per socket:'
#no.of threads each core is having
!lscpu | grep 'Thread(s) per core'
#GPU count and name
!nvidia-smi -L
#use this command to see GPU activity while doing Deep Learning tasks, for this command 'nvidia-smi' and for above one to work, go to 'Runtime > change runtime type > Hardware Accelerator > GPU'
!nvidia-smi
```
Cell-2: Add Google Drive
```
from google.colab import drive
drive.mount('/content/gdrive')
```
Cell-3: Install Required Dependencies
```
!pip install efficientnet_pytorch==0.7.0
!pip install albumentations==0.4.5
!pip install torch==1.6.0+cu101 torchvision==0.7.0+cu101 -f https://download.pytorch.org/whl/torch\_stable.html -q\
```
Cell-4: Run this cell to generate current fold weight ( Estimated Time for training this fold is around 2 hours 48 minutes )
```
import sys
sys.path.insert(0, "/content/gdrive/My Drive/zindi_cgiar_wheat_growth_stage_challenge/src_lq2")
from dataset import *
from model import *
from trainer import *
from utils import *
import numpy as np
from sklearn.model_selection import StratifiedKFold
from torch.utils.data import DataLoader
config = {
'n_folds': 5,
'random_seed': 7200,
'run_fold': 0,
'model_name': 'efficientnet-b4',
'global_dim': 1792,
'batch_size': 48,
'n_core': 2,
'weight_saving_path': '/content/gdrive/My Drive/zindi_cgiar_wheat_growth_stage_challenge/train_lq2_only_effnet_b4_step1/weights/',
'resume_checkpoint_path': None,
'lr': 0.01,
'total_epochs': 100,
}
if __name__ == '__main__':
set_random_state(config['random_seed'])
imgs = np.load('/content/gdrive/My Drive/zindi_cgiar_wheat_growth_stage_challenge/zindi_npy_data/train_imgs.npy')
labels = np.load('/content/gdrive/My Drive/zindi_cgiar_wheat_growth_stage_challenge/zindi_npy_data/train_labels.npy')
labels_quality = np.load('/content/gdrive/My Drive/zindi_cgiar_wheat_growth_stage_challenge/zindi_npy_data/train_labels_quality.npy')
imgs = imgs[labels_quality == 2]
labels = labels[labels_quality == 2]
labels = labels - 1
skf = StratifiedKFold(n_splits=config['n_folds'], shuffle=True, random_state=config['random_seed'])
for fold_number, (train_index, val_index) in enumerate(skf.split(X=imgs, y=labels)):
if fold_number != config['run_fold']:
continue
train_dataset = ZCDataset(
imgs[train_index],
labels[train_index],
transform=get_train_transforms(),
test=False,
)
train_loader = DataLoader(
train_dataset,
batch_size=config['batch_size'],
shuffle=True,
num_workers=config['n_core'],
drop_last=True,
pin_memory=True,
)
val_dataset = ZCDataset(
imgs[val_index],
labels[val_index],
transform=get_val_transforms(),
test=True,
)
val_loader = DataLoader(
val_dataset,
batch_size=config['batch_size'],
shuffle=False,
num_workers=config['n_core'],
pin_memory=True,
)
del imgs, labels
model = CNN_Model(config['model_name'], config['global_dim'])
args = {
'model': model,
'Loaders': [train_loader,val_loader],
'metrics': {'Loss':AverageMeter, 'f1_score':PrintMeter, 'rmse':PrintMeter},
'checkpoint_saving_path': config['weight_saving_path'],
'resume_train_from_checkpoint': False,
'resume_checkpoint_path': config['resume_checkpoint_path'],
'lr': config['lr'],
'fold': fold_number,
'epochsTorun': config['total_epochs'],
'batch_size': config['batch_size'],
'test_run_for_error': False,
'problem_name': 'zindi_cigar',
}
Trainer = ModelTrainer(**args)
Trainer.fit()
```
|
github_jupyter
|
#no.of sockets i.e available slots for physical processors
!lscpu | grep 'Socket(s):'
#no.of cores each processor is having
!lscpu | grep 'Core(s) per socket:'
#no.of threads each core is having
!lscpu | grep 'Thread(s) per core'
#GPU count and name
!nvidia-smi -L
#use this command to see GPU activity while doing Deep Learning tasks, for this command 'nvidia-smi' and for above one to work, go to 'Runtime > change runtime type > Hardware Accelerator > GPU'
!nvidia-smi
from google.colab import drive
drive.mount('/content/gdrive')
!pip install efficientnet_pytorch==0.7.0
!pip install albumentations==0.4.5
!pip install torch==1.6.0+cu101 torchvision==0.7.0+cu101 -f https://download.pytorch.org/whl/torch\_stable.html -q\
import sys
sys.path.insert(0, "/content/gdrive/My Drive/zindi_cgiar_wheat_growth_stage_challenge/src_lq2")
from dataset import *
from model import *
from trainer import *
from utils import *
import numpy as np
from sklearn.model_selection import StratifiedKFold
from torch.utils.data import DataLoader
config = {
'n_folds': 5,
'random_seed': 7200,
'run_fold': 0,
'model_name': 'efficientnet-b4',
'global_dim': 1792,
'batch_size': 48,
'n_core': 2,
'weight_saving_path': '/content/gdrive/My Drive/zindi_cgiar_wheat_growth_stage_challenge/train_lq2_only_effnet_b4_step1/weights/',
'resume_checkpoint_path': None,
'lr': 0.01,
'total_epochs': 100,
}
if __name__ == '__main__':
set_random_state(config['random_seed'])
imgs = np.load('/content/gdrive/My Drive/zindi_cgiar_wheat_growth_stage_challenge/zindi_npy_data/train_imgs.npy')
labels = np.load('/content/gdrive/My Drive/zindi_cgiar_wheat_growth_stage_challenge/zindi_npy_data/train_labels.npy')
labels_quality = np.load('/content/gdrive/My Drive/zindi_cgiar_wheat_growth_stage_challenge/zindi_npy_data/train_labels_quality.npy')
imgs = imgs[labels_quality == 2]
labels = labels[labels_quality == 2]
labels = labels - 1
skf = StratifiedKFold(n_splits=config['n_folds'], shuffle=True, random_state=config['random_seed'])
for fold_number, (train_index, val_index) in enumerate(skf.split(X=imgs, y=labels)):
if fold_number != config['run_fold']:
continue
train_dataset = ZCDataset(
imgs[train_index],
labels[train_index],
transform=get_train_transforms(),
test=False,
)
train_loader = DataLoader(
train_dataset,
batch_size=config['batch_size'],
shuffle=True,
num_workers=config['n_core'],
drop_last=True,
pin_memory=True,
)
val_dataset = ZCDataset(
imgs[val_index],
labels[val_index],
transform=get_val_transforms(),
test=True,
)
val_loader = DataLoader(
val_dataset,
batch_size=config['batch_size'],
shuffle=False,
num_workers=config['n_core'],
pin_memory=True,
)
del imgs, labels
model = CNN_Model(config['model_name'], config['global_dim'])
args = {
'model': model,
'Loaders': [train_loader,val_loader],
'metrics': {'Loss':AverageMeter, 'f1_score':PrintMeter, 'rmse':PrintMeter},
'checkpoint_saving_path': config['weight_saving_path'],
'resume_train_from_checkpoint': False,
'resume_checkpoint_path': config['resume_checkpoint_path'],
'lr': config['lr'],
'fold': fold_number,
'epochsTorun': config['total_epochs'],
'batch_size': config['batch_size'],
'test_run_for_error': False,
'problem_name': 'zindi_cigar',
}
Trainer = ModelTrainer(**args)
Trainer.fit()
| 0.361728 | 0.620248 |
# Projet CentraleSupelec - Dreem
# Machine Learning course 3A OBT
## Alexis Tuil et Adil Bousfiha
### Approche Deep Learning
# Load useful libraries
```
import numpy as np
import pandas as pd
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import StratifiedShuffleSplit, train_test_split
from keras.models import Sequential
from keras.layers import Activation, Flatten, Dense, Input, Dropout, Convolution1D, MaxPool1D, GlobalMaxPool1D, GlobalAveragePooling1D, \
concatenate, SpatialDropout1D, TimeDistributed, Bidirectional, LSTM, Reshape, Conv1D, MaxPooling1D
from keras.optimizers import SGD
from keras.utils import np_utils
import h5py
from sklearn.metrics import f1_score
import operator
```
# Define the network architecture
```
model = Sequential()
model.add(Reshape((1500, 7), input_shape=(10500,) ))
# Depending on the way you decide to stack the input data (cf report part 3.2) you may have
# to change the Reshape layer
model.add(Conv1D(16, 3, activation='relu' ))
model.add(Conv1D(16, 3, activation='relu'))
model.add(MaxPooling1D(3))
model.add(Conv1D(32, 3, activation='relu'))
model.add(Conv1D(32, 3, activation='relu'))
model.add(MaxPooling1D(3))
model.add(Conv1D(64, 3, activation='relu'))
model.add(Conv1D(64, 3, activation='relu'))
model.add(GlobalAveragePooling1D())
model.add(Dense(64, activation='relu'))
model.add(Dense(5, activation='softmax'))
print(model.summary())
```
# Extract the training set
```
data = h5py.File('train.h5', 'r')
y = pd.read_csv ('train_y.csv').sleep_stage
```
### Transform labels to One Hot Encoding representation
```
y_ohe = np_utils.to_categorical(y, 5)
```
### Stack the signals following the approach you choose
```
X = np.hstack((data['eeg_1'], data['eeg_2'], data['eeg_3'],data['eeg_4'],data['eeg_5'],data['eeg_6'],data['eeg_7']))
```
### Compile the model
```
model.compile(loss='categorical_crossentropy',
optimizer='adam', metrics=['accuracy'])
BATCH_SIZE = 32
EPOCHS = 30
callbacks_list = [
ModelCheckpoint(
filepath='best_model.{epoch:02d}-{val_loss:.2f}.h5',
monitor='val_loss', save_best_only=True),
EarlyStopping(monitor='acc', patience=10)
]
history = model.fit(X,
y_ohe,
batch_size=BATCH_SIZE,
epochs=EPOCHS,
callbacks=callbacks_list,
validation_split=0.2,
verbose=1)
model.save('my_model.h5') # creates a HDF5 file 'my_model.h5'
```
# Extract the test set
```
data_test = h5py.File('test.h5', 'r')
X_test = np.hstack((data_test['eeg_1'], data_test['eeg_2'], data_test['eeg_3'],data_test['eeg_4'],
data_test['eeg_5'],data_test['eeg_6'],data_test['eeg_7']))
```
### Predict the test labels
```
y_predicted = model.predict(X_test)
```
### Convert the test labels from One Hot Encoding to scalar representation
```
y_predicted_r = [max(enumerate(score), key=operator.itemgetter(1))[0] for score in y_predicted]
```
### Write the submit CSV
```
res = pd.DataFrame()
res['id'] = range(len(y_predicted_r))
res['sleep_stage'] = y_predicted_r
res.set_index('id').to_csv('submit.csv')
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import StratifiedShuffleSplit, train_test_split
from keras.models import Sequential
from keras.layers import Activation, Flatten, Dense, Input, Dropout, Convolution1D, MaxPool1D, GlobalMaxPool1D, GlobalAveragePooling1D, \
concatenate, SpatialDropout1D, TimeDistributed, Bidirectional, LSTM, Reshape, Conv1D, MaxPooling1D
from keras.optimizers import SGD
from keras.utils import np_utils
import h5py
from sklearn.metrics import f1_score
import operator
model = Sequential()
model.add(Reshape((1500, 7), input_shape=(10500,) ))
# Depending on the way you decide to stack the input data (cf report part 3.2) you may have
# to change the Reshape layer
model.add(Conv1D(16, 3, activation='relu' ))
model.add(Conv1D(16, 3, activation='relu'))
model.add(MaxPooling1D(3))
model.add(Conv1D(32, 3, activation='relu'))
model.add(Conv1D(32, 3, activation='relu'))
model.add(MaxPooling1D(3))
model.add(Conv1D(64, 3, activation='relu'))
model.add(Conv1D(64, 3, activation='relu'))
model.add(GlobalAveragePooling1D())
model.add(Dense(64, activation='relu'))
model.add(Dense(5, activation='softmax'))
print(model.summary())
data = h5py.File('train.h5', 'r')
y = pd.read_csv ('train_y.csv').sleep_stage
y_ohe = np_utils.to_categorical(y, 5)
X = np.hstack((data['eeg_1'], data['eeg_2'], data['eeg_3'],data['eeg_4'],data['eeg_5'],data['eeg_6'],data['eeg_7']))
model.compile(loss='categorical_crossentropy',
optimizer='adam', metrics=['accuracy'])
BATCH_SIZE = 32
EPOCHS = 30
callbacks_list = [
ModelCheckpoint(
filepath='best_model.{epoch:02d}-{val_loss:.2f}.h5',
monitor='val_loss', save_best_only=True),
EarlyStopping(monitor='acc', patience=10)
]
history = model.fit(X,
y_ohe,
batch_size=BATCH_SIZE,
epochs=EPOCHS,
callbacks=callbacks_list,
validation_split=0.2,
verbose=1)
model.save('my_model.h5') # creates a HDF5 file 'my_model.h5'
data_test = h5py.File('test.h5', 'r')
X_test = np.hstack((data_test['eeg_1'], data_test['eeg_2'], data_test['eeg_3'],data_test['eeg_4'],
data_test['eeg_5'],data_test['eeg_6'],data_test['eeg_7']))
y_predicted = model.predict(X_test)
y_predicted_r = [max(enumerate(score), key=operator.itemgetter(1))[0] for score in y_predicted]
res = pd.DataFrame()
res['id'] = range(len(y_predicted_r))
res['sleep_stage'] = y_predicted_r
res.set_index('id').to_csv('submit.csv')
| 0.756717 | 0.853058 |
```
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
%matplotlib inline
import torch
import torchvision
import torchvision.transforms as transforms
import torch.nn as nn
import torch.optim as optim
from torch.nn import functional as F
from torch.utils.data import DataLoader,Dataset
import seaborn as sns
def generate_data(data_size,corruption_percentage):
dims =50
#np.random.seed(1234)
y = np.ones(data_size,dtype="long")
idx = np.random.uniform(size =data_size)>0.5
y[idx] = 0
print("positive_class",sum(y==1),"negative_class",sum(y==0))
idx1 = ~idx #positive class indices
idx2 = idx #negative class indices
x = np.zeros((data_size,dims))
x[idx1,0] = np.random.randn(sum(idx1)) # standard normal
x[idx2,0] = np.random.randn(sum(idx2)) +10 # normal with mean 10 and standard deviation 1
x[:,1:] = np.random.uniform(-1,1,size=(data_size,dims-1))
# plt.figure(figsize=(8,5))
# plt.hist(x[idx1,0],density=True,label = str(data_size)+"_1 class") #positive class histogram
# plt.hist(x[idx2,0],density = True, label = str(data_size)+"_0 class") #negative class histogram
# plt.xlabel("x1")
# plt.ylabel("probability density")
# plt.legend()
# plt.title("corruption_percentage: "+str(corruption_percentage)+"_Data size: "+str(data_size)+"_histogram")
#corruption_percentage = 0.5
mask = np.random.uniform(0,1,data_size) < corruption_percentage
a = np.array(y)
#print("true",a[mask])
a[mask] = np.random.randint(0,2,sum(mask))
#print("randomized",a[mask])
y = list(a)
return x,y,mask
# np.random.seed(1234)
y = np.ones(100,dtype="long")
idx = np.random.uniform(size =100)>0.5
y[idx] = 0
print("positive_class",sum(y==1),"negative_class",sum(y==0))
idx1 = ~idx #positive class indices
idx2 = idx #negative class indices
x = np.zeros((100,50))
x[idx1,0] = np.random.randn(sum(idx1)) # standard normal
x[idx2,0] = np.random.randn(sum(idx2)) +10 # normal with mean 10 and standard deviation 1
x[:,1:] = np.random.uniform(-1,1,size=(100,49))
# plt.hist(x[idx1,0],density=True,label = "+1 class") #positive class histogram
# plt.hist(x[idx2,0],density = True, label = "0 class") #negative class histogram
sns.distplot(x[idx1,0], hist=True, label="+1 class")
sns.distplot(x[idx2,0], hist= True, label="0 class")
plt.xlabel("x1")
plt.ylabel("probability density")
plt.legend()
corruption_percentage = 0.1
mask = np.random.uniform(0,1,100) < corruption_percentage
a = np.array(y)
print("true",a[mask])
a[mask] = np.random.randint(0,2,sum(mask))
print("randomized",a[mask])
y = list(a)
# # cifar_trainset_random.targets[:50000] = np.random.randint(low=0,high=9,size=50000)
# #trainloader_random = torch.utils.data.DataLoader(cifar_trainset_random,batch_size=256,shuffle=False,num_workers=2)
np.unique(y),sum(mask)
def accuracy( a, b):
length = a.shape
#print(a,"dfgfg",length)
correct = a==b
return sum(correct)/length
class Synthetic_data(Dataset):
def __init__(self,x,y):
super(Synthetic_data,self).__init__()
self.x = x
self.y = y
def __len__(self):
return(len(self.y))
def __getitem__(self,idx):
return self.x[idx,:],self.y[idx]
class Net(nn.Module):
def __init__(self):
super(Net,self).__init__()
self.linear1 = nn.Linear(50,16)
self.linear2 = nn.Linear(16,2)
# self.linear3 = nn.Linear(128,64)
# self.linear4 = nn.Linear(64,2)
def forward(self,x):
x = F.relu(self.linear1(x))
x = self.linear2(x)
# x = F.relu(self.linear3(x))
# x = self.linear4(x)
return x
def train(net,data_loader,epochs):
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(net.parameters(),lr =0.01)
for epoch in range(epochs):
running_loss = 0.0
cnt = 0
for i,data in enumerate(data_loader):
x_input ,targets = data
#true.append(targets.cpu().numpy())
optimizer.zero_grad()
outputs = net(x_input)
#out.append(outputs.cpu())
_, predicted = torch.max(outputs, 1)
#pred.append(predicted.cpu().numpy())
loss = criterion(outputs,targets)
loss.backward()
optimizer.step()
running_loss += loss.item()
if cnt % 4 == 3: # print every 50 mini-batches
#print('[%d, %5d] loss: %.3f' %(epoch + 1, cnt + 1, running_loss / 50))
running_loss = 0.0
cnt=cnt+1
return net,criterion,
def evaluate_train(net,dataloader,criterion,mask):
out = []
pred = []
true = []
for i,data in enumerate(trainloader):
x_input ,targets = data
true.append(targets.cpu().numpy())
outputs = net(x_input)
out.append(outputs.cpu())
_, predicted = torch.max(outputs, 1)
pred.append(predicted.cpu().numpy())
true_targets = np.concatenate(true,axis=0)
predicted_targets = np.concatenate(pred,axis =0)
acc_corrupt = accuracy(true_targets[mask],predicted_targets[mask])
acc_uncorrupt = accuracy(true_targets[~mask],predicted_targets[~mask])
acc_full = accuracy(true_targets, predicted_targets)
print("Train accuracy on corrupt data",acc_corrupt)
print("Train accuracy on un-corrupt data",acc_uncorrupt)
print("Train accuracy on full data", accuracy(true_targets, predicted_targets))
l= np.where(mask ==True)
p = np.where(mask == False)
out = torch.cat(out, dim =0)
print("Train cross entropy loss on corrupt data", criterion(out[l], torch.Tensor(true_targets[l]).type(torch.LongTensor)).item())
print("Train cross entropy loss on un-corrupt data",criterion(out[p], torch.Tensor(true_targets[p]).type(torch.LongTensor)).item())
print("Train cross entropy loss on full data",criterion(out, torch.Tensor(true_targets).type(torch.LongTensor)).item())
print("---"*20)
loss_full = criterion(out, torch.Tensor(true_targets).type(torch.LongTensor)).item()
return acc_full, loss_full
def evaluate_test(net,dataloader,criterion):
out = []
pred = []
true = []
for i,data in enumerate(trainloader):
x_input ,targets = data
true.append(targets.cpu().numpy())
outputs = net(x_input)
out.append(outputs.cpu())
_, predicted = torch.max(outputs, 1)
pred.append(predicted.cpu().numpy())
true_targets = np.concatenate(true,axis=0)
predicted_targets = np.concatenate(pred,axis =0)
acc_full = accuracy(true_targets, predicted_targets)
print("Test accuracy on full data", accuracy(true_targets, predicted_targets))
out = torch.cat(out, dim =0)
print("Test cross entropy loss on full data",criterion(out, torch.Tensor(true_targets).type(torch.LongTensor)).item())
print("---"*20)
loss_full = criterion(out, torch.Tensor(true_targets).type(torch.LongTensor)).item()
return acc_full, loss_full
datasizes = [100,500,1000,2000,5000,10000]
corrupt_percent = [0.01,0.1,0.2,0.5]
train_acc=[]
train_ce_loss=[]
test_acc=[]
test_ce_loss=[]
for i in datasizes:
for j in corrupt_percent:
x,y,mask = generate_data(data_size= i , corruption_percentage= j)
x_test,y_test,mask_test = generate_data(data_size= i , corruption_percentage= 0)
#print(sum(mask))
data_set = Synthetic_data(x,y)
test_set = Synthetic_data(x_test,y_test)
trainloader = DataLoader(data_set,batch_size=20,shuffle=False)
testloader = DataLoader(test_set,batch_size=20,shuffle=False)
net = Net().double()
net,criterion = train(net,trainloader,300)
a,b = evaluate_train(net,trainloader,criterion,mask)
c,d = evaluate_test(net , testloader, criterion)
train_acc.append(a)
train_ce_loss.append(b)
test_acc.append(c)
test_ce_loss.append(d)
plt.figure(figsize=(10,6))
plt.plot(datasizes,[train_acc[0],train_acc[4],train_acc[8],train_acc[12],train_acc[16],train_acc[20]],"o--",label="noise 1 %")
plt.plot(datasizes,[train_acc[1],train_acc[5],train_acc[9],train_acc[13],train_acc[17],train_acc[21]],"o--",label="noise 10 %")
plt.plot(datasizes,[train_acc[2],train_acc[6],train_acc[10],train_acc[14],train_acc[18],train_acc[22]],"o--",label="noise 20 %")
plt.plot(datasizes,[train_acc[3],train_acc[7],train_acc[11],train_acc[15],train_acc[19],train_acc[23]],"o--",label="noise 50 %")
plt.legend()
plt.xlabel("Data_set_size")
plt.ylabel("Percentage_Accuracy")
plt.title("Train_Accuracy")
plt.show()
plt.figure(figsize=(10,6))
plt.plot(datasizes,[train_ce_loss[0],train_ce_loss[4],train_ce_loss[8],train_ce_loss[12],train_ce_loss[16],train_ce_loss[20]],"o--",label="noise 1 %")
plt.plot(datasizes,[train_ce_loss[1],train_ce_loss[5],train_ce_loss[9],train_ce_loss[13],train_ce_loss[17],train_ce_loss[21]],"o--",label="noise 10 %")
plt.plot(datasizes,[train_ce_loss[2],train_ce_loss[6],train_ce_loss[10],train_ce_loss[14],train_ce_loss[18],train_ce_loss[22]],"o--",label="noise 20 %")
plt.plot(datasizes,[train_ce_loss[3],train_ce_loss[7],train_ce_loss[11],train_ce_loss[15],train_ce_loss[19],train_ce_loss[23]],"o--",label="noise 50 %")
plt.legend()
plt.xlabel("Data_set_size")
plt.ylabel("CE_Loss")
plt.title("Train_CE_Loss")
plt.show()
plt.figure(figsize=(10,6))
plt.plot(datasizes,[test_acc[0],test_acc[4],test_acc[8],test_acc[12],test_acc[16],test_acc[20]],"o--",label="noise 1 %")
plt.plot(datasizes,[test_acc[1],test_acc[5],test_acc[9],test_acc[13],test_acc[17],test_acc[21]],"o--",label="noise 10 %")
plt.plot(datasizes,[test_acc[2],test_acc[6],test_acc[10],test_acc[14],test_acc[18],test_acc[22]],"o--",label="noise 20 %")
plt.plot(datasizes,[test_acc[3],test_acc[7],test_acc[11],test_acc[15],test_acc[19],test_acc[23]],"o--",label="noise 50 %")
plt.legend()
plt.xlabel("Data_set_size")
plt.ylabel("Percentage_Accuracy")
plt.title("Test_Accuracy")
plt.show()
plt.figure(figsize=(10,6))
plt.plot(datasizes,[test_ce_loss[0],test_ce_loss[4],test_ce_loss[8],test_ce_loss[12],test_ce_loss[16],test_ce_loss[20]],"o--",label="noise 1 %")
plt.plot(datasizes,[test_ce_loss[1],test_ce_loss[5],test_ce_loss[9],test_ce_loss[13],test_ce_loss[17],test_ce_loss[21]],"o--",label="noise 10 %")
plt.plot(datasizes,[test_ce_loss[2],test_ce_loss[6],test_ce_loss[10],test_ce_loss[14],test_ce_loss[18],test_ce_loss[22]],"o--",label="noise 20 %")
plt.plot(datasizes,[test_ce_loss[3],test_ce_loss[7],test_ce_loss[11],test_ce_loss[15],test_ce_loss[19],test_ce_loss[23]],"o--",label="noise 50 %")
plt.legend()
plt.xlabel("Data_set_size")
plt.ylabel("CE_Loss")
plt.title("Test_CE_Loss")
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
%matplotlib inline
import torch
import torchvision
import torchvision.transforms as transforms
import torch.nn as nn
import torch.optim as optim
from torch.nn import functional as F
from torch.utils.data import DataLoader,Dataset
import seaborn as sns
def generate_data(data_size,corruption_percentage):
dims =50
#np.random.seed(1234)
y = np.ones(data_size,dtype="long")
idx = np.random.uniform(size =data_size)>0.5
y[idx] = 0
print("positive_class",sum(y==1),"negative_class",sum(y==0))
idx1 = ~idx #positive class indices
idx2 = idx #negative class indices
x = np.zeros((data_size,dims))
x[idx1,0] = np.random.randn(sum(idx1)) # standard normal
x[idx2,0] = np.random.randn(sum(idx2)) +10 # normal with mean 10 and standard deviation 1
x[:,1:] = np.random.uniform(-1,1,size=(data_size,dims-1))
# plt.figure(figsize=(8,5))
# plt.hist(x[idx1,0],density=True,label = str(data_size)+"_1 class") #positive class histogram
# plt.hist(x[idx2,0],density = True, label = str(data_size)+"_0 class") #negative class histogram
# plt.xlabel("x1")
# plt.ylabel("probability density")
# plt.legend()
# plt.title("corruption_percentage: "+str(corruption_percentage)+"_Data size: "+str(data_size)+"_histogram")
#corruption_percentage = 0.5
mask = np.random.uniform(0,1,data_size) < corruption_percentage
a = np.array(y)
#print("true",a[mask])
a[mask] = np.random.randint(0,2,sum(mask))
#print("randomized",a[mask])
y = list(a)
return x,y,mask
# np.random.seed(1234)
y = np.ones(100,dtype="long")
idx = np.random.uniform(size =100)>0.5
y[idx] = 0
print("positive_class",sum(y==1),"negative_class",sum(y==0))
idx1 = ~idx #positive class indices
idx2 = idx #negative class indices
x = np.zeros((100,50))
x[idx1,0] = np.random.randn(sum(idx1)) # standard normal
x[idx2,0] = np.random.randn(sum(idx2)) +10 # normal with mean 10 and standard deviation 1
x[:,1:] = np.random.uniform(-1,1,size=(100,49))
# plt.hist(x[idx1,0],density=True,label = "+1 class") #positive class histogram
# plt.hist(x[idx2,0],density = True, label = "0 class") #negative class histogram
sns.distplot(x[idx1,0], hist=True, label="+1 class")
sns.distplot(x[idx2,0], hist= True, label="0 class")
plt.xlabel("x1")
plt.ylabel("probability density")
plt.legend()
corruption_percentage = 0.1
mask = np.random.uniform(0,1,100) < corruption_percentage
a = np.array(y)
print("true",a[mask])
a[mask] = np.random.randint(0,2,sum(mask))
print("randomized",a[mask])
y = list(a)
# # cifar_trainset_random.targets[:50000] = np.random.randint(low=0,high=9,size=50000)
# #trainloader_random = torch.utils.data.DataLoader(cifar_trainset_random,batch_size=256,shuffle=False,num_workers=2)
np.unique(y),sum(mask)
def accuracy( a, b):
length = a.shape
#print(a,"dfgfg",length)
correct = a==b
return sum(correct)/length
class Synthetic_data(Dataset):
def __init__(self,x,y):
super(Synthetic_data,self).__init__()
self.x = x
self.y = y
def __len__(self):
return(len(self.y))
def __getitem__(self,idx):
return self.x[idx,:],self.y[idx]
class Net(nn.Module):
def __init__(self):
super(Net,self).__init__()
self.linear1 = nn.Linear(50,16)
self.linear2 = nn.Linear(16,2)
# self.linear3 = nn.Linear(128,64)
# self.linear4 = nn.Linear(64,2)
def forward(self,x):
x = F.relu(self.linear1(x))
x = self.linear2(x)
# x = F.relu(self.linear3(x))
# x = self.linear4(x)
return x
def train(net,data_loader,epochs):
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(net.parameters(),lr =0.01)
for epoch in range(epochs):
running_loss = 0.0
cnt = 0
for i,data in enumerate(data_loader):
x_input ,targets = data
#true.append(targets.cpu().numpy())
optimizer.zero_grad()
outputs = net(x_input)
#out.append(outputs.cpu())
_, predicted = torch.max(outputs, 1)
#pred.append(predicted.cpu().numpy())
loss = criterion(outputs,targets)
loss.backward()
optimizer.step()
running_loss += loss.item()
if cnt % 4 == 3: # print every 50 mini-batches
#print('[%d, %5d] loss: %.3f' %(epoch + 1, cnt + 1, running_loss / 50))
running_loss = 0.0
cnt=cnt+1
return net,criterion,
def evaluate_train(net,dataloader,criterion,mask):
out = []
pred = []
true = []
for i,data in enumerate(trainloader):
x_input ,targets = data
true.append(targets.cpu().numpy())
outputs = net(x_input)
out.append(outputs.cpu())
_, predicted = torch.max(outputs, 1)
pred.append(predicted.cpu().numpy())
true_targets = np.concatenate(true,axis=0)
predicted_targets = np.concatenate(pred,axis =0)
acc_corrupt = accuracy(true_targets[mask],predicted_targets[mask])
acc_uncorrupt = accuracy(true_targets[~mask],predicted_targets[~mask])
acc_full = accuracy(true_targets, predicted_targets)
print("Train accuracy on corrupt data",acc_corrupt)
print("Train accuracy on un-corrupt data",acc_uncorrupt)
print("Train accuracy on full data", accuracy(true_targets, predicted_targets))
l= np.where(mask ==True)
p = np.where(mask == False)
out = torch.cat(out, dim =0)
print("Train cross entropy loss on corrupt data", criterion(out[l], torch.Tensor(true_targets[l]).type(torch.LongTensor)).item())
print("Train cross entropy loss on un-corrupt data",criterion(out[p], torch.Tensor(true_targets[p]).type(torch.LongTensor)).item())
print("Train cross entropy loss on full data",criterion(out, torch.Tensor(true_targets).type(torch.LongTensor)).item())
print("---"*20)
loss_full = criterion(out, torch.Tensor(true_targets).type(torch.LongTensor)).item()
return acc_full, loss_full
def evaluate_test(net,dataloader,criterion):
out = []
pred = []
true = []
for i,data in enumerate(trainloader):
x_input ,targets = data
true.append(targets.cpu().numpy())
outputs = net(x_input)
out.append(outputs.cpu())
_, predicted = torch.max(outputs, 1)
pred.append(predicted.cpu().numpy())
true_targets = np.concatenate(true,axis=0)
predicted_targets = np.concatenate(pred,axis =0)
acc_full = accuracy(true_targets, predicted_targets)
print("Test accuracy on full data", accuracy(true_targets, predicted_targets))
out = torch.cat(out, dim =0)
print("Test cross entropy loss on full data",criterion(out, torch.Tensor(true_targets).type(torch.LongTensor)).item())
print("---"*20)
loss_full = criterion(out, torch.Tensor(true_targets).type(torch.LongTensor)).item()
return acc_full, loss_full
datasizes = [100,500,1000,2000,5000,10000]
corrupt_percent = [0.01,0.1,0.2,0.5]
train_acc=[]
train_ce_loss=[]
test_acc=[]
test_ce_loss=[]
for i in datasizes:
for j in corrupt_percent:
x,y,mask = generate_data(data_size= i , corruption_percentage= j)
x_test,y_test,mask_test = generate_data(data_size= i , corruption_percentage= 0)
#print(sum(mask))
data_set = Synthetic_data(x,y)
test_set = Synthetic_data(x_test,y_test)
trainloader = DataLoader(data_set,batch_size=20,shuffle=False)
testloader = DataLoader(test_set,batch_size=20,shuffle=False)
net = Net().double()
net,criterion = train(net,trainloader,300)
a,b = evaluate_train(net,trainloader,criterion,mask)
c,d = evaluate_test(net , testloader, criterion)
train_acc.append(a)
train_ce_loss.append(b)
test_acc.append(c)
test_ce_loss.append(d)
plt.figure(figsize=(10,6))
plt.plot(datasizes,[train_acc[0],train_acc[4],train_acc[8],train_acc[12],train_acc[16],train_acc[20]],"o--",label="noise 1 %")
plt.plot(datasizes,[train_acc[1],train_acc[5],train_acc[9],train_acc[13],train_acc[17],train_acc[21]],"o--",label="noise 10 %")
plt.plot(datasizes,[train_acc[2],train_acc[6],train_acc[10],train_acc[14],train_acc[18],train_acc[22]],"o--",label="noise 20 %")
plt.plot(datasizes,[train_acc[3],train_acc[7],train_acc[11],train_acc[15],train_acc[19],train_acc[23]],"o--",label="noise 50 %")
plt.legend()
plt.xlabel("Data_set_size")
plt.ylabel("Percentage_Accuracy")
plt.title("Train_Accuracy")
plt.show()
plt.figure(figsize=(10,6))
plt.plot(datasizes,[train_ce_loss[0],train_ce_loss[4],train_ce_loss[8],train_ce_loss[12],train_ce_loss[16],train_ce_loss[20]],"o--",label="noise 1 %")
plt.plot(datasizes,[train_ce_loss[1],train_ce_loss[5],train_ce_loss[9],train_ce_loss[13],train_ce_loss[17],train_ce_loss[21]],"o--",label="noise 10 %")
plt.plot(datasizes,[train_ce_loss[2],train_ce_loss[6],train_ce_loss[10],train_ce_loss[14],train_ce_loss[18],train_ce_loss[22]],"o--",label="noise 20 %")
plt.plot(datasizes,[train_ce_loss[3],train_ce_loss[7],train_ce_loss[11],train_ce_loss[15],train_ce_loss[19],train_ce_loss[23]],"o--",label="noise 50 %")
plt.legend()
plt.xlabel("Data_set_size")
plt.ylabel("CE_Loss")
plt.title("Train_CE_Loss")
plt.show()
plt.figure(figsize=(10,6))
plt.plot(datasizes,[test_acc[0],test_acc[4],test_acc[8],test_acc[12],test_acc[16],test_acc[20]],"o--",label="noise 1 %")
plt.plot(datasizes,[test_acc[1],test_acc[5],test_acc[9],test_acc[13],test_acc[17],test_acc[21]],"o--",label="noise 10 %")
plt.plot(datasizes,[test_acc[2],test_acc[6],test_acc[10],test_acc[14],test_acc[18],test_acc[22]],"o--",label="noise 20 %")
plt.plot(datasizes,[test_acc[3],test_acc[7],test_acc[11],test_acc[15],test_acc[19],test_acc[23]],"o--",label="noise 50 %")
plt.legend()
plt.xlabel("Data_set_size")
plt.ylabel("Percentage_Accuracy")
plt.title("Test_Accuracy")
plt.show()
plt.figure(figsize=(10,6))
plt.plot(datasizes,[test_ce_loss[0],test_ce_loss[4],test_ce_loss[8],test_ce_loss[12],test_ce_loss[16],test_ce_loss[20]],"o--",label="noise 1 %")
plt.plot(datasizes,[test_ce_loss[1],test_ce_loss[5],test_ce_loss[9],test_ce_loss[13],test_ce_loss[17],test_ce_loss[21]],"o--",label="noise 10 %")
plt.plot(datasizes,[test_ce_loss[2],test_ce_loss[6],test_ce_loss[10],test_ce_loss[14],test_ce_loss[18],test_ce_loss[22]],"o--",label="noise 20 %")
plt.plot(datasizes,[test_ce_loss[3],test_ce_loss[7],test_ce_loss[11],test_ce_loss[15],test_ce_loss[19],test_ce_loss[23]],"o--",label="noise 50 %")
plt.legend()
plt.xlabel("Data_set_size")
plt.ylabel("CE_Loss")
plt.title("Test_CE_Loss")
plt.show()
| 0.65368 | 0.596903 |
# 第3回 情報検索の評価
この演習ページでは,既存のツールを使って各種評価指標を計算する方法について説明します.
参考文献
- [情報アクセス評価方法論 -検索エンジンの進歩のために-](https://www.amazon.co.jp/%E6%83%85%E5%A0%B1%E3%82%A2%E3%82%AF%E3%82%BB%E3%82%B9%E8%A9%95%E4%BE%A1%E6%96%B9%E6%B3%95%E8%AB%96-%E9%85%92%E4%BA%95-%E5%93%B2%E4%B9%9F/dp/4339024961), 酒井哲也, コロナ社, 2015.
## ライブラリ
この演習では,情報検索におけるさまざまな評価指標を計算するためのツールキットである NTCIREVAL のPython版である pyNTCIREVAL を使用します.
- [pyNTCIREVAL](https://github.com/mpkato/pyNTCIREVAL/) by 京都大学 加藤 誠 先生
- [NTCIREVAL](http://research.nii.ac.jp/ntcir/tools/ntcireval-ja.html) by 早稲田大学 酒井 哲也 先生
NTCIREVALの説明を上記ページから引用します.
----
```
NTCIREVALは、様々な検索評価指標を計算するためのツールキットです。
NTCIRやTRECのad hoc文書検索タスクの他、diversified search resultsの評価やNTCIR-8コミュニティQAタスクの評価などにも利用できます。
NTCIREVALは例えば以下のような指標を算出できます:
-Average Precision
-Q-measure
-nDCG
-Expected Reciprocal Rank (ERR)
-Graded Average Precision (GAP)
-Rank-Biased Precision (RBP)
-Normalised Cumulative Utility (NCU)
-上記各指標の短縮リスト版
-Bpref
-D-measures and D#-measures (多様性評価用)
-Intent-Aware (IA) metrics (多様性評価用)
```
----
## ライブラリのインストール
`pip`というPythonライブラリ管理ツールを使用してインストールします. ターミナル上で h29iroのフォルダに移動し,下記コマンドで pyNTCIREVAL をインストールしてください.
```
pip install git+https://github.com/mpkato/pyNTCIREVAL.git
```
正しくインストールできれば, notebook上で
!pyNTCIREVAL
と実行すれば,以下の様なメッセージが出力されます.
```
Usage: pyNTCIREVAL [OPTIONS] COMMAND [ARGS]...
Options:
-h, --help Show this message and exit.
Commands:
compute
label
```
なお,notebook上で $!$ の後の文字列はシェル(ターミナル)に対するコマンドと解釈され,シェルの出力がnotebookの画面に出力されます.
# 1. 評価データの準備
NTCIREVALおよびpyNTCIREVALでは,評価用のテキストファイルをプログラムに渡すことで,評価値を計算します.
`../data/eval/` にサンプルデータを置いています.
基本的に,ある手法のある検索課題に対する評価指標を検索するためには,以下の2つのファイルを準備する必要があります.
- 適合性評価ファイル(*.rel)
- 検索結果ファイル(*.res)
## 適合性評価ファイル
適合性評価ファイルは,ある検索課題に対するコレクション中の適合性評価結果を表すテキストファイルです. `../data/eval/q1.rel` にサンプルデータを置いています.このファイル名は,検索課題$q_1$に対する適合性評価ファイルであることを意味しています(NTICREVALではファイル名に形式はありません.単純に利用者が分かりやすいため,山本がこのような名前をつけています).
q1.rel の中身はこのようになっています.
```
!cat ../data/eval/q1.rel
```
このファイルの一行の意味は,
```
文書ID 適合性ラベル
```
となっています. 文書IDは評価データ作成者が適宜付与したIDです.適合性ラベルは慣習上このような書き方をしており,
L0 が不適合を表し,L1, L2 ... , 以降は適合性の度合い(適合度)を表します.今回は,適合度は3段階($\{0,1,2 \}$)のため,ラベルは$\{L0,L1,L2\}$の3種類です.4段階の適合度を用いる場合は,$\{L0,L1,L2,L3\}$をラベルとして用います.
たとえば, q1.relファイルの3行目の
```
d3 L2
```
は,文書ID $d_3$の適合度が $2$ であることを表しています.
## 検索結果ファイル
検索結果ファイルは,ある手法のある検索課題に対する検索結果(つまり,ランク付けされた文書集合)を表すテキストファイルです. `../data/eval/method1.q1.res`にサンプルデータを置いています.
`method1.q1.res` の中身はこのようになっています.
```
!cat ../data/eval/method1.q1.res
```
このように,検索結果ファイルはランキング結果を単純に文書IDで表します.たとえば,このファイルは, 検索課題$q_1$に対して $d_1, d_2, d_3$ の順で文書をランキングしたことを表しています.
```
!pyNTCIREVAL
!python --version
!pyNTCIREVAL label -r ../data/eval/q1.rel < ../data/eval/method1.q1.res
```
# 2. 適合性ラベル付き検索結果ファイルの作成
適合性評価ファイルと検索結果ファイルを準備したら,次に適合性ラベル付き検索結果ファイルを作成します.このファイルはpyNTCIREVALの機能を利用して作成することができます.また,自身でプログラムを書いてこのファイルを作成することもできます. pyNTCIREVAL を用いて適合性ラベル付き検索結果ファイルを作成する場合は,以下のコマンドを実行します.
シェルのパイプを用いているので,シェルについて詳しくない人は上記コマンドの意味がよく分からないかもしれませんが
```
pyNTCIREVAL label
```
は適合性ラベル付き検索結果ファイルを作成するためのコマンドです.
```
-r ../data/eval/q1.rel
```
は適合性評価ファイルの場所を指定しており,
```
< ../data/eval/method1.q1.res
```
はpyNTCIREVALに,ラベルを付与して欲しい検索結果を渡しています.
上記コマンドを実行すると,
```
d1 L1
d2 L0
d3 L2
```
という結果が得られます.つまりこのプログラムは,検索結果ファイル中の各文書IDに対して,適合性評価ファイル中の対応する文書IDの適合性ラベルを付加しています.
ちなみに,上記コマンドだけでは適合性ラベル付き検索結果ファイルの内容が画面に出力されるだけです.この内容をファイルに保存するには,例えば以下の様にします.
```
!pyNTCIREVAL label -r ../data/kadai2/q3.rel < ../data/kadai2/method1.q3.res > ../data/kadai2/method1.q3.rel
```
```
> ../data/eval/method1.q1.rel
```
これもシェルのコマンドで,出力をmethod1.q1.relに書き込むという意味です.
```
!cat ../data/eval/method1.q1.rel
```
# 3. 評価指標の計算
適合性評価ファイルと,適合性評価付き検索結果ファイルが準備できたら,それらをpyNTCIREVALに渡すことで,各種評価指標を計算できます.
```
!pyNTCIREVAL compute -r ../data/kadai2/q1.rel -g 1:3:7:15:31 --cutoffs=3 < ../data/kadai2/method1.q1.rel
```
## コマンドの説明
```
pyNTECIREVAL compute
```
は評価指標を計算するためのコマンドです.
```
-g 1:3
```
は適合度$L1,L2$の文書に対する利得(Gain)を指定しています.この値はたとえばnDCGを計算する際に使用されます.今回は利得関数として, $g(i) = 2^{{\rm rel}_i} -1$を用いるので, $L1 = 1, L2 = 3$という利得を指定しています.
```
--cutoffs 1:3
```
は,指標を計算する際に検索結果上位何件まで考慮するか(@$k$)を指定しています.この場合,上位1,3件における指標がそれぞれ出力されます.
## 結果の見方
各指標の結果が出力されています.たとえば,nERR@0003は上位$3$件まで考慮した際のnERRの値を表しています(すなわち,nERR@$3$).また,@$k$が付与されていない指標は与えられたランキング結果全てを考慮した際の評価値です.
<span style="color:red">注意点として,本講義で示したnDCGの定義は MSnDCG版と呼ばれるnDCGの計算方法です.従って,本講義のnDCG@$3$に対応する指標はMSnDCG@0003になります.</span>
最後に,評価結果をファイルに保存するには,先ほどと同様に出力をファイルに書き込みます.
```
!pyNTCIREVAL compute -r ../data/eval/q1.rel -g 1:3 --cutoffs=1,3 < ../data/eval/method1.q1.rel > ../data/eval/method1.q1.eval
```
pyNTCIREVALの使い方は以上となります. pyNTCIREVALの本体であるNTCIREVALには,複数の検索課題に対する複数の手法の結果を一括で処理するようなプログラムも含まれています.興味がある人,あるいは実際に研究でそのような評価実験を行う可能性がある人は,ぜひNTCIREVALについても調べてください.
----
# 演習課題その2 情報検索の評価
## 必須課題 (1) 動作の確認
このページで用いた検索結果に対するpyNTCIREVALの出力のうち,`MSnDCG@0003`と`nERR@0003`が,講義資料の定義に従った計算と一致していることを確かめよ.つまり,nDCG@3とnERR@3を計算するプログラム書き,その結果がpyNTCIREVALの結果と一致していることを確認せよ.
## 必須課題(2)独自データに対する評価指標の計算
演習課題1で扱った検索課題集合と検索結果に対して各自で評価用データを作成しpyNTCIREVALを用いて評価指標を計算せよ.そして, MRR,nDCG@3およびnERR@3の平均を報告し,それらの値の違いが各指標のどういった要因によるものか考察せよ.なお,演習課題1で扱ったコーパス以外で評価データを作成してもい.ただし,評価データはダミーデータでなく実際の何らかのランキングを評価したものとし,検索課題(クエリ)は3つ以上とする.
## 任意課題 (a)評価指標の調査
情報検索の評価指標にはさまざまなものがあり,今回の講義で扱わなかった指標も多い.たとえば, MAP(Mean Average Precision),RBP(Rank-Biased Precision), Q-measureなどがある.そこで,これらの指標について2つ以上調べ,どのような指標か説明するとともに,それを計算するプログラムを作成し,pyNTCIREVALの結果と一致することを確認せよ.
- Mean Averge Precision
- 参考: Introduction to Informaton Retrieval, Chapter 8: Evaluation in information retrieval.
- Q-measure
- 参考: Sakai, T. and Song, R.: Evaluating Diversified Search Results Using Per-intent Graded Relevance,
SIGIR 2011.
- Rank-Biased Precision:
- 参考: Moffat, A. and Zobel, J.: Rank-biased Precision for Measurement of Retrieval Effectiveness, ACM TOIS 27(1), 2008.
# 課題の提出方法
いずれかの方法で,ipython notebookのページ(.ipynbファイル)とそのhtml版を提出すること.
1. 添付ファイルで山本に送信.
- 送付先 tyamamot at dl.kuis.kyoto-u.ac.jp
2. 各自のgithubやgithub gistにアップロードし,そのURLを山本に送信.この場合はhtml版を用意する必要はない.
3. 上記以外で,山本が実際に.ipynbファイルを確認できる方法.
# 締切
- 2017年11月30日(木)23:59
- 締切に関する個別の相談は``受け付けます``.
|
github_jupyter
|
NTCIREVALは、様々な検索評価指標を計算するためのツールキットです。
NTCIRやTRECのad hoc文書検索タスクの他、diversified search resultsの評価やNTCIR-8コミュニティQAタスクの評価などにも利用できます。
NTCIREVALは例えば以下のような指標を算出できます:
-Average Precision
-Q-measure
-nDCG
-Expected Reciprocal Rank (ERR)
-Graded Average Precision (GAP)
-Rank-Biased Precision (RBP)
-Normalised Cumulative Utility (NCU)
-上記各指標の短縮リスト版
-Bpref
-D-measures and D#-measures (多様性評価用)
-Intent-Aware (IA) metrics (多様性評価用)
pip install git+https://github.com/mpkato/pyNTCIREVAL.git
Usage: pyNTCIREVAL [OPTIONS] COMMAND [ARGS]...
Options:
-h, --help Show this message and exit.
Commands:
compute
label
!cat ../data/eval/q1.rel
文書ID 適合性ラベル
d3 L2
!cat ../data/eval/method1.q1.res
!pyNTCIREVAL
!python --version
!pyNTCIREVAL label -r ../data/eval/q1.rel < ../data/eval/method1.q1.res
pyNTCIREVAL label
-r ../data/eval/q1.rel
< ../data/eval/method1.q1.res
d1 L1
d2 L0
d3 L2
!pyNTCIREVAL label -r ../data/kadai2/q3.rel < ../data/kadai2/method1.q3.res > ../data/kadai2/method1.q3.rel
> ../data/eval/method1.q1.rel
!cat ../data/eval/method1.q1.rel
!pyNTCIREVAL compute -r ../data/kadai2/q1.rel -g 1:3:7:15:31 --cutoffs=3 < ../data/kadai2/method1.q1.rel
pyNTECIREVAL compute
-g 1:3
--cutoffs 1:3
!pyNTCIREVAL compute -r ../data/eval/q1.rel -g 1:3 --cutoffs=1,3 < ../data/eval/method1.q1.rel > ../data/eval/method1.q1.eval
| 0.38885 | 0.815783 |
```
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import time
from scipy.stats import linregress
from citipy import citipy
api_key = "2d7bc754b1f208baf1d97b9f731cceb6"
cl = pd.read_csv("C:/Users/write/Ur_APIHW/Weather Py/CityList.csv")
#drop na values
cl.dropna(inplace = True)
cl
lat_range = (-90, 90)
lng_range = (-180, 180)
lat_lngs = []
cities = []
# Create a set of random lat and lng combinations
lats = np.random.uniform(lat_range[0], lat_range[1], size=1500)
lngs = np.random.uniform(lng_range[0], lng_range[1], size=1500)
lat_lngs = zip(lats, lngs)
# Identify nearest city for each lat, lng combination
for lat_lng in lat_lngs:
city = citipy.nearest_city(lat_lng[0], lat_lng[1])
#get name/country
city_name = city.city_name + "," + city.country_code
# If the city is unique, then add it to a our cities list
if city not in cities:
cities.append(city_name)
# Print the city count to confirm sufficient count
uncities = set(cities)
len(uncities)
uncities
#test for api
url = "http://api.openweathermap.org/data/2.5/weather?"
units = "imperial"
city = cl["City Name"][0]
query_url = f"{url}appid={api_key}&units={units}&q={city}"
weather_response = requests.get(query_url)
weather_json = weather_response.json()
# Get the temperature from the response
print(f"The weather API responded with: {weather_json}.")
params = {"key": api_key}
url = "http://api.openweathermap.org/data/2.5/weather?"
units = "imperial"
##http://api.openweathermap.org/data/2.5/weather?appid=2d7bc754b1f208baf1d97b9f731cceb6&units=imperial&q=ushuaia
city_data = []
# Print to logger
print("Beginning Data Retrieval ")
print("-----------------------------")
# Create counters
record_count = 1
set_count = 1
# Loop through all the cities in our list
for i, city in enumerate(cities):
# Group cities in sets of 50 for logging purposes
if (i % 50 == 0 and i >= 50):
set_count += 1
record_count = 0
# Create endpoint URL with each city
city_url = url + "appid=" + api_key + "&units=" + units + "&q=" + city
# Log the url, record, and set numbers
print("Processing Record %s of Set %s | %s" % (record_count, set_count, city))
# Add 1 to the record count
record_count += 1
# Run an API request for each of the cities
try:
# Parse the JSON and retrieve data
city_weather = requests.get(city_url).json()
# Parse out the max temp, humidity, and cloudiness
city_lat = city_weather["coord"]["lat"]
city_lng = city_weather["coord"]["lon"]
city_max_temp = city_weather["main"]["temp_max"]
city_humidity = city_weather["main"]["humidity"]
city_clouds = city_weather["clouds"]["all"]
city_wind = city_weather["wind"]["speed"]
city_country = city_weather["sys"]["country"]
city_date = city_weather["dt"]
# Append the City information into city_data list
city_data.append({"City": city,
"Lat": city_lat,
"Lng": city_lng,
"Max Temp": city_max_temp,
"Humidity": city_humidity,
"Cloudiness": city_clouds,
"Wind Speed": city_wind,
"Country": city_country,
"Date": city_date})
# If an error is experienced, skip the city
except:
print("Does not exist")
pass
# Indicate that Data Loading is complete
print("-----------------------------")
print("Search Complete ")
print("-----------------------------")
#create DF
cdatapd = pd.DataFrame(city_data)
cdatapd.head()
#Check Humidity
cdatapd.describe()
#create seperate df for Humidity
mhcity = cdatapd[(cdatapd["Humidity"] > 100)].index
mhcity
#Seperate even further
ccity = cdatapd.drop(mhcity, inplace=False)
ccity.head()
ccity.columns
lat = ccity["Lat"]
max_temps = ccity["Max Temp"]
humidity = ccity["Humidity"]
cloudiness = ccity["Cloudiness"]
wind_speed = ccity["Wind Speed"]
#Export to Csv
clean_city = ccity.to_csv("output_data.csv", index_label="City_ID")
lat
#Build Scatterplots Lat V Max Temp
plt.scatter(lat, max_temps, edgecolor="black", linewidths=1, marker="o", alpha=0.8, label="Cities")
plt.title("City Lat vs Max Temp")
plt.ylabel("Max Temperature (F)")
plt.xlabel("Latitude")
plt.grid(True)
plt.savefig(r"Latvtemp.png")
plt.show()
```
Latitude inversely affects the max temperature of the city
```
#Scatterplot Lat V Humidity
plt.scatter(lat, humidity, edgecolor="black", linewidths=1, marker="o", alpha=0.8, label="Cities")
plt.title("City Lat vs Humidity")
plt.ylabel("Humidity (%)")
plt.xlabel("Latitude")
plt.grid(True)
plt.savefig(r"LatvHumidity.png")
plt.show()
```
There is a common grouping of latitude with high humidity
```
#Scatterplot Lat V Cloudiness
plt.scatter(lat, cloudiness, edgecolor="black", linewidths=1, marker="o", alpha=0.8, label="Cities")
plt.title("City Lat vs Cloudiness")
plt.ylabel("Cloudiness (%)")
plt.xlabel("Latitude")
plt.grid(True)
plt.savefig(r"LatvCloud.png")
plt.show()
```
The higher the altitude the greater the cloudiness
```
#Scatterplot Lat V Wind Speed
plt.scatter(lat, wind_speed, edgecolor="black", linewidths=1, marker="o", alpha=0.8, label="Cities")
plt.title("City Lat vs Wind Speed")
plt.ylabel("Wind Speed (mph)")
plt.xlabel("Latitude")
plt.grid(True)
plt.savefig(r"LatvWind.png")
plt.show()
```
The wind speed can have outliers depending on location, but seems to stay within parameters
```
#Create Linear Regression plots
def plot_linear_regression(x_values, y_values, title, text_coordinates):
# Run regresson on southern hemisphere
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
# Plot
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,text_coordinates,fontsize=15,color="red")
plt.xlabel('Latitude')
plt.ylabel(title)
print(f"The r-value is: {rvalue**2}")
plt.show()
# Northern and Southern
nhemidf = cdatapd.loc[(cdatapd["Lat"] >=0)]
shemidf = cdatapd.loc[(cdatapd["Lat"] < 0)]
# N Lat vs Max Temp
x_values = nhemidf["Lat"]
y_values = nhemidf["Max Temp"]
plot_linear_regression(x_values, y_values, 'Max Temp', (6,30))
#S Lat V Temp
x_values = shemidf["Lat"]
y_values = shemidf["Max Temp"]
plot_linear_regression(x_values, y_values, 'Max Temp', (-55,90))
```
There is a correlation between high altitude and low temp
```
# N Lat vs Humidity
x_values = nhemidf["Lat"]
y_values = nhemidf["Humidity"]
plot_linear_regression(x_values, y_values, 'Humidity', (40,10))
# S Lat vs Humidity
x_values = shemidf["Lat"]
y_values = shemidf["Humidity"]
plot_linear_regression(x_values, y_values, 'Humidity', (-50,20))
```
The higher the alitiude the greater the humidity
```
# N Lat vs Cloudiness
x_values = nhemidf["Lat"]
y_values = nhemidf["Cloudiness"]
plot_linear_regression(x_values, y_values, 'Cloudiness', (40,10))
# N Lat vs Cloudiness
x_values = shemidf["Lat"]
y_values = shemidf["Cloudiness"]
plot_linear_regression(x_values, y_values, 'Cloudiness', (-30,30))
```
There is a weak correlation between latidude and cloudiness
```
# N Lat vs Wind Speed
x_values = nhemidf["Lat"]
y_values = nhemidf["Wind Speed"]
plot_linear_regression(x_values, y_values, 'Wind Speed', (40,10))
# N Lat vs Wind Speed
x_values = shemidf["Lat"]
y_values = shemidf["Wind Speed"]
plot_linear_regression(x_values, y_values, 'Wind Speed', (-30,30))
```
The data does not hsow a true correlation between Latitude and Wind Speed
|
github_jupyter
|
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import time
from scipy.stats import linregress
from citipy import citipy
api_key = "2d7bc754b1f208baf1d97b9f731cceb6"
cl = pd.read_csv("C:/Users/write/Ur_APIHW/Weather Py/CityList.csv")
#drop na values
cl.dropna(inplace = True)
cl
lat_range = (-90, 90)
lng_range = (-180, 180)
lat_lngs = []
cities = []
# Create a set of random lat and lng combinations
lats = np.random.uniform(lat_range[0], lat_range[1], size=1500)
lngs = np.random.uniform(lng_range[0], lng_range[1], size=1500)
lat_lngs = zip(lats, lngs)
# Identify nearest city for each lat, lng combination
for lat_lng in lat_lngs:
city = citipy.nearest_city(lat_lng[0], lat_lng[1])
#get name/country
city_name = city.city_name + "," + city.country_code
# If the city is unique, then add it to a our cities list
if city not in cities:
cities.append(city_name)
# Print the city count to confirm sufficient count
uncities = set(cities)
len(uncities)
uncities
#test for api
url = "http://api.openweathermap.org/data/2.5/weather?"
units = "imperial"
city = cl["City Name"][0]
query_url = f"{url}appid={api_key}&units={units}&q={city}"
weather_response = requests.get(query_url)
weather_json = weather_response.json()
# Get the temperature from the response
print(f"The weather API responded with: {weather_json}.")
params = {"key": api_key}
url = "http://api.openweathermap.org/data/2.5/weather?"
units = "imperial"
##http://api.openweathermap.org/data/2.5/weather?appid=2d7bc754b1f208baf1d97b9f731cceb6&units=imperial&q=ushuaia
city_data = []
# Print to logger
print("Beginning Data Retrieval ")
print("-----------------------------")
# Create counters
record_count = 1
set_count = 1
# Loop through all the cities in our list
for i, city in enumerate(cities):
# Group cities in sets of 50 for logging purposes
if (i % 50 == 0 and i >= 50):
set_count += 1
record_count = 0
# Create endpoint URL with each city
city_url = url + "appid=" + api_key + "&units=" + units + "&q=" + city
# Log the url, record, and set numbers
print("Processing Record %s of Set %s | %s" % (record_count, set_count, city))
# Add 1 to the record count
record_count += 1
# Run an API request for each of the cities
try:
# Parse the JSON and retrieve data
city_weather = requests.get(city_url).json()
# Parse out the max temp, humidity, and cloudiness
city_lat = city_weather["coord"]["lat"]
city_lng = city_weather["coord"]["lon"]
city_max_temp = city_weather["main"]["temp_max"]
city_humidity = city_weather["main"]["humidity"]
city_clouds = city_weather["clouds"]["all"]
city_wind = city_weather["wind"]["speed"]
city_country = city_weather["sys"]["country"]
city_date = city_weather["dt"]
# Append the City information into city_data list
city_data.append({"City": city,
"Lat": city_lat,
"Lng": city_lng,
"Max Temp": city_max_temp,
"Humidity": city_humidity,
"Cloudiness": city_clouds,
"Wind Speed": city_wind,
"Country": city_country,
"Date": city_date})
# If an error is experienced, skip the city
except:
print("Does not exist")
pass
# Indicate that Data Loading is complete
print("-----------------------------")
print("Search Complete ")
print("-----------------------------")
#create DF
cdatapd = pd.DataFrame(city_data)
cdatapd.head()
#Check Humidity
cdatapd.describe()
#create seperate df for Humidity
mhcity = cdatapd[(cdatapd["Humidity"] > 100)].index
mhcity
#Seperate even further
ccity = cdatapd.drop(mhcity, inplace=False)
ccity.head()
ccity.columns
lat = ccity["Lat"]
max_temps = ccity["Max Temp"]
humidity = ccity["Humidity"]
cloudiness = ccity["Cloudiness"]
wind_speed = ccity["Wind Speed"]
#Export to Csv
clean_city = ccity.to_csv("output_data.csv", index_label="City_ID")
lat
#Build Scatterplots Lat V Max Temp
plt.scatter(lat, max_temps, edgecolor="black", linewidths=1, marker="o", alpha=0.8, label="Cities")
plt.title("City Lat vs Max Temp")
plt.ylabel("Max Temperature (F)")
plt.xlabel("Latitude")
plt.grid(True)
plt.savefig(r"Latvtemp.png")
plt.show()
#Scatterplot Lat V Humidity
plt.scatter(lat, humidity, edgecolor="black", linewidths=1, marker="o", alpha=0.8, label="Cities")
plt.title("City Lat vs Humidity")
plt.ylabel("Humidity (%)")
plt.xlabel("Latitude")
plt.grid(True)
plt.savefig(r"LatvHumidity.png")
plt.show()
#Scatterplot Lat V Cloudiness
plt.scatter(lat, cloudiness, edgecolor="black", linewidths=1, marker="o", alpha=0.8, label="Cities")
plt.title("City Lat vs Cloudiness")
plt.ylabel("Cloudiness (%)")
plt.xlabel("Latitude")
plt.grid(True)
plt.savefig(r"LatvCloud.png")
plt.show()
#Scatterplot Lat V Wind Speed
plt.scatter(lat, wind_speed, edgecolor="black", linewidths=1, marker="o", alpha=0.8, label="Cities")
plt.title("City Lat vs Wind Speed")
plt.ylabel("Wind Speed (mph)")
plt.xlabel("Latitude")
plt.grid(True)
plt.savefig(r"LatvWind.png")
plt.show()
#Create Linear Regression plots
def plot_linear_regression(x_values, y_values, title, text_coordinates):
# Run regresson on southern hemisphere
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
# Plot
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,text_coordinates,fontsize=15,color="red")
plt.xlabel('Latitude')
plt.ylabel(title)
print(f"The r-value is: {rvalue**2}")
plt.show()
# Northern and Southern
nhemidf = cdatapd.loc[(cdatapd["Lat"] >=0)]
shemidf = cdatapd.loc[(cdatapd["Lat"] < 0)]
# N Lat vs Max Temp
x_values = nhemidf["Lat"]
y_values = nhemidf["Max Temp"]
plot_linear_regression(x_values, y_values, 'Max Temp', (6,30))
#S Lat V Temp
x_values = shemidf["Lat"]
y_values = shemidf["Max Temp"]
plot_linear_regression(x_values, y_values, 'Max Temp', (-55,90))
# N Lat vs Humidity
x_values = nhemidf["Lat"]
y_values = nhemidf["Humidity"]
plot_linear_regression(x_values, y_values, 'Humidity', (40,10))
# S Lat vs Humidity
x_values = shemidf["Lat"]
y_values = shemidf["Humidity"]
plot_linear_regression(x_values, y_values, 'Humidity', (-50,20))
# N Lat vs Cloudiness
x_values = nhemidf["Lat"]
y_values = nhemidf["Cloudiness"]
plot_linear_regression(x_values, y_values, 'Cloudiness', (40,10))
# N Lat vs Cloudiness
x_values = shemidf["Lat"]
y_values = shemidf["Cloudiness"]
plot_linear_regression(x_values, y_values, 'Cloudiness', (-30,30))
# N Lat vs Wind Speed
x_values = nhemidf["Lat"]
y_values = nhemidf["Wind Speed"]
plot_linear_regression(x_values, y_values, 'Wind Speed', (40,10))
# N Lat vs Wind Speed
x_values = shemidf["Lat"]
y_values = shemidf["Wind Speed"]
plot_linear_regression(x_values, y_values, 'Wind Speed', (-30,30))
| 0.391173 | 0.453988 |

# Data Science va Sun'iy Intellekt Praktikum
## Ma'lumotlar tahlili. Pandas kutubxonasi.
### 2.2.3 Foydali funksiyalar
```
import pandas as pd
import numpy as np
data = {'Yil':[2021, 2020, 2019, 2018, 2017, 2016, 2015, 2010],
'Aholi soni':[33.9, 33.5, 32.9, 32.5, 31.9, 31.4, 30.9, 28.5],
'Temp':[1.54, 1.48, 1.56, 1.62, 1.65, 1.66, 1.64, 1.53],
'Zichlik':[79.77, 78.68, 77.53, 76.34, 75.13, 73.91, 72.71, 67.03]}
df = pd.DataFrame(data)
df
df.index = [21, 20, 19, 18, 17, 16, 15, 10]
df
```
## `.reindex()` indekslar o'rnini almashtirish
```
df.reindex([10,15,20])
```
`.reindex` yordamida yangi indekslar qo'shish va ularni turli usullar (method) yordamida to'ldirish ham mumkin
```
df.reindex(index=range(10,22), method="nearest") # method='ffill'
```
Nafaqat indekslarni balki ustunlarni ham qayta indekslash mumkin
```
df.reindex(columns=['Aholi soni','Temp'])
```
### `.loc` yordamida indekslash
```
df.loc[[10,15,20],['Aholi soni','Temp']]
```
**DIQQAT! DataFrame metodlati asl DataFrameni o'zgartirmaydi, balki yangi (o'zgartirilgan) DataFrame qaytaradi.**
## `DataFrame`dan elementlarni o'chirish. `.drop()`
Dataframedan biror qator yoki ustunni o'chirish uchun qator (ustun) indeksini bilish kifoya.
```
df
df_new = df.drop(10) # 10 indeksini o'chiramiz va yangi df yaratamiz
df_new
df.drop([15, 20]) # 15 va 20 qatorlarini tashlab yuboramiz
```
`.drop()` yordamida nafaqat qatorlarni, balki ustunlarni ham o'chirish mumkin. Buning `.drop()` metodining `axis` parametri ishlatiladi.
```
df.drop('Temp', axis=1) # axis=1 ustunni anglatadi, axis=0 esa qatorni
df.drop('Temp', axis='columns') # bu usul ham yuqoridagi natijani beradi
```
Yana bir bor eslatamiz, metodlar asl `DataFrame`ni o'zgartirmaydi.
```
df
```
Lekin buning ham imkoni bor. Odatda `.drop()` kabi metodlar maxsus `inplace` parametri bilan keladi va bu parametr standart `False` qiymatga ega bo'ladi (`inplace=False`)
```
df.drop('Yil', axis=1, inplace=True)
df.head() # E'tibot bering endi Yil ustini yo'q
```
### Indekslash, qator (ustun) tanlash, filtrlash
### `Series`
```
data = df['Aholi soni'] # Series obyekt yaratib olamiz
data
data[0:2] # 0 va 1 elementlar qaytadi
data[[21]]
data[[21,19,17]]
obj = pd.Series(np.arange(8.), index=['a', 'b', 'c', 'd', 'e', 'f', 'g', 'h'])
obj
obj[['b']]
type(obj[['b']])
obj['b']
type(obj['b'])
obj[0:4] # 0,1,2,3 indeksdagi qiymatlar 4 kirmaydi
obj['a':'f'] # a,b,c,d,e,f chiqadi.
obj[['c','d','f']]
obj[[1,5]]
obj['a':'d']=22
obj
obj[obj==22]
obj[obj<22]
```
### `DataFrame`
```
uz = {2020:33.5, 2019:32.9, 2018:32.5, 2017:31.9, 2016:31.4, 2015:30.9}
kz = {2020:18.8, 2019:18.5, 2018:18.3, 2017:18.1, 2016:17.8, 2015:17.6}
tj = {2020:9.5, 2019:9.3, 2018:9.1, 2017:8.9, 2016:8.7, 2015:8.5}
kg = {2020:6.5, 2019:6.4, 2018:6.3, 2017:6.2, 2016:6.1, 2015:6.0}
tr = {2020:6.0, 2019:5.9, 2018:5.9, 2017:5.8, 2016:5.6, 2015:5.6}
df = pd.DataFrame(
{'Uzbekistan':uz,
'Kazakhstan':kz,
'Tajikistan':tj,
'Kyrgizstan':kg,
'Turkmenistan':tr
}
)
df
df['Uzbekistan']
df[['Uzbekistan']]
df[['Uzbekistan','Turkmenistan']]
df[3:]
df>8
df[['Uzbekistan']][df['Uzbekistan']>32]
df2=df.transpose(copy=True)
df2
df2[df2[2015]>6]
df2
df2[df2<8] = 0
df2
df2[[2020]]
```
## `.loc` va `.iloc`
`.loc` yordamida qator va ustunlani ularning nomi bo'yicha tanlash mumkin.
```
df
df.loc[[2020]]
df.loc[[2017,2020]]
df.loc[[2020,2019],['Uzbekistan','Kazakhstan']]
```
`.iloc` yordamida qator va ustunlani ularning tartib raqami bo'yicha tanlash mumkin
```
df.iloc[[0,2,4]]
df.iloc[[0],[0,3,4]]
df.at[2017, 'Uzbekistan']
df.iat[0, 1]
df.loc[:2017,'Uzbekistan']
```
## Sonli indekslar
`pandas` obyektlariga indeks orqali murojaat qilish biroz mujmal tuyulishi mumkin. Bunga sabab biz o'rganib qolgan list va tuple ma'lumot turlari bilan ishlashdagi farq.
Misol uchun, Pythonda ro'yxatning oxirgi elementini ko'rish uchun -1 indeksi orqali murojaat qilishimiz mumkin:
```
nums = list(range(0,11))
nums
nums[-1] # nums oxirgi elementi
```
Yuqoridagi usulni sonli indeksga ega `Series` ma'lumot tuzilmasiga qo'llab ko'ramiz:
```
nums = pd.Series(np.arange(10.))
nums
nums[-1]
```
Ko'rib turganingizdek bu usul raqamli indekslar bilan ishlamas ekan. Agar indesklar raqamli emas, matn bo'lsachi?
```
ser = pd.Series(np.arange(5.), index=['a', 'b', 'c', 'd', 'e'])
ser
ser[-1]
```
Bu holda esa xato yo'q.
Muammoning yechimi doimo `.loc` yoki `.iloc` parametrlaridan foydalanish
```
nums.iloc[-1]
nums.iloc[4]
ser.loc['d']
```
|
github_jupyter
|
import pandas as pd
import numpy as np
data = {'Yil':[2021, 2020, 2019, 2018, 2017, 2016, 2015, 2010],
'Aholi soni':[33.9, 33.5, 32.9, 32.5, 31.9, 31.4, 30.9, 28.5],
'Temp':[1.54, 1.48, 1.56, 1.62, 1.65, 1.66, 1.64, 1.53],
'Zichlik':[79.77, 78.68, 77.53, 76.34, 75.13, 73.91, 72.71, 67.03]}
df = pd.DataFrame(data)
df
df.index = [21, 20, 19, 18, 17, 16, 15, 10]
df
df.reindex([10,15,20])
df.reindex(index=range(10,22), method="nearest") # method='ffill'
df.reindex(columns=['Aholi soni','Temp'])
df.loc[[10,15,20],['Aholi soni','Temp']]
df
df_new = df.drop(10) # 10 indeksini o'chiramiz va yangi df yaratamiz
df_new
df.drop([15, 20]) # 15 va 20 qatorlarini tashlab yuboramiz
df.drop('Temp', axis=1) # axis=1 ustunni anglatadi, axis=0 esa qatorni
df.drop('Temp', axis='columns') # bu usul ham yuqoridagi natijani beradi
df
df.drop('Yil', axis=1, inplace=True)
df.head() # E'tibot bering endi Yil ustini yo'q
data = df['Aholi soni'] # Series obyekt yaratib olamiz
data
data[0:2] # 0 va 1 elementlar qaytadi
data[[21]]
data[[21,19,17]]
obj = pd.Series(np.arange(8.), index=['a', 'b', 'c', 'd', 'e', 'f', 'g', 'h'])
obj
obj[['b']]
type(obj[['b']])
obj['b']
type(obj['b'])
obj[0:4] # 0,1,2,3 indeksdagi qiymatlar 4 kirmaydi
obj['a':'f'] # a,b,c,d,e,f chiqadi.
obj[['c','d','f']]
obj[[1,5]]
obj['a':'d']=22
obj
obj[obj==22]
obj[obj<22]
uz = {2020:33.5, 2019:32.9, 2018:32.5, 2017:31.9, 2016:31.4, 2015:30.9}
kz = {2020:18.8, 2019:18.5, 2018:18.3, 2017:18.1, 2016:17.8, 2015:17.6}
tj = {2020:9.5, 2019:9.3, 2018:9.1, 2017:8.9, 2016:8.7, 2015:8.5}
kg = {2020:6.5, 2019:6.4, 2018:6.3, 2017:6.2, 2016:6.1, 2015:6.0}
tr = {2020:6.0, 2019:5.9, 2018:5.9, 2017:5.8, 2016:5.6, 2015:5.6}
df = pd.DataFrame(
{'Uzbekistan':uz,
'Kazakhstan':kz,
'Tajikistan':tj,
'Kyrgizstan':kg,
'Turkmenistan':tr
}
)
df
df['Uzbekistan']
df[['Uzbekistan']]
df[['Uzbekistan','Turkmenistan']]
df[3:]
df>8
df[['Uzbekistan']][df['Uzbekistan']>32]
df2=df.transpose(copy=True)
df2
df2[df2[2015]>6]
df2
df2[df2<8] = 0
df2
df2[[2020]]
df
df.loc[[2020]]
df.loc[[2017,2020]]
df.loc[[2020,2019],['Uzbekistan','Kazakhstan']]
df.iloc[[0,2,4]]
df.iloc[[0],[0,3,4]]
df.at[2017, 'Uzbekistan']
df.iat[0, 1]
df.loc[:2017,'Uzbekistan']
nums = list(range(0,11))
nums
nums[-1] # nums oxirgi elementi
nums = pd.Series(np.arange(10.))
nums
nums[-1]
ser = pd.Series(np.arange(5.), index=['a', 'b', 'c', 'd', 'e'])
ser
ser[-1]
nums.iloc[-1]
nums.iloc[4]
ser.loc['d']
| 0.169819 | 0.859605 |
```
%matplotlib notebook
from sdss import SDSS
import mikkel_tools.utility as mt_util
import numpy as np
from math import inf
from scipy.optimize import curve_fit
import scipy as sp
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import time
import pyshtools
import pickle
comment = "Synthetic core with tapered ensemble as prior"
nb_name = "nb_case_core_synthetic_Ce1"
shc_g = 30
shc_grid = 30
core = SDSS(comment, N_SH = shc_g, sim_type = "core_ens", sat_height = 350)
core.grid_glq(nmax = shc_grid, r_at = core.r_cmb)
grid_in = np.array([core.grid_phi, 90-core.grid_theta]).T
core.make_grid(core.r_cmb, grid_in, calc_sph_d = False)
core.generate_map()
core.condtab(normsize=10000, quantiles = 1000,
rangn_lim = 3.5, rangn_N = 501, rangv_lim = 2.0, rangv_N = 101, model_hist = "ensemble") #model_hist = "ensemble"
```
# Covariance model
```
core.cov_model_taper(r_at = core.r_cmb, tap_to = 500, tap_exp_p1 = 5, tap_exp_p2 = 2,
tap_scale_start = 0, tap_scale_end = 24, plot_taper = True,
save_fig = False, save_string = "case_core_synthetic", save_dpi = 300)
```
# Synthetic sat
```
C_e_const = 1.0
s_sat = SDSS(comment, N_SH = shc_g, sim_type = "core", sat_height = 350, N_SH_secondary = None)
s_sat.load_swarm("A")
grid_in = np.array([s_sat.grid_phi, 90-s_sat.grid_theta]).T
s_sat.make_grid(s_sat.grid_radial, grid_in, calc_sph_d = False)
s_sat.generate_map(grid_type = "swarm")
s_sat.data += np.random.normal(scale = 2.0, size = s_sat.data.shape)
```
# Source truth
```
s_source = SDSS(comment, N_SH = shc_g, sim_type = "core", sat_height = 350, N_SH_secondary = None)
s_source.grid_glq(nmax = shc_grid, r_at = core.r_cmb)
grid_in = np.array([s_source.grid_phi, 90-s_source.grid_theta]).T
s_source.make_grid(s_source.r_cmb, grid_in, calc_sph_d = False)
s_source.generate_map()
```
# System equations
```
core.integrating_kernel(s_sat, C_e_const = C_e_const, C_mm_supply = core.C_ens_tap)
```
# SDSSIM
```
N_sim = 100
core.target_var = np.max(core.C_ens_tap)
core.run_sim(N_sim, core.grid_N, core.C_mm_all, core.C_dd, core.C_dm_all, core.G,
s_sat.data, core.data, scale_m_i = True, unit_d = False, collect_all = True,
sense_running_error = True, save_string = nb_name, sim_stochastic = False, solve_cho = True)
core.realization_to_sh_coeff(core.r_cmb, set_nmax = shc_grid)
#core.covmod_lsq_equiv(s_sat.data, C_Br_model, core.G, core.r_cmb)
# Reload plot module when making small changes
import importlib
importlib.reload(mt_util)
#truth_obj = s_source
core.grid_glq(nmax = shc_grid, r_at = core.r_cmb)
#m_mode = m_DSS_mode
mt_util.plot_sdssim_reproduce(core, core.m_DSS_res, m_equiv_lsq = None, truth_obj = s_source,
lags_use = 1000, spec_r_at = core.r_cmb, spec_show_differences = False,
spec_ti_ens = True, lwidth = 0.6, lwidth_div = 3, lwidth_mult = 2,
label_fontsize = "small",
res_use = True, sv_use = False, unit_field = "[mT]", hist_ti_ens_limit = [-6,6],
unit_transform_n_to_m = True, patch_legend = True, ens_prior = True,
model_dict = {}, figsize=(9,16), hist_ti_ens = "all", hist_density = False,
hist_bins = 21, res_bins = 21, hist_pos_mean = False,
left=0.08, bottom=0.12, right=0.92, top=0.95, wspace = 0.2, hspace=0.25,
savefig = False, save_string = "case_core_synthetic",
save_dpi = 100, save_path = "images/")
print(core)
core.pickle_save_self(nb_name)
list_coord = np.array([[0,2], [-30,30], [45,-45], [70,-170]])
list_coord[:,0] = 90 - list_coord[:,0]
list_coord[:,1][list_coord[:,1]<0.0] = 360 + list_coord[:,1][list_coord[:,1]<0.0]
m_coord_sph = np.hstack((90 - core.lat.reshape(-1,1), core.lon.reshape(-1,1)))
idx_min = []
for coord in list_coord:
idx_min.append(np.sum(np.abs(m_coord_sph - coord),axis=1).argmin())
print(idx_min)
m_hists_coord = m_coord_sph[idx_min]
m_hists = core.m_DSS[idx_min,:]
left=0.08
bottom=0.12
right=0.92
top=0.95
wspace = 0.2
hspace=0.25
color_rgb_zesty_pos = (1.0, 0.5372549019607843, 0.30196078431372547)
color_rgb_zesty_neg = (0.5019607843137255, 0.6862745098039216, 1.0)
m_hists_scale = m_hists*10**(-6)
tile_size_row = 2
tile_size_column = 2
label_fontsize = 10
fig = plt.figure(figsize=(9,9)) # Initiate figure with constrained layout
# Generate ratio lists
h_ratio = [1]*tile_size_row
w_ratio = [1]*tile_size_column
gs = fig.add_gridspec(tile_size_row, tile_size_column, height_ratios=h_ratio, width_ratios=w_ratio) # Add x-by-y grid
for i in np.arange(0,list_coord.shape[0]):
ax = fig.add_subplot(gs[i])
y,binEdges=np.histogram(m_hists_scale[i,:],bins=11,density=True)
bincenters = 0.5*(binEdges[1:]+binEdges[:-1])
ax.plot(bincenters, y, '-', color = color_rgb_zesty_neg,
label='{}'.format(str(np.round(m_hists_coord[i,:],decimals=1))).lstrip('[').rstrip(']'),
linewidth = 1)
#ax.set_title('test')
#ax.annotate("test", (0.05, 0.5), xycoords='axes fraction', va='center', fontsize = label_fontsize)
ax.set_xlabel("Field value [mT]")
ax.set_ylabel("PDF")
ax.legend(loc='best', fontsize = label_fontsize)
fig.subplots_adjust(left=left, bottom=bottom, right=right, top=top, wspace=wspace, hspace=hspace)
#core.grid_glq(nmax = 256, r_at = core.r_cmb)
#core.grid_glq(nmax = 120, r_at = core.r_cmb)
core.grid_glq(nmax = 400, r_at = core.r_cmb)
set_nmax = shc_grid
core.ensemble_B(core.g_spec, nmax = set_nmax, r_at = core.r_cmb, grid_type = "glq")
zs_eqa = core.B_ensemble[:,0,:].copy()
#core.g_spec_mean = np.mean(core.g_spec,axis=1)
core.ensemble_B(core.g_spec_mean, nmax = set_nmax, r_at = core.r_cmb, grid_type = "glq")
zs_mean_eqa = core.B_ensemble[:,0].copy()
#core.ensemble_B(core.g_prior[:mt_util.shc_vec_len(set_nmax)], nmax = set_nmax, r_at = core.r_cmb, grid_type = "glq")
core.ensemble_B(s_sat.g_prior[:mt_util.shc_vec_len(set_nmax)], nmax = set_nmax, r_at = core.r_cmb, grid_type = "glq")
prior_eqa = core.B_ensemble[:,0].copy()
#core.ensemble_B(core.g_lsq_equiv, nmax = set_nmax, r_at = core.r_cmb, grid_type = "glq")
#lsq_eqa = core.B_ensemble[:,0].copy()
# Reload plot module when making small changes
import importlib
importlib.reload(mt_util)
# ccrs.PlateCarree()
# ccrs.Mollweide()
# ccrs.Orthographic(central_longitude=0.0, central_latitude=0.0)
mt_util.plot_ensemble_map_tiles(core.grid_phi, 90-core.grid_theta, zs_eqa,
field_compare = prior_eqa, field_lsq = None, field_mean = zs_mean_eqa,
tile_size_row = 3, tile_size_column = 2,
figsize=(9,12), limit_for_SF = 10**6, point_size = 0.1, cbar_mm_factor = 1, cbar_limit = [-1.6,1.6],
coast_width = 0.4, coast_color = "grey", unit_transform_n_to_m = True,
cbar_h = 0.1, cbar_text = "mT", cbar_text_color = "black",
left=0.03, bottom=0.12, right=0.97, top=0.95, wspace = 0.05, hspace=0.25,
savefig = False, save_string = "case_core_synthetic",
projection = ccrs.Mollweide(), use_gridlines = True,
gridlines_width = 0.4, gridlines_alpha = 0.4, save_dpi = 100)
```
|
github_jupyter
|
%matplotlib notebook
from sdss import SDSS
import mikkel_tools.utility as mt_util
import numpy as np
from math import inf
from scipy.optimize import curve_fit
import scipy as sp
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import time
import pyshtools
import pickle
comment = "Synthetic core with tapered ensemble as prior"
nb_name = "nb_case_core_synthetic_Ce1"
shc_g = 30
shc_grid = 30
core = SDSS(comment, N_SH = shc_g, sim_type = "core_ens", sat_height = 350)
core.grid_glq(nmax = shc_grid, r_at = core.r_cmb)
grid_in = np.array([core.grid_phi, 90-core.grid_theta]).T
core.make_grid(core.r_cmb, grid_in, calc_sph_d = False)
core.generate_map()
core.condtab(normsize=10000, quantiles = 1000,
rangn_lim = 3.5, rangn_N = 501, rangv_lim = 2.0, rangv_N = 101, model_hist = "ensemble") #model_hist = "ensemble"
core.cov_model_taper(r_at = core.r_cmb, tap_to = 500, tap_exp_p1 = 5, tap_exp_p2 = 2,
tap_scale_start = 0, tap_scale_end = 24, plot_taper = True,
save_fig = False, save_string = "case_core_synthetic", save_dpi = 300)
C_e_const = 1.0
s_sat = SDSS(comment, N_SH = shc_g, sim_type = "core", sat_height = 350, N_SH_secondary = None)
s_sat.load_swarm("A")
grid_in = np.array([s_sat.grid_phi, 90-s_sat.grid_theta]).T
s_sat.make_grid(s_sat.grid_radial, grid_in, calc_sph_d = False)
s_sat.generate_map(grid_type = "swarm")
s_sat.data += np.random.normal(scale = 2.0, size = s_sat.data.shape)
s_source = SDSS(comment, N_SH = shc_g, sim_type = "core", sat_height = 350, N_SH_secondary = None)
s_source.grid_glq(nmax = shc_grid, r_at = core.r_cmb)
grid_in = np.array([s_source.grid_phi, 90-s_source.grid_theta]).T
s_source.make_grid(s_source.r_cmb, grid_in, calc_sph_d = False)
s_source.generate_map()
core.integrating_kernel(s_sat, C_e_const = C_e_const, C_mm_supply = core.C_ens_tap)
N_sim = 100
core.target_var = np.max(core.C_ens_tap)
core.run_sim(N_sim, core.grid_N, core.C_mm_all, core.C_dd, core.C_dm_all, core.G,
s_sat.data, core.data, scale_m_i = True, unit_d = False, collect_all = True,
sense_running_error = True, save_string = nb_name, sim_stochastic = False, solve_cho = True)
core.realization_to_sh_coeff(core.r_cmb, set_nmax = shc_grid)
#core.covmod_lsq_equiv(s_sat.data, C_Br_model, core.G, core.r_cmb)
# Reload plot module when making small changes
import importlib
importlib.reload(mt_util)
#truth_obj = s_source
core.grid_glq(nmax = shc_grid, r_at = core.r_cmb)
#m_mode = m_DSS_mode
mt_util.plot_sdssim_reproduce(core, core.m_DSS_res, m_equiv_lsq = None, truth_obj = s_source,
lags_use = 1000, spec_r_at = core.r_cmb, spec_show_differences = False,
spec_ti_ens = True, lwidth = 0.6, lwidth_div = 3, lwidth_mult = 2,
label_fontsize = "small",
res_use = True, sv_use = False, unit_field = "[mT]", hist_ti_ens_limit = [-6,6],
unit_transform_n_to_m = True, patch_legend = True, ens_prior = True,
model_dict = {}, figsize=(9,16), hist_ti_ens = "all", hist_density = False,
hist_bins = 21, res_bins = 21, hist_pos_mean = False,
left=0.08, bottom=0.12, right=0.92, top=0.95, wspace = 0.2, hspace=0.25,
savefig = False, save_string = "case_core_synthetic",
save_dpi = 100, save_path = "images/")
print(core)
core.pickle_save_self(nb_name)
list_coord = np.array([[0,2], [-30,30], [45,-45], [70,-170]])
list_coord[:,0] = 90 - list_coord[:,0]
list_coord[:,1][list_coord[:,1]<0.0] = 360 + list_coord[:,1][list_coord[:,1]<0.0]
m_coord_sph = np.hstack((90 - core.lat.reshape(-1,1), core.lon.reshape(-1,1)))
idx_min = []
for coord in list_coord:
idx_min.append(np.sum(np.abs(m_coord_sph - coord),axis=1).argmin())
print(idx_min)
m_hists_coord = m_coord_sph[idx_min]
m_hists = core.m_DSS[idx_min,:]
left=0.08
bottom=0.12
right=0.92
top=0.95
wspace = 0.2
hspace=0.25
color_rgb_zesty_pos = (1.0, 0.5372549019607843, 0.30196078431372547)
color_rgb_zesty_neg = (0.5019607843137255, 0.6862745098039216, 1.0)
m_hists_scale = m_hists*10**(-6)
tile_size_row = 2
tile_size_column = 2
label_fontsize = 10
fig = plt.figure(figsize=(9,9)) # Initiate figure with constrained layout
# Generate ratio lists
h_ratio = [1]*tile_size_row
w_ratio = [1]*tile_size_column
gs = fig.add_gridspec(tile_size_row, tile_size_column, height_ratios=h_ratio, width_ratios=w_ratio) # Add x-by-y grid
for i in np.arange(0,list_coord.shape[0]):
ax = fig.add_subplot(gs[i])
y,binEdges=np.histogram(m_hists_scale[i,:],bins=11,density=True)
bincenters = 0.5*(binEdges[1:]+binEdges[:-1])
ax.plot(bincenters, y, '-', color = color_rgb_zesty_neg,
label='{}'.format(str(np.round(m_hists_coord[i,:],decimals=1))).lstrip('[').rstrip(']'),
linewidth = 1)
#ax.set_title('test')
#ax.annotate("test", (0.05, 0.5), xycoords='axes fraction', va='center', fontsize = label_fontsize)
ax.set_xlabel("Field value [mT]")
ax.set_ylabel("PDF")
ax.legend(loc='best', fontsize = label_fontsize)
fig.subplots_adjust(left=left, bottom=bottom, right=right, top=top, wspace=wspace, hspace=hspace)
#core.grid_glq(nmax = 256, r_at = core.r_cmb)
#core.grid_glq(nmax = 120, r_at = core.r_cmb)
core.grid_glq(nmax = 400, r_at = core.r_cmb)
set_nmax = shc_grid
core.ensemble_B(core.g_spec, nmax = set_nmax, r_at = core.r_cmb, grid_type = "glq")
zs_eqa = core.B_ensemble[:,0,:].copy()
#core.g_spec_mean = np.mean(core.g_spec,axis=1)
core.ensemble_B(core.g_spec_mean, nmax = set_nmax, r_at = core.r_cmb, grid_type = "glq")
zs_mean_eqa = core.B_ensemble[:,0].copy()
#core.ensemble_B(core.g_prior[:mt_util.shc_vec_len(set_nmax)], nmax = set_nmax, r_at = core.r_cmb, grid_type = "glq")
core.ensemble_B(s_sat.g_prior[:mt_util.shc_vec_len(set_nmax)], nmax = set_nmax, r_at = core.r_cmb, grid_type = "glq")
prior_eqa = core.B_ensemble[:,0].copy()
#core.ensemble_B(core.g_lsq_equiv, nmax = set_nmax, r_at = core.r_cmb, grid_type = "glq")
#lsq_eqa = core.B_ensemble[:,0].copy()
# Reload plot module when making small changes
import importlib
importlib.reload(mt_util)
# ccrs.PlateCarree()
# ccrs.Mollweide()
# ccrs.Orthographic(central_longitude=0.0, central_latitude=0.0)
mt_util.plot_ensemble_map_tiles(core.grid_phi, 90-core.grid_theta, zs_eqa,
field_compare = prior_eqa, field_lsq = None, field_mean = zs_mean_eqa,
tile_size_row = 3, tile_size_column = 2,
figsize=(9,12), limit_for_SF = 10**6, point_size = 0.1, cbar_mm_factor = 1, cbar_limit = [-1.6,1.6],
coast_width = 0.4, coast_color = "grey", unit_transform_n_to_m = True,
cbar_h = 0.1, cbar_text = "mT", cbar_text_color = "black",
left=0.03, bottom=0.12, right=0.97, top=0.95, wspace = 0.05, hspace=0.25,
savefig = False, save_string = "case_core_synthetic",
projection = ccrs.Mollweide(), use_gridlines = True,
gridlines_width = 0.4, gridlines_alpha = 0.4, save_dpi = 100)
| 0.486332 | 0.688289 |
```
from oas_dev.util.plot.plot_maps import plot_map_diff, fix_axis4map_plot, plot_map_abs_abs_diff, plot_map
from useful_scit.imps import (np, xr, plt, pd)
from oas_dev.util.imports import get_averaged_fields
from IPython.display import clear_output
# load and autoreload
from IPython import get_ipython
# noinspection PyBroadException
try:
_ipython = get_ipython()
_magic = _ipython.magic
_magic('load_ext autoreload')
_magic('autoreload 2')
except:
pass
from oas_dev.util.slice_average.avg_pkg import average_model_var
from oas_dev.data_info import get_nice_name_case
```
## Ideas:
- Root mean square diffence??
- Scatter plots of all values, e.g x-- sectional y-- non sectional color by lat/lev? Or lev lat difference.
# Map plots number concentration:
```
model = 'NorESM'
startyear = '2008-01'
endyear = '2014-12'
pmin = 850. # minimum pressure level
avg_over_lev = True # True#True#False#True
pressure_adjust = True # Can only be false if avg_over_lev false. Plots particular hybrid sigma lev
if avg_over_lev:
pressure_adjust = True
p_levels = [1013.,900., 800., 700., 600.] # used if not avg
```
## Cases
```
cases_sec = ['SECTv21_ctrl_koagD']
cases_orig =['noSECTv21_default']
cases_orig =['noSECTv21_ox_ricc_dd']
cases = cases_orig + cases_sec
cases2 = ['noSECTv21_default_dd']+cases_sec #['noSECTv11_ctrl_fbvoc', 'noSECTv11_noresm2_ctrl']
cases_all = cases_sec + cases_orig + ['noSECTv21_default_dd']
def load_and_plot(var, cases,startyear, endyear,
avg_over_lev=avg_over_lev,
pmin=pmin,
pressure_adjust=pressure_adjust, p_level=None, relative=False, kwargs_diff=None):
maps_dic = get_averaged_fields.get_maps_cases(cases,[var],startyear, endyear,
avg_over_lev=avg_over_lev,
pmin=pmin,
pressure_adjust=pressure_adjust, p_level=p_level)
return plot_map_abs_abs_diff(var, cases, maps_dic, relative=relative, figsize=[18, 3], cbar_equal=True,
kwargs_abs={},
kwargs_diff=kwargs_diff, axs=None, cmap_abs='Reds', cmap_diff='RdBu_r')
```
## Mean to 850hPa weighted by pressure difference:
```
so4_spess_fac = dict(SO4_A1=3.06,
SO4_A2=3.59,
SO4_AC=3.06,
SO4_NA=3.06,
SO4_PR=3.06,
SO4_A1_OCW=3.06,
SO4_A2_OCW=3.59,
SO4_AC_OCW=3.06,
SO4_NA_OCW=3.06,
SO4_PR_OCW=3.06
)
so4_spess = list(so4_spess_fac.keys())
soa_spess = [
'SOA_NA',
'OM_AI',
'OM_AC',
'OM_NI',
'SOA_NA_OCW',
'OM_AI_OCW',
'OM_AC_OCW',
'OM_NI_OCW'
]
soa_spess_fac = {s:1 for s in soa_spess}
import itertools
core_vl = soa_spess + so4_spess
var_ext = ["DDF","SFWET"]
varl = [f'cb_{v}' for v in core_vl]
varl = varl + [f'{v}{ext}' for (v,ext) in itertools.product(core_vl, var_ext)]
varl
#var_ext = [f"{v}DDF",f"{v}SFWET",f"{v}SFSIC",f"{v}SFSBC",f"{v}SFSIS",f"{v}SFSBS"
# , f"{v}_mixnuc1"]
v='SO4_NA'
#varl=[]
for v in ['SOA_NA','SO4_NA']:#, 'SOA_NA_OCW','SO4_NA_OCW']:
varl = [f'{v}coagTend',f'{v}clcoagTend',f'{v}condTend']+ varl
# f"{v}SFSIC",f"{v}SFSBC",f"{v}SFSIS",f"{v}SFSBS", f"{v}_mixnuc1",
"""
for v in [ 'SOA_NA_OCW','SO4_NA_OCW']:
varl=varl+ [f'cb_{v}']#'LWDIR_Ghan']#, 'SO4_NAcondTend']#, 'leaveSecH2SO4','leaveSecSOA']#,'TGCLDCWP']
varl = [f"{v}DDF",f"{v}SFWET"]+ varl
"""
maps_dic = get_averaged_fields.get_maps_cases(cases_all,varl,startyear, endyear,
avg_over_lev=avg_over_lev,
pmin=pmin,
pressure_adjust=pressure_adjust)#, p_level=p_level)
def calc_tot_LR(ds,v):
return (-ds[f'{v}DDF'] + ds[f'{v}SFWET'] + ds[f'{v}coagTend'] + ds[f'{v}clcoagTend'])
def LR_dd_wd(ds,v):
return (-ds[f'{v}DDF'] + ds[f'{v}SFWET'])# + ds[f'{v}coagTend'] + ds[f'{v}clcoagTend'])
def comp_lifetime(ds, which, fac_dic ):
lossrate_OCW_DD = 0
lossrate_OCW_WD = 0
lossrate_nOCW_DD = 0
lossrate_nOCW_WD = 0
cb_OCW = 0
cb_nOCW = 0
for v in fac_dic.keys():
f = fac_dic[v]
if '_OCW' in v:
cb_OCW = f*ds[f'cb_{v}'] + cb_OCW
lossrate_OCW_DD = f*(-ds[f'{v}DDF']) + lossrate_OCW_DD
lossrate_OCW_WD = f*(ds[f'{v}SFWET']) + lossrate_OCW_WD
else:
cb_nOCW = f*ds[f'cb_{v}'] + cb_nOCW
lossrate_nOCW_DD = f*(-ds[f'{v}DDF']) + lossrate_nOCW_DD
lossrate_nOCW_WD = f*(ds[f'{v}SFWET']) + lossrate_nOCW_WD
ds[f'cb_{which}'] = cb_nOCW
ds[f'cb_{which}_OCW'] = cb_OCW
ds[f'cb_{which}_tot'] = cb_nOCW + cb_OCW
ds[f'{which}_OCW_DD'] = lossrate_OCW_DD
ds[f'{which}_OCW_WD'] = lossrate_OCW_WD
ds[f'{which}_OCW_D'] = lossrate_OCW_WD + lossrate_OCW_DD
ds[f'{which}_DD'] = lossrate_nOCW_DD
ds[f'{which}_WD'] = lossrate_nOCW_WD
ds[f'{which}_D'] = lossrate_nOCW_WD + lossrate_nOCW_DD
ds[f'{which}_tot_WD'] = lossrate_nOCW_WD + lossrate_OCW_WD
ds[f'{which}_tot_DD'] = lossrate_nOCW_DD + lossrate_OCW_DD
ds[f'{which}_tot_D'] = lossrate_nOCW_DD + lossrate_OCW_DD + lossrate_nOCW_WD + lossrate_OCW_WD
return ds
for case in cases_all:
comp_lifetime(maps_dic[case], 'OA', soa_spess_fac )
comp_lifetime(maps_dic[case], 'SO4', so4_spess_fac )
def comp_lossr(v, ext, _ds):
cb = average_model_var(_ds, f'cb_{v}', area='Global', dim=None, minp=850., time_mask=None)
lr = average_model_var(_ds, f'{v}{ext}', area='Global', dim=None, minp=850., time_mask=None)
out = cb[f'cb_{v}']/lr[f'{v}{ext}']/(60*60*24)
if out<0:
out=abs(out)
out.attrs['units']='days'
return out
from sectional_v2.data_info import get_nice_name_case
exts_dic = {
'_D':'$\tau_{tot}$',
'_DD':'$\tau_{DDF}$',
'_WD':'$\tau_{WET}$',
#'coagTend':'$\tau_{coag}$',
#'clcoagTend':'$\tau_{clcoag}$'
}
dic_all ={}
for var in ['SO4','SO4_OCW','SO4_tot','OA','OA_OCW','OA_tot',]:
dic_all[var]={}
for case in cases_all:
nncase = get_nice_name_case(case)
dic_all[var][nncase]={}
for ext in exts_dic.keys():
val = comp_lossr(var,ext,maps_dic[case])
dic_all[var][nncase][exts_dic[ext]] = val.values
pd.DataFrame.from_dict(dic_all['SO4'])
pd.DataFrame.from_dict(dic_all['SO4_tot'])
pd.DataFrame.from_dict(dic_all['OA_tot'])
pd.DataFrame.from_dict(dic_all['OA'])
pd.DataFrame.from_dict(dic_all['OA_OCW'])
maps_dic[case]
lss_exts = ['DDF','SFWET','coagTend','clcoagTend']
v = 'SOA_NA'
for v in ['SOA_NA','SO4_NA']:
for case in cases_all:
ds = maps_dic[case]
ds[f'{v}_lr_tot'] = -(-ds[f'{v}DDF'] + ds[f'{v}SFWET'] + ds[f'{v}coagTend'] + ds[f'{v}clcoagTend'])
ds[f'{v}_OCW_lr_tot'] = -(-ds[f'{v}_OCWDDF'] + ds[f'{v}_OCWSFWET'])# + ds[f'{v}coagTend'] + ds[f'{v}clcoagTend'])
ds[f'{v}_lr_tot_inc'] =ds[f'{v}_OCW_lr_tot'] + ds[f'{v}_OCW_lr_tot']
ds[f'tau_new_{v}'] = ds[f'cb_{v}']/ds[f'{v}_lr_tot']
for ex in lss_exts:
ds[f'tau_{ex}_{v}'] = (ds[f'cb_{v}']/ds[f'{v}{ex}'])/60/60/24
ds[f'tau_{ex}_{v}'].attrs['units'] = 'days'
ds[f'tau_prod_{v}'] = ds[f'cb_{v}']/ds[f'{v}condTend']/(60*60*24)
ds[f'tau_prod_{v}'].attrs['units'] = 'days'
ds[f'cb_{v}_tot'] = ds[f'cb_{v}']+ ds[f'cb_{v}_OCW']
from sectional_v2.util.slice_average.avg_pkg import average_model_var
from sectional_v2.data_info import get_nice_name_case
def comp_lossr(v, ext, _ds):
cb = average_model_var(_ds, f'cb_{v}', area='Global', dim=None, minp=850., time_mask=None)
lr = average_model_var(_ds, f'{v}{ext}', area='Global', dim=None, minp=850., time_mask=None)
out = cb[f'cb_{v}']/lr[f'{v}{ext}']/(60*60*24)
if out<0:
out=abs(out)
out.attrs['units']='days'
return out
```
## NA-mode lifetime
```
exts_dic = {
'_lr_tot':'$\tau_{tot}$',
'DDF':'$\tau_{DDF}$',
'SFWET':'$\tau_{WET}$',
'coagTend':'$\tau_{coag}$',
'clcoagTend':'$\tau_{clcoag}$'}
dic_all ={}
for var in ['SOA_NA','SO4_NA']:
dic_all[var]={}
for case in cases_all:
nncase = get_nice_name_case(case)
dic_all[var][nncase]={}
for ext in exts_dic.keys():
val = comp_lossr(var,ext,maps_dic[case])
dic_all[var][nncase][exts_dic[ext]] = val.values
pd.DataFrame.from_dict(dic_all['SOA_NA'])
pd.DataFrame.from_dict(dic_all['SO4_NA'])
```
|
github_jupyter
|
from oas_dev.util.plot.plot_maps import plot_map_diff, fix_axis4map_plot, plot_map_abs_abs_diff, plot_map
from useful_scit.imps import (np, xr, plt, pd)
from oas_dev.util.imports import get_averaged_fields
from IPython.display import clear_output
# load and autoreload
from IPython import get_ipython
# noinspection PyBroadException
try:
_ipython = get_ipython()
_magic = _ipython.magic
_magic('load_ext autoreload')
_magic('autoreload 2')
except:
pass
from oas_dev.util.slice_average.avg_pkg import average_model_var
from oas_dev.data_info import get_nice_name_case
model = 'NorESM'
startyear = '2008-01'
endyear = '2014-12'
pmin = 850. # minimum pressure level
avg_over_lev = True # True#True#False#True
pressure_adjust = True # Can only be false if avg_over_lev false. Plots particular hybrid sigma lev
if avg_over_lev:
pressure_adjust = True
p_levels = [1013.,900., 800., 700., 600.] # used if not avg
cases_sec = ['SECTv21_ctrl_koagD']
cases_orig =['noSECTv21_default']
cases_orig =['noSECTv21_ox_ricc_dd']
cases = cases_orig + cases_sec
cases2 = ['noSECTv21_default_dd']+cases_sec #['noSECTv11_ctrl_fbvoc', 'noSECTv11_noresm2_ctrl']
cases_all = cases_sec + cases_orig + ['noSECTv21_default_dd']
def load_and_plot(var, cases,startyear, endyear,
avg_over_lev=avg_over_lev,
pmin=pmin,
pressure_adjust=pressure_adjust, p_level=None, relative=False, kwargs_diff=None):
maps_dic = get_averaged_fields.get_maps_cases(cases,[var],startyear, endyear,
avg_over_lev=avg_over_lev,
pmin=pmin,
pressure_adjust=pressure_adjust, p_level=p_level)
return plot_map_abs_abs_diff(var, cases, maps_dic, relative=relative, figsize=[18, 3], cbar_equal=True,
kwargs_abs={},
kwargs_diff=kwargs_diff, axs=None, cmap_abs='Reds', cmap_diff='RdBu_r')
so4_spess_fac = dict(SO4_A1=3.06,
SO4_A2=3.59,
SO4_AC=3.06,
SO4_NA=3.06,
SO4_PR=3.06,
SO4_A1_OCW=3.06,
SO4_A2_OCW=3.59,
SO4_AC_OCW=3.06,
SO4_NA_OCW=3.06,
SO4_PR_OCW=3.06
)
so4_spess = list(so4_spess_fac.keys())
soa_spess = [
'SOA_NA',
'OM_AI',
'OM_AC',
'OM_NI',
'SOA_NA_OCW',
'OM_AI_OCW',
'OM_AC_OCW',
'OM_NI_OCW'
]
soa_spess_fac = {s:1 for s in soa_spess}
import itertools
core_vl = soa_spess + so4_spess
var_ext = ["DDF","SFWET"]
varl = [f'cb_{v}' for v in core_vl]
varl = varl + [f'{v}{ext}' for (v,ext) in itertools.product(core_vl, var_ext)]
varl
#var_ext = [f"{v}DDF",f"{v}SFWET",f"{v}SFSIC",f"{v}SFSBC",f"{v}SFSIS",f"{v}SFSBS"
# , f"{v}_mixnuc1"]
v='SO4_NA'
#varl=[]
for v in ['SOA_NA','SO4_NA']:#, 'SOA_NA_OCW','SO4_NA_OCW']:
varl = [f'{v}coagTend',f'{v}clcoagTend',f'{v}condTend']+ varl
# f"{v}SFSIC",f"{v}SFSBC",f"{v}SFSIS",f"{v}SFSBS", f"{v}_mixnuc1",
"""
for v in [ 'SOA_NA_OCW','SO4_NA_OCW']:
varl=varl+ [f'cb_{v}']#'LWDIR_Ghan']#, 'SO4_NAcondTend']#, 'leaveSecH2SO4','leaveSecSOA']#,'TGCLDCWP']
varl = [f"{v}DDF",f"{v}SFWET"]+ varl
"""
maps_dic = get_averaged_fields.get_maps_cases(cases_all,varl,startyear, endyear,
avg_over_lev=avg_over_lev,
pmin=pmin,
pressure_adjust=pressure_adjust)#, p_level=p_level)
def calc_tot_LR(ds,v):
return (-ds[f'{v}DDF'] + ds[f'{v}SFWET'] + ds[f'{v}coagTend'] + ds[f'{v}clcoagTend'])
def LR_dd_wd(ds,v):
return (-ds[f'{v}DDF'] + ds[f'{v}SFWET'])# + ds[f'{v}coagTend'] + ds[f'{v}clcoagTend'])
def comp_lifetime(ds, which, fac_dic ):
lossrate_OCW_DD = 0
lossrate_OCW_WD = 0
lossrate_nOCW_DD = 0
lossrate_nOCW_WD = 0
cb_OCW = 0
cb_nOCW = 0
for v in fac_dic.keys():
f = fac_dic[v]
if '_OCW' in v:
cb_OCW = f*ds[f'cb_{v}'] + cb_OCW
lossrate_OCW_DD = f*(-ds[f'{v}DDF']) + lossrate_OCW_DD
lossrate_OCW_WD = f*(ds[f'{v}SFWET']) + lossrate_OCW_WD
else:
cb_nOCW = f*ds[f'cb_{v}'] + cb_nOCW
lossrate_nOCW_DD = f*(-ds[f'{v}DDF']) + lossrate_nOCW_DD
lossrate_nOCW_WD = f*(ds[f'{v}SFWET']) + lossrate_nOCW_WD
ds[f'cb_{which}'] = cb_nOCW
ds[f'cb_{which}_OCW'] = cb_OCW
ds[f'cb_{which}_tot'] = cb_nOCW + cb_OCW
ds[f'{which}_OCW_DD'] = lossrate_OCW_DD
ds[f'{which}_OCW_WD'] = lossrate_OCW_WD
ds[f'{which}_OCW_D'] = lossrate_OCW_WD + lossrate_OCW_DD
ds[f'{which}_DD'] = lossrate_nOCW_DD
ds[f'{which}_WD'] = lossrate_nOCW_WD
ds[f'{which}_D'] = lossrate_nOCW_WD + lossrate_nOCW_DD
ds[f'{which}_tot_WD'] = lossrate_nOCW_WD + lossrate_OCW_WD
ds[f'{which}_tot_DD'] = lossrate_nOCW_DD + lossrate_OCW_DD
ds[f'{which}_tot_D'] = lossrate_nOCW_DD + lossrate_OCW_DD + lossrate_nOCW_WD + lossrate_OCW_WD
return ds
for case in cases_all:
comp_lifetime(maps_dic[case], 'OA', soa_spess_fac )
comp_lifetime(maps_dic[case], 'SO4', so4_spess_fac )
def comp_lossr(v, ext, _ds):
cb = average_model_var(_ds, f'cb_{v}', area='Global', dim=None, minp=850., time_mask=None)
lr = average_model_var(_ds, f'{v}{ext}', area='Global', dim=None, minp=850., time_mask=None)
out = cb[f'cb_{v}']/lr[f'{v}{ext}']/(60*60*24)
if out<0:
out=abs(out)
out.attrs['units']='days'
return out
from sectional_v2.data_info import get_nice_name_case
exts_dic = {
'_D':'$\tau_{tot}$',
'_DD':'$\tau_{DDF}$',
'_WD':'$\tau_{WET}$',
#'coagTend':'$\tau_{coag}$',
#'clcoagTend':'$\tau_{clcoag}$'
}
dic_all ={}
for var in ['SO4','SO4_OCW','SO4_tot','OA','OA_OCW','OA_tot',]:
dic_all[var]={}
for case in cases_all:
nncase = get_nice_name_case(case)
dic_all[var][nncase]={}
for ext in exts_dic.keys():
val = comp_lossr(var,ext,maps_dic[case])
dic_all[var][nncase][exts_dic[ext]] = val.values
pd.DataFrame.from_dict(dic_all['SO4'])
pd.DataFrame.from_dict(dic_all['SO4_tot'])
pd.DataFrame.from_dict(dic_all['OA_tot'])
pd.DataFrame.from_dict(dic_all['OA'])
pd.DataFrame.from_dict(dic_all['OA_OCW'])
maps_dic[case]
lss_exts = ['DDF','SFWET','coagTend','clcoagTend']
v = 'SOA_NA'
for v in ['SOA_NA','SO4_NA']:
for case in cases_all:
ds = maps_dic[case]
ds[f'{v}_lr_tot'] = -(-ds[f'{v}DDF'] + ds[f'{v}SFWET'] + ds[f'{v}coagTend'] + ds[f'{v}clcoagTend'])
ds[f'{v}_OCW_lr_tot'] = -(-ds[f'{v}_OCWDDF'] + ds[f'{v}_OCWSFWET'])# + ds[f'{v}coagTend'] + ds[f'{v}clcoagTend'])
ds[f'{v}_lr_tot_inc'] =ds[f'{v}_OCW_lr_tot'] + ds[f'{v}_OCW_lr_tot']
ds[f'tau_new_{v}'] = ds[f'cb_{v}']/ds[f'{v}_lr_tot']
for ex in lss_exts:
ds[f'tau_{ex}_{v}'] = (ds[f'cb_{v}']/ds[f'{v}{ex}'])/60/60/24
ds[f'tau_{ex}_{v}'].attrs['units'] = 'days'
ds[f'tau_prod_{v}'] = ds[f'cb_{v}']/ds[f'{v}condTend']/(60*60*24)
ds[f'tau_prod_{v}'].attrs['units'] = 'days'
ds[f'cb_{v}_tot'] = ds[f'cb_{v}']+ ds[f'cb_{v}_OCW']
from sectional_v2.util.slice_average.avg_pkg import average_model_var
from sectional_v2.data_info import get_nice_name_case
def comp_lossr(v, ext, _ds):
cb = average_model_var(_ds, f'cb_{v}', area='Global', dim=None, minp=850., time_mask=None)
lr = average_model_var(_ds, f'{v}{ext}', area='Global', dim=None, minp=850., time_mask=None)
out = cb[f'cb_{v}']/lr[f'{v}{ext}']/(60*60*24)
if out<0:
out=abs(out)
out.attrs['units']='days'
return out
exts_dic = {
'_lr_tot':'$\tau_{tot}$',
'DDF':'$\tau_{DDF}$',
'SFWET':'$\tau_{WET}$',
'coagTend':'$\tau_{coag}$',
'clcoagTend':'$\tau_{clcoag}$'}
dic_all ={}
for var in ['SOA_NA','SO4_NA']:
dic_all[var]={}
for case in cases_all:
nncase = get_nice_name_case(case)
dic_all[var][nncase]={}
for ext in exts_dic.keys():
val = comp_lossr(var,ext,maps_dic[case])
dic_all[var][nncase][exts_dic[ext]] = val.values
pd.DataFrame.from_dict(dic_all['SOA_NA'])
pd.DataFrame.from_dict(dic_all['SO4_NA'])
| 0.397471 | 0.728555 |
# RESULTS OF THE SET OF SIMULATIONS
## Loading results
```
%matplotlib notebook
import numpy as np
import scipy.stats as stats
import matplotlib.pyplot as plt
from thermalspin.data_analysis import *
# Insert here the name of the simulation set
setname = "ferro_critic_set"
final_state_lst, L_lst, t_lst, J_lst, h_lst, T_lst, e_lst, m_lst, snp_lst = load_set_results(setname, load_set_snapshots=False)
L, H, t_whole, J_whole, T_whole, E_whole, m_whole, final_state, snp = arrange_set_results_LH(L_lst, t_lst, J_lst, h_lst, T_lst, e_lst, m_lst, final_state_lst)
L_num = t_whole.shape[0]
H_num = t_whole.shape[1]
t_num = t_whole.shape[2]
```
## Global behaviour
```
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
for i,j in np.ndindex(L_num, H_num):
ax.plot(t_whole[i,j], E_whole[i,j], label=f"L = {L[i]}, H = {H[j]}")
box = ax.get_position()
ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("Step")
plt.ylabel("Energy")
plt.grid()
plt.show()
m_magnitude_whole = np.sqrt(np.sum(m_whole**2, axis = 3))
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
for i,j in np.ndindex(L_num, H_num):
ax.plot(t_whole[i,j], m_magnitude_whole[i,j], label=f"L = {L[i]}, H = {H[j]}")
box = ax.get_position()
ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("Step")
plt.ylabel("Absolute magnetization")
plt.grid()
plt.show()
```
## Single ensemble analysis
```
# Insert here index of the ensemble to be analyzed
L_idx = 0
H_idx = 0
fig = plt.figure(figsize=(10,6))
ax = plt.subplot(111)
ax.plot(t_whole[L_idx, H_idx], E_whole[L_idx, H_idx], label=f"L = {L[L_idx]}, H = {H[H_idx]}")
ax.legend()
plt.ylabel("Energy")
plt.xlabel("Steps")
plt.title("Energy")
plt.grid()
plt.show()
fig = plt.figure(figsize=(10,6))
ax = plt.subplot(111)
ax.plot(t_whole[L_idx, H_idx], m_whole[L_idx, H_idx, :, 0], label = r"$M_x$")
ax.plot(t_whole[L_idx, H_idx], m_whole[L_idx, H_idx, :, 1], label = r"$M_y$")
ax.plot(t_whole[L_idx, H_idx], m_whole[L_idx, H_idx, :, 2], label = r"$M_z$")
plt.legend()
plt.ylabel("Magnetization")
plt.xlabel("Steps")
plt.title("Magnetization")
plt.grid()
plt.show()
fig = plt.figure(figsize=(10,6))
ax = plt.subplot(111)
ax.plot(t_whole[L_idx, H_idx], np.abs(m_whole[L_idx, H_idx, :, 0]), label = r"$|M_x|$")
ax.plot(t_whole[L_idx, H_idx], np.abs(m_whole[L_idx, H_idx, :, 1]), label = r"$|M_y|$")
ax.plot(t_whole[L_idx, H_idx], np.abs(m_whole[L_idx, H_idx, :, 2]), label = r"$|M_z|$")
plt.legend()
plt.ylabel("Absolute magnetization")
plt.xlabel("Steps")
plt.title("Absolute magnetization")
plt.grid()
plt.show()
plot_state(final_state[L_idx][H_idx])
plot_spin_directions(final_state[L_idx][H_idx])
```
## Set results
```
# SELECT WARMUP PERIOD TO LAST UNTIL STEP NUMBER warmup_final_step
warmup_final_step = np.argmax(t_whole[0,0]==1e7)
warmup_final_idx = np.argmax(np.equal(t_whole[0,0], warmup_final_step))
t = t_whole[:, :, warmup_final_idx:]
E = E_whole[:, :, warmup_final_idx:]
m = m_whole[:, :, warmup_final_idx:]
E_mean = np.mean(E, axis=2)
E_std = np.sqrt(np.var(E, axis=2))
m_mean = np.mean(m, axis=2)
m_std = np.sqrt(np.var(m, axis=2))
m_magnitude = np.sqrt(np.sum(m**2, axis = 3))
m_magnitude_mean = np.mean(m_magnitude, axis=2)
m_magnitude_std = np.sqrt(np.var(m_magnitude, axis=2))
```
### Mean energy
```
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
for i in np.ndindex(L_num):
ax.errorbar(H, E_mean[i], yerr=[E_std[i]/2, E_std[i]/2], fmt="o--", label=f"L = {L[i]}")
box = ax.get_position()
ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel(r"$H_z$")
plt.ylabel("Mean energy")
plt.grid()
plt.show()
fig.savefig("./plots/"+setname+"_energy.svg")
```
### Mean magnetization magnitude
```
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
for i in np.ndindex(L_num):
ax.errorbar(H, m_magnitude_mean[i], yerr=m_magnitude_std[i]/2, fmt="o--", label=f"L = {L[i]}")
box = ax.get_position()
ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel(r"$H_z$")
plt.ylabel("Mean magnetization magnitude")
plt.grid()
plt.show()
fig.savefig("./plots/"+setname+"_energy.svg")
def cov(M,i,j):
M_mean = np.mean(M, axis=2)
ret = np.zeros(shape=(L_num, 1))
for l,t in np.ndindex(L_num, 1):
ret[l,t] = np.mean((M[l,t,:,i]-M_mean[l,t,i])*(M[l,t,:,j]-M_mean[l,t,j]))
return ret
T=1.445
chi_xx = np.zeros(shape=(L_num, 1))
chi_yy = np.zeros(shape=(L_num, 1))
chi_zz = np.zeros(shape=(L_num, 1))
chi_xy = np.zeros(shape=(L_num, 1))
chi_yz = np.zeros(shape=(L_num, 1))
chi_zx = np.zeros(shape=(L_num, 1))
for i in np.ndindex(L_num):
chi_xx[i] = cov(m,0,0)[i]/T*L[i]**3
chi_yy[i] = cov(m,1,1)[i]/T*L[i]**3
chi_zz[i] = cov(m,2,2)[i]/T*L[i]**3
chi_xy[i] = cov(m,0,1)[i]/T*L[i]**3
chi_yz[i] = cov(m,1,2)[i]/T*L[i]**3
chi_zx[i] = cov(m,2,0)[i]/T*L[i]**3
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
ax.plot(L, chi_zz, "o--")
plt.xlabel(r"$L$")
plt.ylabel(r"$\chi_{zz}$")
plt.title("System susceptibility")
plt.grid()
plt.show()
```
### Critical indices
```
slope, intercept, r_value, p_value, std_err = stats.linregress(np.log(L), np.log(chi_zz[:,0]))
gamma_nu = slope
gamma_nu_u = std_err
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
ax.set_xscale('log')
ax.set_yscale('log')
plt.scatter(L, chi_zz[:,0])
plt.plot(L, np.exp(intercept)*L**(slope), ":")
slope_str="{0:.3}".format(slope)
std_str="{0:.3}".format(std_err)
plt.text(10,65,fr"$slope = {slope_str}\pm{std_str}$", fontsize=11)
plt.grid(True, which="both")
plt.xlabel(r"$L$")
plt.ylabel(r"$\chi_{zz}$")
plt.title(f"T=1.445")
plt.show()
fig.savefig("./plots/"+setname+"_gamma_nu.svg")
slope, intercept, r_value, p_value, std_err = stats.linregress(np.log(L), np.log(m_magnitude_mean[:,0]))
beta_nu = -slope
beta_nu_u = std_err
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
ax.set_xscale('log')
ax.set_yscale('log')
plt.scatter(L, m_magnitude_mean[:,0])
plt.plot(L, np.exp(intercept)*L**(slope), ":")
slope_str="{0:.4}".format(slope)
std_str="{0:.3}".format(std_err)
plt.text(15,3.1e-1,fr"$slope = {slope_str}\pm{std_str}$", fontsize=11)
plt.grid(True, which="both")
plt.xlabel(r"$L$")
plt.ylabel(r"$\langle m\rangle$")
plt.title(f"T=1.445")
plt.show()
fig.savefig("./plots/"+setname+"_beta_nu.svg")
binder = 1 - (1/3)*np.mean(m_magnitude**4, axis=2)/(np.mean(m_magnitude**2, axis=2)**2)
binder
m2 = np.mean(m_magnitude**2, axis=2)
m4 = np.mean(m_magnitude**4, axis=2)
m2E = np.mean(E*m_magnitude**2, axis=2)
m4E = np.mean(E*m_magnitude**4, axis=2)
dbinder = (1-binder)*(E_mean - 2*m2E/m2 + m4E/m4)
dbinder
slope, intercept, r_value, p_value, std_err = stats.linregress(np.log(L), np.log(dbinder[:,0]))
beta_nu = -slope
beta_nu_u = std_err
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
ax.set_xscale('log')
ax.set_yscale('log')
plt.scatter(L, dbinder[:,0])
plt.plot(L, np.exp(intercept)*L**(slope), ":")
slope_str="{0:.3}".format(slope)
std_str="{0:.3}".format(std_err)
plt.text(10,40,fr"$slope = {slope_str} \pm {std_str}$", fontsize=11)
plt.grid(True, which="both")
plt.xlabel(r"$L$")
plt.ylabel(r"$\langle \frac{d U_L}{d \beta } \rangle$")
plt.title(f"T=1.445")
plt.show()
fig.savefig("./plots/"+setname+"_nu.svg")
```
### Spatial correlation
```
scm = spatial_correlation_matrix(final_state[-1][0])
r,c = radial_distribution(scm)
fig = plt.figure(figsize=(10,6))
ax = plt.subplot(111)
ax.plot(r,c)
plt.xlabel(r"$r$")
plt.ylabel(r"$C(r)$")
plt.title("Spatial correlation")
plt.grid()
plt.show()
```
|
github_jupyter
|
%matplotlib notebook
import numpy as np
import scipy.stats as stats
import matplotlib.pyplot as plt
from thermalspin.data_analysis import *
# Insert here the name of the simulation set
setname = "ferro_critic_set"
final_state_lst, L_lst, t_lst, J_lst, h_lst, T_lst, e_lst, m_lst, snp_lst = load_set_results(setname, load_set_snapshots=False)
L, H, t_whole, J_whole, T_whole, E_whole, m_whole, final_state, snp = arrange_set_results_LH(L_lst, t_lst, J_lst, h_lst, T_lst, e_lst, m_lst, final_state_lst)
L_num = t_whole.shape[0]
H_num = t_whole.shape[1]
t_num = t_whole.shape[2]
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
for i,j in np.ndindex(L_num, H_num):
ax.plot(t_whole[i,j], E_whole[i,j], label=f"L = {L[i]}, H = {H[j]}")
box = ax.get_position()
ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("Step")
plt.ylabel("Energy")
plt.grid()
plt.show()
m_magnitude_whole = np.sqrt(np.sum(m_whole**2, axis = 3))
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
for i,j in np.ndindex(L_num, H_num):
ax.plot(t_whole[i,j], m_magnitude_whole[i,j], label=f"L = {L[i]}, H = {H[j]}")
box = ax.get_position()
ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("Step")
plt.ylabel("Absolute magnetization")
plt.grid()
plt.show()
# Insert here index of the ensemble to be analyzed
L_idx = 0
H_idx = 0
fig = plt.figure(figsize=(10,6))
ax = plt.subplot(111)
ax.plot(t_whole[L_idx, H_idx], E_whole[L_idx, H_idx], label=f"L = {L[L_idx]}, H = {H[H_idx]}")
ax.legend()
plt.ylabel("Energy")
plt.xlabel("Steps")
plt.title("Energy")
plt.grid()
plt.show()
fig = plt.figure(figsize=(10,6))
ax = plt.subplot(111)
ax.plot(t_whole[L_idx, H_idx], m_whole[L_idx, H_idx, :, 0], label = r"$M_x$")
ax.plot(t_whole[L_idx, H_idx], m_whole[L_idx, H_idx, :, 1], label = r"$M_y$")
ax.plot(t_whole[L_idx, H_idx], m_whole[L_idx, H_idx, :, 2], label = r"$M_z$")
plt.legend()
plt.ylabel("Magnetization")
plt.xlabel("Steps")
plt.title("Magnetization")
plt.grid()
plt.show()
fig = plt.figure(figsize=(10,6))
ax = plt.subplot(111)
ax.plot(t_whole[L_idx, H_idx], np.abs(m_whole[L_idx, H_idx, :, 0]), label = r"$|M_x|$")
ax.plot(t_whole[L_idx, H_idx], np.abs(m_whole[L_idx, H_idx, :, 1]), label = r"$|M_y|$")
ax.plot(t_whole[L_idx, H_idx], np.abs(m_whole[L_idx, H_idx, :, 2]), label = r"$|M_z|$")
plt.legend()
plt.ylabel("Absolute magnetization")
plt.xlabel("Steps")
plt.title("Absolute magnetization")
plt.grid()
plt.show()
plot_state(final_state[L_idx][H_idx])
plot_spin_directions(final_state[L_idx][H_idx])
# SELECT WARMUP PERIOD TO LAST UNTIL STEP NUMBER warmup_final_step
warmup_final_step = np.argmax(t_whole[0,0]==1e7)
warmup_final_idx = np.argmax(np.equal(t_whole[0,0], warmup_final_step))
t = t_whole[:, :, warmup_final_idx:]
E = E_whole[:, :, warmup_final_idx:]
m = m_whole[:, :, warmup_final_idx:]
E_mean = np.mean(E, axis=2)
E_std = np.sqrt(np.var(E, axis=2))
m_mean = np.mean(m, axis=2)
m_std = np.sqrt(np.var(m, axis=2))
m_magnitude = np.sqrt(np.sum(m**2, axis = 3))
m_magnitude_mean = np.mean(m_magnitude, axis=2)
m_magnitude_std = np.sqrt(np.var(m_magnitude, axis=2))
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
for i in np.ndindex(L_num):
ax.errorbar(H, E_mean[i], yerr=[E_std[i]/2, E_std[i]/2], fmt="o--", label=f"L = {L[i]}")
box = ax.get_position()
ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel(r"$H_z$")
plt.ylabel("Mean energy")
plt.grid()
plt.show()
fig.savefig("./plots/"+setname+"_energy.svg")
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
for i in np.ndindex(L_num):
ax.errorbar(H, m_magnitude_mean[i], yerr=m_magnitude_std[i]/2, fmt="o--", label=f"L = {L[i]}")
box = ax.get_position()
ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel(r"$H_z$")
plt.ylabel("Mean magnetization magnitude")
plt.grid()
plt.show()
fig.savefig("./plots/"+setname+"_energy.svg")
def cov(M,i,j):
M_mean = np.mean(M, axis=2)
ret = np.zeros(shape=(L_num, 1))
for l,t in np.ndindex(L_num, 1):
ret[l,t] = np.mean((M[l,t,:,i]-M_mean[l,t,i])*(M[l,t,:,j]-M_mean[l,t,j]))
return ret
T=1.445
chi_xx = np.zeros(shape=(L_num, 1))
chi_yy = np.zeros(shape=(L_num, 1))
chi_zz = np.zeros(shape=(L_num, 1))
chi_xy = np.zeros(shape=(L_num, 1))
chi_yz = np.zeros(shape=(L_num, 1))
chi_zx = np.zeros(shape=(L_num, 1))
for i in np.ndindex(L_num):
chi_xx[i] = cov(m,0,0)[i]/T*L[i]**3
chi_yy[i] = cov(m,1,1)[i]/T*L[i]**3
chi_zz[i] = cov(m,2,2)[i]/T*L[i]**3
chi_xy[i] = cov(m,0,1)[i]/T*L[i]**3
chi_yz[i] = cov(m,1,2)[i]/T*L[i]**3
chi_zx[i] = cov(m,2,0)[i]/T*L[i]**3
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
ax.plot(L, chi_zz, "o--")
plt.xlabel(r"$L$")
plt.ylabel(r"$\chi_{zz}$")
plt.title("System susceptibility")
plt.grid()
plt.show()
slope, intercept, r_value, p_value, std_err = stats.linregress(np.log(L), np.log(chi_zz[:,0]))
gamma_nu = slope
gamma_nu_u = std_err
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
ax.set_xscale('log')
ax.set_yscale('log')
plt.scatter(L, chi_zz[:,0])
plt.plot(L, np.exp(intercept)*L**(slope), ":")
slope_str="{0:.3}".format(slope)
std_str="{0:.3}".format(std_err)
plt.text(10,65,fr"$slope = {slope_str}\pm{std_str}$", fontsize=11)
plt.grid(True, which="both")
plt.xlabel(r"$L$")
plt.ylabel(r"$\chi_{zz}$")
plt.title(f"T=1.445")
plt.show()
fig.savefig("./plots/"+setname+"_gamma_nu.svg")
slope, intercept, r_value, p_value, std_err = stats.linregress(np.log(L), np.log(m_magnitude_mean[:,0]))
beta_nu = -slope
beta_nu_u = std_err
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
ax.set_xscale('log')
ax.set_yscale('log')
plt.scatter(L, m_magnitude_mean[:,0])
plt.plot(L, np.exp(intercept)*L**(slope), ":")
slope_str="{0:.4}".format(slope)
std_str="{0:.3}".format(std_err)
plt.text(15,3.1e-1,fr"$slope = {slope_str}\pm{std_str}$", fontsize=11)
plt.grid(True, which="both")
plt.xlabel(r"$L$")
plt.ylabel(r"$\langle m\rangle$")
plt.title(f"T=1.445")
plt.show()
fig.savefig("./plots/"+setname+"_beta_nu.svg")
binder = 1 - (1/3)*np.mean(m_magnitude**4, axis=2)/(np.mean(m_magnitude**2, axis=2)**2)
binder
m2 = np.mean(m_magnitude**2, axis=2)
m4 = np.mean(m_magnitude**4, axis=2)
m2E = np.mean(E*m_magnitude**2, axis=2)
m4E = np.mean(E*m_magnitude**4, axis=2)
dbinder = (1-binder)*(E_mean - 2*m2E/m2 + m4E/m4)
dbinder
slope, intercept, r_value, p_value, std_err = stats.linregress(np.log(L), np.log(dbinder[:,0]))
beta_nu = -slope
beta_nu_u = std_err
fig = plt.figure(figsize=(8,5))
ax = plt.subplot(111)
ax.set_xscale('log')
ax.set_yscale('log')
plt.scatter(L, dbinder[:,0])
plt.plot(L, np.exp(intercept)*L**(slope), ":")
slope_str="{0:.3}".format(slope)
std_str="{0:.3}".format(std_err)
plt.text(10,40,fr"$slope = {slope_str} \pm {std_str}$", fontsize=11)
plt.grid(True, which="both")
plt.xlabel(r"$L$")
plt.ylabel(r"$\langle \frac{d U_L}{d \beta } \rangle$")
plt.title(f"T=1.445")
plt.show()
fig.savefig("./plots/"+setname+"_nu.svg")
scm = spatial_correlation_matrix(final_state[-1][0])
r,c = radial_distribution(scm)
fig = plt.figure(figsize=(10,6))
ax = plt.subplot(111)
ax.plot(r,c)
plt.xlabel(r"$r$")
plt.ylabel(r"$C(r)$")
plt.title("Spatial correlation")
plt.grid()
plt.show()
| 0.372277 | 0.839405 |
```
# %%bash
# pip install tensorflow==1.7
# pip install tensorflow-transform
# pip install tensorflow-hub
```
# Text Classification using TensorFlow and Google Cloud - Part 2
This [bigquery-public-data:hacker_news](https://cloud.google.com/bigquery/public-data/hacker-news) contains all stories and comments from Hacker News from its launch in 2006. Each story contains a story id, url, the title of the story, tthe author that made the post, when it was written, and the number of points the story received.
The objective is, given the title of the story, we want to build an ML model that can predict the source of this story.
## TF DNNClassifier with TF.Hub Sentence Embedding
This notebook illustrates how to build a TF premade estimator, namely DNNClassifier, while the input text will be repesented as sentence embedding, using a [tf.hub text embedding module](https://www.tensorflow.org/hub/modules/text). The model will be using the transformed data produced in part one.
Note that, the tf.hub text embedding module will make use of only the the raw text feature (title). The overall steps are as follows:
1. Define the metadata
2. Define data input function
2. Create feature columns (use the tf.hub text embedding module)
3. Create the premade DNNClassifier estimator
4. Setup experiement
* Hyper-parameters & RunConfig
* Serving function (for exported model)
* TrainSpec & EvalSpec
5. Run experiement
6. Evalute the model
7. Use SavedModel for prediction
### Setting Global Parameters
```
import os
class Params:
pass
# Set to run on GCP
Params.GCP_PROJECT_ID = 'ksalama-gcp-playground'
Params.REGION = 'europe-west1'
Params.BUCKET = 'ksalama-gcs-cloudml'
Params.PLATFORM = 'local' # local | GCP
Params.DATA_DIR = 'data/news' if Params.PLATFORM == 'local' else 'gs://{}/data/news'.format(Params.BUCKET)
Params.TRANSFORMED_DATA_DIR = os.path.join(Params.DATA_DIR, 'transformed')
Params.TRANSFORMED_TRAIN_DATA_FILE_PREFIX = os.path.join(Params.TRANSFORMED_DATA_DIR, 'train')
Params.TRANSFORMED_EVAL_DATA_FILE_PREFIX = os.path.join(Params.TRANSFORMED_DATA_DIR, 'eval')
Params.TEMP_DIR = os.path.join(Params.DATA_DIR, 'tmp')
Params.MODELS_DIR = 'models/news' if Params.PLATFORM == 'local' else 'gs://{}/models/news'.format(Params.BUCKET)
Params.TRANSFORM_ARTEFACTS_DIR = os.path.join(Params.MODELS_DIR,'transform')
Params.TRAIN = True
Params.RESUME_TRAINING = False
Params.EAGER = False
if Params.EAGER:
tf.enable_eager_execution()
```
### Importing libraries
```
import tensorflow as tf
from tensorflow import data
from tensorflow.contrib.learn.python.learn.utils import input_fn_utils
from tensorflow_transform.beam.tft_beam_io import transform_fn_io
from tensorflow_transform.tf_metadata import metadata_io
from tensorflow_transform.tf_metadata import dataset_schema
from tensorflow_transform.tf_metadata import dataset_metadata
from tensorflow_transform.saved import saved_transform_io
print tf.__version__
```
## 1. Define Metadata
```
RAW_HEADER = 'key,title,source'.split(',')
RAW_DEFAULTS = [['NA'],['NA'],['NA']]
TARGET_FEATURE_NAME = 'source'
TARGET_LABELS = ['github', 'nytimes', 'techcrunch']
TEXT_FEATURE_NAME = 'title'
KEY_COLUMN = 'key'
VOCAB_SIZE = 20000
TRAIN_SIZE = 73124
EVAL_SIZE = 23079
DELIMITERS = '.,!?() '
raw_metadata = dataset_metadata.DatasetMetadata(dataset_schema.Schema({
KEY_COLUMN: dataset_schema.ColumnSchema(
tf.string, [], dataset_schema.FixedColumnRepresentation()),
TEXT_FEATURE_NAME: dataset_schema.ColumnSchema(
tf.string, [], dataset_schema.FixedColumnRepresentation()),
TARGET_FEATURE_NAME: dataset_schema.ColumnSchema(
tf.string, [], dataset_schema.FixedColumnRepresentation()),
}))
transformed_metadata = metadata_io.read_metadata(
os.path.join(Params.TRANSFORM_ARTEFACTS_DIR,"transformed_metadata"))
raw_feature_spec = raw_metadata.schema.as_feature_spec()
transformed_feature_spec = transformed_metadata.schema.as_feature_spec()
print transformed_feature_spec
```
## 2. Define Input Function
```
def parse_tf_example(tf_example):
parsed_features = tf.parse_single_example(serialized=tf_example, features=transformed_feature_spec)
target = parsed_features.pop(TARGET_FEATURE_NAME)
return parsed_features, target
def generate_tfrecords_input_fn(files_pattern,
mode=tf.estimator.ModeKeys.EVAL,
num_epochs=1,
batch_size=200):
def _input_fn():
file_names = data.Dataset.list_files(files_pattern)
if Params.EAGER:
print file_names
dataset = data.TFRecordDataset(file_names )
dataset = dataset.apply(
tf.contrib.data.shuffle_and_repeat(count=num_epochs,
buffer_size=batch_size*2)
)
dataset = dataset.apply(
tf.contrib.data.map_and_batch(parse_tf_example,
batch_size=batch_size,
num_parallel_batches=2)
)
datset = dataset.prefetch(batch_size)
if Params.EAGER:
return dataset
iterator = dataset.make_one_shot_iterator()
features, target = iterator.get_next()
return features, target
return _input_fn
```
## 3. Create feature columns
```
import tensorflow_hub as hub
print hub.__version__
def create_feature_columns(hparams):
title_embeding_column = hub.text_embedding_column(
"title", "https://tfhub.dev/google/universal-sentence-encoder/1",
trainable=hparams.trainable_embedding)
feature_columns = [title_embeding_column]
print "feature columns: \n {}".format(feature_columns)
print ""
return feature_columns
```
## 4. Create a model using a premade DNNClassifer
```
def create_estimator(hparams, run_config):
feature_columns = create_feature_columns(hparams)
optimizer = tf.train.AdamOptimizer(learning_rate=hparams.learning_rate)
estimator = tf.estimator.DNNClassifier(
feature_columns=feature_columns,
n_classes =len(TARGET_LABELS),
label_vocabulary=TARGET_LABELS,
hidden_units=hparams.hidden_units,
optimizer=optimizer,
config=run_config
)
return estimator
```
## 5. Setup Experiment
### 5.1 HParams and RunConfig
```
NUM_EPOCHS = 10
BATCH_SIZE = 1000
TOTAL_STEPS = (TRAIN_SIZE/BATCH_SIZE)*NUM_EPOCHS
EVAL_EVERY_SEC = 60
hparams = tf.contrib.training.HParams(
num_epochs = NUM_EPOCHS,
batch_size = BATCH_SIZE,
trainable_embedding = False,
learning_rate = 0.01,
hidden_units=[128, 64],
max_steps = TOTAL_STEPS,
)
MODEL_NAME = 'dnn_estimator_hub'
model_dir = os.path.join(Params.MODELS_DIR, MODEL_NAME)
run_config = tf.estimator.RunConfig(
tf_random_seed=19830610,
log_step_count_steps=1000,
save_checkpoints_secs=EVAL_EVERY_SEC,
keep_checkpoint_max=1,
model_dir=model_dir
)
print(hparams)
print("")
print("Model Directory:", run_config.model_dir)
print("Dataset Size:", TRAIN_SIZE)
print("Batch Size:", BATCH_SIZE)
print("Steps per Epoch:",TRAIN_SIZE/BATCH_SIZE)
print("Total Steps:", TOTAL_STEPS)
```
### 5.2 Serving function
```
def generate_serving_input_fn():
def _serving_fn():
receiver_tensor = {
'title': tf.placeholder(dtype=tf.string, shape=[None])
}
return tf.estimator.export.ServingInputReceiver(
receiver_tensor, receiver_tensor)
return _serving_fn
```
### 5.3 TrainSpec & EvalSpec
```
train_spec = tf.estimator.TrainSpec(
input_fn = generate_tfrecords_input_fn(
Params.TRANSFORMED_TRAIN_DATA_FILE_PREFIX+"*",
mode = tf.estimator.ModeKeys.TRAIN,
num_epochs=hparams.num_epochs,
batch_size=hparams.batch_size
),
max_steps=hparams.max_steps,
hooks=None
)
eval_spec = tf.estimator.EvalSpec(
input_fn = generate_tfrecords_input_fn(
Params.TRANSFORMED_EVAL_DATA_FILE_PREFIX+"*",
mode=tf.estimator.ModeKeys.EVAL,
num_epochs=1,
batch_size=hparams.batch_size
),
exporters=[tf.estimator.LatestExporter(
name="estimate", # the name of the folder in which the model will be exported to under export
serving_input_receiver_fn=generate_serving_input_fn(),
exports_to_keep=1,
as_text=False)],
steps=None,
throttle_secs=EVAL_EVERY_SEC
)
```
## 6. Run experiment
```
from datetime import datetime
import shutil
if Params.TRAIN:
if not Params.RESUME_TRAINING:
print("Removing previous training artefacts...")
shutil.rmtree(model_dir, ignore_errors=True)
else:
print("Resuming training...")
tf.logging.set_verbosity(tf.logging.INFO)
time_start = datetime.utcnow()
print("Experiment started at {}".format(time_start.strftime("%H:%M:%S")))
print(".......................................")
estimator = create_estimator(hparams, run_config)
tf.estimator.train_and_evaluate(
estimator=estimator,
train_spec=train_spec,
eval_spec=eval_spec
)
time_end = datetime.utcnow()
print(".......................................")
print("Experiment finished at {}".format(time_end.strftime("%H:%M:%S")))
print("")
time_elapsed = time_end - time_start
print("Experiment elapsed time: {} seconds".format(time_elapsed.total_seconds()))
else:
print "Training was skipped!"
```
## 7. Evaluate the model
```
tf.logging.set_verbosity(tf.logging.ERROR)
estimator = create_estimator(hparams, run_config)
train_metrics = estimator.evaluate(
input_fn = generate_tfrecords_input_fn(
files_pattern= Params.TRANSFORMED_TRAIN_DATA_FILE_PREFIX+"*",
mode= tf.estimator.ModeKeys.EVAL,
batch_size= TRAIN_SIZE),
steps=1
)
print("############################################################################################")
print("# Train Measures: {}".format(train_metrics))
print("############################################################################################")
eval_metrics = estimator.evaluate(
input_fn=generate_tfrecords_input_fn(
files_pattern= Params.TRANSFORMED_EVAL_DATA_FILE_PREFIX+"*",
mode= tf.estimator.ModeKeys.EVAL,
batch_size= EVAL_SIZE),
steps=1
)
print("")
print("############################################################################################")
print("# Eval Measures: {}".format(eval_metrics))
print("############################################################################################")
```
## 8. Use Saved Model for Predictions
```
import os
export_dir = model_dir +"/export/estimate/"
saved_model_dir = os.path.join(export_dir, os.listdir(export_dir)[0])
print(saved_model_dir)
print("")
predictor_fn = tf.contrib.predictor.from_saved_model(
export_dir = saved_model_dir,
signature_def_key="predict"
)
output = predictor_fn(
{
'title':[
'Microsoft and Google are joining forces for a new AI framework',
'A new version of Python is mind blowing',
'EU is investigating new data privacy policies'
]
}
)
print(output)
```
|
github_jupyter
|
# %%bash
# pip install tensorflow==1.7
# pip install tensorflow-transform
# pip install tensorflow-hub
import os
class Params:
pass
# Set to run on GCP
Params.GCP_PROJECT_ID = 'ksalama-gcp-playground'
Params.REGION = 'europe-west1'
Params.BUCKET = 'ksalama-gcs-cloudml'
Params.PLATFORM = 'local' # local | GCP
Params.DATA_DIR = 'data/news' if Params.PLATFORM == 'local' else 'gs://{}/data/news'.format(Params.BUCKET)
Params.TRANSFORMED_DATA_DIR = os.path.join(Params.DATA_DIR, 'transformed')
Params.TRANSFORMED_TRAIN_DATA_FILE_PREFIX = os.path.join(Params.TRANSFORMED_DATA_DIR, 'train')
Params.TRANSFORMED_EVAL_DATA_FILE_PREFIX = os.path.join(Params.TRANSFORMED_DATA_DIR, 'eval')
Params.TEMP_DIR = os.path.join(Params.DATA_DIR, 'tmp')
Params.MODELS_DIR = 'models/news' if Params.PLATFORM == 'local' else 'gs://{}/models/news'.format(Params.BUCKET)
Params.TRANSFORM_ARTEFACTS_DIR = os.path.join(Params.MODELS_DIR,'transform')
Params.TRAIN = True
Params.RESUME_TRAINING = False
Params.EAGER = False
if Params.EAGER:
tf.enable_eager_execution()
import tensorflow as tf
from tensorflow import data
from tensorflow.contrib.learn.python.learn.utils import input_fn_utils
from tensorflow_transform.beam.tft_beam_io import transform_fn_io
from tensorflow_transform.tf_metadata import metadata_io
from tensorflow_transform.tf_metadata import dataset_schema
from tensorflow_transform.tf_metadata import dataset_metadata
from tensorflow_transform.saved import saved_transform_io
print tf.__version__
RAW_HEADER = 'key,title,source'.split(',')
RAW_DEFAULTS = [['NA'],['NA'],['NA']]
TARGET_FEATURE_NAME = 'source'
TARGET_LABELS = ['github', 'nytimes', 'techcrunch']
TEXT_FEATURE_NAME = 'title'
KEY_COLUMN = 'key'
VOCAB_SIZE = 20000
TRAIN_SIZE = 73124
EVAL_SIZE = 23079
DELIMITERS = '.,!?() '
raw_metadata = dataset_metadata.DatasetMetadata(dataset_schema.Schema({
KEY_COLUMN: dataset_schema.ColumnSchema(
tf.string, [], dataset_schema.FixedColumnRepresentation()),
TEXT_FEATURE_NAME: dataset_schema.ColumnSchema(
tf.string, [], dataset_schema.FixedColumnRepresentation()),
TARGET_FEATURE_NAME: dataset_schema.ColumnSchema(
tf.string, [], dataset_schema.FixedColumnRepresentation()),
}))
transformed_metadata = metadata_io.read_metadata(
os.path.join(Params.TRANSFORM_ARTEFACTS_DIR,"transformed_metadata"))
raw_feature_spec = raw_metadata.schema.as_feature_spec()
transformed_feature_spec = transformed_metadata.schema.as_feature_spec()
print transformed_feature_spec
def parse_tf_example(tf_example):
parsed_features = tf.parse_single_example(serialized=tf_example, features=transformed_feature_spec)
target = parsed_features.pop(TARGET_FEATURE_NAME)
return parsed_features, target
def generate_tfrecords_input_fn(files_pattern,
mode=tf.estimator.ModeKeys.EVAL,
num_epochs=1,
batch_size=200):
def _input_fn():
file_names = data.Dataset.list_files(files_pattern)
if Params.EAGER:
print file_names
dataset = data.TFRecordDataset(file_names )
dataset = dataset.apply(
tf.contrib.data.shuffle_and_repeat(count=num_epochs,
buffer_size=batch_size*2)
)
dataset = dataset.apply(
tf.contrib.data.map_and_batch(parse_tf_example,
batch_size=batch_size,
num_parallel_batches=2)
)
datset = dataset.prefetch(batch_size)
if Params.EAGER:
return dataset
iterator = dataset.make_one_shot_iterator()
features, target = iterator.get_next()
return features, target
return _input_fn
import tensorflow_hub as hub
print hub.__version__
def create_feature_columns(hparams):
title_embeding_column = hub.text_embedding_column(
"title", "https://tfhub.dev/google/universal-sentence-encoder/1",
trainable=hparams.trainable_embedding)
feature_columns = [title_embeding_column]
print "feature columns: \n {}".format(feature_columns)
print ""
return feature_columns
def create_estimator(hparams, run_config):
feature_columns = create_feature_columns(hparams)
optimizer = tf.train.AdamOptimizer(learning_rate=hparams.learning_rate)
estimator = tf.estimator.DNNClassifier(
feature_columns=feature_columns,
n_classes =len(TARGET_LABELS),
label_vocabulary=TARGET_LABELS,
hidden_units=hparams.hidden_units,
optimizer=optimizer,
config=run_config
)
return estimator
NUM_EPOCHS = 10
BATCH_SIZE = 1000
TOTAL_STEPS = (TRAIN_SIZE/BATCH_SIZE)*NUM_EPOCHS
EVAL_EVERY_SEC = 60
hparams = tf.contrib.training.HParams(
num_epochs = NUM_EPOCHS,
batch_size = BATCH_SIZE,
trainable_embedding = False,
learning_rate = 0.01,
hidden_units=[128, 64],
max_steps = TOTAL_STEPS,
)
MODEL_NAME = 'dnn_estimator_hub'
model_dir = os.path.join(Params.MODELS_DIR, MODEL_NAME)
run_config = tf.estimator.RunConfig(
tf_random_seed=19830610,
log_step_count_steps=1000,
save_checkpoints_secs=EVAL_EVERY_SEC,
keep_checkpoint_max=1,
model_dir=model_dir
)
print(hparams)
print("")
print("Model Directory:", run_config.model_dir)
print("Dataset Size:", TRAIN_SIZE)
print("Batch Size:", BATCH_SIZE)
print("Steps per Epoch:",TRAIN_SIZE/BATCH_SIZE)
print("Total Steps:", TOTAL_STEPS)
def generate_serving_input_fn():
def _serving_fn():
receiver_tensor = {
'title': tf.placeholder(dtype=tf.string, shape=[None])
}
return tf.estimator.export.ServingInputReceiver(
receiver_tensor, receiver_tensor)
return _serving_fn
train_spec = tf.estimator.TrainSpec(
input_fn = generate_tfrecords_input_fn(
Params.TRANSFORMED_TRAIN_DATA_FILE_PREFIX+"*",
mode = tf.estimator.ModeKeys.TRAIN,
num_epochs=hparams.num_epochs,
batch_size=hparams.batch_size
),
max_steps=hparams.max_steps,
hooks=None
)
eval_spec = tf.estimator.EvalSpec(
input_fn = generate_tfrecords_input_fn(
Params.TRANSFORMED_EVAL_DATA_FILE_PREFIX+"*",
mode=tf.estimator.ModeKeys.EVAL,
num_epochs=1,
batch_size=hparams.batch_size
),
exporters=[tf.estimator.LatestExporter(
name="estimate", # the name of the folder in which the model will be exported to under export
serving_input_receiver_fn=generate_serving_input_fn(),
exports_to_keep=1,
as_text=False)],
steps=None,
throttle_secs=EVAL_EVERY_SEC
)
from datetime import datetime
import shutil
if Params.TRAIN:
if not Params.RESUME_TRAINING:
print("Removing previous training artefacts...")
shutil.rmtree(model_dir, ignore_errors=True)
else:
print("Resuming training...")
tf.logging.set_verbosity(tf.logging.INFO)
time_start = datetime.utcnow()
print("Experiment started at {}".format(time_start.strftime("%H:%M:%S")))
print(".......................................")
estimator = create_estimator(hparams, run_config)
tf.estimator.train_and_evaluate(
estimator=estimator,
train_spec=train_spec,
eval_spec=eval_spec
)
time_end = datetime.utcnow()
print(".......................................")
print("Experiment finished at {}".format(time_end.strftime("%H:%M:%S")))
print("")
time_elapsed = time_end - time_start
print("Experiment elapsed time: {} seconds".format(time_elapsed.total_seconds()))
else:
print "Training was skipped!"
tf.logging.set_verbosity(tf.logging.ERROR)
estimator = create_estimator(hparams, run_config)
train_metrics = estimator.evaluate(
input_fn = generate_tfrecords_input_fn(
files_pattern= Params.TRANSFORMED_TRAIN_DATA_FILE_PREFIX+"*",
mode= tf.estimator.ModeKeys.EVAL,
batch_size= TRAIN_SIZE),
steps=1
)
print("############################################################################################")
print("# Train Measures: {}".format(train_metrics))
print("############################################################################################")
eval_metrics = estimator.evaluate(
input_fn=generate_tfrecords_input_fn(
files_pattern= Params.TRANSFORMED_EVAL_DATA_FILE_PREFIX+"*",
mode= tf.estimator.ModeKeys.EVAL,
batch_size= EVAL_SIZE),
steps=1
)
print("")
print("############################################################################################")
print("# Eval Measures: {}".format(eval_metrics))
print("############################################################################################")
import os
export_dir = model_dir +"/export/estimate/"
saved_model_dir = os.path.join(export_dir, os.listdir(export_dir)[0])
print(saved_model_dir)
print("")
predictor_fn = tf.contrib.predictor.from_saved_model(
export_dir = saved_model_dir,
signature_def_key="predict"
)
output = predictor_fn(
{
'title':[
'Microsoft and Google are joining forces for a new AI framework',
'A new version of Python is mind blowing',
'EU is investigating new data privacy policies'
]
}
)
print(output)
| 0.469763 | 0.896976 |
### Micromobility
The way we travel in urban areas is changing rapidly with the advent of new types of vehicles i.e scooters, electrical bikes and etc. Although these devices can be privately owned, they are mostly owned by shared mobility service companies.
The most important aspects of these latest services is that there are not any docks for picking up and leaving back the devices. Instead, the vehicles can be left anywhere in designated boundaries in the city center.
In Austin, Texas there are 8 mobility companies that provide more than 15000 scooters and 2000 e-bikes. Austin Transportation provides the dockless vehicle trips dataset records trips taken in the city which can be interesting to explore. Let's explore the following questions:
### Questions we wanna address:
1. When are these devices generally used?
2. Where are these devices generally picked up and left?
3. What is the average distance per trip?
4. What is the average duration per trip?
5. What are the factors that influence the trip distance and duration?
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
from IPython.display import display, HTML, display_html
import seaborn as sns
sns.set()
import datetime
```
Load the mobility dataset
```
# set formatting
pd.set_option('display.max_columns', 50)
pd.set_option('display.max_rows', 50)
# read in CSV file data
df = pd.read_csv('Dockless_Vehicle_Trips.csv')
df = df[(df.loc[:,'Month'] >= 1) &
(df.loc[:, 'Month'] <= 12)]
# ((df.loc[:,'Year'] == 2018) &
# (df.loc[:,'Year'] == 2019) &
```
### Several properties of the data:
1. What are the columns?
2. How many rows and columns
3. What are the data types
4. Are there any null values, incomplete values
5. What are some other caveats to the data?
```
# look at the data
display(df.head())
# shape of the data
display(df.shape)
# look at data types. Ideally look at all rows. Only look at first five here for minimal output.
display(df.iloc[:5,:].dtypes)
# see if any columns have nulls. Ideally look at all rows. Only look at first five here for minimal output.
display(df.iloc[:,:].isnull().any())
# display descriptive statistics
# Trip Distance and Trip Duration percentiles
display(df.iloc[:,3:5].describe(percentiles=[0.01, 0.05, 0.25, 0.5, 0.75, 0.85, 0.95, 0.98, 0.99]))
# display the columns
display(df.columns.values)
```
## Cleaning
1. Drop unwanted columns
2. Remove rows with NaNs
3. Drop duplicates (optional)
4. Remove unwanted columns
5. Remove negative trip duration
6. Remove negative trip distances
7. Check other types
```
# Drop unwanted columns
columns_to_drop = ['Census Tract Start', 'Census Tract End', 'End Time']
df.drop(columns_to_drop, axis=1, inplace=True)
# remove the NaN rows
df.dropna(axis='rows', how='any', inplace=True)
# drop the duplicates
#df.drop_duplicates()
# remove the extreme values
df = df[df.iloc[:, 3] < np.percentile(df.iloc[:,3], 99)]
df = df[df.iloc[:, 3] > np.percentile(df.iloc[:,3], 1)]
df = df[df.iloc[:, 4] < np.percentile(df.iloc[:,4], 99)]
df = df[df.iloc[:, 4] > np.percentile(df.iloc[:,4], 1)]
# Check Vehicle Types
display(df.iloc[:, 2].unique())
# Check districts
display(df.iloc[:, 11].unique())
display(df.iloc[:, 12].unique())
```
## Processing
1. Rename columns for easier processing
2. Remove rows that has zero trip distance and duration
3. Log transform trip distance and trip duration for exploratory analysis
4. Day of Week: Sunday 0 ... Saturday 6
5. Month: January 0 ... December 11
```
# Rename columns
df_example = df.rename(columns = {'Day of Week':'dow','Start Time': 'st', 'Hour': 'h', 'Month': 'm',
'Vehicle Type': 'vt', 'Trip Distance': 'tdis', 'Trip Duration': 'tdur',
'Council District (Start)':'scd', 'Council District (End)':'ecd'})
# more than one second
df = df[df.iloc[:, 3] > 0]
df = df[df.iloc[:, 4] > 0]
# Group data by number of listings per date
fig, ax = plt.subplots(4, figsize=(24, 24))
display(df_example.groupby(['h','m']).count()['tdis'].unstack().apply(lambda x: np.log(x)+0.000005).describe())
df_example.groupby(['h','dow']).sum()['tdis'].apply(lambda x: np.log(x)+0.000005).unstack().plot(ax=ax[0])
df_example.groupby(['m','dow']).sum()['tdis'].apply(lambda x: np.log(x)+0.000005).unstack().plot(ax=ax[1])
df_example.groupby(['m','dow']).mean()['tdur'].unstack().apply(lambda x: np.log(x)+0.000005).plot(ax=ax[2])
df_example.groupby(['h','m']).count()['tdis'].unstack().apply(lambda x: np.log(x)+0.000005).plot(ax=ax[3]).legend(loc='right')
#df_example.groupby(['h','dow']).sum()['tdur'].unstack().apply(lambda x: np.log(x)+0.000005).plot(ax=ax[2])
#df_example.groupby(['h','m']).sum()['tdur'].unstack().apply(lambda x: np.log(x)+0.000005).plot(ax=ax[3])
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
from IPython.display import display, HTML, display_html
import seaborn as sns
sns.set()
import datetime
# set formatting
pd.set_option('display.max_columns', 50)
pd.set_option('display.max_rows', 50)
# read in CSV file data
df = pd.read_csv('Dockless_Vehicle_Trips.csv')
df = df[(df.loc[:,'Month'] >= 1) &
(df.loc[:, 'Month'] <= 12)]
# ((df.loc[:,'Year'] == 2018) &
# (df.loc[:,'Year'] == 2019) &
# look at the data
display(df.head())
# shape of the data
display(df.shape)
# look at data types. Ideally look at all rows. Only look at first five here for minimal output.
display(df.iloc[:5,:].dtypes)
# see if any columns have nulls. Ideally look at all rows. Only look at first five here for minimal output.
display(df.iloc[:,:].isnull().any())
# display descriptive statistics
# Trip Distance and Trip Duration percentiles
display(df.iloc[:,3:5].describe(percentiles=[0.01, 0.05, 0.25, 0.5, 0.75, 0.85, 0.95, 0.98, 0.99]))
# display the columns
display(df.columns.values)
# Drop unwanted columns
columns_to_drop = ['Census Tract Start', 'Census Tract End', 'End Time']
df.drop(columns_to_drop, axis=1, inplace=True)
# remove the NaN rows
df.dropna(axis='rows', how='any', inplace=True)
# drop the duplicates
#df.drop_duplicates()
# remove the extreme values
df = df[df.iloc[:, 3] < np.percentile(df.iloc[:,3], 99)]
df = df[df.iloc[:, 3] > np.percentile(df.iloc[:,3], 1)]
df = df[df.iloc[:, 4] < np.percentile(df.iloc[:,4], 99)]
df = df[df.iloc[:, 4] > np.percentile(df.iloc[:,4], 1)]
# Check Vehicle Types
display(df.iloc[:, 2].unique())
# Check districts
display(df.iloc[:, 11].unique())
display(df.iloc[:, 12].unique())
# Rename columns
df_example = df.rename(columns = {'Day of Week':'dow','Start Time': 'st', 'Hour': 'h', 'Month': 'm',
'Vehicle Type': 'vt', 'Trip Distance': 'tdis', 'Trip Duration': 'tdur',
'Council District (Start)':'scd', 'Council District (End)':'ecd'})
# more than one second
df = df[df.iloc[:, 3] > 0]
df = df[df.iloc[:, 4] > 0]
# Group data by number of listings per date
fig, ax = plt.subplots(4, figsize=(24, 24))
display(df_example.groupby(['h','m']).count()['tdis'].unstack().apply(lambda x: np.log(x)+0.000005).describe())
df_example.groupby(['h','dow']).sum()['tdis'].apply(lambda x: np.log(x)+0.000005).unstack().plot(ax=ax[0])
df_example.groupby(['m','dow']).sum()['tdis'].apply(lambda x: np.log(x)+0.000005).unstack().plot(ax=ax[1])
df_example.groupby(['m','dow']).mean()['tdur'].unstack().apply(lambda x: np.log(x)+0.000005).plot(ax=ax[2])
df_example.groupby(['h','m']).count()['tdis'].unstack().apply(lambda x: np.log(x)+0.000005).plot(ax=ax[3]).legend(loc='right')
#df_example.groupby(['h','dow']).sum()['tdur'].unstack().apply(lambda x: np.log(x)+0.000005).plot(ax=ax[2])
#df_example.groupby(['h','m']).sum()['tdur'].unstack().apply(lambda x: np.log(x)+0.000005).plot(ax=ax[3])
| 0.412175 | 0.986494 |
```
from PW_explorer.load_worlds import load_worlds
from PW_explorer.run_clingo import run_clingo
%load_ext PWE_NB_Extension
%%clingo --donot-display_input -lci qep_eq_check2 --donot-run
% e(X,Y) :- e(Y,X). --> only if undirected
gnode1(X) :- e1(X,_).
gnode1(X) :- e1(_,X).
gnode1(X) :- ne1(X,_).
gnode1(X) :- ne1(_,X).
gnode2(X) :- e2(X,_).
gnode2(X) :- e2(_,X).
gnode2(X) :- ne2(X,_).
gnode2(X) :- ne2(_,X).
vmap(X,Y) ; not vmap(X,Y) :- gnode1(X), gnode2(Y).
:- vmap(X1,Y1), vmap(X2,Y2), e1(X1,X2), not e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not e1(X1,X2), e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), ne1(X1,X2), not ne2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not ne1(X1,X2), ne2(Y1,Y2).
% 3hop Query
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), thop1(X1,X2,X3,X4), not thop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), not thop1(X1,X2,X3,X4), thop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), thop1(X1,X2,X3), not thop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not thop1(X1,X2,X3), thop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), thop1(X1,X2), not thop2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not thop1(X1,X2), thop2(Y1,Y2).
:- vmap(X1,Y1), thop1(X1), not thop2(Y1).
:- vmap(X1,Y1), not thop1(X1), thop2(Y1).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), nthop1(X1,X2,X3,X4), not nthop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), not nthop1(X1,X2,X3,X4), nthop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), nthop1(X1,X2,X3), not nthop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not nthop1(X1,X2,X3), nthop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), nthop1(X1,X2), not nthop2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not nthop1(X1,X2), nthop2(Y1,Y2).
:- vmap(X1,Y1), nthop1(X1), not nthop2(Y1).
:- vmap(X1,Y1), not nthop1(X1), nthop2(Y1).
% Triangle Query
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), tri1(X1,X2,X3), not tri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not tri1(X1,X2,X3), tri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), tri1(X1,X2), not tri2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not tri1(X1,X2), tri2(Y1,Y2).
:- vmap(X1,Y1), tri1(X1), not tri2(Y1).
:- vmap(X1,Y1), not tri1(X1), tri2(Y1).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), ntri1(X1,X2,X3), not ntri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not ntri1(X1,X2,X3), ntri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), ntri1(X1,X2), not ntri2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not ntri1(X1,X2), ntri2(Y1,Y2).
:- vmap(X1,Y1), ntri1(X1), not ntri2(Y1).
:- vmap(X1,Y1), not ntri1(X1), ntri2(Y1).
% used1(X) :- vmap(X,_).
% :- gnode(X), not used1(X).
% :- vmap(X,Y),vmap(X,Z),Y!=Z.
% :- vmap(Y,X),vmap(Z,X),Y!=Z.
:- gnode1(X), #count {Y: vmap(X,Y)} != 1.
:- gnode2(Y), #count {X: vmap(X,Y)} != 1.
#show.
% #show vmap/2.
%%clingo -l qep_eq_check2 --donot-display_input
e1(x,y). e1(y,z). e1(z,x). tri1(x,z).
e2(x,y). e2(y,z). e2(z,x). tri2(x,y).
%%clingo --donot-display_input -lci qep_subsumption_check2 --donot-run
% e(X,Y) :- e(Y,X). --> only if undirected
gnode1(X) :- e1(X,_).
gnode1(X) :- e1(_,X).
gnode1(X) :- ne1(X,_).
gnode1(X) :- ne1(_,X).
gnode2(X) :- e2(X,_).
gnode2(X) :- e2(_,X).
gnode2(X) :- ne2(X,_).
gnode2(X) :- ne2(_,X).
vmap(X,Y) ; not vmap(X,Y) :- gnode1(X), gnode2(Y).
:- vmap(X1,Y1), vmap(X2,Y2), e1(X1,X2), not e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not e1(X1,X2), e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), ne1(X1,X2), not ne2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not ne1(X1,X2), ne2(Y1,Y2).
% 3hop Query
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), thop1(X1,X2,X3,X4), not thop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), not thop1(X1,X2,X3,X4), thop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), thop1(X1,X2,X3), not thop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not thop1(X1,X2,X3), thop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), thop1(X1,X2), not thop2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not thop1(X1,X2), thop2(Y1,Y2).
:- vmap(X1,Y1), thop1(X1), not thop2(Y1).
:- vmap(X1,Y1), not thop1(X1), thop2(Y1).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), nthop1(X1,X2,X3,X4), not nthop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), not nthop1(X1,X2,X3,X4), nthop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), nthop1(X1,X2,X3), not nthop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not nthop1(X1,X2,X3), nthop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), nthop1(X1,X2), not nthop2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not nthop1(X1,X2), nthop2(Y1,Y2).
:- vmap(X1,Y1), nthop1(X1), not nthop2(Y1).
:- vmap(X1,Y1), not nthop1(X1), nthop2(Y1).
% Triangle Query
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), tri1(X1,X2,X3), not tri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not tri1(X1,X2,X3), tri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), tri1(X1,X2), not tri2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not tri1(X1,X2), tri2(Y1,Y2).
:- vmap(X1,Y1), tri1(X1), not tri2(Y1).
:- vmap(X1,Y1), not tri1(X1), tri2(Y1).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), ntri1(X1,X2,X3), not ntri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not ntri1(X1,X2,X3), ntri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), ntri1(X1,X2), not ntri2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not ntri1(X1,X2), ntri2(Y1,Y2).
:- vmap(X1,Y1), ntri1(X1), not ntri2(Y1).
:- vmap(X1,Y1), not ntri1(X1), ntri2(Y1).
% used1(X) :- vmap(X,_).
% :- gnode(X), not used1(X).
% :- vmap(X,Y),vmap(X,Z),Y!=Z.
% :- vmap(Y,X),vmap(Z,X),Y!=Z.
:- gnode1(X), #count {Y: vmap(X,Y)} < 1.
:- gnode2(Y), #count {X: vmap(X,Y)} < 1.
#show.
% #show vmap/2.
%%clingo -l qep_subsumption_check2 --donot-display_input
e1(x,y). e1(y,z). e1(z,x). tri1(x,y).
e2(x,y). e2(y,y). e2(y,x). tri2(x,y).
%%clingo --donot-display_input -lci automorphisms --donot-run
% e(X,Y) :- e(Y,X). --> only if undirected
gnode(X) :- e(X,_).
gnode(X) :- e(_,X).
vmap(X,Y) ; vout(X,Y) :- gnode(X), gnode(Y).
:- vmap(X1,Y1), vmap(X2,Y2), e(X1,X2), not e(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not e(X1,X2), e(Y1,Y2).
% used1(X) :- vmap(X,_).
% :- gnode(X), not used1(X).
% :- vmap(X,Y),vmap(X,Z),Y!=Z.
% :- vmap(Y,X),vmap(Z,X),Y!=Z.
:- gnode(X), #count {Y: vmap(X,Y)} != 1.
:- gnode(X), #count {Y: vmap(Y,X)} != 1.
% #show vmap/2.
#show.
%%clingo --donot-display_input -l automorphisms
e(x,e1). e(z1,e1). e(z1,e2). e(z2,e2). e(z2,e3). e(y,e3).
e(x,head). e(y,head).
e(z1,head). e(z2,head).
%%clingo --donot-display_input -lci qep_eq_check --donot-run
% e(X,Y) :- e(Y,X). --> only if undirected
gnode1(X) :- e1(X,_).
gnode1(X) :- e1(_,X).
gnode1(X) :- ne1(X,_).
gnode1(X) :- ne1(_,X).
gnode2(X) :- e2(X,_).
gnode2(X) :- e2(_,X).
gnode2(X) :- ne2(X,_).
gnode2(X) :- ne2(_,X).
vmap(X,Y) ; not vmap(X,Y) :- gnode1(X), gnode2(Y).
:- vmap(X1,Y1), vmap(X2,Y2), e1(X1,X2), not e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not e1(X1,X2), e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), ne1(X1,X2), not ne2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not ne1(X1,X2), ne2(Y1,Y2).
% used1(X) :- vmap(X,_).
% :- gnode(X), not used1(X).
% :- vmap(X,Y),vmap(X,Z),Y!=Z.
% :- vmap(Y,X),vmap(Z,X),Y!=Z.
:- gnode1(X), #count {Y: vmap(X,Y)} != 1.
:- gnode2(Y), #count {X: vmap(X,Y)} != 1.
#show vmap/2.
%%clingo --donot-display_input -l qep_eq_check -lci thop_4_qep_eq_check --donot-run
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), thop1(X1,X2,X3,X4), not thop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), not thop1(X1,X2,X3,X4), thop2(Y1,Y2,Y3,Y4).
%%clingo --donot-display_input -l qep_eq_check -lci thop_3_qep_eq_check --donot-run
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), thop1(X1,X2,X3), not thop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not thop1(X1,X2,X3), thop2(Y1,Y2,Y3).
%%clingo --donot-display_input -l qep_eq_check -lci thop_2_qep_eq_check --donot-run
:- vmap(X1,Y1), vmap(X2,Y2), thop1(X1,X2), not thop2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not thop1(X1,X2), thop2(Y1,Y2).
%%clingo --donot-display_input -l qep_eq_check -lci thop_1_qep_eq_check --donot-run
:- vmap(X1,Y1), thop1(X1), not thop2(Y1).
:- vmap(X1,Y1), not thop1(X1), thop2(Y1).
%%clingo --donot-display_input -l qep_eq_check -lci thop_0_qep_eq_check --donot-run
%empty
# thop(X,X) :- e(X,X), e(X,Z2), e(Z2,X).
# thop(X,X) :- e(X,Z1), e(Z1,Z1), e(Z1,X).
# thop(X,X) :- e(X,Z1), e(Z1,X), e(X,X).
%%clingo --donot-display_input -l thop_4_qep_eq_check
e1("X","X"). e1("X","Z2"). e1("Z2","X").
thop1("X","X","Z2","X").
e2("X","Z1"). e2("Z1","X"). e2("X","X").
thop2("X","Z1","X","X").
%%clingo --donot-display_input -l thop_2_qep_eq_check
e1("X","X"). e1("X","Z2"). e1("Z2","X").
thop1("X", "X").
e2("X","Z1"). e2("Z1","X"). e2("X","X").
thop2("X","X").
%%clingo --donot-display_input -l thop_1_qep_eq_check
e1("X","X"). e1("X","Z2"). e1("Z2","X").
thop1("X").
e2("X","Z1"). e2("Z1","X"). e2("X","X").
thop2("X").
%%clingo --donot-display_input -l qep_eq_check
e1("X","Y"). ne1("Y","X"). e1("X","Y").
e1("X", head). %e1("Y", head).
ne2("X","Y"). e2("Y","X"). ne2("X","Y").
e2("X", head). %e2("Y", head).
vmap(head, head).
# Should we be computing the isomorphisms on the incidence graph instead?
%%clingo --donot-run
nthop("X","X") :- e("X","Z1"), not e("Z1","Z2"), not e("Z2","X").
nthop("X","X") :- not e("X","Z1"), not e("Z1","Z2"), e("Z2","X").
% nthop("X","X") :- not e("X","Z1"), e("Z1","Z2"), not e("Z2","X")
%%clingo --donot-display_input -l qep_eq_check
e1("X","Z1"). ne1("Z1","Z2"). ne1("Z2","X").
%e1("X", head).
%e1("Z1", head).
%e1("Z2", head).
ne2("X","Z1"). ne2("Z1","Z2"). e2("Z2","X").
%e2("X", head).
%e2("Z1", head).
%e2("Z2", head).
ne2("X","Z1"). e2("Z1","Z2"). ne2("Z2","X").
%e2("X", head).
%e2("Z1", head).
%e2("Z2", head).
vmap(head, head).
%%clingo --donot-display_input -l qep_eq_check
e1("X", e1). e1("Z1", e1). ne1("Z1", e2). ne1("Z2", e2). ne1("Z2", e3). ne1("X", e3).
%ne1("X", head).
%ne1("Z1", head).
%ne1("Z2", head).
ne2("X", e1). ne2("Z1", e1). ne2("Z1", e2). ne2("Z2", e2). e2("Z2", e3). e2("X", e3).
%ne2("X", head).
%ne2("Z1", head).
%ne2("Z2", head).
vmap(head, head).
%%clingo
tri(X,Y,Z) :- e(X,Y), e(Y,Z), e(Z,X), X=Y, X=Z.
%%clingo --donot-display_input -lci query_analysis_starter
% schema var(VAR, DOMAIN). % TO ADD
% schema ruleOcc(ATOM, OCC). % TO ADD
% schema ruleH(HEAD). % TO ADD
% schema ruleOccTrue(ATOM, OCC)
ruleOccTrue(R, OC) ; not ruleOccTrue(R, OC) :- ruleOcc(R, OC).
% schema ruleHTrue(HEAD)
ruleHFalse(H) :- ruleH(H), ruleOcc(R, OCC), not ruleOccTrue(R,OCC).
ruleHTrue(H) :- ruleH(H), not ruleHFalse(H).
% schema eq(VAR1, VAR2, DOMAIN)
eq(A,B,TYPE) ; not eq(A,B,TYPE) :- var(A, TYPE), var(B, TYPE), A!=B.
eq(A,B,TYPE) :- eq(B,A,TYPE).
eq(A,B,TYPE) :- eq(A,C,TYPE), eq(C,B,TYPE), A!=B, B!=C, A!=C.
% schema eqOrd(VAR1, VAR2, DOMAIN)
eqOrd(A,B,TYPE) :- eq(A,B,TYPE), A<B.
% schema arc(VAR, POS, ATOM, OCC). % TO ADD
% schema hArc(VAR, POS, HEAD). % TO ADD
% schema newVar(VAR, DOMAIN)
newVar(VAR, TYPE) :- var(VAR, TYPE), not eqOrd(_,VAR,TYPE).
% schema eqOrdMinimal(VAR1, VAR2, DOMAIN)
eqOrdMinimal(A,B,TYPE) :- eqOrd(A,B,TYPE), newVar(A,TYPE).
% schema neqOrd(VAR1, VAR2, DOMAIN)
neqOrd(A,B,TYPE) :- newVar(A,TYPE), newVar(B,TYPE), A<B.
% Find the new arcs, getting rid of the redundancies
% schema newArc(VAR, POS, ATOM, OCC)
newArc(VAR, POS, ATOM, OCC) :- arc(VAR_ORIG, POS, ATOM, OCC), eqOrd(VAR,VAR_ORIG,TYPE), not eqOrd(_,VAR,TYPE).
newArc(VAR, POS, ATOM, OCC) :- arc(VAR, POS, ATOM, OCC), not eqOrd(_,VAR,_).
% schema newHArc(VAR, POS, HEAD)
newHArc(VAR, POS, H) :- hArc(VAR_ORIG,POS,H), eqOrd(VAR,VAR_ORIG,TYPE), not eqOrd(_,VAR,TYPE).
newHArc(VAR, POS, H) :- hArc(VAR, POS, H), not eqOrd(_,VAR,_).
% It cannot be true that eX and eY have the same variable assignments but one is True while other is False
:- newArc(V1, 1, "e", OCC1), newArc(V2, 2, "e", OCC1), newArc(V1, 1, "e", OCC2), newArc(V2, 2, "e", OCC2), ruleOccTrue("e", OCC1), not ruleOccTrue("e", OCC2).
% eqAtomOccOrd("e", OCC1, OCC2) :- newArc(V1, 1, "e", OCC1), newArc(V2, 2, "e", OCC1), newArc(V1, 1, "e", OCC2), newArc(V2, 2, "e", OCC2), OCC1<OCC2.
% eqAtomOccOrd("e", OCC1, OCC3) :- eqAtomOccOrd("e", OCC1, OCC2), eqAtomOccOrd("e", OCC2, OCC3).
% fAtom("e", V1, V2) :- newArc(V1, 1, "e", OCC1), newArc(V2, 2, "e", OCC1), not eqAtomOccOrd("e",_,OCC1).
% New inferred edges
% schema e(NODE1, NODE2)
e(V1,V2) :- newArc(V1, 1, "e", OCC), newArc(V2, 2, "e", OCC), ruleOccTrue("e", OCC).
% schema e(OCC, NODE1, NODE2)
e(OCC,V1,V2) :- newArc(V1, 1, "e", OCC), newArc(V2, 2, "e", OCC), ruleOccTrue("e", OCC).
% New inferred missing edges
% schema ne(NODE1, NODE2)
ne(V1,V2) :- newArc(V1, 1, "e", OCC), newArc(V2, 2, "e", OCC), not ruleOccTrue("e", OCC).
% schema ne(OCC, NODE1, NODE2)
ne(OCC,V1,V2) :- newArc(V1, 1, "e", OCC), newArc(V2, 2, "e", OCC), not ruleOccTrue("e", OCC).
% Given these inferred edges, can a triangle exist?
% tri(V1,V2,V3) :- e(V1,V2), e(V2,V3), e(V3,V1).
% triExists :- tri(_,_,_).
%graphviz graph graph_type=directed rankdir=LR
%graphviz edge newArc(HEAD, _, TAIL) label=$2
%graphviz node ruleOccTrue(N) color=green
% :- not ruleHTrue("tri"). % TO ADD
#show ruleOccTrue/2.
#show var/2.
#show newVar/2.
#show ruleOcc/2.
#show ruleH/1.
#show eqOrd/3.
#show neqOrd/3.
#show eqOrdMinimal/3.
#show arc/4.
#show newArc/4.
#show hArc/3.
#show newHArc/3.
#show e/2.
#show ne/2.
#show e/3.
#show ne/3.
#show eqAtomOccOrd/3.
#show fAtom/3.
% #show triExists/0.
% #show tri/3.
#show ruleHTrue/1.
```
|
github_jupyter
|
from PW_explorer.load_worlds import load_worlds
from PW_explorer.run_clingo import run_clingo
%load_ext PWE_NB_Extension
%%clingo --donot-display_input -lci qep_eq_check2 --donot-run
% e(X,Y) :- e(Y,X). --> only if undirected
gnode1(X) :- e1(X,_).
gnode1(X) :- e1(_,X).
gnode1(X) :- ne1(X,_).
gnode1(X) :- ne1(_,X).
gnode2(X) :- e2(X,_).
gnode2(X) :- e2(_,X).
gnode2(X) :- ne2(X,_).
gnode2(X) :- ne2(_,X).
vmap(X,Y) ; not vmap(X,Y) :- gnode1(X), gnode2(Y).
:- vmap(X1,Y1), vmap(X2,Y2), e1(X1,X2), not e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not e1(X1,X2), e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), ne1(X1,X2), not ne2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not ne1(X1,X2), ne2(Y1,Y2).
% 3hop Query
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), thop1(X1,X2,X3,X4), not thop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), not thop1(X1,X2,X3,X4), thop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), thop1(X1,X2,X3), not thop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not thop1(X1,X2,X3), thop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), thop1(X1,X2), not thop2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not thop1(X1,X2), thop2(Y1,Y2).
:- vmap(X1,Y1), thop1(X1), not thop2(Y1).
:- vmap(X1,Y1), not thop1(X1), thop2(Y1).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), nthop1(X1,X2,X3,X4), not nthop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), not nthop1(X1,X2,X3,X4), nthop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), nthop1(X1,X2,X3), not nthop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not nthop1(X1,X2,X3), nthop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), nthop1(X1,X2), not nthop2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not nthop1(X1,X2), nthop2(Y1,Y2).
:- vmap(X1,Y1), nthop1(X1), not nthop2(Y1).
:- vmap(X1,Y1), not nthop1(X1), nthop2(Y1).
% Triangle Query
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), tri1(X1,X2,X3), not tri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not tri1(X1,X2,X3), tri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), tri1(X1,X2), not tri2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not tri1(X1,X2), tri2(Y1,Y2).
:- vmap(X1,Y1), tri1(X1), not tri2(Y1).
:- vmap(X1,Y1), not tri1(X1), tri2(Y1).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), ntri1(X1,X2,X3), not ntri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not ntri1(X1,X2,X3), ntri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), ntri1(X1,X2), not ntri2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not ntri1(X1,X2), ntri2(Y1,Y2).
:- vmap(X1,Y1), ntri1(X1), not ntri2(Y1).
:- vmap(X1,Y1), not ntri1(X1), ntri2(Y1).
% used1(X) :- vmap(X,_).
% :- gnode(X), not used1(X).
% :- vmap(X,Y),vmap(X,Z),Y!=Z.
% :- vmap(Y,X),vmap(Z,X),Y!=Z.
:- gnode1(X), #count {Y: vmap(X,Y)} != 1.
:- gnode2(Y), #count {X: vmap(X,Y)} != 1.
#show.
% #show vmap/2.
%%clingo -l qep_eq_check2 --donot-display_input
e1(x,y). e1(y,z). e1(z,x). tri1(x,z).
e2(x,y). e2(y,z). e2(z,x). tri2(x,y).
%%clingo --donot-display_input -lci qep_subsumption_check2 --donot-run
% e(X,Y) :- e(Y,X). --> only if undirected
gnode1(X) :- e1(X,_).
gnode1(X) :- e1(_,X).
gnode1(X) :- ne1(X,_).
gnode1(X) :- ne1(_,X).
gnode2(X) :- e2(X,_).
gnode2(X) :- e2(_,X).
gnode2(X) :- ne2(X,_).
gnode2(X) :- ne2(_,X).
vmap(X,Y) ; not vmap(X,Y) :- gnode1(X), gnode2(Y).
:- vmap(X1,Y1), vmap(X2,Y2), e1(X1,X2), not e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not e1(X1,X2), e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), ne1(X1,X2), not ne2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not ne1(X1,X2), ne2(Y1,Y2).
% 3hop Query
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), thop1(X1,X2,X3,X4), not thop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), not thop1(X1,X2,X3,X4), thop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), thop1(X1,X2,X3), not thop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not thop1(X1,X2,X3), thop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), thop1(X1,X2), not thop2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not thop1(X1,X2), thop2(Y1,Y2).
:- vmap(X1,Y1), thop1(X1), not thop2(Y1).
:- vmap(X1,Y1), not thop1(X1), thop2(Y1).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), nthop1(X1,X2,X3,X4), not nthop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), not nthop1(X1,X2,X3,X4), nthop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), nthop1(X1,X2,X3), not nthop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not nthop1(X1,X2,X3), nthop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), nthop1(X1,X2), not nthop2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not nthop1(X1,X2), nthop2(Y1,Y2).
:- vmap(X1,Y1), nthop1(X1), not nthop2(Y1).
:- vmap(X1,Y1), not nthop1(X1), nthop2(Y1).
% Triangle Query
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), tri1(X1,X2,X3), not tri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not tri1(X1,X2,X3), tri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), tri1(X1,X2), not tri2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not tri1(X1,X2), tri2(Y1,Y2).
:- vmap(X1,Y1), tri1(X1), not tri2(Y1).
:- vmap(X1,Y1), not tri1(X1), tri2(Y1).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), ntri1(X1,X2,X3), not ntri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not ntri1(X1,X2,X3), ntri2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), ntri1(X1,X2), not ntri2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not ntri1(X1,X2), ntri2(Y1,Y2).
:- vmap(X1,Y1), ntri1(X1), not ntri2(Y1).
:- vmap(X1,Y1), not ntri1(X1), ntri2(Y1).
% used1(X) :- vmap(X,_).
% :- gnode(X), not used1(X).
% :- vmap(X,Y),vmap(X,Z),Y!=Z.
% :- vmap(Y,X),vmap(Z,X),Y!=Z.
:- gnode1(X), #count {Y: vmap(X,Y)} < 1.
:- gnode2(Y), #count {X: vmap(X,Y)} < 1.
#show.
% #show vmap/2.
%%clingo -l qep_subsumption_check2 --donot-display_input
e1(x,y). e1(y,z). e1(z,x). tri1(x,y).
e2(x,y). e2(y,y). e2(y,x). tri2(x,y).
%%clingo --donot-display_input -lci automorphisms --donot-run
% e(X,Y) :- e(Y,X). --> only if undirected
gnode(X) :- e(X,_).
gnode(X) :- e(_,X).
vmap(X,Y) ; vout(X,Y) :- gnode(X), gnode(Y).
:- vmap(X1,Y1), vmap(X2,Y2), e(X1,X2), not e(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not e(X1,X2), e(Y1,Y2).
% used1(X) :- vmap(X,_).
% :- gnode(X), not used1(X).
% :- vmap(X,Y),vmap(X,Z),Y!=Z.
% :- vmap(Y,X),vmap(Z,X),Y!=Z.
:- gnode(X), #count {Y: vmap(X,Y)} != 1.
:- gnode(X), #count {Y: vmap(Y,X)} != 1.
% #show vmap/2.
#show.
%%clingo --donot-display_input -l automorphisms
e(x,e1). e(z1,e1). e(z1,e2). e(z2,e2). e(z2,e3). e(y,e3).
e(x,head). e(y,head).
e(z1,head). e(z2,head).
%%clingo --donot-display_input -lci qep_eq_check --donot-run
% e(X,Y) :- e(Y,X). --> only if undirected
gnode1(X) :- e1(X,_).
gnode1(X) :- e1(_,X).
gnode1(X) :- ne1(X,_).
gnode1(X) :- ne1(_,X).
gnode2(X) :- e2(X,_).
gnode2(X) :- e2(_,X).
gnode2(X) :- ne2(X,_).
gnode2(X) :- ne2(_,X).
vmap(X,Y) ; not vmap(X,Y) :- gnode1(X), gnode2(Y).
:- vmap(X1,Y1), vmap(X2,Y2), e1(X1,X2), not e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not e1(X1,X2), e2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), ne1(X1,X2), not ne2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not ne1(X1,X2), ne2(Y1,Y2).
% used1(X) :- vmap(X,_).
% :- gnode(X), not used1(X).
% :- vmap(X,Y),vmap(X,Z),Y!=Z.
% :- vmap(Y,X),vmap(Z,X),Y!=Z.
:- gnode1(X), #count {Y: vmap(X,Y)} != 1.
:- gnode2(Y), #count {X: vmap(X,Y)} != 1.
#show vmap/2.
%%clingo --donot-display_input -l qep_eq_check -lci thop_4_qep_eq_check --donot-run
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), thop1(X1,X2,X3,X4), not thop2(Y1,Y2,Y3,Y4).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), vmap(X4,Y4), not thop1(X1,X2,X3,X4), thop2(Y1,Y2,Y3,Y4).
%%clingo --donot-display_input -l qep_eq_check -lci thop_3_qep_eq_check --donot-run
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), thop1(X1,X2,X3), not thop2(Y1,Y2,Y3).
:- vmap(X1,Y1), vmap(X2,Y2), vmap(X3,Y3), not thop1(X1,X2,X3), thop2(Y1,Y2,Y3).
%%clingo --donot-display_input -l qep_eq_check -lci thop_2_qep_eq_check --donot-run
:- vmap(X1,Y1), vmap(X2,Y2), thop1(X1,X2), not thop2(Y1,Y2).
:- vmap(X1,Y1), vmap(X2,Y2), not thop1(X1,X2), thop2(Y1,Y2).
%%clingo --donot-display_input -l qep_eq_check -lci thop_1_qep_eq_check --donot-run
:- vmap(X1,Y1), thop1(X1), not thop2(Y1).
:- vmap(X1,Y1), not thop1(X1), thop2(Y1).
%%clingo --donot-display_input -l qep_eq_check -lci thop_0_qep_eq_check --donot-run
%empty
# thop(X,X) :- e(X,X), e(X,Z2), e(Z2,X).
# thop(X,X) :- e(X,Z1), e(Z1,Z1), e(Z1,X).
# thop(X,X) :- e(X,Z1), e(Z1,X), e(X,X).
%%clingo --donot-display_input -l thop_4_qep_eq_check
e1("X","X"). e1("X","Z2"). e1("Z2","X").
thop1("X","X","Z2","X").
e2("X","Z1"). e2("Z1","X"). e2("X","X").
thop2("X","Z1","X","X").
%%clingo --donot-display_input -l thop_2_qep_eq_check
e1("X","X"). e1("X","Z2"). e1("Z2","X").
thop1("X", "X").
e2("X","Z1"). e2("Z1","X"). e2("X","X").
thop2("X","X").
%%clingo --donot-display_input -l thop_1_qep_eq_check
e1("X","X"). e1("X","Z2"). e1("Z2","X").
thop1("X").
e2("X","Z1"). e2("Z1","X"). e2("X","X").
thop2("X").
%%clingo --donot-display_input -l qep_eq_check
e1("X","Y"). ne1("Y","X"). e1("X","Y").
e1("X", head). %e1("Y", head).
ne2("X","Y"). e2("Y","X"). ne2("X","Y").
e2("X", head). %e2("Y", head).
vmap(head, head).
# Should we be computing the isomorphisms on the incidence graph instead?
%%clingo --donot-run
nthop("X","X") :- e("X","Z1"), not e("Z1","Z2"), not e("Z2","X").
nthop("X","X") :- not e("X","Z1"), not e("Z1","Z2"), e("Z2","X").
% nthop("X","X") :- not e("X","Z1"), e("Z1","Z2"), not e("Z2","X")
%%clingo --donot-display_input -l qep_eq_check
e1("X","Z1"). ne1("Z1","Z2"). ne1("Z2","X").
%e1("X", head).
%e1("Z1", head).
%e1("Z2", head).
ne2("X","Z1"). ne2("Z1","Z2"). e2("Z2","X").
%e2("X", head).
%e2("Z1", head).
%e2("Z2", head).
ne2("X","Z1"). e2("Z1","Z2"). ne2("Z2","X").
%e2("X", head).
%e2("Z1", head).
%e2("Z2", head).
vmap(head, head).
%%clingo --donot-display_input -l qep_eq_check
e1("X", e1). e1("Z1", e1). ne1("Z1", e2). ne1("Z2", e2). ne1("Z2", e3). ne1("X", e3).
%ne1("X", head).
%ne1("Z1", head).
%ne1("Z2", head).
ne2("X", e1). ne2("Z1", e1). ne2("Z1", e2). ne2("Z2", e2). e2("Z2", e3). e2("X", e3).
%ne2("X", head).
%ne2("Z1", head).
%ne2("Z2", head).
vmap(head, head).
%%clingo
tri(X,Y,Z) :- e(X,Y), e(Y,Z), e(Z,X), X=Y, X=Z.
%%clingo --donot-display_input -lci query_analysis_starter
% schema var(VAR, DOMAIN). % TO ADD
% schema ruleOcc(ATOM, OCC). % TO ADD
% schema ruleH(HEAD). % TO ADD
% schema ruleOccTrue(ATOM, OCC)
ruleOccTrue(R, OC) ; not ruleOccTrue(R, OC) :- ruleOcc(R, OC).
% schema ruleHTrue(HEAD)
ruleHFalse(H) :- ruleH(H), ruleOcc(R, OCC), not ruleOccTrue(R,OCC).
ruleHTrue(H) :- ruleH(H), not ruleHFalse(H).
% schema eq(VAR1, VAR2, DOMAIN)
eq(A,B,TYPE) ; not eq(A,B,TYPE) :- var(A, TYPE), var(B, TYPE), A!=B.
eq(A,B,TYPE) :- eq(B,A,TYPE).
eq(A,B,TYPE) :- eq(A,C,TYPE), eq(C,B,TYPE), A!=B, B!=C, A!=C.
% schema eqOrd(VAR1, VAR2, DOMAIN)
eqOrd(A,B,TYPE) :- eq(A,B,TYPE), A<B.
% schema arc(VAR, POS, ATOM, OCC). % TO ADD
% schema hArc(VAR, POS, HEAD). % TO ADD
% schema newVar(VAR, DOMAIN)
newVar(VAR, TYPE) :- var(VAR, TYPE), not eqOrd(_,VAR,TYPE).
% schema eqOrdMinimal(VAR1, VAR2, DOMAIN)
eqOrdMinimal(A,B,TYPE) :- eqOrd(A,B,TYPE), newVar(A,TYPE).
% schema neqOrd(VAR1, VAR2, DOMAIN)
neqOrd(A,B,TYPE) :- newVar(A,TYPE), newVar(B,TYPE), A<B.
% Find the new arcs, getting rid of the redundancies
% schema newArc(VAR, POS, ATOM, OCC)
newArc(VAR, POS, ATOM, OCC) :- arc(VAR_ORIG, POS, ATOM, OCC), eqOrd(VAR,VAR_ORIG,TYPE), not eqOrd(_,VAR,TYPE).
newArc(VAR, POS, ATOM, OCC) :- arc(VAR, POS, ATOM, OCC), not eqOrd(_,VAR,_).
% schema newHArc(VAR, POS, HEAD)
newHArc(VAR, POS, H) :- hArc(VAR_ORIG,POS,H), eqOrd(VAR,VAR_ORIG,TYPE), not eqOrd(_,VAR,TYPE).
newHArc(VAR, POS, H) :- hArc(VAR, POS, H), not eqOrd(_,VAR,_).
% It cannot be true that eX and eY have the same variable assignments but one is True while other is False
:- newArc(V1, 1, "e", OCC1), newArc(V2, 2, "e", OCC1), newArc(V1, 1, "e", OCC2), newArc(V2, 2, "e", OCC2), ruleOccTrue("e", OCC1), not ruleOccTrue("e", OCC2).
% eqAtomOccOrd("e", OCC1, OCC2) :- newArc(V1, 1, "e", OCC1), newArc(V2, 2, "e", OCC1), newArc(V1, 1, "e", OCC2), newArc(V2, 2, "e", OCC2), OCC1<OCC2.
% eqAtomOccOrd("e", OCC1, OCC3) :- eqAtomOccOrd("e", OCC1, OCC2), eqAtomOccOrd("e", OCC2, OCC3).
% fAtom("e", V1, V2) :- newArc(V1, 1, "e", OCC1), newArc(V2, 2, "e", OCC1), not eqAtomOccOrd("e",_,OCC1).
% New inferred edges
% schema e(NODE1, NODE2)
e(V1,V2) :- newArc(V1, 1, "e", OCC), newArc(V2, 2, "e", OCC), ruleOccTrue("e", OCC).
% schema e(OCC, NODE1, NODE2)
e(OCC,V1,V2) :- newArc(V1, 1, "e", OCC), newArc(V2, 2, "e", OCC), ruleOccTrue("e", OCC).
% New inferred missing edges
% schema ne(NODE1, NODE2)
ne(V1,V2) :- newArc(V1, 1, "e", OCC), newArc(V2, 2, "e", OCC), not ruleOccTrue("e", OCC).
% schema ne(OCC, NODE1, NODE2)
ne(OCC,V1,V2) :- newArc(V1, 1, "e", OCC), newArc(V2, 2, "e", OCC), not ruleOccTrue("e", OCC).
% Given these inferred edges, can a triangle exist?
% tri(V1,V2,V3) :- e(V1,V2), e(V2,V3), e(V3,V1).
% triExists :- tri(_,_,_).
%graphviz graph graph_type=directed rankdir=LR
%graphviz edge newArc(HEAD, _, TAIL) label=$2
%graphviz node ruleOccTrue(N) color=green
% :- not ruleHTrue("tri"). % TO ADD
#show ruleOccTrue/2.
#show var/2.
#show newVar/2.
#show ruleOcc/2.
#show ruleH/1.
#show eqOrd/3.
#show neqOrd/3.
#show eqOrdMinimal/3.
#show arc/4.
#show newArc/4.
#show hArc/3.
#show newHArc/3.
#show e/2.
#show ne/2.
#show e/3.
#show ne/3.
#show eqAtomOccOrd/3.
#show fAtom/3.
% #show triExists/0.
% #show tri/3.
#show ruleHTrue/1.
| 0.081 | 0.670028 |
# Improving the Random Forest Model Part 1
## Using More Data and Feature Reduction
This is the first of three parts that will examine improving the simple random forest model created in a [previous blog post](https://medium.com/@williamkoehrsen/random-forest-in-python-24d0893d51c0). The first post will focus on achieving better performance through additional data and choosing the most important features.
## Three Approaches to Making a Better ML Model
1. More high-quality data
2. Hyperparameter tuning of algorithm
3. Try different algorithm
We will look at approach 1. here with the other two methods to be explored later!
# Recap of Previous Work
We are working on a supervised, regression machine learning task where the goal is to predict the maximum temperature tomorrow (in Seattle, WA) from past historical data. The first attempt used one year of data for training.
The code below recreates the simple random forest model written up in [my article](https://medium.com/@williamkoehrsen/random-forest-in-python-24d0893d51c0) on Medium. We use the default parameters except for increasing the number of decision trees (n_estimators) to 1000. The original model achieves a mean average error of 3.83 degrees and an accuracy of 94%.
```
# Pandas is used for data manipulation
import pandas as pd
# Read in data as pandas dataframe and display first 5 rows
original_features = pd.read_csv('data/temps.csv')
original_features = pd.get_dummies(original_features)
# Use numpy to convert to arrays
import numpy as np
# Labels are the values we want to predict
original_labels = np.array(original_features['actual'])
# Remove the labels from the features
# axis 1 refers to the columns
original_features= original_features.drop('actual', axis = 1)
# Saving feature names for later use
original_feature_list = list(original_features.columns)
# Convert to numpy array
original_features = np.array(original_features)
# Using Skicit-learn to split data into training and testing sets
from sklearn.model_selection import train_test_split
# Split the data into training and testing sets
original_train_features, original_test_features, original_train_labels, original_test_labels = train_test_split(original_features, original_labels, test_size = 0.25, random_state = 42)
# The baseline predictions are the historical averages
baseline_preds = original_test_features[:, original_feature_list.index('average')]
# Baseline errors, and display average baseline error
baseline_errors = abs(baseline_preds - original_test_labels)
print('Average baseline error: ', round(np.mean(baseline_errors), 2), 'degrees.')
# Import the model we are using
from sklearn.ensemble import RandomForestRegressor
# Instantiate model
rf = RandomForestRegressor(n_estimators= 1000, random_state=42)
# Train the model on training data
rf.fit(original_train_features, original_train_labels);
# Use the forest's predict method on the test data
predictions = rf.predict(original_test_features)
# Calculate the absolute errors
errors = abs(predictions - original_test_labels)
# Print out the mean absolute error (mae)
print('Average model error:', round(np.mean(errors), 2), 'degrees.')
# Compare to baseline
improvement_baseline = 100 * abs(np.mean(errors) - np.mean(baseline_errors)) / np.mean(baseline_errors)
print('Improvement over baseline:', round(improvement_baseline, 2), '%.')
# Calculate mean absolute percentage error (MAPE)
mape = 100 * (errors / original_test_labels)
# Calculate and display accuracy
accuracy = 100 - np.mean(mape)
print('Accuracy:', round(accuracy, 2), '%.')
```
# Collect More Data
All data is obtained from the [NOAA climate data online](https://www.ncdc.noaa.gov/cdo-web/) tool. Feel free to check for your city!
This time we will use 6 years of historical data and include some additional variables to augment the temperature features used in the simple model.
```
# Pandas is used for data manipulation
import pandas as pd
# Read in data as a dataframe
features = pd.read_csv('data/temps_extended.csv')
features.head(5)
print('We have {} days of data with {} variables.'.format(*features.shape))
```
## Numerical and Visual Inspection of Data
```
round(features.describe(), 2)
# Use datetime for dealing with dates
import datetime
# Get years, months, and days
years = features['year']
months = features['month']
days = features['day']
# List and then convert to datetime object
dates = [str(int(year)) + '-' + str(int(month)) + '-' + str(int(day)) for year, month, day in zip(years, months, days)]
dates = [datetime.datetime.strptime(date, '%Y-%m-%d') for date in dates]
# Import matplotlib for plotting and use magic command for Jupyter Notebooks
import matplotlib.pyplot as plt
%matplotlib inline
# Set the style
plt.style.use('fivethirtyeight')
# Set up the plotting layout
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(nrows=2, ncols=2, figsize = (15,10))
fig.autofmt_xdate(rotation = 45)
# Actual max temperature measurement
ax1.plot(dates, features['actual'])
ax1.set_xlabel(''); ax1.set_ylabel('Temperature (F)'); ax1.set_title('Max Temp')
# Temperature from 1 day ago
ax2.plot(dates, features['temp_1'])
ax2.set_xlabel(''); ax2.set_ylabel('Temperature (F)'); ax2.set_title('Prior Max Temp')
# Temperature from 2 days ago
ax3.plot(dates, features['temp_2'])
ax3.set_xlabel('Date'); ax3.set_ylabel('Temperature (F)'); ax3.set_title('Two Days Prior Max Temp')
# Friend Estimate
ax4.plot(dates, features['friend'])
ax4.set_xlabel('Date'); ax4.set_ylabel('Temperature (F)'); ax4.set_title('Friend Estimate')
plt.tight_layout(pad=2)
# Set up the plotting layout
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(nrows=2, ncols=2, figsize = (15,10))
fig.autofmt_xdate(rotation = 45)
# Historical Average Max Temp
ax1.plot(dates, features['average'])
ax1.set_xlabel(''); ax1.set_ylabel('Temperature (F)'); ax1.set_title('Historical Avg Max Temp')
# Prior Avg Wind Speed
ax2.plot(dates, features['ws_1'], 'r-')
ax2.set_xlabel(''); ax2.set_ylabel('Wind Speed (mph)'); ax2.set_title('Prior Wind Speed')
# Prior Precipitation
ax3.plot(dates, features['prcp_1'], 'r-')
ax3.set_xlabel('Date'); ax3.set_ylabel('Precipitation (in)'); ax3.set_title('Prior Precipitation')
# Prior Snowdepth
ax4.plot(dates, features['snwd_1'], 'ro')
ax4.set_xlabel('Date'); ax4.set_ylabel('Snow Depth (in)'); ax4.set_title('Prior Snow Depth')
plt.tight_layout(pad=2)
```
## Pairplots
One of my favorite graphs to make is a pairplot which shows all the relationships between variables in a dataset. We can do this with the seaborn library and examine the plots to see which variables are highly correlated. We would suspect those that are more correlated with max temperature would be more useful for prediction.
```
# Create columns of seasons for pair plotting colors
seasons = []
for month in features['month']:
if month in [1, 2, 12]:
seasons.append('winter')
elif month in [3, 4, 5]:
seasons.append('spring')
elif month in [6, 7, 8]:
seasons.append('summer')
elif month in [9, 10, 11]:
seasons.append('fall')
# Will only use six variables for plotting pairs
reduced_features = features[['temp_1', 'prcp_1', 'average', 'actual']]
reduced_features['season'] = seasons
# Use seaborn for pair plots
import seaborn as sns
sns.set(style="ticks", color_codes=True);
# Create a custom color palete
palette = sns.xkcd_palette(['dark blue', 'dark green', 'gold', 'orange'])
# Make the pair plot with a some aesthetic changes
sns.pairplot(reduced_features, hue = 'season', diag_kind = 'kde', palette= palette, plot_kws=dict(alpha = 0.7),
diag_kws=dict(shade=True));
```
## Data Preparation
```
# One Hot Encoding
features = pd.get_dummies(features)
# Extract features and labels
labels = features['actual']
features = features.drop('actual', axis = 1)
# List of features for later use
feature_list = list(features.columns)
# Convert to numpy arrays
import numpy as np
features = np.array(features)
labels = np.array(labels)
# Training and Testing Sets
from sklearn.model_selection import train_test_split
train_features, test_features, train_labels, test_labels = train_test_split(features, labels,
test_size = 0.25, random_state = 42)
print('Training Features Shape:', train_features.shape)
print('Training Labels Shape:', train_labels.shape)
print('Testing Features Shape:', test_features.shape)
print('Testing Labels Shape:', test_labels.shape)
```
## Establish New Baseline
The new baseline will be the predictions of the model trained on only one year of data but tested on the expanded testing set. In order to make predictions, we will need to restrict the features to those in the original one year of data (exclude the wind speed, precipitation, and snow depth). Testing with the same test set allows us to assess the effect of using additional training data.
```
# Find the original feature indices
original_feature_indices = [feature_list.index(feature) for feature in
feature_list if feature not in
['ws_1', 'prcp_1', 'snwd_1']]
# Create a test set of the original features
original_test_features = test_features[:, original_feature_indices]
# Make predictions on test data using the model trained on original data
baseline_predictions = rf.predict(original_test_features)
# Performance metrics
baseline_errors = abs(baseline_predictions - test_labels)
print('Metrics for Random Forest Trained on Original Data')
print('Average absolute error:', round(np.mean(baseline_errors), 2), 'degrees.')
# Calculate mean absolute percentage error (MAPE)
baseline_mape = 100 * np.mean((baseline_errors / test_labels))
# Calculate and display accuracy
baseline_accuracy = 100 - baseline_mape
print('Accuracy:', round(baseline_accuracy, 2), '%.')
```
## Train on Expanded Data and Features
The rf_exp uses the same number of decision trees (n_estimators) but is trained on the longer dataset with 3 additional features.
```
# Instantiate random forest and train on new features
from sklearn.ensemble import RandomForestRegressor
rf_exp = RandomForestRegressor(n_estimators= 1000, random_state=42)
rf_exp.fit(train_features, train_labels);
```
## Metrics for Expanded Data and Features
```
# Make predictions on test data
predictions = rf_exp.predict(test_features)
# Performance metrics
errors = abs(predictions - test_labels)
print('Metrics for Random Forest Trained on Expanded Data')
print('Average absolute error:', round(np.mean(errors), 4), 'degrees.')
# Calculate mean absolute percentage error (MAPE)
mape = np.mean(100 * (errors / test_labels))
# Compare to baseline
improvement_baseline = 100 * abs(mape - baseline_mape) / baseline_mape
print('Improvement over baseline:', round(improvement_baseline, 2), '%.')
# Calculate and display accuracy
accuracy = 100 - mape
print('Accuracy:', round(accuracy, 2), '%.')
```
Using more data (more data points and more features) has decreased the absolute error, increased performance relative to the baseline, and increased accuracy.
Although the exact metrics will depend on the random state used, overall, we can be confident using additional high-quality data improves our model.
At this point, our model can predict the maximum temperature for tomrrow with an average error of __3.7__ degrees resulting in an accuracy of __93.7%__.
# Feature Reduction
From previous experience and the graphs produced at the beginning, we know that some features are not useful for our temperature prediction problem. To reduce the number of features, which will reduce runtime, hopefully without significantly reducing performance, we can examine the feature importances from the random forest.
### Feature Importances
```
# Get numerical feature importances
importances = list(rf_exp.feature_importances_)
# List of tuples with variable and importance
feature_importances = [(feature, round(importance, 2)) for feature, importance in zip(feature_list, importances)]
# Sort the feature importances by most important first
feature_importances = sorted(feature_importances, key = lambda x: x[1], reverse = True)
# Print out the feature and importances
[print('Variable: {:20} Importance: {}'.format(*pair)) for pair in feature_importances];
```
#### Visualize Feature Importances
```
# Reset style
plt.style.use('fivethirtyeight')
# list of x locations for plotting
x_values = list(range(len(importances)))
# Make a bar chart
plt.bar(x_values, importances, orientation = 'vertical', color = 'r', edgecolor = 'k', linewidth = 1.2)
# Tick labels for x axis
plt.xticks(x_values, feature_list, rotation='vertical')
# Axis labels and title
plt.ylabel('Importance'); plt.xlabel('Variable'); plt.title('Variable Importances');
# List of features sorted from most to least important
sorted_importances = [importance[1] for importance in feature_importances]
sorted_features = [importance[0] for importance in feature_importances]
# Cumulative importances
cumulative_importances = np.cumsum(sorted_importances)
# Make a line graph
plt.plot(x_values, cumulative_importances, 'g-')
# Draw line at 95% of importance retained
plt.hlines(y = 0.95, xmin=0, xmax=len(sorted_importances), color = 'r', linestyles = 'dashed')
# Format x ticks and labels
plt.xticks(x_values, sorted_features, rotation = 'vertical')
# Axis labels and title
plt.xlabel('Variable'); plt.ylabel('Cumulative Importance'); plt.title('Cumulative Importances');
```
### Limit Number of Features
We will now reduce the number of features in use by the model to only those required to account for 95% of the importance.
The same number of features must be used in the training and testing sets.
```
# Find number of features for cumulative importance of 95%
# Add 1 because Python is zero-indexed
print('Number of features for 95% importance:', np.where(cumulative_importances > 0.95)[0][0] + 1)
# Extract the names of the most important features
important_feature_names = [feature[0] for feature in feature_importances[0:6]]
# Find the columns of the most important features
important_indices = [feature_list.index(feature) for feature in important_feature_names]
# Create training and testing sets with only the important features
important_train_features = train_features[:, important_indices]
important_test_features = test_features[:, important_indices]
# Sanity check on operations
print('Important train features shape:', important_train_features.shape)
print('Important test features shape:', important_test_features.shape)
```
### Training on Important Features
```
# Train the expanded model on only the important features
rf_exp.fit(important_train_features, train_labels);
```
### Evaluate on Important features
```
# Make predictions on test data
predictions = rf_exp.predict(important_test_features)
# Performance metrics
errors = abs(predictions - test_labels)
print('Average absolute error:', round(np.mean(errors), 4), 'degrees.')
# Calculate mean absolute percentage error (MAPE)
mape = 100 * (errors / test_labels)
# Calculate and display accuracy
accuracy = 100 - np.mean(mape)
print('Accuracy:', round(accuracy, 2), '%.')
```
Using only the eight most important features (instead of all 16) results in a minor decrease in accuracy by 0.17 degrees. For some models, decreasing the number of features can increase performance and therefore should be done. However, in other situations, performance will decrease but run time will also decrease. The final decision on how many features to retain will therefore be a trade-off between accuracy and run time.
## Compare Trade-Offs
```
# Use time library for run time evaluation
import time
# All features training and testing time
all_features_time = []
# Do 10 iterations and take average for all features
for _ in range(10):
start_time = time.time()
rf_exp.fit(train_features, train_labels)
all_features_predictions = rf_exp.predict(test_features)
end_time = time.time()
all_features_time.append(end_time - start_time)
all_features_time = np.mean(all_features_time)
print('All features total training and testing time:', round(all_features_time, 2), 'seconds.')
# Time training and testing for reduced feature set
reduced_features_time = []
# Do 10 iterations and take average
for _ in range(10):
start_time = time.time()
rf_exp.fit(important_train_features, train_labels)
reduced_features_predictions = rf_exp.predict(important_test_features)
end_time = time.time()
reduced_features_time.append(end_time - start_time)
reduced_features_time = np.mean(reduced_features_time)
print('Reduced features total training and testing time:', round(reduced_features_time, 2), 'seconds.')
```
### Accuracy vs Run-Time
```
all_accuracy = 100 * (1- np.mean(abs(all_features_predictions - test_labels) / test_labels))
reduced_accuracy = 100 * (1- np.mean(abs(reduced_features_predictions - test_labels) / test_labels))
comparison = pd.DataFrame({'features': ['all (17)', 'reduced (5)'],
'run_time': [round(all_features_time, 2), round(reduced_features_time, 2)],
'accuracy': [round(all_accuracy, 2), round(reduced_accuracy, 2)]})
comparison[['features', 'accuracy', 'run_time']]
relative_accuracy_decrease = 100 * (all_accuracy - reduced_accuracy) / all_accuracy
print('Relative decrease in accuracy:', round(relative_accuracy_decrease, 3), '%.')
relative_runtime_decrease = 100 * (all_features_time - reduced_features_time) / all_features_time
print('Relative decrease in run time:', round(relative_runtime_decrease, 3), '%.')
```
# Concluding Graphs
```
# Find the original feature indices
original_feature_indices = [feature_list.index(feature) for feature in
feature_list if feature not in
['ws_1', 'prcp_1', 'snwd_1']]
# Create a test set of the original features
original_test_features = test_features[:, original_feature_indices]
# Time to train on original data set (1 year)
original_features_time = []
# Do 10 iterations and take average for all features
for _ in range(10):
start_time = time.time()
rf.fit(original_train_features, original_train_labels)
original_features_predictions = rf.predict(original_test_features)
end_time = time.time()
original_features_time.append(end_time - start_time)
original_features_time = np.mean(original_features_time)
# Calculate mean absolute error for each model
original_mae = np.mean(abs(original_features_predictions - test_labels))
exp_all_mae = np.mean(abs(all_features_predictions - test_labels))
exp_reduced_mae = np.mean(abs(reduced_features_predictions - test_labels))
# Calculate accuracy for model trained on 1 year of data
original_accuracy = 100 * (1 - np.mean(abs(original_features_predictions - test_labels) / test_labels))
# Create a dataframe for comparison
model_comparison = pd.DataFrame({'model': ['original', 'exp_all', 'exp_reduced'],
'error (degrees)': [original_mae, exp_all_mae, exp_reduced_mae],
'accuracy': [original_accuracy, all_accuracy, reduced_accuracy],
'run_time (s)': [original_features_time, all_features_time, reduced_features_time]})
# Order the dataframe
model_comparison = model_comparison[['model', 'error (degrees)', 'accuracy', 'run_time (s)']]
```
### Comparison of Three Models
There are three models to compare: the model trained on a single year of data and the original number of features, the model trained on six years of data with the full set of features, and the model trained on six years of data with only the 6 most important features
```
model_comparison
# Make plots
# Set up the plotting layout
fig, (ax1, ax2, ax3) = plt.subplots(nrows=3, ncols=1, figsize = (8,16), sharex = True)
# Set up x-axis
x_values = [0, 1, 2]
labels = list(model_comparison['model'])
plt.xticks(x_values, labels)
# Set up fonts
fontdict = {'fontsize': 18}
fontdict_yaxis = {'fontsize': 14}
# Error Comparison
ax1.bar(x_values, model_comparison['error (degrees)'], color = ['b', 'r', 'g'], edgecolor = 'k', linewidth = 1.5)
ax1.set_ylim(bottom = 3.5, top = 4.5)
ax1.set_ylabel('Error (degrees) (F)', fontdict = fontdict_yaxis);
ax1.set_title('Model Error Comparison', fontdict= fontdict)
# Accuracy Comparison
ax2.bar(x_values, model_comparison['accuracy'], color = ['b', 'r', 'g'], edgecolor = 'k', linewidth = 1.5)
ax2.set_ylim(bottom = 92, top = 94)
ax2.set_ylabel('Accuracy (%)', fontdict = fontdict_yaxis);
ax2.set_title('Model Accuracy Comparison', fontdict= fontdict)
# Run Time Comparison
ax3.bar(x_values, model_comparison['run_time (s)'], color = ['b', 'r', 'g'], edgecolor = 'k', linewidth = 1.5)
ax3.set_ylim(bottom = 2, top = 12)
ax3.set_ylabel('Run Time (sec)', fontdict = fontdict_yaxis);
ax3.set_title('Model Run-Time Comparison', fontdict= fontdict);
```
Check in on [my blog](https://medium.com/@williamkoehrsen) for more data science/machine learning articles. Additional parts of this machine learning exercise will be out shortly. I appreciate any comments and constructive feedback.
|
github_jupyter
|
# Pandas is used for data manipulation
import pandas as pd
# Read in data as pandas dataframe and display first 5 rows
original_features = pd.read_csv('data/temps.csv')
original_features = pd.get_dummies(original_features)
# Use numpy to convert to arrays
import numpy as np
# Labels are the values we want to predict
original_labels = np.array(original_features['actual'])
# Remove the labels from the features
# axis 1 refers to the columns
original_features= original_features.drop('actual', axis = 1)
# Saving feature names for later use
original_feature_list = list(original_features.columns)
# Convert to numpy array
original_features = np.array(original_features)
# Using Skicit-learn to split data into training and testing sets
from sklearn.model_selection import train_test_split
# Split the data into training and testing sets
original_train_features, original_test_features, original_train_labels, original_test_labels = train_test_split(original_features, original_labels, test_size = 0.25, random_state = 42)
# The baseline predictions are the historical averages
baseline_preds = original_test_features[:, original_feature_list.index('average')]
# Baseline errors, and display average baseline error
baseline_errors = abs(baseline_preds - original_test_labels)
print('Average baseline error: ', round(np.mean(baseline_errors), 2), 'degrees.')
# Import the model we are using
from sklearn.ensemble import RandomForestRegressor
# Instantiate model
rf = RandomForestRegressor(n_estimators= 1000, random_state=42)
# Train the model on training data
rf.fit(original_train_features, original_train_labels);
# Use the forest's predict method on the test data
predictions = rf.predict(original_test_features)
# Calculate the absolute errors
errors = abs(predictions - original_test_labels)
# Print out the mean absolute error (mae)
print('Average model error:', round(np.mean(errors), 2), 'degrees.')
# Compare to baseline
improvement_baseline = 100 * abs(np.mean(errors) - np.mean(baseline_errors)) / np.mean(baseline_errors)
print('Improvement over baseline:', round(improvement_baseline, 2), '%.')
# Calculate mean absolute percentage error (MAPE)
mape = 100 * (errors / original_test_labels)
# Calculate and display accuracy
accuracy = 100 - np.mean(mape)
print('Accuracy:', round(accuracy, 2), '%.')
# Pandas is used for data manipulation
import pandas as pd
# Read in data as a dataframe
features = pd.read_csv('data/temps_extended.csv')
features.head(5)
print('We have {} days of data with {} variables.'.format(*features.shape))
round(features.describe(), 2)
# Use datetime for dealing with dates
import datetime
# Get years, months, and days
years = features['year']
months = features['month']
days = features['day']
# List and then convert to datetime object
dates = [str(int(year)) + '-' + str(int(month)) + '-' + str(int(day)) for year, month, day in zip(years, months, days)]
dates = [datetime.datetime.strptime(date, '%Y-%m-%d') for date in dates]
# Import matplotlib for plotting and use magic command for Jupyter Notebooks
import matplotlib.pyplot as plt
%matplotlib inline
# Set the style
plt.style.use('fivethirtyeight')
# Set up the plotting layout
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(nrows=2, ncols=2, figsize = (15,10))
fig.autofmt_xdate(rotation = 45)
# Actual max temperature measurement
ax1.plot(dates, features['actual'])
ax1.set_xlabel(''); ax1.set_ylabel('Temperature (F)'); ax1.set_title('Max Temp')
# Temperature from 1 day ago
ax2.plot(dates, features['temp_1'])
ax2.set_xlabel(''); ax2.set_ylabel('Temperature (F)'); ax2.set_title('Prior Max Temp')
# Temperature from 2 days ago
ax3.plot(dates, features['temp_2'])
ax3.set_xlabel('Date'); ax3.set_ylabel('Temperature (F)'); ax3.set_title('Two Days Prior Max Temp')
# Friend Estimate
ax4.plot(dates, features['friend'])
ax4.set_xlabel('Date'); ax4.set_ylabel('Temperature (F)'); ax4.set_title('Friend Estimate')
plt.tight_layout(pad=2)
# Set up the plotting layout
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(nrows=2, ncols=2, figsize = (15,10))
fig.autofmt_xdate(rotation = 45)
# Historical Average Max Temp
ax1.plot(dates, features['average'])
ax1.set_xlabel(''); ax1.set_ylabel('Temperature (F)'); ax1.set_title('Historical Avg Max Temp')
# Prior Avg Wind Speed
ax2.plot(dates, features['ws_1'], 'r-')
ax2.set_xlabel(''); ax2.set_ylabel('Wind Speed (mph)'); ax2.set_title('Prior Wind Speed')
# Prior Precipitation
ax3.plot(dates, features['prcp_1'], 'r-')
ax3.set_xlabel('Date'); ax3.set_ylabel('Precipitation (in)'); ax3.set_title('Prior Precipitation')
# Prior Snowdepth
ax4.plot(dates, features['snwd_1'], 'ro')
ax4.set_xlabel('Date'); ax4.set_ylabel('Snow Depth (in)'); ax4.set_title('Prior Snow Depth')
plt.tight_layout(pad=2)
# Create columns of seasons for pair plotting colors
seasons = []
for month in features['month']:
if month in [1, 2, 12]:
seasons.append('winter')
elif month in [3, 4, 5]:
seasons.append('spring')
elif month in [6, 7, 8]:
seasons.append('summer')
elif month in [9, 10, 11]:
seasons.append('fall')
# Will only use six variables for plotting pairs
reduced_features = features[['temp_1', 'prcp_1', 'average', 'actual']]
reduced_features['season'] = seasons
# Use seaborn for pair plots
import seaborn as sns
sns.set(style="ticks", color_codes=True);
# Create a custom color palete
palette = sns.xkcd_palette(['dark blue', 'dark green', 'gold', 'orange'])
# Make the pair plot with a some aesthetic changes
sns.pairplot(reduced_features, hue = 'season', diag_kind = 'kde', palette= palette, plot_kws=dict(alpha = 0.7),
diag_kws=dict(shade=True));
# One Hot Encoding
features = pd.get_dummies(features)
# Extract features and labels
labels = features['actual']
features = features.drop('actual', axis = 1)
# List of features for later use
feature_list = list(features.columns)
# Convert to numpy arrays
import numpy as np
features = np.array(features)
labels = np.array(labels)
# Training and Testing Sets
from sklearn.model_selection import train_test_split
train_features, test_features, train_labels, test_labels = train_test_split(features, labels,
test_size = 0.25, random_state = 42)
print('Training Features Shape:', train_features.shape)
print('Training Labels Shape:', train_labels.shape)
print('Testing Features Shape:', test_features.shape)
print('Testing Labels Shape:', test_labels.shape)
# Find the original feature indices
original_feature_indices = [feature_list.index(feature) for feature in
feature_list if feature not in
['ws_1', 'prcp_1', 'snwd_1']]
# Create a test set of the original features
original_test_features = test_features[:, original_feature_indices]
# Make predictions on test data using the model trained on original data
baseline_predictions = rf.predict(original_test_features)
# Performance metrics
baseline_errors = abs(baseline_predictions - test_labels)
print('Metrics for Random Forest Trained on Original Data')
print('Average absolute error:', round(np.mean(baseline_errors), 2), 'degrees.')
# Calculate mean absolute percentage error (MAPE)
baseline_mape = 100 * np.mean((baseline_errors / test_labels))
# Calculate and display accuracy
baseline_accuracy = 100 - baseline_mape
print('Accuracy:', round(baseline_accuracy, 2), '%.')
# Instantiate random forest and train on new features
from sklearn.ensemble import RandomForestRegressor
rf_exp = RandomForestRegressor(n_estimators= 1000, random_state=42)
rf_exp.fit(train_features, train_labels);
# Make predictions on test data
predictions = rf_exp.predict(test_features)
# Performance metrics
errors = abs(predictions - test_labels)
print('Metrics for Random Forest Trained on Expanded Data')
print('Average absolute error:', round(np.mean(errors), 4), 'degrees.')
# Calculate mean absolute percentage error (MAPE)
mape = np.mean(100 * (errors / test_labels))
# Compare to baseline
improvement_baseline = 100 * abs(mape - baseline_mape) / baseline_mape
print('Improvement over baseline:', round(improvement_baseline, 2), '%.')
# Calculate and display accuracy
accuracy = 100 - mape
print('Accuracy:', round(accuracy, 2), '%.')
# Get numerical feature importances
importances = list(rf_exp.feature_importances_)
# List of tuples with variable and importance
feature_importances = [(feature, round(importance, 2)) for feature, importance in zip(feature_list, importances)]
# Sort the feature importances by most important first
feature_importances = sorted(feature_importances, key = lambda x: x[1], reverse = True)
# Print out the feature and importances
[print('Variable: {:20} Importance: {}'.format(*pair)) for pair in feature_importances];
# Reset style
plt.style.use('fivethirtyeight')
# list of x locations for plotting
x_values = list(range(len(importances)))
# Make a bar chart
plt.bar(x_values, importances, orientation = 'vertical', color = 'r', edgecolor = 'k', linewidth = 1.2)
# Tick labels for x axis
plt.xticks(x_values, feature_list, rotation='vertical')
# Axis labels and title
plt.ylabel('Importance'); plt.xlabel('Variable'); plt.title('Variable Importances');
# List of features sorted from most to least important
sorted_importances = [importance[1] for importance in feature_importances]
sorted_features = [importance[0] for importance in feature_importances]
# Cumulative importances
cumulative_importances = np.cumsum(sorted_importances)
# Make a line graph
plt.plot(x_values, cumulative_importances, 'g-')
# Draw line at 95% of importance retained
plt.hlines(y = 0.95, xmin=0, xmax=len(sorted_importances), color = 'r', linestyles = 'dashed')
# Format x ticks and labels
plt.xticks(x_values, sorted_features, rotation = 'vertical')
# Axis labels and title
plt.xlabel('Variable'); plt.ylabel('Cumulative Importance'); plt.title('Cumulative Importances');
# Find number of features for cumulative importance of 95%
# Add 1 because Python is zero-indexed
print('Number of features for 95% importance:', np.where(cumulative_importances > 0.95)[0][0] + 1)
# Extract the names of the most important features
important_feature_names = [feature[0] for feature in feature_importances[0:6]]
# Find the columns of the most important features
important_indices = [feature_list.index(feature) for feature in important_feature_names]
# Create training and testing sets with only the important features
important_train_features = train_features[:, important_indices]
important_test_features = test_features[:, important_indices]
# Sanity check on operations
print('Important train features shape:', important_train_features.shape)
print('Important test features shape:', important_test_features.shape)
# Train the expanded model on only the important features
rf_exp.fit(important_train_features, train_labels);
# Make predictions on test data
predictions = rf_exp.predict(important_test_features)
# Performance metrics
errors = abs(predictions - test_labels)
print('Average absolute error:', round(np.mean(errors), 4), 'degrees.')
# Calculate mean absolute percentage error (MAPE)
mape = 100 * (errors / test_labels)
# Calculate and display accuracy
accuracy = 100 - np.mean(mape)
print('Accuracy:', round(accuracy, 2), '%.')
# Use time library for run time evaluation
import time
# All features training and testing time
all_features_time = []
# Do 10 iterations and take average for all features
for _ in range(10):
start_time = time.time()
rf_exp.fit(train_features, train_labels)
all_features_predictions = rf_exp.predict(test_features)
end_time = time.time()
all_features_time.append(end_time - start_time)
all_features_time = np.mean(all_features_time)
print('All features total training and testing time:', round(all_features_time, 2), 'seconds.')
# Time training and testing for reduced feature set
reduced_features_time = []
# Do 10 iterations and take average
for _ in range(10):
start_time = time.time()
rf_exp.fit(important_train_features, train_labels)
reduced_features_predictions = rf_exp.predict(important_test_features)
end_time = time.time()
reduced_features_time.append(end_time - start_time)
reduced_features_time = np.mean(reduced_features_time)
print('Reduced features total training and testing time:', round(reduced_features_time, 2), 'seconds.')
all_accuracy = 100 * (1- np.mean(abs(all_features_predictions - test_labels) / test_labels))
reduced_accuracy = 100 * (1- np.mean(abs(reduced_features_predictions - test_labels) / test_labels))
comparison = pd.DataFrame({'features': ['all (17)', 'reduced (5)'],
'run_time': [round(all_features_time, 2), round(reduced_features_time, 2)],
'accuracy': [round(all_accuracy, 2), round(reduced_accuracy, 2)]})
comparison[['features', 'accuracy', 'run_time']]
relative_accuracy_decrease = 100 * (all_accuracy - reduced_accuracy) / all_accuracy
print('Relative decrease in accuracy:', round(relative_accuracy_decrease, 3), '%.')
relative_runtime_decrease = 100 * (all_features_time - reduced_features_time) / all_features_time
print('Relative decrease in run time:', round(relative_runtime_decrease, 3), '%.')
# Find the original feature indices
original_feature_indices = [feature_list.index(feature) for feature in
feature_list if feature not in
['ws_1', 'prcp_1', 'snwd_1']]
# Create a test set of the original features
original_test_features = test_features[:, original_feature_indices]
# Time to train on original data set (1 year)
original_features_time = []
# Do 10 iterations and take average for all features
for _ in range(10):
start_time = time.time()
rf.fit(original_train_features, original_train_labels)
original_features_predictions = rf.predict(original_test_features)
end_time = time.time()
original_features_time.append(end_time - start_time)
original_features_time = np.mean(original_features_time)
# Calculate mean absolute error for each model
original_mae = np.mean(abs(original_features_predictions - test_labels))
exp_all_mae = np.mean(abs(all_features_predictions - test_labels))
exp_reduced_mae = np.mean(abs(reduced_features_predictions - test_labels))
# Calculate accuracy for model trained on 1 year of data
original_accuracy = 100 * (1 - np.mean(abs(original_features_predictions - test_labels) / test_labels))
# Create a dataframe for comparison
model_comparison = pd.DataFrame({'model': ['original', 'exp_all', 'exp_reduced'],
'error (degrees)': [original_mae, exp_all_mae, exp_reduced_mae],
'accuracy': [original_accuracy, all_accuracy, reduced_accuracy],
'run_time (s)': [original_features_time, all_features_time, reduced_features_time]})
# Order the dataframe
model_comparison = model_comparison[['model', 'error (degrees)', 'accuracy', 'run_time (s)']]
model_comparison
# Make plots
# Set up the plotting layout
fig, (ax1, ax2, ax3) = plt.subplots(nrows=3, ncols=1, figsize = (8,16), sharex = True)
# Set up x-axis
x_values = [0, 1, 2]
labels = list(model_comparison['model'])
plt.xticks(x_values, labels)
# Set up fonts
fontdict = {'fontsize': 18}
fontdict_yaxis = {'fontsize': 14}
# Error Comparison
ax1.bar(x_values, model_comparison['error (degrees)'], color = ['b', 'r', 'g'], edgecolor = 'k', linewidth = 1.5)
ax1.set_ylim(bottom = 3.5, top = 4.5)
ax1.set_ylabel('Error (degrees) (F)', fontdict = fontdict_yaxis);
ax1.set_title('Model Error Comparison', fontdict= fontdict)
# Accuracy Comparison
ax2.bar(x_values, model_comparison['accuracy'], color = ['b', 'r', 'g'], edgecolor = 'k', linewidth = 1.5)
ax2.set_ylim(bottom = 92, top = 94)
ax2.set_ylabel('Accuracy (%)', fontdict = fontdict_yaxis);
ax2.set_title('Model Accuracy Comparison', fontdict= fontdict)
# Run Time Comparison
ax3.bar(x_values, model_comparison['run_time (s)'], color = ['b', 'r', 'g'], edgecolor = 'k', linewidth = 1.5)
ax3.set_ylim(bottom = 2, top = 12)
ax3.set_ylabel('Run Time (sec)', fontdict = fontdict_yaxis);
ax3.set_title('Model Run-Time Comparison', fontdict= fontdict);
| 0.625209 | 0.987092 |
################################################################################
**Author**: _Pradip Kumar Das_
**License:** https://github.com/PradipKumarDas/Competitions/blob/main/LICENSE
**Profile & Contact:** [LinkedIn](https://www.linkedin.com/in/daspradipkumar/) | [GitHub](https://github.com/PradipKumarDas) | [Kaggle](https://www.kaggle.com/pradipkumardas) | pradipkumardas@hotmail.com (Email)
################################################################################
# IPL 2021 Match Score Prediction Contest Organized by IIT Madras Online B.Sc. Programme Team
## Few Deep Learning Models
```
# Imports required packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.preprocessing import OneHotEncoder
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
from tensorflow.keras import models
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
from tensorflow.keras import regularizers
import pickle
# Sets Pandas option to show all columns
pd.set_option('display.max_columns', None)
```
#### Downloads dataset
```
# Deletes the following files before downloading the same files
!rm Data/README.txt Data/ll_matches.csv Data/ipl_csv2.zip
# Downloads the dataset from cricsheet.org/downloads (overwrites the file if exists)
!wget https://cricsheet.org/downloads/ipl_csv2.zip -O Data/ipl_csv2.zip
# Unzips the data (overwrites existing files having same name)
!unzip -o -d Data Data/ipl_csv2.zip all_matches.csv README.txt
```
#### Loads dataset and checks how it looks
```
# Load data for all matches
data = pd.read_csv("Data/all_matches.csv")
# Checks top few rows of the data
data.head()
# Checks bottom rows of the data
data.tail()
# Converts all string data into lowercase
data.venue = data.venue.str.lower()
data.batting_team = data.batting_team.str.lower()
data.bowling_team = data.bowling_team.str.lower()
data.striker = data.striker.str.lower()
data.non_striker = data.non_striker.str.lower()
data.bowler = data.bowler.str.lower()
# Checks for missing values
data.isna().sum()
```
#### Checks for venues for duplicates with slightly different names, if any, and updates the rows with the same venue names accordingly
```
data.venue.value_counts().sort_index()
# Updates these venues that are mentioned in different names with same name
data.venue[data.venue.str.contains("arun jaitley",
case=False)] = "arun jaitley stadium"
data.venue[data.venue.str.contains("brabourne",
case=False)] = "brabourne stadium"
data.venue[data.venue.str.contains("chinnaswamy",
case=False)] = "m. chinnaswamy stadium"
data.venue[data.venue.str.contains("chidambaram",
case=False)] = "m. a. chidambaram stadium"
data.venue[data.venue.str.contains(r'narendra modi',
case=False)] = "narendra modi stadium"
data.venue[data.venue.str.contains(r'punjab cricket|is bindra|inderjit singh bindra',
case=False)] = "is bindra stadium"
data.venue[data.venue.str.contains("rajiv gandhi",
case=False)] = "rajiv gandhi international cricket stadium"
data.venue[data.venue.str.contains("wankhede",
case=False)] = "wankhede stadium"
```
#### Checks for teams for duplicates with slightly different names, if any, and updates the rows with the same team names accordingly
```
data.batting_team.append(data.bowling_team).value_counts().sort_index()
# Updates team name from "Delhi Daredevils" with the new name "Delhi Capitals"
data.batting_team[data.batting_team.str.contains("delhi daredevils", case=False)] = "delhi capitals"
data.bowling_team[data.bowling_team.str.contains("delhi daredevils", case=False)] = "delhi capitals"
# Updates team name from "Kings XI Punjab" with the new name "Punjab Kings"
data.batting_team[data.batting_team.str.contains("kings xi punjab", case=False)] = "punjab kings"
data.bowling_team[data.bowling_team.str.contains("kings xi punjab", case=False)] = "punjab kings"
# Updates appropriate team name for "Rising Pune Supergiant"
data.batting_team[data.batting_team.str.contains("rising pune supergiants", case=False)] = "rising pune supergiant"
data.bowling_team[data.bowling_team.str.contains("rising pune supergiants", case=False)] = "rising pune supergiant"
# Inserts a new calculated column called "score_off_ball" which is a sum of values in
# columns "runs_off_bat" and "extras" just after column "extras" to indicate contributing score off the ball
# to make calculating total score at the end of the match easy
data.insert(loc=13, column="score_off_ball", value=data.runs_off_bat + data.extras)
# Removes columsn that are not required
columns_required = ["match_id", "venue", "innings", "ball", "batting_team", "bowling_team",
"striker", "non_striker", "bowler", "score_off_ball"]
data = data[columns_required]
# Considers only first 6 overs
data_6_overs = data[data.ball <= 6.0]
# Checks how the 6 overs data looks
data_6_overs
```
#### Some statistics below to realize the trend
```
# Venue wise average innings runs after 6 overs
data_6_overs.groupby(
["venue", "match_id", "innings"]).score_off_ball.sum().reset_index().groupby(
["venue"]).score_off_ball.mean().sort_values(ascending=False)
# Batting team wise average innings runs after 6 overs
data_6_overs.groupby(["batting_team", "match_id", "innings"]).score_off_ball.sum().reset_index().groupby(
["batting_team"]).score_off_ball.mean().sort_values(ascending=False)
# Striker's inning wise average scores after 6 overs
pd.set_option("display.max_rows", None) # To view all the rows temporarily
data_6_overs.groupby(
["striker", "match_id", "innings"]).score_off_ball.sum().reset_index().groupby(
["striker"]).score_off_ball.mean().sort_values(ascending=False)
pd.reset_option('display.max_rows') # Resets to its default number of visible rows
# Resets its index
data_6_overs.reset_index(drop = True, inplace = True)
# Checks how the 6 overs data looks after reset index and with new column "score_off_ball"
data_6_overs
# Creates initial innings group out of 6 overs to further add other innings related data into
innings = data_6_overs.groupby(
["match_id", "venue", "innings", "batting_team", "bowling_team"])
# Calculates the innings wise score at the end of 6 overs
innings_runs = innings.score_off_ball.sum()
innings_runs.name = "score_6_overs"
# Checks how the innings data looks like
innings_runs
# Resets the index of the group to flatten the grouped data
data_6_overs_agg = innings_runs.reset_index()
# Checks how the flattend innings wise grouped data looks
data_6_overs_agg
# Encodes venues with one-hot encoding technique
venue_count = len(data_6_overs_agg.venue.unique())
venue_encoder = OneHotEncoder(handle_unknown='ignore')
venue_encoded = pd.DataFrame(venue_encoder.fit_transform(data_6_overs_agg[["venue"]]).toarray(),
columns=[("venue_" + str(i)) for i in range(venue_count)])
# Saves the encoder into persistent store for later use
with open("Models/Venue_Encoder.pickle", "wb") as f:
pickle.dump(venue_encoder, f, pickle.HIGHEST_PROTOCOL)
# Joins the encoded venue columns with the dataset
data_6_overs_agg = data_6_overs_agg.join(venue_encoded).drop(["venue"], axis = 1)
# Encodes innings with one-hot encoding technique
innings_count = len(data_6_overs_agg.innings.unique())
innings_encoder = OneHotEncoder(handle_unknown='ignore')
innings_encoded = pd.DataFrame(innings_encoder.fit_transform(data_6_overs_agg[["innings"]]).toarray(),
columns=[("innings_" + str(i)) for i in range(innings_count)])
# Saves the encoder into persistent store for later use
with open("Models/Innings_Encoder.pickle", "wb") as f:
pickle.dump(innings_encoder, f, pickle.HIGHEST_PROTOCOL)
# Joins the encoded innings columns with the dataset
data_6_overs_agg = data_6_overs_agg.join(innings_encoded).drop(["innings"], axis = 1)
# Now, encodes teams with one-hot encoding technique
team = data_6_overs_agg.batting_team.append(data_6_overs_agg.bowling_team).unique()
team_count = len(team)
team_encoder = OneHotEncoder(handle_unknown='ignore')
team_encoder.fit(pd.DataFrame(team))
batting_team_encoded = pd.DataFrame(team_encoder.transform(data_6_overs_agg[["batting_team"]]).toarray(),
columns=[("batting_team_" + str(i)) for i in range(team_count)])
bowling_team_encoded = pd.DataFrame(team_encoder.transform(data_6_overs_agg[["bowling_team"]]).toarray(),
columns=[("bowling_team_" + str(i)) for i in range(team_count)])
# Saves the encoder into persistent store for later use
with open("Models/Team_Encoder.pickle", "wb") as f:
pickle.dump(team_encoder, f, pickle.HIGHEST_PROTOCOL)
# Joins the encoded team columns with the dataset
data_6_overs_agg = data_6_overs_agg.join(batting_team_encoded).drop(["batting_team"], axis = 1)
data_6_overs_agg = data_6_overs_agg.join(bowling_team_encoded).drop(["bowling_team"], axis = 1)
# Now, encodes players with one-hot encoding technique
player = data_6_overs.striker.append(
data_6_overs.non_striker).append(data_6_overs.bowler).unique()
player_count = len(player)
player_encoder = OneHotEncoder(handle_unknown='ignore')
player_encoder.fit(pd.DataFrame(player))
# Saves the encoder into persistent store for later use
with open("Models/Player_Encoder.pickle", "wb") as f:
pickle.dump(player_encoder, f, pickle.HIGHEST_PROTOCOL)
# Transforms both striker and non-striker
striker_encoded_array = player_encoder.transform(data_6_overs[["striker"]]).toarray()
non_striker_encoded_array = player_encoder.transform(data_6_overs[["non_striker"]]).toarray()
# Considers striker and non-striker as batsmen, creates a dataframe out of it,
# then joins with innings data to consider this batsmen information in the main dataset
data_6_overs_batsmen = data_6_overs[["match_id", "innings"]].join(
pd.DataFrame(np.add(striker_encoded_array, non_striker_encoded_array),
columns=[("batsman_" + str(i)) for i in range(player_count)]))
# Transforms bowlers and creates a dataframe out of it, and then joins with innings data to
# consider this bowlers information in the main dataset
data_6_overs_bowlers = data_6_overs[["match_id", "innings"]].join(
pd.DataFrame(player_encoder.transform(data_6_overs[["bowler"]]).toarray(),
columns=[("bowler_" + str(i)) for i in range(player_count)]))
# Combines all the batsmen, both striker and non-striker innings wise, and
# resets index of the aggregation to get tabular data
batsmen_encoded = data_6_overs_batsmen.groupby(["match_id", "innings"]).max().reset_index()
# Drops columns "match_id" and "innings" as these columns already exist in aggregated dataset that this
# data will be joined with
batsmen_encoded.drop(["match_id", "innings"], inplace=True, axis=1)
# Combines all the bowlers innings wise and resets index of the aggregation to get tabular data
bowlers_encoded = data_6_overs_bowlers.groupby(["match_id", "innings"]).max().reset_index()
# Drops columns "match_id" and "innings" as these columns already exist in aggregated dataset that this
# data will be joined with
bowlers_encoded.drop(["match_id", "innings"], inplace=True, axis=1)
# Joins encoded batsmen and bowlers information with aggregated innings data
data_6_overs_agg = data_6_overs_agg.join(batsmen_encoded)
data_6_overs_agg = data_6_overs_agg.join(bowlers_encoded)
# Checks how the final dataset the machine learning model will be trained on looks like
data_6_overs_agg
```
## Let's now first build a Fully Connected Dense Neural Network model.
```
# But, first removes the column "match_id" as it is not required for machine learning model
data_6_overs_agg.drop(["match_id"], axis=1, inplace=True)
# Converts DataFrame into 2D tensor
data_6_overs_agg_array = data_6_overs_agg.to_numpy()
# Seperates training labels
X_train, y_train = data_6_overs_agg_array[:,1:], data_6_overs_agg_array[:,0]
# Splits the available data into train and test data sets
X_train, X_test, y_train, y_test = train_test_split(X_train, y_train, test_size = 0.2, shuffle = True)
# Creates a sequential dense neural network model
model = Sequential()
model.add(Dense(128, activation="relu", input_shape=(X_train.shape[1],)))
model.add(Dense(64, activation="relu"))
model.add(Dense(32, activation="relu"))
model.add(Dense(1))
# Checks the model summary
model.summary()
# Compiles the model
model.compile(optimizer="rmsprop", loss="mse", metrics=["mae"])
# Sets the number of epoch
epoch_count = 10
# Fits the model with training data and performs validation with validation data
history = model.fit(X_train, y_train, batch_size=4, epochs=epoch_count, validation_data=(X_test, y_test))
# Gets referene to model performance metrics over training and validation data
history = history.history
# Plots training and validation performance
plt.plot(range(1, epoch_count+1), history['loss'], "bo", label="Training Loss")
plt.plot(range(1, epoch_count+1), history['val_loss'], "b", label="Validation Loss")
plt.legend()
plt.xlabel = "Epochs"
plt.ylabel = "Loss"
plt.figure()
plt.plot(range(1, epoch_count+1), history['mae'], "bo", label="Training MAE")
plt.plot(range(1, epoch_count+1), history['val_mae'], "b", label="Validation MAE")
plt.legend()
plt.xlabel = "Epochs"
plt.ylabel = "Mean Absolute Error (MAE)"
plt.show()
# Checks for mean of validation loss
np.mean(history["val_mae"])
# Performs predictions on the test data
predictions = model.predict(X_test)
# Calculates mean absolute error for all predictions
mean_absolute_error(y_test, predictions)
# Now with validation and test performance analysed, let's train the model once again with all data so
# that the model can be used for real predictions
X_train, y_train = data_6_overs_agg_array[:,1:], data_6_overs_agg_array[:,0]
model = Sequential()
model.add(Dense(128, activation="relu", input_shape=(X_train.shape[1],)))
model.add(Dense(64, activation="relu"))
model.add(Dense(32, activation="relu"))
model.add(Dense(1))
model.compile(optimizer="rmsprop", loss="mse", metrics=["mae"])
history = model.fit(X_train, y_train, batch_size=4, epochs=epoch_count)
# Saves the model into persistent store for later use
model.save("Models/Dense_ANN_Regressor")
```
## From the above, the observed test data Mean Absolute Error (MAE) for this Dense Neural Network is around 10. Though it was expected that this model's test performance will outperform that of all other models implemented so far on these notebooks, but it could only beat test performance of Common Sense model (10.71) and Decision Tree model (11.12), and scored less than score of Random Forest (9.70) and Gradient Boosted model (8.95)
## Scope of future work on this is
### 1) to refer the dataset once again to check if any other preprocessing is required in order to get new feature(s) and/or optimize any existing feature(s), and
### 2) to try differ models both from shallow and deep learning techniques
|
github_jupyter
|
# Imports required packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.preprocessing import OneHotEncoder
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
from tensorflow.keras import models
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
from tensorflow.keras import regularizers
import pickle
# Sets Pandas option to show all columns
pd.set_option('display.max_columns', None)
# Deletes the following files before downloading the same files
!rm Data/README.txt Data/ll_matches.csv Data/ipl_csv2.zip
# Downloads the dataset from cricsheet.org/downloads (overwrites the file if exists)
!wget https://cricsheet.org/downloads/ipl_csv2.zip -O Data/ipl_csv2.zip
# Unzips the data (overwrites existing files having same name)
!unzip -o -d Data Data/ipl_csv2.zip all_matches.csv README.txt
# Load data for all matches
data = pd.read_csv("Data/all_matches.csv")
# Checks top few rows of the data
data.head()
# Checks bottom rows of the data
data.tail()
# Converts all string data into lowercase
data.venue = data.venue.str.lower()
data.batting_team = data.batting_team.str.lower()
data.bowling_team = data.bowling_team.str.lower()
data.striker = data.striker.str.lower()
data.non_striker = data.non_striker.str.lower()
data.bowler = data.bowler.str.lower()
# Checks for missing values
data.isna().sum()
data.venue.value_counts().sort_index()
# Updates these venues that are mentioned in different names with same name
data.venue[data.venue.str.contains("arun jaitley",
case=False)] = "arun jaitley stadium"
data.venue[data.venue.str.contains("brabourne",
case=False)] = "brabourne stadium"
data.venue[data.venue.str.contains("chinnaswamy",
case=False)] = "m. chinnaswamy stadium"
data.venue[data.venue.str.contains("chidambaram",
case=False)] = "m. a. chidambaram stadium"
data.venue[data.venue.str.contains(r'narendra modi',
case=False)] = "narendra modi stadium"
data.venue[data.venue.str.contains(r'punjab cricket|is bindra|inderjit singh bindra',
case=False)] = "is bindra stadium"
data.venue[data.venue.str.contains("rajiv gandhi",
case=False)] = "rajiv gandhi international cricket stadium"
data.venue[data.venue.str.contains("wankhede",
case=False)] = "wankhede stadium"
data.batting_team.append(data.bowling_team).value_counts().sort_index()
# Updates team name from "Delhi Daredevils" with the new name "Delhi Capitals"
data.batting_team[data.batting_team.str.contains("delhi daredevils", case=False)] = "delhi capitals"
data.bowling_team[data.bowling_team.str.contains("delhi daredevils", case=False)] = "delhi capitals"
# Updates team name from "Kings XI Punjab" with the new name "Punjab Kings"
data.batting_team[data.batting_team.str.contains("kings xi punjab", case=False)] = "punjab kings"
data.bowling_team[data.bowling_team.str.contains("kings xi punjab", case=False)] = "punjab kings"
# Updates appropriate team name for "Rising Pune Supergiant"
data.batting_team[data.batting_team.str.contains("rising pune supergiants", case=False)] = "rising pune supergiant"
data.bowling_team[data.bowling_team.str.contains("rising pune supergiants", case=False)] = "rising pune supergiant"
# Inserts a new calculated column called "score_off_ball" which is a sum of values in
# columns "runs_off_bat" and "extras" just after column "extras" to indicate contributing score off the ball
# to make calculating total score at the end of the match easy
data.insert(loc=13, column="score_off_ball", value=data.runs_off_bat + data.extras)
# Removes columsn that are not required
columns_required = ["match_id", "venue", "innings", "ball", "batting_team", "bowling_team",
"striker", "non_striker", "bowler", "score_off_ball"]
data = data[columns_required]
# Considers only first 6 overs
data_6_overs = data[data.ball <= 6.0]
# Checks how the 6 overs data looks
data_6_overs
# Venue wise average innings runs after 6 overs
data_6_overs.groupby(
["venue", "match_id", "innings"]).score_off_ball.sum().reset_index().groupby(
["venue"]).score_off_ball.mean().sort_values(ascending=False)
# Batting team wise average innings runs after 6 overs
data_6_overs.groupby(["batting_team", "match_id", "innings"]).score_off_ball.sum().reset_index().groupby(
["batting_team"]).score_off_ball.mean().sort_values(ascending=False)
# Striker's inning wise average scores after 6 overs
pd.set_option("display.max_rows", None) # To view all the rows temporarily
data_6_overs.groupby(
["striker", "match_id", "innings"]).score_off_ball.sum().reset_index().groupby(
["striker"]).score_off_ball.mean().sort_values(ascending=False)
pd.reset_option('display.max_rows') # Resets to its default number of visible rows
# Resets its index
data_6_overs.reset_index(drop = True, inplace = True)
# Checks how the 6 overs data looks after reset index and with new column "score_off_ball"
data_6_overs
# Creates initial innings group out of 6 overs to further add other innings related data into
innings = data_6_overs.groupby(
["match_id", "venue", "innings", "batting_team", "bowling_team"])
# Calculates the innings wise score at the end of 6 overs
innings_runs = innings.score_off_ball.sum()
innings_runs.name = "score_6_overs"
# Checks how the innings data looks like
innings_runs
# Resets the index of the group to flatten the grouped data
data_6_overs_agg = innings_runs.reset_index()
# Checks how the flattend innings wise grouped data looks
data_6_overs_agg
# Encodes venues with one-hot encoding technique
venue_count = len(data_6_overs_agg.venue.unique())
venue_encoder = OneHotEncoder(handle_unknown='ignore')
venue_encoded = pd.DataFrame(venue_encoder.fit_transform(data_6_overs_agg[["venue"]]).toarray(),
columns=[("venue_" + str(i)) for i in range(venue_count)])
# Saves the encoder into persistent store for later use
with open("Models/Venue_Encoder.pickle", "wb") as f:
pickle.dump(venue_encoder, f, pickle.HIGHEST_PROTOCOL)
# Joins the encoded venue columns with the dataset
data_6_overs_agg = data_6_overs_agg.join(venue_encoded).drop(["venue"], axis = 1)
# Encodes innings with one-hot encoding technique
innings_count = len(data_6_overs_agg.innings.unique())
innings_encoder = OneHotEncoder(handle_unknown='ignore')
innings_encoded = pd.DataFrame(innings_encoder.fit_transform(data_6_overs_agg[["innings"]]).toarray(),
columns=[("innings_" + str(i)) for i in range(innings_count)])
# Saves the encoder into persistent store for later use
with open("Models/Innings_Encoder.pickle", "wb") as f:
pickle.dump(innings_encoder, f, pickle.HIGHEST_PROTOCOL)
# Joins the encoded innings columns with the dataset
data_6_overs_agg = data_6_overs_agg.join(innings_encoded).drop(["innings"], axis = 1)
# Now, encodes teams with one-hot encoding technique
team = data_6_overs_agg.batting_team.append(data_6_overs_agg.bowling_team).unique()
team_count = len(team)
team_encoder = OneHotEncoder(handle_unknown='ignore')
team_encoder.fit(pd.DataFrame(team))
batting_team_encoded = pd.DataFrame(team_encoder.transform(data_6_overs_agg[["batting_team"]]).toarray(),
columns=[("batting_team_" + str(i)) for i in range(team_count)])
bowling_team_encoded = pd.DataFrame(team_encoder.transform(data_6_overs_agg[["bowling_team"]]).toarray(),
columns=[("bowling_team_" + str(i)) for i in range(team_count)])
# Saves the encoder into persistent store for later use
with open("Models/Team_Encoder.pickle", "wb") as f:
pickle.dump(team_encoder, f, pickle.HIGHEST_PROTOCOL)
# Joins the encoded team columns with the dataset
data_6_overs_agg = data_6_overs_agg.join(batting_team_encoded).drop(["batting_team"], axis = 1)
data_6_overs_agg = data_6_overs_agg.join(bowling_team_encoded).drop(["bowling_team"], axis = 1)
# Now, encodes players with one-hot encoding technique
player = data_6_overs.striker.append(
data_6_overs.non_striker).append(data_6_overs.bowler).unique()
player_count = len(player)
player_encoder = OneHotEncoder(handle_unknown='ignore')
player_encoder.fit(pd.DataFrame(player))
# Saves the encoder into persistent store for later use
with open("Models/Player_Encoder.pickle", "wb") as f:
pickle.dump(player_encoder, f, pickle.HIGHEST_PROTOCOL)
# Transforms both striker and non-striker
striker_encoded_array = player_encoder.transform(data_6_overs[["striker"]]).toarray()
non_striker_encoded_array = player_encoder.transform(data_6_overs[["non_striker"]]).toarray()
# Considers striker and non-striker as batsmen, creates a dataframe out of it,
# then joins with innings data to consider this batsmen information in the main dataset
data_6_overs_batsmen = data_6_overs[["match_id", "innings"]].join(
pd.DataFrame(np.add(striker_encoded_array, non_striker_encoded_array),
columns=[("batsman_" + str(i)) for i in range(player_count)]))
# Transforms bowlers and creates a dataframe out of it, and then joins with innings data to
# consider this bowlers information in the main dataset
data_6_overs_bowlers = data_6_overs[["match_id", "innings"]].join(
pd.DataFrame(player_encoder.transform(data_6_overs[["bowler"]]).toarray(),
columns=[("bowler_" + str(i)) for i in range(player_count)]))
# Combines all the batsmen, both striker and non-striker innings wise, and
# resets index of the aggregation to get tabular data
batsmen_encoded = data_6_overs_batsmen.groupby(["match_id", "innings"]).max().reset_index()
# Drops columns "match_id" and "innings" as these columns already exist in aggregated dataset that this
# data will be joined with
batsmen_encoded.drop(["match_id", "innings"], inplace=True, axis=1)
# Combines all the bowlers innings wise and resets index of the aggregation to get tabular data
bowlers_encoded = data_6_overs_bowlers.groupby(["match_id", "innings"]).max().reset_index()
# Drops columns "match_id" and "innings" as these columns already exist in aggregated dataset that this
# data will be joined with
bowlers_encoded.drop(["match_id", "innings"], inplace=True, axis=1)
# Joins encoded batsmen and bowlers information with aggregated innings data
data_6_overs_agg = data_6_overs_agg.join(batsmen_encoded)
data_6_overs_agg = data_6_overs_agg.join(bowlers_encoded)
# Checks how the final dataset the machine learning model will be trained on looks like
data_6_overs_agg
# But, first removes the column "match_id" as it is not required for machine learning model
data_6_overs_agg.drop(["match_id"], axis=1, inplace=True)
# Converts DataFrame into 2D tensor
data_6_overs_agg_array = data_6_overs_agg.to_numpy()
# Seperates training labels
X_train, y_train = data_6_overs_agg_array[:,1:], data_6_overs_agg_array[:,0]
# Splits the available data into train and test data sets
X_train, X_test, y_train, y_test = train_test_split(X_train, y_train, test_size = 0.2, shuffle = True)
# Creates a sequential dense neural network model
model = Sequential()
model.add(Dense(128, activation="relu", input_shape=(X_train.shape[1],)))
model.add(Dense(64, activation="relu"))
model.add(Dense(32, activation="relu"))
model.add(Dense(1))
# Checks the model summary
model.summary()
# Compiles the model
model.compile(optimizer="rmsprop", loss="mse", metrics=["mae"])
# Sets the number of epoch
epoch_count = 10
# Fits the model with training data and performs validation with validation data
history = model.fit(X_train, y_train, batch_size=4, epochs=epoch_count, validation_data=(X_test, y_test))
# Gets referene to model performance metrics over training and validation data
history = history.history
# Plots training and validation performance
plt.plot(range(1, epoch_count+1), history['loss'], "bo", label="Training Loss")
plt.plot(range(1, epoch_count+1), history['val_loss'], "b", label="Validation Loss")
plt.legend()
plt.xlabel = "Epochs"
plt.ylabel = "Loss"
plt.figure()
plt.plot(range(1, epoch_count+1), history['mae'], "bo", label="Training MAE")
plt.plot(range(1, epoch_count+1), history['val_mae'], "b", label="Validation MAE")
plt.legend()
plt.xlabel = "Epochs"
plt.ylabel = "Mean Absolute Error (MAE)"
plt.show()
# Checks for mean of validation loss
np.mean(history["val_mae"])
# Performs predictions on the test data
predictions = model.predict(X_test)
# Calculates mean absolute error for all predictions
mean_absolute_error(y_test, predictions)
# Now with validation and test performance analysed, let's train the model once again with all data so
# that the model can be used for real predictions
X_train, y_train = data_6_overs_agg_array[:,1:], data_6_overs_agg_array[:,0]
model = Sequential()
model.add(Dense(128, activation="relu", input_shape=(X_train.shape[1],)))
model.add(Dense(64, activation="relu"))
model.add(Dense(32, activation="relu"))
model.add(Dense(1))
model.compile(optimizer="rmsprop", loss="mse", metrics=["mae"])
history = model.fit(X_train, y_train, batch_size=4, epochs=epoch_count)
# Saves the model into persistent store for later use
model.save("Models/Dense_ANN_Regressor")
| 0.758868 | 0.836421 |
# Mass-spring-damper
In this tutorial, we will describe the mechanics and control of the one degree of freedom translational mass-spring-damper system subject to a control input force. We will first derive the dynamic equations by hand. Then, we will derive them using the `sympy.mechanics` python package.
The system on which we will work is depicted below:

Note that in what follows, we use the notation $u(t) = F$.
## 1. Mechanics
### Deriving the dynamical equations by hand
#### 1.1 By using Newton equations
Using Newton's law, we have:
\begin{align}
m \ddot{x}(t) &= \sum F_{ext} \\
&= - b \dot{x}(t) - k x(t) + u(t)
\end{align}
#### 1.2 By using the Lagrange Method
Let's first derive the kinematic and potential energies.
\begin{equation}
T = \frac{1}{2} m \dot{x} \\
V = - \int \vec{F} . \vec{dl} = - \int (-kx \vec{1_x}) . dx \vec{1_x} = \frac{k x^2}{2}
\end{equation}
The Lagrangian is then given by:
\begin{equation}
\mathcal{L} = T - V = \frac{1}{2} m \dot{x} - \frac{k x^2}{2}
\end{equation}
Using the Lagrange's equations we can derive the dynamics of the system:
\begin{equation}
\frac{d}{dt} \frac{\partial \mathcal{L}}{\partial \dot{q}} - \frac{\partial \mathcal{L}}{\partial q} = Q
\end{equation}
where $q$ are the generalized coordinates (in this case $x$), and $Q$ represents the non-conservative forces (input force, dragging or friction forces, etc).
* $\frac{d}{dt} \frac{\partial \mathcal{L}}{\partial \dot{x}} = \frac{d}{dt} m \dot{x}(t) = m \ddot{x}(t) $
* $\frac{\partial \mathcal{L}}{\partial x} = - k x(t) $
* $Q = - b \dot{x}(t) + u(t) $
which when putting everything back together gives us:
\begin{equation}
m \ddot{x}(t) + b \dot{x}(t) + k x(t) = u(t)
\end{equation}
### Deriving the dynamical equations using sympy
```
import sympy
import sympy.physics.mechanics as mechanics
from sympy import init_printing
init_printing(use_latex='mathjax')
from sympy import pprint
# define variables
q = mechanics.dynamicsymbols('q')
dq = mechanics.dynamicsymbols('q', 1)
u = mechanics.dynamicsymbols('u')
# define constants
m, k, b = sympy.symbols('m k b')
# define the inertial frame
N = mechanics.ReferenceFrame('N')
# define a particle for the mass
P = mechanics.Point('P')
P.set_vel(N, dq * N.x) # go in the x direction
Pa = mechanics.Particle('Pa', P, m)
# define the potential energy for the particle (the kinematic one is derived automatically)
Pa.potential_energy = k * q**2 / 2.0
# define the Lagrangian and the non-conservative force applied on the point P
L = mechanics.Lagrangian(N, Pa)
force = [(P, -b * dq * N.x + u * N.x)]
# Lagrange equations
lagrange = mechanics.LagrangesMethod(L, [q], forcelist = force, frame = N)
pprint(lagrange.form_lagranges_equations())
```
## 2. Laplace transform and transfer function
Applying the Laplace transform on the dynamic equation:
\begin{equation}
m \ddot{x}(t) + b \dot{x}(t) + k x(t) = u(t) \stackrel{L}{\rightarrow} m s^2 X(s) + b s X(s) + k X(s) = U(s)
\end{equation}
The transfer equation is given by:
\begin{equation}
H(s) = \frac{X(s)}{U(s)} = \frac{1}{m s^2 + b s + k}
\end{equation}
By calculating the pole:
\begin{equation}
m s^2 + b s + k = 0 \Leftrightarrow s = \frac{-b}{2m} \pm \sqrt{\left(\frac{b}{2m}\right)^2 - \frac{k}{m}}
\end{equation}
Note that $b, k, m > 0$ because they represent real physical quantities.
### LTI system
We can rewrite the above equation as a first-order system of equations. Let's first define the state vector $\pmb{x} = \left[ \begin{array}{c} x(t) \\ \dot{x}(t) \end{array} \right]$ and the control vector $\pmb{u} = \left[ \begin{array}{c} u(t) \end{array} \right]$, then we can rewrite the above equation in the form $\pmb{\dot{x}} = \pmb{Ax} + \pmb{Bu}$, as below:
\begin{equation}
\left[ \begin{array}{c} \dot{x}(t) \\ \ddot{x}(t) \end{array} \right] = \left[ \begin{array}{cc} 0 & 1 \\ -\frac{k}{m} & -\frac{b}{m} \end{array} \right] \left[ \begin{array}{c} x(t) \\ \dot{x}(t) \end{array} \right] + \left[ \begin{array}{c} 0 \\ \frac{1}{m} \end{array} \right] \left[ \begin{array}{c} u(t) \end{array} \right]
\end{equation}
If there is no $u(t)$, i.e. $u(t) = 0 \; \forall t$, then we have $\pmb{\dot{x}} = \pmb{Ax}$. The solution to this system of equation is $\pmb{x}(t) = e^{\pmb{A}t} \pmb{x}_0$.
|
github_jupyter
|
import sympy
import sympy.physics.mechanics as mechanics
from sympy import init_printing
init_printing(use_latex='mathjax')
from sympy import pprint
# define variables
q = mechanics.dynamicsymbols('q')
dq = mechanics.dynamicsymbols('q', 1)
u = mechanics.dynamicsymbols('u')
# define constants
m, k, b = sympy.symbols('m k b')
# define the inertial frame
N = mechanics.ReferenceFrame('N')
# define a particle for the mass
P = mechanics.Point('P')
P.set_vel(N, dq * N.x) # go in the x direction
Pa = mechanics.Particle('Pa', P, m)
# define the potential energy for the particle (the kinematic one is derived automatically)
Pa.potential_energy = k * q**2 / 2.0
# define the Lagrangian and the non-conservative force applied on the point P
L = mechanics.Lagrangian(N, Pa)
force = [(P, -b * dq * N.x + u * N.x)]
# Lagrange equations
lagrange = mechanics.LagrangesMethod(L, [q], forcelist = force, frame = N)
pprint(lagrange.form_lagranges_equations())
| 0.492188 | 0.993009 |
```
# imports
%matplotlib inline
from IPython.display import YouTubeVideo
from IPython.display import Math
import sklearn
from sklearn import datasets, linear_model
from sklearn.linear_model import LinearRegression
import numpy as np
import numpy.random as rng
import matplotlib
import matplotlib.pyplot as plt
from sklearn.datasets import fetch_20newsgroups
# diabetes_data
diabetes_data = datasets.load_diabetes()
print(diabetes_data.DESCR)
```
## Bayes / Navie bayes problem 3
```
diabetes_data.data.shape
N = len(diabetes_data.target)
i = rng.randint(0,N-1,100)
diabetes_data.data[i,:].shape
X_train = diabetes_data.data[i,np.newaxis,i]
y_train = diabetes_data.target[i]
print(X_train.shape)
print(y_train.shape)
# A linear model
model = LinearRegression()
# Train the model using the training sets
# i.e., find parameters that make the model best fit the data
model.fit(X_train, y_train)
# What are the parameters we found to be best?
# print('Optimal parameters: ')
# print('slope: {:.3f}\nintercept: {:.3f}'.format(model.coef_[0], model.intercept_ ))
model.coef_.shape
y_pred = model.predict(X_train)
plt.scatter( X_train, y_train )
plt.plot( X_train, y_pred, color='black', linewidth=2 )
plt.ylabel('Diabetes progression')
# plt.xlabel('Average blood pressure');
# plot a histogram of the errors
plt.hist(y_pred-y_train)
```
## Solve using linear algebra
```
X_train = diabetes_data.data[i,:]
y_train = diabetes_data.target[i]
p = np.linalg.lstsq(X_train,y_train)
p[0].shape
X1 = np.concatenate((X_train, np.ones((X_train.shape[0],1))), axis=1)
X1.shape
p,resid,rank,s = np.linalg.lstsq(X1, y_train) # Note this is not explicitly computing the SVD
y_linsolv_pred = np.matmul( X1, p ); # Compare this to the previous output
y_linsolv_pred - y_pred # output is very tiny, so that's what we want!
```
## Fitting a quadratic function with linear regression
Linear regression doesn't mean fitting lines. It means fitting linear basis functions.
$ y_i = p_0 + p_1 x_{i,1} + p_2 x_{i,2} + p_3 x_{i,1}^2 + p_4 x_{i,2}^2 + p_5 x_{i,1} x_{i,2} $
Regularization is a good idea here, but not necessary. One thing that regularization does is to give preference to some kind of solutions.
```
X_train_poly = X_train[:,[1,2]]
X_poly = np.concatenate((np.ones((X_train_poly.shape[0],1)), X_train_poly), axis=1)
X_poly.shape
# Add quadratic terms
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,0]**2)), axis=1)
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,1]**2)), axis=1)
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,0]*X_train_poly[:,np.newaxis,1])), axis=1)
X_poly.shape
p_poly,resid,rank,s = np.linalg.lstsq(X_poly,y_train)
y_pred_poly_lsq = np.matmul(X_poly,p_poly)
poly_model = Pipeline(
[('poly', PolynomialFeatures(degree=3)),
('linear', LinearRegression())])
```
# Class notes
## Bayes / Navie bayes problem 3
```
diabetes_data.data.shape
N = len(diabetes_data.target)
i = rng.randint(0,N-1,100)
diabetes_data.data[i,:].shape
X_train = diabetes_data.data[i,np.newaxis,i]
y_train = diabetes_data.target[i]
print(X_train.shape)
print(y_train.shape)
# A linear model
model = LinearRegression()
# Train the model using the training sets
# i.e., find parameters that make the model best fit the data
model.fit(X_train, y_train)
# What are the parameters we found to be best?
# print('Optimal parameters: ')
# print('slope: {:.3f}\nintercept: {:.3f}'.format(model.coef_[0], model.intercept_ ))
model.coef_.shape
y_pred = model.predict(X_train)
plt.scatter( X_train, y_train )
plt.plot( X_train, y_pred, color='black', linewidth=2 )
plt.ylabel('Diabetes progression')
# plt.xlabel('Average blood pressure');
# plot a histogram of the errors
plt.hist(y_pred-y_train)
```
## Solve using linear algebra
```
X_train = diabetes_data.data[i,:]
y_train = diabetes_data.target[i]
p = np.linalg.lstsq(X_train,y_train)
p[0].shape
X1 = np.concatenate((X_train, np.ones((X_train.shape[0],1))), axis=1)
X1.shape
p,resid,rank,s = np.linalg.lstsq(X1, y_train) # Note this is not explicitly computing the SVD
y_linsolv_pred = np.matmul( X1, p ); # Compare this to the previous output
y_linsolv_pred - y_pred # output is very tiny, so that's what we want!
```
## Fitting a quadratic function with linear regression
Linear regression doesn't mean fitting lines. It means fitting linear basis functions.
$ y_i = p_0 + p_1 x_{i,1} + p_2 x_{i,2} + p_3 x_{i,1}^2 + p_4 x_{i,2}^2 + p_5 x_{i,1} x_{i,2} $
Regularization is a good idea here, but not necessary. One thing that regularization does is to give preference to some kind of solutions.
```
X_train_poly = X_train[:,[1,2]]
X_poly = np.concatenate((np.ones((X_train_poly.shape[0],1)), X_train_poly), axis=1)
X_poly.shape
# Add quadratic terms
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,0]**2)), axis=1)
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,1]**2)), axis=1)
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,0]*X_train_poly[:,np.newaxis,1])), axis=1)
X_poly.shape
p_poly,resid,rank,s = np.linalg.lstsq(X_poly,y_train)
y_pred_poly_lsq = np.matmul(X_poly,p_poly)
poly_model = Pipeline(
[('poly', PolynomialFeatures(degree=3)),
('linear', LinearRegression())])
```
|
github_jupyter
|
# imports
%matplotlib inline
from IPython.display import YouTubeVideo
from IPython.display import Math
import sklearn
from sklearn import datasets, linear_model
from sklearn.linear_model import LinearRegression
import numpy as np
import numpy.random as rng
import matplotlib
import matplotlib.pyplot as plt
from sklearn.datasets import fetch_20newsgroups
# diabetes_data
diabetes_data = datasets.load_diabetes()
print(diabetes_data.DESCR)
diabetes_data.data.shape
N = len(diabetes_data.target)
i = rng.randint(0,N-1,100)
diabetes_data.data[i,:].shape
X_train = diabetes_data.data[i,np.newaxis,i]
y_train = diabetes_data.target[i]
print(X_train.shape)
print(y_train.shape)
# A linear model
model = LinearRegression()
# Train the model using the training sets
# i.e., find parameters that make the model best fit the data
model.fit(X_train, y_train)
# What are the parameters we found to be best?
# print('Optimal parameters: ')
# print('slope: {:.3f}\nintercept: {:.3f}'.format(model.coef_[0], model.intercept_ ))
model.coef_.shape
y_pred = model.predict(X_train)
plt.scatter( X_train, y_train )
plt.plot( X_train, y_pred, color='black', linewidth=2 )
plt.ylabel('Diabetes progression')
# plt.xlabel('Average blood pressure');
# plot a histogram of the errors
plt.hist(y_pred-y_train)
X_train = diabetes_data.data[i,:]
y_train = diabetes_data.target[i]
p = np.linalg.lstsq(X_train,y_train)
p[0].shape
X1 = np.concatenate((X_train, np.ones((X_train.shape[0],1))), axis=1)
X1.shape
p,resid,rank,s = np.linalg.lstsq(X1, y_train) # Note this is not explicitly computing the SVD
y_linsolv_pred = np.matmul( X1, p ); # Compare this to the previous output
y_linsolv_pred - y_pred # output is very tiny, so that's what we want!
X_train_poly = X_train[:,[1,2]]
X_poly = np.concatenate((np.ones((X_train_poly.shape[0],1)), X_train_poly), axis=1)
X_poly.shape
# Add quadratic terms
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,0]**2)), axis=1)
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,1]**2)), axis=1)
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,0]*X_train_poly[:,np.newaxis,1])), axis=1)
X_poly.shape
p_poly,resid,rank,s = np.linalg.lstsq(X_poly,y_train)
y_pred_poly_lsq = np.matmul(X_poly,p_poly)
poly_model = Pipeline(
[('poly', PolynomialFeatures(degree=3)),
('linear', LinearRegression())])
diabetes_data.data.shape
N = len(diabetes_data.target)
i = rng.randint(0,N-1,100)
diabetes_data.data[i,:].shape
X_train = diabetes_data.data[i,np.newaxis,i]
y_train = diabetes_data.target[i]
print(X_train.shape)
print(y_train.shape)
# A linear model
model = LinearRegression()
# Train the model using the training sets
# i.e., find parameters that make the model best fit the data
model.fit(X_train, y_train)
# What are the parameters we found to be best?
# print('Optimal parameters: ')
# print('slope: {:.3f}\nintercept: {:.3f}'.format(model.coef_[0], model.intercept_ ))
model.coef_.shape
y_pred = model.predict(X_train)
plt.scatter( X_train, y_train )
plt.plot( X_train, y_pred, color='black', linewidth=2 )
plt.ylabel('Diabetes progression')
# plt.xlabel('Average blood pressure');
# plot a histogram of the errors
plt.hist(y_pred-y_train)
X_train = diabetes_data.data[i,:]
y_train = diabetes_data.target[i]
p = np.linalg.lstsq(X_train,y_train)
p[0].shape
X1 = np.concatenate((X_train, np.ones((X_train.shape[0],1))), axis=1)
X1.shape
p,resid,rank,s = np.linalg.lstsq(X1, y_train) # Note this is not explicitly computing the SVD
y_linsolv_pred = np.matmul( X1, p ); # Compare this to the previous output
y_linsolv_pred - y_pred # output is very tiny, so that's what we want!
X_train_poly = X_train[:,[1,2]]
X_poly = np.concatenate((np.ones((X_train_poly.shape[0],1)), X_train_poly), axis=1)
X_poly.shape
# Add quadratic terms
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,0]**2)), axis=1)
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,1]**2)), axis=1)
X_poly = np.concatenate((X_poly, (X_train_poly[:,np.newaxis,0]*X_train_poly[:,np.newaxis,1])), axis=1)
X_poly.shape
p_poly,resid,rank,s = np.linalg.lstsq(X_poly,y_train)
y_pred_poly_lsq = np.matmul(X_poly,p_poly)
poly_model = Pipeline(
[('poly', PolynomialFeatures(degree=3)),
('linear', LinearRegression())])
| 0.586996 | 0.894375 |
# Developing an AI application
Going forward, AI algorithms will be incorporated into more and more everyday applications. For example, you might want to include an image classifier in a smart phone app. To do this, you'd use a deep learning model trained on hundreds of thousands of images as part of the overall application architecture. A large part of software development in the future will be using these types of models as common parts of applications.
In this project, you'll train an image classifier to recognize different species of flowers. You can imagine using something like this in a phone app that tells you the name of the flower your camera is looking at. In practice you'd train this classifier, then export it for use in your application. We'll be using [this dataset](http://www.robots.ox.ac.uk/~vgg/data/flowers/102/index.html) of 102 flower categories, you can see a few examples below.
<img src='assets/Flowers.png' width=500px>
The project is broken down into multiple steps:
* Load and preprocess the image dataset
* Train the image classifier on your dataset
* Use the trained classifier to predict image content
We'll lead you through each part which you'll implement in Python.
When you've completed this project, you'll have an application that can be trained on any set of labeled images. Here your network will be learning about flowers and end up as a command line application. But, what you do with your new skills depends on your imagination and effort in building a dataset. For example, imagine an app where you take a picture of a car, it tells you what the make and model is, then looks up information about it. Go build your own dataset and make something new.
First up is importing the packages you'll need. It's good practice to keep all the imports at the beginning of your code. As you work through this notebook and find you need to import a package, make sure to add the import up here.
```
# Imports here
import matplotlib.pyplot as plt
from matplotlib.ticker import FormatStrFormatter
import numpy as np
import torch
from torch import nn
from torch import optim
import torch.nn.functional as F
from torchvision import datasets, transforms, models
import torchvision.models as models
from PIL import Image
import json
from collections import OrderedDict
import torchvision
```
## Load the data
Here you'll use `torchvision` to load the data ([documentation](http://pytorch.org/docs/0.3.0/torchvision/index.html)). The data should be included alongside this notebook, otherwise you can [download it here](https://s3.amazonaws.com/content.udacity-data.com/nd089/flower_data.tar.gz). The dataset is split into three parts, training, validation, and testing. For the training, you'll want to apply transformations such as random scaling, cropping, and flipping. This will help the network generalize leading to better performance. You'll also need to make sure the input data is resized to 224x224 pixels as required by the pre-trained networks.
The validation and testing sets are used to measure the model's performance on data it hasn't seen yet. For this you don't want any scaling or rotation transformations, but you'll need to resize then crop the images to the appropriate size.
The pre-trained networks you'll use were trained on the ImageNet dataset where each color channel was normalized separately. For all three sets you'll need to normalize the means and standard deviations of the images to what the network expects. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`, calculated from the ImageNet images. These values will shift each color channel to be centered at 0 and range from -1 to 1.
```
data_dir = 'flowers'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
# TODO: Define your transforms for the training, validation, and testing sets
train_transforms = transforms.Compose([transforms.RandomRotation(30),
transforms.RandomResizedCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
test_transforms = transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
valid_transforms = transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
# TODO: Load the datasets with ImageFolder
train_data = datasets.ImageFolder(train_dir, transform = train_transforms)
test_data = datasets.ImageFolder(test_dir, transform = test_transforms)
valid_data = datasets.ImageFolder(valid_dir, transform = valid_transforms)
# TODO: Using the image datasets and the trainforms, define the dataloaders
trainloader = torch.utils.data.DataLoader(train_data, batch_size = 64, shuffle = True)
testloader = torch.utils.data.DataLoader(test_data, batch_size = 64, shuffle = True)
validloader = torch.utils.data.DataLoader(valid_data, batch_size = 64, shuffle = True)
```
### Label mapping
You'll also need to load in a mapping from category label to category name. You can find this in the file `cat_to_name.json`. It's a JSON object which you can read in with the [`json` module](https://docs.python.org/2/library/json.html). This will give you a dictionary mapping the integer encoded categories to the actual names of the flowers.
```
with open('label_map.json', 'r') as f:
cat_to_name = json.load(f)
```
# Building and training the classifier
Now that the data is ready, it's time to build and train the classifier. As usual, you should use one of the pretrained models from `torchvision.models` to get the image features. Build and train a new feed-forward classifier using those features.
We're going to leave this part up to you. Refer to [the rubric](https://review.udacity.com/#!/rubrics/1663/view) for guidance on successfully completing this section. Things you'll need to do:
* Load a [pre-trained network](http://pytorch.org/docs/master/torchvision/models.html) (If you need a starting point, the VGG networks work great and are straightforward to use)
* Define a new, untrained feed-forward network as a classifier, using ReLU activations and dropout
* Train the classifier layers using backpropagation using the pre-trained network to get the features
* Track the loss and accuracy on the validation set to determine the best hyperparameters
We've left a cell open for you below, but use as many as you need. Our advice is to break the problem up into smaller parts you can run separately. Check that each part is doing what you expect, then move on to the next. You'll likely find that as you work through each part, you'll need to go back and modify your previous code. This is totally normal!
When training make sure you're updating only the weights of the feed-forward network. You should be able to get the validation accuracy above 70% if you build everything right. Make sure to try different hyperparameters (learning rate, units in the classifier, epochs, etc) to find the best model. Save those hyperparameters to use as default values in the next part of the project.
One last important tip if you're using the workspace to run your code: To avoid having your workspace disconnect during the long-running tasks in this notebook, please read in the earlier page in this lesson called Intro to
GPU Workspaces about Keeping Your Session Active. You'll want to include code from the workspace_utils.py module.
**Note for Workspace users:** If your network is over 1 GB when saved as a checkpoint, there might be issues with saving backups in your workspace. Typically this happens with wide dense layers after the convolutional layers. If your saved checkpoint is larger than 1 GB (you can open a terminal and check with `ls -lh`), you should reduce the size of your hidden layers and train again.
```
# TODO: Build and train your network
def nn_setup(structure = 'vgg16', lr = 0.001):
model = models.vgg16(pretrained = True)
for param in model.parameters():
param.requires_grad = False
model.classifier = nn.Sequential(OrderedDict([
('fc1', nn.Linear(25088, 2048)),
('relu', nn.ReLU()),
('fc2', nn.Linear(2048, 256)),
('relu', nn.ReLU()),
('fc3', nn.Linear(256, 102)),
('output', nn.LogSoftmax(dim=1))
]))
model = model.to('cuda')
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.classifier.parameters(), lr)
return model, criterion, optimizer
model, criterion, optimizer = nn_setup()
print(model)
epochs = 3
print_every = 5
steps = 0
loss_show = []
for e in range(epochs):
running_loss = 0
for inputs, labels in trainloader:
steps += 1
inputs, labels = inputs.to('cuda'), labels.to('cuda')
optimizer.zero_grad()
outputs = model.forward(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
if steps % print_every == 0:
model.eval()
valid_loss = 0
accuracy = 0
with torch.no_grad():
for inputs, labels in validloader:
inputs, labels = inputs.to('cuda'), labels.to('cuda')
log_ps = model.forward(inputs)
batch_loss = criterion(log_ps, labels)
valid_loss += batch_loss.item()
ps = torch.exp(log_ps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Epoch {e+1}/{epochs}.. "
f"Loss: {running_loss/print_every:.3f}.. "
f"Validation Loss: {valid_loss/len(validloader):.3f}.. "
f"Accuracy: {accuracy/len(validloader):.3f}")
running_loss = 0
model.train()
```
## Testing your network
It's good practice to test your trained network on test data, images the network has never seen either in training or validation. This will give you a good estimate for the model's performance on completely new images. Run the test images through the network and measure the accuracy, the same way you did validation. You should be able to reach around 70% accuracy on the test set if the model has been trained well.
```
# TODO: Do validation on the test set
test_loss = 0
accuracy = 0
model.to('cuda')
with torch.no_grad():
for inputs, labels in testloader:
inputs, labels = inputs.to('cuda'), labels.to('cuda')
log_ps = model.forward(inputs)
batch_loss = criterion(log_ps, labels)
test_loss += batch_loss.item()
ps = torch.exp(log_ps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Test accuracy: {accuracy/len(testloader):.3f}")
```
## Save the checkpoint
Now that your network is trained, save the model so you can load it later for making predictions. You probably want to save other things such as the mapping of classes to indices which you get from one of the image datasets: `image_datasets['train'].class_to_idx`. You can attach this to the model as an attribute which makes inference easier later on.
```model.class_to_idx = image_datasets['train'].class_to_idx```
Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
```
# TODO: Save the checkpoint
model.class_to_idx = train_data.class_to_idx
torch.save({'input_size': 25088,
'output_size': 102,
'structure': 'vgg16',
'learning_rate': 0.001,
'classifier': model.classifier,
'epochs': epochs,
'optimizer': optimizer.state_dict(),
'state_dict': model.state_dict(),
'class_to_idx': model.class_to_idx}, 'checkpoint.pth')
```
## Loading the checkpoint
At this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
```
# TODO: Write a function that loads a checkpoint and rebuilds the model
def load_model(path):
checkpoint = torch.load('checkpoint.pth')
structure = checkpoint['structure']
model,_,_ = nn_setup()
model.class_to_idx = checkpoint['class_to_idx']
model.load_state_dict(checkpoint['state_dict'])
return model
model = load_model('checkpoint.pth')
print(model)
```
# Inference for classification
Now you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like
```python
probs, classes = predict(image_path, model)
print(probs)
print(classes)
> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
> ['70', '3', '45', '62', '55']
```
First you'll need to handle processing the input image such that it can be used in your network.
## Image Preprocessing
You'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training.
First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.
Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.
As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation.
And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
```
def process_image(image):
''' Scales, crops, and normalizes a PIL image for a PyTorch model,
returns an Numpy array
'''
# TODO: Process a PIL image for use in a PyTorch model
img_pil = Image.open(image)
img_transforms = transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
image = img_transforms(img_pil)
return image
```
To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
```
def imshow(image, ax=None, title=None):
"""Imshow for Tensor."""
if ax is None:
fig, ax = plt.subplots()
# PyTorch tensors assume the color channel is the first dimension
# but matplotlib assumes is the third dimension
image = image.numpy().transpose((1, 2, 0))
# Undo preprocessing
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
image = std * image + mean
# Image needs to be clipped between 0 and 1 or it looks like noise when displayed
image = np.clip(image, 0, 1)
ax.imshow(image)
return ax
```
## Class Prediction
Once you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.
To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.
Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.
```python
probs, classes = predict(image_path, model)
print(probs)
print(classes)
> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
> ['70', '3', '45', '62', '55']
```
```
def predict(image_path, model, topk=5):
''' Predict the class (or classes) of an image using a trained deep learning model.
'''
# TODO: Implement the code to predict the class from an image file
model.to('cuda')
model.eval()
img = process_image(image_path).numpy()
img = torch.from_numpy(np.array([img])).float()
with torch.no_grad():
logps = model.forward(img.cuda())
probability = torch.exp(logps).data
return probability.topk(topk)
image_path = "flowers/test/10/image_07090.jpg"
probs, classes = predict(image_path, model)
print (probs)
print (classes)
```
## Sanity Checking
Now that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:
<img src='assets/inference_example.png' width=300px>
You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
```
# TODO: Display an image along with the top 5 classes
plt.rcdefaults()
fig, ax = plt.subplots()
index = 1
path = test_dir + '/1/image_06743.jpg'
ps = predict(path, model)
image = process_image(path)
ax1 = imshow(image, ax = plt)
ax1.axis('off')
ax1.title(cat_to_name[str(index)])
a = np.array(ps[0][0])
b = [cat_to_name[str(index+1)] for index in np.array(ps[1][0])]
fig,ax2 = plt.subplots(figsize=(5,5))
y_pos = np.arange(5)
ax2.set_yticks(y_pos)
ax2.set_yticklabels(b)
ax2.set_xlabel('Probability')
ax2.invert_yaxis()
ax2.barh(y_pos, a)
plt.show()
```
|
github_jupyter
|
# Imports here
import matplotlib.pyplot as plt
from matplotlib.ticker import FormatStrFormatter
import numpy as np
import torch
from torch import nn
from torch import optim
import torch.nn.functional as F
from torchvision import datasets, transforms, models
import torchvision.models as models
from PIL import Image
import json
from collections import OrderedDict
import torchvision
data_dir = 'flowers'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
# TODO: Define your transforms for the training, validation, and testing sets
train_transforms = transforms.Compose([transforms.RandomRotation(30),
transforms.RandomResizedCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
test_transforms = transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
valid_transforms = transforms.Compose([transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
# TODO: Load the datasets with ImageFolder
train_data = datasets.ImageFolder(train_dir, transform = train_transforms)
test_data = datasets.ImageFolder(test_dir, transform = test_transforms)
valid_data = datasets.ImageFolder(valid_dir, transform = valid_transforms)
# TODO: Using the image datasets and the trainforms, define the dataloaders
trainloader = torch.utils.data.DataLoader(train_data, batch_size = 64, shuffle = True)
testloader = torch.utils.data.DataLoader(test_data, batch_size = 64, shuffle = True)
validloader = torch.utils.data.DataLoader(valid_data, batch_size = 64, shuffle = True)
with open('label_map.json', 'r') as f:
cat_to_name = json.load(f)
# TODO: Build and train your network
def nn_setup(structure = 'vgg16', lr = 0.001):
model = models.vgg16(pretrained = True)
for param in model.parameters():
param.requires_grad = False
model.classifier = nn.Sequential(OrderedDict([
('fc1', nn.Linear(25088, 2048)),
('relu', nn.ReLU()),
('fc2', nn.Linear(2048, 256)),
('relu', nn.ReLU()),
('fc3', nn.Linear(256, 102)),
('output', nn.LogSoftmax(dim=1))
]))
model = model.to('cuda')
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.classifier.parameters(), lr)
return model, criterion, optimizer
model, criterion, optimizer = nn_setup()
print(model)
epochs = 3
print_every = 5
steps = 0
loss_show = []
for e in range(epochs):
running_loss = 0
for inputs, labels in trainloader:
steps += 1
inputs, labels = inputs.to('cuda'), labels.to('cuda')
optimizer.zero_grad()
outputs = model.forward(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
if steps % print_every == 0:
model.eval()
valid_loss = 0
accuracy = 0
with torch.no_grad():
for inputs, labels in validloader:
inputs, labels = inputs.to('cuda'), labels.to('cuda')
log_ps = model.forward(inputs)
batch_loss = criterion(log_ps, labels)
valid_loss += batch_loss.item()
ps = torch.exp(log_ps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Epoch {e+1}/{epochs}.. "
f"Loss: {running_loss/print_every:.3f}.. "
f"Validation Loss: {valid_loss/len(validloader):.3f}.. "
f"Accuracy: {accuracy/len(validloader):.3f}")
running_loss = 0
model.train()
# TODO: Do validation on the test set
test_loss = 0
accuracy = 0
model.to('cuda')
with torch.no_grad():
for inputs, labels in testloader:
inputs, labels = inputs.to('cuda'), labels.to('cuda')
log_ps = model.forward(inputs)
batch_loss = criterion(log_ps, labels)
test_loss += batch_loss.item()
ps = torch.exp(log_ps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Test accuracy: {accuracy/len(testloader):.3f}")
Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
## Loading the checkpoint
At this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
# Inference for classification
Now you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like
First you'll need to handle processing the input image such that it can be used in your network.
## Image Preprocessing
You'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training.
First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.
Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.
As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation.
And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
## Class Prediction
Once you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.
To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.
Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.
## Sanity Checking
Now that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:
<img src='assets/inference_example.png' width=300px>
You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
| 0.656438 | 0.987472 |
# Prepare Enviorment
```
from google.colab import drive
drive.mount('/content/drive')
%cd drive/MyDrive/'Colab Notebooks'/
%cd Thesis/PeerRead/code/accept_classify/
import json
import pandas as pd
import os
from tqdm.notebook import tqdm
import os, re, random, time, pickle
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from sklearn.metrics import accuracy_score, roc_curve, auc, confusion_matrix, plot_roc_curve
path = '../../my_data/Figures/Overall Analysis-'
```
# Load the parameter search results
>
```
def returnDF(filename):
with open('../../my_data/'+filename, "rb") as f:
quick_logs=pickle.load(f)
df=pd.DataFrame(quick_logs,columns=['lr', 'seed','auc','accuracy'])
df['avg'] = df.apply(lambda row: (row['auc']+row['accuracy'])/2, axis=1)
return df
names = ['bert_quick_logs-16','scibert_quick_logs-16']
models = ['Bert','SciBert']
dirs = ['01-Paper-Acceptance', '02-Review-Acceptance'] #'03-Paper-Score', '04-Review-Score']
DF = []
for dir in dirs:
temp = []
for name in names:
df = returnDF(dir+'/'+name)
temp.append(df)
DF.append(temp)
sns.set_context("notebook", font_scale=1.7)
fig,axs=plt.subplots(len(dirs),len(names),figsize=(18,18))
for i in range(len(dirs)):
for j in range(len(names)):
sns.boxplot(data=DF[i][j],x='lr',y='accuracy',ax=axs[i,j], palette="BuPu", saturation=100)
sns.boxplot(data=DF[i][j],x='lr',y='auc',ax=axs[i,j], palette="BuPu", saturation=100)
axs[i,j].set_title(dirs[i]+' - '+models[j])
axs[i,j].set_ylim(0.15,0.75)
axs[i,j].set_xlabel('')
axs[i,j].set_ylabel('Average')
sns.despine()
#plt.savefig(path+'Learning Rate', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
```
## Updated hyperparameter plots
```
# get each df
df_01_bert = DF[0][0]
df_01_scibert = DF[0][1]
df_02_bert = DF[1][0]
df_02_scibert = DF[1][1]
# add model name
df_01_bert ['model'] = 'bert'
df_01_scibert ['model'] = 'scibert'
df_02_bert ['model'] = 'bert'
df_02_scibert ['model'] = 'scibert'
# add experiment name
df_01_bert ['exp'] = 'paper'
df_01_scibert ['exp'] = 'paper'
df_02_bert ['exp'] = 'review'
df_02_scibert ['exp'] = 'review'
df_all = pd.concat([df_01_bert, df_01_scibert, df_02_bert, df_02_scibert])
def mask(df,key,value):
return df[df[key] == value]
pd.DataFrame.mask = mask
sns.set_context("notebook", font_scale=2)
fig,ax = plt.subplots(figsize = (10,8))
g=sns.boxplot(data=df_all.mask('exp','paper'), x='lr',y='auc',hue='model', palette="GnBu", saturation=0.80)
sns.despine()
ax.set_ylabel('area-under-curve')
ax.set_xlabel('learning rate')
ax.set_ylim(0.3,0.8)
handles, _ = ax.get_legend_handles_labels() # Get the artists.
ax.legend(handles, ['BERT','SciBERT'], loc="best") # Associate manually the artists to a label.
plt.savefig(path+'Paper_auc_lr', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
sns.set_context("notebook", font_scale=2)
fig,ax = plt.subplots(figsize = (10,8))
g=sns.boxplot(data=df_all.mask('exp','paper'), x='lr',y='accuracy',hue='model', palette="GnBu", saturation=0.80)
sns.despine()
ax.set_ylabel('accuracy')
ax.set_xlabel('learning rate')
ax.set_ylim(0.3,0.8)
handles, _ = ax.get_legend_handles_labels() # Get the artists.
ax.legend(handles, ['BERT','SciBERT'], loc="best") # Associate manually the artists to a label.
plt.savefig(path+'Paper_acc_lr', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
sns.set_context("notebook", font_scale=2)
fig,ax = plt.subplots(figsize = (10,8))
g=sns.boxplot(data=df_all.mask('exp','review'), x='lr',y='auc',hue='model',palette="GnBu", saturation=0.80)
sns.despine()
ax.set_ylabel('area-under-curve')
ax.set_xlabel('learning rate')
ax.set_ylim(0.25, 0.75)
handles, _ = ax.get_legend_handles_labels()
ax.legend(handles, ['BERT','SciBERT'], loc="best")
plt.savefig(path+'Review_auc_lr', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
sns.set_context("notebook", font_scale=2)
fig,ax = plt.subplots(figsize = (10,8))
g=sns.boxplot(data=df_all.mask('exp','review'), x='lr',y='accuracy',hue='model', palette="GnBu", saturation=0.80)
sns.despine()
ax.set_ylabel('accuracy')
ax.set_xlabel('learning rate')
ax.set_ylim(0.25, 0.75)
handles, _ = ax.get_legend_handles_labels()
ax.legend(handles, ['BERT','SciBERT'], loc="best")
plt.savefig(path+'Review_acc_lr', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
```
# Analysis of results
```
def returnDF(filename):
with open('../../my_data/'+filename, "rb") as f:
outcome=pickle.load(f)
df=pd.DataFrame(outcome,columns=['roc_auc' 'accuracy' 'probs' 'fpr tpr])
df['avg'] = df.apply(lambda row: (row['auc']+row['accuracy'])/2, axis=1)
return df
names = ['bert_outcome-16','scibert_outcome-16']
models = ['Bert','SciBert']
dirs = ['01-Paper-Acceptance', '02-Review-Acceptance'] #'03-Paper-Score', '04-Review-Score']
DF = []
for dir in dirs:
temp = []
for name in names:
df = returnDF(dir+'/'+name)
temp.append(df)
DF.append(temp)
DF
sns.set_context("notebook", font_scale=1.7)
fig,axs=plt.subplots(len(dirs),len(names),figsize=(18,18))
for i in range(len(dirs)):
for j in range(len(names)):
sns.boxplot(data=DF[i][j],x='lr',y='avg',ax=axs[i,j], palette="BuPu", saturation=100)
axs[i,j].set_title(dirs[i]+' - '+models[j])
axs[i,j].set_ylim(0.15,0.75)
axs[i,j].set_xlabel('')
axs[i,j].set_ylabel('Average')
sns.despine()
plt.savefig(path+'Learning Rate', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
with open('../../my_data/02-Review-Acceptance/bert_outcome-16', "rb") as f:
outcome_16=pickle.load(f)
df_16=pd.DataFrame(outcome_16)
df_16
PROBS = np.array([out['probs'] for out in outcome_16])
ACC = np.array([out['accuracy'] for out in outcome_16])
FPR = np.array([out['fpr'] for out in outcome_16])
TPR = np.array([out['tpr'] for out in outcome_16])
AUC = np.array([out['roc_auc'] for out in outcome_16])
for i in range(10):
plt.plot(FPR[i],TPR[i],alpha=0.6)
sns.boxplot(data=df,x='seed',y='accuracy', palette="Set2")
```
|
github_jupyter
|
from google.colab import drive
drive.mount('/content/drive')
%cd drive/MyDrive/'Colab Notebooks'/
%cd Thesis/PeerRead/code/accept_classify/
import json
import pandas as pd
import os
from tqdm.notebook import tqdm
import os, re, random, time, pickle
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from sklearn.metrics import accuracy_score, roc_curve, auc, confusion_matrix, plot_roc_curve
path = '../../my_data/Figures/Overall Analysis-'
def returnDF(filename):
with open('../../my_data/'+filename, "rb") as f:
quick_logs=pickle.load(f)
df=pd.DataFrame(quick_logs,columns=['lr', 'seed','auc','accuracy'])
df['avg'] = df.apply(lambda row: (row['auc']+row['accuracy'])/2, axis=1)
return df
names = ['bert_quick_logs-16','scibert_quick_logs-16']
models = ['Bert','SciBert']
dirs = ['01-Paper-Acceptance', '02-Review-Acceptance'] #'03-Paper-Score', '04-Review-Score']
DF = []
for dir in dirs:
temp = []
for name in names:
df = returnDF(dir+'/'+name)
temp.append(df)
DF.append(temp)
sns.set_context("notebook", font_scale=1.7)
fig,axs=plt.subplots(len(dirs),len(names),figsize=(18,18))
for i in range(len(dirs)):
for j in range(len(names)):
sns.boxplot(data=DF[i][j],x='lr',y='accuracy',ax=axs[i,j], palette="BuPu", saturation=100)
sns.boxplot(data=DF[i][j],x='lr',y='auc',ax=axs[i,j], palette="BuPu", saturation=100)
axs[i,j].set_title(dirs[i]+' - '+models[j])
axs[i,j].set_ylim(0.15,0.75)
axs[i,j].set_xlabel('')
axs[i,j].set_ylabel('Average')
sns.despine()
#plt.savefig(path+'Learning Rate', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
# get each df
df_01_bert = DF[0][0]
df_01_scibert = DF[0][1]
df_02_bert = DF[1][0]
df_02_scibert = DF[1][1]
# add model name
df_01_bert ['model'] = 'bert'
df_01_scibert ['model'] = 'scibert'
df_02_bert ['model'] = 'bert'
df_02_scibert ['model'] = 'scibert'
# add experiment name
df_01_bert ['exp'] = 'paper'
df_01_scibert ['exp'] = 'paper'
df_02_bert ['exp'] = 'review'
df_02_scibert ['exp'] = 'review'
df_all = pd.concat([df_01_bert, df_01_scibert, df_02_bert, df_02_scibert])
def mask(df,key,value):
return df[df[key] == value]
pd.DataFrame.mask = mask
sns.set_context("notebook", font_scale=2)
fig,ax = plt.subplots(figsize = (10,8))
g=sns.boxplot(data=df_all.mask('exp','paper'), x='lr',y='auc',hue='model', palette="GnBu", saturation=0.80)
sns.despine()
ax.set_ylabel('area-under-curve')
ax.set_xlabel('learning rate')
ax.set_ylim(0.3,0.8)
handles, _ = ax.get_legend_handles_labels() # Get the artists.
ax.legend(handles, ['BERT','SciBERT'], loc="best") # Associate manually the artists to a label.
plt.savefig(path+'Paper_auc_lr', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
sns.set_context("notebook", font_scale=2)
fig,ax = plt.subplots(figsize = (10,8))
g=sns.boxplot(data=df_all.mask('exp','paper'), x='lr',y='accuracy',hue='model', palette="GnBu", saturation=0.80)
sns.despine()
ax.set_ylabel('accuracy')
ax.set_xlabel('learning rate')
ax.set_ylim(0.3,0.8)
handles, _ = ax.get_legend_handles_labels() # Get the artists.
ax.legend(handles, ['BERT','SciBERT'], loc="best") # Associate manually the artists to a label.
plt.savefig(path+'Paper_acc_lr', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
sns.set_context("notebook", font_scale=2)
fig,ax = plt.subplots(figsize = (10,8))
g=sns.boxplot(data=df_all.mask('exp','review'), x='lr',y='auc',hue='model',palette="GnBu", saturation=0.80)
sns.despine()
ax.set_ylabel('area-under-curve')
ax.set_xlabel('learning rate')
ax.set_ylim(0.25, 0.75)
handles, _ = ax.get_legend_handles_labels()
ax.legend(handles, ['BERT','SciBERT'], loc="best")
plt.savefig(path+'Review_auc_lr', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
sns.set_context("notebook", font_scale=2)
fig,ax = plt.subplots(figsize = (10,8))
g=sns.boxplot(data=df_all.mask('exp','review'), x='lr',y='accuracy',hue='model', palette="GnBu", saturation=0.80)
sns.despine()
ax.set_ylabel('accuracy')
ax.set_xlabel('learning rate')
ax.set_ylim(0.25, 0.75)
handles, _ = ax.get_legend_handles_labels()
ax.legend(handles, ['BERT','SciBERT'], loc="best")
plt.savefig(path+'Review_acc_lr', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
def returnDF(filename):
with open('../../my_data/'+filename, "rb") as f:
outcome=pickle.load(f)
df=pd.DataFrame(outcome,columns=['roc_auc' 'accuracy' 'probs' 'fpr tpr])
df['avg'] = df.apply(lambda row: (row['auc']+row['accuracy'])/2, axis=1)
return df
names = ['bert_outcome-16','scibert_outcome-16']
models = ['Bert','SciBert']
dirs = ['01-Paper-Acceptance', '02-Review-Acceptance'] #'03-Paper-Score', '04-Review-Score']
DF = []
for dir in dirs:
temp = []
for name in names:
df = returnDF(dir+'/'+name)
temp.append(df)
DF.append(temp)
DF
sns.set_context("notebook", font_scale=1.7)
fig,axs=plt.subplots(len(dirs),len(names),figsize=(18,18))
for i in range(len(dirs)):
for j in range(len(names)):
sns.boxplot(data=DF[i][j],x='lr',y='avg',ax=axs[i,j], palette="BuPu", saturation=100)
axs[i,j].set_title(dirs[i]+' - '+models[j])
axs[i,j].set_ylim(0.15,0.75)
axs[i,j].set_xlabel('')
axs[i,j].set_ylabel('Average')
sns.despine()
plt.savefig(path+'Learning Rate', dpi=400, bbox_inches = 'tight', pad_inches = 0 )
with open('../../my_data/02-Review-Acceptance/bert_outcome-16', "rb") as f:
outcome_16=pickle.load(f)
df_16=pd.DataFrame(outcome_16)
df_16
PROBS = np.array([out['probs'] for out in outcome_16])
ACC = np.array([out['accuracy'] for out in outcome_16])
FPR = np.array([out['fpr'] for out in outcome_16])
TPR = np.array([out['tpr'] for out in outcome_16])
AUC = np.array([out['roc_auc'] for out in outcome_16])
for i in range(10):
plt.plot(FPR[i],TPR[i],alpha=0.6)
sns.boxplot(data=df,x='seed',y='accuracy', palette="Set2")
| 0.312475 | 0.411052 |
```
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
from tensorflow.examples.tutorials.mnist import input_data
mnist_data = input_data.read_data_sets('MNIST_data/', one_hot=True)
```
## basic ops
```
input_dim = 784
output_dim = 10
w_value = np.random.uniform(size=(input_dim, output_dim)).astype(np.float32)
b_value = np.zeros(shape=(output_dim)).astype(np.float32)
input = tf.placeholder(dtype=tf.float32, shape=(None, 784), name='input')
w = tf.Variable(w_value)
b = tf.Variable(b_value)
output = tf.nn.softmax(tf.matmul(input, w) + b)
y_true = tf.placeholder(dtype=tf.float32, shape=(None, 10))
loss = tf.reduce_mean(-tf.reduce_sum(y_true * tf.log(output), reduction_indices=[1]))
train_step = tf.train.GradientDescentOptimizer(0.5).minimize(loss)
```
## build model
```
def build_model(x, y_):
W = tf.Variable(tf.zeros(shape=[784, 10]))
b = tf.Variable(tf.zeros(shape=[10]))
y = tf.matmul(x, W) + b
loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(y, y_))
train_step = tf.train.GradientDescentOptimizer(0.5).minimize(loss)
accur = tf.equal(tf.argmax(y, 1), tf.argmax(y_, 1))
accur = tf.reduce_mean(tf.cast(accur, tf.float32))
return train_step, accur
```
## train model
```
x = tf.placeholder(shape=(None, 784), dtype=tf.float32)
y_ = tf.placeholder(shape=[None, 10], dtype=tf.float32)
train_step, accur = build_model(x, y_)
sess = tf.Session()
init = tf.global_variables_initializer()
sess.run(init)
accur_list = []
minibatch_size = 100
report_freq = 20
n_epochs = 1000
for ii in range(n_epochs):
batch = mnist_data.train.next_batch(minibatch_size)
#train_step.run(feed_dict={x: batch[0], y_: batch[1]})
sess.run(train_step, feed_dict={x: batch[0], y_: batch[1]})
if ii % report_freq == 0:
accuracy = sess.run(accur, feed_dict={x: mnist_data.test.images, y_: mnist_data.test.labels})
accur_list.append(accuracy)
# print (accur.eval(feed_dict={x: mnist_data.test.images, y_: mnist_data.test.labels}))
plt.plot(accur_list)
plt.show()
def weight_var(shape):
init = tf.truncated_normal(shape, stddev=0.1)
return tf.Variable(init)
def bias_var(shape):
init = tf.constant(0.1, shape=shape)
return tf.Variable(init)
def conv2d(x, W):
return tf.nn.conv2d(x, W, strides=[1, 1, 1, 1], padding='SAME')
def max_pool_2x2(x):
return tf.nn.max_pool(x, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding='SAME')
print (b_conv1.get_shape())
x = tf.placeholder(tf.float32, [None, 784])
y = tf.placeholder(tf.float32, [None, 10])
x_image = tf.reshape(x, [-1, 28, 28, 1]) #[batch_n, in_h, in_w, in_channels]
W_conv1 = weight_var([5, 5, 1, 32]) #[filter_h, filter_w, in_channels, out_channels]
b_conv1 = bias_var([1, 32]) #[out_channels]
h_conv1 = tf.nn.relu(conv2d(x_image, W_conv1))
h_pool1 = max_pool_2x2(h_conv1)
W_conv2 = weight_var([5, 5, 32, 64])
b_conv2 = bias_var([1, 64])
h_conv2 = tf.nn.relu(conv2d(h_pool1, W_conv2))
h_pool2 = max_pool_2x2(h_conv2)
W_fc1 = weight_var([7*7*64, 1024])
b_fc1 = bias_var([1024])
h_pool2_flat = tf.reshape(h_pool2, [-1, 7*7*64])
h_fc1 = tf.nn.relu(tf.matmul(h_pool2_flat, W_fc1) + b_fc1)
keep_prob = tf.placeholder(tf.float32)
h_fc1_drop = tf.nn.dropout(h_fc1, keep_prob)
W_fc2 = weight_var([1024, 10])
b_fc2 = bias_var([10])
h_conv = tf.matmul(h_fc1_drop, W_fc2) + b_fc2
print (h_conv1.get_shape())
print (h_pool1.get_shape())
print (h_conv2.get_shape())
print (h_pool2.get_shape())
print (h_fc1.get_shape())
print (h_fc1_drop.get_shape())
print (h_conv.get_shape())
loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(h_conv, y))
train_step = tf.train.GradientDescentOptimizer(0.5).minimize(loss)
accur = tf.equal(tf.argmax(h_conv, 1), tf.argmax(y, 1))
accur = tf.reduce_mean(tf.cast(accur, tf.float32))
sess = tf.Session()
init = tf.global_variables_initializer()
sess.run(init)
accur_list = []
minibatch_size = 100
report_freq = 20
n_epochs = 1000
for ii in range(n_epochs):
batch = mnist_data.train.next_batch(minibatch_size)
sess.run(train_step, feed_dict={x: batch[0], y: batch[1], keep_prob: 1.}) ## remember 'keep_prob'
if ii % report_freq == 0:
accuracy = sess.run(accur, feed_dict={x: mnist_data.test.images, y: mnist_data.test.labels, keep_prob: 1.})
accur_list.append(accuracy)
plt.plot(accur_list)
plt.show()
```
|
github_jupyter
|
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
from tensorflow.examples.tutorials.mnist import input_data
mnist_data = input_data.read_data_sets('MNIST_data/', one_hot=True)
input_dim = 784
output_dim = 10
w_value = np.random.uniform(size=(input_dim, output_dim)).astype(np.float32)
b_value = np.zeros(shape=(output_dim)).astype(np.float32)
input = tf.placeholder(dtype=tf.float32, shape=(None, 784), name='input')
w = tf.Variable(w_value)
b = tf.Variable(b_value)
output = tf.nn.softmax(tf.matmul(input, w) + b)
y_true = tf.placeholder(dtype=tf.float32, shape=(None, 10))
loss = tf.reduce_mean(-tf.reduce_sum(y_true * tf.log(output), reduction_indices=[1]))
train_step = tf.train.GradientDescentOptimizer(0.5).minimize(loss)
def build_model(x, y_):
W = tf.Variable(tf.zeros(shape=[784, 10]))
b = tf.Variable(tf.zeros(shape=[10]))
y = tf.matmul(x, W) + b
loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(y, y_))
train_step = tf.train.GradientDescentOptimizer(0.5).minimize(loss)
accur = tf.equal(tf.argmax(y, 1), tf.argmax(y_, 1))
accur = tf.reduce_mean(tf.cast(accur, tf.float32))
return train_step, accur
x = tf.placeholder(shape=(None, 784), dtype=tf.float32)
y_ = tf.placeholder(shape=[None, 10], dtype=tf.float32)
train_step, accur = build_model(x, y_)
sess = tf.Session()
init = tf.global_variables_initializer()
sess.run(init)
accur_list = []
minibatch_size = 100
report_freq = 20
n_epochs = 1000
for ii in range(n_epochs):
batch = mnist_data.train.next_batch(minibatch_size)
#train_step.run(feed_dict={x: batch[0], y_: batch[1]})
sess.run(train_step, feed_dict={x: batch[0], y_: batch[1]})
if ii % report_freq == 0:
accuracy = sess.run(accur, feed_dict={x: mnist_data.test.images, y_: mnist_data.test.labels})
accur_list.append(accuracy)
# print (accur.eval(feed_dict={x: mnist_data.test.images, y_: mnist_data.test.labels}))
plt.plot(accur_list)
plt.show()
def weight_var(shape):
init = tf.truncated_normal(shape, stddev=0.1)
return tf.Variable(init)
def bias_var(shape):
init = tf.constant(0.1, shape=shape)
return tf.Variable(init)
def conv2d(x, W):
return tf.nn.conv2d(x, W, strides=[1, 1, 1, 1], padding='SAME')
def max_pool_2x2(x):
return tf.nn.max_pool(x, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding='SAME')
print (b_conv1.get_shape())
x = tf.placeholder(tf.float32, [None, 784])
y = tf.placeholder(tf.float32, [None, 10])
x_image = tf.reshape(x, [-1, 28, 28, 1]) #[batch_n, in_h, in_w, in_channels]
W_conv1 = weight_var([5, 5, 1, 32]) #[filter_h, filter_w, in_channels, out_channels]
b_conv1 = bias_var([1, 32]) #[out_channels]
h_conv1 = tf.nn.relu(conv2d(x_image, W_conv1))
h_pool1 = max_pool_2x2(h_conv1)
W_conv2 = weight_var([5, 5, 32, 64])
b_conv2 = bias_var([1, 64])
h_conv2 = tf.nn.relu(conv2d(h_pool1, W_conv2))
h_pool2 = max_pool_2x2(h_conv2)
W_fc1 = weight_var([7*7*64, 1024])
b_fc1 = bias_var([1024])
h_pool2_flat = tf.reshape(h_pool2, [-1, 7*7*64])
h_fc1 = tf.nn.relu(tf.matmul(h_pool2_flat, W_fc1) + b_fc1)
keep_prob = tf.placeholder(tf.float32)
h_fc1_drop = tf.nn.dropout(h_fc1, keep_prob)
W_fc2 = weight_var([1024, 10])
b_fc2 = bias_var([10])
h_conv = tf.matmul(h_fc1_drop, W_fc2) + b_fc2
print (h_conv1.get_shape())
print (h_pool1.get_shape())
print (h_conv2.get_shape())
print (h_pool2.get_shape())
print (h_fc1.get_shape())
print (h_fc1_drop.get_shape())
print (h_conv.get_shape())
loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(h_conv, y))
train_step = tf.train.GradientDescentOptimizer(0.5).minimize(loss)
accur = tf.equal(tf.argmax(h_conv, 1), tf.argmax(y, 1))
accur = tf.reduce_mean(tf.cast(accur, tf.float32))
sess = tf.Session()
init = tf.global_variables_initializer()
sess.run(init)
accur_list = []
minibatch_size = 100
report_freq = 20
n_epochs = 1000
for ii in range(n_epochs):
batch = mnist_data.train.next_batch(minibatch_size)
sess.run(train_step, feed_dict={x: batch[0], y: batch[1], keep_prob: 1.}) ## remember 'keep_prob'
if ii % report_freq == 0:
accuracy = sess.run(accur, feed_dict={x: mnist_data.test.images, y: mnist_data.test.labels, keep_prob: 1.})
accur_list.append(accuracy)
plt.plot(accur_list)
plt.show()
| 0.731442 | 0.887497 |
# French electric power consumption
The idea is to make an advanced version of the "Dessous des cartes" TV show in tribute to Jean Christophe Victor.
## Libraries
```
%run "../config/notebook.ipynb"
%run "../config/files.ipynb"
%run "../eda/eda_electric_power_consumption.ipynb"
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import geopandas as gpd
%matplotlib inline
```
## Maps
### Metropolitan France base map
#### Build the dataframe
```
france = gpd.read_file(FRANCE_DEPARTMENTS_BASEMAP_FILE_PATH)
# filter to remove the overseas departments
not_france_overseas = (france.code_insee != "971") & (france.code_insee != "972") & (france.code_insee != "973") \
& (france.code_insee != "974") & (france.code_insee != "975") & (france.code_insee != "976") & (france.code_insee != "977") \
& (france.code_insee != "978") & (france.code_insee != "984") & (france.code_insee != "986") & (france.code_insee != "987") \
& (france.code_insee != "988") & (france.code_insee != "989")
# apply the filter to remove the overseas departments
france_metropolitan = france[not_france_overseas]
# add a column to the map
france_metropolitan = france_metropolitan.set_index('code_insee')
```
#### Display the France metropolitan base map
```
ax = france_metropolitan.plot(figsize=(4,4),column='nom', cmap='jet')
# Turns off-grid on the left Axis.
ax.grid(False)
ax.axis('off')
```
### Merge the base maps with consumption statistics
```
def merge_year(year):
""" Merge the base map and the stats on the given year """
df = df_total_consumptions[df_total_consumptions['year'] == year]
df_result = france_metropolitan.join(df, left_on='code_insee', right_on='code_insee')
return df_result.set_index('code_insee')
test_df = merge_year(2011)
test_df.head()
test_df = merge_year(2011)
fig, axs = plt.subplots(1,1, figsize=(10, 5))
test_df.plot(ax=axs, column='total_residential', cmap='RdPu')
def display_total_row(year):
# get all differents totals
columns = [column for column in list(df_total_consumptions.columns) if column.startswith('total')]
fig, axs = plt.subplots(1, len(columns), figsize=(len(columns)*2, len(years)*2))
geodf = merge_year(year)
for column_index in range(len(columns)):
axs[column_index].set_title(columns[column_index])
geodf.plot(ax=axs[column_index], column=columns[column_index], cmap='RdPu')
axs[column_index].grid(False)
axs[column_index].axis('off')
def display_total_rows():
# get all differents totals
columns = [column for column in list(df_total_consumptions.columns) if column.startswith('total')]
# get distinc years
years = df_total_consumptions['year'].unique()
# build the figure
fig, axs = plt.subplots(len(years), len(columns), figsize=(len(columns)*2, len(years)*2))
# plot each row
for year_index in range(len(years)):
geodf = merge_year(years[year_index])
for column_index in range(len(columns)):
axs[year_index, column_index].set_title(columns[column_index])
geodf.plot(ax=axs[year_index, column_index], column=columns[column_index], cmap='RdPu')
axs[year_index, column_index].grid(False)
axs[year_index, column_index].axis('off')
display_total_row(2011)
```
### Metropolitan France total electric consumption map
## Augment the map with the conso by dep
```
# add a column to the map
france_metropolitan = france_metropolitan.set_index('code_insee')
# join with conso by department
france_metropolitan_conso = france_metropolitan.join(df_conso_by_dep)
# patch 69D and 69M
france_metropolitan_conso.loc['69D', 'conso'] = df_conso_by_dep.loc['69','conso']
france_metropolitan_conso.loc['69M', 'conso'] = df_conso_by_dep.loc['69','conso']
# remove the index
france_metropolitan_conso = france_metropolitan_conso.reset_index()
france_metropolitan_conso.head()
```
### Display France departments electric power consumption
```
ax = france_metropolitan_conso.plot(figsize=(16,16),column='conso', cmap='RdPu', legend=True)
# Turns off-grid on the left Axis.
ax.grid(False)
ax.axis('off')
```
## Electric power consumption by department resident
#### Electric consumption by department for residential purpose
```
sr_residential_electric_conso_by_dep = df_conso[electricity_only].loc[:, ['Code Département', 'Consommation Résidentiel (MWh)']].groupby('Code Département').sum().sort_values(by='Consommation Résidentiel (MWh)'
,ascending=False)
sr_residential_electric_conso_by_dep.reset_index()
# create a DataFrame from the Series
df_residential_electric_conso_by_dep = pd.DataFrame(sr_residential_electric_conso_by_dep)
# Remove the index
df_residential_electric_conso_by_dep = df_residential_electric_conso_by_dep.reset_index()
# Rename the columns
df_residential_electric_conso_by_dep.columns = ['code_insee', 'conso']
# Set the index on the code_insee column
df_residential_electric_conso_by_dep = df_residential_electric_conso_by_dep.set_index('code_insee')
```
#### Geomap of electric consumption by department for residential purpose
```
# join with conso by department
france_metropolitan_conso_resident = france_metropolitan.join(df_residential_electric_conso_by_dep)
# patch 69D and 69M
france_metropolitan_conso_resident.loc['69D', 'conso'] = df_residential_electric_conso_by_dep.loc['69','conso']
france_metropolitan_conso_resident.loc['69M', 'conso'] = df_residential_electric_conso_by_dep.loc['69','conso']
# remove the index
france_metropolitan_conso_resident = france_metropolitan_conso_resident.reset_index()
```
#### Display the Geomap
```
ax = france_metropolitan_conso_resident.plot(figsize=(16,16),column='conso', cmap='RdPu', legend=True)
# Turns off-grid on the left Axis.
ax.grid(False)
ax.axis('off')
```
## Tertiary electric power consumption by department
#### Electric consumption by department for tertiary purpose
```
sr_tertiary_electric_conso_by_dep = df_conso[electricity_only].loc[:, ['Code Département', 'Consommation Tertiaire (MWh)']].groupby('Code Département').sum().sort_values(by='Consommation Tertiaire (MWh)'
,ascending=False)
sr_tertiary_electric_conso_by_dep.reset_index()
# create a DataFrame from the Series
df_tertiary_electric_conso_by_dep = pd.DataFrame(sr_tertiary_electric_conso_by_dep)
# Remove the index
df_tertiary_electric_conso_by_dep = df_tertiary_electric_conso_by_dep.reset_index()
# Rename the columns
df_tertiary_electric_conso_by_dep.columns = ['code_insee', 'conso']
# Set the index on the code_insee column
df_tertiary_electric_conso_by_dep = df_tertiary_electric_conso_by_dep.set_index('code_insee')
```
#### Build the Geomap
```
# join with conso by department
france_metropolitan_conso_tertiary = france_metropolitan.join(df_tertiary_electric_conso_by_dep)
# patch 69D and 69M
france_metropolitan_conso_tertiary.loc['69D', 'conso'] = df_tertiary_electric_conso_by_dep.loc['69','conso']
france_metropolitan_conso_tertiary.loc['69M', 'conso'] = df_tertiary_electric_conso_by_dep.loc['69','conso']
# remove the index
france_metropolitan_conso_tertiary = france_metropolitan_conso_tertiary.reset_index()
```
#### Display the Geomap
```
ax = france_metropolitan_conso_tertiary.plot(figsize=(16,16),column='conso', cmap='RdPu', legend=True)
# Turns off-grid on the left Axis.
ax.grid(False)
ax.axis('off')
```
## Industrial electric power consumption by department
#### Electric consumption by department for industrial purpose
```
sr_industrial_electric_conso_by_dep = df_conso[electricity_only].loc[:, ['Code Département', 'Consommation Industrie (MWh)']].groupby('Code Département').sum().sort_values(by='Consommation Industrie (MWh)'
,ascending=False)
sr_industrial_electric_conso_by_dep.reset_index()
# create a DataFrame from the Series
df_industrial_electric_conso_by_dep = pd.DataFrame(sr_industrial_electric_conso_by_dep)
# Remove the index
df_industrial_electric_conso_by_dep = df_industrial_electric_conso_by_dep.reset_index()
# Rename the columns
df_industrial_electric_conso_by_dep.columns = ['code_insee', 'conso']
# Set the index on the code_insee column
df_industrial_electric_conso_by_dep = df_industrial_electric_conso_by_dep.set_index('code_insee')fig, axs = plt.subplots(1, 3, figsize=(20, 10))
axs[0].set_title('Total consumption')
france_metropolitan_conso.plot(ax=axs[0], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
axs[1].set_title('Tertiary consumption')
france_metropolitan_conso_tertiary.plot(ax=axs[1], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
axs[2].set_title('Residential consumption')
france_metropolitan_conso_resident.plot(ax=axs[2], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
# Turns off-grid on the left Axis.
for ax in axs:
ax.grid(False)
ax.axis('off')
```
#### Build the Geomap
```
# join with conso by department
france_metropolitan_conso_industrial = france_metropolitan.join(df_industrial_electric_conso_by_dep)
# patch 69D and 69M
france_metropolitan_conso_industrial.loc['69D', 'conso'] = df_industrial_electric_conso_by_dep.loc['69','conso']
france_metropolitan_conso_industrial.loc['69M', 'conso'] = df_industrial_electric_conso_by_dep.loc['69','conso']
# remove the index
france_metropolitan_conso_industrial = france_metropolitan_conso_industrial.reset_index()
```
#### Display the Geomap
```
ax = france_metropolitan_conso_industrial.plot(figsize=(16,16),column='conso', cmap='RdPu', legend=True)
# Turns off-grid on the left Axis.
ax.grid(False)
ax.axis('off')
```
## Bringing all together
```
fig, axs = plt.subplots(1, 4, figsize=(20, 10))
axs[0].set_title('Total consumption')
france_metropolitan_conso.plot(ax=axs[0], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
axs[1].set_title('Tertiary consumption')
france_metropolitan_conso_tertiary.plot(ax=axs[1], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
axs[2].set_title('Industrial consumption')
france_metropolitan_conso_industrial.plot(ax=axs[2], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
axs[3].set_title('Residential consumption')
france_metropolitan_conso_resident.plot(ax=axs[3], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
# Turns off-grid on the left Axis.
for ax in axs:
ax.grid(False)
ax.axis('off')
```
|
github_jupyter
|
%run "../config/notebook.ipynb"
%run "../config/files.ipynb"
%run "../eda/eda_electric_power_consumption.ipynb"
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import geopandas as gpd
%matplotlib inline
france = gpd.read_file(FRANCE_DEPARTMENTS_BASEMAP_FILE_PATH)
# filter to remove the overseas departments
not_france_overseas = (france.code_insee != "971") & (france.code_insee != "972") & (france.code_insee != "973") \
& (france.code_insee != "974") & (france.code_insee != "975") & (france.code_insee != "976") & (france.code_insee != "977") \
& (france.code_insee != "978") & (france.code_insee != "984") & (france.code_insee != "986") & (france.code_insee != "987") \
& (france.code_insee != "988") & (france.code_insee != "989")
# apply the filter to remove the overseas departments
france_metropolitan = france[not_france_overseas]
# add a column to the map
france_metropolitan = france_metropolitan.set_index('code_insee')
ax = france_metropolitan.plot(figsize=(4,4),column='nom', cmap='jet')
# Turns off-grid on the left Axis.
ax.grid(False)
ax.axis('off')
def merge_year(year):
""" Merge the base map and the stats on the given year """
df = df_total_consumptions[df_total_consumptions['year'] == year]
df_result = france_metropolitan.join(df, left_on='code_insee', right_on='code_insee')
return df_result.set_index('code_insee')
test_df = merge_year(2011)
test_df.head()
test_df = merge_year(2011)
fig, axs = plt.subplots(1,1, figsize=(10, 5))
test_df.plot(ax=axs, column='total_residential', cmap='RdPu')
def display_total_row(year):
# get all differents totals
columns = [column for column in list(df_total_consumptions.columns) if column.startswith('total')]
fig, axs = plt.subplots(1, len(columns), figsize=(len(columns)*2, len(years)*2))
geodf = merge_year(year)
for column_index in range(len(columns)):
axs[column_index].set_title(columns[column_index])
geodf.plot(ax=axs[column_index], column=columns[column_index], cmap='RdPu')
axs[column_index].grid(False)
axs[column_index].axis('off')
def display_total_rows():
# get all differents totals
columns = [column for column in list(df_total_consumptions.columns) if column.startswith('total')]
# get distinc years
years = df_total_consumptions['year'].unique()
# build the figure
fig, axs = plt.subplots(len(years), len(columns), figsize=(len(columns)*2, len(years)*2))
# plot each row
for year_index in range(len(years)):
geodf = merge_year(years[year_index])
for column_index in range(len(columns)):
axs[year_index, column_index].set_title(columns[column_index])
geodf.plot(ax=axs[year_index, column_index], column=columns[column_index], cmap='RdPu')
axs[year_index, column_index].grid(False)
axs[year_index, column_index].axis('off')
display_total_row(2011)
# add a column to the map
france_metropolitan = france_metropolitan.set_index('code_insee')
# join with conso by department
france_metropolitan_conso = france_metropolitan.join(df_conso_by_dep)
# patch 69D and 69M
france_metropolitan_conso.loc['69D', 'conso'] = df_conso_by_dep.loc['69','conso']
france_metropolitan_conso.loc['69M', 'conso'] = df_conso_by_dep.loc['69','conso']
# remove the index
france_metropolitan_conso = france_metropolitan_conso.reset_index()
france_metropolitan_conso.head()
ax = france_metropolitan_conso.plot(figsize=(16,16),column='conso', cmap='RdPu', legend=True)
# Turns off-grid on the left Axis.
ax.grid(False)
ax.axis('off')
sr_residential_electric_conso_by_dep = df_conso[electricity_only].loc[:, ['Code Département', 'Consommation Résidentiel (MWh)']].groupby('Code Département').sum().sort_values(by='Consommation Résidentiel (MWh)'
,ascending=False)
sr_residential_electric_conso_by_dep.reset_index()
# create a DataFrame from the Series
df_residential_electric_conso_by_dep = pd.DataFrame(sr_residential_electric_conso_by_dep)
# Remove the index
df_residential_electric_conso_by_dep = df_residential_electric_conso_by_dep.reset_index()
# Rename the columns
df_residential_electric_conso_by_dep.columns = ['code_insee', 'conso']
# Set the index on the code_insee column
df_residential_electric_conso_by_dep = df_residential_electric_conso_by_dep.set_index('code_insee')
# join with conso by department
france_metropolitan_conso_resident = france_metropolitan.join(df_residential_electric_conso_by_dep)
# patch 69D and 69M
france_metropolitan_conso_resident.loc['69D', 'conso'] = df_residential_electric_conso_by_dep.loc['69','conso']
france_metropolitan_conso_resident.loc['69M', 'conso'] = df_residential_electric_conso_by_dep.loc['69','conso']
# remove the index
france_metropolitan_conso_resident = france_metropolitan_conso_resident.reset_index()
ax = france_metropolitan_conso_resident.plot(figsize=(16,16),column='conso', cmap='RdPu', legend=True)
# Turns off-grid on the left Axis.
ax.grid(False)
ax.axis('off')
sr_tertiary_electric_conso_by_dep = df_conso[electricity_only].loc[:, ['Code Département', 'Consommation Tertiaire (MWh)']].groupby('Code Département').sum().sort_values(by='Consommation Tertiaire (MWh)'
,ascending=False)
sr_tertiary_electric_conso_by_dep.reset_index()
# create a DataFrame from the Series
df_tertiary_electric_conso_by_dep = pd.DataFrame(sr_tertiary_electric_conso_by_dep)
# Remove the index
df_tertiary_electric_conso_by_dep = df_tertiary_electric_conso_by_dep.reset_index()
# Rename the columns
df_tertiary_electric_conso_by_dep.columns = ['code_insee', 'conso']
# Set the index on the code_insee column
df_tertiary_electric_conso_by_dep = df_tertiary_electric_conso_by_dep.set_index('code_insee')
# join with conso by department
france_metropolitan_conso_tertiary = france_metropolitan.join(df_tertiary_electric_conso_by_dep)
# patch 69D and 69M
france_metropolitan_conso_tertiary.loc['69D', 'conso'] = df_tertiary_electric_conso_by_dep.loc['69','conso']
france_metropolitan_conso_tertiary.loc['69M', 'conso'] = df_tertiary_electric_conso_by_dep.loc['69','conso']
# remove the index
france_metropolitan_conso_tertiary = france_metropolitan_conso_tertiary.reset_index()
ax = france_metropolitan_conso_tertiary.plot(figsize=(16,16),column='conso', cmap='RdPu', legend=True)
# Turns off-grid on the left Axis.
ax.grid(False)
ax.axis('off')
sr_industrial_electric_conso_by_dep = df_conso[electricity_only].loc[:, ['Code Département', 'Consommation Industrie (MWh)']].groupby('Code Département').sum().sort_values(by='Consommation Industrie (MWh)'
,ascending=False)
sr_industrial_electric_conso_by_dep.reset_index()
# create a DataFrame from the Series
df_industrial_electric_conso_by_dep = pd.DataFrame(sr_industrial_electric_conso_by_dep)
# Remove the index
df_industrial_electric_conso_by_dep = df_industrial_electric_conso_by_dep.reset_index()
# Rename the columns
df_industrial_electric_conso_by_dep.columns = ['code_insee', 'conso']
# Set the index on the code_insee column
df_industrial_electric_conso_by_dep = df_industrial_electric_conso_by_dep.set_index('code_insee')fig, axs = plt.subplots(1, 3, figsize=(20, 10))
axs[0].set_title('Total consumption')
france_metropolitan_conso.plot(ax=axs[0], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
axs[1].set_title('Tertiary consumption')
france_metropolitan_conso_tertiary.plot(ax=axs[1], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
axs[2].set_title('Residential consumption')
france_metropolitan_conso_resident.plot(ax=axs[2], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
# Turns off-grid on the left Axis.
for ax in axs:
ax.grid(False)
ax.axis('off')
# join with conso by department
france_metropolitan_conso_industrial = france_metropolitan.join(df_industrial_electric_conso_by_dep)
# patch 69D and 69M
france_metropolitan_conso_industrial.loc['69D', 'conso'] = df_industrial_electric_conso_by_dep.loc['69','conso']
france_metropolitan_conso_industrial.loc['69M', 'conso'] = df_industrial_electric_conso_by_dep.loc['69','conso']
# remove the index
france_metropolitan_conso_industrial = france_metropolitan_conso_industrial.reset_index()
ax = france_metropolitan_conso_industrial.plot(figsize=(16,16),column='conso', cmap='RdPu', legend=True)
# Turns off-grid on the left Axis.
ax.grid(False)
ax.axis('off')
fig, axs = plt.subplots(1, 4, figsize=(20, 10))
axs[0].set_title('Total consumption')
france_metropolitan_conso.plot(ax=axs[0], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
axs[1].set_title('Tertiary consumption')
france_metropolitan_conso_tertiary.plot(ax=axs[1], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
axs[2].set_title('Industrial consumption')
france_metropolitan_conso_industrial.plot(ax=axs[2], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
axs[3].set_title('Residential consumption')
france_metropolitan_conso_resident.plot(ax=axs[3], column='conso', cmap='RdPu', legend=False, vmin=0, vmax=6e7)
# Turns off-grid on the left Axis.
for ax in axs:
ax.grid(False)
ax.axis('off')
| 0.473901 | 0.895202 |
# [Instacart Market Basket Analysis | Kaggle](https://www.kaggle.com/c/instacart-market-basket-analysis)
인스타카트는 식료품 주문 및 배달 앱이다. 냉장고와 팬트리를 개인화된 취향에 맞춰 쉽게 채우는 것을 목표로 한다.
인스타카트의 데이터과학팀은 즐거운 쇼핑겸험을 제공하는데 큰 역할을 한다. 트랜잭션 데이터를 사용하여 이용자가 다시 구매할 제품을 예측하거나 첫 구매를 시도하거나 다음 쇼핑 때 장바구니에 추가 할 제품에 대한 모델을 개발한다. 최근 인스타카트는 이 데이터를 소스로 공개하였다. 오픈 소스로 공개된 300만개의 인스타카드 주문에 대한 블로그 포스트를 참조하길 바란다.
이 경진대회에서 인스타카트는 캐글커뮤니티에 고객의 주문에 대한 익명화된 데이터를 제공하여 이전에 주문했던 데이터를 바탕으로 다음 주문을 예측한다. 인스타카트는 최고의 모델을 찾는 것 뿐만 아니라 그들과 함께 성장할 머신러닝 엔지니어를 찾는다.
이 대회의 수상자는 상금과 리쿠르팅 기회를 함께 얻지만 이 대회는 2017년 8월 14일에 서브미션 제출이 끝난대회다.
### 평가 [mean F1 score - Wikipedia](https://en.wikipedia.org/wiki/F1_score)
테스트 세트의 각 order_id에 대해 해당 주문에 대한 product_ids의 공백으로 구분 된 목록을 예측해야한다. 빈 주문을 예측하려면 명시 적 'None'값을 제출해야한다. 'None'을 product_ids와 결합 할 수 있다. '없음'의 철자는 채점 척도에서 대소문자를 구분한다. 파일에는 헤더가 있고 다음과 같이 표시되어야한다.
```
order_id,products
17,1 2
34,None
137,1 2 3
etc.
```
참고 : https://www.kaggle.com/sudalairajkumar/simple-exploration-notebook-instacart
```
import numpy as np # 수학 계산을 위해
import pandas as pd # CSV 파일을 읽어오고 데이터 처리를 위해
import matplotlib as mpl # 그래프를 그리기 위해
import matplotlib.pyplot as plt
import seaborn as sns # matplotlib을 쉽게 사용하기 위해
color = sns.color_palette()
# 노트북 안에 그래프를 그리기 위해
%matplotlib inline
# 그래프에서 격자로 숫자 범위가 눈에 잘 띄도록 ggplot 스타일을 사용
plt.style.use('ggplot')
# 그래프에서 마이너스 폰트 깨지는 문제에 대한 대처
mpl.rcParams['axes.unicode_minus'] = False
# pd.options.mode.chained_assignment = None # default='warn'
from subprocess import check_output
print(check_output(["ls", "data"]).decode("utf8"))
order_products_train_df = pd.read_csv("data/order_products__train.csv")
order_products_prior_df = pd.read_csv("data/order_products__prior.csv")
orders_df = pd.read_csv("data/orders.csv")
products_df = pd.read_csv("data/products.csv")
aisles_df = pd.read_csv("data/aisles.csv")
departments_df = pd.read_csv("data/departments.csv")
orders_df.head()
order_products_train_df.head()
order_products_prior_df.head()
products_df.head()
aisles_df.head()
departments_df.head()
cnt_srs = orders_df.eval_set.value_counts()
plt.figure(figsize=(8,4))
sns.barplot(cnt_srs.index, cnt_srs.values, alpha=0.8, color=color[1])
plt.ylabel('Number of Occurrences', fontsize=12)
plt.xlabel('Eval set type', fontsize=12)
plt.title('각 데이터셋의 행 갯수', fontsize=15)
plt.xticks(rotation='vertical')
plt.show()
def get_unique_count(x):
return len(np.unique(x))
cnt_srs = orders_df.groupby("eval_set")["user_id"].aggregate(get_unique_count)
cnt_srs
cnt_srs = orders_df.groupby("user_id")["order_number"].aggregate(np.max).reset_index()
cnt_srs = cnt_srs.order_number.value_counts()
cnt_srs.head()
plt.figure(figsize=(12,6))
sns.barplot(cnt_srs.index, cnt_srs.values, alpha=0.8, color=color[2])
plt.ylabel('총 주문 건', fontsize=12)
plt.xlabel('최대 주문 횟수', fontsize=12)
plt.xticks(rotation='vertical')
plt.show()
```
#### 요일별 주문 횟수
토요일과 일요일에 주문이 가장 많고 수요일이 가장 적다.
```
dow = ['토', '일', '월', '화', '수', '목', '금']
plt.figure(figsize=(8,4))
fig, ax = plt.subplots()
sns.countplot(x="order_dow", data=orders_df, color=color[0])
ax.set_xticklabels(dow)
plt.ylabel('주문수량', fontsize=12)
plt.xlabel('요일', fontsize=12)
plt.title("요일별 주문 수량", fontsize=15)
plt.show()
plt.figure(figsize=(12,8))
sns.countplot(x="order_hour_of_day", data=orders_df, color=color[1])
plt.ylabel('주문 량', fontsize=12)
plt.xlabel('시간', fontsize=12)
plt.xticks(rotation='vertical')
plt.title("시간대별 주문 수량", fontsize=15)
plt.show()
```
토요일 오후와 일요일 오전에 주문량이 많다.
```
dow = ['토', '일', '월', '화', '수', '목', '금']
grouped_df = orders_df.groupby(
["order_dow", "order_hour_of_day"]
)["order_number"].aggregate("count").reset_index()
grouped_df = grouped_df.pivot(
'order_dow', 'order_hour_of_day', 'order_number')
plt.figure(figsize=(20,6))
fig, ax = plt.subplots()
sns.heatmap(grouped_df)
ax.set_yticklabels(dow)
plt.ylabel('주문 요일', fontsize=12)
plt.xlabel('주문 시간', fontsize=12)
plt.title("요일별 시간대별 주문량")
plt.show()
products_df.head()
aisles_df.head()
departments_df.head()
order_products_prior_df = pd.merge(order_products_prior_df, products_df, on='product_id', how='left')
order_products_prior_df = pd.merge(order_products_prior_df, aisles_df, on='aisle_id', how='left')
order_products_prior_df = pd.merge(order_products_prior_df, departments_df, on='department_id', how='left')
order_products_prior_df.head()
cnt_srs = order_products_prior_df['product_name'].value_counts().reset_index().head(20)
cnt_srs.columns = ['product_name', 'frequency_count']
cnt_srs
cnt_srs = order_products_prior_df['aisle'].value_counts().head(20)
plt.figure(figsize=(12,8))
sns.barplot(cnt_srs.index, cnt_srs.values, alpha=0.8, color=color[5])
plt.ylabel('Number of Occurrences', fontsize=12)
plt.xlabel('Aisle', fontsize=12)
plt.xticks(rotation='vertical')
plt.show()
plt.figure(figsize=(10,10))
temp_series = order_products_prior_df['department'].value_counts()
labels = (np.array(temp_series.index))
sizes = (np.array((temp_series / temp_series.sum())*100))
plt.pie(sizes, labels=labels,
autopct='%1.1f%%', startangle=200)
plt.title("Departments distribution", fontsize=15)
plt.show()
grouped_df = order_products_prior_df.groupby(
["department"])["reordered"].aggregate("mean").reset_index()
plt.figure(figsize=(12,8))
sns.pointplot(
grouped_df['department'].values,
grouped_df['reordered'].values,
alpha=0.8, color=color[2])
plt.ylabel('재구매 비율', fontsize=12)
plt.xlabel('Department', fontsize=12)
plt.title("Department별 재구매 비율", fontsize=15)
plt.xticks(rotation='vertical')
plt.show()
grouped_df = order_products_prior_df.groupby(
["department_id", "aisle"])["reordered"].aggregate(
"mean").reset_index()
fig, ax = plt.subplots(figsize=(12,20))
ax.scatter(grouped_df.reordered.values, grouped_df.department_id.values)
for i, txt in enumerate(grouped_df.aisle.values):
ax.annotate(txt, (grouped_df.reordered.values[i], grouped_df.department_id.values[i]), rotation=45, ha='center', va='center', color='green')
plt.xlabel('재구매 비율')
plt.ylabel('department_id')
plt.title("각 통로별 재구매 비율", fontsize=15)
plt.show()
```
|
github_jupyter
|
order_id,products
17,1 2
34,None
137,1 2 3
etc.
import numpy as np # 수학 계산을 위해
import pandas as pd # CSV 파일을 읽어오고 데이터 처리를 위해
import matplotlib as mpl # 그래프를 그리기 위해
import matplotlib.pyplot as plt
import seaborn as sns # matplotlib을 쉽게 사용하기 위해
color = sns.color_palette()
# 노트북 안에 그래프를 그리기 위해
%matplotlib inline
# 그래프에서 격자로 숫자 범위가 눈에 잘 띄도록 ggplot 스타일을 사용
plt.style.use('ggplot')
# 그래프에서 마이너스 폰트 깨지는 문제에 대한 대처
mpl.rcParams['axes.unicode_minus'] = False
# pd.options.mode.chained_assignment = None # default='warn'
from subprocess import check_output
print(check_output(["ls", "data"]).decode("utf8"))
order_products_train_df = pd.read_csv("data/order_products__train.csv")
order_products_prior_df = pd.read_csv("data/order_products__prior.csv")
orders_df = pd.read_csv("data/orders.csv")
products_df = pd.read_csv("data/products.csv")
aisles_df = pd.read_csv("data/aisles.csv")
departments_df = pd.read_csv("data/departments.csv")
orders_df.head()
order_products_train_df.head()
order_products_prior_df.head()
products_df.head()
aisles_df.head()
departments_df.head()
cnt_srs = orders_df.eval_set.value_counts()
plt.figure(figsize=(8,4))
sns.barplot(cnt_srs.index, cnt_srs.values, alpha=0.8, color=color[1])
plt.ylabel('Number of Occurrences', fontsize=12)
plt.xlabel('Eval set type', fontsize=12)
plt.title('각 데이터셋의 행 갯수', fontsize=15)
plt.xticks(rotation='vertical')
plt.show()
def get_unique_count(x):
return len(np.unique(x))
cnt_srs = orders_df.groupby("eval_set")["user_id"].aggregate(get_unique_count)
cnt_srs
cnt_srs = orders_df.groupby("user_id")["order_number"].aggregate(np.max).reset_index()
cnt_srs = cnt_srs.order_number.value_counts()
cnt_srs.head()
plt.figure(figsize=(12,6))
sns.barplot(cnt_srs.index, cnt_srs.values, alpha=0.8, color=color[2])
plt.ylabel('총 주문 건', fontsize=12)
plt.xlabel('최대 주문 횟수', fontsize=12)
plt.xticks(rotation='vertical')
plt.show()
dow = ['토', '일', '월', '화', '수', '목', '금']
plt.figure(figsize=(8,4))
fig, ax = plt.subplots()
sns.countplot(x="order_dow", data=orders_df, color=color[0])
ax.set_xticklabels(dow)
plt.ylabel('주문수량', fontsize=12)
plt.xlabel('요일', fontsize=12)
plt.title("요일별 주문 수량", fontsize=15)
plt.show()
plt.figure(figsize=(12,8))
sns.countplot(x="order_hour_of_day", data=orders_df, color=color[1])
plt.ylabel('주문 량', fontsize=12)
plt.xlabel('시간', fontsize=12)
plt.xticks(rotation='vertical')
plt.title("시간대별 주문 수량", fontsize=15)
plt.show()
dow = ['토', '일', '월', '화', '수', '목', '금']
grouped_df = orders_df.groupby(
["order_dow", "order_hour_of_day"]
)["order_number"].aggregate("count").reset_index()
grouped_df = grouped_df.pivot(
'order_dow', 'order_hour_of_day', 'order_number')
plt.figure(figsize=(20,6))
fig, ax = plt.subplots()
sns.heatmap(grouped_df)
ax.set_yticklabels(dow)
plt.ylabel('주문 요일', fontsize=12)
plt.xlabel('주문 시간', fontsize=12)
plt.title("요일별 시간대별 주문량")
plt.show()
products_df.head()
aisles_df.head()
departments_df.head()
order_products_prior_df = pd.merge(order_products_prior_df, products_df, on='product_id', how='left')
order_products_prior_df = pd.merge(order_products_prior_df, aisles_df, on='aisle_id', how='left')
order_products_prior_df = pd.merge(order_products_prior_df, departments_df, on='department_id', how='left')
order_products_prior_df.head()
cnt_srs = order_products_prior_df['product_name'].value_counts().reset_index().head(20)
cnt_srs.columns = ['product_name', 'frequency_count']
cnt_srs
cnt_srs = order_products_prior_df['aisle'].value_counts().head(20)
plt.figure(figsize=(12,8))
sns.barplot(cnt_srs.index, cnt_srs.values, alpha=0.8, color=color[5])
plt.ylabel('Number of Occurrences', fontsize=12)
plt.xlabel('Aisle', fontsize=12)
plt.xticks(rotation='vertical')
plt.show()
plt.figure(figsize=(10,10))
temp_series = order_products_prior_df['department'].value_counts()
labels = (np.array(temp_series.index))
sizes = (np.array((temp_series / temp_series.sum())*100))
plt.pie(sizes, labels=labels,
autopct='%1.1f%%', startangle=200)
plt.title("Departments distribution", fontsize=15)
plt.show()
grouped_df = order_products_prior_df.groupby(
["department"])["reordered"].aggregate("mean").reset_index()
plt.figure(figsize=(12,8))
sns.pointplot(
grouped_df['department'].values,
grouped_df['reordered'].values,
alpha=0.8, color=color[2])
plt.ylabel('재구매 비율', fontsize=12)
plt.xlabel('Department', fontsize=12)
plt.title("Department별 재구매 비율", fontsize=15)
plt.xticks(rotation='vertical')
plt.show()
grouped_df = order_products_prior_df.groupby(
["department_id", "aisle"])["reordered"].aggregate(
"mean").reset_index()
fig, ax = plt.subplots(figsize=(12,20))
ax.scatter(grouped_df.reordered.values, grouped_df.department_id.values)
for i, txt in enumerate(grouped_df.aisle.values):
ax.annotate(txt, (grouped_df.reordered.values[i], grouped_df.department_id.values[i]), rotation=45, ha='center', va='center', color='green')
plt.xlabel('재구매 비율')
plt.ylabel('department_id')
plt.title("각 통로별 재구매 비율", fontsize=15)
plt.show()
| 0.341583 | 0.933854 |
The [PoetryDB API](https://github.com/thundercomb/poetrydb/blob/master/README.md) stores its data in [MongoDB](https://www.mongodb.com/), a popular [NoSQL database](https://en.wikipedia.org/wiki/NoSQL). Indeed, a NoSQL database is a solid choice for the type of data that is stored in PoetryDB (unstructured text, for example). However, what if we wanted to create a more traditional SQL database with the PoetryDB API data for use in other projects where a relational database would be preferred? By extracting the data from the PoetryDB API using a combination of a few Python libraries, we can recreate the NoSQL PoetryDB database as a SQL database which will allow us more freedom to create additional data features and avoid the need to hit the PoetryDB database more than necessary.
## Getting Started
In this example, we walk through a sample use case of extracting data from a database using an API and then structuring that data in a cohesive manner that allows us to create a relational database that we can then query with SQL statements. The database we will create with the extracted data will use [Postgresql](https://www.postgresql.org/).
The Python libraries that will be used in this example are [poetpy](https://github.com/aschleg/poetpy), a Python wrapper for the PoetryDB API written by yours truly, [pandas](https://pandas.pydata.org/) for transforming and cleansing the data as needed, and [sqlalchemy](https://www.sqlalchemy.org/) for handling the SQL side of things. We start by importing the needed libraries as per usual.
```
from poetpy import get_poetry
import pandas as pd
from pandas.io.json import json_normalize
import sqlalchemy
import os
import warnings
warnings.simplefilter('ignore')
```
## Getting the Poetry Data
We can't have a useful database without any data! Before beginning to hit any API, it is often useful to devise a strategy for getting the wanted data in an efficient manner that avoids requesting the API more than needed. According to the [PoetryDB API documentation](https://github.com/thundercomb/poetrydb/blob/master/README.md), we can get a list of authors which we can then use to iterate over to get each author's poetry and other available information from the database.
We can use the `poetpy` function `get_poetry` to return a dictionary object of the available authors in the PoetryDB database.
```
authors = get_poetry('author')
```
The returned dictionary contains a list of the available authors, which we can quickly inspect to make sure our API call was successful.
```
authors['authors'][0:5]
```
To store the extracted authors for later exporting into a flat-file such as a CSV and loading into a database, we convert the returned dictionary into a [pandas DataFrame](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.html) using the [`from_dict`](https://pandas.pydata.org/pandas-docs/version/0.22/generated/pandas.DataFrame.from_dict.html) method.
```
authors_df = pd.DataFrame.from_dict(authors)
```
Now that we have a list of authors to iterate over, we can extract the remaining data from the PoetryDB database! For each of the authors in the database, we extract the titles, content, and linecounts of their poetry, normalize the returned JSON into a `DataFrame` with `pandas`'s handy [`json_normalize`](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.io.json.json_normalize.html) function and append the resulting data to a list. After each author in the list has been iterated over, the list with the appended results are then concatenated into one `DataFrame` with the [`pd.concat`](https://pandas.pydata.org/pandas-docs/version/0.23/generated/pandas.concat.html) function. This operation will give us a complete dataset of all the available information in the PoetryDB API as a pandas `DataFrame`.
```
poems = []
for author in authors['authors']:
author_poems = get_poetry('author', author, 'author,title,lines,linecount')
author_poems_df = json_normalize(author_poems)
poems.append(author_poems_df)
poems_df = pd.concat(poems)
```
The PoetryDB API data is now collected into one `DataFrame`! We can inspect the first few rows of the `DataFrame` to see the resulting data that was returned with the [`head`](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.head.html) method.
```
poems_df.head()
```
We see each value in the `lines` column of the `DataFrame` is still a list of strings that comprise the particular poem. To edit the `lines` column to extract the poetry lines, we can use the [`apply`](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.apply.html) method to apply a `lambda` function over each row in the `DataFrame` to join each string in the list as one string.
```
poems_df['lines'] = poems_df['lines'].apply(lambda x: ' \n '.join(x))
```
Let's inspect the first couple rows of the `lines` column to ensure the operation returned what we expect.
```
poems_df['lines'].head(2)
```
With the data extracted from the PoetryDB database and transformed into a tabular data structure, we then save the datasets into a csv file using the [`to_csv`](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.to_csv.html) method. The exported csv files will be used to insert the data into our Postgresql database.
```
poems_df.to_csv('../data/poetrydb_copy.csv', index=False, encoding='utf-8')
authors_df.to_csv('../data/poetrydb_authors.csv', index=False, encoding='utf-8')
```
## Building the Postgresql database
The installation of Postgresql is beyond the scope of this example; however, there is a handy [tutorial](http://www.postgresqltutorial.com/install-postgresql/) available which details the steps for installing Postgresql for particular operating systems.
During the installation of Postgresql, a `postgres` database is created that we can use for testing our relational database. Postgresql works slightly different than other SQL engines in that it employs the concept of [schemas](https://www.postgresql.org/docs/8.1/static/ddl-schemas.html) for managing data, which in other types of SQL would be a database. The installation will prompt the user to create a master username and password, which we will use to connect to the localhost `postgres` database.
This is the section of the example where SQLAlchemy comes into play. The first step in connecting to a database with SQLAlchemy is to employ the [`create_engine`](http://docs.sqlalchemy.org/en/latest/core/engines.html) function. According to the function's documentation, the `create_engine` function takes a string parameter that details the connection info based on the following structure:
```terminal
dialect+driver://username:password@host:port/database
```
Where `dialect` is a SQL engine such as `postgresql`, `mysql`, `mssql`, and so on. Thus, we use the newly created `postgres` database along with the username and password specified during the installation of `Postgresql`.
```
engine = sqlalchemy.create_engine('postgresql://postgres:root@localhost:5432/postgres')
```
The `engine` variable is then used to create the connection to the `postgres` database.
```
conn = engine.connect()
```
We can now begin working with the `Postgresql` database and insert our extracted data! The first step is to create a [schema](https://www.postgresql.org/docs/8.1/static/ddl-schemas.html) which we will use as our local database for testing purposes. Using our database connection, we can send a query to the `postgres` database to create a schema using the `CREATE SCHEMA` statement.
```
conn.execute('CREATE SCHEMA poetry')
```
The output indicates the operation was successful! The next step is to create the necessary schema tables and load the data we extracted earlier from the PoetryDB API into those tables. There are several approaches to creating the tables and loading the data into those tables. One approach and typically the most general way to load data into Postgresql is to create the tables and then load a flat file such as a csv into the tables using the `psql` command line. Another approach is using `pandas` and `SQLAlchemy` to load the data directly from a `DataFrame` into a Postgresql database or schema.
As the last step before loading the data, let's use our `SQLAlchemy` connection to the database to create the tables that we will use to store our data. For more extended SQL statements, it can be a good idea to write out the statement or query as a multi-line string for clarity.
```
create_author_table = '''
CREATE TABLE poetry.authors
(
id serial PRIMARY KEY,
author VARCHAR(255)
);'''
create_poems_table = '''
CREATE TABLE poetry.poems
(
id serial PRIMARY KEY,
author VARCHAR(255),
linecount INT,
lines TEXT,
title VARCHAR(510)
);
'''
conn.execute(create_author_table)
conn.execute(create_poems_table)
```
## Using `psql` and `\copy` to load the data
[`psql`](https://www.postgresql.org/docs/9.2/static/app-psql.html) is a Postgresql interactive terminal and is very useful for working with Postgresql databases and schemas. For those with a MySQL background, `psql` is quite similar to the `mysql` interactive terminal. The following is used to launch the `psql` terminal.
```terminal
psql -h localhost -U postgres postgres
```
Where the `-h` flag specifies the host to connect, which in this case is `localhost`. The `-U postgres` argument specifies the username `postgres` to use to connect to the `postgres` database. For those having issues launching the `psql` terminal, it is usually due to the command not being set in the PATH. Here is a good [StackOverflow](https://stackoverflow.com/questions/36155219/psql-command-not-found-mac) page that explains in detail how to add the `psql` command to the PATH on Mac.
After launching `psql`, using the `\dt` command will display the current tables and relations in the current schema.
The `\copy` command is used to insert data from a standard flat-file such as a csv into a schema table. The path to the data file to load into the table generally needs to be the absolute path. We specify the columns to insert the data within the parentheses to avoid accidentally inserting the rows into the schema table's `id` column, which acts as its `PRIMARY KEY`.
```terminal
\copy poetry.authors(author) FROM '/Users/aaronschlegel/Dropbox/Projects/poetpy/data/poetrydb_authors.csv' DELIMITER ',' CSV HEADER;
```
```terminal
\copy poetry.poems(author, linecount, lines, title) FROM '/Users/aaronschlegel/Dropbox/Projects/poetpy/data/poetrydb_copy.csv' DELIMITER ',' CSV HEADER;
```
If the `\copy` is successful, the terminal will output the number of rows that were inserted into the table. We can now perform queries on the tables!
## Using SQLAlchemy and pandas to load the data
The `DataFrame` class has a handy method, [`to_sql`](https://pandas.pydata.org/pandas-docs/version/0.22/generated/pandas.DataFrame.to_sql.html) for inserting data into a SQL table directly. As we already created two tables earlier, we will give these a different name to identify the new tables created from the `DataFrame`. The name of the SQL table to insert into is designated by the first argument of the `to_sql` method, while the second required argument is a database connection string, just like the one we created previously with SQLAlchemy! To get a sequential `id` column inserted into the SQL table simultaneously, we will also specify that the `DataFrame` index column is named `id` with the optional `index_label` argument. We also want to be sure to set the `schema` optional argument to the `poetry` schema (since we are working with Postgresql) that we created earlier in the example. Otherwise, the tables will be created in the default `public` schema.
```
poems_df.to_sql('poems_df', conn, schema='poetry', index_label='id')
authors_df.to_sql('authors_df', conn, schema='poetry', index_label='id')
```
There were no errors or warnings issued. Therefore the data insertion should have been successful! In the next section, we perform some sample queries on the newly created SQL tables to ensure the data is what we expect.
## Example Queries
Perhaps unsurprisingly, there are multiple ways to query our Postgresql schema tables. The first is to use the `.execute()` method from our database connection variable `conn`, which we created earlier in the example. Let's say we are interested in finding the first 3 authors from the `author` table in our `poetry` schema. The SQL query can be written as:
```sql
SELECT author FROM poetry.authors LIMIT 3
```
The above query can be passed as an argument to the `.execute()` method as a string to query the database. The `.fetchall()` is chained to the end of the `.execute()` method to extract all the results.
```
conn.execute('SELECT author FROM poetry.authors LIMIT 3').fetchall()
```
We see the statement returned the first three authors as a list of tuples as expected! More information on using SQL queries with SQLAlchemy can be found in [SQLAlchemy's tutorial](http://docs.sqlalchemy.org/en/latest/core/tutorial.html).
Another method for querying a database that can be very useful is to use the `pandas` function [`read_sql_query`](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_sql_query.html). The function [`read_sql`](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_sql.html#pandas.read_sql) can also be used to return the same results. The required arguments for the function are the query and a connection string. The benefit of using the `read_sql_query` function is the results are pulled directly into a pandas `DataFrame`.
```
pd.read_sql_query('SELECT authors FROM poetry.authors_df LIMIT 3', conn)
```
## Conclusion
In this example, we displayed a sample use case for extracting data from a database through an API and then using that data to create the database that we can use for further analysis and more without worrying about hitting the API database more than needed. In further examples, we will enhance the data we extracted from the API with more information collected from different sources and feature engineering on the already available data.
## About PoetryDB
[PoetryDB](http://poetrydb.org/index.html) was created and is currently maintained by
[@thundercomb](https://twitter.com/thundercomb). They blog about poetry and related technology and
other topics at [thecombedthunderclap.blogspot.com](http://thecombedthunderclap.blogspot.com/).
|
github_jupyter
|
from poetpy import get_poetry
import pandas as pd
from pandas.io.json import json_normalize
import sqlalchemy
import os
import warnings
warnings.simplefilter('ignore')
authors = get_poetry('author')
authors['authors'][0:5]
authors_df = pd.DataFrame.from_dict(authors)
poems = []
for author in authors['authors']:
author_poems = get_poetry('author', author, 'author,title,lines,linecount')
author_poems_df = json_normalize(author_poems)
poems.append(author_poems_df)
poems_df = pd.concat(poems)
poems_df.head()
poems_df['lines'] = poems_df['lines'].apply(lambda x: ' \n '.join(x))
poems_df['lines'].head(2)
poems_df.to_csv('../data/poetrydb_copy.csv', index=False, encoding='utf-8')
authors_df.to_csv('../data/poetrydb_authors.csv', index=False, encoding='utf-8')
dialect+driver://username:password@host:port/database
engine = sqlalchemy.create_engine('postgresql://postgres:root@localhost:5432/postgres')
conn = engine.connect()
conn.execute('CREATE SCHEMA poetry')
create_author_table = '''
CREATE TABLE poetry.authors
(
id serial PRIMARY KEY,
author VARCHAR(255)
);'''
create_poems_table = '''
CREATE TABLE poetry.poems
(
id serial PRIMARY KEY,
author VARCHAR(255),
linecount INT,
lines TEXT,
title VARCHAR(510)
);
'''
conn.execute(create_author_table)
conn.execute(create_poems_table)
psql -h localhost -U postgres postgres
\copy poetry.authors(author) FROM '/Users/aaronschlegel/Dropbox/Projects/poetpy/data/poetrydb_authors.csv' DELIMITER ',' CSV HEADER;
\copy poetry.poems(author, linecount, lines, title) FROM '/Users/aaronschlegel/Dropbox/Projects/poetpy/data/poetrydb_copy.csv' DELIMITER ',' CSV HEADER;
poems_df.to_sql('poems_df', conn, schema='poetry', index_label='id')
authors_df.to_sql('authors_df', conn, schema='poetry', index_label='id')
SELECT author FROM poetry.authors LIMIT 3
conn.execute('SELECT author FROM poetry.authors LIMIT 3').fetchall()
pd.read_sql_query('SELECT authors FROM poetry.authors_df LIMIT 3', conn)
| 0.175397 | 0.987911 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.