code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
<a href="https://colab.research.google.com/github/PacktPublishing/Hands-On-Computer-Vision-with-PyTorch/blob/master/Chapter03/Varying_learning_rate_on_non_scaled_data.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
from torchvision import datasets
import torch
data_folder = '../data/FMNIST' # This can be any directory you want to
# download FMNIST to
fmnist = datasets.FashionMNIST(data_folder, download=True, train=True)
tr_images = fmnist.data
tr_targets = fmnist.targets
val_fmnist = datasets.FashionMNIST(data_folder, download=True, train=False)
val_images = val_fmnist.data
val_targets = val_fmnist.targets
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
from torch.utils.data import Dataset, DataLoader
import torch
import torch.nn as nn
device = 'cuda' if torch.cuda.is_available() else 'cpu'
```
### High Learning Rate
```
class FMNISTDataset(Dataset):
def __init__(self, x, y):
x = x.float()
x = x.view(-1,28*28)
self.x, self.y = x, y
def __getitem__(self, ix):
x, y = self.x[ix], self.y[ix]
return x.to(device), y.to(device)
def __len__(self):
return len(self.x)
from torch.optim import SGD, Adam
def get_model():
model = nn.Sequential(
nn.Linear(28 * 28, 1000),
nn.ReLU(),
nn.Linear(1000, 10)
).to(device)
loss_fn = nn.CrossEntropyLoss()
optimizer = Adam(model.parameters(), lr=1e-1)
return model, loss_fn, optimizer
def train_batch(x, y, model, opt, loss_fn):
model.train()
prediction = model(x)
batch_loss = loss_fn(prediction, y)
batch_loss.backward()
optimizer.step()
optimizer.zero_grad()
return batch_loss.item()
def accuracy(x, y, model):
model.eval()
# this is the same as @torch.no_grad
# at the top of function, only difference
# being, grad is not computed in the with scope
with torch.no_grad():
prediction = model(x)
max_values, argmaxes = prediction.max(-1)
is_correct = argmaxes == y
return is_correct.cpu().numpy().tolist()
def get_data():
train = FMNISTDataset(tr_images, tr_targets)
trn_dl = DataLoader(train, batch_size=32, shuffle=True)
val = FMNISTDataset(val_images, val_targets)
val_dl = DataLoader(val, batch_size=len(val_images), shuffle=False)
return trn_dl, val_dl
@torch.no_grad()
def val_loss(x, y, model):
prediction = model(x)
val_loss = loss_fn(prediction, y)
return val_loss.item()
trn_dl, val_dl = get_data()
model, loss_fn, optimizer = get_model()
train_losses, train_accuracies = [], []
val_losses, val_accuracies = [], []
for epoch in range(5):
print(epoch)
train_epoch_losses, train_epoch_accuracies = [], []
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
batch_loss = train_batch(x, y, model, optimizer, loss_fn)
train_epoch_losses.append(batch_loss)
train_epoch_loss = np.array(train_epoch_losses).mean()
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
is_correct = accuracy(x, y, model)
train_epoch_accuracies.extend(is_correct)
train_epoch_accuracy = np.mean(train_epoch_accuracies)
for ix, batch in enumerate(iter(val_dl)):
x, y = batch
val_is_correct = accuracy(x, y, model)
validation_loss = val_loss(x, y, model)
val_epoch_accuracy = np.mean(val_is_correct)
train_losses.append(train_epoch_loss)
train_accuracies.append(train_epoch_accuracy)
val_losses.append(validation_loss)
val_accuracies.append(val_epoch_accuracy)
epochs = np.arange(5)+1
import matplotlib.ticker as mtick
import matplotlib.pyplot as plt
import matplotlib.ticker as mticker
%matplotlib inline
plt.subplot(211)
plt.plot(epochs, train_losses, 'bo', label='Training loss')
plt.plot(epochs, val_losses, 'r', label='Validation loss')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation loss with 0.1 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.grid('off')
plt.show()
plt.subplot(212)
plt.plot(epochs, train_accuracies, 'bo', label='Training accuracy')
plt.plot(epochs, val_accuracies, 'r', label='Validation accuracy')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation accuracy with 0.1 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.gca().set_yticklabels(['{:.0f}%'.format(x*100) for x in plt.gca().get_yticks()])
plt.legend()
plt.grid('off')
plt.show()
for ix, par in enumerate(model.parameters()):
if(ix==0):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting input to hidden layer')
plt.show()
elif(ix ==1):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of hidden layer')
plt.show()
elif(ix==2):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting hidden to output layer')
plt.show()
elif(ix ==3):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of output layer')
plt.show()
```
### Medium learning rate
```
def get_model():
model = nn.Sequential(
nn.Linear(28 * 28, 1000),
nn.ReLU(),
nn.Linear(1000, 10)
).to(device)
loss_fn = nn.CrossEntropyLoss()
optimizer = Adam(model.parameters(), lr=1e-3)
return model, loss_fn, optimizer
trn_dl, val_dl = get_data()
model, loss_fn, optimizer = get_model()
train_losses, train_accuracies = [], []
val_losses, val_accuracies = [], []
for epoch in range(5):
print(epoch)
train_epoch_losses, train_epoch_accuracies = [], []
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
batch_loss = train_batch(x, y, model, optimizer, loss_fn)
train_epoch_losses.append(batch_loss)
train_epoch_loss = np.array(train_epoch_losses).mean()
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
is_correct = accuracy(x, y, model)
train_epoch_accuracies.extend(is_correct)
train_epoch_accuracy = np.mean(train_epoch_accuracies)
for ix, batch in enumerate(iter(val_dl)):
x, y = batch
val_is_correct = accuracy(x, y, model)
validation_loss = val_loss(x, y, model)
val_epoch_accuracy = np.mean(val_is_correct)
train_losses.append(train_epoch_loss)
train_accuracies.append(train_epoch_accuracy)
val_losses.append(validation_loss)
val_accuracies.append(val_epoch_accuracy)
epochs = np.arange(5)+1
import matplotlib.ticker as mtick
import matplotlib.pyplot as plt
import matplotlib.ticker as mticker
%matplotlib inline
plt.subplot(211)
plt.plot(epochs, train_losses, 'bo', label='Training loss')
plt.plot(epochs, val_losses, 'r', label='Validation loss')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation loss with 0.001 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.grid('off')
plt.show()
plt.subplot(212)
plt.plot(epochs, train_accuracies, 'bo', label='Training accuracy')
plt.plot(epochs, val_accuracies, 'r', label='Validation accuracy')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation accuracy with 0.001 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.gca().set_yticklabels(['{:.0f}%'.format(x*100) for x in plt.gca().get_yticks()])
plt.legend()
plt.grid('off')
plt.show()
for ix, par in enumerate(model.parameters()):
if(ix==0):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting input to hidden layer')
plt.show()
elif(ix ==1):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of hidden layer')
plt.show()
elif(ix==2):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting hidden to output layer')
plt.show()
elif(ix ==3):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of output layer')
plt.show()
```
### Low learning rate
```
def get_model():
model = nn.Sequential(
nn.Linear(28 * 28, 1000),
nn.ReLU(),
nn.Linear(1000, 10)
).to(device)
loss_fn = nn.CrossEntropyLoss()
optimizer = Adam(model.parameters(), lr=1e-5)
return model, loss_fn, optimizer
trn_dl, val_dl = get_data()
model, loss_fn, optimizer = get_model()
train_losses, train_accuracies = [], []
val_losses, val_accuracies = [], []
for epoch in range(5):
print(epoch)
train_epoch_losses, train_epoch_accuracies = [], []
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
batch_loss = train_batch(x, y, model, optimizer, loss_fn)
train_epoch_losses.append(batch_loss)
train_epoch_loss = np.array(train_epoch_losses).mean()
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
is_correct = accuracy(x, y, model)
train_epoch_accuracies.extend(is_correct)
train_epoch_accuracy = np.mean(train_epoch_accuracies)
for ix, batch in enumerate(iter(val_dl)):
x, y = batch
val_is_correct = accuracy(x, y, model)
validation_loss = val_loss(x, y, model)
val_epoch_accuracy = np.mean(val_is_correct)
train_losses.append(train_epoch_loss)
train_accuracies.append(train_epoch_accuracy)
val_losses.append(validation_loss)
val_accuracies.append(val_epoch_accuracy)
epochs = np.arange(5)+1
import matplotlib.ticker as mtick
import matplotlib.pyplot as plt
import matplotlib.ticker as mticker
%matplotlib inline
plt.subplot(211)
plt.plot(epochs, train_losses, 'bo', label='Training loss')
plt.plot(epochs, val_losses, 'r', label='Validation loss')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation loss with 0.00001 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.grid('off')
plt.show()
plt.subplot(212)
plt.plot(epochs, train_accuracies, 'bo', label='Training accuracy')
plt.plot(epochs, val_accuracies, 'r', label='Validation accuracy')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation accuracy with 0.00001 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.gca().set_yticklabels(['{:.0f}%'.format(x*100) for x in plt.gca().get_yticks()])
plt.legend()
plt.grid('off')
plt.show()
for ix, par in enumerate(model.parameters()):
if(ix==0):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting input to hidden layer')
plt.show()
elif(ix ==1):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of hidden layer')
plt.show()
elif(ix==2):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting hidden to output layer')
plt.show()
elif(ix ==3):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of output layer')
plt.show()
```
|
github_jupyter
|
from torchvision import datasets
import torch
data_folder = '../data/FMNIST' # This can be any directory you want to
# download FMNIST to
fmnist = datasets.FashionMNIST(data_folder, download=True, train=True)
tr_images = fmnist.data
tr_targets = fmnist.targets
val_fmnist = datasets.FashionMNIST(data_folder, download=True, train=False)
val_images = val_fmnist.data
val_targets = val_fmnist.targets
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
from torch.utils.data import Dataset, DataLoader
import torch
import torch.nn as nn
device = 'cuda' if torch.cuda.is_available() else 'cpu'
class FMNISTDataset(Dataset):
def __init__(self, x, y):
x = x.float()
x = x.view(-1,28*28)
self.x, self.y = x, y
def __getitem__(self, ix):
x, y = self.x[ix], self.y[ix]
return x.to(device), y.to(device)
def __len__(self):
return len(self.x)
from torch.optim import SGD, Adam
def get_model():
model = nn.Sequential(
nn.Linear(28 * 28, 1000),
nn.ReLU(),
nn.Linear(1000, 10)
).to(device)
loss_fn = nn.CrossEntropyLoss()
optimizer = Adam(model.parameters(), lr=1e-1)
return model, loss_fn, optimizer
def train_batch(x, y, model, opt, loss_fn):
model.train()
prediction = model(x)
batch_loss = loss_fn(prediction, y)
batch_loss.backward()
optimizer.step()
optimizer.zero_grad()
return batch_loss.item()
def accuracy(x, y, model):
model.eval()
# this is the same as @torch.no_grad
# at the top of function, only difference
# being, grad is not computed in the with scope
with torch.no_grad():
prediction = model(x)
max_values, argmaxes = prediction.max(-1)
is_correct = argmaxes == y
return is_correct.cpu().numpy().tolist()
def get_data():
train = FMNISTDataset(tr_images, tr_targets)
trn_dl = DataLoader(train, batch_size=32, shuffle=True)
val = FMNISTDataset(val_images, val_targets)
val_dl = DataLoader(val, batch_size=len(val_images), shuffle=False)
return trn_dl, val_dl
@torch.no_grad()
def val_loss(x, y, model):
prediction = model(x)
val_loss = loss_fn(prediction, y)
return val_loss.item()
trn_dl, val_dl = get_data()
model, loss_fn, optimizer = get_model()
train_losses, train_accuracies = [], []
val_losses, val_accuracies = [], []
for epoch in range(5):
print(epoch)
train_epoch_losses, train_epoch_accuracies = [], []
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
batch_loss = train_batch(x, y, model, optimizer, loss_fn)
train_epoch_losses.append(batch_loss)
train_epoch_loss = np.array(train_epoch_losses).mean()
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
is_correct = accuracy(x, y, model)
train_epoch_accuracies.extend(is_correct)
train_epoch_accuracy = np.mean(train_epoch_accuracies)
for ix, batch in enumerate(iter(val_dl)):
x, y = batch
val_is_correct = accuracy(x, y, model)
validation_loss = val_loss(x, y, model)
val_epoch_accuracy = np.mean(val_is_correct)
train_losses.append(train_epoch_loss)
train_accuracies.append(train_epoch_accuracy)
val_losses.append(validation_loss)
val_accuracies.append(val_epoch_accuracy)
epochs = np.arange(5)+1
import matplotlib.ticker as mtick
import matplotlib.pyplot as plt
import matplotlib.ticker as mticker
%matplotlib inline
plt.subplot(211)
plt.plot(epochs, train_losses, 'bo', label='Training loss')
plt.plot(epochs, val_losses, 'r', label='Validation loss')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation loss with 0.1 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.grid('off')
plt.show()
plt.subplot(212)
plt.plot(epochs, train_accuracies, 'bo', label='Training accuracy')
plt.plot(epochs, val_accuracies, 'r', label='Validation accuracy')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation accuracy with 0.1 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.gca().set_yticklabels(['{:.0f}%'.format(x*100) for x in plt.gca().get_yticks()])
plt.legend()
plt.grid('off')
plt.show()
for ix, par in enumerate(model.parameters()):
if(ix==0):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting input to hidden layer')
plt.show()
elif(ix ==1):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of hidden layer')
plt.show()
elif(ix==2):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting hidden to output layer')
plt.show()
elif(ix ==3):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of output layer')
plt.show()
def get_model():
model = nn.Sequential(
nn.Linear(28 * 28, 1000),
nn.ReLU(),
nn.Linear(1000, 10)
).to(device)
loss_fn = nn.CrossEntropyLoss()
optimizer = Adam(model.parameters(), lr=1e-3)
return model, loss_fn, optimizer
trn_dl, val_dl = get_data()
model, loss_fn, optimizer = get_model()
train_losses, train_accuracies = [], []
val_losses, val_accuracies = [], []
for epoch in range(5):
print(epoch)
train_epoch_losses, train_epoch_accuracies = [], []
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
batch_loss = train_batch(x, y, model, optimizer, loss_fn)
train_epoch_losses.append(batch_loss)
train_epoch_loss = np.array(train_epoch_losses).mean()
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
is_correct = accuracy(x, y, model)
train_epoch_accuracies.extend(is_correct)
train_epoch_accuracy = np.mean(train_epoch_accuracies)
for ix, batch in enumerate(iter(val_dl)):
x, y = batch
val_is_correct = accuracy(x, y, model)
validation_loss = val_loss(x, y, model)
val_epoch_accuracy = np.mean(val_is_correct)
train_losses.append(train_epoch_loss)
train_accuracies.append(train_epoch_accuracy)
val_losses.append(validation_loss)
val_accuracies.append(val_epoch_accuracy)
epochs = np.arange(5)+1
import matplotlib.ticker as mtick
import matplotlib.pyplot as plt
import matplotlib.ticker as mticker
%matplotlib inline
plt.subplot(211)
plt.plot(epochs, train_losses, 'bo', label='Training loss')
plt.plot(epochs, val_losses, 'r', label='Validation loss')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation loss with 0.001 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.grid('off')
plt.show()
plt.subplot(212)
plt.plot(epochs, train_accuracies, 'bo', label='Training accuracy')
plt.plot(epochs, val_accuracies, 'r', label='Validation accuracy')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation accuracy with 0.001 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.gca().set_yticklabels(['{:.0f}%'.format(x*100) for x in plt.gca().get_yticks()])
plt.legend()
plt.grid('off')
plt.show()
for ix, par in enumerate(model.parameters()):
if(ix==0):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting input to hidden layer')
plt.show()
elif(ix ==1):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of hidden layer')
plt.show()
elif(ix==2):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting hidden to output layer')
plt.show()
elif(ix ==3):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of output layer')
plt.show()
def get_model():
model = nn.Sequential(
nn.Linear(28 * 28, 1000),
nn.ReLU(),
nn.Linear(1000, 10)
).to(device)
loss_fn = nn.CrossEntropyLoss()
optimizer = Adam(model.parameters(), lr=1e-5)
return model, loss_fn, optimizer
trn_dl, val_dl = get_data()
model, loss_fn, optimizer = get_model()
train_losses, train_accuracies = [], []
val_losses, val_accuracies = [], []
for epoch in range(5):
print(epoch)
train_epoch_losses, train_epoch_accuracies = [], []
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
batch_loss = train_batch(x, y, model, optimizer, loss_fn)
train_epoch_losses.append(batch_loss)
train_epoch_loss = np.array(train_epoch_losses).mean()
for ix, batch in enumerate(iter(trn_dl)):
x, y = batch
is_correct = accuracy(x, y, model)
train_epoch_accuracies.extend(is_correct)
train_epoch_accuracy = np.mean(train_epoch_accuracies)
for ix, batch in enumerate(iter(val_dl)):
x, y = batch
val_is_correct = accuracy(x, y, model)
validation_loss = val_loss(x, y, model)
val_epoch_accuracy = np.mean(val_is_correct)
train_losses.append(train_epoch_loss)
train_accuracies.append(train_epoch_accuracy)
val_losses.append(validation_loss)
val_accuracies.append(val_epoch_accuracy)
epochs = np.arange(5)+1
import matplotlib.ticker as mtick
import matplotlib.pyplot as plt
import matplotlib.ticker as mticker
%matplotlib inline
plt.subplot(211)
plt.plot(epochs, train_losses, 'bo', label='Training loss')
plt.plot(epochs, val_losses, 'r', label='Validation loss')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation loss with 0.00001 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.grid('off')
plt.show()
plt.subplot(212)
plt.plot(epochs, train_accuracies, 'bo', label='Training accuracy')
plt.plot(epochs, val_accuracies, 'r', label='Validation accuracy')
plt.gca().xaxis.set_major_locator(mticker.MultipleLocator(1))
plt.title('Training and validation accuracy with 0.00001 learning rate')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.gca().set_yticklabels(['{:.0f}%'.format(x*100) for x in plt.gca().get_yticks()])
plt.legend()
plt.grid('off')
plt.show()
for ix, par in enumerate(model.parameters()):
if(ix==0):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting input to hidden layer')
plt.show()
elif(ix ==1):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of hidden layer')
plt.show()
elif(ix==2):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of weights conencting hidden to output layer')
plt.show()
elif(ix ==3):
plt.hist(par.cpu().detach().numpy().flatten())
plt.title('Distribution of biases of output layer')
plt.show()
| 0.886463 | 0.983279 |
```
import tensorflow as tf
print(tf.__version__)
# !pip install -q tensorflow-datasets
import tensorflow_datasets as tfds
imdb, info = tfds.load("imdb_reviews", with_info=True, as_supervised=True)
import numpy as np
train_data, test_data = imdb['train'], imdb['test']
training_sentences = []
training_labels = []
testing_sentences = []
testing_labels = []
# str(s.tonumpy()) is needed in Python3 instead of just s.numpy()
for s,l in train_data:
training_sentences.append(str(s.numpy()))
training_labels.append(l.numpy())
for s,l in test_data:
testing_sentences.append(str(s.numpy()))
testing_labels.append(l.numpy())
training_labels_final = np.array(training_labels)
testing_labels_final = np.array(testing_labels)
vocab_size = 10000
embedding_dim = 16
max_length = 120
trunc_type='post'
oov_tok = "<OOV>"
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
tokenizer = Tokenizer(num_words = vocab_size, oov_token=oov_tok)
tokenizer.fit_on_texts(training_sentences)
word_index = tokenizer.word_index
sequences = tokenizer.texts_to_sequences(training_sentences)
padded = pad_sequences(sequences,maxlen=max_length, truncating=trunc_type)
testing_sequences = tokenizer.texts_to_sequences(testing_sentences)
testing_padded = pad_sequences(testing_sequences,maxlen=max_length)
reverse_word_index = dict([(value, key) for (key, value) in word_index.items()])
def decode_review(text):
return ' '.join([reverse_word_index.get(i, '?') for i in text])
print(decode_review(padded[3]))
print(training_sentences[3])
model = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(6, activation='relu'),
tf.keras.layers.Dense(1, activation='sigmoid')
])
model.compile(loss='binary_crossentropy',optimizer='adam',metrics=['accuracy'])
model.summary()
num_epochs = 10
model.fit(padded, training_labels_final, epochs=num_epochs, validation_data=(testing_padded, testing_labels_final))
e = model.layers[0]
weights = e.get_weights()[0]
print(weights.shape) # shape: (vocab_size, embedding_dim)
import io
out_v = io.open('vecs.tsv', 'w', encoding='utf-8')
out_m = io.open('meta.tsv', 'w', encoding='utf-8')
for word_num in range(1, vocab_size):
word = reverse_word_index[word_num]
embeddings = weights[word_num]
out_m.write(word + "\n")
out_v.write('\t'.join([str(x) for x in embeddings]) + "\n")
out_v.close()
out_m.close()
try:
from google.colab import files
except ImportError:
pass
else:
files.download('vecs.tsv')
files.download('meta.tsv')
sentence = "I really think this is amazing. honest."
sequence = tokenizer.texts_to_sequences(sentence)
print(sequence)
```
|
github_jupyter
|
import tensorflow as tf
print(tf.__version__)
# !pip install -q tensorflow-datasets
import tensorflow_datasets as tfds
imdb, info = tfds.load("imdb_reviews", with_info=True, as_supervised=True)
import numpy as np
train_data, test_data = imdb['train'], imdb['test']
training_sentences = []
training_labels = []
testing_sentences = []
testing_labels = []
# str(s.tonumpy()) is needed in Python3 instead of just s.numpy()
for s,l in train_data:
training_sentences.append(str(s.numpy()))
training_labels.append(l.numpy())
for s,l in test_data:
testing_sentences.append(str(s.numpy()))
testing_labels.append(l.numpy())
training_labels_final = np.array(training_labels)
testing_labels_final = np.array(testing_labels)
vocab_size = 10000
embedding_dim = 16
max_length = 120
trunc_type='post'
oov_tok = "<OOV>"
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
tokenizer = Tokenizer(num_words = vocab_size, oov_token=oov_tok)
tokenizer.fit_on_texts(training_sentences)
word_index = tokenizer.word_index
sequences = tokenizer.texts_to_sequences(training_sentences)
padded = pad_sequences(sequences,maxlen=max_length, truncating=trunc_type)
testing_sequences = tokenizer.texts_to_sequences(testing_sentences)
testing_padded = pad_sequences(testing_sequences,maxlen=max_length)
reverse_word_index = dict([(value, key) for (key, value) in word_index.items()])
def decode_review(text):
return ' '.join([reverse_word_index.get(i, '?') for i in text])
print(decode_review(padded[3]))
print(training_sentences[3])
model = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(6, activation='relu'),
tf.keras.layers.Dense(1, activation='sigmoid')
])
model.compile(loss='binary_crossentropy',optimizer='adam',metrics=['accuracy'])
model.summary()
num_epochs = 10
model.fit(padded, training_labels_final, epochs=num_epochs, validation_data=(testing_padded, testing_labels_final))
e = model.layers[0]
weights = e.get_weights()[0]
print(weights.shape) # shape: (vocab_size, embedding_dim)
import io
out_v = io.open('vecs.tsv', 'w', encoding='utf-8')
out_m = io.open('meta.tsv', 'w', encoding='utf-8')
for word_num in range(1, vocab_size):
word = reverse_word_index[word_num]
embeddings = weights[word_num]
out_m.write(word + "\n")
out_v.write('\t'.join([str(x) for x in embeddings]) + "\n")
out_v.close()
out_m.close()
try:
from google.colab import files
except ImportError:
pass
else:
files.download('vecs.tsv')
files.download('meta.tsv')
sentence = "I really think this is amazing. honest."
sequence = tokenizer.texts_to_sequences(sentence)
print(sequence)
| 0.718792 | 0.492066 |
# Tutorial - Unobserved Heterogeneity and Finite Mixture Models
Unobserved heterogeneity is a concern in every econometric application. Keane and Wolpin (1997) face the problem that individuals at the age of sixteen report varying years of schooling. Neglecting the issue of measurement error, it is unlikely that the differences in initial schooling are caused by exogenous factors. Instead, the schooling decision is affected by a variety of endogenous factors such as parental investement, school and teacher quality, intrinsic motivation, and ability. Without correction, estimation methods fail to recover the true parameters.
One solution would be to extend the model and incorporate the whole human capital investement process up to the age where initial schooling was zero. Although such a model would be extremely interesting, it is also almost infeasible to model that many factors in terms of modeling, computation and data.
Another solution is to employ individual fixed-effects. Then, the state space comprises a dimension with has the same number of unique values as there are individuals in the sample. Thus, you have to compute the decision rules for every individual for the whole state space separately which is computationally infeasible.
Keane and Wolpin (1997) resort to model unobserved heterogeneity with a finite mixture. A mixture model can be used to model the presence of subpopulations (types) in the general population without requiring the observed data to identify the affiliation to a group. In contrast to fixed-effects, the number of subpopulations is much lower than the number of individuals. There is also no fixed and unique assignment to one subpopulation, but relations are defined by a probability mass function.
Each type has a preference for a particular choice which is modeled by a constant in the utility functions. For working alternatives, $w$, the constant is in the log wage equation whereas for non-working alternatives, $n$, it is in the nonpecuniary reward. Note that ``respy`` allows for type-specific effects in every utility component. Keane and Wolpin (1997) call it endowment with the symbol $e_{ak}$ for type $k$ and alternative $a$.
$$\begin{align}
\log(W(s_t, a_t)) = x^w\beta^w + e_{ak} + \epsilon_{at}\\
N^n(s_t, a_t) = x^n\beta^n + e_{ak} + \epsilon_{at}
\end{align}$$
To estimate model parameters with maximum likelihood, the likelihood contribution for one individual is defined as the joint probability of choices and wages accumulated over time.
$$
P(\{a_t\}^T_{t=0} \mid s^-_t, e_{ak}, W_t) =
\prod^T_{t = 0} p(a_t, \mid s^-_t, e_{ak}, W_t)
$$
We can weight the contribution for type $k$ with the probability for being the same type to get the unconditioned likelihood contribution of an individual.
$$
P(\{a_t, W_t\}^T_{t=0}) = \sum^K_{k=1} \pi_k
P(\{a_t\}^T_{t=0} \mid s^-_t, e_{ak}, W_t)
$$
To avoid misspecification of the likelihood, $\pi_k$ must be a function of all individual characteristics which are determined before individuals enter the model horizon and are not the result of exogenous factors. The type-specific probability $\pi_k = f(x^\pi \beta^\pi)$ is calculated with softmax function based on a vector of covariates $x^\pi$ and a matrix of coefficients $\beta^\pi$ for each type-covariate combination.
$$
\pi_k = f(x^\pi \beta^\pi_k) =
\frac{\exp{\{x^\pi \beta^\pi_k\}}}{\sum^K_{k=1} \exp \{x^\pi \beta^\pi_k\}}
$$
To implement a finite mixture, we have to express $e_{ak}$ and $\beta^\pi$ in the parameters. As an example, we start with the basic Robinson Crusoe Economy.
```
import io
import pandas as pd
import respy as rp
params, options = rp.get_example_model(
"robinson_crusoe_basic", with_data=False
)
params
```
We extend the model by allowing for different periods of experience in fishing at $t = 0$. Robinsons starts with zero, one or two experience in fishing because of different tastes for fishing.
```
initial_exp_fishing = pd.read_csv(
io.StringIO(
"""
category,name,value
initial_exp_fishing_0,probability,0.33
initial_exp_fishing_1,probability,0.33
initial_exp_fishing_2,probability,0.34
"""
),
index_col=["category", "name"],
)
initial_exp_fishing
```
In the next step, we add type-specific endowment effects $e_{ak}$. We assume that there exist three types and the additional utility is increasing from the first to the third type. For computational simplicity, the benefit of the first type is normalized to zero such that all other types are in relation to the first.
```
endowments = pd.read_csv(
io.StringIO(
"""
category,name,value
wage_fishing,type_1,0.2
wage_fishing,type_2,0.4
"""
),
index_col=["category", "name"],
)
endowments
```
We assume no effect for choosing the hammock.
At last, we need to specify the probability mass function which relates individuals to types. We simply assume that initial experience is positively correlated with a stronger taste for fishing. For a comprehensive overview on how to specify distributions with multinomial coefficients, see the guide on the [initial conditions](tutorial-initial-conditions.ipynb). Note that, the distribution is also only specified for type 1 and 2 and the coefficients for type 1 are left out for a parsimonuous representation. You cannot use probabilities as type assignment cannot be completely random. The following example is designed to specify a certain distribution and recover the pattern in the data. In reality, the distribution of unobservables is unknown.
First, we define that Robinsons without prior experience are of type 0. Thus, we make the coefficients for type 1 and 2 extremely small. Robinsons with one prior experience are of type 1 with probability 0.66 and type 2 with 0.33. For two periods of experience for fishing, the share of type 1 individuals is 0.33 and of type 2 is 0.66. The coefficients for type 1 and 2 are simply the log of the probabilities.
At last, we add a sufficiently large integer to all coefficients. The coefficient of type 0 is implicitly set to zero, so the distribution samples type 0 individuals for one or two experience in fishing. By shifting the parameters with a positive value, this is prevented. At the same time, the softmax function is shift-invariant and the relation of type 1 and type 2 shares is preserved.
```
type_probabilities = pd.read_csv(
io.StringIO(
"""
category,name,value
type_1,initial_exp_fishing_0,-100
type_1,initial_exp_fishing_1,-0.4055
type_1,initial_exp_fishing_2,-1.0986
type_2,initial_exp_fishing_0,-100
type_2,initial_exp_fishing_1,-1.0986
type_2,initial_exp_fishing_2,-0.4055
"""
),
index_col=["category", "name"],
)
type_probabilities += 10
type_probabilities
```
The covariates used for the probabilities are defined below.
```
type_covariates = {
"initial_exp_fishing_0": "exp_fishing == 0",
"initial_exp_fishing_1": "exp_fishing == 1",
"initial_exp_fishing_2": "exp_fishing == 2",
}
type_covariates
```
In the next step, we put all pieces together to get the complete model specification.
```
params = params.append([initial_exp_fishing, endowments, type_probabilities])
params
options["covariates"] = {**options["covariates"], **type_covariates}
options["simulation_agents"] = 10_000
options
```
Let us simulate a dataset to see whether the distribution of types can be recovered from the data.
```
simulate = rp.get_simulate_func(params, options)
df = simulate(params)
df.query("Period == 0").groupby("Experience_Fishing").Type.value_counts(
normalize="rows"
).unstack().fillna(0)
```
We also know that type 1 and 2 experience a higher utility for choosing fishing. Here are the choice probabilities for each type.
```
df.groupby("Type").Choice.value_counts(normalize=True).unstack()
```
|
github_jupyter
|
import io
import pandas as pd
import respy as rp
params, options = rp.get_example_model(
"robinson_crusoe_basic", with_data=False
)
params
initial_exp_fishing = pd.read_csv(
io.StringIO(
"""
category,name,value
initial_exp_fishing_0,probability,0.33
initial_exp_fishing_1,probability,0.33
initial_exp_fishing_2,probability,0.34
"""
),
index_col=["category", "name"],
)
initial_exp_fishing
endowments = pd.read_csv(
io.StringIO(
"""
category,name,value
wage_fishing,type_1,0.2
wage_fishing,type_2,0.4
"""
),
index_col=["category", "name"],
)
endowments
type_probabilities = pd.read_csv(
io.StringIO(
"""
category,name,value
type_1,initial_exp_fishing_0,-100
type_1,initial_exp_fishing_1,-0.4055
type_1,initial_exp_fishing_2,-1.0986
type_2,initial_exp_fishing_0,-100
type_2,initial_exp_fishing_1,-1.0986
type_2,initial_exp_fishing_2,-0.4055
"""
),
index_col=["category", "name"],
)
type_probabilities += 10
type_probabilities
type_covariates = {
"initial_exp_fishing_0": "exp_fishing == 0",
"initial_exp_fishing_1": "exp_fishing == 1",
"initial_exp_fishing_2": "exp_fishing == 2",
}
type_covariates
params = params.append([initial_exp_fishing, endowments, type_probabilities])
params
options["covariates"] = {**options["covariates"], **type_covariates}
options["simulation_agents"] = 10_000
options
simulate = rp.get_simulate_func(params, options)
df = simulate(params)
df.query("Period == 0").groupby("Experience_Fishing").Type.value_counts(
normalize="rows"
).unstack().fillna(0)
df.groupby("Type").Choice.value_counts(normalize=True).unstack()
| 0.377426 | 0.992108 |
# Introduction to machine learning, neural networks and deep learning
## Objectives
- Understand the fundamental goals of machine learning and a bit of the field's history
- Gain familiarity with the mechanics of a neural network, convolutional neural networks, and the U-Net architecture in particular
- Discuss considerations for choosing a deep learning architecture for a particular problem
Below is a video recording of the oral lecture associated with this lesson and the following. It was given by Lilly Thomas, ML Engineer at Development Seed.
```
from IPython.display import YouTubeVideo
def display_yotube_video(url, **kwargs):
id_ = url.split("=")[-1]
return YouTubeVideo(id_, **kwargs)
display_yotube_video("https://www.youtube.com/watch?v=-C3niPVd-zU", width=800, height=600)
```
### What is Machine Learning?
Machine learning (ML) is a subset of artificial intelligence (AI), which in broad terms, is defined as the ability of a machine to simulate intelligent human behavior.
:::{figure-md} ai_ml_dl-fig
<img src="https://human-centered.ai/wordpress/wp-content/uploads/2017/11/Deep-Learning-subset-of-Machine-Learning-subset-of-Artificial-Intelligence.jpg" width="450px">
[AI, ML, DL](https://www.frwebs.top/products.aspx?cname=difference+between+ml+dl+and+ai&cid=7).
:::
Compared to traditional programming, ML offers:
1) time savings on behalf of the human programmer,
2) time savings on behalf of a human manual interpreter,
3) reduction of human error,
4) scalable decision making
ML requires good quality data, and a lot of it, to recognize key patterns and features.
Humans still have a role in this process, by way of supplying the model with data and choosing algorithms and parameters.
There are several subcategories of machine learning:
1) **Supervised machine learning** involves training a model with labeled data sets that explicitly give examples of predictive features and their target attribute(s).
2) **Unsupervised machine learning** involves tasking a model to search for patterns in data without the guidance of labels.
```{important}
There are also some problems where machine learning is uniquely equipped to learn insights and make decisions when a human might not, such as drawing relationships from combined spectral indices in a complex terrain.
```
### What are Neural Networks?
Artificial neural networks (ANNs) are a specific, biologically-inspired class of machine learning algorithms. They are modeled after the structure and function of the human brain.
:::{figure-md} neuron-fig
<img src="https://github.com/developmentseed/tensorflow-eo-training/blob/main/ds_book/docs/images/neuron-structure.jpg?raw=1" width="450px">
Biological neuron (from [https://training.seer.cancer.gov/anatomy/nervous/tissue.html](https://training.seer.cancer.gov/anatomy/nervous/tissue.html)).
:::
ANNs are essentially programs that make decisions by weighing the evidence and responding to feedback. By varying the input data, types of parameters and their values, we can get different models of decision-making.
:::{figure-md} neuralnet_basic-fig
<img src="https://miro.medium.com/max/1100/1*x6KWjKTOBhUYL0MRX4M3oQ.png" width="450px">
Basic neural network from [https://towardsdatascience.com/machine-learning-for-beginners-an-introduction-to-neural-networks-d49f22d238f9](https://towardsdatascience.com/machine-learning-for-beginners-an-introduction-to-neural-networks-d49f22d238f9).
:::
In network architectures, neurons are grouped in layers, with synapses traversing the interstitial space between neurons in one layer and the next.
#### What are Convolutional Neural Networks?
A Convolutional Neural Network (ConvNet/CNN) is a form of deep learning inspired by the organization of the human visual cortex, in which individual neurons respond to stimuli within a constrained region of the visual field known as the receptive field. Several receptive fields overlap to account for the entire visual area.
In artificial CNNs, an input matrix such as an image is given importance per various aspects and objects in the image through a moving, convoling receptive field. Very little pre-processing is required for CNNs relative to other classification methods as the need for upfront feature-engineering is removed. Rather, CNNs learn the correct filters and consequent features on their own, provided enough training time and examples.
:::{figure-md} convolution-fig
<img src="https://miro.medium.com/max/1400/1*Fw-ehcNBR9byHtho-Rxbtw.gif" width="450px">
Convolution of a kernal over an input matrix from [https://towardsdatascience.com/intuitively-understanding-convolutions-for-deep-learning-1f6f42faee1](https://towardsdatascience.com/intuitively-understanding-convolutions-for-deep-learning-1f6f42faee1).
:::
#### What is a kernel/filter?
A kernel is matrix smaller than the input. It acts as a receptive field that moves over the input matrix from left to right and top to bottom and filters for features in the image.
#### What is stride?
Stride refers to the number of pixels that the kernel shifts at each step in its navigation of the input matrix.
#### What is a convolution operation?
The convolution operation is the combination of two functions to produce a third function as a result. In effect, it is a merging of two sets of information, the kernel and the input matrix.
:::{figure-md} convolution-arithmetic-fig
<img src="https://theano-pymc.readthedocs.io/en/latest/_images/numerical_no_padding_no_strides.gif" width="450px">
Convolution of a kernal over an input matrix from [https://theano-pymc.readthedocs.io/en/latest/tutorial/conv_arithmetic.html](https://theano-pymc.readthedocs.io/en/latest/tutorial/conv_arithmetic.html).
:::
#### Convolution operation using 3D filter
An input image is often represented as a 3D matrix with a dimension for width (pixels), height (pixels), and depth (channels). In the case of an optical image with red, green and blue channels, the kernel/filter matrix is shaped with the same channel depth as the input and the weighted sum of dot products is computed across all 3 dimensions.
#### What is padding?
After a convolution operation, the feature map is by default smaller than the original input matrix.
:::{figure-md} multi_layer_CNN-fig
<img src="https://www.researchgate.net/profile/Sheraz-Khan-14/publication/321586653/figure/fig4/AS:568546847014912@1512563539828/The-LeNet-5-Architecture-a-convolutional-neural-network.png" width="450px">
[Progressive downsizing of feature maps in a multi-layer CNN](https://www.researchgate.net/figure/The-LeNet-5-Architecture-a-convolutional-neural-network_fig4_321586653).
:::
To maintain the same spatial dimensions between input matrix and output feature map, we may pad the input matrix with a border of zeroes or ones. There are two types of padding:
1. Same padding: a border of zeroes or ones is added to match the input/output dimensions
2. Valid padding: no border is added and the output dimensions are not matched to the input
:::{figure-md} padding-fig
<img src="https://miro.medium.com/max/666/1*noYcUAa_P8nRilg3Lt_nuA.png" width="450px">
[Padding an input matrix with zeroes](https://ayeshmanthaperera.medium.com/what-is-padding-in-cnns-71b21fb0dd7).
:::
### What is Deep Learning?
Deep learning is defined by neural networks with depth, i.e. many layers and connections. The reason for why deep learning is so highly performant lies in the degree of abstraction made possible by feature extraction across so many layers in which each neuron, or processing unit, is interacting with input from neurons in previous layers and making decisions accordingly. The deepest layers of a network once trained can be capable inferring highly abstract concepts, such as what differentiates a school from a house in satellite imagery.
```{admonition} **Cost of deep learning**
Deep learning requires a lot of data to learn from and usually a significant amount of computing power, so it can be expensive depending on the scope of the problem.
```
#### Training and Testing Data
The dataset (e.g. all images and their labels) are split into training, validation and testing sets. A common ratio is 70:20:10 percent, train:validation:test. If randomly split, it is important to check that all class labels exist in all sets and are well represented.
```{important} Why do we need validation and test data? Are they redundant?
We need separate test data to evaluate the performance of the model because the validation data is used during training to measure error and therefore inform updates to the model parameters. Therefore, validation data is not unbiased to the model. A need for new, wholly unseen data to test with is required.
```
#### Forward and backward propagation, hyper-parameters, and learnable parameters
Neural networks train in cycles, where the input data passes through the network, a relationship between input data and target values is learned, a prediction is made, the prediction value is measured for error relative to its true value, and the errors are used to inform updates to parameters in the network, feeding into the next cycle of learning and prediction using the updated information. This happens through a two-step process called forward propagation and back propagation, in which the first part is used to gather knowledge and the second part is used to correct errors in the model’s knowledge.
:::{figure-md} forward_backprop-fig
<img src="https://thumbs.gfycat.com/BitesizedWeeBlacklemur-max-1mb.gif" width="450px">
[Forward and back propagation](https://gfycat.com/gifs/search/backpropagation).
:::
The **activation function** decides whether or not the output from one neuron is useful or not based on a threshold value, and therefore, whether it will be carried from one layer to the next.
**Weights** control the signal (or the strength of the connection) between two neurons in two consecutive layers.
**Biases** are values which help determine whether or not the activation output from a neuron is going to be passed forward through the network.
In a neural network, neurons in one layer are connected to neurons in the next layer. As information passes from one neuron to the next, the information is conditioned by the weight of the synapse and is subjected to a bias. The weights and biases determine if the information passes further beyond the current neuron.
:::{figure-md} activation-fig
<img src="https://cdn-images-1.medium.com/max/651/1*UA30b0mJUPYoPvN8yJr2iQ.jpeg" width="450px">
[Weights, bias, activation](https://laptrinhx.com/statistics-is-freaking-hard-wtf-is-activation-function-207913705/).
:::
During training, the weights and biases are learned and updated using the training and validation dataset to fit the data and reduce error of prediction values relative to target values.
```{important}
- **Activation function**: decides whether or not the output from one neuron is useful or not
- **Weights**: control the signal between neurons in consecutive layers
- **Biases**: a threshold value that determines the activation of each neuron
- Weights and biases are the learnable parameters of a deep learning model
```
The **learning rate** controls how much we want the model to change in response to the estimated error after each training cycle
:::{figure-md} loss_curve-fig
<img src="https://d1zx6djv3kb1v7.cloudfront.net/wp-content/media/2019/09/Neural-network-32-i2tutorials.png" width="450px">
[Local vs. global minimum (the optimal point to reach)](https://www.i2tutorials.com/what-are-local-minima-and-global-minima-in-gradient-descent/).
:::
The **batch size** determines the portion of our training dataset that can be fed to the model during each cycle. Stated otherwise, batch size controls the number of training samples to work through before the model’s internal parameters are updated.
:::{figure-md} batch_epoch-fig
<img src="https://www.baeldung.com/wp-content/uploads/sites/4/2020/12/epoch-batch-size.png" width="250px">
[Modulating batch size detetmines how many iterations are within one epoch](https://www.baeldung.com/cs/epoch-neural-networks).
:::
An **epoch** is defined as the point when all training samples, aka the entire dataset, has passed through the neural network once. The number of epochs controls how many times the entire dataset is cycled through and analyzed by the neural network. Related, but not necessarily as a parameter is an **iteration**, which is the pass of one batch through the network. If the batch size is smaller than the size of the whole dataset, then there are multiple iterations in one epoch.
The **optimization function** is really important. It’s what we use to change the attributes of your neural network such as weights and biases in order to reduce the losses. The goal of an optimization function is to minimize the error produced by the model.
The **loss function**, also known as the cost function, measures how much the model needs to improve based on the prediction errors relative to the true values during training.
:::{figure-md} loss_curve-fig
<img src="https://miro.medium.com/max/810/1*UUHvSixG7rX2EfNFTtqBDA.gif" width="450px">
[Loss curve](https://towardsdatascience.com/machine-learning-fundamentals-via-linear-regression-41a5d11f5220).
:::
The **accuracy metric** measures the performance of a model. For example, a pixel to pixel comparison for agreement on class.
Note: the **activation function** is also a hyper-parameter.
#### Common Deep Learning Algorithms for Computer Vision
- Image classification: classifying whole images, e.g. image with clouds, image without clouds
- Object detection: identifying locations of objects in an image and classifying them, e.g. identify bounding boxes of cars and planes in satellite imagery
- Semantic segmentation: classifying individual pixels in an image, e.g. land cover classification
- Instance segmentation: classifying individual pixels in an image in terms of both class and individual membership, e.g. detecting unique agricultural field polygons and classifying them
- Generative Adversarial: a type of image generation where synthetic images are created from real ones, e.g. creating synthetic landscapes from real landscape images
#### Semantic Segmentation
To pair with the content of these tutorials, we will demonstrate semantic segmentation (supervised) to map land use categories and illegal gold mining activity.
- Semantic = of or relating to meaning (class)
- Segmentation = division (of image) into separate parts
#### U-Net Segmentation Architecture
Semantic segmentation is often distilled into the combination of an encoder and a decoder. An encoder generates logic or feedback from input data, and a decoder takes that feedback and translates it to output data in the same form as the input.
The U-Net model, which is one of many deep learning segmentation algorithms, has a great illustration of this structure.
:::{figure-md} Unet-fig
<img src="https://developers.arcgis.com/assets/img/python-graphics/unet.png" width="600px">
U-Net architecture (from [Ronneberger et al., 2015](https://arxiv.org/abs/1505.04597)).
:::
In Fig. 13, the encoder is on the left side of the model. It consists of consecutive convolutional layers, each followed by ReLU and a max pooling operation to encode feature representations at multiple scales. The encoder can be represented by most feature extraction networks designed for classification.
The decoder, on the right side of the Fig. 13 diagram, is tasked to semantically project the discriminative features learned by the encoder onto the original pixel space to render a dense classification. The decoder consists of deconvolution and concatenation followed by regular convolution operations.
Following the decoder is the final classification layer, which computes the pixel-wise classification for each cell in the final feature map.
ReLU is an operation, an activation function to be specific, that induces non-linearity. This function intakes the feature map from a convolution operation and remaps it such that any positive value stays exactly the same, and any negative value becomes zero.
:::{figure-md} relu-graph-fig
<img src="https://miro.medium.com/max/3200/1*w48zY6o9_5W9iesSsNabmQ.gif" width="450px">
[ReLU activation function](https://medium.com/ai%C2%B3-theory-practice-business/magic-behind-activation-function-c6fbc5e36a92).
:::
:::{figure-md} relu-maxpooling-fig
<img src="https://miro.medium.com/max/1000/1*cmGESKfSZLH2ksqF_kBgfQ.gif" width="450px">
[ReLU applied to an input matrix](https://towardsdatascience.com/a-laymans-guide-to-building-your-first-image-classification-model-in-r-using-keras-b285deac6572).
:::
Max pooling is used to summarize a feature map and only retain the important structural elements, foregoing the more granular detail that may not be significant to the modeling task. This helps to denoise the signal and helps with computational efficiency. It works similar to convolution in that a kernel with a stride is applied to the feature map and only the maximum value within each patch is reserved.
:::{figure-md} maxpooling-fig
<img src="https://thumbs.gfycat.com/FirstMediumDalmatian-size_restricted.gif" width="450px">
[Max pooling with a kernal over an input matrix](https://gfycat.com/firstmediumdalmatian).
:::
|
github_jupyter
|
from IPython.display import YouTubeVideo
def display_yotube_video(url, **kwargs):
id_ = url.split("=")[-1]
return YouTubeVideo(id_, **kwargs)
display_yotube_video("https://www.youtube.com/watch?v=-C3niPVd-zU", width=800, height=600)
### What are Neural Networks?
Artificial neural networks (ANNs) are a specific, biologically-inspired class of machine learning algorithms. They are modeled after the structure and function of the human brain.
:::{figure-md} neuron-fig
<img src="https://github.com/developmentseed/tensorflow-eo-training/blob/main/ds_book/docs/images/neuron-structure.jpg?raw=1" width="450px">
Biological neuron (from [https://training.seer.cancer.gov/anatomy/nervous/tissue.html](https://training.seer.cancer.gov/anatomy/nervous/tissue.html)).
:::
ANNs are essentially programs that make decisions by weighing the evidence and responding to feedback. By varying the input data, types of parameters and their values, we can get different models of decision-making.
:::{figure-md} neuralnet_basic-fig
<img src="https://miro.medium.com/max/1100/1*x6KWjKTOBhUYL0MRX4M3oQ.png" width="450px">
Basic neural network from [https://towardsdatascience.com/machine-learning-for-beginners-an-introduction-to-neural-networks-d49f22d238f9](https://towardsdatascience.com/machine-learning-for-beginners-an-introduction-to-neural-networks-d49f22d238f9).
:::
In network architectures, neurons are grouped in layers, with synapses traversing the interstitial space between neurons in one layer and the next.
#### What are Convolutional Neural Networks?
A Convolutional Neural Network (ConvNet/CNN) is a form of deep learning inspired by the organization of the human visual cortex, in which individual neurons respond to stimuli within a constrained region of the visual field known as the receptive field. Several receptive fields overlap to account for the entire visual area.
In artificial CNNs, an input matrix such as an image is given importance per various aspects and objects in the image through a moving, convoling receptive field. Very little pre-processing is required for CNNs relative to other classification methods as the need for upfront feature-engineering is removed. Rather, CNNs learn the correct filters and consequent features on their own, provided enough training time and examples.
:::{figure-md} convolution-fig
<img src="https://miro.medium.com/max/1400/1*Fw-ehcNBR9byHtho-Rxbtw.gif" width="450px">
Convolution of a kernal over an input matrix from [https://towardsdatascience.com/intuitively-understanding-convolutions-for-deep-learning-1f6f42faee1](https://towardsdatascience.com/intuitively-understanding-convolutions-for-deep-learning-1f6f42faee1).
:::
#### What is a kernel/filter?
A kernel is matrix smaller than the input. It acts as a receptive field that moves over the input matrix from left to right and top to bottom and filters for features in the image.
#### What is stride?
Stride refers to the number of pixels that the kernel shifts at each step in its navigation of the input matrix.
#### What is a convolution operation?
The convolution operation is the combination of two functions to produce a third function as a result. In effect, it is a merging of two sets of information, the kernel and the input matrix.
:::{figure-md} convolution-arithmetic-fig
<img src="https://theano-pymc.readthedocs.io/en/latest/_images/numerical_no_padding_no_strides.gif" width="450px">
Convolution of a kernal over an input matrix from [https://theano-pymc.readthedocs.io/en/latest/tutorial/conv_arithmetic.html](https://theano-pymc.readthedocs.io/en/latest/tutorial/conv_arithmetic.html).
:::
#### Convolution operation using 3D filter
An input image is often represented as a 3D matrix with a dimension for width (pixels), height (pixels), and depth (channels). In the case of an optical image with red, green and blue channels, the kernel/filter matrix is shaped with the same channel depth as the input and the weighted sum of dot products is computed across all 3 dimensions.
#### What is padding?
After a convolution operation, the feature map is by default smaller than the original input matrix.
:::{figure-md} multi_layer_CNN-fig
<img src="https://www.researchgate.net/profile/Sheraz-Khan-14/publication/321586653/figure/fig4/AS:568546847014912@1512563539828/The-LeNet-5-Architecture-a-convolutional-neural-network.png" width="450px">
[Progressive downsizing of feature maps in a multi-layer CNN](https://www.researchgate.net/figure/The-LeNet-5-Architecture-a-convolutional-neural-network_fig4_321586653).
:::
To maintain the same spatial dimensions between input matrix and output feature map, we may pad the input matrix with a border of zeroes or ones. There are two types of padding:
1. Same padding: a border of zeroes or ones is added to match the input/output dimensions
2. Valid padding: no border is added and the output dimensions are not matched to the input
:::{figure-md} padding-fig
<img src="https://miro.medium.com/max/666/1*noYcUAa_P8nRilg3Lt_nuA.png" width="450px">
[Padding an input matrix with zeroes](https://ayeshmanthaperera.medium.com/what-is-padding-in-cnns-71b21fb0dd7).
:::
### What is Deep Learning?
Deep learning is defined by neural networks with depth, i.e. many layers and connections. The reason for why deep learning is so highly performant lies in the degree of abstraction made possible by feature extraction across so many layers in which each neuron, or processing unit, is interacting with input from neurons in previous layers and making decisions accordingly. The deepest layers of a network once trained can be capable inferring highly abstract concepts, such as what differentiates a school from a house in satellite imagery.
#### Training and Testing Data
The dataset (e.g. all images and their labels) are split into training, validation and testing sets. A common ratio is 70:20:10 percent, train:validation:test. If randomly split, it is important to check that all class labels exist in all sets and are well represented.
#### Forward and backward propagation, hyper-parameters, and learnable parameters
Neural networks train in cycles, where the input data passes through the network, a relationship between input data and target values is learned, a prediction is made, the prediction value is measured for error relative to its true value, and the errors are used to inform updates to parameters in the network, feeding into the next cycle of learning and prediction using the updated information. This happens through a two-step process called forward propagation and back propagation, in which the first part is used to gather knowledge and the second part is used to correct errors in the model’s knowledge.
:::{figure-md} forward_backprop-fig
<img src="https://thumbs.gfycat.com/BitesizedWeeBlacklemur-max-1mb.gif" width="450px">
[Forward and back propagation](https://gfycat.com/gifs/search/backpropagation).
:::
The **activation function** decides whether or not the output from one neuron is useful or not based on a threshold value, and therefore, whether it will be carried from one layer to the next.
**Weights** control the signal (or the strength of the connection) between two neurons in two consecutive layers.
**Biases** are values which help determine whether or not the activation output from a neuron is going to be passed forward through the network.
In a neural network, neurons in one layer are connected to neurons in the next layer. As information passes from one neuron to the next, the information is conditioned by the weight of the synapse and is subjected to a bias. The weights and biases determine if the information passes further beyond the current neuron.
:::{figure-md} activation-fig
<img src="https://cdn-images-1.medium.com/max/651/1*UA30b0mJUPYoPvN8yJr2iQ.jpeg" width="450px">
[Weights, bias, activation](https://laptrinhx.com/statistics-is-freaking-hard-wtf-is-activation-function-207913705/).
:::
During training, the weights and biases are learned and updated using the training and validation dataset to fit the data and reduce error of prediction values relative to target values.
| 0.926499 | 0.987876 |
# Train a SMILES language model from scratch
> Tutorial how to train a reaction language model
```
# optional
import os
import numpy as np
import pandas as pd
import torch
import logging
import random
from rxnfp.models import SmilesLanguageModelingModel
logger = logging.getLogger(__name__)
```
## Track the training
We will be using wandb to keep track of our training. You can use the an account on [wandb](https://www.wandb.com) or create an own instance following the instruction in the [documentation](https://docs.wandb.com/self-hosted).
If you then create an `.env` file in the root folder and specify the `WANDB_API_KEY=` (and the `WANDB_BASE_URL=`), you can use dotenv to load those enviroment variables.
```
# optional
# !pip install python-dotenv
from dotenv import load_dotenv, find_dotenv
load_dotenv(find_dotenv())
```
## Setup MLM training
Choose the hyperparameters you want and start the training. The default parameters will train a BERT model with 12 layers and 4 attention heads per layer. The training task is Masked Language Modeling (MLM), where tokens from the input reactions are randomly masked and predicted by the model given the context.
After defining the config, the training is launched in 3 lines of code using our adapter written for the [SimpleTransformers](https://simpletransformers.ai) library (based on huggingface [Transformers](https://github.com/huggingface/transformers)).
To make it work you will have to install simpletransformers:
```bash
pip install simpletransformers
```
```
# optional
config = {
"architectures": [
"BertForMaskedLM"
],
"attention_probs_dropout_prob": 0.1,
"hidden_act": "gelu",
"hidden_dropout_prob": 0.1,
"hidden_size": 256,
"initializer_range": 0.02,
"intermediate_size": 512,
"layer_norm_eps": 1e-12,
"max_position_embeddings": 512,
"model_type": "bert",
"num_attention_heads": 4,
"num_hidden_layers": 12,
"pad_token_id": 0,
"type_vocab_size": 2,
}
vocab_path = '../data/uspto_1k_TPL/individual_files/vocab.txt'
args = {'config': config,
'vocab_path': vocab_path,
'wandb_project': 'uspto_mlm_temp_1000',
'train_batch_size': 32,
'manual_seed': 42,
"fp16": False,
"num_train_epochs": 50,
'max_seq_length': 256,
'evaluate_during_training': True,
'overwrite_output_dir': True,
'output_dir': '../out/bert_mlm_1k_tpl',
'learning_rate': 1e-4
}
# optional
model = SmilesLanguageModelingModel(model_type='bert', model_name=None, args=args)
# optional
# !unzip ../data/uspto_1k_TPL/individual_files/mlm_training.zip -d ../data/uspto_1k_TPL/individual_files/
train_file = '../data/uspto_1k_TPL/individual_files/mlm_train_file.txt'
eval_file = '../data/uspto_1k_TPL/individual_files/mlm_eval_file_1k.txt'
model.train_model(train_file=train_file, eval_file=eval_file)
```
|
github_jupyter
|
# optional
import os
import numpy as np
import pandas as pd
import torch
import logging
import random
from rxnfp.models import SmilesLanguageModelingModel
logger = logging.getLogger(__name__)
# optional
# !pip install python-dotenv
from dotenv import load_dotenv, find_dotenv
load_dotenv(find_dotenv())
pip install simpletransformers
# optional
config = {
"architectures": [
"BertForMaskedLM"
],
"attention_probs_dropout_prob": 0.1,
"hidden_act": "gelu",
"hidden_dropout_prob": 0.1,
"hidden_size": 256,
"initializer_range": 0.02,
"intermediate_size": 512,
"layer_norm_eps": 1e-12,
"max_position_embeddings": 512,
"model_type": "bert",
"num_attention_heads": 4,
"num_hidden_layers": 12,
"pad_token_id": 0,
"type_vocab_size": 2,
}
vocab_path = '../data/uspto_1k_TPL/individual_files/vocab.txt'
args = {'config': config,
'vocab_path': vocab_path,
'wandb_project': 'uspto_mlm_temp_1000',
'train_batch_size': 32,
'manual_seed': 42,
"fp16": False,
"num_train_epochs": 50,
'max_seq_length': 256,
'evaluate_during_training': True,
'overwrite_output_dir': True,
'output_dir': '../out/bert_mlm_1k_tpl',
'learning_rate': 1e-4
}
# optional
model = SmilesLanguageModelingModel(model_type='bert', model_name=None, args=args)
# optional
# !unzip ../data/uspto_1k_TPL/individual_files/mlm_training.zip -d ../data/uspto_1k_TPL/individual_files/
train_file = '../data/uspto_1k_TPL/individual_files/mlm_train_file.txt'
eval_file = '../data/uspto_1k_TPL/individual_files/mlm_eval_file_1k.txt'
model.train_model(train_file=train_file, eval_file=eval_file)
| 0.371935 | 0.922273 |
# Ecommerce Data Project
Based on https://github.com/tinybirdco/ecommerce_data_project:
If you have opened the notebook in Google Colab then `Copy to Drive` (see above).
```
#@title Mount your Google Drive to save and use local files
from google.colab import drive
drive.mount('/content/gdrive', force_remount=False)
% cd "/content/gdrive/My Drive/Colab Notebooks/Tinybird/tb_examples"
#@title Install Tinybird CLI, libraries and your token
!pip install tinybird-cli -q
!sudo apt-get install jq
import os
import re
if not os.path.isfile('.tinyb'):
!tb auth
if not os.path.isdir('./datasources'):
!tb init
#@title Helper function to write to files
def write_text_to_file(filename, text):
with open(filename, 'w') as f: f.write(text)
```
# Create Data Sources
## 1. Events Data Source
```
filename="./datasources/events.datasource"
text='''
DESCRIPTION > # Events from users
this contains all the events produced by kafka, there are 4 fixed columns
plus a `json` column which contains the rest of the data for that event
SCHEMA >
date DateTime,
product_id String,
user_id String,
event String,
extra_data String
ENGINE MergeTree
ENGINE_SORTING_KEY timestamp
'''
write_text_to_file(filename, text)
!tb datasource generate datasources/events.datasource --force
!tb datasource append events https://storage.googleapis.com/tinybird-assets/datasets/guides/events_50M_1.csv
!tb datasource append events https://storage.googleapis.com/tinybird-assets/datasets/guides/events_50M_2.csv
!tb sql "SELECT count() FROM events"
!tb sql "SELECT * FROM events LIMIT 1"
```
## 2. Products Data Source
```
filename="datasources/products_join_sku.datasource"
text='''
SCHEMA >
sku String,
color String,
section_id String,
title String
# this creates a join table ready to access by sku
# using joinGet('products_join_by_id', 'color', sku)
ENGINE Join
ENGINE_JOIN_STRICTNESS ANY
ENGINE_JOIN_TYPE LEFT
ENGINE_KEY_COLUMNS sku
'''
write_text_to_file(filename, text)
!tb push datasources/products_join_sku.datasource
!tb datasource append products_join_sku https://storage.googleapis.com/tinybird-assets/datasets/guides/products_1.csv
!tb datasource append products_join_sku https://storage.googleapis.com/tinybird-assets/datasets/guides/products_2.csv
!tb sql "SELECT count() FROM products_join_sku"
!tb sql "SELECT * FROM products_join_sku LIMIT 1"
```
## 3. Top Products View Data Source
```
filename="datasources/top_products_view.datasource"
text='''
SCHEMA >
date Date,
top_10 AggregateFunction(topK(10), String),
total_sales AggregateFunction(sum, Float64)
ENGINE AggregatingMergeTree
ENGINE_SORTING_KEY date
'''
write_text_to_file(filename, text)
!tb push datasources/top_products_view.datasource
```
# Create Pipes
## Top Product Per Day Pipe
```
filename="pipes/top_product_per_day.pipe"
text='''
NODE only_buy_events
DESCRIPTION >
filters all the buy events
SQL >
SELECT
toDate(date) date,
product_id,
JSONExtractFloat(extra_data, 'price') as price
FROM events
where event = 'buy'
NODE top_per_day
SQL >
SELECT date,
topKState(10)(product_id) top_10,
sumState(price) total_sales
from only_buy_events
group by date
TYPE materialized
DATASOURCE top_products_view
'''
write_text_to_file(filename, text)
!tb push 'pipes/top_product_per_day.pipe' --force --populate
!tb sql "SELECT date, topKMerge(top_10), sumMerge(total_sales) \
FROM top_products_view \
GROUP BY date LIMIT 3"
```
# Create Endpoints
```
filename="endpoints/sales.pipe"
text='''
DESCRIPTION >
return sales for a product with color filter
NODE only_buy_events
SQL >
SELECT
toDate(date) date,
product_id,
joinGet('products_join_sku', 'color', product_id) as color,
JSONExtractFloat(extra_data, 'price') as price
FROM events
WHERE event = 'buy'
NODE endpoint
DESCRIPTION >
return sales for a product with color filter
SQL >
%
select date, sum(price) total_sales
from only_buy_events
where color = 'dark green'
group by date
'''
write_text_to_file(filename, text)
!tb push 'endpoints/sales.pipe' --force --populate
!tb sql "SELECT * FROM sales LIMIT 10"
filename="endpoints/top_products_params.pipe"
text='''
NODE endpoint
DESCRIPTION >
returns top 10 products given start and end dates
SQL >
%
select
date,
topKMerge(10)(top_10) as top_10
from top_product_per_day
where date between {{Date(start)}} and {{Date(end)}}
group by date
'''
write_text_to_file(filename, text)
!tb push 'endpoints/top_products_params.pipe' --force --populate
```
https://api.tinybird.co/v0/pipes/top_products_params.json?start=2019-01-01&end=2019-01-05
|
github_jupyter
|
#@title Mount your Google Drive to save and use local files
from google.colab import drive
drive.mount('/content/gdrive', force_remount=False)
% cd "/content/gdrive/My Drive/Colab Notebooks/Tinybird/tb_examples"
#@title Install Tinybird CLI, libraries and your token
!pip install tinybird-cli -q
!sudo apt-get install jq
import os
import re
if not os.path.isfile('.tinyb'):
!tb auth
if not os.path.isdir('./datasources'):
!tb init
#@title Helper function to write to files
def write_text_to_file(filename, text):
with open(filename, 'w') as f: f.write(text)
filename="./datasources/events.datasource"
text='''
DESCRIPTION > # Events from users
this contains all the events produced by kafka, there are 4 fixed columns
plus a `json` column which contains the rest of the data for that event
SCHEMA >
date DateTime,
product_id String,
user_id String,
event String,
extra_data String
ENGINE MergeTree
ENGINE_SORTING_KEY timestamp
'''
write_text_to_file(filename, text)
!tb datasource generate datasources/events.datasource --force
!tb datasource append events https://storage.googleapis.com/tinybird-assets/datasets/guides/events_50M_1.csv
!tb datasource append events https://storage.googleapis.com/tinybird-assets/datasets/guides/events_50M_2.csv
!tb sql "SELECT count() FROM events"
!tb sql "SELECT * FROM events LIMIT 1"
filename="datasources/products_join_sku.datasource"
text='''
SCHEMA >
sku String,
color String,
section_id String,
title String
# this creates a join table ready to access by sku
# using joinGet('products_join_by_id', 'color', sku)
ENGINE Join
ENGINE_JOIN_STRICTNESS ANY
ENGINE_JOIN_TYPE LEFT
ENGINE_KEY_COLUMNS sku
'''
write_text_to_file(filename, text)
!tb push datasources/products_join_sku.datasource
!tb datasource append products_join_sku https://storage.googleapis.com/tinybird-assets/datasets/guides/products_1.csv
!tb datasource append products_join_sku https://storage.googleapis.com/tinybird-assets/datasets/guides/products_2.csv
!tb sql "SELECT count() FROM products_join_sku"
!tb sql "SELECT * FROM products_join_sku LIMIT 1"
filename="datasources/top_products_view.datasource"
text='''
SCHEMA >
date Date,
top_10 AggregateFunction(topK(10), String),
total_sales AggregateFunction(sum, Float64)
ENGINE AggregatingMergeTree
ENGINE_SORTING_KEY date
'''
write_text_to_file(filename, text)
!tb push datasources/top_products_view.datasource
filename="pipes/top_product_per_day.pipe"
text='''
NODE only_buy_events
DESCRIPTION >
filters all the buy events
SQL >
SELECT
toDate(date) date,
product_id,
JSONExtractFloat(extra_data, 'price') as price
FROM events
where event = 'buy'
NODE top_per_day
SQL >
SELECT date,
topKState(10)(product_id) top_10,
sumState(price) total_sales
from only_buy_events
group by date
TYPE materialized
DATASOURCE top_products_view
'''
write_text_to_file(filename, text)
!tb push 'pipes/top_product_per_day.pipe' --force --populate
!tb sql "SELECT date, topKMerge(top_10), sumMerge(total_sales) \
FROM top_products_view \
GROUP BY date LIMIT 3"
filename="endpoints/sales.pipe"
text='''
DESCRIPTION >
return sales for a product with color filter
NODE only_buy_events
SQL >
SELECT
toDate(date) date,
product_id,
joinGet('products_join_sku', 'color', product_id) as color,
JSONExtractFloat(extra_data, 'price') as price
FROM events
WHERE event = 'buy'
NODE endpoint
DESCRIPTION >
return sales for a product with color filter
SQL >
%
select date, sum(price) total_sales
from only_buy_events
where color = 'dark green'
group by date
'''
write_text_to_file(filename, text)
!tb push 'endpoints/sales.pipe' --force --populate
!tb sql "SELECT * FROM sales LIMIT 10"
filename="endpoints/top_products_params.pipe"
text='''
NODE endpoint
DESCRIPTION >
returns top 10 products given start and end dates
SQL >
%
select
date,
topKMerge(10)(top_10) as top_10
from top_product_per_day
where date between {{Date(start)}} and {{Date(end)}}
group by date
'''
write_text_to_file(filename, text)
!tb push 'endpoints/top_products_params.pipe' --force --populate
| 0.4206 | 0.618723 |
<a href="https://colab.research.google.com/drive/1GH_h7TFmZJGZTtUreTLb2JSvGj6D50pd?usp=sharing" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Outline
This tutorial will demonstrate the application of soundscape_IR in detecting sika deer (*Cervus nippon*) calls from tropical forest recordings and learning their spectral variations. Please contact Yi-Jen Sun (elainesun442@gmail.com) or Tzu-Hao Harry Lin (schonkopf@gmail.com) for questions or suggestions.
<img src="https://raw.githubusercontent.com/yijensun/soundscape_IR/master/docs/images/workflow_case1_v5.png" width="900"/>
This tutorial contains four sections:
1. Preparation of a training spectrogram
- Concatenation
2. Weekly-supervised model training
- Periodicity-coded nonnegative matrix factorization (PC-NMF)
3. Target detection
- Model prediction using adapative and semi-supervised learning
- Spectrogram reconstrunction and occurrence detection
4. Feature learning
- Extracting call-specific spectral variation using adaptive learning
# Installation
```
# Clone soundscape_IR from GitHub @schonkopf
!git clone https://github.com/schonkopf/soundscape_IR.git
# Install required packages
%cd soundscape_IR
%pip install -r requirements.txt
```
# 1. Training spectrogram
In the beginning, use the function ```audio_visualization``` to generate a spectrogram of sika deer calls. Most sika deer calls were recorded in frequencies below 4 kHz. In addition to sika deer calls, this audio also recorded many insect calls (> 3.5 kHz).
```
from soundscape_IR.soundscape_viewer import audio_visualization
# Generate a spectrogram, using 25 perentile (for each frequency bin) as the noise baseline to prewhiten the spectrogram
sound_train=audio_visualization(filename='case1_train.wav', path='./data/wav', FFT_size=512, time_resolution=0.1, window_overlap=0.5, plot_type='Spectrogram', prewhiten_percent=25, f_range=[0,8000])
```
## Concatenation
Based on the ```audio_visualization```, we can import a txt file contains manual annotations (generated by using [Raven software](https://ravensoundsoftware.com/) of Cornell Lab of Ornithology) to produce a concatenated spectrogram of deer calls. The concatenated spectrogram can reduce the amount of noise displayed in the training spectrogram and enhance the occuring periodicity of deer calls for training a source separation model in a weakly-supervised manner.
```
# Generate a spectrogram
sound_train=audio_visualization(filename='case1_train.wav', path='./data/wav', FFT_size=512, time_resolution=0.1, window_overlap=0.5, plot_type='Spectrogram', prewhiten_percent=25, f_range=[0,8000], annotation='./data/txt/case1_anno.txt', padding=0.5)
```
# 2. Model training
## PC-NMF
PC-NMF is a machine learning-based source separation model (Lin et al. 2017). It learns a set of basis functions essential for reconstructing the input spectrogram and separates the basis functions into groups according to their specific periodicity. In the function ```source_separation```, we use PC-NMF to learn two groups of basis functions for separating deer calls and noise.
After model training, we can visualize the two groups of basis functions and their source numbers (*source=1 or 2*). If the procedure of weakly-supervised learning works well, the model can be saved for further applications.
* Tzu-Hao Lin, Shih-Hua Fang, Yu Tsao. (2017) Improving biodiversity assessment via unsupervised separation of biological sounds from long-duration recordings. Scientific Reports, 7: 4547. https://doi.org/10.1038/s41598-017-04790-7
```
from soundscape_IR.soundscape_viewer import source_separation
# Define NMF parameters and train the model
model=source_separation(feature_length=20, basis_num=10)
model.learn_feature(input_data=sound_train.data, f=sound_train.f, method='PCNMF')
# Plot the basis functions of each source
model.plot_nmf(plot_type='W', source=1)
model.plot_nmf(plot_type='W', source=2)
# Save the model
model.save_model(filename='./data/model/case1_model.mat')
```
# 3. Target detection
Let us load another audio for testing the source separation model. Again, use audio_visualization to generate a spectrogram.
```
# Generate a spectrogram
sound_predict=audio_visualization(filename='case1_predict.wav', path='./data/wav', FFT_size=512, time_resolution=0.1, window_overlap=0.5, plot_type='Spectrogram', prewhiten_percent=25, f_range=[0,8000])
```
## Model prediction
Load the model by using ```source_separation```. Run model prediction with adaptive (*adaptive_alpha*) and semi-supervised (*additional_basis*) learning. After prediction, plot the basis functions to see how they adapt to the new spectrogram. With semi-supervised learning, the model also learns a new set of basis functions associated with unseen sound sources (*source=3*).
```
# Load the model and run prediction with adaptive and semi-supervised separation
model=source_separation()
model.load_model(filename = './data/model/case1_model.mat')
model.prediction(input_data=sound_predict.data, f=sound_predict.f, adaptive_alpha = [0.05,0.05], additional_basis = 2)
# Plot the adapted and newly learned features
for i in range(1, model.source_num+1):
model.plot_nmf(plot_type='W', source=i)
```
## Spectrogram reconstruction and detection
After the prediction procedure, the reconstructed spectrogram of deer calls is saved in ```model.separation```. The reconstructed spectrogram is no longer interfered by noise. A simple energy detector can help identify the occurrence of deer calls. Use the function ```spectrogram_detection``` to process the reconstructed spectrogram for visualizing the detection result. Save the detection result in txt format and use Raven to explore.
```
from soundscape_IR.soundscape_viewer import spectrogram_detection
# Detect the occurrence of target signals
source_num=1
sp=spectrogram_detection(input=model.separation[source_num-1], f=model.f, threshold=4.5, smooth=1, minimum_interval=0.5, pad_size=0, filename='sika_case1_detection.txt', path='./')
```
# 4. Feature learning
## Extracting call-specific spectral variation
In the final section, we will demonstrate the use of adaptive learning in extracting call-specific spectral variation. At first, select one deer call from the detection result and generate its spectrogram. Then, run model prediction but this time use a higher alpha value for the basis functions of deer calls to learn the spectral variation specific to the detected deer call. The result is a set of adapted basis functions (generally displaying different phases), we can select one adapted basis function to represent the selected deer call.
```
# Assign which call to extract
call_num=4
offset=float(sp.output[call_num][3]) # beginning time of the detected call
duration=float(sp.output[call_num][4]) - float(sp.output[call_num][3]) # duration of the detected call
# Generate a spectrogram
sound_call=audio_visualization(filename='case1_predict.wav', path='./data/wav', FFT_size=512, offset_read=offset, duration_read=duration, time_resolution=0.1, window_overlap=0.5, plot_type='Spectrogram', prewhiten_percent=25, f_range=[0,8000])
# Deploy the model with adaptive NMF
model.prediction(input_data=sound_call.data, f=sound_call.f, adaptive_alpha=[0.2,0,0])
# Plot the adapted features
model.plot_nmf(plot_type='W', source=source_num)
```
|
github_jupyter
|
# Clone soundscape_IR from GitHub @schonkopf
!git clone https://github.com/schonkopf/soundscape_IR.git
# Install required packages
%cd soundscape_IR
%pip install -r requirements.txt
from soundscape_IR.soundscape_viewer import audio_visualization
# Generate a spectrogram, using 25 perentile (for each frequency bin) as the noise baseline to prewhiten the spectrogram
sound_train=audio_visualization(filename='case1_train.wav', path='./data/wav', FFT_size=512, time_resolution=0.1, window_overlap=0.5, plot_type='Spectrogram', prewhiten_percent=25, f_range=[0,8000])
# Generate a spectrogram
sound_train=audio_visualization(filename='case1_train.wav', path='./data/wav', FFT_size=512, time_resolution=0.1, window_overlap=0.5, plot_type='Spectrogram', prewhiten_percent=25, f_range=[0,8000], annotation='./data/txt/case1_anno.txt', padding=0.5)
from soundscape_IR.soundscape_viewer import source_separation
# Define NMF parameters and train the model
model=source_separation(feature_length=20, basis_num=10)
model.learn_feature(input_data=sound_train.data, f=sound_train.f, method='PCNMF')
# Plot the basis functions of each source
model.plot_nmf(plot_type='W', source=1)
model.plot_nmf(plot_type='W', source=2)
# Save the model
model.save_model(filename='./data/model/case1_model.mat')
# Generate a spectrogram
sound_predict=audio_visualization(filename='case1_predict.wav', path='./data/wav', FFT_size=512, time_resolution=0.1, window_overlap=0.5, plot_type='Spectrogram', prewhiten_percent=25, f_range=[0,8000])
# Load the model and run prediction with adaptive and semi-supervised separation
model=source_separation()
model.load_model(filename = './data/model/case1_model.mat')
model.prediction(input_data=sound_predict.data, f=sound_predict.f, adaptive_alpha = [0.05,0.05], additional_basis = 2)
# Plot the adapted and newly learned features
for i in range(1, model.source_num+1):
model.plot_nmf(plot_type='W', source=i)
from soundscape_IR.soundscape_viewer import spectrogram_detection
# Detect the occurrence of target signals
source_num=1
sp=spectrogram_detection(input=model.separation[source_num-1], f=model.f, threshold=4.5, smooth=1, minimum_interval=0.5, pad_size=0, filename='sika_case1_detection.txt', path='./')
# Assign which call to extract
call_num=4
offset=float(sp.output[call_num][3]) # beginning time of the detected call
duration=float(sp.output[call_num][4]) - float(sp.output[call_num][3]) # duration of the detected call
# Generate a spectrogram
sound_call=audio_visualization(filename='case1_predict.wav', path='./data/wav', FFT_size=512, offset_read=offset, duration_read=duration, time_resolution=0.1, window_overlap=0.5, plot_type='Spectrogram', prewhiten_percent=25, f_range=[0,8000])
# Deploy the model with adaptive NMF
model.prediction(input_data=sound_call.data, f=sound_call.f, adaptive_alpha=[0.2,0,0])
# Plot the adapted features
model.plot_nmf(plot_type='W', source=source_num)
| 0.876489 | 0.983198 |
## Dependencies
```
import json, glob
from tweet_utility_scripts import *
from tweet_utility_preprocess_roberta_scripts_aux import *
from transformers import TFRobertaModel, RobertaConfig
from tokenizers import ByteLevelBPETokenizer
from tensorflow.keras import layers
from tensorflow.keras.models import Model
```
# Load data
```
test = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/test.csv')
print('Test samples: %s' % len(test))
display(test.head())
```
# Model parameters
```
input_base_path = '/kaggle/input/245-robertabase/'
with open(input_base_path + 'config.json') as json_file:
config = json.load(json_file)
config
# vocab_path = input_base_path + 'vocab.json'
# merges_path = input_base_path + 'merges.txt'
base_path = '/kaggle/input/qa-transformers/roberta/'
vocab_path = base_path + 'roberta-base-vocab.json'
merges_path = base_path + 'roberta-base-merges.txt'
config['base_model_path'] = base_path + 'roberta-base-tf_model.h5'
config['config_path'] = base_path + 'roberta-base-config.json'
model_path_list = glob.glob(input_base_path + '*.h5')
model_path_list.sort()
print('Models to predict:')
print(*model_path_list, sep='\n')
```
# Tokenizer
```
tokenizer = ByteLevelBPETokenizer(vocab_file=vocab_path, merges_file=merges_path,
lowercase=True, add_prefix_space=True)
```
# Pre process
```
test['text'].fillna('', inplace=True)
test['text'] = test['text'].apply(lambda x: x.lower())
test['text'] = test['text'].apply(lambda x: x.strip())
x_test, x_test_aux, x_test_aux_2 = get_data_test(test, tokenizer, config['MAX_LEN'], preprocess_fn=preprocess_roberta_test)
```
# Model
```
module_config = RobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)
def model_fn(MAX_LEN):
input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')
attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')
base_model = TFRobertaModel.from_pretrained(config['base_model_path'], config=module_config, name="base_model")
last_hidden_state, _ = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})
logits = layers.Dense(2, name="qa_outputs", use_bias=False)(last_hidden_state)
start_logits, end_logits = tf.split(logits, 2, axis=-1)
start_logits = tf.squeeze(start_logits, axis=-1)
end_logits = tf.squeeze(end_logits, axis=-1)
model = Model(inputs=[input_ids, attention_mask], outputs=[start_logits, end_logits])
return model
```
# Make predictions
```
NUM_TEST_IMAGES = len(test)
test_start_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
test_end_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
for model_path in model_path_list:
print(model_path)
model = model_fn(config['MAX_LEN'])
model.load_weights(model_path)
test_preds = model.predict(get_test_dataset(x_test, config['BATCH_SIZE']))
test_start_preds += test_preds[0]
test_end_preds += test_preds[1]
```
# Post process
```
test['start'] = test_start_preds.argmax(axis=-1)
test['end'] = test_end_preds.argmax(axis=-1)
test['selected_text'] = test.apply(lambda x: decode(x['start'], x['end'], x['text'], config['question_size'], tokenizer), axis=1)
# Post-process
test["selected_text"] = test.apply(lambda x: ' '.join([word for word in x['selected_text'].split() if word in x['text'].split()]), axis=1)
test['selected_text'] = test.apply(lambda x: x['text'] if (x['selected_text'] == '') else x['selected_text'], axis=1)
test['selected_text'].fillna(test['text'], inplace=True)
```
# Visualize predictions
```
test['text_len'] = test['text'].apply(lambda x : len(x))
test['label_len'] = test['selected_text'].apply(lambda x : len(x))
test['text_wordCnt'] = test['text'].apply(lambda x : len(x.split(' ')))
test['label_wordCnt'] = test['selected_text'].apply(lambda x : len(x.split(' ')))
test['text_tokenCnt'] = test['text'].apply(lambda x : len(tokenizer.encode(x).ids))
test['label_tokenCnt'] = test['selected_text'].apply(lambda x : len(tokenizer.encode(x).ids))
test['jaccard'] = test.apply(lambda x: jaccard(x['text'], x['selected_text']), axis=1)
display(test.head(10))
display(test.describe())
```
# Test set predictions
```
submission = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/sample_submission.csv')
submission['selected_text'] = test['selected_text']
submission.to_csv('submission.csv', index=False)
submission.head(10)
```
|
github_jupyter
|
import json, glob
from tweet_utility_scripts import *
from tweet_utility_preprocess_roberta_scripts_aux import *
from transformers import TFRobertaModel, RobertaConfig
from tokenizers import ByteLevelBPETokenizer
from tensorflow.keras import layers
from tensorflow.keras.models import Model
test = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/test.csv')
print('Test samples: %s' % len(test))
display(test.head())
input_base_path = '/kaggle/input/245-robertabase/'
with open(input_base_path + 'config.json') as json_file:
config = json.load(json_file)
config
# vocab_path = input_base_path + 'vocab.json'
# merges_path = input_base_path + 'merges.txt'
base_path = '/kaggle/input/qa-transformers/roberta/'
vocab_path = base_path + 'roberta-base-vocab.json'
merges_path = base_path + 'roberta-base-merges.txt'
config['base_model_path'] = base_path + 'roberta-base-tf_model.h5'
config['config_path'] = base_path + 'roberta-base-config.json'
model_path_list = glob.glob(input_base_path + '*.h5')
model_path_list.sort()
print('Models to predict:')
print(*model_path_list, sep='\n')
tokenizer = ByteLevelBPETokenizer(vocab_file=vocab_path, merges_file=merges_path,
lowercase=True, add_prefix_space=True)
test['text'].fillna('', inplace=True)
test['text'] = test['text'].apply(lambda x: x.lower())
test['text'] = test['text'].apply(lambda x: x.strip())
x_test, x_test_aux, x_test_aux_2 = get_data_test(test, tokenizer, config['MAX_LEN'], preprocess_fn=preprocess_roberta_test)
module_config = RobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)
def model_fn(MAX_LEN):
input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')
attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')
base_model = TFRobertaModel.from_pretrained(config['base_model_path'], config=module_config, name="base_model")
last_hidden_state, _ = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})
logits = layers.Dense(2, name="qa_outputs", use_bias=False)(last_hidden_state)
start_logits, end_logits = tf.split(logits, 2, axis=-1)
start_logits = tf.squeeze(start_logits, axis=-1)
end_logits = tf.squeeze(end_logits, axis=-1)
model = Model(inputs=[input_ids, attention_mask], outputs=[start_logits, end_logits])
return model
NUM_TEST_IMAGES = len(test)
test_start_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
test_end_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
for model_path in model_path_list:
print(model_path)
model = model_fn(config['MAX_LEN'])
model.load_weights(model_path)
test_preds = model.predict(get_test_dataset(x_test, config['BATCH_SIZE']))
test_start_preds += test_preds[0]
test_end_preds += test_preds[1]
test['start'] = test_start_preds.argmax(axis=-1)
test['end'] = test_end_preds.argmax(axis=-1)
test['selected_text'] = test.apply(lambda x: decode(x['start'], x['end'], x['text'], config['question_size'], tokenizer), axis=1)
# Post-process
test["selected_text"] = test.apply(lambda x: ' '.join([word for word in x['selected_text'].split() if word in x['text'].split()]), axis=1)
test['selected_text'] = test.apply(lambda x: x['text'] if (x['selected_text'] == '') else x['selected_text'], axis=1)
test['selected_text'].fillna(test['text'], inplace=True)
test['text_len'] = test['text'].apply(lambda x : len(x))
test['label_len'] = test['selected_text'].apply(lambda x : len(x))
test['text_wordCnt'] = test['text'].apply(lambda x : len(x.split(' ')))
test['label_wordCnt'] = test['selected_text'].apply(lambda x : len(x.split(' ')))
test['text_tokenCnt'] = test['text'].apply(lambda x : len(tokenizer.encode(x).ids))
test['label_tokenCnt'] = test['selected_text'].apply(lambda x : len(tokenizer.encode(x).ids))
test['jaccard'] = test.apply(lambda x: jaccard(x['text'], x['selected_text']), axis=1)
display(test.head(10))
display(test.describe())
submission = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/sample_submission.csv')
submission['selected_text'] = test['selected_text']
submission.to_csv('submission.csv', index=False)
submission.head(10)
| 0.396068 | 0.339636 |
# **Image Classification**
By Kevin Leonard Sugiman
[GitHub](https://github.com/TGasG)
# **Import Libraries**
```
import numpy as np
import PIL
import PIL.Image
import tensorflow as tf
import matplotlib.pyplot as plt
from tensorflow import keras
from tensorflow.keras import layers
# Tensorflow Version
print(tf.__version__)
```
# **Dowload Dataset**
```
!wget --no-check-certificate \
https://dicodingacademy.blob.core.windows.net/picodiploma/ml_pemula_academy/rockpaperscissors.zip \
-O /tmp/rockpaperscissors.zip
# Extract zip file
import zipfile,os
local_zip = '/tmp/rockpaperscissors.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp')
zip_ref.close()
import pathlib
data_dir = '/tmp/rockpaperscissors/rps-cv-images'
data_dir = pathlib.Path(data_dir)
image_count = len(list(data_dir.glob('*/*.png')))
print(image_count)
```
Rock image sample
```
rock = list(data_dir.glob('rock/*'))
PIL.Image.open(str(rock[0]))
```
Paper image sample
```
paper = list(data_dir.glob('paper/*'))
PIL.Image.open(str(paper[0]))
```
Scissor image sample
```
scissors = list(data_dir.glob('scissors/*'))
PIL.Image.open(str(scissors[0]))
```
# **Load data with keras.preprocessing**
Define loader parameter
```
batch_size = 32
img_height = 200
img_width = 200
```
Divide 60% dataset for training
```
train_ds = tf.keras.preprocessing.image_dataset_from_directory(
data_dir,
validation_split=0.4,
subset="training",
seed=1,
image_size=(img_height, img_width),
batch_size=batch_size,
color_mode='grayscale'
)
```
Divide 40% dataset for validation
```
val_ds = tf.keras.preprocessing.image_dataset_from_directory(
data_dir,
validation_split=0.4,
subset="validation",
seed=1,
image_size=(img_height, img_width),
batch_size=batch_size,
color_mode='grayscale'
)
```
# **Check classess**
```
class_names = train_ds.class_names
print(class_names)
for image_batch, labels_batch in train_ds:
print(image_batch.shape)
print(labels_batch.shape)
break
```
# **Train model**
```
num_classes = 3
model = tf.keras.models.Sequential([
layers.experimental.preprocessing.Rescaling(1./255),
tf.keras.layers.Conv2D(32, (3,3), activation='relu', input_shape=(200, 200, 1)),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(128, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(128, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(256, activation='relu'),
tf.keras.layers.Dense(256, activation='relu'),
tf.keras.layers.Dense(num_classes)
])
model.compile(
optimizer='adam',
loss=tf.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=['accuracy'])
model.fit(
train_ds,
validation_data=val_ds,
epochs=15
)
```
# **Image Prediction**
```
from google.colab import files
from keras.preprocessing import image
import matplotlib.image as mpimg
%matplotlib inline
uploaded = files.upload()
for fn in uploaded.keys():
path = fn
image = tf.keras.preprocessing.image.load_img(
path, color_mode='grayscale', target_size=(200, 200),
interpolation='nearest'
)
imgplot = plt.imshow(image)
input_arr = keras.preprocessing.image.img_to_array(image)
input_arr = np.array([input_arr])
print(fn)
predictions = model.predict(input_arr)
score = tf.nn.softmax(predictions[0])
print(
"This picture is {} with {:.2f} % probability."
.format(class_names[np.argmax(score)], 100 * np.max(score))
)
```
|
github_jupyter
|
import numpy as np
import PIL
import PIL.Image
import tensorflow as tf
import matplotlib.pyplot as plt
from tensorflow import keras
from tensorflow.keras import layers
# Tensorflow Version
print(tf.__version__)
!wget --no-check-certificate \
https://dicodingacademy.blob.core.windows.net/picodiploma/ml_pemula_academy/rockpaperscissors.zip \
-O /tmp/rockpaperscissors.zip
# Extract zip file
import zipfile,os
local_zip = '/tmp/rockpaperscissors.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp')
zip_ref.close()
import pathlib
data_dir = '/tmp/rockpaperscissors/rps-cv-images'
data_dir = pathlib.Path(data_dir)
image_count = len(list(data_dir.glob('*/*.png')))
print(image_count)
rock = list(data_dir.glob('rock/*'))
PIL.Image.open(str(rock[0]))
paper = list(data_dir.glob('paper/*'))
PIL.Image.open(str(paper[0]))
scissors = list(data_dir.glob('scissors/*'))
PIL.Image.open(str(scissors[0]))
batch_size = 32
img_height = 200
img_width = 200
train_ds = tf.keras.preprocessing.image_dataset_from_directory(
data_dir,
validation_split=0.4,
subset="training",
seed=1,
image_size=(img_height, img_width),
batch_size=batch_size,
color_mode='grayscale'
)
val_ds = tf.keras.preprocessing.image_dataset_from_directory(
data_dir,
validation_split=0.4,
subset="validation",
seed=1,
image_size=(img_height, img_width),
batch_size=batch_size,
color_mode='grayscale'
)
class_names = train_ds.class_names
print(class_names)
for image_batch, labels_batch in train_ds:
print(image_batch.shape)
print(labels_batch.shape)
break
num_classes = 3
model = tf.keras.models.Sequential([
layers.experimental.preprocessing.Rescaling(1./255),
tf.keras.layers.Conv2D(32, (3,3), activation='relu', input_shape=(200, 200, 1)),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(128, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(128, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(256, activation='relu'),
tf.keras.layers.Dense(256, activation='relu'),
tf.keras.layers.Dense(num_classes)
])
model.compile(
optimizer='adam',
loss=tf.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=['accuracy'])
model.fit(
train_ds,
validation_data=val_ds,
epochs=15
)
from google.colab import files
from keras.preprocessing import image
import matplotlib.image as mpimg
%matplotlib inline
uploaded = files.upload()
for fn in uploaded.keys():
path = fn
image = tf.keras.preprocessing.image.load_img(
path, color_mode='grayscale', target_size=(200, 200),
interpolation='nearest'
)
imgplot = plt.imshow(image)
input_arr = keras.preprocessing.image.img_to_array(image)
input_arr = np.array([input_arr])
print(fn)
predictions = model.predict(input_arr)
score = tf.nn.softmax(predictions[0])
print(
"This picture is {} with {:.2f} % probability."
.format(class_names[np.argmax(score)], 100 * np.max(score))
)
| 0.733833 | 0.883437 |
```
# ! wget http://www.cs.cornell.edu/~cristian/data/cornell_movie_dialogs_corpus.zip
# ! ls
# ! apt install unzip
# ! unzip cornell_movie_dialogs_corpus.zip
# ! ls
# ! wget http://opus.nlpl.eu/download.php?f=OpenSubtitles/v2018/mono/OpenSubtitles.raw.en.gz
# ! ls
# !tar -xvf 'download.php?f=OpenSubtitles%2Fv2018%2Fmono%2FOpenSubtitles.raw.en.gz' -C 'data'
! wget https://object.pouta.csc.fi/OPUS-OpenSubtitles/v2018/mono/en.txt.gz
! apt install gunzip
! gunzip en.txt.gz
! mkdir lines
! split -a 3 -l 100000 --verbose en.txt lines/lines-
# % cd ..
# ! wget https://github.com/PolyAI-LDN/conversational-datasets/blob/master/opensubtitles/create_data.py
import os
import re
from os import path
import tensorflow as tf
import numpy as np
def _should_skip(line, min_length, max_length):
"""Whether a line should be skipped depending on the length."""
return len(line) < min_length or len(line) > max_length
def create_example(previous_lines, line, file_id):
"""Creates examples with multi-line context
The examples will include:
file_id: the name of the file where these lines were obtained.
response: the current line text
context: the previous line text
context/0: 2 lines before
context/1: 3 lines before, etc.
"""
example = {
'file_id': file_id,
'context': previous_lines[-1],
'response': line,
}
example['file_id'] = file_id
print(file_id)
example['context'] = previous_lines[-1]
print(previous_lines[-1])
extra_contexts = previous_lines[:-1]
example.update({
'context/{}'.format(i): context
for i, context in enumerate(extra_contexts[::-1])
})
return example
def _preprocess_line(line):
line = line.decode("utf-8")
# Remove the first word if it is followed by colon (speaker names)
# NOTE: this wont work if the speaker's name has more than one word
line = re.sub('(?:^|(?:[.!?]\\s))(\\w+):', "", line)
# Remove anything between brackets (corresponds to acoustic events).
line = re.sub("[\\[(](.*?)[\\])]", "", line)
# Strip blanks hyphens and line breaks
line = line.strip(" -\n")
return line
import itertools
import collections
import numpy as np
import nltk
SENT_START_TOKEN = "SENTENCE_START"
SENT_END_TOKEN = "SENTENCE_END"
UNKNOWN_TOKEN = "UNKNOWN_TOKEN"
PADDING_TOKEN = "PADDING"
def tokenize_text(text_lines):
"""
Split text into sentences, append start and end tokens to each and tokenize
:param text_lines: list of text lines or list of length one containing all text
:return: list of sentences
"""
sentences = itertools.chain(*[nltk.sent_tokenize(line.lower()) for line in text_lines])
sentences = ["{} {} {}".format(SENT_START_TOKEN, x, SENT_END_TOKEN) for x in sentences]
tokenized_sentences = [nltk.word_tokenize(sent) for sent in sentences]
return tokenized_sentences
def _create_examples_from_file(file_name,file_id, min_length=0, max_length=24,
num_extra_contexts=2):
# _, file_id = path.split(file_name)
#print(file_id,"#")
previous_lines = []
for line in open(file_name,'rb'):
line = _preprocess_line(line)
if not line:
continue
should_skip = _should_skip(
line,
min_length=min_length,
max_length=max_length)
if previous_lines:
should_skip |= _should_skip(
previous_lines[-1],
min_length=min_length,
max_length=max_length)
if not should_skip:
yield create_example(previous_lines, line, file_id)
previous_lines.append(line)
if len(previous_lines) > num_extra_contexts + 1:
del previous_lines[0]
def _features_to_serialized_tf_example(features):
"""Convert a string dict to a serialized TF example.
The dictionary maps feature names (strings) to feature values (strings).
"""
#print("hello")
example = tf.train.Example()
for feature_name, feature_value in features.items():
example.features.feature[feature_name].bytes_list.value.append(
feature_value.encode("utf-8"))
return example.SerializeToString()
def _shuffle_examples(examples):
examples |= ("add random key" >> beam.Map(
lambda example: (uuid.uuid4(), example)))
examples |= ("group by key" >> beam.GroupByKey())
examples |= ("get shuffled values" >> beam.FlatMap(lambda t: t[1]))
return examples
# % cd ..
# ! ls
# ! pwd
# ! ( head -1000000 en.txt ; ) > million.txt
test=_create_examples_from_file(file_name='lines-aaa',file_id='lines-aaa')
for x in test:
print(x)
!wget http://nlp.stanford.edu/data/glove.6B.zip
!unzip glove.6B.zip
glove_dir = './'
embeddings_index = {} #initialize dictionary
f = open(os.path.join(glove_dir, 'glove.6B.100d.txt'))
for line in f:
values = line.split()
word = values[0]
coefs = np.asarray(values[1:], dtype='float32')
embeddings_index[word] = coefs
f.close()
print('Found %s word vectors.' % len(embeddings_index))
embedding_dim = 100
num_words= 72827
embedding_matrix = np.zeros((num_words, embedding_dim)) #create an array of zeros with word_num rows and embedding_dim columns
for word, i in word_index.items():
embedding_vector = embeddings_index.get(word)
if i < num_words:
if embedding_vector is not None:
# Words not found in embedding index will be all-zeros.
embedding_matrix[i] = embedding_vector
from keras import initializers, models, regularizers
from keras.layers import Dense, Dropout, Embedding, SeparableConv1D, MaxPooling1D, GlobalAveragePooling1D
toyModel = models.Sequential()
toyModel.add(Embedding(num_words,
embedding_dim,
input_length=max_len,
weights=[embedding_matrix],
trainable=False))
```
|
github_jupyter
|
# ! wget http://www.cs.cornell.edu/~cristian/data/cornell_movie_dialogs_corpus.zip
# ! ls
# ! apt install unzip
# ! unzip cornell_movie_dialogs_corpus.zip
# ! ls
# ! wget http://opus.nlpl.eu/download.php?f=OpenSubtitles/v2018/mono/OpenSubtitles.raw.en.gz
# ! ls
# !tar -xvf 'download.php?f=OpenSubtitles%2Fv2018%2Fmono%2FOpenSubtitles.raw.en.gz' -C 'data'
! wget https://object.pouta.csc.fi/OPUS-OpenSubtitles/v2018/mono/en.txt.gz
! apt install gunzip
! gunzip en.txt.gz
! mkdir lines
! split -a 3 -l 100000 --verbose en.txt lines/lines-
# % cd ..
# ! wget https://github.com/PolyAI-LDN/conversational-datasets/blob/master/opensubtitles/create_data.py
import os
import re
from os import path
import tensorflow as tf
import numpy as np
def _should_skip(line, min_length, max_length):
"""Whether a line should be skipped depending on the length."""
return len(line) < min_length or len(line) > max_length
def create_example(previous_lines, line, file_id):
"""Creates examples with multi-line context
The examples will include:
file_id: the name of the file where these lines were obtained.
response: the current line text
context: the previous line text
context/0: 2 lines before
context/1: 3 lines before, etc.
"""
example = {
'file_id': file_id,
'context': previous_lines[-1],
'response': line,
}
example['file_id'] = file_id
print(file_id)
example['context'] = previous_lines[-1]
print(previous_lines[-1])
extra_contexts = previous_lines[:-1]
example.update({
'context/{}'.format(i): context
for i, context in enumerate(extra_contexts[::-1])
})
return example
def _preprocess_line(line):
line = line.decode("utf-8")
# Remove the first word if it is followed by colon (speaker names)
# NOTE: this wont work if the speaker's name has more than one word
line = re.sub('(?:^|(?:[.!?]\\s))(\\w+):', "", line)
# Remove anything between brackets (corresponds to acoustic events).
line = re.sub("[\\[(](.*?)[\\])]", "", line)
# Strip blanks hyphens and line breaks
line = line.strip(" -\n")
return line
import itertools
import collections
import numpy as np
import nltk
SENT_START_TOKEN = "SENTENCE_START"
SENT_END_TOKEN = "SENTENCE_END"
UNKNOWN_TOKEN = "UNKNOWN_TOKEN"
PADDING_TOKEN = "PADDING"
def tokenize_text(text_lines):
"""
Split text into sentences, append start and end tokens to each and tokenize
:param text_lines: list of text lines or list of length one containing all text
:return: list of sentences
"""
sentences = itertools.chain(*[nltk.sent_tokenize(line.lower()) for line in text_lines])
sentences = ["{} {} {}".format(SENT_START_TOKEN, x, SENT_END_TOKEN) for x in sentences]
tokenized_sentences = [nltk.word_tokenize(sent) for sent in sentences]
return tokenized_sentences
def _create_examples_from_file(file_name,file_id, min_length=0, max_length=24,
num_extra_contexts=2):
# _, file_id = path.split(file_name)
#print(file_id,"#")
previous_lines = []
for line in open(file_name,'rb'):
line = _preprocess_line(line)
if not line:
continue
should_skip = _should_skip(
line,
min_length=min_length,
max_length=max_length)
if previous_lines:
should_skip |= _should_skip(
previous_lines[-1],
min_length=min_length,
max_length=max_length)
if not should_skip:
yield create_example(previous_lines, line, file_id)
previous_lines.append(line)
if len(previous_lines) > num_extra_contexts + 1:
del previous_lines[0]
def _features_to_serialized_tf_example(features):
"""Convert a string dict to a serialized TF example.
The dictionary maps feature names (strings) to feature values (strings).
"""
#print("hello")
example = tf.train.Example()
for feature_name, feature_value in features.items():
example.features.feature[feature_name].bytes_list.value.append(
feature_value.encode("utf-8"))
return example.SerializeToString()
def _shuffle_examples(examples):
examples |= ("add random key" >> beam.Map(
lambda example: (uuid.uuid4(), example)))
examples |= ("group by key" >> beam.GroupByKey())
examples |= ("get shuffled values" >> beam.FlatMap(lambda t: t[1]))
return examples
# % cd ..
# ! ls
# ! pwd
# ! ( head -1000000 en.txt ; ) > million.txt
test=_create_examples_from_file(file_name='lines-aaa',file_id='lines-aaa')
for x in test:
print(x)
!wget http://nlp.stanford.edu/data/glove.6B.zip
!unzip glove.6B.zip
glove_dir = './'
embeddings_index = {} #initialize dictionary
f = open(os.path.join(glove_dir, 'glove.6B.100d.txt'))
for line in f:
values = line.split()
word = values[0]
coefs = np.asarray(values[1:], dtype='float32')
embeddings_index[word] = coefs
f.close()
print('Found %s word vectors.' % len(embeddings_index))
embedding_dim = 100
num_words= 72827
embedding_matrix = np.zeros((num_words, embedding_dim)) #create an array of zeros with word_num rows and embedding_dim columns
for word, i in word_index.items():
embedding_vector = embeddings_index.get(word)
if i < num_words:
if embedding_vector is not None:
# Words not found in embedding index will be all-zeros.
embedding_matrix[i] = embedding_vector
from keras import initializers, models, regularizers
from keras.layers import Dense, Dropout, Embedding, SeparableConv1D, MaxPooling1D, GlobalAveragePooling1D
toyModel = models.Sequential()
toyModel.add(Embedding(num_words,
embedding_dim,
input_length=max_len,
weights=[embedding_matrix],
trainable=False))
| 0.535341 | 0.21767 |
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib notebook
```
# Real World Considerations for the Lomb-Scargle Periodogram
**Version 0.2**
* * *
By AA Miller (Northwestern/CIERA)
23 Sep 2021
In [Lecture III](https://github.com/LSSTC-DSFP/LSSTC-DSFP-Sessions/blob/main/Session13/Day3/IntroductionToTheLombScarglePeriodogram.ipynb) we built the software necessary to estimate the power spectrum via the Lomb-Scargle periodogram.
We also discovered that LS is somewhat slow. We will now leverage the faster implementation in `astropy`, while exploring some specific challenges related to real astrophysical light curves.
The helper functions from [Lecture III](https://github.com/LSSTC-DSFP/LSSTC-DSFP-Sessions/blob/main/Session13/Day3/IntroductionToTheLombScarglePeriodogram.ipynb) are recreated at the end of this notebook - execute those cells and then the cell below to recreate the simulated data from Lecture III.
```
def gen_periodic_data(x, period=1, amplitude=1, phase=0, noise=0):
'''Generate periodic data given the function inputs
y = A*sin(2*pi*x/p - phase) + noise
Parameters
----------
x : array-like
input values to evaluate the array
period : float (default=1)
period of the periodic signal
amplitude : float (default=1)
amplitude of the periodic signal
phase : float (default=0)
phase offset of the periodic signal
noise : float (default=0)
variance of the noise term added to the periodic signal
Returns
-------
y : array-like
Periodic signal evaluated at all points x
'''
y = amplitude*np.sin(2*np.pi*x/period - phase) # complete
dy = np.random.normal(0, np.sqrt(noise), size=len(x)) # complete
return y + dy
def ls_periodogram(y, y_unc, x, f_grid):
psd = np.empty_like(f_grid)
chi2_0 = np.sum(((y - np.mean(y))/y_unc)**2)
for f_num, f in enumerate(f_grid):
psd[f_num] = 0.5*(chi2_0 - min_chi2([0,0], y, y_unc, x, f))
return psd
np.random.seed(185)
# calculate the periodogram
x = 10*np.random.rand(100)
y = gen_periodic_data(x, period=5.25, amplitude=7.4, noise=0.8)
y_unc = np.ones_like(x)*np.sqrt(0.8)
```
## Problem 1) Other Considerations and Faster Implementations
While our "home-grown" `ls_periodogram` works, it would take a loooooong time to evaluate $\sim4\times 10^5$ frequencies for $\sim2\times 10^7$ variable LSST sources. (as is often the case...) `astropy` to the rescue!
**Problem 1a**
[`LombScargle`](https://docs.astropy.org/en/stable/timeseries/lombscargle.html) in `astropy.timeseries` is fast. Run it below to compare to `ls_periodogram`.
```
from astropy.timeseries import LombScargle
frequency, power = LombScargle(x, y, y_unc).autopower()
```
Unlike `ls_periodogram`, `LombScargle` effectively takes no time to run on the simulated data.
**Problem 1b**
Plot the periodogram for the simulated data.
```
fig, ax = plt.subplots()
ax.plot(frequency, power)
ax.set_xlim(0,15)
# complete
# complete
fig.tight_layout()
```
There are many choices regarding the calculation of the periodogram, so [read the docs](http://docs.astropy.org/en/stable/api/astropy.stats.LombScargle.html#astropy.stats.LombScargle).
### Floating Mean Periodogram
A basic assumption that we preivously made is that the data are "centered" - in other words, our model explicitly assumes that the signal oscillates about a mean of 0.
For astronomical applications, this assumption can be harmful. Instead, it is useful to fit for the mean of the signal in addition to the periodic component (as is the default in `LombScargle`):
$$y(t;f) = y_0(f) + A_f \sin(2\pi f(t - \phi_f).$$
To illustrate why this is important for astronomy, assume that any signal fainter than $-2$ in our simulated data cannot be detected.
**Problem 1c**
Remove the observations from `x` and `y` where $y \le -2$ and calculate the periodogram both with and without fitting the mean (`fit_mean = False` in the call to `LombScargle`). Plot the periodograms. Do both methods recover the correct period?
```
x = 10*np.random.rand(100)
y = gen_periodic_data(x, period=5.25, amplitude=7.4, noise=0.8)
y_unc = np.ones_like(x)*np.sqrt(0.8)
mask = y > -2
bright = mask
xmask = x[mask]
ymask = y[mask]
y_uncmask = y_unc[mask]
freq_no_mean, power_no_mean = LombScargle(xmask, ymask, y_uncmask, fit_mean = False).autopower()
freq_fit_mean, power_fit_mean = LombScargle(xmask, ymask, y_uncmask).autopower()
fig, (ax1, ax2) = plt.subplots(2, 1, sharex=True)
ax1.plot(freq_no_mean, power_no_mean) # complete
ax2.plot(freq_fit_mean, power_fit_mean) # complete
ax1.set_xlim(0,15)
fig.tight_layout()
```
We can see that the best fit model doesn't match the signal in the case where we do not allow a floating mean.
```
fit_mean_model = LombScargle(x[bright], y[bright], y_unc[bright],
fit_mean=True).model(np.linspace(0,10,1000),
freq_fit_mean[np.argmax(power_fit_mean)])
no_mean_model = LombScargle(x[bright], y[bright], y_unc[bright],
fit_mean=False).model(np.linspace(0,10,1000),
freq_no_mean[np.argmax(power_no_mean)])
fig, ax = plt.subplots()
ax.errorbar(x[bright], y[bright], y_unc[bright], fmt='o', label='data')
ax.plot(np.linspace(0,10,1000), fit_mean_model, label='fit mean')
ax.plot(np.linspace(0,10,1000), no_mean_model, label='no mean')
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.legend()
fig.tight_layout()
```
### Window Functions
Recall that the convolution theorem tells us that:
$$\mathcal{F}[f\cdot g] = \mathcal{F}(f) \ast \mathcal{F}(g)$$
Telescope observations are effectively the product of a continous signal with several delta functions (corresponding to the times of observations). As a result, the convolution that produces the periodogram will retain signal from both the source and the observational cadence.
To illustrate this effect, let us simulate "realistic" observations for a 10 year telescope survey. We do this by assuming that a source is observed every 3 nights (the LSST cadence) within $\pm 4\,\mathrm{hr}$ of the same time, and that $\sim 30\%$ of the observations did not occur due to bad weather. We further assume that the source cannot be observed for 40% of the year because it is behind the sun.
Simulate a periodic signal with this cadence, a period = 220 days (typical for Miras), amplitude = 12.4, and noise = 1. Plot the simulated light curve.
**Problem 1d**
Simulate a periodic signal with 3 day cadence (and the observing conditions described above), a period = 220 days (typical for Miras), amplitude = 12.4, and variance of the noise = 1. Plot the simulated light curve.
```
# set up simulated observations
t_obs = np.arange(0, 10*365, 3, dtype = float) # 3d cadence
t_obs += np.random.normal(0, scale = 1/6, size = len(t_obs))
t_sun = np.ones(365)
t_sun[0:147] = 0
t_sun = np.array(list(t_sun) * 10, dtype = bool)[::3]
period = 220
amplitude = 12.4
noise = 1
t = np.linspace(0, 3650, len(t_sun))
y_obs = gen_periodic_data(t, period = period, amplitude = amplitude, noise = noise)
# Sun mask incl. random uncertainty
t_obs = t_obs[t_sun]
y_obs = y_obs[t_sun]
# Weather mask
w_mask = np.ones(len(t_obs), dtype = bool)
nights_lost = int(0.3*len(t_obs))
lost_ind = np.random.choice(np.arange(len(t_obs)), size = nights_lost, replace = False)
w_mask[lost_ind] = 0
# Final mask
t_obs_f = t_obs[w_mask]
y_obs_f = y_obs[w_mask]
#(x, period=1, amplitude=1, phase=0, noise=0)
y = gen_periodic_data(t, period = period, amplitude = amplitude, noise = noise)
fig, ax = plt.subplots()
ax.errorbar(t_obs_f, y_obs_f, yerr = 1.0, label = "Flux", fmt = '.')#, xerr = 1/6)# complete
ax.plot(t, y, zorder = 0, label = "Signal")
ax.plot(t_obs[~w_mask], y_obs[~w_mask], 'x')
ax.legend()
ax.set_xlabel("Time (d)")
ax.set_ylabel("Flux (arbitrary units)")
```
**Problem 1e**
Calculate and plot the periodogram for the window function (i.e., set `y = 1` in `LombScargle`) of the observations. Do you notice any significant power?
*Hint* - you may need to zoom in on the plot to see all the relevant features.
```
ls = LombScargle(t_obs, 1, fit_mean=False).autopower(maximum_frequency = 5)
freq_window, power_window = # complete
fig, ax = plt.subplots()
ax.plot( # complete
ax.set_ylabel("Power")
ax.set_xlabel("Period (d)")
ax.set_xlim(0,500)
axins = plt.axes([.2, .65, .5, .2])
axins.plot( # complete
axins.set_xlim(0,5)
```
Interestingly, there are very strong peaks in the data at $P \approx 3\,\mathrm{d} \;\&\; 365\,\mathrm{d}$.
What is this telling us? Essentially that observations are likely to be repeated at intervals of 3 or 365 days (shorter period spikes are aliases of the 3 d peak).
This is important to understand, however, because this same power will be present in the periodogram where we search for the periodic signal.
**Problem 1f**
Calculate the periodogram for the data and compare it to the periodogram for the window function.
```
ls = LombScargle( # complete
frequency, power = # complete
fig, (ax,ax2) = plt.subplots(2,1, sharex=True)
ax.plot( # complete
ax.set_ylabel("Power")
ax.set_ylim(0,1)
ax2.plot( # complete
ax2.set_ylabel("Power")
ax2.set_xlabel("Period (d)")
ax2.set_xlim(0,10)
fig.tight_layout()
```
### Uncertainty on the best-fit period
How do we report uncertainties on the best-fit period from LS? For example, for the previously simulated LSST light curve we would want to report something like $P = 102 \pm 4\,\mathrm{d}$. However, the uncertainty from LS periodograms cannot be determined in this way.
Naively, one could report the width of the peak in the periodogram as the uncertainty in the fit. However, we previously saw that the peak width $\propto 1/T$ (the peak width does not decrease as the number of observations or their S/N increases; see Vander Plas 2017). Reporting such an uncertainty is particularly ridiculous for long duration surveys, whereby the peaks become very very narrow.
An alternative approach is to report the False Alarm Probability (FAP), which estimates the probability that a dataset with no periodic signal could produce a peak of similar magnitude, due to random gaussian fluctuations, as the data.
There are a few different methods to calculate the FAP. Perhaps the most useful, however, is the bootstrap method. To obtain a bootstrap estimate of the LS FAP one leaves the observation times fixed, and then draws new observation values with replacement from the actual set of observations. This procedure is then repeated many times to determine the FAP.
One nice advantage of this procedure is that any effects due to the window function will be imprinted in each iteration of the bootstrap resampling.
The major disadvantage is that many many periodograms must be calculated. The rule of thumb is that to acieve a FAP $= p_\mathrm{false}$, one must run $n_\mathrm{boot} \approx 10/p_\mathrm{false}$ bootstrap periodogram calculations. Thus, an FAP $\approx 0.1\%$ requires an increase of 1000 in computational time.
`LombScargle` provides the [`false_alarm_probability`](One nice advantage of this procedure is that any effects due to the window function will be imprinted in each iteration of the bootstrap resampling. The major disadvantage is that many many periodograms must be calculated) method, including a bootstrap option. We skip that for now in the interest of time.
As a final note of caution - be weary of over-interpreting the FAP. The specific question answered by the FAP is, what is the probability that gaussian fluctations could produce a signal of equivalent magnitude? Whereas, the question we generally want to answer is: did a periodic signal produce these data?
These questions are very different, and thus, the FAP cannot be used to *prove* that a source is periodic.
## Problem 2) Real-world considerations
We have covered many, though not all, considerations that are necessary when employing a Lomb Scargle periodogram. We have not yet, however, encountered real world data. Here we highlight some of the issues associated with astronomical light curves.
We will now use LS to analyze actual data from the [All Sky Automated Survey (ASAS)](http://www.astrouw.edu.pl/asas/?page=catalogues). Download the [example light curve](https://northwestern.box.com/s/rclcz4lkcdfjn4829p8pa5ddfmcyd0gm).
**Problem 2a**
Read in the light curve from example_asas_lc.dat. Plot the light curve.
*Hint* - I recommend using `astropy Tables` or `pandas dataframe`.
```
data = # complete
fig, ax = plt.subplots()
ax.errorbar( # complete
ax.set_xlabel('HJD (d)')
ax.set_ylabel('V (mag)')
ax.set_ylim(ax.get_ylim()[::-1])
fig.tight_layout()
```
**Problem 2b**
Use `LombScargle` to measure the periodogram. Then plot the periodogram and the phase folded light curve at the best-fit period.
*Hint* - search periods longer than 2 hr.
```
frequency, power = # complete
# complete
fig,ax = plt.subplots()
ax.plot(# complete
ax.set_ylabel("Power")
ax.set_xlabel("Period (d)")
ax.set_xlim(0, 800)
axins = plt.axes([.25, .55, .6, .3])
axins.plot( # complete
axins.set_xlim(0,5)
fig.tight_layout()
# plot the phase folded light curve
phase_plot( # complete
```
**Problem 2c**
Now plot the light curve folded at twice the best LS period.
Which of these 2 is better?
```
phase_plot( # complete
```
Herein lies a fundamental issue regarding the LS periodogram: the model does not search for "periodicity." The LS model asks if the data support a sinusoidal signal. As astronomers we typically assume this question is good enough, but as we can see in the example of this eclipsing binary that is not the case [and this is not limited to eclipsing binaries].
We can see why LS is not sufficient for an EB by comparing the model to the phase-folded light curve:
**Problem 2d**
Overplot the model on top of the phase folded light curve.
*Hint* – you can access the best LS fit via the [`.model()`](https://docs.astropy.org/en/stable/timeseries/lombscargle.html#the-lomb-scargle-model) method on `LombScargle` objects in astropy.
```
phase_plot( # complete
phase_grid = np.linspace(0,1,1000)
plt.plot( # complete
plt.tight_layout()
```
One way to combat this very specific issue is to include more Fourier terms at the harmonic of the best fit period. This is easy to implement in `LombScargle` with the `nterms` keyword. [Though always be weary of adding degrees of freedom to a model, especially at the large pipeline level of analysis.]
**Problem 2e**
Calculate the LS periodogram for the eclipsing binary, with `nterms` = 1, 2, 3, 4, 5. Report the best-fit period for each of these models.
*Hint* - we have good reason to believe that the best fit frequency is < 3 in this case, so set `maximum_frequency = 3`.
```
for i in np.arange(1,6):
frequency, power = # complete
# complete
print('For {:d} harmonics, P_LS = {:.8f}'.format( # complete
```
Interestingly, for the $n=2, 3, 4$ harmonics, it appears as though we get the period that we have visually confirmed. However, by $n=5$ harmonics we no longer get a reasonable answer. Again - be very careful about adding harmonics, especially in large analysis pipelines.
**Problem 2f**
Plot the the $n = 4$ model on top of the light curve folded at the correct period.
```
best_period = # complete
phase_plot( # complete
phase_grid = np.linspace(0,1,1000)
plt.plot( # complete
plt.tight_layout()
```
This example also shows why it is somewhat strange to provide an uncertainty with a LS best-fit period. Errors tend to be catastrophic, and not some small fractional percentage, with the LS periodogram.
In the case of the above EB, the "best-fit" period was off by a factor 2. This is not isolated to EBs, however, LS periodograms frequently identify a correct harmonic of the true period, but not the actual period of variability.
## Problem 3) The "Real" World
**Problem 3a**
Re-write `gen_periodic_data` to create periodic signals using the first 4 harmonics in a Fourier series. The $n > 1$ harmonics should have random phase offsets, and the amplitude of the $n > 1$ harmonics should be drawn randomly from a uniform distribution between 0 and `amplitude` the amplitude of the first harmonic.
```
def gen_periodic_data(x, period=1, amplitude=1, phase=0, noise=0):
'''Generate periodic data
Parameters
----------
x : array-like
input values to evaluate the array
period : float (default=1)
period of the periodic signal
amplitude : float (default=1)
amplitude of the periodic signal
phase : float (default=0)
phase offset of the periodic signal
noise : float (default=0)
variance of the noise term added to the periodic signal
Returns
-------
y : array-like
Periodic signal evaluated at all points x
'''
y1 = # complete
amp2 = # complete
phase2 = # complete
y2 = # complete
amp3 = # complete
phase3 = # complete
y3 = # complete
amp4 = # complete
phase4 = # complete
y4 = # complete
dy = # complete
return y1 + y2 + y3 + y4 + dy
```
**Problem 3b**
Confirm the updated version of `gen_periodic_data` works by creating a phase plot for a simulated signal with `amplitude = 4`, `period = 1.234`, and `noise=0.81`, and 100 observations obtained on a regular grid from 0 to 50.
```
np.random.seed(185)
x_grid = np.linspace( # complete
y = gen_periodic_data( # complete
phase_plot( # complete
```
**Problem 3c**
Simulate 1000 "realistic" light curves using the astronomical cadence from **1d** for a full survey duration of 2 years.
For each light curve draw the period randomly from [0.2, 10], and the amplitude randomly from [1, 5], and the noise randomly from [1,2].
Record the period in an array `true_p` and estimate the period for the simulated data via LS and store the result in an array `ls_p`.
```
n_lc = # complete
true_p = np.zeros(n_lc)
ls_p = np.zeros_like(true_p)
for lc in range(n_lc):
# set up simulated observations
t_obs = np.arange(0, 2*365, 3) # 3d cadence
# complete
# complete
# complete
period = # complete
true_p[lc] = # complete
amp = # complete
noise = # complete
y = gen_periodic_data( # complete
freq, power = LombScargle( # complete
ls_p[lc] = 1/freq[np.argmax(power)]
```
**Problem 3d**
Plot the LS recovered period vs. the true period for the simulated sources.
Do you notice anything interesting? Do you manage to recover the correct period most of the time?
```
fig, ax = plt.subplots()
ax.plot( # complete
ax.set_ylim(0, 20)
ax.set_xlabel('True period (d)')
ax.set_ylabel('LS peak (d)')
fig.tight_layout()
```
**Problem 3e**
For how many of the simulated sources do you recover the correct period? Consider a period estimate "correct" if the LS estimate is within 10% of the true period.
```
# complete
```
The results are a bit disappointing. However, it is also clear that there is strong structure in the plot off the 1:1 line. That structure can be understood in terms of the window function that was discussed in **1d**, **1e**, and **1f**.
**Problem 3f**
Recreate the plot in **3d** and overplot the line
$$P_\mathrm{LS} = \left(\frac{1}{P_\mathrm{true}} + \frac{n}{3}\right)^{-1}$$
for $n = -2, -1, 1, 2$.
*Hint* - only plot the values where $P_\mathrm{LS} > 0$ since, by definition, we do not search for negative periods.
```
p_grid = np.linspace(1e-1,10,100)
fig, ax = plt.subplots()
ax.plot(# complete
# complete
# complete
# complete
ax.set_ylim(0, 9)
ax.set_xlabel('True period (d)')
ax.set_ylabel('LS peak (d)')
fig.tight_layout()
```
What in the...
We see that these lines account for a lot of the off-diagonal structure in this plot.
In this case, what is happening is that the true frequency of the signal $f_\mathrm{true}$ is being aliased by the window function and it's harmonics. In other words LS is pulling out $f_\mathrm{true} + n\delta{f}$, where $n$ is an integer and $\delta{f}$ is the observational cadence $3\,\mathrm{d}$. Many of the false positives can be explained via the window function.
Similarly, LS might be recovering higher order harmonics of the true period since we aren't trying to recover pure sinusoidal signals in this simulation. These harmonics would also be aliased by the window function, so LS will pull out $f_\mathrm{true}/m + n\delta{f}$, where $m$ is a postive integer.
**Problem 3g**
Recreate the plot in **3d** and overplot lines for the $m = 2$ harmonic aliased with $n = -1, 1, 2$.
*Hint* - only plot the values where $P_\mathrm{LS} > 0$ since, by definition, we do not search for negative periods.
```
p_grid = np.linspace(1e-1,10,100)
fig, ax = plt.subplots()
ax.plot(# complete
# complete
# complete
# complete
ax.set_ylim(0, 2)
ax.set_xlabel('True period (d)')
ax.set_ylabel('LS peak (d)')
fig.tight_layout()
```
The last bit of structure can be understood via the symmetry in the LS periodogram about 0. In particular, if there is an aliased frequency that is less than 0, which will occur for $n < 0$ in the equations above, then there will also be power at the positive value of that frequency. In other words, LS will pull out $|f_\mathrm{true}/m + n\delta{f}|$.
**Problem 3h**
Recreate the plot in **3d** and overplot lines for the "reflected" $m = 1$ harmonic aliased with $n = -3, -2, -1$.
*Hint* - only plot the values where $P_\mathrm{LS} < 0$ since we are looking for "reflected" peaks in the periodogram in this case.
```
p_grid = np.linspace(1e-1,10,1000)
fig, ax = plt.subplots()
ax.plot(# complete
# complete
# complete
# complete
ax.set_ylim(0, 15)
ax.set_xlabel('True period (d)')
ax.set_ylabel('LS peak (d)')
fig.tight_layout()
```
Now we have seen that nearly all the structure in the LS period vs. true period plot can be explained via aliasing with the window function! This is good (we understand why the results do not conform to what we expect), but also bad, (we were not able to recover the correct period for the majority of our sources). Ultimately, this means - be careful when inspecting the results of the LS periodogram as the peaks aren't driven solely by the signal from the source in question!
(If only there were some way to get rid of the sun, then we'd never have these problems...)
## Conclusions
The Lomb-Scargle periodogram is a useful tool to search for *sinusoidal* signals in noisy, irregular data.
However, as highlighted throughout, there are many ways in which the methodology can run awry.
In closing, I will summarize some practical considerations from VanderPlas (2017):
1. Choose an appropriate frequency grid (defaults in `LombScargle` are not sufficient)
2. Calculate the LS periodogram for the observation times to search for dominant signals (e.g., 1 day in astro)
3. Compute LS periodogram for data (avoid multi-Fourier models if signal unknown)
4. Plot periodogram and various FAP levels (do not over-interpret FAP)
5. If window function shows strong aliasing, plot phased light curve at each peak (now add more Fourier terms if necessary)
6. If looking for a particular signal (e.g., detatched EBs), consider different methods that better match expected signal
7. Inject fake signals into data to understand systematics if using LS in a survey pipeline
### Finally, Finally
As a very last note: know that there are many different ways to search for periodicity in astronomical data. Depending on your application (and computational resources), LS may be a poor choice (even though this is often the default choice by all astronomers!) [Graham et al. (2013)](http://adsabs.harvard.edu/abs/2013MNRAS.434.3423G) provides a summary of several methods using actual astronomical data. The results of that study show that no single method is best. However, they also show that no single method performs particularly well: the detection efficiences in Graham et al. (2013) are disappointing given the importance of periodicity in astronomical signals.
Period detection is a fundamental problem for astronomical time-series, but it is especially difficult in "production" mode. Be careful when setting up pipelines to analyze large datasets.
**Challenge Problem**
Re-create problem 4, but include additional terms in the fit with the LS periodogram. What differences do you notice when comparing the true period to the best-fit LS periods?
## Helper Functions
We developed useful helper functions as part of [Lecture III](https://github.com/LSSTC-DSFP/LSSTC-DSFP-Sessions/tree/main/Session13/Day3/IntroductionToTheLombScarglePeriodogram.ipynb) from this session.
These functions generate periodic data, and phase fold light curves on a specified period. These functions will once again prove useful, so we include them here in order to simulate data above.
**Helper 1**
Create a function, `gen_periodic_data`, that creates simulated data (including noise) over a grid of user supplied positions:
$$ y = A\,cos\left(\frac{x}{P} - \phi\right) + \sigma_y$$
where $A, P, \phi$ are inputs to the function. `gen_periodic_data` should include Gaussian noise, $\sigma_y$, for each output $y_i$.
```
def gen_periodic_data(x, period=1, amplitude=1, phase=0, noise=0):
'''Generate periodic data given the function inputs
y = A*cos(x/p - phase) + noise
Parameters
----------
x : array-like
input values to evaluate the array
period : float (default=1)
period of the periodic signal
amplitude : float (default=1)
amplitude of the periodic signal
phase : float (default=0)
phase offset of the periodic signal
noise : float (default=0)
variance of the noise term added to the periodic signal
Returns
-------
y : array-like
Periodic signal evaluated at all points x
'''
y = amplitude*np.sin(2*np.pi*x/(period) - phase) + np.random.normal(0, np.sqrt(noise), size=len(x))
return y
```
**Helper 2**
Create a function, `phase_plot`, that takes x, y, and $P$ as inputs to create a phase-folded light curve (i.e., plot the data at their respective phase values given the period $P$).
Include an optional argument, `y_unc`, to include uncertainties on the `y` values, when available.
```
def phase_plot(x, y, period, y_unc = 0.0, mag_plot=False):
'''Create phase-folded plot of input data x, y
Parameters
----------
x : array-like
data values along abscissa
y : array-like
data values along ordinate
period : float
period to fold the data
y_unc : array-like
uncertainty of the
'''
phases = (x/period) % 1
if isinstance(y_unc, (np.floating, float)):
y_unc = np.ones_like(x)*y_unc
plot_order = np.argsort(phases)
fig, ax = plt.subplots()
ax.errorbar(phases[plot_order], y[plot_order], y_unc[plot_order],
fmt='o', mec="0.2", mew=0.1)
ax.set_xlabel("phase")
ax.set_ylabel("signal")
if mag_plot:
ax.set_ylim(ax.get_ylim()[::-1])
fig.tight_layout()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib notebook
def gen_periodic_data(x, period=1, amplitude=1, phase=0, noise=0):
'''Generate periodic data given the function inputs
y = A*sin(2*pi*x/p - phase) + noise
Parameters
----------
x : array-like
input values to evaluate the array
period : float (default=1)
period of the periodic signal
amplitude : float (default=1)
amplitude of the periodic signal
phase : float (default=0)
phase offset of the periodic signal
noise : float (default=0)
variance of the noise term added to the periodic signal
Returns
-------
y : array-like
Periodic signal evaluated at all points x
'''
y = amplitude*np.sin(2*np.pi*x/period - phase) # complete
dy = np.random.normal(0, np.sqrt(noise), size=len(x)) # complete
return y + dy
def ls_periodogram(y, y_unc, x, f_grid):
psd = np.empty_like(f_grid)
chi2_0 = np.sum(((y - np.mean(y))/y_unc)**2)
for f_num, f in enumerate(f_grid):
psd[f_num] = 0.5*(chi2_0 - min_chi2([0,0], y, y_unc, x, f))
return psd
np.random.seed(185)
# calculate the periodogram
x = 10*np.random.rand(100)
y = gen_periodic_data(x, period=5.25, amplitude=7.4, noise=0.8)
y_unc = np.ones_like(x)*np.sqrt(0.8)
from astropy.timeseries import LombScargle
frequency, power = LombScargle(x, y, y_unc).autopower()
fig, ax = plt.subplots()
ax.plot(frequency, power)
ax.set_xlim(0,15)
# complete
# complete
fig.tight_layout()
x = 10*np.random.rand(100)
y = gen_periodic_data(x, period=5.25, amplitude=7.4, noise=0.8)
y_unc = np.ones_like(x)*np.sqrt(0.8)
mask = y > -2
bright = mask
xmask = x[mask]
ymask = y[mask]
y_uncmask = y_unc[mask]
freq_no_mean, power_no_mean = LombScargle(xmask, ymask, y_uncmask, fit_mean = False).autopower()
freq_fit_mean, power_fit_mean = LombScargle(xmask, ymask, y_uncmask).autopower()
fig, (ax1, ax2) = plt.subplots(2, 1, sharex=True)
ax1.plot(freq_no_mean, power_no_mean) # complete
ax2.plot(freq_fit_mean, power_fit_mean) # complete
ax1.set_xlim(0,15)
fig.tight_layout()
fit_mean_model = LombScargle(x[bright], y[bright], y_unc[bright],
fit_mean=True).model(np.linspace(0,10,1000),
freq_fit_mean[np.argmax(power_fit_mean)])
no_mean_model = LombScargle(x[bright], y[bright], y_unc[bright],
fit_mean=False).model(np.linspace(0,10,1000),
freq_no_mean[np.argmax(power_no_mean)])
fig, ax = plt.subplots()
ax.errorbar(x[bright], y[bright], y_unc[bright], fmt='o', label='data')
ax.plot(np.linspace(0,10,1000), fit_mean_model, label='fit mean')
ax.plot(np.linspace(0,10,1000), no_mean_model, label='no mean')
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.legend()
fig.tight_layout()
# set up simulated observations
t_obs = np.arange(0, 10*365, 3, dtype = float) # 3d cadence
t_obs += np.random.normal(0, scale = 1/6, size = len(t_obs))
t_sun = np.ones(365)
t_sun[0:147] = 0
t_sun = np.array(list(t_sun) * 10, dtype = bool)[::3]
period = 220
amplitude = 12.4
noise = 1
t = np.linspace(0, 3650, len(t_sun))
y_obs = gen_periodic_data(t, period = period, amplitude = amplitude, noise = noise)
# Sun mask incl. random uncertainty
t_obs = t_obs[t_sun]
y_obs = y_obs[t_sun]
# Weather mask
w_mask = np.ones(len(t_obs), dtype = bool)
nights_lost = int(0.3*len(t_obs))
lost_ind = np.random.choice(np.arange(len(t_obs)), size = nights_lost, replace = False)
w_mask[lost_ind] = 0
# Final mask
t_obs_f = t_obs[w_mask]
y_obs_f = y_obs[w_mask]
#(x, period=1, amplitude=1, phase=0, noise=0)
y = gen_periodic_data(t, period = period, amplitude = amplitude, noise = noise)
fig, ax = plt.subplots()
ax.errorbar(t_obs_f, y_obs_f, yerr = 1.0, label = "Flux", fmt = '.')#, xerr = 1/6)# complete
ax.plot(t, y, zorder = 0, label = "Signal")
ax.plot(t_obs[~w_mask], y_obs[~w_mask], 'x')
ax.legend()
ax.set_xlabel("Time (d)")
ax.set_ylabel("Flux (arbitrary units)")
ls = LombScargle(t_obs, 1, fit_mean=False).autopower(maximum_frequency = 5)
freq_window, power_window = # complete
fig, ax = plt.subplots()
ax.plot( # complete
ax.set_ylabel("Power")
ax.set_xlabel("Period (d)")
ax.set_xlim(0,500)
axins = plt.axes([.2, .65, .5, .2])
axins.plot( # complete
axins.set_xlim(0,5)
ls = LombScargle( # complete
frequency, power = # complete
fig, (ax,ax2) = plt.subplots(2,1, sharex=True)
ax.plot( # complete
ax.set_ylabel("Power")
ax.set_ylim(0,1)
ax2.plot( # complete
ax2.set_ylabel("Power")
ax2.set_xlabel("Period (d)")
ax2.set_xlim(0,10)
fig.tight_layout()
data = # complete
fig, ax = plt.subplots()
ax.errorbar( # complete
ax.set_xlabel('HJD (d)')
ax.set_ylabel('V (mag)')
ax.set_ylim(ax.get_ylim()[::-1])
fig.tight_layout()
frequency, power = # complete
# complete
fig,ax = plt.subplots()
ax.plot(# complete
ax.set_ylabel("Power")
ax.set_xlabel("Period (d)")
ax.set_xlim(0, 800)
axins = plt.axes([.25, .55, .6, .3])
axins.plot( # complete
axins.set_xlim(0,5)
fig.tight_layout()
# plot the phase folded light curve
phase_plot( # complete
phase_plot( # complete
phase_plot( # complete
phase_grid = np.linspace(0,1,1000)
plt.plot( # complete
plt.tight_layout()
for i in np.arange(1,6):
frequency, power = # complete
# complete
print('For {:d} harmonics, P_LS = {:.8f}'.format( # complete
best_period = # complete
phase_plot( # complete
phase_grid = np.linspace(0,1,1000)
plt.plot( # complete
plt.tight_layout()
def gen_periodic_data(x, period=1, amplitude=1, phase=0, noise=0):
'''Generate periodic data
Parameters
----------
x : array-like
input values to evaluate the array
period : float (default=1)
period of the periodic signal
amplitude : float (default=1)
amplitude of the periodic signal
phase : float (default=0)
phase offset of the periodic signal
noise : float (default=0)
variance of the noise term added to the periodic signal
Returns
-------
y : array-like
Periodic signal evaluated at all points x
'''
y1 = # complete
amp2 = # complete
phase2 = # complete
y2 = # complete
amp3 = # complete
phase3 = # complete
y3 = # complete
amp4 = # complete
phase4 = # complete
y4 = # complete
dy = # complete
return y1 + y2 + y3 + y4 + dy
np.random.seed(185)
x_grid = np.linspace( # complete
y = gen_periodic_data( # complete
phase_plot( # complete
n_lc = # complete
true_p = np.zeros(n_lc)
ls_p = np.zeros_like(true_p)
for lc in range(n_lc):
# set up simulated observations
t_obs = np.arange(0, 2*365, 3) # 3d cadence
# complete
# complete
# complete
period = # complete
true_p[lc] = # complete
amp = # complete
noise = # complete
y = gen_periodic_data( # complete
freq, power = LombScargle( # complete
ls_p[lc] = 1/freq[np.argmax(power)]
fig, ax = plt.subplots()
ax.plot( # complete
ax.set_ylim(0, 20)
ax.set_xlabel('True period (d)')
ax.set_ylabel('LS peak (d)')
fig.tight_layout()
# complete
p_grid = np.linspace(1e-1,10,100)
fig, ax = plt.subplots()
ax.plot(# complete
# complete
# complete
# complete
ax.set_ylim(0, 9)
ax.set_xlabel('True period (d)')
ax.set_ylabel('LS peak (d)')
fig.tight_layout()
p_grid = np.linspace(1e-1,10,100)
fig, ax = plt.subplots()
ax.plot(# complete
# complete
# complete
# complete
ax.set_ylim(0, 2)
ax.set_xlabel('True period (d)')
ax.set_ylabel('LS peak (d)')
fig.tight_layout()
p_grid = np.linspace(1e-1,10,1000)
fig, ax = plt.subplots()
ax.plot(# complete
# complete
# complete
# complete
ax.set_ylim(0, 15)
ax.set_xlabel('True period (d)')
ax.set_ylabel('LS peak (d)')
fig.tight_layout()
def gen_periodic_data(x, period=1, amplitude=1, phase=0, noise=0):
'''Generate periodic data given the function inputs
y = A*cos(x/p - phase) + noise
Parameters
----------
x : array-like
input values to evaluate the array
period : float (default=1)
period of the periodic signal
amplitude : float (default=1)
amplitude of the periodic signal
phase : float (default=0)
phase offset of the periodic signal
noise : float (default=0)
variance of the noise term added to the periodic signal
Returns
-------
y : array-like
Periodic signal evaluated at all points x
'''
y = amplitude*np.sin(2*np.pi*x/(period) - phase) + np.random.normal(0, np.sqrt(noise), size=len(x))
return y
def phase_plot(x, y, period, y_unc = 0.0, mag_plot=False):
'''Create phase-folded plot of input data x, y
Parameters
----------
x : array-like
data values along abscissa
y : array-like
data values along ordinate
period : float
period to fold the data
y_unc : array-like
uncertainty of the
'''
phases = (x/period) % 1
if isinstance(y_unc, (np.floating, float)):
y_unc = np.ones_like(x)*y_unc
plot_order = np.argsort(phases)
fig, ax = plt.subplots()
ax.errorbar(phases[plot_order], y[plot_order], y_unc[plot_order],
fmt='o', mec="0.2", mew=0.1)
ax.set_xlabel("phase")
ax.set_ylabel("signal")
if mag_plot:
ax.set_ylim(ax.get_ylim()[::-1])
fig.tight_layout()
| 0.788461 | 0.934035 |
# Deep Learning for Predictive Maintenance
Deep learning has proven to show superior performance in certain domains such as object recognition and image classification. It has also gained popularity in domains such as finance where time-series data plays an important role. Predictive Maintenance is also a domain where data is collected over time to monitor the state of an asset with the goal of finding patterns to predict failures which can also benefit from certain deep learning algorithms. Among the deep learning methods, Long Short Term Memory [(LSTM)](http://colah.github.io/posts/2015-08-Understanding-LSTMs/) networks are especially appealing to the predictive maintenance domain due to the fact that they are very good at learning from sequences. This fact lends itself to their applications using time series data by making it possible to look back for longer periods of time to detect failure patterns. In this notebook, we build an LSTM network for the data set and scenerio described at [Predictive Maintenance Template](https://gallery.cortanaintelligence.com/Collection/Predictive-Maintenance-Template-3) to predict remaining useful life of aircraft engines. In summary, the template uses simulated aircraft sensor values to predict when an aircraft engine will fail in the future so that maintenance can be planned in advance.
This notebook serves as a tutorial for beginners looking to apply deep learning in predictive maintenance domain and uses a simple scenario where only one data source (sensor values) is used to make predictions. In more advanced predictive maintenance scenarios such as in [Predictive Maintenance Modelling Guide](https://gallery.cortanaintelligence.com/Notebook/Predictive-Maintenance-Modelling-Guide-R-Notebook-1), there are many other data sources (i.e. historical maintenance records, error logs, machine and operator features etc.) which may require different types of treatments to be used in the deep learning networks. Since predictive maintenance is not a typical domain for deep learning, its application is an open area of research.
This notebook uses [keras](https://keras.io/) deep learning library with Microsoft Cognitive Toolkit [CNTK](https://docs.microsoft.com/en-us/cognitive-toolkit/Using-CNTK-with-Keras) as backend.
```
import keras
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# Setting seed for reproducability
np.random.seed(1234)
PYTHONHASHSEED = 0
from sklearn import preprocessing
from sklearn.metrics import confusion_matrix, recall_score, precision_score
from keras.models import Sequential
from keras.layers import Dense, Dropout, LSTM, Activation
%matplotlib inline
```
## Data Ingestion
In the following section, we ingest the training, test and ground truth datasets from azure storage. The training data consists of multiple multivariate time series with "cycle" as the time unit, together with 21 sensor readings for each cycle. Each time series can be assumed as being generated from a different engine of the same type. The testing data has the same data schema as the training data. The only difference is that the data does not indicate when the failure occurs. Finally, the ground truth data provides the number of remaining working cycles for the engines in the testing data. You can find more details about the type of data used for this notebook at [Predictive Maintenance Template](https://gallery.cortanaintelligence.com/Collection/Predictive-Maintenance-Template-3).
```
# Data ingestion - reading the datasets from Azure blob
!wget http://azuremlsamples.azureml.net/templatedata/PM_train.txt
!wget http://azuremlsamples.azureml.net/templatedata/PM_test.txt
!wget http://azuremlsamples.azureml.net/templatedata/PM_truth.txt
# read training data
train_df = pd.read_csv('PM_train.txt', sep=" ", header=None)
train_df.drop(train_df.columns[[26, 27]], axis=1, inplace=True)
train_df.columns = ['id', 'cycle', 'setting1', 'setting2', 'setting3', 's1', 's2', 's3',
's4', 's5', 's6', 's7', 's8', 's9', 's10', 's11', 's12', 's13', 's14',
's15', 's16', 's17', 's18', 's19', 's20', 's21']
# read test data
test_df = pd.read_csv('PM_test.txt', sep=" ", header=None)
test_df.drop(test_df.columns[[26, 27]], axis=1, inplace=True)
test_df.columns = ['id', 'cycle', 'setting1', 'setting2', 'setting3', 's1', 's2', 's3',
's4', 's5', 's6', 's7', 's8', 's9', 's10', 's11', 's12', 's13', 's14',
's15', 's16', 's17', 's18', 's19', 's20', 's21']
# read ground truth data
truth_df = pd.read_csv('PM_truth.txt', sep=" ", header=None)
truth_df.drop(truth_df.columns[[1]], axis=1, inplace=True)
train_df = train_df.sort_values(['id','cycle'])
train_df.head()
```
## Data Preprocessing
First step is to generate labels for the training data which are Remaining Useful Life (RUL), label1 and label2 as was done in the [Predictive Maintenance Template](https://gallery.cortanaintelligence.com/Collection/Predictive-Maintenance-Template-3). Here, we will only make use of "label1" for binary clasification, while trying to answer the question: is a specific engine going to fail within w1 cycles?
```
# Data Labeling - generate column RUL
rul = pd.DataFrame(train_df.groupby('id')['cycle'].max()).reset_index()
rul.columns = ['id', 'max']
train_df = train_df.merge(rul, on=['id'], how='left')
train_df['RUL'] = train_df['max'] - train_df['cycle']
train_df.drop('max', axis=1, inplace=True)
train_df.head()
# generate label columns for training data
w1 = 30
w0 = 15
train_df['label1'] = np.where(train_df['RUL'] <= w1, 1, 0 )
train_df['label2'] = train_df['label1']
train_df.loc[train_df['RUL'] <= w0, 'label2'] = 2
train_df.head()
```
In the [Predictive Maintenance Template](https://gallery.cortanaintelligence.com/Collection/Predictive-Maintenance-Template-3) , cycle column is also used for training so we will also include the cycle column. Here, we normalize the columns in the training data.
```
# MinMax normalization
train_df['cycle_norm'] = train_df['cycle']
cols_normalize = train_df.columns.difference(['id','cycle','RUL','label1','label2'])
min_max_scaler = preprocessing.MinMaxScaler()
norm_train_df = pd.DataFrame(min_max_scaler.fit_transform(train_df[cols_normalize]),
columns=cols_normalize,
index=train_df.index)
join_df = train_df[train_df.columns.difference(cols_normalize)].join(norm_train_df)
train_df = join_df.reindex(columns = train_df.columns)
train_df.head()
```
Next, we prepare the test data. We first normalize the test data using the parameters from the MinMax normalization applied on the training data.
```
test_df['cycle_norm'] = test_df['cycle']
norm_test_df = pd.DataFrame(min_max_scaler.transform(test_df[cols_normalize]),
columns=cols_normalize,
index=test_df.index)
test_join_df = test_df[test_df.columns.difference(cols_normalize)].join(norm_test_df)
test_df = test_join_df.reindex(columns = test_df.columns)
test_df = test_df.reset_index(drop=True)
test_df.head()
```
Next, we use the ground truth dataset to generate labels for the test data.
```
# generate column max for test data
rul = pd.DataFrame(test_df.groupby('id')['cycle'].max()).reset_index()
rul.columns = ['id', 'max']
truth_df.columns = ['more']
truth_df['id'] = truth_df.index + 1
truth_df['max'] = rul['max'] + truth_df['more']
truth_df.drop('more', axis=1, inplace=True)
# generate RUL for test data
test_df = test_df.merge(truth_df, on=['id'], how='left')
test_df['RUL'] = test_df['max'] - test_df['cycle']
test_df.drop('max', axis=1, inplace=True)
test_df.head()
# generate label columns w0 and w1 for test data
test_df['label1'] = np.where(test_df['RUL'] <= w1, 1, 0 )
test_df['label2'] = test_df['label1']
test_df.loc[test_df['RUL'] <= w0, 'label2'] = 2
test_df.head()
```
In the rest of the notebook, we train an LSTM network that we will compare to the results in [Predictive Maintenance Template Step 2B of 3](https://gallery.cortanaintelligence.com/Experiment/Predictive-Maintenance-Step-2B-of-3-train-and-evaluate-binary-classification-models-2) where a series of machine learning models are used to train and evaluate the binary classification model that uses column "label1" as the label.
## Modelling
The traditional predictive maintenance machine learning models are based on feature engineering which is manual construction of right features using domain expertise and similar methods. This usually makes these models hard to reuse since feature engineering is specific to the problem scenario and the available data which varies from one business to the other. Perhaps the most attractive part of applying deep learning in the predictive maintenance domain is the fact that these networks can automatically extract the right features from the data, eliminating the need for manual feature engineering.
When using LSTMs in the time-series domain, one important parameter to pick is the sequence length which is the window for LSTMs to look back. This may be viewed as similar to picking window_size = 5 cycles for calculating the rolling features in the [Predictive Maintenance Template](https://gallery.cortanaintelligence.com/Collection/Predictive-Maintenance-Template-3) which are rolling mean and rolling standard deviation for 21 sensor values. The idea of using LSTMs is to let the model extract abstract features out of the sequence of sensor values in the window rather than engineering those manually. The expectation is that if there is a pattern in these sensor values within the window prior to failure, the pattern should be encoded by the LSTM.
One critical advantage of LSTMs is their ability to remember from long-term sequences (window sizes) which is hard to achieve by traditional feature engineering. For example, computing rolling averages over a window size of 50 cycles may lead to loss of information due to smoothing and abstracting of values over such a long period, instead, using all 50 values as input may provide better results. While feature engineering over large window sizes may not make sense, LSTMs are able to use larger window sizes and use all the information in the window as input. Below, we illustrate the approach.
```
# pick a large window size of 50 cycles
sequence_length = 50
```
Let's first look at an example of the sensor values 50 cycles prior to the failure for engine id 3. We will be feeding LSTM network this type of data for each time step for each engine id.
```
# preparing data for visualizations
# window of 50 cycles prior to a failure point for engine id 3
engine_id3 = test_df[test_df['id'] == 3]
engine_id3_50cycleWindow = engine_id3[engine_id3['RUL'] <= engine_id3['RUL'].min() + 50]
cols1 = ['s1', 's2', 's3', 's4', 's5', 's6', 's7', 's8', 's9', 's10']
engine_id3_50cycleWindow1 = engine_id3_50cycleWindow[cols1]
cols2 = ['s11', 's12', 's13', 's14', 's15', 's16', 's17', 's18', 's19', 's20', 's21']
engine_id3_50cycleWindow2 = engine_id3_50cycleWindow[cols2]
# plotting sensor data for engine ID 3 prior to a failure point - sensors 1-10
ax1 = engine_id3_50cycleWindow1.plot(subplots=True, sharex=True, figsize=(20,20))
# plotting sensor data for engine ID 3 prior to a failure point - sensors 11-21
ax2 = engine_id3_50cycleWindow2.plot(subplots=True, sharex=True, figsize=(20,20))
```
[Keras LSTM](https://keras.io/layers/recurrent/) layers expect an input in the shape of a numpy array of 3 dimensions (samples, time steps, features) where samples is the number of training sequences, time steps is the look back window or sequence length and features is the number of features of each sequence at each time step.
```
# function to reshape features into (samples, time steps, features)
def gen_sequence(id_df, seq_length, seq_cols):
""" Only sequences that meet the window-length are considered, no padding is used. This means for testing
we need to drop those which are below the window-length. An alternative would be to pad sequences so that
we can use shorter ones """
data_array = id_df[seq_cols].values
num_elements = data_array.shape[0]
for start, stop in zip(range(0, num_elements-seq_length), range(seq_length, num_elements)):
yield data_array[start:stop, :]
# pick the feature columns
sensor_cols = ['s' + str(i) for i in range(1,22)]
sequence_cols = ['setting1', 'setting2', 'setting3', 'cycle_norm']
sequence_cols.extend(sensor_cols)
# generator for the sequences
seq_gen = (list(gen_sequence(train_df[train_df['id']==id], sequence_length, sequence_cols))
for id in train_df['id'].unique())
# generate sequences and convert to numpy array
seq_array = np.concatenate(list(seq_gen)).astype(np.float32)
seq_array.shape
# function to generate labels
def gen_labels(id_df, seq_length, label):
data_array = id_df[label].values
num_elements = data_array.shape[0]
return data_array[seq_length:num_elements, :]
# generate labels
label_gen = [gen_labels(train_df[train_df['id']==id], sequence_length, ['label1'])
for id in train_df['id'].unique()]
label_array = np.concatenate(label_gen).astype(np.float32)
label_array.shape
```
## LSTM Network
Next, we build a deep network. The first layer is an LSTM layer with 100 units followed by another LSTM layer with 50 units. Dropout is also applied after each LSTM layer to control overfitting. Final layer is a Dense output layer with single unit and sigmoid activation since this is a binary classification problem.
```
# build the network
nb_features = seq_array.shape[2]
nb_out = label_array.shape[1]
model = Sequential()
model.add(LSTM(
input_shape=(sequence_length, nb_features),
units=100,
return_sequences=True))
model.add(Dropout(0.2))
model.add(LSTM(
units=50,
return_sequences=False))
model.add(Dropout(0.2))
model.add(Dense(units=nb_out, activation='sigmoid'))
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
print(model.summary())
%%time
# fit the network
model.fit(seq_array, label_array, epochs=10, batch_size=200, validation_split=0.05, verbose=1,
callbacks = [keras.callbacks.EarlyStopping(monitor='val_loss', min_delta=0, patience=0, verbose=0, mode='auto')])
# training metrics
scores = model.evaluate(seq_array, label_array, verbose=1, batch_size=200)
print('Accurracy: {}'.format(scores[1]))
# make predictions and compute confusion matrix
y_pred = model.predict_classes(seq_array,verbose=1, batch_size=200)
y_true = label_array
print('Confusion matrix\n- x-axis is true labels.\n- y-axis is predicted labels')
cm = confusion_matrix(y_true, y_pred)
cm
# compute precision and recall
precision = precision_score(y_true, y_pred)
recall = recall_score(y_true, y_pred)
print( 'precision = ', precision, '\n', 'recall = ', recall)
```
Next, we look at the performance on the test data. In the [Predictive Maintenance Template Step 1 of 3](https://gallery.cortanaintelligence.com/Experiment/Predictive-Maintenance-Step-1-of-3-data-preparation-and-feature-engineering-2), only the last cycle data for each engine id in the test data is kept for testing purposes. In order to compare the results to the template, we pick the last sequence for each id in the test data.
```
seq_array_test_last = [test_df[test_df['id']==id][sequence_cols].values[-sequence_length:]
for id in test_df['id'].unique() if len(test_df[test_df['id']==id]) >= sequence_length]
seq_array_test_last = np.asarray(seq_array_test_last).astype(np.float32)
seq_array_test_last.shape
y_mask = [len(test_df[test_df['id']==id]) >= sequence_length for id in test_df['id'].unique()]
```
Similarly, we pick the labels.
```
label_array_test_last = test_df.groupby('id')['label1'].nth(-1)[y_mask].values
label_array_test_last = label_array_test_last.reshape(label_array_test_last.shape[0],1).astype(np.float32)
label_array_test_last.shape
print(seq_array_test_last.shape)
print(label_array_test_last.shape)
# test metrics
scores_test = model.evaluate(seq_array_test_last, label_array_test_last, verbose=2)
print('Accurracy: {}'.format(scores_test[1]))
# make predictions and compute confusion matrix
y_pred_test = model.predict_classes(seq_array_test_last)
y_true_test = label_array_test_last
print('Confusion matrix\n- x-axis is true labels.\n- y-axis is predicted labels')
cm = confusion_matrix(y_true_test, y_pred_test)
cm
# compute precision and recall
precision_test = precision_score(y_true_test, y_pred_test)
recall_test = recall_score(y_true_test, y_pred_test)
f1_test = 2 * (precision_test * recall_test) / (precision_test + recall_test)
print( 'Precision: ', precision_test, '\n', 'Recall: ', recall_test,'\n', 'F1-score:', f1_test )
results_df = pd.DataFrame([[scores_test[1],precision_test,recall_test,f1_test],
[0.94, 0.952381, 0.8, 0.869565]],
columns = ['Accuracy', 'Precision', 'Recall', 'F1-score'],
index = ['LSTM',
'Template Best Model'])
results_df
```
Comparing the above test results to the predictive maintenance template, we see that the LSTM results are better than the template. It should be noted that the data set used here is very small and deep learning models are known to perform superior with large datasets so for a more fair comparison larger datasets should be used.
## Future Directions and Improvements
This tutorial covers the basics of using deep learning in predictive maintenance and many predictive maintenance problems usually involve a variety of data sources that needs to be taken into account when applying deep learning in this domain. Additionally, it is important to tune the models for the right parameters such as window size. Here are some suggestions on future directions on improvements:
- Try different window sizes.
- Try different architectures with different number of layers and nodes.
- Try tuning hyperparmeters of the network.
- Try predicting RUL (regression) such as in [Predictive Maintenance Template Step 2A of 3](https://gallery.cortanaintelligence.com/Experiment/Predictive-Maintenance-Step-2A-of-3-train-and-evaluate-regression-models-2) and label2 (multi-class classification) such as in [Predictive Maintenance Template Step 2C of 3](https://gallery.cortanaintelligence.com/Experiment/Predictive-Maintenance-Step-2C-of-3-train-and-evaluation-multi-class-classification-models-2).
- Try on larger data sets with more records.
- Try a different problem scenario such as in [Predictive Maintenance Modelling Guide](https://gallery.cortanaintelligence.com/Notebook/Predictive-Maintenance-Modelling-Guide-R-Notebook-1) where multiple other data sources are involved such as maintenance records.
|
github_jupyter
|
import keras
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# Setting seed for reproducability
np.random.seed(1234)
PYTHONHASHSEED = 0
from sklearn import preprocessing
from sklearn.metrics import confusion_matrix, recall_score, precision_score
from keras.models import Sequential
from keras.layers import Dense, Dropout, LSTM, Activation
%matplotlib inline
# Data ingestion - reading the datasets from Azure blob
!wget http://azuremlsamples.azureml.net/templatedata/PM_train.txt
!wget http://azuremlsamples.azureml.net/templatedata/PM_test.txt
!wget http://azuremlsamples.azureml.net/templatedata/PM_truth.txt
# read training data
train_df = pd.read_csv('PM_train.txt', sep=" ", header=None)
train_df.drop(train_df.columns[[26, 27]], axis=1, inplace=True)
train_df.columns = ['id', 'cycle', 'setting1', 'setting2', 'setting3', 's1', 's2', 's3',
's4', 's5', 's6', 's7', 's8', 's9', 's10', 's11', 's12', 's13', 's14',
's15', 's16', 's17', 's18', 's19', 's20', 's21']
# read test data
test_df = pd.read_csv('PM_test.txt', sep=" ", header=None)
test_df.drop(test_df.columns[[26, 27]], axis=1, inplace=True)
test_df.columns = ['id', 'cycle', 'setting1', 'setting2', 'setting3', 's1', 's2', 's3',
's4', 's5', 's6', 's7', 's8', 's9', 's10', 's11', 's12', 's13', 's14',
's15', 's16', 's17', 's18', 's19', 's20', 's21']
# read ground truth data
truth_df = pd.read_csv('PM_truth.txt', sep=" ", header=None)
truth_df.drop(truth_df.columns[[1]], axis=1, inplace=True)
train_df = train_df.sort_values(['id','cycle'])
train_df.head()
# Data Labeling - generate column RUL
rul = pd.DataFrame(train_df.groupby('id')['cycle'].max()).reset_index()
rul.columns = ['id', 'max']
train_df = train_df.merge(rul, on=['id'], how='left')
train_df['RUL'] = train_df['max'] - train_df['cycle']
train_df.drop('max', axis=1, inplace=True)
train_df.head()
# generate label columns for training data
w1 = 30
w0 = 15
train_df['label1'] = np.where(train_df['RUL'] <= w1, 1, 0 )
train_df['label2'] = train_df['label1']
train_df.loc[train_df['RUL'] <= w0, 'label2'] = 2
train_df.head()
# MinMax normalization
train_df['cycle_norm'] = train_df['cycle']
cols_normalize = train_df.columns.difference(['id','cycle','RUL','label1','label2'])
min_max_scaler = preprocessing.MinMaxScaler()
norm_train_df = pd.DataFrame(min_max_scaler.fit_transform(train_df[cols_normalize]),
columns=cols_normalize,
index=train_df.index)
join_df = train_df[train_df.columns.difference(cols_normalize)].join(norm_train_df)
train_df = join_df.reindex(columns = train_df.columns)
train_df.head()
test_df['cycle_norm'] = test_df['cycle']
norm_test_df = pd.DataFrame(min_max_scaler.transform(test_df[cols_normalize]),
columns=cols_normalize,
index=test_df.index)
test_join_df = test_df[test_df.columns.difference(cols_normalize)].join(norm_test_df)
test_df = test_join_df.reindex(columns = test_df.columns)
test_df = test_df.reset_index(drop=True)
test_df.head()
# generate column max for test data
rul = pd.DataFrame(test_df.groupby('id')['cycle'].max()).reset_index()
rul.columns = ['id', 'max']
truth_df.columns = ['more']
truth_df['id'] = truth_df.index + 1
truth_df['max'] = rul['max'] + truth_df['more']
truth_df.drop('more', axis=1, inplace=True)
# generate RUL for test data
test_df = test_df.merge(truth_df, on=['id'], how='left')
test_df['RUL'] = test_df['max'] - test_df['cycle']
test_df.drop('max', axis=1, inplace=True)
test_df.head()
# generate label columns w0 and w1 for test data
test_df['label1'] = np.where(test_df['RUL'] <= w1, 1, 0 )
test_df['label2'] = test_df['label1']
test_df.loc[test_df['RUL'] <= w0, 'label2'] = 2
test_df.head()
# pick a large window size of 50 cycles
sequence_length = 50
# preparing data for visualizations
# window of 50 cycles prior to a failure point for engine id 3
engine_id3 = test_df[test_df['id'] == 3]
engine_id3_50cycleWindow = engine_id3[engine_id3['RUL'] <= engine_id3['RUL'].min() + 50]
cols1 = ['s1', 's2', 's3', 's4', 's5', 's6', 's7', 's8', 's9', 's10']
engine_id3_50cycleWindow1 = engine_id3_50cycleWindow[cols1]
cols2 = ['s11', 's12', 's13', 's14', 's15', 's16', 's17', 's18', 's19', 's20', 's21']
engine_id3_50cycleWindow2 = engine_id3_50cycleWindow[cols2]
# plotting sensor data for engine ID 3 prior to a failure point - sensors 1-10
ax1 = engine_id3_50cycleWindow1.plot(subplots=True, sharex=True, figsize=(20,20))
# plotting sensor data for engine ID 3 prior to a failure point - sensors 11-21
ax2 = engine_id3_50cycleWindow2.plot(subplots=True, sharex=True, figsize=(20,20))
# function to reshape features into (samples, time steps, features)
def gen_sequence(id_df, seq_length, seq_cols):
""" Only sequences that meet the window-length are considered, no padding is used. This means for testing
we need to drop those which are below the window-length. An alternative would be to pad sequences so that
we can use shorter ones """
data_array = id_df[seq_cols].values
num_elements = data_array.shape[0]
for start, stop in zip(range(0, num_elements-seq_length), range(seq_length, num_elements)):
yield data_array[start:stop, :]
# pick the feature columns
sensor_cols = ['s' + str(i) for i in range(1,22)]
sequence_cols = ['setting1', 'setting2', 'setting3', 'cycle_norm']
sequence_cols.extend(sensor_cols)
# generator for the sequences
seq_gen = (list(gen_sequence(train_df[train_df['id']==id], sequence_length, sequence_cols))
for id in train_df['id'].unique())
# generate sequences and convert to numpy array
seq_array = np.concatenate(list(seq_gen)).astype(np.float32)
seq_array.shape
# function to generate labels
def gen_labels(id_df, seq_length, label):
data_array = id_df[label].values
num_elements = data_array.shape[0]
return data_array[seq_length:num_elements, :]
# generate labels
label_gen = [gen_labels(train_df[train_df['id']==id], sequence_length, ['label1'])
for id in train_df['id'].unique()]
label_array = np.concatenate(label_gen).astype(np.float32)
label_array.shape
# build the network
nb_features = seq_array.shape[2]
nb_out = label_array.shape[1]
model = Sequential()
model.add(LSTM(
input_shape=(sequence_length, nb_features),
units=100,
return_sequences=True))
model.add(Dropout(0.2))
model.add(LSTM(
units=50,
return_sequences=False))
model.add(Dropout(0.2))
model.add(Dense(units=nb_out, activation='sigmoid'))
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
print(model.summary())
%%time
# fit the network
model.fit(seq_array, label_array, epochs=10, batch_size=200, validation_split=0.05, verbose=1,
callbacks = [keras.callbacks.EarlyStopping(monitor='val_loss', min_delta=0, patience=0, verbose=0, mode='auto')])
# training metrics
scores = model.evaluate(seq_array, label_array, verbose=1, batch_size=200)
print('Accurracy: {}'.format(scores[1]))
# make predictions and compute confusion matrix
y_pred = model.predict_classes(seq_array,verbose=1, batch_size=200)
y_true = label_array
print('Confusion matrix\n- x-axis is true labels.\n- y-axis is predicted labels')
cm = confusion_matrix(y_true, y_pred)
cm
# compute precision and recall
precision = precision_score(y_true, y_pred)
recall = recall_score(y_true, y_pred)
print( 'precision = ', precision, '\n', 'recall = ', recall)
seq_array_test_last = [test_df[test_df['id']==id][sequence_cols].values[-sequence_length:]
for id in test_df['id'].unique() if len(test_df[test_df['id']==id]) >= sequence_length]
seq_array_test_last = np.asarray(seq_array_test_last).astype(np.float32)
seq_array_test_last.shape
y_mask = [len(test_df[test_df['id']==id]) >= sequence_length for id in test_df['id'].unique()]
label_array_test_last = test_df.groupby('id')['label1'].nth(-1)[y_mask].values
label_array_test_last = label_array_test_last.reshape(label_array_test_last.shape[0],1).astype(np.float32)
label_array_test_last.shape
print(seq_array_test_last.shape)
print(label_array_test_last.shape)
# test metrics
scores_test = model.evaluate(seq_array_test_last, label_array_test_last, verbose=2)
print('Accurracy: {}'.format(scores_test[1]))
# make predictions and compute confusion matrix
y_pred_test = model.predict_classes(seq_array_test_last)
y_true_test = label_array_test_last
print('Confusion matrix\n- x-axis is true labels.\n- y-axis is predicted labels')
cm = confusion_matrix(y_true_test, y_pred_test)
cm
# compute precision and recall
precision_test = precision_score(y_true_test, y_pred_test)
recall_test = recall_score(y_true_test, y_pred_test)
f1_test = 2 * (precision_test * recall_test) / (precision_test + recall_test)
print( 'Precision: ', precision_test, '\n', 'Recall: ', recall_test,'\n', 'F1-score:', f1_test )
results_df = pd.DataFrame([[scores_test[1],precision_test,recall_test,f1_test],
[0.94, 0.952381, 0.8, 0.869565]],
columns = ['Accuracy', 'Precision', 'Recall', 'F1-score'],
index = ['LSTM',
'Template Best Model'])
results_df
| 0.595728 | 0.97959 |
- Reference
- Blog: https://work-in-progress.hatenablog.com/entry/2019/04/06/113629
- Source: https://github.com/eriklindernoren/Keras-GAN/blob/master/gan/gan.py
- Source: https://github.com/eriklindernoren/Keras-GAN/pull/117
- Added functionality to save/load Keras model for intermittent training.
```
import os
os.makedirs('data/images', exist_ok=True)
os.makedirs('data/saved_models', exist_ok=True)
from tensorflow.keras.datasets import mnist
from tensorflow.keras.layers import Input, Dense, Reshape, Flatten
from tensorflow.keras.layers import BatchNormalization
from tensorflow.keras.layers import LeakyReLU
from tensorflow.keras.models import Sequential, Model, model_from_json
from tensorflow.keras.optimizers import Adam
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import datetime
class GAN():
def __init__(self):
self.history = pd.DataFrame({}, columns=['d_loss', 'acc', 'g_loss'])
self.img_save_dir = 'data/images'
self.model_save_dir = 'data/saved_models'
self.discriminator_name = 'discriminator_model'
self.generator_name = 'generator_model'
self.combined_name = 'combined_model'
self.discriminator = None
self.generator = None
self.combined = None
self.img_rows = 28
self.img_cols = 28
self.channels = 1
self.img_shape = (self.img_rows, self.img_cols, self.channels)
self.latent_dim = 100
def init(self, loading=False):
optimizer = Adam(0.0002, 0.5)
self.discriminator = self.build_discriminator()
self.discriminator.compile(loss='binary_crossentropy',
optimizer=optimizer,
metrics=['accuracy'])
if loading:
self.load_model_weight(self.discriminator_name)
self.generator = self.build_generator()
if loading:
self.load_model_weight(self.generator_name)
z = Input(shape=(self.latent_dim,))
img = self.generator(z)
# For the combined model we will only train the generator
self.discriminator.trainable = False
# The discriminator takes generated images as input and determines validity
validity = self.discriminator(img)
# The combined model (stacked generator and discriminator)
# Trains the generator to fool the discriminator
self.combined = Model(z, validity)
self.combined.summary()
self.combined.compile(loss='binary_crossentropy', optimizer=optimizer)
if loading:
self.load_model_weight(self.combined_name)
def build_generator(self):
model = Sequential()
model.add(Dense(256, input_dim=self.latent_dim))
model.add(LeakyReLU(alpha=0.2))
model.add(BatchNormalization(momentum=0.8))
model.add(Dense(512))
model.add(LeakyReLU(alpha=0.2))
model.add(BatchNormalization(momentum=0.8))
model.add(Dense(1024))
model.add(LeakyReLU(alpha=0.2))
model.add(BatchNormalization(momentum=0.8))
model.add(Dense(np.prod(self.img_shape), activation='tanh'))
model.add(Reshape(self.img_shape))
model.summary()
noise = Input(shape=(self.latent_dim,))
img = model(noise)
return Model(noise, img)
def build_discriminator(self):
model = Sequential()
model.add(Flatten(input_shape=self.img_shape))
model.add(Dense(512))
model.add(LeakyReLU(alpha=0.2))
model.add(Dense(256))
model.add(LeakyReLU(alpha=0.2))
model.add(Dense(1, activation='sigmoid'))
model.summary()
img = Input(shape=self.img_shape)
validity = model(img)
return Model(img, validity)
def train(self, epochs, batch_size=128, sample_interval=-1, save_interval=-1):
# Load the dataset
(X_train, _), (_, _) = mnist.load_data()
#print(X_train.shape)
# Rescale -1 to 1
X_train = X_train / 127.5 - 1.
X_train = np.expand_dims(X_train, axis=3)
#print(X_train.shape)
# Adversarial ground truths
valid = np.ones((batch_size, 1))
fake = np.zeros((batch_size, 1))
print(datetime.datetime.now().isoformat(), 'Epoch Start')
for epoch in range(epochs):
# Select a random batch of images
idx = np.random.randint(0, X_train.shape[0], batch_size)
imgs = X_train[idx]
noise = np.random.normal(0, 1, (batch_size, self.latent_dim))
gen_imgs = self.generator.predict(noise)
d_loss_real = self.discriminator.train_on_batch(imgs, valid)
d_loss_fake = self.discriminator.train_on_batch(gen_imgs, fake)
d_loss = 0.5 * np.add(d_loss_real, d_loss_fake)
noise = np.random.normal(0, 1, (batch_size, self.latent_dim))
g_loss = self.combined.train_on_batch(noise, valid)
self.history = self.history.append({'d_loss': d_loss[0], 'acc': d_loss[1], 'g_loss': g_loss}, ignore_index=True)
if sample_interval > 0 and epoch % sample_interval == 0:
print(datetime.datetime.now().isoformat(), '%d [D loss: %f, acc.: %.2f%%] [G loss: %f]' % (epoch, d_loss[0], 100*d_loss[1], g_loss))
self.sample_images(epoch)
if save_interval > 0 and epoch != 0 and epoch % save_interval == 0:
self.save_model_weights_all()
print(datetime.datetime.now().isoformat(), 'Epoch End')
def generate_image(self):
noise = np.random.normal(0, 1, (1, self.latent_dim))
return self.generator.predict(noise)
def sample_images(self, epoch):
r, c = 5, 5
noise = np.random.normal(0, 1, (r * c, self.latent_dim))
gen_imgs = self.generator.predict(noise)
# Rescale images 0 - 1
gen_imgs = 0.5 * gen_imgs + 0.5
fig, axs = plt.subplots(r, c)
cnt = 0
for i in range(r):
for j in range(c):
axs[i, j].imshow(gen_imgs[cnt, :, :, 0], cmap='gray')
axs[i, j].axis('off')
cnt += 1
file_name = '{}.png'.format(epoch)
file_path = os.path.join(self.img_save_dir, file_name)
fig.savefig(file_path)
plt.close()
def plot_hisotry(self, columns=[]):
if len(columns) == 0:
columns = ['d_loss', 'acc', 'g_loss']
self.history[columns].plot()
def save_model_weights_all(self):
self.save_model_weights(self.discriminator, self.discriminator_name)
self.save_model_weights(self.generator, self.generator_name)
self.save_model_weights(self.combined, self.combined_name)
def save_model_weights(self, model, model_name):
weights_path = os.path.join(self.model_save_dir, '{}.h5'.format(model_name))
model.save_weights(weights_path)
print('Weights saved.', model_name)
def load_model_weight(self, model_name):
model = None
if model_name == self.discriminator_name:
model = self.discriminator
elif model_name == self.generator_name:
model = self.generator
elif model_name == self.combined_name:
model = self.combined
if not model:
print('Model is not initialized.', model_name)
return
weights_path = os.path.join(self.model_save_dir, '{}.h5'.format(model_name))
if not os.path.exists(weights_path):
print('Not found h5 file.', model_name)
return
model.load_weights(weights_path)
print('Weights loaded.', model_name)
gan = GAN()
gan.init(loading=True)
#gan.train(epochs=500, batch_size=32, sample_interval=50, save_interval=50)
#gan.train(epochs=500, batch_size=32, sample_interval=50)
#gan.save_model_weights_all()
gen_img = gan.generate_image()
gen_img.shape
plt.imshow(gen_img[0], cmap='gray')
plt.imshow((gan.generate_image())[0], cmap='gray')
```
|
github_jupyter
|
import os
os.makedirs('data/images', exist_ok=True)
os.makedirs('data/saved_models', exist_ok=True)
from tensorflow.keras.datasets import mnist
from tensorflow.keras.layers import Input, Dense, Reshape, Flatten
from tensorflow.keras.layers import BatchNormalization
from tensorflow.keras.layers import LeakyReLU
from tensorflow.keras.models import Sequential, Model, model_from_json
from tensorflow.keras.optimizers import Adam
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import datetime
class GAN():
def __init__(self):
self.history = pd.DataFrame({}, columns=['d_loss', 'acc', 'g_loss'])
self.img_save_dir = 'data/images'
self.model_save_dir = 'data/saved_models'
self.discriminator_name = 'discriminator_model'
self.generator_name = 'generator_model'
self.combined_name = 'combined_model'
self.discriminator = None
self.generator = None
self.combined = None
self.img_rows = 28
self.img_cols = 28
self.channels = 1
self.img_shape = (self.img_rows, self.img_cols, self.channels)
self.latent_dim = 100
def init(self, loading=False):
optimizer = Adam(0.0002, 0.5)
self.discriminator = self.build_discriminator()
self.discriminator.compile(loss='binary_crossentropy',
optimizer=optimizer,
metrics=['accuracy'])
if loading:
self.load_model_weight(self.discriminator_name)
self.generator = self.build_generator()
if loading:
self.load_model_weight(self.generator_name)
z = Input(shape=(self.latent_dim,))
img = self.generator(z)
# For the combined model we will only train the generator
self.discriminator.trainable = False
# The discriminator takes generated images as input and determines validity
validity = self.discriminator(img)
# The combined model (stacked generator and discriminator)
# Trains the generator to fool the discriminator
self.combined = Model(z, validity)
self.combined.summary()
self.combined.compile(loss='binary_crossentropy', optimizer=optimizer)
if loading:
self.load_model_weight(self.combined_name)
def build_generator(self):
model = Sequential()
model.add(Dense(256, input_dim=self.latent_dim))
model.add(LeakyReLU(alpha=0.2))
model.add(BatchNormalization(momentum=0.8))
model.add(Dense(512))
model.add(LeakyReLU(alpha=0.2))
model.add(BatchNormalization(momentum=0.8))
model.add(Dense(1024))
model.add(LeakyReLU(alpha=0.2))
model.add(BatchNormalization(momentum=0.8))
model.add(Dense(np.prod(self.img_shape), activation='tanh'))
model.add(Reshape(self.img_shape))
model.summary()
noise = Input(shape=(self.latent_dim,))
img = model(noise)
return Model(noise, img)
def build_discriminator(self):
model = Sequential()
model.add(Flatten(input_shape=self.img_shape))
model.add(Dense(512))
model.add(LeakyReLU(alpha=0.2))
model.add(Dense(256))
model.add(LeakyReLU(alpha=0.2))
model.add(Dense(1, activation='sigmoid'))
model.summary()
img = Input(shape=self.img_shape)
validity = model(img)
return Model(img, validity)
def train(self, epochs, batch_size=128, sample_interval=-1, save_interval=-1):
# Load the dataset
(X_train, _), (_, _) = mnist.load_data()
#print(X_train.shape)
# Rescale -1 to 1
X_train = X_train / 127.5 - 1.
X_train = np.expand_dims(X_train, axis=3)
#print(X_train.shape)
# Adversarial ground truths
valid = np.ones((batch_size, 1))
fake = np.zeros((batch_size, 1))
print(datetime.datetime.now().isoformat(), 'Epoch Start')
for epoch in range(epochs):
# Select a random batch of images
idx = np.random.randint(0, X_train.shape[0], batch_size)
imgs = X_train[idx]
noise = np.random.normal(0, 1, (batch_size, self.latent_dim))
gen_imgs = self.generator.predict(noise)
d_loss_real = self.discriminator.train_on_batch(imgs, valid)
d_loss_fake = self.discriminator.train_on_batch(gen_imgs, fake)
d_loss = 0.5 * np.add(d_loss_real, d_loss_fake)
noise = np.random.normal(0, 1, (batch_size, self.latent_dim))
g_loss = self.combined.train_on_batch(noise, valid)
self.history = self.history.append({'d_loss': d_loss[0], 'acc': d_loss[1], 'g_loss': g_loss}, ignore_index=True)
if sample_interval > 0 and epoch % sample_interval == 0:
print(datetime.datetime.now().isoformat(), '%d [D loss: %f, acc.: %.2f%%] [G loss: %f]' % (epoch, d_loss[0], 100*d_loss[1], g_loss))
self.sample_images(epoch)
if save_interval > 0 and epoch != 0 and epoch % save_interval == 0:
self.save_model_weights_all()
print(datetime.datetime.now().isoformat(), 'Epoch End')
def generate_image(self):
noise = np.random.normal(0, 1, (1, self.latent_dim))
return self.generator.predict(noise)
def sample_images(self, epoch):
r, c = 5, 5
noise = np.random.normal(0, 1, (r * c, self.latent_dim))
gen_imgs = self.generator.predict(noise)
# Rescale images 0 - 1
gen_imgs = 0.5 * gen_imgs + 0.5
fig, axs = plt.subplots(r, c)
cnt = 0
for i in range(r):
for j in range(c):
axs[i, j].imshow(gen_imgs[cnt, :, :, 0], cmap='gray')
axs[i, j].axis('off')
cnt += 1
file_name = '{}.png'.format(epoch)
file_path = os.path.join(self.img_save_dir, file_name)
fig.savefig(file_path)
plt.close()
def plot_hisotry(self, columns=[]):
if len(columns) == 0:
columns = ['d_loss', 'acc', 'g_loss']
self.history[columns].plot()
def save_model_weights_all(self):
self.save_model_weights(self.discriminator, self.discriminator_name)
self.save_model_weights(self.generator, self.generator_name)
self.save_model_weights(self.combined, self.combined_name)
def save_model_weights(self, model, model_name):
weights_path = os.path.join(self.model_save_dir, '{}.h5'.format(model_name))
model.save_weights(weights_path)
print('Weights saved.', model_name)
def load_model_weight(self, model_name):
model = None
if model_name == self.discriminator_name:
model = self.discriminator
elif model_name == self.generator_name:
model = self.generator
elif model_name == self.combined_name:
model = self.combined
if not model:
print('Model is not initialized.', model_name)
return
weights_path = os.path.join(self.model_save_dir, '{}.h5'.format(model_name))
if not os.path.exists(weights_path):
print('Not found h5 file.', model_name)
return
model.load_weights(weights_path)
print('Weights loaded.', model_name)
gan = GAN()
gan.init(loading=True)
#gan.train(epochs=500, batch_size=32, sample_interval=50, save_interval=50)
#gan.train(epochs=500, batch_size=32, sample_interval=50)
#gan.save_model_weights_all()
gen_img = gan.generate_image()
gen_img.shape
plt.imshow(gen_img[0], cmap='gray')
plt.imshow((gan.generate_image())[0], cmap='gray')
| 0.807233 | 0.776686 |
REGON strings can be converted to the following formats via the `output_format` parameter:
* `compact`: only number strings without any seperators or whitespace, like "192598184"
* `standard`: REGON strings with proper whitespace in the proper places. Note that in the case of REGON, the compact format is the same as the standard one.
Invalid parsing is handled with the `errors` parameter:
* `coerce` (default): invalid parsing will be set to NaN
* `ignore`: invalid parsing will return the input
* `raise`: invalid parsing will raise an exception
The following sections demonstrate the functionality of `clean_pl_regon()` and `validate_pl_regon()`.
### An example dataset containing REGON strings
```
import pandas as pd
import numpy as np
df = pd.DataFrame(
{
"regon": [
'192598184',
'192598183',
'BE 428759497',
'BE431150351',
"002 724 334",
"hello",
np.nan,
"NULL",
],
"address": [
"123 Pine Ave.",
"main st",
"1234 west main heights 57033",
"apt 1 789 s maple rd manhattan",
"robie house, 789 north main street",
"1111 S Figueroa St, Los Angeles, CA 90015",
"(staples center) 1111 S Figueroa St, Los Angeles",
"hello",
]
}
)
df
```
## 1. Default `clean_pl_regon`
By default, `clean_pl_regon` will clean regon strings and output them in the standard format with proper separators.
```
from dataprep.clean import clean_pl_regon
clean_pl_regon(df, column = "regon")
```
## 2. Output formats
This section demonstrates the output parameter.
### `standard` (default)
```
clean_pl_regon(df, column = "regon", output_format="standard")
```
### `compact`
```
clean_pl_regon(df, column = "regon", output_format="compact")
```
## 3. `inplace` parameter
This deletes the given column from the returned DataFrame.
A new column containing cleaned REGON strings is added with a title in the format `"{original title}_clean"`.
```
clean_pl_regon(df, column="regon", inplace=True)
```
## 4. `errors` parameter
### `coerce` (default)
```
clean_pl_regon(df, "regon", errors="coerce")
```
### `ignore`
```
clean_pl_regon(df, "regon", errors="ignore")
```
## 4. `validate_pl_regon()`
`validate_pl_regon()` returns `True` when the input is a valid REGON. Otherwise it returns `False`.
The input of `validate_pl_regon()` can be a string, a Pandas DataSeries, a Dask DataSeries, a Pandas DataFrame and a dask DataFrame.
When the input is a string, a Pandas DataSeries or a Dask DataSeries, user doesn't need to specify a column name to be validated.
When the input is a Pandas DataFrame or a dask DataFrame, user can both specify or not specify a column name to be validated. If user specify the column name, `validate_pl_regon()` only returns the validation result for the specified column. If user doesn't specify the column name, `validate_pl_regon()` returns the validation result for the whole DataFrame.
```
from dataprep.clean import validate_pl_regon
print(validate_pl_regon("192598184"))
print(validate_pl_regon("192598183"))
print(validate_pl_regon('BE 428759497'))
print(validate_pl_regon('BE431150351'))
print(validate_pl_regon("004085616"))
print(validate_pl_regon("hello"))
print(validate_pl_regon(np.nan))
print(validate_pl_regon("NULL"))
```
### Series
```
validate_pl_regon(df["regon"])
```
### DataFrame + Specify Column
```
validate_pl_regon(df, column="regon")
```
### Only DataFrame
```
validate_pl_regon(df)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
df = pd.DataFrame(
{
"regon": [
'192598184',
'192598183',
'BE 428759497',
'BE431150351',
"002 724 334",
"hello",
np.nan,
"NULL",
],
"address": [
"123 Pine Ave.",
"main st",
"1234 west main heights 57033",
"apt 1 789 s maple rd manhattan",
"robie house, 789 north main street",
"1111 S Figueroa St, Los Angeles, CA 90015",
"(staples center) 1111 S Figueroa St, Los Angeles",
"hello",
]
}
)
df
from dataprep.clean import clean_pl_regon
clean_pl_regon(df, column = "regon")
clean_pl_regon(df, column = "regon", output_format="standard")
clean_pl_regon(df, column = "regon", output_format="compact")
clean_pl_regon(df, column="regon", inplace=True)
clean_pl_regon(df, "regon", errors="coerce")
clean_pl_regon(df, "regon", errors="ignore")
from dataprep.clean import validate_pl_regon
print(validate_pl_regon("192598184"))
print(validate_pl_regon("192598183"))
print(validate_pl_regon('BE 428759497'))
print(validate_pl_regon('BE431150351'))
print(validate_pl_regon("004085616"))
print(validate_pl_regon("hello"))
print(validate_pl_regon(np.nan))
print(validate_pl_regon("NULL"))
validate_pl_regon(df["regon"])
validate_pl_regon(df, column="regon")
validate_pl_regon(df)
| 0.403332 | 0.990006 |
# Download and Mosaic Multiple 3DEP DTM Tiles
This code is adapted (by Hannah Besso) from David Shean's geospatial_cookbook_rendered.ipynb notebook created for UW's SnowEx Hackweek 2021
### This notebook does the following:
* Read in a list of urls linking to individual 3DEP tiles that cover Grand Mesa
* Downloads, reprojects, and mosaics the tiles using gdal
* Clips the mosaic'd image to the flat top of Grand Mesa
```
# Import necessary packages
import os
import rasterio as rio
import rasterio.plot
import numpy as np
import matplotlib.pyplot as plt
import geopandas as gpd
import fiona
```
### Open the text file listing urls of all the tiles intersecting Grand Mesa
```
url_fn_3DEP = 'gm_3dep_1m_lidar_tiles.txt'
with open(url_fn_3DEP) as f:
url_list = f.read().splitlines()
url_list.sort()
path_list = []
for url in url_list:
path = f'/vsicurl/{url}'
path_list.append(path)
path_list_str = ' '.join(path_list)
url = open("url_list.txt", "w")
for element in url_list:
url.write(element + "\n")
url.close()
vrt_fn = os.path.splitext(url_fn_3DEP)[0]+'.vrt'
tif_fn = os.path.splitext(url_fn_3DEP)[0]+'.tif'
```
### Build a Virtual Dataset using GDAL and the list of urls
https://gdal.org/programs/gdalbuildvrt.html
```
#This actually takes some time as file must be downloaded and unzipped to read img header
!gdalbuildvrt -quiet $vrt_fn $path_list_str
# Set the destination crs to UTM Zone 12N
dst_crs = 'EPSG:32612'
```
### Download, Reproject, and Mosaic the 3DEP Tiles
https://gdal.org/programs/gdalwarp.html
```
#Since these tiles are mixed projection, can download, reproject and mosaic in one go
!gdalwarp -q -r cubic -tr 3.0 3.0 -dstnodata -9999 -t_srs $dst_crs \
-co COMPRESS=LZW -co TILED=YES -co BIGTIFF=IF_SAFER \
$path_list_str $tif_fn
```
### Upload the Mosaic'd File and Clip to GM Flat Top
```
# Open the new .tif file containing the mosaic'd DTMs
src = rio.open('/home/jovyan/space_lasers/notebooks/gm_3dep_1m_lidar_tiles.tif')
# Inspect the file, confirming that the crs is 32612 (UTM Zone 12N)
src.profile
# Plot the mosaic'd file
fig, ax = plt.subplots(figsize=(10,10))
rio.plot.show(src, cmap='gray')
# Load in a polygon of the flat top of the Mesa
# Can create a polygon of a different section using:
# https://geojson.io/
poly_fn = '/home/jovyan/space_lasers/notebooks/map.geojson'
gm_poly = gpd.read_file(poly_fn)
```
**Check the CRS of the polygon. If it does not match that of the .tif file, complete the following steps to reproject:**
```
gm_poly.crs
# Reproject the polygon so that it matches the crs of the 3DEP .tif file
gm_poly = gm_poly.to_crs(crs=dst_crs)
# Save the reprojected polygon:
gm_poly.to_file("/home/jovyan/space_lasers/notebooks/GM_tight_poly_32612.geojson", driver='GeoJSON')
# Define the file path to the new projected polygon, and the mosaic'd tif output in earlier steps:
poly_fn = "/home/jovyan/space_lasers/notebooks/GM_tight_poly_32612.geojson"
tif_fn = '/home/jovyan/space_lasers/notebooks/gm_3dep_1m_lidar_tiles.tif'
```
**Clip the .tif using the polygon:**
```
# Open the polygon using fiona and make a list of features in the polygon (in this case the list will contain one feature)
with fiona.open(poly_fn, "r") as shapefile:
shapes = [feature["geometry"] for feature in shapefile]
with rio.open(tif_fn) as src:
out_image, out_transform = rio.mask.mask(src, shapes, crop=True)
out_meta = src.meta
out_meta.update({"driver": "GTiff",
"height": out_image.shape[1],
"width": out_image.shape[2],
"transform": out_transform})
with rasterio.open(tif_fn, "w", **out_meta) as dest:
dest.write(out_image)
rio.plot.show(out_image)
```
**The clipped 3DEP tif should have now overwritten the original larger mosaic'd tif created in earlier steps.**
|
github_jupyter
|
# Import necessary packages
import os
import rasterio as rio
import rasterio.plot
import numpy as np
import matplotlib.pyplot as plt
import geopandas as gpd
import fiona
url_fn_3DEP = 'gm_3dep_1m_lidar_tiles.txt'
with open(url_fn_3DEP) as f:
url_list = f.read().splitlines()
url_list.sort()
path_list = []
for url in url_list:
path = f'/vsicurl/{url}'
path_list.append(path)
path_list_str = ' '.join(path_list)
url = open("url_list.txt", "w")
for element in url_list:
url.write(element + "\n")
url.close()
vrt_fn = os.path.splitext(url_fn_3DEP)[0]+'.vrt'
tif_fn = os.path.splitext(url_fn_3DEP)[0]+'.tif'
#This actually takes some time as file must be downloaded and unzipped to read img header
!gdalbuildvrt -quiet $vrt_fn $path_list_str
# Set the destination crs to UTM Zone 12N
dst_crs = 'EPSG:32612'
#Since these tiles are mixed projection, can download, reproject and mosaic in one go
!gdalwarp -q -r cubic -tr 3.0 3.0 -dstnodata -9999 -t_srs $dst_crs \
-co COMPRESS=LZW -co TILED=YES -co BIGTIFF=IF_SAFER \
$path_list_str $tif_fn
# Open the new .tif file containing the mosaic'd DTMs
src = rio.open('/home/jovyan/space_lasers/notebooks/gm_3dep_1m_lidar_tiles.tif')
# Inspect the file, confirming that the crs is 32612 (UTM Zone 12N)
src.profile
# Plot the mosaic'd file
fig, ax = plt.subplots(figsize=(10,10))
rio.plot.show(src, cmap='gray')
# Load in a polygon of the flat top of the Mesa
# Can create a polygon of a different section using:
# https://geojson.io/
poly_fn = '/home/jovyan/space_lasers/notebooks/map.geojson'
gm_poly = gpd.read_file(poly_fn)
gm_poly.crs
# Reproject the polygon so that it matches the crs of the 3DEP .tif file
gm_poly = gm_poly.to_crs(crs=dst_crs)
# Save the reprojected polygon:
gm_poly.to_file("/home/jovyan/space_lasers/notebooks/GM_tight_poly_32612.geojson", driver='GeoJSON')
# Define the file path to the new projected polygon, and the mosaic'd tif output in earlier steps:
poly_fn = "/home/jovyan/space_lasers/notebooks/GM_tight_poly_32612.geojson"
tif_fn = '/home/jovyan/space_lasers/notebooks/gm_3dep_1m_lidar_tiles.tif'
# Open the polygon using fiona and make a list of features in the polygon (in this case the list will contain one feature)
with fiona.open(poly_fn, "r") as shapefile:
shapes = [feature["geometry"] for feature in shapefile]
with rio.open(tif_fn) as src:
out_image, out_transform = rio.mask.mask(src, shapes, crop=True)
out_meta = src.meta
out_meta.update({"driver": "GTiff",
"height": out_image.shape[1],
"width": out_image.shape[2],
"transform": out_transform})
with rasterio.open(tif_fn, "w", **out_meta) as dest:
dest.write(out_image)
rio.plot.show(out_image)
| 0.581541 | 0.81335 |
<a id="title_ID"></a>
# JWST Pipeline Validation Notebook:
# calwebb_detector1, firstframe unit tests
<span style="color:red"> **Instruments Affected**</span>: MIRI
### Table of Contents
<div style="text-align: left">
<br> [Introduction](#intro)
<br> [JWST Unit Tests](#unit)
<br> [Defining Terms](#terms)
<br> [Test Description](#description)
<br> [Data Description](#data_descr)
<br> [Imports](#imports)
<br> [Convenience Functions](#functions)
<br> [Perform Tests](#testing)
<br> [About This Notebook](#about)
<br>
</div>
<a id="intro"></a>
# Introduction
This is the validation notebook that displays the unit tests for the Firstframe step in calwebb_detector1. This notebook runs and displays the unit tests that are performed as a part of the normal software continuous integration process. For more information on the pipeline visit the links below.
* Pipeline description: https://jwst-pipeline.readthedocs.io/en/latest/jwst/firstframe/index.html
* Pipeline code: https://github.com/spacetelescope/jwst/tree/master/jwst/
[Top of Page](#title_ID)
<a id="unit"></a>
# JWST Unit Tests
JWST unit tests are located in the "tests" folder for each pipeline step within the [GitHub repository](https://github.com/spacetelescope/jwst/tree/master/jwst/), e.g., ```jwst/firstframe/tests```.
* Unit test README: https://github.com/spacetelescope/jwst#unit-tests
[Top of Page](#title_ID)
<a id="terms"></a>
# Defining Terms
These are terms or acronymns used in this notebook that may not be known a general audience.
* JWST: James Webb Space Telescope
* NIRCam: Near-Infrared Camera
[Top of Page](#title_ID)
<a id="description"></a>
# Test Description
Unit testing is a software testing method by which individual units of source code are tested to determine whether they are working sufficiently well. Unit tests do not require a separate data file; the test creates the necessary test data and parameters as a part of the test code.
[Top of Page](#title_ID)
<a id="data_descr"></a>
# Data Description
Data used for unit tests is created on the fly within the test itself, and is typically an array in the expected format of JWST data with added metadata needed to run through the pipeline.
[Top of Page](#title_ID)
<a id="imports"></a>
# Imports
* tempfile for creating temporary output products
* pytest for unit test functions
* jwst for the JWST Pipeline
* IPython.display for display pytest reports
[Top of Page](#title_ID)
```
import tempfile
import pytest
import jwst
from IPython.display import IFrame
```
<a id="functions"></a>
# Convenience Functions
Here we define any convenience functions to help with running the unit tests.
[Top of Page](#title_ID)
```
def display_report(fname):
'''Convenience function to display pytest report.'''
return IFrame(src=fname, width=700, height=600)
```
<a id="testing"></a>
# Perform Tests
Below we run the unit tests for the Firstframe step.
[Top of Page](#title_ID)
```
with tempfile.TemporaryDirectory() as tmpdir:
!pytest jwst/firstframe -v --ignore=jwst/associations --ignore=jwst/datamodels --ignore=jwst/stpipe --ignore=jwst/regtest --html=tmpdir/unit_report.html --self-contained-html
report = display_report('tmpdir/unit_report.html')
report
```
<a id="about"></a>
## About This Notebook
**Author:** Alicia Canipe, Staff Scientist, NIRCam
<br>**Updated On:** 01/07/2021
[Top of Page](#title_ID)
<img style="float: right;" src="./stsci_pri_combo_mark_horizonal_white_bkgd.png" alt="stsci_pri_combo_mark_horizonal_white_bkgd" width="200px"/>
|
github_jupyter
|
import tempfile
import pytest
import jwst
from IPython.display import IFrame
def display_report(fname):
'''Convenience function to display pytest report.'''
return IFrame(src=fname, width=700, height=600)
with tempfile.TemporaryDirectory() as tmpdir:
!pytest jwst/firstframe -v --ignore=jwst/associations --ignore=jwst/datamodels --ignore=jwst/stpipe --ignore=jwst/regtest --html=tmpdir/unit_report.html --self-contained-html
report = display_report('tmpdir/unit_report.html')
report
| 0.191517 | 0.959649 |
<a href="https://colab.research.google.com/github/ashesm/ISYS5002_PORTFOLIO1/blob/main/Sale_Price_LIVE.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
A local department store needs to develop a program which when given an items original price and percentage it has been discounted, will computer the total price (including goods and services tax) of the item on sale.
* What the results I am try to obtain?
* What data does needs to be provided?
* How will we obtain the output form the given input?
Input -> Processing -> Output
A local department store needs to develop a program which when given an **items** **original price** and percentage it has been **discounted**, will computer the **total price** (including goods and services tax **GST**) of the item on sale.
* Item Name. - create *item_name*
* Discounted Price - crate *sale_price*
* GST - create *gst*
* Total Price - create *total_price*
### Input
* item_name
* gst (?)
* original_price
* discount_rate ( assume in percent, will need to divide by 100)
### Output
* item_name
* sale_price
### Calculations (intial thoughts on processing)
* Calclate *sale_price*
* Calculate *tax*
* What is the discount? Amount saved?
* Calculate *total_price*
### Worked Examples
Assume I have a book, original price is $50. I need to discount the book by 20%
Amount Saved (Savings)
\$50 X 20% = $50 X 20/100
```
50 * 20 / 100
```
Sale price = \$50 - \$10 = \$40
Suggested formula is:
sale_price = original_price - amount_saved
Where
amount_saved = original_price * ( discount_rate / 100)
```
original_price = 50
discount_rate = 20
amount_saved = original_price * (discount_rate / 100)
sale_price = original_price - amount_saved
print("Amount Saved: ", amount_saved)
print("Sale Price: ", sale_price)
```
What about the tax?
Tax is calculated on the sale price
tax = sale_price * gst
let gst = 10%
total_price = sale_price + tax
```
gst = 10
tax = sale_price * gst/100
total_price = sale_price + tax
print("Total Price: ", total_price )
```
## Processing/Algorithm
1. Input data
2. Perform the Calculations
3. Output the results
# Modular
* easier to read
* think about each step
* may be ble to reuse "module"
* may be build library of common tasks
### Input Data Module (Step 1)
Input Item Name
Input Original Price
Input Discount Rate
```
def inputData():
item_name = input("Please enter the item name: ")
original_price = int(input("Pease enter the original price: "))
discount_rate = int(input("Please enter the discount rate: "))
return item_name, original_price, discount_rate
```
### Input Data Module (Step 1). v0.2
Write "Please input the item name"
Input Item Name
Write "Please enter the original price"
Input Original Price
Write "Please enter the discount rate"
Input Discount Rate
### Input Data Module (Step 1) v0.3
display to the screen "Please intput the item name"
store the input into a variable called item_name
display to the screen "Please enter the original price"
store the input into a variable called original_price
display to the screen "Please enter the discount rate"
store the input into a variable called discount_rate
### Output Results Module (Step 3)
Write "The item is:" +. item_name
Write "Pre sale price was: " + original_price
etc....
```
def outputResults(item_name, original_price, total_price):
'''This function will output the results of the discount in a nice format'''
print("The item is:" , item_name)
print("Pre-sale item is:" , original_price)
# print("Tax", tax)
print("Total Price is:" , total_price)
# Nedd to add more here!
outputResults("Python Programming Book", 50, 44)
```
## Perform the Calculations (Step 2)
amount_saved = original_price * ( discount_rate / 100)
sale_price = original_price - amount_saved
let gst = 10%
tax = sale_price * gst
total_price = sale_price + tax
```
def calculateTotalPrice(original_price, discount_rate):
amount_saved = original_price * ( discount_rate / 100)
sale_price = original_price - amount_saved
gst = 10
tax = sale_price * gst/100
total_price = sale_price + tax
return total_price
calculateTotalPrice(50, 20)
# Main Program
# Step 1: Input data
name, price, discount = inputData()
# Step 2: Perform the Calculations
total = calculateTotalPrice(price, discount)
# Step 3: Output the results
outputResults(name, price, total)
help(outputResults)
```
```
```
|
github_jupyter
|
50 * 20 / 100
original_price = 50
discount_rate = 20
amount_saved = original_price * (discount_rate / 100)
sale_price = original_price - amount_saved
print("Amount Saved: ", amount_saved)
print("Sale Price: ", sale_price)
gst = 10
tax = sale_price * gst/100
total_price = sale_price + tax
print("Total Price: ", total_price )
def inputData():
item_name = input("Please enter the item name: ")
original_price = int(input("Pease enter the original price: "))
discount_rate = int(input("Please enter the discount rate: "))
return item_name, original_price, discount_rate
def outputResults(item_name, original_price, total_price):
'''This function will output the results of the discount in a nice format'''
print("The item is:" , item_name)
print("Pre-sale item is:" , original_price)
# print("Tax", tax)
print("Total Price is:" , total_price)
# Nedd to add more here!
outputResults("Python Programming Book", 50, 44)
def calculateTotalPrice(original_price, discount_rate):
amount_saved = original_price * ( discount_rate / 100)
sale_price = original_price - amount_saved
gst = 10
tax = sale_price * gst/100
total_price = sale_price + tax
return total_price
calculateTotalPrice(50, 20)
# Main Program
# Step 1: Input data
name, price, discount = inputData()
# Step 2: Perform the Calculations
total = calculateTotalPrice(price, discount)
# Step 3: Output the results
outputResults(name, price, total)
help(outputResults)
| 0.212069 | 0.97859 |
# Upload video stimuli to s3
```
#Which experiment? bucket_name is the name of the experiment and will be name of the databases both on mongoDB and S3
bucket_name = 'human-physics-benchmarking-XXX-pilot' #CHANGE THIS ⚡️
import os
from glob import glob
import boto3
import botocore
from IPython.display import clear_output
import json
import pandas as pd
from PIL import Image
def list_files(paths, ext='mp4'):
"""Pass list of folders if there are stimuli in multiple folders.
Make sure that the containing folder is informative, as the rest of the path is ignored in naming.
Also returns filenames as uploaded to S3"""
if type(paths) is not list:
paths = [paths]
results = []
names = []
for path in paths:
results += [y for x in os.walk(path) for y in glob(os.path.join(x[0], '*.%s' % ext))]
names += [os.path.basename(os.path.dirname(y))+'_'+os.path.split(y)[1] for x in os.walk(path) for y in glob(os.path.join(x[0], '*.%s' % ext))]
return results,names
## helper to speed things up by not uploading images if they already exist, can be overriden
def check_exists(s3, bucket_name, stim_name):
try:
s3.Object(bucket_name,stim_name).load()
return True
except botocore.exceptions.ClientError as e:
if (e.response['Error']['Code'] == "404"):
print('The object does not exist.')
return False
else:
print('Something else has gone wrong with {}'.format(stim_name))
```
Pass list of folders if there are stimuli in multiple folders. Make sure that the containing folder is informative, as the rest of the path is ignored in naming.
```
## provide a stem directory
local_stem = 'XXX' #CHANGE THIS ⚡️
dirnames = [d.split('/')[-1] for d in glob(local_stem+'/*')]
paths_to_stim = [local_stem + d for d in dirnames]
full_stim_paths, filenames = [x for x in list_files(paths_to_stim) if x !='.DS_Store'] #generate filenames and stimpaths
full_map_paths, mapnames = [x for x in list_files(paths_to_stim, ext = 'png') if x !='.DS_Store'] #generate filenames and stimpaths for target/zone map
full_hdf5_paths, hdf5names = [x for x in list_files(paths_to_stim, ext = 'hdf5') if x !='.DS_Store'] #generate filenames and stimpaths for hdf5
print('We have {} stimuli to upload.'.format(len(full_stim_paths)))
# make sure to only up the _img pass
full_stim_paths = [p for p in full_stim_paths if '_img' in p]
filenames = [p for p in filenames if '_img' in p]
print('We have {} stimuli to upload.'.format(len(full_stim_paths)))
```
Upload to S3. This expects the `.aws/credentials` file in your home directory.
```
reallyRun = True
upload_hdf5s = True
if reallyRun:
## establish connection to s3
s3 = boto3.resource('s3')
## create a bucket with the appropriate bucket name
try:
b = s3.create_bucket(Bucket=bucket_name)
print('Created new bucket.')
# except NoCredentialsError:
# print("Credential missing") #.aws/credentials should be in home folder, not in repo folder
except Exception as e:
b = s3.Bucket(bucket_name)
print('Bucket already exists.',e)
## do we want to overwrite files on s3?
overwrite = True
## set bucket and objects to public
b.Acl().put(ACL='public-read') ## sets bucket to public
## now let's loop through stim paths and actually upload to s3 (woot!)
for i,path_to_file in enumerate(full_stim_paths):
stim_name = filenames[i]
if ((check_exists(s3, bucket_name, stim_name)==False) | (overwrite==True)):
print('Now uploading {} as {} | {} of {}'.format(os.path.split(path_to_file)[-1],stim_name,(i+1),len(full_stim_paths)))
s3.Object(bucket_name,stim_name).put(Body=open(path_to_file,'rb')) ## upload stimuli
s3.Object(bucket_name,stim_name).Acl().put(ACL='public-read') ## set access controls
else:
print('Skipping {} | {} of {} because it already exists.'.format(os.path.split(path_to_file)[-1],(i+1),len(full_stim_paths)))
clear_output(wait=True)
print('Done uploading videos')
for i,path_to_file in enumerate(full_map_paths):
stim_name = mapnames[i]
if ((check_exists(s3, bucket_name, stim_name)==False) | (overwrite==True)):
print('Now uploading {} as {} | {} of {}'.format(os.path.split(path_to_file)[-1],stim_name,(i+1),len(full_map_paths)))
s3.Object(bucket_name,stim_name).put(Body=open(path_to_file,'rb')) ## upload stimuli
s3.Object(bucket_name,stim_name).Acl().put(ACL='public-read') ## set access controls
else:
print('Skipping {} | {} of {} because it already exists.'.format(os.path.split(path_to_file)[-1],(i+1),len(full_map_paths)))
clear_output(wait=True)
print('Done uploading target/zone maps')
if upload_hdf5s:
for i,path_to_file in enumerate(full_hdf5_paths):
stim_name = hdf5names[i]
if ((check_exists(s3, bucket_name, stim_name)==False) | (overwrite==True)):
print('Now uploading {} as {} | {} of {}'.format(os.path.split(path_to_file)[-1],stim_name,(i+1),len(full_hdf5_paths)))
s3.Object(bucket_name,stim_name).put(Body=open(path_to_file,'rb')) ## upload stimuli
s3.Object(bucket_name,stim_name).Acl().put(ACL='public-read') ## set access controls
else:
print('Skipping {} | {} of {} because it already exists.'.format(os.path.split(path_to_file)[-1],(i+1),len(full_hdf5_paths)))
clear_output(wait=True)
print('Done uploading hdf5s')
print('Done!')
for my_bucket_object in b.objects.all():
print(my_bucket_object)
```
|
github_jupyter
|
#Which experiment? bucket_name is the name of the experiment and will be name of the databases both on mongoDB and S3
bucket_name = 'human-physics-benchmarking-XXX-pilot' #CHANGE THIS ⚡️
import os
from glob import glob
import boto3
import botocore
from IPython.display import clear_output
import json
import pandas as pd
from PIL import Image
def list_files(paths, ext='mp4'):
"""Pass list of folders if there are stimuli in multiple folders.
Make sure that the containing folder is informative, as the rest of the path is ignored in naming.
Also returns filenames as uploaded to S3"""
if type(paths) is not list:
paths = [paths]
results = []
names = []
for path in paths:
results += [y for x in os.walk(path) for y in glob(os.path.join(x[0], '*.%s' % ext))]
names += [os.path.basename(os.path.dirname(y))+'_'+os.path.split(y)[1] for x in os.walk(path) for y in glob(os.path.join(x[0], '*.%s' % ext))]
return results,names
## helper to speed things up by not uploading images if they already exist, can be overriden
def check_exists(s3, bucket_name, stim_name):
try:
s3.Object(bucket_name,stim_name).load()
return True
except botocore.exceptions.ClientError as e:
if (e.response['Error']['Code'] == "404"):
print('The object does not exist.')
return False
else:
print('Something else has gone wrong with {}'.format(stim_name))
## provide a stem directory
local_stem = 'XXX' #CHANGE THIS ⚡️
dirnames = [d.split('/')[-1] for d in glob(local_stem+'/*')]
paths_to_stim = [local_stem + d for d in dirnames]
full_stim_paths, filenames = [x for x in list_files(paths_to_stim) if x !='.DS_Store'] #generate filenames and stimpaths
full_map_paths, mapnames = [x for x in list_files(paths_to_stim, ext = 'png') if x !='.DS_Store'] #generate filenames and stimpaths for target/zone map
full_hdf5_paths, hdf5names = [x for x in list_files(paths_to_stim, ext = 'hdf5') if x !='.DS_Store'] #generate filenames and stimpaths for hdf5
print('We have {} stimuli to upload.'.format(len(full_stim_paths)))
# make sure to only up the _img pass
full_stim_paths = [p for p in full_stim_paths if '_img' in p]
filenames = [p for p in filenames if '_img' in p]
print('We have {} stimuli to upload.'.format(len(full_stim_paths)))
reallyRun = True
upload_hdf5s = True
if reallyRun:
## establish connection to s3
s3 = boto3.resource('s3')
## create a bucket with the appropriate bucket name
try:
b = s3.create_bucket(Bucket=bucket_name)
print('Created new bucket.')
# except NoCredentialsError:
# print("Credential missing") #.aws/credentials should be in home folder, not in repo folder
except Exception as e:
b = s3.Bucket(bucket_name)
print('Bucket already exists.',e)
## do we want to overwrite files on s3?
overwrite = True
## set bucket and objects to public
b.Acl().put(ACL='public-read') ## sets bucket to public
## now let's loop through stim paths and actually upload to s3 (woot!)
for i,path_to_file in enumerate(full_stim_paths):
stim_name = filenames[i]
if ((check_exists(s3, bucket_name, stim_name)==False) | (overwrite==True)):
print('Now uploading {} as {} | {} of {}'.format(os.path.split(path_to_file)[-1],stim_name,(i+1),len(full_stim_paths)))
s3.Object(bucket_name,stim_name).put(Body=open(path_to_file,'rb')) ## upload stimuli
s3.Object(bucket_name,stim_name).Acl().put(ACL='public-read') ## set access controls
else:
print('Skipping {} | {} of {} because it already exists.'.format(os.path.split(path_to_file)[-1],(i+1),len(full_stim_paths)))
clear_output(wait=True)
print('Done uploading videos')
for i,path_to_file in enumerate(full_map_paths):
stim_name = mapnames[i]
if ((check_exists(s3, bucket_name, stim_name)==False) | (overwrite==True)):
print('Now uploading {} as {} | {} of {}'.format(os.path.split(path_to_file)[-1],stim_name,(i+1),len(full_map_paths)))
s3.Object(bucket_name,stim_name).put(Body=open(path_to_file,'rb')) ## upload stimuli
s3.Object(bucket_name,stim_name).Acl().put(ACL='public-read') ## set access controls
else:
print('Skipping {} | {} of {} because it already exists.'.format(os.path.split(path_to_file)[-1],(i+1),len(full_map_paths)))
clear_output(wait=True)
print('Done uploading target/zone maps')
if upload_hdf5s:
for i,path_to_file in enumerate(full_hdf5_paths):
stim_name = hdf5names[i]
if ((check_exists(s3, bucket_name, stim_name)==False) | (overwrite==True)):
print('Now uploading {} as {} | {} of {}'.format(os.path.split(path_to_file)[-1],stim_name,(i+1),len(full_hdf5_paths)))
s3.Object(bucket_name,stim_name).put(Body=open(path_to_file,'rb')) ## upload stimuli
s3.Object(bucket_name,stim_name).Acl().put(ACL='public-read') ## set access controls
else:
print('Skipping {} | {} of {} because it already exists.'.format(os.path.split(path_to_file)[-1],(i+1),len(full_hdf5_paths)))
clear_output(wait=True)
print('Done uploading hdf5s')
print('Done!')
for my_bucket_object in b.objects.all():
print(my_bucket_object)
| 0.135061 | 0.641057 |
# 6.1 Pig
## Что такое Pig
<img src="slides/le06/pig/pig-03.png" title="Что такое Pig" width="400" height="400"/>
## Для чего нужен Pig
<img src="slides/le06/pig/pig-04.png" title="Для чего нужен Pig" width="400" height="400"/>
## Основные возможности Pig
<img src="slides/le06/pig/pig-06.png" title="Основные возможности Pig" width="400" height="400"/>
## Основные возможности Pig
<img src="slides/le06/pig/pig-07.png" title="КОмпоненты Pig" width="400" height="400"/>
## Режим выполнения
<img src="slides/le06/pig/pig-08.png" title="Режим выполнения" width="400" height="400"/>
## Запуск Pig
<img src="slides/le06/pig/pig-09.png" title="Запуск Pig" width="400" height="400"/>
## Pig Latin
<img src="slides/le06/pig/pig-10.png" title="Pig Latin" width="400" height="400"/>
Pig - типизированный язык.
- **field** - минимальный блок (число, строка и тд).
- Объединение field - **tuple** (кортеж), заключенный в `(` и `)`.
- **Bag** (мешок) - набор кортежей, заключенных в `{` и `}`. Кортежи могут отличаться по своему наполнению.
<img src="slides/le06/pig/pig-11.png" title="Pig Latin" width="400" height="400"/>
Схожесть с реляционной моделью:
- Bag vs Таблица БД
- Tuple vs Строка таблицы БД
- каждый tuple Bag **может имеет различную** структуру
- каждая строка БД **имеет одинаковую** структуру
- Field vs Поля строки
## Операции Dump и Store
<img src="slides/le06/pig/pig-13.png" title="Операции Dump и Store" width="400" height="400"/>
Работает режим отложенного запуска. Данные обрабатываются не в момент написания команд, а в момент необходимости вывода. Компилятор следит за синтаксисом.
## Большой объем данных
<img src="slides/le06/pig/pig-14.png" title="Большой объем данных" width="400" height="400"/>
## Операция Load
<img src="slides/le06/pig/pig-15.png" title="Операция Load" width="400" height="400"/>
По-умолчанию в using используется функция `PigStorage`, которая разделяет строки по табуляции. Можно изменить разделитель указав `PigStorage(';')`.
# 6.2 Основные операторы PigLatin
<img src="slides/le06/pig/pig-18.png" title="Пример" width="400" height="400"/>
- Сначала загружаем данные из `b.txt` файла в переменную `chars`. Файл предположительно состоит из одного поля `c`.
- Вызываем `DESCRIBE` чтобы посмотреть описание переменной.
- Вызываем `DUMP`, чтобы посмотреть, что содержит переменная.
- Делаем группировку `GROUP`, указав данные какой переменной мы хотим сгруппировать и по какому полю.
- Вызываем `DESCRIBE` чтобы посмотреть описание переменной после группировки. Получаем некий bag, который содержит первым полем строку (значение ключа), второе поле тоже bag, который содержит все tuples из bag `chars`.
<img src="slides/le06/pig/pig-19.png" title="Пример" width="400" height="400"/>
`ILLUSTRATE`
## FOREACH
<img src="slides/le06/pig/pig-21.png" title="FOREACH" width="400" height="400"/>
`FOREACH <bag> GENERATE <data>` - перебирает все кортежи из некоторого bag и определяем, что делать с данными через `GENERATE`.
```
# Получаем все поля i из кортежа
counts = FOREACH records GENERATE i;
```
<img src="slides/le06/pig/pig-22.png" title="FOREACH с функцией" width="400" height="400"/>
Обычно `FOREACH` используют с функцией. В Pig стандартные функции:
- `COUNT` количество объектов
- `FLATTEN`
- `CONCAT`.
Можно и пользовательские функции реализовывать.
<img src="slides/le06/pig/pig-23.png" title="Пример FOREACH с функцией" width="400" height="400"/>
Считаем количество объектов.
## TOKENIZE
<img src="slides/le06/pig/pig-24.png" title="TOKENIZE" width="400" height="400"/>
Функция разбивает строку на токены.
<img src="slides/le06/pig/pig-25.png" title="Пример TOKENIZE" width="400" height="400"/>
Зачитываем файл `c.txt` как одно большое поле `c`. Далее перебираем эти данные и применяем к каждой строке функцию `TOKENIZE` (аналогия со `split` строки). Было поле `(line)`, а мы его разбили на bag `{(token), (token), (token)}`.
**Дополнение**: запятые `TOKENIZE` отбрасывает. Он делит по пробелам, кавычкам, скобкам и запятым и звездочкам.
```
# Veni, vidi, vici
grunt> text = LOAD 'latin.txt' AS (line:chararray);
grunt> tokens = FOREACH text GENERATE TOKENIZE(line);
grunt> DUMP tokens;
({(veni), (vidi), (vici)})
```
<img src="slides/le06/pig/pig-26.png" title="Пример TOKENIZE" width="400" height="400"/>
Чтобы обратно собрать все токены используют функцию `flatten`. Из `{(t), (t), (t)}` -> `(f) (f) (f)`. `flatten` принимает индекс строки. `$0` это индекс поля в кортеже.
## Пример решения WordCount на Pig
<img src="slides/le06/pig/pig-27.png" title="Пример WordCount" width="400" height="400"/>
## JOINS
<img src="slides/le06/pig/pig-28.png" title="JOINS" width="400" height="400"/>
<img src="slides/le06/pig/pig-29.png" title="INNER JOINS" width="400" height="400"/>
`JOIN posts BY user, likes BY user` мы смотрим пересечение таблицы `posts` по полю `user` с таблицей `likes` то же по полю `user`.
<img src="slides/le06/pig/pig-30.png" title="INNER JOINS" width="400" height="400"/>
Результат объединения таков, что сначала будут идти данные из 1 файла для **user1**, а потом из второго. **User5** есть только во 2 файле, поэтому информацию по нему в итоговое пересечение не попадет. **User3** есть только в 1 файле, поэтому данные тоже не попадут.
<img src="slides/le06/pig/pig-31.png" title="Структура данных INNER JOINS" width="400" height="400"/>
Так как мы имеем 2 поля **user**, то необходимо указывать префикс, чтобы обозначить к какому полю мы обращаемся.
<img src="slides/le06/pig/pig-32.png" title="OUTER JOINS" width="400" height="400"/>
- Поле из 1 источника остается в любом случае
- Поле из 2 источника остается в любом случае
- Поля из обеих полей остаются в любом случае
<img src="slides/le06/pig/pig-33.png" title="LEFT JOINS" width="400" height="400"/>
<img src="slides/le06/pig/pig-34.png" title="LEFT JOINS" width="400" height="400"/>
# 6.3 Hive
<img src="slides/le06/hive/hive-02.png" title="Hive" width="400" height="400"/>
Hive похож на Pig. Они используются для одних и тех же целей. Hive работает поверх Hadoop. Hive не БД.
Hive online:
https://demo.gethue.com/hue/editor/?type=hive
<img src="slides/le06/hive/hive-03.png" title="Hive" width="400" height="400"/>
SQL-подобный язык HiveQL.
<img src="slides/le06/hive/hive-04.png" title="Hive" width="400" height="400"/>
Low latency запросы, которые предоставляются достаточно быстро. Хорошо масштабируемая система: хорошо работает как на малых объемах, так и на больших.
## Общая схема работы Hive
<img src="slides/le06/hive/hive-05.png" title="Общая схема работы Hive" width="400" height="400"/>
Пользователь пишет некоторый скрипт на HiveQL. Client Hive транслирует задачу в MapReduce задачи и выполняет на кластере Hadoop. По выполнению задачи результ отправляется обратно клиенту.
## Общая архитектура Hive
<img src="slides/le06/hive/hive-07.png" title="Общая архитектура Hive" width="400" height="400"/>
Центральное место занимает ядро **Hive**, которое состоит из **Query Parser** (раскладывает команды, которые клиент отрпавил для запуска). **Executor** запускает MapReduce задачи на кластере Hadoop.
В отличии от Pig, Hive использует фиксированную структуру данных и meta информацию он хранит в некотором хранилище **Metastore** (реляционная БД).
Клиент может взаимодействовать через CLI, через Java API, web интерфейс. Обычно клиенты выполняют 2 типа действий:
- Определяют структуру и взаимодействуют с Metastore.
- Запуск происходит с помощью обращения к ядру, которое распарсит запрос и транслирует в MapReduce задачи.
## Модель данных Hive
<img src="slides/le06/hive/hive-09.png" title="Модель данных Hive" width="400" height="400"/>
Модель данных очень похожа на обычную реляционную. В отличии от Pig строка имеет определенный набор полей. В начале пользователь должен описать структуру таблицы.
## Пример
<img src="slides/le06/hive/hive-10.png" title="Пример задачи Hive" width="400" height="400"/>
<img src="slides/le06/hive/hive-11.png" title="Пример задачи Hive" width="400" height="400"/>
Выполняем команду `hive`, чтобы войти в оболочку. В Hive мы можем выполнять стандартные Linux команды, только мы должны указать `!cat`.
<img src="slides/le06/hive/hive-12.png" title="Пример задачи Hive" width="400" height="400"/>
Далее описываем структуру таблицы.
- `CREATE TABLE post ...` - объявление таблицы
- `ROW FORMAT DELIMITED`
- `FIELDS TERMINATED BY ','` разделитель
- `STORED AS TEXTFILE` как сохранять данные
```sql
CREATE TABLE posts
(
user STRING,
post STRING,
time BIGINT
)
ROW FORMAT DELIMITED
FIELDS TERMITED BY ','
STORED AS TEXTFILE;
```
```bash
# Какие таблицы есть
show tables
# Описание таблицы
describe posts
```
<img src="slides/le06/hive/hive-14.png" title="Пример задачи Hive" width="400" height="400"/>
`select count(1) from posts` - считает количество записей в таблице posts. HiveQL преобразуется в 1 MapReduce задачу.
<img src="slides/le06/hive/hive-15.png" title="Пример задачи Hive" width="400" height="400"/>
Мы можем выбрать все записи пользователя:
`select * from posts where user="user2"`
## Hive: Joins
<img src="slides/le06/hive/hive-19.png" title="Hive Joins" width="400" height="400"/>
По-умолчанию Inner Join. В отличие от Pig Hive умеет объединять несколько таблиц.
<img src="slides/le06/hive/hive-20.png" title="Пример Hive Join" width="400" height="400"/>
<img src="slides/le06/hive/hive-21.png" title="Пример Hive Join" width="400" height="400"/>
Заполняем таблицу **posts_likes** пересечением двух таблиц:
```SQL
INSERT OVERWRITE TABLE posts_likes
SELECT p.user, p.post, l.count
FROM posts p
JOIN likes l
ON (p.user = l.user)
```
## Пример WordCounts
<img src="slides/le06/hive/hive-23.png" title="Пример Hive WordCounts" width="400" height="400"/>
Для подсчета слов используем временную таблицу
```sql
/* Сначала объявляем таблицу, в которой будем хранить весь текст в одной колонке типа STRING. */
CREATE TABLE docs (line STRING);
/*Загружаем данные из файла docks в таблицу docks путем перезаписи.*/
LOAD DATA INPATH 'docs' OVERWRITE INTO TABLE docs;
/*
Создаем временную таблицу word_counts (аналогия WITH).
Выбираем из нее слово и считаем количество
В подзапросе разбиваем строку на слова
explode - каждое слово, как отдельная строка
w - название подзапроса
*/
CREATE TABLE word_counts AS
SELECT word,
count(1) AS count
FROM
(SELECT explode(split(line, '\s')) AS word
FROM docs) w)
GROUP BY word
ORDER BY word;
```
<img src="slides/le06/hive/hive-24.png" title="Пример Hive WordCounts" width="400" height="400"/>
Hive для любителей SQL.
|
github_jupyter
|
# Получаем все поля i из кортежа
counts = FOREACH records GENERATE i;
# Veni, vidi, vici
grunt> text = LOAD 'latin.txt' AS (line:chararray);
grunt> tokens = FOREACH text GENERATE TOKENIZE(line);
grunt> DUMP tokens;
({(veni), (vidi), (vici)})
CREATE TABLE posts
(
user STRING,
post STRING,
time BIGINT
)
ROW FORMAT DELIMITED
FIELDS TERMITED BY ','
STORED AS TEXTFILE;
# Какие таблицы есть
show tables
# Описание таблицы
describe posts
INSERT OVERWRITE TABLE posts_likes
SELECT p.user, p.post, l.count
FROM posts p
JOIN likes l
ON (p.user = l.user)
/* Сначала объявляем таблицу, в которой будем хранить весь текст в одной колонке типа STRING. */
CREATE TABLE docs (line STRING);
/*Загружаем данные из файла docks в таблицу docks путем перезаписи.*/
LOAD DATA INPATH 'docs' OVERWRITE INTO TABLE docs;
/*
Создаем временную таблицу word_counts (аналогия WITH).
Выбираем из нее слово и считаем количество
В подзапросе разбиваем строку на слова
explode - каждое слово, как отдельная строка
w - название подзапроса
*/
CREATE TABLE word_counts AS
SELECT word,
count(1) AS count
FROM
(SELECT explode(split(line, '\s')) AS word
FROM docs) w)
GROUP BY word
ORDER BY word;
| 0.084276 | 0.782766 |
# Comparing currency trading strategies
[](https://mybinder.org/v2/gh/joepatmckenna/fem/master?filepath=doc%2Fnotebooks%2Fdiscrete%2F04_currency_trading.ipynb)
In this example, we compare two strategies for trading currencies that differ only in the degree of the underlying model. We present an example in which the higher degree model outperforms the lower degree model.
We start by loading the necessary modules and functions.
```
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import numpy as np
import pandas as pd
import os, fem, time
data_dir = '../../../data/currency'
cache = False
print 'number of processors: %i' % (fem.fortran_module.fortran_module.num_threads(),)
```
We use data of the currency exchange rates relative to the Euro from 2000 to 2018 for $n=11$ currencies (USD, CAD, MXN, GBP, NOK, CHF, SEK, AUD, JPY, KRW, and SGD) plotted below.
```
# load data
currency = pd.read_csv(os.path.join(data_dir, 'currency.csv'), index_col=0)
x = currency.values.T
# plot data
fig, ax = plt.subplots(x.shape[0], 1, figsize=(16,4))
date2num = mdates.strpdate2num(fmt='%Y-%m-%d')
dates = [date2num(date) for date in currency.index]
for i, xi in enumerate(x):
ax[i].plot_date(dates, xi, 'k-')
ax[i].set_ylabel(currency.columns[i], rotation=0, ha='right')
ax[i].set_yticks([])
for i in range(x.shape[0]-1):
for spine in ['left', 'right', 'top', 'bottom']:
ax[i].spines[spine].set_visible(False)
for spine in ['left', 'right', 'top']:
ax[-1].spines[spine].set_visible(False)
ax[-1].xaxis.set_major_locator(mdates.YearLocator())
ax[-1].xaxis.set_major_formatter(mdates.DateFormatter('%Y'))
ax[-1].xaxis.set_minor_locator(mdates.MonthLocator())
fig.autofmt_xdate()
plt.show()
```
We discretize each currency exchange rate sequence $\{x_i(t)\}_{t=t_1}^{t_{\ell}}$, where $\Delta t=t_{k+1}-t_k=1$ day, by recording the sign of the day-to-day change $\{s_i(t)\}_{t=t_2}^{t_{\ell}}$ with stagnation settled by the sign of the mean change: $$s_i(t_{k+1}) = \begin{cases}1&\text{ if }x_i(t_{k+1})-x_i(t_k)>0\\-1&\text{ if }x_i(t_{k+1})-x_i(t_k)<0\\\text{sign}\left({1\over\ell-1}\sum_{k=1}^{\ell-1}(x_i(t_{k+1})-x_i(t_k))\right)&\text{ if }x_i(t_{k+1})-x_i(t_k)=0 \end{cases}.$$ The first 200 fluctuations for each currency are plotted below.
```
# daily movement
dx = np.diff(x, axis=1)
# sign of daily movement
s = np.sign(dx).astype(int)
for i, si in enumerate(s):
s[i][si==0] = np.sign(dx[i].mean())
fig, ax = plt.subplots(s.shape[0], 1, figsize=(16,4))
for i, si in enumerate(s):
ax[i].plot_date(dates[1:201], si[:200], 'k-')
ax[i].set_ylabel(currency.columns[i], rotation=0, ha='right')
ax[i].set_yticks([])
for i in range(s.shape[0]-1):
for spine in ['left', 'right', 'top', 'bottom']:
ax[i].spines[spine].set_visible(False)
for spine in ['left', 'right', 'top']:
ax[-1].spines[spine].set_visible(False)
ax[-1].xaxis.set_major_locator(mdates.MonthLocator())
ax[-1].xaxis.set_major_formatter(mdates.DateFormatter('%B %Y'))
ax[-1].xaxis.set_minor_locator(mdates.DayLocator())
fig.autofmt_xdate()
plt.show()
```
We fit two different models to the one-hot encodings of the binary data $\{s_i(t)\}_{t=t_2}^{t_{\ell}}$. The one-hot encoding of $s(t_k)=(s_1(t_k),\ldots,s_{n}(t_k))^T\in\{-1,1\}^n$ is a binary vector $\sigma(t_k)=(\sigma_1(t_k),\ldots,\sigma_n(t_k))^T\in\{0,1\}^{2n}$ where $\sigma_i(t_k)=(1,0)^T$ if $s_i(t_k)=1$ and $\sigma_i(t_k)=(0,1)^T$ if $s_i(t_k)=-1$. In either model, the probability that $x_i$ increases from $t_k$ to $t_{k+1}$ is assumed to be
$$p(s_i(t_{k+1})~|~s(t_k)) = {\exp e_{2i}^T h(\sigma(t_k))\over\sum_{i=1}^n\exp e_{2i}^Th(\sigma(t_k))}.$$
The two models differ in their definition of $h$; in the first model, $h(\sigma(t_k)) = W_1\sigma(t_k)$, but in the second model, $h(\sigma(t_k)) = W_1\sigma(t_k) + W_2\sigma^2(t_k)$. The quadratic term $\sigma^2$ consists of distinct nonzero products of the form $\sigma_{j_2}\sigma_{j_1}$, $1\leq j_1, j_2\leq 2n$ (see [FEM for discrete data](../../../discrete) for more information).
For demonstration, we instantiate two models, one with `degs=[1]` and one with `degs=[1,2]`, and fit them to the whole currency data set. We plot the heat maps of the fitted model parameters and the running discrepancies during the fit for both models below.
```
# create two models
models = [fem.discrete.fit.model(degs=[1]), fem.discrete.fit.model(degs=[1, 2])]
# fit model to whole currency data set
for i, model in enumerate(models):
start = time.time()
model.fit(s[:,:-1], s[:, 1:], overfit=False)
end = time.time()
print 'model %i fit time: %.02f seconds' % (i+1, end-start)
# plot model parameter heat maps and running discrepancies
fig, ax = plt.subplots(1, 4, figsize=(12, 3))
for i, model in enumerate(models):
w = np.hstack(model.w.itervalues())
scale = np.abs(w).max()
ax[2*i].matshow(w, cmap='seismic', vmin=-scale, vmax=scale, aspect='auto')
ax[2*i].xaxis.set_ticks_position('bottom')
for d in model.d:
ax[2*i+1].plot(1 + np.arange(len(d)), d, 'k-', lw=0.1)
ax[2*i+1].set_xlabel('iteration')
ax[2*i+1].set_ylabel('discrepancy')
ax[0].set_title('model 1 $W_1$', fontsize=14)
ax[2].set_title('model 2 $[W_1, W_2]$', fontsize=14)
plt.tight_layout()
plt.show()
```
Next, we devise two simple trading strategies that train the models on a moving time window then predict $s$ one day ahead of the window. We keep track of our account balances and prediction accuracies in the `acccount` and `accuracy` variables. The trading strategies work as follows. The models are trained on the data in the time window $[$ `t1` $,$ `t1` $+$ `tw` $-1]$ to predict $s($ `t1+tw` $)$. Initially, `t1` $=$ `t0`, the initial time of the data set, and `t1` is incremented by 1 while `t1+tw` is less than `tn`, the final time of the data set. On each day, we pass the data at `t1` $+$ `tw` to `model.predict`, which returns a prediction, 1 or -1, and probability greater than 0.5 of the price movement tomorrow. We select to trade only those currencies whose probabilities are greater than `threshold`, and we invest our whole account value weighted in the chosen currencies according the probabilities leveraged by a constant factor `leverage`. The models are retrained every `dt` days.
```
account_file = os.path.join(data_dir, 'account.npy')
accuracy_file = os.path.join(data_dir, 'accuracy.npy')
if cache and os.path.exists(account_file) and os.path.exists(accuracy_file):
account = np.load(account_file)
accuracy = np.load(accuracy_file)
else:
# t0, tn: initial and final time
t0, tn = 0, s.shape[1]
# daily balance
account = np.ones((2, tn))
# daily prediction accuracy
accuracy = np.zeros((2, tn))
# tw: training time window width
# dt: retraining period
tw, dt = 200, 1
# t1, t2: limits of moving training window
t1, t2 = t0, t0+tw
# maximum position weight
max_weight = 1.0 / 3.0
# trade if probability above threshold
threshold = 0.75
# percentage of account to bid
leverage = 1.0
start = time.time()
while t2 < tn:
# todays price
price = x[:, t2]
# tomorrows change
realized_dx = dx[:, t2]
realized_s = s[:, t2]
percent_change = realized_dx / price
for i, model in enumerate(models):
# start todays account with yesterdays balance
account[i, t2] = account[i, t2-1]
# retrain the model
if not (t1-t0) % dt:
s_train = s[:,t1:t2]
model.fit(s_train[:,:-1], s_train[:, 1:], overfit=False)
# predict which currencies will increase
pred, prob = model.predict(s_train[:, -1])
# trade if probability above threshold
portfolio = prob > threshold
if not portfolio.any():
continue
# position weights proportional to probability of prediction
weights = (prob * pred)[portfolio]
weights /= np.abs(weights).sum()
weights[weights > max_weight] = max_weight
# take trading positions
position = leverage * account[i,t2] * weights
# calculuate returns and accuracy
account[i, t2] += (position * percent_change[portfolio]).sum()
accuracy[i, t2] = (pred == realized_s)[portfolio].mean()
t1 += 1
t2 += 1
end = time.time()
print 'backtest time: %.02f minutes' % ((end-start)/60.,)
np.save(account_file, account)
np.save(accuracy_file, accuracy)
accuracy_rolling_mean = pd.DataFrame(accuracy.T).rolling(100).mean().values.T
print 'accuracy: model 1: %.02f, model 2: %.02f' % (accuracy[0].mean(), accuracy[1].mean())
fig, ax = plt.subplots(2, 1, figsize=(14, 5))
ax[0].plot_date(dates[1:], account[0], 'k-', label='linear')
ax[0].plot_date(dates[1:], account[1], 'r-', label='quadratic')
ax[1].plot_date(dates[1:], accuracy_rolling_mean[0], 'k-')
ax[1].plot_date(dates[1:], accuracy_rolling_mean[1], 'r-')
ax[0].set_ylabel('account %% change')
ax[1].set_ylabel('accuracy')
plt.show()
```
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import numpy as np
import pandas as pd
import os, fem, time
data_dir = '../../../data/currency'
cache = False
print 'number of processors: %i' % (fem.fortran_module.fortran_module.num_threads(),)
# load data
currency = pd.read_csv(os.path.join(data_dir, 'currency.csv'), index_col=0)
x = currency.values.T
# plot data
fig, ax = plt.subplots(x.shape[0], 1, figsize=(16,4))
date2num = mdates.strpdate2num(fmt='%Y-%m-%d')
dates = [date2num(date) for date in currency.index]
for i, xi in enumerate(x):
ax[i].plot_date(dates, xi, 'k-')
ax[i].set_ylabel(currency.columns[i], rotation=0, ha='right')
ax[i].set_yticks([])
for i in range(x.shape[0]-1):
for spine in ['left', 'right', 'top', 'bottom']:
ax[i].spines[spine].set_visible(False)
for spine in ['left', 'right', 'top']:
ax[-1].spines[spine].set_visible(False)
ax[-1].xaxis.set_major_locator(mdates.YearLocator())
ax[-1].xaxis.set_major_formatter(mdates.DateFormatter('%Y'))
ax[-1].xaxis.set_minor_locator(mdates.MonthLocator())
fig.autofmt_xdate()
plt.show()
# daily movement
dx = np.diff(x, axis=1)
# sign of daily movement
s = np.sign(dx).astype(int)
for i, si in enumerate(s):
s[i][si==0] = np.sign(dx[i].mean())
fig, ax = plt.subplots(s.shape[0], 1, figsize=(16,4))
for i, si in enumerate(s):
ax[i].plot_date(dates[1:201], si[:200], 'k-')
ax[i].set_ylabel(currency.columns[i], rotation=0, ha='right')
ax[i].set_yticks([])
for i in range(s.shape[0]-1):
for spine in ['left', 'right', 'top', 'bottom']:
ax[i].spines[spine].set_visible(False)
for spine in ['left', 'right', 'top']:
ax[-1].spines[spine].set_visible(False)
ax[-1].xaxis.set_major_locator(mdates.MonthLocator())
ax[-1].xaxis.set_major_formatter(mdates.DateFormatter('%B %Y'))
ax[-1].xaxis.set_minor_locator(mdates.DayLocator())
fig.autofmt_xdate()
plt.show()
# create two models
models = [fem.discrete.fit.model(degs=[1]), fem.discrete.fit.model(degs=[1, 2])]
# fit model to whole currency data set
for i, model in enumerate(models):
start = time.time()
model.fit(s[:,:-1], s[:, 1:], overfit=False)
end = time.time()
print 'model %i fit time: %.02f seconds' % (i+1, end-start)
# plot model parameter heat maps and running discrepancies
fig, ax = plt.subplots(1, 4, figsize=(12, 3))
for i, model in enumerate(models):
w = np.hstack(model.w.itervalues())
scale = np.abs(w).max()
ax[2*i].matshow(w, cmap='seismic', vmin=-scale, vmax=scale, aspect='auto')
ax[2*i].xaxis.set_ticks_position('bottom')
for d in model.d:
ax[2*i+1].plot(1 + np.arange(len(d)), d, 'k-', lw=0.1)
ax[2*i+1].set_xlabel('iteration')
ax[2*i+1].set_ylabel('discrepancy')
ax[0].set_title('model 1 $W_1$', fontsize=14)
ax[2].set_title('model 2 $[W_1, W_2]$', fontsize=14)
plt.tight_layout()
plt.show()
account_file = os.path.join(data_dir, 'account.npy')
accuracy_file = os.path.join(data_dir, 'accuracy.npy')
if cache and os.path.exists(account_file) and os.path.exists(accuracy_file):
account = np.load(account_file)
accuracy = np.load(accuracy_file)
else:
# t0, tn: initial and final time
t0, tn = 0, s.shape[1]
# daily balance
account = np.ones((2, tn))
# daily prediction accuracy
accuracy = np.zeros((2, tn))
# tw: training time window width
# dt: retraining period
tw, dt = 200, 1
# t1, t2: limits of moving training window
t1, t2 = t0, t0+tw
# maximum position weight
max_weight = 1.0 / 3.0
# trade if probability above threshold
threshold = 0.75
# percentage of account to bid
leverage = 1.0
start = time.time()
while t2 < tn:
# todays price
price = x[:, t2]
# tomorrows change
realized_dx = dx[:, t2]
realized_s = s[:, t2]
percent_change = realized_dx / price
for i, model in enumerate(models):
# start todays account with yesterdays balance
account[i, t2] = account[i, t2-1]
# retrain the model
if not (t1-t0) % dt:
s_train = s[:,t1:t2]
model.fit(s_train[:,:-1], s_train[:, 1:], overfit=False)
# predict which currencies will increase
pred, prob = model.predict(s_train[:, -1])
# trade if probability above threshold
portfolio = prob > threshold
if not portfolio.any():
continue
# position weights proportional to probability of prediction
weights = (prob * pred)[portfolio]
weights /= np.abs(weights).sum()
weights[weights > max_weight] = max_weight
# take trading positions
position = leverage * account[i,t2] * weights
# calculuate returns and accuracy
account[i, t2] += (position * percent_change[portfolio]).sum()
accuracy[i, t2] = (pred == realized_s)[portfolio].mean()
t1 += 1
t2 += 1
end = time.time()
print 'backtest time: %.02f minutes' % ((end-start)/60.,)
np.save(account_file, account)
np.save(accuracy_file, accuracy)
accuracy_rolling_mean = pd.DataFrame(accuracy.T).rolling(100).mean().values.T
print 'accuracy: model 1: %.02f, model 2: %.02f' % (accuracy[0].mean(), accuracy[1].mean())
fig, ax = plt.subplots(2, 1, figsize=(14, 5))
ax[0].plot_date(dates[1:], account[0], 'k-', label='linear')
ax[0].plot_date(dates[1:], account[1], 'r-', label='quadratic')
ax[1].plot_date(dates[1:], accuracy_rolling_mean[0], 'k-')
ax[1].plot_date(dates[1:], accuracy_rolling_mean[1], 'r-')
ax[0].set_ylabel('account %% change')
ax[1].set_ylabel('accuracy')
plt.show()
| 0.40204 | 0.980053 |
```
import gc
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import torch
import torch.nn as nn
import torchvision
import sys
# To view tensorboard metrics
# tensorboard --logdir=logs --port=6006 --bind_all
from torch.utils.tensorboard import SummaryWriter
from functools import partial
from evolver import CrossoverType, MutationType, InitType, MatrixEvolver, VectorEvolver
from unet import UNet
from dataset_utils import PartitionType
from cuda_utils import maybe_get_cuda_device, clear_cuda
from landcover_dataloader import get_landcover_dataloaders, get_landcover_dataloader
from ignite.contrib.handlers.tensorboard_logger import *
from ignite.engine import Events, create_supervised_trainer, create_supervised_evaluator
from ignite.metrics import Accuracy, Loss, ConfusionMatrix, mIoU
from ignite.handlers import ModelCheckpoint
from ignite.utils import setup_logger
from ignite.engine import Engine
# Define directories for data, logging and model saving.
base_dir = os.getcwd()
dataset_name = "landcover_large"
dataset_dir = os.path.join(base_dir, "data/" + dataset_name)
experiment_name = "dropout_single_point_finetuning_100_children"
model_name = "best_model_9_validation_accuracy=0.8940.pt"
model_path = os.path.join(base_dir, "logs/" + dataset_name + "/" + model_name)
log_dir = os.path.join(base_dir, "logs/" + dataset_name + "_" + experiment_name)
# Create DataLoaders for each partition of Landcover data.
dataloader_params = {
'batch_size': 16,
'shuffle': True,
'num_workers': 6,
'pin_memory': True}
partition_types = [PartitionType.TRAIN, PartitionType.VALIDATION,
PartitionType.FINETUNING, PartitionType.TEST]
data_loaders = get_landcover_dataloaders(dataset_dir,
partition_types,
dataloader_params,
force_create_dataset=False)
train_loader = data_loaders[0]
finetuning_loader = data_loaders[2]
dataloader_params['shuffle'] = False
test_loader = get_landcover_dataloader(dataset_dir, PartitionType.TEST, dataloader_params)
# Get GPU device if available.
device = maybe_get_cuda_device()
# Determine model and training params.
params = {
'max_epochs': 10,
'n_classes': 4,
'in_channels': 4,
'depth': 5,
'learning_rate': 0.001,
'log_steps': 1,
'save_top_n_models': 4,
'num_children': 100
}
clear_cuda()
model = UNet(in_channels = params['in_channels'],
n_classes = params['n_classes'],
depth = params['depth'])
model.load_state_dict(torch.load(model_path))
# Create Trainer or Evaluators
criterion = nn.NLLLoss()
optimizer = torch.optim.Adam(model.parameters(),
lr=params['learning_rate'])
# Determine metrics for evaluation.
metrics = {
"accuracy": Accuracy(),
"loss": Loss(criterion),
"mean_iou": mIoU(ConfusionMatrix(num_classes = params['n_classes'])),
}
for batch in train_loader:
batch_x = batch[0]
_ = model(batch_x)
break
drop_out_layers = model.get_dropout_layers()
del model, batch_x
clear_cuda()
for layer in drop_out_layers:
layer_name = layer.name
size = layer.x_size[1:]
sizes = [size]
clear_cuda()
model = UNet(in_channels = params['in_channels'],
n_classes = params['n_classes'],
depth = params['depth'])
model.load_state_dict(torch.load(model_path))
model.to(device)
criterion = nn.NLLLoss()
optimizer = torch.optim.Adam(model.parameters(),
lr=params['learning_rate'])
num_channels = size[0]
evolver = VectorEvolver(num_channels,
CrossoverType.UNIFORM,
MutationType.FLIP_BIT,
InitType.RANDOM,
flip_bit_prob=0.25,
flip_bit_decay=0.5)
log_dir_test = log_dir + "_" + layer_name
def mask_from_vec(vec, matrix_size):
mask = np.ones(matrix_size)
for i in range(len(vec)):
if vec[i] == 0:
mask[i, :, :] = 0
elif vec[i] == 1:
mask[i, :, :] = 1
return mask
def dropout_finetune_step(engine, batch):
model.eval()
with torch.no_grad():
batch_x, batch_y = batch
batch_x = batch_x.to(device)
batch_y = batch_y.to(device)
loss = sys.float_info.max
for i in range(params['num_children']):
model.zero_grad()
child_vec = evolver.spawn_child()
child_mask = mask_from_vec(child_vec, size)
model.set_dropout_masks({layer_name: torch.tensor(child_mask, dtype=torch.float32).to(device)})
outputs = model(batch_x)
current_loss = criterion(outputs[:, :, 127:128,127:128], batch_y[:,127:128,127:128]).item()
loss = min(loss, current_loss)
if current_loss == 0.0:
current_loss = sys.float_info.max
else:
current_loss = 1.0 / current_loss
evolver.add_child(child_vec, current_loss)
priority, best_child = evolver.get_best_child()
best_mask = mask_from_vec(best_child, size)
model.set_dropout_masks({layer_name: torch.tensor(best_mask, dtype=torch.float32).to(device)})
return loss
# Create Trainer or Evaluators
trainer = Engine(dropout_finetune_step)
train_evaluator = create_supervised_evaluator(model, metrics=metrics, device=device)
validation_evaluator = create_supervised_evaluator(model, metrics=metrics, device=device)
trainer.logger = setup_logger("Trainer")
train_evaluator.logger = setup_logger("Train Evaluator")
validation_evaluator.logger = setup_logger("Validation Evaluator")
@trainer.on(Events.ITERATION_COMPLETED(every=1))
def report_evolver_stats(engine):
priorities = np.array(evolver.get_generation_priorities())
# Take reciprocal since we needed to store priorities in min heap.
priorities = 1.0 / priorities
tb_logger.writer.add_scalar("training/evolver_count",
priorities.shape[0], engine.state.iteration)
tb_logger.writer.add_scalar("training/evolver_mean",
np.mean(priorities), engine.state.iteration)
tb_logger.writer.add_scalar("training/evolver_std",
np.std(priorities), engine.state.iteration)
evolver.update_parents()
@trainer.on(Events.EPOCH_COMPLETED)
def visualize_validation_predictions(engine):
for i, batch in enumerate(test_loader):
batch_x, batch_y = batch
batch_x = batch_x.to(device)
batch_y = batch_y.to(device)
outputs = model(batch_x)
num_images = batch_x.shape[0]
batch_y_detach = batch_y.detach().cpu().numpy()
batch_x_detach = batch_x.detach().cpu().numpy()
outputs_detach = outputs.detach().cpu().numpy()
for j in range(num_images):
f, ax = plt.subplots(1, 3, figsize=(10, 4))
ax[0].imshow(np.moveaxis(batch_x_detach[j, :, :, :], [0], [2]) / 255.0)
ax[1].imshow((np.array(batch_y_detach[j, :, :])))
ax[2].imshow(np.argmax(np.moveaxis(np.array(outputs_detach[j, :, :, :]), [0],[ 2]), axis=2))
ax[0].set_title("X")
ax[1].set_title("Y")
ax[2].set_title("Predict")
f.suptitle("Layer: " + layer_name + " Itteration: " + str(engine.state.iteration) + " Image: " + str(j))
plt.show()
if i > 5:
break
break
# Tensorboard Logger setup below based on pytorch ignite example
# https://github.com/pytorch/ignite/blob/master/examples/contrib/mnist/mnist_with_tensorboard_logger.py
@trainer.on(Events.EPOCH_COMPLETED)
def compute_metrics(engine):
"""Callback to compute metrics on the train and validation data."""
train_evaluator.run(finetuning_loader)
validation_evaluator.run(test_loader)
def score_function(engine):
"""Function to determine the metric upon which to compare model."""
return engine.state.metrics["accuracy"]
# Setup Tensor Board Logging
tb_logger = TensorboardLogger(log_dir=log_dir_test)
tb_logger.attach_output_handler(
trainer,
event_name=Events.ITERATION_COMPLETED(every=params['log_steps']),
tag="training",
output_transform=lambda loss: {"batchloss": loss},
metric_names="all",
)
for tag, evaluator in [("training", train_evaluator), ("validation", validation_evaluator)]:
tb_logger.attach_output_handler(
evaluator,
event_name=Events.EPOCH_COMPLETED,
tag=tag,
metric_names="all",
global_step_transform=global_step_from_engine(trainer),
)
tb_logger.attach_opt_params_handler(trainer,
event_name=Events.ITERATION_COMPLETED(every=params['log_steps']),
optimizer=optimizer)
model_checkpoint = ModelCheckpoint(
log_dir_test,
n_saved=params['save_top_n_models'],
filename_prefix="best",
score_function=score_function,
score_name="validation_accuracy",
global_step_transform=global_step_from_engine(trainer),
)
validation_evaluator.add_event_handler(Events.COMPLETED, model_checkpoint, {"model": model})
trainer.run(finetuning_loader, max_epochs=params['max_epochs'])
tb_logger.close()
```
|
github_jupyter
|
import gc
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import torch
import torch.nn as nn
import torchvision
import sys
# To view tensorboard metrics
# tensorboard --logdir=logs --port=6006 --bind_all
from torch.utils.tensorboard import SummaryWriter
from functools import partial
from evolver import CrossoverType, MutationType, InitType, MatrixEvolver, VectorEvolver
from unet import UNet
from dataset_utils import PartitionType
from cuda_utils import maybe_get_cuda_device, clear_cuda
from landcover_dataloader import get_landcover_dataloaders, get_landcover_dataloader
from ignite.contrib.handlers.tensorboard_logger import *
from ignite.engine import Events, create_supervised_trainer, create_supervised_evaluator
from ignite.metrics import Accuracy, Loss, ConfusionMatrix, mIoU
from ignite.handlers import ModelCheckpoint
from ignite.utils import setup_logger
from ignite.engine import Engine
# Define directories for data, logging and model saving.
base_dir = os.getcwd()
dataset_name = "landcover_large"
dataset_dir = os.path.join(base_dir, "data/" + dataset_name)
experiment_name = "dropout_single_point_finetuning_100_children"
model_name = "best_model_9_validation_accuracy=0.8940.pt"
model_path = os.path.join(base_dir, "logs/" + dataset_name + "/" + model_name)
log_dir = os.path.join(base_dir, "logs/" + dataset_name + "_" + experiment_name)
# Create DataLoaders for each partition of Landcover data.
dataloader_params = {
'batch_size': 16,
'shuffle': True,
'num_workers': 6,
'pin_memory': True}
partition_types = [PartitionType.TRAIN, PartitionType.VALIDATION,
PartitionType.FINETUNING, PartitionType.TEST]
data_loaders = get_landcover_dataloaders(dataset_dir,
partition_types,
dataloader_params,
force_create_dataset=False)
train_loader = data_loaders[0]
finetuning_loader = data_loaders[2]
dataloader_params['shuffle'] = False
test_loader = get_landcover_dataloader(dataset_dir, PartitionType.TEST, dataloader_params)
# Get GPU device if available.
device = maybe_get_cuda_device()
# Determine model and training params.
params = {
'max_epochs': 10,
'n_classes': 4,
'in_channels': 4,
'depth': 5,
'learning_rate': 0.001,
'log_steps': 1,
'save_top_n_models': 4,
'num_children': 100
}
clear_cuda()
model = UNet(in_channels = params['in_channels'],
n_classes = params['n_classes'],
depth = params['depth'])
model.load_state_dict(torch.load(model_path))
# Create Trainer or Evaluators
criterion = nn.NLLLoss()
optimizer = torch.optim.Adam(model.parameters(),
lr=params['learning_rate'])
# Determine metrics for evaluation.
metrics = {
"accuracy": Accuracy(),
"loss": Loss(criterion),
"mean_iou": mIoU(ConfusionMatrix(num_classes = params['n_classes'])),
}
for batch in train_loader:
batch_x = batch[0]
_ = model(batch_x)
break
drop_out_layers = model.get_dropout_layers()
del model, batch_x
clear_cuda()
for layer in drop_out_layers:
layer_name = layer.name
size = layer.x_size[1:]
sizes = [size]
clear_cuda()
model = UNet(in_channels = params['in_channels'],
n_classes = params['n_classes'],
depth = params['depth'])
model.load_state_dict(torch.load(model_path))
model.to(device)
criterion = nn.NLLLoss()
optimizer = torch.optim.Adam(model.parameters(),
lr=params['learning_rate'])
num_channels = size[0]
evolver = VectorEvolver(num_channels,
CrossoverType.UNIFORM,
MutationType.FLIP_BIT,
InitType.RANDOM,
flip_bit_prob=0.25,
flip_bit_decay=0.5)
log_dir_test = log_dir + "_" + layer_name
def mask_from_vec(vec, matrix_size):
mask = np.ones(matrix_size)
for i in range(len(vec)):
if vec[i] == 0:
mask[i, :, :] = 0
elif vec[i] == 1:
mask[i, :, :] = 1
return mask
def dropout_finetune_step(engine, batch):
model.eval()
with torch.no_grad():
batch_x, batch_y = batch
batch_x = batch_x.to(device)
batch_y = batch_y.to(device)
loss = sys.float_info.max
for i in range(params['num_children']):
model.zero_grad()
child_vec = evolver.spawn_child()
child_mask = mask_from_vec(child_vec, size)
model.set_dropout_masks({layer_name: torch.tensor(child_mask, dtype=torch.float32).to(device)})
outputs = model(batch_x)
current_loss = criterion(outputs[:, :, 127:128,127:128], batch_y[:,127:128,127:128]).item()
loss = min(loss, current_loss)
if current_loss == 0.0:
current_loss = sys.float_info.max
else:
current_loss = 1.0 / current_loss
evolver.add_child(child_vec, current_loss)
priority, best_child = evolver.get_best_child()
best_mask = mask_from_vec(best_child, size)
model.set_dropout_masks({layer_name: torch.tensor(best_mask, dtype=torch.float32).to(device)})
return loss
# Create Trainer or Evaluators
trainer = Engine(dropout_finetune_step)
train_evaluator = create_supervised_evaluator(model, metrics=metrics, device=device)
validation_evaluator = create_supervised_evaluator(model, metrics=metrics, device=device)
trainer.logger = setup_logger("Trainer")
train_evaluator.logger = setup_logger("Train Evaluator")
validation_evaluator.logger = setup_logger("Validation Evaluator")
@trainer.on(Events.ITERATION_COMPLETED(every=1))
def report_evolver_stats(engine):
priorities = np.array(evolver.get_generation_priorities())
# Take reciprocal since we needed to store priorities in min heap.
priorities = 1.0 / priorities
tb_logger.writer.add_scalar("training/evolver_count",
priorities.shape[0], engine.state.iteration)
tb_logger.writer.add_scalar("training/evolver_mean",
np.mean(priorities), engine.state.iteration)
tb_logger.writer.add_scalar("training/evolver_std",
np.std(priorities), engine.state.iteration)
evolver.update_parents()
@trainer.on(Events.EPOCH_COMPLETED)
def visualize_validation_predictions(engine):
for i, batch in enumerate(test_loader):
batch_x, batch_y = batch
batch_x = batch_x.to(device)
batch_y = batch_y.to(device)
outputs = model(batch_x)
num_images = batch_x.shape[0]
batch_y_detach = batch_y.detach().cpu().numpy()
batch_x_detach = batch_x.detach().cpu().numpy()
outputs_detach = outputs.detach().cpu().numpy()
for j in range(num_images):
f, ax = plt.subplots(1, 3, figsize=(10, 4))
ax[0].imshow(np.moveaxis(batch_x_detach[j, :, :, :], [0], [2]) / 255.0)
ax[1].imshow((np.array(batch_y_detach[j, :, :])))
ax[2].imshow(np.argmax(np.moveaxis(np.array(outputs_detach[j, :, :, :]), [0],[ 2]), axis=2))
ax[0].set_title("X")
ax[1].set_title("Y")
ax[2].set_title("Predict")
f.suptitle("Layer: " + layer_name + " Itteration: " + str(engine.state.iteration) + " Image: " + str(j))
plt.show()
if i > 5:
break
break
# Tensorboard Logger setup below based on pytorch ignite example
# https://github.com/pytorch/ignite/blob/master/examples/contrib/mnist/mnist_with_tensorboard_logger.py
@trainer.on(Events.EPOCH_COMPLETED)
def compute_metrics(engine):
"""Callback to compute metrics on the train and validation data."""
train_evaluator.run(finetuning_loader)
validation_evaluator.run(test_loader)
def score_function(engine):
"""Function to determine the metric upon which to compare model."""
return engine.state.metrics["accuracy"]
# Setup Tensor Board Logging
tb_logger = TensorboardLogger(log_dir=log_dir_test)
tb_logger.attach_output_handler(
trainer,
event_name=Events.ITERATION_COMPLETED(every=params['log_steps']),
tag="training",
output_transform=lambda loss: {"batchloss": loss},
metric_names="all",
)
for tag, evaluator in [("training", train_evaluator), ("validation", validation_evaluator)]:
tb_logger.attach_output_handler(
evaluator,
event_name=Events.EPOCH_COMPLETED,
tag=tag,
metric_names="all",
global_step_transform=global_step_from_engine(trainer),
)
tb_logger.attach_opt_params_handler(trainer,
event_name=Events.ITERATION_COMPLETED(every=params['log_steps']),
optimizer=optimizer)
model_checkpoint = ModelCheckpoint(
log_dir_test,
n_saved=params['save_top_n_models'],
filename_prefix="best",
score_function=score_function,
score_name="validation_accuracy",
global_step_transform=global_step_from_engine(trainer),
)
validation_evaluator.add_event_handler(Events.COMPLETED, model_checkpoint, {"model": model})
trainer.run(finetuning_loader, max_epochs=params['max_epochs'])
tb_logger.close()
| 0.654564 | 0.285434 |
```
#!/usr/bin/env python
# vim:fileencoding=utf-8
import sys
import scipy.signal as sg
import matplotlib.pyplot as plt
import soundfile as sf
import matplotlib
import pandas as pd
#可視化ライブラリ
import seaborn as sns
#距離計算
from scipy.spatial import distance
#音楽ファイル
NHKRadio_file = './Input/01_Radio/NHKRadio.wav'
NHKBusiness_file = './Input/02_Business/NHKBusiness.wav'
Classic_file = './Input/03_Classic/Classic.wav'
#このノイズファイルは、ノイズが追加された信号ではなく、
#音楽ファイルと長さが同じノイズファイル
NHKRadio_noise_file = './Output/Signal_Reconstruction_0.18.1/NHKRadio_tpdfnoiseWH_write_Matrix_50.wav'
NHKBusiness_noise_file = './Output/Signal_Reconstruction_0.18.1/NHKBusiness_tpdfnoiseWH_write_Matrix_50.wav'
Classic_noise_file = './Output/Signal_Reconstruction_0.18.1/Classic_tpdfnoiseWH_write_Matrix_50.wav'
NoiseType = "Tpdfnoise"
plt.close("all")
# wavファイル読み込み
NHKRadio_wav, NHKRadio_fs = sf.read(NHKRadio_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(NHKRadio_wav.shape[1] == 1):
NHKRadio_wavdata = NHKRadio_wav
print(NHKRadio_wav.shape[1])
else:
NHKRadio_wavdata = (0.5 * NHKRadio_wav[:, 1]) + (0.5 * NHKRadio_wav[:, 0])
# wavファイル読み込み
NHKRadio_noise_wav, NHKRadio_noise_fs = sf.read(NHKRadio_noise_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(NHKRadio_noise_wav.shape[1] == 1):
NHKRadio_noise_wavdata = NHKRadio_noise_wav
print(NHKRadio_noise_wav.shape[1])
else:
NHKRadio_noise_wavdata = (0.5 * NHKRadio_noise_wav[:, 1]) + (0.5 * NHKRadio_noise_wav[:, 0])
# wavファイル読み込み
NHKBusiness_wav, NHKBusiness_fs = sf.read(NHKBusiness_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(NHKBusiness_wav.shape[1] == 1):
NHKBusiness_wavdata = NHKBusiness_wav
print(NHKBusiness_wav.shape[1])
else:
NHKBusiness_wavdata = (0.5 * NHKBusiness_wav[:, 1]) + (0.5 * NHKBusiness_wav[:, 0])
# wavファイル読み込み
NHKBusiness_noise_wav, NHKBusiness_noise_fs = sf.read(NHKBusiness_noise_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(NHKBusiness_noise_wav.shape[1] == 1):
NHKBusiness_noise_wavdata = NHKBusiness_noise_wav
print(NHKBusiness_noise_wav.shape[1])
else:
NHKBusiness_noise_wavdata = (0.5 * NHKBusiness_noise_wav[:, 1]) + (0.5 * NHKBusiness_noise_wav[:, 0])
# wavファイル読み込み
Classic_wav, Classic_fs = sf.read(Classic_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(Classic_wav.shape[1] == 1):
Classic_wavdata = Classic_wav
print(Classic_wav.shape[1])
else:
Classic_wavdata = (0.5 * Classic_wav[:, 1]) + (0.5 * Classic_wav[:, 0])
# wavファイル読み込み
Classic_noise_wav, Classic_noise_fs = sf.read(Classic_noise_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(Classic_noise_wav.shape[1] == 1):
Classic_noise_wavdata = Classic_noise_wav
print(Classic_noise_wav.shape[1])
else:
Classic_noise_wavdata = (0.5 * Classic_noise_wav[:, 1]) + (0.5 * Classic_noise_wav[:, 0])
#NHKRadio_wavdata
#NHKRadio_noise_wavdata
#NHKBusiness_wavdata
#NHKBusiness_noise_wavdata
#Classic_wavdata
#Classic_noise_wavdata
#Distance
NHKRadio_euclidean = distance.euclidean(NHKRadio_wavdata,NHKRadio_noise_wavdata)
NHKRadio_cosine = distance.cosine(NHKRadio_wavdata,NHKRadio_noise_wavdata)
NHKBusiness_euclidean = distance.euclidean(NHKBusiness_wavdata,NHKBusiness_noise_wavdata)
NHKBusiness_cosine = distance.cosine(NHKBusiness_wavdata,NHKBusiness_noise_wavdata)
Classic_euclidean = distance.euclidean(Classic_wavdata,Classic_noise_wavdata)
Classic_cosine = distance.cosine(Classic_wavdata,Classic_noise_wavdata)
Wavdata_Euclidean = pd.DataFrame([NHKRadio_euclidean,NHKBusiness_euclidean,Classic_euclidean],
columns=['Euclidean'],
index=['NHKRadio', 'NHKBusiness', 'Classic'])
Wavdata_Cosine = pd.DataFrame([NHKRadio_cosine,NHKBusiness_cosine,Classic_cosine],
columns=['Cosine'],
index=['NHKRadio', 'NHKBusiness', 'Classic'])
Wavdata_Euclidean.to_csv('./Output/Noise_Computation_Signal_Reconstruction/Wavdata_Euclidean_' + NoiseType + '.tsv', index=True, sep='\t')
Wavdata_Cosine.to_csv('./Output/Noise_Computation_Signal_Reconstruction/Wavdata_Cosine_' + NoiseType + '.tsv', index=True, sep='\t')
```
|
github_jupyter
|
#!/usr/bin/env python
# vim:fileencoding=utf-8
import sys
import scipy.signal as sg
import matplotlib.pyplot as plt
import soundfile as sf
import matplotlib
import pandas as pd
#可視化ライブラリ
import seaborn as sns
#距離計算
from scipy.spatial import distance
#音楽ファイル
NHKRadio_file = './Input/01_Radio/NHKRadio.wav'
NHKBusiness_file = './Input/02_Business/NHKBusiness.wav'
Classic_file = './Input/03_Classic/Classic.wav'
#このノイズファイルは、ノイズが追加された信号ではなく、
#音楽ファイルと長さが同じノイズファイル
NHKRadio_noise_file = './Output/Signal_Reconstruction_0.18.1/NHKRadio_tpdfnoiseWH_write_Matrix_50.wav'
NHKBusiness_noise_file = './Output/Signal_Reconstruction_0.18.1/NHKBusiness_tpdfnoiseWH_write_Matrix_50.wav'
Classic_noise_file = './Output/Signal_Reconstruction_0.18.1/Classic_tpdfnoiseWH_write_Matrix_50.wav'
NoiseType = "Tpdfnoise"
plt.close("all")
# wavファイル読み込み
NHKRadio_wav, NHKRadio_fs = sf.read(NHKRadio_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(NHKRadio_wav.shape[1] == 1):
NHKRadio_wavdata = NHKRadio_wav
print(NHKRadio_wav.shape[1])
else:
NHKRadio_wavdata = (0.5 * NHKRadio_wav[:, 1]) + (0.5 * NHKRadio_wav[:, 0])
# wavファイル読み込み
NHKRadio_noise_wav, NHKRadio_noise_fs = sf.read(NHKRadio_noise_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(NHKRadio_noise_wav.shape[1] == 1):
NHKRadio_noise_wavdata = NHKRadio_noise_wav
print(NHKRadio_noise_wav.shape[1])
else:
NHKRadio_noise_wavdata = (0.5 * NHKRadio_noise_wav[:, 1]) + (0.5 * NHKRadio_noise_wav[:, 0])
# wavファイル読み込み
NHKBusiness_wav, NHKBusiness_fs = sf.read(NHKBusiness_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(NHKBusiness_wav.shape[1] == 1):
NHKBusiness_wavdata = NHKBusiness_wav
print(NHKBusiness_wav.shape[1])
else:
NHKBusiness_wavdata = (0.5 * NHKBusiness_wav[:, 1]) + (0.5 * NHKBusiness_wav[:, 0])
# wavファイル読み込み
NHKBusiness_noise_wav, NHKBusiness_noise_fs = sf.read(NHKBusiness_noise_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(NHKBusiness_noise_wav.shape[1] == 1):
NHKBusiness_noise_wavdata = NHKBusiness_noise_wav
print(NHKBusiness_noise_wav.shape[1])
else:
NHKBusiness_noise_wavdata = (0.5 * NHKBusiness_noise_wav[:, 1]) + (0.5 * NHKBusiness_noise_wav[:, 0])
# wavファイル読み込み
Classic_wav, Classic_fs = sf.read(Classic_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(Classic_wav.shape[1] == 1):
Classic_wavdata = Classic_wav
print(Classic_wav.shape[1])
else:
Classic_wavdata = (0.5 * Classic_wav[:, 1]) + (0.5 * Classic_wav[:, 0])
# wavファイル読み込み
Classic_noise_wav, Classic_noise_fs = sf.read(Classic_noise_file)
# ステレオ2chの場合、モノラル音源に変換(左右の各音を2で割った音を足して作成.)
if(Classic_noise_wav.shape[1] == 1):
Classic_noise_wavdata = Classic_noise_wav
print(Classic_noise_wav.shape[1])
else:
Classic_noise_wavdata = (0.5 * Classic_noise_wav[:, 1]) + (0.5 * Classic_noise_wav[:, 0])
#NHKRadio_wavdata
#NHKRadio_noise_wavdata
#NHKBusiness_wavdata
#NHKBusiness_noise_wavdata
#Classic_wavdata
#Classic_noise_wavdata
#Distance
NHKRadio_euclidean = distance.euclidean(NHKRadio_wavdata,NHKRadio_noise_wavdata)
NHKRadio_cosine = distance.cosine(NHKRadio_wavdata,NHKRadio_noise_wavdata)
NHKBusiness_euclidean = distance.euclidean(NHKBusiness_wavdata,NHKBusiness_noise_wavdata)
NHKBusiness_cosine = distance.cosine(NHKBusiness_wavdata,NHKBusiness_noise_wavdata)
Classic_euclidean = distance.euclidean(Classic_wavdata,Classic_noise_wavdata)
Classic_cosine = distance.cosine(Classic_wavdata,Classic_noise_wavdata)
Wavdata_Euclidean = pd.DataFrame([NHKRadio_euclidean,NHKBusiness_euclidean,Classic_euclidean],
columns=['Euclidean'],
index=['NHKRadio', 'NHKBusiness', 'Classic'])
Wavdata_Cosine = pd.DataFrame([NHKRadio_cosine,NHKBusiness_cosine,Classic_cosine],
columns=['Cosine'],
index=['NHKRadio', 'NHKBusiness', 'Classic'])
Wavdata_Euclidean.to_csv('./Output/Noise_Computation_Signal_Reconstruction/Wavdata_Euclidean_' + NoiseType + '.tsv', index=True, sep='\t')
Wavdata_Cosine.to_csv('./Output/Noise_Computation_Signal_Reconstruction/Wavdata_Cosine_' + NoiseType + '.tsv', index=True, sep='\t')
| 0.187579 | 0.178669 |
# Задание 6: Рекуррентные нейронные сети (RNNs)
Это задание адаптиповано из Deep NLP Course at ABBYY (https://github.com/DanAnastasyev/DeepNLP-Course) с разрешения автора - Даниила Анастасьева. Спасибо ему огромное!
```
# !pip3 -qq install torch==0.4.1
# !pip3 -qq install bokeh==0.13.0
# !pip3 -qq install gensim==3.6.0
# !pip3 -qq install nltk
# !pip3 -qq install scikit-learn==0.20.2
pip list
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
if torch.cuda.is_available():
from torch.cuda import FloatTensor, LongTensor
else:
from torch import FloatTensor, LongTensor
np.random.seed(42)
```
# Рекуррентные нейронные сети (RNNs)
## POS Tagging
Мы рассмотрим применение рекуррентных сетей к задаче sequence labeling (последняя картинка).
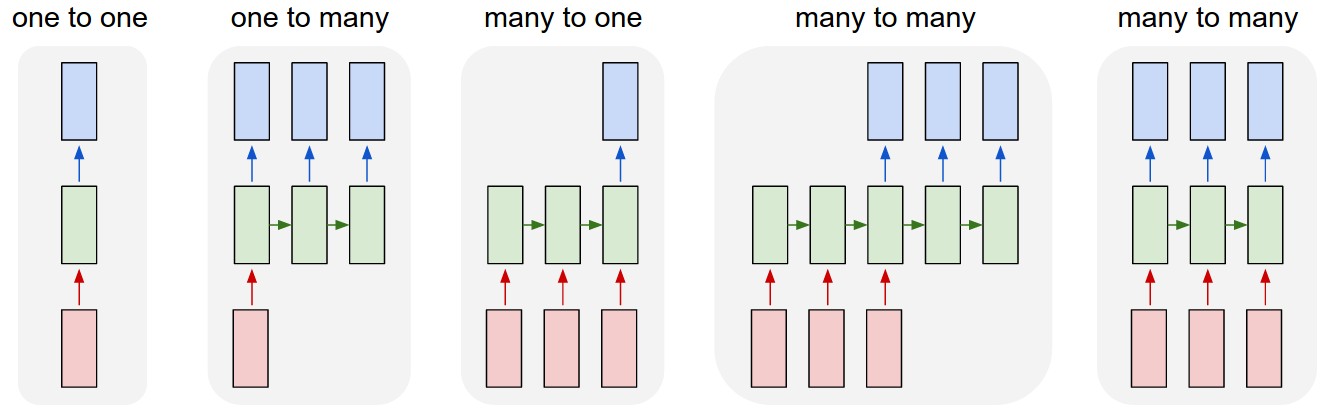
*From [The Unreasonable Effectiveness of Recurrent Neural Networks](http://karpathy.github.io/2015/05/21/rnn-effectiveness/)*
Самые популярные примеры для такой постановки задачи - Part-of-Speech Tagging и Named Entity Recognition.
Мы порешаем сейчас POS Tagging для английского.
Будем работать с таким набором тегов:
- ADJ - adjective (new, good, high, ...)
- ADP - adposition (on, of, at, ...)
- ADV - adverb (really, already, still, ...)
- CONJ - conjunction (and, or, but, ...)
- DET - determiner, article (the, a, some, ...)
- NOUN - noun (year, home, costs, ...)
- NUM - numeral (twenty-four, fourth, 1991, ...)
- PRT - particle (at, on, out, ...)
- PRON - pronoun (he, their, her, ...)
- VERB - verb (is, say, told, ...)
- . - punctuation marks (. , ;)
- X - other (ersatz, esprit, dunno, ...)
Скачаем данные:
```
import nltk
from sklearn.model_selection import train_test_split
from nltk.corpus import brown
nltk.download('brown')
nltk.download('universal_tagset')
data = nltk.corpus.brown.tagged_sents(tagset='universal')
```
Пример размеченного предложения:
```
for word, tag in data[0]:
print('{:19}\t{}'.format(word, tag))
```
Построим разбиение на train/val/test - наконец-то, всё как у нормальных людей.
На train будем учиться, по val - подбирать параметры и делать всякие early stopping, а на test - принимать модель по ее финальному качеству.
```
train_data, test_data = train_test_split(data, test_size=0.25, random_state=42)
train_data, val_data = train_test_split(train_data, test_size=0.15, random_state=42)
print('Words count in train set:', sum(len(sent) for sent in train_data))
print('Words count in val set:', sum(len(sent) for sent in val_data))
print('Words count in test set:', sum(len(sent) for sent in test_data))
```
Построим маппинги из слов в индекс и из тега в индекс:
```
words = {word for sample in train_data for word, tag in sample}
word2ind = {word: ind + 1 for ind, word in enumerate(words)}
word2ind['<pad>'] = 0
tags = {tag for sample in train_data for word, tag in sample}
tag2ind = {tag: ind + 1 for ind, tag in enumerate(tags)}
tag2ind['<pad>'] = 0
print('Unique words in train = {}. Tags = {}'.format(len(word2ind), tags))
import matplotlib.pyplot as plt
%matplotlib inline
from collections import Counter
tag_distribution = Counter(tag for sample in train_data for _, tag in sample)
tag_distribution = [tag_distribution[tag] for tag in tags]
plt.figure(figsize=(10, 5))
bar_width = 0.35
plt.bar(np.arange(len(tags)), tag_distribution, bar_width, align='center', alpha=0.5)
plt.xticks(np.arange(len(tags)), tags)
plt.show()
```
## Бейзлайн
Какой самый простой теггер можно придумать? Давайте просто запоминать, какие теги самые вероятные для слова (или для последовательности):
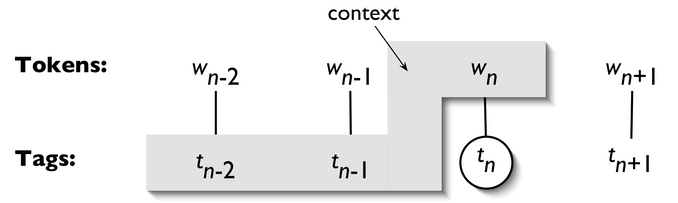
*From [Categorizing and Tagging Words, nltk](https://www.nltk.org/book/ch05.html)*
На картинке показано, что для предсказания $t_n$ используются два предыдущих предсказанных тега + текущее слово. По корпусу считаются вероятность для $P(t_n| w_n, t_{n-1}, t_{n-2})$, выбирается тег с максимальной вероятностью.
Более аккуратно такая идея реализована в Hidden Markov Models: по тренировочному корпусу вычисляются вероятности $P(w_n| t_n), P(t_n|t_{n-1}, t_{n-2})$ и максимизируется их произведение.
Простейший вариант - униграммная модель, учитывающая только слово:
```
import nltk
default_tagger = nltk.DefaultTagger('NN')
unigram_tagger = nltk.UnigramTagger(train_data, backoff=default_tagger)
print('Accuracy of unigram tagger = {:.2%}'.format(unigram_tagger.evaluate(test_data)))
```
Добавим вероятности переходов:
```
bigram_tagger = nltk.BigramTagger(train_data, backoff=unigram_tagger)
print('Accuracy of bigram tagger = {:.2%}'.format(bigram_tagger.evaluate(test_data)))
```
Обратите внимание, что `backoff` важен:
```
trigram_tagger = nltk.TrigramTagger(train_data)
print('Accuracy of trigram tagger = {:.2%}'.format(trigram_tagger.evaluate(test_data)))
```
## Увеличиваем контекст с рекуррентными сетями
Униграмная модель работает на удивление хорошо, но мы же собрались учить сеточки.
Омонимия - основная причина, почему униграмная модель плоха:
*“he cashed a check at the **bank**”*
vs
*“he sat on the **bank** of the river”*
Поэтому нам очень полезно учитывать контекст при предсказании тега.
Воспользуемся LSTM - он умеет работать с контекстом очень даже хорошо:

Синим показано выделение фичей из слова, LSTM оранжевенький - он строит эмбеддинги слов с учетом контекста, а дальше зелененькая логистическая регрессия делает предсказания тегов.
```
def convert_data(data, word2ind, tag2ind):
X = [[word2ind.get(word, 0) for word, _ in sample] for sample in data]
y = [[tag2ind[tag] for _, tag in sample] for sample in data]
return X, y
X_train, y_train = convert_data(train_data, word2ind, tag2ind)
X_val, y_val = convert_data(val_data, word2ind, tag2ind)
X_test, y_test = convert_data(test_data, word2ind, tag2ind)
def iterate_batches(data, batch_size):
X, y = data
n_samples = len(X)
indices = np.arange(n_samples)
np.random.shuffle(indices)
for start in range(0, n_samples, batch_size):
end = min(start + batch_size, n_samples)
batch_indices = indices[start:end]
max_sent_len = max(len(X[ind]) for ind in batch_indices)
X_batch = np.zeros((max_sent_len, len(batch_indices)))
y_batch = np.zeros((max_sent_len, len(batch_indices)))
for batch_ind, sample_ind in enumerate(batch_indices):
X_batch[:len(X[sample_ind]), batch_ind] = X[sample_ind]
y_batch[:len(y[sample_ind]), batch_ind] = y[sample_ind]
yield X_batch, y_batch
X_batch, y_batch = next(iterate_batches((X_train, y_train), 4))
X_batch, X_batch.shape
```
**Задание** Реализуйте `LSTMTagger`:
```
class LSTMTagger(nn.Module):
def __init__(self, vocab_size, tagset_size, word_emb_dim=100, lstm_hidden_dim=128, lstm_layers_count=1):
super().__init__()
self.hidden_dim = lstm_hidden_dim
self.word_embeddings = nn.Embedding(vocab_size, word_emb_dim)
self.lstm = nn.LSTM(word_emb_dim, lstm_hidden_dim, lstm_layers_count)
self.hidden2tag = nn.Linear(lstm_hidden_dim, tagset_size)
def forward(self, inputs):
embeds = self.word_embeddings(inputs)
lstm_out, _ = self.lstm(embeds)
tag_space = self.hidden2tag(lstm_out)
return tag_space
```
**Задание** Научитесь считать accuracy и loss (а заодно проверьте, что модель работает)
```
model = LSTMTagger(
vocab_size=len(word2ind),
tagset_size=len(tag2ind)
)
X_batch, y_batch = torch.LongTensor(X_batch), torch.LongTensor(y_batch)
tag_space = model(X_batch)
tag_space_resh = tag_space.view(-1, tag_space.shape[-1])
y_batch_resh = y_batch.view(-1)
criterion = nn.CrossEntropyLoss(ignore_index=0)
loss = criterion(tag_space_resh, y_batch_resh)
print(loss.item())
mask = (y_batch != 0).type('torch.LongTensor')
y_pred = torch.argmax(tag_space, 2)
cur_correct_count, cur_sum_count = torch.sum(mask*torch.eq(y_batch, y_pred)).item(), torch.sum(mask).item()
accuracy = cur_correct_count*100/cur_sum_count
print(accuracy)
```
**Задание** Вставьте эти вычисление в функцию:
```
import math
from tqdm import tqdm
def do_epoch(model, criterion, data, batch_size, optimizer=None, name=None):
epoch_loss = 0
correct_count = 0
sum_count = 0
is_train = not optimizer is None
name = name or ''
model.train(is_train)
batches_count = math.ceil(len(data[0]) / batch_size)
with torch.autograd.set_grad_enabled(is_train):
with tqdm(total=batches_count) as progress_bar:
for i, (X_batch, y_batch) in enumerate(iterate_batches(data, batch_size)):
X_batch, y_batch = LongTensor(X_batch), LongTensor(y_batch)
tag_space = model(X_batch)
tag_space_resh = tag_space.view(-1, tag_space.shape[-1])
y_batch_resh = y_batch.view(-1)
loss = criterion(tag_space_resh, y_batch_resh)
if torch.cuda.is_available():
mask = (y_batch != 0).type('torch.LongTensor').cuda()
else:
mask = (y_batch != 0).type('torch.LongTensor')
epoch_loss += loss.item()
if optimizer:
optimizer.zero_grad()
loss.backward()
optimizer.step()
y_pred = torch.argmax(tag_space, 2)
cur_correct_count, cur_sum_count = torch.sum(mask*torch.eq(y_batch, y_pred)).item(), torch.sum(mask).item()
correct_count += cur_correct_count
sum_count += cur_sum_count
progress_bar.update()
progress_bar.set_description('{:>5s} Loss = {:.5f}, Accuracy = {:.2%}'.format(
name, loss.item(), cur_correct_count / cur_sum_count)
)
progress_bar.set_description('{:>5s} Loss = {:.5f}, Accuracy = {:.2%}'.format(
name, epoch_loss / batches_count, correct_count / sum_count)
)
return epoch_loss / batches_count, correct_count / sum_count
def fit(model, criterion, optimizer, train_data, epochs_count=1, batch_size=32,
val_data=None, val_batch_size=None):
if not val_data is None and val_batch_size is None:
val_batch_size = batch_size
for epoch in range(epochs_count):
name_prefix = '[{} / {}] '.format(epoch + 1, epochs_count)
train_loss, train_acc = do_epoch(model, criterion, train_data, batch_size, optimizer, name_prefix + 'Train:')
if not val_data is None:
val_loss, val_acc = do_epoch(model, criterion, val_data, val_batch_size, None, name_prefix + ' Val:')
lr = 0.001
if torch.cuda.is_available():
model = LSTMTagger(
vocab_size=len(word2ind),
tagset_size=len(tag2ind)
).cuda()
criterion = nn.CrossEntropyLoss(ignore_index = 0).cuda()
optimizer = optim.Adam(model.parameters(), lr = lr)
fit(model, criterion, optimizer, train_data=(X_train, y_train), epochs_count=50,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
else:
model = LSTMTagger(
vocab_size=len(word2ind),
tagset_size=len(tag2ind))
criterion = nn.CrossEntropyLoss(ignore_index = 0)
optimizer = optim.Adam(model.parameters(), lr = lr)
fit(model, criterion, optimizer, train_data=(X_train, y_train), epochs_count=50, batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
```
### Masking
**Задание** Проверьте себя - не считаете ли вы потери и accuracy на паддингах - очень легко получить высокое качество за счет этого.
У функции потерь есть параметр `ignore_index`, для таких целей. Для accuracy нужно использовать маскинг - умножение на маску из нулей и единиц, где нули на позициях паддингов (а потом усреднение по ненулевым позициям в маске).
**Задание** Посчитайте качество модели на тесте. Ожидается результат лучше бейзлайна!
```
loss_test, accuracy_test = do_epoch(model, criterion, data = (X_test, y_test), batch_size = 256, optimizer=None, name=None)
```
### Bidirectional LSTM
Благодаря BiLSTM можно использовать сразу оба контеста при предсказании тега слова. Т.е. для каждого токена $w_i$ forward LSTM будет выдавать представление $\mathbf{f_i} \sim (w_1, \ldots, w_i)$ - построенное по всему левому контексту - и $\mathbf{b_i} \sim (w_n, \ldots, w_i)$ - представление правого контекста. Их конкатенация автоматически захватит весь доступный контекст слова: $\mathbf{h_i} = [\mathbf{f_i}, \mathbf{b_i}] \sim (w_1, \ldots, w_n)$.

*From [Finding Function in Form: Compositional Character Models for Open Vocabulary Word Representation](https://arxiv.org/abs/1508.02096)*
**Задание** Добавьте Bidirectional LSTM.
```
class BidirectionalLSTMTagger(nn.Module):
def __init__(self, vocab_size, tagset_size, word_emb_dim=100, lstm_hidden_dim=128, lstm_layers_count=1, bidirectional=True):
super().__init__()
self.hidden_dim = lstm_hidden_dim
self.word_embeddings = nn.Embedding(vocab_size, word_emb_dim)
self.lstm = nn.LSTM(word_emb_dim, lstm_hidden_dim, lstm_layers_count, bidirectional=True)
self.hidden2tag = nn.Linear(lstm_hidden_dim * 2 if bidirectional else lstm_hidden_dim, tagset_size)
def forward(self, inputs):
embeds = self.word_embeddings(inputs)
lstm_out, _ = self.lstm(embeds)
tag_space = self.hidden2tag(lstm_out)
return tag_space
lr = 0.001
if torch.cuda.is_available():
model_2 = BidirectionalLSTMTagger(
vocab_size=len(word2ind),
tagset_size=len(tag2ind)
).cuda()
criterion = nn.CrossEntropyLoss(ignore_index = 0).cuda()
optimizer = optim.Adam(model_2.parameters(), lr = lr)
fit(model_2, criterion, optimizer, train_data=(X_train, y_train), epochs_count=30,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
else:
model_2 = BidirectionalLSTMTagger(
vocab_size=len(word2ind),
tagset_size=len(tag2ind))
criterion = nn.CrossEntropyLoss(ignore_index = 0)
optimizer = optim.Adam(model_2.parameters(), lr = lr)
fit(model_2, criterion, optimizer, train_data=(X_train, y_train), epochs_count=30, batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
loss_test, accuracy_test = do_epoch(model_2, criterion, data = (X_test, y_test), batch_size = 256, optimizer=None, name=None)
```
### Предобученные эмбеддинги
Мы знаем, какая клёвая вещь - предобученные эмбеддинги. При текущем размере обучающей выборки еще можно было учить их и с нуля - с меньшей было бы совсем плохо.
Поэтому стандартный пайплайн - скачать эмбеддинги, засунуть их в сеточку. Запустим его:
```
import gensim.downloader as api
w2v_model = api.load('glove-wiki-gigaword-100')
w2v_model.vectors.shape
```
Построим подматрицу для слов из нашей тренировочной выборки:
```
embeddings_random = nn.Embedding(len(word2ind), w2v_model.vectors.shape[1])
embeddings_random.weight[0]
known_count = 0
embeddings = embeddings_random.weight.detach().numpy()
for word, ind in word2ind.items():
word = word.lower()
if word in w2v_model.vocab:
embeddings[ind] = w2v_model.get_vector(word)
known_count += 1
print('Know {} out of {} word embeddings'.format(known_count, len(word2ind)))
embeddings[0]
```
**Задание** Сделайте модель с предобученной матрицей. Используйте `nn.Embedding.from_pretrained`.
```
class LSTMTaggerWithPretrainedEmbs(nn.Module):
def __init__(self, embeddings, tagset_size, lstm_hidden_dim=128, lstm_layers_count=1):
super().__init__()
self.hidden_dim = lstm_hidden_dim
self.word_embeddings = nn.Embedding.from_pretrained(torch.Tensor(embeddings))
self.lstm = nn.LSTM(embeddings.shape[1], lstm_hidden_dim, lstm_layers_count)
self.hidden2tag = nn.Linear(lstm_hidden_dim, tagset_size)
def forward(self, inputs):
embeds = self.word_embeddings(inputs)
lstm_out, _ = self.lstm(embeds)
tag_space = self.hidden2tag(lstm_out)
return tag_space
if torch.cuda.is_available():
model_3 = LSTMTaggerWithPretrainedEmbs(
embeddings=embeddings,
tagset_size=len(tag2ind)
).cuda()
criterion = nn.CrossEntropyLoss(ignore_index=0)
optimizer = optim.Adam(model_3.parameters())
fit(model_3, criterion, optimizer, train_data=(X_train, y_train), epochs_count=100,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
else:
model_3 = LSTMTaggerWithPretrainedEmbs(
embeddings=embeddings,
tagset_size=len(tag2ind)
)
criterion = nn.CrossEntropyLoss(ignore_index=0)
optimizer = optim.Adam(model_3.parameters())
fit(model_3, criterion, optimizer, train_data=(X_train, y_train), epochs_count=100,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
```
**Задание** Оцените качество модели на тестовой выборке. Обратите внимание, вовсе не обязательно ограничиваться векторами из урезанной матрицы - вполне могут найтись слова в тесте, которых не было в трейне и для которых есть эмбеддинги.
Добейтесь качества лучше прошлых моделей.
```
loss_test, accuracy_test = do_epoch(model_3, criterion, data = (X_test, y_test), batch_size = 256, optimizer=None, name=None)
class BidirectionalLSTMTaggerWithPretrainedEmbs(nn.Module):
def __init__(self, embeddings, tagset_size, lstm_hidden_dim=128, lstm_layers_count=1):
super().__init__()
self.hidden_dim = lstm_hidden_dim
self.word_embeddings = nn.Embedding.from_pretrained(torch.Tensor(embeddings))
self.lstm = nn.LSTM(embeddings.shape[1], lstm_hidden_dim, lstm_layers_count, bidirectional=True)
self.hidden2tag = nn.Linear(lstm_hidden_dim * 2, tagset_size)
def forward(self, inputs):
embeds = self.word_embeddings(inputs)
lstm_out, _ = self.lstm(embeds)
tag_space = self.hidden2tag(lstm_out)
return tag_space
if torch.cuda.is_available():
model_4 = BidirectionalLSTMTaggerWithPretrainedEmbs(
embeddings=embeddings,
tagset_size=len(tag2ind)
).cuda()
criterion = nn.CrossEntropyLoss(ignore_index=0)
optimizer = optim.Adam(model_4.parameters())
fit(model_4, criterion, optimizer, train_data=(X_train, y_train), epochs_count=100,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
else:
model_4 = BidirectionalLSTMTaggerWithPretrainedEmbs(
embeddings=embeddings,
tagset_size=len(tag2ind)
)
criterion = nn.CrossEntropyLoss(ignore_index=0)
optimizer = optim.Adam(model_4.parameters())
fit(model_4, criterion, optimizer, train_data=(X_train, y_train), epochs_count=11,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
loss_test, accuracy_test = do_epoch(model_4, criterion, data = (X_test, y_test), batch_size = 256, optimizer=None, name=None)
```
|
github_jupyter
|
# !pip3 -qq install torch==0.4.1
# !pip3 -qq install bokeh==0.13.0
# !pip3 -qq install gensim==3.6.0
# !pip3 -qq install nltk
# !pip3 -qq install scikit-learn==0.20.2
pip list
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
if torch.cuda.is_available():
from torch.cuda import FloatTensor, LongTensor
else:
from torch import FloatTensor, LongTensor
np.random.seed(42)
import nltk
from sklearn.model_selection import train_test_split
from nltk.corpus import brown
nltk.download('brown')
nltk.download('universal_tagset')
data = nltk.corpus.brown.tagged_sents(tagset='universal')
for word, tag in data[0]:
print('{:19}\t{}'.format(word, tag))
train_data, test_data = train_test_split(data, test_size=0.25, random_state=42)
train_data, val_data = train_test_split(train_data, test_size=0.15, random_state=42)
print('Words count in train set:', sum(len(sent) for sent in train_data))
print('Words count in val set:', sum(len(sent) for sent in val_data))
print('Words count in test set:', sum(len(sent) for sent in test_data))
words = {word for sample in train_data for word, tag in sample}
word2ind = {word: ind + 1 for ind, word in enumerate(words)}
word2ind['<pad>'] = 0
tags = {tag for sample in train_data for word, tag in sample}
tag2ind = {tag: ind + 1 for ind, tag in enumerate(tags)}
tag2ind['<pad>'] = 0
print('Unique words in train = {}. Tags = {}'.format(len(word2ind), tags))
import matplotlib.pyplot as plt
%matplotlib inline
from collections import Counter
tag_distribution = Counter(tag for sample in train_data for _, tag in sample)
tag_distribution = [tag_distribution[tag] for tag in tags]
plt.figure(figsize=(10, 5))
bar_width = 0.35
plt.bar(np.arange(len(tags)), tag_distribution, bar_width, align='center', alpha=0.5)
plt.xticks(np.arange(len(tags)), tags)
plt.show()
import nltk
default_tagger = nltk.DefaultTagger('NN')
unigram_tagger = nltk.UnigramTagger(train_data, backoff=default_tagger)
print('Accuracy of unigram tagger = {:.2%}'.format(unigram_tagger.evaluate(test_data)))
bigram_tagger = nltk.BigramTagger(train_data, backoff=unigram_tagger)
print('Accuracy of bigram tagger = {:.2%}'.format(bigram_tagger.evaluate(test_data)))
trigram_tagger = nltk.TrigramTagger(train_data)
print('Accuracy of trigram tagger = {:.2%}'.format(trigram_tagger.evaluate(test_data)))
def convert_data(data, word2ind, tag2ind):
X = [[word2ind.get(word, 0) for word, _ in sample] for sample in data]
y = [[tag2ind[tag] for _, tag in sample] for sample in data]
return X, y
X_train, y_train = convert_data(train_data, word2ind, tag2ind)
X_val, y_val = convert_data(val_data, word2ind, tag2ind)
X_test, y_test = convert_data(test_data, word2ind, tag2ind)
def iterate_batches(data, batch_size):
X, y = data
n_samples = len(X)
indices = np.arange(n_samples)
np.random.shuffle(indices)
for start in range(0, n_samples, batch_size):
end = min(start + batch_size, n_samples)
batch_indices = indices[start:end]
max_sent_len = max(len(X[ind]) for ind in batch_indices)
X_batch = np.zeros((max_sent_len, len(batch_indices)))
y_batch = np.zeros((max_sent_len, len(batch_indices)))
for batch_ind, sample_ind in enumerate(batch_indices):
X_batch[:len(X[sample_ind]), batch_ind] = X[sample_ind]
y_batch[:len(y[sample_ind]), batch_ind] = y[sample_ind]
yield X_batch, y_batch
X_batch, y_batch = next(iterate_batches((X_train, y_train), 4))
X_batch, X_batch.shape
class LSTMTagger(nn.Module):
def __init__(self, vocab_size, tagset_size, word_emb_dim=100, lstm_hidden_dim=128, lstm_layers_count=1):
super().__init__()
self.hidden_dim = lstm_hidden_dim
self.word_embeddings = nn.Embedding(vocab_size, word_emb_dim)
self.lstm = nn.LSTM(word_emb_dim, lstm_hidden_dim, lstm_layers_count)
self.hidden2tag = nn.Linear(lstm_hidden_dim, tagset_size)
def forward(self, inputs):
embeds = self.word_embeddings(inputs)
lstm_out, _ = self.lstm(embeds)
tag_space = self.hidden2tag(lstm_out)
return tag_space
model = LSTMTagger(
vocab_size=len(word2ind),
tagset_size=len(tag2ind)
)
X_batch, y_batch = torch.LongTensor(X_batch), torch.LongTensor(y_batch)
tag_space = model(X_batch)
tag_space_resh = tag_space.view(-1, tag_space.shape[-1])
y_batch_resh = y_batch.view(-1)
criterion = nn.CrossEntropyLoss(ignore_index=0)
loss = criterion(tag_space_resh, y_batch_resh)
print(loss.item())
mask = (y_batch != 0).type('torch.LongTensor')
y_pred = torch.argmax(tag_space, 2)
cur_correct_count, cur_sum_count = torch.sum(mask*torch.eq(y_batch, y_pred)).item(), torch.sum(mask).item()
accuracy = cur_correct_count*100/cur_sum_count
print(accuracy)
import math
from tqdm import tqdm
def do_epoch(model, criterion, data, batch_size, optimizer=None, name=None):
epoch_loss = 0
correct_count = 0
sum_count = 0
is_train = not optimizer is None
name = name or ''
model.train(is_train)
batches_count = math.ceil(len(data[0]) / batch_size)
with torch.autograd.set_grad_enabled(is_train):
with tqdm(total=batches_count) as progress_bar:
for i, (X_batch, y_batch) in enumerate(iterate_batches(data, batch_size)):
X_batch, y_batch = LongTensor(X_batch), LongTensor(y_batch)
tag_space = model(X_batch)
tag_space_resh = tag_space.view(-1, tag_space.shape[-1])
y_batch_resh = y_batch.view(-1)
loss = criterion(tag_space_resh, y_batch_resh)
if torch.cuda.is_available():
mask = (y_batch != 0).type('torch.LongTensor').cuda()
else:
mask = (y_batch != 0).type('torch.LongTensor')
epoch_loss += loss.item()
if optimizer:
optimizer.zero_grad()
loss.backward()
optimizer.step()
y_pred = torch.argmax(tag_space, 2)
cur_correct_count, cur_sum_count = torch.sum(mask*torch.eq(y_batch, y_pred)).item(), torch.sum(mask).item()
correct_count += cur_correct_count
sum_count += cur_sum_count
progress_bar.update()
progress_bar.set_description('{:>5s} Loss = {:.5f}, Accuracy = {:.2%}'.format(
name, loss.item(), cur_correct_count / cur_sum_count)
)
progress_bar.set_description('{:>5s} Loss = {:.5f}, Accuracy = {:.2%}'.format(
name, epoch_loss / batches_count, correct_count / sum_count)
)
return epoch_loss / batches_count, correct_count / sum_count
def fit(model, criterion, optimizer, train_data, epochs_count=1, batch_size=32,
val_data=None, val_batch_size=None):
if not val_data is None and val_batch_size is None:
val_batch_size = batch_size
for epoch in range(epochs_count):
name_prefix = '[{} / {}] '.format(epoch + 1, epochs_count)
train_loss, train_acc = do_epoch(model, criterion, train_data, batch_size, optimizer, name_prefix + 'Train:')
if not val_data is None:
val_loss, val_acc = do_epoch(model, criterion, val_data, val_batch_size, None, name_prefix + ' Val:')
lr = 0.001
if torch.cuda.is_available():
model = LSTMTagger(
vocab_size=len(word2ind),
tagset_size=len(tag2ind)
).cuda()
criterion = nn.CrossEntropyLoss(ignore_index = 0).cuda()
optimizer = optim.Adam(model.parameters(), lr = lr)
fit(model, criterion, optimizer, train_data=(X_train, y_train), epochs_count=50,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
else:
model = LSTMTagger(
vocab_size=len(word2ind),
tagset_size=len(tag2ind))
criterion = nn.CrossEntropyLoss(ignore_index = 0)
optimizer = optim.Adam(model.parameters(), lr = lr)
fit(model, criterion, optimizer, train_data=(X_train, y_train), epochs_count=50, batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
loss_test, accuracy_test = do_epoch(model, criterion, data = (X_test, y_test), batch_size = 256, optimizer=None, name=None)
class BidirectionalLSTMTagger(nn.Module):
def __init__(self, vocab_size, tagset_size, word_emb_dim=100, lstm_hidden_dim=128, lstm_layers_count=1, bidirectional=True):
super().__init__()
self.hidden_dim = lstm_hidden_dim
self.word_embeddings = nn.Embedding(vocab_size, word_emb_dim)
self.lstm = nn.LSTM(word_emb_dim, lstm_hidden_dim, lstm_layers_count, bidirectional=True)
self.hidden2tag = nn.Linear(lstm_hidden_dim * 2 if bidirectional else lstm_hidden_dim, tagset_size)
def forward(self, inputs):
embeds = self.word_embeddings(inputs)
lstm_out, _ = self.lstm(embeds)
tag_space = self.hidden2tag(lstm_out)
return tag_space
lr = 0.001
if torch.cuda.is_available():
model_2 = BidirectionalLSTMTagger(
vocab_size=len(word2ind),
tagset_size=len(tag2ind)
).cuda()
criterion = nn.CrossEntropyLoss(ignore_index = 0).cuda()
optimizer = optim.Adam(model_2.parameters(), lr = lr)
fit(model_2, criterion, optimizer, train_data=(X_train, y_train), epochs_count=30,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
else:
model_2 = BidirectionalLSTMTagger(
vocab_size=len(word2ind),
tagset_size=len(tag2ind))
criterion = nn.CrossEntropyLoss(ignore_index = 0)
optimizer = optim.Adam(model_2.parameters(), lr = lr)
fit(model_2, criterion, optimizer, train_data=(X_train, y_train), epochs_count=30, batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
loss_test, accuracy_test = do_epoch(model_2, criterion, data = (X_test, y_test), batch_size = 256, optimizer=None, name=None)
import gensim.downloader as api
w2v_model = api.load('glove-wiki-gigaword-100')
w2v_model.vectors.shape
embeddings_random = nn.Embedding(len(word2ind), w2v_model.vectors.shape[1])
embeddings_random.weight[0]
known_count = 0
embeddings = embeddings_random.weight.detach().numpy()
for word, ind in word2ind.items():
word = word.lower()
if word in w2v_model.vocab:
embeddings[ind] = w2v_model.get_vector(word)
known_count += 1
print('Know {} out of {} word embeddings'.format(known_count, len(word2ind)))
embeddings[0]
class LSTMTaggerWithPretrainedEmbs(nn.Module):
def __init__(self, embeddings, tagset_size, lstm_hidden_dim=128, lstm_layers_count=1):
super().__init__()
self.hidden_dim = lstm_hidden_dim
self.word_embeddings = nn.Embedding.from_pretrained(torch.Tensor(embeddings))
self.lstm = nn.LSTM(embeddings.shape[1], lstm_hidden_dim, lstm_layers_count)
self.hidden2tag = nn.Linear(lstm_hidden_dim, tagset_size)
def forward(self, inputs):
embeds = self.word_embeddings(inputs)
lstm_out, _ = self.lstm(embeds)
tag_space = self.hidden2tag(lstm_out)
return tag_space
if torch.cuda.is_available():
model_3 = LSTMTaggerWithPretrainedEmbs(
embeddings=embeddings,
tagset_size=len(tag2ind)
).cuda()
criterion = nn.CrossEntropyLoss(ignore_index=0)
optimizer = optim.Adam(model_3.parameters())
fit(model_3, criterion, optimizer, train_data=(X_train, y_train), epochs_count=100,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
else:
model_3 = LSTMTaggerWithPretrainedEmbs(
embeddings=embeddings,
tagset_size=len(tag2ind)
)
criterion = nn.CrossEntropyLoss(ignore_index=0)
optimizer = optim.Adam(model_3.parameters())
fit(model_3, criterion, optimizer, train_data=(X_train, y_train), epochs_count=100,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
loss_test, accuracy_test = do_epoch(model_3, criterion, data = (X_test, y_test), batch_size = 256, optimizer=None, name=None)
class BidirectionalLSTMTaggerWithPretrainedEmbs(nn.Module):
def __init__(self, embeddings, tagset_size, lstm_hidden_dim=128, lstm_layers_count=1):
super().__init__()
self.hidden_dim = lstm_hidden_dim
self.word_embeddings = nn.Embedding.from_pretrained(torch.Tensor(embeddings))
self.lstm = nn.LSTM(embeddings.shape[1], lstm_hidden_dim, lstm_layers_count, bidirectional=True)
self.hidden2tag = nn.Linear(lstm_hidden_dim * 2, tagset_size)
def forward(self, inputs):
embeds = self.word_embeddings(inputs)
lstm_out, _ = self.lstm(embeds)
tag_space = self.hidden2tag(lstm_out)
return tag_space
if torch.cuda.is_available():
model_4 = BidirectionalLSTMTaggerWithPretrainedEmbs(
embeddings=embeddings,
tagset_size=len(tag2ind)
).cuda()
criterion = nn.CrossEntropyLoss(ignore_index=0)
optimizer = optim.Adam(model_4.parameters())
fit(model_4, criterion, optimizer, train_data=(X_train, y_train), epochs_count=100,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
else:
model_4 = BidirectionalLSTMTaggerWithPretrainedEmbs(
embeddings=embeddings,
tagset_size=len(tag2ind)
)
criterion = nn.CrossEntropyLoss(ignore_index=0)
optimizer = optim.Adam(model_4.parameters())
fit(model_4, criterion, optimizer, train_data=(X_train, y_train), epochs_count=11,
batch_size=64, val_data=(X_val, y_val), val_batch_size=512)
loss_test, accuracy_test = do_epoch(model_4, criterion, data = (X_test, y_test), batch_size = 256, optimizer=None, name=None)
| 0.753376 | 0.936692 |
# MNIST
## Package
```
import torch
import torch.nn as nn
from torchvision import datasets
from torchvision import transforms
import numpy as np
import matplotlib.pyplot as plt
from torch.utils.tensorboard import SummaryWriter
import torch.optim as optim
```
## Data set
```
traindata = datasets.MNIST('data/mnist_train', train=True, download=False, transform=transforms.ToTensor()) # download = True to download
len(traindata)
```
## Sample
```
img, label = traindata[np.random.randint(0,60000-1)]
print(label)
print(img)
img.size()
plt.imshow(img.reshape(28,28), cmap='gray')
img_train = img.view(-1).unsqueeze(0)
img_train.size()
```
## Model
```
model = nn.Sequential(
nn.Linear(784, 100),
nn.ReLU(),
nn.Linear(100, 10),
nn.ReLU(),
nn.Softmax(dim=1))
predict = model(img_train)
print(predict)
predict.size()
```
## Loss
### one-hot encoding
```
label_one_hot = torch.zeros(10).scatter_(0, torch.tensor(label), 1.0).unsqueeze(0)
label, label_one_hot, label_one_hot.size()
```
### Mean square loss
```
loss = torch.nn.MSELoss()
loss(predict, label_one_hot)
```
## Train
### Data loader
```
batch_sz = 600
train_loader = torch.utils.data.DataLoader(traindata, batch_size=batch_sz, shuffle=True)
#img, label = next(iter(train_loader))
```
### Learn rate
```
learning_rate = 0.5
```
### Optimizer
```
optimizer = optim.SGD(model.parameters(), lr=learning_rate)
```
### Batch training
```
n_epochs = 100
for epoch in range(n_epochs):
epoch_loss = 0
for img, label in train_loader:
label_one_hot = torch.zeros(batch_sz, 10).scatter_(1, label.view(batch_sz,1), 1.0)
predict = model(img.view(batch_sz, -1))
curr_loss = loss(predict, label_one_hot)
optimizer.zero_grad()
curr_loss.backward() ## gradient
optimizer.step()
epoch_loss += curr_loss
print("Epoch: %d, Loss: %f" % (epoch, float(epoch_loss)))
```
## Test accuracy
### Test set
```
testdata = datasets.MNIST('data/mnist_test', train=False, download=False, transform=transforms.ToTensor()) # download=True to download, train=False means test set
test_loader = torch.utils.data.DataLoader(testdata, batch_size=1, shuffle=True)
img, label = next(iter(test_loader))
predict = model(img.view(-1).unsqueeze(0))
_, predicted_label = torch.max(predict, dim=1)
print(predicted_label.item())
plt.imshow(img.reshape(28,28), cmap='gray')
```
## Visualization
```
!rm -rf runs
writer = SummaryWriter('runs/mnist')
```
### add loss
```
n_epochs = 10
learning_rate = 0.1
for epoch in range(n_epochs):
epoch_loss = 0
for img, label in train_loader:
label_one_hot = torch.zeros(batch_sz, 10).scatter_(1, label.view(batch_sz,1), 1.0)
predict = model(img.view(batch_sz, -1))
curr_loss = loss(predict, label_one_hot)
optimizer.zero_grad()
curr_loss.backward()
optimizer.step()
epoch_loss += curr_loss
writer.add_scalar("Loss/train", epoch_loss, epoch)
print("Epoch: %d, Loss: %f" % (epoch, float(epoch_loss)))
```
### add model
```
img, _ = next(iter(train_loader))
writer.add_graph(model, img.view(batch_sz,-1))
writer.flush()
writer.close()
!tensorboard --logdir=runs/mnist
```
|
github_jupyter
|
import torch
import torch.nn as nn
from torchvision import datasets
from torchvision import transforms
import numpy as np
import matplotlib.pyplot as plt
from torch.utils.tensorboard import SummaryWriter
import torch.optim as optim
traindata = datasets.MNIST('data/mnist_train', train=True, download=False, transform=transforms.ToTensor()) # download = True to download
len(traindata)
img, label = traindata[np.random.randint(0,60000-1)]
print(label)
print(img)
img.size()
plt.imshow(img.reshape(28,28), cmap='gray')
img_train = img.view(-1).unsqueeze(0)
img_train.size()
model = nn.Sequential(
nn.Linear(784, 100),
nn.ReLU(),
nn.Linear(100, 10),
nn.ReLU(),
nn.Softmax(dim=1))
predict = model(img_train)
print(predict)
predict.size()
label_one_hot = torch.zeros(10).scatter_(0, torch.tensor(label), 1.0).unsqueeze(0)
label, label_one_hot, label_one_hot.size()
loss = torch.nn.MSELoss()
loss(predict, label_one_hot)
batch_sz = 600
train_loader = torch.utils.data.DataLoader(traindata, batch_size=batch_sz, shuffle=True)
#img, label = next(iter(train_loader))
learning_rate = 0.5
optimizer = optim.SGD(model.parameters(), lr=learning_rate)
n_epochs = 100
for epoch in range(n_epochs):
epoch_loss = 0
for img, label in train_loader:
label_one_hot = torch.zeros(batch_sz, 10).scatter_(1, label.view(batch_sz,1), 1.0)
predict = model(img.view(batch_sz, -1))
curr_loss = loss(predict, label_one_hot)
optimizer.zero_grad()
curr_loss.backward() ## gradient
optimizer.step()
epoch_loss += curr_loss
print("Epoch: %d, Loss: %f" % (epoch, float(epoch_loss)))
testdata = datasets.MNIST('data/mnist_test', train=False, download=False, transform=transforms.ToTensor()) # download=True to download, train=False means test set
test_loader = torch.utils.data.DataLoader(testdata, batch_size=1, shuffle=True)
img, label = next(iter(test_loader))
predict = model(img.view(-1).unsqueeze(0))
_, predicted_label = torch.max(predict, dim=1)
print(predicted_label.item())
plt.imshow(img.reshape(28,28), cmap='gray')
!rm -rf runs
writer = SummaryWriter('runs/mnist')
n_epochs = 10
learning_rate = 0.1
for epoch in range(n_epochs):
epoch_loss = 0
for img, label in train_loader:
label_one_hot = torch.zeros(batch_sz, 10).scatter_(1, label.view(batch_sz,1), 1.0)
predict = model(img.view(batch_sz, -1))
curr_loss = loss(predict, label_one_hot)
optimizer.zero_grad()
curr_loss.backward()
optimizer.step()
epoch_loss += curr_loss
writer.add_scalar("Loss/train", epoch_loss, epoch)
print("Epoch: %d, Loss: %f" % (epoch, float(epoch_loss)))
img, _ = next(iter(train_loader))
writer.add_graph(model, img.view(batch_sz,-1))
writer.flush()
writer.close()
!tensorboard --logdir=runs/mnist
| 0.888287 | 0.965835 |
<a href="https://colab.research.google.com/github/OODA-OPS/DS-Unit-1-Sprint-1-Dealing-With-Data/blob/master/Copy_of_Final_Assignment_Jason_Meil.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
df = pd.read_csv('https://raw.githubusercontent.com/OODA-OPS/IBM-Data-Science-Professional-Certificate/master/6.%20Data%20Visualization/Final%20Assignment/Data_Science_Topics_Survey.csv')
df.head(3)
```
# Problem 1
```
df.drop(columns='Timestamp', axis = 0, inplace = True)
df.rename(columns=lambda x: x.split('[')[-1][:-1], inplace=True)
df.head()
result = df.transpose()
result['Not interested'] = np.sum(result == 'Not interested', axis = 1)
result['Somewhat interested'] = np.sum(result == 'Somewhat interested', axis = 1)
result['Very interested'] = np.sum(result == 'Very interested', axis = 1)
result.drop(result.columns[:-3],axis=1,inplace=True)
result.sort_index()
```
# Problem 2
```
result = result.sort_index(axis=1, ascending=False)
result = result.sort_values('Very interested', ascending=False)
result = result / len(df) * 100
result.round(2)
ax = result.plot(kind='bar',
figsize=(20, 8),
width=0.8,
color=['#5cb85c', '#5bc0de', '#d9534f'],
)
ax.spines['left'].set_visible(False)
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.axes.get_yaxis().set_visible(False)
ax.tick_params(labelsize=14)
ax.legend(fontsize=14)
ax.set_title("Percentage of Respondents' Interest in Data Science Area", fontsize=16)
for p in ax.patches:
ax.annotate(np.round(p.get_height(),decimals=2),
(p.get_x()+p.get_width()/2., p.get_height()),
ha='center',
va='center',
xytext=(0, 10),
textcoords='offset points',
fontsize = 14
)
plt.show()
```
# Problem 3
```
df = pd.read_csv('https://raw.githubusercontent.com/OODA-OPS/IBM-Data-Science-Professional-Certificate/master/6.%20Data%20Visualization/Final%20Assignment/Police_Department_Incidents_-_Previous_Year__2016_.csv')
df.head()
a = df.PdDistrict.value_counts()
result = pd.DataFrame(data=a.values, index=a.index, columns=['Count'])
result = result.reindex(["CENTRAL", "NORTHERN", "PARK", "SOUTHERN", "MISSION", "TENDERLOIN", "RICHMOND", "TARAVAL", "INGLESIDE", "BAYVIEW"])
result = result.reset_index()
result.rename({'index': 'Neighborhood'}, axis='columns', inplace=True)
result
```
# Problem 4
```
SF_geo = r'https://raw.githubusercontent.com/OODA-OPS/IBM-Data-Science-Professional-Certificate/master/6.%20Data%20Visualization/Final%20Assignment/san-francisco.geojson'
import folium
# San Francisco latitude and longitude values
latitude = 37.77
longitude = -122.42
sanfran_map = folium.Map(location = [latitude, longitude], zoom_start = 12)
# generate choropleth map using the total immigration of each country to Canada from 1980 to 2013
sanfran_map.choropleth(
geo_str=SF_geo,
columns=['Neighborhood', 'Count'],
key_on='feature.properties.DISTRICT',
fill_color='YlOrRd',
fill_opacity=0.7,
line_opacity=0.2,
legend_name='Crime Rate in San Francisco'
)
# display map
sanfran_map
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
df = pd.read_csv('https://raw.githubusercontent.com/OODA-OPS/IBM-Data-Science-Professional-Certificate/master/6.%20Data%20Visualization/Final%20Assignment/Data_Science_Topics_Survey.csv')
df.head(3)
df.drop(columns='Timestamp', axis = 0, inplace = True)
df.rename(columns=lambda x: x.split('[')[-1][:-1], inplace=True)
df.head()
result = df.transpose()
result['Not interested'] = np.sum(result == 'Not interested', axis = 1)
result['Somewhat interested'] = np.sum(result == 'Somewhat interested', axis = 1)
result['Very interested'] = np.sum(result == 'Very interested', axis = 1)
result.drop(result.columns[:-3],axis=1,inplace=True)
result.sort_index()
result = result.sort_index(axis=1, ascending=False)
result = result.sort_values('Very interested', ascending=False)
result = result / len(df) * 100
result.round(2)
ax = result.plot(kind='bar',
figsize=(20, 8),
width=0.8,
color=['#5cb85c', '#5bc0de', '#d9534f'],
)
ax.spines['left'].set_visible(False)
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.axes.get_yaxis().set_visible(False)
ax.tick_params(labelsize=14)
ax.legend(fontsize=14)
ax.set_title("Percentage of Respondents' Interest in Data Science Area", fontsize=16)
for p in ax.patches:
ax.annotate(np.round(p.get_height(),decimals=2),
(p.get_x()+p.get_width()/2., p.get_height()),
ha='center',
va='center',
xytext=(0, 10),
textcoords='offset points',
fontsize = 14
)
plt.show()
df = pd.read_csv('https://raw.githubusercontent.com/OODA-OPS/IBM-Data-Science-Professional-Certificate/master/6.%20Data%20Visualization/Final%20Assignment/Police_Department_Incidents_-_Previous_Year__2016_.csv')
df.head()
a = df.PdDistrict.value_counts()
result = pd.DataFrame(data=a.values, index=a.index, columns=['Count'])
result = result.reindex(["CENTRAL", "NORTHERN", "PARK", "SOUTHERN", "MISSION", "TENDERLOIN", "RICHMOND", "TARAVAL", "INGLESIDE", "BAYVIEW"])
result = result.reset_index()
result.rename({'index': 'Neighborhood'}, axis='columns', inplace=True)
result
SF_geo = r'https://raw.githubusercontent.com/OODA-OPS/IBM-Data-Science-Professional-Certificate/master/6.%20Data%20Visualization/Final%20Assignment/san-francisco.geojson'
import folium
# San Francisco latitude and longitude values
latitude = 37.77
longitude = -122.42
sanfran_map = folium.Map(location = [latitude, longitude], zoom_start = 12)
# generate choropleth map using the total immigration of each country to Canada from 1980 to 2013
sanfran_map.choropleth(
geo_str=SF_geo,
columns=['Neighborhood', 'Count'],
key_on='feature.properties.DISTRICT',
fill_color='YlOrRd',
fill_opacity=0.7,
line_opacity=0.2,
legend_name='Crime Rate in San Francisco'
)
# display map
sanfran_map
| 0.399694 | 0.944382 |
# Linear Shooting Method
To numerically approximate the Boundary Value Problem
$$
y^{''}=p(x)y^{'}+q(x)y+g(x) \ \ \ a < x < b $$
$$y(a)=\alpha$$
$$y(b) =\beta$$
The Boundary Value Problem is divided into two
Initial Value Problems:
1. The first 2nd order Intial Value Problem is the same as the original Boundary Value Problem with an extra initial condtion $y_1^{'}(a)=0$.
\begin{equation}
y^{''}_1=p(x)y^{'}_1+q(x)y_1+r(x), \ \ y_1(a)=\alpha, \ \ \color{green}{y^{'}_1(a)=0},\\
\end{equation}
2. The second 2nd order Intial Value Problem is the homogenous form of the original Boundary Value Problem with the initial condtions $y_2(a)=0$ and $y_2^{'}(a)=1$.
\begin{equation}
y^{''}_2=p(x)y^{'}_2+q(x)y_2, \ \ \color{green}{y_2(a)=0, \ \ y^{'}_2(a)=1}.
\end{equation}
combining these results together to get the unique solution
\begin{equation}
y(x)=y_1(x)+\frac{\beta-y_1(b)}{y_2(b)}y_2(x)
\end{equation}
provided that $y_2(b)\not=0$.
The truncation error for the shooting method is
$$ |y_i - y(x_i)| \leq K h^n\left|1+\frac{w_{1 i}}{u_{1 i}}\right| $$
$O(h^n)$ is the order of the numerical method used to approximate the solution of the Initial Value Problems.
```
import numpy as np
import math
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
class ListTable(list):
""" Overridden list class which takes a 2-dimensional list of
the form [[1,2,3],[4,5,6]], and renders an HTML Table in
IPython Notebook. """
def _repr_html_(self):
html = ["<table>"]
for row in self:
html.append("<tr>")
for col in row:
html.append("<td>{0}</td>".format(col))
html.append("</tr>")
html.append("</table>")
return ''.join(html)
from IPython.display import HTML
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<form action="javascript:code_toggle()"><input type="submit" value="Click here to toggle on/off the raw code."></form>''')
```
## Example Boundary Value Problem
To illustrate the shooting method we shall apply it to the Boundary Value Problem:
$$ y^{''}=2y^{'}+3y-6, $$
with boundary conditions
$$y(0) = 3, $$
$$y(1) = e^3+2, $$
with the exact solution is
$$y=e^{3x}+2. $$
The __boundary value problem__ is broken into two second order __Initial Value Problems:__
1. The first 2nd order Intial Value Problem is the same as the original Boundary Value Problem with an extra initial condtion $u^{'}(0)=0$.
\begin{equation}
u^{''} =2u'+3u-6, \ \ \ \ u(0)=3, \ \ \ \color{green}{u^{'}(0)=0}
\end{equation}
2. The second 2nd order Intial Value Problem is the homogenous form of the original Boundary Value Problem with the initial condtions $w^{'}(0)=0$ and $w^{'}(0)=1$.
\begin{equation}
w^{''} =2w^{'}+3w, \ \ \ \ \color{green}{w(1)=0}, \ \ \ \color{green}{w^{'}(1)=1}
\end{equation}
combining these results of these two intial value problems as a linear sum
\begin{equation}
y(x)=u(x)+\frac{e^{3x}+2-u(1)}{w(1)}w(x)
\end{equation}
gives the solution of the Boundary Value Problem.
## Discrete Axis
The stepsize is defined as
$$h=\frac{b-a}{N}$$
here it is
$$h=\frac{1-0}{10}$$
giving
$$x_i=0+0.1 i$$
for $i=0,1,...10.$
```
## BVP
N=10
h=1/N
x=np.linspace(0,1,N+1)
fig = plt.figure(figsize=(10,4))
plt.plot(x,0*x,'o:',color='red')
plt.xlim((0,1))
plt.title('Illustration of discrete time points for h=%s'%(h))
plt.show()
```
## Initial conditions
The initial conditions for the discrete equations are:
$$ u_1[0]=3$$
$$ \color{green}{u_2[0]=0}$$
$$ \color{green}{w_1[0]=0}$$
$$ \color{green}{w_2[0]=1}$$
```
U1=np.zeros(N+1)
U2=np.zeros(N+1)
W1=np.zeros(N+1)
W2=np.zeros(N+1)
U1[0]=3
U2[0]=0
W1[0]=0
W2[0]=1
```
## Numerical method
The Euler method is applied to numerically approximate the solution of the system of the two second order initial value problems they are converted in to two pairs of two first order initial value problems:
### 1. Inhomogenous Approximation
The plot below shows the numerical approximation for the two first order Intial Value Problems
\begin{equation}
u_1^{'} =u_2, \ \ \ \ u_1(0)=3,
\end{equation}
\begin{equation}
u_2^{'} =2u_2+3u_1-6, \ \ \ \color{green}{u_2(0)=0},
\end{equation}
that Euler approximate of the inhomogeneous two Initial Value Problems is :
$$u_{1}[i+1]=u_{1}[i] + h u_{2}[i]$$
$$u_{2}[i+1]=u_{2}[i] + h (2u_{2}[i]+3u_{1}[i] -6)$$
with $u_1[0]=3$ and $\color{green}{u_2[0]=0}$.
```
for i in range (0,N):
U1[i+1]=U1[i]+h*(U2[i])
U2[i+1]=U2[i]+h*(2*U2[i]+3*U1[i]-6)
```
### Plots
The plot below shows the Euler approximation of the two intial value problems $u_1$ on the left and $u2$ on the right.
```
fig = plt.figure(figsize=(12,4))
ax = fig.add_subplot(1,2,1)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,U1,'^')
plt.title(r"$u_1'=u_2, \ \ u_1(0)=3$",fontsize=16)
plt.grid(True)
ax = fig.add_subplot(1,2,2)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,U2,'v')
plt.title(r"$u_2'=2u_2+3u_1-6, \ \ u_2(0)=0$", fontsize=16)
plt.grid(True)
plt.show()
```
### 2. Homogenous Approximation
The homogeneous Bounday Value Problem is divided into two first order Intial Value Problems
\begin{equation}
w_1^{'} =w_2, \ \ \ \ \color{green}{w_1(1)=0}
\end{equation}
\begin{equation}
w_2^{'} =2w_2+3w_1, \ \ \ \color{green}{w_2(1)=1}
\end{equation}
The Euler approximation of the homogeneous of the two Initial Value Problem is
$$w_{1}[i+1]=w_{1}[i] + h w_{2}[i]$$
$$w_{2}[i+1]=w_{2}[i] + h (2w_{2}[i]+3w_{1}[i])$$
with $\color{green}{w_1[0]=0}$ and $\color{green}{w_2[1]=1}$.
```
for i in range (0,N):
W1[i+1]=W1[i]+h*(W2[i])
W2[i+1]=W2[i]+h*(2*W2[i]+3*W1[i])
```
### Homogenous Approximation
### Plots
The plot below shows the Euler approximation of the two intial value problems $u_1$ on the left and $u2$ on the right.
```
fig = plt.figure(figsize=(12,4))
ax = fig.add_subplot(1,2,1)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,W1,'^')
plt.grid(True)
plt.title(r"$w_1'=w_2, \ \ w_1(0)=0$",fontsize=16)
ax = fig.add_subplot(1,2,2)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,W2,'v')
plt.grid(True)
plt.title(r"$w_2'=2w_2+3w_1, \ \ w_2(0)=1$",fontsize=16)
plt.tight_layout()
plt.subplots_adjust(top=0.85)
plt.show()
beta=math.exp(3)+2
y=U1+(beta-U1[N])/W1[N]*W1
```
## Approximate Solution
Combining together the numerical approximation of $u_1$ and $u_2$ as a weighted sum
$$y(x[i])\approx u_{1}[i] + \frac{e^3+2-u_{1}[N]}{w_1[N]}w_{1}[i]$$
gives the approximate solution of the Boundary Value Problem.
The truncation error for the shooting method using the Euler method is
$$ |y_i - y(x[i])| \leq K h\left|1+\frac{w_{1}[i]}{u_{1}[i]}\right| $$
$O(h)$ is the order of the method.
The plot below shows the approximate solution of the Boundary Value Problem (left), the exact solution (middle) and the error (right)
```
Exact=np.exp(3*x)+2
fig = plt.figure(figsize=(12,4))
ax = fig.add_subplot(2,3,1)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,y,'o')
plt.grid(True)
plt.title(r"Numerical: $u_1+\frac{e^3+2-u_1(N)}{w_1(N)}w_1$",
fontsize=16)
ax = fig.add_subplot(2,3,2)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,Exact,'ks-')
plt.grid(True)
plt.title(r"Exact: $y=e^{3x}+2$",
fontsize=16)
ax = fig.add_subplot(2,3,3)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,abs(y-Exact),'ro')
plt.grid(True)
plt.title(r"Error ",fontsize=16)
plt.tight_layout()
plt.subplots_adjust(top=0.85)
plt.show()
```
### Data
The Table below shows that output for $x$, the Euler numerical approximations $U1$, $U2$, $W1$ and $W2$ of the system of four Intial Value Problems, the shooting methods approximate solution $y_i=u_{1 i} + \frac{e^3+2-u_{1}(x_N)}{w_1(x_N)}w_{1 i}$ and the exact solution of the Boundary Value Problem.
```
table = ListTable()
table.append(['x', 'U1','U2','W1','W2','Approx','Exact'])
for i in range (0,len(x)):
table.append([round(x[i],3), round(U1[i],3), round(U2[i],3),
round(W1[i],5),round(W2[i],3),
round(y[i],5),
round(Exact[i],5)])
table
```
|
github_jupyter
|
import numpy as np
import math
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
class ListTable(list):
""" Overridden list class which takes a 2-dimensional list of
the form [[1,2,3],[4,5,6]], and renders an HTML Table in
IPython Notebook. """
def _repr_html_(self):
html = ["<table>"]
for row in self:
html.append("<tr>")
for col in row:
html.append("<td>{0}</td>".format(col))
html.append("</tr>")
html.append("</table>")
return ''.join(html)
from IPython.display import HTML
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<form action="javascript:code_toggle()"><input type="submit" value="Click here to toggle on/off the raw code."></form>''')
## BVP
N=10
h=1/N
x=np.linspace(0,1,N+1)
fig = plt.figure(figsize=(10,4))
plt.plot(x,0*x,'o:',color='red')
plt.xlim((0,1))
plt.title('Illustration of discrete time points for h=%s'%(h))
plt.show()
U1=np.zeros(N+1)
U2=np.zeros(N+1)
W1=np.zeros(N+1)
W2=np.zeros(N+1)
U1[0]=3
U2[0]=0
W1[0]=0
W2[0]=1
for i in range (0,N):
U1[i+1]=U1[i]+h*(U2[i])
U2[i+1]=U2[i]+h*(2*U2[i]+3*U1[i]-6)
fig = plt.figure(figsize=(12,4))
ax = fig.add_subplot(1,2,1)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,U1,'^')
plt.title(r"$u_1'=u_2, \ \ u_1(0)=3$",fontsize=16)
plt.grid(True)
ax = fig.add_subplot(1,2,2)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,U2,'v')
plt.title(r"$u_2'=2u_2+3u_1-6, \ \ u_2(0)=0$", fontsize=16)
plt.grid(True)
plt.show()
for i in range (0,N):
W1[i+1]=W1[i]+h*(W2[i])
W2[i+1]=W2[i]+h*(2*W2[i]+3*W1[i])
fig = plt.figure(figsize=(12,4))
ax = fig.add_subplot(1,2,1)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,W1,'^')
plt.grid(True)
plt.title(r"$w_1'=w_2, \ \ w_1(0)=0$",fontsize=16)
ax = fig.add_subplot(1,2,2)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,W2,'v')
plt.grid(True)
plt.title(r"$w_2'=2w_2+3w_1, \ \ w_2(0)=1$",fontsize=16)
plt.tight_layout()
plt.subplots_adjust(top=0.85)
plt.show()
beta=math.exp(3)+2
y=U1+(beta-U1[N])/W1[N]*W1
Exact=np.exp(3*x)+2
fig = plt.figure(figsize=(12,4))
ax = fig.add_subplot(2,3,1)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,y,'o')
plt.grid(True)
plt.title(r"Numerical: $u_1+\frac{e^3+2-u_1(N)}{w_1(N)}w_1$",
fontsize=16)
ax = fig.add_subplot(2,3,2)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,Exact,'ks-')
plt.grid(True)
plt.title(r"Exact: $y=e^{3x}+2$",
fontsize=16)
ax = fig.add_subplot(2,3,3)
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.plot(x,abs(y-Exact),'ro')
plt.grid(True)
plt.title(r"Error ",fontsize=16)
plt.tight_layout()
plt.subplots_adjust(top=0.85)
plt.show()
table = ListTable()
table.append(['x', 'U1','U2','W1','W2','Approx','Exact'])
for i in range (0,len(x)):
table.append([round(x[i],3), round(U1[i],3), round(U2[i],3),
round(W1[i],5),round(W2[i],3),
round(y[i],5),
round(Exact[i],5)])
table
| 0.232659 | 0.988335 |
```
import numpy
from diatom import Hamiltonian
from diatom import Calculate
from matplotlib import pyplot
from scipy import constants
import os,time
cwd = os.path.abspath('')
```
lets start by choosing a molecule. We will use the bialkali $^{87}$Rb$^{133}$Cs, this is one of the preset molecules. Lets also use this opportunity to set some universal constants.
```
Constants = Hamiltonian.RbCs
print(Constants)
h = constants.h #Planck's Constant
c = constants.c #Speed of Light
eps0 = constants.epsilon_0 #permittivity of free space (electric constant)
pi = numpy.pi #ratio of circumference to diameter
bohr = constants.physical_constants['Bohr radius'][0] #Bohr radius
```
The dictionary "Constants" contains all of the parameters needed to fully construct the hyperfine Hamiltonian in SI units. First lets do a single calculation of the hyperfine structure at a fixed magnetic field of 181.5 G
## Energy and TDMs at 181.5 G
First define some of the constants in the problem. As well as a location to store the output.
```
B = 181.5*1e-4 #Magnetic field in Tesla
Nmax = 3 #Maximum Rotation quantum number to include
filepath = cwd+"\\Outputs\\"
FileName_Suffix ="B_{:.1f}G Nmax_{:d}".format(B*1e4,Nmax)
print("Output files will be:",filepath,"<Var>",FileName_Suffix)
```
Now to generate the Hyperfine Hamiltonian
```
then = time.time()
H0,Hz,HDC,HAC = Hamiltonian.Build_Hamiltonians(Nmax,Constants,zeeman=True)
now=time.time()
print("Took {:.3f} seconds".format(now-then))
```
Now we need to calculate the eigenstates and eigenvalues. This is best done using numpy.linalg.eigh
```
then = time.time()
H = H0+B*Hz
eigvals,eigstates = numpy.linalg.eigh(H)
now = time.time()
print("Took {:.3f} seconds".format(now-then))
```
We now have a list of all of the energies and eigenstates at this magnetic field. Lets say that we want to calculate the transition dipole moment from the lowest state with $N=0$ ,$M_F =5$. First we need to label each state with $N$ and $M_F$.
```
then = time.time()
N,MN = Calculate.LabelStates_N_MN(eigstates,Nmax,Constants['I1'],Constants['I2'])
F,MF = Calculate.LabelStates_F_MF(eigstates,Nmax,Constants['I1'],Constants['I2'])
now = time.time()
print("Took {:.3f} seconds".format(now-then))
```
Now we need to find the states where N=0 and where MF = 5
```
then = time.time()
loc = numpy.where(numpy.logical_and(N==0,MF==5))[0][0]
gs = eigstates[:,loc]
now = time.time()
print("Took {:.3f} seconds".format(now-then))
```
Now to calculate the TDM, by deafult this is in units of the permanent dipole moment
```
then = time.time()
TDM_pi = Calculate.TDM(Nmax,Constants['I1'],Constants['I2'],0,eigstates,loc)
TDM_Sigma_plus = Calculate.TDM(Nmax,Constants['I1'],Constants['I2'],-1,eigstates,loc)
TDM_Sigma_minus = Calculate.TDM(Nmax,Constants['I1'],Constants['I2'],+1,eigstates,loc)
now = time.time()
print("Took {:.3f} seconds".format(now-then))
```
Now we want to save this result so that we can read it. We will save this along with the energy. In a separate file we can save the state compositions
```
then = time.time()
file = filepath + "TDM" + FileName_Suffix
fmt = ['%.0f',"%.0f","%.6f","%.6f","%.6f"]
Calculate.Export_Energy(file,eigvals/h,labels=[N,MF,TDM_pi,TDM_Sigma_plus,TDM_Sigma_minus],headers=["N","MF","d_pi(d0)","d_plus(d0)","d_minus(d0)"],format = fmt)
file = filepath + "States" + FileName_Suffix
Calculate.Export_State_Comp(file,Nmax,Constants['I1'],Constants['I2'],eigstates,labels=[N,MF,eigvals/h],headers=["N","MF","Energy (Hz)"])
now = time.time()
print("Took {:.3f} seconds".format(now-then))
```
What if we want a more visual representation of these numbers however? The plotting module includes a useful function just for this purpose
```
from diatom import Plotting
figure = pyplot.figure()
TDMs =[TDM_Sigma_minus,TDM_pi,TDM_Sigma_plus]
Plotting.TDM_plot(eigvals,eigstates,loc,Nmax,Constants['I1'],Constants['I2'],Offset=980e6)
pyplot.show()
```
## Calculate a Breit-Rabi Diagram
The majority of the things we want to calculate are maps. Showing the variation of the molecular structure with a given parameter. For a demonstration lets plot a Breit-Rabi diagram showing the variation of the hyperfine structure with magnetic field.
```
Bmax = 181.5*1e-4 #Maximum Magnetic field in Tesla
Bmin = 1e-9 #Minimum Magnetic field in Tesla
Nmax = 3 #Maximum Rotation quantum number to include
filepath = cwd+"\\Outputs\\"
FileName_Suffix ="B_{:.1f}G Nmax_{:d}".format(B*1e4,Nmax)
print("Output files will be:",filepath,"<Var>",FileName_Suffix)
```
Lets solve this problem, using the same method as for the single magnetic field. We will build a list of Hamiltonians using pythonic list comprehension
```
then = time.time()
fields = numpy.linspace(Bmin,Bmax,250)
H = [H0+B*Hz for B in fields]
eigvals,eigstates = numpy.linalg.eigh(H)
now = time.time()
print("Took {:.3f} seconds".format(now-then))
```
Lets plot this!
```
then = time.time()
figure = pyplot.figure()
ax = figure.add_subplot(111)
for x in range(eigvals.shape[1]):
ax.plot(fields*1e4,1e-6*eigvals[:,x]/h)
ax.set_xlabel("Magnetic Field (G)")
ax.set_ylabel("Energy/$h$ (MHz)")
ax.set_ylim(-0.5,0.25)
ax.set_xlim(0,181.5)
pyplot.show()
now = time.time()
print("Took {:.3f} seconds".format(now-then))
```
At first glance this looks great, but looking closer the states "hop" across one another, this won't do as we can't identify an energy level in any useful manner! We can solve this by sorting the states to ensure smooth variation of the energy.
```
eigvals,eigstates = Calculate.Sort_Smooth(eigvals/h,eigstates,pb=True)
```
Let's look to see if this is worthwhile:
```
then = time.time()
figure = pyplot.figure()
ax = figure.add_subplot(111)
for x in range(eigvals.shape[1]):
ax.plot(fields*1e4,1e-6*eigvals[:,x])
ax.set_xlabel("Magnetic Field (G)")
ax.set_ylabel("Energy/$h$ (MHz)")
ax.set_ylim(-0.5,0.25)
ax.set_xlim(0,181.5)
pyplot.show()
now = time.time()
print("Took {:.3f} seconds".format(now-then))
```
Some labels would be handy, lets label by the last state
```
then = time.time()
N,MN = Calculate.LabelStates_N_MN(eigstates[-1,:,:],Nmax,Constants['I1'],Constants['I2'])
F,MF = Calculate.LabelStates_F_MF(eigstates[-1,:,:],Nmax,Constants['I1'],Constants['I2'])
now = time.time()
print("Took {:.3f} seconds".format(now-then))
```
And as before lets save this file
```
then = time.time()
file = filepath + "Energies" + FileName_Suffix
Calculate.Export_Energy(file,eigvals,Fields= 1e4*fields,labels=[N,MF],
headers=["N","MF"])
now = time.time()
print("Took {:.3f} seconds".format(now-then))
```
|
github_jupyter
|
import numpy
from diatom import Hamiltonian
from diatom import Calculate
from matplotlib import pyplot
from scipy import constants
import os,time
cwd = os.path.abspath('')
Constants = Hamiltonian.RbCs
print(Constants)
h = constants.h #Planck's Constant
c = constants.c #Speed of Light
eps0 = constants.epsilon_0 #permittivity of free space (electric constant)
pi = numpy.pi #ratio of circumference to diameter
bohr = constants.physical_constants['Bohr radius'][0] #Bohr radius
B = 181.5*1e-4 #Magnetic field in Tesla
Nmax = 3 #Maximum Rotation quantum number to include
filepath = cwd+"\\Outputs\\"
FileName_Suffix ="B_{:.1f}G Nmax_{:d}".format(B*1e4,Nmax)
print("Output files will be:",filepath,"<Var>",FileName_Suffix)
then = time.time()
H0,Hz,HDC,HAC = Hamiltonian.Build_Hamiltonians(Nmax,Constants,zeeman=True)
now=time.time()
print("Took {:.3f} seconds".format(now-then))
then = time.time()
H = H0+B*Hz
eigvals,eigstates = numpy.linalg.eigh(H)
now = time.time()
print("Took {:.3f} seconds".format(now-then))
then = time.time()
N,MN = Calculate.LabelStates_N_MN(eigstates,Nmax,Constants['I1'],Constants['I2'])
F,MF = Calculate.LabelStates_F_MF(eigstates,Nmax,Constants['I1'],Constants['I2'])
now = time.time()
print("Took {:.3f} seconds".format(now-then))
then = time.time()
loc = numpy.where(numpy.logical_and(N==0,MF==5))[0][0]
gs = eigstates[:,loc]
now = time.time()
print("Took {:.3f} seconds".format(now-then))
then = time.time()
TDM_pi = Calculate.TDM(Nmax,Constants['I1'],Constants['I2'],0,eigstates,loc)
TDM_Sigma_plus = Calculate.TDM(Nmax,Constants['I1'],Constants['I2'],-1,eigstates,loc)
TDM_Sigma_minus = Calculate.TDM(Nmax,Constants['I1'],Constants['I2'],+1,eigstates,loc)
now = time.time()
print("Took {:.3f} seconds".format(now-then))
then = time.time()
file = filepath + "TDM" + FileName_Suffix
fmt = ['%.0f',"%.0f","%.6f","%.6f","%.6f"]
Calculate.Export_Energy(file,eigvals/h,labels=[N,MF,TDM_pi,TDM_Sigma_plus,TDM_Sigma_minus],headers=["N","MF","d_pi(d0)","d_plus(d0)","d_minus(d0)"],format = fmt)
file = filepath + "States" + FileName_Suffix
Calculate.Export_State_Comp(file,Nmax,Constants['I1'],Constants['I2'],eigstates,labels=[N,MF,eigvals/h],headers=["N","MF","Energy (Hz)"])
now = time.time()
print("Took {:.3f} seconds".format(now-then))
from diatom import Plotting
figure = pyplot.figure()
TDMs =[TDM_Sigma_minus,TDM_pi,TDM_Sigma_plus]
Plotting.TDM_plot(eigvals,eigstates,loc,Nmax,Constants['I1'],Constants['I2'],Offset=980e6)
pyplot.show()
Bmax = 181.5*1e-4 #Maximum Magnetic field in Tesla
Bmin = 1e-9 #Minimum Magnetic field in Tesla
Nmax = 3 #Maximum Rotation quantum number to include
filepath = cwd+"\\Outputs\\"
FileName_Suffix ="B_{:.1f}G Nmax_{:d}".format(B*1e4,Nmax)
print("Output files will be:",filepath,"<Var>",FileName_Suffix)
then = time.time()
fields = numpy.linspace(Bmin,Bmax,250)
H = [H0+B*Hz for B in fields]
eigvals,eigstates = numpy.linalg.eigh(H)
now = time.time()
print("Took {:.3f} seconds".format(now-then))
then = time.time()
figure = pyplot.figure()
ax = figure.add_subplot(111)
for x in range(eigvals.shape[1]):
ax.plot(fields*1e4,1e-6*eigvals[:,x]/h)
ax.set_xlabel("Magnetic Field (G)")
ax.set_ylabel("Energy/$h$ (MHz)")
ax.set_ylim(-0.5,0.25)
ax.set_xlim(0,181.5)
pyplot.show()
now = time.time()
print("Took {:.3f} seconds".format(now-then))
eigvals,eigstates = Calculate.Sort_Smooth(eigvals/h,eigstates,pb=True)
then = time.time()
figure = pyplot.figure()
ax = figure.add_subplot(111)
for x in range(eigvals.shape[1]):
ax.plot(fields*1e4,1e-6*eigvals[:,x])
ax.set_xlabel("Magnetic Field (G)")
ax.set_ylabel("Energy/$h$ (MHz)")
ax.set_ylim(-0.5,0.25)
ax.set_xlim(0,181.5)
pyplot.show()
now = time.time()
print("Took {:.3f} seconds".format(now-then))
then = time.time()
N,MN = Calculate.LabelStates_N_MN(eigstates[-1,:,:],Nmax,Constants['I1'],Constants['I2'])
F,MF = Calculate.LabelStates_F_MF(eigstates[-1,:,:],Nmax,Constants['I1'],Constants['I2'])
now = time.time()
print("Took {:.3f} seconds".format(now-then))
then = time.time()
file = filepath + "Energies" + FileName_Suffix
Calculate.Export_Energy(file,eigvals,Fields= 1e4*fields,labels=[N,MF],
headers=["N","MF"])
now = time.time()
print("Took {:.3f} seconds".format(now-then))
| 0.482917 | 0.880746 |
<a href="https://colab.research.google.com/github/TerradasExatas/Controle_em_python/blob/main/Controle_python_11_sintonia_de_PID_tudo.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
pip install control
```

```
#circuito de 3ª ordem, saida no capacitor
#****parte 1: o sistema*****
#importa a biblioteca simbólica
import sympy
#define as variáveis
s,vc1,vout,il,iR1,iR2,vin,R1,R2,C1,C2,L = sympy.symbols(
's,vc1,vout,il,iR1,iR2,vin,R1,R2,C1,C2,L')
#valor dos componentes
R1=1; R2=1; C1=1; C2=1; L=1 #comente essa linha para solução geral!
#equações
eq1 = vc1 + R1*iR1 - vin
eq2 = vout-vc1 + L*s*il + R2*iR2
eq3 = -C1*s*vc1 + iR1 - iR2
eq4 = iR2 - il
eq5 = C2*s*vout - il
#resolve o sistema
solucao=sympy.solve([eq1, eq2,eq3,eq4,eq5],(vout,vc1,il,iR1,iR2))
#mostra a solução
print('\n Vout=')
sympy.pprint(solucao[vout])
#****parte 2: O controle****
#calcula a saída sobre a entrada simbólica
tf=sympy.simplify(solucao[vout]/vin)
print('\n Vout/Vin=',tf)
#importa as demais bibliotecas
import control as ctl
import matplotlib.pyplot as plt
import numpy as np
#transforma Vout/Vin simbólica em FT
s = ctl.TransferFunction.s
P_s=eval(str(tf))
print("FT=",P_s)
#calcula o ganho crítico para oscilação
out_st_magn=ctl.stability_margins(P_s)
K_cr=np.round(out_st_magn[0]*1000)/1000
W_cr=np.round(out_st_magn[3]*1000)/1000
T_cr=2*np.pi/W_cr
print('K de oscilação ',K_cr)
print('T de oscilação ',T_cr)
# controlador PID Ziegler-Nichols da frequencia
Ki=1.2*K_cr/T_cr;Kp=K_cr*0.6;Kd=0.075*K_cr*T_cr;
print('Kp=' f'{Kp:6.2f}, Ki=' f'{Ki:6.2f}, Kd=' f'{Kd:6.2f}')
C_s=(Ki+Kp*s+Kd*s**2)/(s)
# calcula FT em malha fechada
H_s=1
G_s=ctl.series(C_s, P_s);
G1_s=ctl.feedback(G_s, H_s, sign=-1);
#calcula a resposta ao degrau do PID sintonizado
Tsim=10
T, yout = ctl.step_response(G1_s, Tsim)
plt.rcParams.update({'font.size': 14})
plt.figure()
plt.plot(T,yout,'k-',label="resposta em malha fechada");plt.grid()
T2=np.linspace(-0.2,Tsim,1000)
degrau=np.ones_like(T2)
degrau[T2<0]=0;
plt.plot(T2,degrau,'r-',label="entrada: degrau unitário")
plt.legend()
```
|
github_jupyter
|
pip install control
#circuito de 3ª ordem, saida no capacitor
#****parte 1: o sistema*****
#importa a biblioteca simbólica
import sympy
#define as variáveis
s,vc1,vout,il,iR1,iR2,vin,R1,R2,C1,C2,L = sympy.symbols(
's,vc1,vout,il,iR1,iR2,vin,R1,R2,C1,C2,L')
#valor dos componentes
R1=1; R2=1; C1=1; C2=1; L=1 #comente essa linha para solução geral!
#equações
eq1 = vc1 + R1*iR1 - vin
eq2 = vout-vc1 + L*s*il + R2*iR2
eq3 = -C1*s*vc1 + iR1 - iR2
eq4 = iR2 - il
eq5 = C2*s*vout - il
#resolve o sistema
solucao=sympy.solve([eq1, eq2,eq3,eq4,eq5],(vout,vc1,il,iR1,iR2))
#mostra a solução
print('\n Vout=')
sympy.pprint(solucao[vout])
#****parte 2: O controle****
#calcula a saída sobre a entrada simbólica
tf=sympy.simplify(solucao[vout]/vin)
print('\n Vout/Vin=',tf)
#importa as demais bibliotecas
import control as ctl
import matplotlib.pyplot as plt
import numpy as np
#transforma Vout/Vin simbólica em FT
s = ctl.TransferFunction.s
P_s=eval(str(tf))
print("FT=",P_s)
#calcula o ganho crítico para oscilação
out_st_magn=ctl.stability_margins(P_s)
K_cr=np.round(out_st_magn[0]*1000)/1000
W_cr=np.round(out_st_magn[3]*1000)/1000
T_cr=2*np.pi/W_cr
print('K de oscilação ',K_cr)
print('T de oscilação ',T_cr)
# controlador PID Ziegler-Nichols da frequencia
Ki=1.2*K_cr/T_cr;Kp=K_cr*0.6;Kd=0.075*K_cr*T_cr;
print('Kp=' f'{Kp:6.2f}, Ki=' f'{Ki:6.2f}, Kd=' f'{Kd:6.2f}')
C_s=(Ki+Kp*s+Kd*s**2)/(s)
# calcula FT em malha fechada
H_s=1
G_s=ctl.series(C_s, P_s);
G1_s=ctl.feedback(G_s, H_s, sign=-1);
#calcula a resposta ao degrau do PID sintonizado
Tsim=10
T, yout = ctl.step_response(G1_s, Tsim)
plt.rcParams.update({'font.size': 14})
plt.figure()
plt.plot(T,yout,'k-',label="resposta em malha fechada");plt.grid()
T2=np.linspace(-0.2,Tsim,1000)
degrau=np.ones_like(T2)
degrau[T2<0]=0;
plt.plot(T2,degrau,'r-',label="entrada: degrau unitário")
plt.legend()
| 0.295535 | 0.710481 |
# Winnining Wrestlers Entertainment
In this activity you will be taking four seperate csvs that were scraped down from a wrestling database, merging them together, and then creating charts to visualize a wrestler's wins and losses over the course of four years.
### Part 1 - Macho Merging
* You will likely need to perform three different merges over the course of this activity, changing the names of your columns as you go along.
```
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
# Take in all of our wrestling data and read it into pandas
wrestling_2013 = "../Resources/WWE-Data-2013.csv"
wrestling_2014 = "../Resources/WWE-Data-2014.csv"
wrestling_2015 = "../Resources/WWE-Data-2015.csv"
wrestling_2016 = "../Resources/WWE-Data-2016.csv"
wrestlers_2013_df = pd.read_csv(wrestling_2013)
wrestlers_2014_df = pd.read_csv(wrestling_2014)
wrestlers_2015_df = pd.read_csv(wrestling_2015)
wrestlers_2016_df = pd.read_csv(wrestling_2016)
# Merge the first two datasets on "Wrestler" so that no data is lost (should be 182 rows)
combined_wrestlers_df = pd.merge(wrestlers_2013_df, wrestlers_2014_df,
how='outer', on='Wrestler')
combined_wrestlers_df.head()
# Rename our _x columns to "2013 Wins", "2013 Losses", and "2013 Draws"
combined_wrestlers_df = combined_wrestlers_df.rename(columns={"Wins_x":"2013 Wins",
"Losses_x":"2013 Losses",
"Draws_x":"2013 Draws"})
# Rename our _y columns to "2014 Wins", "2014 Losses", and "2014 Draws"
combined_wrestlers_df = combined_wrestlers_df.rename(columns={"Wins_y":"2014 Wins",
"Losses_y":"2014 Losses",
"Draws_y":"2014 Draws"})
combined_wrestlers_df.head()
# Merge our newly combined dataframe with the 2015 dataframe
combined_wrestlers_df = pd.merge(combined_wrestlers_df, wrestlers_2015_df, how="outer", on="Wrestler")
combined_wrestlers_df
# Rename "wins", "losses", and "draws" to "2015 Wins", "2015 Losses", and "2015 Draws"
combined_wrestlers_df = combined_wrestlers_df.rename(columns={"Wins":"2015 Wins","Losses":"2015 Losses","Draws":"2015 Draws"})
combined_wrestlers_df.head()
# Merge our newly combined dataframe with the 2016 dataframe
combined_wrestlers_df = pd.merge(combined_wrestlers_df, wrestlers_2016_df, how="outer", on="Wrestler")
combined_wrestlers_df
# Rename "wins", "losses", and "draws" to "2016 Wins", "2016 Losses", and "2016 Draws"
combined_wrestlers_df = combined_wrestlers_df.rename(columns={"Wins":"2016 Wins","Losses":"2016 Losses","Draws":"2016 Draws"})
combined_wrestlers_df.head(10)
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
# Take in all of our wrestling data and read it into pandas
wrestling_2013 = "../Resources/WWE-Data-2013.csv"
wrestling_2014 = "../Resources/WWE-Data-2014.csv"
wrestling_2015 = "../Resources/WWE-Data-2015.csv"
wrestling_2016 = "../Resources/WWE-Data-2016.csv"
wrestlers_2013_df = pd.read_csv(wrestling_2013)
wrestlers_2014_df = pd.read_csv(wrestling_2014)
wrestlers_2015_df = pd.read_csv(wrestling_2015)
wrestlers_2016_df = pd.read_csv(wrestling_2016)
# Merge the first two datasets on "Wrestler" so that no data is lost (should be 182 rows)
combined_wrestlers_df = pd.merge(wrestlers_2013_df, wrestlers_2014_df,
how='outer', on='Wrestler')
combined_wrestlers_df.head()
# Rename our _x columns to "2013 Wins", "2013 Losses", and "2013 Draws"
combined_wrestlers_df = combined_wrestlers_df.rename(columns={"Wins_x":"2013 Wins",
"Losses_x":"2013 Losses",
"Draws_x":"2013 Draws"})
# Rename our _y columns to "2014 Wins", "2014 Losses", and "2014 Draws"
combined_wrestlers_df = combined_wrestlers_df.rename(columns={"Wins_y":"2014 Wins",
"Losses_y":"2014 Losses",
"Draws_y":"2014 Draws"})
combined_wrestlers_df.head()
# Merge our newly combined dataframe with the 2015 dataframe
combined_wrestlers_df = pd.merge(combined_wrestlers_df, wrestlers_2015_df, how="outer", on="Wrestler")
combined_wrestlers_df
# Rename "wins", "losses", and "draws" to "2015 Wins", "2015 Losses", and "2015 Draws"
combined_wrestlers_df = combined_wrestlers_df.rename(columns={"Wins":"2015 Wins","Losses":"2015 Losses","Draws":"2015 Draws"})
combined_wrestlers_df.head()
# Merge our newly combined dataframe with the 2016 dataframe
combined_wrestlers_df = pd.merge(combined_wrestlers_df, wrestlers_2016_df, how="outer", on="Wrestler")
combined_wrestlers_df
# Rename "wins", "losses", and "draws" to "2016 Wins", "2016 Losses", and "2016 Draws"
combined_wrestlers_df = combined_wrestlers_df.rename(columns={"Wins":"2016 Wins","Losses":"2016 Losses","Draws":"2016 Draws"})
combined_wrestlers_df.head(10)
| 0.404625 | 0.653721 |
# Imports
```
import math
import pandas as pd
import pennylane as qml
import time
from keras.datasets import mnist
from matplotlib import pyplot as plt
from pennylane import numpy as np
from pennylane.templates import AmplitudeEmbedding, AngleEmbedding
from pennylane.templates.subroutines import ArbitraryUnitary
from sklearn.decomposition import PCA
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
```
# Model Params
```
np.random.seed(131)
initial_params = np.random.random([15])
INITIALIZATION_METHOD = 'Amplitude'
BATCH_SIZE = 20
EPOCHS = 400
STEP_SIZE = 0.01
BETA_1 = 0.9
BETA_2 = 0.99
EPSILON = 0.00000001
TRAINING_SIZE = 0.78
VALIDATION_SIZE = 0.07
TEST_SIZE = 1-TRAINING_SIZE-VALIDATION_SIZE
initial_time = time.time()
```
# Import dataset
```
(train_X, train_y), (test_X, test_y) = mnist.load_data()
examples = np.append(train_X, test_X, axis=0)
examples = examples.reshape(70000, 28*28)
classes = np.append(train_y, test_y)
x = []
y = []
for (example, label) in zip(examples, classes):
if label == 2:
x.append(example)
y.append(-1)
elif label == 7:
x.append(example)
y.append(1)
x = np.array(x)
y = np.array(y)
# Normalize pixels values
x = x / 255
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=TEST_SIZE, shuffle=True)
validation_indexes = np.random.random_integers(len(X_train), size=(math.floor(len(X_train)*VALIDATION_SIZE),))
X_validation = [X_train[n] for n in validation_indexes]
y_validation = [y_train[n] for n in validation_indexes]
pca = PCA(n_components=8)
pca.fit(X_train)
X_train = pca.transform(X_train)
X_validation = pca.transform(X_validation)
X_test = pca.transform(X_test)
preprocessing_time = time.time()
```
# Circuit creation
```
device = qml.device("default.qubit", wires=3)
@qml.qnode(device)
def circuit(features, params):
# Load state
if INITIALIZATION_METHOD == 'Amplitude':
AmplitudeEmbedding(features=features, wires=range(3), normalize=True, pad_with=0.)
else:
AngleEmbedding(features=features, wires=range(3), rotation='Y')
# First layer
qml.U3(params[0], params[1], params[2], wires=0)
qml.U3(params[3], params[4], params[5], wires=1)
qml.CNOT(wires=[0, 1])
# Second layer
qml.U3(params[6], params[7], params[8], wires=1)
qml.U3(params[9], params[10], params[11], wires=2)
qml.CNOT(wires=[1, 2])
# Third layer
qml.U3(params[12], params[13], params[14], wires=2)
# Measurement
return qml.expval(qml.PauliZ(2))
```
## Circuit example
```
features = X_train[0]
print(f"Inital parameters: {initial_params}\n")
print(f"Example features: {features}\n")
print(f"Expectation value: {circuit(features, initial_params)}\n")
print(circuit.draw())
```
# Accuracy test definition
```
def measure_accuracy(x, y, circuit_params):
class_errors = 0
for example, example_class in zip(x, y):
predicted_value = circuit(example, circuit_params)
if (example_class > 0 and predicted_value <= 0) or (example_class <= 0 and predicted_value > 0):
class_errors += 1
return 1 - (class_errors/len(y))
```
# Training
```
params = initial_params
opt = qml.AdamOptimizer(stepsize=STEP_SIZE, beta1=BETA_1, beta2=BETA_2, eps=EPSILON)
test_accuracies = []
best_validation_accuracy = 0.0
best_params = []
for i in range(len(X_train)):
features = X_train[i]
expected_value = y_train[i]
def cost(circuit_params):
value = circuit(features, circuit_params)
return ((expected_value - value) ** 2)/len(X_train)
params = opt.step(cost, params)
if i % BATCH_SIZE == 0:
print(f"epoch {i//BATCH_SIZE}")
if i % (10*BATCH_SIZE) == 0:
current_accuracy = measure_accuracy(X_validation, y_validation, params)
test_accuracies.append(current_accuracy)
print(f"accuracy: {current_accuracy}")
if current_accuracy > best_validation_accuracy:
print("best accuracy so far!")
best_validation_accuracy = current_accuracy
best_params = params
if len(test_accuracies) == 30:
print(f"test_accuracies: {test_accuracies}")
if np.allclose(best_validation_accuracy, test_accuracies[0]):
params = best_params
break
del test_accuracies[0]
print("Optimized rotation angles: {}".format(params))
training_time = time.time()
```
# Testing
```
accuracy = measure_accuracy(X_test, y_test, params)
print(accuracy)
test_time = time.time()
print(f"pre-processing time: {preprocessing_time-initial_time}")
print(f"training time: {training_time - preprocessing_time}")
print(f"test time: {test_time - training_time}")
print(f"total time: {test_time - initial_time}")
```
|
github_jupyter
|
import math
import pandas as pd
import pennylane as qml
import time
from keras.datasets import mnist
from matplotlib import pyplot as plt
from pennylane import numpy as np
from pennylane.templates import AmplitudeEmbedding, AngleEmbedding
from pennylane.templates.subroutines import ArbitraryUnitary
from sklearn.decomposition import PCA
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
np.random.seed(131)
initial_params = np.random.random([15])
INITIALIZATION_METHOD = 'Amplitude'
BATCH_SIZE = 20
EPOCHS = 400
STEP_SIZE = 0.01
BETA_1 = 0.9
BETA_2 = 0.99
EPSILON = 0.00000001
TRAINING_SIZE = 0.78
VALIDATION_SIZE = 0.07
TEST_SIZE = 1-TRAINING_SIZE-VALIDATION_SIZE
initial_time = time.time()
(train_X, train_y), (test_X, test_y) = mnist.load_data()
examples = np.append(train_X, test_X, axis=0)
examples = examples.reshape(70000, 28*28)
classes = np.append(train_y, test_y)
x = []
y = []
for (example, label) in zip(examples, classes):
if label == 2:
x.append(example)
y.append(-1)
elif label == 7:
x.append(example)
y.append(1)
x = np.array(x)
y = np.array(y)
# Normalize pixels values
x = x / 255
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=TEST_SIZE, shuffle=True)
validation_indexes = np.random.random_integers(len(X_train), size=(math.floor(len(X_train)*VALIDATION_SIZE),))
X_validation = [X_train[n] for n in validation_indexes]
y_validation = [y_train[n] for n in validation_indexes]
pca = PCA(n_components=8)
pca.fit(X_train)
X_train = pca.transform(X_train)
X_validation = pca.transform(X_validation)
X_test = pca.transform(X_test)
preprocessing_time = time.time()
device = qml.device("default.qubit", wires=3)
@qml.qnode(device)
def circuit(features, params):
# Load state
if INITIALIZATION_METHOD == 'Amplitude':
AmplitudeEmbedding(features=features, wires=range(3), normalize=True, pad_with=0.)
else:
AngleEmbedding(features=features, wires=range(3), rotation='Y')
# First layer
qml.U3(params[0], params[1], params[2], wires=0)
qml.U3(params[3], params[4], params[5], wires=1)
qml.CNOT(wires=[0, 1])
# Second layer
qml.U3(params[6], params[7], params[8], wires=1)
qml.U3(params[9], params[10], params[11], wires=2)
qml.CNOT(wires=[1, 2])
# Third layer
qml.U3(params[12], params[13], params[14], wires=2)
# Measurement
return qml.expval(qml.PauliZ(2))
features = X_train[0]
print(f"Inital parameters: {initial_params}\n")
print(f"Example features: {features}\n")
print(f"Expectation value: {circuit(features, initial_params)}\n")
print(circuit.draw())
def measure_accuracy(x, y, circuit_params):
class_errors = 0
for example, example_class in zip(x, y):
predicted_value = circuit(example, circuit_params)
if (example_class > 0 and predicted_value <= 0) or (example_class <= 0 and predicted_value > 0):
class_errors += 1
return 1 - (class_errors/len(y))
params = initial_params
opt = qml.AdamOptimizer(stepsize=STEP_SIZE, beta1=BETA_1, beta2=BETA_2, eps=EPSILON)
test_accuracies = []
best_validation_accuracy = 0.0
best_params = []
for i in range(len(X_train)):
features = X_train[i]
expected_value = y_train[i]
def cost(circuit_params):
value = circuit(features, circuit_params)
return ((expected_value - value) ** 2)/len(X_train)
params = opt.step(cost, params)
if i % BATCH_SIZE == 0:
print(f"epoch {i//BATCH_SIZE}")
if i % (10*BATCH_SIZE) == 0:
current_accuracy = measure_accuracy(X_validation, y_validation, params)
test_accuracies.append(current_accuracy)
print(f"accuracy: {current_accuracy}")
if current_accuracy > best_validation_accuracy:
print("best accuracy so far!")
best_validation_accuracy = current_accuracy
best_params = params
if len(test_accuracies) == 30:
print(f"test_accuracies: {test_accuracies}")
if np.allclose(best_validation_accuracy, test_accuracies[0]):
params = best_params
break
del test_accuracies[0]
print("Optimized rotation angles: {}".format(params))
training_time = time.time()
accuracy = measure_accuracy(X_test, y_test, params)
print(accuracy)
test_time = time.time()
print(f"pre-processing time: {preprocessing_time-initial_time}")
print(f"training time: {training_time - preprocessing_time}")
print(f"test time: {test_time - training_time}")
print(f"total time: {test_time - initial_time}")
| 0.566378 | 0.882731 |
# Линейная регрессия и основные библиотеки Python для анализа данных и научных вычислений
Это задание посвящено линейной регрессии. На примере прогнозирования роста человека по его весу Вы увидите, какая математика за этим стоит, а заодно познакомитесь с основными библиотеками Python, необходимыми для дальнейшего прохождения курса.
**Материалы**
- Лекции данного курса по линейным моделям и градиентному спуску
- [Документация](http://docs.scipy.org/doc/) по библиотекам NumPy и SciPy
- [Документация](http://matplotlib.org/) по библиотеке Matplotlib
- [Документация](http://pandas.pydata.org/pandas-docs/stable/tutorials.html) по библиотеке Pandas
- [Pandas Cheat Sheet](http://www.analyticsvidhya.com/blog/2015/07/11-steps-perform-data-analysis-pandas-python/)
- [Документация](http://stanford.edu/~mwaskom/software/seaborn/) по библиотеке Seaborn
## Задание 1. Первичный анализ данных c Pandas
В этом заданиии мы будем использовать данные [SOCR](http://wiki.stat.ucla.edu/socr/index.php/SOCR_Data_Dinov_020108_HeightsWeights) по росту и весу 25 тысяч подростков.
**[1].** Если у Вас не установлена библиотека Seaborn - выполните в терминале команду *conda install seaborn*. (Seaborn не входит в сборку Anaconda, но эта библиотека предоставляет удобную высокоуровневую функциональность для визуализации данных).
```
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
```
Считаем данные по росту и весу (*weights_heights.csv*, приложенный в задании) в объект Pandas DataFrame:
```
data = pd.read_csv('weights_heights.csv', index_col='Index')
```
Чаще всего первое, что надо надо сделать после считывания данных - это посмотреть на первые несколько записей. Так можно отловить ошибки чтения данных (например, если вместо 10 столбцов получился один, в названии которого 9 точек с запятой). Также это позволяет познакомиться с данными, как минимум, посмотреть на признаки и их природу (количественный, категориальный и т.д.).
После этого стоит построить гистограммы распределения признаков - это опять-таки позволяет понять природу признака (степенное у него распределение, или нормальное, или какое-то еще). Также благодаря гистограмме можно найти какие-то значения, сильно не похожие на другие - "выбросы" в данных.
Гистограммы удобно строить методом *plot* Pandas DataFrame с аргументом *kind='hist'*.
**Пример.** Построим гистограмму распределения роста подростков из выборки *data*. Используем метод *plot* для DataFrame *data* c аргументами *y='Height'* (это тот признак, распределение которого мы строим)
```
data.plot(y='Height', kind='hist',
color='red', title='Height (inch.) distribution')
```
Аргументы:
- *y='Height'* - тот признак, распределение которого мы строим
- *kind='hist'* - означает, что строится гистограмма
- *color='red'* - цвет
**[2]**. Посмотрите на первые 5 записей с помощью метода *head* Pandas DataFrame. Нарисуйте гистограмму распределения веса с помощью метода *plot* Pandas DataFrame. Сделайте гистограмму зеленой, подпишите картинку.
```
# Ваш код здесь
print(data.head(5))
# Ваш код здесь
data.plot(y='Weight', kind='hist',
color='green', title='Weight (pd.) distribution')
```
Один из эффективных методов первичного анализа данных - отображение попарных зависимостей признаков. Создается $m \times m$ графиков (*m* - число признаков), где по диагонали рисуются гистограммы распределения признаков, а вне диагонали - scatter plots зависимости двух признаков. Это можно делать с помощью метода $scatter\_matrix$ Pandas Data Frame или *pairplot* библиотеки Seaborn.
Чтобы проиллюстрировать этот метод, интересней добавить третий признак. Создадим признак *Индекс массы тела* ([BMI](https://en.wikipedia.org/wiki/Body_mass_index)). Для этого воспользуемся удобной связкой метода *apply* Pandas DataFrame и lambda-функций Python.
```
def make_bmi(height_inch, weight_pound):
METER_TO_INCH, KILO_TO_POUND = 39.37, 2.20462
return (weight_pound / KILO_TO_POUND) / \
(height_inch / METER_TO_INCH) ** 2
data['BMI'] = data.apply(lambda row: make_bmi(row['Height'],
row['Weight']), axis=1)
```
**[3].** Постройте картинку, на которой будут отображены попарные зависимости признаков , 'Height', 'Weight' и 'BMI' друг от друга. Используйте метод *pairplot* библиотеки Seaborn.
```
# Ваш код здесь
sns.pairplot(data)
```
Часто при первичном анализе данных надо исследовать зависимость какого-то количественного признака от категориального (скажем, зарплаты от пола сотрудника). В этом помогут "ящики с усами" - boxplots библиотеки Seaborn. Box plot - это компактный способ показать статистики вещественного признака (среднее и квартили) по разным значениям категориального признака. Также помогает отслеживать "выбросы" - наблюдения, в которых значение данного вещественного признака сильно отличается от других.
**[4]**. Создайте в DataFrame *data* новый признак *weight_category*, который будет иметь 3 значения: 1 – если вес меньше 120 фунтов. (~ 54 кг.), 3 - если вес больше или равен 150 фунтов (~68 кг.), 2 – в остальных случаях. Постройте «ящик с усами» (boxplot), демонстрирующий зависимость роста от весовой категории. Используйте метод *boxplot* библиотеки Seaborn и метод *apply* Pandas DataFrame. Подпишите ось *y* меткой «Рост», ось *x* – меткой «Весовая категория».
```
def weight_category(weight):
pass
# Ваш код здесь
data['weight_cat'] = data['Weight'].apply(weight_category)
# Ваш код здесь
```
**[5].** Постройте scatter plot зависимости роста от веса, используя метод *plot* для Pandas DataFrame с аргументом *kind='scatter'*. Подпишите картинку.
```
# Ваш код здесь
```
## Задание 2. Минимизация квадратичной ошибки
В простейшей постановке задача прогноза значения вещественного признака по прочим признакам (задача восстановления регрессии) решается минимизацией квадратичной функции ошибки.
**[6].** Напишите функцию, которая по двум параметрам $w_0$ и $w_1$ вычисляет квадратичную ошибку приближения зависимости роста $y$ от веса $x$ прямой линией $y = w_0 + w_1 * x$:
$$error(w_0, w_1) = \sum_{i=1}^n {(y_i - (w_0 + w_1 * x_i))}^2 $$
Здесь $n$ – число наблюдений в наборе данных, $y_i$ и $x_i$ – рост и вес $i$-ого человека в наборе данных.
```
# Ваш код здесь
```
Итак, мы решаем задачу: как через облако точек, соответсвующих наблюдениям в нашем наборе данных, в пространстве признаков "Рост" и "Вес" провести прямую линию так, чтобы минимизировать функционал из п. 6. Для начала давайте отобразим хоть какие-то прямые и убедимся, что они плохо передают зависимость роста от веса.
**[7].** Проведите на графике из п. 5 Задания 1 две прямые, соответствующие значениям параметров ($w_0, w_1) = (60, 0.05)$ и ($w_0, w_1) = (50, 0.16)$. Используйте метод *plot* из *matplotlib.pyplot*, а также метод *linspace* библиотеки NumPy. Подпишите оси и график.
```
# Ваш код здесь
```
Минимизация квадратичной функции ошибки - относительная простая задача, поскольку функция выпуклая. Для такой задачи существует много методов оптимизации. Посмотрим, как функция ошибки зависит от одного параметра (наклон прямой), если второй параметр (свободный член) зафиксировать.
**[8].** Постройте график зависимости функции ошибки, посчитанной в п. 6, от параметра $w_1$ при $w_0$ = 50. Подпишите оси и график.
```
# Ваш код здесь
```
Теперь методом оптимизации найдем "оптимальный" наклон прямой, приближающей зависимость роста от веса, при фиксированном коэффициенте $w_0 = 50$.
**[9].** С помощью метода *minimize_scalar* из *scipy.optimize* найдите минимум функции, определенной в п. 6, для значений параметра $w_1$ в диапазоне [-5,5]. Проведите на графике из п. 5 Задания 1 прямую, соответствующую значениям параметров ($w_0$, $w_1$) = (50, $w_1\_opt$), где $w_1\_opt$ – найденное в п. 8 оптимальное значение параметра $w_1$.
```
# Ваш код здесь
# Ваш код здесь
```
При анализе многомерных данных человек часто хочет получить интуитивное представление о природе данных с помощью визуализации. Увы, при числе признаков больше 3 такие картинки нарисовать невозможно. На практике для визуализации данных в 2D и 3D в данных выделаяют 2 или, соответственно, 3 главные компоненты (как именно это делается - мы увидим далее в курсе) и отображают данные на плоскости или в объеме.
Посмотрим, как в Python рисовать 3D картинки, на примере отображения функции $z(x,y) = sin(\sqrt{x^2+y^2})$ для значений $x$ и $y$ из интервала [-5,5] c шагом 0.25.
```
from mpl_toolkits.mplot3d import Axes3D
```
Создаем объекты типа matplotlib.figure.Figure (рисунок) и matplotlib.axes._subplots.Axes3DSubplot (ось).
```
fig = plt.figure()
ax = fig.gca(projection='3d') # get current axis
# Создаем массивы NumPy с координатами точек по осям X и У.
# Используем метод meshgrid, при котором по векторам координат
# создается матрица координат. Задаем нужную функцию Z(x, y).
X = np.arange(-5, 5, 0.25)
Y = np.arange(-5, 5, 0.25)
X, Y = np.meshgrid(X, Y)
Z = np.sin(np.sqrt(X**2 + Y**2))
# Наконец, используем метод *plot_surface* объекта
# типа Axes3DSubplot. Также подписываем оси.
surf = ax.plot_surface(X, Y, Z)
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_zlabel('Z')
plt.show()
```
**[10].** Постройте 3D-график зависимости функции ошибки, посчитанной в п.6 от параметров $w_0$ и $w_1$. Подпишите ось $x$ меткой «Intercept», ось $y$ – меткой «Slope», a ось $z$ – меткой «Error».
```
# Ваш код здесь
# Ваш код здесь
# Ваш код здесь
```
**[11].** С помощью метода *minimize* из scipy.optimize найдите минимум функции, определенной в п. 6, для значений параметра $w_0$ в диапазоне [-100,100] и $w_1$ - в диапазоне [-5, 5]. Начальная точка – ($w_0$, $w_1$) = (0, 0). Используйте метод оптимизации L-BFGS-B (аргумент method метода minimize). Проведите на графике из п. 5 Задания 1 прямую, соответствующую найденным оптимальным значениям параметров $w_0$ и $w_1$. Подпишите оси и график.
```
# Ваш код здесь
# Ваш код здесь
```
## Критерии оценки работы
- Выполняется ли тетрадка IPython без ошибок? (15 баллов)
- Верно ли отображена гистограмма распределения роста из п. 2? (3 балла). Правильно ли оформлены подписи? (1 балл)
- Верно ли отображены попарные зависимости признаков из п. 3? (3 балла). Правильно ли оформлены подписи? (1 балл)
- Верно ли отображена зависимость роста от весовой категории из п. 4? (3 балла). Правильно ли оформлены подписи? (1 балл)
- Верно ли отображен scatter plot роста от веса из п. 5? (3 балла). Правильно ли оформлены подписи? (1 балл)
- Правильно ли реализована функция подсчета квадратичной ошибки из п. 6? (10 баллов)
- Правильно ли нарисован график из п. 7? (3 балла) Правильно ли оформлены подписи? (1 балл)
- Правильно ли нарисован график из п. 8? (3 балла) Правильно ли оформлены подписи? (1 балл)
- Правильно ли используется метод minimize\_scalar из scipy.optimize? (6 баллов). Правильно ли нарисован график из п. 9? (3 балла) Правильно ли оформлены подписи? (1 балл)
- Правильно ли нарисован 3D-график из п. 10? (6 баллов) Правильно ли оформлены подписи? (1 балл)
- Правильно ли используется метод minimize из scipy.optimize? (6 баллов). Правильно ли нарисован график из п. 11? (3 балла). Правильно ли оформлены подписи? (1 балл)
|
github_jupyter
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
data = pd.read_csv('weights_heights.csv', index_col='Index')
data.plot(y='Height', kind='hist',
color='red', title='Height (inch.) distribution')
# Ваш код здесь
print(data.head(5))
# Ваш код здесь
data.plot(y='Weight', kind='hist',
color='green', title='Weight (pd.) distribution')
def make_bmi(height_inch, weight_pound):
METER_TO_INCH, KILO_TO_POUND = 39.37, 2.20462
return (weight_pound / KILO_TO_POUND) / \
(height_inch / METER_TO_INCH) ** 2
data['BMI'] = data.apply(lambda row: make_bmi(row['Height'],
row['Weight']), axis=1)
# Ваш код здесь
sns.pairplot(data)
def weight_category(weight):
pass
# Ваш код здесь
data['weight_cat'] = data['Weight'].apply(weight_category)
# Ваш код здесь
# Ваш код здесь
# Ваш код здесь
# Ваш код здесь
# Ваш код здесь
# Ваш код здесь
# Ваш код здесь
from mpl_toolkits.mplot3d import Axes3D
fig = plt.figure()
ax = fig.gca(projection='3d') # get current axis
# Создаем массивы NumPy с координатами точек по осям X и У.
# Используем метод meshgrid, при котором по векторам координат
# создается матрица координат. Задаем нужную функцию Z(x, y).
X = np.arange(-5, 5, 0.25)
Y = np.arange(-5, 5, 0.25)
X, Y = np.meshgrid(X, Y)
Z = np.sin(np.sqrt(X**2 + Y**2))
# Наконец, используем метод *plot_surface* объекта
# типа Axes3DSubplot. Также подписываем оси.
surf = ax.plot_surface(X, Y, Z)
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_zlabel('Z')
plt.show()
# Ваш код здесь
# Ваш код здесь
# Ваш код здесь
# Ваш код здесь
# Ваш код здесь
| 0.228845 | 0.990413 |
# Cases for Nurse Availability Levels with different Utility Functions
Task:
Evaluate Shift Coverage, Agent Satisfaction and Agent Productivity over changes in nurse availability
```
import abm_scheduling
from abm_scheduling import Schedule as Schedule
from abm_scheduling import Nurse as Nurse
import time
from datetime import datetime
import abm_scheduling.Log
from abm_scheduling.Log import Log as Log
import matplotlib.pylab as plt
%matplotlib inline
log = Log()
```
## Shift coverage on number of nurses
```
beta = 0.8
p_to_accept_negative_change = .001
degree_of_agent_availability = 0.7
min_number_of_runs_with_shift_coverage_1 = 3
works_weekends = True
num_nurses_per_shift = 5
number_of_runs_with_shift_coverage_1 = 0
nurses = []
run_results_SC_over_NN_default_UF = []
run_results_SC_over_NN_AS_UF = []
schedule = Schedule(num_nurses_needed=num_nurses_per_shift, is_random=True)
model = abm_scheduling.NSP_AB_Model()
# run model with default utility function
num_nurses_D_UF = 0
while number_of_runs_with_shift_coverage_1 < min_number_of_runs_with_shift_coverage_1:
nurse = Nurse(id_name=num_nurses)
nurse.generate_shift_preferences(degree_of_agent_availability=0.7, works_weekends=True)
nurses.append(nurse)
num_nurses_D_UF += 1
results = model.run(schedule_org=schedule,
nurses_org=nurses,
p_to_accept_negative_change=p_to_accept_negative_change,
utility_function_parameters=None,
print_stats=False)
run_results_SC_over_NN_default_UF.append(results)
if results.shift_coverage >= 1:
number_of_runs_with_shift_coverage_1 += 1
# run model with agetn satisfaction utility function
utility_function_parameters = abm_scheduling.Utility_Function_Parameters()
utility_function_parameters.utility_function = 'agent_satisfaction'
num_nurses_AS_UF = 0
number_of_runs_with_shift_coverage_1 = 0
while number_of_runs_with_shift_coverage_1 < min_number_of_runs_with_shift_coverage_1:
nurse = Nurse(id_name=num_nurses)
nurse.generate_shift_preferences(degree_of_agent_availability=0.7, works_weekends=True)
nurses.append(nurse)
num_nurses_AS_UF += 1
results = model.run(schedule_org=schedule,
nurses_org=nurses,
p_to_accept_negative_change=p_to_accept_negative_change,
utility_function_parameters = utility_function_parameters,
print_stats=False)
run_results_SC_over_NN_AS_UF.append(results)
if results.shift_coverage >= 1:
number_of_runs_with_shift_coverage_1 += 1
plt.figure()
plt.plot(range(num_nurses_D_UF), [r.shift_coverage for r in run_results_SC_over_NN_default_UF], label="Default Util.function")
plt.plot(range(num_nurses), [r.shift_coverage for r in run_results_SC_over_NN_AS_UF], label="A.Satisf.Util.function")
plt.title(f'Shift coverage as a function of number of nurses', y=1.15, fontsize=14)
plt.suptitle(f'Av:0.7, WW = True', y=1.0)
plt.xlabel("Number of nurses")
plt.ylabel("Shift Coverage")
plt.legend()
plt.show()
print(run_results_1[19].shift_coverage)
print(run_results_1[20].shift_coverage)
print(run_results_1[21].shift_coverage)
plt.figure()
plt.plot(range(num_nurses), [r.total_agent_satisfaction for r in run_results_1], label="Agent Satisfaction")
plt.title(f'Agent satisfaction as a function of Number of nurses')
plt.xlabel("Number of nurses")
plt.ylabel("Agent Satisfaction")
plt.legend()
plt.show()
detail_productivity_over_nr_nurses = []
avg_productivity_over_nr_nurses = []
for r in run_results_1:
data = []
for nurse in r.nurses:
assigned_shifts = len(nurse.shifts)
data.append(assigned_shifts/ nurse.minimum_shifts)
detail_productivity_over_nr_nurses.append(data)
avg_productivity_over_nr_nurses.append(sum(data)/len(data))
plt.figure()
plt.plot(range(num_nurses), avg_productivity_over_nr_nurses, label="Average productivity")
plt.title(f'Average nurse productivity (assigned_shifts/minimum_shifts)')
plt.xlabel("Number of nurses")
plt.ylabel("Agent productivity")
plt.legend()
plt.show()
plt.figure()
w = plt.boxplot(detail_productivity_over_nr_nurses)
plt.title(f'Nurse productivity (assigned_shifts/minimum_shifts)')
plt.xlabel("Nurses availability")
plt.ylabel("Agent productivity")
plt.show()
print(avg_productivity_over_nr_nurses)
```
## Shift coverage over nurse availability
```
beta = 0.8
p_to_accept_negative_change = .001
min_number_of_runs_with_shift_coverage_1 = 3
works_weekends = True
num_nurses_per_shift = 5
num_nurses = 22
values_degree_of_agent_availability = [i * 0.1 for i in range(2, 10)]
nurses = []
run_results_2 = []
schedule = Schedule(num_nurses_needed=num_nurses_per_shift, is_random=True)
model = abm_scheduling.NSP_AB_Model()
utility_function_parameters = abm_scheduling.Utility_Function_Parameters()
utility_function_parameters.utility_function = 'agent_satisfaction'
for degree_of_agent_availability in values_degree_of_agent_availability:
nurses = model.generate_nurses(num_nurses=num_nurses,
degree_of_agent_availability=degree_of_agent_availability,
works_weekends=works_weekends)
results = model.run(schedule_org=schedule,
nurses_org=nurses,
beta=beta,
p_to_accept_negative_change=p_to_accept_negative_change,
utility_function_parameters=utility_function_parameters,
print_stats=False)
run_results_2.append(results)
plt.figure()
plt.plot(values_degree_of_agent_availability, [r.shift_coverage for r in run_results_2], label="Shift Coverage")
plt.title(f'Shift coverage as a function of nurses availability')
plt.xlabel("Nurses availability")
plt.ylabel("Shift Coverage")
plt.legend()
plt.show()
plt.figure()
plt.plot(values_degree_of_agent_availability, [r.total_agent_satisfaction for r in run_results_2], label="Agent Satisfaction")
plt.title(f'Agent satisfaction as a function of nurses availability')
plt.xlabel("Nurses availability")
plt.ylabel("Agent Satisfaction")
plt.legend()
plt.show()
detail_productivity_over_nr_nurses = []
avg_productivity_over_nr_nurses = []
for r in run_results_2:
data = []
for nurse in r.nurses:
assigned_shifts = len(nurse.shifts)
data.append(assigned_shifts/ nurse.minimum_shifts)
detail_productivity_over_nr_nurses.append(data)
avg_productivity_over_nr_nurses.append(sum(data)/len(data))
plt.figure()
plt.plot(values_degree_of_agent_availability, avg_productivity_over_nr_nurses, label="Average productivity")
plt.title(f'Average nurse productivity (assigned_shifts/minimum_shifts)')
plt.xlabel("Nurses availability")
plt.ylabel("Agent productivity")
plt.legend()
plt.show()
plt.figure()
w = plt.boxplot(detail_productivity_over_nr_nurses)
plt.title(f'Nurse productivity (assigned_shifts/minimum_shifts)')
plt.xlabel("Nurses availability")
plt.ylabel("Agent productivity")
ww = plt.xticks([i for i in range(1, 9)], [f'{i*0.1:1.2f}' for i in range(2, 10)])
print(avg_productivity_over_nr_nurses)
```
|
github_jupyter
|
import abm_scheduling
from abm_scheduling import Schedule as Schedule
from abm_scheduling import Nurse as Nurse
import time
from datetime import datetime
import abm_scheduling.Log
from abm_scheduling.Log import Log as Log
import matplotlib.pylab as plt
%matplotlib inline
log = Log()
beta = 0.8
p_to_accept_negative_change = .001
degree_of_agent_availability = 0.7
min_number_of_runs_with_shift_coverage_1 = 3
works_weekends = True
num_nurses_per_shift = 5
number_of_runs_with_shift_coverage_1 = 0
nurses = []
run_results_SC_over_NN_default_UF = []
run_results_SC_over_NN_AS_UF = []
schedule = Schedule(num_nurses_needed=num_nurses_per_shift, is_random=True)
model = abm_scheduling.NSP_AB_Model()
# run model with default utility function
num_nurses_D_UF = 0
while number_of_runs_with_shift_coverage_1 < min_number_of_runs_with_shift_coverage_1:
nurse = Nurse(id_name=num_nurses)
nurse.generate_shift_preferences(degree_of_agent_availability=0.7, works_weekends=True)
nurses.append(nurse)
num_nurses_D_UF += 1
results = model.run(schedule_org=schedule,
nurses_org=nurses,
p_to_accept_negative_change=p_to_accept_negative_change,
utility_function_parameters=None,
print_stats=False)
run_results_SC_over_NN_default_UF.append(results)
if results.shift_coverage >= 1:
number_of_runs_with_shift_coverage_1 += 1
# run model with agetn satisfaction utility function
utility_function_parameters = abm_scheduling.Utility_Function_Parameters()
utility_function_parameters.utility_function = 'agent_satisfaction'
num_nurses_AS_UF = 0
number_of_runs_with_shift_coverage_1 = 0
while number_of_runs_with_shift_coverage_1 < min_number_of_runs_with_shift_coverage_1:
nurse = Nurse(id_name=num_nurses)
nurse.generate_shift_preferences(degree_of_agent_availability=0.7, works_weekends=True)
nurses.append(nurse)
num_nurses_AS_UF += 1
results = model.run(schedule_org=schedule,
nurses_org=nurses,
p_to_accept_negative_change=p_to_accept_negative_change,
utility_function_parameters = utility_function_parameters,
print_stats=False)
run_results_SC_over_NN_AS_UF.append(results)
if results.shift_coverage >= 1:
number_of_runs_with_shift_coverage_1 += 1
plt.figure()
plt.plot(range(num_nurses_D_UF), [r.shift_coverage for r in run_results_SC_over_NN_default_UF], label="Default Util.function")
plt.plot(range(num_nurses), [r.shift_coverage for r in run_results_SC_over_NN_AS_UF], label="A.Satisf.Util.function")
plt.title(f'Shift coverage as a function of number of nurses', y=1.15, fontsize=14)
plt.suptitle(f'Av:0.7, WW = True', y=1.0)
plt.xlabel("Number of nurses")
plt.ylabel("Shift Coverage")
plt.legend()
plt.show()
print(run_results_1[19].shift_coverage)
print(run_results_1[20].shift_coverage)
print(run_results_1[21].shift_coverage)
plt.figure()
plt.plot(range(num_nurses), [r.total_agent_satisfaction for r in run_results_1], label="Agent Satisfaction")
plt.title(f'Agent satisfaction as a function of Number of nurses')
plt.xlabel("Number of nurses")
plt.ylabel("Agent Satisfaction")
plt.legend()
plt.show()
detail_productivity_over_nr_nurses = []
avg_productivity_over_nr_nurses = []
for r in run_results_1:
data = []
for nurse in r.nurses:
assigned_shifts = len(nurse.shifts)
data.append(assigned_shifts/ nurse.minimum_shifts)
detail_productivity_over_nr_nurses.append(data)
avg_productivity_over_nr_nurses.append(sum(data)/len(data))
plt.figure()
plt.plot(range(num_nurses), avg_productivity_over_nr_nurses, label="Average productivity")
plt.title(f'Average nurse productivity (assigned_shifts/minimum_shifts)')
plt.xlabel("Number of nurses")
plt.ylabel("Agent productivity")
plt.legend()
plt.show()
plt.figure()
w = plt.boxplot(detail_productivity_over_nr_nurses)
plt.title(f'Nurse productivity (assigned_shifts/minimum_shifts)')
plt.xlabel("Nurses availability")
plt.ylabel("Agent productivity")
plt.show()
print(avg_productivity_over_nr_nurses)
beta = 0.8
p_to_accept_negative_change = .001
min_number_of_runs_with_shift_coverage_1 = 3
works_weekends = True
num_nurses_per_shift = 5
num_nurses = 22
values_degree_of_agent_availability = [i * 0.1 for i in range(2, 10)]
nurses = []
run_results_2 = []
schedule = Schedule(num_nurses_needed=num_nurses_per_shift, is_random=True)
model = abm_scheduling.NSP_AB_Model()
utility_function_parameters = abm_scheduling.Utility_Function_Parameters()
utility_function_parameters.utility_function = 'agent_satisfaction'
for degree_of_agent_availability in values_degree_of_agent_availability:
nurses = model.generate_nurses(num_nurses=num_nurses,
degree_of_agent_availability=degree_of_agent_availability,
works_weekends=works_weekends)
results = model.run(schedule_org=schedule,
nurses_org=nurses,
beta=beta,
p_to_accept_negative_change=p_to_accept_negative_change,
utility_function_parameters=utility_function_parameters,
print_stats=False)
run_results_2.append(results)
plt.figure()
plt.plot(values_degree_of_agent_availability, [r.shift_coverage for r in run_results_2], label="Shift Coverage")
plt.title(f'Shift coverage as a function of nurses availability')
plt.xlabel("Nurses availability")
plt.ylabel("Shift Coverage")
plt.legend()
plt.show()
plt.figure()
plt.plot(values_degree_of_agent_availability, [r.total_agent_satisfaction for r in run_results_2], label="Agent Satisfaction")
plt.title(f'Agent satisfaction as a function of nurses availability')
plt.xlabel("Nurses availability")
plt.ylabel("Agent Satisfaction")
plt.legend()
plt.show()
detail_productivity_over_nr_nurses = []
avg_productivity_over_nr_nurses = []
for r in run_results_2:
data = []
for nurse in r.nurses:
assigned_shifts = len(nurse.shifts)
data.append(assigned_shifts/ nurse.minimum_shifts)
detail_productivity_over_nr_nurses.append(data)
avg_productivity_over_nr_nurses.append(sum(data)/len(data))
plt.figure()
plt.plot(values_degree_of_agent_availability, avg_productivity_over_nr_nurses, label="Average productivity")
plt.title(f'Average nurse productivity (assigned_shifts/minimum_shifts)')
plt.xlabel("Nurses availability")
plt.ylabel("Agent productivity")
plt.legend()
plt.show()
plt.figure()
w = plt.boxplot(detail_productivity_over_nr_nurses)
plt.title(f'Nurse productivity (assigned_shifts/minimum_shifts)')
plt.xlabel("Nurses availability")
plt.ylabel("Agent productivity")
ww = plt.xticks([i for i in range(1, 9)], [f'{i*0.1:1.2f}' for i in range(2, 10)])
print(avg_productivity_over_nr_nurses)
| 0.408631 | 0.808823 |
```
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/horse-or-human.zip \
-O /tmp/horse-or-human.zip
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/validation-horse-or-human.zip \
-O /tmp/validation-horse-or-human.zip
import os
import zipfile
local_zip = '/tmp/horse-or-human.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp/horse-or-human')
local_zip = '/tmp/validation-horse-or-human.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp/validation-horse-or-human')
zip_ref.close()
# Directory with our training horse pictures
train_horse_dir = os.path.join('/tmp/horse-or-human/horses')
# Directory with our training human pictures
train_human_dir = os.path.join('/tmp/horse-or-human/humans')
# Directory with our training horse pictures
validation_horse_dir = os.path.join('/tmp/validation-horse-or-human/horses')
# Directory with our training human pictures
validation_human_dir = os.path.join('/tmp/validation-horse-or-human/humans')
train_horse_names = os.listdir(train_horse_dir)
train_human_names = os.listdir(train_human_dir)
validation_horse_hames = os.listdir(validation_horse_dir)
validation_human_names = os.listdir(validation_human_dir)
import tensorflow as tf
model = tf.keras.models.Sequential([
# Note the input shape is the desired size of the image 150x150 with 3 bytes color
# This is the first convolution
tf.keras.layers.Conv2D(16, (3,3), activation='relu', input_shape=(150, 150, 3)),
tf.keras.layers.MaxPooling2D(2, 2),
# The second convolution
tf.keras.layers.Conv2D(32, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# The third convolution
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# The fourth convolution
#tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
#tf.keras.layers.MaxPooling2D(2,2),
# The fifth convolution
#tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
#tf.keras.layers.MaxPooling2D(2,2),
# Flatten the results to feed into a DNN
tf.keras.layers.Flatten(),
# 512 neuron hidden layer
tf.keras.layers.Dense(512, activation='relu'),
# Only 1 output neuron. It will contain a value from 0-1 where 0 for 1 class ('horses') and 1 for the other ('humans')
tf.keras.layers.Dense(1, activation='sigmoid')
])
model.summary()
from tensorflow.keras.optimizers import RMSprop
model.compile(loss='binary_crossentropy',
optimizer=RMSprop(lr=0.001),
metrics=['accuracy'])
from tensorflow.keras.preprocessing.image import ImageDataGenerator
# All images will be rescaled by 1./255
train_datagen = ImageDataGenerator(rescale=1/255)
validation_datagen = ImageDataGenerator(rescale=1/255)
# Flow training images in batches of 128 using train_datagen generator
train_generator = train_datagen.flow_from_directory(
'/tmp/horse-or-human/', # This is the source directory for training images
target_size=(150, 150), # All images will be resized to 150x150
batch_size=128,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
# Flow training images in batches of 128 using train_datagen generator
validation_generator = validation_datagen.flow_from_directory(
'/tmp/validation-horse-or-human/', # This is the source directory for training images
target_size=(150, 150), # All images will be resized to 150x150
batch_size=32,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
history = model.fit(
train_generator,
steps_per_epoch=8,
epochs=15,
verbose=1,
validation_data = validation_generator,
validation_steps=8)
# Try yourself
#
import numpy as np
from tensorflow.keras.preprocessing import image
path='./HorseOrHuman/Horse1.jpg'
def HorseOrHuman(path):
fn=path.split('/')[-1]
img = image.load_img(path, target_size=(150, 150))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
images = np.vstack([x])
classes = model.predict(images, batch_size=10)
print(classes[0])
if classes[0]>0.5:
print(fn + " is a human")
else:
print(fn + " is a horse")
HorseOrHuman(path)
```
|
github_jupyter
|
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/horse-or-human.zip \
-O /tmp/horse-or-human.zip
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/validation-horse-or-human.zip \
-O /tmp/validation-horse-or-human.zip
import os
import zipfile
local_zip = '/tmp/horse-or-human.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp/horse-or-human')
local_zip = '/tmp/validation-horse-or-human.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp/validation-horse-or-human')
zip_ref.close()
# Directory with our training horse pictures
train_horse_dir = os.path.join('/tmp/horse-or-human/horses')
# Directory with our training human pictures
train_human_dir = os.path.join('/tmp/horse-or-human/humans')
# Directory with our training horse pictures
validation_horse_dir = os.path.join('/tmp/validation-horse-or-human/horses')
# Directory with our training human pictures
validation_human_dir = os.path.join('/tmp/validation-horse-or-human/humans')
train_horse_names = os.listdir(train_horse_dir)
train_human_names = os.listdir(train_human_dir)
validation_horse_hames = os.listdir(validation_horse_dir)
validation_human_names = os.listdir(validation_human_dir)
import tensorflow as tf
model = tf.keras.models.Sequential([
# Note the input shape is the desired size of the image 150x150 with 3 bytes color
# This is the first convolution
tf.keras.layers.Conv2D(16, (3,3), activation='relu', input_shape=(150, 150, 3)),
tf.keras.layers.MaxPooling2D(2, 2),
# The second convolution
tf.keras.layers.Conv2D(32, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# The third convolution
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# The fourth convolution
#tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
#tf.keras.layers.MaxPooling2D(2,2),
# The fifth convolution
#tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
#tf.keras.layers.MaxPooling2D(2,2),
# Flatten the results to feed into a DNN
tf.keras.layers.Flatten(),
# 512 neuron hidden layer
tf.keras.layers.Dense(512, activation='relu'),
# Only 1 output neuron. It will contain a value from 0-1 where 0 for 1 class ('horses') and 1 for the other ('humans')
tf.keras.layers.Dense(1, activation='sigmoid')
])
model.summary()
from tensorflow.keras.optimizers import RMSprop
model.compile(loss='binary_crossentropy',
optimizer=RMSprop(lr=0.001),
metrics=['accuracy'])
from tensorflow.keras.preprocessing.image import ImageDataGenerator
# All images will be rescaled by 1./255
train_datagen = ImageDataGenerator(rescale=1/255)
validation_datagen = ImageDataGenerator(rescale=1/255)
# Flow training images in batches of 128 using train_datagen generator
train_generator = train_datagen.flow_from_directory(
'/tmp/horse-or-human/', # This is the source directory for training images
target_size=(150, 150), # All images will be resized to 150x150
batch_size=128,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
# Flow training images in batches of 128 using train_datagen generator
validation_generator = validation_datagen.flow_from_directory(
'/tmp/validation-horse-or-human/', # This is the source directory for training images
target_size=(150, 150), # All images will be resized to 150x150
batch_size=32,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
history = model.fit(
train_generator,
steps_per_epoch=8,
epochs=15,
verbose=1,
validation_data = validation_generator,
validation_steps=8)
# Try yourself
#
import numpy as np
from tensorflow.keras.preprocessing import image
path='./HorseOrHuman/Horse1.jpg'
def HorseOrHuman(path):
fn=path.split('/')[-1]
img = image.load_img(path, target_size=(150, 150))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
images = np.vstack([x])
classes = model.predict(images, batch_size=10)
print(classes[0])
if classes[0]>0.5:
print(fn + " is a human")
else:
print(fn + " is a horse")
HorseOrHuman(path)
| 0.599368 | 0.713357 |
# CLEAN TEXT DATA AND GENERATE WORD2VEC
## 1. CLEAN TEXT DATA
### 1.1 Import libraries
**Import neccessary packages and modules**
```
import time
import os
t = time.time()
import json
import string
import random
import math
import random
import pandas as pd
import numpy as np
import tensorflow as tf
import matplotlib
from matplotlib import pyplot as plt
%matplotlib inline
```
**Import nlp packages and modules**
```
import nltk
# nltk.download()
import nltk, re, time
from nltk.corpus import stopwords
from nltk.stem.snowball import SnowballStemmer
from gensim.models import Word2Vec
```
**Count the number of workers**
```
import multiprocessing
WORKERS = multiprocessing.cpu_count()
print("Number of workers:",WORKERS)
```
### 1.2 Load and Inspect Data
**Set directory**
```
input_dir = "../input/"
word2vec_model_dir = "./word2vec_models/"
```
**Load train and test data**
```
train_data = pd.read_csv(input_dir+'train.csv')
test_data = pd.read_csv(input_dir+'test.csv')
train_data.head(20)
print("Shape of train data:", train_data.shape)
test_data.head(20)
print("Shape of test data:", test_data.shape)
```
### 1.3 3 Preprocess the text data
```
# load list of stopwords
sw = set(stopwords.words("english"))
# load teh snowball stemmer
stemmer = SnowballStemmer("english")
# translator object to replace punctuations with space
translator = str.maketrans(string.punctuation, ' '*len(string.punctuation))
print(sw)
```
**Function for preprocessing text**
```
def clean_text(text):
"""
A function for preprocessing text
"""
text = str(text)
# replacing the punctuations with no space,which in effect deletes the punctuation marks
text = text.translate(translator)
# remove stop word
text = [word.lower() for word in text.split() if word.lower() not in sw]
text = " ".join(text)
# stemming
text = [stemmer.stem(word) for word in text.split()]
text = " ".join(text)
# Clean the text
text = re.sub(r"<br />", " ", text)
text = re.sub(r"[^a-z]", " ", text)
text = re.sub(r" ", " ", text) # Remove any extra spaces
text = re.sub(r" ", " ", text)
return(text)
```
**Clean train and test data**
```
t1 = time.time()
train_data['text'] = train_data['text'].apply(clean_text)
print("Finished cleaning the train set.", "Time needed:", time.time()-t1,"sec")
t2 = time.time()
test_data['text'] = test_data['text'].apply(clean_text)
print("Finished cleaning the test set.", "Time needed:", time.time()-t2,"sec")
```
**Inspect the cleaned train and test data**
```
train_data.head(10)
test_data.head(10)
```
### 1.4 Create columns for length of the data
**A function for finding the length of text**
```
def find_length(text):
"""
A function to find the length
"""
text = str(text)
return len(text.split())
```
**Create the column of text length in train and test data**
```
train_data['length'] = train_data['text'].apply(find_length)
train_data.head(10)
test_data['length'] = test_data['text'].apply(find_length)
test_data.head(10)
```
### 1.5 One hot encode author's column of train and test data
```
train_data = pd.concat([train_data, pd.get_dummies(train_data['author'])], axis=1)
train_data.drop("author", axis = 1)
train_data.head(10)
```
### 1.6 Save the processed train and test data
```
train_data.to_csv(input_dir+"modified_train_data.csv",header=False, index=False)
test_data.to_csv(input_dir+"modified_test_data.csv",header=False, index=False)
```
## 2. Create word2Vec models
### 2.1 Create Word2Vec Model
**Aggregrate all the comments from train and test data in a list**
```
train_comment = train_data["text"].values
test_comment = test_data["text"].values
all_comments = np.concatenate((train_comment, test_comment), axis = 0)
print("Shape of all comments:",all_comments.shape)
del(train_comment, test_comment)
all_comments = all_comments.tolist()
```
**Break each comment into list of words**
```
sentences = []
for comment in all_comments:
comment = str(comment)
sentences.append(comment.split())
del(all_comments)
```
**Create Word2Vec model using gensim**
```
t = time.time()
size = 300
window = 15
model = Word2Vec(sentences, size=size, window=window, sg= 1, workers=WORKERS, min_count=1)
model.train(sentences, total_examples=len(sentences), epochs=5)
print("Finished creating word2vec model.", "Time needed:", time.time()-t,"sec")
```
### 2.2 Save the model to the kernel
**Save the Word2Vec model in the designated directory**
```
fname = "word2vec_model.mdl"
model.wv.save_word2vec_format(word2vec_model_dir+fname)
```
**Save the Word2Vec model info in a json file in the designated directory**
```
data = {"size": size, "window":window}
with open(word2vec_model_dir+'data.json', 'w') as outfile:
json.dump(data, outfile)
```
|
github_jupyter
|
import time
import os
t = time.time()
import json
import string
import random
import math
import random
import pandas as pd
import numpy as np
import tensorflow as tf
import matplotlib
from matplotlib import pyplot as plt
%matplotlib inline
import nltk
# nltk.download()
import nltk, re, time
from nltk.corpus import stopwords
from nltk.stem.snowball import SnowballStemmer
from gensim.models import Word2Vec
import multiprocessing
WORKERS = multiprocessing.cpu_count()
print("Number of workers:",WORKERS)
input_dir = "../input/"
word2vec_model_dir = "./word2vec_models/"
train_data = pd.read_csv(input_dir+'train.csv')
test_data = pd.read_csv(input_dir+'test.csv')
train_data.head(20)
print("Shape of train data:", train_data.shape)
test_data.head(20)
print("Shape of test data:", test_data.shape)
# load list of stopwords
sw = set(stopwords.words("english"))
# load teh snowball stemmer
stemmer = SnowballStemmer("english")
# translator object to replace punctuations with space
translator = str.maketrans(string.punctuation, ' '*len(string.punctuation))
print(sw)
def clean_text(text):
"""
A function for preprocessing text
"""
text = str(text)
# replacing the punctuations with no space,which in effect deletes the punctuation marks
text = text.translate(translator)
# remove stop word
text = [word.lower() for word in text.split() if word.lower() not in sw]
text = " ".join(text)
# stemming
text = [stemmer.stem(word) for word in text.split()]
text = " ".join(text)
# Clean the text
text = re.sub(r"<br />", " ", text)
text = re.sub(r"[^a-z]", " ", text)
text = re.sub(r" ", " ", text) # Remove any extra spaces
text = re.sub(r" ", " ", text)
return(text)
t1 = time.time()
train_data['text'] = train_data['text'].apply(clean_text)
print("Finished cleaning the train set.", "Time needed:", time.time()-t1,"sec")
t2 = time.time()
test_data['text'] = test_data['text'].apply(clean_text)
print("Finished cleaning the test set.", "Time needed:", time.time()-t2,"sec")
train_data.head(10)
test_data.head(10)
def find_length(text):
"""
A function to find the length
"""
text = str(text)
return len(text.split())
train_data['length'] = train_data['text'].apply(find_length)
train_data.head(10)
test_data['length'] = test_data['text'].apply(find_length)
test_data.head(10)
train_data = pd.concat([train_data, pd.get_dummies(train_data['author'])], axis=1)
train_data.drop("author", axis = 1)
train_data.head(10)
train_data.to_csv(input_dir+"modified_train_data.csv",header=False, index=False)
test_data.to_csv(input_dir+"modified_test_data.csv",header=False, index=False)
train_comment = train_data["text"].values
test_comment = test_data["text"].values
all_comments = np.concatenate((train_comment, test_comment), axis = 0)
print("Shape of all comments:",all_comments.shape)
del(train_comment, test_comment)
all_comments = all_comments.tolist()
sentences = []
for comment in all_comments:
comment = str(comment)
sentences.append(comment.split())
del(all_comments)
t = time.time()
size = 300
window = 15
model = Word2Vec(sentences, size=size, window=window, sg= 1, workers=WORKERS, min_count=1)
model.train(sentences, total_examples=len(sentences), epochs=5)
print("Finished creating word2vec model.", "Time needed:", time.time()-t,"sec")
fname = "word2vec_model.mdl"
model.wv.save_word2vec_format(word2vec_model_dir+fname)
data = {"size": size, "window":window}
with open(word2vec_model_dir+'data.json', 'w') as outfile:
json.dump(data, outfile)
| 0.234407 | 0.714292 |
# HW1
```
from matplotlib import pyplot as plt
import Blur
import UnBlur
import ImageWrapper
import ImgUtils
import PSNR
import Trajectory
ORIGINAL_IMAGE_PATH = "./DIPSourceHW1.jpg"
TRAJACTORY_PATH = "./100_motion_paths.mat"
TRAJECTORIES_FOLDER = "./TRAJECTORIES"
PSF_FOLDER = "./PSF"
BLURRED_FOLDER = "./Blurred"
image = ImageWrapper.ImageWrapper(ORIGINAL_IMAGE_PATH )
display(plt.imshow(image.get_image(), cmap='gray'))
trajactoryController = Trajectory.Trajectories(TRAJACTORY_PATH)
trajactoryController.plot_trajectory(1)
trajactoryController.save_trajectories(TRAJECTORIES_FOLDER)
```
## Plot Trajectories, PSFs and Blurred images
```
blurController = Blur.Blur(trajactoryController,image)
blurController.save_psfs(PSF_FOLDER)
blurController.plot_blured_batch(5)
blurController.save_blurred_images(BLURRED_FOLDER)
unblurController = UnBlur.Blurr_Fixer(blurController.get_blurred_images(),power=10,ifft_scale=995,
original_size=256, margin=6)
fixed = unblurController.unblur_images()
unblurController.show_unblur_image(image)
print_every = 5
num_samples = [i for i in range(100)]
PSNR_results = []
fixed_images = []
for sample in num_samples:
if sample % print_every == 1:
print("Deblurring for ",sample," samples...")
fixed = unblurController.unblur_images()
fixed_images.append(fixed)
my_calc = PSNR.PSNR_calculator(image.get_image(), fixed)
PSNR_results.append(my_calc.evaluate_PSNR())
plt.plot(num_samples, PSNR_results)
plt.xlabel("Number of blurred samples")
plt.ylabel("PSNR [dB]")
plt.savefig("./psnr_graph.png")
plt.show()
```
## save to file PSNR + images
```
for i in range(len(fixed_images)):
cropped = fixed_images[i]
plt.imshow(cropped, cmap='gray')
plt.title("n={0}, PSNR={1}".format(i+1,PSNR_results[i]), fontsize=20)
plt.tick_params(labelbottom=False)
plt.tick_params(labelleft=False)
plt.savefig( "./psnr_images/" + str(i) + '.png')
for i in range(len(fixed_images)):
plt.subplot(10, 10, i+1)
cropped = fixed_images[i][25:100, 100:175]
plt.imshow(cropped, cmap='gray')
k = i+1
plt.ylabel("n=%i" % k, fontsize=5)
plt.tick_params(labelbottom=False)
plt.tick_params(labelleft=False)
plt.show()
```
### Show first and last iteration for comparison.
```
plt.subplot(1, 3, 1)
cropped = blurred_images[0]
plt.imshow(cropped, cmap='gray')
plt.title("First Blurred image")
plt.subplot(1, 3, 2)
cropped = fixed_images[0]
plt.imshow(cropped, cmap='gray')
plt.title("First iteration")
plt.subplot(1, 3, 3)
cropped = fixed
plt.imshow(cropped, cmap='gray')
plt.title("100th iteration")
plt.show()
```
|
github_jupyter
|
from matplotlib import pyplot as plt
import Blur
import UnBlur
import ImageWrapper
import ImgUtils
import PSNR
import Trajectory
ORIGINAL_IMAGE_PATH = "./DIPSourceHW1.jpg"
TRAJACTORY_PATH = "./100_motion_paths.mat"
TRAJECTORIES_FOLDER = "./TRAJECTORIES"
PSF_FOLDER = "./PSF"
BLURRED_FOLDER = "./Blurred"
image = ImageWrapper.ImageWrapper(ORIGINAL_IMAGE_PATH )
display(plt.imshow(image.get_image(), cmap='gray'))
trajactoryController = Trajectory.Trajectories(TRAJACTORY_PATH)
trajactoryController.plot_trajectory(1)
trajactoryController.save_trajectories(TRAJECTORIES_FOLDER)
blurController = Blur.Blur(trajactoryController,image)
blurController.save_psfs(PSF_FOLDER)
blurController.plot_blured_batch(5)
blurController.save_blurred_images(BLURRED_FOLDER)
unblurController = UnBlur.Blurr_Fixer(blurController.get_blurred_images(),power=10,ifft_scale=995,
original_size=256, margin=6)
fixed = unblurController.unblur_images()
unblurController.show_unblur_image(image)
print_every = 5
num_samples = [i for i in range(100)]
PSNR_results = []
fixed_images = []
for sample in num_samples:
if sample % print_every == 1:
print("Deblurring for ",sample," samples...")
fixed = unblurController.unblur_images()
fixed_images.append(fixed)
my_calc = PSNR.PSNR_calculator(image.get_image(), fixed)
PSNR_results.append(my_calc.evaluate_PSNR())
plt.plot(num_samples, PSNR_results)
plt.xlabel("Number of blurred samples")
plt.ylabel("PSNR [dB]")
plt.savefig("./psnr_graph.png")
plt.show()
for i in range(len(fixed_images)):
cropped = fixed_images[i]
plt.imshow(cropped, cmap='gray')
plt.title("n={0}, PSNR={1}".format(i+1,PSNR_results[i]), fontsize=20)
plt.tick_params(labelbottom=False)
plt.tick_params(labelleft=False)
plt.savefig( "./psnr_images/" + str(i) + '.png')
for i in range(len(fixed_images)):
plt.subplot(10, 10, i+1)
cropped = fixed_images[i][25:100, 100:175]
plt.imshow(cropped, cmap='gray')
k = i+1
plt.ylabel("n=%i" % k, fontsize=5)
plt.tick_params(labelbottom=False)
plt.tick_params(labelleft=False)
plt.show()
plt.subplot(1, 3, 1)
cropped = blurred_images[0]
plt.imshow(cropped, cmap='gray')
plt.title("First Blurred image")
plt.subplot(1, 3, 2)
cropped = fixed_images[0]
plt.imshow(cropped, cmap='gray')
plt.title("First iteration")
plt.subplot(1, 3, 3)
cropped = fixed
plt.imshow(cropped, cmap='gray')
plt.title("100th iteration")
plt.show()
| 0.321993 | 0.583886 |
# Modeling 07
Idea:
- build features up to lag 52 (one year)
- select 50, 100 most informative features (as per mutual information with target) to build a linear model
- more sophisticated feature selection schemes may be used if the mutual information approach does not yield the expected improvements
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib as mpl
import sklearn
from os.path import join
from sklearn.feature_selection import mutual_info_regression
from sklearn.linear_model import Lasso, Ridge
from sklearn.preprocessing import StandardScaler, PolynomialFeatures
from sklearn.pipeline import Pipeline
from sklearn.model_selection import cross_validate, TimeSeriesSplit, GridSearchCV
from xgboost import XGBRegressor
cd ..
from src.features.build_lagged_features import add_lagged_features
mpl.rcParams.update({
'figure.autolayout': True,
'figure.dpi': 150
})
sns.set()
RAW_PATH = 'data/raw'
```
## Reading the data
```
train_features_o = pd.read_csv(join(RAW_PATH, 'dengue_features_train.csv'))
test_features_o = pd.read_csv(join(RAW_PATH, 'dengue_features_test.csv'))
train_labels = pd.read_csv(join(RAW_PATH, 'dengue_labels_train.csv'))
```
## Feature Engineering
Concatenate training and testing data to create lagged features as test data is in the immediate
future of the training data for each of the cities
```
train_features_sj = train_features_o[train_features_o['city'] == 'sj'].drop('city', axis = 1)
train_features_iq = train_features_o[train_features_o['city'] == 'iq'].drop('city', axis = 1)
test_features_sj = test_features_o[test_features_o['city'] == 'sj'].drop('city', axis = 1)
test_features_iq = test_features_o[test_features_o['city'] == 'iq'].drop('city', axis = 1)
features_sj = pd.concat([train_features_sj, test_features_sj])
features_iq = pd.concat([train_features_iq, test_features_iq])
len(test_features_sj)
len(test_features_iq)
```
Function used in previous notebook (`apoirel_exploration_02`) to build lagged features
```
def make_dataset(features):
features = (features
.drop( # correlated features
['reanalysis_sat_precip_amt_mm', 'reanalysis_dew_point_temp_k',
'reanalysis_air_temp_k', 'reanalysis_tdtr_k'],
axis = 1
)
.fillna(method = 'backfill')
.drop( # unused features
['year', 'weekofyear','week_start_date'],
axis = 1
)
)
ts_features = list(features.loc[:, 'ndvi_ne' :].columns.values)
features = add_lagged_features(
features, 52, ts_features)
return features
features_sj = make_dataset(features_sj)
features_iq = make_dataset(features_iq)
```
Seperate the testing features from training
```
test_features_sj = features_sj.iloc[-260:, :]
test_features_iq = features_iq.iloc[-156:, :]
```
Remove the first year of values (number of NAs is too high due to unavailability of
lagged features)
```
train_features_sj = features_sj.iloc[52:-260,:]
train_features_iq = features_iq.iloc[52:-156,:]
train_labels_iq = train_labels[
train_labels['city'] == 'iq']['total_cases'].astype('float')[52:]
train_labels_sj = train_labels[
train_labels['city'] == 'sj']['total_cases'].astype('float')[52:]
```
## Features selection
From research papers I've read, local particularities have a large bearing on the impact of
climactic factors on dengue outbreaks, so it makes sense to select features seperately
for each city
### San Juan
```
sj_corrs = train_features_sj.corrwith(train_labels_sj).abs().sort_values()
train_features_sj = train_features_sj[list(sj_corrs.iloc[-100:].index)]
test_features_sj = test_features_sj[list(sj_corrs.iloc[-100:].index)]
```
### Iquitos
```
iq_corrs = train_features_iq.corrwith(train_labels_iq).abs().sort_values()
train_features_iq = train_features_iq[list(iq_corrs.iloc[-100:].index)]
test_features_iq = test_features_iq[list(iq_corrs.iloc[-100:].index)]
```
## Modeling
### San Juan
#### Linear
```
model_sj = Pipeline([
('scale', StandardScaler()),
('lasso', Ridge(10000))
])
cv_res_sj = cross_validate(
estimator = model_sj,
X = train_features_sj,
y = train_labels_sj,
cv = TimeSeriesSplit(n_splits = 10),
scoring = 'neg_mean_absolute_error',
n_jobs = -1
)
sj_score = np.mean(cv_res_sj['test_score'])
sj_score
```
##### Visualization
```
model_sj.fit(train_features_sj, train_labels_sj)
y_val_sj = model_sj.predict(train_features_sj).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_sj, x = train_features_sj.index.values, ax = ax)
sns.lineplot(y = y_val_sj, x = train_features_sj.index.values, ax = ax)
ax.set(title = 'San Juan predictions on training data')
ax.legend(['True', 'Predicted'])
```
### Gradient boosting
```
model_sj = XGBRegressor(
max_depth = 10,
learning_rate = 0.01,
n_estimators = 200,
reg_lambda = 1,
)
cv_res_sj = cross_validate(
estimator = model_sj,
X = train_features_sj,
y = train_labels_sj,
cv = TimeSeriesSplit(n_splits = 10),
scoring = 'neg_mean_absolute_error',
n_jobs = -1
)
sj_score = np.mean(cv_res_sj['test_score'])
sj_score
np.std(cv_res_sj['test_score'])
```
##### Visualization
```
model_sj.fit(train_features_sj, train_labels_sj)
y_val_sj = model_sj.predict(train_features_sj).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_sj, x = train_features_sj.index.values, ax = ax)
sns.lineplot(y = y_val_sj, x = train_features_sj.index.values, ax = ax)
ax.set(title = 'San Juan predictions on training data')
ax.legend(['True', 'Predicted'])
```
This looks seriously overfitting, yet the CV score is pretty good. What's up with that?
#### Tuned gradient boosting
```
params = {
'learning_rate' : [0.001 , 0.01, 0.1],
'reg_lambda' : [0.1, 1, 10],
'max_depth': [2, 5, 8, 10],
'n_estimators': [50, 100 , 200]
}
tuned_model_sj = GridSearchCV(
estimator = XGBRegressor(),
param_grid = params,
scoring = 'neg_mean_absolute_error',
cv = TimeSeriesSplit(10),
n_jobs = -1
)
tuned_model_sj.fit(train_features_sj, train_labels_sj)
tuned_model_sj.best_score_
tuned_model_sj.best_params_
cv_res_sj = cross_validate(
estimator = tuned_model_sj.best_estimator_,
X = train_features_sj,
y = train_labels_sj,
cv = TimeSeriesSplit(n_splits = 10),
scoring = 'neg_mean_absolute_error',
n_jobs = -1
)
sj_score = np.mean(cv_res_sj['test_score'])
sj_score
np.std(cv_res_sj['test_score'])
```
##### Visualization
```
y_val_sj = tuned_model_sj.predict(train_features_sj).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_sj, x = train_features_sj.index.values, ax = ax)
sns.lineplot(y = y_val_sj, x = train_features_sj.index.values, ax = ax)
ax.set(title = 'San Juan predictions on training data')
ax.legend(['True', 'Predicted'])
```
### Iquitos
#### Linear model
```
model_iq = Pipeline([
('scale', StandardScaler()),
('lasso', Lasso(2))
])
cv_res_iq = cross_validate(
estimator = model_iq,
X = train_features_iq,
y = train_labels_iq,
cv = TimeSeriesSplit(n_splits = 10),
scoring = 'neg_mean_absolute_error',
n_jobs = -1
)
iq_score = np.mean(cv_res_iq['test_score'])
iq_score
```
##### Visualization
```
model_iq.fit(train_features_iq, train_labels_iq)
y_val_iq = model_iq.predict(train_features_iq).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_iq, x = train_features_iq.index.values, ax = ax)
sns.lineplot(y = y_val_iq, x = train_features_iq.index.values, ax = ax)
ax.set(title = 'Iquitos predictions on training data')
ax.legend(['True', 'Predicted'])
```
### Gradient boosting
```
model_iq = XGBRegressor(
max_depth = 10,
learning_rate = 0.02,
n_estimators = 200,
reg_lambda = 20,
)
cv_res_iq = cross_validate(
estimator = model_iq,
X = train_features_iq,
y = train_labels_iq,
cv = TimeSeriesSplit(n_splits = 10),
scoring = 'neg_mean_absolute_error',
n_jobs = -1
)
iq_score = np.mean(cv_res_iq['test_score'])
iq_score
```
##### Visualization
```
model_iq.fit(train_features_iq, train_labels_iq)
y_val_iq = model_iq.predict(train_features_iq).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_iq, x = train_features_iq.index.values, ax = ax)
sns.lineplot(y = y_val_iq, x = train_features_iq.index.values, ax = ax)
ax.set(title = 'Iquitos predictions on training data')
ax.legend(['True', 'Predicted'])
```
Same here, this seems to overfit a lot, yet the CV score looks good
#### Tuned gradient boosting
```
params = {
'learning_rate' : [0.001 , 0.01, 0.1],
'reg_lambda' : [0.1, 1, 10],
'max_depth': [2, 5, 8, 10],
'n_estimators': [50, 100 , 200]
}
tuned_model_iq = GridSearchCV(
estimator = XGBRegressor(),
param_grid = params,
scoring = 'neg_mean_absolute_error',
cv = TimeSeriesSplit(10),
n_jobs = -1
)
tuned_model_iq.fit(train_features_iq, train_labels_iq)
tuned_model_iq.best_score_
tuned_model_iq.best_params_
```
##### Visualization
```
y_val_iq = tuned_model_iq.predict(train_features_iq).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_iq, x = train_features_iq.index.values, ax = ax)
sns.lineplot(y = y_val_iq, x = train_features_iq.index.values, ax = ax)
ax.set(title = 'Iquitos predictions on training data')
ax.legend(['True', 'Predicted'])
```
## Overall performance
```
sj_ratio = len(train_features_sj) / (len(train_features_sj) + len(train_features_iq))
iq_ratio = len(train_features_iq) / (len(train_features_sj) + len(train_features_iq))
```
Gradient boosting
```
sj_ratio * sj_score + iq_ratio * iq_score
```
Tuned gradient boosting
```
sj_ratio * tuned_model_sj.best_score_ + iq_ratio * tuned_model_iq.best_score_
```
Much better than our previous best, this warrants a submission!
## Model predictions
### XGB First pass
```
lagyear_sub = pd.read_csv(join(RAW_PATH, 'submission_format.csv'))
y_pred_sj = model_sj.predict(test_features_sj)
y_pred_iq = model_iq.predict(test_features_iq)
y_pred = np.concatenate((y_pred_sj, y_pred_iq))
lagyear_sub['total_cases'] = np.round(y_pred).astype(int)
lagyear_sub.to_csv('models/lagyear_xgb.csv', index = None)
```
#### Results
25.1 MAE on leaderboard, not an improvement over our best score (~24 MAE) but still our second best model overall
### Tuned XGB
```
lagyear_tuned_sub = pd.read_csv(join(RAW_PATH, 'submission_format.csv'))
y_pred_sj = tuned_model_sj.predict(test_features_sj)
y_pred_iq = tuned_model_iq.predict(test_features_iq)
y_pred = np.concatenate((y_pred_sj, y_pred_iq))
lagyear_tuned_sub['total_cases'] = np.round(y_pred).astype(int)
lagyear_tuned_sub.to_csv('models/lagyear_tuned_xgb.csv', index = None)
```
#### Results
29.6 MAE on leaderbpard, which is far worse than our previous bests. After examination, it seems like
the tuned model has much higher variance in error, which may help explain the poor final scores
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib as mpl
import sklearn
from os.path import join
from sklearn.feature_selection import mutual_info_regression
from sklearn.linear_model import Lasso, Ridge
from sklearn.preprocessing import StandardScaler, PolynomialFeatures
from sklearn.pipeline import Pipeline
from sklearn.model_selection import cross_validate, TimeSeriesSplit, GridSearchCV
from xgboost import XGBRegressor
cd ..
from src.features.build_lagged_features import add_lagged_features
mpl.rcParams.update({
'figure.autolayout': True,
'figure.dpi': 150
})
sns.set()
RAW_PATH = 'data/raw'
train_features_o = pd.read_csv(join(RAW_PATH, 'dengue_features_train.csv'))
test_features_o = pd.read_csv(join(RAW_PATH, 'dengue_features_test.csv'))
train_labels = pd.read_csv(join(RAW_PATH, 'dengue_labels_train.csv'))
train_features_sj = train_features_o[train_features_o['city'] == 'sj'].drop('city', axis = 1)
train_features_iq = train_features_o[train_features_o['city'] == 'iq'].drop('city', axis = 1)
test_features_sj = test_features_o[test_features_o['city'] == 'sj'].drop('city', axis = 1)
test_features_iq = test_features_o[test_features_o['city'] == 'iq'].drop('city', axis = 1)
features_sj = pd.concat([train_features_sj, test_features_sj])
features_iq = pd.concat([train_features_iq, test_features_iq])
len(test_features_sj)
len(test_features_iq)
def make_dataset(features):
features = (features
.drop( # correlated features
['reanalysis_sat_precip_amt_mm', 'reanalysis_dew_point_temp_k',
'reanalysis_air_temp_k', 'reanalysis_tdtr_k'],
axis = 1
)
.fillna(method = 'backfill')
.drop( # unused features
['year', 'weekofyear','week_start_date'],
axis = 1
)
)
ts_features = list(features.loc[:, 'ndvi_ne' :].columns.values)
features = add_lagged_features(
features, 52, ts_features)
return features
features_sj = make_dataset(features_sj)
features_iq = make_dataset(features_iq)
test_features_sj = features_sj.iloc[-260:, :]
test_features_iq = features_iq.iloc[-156:, :]
train_features_sj = features_sj.iloc[52:-260,:]
train_features_iq = features_iq.iloc[52:-156,:]
train_labels_iq = train_labels[
train_labels['city'] == 'iq']['total_cases'].astype('float')[52:]
train_labels_sj = train_labels[
train_labels['city'] == 'sj']['total_cases'].astype('float')[52:]
sj_corrs = train_features_sj.corrwith(train_labels_sj).abs().sort_values()
train_features_sj = train_features_sj[list(sj_corrs.iloc[-100:].index)]
test_features_sj = test_features_sj[list(sj_corrs.iloc[-100:].index)]
iq_corrs = train_features_iq.corrwith(train_labels_iq).abs().sort_values()
train_features_iq = train_features_iq[list(iq_corrs.iloc[-100:].index)]
test_features_iq = test_features_iq[list(iq_corrs.iloc[-100:].index)]
model_sj = Pipeline([
('scale', StandardScaler()),
('lasso', Ridge(10000))
])
cv_res_sj = cross_validate(
estimator = model_sj,
X = train_features_sj,
y = train_labels_sj,
cv = TimeSeriesSplit(n_splits = 10),
scoring = 'neg_mean_absolute_error',
n_jobs = -1
)
sj_score = np.mean(cv_res_sj['test_score'])
sj_score
model_sj.fit(train_features_sj, train_labels_sj)
y_val_sj = model_sj.predict(train_features_sj).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_sj, x = train_features_sj.index.values, ax = ax)
sns.lineplot(y = y_val_sj, x = train_features_sj.index.values, ax = ax)
ax.set(title = 'San Juan predictions on training data')
ax.legend(['True', 'Predicted'])
model_sj = XGBRegressor(
max_depth = 10,
learning_rate = 0.01,
n_estimators = 200,
reg_lambda = 1,
)
cv_res_sj = cross_validate(
estimator = model_sj,
X = train_features_sj,
y = train_labels_sj,
cv = TimeSeriesSplit(n_splits = 10),
scoring = 'neg_mean_absolute_error',
n_jobs = -1
)
sj_score = np.mean(cv_res_sj['test_score'])
sj_score
np.std(cv_res_sj['test_score'])
model_sj.fit(train_features_sj, train_labels_sj)
y_val_sj = model_sj.predict(train_features_sj).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_sj, x = train_features_sj.index.values, ax = ax)
sns.lineplot(y = y_val_sj, x = train_features_sj.index.values, ax = ax)
ax.set(title = 'San Juan predictions on training data')
ax.legend(['True', 'Predicted'])
params = {
'learning_rate' : [0.001 , 0.01, 0.1],
'reg_lambda' : [0.1, 1, 10],
'max_depth': [2, 5, 8, 10],
'n_estimators': [50, 100 , 200]
}
tuned_model_sj = GridSearchCV(
estimator = XGBRegressor(),
param_grid = params,
scoring = 'neg_mean_absolute_error',
cv = TimeSeriesSplit(10),
n_jobs = -1
)
tuned_model_sj.fit(train_features_sj, train_labels_sj)
tuned_model_sj.best_score_
tuned_model_sj.best_params_
cv_res_sj = cross_validate(
estimator = tuned_model_sj.best_estimator_,
X = train_features_sj,
y = train_labels_sj,
cv = TimeSeriesSplit(n_splits = 10),
scoring = 'neg_mean_absolute_error',
n_jobs = -1
)
sj_score = np.mean(cv_res_sj['test_score'])
sj_score
np.std(cv_res_sj['test_score'])
y_val_sj = tuned_model_sj.predict(train_features_sj).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_sj, x = train_features_sj.index.values, ax = ax)
sns.lineplot(y = y_val_sj, x = train_features_sj.index.values, ax = ax)
ax.set(title = 'San Juan predictions on training data')
ax.legend(['True', 'Predicted'])
model_iq = Pipeline([
('scale', StandardScaler()),
('lasso', Lasso(2))
])
cv_res_iq = cross_validate(
estimator = model_iq,
X = train_features_iq,
y = train_labels_iq,
cv = TimeSeriesSplit(n_splits = 10),
scoring = 'neg_mean_absolute_error',
n_jobs = -1
)
iq_score = np.mean(cv_res_iq['test_score'])
iq_score
model_iq.fit(train_features_iq, train_labels_iq)
y_val_iq = model_iq.predict(train_features_iq).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_iq, x = train_features_iq.index.values, ax = ax)
sns.lineplot(y = y_val_iq, x = train_features_iq.index.values, ax = ax)
ax.set(title = 'Iquitos predictions on training data')
ax.legend(['True', 'Predicted'])
model_iq = XGBRegressor(
max_depth = 10,
learning_rate = 0.02,
n_estimators = 200,
reg_lambda = 20,
)
cv_res_iq = cross_validate(
estimator = model_iq,
X = train_features_iq,
y = train_labels_iq,
cv = TimeSeriesSplit(n_splits = 10),
scoring = 'neg_mean_absolute_error',
n_jobs = -1
)
iq_score = np.mean(cv_res_iq['test_score'])
iq_score
model_iq.fit(train_features_iq, train_labels_iq)
y_val_iq = model_iq.predict(train_features_iq).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_iq, x = train_features_iq.index.values, ax = ax)
sns.lineplot(y = y_val_iq, x = train_features_iq.index.values, ax = ax)
ax.set(title = 'Iquitos predictions on training data')
ax.legend(['True', 'Predicted'])
params = {
'learning_rate' : [0.001 , 0.01, 0.1],
'reg_lambda' : [0.1, 1, 10],
'max_depth': [2, 5, 8, 10],
'n_estimators': [50, 100 , 200]
}
tuned_model_iq = GridSearchCV(
estimator = XGBRegressor(),
param_grid = params,
scoring = 'neg_mean_absolute_error',
cv = TimeSeriesSplit(10),
n_jobs = -1
)
tuned_model_iq.fit(train_features_iq, train_labels_iq)
tuned_model_iq.best_score_
tuned_model_iq.best_params_
y_val_iq = tuned_model_iq.predict(train_features_iq).flatten()
fig, ax = plt.subplots(figsize = (10, 5))
sns.lineplot(y = train_labels_iq, x = train_features_iq.index.values, ax = ax)
sns.lineplot(y = y_val_iq, x = train_features_iq.index.values, ax = ax)
ax.set(title = 'Iquitos predictions on training data')
ax.legend(['True', 'Predicted'])
sj_ratio = len(train_features_sj) / (len(train_features_sj) + len(train_features_iq))
iq_ratio = len(train_features_iq) / (len(train_features_sj) + len(train_features_iq))
sj_ratio * sj_score + iq_ratio * iq_score
sj_ratio * tuned_model_sj.best_score_ + iq_ratio * tuned_model_iq.best_score_
lagyear_sub = pd.read_csv(join(RAW_PATH, 'submission_format.csv'))
y_pred_sj = model_sj.predict(test_features_sj)
y_pred_iq = model_iq.predict(test_features_iq)
y_pred = np.concatenate((y_pred_sj, y_pred_iq))
lagyear_sub['total_cases'] = np.round(y_pred).astype(int)
lagyear_sub.to_csv('models/lagyear_xgb.csv', index = None)
lagyear_tuned_sub = pd.read_csv(join(RAW_PATH, 'submission_format.csv'))
y_pred_sj = tuned_model_sj.predict(test_features_sj)
y_pred_iq = tuned_model_iq.predict(test_features_iq)
y_pred = np.concatenate((y_pred_sj, y_pred_iq))
lagyear_tuned_sub['total_cases'] = np.round(y_pred).astype(int)
lagyear_tuned_sub.to_csv('models/lagyear_tuned_xgb.csv', index = None)
| 0.48438 | 0.918809 |
___
<a href='http://www.pieriandata.com'> <img src='../Pierian_Data_Logo.png' /></a>
___
<center>*Copyright Pierian Data 2017*</center>
<center>*For more information, visit us at www.pieriandata.com*</center>
# Time Resampling
Let's learn how to sample time series data! This will be useful later on in the course!
```
import numpy as np
import pandas as pd
%matplotlib inline
import matplotlib.pyplot as plt
# Grab data
# Faster alternative
# df = pd.read_csv('time_data/walmart_stock.csv',index_col='Date')
df = pd.read_csv('time_data/walmart_stock.csv')
df.head()
```
Create a date index from the date column
```
df['Date'] = df['Date'].apply(pd.to_datetime)
df.head()
df.set_index('Date',inplace=True)
df.head()
```
## resample()
A common operation with time series data is resamplling based on the time series index. Let see how to use the resample() method.
#### All possible time series offest strings
<table border="1" class="docutils">
<colgroup>
<col width="13%" />
<col width="87%" />
</colgroup>
<thead valign="bottom">
<tr class="row-odd"><th class="head">Alias</th>
<th class="head">Description</th>
</tr>
</thead>
<tbody valign="top">
<tr class="row-even"><td>B</td>
<td>business day frequency</td>
</tr>
<tr class="row-odd"><td>C</td>
<td>custom business day frequency (experimental)</td>
</tr>
<tr class="row-even"><td>D</td>
<td>calendar day frequency</td>
</tr>
<tr class="row-odd"><td>W</td>
<td>weekly frequency</td>
</tr>
<tr class="row-even"><td>M</td>
<td>month end frequency</td>
</tr>
<tr class="row-odd"><td>SM</td>
<td>semi-month end frequency (15th and end of month)</td>
</tr>
<tr class="row-even"><td>BM</td>
<td>business month end frequency</td>
</tr>
<tr class="row-odd"><td>CBM</td>
<td>custom business month end frequency</td>
</tr>
<tr class="row-even"><td>MS</td>
<td>month start frequency</td>
</tr>
<tr class="row-odd"><td>SMS</td>
<td>semi-month start frequency (1st and 15th)</td>
</tr>
<tr class="row-even"><td>BMS</td>
<td>business month start frequency</td>
</tr>
<tr class="row-odd"><td>CBMS</td>
<td>custom business month start frequency</td>
</tr>
<tr class="row-even"><td>Q</td>
<td>quarter end frequency</td>
</tr>
<tr class="row-odd"><td>BQ</td>
<td>business quarter endfrequency</td>
</tr>
<tr class="row-even"><td>QS</td>
<td>quarter start frequency</td>
</tr>
<tr class="row-odd"><td>BQS</td>
<td>business quarter start frequency</td>
</tr>
<tr class="row-even"><td>A</td>
<td>year end frequency</td>
</tr>
<tr class="row-odd"><td>BA</td>
<td>business year end frequency</td>
</tr>
<tr class="row-even"><td>AS</td>
<td>year start frequency</td>
</tr>
<tr class="row-odd"><td>BAS</td>
<td>business year start frequency</td>
</tr>
<tr class="row-even"><td>BH</td>
<td>business hour frequency</td>
</tr>
<tr class="row-odd"><td>H</td>
<td>hourly frequency</td>
</tr>
<tr class="row-even"><td>T, min</td>
<td>minutely frequency</td>
</tr>
<tr class="row-odd"><td>S</td>
<td>secondly frequency</td>
</tr>
<tr class="row-even"><td>L, ms</td>
<td>milliseconds</td>
</tr>
<tr class="row-odd"><td>U, us</td>
<td>microseconds</td>
</tr>
<tr class="row-even"><td>N</td>
<td>nanoseconds</td>
</tr>
</tbody>
</table>
```
# Our index
df.index
```
You need to call resample with the rule parameter, then you need to call some sort of aggregation function. This is because due to resampling, we need some sort of mathematical rule to join the rows by (mean,sum,count,etc...)
```
# Yearly Means
df.resample(rule='A').mean()
```
### Custom Resampling
You could technically also create your own custom resampling function:
```
def first_day(entry):
"""
Returns the first instance of the period, regardless of samplling rate.
"""
return entry[0]
df.resample(rule='A').apply(first_day)
df['Close'].resample('A').mean().plot(kind='bar')
plt.title('Yearly Mean Close Price for Walmart')
df['Open'].resample('M').max().plot(kind='bar',figsize=(16,6))
plt.title('Monthly Max Opening Price for Walmart')
```
That is it! Up next we'll learn about time shifts!
|
github_jupyter
|
import numpy as np
import pandas as pd
%matplotlib inline
import matplotlib.pyplot as plt
# Grab data
# Faster alternative
# df = pd.read_csv('time_data/walmart_stock.csv',index_col='Date')
df = pd.read_csv('time_data/walmart_stock.csv')
df.head()
df['Date'] = df['Date'].apply(pd.to_datetime)
df.head()
df.set_index('Date',inplace=True)
df.head()
# Our index
df.index
# Yearly Means
df.resample(rule='A').mean()
def first_day(entry):
"""
Returns the first instance of the period, regardless of samplling rate.
"""
return entry[0]
df.resample(rule='A').apply(first_day)
df['Close'].resample('A').mean().plot(kind='bar')
plt.title('Yearly Mean Close Price for Walmart')
df['Open'].resample('M').max().plot(kind='bar',figsize=(16,6))
plt.title('Monthly Max Opening Price for Walmart')
| 0.482673 | 0.970296 |
# Particle Autoencoder Analysis Example
```
import torch
import os, math, time
import numpy as np
from sklearn.cluster import KMeans
from matplotlib import pyplot as plt
from train import inference_latent, reconstruction
from process_data import collect_file, data_reader, PointData, vtk_write, numpy_to_vtp, collate_ball
from mean_shift import mean_shift_track
from model.GeoConvNet import GeoConvNet
from torch.utils.data import DataLoader
try:
data_path = os.environ['data']
except KeyError:
data_path = './data/'
```
## Load trained model and prepare the data
```
load_filename = './example/final_model.pth'
use_cuda = torch.cuda.is_available()
state_dict = torch.load(load_filename,map_location='cuda' if use_cuda else 'cpu')
state = state_dict['state']
# load model related arguments
config = state_dict['config']
args = config
device = torch.device('cuda') if use_cuda else torch.device('cpu')
print(args)
if args.source == "fpm":
file_list = collect_file(os.path.join(data_path, "2016_scivis_fpm/0.44/run41"),args.source,shuffle=False)
fileName = os.path.join(data_path,"2016_scivis_fpm/0.44/run41/025.vtu")
# fileName = os.path.join(data_path,"/2016_scivis_fpm/0.44/run41/035.vtu")
input_dim = 7
elif args.source == "cos":
file_list = collect_file(os.path.join(data_path,"ds14_scivis_0128/raw"),args.source,shuffle=False)
fileName = os.path.join(data_path,'ds14_scivis_0128/raw/ds14_scivis_0128_e4_dt04_0.4900')
input_dim = 10
elif args.source == 'jet3b':
file_list = collect_file(os.path.join(data_path,"jet3b/run3g_50Am_jet3b_sph.3400"),args.source,shuffle=False)
fileName = os.path.join(data_path,"jet3b/run3g_50Am_jet3b_sph.3400")
input_dim = 5
print(fileName)
model = GeoConvNet(args.lat_dim, input_dim, args.ball, args.enc_out, args.r).float().to(device)
model.load_state_dict(state)
model.eval()
torch.set_grad_enabled(False)
data_source = data_reader(fileName, args.source)
pd = PointData(data_source, args.k, args.r, args.ball, np.arange(len(data_source)))
kwargs = {'pin_memory': True} if use_cuda else {}
batch_size = 1024
loader = DataLoader(pd, batch_size=batch_size, shuffle=False, drop_last=False, collate_fn=collate_ball if args.ball else None, **kwargs)
```
## Plot training loss
```
with open(os.path.join(args.result_dir,'epoch_loss_log.txt'), 'r') as f:
lines = f.readlines()
x = [float(line) for line in lines]
fig=plt.figure(figsize=(10,6), dpi= 100, facecolor='w', edgecolor='k')
plt.plot(np.arange(1,len(x)+1),x)
plt.show()
```
## Calculate PSNR
```
output = reconstruction(model,loader,input_dim,args.ball,device)
output = output.numpy()
mse = np.mean((output - data_source[:,3:]) ** 2)
psnr = 10 * math.log10(0.64/mse)
print('psnr:',psnr)
```
# Inference latent vectors
```
# inference latent vectors
latent = inference_latent(model,loader,args.lat_dim,args.ball,device)
print(latent.shape)
np.save(os.path.join(args.result_dir,'latent.npy'),latent.numpy())
```
### Cluster the latent vectors and save to vtk format
```
# clustering
km = KMeans(8,n_init=10,n_jobs=8)
clu = km.fit_predict(latent)
print(clu.shape)
# write npy output to vti format
coord = pd.data[:,:3]
if args.source == 'jet3b':
data_dict = {
'pred_rho': output[:,0],
'pred_temp': output[:,1],
'rho': pd.data[:,3],
'temp': pd.data[:,4],
'clu': clu
}
elif args.source == 'fpm':
data_dict = {
'pred_concentration': output[:,0],
'pred_velocity': output[:,1:],
'concentration': pd.data[:,3],
'velocity': pd.data[:,4:],
'clu': clu
}
elif args.source == 'cos':
data_dict = {
"pred_phi":output[:,-1],
"pred_velocity":output[:,0:3],
"pred_acceleration":output[:,3:6],
"phi":pd.data[:,-1],
"velocity":pd.data[:,3:6],
"acceleration":pd.data[:,6:9],
'clu': clu
}
vtk_data = numpy_to_vtp(coord,data_dict)
vtk_write( vtk_data, args.result_dir + "/predict.vtp")
```
|
github_jupyter
|
import torch
import os, math, time
import numpy as np
from sklearn.cluster import KMeans
from matplotlib import pyplot as plt
from train import inference_latent, reconstruction
from process_data import collect_file, data_reader, PointData, vtk_write, numpy_to_vtp, collate_ball
from mean_shift import mean_shift_track
from model.GeoConvNet import GeoConvNet
from torch.utils.data import DataLoader
try:
data_path = os.environ['data']
except KeyError:
data_path = './data/'
load_filename = './example/final_model.pth'
use_cuda = torch.cuda.is_available()
state_dict = torch.load(load_filename,map_location='cuda' if use_cuda else 'cpu')
state = state_dict['state']
# load model related arguments
config = state_dict['config']
args = config
device = torch.device('cuda') if use_cuda else torch.device('cpu')
print(args)
if args.source == "fpm":
file_list = collect_file(os.path.join(data_path, "2016_scivis_fpm/0.44/run41"),args.source,shuffle=False)
fileName = os.path.join(data_path,"2016_scivis_fpm/0.44/run41/025.vtu")
# fileName = os.path.join(data_path,"/2016_scivis_fpm/0.44/run41/035.vtu")
input_dim = 7
elif args.source == "cos":
file_list = collect_file(os.path.join(data_path,"ds14_scivis_0128/raw"),args.source,shuffle=False)
fileName = os.path.join(data_path,'ds14_scivis_0128/raw/ds14_scivis_0128_e4_dt04_0.4900')
input_dim = 10
elif args.source == 'jet3b':
file_list = collect_file(os.path.join(data_path,"jet3b/run3g_50Am_jet3b_sph.3400"),args.source,shuffle=False)
fileName = os.path.join(data_path,"jet3b/run3g_50Am_jet3b_sph.3400")
input_dim = 5
print(fileName)
model = GeoConvNet(args.lat_dim, input_dim, args.ball, args.enc_out, args.r).float().to(device)
model.load_state_dict(state)
model.eval()
torch.set_grad_enabled(False)
data_source = data_reader(fileName, args.source)
pd = PointData(data_source, args.k, args.r, args.ball, np.arange(len(data_source)))
kwargs = {'pin_memory': True} if use_cuda else {}
batch_size = 1024
loader = DataLoader(pd, batch_size=batch_size, shuffle=False, drop_last=False, collate_fn=collate_ball if args.ball else None, **kwargs)
with open(os.path.join(args.result_dir,'epoch_loss_log.txt'), 'r') as f:
lines = f.readlines()
x = [float(line) for line in lines]
fig=plt.figure(figsize=(10,6), dpi= 100, facecolor='w', edgecolor='k')
plt.plot(np.arange(1,len(x)+1),x)
plt.show()
output = reconstruction(model,loader,input_dim,args.ball,device)
output = output.numpy()
mse = np.mean((output - data_source[:,3:]) ** 2)
psnr = 10 * math.log10(0.64/mse)
print('psnr:',psnr)
# inference latent vectors
latent = inference_latent(model,loader,args.lat_dim,args.ball,device)
print(latent.shape)
np.save(os.path.join(args.result_dir,'latent.npy'),latent.numpy())
# clustering
km = KMeans(8,n_init=10,n_jobs=8)
clu = km.fit_predict(latent)
print(clu.shape)
# write npy output to vti format
coord = pd.data[:,:3]
if args.source == 'jet3b':
data_dict = {
'pred_rho': output[:,0],
'pred_temp': output[:,1],
'rho': pd.data[:,3],
'temp': pd.data[:,4],
'clu': clu
}
elif args.source == 'fpm':
data_dict = {
'pred_concentration': output[:,0],
'pred_velocity': output[:,1:],
'concentration': pd.data[:,3],
'velocity': pd.data[:,4:],
'clu': clu
}
elif args.source == 'cos':
data_dict = {
"pred_phi":output[:,-1],
"pred_velocity":output[:,0:3],
"pred_acceleration":output[:,3:6],
"phi":pd.data[:,-1],
"velocity":pd.data[:,3:6],
"acceleration":pd.data[:,6:9],
'clu': clu
}
vtk_data = numpy_to_vtp(coord,data_dict)
vtk_write( vtk_data, args.result_dir + "/predict.vtp")
| 0.451568 | 0.791902 |
# Now You Code 2: Shopping List
Write a program to input a list of grocery items for your shopping list and then writes them to a file a line at a time. The program should keep asking you to enter grocery items until you type `'done'`.
After you complete the list, the program should then load the file and read the list back to you by printing each item out.
Sample Run:
```
Let's make a shopping list. Type 'done' when you're finished:
Enter Item: milk
Enter Item: cheese
Enter Item: eggs
Enter Item: beer
Enter Item: apples
Enter Item: done
Your shopping list:
milk
cheese
eggs
beer
apples
```
## Step 1: Problem Analysis
Inputs:
grocery list
or doene to finish
Outputs:
"Your shopping list:"
followed by your shopping list
Algorithm (Steps in Program):
take the input
some black box magic
make it the output
```
filename = "NYC2-shopping-list.txt"
shopping_list = 'O'
## Step 2: write code here
with open(filename, 'w') as grocery:
print("Let's make a shopping list. Type 'done' when you're finished: ")
grocery.write("Your shopping list: ")
grocery.write("\n")
while shopping_list != 'done':
shopping_list = input("Enter Item: " )
grocery.write(shopping_list)
grocery.write("\n")
with open(filename, 'r') as grocery:
for line in grocery.readlines():
print(line)
```
## Step 3: Refactoring
Refactor the part of your program which reads the shopping list from the file into a separate user-defined function called `readShoppingList`
re-write your program to use this function.
## ReadShoppingList function
Inputs:
Outputs:
Algorithm (Steps in Program):
```
## Step 4: Write program again with refactored code.
def readShoppingList(filename):
shoppingList = []
# todo read shopping list here
with open(filename, 'r') as grocery:
for line in grocery.readlines():
shoppingList = line
return shoppingList
# TODO Main code here
filename = "NYC2-shopping-list.txt"
shopping_list = 'o'
with open(filename, 'w') as grocery:
print("Let's make a shopping list. Type 'done' when you're finished: ")
grocery.write("Your shopping list: ")
grocery.write("\n")
while shopping_list != 'done':
shopping_list = input("Enter Item: " )
grocery.write(shopping_list)
grocery.write("\n")
readShoppingList(filename)
```
## Step 5: Questions
1. Is the refactored code in step 4 easier to read? Why or why not?\
they're about equal because we are stating what the functuion does
2. Explain how this program could be refarctored further (there's one thing that's obvious).
we could make a function for the writing poriton
3. Describe how this program can be modified to support multiple shopping lists?
we could make it in a loop where you could write multiple shopping lists
## Reminder of Evaluation Criteria
1. What the problem attempted (analysis, code, and answered questions) ?
2. What the problem analysis thought out? (does the program match the plan?)
3. Does the code execute without syntax error?
4. Does the code solve the intended problem?
5. Is the code well written? (easy to understand, modular, and self-documenting, handles errors)
|
github_jupyter
|
Let's make a shopping list. Type 'done' when you're finished:
Enter Item: milk
Enter Item: cheese
Enter Item: eggs
Enter Item: beer
Enter Item: apples
Enter Item: done
Your shopping list:
milk
cheese
eggs
beer
apples
filename = "NYC2-shopping-list.txt"
shopping_list = 'O'
## Step 2: write code here
with open(filename, 'w') as grocery:
print("Let's make a shopping list. Type 'done' when you're finished: ")
grocery.write("Your shopping list: ")
grocery.write("\n")
while shopping_list != 'done':
shopping_list = input("Enter Item: " )
grocery.write(shopping_list)
grocery.write("\n")
with open(filename, 'r') as grocery:
for line in grocery.readlines():
print(line)
## Step 4: Write program again with refactored code.
def readShoppingList(filename):
shoppingList = []
# todo read shopping list here
with open(filename, 'r') as grocery:
for line in grocery.readlines():
shoppingList = line
return shoppingList
# TODO Main code here
filename = "NYC2-shopping-list.txt"
shopping_list = 'o'
with open(filename, 'w') as grocery:
print("Let's make a shopping list. Type 'done' when you're finished: ")
grocery.write("Your shopping list: ")
grocery.write("\n")
while shopping_list != 'done':
shopping_list = input("Enter Item: " )
grocery.write(shopping_list)
grocery.write("\n")
readShoppingList(filename)
| 0.165762 | 0.78789 |
## Generative Adversarial Networks
In this notebook we'll see how to build a very popular type of deep learning model called a generative adversarial network, or GAN for short.
<img src="figures/LweaD1s.png" width="600px">
Interest in GANs has exploded in the last few years thanks to their unique ability to generate complex human-like results. GANs have been used for image synthesis, style transfer, music generation, and many other novel applications. We'll build a particular type of GAN called the Deep Convolutional GAN (DCGAN) from scratch and train it to generate MNIST images.
Much of the code used in this notebook is adapted from the excellent [Keras-GAN](https://github.com/eriklindernoren/Keras-GAN) library created by Erik Linder-Noren.
Let's get started by importing a few libraries and loading up MNIST into an array.
```
import time
import matplotlib.pyplot as plt
import numpy as np
from keras.datasets import mnist
(X, _), (_, _) = mnist.load_data()
X = X / 127.5 - 1.
X = np.expand_dims(X, axis=3)
X.shape
```
Note that we've added another dimension for the channel which is usually for the color value (red, green, blue). Since these images are greyscale it's not strictly necessary but Keras expects an input array in this shape.
The first thing we need to do is define the generator. This is the model that will learn how to produce new images. The architecture looks a lot like a normal convolutional network except in reverse. We start with an input that is essentially random noise and mold it into an image through a series of convolution blocks. All of these layers should be familiar to anyone that's worked with convolutional networks before with the possible exception of the "upsampling" layer, which essentially doubles the size of the tensor along two axes by repeating rows and columns.
```
from keras.models import Model
from keras.layers import Activation, BatchNormalization, Conv2D
from keras.layers import Dense, Input, Reshape, UpSampling2D
def get_generator(noise_shape):
i = Input(noise_shape)
x = Dense(128 * 7 * 7)(i)
x = Activation("relu")(x)
x = Reshape((7, 7, 128))(x)
x = UpSampling2D()(x)
x = Conv2D(128, kernel_size=3, padding="same")(x)
x = BatchNormalization(momentum=0.8)(x)
x = Activation("relu")(x)
x = UpSampling2D()(x)
x = Conv2D(64, kernel_size=3, padding="same")(x)
x = BatchNormalization(momentum=0.8)(x)
x = Activation("relu")(x)
x = Conv2D(1, kernel_size=3, padding="same")(x)
o = Activation("tanh")(x)
return Model(i, o)
```
It helps to see the summary output for the model and follow along as the shape changes. One interesting thing to note is how many of the parameters in the model are in that initial dense layer.
```
g = get_generator(noise_shape=(100,))
g.summary()
```
Next up is the discriminator. This model will be tasked with looking at an image and deciding if it's really an MNIST image or if it's a fake. The discriminator looks a lot more like a standard convolutional network. It takes a tensor of an image as it's input and runs it through a series of convolution blocks, decreasing in size until it outputs a single activation representing the probability of the tensor being a real MNIST image.
```
from keras.layers import Dropout, Flatten, LeakyReLU, ZeroPadding2D
def get_discriminator(img_shape):
i = Input(img_shape)
x = Conv2D(32, kernel_size=3, strides=2, padding="same")(i)
x = LeakyReLU(alpha=0.2)(x)
x = Dropout(0.25)(x)
x = Conv2D(64, kernel_size=3, strides=2, padding="same")(i)
x = ZeroPadding2D(padding=((0, 1), (0, 1)))(x)
x = BatchNormalization(momentum=0.8)(x)
x = LeakyReLU(alpha=0.2)(x)
x = Dropout(0.25)(x)
x = Conv2D(128, kernel_size=3, strides=2, padding="same")(x)
x = BatchNormalization(momentum=0.8)(x)
x = LeakyReLU(alpha=0.2)(x)
x = Dropout(0.25)(x)
x = Conv2D(256, kernel_size=3, strides=1, padding="same")(x)
x = BatchNormalization(momentum=0.8)(x)
x = LeakyReLU(alpha=0.2)(x)
x = Dropout(0.25)(x)
x = Flatten()(x)
o = Dense(1)(x)
o = Activation("sigmoid")(o)
return Model(i, o)
d = get_discriminator(img_shape=(28, 28, 1))
d.summary()
```
With both components defined, we can now bring it all together and create the GAN model. It's worth spending some time examining this part closely as it can be confusing at first. Notice that we first compile the distriminator by itself. Then we define the generator and feed its output to the discriminator, but only after setting the discriminator so that training is disabled. This is because we don't want the discriminator to update its weights during this part, only the generator. Finally, a third model is created combining the original input to the generator and the output of the discriminator. Don't worry if you find this confusing, it takes some time to fully comprehend.
```
from keras.optimizers import Adam
def DCGAN():
optimizer = Adam(lr=0.0002, beta_1=0.5)
discriminator = get_discriminator(img_shape=(28, 28, 1))
discriminator.compile(loss="binary_crossentropy", optimizer=optimizer, metrics=["accuracy"])
generator = get_generator(noise_shape=(100,))
z = Input(shape=(100,))
g_out = generator(z)
discriminator.trainable = False
d_out = discriminator(g_out)
combined = Model(z, d_out)
combined.compile(loss="binary_crossentropy", optimizer=optimizer)
return generator, discriminator, combined
generator, discriminator, combined = DCGAN()
combined.summary()
```
Create a function to save the generator's output so we can visually see the results. We'll use this in the training loop below.
```
def save_image(generator, batch):
r, c = 5, 5
noise = np.random.normal(0, 1, (r * c, 100))
gen_images = generator.predict(noise)
gen_images = 0.5 * gen_images + 0.5
fig, axs = plt.subplots(r, c)
cnt = 0
for i in range(r):
for j in range(c):
axs[i, j].imshow(gen_images[cnt, :, :, 0], cmap="gray")
axs[i, j].axis('off')
cnt += 1
fig.savefig("mnist_%d.png" % (batch))
plt.close()
```
The last challenge with building a GAN is training the whole thing. With something this complex we can't just call the standard fit function because there are multiple steps involed. Let's roll our own and see how the algorithm works. Start by generating some random noise and feeding it through the generator to produce a batch of "fake" images. Next, select a batch of real images from the training data. Now train the distriminator on both the fake image batch and the real image batch, setting labels appropriately (0 for fake and 1 for real). Finally, train the generator by feeding the "combined" model a batch of noise with the "real" label.
You may find this last step confusing. Remember what this "combined" model is doing. It runs the input (random noise) through the generator, then runs that output through the discriminator (which is frozen), and gets a loss. What this ends of doing in essence is saying "train the generator to minimize the discriminator's ability to discern this output from a real image". This has the effect, over time, of causing the generator to produce realistic images!
```
def train(generator, discriminator, combined, X, batch_size, n_batches):
t0 = time.time()
for batch in range(n_batches + 1):
t1 = time.time()
noise = np.random.normal(0, 1, (batch_size, 100))
# Create fake images
fake_images = generator.predict(noise)
fake_labels = np.zeros((batch_size, 1))
# Select real images
idx = np.random.randint(0, X.shape[0], batch_size)
real_images = X[idx]
real_labels = np.ones((batch_size, 1))
# Train the discriminator
d_loss_real = discriminator.train_on_batch(real_images, real_labels)
d_loss_fake = discriminator.train_on_batch(fake_images, fake_labels)
d_loss = 0.5 * np.add(d_loss_real, d_loss_fake)
# Train the generator
noise = np.random.normal(0, 1, (batch_size, 100))
g_loss = combined.train_on_batch(noise, real_labels)
t2 = time.time()
# Report progress
if batch % 100 == 0:
print("Batch %d/%d [Batch time: %.2f s, Total: %.2f m] [D loss: %f, Acc: %.2f%%] [G loss: %f]" %
(batch, (n_batches), (t2 - t1), ((t2 - t0) / 60), d_loss[0], 100 * d_loss[1], g_loss))
if batch % 500 == 0:
save_image(generator, batch)
```
Run the training loop for a while. We can't read too much into the losses reported by this process because the models are competing with each other. They both end up being pretty stable over the training period but it doesn't really capture what's going on because both models are getting better at their task by roughly equal amounts. If either model went to zero loss that would likely indicate there's a problem that needs fixed.
```
train(generator, discriminator, combined, X=X, batch_size=32, n_batches=3000)
```
Take a look at the generator's output at various stages of training. In the beginning it's clear that the model is producing noise, but after just a few hundred batches it's already making progress. After a couple thousand batches the images look quite realistic.
```
from IPython.display import Image, display
names = ["mnist_0.png",
"mnist_500.png",
"mnist_1000.png",
"mnist_1500.png",
"mnist_2000.png",
"mnist_2500.png",
"mnist_3000.png"]
for name in names:
display(Image(filename=name))
```
|
github_jupyter
|
import time
import matplotlib.pyplot as plt
import numpy as np
from keras.datasets import mnist
(X, _), (_, _) = mnist.load_data()
X = X / 127.5 - 1.
X = np.expand_dims(X, axis=3)
X.shape
from keras.models import Model
from keras.layers import Activation, BatchNormalization, Conv2D
from keras.layers import Dense, Input, Reshape, UpSampling2D
def get_generator(noise_shape):
i = Input(noise_shape)
x = Dense(128 * 7 * 7)(i)
x = Activation("relu")(x)
x = Reshape((7, 7, 128))(x)
x = UpSampling2D()(x)
x = Conv2D(128, kernel_size=3, padding="same")(x)
x = BatchNormalization(momentum=0.8)(x)
x = Activation("relu")(x)
x = UpSampling2D()(x)
x = Conv2D(64, kernel_size=3, padding="same")(x)
x = BatchNormalization(momentum=0.8)(x)
x = Activation("relu")(x)
x = Conv2D(1, kernel_size=3, padding="same")(x)
o = Activation("tanh")(x)
return Model(i, o)
g = get_generator(noise_shape=(100,))
g.summary()
from keras.layers import Dropout, Flatten, LeakyReLU, ZeroPadding2D
def get_discriminator(img_shape):
i = Input(img_shape)
x = Conv2D(32, kernel_size=3, strides=2, padding="same")(i)
x = LeakyReLU(alpha=0.2)(x)
x = Dropout(0.25)(x)
x = Conv2D(64, kernel_size=3, strides=2, padding="same")(i)
x = ZeroPadding2D(padding=((0, 1), (0, 1)))(x)
x = BatchNormalization(momentum=0.8)(x)
x = LeakyReLU(alpha=0.2)(x)
x = Dropout(0.25)(x)
x = Conv2D(128, kernel_size=3, strides=2, padding="same")(x)
x = BatchNormalization(momentum=0.8)(x)
x = LeakyReLU(alpha=0.2)(x)
x = Dropout(0.25)(x)
x = Conv2D(256, kernel_size=3, strides=1, padding="same")(x)
x = BatchNormalization(momentum=0.8)(x)
x = LeakyReLU(alpha=0.2)(x)
x = Dropout(0.25)(x)
x = Flatten()(x)
o = Dense(1)(x)
o = Activation("sigmoid")(o)
return Model(i, o)
d = get_discriminator(img_shape=(28, 28, 1))
d.summary()
from keras.optimizers import Adam
def DCGAN():
optimizer = Adam(lr=0.0002, beta_1=0.5)
discriminator = get_discriminator(img_shape=(28, 28, 1))
discriminator.compile(loss="binary_crossentropy", optimizer=optimizer, metrics=["accuracy"])
generator = get_generator(noise_shape=(100,))
z = Input(shape=(100,))
g_out = generator(z)
discriminator.trainable = False
d_out = discriminator(g_out)
combined = Model(z, d_out)
combined.compile(loss="binary_crossentropy", optimizer=optimizer)
return generator, discriminator, combined
generator, discriminator, combined = DCGAN()
combined.summary()
def save_image(generator, batch):
r, c = 5, 5
noise = np.random.normal(0, 1, (r * c, 100))
gen_images = generator.predict(noise)
gen_images = 0.5 * gen_images + 0.5
fig, axs = plt.subplots(r, c)
cnt = 0
for i in range(r):
for j in range(c):
axs[i, j].imshow(gen_images[cnt, :, :, 0], cmap="gray")
axs[i, j].axis('off')
cnt += 1
fig.savefig("mnist_%d.png" % (batch))
plt.close()
def train(generator, discriminator, combined, X, batch_size, n_batches):
t0 = time.time()
for batch in range(n_batches + 1):
t1 = time.time()
noise = np.random.normal(0, 1, (batch_size, 100))
# Create fake images
fake_images = generator.predict(noise)
fake_labels = np.zeros((batch_size, 1))
# Select real images
idx = np.random.randint(0, X.shape[0], batch_size)
real_images = X[idx]
real_labels = np.ones((batch_size, 1))
# Train the discriminator
d_loss_real = discriminator.train_on_batch(real_images, real_labels)
d_loss_fake = discriminator.train_on_batch(fake_images, fake_labels)
d_loss = 0.5 * np.add(d_loss_real, d_loss_fake)
# Train the generator
noise = np.random.normal(0, 1, (batch_size, 100))
g_loss = combined.train_on_batch(noise, real_labels)
t2 = time.time()
# Report progress
if batch % 100 == 0:
print("Batch %d/%d [Batch time: %.2f s, Total: %.2f m] [D loss: %f, Acc: %.2f%%] [G loss: %f]" %
(batch, (n_batches), (t2 - t1), ((t2 - t0) / 60), d_loss[0], 100 * d_loss[1], g_loss))
if batch % 500 == 0:
save_image(generator, batch)
train(generator, discriminator, combined, X=X, batch_size=32, n_batches=3000)
from IPython.display import Image, display
names = ["mnist_0.png",
"mnist_500.png",
"mnist_1000.png",
"mnist_1500.png",
"mnist_2000.png",
"mnist_2500.png",
"mnist_3000.png"]
for name in names:
display(Image(filename=name))
| 0.87127 | 0.98484 |
# Pandas_Alive
Animated plotting extension for Pandas with Matplotlib
**Pandas_Alive** is intended to provide a plotting backend for animated [matplotlib](https://matplotlib.org/) charts for [Pandas](https://pandas.pydata.org/) DataFrames, similar to the already [existing Visualization feature of Pandas](https://pandas.pydata.org/pandas-docs/stable/visualization.html).
With **Pandas_Alive**, creating stunning, animated visualisations is as easy as calling:
`df.plot_animated()`

## Table of Contents
<!-- START doctoc -->
<!-- END doctoc -->
## Installation
Install with `pip install pandas_alive`
## Usage
As this package builds upon [`bar_chart_race`](https://github.com/dexplo/bar_chart_race), the example data set is sourced from there.
Must begin with a pandas DataFrame containing 'wide' data where:
- Every row represents a single period of time
- Each column holds the value for a particular category
- The index contains the time component (optional)
The data below is an example of properly formatted data. It shows total deaths from COVID-19 for the highest 20 countries by date.

[Example Table](examples/example_dataset_table.md)
To produce the above visualisation:
- Check [Requirements](#requirements) first to ensure you have the tooling installed!
- Call `plot_animated()` on the DataFrame
- Either specify a file name to write to with `df.plot_animated(filename='example.mp4')` or use `df.plot_animated().get_html5_video` to return a HTML5 video
- Done!
```
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.plot_animated(filename='examples/example-barh-chart.gif')
```
### Currently Supported Chart Types
`pandas_alive` current supports:
- [Horizontal Bar Charts](#horizontal-bar-charts)
- [Vertical Bar Charts](#vertical-bar-charts)
- [Line Charts](#line-charts)
- [Scatter Charts](#scatter-charts)
- [Pie Charts](#pie-charts)
#### Horizontal Bar Charts
```
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.plot_animated(filename='example-barh-chart.gif')
```

```
import pandas as pd
import pandas_alive
elec_df = pd.read_csv("data/Aus_Elec_Gen_1980_2018.csv",index_col=0,parse_dates=[0],thousands=',')
elec_df.fillna(0).plot_animated('examples/example-electricity-generated-australia.gif',period_fmt="%Y",title='Australian Electricity Generation Sources 1980-2018')
```

```
import pandas_alive
covid_df = pandas_alive.load_dataset()
def current_total(values):
total = values.sum()
s = f'Total : {int(total)}'
return {'x': .85, 'y': .2, 's': s, 'ha': 'right', 'size': 11}
covid_df.plot_animated(filename='examples/summary-func-example.gif',period_summary_func=current_total)
```

```
import pandas as pd
import pandas_alive
elec_df = pd.read_csv("data/Aus_Elec_Gen_1980_2018.csv",index_col=0,parse_dates=[0],thousands=',')
elec_df.fillna(0).plot_animated('examples/fixed-example.gif',period_fmt="%Y",title='Australian Electricity Generation Sources 1980-2018',fixed_max=True,fixed_order=True)
```

```
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.plot_animated(filename='examples/perpendicular-example.gif',perpendicular_bar_func='mean')
```

#### Vertical Bar Charts
```
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.plot_animated(filename='examples/example-barv-chart.gif',orientation='v')
```

#### Line Charts
With as many lines as data columns in the DataFrame.
```
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.diff().fillna(0).plot_animated(filename='examples/example-line-chart.gif',kind='line',period_label={'x':0.1,'y':0.9})
```

#### Scatter Charts
```
import pandas as pd
import pandas_alive
max_temp_df = pd.read_csv(
"data/Newcastle_Australia_Max_Temps.csv",
parse_dates={"Timestamp": ["Year", "Month", "Day"]},
)
min_temp_df = pd.read_csv(
"data/Newcastle_Australia_Min_Temps.csv",
parse_dates={"Timestamp": ["Year", "Month", "Day"]},
)
merged_temp_df = pd.merge_asof(max_temp_df, min_temp_df, on="Timestamp")
merged_temp_df.index = pd.to_datetime(merged_temp_df["Timestamp"].dt.strftime('%Y/%m/%d'))
keep_columns = ["Minimum temperature (Degree C)", "Maximum temperature (Degree C)"]
merged_temp_df[keep_columns].resample("Y").mean().plot_animated(filename='examples/example-scatter-chart.gif',kind="scatter",title='Max & Min Temperature Newcastle, Australia')
```

#### Pie Charts
```
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.plot_animated(filename='examples/example-pie-chart.gif',kind="pie",rotatelabels=True)
```

### Multiple Charts
`pandas_alive` supports multiple animated charts in a single visualisation.
- Create a list of all charts to include in animation
- Use `animate_multiple_plots` with a `filename` and the list of charts (this will use `matplotlib.subplots`)
- Done!
```
import pandas_alive
covid_df = pandas_alive.load_dataset()
animated_line_chart = covid_df.diff().fillna(0).plot_animated(kind='line',period_label=False)
animated_bar_chart = covid_df.plot_animated(n_visible=10)
pandas_alive.animate_multiple_plots('examples/example-bar-and-line-chart.gif',[animated_bar_chart,animated_line_chart])
```

#### Urban Population
```
import pandas_alive
urban_df = pandas_alive.load_dataset("urban_pop")
animated_line_chart = (
urban_df.sum(axis=1)
.pct_change()
.dropna()
.mul(100)
.plot_animated(kind="line", title="Total % Change in Population",period_label=False)
)
animated_bar_chart = urban_df.plot_animated(n_visible=10,title='Top 10 Populous Countries',period_fmt="%Y")
pandas_alive.animate_multiple_plots('examples/example-bar-and-line-urban-chart.gif',[animated_bar_chart,animated_line_chart],title='Urban Population 1977 - 2018',adjust_subplot_top=0.85)
```

#### Life Expectancy in G7 Countries
```
import pandas_alive
import pandas as pd
data_raw = pd.read_csv(
"https://raw.githubusercontent.com/owid/owid-datasets/master/datasets/Long%20run%20life%20expectancy%20-%20Gapminder%2C%20UN/Long%20run%20life%20expectancy%20-%20Gapminder%2C%20UN.csv"
)
list_G7 = [
"Canada",
"France",
"Germany",
"Italy",
"Japan",
"United Kingdom",
"United States",
]
data_raw = data_raw.pivot(
index="Year", columns="Entity", values="Life expectancy (Gapminder, UN)"
)
data = pd.DataFrame()
data["Year"] = data_raw.reset_index()["Year"]
for country in list_G7:
data[country] = data_raw[country].values
data = data.fillna(method="pad")
data = data.fillna(0)
data = data.set_index("Year").loc[1900:].reset_index()
data["Year"] = pd.to_datetime(data.reset_index()["Year"].astype(str))
data = data.set_index("Year")
animated_bar_chart = data.plot_animated(
period_fmt="%Y",perpendicular_bar_func="mean", period_length=200,fixed_max=True
)
animated_line_chart = data.plot_animated(
kind="line", period_fmt="%Y", period_length=200,fixed_max=True
)
pandas_alive.animate_multiple_plots(
"examples/life-expectancy.gif",
plots=[animated_bar_chart, animated_line_chart],
title="Life expectancy in G7 countries up to 2015",
adjust_subplot_left=0.2,
)
```

## Future Features
A list of future features that may/may not be developed is:
- Multiple dimension plots (with multi indexed dataframes)
- Bubble charts
- Geographic charts (currently using OSM export image, potential [cartopy](https://github.com/SciTools/cartopy))
A chart that was built using a development branch of Pandas_Alive is:
[](https://www.youtube.com/watch?v=qyqiYrtpxRE)
## Inspiration
The inspiration for this project comes from:
- [bar_chart_race](https://github.com/dexplo/bar_chart_race) by [Ted Petrou](https://github.com/tdpetrou)
- [Pandas-Bokeh](https://github.com/PatrikHlobil/Pandas-Bokeh) by [Patrik Hlobil](https://github.com/PatrikHlobil)
## Requirements
If you get an error such as `TypeError: 'MovieWriterRegistry' object is not an iterator`, this signals there isn't a writer library installed on your machine.
This package utilises the [matplotlib.animation function](https://matplotlib.org/3.2.1/api/animation_api.html), thus requiring a writer library.
Ensure to have one of the supported tooling software installed prior to use!
- [ffmpeg](https://ffmpeg.org/)
- [ImageMagick](https://imagemagick.org/index.php)
- [Pillow](https://pillow.readthedocs.io/en/stable/)
- See more at <https://matplotlib.org/3.2.1/api/animation_api.html#writer-classes>
## Documentation
Documentation is provided at <https://jackmckew.github.io/pandas_alive/>
## Contributing
Pull requests are welcome! Please help to cover more and more chart types!
## [Changelog](CHANGELOG.md)
|
github_jupyter
|
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.plot_animated(filename='examples/example-barh-chart.gif')
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.plot_animated(filename='example-barh-chart.gif')
import pandas as pd
import pandas_alive
elec_df = pd.read_csv("data/Aus_Elec_Gen_1980_2018.csv",index_col=0,parse_dates=[0],thousands=',')
elec_df.fillna(0).plot_animated('examples/example-electricity-generated-australia.gif',period_fmt="%Y",title='Australian Electricity Generation Sources 1980-2018')
import pandas_alive
covid_df = pandas_alive.load_dataset()
def current_total(values):
total = values.sum()
s = f'Total : {int(total)}'
return {'x': .85, 'y': .2, 's': s, 'ha': 'right', 'size': 11}
covid_df.plot_animated(filename='examples/summary-func-example.gif',period_summary_func=current_total)
import pandas as pd
import pandas_alive
elec_df = pd.read_csv("data/Aus_Elec_Gen_1980_2018.csv",index_col=0,parse_dates=[0],thousands=',')
elec_df.fillna(0).plot_animated('examples/fixed-example.gif',period_fmt="%Y",title='Australian Electricity Generation Sources 1980-2018',fixed_max=True,fixed_order=True)
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.plot_animated(filename='examples/perpendicular-example.gif',perpendicular_bar_func='mean')
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.plot_animated(filename='examples/example-barv-chart.gif',orientation='v')
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.diff().fillna(0).plot_animated(filename='examples/example-line-chart.gif',kind='line',period_label={'x':0.1,'y':0.9})
import pandas as pd
import pandas_alive
max_temp_df = pd.read_csv(
"data/Newcastle_Australia_Max_Temps.csv",
parse_dates={"Timestamp": ["Year", "Month", "Day"]},
)
min_temp_df = pd.read_csv(
"data/Newcastle_Australia_Min_Temps.csv",
parse_dates={"Timestamp": ["Year", "Month", "Day"]},
)
merged_temp_df = pd.merge_asof(max_temp_df, min_temp_df, on="Timestamp")
merged_temp_df.index = pd.to_datetime(merged_temp_df["Timestamp"].dt.strftime('%Y/%m/%d'))
keep_columns = ["Minimum temperature (Degree C)", "Maximum temperature (Degree C)"]
merged_temp_df[keep_columns].resample("Y").mean().plot_animated(filename='examples/example-scatter-chart.gif',kind="scatter",title='Max & Min Temperature Newcastle, Australia')
import pandas_alive
covid_df = pandas_alive.load_dataset()
covid_df.plot_animated(filename='examples/example-pie-chart.gif',kind="pie",rotatelabels=True)
import pandas_alive
covid_df = pandas_alive.load_dataset()
animated_line_chart = covid_df.diff().fillna(0).plot_animated(kind='line',period_label=False)
animated_bar_chart = covid_df.plot_animated(n_visible=10)
pandas_alive.animate_multiple_plots('examples/example-bar-and-line-chart.gif',[animated_bar_chart,animated_line_chart])
import pandas_alive
urban_df = pandas_alive.load_dataset("urban_pop")
animated_line_chart = (
urban_df.sum(axis=1)
.pct_change()
.dropna()
.mul(100)
.plot_animated(kind="line", title="Total % Change in Population",period_label=False)
)
animated_bar_chart = urban_df.plot_animated(n_visible=10,title='Top 10 Populous Countries',period_fmt="%Y")
pandas_alive.animate_multiple_plots('examples/example-bar-and-line-urban-chart.gif',[animated_bar_chart,animated_line_chart],title='Urban Population 1977 - 2018',adjust_subplot_top=0.85)
import pandas_alive
import pandas as pd
data_raw = pd.read_csv(
"https://raw.githubusercontent.com/owid/owid-datasets/master/datasets/Long%20run%20life%20expectancy%20-%20Gapminder%2C%20UN/Long%20run%20life%20expectancy%20-%20Gapminder%2C%20UN.csv"
)
list_G7 = [
"Canada",
"France",
"Germany",
"Italy",
"Japan",
"United Kingdom",
"United States",
]
data_raw = data_raw.pivot(
index="Year", columns="Entity", values="Life expectancy (Gapminder, UN)"
)
data = pd.DataFrame()
data["Year"] = data_raw.reset_index()["Year"]
for country in list_G7:
data[country] = data_raw[country].values
data = data.fillna(method="pad")
data = data.fillna(0)
data = data.set_index("Year").loc[1900:].reset_index()
data["Year"] = pd.to_datetime(data.reset_index()["Year"].astype(str))
data = data.set_index("Year")
animated_bar_chart = data.plot_animated(
period_fmt="%Y",perpendicular_bar_func="mean", period_length=200,fixed_max=True
)
animated_line_chart = data.plot_animated(
kind="line", period_fmt="%Y", period_length=200,fixed_max=True
)
pandas_alive.animate_multiple_plots(
"examples/life-expectancy.gif",
plots=[animated_bar_chart, animated_line_chart],
title="Life expectancy in G7 countries up to 2015",
adjust_subplot_left=0.2,
)
| 0.54359 | 0.969671 |
# Exemplo dados do ***Big Query***
## Carregar pacotes
```
import numpy as np
import pandas as pd
from google.colab import auth
from google.cloud import bigquery
import matplotlib.pyplot as plt
%matplotlib inline
```
## Autenticar projeto
```
auth.authenticate_user()
project_id = 'aula01in'
client = bigquery.Client(project=project_id)
```
## Dados
### Partidas
buscar dados do *Big Query*
```
dados_partidas_bq = client.query('''
select *
from `cartola_partidas.partidas_*` ''')
```
Transformar em pandas dataframe
```
dados_partidas = dados_partidas_bq.to_dataframe()
dados_partidas.ano
```
### Classificação
```
dados_classificacao_bq = client.query('''
select *
from `cartola_classificacao.classificacao_*` ''')
dados_classificacao = dados_classificacao_bq.to_dataframe()
dados_classificacao.ano
```
### Jogadores
```
dados_jogadores_bq = client.query('''
select *
from `cartola_scouts.scouts_*` ''')
dados_jogadores = dados_jogadores_bq.to_dataframe()
dados_jogadores.head()
```
## Selecionar dados para criação da *view*
```
colunas_partidas = ['rodada_id', 'ano', 'clube_casa_id', 'clube_visitante_id', 'partidas_local']
partidas_view = dados_partidas[colunas_partidas]
partidas_view.head()
colunas_jogadores = ['atleta_id', 'apelido', 'rodada_id', 'ano', 'clube_id', 'clube',
'posicao', 'pontos_num', 'preco_num']
players_view = dados_jogadores[colunas_jogadores]
players_view.head()
colunas_classificacao = ['clube_id', 'rodada_id', 'ano', 'clube_rank']
classif_view = dados_classificacao[colunas_classificacao]
classif_view.head()
players_view = pd.merge(players_view, classif_view, how = 'left', on=['clube_id', 'rodada_id', 'ano'])
players_view.head()
players_casa = pd.merge(players_view, partidas_view, how = 'right',
left_on=['clube_id', 'rodada_id', 'ano'],
right_on=['clube_casa_id', 'rodada_id', 'ano'])
players_casa.head()
players_fora = pd.merge(players_view, partidas_view, how = 'right',
left_on=['clube_id', 'rodada_id', 'ano'],
right_on=['clube_visitante_id', 'rodada_id', 'ano'])
players_fora.head()
```
Ajustes casa e fora
```
players_view = players_casa.append(players_fora)
players_view.head()
players_view['mandante'] = np.where(players_view['clube_id'] == players_view['clube_casa_id'], 'S', 'N')
players_view.head()
players_view.sort_values(['atleta_id', 'rodada_id', 'ano'], inplace=True)
players_view.head()
colunas_view = ['apelido', 'posicao', 'rodada_id', 'ano', 'clube', 'clube_rank',
'mandante', 'partidas_local', 'pontos_num', 'preco_num']
players_view = players_view[colunas_view].reset_index(drop=True)
players_view['apelido'] = players_view['apelido'].astype(str)
players_view.head()
```
##Gravar a View
```
dataset_ref = client.dataset('TEMP')
table_ref = dataset_ref.table("players_view")
```
Tipo 1 - somente atualização
```
#job = client.load_table_from_dataframe(players_view, table_ref,
#job_config = bigquery.LoadJobConfig(write_disposition="WRITE_TRUNCATE"))
#job.result()
```
Tipo 2 - Atualiza e mantém adicionando as novas linhas
```
job = client.load_table_from_dataframe(players_view, table_ref)
job.result()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from google.colab import auth
from google.cloud import bigquery
import matplotlib.pyplot as plt
%matplotlib inline
auth.authenticate_user()
project_id = 'aula01in'
client = bigquery.Client(project=project_id)
dados_partidas_bq = client.query('''
select *
from `cartola_partidas.partidas_*` ''')
dados_partidas = dados_partidas_bq.to_dataframe()
dados_partidas.ano
dados_classificacao_bq = client.query('''
select *
from `cartola_classificacao.classificacao_*` ''')
dados_classificacao = dados_classificacao_bq.to_dataframe()
dados_classificacao.ano
dados_jogadores_bq = client.query('''
select *
from `cartola_scouts.scouts_*` ''')
dados_jogadores = dados_jogadores_bq.to_dataframe()
dados_jogadores.head()
colunas_partidas = ['rodada_id', 'ano', 'clube_casa_id', 'clube_visitante_id', 'partidas_local']
partidas_view = dados_partidas[colunas_partidas]
partidas_view.head()
colunas_jogadores = ['atleta_id', 'apelido', 'rodada_id', 'ano', 'clube_id', 'clube',
'posicao', 'pontos_num', 'preco_num']
players_view = dados_jogadores[colunas_jogadores]
players_view.head()
colunas_classificacao = ['clube_id', 'rodada_id', 'ano', 'clube_rank']
classif_view = dados_classificacao[colunas_classificacao]
classif_view.head()
players_view = pd.merge(players_view, classif_view, how = 'left', on=['clube_id', 'rodada_id', 'ano'])
players_view.head()
players_casa = pd.merge(players_view, partidas_view, how = 'right',
left_on=['clube_id', 'rodada_id', 'ano'],
right_on=['clube_casa_id', 'rodada_id', 'ano'])
players_casa.head()
players_fora = pd.merge(players_view, partidas_view, how = 'right',
left_on=['clube_id', 'rodada_id', 'ano'],
right_on=['clube_visitante_id', 'rodada_id', 'ano'])
players_fora.head()
players_view = players_casa.append(players_fora)
players_view.head()
players_view['mandante'] = np.where(players_view['clube_id'] == players_view['clube_casa_id'], 'S', 'N')
players_view.head()
players_view.sort_values(['atleta_id', 'rodada_id', 'ano'], inplace=True)
players_view.head()
colunas_view = ['apelido', 'posicao', 'rodada_id', 'ano', 'clube', 'clube_rank',
'mandante', 'partidas_local', 'pontos_num', 'preco_num']
players_view = players_view[colunas_view].reset_index(drop=True)
players_view['apelido'] = players_view['apelido'].astype(str)
players_view.head()
dataset_ref = client.dataset('TEMP')
table_ref = dataset_ref.table("players_view")
#job = client.load_table_from_dataframe(players_view, table_ref,
#job_config = bigquery.LoadJobConfig(write_disposition="WRITE_TRUNCATE"))
#job.result()
job = client.load_table_from_dataframe(players_view, table_ref)
job.result()
| 0.19046 | 0.764122 |
# <center>Econometrics HW_07</center>
**<center>11510691 程远星$\DeclareMathOperator*{\argmin}{argmin}
\DeclareMathOperator*{\argmax}{argmax}
\DeclareMathOperator*{\plim}{plim}
\newcommand{\using}[1]{\stackrel{\mathrm{#1}}{=}}
\newcommand{\ffrac}{\displaystyle \frac}
\newcommand{\asim}{\overset{\text{a}}{\sim}}
\newcommand{\space}{\text{ }}
\newcommand{\bspace}{\;\;\;\;}
\newcommand{\QQQ}{\boxed{?\:}}
\newcommand{\void}{\left.\right.}
\newcommand{\Tran}[1]{{#1}^{\mathrm{T}}}
\newcommand{\d}[1]{\displaystyle{#1}}
\newcommand{\CB}[1]{\left\{ #1 \right\}}
\newcommand{\SB}[1]{\left[ #1 \right]}
\newcommand{\P}[1]{\left( #1 \right)}
\newcommand{\abs}[1]{\left| #1 \right|}
\newcommand{\norm}[1]{\left\| #1 \right\|}
\newcommand{\dd}{\mathrm{d}}
\newcommand{\Exp}{\mathrm{E}}
\newcommand{\RR}{\mathbb{R}}
\newcommand{\EE}{\mathbb{E}}
\newcommand{\NN}{\mathbb{N}}
\newcommand{\ZZ}{\mathbb{Z}}
\newcommand{\QQ}{\mathbb{Q}}
\newcommand{\AcA}{\mathscr{A}}
\newcommand{\FcF}{\mathscr{F}}
\newcommand{\Var}[2][\,\!]{\mathrm{Var}_{#1}\left[#2\right]}
\newcommand{\Avar}[2][\,\!]{\mathrm{Avar}_{#1}\left[#2\right]}
\newcommand{\Cov}[2][\,\!]{\mathrm{Cov}_{#1}\left(#2\right)}
\newcommand{\Corr}[2][\,\!]{\mathrm{Corr}_{#1}\left(#2\right)}
\newcommand{\I}[1]{\mathrm{I}\left( #1 \right)}
\newcommand{\N}[1]{\mathcal{N} \left( #1 \right)}
\newcommand{\ow}{\text{otherwise}}
\void^\dagger$</center>**
## Question 2
$\P{1}$
$\bspace$Fix all other variables, we have $\Delta \log\P{\text{bwght}} = -\beta_{\text{cigs}}\Delta \text{cigs} = -0.0044\Delta \text{cigs}$ meaning that the percent change of $\text{bwght}$ is $-0.0044$ times of the change of $\text{cigs}$. So for smoking $10$ more per day, we have $\Delta \text{cigs} = 10$ and thus we have a $4.4\%$ lower $\text{bwght}$.
$\P{2}$
$\bspace$Since the parameter for the dummy variable $\text{white}$ is $0.055$ we assert that a white baby is to weigh about $5.5\%$ more than nonwhite baby, holding all other variables fixed.
$\bspace$The $t$-statistic is $55/13 = 4.230769230769231$. Well, big enough for the critical value, so the difference is statistically significant.
$\P{3}$
$\bspace$The $t$-statistic is just $1$. Not statistically significant. If it do increase by one unit, the $\text{bwght}$ will decrease by $0.3\%$.
$\P{4}$
$\bspace$The restricted model where $\text{motheduc} = \text{fatheduc} = 0$, has less samples meaning that we can't use the result in the first estimation to obtain the $F$ statistic when conducting joint significance test for $\text{motheduc}$ and $\text{fatheduc}$. To do so, we need to limit the sample who has the data for $\text{motheduc}$ and $\text{fatheduc}$ in the first estimation and redo the estimation with less samples.
## Question 3
$\P{1}$
$\bspace$Surely there is since the $t$ statistic for $\text{hsize}^2$ is $2.19/0.53=4.132075471698113$, way more than the critical value. And the optimal $\text{hsize}$ can be solved by:
$$\text{hsize}^* = \abs{\ffrac{19.30} {2\times2.19}} = 4.4063926940639275$$
$\bspace$And since it's *measured in hundreds*, the optimal size of graduating class is about $441$.
$\P{2}$
$\bspace$The difference between these two groups lies only in their sex so the different, according to the estimated model, is $45.09$. The $t$ statistic for this difference is $-45.09/4.29=-10.510489510489512$ which means that, well, statistically significant.
$\P{3}$
$\bspace$Similarly, we have the difference is $169.81$ and the $t$ statistic is $-169.81/12.71=-
13.360346184107001$. So, statistically significant.
$\P{4}$
$\bspace$Now the situation is different in that we have add the last parameter, and their difference is $62.31-169.81=-107.5$. However there's no $t$ statistic for testing the statistical significance. We can change the third composite dummy variable $\text{female}\cdot\text{black}$ to $\text{male}\cdot\text{black}$ then do the estimation again. Done!
## Question 8
$\P{1}$
$$\log\P{\text{wag}} = \beta_0 + \beta_1 \text{mus} + \beta_2\text{edu} + \beta_3\text{exp} + \beta_4\text{male} + u$$
$\P{2}$
$$\log\P{\text{wag}} = \beta_0 + \beta_1 \text{mus} + \beta_2\text{edu} + \beta_3\text{exp} + \beta_4\text{male} + \beta_5\text{male}\cdot\text{mus} + u$$
$\bspace$Add $\text{male}\cdot\text{mus}$ and things can be done. And $H_0:\beta_5 = 0$.
$\P{3}$
$$\log\P{\text{wag}} = \beta_0 + \beta_{11} \text{lmu} + \beta_{12} \text{mmu} + \beta_{13} \text{hmu} + \beta_2\text{edu} + \beta_3\text{exp} + \beta_4\text{male} + u$$
$\P{4}$
$\bspace H_0:\beta_{11} = \beta_{12} = \beta_{13} = 0$. Suppose that there're $n$ samples then $df_r = n-4$ and $df_{ur} = n-7$. The $F$ statistic is then
$$F = \ffrac{\ffrac{\text{SSR}_r - \text{SSR}_{ur}} {3}} {\ffrac{\text{SSR}_{ur}} {n-7}}$$
$\bspace$And then compare this to the critical value $F_{3,n-7}$. If it's greater than the critical value, then we reject the null hypothesis and claim that the variables indicating the four groups of marijuana user are jointly statistically significant. Otherwise, nothing can we say.
$\P{5}$
$\bspace$How could everyone be so honest that they are drug users, or addicts. So data from the survey will, as far as I can guess, will be far less than the real data, which result in a rather high significance in the model, comparing to the real one.
|
github_jupyter
|
# <center>Econometrics HW_07</center>
**<center>11510691 程远星$\DeclareMathOperator*{\argmin}{argmin}
\DeclareMathOperator*{\argmax}{argmax}
\DeclareMathOperator*{\plim}{plim}
\newcommand{\using}[1]{\stackrel{\mathrm{#1}}{=}}
\newcommand{\ffrac}{\displaystyle \frac}
\newcommand{\asim}{\overset{\text{a}}{\sim}}
\newcommand{\space}{\text{ }}
\newcommand{\bspace}{\;\;\;\;}
\newcommand{\QQQ}{\boxed{?\:}}
\newcommand{\void}{\left.\right.}
\newcommand{\Tran}[1]{{#1}^{\mathrm{T}}}
\newcommand{\d}[1]{\displaystyle{#1}}
\newcommand{\CB}[1]{\left\{ #1 \right\}}
\newcommand{\SB}[1]{\left[ #1 \right]}
\newcommand{\P}[1]{\left( #1 \right)}
\newcommand{\abs}[1]{\left| #1 \right|}
\newcommand{\norm}[1]{\left\| #1 \right\|}
\newcommand{\dd}{\mathrm{d}}
\newcommand{\Exp}{\mathrm{E}}
\newcommand{\RR}{\mathbb{R}}
\newcommand{\EE}{\mathbb{E}}
\newcommand{\NN}{\mathbb{N}}
\newcommand{\ZZ}{\mathbb{Z}}
\newcommand{\QQ}{\mathbb{Q}}
\newcommand{\AcA}{\mathscr{A}}
\newcommand{\FcF}{\mathscr{F}}
\newcommand{\Var}[2][\,\!]{\mathrm{Var}_{#1}\left[#2\right]}
\newcommand{\Avar}[2][\,\!]{\mathrm{Avar}_{#1}\left[#2\right]}
\newcommand{\Cov}[2][\,\!]{\mathrm{Cov}_{#1}\left(#2\right)}
\newcommand{\Corr}[2][\,\!]{\mathrm{Corr}_{#1}\left(#2\right)}
\newcommand{\I}[1]{\mathrm{I}\left( #1 \right)}
\newcommand{\N}[1]{\mathcal{N} \left( #1 \right)}
\newcommand{\ow}{\text{otherwise}}
\void^\dagger$</center>**
## Question 2
$\P{1}$
$\bspace$Fix all other variables, we have $\Delta \log\P{\text{bwght}} = -\beta_{\text{cigs}}\Delta \text{cigs} = -0.0044\Delta \text{cigs}$ meaning that the percent change of $\text{bwght}$ is $-0.0044$ times of the change of $\text{cigs}$. So for smoking $10$ more per day, we have $\Delta \text{cigs} = 10$ and thus we have a $4.4\%$ lower $\text{bwght}$.
$\P{2}$
$\bspace$Since the parameter for the dummy variable $\text{white}$ is $0.055$ we assert that a white baby is to weigh about $5.5\%$ more than nonwhite baby, holding all other variables fixed.
$\bspace$The $t$-statistic is $55/13 = 4.230769230769231$. Well, big enough for the critical value, so the difference is statistically significant.
$\P{3}$
$\bspace$The $t$-statistic is just $1$. Not statistically significant. If it do increase by one unit, the $\text{bwght}$ will decrease by $0.3\%$.
$\P{4}$
$\bspace$The restricted model where $\text{motheduc} = \text{fatheduc} = 0$, has less samples meaning that we can't use the result in the first estimation to obtain the $F$ statistic when conducting joint significance test for $\text{motheduc}$ and $\text{fatheduc}$. To do so, we need to limit the sample who has the data for $\text{motheduc}$ and $\text{fatheduc}$ in the first estimation and redo the estimation with less samples.
## Question 3
$\P{1}$
$\bspace$Surely there is since the $t$ statistic for $\text{hsize}^2$ is $2.19/0.53=4.132075471698113$, way more than the critical value. And the optimal $\text{hsize}$ can be solved by:
$$\text{hsize}^* = \abs{\ffrac{19.30} {2\times2.19}} = 4.4063926940639275$$
$\bspace$And since it's *measured in hundreds*, the optimal size of graduating class is about $441$.
$\P{2}$
$\bspace$The difference between these two groups lies only in their sex so the different, according to the estimated model, is $45.09$. The $t$ statistic for this difference is $-45.09/4.29=-10.510489510489512$ which means that, well, statistically significant.
$\P{3}$
$\bspace$Similarly, we have the difference is $169.81$ and the $t$ statistic is $-169.81/12.71=-
13.360346184107001$. So, statistically significant.
$\P{4}$
$\bspace$Now the situation is different in that we have add the last parameter, and their difference is $62.31-169.81=-107.5$. However there's no $t$ statistic for testing the statistical significance. We can change the third composite dummy variable $\text{female}\cdot\text{black}$ to $\text{male}\cdot\text{black}$ then do the estimation again. Done!
## Question 8
$\P{1}$
$$\log\P{\text{wag}} = \beta_0 + \beta_1 \text{mus} + \beta_2\text{edu} + \beta_3\text{exp} + \beta_4\text{male} + u$$
$\P{2}$
$$\log\P{\text{wag}} = \beta_0 + \beta_1 \text{mus} + \beta_2\text{edu} + \beta_3\text{exp} + \beta_4\text{male} + \beta_5\text{male}\cdot\text{mus} + u$$
$\bspace$Add $\text{male}\cdot\text{mus}$ and things can be done. And $H_0:\beta_5 = 0$.
$\P{3}$
$$\log\P{\text{wag}} = \beta_0 + \beta_{11} \text{lmu} + \beta_{12} \text{mmu} + \beta_{13} \text{hmu} + \beta_2\text{edu} + \beta_3\text{exp} + \beta_4\text{male} + u$$
$\P{4}$
$\bspace H_0:\beta_{11} = \beta_{12} = \beta_{13} = 0$. Suppose that there're $n$ samples then $df_r = n-4$ and $df_{ur} = n-7$. The $F$ statistic is then
$$F = \ffrac{\ffrac{\text{SSR}_r - \text{SSR}_{ur}} {3}} {\ffrac{\text{SSR}_{ur}} {n-7}}$$
$\bspace$And then compare this to the critical value $F_{3,n-7}$. If it's greater than the critical value, then we reject the null hypothesis and claim that the variables indicating the four groups of marijuana user are jointly statistically significant. Otherwise, nothing can we say.
$\P{5}$
$\bspace$How could everyone be so honest that they are drug users, or addicts. So data from the survey will, as far as I can guess, will be far less than the real data, which result in a rather high significance in the model, comparing to the real one.
| 0.52975 | 0.957675 |
```
'''讀取每行txt檔案,並將每行文字以逗號分隔,並放入data[]陣列,所需取得的資料:時間在GPGGA,經緯度在GPFLL,衛星編號與強度在GPGSV,
因此透過data陣列的第一個值進行判斷進入想取值的那行。由於要取得台灣時間,但GPGGA的值是UTC時間,故須將時間+8小時;
經緯度的部分則是依照公式將GPGLL的值轉換成google map可讀之格式,緯度公式為前兩個值+上後面值/60,經度公式為前三個值+上後面值/60;
而衛星編號與訊號強度則是在GPGSV,但因為*之前的格式有5種,為了不讓數值被順序影響所以先將GPGSV的行以*分隔,並透過一個變數cn,
計算每行當中包含多少衛星,接著再透過GPGSV的第一個數字與第二個數字判斷現在在第幾行,以輸出正確的編號與強度,並且可以透過第三個數字,
確認衛星數量是否正確,最後將每個部份所獲得的值都放在lines[]陣列裡面,並打開一個可寫的txt文件將輸出都寫入。
其中有透過folium地圖視覺化工具,先設定map畫面位置與zoom in大小,再把抓到的經緯度標記到map上,最後把標記完的map輸出到html,完成加分題。'''
import folium #地圖視覺化工具
myMap = folium.Map([24.970097, 121.26733516666667], zoom_start=50) #設定map畫面位置與zoom in大小
f = open('GPS_Data.txt', 'r') #讀檔
path = 'output.txt' #寫檔用
f1 = open(path, 'w')
lines= [] #用來放每個部份所獲得的值
for line in f.readlines(): #以每行方式進行讀取
data= []
#print(line)
data2=[]
data3=[]
for num in line.strip().split(','): #以逗號分割
data.append(num) #將分割完字串放入data陣列
if data[0]=='$GPGGA': #從GPGGA取得時間
count = 0
times=int(data[1][0:2])+8 #GPGGA的值是UTC時間,故須將時間+8小時
time = ['台灣時區時間: ',str(times),':',data[1][2:4],':',data[1][4:6],'\n'] #打時間輸出格式
lines.append(time)
f1.writelines(time) #寫入output.txt
elif data[0]=='$GPGLL': #從GPGLL取得經緯度
if data[1] != '' and data[3] != '':
map1= float(data[1][0:2])+float(data[1][2:])/60 #緯度公式為前兩個值+上後面值/60
#print(float(data[1][0:2]))
#print(float(data[1][2:]))
#print(map1)
map2= float(data[3][0:3])+float(data[3][3:])/60 #經度公式為前三個值+上後面值/60
#print(float(data[3][0:3]))
#print(float(data[3][3:]))
#print(map2)
myMap.add_child(folium.Marker(location=[map1,map2],popup='')) #把抓到的經緯度標記到map上
loc = ['緯度: ',str(map1),data[2],' 經度: ',str(map2),data[4],'\n\n'] #打經緯度輸出格式
lines.append(loc)
f1.writelines(loc) #寫入output.txt
elif data[0]=='$GPGSV': #從GPSV取得衛星編號、訊號強度
for gsv in line.strip().split('*'): #為了不讓數值被順序影響所以先將GPGSV的行以*分隔
data2.append(gsv) #將分割完字串放入data2陣列
#print(data2[0])
c=0
for num2 in data2[0].strip().split(','): #以逗號分割
c+=1
data3.append(num2) #將分割完字串放入data3陣列
#print(data3[0])
#print(c)
c1=int(data3[1]) #GPGSV的第一個數值,此回總共有幾行GPGSV,放在c1
c2=int(data3[2]) #GPGSV的第二個數值,此行為這回的第幾行GPGSV,放在c2
cn=c/4-1 #每行GPGSV最多只有4個衛星,故透過公式計算這行要幾個衛星
#print(cn)
if data3[0]=='$GPGSV' and c2<c1: #若此GPGSV不是最後一行則進入判斷
sv = ['衛星編號: ',data3[4],' ','訊號強度: ',data3[7],'\n',
'衛星編號: ',data3[8],' ','訊號強度: ',data3[11],'\n',
'衛星編號: ',data3[12],' ','訊號強度: ',data3[15],'\n',
'衛星編號: ',data3[16],' ','訊號強度: ',data3[19],'\n'] #輸出每顆衛星的編號與強度
lines.append(sv)
f1.writelines(sv) #寫入output.txt
if data3[0]=='$GPGSV' and c2==c1 and cn==4: #若此GPGSV是最後一行而衛星數有4個
sv1 = ['衛星編號: ',data3[4],' ','訊號強度: ',data3[7],'\n',
'衛星編號: ',data3[8],' ','訊號強度: ',data3[11],'\n',
'衛星編號: ',data3[12],' ','訊號強度: ',data3[15],'\n',
'衛星編號: ',data3[16],' ','訊號強度: ',data3[19],'\n'] #輸出每顆衛星的編號與強度
lines.append(sv1)
f1.writelines(sv1) #寫入output.txt
if data3[0]=='$GPGSV' and c2==c1 and cn==3: #若此GPGSV是最後一行而衛星數有3個
sv1 = ['衛星編號: ',data3[4],' ','訊號強度: ',data3[7],'\n',
'衛星編號: ',data3[8],' ','訊號強度: ',data3[11],'\n',
'衛星編號: ',data3[12],' ','訊號強度: ',data3[15],'\n',] #輸出每顆衛星的編號與強度
lines.append(sv1)
f1.writelines(sv1) #寫入output.txt
if data3[0]=='$GPGSV' and c2==c1 and cn==2: #若此GPGSV是最後一行而衛星數有2個
sv1 = ['衛星編號: ',data3[4],' ','訊號強度: ',data3[7],'\n',
'衛星編號: ',data3[8],' ','訊號強度: ',data3[11],'\n',] #輸出每顆衛星的編號與強度
lines.append(sv1)
f1.writelines(sv1) #寫入output.txt
if data3[0]=='$GPGSV' and c2==c1 and cn==1: #若此GPGSV是最後一行而衛星數有1個
sv1 = ['衛星編號: ',data3[4],' ','訊號強度: ',data3[7],'\n',] #輸出每顆衛星的編號與強度
lines.append(sv1)
f1.writelines(sv1) #寫入output.txt
myMap.save('myMap.html') #把標記完的map輸出到html
f1.close() #關閉寫入檔
f.close #關閉讀取檔
```
|
github_jupyter
|
'''讀取每行txt檔案,並將每行文字以逗號分隔,並放入data[]陣列,所需取得的資料:時間在GPGGA,經緯度在GPFLL,衛星編號與強度在GPGSV,
因此透過data陣列的第一個值進行判斷進入想取值的那行。由於要取得台灣時間,但GPGGA的值是UTC時間,故須將時間+8小時;
經緯度的部分則是依照公式將GPGLL的值轉換成google map可讀之格式,緯度公式為前兩個值+上後面值/60,經度公式為前三個值+上後面值/60;
而衛星編號與訊號強度則是在GPGSV,但因為*之前的格式有5種,為了不讓數值被順序影響所以先將GPGSV的行以*分隔,並透過一個變數cn,
計算每行當中包含多少衛星,接著再透過GPGSV的第一個數字與第二個數字判斷現在在第幾行,以輸出正確的編號與強度,並且可以透過第三個數字,
確認衛星數量是否正確,最後將每個部份所獲得的值都放在lines[]陣列裡面,並打開一個可寫的txt文件將輸出都寫入。
其中有透過folium地圖視覺化工具,先設定map畫面位置與zoom in大小,再把抓到的經緯度標記到map上,最後把標記完的map輸出到html,完成加分題。'''
import folium #地圖視覺化工具
myMap = folium.Map([24.970097, 121.26733516666667], zoom_start=50) #設定map畫面位置與zoom in大小
f = open('GPS_Data.txt', 'r') #讀檔
path = 'output.txt' #寫檔用
f1 = open(path, 'w')
lines= [] #用來放每個部份所獲得的值
for line in f.readlines(): #以每行方式進行讀取
data= []
#print(line)
data2=[]
data3=[]
for num in line.strip().split(','): #以逗號分割
data.append(num) #將分割完字串放入data陣列
if data[0]=='$GPGGA': #從GPGGA取得時間
count = 0
times=int(data[1][0:2])+8 #GPGGA的值是UTC時間,故須將時間+8小時
time = ['台灣時區時間: ',str(times),':',data[1][2:4],':',data[1][4:6],'\n'] #打時間輸出格式
lines.append(time)
f1.writelines(time) #寫入output.txt
elif data[0]=='$GPGLL': #從GPGLL取得經緯度
if data[1] != '' and data[3] != '':
map1= float(data[1][0:2])+float(data[1][2:])/60 #緯度公式為前兩個值+上後面值/60
#print(float(data[1][0:2]))
#print(float(data[1][2:]))
#print(map1)
map2= float(data[3][0:3])+float(data[3][3:])/60 #經度公式為前三個值+上後面值/60
#print(float(data[3][0:3]))
#print(float(data[3][3:]))
#print(map2)
myMap.add_child(folium.Marker(location=[map1,map2],popup='')) #把抓到的經緯度標記到map上
loc = ['緯度: ',str(map1),data[2],' 經度: ',str(map2),data[4],'\n\n'] #打經緯度輸出格式
lines.append(loc)
f1.writelines(loc) #寫入output.txt
elif data[0]=='$GPGSV': #從GPSV取得衛星編號、訊號強度
for gsv in line.strip().split('*'): #為了不讓數值被順序影響所以先將GPGSV的行以*分隔
data2.append(gsv) #將分割完字串放入data2陣列
#print(data2[0])
c=0
for num2 in data2[0].strip().split(','): #以逗號分割
c+=1
data3.append(num2) #將分割完字串放入data3陣列
#print(data3[0])
#print(c)
c1=int(data3[1]) #GPGSV的第一個數值,此回總共有幾行GPGSV,放在c1
c2=int(data3[2]) #GPGSV的第二個數值,此行為這回的第幾行GPGSV,放在c2
cn=c/4-1 #每行GPGSV最多只有4個衛星,故透過公式計算這行要幾個衛星
#print(cn)
if data3[0]=='$GPGSV' and c2<c1: #若此GPGSV不是最後一行則進入判斷
sv = ['衛星編號: ',data3[4],' ','訊號強度: ',data3[7],'\n',
'衛星編號: ',data3[8],' ','訊號強度: ',data3[11],'\n',
'衛星編號: ',data3[12],' ','訊號強度: ',data3[15],'\n',
'衛星編號: ',data3[16],' ','訊號強度: ',data3[19],'\n'] #輸出每顆衛星的編號與強度
lines.append(sv)
f1.writelines(sv) #寫入output.txt
if data3[0]=='$GPGSV' and c2==c1 and cn==4: #若此GPGSV是最後一行而衛星數有4個
sv1 = ['衛星編號: ',data3[4],' ','訊號強度: ',data3[7],'\n',
'衛星編號: ',data3[8],' ','訊號強度: ',data3[11],'\n',
'衛星編號: ',data3[12],' ','訊號強度: ',data3[15],'\n',
'衛星編號: ',data3[16],' ','訊號強度: ',data3[19],'\n'] #輸出每顆衛星的編號與強度
lines.append(sv1)
f1.writelines(sv1) #寫入output.txt
if data3[0]=='$GPGSV' and c2==c1 and cn==3: #若此GPGSV是最後一行而衛星數有3個
sv1 = ['衛星編號: ',data3[4],' ','訊號強度: ',data3[7],'\n',
'衛星編號: ',data3[8],' ','訊號強度: ',data3[11],'\n',
'衛星編號: ',data3[12],' ','訊號強度: ',data3[15],'\n',] #輸出每顆衛星的編號與強度
lines.append(sv1)
f1.writelines(sv1) #寫入output.txt
if data3[0]=='$GPGSV' and c2==c1 and cn==2: #若此GPGSV是最後一行而衛星數有2個
sv1 = ['衛星編號: ',data3[4],' ','訊號強度: ',data3[7],'\n',
'衛星編號: ',data3[8],' ','訊號強度: ',data3[11],'\n',] #輸出每顆衛星的編號與強度
lines.append(sv1)
f1.writelines(sv1) #寫入output.txt
if data3[0]=='$GPGSV' and c2==c1 and cn==1: #若此GPGSV是最後一行而衛星數有1個
sv1 = ['衛星編號: ',data3[4],' ','訊號強度: ',data3[7],'\n',] #輸出每顆衛星的編號與強度
lines.append(sv1)
f1.writelines(sv1) #寫入output.txt
myMap.save('myMap.html') #把標記完的map輸出到html
f1.close() #關閉寫入檔
f.close #關閉讀取檔
| 0.087107 | 0.627523 |
# FUNCTIONS FOR DIFFERENT METRICS <a name="Top"></a>
Levels of metrics:
1. Difference between latitude and longitude values (Zindi challenge)
2. Air distance in kilometers
3. Road distance in kilometers
4. Driving distance in minutes
5. Evaluation of driving distance with threshold ("Golden Hour")
In this notebook we will use the Uber movement data to find distances between hexbins. To map the Uber movement data, Open Street Map data is used. For more on the Uber data read:
https://medium.com/uber-movement/working-with-uber-movement-speeds-data-cc01d35937b3
More on Open Street Map:
https://wiki.openstreetmap.org/wiki/Downloading_data
## Table of contents
***
[Imports and setup](#Imports_setup)<br>
[Extract, transform and load the data](#ETL)<br>
[Data analysis](#Data_analysis)<br>
</br>
</br>
</br>
## Imports and setup <a name="Imports_setup"></a>
***
### Importing packages
```
import pandas as pd
import json
import numpy as np
import math
import matplotlib.pyplot as plt
import h3
from geopy.distance import geodesic
import sys
sys.path.insert(0, '../Scripts')
import capstone_functions as cf
```
### Setup
```
pd.set_option('display.max_columns', None)
#pd.set_option('display.max_rows', None)
pd.options.display.float_format = '{:,.3f}'.format
```
### Functions
```
def import_uber_data():
'''Imports the hourly travel times from Uber movement data.
In addition, the hexlusters used by Uber in Nairobi are imported. '''
# Read the JSON file with the hexclusters
file = open('../Inputs/540_hexclusters.json',)
hexclusters = json.load(file)
file.close()
# Find the centroids of the hexbin clusters
cluster_id = []
longitude = []
latitude = []
for i in range(len(hexclusters['features'])):
coords = hexclusters['features'][i]['geometry']['coordinates'][0]
x = [long for long, lat in coords]
y = [lat for long, lat in coords]
x_c = sum(x) / len(x)
y_c = sum(y) / len(y)
cluster_id.append(hexclusters['features'][i]['properties']['MOVEMENT_ID'])
longitude.append(x_c)
latitude.append(y_c)
# Create DataFrame with hexcluster ids and the lat and long values of the centroids
global df_hexclusters
df_hexclusters = pd.DataFrame([cluster_id, longitude, latitude]).transpose()
df_hexclusters.columns = ['cluster_id', 'longitude', 'latitude']
df_hexclusters['cluster_id'] = df_hexclusters['cluster_id'].astype('int')
df_hexclusters = cf.assign_hex_bin(df_hexclusters, 'latitude', 'longitude')
# Read the travel times for weekdays
df_tt_hourly_wd = pd.read_csv('../Inputs/nairobi-hexclusters-2018-3-OnlyWeekdays-HourlyAggregate.csv')
# Add lat and long values to the tavel time data
global df_combined_wd
df_combined_wd = df_tt_hourly_wd.merge(df_hexclusters, how='left', left_on='sourceid', right_on='cluster_id')
df_combined_wd.drop(['cluster_id'], axis=1, inplace=True)
df_combined_wd = df_combined_wd.merge(df_hexclusters, how='left', left_on='dstid', right_on='cluster_id', suffixes=('_source', '_dst'))
df_combined_wd.drop(['cluster_id'], axis=1, inplace=True)
df_combined_wd['dist_air'] = df_combined_wd[['latitude_source', 'longitude_source', 'latitude_dst', 'longitude_dst']].apply(lambda x: get_distance_air(x.latitude_source, x.longitude_source, x.latitude_dst, x.longitude_dst, h3res), axis=1)
df_combined_wd['avg_speed'] = df_combined_wd['dist_air'] / df_combined_wd['mean_travel_time'] * 3600
# Get average speeds per hour
global avg_speeds_wd
avg_speeds_wd = df_combined_wd.groupby('hod').mean()['avg_speed']
# Read the travel times for weekends
df_tt_hourly_we = pd.read_csv('../Inputs/nairobi-hexclusters-2018-3-OnlyWeekends-HourlyAggregate.csv')
# Add lat and long values to the tavel time data
global df_combined_we
df_combined_we = df_tt_hourly_we.merge(df_hexclusters, how='left', left_on='sourceid', right_on='cluster_id')
df_combined_we.drop(['cluster_id'], axis=1, inplace=True)
df_combined_we = df_combined_we.merge(df_hexclusters, how='left', left_on='dstid', right_on='cluster_id', suffixes=('_source', '_dst'))
df_combined_we.drop(['cluster_id'], axis=1, inplace=True)
df_combined_we['dist_air'] = df_combined_we[['latitude_source', 'longitude_source', 'latitude_dst', 'longitude_dst']].apply(lambda x: get_distance_air(x.latitude_source, x.longitude_source, x.latitude_dst, x.longitude_dst, h3res), axis=1)
df_combined_we['avg_speed'] = df_combined_we['dist_air'] / df_combined_we['mean_travel_time'] * 3600
# Get average speeds per hour
global avg_speeds_we
avg_speeds_we = df_combined_we.groupby('hod').mean()['avg_speed']
def get_metrics(coord_src, coord_dst, weekend, hour):
'''
Inputs:
* coord_src: H3 hexbin or coordinate as list or tuple of the origin of the trip
* coord_dst: H3 hexbin or coordinate as list or tuple of the destination of the trip
* weekend: 1 if weekend, 0 if weekday
* hour: Hour of the day as integer
Output: Returns a list with the five levels of metics:
* Zindi: Euklidean distance between latitude and longitude values (Zindi challenge)
* Air: Air distance in kilometers
* Road: Road distance in kilometers
* Time: Driving distance in minutes
* Golden: Binary value: False if driving distance below threshold ("Golden Hour"), True if above
'''
if type(coord_src) == str:
lat_src = h3.h3_to_geo(coord_src)[0]
long_src = h3.h3_to_geo(coord_src)[1]
h3res = h3.h3_get_resolution(coord_src)
elif type(coord_src) == list or tuple:
lat_src = coord_src[0]
long_src = coord_src[1]
h3res = 0
if type(coord_dst) == str:
lat_dst = h3.h3_to_geo(coord_dst)[0]
long_dst = h3.h3_to_geo(coord_dst)[1]
elif type(coord_dst) == list or tuple:
lat_dst = coord_dst[0]
long_dst = coord_dst[1]
metric = {}
# Zindi score
metric['Zindi'] = get_distance_zindi(lat_src, long_src, lat_dst, long_dst)
# Air distance
distance_air = get_distance_air(lat_src, long_src, lat_dst, long_dst, h3res)
metric['Air'] = distance_air
# Approximated road distance
detour_coef = 1.3 # Known as Henning- or Hanno-coefficient
metric['Road'] = distance_air * detour_coef
# Travel time from Uber movement data
travel_time = get_distance_time2(lat_src, long_src, lat_dst, long_dst, weekend, hour, h3res)
metric['Time'] = travel_time
# 'Golden hour'-threshold classification
golden_hour = 60 # Minutes
metric['Golden'] = travel_time > golden_hour
return metric
def get_distance_zindi(lat_src, long_src, lat_dst, long_dst):
'''
Returns the Euklidean distance between latitude and longitude values like in the Zindi-score.
'''
return ((lat_src - lat_dst)**2 + (long_src - long_dst)**2) ** 0.5
def get_distance_air(lat_src, long_src, lat_dst, long_dst, h3res):
'''
Returns the Euklidean distance between two pairs of coordinates in km.
If a distance between two points within a single cluster has to be calculated,
the average distance of all possible distances within one cluster is returned.
'''
distance_air = geodesic((lat_src, long_src), (lat_dst, long_dst)).km
if distance_air == 0:
area = h3.hex_area(resolution = h3res)
radius = (area / math.pi) ** 0.5
distance_air = 128 / (45 * math.pi) * radius
return distance_air
def get_distance_time(lat_src, long_src, lat_dst, long_dst, weekend, hour, h3res):
'''
Returns the time that is needed to cover the road distance between two pairs of coordinates in minutes based on the Uber movement data.
'''
hex_src = h3.geo_to_h3(lat=lat_src, lng=long_src, resolution=h3res)
hex_dst = h3.geo_to_h3(lat=lat_dst, lng=long_dst, resolution=h3res)
if weekend == 1:
travel_times = df_combined_we[(df_combined_we['h3_zone_6_source'] == hex_src) & \
(df_combined_we['h3_zone_6_dst'] == hex_dst) & \
(df_combined_we['hod'] == hour) \
]['mean_travel_time']
else:
travel_times = df_combined_wd[(df_combined_wd['h3_zone_6_source'] == hex_src) & \
(df_combined_wd['h3_zone_6_dst'] == hex_dst) & \
(df_combined_wd['hod'] == hour) \
]['mean_travel_time']
if len(travel_times) > 0:
travel_time = sum(travel_times) / len(travel_times) / 60
else:
#print('Not in Uber movement data.')
# len(travel_times) == 0 means that no travel times exist for this connection in the Uber movement data
# Create list of closest hex-bin clusters to source
if weekend == 1:
valid_src = df_combined_we['h3_zone_6_source'].unique()
else:
valid_src = df_combined_wd['h3_zone_6_source'].unique()
closest_hex_src = [(h3.geo_to_h3(lat_src, long_src, h3res), 0)]
for src in valid_src:
lat = h3.h3_to_geo(src)[0]
long = h3.h3_to_geo(src)[1]
dist = get_distance_air(lat_src, long_src, lat, long, h3res)
closest_hex_src.append((src, dist))
closest_hex_src.sort(key=lambda tup: tup[1])
# Create list of closest hex-bin clusters to destination
if weekend == 1:
valid_dst = df_combined_we['h3_zone_6_dst'].unique()
else:
valid_dst = df_combined_wd['h3_zone_6_dst'].unique()
closest_hex_dst = [(h3.geo_to_h3(lat_dst, long_dst, h3res), 0)]
for dst in valid_dst:
lat = h3.h3_to_geo(dst)[0]
long = h3.h3_to_geo(dst)[1]
dist = get_distance_air(lat_dst, long_dst, lat, long, h3res)
closest_hex_dst.append((dst, dist))
closest_hex_dst.sort(key=lambda tup: tup[1])
# Run through the closest clusters and check if there is connection between them
i_src = 0
i_dst = 0
if (closest_hex_src[1][1] - closest_hex_src[0][1]) > (closest_hex_dst[1][1] - closest_hex_dst[0][1]):
i_src += 1
switcher = 1
else:
i_dst += 1
switcher = 0
looking = True
while looking:
if weekend == 1:
alt_times = df_combined_we[(df_combined_we['h3_zone_6_source'] == closest_hex_src[i_src][0]) & \
(df_combined_we['h3_zone_6_dst'] == closest_hex_dst[i_dst][0]) & \
(df_combined_we['hod'] == hour) \
]['mean_travel_time']
else:
alt_times = df_combined_wd[(df_combined_wd['h3_zone_6_source'] == closest_hex_src[i_src][0]) & \
(df_combined_wd['h3_zone_6_dst'] == closest_hex_dst[i_dst][0]) & \
(df_combined_wd['hod'] == hour) \
]['mean_travel_time']
if len(alt_times) > 0:
# Get average speed from this connection (based on air distance)
alt_lat_src = h3.h3_to_geo(closest_hex_src[i_src][0])[0]
alt_long_src = h3.h3_to_geo(closest_hex_src[i_src][0])[1]
alt_lat_dst = h3.h3_to_geo(closest_hex_dst[i_dst][0])[0]
alt_long_dst = h3.h3_to_geo(closest_hex_dst[i_dst][0])[1]
alt_distance_air = get_distance_air(alt_lat_src, alt_long_src, alt_lat_dst, alt_long_dst, h3res)
#print('alt_distance_air:', alt_distance_air, 'km')
alt_time = sum(alt_times) / len(alt_times) / 60
#print('alt_time:', alt_time, 'min')
alt_speed = alt_distance_air / alt_time
#print('alt_speed:', alt_speed*60, 'km/h')
looking = False
if switcher % 2 == 0:
i_src += 1
else:
i_dst += 1
switcher += 1
# Get air distance between two original coordinates
orig_dist = get_distance_air(lat_src, long_src, lat_dst, long_dst, h3res)
#print('orig_dist:', orig_dist, 'km')
# Divide air distance through average speed
travel_time = orig_dist / alt_speed
return travel_time
def get_distance_time2(lat_src, long_src, lat_dst, long_dst, weekend, hour, h3res):
'''
Returns the time that is needed to cover the road distance between two pairs of coordinates in minutes based on the Uber movement data.
'''
hex_src = h3.geo_to_h3(lat=lat_src, lng=long_src, resolution=h3res)
hex_dst = h3.geo_to_h3(lat=lat_dst, lng=long_dst, resolution=h3res)
if weekend == 1:
travel_times = df_combined_we[(df_combined_we['h3_zone_6_source'] == hex_src) & \
(df_combined_we['h3_zone_6_dst'] == hex_dst) & \
(df_combined_we['hod'] == hour) \
]['mean_travel_time']
else:
travel_times = df_combined_wd[(df_combined_wd['h3_zone_6_source'] == hex_src) & \
(df_combined_wd['h3_zone_6_dst'] == hex_dst) & \
(df_combined_wd['hod'] == hour) \
]['mean_travel_time']
if len(travel_times) > 0:
travel_time = sum(travel_times) / len(travel_times) / 60
else:
# len(travel_times) == 0 means that no travel times exist for this connection in the Uber movement data
# Get air distance between two original coordinates
orig_dist = get_distance_air(lat_src, long_src, lat_dst, long_dst, h3res)
# Divide air distance through average speed
if weekend == 1:
travel_time = orig_dist / avg_speeds_we[hour] * 60
else:
travel_time = orig_dist / avg_speeds_wd[hour] * 60
return travel_time
```
[Back to top](#Top)<br>
</br>
</br>
</br>
## Extract, transform and load the data <a name="ETL"></a>
***
```
df_tt_hourly_wd = pd.read_csv('../Inputs/nairobi-hexclusters-2018-3-OnlyWeekdays-HourlyAggregate.csv')
print(df_tt_hourly_wd.shape)
df_tt_hourly_wd.head()
df_tt_hourly_we = pd.read_csv('../Inputs/nairobi-hexclusters-2018-3-OnlyWeekends-HourlyAggregate.csv')
print(df_tt_hourly_we.shape)
df_tt_hourly_we.head()
file = open('../Inputs/540_hexclusters.json',)
hexclusters = json.load(file)
file.close()
print(hexclusters['features'][0])
coords = hexclusters['features'][0]['geometry']['coordinates'][0]
x = [long for long, lat in coords]
y = [lat for long, lat in coords]
x_c = sum(x) / len(x)
y_c = sum(y) / len(y)
plt.scatter(x=x, y=y)
plt.scatter(x=x_c, y=y_c);
cluster_id = []
longitude = []
latitude = []
for i in range(len(hexclusters['features'])):
coords = hexclusters['features'][i]['geometry']['coordinates'][0]
x = [long for long, lat in coords]
y = [lat for long, lat in coords]
x_c = sum(x) / len(x)
y_c = sum(y) / len(y)
cluster_id.append(hexclusters['features'][i]['properties']['MOVEMENT_ID'])
longitude.append(x_c)
latitude.append(y_c)
plt.scatter(x=longitude, y=latitude)
df_hexclusters = pd.DataFrame([cluster_id, longitude, latitude]).transpose()
df_hexclusters.columns = ['cluster_id', 'longitude', 'latitude']
df_hexclusters['cluster_id'] = df_hexclusters['cluster_id'].astype('int')
df_hexclusters = cf.assign_hex_bin(df_hexclusters, 'latitude', 'longitude')
print(df_hexclusters.shape)
df_hexclusters.head()
df_hexclusters.nunique()
```
[Back to top](#Top)<br>
</br>
</br>
</br>
## Data analysis <a name="Data_analysis"></a>
***
```
h3res = h3.h3_get_resolution(df_hexclusters.loc[0, 'h3_zone_6'])
df_combined_wd = df_tt_hourly_wd.merge(df_hexclusters, how='left', left_on='sourceid', right_on='cluster_id')
df_combined_wd.drop(['cluster_id'], axis=1, inplace=True)
df_combined_wd = df_combined_wd.merge(df_hexclusters, how='left', left_on='dstid', right_on='cluster_id', suffixes=('_source', '_dst'))
df_combined_wd.drop(['cluster_id'], axis=1, inplace=True)
df_combined_wd['dist_air'] = df_combined_wd[['latitude_source', 'longitude_source', 'latitude_dst', 'longitude_dst']].apply(lambda x: get_distance_air(x.latitude_source, x.longitude_source, x.latitude_dst, x.longitude_dst, h3res), axis=1)
df_combined_wd['avg_speed'] = df_combined_wd['dist_air'] / df_combined_wd['mean_travel_time'] * 3600
print(df_combined_wd.shape)
df_combined_wd.head()
avg_speeds_wd = df_combined_wd.groupby('hod').mean()['avg_speed']
avg_speeds_wd
df_combined_we = df_tt_hourly_we.merge(df_hexclusters, how='left', left_on='sourceid', right_on='cluster_id')
df_combined_we.drop(['cluster_id'], axis=1, inplace=True)
df_combined_we = df_combined_we.merge(df_hexclusters, how='left', left_on='dstid', right_on='cluster_id', suffixes=('_source', '_dst'))
df_combined_we.drop(['cluster_id'], axis=1, inplace=True)
df_combined_we['dist_air'] = df_combined_we[['latitude_source', 'longitude_source', 'latitude_dst', 'longitude_dst']].apply(lambda x: get_distance_air(x.latitude_source, x.longitude_source, x.latitude_dst, x.longitude_dst, h3res), axis=1)
df_combined_we['avg_speed'] = df_combined_we['dist_air'] / df_combined_we['mean_travel_time'] * 3600
print(df_combined_we.shape)
df_combined_we.head()
avg_speeds_we = df_combined_we.groupby('hod').mean()['avg_speed']
avg_speeds_we
get_distance_time(lat_src=53.561468372732406, long_src=9.915019036741262, lat_dst=-1.188850, long_dst=36.931382, weekend=1, hour=7, h3res=6)
get_distance_time2(lat_src=53.561468372732406, long_src=9.915019036741262, lat_dst=-1.188850, long_dst=36.931382, weekend=1, hour=7, h3res=6)
get_metrics((64.561468372732406, 9.915019036741262), (-1.188850, 36.931382), weekend=1, hour=7)
get_metrics('867a6e55fffffff', '867a6e55fffffff', weekend=0, hour=1)
lat_src = -1.328
long_src = 36.716
lat_dst = -1.188850
long_dst = 36.931382
hour = 5
h3res = 6
hex_src = h3.geo_to_h3(lat=lat_src, lng=long_src, resolution=h3res)
hex_dst = h3.geo_to_h3(lat=lat_dst, lng=long_dst, resolution=h3res)
df_combined_we[(df_combined_we['h3_zone_6_source'] == hex_src) & \
(df_combined_we['h3_zone_6_dst'] == hex_dst) & \
(df_combined_we['hod'] == hour) \
]['mean_travel_time']
```
[Back to top](#Top)<br>
|
github_jupyter
|
import pandas as pd
import json
import numpy as np
import math
import matplotlib.pyplot as plt
import h3
from geopy.distance import geodesic
import sys
sys.path.insert(0, '../Scripts')
import capstone_functions as cf
pd.set_option('display.max_columns', None)
#pd.set_option('display.max_rows', None)
pd.options.display.float_format = '{:,.3f}'.format
def import_uber_data():
'''Imports the hourly travel times from Uber movement data.
In addition, the hexlusters used by Uber in Nairobi are imported. '''
# Read the JSON file with the hexclusters
file = open('../Inputs/540_hexclusters.json',)
hexclusters = json.load(file)
file.close()
# Find the centroids of the hexbin clusters
cluster_id = []
longitude = []
latitude = []
for i in range(len(hexclusters['features'])):
coords = hexclusters['features'][i]['geometry']['coordinates'][0]
x = [long for long, lat in coords]
y = [lat for long, lat in coords]
x_c = sum(x) / len(x)
y_c = sum(y) / len(y)
cluster_id.append(hexclusters['features'][i]['properties']['MOVEMENT_ID'])
longitude.append(x_c)
latitude.append(y_c)
# Create DataFrame with hexcluster ids and the lat and long values of the centroids
global df_hexclusters
df_hexclusters = pd.DataFrame([cluster_id, longitude, latitude]).transpose()
df_hexclusters.columns = ['cluster_id', 'longitude', 'latitude']
df_hexclusters['cluster_id'] = df_hexclusters['cluster_id'].astype('int')
df_hexclusters = cf.assign_hex_bin(df_hexclusters, 'latitude', 'longitude')
# Read the travel times for weekdays
df_tt_hourly_wd = pd.read_csv('../Inputs/nairobi-hexclusters-2018-3-OnlyWeekdays-HourlyAggregate.csv')
# Add lat and long values to the tavel time data
global df_combined_wd
df_combined_wd = df_tt_hourly_wd.merge(df_hexclusters, how='left', left_on='sourceid', right_on='cluster_id')
df_combined_wd.drop(['cluster_id'], axis=1, inplace=True)
df_combined_wd = df_combined_wd.merge(df_hexclusters, how='left', left_on='dstid', right_on='cluster_id', suffixes=('_source', '_dst'))
df_combined_wd.drop(['cluster_id'], axis=1, inplace=True)
df_combined_wd['dist_air'] = df_combined_wd[['latitude_source', 'longitude_source', 'latitude_dst', 'longitude_dst']].apply(lambda x: get_distance_air(x.latitude_source, x.longitude_source, x.latitude_dst, x.longitude_dst, h3res), axis=1)
df_combined_wd['avg_speed'] = df_combined_wd['dist_air'] / df_combined_wd['mean_travel_time'] * 3600
# Get average speeds per hour
global avg_speeds_wd
avg_speeds_wd = df_combined_wd.groupby('hod').mean()['avg_speed']
# Read the travel times for weekends
df_tt_hourly_we = pd.read_csv('../Inputs/nairobi-hexclusters-2018-3-OnlyWeekends-HourlyAggregate.csv')
# Add lat and long values to the tavel time data
global df_combined_we
df_combined_we = df_tt_hourly_we.merge(df_hexclusters, how='left', left_on='sourceid', right_on='cluster_id')
df_combined_we.drop(['cluster_id'], axis=1, inplace=True)
df_combined_we = df_combined_we.merge(df_hexclusters, how='left', left_on='dstid', right_on='cluster_id', suffixes=('_source', '_dst'))
df_combined_we.drop(['cluster_id'], axis=1, inplace=True)
df_combined_we['dist_air'] = df_combined_we[['latitude_source', 'longitude_source', 'latitude_dst', 'longitude_dst']].apply(lambda x: get_distance_air(x.latitude_source, x.longitude_source, x.latitude_dst, x.longitude_dst, h3res), axis=1)
df_combined_we['avg_speed'] = df_combined_we['dist_air'] / df_combined_we['mean_travel_time'] * 3600
# Get average speeds per hour
global avg_speeds_we
avg_speeds_we = df_combined_we.groupby('hod').mean()['avg_speed']
def get_metrics(coord_src, coord_dst, weekend, hour):
'''
Inputs:
* coord_src: H3 hexbin or coordinate as list or tuple of the origin of the trip
* coord_dst: H3 hexbin or coordinate as list or tuple of the destination of the trip
* weekend: 1 if weekend, 0 if weekday
* hour: Hour of the day as integer
Output: Returns a list with the five levels of metics:
* Zindi: Euklidean distance between latitude and longitude values (Zindi challenge)
* Air: Air distance in kilometers
* Road: Road distance in kilometers
* Time: Driving distance in minutes
* Golden: Binary value: False if driving distance below threshold ("Golden Hour"), True if above
'''
if type(coord_src) == str:
lat_src = h3.h3_to_geo(coord_src)[0]
long_src = h3.h3_to_geo(coord_src)[1]
h3res = h3.h3_get_resolution(coord_src)
elif type(coord_src) == list or tuple:
lat_src = coord_src[0]
long_src = coord_src[1]
h3res = 0
if type(coord_dst) == str:
lat_dst = h3.h3_to_geo(coord_dst)[0]
long_dst = h3.h3_to_geo(coord_dst)[1]
elif type(coord_dst) == list or tuple:
lat_dst = coord_dst[0]
long_dst = coord_dst[1]
metric = {}
# Zindi score
metric['Zindi'] = get_distance_zindi(lat_src, long_src, lat_dst, long_dst)
# Air distance
distance_air = get_distance_air(lat_src, long_src, lat_dst, long_dst, h3res)
metric['Air'] = distance_air
# Approximated road distance
detour_coef = 1.3 # Known as Henning- or Hanno-coefficient
metric['Road'] = distance_air * detour_coef
# Travel time from Uber movement data
travel_time = get_distance_time2(lat_src, long_src, lat_dst, long_dst, weekend, hour, h3res)
metric['Time'] = travel_time
# 'Golden hour'-threshold classification
golden_hour = 60 # Minutes
metric['Golden'] = travel_time > golden_hour
return metric
def get_distance_zindi(lat_src, long_src, lat_dst, long_dst):
'''
Returns the Euklidean distance between latitude and longitude values like in the Zindi-score.
'''
return ((lat_src - lat_dst)**2 + (long_src - long_dst)**2) ** 0.5
def get_distance_air(lat_src, long_src, lat_dst, long_dst, h3res):
'''
Returns the Euklidean distance between two pairs of coordinates in km.
If a distance between two points within a single cluster has to be calculated,
the average distance of all possible distances within one cluster is returned.
'''
distance_air = geodesic((lat_src, long_src), (lat_dst, long_dst)).km
if distance_air == 0:
area = h3.hex_area(resolution = h3res)
radius = (area / math.pi) ** 0.5
distance_air = 128 / (45 * math.pi) * radius
return distance_air
def get_distance_time(lat_src, long_src, lat_dst, long_dst, weekend, hour, h3res):
'''
Returns the time that is needed to cover the road distance between two pairs of coordinates in minutes based on the Uber movement data.
'''
hex_src = h3.geo_to_h3(lat=lat_src, lng=long_src, resolution=h3res)
hex_dst = h3.geo_to_h3(lat=lat_dst, lng=long_dst, resolution=h3res)
if weekend == 1:
travel_times = df_combined_we[(df_combined_we['h3_zone_6_source'] == hex_src) & \
(df_combined_we['h3_zone_6_dst'] == hex_dst) & \
(df_combined_we['hod'] == hour) \
]['mean_travel_time']
else:
travel_times = df_combined_wd[(df_combined_wd['h3_zone_6_source'] == hex_src) & \
(df_combined_wd['h3_zone_6_dst'] == hex_dst) & \
(df_combined_wd['hod'] == hour) \
]['mean_travel_time']
if len(travel_times) > 0:
travel_time = sum(travel_times) / len(travel_times) / 60
else:
#print('Not in Uber movement data.')
# len(travel_times) == 0 means that no travel times exist for this connection in the Uber movement data
# Create list of closest hex-bin clusters to source
if weekend == 1:
valid_src = df_combined_we['h3_zone_6_source'].unique()
else:
valid_src = df_combined_wd['h3_zone_6_source'].unique()
closest_hex_src = [(h3.geo_to_h3(lat_src, long_src, h3res), 0)]
for src in valid_src:
lat = h3.h3_to_geo(src)[0]
long = h3.h3_to_geo(src)[1]
dist = get_distance_air(lat_src, long_src, lat, long, h3res)
closest_hex_src.append((src, dist))
closest_hex_src.sort(key=lambda tup: tup[1])
# Create list of closest hex-bin clusters to destination
if weekend == 1:
valid_dst = df_combined_we['h3_zone_6_dst'].unique()
else:
valid_dst = df_combined_wd['h3_zone_6_dst'].unique()
closest_hex_dst = [(h3.geo_to_h3(lat_dst, long_dst, h3res), 0)]
for dst in valid_dst:
lat = h3.h3_to_geo(dst)[0]
long = h3.h3_to_geo(dst)[1]
dist = get_distance_air(lat_dst, long_dst, lat, long, h3res)
closest_hex_dst.append((dst, dist))
closest_hex_dst.sort(key=lambda tup: tup[1])
# Run through the closest clusters and check if there is connection between them
i_src = 0
i_dst = 0
if (closest_hex_src[1][1] - closest_hex_src[0][1]) > (closest_hex_dst[1][1] - closest_hex_dst[0][1]):
i_src += 1
switcher = 1
else:
i_dst += 1
switcher = 0
looking = True
while looking:
if weekend == 1:
alt_times = df_combined_we[(df_combined_we['h3_zone_6_source'] == closest_hex_src[i_src][0]) & \
(df_combined_we['h3_zone_6_dst'] == closest_hex_dst[i_dst][0]) & \
(df_combined_we['hod'] == hour) \
]['mean_travel_time']
else:
alt_times = df_combined_wd[(df_combined_wd['h3_zone_6_source'] == closest_hex_src[i_src][0]) & \
(df_combined_wd['h3_zone_6_dst'] == closest_hex_dst[i_dst][0]) & \
(df_combined_wd['hod'] == hour) \
]['mean_travel_time']
if len(alt_times) > 0:
# Get average speed from this connection (based on air distance)
alt_lat_src = h3.h3_to_geo(closest_hex_src[i_src][0])[0]
alt_long_src = h3.h3_to_geo(closest_hex_src[i_src][0])[1]
alt_lat_dst = h3.h3_to_geo(closest_hex_dst[i_dst][0])[0]
alt_long_dst = h3.h3_to_geo(closest_hex_dst[i_dst][0])[1]
alt_distance_air = get_distance_air(alt_lat_src, alt_long_src, alt_lat_dst, alt_long_dst, h3res)
#print('alt_distance_air:', alt_distance_air, 'km')
alt_time = sum(alt_times) / len(alt_times) / 60
#print('alt_time:', alt_time, 'min')
alt_speed = alt_distance_air / alt_time
#print('alt_speed:', alt_speed*60, 'km/h')
looking = False
if switcher % 2 == 0:
i_src += 1
else:
i_dst += 1
switcher += 1
# Get air distance between two original coordinates
orig_dist = get_distance_air(lat_src, long_src, lat_dst, long_dst, h3res)
#print('orig_dist:', orig_dist, 'km')
# Divide air distance through average speed
travel_time = orig_dist / alt_speed
return travel_time
def get_distance_time2(lat_src, long_src, lat_dst, long_dst, weekend, hour, h3res):
'''
Returns the time that is needed to cover the road distance between two pairs of coordinates in minutes based on the Uber movement data.
'''
hex_src = h3.geo_to_h3(lat=lat_src, lng=long_src, resolution=h3res)
hex_dst = h3.geo_to_h3(lat=lat_dst, lng=long_dst, resolution=h3res)
if weekend == 1:
travel_times = df_combined_we[(df_combined_we['h3_zone_6_source'] == hex_src) & \
(df_combined_we['h3_zone_6_dst'] == hex_dst) & \
(df_combined_we['hod'] == hour) \
]['mean_travel_time']
else:
travel_times = df_combined_wd[(df_combined_wd['h3_zone_6_source'] == hex_src) & \
(df_combined_wd['h3_zone_6_dst'] == hex_dst) & \
(df_combined_wd['hod'] == hour) \
]['mean_travel_time']
if len(travel_times) > 0:
travel_time = sum(travel_times) / len(travel_times) / 60
else:
# len(travel_times) == 0 means that no travel times exist for this connection in the Uber movement data
# Get air distance between two original coordinates
orig_dist = get_distance_air(lat_src, long_src, lat_dst, long_dst, h3res)
# Divide air distance through average speed
if weekend == 1:
travel_time = orig_dist / avg_speeds_we[hour] * 60
else:
travel_time = orig_dist / avg_speeds_wd[hour] * 60
return travel_time
df_tt_hourly_wd = pd.read_csv('../Inputs/nairobi-hexclusters-2018-3-OnlyWeekdays-HourlyAggregate.csv')
print(df_tt_hourly_wd.shape)
df_tt_hourly_wd.head()
df_tt_hourly_we = pd.read_csv('../Inputs/nairobi-hexclusters-2018-3-OnlyWeekends-HourlyAggregate.csv')
print(df_tt_hourly_we.shape)
df_tt_hourly_we.head()
file = open('../Inputs/540_hexclusters.json',)
hexclusters = json.load(file)
file.close()
print(hexclusters['features'][0])
coords = hexclusters['features'][0]['geometry']['coordinates'][0]
x = [long for long, lat in coords]
y = [lat for long, lat in coords]
x_c = sum(x) / len(x)
y_c = sum(y) / len(y)
plt.scatter(x=x, y=y)
plt.scatter(x=x_c, y=y_c);
cluster_id = []
longitude = []
latitude = []
for i in range(len(hexclusters['features'])):
coords = hexclusters['features'][i]['geometry']['coordinates'][0]
x = [long for long, lat in coords]
y = [lat for long, lat in coords]
x_c = sum(x) / len(x)
y_c = sum(y) / len(y)
cluster_id.append(hexclusters['features'][i]['properties']['MOVEMENT_ID'])
longitude.append(x_c)
latitude.append(y_c)
plt.scatter(x=longitude, y=latitude)
df_hexclusters = pd.DataFrame([cluster_id, longitude, latitude]).transpose()
df_hexclusters.columns = ['cluster_id', 'longitude', 'latitude']
df_hexclusters['cluster_id'] = df_hexclusters['cluster_id'].astype('int')
df_hexclusters = cf.assign_hex_bin(df_hexclusters, 'latitude', 'longitude')
print(df_hexclusters.shape)
df_hexclusters.head()
df_hexclusters.nunique()
h3res = h3.h3_get_resolution(df_hexclusters.loc[0, 'h3_zone_6'])
df_combined_wd = df_tt_hourly_wd.merge(df_hexclusters, how='left', left_on='sourceid', right_on='cluster_id')
df_combined_wd.drop(['cluster_id'], axis=1, inplace=True)
df_combined_wd = df_combined_wd.merge(df_hexclusters, how='left', left_on='dstid', right_on='cluster_id', suffixes=('_source', '_dst'))
df_combined_wd.drop(['cluster_id'], axis=1, inplace=True)
df_combined_wd['dist_air'] = df_combined_wd[['latitude_source', 'longitude_source', 'latitude_dst', 'longitude_dst']].apply(lambda x: get_distance_air(x.latitude_source, x.longitude_source, x.latitude_dst, x.longitude_dst, h3res), axis=1)
df_combined_wd['avg_speed'] = df_combined_wd['dist_air'] / df_combined_wd['mean_travel_time'] * 3600
print(df_combined_wd.shape)
df_combined_wd.head()
avg_speeds_wd = df_combined_wd.groupby('hod').mean()['avg_speed']
avg_speeds_wd
df_combined_we = df_tt_hourly_we.merge(df_hexclusters, how='left', left_on='sourceid', right_on='cluster_id')
df_combined_we.drop(['cluster_id'], axis=1, inplace=True)
df_combined_we = df_combined_we.merge(df_hexclusters, how='left', left_on='dstid', right_on='cluster_id', suffixes=('_source', '_dst'))
df_combined_we.drop(['cluster_id'], axis=1, inplace=True)
df_combined_we['dist_air'] = df_combined_we[['latitude_source', 'longitude_source', 'latitude_dst', 'longitude_dst']].apply(lambda x: get_distance_air(x.latitude_source, x.longitude_source, x.latitude_dst, x.longitude_dst, h3res), axis=1)
df_combined_we['avg_speed'] = df_combined_we['dist_air'] / df_combined_we['mean_travel_time'] * 3600
print(df_combined_we.shape)
df_combined_we.head()
avg_speeds_we = df_combined_we.groupby('hod').mean()['avg_speed']
avg_speeds_we
get_distance_time(lat_src=53.561468372732406, long_src=9.915019036741262, lat_dst=-1.188850, long_dst=36.931382, weekend=1, hour=7, h3res=6)
get_distance_time2(lat_src=53.561468372732406, long_src=9.915019036741262, lat_dst=-1.188850, long_dst=36.931382, weekend=1, hour=7, h3res=6)
get_metrics((64.561468372732406, 9.915019036741262), (-1.188850, 36.931382), weekend=1, hour=7)
get_metrics('867a6e55fffffff', '867a6e55fffffff', weekend=0, hour=1)
lat_src = -1.328
long_src = 36.716
lat_dst = -1.188850
long_dst = 36.931382
hour = 5
h3res = 6
hex_src = h3.geo_to_h3(lat=lat_src, lng=long_src, resolution=h3res)
hex_dst = h3.geo_to_h3(lat=lat_dst, lng=long_dst, resolution=h3res)
df_combined_we[(df_combined_we['h3_zone_6_source'] == hex_src) & \
(df_combined_we['h3_zone_6_dst'] == hex_dst) & \
(df_combined_we['hod'] == hour) \
]['mean_travel_time']
| 0.403097 | 0.835484 |
# Fig. 3 - Bifurcation diagram with respect to $e$
Here we compute the bifurcation diagrams with respect to $e$ variable.
```
%matplotlib inline
from functools import partial
from itertools import chain
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
import xarray as xr
from neurolib.models.multimodel import MultiModel
from neurolib.utils.stimulus import ZeroInput
from hippocampus import HippocampalCA3Node
from utils import run_in_parallel
plt.style.use("default_light")
```
## Helper function
- `compute_wrt_e`: computes steady state solution of the model for a given value of $e$ for two initial conditions: one is starting in the SWR state, while the second one is starting in the non-SWR state; computation is done with both backends available in the `MultiModel` framework of `neurolib`
```
def compute_wrt_e(e, backend="numba"):
results = []
for init_c in [0, 1]:
hc = HippocampalCA3Node(constant_depression=True)
for mass in hc:
mass._noise_input = [ZeroInput()]
m = MultiModel.init_node(hc)
if init_c == 0:
# SWR state
m.model_instance.initial_state = np.array([0.044, 0.092, 0.0])
else:
# non-SWR state
m.model_instance.initial_state = np.array([0.0, 0.0, 0.0125])
m.params["backend"] = backend
m.params["duration"] = 2000
m.params["sampling_dt"] = 1.0
m.params["dt"] = 0.05
m.params["*aSWR*e|noise"] = e
m.run()
arr = xr.DataArray(
np.array(
[
float(m.r_mean_EXC.T[-1]),
float(m.r_mean_INH.T[-1]),
float(m.r_mean_aSWR.T[-1]),
]
),
dims=["node"],
coords={"node": ["P", "B", "A"]},
).assign_coords({"e": e, "init_c": init_c})
results.append(arr)
return results
```
## Compute
Compute steady state solutions in parallel and stack them using `xr.DataArray`
```
es = np.linspace(0.0, 1.0, 151)
res = run_in_parallel(
partial(compute_wrt_e, backend="numba"), es, workers=6
)
bif_wrt_e = (
xr.concat(list(chain.from_iterable(res)), dim="new")
.set_index(new=["e", "init_c"])
.unstack("new")
)
```
## Plot
```
fig, axs = plt.subplots(ncols=1, nrows=3, figsize=(6, 10), sharex=True)
colors = ["C0", "C1", "C5"]
maxs = [0.07, 0.12, 0.015]
for i, var in enumerate(bif_wrt_e["node"]):
axs[i].plot(
es, bif_wrt_e.sel({"node": var}).values, "o", color=colors[i], markersize=4
)
axs[i].set_ylim([-0.002, maxs[i]])
axs[i].set_ylabel(f"{var.values} [kHz]")
axs[i].axvline(0.4, linestyle="--", color="#555555")
axs[i].axvline(0.5, linestyle="--", color="#555555")
sns.despine(ax=axs[i])
axs[-1].set_xlabel("e")
plt.savefig("../figs/bifurcation_wrt_e.pdf", transparent=True, bbox_inches="tight")
```
## Repeat for `jitcdde` backend
```
es = np.linspace(0.0, 1.0, 151)
res = run_in_parallel(
partial(compute_wrt_e, backend="jitcdde"), es, workers=6
)
bif_wrt_e = (
xr.concat(list(chain.from_iterable(res)), dim="new")
.set_index(new=["e", "init_c"])
.unstack("new")
)
fig, axs = plt.subplots(ncols=1, nrows=3, figsize=(6, 10), sharex=True)
colors = ["C0", "C1", "C5"]
maxs = [0.07, 0.12, 0.015]
for i, var in enumerate(bif_wrt_e["node"]):
axs[i].plot(
es, bif_wrt_e.sel({"node": var}).values, "o", color=colors[i], markersize=4
)
axs[i].set_ylim([-0.002, maxs[i]])
axs[i].set_ylabel(f"{var.values} [kHz]")
axs[i].axvline(0.4, linestyle="--", color="#555555")
axs[i].axvline(0.5, linestyle="--", color="#555555")
sns.despine(ax=axs[i])
axs[-1].set_xlabel("e")
```
## Profit
We got same figures, all good.
|
github_jupyter
|
%matplotlib inline
from functools import partial
from itertools import chain
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
import xarray as xr
from neurolib.models.multimodel import MultiModel
from neurolib.utils.stimulus import ZeroInput
from hippocampus import HippocampalCA3Node
from utils import run_in_parallel
plt.style.use("default_light")
def compute_wrt_e(e, backend="numba"):
results = []
for init_c in [0, 1]:
hc = HippocampalCA3Node(constant_depression=True)
for mass in hc:
mass._noise_input = [ZeroInput()]
m = MultiModel.init_node(hc)
if init_c == 0:
# SWR state
m.model_instance.initial_state = np.array([0.044, 0.092, 0.0])
else:
# non-SWR state
m.model_instance.initial_state = np.array([0.0, 0.0, 0.0125])
m.params["backend"] = backend
m.params["duration"] = 2000
m.params["sampling_dt"] = 1.0
m.params["dt"] = 0.05
m.params["*aSWR*e|noise"] = e
m.run()
arr = xr.DataArray(
np.array(
[
float(m.r_mean_EXC.T[-1]),
float(m.r_mean_INH.T[-1]),
float(m.r_mean_aSWR.T[-1]),
]
),
dims=["node"],
coords={"node": ["P", "B", "A"]},
).assign_coords({"e": e, "init_c": init_c})
results.append(arr)
return results
es = np.linspace(0.0, 1.0, 151)
res = run_in_parallel(
partial(compute_wrt_e, backend="numba"), es, workers=6
)
bif_wrt_e = (
xr.concat(list(chain.from_iterable(res)), dim="new")
.set_index(new=["e", "init_c"])
.unstack("new")
)
fig, axs = plt.subplots(ncols=1, nrows=3, figsize=(6, 10), sharex=True)
colors = ["C0", "C1", "C5"]
maxs = [0.07, 0.12, 0.015]
for i, var in enumerate(bif_wrt_e["node"]):
axs[i].plot(
es, bif_wrt_e.sel({"node": var}).values, "o", color=colors[i], markersize=4
)
axs[i].set_ylim([-0.002, maxs[i]])
axs[i].set_ylabel(f"{var.values} [kHz]")
axs[i].axvline(0.4, linestyle="--", color="#555555")
axs[i].axvline(0.5, linestyle="--", color="#555555")
sns.despine(ax=axs[i])
axs[-1].set_xlabel("e")
plt.savefig("../figs/bifurcation_wrt_e.pdf", transparent=True, bbox_inches="tight")
es = np.linspace(0.0, 1.0, 151)
res = run_in_parallel(
partial(compute_wrt_e, backend="jitcdde"), es, workers=6
)
bif_wrt_e = (
xr.concat(list(chain.from_iterable(res)), dim="new")
.set_index(new=["e", "init_c"])
.unstack("new")
)
fig, axs = plt.subplots(ncols=1, nrows=3, figsize=(6, 10), sharex=True)
colors = ["C0", "C1", "C5"]
maxs = [0.07, 0.12, 0.015]
for i, var in enumerate(bif_wrt_e["node"]):
axs[i].plot(
es, bif_wrt_e.sel({"node": var}).values, "o", color=colors[i], markersize=4
)
axs[i].set_ylim([-0.002, maxs[i]])
axs[i].set_ylabel(f"{var.values} [kHz]")
axs[i].axvline(0.4, linestyle="--", color="#555555")
axs[i].axvline(0.5, linestyle="--", color="#555555")
sns.despine(ax=axs[i])
axs[-1].set_xlabel("e")
| 0.382372 | 0.868548 |
# Credit Risk Resampling Techniques
```
import warnings
warnings.filterwarnings('ignore')
import numpy as np
import pandas as pd
from pathlib import Path
from collections import Counter
```
# Read the CSV and Perform Basic Data Cleaning
```
columns = [
"loan_amnt", "int_rate", "installment", "home_ownership",
"annual_inc", "verification_status", "issue_d", "loan_status",
"pymnt_plan", "dti", "delinq_2yrs", "inq_last_6mths",
"open_acc", "pub_rec", "revol_bal", "total_acc",
"initial_list_status", "out_prncp", "out_prncp_inv", "total_pymnt",
"total_pymnt_inv", "total_rec_prncp", "total_rec_int", "total_rec_late_fee",
"recoveries", "collection_recovery_fee", "last_pymnt_amnt", "next_pymnt_d",
"collections_12_mths_ex_med", "policy_code", "application_type", "acc_now_delinq",
"tot_coll_amt", "tot_cur_bal", "open_acc_6m", "open_act_il",
"open_il_12m", "open_il_24m", "mths_since_rcnt_il", "total_bal_il",
"il_util", "open_rv_12m", "open_rv_24m", "max_bal_bc",
"all_util", "total_rev_hi_lim", "inq_fi", "total_cu_tl",
"inq_last_12m", "acc_open_past_24mths", "avg_cur_bal", "bc_open_to_buy",
"bc_util", "chargeoff_within_12_mths", "delinq_amnt", "mo_sin_old_il_acct",
"mo_sin_old_rev_tl_op", "mo_sin_rcnt_rev_tl_op", "mo_sin_rcnt_tl", "mort_acc",
"mths_since_recent_bc", "mths_since_recent_inq", "num_accts_ever_120_pd", "num_actv_bc_tl",
"num_actv_rev_tl", "num_bc_sats", "num_bc_tl", "num_il_tl",
"num_op_rev_tl", "num_rev_accts", "num_rev_tl_bal_gt_0",
"num_sats", "num_tl_120dpd_2m", "num_tl_30dpd", "num_tl_90g_dpd_24m",
"num_tl_op_past_12m", "pct_tl_nvr_dlq", "percent_bc_gt_75", "pub_rec_bankruptcies",
"tax_liens", "tot_hi_cred_lim", "total_bal_ex_mort", "total_bc_limit",
"total_il_high_credit_limit", "hardship_flag", "debt_settlement_flag"
]
target = ["loan_status"]
# Load the data
file_path = Path('LoanStats_2019Q1.csv')
df = pd.read_csv(file_path, skiprows=1)[:-2]
df = df.loc[:, columns].copy()
# Drop the null columns where all values are null
df = df.dropna(axis='columns', how='all')
# Drop the null rows
df = df.dropna()
# Remove the `Issued` loan status
issued_mask = df['loan_status'] != 'Issued'
df = df.loc[issued_mask]
# convert interest rate to numerical
df['int_rate'] = df['int_rate'].str.replace('%', '')
df['int_rate'] = df['int_rate'].astype('float') / 100
# Convert the target column values to low_risk and high_risk based on their values
x = {'Current': 'low_risk'}
df = df.replace(x)
x = dict.fromkeys(['Late (31-120 days)', 'Late (16-30 days)', 'Default', 'In Grace Period'], 'high_risk')
df = df.replace(x)
df.reset_index(inplace=True, drop=True)
df.head()
```
# Split the Data into Training and Testing
```
# Create our features
X = pd.get_dummies(df, columns=['home_ownership', 'verification_status', 'issue_d', 'pymnt_plan',
'initial_list_status', 'next_pymnt_d', 'application_type', 'hardship_flag',
'debt_settlement_flag']).drop(columns="loan_status")
# Create our target
y = df["loan_status"]
X.describe()
# Check the balance of our target values
y.value_counts()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1, stratify=y)
```
# Oversampling
In this section, you will compare two oversampling algorithms to determine which algorithm results in the best performance. You will oversample the data using the naive random oversampling algorithm and the SMOTE algorithm. For each algorithm, be sure to complete the folliowing steps:
1. View the count of the target classes using `Counter` from the collections library.
3. Use the resampled data to train a logistic regression model.
3. Calculate the balanced accuracy score from sklearn.metrics.
4. Print the confusion matrix from sklearn.metrics.
5. Generate a classication report using the `imbalanced_classification_report` from imbalanced-learn.
Note: Use a random state of 1 for each sampling algorithm to ensure consistency between tests
### Naive Random Oversampling
```
# Resample the training data with the RandomOversampler
from imblearn.over_sampling import RandomOverSampler
ros = RandomOverSampler(random_state=1)
X_resampled, y_resampled = ros.fit_resample(X_train, y_train)
Counter(y_resampled)
# Train the Logistic Regression model using the resampled data
# Logistic regression using random oversampled data
from sklearn.linear_model import LogisticRegression
model = LogisticRegression(solver='lbfgs', random_state=1)
model.fit(X_resampled, y_resampled)
#Make prediction
y_pred = model.predict(X_test)
# Calculated the balanced accuracy score
from sklearn.metrics import balanced_accuracy_score
balanced_accuracy_score(y_test, y_pred)
# Display the confusion matrix
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test, y_pred)
# Print the imbalanced classification report
from imblearn.metrics import classification_report_imbalanced
print(classification_report_imbalanced(y_test, y_pred))
```
### SMOTE Oversampling
```
# Resample the training data with SMOTE
from imblearn.over_sampling import SMOTE
smote = SMOTE(random_state=1, sampling_strategy='auto')
X_resampled, y_resampled = smote.fit_resample(X_train, y_train)
Counter(y_resampled)
# Train the Logistic Regression model using the resampled data
model = LogisticRegression(solver='lbfgs', random_state=1)
model.fit(X_resampled, y_resampled)
# Calculated the balanced accuracy score
y_pred = model.predict(X_test)
balanced_accuracy_score(y_test, y_pred)
# Display the confusion matrix
confusion_matrix(y_test, y_pred)
# Print the imbalanced classification report
print(classification_report_imbalanced(y_test, y_pred))
```
# Undersampling
In this section, you will test an undersampling algorithms to determine which algorithm results in the best performance compared to the oversampling algorithms above. You will undersample the data using the Cluster Centroids algorithm and complete the folliowing steps:
1. View the count of the target classes using `Counter` from the collections library.
3. Use the resampled data to train a logistic regression model.
3. Calculate the balanced accuracy score from sklearn.metrics.
4. Print the confusion matrix from sklearn.metrics.
5. Generate a classication report using the `imbalanced_classification_report` from imbalanced-learn.
Note: Use a random state of 1 for each sampling algorithm to ensure consistency between tests
```
# Resample the data using the ClusterCentroids resampler
# Warning: This is a large dataset, and this step may take some time to complete
from imblearn.under_sampling import ClusterCentroids
cc = ClusterCentroids(random_state=1)
X_resampled, y_resampled = cc.fit_resample(X_train, y_train)
Counter(y_resampled)
# Train the Logistic Regression model using the resampled data
model.fit(X_resampled, y_resampled)
y_pred = model.predict(X_test)
# Calculated the balanced accuracy score
balanced_accuracy_score(y_test, y_pred)
# Display the confusion matrix
confusion_matrix(y_test, y_pred)
# Print the imbalanced classification report
print(classification_report_imbalanced(y_test, y_pred))
```
# Combination (Over and Under) Sampling
In this section, you will test a combination over- and under-sampling algorithm to determine if the algorithm results in the best performance compared to the other sampling algorithms above. You will resample the data using the SMOTEENN algorithm and complete the folliowing steps:
1. View the count of the target classes using `Counter` from the collections library.
3. Use the resampled data to train a logistic regression model.
3. Calculate the balanced accuracy score from sklearn.metrics.
4. Print the confusion matrix from sklearn.metrics.
5. Generate a classication report using the `imbalanced_classification_report` from imbalanced-learn.
Note: Use a random state of 1 for each sampling algorithm to ensure consistency between tests
```
# Resample the training data with SMOTEENN
# Warning: This is a large dataset, and this step may take some time to complete
from imblearn.combine import SMOTEENN
smoteenn = SMOTEENN(random_state=1)
X_resampled, y_resampled = smoteenn.fit_resample(X_train, y_train)
Counter(y_resampled)
# Train the Logistic Regression model using the resampled data
model.fit(X_resampled, y_resampled)
# Calculated the balanced accuracy score
y_pred = model.predict(X_test)
balanced_accuracy_score(y_test, y_pred)
# Display the confusion matrix
confusion_matrix(y_test, y_pred)
# Print the imbalanced classification report
print(classification_report_imbalanced(y_test, y_pred))
```
|
github_jupyter
|
import warnings
warnings.filterwarnings('ignore')
import numpy as np
import pandas as pd
from pathlib import Path
from collections import Counter
columns = [
"loan_amnt", "int_rate", "installment", "home_ownership",
"annual_inc", "verification_status", "issue_d", "loan_status",
"pymnt_plan", "dti", "delinq_2yrs", "inq_last_6mths",
"open_acc", "pub_rec", "revol_bal", "total_acc",
"initial_list_status", "out_prncp", "out_prncp_inv", "total_pymnt",
"total_pymnt_inv", "total_rec_prncp", "total_rec_int", "total_rec_late_fee",
"recoveries", "collection_recovery_fee", "last_pymnt_amnt", "next_pymnt_d",
"collections_12_mths_ex_med", "policy_code", "application_type", "acc_now_delinq",
"tot_coll_amt", "tot_cur_bal", "open_acc_6m", "open_act_il",
"open_il_12m", "open_il_24m", "mths_since_rcnt_il", "total_bal_il",
"il_util", "open_rv_12m", "open_rv_24m", "max_bal_bc",
"all_util", "total_rev_hi_lim", "inq_fi", "total_cu_tl",
"inq_last_12m", "acc_open_past_24mths", "avg_cur_bal", "bc_open_to_buy",
"bc_util", "chargeoff_within_12_mths", "delinq_amnt", "mo_sin_old_il_acct",
"mo_sin_old_rev_tl_op", "mo_sin_rcnt_rev_tl_op", "mo_sin_rcnt_tl", "mort_acc",
"mths_since_recent_bc", "mths_since_recent_inq", "num_accts_ever_120_pd", "num_actv_bc_tl",
"num_actv_rev_tl", "num_bc_sats", "num_bc_tl", "num_il_tl",
"num_op_rev_tl", "num_rev_accts", "num_rev_tl_bal_gt_0",
"num_sats", "num_tl_120dpd_2m", "num_tl_30dpd", "num_tl_90g_dpd_24m",
"num_tl_op_past_12m", "pct_tl_nvr_dlq", "percent_bc_gt_75", "pub_rec_bankruptcies",
"tax_liens", "tot_hi_cred_lim", "total_bal_ex_mort", "total_bc_limit",
"total_il_high_credit_limit", "hardship_flag", "debt_settlement_flag"
]
target = ["loan_status"]
# Load the data
file_path = Path('LoanStats_2019Q1.csv')
df = pd.read_csv(file_path, skiprows=1)[:-2]
df = df.loc[:, columns].copy()
# Drop the null columns where all values are null
df = df.dropna(axis='columns', how='all')
# Drop the null rows
df = df.dropna()
# Remove the `Issued` loan status
issued_mask = df['loan_status'] != 'Issued'
df = df.loc[issued_mask]
# convert interest rate to numerical
df['int_rate'] = df['int_rate'].str.replace('%', '')
df['int_rate'] = df['int_rate'].astype('float') / 100
# Convert the target column values to low_risk and high_risk based on their values
x = {'Current': 'low_risk'}
df = df.replace(x)
x = dict.fromkeys(['Late (31-120 days)', 'Late (16-30 days)', 'Default', 'In Grace Period'], 'high_risk')
df = df.replace(x)
df.reset_index(inplace=True, drop=True)
df.head()
# Create our features
X = pd.get_dummies(df, columns=['home_ownership', 'verification_status', 'issue_d', 'pymnt_plan',
'initial_list_status', 'next_pymnt_d', 'application_type', 'hardship_flag',
'debt_settlement_flag']).drop(columns="loan_status")
# Create our target
y = df["loan_status"]
X.describe()
# Check the balance of our target values
y.value_counts()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1, stratify=y)
# Resample the training data with the RandomOversampler
from imblearn.over_sampling import RandomOverSampler
ros = RandomOverSampler(random_state=1)
X_resampled, y_resampled = ros.fit_resample(X_train, y_train)
Counter(y_resampled)
# Train the Logistic Regression model using the resampled data
# Logistic regression using random oversampled data
from sklearn.linear_model import LogisticRegression
model = LogisticRegression(solver='lbfgs', random_state=1)
model.fit(X_resampled, y_resampled)
#Make prediction
y_pred = model.predict(X_test)
# Calculated the balanced accuracy score
from sklearn.metrics import balanced_accuracy_score
balanced_accuracy_score(y_test, y_pred)
# Display the confusion matrix
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test, y_pred)
# Print the imbalanced classification report
from imblearn.metrics import classification_report_imbalanced
print(classification_report_imbalanced(y_test, y_pred))
# Resample the training data with SMOTE
from imblearn.over_sampling import SMOTE
smote = SMOTE(random_state=1, sampling_strategy='auto')
X_resampled, y_resampled = smote.fit_resample(X_train, y_train)
Counter(y_resampled)
# Train the Logistic Regression model using the resampled data
model = LogisticRegression(solver='lbfgs', random_state=1)
model.fit(X_resampled, y_resampled)
# Calculated the balanced accuracy score
y_pred = model.predict(X_test)
balanced_accuracy_score(y_test, y_pred)
# Display the confusion matrix
confusion_matrix(y_test, y_pred)
# Print the imbalanced classification report
print(classification_report_imbalanced(y_test, y_pred))
# Resample the data using the ClusterCentroids resampler
# Warning: This is a large dataset, and this step may take some time to complete
from imblearn.under_sampling import ClusterCentroids
cc = ClusterCentroids(random_state=1)
X_resampled, y_resampled = cc.fit_resample(X_train, y_train)
Counter(y_resampled)
# Train the Logistic Regression model using the resampled data
model.fit(X_resampled, y_resampled)
y_pred = model.predict(X_test)
# Calculated the balanced accuracy score
balanced_accuracy_score(y_test, y_pred)
# Display the confusion matrix
confusion_matrix(y_test, y_pred)
# Print the imbalanced classification report
print(classification_report_imbalanced(y_test, y_pred))
# Resample the training data with SMOTEENN
# Warning: This is a large dataset, and this step may take some time to complete
from imblearn.combine import SMOTEENN
smoteenn = SMOTEENN(random_state=1)
X_resampled, y_resampled = smoteenn.fit_resample(X_train, y_train)
Counter(y_resampled)
# Train the Logistic Regression model using the resampled data
model.fit(X_resampled, y_resampled)
# Calculated the balanced accuracy score
y_pred = model.predict(X_test)
balanced_accuracy_score(y_test, y_pred)
# Display the confusion matrix
confusion_matrix(y_test, y_pred)
# Print the imbalanced classification report
print(classification_report_imbalanced(y_test, y_pred))
| 0.524638 | 0.804713 |
# Enemy Survivability Detailed
How long it takes for each weapon to kill an enemy.
## Setting Up
```
import math
import pandas as pd
%run weapon_full_data.ipynb
```
## Alien Details
### Sectoid
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Sectoid")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to sectoid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Sectoid")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill sectoid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
### Snakeman
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Snakeman")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to snakeman", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Snakeman")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill snakeman", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
### Floater
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Floater")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to floater", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Floater")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill floater", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
### Muton
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Muton")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to muton", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Muton")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
hits_to_kill_graph = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 50]
ax = hits_to_kill_graph.plot.bar(title="Hits to kill muton", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
### Ethereal
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Ethereal")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to ethereal", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Ethereal")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
hits_to_kill_graph = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 50]
ax = hits_to_kill_graph.plot.bar(title="Hits to kill ethereal", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
### Reaper
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Reaper")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to reaper", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Reaper")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill reaper", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
### Chryssalid
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Chryssalid")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to chryssalid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Chryssalid")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill chryssalid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
### Zombie
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Zombie")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to zombie", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Zombie")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 1000]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill zombie", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
### Cyberdisk
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Cyberdisc")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to cyberdisc", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Cyberdisc")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 50]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill cyberdisc", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
### Sectopod
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Sectopod")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to sectopod", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Sectopod")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 150].plot.bar(title="Hits to kill sectopod", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
### Silacoid
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Silacoid")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to silacoid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Silacoid")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 50]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill silacoid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
### Celatid
```
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Celatid")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to celatid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Celatid")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill celatid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
```
|
github_jupyter
|
import math
import pandas as pd
%run weapon_full_data.ipynb
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Sectoid")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to sectoid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Sectoid")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill sectoid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Snakeman")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to snakeman", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Snakeman")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill snakeman", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Floater")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to floater", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Floater")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill floater", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Muton")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to muton", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Muton")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
hits_to_kill_graph = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 50]
ax = hits_to_kill_graph.plot.bar(title="Hits to kill muton", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Ethereal")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to ethereal", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Ethereal")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
hits_to_kill_graph = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 50]
ax = hits_to_kill_graph.plot.bar(title="Hits to kill ethereal", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Reaper")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to reaper", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Reaper")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill reaper", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Chryssalid")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to chryssalid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Chryssalid")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill chryssalid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Zombie")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to zombie", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Zombie")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 1000]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill zombie", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Cyberdisc")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to cyberdisc", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Cyberdisc")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 50]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill cyberdisc", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Sectopod")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to sectopod", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Sectopod")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 150].plot.bar(title="Hits to kill sectopod", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Silacoid")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to silacoid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Silacoid")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph = hits_to_kill_graph[hits_to_kill_graph["hits_to_kill"] < 50]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill silacoid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
weapon_damages_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Celatid")]
weapon_damages_graph = weapon_damages_graph.groupby(["weapon"]).mean()
weapon_damages_graph = weapon_damages_graph[["turn_penetrating_damage_expected"]]
weapon_damages_graph = weapon_damages_graph.rename(columns={"turn_penetrating_damage_expected": "damage"})
weapon_damages_graph = weapon_damages_graph.sort_values(by=["damage"])
ax = weapon_damages_graph.plot.bar(title="Damage to celatid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Damage")
hits_to_kill_graph = weapon_data_aliens[weapon_data_aliens["alien"].str.contains("Celatid")]
hits_to_kill_graph = hits_to_kill_graph.groupby(["weapon"]).mean()
hits_to_kill_graph = hits_to_kill_graph[["hits_to_kill"]]
hits_to_kill_graph["hits_to_kill"] = hits_to_kill_graph.apply(lambda x: math.ceil(x["hits_to_kill"]), axis=1)
hits_to_kill_graph = hits_to_kill_graph.sort_values(by=["hits_to_kill"])
ax = hits_to_kill_graph.plot.bar(title="Hits to kill celatid", legend=False)
ax.xaxis.get_label().set_visible(False)
ax.set_ylabel("Hits")
| 0.362066 | 0.696494 |
```
%reload_ext autoreload
%autoreload 2
%matplotlib inline
import torch
import torch.nn as nn
import torch.nn.functional as F
nb_batches = 4
in_chan, out_chan = 2, 6
in_dim = (8,)
k_size = (3,)
dtype = torch.float32
input_1d = torch.rand((nb_batches, in_chan, *in_dim), dtype=dtype)
kernel_1d = torch.rand((out_chan, in_chan, *k_size), dtype=dtype)
in_dim = (8,) * 2
k_size = (3,) * 2
input_2d = torch.rand((nb_batches, in_chan, *in_dim), dtype=dtype)
kernel_2d = torch.rand((out_chan, in_chan, *k_size), dtype=dtype)
in_dim = (8,) * 3
k_size = (3,) * 3
input_3d = torch.rand((nb_batches, in_chan, *in_dim), dtype=dtype)
kernel_3d = torch.rand((out_chan, in_chan, *k_size), dtype=dtype)
print(input_2d.shape, kernel_2d.shape)
# By batch, for each window, flatten / unfold the matmul to perform
# B * I * H * W --> B * (I * K1 * K2) * -1
tmp_in = F.unfold(input_2d, kernel_2d.shape[-2:])
print(tmp_in.shape)
print(tmp_in.shape)
# B * ops_per_window * num_windows --> B * num_windows * ops_per_window
tmp_in = tmp_in.transpose(1, 2)
print(tmp_in.shape)
print(kernel_2d.shape)
# O * I * K1 * K2 --> O * ops_per_window
tmp_k = kernel_2d.view(kernel_2d.shape[0], -1)
print(tmp_k.shape)
print(tmp_k.shape)
# O * ops_per_window --> ops_per_window * O
tmp_k = tmp_k.t()
print(tmp_k.shape)
print(tmp_in.shape, tmp_k.shape)
# B * num_windows * ops_per_window @ ops_per_window * O --> B * num_windows * O
tmp = tmp_in.matmul(tmp_k)
print(tmp.shape)
print(tmp.shape)
# B * num_windows * O --> B * O * num_windows
tmp = tmp.transpose(1, 2)
print(tmp.shape)
print(tmp.shape)
# B * O * num_windows --> B * O * H * W
tmp = tmp.view(input_2d.shape[0], kernel_2d.shape[0], input_2d.shape[-2] - (kernel_2d.shape[-2] - 1), input_2d.shape[-1] - (kernel_2d.shape[-1] - 1))
print(tmp.shape)
res = F.unfold(input_2d, kernel_2d.shape[-2:]).transpose(1, 2).matmul(kernel_2d.view(kernel_2d.shape[0], -1).t()).transpose(1, 2).view(input_2d.shape[0], kernel_2d.shape[0], input_2d.shape[-2] - (kernel_2d.shape[-2] - 1), input_2d.shape[-1] - (kernel_2d.shape[-1] - 1))
print(res.shape)
%timeit -n 10 _ = F.unfold(input_2d, kernel_2d.shape[-2:]).transpose(1, 2).matmul(kernel_2d.view(kernel_2d.shape[0], -1).t()).transpose(1, 2).view(input_2d.shape[0], kernel_2d.shape[0], input_2d.shape[-2] - (kernel_2d.shape[-2] - 1), input_2d.shape[-1] - (kernel_2d.shape[-1] - 1))
```
## SlidingND
```
def naive_sliding1D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, L]):
kernel (torch.Tensor[O, I, K]):
padding (int, optional):
Returns:
torch.Tensor[N, O, L]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
for bidx in range(res.shape[0]):
for oidx in range(kernel.shape[0]):
for idx in range(res.shape[-1]):
res[bidx, oidx, idx] = fn(_pad[bidx, ..., idx: idx + kernel.shape[-1]], kernel[oidx, ...])
return res
def mid_sliding1D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, L]):
kernel (torch.Tensor[O, I, K]):
padding (int, optional):
Returns:
torch.Tensor[N, O, L]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
for idx in range(res.shape[-1]):
# N, O <-- (N, I, K - O, I, K)
res[..., idx] = fn(_pad[..., idx: idx + kernel.shape[-1]], kernel)
return res
def sliding1D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, L]):
kernel (torch.Tensor[O, I, K]):
padding (int, optional):
Returns:
torch.Tensor[N, O, L]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
for kidx in range(kernel.shape[-1]):
# N, O, L <-- N, I, L @ O, I
res += fn(_pad[..., kidx: kidx + input.shape[-1]], kernel[..., kidx])
return res
def naive_sliding2D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W]):
kernel (torch.Tensor[O, I, K1, K2]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
for bidx in range(res.shape[0]):
for oidx in range(kernel.shape[0]):
for row in range(res.shape[-2]):
for col in range(res.shape[-1]):
# N, O <-- (N, I, H, W - O, I, K1, K2)
res[bidx, oidx, row, col] = fn(_pad[bidx, ..., row: row + kernel.shape[-2], col: col + kernel.shape[-1]],
kernel[oidx, ...])
return res
def mid_sliding2D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W]):
kernel (torch.Tensor[O, I, K1, K2]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
# Loop on input spatially
for row in range(res.shape[-2]):
for col in range(res.shape[-1]):
# N, O <-- (N, I, H, W - O, I, K1, K2)
res[..., row, col] = fn(_pad[..., row: row + kernel.shape[-2], col: col + kernel.shape[-1]],
kernel)
return res
def sliding2D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W]):
kernel (torch.Tensor[O, I, K1, K2]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
# Loop on kernel spatially
for krow in range(kernel.shape[-2]):
for kcol in range(kernel.shape[-1]):
# N, O, ... <-- N, I, H, W @ O, I
res += fn(_pad[..., krow: krow + input.shape[-2], kcol: kcol + input.shape[-1]], kernel[..., krow, kcol])
return res
def naive_sliding3D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W, D]):
kernel (torch.Tensor[O, I, K1, K2, K3]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W, D]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
for bidx in range(res.shape[0]):
for oidx in range(kernel.shape[0]):
for row in range(res.shape[-3]):
for col in range(res.shape[-2]):
for depth in range(res.shape[-1]):
# . <-- (I, K1, K2, K3 - I, K1, K2, K3)
res[bidx, oidx, row, col] = fn(_pad[bidx, ..., row: row + kernel.shape[-3], col: col + kernel.shape[-2], depth: depth + kernel.shape[-1]],
kernel[oidx, ...])
return res
def mid_sliding3D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W, D]):
kernel (torch.Tensor[O, I, K1, K2, K3]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W, D]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
# Loop on input spatially
for row in range(res.shape[-3]):
for col in range(res.shape[-2]):
for depth in range(res.shape[-1]):
# N, O <-- (N, I, H, W, D - O, I, K1, K2, K3)
res[..., row, col, depth] = fn(_pad[..., row: row + kernel.shape[-3], col: col + kernel.shape[-2], depth: depth + kernel.shape[-1]],
kernel)
return res
def sliding3D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W, D]):
kernel (torch.Tensor[O, I, K1, K2, K3]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W, D]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
# Loop on kernel spatially
for krow in range(kernel.shape[-3]):
for kcol in range(kernel.shape[-2]):
for kdep in range(kernel.shape[-1]):
# N, O, ... <-- N, I, H, W @ O, I
res += fn(_pad[..., krow: krow + input.shape[-3], kcol: kcol + input.shape[-2], kdep: kdep + input.shape[-1]],
kernel[..., krow, kcol, kdep])
return res
```
## OpND
```
def naive_opND(fn, input, kernel):
"""Apply Conv1D locally
Args:
input (torch.Tensor[I, K1, ..., Kn]):
kernel (torch.Tensor[I, K1, ..., Kn]):
Returns:
torch.Tensor[]
"""
if input.shape != kernel.shape:
raise AssertionError("expected input and kernel to have identical shape")
return fn(input, kernel)
def mid_opND(fn, input, kernel):
"""Apply Conv1D locally
Args:
input (torch.Tensor[N, I, K1, ..., Kn]):
kernel (torch.Tensor[O, I, K1, ..., Kn]):
Returns:
torch.Tensor[N, O]
"""
if input.ndim != kernel.ndim:
raise AssertionError("wrong number of dimensions")
if input.shape[1:] != kernel.shape[1:]:
raise AssertionError("wrong shapes")
return fn(input.unsqueeze(1), kernel.unsqueeze(0))
def opND(fn, input, kernel, n):
"""Apply Conv1D locally
Args:
input (torch.Tensor[N, I, ...]):
kernel (torch.Tensor[O, I]):
Returns:
torch.Tensor[N, O, ...]
"""
if input.ndim != kernel.ndim + n:
raise AssertionError("wrong number of dimensions")
if input.shape[1] != kernel.shape[1]:
raise AssertionError("expected input and kernel to share same second axis size")
return fn(input.unsqueeze(1), kernel.unsqueeze(0)[(...,) + (None,) * n])
```
## ConvND
```
def n_convND(a, b):
"""Apply ConvND locally
Args:
a (torch.Tensor[I, K1, ..., Kn]):
b (torch.Tensor[I, K1, ..., Kn]):
Returns:
torch.Tensor[1]
"""
return a.mul(b).sum()
def m_convND(a, b):
"""Apply ConvND locally
Args:
a (torch.Tensor[N, O, I, K1, ..., Kn]):
b (torch.Tensor[N, O, I, K1, ..., Kn]):
Returns:
torch.Tensor[N, O]
"""
return a.mul(b).flatten(2).sum(2)
def convND(a, b):
"""Apply ConvND locally
Args:
a (torch.Tensor[N, 1, I, ...]):
b (torch.Tensor[1, O, I, ...]):
Returns:
torch.Tensor[N, O, ...]
"""
return a.mul(b).sum(2)
def naive_convND(input, kernel):
return naive_opND(n_convND, input, kernel)
def mid_convND(input, kernel):
return mid_opND(m_convND, input, kernel)
def conv1D(input, kernel):
return opND(convND, input, kernel, n=1)
%timeit -n 10 _ = naive_sliding1D(naive_convND, input_1d, kernel_1d)
%timeit -n 10 _ = mid_sliding1D(mid_convND, input_1d, kernel_1d)
%timeit -n 10 _ = sliding1D(conv1D, input_1d, kernel_1d)
%timeit -n 10 _ = F.conv1d(input_1d, kernel_1d, padding=1)
def conv2D(input, kernel):
return opND(convND, input, kernel, n=2)
%timeit -n 10 _ = naive_sliding2D(naive_convND, input_2d, kernel_2d)
%timeit -n 10 _ = mid_sliding2D(mid_convND, input_2d, kernel_2d)
%timeit -n 10 _ = sliding2D(conv2D, input_2d, kernel_2d)
%timeit -n 10 _ = F.conv2d(input_2d, kernel_2d, padding=1)
def conv3D(input, kernel):
return opND(convND, input, kernel, n=3)
%timeit -n 10 _ = naive_sliding3D(naive_convND, input_3d, kernel_3d)
%timeit -n 10 _ = mid_sliding3D(mid_convND, input_3d, kernel_3d)
%timeit -n 10 _ = sliding3D(conv3D, input_3d, kernel_3d)
%timeit -n 10 _ = F.conv3d(input_3d, kernel_3d, padding=1)
```
## AdderND
```
def n_adderND(a, b):
"""Apply ConvND locally
Args:
a (torch.Tensor[I, K1, ..., Kn]):
b (torch.Tensor[I, K1, ..., Kn]):
Returns:
torch.Tensor[1]
"""
return a.sub(b).abs_().sum()
def m_adderND(a, b):
"""Apply ConvND locally
Args:
a (torch.Tensor[N, O, I, K1, ..., Kn]):
b (torch.Tensor[N, O, I, K1, ..., Kn]):
Returns:
torch.Tensor[N, O]
"""
return a.sub(b).abs_().flatten(2).sum(2)
def adderND(a, b):
"""Apply AdderND locally
Args:
a (torch.Tensor[N, 1, I, ...]):
b (torch.Tensor[1, O, I, ...]):
Returns:
torch.Tensor[N, O, ...]
"""
return a.sub(b).abs_().sum(2)
def naive_adderND(input, kernel):
return naive_opND(n_adderND, input, kernel)
def mid_adderND(input, kernel):
return mid_opND(m_adderND, input, kernel)
def adder1D(input, kernel):
return opND(adderND, input, kernel, n=1)
%timeit -n 10 _ = naive_sliding1D(naive_adderND, input_1d, kernel_1d)
%timeit -n 10 _ = mid_sliding1D(mid_adderND, input_1d, kernel_1d)
%timeit -n 10 _ = sliding1D(conv1D, input_1d, kernel_1d)
def adder2D(input, kernel):
return opND(adderND, input, kernel, n=2)
%timeit -n 10 _ = naive_sliding2D(naive_adderND, input_2d, kernel_2d)
%timeit -n 10 _ = mid_sliding2D(mid_adderND, input_2d, kernel_2d)
%timeit -n 10 _ = sliding2D(adder2D, input_2d, kernel_2d)
def adder3D(input, kernel):
return opND(adderND, input, kernel, n=3)
%timeit -n 10 _ = naive_sliding3D(naive_adderND, input_3d, kernel_3d)
%timeit -n 10 _ = mid_sliding3D(mid_adderND, input_3d, kernel_3d)
%timeit -n 10 _ = sliding3D(adder3D, input_3d, kernel_3d)
```
## CosimND
```
def n_cosimND(a, b, q=0):
"""Apply ConvND locally
Args:
a (torch.Tensor[I, K1, ..., Kn]):
b (torch.Tensor[I, K1, ..., Kn]):
Returns:
torch.Tensor[1]
"""
return n_convND(a, b).div_(a.norm().add_(q) * b.norm().add_(q))
def m_cosimND(a, b, q=0):
"""Apply ConvND locally
Args:
a (torch.Tensor[N, O, I, K1, ..., Kn]):
b (torch.Tensor[N, O, I, K1, ..., Kn]):
Returns:
torch.Tensor[N, O]
"""
return m_convND(a, b).div_(a.pow(2).flatten(2).sum(2).add_(q) * b.pow(2).flatten(2).sum(2).add_(q))
def cosimND(a, b, q=0):
"""Apply CosimND locally
Args:
a (torch.Tensor[N, 1, I, ...]):
b (torch.Tensor[1, O, I, ...]):
Returns:
torch.Tensor[N, O, ...]
"""
return convND(a, b).div_(a.norm(dim=2).add_(q) * b.norm(dim=2).add_(q))
def naive_cosimND(input, kernel):
return naive_opND(n_cosimND, input, kernel)
def mid_cosimND(input, kernel):
return mid_opND(m_cosimND, input, kernel)
def cosim1D(input, kernel):
return opND(cosimND, input, kernel, n=1)
%timeit -n 10 _ = naive_sliding1D(naive_cosimND, input_1d, kernel_1d)
%timeit -n 10 _ = mid_sliding1D(mid_cosimND, input_1d, kernel_1d)
%timeit -n 10 _ = sliding1D(cosim1D, input_1d, kernel_1d)
def cosim2D(input, kernel):
return opND(cosimND, input, kernel, n=2)
%timeit -n 10 _ = naive_sliding2D(naive_cosimND, input_2d, kernel_2d)
%timeit -n 10 _ = mid_sliding2D(mid_cosimND, input_2d, kernel_2d)
%timeit -n 10 _ = sliding2D(cosim2D, input_2d, kernel_2d)
def cosim3D(input, kernel):
return opND(cosimND, input, kernel, n=3)
%timeit -n 10 _ = naive_sliding3D(naive_cosimND, input_3d, kernel_3d)
%timeit -n 10 _ = mid_sliding3D(mid_cosimND, input_3d, kernel_3d)
%timeit -n 10 _ = sliding3D(cosim3D, input_3d, kernel_3d)
```
## SharpCosimND
```
def n_scosimND(a, b, p=2, q=1e-3):
"""Apply ConvND locally
Args:
a (torch.Tensor[I, K1, ..., Kn]):
b (torch.Tensor[I, K1, ..., Kn]):
Returns:
torch.Tensor[1]
"""
num = n_convND(a, b)
return torch.sign(num) * num.div_(a.norm().add_(q) * b.norm().add_(q)).pow_(p)
def m_scosimND(a, b, p=2, q=1e-3):
"""Apply ConvND locally
Args:
a (torch.Tensor[N, O, I, K1, ..., Kn]):
b (torch.Tensor[N, O, I, K1, ..., Kn]):
Returns:
torch.Tensor[N, O]
"""
num = m_convND(a, b)
return torch.sign(num) * num.div_(a.pow(2).flatten(2).sum(2).add_(q) * b.pow(2).flatten(2).sum(2).add_(q)).pow_(p)
def scosimND(a, b, p=2, q=1e-3):
"""Apply Conv1D locally
Args:
a (torch.Tensor[N, 1, I, ...]):
b (torch.Tensor[1, O, I, ...]):
Returns:
torch.Tensor[N, O, ...]
"""
num = convND(a, b)
return torch.sign(num) * num.div_(a.norm(dim=2).add_(q) * b.norm(dim=2).add_(q)).pow_(p)
def naive_scosimND(input, kernel):
return naive_opND(n_scosimND, input, kernel)
def mid_scosimND(input, kernel):
return mid_opND(m_scosimND, input, kernel)
def scosim1D(input, kernel):
return opND(scosimND, input, kernel, n=1)
%timeit -n 10 _ = naive_sliding1D(naive_scosimND, input_1d, kernel_1d)
%timeit -n 10 _ = mid_sliding1D(mid_scosimND, input_1d, kernel_1d)
%timeit -n 10 _ = sliding1D(scosim1D, input_1d, kernel_1d)
def scosim2D(input, kernel):
return opND(scosimND, input, kernel, n=2)
%timeit -n 10 _ = naive_sliding2D(naive_scosimND, input_2d, kernel_2d)
%timeit -n 10 _ = mid_sliding2D(mid_scosimND, input_2d, kernel_2d)
%timeit -n 10 _ = sliding2D(scosim2D, input_2d, kernel_2d)
def scosim3D(input, kernel):
return opND(scosimND, input, kernel, n=3)
%timeit -n 10 _ = naive_sliding3D(naive_scosimND, input_3d, kernel_3d)
%timeit -n 10 _ = mid_sliding3D(mid_scosimND, input_3d, kernel_3d)
%timeit -n 10 _ = sliding3D(scosim3D, input_3d, kernel_3d)
```
|
github_jupyter
|
%reload_ext autoreload
%autoreload 2
%matplotlib inline
import torch
import torch.nn as nn
import torch.nn.functional as F
nb_batches = 4
in_chan, out_chan = 2, 6
in_dim = (8,)
k_size = (3,)
dtype = torch.float32
input_1d = torch.rand((nb_batches, in_chan, *in_dim), dtype=dtype)
kernel_1d = torch.rand((out_chan, in_chan, *k_size), dtype=dtype)
in_dim = (8,) * 2
k_size = (3,) * 2
input_2d = torch.rand((nb_batches, in_chan, *in_dim), dtype=dtype)
kernel_2d = torch.rand((out_chan, in_chan, *k_size), dtype=dtype)
in_dim = (8,) * 3
k_size = (3,) * 3
input_3d = torch.rand((nb_batches, in_chan, *in_dim), dtype=dtype)
kernel_3d = torch.rand((out_chan, in_chan, *k_size), dtype=dtype)
print(input_2d.shape, kernel_2d.shape)
# By batch, for each window, flatten / unfold the matmul to perform
# B * I * H * W --> B * (I * K1 * K2) * -1
tmp_in = F.unfold(input_2d, kernel_2d.shape[-2:])
print(tmp_in.shape)
print(tmp_in.shape)
# B * ops_per_window * num_windows --> B * num_windows * ops_per_window
tmp_in = tmp_in.transpose(1, 2)
print(tmp_in.shape)
print(kernel_2d.shape)
# O * I * K1 * K2 --> O * ops_per_window
tmp_k = kernel_2d.view(kernel_2d.shape[0], -1)
print(tmp_k.shape)
print(tmp_k.shape)
# O * ops_per_window --> ops_per_window * O
tmp_k = tmp_k.t()
print(tmp_k.shape)
print(tmp_in.shape, tmp_k.shape)
# B * num_windows * ops_per_window @ ops_per_window * O --> B * num_windows * O
tmp = tmp_in.matmul(tmp_k)
print(tmp.shape)
print(tmp.shape)
# B * num_windows * O --> B * O * num_windows
tmp = tmp.transpose(1, 2)
print(tmp.shape)
print(tmp.shape)
# B * O * num_windows --> B * O * H * W
tmp = tmp.view(input_2d.shape[0], kernel_2d.shape[0], input_2d.shape[-2] - (kernel_2d.shape[-2] - 1), input_2d.shape[-1] - (kernel_2d.shape[-1] - 1))
print(tmp.shape)
res = F.unfold(input_2d, kernel_2d.shape[-2:]).transpose(1, 2).matmul(kernel_2d.view(kernel_2d.shape[0], -1).t()).transpose(1, 2).view(input_2d.shape[0], kernel_2d.shape[0], input_2d.shape[-2] - (kernel_2d.shape[-2] - 1), input_2d.shape[-1] - (kernel_2d.shape[-1] - 1))
print(res.shape)
%timeit -n 10 _ = F.unfold(input_2d, kernel_2d.shape[-2:]).transpose(1, 2).matmul(kernel_2d.view(kernel_2d.shape[0], -1).t()).transpose(1, 2).view(input_2d.shape[0], kernel_2d.shape[0], input_2d.shape[-2] - (kernel_2d.shape[-2] - 1), input_2d.shape[-1] - (kernel_2d.shape[-1] - 1))
def naive_sliding1D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, L]):
kernel (torch.Tensor[O, I, K]):
padding (int, optional):
Returns:
torch.Tensor[N, O, L]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
for bidx in range(res.shape[0]):
for oidx in range(kernel.shape[0]):
for idx in range(res.shape[-1]):
res[bidx, oidx, idx] = fn(_pad[bidx, ..., idx: idx + kernel.shape[-1]], kernel[oidx, ...])
return res
def mid_sliding1D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, L]):
kernel (torch.Tensor[O, I, K]):
padding (int, optional):
Returns:
torch.Tensor[N, O, L]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
for idx in range(res.shape[-1]):
# N, O <-- (N, I, K - O, I, K)
res[..., idx] = fn(_pad[..., idx: idx + kernel.shape[-1]], kernel)
return res
def sliding1D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, L]):
kernel (torch.Tensor[O, I, K]):
padding (int, optional):
Returns:
torch.Tensor[N, O, L]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
for kidx in range(kernel.shape[-1]):
# N, O, L <-- N, I, L @ O, I
res += fn(_pad[..., kidx: kidx + input.shape[-1]], kernel[..., kidx])
return res
def naive_sliding2D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W]):
kernel (torch.Tensor[O, I, K1, K2]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
for bidx in range(res.shape[0]):
for oidx in range(kernel.shape[0]):
for row in range(res.shape[-2]):
for col in range(res.shape[-1]):
# N, O <-- (N, I, H, W - O, I, K1, K2)
res[bidx, oidx, row, col] = fn(_pad[bidx, ..., row: row + kernel.shape[-2], col: col + kernel.shape[-1]],
kernel[oidx, ...])
return res
def mid_sliding2D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W]):
kernel (torch.Tensor[O, I, K1, K2]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
# Loop on input spatially
for row in range(res.shape[-2]):
for col in range(res.shape[-1]):
# N, O <-- (N, I, H, W - O, I, K1, K2)
res[..., row, col] = fn(_pad[..., row: row + kernel.shape[-2], col: col + kernel.shape[-1]],
kernel)
return res
def sliding2D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W]):
kernel (torch.Tensor[O, I, K1, K2]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
# Loop on kernel spatially
for krow in range(kernel.shape[-2]):
for kcol in range(kernel.shape[-1]):
# N, O, ... <-- N, I, H, W @ O, I
res += fn(_pad[..., krow: krow + input.shape[-2], kcol: kcol + input.shape[-1]], kernel[..., krow, kcol])
return res
def naive_sliding3D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W, D]):
kernel (torch.Tensor[O, I, K1, K2, K3]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W, D]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
for bidx in range(res.shape[0]):
for oidx in range(kernel.shape[0]):
for row in range(res.shape[-3]):
for col in range(res.shape[-2]):
for depth in range(res.shape[-1]):
# . <-- (I, K1, K2, K3 - I, K1, K2, K3)
res[bidx, oidx, row, col] = fn(_pad[bidx, ..., row: row + kernel.shape[-3], col: col + kernel.shape[-2], depth: depth + kernel.shape[-1]],
kernel[oidx, ...])
return res
def mid_sliding3D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W, D]):
kernel (torch.Tensor[O, I, K1, K2, K3]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W, D]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
# Loop on input spatially
for row in range(res.shape[-3]):
for col in range(res.shape[-2]):
for depth in range(res.shape[-1]):
# N, O <-- (N, I, H, W, D - O, I, K1, K2, K3)
res[..., row, col, depth] = fn(_pad[..., row: row + kernel.shape[-3], col: col + kernel.shape[-2], depth: depth + kernel.shape[-1]],
kernel)
return res
def sliding3D(fn, input, kernel, padding=1, dtype=torch.float32):
"""Apply fn in a convolutioned fashion
Args:
fn (callable)
input (torch.Tensor[N, I, H, W, D]):
kernel (torch.Tensor[O, I, K1, K2, K3]):
padding (int, optional):
Returns:
torch.Tensor[N, O, H, W, D]
"""
if isinstance(padding, int):
padding = (padding, ) * 2 * (kernel.ndim - 2)
if input.ndim != kernel.ndim:
raise AssertionError(f"expected (2+N)D input, received {input.ndim}")
# N, O, ...
res = torch.zeros((input.shape[0], kernel.shape[0], *input.shape[2:]), dtype=dtype)
_pad = F.pad(input, padding, mode='constant', value=0.)
# Loop on kernel spatially
for krow in range(kernel.shape[-3]):
for kcol in range(kernel.shape[-2]):
for kdep in range(kernel.shape[-1]):
# N, O, ... <-- N, I, H, W @ O, I
res += fn(_pad[..., krow: krow + input.shape[-3], kcol: kcol + input.shape[-2], kdep: kdep + input.shape[-1]],
kernel[..., krow, kcol, kdep])
return res
def naive_opND(fn, input, kernel):
"""Apply Conv1D locally
Args:
input (torch.Tensor[I, K1, ..., Kn]):
kernel (torch.Tensor[I, K1, ..., Kn]):
Returns:
torch.Tensor[]
"""
if input.shape != kernel.shape:
raise AssertionError("expected input and kernel to have identical shape")
return fn(input, kernel)
def mid_opND(fn, input, kernel):
"""Apply Conv1D locally
Args:
input (torch.Tensor[N, I, K1, ..., Kn]):
kernel (torch.Tensor[O, I, K1, ..., Kn]):
Returns:
torch.Tensor[N, O]
"""
if input.ndim != kernel.ndim:
raise AssertionError("wrong number of dimensions")
if input.shape[1:] != kernel.shape[1:]:
raise AssertionError("wrong shapes")
return fn(input.unsqueeze(1), kernel.unsqueeze(0))
def opND(fn, input, kernel, n):
"""Apply Conv1D locally
Args:
input (torch.Tensor[N, I, ...]):
kernel (torch.Tensor[O, I]):
Returns:
torch.Tensor[N, O, ...]
"""
if input.ndim != kernel.ndim + n:
raise AssertionError("wrong number of dimensions")
if input.shape[1] != kernel.shape[1]:
raise AssertionError("expected input and kernel to share same second axis size")
return fn(input.unsqueeze(1), kernel.unsqueeze(0)[(...,) + (None,) * n])
def n_convND(a, b):
"""Apply ConvND locally
Args:
a (torch.Tensor[I, K1, ..., Kn]):
b (torch.Tensor[I, K1, ..., Kn]):
Returns:
torch.Tensor[1]
"""
return a.mul(b).sum()
def m_convND(a, b):
"""Apply ConvND locally
Args:
a (torch.Tensor[N, O, I, K1, ..., Kn]):
b (torch.Tensor[N, O, I, K1, ..., Kn]):
Returns:
torch.Tensor[N, O]
"""
return a.mul(b).flatten(2).sum(2)
def convND(a, b):
"""Apply ConvND locally
Args:
a (torch.Tensor[N, 1, I, ...]):
b (torch.Tensor[1, O, I, ...]):
Returns:
torch.Tensor[N, O, ...]
"""
return a.mul(b).sum(2)
def naive_convND(input, kernel):
return naive_opND(n_convND, input, kernel)
def mid_convND(input, kernel):
return mid_opND(m_convND, input, kernel)
def conv1D(input, kernel):
return opND(convND, input, kernel, n=1)
%timeit -n 10 _ = naive_sliding1D(naive_convND, input_1d, kernel_1d)
%timeit -n 10 _ = mid_sliding1D(mid_convND, input_1d, kernel_1d)
%timeit -n 10 _ = sliding1D(conv1D, input_1d, kernel_1d)
%timeit -n 10 _ = F.conv1d(input_1d, kernel_1d, padding=1)
def conv2D(input, kernel):
return opND(convND, input, kernel, n=2)
%timeit -n 10 _ = naive_sliding2D(naive_convND, input_2d, kernel_2d)
%timeit -n 10 _ = mid_sliding2D(mid_convND, input_2d, kernel_2d)
%timeit -n 10 _ = sliding2D(conv2D, input_2d, kernel_2d)
%timeit -n 10 _ = F.conv2d(input_2d, kernel_2d, padding=1)
def conv3D(input, kernel):
return opND(convND, input, kernel, n=3)
%timeit -n 10 _ = naive_sliding3D(naive_convND, input_3d, kernel_3d)
%timeit -n 10 _ = mid_sliding3D(mid_convND, input_3d, kernel_3d)
%timeit -n 10 _ = sliding3D(conv3D, input_3d, kernel_3d)
%timeit -n 10 _ = F.conv3d(input_3d, kernel_3d, padding=1)
def n_adderND(a, b):
"""Apply ConvND locally
Args:
a (torch.Tensor[I, K1, ..., Kn]):
b (torch.Tensor[I, K1, ..., Kn]):
Returns:
torch.Tensor[1]
"""
return a.sub(b).abs_().sum()
def m_adderND(a, b):
"""Apply ConvND locally
Args:
a (torch.Tensor[N, O, I, K1, ..., Kn]):
b (torch.Tensor[N, O, I, K1, ..., Kn]):
Returns:
torch.Tensor[N, O]
"""
return a.sub(b).abs_().flatten(2).sum(2)
def adderND(a, b):
"""Apply AdderND locally
Args:
a (torch.Tensor[N, 1, I, ...]):
b (torch.Tensor[1, O, I, ...]):
Returns:
torch.Tensor[N, O, ...]
"""
return a.sub(b).abs_().sum(2)
def naive_adderND(input, kernel):
return naive_opND(n_adderND, input, kernel)
def mid_adderND(input, kernel):
return mid_opND(m_adderND, input, kernel)
def adder1D(input, kernel):
return opND(adderND, input, kernel, n=1)
%timeit -n 10 _ = naive_sliding1D(naive_adderND, input_1d, kernel_1d)
%timeit -n 10 _ = mid_sliding1D(mid_adderND, input_1d, kernel_1d)
%timeit -n 10 _ = sliding1D(conv1D, input_1d, kernel_1d)
def adder2D(input, kernel):
return opND(adderND, input, kernel, n=2)
%timeit -n 10 _ = naive_sliding2D(naive_adderND, input_2d, kernel_2d)
%timeit -n 10 _ = mid_sliding2D(mid_adderND, input_2d, kernel_2d)
%timeit -n 10 _ = sliding2D(adder2D, input_2d, kernel_2d)
def adder3D(input, kernel):
return opND(adderND, input, kernel, n=3)
%timeit -n 10 _ = naive_sliding3D(naive_adderND, input_3d, kernel_3d)
%timeit -n 10 _ = mid_sliding3D(mid_adderND, input_3d, kernel_3d)
%timeit -n 10 _ = sliding3D(adder3D, input_3d, kernel_3d)
def n_cosimND(a, b, q=0):
"""Apply ConvND locally
Args:
a (torch.Tensor[I, K1, ..., Kn]):
b (torch.Tensor[I, K1, ..., Kn]):
Returns:
torch.Tensor[1]
"""
return n_convND(a, b).div_(a.norm().add_(q) * b.norm().add_(q))
def m_cosimND(a, b, q=0):
"""Apply ConvND locally
Args:
a (torch.Tensor[N, O, I, K1, ..., Kn]):
b (torch.Tensor[N, O, I, K1, ..., Kn]):
Returns:
torch.Tensor[N, O]
"""
return m_convND(a, b).div_(a.pow(2).flatten(2).sum(2).add_(q) * b.pow(2).flatten(2).sum(2).add_(q))
def cosimND(a, b, q=0):
"""Apply CosimND locally
Args:
a (torch.Tensor[N, 1, I, ...]):
b (torch.Tensor[1, O, I, ...]):
Returns:
torch.Tensor[N, O, ...]
"""
return convND(a, b).div_(a.norm(dim=2).add_(q) * b.norm(dim=2).add_(q))
def naive_cosimND(input, kernel):
return naive_opND(n_cosimND, input, kernel)
def mid_cosimND(input, kernel):
return mid_opND(m_cosimND, input, kernel)
def cosim1D(input, kernel):
return opND(cosimND, input, kernel, n=1)
%timeit -n 10 _ = naive_sliding1D(naive_cosimND, input_1d, kernel_1d)
%timeit -n 10 _ = mid_sliding1D(mid_cosimND, input_1d, kernel_1d)
%timeit -n 10 _ = sliding1D(cosim1D, input_1d, kernel_1d)
def cosim2D(input, kernel):
return opND(cosimND, input, kernel, n=2)
%timeit -n 10 _ = naive_sliding2D(naive_cosimND, input_2d, kernel_2d)
%timeit -n 10 _ = mid_sliding2D(mid_cosimND, input_2d, kernel_2d)
%timeit -n 10 _ = sliding2D(cosim2D, input_2d, kernel_2d)
def cosim3D(input, kernel):
return opND(cosimND, input, kernel, n=3)
%timeit -n 10 _ = naive_sliding3D(naive_cosimND, input_3d, kernel_3d)
%timeit -n 10 _ = mid_sliding3D(mid_cosimND, input_3d, kernel_3d)
%timeit -n 10 _ = sliding3D(cosim3D, input_3d, kernel_3d)
def n_scosimND(a, b, p=2, q=1e-3):
"""Apply ConvND locally
Args:
a (torch.Tensor[I, K1, ..., Kn]):
b (torch.Tensor[I, K1, ..., Kn]):
Returns:
torch.Tensor[1]
"""
num = n_convND(a, b)
return torch.sign(num) * num.div_(a.norm().add_(q) * b.norm().add_(q)).pow_(p)
def m_scosimND(a, b, p=2, q=1e-3):
"""Apply ConvND locally
Args:
a (torch.Tensor[N, O, I, K1, ..., Kn]):
b (torch.Tensor[N, O, I, K1, ..., Kn]):
Returns:
torch.Tensor[N, O]
"""
num = m_convND(a, b)
return torch.sign(num) * num.div_(a.pow(2).flatten(2).sum(2).add_(q) * b.pow(2).flatten(2).sum(2).add_(q)).pow_(p)
def scosimND(a, b, p=2, q=1e-3):
"""Apply Conv1D locally
Args:
a (torch.Tensor[N, 1, I, ...]):
b (torch.Tensor[1, O, I, ...]):
Returns:
torch.Tensor[N, O, ...]
"""
num = convND(a, b)
return torch.sign(num) * num.div_(a.norm(dim=2).add_(q) * b.norm(dim=2).add_(q)).pow_(p)
def naive_scosimND(input, kernel):
return naive_opND(n_scosimND, input, kernel)
def mid_scosimND(input, kernel):
return mid_opND(m_scosimND, input, kernel)
def scosim1D(input, kernel):
return opND(scosimND, input, kernel, n=1)
%timeit -n 10 _ = naive_sliding1D(naive_scosimND, input_1d, kernel_1d)
%timeit -n 10 _ = mid_sliding1D(mid_scosimND, input_1d, kernel_1d)
%timeit -n 10 _ = sliding1D(scosim1D, input_1d, kernel_1d)
def scosim2D(input, kernel):
return opND(scosimND, input, kernel, n=2)
%timeit -n 10 _ = naive_sliding2D(naive_scosimND, input_2d, kernel_2d)
%timeit -n 10 _ = mid_sliding2D(mid_scosimND, input_2d, kernel_2d)
%timeit -n 10 _ = sliding2D(scosim2D, input_2d, kernel_2d)
def scosim3D(input, kernel):
return opND(scosimND, input, kernel, n=3)
%timeit -n 10 _ = naive_sliding3D(naive_scosimND, input_3d, kernel_3d)
%timeit -n 10 _ = mid_sliding3D(mid_scosimND, input_3d, kernel_3d)
%timeit -n 10 _ = sliding3D(scosim3D, input_3d, kernel_3d)
| 0.783077 | 0.771047 |
# Character-Level LSTM in PyTorch
In this notebook, I'll construct a character-level LSTM with PyTorch. The network will train character by character on some text, then generate new text character by character. As an example, I will train on Anna Karenina. **This model will be able to generate new text based on the text from the book!**
This network is based off of Andrej Karpathy's [post on RNNs](http://karpathy.github.io/2015/05/21/rnn-effectiveness/) and [implementation in Torch](https://github.com/karpathy/char-rnn). Below is the general architecture of the character-wise RNN.
<img src="assets/charseq.jpeg" width="500">
First let's load in our required resources for data loading and model creation.
```
import numpy as np
import torch
from torch import nn
import torch.nn.functional as F
```
## Load in Data
Then, we'll load the Anna Karenina text file and convert it into integers for our network to use.
```
# open text file and read in data as `text`
with open('data/anna.txt', 'r') as f:
text = f.read()
```
Let's check out the first 100 characters, make sure everything is peachy. According to the [American Book Review](http://americanbookreview.org/100bestlines.asp), this is the 6th best first line of a book ever.
```
text[:100]
```
### Tokenization
In the cells, below, I'm creating a couple **dictionaries** to convert the characters to and from integers. Encoding the characters as integers makes it easier to use as input in the network.
```
# encode the text and map each character to an integer and vice versa
# we create two dictionaries:
# 1. int2char, which maps integers to characters
# 2. char2int, which maps characters to unique integers
chars = tuple(set(text))
int2char = dict(enumerate(chars))
char2int = {ch: ii for ii, ch in int2char.items()}
# encode the text
encoded = np.array([char2int[ch] for ch in text])
```
And we can see those same characters from above, encoded as integers.
```
encoded[:100]
```
## Pre-processing the data
As you can see in our char-RNN image above, our LSTM expects an input that is **one-hot encoded** meaning that each character is converted into an integer (via our created dictionary) and *then* converted into a column vector where only it's corresponding integer index will have the value of 1 and the rest of the vector will be filled with 0's. Since we're one-hot encoding the data, let's make a function to do that!
```
def one_hot_encode(arr, n_labels):
# Initialize the the encoded array
one_hot = np.zeros((np.multiply(*arr.shape), n_labels), dtype=np.float32)
# Fill the appropriate elements with ones
one_hot[np.arange(one_hot.shape[0]), arr.flatten()] = 1.
# Finally reshape it to get back to the original array
one_hot = one_hot.reshape((*arr.shape, n_labels))
return one_hot
# check that the function works as expected
test_seq = np.array([[3, 5, 1]])
one_hot = one_hot_encode(test_seq, 8)
print(one_hot)
```
## Making training mini-batches
To train on this data, we also want to create mini-batches for training. Remember that we want our batches to be multiple sequences of some desired number of sequence steps. Considering a simple example, our batches would look like this:
<img src="assets/sequence_batching@1x.png" width=500px>
<br>
In this example, we'll take the encoded characters (passed in as the `arr` parameter) and split them into multiple sequences, given by `batch_size`. Each of our sequences will be `seq_length` long.
### Creating Batches
**1. The first thing we need to do is discard some of the text so we only have completely full mini-batches. **
Each batch contains $N \times M$ characters, where $N$ is the batch size (the number of sequences in a batch) and $M$ is the seq_length or number of time steps in a sequence. Then, to get the total number of batches, $K$, that we can make from the array `arr`, you divide the length of `arr` by the number of characters per batch. Once you know the number of batches, you can get the total number of characters to keep from `arr`, $N * M * K$.
**2. After that, we need to split `arr` into $N$ batches. **
You can do this using `arr.reshape(size)` where `size` is a tuple containing the dimensions sizes of the reshaped array. We know we want $N$ sequences in a batch, so let's make that the size of the first dimension. For the second dimension, you can use `-1` as a placeholder in the size, it'll fill up the array with the appropriate data for you. After this, you should have an array that is $N \times (M * K)$.
**3. Now that we have this array, we can iterate through it to get our mini-batches. **
The idea is each batch is a $N \times M$ window on the $N \times (M * K)$ array. For each subsequent batch, the window moves over by `seq_length`. We also want to create both the input and target arrays. Remember that the targets are just the inputs shifted over by one character. The way I like to do this window is use `range` to take steps of size `n_steps` from $0$ to `arr.shape[1]`, the total number of tokens in each sequence. That way, the integers you get from `range` always point to the start of a batch, and each window is `seq_length` wide.
We will implement the code for creating batches in the function below.
```
def get_batches(arr, batch_size, seq_length):
'''Create a generator that returns batches of size
batch_size x seq_length from arr.
Arguments
---------
arr: Array you want to make batches from
batch_size: Batch size, the number of sequences per batch
seq_length: Number of encoded chars in a sequence
'''
batch_size_total = batch_size * seq_length
## Get the number of batches we can make
n_batches = len(arr)//batch_size_total
## Keep only enough characters to make full batches
arr = arr[:n_batches * batch_size_total]
## Reshape into batch_size rows
arr = arr.reshape((batch_size, -1))
## Iterate over the batches using a window of size seq_length
for n in range(0, arr.shape[1], seq_length):
# The features
x = arr[:, n:n+seq_length]
# The targets, shifted by one
y = np.zeros_like(x)
try:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, n+seq_length]
except IndexError:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, 0]
yield x, y
```
### Test Your Implementation
Now I'll make some data sets and we can check out what's going on as we batch data. Here, as an example, I'm going to use a batch size of 8 and 50 sequence steps.
```
batches = get_batches(encoded, 8, 50)
x, y = next(batches)
# printing out the first 10 items in a sequence
print('x\n', x[:10, :10])
print('\ny\n', y[:10, :10])
```
If you implemented `get_batches` correctly, the above output should look something like
```
x
[[25 8 60 11 45 27 28 73 1 2]
[17 7 20 73 45 8 60 45 73 60]
[27 20 80 73 7 28 73 60 73 65]
[17 73 45 8 27 73 66 8 46 27]
[73 17 60 12 73 8 27 28 73 45]
[66 64 17 17 46 7 20 73 60 20]
[73 76 20 20 60 73 8 60 80 73]
[47 35 43 7 20 17 24 50 37 73]]
y
[[ 8 60 11 45 27 28 73 1 2 2]
[ 7 20 73 45 8 60 45 73 60 45]
[20 80 73 7 28 73 60 73 65 7]
[73 45 8 27 73 66 8 46 27 65]
[17 60 12 73 8 27 28 73 45 27]
[64 17 17 46 7 20 73 60 20 80]
[76 20 20 60 73 8 60 80 73 17]
[35 43 7 20 17 24 50 37 73 36]]
```
although the exact numbers may be different. Check to make sure the data is shifted over one step for `y`.
---
## Defining the network with PyTorch
Below is where you'll define the network.
<img src="assets/charRNN.png" width=500px>
Next, you'll use PyTorch to define the architecture of the network. We start by defining the layers and operations we want. Then, define a method for the forward pass. You've also been given a method for predicting characters.
### Model Structure
In `__init__` the suggested structure is as follows:
* Create and store the necessary dictionaries (this has been done for you)
* Define an LSTM layer that takes as params: an input size (the number of characters), a hidden layer size `n_hidden`, a number of layers `n_layers`, a dropout probability `drop_prob`, and a batch_first boolean (True, since we are batching)
* Define a dropout layer with `dropout_prob`
* Define a fully-connected layer with params: input size `n_hidden` and output size (the number of characters)
* Finally, initialize the weights (again, this has been given)
Note that some parameters have been named and given in the `__init__` function, and we use them and store them by doing something like `self.drop_prob = drop_prob`.
---
### LSTM Inputs/Outputs
You can create a basic [LSTM layer](https://pytorch.org/docs/stable/nn.html#lstm) as follows
```python
self.lstm = nn.LSTM(input_size, n_hidden, n_layers,
dropout=drop_prob, batch_first=True)
```
where `input_size` is the number of characters this cell expects to see as sequential input, and `n_hidden` is the number of units in the hidden layers in the cell. And we can add dropout by adding a dropout parameter with a specified probability; this will automatically add dropout to the inputs or outputs. Finally, in the `forward` function, we can stack up the LSTM cells into layers using `.view`. With this, you pass in a list of cells and it will send the output of one cell into the next cell.
We also need to create an initial hidden state of all zeros. This is done like so
```python
self.init_hidden()
```
```
# check if GPU is available
train_on_gpu = torch.cuda.is_available()
if(train_on_gpu):
print('Training on GPU!')
else:
print('No GPU available, training on CPU; consider making n_epochs very small.')
class CharRNN(nn.Module):
def __init__(self, tokens, n_hidden=256, n_layers=2,
drop_prob=0.5, lr=0.001):
super().__init__()
self.drop_prob = drop_prob
self.n_layers = n_layers
self.n_hidden = n_hidden
self.lr = lr
# creating character dictionaries
self.chars = tokens
self.int2char = dict(enumerate(self.chars))
self.char2int = {ch: ii for ii, ch in self.int2char.items()}
## Define the layers of the model
## Define the LSTM
self.lstm = nn.LSTM(len(self.chars), n_hidden, n_layers,
dropout=drop_prob, batch_first=True)
## Define a dropout layer
self.dropout = nn.Dropout(drop_prob)
## Define the final, fully-connected output layer
self.fc = nn.Linear(n_hidden, len(self.chars))
def forward(self, x, hidden):
''' Forward pass through the network.
These inputs are x, and the hidden/cell state `hidden`. '''
## Get the outputs and the new hidden state from the lstm
r_output, hidden = self.lstm(x, hidden)
## Pass through a dropout layer
out = self.dropout(r_output)
# Stack up LSTM outputs using view
# you may need to use contiguous to reshape the output
out = out.contiguous().view(-1, self.n_hidden)
## Put x through the fully-connected layer
out = self.fc(out)
# return the final output and the hidden state
return out, hidden
def init_hidden(self, batch_size):
''' Initializes hidden state '''
# Create two new tensors with sizes n_layers x batch_size x n_hidden,
# initialized to zero, for hidden state and cell state of LSTM
weight = next(self.parameters()).data
if (train_on_gpu):
hidden = (weight.new(self.n_layers, batch_size, self.n_hidden).zero_().cuda(),
weight.new(self.n_layers, batch_size, self.n_hidden).zero_().cuda())
else:
hidden = (weight.new(self.n_layers, batch_size, self.n_hidden).zero_(),
weight.new(self.n_layers, batch_size, self.n_hidden).zero_())
return hidden
```
## Time to train
The train function gives us the ability to set the number of epochs, the learning rate, and other parameters.
Below we're using an Adam optimizer and cross entropy loss since we are looking at character class scores as output. We calculate the loss and perform backpropagation, as usual!
A couple of details about training:
>* Within the batch loop, we detach the hidden state from its history; this time setting it equal to a new *tuple* variable because an LSTM has a hidden state that is a tuple of the hidden and cell states.
* We use [`clip_grad_norm_`](https://pytorch.org/docs/stable/_modules/torch/nn/utils/clip_grad.html) to help prevent exploding gradients.
```
def train(net, data, epochs=10, batch_size=10, seq_length=50, lr=0.001, clip=5, val_frac=0.1, print_every=10):
''' Training a network
Arguments
---------
net: CharRNN network
data: text data to train the network
epochs: Number of epochs to train
batch_size: Number of mini-sequences per mini-batch, aka batch size
seq_length: Number of character steps per mini-batch
lr: learning rate
clip: gradient clipping
val_frac: Fraction of data to hold out for validation
print_every: Number of steps for printing training and validation loss
'''
net.train()
opt = torch.optim.Adam(net.parameters(), lr=lr)
criterion = nn.CrossEntropyLoss()
# create training and validation data
val_idx = int(len(data)*(1-val_frac))
data, val_data = data[:val_idx], data[val_idx:]
if(train_on_gpu):
net.cuda()
counter = 0
n_chars = len(net.chars)
for e in range(epochs):
# initialize hidden state
h = net.init_hidden(batch_size)
for x, y in get_batches(data, batch_size, seq_length):
counter += 1
# One-hot encode our data and make them Torch tensors
x = one_hot_encode(x, n_chars)
inputs, targets = torch.from_numpy(x), torch.from_numpy(y)
if(train_on_gpu):
inputs, targets = inputs.cuda(), targets.cuda()
# Creating new variables for the hidden state, otherwise
# we'd backprop through the entire training history
h = tuple([each.data for each in h])
# zero accumulated gradients
net.zero_grad()
# get the output from the model
output, h = net(inputs, h)
# calculate the loss and perform backprop
loss = criterion(output, targets.view(batch_size*seq_length))
loss.backward()
# `clip_grad_norm` helps prevent the exploding gradient problem in RNNs / LSTMs.
nn.utils.clip_grad_norm_(net.parameters(), clip)
opt.step()
# loss stats
if counter % print_every == 0:
# Get validation loss
val_h = net.init_hidden(batch_size)
val_losses = []
net.eval()
for x, y in get_batches(val_data, batch_size, seq_length):
# One-hot encode our data and make them Torch tensors
x = one_hot_encode(x, n_chars)
x, y = torch.from_numpy(x), torch.from_numpy(y)
# Creating new variables for the hidden state, otherwise
# we'd backprop through the entire training history
val_h = tuple([each.data for each in val_h])
inputs, targets = x, y
if(train_on_gpu):
inputs, targets = inputs.cuda(), targets.cuda()
output, val_h = net(inputs, val_h)
val_loss = criterion(output, targets.view(batch_size*seq_length))
val_losses.append(val_loss.item())
net.train() # reset to train mode after iterationg through validation data
print("Epoch: {}/{}...".format(e+1, epochs),
"Step: {}...".format(counter),
"Loss: {:.4f}...".format(loss.item()),
"Val Loss: {:.4f}".format(np.mean(val_losses)))
```
## Instantiating the model
Now we can actually train the network. First we'll create the network itself, with some given hyperparameters. Then, define the mini-batches sizes, and start training!
```
## Set you model hyperparameters
# Define and print the net
n_hidden=512
n_layers=2
net = CharRNN(chars, n_hidden, n_layers)
print(net)
```
### Set your training hyperparameters!
```
batch_size = 128
seq_length = 100
n_epochs = 20 # start small if you are just testing initial behavior
# train the model
train(net, encoded, epochs=n_epochs, batch_size=batch_size, seq_length=seq_length, lr=0.001, print_every=10)
```
## Getting the best model
To set your hyperparameters to get the best performance, you'll want to watch the training and validation losses. If your training loss is much lower than the validation loss, you're overfitting. Increase regularization (more dropout) or use a smaller network. If the training and validation losses are close, you're underfitting so you can increase the size of the network.
## Hyperparameters
Here are the hyperparameters for the network.
In defining the model:
* `n_hidden` - The number of units in the hidden layers.
* `n_layers` - Number of hidden LSTM layers to use.
We assume that dropout probability and learning rate will be kept at the default, in this example.
And in training:
* `batch_size` - Number of sequences running through the network in one pass.
* `seq_length` - Number of characters in the sequence the network is trained on. Larger is better typically, the network will learn more long range dependencies. But it takes longer to train. 100 is typically a good number here.
* `lr` - Learning rate for training
Here's some good advice from Andrej Karpathy on training the network. I'm going to copy it in here for your benefit, but also link to [where it originally came from](https://github.com/karpathy/char-rnn#tips-and-tricks).
> ## Tips and Tricks
>### Monitoring Validation Loss vs. Training Loss
>If you're somewhat new to Machine Learning or Neural Networks it can take a bit of expertise to get good models. The most important quantity to keep track of is the difference between your training loss (printed during training) and the validation loss (printed once in a while when the RNN is run on the validation data (by default every 1000 iterations)). In particular:
> - If your training loss is much lower than validation loss then this means the network might be **overfitting**. Solutions to this are to decrease your network size, or to increase dropout. For example you could try dropout of 0.5 and so on.
> - If your training/validation loss are about equal then your model is **underfitting**. Increase the size of your model (either number of layers or the raw number of neurons per layer)
> ### Approximate number of parameters
> The two most important parameters that control the model are `n_hidden` and `n_layers`. I would advise that you always use `n_layers` of either 2/3. The `n_hidden` can be adjusted based on how much data you have. The two important quantities to keep track of here are:
> - The number of parameters in your model. This is printed when you start training.
> - The size of your dataset. 1MB file is approximately 1 million characters.
>These two should be about the same order of magnitude. It's a little tricky to tell. Here are some examples:
> - I have a 100MB dataset and I'm using the default parameter settings (which currently print 150K parameters). My data size is significantly larger (100 mil >> 0.15 mil), so I expect to heavily underfit. I am thinking I can comfortably afford to make `n_hidden` larger.
> - I have a 10MB dataset and running a 10 million parameter model. I'm slightly nervous and I'm carefully monitoring my validation loss. If it's larger than my training loss then I may want to try to increase dropout a bit and see if that helps the validation loss.
> ### Best models strategy
>The winning strategy to obtaining very good models (if you have the compute time) is to always err on making the network larger (as large as you're willing to wait for it to compute) and then try different dropout values (between 0,1). Whatever model has the best validation performance (the loss, written in the checkpoint filename, low is good) is the one you should use in the end.
>It is very common in deep learning to run many different models with many different hyperparameter settings, and in the end take whatever checkpoint gave the best validation performance.
>By the way, the size of your training and validation splits are also parameters. Make sure you have a decent amount of data in your validation set or otherwise the validation performance will be noisy and not very informative.
## Checkpoint
After training, we'll save the model so we can load it again later if we need too. Here I'm saving the parameters needed to create the same architecture, the hidden layer hyperparameters and the text characters.
```
# change the name, for saving multiple files
model_name = 'rnn_x_epoch.net'
checkpoint = {'n_hidden': net.n_hidden,
'n_layers': net.n_layers,
'state_dict': net.state_dict(),
'tokens': net.chars}
with open(model_name, 'wb') as f:
torch.save(checkpoint, f)
```
---
## Making Predictions
Now that the model is trained, we'll want to sample from it and make predictions about next characters! To sample, we pass in a character and have the network predict the next character. Then we take that character, pass it back in, and get another predicted character. Just keep doing this and you'll generate a bunch of text!
### A note on the `predict` function
The output of our RNN is from a fully-connected layer and it outputs a **distribution of next-character scores**.
> To actually get the next character, we apply a softmax function, which gives us a *probability* distribution that we can then sample to predict the next character.
### Top K sampling
Our predictions come from a categorical probability distribution over all the possible characters. We can make the sample text and make it more reasonable to handle (with less variables) by only considering some $K$ most probable characters. This will prevent the network from giving us completely absurd characters while allowing it to introduce some noise and randomness into the sampled text. Read more about [topk, here](https://pytorch.org/docs/stable/torch.html#torch.topk).
```
def predict(net, char, h=None, top_k=None):
''' Given a character, predict the next character.
Returns the predicted character and the hidden state.
'''
# tensor inputs
x = np.array([[net.char2int[char]]])
x = one_hot_encode(x, len(net.chars))
inputs = torch.from_numpy(x)
if(train_on_gpu):
inputs = inputs.cuda()
# detach hidden state from history
h = tuple([each.data for each in h])
# get the output of the model
out, h = net(inputs, h)
# get the character probabilities
p = F.softmax(out, dim=1).data
if(train_on_gpu):
p = p.cpu() # move to cpu
# get top characters
if top_k is None:
top_ch = np.arange(len(net.chars))
else:
p, top_ch = p.topk(top_k)
top_ch = top_ch.numpy().squeeze()
# select the likely next character with some element of randomness
p = p.numpy().squeeze()
char = np.random.choice(top_ch, p=p/p.sum())
# return the encoded value of the predicted char and the hidden state
return net.int2char[char], h
```
### Priming and generating text
Typically you'll want to prime the network so you can build up a hidden state. Otherwise the network will start out generating characters at random. In general the first bunch of characters will be a little rough since it hasn't built up a long history of characters to predict from.
```
def sample(net, size, prime='The', top_k=None):
if(train_on_gpu):
net.cuda()
else:
net.cpu()
net.eval() # eval mode
# First off, run through the prime characters
chars = [ch for ch in prime]
h = net.init_hidden(1)
for ch in prime:
char, h = predict(net, ch, h, top_k=top_k)
chars.append(char)
# Now pass in the previous character and get a new one
for ii in range(size):
char, h = predict(net, chars[-1], h, top_k=top_k)
chars.append(char)
return ''.join(chars)
print(sample(net, 1000, prime='Anna', top_k=5))
```
## Loading a checkpoint
```
# Here we have loaded in a model that trained over 20 epochs `rnn_20_epoch.net`
with open('rnn_x_epoch.net', 'rb') as f:
checkpoint = torch.load(f)
loaded = CharRNN(checkpoint['tokens'], n_hidden=checkpoint['n_hidden'], n_layers=checkpoint['n_layers'])
loaded.load_state_dict(checkpoint['state_dict'])
# Sample using a loaded model
print(sample(loaded, 2000, top_k=5, prime="And Levin said"))
```
|
github_jupyter
|
import numpy as np
import torch
from torch import nn
import torch.nn.functional as F
# open text file and read in data as `text`
with open('data/anna.txt', 'r') as f:
text = f.read()
text[:100]
# encode the text and map each character to an integer and vice versa
# we create two dictionaries:
# 1. int2char, which maps integers to characters
# 2. char2int, which maps characters to unique integers
chars = tuple(set(text))
int2char = dict(enumerate(chars))
char2int = {ch: ii for ii, ch in int2char.items()}
# encode the text
encoded = np.array([char2int[ch] for ch in text])
encoded[:100]
def one_hot_encode(arr, n_labels):
# Initialize the the encoded array
one_hot = np.zeros((np.multiply(*arr.shape), n_labels), dtype=np.float32)
# Fill the appropriate elements with ones
one_hot[np.arange(one_hot.shape[0]), arr.flatten()] = 1.
# Finally reshape it to get back to the original array
one_hot = one_hot.reshape((*arr.shape, n_labels))
return one_hot
# check that the function works as expected
test_seq = np.array([[3, 5, 1]])
one_hot = one_hot_encode(test_seq, 8)
print(one_hot)
def get_batches(arr, batch_size, seq_length):
'''Create a generator that returns batches of size
batch_size x seq_length from arr.
Arguments
---------
arr: Array you want to make batches from
batch_size: Batch size, the number of sequences per batch
seq_length: Number of encoded chars in a sequence
'''
batch_size_total = batch_size * seq_length
## Get the number of batches we can make
n_batches = len(arr)//batch_size_total
## Keep only enough characters to make full batches
arr = arr[:n_batches * batch_size_total]
## Reshape into batch_size rows
arr = arr.reshape((batch_size, -1))
## Iterate over the batches using a window of size seq_length
for n in range(0, arr.shape[1], seq_length):
# The features
x = arr[:, n:n+seq_length]
# The targets, shifted by one
y = np.zeros_like(x)
try:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, n+seq_length]
except IndexError:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, 0]
yield x, y
batches = get_batches(encoded, 8, 50)
x, y = next(batches)
# printing out the first 10 items in a sequence
print('x\n', x[:10, :10])
print('\ny\n', y[:10, :10])
x
[[25 8 60 11 45 27 28 73 1 2]
[17 7 20 73 45 8 60 45 73 60]
[27 20 80 73 7 28 73 60 73 65]
[17 73 45 8 27 73 66 8 46 27]
[73 17 60 12 73 8 27 28 73 45]
[66 64 17 17 46 7 20 73 60 20]
[73 76 20 20 60 73 8 60 80 73]
[47 35 43 7 20 17 24 50 37 73]]
y
[[ 8 60 11 45 27 28 73 1 2 2]
[ 7 20 73 45 8 60 45 73 60 45]
[20 80 73 7 28 73 60 73 65 7]
[73 45 8 27 73 66 8 46 27 65]
[17 60 12 73 8 27 28 73 45 27]
[64 17 17 46 7 20 73 60 20 80]
[76 20 20 60 73 8 60 80 73 17]
[35 43 7 20 17 24 50 37 73 36]]
```
although the exact numbers may be different. Check to make sure the data is shifted over one step for `y`.
---
## Defining the network with PyTorch
Below is where you'll define the network.
<img src="assets/charRNN.png" width=500px>
Next, you'll use PyTorch to define the architecture of the network. We start by defining the layers and operations we want. Then, define a method for the forward pass. You've also been given a method for predicting characters.
### Model Structure
In `__init__` the suggested structure is as follows:
* Create and store the necessary dictionaries (this has been done for you)
* Define an LSTM layer that takes as params: an input size (the number of characters), a hidden layer size `n_hidden`, a number of layers `n_layers`, a dropout probability `drop_prob`, and a batch_first boolean (True, since we are batching)
* Define a dropout layer with `dropout_prob`
* Define a fully-connected layer with params: input size `n_hidden` and output size (the number of characters)
* Finally, initialize the weights (again, this has been given)
Note that some parameters have been named and given in the `__init__` function, and we use them and store them by doing something like `self.drop_prob = drop_prob`.
---
### LSTM Inputs/Outputs
You can create a basic [LSTM layer](https://pytorch.org/docs/stable/nn.html#lstm) as follows
where `input_size` is the number of characters this cell expects to see as sequential input, and `n_hidden` is the number of units in the hidden layers in the cell. And we can add dropout by adding a dropout parameter with a specified probability; this will automatically add dropout to the inputs or outputs. Finally, in the `forward` function, we can stack up the LSTM cells into layers using `.view`. With this, you pass in a list of cells and it will send the output of one cell into the next cell.
We also need to create an initial hidden state of all zeros. This is done like so
## Time to train
The train function gives us the ability to set the number of epochs, the learning rate, and other parameters.
Below we're using an Adam optimizer and cross entropy loss since we are looking at character class scores as output. We calculate the loss and perform backpropagation, as usual!
A couple of details about training:
>* Within the batch loop, we detach the hidden state from its history; this time setting it equal to a new *tuple* variable because an LSTM has a hidden state that is a tuple of the hidden and cell states.
* We use [`clip_grad_norm_`](https://pytorch.org/docs/stable/_modules/torch/nn/utils/clip_grad.html) to help prevent exploding gradients.
## Instantiating the model
Now we can actually train the network. First we'll create the network itself, with some given hyperparameters. Then, define the mini-batches sizes, and start training!
### Set your training hyperparameters!
## Getting the best model
To set your hyperparameters to get the best performance, you'll want to watch the training and validation losses. If your training loss is much lower than the validation loss, you're overfitting. Increase regularization (more dropout) or use a smaller network. If the training and validation losses are close, you're underfitting so you can increase the size of the network.
## Hyperparameters
Here are the hyperparameters for the network.
In defining the model:
* `n_hidden` - The number of units in the hidden layers.
* `n_layers` - Number of hidden LSTM layers to use.
We assume that dropout probability and learning rate will be kept at the default, in this example.
And in training:
* `batch_size` - Number of sequences running through the network in one pass.
* `seq_length` - Number of characters in the sequence the network is trained on. Larger is better typically, the network will learn more long range dependencies. But it takes longer to train. 100 is typically a good number here.
* `lr` - Learning rate for training
Here's some good advice from Andrej Karpathy on training the network. I'm going to copy it in here for your benefit, but also link to [where it originally came from](https://github.com/karpathy/char-rnn#tips-and-tricks).
> ## Tips and Tricks
>### Monitoring Validation Loss vs. Training Loss
>If you're somewhat new to Machine Learning or Neural Networks it can take a bit of expertise to get good models. The most important quantity to keep track of is the difference between your training loss (printed during training) and the validation loss (printed once in a while when the RNN is run on the validation data (by default every 1000 iterations)). In particular:
> - If your training loss is much lower than validation loss then this means the network might be **overfitting**. Solutions to this are to decrease your network size, or to increase dropout. For example you could try dropout of 0.5 and so on.
> - If your training/validation loss are about equal then your model is **underfitting**. Increase the size of your model (either number of layers or the raw number of neurons per layer)
> ### Approximate number of parameters
> The two most important parameters that control the model are `n_hidden` and `n_layers`. I would advise that you always use `n_layers` of either 2/3. The `n_hidden` can be adjusted based on how much data you have. The two important quantities to keep track of here are:
> - The number of parameters in your model. This is printed when you start training.
> - The size of your dataset. 1MB file is approximately 1 million characters.
>These two should be about the same order of magnitude. It's a little tricky to tell. Here are some examples:
> - I have a 100MB dataset and I'm using the default parameter settings (which currently print 150K parameters). My data size is significantly larger (100 mil >> 0.15 mil), so I expect to heavily underfit. I am thinking I can comfortably afford to make `n_hidden` larger.
> - I have a 10MB dataset and running a 10 million parameter model. I'm slightly nervous and I'm carefully monitoring my validation loss. If it's larger than my training loss then I may want to try to increase dropout a bit and see if that helps the validation loss.
> ### Best models strategy
>The winning strategy to obtaining very good models (if you have the compute time) is to always err on making the network larger (as large as you're willing to wait for it to compute) and then try different dropout values (between 0,1). Whatever model has the best validation performance (the loss, written in the checkpoint filename, low is good) is the one you should use in the end.
>It is very common in deep learning to run many different models with many different hyperparameter settings, and in the end take whatever checkpoint gave the best validation performance.
>By the way, the size of your training and validation splits are also parameters. Make sure you have a decent amount of data in your validation set or otherwise the validation performance will be noisy and not very informative.
## Checkpoint
After training, we'll save the model so we can load it again later if we need too. Here I'm saving the parameters needed to create the same architecture, the hidden layer hyperparameters and the text characters.
---
## Making Predictions
Now that the model is trained, we'll want to sample from it and make predictions about next characters! To sample, we pass in a character and have the network predict the next character. Then we take that character, pass it back in, and get another predicted character. Just keep doing this and you'll generate a bunch of text!
### A note on the `predict` function
The output of our RNN is from a fully-connected layer and it outputs a **distribution of next-character scores**.
> To actually get the next character, we apply a softmax function, which gives us a *probability* distribution that we can then sample to predict the next character.
### Top K sampling
Our predictions come from a categorical probability distribution over all the possible characters. We can make the sample text and make it more reasonable to handle (with less variables) by only considering some $K$ most probable characters. This will prevent the network from giving us completely absurd characters while allowing it to introduce some noise and randomness into the sampled text. Read more about [topk, here](https://pytorch.org/docs/stable/torch.html#torch.topk).
### Priming and generating text
Typically you'll want to prime the network so you can build up a hidden state. Otherwise the network will start out generating characters at random. In general the first bunch of characters will be a little rough since it hasn't built up a long history of characters to predict from.
## Loading a checkpoint
| 0.885977 | 0.962918 |
<a href="https://colab.research.google.com/github/AI4Finance-Foundation/ElegantRL/blob/master/tutorial_BipedalWalker.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# **BipedalWalker-v3 Example in ElegantRL**
# **Part 1: Testing Task Description**
[BipedalWalker-v3](https://gym.openai.com/envs/BipedalWalker-v2/) is a robotic task in OpenAI Gym since it performs one of the most fundamental skills: moving. In this task, our goal is to make a 2D biped walker to walk through rough terrain. BipedalWalker is a difficult task in continuous action space, and there are only a few RL implementations can reach the target reward.
# **Part 2: Install ElegantRL**
```
# install elegantrl library
!pip install git+https://github.com/AI4Finance-LLC/ElegantRL.git
```
# **Part 3: Import Packages**
* **elegantrl**
* **OpenAI Gym**: a toolkit for developing and comparing reinforcement learning algorithms.
* **PyBullet Gym**: an open-source implementation of the OpenAI Gym MuJoCo environments.
```
from elegantrl.train.run_tutorial import *
from elegantrl.train.config import Arguments
from elegantrl.agents.AgentTD3 import AgentTD3
from elegantrl.envs.Gym import build_env
import gym
gym.logger.set_level(40) # Block warning
```
# **Part 4: Specify Agent and Environment**
* **args.agent**: firstly chooses one DRL algorithm to use, and the user is able to choose any agent from agent.py
* **args.env**: creates and preprocesses the environment, and the user can either customize own environment or preprocess environments from OpenAI Gym and PyBullet Gym from env.py.
> Before finishing initialization of **args**, please see Arguments() in run.py for more details about adjustable hyper-parameters.
```
agent = AgentTD3() # AgentSAC(), AgentTD3(), AgentDDPG()
env = build_env('BipedalWalker-v3')
args = Arguments(env, agent)
args.eval_times1 = 2 ** 3
args.eval_times2 = 2 ** 5
args.gamma = 0.98
args.target_step = args.env.max_step
```
# **Part 5: Train and Evaluate the Agent**
> The training and evaluating processes are all finished inside function **train_and_evaluate()**, and the only parameter for it is **args**. It includes the fundamental objects in DRL:
* agent,
* environment.
> And it also includes the parameters for training-control:
* batch_size,
* target_step,
* reward_scale,
* gamma, etc.
> The parameters for evaluation-control:
* break_step,
* random_seed, etc.
```
train_and_evaluate(args) # the training process will terminate once it reaches the target reward.
```
Understanding the above results::
* **Step**: the total training steps.
* **MaxR**: the maximum reward.
* **avgR**: the average of the rewards.
* **stdR**: the standard deviation of the rewards.
* **objA**: the objective function value of Actor Network (Policy Network).
* **objC**: the objective function value (Q-value) of Critic Network (Value Network).
```
from google.colab.patches import cv2_imshow
import cv2
img = cv2.imread(f"/content/{args.cwd}/plot_learning_curve.jpg", cv2.IMREAD_UNCHANGED)
cv2_imshow(img)
```
|
github_jupyter
|
# install elegantrl library
!pip install git+https://github.com/AI4Finance-LLC/ElegantRL.git
from elegantrl.train.run_tutorial import *
from elegantrl.train.config import Arguments
from elegantrl.agents.AgentTD3 import AgentTD3
from elegantrl.envs.Gym import build_env
import gym
gym.logger.set_level(40) # Block warning
agent = AgentTD3() # AgentSAC(), AgentTD3(), AgentDDPG()
env = build_env('BipedalWalker-v3')
args = Arguments(env, agent)
args.eval_times1 = 2 ** 3
args.eval_times2 = 2 ** 5
args.gamma = 0.98
args.target_step = args.env.max_step
train_and_evaluate(args) # the training process will terminate once it reaches the target reward.
from google.colab.patches import cv2_imshow
import cv2
img = cv2.imread(f"/content/{args.cwd}/plot_learning_curve.jpg", cv2.IMREAD_UNCHANGED)
cv2_imshow(img)
| 0.444565 | 0.982339 |
```
import pandas as pd
data=pd.read_csv(r'C:\Users\kshitij\Downloads\ContextuaRecommendationSystem.csv')
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
data.head()
data["Would you like to get recommended Holiday Theme Movies On Holidays (Ex : Border on Independence Day and Dangal /Marry Kom on Women's Day) -:"]=data["Would you like to get recommended Holiday Theme Movies On Holidays (Ex : Border on Independence Day and Dangal /Marry Kom on Women's Day) -:"].fillna(0)
data.shape
for i,j in data.iterrows():
for k in j[2].split(";"):
genre[k]=genre[k]+1
print(genre)
genre.values
```
# Genre preference in morning
```
import matplotlib.pyplot as plt
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[2].split(";"):
genre[k]=genre[k]+1
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
# Genre preference during afternoon
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[3].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
## Genre preference in evening
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[4].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
## Genre preference at night
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[5].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
## Genre Preference during Winter
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[6].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
# Genre Preference during Summer
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[7].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
## Genre Preference during Rainy/Monsoon
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[8].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
# Genre Preference during Autumn
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[9].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
# Genre Preference when on Large Screen Device
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime and Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[10].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
# Genre Preference when on Tablet-Size Screen
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime and Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[11].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
data.columns
data.shape
```
# Genre Preference when on Phone-Size Screen
```
data.dropna(subset=["Genre Preference when on Phone-Size Screen -: ( Choose all that apply )",inplace=True)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime and Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[12].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
data.shape
```
## Genre Preference when you are happy
```
data.dropna(subset=["Movie Genre when you are Happy -: ( Choose all that apply )"],inplace=True)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime and Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[13].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
## Movie Genre when you are Sad
```
data.dropna(subset=['Movie Genre when you are Sad -: ( Choose all that apply )','Movie Genre when you are Angry -: ( Choose all that apply )','Movie Genre when you are Angry -: ( Choose all that apply )','Movie Genre when you are Normal -: ( Choose all that apply )','Genre Preference when watching with Family -: ( Choose all that apply )','Genre Preference when watching with Friends -: ( Choose all that apply )'],inplace=True)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime and Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[14].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
## Movie Genre when you are Angry
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[15].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
## Movie Genre when you are Normal
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[16].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
```
## Genre Preference when watching with Friends
```
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[17].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
data.columns
```
## Genre Preference when watching with Family
```
i=0
for j in data['Genre Preference when watching with Family -: ( Choose all that apply )'].isnull():
if(j):
data=data.drop(i)
i=i+1
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[18].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
data.shape
```
## Would you like to get recommended Holiday Theme Movies On Holidays
```
import numpy as np
import statistics
x=np.arange(1,64)
y=[]
import matplotlib.pyplot as plt
for i,j in data.iterrows():
y.append(j[21])
plt.bar(x,y)
print("Average rating :")
print(statistics.mean(y))
if(statistics.mean(y)>=5):
print("YES , users would like to get recommended holiday theme movies in holidays")
data.head()
```
# Chi Square Test
```
genremorning=[]
genreafternoon=[]
genreevening=[]
genrenight=[]
genrewinter,genresummer,genrerainy,genreautumn,genreweekend,genreweekdays,genrelargescreen,genresmallscreen,genretabletscreen,genrehappy,genresad,genreangry,genrenormal,genrewithfamily,genrewithoutfamily=[],[],[],[],[],[],[],[],[],[],[],[],[],[],[]
genre=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"]
for i,j in data.iterrows():
genremorning=genremorning+j[2].split(";")
genreafternoon=genreafternoon+j[3].split(";")
genreevening=genreevening+j[4].split(";")
genrenight=genrenight+j[5].split(";")
genrewinter=genrewinter+j[6].split(";")
genresummer=genresummer+j[7].split(";")
genrerainy=genrerainy+j[8].split(";")
genreautumn=genreautumn+j[9].split(";")
genreweekend=genreweekend+j[10].split(";")
genreweekdays=genreweekdays+j[11].split(";")
genrelargescreen=genrelargescreen+j[12].split(";")
#genresmallscreen=genresmallscreen+j[13].split(";")
genretabletscreen=genretabletscreen+j[14].split(";")
genrehappy=genrehappy+j[15].split(";")
genresad=genresad+j[16].split(";")
genreangry=genreangry+j[17].split(";")
genrenormal=genrenormal+j[18].split(";")
genrewithfamily=genrewithfamily+j[19].split(";")
genrewithoutfamily=genrewithoutfamily+j[20].split(";")
df=pd.DataFrame(pd.Series(genremorning).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["afternooncount"]=pd.Series(pd.Series(genreafternoon).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["eveningnooncount"]=pd.Series(pd.Series(genreevening).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["nightooncount"]=pd.Series(pd.Series(genrenight).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["winter"]=pd.Series(pd.Series(genrewinter).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["summer"]=pd.Series(pd.Series(genresummer).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["rainy"]=pd.Series(pd.Series(genrerainy).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["autumn"]=pd.Series(pd.Series(genreautumn).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["weekend"]=pd.Series(pd.Series(genreweekend).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["weekdays"]=pd.Series(pd.Series(genreweekdays).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["largscreen"]=pd.Series(pd.Series(genrelargescreen).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["tablet"]=pd.Series(pd.Series(genretabletscreen).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["happy"]=pd.Series(pd.Series(genrehappy).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["sad"]=pd.Series(pd.Series(genresad).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["angry"]=pd.Series(pd.Series(genreangry).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["normal"]=pd.Series(pd.Series(genrenormal).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["with family"]=pd.Series(pd.Series(genrewithfamily).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["without family"]=pd.Series(pd.Series(genrewithoutfamily).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
```
## Contingency Table
```
df.columns=["Morning","Afternoon","Evening","Night","Winter","Summer","Rainy","Autumn","Weekend","Weekdays","Largscreen","Tablet","Happy","Sad","Angry","Normal","With Family","Without Family"]
df.head()
df.loc['Total',:]= df.sum(axis=0)
df.loc[:,'Total'] = df.sum(axis=1)
df
expected=np.outer(df["Total"][0:7],
df.loc["Total"][0:18])/2868
print(expected)
expected_df=pd.DataFrame(expected)
expected_df
expected_df=expected_df.fillna(0)
df=df.fillna(0)
expected_df
chisq=0
for i in range(0,7):
for j in range(0,18):
x=(df.iloc[i,j]-expected_df.iloc[i,j])*(df.iloc[i,j]-expected_df.iloc[i,j])
x=x/expected_df.iloc[i,j]
chisq=chisq+x
print(chisq)
import scipy.stats as stats
dfreedom=17*6
crit = stats.chi2.ppf(q = 0.95, df = dfreedom)
print(crit)
observe=df.iloc[0:7,0:18]
stats.chi2_contingency(observed=observe)
print("chisquare is greater than pvalue which indicates attributes are corelated")
```
|
github_jupyter
|
import pandas as pd
data=pd.read_csv(r'C:\Users\kshitij\Downloads\ContextuaRecommendationSystem.csv')
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
data.head()
data["Would you like to get recommended Holiday Theme Movies On Holidays (Ex : Border on Independence Day and Dangal /Marry Kom on Women's Day) -:"]=data["Would you like to get recommended Holiday Theme Movies On Holidays (Ex : Border on Independence Day and Dangal /Marry Kom on Women's Day) -:"].fillna(0)
data.shape
for i,j in data.iterrows():
for k in j[2].split(";"):
genre[k]=genre[k]+1
print(genre)
genre.values
import matplotlib.pyplot as plt
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[2].split(";"):
genre[k]=genre[k]+1
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[3].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[4].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[5].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[6].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[7].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[8].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[9].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime and Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[10].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime and Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[11].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
data.columns
data.shape
data.dropna(subset=["Genre Preference when on Phone-Size Screen -: ( Choose all that apply )",inplace=True)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime and Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[12].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
data.shape
data.dropna(subset=["Movie Genre when you are Happy -: ( Choose all that apply )"],inplace=True)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime and Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[13].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
data.dropna(subset=['Movie Genre when you are Sad -: ( Choose all that apply )','Movie Genre when you are Angry -: ( Choose all that apply )','Movie Genre when you are Angry -: ( Choose all that apply )','Movie Genre when you are Normal -: ( Choose all that apply )','Genre Preference when watching with Family -: ( Choose all that apply )','Genre Preference when watching with Friends -: ( Choose all that apply )'],inplace=True)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime and Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[14].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[15].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[16].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[17].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
data.columns
i=0
for j in data['Genre Preference when watching with Family -: ( Choose all that apply )'].isnull():
if(j):
data=data.drop(i)
i=i+1
genre={"Comedy":0,"Romance":0,"Drama":0,"Sci-Fi":0,"Action":0,"Crime And Thriller":0,"Documentary":0}
for i,j in data.iterrows():
for k in j[18].split(";"):
genre[k]=genre[k]+1
import matplotlib.pyplot as plt
plt.pie(genre.values(),labels=genre.keys())
#plt.legend(loc="right")
plt.show()
print(genre)
data.shape
import numpy as np
import statistics
x=np.arange(1,64)
y=[]
import matplotlib.pyplot as plt
for i,j in data.iterrows():
y.append(j[21])
plt.bar(x,y)
print("Average rating :")
print(statistics.mean(y))
if(statistics.mean(y)>=5):
print("YES , users would like to get recommended holiday theme movies in holidays")
data.head()
genremorning=[]
genreafternoon=[]
genreevening=[]
genrenight=[]
genrewinter,genresummer,genrerainy,genreautumn,genreweekend,genreweekdays,genrelargescreen,genresmallscreen,genretabletscreen,genrehappy,genresad,genreangry,genrenormal,genrewithfamily,genrewithoutfamily=[],[],[],[],[],[],[],[],[],[],[],[],[],[],[]
genre=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"]
for i,j in data.iterrows():
genremorning=genremorning+j[2].split(";")
genreafternoon=genreafternoon+j[3].split(";")
genreevening=genreevening+j[4].split(";")
genrenight=genrenight+j[5].split(";")
genrewinter=genrewinter+j[6].split(";")
genresummer=genresummer+j[7].split(";")
genrerainy=genrerainy+j[8].split(";")
genreautumn=genreautumn+j[9].split(";")
genreweekend=genreweekend+j[10].split(";")
genreweekdays=genreweekdays+j[11].split(";")
genrelargescreen=genrelargescreen+j[12].split(";")
#genresmallscreen=genresmallscreen+j[13].split(";")
genretabletscreen=genretabletscreen+j[14].split(";")
genrehappy=genrehappy+j[15].split(";")
genresad=genresad+j[16].split(";")
genreangry=genreangry+j[17].split(";")
genrenormal=genrenormal+j[18].split(";")
genrewithfamily=genrewithfamily+j[19].split(";")
genrewithoutfamily=genrewithoutfamily+j[20].split(";")
df=pd.DataFrame(pd.Series(genremorning).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["afternooncount"]=pd.Series(pd.Series(genreafternoon).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["eveningnooncount"]=pd.Series(pd.Series(genreevening).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["nightooncount"]=pd.Series(pd.Series(genrenight).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["winter"]=pd.Series(pd.Series(genrewinter).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["summer"]=pd.Series(pd.Series(genresummer).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["rainy"]=pd.Series(pd.Series(genrerainy).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["autumn"]=pd.Series(pd.Series(genreautumn).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["weekend"]=pd.Series(pd.Series(genreweekend).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["weekdays"]=pd.Series(pd.Series(genreweekdays).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["largscreen"]=pd.Series(pd.Series(genrelargescreen).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["tablet"]=pd.Series(pd.Series(genretabletscreen).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["happy"]=pd.Series(pd.Series(genrehappy).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["sad"]=pd.Series(pd.Series(genresad).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["angry"]=pd.Series(pd.Series(genreangry).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["normal"]=pd.Series(pd.Series(genrenormal).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["with family"]=pd.Series(pd.Series(genrewithfamily).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df["without family"]=pd.Series(pd.Series(genrewithoutfamily).value_counts(),index=["Comedy","Romance","Drama","Sci-Fi","Action","Crime And Thriller","Documentary"])
df.columns=["Morning","Afternoon","Evening","Night","Winter","Summer","Rainy","Autumn","Weekend","Weekdays","Largscreen","Tablet","Happy","Sad","Angry","Normal","With Family","Without Family"]
df.head()
df.loc['Total',:]= df.sum(axis=0)
df.loc[:,'Total'] = df.sum(axis=1)
df
expected=np.outer(df["Total"][0:7],
df.loc["Total"][0:18])/2868
print(expected)
expected_df=pd.DataFrame(expected)
expected_df
expected_df=expected_df.fillna(0)
df=df.fillna(0)
expected_df
chisq=0
for i in range(0,7):
for j in range(0,18):
x=(df.iloc[i,j]-expected_df.iloc[i,j])*(df.iloc[i,j]-expected_df.iloc[i,j])
x=x/expected_df.iloc[i,j]
chisq=chisq+x
print(chisq)
import scipy.stats as stats
dfreedom=17*6
crit = stats.chi2.ppf(q = 0.95, df = dfreedom)
print(crit)
observe=df.iloc[0:7,0:18]
stats.chi2_contingency(observed=observe)
print("chisquare is greater than pvalue which indicates attributes are corelated")
| 0.083718 | 0.718533 |
# Data Wrangling with Pandas
Now that we have been exposed to the basic functionality of Pandas, lets explore some more advanced features that will be useful when addressing more complex data management tasks.
As most statisticians/data analysts will admit, often the lion's share of the time spent implementing an analysis is devoted to preparing the data itself, rather than to coding or running a particular model that uses the data. This is where Pandas and Python's standard library are beneficial, providing high-level, flexible, and efficient tools for manipulating your data as needed.
```
%matplotlib inline
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# Set some Pandas options
pd.set_option('display.notebook_repr_html', False)
pd.set_option('display.max_columns', 20)
pd.set_option('display.max_rows', 25)
```
## Date/Time data handling
Date and time data are inherently problematic. There are an unequal number of days in every month, an unequal number of days in a year (due to leap years), and time zones that vary over space. Yet information about time is essential in many analyses, particularly in the case of time series analysis.
The `datetime` built-in library handles temporal information down to the nanosecond.
```
from datetime import datetime
now = datetime.now()
now
now.day
now.weekday()
```
In addition to `datetime` there are simpler objects for date and time information only, respectively.
```
from datetime import date, time
time(3, 24)
date(1970, 9, 3)
```
Having a custom data type for dates and times is convenient because we can perform operations on them easily. For example, we may want to calculate the difference between two times:
```
my_age = now - datetime(1970, 9, 3)
my_age
my_age.days/365.
```
In this section, we will manipulate data collected from ocean-going vessels on the eastern seaboard. Vessel operations are monitored using the Automatic Identification System (AIS), a safety at sea navigation technology which vessels are required to maintain and that uses transponders to transmit very high frequency (VHF) radio signals containing static information including ship name, call sign, and country of origin, as well as dynamic information unique to a particular voyage such as vessel location, heading, and speed.
The International Maritime Organization’s (IMO) International Convention for the Safety of Life at Sea requires functioning AIS capabilities on all vessels 300 gross tons or greater and the US Coast Guard requires AIS on nearly all vessels sailing in U.S. waters. The Coast Guard has established a national network of AIS receivers that provides coverage of nearly all U.S. waters. AIS signals are transmitted several times each minute and the network is capable of handling thousands of reports per minute and updates as often as every two seconds. Therefore, a typical voyage in our study might include the transmission of hundreds or thousands of AIS encoded signals. This provides a rich source of spatial data that includes both spatial and temporal information.
For our purposes, we will use summarized data that describes the transit of a given vessel through a particular administrative area. The data includes the start and end time of the transit segment, as well as information about the speed of the vessel, how far it travelled, etc.
```
segments = pd.read_csv("data/AIS/transit_segments.csv")
segments.head()
```
For example, we might be interested in the distribution of transit lengths, so we can plot them as a histogram:
```
segments.seg_length.hist(bins=500)
```
Though most of the transits appear to be short, there are a few longer distances that make the plot difficult to read. This is where a transformation is useful:
```
segments.seg_length.apply(np.log).hist(bins=500)
```
We can see that although there are date/time fields in the dataset, they are not in any specialized format, such as `datetime`.
```
segments.st_time.dtype
```
Our first order of business will be to convert these data to `datetime`. The `strptime` method parses a string representation of a date and/or time field, according to the expected format of this information.
```
datetime.strptime(segments.st_time.ix[0], '%m/%d/%y %H:%M')
```
The `dateutil` package includes a parser that attempts to detect the format of the date strings, and convert them automatically.
```
from dateutil.parser import parse
parse(segments.st_time.ix[0])
```
We can convert all the dates in a particular column by using the `apply` method.
```
segments.st_time.apply(lambda d: datetime.strptime(d, '%m/%d/%y %H:%M'))
```
As a convenience, Pandas has a `to_datetime` method that will parse and convert an entire Series of formatted strings into `datetime` objects.
```
pd.to_datetime(segments.st_time)
```
Pandas also has a custom NA value for missing datetime objects, `NaT`.
```
pd.to_datetime([None])
```
Also, if `to_datetime()` has problems parsing any particular date/time format, you can pass the spec in using the `format=` argument.
## Merging and joining DataFrame objects
Now that we have the vessel transit information as we need it, we may want a little more information regarding the vessels themselves. In the `data/AIS` folder there is a second table that contains information about each of the ships that traveled the segments in the `segments` table.
```
vessels = pd.read_csv("data/AIS/vessel_information.csv", index_col='mmsi')
vessels.head()
[v for v in vessels.type.unique() if v.find('/')==-1]
vessels.type.value_counts()
```
The challenge, however, is that several ships have travelled multiple segments, so there is not a one-to-one relationship between the rows of the two tables. The table of vessel information has a *one-to-many* relationship with the segments.
In Pandas, we can combine tables according to the value of one or more *keys* that are used to identify rows, much like an index. Using a trivial example:
```
df1 = pd.DataFrame(dict(id=range(4), age=np.random.randint(18, 31, size=4)))
df2 = pd.DataFrame(dict(id=range(3)+range(3), score=np.random.random(size=6)))
df1, df2
pd.merge(df1, df2)
```
Notice that without any information about which column to use as a key, Pandas did the right thing and used the `id` column in both tables. Unless specified otherwise, `merge` will used any common column names as keys for merging the tables.
Notice also that `id=3` from `df1` was omitted from the merged table. This is because, by default, `merge` performs an **inner join** on the tables, meaning that the merged table represents an intersection of the two tables.
```
pd.merge(df1, df2, how='outer')
```
The **outer join** above yields the union of the two tables, so all rows are represented, with missing values inserted as appropriate. One can also perform **right** and **left** joins to include all rows of the right or left table (*i.e.* first or second argument to `merge`), but not necessarily the other.
Looking at the two datasets that we wish to merge:
```
segments.head(1)
vessels.head(1)
```
we see that there is a `mmsi` value (a vessel identifier) in each table, but it is used as an index for the `vessels` table. In this case, we have to specify to join on the index for this table, and on the `mmsi` column for the other.
```
segments_merged = pd.merge(vessels, segments, left_index=True, right_on='mmsi')
segments_merged.head()
```
In this case, the default inner join is suitable; we are not interested in observations from either table that do not have corresponding entries in the other.
Notice that `mmsi` field that was an index on the `vessels` table is no longer an index on the merged table.
Here, we used the `merge` function to perform the merge; we could also have used the `merge` method for either of the tables:
```
vessels.merge(segments, left_index=True, right_on='mmsi').head()
```
Occasionally, there will be fields with the same in both tables that we do not wish to use to join the tables; they may contain different information, despite having the same name. In this case, Pandas will by default append suffixes `_x` and `_y` to the columns to uniquely identify them.
```
segments['type'] = 'foo'
pd.merge(vessels, segments, left_index=True, right_on='mmsi').head()
```
This behavior can be overridden by specifying a `suffixes` argument, containing a list of the suffixes to be used for the columns of the left and right columns, respectively.
## Concatenation
A common data manipulation is appending rows or columns to a dataset that already conform to the dimensions of the exsiting rows or colums, respectively. In NumPy, this is done either with `concatenate` or the convenience functions `c_` and `r_`:
```
np.concatenate([np.random.random(5), np.random.random(5)])
np.r_[np.random.random(5), np.random.random(5)]
np.c_[np.random.random(5), np.random.random(5)]
```
This operation is also called *binding* or *stacking*.
With Pandas' indexed data structures, there are additional considerations as the overlap in index values between two data structures affects how they are concatenate.
Lets import two microbiome datasets, each consisting of counts of microorganiams from a particular patient. We will use the first column of each dataset as the index.
```
mb1 = pd.read_excel('data/microbiome/MID1.xls', 'Sheet 1', index_col=0, header=None)
mb2 = pd.read_excel('data/microbiome/MID2.xls', 'Sheet 1', index_col=0, header=None)
mb1.shape, mb2.shape
mb1.head()
```
Let's give the index and columns meaningful labels:
```
mb1.columns = mb2.columns = ['Count']
mb1.index.name = mb2.index.name = 'Taxon'
mb1.head()
```
The index of these data is the unique biological classification of each organism, beginning with *domain*, *phylum*, *class*, and for some organisms, going all the way down to the genus level.

```
mb1.index[:3]
mb1.index.is_unique
```
If we concatenate along `axis=0` (the default), we will obtain another data frame with the the rows concatenated:
```
pd.concat([mb1, mb2], axis=0).shape
```
However, the index is no longer unique, due to overlap between the two DataFrames.
```
pd.concat([mb1, mb2], axis=0).index.is_unique
```
Concatenating along `axis=1` will concatenate column-wise, but respecting the indices of the two DataFrames.
```
pd.concat([mb1, mb2], axis=1).shape
pd.concat([mb1, mb2], axis=1).head()
pd.concat([mb1, mb2], axis=1).values[:5]
```
If we are only interested in taxa that are included in both DataFrames, we can specify a `join=inner` argument.
```
pd.concat([mb1, mb2], axis=1, join='inner').head()
```
If we wanted to use the second table to fill values absent from the first table, we could use `combine_first`.
```
mb1.combine_first(mb2).head()
```
We can also create a hierarchical index based on keys identifying the original tables.
```
pd.concat([mb1, mb2], keys=['patient1', 'patient2']).head()
pd.concat([mb1, mb2], keys=['patient1', 'patient2']).index.is_unique
```
Alternatively, you can pass keys to the concatenation by supplying the DataFrames (or Series) as a dict.
```
pd.concat(dict(patient1=mb1, patient2=mb2), axis=1).head()
```
If you want `concat` to work like `numpy.concatanate`, you may provide the `ignore_index=True` argument.
## Exercise
In the *data/microbiome* subdirectory, there are 9 spreadsheets of microbiome data that was acquired from high-throughput RNA sequencing procedures, along with a 10th file that describes the content of each. Write code that imports each of the data spreadsheets and combines them into a single `DataFrame`, adding the identifying information from the metadata spreadsheet as columns in the combined `DataFrame`.
```
# Write your answer here
```
## Reshaping DataFrame objects
In the context of a single DataFrame, we are often interested in re-arranging the layout of our data.
This dataset in from Table 6.9 of [Statistical Methods for the Analysis of Repeated Measurements](http://www.amazon.com/Statistical-Methods-Analysis-Repeated-Measurements/dp/0387953701) by Charles S. Davis, pp. 161-163 (Springer, 2002). These data are from a multicenter, randomized controlled trial of botulinum toxin type B (BotB) in patients with cervical dystonia from nine U.S. sites.
* Randomized to placebo (N=36), 5000 units of BotB (N=36), 10,000 units of BotB (N=37)
* Response variable: total score on Toronto Western Spasmodic Torticollis Rating Scale (TWSTRS), measuring severity, pain, and disability of cervical dystonia (high scores mean more impairment)
* TWSTRS measured at baseline (week 0) and weeks 2, 4, 8, 12, 16 after treatment began
```
cdystonia = pd.read_csv("data/cdystonia.csv", index_col=None)
cdystonia.head()
```
This dataset includes repeated measurements of the same individuals (longitudinal data). Its possible to present such information in (at least) two ways: showing each repeated measurement in their own row, or in multiple columns representing mutliple measurements.
The `stack` method rotates the data frame so that columns are represented in rows:
```
stacked = cdystonia.stack()
stacked
```
To complement this, `unstack` pivots from rows back to columns.
```
stacked.unstack().head()
```
For this dataset, it makes sense to create a hierarchical index based on the patient and observation:
```
cdystonia2 = cdystonia.set_index(['patient','obs'])
cdystonia2.head()
cdystonia2.index.is_unique
```
If we want to transform this data so that repeated measurements are in columns, we can `unstack` the `twstrs` measurements according to `obs`.
```
twstrs_wide = cdystonia2['twstrs'].unstack('obs')
twstrs_wide.head()
cdystonia_long = cdystonia[['patient','site','id','treat','age','sex']].drop_duplicates().merge(
twstrs_wide, right_index=True, left_on='patient', how='inner').head()
cdystonia_long
```
A slightly cleaner way of doing this is to set the patient-level information as an index before unstacking:
```
cdystonia.set_index(['patient','site','id','treat','age','sex','week'])['twstrs'].unstack('week').head()
```
To convert our "wide" format back to long, we can use the `melt` function, appropriately parameterized:
```
pd.melt(cdystonia_long, id_vars=['patient','site','id','treat','age','sex'],
var_name='obs', value_name='twsters').head()
```
This illustrates the two formats for longitudinal data: **long** and **wide** formats. Its typically better to store data in long format because additional data can be included as additional rows in the database, while wide format requires that the entire database schema be altered by adding columns to every row as data are collected.
The preferable format for analysis depends entirely on what is planned for the data, so it is imporant to be able to move easily between them.
## Pivoting
The `pivot` method allows a DataFrame to be transformed easily between long and wide formats in the same way as a pivot table is created in a spreadsheet. It takes three arguments: `index`, `columns` and `values`, corresponding to the DataFrame index (the row headers), columns and cell values, respectively.
For example, we may want the `twstrs` variable (the response variable) in wide format according to patient:
```
cdystonia.pivot(index='patient', columns='obs', values='twstrs').head()
```
If we omit the `values` argument, we get a `DataFrame` with hierarchical columns, just as when we applied `unstack` to the hierarchically-indexed table:
```
cdystonia.pivot('patient', 'obs')
```
A related method, `pivot_table`, creates a spreadsheet-like table with a hierarchical index, and allows the values of the table to be populated using an arbitrary aggregation function.
```
cdystonia.pivot_table(rows=['site', 'treat'], cols='week', values='twstrs', aggfunc=max).head(20)
```
For a simple cross-tabulation of group frequencies, the `crosstab` function (not a method) aggregates counts of data according to factors in rows and columns. The factors may be hierarchical if desired.
```
pd.crosstab(cdystonia.sex, cdystonia.site)
```
## Data transformation
There are a slew of additional operations for DataFrames that we would collectively refer to as "transformations" that include tasks such as removing duplicate values, replacing values, and grouping values.
### Dealing with duplicates
We can easily identify and remove duplicate values from `DataFrame` objects. For example, say we want to removed ships from our `vessels` dataset that have the same name:
```
vessels.duplicated(cols='names')
vessels.drop_duplicates(['names'])
```
### Value replacement
Frequently, we get data columns that are encoded as strings that we wish to represent numerically for the purposes of including it in a quantitative analysis. For example, consider the treatment variable in the cervical dystonia dataset:
```
cdystonia.treat.value_counts()
```
A logical way to specify these numerically is to change them to integer values, perhaps using "Placebo" as a baseline value. If we create a dict with the original values as keys and the replacements as values, we can pass it to the `map` method to implement the changes.
```
treatment_map = {'Placebo': 0, '5000U': 1, '10000U': 2}
cdystonia['treatment'] = cdystonia.treat.map(treatment_map)
cdystonia.treatment
```
Alternately, if we simply want to replace particular values in a `Series` or `DataFrame`, we can use the `replace` method.
An example where replacement is useful is dealing with zeros in certain transformations. For example, if we try to take the log of a set of values:
```
vals = pd.Series([float(i)**10 for i in range(10)])
vals
np.log(vals)
```
In such situations, we can replace the zero with a value so small that it makes no difference to the ensuing analysis. We can do this with `replace`.
```
vals = vals.replace(0, 1e-6)
np.log(vals)
```
We can also perform the same replacement that we used `map` for with `replace`:
```
cdystonia2.treat.replace({'Placebo': 0, '5000U': 1, '10000U': 2})
```
### Inidcator variables
For some statistical analyses (*e.g.* regression models or analyses of variance), categorical or group variables need to be converted into columns of indicators--zeros and ones--to create a so-called **design matrix**. The Pandas function `get_dummies` (indicator variables are also known as *dummy variables*) makes this transformation straightforward.
Let's consider the DataFrame containing the ships corresponding to the transit segments on the eastern seaboard. The `type` variable denotes the class of vessel; we can create a matrix of indicators for this. For simplicity, lets filter out the 5 most common types of ships:
```
top5 = vessels.type.apply(lambda s: s in vessels.type.value_counts().index[:5])
vessels5 = vessels[top5]
pd.get_dummies(vessels5.type).head(10)
```
### Discretization
Pandas' `cut` function can be used to group continuous or countable data in to bins. Discretization is generally a very **bad idea** for statistical analysis, so use this function responsibly!
Lets say we want to bin the ages of the cervical dystonia patients into a smaller number of groups:
```
cdystonia.age.describe()
```
Let's transform these data into decades, beginnnig with individuals in their 20's and ending with those in their 90's:
```
pd.cut(cdystonia.age, [20,30,40,50,60,70,80,90])[:30]
```
The parentheses indicate an open interval, meaning that the interval includes values up to but *not including* the endpoint, whereas the square bracket is a closed interval, where the endpoint is included in the interval. We can switch the closure to the left side by setting the `right` flag to `False`:
```
pd.cut(cdystonia.age, [20,30,40,50,60,70,80,90], right=False)[:30]
```
Since the data are now **ordinal**, rather than numeric, we can give them labels:
```
pd.cut(cdystonia.age, [20,40,60,80,90], labels=['young','middle-aged','old','ancient'])[:30]
```
A related function `qcut` uses empirical quantiles to divide the data. If, for example, we want the quartiles -- (0-25%], (25-50%], (50-70%], (75-100%] -- we can just specify 4 intervals, which will be equally-spaced by default:
```
pd.qcut(cdystonia.age, 4)[:30]
```
Alternatively, one can specify custom quantiles to act as cut points:
```
quantiles = pd.qcut(segments.seg_length, [0, 0.01, 0.05, 0.95, 0.99, 1])
quantiles[:30]
```
Note that you can easily combine discretiztion with the generation of indicator variables shown above:
```
pd.get_dummies(quantiles).head(10)
```
### Permutation and sampling
For some data analysis tasks, such as simulation, we need to be able to randomly reorder our data, or draw random values from it. Calling NumPy's `permutation` function with the length of the sequence you want to permute generates an array with a permuted sequence of integers, which can be used to re-order the sequence.
```
new_order = np.random.permutation(len(segments))
new_order[:30]
```
Using this sequence as an argument to the `take` method results in a reordered DataFrame:
```
segments.take(new_order).head()
```
Compare this ordering with the original:
```
segments.head()
```
## Exercise
Its easy to see how this permutation approach allows us to draw a random sample **without replacement**. How would you sample **with replacement**? Generate a random sample of 5 ships from the `vessels` DataFrame using this scheme.
```
# Write your answer here
```
## Data aggregation and GroupBy operations
One of the most powerful features of Pandas is its **GroupBy** functionality. On occasion we may want to perform operations on *groups* of observations within a dataset. For exmaple:
* **aggregation**, such as computing the sum of mean of each group, which involves applying a function to each group and returning the aggregated results
* **slicing** the DataFrame into groups and then doing something with the resulting slices (*e.g.* plotting)
* group-wise **transformation**, such as standardization/normalization
```
cdystonia_grouped = cdystonia.groupby(cdystonia.patient)
```
This *grouped* dataset is hard to visualize
```
cdystonia_grouped
```
However, the grouping is only an intermediate step; for example, we may want to **iterate** over each of the patient groups:
```
for patient, group in cdystonia_grouped:
print patient
print group
print
```
A common data analysis procedure is the **split-apply-combine** operation, which groups subsets of data together, applies a function to each of the groups, then recombines them into a new data table.
For example, we may want to aggregate our data with with some function.

<div align="right">*(figure taken from "Python for Data Analysis", p.251)*</div>
We can aggregate in Pandas using the `aggregate` (or `agg`, for short) method:
```
cdystonia_grouped.agg(np.mean).head()
```
Notice that the `treat` and `sex` variables are not included in the aggregation. Since it does not make sense to aggregate non-string variables, these columns are simply ignored by the method.
Some aggregation functions are so common that Pandas has a convenience method for them, such as `mean`:
```
cdystonia_grouped.mean().head()
```
The `add_prefix` and `add_suffix` methods can be used to give the columns of the resulting table labels that reflect the transformation:
```
cdystonia_grouped.mean().add_suffix('_mean').head()
# The median of the `twstrs` variable
cdystonia_grouped['twstrs'].quantile(0.5)
```
If we wish, we can easily aggregate according to multiple keys:
```
cdystonia.groupby(['week','site']).mean().head()
```
Alternately, we can **transform** the data, using a function of our choice with the `transform` method:
```
normalize = lambda x: (x - x.mean())/x.std()
cdystonia_grouped.transform(normalize).head()
```
It is easy to do column selection within `groupby` operations, if we are only interested split-apply-combine operations on a subset of columns:
```
cdystonia_grouped['twstrs'].mean().head()
# This gives the same result as a DataFrame
cdystonia_grouped[['twstrs']].mean().head()
```
If you simply want to divide your DataFrame into chunks for later use, its easy to convert them into a dict so that they can be easily indexed out as needed:
```
chunks = dict(list(cdystonia_grouped))
chunks[4]
```
By default, `groupby` groups by row, but we can specify the `axis` argument to change this. For example, we can group our columns by type this way:
```
dict(list(cdystonia.groupby(cdystonia.dtypes, axis=1)))
```
Its also possible to group by one or more levels of a hierarchical index. Recall `cdystonia2`, which we created with a hierarchical index:
```
cdystonia2.head(10)
cdystonia2.groupby(level='obs', axis=0)['twstrs'].mean()
```
### Apply
We can generalize the split-apply-combine methodology by using `apply` function. This allows us to invoke any function we wish on a grouped dataset and recombine them into a DataFrame.
The function below takes a DataFrame and a column name, sorts by the column, and takes the `n` largest values of that column. We can use this with `apply` to return the largest values from every group in a DataFrame in a single call.
```
def top(df, column, n=5):
return df.sort_index(by=column, ascending=False)[:n]
```
To see this in action, consider the vessel transit segments dataset (which we merged with the vessel information to yield `segments_merged`). Say we wanted to return the 3 longest segments travelled by each ship:
```
top3segments = segments_merged.groupby('mmsi').apply(top, column='seg_length', n=3)[['names', 'seg_length']]
top3segments
```
Notice that additional arguments for the applied function can be passed via `apply` after the function name. It assumes that the DataFrame is the first argument.
```
top3segments.head(20)
```
Recall the microbiome data sets that we used previously for the concatenation example. Suppose that we wish to aggregate the data at a higher biological classification than genus. For example, we can identify samples down to *class*, which is the 3rd level of organization in each index.
```
mb1.index[:3]
```
Using the string methods `split` and `join` we can create an index that just uses the first three classifications: domain, phylum and class.
```
class_index = mb1.index.map(lambda x: ' '.join(x.split(' ')[:3]))
mb_class = mb1.copy()
mb_class.index = class_index
```
However, since there are multiple taxonomic units with the same class, our index is no longer unique:
```
mb_class.head()
```
We can re-establish a unique index by summing all rows with the same class, using `groupby`:
```
mb_class.groupby(level=0).sum().head(10)
```
### Exercise
Load the dataset in `titanic.xls`. It contains data on all the passengers that travelled on the Titanic.
```
from IPython.core.display import HTML
HTML(filename='data/titanic.html')
```
Women and children first?
1. Use the `groupby` method to calculate the proportion of passengers that survived by sex.
2. Calculate the same proportion, but by class and sex.
3. Create age categories: children (under 14 years), adolescents (14-20), adult (21-64), and senior(65+), and calculate survival proportions by age category, class and sex.
```
# Write your answer here
```
|
github_jupyter
|
%matplotlib inline
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# Set some Pandas options
pd.set_option('display.notebook_repr_html', False)
pd.set_option('display.max_columns', 20)
pd.set_option('display.max_rows', 25)
from datetime import datetime
now = datetime.now()
now
now.day
now.weekday()
from datetime import date, time
time(3, 24)
date(1970, 9, 3)
my_age = now - datetime(1970, 9, 3)
my_age
my_age.days/365.
segments = pd.read_csv("data/AIS/transit_segments.csv")
segments.head()
segments.seg_length.hist(bins=500)
segments.seg_length.apply(np.log).hist(bins=500)
segments.st_time.dtype
datetime.strptime(segments.st_time.ix[0], '%m/%d/%y %H:%M')
from dateutil.parser import parse
parse(segments.st_time.ix[0])
segments.st_time.apply(lambda d: datetime.strptime(d, '%m/%d/%y %H:%M'))
pd.to_datetime(segments.st_time)
pd.to_datetime([None])
vessels = pd.read_csv("data/AIS/vessel_information.csv", index_col='mmsi')
vessels.head()
[v for v in vessels.type.unique() if v.find('/')==-1]
vessels.type.value_counts()
df1 = pd.DataFrame(dict(id=range(4), age=np.random.randint(18, 31, size=4)))
df2 = pd.DataFrame(dict(id=range(3)+range(3), score=np.random.random(size=6)))
df1, df2
pd.merge(df1, df2)
pd.merge(df1, df2, how='outer')
segments.head(1)
vessels.head(1)
segments_merged = pd.merge(vessels, segments, left_index=True, right_on='mmsi')
segments_merged.head()
vessels.merge(segments, left_index=True, right_on='mmsi').head()
segments['type'] = 'foo'
pd.merge(vessels, segments, left_index=True, right_on='mmsi').head()
np.concatenate([np.random.random(5), np.random.random(5)])
np.r_[np.random.random(5), np.random.random(5)]
np.c_[np.random.random(5), np.random.random(5)]
mb1 = pd.read_excel('data/microbiome/MID1.xls', 'Sheet 1', index_col=0, header=None)
mb2 = pd.read_excel('data/microbiome/MID2.xls', 'Sheet 1', index_col=0, header=None)
mb1.shape, mb2.shape
mb1.head()
mb1.columns = mb2.columns = ['Count']
mb1.index.name = mb2.index.name = 'Taxon'
mb1.head()
mb1.index[:3]
mb1.index.is_unique
pd.concat([mb1, mb2], axis=0).shape
pd.concat([mb1, mb2], axis=0).index.is_unique
pd.concat([mb1, mb2], axis=1).shape
pd.concat([mb1, mb2], axis=1).head()
pd.concat([mb1, mb2], axis=1).values[:5]
pd.concat([mb1, mb2], axis=1, join='inner').head()
mb1.combine_first(mb2).head()
pd.concat([mb1, mb2], keys=['patient1', 'patient2']).head()
pd.concat([mb1, mb2], keys=['patient1', 'patient2']).index.is_unique
pd.concat(dict(patient1=mb1, patient2=mb2), axis=1).head()
# Write your answer here
cdystonia = pd.read_csv("data/cdystonia.csv", index_col=None)
cdystonia.head()
stacked = cdystonia.stack()
stacked
stacked.unstack().head()
cdystonia2 = cdystonia.set_index(['patient','obs'])
cdystonia2.head()
cdystonia2.index.is_unique
twstrs_wide = cdystonia2['twstrs'].unstack('obs')
twstrs_wide.head()
cdystonia_long = cdystonia[['patient','site','id','treat','age','sex']].drop_duplicates().merge(
twstrs_wide, right_index=True, left_on='patient', how='inner').head()
cdystonia_long
cdystonia.set_index(['patient','site','id','treat','age','sex','week'])['twstrs'].unstack('week').head()
pd.melt(cdystonia_long, id_vars=['patient','site','id','treat','age','sex'],
var_name='obs', value_name='twsters').head()
cdystonia.pivot(index='patient', columns='obs', values='twstrs').head()
cdystonia.pivot('patient', 'obs')
cdystonia.pivot_table(rows=['site', 'treat'], cols='week', values='twstrs', aggfunc=max).head(20)
pd.crosstab(cdystonia.sex, cdystonia.site)
vessels.duplicated(cols='names')
vessels.drop_duplicates(['names'])
cdystonia.treat.value_counts()
treatment_map = {'Placebo': 0, '5000U': 1, '10000U': 2}
cdystonia['treatment'] = cdystonia.treat.map(treatment_map)
cdystonia.treatment
vals = pd.Series([float(i)**10 for i in range(10)])
vals
np.log(vals)
vals = vals.replace(0, 1e-6)
np.log(vals)
cdystonia2.treat.replace({'Placebo': 0, '5000U': 1, '10000U': 2})
top5 = vessels.type.apply(lambda s: s in vessels.type.value_counts().index[:5])
vessels5 = vessels[top5]
pd.get_dummies(vessels5.type).head(10)
cdystonia.age.describe()
pd.cut(cdystonia.age, [20,30,40,50,60,70,80,90])[:30]
pd.cut(cdystonia.age, [20,30,40,50,60,70,80,90], right=False)[:30]
pd.cut(cdystonia.age, [20,40,60,80,90], labels=['young','middle-aged','old','ancient'])[:30]
pd.qcut(cdystonia.age, 4)[:30]
quantiles = pd.qcut(segments.seg_length, [0, 0.01, 0.05, 0.95, 0.99, 1])
quantiles[:30]
pd.get_dummies(quantiles).head(10)
new_order = np.random.permutation(len(segments))
new_order[:30]
segments.take(new_order).head()
segments.head()
# Write your answer here
cdystonia_grouped = cdystonia.groupby(cdystonia.patient)
cdystonia_grouped
for patient, group in cdystonia_grouped:
print patient
print group
print
cdystonia_grouped.agg(np.mean).head()
cdystonia_grouped.mean().head()
cdystonia_grouped.mean().add_suffix('_mean').head()
# The median of the `twstrs` variable
cdystonia_grouped['twstrs'].quantile(0.5)
cdystonia.groupby(['week','site']).mean().head()
normalize = lambda x: (x - x.mean())/x.std()
cdystonia_grouped.transform(normalize).head()
cdystonia_grouped['twstrs'].mean().head()
# This gives the same result as a DataFrame
cdystonia_grouped[['twstrs']].mean().head()
chunks = dict(list(cdystonia_grouped))
chunks[4]
dict(list(cdystonia.groupby(cdystonia.dtypes, axis=1)))
cdystonia2.head(10)
cdystonia2.groupby(level='obs', axis=0)['twstrs'].mean()
def top(df, column, n=5):
return df.sort_index(by=column, ascending=False)[:n]
top3segments = segments_merged.groupby('mmsi').apply(top, column='seg_length', n=3)[['names', 'seg_length']]
top3segments
top3segments.head(20)
mb1.index[:3]
class_index = mb1.index.map(lambda x: ' '.join(x.split(' ')[:3]))
mb_class = mb1.copy()
mb_class.index = class_index
mb_class.head()
mb_class.groupby(level=0).sum().head(10)
from IPython.core.display import HTML
HTML(filename='data/titanic.html')
# Write your answer here
| 0.256366 | 0.993359 |
The Stableswap equation given in the Curve whitepaper is
$$ An^n\sum_i x_i + D = An^n D + \frac{D^{n+1}}{n^n\prod\limits_i x_i}$$
$D$ is the stableswap invariant.
$A$ is the amplification coefficient.
The $x_i$'s are the pool balances for each token.
To solve for $D$ in the stableswap equation, we can use the auxiliary form of the equation
$$ f(D) = 0 $$
where
$$ f(D) = An^n S + D - An^n D - \frac{D^{n+1}}{n^n\prod x_i} $$
Note $f(P) > 0$ and $f(S) < 0$ (this is simple to see using that $P <= S$ with equality only when $x_1 = x_2$). Since $P < S$ in the generic case, we expect that somewhere in-between, there is a $D$ such that $f(D) = 0$. In fact, the situation is much better than that.
The derivative of $f$ is
$$ f'(D) = 1 - An^n - (n+1) \frac{D^n}{n^n\prod x_i} $$
Since $f'$ is always be negative (as long as $A > 1$), $f$ is strictly decreasing and Newton's method will rapidly find a solution.
Newton's method gives the iteration:
$$ x_{k+1} = x_k - \frac{f(x_k)}{f'(x_k)} $$
After some cleanup, this gives the iterative formula:
$$ D = \frac{(nD_p + An^nS)D}{(n+1)D_p + (An^n - 1)D} $$
where $D_p = \frac{D^{n+1}}{n^n\prod x_i}$
```
import matplotlib.pyplot as plt
import numpy as np
# Amplification coefficient
A = 10
# number of token types in pool
n = 2
# token balances
x_1 = 85
x_2 = 15
# "unnormalized" arithmetic mean
S = x_1 + x_2
print("S:", S)
# "unnormalized" geometric mean
P = n * np.sqrt(x_1*x_2)
print("P:", P)
# iterative calculation for D invariant
D = S
for i in range(256):
D_p = (D/n)**n / (x_1 * x_2) * D
D = (A* n**n * S + n * D_p) * D / ((A*n**n - 1) * D + (n+ 1) * D_p)
print("D:", D)
```
For $n=2$, the stableswap equation becomes
$$ 4 A (x_1 + x_2) + D = 4 AD + \frac{D^3}{4x_1x_2} $$
which can be rearranged to
$$ 4Ax_2^2 + (4Ax_1 + D - 4AD)x_2 - \frac{D^3}{4x_1} = 0 $$
Given $x_1$, we can solve for $x_2$ via the quadratic formula (see the function `get_other_token_balance`)
```
def get_other_token_balance(D, A, x):
"""
For invariant D and amplification coefficient A,
given the balance of one token, x, return the
requisite balance of the other token.
"""
a = 4*A
b = 4*A*x_ + D - 4 * A * D
c = -D**3/(4*x_)
# x_minus = (-b - np.sqrt(b**2 - 4*a*c))/(2*a)
x_plus = (-b + np.sqrt(b**2 - 4*a*c))/(2*a)
# print(x_minus, x_plus)
return x_plus
x_end = 115.0
num_steps = 75
fig, ax = plt.subplots()
ax.set_xlim(0, x_end)
ax.set_ylim(0, x_end)
ax.set(xlabel='Token X', ylabel='Token Y',
title='Stableswap interpolation')
ax.grid()
ax.annotate('(x_1, x_2)', xy=(x_1, x_2), xytext=(20,20),
textcoords='offset points', ha='center', va='bottom',
bbox=dict(boxstyle='round,pad=0.2', fc='yellow', alpha=0.3),
arrowprops=dict(arrowstyle='->', connectionstyle='arc3,rad=0.5',
color='black'))
x = np.linspace(0.0, x_end, num_steps)[1:]
y_uni = (P**2/4) / x
ax.plot(x, y_uni, color='b', label='uniswap')
y_mstable = S - x
ax.plot(x, y_mstable, color='r', label='mstable')
ax.plot(x, x, ls='--', color='g')
y_sswap = []
for x_ in x:
y = get_other_token_balance(D, A, x_)
y_sswap.append(y)
ax.plot(x, y_sswap, 'black', label='stableswap')
plt.legend(loc='upper right');
# fig.savefig("example.png")
plt.show()
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
# Amplification coefficient
A = 10
# number of token types in pool
n = 2
# token balances
x_1 = 85
x_2 = 15
# "unnormalized" arithmetic mean
S = x_1 + x_2
print("S:", S)
# "unnormalized" geometric mean
P = n * np.sqrt(x_1*x_2)
print("P:", P)
# iterative calculation for D invariant
D = S
for i in range(256):
D_p = (D/n)**n / (x_1 * x_2) * D
D = (A* n**n * S + n * D_p) * D / ((A*n**n - 1) * D + (n+ 1) * D_p)
print("D:", D)
def get_other_token_balance(D, A, x):
"""
For invariant D and amplification coefficient A,
given the balance of one token, x, return the
requisite balance of the other token.
"""
a = 4*A
b = 4*A*x_ + D - 4 * A * D
c = -D**3/(4*x_)
# x_minus = (-b - np.sqrt(b**2 - 4*a*c))/(2*a)
x_plus = (-b + np.sqrt(b**2 - 4*a*c))/(2*a)
# print(x_minus, x_plus)
return x_plus
x_end = 115.0
num_steps = 75
fig, ax = plt.subplots()
ax.set_xlim(0, x_end)
ax.set_ylim(0, x_end)
ax.set(xlabel='Token X', ylabel='Token Y',
title='Stableswap interpolation')
ax.grid()
ax.annotate('(x_1, x_2)', xy=(x_1, x_2), xytext=(20,20),
textcoords='offset points', ha='center', va='bottom',
bbox=dict(boxstyle='round,pad=0.2', fc='yellow', alpha=0.3),
arrowprops=dict(arrowstyle='->', connectionstyle='arc3,rad=0.5',
color='black'))
x = np.linspace(0.0, x_end, num_steps)[1:]
y_uni = (P**2/4) / x
ax.plot(x, y_uni, color='b', label='uniswap')
y_mstable = S - x
ax.plot(x, y_mstable, color='r', label='mstable')
ax.plot(x, x, ls='--', color='g')
y_sswap = []
for x_ in x:
y = get_other_token_balance(D, A, x_)
y_sswap.append(y)
ax.plot(x, y_sswap, 'black', label='stableswap')
plt.legend(loc='upper right');
# fig.savefig("example.png")
plt.show()
| 0.41052 | 0.975992 |
```
!nvidia-smi
```
### Install the necessary stuff
```
!sudo apt-get install xvfb
pip install xagents
pip install matplotlib==3.1.3
```
### Training (trial 1)
We will train A2C and PPO agents on the CartPole-v1 environment. Since no hyperparameter optimization was conducted, both agents will yield suboptimal results.
### PPO (training)
```
!xagents train ppo --env CartPole-v1 --max-steps 300000 --n-envs 16 --seed 55 --checkpoints ppo-cartpole.tf --history-checkpoint ppo-cartpole.parquet
```
### A2C (training)
```
!xagents train a2c --env CartPole-v1 --max-steps 300000 --n-envs 16 --checkpoints a2c-cartpole.tf \
--seed 55 --history-checkpoint a2c-cartpole.parquet --n-steps 128
```
### Tuning
In this section, we are going to tune hyperparameters for A2C and PPO.
**Notes:**
* The `xagents <command> <agent>` syntax displays the available options for the given command and agent. We will use this syntax for displaying tuning options for both agents.
* There are multiple hyperparameter types, which you can find under `hp_type` column in the menu displayed below. We will be using the 2 below ...
1. `log_uniform` hp_type, accepts a minimum and maximum bound. Therefore, you will need to pass either 1 value or 2 values in the following fashion:
`xagents tune <agent> --<log-uniform-hp> <min-val> <max-val>`
2. `categorical` hp_type, accepts n number of values.
`xagents tune <agent> --<categorical-hp> <val1> <val2> <val3> ...`
```
!xagents tune a2c
```
### A2C (tuning)
```
!xagents tune a2c --study a2c-cartpole --env CartPole-v1 --trial-steps 200000 --warmup-trials 4 \
--n-trials 20 --n-jobs 2 --storage sqlite:///a2c-cartpole.sqlite --entropy-coef 1e-5 0.5 --gamma 0.9 0.99 \
--grad-norm 0.1 10 --lr 1e-5 1e-2 --n-envs 16 --n-steps 8 16 32 64 128 256 512 1024 \
--opt-epsilon 1e-7 1e-3
```
We can use [optuna.visualization.matplotlib](https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&cad=rja&uact=8&ved=2ahUKEwi5p9239uDyAhXb_rsIHZ9EDPYQFnoECAMQAQ&url=https%3A%2F%2Foptuna.readthedocs.io%2Fen%2Flatest%2Freference%2Fvisualization%2Fmatplotlib.html&usg=AOvVaw20M4GHVSpQJQAkegkfJttS) API to visualize hyperparameter importances.
```
import optuna
import matplotlib.pyplot as plt
a2c_study = optuna.load_study('a2c-cartpole', storage='sqlite:///a2c-cartpole.sqlite')
optuna.visualization.matplotlib.plot_param_importances(a2c_study)
plt.show()
!xagents tune ppo
```
### PPO (tuning)
```
!xagents tune ppo --env CartPole-v1 --study ppo-cartpole --storage sqlite:///ppo-cartpole.sqlite \
--trial-steps 200000 --warmup-trials 4 --n-trials 20 --advantage-epsilon 1e-8 1e-5 --clip-norm 0.01 0.5 \
--entropy-coef 1e-4 0.3 --gamma 0.9 0.999 --grad-norm 0.1 10 --lam 0.7 0.99 --lr 1e-5 1e-2 \
--n-steps 16 32 64 128 256 512 1024 --opt-epsilon 1e-7 1e-4 --n-envs 16 32 \
--opt-epsilon 1e-7 1e-3 --n-jobs 2
ppo_study = optuna.load_study('ppo-cartpole', 'sqlite:///ppo-cartpole.sqlite')
optuna.visualization.matplotlib.plot_param_importances(ppo_study)
plt.show()
```
Display A2C best parameters, and use them to re-train.
```
a2c_study.best_params
```
### A2C (training using tuned hyperparameters)
```
!xagents train a2c --env CartPole-v1 --max-steps 300000 --n-envs 16 --checkpoints a2c-cartpole-tuned.tf \
--seed 55 --history-checkpoint a2c-cartpole-tuned.parquet --n-steps 8 --opt-epsilon 0.0009386796496510724 \
--lr 0.0012985885268425004 --grad-norm 0.9964628998438626 --gamma 0.9387388102974632 \
--entropy-coef 0.010565924673903932
ppo_study.best_params
```
### PPO (training using tuned hyperparameters)
```
!xagents train ppo --env CartPole-v1 --max-steps 300000 --n-envs 32 --seed 55 \
--checkpoints ppo-cartpole-tuned.tf --history-checkpoint ppo-cartpole-tuned.parquet \
--advantage-epsilon 1.3475350681876062e-08 --clip-norm 0.0503693625084303 \
--entropy-coef 0.06363366133416302 --gamma 0.93959608546301 --grad-norm 6.2465542151066495 \
--lam 0.9818834679479003 --lr 0.001549335940636045 --n-steps 16 --opt-epsilon 8.539506175014364e-07
```
### Benchmarks
```
from xagents.utils.common import plot_history
plot_history(
['a2c-cartpole.parquet', 'a2c-cartpole-tuned.parquet', 'ppo-cartpole.parquet', 'ppo-cartpole-tuned.parquet'],
['A2C', 'A2C(tuned)', 'PPO', 'PPO(tuned)'],
'CartPole-v1',
history_interval=50
)
plt.show()
```
### Play and save episode video for A2C and PPO
```
!xvfb-run xagents play a2c --env CartPole-v1 --weights a2c-cartpole-tuned.tf --video-dir a2c-vid
from IPython.display import HTML
from base64 import b64encode
import glob
def get_vid_url(vid_folder):
vid = open(glob.glob(f'{vid_folder}/*.mp4')[0],'rb').read()
return "data:video/mp4;base64," + b64encode(vid).decode()
ppo_url = get_vid_url('ppo-vid')
HTML("""
<video width=400 controls>
<source src="%s" type="video/mp4">
</video>
""" % ppo_url)
!xvfb-run xagents play ppo --env CartPole-v1 --weights ppo-cartpole-tuned.tf --video-dir ppo-vid
a2c_url = get_vid_url('a2c-vid')
HTML("""
<video width=400 controls>
<source src="%s" type="video/mp4">
</video>
""" % a2c_url)
```
|
github_jupyter
|
!nvidia-smi
!sudo apt-get install xvfb
pip install xagents
pip install matplotlib==3.1.3
!xagents train ppo --env CartPole-v1 --max-steps 300000 --n-envs 16 --seed 55 --checkpoints ppo-cartpole.tf --history-checkpoint ppo-cartpole.parquet
!xagents train a2c --env CartPole-v1 --max-steps 300000 --n-envs 16 --checkpoints a2c-cartpole.tf \
--seed 55 --history-checkpoint a2c-cartpole.parquet --n-steps 128
!xagents tune a2c
!xagents tune a2c --study a2c-cartpole --env CartPole-v1 --trial-steps 200000 --warmup-trials 4 \
--n-trials 20 --n-jobs 2 --storage sqlite:///a2c-cartpole.sqlite --entropy-coef 1e-5 0.5 --gamma 0.9 0.99 \
--grad-norm 0.1 10 --lr 1e-5 1e-2 --n-envs 16 --n-steps 8 16 32 64 128 256 512 1024 \
--opt-epsilon 1e-7 1e-3
import optuna
import matplotlib.pyplot as plt
a2c_study = optuna.load_study('a2c-cartpole', storage='sqlite:///a2c-cartpole.sqlite')
optuna.visualization.matplotlib.plot_param_importances(a2c_study)
plt.show()
!xagents tune ppo
!xagents tune ppo --env CartPole-v1 --study ppo-cartpole --storage sqlite:///ppo-cartpole.sqlite \
--trial-steps 200000 --warmup-trials 4 --n-trials 20 --advantage-epsilon 1e-8 1e-5 --clip-norm 0.01 0.5 \
--entropy-coef 1e-4 0.3 --gamma 0.9 0.999 --grad-norm 0.1 10 --lam 0.7 0.99 --lr 1e-5 1e-2 \
--n-steps 16 32 64 128 256 512 1024 --opt-epsilon 1e-7 1e-4 --n-envs 16 32 \
--opt-epsilon 1e-7 1e-3 --n-jobs 2
ppo_study = optuna.load_study('ppo-cartpole', 'sqlite:///ppo-cartpole.sqlite')
optuna.visualization.matplotlib.plot_param_importances(ppo_study)
plt.show()
a2c_study.best_params
!xagents train a2c --env CartPole-v1 --max-steps 300000 --n-envs 16 --checkpoints a2c-cartpole-tuned.tf \
--seed 55 --history-checkpoint a2c-cartpole-tuned.parquet --n-steps 8 --opt-epsilon 0.0009386796496510724 \
--lr 0.0012985885268425004 --grad-norm 0.9964628998438626 --gamma 0.9387388102974632 \
--entropy-coef 0.010565924673903932
ppo_study.best_params
!xagents train ppo --env CartPole-v1 --max-steps 300000 --n-envs 32 --seed 55 \
--checkpoints ppo-cartpole-tuned.tf --history-checkpoint ppo-cartpole-tuned.parquet \
--advantage-epsilon 1.3475350681876062e-08 --clip-norm 0.0503693625084303 \
--entropy-coef 0.06363366133416302 --gamma 0.93959608546301 --grad-norm 6.2465542151066495 \
--lam 0.9818834679479003 --lr 0.001549335940636045 --n-steps 16 --opt-epsilon 8.539506175014364e-07
from xagents.utils.common import plot_history
plot_history(
['a2c-cartpole.parquet', 'a2c-cartpole-tuned.parquet', 'ppo-cartpole.parquet', 'ppo-cartpole-tuned.parquet'],
['A2C', 'A2C(tuned)', 'PPO', 'PPO(tuned)'],
'CartPole-v1',
history_interval=50
)
plt.show()
!xvfb-run xagents play a2c --env CartPole-v1 --weights a2c-cartpole-tuned.tf --video-dir a2c-vid
from IPython.display import HTML
from base64 import b64encode
import glob
def get_vid_url(vid_folder):
vid = open(glob.glob(f'{vid_folder}/*.mp4')[0],'rb').read()
return "data:video/mp4;base64," + b64encode(vid).decode()
ppo_url = get_vid_url('ppo-vid')
HTML("""
<video width=400 controls>
<source src="%s" type="video/mp4">
</video>
""" % ppo_url)
!xvfb-run xagents play ppo --env CartPole-v1 --weights ppo-cartpole-tuned.tf --video-dir ppo-vid
a2c_url = get_vid_url('a2c-vid')
HTML("""
<video width=400 controls>
<source src="%s" type="video/mp4">
</video>
""" % a2c_url)
| 0.399226 | 0.853669 |
```
import pandas as pd
import seaborn as sns
from datetime import timezone
# Contents
```
# Data Input
<hr/>
```
columns_names = ['IMEI', 'model','android_vers','bat_tech','bat_cap','timestamp','scren_status',
'bat_prec','app_running','cpu_usage','bat_temp','bat_volt','bat_current','network',
'plugged_in']
df1 = pd.read_csv('../mobileBatterywithtime/data/352944080639365.csv', names = columns_names,
usecols=[0,1,2,3,4,5,6,7,8,9,10,11,12,13,14] )
print(df1.shape)
df1.head(3)
df2 = pd.read_csv('../mobileBatterywithtime/data/358057080902248.csv', names = columns_names,
usecols=[0,1,2,3,4,5,6,7,8,9,10,11,12,13,14] )
print(df2.shape)
df1.head(2)
df3 = pd.read_csv('../mobileBatterywithtime/data/866558034754119.csv',names = columns_names,
usecols=[0,1,2,3,4,5,6,7,8,9,10,11,12,13,14] )
print(df3.shape)
df3.head(2)
df4 = pd.read_csv('../mobileBatterywithtime/data/866817034014258.csv', names = columns_names,
usecols=[0,1,2,3,4,5,6,7,8,9,10,11,12,13,14] )
print(df4.shape)
df4.head(2)
df5 = pd.read_csv('../mobileBatterywithtime/data/867194030192184.csv', names = columns_names,
usecols=[0,1,2,3,4,5,6,7,8,9,10,11,12,13,14] )
print(df5.shape)
df5.head(2)
df6 = pd.read_excel('../mobileBatterywithtime/data/Activation_Date_Phone.xlsx')
print(df6.shape)
df6
df6.info()
df6.rename(columns={'IMEI Number ':'IMEI','Activation Date (YYYY-MM-DD)': 'act_date'}, inplace = True)
df6.head(1)
pd.to_datetime(df6['act_date'], unit='s')
# convert timestamp to Unix Timestamp Conversion
for c in range(0, len(df6['act_date'])):
df6.iloc[c,1] = df6.iloc[c,1].timestamp() #.replace(tzinfo=timezone.utc)
df6.head()
df6.iloc[3,1]
# NOTES - df6 to be used to create feature on df timestamp
"""
Convert activation date to timestamp
Calculate difference between
"""
```
# Concat
<hr/>
```
df = pd.concat([df1,df2,df3,df4,df5], axis = 0)
df.head()
df.info()
# read IMEI in DF
for index, row in df.iterrows():
IMEI_time = row['IMEI'].map({ 352944080639365:1508284800.0, 358057080902248:1499644800.0,
866558034754119:1508889600.0, 867194030192184:1520640000.0 })
print( row['timestamp'] - IMEI_time )
if index == 5 : break
#print(row['c1'], row['c2'])
# match IMEI to df6 sheet
# create feature = timestamp-df1 - timestamp-
# print into feature
```
# EDA
<hr/>
```
# Look at - Unique Values for each column
for df_columns in df:
print()
print()
print(df_columns)
print('--------------')
print( df[df_columns].nunique() )
print( df[df_columns].unique() )
"""
"""
df.plot(kind = 'bar');
battery tech
battery capacity
timestamp -
scren_status -
bat_prec - %
cpu_usage - cpu
bat_temp - temputure
bat_volt - volt
bat_amp - curret
network - 3x
plugged_in - bool
df.head(2)
sns
# drop app_running
# timestamp
# do boolian values to 1 & 0
# screen_status
# plugged_in
# Map IMEI
df['timestamp'] = map({352944080639365:1, 358057080902248:2, 866558034754119:3, 866817034014258:4,
867194030192184:5 })
# Map model
{'samsung SM-A910F':1, 'samsung SM-G950F':2, nan:0, 'OPPO CPH1723':3,
'OnePlus ONEPLUS A5010':4 'Xiaomi Redmi Note 5 Pro':5}
# map battery tech
{'Li-ion':1 , 'Li-poly':2}
# Map network
{'none':0, 'wi-fi':1, 'mobile':2}
```
|
github_jupyter
|
import pandas as pd
import seaborn as sns
from datetime import timezone
# Contents
columns_names = ['IMEI', 'model','android_vers','bat_tech','bat_cap','timestamp','scren_status',
'bat_prec','app_running','cpu_usage','bat_temp','bat_volt','bat_current','network',
'plugged_in']
df1 = pd.read_csv('../mobileBatterywithtime/data/352944080639365.csv', names = columns_names,
usecols=[0,1,2,3,4,5,6,7,8,9,10,11,12,13,14] )
print(df1.shape)
df1.head(3)
df2 = pd.read_csv('../mobileBatterywithtime/data/358057080902248.csv', names = columns_names,
usecols=[0,1,2,3,4,5,6,7,8,9,10,11,12,13,14] )
print(df2.shape)
df1.head(2)
df3 = pd.read_csv('../mobileBatterywithtime/data/866558034754119.csv',names = columns_names,
usecols=[0,1,2,3,4,5,6,7,8,9,10,11,12,13,14] )
print(df3.shape)
df3.head(2)
df4 = pd.read_csv('../mobileBatterywithtime/data/866817034014258.csv', names = columns_names,
usecols=[0,1,2,3,4,5,6,7,8,9,10,11,12,13,14] )
print(df4.shape)
df4.head(2)
df5 = pd.read_csv('../mobileBatterywithtime/data/867194030192184.csv', names = columns_names,
usecols=[0,1,2,3,4,5,6,7,8,9,10,11,12,13,14] )
print(df5.shape)
df5.head(2)
df6 = pd.read_excel('../mobileBatterywithtime/data/Activation_Date_Phone.xlsx')
print(df6.shape)
df6
df6.info()
df6.rename(columns={'IMEI Number ':'IMEI','Activation Date (YYYY-MM-DD)': 'act_date'}, inplace = True)
df6.head(1)
pd.to_datetime(df6['act_date'], unit='s')
# convert timestamp to Unix Timestamp Conversion
for c in range(0, len(df6['act_date'])):
df6.iloc[c,1] = df6.iloc[c,1].timestamp() #.replace(tzinfo=timezone.utc)
df6.head()
df6.iloc[3,1]
# NOTES - df6 to be used to create feature on df timestamp
"""
Convert activation date to timestamp
Calculate difference between
"""
df = pd.concat([df1,df2,df3,df4,df5], axis = 0)
df.head()
df.info()
# read IMEI in DF
for index, row in df.iterrows():
IMEI_time = row['IMEI'].map({ 352944080639365:1508284800.0, 358057080902248:1499644800.0,
866558034754119:1508889600.0, 867194030192184:1520640000.0 })
print( row['timestamp'] - IMEI_time )
if index == 5 : break
#print(row['c1'], row['c2'])
# match IMEI to df6 sheet
# create feature = timestamp-df1 - timestamp-
# print into feature
# Look at - Unique Values for each column
for df_columns in df:
print()
print()
print(df_columns)
print('--------------')
print( df[df_columns].nunique() )
print( df[df_columns].unique() )
"""
"""
df.plot(kind = 'bar');
battery tech
battery capacity
timestamp -
scren_status -
bat_prec - %
cpu_usage - cpu
bat_temp - temputure
bat_volt - volt
bat_amp - curret
network - 3x
plugged_in - bool
df.head(2)
sns
# drop app_running
# timestamp
# do boolian values to 1 & 0
# screen_status
# plugged_in
# Map IMEI
df['timestamp'] = map({352944080639365:1, 358057080902248:2, 866558034754119:3, 866817034014258:4,
867194030192184:5 })
# Map model
{'samsung SM-A910F':1, 'samsung SM-G950F':2, nan:0, 'OPPO CPH1723':3,
'OnePlus ONEPLUS A5010':4 'Xiaomi Redmi Note 5 Pro':5}
# map battery tech
{'Li-ion':1 , 'Li-poly':2}
# Map network
{'none':0, 'wi-fi':1, 'mobile':2}
| 0.290276 | 0.371849 |
SHIOBAM VALENTINA ESPITIA PRADA
PRIMER PUNTO
```
def datos():
primern = (input("Escriba su primer nombre: "))
segundon = (input("Si tiene segundo nombre diga Si de lo contrario diga No: "))
if segundon == "Si":
segundo = (input("Escriba su segundo nombre: "))
PrimerA = input("Escriba su primer apellido: ")
seg = (input("Si tiene segundo apellido diga Si de lo contrario diga No: "))
if seg == "Si":
SegAp =str((input("Escriba su segundo apellido: ")))
Edad = int(input("Escriba su edad: "))
iden_via = input("Escriba la identificacion de la vida: ")
num_via = int(input("Escriba el numero que acompaña a la via: "))
marca_num1 = int(input("Escriba el numero de marca 1: "))
letra = input("Escriba la letra luego del numero: ")
marca_num2 = int(input("Escriba el numero de marca 2: "))
casa = (input("Escriba numero de casa: "))
if segundon == "No" and seg == "No":
print(f"Su hombre es {primern} {PrimerA} su edad es {Edad} y\nLa direccion es {iden_via} {num_via} # {marca_num1}{letra} - {marca_num2} y la casa es {casa}" )
if segundon == "No":
print(f"Su hombre es {primern} {PrimerA} {SegAp} su edad es {Edad} y\nLa direccion es {iden_via} {num_via} # {marca_num1}{letra} - {marca_num2} y la casa es {casa}" )
if seg == "No":
print(f"Su hombre es {primern} {segundo} {PrimerA} su edad es {Edad} y\nLa direccion es {iden_via} {num_via} # {marca_num1}{letra} - {marca_num2} y la casa es {casa}" )
else:
print(f"Su hombre es {primern} {segundo} {PrimerA} {SegAp} su edad es {Edad} y\nLa direccion es {iden_via} {num_via} # {marca_num1}{letra} - {marca_num2} y la casa es {casa}" )
datos()
```
SEGUNDO PUNTO
```
def Nombre():
n = (input("Escriba su nombre: "))
resul = n
return f"Hola {resul} "
Nombre()
```
TERCER PUNTO
```
def Area():
CmLados = int(input("Escriba cuanto mide uno de los lados del cuadrado: "))
resul = CmLados**2
return f"El area del cuadrado es: {resul} centimetros cuadrados"
Area()
```
CUARTO PUNTO
```
def AreaRec():
Base = int(input("Escriba en cm la base del rectangulo: "))
Altura = int(input("Escriba en cm la altura del rectangulo: "))
resul = Base * Altura
return f"El area del rectangulo es: {resul} centimetros cuadrados"
AreaRec()
```
QUINTO PUNTO
```
def AreaTria():
Base = int(input("Escriba en cm la base del triangulo: "))
Altura = int(input("Escriba en cm la altura del triangulo: "))
resul = int((Base * Altura)/2)
return f"El area del triangulo es: {resul} centimetros cuadrados"
AreaTria()
```
SEXTO PUNTO
```
def Botellas():
Bot1L = int(input("Escriba cuantas botellas de 1 litro reciclo: "))
Bot1mL = int(input("Escriba cuantas botellas de 1.5 litros reciclo: "))
Bot2L = int(input("Escriba cuantas botellas de 2 litros reciclo: "))
resul = Bot1L * 1000 + Bot1mL * 2000 + Bot2L * 3000
return f"Lo que el usuario debe recibir es: {resul}"
Botellas()
```
SEPTIMO PUNTO
```
def Comida():
Valor = int(input("Escriba el costo de su comida: "))
Propi = int(input("Escriba el valor de propina: "))
resul = ((Propi/100)* Valor)+(Valor*0.08)+Valor
return f"Su valor total es {resul}"
Comida()
```
OCTAVO PUNTO
```
def producto():
A = int(input("Escriba cuantos productos del A compro: "))
B = int(input("Escriba cuantos productos del B compro: "))
Peso = A*123 + B*35
if Peso%2 == 0:
print("Su peso es par y es: " , Peso )
else:
print("No es par y no le podemos vender si no es par, su peso es:", Peso)
producto()
```
NOVENO PUNTO
```
def parqueadero():
vehiculo = (input("¿Que tipo de vehiculo tiene: "))
if vehiculo == "carro":
vcarro = int(input("¿Cuantos minutos lleva su carro estacionado?: "))
print("Su valor a pagar es:", vcarro*70)
pago = int(input("¿Con cuanto dinero va a pagar?: "))
print("Su cambio es: ", pago - (vcarro*70) )
elif vehiculo == "moto":
vmoto = int(input("¿Cuantos minutos lleva su moto estacionada?: "))
print("Su valor a pagar es: ", vmoto*42)
pago1 = int(input("¿Con cuanto dinero va a pagar?: "))
print("Su cambio es: ", pago1 - (vmoto*42) )
elif vehiculo == "bicicleta":
vbici = int(input("¿Cuantos minutos lleva su bicicleta estacionada?: "))
print("Su valor a pagar es: ", vbici*10)
pago2 = int(input("¿Con cuanto dinero va a pagar?: "))
print("Su cambio es: ", pago2 - (vbici*10) )
parqueadero()
```
DECIMO PUNTO
```
import numpy as np
def Circulo():
radio = int(input("Escriba el radio de el circulo: "))
perimetro = 2*np.pi*radio
area = np.pi*radio**2
return f"El perimetro del circulo es: {perimetro} centimetros y su area es: {area} centimetros cuadrados"
Circulo()
```
DECIMO PRIMER PUNTO
```
from datetime import datetime
def Años():
fecha_nacimiento=(input("ingresa le fecha de tu nacimiento con el siguiente formato: DD/MM/YYYY"))
fecha_actual=datetime.today().strftime('%d/%m/%Y')
fecha_nacimiento=fecha_nacimiento.split('/')
fecha_actual=fecha_actual.split('/')
if int(fecha_actual[1]) > int(fecha_nacimiento[1]) or int(fecha_actual[1]) == int(fecha_nacimiento[1]) and int(fecha_actual[0]) >= int(fecha_nacimiento[0]):
years=int(fecha_actual[2])-int(fecha_nacimiento[2])
print('Tienes ' + str(years) + ' años')
else:
years=int(fecha_actual[2])-int(fecha_nacimiento[2])-1
print('Tienes ' + str(years) + ' años')
Años()
```
DECIMO SEGUNDO PUNTO
```
def Temperatura():
Cel = int(input("Escriba los grados en Celsius: "))
Fahrenheit = Cel * 1.8 + 32
Kelvin = Cel + 273.15
return f"Loa grados de Celsius a Fahrenheit son: {Fahrenheit} y de Celsius a Kelvin son: {Kelvin}"
Temperatura()
```
DECIMO TERCER PUNTO
```
lista= []
cantidad = int(input("Cuantos datos desea agregar: "))
while cantidad>0:
dato = input("Ingrese sus datos: ")
lista.append(dato)
cantidad-=1
print("Contenido lista",lista)
for i in range(len(lista)):
lista[i] = int(lista[i])
lista.sort()
Max = (max(lista))
Min = (min(lista))
Sum = (sum(lista))
print(f"El valor maximo es: {Max} el valor minimo es: {Min} y la suma de todos los elementos es {Sum}")
```
DECIMO CUARTO PUNTO
```
def dias(mes):
if mes.lower() in ("enero", "marzo","mayo","julio","agosto","octubre","diciembre"):
return "31"
elif mes.lower() == "febrero":
return "28/29"
else:
return "30"
meses = input("Ingrese el mes: ")
print(dias(meses))
```
DECIMO QUINTO
```
def edades():
n= int(input("Escriba su edad: "))
if n<18:
print("MENOR DE EDAD")
elif n > 18 and n< 45:
print("ADULTO JOVEN")
elif n > 45 and n< 60:
print("ADULTO")
elif n>60:
print("ADULTO MAYOR")
edades()
```
DECIMO SEXTO
```
def Caras():
valor= int(input("Ingrese el valor de su billete: "))
if valor == 1000:
print("La cara es: Jorge Eliecer Gaitan")
elif valor == 2000:
print("La cara es: Francisco de Paula Santander")
elif valor == 5000:
print("La cara es: Jose Asuncion Silva")
elif valor == 10000:
print("La cara es: Policarpa Salavarrieta")
elif valor == 20000:
print("La cara es: Julio Garavito Armero")
elif valor == 50000:
print("La cara es: Jorge Isaacs")
elif valor == 100000:
print("La cara es: Carlos Lleras Restrepo")
Caras()
```
DECIMO SEPTIMO
```
New = [3,5,1,9,10,11,32,21,5,1,209,432,1,32,45]
#Agregar un elemento
New.append(10)
#Agregar un elemento
New.append(3)
#Agregar varios elementos
New.extend([5,6,7])
#Eliminar el ultimo elemento
New.pop()
#Ordenar lista ascendente
New.sort()
#Eliminar el ultimo elemento
New.pop()
#Ordenar lista descendente
New.sort(reverse=True)
#Eliminar posicion 10
New.pop(10)
#Agrege el 10
New.append(10)
#Agrege el 345
New.append(345)
#Agrege el 1
New.append(1)
#Elimine el 9
New.remove(9)
#Invierta el orden de la lista
New.reverse()
#Organice la lista
New.sort()
New
#Numeros pares de la lista al cuadrado
for num in New:
if num % 2 == 0:
print(num**2, end = " ")
#Numeros multiplos de 3 al cubo
for num in New:
if num % 3 == 0:
print(num**3, end = " ")
#Elimine ultimo elemento
New.pop()
#Elimine ultimo elemento
New.pop()
New
```
|
github_jupyter
|
def datos():
primern = (input("Escriba su primer nombre: "))
segundon = (input("Si tiene segundo nombre diga Si de lo contrario diga No: "))
if segundon == "Si":
segundo = (input("Escriba su segundo nombre: "))
PrimerA = input("Escriba su primer apellido: ")
seg = (input("Si tiene segundo apellido diga Si de lo contrario diga No: "))
if seg == "Si":
SegAp =str((input("Escriba su segundo apellido: ")))
Edad = int(input("Escriba su edad: "))
iden_via = input("Escriba la identificacion de la vida: ")
num_via = int(input("Escriba el numero que acompaña a la via: "))
marca_num1 = int(input("Escriba el numero de marca 1: "))
letra = input("Escriba la letra luego del numero: ")
marca_num2 = int(input("Escriba el numero de marca 2: "))
casa = (input("Escriba numero de casa: "))
if segundon == "No" and seg == "No":
print(f"Su hombre es {primern} {PrimerA} su edad es {Edad} y\nLa direccion es {iden_via} {num_via} # {marca_num1}{letra} - {marca_num2} y la casa es {casa}" )
if segundon == "No":
print(f"Su hombre es {primern} {PrimerA} {SegAp} su edad es {Edad} y\nLa direccion es {iden_via} {num_via} # {marca_num1}{letra} - {marca_num2} y la casa es {casa}" )
if seg == "No":
print(f"Su hombre es {primern} {segundo} {PrimerA} su edad es {Edad} y\nLa direccion es {iden_via} {num_via} # {marca_num1}{letra} - {marca_num2} y la casa es {casa}" )
else:
print(f"Su hombre es {primern} {segundo} {PrimerA} {SegAp} su edad es {Edad} y\nLa direccion es {iden_via} {num_via} # {marca_num1}{letra} - {marca_num2} y la casa es {casa}" )
datos()
def Nombre():
n = (input("Escriba su nombre: "))
resul = n
return f"Hola {resul} "
Nombre()
def Area():
CmLados = int(input("Escriba cuanto mide uno de los lados del cuadrado: "))
resul = CmLados**2
return f"El area del cuadrado es: {resul} centimetros cuadrados"
Area()
def AreaRec():
Base = int(input("Escriba en cm la base del rectangulo: "))
Altura = int(input("Escriba en cm la altura del rectangulo: "))
resul = Base * Altura
return f"El area del rectangulo es: {resul} centimetros cuadrados"
AreaRec()
def AreaTria():
Base = int(input("Escriba en cm la base del triangulo: "))
Altura = int(input("Escriba en cm la altura del triangulo: "))
resul = int((Base * Altura)/2)
return f"El area del triangulo es: {resul} centimetros cuadrados"
AreaTria()
def Botellas():
Bot1L = int(input("Escriba cuantas botellas de 1 litro reciclo: "))
Bot1mL = int(input("Escriba cuantas botellas de 1.5 litros reciclo: "))
Bot2L = int(input("Escriba cuantas botellas de 2 litros reciclo: "))
resul = Bot1L * 1000 + Bot1mL * 2000 + Bot2L * 3000
return f"Lo que el usuario debe recibir es: {resul}"
Botellas()
def Comida():
Valor = int(input("Escriba el costo de su comida: "))
Propi = int(input("Escriba el valor de propina: "))
resul = ((Propi/100)* Valor)+(Valor*0.08)+Valor
return f"Su valor total es {resul}"
Comida()
def producto():
A = int(input("Escriba cuantos productos del A compro: "))
B = int(input("Escriba cuantos productos del B compro: "))
Peso = A*123 + B*35
if Peso%2 == 0:
print("Su peso es par y es: " , Peso )
else:
print("No es par y no le podemos vender si no es par, su peso es:", Peso)
producto()
def parqueadero():
vehiculo = (input("¿Que tipo de vehiculo tiene: "))
if vehiculo == "carro":
vcarro = int(input("¿Cuantos minutos lleva su carro estacionado?: "))
print("Su valor a pagar es:", vcarro*70)
pago = int(input("¿Con cuanto dinero va a pagar?: "))
print("Su cambio es: ", pago - (vcarro*70) )
elif vehiculo == "moto":
vmoto = int(input("¿Cuantos minutos lleva su moto estacionada?: "))
print("Su valor a pagar es: ", vmoto*42)
pago1 = int(input("¿Con cuanto dinero va a pagar?: "))
print("Su cambio es: ", pago1 - (vmoto*42) )
elif vehiculo == "bicicleta":
vbici = int(input("¿Cuantos minutos lleva su bicicleta estacionada?: "))
print("Su valor a pagar es: ", vbici*10)
pago2 = int(input("¿Con cuanto dinero va a pagar?: "))
print("Su cambio es: ", pago2 - (vbici*10) )
parqueadero()
import numpy as np
def Circulo():
radio = int(input("Escriba el radio de el circulo: "))
perimetro = 2*np.pi*radio
area = np.pi*radio**2
return f"El perimetro del circulo es: {perimetro} centimetros y su area es: {area} centimetros cuadrados"
Circulo()
from datetime import datetime
def Años():
fecha_nacimiento=(input("ingresa le fecha de tu nacimiento con el siguiente formato: DD/MM/YYYY"))
fecha_actual=datetime.today().strftime('%d/%m/%Y')
fecha_nacimiento=fecha_nacimiento.split('/')
fecha_actual=fecha_actual.split('/')
if int(fecha_actual[1]) > int(fecha_nacimiento[1]) or int(fecha_actual[1]) == int(fecha_nacimiento[1]) and int(fecha_actual[0]) >= int(fecha_nacimiento[0]):
years=int(fecha_actual[2])-int(fecha_nacimiento[2])
print('Tienes ' + str(years) + ' años')
else:
years=int(fecha_actual[2])-int(fecha_nacimiento[2])-1
print('Tienes ' + str(years) + ' años')
Años()
def Temperatura():
Cel = int(input("Escriba los grados en Celsius: "))
Fahrenheit = Cel * 1.8 + 32
Kelvin = Cel + 273.15
return f"Loa grados de Celsius a Fahrenheit son: {Fahrenheit} y de Celsius a Kelvin son: {Kelvin}"
Temperatura()
lista= []
cantidad = int(input("Cuantos datos desea agregar: "))
while cantidad>0:
dato = input("Ingrese sus datos: ")
lista.append(dato)
cantidad-=1
print("Contenido lista",lista)
for i in range(len(lista)):
lista[i] = int(lista[i])
lista.sort()
Max = (max(lista))
Min = (min(lista))
Sum = (sum(lista))
print(f"El valor maximo es: {Max} el valor minimo es: {Min} y la suma de todos los elementos es {Sum}")
def dias(mes):
if mes.lower() in ("enero", "marzo","mayo","julio","agosto","octubre","diciembre"):
return "31"
elif mes.lower() == "febrero":
return "28/29"
else:
return "30"
meses = input("Ingrese el mes: ")
print(dias(meses))
def edades():
n= int(input("Escriba su edad: "))
if n<18:
print("MENOR DE EDAD")
elif n > 18 and n< 45:
print("ADULTO JOVEN")
elif n > 45 and n< 60:
print("ADULTO")
elif n>60:
print("ADULTO MAYOR")
edades()
def Caras():
valor= int(input("Ingrese el valor de su billete: "))
if valor == 1000:
print("La cara es: Jorge Eliecer Gaitan")
elif valor == 2000:
print("La cara es: Francisco de Paula Santander")
elif valor == 5000:
print("La cara es: Jose Asuncion Silva")
elif valor == 10000:
print("La cara es: Policarpa Salavarrieta")
elif valor == 20000:
print("La cara es: Julio Garavito Armero")
elif valor == 50000:
print("La cara es: Jorge Isaacs")
elif valor == 100000:
print("La cara es: Carlos Lleras Restrepo")
Caras()
New = [3,5,1,9,10,11,32,21,5,1,209,432,1,32,45]
#Agregar un elemento
New.append(10)
#Agregar un elemento
New.append(3)
#Agregar varios elementos
New.extend([5,6,7])
#Eliminar el ultimo elemento
New.pop()
#Ordenar lista ascendente
New.sort()
#Eliminar el ultimo elemento
New.pop()
#Ordenar lista descendente
New.sort(reverse=True)
#Eliminar posicion 10
New.pop(10)
#Agrege el 10
New.append(10)
#Agrege el 345
New.append(345)
#Agrege el 1
New.append(1)
#Elimine el 9
New.remove(9)
#Invierta el orden de la lista
New.reverse()
#Organice la lista
New.sort()
New
#Numeros pares de la lista al cuadrado
for num in New:
if num % 2 == 0:
print(num**2, end = " ")
#Numeros multiplos de 3 al cubo
for num in New:
if num % 3 == 0:
print(num**3, end = " ")
#Elimine ultimo elemento
New.pop()
#Elimine ultimo elemento
New.pop()
New
| 0.212886 | 0.804137 |
<a href="https://colab.research.google.com/github/joshsbloom/swabseq/blob/master/kb/notebooks/comparison_v18.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
!git clone https://github.com/joshsbloom/swabseq.git
!wget http://131.215.54.5:8080/media/core/2/matrix.h5ad
!wget http://131.215.54.5:8080/media/core/2/samplesheet.csv
```
# Install packages
```
!pip install anndata
# Convert samplesheet to csv for loading
def samplesheet_to_csv(samplesheet, csv):
with open(samplesheet, 'r') as ss, open(csv,'w') as c:
line = ss.readline().strip()
while line != '[Data]':
line = ss.readline().strip()
# the rest of the lines are data
for line in ss:
c.write(line)
samplesheet_to_csv("samplesheet.csv", "ss.csv")
!head ss.csv
```
# Python analysis
```
#@title Imports
import anndata
import pandas as pd
import numpy as np
from collections import defaultdict
import matplotlib.pyplot as plt
import matplotlib as mpl
from mpl_toolkits.axes_grid1 import make_axes_locatable
import matplotlib.gridspec as gridspec
def nd(arr):
return np.asarray(arr).reshape(-1)
def yex(ax, offset=0):
lims = [
np.min([ax.get_xlim(), ax.get_ylim()]), # min of both axes
np.max([ax.get_xlim(), ax.get_ylim()]), # max of both axes
]
# now plot both limits against eachother
ax.plot(lims, np.array(lims)+offset, 'k-', alpha=0.75, zorder=0)
ax.set_aspect('equal')
ax.set_xlim(lims)
ax.set_ylim(lims)
return ax
fsize=20
plt.rcParams.update({'font.size': fsize})
%config InlineBackend.figure_format = 'retina'
```
# Load data
```
adata = anndata.read_h5ad("matrix.h5ad")
adata.obs['index'] = adata.obs.index.str.slice(0, 10)
adata.obs['index2'] = adata.obs.index.str.slice(10)
ss = pd.read_csv('ss.csv')
ss['bcs'] = ss['index'] + ss['index2']
ss.index = ss['bcs']
ss["sample_id"] = ss["Plate_ID"] + "_" + ss["Sample_Well"]
print(ss.bcs.nunique())
samples = ss['Sample_ID'].values
index = ss['index'].values
index2 = ss['index2'].values
```
# Collapse the Rpp30 and S and S spike
```
data = adata[adata.obs.index.isin(ss.bcs.values)]
data = data[data.obs.sort_values("Sample_ID").index]
data.obs["sample_id"] = data.obs.Plate_ID.astype(str) + "_" + data.obs.Sample_Well.astype(str)
bcs_lst = data.obs.bcs.values.astype(str)
sample_id_lst = data.obs.sample_id.values.astype(str)
mtx = []
data.obs.groupby(["Plate_ID", "Sample_Well"]).apply(lambda x: mtx.append(nd(data.X[np.squeeze([np.where(bcs_lst == i)[0] for i in x.index.values] ) ].sum(axis=0))))
mtx = np.squeeze(mtx).astype(int)
mtx.astype(int)
mtx.shape
data.X.todense().astype(int)
obs = data.obs.drop_duplicates("sample_id")
var = data.var
ndata = anndata.AnnData(X=mtx, obs=obs, var=var)
ndata.obs['sample_id'] = ndata.obs.Plate_ID.astype(str) + "_" + ndata.obs.Sample_Well.astype(str)
```
# Load simple
```
def make_mtx(bcs, ecs, cnt, unique_ecs):
bold = bcs[0]
eold = ecs[0]
cold = cnt[0]
mtx = []
d = defaultdict()
#d[eold] = cold
bold = 0
for idx, b in enumerate(bcs):
if b != bold and idx > 0:
count = []
for e in unique_ecs:
count.append(d.get(e, 0))
mtx.append(count)
d = defaultdict()
d[ecs[idx]] = cnt[idx]
bold = b
count = []
for e in unique_ecs:
count.append(d.get(e, 0))
mtx.append(count)
return np.asarray(mtx)
simple = pd.read_csv("swabseq/runs/v18/countTable.csv")
simple["sample_id"] = simple.Plate_ID + "_" + simple.Sample_Well
var = simple.amplicon.unique()
obs = simple.mergedIndex.unique()
df = simple.groupby(["sample_id","amplicon"])["Count"].sum().reset_index()
bcs = df.sample_id
var = df.amplicon
cnt = df.Count
unique_var = np.unique(var)
mtx = make_mtx(bcs, var, cnt, unique_var)
sm = anndata.AnnData(X=mtx.astype(int), obs = {"sample_id": df.drop_duplicates("sample_id").sample_id.values}, var = {"amplicon": unique_var})
sm.X = sm.X.astype(int)
```
# Do the actual comparison
```
gene_map = {
"S2": "S2",
"S2_spikein": "S2_spike",
"RPP30": "RPP30"
}
m = [True, True, False, False, True]
kb = ndata[:,m].copy()
kb.var.index = kb.var.index.map(gene_map).values
kb.var
kb_bcs = kb.obs.sample_id.values
sm_bcs = sm.obs.sample_id.values
common = np.intersect1d(kb_bcs, sm_bcs)
common.shape
kb = kb[kb.obs.sample_id.isin(common).values]
sm = sm[sm.obs.sample_id.isin(common).values]
sm = sm[sm.obs.sort_values("sample_id").index]
sm = sm[:,sm.var.sort_index().index]
kb = kb[kb.obs.sort_values("sample_id").index]
kb = kb[:,kb.var.sort_index().index]
g = ["RPP30",
"S2",
"S2_spike"]
def trim_axs(axs, N):
"""little helper to massage the axs list to have correct length..."""
axs = axs.flat
for ax in axs[N:]:
ax.remove()
return axs[:N]
import matplotlib as mpl
from scipy import stats
fig, ax = plt.subplots(figsize=(7*3,7), ncols=3)
axs = trim_axs(ax, len(kb.var.index.values))
for gidx, (ax, gene) in enumerate(zip(axs, g)):
kb_gidx = np.where(kb.var.index.values==gene)[0][0]
sm_gidx = np.where(sm.var.amplicon.values==gene)[0][0]
x = nd(sm.X[:,sm_gidx])
y = nd(kb.X[:,kb_gidx])
#c = (y>250000).astype(int)
r, p = stats.pearsonr(x, y)
if gene=="S2":
gene = "S"
elif gene == "S2_spike":
gene="S_spike"
ax.scatter(x, y, color="k", s=100, label="{}\nr$^2$:{:,.2f}".format(gene, r**2))
yex(ax)
#ax.set_axis_off()
ax.legend(loc="upper left")
ax.xaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
for label in ax.get_xticklabels():
label.set_ha("right")
label.set_rotation(45)
fig.text(0.5, 0, 'simple counts', ha='center', va='center', fontsize=20)
fig.text(0, 0.5, 'kallisto | bustools counts', ha='center', va='center', rotation='vertical', fontsize=20)
plt.tight_layout()
#plt.savefig("./figs/kb_v_starcode.png",bbox_inches='tight', dpi=300)
plt.show()
ntc = ss.query("virus_copy=='0.0'")["sample_id"]
ntc.shape
kb_ntc = kb[kb.obs.sample_id.isin(ntc).values].copy()
sm_ntc = sm[sm.obs.sample_id.isin(ntc).values].copy()
kb_ntc.shape, sm_ntc.shape
```
# Counts for NTC
```
fig, ax = plt.subplots(figsize=(7*3,7), ncols=3)
axs = trim_axs(ax, len(kb_ntc.var.index.values))
for gidx, (ax, gene) in enumerate(zip(axs, g)):
kb_gidx = np.where(kb_ntc.var.index.values==gene)[0][0]
sm_gidx = np.where(sm_ntc.var.amplicon.values==gene)[0][0]
x = nd(sm_ntc.X[:,sm_gidx])
y = nd(kb_ntc.X[:,kb_gidx])
#c = (y>250000).astype(int)
r, p = stats.pearsonr(x, y)
if gene=="S2":
gene = "S"
elif gene == "S2_spike":
gene="S_spike"
ax.scatter(x, y, color="k", s=100, label="{}\nr$^2$:{:,.2f}".format(gene, r**2))
yex(ax)
#ax.set_axis_off()
ax.legend()
ax.xaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
for label in ax.get_xticklabels():
label.set_ha("right")
label.set_rotation(45)
fig.text(0.5, 0, 'simple counts', ha='center', va='center', fontsize=20)
fig.text(0, 0.5, 'kallisto | bustools counts', ha='center', va='center', rotation='vertical', fontsize=20)
plt.tight_layout()
#plt.savefig("./figs/kb_v_starcode.png",bbox_inches='tight', dpi=300)
plt.show()
fig, ax = plt.subplots(figsize=(10, 10))
sm_vals = nd(sm_ntc.X[:,1:][:,0]/sm_ntc.X[:,1:][:,1])
kb_vals = nd(kb_ntc.X[:,1:][:,0]/kb_ntc.X[:,1:][:,1])
x = sm_vals[~np.isnan(sm_vals)]
y = kb_vals[~np.isnan(kb_vals)]
ax.scatter(x, y, s=50)
kwd = {
"xlabel": "simple S/Spike",
"ylabel":"kb S/Spike",
}
ax.set(**kwd)
yex(ax)
# create new axes on the right and on the top of the current axes
# The first argument of the new_vertical(new_horizontal) method is
# the height (width) of the axes to be created in inches.
divider = make_axes_locatable(ax)
axHistx = divider.append_axes("top", 1.2, pad=0.4, sharex=ax)
axHisty = divider.append_axes("right", 1.2, pad=0.4, sharey=ax)
# make some labels invisible
axHistx.xaxis.set_tick_params(labelbottom=False)
axHisty.yaxis.set_tick_params(labelleft=False)
# now determine nice limits by hand:
binwidth = 0.00025
xymax = max(np.max(np.abs(x)), np.max(np.abs(y)))
lim = (int(xymax/binwidth) + 1)*binwidth
bins = np.arange(-lim, lim + binwidth, binwidth)
axHistx.hist(x, bins=bins)
axHisty.hist(y, bins=bins, orientation='horizontal')
axHistx.axvline(x = x.mean(), color="lightgray", linestyle="--")
axHisty.axhline(y = y.mean(), color="lightgray", linestyle="--")
# the xaxis of axHistx and yaxis of axHisty are shared with ax,
# thus there is no need to manually adjust the xlim and ylim of these
# axis.
ax.axvline(x=x.mean(), color="lightgray", linestyle="--")
ax.axhline(y=y.mean(), color="lightgray", linestyle="--")
fig.show()
print(x.mean(), y.mean())
print(x.var(), y.var())
print(f"{kb.X.sum():,.0f}")
print(f"{sm.X.sum():,.0f}")
```
|
github_jupyter
|
!git clone https://github.com/joshsbloom/swabseq.git
!wget http://131.215.54.5:8080/media/core/2/matrix.h5ad
!wget http://131.215.54.5:8080/media/core/2/samplesheet.csv
!pip install anndata
# Convert samplesheet to csv for loading
def samplesheet_to_csv(samplesheet, csv):
with open(samplesheet, 'r') as ss, open(csv,'w') as c:
line = ss.readline().strip()
while line != '[Data]':
line = ss.readline().strip()
# the rest of the lines are data
for line in ss:
c.write(line)
samplesheet_to_csv("samplesheet.csv", "ss.csv")
!head ss.csv
#@title Imports
import anndata
import pandas as pd
import numpy as np
from collections import defaultdict
import matplotlib.pyplot as plt
import matplotlib as mpl
from mpl_toolkits.axes_grid1 import make_axes_locatable
import matplotlib.gridspec as gridspec
def nd(arr):
return np.asarray(arr).reshape(-1)
def yex(ax, offset=0):
lims = [
np.min([ax.get_xlim(), ax.get_ylim()]), # min of both axes
np.max([ax.get_xlim(), ax.get_ylim()]), # max of both axes
]
# now plot both limits against eachother
ax.plot(lims, np.array(lims)+offset, 'k-', alpha=0.75, zorder=0)
ax.set_aspect('equal')
ax.set_xlim(lims)
ax.set_ylim(lims)
return ax
fsize=20
plt.rcParams.update({'font.size': fsize})
%config InlineBackend.figure_format = 'retina'
adata = anndata.read_h5ad("matrix.h5ad")
adata.obs['index'] = adata.obs.index.str.slice(0, 10)
adata.obs['index2'] = adata.obs.index.str.slice(10)
ss = pd.read_csv('ss.csv')
ss['bcs'] = ss['index'] + ss['index2']
ss.index = ss['bcs']
ss["sample_id"] = ss["Plate_ID"] + "_" + ss["Sample_Well"]
print(ss.bcs.nunique())
samples = ss['Sample_ID'].values
index = ss['index'].values
index2 = ss['index2'].values
data = adata[adata.obs.index.isin(ss.bcs.values)]
data = data[data.obs.sort_values("Sample_ID").index]
data.obs["sample_id"] = data.obs.Plate_ID.astype(str) + "_" + data.obs.Sample_Well.astype(str)
bcs_lst = data.obs.bcs.values.astype(str)
sample_id_lst = data.obs.sample_id.values.astype(str)
mtx = []
data.obs.groupby(["Plate_ID", "Sample_Well"]).apply(lambda x: mtx.append(nd(data.X[np.squeeze([np.where(bcs_lst == i)[0] for i in x.index.values] ) ].sum(axis=0))))
mtx = np.squeeze(mtx).astype(int)
mtx.astype(int)
mtx.shape
data.X.todense().astype(int)
obs = data.obs.drop_duplicates("sample_id")
var = data.var
ndata = anndata.AnnData(X=mtx, obs=obs, var=var)
ndata.obs['sample_id'] = ndata.obs.Plate_ID.astype(str) + "_" + ndata.obs.Sample_Well.astype(str)
def make_mtx(bcs, ecs, cnt, unique_ecs):
bold = bcs[0]
eold = ecs[0]
cold = cnt[0]
mtx = []
d = defaultdict()
#d[eold] = cold
bold = 0
for idx, b in enumerate(bcs):
if b != bold and idx > 0:
count = []
for e in unique_ecs:
count.append(d.get(e, 0))
mtx.append(count)
d = defaultdict()
d[ecs[idx]] = cnt[idx]
bold = b
count = []
for e in unique_ecs:
count.append(d.get(e, 0))
mtx.append(count)
return np.asarray(mtx)
simple = pd.read_csv("swabseq/runs/v18/countTable.csv")
simple["sample_id"] = simple.Plate_ID + "_" + simple.Sample_Well
var = simple.amplicon.unique()
obs = simple.mergedIndex.unique()
df = simple.groupby(["sample_id","amplicon"])["Count"].sum().reset_index()
bcs = df.sample_id
var = df.amplicon
cnt = df.Count
unique_var = np.unique(var)
mtx = make_mtx(bcs, var, cnt, unique_var)
sm = anndata.AnnData(X=mtx.astype(int), obs = {"sample_id": df.drop_duplicates("sample_id").sample_id.values}, var = {"amplicon": unique_var})
sm.X = sm.X.astype(int)
gene_map = {
"S2": "S2",
"S2_spikein": "S2_spike",
"RPP30": "RPP30"
}
m = [True, True, False, False, True]
kb = ndata[:,m].copy()
kb.var.index = kb.var.index.map(gene_map).values
kb.var
kb_bcs = kb.obs.sample_id.values
sm_bcs = sm.obs.sample_id.values
common = np.intersect1d(kb_bcs, sm_bcs)
common.shape
kb = kb[kb.obs.sample_id.isin(common).values]
sm = sm[sm.obs.sample_id.isin(common).values]
sm = sm[sm.obs.sort_values("sample_id").index]
sm = sm[:,sm.var.sort_index().index]
kb = kb[kb.obs.sort_values("sample_id").index]
kb = kb[:,kb.var.sort_index().index]
g = ["RPP30",
"S2",
"S2_spike"]
def trim_axs(axs, N):
"""little helper to massage the axs list to have correct length..."""
axs = axs.flat
for ax in axs[N:]:
ax.remove()
return axs[:N]
import matplotlib as mpl
from scipy import stats
fig, ax = plt.subplots(figsize=(7*3,7), ncols=3)
axs = trim_axs(ax, len(kb.var.index.values))
for gidx, (ax, gene) in enumerate(zip(axs, g)):
kb_gidx = np.where(kb.var.index.values==gene)[0][0]
sm_gidx = np.where(sm.var.amplicon.values==gene)[0][0]
x = nd(sm.X[:,sm_gidx])
y = nd(kb.X[:,kb_gidx])
#c = (y>250000).astype(int)
r, p = stats.pearsonr(x, y)
if gene=="S2":
gene = "S"
elif gene == "S2_spike":
gene="S_spike"
ax.scatter(x, y, color="k", s=100, label="{}\nr$^2$:{:,.2f}".format(gene, r**2))
yex(ax)
#ax.set_axis_off()
ax.legend(loc="upper left")
ax.xaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
for label in ax.get_xticklabels():
label.set_ha("right")
label.set_rotation(45)
fig.text(0.5, 0, 'simple counts', ha='center', va='center', fontsize=20)
fig.text(0, 0.5, 'kallisto | bustools counts', ha='center', va='center', rotation='vertical', fontsize=20)
plt.tight_layout()
#plt.savefig("./figs/kb_v_starcode.png",bbox_inches='tight', dpi=300)
plt.show()
ntc = ss.query("virus_copy=='0.0'")["sample_id"]
ntc.shape
kb_ntc = kb[kb.obs.sample_id.isin(ntc).values].copy()
sm_ntc = sm[sm.obs.sample_id.isin(ntc).values].copy()
kb_ntc.shape, sm_ntc.shape
fig, ax = plt.subplots(figsize=(7*3,7), ncols=3)
axs = trim_axs(ax, len(kb_ntc.var.index.values))
for gidx, (ax, gene) in enumerate(zip(axs, g)):
kb_gidx = np.where(kb_ntc.var.index.values==gene)[0][0]
sm_gidx = np.where(sm_ntc.var.amplicon.values==gene)[0][0]
x = nd(sm_ntc.X[:,sm_gidx])
y = nd(kb_ntc.X[:,kb_gidx])
#c = (y>250000).astype(int)
r, p = stats.pearsonr(x, y)
if gene=="S2":
gene = "S"
elif gene == "S2_spike":
gene="S_spike"
ax.scatter(x, y, color="k", s=100, label="{}\nr$^2$:{:,.2f}".format(gene, r**2))
yex(ax)
#ax.set_axis_off()
ax.legend()
ax.xaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
for label in ax.get_xticklabels():
label.set_ha("right")
label.set_rotation(45)
fig.text(0.5, 0, 'simple counts', ha='center', va='center', fontsize=20)
fig.text(0, 0.5, 'kallisto | bustools counts', ha='center', va='center', rotation='vertical', fontsize=20)
plt.tight_layout()
#plt.savefig("./figs/kb_v_starcode.png",bbox_inches='tight', dpi=300)
plt.show()
fig, ax = plt.subplots(figsize=(10, 10))
sm_vals = nd(sm_ntc.X[:,1:][:,0]/sm_ntc.X[:,1:][:,1])
kb_vals = nd(kb_ntc.X[:,1:][:,0]/kb_ntc.X[:,1:][:,1])
x = sm_vals[~np.isnan(sm_vals)]
y = kb_vals[~np.isnan(kb_vals)]
ax.scatter(x, y, s=50)
kwd = {
"xlabel": "simple S/Spike",
"ylabel":"kb S/Spike",
}
ax.set(**kwd)
yex(ax)
# create new axes on the right and on the top of the current axes
# The first argument of the new_vertical(new_horizontal) method is
# the height (width) of the axes to be created in inches.
divider = make_axes_locatable(ax)
axHistx = divider.append_axes("top", 1.2, pad=0.4, sharex=ax)
axHisty = divider.append_axes("right", 1.2, pad=0.4, sharey=ax)
# make some labels invisible
axHistx.xaxis.set_tick_params(labelbottom=False)
axHisty.yaxis.set_tick_params(labelleft=False)
# now determine nice limits by hand:
binwidth = 0.00025
xymax = max(np.max(np.abs(x)), np.max(np.abs(y)))
lim = (int(xymax/binwidth) + 1)*binwidth
bins = np.arange(-lim, lim + binwidth, binwidth)
axHistx.hist(x, bins=bins)
axHisty.hist(y, bins=bins, orientation='horizontal')
axHistx.axvline(x = x.mean(), color="lightgray", linestyle="--")
axHisty.axhline(y = y.mean(), color="lightgray", linestyle="--")
# the xaxis of axHistx and yaxis of axHisty are shared with ax,
# thus there is no need to manually adjust the xlim and ylim of these
# axis.
ax.axvline(x=x.mean(), color="lightgray", linestyle="--")
ax.axhline(y=y.mean(), color="lightgray", linestyle="--")
fig.show()
print(x.mean(), y.mean())
print(x.var(), y.var())
print(f"{kb.X.sum():,.0f}")
print(f"{sm.X.sum():,.0f}")
| 0.460532 | 0.920576 |
```
from Bio import SeqIO
for seq_record in SeqIO.parse("datas/example.fa","fasta"):
print(seq_record.id)
#print(seq_record.seq)
```
## Methods to Use in Machine Learning Seq data
#### 1.Encode the seq informatin as an ordinal Vector and work with that directly,<br> 2.One-hot encode the sequence letters and use the resulting array and <br>3. treat the DNA sequence as a language(text) and use various "language Processing" methods
```
# Function to convert a DNA sequence string to a numpy array
# converts lower case , changes any non **acgt** character to "n"
import numpy as np
import re
def string_to_array(my_string):
my_string = my_string.lower()
my_string = re.sub('[^acgt]',"z",my_string)
my_array = np.array(list(my_string))
return my_array
#testing Our Function
#string_to_array("actgmamnklhh")
```
#### Label Encoder
```
from sklearn.preprocessing import LabelEncoder
label_encoder = LabelEncoder()
label_encoder.fit(np.array(["a","c","g","t","z"]))
```
It returns a numpy array with a =0.25 , c =0.5 ,g =0.75 , t =1.0 , z =0
```
def ordinal_encoder(my_array):
integer_encoded = label_encoder.transform(my_array)
float_encoded = integer_encoded.astype(float)
float_encoded[float_encoded == 0] = 0.25 #A
float_encoded[float_encoded == 1] = 0.5 #C
float_encoded[float_encoded == 2] = 0.75 #T
float_encoded[float_encoded == 3] = 1.00 #G
float_encoded[float_encoded==4] = 0 # Other character zero
return float_encoded
# testing
test_seq = "zzACTACGMNCC"
ordinal_encoder(string_to_array(test_seq))
```
### One Hot encoding DNA Sequence data
#### Another approach is to use one hot encoding to represent the DNA sequence. This is widely used in deep learning methods and lends itself well to algorithms like convolutional neural nerworks. In this example, "ATCG" would become[0,0,0,1],[0,0,1,0],[0,1,0,0],[1,0,0,0]
```
# Function to one-hot encode a DNA sequence String
# non "acgt" bases (n) are 0000
# returnsa LX4 numpy array
from sklearn.preprocessing import OneHotEncoder
def one_hot_encoder(my_array):
integer_encoded = label_encoder.transform(my_array)
onehot_encoder = OneHotEncoder(sparse = False , dtype = int , n_values = 5)
integer_encoded = integer_encoded.reshape(len(integer_encoded),1)
onehot_encoded = onehot_encoder.fit_transform(integer_encoded)
onehot_encoded = np.delete(onehot_encoded,-1,1)
return onehot_encoded
# test the above function
test_sequence = "AACGCGGTTNM"
one_hot_encoder(string_to_array(test_sequence))
```
### Treating DNA Sequence as a "Language" otherwise known as k-mer counting
```
def getkmers(seq ,size):
return [seq[x:x+size].lower() for x in range(len(seq)-size +1)]
my_seq = "CATGGCCATCCCCCCCCGAGCGGGGGGGGGG"
#getkmers(my_seq, size=10)
```
It returns a list of K-mer "words". You can then join the "words" into a "sentence" then apply your favorite natural language processing methods
```
words = getkmers(my_seq, size = 6)
sentence = " ".join(words)
sentence[:30]
###>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
my_seq2 = 'GATGGCCATCCCCGCCCGAGCGGGGGGGG'
my_seq3 = 'CATGGCCATCCCCGCCCGAGCGGGCGGGG'
sentence2 = " ".join(getkmers(my_seq2,size =6))
sentence3 = " ".join(getkmers(my_seq3, size = 6))
## Creating the Bag of Words Model\
from sklearn.feature_extraction.text import CountVectorizer
cv = CountVectorizer()
x = cv.fit_transform([sentence, sentence2 , sentence3]).toarray()
```
# Classification of gene function
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
```
### Let's open the data for human and see what we have
```
human = pd.read_table("datas/human_data/human_data.txt")
human.head()
```
#### We have some data for human DNA sequence coding regions and a class label. We also have data for Chimpanzee and a more divergent species, the dog. Let's get that.
```
chimp = pd.read_table("datas/chimp_data/chimp_data.txt")
dog = pd.read_table("datas/dog_data/dog_data.txt")
chimp.head() , dog.head()
```
### let's define a function to collect all possible overlapping k-mers of a specified length from any sequence string
```
# Function to convert sequence strings into k-mer words, default size =6 (hexamer words)
def getkmers(sequence, size = 6):
return [sequence [x:x+size].lower() for x in range(len(sequence)-size+1)]
```
#### Now we can convert our training data sequences into short overlapping k-mers of legth 6. lets do that for each species of data we have using our getKmers function.
```
human["words"] = human.apply(lambda x : getkmers(x["sequence"]), axis =1)
human = human = human.drop("sequence", axis = 1)
chimp["words"] = chimp.apply(lambda x : getkmers(x["sequence"]),axis =1)
chimp = chimp.drop("sequence", axis=1)
dog["words"] = dog.apply(lambda x:getkmers(x["sequence"]),axis =1)
dog = dog.drop("sequence", axis = 1)
```
### Now our coding sequence data is changed to lowercase, split up into all possible k-mer words of length 6 and ready for the next step. Let's take a look
```
human.head()
human.columns
len(human.words[1])
human.shape, len(human.words[44][5])
```
Since we are going to use scikit-learn natural language processing tools to do the k-mer , we need to now convert the lists of k-mers for each gene into string sentences of words that the count vectorizeer can use. We can also make a y - variable to hold the class labels. Lets do that now.
```
human_texts = list(human["words"])
for item in range(len(human_texts)):
human_texts[item] = " ".join(human_texts[item])
y_h = human.iloc[:,0].values
y_h
#human_texts[1]
```
### Now let's do the same for chimp and dog.
```
chimp_text = list(chimp["words"])
for item in range(len(chimp_text)):
chimp_text[item] = " ".join(chimp_text[item])
y_c = chimp.iloc[: , 0].values # y_c for chimp
dog_texts = list(dog["words"])
for item in range(len(dog_texts)):
dog_texts[item] = " ".join(dog_texts[item])
y_d = dog.iloc[: , 0].values # y_d for dog
#y_c , y_d
```
### Now let's review how to use sklearn's "Natural Language " Processing tools to convert out k-mer words into uniform length numerical vectors that represent counts for every k-mer in the vocabulary.
```
# Creating the Bag of Words model using CountVectorizer()
# This is equivalent to k-mer counting
# The n-gram size of 4 was previously determined by testing
from sklearn.feature_extraction.text import CountVectorizer
cv = CountVectorizer(ngram_range = (4,4))
x= cv.fit_transform(human_texts)
x_chimp = cv.transform(chimp_text)
x_dog = cv.transform(dog_texts)
```
Let's see what we have<br>
for human we have 4380 genes converted into uniform length feature vectors of 4-gram k-mer (length 6 ) counts. For chimp and dog we have the expected same number of features with 1682 and 820 genes respectively.
```
print(x.shape,x_chimp.shape , x_dog.shape)
human["class"].value_counts().sort_index().plot.bar()
chimp["class"].value_counts().sort_index().plot.bar()
dog["class"].value_counts().sort_index().plot.bar()
```
|
github_jupyter
|
from Bio import SeqIO
for seq_record in SeqIO.parse("datas/example.fa","fasta"):
print(seq_record.id)
#print(seq_record.seq)
# Function to convert a DNA sequence string to a numpy array
# converts lower case , changes any non **acgt** character to "n"
import numpy as np
import re
def string_to_array(my_string):
my_string = my_string.lower()
my_string = re.sub('[^acgt]',"z",my_string)
my_array = np.array(list(my_string))
return my_array
#testing Our Function
#string_to_array("actgmamnklhh")
from sklearn.preprocessing import LabelEncoder
label_encoder = LabelEncoder()
label_encoder.fit(np.array(["a","c","g","t","z"]))
def ordinal_encoder(my_array):
integer_encoded = label_encoder.transform(my_array)
float_encoded = integer_encoded.astype(float)
float_encoded[float_encoded == 0] = 0.25 #A
float_encoded[float_encoded == 1] = 0.5 #C
float_encoded[float_encoded == 2] = 0.75 #T
float_encoded[float_encoded == 3] = 1.00 #G
float_encoded[float_encoded==4] = 0 # Other character zero
return float_encoded
# testing
test_seq = "zzACTACGMNCC"
ordinal_encoder(string_to_array(test_seq))
# Function to one-hot encode a DNA sequence String
# non "acgt" bases (n) are 0000
# returnsa LX4 numpy array
from sklearn.preprocessing import OneHotEncoder
def one_hot_encoder(my_array):
integer_encoded = label_encoder.transform(my_array)
onehot_encoder = OneHotEncoder(sparse = False , dtype = int , n_values = 5)
integer_encoded = integer_encoded.reshape(len(integer_encoded),1)
onehot_encoded = onehot_encoder.fit_transform(integer_encoded)
onehot_encoded = np.delete(onehot_encoded,-1,1)
return onehot_encoded
# test the above function
test_sequence = "AACGCGGTTNM"
one_hot_encoder(string_to_array(test_sequence))
def getkmers(seq ,size):
return [seq[x:x+size].lower() for x in range(len(seq)-size +1)]
my_seq = "CATGGCCATCCCCCCCCGAGCGGGGGGGGGG"
#getkmers(my_seq, size=10)
words = getkmers(my_seq, size = 6)
sentence = " ".join(words)
sentence[:30]
###>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
my_seq2 = 'GATGGCCATCCCCGCCCGAGCGGGGGGGG'
my_seq3 = 'CATGGCCATCCCCGCCCGAGCGGGCGGGG'
sentence2 = " ".join(getkmers(my_seq2,size =6))
sentence3 = " ".join(getkmers(my_seq3, size = 6))
## Creating the Bag of Words Model\
from sklearn.feature_extraction.text import CountVectorizer
cv = CountVectorizer()
x = cv.fit_transform([sentence, sentence2 , sentence3]).toarray()
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
human = pd.read_table("datas/human_data/human_data.txt")
human.head()
chimp = pd.read_table("datas/chimp_data/chimp_data.txt")
dog = pd.read_table("datas/dog_data/dog_data.txt")
chimp.head() , dog.head()
# Function to convert sequence strings into k-mer words, default size =6 (hexamer words)
def getkmers(sequence, size = 6):
return [sequence [x:x+size].lower() for x in range(len(sequence)-size+1)]
human["words"] = human.apply(lambda x : getkmers(x["sequence"]), axis =1)
human = human = human.drop("sequence", axis = 1)
chimp["words"] = chimp.apply(lambda x : getkmers(x["sequence"]),axis =1)
chimp = chimp.drop("sequence", axis=1)
dog["words"] = dog.apply(lambda x:getkmers(x["sequence"]),axis =1)
dog = dog.drop("sequence", axis = 1)
human.head()
human.columns
len(human.words[1])
human.shape, len(human.words[44][5])
human_texts = list(human["words"])
for item in range(len(human_texts)):
human_texts[item] = " ".join(human_texts[item])
y_h = human.iloc[:,0].values
y_h
#human_texts[1]
chimp_text = list(chimp["words"])
for item in range(len(chimp_text)):
chimp_text[item] = " ".join(chimp_text[item])
y_c = chimp.iloc[: , 0].values # y_c for chimp
dog_texts = list(dog["words"])
for item in range(len(dog_texts)):
dog_texts[item] = " ".join(dog_texts[item])
y_d = dog.iloc[: , 0].values # y_d for dog
#y_c , y_d
# Creating the Bag of Words model using CountVectorizer()
# This is equivalent to k-mer counting
# The n-gram size of 4 was previously determined by testing
from sklearn.feature_extraction.text import CountVectorizer
cv = CountVectorizer(ngram_range = (4,4))
x= cv.fit_transform(human_texts)
x_chimp = cv.transform(chimp_text)
x_dog = cv.transform(dog_texts)
print(x.shape,x_chimp.shape , x_dog.shape)
human["class"].value_counts().sort_index().plot.bar()
chimp["class"].value_counts().sort_index().plot.bar()
dog["class"].value_counts().sort_index().plot.bar()
| 0.429429 | 0.898988 |
```
import cairo
import cv2
from math import pi
import random
import math
from PIL import Image
import numpy as np
from scipy.spatial import distance
import matplotlib.pyplot as plt
from sklearn.neighbors import KDTree
def polar_to_cart(theta, dist):
x = 1 + dist * math.cos(theta)
y = 1 + dist * math.sin(theta)
return x,y
t = math.pi/180.0
def remap(old_val, old_min, old_max, new_min, new_max):
return (new_max - new_min)*(old_val - old_min) / (old_max - old_min) + new_min
def make_hashable(array):
return tuple(map(float, array))
def draw(geno):
surface = cairo.ImageSurface(cairo.FORMAT_ARGB32, 100, 100)
ctx = cairo.Context(surface)
#ctx.set_antialias(cairo.ANTIALIAS_NONE)
ctx.scale(50, 50)
# Paint the background
ctx.set_source_rgb(0, 0 , 0)
ctx.paint()
r1 = remap(geno[8], 0, 1,0.1, 1)
r2 = remap(geno[9], 0, 1,0.1, 1)
r3 = remap(geno[10], 0, 1,0.1, 1)
r4 = remap(geno[11], 0, 1,0.1, 1)
r5 = remap(geno[12], 0, 1,0.1, 1)
r6 = remap(geno[13], 0, 1,0.1, 1)
r7 = remap(geno[14], 0, 1,0.1, 1)
r8 = remap(geno[15], 0, 1,0.1, 1)
# Draw the image
firstx, firsty = polar_to_cart((0 + geno[0])*45*t, r1)
secondx, secondy = polar_to_cart((1 + geno[1])*45*t, r2)
thirdx, thirdy = polar_to_cart((2 + geno[2])*45*t, r3)
forthx, forthy = polar_to_cart((3 + geno[3])*45*t, r4)
fifthx, fifthy = polar_to_cart((4 + geno[4])*45*t, r5)
sixthx, sixthy = polar_to_cart((5 + geno[5])*45*t, r6)
seventhx, seventhy = polar_to_cart((6 + geno[6])*45*t, r7)
eigthx, eigthy = polar_to_cart((7 + geno[7])*45*t, r8)
ctx.move_to(firstx, firsty)
ctx.line_to(secondx, secondy)
ctx.line_to(thirdx, thirdy)
ctx.line_to(forthx, forthy)
ctx.line_to(fifthx, fifthy)
ctx.line_to(sixthx, sixthy)
ctx.line_to(seventhx, seventhy)
ctx.line_to(eigthx, eigthy)
ctx.close_path()
ctx.set_source_rgb(1, 1, 1)
ctx.fill_preserve()
return surface
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_avg3.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_avg3 = []
spread_avg3 = []
centroids_avg3 = load_centroids("centroids_avg3.dat")
data_avg3 = np.loadtxt("archive_avg3.dat")
fit_avg3 = data_avg3[:,0:1]
cent_avg3 = data_avg3[:, 1:3]
desc_avg3 = data_avg3[:, 3: 5]
geno_avg3 = data_avg3[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_avg3 = []
archive = {}
for j in range(0, fit_avg3.shape[0]):
archive[tuple(cent_avg3[j])] = [geno_avg3[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_avg3.shape[0]):
try:
test = kdt.query([np.array([desc_avg3[i][0], desc_avg3[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten()
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten()
dist = distance.hamming(flat1,flat2)
distances.append(dist)
except:
pass
spread_avg3 = sum(distances)/len(distances)
print("Spread avg3 : {}".format(spread_avg3))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_avg7.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_avg7 = []
spread_avg7 = []
centroids_avg7 = load_centroids("centroids_avg7.dat")
data_avg7 = np.loadtxt("archive_avg7.dat")
fit_avg7 = data_avg7[:,0:1]
cent_avg7 = data_avg7[:, 1:3]
desc_avg7 = data_avg7[:, 3: 5]
geno_avg7 = data_avg7[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_avg7 = []
archive = {}
for j in range(0, fit_avg7.shape[0]):
archive[tuple(cent_avg7[j])] = [geno_avg7[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_avg7.shape[0]):
try:
test = kdt.query([np.array([desc_avg7[i][0], desc_avg7[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten()
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten()
dist = distance.hamming(flat1,flat2)
distances.append(dist)
except:
pass
spread_avg7 = sum(distances)/len(distances)
print("Spread avg7 : {}".format(spread_avg7))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_myversion3.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_myversion3 = []
spread_myversion3 = []
centroids_myversion3 = load_centroids("centroids_myversion3.dat")
data_myversion3 = np.loadtxt("archive_myversion3.dat")
fit_myversion3 = data_myversion3[:,0:1]
cent_myversion3 = data_myversion3[:, 1:3]
desc_myversion3 = data_myversion3[:, 3: 5]
geno_myversion3 = data_myversion3[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_myversion3 = []
archive = {}
for j in range(0, fit_myversion3.shape[0]):
archive[tuple(cent_myversion3[j])] = [geno_myversion3[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_myversion3.shape[0]):
try:
test = kdt.query([np.array([desc_myversion3[i][0], desc_myversion3[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten()
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten()
dist = distance.hamming(flat1,flat2)
distances.append(dist)
except:
pass
spread_myversion3 = sum(distances)/len(distances)
print("Spread myversion3 : {}".format(spread_myversion3))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_myversion7.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_myversion7 = []
spread_myversion7 = []
centroids_myversion7 = load_centroids("centroids_myversion7.dat")
data_myversion7 = np.loadtxt("archive_myversion7.dat")
fit_myversion7 = data_myversion7[:,0:1]
cent_myversion7 = data_myversion7[:, 1:3]
desc_myversion7 = data_myversion7[:, 3: 5]
geno_myversion7 = data_myversion7[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_myversion7 = []
archive = {}
for j in range(0, fit_myversion7.shape[0]):
archive[tuple(cent_myversion7[j])] = [geno_myversion7[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_myversion7.shape[0]):
try:
test = kdt.query([np.array([desc_myversion7[i][0], desc_myversion7[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten()
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten()
dist = distance.hamming(flat1,flat2)
distances.append(dist)
except:
pass
spread_myversion7 = sum(distances)/len(distances)
print("Spread myversion7 : {}".format(spread_myversion7))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_standard.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_standard = []
spread_standard = []
centroids_standard = load_centroids("centroids_standard.dat")
data_standard = np.loadtxt("archive_standard.dat")
fit_standard = data_standard[:,0:1]
cent_standard = data_standard[:, 1:3]
desc_standard = data_standard[:, 3: 5]
geno_standard = data_standard[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_standard = []
archive = {}
for j in range(0, fit_standard.shape[0]):
archive[tuple(cent_standard[j])] = [geno_standard[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_standard.shape[0]):
try:
test = kdt.query([np.array([desc_standard[i][0], desc_standard[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten()
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten()
dist = distance.hamming(flat1,flat2)
distances.append(dist)
except:
pass
spread_standard = sum(distances)/len(distances)
print("Spread standard : {}".format(spread_standard))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_standard.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_standard = []
spread_standard = []
t1 = np.zeros(10000)
t2 = np.ones(10000)
max_d = np.linalg.norm(t1 - t2)
print(max_d)
centroids_standard = load_centroids("centroids_standard.dat")
data_standard = np.loadtxt("archive_standard.dat")
fit_standard = data_standard[:,0:1]
cent_standard = data_standard[:, 1:3]
desc_standard = data_standard[:, 3: 5]
geno_standard = data_standard[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_standard = []
archive = {}
for j in range(0, fit_standard.shape[0]):
archive[tuple(cent_standard[j])] = [geno_standard[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_standard.shape[0]):
try:
test = kdt.query([np.array([desc_standard[i][0], desc_standard[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten() / 255
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten() / 255
dist = np.linalg.norm(flat1 - flat2)
distances.append(dist / max_d)
except:
pass
spread_standard = sum(distances)/len(distances)
print("Spread standard Euclidean : {}".format(spread_standard))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_avg3.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_avg3 = []
spread_avg3 = []
t1 = np.zeros(10000)
t2 = np.ones(10000)
max_d = np.linalg.norm(t1 - t2)
print(max_d)
centroids_avg3 = load_centroids("centroids_avg3.dat")
data_avg3 = np.loadtxt("archive_avg3.dat")
fit_avg3 = data_avg3[:,0:1]
cent_avg3 = data_avg3[:, 1:3]
desc_avg3 = data_avg3[:, 3: 5]
geno_avg3 = data_avg3[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_avg3 = []
archive = {}
for j in range(0, fit_avg3.shape[0]):
archive[tuple(cent_avg3[j])] = [geno_avg3[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_avg3.shape[0]):
try:
test = kdt.query([np.array([desc_avg3[i][0], desc_avg3[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten() / 255
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten() / 255
dist = np.linalg.norm(flat1 - flat2)
distances.append(dist / max_d)
except:
pass
spread_avg3 = sum(distances)/len(distances)
print("Spread avg3 Euclidean : {}".format(spread_avg3))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_avg7.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_avg7 = []
spread_avg7 = []
t1 = np.zeros(10000)
t2 = np.ones(10000)
max_d = np.linalg.norm(t1 - t2)
print(max_d)
centroids_avg7 = load_centroids("centroids_avg7.dat")
data_avg7 = np.loadtxt("archive_avg7.dat")
fit_avg7 = data_avg7[:,0:1]
cent_avg7 = data_avg7[:, 1:3]
desc_avg7 = data_avg7[:, 3: 5]
geno_avg7 = data_avg7[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_avg7 = []
archive = {}
for j in range(0, fit_avg7.shape[0]):
archive[tuple(cent_avg7[j])] = [geno_avg7[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_avg7.shape[0]):
try:
test = kdt.query([np.array([desc_avg7[i][0], desc_avg7[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten() / 255
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten() / 255
dist = np.linalg.norm(flat1 - flat2)
distances.append(dist / max_d)
except:
pass
spread_avg7 = sum(distances)/len(distances)
print("Spread avg7 Euclidean : {}".format(spread_avg7))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_myversion3.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_myversion3 = []
spread_myversion3 = []
t1 = np.zeros(10000)
t2 = np.ones(10000)
max_d = np.linalg.norm(t1 - t2)
print(max_d)
centroids_myversion3 = load_centroids("centroids_myversion3.dat")
data_myversion3 = np.loadtxt("archive_myversion3.dat")
fit_myversion3 = data_myversion3[:,0:1]
cent_myversion3 = data_myversion3[:, 1:3]
desc_myversion3 = data_myversion3[:, 3: 5]
geno_myversion3 = data_myversion3[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_myversion3 = []
archive = {}
for j in range(0, fit_myversion3.shape[0]):
archive[tuple(cent_myversion3[j])] = [geno_myversion3[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_myversion3.shape[0]):
try:
test = kdt.query([np.array([desc_myversion3[i][0], desc_myversion3[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten() / 255
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten() / 255
dist = np.linalg.norm(flat1 - flat2)
distances.append(dist / max_d)
except:
pass
spread_myversion3 = sum(distances)/len(distances)
print("Spread myversion3 Euclidean : {}".format(spread_myversion3))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_myversion7.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_myversion7 = []
spread_myversion7 = []
t1 = np.zeros(10000)
t2 = np.ones(10000)
max_d = np.linalg.norm(t1 - t2)
print(max_d)
centroids_myversion7 = load_centroids("centroids_myversion7.dat")
data_myversion7 = np.loadtxt("archive_myversion7.dat")
fit_myversion7 = data_myversion7[:,0:1]
cent_myversion7 = data_myversion7[:, 1:3]
desc_myversion7 = data_myversion7[:, 3: 5]
geno_myversion7 = data_myversion7[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_myversion7 = []
archive = {}
for j in range(0, fit_myversion7.shape[0]):
archive[tuple(cent_myversion7[j])] = [geno_myversion7[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_myversion7.shape[0]):
try:
test = kdt.query([np.array([desc_myversion7[i][0], desc_myversion7[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten() / 255
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten() / 255
dist = np.linalg.norm(flat1 - flat2)
distances.append(dist / max_d)
except:
pass
spread_myversion7 = sum(distances)/len(distances)
print("Spread myversion7 Euclidean : {}".format(spread_myversion7))
```
|
github_jupyter
|
import cairo
import cv2
from math import pi
import random
import math
from PIL import Image
import numpy as np
from scipy.spatial import distance
import matplotlib.pyplot as plt
from sklearn.neighbors import KDTree
def polar_to_cart(theta, dist):
x = 1 + dist * math.cos(theta)
y = 1 + dist * math.sin(theta)
return x,y
t = math.pi/180.0
def remap(old_val, old_min, old_max, new_min, new_max):
return (new_max - new_min)*(old_val - old_min) / (old_max - old_min) + new_min
def make_hashable(array):
return tuple(map(float, array))
def draw(geno):
surface = cairo.ImageSurface(cairo.FORMAT_ARGB32, 100, 100)
ctx = cairo.Context(surface)
#ctx.set_antialias(cairo.ANTIALIAS_NONE)
ctx.scale(50, 50)
# Paint the background
ctx.set_source_rgb(0, 0 , 0)
ctx.paint()
r1 = remap(geno[8], 0, 1,0.1, 1)
r2 = remap(geno[9], 0, 1,0.1, 1)
r3 = remap(geno[10], 0, 1,0.1, 1)
r4 = remap(geno[11], 0, 1,0.1, 1)
r5 = remap(geno[12], 0, 1,0.1, 1)
r6 = remap(geno[13], 0, 1,0.1, 1)
r7 = remap(geno[14], 0, 1,0.1, 1)
r8 = remap(geno[15], 0, 1,0.1, 1)
# Draw the image
firstx, firsty = polar_to_cart((0 + geno[0])*45*t, r1)
secondx, secondy = polar_to_cart((1 + geno[1])*45*t, r2)
thirdx, thirdy = polar_to_cart((2 + geno[2])*45*t, r3)
forthx, forthy = polar_to_cart((3 + geno[3])*45*t, r4)
fifthx, fifthy = polar_to_cart((4 + geno[4])*45*t, r5)
sixthx, sixthy = polar_to_cart((5 + geno[5])*45*t, r6)
seventhx, seventhy = polar_to_cart((6 + geno[6])*45*t, r7)
eigthx, eigthy = polar_to_cart((7 + geno[7])*45*t, r8)
ctx.move_to(firstx, firsty)
ctx.line_to(secondx, secondy)
ctx.line_to(thirdx, thirdy)
ctx.line_to(forthx, forthy)
ctx.line_to(fifthx, fifthy)
ctx.line_to(sixthx, sixthy)
ctx.line_to(seventhx, seventhy)
ctx.line_to(eigthx, eigthy)
ctx.close_path()
ctx.set_source_rgb(1, 1, 1)
ctx.fill_preserve()
return surface
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_avg3.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_avg3 = []
spread_avg3 = []
centroids_avg3 = load_centroids("centroids_avg3.dat")
data_avg3 = np.loadtxt("archive_avg3.dat")
fit_avg3 = data_avg3[:,0:1]
cent_avg3 = data_avg3[:, 1:3]
desc_avg3 = data_avg3[:, 3: 5]
geno_avg3 = data_avg3[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_avg3 = []
archive = {}
for j in range(0, fit_avg3.shape[0]):
archive[tuple(cent_avg3[j])] = [geno_avg3[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_avg3.shape[0]):
try:
test = kdt.query([np.array([desc_avg3[i][0], desc_avg3[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten()
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten()
dist = distance.hamming(flat1,flat2)
distances.append(dist)
except:
pass
spread_avg3 = sum(distances)/len(distances)
print("Spread avg3 : {}".format(spread_avg3))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_avg7.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_avg7 = []
spread_avg7 = []
centroids_avg7 = load_centroids("centroids_avg7.dat")
data_avg7 = np.loadtxt("archive_avg7.dat")
fit_avg7 = data_avg7[:,0:1]
cent_avg7 = data_avg7[:, 1:3]
desc_avg7 = data_avg7[:, 3: 5]
geno_avg7 = data_avg7[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_avg7 = []
archive = {}
for j in range(0, fit_avg7.shape[0]):
archive[tuple(cent_avg7[j])] = [geno_avg7[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_avg7.shape[0]):
try:
test = kdt.query([np.array([desc_avg7[i][0], desc_avg7[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten()
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten()
dist = distance.hamming(flat1,flat2)
distances.append(dist)
except:
pass
spread_avg7 = sum(distances)/len(distances)
print("Spread avg7 : {}".format(spread_avg7))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_myversion3.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_myversion3 = []
spread_myversion3 = []
centroids_myversion3 = load_centroids("centroids_myversion3.dat")
data_myversion3 = np.loadtxt("archive_myversion3.dat")
fit_myversion3 = data_myversion3[:,0:1]
cent_myversion3 = data_myversion3[:, 1:3]
desc_myversion3 = data_myversion3[:, 3: 5]
geno_myversion3 = data_myversion3[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_myversion3 = []
archive = {}
for j in range(0, fit_myversion3.shape[0]):
archive[tuple(cent_myversion3[j])] = [geno_myversion3[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_myversion3.shape[0]):
try:
test = kdt.query([np.array([desc_myversion3[i][0], desc_myversion3[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten()
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten()
dist = distance.hamming(flat1,flat2)
distances.append(dist)
except:
pass
spread_myversion3 = sum(distances)/len(distances)
print("Spread myversion3 : {}".format(spread_myversion3))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_myversion7.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_myversion7 = []
spread_myversion7 = []
centroids_myversion7 = load_centroids("centroids_myversion7.dat")
data_myversion7 = np.loadtxt("archive_myversion7.dat")
fit_myversion7 = data_myversion7[:,0:1]
cent_myversion7 = data_myversion7[:, 1:3]
desc_myversion7 = data_myversion7[:, 3: 5]
geno_myversion7 = data_myversion7[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_myversion7 = []
archive = {}
for j in range(0, fit_myversion7.shape[0]):
archive[tuple(cent_myversion7[j])] = [geno_myversion7[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_myversion7.shape[0]):
try:
test = kdt.query([np.array([desc_myversion7[i][0], desc_myversion7[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten()
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten()
dist = distance.hamming(flat1,flat2)
distances.append(dist)
except:
pass
spread_myversion7 = sum(distances)/len(distances)
print("Spread myversion7 : {}".format(spread_myversion7))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_standard.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_standard = []
spread_standard = []
centroids_standard = load_centroids("centroids_standard.dat")
data_standard = np.loadtxt("archive_standard.dat")
fit_standard = data_standard[:,0:1]
cent_standard = data_standard[:, 1:3]
desc_standard = data_standard[:, 3: 5]
geno_standard = data_standard[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_standard = []
archive = {}
for j in range(0, fit_standard.shape[0]):
archive[tuple(cent_standard[j])] = [geno_standard[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_standard.shape[0]):
try:
test = kdt.query([np.array([desc_standard[i][0], desc_standard[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten()
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten()
dist = distance.hamming(flat1,flat2)
distances.append(dist)
except:
pass
spread_standard = sum(distances)/len(distances)
print("Spread standard : {}".format(spread_standard))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_standard.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_standard = []
spread_standard = []
t1 = np.zeros(10000)
t2 = np.ones(10000)
max_d = np.linalg.norm(t1 - t2)
print(max_d)
centroids_standard = load_centroids("centroids_standard.dat")
data_standard = np.loadtxt("archive_standard.dat")
fit_standard = data_standard[:,0:1]
cent_standard = data_standard[:, 1:3]
desc_standard = data_standard[:, 3: 5]
geno_standard = data_standard[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_standard = []
archive = {}
for j in range(0, fit_standard.shape[0]):
archive[tuple(cent_standard[j])] = [geno_standard[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_standard.shape[0]):
try:
test = kdt.query([np.array([desc_standard[i][0], desc_standard[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten() / 255
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten() / 255
dist = np.linalg.norm(flat1 - flat2)
distances.append(dist / max_d)
except:
pass
spread_standard = sum(distances)/len(distances)
print("Spread standard Euclidean : {}".format(spread_standard))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_avg3.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_avg3 = []
spread_avg3 = []
t1 = np.zeros(10000)
t2 = np.ones(10000)
max_d = np.linalg.norm(t1 - t2)
print(max_d)
centroids_avg3 = load_centroids("centroids_avg3.dat")
data_avg3 = np.loadtxt("archive_avg3.dat")
fit_avg3 = data_avg3[:,0:1]
cent_avg3 = data_avg3[:, 1:3]
desc_avg3 = data_avg3[:, 3: 5]
geno_avg3 = data_avg3[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_avg3 = []
archive = {}
for j in range(0, fit_avg3.shape[0]):
archive[tuple(cent_avg3[j])] = [geno_avg3[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_avg3.shape[0]):
try:
test = kdt.query([np.array([desc_avg3[i][0], desc_avg3[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten() / 255
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten() / 255
dist = np.linalg.norm(flat1 - flat2)
distances.append(dist / max_d)
except:
pass
spread_avg3 = sum(distances)/len(distances)
print("Spread avg3 Euclidean : {}".format(spread_avg3))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_avg7.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_avg7 = []
spread_avg7 = []
t1 = np.zeros(10000)
t2 = np.ones(10000)
max_d = np.linalg.norm(t1 - t2)
print(max_d)
centroids_avg7 = load_centroids("centroids_avg7.dat")
data_avg7 = np.loadtxt("archive_avg7.dat")
fit_avg7 = data_avg7[:,0:1]
cent_avg7 = data_avg7[:, 1:3]
desc_avg7 = data_avg7[:, 3: 5]
geno_avg7 = data_avg7[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_avg7 = []
archive = {}
for j in range(0, fit_avg7.shape[0]):
archive[tuple(cent_avg7[j])] = [geno_avg7[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_avg7.shape[0]):
try:
test = kdt.query([np.array([desc_avg7[i][0], desc_avg7[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten() / 255
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten() / 255
dist = np.linalg.norm(flat1 - flat2)
distances.append(dist / max_d)
except:
pass
spread_avg7 = sum(distances)/len(distances)
print("Spread avg7 Euclidean : {}".format(spread_avg7))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_myversion3.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_myversion3 = []
spread_myversion3 = []
t1 = np.zeros(10000)
t2 = np.ones(10000)
max_d = np.linalg.norm(t1 - t2)
print(max_d)
centroids_myversion3 = load_centroids("centroids_myversion3.dat")
data_myversion3 = np.loadtxt("archive_myversion3.dat")
fit_myversion3 = data_myversion3[:,0:1]
cent_myversion3 = data_myversion3[:, 1:3]
desc_myversion3 = data_myversion3[:, 3: 5]
geno_myversion3 = data_myversion3[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_myversion3 = []
archive = {}
for j in range(0, fit_myversion3.shape[0]):
archive[tuple(cent_myversion3[j])] = [geno_myversion3[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_myversion3.shape[0]):
try:
test = kdt.query([np.array([desc_myversion3[i][0], desc_myversion3[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten() / 255
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten() / 255
dist = np.linalg.norm(flat1 - flat2)
distances.append(dist / max_d)
except:
pass
spread_myversion3 = sum(distances)/len(distances)
print("Spread myversion3 Euclidean : {}".format(spread_myversion3))
def load_data(filename, dim,dim_x):
print("Loading ",filename)
data = np.loadtxt(filename)
fit = data[:, 0:1]
cent = data[:,1: dim+1]
desc = data[:,dim+1: 2*dim+1]
x = data[:,2*dim+1:2*dim+1+dim_x]
def load_centroids(filename):
points = np.loadtxt(filename)
return points
def getKDT(n_niches, dim_map):
fname = "centroids_myversion7.dat"
c = np.loadtxt(fname)
kdt = KDTree(c, leaf_size=30, metric='euclidean')
return kdt
diversity_myversion7 = []
spread_myversion7 = []
t1 = np.zeros(10000)
t2 = np.ones(10000)
max_d = np.linalg.norm(t1 - t2)
print(max_d)
centroids_myversion7 = load_centroids("centroids_myversion7.dat")
data_myversion7 = np.loadtxt("archive_myversion7.dat")
fit_myversion7 = data_myversion7[:,0:1]
cent_myversion7 = data_myversion7[:, 1:3]
desc_myversion7 = data_myversion7[:, 3: 5]
geno_myversion7 = data_myversion7[:, 5: 21]
#print("Fit: {}".format(fit[1]))
#print("Cent: {}".format(cent[1]))
#print("Behavior: {}".format(desc[1]))
#print("Geno: {}".format(geno[1]))
#Spread
spread_myversion7 = []
archive = {}
for j in range(0, fit_myversion7.shape[0]):
archive[tuple(cent_myversion7[j])] = [geno_myversion7[j]]
kdt = getKDT(1000, 2)
distances = []
for i in range(0, fit_myversion7.shape[0]):
try:
test = kdt.query([np.array([desc_myversion7[i][0], desc_myversion7[i][1]])], k=2)[1][0]
niche_1= kdt.data[test[0]]
niche_2= kdt.data[test[1]]
n1 = make_hashable(niche_1)
n2 = make_hashable(niche_2)
uno = np.array(archive[n1][0])
due = np.array(archive[n2][0])
img1 = draw(uno)
imgP1 = Image.frombuffer("RGBA",( img1.get_width(),img1.get_height() ),img1.get_data(),"raw","RGBA",0,1)
img_arr1 = np.array(imgP1)
flat1 = img_arr1[:,:,0].flatten() / 255
img2 = draw(due)
imgP2 = Image.frombuffer("RGBA",( img2.get_width(),img2.get_height() ),img2.get_data(),"raw","RGBA",0,1)
img_arr2 = np.array(imgP2)
flat2 = img_arr2[:,:,0].flatten() / 255
dist = np.linalg.norm(flat1 - flat2)
distances.append(dist / max_d)
except:
pass
spread_myversion7 = sum(distances)/len(distances)
print("Spread myversion7 Euclidean : {}".format(spread_myversion7))
| 0.356335 | 0.47317 |
# SciPy
Heavily depends on the following libraries:
1. matplotlib
2. numpy
"SciPy is organized into subpackages covering different scientific computing domains."
**Subpackages**
1. cluster: Clustering algorithms
2. constants: Physical and mathematical constants
3. fftpack: Fast Fourier Transform routines
4. **integrate**: Integration and ordinary differential equation solvers
5. **interpolate**: Interpolation and smoothing splines
6. io: Input and Output
7. linalg: Linear algebra
8. ndimage: N-dimensional image processing
9. odr: Orthogonal distance regression
10. **optimize**: Optimization and root-finding routines
11. signal: Signal processing
12. sparse: Sparse matrices and associated routines
13. spatial: Spatial data structures and algorithms
14. special: Special functions
15. stats: Statistical distributions and functions
One fo the strengths of SciPy is that it can provide **numerical solutions** (i.e. approximated). The opposite of numerical solutions are **analytic solutions** (i.e. exact; `f(2) = x^2 = 4`).
Sources:
https://docs.scipy.org/doc/scipy/reference/
https://docs.scipy.org/doc/
https://docs.scipy.org/doc/scipy/reference/tutorial/general.html
https://scipy-lectures.org/intro/scipy.html
```
#help(scipy)
```
---
## Integration
Let's start with integration.
What can integration do for us? For something that is defined by a mathematical function (i.e. equation), we can obtain the following:
1. areas (2D) (e.g. the area between two curves that cross each other),
2. volumes (3D),
3. surface area (e.g. of a protein)
3. displacements (i.e. distance) (w.r.t. time)
4. center (e.g. of mass)
5. probability
- integrate https://docs.scipy.org/doc/scipy/reference/tutorial/integrate.html
Graphical when we integrate a function f(x), we obtain the "area under the curve."
<img src="00_images/integral_example.png" alt="integral" style="width: 200px;"/>
<center>Figure 1: Depiction that shows the "area under the curve" determined through integration of function `f(x)` with limits from `a` to `b`.</center>
It is kinda like doing addition, but for something that is continuous (i.e. not finite).
Image Source: https://en.wikipedia.org/wiki/Integral#/media/File:Integral_example.svg
```
import matplotlib.pyplot as plt
import numpy as np
import scipy
from scipy.integrate import quad
scipy.__version__
```
Let's define a simple function:
$$\int_0^1 (mx^2 + n) dx$$
I'm going to stick with variable names that match the equation given above for consistency. We will focus on the equation within the integration. (Recall, that Sympy can also do what we do below).
```
def simple_function(x: float=None, m: float=None, n: float=None):
return m*x**2 + n
```
---
**Sidenote**: Numpy's linspace vs arrange:
- linspace (i.e. `numpy.linspace(start, stop, num`): "Return evenly spaced numbers over a specified interval."
- https://numpy.org/devdocs/reference/generated/numpy.linspace.html
- the stepsize is created
- the number of steps must be given
Versus
- arange (i.e. `numpy.arange(start, stop, step)`: "Return evenly spaced values within a given interval."
- https://numpy.org/doc/stable/reference/generated/numpy.arange.html
- the stepsize is specified
- the number of steps is created
---
Let's generate the starting data:
```
m = 3
n = 5
x_data = np.linspace(-1, 2, 20)
```
We can plot the curve:
- x range = -1 to 2 (i.e. integration limits $\pm 1$), and then
- visualize the area between the integration limits
```
plt.figure()
plt.plot(x_data, simple_function(x_data, m, n), color='orange', linewidth=5)
plt.hlines(y=0.0, xmin=0.0, xmax=1.0, linewidth=5)
plt.hlines(y=5.0, xmin=0.0, xmax=1.0, linewidth=5, linestyle='dashed')
plt.vlines(x=0.0, ymin=0.0, ymax=5.0, linewidth=5)
plt.vlines(x=1.0, ymin=0.0, ymax=7.85, linewidth=5)
plt.show()
```
We can approximate the area under the orange curve and within the blue region to be:
rectangle + ca. triangle
`1*5 + [(1*1.5)/2] = 5 + 0.75 = 5.75`
Okay, Good. Now let's integrate that function.
`quad`: general purpose single integration a function containing
- one variable (e.g. x), and
- evaluated between two points (e.g. 0 to 1)
https://docs.scipy.org/doc/scipy/reference/generated/scipy.integrate.quad.html#scipy.integrate.quad
<br><br>
`quad(func, a, b, args=() ...)`:
where
- `func` = **simple_function** (i.e. a “callable” Python object)
- `a` (lower integration limit) = **0**
- `b` (upper inegration limit) = **1**
- `args` (additional arguments to pass) = **(3, 5)**
(i.e. quad(function, lower limit, upper limit, what to pass to our simple_function)
The **return value** is a tuple:
- 1$^{st}$: the **estimated value of the integral**, and
- 2$^{nd}$: an **upper bound on the error**
```
result = quad(func=simple_function, a=0, b=1, args=(m, n))
result
```
###### Accessing value and error (plus, remembering string formatting):
- f: Fixed-point notation. Displays the number as a fixed-point number. The default precision is 6.
- e: Exponent notation. Prints the number in scientific notation using the letter ‘e’ to indicate the exponent. The default precision is 6.
(Rounding for simplicity of reading, not due to accuracy.)
```
print('Full answer: {:0.2f} ± {:0.2e}'.format(result[0], result[1]))
```
---
## A more complicated example
1. Handeling infinity limits (i.e. indefinite integrals)
2. Python's built in function `eval` (evaluate)
- https://docs.python.org/3/library/functions.html#eval
Let's first look at each piece, and then we will put it together.
`eval` works on single functions (note the use of quotes here):
```
number = 2
eval('number**2')
```
The `eval` function also works on np.arrays
Example function will be the following:
$$\frac{1}{x^2}$$
First create some x-data:
```
x_data_array = np.linspace(1, 11, 30)
x_data_array
```
Now evaluate the function at those points (i.e. determine the y-values):
```
y_data_array = eval('1/(x_data_array**2)')
y_data_array
```
Let's plot this to visualize the data:
```
plt.plot()
plt.plot(x_data_array, y_data_array, linewidth=5, color='orange')
plt.hlines(y=0.0, xmin=1.0, xmax=9.0, linestyle='dashed', linewidth=3)
plt.vlines(x=1.0, ymin=0.0, ymax=0.9, linestyle='dashed', linewidth=3)
plt.show()
```
Imagine this plot going to **infinity** on the **x-axis**.
What is the area from x=1 to x=infinity?
Hard to say right?
---
Okay, let's create a callable function that we will pass to SciPy's `quad` function for integration:
```
def function(x: float=None):
return 1/x**2
```
Let's focus now upon an "improper" integral (i.e. the upper integration limit is infinity.
$$\int_1^{\infty} \frac{1}{x^2} dx$$
```
result = quad(func=function, a=1, b=np.inf)
result
```
Therefore, the area under the $\frac{1}{x^2}$ curve from x=1 to infinity is 1.0.
(What is the area under the curve from x=2 to infinity?)
**Note**: if we try to do this all in one step where we provide the function directly, we get an error. That is the practical reason why one must create a function for quad to call.
```
#result = quad(1/x**2, 1, np.inf)
```
---
## Interpolation
- A method for **generating new data** using a discrete set of **known data** points.
- Good for filling in some missing data points within a **continuous** data set
- https://docs.scipy.org/doc/scipy/reference/interpolate.html
---
### A simple example
First things to do is create a **hypothetical set of known** x- and y-data points
```
x_data_array = np.arange(0, 10, 1)
x_data_array
```
Create a corresponding range of y values
- exponential via `np.exp()`: https://numpy.org/doc/stable/reference/generated/numpy.exp.html
```
y_data_array = np.exp(-x_data_array/3.0)
y_data_array
```
Now plot to visualize what the data looks like, and highlight the third data point in the series (i.e. **(x,y) = (2, 0.51341712))** as an ideal value to reference later.
```
plt.plot()
plt.plot(x_data_array, y_data_array, linestyle='solid', linewidth=5, marker='o', markersize=15)
plt.hlines(y=y_data_array[2], xmin=0, xmax=9, colors='#1f77b4', linewidth=5)
plt.show()
```
#### Create an interprelated function from the existing data points
1-dimensional function
- interp1d: https://docs.scipy.org/doc/scipy/reference/generated/scipy.interpolate.interp1d.html#scipy.interpolate.interp1d
```
from scipy.interpolate import interp1d
interp_function = interp1d(x_data_array, y_data_array)
```
First let's see if we can reproduce a **known** data point (i.e. a simple idea)
- x = 2.0 should give a value of 0.51341712 (see above hypothetical data set)
```
interp_function(2.0)
plt.figure()
plt.plot(x_data_array, y_data_array, 'o', markersize=15)
plt.hlines(y=0.51341712, xmin=0, xmax=9, colors='#1f77b4', linewidth=5)
plt.hlines(y=interp_function(2.0), xmin=0, xmax=9, colors='#ff7f0e', linestyles='dashed', linewidth=5)
plt.show()
```
We can also do this for lots of new x-values that fit between 0 and 9 (i.e. interpolated data).
First, we need to create a new range of x values that we want to fill in -- for example, from 1 to 7 in 0.2 increments (step size):
```
x_values_new = np.arange(1, 8.2, 0.2)
print(x_values_new)
```
Now, using the user-defined function that imploys `interp1d`, solve for the interpolated y-values:
```
y_values_new = interp_function(x_values_new)
y_values_new
print(y_values_new)
plt.figure()
plt.plot(x_data_array, y_data_array, marker='o', markersize=15)
plt.hlines(y=0.51341712, xmin=0, xmax=9, colors='#1f77b4', linewidth=5)
plt.plot(x_values_new, y_values_new, marker='o', markersize=5)
plt.hlines(y=interp_function(2.0), xmin=0, xmax=9, colors='#ff7f0e', linestyles='dashed', linewidth=5)
plt.show()
```
We see that the interpolated **new data** points (orange) fall nicely onto the known data.
### A more complicated (and practical) example
```
x_values = np.linspace(0, 1, 10)
x_values
```
##### Create some noise that will allow us to better mimic what real data looks like
"Noise" refers to how much the **real** data varies from (hypothetical) **ideal** data. Understanding the noise in data is understanding the data's stability (e.g. reproducibility, predictable). Noise is often coming from unaccounted sources (and represent possible areas to learn from).
**Side Note**: The following **np.random.seed()** statement will allow us to reproduce the random number generation (e.g. allows for reproducibility in examples). This isn't necessary here, but it is nice to know about.
- `np.random.random(n)`: https://numpy.org/doc/stable/reference/random/generated/numpy.random.random.html
- create n random numbers that **range from 0 to 1**
```
np.random.seed(30)
np.random.random(10)
```
Now let's create the noise by adding in some math to the random values:
```
noise = (np.random.random(10)**2 - 1) * 2e-1
print(noise)
```
Now generate some two types of **y-data** that is a **function of the x-axis data**:
1. ideal y data
- perfect data that arrises from an equation
2. ideal y data with noise
- we will call this **simulated real data**, which suggest that it was obtained using **experiments**
#### 1. ideal data
```
y_values_ideal = np.sin(2 * np.pi * x_values)
y_values_ideal
```
##### 2. ideal data with noise (i.e. simulated "real" data)
```
y_values_sim = np.sin(2 * np.pi * x_values) + noise
y_values_sim
```
Plot the "idea" (blue) and "simulated real" (orange) data, and highlight the 6$^{th}$ data point:
```
plt.figure()
plt.plot(x_values, y_values_ideal, marker='o', color='#1f77b4', markersize=7, linewidth=2, alpha=0.5) #ideal, blue
plt.plot(x_values, y_values_sim, marker='o', color='#ff7f0e', markersize=15, linewidth=5) #simulated, orange
plt.hlines(y=y_values_sim[5], xmin=0, xmax=1, colors='#ff7f0e', linewidth=3)
plt.show()
```
Create a **new function** that is an **interpolation** of the existing (i.e. known, but non-ideal) data points
```
interp_function = interp1d(x_values, y_values_sim)
```
First let's see if we can reproduce an "known" point
- We want to reproduce the sixth data point: x_value[5]
- interp_function(x_value[5]) should give y_value[5] of the original function
<br>
**Simulated** (i.e. ideal+noise) **y-value** at the 6$^{th}$ data point:
```
y_values_sim[5]
```
Now for the **interpolated y-value** at the 6$^{th}$ data point:
```
interp_function(x_values[5])
```
Quantify the difference between the interpolated and true value for the 6$^{th}$ data point:
```
interp_function(x_values[5]) - y_values_sim[5]
```
Let's also fill in some of the space between the data points by creating a new range of x-data:
```
x_data_new = np.arange(0, 1, 0.02)
x_data_new
y_data_new = interp_function(x_data_new)
y_data_new
```
Create and overlay plot that shows
1. ideal values,
2. simulated values (i.e. idea+noise),
3. interpolated values (shown in green)
```
plt.figure()
plt.plot(x_values, y_values_ideal, marker='o', color='#1f77b4', markersize=7, linewidth=2, alpha=0.5)
plt.plot(x_values, y_values_sim, marker='o', color='#ff7f0e', markersize=15, linewidth=5)
plt.hlines(y=y_values_sim[5], xmin=0, xmax=1, colors='#ff7f0e', linewidth=5)
## plot the interpolated curve (green)
plt.plot(x_data_new, y_data_new, marker='o', color='#2ca02c', markersize=10, linewidth=2, alpha=0.5)
plt.hlines(y=interp_function(x_values[5]), xmin=0, xmax=1, colors='#2ca02c',
linestyles='dashed', linewidth=2, alpha=0.5)
plt.show()
```
---
**side note**: Percent Relative Error
The **percent relative error** is often calculated in the natural sciences, whose formuala is the following:
$$\text{Percentage Relative Error} = \frac{\text{estimated}-\text{actual}}{\text{actual}}*100$$
What is the PRE between the interpolated vs. simulated (i.e. ideal+noise):
```
def percentage_rel_error(estimated: float=None, actual: float=None) -> float:
return ((estimated - actual)/actual)*100
pce = percentage_rel_error(estimated=(interp_function(x_values[5])), actual=y_values_sim[5])
print(f'{pce:.2}')
```
So the percentage relative error is 0%.
How about the interpolated versus ideal (i.e. noiseless)?
This shows how the addition of noise to the ideal data impacted our "modeling building":
```
pce = percentage_rel_error(estimated=(interp_function(x_values[5])), actual=y_values_ideal[5])
pce
```
So, the addition of noise significantly changed the ideal data, which is what we wanted.
**Final note**:
There is a relatively simple alternative to `interp1d` that is easy to use: `Akima1DInterpolator`
- https://docs.scipy.org/doc/scipy/reference/generated/scipy.interpolate.Akima1DInterpolator.html
---
## Curve Fitting
- curve_fit: https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.curve_fit.html
Curve fitting is the act of fitting a function to provided data point. The result is an optmized function that best models the data points.
**Example**:
We obtain data that follows a **sine wave**, but we don't know the amplitude or period that the data has. Thus, we need to **curve fit** the **data** to provide a the **amplitude and period**.
Recall some basic math
$$y = Asin(Bx + C) + D$$
<img src="00_images/Wave_sine.png" alt="sine" style="width: 600px;"/>
```
from scipy import optimize
```
Create 50 equally spaced data points from -5 to +5:
```
x_values = np.linspace(-5, 5, num=50)
x_values
```
Create some noise (for more realistic data):
```
noise = np.random.random(50)
noise
```
Create our y-target data (i.e. simulated experimental data) that follows a sine wave by adding some noise
Amplitude: 1.7
Period: 2π/2.5
C=D=0
```
y_values = 1.7*np.sin(2.5 * x_values) + noise
y_values
plt.figure()
plt.plot(x_values, y_values, '-o', markersize=15, linewidth=5)
plt.show()
```
Setup our simple test function that we can solve for the amplitude and period (i.e. a test function with two variables only: a and b).
(**Note**: I'm not including any internal test (e.g. isinstance, assert) in order to keep the teaching aspects clear here.)
```
def sine_func(x=None, a=None, b=None):
return a * np.sin(b * x)
```
Use SciPy's optimize.curve_fit to find the solutions
- https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.curve_fit.html?highlight=curve_fit
What is needed:
1. a function
2. x and y target data values, and
3. and initial guesses (i.e. `p0` below) - we need a total of two (i.e. one for `a` and one for `b`)
What is returned:
1. **solution values**
2. covariance: estimate of how much 2 variables vary (i.e. change) together (e.g. smoking and lifespan), or
- in other words, how correlated they are to one another (i.e. the off diagonal of the resulting matrix, which includes the concept of positive or negative correlation)
- the square of the diagonals of the covariance matrix gives the standard deviation for each of the solution values
Will use `p0=[2.0, 2.0]` as the initial guess:
```
solution, solution_covariance = optimize.curve_fit(sine_func, x_values, y_values, p0=[2.0, 2.0])
```
The ideal values are: amplitude (a) = 1.7, period (b) = 2.5 with C=D=0
But remember, we added noise, so our solution will be close to these values solution
```
solution
solution_covariance
std_dev = np.sqrt(np.diag(solution_covariance))
std_dev
plt.plot()
plt.plot(x_values, y_values, '-o', markersize=15, linewidth=5) # blue (simulated experimental date)
plt.plot(x_values, sine_func(x_values, solution[0], solution[1]),
'-o', markersize=15, linewidth=5, alpha=0.7) # orange
plt.show()
```
Note: The **solution** will **depend** on the **intial guess**. There are several possible "local" solutions that can can be found.
We **artifically knew** the solution before hand, to be near **a=1.7 and b=2.5**...so p0=[2.0, 2.0] was a good starting point.
Exploration is needed when we don't know the approximate (or exact) solution before. Visualization of the results helps you interpret them (i.e. build your understanding of what the results are).
Demonstrate by redoing the above steps, and plot the results using:
- p0=[1.0, 1.0] --> should give a different result
- p0=[3.0, 3.0] --> should give you the "correct" solution
- p0=[5.0, 5.0] --> should give a different result
Example: `solution, solution_covariance = optimize.curve_fit(sine_func, x_values, y_values, p0=[1.0, 1.0])`
---
## Optimization
Finding a numerical solution for maximizing or minimizing a function.
In other words, if we start with an arbitrary point on a function's curve or surface, an optimization algorithm will locate (i.e. optimize) the lowest energy value with respect to that starting position (see Figure 2).
<img src="00_images/Gradient_descent.gif" alt="gradient_opt" style="width: 400px;"/>
<center>Figure 2: Three starting points on a mathematically defined surface that are optimized to two local minima.</center>
(Image source: https://commons.wikimedia.org/wiki/File:Gradient_descent.gif)
- optimize: https://docs.scipy.org/doc/scipy/reference/optimize.html#module-scipy.optimize
- https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.minimize.html#scipy.optimize.minimize
##### scipy.optimize.minimize() and its output **type**
**Input**
func: a function that will be minimized
x0: an initial guess
**Output**
The output is a compound object containing lot of information regarding the convergence (see example below for what it looks like).
##### Solvers
- Nelder-Mead
- Powell
- CG
- **BFGS**
- Newton-CG
- L-BFGS-B
- TNC
- COBYLA
- SLSQP
- trust-constr
- dogleg
- trust-ncg
- trust-exact
- trust-krylov
- **Default solver**: quasi-Newton method of Broyden, Fletcher, Goldfarb, and Shanno (**BFGS**)
- https://docs.scipy.org/doc/scipy/reference/tutorial/optimize.html#broyden-fletcher-goldfarb-shanno-algorithm-method-bfgs
- More background on minimization: http://scipy-lectures.org/advanced/mathematical_optimization
---
**Example**: Find the minimum of a 1D function (i.e. a scalar function; a function that return a single value from input values)
$$ x^2 + 25sin(x) $$
```
def scalar_func(x: float=None) -> float:
return x**2 + 25*np.sin(x)
x_values = np.arange(-10, 10, 0.1)
y_values = scalar_func(x_values)
```
View what the x- and y-data look like:
```
plt.figure()
plt.plot(x_values, y_values, 'o')
plt.show()
```
Notice the **three significant minima** that are present (i.e. one **global**, and two **local**)
Use `optimize.minimier` to find a minimum
- https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.minimize.html#scipy.optimize.minimize
Let's start with an **inital guess** near the global minimum (i.e `x0 = 0.0`):
```
result_global = optimize.minimize(scalar_func, x0=0.0, method="BFGS")
result_global
type(result_global)
```
You can retrieve each of these items, as demonstrated by the following:
Position of the found **minimum** on the **x-axis**:
```
result_global.x
```
Value of the found **minimum** on the **y-axis**:
```
result_global.fun
```
Now let's set an **initial guess** closer to one of the **local minimum** (i.e. `x0 = 3.0`)
```
result_local = optimize.minimize(scalar_func, x0=3.0, method="BFGS")
result_local
```
Notice that it finds the local minimum at x=4.4 (i.e. NOT the global minimia). Thus, BFGS apears to be a **local optimizer**.
#### Overcoming the dependency on the initial guess (the idea of a global optimizer)
- fminbound: a minimization within boundaries
- brute: minimize a function over a given range through lots of sampling
- differential_evolution: global minimum a multivariate function
- shgo: global minimum using SHG optimization
- dual_annealing: global minimum using dual annealing
Let's try these out:
**fminbound** https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.fminbound.html
`scipy.optimize.fminbound(func, x1, x2, ...)`
- **no startign guess** is used as input
```
optimize.fminbound(func=scalar_func, x1=-10, x2=10)
```
Therefore, `fminbound` finds the global minimum.
**brute force** https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.brute.html#scipy.optimize.brute
`scipy.optimize.brute(func, ranges,...)`
- ranges (tuple): "Each component of the ranges **tuple** must be either a **“slice object”** or a **range tuple of the form (low, high)**. The program uses these to create the grid of points on which the objective function will be computed."
Built-in function: `slice(start, stop[, step])`
Since we have only one variable (i.e. `x`), we only need to "fill-in" the first part of the tuple (e.g. `(slice(-10, 10, 1), )`:
Slice object:
```
optimize.brute(func=scalar_func, ranges=(slice(-10, 10, 0.1), ))
```
Range of tuple (low, high):
```
optimize.brute(func=scalar_func, ranges=((-10, 10), ))
```
**basin hopping** https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.basinhopping.html
`scipy.optimize.basinhopping(func, x0, niter=100, T=1.0, stepsize=0.5, ...)`
- combines global stepping with local minimization
- rugged, funnel-like surfaces (e.g. molecular potential energy surfaces)
- requires: a function and an initial guess (so not a "perfect" method)
- sensitive to stepsize
- stepsize=0.5 (i.e. default value) will find in a local minmium
- stepsize=2.5 will find the global mimimum
Recall that `x0 = 3.0` gave a **local minimum** at `x = 4.454335797238624` when using `optimize.minimize`.
```
optimize.basinhopping(func=scalar_func, x0=3.0, stepsize=0.5)
```
Basin hopping with the **small stepsize did not find the global minimum**.
Let's make the **stepsize larger**:
```
optimize.basinhopping(func=scalar_func, x0=3.0, stepsize=2.5)
```
Now Basin Hopping found the global minimum.
## Finding the **Roots**
- Roots: points where f(x) = 0
- For example, the values of x that satisfies the equation $ x^2 + 25sin(x) = 0$
- Finding the roots of a function provides you a solution to that function, which can be useful depending on the problem at hand.
```
plt.figure()
plt.plot(x_values, scalar_func(x_values), linestyle='solid', linewidth=5, alpha=0.5)
plt.hlines(y=0, xmin=-10, xmax=10, colors='red')
plt.show()
```
#### Through visualization, we see that there should be four roots (ca. -3.0, 0.0, 4.0 and 5.0)
`scipy.optimize.root(fun, x0, ...)`
- `x0`: initial starting point (i.e. guess)
- https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.root.html
```
root1 = optimize.root(fun=scalar_func, x0=-4)
root1
```
Therefore, one root is at x=-2.8.
```
root2 = optimize.root(fun=scalar_func, x0=1)
root2
```
A second root is at x=-0.0.
```
root3 = optimize.root(scalar_func, x0=4)
root3
```
A second root is at x=-3.7.
```
root4 = optimize.root(scalar_func, x0=7)
root4
```
A second root is at x=-4.9.
```
root4.x
my_x = root4.x[0]
my_x**2 + 25*np.sin(my_x)
```
---
### SciPy Summary:
1. Integration of a function
2. Interpolation of data points (e.g. filling in missing data)
3. Curve fitting - optimizing a function to best fit a data set
4. Optimization to find local and global minima positions and values of a function
5. Finding the roots of an equation (e.g. f(x)=0)
|
github_jupyter
|
#help(scipy)
import matplotlib.pyplot as plt
import numpy as np
import scipy
from scipy.integrate import quad
scipy.__version__
def simple_function(x: float=None, m: float=None, n: float=None):
return m*x**2 + n
m = 3
n = 5
x_data = np.linspace(-1, 2, 20)
plt.figure()
plt.plot(x_data, simple_function(x_data, m, n), color='orange', linewidth=5)
plt.hlines(y=0.0, xmin=0.0, xmax=1.0, linewidth=5)
plt.hlines(y=5.0, xmin=0.0, xmax=1.0, linewidth=5, linestyle='dashed')
plt.vlines(x=0.0, ymin=0.0, ymax=5.0, linewidth=5)
plt.vlines(x=1.0, ymin=0.0, ymax=7.85, linewidth=5)
plt.show()
result = quad(func=simple_function, a=0, b=1, args=(m, n))
result
print('Full answer: {:0.2f} ± {:0.2e}'.format(result[0], result[1]))
number = 2
eval('number**2')
x_data_array = np.linspace(1, 11, 30)
x_data_array
y_data_array = eval('1/(x_data_array**2)')
y_data_array
plt.plot()
plt.plot(x_data_array, y_data_array, linewidth=5, color='orange')
plt.hlines(y=0.0, xmin=1.0, xmax=9.0, linestyle='dashed', linewidth=3)
plt.vlines(x=1.0, ymin=0.0, ymax=0.9, linestyle='dashed', linewidth=3)
plt.show()
def function(x: float=None):
return 1/x**2
result = quad(func=function, a=1, b=np.inf)
result
#result = quad(1/x**2, 1, np.inf)
x_data_array = np.arange(0, 10, 1)
x_data_array
y_data_array = np.exp(-x_data_array/3.0)
y_data_array
plt.plot()
plt.plot(x_data_array, y_data_array, linestyle='solid', linewidth=5, marker='o', markersize=15)
plt.hlines(y=y_data_array[2], xmin=0, xmax=9, colors='#1f77b4', linewidth=5)
plt.show()
from scipy.interpolate import interp1d
interp_function = interp1d(x_data_array, y_data_array)
interp_function(2.0)
plt.figure()
plt.plot(x_data_array, y_data_array, 'o', markersize=15)
plt.hlines(y=0.51341712, xmin=0, xmax=9, colors='#1f77b4', linewidth=5)
plt.hlines(y=interp_function(2.0), xmin=0, xmax=9, colors='#ff7f0e', linestyles='dashed', linewidth=5)
plt.show()
x_values_new = np.arange(1, 8.2, 0.2)
print(x_values_new)
y_values_new = interp_function(x_values_new)
y_values_new
print(y_values_new)
plt.figure()
plt.plot(x_data_array, y_data_array, marker='o', markersize=15)
plt.hlines(y=0.51341712, xmin=0, xmax=9, colors='#1f77b4', linewidth=5)
plt.plot(x_values_new, y_values_new, marker='o', markersize=5)
plt.hlines(y=interp_function(2.0), xmin=0, xmax=9, colors='#ff7f0e', linestyles='dashed', linewidth=5)
plt.show()
x_values = np.linspace(0, 1, 10)
x_values
np.random.seed(30)
np.random.random(10)
noise = (np.random.random(10)**2 - 1) * 2e-1
print(noise)
y_values_ideal = np.sin(2 * np.pi * x_values)
y_values_ideal
y_values_sim = np.sin(2 * np.pi * x_values) + noise
y_values_sim
plt.figure()
plt.plot(x_values, y_values_ideal, marker='o', color='#1f77b4', markersize=7, linewidth=2, alpha=0.5) #ideal, blue
plt.plot(x_values, y_values_sim, marker='o', color='#ff7f0e', markersize=15, linewidth=5) #simulated, orange
plt.hlines(y=y_values_sim[5], xmin=0, xmax=1, colors='#ff7f0e', linewidth=3)
plt.show()
interp_function = interp1d(x_values, y_values_sim)
y_values_sim[5]
interp_function(x_values[5])
interp_function(x_values[5]) - y_values_sim[5]
x_data_new = np.arange(0, 1, 0.02)
x_data_new
y_data_new = interp_function(x_data_new)
y_data_new
plt.figure()
plt.plot(x_values, y_values_ideal, marker='o', color='#1f77b4', markersize=7, linewidth=2, alpha=0.5)
plt.plot(x_values, y_values_sim, marker='o', color='#ff7f0e', markersize=15, linewidth=5)
plt.hlines(y=y_values_sim[5], xmin=0, xmax=1, colors='#ff7f0e', linewidth=5)
## plot the interpolated curve (green)
plt.plot(x_data_new, y_data_new, marker='o', color='#2ca02c', markersize=10, linewidth=2, alpha=0.5)
plt.hlines(y=interp_function(x_values[5]), xmin=0, xmax=1, colors='#2ca02c',
linestyles='dashed', linewidth=2, alpha=0.5)
plt.show()
def percentage_rel_error(estimated: float=None, actual: float=None) -> float:
return ((estimated - actual)/actual)*100
pce = percentage_rel_error(estimated=(interp_function(x_values[5])), actual=y_values_sim[5])
print(f'{pce:.2}')
pce = percentage_rel_error(estimated=(interp_function(x_values[5])), actual=y_values_ideal[5])
pce
from scipy import optimize
x_values = np.linspace(-5, 5, num=50)
x_values
noise = np.random.random(50)
noise
y_values = 1.7*np.sin(2.5 * x_values) + noise
y_values
plt.figure()
plt.plot(x_values, y_values, '-o', markersize=15, linewidth=5)
plt.show()
def sine_func(x=None, a=None, b=None):
return a * np.sin(b * x)
solution, solution_covariance = optimize.curve_fit(sine_func, x_values, y_values, p0=[2.0, 2.0])
solution
solution_covariance
std_dev = np.sqrt(np.diag(solution_covariance))
std_dev
plt.plot()
plt.plot(x_values, y_values, '-o', markersize=15, linewidth=5) # blue (simulated experimental date)
plt.plot(x_values, sine_func(x_values, solution[0], solution[1]),
'-o', markersize=15, linewidth=5, alpha=0.7) # orange
plt.show()
def scalar_func(x: float=None) -> float:
return x**2 + 25*np.sin(x)
x_values = np.arange(-10, 10, 0.1)
y_values = scalar_func(x_values)
plt.figure()
plt.plot(x_values, y_values, 'o')
plt.show()
result_global = optimize.minimize(scalar_func, x0=0.0, method="BFGS")
result_global
type(result_global)
result_global.x
result_global.fun
result_local = optimize.minimize(scalar_func, x0=3.0, method="BFGS")
result_local
optimize.fminbound(func=scalar_func, x1=-10, x2=10)
optimize.brute(func=scalar_func, ranges=(slice(-10, 10, 0.1), ))
optimize.brute(func=scalar_func, ranges=((-10, 10), ))
optimize.basinhopping(func=scalar_func, x0=3.0, stepsize=0.5)
optimize.basinhopping(func=scalar_func, x0=3.0, stepsize=2.5)
plt.figure()
plt.plot(x_values, scalar_func(x_values), linestyle='solid', linewidth=5, alpha=0.5)
plt.hlines(y=0, xmin=-10, xmax=10, colors='red')
plt.show()
root1 = optimize.root(fun=scalar_func, x0=-4)
root1
root2 = optimize.root(fun=scalar_func, x0=1)
root2
root3 = optimize.root(scalar_func, x0=4)
root3
root4 = optimize.root(scalar_func, x0=7)
root4
root4.x
my_x = root4.x[0]
my_x**2 + 25*np.sin(my_x)
| 0.690142 | 0.988142 |
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
automobile_data = pd.read_csv('datasets/auto-mpg.csv')
automobile_data.head(10)
automobile_data.shape
automobile_data = automobile_data.replace('?', np.nan)
automobile_data = automobile_data.dropna()
automobile_data.shape
automobile_data.drop(['origin', 'car name'], axis=1, inplace=True)
automobile_data.head()
automobile_data['model year'] = '19' + automobile_data['model year'].astype(str)
automobile_data.sample(5)
import datetime
automobile_data['age'] = datetime.datetime.now().year - \
pd.to_numeric(automobile_data['model year'])
automobile_data.drop(['model year'], axis=1, inplace=True)
automobile_data.sample(5)
automobile_data.dtypes
automobile_data['horsepower'] = pd.to_numeric(automobile_data['horsepower'], errors='coerce')
automobile_data.describe()
automobile_data.to_csv('datasets/automobile_data_processed.csv', index=False)
!ls datasets/
automobile_data.plot.scatter(x='displacement', y='mpg', figsize=(12, 8))
plt.show()
automobile_data.plot.scatter(x='horsepower', y='mpg', figsize=(12, 8))
plt.show()
automobile_data.plot.hexbin(x='acceleration', y='mpg', gridsize=20, figsize=(12, 8))
plt.show()
automobile_grouped = automobile_data.groupby(['cylinders']).mean()[['mpg', 'horsepower',
'acceleration', 'displacement']]
automobile_grouped
automobile_grouped.plot.line(figsize=(12, 8))
plt.show()
fig, ax = plt.subplots()
automobile_data.plot(x='horsepower', y='mpg',
kind='scatter', s=60, c='cylinders',
cmap='magma_r', title='Automobile Data',
figsize=(12, 8), ax=ax)
plt.show()
fig, ax = plt.subplots()
automobile_data.plot(x='acceleration', y='mpg',
kind='scatter', s=60, c='cylinders',
cmap='magma_r', title='Automobile Data',
figsize=(12, 8), ax=ax)
plt.show()
fig, ax = plt.subplots()
automobile_data.plot(x='displacement', y='mpg',
kind='scatter', s=60, c='cylinders',
cmap='viridis', title='Automobile Data',
figsize=(12, 8), ax=ax)
plt.show()
automobile_data['acceleration'].cov(automobile_data['mpg'])
automobile_data['acceleration'].corr(automobile_data['mpg'])
automobile_data['horsepower'].cov(automobile_data['mpg'])
automobile_data['horsepower'].corr(automobile_data['mpg'])
automobile_data['horsepower'].cov(automobile_data['displacement'])
automobile_data['horsepower'].corr(automobile_data['displacement'])
automobile_data_cov = automobile_data.cov()
automobile_data_cov
automobile_data_corr = automobile_data.corr()
automobile_data_corr
plt.figure(figsize=(12, 8))
sns.heatmap(automobile_data_corr, annot=True)
mpg_mean = automobile_data['mpg'].mean()
mpg_mean
horsepower_mean = automobile_data['horsepower'].mean()
horsepower_mean
automobile_data['horsepower_mpg_cov'] = (automobile_data['horsepower'] - horsepower_mean) * \
(automobile_data['mpg'] - mpg_mean)
automobile_data['horsepower_var'] = (automobile_data['horsepower'] - horsepower_mean)**2
automobile_data['horsepower_mpg_cov']
automobile_data['horsepower_var']
beta = automobile_data['horsepower_mpg_cov'].sum() / automobile_data['horsepower_var'].sum()
print(f'beta = {beta}')
alpha = mpg_mean - (beta * horsepower_mean)
print(f'alpha = {alpha}')
y_pred = alpha + beta * automobile_data['horsepower']
print(y_pred)
automobile_data.plot(x='horsepower', y='mpg',
kind='scatter', s=50, figsize=(12, 8))
plt.plot(automobile_data['horsepower'], y_pred, color='red')
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
automobile_data = pd.read_csv('datasets/auto-mpg.csv')
automobile_data.head(10)
automobile_data.shape
automobile_data = automobile_data.replace('?', np.nan)
automobile_data = automobile_data.dropna()
automobile_data.shape
automobile_data.drop(['origin', 'car name'], axis=1, inplace=True)
automobile_data.head()
automobile_data['model year'] = '19' + automobile_data['model year'].astype(str)
automobile_data.sample(5)
import datetime
automobile_data['age'] = datetime.datetime.now().year - \
pd.to_numeric(automobile_data['model year'])
automobile_data.drop(['model year'], axis=1, inplace=True)
automobile_data.sample(5)
automobile_data.dtypes
automobile_data['horsepower'] = pd.to_numeric(automobile_data['horsepower'], errors='coerce')
automobile_data.describe()
automobile_data.to_csv('datasets/automobile_data_processed.csv', index=False)
!ls datasets/
automobile_data.plot.scatter(x='displacement', y='mpg', figsize=(12, 8))
plt.show()
automobile_data.plot.scatter(x='horsepower', y='mpg', figsize=(12, 8))
plt.show()
automobile_data.plot.hexbin(x='acceleration', y='mpg', gridsize=20, figsize=(12, 8))
plt.show()
automobile_grouped = automobile_data.groupby(['cylinders']).mean()[['mpg', 'horsepower',
'acceleration', 'displacement']]
automobile_grouped
automobile_grouped.plot.line(figsize=(12, 8))
plt.show()
fig, ax = plt.subplots()
automobile_data.plot(x='horsepower', y='mpg',
kind='scatter', s=60, c='cylinders',
cmap='magma_r', title='Automobile Data',
figsize=(12, 8), ax=ax)
plt.show()
fig, ax = plt.subplots()
automobile_data.plot(x='acceleration', y='mpg',
kind='scatter', s=60, c='cylinders',
cmap='magma_r', title='Automobile Data',
figsize=(12, 8), ax=ax)
plt.show()
fig, ax = plt.subplots()
automobile_data.plot(x='displacement', y='mpg',
kind='scatter', s=60, c='cylinders',
cmap='viridis', title='Automobile Data',
figsize=(12, 8), ax=ax)
plt.show()
automobile_data['acceleration'].cov(automobile_data['mpg'])
automobile_data['acceleration'].corr(automobile_data['mpg'])
automobile_data['horsepower'].cov(automobile_data['mpg'])
automobile_data['horsepower'].corr(automobile_data['mpg'])
automobile_data['horsepower'].cov(automobile_data['displacement'])
automobile_data['horsepower'].corr(automobile_data['displacement'])
automobile_data_cov = automobile_data.cov()
automobile_data_cov
automobile_data_corr = automobile_data.corr()
automobile_data_corr
plt.figure(figsize=(12, 8))
sns.heatmap(automobile_data_corr, annot=True)
mpg_mean = automobile_data['mpg'].mean()
mpg_mean
horsepower_mean = automobile_data['horsepower'].mean()
horsepower_mean
automobile_data['horsepower_mpg_cov'] = (automobile_data['horsepower'] - horsepower_mean) * \
(automobile_data['mpg'] - mpg_mean)
automobile_data['horsepower_var'] = (automobile_data['horsepower'] - horsepower_mean)**2
automobile_data['horsepower_mpg_cov']
automobile_data['horsepower_var']
beta = automobile_data['horsepower_mpg_cov'].sum() / automobile_data['horsepower_var'].sum()
print(f'beta = {beta}')
alpha = mpg_mean - (beta * horsepower_mean)
print(f'alpha = {alpha}')
y_pred = alpha + beta * automobile_data['horsepower']
print(y_pred)
automobile_data.plot(x='horsepower', y='mpg',
kind='scatter', s=50, figsize=(12, 8))
plt.plot(automobile_data['horsepower'], y_pred, color='red')
plt.show()
| 0.474631 | 0.685594 |
# Pyber Ride Share Analysis
```
# Importing libraries
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
# Importing data, merging into one main dataframe
city_df = pd.read_csv("data/city_data.csv")
ride_df = pd.read_csv("data/ride_data.csv")
main_df = pd.merge(ride_df, city_df, how='left', on='city')
main_df.head()
# Gathering data for Urban plot
urban_df = main_df[main_df['type'] == "Urban"]
urban_sort = urban_df.groupby('city')
urban_x = urban_sort['ride_id'].count()
urban_y = urban_sort['fare'].mean()
urban_z = urban_sort['driver_count'].mean()
# Gathering data for Suburban plot
suburb_df = main_df[main_df['type'] == "Suburban"]
suburb_sort = suburb_df.groupby('city')
suburb_x = suburb_sort['ride_id'].count()
suburb_y = suburb_sort['fare'].mean()
suburb_z = suburb_sort['driver_count'].mean()
# Gathering data for Rural plot
rural_df = main_df[main_df['type'] == "Rural"]
rural_sort = rural_df.groupby('city')
rural_x = rural_sort['ride_id'].count()
rural_y = rural_sort['fare'].mean()
rural_z = rural_sort['driver_count'].mean()
# Plotting each area type, formatting bubble plot
ax1 = plt.scatter(urban_x, urban_y, s=urban_z*10,
facecolors='xkcd:coral', edgecolors='black', alpha=.7, label="Urban")
ax2 = plt.scatter(suburb_x, suburb_y, s=suburb_z*10,
facecolors='xkcd:lightblue', edgecolors='black', alpha=.7, label="Suburban")
ax3 = plt.scatter(rural_x, rural_y, s=rural_z*10,
facecolors='xkcd:gold', edgecolors='black', alpha=.7, label="Rural")
plt.title('Pyber Ride Sharing Data (2016)')
plt.ylabel('Avg. Cost of Ride ($)')
plt.xlabel('Total Number of Rides (Per City)')
plt.text(41.2, 42, "Note:")
plt.text(41.2, 40.5, "Marker sizes represent the")
plt.text(41.2, 39, "number of drivers per city.")
plt.grid()
plt.legend(handles=[ax1, ax2, ax3], loc="best", markerscale=.7)
plt.show()
```
### Bubble Plot Analysis
Looking at the above bubble plot, we can see that the largest share of Pyber rides are taking place within the urban areas and that each city has a much larger amount of drivers per city. This follows common sense, as customers in rural/suburban areas are much more likely to rely on their own vehicles for the majority of their transportation needs while urban users are less likely to own or use their own cars because of a number of factors such as cost, traffic congestion, limited parking, etc..
Another obivous trend is that ride prices decrease from rural to urban areas. The most likely explanation for this trend is that travel times/distances are likely shorter in urban areas (denser populations) while travel distances in rural areas are likely much longer given how spread out the populations tend to be. Also increased competition in urban areas could drive down average prices in comparison to the rural areas with a small number of available drivers.
```
# Creating pie chart of total fares share
types = ['Rural', 'Suburban', 'Urban']
explode = [0, 0, .1]
colors = ['xkcd:gold', 'xkcd:lightblue', 'xkcd:coral']
type_sort = main_df.groupby('type')
fare_sum = type_sort.fare.sum()
fare_sum.sort_values(ascending=False)
plt.pie(fare_sum, explode=explode, labels=types, colors=colors, autopct="%1.1f%%", shadow=True, startangle=120)
plt.title("% of Total Fares by Area Type")
plt.show()
# Creating pie chart of total rides share
types = ['Rural', 'Suburban', 'Urban']
explode = [0, 0, .1]
colors = ['xkcd:gold', 'xkcd:lightblue', 'xkcd:coral']
type_sort = main_df.groupby('type')
fare_count = type_sort.fare.count()
fare_count.sort_values(ascending=False)
plt.pie(fare_count, explode=explode, labels=types, colors=colors, autopct="%1.1f%%", shadow=True, startangle=120)
plt.title("% of Total Rides by Area Type")
plt.show()
# Creating pie chart of total drivers share
types = ['Rural', 'Suburban', 'Urban']
explode = [0, 0, .1]
colors = ['xkcd:gold', 'xkcd:lightblue', 'xkcd:coral']
type_city_sort = city_df.groupby('type')
drivers_count = type_city_sort['driver_count'].sum()
plt.pie(drivers_count, explode=explode, labels=types, colors=colors, autopct="%1.1f%%", shadow=True, startangle=120)
plt.title("% of Total Drivers by Area Type")
plt.show()
```
### Pie Charts Analysis
In the pie charts above, we can really see how much of the total Pyber business is concentrated in urban areas. But an important trend to notice is that while 80.9% of Pyber drivers are working within urban areas, they take a significantly smaller portion of the fares (62.7%). This shows that drivers in rural and suburban areas are taking much higher fares from their customers, whether it be from longer drive times or decreased driver competition. It could be additionally insightful to factor in average driver costs to see if it is truly more lucrative to be a driver in a rural or suburban area or if in fact driver costs (gas, maintenence, time) mitigate the larger share of total fares.
|
github_jupyter
|
# Importing libraries
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
# Importing data, merging into one main dataframe
city_df = pd.read_csv("data/city_data.csv")
ride_df = pd.read_csv("data/ride_data.csv")
main_df = pd.merge(ride_df, city_df, how='left', on='city')
main_df.head()
# Gathering data for Urban plot
urban_df = main_df[main_df['type'] == "Urban"]
urban_sort = urban_df.groupby('city')
urban_x = urban_sort['ride_id'].count()
urban_y = urban_sort['fare'].mean()
urban_z = urban_sort['driver_count'].mean()
# Gathering data for Suburban plot
suburb_df = main_df[main_df['type'] == "Suburban"]
suburb_sort = suburb_df.groupby('city')
suburb_x = suburb_sort['ride_id'].count()
suburb_y = suburb_sort['fare'].mean()
suburb_z = suburb_sort['driver_count'].mean()
# Gathering data for Rural plot
rural_df = main_df[main_df['type'] == "Rural"]
rural_sort = rural_df.groupby('city')
rural_x = rural_sort['ride_id'].count()
rural_y = rural_sort['fare'].mean()
rural_z = rural_sort['driver_count'].mean()
# Plotting each area type, formatting bubble plot
ax1 = plt.scatter(urban_x, urban_y, s=urban_z*10,
facecolors='xkcd:coral', edgecolors='black', alpha=.7, label="Urban")
ax2 = plt.scatter(suburb_x, suburb_y, s=suburb_z*10,
facecolors='xkcd:lightblue', edgecolors='black', alpha=.7, label="Suburban")
ax3 = plt.scatter(rural_x, rural_y, s=rural_z*10,
facecolors='xkcd:gold', edgecolors='black', alpha=.7, label="Rural")
plt.title('Pyber Ride Sharing Data (2016)')
plt.ylabel('Avg. Cost of Ride ($)')
plt.xlabel('Total Number of Rides (Per City)')
plt.text(41.2, 42, "Note:")
plt.text(41.2, 40.5, "Marker sizes represent the")
plt.text(41.2, 39, "number of drivers per city.")
plt.grid()
plt.legend(handles=[ax1, ax2, ax3], loc="best", markerscale=.7)
plt.show()
# Creating pie chart of total fares share
types = ['Rural', 'Suburban', 'Urban']
explode = [0, 0, .1]
colors = ['xkcd:gold', 'xkcd:lightblue', 'xkcd:coral']
type_sort = main_df.groupby('type')
fare_sum = type_sort.fare.sum()
fare_sum.sort_values(ascending=False)
plt.pie(fare_sum, explode=explode, labels=types, colors=colors, autopct="%1.1f%%", shadow=True, startangle=120)
plt.title("% of Total Fares by Area Type")
plt.show()
# Creating pie chart of total rides share
types = ['Rural', 'Suburban', 'Urban']
explode = [0, 0, .1]
colors = ['xkcd:gold', 'xkcd:lightblue', 'xkcd:coral']
type_sort = main_df.groupby('type')
fare_count = type_sort.fare.count()
fare_count.sort_values(ascending=False)
plt.pie(fare_count, explode=explode, labels=types, colors=colors, autopct="%1.1f%%", shadow=True, startangle=120)
plt.title("% of Total Rides by Area Type")
plt.show()
# Creating pie chart of total drivers share
types = ['Rural', 'Suburban', 'Urban']
explode = [0, 0, .1]
colors = ['xkcd:gold', 'xkcd:lightblue', 'xkcd:coral']
type_city_sort = city_df.groupby('type')
drivers_count = type_city_sort['driver_count'].sum()
plt.pie(drivers_count, explode=explode, labels=types, colors=colors, autopct="%1.1f%%", shadow=True, startangle=120)
plt.title("% of Total Drivers by Area Type")
plt.show()
| 0.717012 | 0.727201 |
# OGGM-Shop and Glacier Directories in OGGM
## Set-up
### Input data folders
**If you are using your own computer**: before you start, make sure that you have set-up the [input data configuration file](https://docs.oggm.org/en/stable/input-data.html) at your wish. In the course of this tutorial, we will need to download data needed for each glacier (a couple of mb at max, depending on the chosen glaciers), so make sure you have an internet connection.
### cfg.initialize() and cfg.PARAMS
An OGGM simulation script will always start with the following commands:
```
from oggm import cfg, utils
cfg.initialize(logging_level='WARNING')
```
A call to [cfg.initialize()](https://docs.oggm.org/en/stable/generated/oggm.cfg.initialize.html) will read the default parameter file (or any user-provided file) and make them available to all other OGGM tools via the `cfg.PARAMS` dictionary. Here are some examples of these parameters:
```
cfg.PARAMS['continue_on_error']
cfg.PARAMS['border']
cfg.PARAMS['has_internet']
```
## Workflow
```
import os
from oggm import workflow, tasks
```
### Working directory
Each OGGM run needs a **single folder** where to store the results of the computations for all glaciers. This is called a "working directory" and needs to be specified before each run. Here we create a temporary folder for you:
```
cfg.PATHS['working_dir'] = utils.gettempdir(dirname='OGGM-Shop', reset=True)
cfg.PATHS['working_dir']
```
We use a temporary directory for this example, but in practice you will set this working directory yourself (for example: `/home/john/OGGM_output`. The size of this directory will depend on how many glaciers you'll simulate!
**This working directory is meant to be persistent**, i.e. you can stop your processing workflow after any task, and restart from an existing working directory at a later stage.
### Define the glaciers for the run
```
rgi_ids = ['RGI60-01.13696'] # Malaspina glacier (large - hungry in memory)
rgi_ids = ['RGI60-05.00800'] # Glacier in Greenland
```
You can provide any number of glacier identifiers. You can find other glacier identifiers by exploring the [GLIMS viewer](https://www.glims.org/maps/glims).
For an operational run on an RGI region, you might want to download the [Randolph Glacier Inventory](https://www.glims.org/RGI/) dataset instead, and start from it. This case is covered in the [working with the RGI](working_with_rgi.ipynb) tutorial and in the "Starting from scratch" section below.
### Starting from RGItopo
The OGGM workflow is organized as a list of **tasks** that have to be applied to a list of glaciers. The vast majority of tasks are called **entity tasks**: they are standalone operations to be realized on one single glacier entity. These tasks are executed sequentially (one after another): they often need input generated by the previous task(s): for example, the glacier mask task needs the glacier topography data.
To handle this situation, OGGM uses a workflow based on data persistence on disk: instead of passing data as python variables from one task to another, each task will read the data from disk and then write the computation results back to the disk, making these new data available for the next task in the queue.
These glacier specific data are located in [glacier directories](https://docs.oggm.org/en/stable/glacierdir.html#glacier-directories). These directories are initialized with the following command (this can take a little while on the first call, as OGGM needs to download some data):
```
# The RGI version to use
# V62 is an unofficial modification of V6 with only minor, backwards compatible modifications
prepro_rgi_version = 62
# Size of the map around the glacier.
prepro_border = 10
# Degree of processing level. This is OGGM specific and for the shop 1 is the one you want
from_prepro_level = 1
# URL of the preprocessed Gdirs
base_url = 'https://cluster.klima.uni-bremen.de/data/gdirs/dems_v1/default/'
gdirs = workflow.init_glacier_directories(rgi_ids,
from_prepro_level=from_prepro_level,
prepro_base_url=base_url,
prepro_rgi_version=prepro_rgi_version,
prepro_border=prepro_border)
```
`gdirs` is a list of [GlacierDirectory](https://docs.oggm.org/en/stable/generated/oggm.GlacierDirectory.html#oggm.GlacierDirectory) objects (one for each glacier). **Glacier directories are used by OGGM as "file and attribute manager"** for single glaciers.
For example, we now know where to find the glacier mask files for this glacier:
```
gdir = gdirs[0] # take Unteraar
print('Path to the DEM:', gdir.get_filepath('glacier_mask'))
```
And we can also access some attributes of this glacier:
```
gdir
gdir.rgi_date # date at which the outlines are valid
```
The advantage of this Glacier Directory data model is that it simplifies greatly the data transfer between tasks. **The single mandatory argument of all entity tasks will allways be a glacier directory**. With the glacier directory, each task will find the input it needs: for example, the glacier outlines are needed for the next plotting function, and are available via the `gdir` argument:
```
from oggm import graphics
graphics.plot_googlemap(gdir, figsize=(8, 7))
```
For most glaciers in the world there are several digital elevation models (DEM) which cover the respective glacier. In OGGM we have currently implemented many different open access DEMs to choose from. For some, you need to register to get access, see [dem_sources.ipynb/register](dem_sources.ipynb#register). Some are regional and only available in certain areas (e.g. Greenland or Antarctica) and some cover almost the entire globe. For more information, visit the [rgitools documentation about DEMs](https://rgitools.readthedocs.io/en/latest/dems.html).
### RGItopo data
```
sources = [src for src in os.listdir(gdir.dir) if src in utils.DEM_SOURCES]
print('RGI ID:', gdir.rgi_id)
print('Available DEM sources:', sources)
# We use xarray to store the data
import xarray as xr
import numpy as np
ods = xr.Dataset()
for src in sources:
demfile = os.path.join(gdir.dir, src) + '/dem.tif'
with xr.open_rasterio(demfile) as ds:
data = ds.sel(band=1).load() * 1.
ods[src] = data.where(data > -100, np.NaN)
sy, sx = np.gradient(ods[src], gdir.grid.dx, gdir.grid.dx)
ods[src + '_slope'] = ('y', 'x'), np.arctan(np.sqrt(sy**2 + sx**2))
with xr.open_rasterio(gdir.get_filepath('glacier_mask')) as ds:
ods['mask'] = ds.sel(band=1).load()
# Decide on the number of plots and figure size
ns = len(sources)
n_col = 3
x_size = 12
n_rows = -(-ns // n_col)
y_size = x_size / n_col * n_rows
from mpl_toolkits.axes_grid1 import AxesGrid
import salem
import matplotlib.pyplot as plt
smap = salem.graphics.Map(gdir.grid, countries=False)
smap.set_shapefile(gdir.read_shapefile('outlines'))
smap.set_plot_params(cmap='topo')
smap.set_lonlat_contours(add_tick_labels=False)
smap.set_plot_params(vmin=np.nanquantile([ods[s].min() for s in sources], 0.25),
vmax=np.nanquantile([ods[s].max() for s in sources], 0.75))
fig = plt.figure(figsize=(x_size, y_size))
grid = AxesGrid(fig, 111,
nrows_ncols=(n_rows, n_col),
axes_pad=0.7,
cbar_mode='each',
cbar_location='right',
cbar_pad=0.1
)
for i, s in enumerate(sources):
data = ods[s]
smap.set_data(data)
ax = grid[i]
smap.visualize(ax=ax, addcbar=False, title=s)
if np.isnan(data).all():
grid[i].cax.remove()
continue
cax = grid.cbar_axes[i]
smap.colorbarbase(cax)
# take care of uneven grids
if ax != grid[-1]:
grid[-1].remove()
grid[-1].cax.remove()
```
## Original (raw) topography data
See [dem_sources.ipynb](dem_sources.ipynb).
## OGGM-Shop: ITS-live
This is an example on how to extract velocity fields from the [ITS_live](https://its-live.jpl.nasa.gov/) Regional Glacier and Ice Sheet Surface Velocities Mosaic ([Gardner, A. et al 2019](http://its-live-data.jpl.nasa.gov.s3.amazonaws.com/documentation/ITS_LIVE-Regional-Glacier-and-Ice-Sheet-Surface-Velocities.pdf)) at 120 m resolution and reproject this data to the OGGM-glacier grid. This only works where ITS-live data is available! (not in the Alps).
The data source used is https://its-live.jpl.nasa.gov/#data
Currently the only data downloaded is the 120m composite for both
(u, v) and their uncertainty. The composite is computed from the
1985 to 2018 average. If you want more velocity products, feel free to open a new topic on the OGGM issue tracker!
```
# this will download severals large dataset (2*~800MB)
from oggm.shop import its_live, rgitopo
workflow.execute_entity_task(rgitopo.select_dem_from_dir, gdirs, dem_source='COPDEM', keep_dem_folders=True);
workflow.execute_entity_task(tasks.glacier_masks, gdirs);
workflow.execute_entity_task(its_live.velocity_to_gdir, gdirs);
```
By applying the entity task [its_live.velocity_to_gdir()](https://github.com/OGGM/oggm/blob/master/oggm/shop/its_live.py#L185) the model downloads and reprojects the ITS_live files to a given glacier map.
The velocity components (**vx**, **vy**) are added to the `gridded_data` nc file stored on each glacier directory.
According to the [ITS_LIVE documentation](http://its-live-data.jpl.nasa.gov.s3.amazonaws.com/documentation/ITS_LIVE-Regional-Glacier-and-Ice-Sheet-Surface-Velocities.pdf) velocities are given in ground units (i.e. absolute velocities). We then use bilinear interpolation to reproject the velocities to the local glacier map by re-projecting the vector distances.
By specifying `add_error=True`, we also reproject and scale the error for each component (**evx**, **evy**).
Now we can read in all the gridded data that comes with OGGM, including the ITS_Live velocity components.
```
with xr.open_dataset(gdir.get_filepath('gridded_data')) as ds:
ds = ds.load()
ds
# plot the salem map background, make countries in grey
smap = ds.salem.get_map(countries=False)
smap.set_shapefile(gdir.read_shapefile('outlines'))
smap.set_topography(ds.topo.data);
# get the velocity data
u = ds.obs_icevel_x.where(ds.glacier_mask == 1)
v = ds.obs_icevel_y.where(ds.glacier_mask == 1)
ws = (u**2 + v**2)**0.5
```
The `ds.glacier_mask == 1` command will remove the data outside of the glacier outline.
```
# get the axes ready
f, ax = plt.subplots(figsize=(9, 9))
# Quiver only every 3rd grid point
us = u[1::3, 1::3]
vs = v[1::3, 1::3]
smap.set_data(ws)
smap.set_cmap('Blues')
smap.plot(ax=ax)
smap.append_colorbar(ax=ax, label = 'ice velocity (m yr$^{-1}$)')
# transform their coordinates to the map reference system and plot the arrows
xx, yy = smap.grid.transform(us.x.values, us.y.values, crs=gdir.grid.proj)
xx, yy = np.meshgrid(xx, yy)
qu = ax.quiver(xx, yy, us.values, vs.values)
qk = ax.quiverkey(qu, 0.82, 0.97, 100, '100 m yr$^{-1}$',
labelpos='E', coordinates='axes')
```
## OGGM-Shop: bed topography data
OGGM can also download data from the [Farinotti et al., (2019) consensus estimate](https://www.nature.com/articles/s41561-019-0300-3) and reproject it to the glacier directories map:
```
from oggm.shop import bedtopo
workflow.execute_entity_task(bedtopo.add_consensus_thickness, gdirs);
with xr.open_dataset(gdir.get_filepath('gridded_data')) as ds:
ds = ds.load()
# plot the salem map background, make countries in grey
smap = ds.salem.get_map(countries=False)
smap.set_shapefile(gdir.read_shapefile('outlines'))
smap.set_topography(ds.topo.data);
f, ax = plt.subplots(figsize=(9, 9))
smap.set_data(ds.consensus_ice_thickness)
smap.set_cmap('Blues')
smap.plot(ax=ax)
smap.append_colorbar(ax=ax, label='ice thickness (m)');
```
## OGGM-Shop: climate data
```
# TODO
```
## What's next?
- look at the [OGGM-Shop documentation](https://docs.oggm.org/en/stable/input-data.html#)
- return to the [OGGM documentation](https://docs.oggm.org)
- back to the [table of contents](welcome.ipynb)
|
github_jupyter
|
from oggm import cfg, utils
cfg.initialize(logging_level='WARNING')
cfg.PARAMS['continue_on_error']
cfg.PARAMS['border']
cfg.PARAMS['has_internet']
import os
from oggm import workflow, tasks
cfg.PATHS['working_dir'] = utils.gettempdir(dirname='OGGM-Shop', reset=True)
cfg.PATHS['working_dir']
rgi_ids = ['RGI60-01.13696'] # Malaspina glacier (large - hungry in memory)
rgi_ids = ['RGI60-05.00800'] # Glacier in Greenland
# The RGI version to use
# V62 is an unofficial modification of V6 with only minor, backwards compatible modifications
prepro_rgi_version = 62
# Size of the map around the glacier.
prepro_border = 10
# Degree of processing level. This is OGGM specific and for the shop 1 is the one you want
from_prepro_level = 1
# URL of the preprocessed Gdirs
base_url = 'https://cluster.klima.uni-bremen.de/data/gdirs/dems_v1/default/'
gdirs = workflow.init_glacier_directories(rgi_ids,
from_prepro_level=from_prepro_level,
prepro_base_url=base_url,
prepro_rgi_version=prepro_rgi_version,
prepro_border=prepro_border)
gdir = gdirs[0] # take Unteraar
print('Path to the DEM:', gdir.get_filepath('glacier_mask'))
gdir
gdir.rgi_date # date at which the outlines are valid
from oggm import graphics
graphics.plot_googlemap(gdir, figsize=(8, 7))
sources = [src for src in os.listdir(gdir.dir) if src in utils.DEM_SOURCES]
print('RGI ID:', gdir.rgi_id)
print('Available DEM sources:', sources)
# We use xarray to store the data
import xarray as xr
import numpy as np
ods = xr.Dataset()
for src in sources:
demfile = os.path.join(gdir.dir, src) + '/dem.tif'
with xr.open_rasterio(demfile) as ds:
data = ds.sel(band=1).load() * 1.
ods[src] = data.where(data > -100, np.NaN)
sy, sx = np.gradient(ods[src], gdir.grid.dx, gdir.grid.dx)
ods[src + '_slope'] = ('y', 'x'), np.arctan(np.sqrt(sy**2 + sx**2))
with xr.open_rasterio(gdir.get_filepath('glacier_mask')) as ds:
ods['mask'] = ds.sel(band=1).load()
# Decide on the number of plots and figure size
ns = len(sources)
n_col = 3
x_size = 12
n_rows = -(-ns // n_col)
y_size = x_size / n_col * n_rows
from mpl_toolkits.axes_grid1 import AxesGrid
import salem
import matplotlib.pyplot as plt
smap = salem.graphics.Map(gdir.grid, countries=False)
smap.set_shapefile(gdir.read_shapefile('outlines'))
smap.set_plot_params(cmap='topo')
smap.set_lonlat_contours(add_tick_labels=False)
smap.set_plot_params(vmin=np.nanquantile([ods[s].min() for s in sources], 0.25),
vmax=np.nanquantile([ods[s].max() for s in sources], 0.75))
fig = plt.figure(figsize=(x_size, y_size))
grid = AxesGrid(fig, 111,
nrows_ncols=(n_rows, n_col),
axes_pad=0.7,
cbar_mode='each',
cbar_location='right',
cbar_pad=0.1
)
for i, s in enumerate(sources):
data = ods[s]
smap.set_data(data)
ax = grid[i]
smap.visualize(ax=ax, addcbar=False, title=s)
if np.isnan(data).all():
grid[i].cax.remove()
continue
cax = grid.cbar_axes[i]
smap.colorbarbase(cax)
# take care of uneven grids
if ax != grid[-1]:
grid[-1].remove()
grid[-1].cax.remove()
# this will download severals large dataset (2*~800MB)
from oggm.shop import its_live, rgitopo
workflow.execute_entity_task(rgitopo.select_dem_from_dir, gdirs, dem_source='COPDEM', keep_dem_folders=True);
workflow.execute_entity_task(tasks.glacier_masks, gdirs);
workflow.execute_entity_task(its_live.velocity_to_gdir, gdirs);
with xr.open_dataset(gdir.get_filepath('gridded_data')) as ds:
ds = ds.load()
ds
# plot the salem map background, make countries in grey
smap = ds.salem.get_map(countries=False)
smap.set_shapefile(gdir.read_shapefile('outlines'))
smap.set_topography(ds.topo.data);
# get the velocity data
u = ds.obs_icevel_x.where(ds.glacier_mask == 1)
v = ds.obs_icevel_y.where(ds.glacier_mask == 1)
ws = (u**2 + v**2)**0.5
# get the axes ready
f, ax = plt.subplots(figsize=(9, 9))
# Quiver only every 3rd grid point
us = u[1::3, 1::3]
vs = v[1::3, 1::3]
smap.set_data(ws)
smap.set_cmap('Blues')
smap.plot(ax=ax)
smap.append_colorbar(ax=ax, label = 'ice velocity (m yr$^{-1}$)')
# transform their coordinates to the map reference system and plot the arrows
xx, yy = smap.grid.transform(us.x.values, us.y.values, crs=gdir.grid.proj)
xx, yy = np.meshgrid(xx, yy)
qu = ax.quiver(xx, yy, us.values, vs.values)
qk = ax.quiverkey(qu, 0.82, 0.97, 100, '100 m yr$^{-1}$',
labelpos='E', coordinates='axes')
from oggm.shop import bedtopo
workflow.execute_entity_task(bedtopo.add_consensus_thickness, gdirs);
with xr.open_dataset(gdir.get_filepath('gridded_data')) as ds:
ds = ds.load()
# plot the salem map background, make countries in grey
smap = ds.salem.get_map(countries=False)
smap.set_shapefile(gdir.read_shapefile('outlines'))
smap.set_topography(ds.topo.data);
f, ax = plt.subplots(figsize=(9, 9))
smap.set_data(ds.consensus_ice_thickness)
smap.set_cmap('Blues')
smap.plot(ax=ax)
smap.append_colorbar(ax=ax, label='ice thickness (m)');
# TODO
| 0.395951 | 0.923247 |
# Nanodegree Engenheiro de Machine Learning
## Aprendizado Supervisionado
## Projeto: Encontrando doadores para a *CharityML*
Seja bem-vindo ao segundo projeto do Nanodegree Engenheiro de Machine Learning! Neste notebook, você receberá alguns códigos de exemplo e será seu trabalho implementar as funcionalidades adicionais necessárias para a conclusão do projeto. As seções cujo cabeçalho começa com **'Implementação'** indicam que o bloco de código posterior requer funcionalidades adicionais que você deve desenvolver. Para cada parte do projeto serão fornecidas instruções e as diretrizes da implementação estarão marcadas no bloco de código com uma expressão `'TODO'`.
Por favor, leia cuidadosamente as instruções!
Além de implementações de código, você terá de responder questões relacionadas ao projeto e à sua implementação. Cada seção onde você responderá uma questão terá um cabeçalho com o termo **'Questão X'**. Leia com atenção as questões e forneça respostas completas nas caixas de texto que começam com o termo **'Resposta:'**. A submissão do seu projeto será avaliada baseada nas suas resostas para cada uma das questões além das implementações que você disponibilizar.
>**Nota:** Por favor, especifique QUAL A VERSÃO DO PYTHON utilizada por você para a submissão deste notebook. As células "Code" e "Markdown" podem ser executadas utilizando o atalho do teclado **Shift + Enter**. Além disso, as células "Markdown" podem ser editadas clicando-se duas vezes na célula.
## Iniciando
Neste projeto, você utilizará diversos algoritmos de aprendizado supervisionado para modelar com precisão a remuneração de indivíduos utilizando dados coletados no censo americano de 1994. Você escolherá o algoritmo mais adequado através dos resultados preliminares e irá otimizá-lo para modelagem dos dados. O seu objetivo com esta implementação é construir um modelo que pode predizer com precisão se um indivíduo possui uma remuneração superior a $50,000. Este tipo de tarefa pode surgir em organizações sem fins lucrativos que sobrevivem de doações. Entender a remuneração de um indivíduo pode ajudar a organização o montante mais adequado para uma solicitação de doação, ou ainda se eles realmente deveriam entrar em contato com a pessoa. Enquanto pode ser uma tarefa difícil determinar a faixa de renda de uma pesssoa de maneira direta, nós podemos inferir estes valores através de outros recursos disponíveis publicamente.
O conjunto de dados para este projeto se origina do [Repositório de Machine Learning UCI](https://archive.ics.uci.edu/ml/datasets/Census+Income) e foi cedido por Ron Kohavi e Barry Becker, após a sua publicação no artigo _"Scaling Up the Accuracy of Naive-Bayes Classifiers: A Decision-Tree Hybrid"_. Você pode encontrar o artigo de Ron Kohavi [online](https://www.aaai.org/Papers/KDD/1996/KDD96-033.pdf). Os dados que investigaremos aqui possuem algumas pequenas modificações se comparados com os dados originais, como por exemplo a remoção da funcionalidade `'fnlwgt'` e a remoção de registros inconsistentes.
----
## Explorando os dados
Execute a célula de código abaixo para carregas as bibliotecas Python necessárias e carregas os dados do censo. Perceba que a última coluna deste conjunto de dados, `'income'`, será o rótulo do nosso alvo (se um indivíduo possui remuneração igual ou maior do que $50,000 anualmente). Todas as outras colunas são dados de cada indívduo na base de dados do censo.
```
# Importe as bibliotecas necessárias para o projeto.
import numpy as np
import pandas as pd
from time import time
from IPython.display import display # Permite a utilização da função display() para DataFrames.
import warnings
warnings.filterwarnings('ignore')
# Importação da biblioteca de visualização visuals.py
import visuals as vs
# Exibição amigável para notebooks
%matplotlib inline
# Carregando os dados do Censo
data = pd.read_csv("census.csv")
# Sucesso - Exibindo o primeiro registro
display(data.head(n=1))
```
### Implementação: Explorando os Dados
Uma investigação superficial da massa de dados determinará quantos indivíduos se enquadram em cada grupo e nos dirá sobre o percentual destes indivúdos com remuneração anual superior à \$50,000. No código abaixo, você precisará calcular o seguinte:
- O número total de registros, `'n_records'`
- O número de indivíduos com remuneração anual superior à \$50,000, `'n_greater_50k'`.
- O número de indivíduos com remuneração anual até \$50,000, `'n_at_most_50k'`.
- O percentual de indivíduos com remuneração anual superior à \$50,000, `'greater_percent'`.
** DICA: ** Você pode precisar olhar a tabela acima para entender como os registros da coluna `'income'` estão formatados.
```
# TODO: Número total de registros.
n_records = data.shape[0]
# TODO: Número de registros com remuneração anual superior à $50,000
n_greater_50k = data['income'].value_counts()['>50K']
# TODO: O número de registros com remuneração anual até $50,000
n_at_most_50k = data['income'].value_counts()['<=50K']
# TODO: O percentual de indivíduos com remuneração anual superior à $50,000
greater_percent = n_greater_50k / n_records * 100
# Exibindo os resultados
print("Total number of records: {}".format(n_records))
print("Individuals making more than $50,000: {}".format(n_greater_50k))
print("Individuals making at most $50,000: {}".format(n_at_most_50k))
print("Percentage of individuals making more than $50,000: {:.2f}%".format(greater_percent))
```
**Explorando as colunas**
* **age**: contínuo.
* **workclass**: Private, Self-emp-not-inc, Self-emp-inc, Federal-gov, Local-gov, State-gov, Without-pay, Never-worked.
* **education**: Bachelors, Some-college, 11th, HS-grad, Prof-school, Assoc-acdm, Assoc-voc, 9th, 7th-8th, 12th, Masters, 1st-4th, 10th, Doctorate, 5th-6th, Preschool.
* **education-num**: contínuo.
* **marital-status**: Married-civ-spouse, Divorced, Never-married, Separated, Widowed, Married-spouse-absent, Married-AF-spouse.
* **occupation**: Tech-support, Craft-repair, Other-service, Sales, Exec-managerial, Prof-specialty, Handlers-cleaners, Machine-op-inspct, Adm-clerical, Farming-fishing, Transport-moving, Priv-house-serv, Protective-serv, Armed-Forces.
* **relationship**: Wife, Own-child, Husband, Not-in-family, Other-relative, Unmarried.
* **race**: Black, White, Asian-Pac-Islander, Amer-Indian-Eskimo, Other.
* **sex**: Female, Male.
* **capital-gain**: contínuo.
* **capital-loss**: contínuo.
* **hours-per-week**: contínuo.
* **native-country**: United-States, Cambodia, England, Puerto-Rico, Canada, Germany, Outlying-US(Guam-USVI-etc), India, Japan, Greece, South, China, Cuba, Iran, Honduras, Philippines, Italy, Poland, Jamaica, Vietnam, Mexico, Portugal, Ireland, France, Dominican-Republic, Laos, Ecuador, Taiwan, Haiti, Columbia, Hungary, Guatemala, Nicaragua, Scotland, Thailand, Yugoslavia, El-Salvador, Trinadad&Tobago, Peru, Hong, Holand-Netherlands.
----
## Preparando os dados
Antes de que os dados possam ser utilizados como input para algoritmos de machine learning, muitas vezes eles precisam ser tratados, formatados e reestruturados — este processo é conhecido como **pré-processamento**. Felizmente neste conjunto de dados não existem registros inconsistentes para tratamento, porém algumas colunas precisam ser ajustadas. Este pré-processamento pode ajudar muito com o resultado e poder de predição de quase todos os algoritmos de aprendizado.
### Transformando os principais desvios das colunas contínuas
Um conjunto de dados pode conter ao menos uma coluna onde os valores tendem a se próximar para um único número, mas também podem conter registros com o mesmo atributo contendo um valor muito maior ou muito menor do que esta tendência. Algoritmos podem ser sensíveis para estes casos de distribuição de valores e este fator pode prejudicar sua performance se a distribuição não estiver normalizada de maneira adequada. Com o conjunto de dados do censo, dois atributos se encaixam nesta descrição: '`capital-gain'` e `'capital-loss'`.
Execute o código da célula abaixo para plotar um histograma destes dois atributos. Repare na distribuição destes valores.
```
# Dividindo os dados entre features e coluna alvo
income_raw = data['income']
features_raw = data.drop('income', axis = 1)
# Visualizando os principais desvios das colunas contínuas entre os dados
vs.distribution(data)
```
Para atributos com distribuição muito distorcida, tais como `'capital-gain'` e `'capital-loss'`, é uma prática comum aplicar uma <a href="https://en.wikipedia.org/wiki/Data_transformation_(statistics)">transformação logarítmica</a> nos dados para que os valores muito grandes e muito pequenos não afetem a performance do algoritmo de aprendizado. Usar a transformação logarítmica reduz significativamente os limites dos valores afetados pelos outliers (valores muito grandes ou muito pequenos). Deve-se tomar cuidado ao aplicar esta transformação, pois o logaritmo de `0` é indefinido, portanto temos que incrementar os valores em uma pequena quantia acima de `0` para aplicar o logaritmo adequadamente.
Execute o código da célula abaixo para realizar a transformação nos dados e visualizar os resultados. De novo, note os valores limite e como os valores estão distribuídos.
```
# Aplicando a transformação de log nos registros distorcidos.
skewed = ['capital-gain', 'capital-loss']
features_log_transformed = pd.DataFrame(data = features_raw)
features_log_transformed[skewed] = features_raw[skewed].apply(lambda x: np.log(x + 1))
# Visualizando as novas distribuições após a transformação.
vs.distribution(features_log_transformed, transformed = True)
```
### Normalizando atributos numéricos
Além das transformações em atributos distorcidos, é uma boa prática comum realizar algum tipo de adaptação de escala nos atributos numéricos. Ajustar a escala nos dados não modifica o formato da distribuição de cada coluna (tais como `'capital-gain'` ou `'capital-loss'` acima); no entanto, a normalização garante que cada atributo será tratado com o mesmo peso durante a aplicação de aprendizado supervisionado. Note que uma vez aplicada a escala, a observação dos dados não terá o significado original, como exemplificado abaixo.
Execute o código da célula abaixo para normalizar cada atributo numérico, nós usaremos para isso a [`sklearn.preprocessing.MinMaxScaler`](http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.MinMaxScaler.html).
```
# Importando sklearn.preprocessing.StandardScaler
from sklearn.preprocessing import MinMaxScaler
# Inicializando um aplicador de escala e aplicando em seguida aos atributos
scaler = MinMaxScaler() # default=(0, 1)
numerical = ['age', 'education-num', 'capital-gain', 'capital-loss', 'hours-per-week']
features_log_minmax_transform = pd.DataFrame(data = features_log_transformed)
features_log_minmax_transform[numerical] = scaler.fit_transform(features_log_transformed[numerical])
# Exibindo um exemplo de registro com a escala aplicada
display(features_log_minmax_transform.head(n=5))
```
### Implementação: Pré-processamento dos dados
A partir da tabela em **Explorando os dados** acima, nós podemos observar que existem diversos atributos não-numéricos para cada registro. Usualmente, algoritmos de aprendizado esperam que os inputs sejam numéricos, o que requer que os atributos não numéricos (chamados de *variáveis de categoria*) sejam convertidos. Uma maneira popular de converter as variáveis de categoria é utilizar a estratégia **one-hot encoding**. Esta estratégia cria uma variável para cada categoria possível de cada atributo não numérico. Por exemplo, assuma que `algumAtributo` possuí três valores possíveis: `A`, `B`, ou `C`. Nós então transformamos este atributo em três novos atributos: `algumAtributo_A`, `algumAtributo_B` e `algumAtributo_C`.
| | algumAtributo | | algumAtributo_A | algumAtributo_B | algumAtributo_C |
| :-: | :-: | | :-: | :-: | :-: |
| 0 | B | | 0 | 1 | 0 |
| 1 | C | ----> one-hot encode ----> | 0 | 0 | 1 |
| 2 | A | | 1 | 0 | 0 |
Além disso, assim como os atributos não-numéricos, precisaremos converter a coluna alvo não-numérica, `'income'`, para valores numéricos para que o algoritmo de aprendizado funcione. Uma vez que só existem duas categorias possíveis para esta coluna ("<=50K" e ">50K"), nós podemos evitar a utilização do one-hot encoding e simplesmente transformar estas duas categorias para `0` e `1`, respectivamente. No trecho de código abaixo, você precisará implementar o seguinte:
- Utilizar [`pandas.get_dummies()`](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.get_dummies.html?highlight=get_dummies#pandas.get_dummies) para realizar o one-hot encoding nos dados da `'features_log_minmax_transform'`.
- Converter a coluna alvo `'income_raw'` para re.
- Transforme os registros com "<=50K" para `0` e os registros com ">50K" para `1`.
```
# TODO: Utilize o one-hot encoding nos dados em 'features_log_minmax_transform' utilizando pandas.get_dummies()
features_final = pd.get_dummies(features_log_minmax_transform)
# TODO: Faça o encode da coluna 'income_raw' para valores numéricos
income = income_raw.replace("<=50K", 0).replace(">50K", 1)
print("{} income registers '<=50K' converted to 0.".format(income.value_counts()[0]))
print("{} income registers '>50K' converted to 1.".format(income.value_counts()[1]))
# Exiba o número de colunas depois do one-hot encoding
encoded = list(features_final.columns)
print("{} total features after one-hot encoding.".format(len(encoded)))
# Descomente a linha abaixo para ver as colunas após o encode
# print(encoded)
```
### Embaralhar e dividir os dados
Agora todas as _variáveis de categoria_ foram convertidas em atributos numéricos e todos os atributos numéricos foram normalizados. Como sempre, nós agora dividiremos os dados entre conjuntos de treinamento e de teste. 80% dos dados serão utilizados para treinamento e 20% para teste.
Execute o código da célula abaixo para realizar divisão.
```
# Importar train_test_split
from sklearn.model_selection import train_test_split
# Dividir os 'atributos' e 'income' entre conjuntos de treinamento e de testes.
X_train, X_test, y_train, y_test = train_test_split(features_final,
income,
test_size = 0.2,
random_state = 0)
# Show the results of the split
print("Training set has {} samples.".format(X_train.shape[0]))
print("Testing set has {} samples.".format(X_test.shape[0]))
```
----
## Avaliando a performance do modelo
Nesta seção nós investigaremos quatro algoritmos diferentes e determinaremos qual deles é melhor para a modelagem dos dados. Três destes algoritmos serão algoritmos de aprendizado supervisionado de sua escolha e o quarto algoritmo é conhecido como *naive predictor*.
### Métricas e o Naive predictor
*CharityML*, equipada com sua pesquisa, sabe que os indivíduos que fazem mais do que \$50,000 possuem maior probabilidade de doar para a sua campanha de caridade. Por conta disto, a *CharityML* está particularmente interessada em predizer com acurácia quais indivíduos possuem remuneração acima de \$50,000. Parece que utilizar **acurácia (accuracy)** como uma métrica para avaliar a performance de um modelo é um parâmetro adequado. Além disso, identificar alguém que *não possui* remuneração acima de \$50,000 como alguém que recebe acima deste valor seria ruim para a *CharityML*, uma vez que eles estão procurando por indivíduos que desejam doar. Com isso, a habilidade do modelo em predizer com preisão aqueles que possuem a remuneração acima dos \$50,000 é *mais importante* do que a habilidade de realizar o **recall** destes indivíduos. Nós podemos utilizar a fórmula **F-beta score** como uma métrica que considera ambos: precision e recall.
$$ F_{\beta} = (1 + \beta^2) \cdot \frac{precision \cdot recall}{\left( \beta^2 \cdot precision \right) + recall} $$
Em particular, quando $\beta = 0.5$, maior ênfase é atribuída para a variável precision. Isso é chamado de **F$_{0.5}$ score** (ou F-score, simplificando).
Analisando a distribuição de classes (aqueles que possuem remuneração até \$50,000 e aqueles que possuem remuneração superior), fica claro que a maioria dos indivíduos não possui remuneração acima de \$50,000. Isto pode ter grande impacto na **acurácia (accuracy)**, uma vez que nós poderíamos simplesmente dizer *"Esta pessoa não possui remuneração acima de \$50,000"* e estar certos em boa parte das vezes, sem ao menos olhar os dados! Fazer este tipo de afirmação seria chamado de **naive**, uma vez que não consideramos nenhuma informação para balisar este argumento. É sempre importante considerar a *naive prediction* para seu conjunto de dados, para ajudar a estabelecer um benchmark para análise da performance dos modelos. Com isso, sabemos que utilizar a naive prediction não traria resultado algum: Se a predição apontasse que todas as pessoas possuem remuneração inferior à \$50,000, a *CharityML* não identificaria ninguém como potencial doador.
#### Nota: Revisando: accuracy, precision e recall
**Accuracy** mede com que frequência o classificador faz a predição correta. É a proporção entre o número de predições corretas e o número total de predições (o número de registros testados).
**Precision** informa qual a proporção de mensagens classificamos como spam eram realmente spam. Ou seja, é a proporção de verdadeiros positivos (mensagens classificadas como spam que eram realmente spam) sobre todos os positivos (todas as palavras classificadas como spam, independente se a classificação estava correta), em outras palavras, é a proporção
`[Verdadeiros positivos/(Verdadeiros positivos + Falso positivos)]`
**Recall (sensibilidade)** nos informa qual a proporção das mensagens que eram spam que foram corretamente classificadas como spam. É a proporção entre os verdadeiros positivos (classificados como spam, que realmente eram spam) sobre todas as palavras que realmente eram spam. Em outras palavras, é a proporção entre
`[Verdadeiros positivos/(Verdadeiros positivos + Falso negativos)]`
Para problemas de classificação distorcidos em suas distribuições, como no nosso caso, por exemplo, se tivéssemos 100 mensagems de texto e apenas 2 fossem spam e todas as outras não fossem, a "accuracy" por si só não seria uma métrica tão boa. Nós poderiamos classificar 90 mensagems como "não-spam" (incluindo as 2 que eram spam mas que teriam sido classificadas como não-spam e, por tanto, seriam falso negativas.) e 10 mensagems como spam (todas as 10 falso positivas) e ainda assim teriamos uma boa pontuação de accuracy. Para estess casos, precision e recall são muito úteis. Estas duas métricas podem ser combinadas para resgatar o F1 score, que é calculado através da média(harmônica) dos valores de precision e de recall. Este score pode variar entre 0 e 1, sendo 1 o melhor resultado possível para o F1 score (consideramos a média harmônica pois estamos lidando com proporções).
### Questão 1 - Performance do Naive Predictor
Se escolhessemos um modelo que sempre prediz que um indivíduo possui remuneração acima de $50,000, qual seria a accuracy e o F-score considerando este conjunto de dados? Você deverá utilizar o código da célula abaixo e atribuir os seus resultados para as variáveis `'accuracy'` e `'fscore'` que serão usadas posteriormente.
Por favor, note que o propósito ao gerar um naive predictor é simplesmente exibir como um modelo sem nenhuma inteligência se comportaria. No mundo real, idealmente o seu modelo de base será o resultado de um modelo anterior ou poderia ser baseado em um paper no qual você se basearia para melhorar. Quando não houver qualquer benchmark de modelo, utilizar um naive predictor será melhor do que uma escolha aleatória.
**DICA:**
* Quando temos um modelo que sempre prediz '1' (e.x o indivíduo possui remuneração superior à 50k) então nosso modelo não terá Verdadeiros Negativos ou Falso Negativos, pois nós não estaremos afirmando que qualquer dos valores é negativo (ou '0') durante a predição. Com isso, nossa accuracy neste caso se torna o mesmo valor da precision (Verdadeiros positivos/ (Verdadeiros positivos + Falso positivos)) pois cada predição que fizemos com o valor '1' que deveria ter o valor '0' se torna um falso positivo; nosso denominador neste caso é o número total de registros.
* Nossa pontuação de Recall(Verdadeiros positivos/(Verdadeiros Positivos + Falsos negativos)) será 1 pois não teremos Falsos negativos.
```
'''
TP = np.sum(income) # Contando pois este é o caso "naive". Note que 'income' são os dados 'income_raw' convertidos
para valores numéricos durante o passo de pré-processamento de dados.
FP = income.count() - TP # Específico para o caso naive
TN = 0 # Sem predições negativas para o caso naive
FN = 0 # Sem predições negativas para o caso naive
'''
TP = income.sum()
FP = income.count() - TP
TN = 0
FN = 0
# TODO: Calcular accuracy, precision e recall
accuracy = (TP + TN) / (TP + TN + FP + FN)
recall = TP / (TP + FN)
precision = TP / (TP + FP)
# TODO: Calcular o F-score utilizando a fórmula acima para o beta = 0.5 e os valores corretos de precision e recall.
beta = 0.5
fscore = (1 + beta**2) * (precision * recall) / ((beta**2) * precision + recall)
# Exibir os resultados
print("Naive Predictor: [Accuracy score: {:.4f}, F-score: {:.4f}]".format(accuracy, fscore))
```
### Modelos de Aprendizado Supervisionado
**Estes são alguns dos modelos de aprendizado supervisionado disponíveis em** [`scikit-learn`](http://scikit-learn.org/stable/supervised_learning.html)
- Gaussian Naive Bayes (GaussianNB)
- Decision Trees (Árvores de decisão)
- Ensemble Methods (Bagging, AdaBoost, Random Forest, Gradient Boosting)
- K-Nearest Neighbors (KNeighbors)
- Stochastic Gradient Descent Classifier (SGDC)
- Support Vector Machines (SVM)
- Logistic Regression
### Questão 2 - Aplicação do Modelo
Liste três dos modelos de aprendizado supervisionado acima que são apropriados para este problema que você irá testar nos dados do censo. Para cada modelo escolhido
- Descreva uma situação do mundo real onde este modelo pode ser utilizado.
- Quais são as vantagems da utilização deste modelo; quando ele performa bem?
- Quais são as fraquezas do modelo; quando ele performa mal?
- O que torna este modelo um bom candidato para o problema, considerando o que você sabe sobre o conjunto de dados?
**DICA:**
Estruture sua resposta no mesmo formato acima, com 4 partes para cada um dos modelos que você escolher. Por favor, inclua referências em cada uma das respostas.
**Resposta:**
A princípio, o nosso problema é um caso de classificação binária, em que a variável alvo (*income*) ou possui um valor superior a $50k ou um valor menor ou igual a este. Dentre as variáveis independentes, temos dados contínuos e categóricos. Para resolver este problema, vários algoritmos poderiam ser adotados, mas neste trabalho escolhemos os seguintes:
- Regressão Logística
- Árvore de Decisão
- AdaBoost
O índices entre colchetes indicam as fontes e referências bibliográficas utlizadas para obter ou complementar o conhecimento para construção do parágrafo que o acompanha. Todas as referências utilizadas estão no final deste documento.
**Regressão Logística**
É um algoritmo de classificação linear amplamente empregado em várias áreas, como na saúde predizendo o risco de desenvolver determinada doença a partir de várias características do paciente. [1]
Dentre suas vantagens: além de predizer a qual classe pertence, ele pode estimar a probabilidade desse pertencimento; é um algoritmo baseado em funções matemáticas que cria uma linha, plano ou hiperplano de segmentação simples e flexível entre classes, podendo assumir qualquer direção ou orientação do espaço. [1][8]
Por outro lado, entre as desvantagens: este algoritmo se aplica apenas a problemas de classificação binários ou dicotômicos; embora o plano de classificação seja simples, sua interpretação e entendimento pode ser mais complexa para aqueles que não tem uma boa base matemática; não se ajusta muito bem quando a relação entre as variáveis dependentes e independentes for complexa, com uma alta não linearidade. [1][8]
É um método computacionalmente barato e muito poderoso, considerando que ele se restringe a problemas de classificação binárias, o que é o nosso caso, é um algoritmo que vale muito tentar antes de partir para outros mais complexos.
**Árvore de Decisão**
Este é outro algoritmo amplamente usado em várias áreas, mas tem um grande espaço conquistado nas áreas e processos de negócio que usam decisões analíticas por dados, principalmente por sua facilidade de compreensão. [2][8]
Podemos citar como as vantagens: é um método poderoso e flexível, se enquadrando tanto em problemas de classificação quanto de regressão, atua com variáveis dependentes e independentes categóricas e contínuas, podendo ser utilizado em classificações binárias e não-binárias; como em regressão logística, além da classificação pode também estimar a probabilidade de pertencimento à classe; os limites de decisão criados para segmentar as classes são sempre perpendiculares aos eixos, podendo criar complexas regiões de classificação, o que torna um bom algoritmo para lidar com a não linearidade; como sua classificação não se dá por meio de funções matemáticas, mas por declarações lógicas, é um modelo muito mais simples e intuitivo de entender e explicar, principalmente para pessoas sem sólida fundamentação matemática. [2][4][8]
Como desvantagens: por sua possibilidade de criar complexas regiões de classificação, é um método que pode facilmente sobreajustar o modelo, exigindo maior atenção na seleção dos hiperparâmetros; comparado com outros algoritmos, ele tende a produzir modelos com menor acurácia; é instável, significando que pequenas mudanças nos dados pode levar a grandes mudanças na estrutura de otimização do modelo. [2][4][8]
É um modelo também computacionalmente barato, flexível e trabalha bem com problemas não lineares, fazendo um contraponto ao primeiro algoritmo escolhido, a regressão logística. Logo, vale muito a pena avaliar a performance deste algoritmo antes de partirmos para outros ainda mais complexos.
**AdaBoost**
Os chamados *ensemble methods* tem ganhado cada vez mais espaço, principalmente em competições de machine learning como o Kaggle. Dentre eles, o Adaboost é um dos mais famosos e tem sido utilizado em aplicações de visão computacional, de detecção e reconhecimento de objetos. [3][5]
Seus pontos fortes: de modo geral, os ensemble methods utilizam vários estimadores fracos para alcançar um modelo forte e os métodos de boosting, o Adaboot é um deles, fazem isso de forma iterativa, penalizando com maior intensidade os dados que na etapa anterior foram classificados erroneamente. Com isso, a tendência é criar modelos com maior acurácia e redução significativa dos erros com o crescimento da quantidade de estimadores; realiza uma boa generalização do modelo, reduzindos os erros de viés e variância; tem a capacidade de realizar classificações não binárias. [3][6][7]
E suas fraquezas: naturalmente, como precisa de vários estimadores fracos para compor um modelo forte, seu custo computacional é mais elevado; é um modelo menos intuitivo e intelegível para profissionais fora do universo de data science; é um método sensível a ruídos e outliers. [3][6][7]
É um metodo mais complexo e menos intuitivo, contudo tende a apresentar excelente performance com o incremento de estimadores. Com os tratamentos e as transformações dos dados na etapa de preparação, mitigando o efeito dos outliers, avaliamos que esta é uma excelente opção e complementa bem o arcabouço de algoritmos escolhidos para este problema.
### Implementação - Criando um Pipeline de Treinamento e Predição
Para avaliar adequadamente a performance de cada um dos modelos que você escolheu é importante que você crie um pipeline de treinamento e predição que te permite de maneira rápida e eficiente treinar os modelos utilizando vários tamanhos de conjuntos de dados para treinamento, além de performar predições nos dados de teste. Sua implementação aqui será utilizada na próxima seção. No bloco de código abaixo, você precisará implementar o seguinte:
- Importar `fbeta_score` e `accuracy_score` de [`sklearn.metrics`](http://scikit-learn.org/stable/modules/classes.html#sklearn-metrics-metrics).
- Adapte o algoritmo para os dados de treinamento e registre o tempo de treinamento.
- Realize predições nos dados de teste `X_test`, e também nos 300 primeiros pontos de treinamento `X_train[:300]`.
- Registre o tempo total de predição.
- Calcule a acurácia tanto para o conjundo de dados de treino quanto para o conjunto de testes.
- Calcule o F-score para os dois conjuntos de dados: treino e testes.
- Garanta que você configurou o parâmetro `beta`!
```
# TODO: Import two metrics from sklearn - fbeta_score and accuracy_score
from sklearn.metrics import accuracy_score, fbeta_score
def train_predict(learner, sample_size, X_train, y_train, X_test, y_test):
'''
inputs:
- learner: the learning algorithm to be trained and predicted on
- sample_size: the size of samples (number) to be drawn from training set
- X_train: features training set
- y_train: income training set
- X_test: features testing set
- y_test: income testing set
'''
results = {}
# TODO: Fit the learner to the training data using slicing with 'sample_size' using .fit(training_features[:], training_labels[:])
start = time() # Get start time
step = round(len(y_train) / sample_size) # Calcula o passo necessário para selecionar a quantidade de amostras definida por sample_size
learner.fit(X_train[::step], y_train[::step]) # Aplica o passo na seleção das amostras para treinamento do modelo
end = time() # Get end time
# TODO: Calculate the training time
results['train_time'] = end - start
# TODO: Get the predictions on the test set(X_test),
# then get predictions on the first 300 training samples(X_train) using .predict()
start = time() # Get start time
predictions_test = learner.predict(X_test)
predictions_train = learner.predict(X_train[:300])
end = time() # Get end time
# TODO: Calculate the total prediction time
results['pred_time'] = end - start
# TODO: Compute accuracy on the first 300 training samples which is y_train[:300]
results['acc_train'] = accuracy_score(predictions_train, y_train[:300])
# TODO: Compute accuracy on test set using accuracy_score()
results['acc_test'] = accuracy_score(predictions_test, y_test)
# TODO: Compute F-score on the the first 300 training samples using fbeta_score()
results['f_train'] = fbeta_score(predictions_train, y_train[:300], beta=0.5)
# TODO: Compute F-score on the test set which is y_test
results['f_test'] = fbeta_score(predictions_test, y_test, beta=0.5)
# Success
print("{} trained on {} samples.".format(learner.__class__.__name__, sample_size))
# Return the results
return results
```
### Implementação: Validação inicial do modelo
No código da célula, você precisará implementar o seguinte:
- Importar os três modelos de aprendizado supervisionado que você escolheu na seção anterior
- Inicializar os três modelos e armazená-los em `'clf_A'`, `'clf_B'`, e `'clf_C'`.
- Utilize um `'random_state'` para cada modelo que você utilizar, caso seja fornecido.
- **Nota:** Utilize as configurações padrão para cada modelo - você otimizará um modelo específico em uma seção posterior
- Calcule o número de registros equivalentes à 1%, 10%, e 100% dos dados de treinamento.
- Armazene estes valores em `'samples_1'`, `'samples_10'`, e `'samples_100'` respectivamente.
**Nota:** Dependendo do algoritmo de sua escolha, a implementação abaixo pode demorar algum tempo para executar!
```
# TODO: Importe os três modelos de aprendizado supervisionado da sklearn
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import AdaBoostClassifier
# TODO: Inicialize os três modelos
clf_A = LogisticRegression(random_state=42)
clf_B = DecisionTreeClassifier(random_state=42)
clf_C = AdaBoostClassifier(random_state=42)
# TODO: Calcule o número de amostras para 1%, 10%, e 100% dos dados de treinamento
# HINT: samples_100 é todo o conjunto de treinamento e.x.: len(y_train)
# HINT: samples_10 é 10% de samples_100
# HINT: samples_1 é 1% de samples_100
samples_100 = len(y_train)
samples_10 = round(0.1 * samples_100)
samples_1 = round(0.01 * samples_100)
# Colete os resultados dos algoritmos de aprendizado
results = {}
for clf in [clf_A, clf_B, clf_C]:
clf_name = clf.__class__.__name__
results[clf_name] = {}
for i, samples in enumerate([samples_1, samples_10, samples_100]):
results[clf_name][i] = \
train_predict(clf, samples, X_train, y_train, X_test, y_test)
# Run metrics visualization for the three supervised learning models chosen
vs.evaluate(results, accuracy, fscore)
```
----
## Melhorando os resultados
Nesta seção final, você irá escolher o melhor entre os três modelos de aprendizado supervisionado para utilizar nos dados dos estudantes. Você irá então realizar uma busca grid para otimização em todo o conjunto de dados de treino (`X_train` e `y_train`) fazendo o tuning de pelo menos um parâmetro para melhorar o F-score anterior do modelo.
### Questão 3 - Escolhendo o melhor modelo
Baseado na validação anterior, em um ou dois parágrafos explique para a *CharityML* qual dos três modelos você acredita ser o mais apropriado para a tarefa de identificar indivíduos com remuneração anual superior à \$50,000.
**DICA:**
Analise o gráfico do canto inferior esquerdo da célula acima(a visualização criada através do comando `vs.evaluate(results, accuracy, fscore)`) e verifique o F score para o conjunto de testes quando 100% do conjunto de treino é utilizado. Qual modelo possui o maior score? Sua resposta deve abranger os seguintes pontos:
* métricas - F score no conjunto de testes quando 100% dos dados de treino são utilizados,
* tempo de predição/treinamento
* a adequação do algoritmo para este conjunto de dados.
**Resposta:**
Acreditamos que o algoritmo mais adequado para solução do nosso problema é o **Adaboost**. Utilizando os classificadores com os parâmetros padrões, a Árvore de Decisão tem um bom desempenho com os dados de treinamento, mas não generaliza bem como pode ser observado nos resultados com os dados de teste, indicando um sobreajuste do modelo. Observando apenas os resultados gráficos com os dados de teste, notamos uma acurácia e um f-score aproximado entre os 3 modelos, com um desempenho superior para o Adaboost. Por outro lado, embora tenha um desempenho melhor, ele precisa de mais tempo para treinar o modelo e realizar predições.
O nosso caso de uso não é uma aplicação em tempo real, então não é necessário criar um streaming de dados que serão processados e utlizados para tomada de decisão e ação no momento que chegam. Como parte de uma estratégia de campanha de marketing, o nosso modelo tem o papel de identificar potenciais indíviduos com maiores chances de serem impactados por nossa campanha e realizarem doações para nossa ONG. Desta forma, o critério *tempo* pode ser secundarizado ou preterido diante um melhor desempenho de classificação dos indivíduos.
### Questão 4 - Descrevendo o modelo nos termos de Layman
Em um ou dois parágrafos, explique para a *CharityML*, nos termos de layman, como o modelo final escolhido deveria funcionar. Garanta que você está descrevendo as principais vantagens do modelo, tais como o modo de treinar o modelo e como o modelo realiza a predição. Evite a utilização de jargões matemáticos avançados, como por exemplo a descrição de equações.
**DICA:**
Quando estiver explicando seu modelo, cite as fontes externas utilizadas, caso utilize alguma.
**Resposta:**
O Adaboost é o modelo que escolhemos para a seleção dos melhores indíviduos, ou seja, aqueles com o maior potencial de responderem às nossas campanhas e converterem em doações. Ele é um modelo muito simples de ser implementado e muito poderoso. Mas como funciona?
Diferente de vários outros métodos que criam apenas um modelo de classificação, o Adaboost divide aleatoriamente o nosso conjunto de dados em vários conjuntos menores e a partir de cada conjunto reduzido é criado um modelo "fraco" de classificação. Por fraco, estamos dizendo que é um modelo com baixa precisão, que não acerta tanto quanto gostaríamos. Contudo, combinando de forma iterativa vários desses modelos fracos, é possível criar um modelo forte, com esteróides.
Em uma analogia, seria como se pegássemos um grande projeto de um prédio e entregássemos para vários especialistas (arquiteto, paisagista, engenheiro civil, engenheiro eletricista, entre outros) para projetar nossa estrutura. Cada projeto individualmente seria fraco para construir o nosso prédio, mas pegando o melhor de cada é possível alcançarmos o melhor projeto. É isso que o Adaboost faz.
Como é preciso treinar e combinar vários modelos em apenas um, normalmente o Adaboost leva mais tempo que outros algoritmos nas tarefas de treinamento e até mesmo de predição, mas ele apresenta bons resultados em termos de precisão da predição. Então, com certeza é um método que vale a pena investirmos.
### Implementação: Tuning do modelo
Refine o modelo escolhido. Utilize uma busca grid (`GridSearchCV`) com pleo menos um parâmetro importante refinado com pelo menos 3 valores diferentes. Você precisará utilizar todo o conjunto de treinamento para isso. Na célula de código abaixo, você precisará implementar o seguinte:
- Importar [`sklearn.model_selection.GridSearchCV`](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GridSearchCV.html) e [`sklearn.metrics.make_scorer`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.make_scorer.html).
- Inicializar o classificador escolhido por você e armazená-lo em `clf`.
- Configurar um `random_state` se houver um disponível para o mesmo estado que você configurou anteriormente.
- Criar um dicionário dos parâmetros que você quer otimizar para o modelo escolhido.
- Exemplo: `parâmetro = {'parâmetro' : [lista de valores]}`.
- **Nota:** Evite otimizar o parâmetro `max_features` se este parâmetro estiver disponível!
- Utilize `make_scorer` para criar um objeto de pontuação `fbeta_score` (com $\beta = 0.5$).
- Realize a busca gride no classificador `clf` utilizando o `'scorer'` e armazene-o na variável `grid_obj`.
- Adeque o objeto da busca grid aos dados de treino (`X_train`, `y_train`) e armazene em `grid_fit`.
**Nota:** Dependendo do algoritmo escolhido e da lista de parâmetros, a implementação a seguir pode levar algum tempo para executar!
```
# TODO: Importar 'GridSearchCV', 'make_scorer', e qualquer biblioteca necessária
from sklearn.ensemble import AdaBoostClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import make_scorer
from sklearn.metrics import fbeta_score
# TODO: Inicializar o classificador
clf = AdaBoostClassifier(random_state=42)
# TODO: Criar a lista de parâmetros que você quer otimizar, utilizando um dicionário, caso necessário.
# HINT: parameters = {'parameter_1': [value1, value2], 'parameter_2': [value1, value2]}
parameters = {'n_estimators': [50, 100, 250, 500],
'learning_rate': [0.25, 0.5, 1, 1.5, 2]}
# TODO: Criar um objeto fbeta_score utilizando make_scorer()
scorer = make_scorer(fbeta_score, beta=0.5)
# TODO: Realizar uma busca grid no classificador utilizando o 'scorer' como o método de score no GridSearchCV()
grid_obj = GridSearchCV(estimator=clf, param_grid=parameters, scoring=scorer)
# TODO: Adequar o objeto da busca grid como os dados para treinamento e encontrar os parâmetros ótimos utilizando fit()
start = time()
grid_fit = grid_obj.fit(X_train, y_train)
end = time()
train_time = end - start
# Recuperar o estimador
best_clf = grid_fit.best_estimator_
# Realizar predições utilizando o modelo não otimizado e modelar
start = time()
predictions = (clf.fit(X_train, y_train)).predict(X_test)
best_predictions = best_clf.predict(X_test)
end = time()
pred_time = end - start
# Reportar os scores de antes e de depois
print("Train/Optimize and Predict Time\n------")
print("Time to train: ", train_time)
print("Time to predict: ", pred_time)
print("\nUnoptimized model\n------")
print("Accuracy score on testing data: {:.4f}".format(accuracy_score(y_test, predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, predictions, beta = 0.5)))
print("\nOptimized Model\n------")
print("Final accuracy score on the testing data: {:.4f}".format(accuracy_score(y_test, best_predictions)))
print("Final F-score on the testing data: {:.4f}".format(fbeta_score(y_test, best_predictions, beta = 0.5)))
```
### Questão 5 - Validação final do modelo
* Qual é a accuracy e o F-score do modelo otimizado utilizando os dados de testes?
* Estes scores são melhores ou piores do que o modelo antes da otimização?
* Como os resultados do modelo otimizado se comparam aos benchmarks do naive predictor que você encontrou na **Questão 1**?
**Nota:** Preencha a tabela abaixo com seus resultados e então responda as questões no campo **Resposta**
#### Resultados:
| Metric | Naive Predictor | Unoptimized Model | Optimized Model |
|---------------|-----------------|-------------------|-----------------|
|Accuracy Score |0.2478 |0.8576 |0.8677
|F-score |0.2917 |0.7246 |0.7452
**Resposta:**
Observamos que os nossos modelos não otimizados e otimizados apresentam uma performance bem superior ao nosso naive predictor. Observamos também que o Adaboost sem qualquer otimização, utilizando seus hiperparâmetros padrões, já apresenta uma performance considerável, sendo aproximadamente 85,8% de acurácia e 72,5% de F-score. Com a otimização, tivemos um ganho em torno de 1% na acurácia e 2% no F-score, o que pode ser muito significativo, já que a nossa base tem mais de 45 mil pessoas e qualquer percentual pode significar um impacto em 450 pessoas.
----
## Importância dos atributos
Uma tarefa importante quando realizamos aprendizado supervisionado em um conjunto de dados como os dados do censo que estudamos aqui é determinar quais atributos fornecem maior poder de predição. Focando no relacionamento entre alguns poucos atributos mais importantes e na label alvo nós simplificamos muito o nosso entendimento do fenômeno, que é a coisa mais importante a se fazer. No caso deste projeto, isso significa que nós queremos identificar um pequeno número de atributos que possuem maior chance de predizer se um indivíduo possui renda anual superior à \$50,000.
Escolha um classificador da scikit-learn (e.x.: adaboost, random forests) que possua o atributo `feature_importance_`, que é uma função que calcula o ranking de importância dos atributos de acordo com o classificador escolhido. Na próxima célula python ajuste este classificador para o conjunto de treinamento e utilize este atributo para determinar os 5 atributos mais importantes do conjunto de dados do censo.
### Questão 6 - Observação da Relevância dos Atributos
Quando **Exploramos os dados**, vimos que existem treze atributos disponíveis para cada registro nos dados do censo. Destes treze atributos, quais os 5 atributos que você acredita que são os mais importantes para predição e em que ordem você os ranquearia? Por quê?
**Resposta:**
Intuitivamente falando, acreditamos que os cinco atributos abaixo sejam os mais significativos para a determinação do nível de renda (income) dos indívíduos:
- **capital-gain e capita-loss**: o montante de ganho e perda de capital, definitivamente, devem ser dois atribuitos importantes para determinação do nível de renda.
- **education**: níveis maiores de educação normalmente implica em melhores empregos com melhores condições de salário e benefícios.
- **workclass**: a estabilidade financeira também deve outro aspecto importante para indivíduos realizarem doações. Acreditamos que empregos/condições mais estáveis e com maiores direitos/benefícios, traz uma maior segurança e tranquilidade para indíviduos, podendo realizar gastos e investimentos para além da subsistência.
- **age**: acreditamos que pessoas mais jovens e que estão iniciando suas carreiras e construções de patrimônios realizem menos doações do que pessoas mais velhas e com maior estabilidade.
### Implementação - Extraindo a importância do atributo
Escolha um algoritmo de aprendizado supervisionado da `sciki-learn` que possui o atributo `feature_importance_` disponível. Este atributo é uma função que ranqueia a importância de cada atributo dos registros do conjunto de dados quando realizamos predições baseadas no algoritmo escolhido.
Na célula de código abaixo, você precisará implementar o seguinte:
- Importar um modelo de aprendizado supervisionado da sklearn se este for diferente dos três usados anteriormente.
- Treinar o modelo supervisionado com todo o conjunto de treinamento.
- Extrair a importância dos atributos utilizando `'.feature_importances_'`.
```
# TODO: Importar um modelo de aprendizado supervisionado que tenha 'feature_importances_'
from sklearn.ensemble import AdaBoostClassifier
# TODO: Treinar o modelo utilizando o conjunto de treinamento com .fit(X_train, y_train)
model = AdaBoostClassifier(n_estimators=500, learning_rate=1.5, random_state=42)
model.fit(X_train, y_train)
# TODO: Extrair a importância dos atributos utilizando .feature_importances_
importances = model.feature_importances_
# Plotar
vs.feature_plot(importances, X_train, y_train)
```
### Questão 7 - Extraindo importância dos atributos
Observe a visualização criada acima que exibe os cinco atributos mais relevantes para predizer se um indivíduo possui remuneração igual ou superior à \$50,000 por ano.
* Como estes cinco atributos se comparam com os 5 atributos que você discutiu na **Questão 6**?
* Se você estivesse próximo da mesma resposta, como esta visualização confirma o seu raciocínio?
* Se você não estava próximo, por que você acha que estes atributos são mais relevantes?
**Resposta:**
A avaliação dos cinco atributos mais importantes mostra que nossa intuição estava parcialmente certa. De fato, as características *capital-gain* e *capital-loss* são determinantes para estimar a renda sendo, reespectivamente, o primeiro e segunto atributos mais importantes. A idade (*age*) também se mostrou um forte atributo, embora tenha um impacto superior ao esperado, sendo o terceiro atributo mais significativo. Nossa intuição apontava que o nível educacional também seria relevante no processo de classificação e isso se confirma em partes, embora seja importante, esta característica é melhor representada pelo atributo *education-num* ao invés de *education*, que foi a nossa suposição inicial. Por fim, um atributo inicialmente inesperado, sendo o quarto mais relevante, é o *hours-per-week* e faz muito sentido. Naturalmente, a tendência é de que quanto mais horas trabalhadas por dia, semana ou mês, maior será remuneração e consequentemente, mais fácil será atingir o patamar de $50k de renda, o que qualifica o nosso indíviduo com maior propensão para realizar doações.
### Selecionando atributos
Como um modelo performa se nós só utilizamos um subconjunto de todos os atributos disponíveis nos dados? Com menos atributos necessários para treinar, a expectativa é que o treinamento e a predição sejam executados em um tempo muito menor — com o custo da redução nas métricas de performance. A partir da visualização acima, nós vemos que os cinco atributos mais importantes contribuem para mais de 50% da importância de **todos** os atributos presentes nos dados. Isto indica que nós podemos tentar *reduzir os atributos* e simplificar a informação necessária para o modelo aprender. O código abaixo utilizará o mesmo modelo otimizado que você encontrou anteriormente e treinará o modelo com o mesmo conjunto de dados de treinamento, porém apenas com *os cinco atributos mais importantes*
```
# Importar a funcionalidade para clonar um modelo
from sklearn.base import clone
# Reduzir a quantidade de atributos
X_train_reduced = X_train[X_train.columns.values[(np.argsort(importances)[::-1])[:5]]]
X_test_reduced = X_test[X_test.columns.values[(np.argsort(importances)[::-1])[:5]]]
# Treinar o melhor modelo encontrado com a busca grid anterior
clf = (clone(best_clf)).fit(X_train_reduced, y_train)
# Fazer novas predições
reduced_predictions = clf.predict(X_test_reduced)
# Reportar os scores do modelo final utilizando as duas versões dos dados.
print("Final Model trained on full data\n------")
print("Accuracy on testing data: {:.4f}".format(accuracy_score(y_test, best_predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, best_predictions, beta = 0.5)))
print("\nFinal Model trained on reduced data\n------")
print("Accuracy on testing data: {:.4f}".format(accuracy_score(y_test, reduced_predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, reduced_predictions, beta = 0.5)))
```
### Questão 8 - Efeitos da seleção de atributos
* Como o F-score do modelo final e o accuracy score do conjunto de dados reduzido utilizando apenas cinco atributos se compara aos mesmos indicadores utilizando todos os atributos?
* Se o tempo de treinamento é uma variável importante, você consideraria utilizar os dados enxutos como seu conjunto de treinamento?
**Resposta:**
Com o classificador otimizado e utilizando apenas as 5 características mais importantes para treiná-lo, temos uma redução aproximada de 2,5% na acurácia e 4,5% no F-Score quando comparado com o modelo treinado com todas as características. Embora a performance ainda seja muito boa, avaliamos que ela reduz significativamente a eficácia do modelo já que, conforme apontado anteriormente, considerando uma base de 45 mil pessoas qualquer percentual pode impactar em torno de 450 pessoas, o que não pode ser desprezado.
Desta forma, considerando que nosso modelo não é parte de uma aplicação em tempo real, onde cada microssegundo é algo precioso, podemos nos dar ao luxo de gastar mais tempo treinando o nosso modelo com algoritmos mais complexos e uma maior quantidade de atributos, desde que se converta em resultados.
> **Nota**: Uma vez que você tenha concluído toda a implementação de código e respondido cada uma das questões acima, você poderá finalizar o seu trabalho exportando o iPython Notebook como um documento HTML. Você pode fazer isso utilizando o menu acima navegando para
**File -> Download as -> HTML (.html)**. Inclua este documento junto do seu notebook como sua submissão.
## Referências Bibliográficas
[1] [Wikipedia. Logistic Regression.](https://en.wikipedia.org/wiki/Logistic_regression)
[2] [Wikipedia. Decision Tree.](https://en.wikipedia.org/wiki/Decision_tree)
[3] [Wikipedia. Adaboost.](https://en.wikipedia.org/wiki/AdaBoost)
[4] [A Complete Tutorial on Tree Based Modeling from Scratch (in R & Python).](https://www.analyticsvidhya.com/blog/2016/04/complete-tutorial-tree-based-modeling-scratch-in-python/)
[5] [Application of AdaBoost Algorithm in Basketball Player Detection.](https://www.uni-obuda.hu/journal/Markoski_Ivankovic_Ratgeber_Pecev_Glusac_57.pdf)
[6] [Ensemble methods: bagging, boosting and stacking.](https://towardsdatascience.com/ensemble-methods-bagging-boosting-and-stacking-c9214a10a205)
[7] [Freund, Y. Schapire R. E. A Short Introduction to Boosting.](https://cseweb.ucsd.edu/~yfreund/papers/IntroToBoosting.pdf)
[8] Provost, F. Fawcett, T. Data Science para Negócios.
|
github_jupyter
|
# Importe as bibliotecas necessárias para o projeto.
import numpy as np
import pandas as pd
from time import time
from IPython.display import display # Permite a utilização da função display() para DataFrames.
import warnings
warnings.filterwarnings('ignore')
# Importação da biblioteca de visualização visuals.py
import visuals as vs
# Exibição amigável para notebooks
%matplotlib inline
# Carregando os dados do Censo
data = pd.read_csv("census.csv")
# Sucesso - Exibindo o primeiro registro
display(data.head(n=1))
# TODO: Número total de registros.
n_records = data.shape[0]
# TODO: Número de registros com remuneração anual superior à $50,000
n_greater_50k = data['income'].value_counts()['>50K']
# TODO: O número de registros com remuneração anual até $50,000
n_at_most_50k = data['income'].value_counts()['<=50K']
# TODO: O percentual de indivíduos com remuneração anual superior à $50,000
greater_percent = n_greater_50k / n_records * 100
# Exibindo os resultados
print("Total number of records: {}".format(n_records))
print("Individuals making more than $50,000: {}".format(n_greater_50k))
print("Individuals making at most $50,000: {}".format(n_at_most_50k))
print("Percentage of individuals making more than $50,000: {:.2f}%".format(greater_percent))
# Dividindo os dados entre features e coluna alvo
income_raw = data['income']
features_raw = data.drop('income', axis = 1)
# Visualizando os principais desvios das colunas contínuas entre os dados
vs.distribution(data)
# Aplicando a transformação de log nos registros distorcidos.
skewed = ['capital-gain', 'capital-loss']
features_log_transformed = pd.DataFrame(data = features_raw)
features_log_transformed[skewed] = features_raw[skewed].apply(lambda x: np.log(x + 1))
# Visualizando as novas distribuições após a transformação.
vs.distribution(features_log_transformed, transformed = True)
# Importando sklearn.preprocessing.StandardScaler
from sklearn.preprocessing import MinMaxScaler
# Inicializando um aplicador de escala e aplicando em seguida aos atributos
scaler = MinMaxScaler() # default=(0, 1)
numerical = ['age', 'education-num', 'capital-gain', 'capital-loss', 'hours-per-week']
features_log_minmax_transform = pd.DataFrame(data = features_log_transformed)
features_log_minmax_transform[numerical] = scaler.fit_transform(features_log_transformed[numerical])
# Exibindo um exemplo de registro com a escala aplicada
display(features_log_minmax_transform.head(n=5))
# TODO: Utilize o one-hot encoding nos dados em 'features_log_minmax_transform' utilizando pandas.get_dummies()
features_final = pd.get_dummies(features_log_minmax_transform)
# TODO: Faça o encode da coluna 'income_raw' para valores numéricos
income = income_raw.replace("<=50K", 0).replace(">50K", 1)
print("{} income registers '<=50K' converted to 0.".format(income.value_counts()[0]))
print("{} income registers '>50K' converted to 1.".format(income.value_counts()[1]))
# Exiba o número de colunas depois do one-hot encoding
encoded = list(features_final.columns)
print("{} total features after one-hot encoding.".format(len(encoded)))
# Descomente a linha abaixo para ver as colunas após o encode
# print(encoded)
# Importar train_test_split
from sklearn.model_selection import train_test_split
# Dividir os 'atributos' e 'income' entre conjuntos de treinamento e de testes.
X_train, X_test, y_train, y_test = train_test_split(features_final,
income,
test_size = 0.2,
random_state = 0)
# Show the results of the split
print("Training set has {} samples.".format(X_train.shape[0]))
print("Testing set has {} samples.".format(X_test.shape[0]))
'''
TP = np.sum(income) # Contando pois este é o caso "naive". Note que 'income' são os dados 'income_raw' convertidos
para valores numéricos durante o passo de pré-processamento de dados.
FP = income.count() - TP # Específico para o caso naive
TN = 0 # Sem predições negativas para o caso naive
FN = 0 # Sem predições negativas para o caso naive
'''
TP = income.sum()
FP = income.count() - TP
TN = 0
FN = 0
# TODO: Calcular accuracy, precision e recall
accuracy = (TP + TN) / (TP + TN + FP + FN)
recall = TP / (TP + FN)
precision = TP / (TP + FP)
# TODO: Calcular o F-score utilizando a fórmula acima para o beta = 0.5 e os valores corretos de precision e recall.
beta = 0.5
fscore = (1 + beta**2) * (precision * recall) / ((beta**2) * precision + recall)
# Exibir os resultados
print("Naive Predictor: [Accuracy score: {:.4f}, F-score: {:.4f}]".format(accuracy, fscore))
# TODO: Import two metrics from sklearn - fbeta_score and accuracy_score
from sklearn.metrics import accuracy_score, fbeta_score
def train_predict(learner, sample_size, X_train, y_train, X_test, y_test):
'''
inputs:
- learner: the learning algorithm to be trained and predicted on
- sample_size: the size of samples (number) to be drawn from training set
- X_train: features training set
- y_train: income training set
- X_test: features testing set
- y_test: income testing set
'''
results = {}
# TODO: Fit the learner to the training data using slicing with 'sample_size' using .fit(training_features[:], training_labels[:])
start = time() # Get start time
step = round(len(y_train) / sample_size) # Calcula o passo necessário para selecionar a quantidade de amostras definida por sample_size
learner.fit(X_train[::step], y_train[::step]) # Aplica o passo na seleção das amostras para treinamento do modelo
end = time() # Get end time
# TODO: Calculate the training time
results['train_time'] = end - start
# TODO: Get the predictions on the test set(X_test),
# then get predictions on the first 300 training samples(X_train) using .predict()
start = time() # Get start time
predictions_test = learner.predict(X_test)
predictions_train = learner.predict(X_train[:300])
end = time() # Get end time
# TODO: Calculate the total prediction time
results['pred_time'] = end - start
# TODO: Compute accuracy on the first 300 training samples which is y_train[:300]
results['acc_train'] = accuracy_score(predictions_train, y_train[:300])
# TODO: Compute accuracy on test set using accuracy_score()
results['acc_test'] = accuracy_score(predictions_test, y_test)
# TODO: Compute F-score on the the first 300 training samples using fbeta_score()
results['f_train'] = fbeta_score(predictions_train, y_train[:300], beta=0.5)
# TODO: Compute F-score on the test set which is y_test
results['f_test'] = fbeta_score(predictions_test, y_test, beta=0.5)
# Success
print("{} trained on {} samples.".format(learner.__class__.__name__, sample_size))
# Return the results
return results
# TODO: Importe os três modelos de aprendizado supervisionado da sklearn
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import AdaBoostClassifier
# TODO: Inicialize os três modelos
clf_A = LogisticRegression(random_state=42)
clf_B = DecisionTreeClassifier(random_state=42)
clf_C = AdaBoostClassifier(random_state=42)
# TODO: Calcule o número de amostras para 1%, 10%, e 100% dos dados de treinamento
# HINT: samples_100 é todo o conjunto de treinamento e.x.: len(y_train)
# HINT: samples_10 é 10% de samples_100
# HINT: samples_1 é 1% de samples_100
samples_100 = len(y_train)
samples_10 = round(0.1 * samples_100)
samples_1 = round(0.01 * samples_100)
# Colete os resultados dos algoritmos de aprendizado
results = {}
for clf in [clf_A, clf_B, clf_C]:
clf_name = clf.__class__.__name__
results[clf_name] = {}
for i, samples in enumerate([samples_1, samples_10, samples_100]):
results[clf_name][i] = \
train_predict(clf, samples, X_train, y_train, X_test, y_test)
# Run metrics visualization for the three supervised learning models chosen
vs.evaluate(results, accuracy, fscore)
# TODO: Importar 'GridSearchCV', 'make_scorer', e qualquer biblioteca necessária
from sklearn.ensemble import AdaBoostClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import make_scorer
from sklearn.metrics import fbeta_score
# TODO: Inicializar o classificador
clf = AdaBoostClassifier(random_state=42)
# TODO: Criar a lista de parâmetros que você quer otimizar, utilizando um dicionário, caso necessário.
# HINT: parameters = {'parameter_1': [value1, value2], 'parameter_2': [value1, value2]}
parameters = {'n_estimators': [50, 100, 250, 500],
'learning_rate': [0.25, 0.5, 1, 1.5, 2]}
# TODO: Criar um objeto fbeta_score utilizando make_scorer()
scorer = make_scorer(fbeta_score, beta=0.5)
# TODO: Realizar uma busca grid no classificador utilizando o 'scorer' como o método de score no GridSearchCV()
grid_obj = GridSearchCV(estimator=clf, param_grid=parameters, scoring=scorer)
# TODO: Adequar o objeto da busca grid como os dados para treinamento e encontrar os parâmetros ótimos utilizando fit()
start = time()
grid_fit = grid_obj.fit(X_train, y_train)
end = time()
train_time = end - start
# Recuperar o estimador
best_clf = grid_fit.best_estimator_
# Realizar predições utilizando o modelo não otimizado e modelar
start = time()
predictions = (clf.fit(X_train, y_train)).predict(X_test)
best_predictions = best_clf.predict(X_test)
end = time()
pred_time = end - start
# Reportar os scores de antes e de depois
print("Train/Optimize and Predict Time\n------")
print("Time to train: ", train_time)
print("Time to predict: ", pred_time)
print("\nUnoptimized model\n------")
print("Accuracy score on testing data: {:.4f}".format(accuracy_score(y_test, predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, predictions, beta = 0.5)))
print("\nOptimized Model\n------")
print("Final accuracy score on the testing data: {:.4f}".format(accuracy_score(y_test, best_predictions)))
print("Final F-score on the testing data: {:.4f}".format(fbeta_score(y_test, best_predictions, beta = 0.5)))
# TODO: Importar um modelo de aprendizado supervisionado que tenha 'feature_importances_'
from sklearn.ensemble import AdaBoostClassifier
# TODO: Treinar o modelo utilizando o conjunto de treinamento com .fit(X_train, y_train)
model = AdaBoostClassifier(n_estimators=500, learning_rate=1.5, random_state=42)
model.fit(X_train, y_train)
# TODO: Extrair a importância dos atributos utilizando .feature_importances_
importances = model.feature_importances_
# Plotar
vs.feature_plot(importances, X_train, y_train)
# Importar a funcionalidade para clonar um modelo
from sklearn.base import clone
# Reduzir a quantidade de atributos
X_train_reduced = X_train[X_train.columns.values[(np.argsort(importances)[::-1])[:5]]]
X_test_reduced = X_test[X_test.columns.values[(np.argsort(importances)[::-1])[:5]]]
# Treinar o melhor modelo encontrado com a busca grid anterior
clf = (clone(best_clf)).fit(X_train_reduced, y_train)
# Fazer novas predições
reduced_predictions = clf.predict(X_test_reduced)
# Reportar os scores do modelo final utilizando as duas versões dos dados.
print("Final Model trained on full data\n------")
print("Accuracy on testing data: {:.4f}".format(accuracy_score(y_test, best_predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, best_predictions, beta = 0.5)))
print("\nFinal Model trained on reduced data\n------")
print("Accuracy on testing data: {:.4f}".format(accuracy_score(y_test, reduced_predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, reduced_predictions, beta = 0.5)))
| 0.243193 | 0.951459 |
# Estimation des émissions CO₂ d'une conférence en fonction de sa localisation
```
import csv
import geopy.distance
import matplotlib.pyplot as plt
import numpy as np
import unidecode
```
Lecture du nombre d'enseignants-chercheurs (MCF et PU) par académie. Pour chaque académie une seule ville a été retenue (Aix–Marseille ➡️ Marseille). La Corse, les DOMs et les personnels hors-académie ne sont pas pris en compte.
_Source de données : [“Note d'information DGRH relative aux personnels enseignants du supérieur pour l'année universitaire 2012-2013“](https://www.enseignementsup-recherche.gouv.fr/cid22654/demographie-desenseignants-de-l-enseignement-superieur.html)_
```
academies = {}
with open('academies.txt', 'r') as f:
for line in f:
if line[0] != '#':
data = line.split()
academies[data[0]] = int(data[1])
totacademies = sum(academies.values())
academies
```
Coordonnées (latitude et longitude) des grandes villes françaises
_Source : [FRENCH DATASET: POPULATION AND GPS COORDINATES](https://freakonometrics.hypotheses.org/1125)_
```
villes = {}
with open('villes.txt', 'r') as f:
for line in f:
if line[0] != '#':
data = line.split('\t')
villes[data[0]] = tuple(map(float, data[1:]))
villes['TOULOUSE']
def coord(acad):
return villes[unidecode.unidecode(acad.upper())][0:2]
for k in academies.keys():
coord(k)
coord('Besançon')
def distance(ac1, ac2):
c1 = coord(ac1)
c2 = coord(ac2)
return geopy.distance.distance(c1, c2).km
print(distance('Lille', 'Marseille'))
print(distance('Paris', 'Versailles'))
def all_travel_to(center):
sum = 0
for acad in academies.keys():
d = distance(center, acad)
frac = float(academies[acad]) / totacademies
sum += frac * d
return sum
labels = sorted(academies.keys())
dist = [all_travel_to(acad) for acad in labels]
fig = plt.figure()
ax = fig.add_axes([0.1, 0.35, 0.87, 0.6])
ax.bar(labels, dist)
plt.xticks(range(len(labels)), labels, rotation=90)
plt.ylabel('Distance moyenne par participant (km)')
#plt.show()
plt.savefig("distance.png", dpi=300)
```
## Calculs pour le train
```
with open('matrice_train.csv', 'r') as f:
reader = csv.reader(f, delimiter=';')
rows = [row for row in reader]
trainlabels = rows[0][1:]
traintime = dict()
traintime_arr = np.zeros((len(trainlabels), len(trainlabels)), np.int)
for irow, row in enumerate(rows[1:]):
traintime[(trainlabels[irow], trainlabels[irow])] = 0
for icol, cell in enumerate(row[1:]):
if len(cell) > 0:
traintime[(trainlabels[irow], trainlabels[icol])] = int(cell)
traintime[(trainlabels[icol], trainlabels[irow])] = int(cell)
traintime_arr[icol,irow] = int(cell)
traintime_arr[irow,icol] = int(cell)
traintime[('Lyon', 'Bordeaux')]/60
fig, ax = plt.subplots()
ax.imshow(traintime_arr, cmap='cool', interpolation='nearest')
ax.set_xticks([])
ax.set_yticks(np.arange(len(trainlabels)))
ax.set_yticklabels(trainlabels)
plt.show()
def all_train_to(center):
sum = 0
for acad in academies.keys():
d = traintime[(center, acad)]
frac = float(academies[acad]) / totacademies
sum += frac * d
return sum
labels = sorted(academies.keys())
dist = [all_train_to(acad) for acad in labels]
fig = plt.figure()
ax = fig.add_axes([0.1, 0.35, 0.87, 0.6])
ax.bar(labels, dist)
plt.xticks(range(len(labels)), labels, rotation=90)
plt.ylabel('Durée de train moyenne par participant (minutes)')
#plt.show()
plt.savefig("distance.png", dpi=300)
```
|
github_jupyter
|
import csv
import geopy.distance
import matplotlib.pyplot as plt
import numpy as np
import unidecode
academies = {}
with open('academies.txt', 'r') as f:
for line in f:
if line[0] != '#':
data = line.split()
academies[data[0]] = int(data[1])
totacademies = sum(academies.values())
academies
villes = {}
with open('villes.txt', 'r') as f:
for line in f:
if line[0] != '#':
data = line.split('\t')
villes[data[0]] = tuple(map(float, data[1:]))
villes['TOULOUSE']
def coord(acad):
return villes[unidecode.unidecode(acad.upper())][0:2]
for k in academies.keys():
coord(k)
coord('Besançon')
def distance(ac1, ac2):
c1 = coord(ac1)
c2 = coord(ac2)
return geopy.distance.distance(c1, c2).km
print(distance('Lille', 'Marseille'))
print(distance('Paris', 'Versailles'))
def all_travel_to(center):
sum = 0
for acad in academies.keys():
d = distance(center, acad)
frac = float(academies[acad]) / totacademies
sum += frac * d
return sum
labels = sorted(academies.keys())
dist = [all_travel_to(acad) for acad in labels]
fig = plt.figure()
ax = fig.add_axes([0.1, 0.35, 0.87, 0.6])
ax.bar(labels, dist)
plt.xticks(range(len(labels)), labels, rotation=90)
plt.ylabel('Distance moyenne par participant (km)')
#plt.show()
plt.savefig("distance.png", dpi=300)
with open('matrice_train.csv', 'r') as f:
reader = csv.reader(f, delimiter=';')
rows = [row for row in reader]
trainlabels = rows[0][1:]
traintime = dict()
traintime_arr = np.zeros((len(trainlabels), len(trainlabels)), np.int)
for irow, row in enumerate(rows[1:]):
traintime[(trainlabels[irow], trainlabels[irow])] = 0
for icol, cell in enumerate(row[1:]):
if len(cell) > 0:
traintime[(trainlabels[irow], trainlabels[icol])] = int(cell)
traintime[(trainlabels[icol], trainlabels[irow])] = int(cell)
traintime_arr[icol,irow] = int(cell)
traintime_arr[irow,icol] = int(cell)
traintime[('Lyon', 'Bordeaux')]/60
fig, ax = plt.subplots()
ax.imshow(traintime_arr, cmap='cool', interpolation='nearest')
ax.set_xticks([])
ax.set_yticks(np.arange(len(trainlabels)))
ax.set_yticklabels(trainlabels)
plt.show()
def all_train_to(center):
sum = 0
for acad in academies.keys():
d = traintime[(center, acad)]
frac = float(academies[acad]) / totacademies
sum += frac * d
return sum
labels = sorted(academies.keys())
dist = [all_train_to(acad) for acad in labels]
fig = plt.figure()
ax = fig.add_axes([0.1, 0.35, 0.87, 0.6])
ax.bar(labels, dist)
plt.xticks(range(len(labels)), labels, rotation=90)
plt.ylabel('Durée de train moyenne par participant (minutes)')
#plt.show()
plt.savefig("distance.png", dpi=300)
| 0.328422 | 0.884239 |
# **Template OP on salish**
```
%matplotlib inline
import sys
import xarray as xr
import numpy as np
import os
import math
from datetime import datetime, timedelta
from parcels import FieldSet, Field, VectorField, ParticleSet, JITParticle, ErrorCode, ParcelsRandom
sys.path.append('/home/jvalenti/MOAD/analysis-jose/notebooks/parcels')
from Kernels_sink import DeleteParticle, Buoyancy, AdvectionRK4_3D, Stokes_drift
from OP_functions import *
# Define paths
local = 0 #Set to 0 when working on server
paths = path(local)
Dat=xr.open_dataset(get_WW3_path(datetime(2018, 12, 23)))
path_NEMO = make_prefix(datetime(2018, 12, 23), paths['NEMO'])
Dat0=xr.open_dataset(path_NEMO + '_grid_W.nc')
Dat=xr.open_dataset(path_NEMO + '_grid_T.nc')
coord=xr.open_dataset(paths['mask'],decode_times=False)
WW3 = xr.open_dataset(get_WW3_path(datetime(2018, 12, 23)))
```
## Definitions
```
start = datetime(2018, 8, 23) #Start date
# Set Time length [days] and timestep [seconds]
length = 25
duration = timedelta(days=length)
dt = 90 #toggle between - or + to pick backwards or forwards
N = 6 # number of deploying locations
n = 50 # 1000 # number of particles per location
dmin = [0,0,0,0,0,70] #minimum depth
dd = 5 #max depth difference from dmin
x_offset, y_offset, zvals = p_deploy(N,n,dmin,dd)
from parcels import Variable
class MPParticle(JITParticle):
ro = Variable('ro', initial = 1370)
diameter = Variable('diameter', initial = 1.6e-5)
length = Variable('length', initial = 61e-5)
sediment = Variable('sediment', initial = 0)
Dat=xr.open_dataset(paths['coords'],decode_times=False)
outf_lat=Dat['nav_lat'][445,304]
outf_lon=Dat['nav_lon'][445,304]
lon = np.zeros([N,n])
lat = np.zeros([N,n])
# Execute run
clon, clat = [-123.901172,-125.155849,-123.207648,-122.427508,-123.399769,float(outf_lon)], [49.186308,49.975326,49.305448,47.622403,48.399420,float(outf_lat)] # choose horizontal centre of the particle cloud
for i in range(N):
lon[i,:]=(clon[i] + x_offset[i,:])
lat[i,:]=(clat[i] + y_offset[i,:])
z = zvals
#Set start date time and the name of the output file
name = 'Fibers_summer' #name output file
daterange = [start+timedelta(days=i) for i in range(length)]
fn = name + '_'.join(d.strftime('%Y%m%d')+'_1n' for d in [start, start+duration]) + '.nc'
outfile = os.path.join(paths['out'], fn)
print(outfile)
```
## Simulation
```
#Fill in the list of variables that you want to use as fields
varlist=['U','V','W','R']
filenames,variables,dimensions=filename_set(start,length,varlist,local)
field_set=FieldSet.from_nemo(filenames, variables, dimensions, allow_time_extrapolation=True)
varlist=['US','VS','WL']
filenames,variables,dimensions=filename_set(start,length,varlist,local)
us = Field.from_netcdf(filenames['US'], variables['US'], dimensions,allow_time_extrapolation=True)
vs = Field.from_netcdf(filenames['VS'], variables['VS'], dimensions,allow_time_extrapolation=True)
wl = Field.from_netcdf(filenames['WL'], variables['WL'], dimensions,allow_time_extrapolation=True)
field_set.add_field(us)
field_set.add_field(vs)
field_set.add_field(wl)
field_set.add_vector_field(VectorField("stokes", us, vs, wl))
filenames,variables,dimensions=filename_set(start,length,['Bathy'],local)
Bth = Field.from_netcdf(filenames['Bathy'], variables['Bathy'], dimensions,allow_time_extrapolation=True)
field_set.add_field(Bth)
# #Load Salish output as fields
#field_set = FieldSet.from_nemo(filenames, variables, dimensions, allow_time_extrapolation=True)
pset = ParticleSet.from_list(field_set, MPParticle, lon=lon, lat=lat, depth=z, time=start+timedelta(hours=2))
k_sink = pset.Kernel(Buoyancy)
k_waves = pset.Kernel(Stokes_drift)
pset.execute(AdvectionRK4_3D + k_sink + k_waves,
runtime=duration,
dt=dt,
output_file=pset.ParticleFile(name=outfile, outputdt=timedelta(hours=1)),
recovery={ErrorCode.ErrorOutOfBounds: DeleteParticle})
```
|
github_jupyter
|
%matplotlib inline
import sys
import xarray as xr
import numpy as np
import os
import math
from datetime import datetime, timedelta
from parcels import FieldSet, Field, VectorField, ParticleSet, JITParticle, ErrorCode, ParcelsRandom
sys.path.append('/home/jvalenti/MOAD/analysis-jose/notebooks/parcels')
from Kernels_sink import DeleteParticle, Buoyancy, AdvectionRK4_3D, Stokes_drift
from OP_functions import *
# Define paths
local = 0 #Set to 0 when working on server
paths = path(local)
Dat=xr.open_dataset(get_WW3_path(datetime(2018, 12, 23)))
path_NEMO = make_prefix(datetime(2018, 12, 23), paths['NEMO'])
Dat0=xr.open_dataset(path_NEMO + '_grid_W.nc')
Dat=xr.open_dataset(path_NEMO + '_grid_T.nc')
coord=xr.open_dataset(paths['mask'],decode_times=False)
WW3 = xr.open_dataset(get_WW3_path(datetime(2018, 12, 23)))
start = datetime(2018, 8, 23) #Start date
# Set Time length [days] and timestep [seconds]
length = 25
duration = timedelta(days=length)
dt = 90 #toggle between - or + to pick backwards or forwards
N = 6 # number of deploying locations
n = 50 # 1000 # number of particles per location
dmin = [0,0,0,0,0,70] #minimum depth
dd = 5 #max depth difference from dmin
x_offset, y_offset, zvals = p_deploy(N,n,dmin,dd)
from parcels import Variable
class MPParticle(JITParticle):
ro = Variable('ro', initial = 1370)
diameter = Variable('diameter', initial = 1.6e-5)
length = Variable('length', initial = 61e-5)
sediment = Variable('sediment', initial = 0)
Dat=xr.open_dataset(paths['coords'],decode_times=False)
outf_lat=Dat['nav_lat'][445,304]
outf_lon=Dat['nav_lon'][445,304]
lon = np.zeros([N,n])
lat = np.zeros([N,n])
# Execute run
clon, clat = [-123.901172,-125.155849,-123.207648,-122.427508,-123.399769,float(outf_lon)], [49.186308,49.975326,49.305448,47.622403,48.399420,float(outf_lat)] # choose horizontal centre of the particle cloud
for i in range(N):
lon[i,:]=(clon[i] + x_offset[i,:])
lat[i,:]=(clat[i] + y_offset[i,:])
z = zvals
#Set start date time and the name of the output file
name = 'Fibers_summer' #name output file
daterange = [start+timedelta(days=i) for i in range(length)]
fn = name + '_'.join(d.strftime('%Y%m%d')+'_1n' for d in [start, start+duration]) + '.nc'
outfile = os.path.join(paths['out'], fn)
print(outfile)
#Fill in the list of variables that you want to use as fields
varlist=['U','V','W','R']
filenames,variables,dimensions=filename_set(start,length,varlist,local)
field_set=FieldSet.from_nemo(filenames, variables, dimensions, allow_time_extrapolation=True)
varlist=['US','VS','WL']
filenames,variables,dimensions=filename_set(start,length,varlist,local)
us = Field.from_netcdf(filenames['US'], variables['US'], dimensions,allow_time_extrapolation=True)
vs = Field.from_netcdf(filenames['VS'], variables['VS'], dimensions,allow_time_extrapolation=True)
wl = Field.from_netcdf(filenames['WL'], variables['WL'], dimensions,allow_time_extrapolation=True)
field_set.add_field(us)
field_set.add_field(vs)
field_set.add_field(wl)
field_set.add_vector_field(VectorField("stokes", us, vs, wl))
filenames,variables,dimensions=filename_set(start,length,['Bathy'],local)
Bth = Field.from_netcdf(filenames['Bathy'], variables['Bathy'], dimensions,allow_time_extrapolation=True)
field_set.add_field(Bth)
# #Load Salish output as fields
#field_set = FieldSet.from_nemo(filenames, variables, dimensions, allow_time_extrapolation=True)
pset = ParticleSet.from_list(field_set, MPParticle, lon=lon, lat=lat, depth=z, time=start+timedelta(hours=2))
k_sink = pset.Kernel(Buoyancy)
k_waves = pset.Kernel(Stokes_drift)
pset.execute(AdvectionRK4_3D + k_sink + k_waves,
runtime=duration,
dt=dt,
output_file=pset.ParticleFile(name=outfile, outputdt=timedelta(hours=1)),
recovery={ErrorCode.ErrorOutOfBounds: DeleteParticle})
| 0.240685 | 0.614423 |
___
<a href='http://www.pieriandata.com'><img src='../Pierian_Data_Logo.png'/></a>
___
<center><em>Copyright Pierian Data</em></center>
<center><em>For more information, visit us at <a href='http://www.pieriandata.com'>www.pieriandata.com</a></em></center>
# Holt-Winters Methods
In the previous section on <strong>Exponentially Weighted Moving Averages</strong> (EWMA) we applied <em>Simple Exponential Smoothing</em> using just one smoothing factor $\alpha$ (alpha). This failed to account for other contributing factors like trend and seasonality.
In this section we'll look at <em>Double</em> and <em>Triple Exponential Smoothing</em> with the <a href='https://otexts.com/fpp2/holt-winters.html'>Holt-Winters Methods</a>.
In <strong>Double Exponential Smoothing</strong> (aka Holt's Method) we introduce a new smoothing factor $\beta$ (beta) that addresses trend:
\begin{split}l_t &= (1 - \alpha) l_{t-1} + \alpha x_t, & \text{ level}\\
b_t &= (1-\beta)b_{t-1} + \beta(l_t-l_{t-1}) & \text{ trend}\\
y_t &= l_t + b_t & \text{ fitted model}\\
\hat y_{t+h} &= l_t + hb_t & \text{ forecasting model (} h = \text{# periods into the future)}\end{split}
Because we haven't yet considered seasonal fluctuations, the forecasting model is simply a straight sloped line extending from the most recent data point. We'll see an example of this in upcoming lectures.
With <strong>Triple Exponential Smoothing</strong> (aka the Holt-Winters Method) we introduce a smoothing factor $\gamma$ (gamma) that addresses seasonality:
\begin{split}l_t &= (1 - \alpha) l_{t-1} + \alpha x_t, & \text{ level}\\
b_t &= (1-\beta)b_{t-1} + \beta(l_t-l_{t-1}) & \text{ trend}\\
c_t &= (1-\gamma)c_{t-L} + \gamma(x_t-l_{t-1}-b_{t-1}) & \text{ seasonal}\\
y_t &= (l_t + b_t) c_t & \text{ fitted model}\\
\hat y_{t+m} &= (l_t + mb_t)c_{t-L+1+(m-1)modL} & \text{ forecasting model (} m = \text{# periods into the future)}\end{split}
Here $L$ represents the number of divisions per cycle. In our case looking at monthly data that displays a repeating pattern each year, we would use $L=12$.
In general, higher values for $\alpha$, $\beta$ and $\gamma$ (values closer to 1), place more emphasis on recent data.
<div class="alert alert-info"><h3>Related Functions:</h3>
<tt><strong><a href='https://www.statsmodels.org/stable/generated/statsmodels.tsa.holtwinters.SimpleExpSmoothing.html'>statsmodels.tsa.holtwinters.SimpleExpSmoothing</a></strong><font color=black>(endog)</font>
Simple Exponential Smoothing<br>
<strong><a href='https://www.statsmodels.org/stable/generated/statsmodels.tsa.holtwinters.ExponentialSmoothing.html'>statsmodels.tsa.holtwinters.ExponentialSmoothing</a></strong><font color=black>(endog)</font>
Holt-Winters Exponential Smoothing</tt>
<h3>For Further Reading:</h3>
<tt>
<strong>
<a href='https://www.itl.nist.gov/div898/handbook/pmc/section4/pmc43.htm'>NIST/SEMATECH e-Handbook of Statistical Methods</a></strong> <font color=black>What is Exponential Smoothing?</font></tt></div>
### Perform standard imports and load the dataset
For these examples we'll continue to use the International Airline Passengers dataset, which gives monthly totals in thousands from January 1949 to December 1960.
```
import pandas as pd
import numpy as np
%matplotlib inline
df = pd.read_csv('../Data/airline_passengers.csv',index_col='Month',parse_dates=True)
df.dropna(inplace=True)
df.index
```
### Setting a DatetimeIndex Frequency
Note that our DatetimeIndex does not have a frequency. In order to build a Holt-Winters smoothing model, statsmodels needs to know the frequency of the data (whether it's daily, monthly etc.). Since observations occur at the start of each month, we'll use MS.<br>A full list of time series offset aliases can be found <a href='http://pandas.pydata.org/pandas-docs/stable/user_guide/timeseries.html#offset-aliases'>here</a>.
```
df.index.freq = 'MS'
df.index
df.head()
```
___
## Simple Exponential Smoothing
A variation of the statmodels Holt-Winters function provides Simple Exponential Smoothing. We'll show that it performs the same calculation of the weighted moving average as the pandas <tt>.ewm()</tt> method:<br>
$\begin{split}y_0 &= x_0 \\
y_t &= (1 - \alpha) y_{t-1} + \alpha x_t,\end{split}$
```
from statsmodels.tsa.holtwinters import SimpleExpSmoothing
span = 12
alpha = 2/(span+1)
df['EWMA12'] = df['Thousands of Passengers'].ewm(alpha=alpha,adjust=False).mean()
df['SES12']=SimpleExpSmoothing(df['Thousands of Passengers']).fit(smoothing_level=alpha,optimized=False).fittedvalues.shift(-1)
df.head()
```
<div class="alert alert-danger"><strong>NOTE:</strong> For some reason, when <tt>optimized=False</tt> is passed into <tt>.fit()</tt>, the statsmodels <tt>SimpleExpSmoothing</tt> function shifts fitted values down one row. We fix this by adding <tt>.shift(-1)</tt> after <tt>.fittedvalues</tt></div>
___
## Double Exponential Smoothing
Where Simple Exponential Smoothing employs just one smoothing factor $\alpha$ (alpha), Double Exponential Smoothing adds a second smoothing factor $\beta$ (beta) that addresses trends in the data. Like the alpha factor, values for the beta factor fall between zero and one ($0<\beta≤1$). The benefit here is that the model can anticipate future increases or decreases where the level model would only work from recent calculations.
We can also address different types of change (growth/decay) in the trend. If a time series displays a straight-line sloped trend, you would use an <strong>additive</strong> adjustment. If the time series displays an exponential (curved) trend, you would use a <strong>multiplicative</strong> adjustment.
As we move toward forecasting, it's worth noting that both additive and multiplicative adjustments may become exaggerated over time, and require <em>damping</em> that reduces the size of the trend over future periods until it reaches a flat line.
```
from statsmodels.tsa.holtwinters import ExponentialSmoothing
df['DESadd12'] = ExponentialSmoothing(df['Thousands of Passengers'], trend='add').fit().fittedvalues.shift(-1)
df.head()
df[['Thousands of Passengers','EWMA12','DESadd12']].iloc[:24].plot(figsize=(12,6)).autoscale(axis='x',tight=True);
```
Here we can see that Double Exponential Smoothing is a much better representation of the time series data.<br>
Let's see if using a multiplicative seasonal adjustment helps.
```
df['DESmul12'] = ExponentialSmoothing(df['Thousands of Passengers'], trend='mul').fit().fittedvalues.shift(-1)
df.head()
df[['Thousands of Passengers','DESadd12','DESmul12']].iloc[:24].plot(figsize=(12,6)).autoscale(axis='x',tight=True);
```
Although minor, it does appear that a multiplicative adjustment gives better results. Note that the green line almost completely overlaps the original data.
___
## Triple Exponential Smoothing
Triple Exponential Smoothing, the method most closely associated with Holt-Winters, adds support for both trends and seasonality in the data.
```
df['TESadd12'] = ExponentialSmoothing(df['Thousands of Passengers'],trend='add',seasonal='add',seasonal_periods=12).fit().fittedvalues
df.head()
df['TESmul12'] = ExponentialSmoothing(df['Thousands of Passengers'],trend='mul',seasonal='mul',seasonal_periods=12).fit().fittedvalues
df.head()
df[['Thousands of Passengers','TESadd12','TESmul12']].plot(figsize=(12,6)).autoscale(axis='x',tight=True);
df[['Thousands of Passengers','TESadd12','TESmul12']].iloc[:24].plot(figsize=(12,6)).autoscale(axis='x',tight=True);
```
Based on the plot above, you might think that Triple Exponential Smoothing does a poorer job of fitting than Double Exponential Smoothing. The key here is to consider what comes next - <em>forecasting</em>. We'll see that having the ability to predict fluctuating seasonal patterns greatly improves our forecast.
But first, we'll work out some time series exercises before moving on to forecasting.
|
github_jupyter
|
import pandas as pd
import numpy as np
%matplotlib inline
df = pd.read_csv('../Data/airline_passengers.csv',index_col='Month',parse_dates=True)
df.dropna(inplace=True)
df.index
df.index.freq = 'MS'
df.index
df.head()
from statsmodels.tsa.holtwinters import SimpleExpSmoothing
span = 12
alpha = 2/(span+1)
df['EWMA12'] = df['Thousands of Passengers'].ewm(alpha=alpha,adjust=False).mean()
df['SES12']=SimpleExpSmoothing(df['Thousands of Passengers']).fit(smoothing_level=alpha,optimized=False).fittedvalues.shift(-1)
df.head()
from statsmodels.tsa.holtwinters import ExponentialSmoothing
df['DESadd12'] = ExponentialSmoothing(df['Thousands of Passengers'], trend='add').fit().fittedvalues.shift(-1)
df.head()
df[['Thousands of Passengers','EWMA12','DESadd12']].iloc[:24].plot(figsize=(12,6)).autoscale(axis='x',tight=True);
df['DESmul12'] = ExponentialSmoothing(df['Thousands of Passengers'], trend='mul').fit().fittedvalues.shift(-1)
df.head()
df[['Thousands of Passengers','DESadd12','DESmul12']].iloc[:24].plot(figsize=(12,6)).autoscale(axis='x',tight=True);
df['TESadd12'] = ExponentialSmoothing(df['Thousands of Passengers'],trend='add',seasonal='add',seasonal_periods=12).fit().fittedvalues
df.head()
df['TESmul12'] = ExponentialSmoothing(df['Thousands of Passengers'],trend='mul',seasonal='mul',seasonal_periods=12).fit().fittedvalues
df.head()
df[['Thousands of Passengers','TESadd12','TESmul12']].plot(figsize=(12,6)).autoscale(axis='x',tight=True);
df[['Thousands of Passengers','TESadd12','TESmul12']].iloc[:24].plot(figsize=(12,6)).autoscale(axis='x',tight=True);
| 0.360377 | 0.987302 |
# Lake model continued
In the previous week you used the lake problem as a means of getting aquinted with the workbench. In this assignment we will continue with the lake problem, focussing explicitly on using it for open exploration. You can use the second part of [this tutoria](https://emaworkbench.readthedocs.io/en/latest/indepth_tutorial/open-exploration.html) for help.
**It is paramount that you are using the lake problem with 100 decision variables, rather than the one found on the website with the seperate anthropogenic release decision**
## Apply sensitivity analysis
There is substantial support in the ema_workbench for global sensitivity. For this, the workbench relies on [SALib](https://salib.readthedocs.io/en/latest/) and feature scoring which is a machine learning alternative for global sensitivity analysis.
1. Apply Sobol with 3 seperate release policies (0, 0.05, and 0.1) and analyse the results for each release policy seperately focusing on the reliability objective. Do the sensitivities change depending on the release policy? Can you explain why or why not?
*hint: you can use sobol sampling for the uncertainties, and set policies to a list with the 3 different release policies. Next, for the analysis using logical indexing on the experiment.policy column you can select the results for each seperate release policy and apply sobol to each of the three seperate release policies. If this sounds too complicated, just do it on each release policy seperately.*
```
from scipy.optimize import brentq
from SALib.analyze import sobol
from ema_workbench import (Model, RealParameter, ScalarOutcome, Constant,
ema_logging, MultiprocessingEvaluator, Policy, SequentialEvaluator)
from ema_workbench.em_framework.evaluators import SOBOL, LHS
from ema_workbench.em_framework import get_SALib_problem
from ema_workbench import Policy, perform_experiments
import pandas as pd
from SALib.analyze.sobol import analyze
from ema_workbench.analysis import feature_scoring
import seaborn as sns
import matplotlib.pyplot as plt
from lakemodel_function import lake_problem
from ema_workbench import (Model, RealParameter, ScalarOutcome)
#instantiate the model
lake_model = Model('lakeproblem', function=lake_problem)
lake_model.time_horizon = 100 # used to specify the number of timesteps
#specify uncertainties
lake_model.uncertainties = [RealParameter('mean', 0.01, 0.05),
RealParameter('stdev', 0.001, 0.005),
RealParameter('b', 0.1, 0.45),
RealParameter('q', 2.0, 4.5),
RealParameter('delta', 0.93, 0.99)]
# set levers, one for each time step
lake_model.levers = [RealParameter(f"l{i}", 0, 0.1) for i in
range(lake_model.time_horizon)] # we use time_horizon here
#specify outcomes
lake_model.outcomes = [ScalarOutcome('max_P'),
ScalarOutcome('utility'),
ScalarOutcome('inertia'),
ScalarOutcome('reliability')]
policy = [Policy('0', **{l.name:0 for l in lake_model.levers}),
Policy('0.05', **{l.name:0.05 for l in lake_model.levers}),
Policy('0.1', **{l.name:0.1 for l in lake_model.levers})]
n_scenarios = 1000
ema_logging.log_to_stderr(ema_logging.INFO)
with MultiprocessingEvaluator(lake_model) as evaluator:
results = evaluator.perform_experiments(n_scenarios, policy,
uncertainty_sampling=SOBOL)
experiments_sobol, outcomes_sobol = results
problem = get_SALib_problem(lake_model.uncertainties)
y = outcomes_sobol['reliability']
sobol_indices = sobol.analyze(problem, y)
sobol_indices.to_df()[0]
```
## Analysing all policies using Sobol
```
sobol_results = {}
for policy in experiments_sobol.policy.unique():
logical = experiments_sobol.policy == policy
y = outcomes_sobol['reliability'][logical]
indices = analyze(problem,y)
sobol_results[policy] = indices
sobol_p0 = pd.concat([sobol_results['0'].to_df()[0],sobol_results['0'].to_df()[1]], axis = 1)
sobol_p005 = pd.concat([sobol_results['0.05'].to_df()[0],sobol_results['0.05'].to_df()[1]], axis = 1)
sobol_p01 = pd.concat([sobol_results['0.1'].to_df()[0],sobol_results['0.1'].to_df()[1]], axis = 1)
sobol_p0.columns = ['ST0', 'ST_conf0', 'S10', 'S1_conf0']
sobol_p005.columns = ['ST005', 'ST_conf005', 'S1005', 'S1_conf005']
sobol_p01.columns = ['ST01', 'ST_conf01', 'S101', 'S1_conf01']
sobol_results_df = pd.concat([sobol_p0,sobol_p005,sobol_p01], axis = 1)
#sobol_results_df = sobol_results_df[['ST0', 'S10', 'ST005', 'S1005', 'ST01', 'S101']]
sobol_results_df
sns.heatmap(sobol_results_df[['ST0', 'ST005', 'ST01']], annot=True, cmap='viridis')
```
#### Conclusions
Total sensitivity
- The increase in release changes the sensitivity to each of the uncertainties, albiet very small.
- Overall, the sensitivity increases to b
- The sensitivity to std deviation and q falls and rises again
- The sensitivity to mean and delta decreases
Individual Indices
- The individual indices don't show the same pattern
- The interactiion effect has increased even if it is due to one uncertainty
The confidence intervals of all the policies remain similar to each other. It is possible that the sensitivities don't change drastically because the release does not change a lot across these policies.
The change however could be attrubuted to the fact that the policy brings with additional uncertainty/changes to the system and the outcome is sensitive to this change.
b : lakes' natural removal rate
delta : discount rate for economic activity
mean : mean of natural pollution
q : lakes' natural recycling rate
stdev : std dev of natural pollution
```
sns.heatmap(sobol_results_df[['S10', 'S1005', 'S101']], annot=True, cmap='viridis')
```
2. Repeat the above analysis for the 3 release policies but now with extra trees feature scoring and for all outcomes of interest. As a bonus, use the sobol experiment results as input for extra trees, and compare the results with those resulting from latin hypercube sampling.
*hint: you can use [seaborn heatmaps](https://seaborn.pydata.org/generated/seaborn.heatmap.html) for a nice figure of the results*
## Analysing using extra trees
```
n_scenarios = 1000
ema_logging.log_to_stderr(ema_logging.INFO)
with MultiprocessingEvaluator(lake_model) as evaluator:
results_lhs = evaluator.perform_experiments(n_scenarios, policy,
uncertainty_sampling=LHS)
experiments_lhs, outcomes_lhs = results_lhs
cleaned_experiments_lhs = experiments_lhs.drop(columns=[l.name for l in lake_model.levers])
scores_lhs = {}
for key in outcomes_lhs.keys():
for policy in experiments_lhs.policy.unique():
logical = experiments_lhs.policy == policy
subset_results = {k:v[logical] for k,v in outcomes_lhs.items()}
scores_lhs[policy] = feature_scoring.get_feature_scores_all(cleaned_experiments_lhs[logical], subset_results)
cleaned_experiments_sobol = experiments_sobol.drop(columns=[l.name for l in lake_model.levers])
scores_sobol = {}
for key in outcomes_sobol.keys():
for policy in experiments_sobol.policy.unique():
logical = experiments_sobol.policy == policy
subset_results = {k:v[logical] for k,v in outcomes_sobol.items()}
scores_sobol[policy] = feature_scoring.get_feature_scores_all(cleaned_experiments_sobol[logical], subset_results)
scores_lhs['0']
```
### Heat maps comparing all three policies using extra trees and Sobol
```
sns.heatmap(scores_lhs['0'], annot=True, cmap='viridis')
plt.show()
sns.heatmap(scores_lhs['0.05'], annot=True, cmap='viridis')
plt.show()
sns.heatmap(scores_lhs['0.1'], annot=True, cmap='viridis')
plt.show()
sns.heatmap(scores_sobol['0.1'], annot=True, cmap='viridis')
plt.show()
```
|
github_jupyter
|
from scipy.optimize import brentq
from SALib.analyze import sobol
from ema_workbench import (Model, RealParameter, ScalarOutcome, Constant,
ema_logging, MultiprocessingEvaluator, Policy, SequentialEvaluator)
from ema_workbench.em_framework.evaluators import SOBOL, LHS
from ema_workbench.em_framework import get_SALib_problem
from ema_workbench import Policy, perform_experiments
import pandas as pd
from SALib.analyze.sobol import analyze
from ema_workbench.analysis import feature_scoring
import seaborn as sns
import matplotlib.pyplot as plt
from lakemodel_function import lake_problem
from ema_workbench import (Model, RealParameter, ScalarOutcome)
#instantiate the model
lake_model = Model('lakeproblem', function=lake_problem)
lake_model.time_horizon = 100 # used to specify the number of timesteps
#specify uncertainties
lake_model.uncertainties = [RealParameter('mean', 0.01, 0.05),
RealParameter('stdev', 0.001, 0.005),
RealParameter('b', 0.1, 0.45),
RealParameter('q', 2.0, 4.5),
RealParameter('delta', 0.93, 0.99)]
# set levers, one for each time step
lake_model.levers = [RealParameter(f"l{i}", 0, 0.1) for i in
range(lake_model.time_horizon)] # we use time_horizon here
#specify outcomes
lake_model.outcomes = [ScalarOutcome('max_P'),
ScalarOutcome('utility'),
ScalarOutcome('inertia'),
ScalarOutcome('reliability')]
policy = [Policy('0', **{l.name:0 for l in lake_model.levers}),
Policy('0.05', **{l.name:0.05 for l in lake_model.levers}),
Policy('0.1', **{l.name:0.1 for l in lake_model.levers})]
n_scenarios = 1000
ema_logging.log_to_stderr(ema_logging.INFO)
with MultiprocessingEvaluator(lake_model) as evaluator:
results = evaluator.perform_experiments(n_scenarios, policy,
uncertainty_sampling=SOBOL)
experiments_sobol, outcomes_sobol = results
problem = get_SALib_problem(lake_model.uncertainties)
y = outcomes_sobol['reliability']
sobol_indices = sobol.analyze(problem, y)
sobol_indices.to_df()[0]
sobol_results = {}
for policy in experiments_sobol.policy.unique():
logical = experiments_sobol.policy == policy
y = outcomes_sobol['reliability'][logical]
indices = analyze(problem,y)
sobol_results[policy] = indices
sobol_p0 = pd.concat([sobol_results['0'].to_df()[0],sobol_results['0'].to_df()[1]], axis = 1)
sobol_p005 = pd.concat([sobol_results['0.05'].to_df()[0],sobol_results['0.05'].to_df()[1]], axis = 1)
sobol_p01 = pd.concat([sobol_results['0.1'].to_df()[0],sobol_results['0.1'].to_df()[1]], axis = 1)
sobol_p0.columns = ['ST0', 'ST_conf0', 'S10', 'S1_conf0']
sobol_p005.columns = ['ST005', 'ST_conf005', 'S1005', 'S1_conf005']
sobol_p01.columns = ['ST01', 'ST_conf01', 'S101', 'S1_conf01']
sobol_results_df = pd.concat([sobol_p0,sobol_p005,sobol_p01], axis = 1)
#sobol_results_df = sobol_results_df[['ST0', 'S10', 'ST005', 'S1005', 'ST01', 'S101']]
sobol_results_df
sns.heatmap(sobol_results_df[['ST0', 'ST005', 'ST01']], annot=True, cmap='viridis')
sns.heatmap(sobol_results_df[['S10', 'S1005', 'S101']], annot=True, cmap='viridis')
n_scenarios = 1000
ema_logging.log_to_stderr(ema_logging.INFO)
with MultiprocessingEvaluator(lake_model) as evaluator:
results_lhs = evaluator.perform_experiments(n_scenarios, policy,
uncertainty_sampling=LHS)
experiments_lhs, outcomes_lhs = results_lhs
cleaned_experiments_lhs = experiments_lhs.drop(columns=[l.name for l in lake_model.levers])
scores_lhs = {}
for key in outcomes_lhs.keys():
for policy in experiments_lhs.policy.unique():
logical = experiments_lhs.policy == policy
subset_results = {k:v[logical] for k,v in outcomes_lhs.items()}
scores_lhs[policy] = feature_scoring.get_feature_scores_all(cleaned_experiments_lhs[logical], subset_results)
cleaned_experiments_sobol = experiments_sobol.drop(columns=[l.name for l in lake_model.levers])
scores_sobol = {}
for key in outcomes_sobol.keys():
for policy in experiments_sobol.policy.unique():
logical = experiments_sobol.policy == policy
subset_results = {k:v[logical] for k,v in outcomes_sobol.items()}
scores_sobol[policy] = feature_scoring.get_feature_scores_all(cleaned_experiments_sobol[logical], subset_results)
scores_lhs['0']
sns.heatmap(scores_lhs['0'], annot=True, cmap='viridis')
plt.show()
sns.heatmap(scores_lhs['0.05'], annot=True, cmap='viridis')
plt.show()
sns.heatmap(scores_lhs['0.1'], annot=True, cmap='viridis')
plt.show()
sns.heatmap(scores_sobol['0.1'], annot=True, cmap='viridis')
plt.show()
| 0.612078 | 0.922622 |
# Risk sharing, moral hazard and credit rationing
**Note:** this jupyter notebook mixes text, math, visualizations and python code. To keep things uncluttered most of the code was placed in a [code section](#codesection) at the end. If you are running on a jupyter server and want to recreate or modify content or run interactive widgets, navigate to the code section for instructions first.
**Slideshow mode**: this notebook can be viewed as a slideshow by pressing Alt-R if run on a server.
A farmer has access to a risky asset, say a pineapple tree.
If diligent (e.g. tree is watered/fertilized) it will generate state-contingent claims:
- $x_1$ in state of the world 1 with probability $p$
- $x_0<x_1$ in state of the world 0, with probability $(1-p)$
- For expected return:
$$E(x|p) = p \cdot x_1 + (1-p) \cdot x_0$$
Note: we motivate with the two-outcome case but the notation allows for adaptation to the more general case of $N$ outcomes $x_i$ and a finite number $M$ (or infinite continuum) of effort or action/diligence levels $e$ that shift the probability density function of $x_i$:
$$E(x|e) = \sum_i {x_i \cdot f(x_i|e)}$$
### Financial Contracting with action-contingent contracts
Action/diligence is observable and verifiable.
Contracts include enforceable clauses of the form: if farmer fails to be diligent they pay a heavy penalty.
### Financial contracting under Competition
- Risk averse farmer wants to smooth consumption.
- Wants to sell state-contingent claims $x_i$ in exchange for a smoother consumption bundle $(c_1, c_0)$.
- Financial Intermediaries (FIs) compete to offer contract
Competitive market contract maxes farmer's von-Neumann Morgenstern expected utility:
$$\max_{c_0,c_1} EU(c|p)$$
subject to FI's participation (or zero-profit) constraint:
$$E(x|p)-E(c|p) \geq 0$$
where $EU(c|p) = p u(c_1) + (1-p) u(c_0)$
#### Investment lending and risk
More interesting (but hardly changing the math that follows) assume also:
- Farmer must pay lump sum $I$ to initiate project (e.g. purchase fertilizer or pay land rent).
- Assume must borrow entirety from a FI.
**FI's participation (zero profit) constraint:$$
$$E(x|p)-E(c|p) \geq I(1+r)$$
where $r$ expresses the FI's opportunity cost of funds.
As a Lagrangean:
$$\mathcal{L}(c_1,c_2,\lambda) = EU(c|P) + \lambda [E(x|p) - E(c|p) + I(1+r)]$$
First-order necessary conditions (FOCs) for an optimum:
$$p \cdot u'(c_i) = p \cdot \lambda \ \ \ \forall i$$
- Implies $u'(c_i) = \lambda$ for each $i$
- optimum contract offers constant consumption $c_1 = c_2 = \bar c$.
### Financial contracting under Monopoly
- As above but objective and constraint reversed. Maximize FI's profits
$$\max_{c_0,c_1} E(x|p)-E(c|p) $$
subject to farmers participation constraint:
$$ EU(c|p) \geq \bar u$$
where $bar u$ is their reservation utility, or expected utility from next best choice.
FOCs essentially same as competitive case but now substituted into different constraint.
## The consumer's optimum
$$L(c_1,c_2) = U(c_1,c_2) + \lambda (I - p_1 c_1 - p_2 c_2) $$
Differentiate with respect to $c_1$ and $c_2$ and $\lambda$ to get:
$$ U_1 = \lambda{p_1}$$
$$ U_2 = \lambda{p_2}$$
$$ I = p_1 c_1 + p_2 c_2$$
Dividing the first equation by the second we get the familiar necessary tangency condition for an interior optimum:
$$MRS = \frac{U_1}{U_2} =\frac{p_1}{p_2}$$
Using our earlier expression for the MRS of a Cobb-Douglas indifference curve, substituting this into the budget constraint and rearranging then allows us to solve for the Marshallian demands:
$$c_1(p_1,p_2,I)=\alpha \frac{I}{p_1}$$
$$c_1(p_1,p_2,I)=(1-\alpha) \frac{I}{p_2}$$
# Moral Hazard
## Incentive Compatibility Constraint
Now two effort levels: Low $e_L$ (probability of success q) or High $e_H$ (probability of success p).
Agent's private benefit from avoiding diligence or effort is B.
Effort is non-contractible and B cannot be observed/seized. Incentive compatibility constraint:
$$EU(c|p) \geq EU(c|q) + B$$
In 2 outcome case can be re-arranged to:
$$u(c_1) \geq u(c_0) + \frac{B}{p-q}$$
```
consume_plot(p,q,B, 0,ic=True)
```
The following interactive widget will be visible and active only if you are running this on a jupyter server.
```
interact(consume_plot,p=fixed(0.5),q=(0.1,0.5,0.01),B=(0,3,0.1),oassets=(0,60,10));
```
If we set this up and solve it as a Lagrangean (loosely following Holmstrom, 1979) we get a condition like this:
$$\frac{1}{u'(c_i)} = \lambda + \mu \cdot
\left [ {1-\frac{f(x_i,e_L)}{f(x_i,e_H)}} \right ] \text{ }\forall i
$$
In our two outcome case $p=f(x_1|e_H)$ and $q=f(x_1|e_L)$ and this becomes:
$$\frac{1}{u'(c_1)} = \lambda + \mu \cdot
\left [ {1-\frac{q}{p}} \right ]
$$
$$\frac{1}{u'(c_0)} = \lambda + \mu \cdot
\left [ {1-\frac{1-q}{1-p}} \right ]
$$
TODO:
- Functions to solve the two outcome cases (closed form possible, substitute IC into binding PC; or 2 FOC plus IC plus PC for $c_0, c_1, \lambda \text{ and } \mu$).
- Function to solve numerically for N outcomes (N FOCs and one participation constraint).
- discuss how sensitive to distribution
Holmstrom's sufficient statistic
$$\frac{1}{u'(c)} = \lambda + \mu \cdot
\left [ {1-\frac{f(x,y,e_L)}{f(x,y,e_H)}} \right ] \text{ }\forall i
$$
<a id='codesection'></a>
## Code Section
**Note:** To re-create or modify any content go to the 'Cell' menu above run all code cells below by choosing 'Run All Below'. Then 'Run all Above' to recreate all output above (or go to the top and step through each code cell manually).
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
from ipywidgets import interact, fixed
```
### Assumed parameters
```
alpha = 0.25
def u(c, alpha=alpha):
return (1/alpha)*c**alpha
def E(x,p):
return p*x[1] + (1-p)*x[0]
def EU(c, p):
return p*u(c[1]) + (1-p)*u(c[0])
def budgetc(c0, p, x):
return E(x,p)/p - ((1-p)/p)*c0
def indif(c0, p, ubar):
return (alpha*(ubar-(1-p)*u(c0))/p)**(1/alpha)
def IC(c0,p,q,B):
'''incentive compatibility line'''
return (alpha*(u(c0)+B/(p-q)))**(1/alpha)
def Bopt(p,x):
'''Bank profit maximum'''
return (alpha*EU(x,p))**(1/alpha)
def Copt(p,x):
'''Consumer utility maximum'''
return E(x,p)
x = [15,90]
p = 0.6
```
## Interactive indifference curve diagram
Default parameters for example
```
p = 0.5
q = 0.4
cmax = 100
B = 1
def consume_plot(p,q,B, oassets,ic=False):
c0 = np.linspace(0.1,200,num=100)
#bank optimum
c0e = Bopt(p,x)
c1e = c0e
uebar = EU([c0e,c1e],p)
idfc = indif(c0, p, uebar)
budg = budgetc(c0, p, [c0e,c1e])
#consumer optimum
c0ee = Copt(p,x)
c1ee = c0ee
uemax = EU([c0ee,c1ee],p)
idfcmax = indif(c0, p, uemax)
zerop = budgetc(c0, p, x)
icline = IC(c0,p,q,B)
fig, ax = plt.subplots(figsize=(8,8))
if ic:
ax.plot(c0,icline)
ax.plot(c0, budg, lw=2.5)
ax.plot(c0, zerop, lw=2.5)
ax.plot(c0, idfc, lw=2.5)
ax.plot(c0, idfcmax, lw=2.5)
ax.plot(c0,c0)
#ax.vlines(c0e,0,c2e, linestyles="dashed")
#ax.hlines(c1e,0,c1e, linestyles="dashed")
ax.plot(c0e,c1e,'ob')
ax.plot(c0ee,c1ee,'ob')
ax.plot(x[0],x[1],'ob')
ax.set_xlim(0, 110)
ax.set_ylim(0, 110)
ax.set_xlabel(r'$c_0$', fontsize=16)
ax.set_ylabel('$c_1$', fontsize=16)
ax.spines['right'].set_visible(False)
ax.spines['top'].set_visible(False)
ax.grid()
xticks = ax.get_xticks()-oassets
ax.set_xticklabels(xticks)
ax.set_yticklabels(xticks)
plt.show()
consume_plot(p,q,B, 0,ic=True)
```
|
github_jupyter
|
consume_plot(p,q,B, 0,ic=True)
interact(consume_plot,p=fixed(0.5),q=(0.1,0.5,0.01),B=(0,3,0.1),oassets=(0,60,10));
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
from ipywidgets import interact, fixed
alpha = 0.25
def u(c, alpha=alpha):
return (1/alpha)*c**alpha
def E(x,p):
return p*x[1] + (1-p)*x[0]
def EU(c, p):
return p*u(c[1]) + (1-p)*u(c[0])
def budgetc(c0, p, x):
return E(x,p)/p - ((1-p)/p)*c0
def indif(c0, p, ubar):
return (alpha*(ubar-(1-p)*u(c0))/p)**(1/alpha)
def IC(c0,p,q,B):
'''incentive compatibility line'''
return (alpha*(u(c0)+B/(p-q)))**(1/alpha)
def Bopt(p,x):
'''Bank profit maximum'''
return (alpha*EU(x,p))**(1/alpha)
def Copt(p,x):
'''Consumer utility maximum'''
return E(x,p)
x = [15,90]
p = 0.6
p = 0.5
q = 0.4
cmax = 100
B = 1
def consume_plot(p,q,B, oassets,ic=False):
c0 = np.linspace(0.1,200,num=100)
#bank optimum
c0e = Bopt(p,x)
c1e = c0e
uebar = EU([c0e,c1e],p)
idfc = indif(c0, p, uebar)
budg = budgetc(c0, p, [c0e,c1e])
#consumer optimum
c0ee = Copt(p,x)
c1ee = c0ee
uemax = EU([c0ee,c1ee],p)
idfcmax = indif(c0, p, uemax)
zerop = budgetc(c0, p, x)
icline = IC(c0,p,q,B)
fig, ax = plt.subplots(figsize=(8,8))
if ic:
ax.plot(c0,icline)
ax.plot(c0, budg, lw=2.5)
ax.plot(c0, zerop, lw=2.5)
ax.plot(c0, idfc, lw=2.5)
ax.plot(c0, idfcmax, lw=2.5)
ax.plot(c0,c0)
#ax.vlines(c0e,0,c2e, linestyles="dashed")
#ax.hlines(c1e,0,c1e, linestyles="dashed")
ax.plot(c0e,c1e,'ob')
ax.plot(c0ee,c1ee,'ob')
ax.plot(x[0],x[1],'ob')
ax.set_xlim(0, 110)
ax.set_ylim(0, 110)
ax.set_xlabel(r'$c_0$', fontsize=16)
ax.set_ylabel('$c_1$', fontsize=16)
ax.spines['right'].set_visible(False)
ax.spines['top'].set_visible(False)
ax.grid()
xticks = ax.get_xticks()-oassets
ax.set_xticklabels(xticks)
ax.set_yticklabels(xticks)
plt.show()
consume_plot(p,q,B, 0,ic=True)
| 0.386416 | 0.978753 |
# Polynomial Part 2
```
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import warnings
warnings.filterwarnings("ignore")
# fix_yahoo_finance is used to fetch data
import fix_yahoo_finance as yf
yf.pdr_override()
# input
symbol = 'AMD'
start = '2014-01-01'
end = '2019-01-01'
# Read data
dataset = yf.download(symbol,start,end)
# View Columns
dataset.head()
dataset['Increase_Decrease'] = np.where(dataset['Volume'].shift(-1) > dataset['Volume'],1,0)
dataset['Buy_Sell_on_Open'] = np.where(dataset['Open'].shift(-1) > dataset['Open'],1,0)
dataset['Buy_Sell'] = np.where(dataset['Adj Close'].shift(-1) > dataset['Adj Close'],1,0)
dataset['Returns'] = dataset['Adj Close'].pct_change()
dataset = dataset.dropna()
dataset.head()
dataset.shape
X = dataset['Open'].values
y = dataset['Adj Close'].values
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25)
plt.plot(X_train, y_train, 'bo')
plt.plot(X_test, np.zeros_like(X_test), 'r+')
from sklearn.preprocessing import PolynomialFeatures
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(X_train.reshape(-1, 1), y_train)
preds = model.predict(X_test.reshape(-1, 1))
preds[:3]
# fit-predict-evaluate a 1D polynomial (a line)
model_one = np.poly1d(np.polyfit(X_train, y_train,
1))
preds_one = model_one(X_test)
print(preds_one[:3])
```
# Polynomial
```
from sklearn.metrics import mean_squared_error
# the predictions come back the same
print("all close?", np.allclose(preds, preds_one))
# and we can still use sklearn to evaluate it
mse = mean_squared_error
print("RMSE:", np.sqrt(mse(y_test, preds_one)))
# fit-predict-evaluate a 2D polynomial (a parabola)
model_two = np.poly1d(np.polyfit(X_train, y_train,
2))
preds_two = model_two(X_test)
print("RMSE:", np.sqrt(mse(y_test, preds_two)))
model_three = np.poly1d(np.polyfit(X_train, y_train, 9))
preds_three = model_three(X_test)
print("RMSE:", np.sqrt(mse(y_test, preds_three)))
fig, axes = plt.subplots(1, 2, figsize=(6, 3),sharey=True)
labels = ['line', 'parabola', 'nonic']
models = [model_one, model_two, model_three]
train = (X_train, y_train)
test = (X_test, y_test)
for ax, (ftr, tgt) in zip(axes, [train, test]):
ax.plot(ftr, tgt, 'k+')
for m, lbl in zip(models, labels):
ftr = sorted(ftr)
ax.plot(ftr, m(ftr), '-', label=lbl)
axes[1].set_ylim(-20, 200)
axes[0].set_title("Train")
axes[1].set_title("Test");
axes[0].legend(loc='upper center');
results = []
for complexity in [1, 2, 3, 4, 5, 6,7,8, 9]:
model = np.poly1d(np.polyfit(X_train, y_train, complexity))
train_error = np.sqrt(mse(y_train, model(X_train)))
test_error = np.sqrt(mse(y_test,model(X_test)))
results.append((complexity, train_error, test_error))
columns = ["Complexity", "Train Error", "Test Error"]
results_df = pd.DataFrame.from_records(results, columns=columns,index="Complexity")
results_df
results_df.plot()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import warnings
warnings.filterwarnings("ignore")
# fix_yahoo_finance is used to fetch data
import fix_yahoo_finance as yf
yf.pdr_override()
# input
symbol = 'AMD'
start = '2014-01-01'
end = '2019-01-01'
# Read data
dataset = yf.download(symbol,start,end)
# View Columns
dataset.head()
dataset['Increase_Decrease'] = np.where(dataset['Volume'].shift(-1) > dataset['Volume'],1,0)
dataset['Buy_Sell_on_Open'] = np.where(dataset['Open'].shift(-1) > dataset['Open'],1,0)
dataset['Buy_Sell'] = np.where(dataset['Adj Close'].shift(-1) > dataset['Adj Close'],1,0)
dataset['Returns'] = dataset['Adj Close'].pct_change()
dataset = dataset.dropna()
dataset.head()
dataset.shape
X = dataset['Open'].values
y = dataset['Adj Close'].values
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25)
plt.plot(X_train, y_train, 'bo')
plt.plot(X_test, np.zeros_like(X_test), 'r+')
from sklearn.preprocessing import PolynomialFeatures
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(X_train.reshape(-1, 1), y_train)
preds = model.predict(X_test.reshape(-1, 1))
preds[:3]
# fit-predict-evaluate a 1D polynomial (a line)
model_one = np.poly1d(np.polyfit(X_train, y_train,
1))
preds_one = model_one(X_test)
print(preds_one[:3])
from sklearn.metrics import mean_squared_error
# the predictions come back the same
print("all close?", np.allclose(preds, preds_one))
# and we can still use sklearn to evaluate it
mse = mean_squared_error
print("RMSE:", np.sqrt(mse(y_test, preds_one)))
# fit-predict-evaluate a 2D polynomial (a parabola)
model_two = np.poly1d(np.polyfit(X_train, y_train,
2))
preds_two = model_two(X_test)
print("RMSE:", np.sqrt(mse(y_test, preds_two)))
model_three = np.poly1d(np.polyfit(X_train, y_train, 9))
preds_three = model_three(X_test)
print("RMSE:", np.sqrt(mse(y_test, preds_three)))
fig, axes = plt.subplots(1, 2, figsize=(6, 3),sharey=True)
labels = ['line', 'parabola', 'nonic']
models = [model_one, model_two, model_three]
train = (X_train, y_train)
test = (X_test, y_test)
for ax, (ftr, tgt) in zip(axes, [train, test]):
ax.plot(ftr, tgt, 'k+')
for m, lbl in zip(models, labels):
ftr = sorted(ftr)
ax.plot(ftr, m(ftr), '-', label=lbl)
axes[1].set_ylim(-20, 200)
axes[0].set_title("Train")
axes[1].set_title("Test");
axes[0].legend(loc='upper center');
results = []
for complexity in [1, 2, 3, 4, 5, 6,7,8, 9]:
model = np.poly1d(np.polyfit(X_train, y_train, complexity))
train_error = np.sqrt(mse(y_train, model(X_train)))
test_error = np.sqrt(mse(y_test,model(X_test)))
results.append((complexity, train_error, test_error))
columns = ["Complexity", "Train Error", "Test Error"]
results_df = pd.DataFrame.from_records(results, columns=columns,index="Complexity")
results_df
results_df.plot()
| 0.57093 | 0.851459 |
```
#hide
# default_exp conda
```
# Create conda packages
> Pure python packages created from nbdev settings.ini
```
#export
from fastscript import *
from fastcore.all import *
from fastrelease.core import find_config
import yaml,subprocess
from copy import deepcopy
try: from packaging.version import parse
except ImportError: from pip._vendor.packaging.version import parse
_PYPI_URL = 'https://pypi.org/pypi/'
#export
def pypi_json(s):
"Dictionary decoded JSON for PYPI path `s`"
return urljson(f'{_PYPI_URL}{s}/json')
#export
def latest_pypi(name):
"Latest version of `name` on pypi"
return max(parse(r) for r,o in pypi_json(name)['releases'].items()
if not parse(r).is_prerelease and not o[0]['yanked'])
#export
def _pip_conda_meta(name):
ver = str(latest_pypi('sentencepiece'))
pypi = pypi_json(f'{name}/{ver}')
info = pypi['info']
rel = [o for o in pypi['urls'] if o['packagetype']=='sdist'][0]
reqs = ['pip', 'python', 'packaging']
# Work around conda build bug - 'package' and 'source' must be first
d1 = {
'package': {'name': name, 'version': ver},
'source': {'url':rel['url'], 'sha256':rel['digests']['sha256']}
}
d2 = {
'build': {'number': '0', 'noarch': 'python',
'script': '{{ PYTHON }} -m pip install . -vv'},
'test': {'imports': [name]},
'requirements': {'host':reqs, 'run':reqs},
'about': {'license': info['license'], 'home': info['project_url'], 'summary': info['summary']}
}
return d1,d2
#export
def _write_yaml(path, name, d1, d2):
path = Path(path)
p = path/name
p.mkdir(exist_ok=True, parents=True)
yaml.SafeDumper.ignore_aliases = lambda *args : True
with (p/'meta.yaml').open('w') as f:
yaml.safe_dump(d1, f)
yaml.safe_dump(d2, f)
#export
def write_pip_conda_meta(name, path='conda'):
"Writes a `meta.yaml` file for `name` to the `conda` directory of the current directory"
_write_yaml(path, name, *_pip_conda_meta(name))
#export
def _get_conda_meta():
cfg,cfg_path = find_config()
name,ver = cfg.get('lib_name'),cfg.get('version')
url = cfg.get('doc_host') or cfg.get('git_url')
reqs = ['pip', 'python', 'packaging']
if cfg.get('requirements'): reqs += cfg.get('requirements').split()
if cfg.get('conda_requirements'): reqs += cfg.get('conda_requirements').split()
pypi = pypi_json(f'{name}/{ver}')
rel = [o for o in pypi['urls'] if o['packagetype']=='sdist'][0]
# Work around conda build bug - 'package' and 'source' must be first
d1 = {
'package': {'name': name, 'version': ver},
'source': {'url':rel['url'], 'sha256':rel['digests']['sha256']}
}
d2 = {
'build': {'number': '0', 'noarch': 'python',
'script': '{{ PYTHON }} -m pip install . -vv'},
'requirements': {'host':reqs, 'run':reqs},
'test': {'imports': [name]},
'about': {
'license': 'Apache Software',
'license_family': 'APACHE',
'home': url, 'doc_url': url, 'dev_url': url,
'summary': cfg.get('description')
},
'extra': {'recipe-maintainers': [cfg.get('user')]}
}
return name,d1,d2
#export
def write_conda_meta(path='conda'):
"Writes a `meta.yaml` file to the `conda` directory of the current directory"
_write_yaml(path, *_get_conda_meta())
```
This function is used in the `fastrelease_conda_package` CLI command.
**NB**: you need to first of all upload your package to PyPi, before creating the conda package.
```
#export
@call_parse
def fastrelease_conda_package(path:Param("Path where package will be created", str)='conda',
do_build:Param("Run `conda build` step", bool_arg)=True,
build_args:Param("Additional args (as str) to send to `conda build`", str)='',
do_upload:Param("Run `anaconda upload` step", bool_arg)=True,
upload_user:Param("Optional user to upload package to")=None):
"Create a `meta.yaml` file ready to be built into a package, and optionally build and upload it"
write_conda_meta(path)
cfg,cfg_path = find_config()
name = cfg.get('lib_name')
out = f"Done. Next steps:\n```\`cd {path}\n"""
out_upl = f"anaconda upload build/noarch/{name}-{cfg.get('version')}-py_0.tar.bz2"
if not do_build:
print(f"{out}conda build .\n{out_upl}\n```")
return
os.chdir(path)
try: res = subprocess.check_output(f"conda build --output-folder build {build_args} .".split()).decode()
except subprocess.CalledProcessError as e: print(f"{e.output}\n\nBuild failed.")
if 'to anaconda.org' in res: return
if 'anaconda upload' not in res:
print(f"{res}\n\Build failed.")
return
upload_str = re.findall('(anaconda upload .*)', res)[0]
if upload_user: upload_str = upload_str.replace('anaconda upload ', f'anaconda upload -u {upload_user} ')
try: res = subprocess.check_output(upload_str.split(), stderr=STDOUT).decode()
except subprocess.CalledProcessError as e: print(f"{e.output}\n\nUpload failed.")
if 'Upload complete' not in res: print(f"{res}\n\nUpload failed.")
```
To build and upload a conda package, cd to the root of your repo, and then:
fastrelease_conda_package
Or to do things more manually:
```
fastrelease_conda_package --do_build false
cd conda
conda build {name}
anaconda upload $CONDA_PREFIX/conda-bld/noarch/{name}-{ver}-*.tar.bz2
```
Add `--debug` to the `conda build command` to debug any problems that occur. Note that the build step takes a few minutes. Add `-u {org_name}` to the `anaconda upload` command if you wish to upload to an organization, or pass `upload_user` to `fastrelease_conda_package`.
**NB**: you need to first of all upload your package to PyPi, before creating the conda package.
## Export-
```
#hide
from nbdev.export import notebook2script
notebook2script()
```
|
github_jupyter
|
#hide
# default_exp conda
#export
from fastscript import *
from fastcore.all import *
from fastrelease.core import find_config
import yaml,subprocess
from copy import deepcopy
try: from packaging.version import parse
except ImportError: from pip._vendor.packaging.version import parse
_PYPI_URL = 'https://pypi.org/pypi/'
#export
def pypi_json(s):
"Dictionary decoded JSON for PYPI path `s`"
return urljson(f'{_PYPI_URL}{s}/json')
#export
def latest_pypi(name):
"Latest version of `name` on pypi"
return max(parse(r) for r,o in pypi_json(name)['releases'].items()
if not parse(r).is_prerelease and not o[0]['yanked'])
#export
def _pip_conda_meta(name):
ver = str(latest_pypi('sentencepiece'))
pypi = pypi_json(f'{name}/{ver}')
info = pypi['info']
rel = [o for o in pypi['urls'] if o['packagetype']=='sdist'][0]
reqs = ['pip', 'python', 'packaging']
# Work around conda build bug - 'package' and 'source' must be first
d1 = {
'package': {'name': name, 'version': ver},
'source': {'url':rel['url'], 'sha256':rel['digests']['sha256']}
}
d2 = {
'build': {'number': '0', 'noarch': 'python',
'script': '{{ PYTHON }} -m pip install . -vv'},
'test': {'imports': [name]},
'requirements': {'host':reqs, 'run':reqs},
'about': {'license': info['license'], 'home': info['project_url'], 'summary': info['summary']}
}
return d1,d2
#export
def _write_yaml(path, name, d1, d2):
path = Path(path)
p = path/name
p.mkdir(exist_ok=True, parents=True)
yaml.SafeDumper.ignore_aliases = lambda *args : True
with (p/'meta.yaml').open('w') as f:
yaml.safe_dump(d1, f)
yaml.safe_dump(d2, f)
#export
def write_pip_conda_meta(name, path='conda'):
"Writes a `meta.yaml` file for `name` to the `conda` directory of the current directory"
_write_yaml(path, name, *_pip_conda_meta(name))
#export
def _get_conda_meta():
cfg,cfg_path = find_config()
name,ver = cfg.get('lib_name'),cfg.get('version')
url = cfg.get('doc_host') or cfg.get('git_url')
reqs = ['pip', 'python', 'packaging']
if cfg.get('requirements'): reqs += cfg.get('requirements').split()
if cfg.get('conda_requirements'): reqs += cfg.get('conda_requirements').split()
pypi = pypi_json(f'{name}/{ver}')
rel = [o for o in pypi['urls'] if o['packagetype']=='sdist'][0]
# Work around conda build bug - 'package' and 'source' must be first
d1 = {
'package': {'name': name, 'version': ver},
'source': {'url':rel['url'], 'sha256':rel['digests']['sha256']}
}
d2 = {
'build': {'number': '0', 'noarch': 'python',
'script': '{{ PYTHON }} -m pip install . -vv'},
'requirements': {'host':reqs, 'run':reqs},
'test': {'imports': [name]},
'about': {
'license': 'Apache Software',
'license_family': 'APACHE',
'home': url, 'doc_url': url, 'dev_url': url,
'summary': cfg.get('description')
},
'extra': {'recipe-maintainers': [cfg.get('user')]}
}
return name,d1,d2
#export
def write_conda_meta(path='conda'):
"Writes a `meta.yaml` file to the `conda` directory of the current directory"
_write_yaml(path, *_get_conda_meta())
#export
@call_parse
def fastrelease_conda_package(path:Param("Path where package will be created", str)='conda',
do_build:Param("Run `conda build` step", bool_arg)=True,
build_args:Param("Additional args (as str) to send to `conda build`", str)='',
do_upload:Param("Run `anaconda upload` step", bool_arg)=True,
upload_user:Param("Optional user to upload package to")=None):
"Create a `meta.yaml` file ready to be built into a package, and optionally build and upload it"
write_conda_meta(path)
cfg,cfg_path = find_config()
name = cfg.get('lib_name')
out = f"Done. Next steps:\n```\`cd {path}\n"""
out_upl = f"anaconda upload build/noarch/{name}-{cfg.get('version')}-py_0.tar.bz2"
if not do_build:
print(f"{out}conda build .\n{out_upl}\n```")
return
os.chdir(path)
try: res = subprocess.check_output(f"conda build --output-folder build {build_args} .".split()).decode()
except subprocess.CalledProcessError as e: print(f"{e.output}\n\nBuild failed.")
if 'to anaconda.org' in res: return
if 'anaconda upload' not in res:
print(f"{res}\n\Build failed.")
return
upload_str = re.findall('(anaconda upload .*)', res)[0]
if upload_user: upload_str = upload_str.replace('anaconda upload ', f'anaconda upload -u {upload_user} ')
try: res = subprocess.check_output(upload_str.split(), stderr=STDOUT).decode()
except subprocess.CalledProcessError as e: print(f"{e.output}\n\nUpload failed.")
if 'Upload complete' not in res: print(f"{res}\n\nUpload failed.")
fastrelease_conda_package --do_build false
cd conda
conda build {name}
anaconda upload $CONDA_PREFIX/conda-bld/noarch/{name}-{ver}-*.tar.bz2
#hide
from nbdev.export import notebook2script
notebook2script()
| 0.395484 | 0.564819 |
# **Exemples d'applications de la notion de graphe orienté**
Cette section du cours te permet de découvrir des applications de la notion de graphe orienté pour des problèmes concrets.
## **Exemple 1** Organisation de métiers
Sur le chantier d'un bâtiment plusieurs corps de métier doivent intervenir. Il y a des impératifs d'enchainement des différents métiers. Par exemple l'électricien veut intervenir après le peintre et le peintre veut intervenir après le plaquiste et le maçon. Pour l'organisateur du chantier il faut voir si les exigences de chacun sont compatibles et proposer une solution simple pour régler les conflits.
| métier | intervient après |
|:- |:- |
|électricien | peintre, carreleur, couvreur |
|peintre | maçon ,plaquiste |
|menuisier | maçon, carreleur, couvreur |
|maçon | couvreur |
|couvreur | charpentier |
|charpentier | maçon |
|carreleur | maçon, peintre, plombier |
|plombier| maçon , electricien |
|plaquiste | maçon, couvreur |
|soliste | tous les autres |
1. Dessine le graphe orienté dont les sommets sont les différents métiers et où un arc $A \rightarrow B$ signifie que $B$ veut intervenir après $A$.
2. Comment voit-on sur le graphe
* le métier qui intervient en premier ?
* le métier qui intervient en dernier ?
* les conflits ?
---
## **Exemple 2** Le problème des bidons
Dans le film Die Hard 3 (Une journée en enfer), les deux héros, John McClane et Zeus Carver, doivent résoudre l’énigme de Simon Gruber pour arrêter le compte à rebours d’une bombe. Voici l’énigme : « Sur la fontaine, il y a deux bidons : l’un a une contenance de 5 gallons, l’autre de 3 gallons. Remplissez l’un des bidons de 4 gallons d’eau exactement et placez-le sur la balance. La minuterie s’arrêtera. Soyez extrêmement précis : un gramme de plus ou de moins et c’est l’explosion ! ». Les nerfs de John McClane sont alors mis à rude épreuve pour trouver une solution. Il commence par remarquer très justement qu’on ne peut pas remplir le bidon de 3 gallons avec 4 gallons d’eau. Il faut donc trouver le moyen de mettre exactement 4 gallons d’eau dans le bidon de 5 gallons. Dans la scène du film (que vous pouvez consulter en français sur
https://www.youtube.com/watch?v=pmk2mNf9iqE), John commence par donner une première idée peu convaincante puisqu’elle termine par la nécessité de remplir le bidon de 3 gallons au tiers, ce qu’on ne sait faire précisément... Le film propose ensuite une
solution très partielle, coupée au montage. Appliquons donc des méthodes de graphes
orientés pour retrouver la meilleure solution possible que les héros appliquent pour
s’en sortir.
Une configuration du système correspond au volume d’eau contenu dans chacun des deux bidons. On peut donc représenter une telle configuration par une paire $(a , b)$ où a est le volume d’eau contenu dans le bidon de 5 gallons et b le volume d’eau contenu dans le bidon de 3 gallons, avec $0 \le a \le 5$ et $0 \le b \le 3$. Les actions élémentaires possibles du système sont de remplir un des deux bidons (qu’il soit initialement vide ou pas), vider un des deux bidons (qu’il soit initialement plein ou pas) et transférer le contenu d’un des bidons dans l’autre jusqu’à ce que ce dernier soit plein.
1. Ecrire en Python les fonctions qui correspondent à chacune des actions.
Chaque fonction a pour argument une configuration qui est un tuple $(a,b)$ et doit renvoyer la configuration obtenue après l'action.
```python
remplir_A (config)
remplir_B (config)
vider_A(config)
vider_B(config)
transfere_A_B(config)
transfere_B_A(config)
```
Ecris aussi une fonction
```python
est_terminale(config)
```
qui renvoie `True`si la configuration est de la forme $(4,b)$
```
# code à effacer version élèves
def remplir_A(config) :
a,b = config
return 5, b
def remplir_B(config) :
a,b = config
return a, 3
def vider_A(config) :
a,b = config
return 0, b
def vider_B(config) :
a,b = config
return a, 0
def transfere_A_B(config) :
a,b = config
place_dans_b = 3-b
transfert = min (a, place_dans_b)
return a-transfert, b+transfert
def transfere_B_A(config) :
a,b = config
place_dans_a = 5-a
transfert = min (b, place_dans_a)
return a+transfert, b-transfert
def est_terminale(config) :
a,b = config
return a == 4
```
**Il est important que tes fonctions soient correctes. Voici un jeu de tests, à toi de le compléter pour tester chaque fonction.**
```
initial = (0,0)
config1 = (5,0)
config2 = (0,3)
config3 = (2,3)
# test de remplir_A
assert( remplir_A(initial) == (5,0) )
assert( remplir_A(config1) == (5,0) )
assert( remplir_A(config2) == (5,3) )
# test de remplir_B
assert( remplir_B(initial) == (0,3) )
# test de vider_A
assert( vider_A(config1) == (0,0) )
# test de vider_B
assert( vider_B(config1) == (5,0) )
#test de transfere_A_B
assert( transfere_A_B(config1) == (2,3)
#test de transfere_B_A
```
Les configurations possibles du système en partant de (0,0) s'organisent selon un graphe orienté dont voici le début (on a écrit sur les arètes les actions effectuées)
<img src="images/graphe_bidons.png" alt="drawing" width="200"/>
Nous allons écrire un algorithme qui construit le graphe de toutes les configurations possibles, en conservant l'information de l'action sur les arètes.
2. **Recherche des successeurs**
La liste des actions possibles est
```
actions = [remplir_A, remplir_B, vider_A, vider_B, transfere_A_B, transfere_B_A]
```
Le graphe sera stocké par liste d'adjacence selon un **dictionnaire** dont les clés sont des tuples (configurations) et chaque valeur est un dictionnaire { tuple : str , tuple : str, ...} des configurations atteignables.
Exemple, en partant de la configuration $(0,0)$ on a
$(0,0) \stackrel{\text{vider_A }}{\longrightarrow} (0,0) $
$(0,0) \stackrel{\text{remplir_A }}{\longrightarrow} (5,0)$
$ (0,0) \stackrel{\text{remplir_B }}{\longrightarrow} (0,3)$.
On doit avoir dans le dictionnaire :
```python
{ (0,0) : {(0,0): "vider_A",
(5,0): "remplir_A",
(0,3): "remplir_B" } ,
#
}
```
**Ecrire** une fonction `successeurs(config)` qui renvoie le dictionnaire des configurations atteignables depuis config.
`successeurs((0,0))`
doit renvoyer
`{(5, 0): 'remplir_A', (0, 3): 'remplir_B', (0, 0): 'vider_A'}`
Aide : si action est un élément de la liste `actions` on a le code
```python
s = action(config) # s est le successeur de config avec l'action
nom = action.__name__ # name est une (str) contenant le nom de la fonction
```
def successeurs(config) :
# code à effacer version élèves
dict_succ = dict()
for action in actions :
succ = action(config)
if succ not in dict_succ : dict_succ[succ] = action.__name__
return dict_succ
```
N'oublie pas de tester cette fonction
```
successeurs((0,0))
successeurs((1,1))
```
3. **Construction du graphe**
Sachant que l'on a $0 \le a \ le 5 $ et $0 \le b \le 3 $, combien de configurations possibles y a t-il ?
Ecris un script Python qui renvoie la liste de toutes les configurations.
```
# code à supprimer dans la version élèves
configurations = [ (a,b) for a in range(6) for b in range(4)]
configurations
```
Ecris ensuite un script qui construit le dictionnaire { config : dictionnaire des configurations atteignables }
```
# code à supprimer dans la version élèves
graphe = { config : successeurs(config) for config in configurations}
graphe
```
4. **Parcours du graphe**
Ecris un algorithme qui parcourt le graphe en largeur en partant de la configuration initiale (0,0) jusqu'à arriver à une configuration de la forme (4,b).
Lorsque l'algorithme rencontre une telle configuration il renvoie True.
Si le parcours se termine sans avoir rencontré une configuration de la forme (4,b) l'algorithme renvoie False.
Au fur et à mesure de l'exploration, l'algorithme complète un dictionnaire des predecesseurs permettant
```
def explore_graphe(g,s):
""" explore le graphe g à partir du sommet s à la recherche d'un sommet terminal
renvoie le sommet trouve ou None si l'exploration ne trouve pas """
visites = [s] # liste des sommets visités
file = [s] # File FIFO des sommets à traiter
while file != [] :
# code à supprimer en version élèves
sommet = file.pop()
for successeur in g[sommet] :
if successeur not in visites :
visites.append(successeur)
if est_terminale(successeur): return successeur
file.insert(0,successeur)
explore_graphe(graphe, (0,0))
```
Modifie ce code pour qu'il construise un dictionnaire des predecesseurs de la forme
`{ tuple1 : (tuple2 ,str_action) }` tel que l'on ait $ tuple_1 \stackrel{\text{str_action }}{\longrightarrow} tuple_2 $
et qu'il renvoie aussi ce dictionnaire.
```
def explore_graphe2(g,s):
""" explore le graphe g à partir du sommet s à la recherche d'un sommet terminal
renvoie le sommet trouve ou None si l'exploration ne trouve pas """
visites = [s] # liste des sommets visités
file = [s] # File FIFO des sommets à traiter
predecesseurs = { s : None} # dictionnaire des predecesseurs
while file != [] :
# code à supprimer en version élèves
sommet = file.pop()
for successeur in g[sommet] :
if successeur not in visites :
visites.append(successeur)
predecesseurs[successeur] = (sommet ,g[sommet][successeur] )
if est_terminale(successeur) : return successeur , predecesseurs
file.insert(0,successeur)
return None, predecesseurs
final , predecesseurs = explore_graphe2(graphe, (0,0))
print(final)
predecesseurs
```
5. **Reconstruction du chemin**
L'algorithme précédent a trouvé un sommet terminal a partir de la configuration $(0,0)$, renvoie ce sommet ainsi qu'un dictionnaire contenant les prédécesseurs des sommets visités.
Ecris une fonction Python `chemin(predecesseurs, initial, final)` qui renvoie la liste des actions à effectuer pour aller de initial à final.
```
def chemin(predecesseurs, initial, final) :
liste_actions = []
sommet = final
# code à supprimer en version élèves
while predecesseurs[sommet] != None :
pred , action = predecesseurs[sommet]
liste_actions.insert(0, action)
sommet = pred
return liste_actions
chemin(predecesseurs,(0,0) , (4,3))
```
---
## Exemple 3. Séquençage de l'ADN
_section à compléter_
L'idée est de faire construire au élèves le graphe orienté sur l'ensemble des mots de longueur 3 d'une séquence ADN, avec la relation $u \rightarrow v$ si et seulement si le suffixe de longueur 2 de $u$ est égal au préfixe de longueur 2 de $v$.
Voir ensuite le problème de la reconstruction de la séquence comme la recherche d'un chemin passant par chaque sommet.
On peut se contenter de rechercher des cycles.
(Voir le sujet du Capes de NSI)
---
|
github_jupyter
|
remplir_A (config)
remplir_B (config)
vider_A(config)
vider_B(config)
transfere_A_B(config)
transfere_B_A(config)
est_terminale(config)
# code à effacer version élèves
def remplir_A(config) :
a,b = config
return 5, b
def remplir_B(config) :
a,b = config
return a, 3
def vider_A(config) :
a,b = config
return 0, b
def vider_B(config) :
a,b = config
return a, 0
def transfere_A_B(config) :
a,b = config
place_dans_b = 3-b
transfert = min (a, place_dans_b)
return a-transfert, b+transfert
def transfere_B_A(config) :
a,b = config
place_dans_a = 5-a
transfert = min (b, place_dans_a)
return a+transfert, b-transfert
def est_terminale(config) :
a,b = config
return a == 4
initial = (0,0)
config1 = (5,0)
config2 = (0,3)
config3 = (2,3)
# test de remplir_A
assert( remplir_A(initial) == (5,0) )
assert( remplir_A(config1) == (5,0) )
assert( remplir_A(config2) == (5,3) )
# test de remplir_B
assert( remplir_B(initial) == (0,3) )
# test de vider_A
assert( vider_A(config1) == (0,0) )
# test de vider_B
assert( vider_B(config1) == (5,0) )
#test de transfere_A_B
assert( transfere_A_B(config1) == (2,3)
#test de transfere_B_A
actions = [remplir_A, remplir_B, vider_A, vider_B, transfere_A_B, transfere_B_A]
{ (0,0) : {(0,0): "vider_A",
(5,0): "remplir_A",
(0,3): "remplir_B" } ,
#
}
def successeurs(config) :
# code à effacer version élèves
dict_succ = dict()
for action in actions :
succ = action(config)
if succ not in dict_succ : dict_succ[succ] = action.__name__
return dict_succ
successeurs((0,0))
successeurs((1,1))
# code à supprimer dans la version élèves
configurations = [ (a,b) for a in range(6) for b in range(4)]
configurations
# code à supprimer dans la version élèves
graphe = { config : successeurs(config) for config in configurations}
graphe
def explore_graphe(g,s):
""" explore le graphe g à partir du sommet s à la recherche d'un sommet terminal
renvoie le sommet trouve ou None si l'exploration ne trouve pas """
visites = [s] # liste des sommets visités
file = [s] # File FIFO des sommets à traiter
while file != [] :
# code à supprimer en version élèves
sommet = file.pop()
for successeur in g[sommet] :
if successeur not in visites :
visites.append(successeur)
if est_terminale(successeur): return successeur
file.insert(0,successeur)
explore_graphe(graphe, (0,0))
def explore_graphe2(g,s):
""" explore le graphe g à partir du sommet s à la recherche d'un sommet terminal
renvoie le sommet trouve ou None si l'exploration ne trouve pas """
visites = [s] # liste des sommets visités
file = [s] # File FIFO des sommets à traiter
predecesseurs = { s : None} # dictionnaire des predecesseurs
while file != [] :
# code à supprimer en version élèves
sommet = file.pop()
for successeur in g[sommet] :
if successeur not in visites :
visites.append(successeur)
predecesseurs[successeur] = (sommet ,g[sommet][successeur] )
if est_terminale(successeur) : return successeur , predecesseurs
file.insert(0,successeur)
return None, predecesseurs
final , predecesseurs = explore_graphe2(graphe, (0,0))
print(final)
predecesseurs
def chemin(predecesseurs, initial, final) :
liste_actions = []
sommet = final
# code à supprimer en version élèves
while predecesseurs[sommet] != None :
pred , action = predecesseurs[sommet]
liste_actions.insert(0, action)
sommet = pred
return liste_actions
chemin(predecesseurs,(0,0) , (4,3))
| 0.233444 | 0.866189 |
```
%pylab inline
from solveMDP_poorHigh import *
Vgrid = np.load("poorHigh.npy")
matplotlib.rcParams['figure.figsize'] = [16, 8]
plt.rcParams.update({'font.size': 15})
'''
Policy plot:
Input:
x = [w,ab,s,e,o,z] single action
x = [0,1, 2,3,4,5]
a = [c,b,k,h,action] single state
a = [0,1,2,3,4]
'''
wealthLevel = [100, 150, 200, 250]
ageLevel = [30, 45, 60, 75]
savingsRatio = []
investmentsRatio = []
for wealth in wealthLevel:
savingR = []
investmentR = []
for age in ageLevel:
t = age - 20
x = [wealth, 0, 1, 0, 0, 1]
_,a = V_solve(t,Vgrid[:,:,:,:,:,:,t],x)
savingR.append((a[1]+a[2])/wealth)
investmentR.append(a[2]/(a[1]+a[2]))
savingsRatio.append(savingR)
investmentsRatio.append(investmentR)
import pandas as pd
df_saving = pd.DataFrame(np.array(savingsRatio), columns = ['age '+ str(age) for age in ageLevel], index= ['wealth ' + str(wealth) for wealth in wealthLevel])
df_investment = pd.DataFrame(np.array(investmentsRatio), columns = ['age '+ str(age) for age in ageLevel], index= ['wealth ' + str(wealth) for wealth in wealthLevel])
print("savingRatio:")
display(df_saving)
print("investmentRatio:")
display(df_investment)
```
### Simulation Part
```
%%time
# total number of agents
num = 10000
'''
x = [w,ab,s,e,o,z]
x = [5,0, 0,0,0,0]
'''
from jax import random
from quantecon import MarkovChain
# number of economies and each economy has 100 agents
numEcon = 100
numAgents = 100
mc = MarkovChain(Ps)
econStates = mc.simulate(ts_length=T_max-T_min,init=0,num_reps=numEcon)
econStates = jnp.array(econStates,dtype = int)
@partial(jit, static_argnums=(0,))
def transition_real(t,a,x, s_prime):
'''
Input:
x = [w,ab,s,e,o,z] single action
x = [0,1, 2,3,4,5]
a = [c,b,k,h,action] single state
a = [0,1,2,3,4]
Output:
w_next
ab_next
s_next
e_next
o_next
z_next
prob_next
'''
s = jnp.array(x[2], dtype = jnp.int8)
e = jnp.array(x[3], dtype = jnp.int8)
# actions taken
b = a[1]
k = a[2]
action = a[4]
w_next = ((1+r_b[s])*b + (1+r_k[s_prime])*k).repeat(nE)
ab_next = (1-x[4])*(t*(action == 1)).repeat(nE) + x[4]*(x[1]*jnp.ones(nE))
s_next = s_prime.repeat(nE)
e_next = jnp.array([e,(1-e)])
z_next = x[5]*jnp.ones(nE) + ((1-x[5]) * (k > 0)).repeat(nE)
# job status changing probability and econ state transition probability
pe = Pe[s, e]
prob_next = jnp.array([1-pe, pe])
# owner
o_next_own = (x[4] - action).repeat(nE)
# renter
o_next_rent = action.repeat(nE)
o_next = x[4] * o_next_own + (1-x[4]) * o_next_rent
return jnp.column_stack((w_next,ab_next,s_next,e_next,o_next,z_next,prob_next))
def simulation(key):
initE = random.choice(a = nE, p=E_distribution, key = key)
initS = random.choice(a = nS, p=S_distribution, key = key)
x = [5, 0, initS, initE, 0, 0]
path = []
move = []
# first 100 agents are in the 1st economy and second 100 agents are in the 2nd economy
econ = econStates[key.sum()//numAgents,:]
for t in range(T_min, T_max):
_, key = random.split(key)
if t == T_max-1:
_,a = V_solve(t,Vgrid[:,:,:,:,:,:,t],x)
else:
_,a = V_solve(t,Vgrid[:,:,:,:,:,:,t+1],x)
xp = transition_real(t,a,x, econ[t])
p = xp[:,-1]
x_next = xp[:,:-1]
path.append(x)
move.append(a)
x = x_next[random.choice(a = nE, p=p, key = key)]
path.append(x)
return jnp.array(path), jnp.array(move)
# simulation part
keys = vmap(random.PRNGKey)(jnp.arange(num))
Paths, Moves = vmap(simulation)(keys)
# x = [w,ab,s,e,o,z]
# x = [0,1, 2,3,4,5]
ws = Paths[:,:,0].T
ab = Paths[:,:,1].T
ss = Paths[:,:,2].T
es = Paths[:,:,3].T
os = Paths[:,:,4].T
zs = Paths[:,:,5].T
cs = Moves[:,:,0].T
bs = Moves[:,:,1].T
ks = Moves[:,:,2].T
hs = Moves[:,:,3].T
ms = Ms[jnp.append(jnp.array([0]),jnp.arange(T_max)).reshape(-1,1) - jnp.array(ab, dtype = jnp.int8)]*os
```
### Graph and Table
```
plt.figure(1)
plt.title("The mean values of simulation")
startAge = 20
# value of states, state has one more value, since the terminal state does not have associated action
plt.plot(range(startAge, T_max + startAge + 1),jnp.mean(ws + H*pt*os - ms,axis = 1), label = "wealth + home equity")
plt.plot(range(startAge, T_max + startAge + 1),jnp.mean(H*pt*os - ms,axis = 1), label = "home equity")
plt.plot(range(startAge, T_max + startAge + 1),jnp.mean(ws,axis = 1), label = "wealth")
# value of actions
plt.plot(range(startAge, T_max + startAge),jnp.mean(cs,axis = 1), label = "consumption")
plt.plot(range(startAge, T_max + startAge),jnp.mean(bs,axis = 1), label = "bond")
plt.plot(range(startAge, T_max + startAge),jnp.mean(ks,axis = 1), label = "stock")
plt.legend()
plt.figure(2)
plt.title("Stock Participation Ratio through Different Age Periods")
plt.plot(range(20, T_max + 21),jnp.mean(zs,axis = 1), label = "experience")
plt.legend()
plt.figure(3)
plt.title("house ownership ratio in the population")
plt.plot(range(startAge, T_max + startAge + 1),(os).mean(axis = 1), label = "ownership ratio")
plt.legend()
# agent buying time collection
agentTime = []
for t in range(30):
if ((os[t,:] == 0) & (os[t+1,:] == 1)).sum()>0:
for agentNum in jnp.where((os[t,:] == 0) & (os[t+1,:] == 1))[0]:
agentTime.append([t, agentNum])
agentTime = jnp.array(agentTime)
# agent hold time collection
agentHold = []
for t in range(30):
if ((os[t,:] == 0) & (os[t+1,:] == 0)).sum()>0:
for agentNum in jnp.where((os[t,:] == 0) & (os[t+1,:] == 0))[0]:
agentHold.append([t, agentNum])
agentHold = jnp.array(agentHold)
plt.figure(4)
plt.title("weath level for buyer, owner and renter")
www = (os*(ws+H*pt - ms)).sum(axis = 1)/(os).sum(axis = 1)
for age in range(30):
buyer = agentTime[agentTime[:,0] == age]
renter = agentHold[agentHold[:,0] == age]
bp = plt.scatter(age, ws[buyer[:,0], buyer[:,1]].mean(),color = "b")
hp = plt.scatter(age, www[age], color = "green")
rp = plt.scatter(age, ws[renter[:,0], renter[:,1]].mean(),color = "r")
plt.legend((bp,hp,rp), ("FirstTimeBuyer", "HomeOwner", "Renter"))
plt.figure(5)
plt.title("employement status for buyer and renter")
for age in range(31):
buyer = agentTime[agentTime[:,0] == age]
renter = agentHold[agentHold[:,0] == age]
bp = plt.scatter(age, es[buyer[:,0], buyer[:,1]].mean(),color = "b")
rp = plt.scatter(age, es[renter[:,0], renter[:,1]].mean(),color = "r")
plt.legend((bp, rp), ("FirstTimeBuyer", "Renter"))
# agent participate time collection
agentTimep = []
for t in range(30):
if ((zs[t,:] == 0) & (zs[t+1,:] == 1)).sum()>0:
for agentNum in jnp.where((zs[t,:] == 0) & (zs[t+1,:] == 1))[0]:
agentTimep.append([t, agentNum])
agentTimep = jnp.array(agentTimep)
# agent nonparticipate time collection
agentHoldp = []
for t in range(30):
if ((zs[t,:] == 0) & (zs[t+1,:] == 0)).sum()>0:
for agentNum in jnp.where((zs[t,:] == 0) & (zs[t+1,:] == 0))[0]:
agentHoldp.append([t, agentNum])
agentHoldp = jnp.array(agentHoldp)
plt.figure(6)
plt.title("weath level for FirstTimeTrader, ExperiencedTrader and Nonparticipant")
www = (zs*(ws+H*pt - ms)).sum(axis = 1)/(zs).sum(axis = 1)
for age in range(30):
trader = agentTimep[agentTimep[:,0] == age]
noneTrader = agentHoldp[agentHoldp[:,0] == age]
tp = plt.scatter(age, ws[trader[:,0], trader[:,1]].mean(),color = "b")
ep = plt.scatter(age, www[age], color = "green")
ip = plt.scatter(age, ws[noneTrader[:,0], noneTrader[:,1]].mean(),color = "r")
plt.legend((tp,ep,ip), ("FirstTimeTrader", "ExperiencedTrader", "Nonparticipant"))
plt.figure(7)
plt.title("employement status for FirstTimeTrader and Nonparticipant")
for age in range(30):
trader = agentTimep[agentTimep[:,0] == age]
noneTrader = agentHoldp[agentHoldp[:,0] == age]
tp = plt.scatter(age, es[trader[:,0], trader[:,1]].mean(),color = "b")
ip = plt.scatter(age, es[noneTrader[:,0], noneTrader[:,1]].mean(),color = "r")
plt.legend((tp,ip), ("FirstTimeTrader", "Nonparticipant"))
plt.figure(8)
# At every age
plt.title("Stock Investment Percentage as StockInvestmentAmount/(StockInvestmentAmount + BondInvestmentAmount)")
plt.plot((os[:T_max,:]*ks/(ks+bs)).sum(axis = 1)/os[:T_max,:].sum(axis = 1), label = "owner")
plt.plot(((1-os[:T_max,:])*ks/(ks+bs)).sum(axis = 1)/(1-os)[:T_max,:].sum(axis = 1), label = "renter")
plt.legend()
plt.figure(9)
# At every age
plt.title("Stock Investment Percentage")
plt.plot(range(startAge, startAge+T_max), (ks/(ks+bs)).mean(axis = 1), label = "ks/(ks+bs)")
plt.legend()
# # agent number, x = [w,n,m,s,e,o]
# agentNum = 35
# plt.plot(range(20, T_max + 21),(ws + os*(H*pt - ms))[:,agentNum], label = "wealth + home equity")
# plt.plot(range(20, T_max + 21),ms[:,agentNum], label = "mortgage")
# plt.plot(range(20, T_max + 20),cs[:,agentNum], label = "consumption")
# plt.plot(range(20, T_max + 20),bs[:,agentNum], label = "bond")
# plt.plot(range(20, T_max + 20),ks[:,agentNum], label = "stock")
# plt.plot(range(20, T_max + 21),os[:,agentNum]*100, label = "ownership", color = "k")
# plt.legend()
ws.mean(axis = 1)
```
|
github_jupyter
|
%pylab inline
from solveMDP_poorHigh import *
Vgrid = np.load("poorHigh.npy")
matplotlib.rcParams['figure.figsize'] = [16, 8]
plt.rcParams.update({'font.size': 15})
'''
Policy plot:
Input:
x = [w,ab,s,e,o,z] single action
x = [0,1, 2,3,4,5]
a = [c,b,k,h,action] single state
a = [0,1,2,3,4]
'''
wealthLevel = [100, 150, 200, 250]
ageLevel = [30, 45, 60, 75]
savingsRatio = []
investmentsRatio = []
for wealth in wealthLevel:
savingR = []
investmentR = []
for age in ageLevel:
t = age - 20
x = [wealth, 0, 1, 0, 0, 1]
_,a = V_solve(t,Vgrid[:,:,:,:,:,:,t],x)
savingR.append((a[1]+a[2])/wealth)
investmentR.append(a[2]/(a[1]+a[2]))
savingsRatio.append(savingR)
investmentsRatio.append(investmentR)
import pandas as pd
df_saving = pd.DataFrame(np.array(savingsRatio), columns = ['age '+ str(age) for age in ageLevel], index= ['wealth ' + str(wealth) for wealth in wealthLevel])
df_investment = pd.DataFrame(np.array(investmentsRatio), columns = ['age '+ str(age) for age in ageLevel], index= ['wealth ' + str(wealth) for wealth in wealthLevel])
print("savingRatio:")
display(df_saving)
print("investmentRatio:")
display(df_investment)
%%time
# total number of agents
num = 10000
'''
x = [w,ab,s,e,o,z]
x = [5,0, 0,0,0,0]
'''
from jax import random
from quantecon import MarkovChain
# number of economies and each economy has 100 agents
numEcon = 100
numAgents = 100
mc = MarkovChain(Ps)
econStates = mc.simulate(ts_length=T_max-T_min,init=0,num_reps=numEcon)
econStates = jnp.array(econStates,dtype = int)
@partial(jit, static_argnums=(0,))
def transition_real(t,a,x, s_prime):
'''
Input:
x = [w,ab,s,e,o,z] single action
x = [0,1, 2,3,4,5]
a = [c,b,k,h,action] single state
a = [0,1,2,3,4]
Output:
w_next
ab_next
s_next
e_next
o_next
z_next
prob_next
'''
s = jnp.array(x[2], dtype = jnp.int8)
e = jnp.array(x[3], dtype = jnp.int8)
# actions taken
b = a[1]
k = a[2]
action = a[4]
w_next = ((1+r_b[s])*b + (1+r_k[s_prime])*k).repeat(nE)
ab_next = (1-x[4])*(t*(action == 1)).repeat(nE) + x[4]*(x[1]*jnp.ones(nE))
s_next = s_prime.repeat(nE)
e_next = jnp.array([e,(1-e)])
z_next = x[5]*jnp.ones(nE) + ((1-x[5]) * (k > 0)).repeat(nE)
# job status changing probability and econ state transition probability
pe = Pe[s, e]
prob_next = jnp.array([1-pe, pe])
# owner
o_next_own = (x[4] - action).repeat(nE)
# renter
o_next_rent = action.repeat(nE)
o_next = x[4] * o_next_own + (1-x[4]) * o_next_rent
return jnp.column_stack((w_next,ab_next,s_next,e_next,o_next,z_next,prob_next))
def simulation(key):
initE = random.choice(a = nE, p=E_distribution, key = key)
initS = random.choice(a = nS, p=S_distribution, key = key)
x = [5, 0, initS, initE, 0, 0]
path = []
move = []
# first 100 agents are in the 1st economy and second 100 agents are in the 2nd economy
econ = econStates[key.sum()//numAgents,:]
for t in range(T_min, T_max):
_, key = random.split(key)
if t == T_max-1:
_,a = V_solve(t,Vgrid[:,:,:,:,:,:,t],x)
else:
_,a = V_solve(t,Vgrid[:,:,:,:,:,:,t+1],x)
xp = transition_real(t,a,x, econ[t])
p = xp[:,-1]
x_next = xp[:,:-1]
path.append(x)
move.append(a)
x = x_next[random.choice(a = nE, p=p, key = key)]
path.append(x)
return jnp.array(path), jnp.array(move)
# simulation part
keys = vmap(random.PRNGKey)(jnp.arange(num))
Paths, Moves = vmap(simulation)(keys)
# x = [w,ab,s,e,o,z]
# x = [0,1, 2,3,4,5]
ws = Paths[:,:,0].T
ab = Paths[:,:,1].T
ss = Paths[:,:,2].T
es = Paths[:,:,3].T
os = Paths[:,:,4].T
zs = Paths[:,:,5].T
cs = Moves[:,:,0].T
bs = Moves[:,:,1].T
ks = Moves[:,:,2].T
hs = Moves[:,:,3].T
ms = Ms[jnp.append(jnp.array([0]),jnp.arange(T_max)).reshape(-1,1) - jnp.array(ab, dtype = jnp.int8)]*os
plt.figure(1)
plt.title("The mean values of simulation")
startAge = 20
# value of states, state has one more value, since the terminal state does not have associated action
plt.plot(range(startAge, T_max + startAge + 1),jnp.mean(ws + H*pt*os - ms,axis = 1), label = "wealth + home equity")
plt.plot(range(startAge, T_max + startAge + 1),jnp.mean(H*pt*os - ms,axis = 1), label = "home equity")
plt.plot(range(startAge, T_max + startAge + 1),jnp.mean(ws,axis = 1), label = "wealth")
# value of actions
plt.plot(range(startAge, T_max + startAge),jnp.mean(cs,axis = 1), label = "consumption")
plt.plot(range(startAge, T_max + startAge),jnp.mean(bs,axis = 1), label = "bond")
plt.plot(range(startAge, T_max + startAge),jnp.mean(ks,axis = 1), label = "stock")
plt.legend()
plt.figure(2)
plt.title("Stock Participation Ratio through Different Age Periods")
plt.plot(range(20, T_max + 21),jnp.mean(zs,axis = 1), label = "experience")
plt.legend()
plt.figure(3)
plt.title("house ownership ratio in the population")
plt.plot(range(startAge, T_max + startAge + 1),(os).mean(axis = 1), label = "ownership ratio")
plt.legend()
# agent buying time collection
agentTime = []
for t in range(30):
if ((os[t,:] == 0) & (os[t+1,:] == 1)).sum()>0:
for agentNum in jnp.where((os[t,:] == 0) & (os[t+1,:] == 1))[0]:
agentTime.append([t, agentNum])
agentTime = jnp.array(agentTime)
# agent hold time collection
agentHold = []
for t in range(30):
if ((os[t,:] == 0) & (os[t+1,:] == 0)).sum()>0:
for agentNum in jnp.where((os[t,:] == 0) & (os[t+1,:] == 0))[0]:
agentHold.append([t, agentNum])
agentHold = jnp.array(agentHold)
plt.figure(4)
plt.title("weath level for buyer, owner and renter")
www = (os*(ws+H*pt - ms)).sum(axis = 1)/(os).sum(axis = 1)
for age in range(30):
buyer = agentTime[agentTime[:,0] == age]
renter = agentHold[agentHold[:,0] == age]
bp = plt.scatter(age, ws[buyer[:,0], buyer[:,1]].mean(),color = "b")
hp = plt.scatter(age, www[age], color = "green")
rp = plt.scatter(age, ws[renter[:,0], renter[:,1]].mean(),color = "r")
plt.legend((bp,hp,rp), ("FirstTimeBuyer", "HomeOwner", "Renter"))
plt.figure(5)
plt.title("employement status for buyer and renter")
for age in range(31):
buyer = agentTime[agentTime[:,0] == age]
renter = agentHold[agentHold[:,0] == age]
bp = plt.scatter(age, es[buyer[:,0], buyer[:,1]].mean(),color = "b")
rp = plt.scatter(age, es[renter[:,0], renter[:,1]].mean(),color = "r")
plt.legend((bp, rp), ("FirstTimeBuyer", "Renter"))
# agent participate time collection
agentTimep = []
for t in range(30):
if ((zs[t,:] == 0) & (zs[t+1,:] == 1)).sum()>0:
for agentNum in jnp.where((zs[t,:] == 0) & (zs[t+1,:] == 1))[0]:
agentTimep.append([t, agentNum])
agentTimep = jnp.array(agentTimep)
# agent nonparticipate time collection
agentHoldp = []
for t in range(30):
if ((zs[t,:] == 0) & (zs[t+1,:] == 0)).sum()>0:
for agentNum in jnp.where((zs[t,:] == 0) & (zs[t+1,:] == 0))[0]:
agentHoldp.append([t, agentNum])
agentHoldp = jnp.array(agentHoldp)
plt.figure(6)
plt.title("weath level for FirstTimeTrader, ExperiencedTrader and Nonparticipant")
www = (zs*(ws+H*pt - ms)).sum(axis = 1)/(zs).sum(axis = 1)
for age in range(30):
trader = agentTimep[agentTimep[:,0] == age]
noneTrader = agentHoldp[agentHoldp[:,0] == age]
tp = plt.scatter(age, ws[trader[:,0], trader[:,1]].mean(),color = "b")
ep = plt.scatter(age, www[age], color = "green")
ip = plt.scatter(age, ws[noneTrader[:,0], noneTrader[:,1]].mean(),color = "r")
plt.legend((tp,ep,ip), ("FirstTimeTrader", "ExperiencedTrader", "Nonparticipant"))
plt.figure(7)
plt.title("employement status for FirstTimeTrader and Nonparticipant")
for age in range(30):
trader = agentTimep[agentTimep[:,0] == age]
noneTrader = agentHoldp[agentHoldp[:,0] == age]
tp = plt.scatter(age, es[trader[:,0], trader[:,1]].mean(),color = "b")
ip = plt.scatter(age, es[noneTrader[:,0], noneTrader[:,1]].mean(),color = "r")
plt.legend((tp,ip), ("FirstTimeTrader", "Nonparticipant"))
plt.figure(8)
# At every age
plt.title("Stock Investment Percentage as StockInvestmentAmount/(StockInvestmentAmount + BondInvestmentAmount)")
plt.plot((os[:T_max,:]*ks/(ks+bs)).sum(axis = 1)/os[:T_max,:].sum(axis = 1), label = "owner")
plt.plot(((1-os[:T_max,:])*ks/(ks+bs)).sum(axis = 1)/(1-os)[:T_max,:].sum(axis = 1), label = "renter")
plt.legend()
plt.figure(9)
# At every age
plt.title("Stock Investment Percentage")
plt.plot(range(startAge, startAge+T_max), (ks/(ks+bs)).mean(axis = 1), label = "ks/(ks+bs)")
plt.legend()
# # agent number, x = [w,n,m,s,e,o]
# agentNum = 35
# plt.plot(range(20, T_max + 21),(ws + os*(H*pt - ms))[:,agentNum], label = "wealth + home equity")
# plt.plot(range(20, T_max + 21),ms[:,agentNum], label = "mortgage")
# plt.plot(range(20, T_max + 20),cs[:,agentNum], label = "consumption")
# plt.plot(range(20, T_max + 20),bs[:,agentNum], label = "bond")
# plt.plot(range(20, T_max + 20),ks[:,agentNum], label = "stock")
# plt.plot(range(20, T_max + 21),os[:,agentNum]*100, label = "ownership", color = "k")
# plt.legend()
ws.mean(axis = 1)
| 0.230573 | 0.835819 |
R-CNN. Первая модель для решения данной задачи. Работает как обычный классификатор изображений. На вход сети подаются разные регионы изображения и для них делается предсказание. Очень медленная так как прогоняет одно изображение несколько тысяч раз.
Fast R-CNN. Улучненная и более быстрая модель R-CNN, работает по похожему принципу, но сначала все изображения подаются на вход CNN, потом из полученного внутреннего представления генерируются регионы. (медленна для задач реального времени)
Faster R-CNN. Главное отличие от предыдущих в том, что вместо selective search алгоритма для выбора регионов использует нейронную сеть для их заучивания.
YOLO. Совсем другой прицнип работы по сравнению с предыдущим, не использует регионы вообще. Наиболее быстрая.
SSD. По принципу похожа на YOLO, но в качестве сети для извлечения признаков использует VGG16. Тоже довольно быстрая и пригодная для работы в реальном времени.
Feature Pyramid Networks (FPN). Ещё одна разновидность сети типа Single Shot Detector, из за особенности извлечения признаков, лучше чем SSD распознаёт мелкие объекты.
RetinaNet. Использует комбинацию FPN+ResNet и благодаря специальной функции ошибки (focal loss) дает более высокую точность (аccuracy).
Главная особенность этой архитектуры по сравнению с другими состоит в том, что большинство систем применяют CNN несколько раз к разным регионам изображения, в YOLO CNN применяется один раз ко всему изображению сразу. Сеть делит изображение на своеобразную сетку и предсказывает bounding boxes и вероятности того, что там есть искомый объект для каждого участка.
Плюсы данного подхода состоит в том, что сеть смотрит на все изображение сразу и учитывает контекст при детектировании и распознавании объекта. Так же YOLO в 1000 раз быстрее чем R-CNN и около 100x быстрее чем Fast R-CNN. В данной статье мы будем запускать сеть на мобильном устройстве для онлайн обработки, поэтому это для нас это самое главное качество.
YOLOv3
YOLOv3 — это усовершенствованная версия архитектуры YOLO. Она состоит из 106-ти свёрточных слоев и лучше детектирует небольшие объекты по сравнению с её предшествиницей YOLOv2. Основная особенность YOLOv3 состоит в том, что на выходе есть три слоя каждый из которых расчитан на обнаружения объектов разного размера.
Модель YOLOv3 в качестве выхода использует три слоя для разбиения изображения на различную сетку, размеры ячеек этих сеток имеют такие значения: 8, 16 и 32. Допустим на входе у нас есть изображение размером 416x416 пикселей, тогда выходные матрицы (сетки) будут иметь размер 52x52, 26x26 и 13x13 (416/8 = 52, 416/16 = 26 и 416/32 = 13). В случае с YOLOv3-tiny все тоже самое только вместо трех сеток имеем две: 16 и 32, то есть матрицы размерностью 26x26 и 13x13.
Размерность Detection kernel: 1 x 1 x (B x (5 + C) )
|
github_jupyter
|
R-CNN. Первая модель для решения данной задачи. Работает как обычный классификатор изображений. На вход сети подаются разные регионы изображения и для них делается предсказание. Очень медленная так как прогоняет одно изображение несколько тысяч раз.
Fast R-CNN. Улучненная и более быстрая модель R-CNN, работает по похожему принципу, но сначала все изображения подаются на вход CNN, потом из полученного внутреннего представления генерируются регионы. (медленна для задач реального времени)
Faster R-CNN. Главное отличие от предыдущих в том, что вместо selective search алгоритма для выбора регионов использует нейронную сеть для их заучивания.
YOLO. Совсем другой прицнип работы по сравнению с предыдущим, не использует регионы вообще. Наиболее быстрая.
SSD. По принципу похожа на YOLO, но в качестве сети для извлечения признаков использует VGG16. Тоже довольно быстрая и пригодная для работы в реальном времени.
Feature Pyramid Networks (FPN). Ещё одна разновидность сети типа Single Shot Detector, из за особенности извлечения признаков, лучше чем SSD распознаёт мелкие объекты.
RetinaNet. Использует комбинацию FPN+ResNet и благодаря специальной функции ошибки (focal loss) дает более высокую точность (аccuracy).
Главная особенность этой архитектуры по сравнению с другими состоит в том, что большинство систем применяют CNN несколько раз к разным регионам изображения, в YOLO CNN применяется один раз ко всему изображению сразу. Сеть делит изображение на своеобразную сетку и предсказывает bounding boxes и вероятности того, что там есть искомый объект для каждого участка.
Плюсы данного подхода состоит в том, что сеть смотрит на все изображение сразу и учитывает контекст при детектировании и распознавании объекта. Так же YOLO в 1000 раз быстрее чем R-CNN и около 100x быстрее чем Fast R-CNN. В данной статье мы будем запускать сеть на мобильном устройстве для онлайн обработки, поэтому это для нас это самое главное качество.
YOLOv3
YOLOv3 — это усовершенствованная версия архитектуры YOLO. Она состоит из 106-ти свёрточных слоев и лучше детектирует небольшие объекты по сравнению с её предшествиницей YOLOv2. Основная особенность YOLOv3 состоит в том, что на выходе есть три слоя каждый из которых расчитан на обнаружения объектов разного размера.
Модель YOLOv3 в качестве выхода использует три слоя для разбиения изображения на различную сетку, размеры ячеек этих сеток имеют такие значения: 8, 16 и 32. Допустим на входе у нас есть изображение размером 416x416 пикселей, тогда выходные матрицы (сетки) будут иметь размер 52x52, 26x26 и 13x13 (416/8 = 52, 416/16 = 26 и 416/32 = 13). В случае с YOLOv3-tiny все тоже самое только вместо трех сеток имеем две: 16 и 32, то есть матрицы размерностью 26x26 и 13x13.
Размерность Detection kernel: 1 x 1 x (B x (5 + C) )
| 0.199542 | 0.936052 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
pd.options.display.max_rows = 999
pd.options.display.max_columns = 999
bncc_raw = pd.read_csv('C:/Users/Danilo/Desktop/Documentos Acadêmicos/TERA/TERAprojetos/Projeto final/BNCC/bncc_db.csv', encoding = 'utf8')
bncc_db = bncc_raw.copy()
bncc_db.head()
```
## Explorando os dados
```
bncc_db.info()
name1 = bncc_db['name.1'].nunique()
d = ('São um total de %d Áreas do Conhecimento' % (name1))
display(
d,
bncc_db.iloc[:, 5].agg(['value_counts']).head()
)
### Coluna code
bncc_db.iloc[:, 8].unique()
code = bncc_db['code'].nunique()
d = ('São um total de %d códigos da BNCC presentes no dataset' % (code))
display(
d,
bncc_db.iloc[:, 8].agg(['value_counts']).head()
)
description = bncc_db['description'].nunique()
d = ('São um total de %d descrições' % (description))
display(
d,
bncc_db['description'].agg(['value_counts']).head()
)
question = bncc_db['question'].nunique()
d = ('São um total de %d questões' % (question))
display(
d,
bncc_db['question'].agg(['value_counts']).head()
)
```
## Limpando as questões
```
### Resolvendo problema de codificação de caracteres presentes nas Questões
import html
data_quest = bncc_db['question'].astype('str').apply(html.unescape)
### Resolvendo problema de tags html
import regex as reg
CLEANR = reg.compile('<.*?>')
def cleanhtml(raw_html):
cleantext = reg.sub(CLEANR, '', raw_html)
return cleantext
text = data_quest.map(lambda x: cleanhtml(x))
bncc_db.insert(1, 'question_clean', text, allow_duplicates=False)
bncc_db.head()
```
## Instalando Bibliotecas necessárias para NLP
- `!pip install regex`
- `!pip install html`
- `!pip install lxml`
- `!pip install nltk`
- `!pip install gensim`
- `!pip install pyldavis`
- `!pip install wordcloud`
- `!pip install textblob`
- `!pip install spacy`
- `!pip install textstat`
### Número de Caracteres por Sentença
```
max = bncc_db['question_clean'].str.len().max()
min = bncc_db['question_clean'].str.len().min()
median = bncc_db['question_clean'].str.len().median()
mean = bncc_db['question_clean'].str.len().mean()
print('As questões vão de %d à %d caracteres por questão' % (min, max))
print('O valor mediano e médio de caracteres por questão é de %d e de %d, respectivamente.' %
(median, mean))
fig, ax = plt.subplots(figsize=(20, 10))
sns.histplot(bncc_db['question_clean'].str.len(), ax = ax)
```
- Verificando Questões vazias
```
np.where(bncc_db['question_clean'].str.len() == 0)
```
### Número de Palavras em cada questão:
```
text = bncc_db['question_clean']
max = text.str.split().map(lambda x: len(x)).max()
min = text.str.split().map(lambda x: len(x)).min()
median = text.str.split().map(lambda x: len(x)).median()
mean = text.str.split().map(lambda x: len(x)).mean()
print('O número de palavras vão de %d à %d por questão' % (min, max))
print('O valor mediano e médio de palavras por questão é de %d e de %d, respectivamente.' %
(median, mean))
fig, ax = plt.subplots(figsize=(20, 10))
sns.histplot(text.str.split().map(lambda x: len(x)), ax = ax)
```
- Média do tamanho das palavras em cada questão
```
mean_words = bncc_db['question_clean'].str.split().apply(lambda x : [len(i) for i in x]).map(lambda x: np.mean(x))
mean_words
```
- Valor médio máximo e médio do tamanho de palavras por questão:
```
max_len_words = bncc_db['question_clean'].str.split().apply(lambda x : [len(i) for i in x]).map(lambda x: np.mean(x)).max()
print('O valor máximo do tamanho médio das palavras por questão é de %d (Algo está errado)'%(max_len_words))
mean_len_words = bncc_db['question_clean'].str.split().apply(lambda x : [len(i) for i in x]).map(lambda x: np.mean(x)).mean()
print('O valor médio do tamanho de uma palavra por questão é de %d'%(mean_len_words))
```
### Possíveis problemas relacionados a palavras compridas;
- Palavras distintas não separadas por espaço
- Ausencia de espaço após finalizar uma frase.
- Une uma palavra do final de uma frase com a palavra do início de uma nova frase (Acabou.Começou != Acabou. Começou)
Apesar de termos limpados as tag html, a própria estrutura das questões deixam alguns erros. Por ex.:
- Tópicos enumerados a serem cumpridos na questão estão grudados. ex.:
- Deveria ser:
- 1. alternativa
- 2. alternativa
- 3. alternativa
- 4. alternativa
- Como está:
- -1. alternativa-2. alternativa- 3. alternativa-4. alternativa
```
bncc_db['question_clean'][4]
## Verificando o tamanho médio das palavras
fig, ax = plt.subplots(figsize=(20, 10))
sns.histplot(bncc_db['question_clean'].str.split().apply(lambda x : [len(i) for i in x]).map(lambda x: np.mean(x)),
ax = ax)
## Vale notar que existe umas palavras com 40, 60, 80 ... letras, o que é inverossímel
```
## Verificar 'stopwords' nas questões
```
import nltk
nltk.download('stopwords')
stop = nltk.corpus.stopwords.words('portuguese')
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
pd.options.display.max_rows = 999
pd.options.display.max_columns = 999
bncc_raw = pd.read_csv('C:/Users/Danilo/Desktop/Documentos Acadêmicos/TERA/TERAprojetos/Projeto final/BNCC/bncc_db.csv', encoding = 'utf8')
bncc_db = bncc_raw.copy()
bncc_db.head()
bncc_db.info()
name1 = bncc_db['name.1'].nunique()
d = ('São um total de %d Áreas do Conhecimento' % (name1))
display(
d,
bncc_db.iloc[:, 5].agg(['value_counts']).head()
)
### Coluna code
bncc_db.iloc[:, 8].unique()
code = bncc_db['code'].nunique()
d = ('São um total de %d códigos da BNCC presentes no dataset' % (code))
display(
d,
bncc_db.iloc[:, 8].agg(['value_counts']).head()
)
description = bncc_db['description'].nunique()
d = ('São um total de %d descrições' % (description))
display(
d,
bncc_db['description'].agg(['value_counts']).head()
)
question = bncc_db['question'].nunique()
d = ('São um total de %d questões' % (question))
display(
d,
bncc_db['question'].agg(['value_counts']).head()
)
### Resolvendo problema de codificação de caracteres presentes nas Questões
import html
data_quest = bncc_db['question'].astype('str').apply(html.unescape)
### Resolvendo problema de tags html
import regex as reg
CLEANR = reg.compile('<.*?>')
def cleanhtml(raw_html):
cleantext = reg.sub(CLEANR, '', raw_html)
return cleantext
text = data_quest.map(lambda x: cleanhtml(x))
bncc_db.insert(1, 'question_clean', text, allow_duplicates=False)
bncc_db.head()
max = bncc_db['question_clean'].str.len().max()
min = bncc_db['question_clean'].str.len().min()
median = bncc_db['question_clean'].str.len().median()
mean = bncc_db['question_clean'].str.len().mean()
print('As questões vão de %d à %d caracteres por questão' % (min, max))
print('O valor mediano e médio de caracteres por questão é de %d e de %d, respectivamente.' %
(median, mean))
fig, ax = plt.subplots(figsize=(20, 10))
sns.histplot(bncc_db['question_clean'].str.len(), ax = ax)
np.where(bncc_db['question_clean'].str.len() == 0)
text = bncc_db['question_clean']
max = text.str.split().map(lambda x: len(x)).max()
min = text.str.split().map(lambda x: len(x)).min()
median = text.str.split().map(lambda x: len(x)).median()
mean = text.str.split().map(lambda x: len(x)).mean()
print('O número de palavras vão de %d à %d por questão' % (min, max))
print('O valor mediano e médio de palavras por questão é de %d e de %d, respectivamente.' %
(median, mean))
fig, ax = plt.subplots(figsize=(20, 10))
sns.histplot(text.str.split().map(lambda x: len(x)), ax = ax)
mean_words = bncc_db['question_clean'].str.split().apply(lambda x : [len(i) for i in x]).map(lambda x: np.mean(x))
mean_words
max_len_words = bncc_db['question_clean'].str.split().apply(lambda x : [len(i) for i in x]).map(lambda x: np.mean(x)).max()
print('O valor máximo do tamanho médio das palavras por questão é de %d (Algo está errado)'%(max_len_words))
mean_len_words = bncc_db['question_clean'].str.split().apply(lambda x : [len(i) for i in x]).map(lambda x: np.mean(x)).mean()
print('O valor médio do tamanho de uma palavra por questão é de %d'%(mean_len_words))
bncc_db['question_clean'][4]
## Verificando o tamanho médio das palavras
fig, ax = plt.subplots(figsize=(20, 10))
sns.histplot(bncc_db['question_clean'].str.split().apply(lambda x : [len(i) for i in x]).map(lambda x: np.mean(x)),
ax = ax)
## Vale notar que existe umas palavras com 40, 60, 80 ... letras, o que é inverossímel
import nltk
nltk.download('stopwords')
stop = nltk.corpus.stopwords.words('portuguese')
| 0.229018 | 0.472136 |
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc" style="margin-top: 1em;"><ul class="toc-item"><li><span><a href="#Name" data-toc-modified-id="Name-1"><span class="toc-item-num">1 </span>Name</a></span></li><li><span><a href="#Search" data-toc-modified-id="Search-2"><span class="toc-item-num">2 </span>Search</a></span><ul class="toc-item"><li><span><a href="#Load-Cached-Results" data-toc-modified-id="Load-Cached-Results-2.1"><span class="toc-item-num">2.1 </span>Load Cached Results</a></span></li><li><span><a href="#Build-Model-From-Google-Images" data-toc-modified-id="Build-Model-From-Google-Images-2.2"><span class="toc-item-num">2.2 </span>Build Model From Google Images</a></span></li></ul></li><li><span><a href="#Analysis" data-toc-modified-id="Analysis-3"><span class="toc-item-num">3 </span>Analysis</a></span><ul class="toc-item"><li><span><a href="#Gender-cross-validation" data-toc-modified-id="Gender-cross-validation-3.1"><span class="toc-item-num">3.1 </span>Gender cross validation</a></span></li><li><span><a href="#Face-Sizes" data-toc-modified-id="Face-Sizes-3.2"><span class="toc-item-num">3.2 </span>Face Sizes</a></span></li><li><span><a href="#Screen-Time-Across-All-Shows" data-toc-modified-id="Screen-Time-Across-All-Shows-3.3"><span class="toc-item-num">3.3 </span>Screen Time Across All Shows</a></span></li><li><span><a href="#Appearances-on-a-Single-Show" data-toc-modified-id="Appearances-on-a-Single-Show-3.4"><span class="toc-item-num">3.4 </span>Appearances on a Single Show</a></span></li></ul></li><li><span><a href="#Persist-to-Cloud" data-toc-modified-id="Persist-to-Cloud-4"><span class="toc-item-num">4 </span>Persist to Cloud</a></span><ul class="toc-item"><li><span><a href="#Save-Model-to-Google-Cloud-Storage" data-toc-modified-id="Save-Model-to-Google-Cloud-Storage-4.1"><span class="toc-item-num">4.1 </span>Save Model to Google Cloud Storage</a></span></li><li><span><a href="#Save-Labels-to-DB" data-toc-modified-id="Save-Labels-to-DB-4.2"><span class="toc-item-num">4.2 </span>Save Labels to DB</a></span><ul class="toc-item"><li><span><a href="#Commit-the-person-and-labeler" data-toc-modified-id="Commit-the-person-and-labeler-4.2.1"><span class="toc-item-num">4.2.1 </span>Commit the person and labeler</a></span></li><li><span><a href="#Commit-the-FaceIdentity-labels" data-toc-modified-id="Commit-the-FaceIdentity-labels-4.2.2"><span class="toc-item-num">4.2.2 </span>Commit the FaceIdentity labels</a></span></li></ul></li></ul></li></ul></div>
```
from esper.prelude import *
from esper.identity import *
from esper import embed_google_images
```
# Name
Please add the person's name and their expected gender below (Male/Female).
```
name = 'Jesse Watters'
gender = 'Male'
```
# Search
## Load Cached Results
Reads cached identity model from local disk. Run this if the person has been labelled before and you only wish to regenerate the graphs. Otherwise, if you have never created a model for this person, please see the next section.
```
assert name != ''
results = FaceIdentityModel.load(name=name)
imshow(tile_imgs([cv2.resize(x[1][0], (200, 200)) for x in results.model_params['images']], cols=10))
plt.show()
plot_precision_and_cdf(results)
```
## Build Model From Google Images
Run this section if you do not have a cached model and precision curve estimates. This section will grab images using Google Image Search and score each of the faces in the dataset. We will interactively build the precision vs score curve.
It is important that the images that you select are accurate. If you make a mistake, rerun the cell below.
```
assert name != ''
# Grab face images from Google
img_dir = embed_google_images.fetch_images(name)
# If the images returned are not satisfactory, rerun the above with extra params:
# query_extras='' # additional keywords to add to search
# force=True # ignore cached images
face_imgs = load_and_select_faces_from_images(img_dir)
face_embs = embed_google_images.embed_images(face_imgs)
assert(len(face_embs) == len(face_imgs))
reference_imgs = tile_imgs([cv2.resize(x[0], (200, 200)) for x in face_imgs if x], cols=10)
def show_reference_imgs():
print('User selected reference images for {}.'.format(name))
imshow(reference_imgs)
plt.show()
show_reference_imgs()
# Score all of the faces in the dataset (this can take a minute)
face_ids_by_bucket, face_ids_to_score = face_search_by_embeddings(face_embs)
precision_model = PrecisionModel(face_ids_by_bucket)
```
Now we will validate which of the images in the dataset are of the target identity.
__Hover over with mouse and press S to select a face. Press F to expand the frame.__
```
show_reference_imgs()
print(('Mark all images that ARE NOT {}. Thumbnails are ordered by DESCENDING distance '
'to your selected images. (The first page is more likely to have non "{}" images.) '
'There are a total of {} frames. (CLICK THE DISABLE JUPYTER KEYBOARD BUTTON '
'BEFORE PROCEEDING.)').format(
name, name, precision_model.get_lower_count()))
lower_widget = precision_model.get_lower_widget()
lower_widget
show_reference_imgs()
print(('Mark all images that ARE {}. Thumbnails are ordered by ASCENDING distance '
'to your selected images. (The first page is more likely to have "{}" images.) '
'There are a total of {} frames. (CLICK THE DISABLE JUPYTER KEYBOARD BUTTON '
'BEFORE PROCEEDING.)').format(
name, name, precision_model.get_lower_count()))
upper_widget = precision_model.get_upper_widget()
upper_widget
```
Run the following cell after labelling to compute the precision curve. Do not forget to re-enable jupyter shortcuts.
```
# Compute the precision from the selections
lower_precision = precision_model.compute_precision_for_lower_buckets(lower_widget.selected)
upper_precision = precision_model.compute_precision_for_upper_buckets(upper_widget.selected)
precision_by_bucket = {**lower_precision, **upper_precision}
results = FaceIdentityModel(
name=name,
face_ids_by_bucket=face_ids_by_bucket,
face_ids_to_score=face_ids_to_score,
precision_by_bucket=precision_by_bucket,
model_params={
'images': list(zip(face_embs, face_imgs))
}
)
plot_precision_and_cdf(results)
```
The next cell persists the model locally.
```
results.save()
```
# Analysis
## Gender cross validation
Situations where the identity model disagrees with the gender classifier may be cause for alarm. We would like to check that instances of the person have the expected gender as a sanity check. This section shows the breakdown of the identity instances and their labels from the gender classifier.
```
gender_breakdown = compute_gender_breakdown(results)
print('Expected counts by gender:')
for k, v in gender_breakdown.items():
print(' {} : {}'.format(k, int(v)))
print()
print('Percentage by gender:')
denominator = sum(v for v in gender_breakdown.values())
for k, v in gender_breakdown.items():
print(' {} : {:0.1f}%'.format(k, 100 * v / denominator))
print()
```
Situations where the identity detector returns high confidence, but where the gender is not the expected gender indicate either an error on the part of the identity detector or the gender detector. The following visualization shows randomly sampled images, where the identity detector returns high confidence, grouped by the gender label.
```
high_probability_threshold = 0.8
show_gender_examples(results, high_probability_threshold)
```
## Face Sizes
Faces shown on-screen vary in size. For a person such as a host, they may be shown in a full body shot or as a face in a box. Faces in the background or those part of side graphics might be smaller than the rest. When calculuating screentime for a person, we would like to know whether the results represent the time the person was featured as opposed to merely in the background or as a tiny thumbnail in some graphic.
The next cell, plots the distribution of face sizes. Some possible anomalies include there only being very small faces or large faces.
```
plot_histogram_of_face_sizes(results)
```
The histogram above shows the distribution of face sizes, but not how those sizes occur in the dataset. For instance, one might ask why some faces are so large or whhether the small faces are actually errors. The following cell groups example faces, which are of the target identity with probability, by their sizes in terms of screen area.
```
high_probability_threshold = 0.8
show_faces_by_size(results, high_probability_threshold, n=10)
```
## Screen Time Across All Shows
One question that we might ask about a person is whether they received a significantly different amount of screentime on different shows. The following section visualizes the amount of screentime by show in total minutes and also in proportion of the show's total time. For a celebrity or political figure such as Donald Trump, we would expect significant screentime on many shows. For a show host such as Wolf Blitzer, we expect that the screentime be high for shows hosted by Wolf Blitzer.
```
screen_time_by_show = get_screen_time_by_show(results)
plot_screen_time_by_show(name, screen_time_by_show)
```
## Appearances on a Single Show
For people such as hosts, we would like to examine in greater detail the screen time allotted for a single show. First, fill in a show below.
```
show_name = 'The Five'
# Compute the screen time for each video of the show
screen_time_by_video_id = compute_screen_time_by_video(results, show_name)
```
One question we might ask about a host is "how long they are show on screen" for an episode. Likewise, we might also ask for how many episodes is the host not present due to being on vacation or on assignment elsewhere. The following cell plots a histogram of the distribution of the length of the person's appearances in videos of the chosen show.
```
plot_histogram_of_screen_times_by_video(name, show_name, screen_time_by_video_id)
```
For a host, we expect screentime over time to be consistent as long as the person remains a host. For figures such as Hilary Clinton, we expect the screentime to track events in the real world such as the lead-up to 2016 election and then to drop afterwards. The following cell plots a time series of the person's screentime over time. Each dot is a video of the chosen show. Red Xs are videos for which the face detector did not run.
```
plot_screentime_over_time(name, show_name, screen_time_by_video_id)
```
We hypothesized that a host is more likely to appear at the beginning of a video and then also appear throughout the video. The following plot visualizes the distibution of shot beginning times for videos of the show.
```
plot_distribution_of_appearance_times_by_video(results, show_name)
```
In the section 3.3, we see that some shows may have much larger variance in the screen time estimates than others. This may be because a host or frequent guest appears similar to the target identity. Alternatively, the images of the identity may be consistently low quality, leading to lower scores. The next cell plots a histogram of the probabilites for for faces in a show.
```
plot_distribution_of_identity_probabilities(results, show_name)
```
# Persist to Cloud
The remaining code in this notebook uploads the built identity model to Google Cloud Storage and adds the FaceIdentity labels to the database.
## Save Model to Google Cloud Storage
```
gcs_model_path = results.save_to_gcs()
```
To ensure that the model stored to Google Cloud is valid, we load it and print the precision and cdf curve below.
```
gcs_results = FaceIdentityModel.load_from_gcs(name=name)
imshow(tile_imgs([cv2.resize(x[1][0], (200, 200)) for x in gcs_results.model_params['images']], cols=10))
plt.show()
plot_precision_and_cdf(gcs_results)
```
## Save Labels to DB
If you are satisfied with the model, we can commit the labels to the database.
```
from django.core.exceptions import ObjectDoesNotExist
def standardize_name(name):
return name.lower()
person_type = ThingType.objects.get(name='person')
try:
person = Thing.objects.get(name=standardize_name(name), type=person_type)
print('Found person:', person.name)
except ObjectDoesNotExist:
person = Thing(name=standardize_name(name), type=person_type)
print('Creating person:', person.name)
labeler = Labeler(name='face-identity-{}'.format(person.name), data_path=gcs_model_path)
```
### Commit the person and labeler
The labeler and person have been created but not set saved to the database. If a person was created, please make sure that the name is correct before saving.
```
person.save()
labeler.save()
```
### Commit the FaceIdentity labels
Now, we are ready to add the labels to the database. We will create a FaceIdentity for each face whose probability exceeds the minimum threshold.
```
commit_face_identities_to_db(results, person, labeler, min_threshold=0.001)
print('Committed {} labels to the db'.format(FaceIdentity.objects.filter(labeler=labeler).count()))
```
|
github_jupyter
|
from esper.prelude import *
from esper.identity import *
from esper import embed_google_images
name = 'Jesse Watters'
gender = 'Male'
assert name != ''
results = FaceIdentityModel.load(name=name)
imshow(tile_imgs([cv2.resize(x[1][0], (200, 200)) for x in results.model_params['images']], cols=10))
plt.show()
plot_precision_and_cdf(results)
assert name != ''
# Grab face images from Google
img_dir = embed_google_images.fetch_images(name)
# If the images returned are not satisfactory, rerun the above with extra params:
# query_extras='' # additional keywords to add to search
# force=True # ignore cached images
face_imgs = load_and_select_faces_from_images(img_dir)
face_embs = embed_google_images.embed_images(face_imgs)
assert(len(face_embs) == len(face_imgs))
reference_imgs = tile_imgs([cv2.resize(x[0], (200, 200)) for x in face_imgs if x], cols=10)
def show_reference_imgs():
print('User selected reference images for {}.'.format(name))
imshow(reference_imgs)
plt.show()
show_reference_imgs()
# Score all of the faces in the dataset (this can take a minute)
face_ids_by_bucket, face_ids_to_score = face_search_by_embeddings(face_embs)
precision_model = PrecisionModel(face_ids_by_bucket)
show_reference_imgs()
print(('Mark all images that ARE NOT {}. Thumbnails are ordered by DESCENDING distance '
'to your selected images. (The first page is more likely to have non "{}" images.) '
'There are a total of {} frames. (CLICK THE DISABLE JUPYTER KEYBOARD BUTTON '
'BEFORE PROCEEDING.)').format(
name, name, precision_model.get_lower_count()))
lower_widget = precision_model.get_lower_widget()
lower_widget
show_reference_imgs()
print(('Mark all images that ARE {}. Thumbnails are ordered by ASCENDING distance '
'to your selected images. (The first page is more likely to have "{}" images.) '
'There are a total of {} frames. (CLICK THE DISABLE JUPYTER KEYBOARD BUTTON '
'BEFORE PROCEEDING.)').format(
name, name, precision_model.get_lower_count()))
upper_widget = precision_model.get_upper_widget()
upper_widget
# Compute the precision from the selections
lower_precision = precision_model.compute_precision_for_lower_buckets(lower_widget.selected)
upper_precision = precision_model.compute_precision_for_upper_buckets(upper_widget.selected)
precision_by_bucket = {**lower_precision, **upper_precision}
results = FaceIdentityModel(
name=name,
face_ids_by_bucket=face_ids_by_bucket,
face_ids_to_score=face_ids_to_score,
precision_by_bucket=precision_by_bucket,
model_params={
'images': list(zip(face_embs, face_imgs))
}
)
plot_precision_and_cdf(results)
results.save()
gender_breakdown = compute_gender_breakdown(results)
print('Expected counts by gender:')
for k, v in gender_breakdown.items():
print(' {} : {}'.format(k, int(v)))
print()
print('Percentage by gender:')
denominator = sum(v for v in gender_breakdown.values())
for k, v in gender_breakdown.items():
print(' {} : {:0.1f}%'.format(k, 100 * v / denominator))
print()
high_probability_threshold = 0.8
show_gender_examples(results, high_probability_threshold)
plot_histogram_of_face_sizes(results)
high_probability_threshold = 0.8
show_faces_by_size(results, high_probability_threshold, n=10)
screen_time_by_show = get_screen_time_by_show(results)
plot_screen_time_by_show(name, screen_time_by_show)
show_name = 'The Five'
# Compute the screen time for each video of the show
screen_time_by_video_id = compute_screen_time_by_video(results, show_name)
plot_histogram_of_screen_times_by_video(name, show_name, screen_time_by_video_id)
plot_screentime_over_time(name, show_name, screen_time_by_video_id)
plot_distribution_of_appearance_times_by_video(results, show_name)
plot_distribution_of_identity_probabilities(results, show_name)
gcs_model_path = results.save_to_gcs()
gcs_results = FaceIdentityModel.load_from_gcs(name=name)
imshow(tile_imgs([cv2.resize(x[1][0], (200, 200)) for x in gcs_results.model_params['images']], cols=10))
plt.show()
plot_precision_and_cdf(gcs_results)
from django.core.exceptions import ObjectDoesNotExist
def standardize_name(name):
return name.lower()
person_type = ThingType.objects.get(name='person')
try:
person = Thing.objects.get(name=standardize_name(name), type=person_type)
print('Found person:', person.name)
except ObjectDoesNotExist:
person = Thing(name=standardize_name(name), type=person_type)
print('Creating person:', person.name)
labeler = Labeler(name='face-identity-{}'.format(person.name), data_path=gcs_model_path)
person.save()
labeler.save()
commit_face_identities_to_db(results, person, labeler, min_threshold=0.001)
print('Committed {} labels to the db'.format(FaceIdentity.objects.filter(labeler=labeler).count()))
| 0.63477 | 0.942981 |
## BiRNN Overview
<img src="https://ai2-s2-public.s3.amazonaws.com/figures/2016-11-08/191dd7df9cb91ac22f56ed0dfa4a5651e8767a51/1-Figure2-1.png" alt="nn" style="width: 600px;"/>
References:
- [Long Short Term Memory](http://deeplearning.cs.cmu.edu/pdfs/Hochreiter97_lstm.pdf), Sepp Hochreiter & Jurgen Schmidhuber, Neural Computation 9(8): 1735-1780, 1997.
## MNIST Dataset Overview
This example is using MNIST handwritten digits. The dataset contains 60,000 examples for training and 10,000 examples for testing. The digits have been size-normalized and centered in a fixed-size image (28x28 pixels) with values from 0 to 1. For simplicity, each image has been flattened and converted to a 1-D numpy array of 784 features (28*28).

To classify images using a recurrent neural network, we consider every image row as a sequence of pixels. Because MNIST image shape is 28*28px, we will then handle 28 sequences of 28 timesteps for every sample.
More info: http://yann.lecun.com/exdb/mnist/
```
from __future__ import print_function
import tensorflow as tf
from tensorflow.contrib import rnn
import numpy as np
# Import MNIST data
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("/tmp/data/", one_hot=True)
# Training Parameters
learning_rate = 0.001
training_steps = 100
batch_size = 128
display_step = 20
# Network Parameters
num_input = 28 # MNIST data input (img shape: 28*28)
timesteps = 28 # timesteps
num_hidden = 128 # hidden layer num of features
num_classes = 10 # MNIST total classes (0-9 digits)
# tf Graph input
X = tf.placeholder("float", [None, timesteps, num_input])
Y = tf.placeholder("float", [None, num_classes])
# Define weights
weights = {
# Hidden layer weights => 2*n_hidden because of forward + backward cells
'out': tf.Variable(tf.random_normal([2*num_hidden, num_classes]))
}
biases = {
'out': tf.Variable(tf.random_normal([num_classes]))
}
def BiRNN(x, weights, biases):
# Prepare data shape to match `rnn` function requirements
# Current data input shape: (batch_size, timesteps, n_input)
# Required shape: 'timesteps' tensors list of shape (batch_size, num_input)
# Unstack to get a list of 'timesteps' tensors of shape (batch_size, num_input)
x = tf.unstack(x, timesteps, 1)
# Define lstm cells with tensorflow
# Forward direction cell
lstm_fw_cell = rnn.BasicLSTMCell(num_hidden, forget_bias=1.0)
# Backward direction cell
lstm_bw_cell = rnn.BasicLSTMCell(num_hidden, forget_bias=1.0)
# Get lstm cell output
try:
outputs, _, _ = rnn.static_bidirectional_rnn(lstm_fw_cell, lstm_bw_cell, x,
dtype=tf.float32)
except Exception: # Old TensorFlow version only returns outputs not states
outputs = rnn.static_bidirectional_rnn(lstm_fw_cell, lstm_bw_cell, x,
dtype=tf.float32)
# Linear activation, using rnn inner loop last output
return tf.matmul(outputs[-1], weights['out']) + biases['out']
logits = BiRNN(X, weights, biases)
prediction = tf.nn.softmax(logits)
# Define loss and optimizer
loss_op = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(
logits=logits, labels=Y))
optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)
train_op = optimizer.minimize(loss_op)
# Evaluate model (with test logits, for dropout to be disabled)
correct_pred = tf.equal(tf.argmax(prediction, 1), tf.argmax(Y, 1))
accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
# Initialize the variables (i.e. assign their default value)
init = tf.global_variables_initializer()
# Start training
with tf.Session() as sess:
# Run the initializer
sess.run(init)
for step in range(1, training_steps+1):
batch_x, batch_y = mnist.train.next_batch(batch_size)
# Reshape data to get 28 seq of 28 elements
batch_x = batch_x.reshape((batch_size, timesteps, num_input))
# Run optimization op (backprop)
sess.run(train_op, feed_dict={X: batch_x, Y: batch_y})
if step % display_step == 0 or step == 1:
# Calculate batch loss and accuracy
loss, acc = sess.run([loss_op, accuracy], feed_dict={X: batch_x,
Y: batch_y})
print("Step " + str(step) + ", Minibatch Loss= " + \
"{:.4f}".format(loss) + ", Training Accuracy= " + \
"{:.3f}".format(acc))
print("Optimization Finished!")
# Calculate accuracy for 128 mnist test images
test_len = 128
test_data = mnist.test.images[:test_len].reshape((-1, timesteps, num_input))
test_label = mnist.test.labels[:test_len]
print("Testing Accuracy:", \
sess.run(accuracy, feed_dict={X: test_data, Y: test_label}))
```
|
github_jupyter
|
from __future__ import print_function
import tensorflow as tf
from tensorflow.contrib import rnn
import numpy as np
# Import MNIST data
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("/tmp/data/", one_hot=True)
# Training Parameters
learning_rate = 0.001
training_steps = 100
batch_size = 128
display_step = 20
# Network Parameters
num_input = 28 # MNIST data input (img shape: 28*28)
timesteps = 28 # timesteps
num_hidden = 128 # hidden layer num of features
num_classes = 10 # MNIST total classes (0-9 digits)
# tf Graph input
X = tf.placeholder("float", [None, timesteps, num_input])
Y = tf.placeholder("float", [None, num_classes])
# Define weights
weights = {
# Hidden layer weights => 2*n_hidden because of forward + backward cells
'out': tf.Variable(tf.random_normal([2*num_hidden, num_classes]))
}
biases = {
'out': tf.Variable(tf.random_normal([num_classes]))
}
def BiRNN(x, weights, biases):
# Prepare data shape to match `rnn` function requirements
# Current data input shape: (batch_size, timesteps, n_input)
# Required shape: 'timesteps' tensors list of shape (batch_size, num_input)
# Unstack to get a list of 'timesteps' tensors of shape (batch_size, num_input)
x = tf.unstack(x, timesteps, 1)
# Define lstm cells with tensorflow
# Forward direction cell
lstm_fw_cell = rnn.BasicLSTMCell(num_hidden, forget_bias=1.0)
# Backward direction cell
lstm_bw_cell = rnn.BasicLSTMCell(num_hidden, forget_bias=1.0)
# Get lstm cell output
try:
outputs, _, _ = rnn.static_bidirectional_rnn(lstm_fw_cell, lstm_bw_cell, x,
dtype=tf.float32)
except Exception: # Old TensorFlow version only returns outputs not states
outputs = rnn.static_bidirectional_rnn(lstm_fw_cell, lstm_bw_cell, x,
dtype=tf.float32)
# Linear activation, using rnn inner loop last output
return tf.matmul(outputs[-1], weights['out']) + biases['out']
logits = BiRNN(X, weights, biases)
prediction = tf.nn.softmax(logits)
# Define loss and optimizer
loss_op = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(
logits=logits, labels=Y))
optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)
train_op = optimizer.minimize(loss_op)
# Evaluate model (with test logits, for dropout to be disabled)
correct_pred = tf.equal(tf.argmax(prediction, 1), tf.argmax(Y, 1))
accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
# Initialize the variables (i.e. assign their default value)
init = tf.global_variables_initializer()
# Start training
with tf.Session() as sess:
# Run the initializer
sess.run(init)
for step in range(1, training_steps+1):
batch_x, batch_y = mnist.train.next_batch(batch_size)
# Reshape data to get 28 seq of 28 elements
batch_x = batch_x.reshape((batch_size, timesteps, num_input))
# Run optimization op (backprop)
sess.run(train_op, feed_dict={X: batch_x, Y: batch_y})
if step % display_step == 0 or step == 1:
# Calculate batch loss and accuracy
loss, acc = sess.run([loss_op, accuracy], feed_dict={X: batch_x,
Y: batch_y})
print("Step " + str(step) + ", Minibatch Loss= " + \
"{:.4f}".format(loss) + ", Training Accuracy= " + \
"{:.3f}".format(acc))
print("Optimization Finished!")
# Calculate accuracy for 128 mnist test images
test_len = 128
test_data = mnist.test.images[:test_len].reshape((-1, timesteps, num_input))
test_label = mnist.test.labels[:test_len]
print("Testing Accuracy:", \
sess.run(accuracy, feed_dict={X: test_data, Y: test_label}))
| 0.879974 | 0.95511 |
# Model 1
## haarcascade_frontalface_default.xml
```
#importing libraries
import cv2
import os
import requests
import numpy as np
import pandas as pd
from IPython.display import display
#starting video
cap=cv2.VideoCapture(0)
#loading default cascade
face=cv2.CascadeClassifier("haarcascade_frontalface_default.xml")
#variable to be used
skip=0
face_data=[]
dataset_path='./data/'
#getting required info from user
file_roll_person=input("enter the roll number:")
stud_phone = input("enter the Phone Number :")
#saving the info in the file
df = pd.read_csv('students.csv')
data = {
"Phone Number" : [str(stud_phone)],
"Roll Number" :[ str(file_roll_person)]
}
add_df = pd.DataFrame(data)
new_df = df.append(add_df)
new_df.to_csv('students.csv',index=False)
#setting file name to roll number of user
file_name = str(file_roll_person)
#recording the face through webcam
while True:
ret,frame=cap.read()
#converting into gray
gray=cv2.cvtColor(frame,cv2.COLOR_BGR2GRAY)
if ret==False:
continue
#detection of face
faces=face.detectMultiScale(frame,1.3,5)
#sort them in order to achieve highest face ratio
faces=sorted(faces,key=lambda f:f[2]*f[3])
#lopping the faces and appending face data
for (x,y,w,h) in faces[-1:]:
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+w+offset]
face_section=cv2.resize(face_section,(100,100))
skip+=1
if skip%10==0:
face_data.append(face_section)
print(face_data)
cv2.imshow("frame",frame)
#cv2.imshow("face_section",face_section)
key=cv2.waitKey(30) & 0xFF
if key==ord('q'):
break
#converting data into face
face_data=np.asarray(face_data)
face_data=face_data.reshape((face_data.shape[0],-1))
#save the data
np.save(dataset_path+file_name+".npy",face_data)
#turn of the webcam
cap.release()
cv2.destroyAllWindows()
#importing the libraries
import cv2
import requests
import os
import numpy as np
import pandas as pd
from IPython.display import display
def knn(X,Y,k=5):
"""
It takes trainset,face section and nearest neighbour and based on
data it has it return highest probability prediction.
-Args: trainset,face section and nearest neighbour
-return: prediction
"""
val=[]
m=X.shape[0]
for i in range(m):
ix=X[i,:-1]
iy=X[i,-1]
d=dist(Y,ix)
val.append((d,iy))
vals=sorted(val,key=lambda x:x[0])[:k]
vals=np.array(vals)[:,-1]
new_val=np.unique(vals,return_counts=True)
index=np.argmax(new_val[1])
pred=new_val[0][index]
return pred
def dist(x1,x2):
"""
It takes X1 and X2 and it return the square root distance between them.
-Args: X1,X2
-return: distance between them
"""
return np.sqrt(sum(((x1-x2)**2)))
def mark_attendance(ids):
"""
It takes id , save the ids in attendance.csv file and send them notification on their
phone number .
-Args: ids
-return: None
"""
df = pd.DataFrame({
'Roll Number' : ids
})
df.to_csv('attendance.csv')
#saving the roll number and dropping un necessary columns
unique_phone_ = []
new_df = pd.read_csv('attendance.csv')
columns_list = np.array(new_df.columns)
drop_col = []
for col in columns_list:
if "Unnamed:" in col:
drop_col.append(col)
new_df.drop(drop_col,axis = 1,inplace=True)
new_df.fillna(0,inplace=True)
new_df.to_csv('attendance.csv',index=False)
#sending them notification using fast 2 sms service
df = pd.read_csv('students.csv')
phone_numbers = []
for idi in ids:
if int(idi) in df['Roll Number'].unique():
phone_numbers.append((df[df['Roll Number']==idi]['Phone Number'].values[0]))
url = "https://www.fast2sms.com/dev/bulk"
headers = {'authorization': "AUTHORIZATION_KEY",
'Content-Type': "application/x-www-form-urlencoded",
'Cache-Control': "no-cache",
}
print("before sending messages")
print(phone_numbers)
for num in phone_numbers:
if num not in unique_phone_:
unique_phone_.append(num)
for numbers in unique_phone_:
print(numbers)
payload = "sender_id=FSTSMS&message= Your Attendance is marked &language=english&route=p&numbers="+str(numbers)
response = requests.request("POST", url, data=payload, headers=headers)
print(response.text)
cap=cv2.VideoCapture(0)
face_cascade=cv2.CascadeClassifier("haarcascade_frontalface_default.xml")
skip=0
face_data=[]
dataset_path='./data/'
label=[]
class_id=0
uniq_student_ids = []
names={}
students_ids = [ ]
stud_df = pd.read_csv('students.csv')
current_students = [ ]
student_id = ' '
for i in range(stud_df.shape[0]):
student_id = str(stud_df['Roll Number'].values[i])
current_students.append(student_id)
for fx in os.listdir(dataset_path):
if fx.endswith('.npy'):
names[class_id]=fx[:-4]
data_item=np.load(dataset_path+fx)
face_data.append(data_item)
#Create labels for class
target=class_id*np.ones((data_item.shape[0],))
class_id+=1
label.append(target)
face_dataset=np.concatenate(face_data,axis=0)
labels_dataset=np.concatenate(label,axis=0).reshape((-1,1))
trainset=np.concatenate((face_dataset,labels_dataset),axis=1)
while True:
ret,frame=cap.read()
if ret==False:
continue
faces=face_cascade.detectMultiScale(frame,1.3,5)
for face in faces:
x,y,w,h=face
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+offset+w]
face_section=cv2.resize(face_section,(100,100))
out=knn(trainset,face_section.flatten())
pred=names[int(out)]
students_ids.append(pred)
cv2.putText(frame,pred,(x,y-10),cv2.FONT_HERSHEY_SIMPLEX,0.5,(0,255,1),1,cv2.LINE_AA)
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
cv2.imshow("frame",frame)
key=cv2.waitKey(1) & 0xFF
if key==ord('q'):
break
for ids in students_ids:
if ids not in uniq_student_ids:
uniq_student_ids.append(int(ids))
print(uniq_student_ids )
mark_attendance(uniq_student_ids)
cap.release()
cv2.destroyAllWindows()
```
# Model 2
## haarcascade_frontalface_alt .xml
```
#importing libraries
import cv2
import os
import requests
import numpy as np
import pandas as pd
from IPython.display import display
#starting video
cap=cv2.VideoCapture(0)
#loading default cascade
face=cv2.CascadeClassifier("haarcascade_frontalface_alt.xml")
#variable to be used
skip=0
face_data=[]
dataset_path='./data/'
#getting required info from user
file_roll_person=input("enter the roll number:")
stud_phone = input("enter the Phone Number :")
#saving the info in the file
df = pd.read_csv('students.csv')
data = {
"Phone Number" : [str(stud_phone)],
"Roll Number" :[ str(file_roll_person)]
}
add_df = pd.DataFrame(data)
new_df = df.append(add_df)
new_df.to_csv('students.csv',index=False)
#setting file name to roll number of user
file_name = str(file_roll_person)
#recording the face through webcam
while True:
ret,frame=cap.read()
#converting into gray
gray=cv2.cvtColor(frame,cv2.COLOR_BGR2GRAY)
if ret==False:
continue
#detection of face
faces=face.detectMultiScale(frame,1.3,5)
#sort them in order to achieve highest face ratio
faces=sorted(faces,key=lambda f:f[2]*f[3])
#lopping the faces and appending face data
for (x,y,w,h) in faces[-1:]:
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+w+offset]
face_section=cv2.resize(face_section,(100,100))
skip+=1
if skip%10==0:
face_data.append(face_section)
print(face_data)
cv2.imshow("frame",frame)
#cv2.imshow("face_section",face_section)
key=cv2.waitKey(30) & 0xFF
if key==ord('q'):
break
#converting data into face
face_data=np.asarray(face_data)
face_data=face_data.reshape((face_data.shape[0],-1))
#save the data
np.save(dataset_path+file_name+".npy",face_data)
#turn of the webcam
cap.release()
cv2.destroyAllWindows()
#importing the libraries
import cv2
import requests
import os
import numpy as np
import pandas as pd
from IPython.display import display
def knn(X,Y,k=5):
"""
It takes trainset,face section and nearest neighbour and based on
data it has it return highest probability prediction.
-Args: trainset,face section and nearest neighbour
-return: prediction
"""
val=[]
m=X.shape[0]
for i in range(m):
ix=X[i,:-1]
iy=X[i,-1]
d=dist(Y,ix)
val.append((d,iy))
vals=sorted(val,key=lambda x:x[0])[:k]
vals=np.array(vals)[:,-1]
new_val=np.unique(vals,return_counts=True)
index=np.argmax(new_val[1])
pred=new_val[0][index]
return pred
def dist(x1,x2):
"""
It takes X1 and X2 and it return the square root distance between them.
-Args: X1,X2
-return: distance between them
"""
return np.sqrt(sum(((x1-x2)**2)))
def mark_attendance(ids):
"""
It takes id , save the ids in attendance.csv file and send them notification on their
phone number .
-Args: ids
-return: None
"""
df = pd.DataFrame({
'Roll Number' : ids
})
df.to_csv('attendance.csv')
#saving the roll number and dropping un necessary columns
unique_phone_ = []
new_df = pd.read_csv('attendance.csv')
columns_list = np.array(new_df.columns)
drop_col = []
for col in columns_list:
if "Unnamed:" in col:
drop_col.append(col)
new_df.drop(drop_col,axis = 1,inplace=True)
new_df.fillna(0,inplace=True)
new_df.to_csv('attendance.csv',index=False)
#sending them notification using fast 2 sms service
df = pd.read_csv('students.csv')
phone_numbers = []
for idi in ids:
if int(idi) in df['Roll Number'].unique():
phone_numbers.append((df[df['Roll Number']==idi]['Phone Number'].values[0]))
url = "https://www.fast2sms.com/dev/bulk"
headers = {'authorization': "AUTHORIZATION_KEY",
'Content-Type': "application/x-www-form-urlencoded",
'Cache-Control': "no-cache",
}
print("before sending messages")
print(phone_numbers)
for num in phone_numbers:
if num not in unique_phone_:
unique_phone_.append(num)
for numbers in unique_phone_:
print(numbers)
payload = "sender_id=FSTSMS&message= Your Attendance is marked &language=english&route=p&numbers="+str(numbers)
response = requests.request("POST", url, data=payload, headers=headers)
print(response.text)
cap=cv2.VideoCapture(0)
face_cascade=cv2.CascadeClassifier("haarcascade_frontalface_alt.xml")
skip=0
face_data=[]
dataset_path='./data/'
label=[]
class_id=0
uniq_student_ids = []
names={}
students_ids = [ ]
stud_df = pd.read_csv('students.csv')
current_students = [ ]
student_id = ' '
for i in range(stud_df.shape[0]):
student_id = str(stud_df['Roll Number'].values[i])
current_students.append(student_id)
for fx in os.listdir(dataset_path):
if fx.endswith('.npy'):
names[class_id]=fx[:-4]
data_item=np.load(dataset_path+fx)
face_data.append(data_item)
#Create labels for class
target=class_id*np.ones((data_item.shape[0],))
class_id+=1
label.append(target)
face_dataset=np.concatenate(face_data,axis=0)
labels_dataset=np.concatenate(label,axis=0).reshape((-1,1))
trainset=np.concatenate((face_dataset,labels_dataset),axis=1)
while True:
ret,frame=cap.read()
if ret==False:
continue
faces=face_cascade.detectMultiScale(frame,1.3,5)
for face in faces:
x,y,w,h=face
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+offset+w]
face_section=cv2.resize(face_section,(100,100))
out=knn(trainset,face_section.flatten())
pred=names[int(out)]
students_ids.append(pred)
cv2.putText(frame,pred,(x,y-10),cv2.FONT_HERSHEY_SIMPLEX,0.5,(0,255,1),1,cv2.LINE_AA)
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
cv2.imshow("frame",frame)
key=cv2.waitKey(1) & 0xFF
if key==ord('q'):
break
for ids in students_ids:
if ids not in uniq_student_ids:
uniq_student_ids.append(int(ids))
print(uniq_student_ids )
mark_attendance(uniq_student_ids)
cap.release()
cv2.destroyAllWindows()
```
# Model 3
## haarcascade_frontalface_alt2 .xml
```
#importing libraries
import cv2
import os
import requests
import numpy as np
import pandas as pd
from IPython.display import display
#starting video
cap=cv2.VideoCapture(0)
#loading default cascade
face=cv2.CascadeClassifier("haarcascade_frontalface_alt2.xml")
#variable to be used
skip=0
face_data=[]
dataset_path='./data/'
#getting required info from user
file_roll_person=input("enter the roll number:")
stud_phone = input("enter the Phone Number :")
#saving the info in the file
df = pd.read_csv('students.csv')
data = {
"Phone Number" : [str(stud_phone)],
"Roll Number" :[ str(file_roll_person)]
}
add_df = pd.DataFrame(data)
new_df = df.append(add_df)
new_df.to_csv('students.csv',index=False)
#setting file name to roll number of user
file_name = str(file_roll_person)
#recording the face through webcam
while True:
ret,frame=cap.read()
#converting into gray
gray=cv2.cvtColor(frame,cv2.COLOR_BGR2GRAY)
if ret==False:
continue
#detection of face
faces=face.detectMultiScale(frame,1.3,5)
#sort them in order to achieve highest face ratio
faces=sorted(faces,key=lambda f:f[2]*f[3])
#lopping the faces and appending face data
for (x,y,w,h) in faces[-1:]:
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+w+offset]
face_section=cv2.resize(face_section,(100,100))
skip+=1
if skip%10==0:
face_data.append(face_section)
print(face_data)
cv2.imshow("frame",frame)
#cv2.imshow("face_section",face_section)
key=cv2.waitKey(30) & 0xFF
if key==ord('q'):
break
#converting data into face
face_data=np.asarray(face_data)
face_data=face_data.reshape((face_data.shape[0],-1))
#save the data
np.save(dataset_path+file_name+".npy",face_data)
#turn of the webcam
cap.release()
cv2.destroyAllWindows()
#importing the libraries
import cv2
import requests
import os
import numpy as np
import pandas as pd
from IPython.display import display
def knn(X,Y,k=5):
"""
It takes trainset,face section and nearest neighbour and based on
data it has it return highest probability prediction.
-Args: trainset,face section and nearest neighbour
-return: prediction
"""
val=[]
m=X.shape[0]
for i in range(m):
ix=X[i,:-1]
iy=X[i,-1]
d=dist(Y,ix)
val.append((d,iy))
vals=sorted(val,key=lambda x:x[0])[:k]
vals=np.array(vals)[:,-1]
new_val=np.unique(vals,return_counts=True)
index=np.argmax(new_val[1])
pred=new_val[0][index]
return pred
def dist(x1,x2):
"""
It takes X1 and X2 and it return the square root distance between them.
-Args: X1,X2
-return: distance between them
"""
return np.sqrt(sum(((x1-x2)**2)))
def mark_attendance(ids):
"""
It takes id , save the ids in attendance.csv file and send them notification on their
phone number .
-Args: ids
-return: None
"""
df = pd.DataFrame({
'Roll Number' : ids
})
df.to_csv('attendance.csv')
#saving the roll number and dropping un necessary columns
unique_phone_ = []
new_df = pd.read_csv('attendance.csv')
columns_list = np.array(new_df.columns)
drop_col = []
for col in columns_list:
if "Unnamed:" in col:
drop_col.append(col)
new_df.drop(drop_col,axis = 1,inplace=True)
new_df.fillna(0,inplace=True)
new_df.to_csv('attendance.csv',index=False)
#sending them notification using fast 2 sms service
df = pd.read_csv('students.csv')
phone_numbers = []
for idi in ids:
if int(idi) in df['Roll Number'].unique():
phone_numbers.append((df[df['Roll Number']==idi]['Phone Number'].values[0]))
url = "https://www.fast2sms.com/dev/bulk"
headers = {'authorization': "AUTHORIZATION_KEY",
'Content-Type': "application/x-www-form-urlencoded",
'Cache-Control': "no-cache",
}
print("before sending messages")
print(phone_numbers)
for num in phone_numbers:
if num not in unique_phone_:
unique_phone_.append(num)
for numbers in unique_phone_:
print(numbers)
payload = "sender_id=FSTSMS&message= Your Attendance is marked &language=english&route=p&numbers="+str(numbers)
response = requests.request("POST", url, data=payload, headers=headers)
print(response.text)
cap=cv2.VideoCapture(0)
face_cascade=cv2.CascadeClassifier("haarcascade_frontalface_alt2.xml")
skip=0
face_data=[]
dataset_path='./data/'
label=[]
class_id=0
uniq_student_ids = []
names={}
students_ids = [ ]
stud_df = pd.read_csv('students.csv')
current_students = [ ]
student_id = ' '
for i in range(stud_df.shape[0]):
student_id = str(stud_df['Roll Number'].values[i])
current_students.append(student_id)
for fx in os.listdir(dataset_path):
if fx.endswith('.npy'):
names[class_id]=fx[:-4]
data_item=np.load(dataset_path+fx)
face_data.append(data_item)
#Create labels for class
target=class_id*np.ones((data_item.shape[0],))
class_id+=1
label.append(target)
face_dataset=np.concatenate(face_data,axis=0)
labels_dataset=np.concatenate(label,axis=0).reshape((-1,1))
trainset=np.concatenate((face_dataset,labels_dataset),axis=1)
while True:
ret,frame=cap.read()
if ret==False:
continue
faces=face_cascade.detectMultiScale(frame,1.3,5)
for face in faces:
x,y,w,h=face
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+offset+w]
face_section=cv2.resize(face_section,(100,100))
out=knn(trainset,face_section.flatten())
pred=names[int(out)]
students_ids.append(pred)
cv2.putText(frame,pred,(x,y-10),cv2.FONT_HERSHEY_SIMPLEX,0.5,(0,255,1),1,cv2.LINE_AA)
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
cv2.imshow("frame",frame)
key=cv2.waitKey(1) & 0xFF
if key==ord('q'):
break
for ids in students_ids:
if ids not in uniq_student_ids:
uniq_student_ids.append(int(ids))
print(uniq_student_ids )
mark_attendance(uniq_student_ids)
cap.release()
cv2.destroyAllWindows()
```
|
github_jupyter
|
#importing libraries
import cv2
import os
import requests
import numpy as np
import pandas as pd
from IPython.display import display
#starting video
cap=cv2.VideoCapture(0)
#loading default cascade
face=cv2.CascadeClassifier("haarcascade_frontalface_default.xml")
#variable to be used
skip=0
face_data=[]
dataset_path='./data/'
#getting required info from user
file_roll_person=input("enter the roll number:")
stud_phone = input("enter the Phone Number :")
#saving the info in the file
df = pd.read_csv('students.csv')
data = {
"Phone Number" : [str(stud_phone)],
"Roll Number" :[ str(file_roll_person)]
}
add_df = pd.DataFrame(data)
new_df = df.append(add_df)
new_df.to_csv('students.csv',index=False)
#setting file name to roll number of user
file_name = str(file_roll_person)
#recording the face through webcam
while True:
ret,frame=cap.read()
#converting into gray
gray=cv2.cvtColor(frame,cv2.COLOR_BGR2GRAY)
if ret==False:
continue
#detection of face
faces=face.detectMultiScale(frame,1.3,5)
#sort them in order to achieve highest face ratio
faces=sorted(faces,key=lambda f:f[2]*f[3])
#lopping the faces and appending face data
for (x,y,w,h) in faces[-1:]:
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+w+offset]
face_section=cv2.resize(face_section,(100,100))
skip+=1
if skip%10==0:
face_data.append(face_section)
print(face_data)
cv2.imshow("frame",frame)
#cv2.imshow("face_section",face_section)
key=cv2.waitKey(30) & 0xFF
if key==ord('q'):
break
#converting data into face
face_data=np.asarray(face_data)
face_data=face_data.reshape((face_data.shape[0],-1))
#save the data
np.save(dataset_path+file_name+".npy",face_data)
#turn of the webcam
cap.release()
cv2.destroyAllWindows()
#importing the libraries
import cv2
import requests
import os
import numpy as np
import pandas as pd
from IPython.display import display
def knn(X,Y,k=5):
"""
It takes trainset,face section and nearest neighbour and based on
data it has it return highest probability prediction.
-Args: trainset,face section and nearest neighbour
-return: prediction
"""
val=[]
m=X.shape[0]
for i in range(m):
ix=X[i,:-1]
iy=X[i,-1]
d=dist(Y,ix)
val.append((d,iy))
vals=sorted(val,key=lambda x:x[0])[:k]
vals=np.array(vals)[:,-1]
new_val=np.unique(vals,return_counts=True)
index=np.argmax(new_val[1])
pred=new_val[0][index]
return pred
def dist(x1,x2):
"""
It takes X1 and X2 and it return the square root distance between them.
-Args: X1,X2
-return: distance between them
"""
return np.sqrt(sum(((x1-x2)**2)))
def mark_attendance(ids):
"""
It takes id , save the ids in attendance.csv file and send them notification on their
phone number .
-Args: ids
-return: None
"""
df = pd.DataFrame({
'Roll Number' : ids
})
df.to_csv('attendance.csv')
#saving the roll number and dropping un necessary columns
unique_phone_ = []
new_df = pd.read_csv('attendance.csv')
columns_list = np.array(new_df.columns)
drop_col = []
for col in columns_list:
if "Unnamed:" in col:
drop_col.append(col)
new_df.drop(drop_col,axis = 1,inplace=True)
new_df.fillna(0,inplace=True)
new_df.to_csv('attendance.csv',index=False)
#sending them notification using fast 2 sms service
df = pd.read_csv('students.csv')
phone_numbers = []
for idi in ids:
if int(idi) in df['Roll Number'].unique():
phone_numbers.append((df[df['Roll Number']==idi]['Phone Number'].values[0]))
url = "https://www.fast2sms.com/dev/bulk"
headers = {'authorization': "AUTHORIZATION_KEY",
'Content-Type': "application/x-www-form-urlencoded",
'Cache-Control': "no-cache",
}
print("before sending messages")
print(phone_numbers)
for num in phone_numbers:
if num not in unique_phone_:
unique_phone_.append(num)
for numbers in unique_phone_:
print(numbers)
payload = "sender_id=FSTSMS&message= Your Attendance is marked &language=english&route=p&numbers="+str(numbers)
response = requests.request("POST", url, data=payload, headers=headers)
print(response.text)
cap=cv2.VideoCapture(0)
face_cascade=cv2.CascadeClassifier("haarcascade_frontalface_default.xml")
skip=0
face_data=[]
dataset_path='./data/'
label=[]
class_id=0
uniq_student_ids = []
names={}
students_ids = [ ]
stud_df = pd.read_csv('students.csv')
current_students = [ ]
student_id = ' '
for i in range(stud_df.shape[0]):
student_id = str(stud_df['Roll Number'].values[i])
current_students.append(student_id)
for fx in os.listdir(dataset_path):
if fx.endswith('.npy'):
names[class_id]=fx[:-4]
data_item=np.load(dataset_path+fx)
face_data.append(data_item)
#Create labels for class
target=class_id*np.ones((data_item.shape[0],))
class_id+=1
label.append(target)
face_dataset=np.concatenate(face_data,axis=0)
labels_dataset=np.concatenate(label,axis=0).reshape((-1,1))
trainset=np.concatenate((face_dataset,labels_dataset),axis=1)
while True:
ret,frame=cap.read()
if ret==False:
continue
faces=face_cascade.detectMultiScale(frame,1.3,5)
for face in faces:
x,y,w,h=face
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+offset+w]
face_section=cv2.resize(face_section,(100,100))
out=knn(trainset,face_section.flatten())
pred=names[int(out)]
students_ids.append(pred)
cv2.putText(frame,pred,(x,y-10),cv2.FONT_HERSHEY_SIMPLEX,0.5,(0,255,1),1,cv2.LINE_AA)
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
cv2.imshow("frame",frame)
key=cv2.waitKey(1) & 0xFF
if key==ord('q'):
break
for ids in students_ids:
if ids not in uniq_student_ids:
uniq_student_ids.append(int(ids))
print(uniq_student_ids )
mark_attendance(uniq_student_ids)
cap.release()
cv2.destroyAllWindows()
#importing libraries
import cv2
import os
import requests
import numpy as np
import pandas as pd
from IPython.display import display
#starting video
cap=cv2.VideoCapture(0)
#loading default cascade
face=cv2.CascadeClassifier("haarcascade_frontalface_alt.xml")
#variable to be used
skip=0
face_data=[]
dataset_path='./data/'
#getting required info from user
file_roll_person=input("enter the roll number:")
stud_phone = input("enter the Phone Number :")
#saving the info in the file
df = pd.read_csv('students.csv')
data = {
"Phone Number" : [str(stud_phone)],
"Roll Number" :[ str(file_roll_person)]
}
add_df = pd.DataFrame(data)
new_df = df.append(add_df)
new_df.to_csv('students.csv',index=False)
#setting file name to roll number of user
file_name = str(file_roll_person)
#recording the face through webcam
while True:
ret,frame=cap.read()
#converting into gray
gray=cv2.cvtColor(frame,cv2.COLOR_BGR2GRAY)
if ret==False:
continue
#detection of face
faces=face.detectMultiScale(frame,1.3,5)
#sort them in order to achieve highest face ratio
faces=sorted(faces,key=lambda f:f[2]*f[3])
#lopping the faces and appending face data
for (x,y,w,h) in faces[-1:]:
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+w+offset]
face_section=cv2.resize(face_section,(100,100))
skip+=1
if skip%10==0:
face_data.append(face_section)
print(face_data)
cv2.imshow("frame",frame)
#cv2.imshow("face_section",face_section)
key=cv2.waitKey(30) & 0xFF
if key==ord('q'):
break
#converting data into face
face_data=np.asarray(face_data)
face_data=face_data.reshape((face_data.shape[0],-1))
#save the data
np.save(dataset_path+file_name+".npy",face_data)
#turn of the webcam
cap.release()
cv2.destroyAllWindows()
#importing the libraries
import cv2
import requests
import os
import numpy as np
import pandas as pd
from IPython.display import display
def knn(X,Y,k=5):
"""
It takes trainset,face section and nearest neighbour and based on
data it has it return highest probability prediction.
-Args: trainset,face section and nearest neighbour
-return: prediction
"""
val=[]
m=X.shape[0]
for i in range(m):
ix=X[i,:-1]
iy=X[i,-1]
d=dist(Y,ix)
val.append((d,iy))
vals=sorted(val,key=lambda x:x[0])[:k]
vals=np.array(vals)[:,-1]
new_val=np.unique(vals,return_counts=True)
index=np.argmax(new_val[1])
pred=new_val[0][index]
return pred
def dist(x1,x2):
"""
It takes X1 and X2 and it return the square root distance between them.
-Args: X1,X2
-return: distance between them
"""
return np.sqrt(sum(((x1-x2)**2)))
def mark_attendance(ids):
"""
It takes id , save the ids in attendance.csv file and send them notification on their
phone number .
-Args: ids
-return: None
"""
df = pd.DataFrame({
'Roll Number' : ids
})
df.to_csv('attendance.csv')
#saving the roll number and dropping un necessary columns
unique_phone_ = []
new_df = pd.read_csv('attendance.csv')
columns_list = np.array(new_df.columns)
drop_col = []
for col in columns_list:
if "Unnamed:" in col:
drop_col.append(col)
new_df.drop(drop_col,axis = 1,inplace=True)
new_df.fillna(0,inplace=True)
new_df.to_csv('attendance.csv',index=False)
#sending them notification using fast 2 sms service
df = pd.read_csv('students.csv')
phone_numbers = []
for idi in ids:
if int(idi) in df['Roll Number'].unique():
phone_numbers.append((df[df['Roll Number']==idi]['Phone Number'].values[0]))
url = "https://www.fast2sms.com/dev/bulk"
headers = {'authorization': "AUTHORIZATION_KEY",
'Content-Type': "application/x-www-form-urlencoded",
'Cache-Control': "no-cache",
}
print("before sending messages")
print(phone_numbers)
for num in phone_numbers:
if num not in unique_phone_:
unique_phone_.append(num)
for numbers in unique_phone_:
print(numbers)
payload = "sender_id=FSTSMS&message= Your Attendance is marked &language=english&route=p&numbers="+str(numbers)
response = requests.request("POST", url, data=payload, headers=headers)
print(response.text)
cap=cv2.VideoCapture(0)
face_cascade=cv2.CascadeClassifier("haarcascade_frontalface_alt.xml")
skip=0
face_data=[]
dataset_path='./data/'
label=[]
class_id=0
uniq_student_ids = []
names={}
students_ids = [ ]
stud_df = pd.read_csv('students.csv')
current_students = [ ]
student_id = ' '
for i in range(stud_df.shape[0]):
student_id = str(stud_df['Roll Number'].values[i])
current_students.append(student_id)
for fx in os.listdir(dataset_path):
if fx.endswith('.npy'):
names[class_id]=fx[:-4]
data_item=np.load(dataset_path+fx)
face_data.append(data_item)
#Create labels for class
target=class_id*np.ones((data_item.shape[0],))
class_id+=1
label.append(target)
face_dataset=np.concatenate(face_data,axis=0)
labels_dataset=np.concatenate(label,axis=0).reshape((-1,1))
trainset=np.concatenate((face_dataset,labels_dataset),axis=1)
while True:
ret,frame=cap.read()
if ret==False:
continue
faces=face_cascade.detectMultiScale(frame,1.3,5)
for face in faces:
x,y,w,h=face
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+offset+w]
face_section=cv2.resize(face_section,(100,100))
out=knn(trainset,face_section.flatten())
pred=names[int(out)]
students_ids.append(pred)
cv2.putText(frame,pred,(x,y-10),cv2.FONT_HERSHEY_SIMPLEX,0.5,(0,255,1),1,cv2.LINE_AA)
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
cv2.imshow("frame",frame)
key=cv2.waitKey(1) & 0xFF
if key==ord('q'):
break
for ids in students_ids:
if ids not in uniq_student_ids:
uniq_student_ids.append(int(ids))
print(uniq_student_ids )
mark_attendance(uniq_student_ids)
cap.release()
cv2.destroyAllWindows()
#importing libraries
import cv2
import os
import requests
import numpy as np
import pandas as pd
from IPython.display import display
#starting video
cap=cv2.VideoCapture(0)
#loading default cascade
face=cv2.CascadeClassifier("haarcascade_frontalface_alt2.xml")
#variable to be used
skip=0
face_data=[]
dataset_path='./data/'
#getting required info from user
file_roll_person=input("enter the roll number:")
stud_phone = input("enter the Phone Number :")
#saving the info in the file
df = pd.read_csv('students.csv')
data = {
"Phone Number" : [str(stud_phone)],
"Roll Number" :[ str(file_roll_person)]
}
add_df = pd.DataFrame(data)
new_df = df.append(add_df)
new_df.to_csv('students.csv',index=False)
#setting file name to roll number of user
file_name = str(file_roll_person)
#recording the face through webcam
while True:
ret,frame=cap.read()
#converting into gray
gray=cv2.cvtColor(frame,cv2.COLOR_BGR2GRAY)
if ret==False:
continue
#detection of face
faces=face.detectMultiScale(frame,1.3,5)
#sort them in order to achieve highest face ratio
faces=sorted(faces,key=lambda f:f[2]*f[3])
#lopping the faces and appending face data
for (x,y,w,h) in faces[-1:]:
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+w+offset]
face_section=cv2.resize(face_section,(100,100))
skip+=1
if skip%10==0:
face_data.append(face_section)
print(face_data)
cv2.imshow("frame",frame)
#cv2.imshow("face_section",face_section)
key=cv2.waitKey(30) & 0xFF
if key==ord('q'):
break
#converting data into face
face_data=np.asarray(face_data)
face_data=face_data.reshape((face_data.shape[0],-1))
#save the data
np.save(dataset_path+file_name+".npy",face_data)
#turn of the webcam
cap.release()
cv2.destroyAllWindows()
#importing the libraries
import cv2
import requests
import os
import numpy as np
import pandas as pd
from IPython.display import display
def knn(X,Y,k=5):
"""
It takes trainset,face section and nearest neighbour and based on
data it has it return highest probability prediction.
-Args: trainset,face section and nearest neighbour
-return: prediction
"""
val=[]
m=X.shape[0]
for i in range(m):
ix=X[i,:-1]
iy=X[i,-1]
d=dist(Y,ix)
val.append((d,iy))
vals=sorted(val,key=lambda x:x[0])[:k]
vals=np.array(vals)[:,-1]
new_val=np.unique(vals,return_counts=True)
index=np.argmax(new_val[1])
pred=new_val[0][index]
return pred
def dist(x1,x2):
"""
It takes X1 and X2 and it return the square root distance between them.
-Args: X1,X2
-return: distance between them
"""
return np.sqrt(sum(((x1-x2)**2)))
def mark_attendance(ids):
"""
It takes id , save the ids in attendance.csv file and send them notification on their
phone number .
-Args: ids
-return: None
"""
df = pd.DataFrame({
'Roll Number' : ids
})
df.to_csv('attendance.csv')
#saving the roll number and dropping un necessary columns
unique_phone_ = []
new_df = pd.read_csv('attendance.csv')
columns_list = np.array(new_df.columns)
drop_col = []
for col in columns_list:
if "Unnamed:" in col:
drop_col.append(col)
new_df.drop(drop_col,axis = 1,inplace=True)
new_df.fillna(0,inplace=True)
new_df.to_csv('attendance.csv',index=False)
#sending them notification using fast 2 sms service
df = pd.read_csv('students.csv')
phone_numbers = []
for idi in ids:
if int(idi) in df['Roll Number'].unique():
phone_numbers.append((df[df['Roll Number']==idi]['Phone Number'].values[0]))
url = "https://www.fast2sms.com/dev/bulk"
headers = {'authorization': "AUTHORIZATION_KEY",
'Content-Type': "application/x-www-form-urlencoded",
'Cache-Control': "no-cache",
}
print("before sending messages")
print(phone_numbers)
for num in phone_numbers:
if num not in unique_phone_:
unique_phone_.append(num)
for numbers in unique_phone_:
print(numbers)
payload = "sender_id=FSTSMS&message= Your Attendance is marked &language=english&route=p&numbers="+str(numbers)
response = requests.request("POST", url, data=payload, headers=headers)
print(response.text)
cap=cv2.VideoCapture(0)
face_cascade=cv2.CascadeClassifier("haarcascade_frontalface_alt2.xml")
skip=0
face_data=[]
dataset_path='./data/'
label=[]
class_id=0
uniq_student_ids = []
names={}
students_ids = [ ]
stud_df = pd.read_csv('students.csv')
current_students = [ ]
student_id = ' '
for i in range(stud_df.shape[0]):
student_id = str(stud_df['Roll Number'].values[i])
current_students.append(student_id)
for fx in os.listdir(dataset_path):
if fx.endswith('.npy'):
names[class_id]=fx[:-4]
data_item=np.load(dataset_path+fx)
face_data.append(data_item)
#Create labels for class
target=class_id*np.ones((data_item.shape[0],))
class_id+=1
label.append(target)
face_dataset=np.concatenate(face_data,axis=0)
labels_dataset=np.concatenate(label,axis=0).reshape((-1,1))
trainset=np.concatenate((face_dataset,labels_dataset),axis=1)
while True:
ret,frame=cap.read()
if ret==False:
continue
faces=face_cascade.detectMultiScale(frame,1.3,5)
for face in faces:
x,y,w,h=face
offset=10
face_section=frame[y-offset:y+h+offset,x-offset:x+offset+w]
face_section=cv2.resize(face_section,(100,100))
out=knn(trainset,face_section.flatten())
pred=names[int(out)]
students_ids.append(pred)
cv2.putText(frame,pred,(x,y-10),cv2.FONT_HERSHEY_SIMPLEX,0.5,(0,255,1),1,cv2.LINE_AA)
cv2.rectangle(frame,(x,y),(x+w,y+h),(0,255,255),2)
cv2.imshow("frame",frame)
key=cv2.waitKey(1) & 0xFF
if key==ord('q'):
break
for ids in students_ids:
if ids not in uniq_student_ids:
uniq_student_ids.append(int(ids))
print(uniq_student_ids )
mark_attendance(uniq_student_ids)
cap.release()
cv2.destroyAllWindows()
| 0.166981 | 0.488344 |
# TfIdf Vectorization of a large corpus
Usually Tfidf vectors need to be trained on a domain-specific corpus. However, in many cases, a generic baseline of idf values can be good enough, and helpful for computing generic tasks like weighting sentence embeddings. Besides the obvious memory challenges with processing a large corpus, there are important questions that need to be resolved when organizing a collection of documents:
* What is considered a document?
* is one epistle one document?
* is one section or chapter of one speech one document?
* is one poem a one document? ranging from epigram to a book of epic poetry?
* is one chapter in a prose book one document?
* Disagree with any of these? then you'll want to train your own word idf mapping and compare results.
* How can we compare TfIdf vectors, what are some simple baselines?
In this notebook we'll work towards creating a generic tfidf vector for a discrete but general purpose corpus.
Of course, any time you can limit the scope of your documents to a particular domain and train on those, then you will get better results, but to handle unseen data in a robust manner, a general idf mapping is better than assuming a uniform distribution!
We'll look at the Tessearae corpus, and generate a word : idf mapping that we can use elsewhere for computing sentence embeddings.
We'll generate and assess tfidf vectors of the Tesserae corpus broken into (by turns):
* 762 files
* 49,938 docs
```
import os
import pickle
import re
import sys
from collections import Counter, defaultdict
from glob import glob
from pathlib import Path
currentdir = Path.cwd()
parentdir = os.path.dirname(currentdir)
sys.path.insert(0,parentdir)
from tqdm import tqdm
from cltk.alphabet.lat import normalize_lat
from cltk.sentence.lat import LatinPunktSentenceTokenizer
from cltk.tokenizers.lat.lat import LatinWordTokenizer
from mlyoucanuse.text_cleaners import swallow
from scipy.spatial.distance import cosine
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics import mean_squared_error as mse
import matplotlib.pyplot as plt
tesserae = glob(os.path.expanduser('~/cltk_data/latin/text/latin_text_tesserae/texts/*.tess'))
print(f"Tesserae corpus contains: {len(tesserae)} files")
```
# Conversions and helper functions
```
ANY_ANGLE = re.compile("<.[^>]+>") # used to remove tesserae metadata
toker = LatinWordTokenizer()
sent_toker = LatinPunktSentenceTokenizer()
def toker_call(text):
# skip blank lines
if text.strip() is None:
return []
text = swallow(text, ANY_ANGLE)
# normalize effectively reduces our corpus diversity by 0.028%
text = normalize_lat(text, drop_accents=True,
drop_macrons=True,
jv_replacement=True,
ligature_replacement=True)
return toker.tokenize(text)
vectorizer = TfidfVectorizer(input='filename', tokenizer=toker_call)
vectorizer.fit(tesserae)
print(f"size of vocab: {len(vectorizer.vocabulary_):,}")
word_idf_files = {key: vectorizer.idf_[idx]
for key,idx in tqdm(vectorizer.vocabulary_.items(), total=len(vectorizer.idf_))}
del vectorizer
```
# Corpus to Documents functions
```
def count_numbers(text):
"""
Count the numbers groups in a line of text
>>> count_numbers ('<caes. gal. 8.0.4>')
3
>>> count_numbers('<caes. gal. 1.10.1>')
3
>>> count_numbers("<ov. her. 1.116> Protinus")
2
>>> count_numbers("<cic. arch. 1> si quid est in me ingeni")
1
"""
if re.search(r'\d+\.\d+\.\d+', text):
return 3
if re.search(r'\d+\.\d+', text):
return 2
if re.search(r'\d+', text):
return 1
return 0
def make_file_docs(filename):
"""given a filename return a dictionary with a list of docs.
if two numbers found, join on the first one
<verg. aen. 9.10> Nec satis: extremas Corythi penetravit ad urbes
<verg. ecl. 1.2> silvestrem tenui Musam meditaris avena;
if 3 numbers found, create a doc for each cluster of the first two numbers
<livy. urbe. 31.1.3> tot enim sunt a primo Punico ad secundum bellum finitum—
if just one number split on that
"<cic. arch. 1> si quid est in me ingeni"
"""
file_docs =defaultdict(list)
file_stats = {}
file = os.path.basename(filename)
ibook = None
ichapter = None
with open(filename, 'rt') as fin:
prev_ch= None
lines =[]
all_text=""
for line in fin:
numbers_found = count_numbers(line)
if numbers_found == 0:
if line.strip():
text = swallow(line, ANY_ANGLE)
file_docs[f"{file}"].append(text)
continue
if numbers_found == 3:
match = re.search(r'\d+\.\d+\.\d+', line)
if not match:
continue
start, end = match.span()
num_section = line[start:end]
book, chapter, sent = num_section.split(".")
ibook = int(book)
ichapter = int(chapter)
text = swallow(line, ANY_ANGLE)
if prev_ch == None:
lines.append(text)
prev_ch = ichapter
continue
if prev_ch != ichapter:
file_docs[f"{file}.{ibook}.{prev_ch}"].extend(lines)
lines = []
lines.append(text)
prev_ch = ichapter
else:
lines.append(text)
if numbers_found ==2:
if line.strip():
match = re.search(r'\d+\.\d+', line)
if not match:
continue
start, end = match.span()
num_section = line[start:end]
book, chapter = num_section.split(".")
ibook = int(book)
ichapter = int(chapter)
text = swallow(line, ANY_ANGLE)
file_docs[f"{file}.{ibook}"].append(text)
continue
if numbers_found ==1:
if line.strip():
match = re.search(r'\d+', line)
start, end = match.span()
num_section = line[start:end]
ibook = int(num_section)
text = swallow(line, ANY_ANGLE)
file_docs[f"{file}.{ibook}"].append(text)
continue
if ibook and ichapter and lines:
all_text = ' '.join(lines)
file_docs[f"{file}.{ibook}.{ichapter}"].append(all_text)
prev_ch = None
return file_docs
def make_docs(files):
docs = []
for file in files:
try:
file_docs = make_file_docs( file )
for key in file_docs:
docs.append(' '.join(file_docs[key]))
except Exception as ex:
print("fail with", file)
raise(ex)
return docs
```
## Tests of corpus processing
```
base = os.path.expanduser("~/cltk_data/latin/text/latin_text_tesserae/texts/")
file_docs = make_file_docs(f"{base}caesar.de_bello_gallico.part.1.tess")
assert(len(file_docs)==54)
file_docs = make_file_docs(f"{base}vergil.eclogues.tess")
assert(len(file_docs)==10)
file_docs = make_file_docs(f"{base}ovid.fasti.part.1.tess")
assert(len(file_docs)==1)
# print(len(file_docs))
# file_docs
test_files = [ f"{base}caesar.de_bello_gallico.part.1.tess" ,
f"{base}vergil.eclogues.tess",
f"{base}ovid.fasti.part.1.tess"]
docs = make_docs(test_files)
assert(len(docs)==65)
docs = make_docs(tesserae)
print(f"{len(tesserae)} corpus files broken up into {len(docs):,} documents")
vectorizer = TfidfVectorizer(tokenizer=toker_call)
vectorizer.fit(docs)
word_idf = {key: vectorizer.idf_[idx]
for key,idx in tqdm(vectorizer.vocabulary_.items(), total=len(vectorizer.idf_))}
del vectorizer
print(f"distinct words {len(word_idf):,}")
token_lengths = [len(tmp.split()) for tmp in docs]
counter = Counter(token_lengths)
indices_counts = list(counter.items())
indices_counts.sort(key=lambda x:x[0])
indices, counts = zip(*indices_counts )
fig = plt.figure()
ax = fig.add_subplot(2, 1, 1)
line, = ax.plot(counts, color='blue', lw=2)
ax.set_yscale('log')
plt.title("Document Token Counts")
plt.xlabel("# Tokens per Doc")
plt.ylabel("# of Docs")
plt.show()
```
## This word : idf mapping we'll save for sentence vectorization
```
latin_idf_dict_file = "word_idf.latin.pkl"
with open(latin_idf_dict_file, 'wb') as fout:
pickle.dump(word_idf, fout)
```
## Compare the idf values using Mean Square Error, Cosine
These values become more meaningful as the ETL processes are changed; the measurements may well indicate how much value have shifted.
```
words_idfs = list(word_idf.items())
words_idfs.sort(key=lambda x: x[0])
words_idf_files = list(word_idf_files.items())
words_idf_files.sort(key=lambda x: x[0])
print(f"Words Idfs vocab size: {len(words_idfs):,}, Words Idf from files {len(words_idf_files):,}")
words_idfs = [(key, word_idf.get(key)) for key,val in words_idfs
if key in word_idf_files]
words_idf_files = [(key, word_idf_files.get(key)) for key,val in words_idf_files
if key in word_idf]
assert( len(words_idfs) == len(words_idf_files))
print(f"Total # shared vocab: {len(words_idfs):,}")
_, idfs = zip(*words_idfs)
_, idfs2 = zip(*words_idf_files)
print(f"MSE: {mse(idfs, idfs2)}")
print(f"Cosine: {cosine(idfs, idfs2)}")
```
|
github_jupyter
|
import os
import pickle
import re
import sys
from collections import Counter, defaultdict
from glob import glob
from pathlib import Path
currentdir = Path.cwd()
parentdir = os.path.dirname(currentdir)
sys.path.insert(0,parentdir)
from tqdm import tqdm
from cltk.alphabet.lat import normalize_lat
from cltk.sentence.lat import LatinPunktSentenceTokenizer
from cltk.tokenizers.lat.lat import LatinWordTokenizer
from mlyoucanuse.text_cleaners import swallow
from scipy.spatial.distance import cosine
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics import mean_squared_error as mse
import matplotlib.pyplot as plt
tesserae = glob(os.path.expanduser('~/cltk_data/latin/text/latin_text_tesserae/texts/*.tess'))
print(f"Tesserae corpus contains: {len(tesserae)} files")
ANY_ANGLE = re.compile("<.[^>]+>") # used to remove tesserae metadata
toker = LatinWordTokenizer()
sent_toker = LatinPunktSentenceTokenizer()
def toker_call(text):
# skip blank lines
if text.strip() is None:
return []
text = swallow(text, ANY_ANGLE)
# normalize effectively reduces our corpus diversity by 0.028%
text = normalize_lat(text, drop_accents=True,
drop_macrons=True,
jv_replacement=True,
ligature_replacement=True)
return toker.tokenize(text)
vectorizer = TfidfVectorizer(input='filename', tokenizer=toker_call)
vectorizer.fit(tesserae)
print(f"size of vocab: {len(vectorizer.vocabulary_):,}")
word_idf_files = {key: vectorizer.idf_[idx]
for key,idx in tqdm(vectorizer.vocabulary_.items(), total=len(vectorizer.idf_))}
del vectorizer
def count_numbers(text):
"""
Count the numbers groups in a line of text
>>> count_numbers ('<caes. gal. 8.0.4>')
3
>>> count_numbers('<caes. gal. 1.10.1>')
3
>>> count_numbers("<ov. her. 1.116> Protinus")
2
>>> count_numbers("<cic. arch. 1> si quid est in me ingeni")
1
"""
if re.search(r'\d+\.\d+\.\d+', text):
return 3
if re.search(r'\d+\.\d+', text):
return 2
if re.search(r'\d+', text):
return 1
return 0
def make_file_docs(filename):
"""given a filename return a dictionary with a list of docs.
if two numbers found, join on the first one
<verg. aen. 9.10> Nec satis: extremas Corythi penetravit ad urbes
<verg. ecl. 1.2> silvestrem tenui Musam meditaris avena;
if 3 numbers found, create a doc for each cluster of the first two numbers
<livy. urbe. 31.1.3> tot enim sunt a primo Punico ad secundum bellum finitum—
if just one number split on that
"<cic. arch. 1> si quid est in me ingeni"
"""
file_docs =defaultdict(list)
file_stats = {}
file = os.path.basename(filename)
ibook = None
ichapter = None
with open(filename, 'rt') as fin:
prev_ch= None
lines =[]
all_text=""
for line in fin:
numbers_found = count_numbers(line)
if numbers_found == 0:
if line.strip():
text = swallow(line, ANY_ANGLE)
file_docs[f"{file}"].append(text)
continue
if numbers_found == 3:
match = re.search(r'\d+\.\d+\.\d+', line)
if not match:
continue
start, end = match.span()
num_section = line[start:end]
book, chapter, sent = num_section.split(".")
ibook = int(book)
ichapter = int(chapter)
text = swallow(line, ANY_ANGLE)
if prev_ch == None:
lines.append(text)
prev_ch = ichapter
continue
if prev_ch != ichapter:
file_docs[f"{file}.{ibook}.{prev_ch}"].extend(lines)
lines = []
lines.append(text)
prev_ch = ichapter
else:
lines.append(text)
if numbers_found ==2:
if line.strip():
match = re.search(r'\d+\.\d+', line)
if not match:
continue
start, end = match.span()
num_section = line[start:end]
book, chapter = num_section.split(".")
ibook = int(book)
ichapter = int(chapter)
text = swallow(line, ANY_ANGLE)
file_docs[f"{file}.{ibook}"].append(text)
continue
if numbers_found ==1:
if line.strip():
match = re.search(r'\d+', line)
start, end = match.span()
num_section = line[start:end]
ibook = int(num_section)
text = swallow(line, ANY_ANGLE)
file_docs[f"{file}.{ibook}"].append(text)
continue
if ibook and ichapter and lines:
all_text = ' '.join(lines)
file_docs[f"{file}.{ibook}.{ichapter}"].append(all_text)
prev_ch = None
return file_docs
def make_docs(files):
docs = []
for file in files:
try:
file_docs = make_file_docs( file )
for key in file_docs:
docs.append(' '.join(file_docs[key]))
except Exception as ex:
print("fail with", file)
raise(ex)
return docs
base = os.path.expanduser("~/cltk_data/latin/text/latin_text_tesserae/texts/")
file_docs = make_file_docs(f"{base}caesar.de_bello_gallico.part.1.tess")
assert(len(file_docs)==54)
file_docs = make_file_docs(f"{base}vergil.eclogues.tess")
assert(len(file_docs)==10)
file_docs = make_file_docs(f"{base}ovid.fasti.part.1.tess")
assert(len(file_docs)==1)
# print(len(file_docs))
# file_docs
test_files = [ f"{base}caesar.de_bello_gallico.part.1.tess" ,
f"{base}vergil.eclogues.tess",
f"{base}ovid.fasti.part.1.tess"]
docs = make_docs(test_files)
assert(len(docs)==65)
docs = make_docs(tesserae)
print(f"{len(tesserae)} corpus files broken up into {len(docs):,} documents")
vectorizer = TfidfVectorizer(tokenizer=toker_call)
vectorizer.fit(docs)
word_idf = {key: vectorizer.idf_[idx]
for key,idx in tqdm(vectorizer.vocabulary_.items(), total=len(vectorizer.idf_))}
del vectorizer
print(f"distinct words {len(word_idf):,}")
token_lengths = [len(tmp.split()) for tmp in docs]
counter = Counter(token_lengths)
indices_counts = list(counter.items())
indices_counts.sort(key=lambda x:x[0])
indices, counts = zip(*indices_counts )
fig = plt.figure()
ax = fig.add_subplot(2, 1, 1)
line, = ax.plot(counts, color='blue', lw=2)
ax.set_yscale('log')
plt.title("Document Token Counts")
plt.xlabel("# Tokens per Doc")
plt.ylabel("# of Docs")
plt.show()
latin_idf_dict_file = "word_idf.latin.pkl"
with open(latin_idf_dict_file, 'wb') as fout:
pickle.dump(word_idf, fout)
words_idfs = list(word_idf.items())
words_idfs.sort(key=lambda x: x[0])
words_idf_files = list(word_idf_files.items())
words_idf_files.sort(key=lambda x: x[0])
print(f"Words Idfs vocab size: {len(words_idfs):,}, Words Idf from files {len(words_idf_files):,}")
words_idfs = [(key, word_idf.get(key)) for key,val in words_idfs
if key in word_idf_files]
words_idf_files = [(key, word_idf_files.get(key)) for key,val in words_idf_files
if key in word_idf]
assert( len(words_idfs) == len(words_idf_files))
print(f"Total # shared vocab: {len(words_idfs):,}")
_, idfs = zip(*words_idfs)
_, idfs2 = zip(*words_idf_files)
print(f"MSE: {mse(idfs, idfs2)}")
print(f"Cosine: {cosine(idfs, idfs2)}")
| 0.177704 | 0.834069 |
# Convert data to NILMTK format and load into NILMTK
NILMTK uses an open file format based on the HDF5 binary file format to store both the power data and the metadata. The very first step when using NILMTK is to convert your dataset to the NILMTK HDF5 file format.
## REDD
Converting the REDD dataset is easy:
```
from nilmtk.dataset_converters import convert_redd
convert_redd('/data/REDD/low_freq', '/data/REDD/redd.h5')
```
Now `redd.h5` holds all the REDD power data and all the relevant metadata. In NILMTK v0.2 this conversion only uses a tiny fraction of the system memory (unlike NILMTK v0.1 which would guzzle ~1 GByte of RAM just to do the dataset conversion!).
Of course, if you want to run `convert_redd` on your own machine then you first need to download [REDD](http://redd.csail.mit.edu), decompress it and pass the relevant `source_directory` and `output_filename` to `convert_redd()`.
## Other datasets
At the time of writing, [NILMTK contains converters for 8 datasets](https://github.com/nilmtk/nilmtk/tree/master/nilmtk/dataset_converters).
Contributing a new converter is easy and highly encouraged! [Learn how to write a dataset converter](https://github.com/nilmtk/nilmtk/blob/master/docs/manual/development_guide/writing_a_dataset_converter.md).
## Open HDF5 in NILMTK
```
from nilmtk import DataSet
from nilmtk.utils import print_dict
redd = DataSet('/data/REDD/redd.h5')
```
At this point, all the metadata has been loaded into memory but none of the power data has been loaded. This is our first encounter with a fundamental difference between NILMTK v0.1 and v0.2: NILMTK v0.1 used to eagerly load the entire dataset into memory before you did any actual work on the data. NILMTK v0.2 is lazy! It won't load data into memory until you tell it what you want to do with the data (and, even then, large dataset will be loaded in chunks that fit into memory). This allows NILMTK v0.2 to work with arbitrarily large datasets (datasets too large to fit into memory) without choking your system.
### Exploring the `DataSet` object
Let's have a quick poke around to see what's in this `redd` object...
There is a lot of metadata associated with the dataset, including information about the two models of meter device the authors used to record REDD:
```
print_dict(redd.metadata)
```
We also have all the buildings available as an [OrderedDict](https://docs.python.org/2/library/collections.html#collections.OrderedDict) (indexed from 1 not 0 because every dataset we are aware of starts numbering buildings from 1 not 0)
```
print_dict(redd.buildings)
```
Each building has a little bit of metadata associated with it (there isn't much building-specific metadata in REDD):
```
print_dict(redd.buildings[1].metadata)
```
Each building has an `elec` attribute which is a `MeterGroup` object (much more about those soon!)
```
redd.buildings[1].elec
```
Yup, that's where all the meat lies!
|
github_jupyter
|
from nilmtk.dataset_converters import convert_redd
convert_redd('/data/REDD/low_freq', '/data/REDD/redd.h5')
from nilmtk import DataSet
from nilmtk.utils import print_dict
redd = DataSet('/data/REDD/redd.h5')
print_dict(redd.metadata)
print_dict(redd.buildings)
print_dict(redd.buildings[1].metadata)
redd.buildings[1].elec
| 0.187281 | 0.97567 |
```
import pandas as pd
import numpy as np
from collections import Counter
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_pickle('childrens_book_df.pkl')
df.head()
#find top 20 words in corpus
common = Counter(" ".join(df["NLP_Text"]).split()).most_common(100)
common[:20]
#plot
top_words = [item[0] for item in common]
top_word_counts = [item[1] for item in common]
sns.set_context('paper')
f, ax = plt.subplots(figsize = (15,6))
sns.set_color_codes('pastel')
sns.barplot(x = top_word_counts[2:12], y = top_words[2:12],
label = '', color = 'b', edgecolor = 'w')
sns.set_color_codes('muted')
ax.set_xlabel('Word Frequency',fontsize=15)
ax.set_ylabel('Word',fontsize=15)
ax.set_title('Top 10 Words in Childrens Books', fontsize=20)
sns.despine(left = True, bottom = True)
plt.show()
plt.savefig('top_10.png')
#initialize dict to store relevant letter combinations for sounds
phonics_dict = {
"diphthongs": ["oo", "ou", "ow", "oi", "oy", "aw", "au", "ie", "igh", "ay", "ee"],
"ending blends":["mp", "ng", "nt", "nk", "nd", "sk", "st"],
"l blends": ["bl", "cl", "gl", "pl", "sl"],
"r blends": ["br", "cr", "dr", "fr", "gr", "pr", "tr"],
"s blends": ["sc", "sk", "sl", "sm", "sn", "sp", "st", "str", "scr", "spl"],
"digraphs": ["ch", "ph", "sh", "wh", "tch", "th", "gh"],
"soft c": ["ce", "ci", "cy"],
"soft g": ["ge", "dge", "gi", "gy"],
"silent letters": ["kn", "wr", "gn"]
}
#get all dictionary keys into list to loop through
phonics = phonics_dict.values()
phonics_flat = [item for sublist in phonics for item in sublist]
#get frequency of all sounds
count = {}.fromkeys(phonics_flat, 0) #initialize dict
#loop through list of sounds and count all occurences in column, update dict
for sound in phonics_flat:
sum = df.Readable_Text.str.count(sound).sum()
count[sound] = sum
#count individual sounds
count_ind = count
count_ind
#get totals by group
count_group = {}.fromkeys(phonics_dict, 0)
for group in phonics_dict:
for sound in phonics_dict[group]:
count_group[group] += count_ind[sound]
count_group
keys = list(count_group.keys())
counts = [int(count_group[k] / 1000) for k in keys]
sns.set_context('paper')
f, ax = plt.subplots(figsize = (15,6))
sns.set_color_codes('pastel')
sns.barplot(x = keys , y = counts,
label = '', color = 'b', edgecolor = 'w')
sns.set_color_codes('muted')
ax.set_xlabel('Sound Group',fontsize=18)
ax.set_ylabel('Count (in thousands)',fontsize=18)
ax.set_yticklabels(ax.get_yticks(), size = 15)
ax.set_yticklabels(ax.get_yticks().astype(int))
ax.set_xticklabels(count_group.keys(), size = 15)
ax.set_title('Frequency of Phonic Groups', fontsize=20)
sns.despine(left = True, bottom = True)
plt.savefig('Count_Groups.jpg')
plt.show()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from collections import Counter
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_pickle('childrens_book_df.pkl')
df.head()
#find top 20 words in corpus
common = Counter(" ".join(df["NLP_Text"]).split()).most_common(100)
common[:20]
#plot
top_words = [item[0] for item in common]
top_word_counts = [item[1] for item in common]
sns.set_context('paper')
f, ax = plt.subplots(figsize = (15,6))
sns.set_color_codes('pastel')
sns.barplot(x = top_word_counts[2:12], y = top_words[2:12],
label = '', color = 'b', edgecolor = 'w')
sns.set_color_codes('muted')
ax.set_xlabel('Word Frequency',fontsize=15)
ax.set_ylabel('Word',fontsize=15)
ax.set_title('Top 10 Words in Childrens Books', fontsize=20)
sns.despine(left = True, bottom = True)
plt.show()
plt.savefig('top_10.png')
#initialize dict to store relevant letter combinations for sounds
phonics_dict = {
"diphthongs": ["oo", "ou", "ow", "oi", "oy", "aw", "au", "ie", "igh", "ay", "ee"],
"ending blends":["mp", "ng", "nt", "nk", "nd", "sk", "st"],
"l blends": ["bl", "cl", "gl", "pl", "sl"],
"r blends": ["br", "cr", "dr", "fr", "gr", "pr", "tr"],
"s blends": ["sc", "sk", "sl", "sm", "sn", "sp", "st", "str", "scr", "spl"],
"digraphs": ["ch", "ph", "sh", "wh", "tch", "th", "gh"],
"soft c": ["ce", "ci", "cy"],
"soft g": ["ge", "dge", "gi", "gy"],
"silent letters": ["kn", "wr", "gn"]
}
#get all dictionary keys into list to loop through
phonics = phonics_dict.values()
phonics_flat = [item for sublist in phonics for item in sublist]
#get frequency of all sounds
count = {}.fromkeys(phonics_flat, 0) #initialize dict
#loop through list of sounds and count all occurences in column, update dict
for sound in phonics_flat:
sum = df.Readable_Text.str.count(sound).sum()
count[sound] = sum
#count individual sounds
count_ind = count
count_ind
#get totals by group
count_group = {}.fromkeys(phonics_dict, 0)
for group in phonics_dict:
for sound in phonics_dict[group]:
count_group[group] += count_ind[sound]
count_group
keys = list(count_group.keys())
counts = [int(count_group[k] / 1000) for k in keys]
sns.set_context('paper')
f, ax = plt.subplots(figsize = (15,6))
sns.set_color_codes('pastel')
sns.barplot(x = keys , y = counts,
label = '', color = 'b', edgecolor = 'w')
sns.set_color_codes('muted')
ax.set_xlabel('Sound Group',fontsize=18)
ax.set_ylabel('Count (in thousands)',fontsize=18)
ax.set_yticklabels(ax.get_yticks(), size = 15)
ax.set_yticklabels(ax.get_yticks().astype(int))
ax.set_xticklabels(count_group.keys(), size = 15)
ax.set_title('Frequency of Phonic Groups', fontsize=20)
sns.despine(left = True, bottom = True)
plt.savefig('Count_Groups.jpg')
plt.show()
| 0.356111 | 0.362518 |
```
import torch
import numpy as np
import matplotlib.pyplot as plt
from utils import *
from models import *
plt.style.use('ggplot')
%matplotlib inline
```
# Training Curves
Use this in a similar manner to tensorboard, to check that things are training.
```
checkpoints = ['resnet9','resnet18','resnet34','resnet50'] # put your checkpoint files in here
cols = 2
rows = len(checkpoints) // cols
fig, axs = plt.subplots(rows,cols,figsize=(20, rows*6)) # width=20cm, height=6cm per row
axs = axs.ravel()
for checkpoint, ax in zip(checkpoints, axs):
sd = torch.load('checkpoints/%s.t7' % checkpoint, map_location='cpu')
ax.plot(list(range(len(sd['error_history']))), sd['error_history'])
ax.set_xlabel('Epoch')
ax.set_ylabel('CIFAR-10 top-1 err')
ax.legend([checkpoint])
plt.show()
```
Given a family of ResNets we can extract a Pareto curve of accuracy vs. number of parameters:
```
fig, ax = plt.subplots(figsize=(12,6))
x = []
y = []
for checkpoint in checkpoints:
sd = torch.load('checkpoints/%s.t7' % checkpoint, map_location='cpu')
x_ = get_no_params(sd['net'])
y_ = sd['error_history'][-1]
x.append(x_)
y.append(y_)
for i, net in enumerate(checkpoints):
offset = 0.05
ax.annotate(net, (x[i]+offset, y[i]+offset))
ax.set_xlabel('Number of parameters')
ax.set_ylabel('CIFAR-10 top-1 err')
ax.plot(x,y, marker='o')
plt.show()
```
You can see immediately that ResNet-50 is over-parameterised for a small task like CIFAR-10. One could employ some more training tricks, but for fairness I will just consider the "Pareto frontier" to be ResNet-9-18-34.
The idea proposed in Han et al. is that we can beat this Pareto frontier by keeping the network structure the same but removing individual weights.
# Pruning Curves
Check that the model is pruning correctly.
```
fig,ax = plt.subplots(figsize=(12,6))
prune_rates = np.arange(0,100,step=5)
checkpoints = ['resnet9','resnet18','resnet34']
def get_nonzeros(sd):
tot = 0
for mask in sd['masks']:
tot += torch.sum(mask!=0)
return tot
for checkpoint in checkpoints:
x = []
y = []
for prune_rate in prune_rates:
sd = torch.load('checkpoints/%s.t7' % str(checkpoint + '_l1_' + str(prune_rate)), map_location='cpu')
x.append(get_nonzeros(sd))
y.append(sd['error_history'][-1])
ax.plot(x,y,marker='o')
x = []
y = []
for checkpoint in checkpoints:
sd = torch.load('checkpoints/%s.t7' % checkpoint, map_location='cpu')
x_ = get_no_params(sd['net'])
y_ = sd['error_history'][-1]
x.append(x_)
y.append(y_)
for i, net in enumerate(checkpoints):
offset = 0.1
ax.annotate(net, (x[i]+offset, y[i]+offset))
ax.plot(x,y, marker='o')
ax.set_xlabel('Num nonzero params')
ax.set_ylabel('Top-1 Error (%)')
plt.show()
```
# Structured vs. Unstructured Pruning
As far as I am aware, the question of whether sparse networks learn better feature extractors than dense ones remains open.
Here we briefly compare Fisher-pruned models to our L1-pruned models.
```
def get_inf_params(net, verbose=True, sd=False):
if sd:
params = net
else:
params = net.state_dict()
tot = 0
conv_tot = 0
for p in params:
no = params[p].view(-1).__len__()
if ('num_batches_tracked' not in p) and ('running' not in p) and ('mask' not in p):
tot += no
if verbose:
print('%s has %d params' % (p, no))
if 'conv' in p:
conv_tot += no
if verbose:
print('Net has %d conv params' % conv_tot)
print('Net has %d params in total' % tot)
return tot
fig, ax = plt.subplots(figsize=(12,6))
baselines = ['wrn_40_2_1'] # ,'wrn_40_2_2','wrn_40_2_3','wrn_40_2_4','wrn_40_2_5']
sparse_prune_rates = np.arange(0,100,step=5)
## sparse prune
for baseline in baselines:
x = []
y = []
for prune_rate in sparse_prune_rates:
sd = torch.load('checkpoints/%s.t7' % str(baseline + '_l1_' + str(prune_rate)), map_location='cpu')
x.append(get_nonzeros(sd))
y.append(sd['error_history'][-1])
ax.plot(x,y,marker='o',label='Unstructured L1 prune')
dense_prune_rates = np.arange(100,1300,step=100)
x = [77202, 223274, 367618, 510234, 669122, 825850, 989778, 1154570, 1306608, 1486746, 1658594, 1835050, 2026338]
y = [19.760, 9.450, 7.590, 6.180, 5.790, 5.810, 5.060, 4.640, 4.620, 4.460, 4.400, 4.300, 4.050]
ax.plot(x,y, marker='o', label='Structured Fisher prune')
ax.set_xlabel('# of parameters')
ax.set_ylabel('Top-1 CIFAR error')
ax.legend()
plt.show()
sd = torch.load('checkpoints/spruning_paper/wrn_402_1.t7', map_location='cpu')
model = WideResNet(40,2)
new_sd = model.state_dict()
old_sd = sd['state_dict']
k_new = [k for k in new_sd.keys() if 'mask' not in k]
k_new = [k for k in k_new if 'num_batches_tracked' not in k]
for o, n in zip(old_sd.keys(), k_new):
new_sd[n] = old_sd[o]
model.load_state_dict(new_sd)
```
|
github_jupyter
|
import torch
import numpy as np
import matplotlib.pyplot as plt
from utils import *
from models import *
plt.style.use('ggplot')
%matplotlib inline
checkpoints = ['resnet9','resnet18','resnet34','resnet50'] # put your checkpoint files in here
cols = 2
rows = len(checkpoints) // cols
fig, axs = plt.subplots(rows,cols,figsize=(20, rows*6)) # width=20cm, height=6cm per row
axs = axs.ravel()
for checkpoint, ax in zip(checkpoints, axs):
sd = torch.load('checkpoints/%s.t7' % checkpoint, map_location='cpu')
ax.plot(list(range(len(sd['error_history']))), sd['error_history'])
ax.set_xlabel('Epoch')
ax.set_ylabel('CIFAR-10 top-1 err')
ax.legend([checkpoint])
plt.show()
fig, ax = plt.subplots(figsize=(12,6))
x = []
y = []
for checkpoint in checkpoints:
sd = torch.load('checkpoints/%s.t7' % checkpoint, map_location='cpu')
x_ = get_no_params(sd['net'])
y_ = sd['error_history'][-1]
x.append(x_)
y.append(y_)
for i, net in enumerate(checkpoints):
offset = 0.05
ax.annotate(net, (x[i]+offset, y[i]+offset))
ax.set_xlabel('Number of parameters')
ax.set_ylabel('CIFAR-10 top-1 err')
ax.plot(x,y, marker='o')
plt.show()
fig,ax = plt.subplots(figsize=(12,6))
prune_rates = np.arange(0,100,step=5)
checkpoints = ['resnet9','resnet18','resnet34']
def get_nonzeros(sd):
tot = 0
for mask in sd['masks']:
tot += torch.sum(mask!=0)
return tot
for checkpoint in checkpoints:
x = []
y = []
for prune_rate in prune_rates:
sd = torch.load('checkpoints/%s.t7' % str(checkpoint + '_l1_' + str(prune_rate)), map_location='cpu')
x.append(get_nonzeros(sd))
y.append(sd['error_history'][-1])
ax.plot(x,y,marker='o')
x = []
y = []
for checkpoint in checkpoints:
sd = torch.load('checkpoints/%s.t7' % checkpoint, map_location='cpu')
x_ = get_no_params(sd['net'])
y_ = sd['error_history'][-1]
x.append(x_)
y.append(y_)
for i, net in enumerate(checkpoints):
offset = 0.1
ax.annotate(net, (x[i]+offset, y[i]+offset))
ax.plot(x,y, marker='o')
ax.set_xlabel('Num nonzero params')
ax.set_ylabel('Top-1 Error (%)')
plt.show()
def get_inf_params(net, verbose=True, sd=False):
if sd:
params = net
else:
params = net.state_dict()
tot = 0
conv_tot = 0
for p in params:
no = params[p].view(-1).__len__()
if ('num_batches_tracked' not in p) and ('running' not in p) and ('mask' not in p):
tot += no
if verbose:
print('%s has %d params' % (p, no))
if 'conv' in p:
conv_tot += no
if verbose:
print('Net has %d conv params' % conv_tot)
print('Net has %d params in total' % tot)
return tot
fig, ax = plt.subplots(figsize=(12,6))
baselines = ['wrn_40_2_1'] # ,'wrn_40_2_2','wrn_40_2_3','wrn_40_2_4','wrn_40_2_5']
sparse_prune_rates = np.arange(0,100,step=5)
## sparse prune
for baseline in baselines:
x = []
y = []
for prune_rate in sparse_prune_rates:
sd = torch.load('checkpoints/%s.t7' % str(baseline + '_l1_' + str(prune_rate)), map_location='cpu')
x.append(get_nonzeros(sd))
y.append(sd['error_history'][-1])
ax.plot(x,y,marker='o',label='Unstructured L1 prune')
dense_prune_rates = np.arange(100,1300,step=100)
x = [77202, 223274, 367618, 510234, 669122, 825850, 989778, 1154570, 1306608, 1486746, 1658594, 1835050, 2026338]
y = [19.760, 9.450, 7.590, 6.180, 5.790, 5.810, 5.060, 4.640, 4.620, 4.460, 4.400, 4.300, 4.050]
ax.plot(x,y, marker='o', label='Structured Fisher prune')
ax.set_xlabel('# of parameters')
ax.set_ylabel('Top-1 CIFAR error')
ax.legend()
plt.show()
sd = torch.load('checkpoints/spruning_paper/wrn_402_1.t7', map_location='cpu')
model = WideResNet(40,2)
new_sd = model.state_dict()
old_sd = sd['state_dict']
k_new = [k for k in new_sd.keys() if 'mask' not in k]
k_new = [k for k in k_new if 'num_batches_tracked' not in k]
for o, n in zip(old_sd.keys(), k_new):
new_sd[n] = old_sd[o]
model.load_state_dict(new_sd)
| 0.360039 | 0.759359 |
# 합성곱 신경망의 시각화
<table align="left">
<td>
<a target="_blank" href="https://colab.research.google.com/github/rickiepark/hg-mldl/blob/master/8-3.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />구글 코랩에서 실행하기</a>
</td>
</table>
## 가중치 시각화
```
from tensorflow import keras
# 코랩에서 실행하는 경우에는 다음 명령을 실행하여 best-cnn-model.h5 파일을 다운로드받아 사용하세요.
!wget https://github.com/rickiepark/hg-mldl/raw/master/best-cnn-model.h5
model = keras.models.load_model('best-cnn-model.h5')
model.layers
conv = model.layers[0]
print(conv.weights[0].shape, conv.weights[1].shape)
conv_weights = conv.weights[0].numpy()
print(conv_weights.mean(), conv_weights.std())
import matplotlib.pyplot as plt
plt.hist(conv_weights.reshape(-1, 1))
plt.xlabel('weight')
plt.ylabel('count')
plt.show()
fig, axs = plt.subplots(2, 16, figsize=(15,2))
for i in range(2):
for j in range(16):
axs[i, j].imshow(conv_weights[:,:,0,i*16 + j], vmin=-0.5, vmax=0.5)
axs[i, j].axis('off')
plt.show()
no_training_model = keras.Sequential()
no_training_model.add(keras.layers.Conv2D(32, kernel_size=3, activation='relu',
padding='same', input_shape=(28,28,1)))
no_training_conv = no_training_model.layers[0]
print(no_training_conv.weights[0].shape)
no_training_weights = no_training_conv.weights[0].numpy()
print(no_training_weights.mean(), no_training_weights.std())
plt.hist(no_training_weights.reshape(-1, 1))
plt.xlabel('weight')
plt.ylabel('count')
plt.show()
fig, axs = plt.subplots(2, 16, figsize=(15,2))
for i in range(2):
for j in range(16):
axs[i, j].imshow(no_training_weights[:,:,0,i*16 + j], vmin=-0.5, vmax=0.5)
axs[i, j].axis('off')
plt.show()
```
## 함수형 API
```
print(model.input)
conv_acti = keras.Model(model.input, model.layers[0].output)
```
## 특성 맵 시각화
```
(train_input, train_target), (test_input, test_target) = keras.datasets.fashion_mnist.load_data()
plt.imshow(train_input[0], cmap='gray_r')
plt.show()
inputs = train_input[0:1].reshape(-1, 28, 28, 1)/255.0
feature_maps = conv_acti.predict(inputs)
print(feature_maps.shape)
fig, axs = plt.subplots(4, 8, figsize=(15,8))
for i in range(4):
for j in range(8):
axs[i, j].imshow(feature_maps[0,:,:,i*8 + j])
axs[i, j].axis('off')
plt.show()
conv2_acti = keras.Model(model.input, model.layers[2].output)
feature_maps = conv2_acti.predict(train_input[0:1].reshape(-1, 28, 28, 1)/255.0)
print(feature_maps.shape)
fig, axs = plt.subplots(8, 8, figsize=(12,12))
for i in range(8):
for j in range(8):
axs[i, j].imshow(feature_maps[0,:,:,i*8 + j])
axs[i, j].axis('off')
plt.show()
```
|
github_jupyter
|
from tensorflow import keras
# 코랩에서 실행하는 경우에는 다음 명령을 실행하여 best-cnn-model.h5 파일을 다운로드받아 사용하세요.
!wget https://github.com/rickiepark/hg-mldl/raw/master/best-cnn-model.h5
model = keras.models.load_model('best-cnn-model.h5')
model.layers
conv = model.layers[0]
print(conv.weights[0].shape, conv.weights[1].shape)
conv_weights = conv.weights[0].numpy()
print(conv_weights.mean(), conv_weights.std())
import matplotlib.pyplot as plt
plt.hist(conv_weights.reshape(-1, 1))
plt.xlabel('weight')
plt.ylabel('count')
plt.show()
fig, axs = plt.subplots(2, 16, figsize=(15,2))
for i in range(2):
for j in range(16):
axs[i, j].imshow(conv_weights[:,:,0,i*16 + j], vmin=-0.5, vmax=0.5)
axs[i, j].axis('off')
plt.show()
no_training_model = keras.Sequential()
no_training_model.add(keras.layers.Conv2D(32, kernel_size=3, activation='relu',
padding='same', input_shape=(28,28,1)))
no_training_conv = no_training_model.layers[0]
print(no_training_conv.weights[0].shape)
no_training_weights = no_training_conv.weights[0].numpy()
print(no_training_weights.mean(), no_training_weights.std())
plt.hist(no_training_weights.reshape(-1, 1))
plt.xlabel('weight')
plt.ylabel('count')
plt.show()
fig, axs = plt.subplots(2, 16, figsize=(15,2))
for i in range(2):
for j in range(16):
axs[i, j].imshow(no_training_weights[:,:,0,i*16 + j], vmin=-0.5, vmax=0.5)
axs[i, j].axis('off')
plt.show()
print(model.input)
conv_acti = keras.Model(model.input, model.layers[0].output)
(train_input, train_target), (test_input, test_target) = keras.datasets.fashion_mnist.load_data()
plt.imshow(train_input[0], cmap='gray_r')
plt.show()
inputs = train_input[0:1].reshape(-1, 28, 28, 1)/255.0
feature_maps = conv_acti.predict(inputs)
print(feature_maps.shape)
fig, axs = plt.subplots(4, 8, figsize=(15,8))
for i in range(4):
for j in range(8):
axs[i, j].imshow(feature_maps[0,:,:,i*8 + j])
axs[i, j].axis('off')
plt.show()
conv2_acti = keras.Model(model.input, model.layers[2].output)
feature_maps = conv2_acti.predict(train_input[0:1].reshape(-1, 28, 28, 1)/255.0)
print(feature_maps.shape)
fig, axs = plt.subplots(8, 8, figsize=(12,12))
for i in range(8):
for j in range(8):
axs[i, j].imshow(feature_maps[0,:,:,i*8 + j])
axs[i, j].axis('off')
plt.show()
| 0.801159 | 0.938463 |
## Tehtävä 1: Blochin pallo
Blochin pallo on tapa hahmottaa yhden kubitin kvanttisysteemin tilaa kolmiulotteisella pallolla.
<img src="https://upload.wikimedia.org/wikipedia/commons/6/6b/Bloch_sphere.svg" />
Yksi tapa määrittää kvanttitilaa vastaava piste Blochin pallolla on valita kuvan kulmat $\theta$ ja $\phi$ siten, että ne vastaavat kubitin tilaa $\ket \psi$ seuraavasti:
$$\ket \psi = cos(\theta / 2)\ket 0 + e^{i*\phi}sin(\theta / 2)\ket 1$$
Bloch-vektorin ja z-akselin välinen kulma $\theta$ kertoo siis $\ket 0$-kertoimen ja $\ket 1$-kertoimen itseisarvojen välisestä suhteesta. Kuvaan merkitäänkin usein pisteet, joissa tila on kokonaan $\ket 0$ (ylhäällä) tai $\ket 1$ (alhaalla). Toisaalta Bloch-vektorin ja x-akselin välinen kulma $\phi$ kuvastaa $\ket 1$:n amplitudin vaihetta eli suuntaa.
Tilavektorin $\ket 0$-komponentin kerroin on sovitusti aina positiivinen reaaliluku. Tämä voidaan tehdä siksi, että systeemin ns. globaali vaihe ei vaikuta mittaustuloksiin, myöskään unitaaristen porttien käytön jälkeen.
Unitaarinen portti on Blochin pallolla aina jokin kierto tietyn akselin ympäri tietyn kulman verran. Esimerkkejä tavallisten porttien operaatioista Blochin pallon kääntöinä:
- $X$: Kääntää X-akselin ympäri kulman $\pi = 180^o$ verran. Käännön suunnalla ei ole merkitystä, koska puolikas kierto tuottaa saman tuloksen molempiin suuntiin.
- $Y$: Kääntää Y-akselin ympäri kulman $\pi$ verran.
- $Z$: Kääntää Z-akselin ympäri kulman $\pi$ verran.
- $H$: Kääntää X+Z-akselin (eli puolivälin X ja Z -akselien välillä) ympäri kulman $\pi$ verran.
- $S$: Kääntää Z-akselin ympäri kulman $\pi/2$ verran, vastapäivään ylhäältä katsottuna.
- $T$: Kääntää Z-akselin ympäri kulman $\pi/4$ verran, jälleen vastapäivään ylhäältä katsottuna.
Demo Blochin pallon toiminnasta: https://bloch.ollpu.fi/
Näkymää voi kääntää vetämällä palloa hiirellä. Zoom toimii myös. Kuvassa akselit $x, y, z$ on piirretty punaisella, vihreällä ja sinisellä.
Systeemin alkutila on $\ket 0$ ja voit käyttää portteja oikealla olevista napeista. Kokeile kaikkien porttien toimintaa, ja totea, että ne toimivat kuten yllä on kuvailtu. Kiinnitä huomiota myös siihen, miten portit vaikuttavat tilavektoriin, joka on esitetty näkymän yläpuolella.
$$\newcommand{\ket}[1]{\left|{#1}\right\rangle}$$
$$\newcommand{\bra}[1]{\left\langle{#1}\right|}$$
## Tehtävä 2: Z-portin toteus kvanttitietokoneesa
### Johdanto
Aiemmin kubittien tilat $\ket +$ ja $\ket -$ määritettiin seuraavasti:
$$
\ket + = \frac{1}{\sqrt 2} (\ket 0 + \ket 1), \\
\ket - = \frac{1}{\sqrt 2} (\ket 0 - \ket 1).
$$
Z-portin (ns. phase-flip gate) matriisiesitys on seuraava:
$$
Z= \pmatrix {1 & 0 \\ 0 & -1}
$$
Z-portin vaikutus esimerkiksi tiloihin $\ket +$ ja $\ket -$ on seuraava:
$$
\begin{align}
Z\ket + &= \ket - \text{ja}\\
Z\ket - &= \ket +
\end{align}
$$
### Tehtävä
Professori Mikko Möttöinen kertoi luennolla, ettei kvanttitietokoneen valmistajan tarvitse realisoida kaikkia kvanttitietokoneen portteja, koska useiden porttien toiminta on korvattavissa muilla porteilla. Miten Z-portin operaatio on toteutettavissa Hadamard H-portilla ja X-portilla eli not portilla?
Ohje: Tutki 1. tehtävän demolla kuinka Z-portti kiertää Blochin pallolle piirrettyä tilavektoria, tai päättele miten Z-portin matriisiesitys saadaan laskettua H- ja X-porttien matriisiesityksestä. Kertauksena:
$$
X= \pmatrix {0 & 1 \\ 1 & 0} \quad \text{ja} \quad H=\frac 1 {\sqrt{2}}\pmatrix {1 & 1 \\ 1 & -1}
$$
## Tehtävä 3: Piirin tilavektorin tarkastelua
### Johdanto
Seuraavassa esimerkissä luodaan yhden kubitin kvanttipiiri, ja tulostetaan piirin tilavektori ja sen esitys Blochin pallolla. Koodin tulosteessa merkintä `1+0j` tarkoittaa, että kompleksiluvun reaaliosa on 1 ja imaginaariosa 0, ja `j` merkitsee imaginääriyksikköä. Koodissa tulostuksen muotoiluun käytettävää `enumerate(statevector)` syntaksia ei tällä kurssilla tarvitse ymmärtää.
```
from qiskit import *
qreg = QuantumRegister(1)
creg = ClassicalRegister(1)
circuit = QuantumCircuit(qreg, creg)
circuit.draw(output='mpl')
simulator = Aer.get_backend('statevector_simulator')
# Suoritetaan
result = execute(circuit, simulator).result()
statevector = result.get_statevector(circuit)
# Tulostetaan saatu tila ket-notaation kera
for i, c in enumerate(statevector):
print(c, f"|{i:0b}>")
visualization.plot_bloch_multivector(statevector, title="Alkutila")
```
**a)** Ohjelmoi yllä olevan esimerkin mukaisesti yhden kubitin kvanttipiiri, jossa kubitin alkutila on alussa $\ket 1$, jonka jälkeen kubittiin operoidaan H-portilla. Simuloi piiri, ja tulosta tämän jälkeen tilavektori käyttäen komentoa `result.get_statevector(circuit)` ja piirrä yhden kubitin systeemin tilavektori Blochin pallolla.
Sinun tulisi saada lopputulosteeksi seuraava:
<img src="https://kvanttilaskenta.net/hub/static/content/kierros5/bloch_k1.png" width="400">
Huomaa, tulosteessa, että kompleksiluvun kerroin $1 / \sqrt 2 \approx 0.707$ ja että imagarinääriosa $10^{-17} \approx 0$. Numeerisesta virheestä johtuen tuloksen imaginääriosa ei ole täsmälleen nolla, mutta äärimmäisen lähellä nollaa. Kuvan tila on siis saavutettu operoimalla tilaan $\ket 1$ portilla H:
$$
H\ket 1 = \frac 1 {\sqrt{2}}(\ket 0 - \ket 1) \quad \text{eli} \quad H\ket 1 = \ket -
$$
Blochin pallo sopii havainnollistamisvälineeksi yhden kubitin systemeissä.
```
#kirjoita vastauksesi tähän
```
**b)** Jatka a-osaa ja tutki simuloimalla, että $H \ket - = \ket 1$. Esitä ratkaisuna lopputuloksen tilavektori ja sen esitys Blochin pallolla.
```
# Ohjelmoi ratkaisusi tähän
```
**c)** Tarkastellaan johdantona seuraavaa kahden kubitin kvanttipiriä.
<img src="https://kvanttilaskenta.net/hub/static/content/kierros5/bloch_k2.png" width="400">
Systeemin tila H-portin jälkeen:
$$
\frac{1}{\sqrt 2} (\ket 0 + \ket 1) \otimes \ket 0=\frac{1}{\sqrt 2}(\ket{00}+\ket{10})
$$
Systeemin tila CNOT-portin jälkeen:
$$
\frac{1}{\sqrt 2}(\ket{00}+\ket{11})
$$
Systeemin tila X-portin jälkeen:
$$
\frac{1}{\sqrt 2}(\ket{10}+\ket{01})
$$
Alla piirin rakentamisessa käytetty koodi:
```
from qiskit import *
qreg = QuantumRegister(2)
creg = ClassicalRegister(2)
circuit = QuantumCircuit(qreg, creg)
circuit.h(qreg[0])
circuit.cx(qreg[0],qreg[1])
circuit.x(qreg[0])
circuit.draw(output='mpl')
```
Simuloidaan piirin toiminta ja tulostetaan tilavektori:
```
simulator = Aer.get_backend('statevector_simulator')
# Suoritetaan
result = execute(circuit, simulator).result()
statevector = result.get_statevector(circuit)
# Tulostetaan saatu tila ket-notaation kera
for i, c in enumerate(statevector):
print(c, f"|{i:02b}>")
```
**Tehtävä.** Simuloi kuvan piirin toiminta ja tulosta tilavektori:
<img src="https://kvanttilaskenta.net/hub/static/content/kierros5/bloch_k3.png" width="400">
Perustele piirin toiminta laskemalle ket-esitystä käyttäen.
```
#kirjoita vastauksesi tähän
```
## Tehtävä 4
Luennolla käsiteltiin Fourier-muunnosta. Katso halutessasi havainnollistava video siihen liittyen:
https://www.youtube.com/watch?v=spUNpyF58BY
|
github_jupyter
|
from qiskit import *
qreg = QuantumRegister(1)
creg = ClassicalRegister(1)
circuit = QuantumCircuit(qreg, creg)
circuit.draw(output='mpl')
simulator = Aer.get_backend('statevector_simulator')
# Suoritetaan
result = execute(circuit, simulator).result()
statevector = result.get_statevector(circuit)
# Tulostetaan saatu tila ket-notaation kera
for i, c in enumerate(statevector):
print(c, f"|{i:0b}>")
visualization.plot_bloch_multivector(statevector, title="Alkutila")
#kirjoita vastauksesi tähän
# Ohjelmoi ratkaisusi tähän
from qiskit import *
qreg = QuantumRegister(2)
creg = ClassicalRegister(2)
circuit = QuantumCircuit(qreg, creg)
circuit.h(qreg[0])
circuit.cx(qreg[0],qreg[1])
circuit.x(qreg[0])
circuit.draw(output='mpl')
simulator = Aer.get_backend('statevector_simulator')
# Suoritetaan
result = execute(circuit, simulator).result()
statevector = result.get_statevector(circuit)
# Tulostetaan saatu tila ket-notaation kera
for i, c in enumerate(statevector):
print(c, f"|{i:02b}>")
#kirjoita vastauksesi tähän
| 0.42931 | 0.982987 |
## Saving Profiles to S3
---
```
from whylogs import get_or_create_session
import pandas as pd
%load_ext autoreload
%autoreload 2
```
## Create a mock s3 server
For this example we will create a fake s3 server using moto lib. You should remove this section if you have you own bucket setup on aws. Make sure you have your aws configuration set. By default this mock server creates a server in region "us-east-1"
```
BUCKET="super_awesome_bucket"
from moto import mock_s3
from moto.s3.responses import DEFAULT_REGION_NAME
import boto3
mocks3 = mock_s3()
mocks3.start()
res = boto3.resource('s3', region_name=DEFAULT_REGION_NAME)
res.create_bucket(Bucket=BUCKET)
```
## Load Data
We can go by our usual way, load a example csv data
```
df = pd.read_csv("data/lending_club_1000.csv")
```
## Config File Example
---
Seting up whylogs to save your data on s3 can be in several ways. Simplest is to simply create a config file,where each data format can be saved to a specific location. As shown below
```
CONFIG = """
project: s3_example_project
pipeline: latest_results
verbose: false
writers:
- formats:
- protobuf
output_path: s3://super_awesome_bucket/
path_template: $name/dataset_summary
filename_template: dataset_summary
type: s3
- formats:
- flat
output_path: s3://super_awesome_bucket/
path_template: $name/dataset_summary
filename_template: dataset_summary
type: s3
- formats:
- json
output_path: s3://super_awesome_bucket/
path_template: $name/dataset_summary
filename_template: dataset_summary
type: s3
"""
config_path=".whylogs_s3.yaml"
with open(".whylogs_s3.yaml","w") as file:
file.write(CONFIG)
```
Checking the content:
```
%cat .whylogs_s3.yaml
```
If you have a custom name for your config file or place it in a special location you can use the helper function
```
from whylogs.app.session import load_config, session_from_config
config = load_config(".whylogs_s3.yaml")
session = session_from_config(config)
print(session.get_config().to_yaml())
```
Otherwise if the file is located in your home directory or current location you are running, you can simply run `get_or_create_session()`
```
session= get_or_create_session()
print(session.get_config().to_yaml())
```
## Loggin Data
---
The data can be save by simply closing a logger, or one a logger is out of scope.
```
with session.logger("dataset_test_s3") as logger:
logger.log_dataframe(df)
client = boto3.client('s3')
objects = client.list_objects(Bucket=BUCKET)
[obj["Key"] for obj in objects.get("Contents",[])]
```
You can define the configure for were the data is save through a configuration file or creating a custom writer.
```
mocks3.stop()
```
## Without Config File
---
```
mocks3.start()
res = boto3.resource('s3', region_name=DEFAULT_REGION_NAME)
res.create_bucket(Bucket=BUCKET)
from whylogs.app.session import load_config, session_from_config
from whylogs.app.config import WriterConfig, SessionConfig
s3_writer_config= WriterConfig(type="s3",formats=["json","flat","protobuf"],
output_path="s3://super_awesome_bucket/",
path_template="$name/dataset_summary",
filename_template="dataset_profile")
#you can also create a local, so you have a local version of the data.
session_config=SessionConfig(project="my_super_duper_project_name",
pipeline="latest_results",
writers=[s3_writer_config])
session = session_from_config(session_config)
print(session.get_config().to_yaml())
with session.logger("dataset_test_s3_config_as_code") as logger:
logger.log_dataframe(df)
client = boto3.client('s3')
objects = client.list_objects(Bucket=BUCKET)
[obj["Key"] for obj in objects.get("Contents",[])]
```
### Close mock s3 server
```
mocks3.stop()
```
|
github_jupyter
|
from whylogs import get_or_create_session
import pandas as pd
%load_ext autoreload
%autoreload 2
BUCKET="super_awesome_bucket"
from moto import mock_s3
from moto.s3.responses import DEFAULT_REGION_NAME
import boto3
mocks3 = mock_s3()
mocks3.start()
res = boto3.resource('s3', region_name=DEFAULT_REGION_NAME)
res.create_bucket(Bucket=BUCKET)
df = pd.read_csv("data/lending_club_1000.csv")
CONFIG = """
project: s3_example_project
pipeline: latest_results
verbose: false
writers:
- formats:
- protobuf
output_path: s3://super_awesome_bucket/
path_template: $name/dataset_summary
filename_template: dataset_summary
type: s3
- formats:
- flat
output_path: s3://super_awesome_bucket/
path_template: $name/dataset_summary
filename_template: dataset_summary
type: s3
- formats:
- json
output_path: s3://super_awesome_bucket/
path_template: $name/dataset_summary
filename_template: dataset_summary
type: s3
"""
config_path=".whylogs_s3.yaml"
with open(".whylogs_s3.yaml","w") as file:
file.write(CONFIG)
%cat .whylogs_s3.yaml
from whylogs.app.session import load_config, session_from_config
config = load_config(".whylogs_s3.yaml")
session = session_from_config(config)
print(session.get_config().to_yaml())
session= get_or_create_session()
print(session.get_config().to_yaml())
with session.logger("dataset_test_s3") as logger:
logger.log_dataframe(df)
client = boto3.client('s3')
objects = client.list_objects(Bucket=BUCKET)
[obj["Key"] for obj in objects.get("Contents",[])]
mocks3.stop()
mocks3.start()
res = boto3.resource('s3', region_name=DEFAULT_REGION_NAME)
res.create_bucket(Bucket=BUCKET)
from whylogs.app.session import load_config, session_from_config
from whylogs.app.config import WriterConfig, SessionConfig
s3_writer_config= WriterConfig(type="s3",formats=["json","flat","protobuf"],
output_path="s3://super_awesome_bucket/",
path_template="$name/dataset_summary",
filename_template="dataset_profile")
#you can also create a local, so you have a local version of the data.
session_config=SessionConfig(project="my_super_duper_project_name",
pipeline="latest_results",
writers=[s3_writer_config])
session = session_from_config(session_config)
print(session.get_config().to_yaml())
with session.logger("dataset_test_s3_config_as_code") as logger:
logger.log_dataframe(df)
client = boto3.client('s3')
objects = client.list_objects(Bucket=BUCKET)
[obj["Key"] for obj in objects.get("Contents",[])]
mocks3.stop()
| 0.375134 | 0.702064 |
# d3viz: Interactive visualization of Aesara compute graphs
## Requirements
``d3viz`` requires the [pydot](https://pypi.python.org/pypi/pydot)
package. [pydot-ng](https://github.com/pydot/pydot-ng) fork is better
maintained, and it works both in Python 2.x and 3.x. Install it with pip::
```
!pip install pydot-ng
```
Like Aesara’s [printing module](http://deeplearning.net/software/aesara/library/printing.html), ``d3viz``
requires [graphviz](http://www.graphviz.org) binary to be available.
## Overview
`d3viz` extends Aesara’s [printing module](http://deeplearning.net/software/aesara/library/printing.html) to interactively visualize compute graphs. Instead of creating a static picture, it creates an HTML file, which can be opened with current web-browsers. `d3viz` allows
* to zoom to different regions and to move graphs via drag and drop,
* to position nodes both manually and automatically,
* to retrieve additional information about nodes and edges such as their data type or definition in the source code,
* to edit node labels,
* to visualizing profiling information, and
* to explore nested graphs such as OpFromGraph nodes.
```
import aesara as th
import aesara.tensor as tt
import numpy as np
```
As an example, consider the following multilayer perceptron with one hidden layer and a softmax output layer.
```
ninputs = 1000
nfeatures = 100
noutputs = 10
nhiddens = 50
rng = np.random.RandomState(0)
x = tt.dmatrix('x')
wh = th.shared(rng.normal(0, 1, (nfeatures, nhiddens)), borrow=True)
bh = th.shared(np.zeros(nhiddens), borrow=True)
h = tt.nnet.sigmoid(tt.dot(x, wh) + bh)
wy = th.shared(rng.normal(0, 1, (nhiddens, noutputs)))
by = th.shared(np.zeros(noutputs), borrow=True)
y = tt.nnet.softmax(tt.dot(h, wy) + by)
predict = th.function([x], y)
```
The function `predict` outputs the probability of 10 classes. You can visualize it with `pydotprint` as follows:
```
from aesara.printing import pydotprint
import os
if not os.path.exists('examples'):
os.makedirs('examples')
pydotprint(predict, 'examples/mlp.png')
from IPython.display import Image
Image('examples/mlp.png', width='80%')
```
To visualize it interactively, import the `d3viz` function from the `d3viz` module, which can be called as before:
```
import aesara.d3viz as d3v
d3v.d3viz(predict, 'examples/mlp.html')
```
[Open visualization!](examples/mlp.html)
When you open the output file `mlp.html` in your web-browser, you will see an interactive visualization of the compute graph. You can move the whole graph or single nodes via drag and drop, and zoom via the mouse wheel. When you move the mouse cursor over a node, a window will pop up that displays detailed information about the node, such as its data type or definition in the source code. When you left-click on a node and select `Edit`, you can change the predefined node label. If you are dealing with a complex graph with many nodes, the default node layout may not be perfect. In this case, you can press the `Release node` button in the top-left corner to automatically arrange nodes. To reset nodes to their default position, press the `Reset nodes` button.
You can also display the interactive graph inline in
IPython using ``IPython.display.IFrame``:
```
from IPython.display import IFrame
d3v.d3viz(predict, 'examples/mlp.html')
IFrame('examples/mlp.html', width=700, height=500)
```
Currently if you use display.IFrame you still have to create a file,
and this file can't be outside notebooks root (e.g. usually it can't be
in /tmp/).
## Profiling
Aesara allows [function profiling](http://deeplearning.net/software/aesara/tutorial/profiling.html) via the `profile=True` flag. After at least one function call, the compute time of each node can be printed in text form with `debugprint`. However, analyzing complex graphs in this way can be cumbersome.
`d3viz` can visualize the same timing information graphically, and hence help to spot bottlenecks in the compute graph more easily! To begin with, we will redefine the `predict` function, this time by using `profile=True` flag. Afterwards, we capture the runtime on random data:
```
predict_profiled = th.function([x], y, profile=True)
x_val = rng.normal(0, 1, (ninputs, nfeatures))
y_val = predict_profiled(x_val)
d3v.d3viz(predict_profiled, 'examples/mlp2.html')
```
[Open visualization!](./examples/mlp2.html)
When you open the HTML file in your browser, you will find an additional `Toggle profile colors` button in the menu bar. By clicking on it, nodes will be colored by their compute time, where red corresponds to a high compute time. You can read out the exact timing information of a node by moving the cursor over it.
## Different output formats
Internally, `d3viz` represents a compute graph in the [Graphviz DOT language](http://www.graphviz.org/), using the [pydot](https://pypi.python.org/pypi/pydot) package, and defines a front-end based on the [d3.js](http://d3js.org/) library to visualize it. However, any other Graphviz front-end can be used, which allows to export graphs to different formats.
```
formatter = d3v.formatting.PyDotFormatter()
pydot_graph = formatter(predict_profiled)
pydot_graph.write_png('examples/mlp2.png');
pydot_graph.write_pdf('examples/mlp2.pdf');
Image('./examples/mlp2.png')
```
Here, we used the `PyDotFormatter` class to convert the compute graph into a `pydot` graph, and created a [PNG](./examples/mlp2.png) and [PDF](./examples/mlp2.pdf) file. You can find all output formats supported by Graphviz [here](http://www.graphviz.org/doc/info/output.html).
## OpFromGraph nodes
An [OpFromGraph](http://deeplearning.net/software/aesara/library/compile/opfromgraph.html) node defines a new operation, which can be called with different inputs at different places in the compute graph. Each `OpFromGraph` node defines a nested graph, which will be visualized accordingly by `d3viz`.
```
x, y, z = tt.scalars('xyz')
e = tt.nnet.sigmoid((x + y + z)**2)
op = th.OpFromGraph([x, y, z], [e])
e2 = op(x, y, z) + op(z, y, x)
f = th.function([x, y, z], e2)
d3v.d3viz(f, 'examples/ofg.html')
```
[Open visualization!](./examples/ofg.html)
In this example, an operation with three inputs is defined, which is used to build a function that calls this operations twice, each time with different input arguments.
In the `d3viz` visualization, you will find two OpFromGraph nodes, which correspond to the two OpFromGraph calls. When you double click on one of them, the nested graph appears with the correct mapping of its input arguments. You can move it around by drag and drop in the shaded area, and close it again by double-click.
An OpFromGraph operation can be composed of further OpFromGraph operations, which will be visualized as nested graphs as you can see in the following example.
```
x, y, z = tt.scalars('xyz')
e = x * y
op = th.OpFromGraph([x, y], [e])
e2 = op(x, y) + z
op2 = th.OpFromGraph([x, y, z], [e2])
e3 = op2(x, y, z) + z
f = th.function([x, y, z], [e3])
d3v.d3viz(f, 'examples/ofg2.html')
```
[Open visualization!](./examples/ofg2.html)
## Feedback
If you have any problems or great ideas on how to improve `d3viz`, please let me know!
* Christof Angermueller
* <cangermueller@gmail.com>
* https://cangermueller.com
|
github_jupyter
|
!pip install pydot-ng
import aesara as th
import aesara.tensor as tt
import numpy as np
ninputs = 1000
nfeatures = 100
noutputs = 10
nhiddens = 50
rng = np.random.RandomState(0)
x = tt.dmatrix('x')
wh = th.shared(rng.normal(0, 1, (nfeatures, nhiddens)), borrow=True)
bh = th.shared(np.zeros(nhiddens), borrow=True)
h = tt.nnet.sigmoid(tt.dot(x, wh) + bh)
wy = th.shared(rng.normal(0, 1, (nhiddens, noutputs)))
by = th.shared(np.zeros(noutputs), borrow=True)
y = tt.nnet.softmax(tt.dot(h, wy) + by)
predict = th.function([x], y)
from aesara.printing import pydotprint
import os
if not os.path.exists('examples'):
os.makedirs('examples')
pydotprint(predict, 'examples/mlp.png')
from IPython.display import Image
Image('examples/mlp.png', width='80%')
import aesara.d3viz as d3v
d3v.d3viz(predict, 'examples/mlp.html')
from IPython.display import IFrame
d3v.d3viz(predict, 'examples/mlp.html')
IFrame('examples/mlp.html', width=700, height=500)
predict_profiled = th.function([x], y, profile=True)
x_val = rng.normal(0, 1, (ninputs, nfeatures))
y_val = predict_profiled(x_val)
d3v.d3viz(predict_profiled, 'examples/mlp2.html')
formatter = d3v.formatting.PyDotFormatter()
pydot_graph = formatter(predict_profiled)
pydot_graph.write_png('examples/mlp2.png');
pydot_graph.write_pdf('examples/mlp2.pdf');
Image('./examples/mlp2.png')
x, y, z = tt.scalars('xyz')
e = tt.nnet.sigmoid((x + y + z)**2)
op = th.OpFromGraph([x, y, z], [e])
e2 = op(x, y, z) + op(z, y, x)
f = th.function([x, y, z], e2)
d3v.d3viz(f, 'examples/ofg.html')
x, y, z = tt.scalars('xyz')
e = x * y
op = th.OpFromGraph([x, y], [e])
e2 = op(x, y) + z
op2 = th.OpFromGraph([x, y, z], [e2])
e3 = op2(x, y, z) + z
f = th.function([x, y, z], [e3])
d3v.d3viz(f, 'examples/ofg2.html')
| 0.361616 | 0.964288 |
```
import os
import math
import numpy as np
import time
from PIL import Image
import fnmatch
from keras.preprocessing.image import ImageDataGenerator, array_to_img, img_to_array, load_img
import h5py
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import mod3
plt.rcParams.update({'figure.figsize': (5.0, 4.0), 'lines.linewidth': 2.0})
label_counter = 0
index = 0
tst_images = []
categ_labels = np.zeros(shape=80)
bllvl_labels = np.zeros(shape=80)
fn_pattern = ''
B = np.zeros(shape=(80, 32, 32, 3))
for subdir, dirs, files in os.walk('C:\\Users\\laptomon\\Documents\\fmri_data_temp\\imgs\\blur\\ImgSet170114'):
for folder in dirs:
for folder_subdir, folder_dirs, folder_files in os.walk(os.path.join(subdir, folder)):
for bllvl in range(4):
fn_pattern = '*' + str(bllvl) + '.jpg'
for fn in fnmatch.filter(folder_files, fn_pattern):
# print(fn)
tst_images.append(os.path.join(folder_subdir, fn))
img = load_img(os.path.join(folder_subdir, fn),target_size=(32, 32))
B[index] = img_to_array(img)
B[index] /= 255
# print(label_counter)
categ_labels[index] = label_counter
# print(label_counter)
bllvl_labels[index] = bllvl
index = index + 1
# print(label_counter)
label_counter = label_counter + 1
print(len(tst_images))
print(categ_labels)
print(len(bllvl_labels))
print(B.shape)
hf = h5py.File('blurred_images_180713.h5', 'w')
hf.create_dataset('images', data=B)
hf.create_dataset('blur_lvl', data=bllvl_labels)
hf.create_dataset('category', data=categ_labels)
hf.close()
from keras.models import Model, load_model
ff_model = load_model('cifar_feedforward_180711.h5');
rcs_model = load_model('cifar_recurrentsimple_180713.h5');
rcc_model = load_model('cifar_recurrentcomplex_180712.h5');
tdc_model = load_model('cifar_rectopdowncomplex_180716.h5');
tds_model = load_model('cifar_rectopdownsimple_180717.h5');
import keras
n_out = 10
oldlab = categ_labels.copy()
print(oldlab)
categ_labels[oldlab == 1] = 2
categ_labels[oldlab == 2] = 1
categ_labels[oldlab == 4] = 5
# print(categ_labels)
# print(tst_images)
labels_test = keras.utils.to_categorical(categ_labels, n_out)
# print(labels_test)
B1 = B[bllvl_labels == 1]
print(B1.shape)
labels_test1 = labels_test[bllvl_labels == 1]
evaluation = ff_model.evaluate(B1, labels_test1, verbose=0)
print('\n[Model evaluation]')
print( format(np.mean(evaluation[0]), '.4f'))
print( format(np.mean(evaluation[1]), '.4f'))
evaluation = rcs_model.evaluate(B1, labels_test1, verbose=0)
print('\n[Model evaluation]')
print( format(np.mean(evaluation[0]), '.4f'))
print( format(np.mean(evaluation[1]), '.4f'))
evaluation = rcc_model.evaluate(B1, labels_test1, verbose=0)
print('\n[Model evaluation]')
print( format(np.mean(evaluation[0]), '.4f'))
print( format(np.mean(evaluation[1]), '.4f'))
evaluation = tdc_model.evaluate(B1, labels_test1, verbose=0)
print('\n[Model evaluation]')
print( format(np.mean(evaluation[0]), '.4f'))
print( format(np.mean(evaluation[1]), '.4f'))
evaluation = tds_model.evaluate(B1, labels_test1, verbose=0)
print('\n[Model evaluation]')
print( format(np.mean(evaluation[0]), '.4f'))
print( format(np.mean(evaluation[1]), '.4f'))
# select n_show samples randomly
plt.figure(figsize=(10, 4))
for bl in range(4):
n_show = 10
B1 = B[bllvl_labels == bl]
selected = np.arange(0,20,2)
categ_labels1 = categ_labels[bllvl_labels == 0]
# print(selected)
# print(labels_test1[0])
# print(categ_labels1[selected])
# plot samples
for idx, img in enumerate(B1[selected]):
plt.subplot(4, n_show, bl*10+idx+1)
plt.imshow(img, cmap=plt.cm.gray)
plt.gca().get_xaxis().set_visible(False)
plt.gca().get_yaxis().set_visible(False)
plt.tight_layout()
plt.show()
lossmat=np.zeros((5,4))
accmat=np.zeros((5,4))
for bl in range(4):
B1 = B[bllvl_labels == bl]
labels_test1 = labels_test[bllvl_labels == bl]
evaluation = ff_model.evaluate(B1, labels_test1, verbose=0)
accmat[0][bl] = evaluation[1]
lossmat[0][bl] = evaluation[0]
evaluation = rcs_model.evaluate(B1, labels_test1, verbose=0)
accmat[1][bl] = evaluation[1]
lossmat[1][bl] = evaluation[0]
evaluation = rcc_model.evaluate(B1, labels_test1, verbose=0)
accmat[2][bl] = evaluation[1]
lossmat[2][bl] = evaluation[0]
evaluation = tds_model.evaluate(B1, labels_test1, verbose=0)
accmat[3][bl] = evaluation[1]
lossmat[3][bl] = evaluation[0]
evaluation = tdc_model.evaluate(B1, labels_test1, verbose=0)
accmat[4][bl] = evaluation[1]
lossmat[4][bl] = evaluation[0]
print (lossmat)
plt.figure(figsize=(10, 4))
plt.plot(lossmat[0], label='feedforward')
plt.plot(lossmat[1], label='recurrent simple')
plt.plot(lossmat[2], label='recurrent complex')
plt.plot(lossmat[3], label='top-down simple')
plt.plot(lossmat[4], label='top-down complex')
plt.xticks(np.arange(4), ('0%', '6%', '12%', '25%'))
plt.legend()
plt.ylabel('test loss')
plt.show()
plt.figure(figsize=(10, 4))
plt.plot(accmat[0], label='feedforward')
plt.plot(accmat[1], label='recurrent simple')
plt.plot(accmat[2], label='recurrent complex')
plt.plot(accmat[3], label='top-down simple')
plt.plot(accmat[4], label='top-down complex')
plt.xticks(np.arange(4), ('0%', '6%', '12%', '25%'))
plt.legend()
plt.ylabel('accuracy')
plt.show()
```
|
github_jupyter
|
import os
import math
import numpy as np
import time
from PIL import Image
import fnmatch
from keras.preprocessing.image import ImageDataGenerator, array_to_img, img_to_array, load_img
import h5py
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import mod3
plt.rcParams.update({'figure.figsize': (5.0, 4.0), 'lines.linewidth': 2.0})
label_counter = 0
index = 0
tst_images = []
categ_labels = np.zeros(shape=80)
bllvl_labels = np.zeros(shape=80)
fn_pattern = ''
B = np.zeros(shape=(80, 32, 32, 3))
for subdir, dirs, files in os.walk('C:\\Users\\laptomon\\Documents\\fmri_data_temp\\imgs\\blur\\ImgSet170114'):
for folder in dirs:
for folder_subdir, folder_dirs, folder_files in os.walk(os.path.join(subdir, folder)):
for bllvl in range(4):
fn_pattern = '*' + str(bllvl) + '.jpg'
for fn in fnmatch.filter(folder_files, fn_pattern):
# print(fn)
tst_images.append(os.path.join(folder_subdir, fn))
img = load_img(os.path.join(folder_subdir, fn),target_size=(32, 32))
B[index] = img_to_array(img)
B[index] /= 255
# print(label_counter)
categ_labels[index] = label_counter
# print(label_counter)
bllvl_labels[index] = bllvl
index = index + 1
# print(label_counter)
label_counter = label_counter + 1
print(len(tst_images))
print(categ_labels)
print(len(bllvl_labels))
print(B.shape)
hf = h5py.File('blurred_images_180713.h5', 'w')
hf.create_dataset('images', data=B)
hf.create_dataset('blur_lvl', data=bllvl_labels)
hf.create_dataset('category', data=categ_labels)
hf.close()
from keras.models import Model, load_model
ff_model = load_model('cifar_feedforward_180711.h5');
rcs_model = load_model('cifar_recurrentsimple_180713.h5');
rcc_model = load_model('cifar_recurrentcomplex_180712.h5');
tdc_model = load_model('cifar_rectopdowncomplex_180716.h5');
tds_model = load_model('cifar_rectopdownsimple_180717.h5');
import keras
n_out = 10
oldlab = categ_labels.copy()
print(oldlab)
categ_labels[oldlab == 1] = 2
categ_labels[oldlab == 2] = 1
categ_labels[oldlab == 4] = 5
# print(categ_labels)
# print(tst_images)
labels_test = keras.utils.to_categorical(categ_labels, n_out)
# print(labels_test)
B1 = B[bllvl_labels == 1]
print(B1.shape)
labels_test1 = labels_test[bllvl_labels == 1]
evaluation = ff_model.evaluate(B1, labels_test1, verbose=0)
print('\n[Model evaluation]')
print( format(np.mean(evaluation[0]), '.4f'))
print( format(np.mean(evaluation[1]), '.4f'))
evaluation = rcs_model.evaluate(B1, labels_test1, verbose=0)
print('\n[Model evaluation]')
print( format(np.mean(evaluation[0]), '.4f'))
print( format(np.mean(evaluation[1]), '.4f'))
evaluation = rcc_model.evaluate(B1, labels_test1, verbose=0)
print('\n[Model evaluation]')
print( format(np.mean(evaluation[0]), '.4f'))
print( format(np.mean(evaluation[1]), '.4f'))
evaluation = tdc_model.evaluate(B1, labels_test1, verbose=0)
print('\n[Model evaluation]')
print( format(np.mean(evaluation[0]), '.4f'))
print( format(np.mean(evaluation[1]), '.4f'))
evaluation = tds_model.evaluate(B1, labels_test1, verbose=0)
print('\n[Model evaluation]')
print( format(np.mean(evaluation[0]), '.4f'))
print( format(np.mean(evaluation[1]), '.4f'))
# select n_show samples randomly
plt.figure(figsize=(10, 4))
for bl in range(4):
n_show = 10
B1 = B[bllvl_labels == bl]
selected = np.arange(0,20,2)
categ_labels1 = categ_labels[bllvl_labels == 0]
# print(selected)
# print(labels_test1[0])
# print(categ_labels1[selected])
# plot samples
for idx, img in enumerate(B1[selected]):
plt.subplot(4, n_show, bl*10+idx+1)
plt.imshow(img, cmap=plt.cm.gray)
plt.gca().get_xaxis().set_visible(False)
plt.gca().get_yaxis().set_visible(False)
plt.tight_layout()
plt.show()
lossmat=np.zeros((5,4))
accmat=np.zeros((5,4))
for bl in range(4):
B1 = B[bllvl_labels == bl]
labels_test1 = labels_test[bllvl_labels == bl]
evaluation = ff_model.evaluate(B1, labels_test1, verbose=0)
accmat[0][bl] = evaluation[1]
lossmat[0][bl] = evaluation[0]
evaluation = rcs_model.evaluate(B1, labels_test1, verbose=0)
accmat[1][bl] = evaluation[1]
lossmat[1][bl] = evaluation[0]
evaluation = rcc_model.evaluate(B1, labels_test1, verbose=0)
accmat[2][bl] = evaluation[1]
lossmat[2][bl] = evaluation[0]
evaluation = tds_model.evaluate(B1, labels_test1, verbose=0)
accmat[3][bl] = evaluation[1]
lossmat[3][bl] = evaluation[0]
evaluation = tdc_model.evaluate(B1, labels_test1, verbose=0)
accmat[4][bl] = evaluation[1]
lossmat[4][bl] = evaluation[0]
print (lossmat)
plt.figure(figsize=(10, 4))
plt.plot(lossmat[0], label='feedforward')
plt.plot(lossmat[1], label='recurrent simple')
plt.plot(lossmat[2], label='recurrent complex')
plt.plot(lossmat[3], label='top-down simple')
plt.plot(lossmat[4], label='top-down complex')
plt.xticks(np.arange(4), ('0%', '6%', '12%', '25%'))
plt.legend()
plt.ylabel('test loss')
plt.show()
plt.figure(figsize=(10, 4))
plt.plot(accmat[0], label='feedforward')
plt.plot(accmat[1], label='recurrent simple')
plt.plot(accmat[2], label='recurrent complex')
plt.plot(accmat[3], label='top-down simple')
plt.plot(accmat[4], label='top-down complex')
plt.xticks(np.arange(4), ('0%', '6%', '12%', '25%'))
plt.legend()
plt.ylabel('accuracy')
plt.show()
| 0.241221 | 0.432243 |
# Training Amazon SageMaker models for molecular property prediction by using DGL with PyTorch backend
The **Amazon SageMaker Python SDK** makes it easy to train Deep Graph Library (DGL) models. In this example, you train a simple graph neural network for molecular toxicity prediction by using [DGL](https://github.com/dmlc/dgl) and the Tox21 dataset.
The dataset contains qualitative toxicity measurements for 8,014 compounds on 12 different targets, including nuclear
receptors and stress-response pathways. Each target yields a binary classification problem. You can model the problem as a graph classification problem.
## Setup
Define a few variables that you need later in the example.
```
import sagemaker
from sagemaker import get_execution_role
from sagemaker.session import Session
# Setup session
sess = sagemaker.Session()
# S3 bucket for saving code and model artifacts.
# Feel free to specify a different bucket here if you wish.
bucket = sess.default_bucket()
# Location to put your custom code.
custom_code_upload_location = 'customcode'
# IAM execution role that gives Amazon SageMaker access to resources in your AWS account.
# You can use the Amazon SageMaker Python SDK to get the role from the notebook environment.
role = get_execution_role()
```
## Training Script
`main.py` provides all the code you need for training a molecular property prediction model by using Amazon SageMaker.
```
!cat main.py
```
## Bring Your Own Image for Amazon SageMaker
In this example, you need rdkit library to handle the tox21 dataset. The DGL CPU and GPU Docker has the rdkit library pre-installed at Dockerhub under dgllib registry (namely, dgllib/dgl-sagemaker-cpu:dgl_0.4_pytorch_1.2.0_rdkit for CPU and dgllib/dgl-sagemaker-gpu:dgl_0.4_pytorch_1.2.0_rdkit for GPU). You can pull the image yourself according to your requirement and push it into your AWS ECR. Following script helps you to do so. You can skip this step if you have already prepared your DGL Docker image in your Amazon Elastic Container Registry (Amazon ECR).
```
%%sh
# For CPU default_docker_name="dgllib/dgl-sagemaker-cpu:dgl_0.4_pytorch_1.2.0_rdkit"
default_docker_name="dgllib/dgl-sagemaker-gpu:dgl_0.4_pytorch_1.2.0_rdkit"
docker pull $default_docker_name
docker_name=sagemaker-dgl-pytorch-gcn-tox21
# For CPU docker build -t $docker_name -f gcn_tox21_cpu.Dockerfile .
docker build -t $docker_name -f gcn_tox21_gpu.Dockerfile .
account=$(aws sts get-caller-identity --query Account --output text)
echo $account
region=$(aws configure get region)
fullname="${account}.dkr.ecr.${region}.amazonaws.com/${docker_name}:latest"
# If the repository doesn't exist in ECR, create it.
aws ecr describe-repositories --repository-names "${docker_name}" > /dev/null 2>&1
if [ $? -ne 0 ]
then
aws ecr create-repository --repository-name "${docker_name}" > /dev/null
fi
# Get the login command from ECR and execute it directly
$(aws ecr get-login --region ${region} --no-include-email)
docker tag ${docker_name} ${fullname}
docker push ${fullname}
```
## The Amazon SageMaker Estimator class
The Amazon SageMaker Estimator allows you to run a single machine in Amazon SageMaker, using CPU or GPU-based instances.
When you create the estimator, pass in the file name of the training script and the name of the IAM execution role. Also provide a few other parameters. `train_instance_count` and `train_instance_type` determine the number and type of SageMaker instances that will be used for the training job. The hyperparameters can be passed to the training script via a dict of values. See `main.py` for how they are handled.
The entrypoint of Amazon SageMaker Docker (e.g., dgllib/dgl-sagemaker-gpu:dgl_0.4_pytorch_1.2.0_rdkit) is a train script under /usr/bin/. The train script inside dgl docker image provided above will try to get the real entrypoint from the hyperparameters (with the key 'entrypoint') and run the real entrypoint under 'training-code' data channel (/opt/ml/input/data/training-code/) .
For this example, choose one ml.p3.2xlarge instance. You can also use a CPU instance such as ml.c4.2xlarge for the CPU image.
```
import boto3
# Set target dgl-docker name
docker_name='sagemaker-dgl-pytorch-gcn-tox21'
CODE_PATH = 'main.py'
code_location = sess.upload_data(CODE_PATH, bucket=bucket, key_prefix=custom_code_upload_location)
account = sess.boto_session.client('sts').get_caller_identity()['Account']
region = sess.boto_session.region_name
image = '{}.dkr.ecr.{}.amazonaws.com/{}:latest'.format(account, region, docker_name)
print(image)
estimator = sagemaker.estimator.Estimator(image,
role,
train_instance_count=1,
train_instance_type= 'ml.p3.2xlarge', #'ml.c4.2xlarge'
hyperparameters={'entrypoint': CODE_PATH},
sagemaker_session=sess)
```
## Running the Training Job
After you construct an Estimator object, fit it by using Amazon SageMaker.
```
estimator.fit({'training-code': code_location})
```
## Output
You can get the model training output from the Amazon Sagemaker console by searching for the training task and looking for the address of 'S3 model artifact'
|
github_jupyter
|
import sagemaker
from sagemaker import get_execution_role
from sagemaker.session import Session
# Setup session
sess = sagemaker.Session()
# S3 bucket for saving code and model artifacts.
# Feel free to specify a different bucket here if you wish.
bucket = sess.default_bucket()
# Location to put your custom code.
custom_code_upload_location = 'customcode'
# IAM execution role that gives Amazon SageMaker access to resources in your AWS account.
# You can use the Amazon SageMaker Python SDK to get the role from the notebook environment.
role = get_execution_role()
!cat main.py
%%sh
# For CPU default_docker_name="dgllib/dgl-sagemaker-cpu:dgl_0.4_pytorch_1.2.0_rdkit"
default_docker_name="dgllib/dgl-sagemaker-gpu:dgl_0.4_pytorch_1.2.0_rdkit"
docker pull $default_docker_name
docker_name=sagemaker-dgl-pytorch-gcn-tox21
# For CPU docker build -t $docker_name -f gcn_tox21_cpu.Dockerfile .
docker build -t $docker_name -f gcn_tox21_gpu.Dockerfile .
account=$(aws sts get-caller-identity --query Account --output text)
echo $account
region=$(aws configure get region)
fullname="${account}.dkr.ecr.${region}.amazonaws.com/${docker_name}:latest"
# If the repository doesn't exist in ECR, create it.
aws ecr describe-repositories --repository-names "${docker_name}" > /dev/null 2>&1
if [ $? -ne 0 ]
then
aws ecr create-repository --repository-name "${docker_name}" > /dev/null
fi
# Get the login command from ECR and execute it directly
$(aws ecr get-login --region ${region} --no-include-email)
docker tag ${docker_name} ${fullname}
docker push ${fullname}
import boto3
# Set target dgl-docker name
docker_name='sagemaker-dgl-pytorch-gcn-tox21'
CODE_PATH = 'main.py'
code_location = sess.upload_data(CODE_PATH, bucket=bucket, key_prefix=custom_code_upload_location)
account = sess.boto_session.client('sts').get_caller_identity()['Account']
region = sess.boto_session.region_name
image = '{}.dkr.ecr.{}.amazonaws.com/{}:latest'.format(account, region, docker_name)
print(image)
estimator = sagemaker.estimator.Estimator(image,
role,
train_instance_count=1,
train_instance_type= 'ml.p3.2xlarge', #'ml.c4.2xlarge'
hyperparameters={'entrypoint': CODE_PATH},
sagemaker_session=sess)
estimator.fit({'training-code': code_location})
| 0.349644 | 0.930395 |
```
import pandas as pd
import datetime
from github import Github
g = Github("jamieshq", "1ca9cfcf3a0f1922b95c582ff5fe5273d4c2a9a6")
cutoff = datetime.datetime(2015, 3, 30, 11, 38, 5, 291165)
def get_prs(org, repo):
prs = []
print("Getting PRs for {}/{}".format(org.login, repo.name))
for pr in repo.get_pulls(state="all"):
if pr.created_at < cutoff:
continue
prs.append({
'date': pr.created_at,
'user': pr.user.login,
'number': pr.number,
'org': org.login,
'repo': repo.name,
'is_merged': pr.is_merged(),
'state': pr.state
})
return prs
def get_pr_comments(org, repo):
comments = []
print("Getting PR comments for {}/{}".format(org.login, repo.name))
for pr in repo.get_pulls(state="all"):
if pr.created_at < cutoff:
continue
for comment in pr.get_comments():
comments.append({
'date': comment.created_at,
'user': comment.user.login,
'number': pr.number,
'org': org.login,
'repo': repo.name
})
return comments
def get_issues(org, repo):
issues = []
print("Getting issues for {}/{}".format(org.login, repo.name))
for issue in repo.get_issues(state="all"):
if issue.created_at < cutoff:
continue
issues.append({
'date': issue.created_at,
'user': issue.user.login,
'number': issue.number,
'org': org.login,
'repo': repo.name,
'state': issue.state
})
return issues
def get_issue_comments(org, repo):
comments = []
print("Getting issue comments for {}/{}".format(org.login, repo.name))
for issue in repo.get_issues(state="all"):
if issue.created_at < cutoff:
continue
for comment in issue.get_comments():
comments.append({
'issue_date': issue.created_at,
'user': issue.user.login,
'number': issue.number,
'org': org.login,
'repo': repo.name,
'comments': comment.body,
'comment_creation_date' : comment.created_at
})
return comments
#prs = []
#pr_comments = []
issues = []
issue_comments = []
test_orgs = ["jupyter-resources"]
real_orgs = ["jupyterlab", "jupyterhub", "jupyter-widgets", "jupyter-incubator"]
for org_name in real_orgs:
org = g.get_organization(org_name)
for repo in org.get_repos():
#prs.extend(get_prs(org, repo))
#pr_comments.extend(get_pr_comments(org, repo))
#issues.extend(get_issues(org, repo))
issue_comments.extend(get_issue_comments(org, repo))
issue_comments = pd.DataFrame(issue_comments).set_index(['org', 'repo', 'number', 'issue_date','comment_creation_date', 'comments']).sortlevel()
issue_comments.to_csv("issue_comments.csv", encoding="utf-8")
```
Notes
http://stackoverflow.com/questions/16923281/pandas-writing-dataframe-to-csv-file
https://developer.github.com/v3/issues/#get-a-single-issue
https://developer.github.com/early-access/graphql/object/issuecomment/
https://developer.github.com/v3/issues/comments/#list-comments-on-an-issue
http://pygithub.readthedocs.io/en/latest/introduction.html
Stuart's
https://github.com/staeiou/github-analytics/blob/master/github-organizations-intro.ipynb
https://github.com/getorg/getorg/blob/master/examples/orgevents/issues.ipynb
Jessica's
##### Converting an epoch timestamp to datetime after querying for ratelimit reset time
I hit my GitHub ratelimit after reading through the first organization listed (Jupyter). I used the below curl command to learn my ratelimit reset time, which was about an hour after receiving the timeout.
`curl -i https://api.github.com/users/username`
HTTP/1.1 200 OK
Date: Mon, 01 Jul 2013 17:27:06 GMT
Status: 200 OK
X-RateLimit-Limit: 60
X-RateLimit-Remaining: 56
X-RateLimit-Reset: 1372700873
Sources:
https://developer.github.com/v3/#rate-limiting
http://stackoverflow.com/questions/12400256/python-converting-epoch-time-into-the-datetime
```
import datetime
datetime.datetime.fromtimestamp(1491196314).strftime('%c')
prs = []
pr_comments = []
issues = []
issue_comments = []
for org_name in ["jupyter", "jupyterlab", "jupyterhub", "jupyter-widgets", "jupyter-incubator"]:
org = g.get_organization(org_name)
for repo in org.get_repos():
prs.extend(get_prs(org, repo))
pr_comments.extend(get_pr_comments(org, repo))
issues.extend(get_issues(org, repo))
issue_comments.extend(get_issue_comments(org, repo))
prs = pd.DataFrame(prs).set_index(['org', 'repo', 'number']).sortlevel()
pr_comments = pd.DataFrame(pr_comments).set_index(['org', 'repo', 'number', 'date']).sortlevel()
issues = pd.DataFrame(issues).set_index(['org', 'repo', 'number']).sortlevel()
issue_comments = pd.DataFrame(issue_comments).set_index(['org', 'repo', 'number', 'date']).sortlevel()
prs.to_csv("prs.csv")
pr_comments.to_csv("pr_comments.csv")
issues.to_csv("issues.csv")
issue_comments.to_csv("issue_comments.csv")
```
prs = pd.read_csv("prs.csv")
pr_comments = pd.read_csv("pr_comments.csv")
issues = pd.read_csv("issues.csv")
issue_comments = pd.read_csv("issue_comments.csv")
```
issue_comments.head()
prs = []
pr_comments = []
issues = []
issue_comments = []
for org_name in ["jupyterlab", "jupyterhub", "jupyter-widgets", "jupyter-incubator"]:
org = g.get_organization(org_name)
for repo in org.get_repos():
prs.extend(get_prs(org, repo))
pr_comments.extend(get_pr_comments(org, repo))
issues.extend(get_issues(org, repo))
issue_comments.extend(get_issue_comments(org, repo))
prs = pd.DataFrame(prs).set_index(['org', 'repo', 'number']).sortlevel()
pr_comments = pd.DataFrame(pr_comments).set_index(['org', 'repo', 'number', 'date']).sortlevel()
issues = pd.DataFrame(issues).set_index(['org', 'repo', 'number']).sortlevel()
issue_comments = pd.DataFrame(issue_comments).set_index(['org', 'repo', 'number', 'date']).sortlevel()
prs.to_csv("prs2.csv")
pr_comments.to_csv("pr_comments2.csv")
issues.to_csv("issues2.csv")
issue_comments.to_csv("issue_comments2.csv")
prs = pd.read_csv("prs.csv")
pr_comments = pd.read_csv("pr_comments.csv")
issues = pd.read_csv("issues.csv")
issue_comments = pd.read_csv("issue_comments.csv")
coreteam = set([
"jasongrout",
"willingc",
"jdfreder",
"minrk",
"Carreau",
"SylvainCorlay",
"blink1073",
"bollwyvl",
"ellisonbg",
"fperez",
"jhamrick",
"parente",
"rgbkrk",
"takluyver",
"damianavila",
"Ruv7",
"sccolbert"
])
df = pd.read_csv("jupyter_prs.csv")
df['is_core'] = df['user'].apply(lambda x: x in coreteam)
df['date'] = pd.to_datetime(df['date'])
df.head()
import datetime
cutoff = datetime.datetime(2015, 3, 30, 11, 38, 5, 291165)
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
recent = df.ix[df['date'] > cutoff]
is_core = recent.groupby(['org'])['is_core'].mean()
is_core
counts = recent\
.ix[recent['org'] != 'jupyter-incubator']\
.groupby('is_core')\
.get_group(False)\
.groupby('user')['number']\
.apply(lambda x: len(x))\
.sort_values(inplace=False)[::-1]
(counts == 1).mean()
plt.hist(counts, bins=100)
sns.factorplot('repo', 'is_core', data=df.ix[df['date'] > cutoff], kind='bar', col='org')
```
|
github_jupyter
|
import pandas as pd
import datetime
from github import Github
g = Github("jamieshq", "1ca9cfcf3a0f1922b95c582ff5fe5273d4c2a9a6")
cutoff = datetime.datetime(2015, 3, 30, 11, 38, 5, 291165)
def get_prs(org, repo):
prs = []
print("Getting PRs for {}/{}".format(org.login, repo.name))
for pr in repo.get_pulls(state="all"):
if pr.created_at < cutoff:
continue
prs.append({
'date': pr.created_at,
'user': pr.user.login,
'number': pr.number,
'org': org.login,
'repo': repo.name,
'is_merged': pr.is_merged(),
'state': pr.state
})
return prs
def get_pr_comments(org, repo):
comments = []
print("Getting PR comments for {}/{}".format(org.login, repo.name))
for pr in repo.get_pulls(state="all"):
if pr.created_at < cutoff:
continue
for comment in pr.get_comments():
comments.append({
'date': comment.created_at,
'user': comment.user.login,
'number': pr.number,
'org': org.login,
'repo': repo.name
})
return comments
def get_issues(org, repo):
issues = []
print("Getting issues for {}/{}".format(org.login, repo.name))
for issue in repo.get_issues(state="all"):
if issue.created_at < cutoff:
continue
issues.append({
'date': issue.created_at,
'user': issue.user.login,
'number': issue.number,
'org': org.login,
'repo': repo.name,
'state': issue.state
})
return issues
def get_issue_comments(org, repo):
comments = []
print("Getting issue comments for {}/{}".format(org.login, repo.name))
for issue in repo.get_issues(state="all"):
if issue.created_at < cutoff:
continue
for comment in issue.get_comments():
comments.append({
'issue_date': issue.created_at,
'user': issue.user.login,
'number': issue.number,
'org': org.login,
'repo': repo.name,
'comments': comment.body,
'comment_creation_date' : comment.created_at
})
return comments
#prs = []
#pr_comments = []
issues = []
issue_comments = []
test_orgs = ["jupyter-resources"]
real_orgs = ["jupyterlab", "jupyterhub", "jupyter-widgets", "jupyter-incubator"]
for org_name in real_orgs:
org = g.get_organization(org_name)
for repo in org.get_repos():
#prs.extend(get_prs(org, repo))
#pr_comments.extend(get_pr_comments(org, repo))
#issues.extend(get_issues(org, repo))
issue_comments.extend(get_issue_comments(org, repo))
issue_comments = pd.DataFrame(issue_comments).set_index(['org', 'repo', 'number', 'issue_date','comment_creation_date', 'comments']).sortlevel()
issue_comments.to_csv("issue_comments.csv", encoding="utf-8")
import datetime
datetime.datetime.fromtimestamp(1491196314).strftime('%c')
prs = []
pr_comments = []
issues = []
issue_comments = []
for org_name in ["jupyter", "jupyterlab", "jupyterhub", "jupyter-widgets", "jupyter-incubator"]:
org = g.get_organization(org_name)
for repo in org.get_repos():
prs.extend(get_prs(org, repo))
pr_comments.extend(get_pr_comments(org, repo))
issues.extend(get_issues(org, repo))
issue_comments.extend(get_issue_comments(org, repo))
prs = pd.DataFrame(prs).set_index(['org', 'repo', 'number']).sortlevel()
pr_comments = pd.DataFrame(pr_comments).set_index(['org', 'repo', 'number', 'date']).sortlevel()
issues = pd.DataFrame(issues).set_index(['org', 'repo', 'number']).sortlevel()
issue_comments = pd.DataFrame(issue_comments).set_index(['org', 'repo', 'number', 'date']).sortlevel()
prs.to_csv("prs.csv")
pr_comments.to_csv("pr_comments.csv")
issues.to_csv("issues.csv")
issue_comments.to_csv("issue_comments.csv")
issue_comments.head()
prs = []
pr_comments = []
issues = []
issue_comments = []
for org_name in ["jupyterlab", "jupyterhub", "jupyter-widgets", "jupyter-incubator"]:
org = g.get_organization(org_name)
for repo in org.get_repos():
prs.extend(get_prs(org, repo))
pr_comments.extend(get_pr_comments(org, repo))
issues.extend(get_issues(org, repo))
issue_comments.extend(get_issue_comments(org, repo))
prs = pd.DataFrame(prs).set_index(['org', 'repo', 'number']).sortlevel()
pr_comments = pd.DataFrame(pr_comments).set_index(['org', 'repo', 'number', 'date']).sortlevel()
issues = pd.DataFrame(issues).set_index(['org', 'repo', 'number']).sortlevel()
issue_comments = pd.DataFrame(issue_comments).set_index(['org', 'repo', 'number', 'date']).sortlevel()
prs.to_csv("prs2.csv")
pr_comments.to_csv("pr_comments2.csv")
issues.to_csv("issues2.csv")
issue_comments.to_csv("issue_comments2.csv")
prs = pd.read_csv("prs.csv")
pr_comments = pd.read_csv("pr_comments.csv")
issues = pd.read_csv("issues.csv")
issue_comments = pd.read_csv("issue_comments.csv")
coreteam = set([
"jasongrout",
"willingc",
"jdfreder",
"minrk",
"Carreau",
"SylvainCorlay",
"blink1073",
"bollwyvl",
"ellisonbg",
"fperez",
"jhamrick",
"parente",
"rgbkrk",
"takluyver",
"damianavila",
"Ruv7",
"sccolbert"
])
df = pd.read_csv("jupyter_prs.csv")
df['is_core'] = df['user'].apply(lambda x: x in coreteam)
df['date'] = pd.to_datetime(df['date'])
df.head()
import datetime
cutoff = datetime.datetime(2015, 3, 30, 11, 38, 5, 291165)
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
recent = df.ix[df['date'] > cutoff]
is_core = recent.groupby(['org'])['is_core'].mean()
is_core
counts = recent\
.ix[recent['org'] != 'jupyter-incubator']\
.groupby('is_core')\
.get_group(False)\
.groupby('user')['number']\
.apply(lambda x: len(x))\
.sort_values(inplace=False)[::-1]
(counts == 1).mean()
plt.hist(counts, bins=100)
sns.factorplot('repo', 'is_core', data=df.ix[df['date'] > cutoff], kind='bar', col='org')
| 0.165054 | 0.231533 |
<img src="images/usm.jpg" width="480" height="240" align="left"/>
# MAT281 - Laboratorio N°03
## Objetivos de la clase
* Reforzar los conceptos básicos de pandas.
## Contenidos
* [Problema 01](#p1)
## Problema 01
<img src="https://imagenes.universia.net/gc/net/images/practicas-empleo/p/pr/pro/profesiones-con-el-avance-de-la-tecnologia.jpg" width="480" height="360" align="center"/>
EL conjunto de datos se denomina `ocupation.csv`, el cual contiene información tal como: edad ,sexo, profesión, etc.
Lo primero es cargar el conjunto de datos y ver las primeras filas que lo componen:
```
import pandas as pd
import os
import numpy as np
# cargar datos
df = pd.read_csv("data/ocupation.csv", sep="|").set_index('user_id')
df.head()
```
El objetivo es tratar de obtener la mayor información posible de este conjunto de datos. Para cumplir este objetivo debe resolver las siguientes problemáticas:
1. ¿Cuál es el número de observaciones en el conjunto de datos?
```
print(len(df.index)*(len(df.head())-1))
```
2. ¿Cuál es el número de columnas en el conjunto de datos?
```
print(len(df.index))
```
3. Imprime el nombre de todas las columnas
```
print(df.columns)
```
4. Imprima el índice del dataframe
```
print(df.index)
```
5. ¿Cuál es el tipo de datos de cada columna?
```
df.dtypes
```
6. Resumir el conjunto de datos
```
df.describe()
```
7. Resume conjunto de datos con todas las columnas
```
df.describe(include='all')
```
8. Imprimir solo la columna de **occupation**.
```
print(df['occupation'])
```
9. ¿Cuántas ocupaciones diferentes hay en este conjunto de datos?
```
lista1=[df.occupation[1]]
for index, row in df.iterrows():
n=len(lista1)
k=0
for i in range(0,n):
if df.occupation[index]==lista1[i]:
k=1
if k==0:
lista1.append(df.occupation[index])
print(len(lista1))
```
10. ¿Cuál es la ocupación más frecuente?
```
lista2=[0]*len(lista1)
maximo=0
for index, row in df.iterrows():
lista2[lista1.index(df.occupation[index])]=lista2[lista1.index(df.occupation[index])]+1
for i in range(0,n):
if maximo <= lista2[i]:
maximo=lista2[i]
print(lista1[lista2.index(maximo)])
```
11. ¿Cuál es la edad media de los usuarios?
```
print(np.mean(df['age']))
```
12. ¿Cuál es la edad con menos ocurrencia?
```
lista3=[df.age[1]]
for index, row in df.iterrows():
n=len(lista3)
k=0
for i in range(0,n):
if df.age[index]==lista3[i]:
k=1
if k==0:
lista3.append(df.age[index])
lista4=[0]*len(lista3)
minimo=len(df.index)-1
for index, row in df.iterrows():
lista4[lista3.index(df.age[index])]=lista4[lista3.index(df.age[index])]+1
for i in range(0,n):
if minimo >= lista4[i]:
minimo=lista4[i]
print(lista3[lista4.index(minimo)])
```
|
github_jupyter
|
import pandas as pd
import os
import numpy as np
# cargar datos
df = pd.read_csv("data/ocupation.csv", sep="|").set_index('user_id')
df.head()
print(len(df.index)*(len(df.head())-1))
print(len(df.index))
print(df.columns)
print(df.index)
df.dtypes
df.describe()
df.describe(include='all')
print(df['occupation'])
lista1=[df.occupation[1]]
for index, row in df.iterrows():
n=len(lista1)
k=0
for i in range(0,n):
if df.occupation[index]==lista1[i]:
k=1
if k==0:
lista1.append(df.occupation[index])
print(len(lista1))
lista2=[0]*len(lista1)
maximo=0
for index, row in df.iterrows():
lista2[lista1.index(df.occupation[index])]=lista2[lista1.index(df.occupation[index])]+1
for i in range(0,n):
if maximo <= lista2[i]:
maximo=lista2[i]
print(lista1[lista2.index(maximo)])
print(np.mean(df['age']))
lista3=[df.age[1]]
for index, row in df.iterrows():
n=len(lista3)
k=0
for i in range(0,n):
if df.age[index]==lista3[i]:
k=1
if k==0:
lista3.append(df.age[index])
lista4=[0]*len(lista3)
minimo=len(df.index)-1
for index, row in df.iterrows():
lista4[lista3.index(df.age[index])]=lista4[lista3.index(df.age[index])]+1
for i in range(0,n):
if minimo >= lista4[i]:
minimo=lista4[i]
print(lista3[lista4.index(minimo)])
| 0.041715 | 0.904651 |
```
import numpy as np
from keras import models
import pandas as pd
import pylab
from keras import layers
from keras.callbacks import History
from keras.models import model_from_json
from keras.utils import to_categorical
import matplotlib.pyplot as plt
from keras.datasets import mnist
from keras.models import model_from_json
from keras import optimizers
from keras.callbacks import CSVLogger
(train_images,train_labels),(test_images,test_labels)= mnist.load_data()
#Pre - Processing
train_images=train_images.reshape((60000,28*28))
test_images=test_images.reshape((10000,28*28))
train_images=train_images.astype('float32')/255
test_images=test_images.astype('float32')/255
#Labels
train_labels=to_categorical(train_labels)
test_labels=to_categorical(test_labels)
adam1 = optimizers.Adam(lr = 0.0001)
adam2 = optimizers.Adam(lr = 0.0005)
adam3 = optimizers.Adam(lr = 0.001)
adam4 = optimizers.Adam(lr = 0.005)
network1=models.Sequential()
network1.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
#network1.add(layers.Dense(512, activation = 'relu'))
#network1.add(layers.Dense(256, activation = 'relu'))
network1.add(layers.Dense(10,activation='softmax'))
network1.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network1.summary()
csv_logger1 = CSVLogger('training1.csv')
h1=network1.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger1])
network2=models.Sequential()
network2.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network2.add(layers.Dense(512, activation = 'relu'))
#network1.add(layers.Dense(256, activation = 'relu'))
network2.add(layers.Dense(10,activation='softmax'))
network2.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network2.summary()
csv_logger2 = CSVLogger('training2.csv')
h2=network2.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger2])
network3=models.Sequential()
network3.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network3.add(layers.Dense(512, activation = 'relu'))
network3.add(layers.Dense(256, activation = 'relu'))
network3.add(layers.Dense(10,activation='softmax'))
network3.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network3.summary()
csv_logger3 = CSVLogger('training3.csv')
h3=network3.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger3])
network4=models.Sequential()
network4.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network4.add(layers.Dense(512, activation = 'relu'))
network4.add(layers.Dense(256, activation = 'relu'))
network4.add(layers.Dense(128, activation = 'relu'))
network4.add(layers.Dense(10,activation='softmax'))
network4.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network4.summary()
csv_logger4 = CSVLogger('training4.csv')
h4=network4.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger4])
network5=models.Sequential()
network5.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network5.add(layers.Dense(512, activation = 'relu'))
network5.add(layers.Dense(256, activation = 'relu'))
network5.add(layers.Dense(128, activation = 'relu'))
network5.add(layers.Dense(64, activation = 'relu'))
network5.add(layers.Dense(10,activation='softmax'))
network5.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network5.summary()
csv_logger5 = CSVLogger('training5.csv')
h5=network5.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger5])
network6=models.Sequential()
network6.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network6.add(layers.Dense(512, activation = 'relu'))
network6.add(layers.Dense(256, activation = 'relu'))
network6.add(layers.Dense(128, activation = 'relu'))
network6.add(layers.Dense(64, activation = 'relu'))
network6.add(layers.Dense(32, activation = 'relu'))
network6.add(layers.Dense(10,activation='softmax'))
network6.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network6.summary()
csv_logger6 = CSVLogger('training6.csv')
h6=network6.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger6])
no_layer = pd.read_csv('training1.csv')
one_layer = pd.read_csv('training2.csv')
two_layer = pd.read_csv('training3.csv')
three_layer = pd.read_csv('training4.csv')
four_layer = pd.read_csv('training5.csv')
five_layer = pd.read_csv('training6.csv')
pylab.plot(no_layer['epoch'],no_layer['val_acc'],label = 'No_hidden')
pylab.plot(one_layer['epoch'], one_layer['val_acc'],label = 'One_hidden')
pylab.plot(two_layer['epoch'], two_layer['val_acc'],label = 'Two_hidden')
pylab.plot(three_layer['epoch'],three_layer['val_acc'],label = 'Three_hidden')
pylab.plot(four_layer['epoch'], four_layer['val_acc'],label = 'four_hidden')
pylab.plot(five_layer['epoch'],five_layer['val_acc'],label = 'five_hidden')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_number_of_hidden_layers')
pylab.savefig("Accuracy_with_different_number_of_hidden_layers")
pylab.show()
```
```
network7=models.Sequential()
network7.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network7.add(layers.Dense(512, activation = 'relu'))
network7.add(layers.Dense(10,activation='softmax'))
network7.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network7.summary()
csv_logger7 = CSVLogger('training7.csv')
h7=network7.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger7])
network8=models.Sequential()
network8.add(layers.Dense(784,activation='sigmoid',input_shape=(28*28,)))
network8.add(layers.Dense(512, activation = 'sigmoid'))
network8.add(layers.Dense(10,activation='softmax'))
network8.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network8.summary()
csv_logger8 = CSVLogger('training8.csv')
h8=network8.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger8])
network9=models.Sequential()
network9.add(layers.Dense(784,activation='tanh',input_shape=(28*28,)))
network9.add(layers.Dense(512, activation = 'tanh'))
network9.add(layers.Dense(10,activation='softmax'))
network9.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network9.summary()
csv_logger9 = CSVLogger('training9.csv')
h9=network9.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger9])
# read all csvs and compare activations
relu_act = pd.read_csv('training7.csv')
sigmoid_act = pd.read_csv('training8.csv')
tanh_act = pd.read_csv('training9.csv')
pylab.plot(relu_act['epoch'],relu_act['val_acc'],label = 'ReLU')
pylab.plot(sigmoid_act['epoch'], sigmoid_act['val_acc'],label = 'sigmoid')
pylab.plot(tanh_act['epoch'], tanh_act['val_acc'],label = 'tanh')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_activations')
pylab.savefig("Accuracy_with_different_activations")
pylab.show()
network10=models.Sequential()
network10.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network10.add(layers.Dense(512, activation = 'relu'))
network10.add(layers.Dense(10,activation='softmax'))
network10.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network10.summary()
csv_logger10 = CSVLogger('training10.csv')
h10=network10.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger10])
network11=models.Sequential()
network11.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network11.add(layers.Dense(512, activation = 'relu'))
network11.add(layers.Dense(10,activation='softmax'))
network11.compile(optimizer='adam',loss='mean_squared_error',metrics=['accuracy'])
network11.summary()
csv_logger11 = CSVLogger('training11.csv')
h11=network11.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger11])
network12=models.Sequential()
network12.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network12.add(layers.Dense(512, activation = 'relu'))
network12.add(layers.Dense(10,activation='softmax'))
network12.compile(optimizer='adagrad',loss='mean_squared_error',metrics=['accuracy'])
network12.summary()
csv_logger12 = CSVLogger('training12.csv')
h12=network12.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger12])
#read csvs and compare optimizers
sgd_opt = pd.read_csv('training10.csv')
adam_opt = pd.read_csv('training11.csv')
adagrad_opt = pd.read_csv('training12.csv')
pylab.plot(sgd_opt['epoch'],sgd_opt['val_acc'],label = 'SGD')
pylab.plot(adam_opt['epoch'], adam_opt['val_acc'],label = 'adam')
pylab.plot(adagrad_opt['epoch'], adagrad_opt['val_acc'],label = 'adagrad')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_optimizers')
pylab.savefig("Accuracy_with_different_optimizers")
pylab.show()
network13=models.Sequential()
network13.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network13.add(layers.Dense(512, activation = 'relu'))
network13.add(layers.Dense(10,activation='softmax'))
network13.compile(optimizer=adam1,loss='mean_squared_error',metrics=['accuracy'])
network13.summary()
csv_logger13 = CSVLogger('training13.csv')
h10=network13.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger13])
network14=models.Sequential()
network14.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network14.add(layers.Dense(512, activation = 'relu'))
network14.add(layers.Dense(10,activation='softmax'))
network14.compile(optimizer=adam2,loss='mean_squared_error',metrics=['accuracy'])
network14.summary()
csv_logger14 = CSVLogger('training14.csv')
h14=network14.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger14])
network15=models.Sequential()
network15.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network15.add(layers.Dense(512, activation = 'relu'))
network15.add(layers.Dense(10,activation='softmax'))
network15.compile(optimizer=adam3,loss='mean_squared_error',metrics=['accuracy'])
network15.summary()
csv_logger15 = CSVLogger('training15.csv')
h15=network15.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger15])
network16=models.Sequential()
network16.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network16.add(layers.Dense(512, activation = 'relu'))
network16.add(layers.Dense(10,activation='softmax'))
network16.compile(optimizer=adam4,loss='mean_squared_error',metrics=['accuracy'])
network16.summary()
csv_logger16 = CSVLogger('training16.csv')
h16=network16.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger16])
#read csv's and compare learning rates
adam1_lr = pd.read_csv('training13.csv')
adam2_lr = pd.read_csv('training14.csv')
adam3_lr = pd.read_csv('training15.csv')
adam4_lr = pd.read_csv('training16.csv')
pylab.plot(adam1_lr['epoch'],adam1_lr['val_acc'],label = '0.0001')
pylab.plot(adam2_lr['epoch'], adam2_lr['val_acc'],label = '0.005')
pylab.plot(adam3_lr['epoch'], adam3_lr['val_acc'],label = '0.001')
pylab.plot(adam4_lr['epoch'], adam4_lr['val_acc'],label = '0.005')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_learning_rates')
pylab.savefig("Accuracy_with_different_learning_rates")
pylab.show()
network17=models.Sequential()
network17.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network17.add(layers.Dense(512, activation = 'relu'))
network17.add(layers.Dense(10,activation='softmax'))
network17.compile(optimizer='adam',loss='mean_squared_error',metrics=['accuracy'])
network17.summary()
csv_logger17 = CSVLogger('training17.csv')
h17=network17.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger17])
network18=models.Sequential()
network18.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network18.add(layers.Dense(512, activation = 'relu'))
network18.add(layers.Dense(10,activation='softmax'))
network18.compile(optimizer='adam',loss='categorical_crossentropy',metrics=['accuracy'])
network18.summary()
csv_logger18 = CSVLogger('training18.csv')
h18=network18.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger18])
network19=models.Sequential()
network19.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network19.add(layers.Dense(512, activation = 'relu'))
network19.add(layers.Dense(10,activation='softmax'))
network19.compile(optimizer='adam',loss='categorical_hinge',metrics=['accuracy'])
network19.summary()
csv_logger19 = CSVLogger('training19.csv')
h19=network19.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger19])
#read csv and graph for loss
mse = pd.read_csv('training17.csv')
cat_cr = pd.read_csv('training18.csv')
cat_hinge = pd.read_csv('training19.csv')
pylab.plot(mse['epoch'],mse['val_acc'],label = 'MSE')
pylab.plot(cat_cr['epoch'], cat_cr['val_acc'],label = 'categorical_cross_entropy')
pylab.plot(cat_hinge['epoch'], cat_hinge['val_acc'],label = 'categorical_hinge')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_losses')
pylab.savefig("Accuracy_with_different_losses")
pylab.show()
network20=models.Sequential()
network20.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network20.add(layers.Dense(128, activation = 'relu'))
network20.add(layers.Dense(10,activation='softmax'))
network20.compile(optimizer='adam',loss='categorical_crossentropy',metrics=['accuracy'])
network20.summary()
csv_logger20 = CSVLogger('training20.csv')
h20=network20.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger20])
network21=models.Sequential()
network21.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network21.add(layers.Dense(784, activation = 'relu'))
network21.add(layers.Dense(10,activation='softmax'))
network21.compile(optimizer='adam',loss='categorical_crossentropy',metrics=['accuracy'])
network21.summary()
csv_logger21 = CSVLogger('training21.csv')
h21=network21.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger21])
network22=models.Sequential()
network22.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network22.add(layers.Dense(1024, activation = 'relu'))
network22.add(layers.Dense(10,activation='softmax'))
network22.compile(optimizer='adam',loss='categorical_crossentropy',metrics=['accuracy'])
network22.summary()
csv_logger22 = CSVLogger('training22.csv')
h22=network22.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger22])
no_128 = pd.read_csv('training20.csv')
no_784 = pd.read_csv('training21.csv')
no_1024 = pd.read_csv('training22.csv')
pylab.plot(no_128['epoch'],no_128['val_acc'],label = '128_nuerons')
pylab.plot(no_784['epoch'], no_784['val_acc'],label = '784_nuerons')
pylab.plot(no_1024['epoch'], no_1024['val_acc'],label = '1024_nuerons')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_nuerons')
pylab.savefig("Accuracy_with_different_nuerons")
pylab.show()
```
|
github_jupyter
|
import numpy as np
from keras import models
import pandas as pd
import pylab
from keras import layers
from keras.callbacks import History
from keras.models import model_from_json
from keras.utils import to_categorical
import matplotlib.pyplot as plt
from keras.datasets import mnist
from keras.models import model_from_json
from keras import optimizers
from keras.callbacks import CSVLogger
(train_images,train_labels),(test_images,test_labels)= mnist.load_data()
#Pre - Processing
train_images=train_images.reshape((60000,28*28))
test_images=test_images.reshape((10000,28*28))
train_images=train_images.astype('float32')/255
test_images=test_images.astype('float32')/255
#Labels
train_labels=to_categorical(train_labels)
test_labels=to_categorical(test_labels)
adam1 = optimizers.Adam(lr = 0.0001)
adam2 = optimizers.Adam(lr = 0.0005)
adam3 = optimizers.Adam(lr = 0.001)
adam4 = optimizers.Adam(lr = 0.005)
network1=models.Sequential()
network1.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
#network1.add(layers.Dense(512, activation = 'relu'))
#network1.add(layers.Dense(256, activation = 'relu'))
network1.add(layers.Dense(10,activation='softmax'))
network1.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network1.summary()
csv_logger1 = CSVLogger('training1.csv')
h1=network1.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger1])
network2=models.Sequential()
network2.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network2.add(layers.Dense(512, activation = 'relu'))
#network1.add(layers.Dense(256, activation = 'relu'))
network2.add(layers.Dense(10,activation='softmax'))
network2.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network2.summary()
csv_logger2 = CSVLogger('training2.csv')
h2=network2.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger2])
network3=models.Sequential()
network3.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network3.add(layers.Dense(512, activation = 'relu'))
network3.add(layers.Dense(256, activation = 'relu'))
network3.add(layers.Dense(10,activation='softmax'))
network3.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network3.summary()
csv_logger3 = CSVLogger('training3.csv')
h3=network3.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger3])
network4=models.Sequential()
network4.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network4.add(layers.Dense(512, activation = 'relu'))
network4.add(layers.Dense(256, activation = 'relu'))
network4.add(layers.Dense(128, activation = 'relu'))
network4.add(layers.Dense(10,activation='softmax'))
network4.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network4.summary()
csv_logger4 = CSVLogger('training4.csv')
h4=network4.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger4])
network5=models.Sequential()
network5.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network5.add(layers.Dense(512, activation = 'relu'))
network5.add(layers.Dense(256, activation = 'relu'))
network5.add(layers.Dense(128, activation = 'relu'))
network5.add(layers.Dense(64, activation = 'relu'))
network5.add(layers.Dense(10,activation='softmax'))
network5.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network5.summary()
csv_logger5 = CSVLogger('training5.csv')
h5=network5.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger5])
network6=models.Sequential()
network6.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network6.add(layers.Dense(512, activation = 'relu'))
network6.add(layers.Dense(256, activation = 'relu'))
network6.add(layers.Dense(128, activation = 'relu'))
network6.add(layers.Dense(64, activation = 'relu'))
network6.add(layers.Dense(32, activation = 'relu'))
network6.add(layers.Dense(10,activation='softmax'))
network6.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network6.summary()
csv_logger6 = CSVLogger('training6.csv')
h6=network6.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger6])
no_layer = pd.read_csv('training1.csv')
one_layer = pd.read_csv('training2.csv')
two_layer = pd.read_csv('training3.csv')
three_layer = pd.read_csv('training4.csv')
four_layer = pd.read_csv('training5.csv')
five_layer = pd.read_csv('training6.csv')
pylab.plot(no_layer['epoch'],no_layer['val_acc'],label = 'No_hidden')
pylab.plot(one_layer['epoch'], one_layer['val_acc'],label = 'One_hidden')
pylab.plot(two_layer['epoch'], two_layer['val_acc'],label = 'Two_hidden')
pylab.plot(three_layer['epoch'],three_layer['val_acc'],label = 'Three_hidden')
pylab.plot(four_layer['epoch'], four_layer['val_acc'],label = 'four_hidden')
pylab.plot(five_layer['epoch'],five_layer['val_acc'],label = 'five_hidden')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_number_of_hidden_layers')
pylab.savefig("Accuracy_with_different_number_of_hidden_layers")
pylab.show()
network7=models.Sequential()
network7.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network7.add(layers.Dense(512, activation = 'relu'))
network7.add(layers.Dense(10,activation='softmax'))
network7.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network7.summary()
csv_logger7 = CSVLogger('training7.csv')
h7=network7.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger7])
network8=models.Sequential()
network8.add(layers.Dense(784,activation='sigmoid',input_shape=(28*28,)))
network8.add(layers.Dense(512, activation = 'sigmoid'))
network8.add(layers.Dense(10,activation='softmax'))
network8.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network8.summary()
csv_logger8 = CSVLogger('training8.csv')
h8=network8.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger8])
network9=models.Sequential()
network9.add(layers.Dense(784,activation='tanh',input_shape=(28*28,)))
network9.add(layers.Dense(512, activation = 'tanh'))
network9.add(layers.Dense(10,activation='softmax'))
network9.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network9.summary()
csv_logger9 = CSVLogger('training9.csv')
h9=network9.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger9])
# read all csvs and compare activations
relu_act = pd.read_csv('training7.csv')
sigmoid_act = pd.read_csv('training8.csv')
tanh_act = pd.read_csv('training9.csv')
pylab.plot(relu_act['epoch'],relu_act['val_acc'],label = 'ReLU')
pylab.plot(sigmoid_act['epoch'], sigmoid_act['val_acc'],label = 'sigmoid')
pylab.plot(tanh_act['epoch'], tanh_act['val_acc'],label = 'tanh')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_activations')
pylab.savefig("Accuracy_with_different_activations")
pylab.show()
network10=models.Sequential()
network10.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network10.add(layers.Dense(512, activation = 'relu'))
network10.add(layers.Dense(10,activation='softmax'))
network10.compile(optimizer='SGD',loss='mean_squared_error',metrics=['accuracy'])
network10.summary()
csv_logger10 = CSVLogger('training10.csv')
h10=network10.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger10])
network11=models.Sequential()
network11.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network11.add(layers.Dense(512, activation = 'relu'))
network11.add(layers.Dense(10,activation='softmax'))
network11.compile(optimizer='adam',loss='mean_squared_error',metrics=['accuracy'])
network11.summary()
csv_logger11 = CSVLogger('training11.csv')
h11=network11.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger11])
network12=models.Sequential()
network12.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network12.add(layers.Dense(512, activation = 'relu'))
network12.add(layers.Dense(10,activation='softmax'))
network12.compile(optimizer='adagrad',loss='mean_squared_error',metrics=['accuracy'])
network12.summary()
csv_logger12 = CSVLogger('training12.csv')
h12=network12.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger12])
#read csvs and compare optimizers
sgd_opt = pd.read_csv('training10.csv')
adam_opt = pd.read_csv('training11.csv')
adagrad_opt = pd.read_csv('training12.csv')
pylab.plot(sgd_opt['epoch'],sgd_opt['val_acc'],label = 'SGD')
pylab.plot(adam_opt['epoch'], adam_opt['val_acc'],label = 'adam')
pylab.plot(adagrad_opt['epoch'], adagrad_opt['val_acc'],label = 'adagrad')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_optimizers')
pylab.savefig("Accuracy_with_different_optimizers")
pylab.show()
network13=models.Sequential()
network13.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network13.add(layers.Dense(512, activation = 'relu'))
network13.add(layers.Dense(10,activation='softmax'))
network13.compile(optimizer=adam1,loss='mean_squared_error',metrics=['accuracy'])
network13.summary()
csv_logger13 = CSVLogger('training13.csv')
h10=network13.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger13])
network14=models.Sequential()
network14.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network14.add(layers.Dense(512, activation = 'relu'))
network14.add(layers.Dense(10,activation='softmax'))
network14.compile(optimizer=adam2,loss='mean_squared_error',metrics=['accuracy'])
network14.summary()
csv_logger14 = CSVLogger('training14.csv')
h14=network14.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger14])
network15=models.Sequential()
network15.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network15.add(layers.Dense(512, activation = 'relu'))
network15.add(layers.Dense(10,activation='softmax'))
network15.compile(optimizer=adam3,loss='mean_squared_error',metrics=['accuracy'])
network15.summary()
csv_logger15 = CSVLogger('training15.csv')
h15=network15.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger15])
network16=models.Sequential()
network16.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network16.add(layers.Dense(512, activation = 'relu'))
network16.add(layers.Dense(10,activation='softmax'))
network16.compile(optimizer=adam4,loss='mean_squared_error',metrics=['accuracy'])
network16.summary()
csv_logger16 = CSVLogger('training16.csv')
h16=network16.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger16])
#read csv's and compare learning rates
adam1_lr = pd.read_csv('training13.csv')
adam2_lr = pd.read_csv('training14.csv')
adam3_lr = pd.read_csv('training15.csv')
adam4_lr = pd.read_csv('training16.csv')
pylab.plot(adam1_lr['epoch'],adam1_lr['val_acc'],label = '0.0001')
pylab.plot(adam2_lr['epoch'], adam2_lr['val_acc'],label = '0.005')
pylab.plot(adam3_lr['epoch'], adam3_lr['val_acc'],label = '0.001')
pylab.plot(adam4_lr['epoch'], adam4_lr['val_acc'],label = '0.005')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_learning_rates')
pylab.savefig("Accuracy_with_different_learning_rates")
pylab.show()
network17=models.Sequential()
network17.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network17.add(layers.Dense(512, activation = 'relu'))
network17.add(layers.Dense(10,activation='softmax'))
network17.compile(optimizer='adam',loss='mean_squared_error',metrics=['accuracy'])
network17.summary()
csv_logger17 = CSVLogger('training17.csv')
h17=network17.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger17])
network18=models.Sequential()
network18.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network18.add(layers.Dense(512, activation = 'relu'))
network18.add(layers.Dense(10,activation='softmax'))
network18.compile(optimizer='adam',loss='categorical_crossentropy',metrics=['accuracy'])
network18.summary()
csv_logger18 = CSVLogger('training18.csv')
h18=network18.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger18])
network19=models.Sequential()
network19.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network19.add(layers.Dense(512, activation = 'relu'))
network19.add(layers.Dense(10,activation='softmax'))
network19.compile(optimizer='adam',loss='categorical_hinge',metrics=['accuracy'])
network19.summary()
csv_logger19 = CSVLogger('training19.csv')
h19=network19.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger19])
#read csv and graph for loss
mse = pd.read_csv('training17.csv')
cat_cr = pd.read_csv('training18.csv')
cat_hinge = pd.read_csv('training19.csv')
pylab.plot(mse['epoch'],mse['val_acc'],label = 'MSE')
pylab.plot(cat_cr['epoch'], cat_cr['val_acc'],label = 'categorical_cross_entropy')
pylab.plot(cat_hinge['epoch'], cat_hinge['val_acc'],label = 'categorical_hinge')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_losses')
pylab.savefig("Accuracy_with_different_losses")
pylab.show()
network20=models.Sequential()
network20.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network20.add(layers.Dense(128, activation = 'relu'))
network20.add(layers.Dense(10,activation='softmax'))
network20.compile(optimizer='adam',loss='categorical_crossentropy',metrics=['accuracy'])
network20.summary()
csv_logger20 = CSVLogger('training20.csv')
h20=network20.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger20])
network21=models.Sequential()
network21.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network21.add(layers.Dense(784, activation = 'relu'))
network21.add(layers.Dense(10,activation='softmax'))
network21.compile(optimizer='adam',loss='categorical_crossentropy',metrics=['accuracy'])
network21.summary()
csv_logger21 = CSVLogger('training21.csv')
h21=network21.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger21])
network22=models.Sequential()
network22.add(layers.Dense(784,activation='relu',input_shape=(28*28,)))
network22.add(layers.Dense(1024, activation = 'relu'))
network22.add(layers.Dense(10,activation='softmax'))
network22.compile(optimizer='adam',loss='categorical_crossentropy',metrics=['accuracy'])
network22.summary()
csv_logger22 = CSVLogger('training22.csv')
h22=network22.fit(train_images,train_labels, verbose = True, validation_data=(test_images,test_labels),epochs=10,batch_size=32, callbacks=[csv_logger22])
no_128 = pd.read_csv('training20.csv')
no_784 = pd.read_csv('training21.csv')
no_1024 = pd.read_csv('training22.csv')
pylab.plot(no_128['epoch'],no_128['val_acc'],label = '128_nuerons')
pylab.plot(no_784['epoch'], no_784['val_acc'],label = '784_nuerons')
pylab.plot(no_1024['epoch'], no_1024['val_acc'],label = '1024_nuerons')
figure = pylab.legend(loc = 'upper right')
pylab.xlabel("No of Epochs")
pylab.ylabel("Validation Accuracy")
plt.title('Accuracy_with_different_nuerons')
pylab.savefig("Accuracy_with_different_nuerons")
pylab.show()
| 0.860222 | 0.437944 |
```
%matplotlib inline
```
Deploying PyTorch in Python via a REST API with Flask
========================================================
**Author**: `Avinash Sajjanshetty <https://avi.im>`_
In this tutorial, we will deploy a PyTorch model using Flask and expose a
REST API for model inference. In particular, we will deploy a pretrained
DenseNet 121 model which detects the image.
.. tip:: All the code used here is released under MIT license and is available on `Github <https://github.com/avinassh/pytorch-flask-api>`_.
This represents the first in a series of tutorials on deploying PyTorch models
in production. Using Flask in this way is by far the easiest way to start
serving your PyTorch models, but it will not work for a use case
with high performance requirements. For that:
- If you're already familiar with TorchScript, you can jump straight into our
`Loading a TorchScript Model in C++ <https://pytorch.org/tutorials/advanced/cpp_export.html>`_ tutorial.
- If you first need a refresher on TorchScript, check out our
`Intro a TorchScript <https://pytorch.org/tutorials/beginner/Intro_to_TorchScript_tutorial.html>`_ tutorial.
API Definition
--------------
We will first define our API endpoints, the request and response types. Our
API endpoint will be at ``/predict`` which takes HTTP POST requests with a
``file`` parameter which contains the image. The response will be of JSON
response containing the prediction:
::
{"class_id": "n02124075", "class_name": "Egyptian_cat"}
Dependencies
------------
Install the required dependenices by running the following command:
::
$ pip install Flask==1.0.3 torchvision-0.3.0
Simple Web Server
-----------------
Following is a simple webserver, taken from Flask's documentaion
```
from flask import Flask
app = Flask(__name__)
@app.route('/')
def hello():
return 'Hello World!'
```
Save the above snippet in a file called ``app.py`` and you can now run a
Flask development server by typing:
::
$ FLASK_ENV=development FLASK_APP=app.py flask run
When you visit ``http://localhost:5000/`` in your web browser, you will be
greeted with ``Hello World!`` text
We will make slight changes to the above snippet, so that it suits our API
definition. First, we will rename the method to ``predict``. We will update
the endpoint path to ``/predict``. Since the image files will be sent via
HTTP POST requests, we will update it so that it also accepts only POST
requests:
```
@app.route('/predict', methods=['POST'])
def predict():
return 'Hello World!'
```
We will also change the response type, so that it returns a JSON response
containing ImageNet class id and name. The updated ``app.py`` file will
be now:
```
from flask import Flask, jsonify
app = Flask(__name__)
@app.route('/predict', methods=['POST'])
def predict():
return jsonify({'class_id': 'IMAGE_NET_XXX', 'class_name': 'Cat'})
```
Inference
-----------------
In the next sections we will focus on writing the inference code. This will
involve two parts, one where we prepare the image so that it can be fed
to DenseNet and next, we will write the code to get the actual prediction
from the model.
Preparing the image
~~~~~~~~~~~~~~~~~~~
DenseNet model requires the image to be of 3 channel RGB image of size
224 x 224. We will also normalise the image tensor with the required mean
and standard deviation values. You can read more about it
`here <https://pytorch.org/docs/stable/torchvision/models.html>`_.
We will use ``transforms`` from ``torchvision`` library and build a
transform pipeline, which transforms our images as required. You
can read more about transforms `here <https://pytorch.org/docs/stable/torchvision/transforms.html>`_.
```
import io
import torchvision.transforms as transforms
from PIL import Image
def transform_image(image_bytes):
my_transforms = transforms.Compose([transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(
[0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
image = Image.open(io.BytesIO(image_bytes))
return my_transforms(image).unsqueeze(0)
```
The above method takes image data in bytes, applies the series of transforms
and returns a tensor. To test the above method, read an image file in
bytes mode (first replacing `../_static/img/sample_file.jpeg` with the actual
path to the file on your computer) and see if you get a tensor back:
```
with open("../_static/img/sample_file.jpeg", 'rb') as f:
image_bytes = f.read()
tensor = transform_image(image_bytes=image_bytes)
print(tensor)
```
Prediction
~~~~~~~~~~~~~~~~~~~
Now will use a pretrained DenseNet 121 model to predict the image class. We
will use one from ``torchvision`` library, load the model and get an
inference. While we'll be using a pretrained model in this example, you can
use this same approach for your own models. See more about loading your
models in this :doc:`tutorial </beginner/saving_loading_models>`.
```
from torchvision import models
# Make sure to pass `pretrained` as `True` to use the pretrained weights:
model = models.densenet121(pretrained=True)
# Since we are using our model only for inference, switch to `eval` mode:
model.eval()
def get_prediction(image_bytes):
tensor = transform_image(image_bytes=image_bytes)
outputs = model.forward(tensor)
_, y_hat = outputs.max(1)
return y_hat
```
The tensor ``y_hat`` will contain the index of the predicted class id.
However, we need a human readable class name. For that we need a class id
to name mapping. Download
`this file <https://s3.amazonaws.com/deep-learning-models/image-models/imagenet_class_index.json>`_
as ``imagenet_class_index.json`` and remember where you saved it (or, if you
are following the exact steps in this tutorial, save it in
`tutorials/_static`). This file contains the mapping of ImageNet class id to
ImageNet class name. We will load this JSON file and get the class name of
the predicted index.
```
import json
imagenet_class_index = json.load(open('../_static/imagenet_class_index.json'))
def get_prediction(image_bytes):
tensor = transform_image(image_bytes=image_bytes)
outputs = model.forward(tensor)
_, y_hat = outputs.max(1)
predicted_idx = str(y_hat.item())
return imagenet_class_index[predicted_idx]
```
Before using ``imagenet_class_index`` dictionary, first we will convert
tensor value to a string value, since the keys in the
``imagenet_class_index`` dictionary are strings.
We will test our above method:
```
with open("../_static/img/sample_file.jpeg", 'rb') as f:
image_bytes = f.read()
print(get_prediction(image_bytes=image_bytes))
```
You should get a response like this:
```
['n02124075', 'Egyptian_cat']
```
The first item in array is ImageNet class id and second item is the human
readable name.
.. Note ::
Did you notice that ``model`` variable is not part of ``get_prediction``
method? Or why is model a global variable? Loading a model can be an
expensive operation in terms of memory and compute. If we loaded the model in the
``get_prediction`` method, then it would get unnecessarily loaded every
time the method is called. Since, we are building a web server, there
could be thousands of requests per second, we should not waste time
redundantly loading the model for every inference. So, we keep the model
loaded in memory just once. In
production systems, it's necessary to be efficient about your use of
compute to be able to serve requests at scale, so you should generally
load your model before serving requests.
Integrating the model in our API Server
---------------------------------------
In this final part we will add our model to our Flask API server. Since
our API server is supposed to take an image file, we will update our ``predict``
method to read files from the requests:
.. code-block:: python
from flask import request
@app.route('/predict', methods=['POST'])
def predict():
if request.method == 'POST':
# we will get the file from the request
file = request.files['file']
# convert that to bytes
img_bytes = file.read()
class_id, class_name = get_prediction(image_bytes=img_bytes)
return jsonify({'class_id': class_id, 'class_name': class_name})
The ``app.py`` file is now complete. Following is the full version; replace
the paths with the paths where you saved your files and it should run:
.. code-block:: python
import io
import json
from torchvision import models
import torchvision.transforms as transforms
from PIL import Image
from flask import Flask, jsonify, request
app = Flask(__name__)
imagenet_class_index = json.load(open('<PATH/TO/.json/FILE>/imagenet_class_index.json'))
model = models.densenet121(pretrained=True)
model.eval()
def transform_image(image_bytes):
my_transforms = transforms.Compose([transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(
[0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
image = Image.open(io.BytesIO(image_bytes))
return my_transforms(image).unsqueeze(0)
def get_prediction(image_bytes):
tensor = transform_image(image_bytes=image_bytes)
outputs = model.forward(tensor)
_, y_hat = outputs.max(1)
predicted_idx = str(y_hat.item())
return imagenet_class_index[predicted_idx]
@app.route('/predict', methods=['POST'])
def predict():
if request.method == 'POST':
file = request.files['file']
img_bytes = file.read()
class_id, class_name = get_prediction(image_bytes=img_bytes)
return jsonify({'class_id': class_id, 'class_name': class_name})
if __name__ == '__main__':
app.run()
Let's test our web server! Run:
::
$ FLASK_ENV=development FLASK_APP=app.py flask run
We can use the
`requests <https://pypi.org/project/requests/>`_
library to send a POST request to our app:
.. code-block:: python
import requests
resp = requests.post("http://localhost:5000/predict",
files={"file": open('<PATH/TO/.jpg/FILE>/cat.jpg','rb')})
Printing `resp.json()` will now show the following:
::
{"class_id": "n02124075", "class_name": "Egyptian_cat"}
Next steps
--------------
The server we wrote is quite trivial and and may not do everything
you need for your production application. So, here are some things you
can do to make it better:
- The endpoint ``/predict`` assumes that always there will be a image file
in the request. This may not hold true for all requests. Our user may
send image with a different parameter or send no images at all.
- The user may send non-image type files too. Since we are not handling
errors, this will break our server. Adding an explicit error handing
path that will throw an exception would allow us to better handle
the bad inputs
- Even though the model can recognize a large number of classes of images,
it may not be able to recognize all images. Enhance the implementation
to handle cases when the model does not recognize anything in the image.
- We run the Flask server in the development mode, which is not suitable for
deploying in production. You can check out `this tutorial <https://flask.palletsprojects.com/en/1.1.x/tutorial/deploy/>`_
for deploying a Flask server in production.
- You can also add a UI by creating a page with a form which takes the image and
displays the prediction. Check out the `demo <https://pytorch-imagenet.herokuapp.com/>`_
of a similar project and its `source code <https://github.com/avinassh/pytorch-flask-api-heroku>`_.
- In this tutorial, we only showed how to build a service that could return predictions for
a single image at a time. We could modify our service to be able to return predictions for
multiple images at once. In addition, the `service-streamer <https://github.com/ShannonAI/service-streamer>`_
library automatically queues requests to your service and samples them into mini-batches
that can be fed into your model. You can check out `this tutorial <https://github.com/ShannonAI/service-streamer/wiki/Vision-Recognition-Service-with-Flask-and-service-streamer>`_.
- Finally, we encourage you to check out our other tutorials on deploying PyTorch models
linked-to at the top of the page.
|
github_jupyter
|
%matplotlib inline
from flask import Flask
app = Flask(__name__)
@app.route('/')
def hello():
return 'Hello World!'
@app.route('/predict', methods=['POST'])
def predict():
return 'Hello World!'
from flask import Flask, jsonify
app = Flask(__name__)
@app.route('/predict', methods=['POST'])
def predict():
return jsonify({'class_id': 'IMAGE_NET_XXX', 'class_name': 'Cat'})
import io
import torchvision.transforms as transforms
from PIL import Image
def transform_image(image_bytes):
my_transforms = transforms.Compose([transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(
[0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
image = Image.open(io.BytesIO(image_bytes))
return my_transforms(image).unsqueeze(0)
with open("../_static/img/sample_file.jpeg", 'rb') as f:
image_bytes = f.read()
tensor = transform_image(image_bytes=image_bytes)
print(tensor)
from torchvision import models
# Make sure to pass `pretrained` as `True` to use the pretrained weights:
model = models.densenet121(pretrained=True)
# Since we are using our model only for inference, switch to `eval` mode:
model.eval()
def get_prediction(image_bytes):
tensor = transform_image(image_bytes=image_bytes)
outputs = model.forward(tensor)
_, y_hat = outputs.max(1)
return y_hat
import json
imagenet_class_index = json.load(open('../_static/imagenet_class_index.json'))
def get_prediction(image_bytes):
tensor = transform_image(image_bytes=image_bytes)
outputs = model.forward(tensor)
_, y_hat = outputs.max(1)
predicted_idx = str(y_hat.item())
return imagenet_class_index[predicted_idx]
with open("../_static/img/sample_file.jpeg", 'rb') as f:
image_bytes = f.read()
print(get_prediction(image_bytes=image_bytes))
['n02124075', 'Egyptian_cat']
| 0.680454 | 0.972675 |
```
%matplotlib inline
```
500 hPa Geopotential Heights and Winds
======================================
Classic 500-hPa plot using NAM analysis file.
This example uses example data from the NAM anlysis for 12 UTC 31
October 2016 and uses xarray as the main read source with using Cartopy
for plotting a CONUS view of the 500-hPa geopotential heights, wind
speed, and wind barbs.
Import the needed modules.
```
from datetime import datetime
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
import metpy.calc as mpcalc
from metpy.units import units
import numpy as np
from scipy.ndimage import gaussian_filter
import xarray as xr
```
The following code reads the example data using the xarray open_dataset
function and prints the coordinate values that are associated with the
various variables contained within the file.
```
ds = xr.open_dataset('https://thredds.ucar.edu/thredds/dodsC/casestudies/'
'python-gallery/NAM_20161031_1200.nc')
ds.coords
```
Data Retrieval
--------------
This code retrieves the necessary data from the file and completes some
smoothing of the geopotential height and wind fields using the SciPy
function gaussian_filter. A nicely formated valid time (vtime) variable
is also created.
```
# Grab lat/lon values (NAM will be 2D)
lats = ds.lat.data
lons = ds.lon.data
# Select and grab data
hght = ds['Geopotential_height_isobaric']
uwnd = ds['u-component_of_wind_isobaric']
vwnd = ds['v-component_of_wind_isobaric']
# Select and grab 500-hPa geopotential heights and wind components, smooth with gaussian_filter
hght_500 = gaussian_filter(hght.sel(isobaric=500).data[0], sigma=3.0)
uwnd_500 = gaussian_filter(uwnd.sel(isobaric=500).data[0], sigma=3.0) * units('m/s')
vwnd_500 = gaussian_filter(vwnd.sel(isobaric=500).data[0], sigma=3.0) * units('m/s')
# Use MetPy to calculate the wind speed for colorfill plot, change units to knots from m/s
sped_500 = mpcalc.wind_speed(uwnd_500, vwnd_500).to('kt')
# Create a clean datetime object for plotting based on time of Geopotential heights
vtime = datetime.strptime(str(ds.time.data[0].astype('datetime64[ms]')),
'%Y-%m-%dT%H:%M:%S.%f')
```
Map Creation
------------
This next set of code creates the plot and draws contours on a Lambert
Conformal map centered on -100 E longitude. The main view is over the
CONUS with geopotential heights contoured every 60 m and wind speed in
knots every 20 knots starting at 30 kt.
```
# Set up the projection that will be used for plotting
mapcrs = ccrs.LambertConformal(central_longitude=-100,
central_latitude=35,
standard_parallels=(30, 60))
# Set up the projection of the data; if lat/lon then PlateCarree is what you want
datacrs = ccrs.PlateCarree()
# Start the figure and create plot axes with proper projection
fig = plt.figure(1, figsize=(14, 12))
ax = plt.subplot(111, projection=mapcrs)
ax.set_extent([-130, -72, 20, 55], ccrs.PlateCarree())
# Add geopolitical boundaries for map reference
ax.add_feature(cfeature.COASTLINE.with_scale('50m'))
ax.add_feature(cfeature.STATES.with_scale('50m'))
# Plot 500-hPa Colorfill Wind Speeds in knots
clevs_500_sped = np.arange(30, 150, 20)
cf = ax.contourf(lons, lats, sped_500, clevs_500_sped, cmap=plt.cm.BuPu,
transform=datacrs)
plt.colorbar(cf, orientation='horizontal', pad=0, aspect=50)
# Plot 500-hPa Geopotential Heights in meters
clevs_500_hght = np.arange(0, 8000, 60)
cs = ax.contour(lons, lats, hght_500, clevs_500_hght, colors='black',
transform=datacrs)
plt.clabel(cs, fmt='%d')
# Plot 500-hPa wind barbs in knots, regrid to reduce number of barbs
ax.barbs(lons, lats, uwnd_500.to('kt').m, vwnd_500.to('kt').m, pivot='middle',
color='black', regrid_shape=20, transform=datacrs)
# Make some nice titles for the plot (one right, one left)
plt.title('500-hPa NAM Geopotential Heights (m), Wind Speed (kt),'
' and Wind Barbs (kt)', loc='left')
plt.title('Valid Time: {}'.format(vtime), loc='right')
# Adjust image and show
plt.subplots_adjust(bottom=0, top=1)
plt.show()
```
|
github_jupyter
|
%matplotlib inline
from datetime import datetime
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
import metpy.calc as mpcalc
from metpy.units import units
import numpy as np
from scipy.ndimage import gaussian_filter
import xarray as xr
ds = xr.open_dataset('https://thredds.ucar.edu/thredds/dodsC/casestudies/'
'python-gallery/NAM_20161031_1200.nc')
ds.coords
# Grab lat/lon values (NAM will be 2D)
lats = ds.lat.data
lons = ds.lon.data
# Select and grab data
hght = ds['Geopotential_height_isobaric']
uwnd = ds['u-component_of_wind_isobaric']
vwnd = ds['v-component_of_wind_isobaric']
# Select and grab 500-hPa geopotential heights and wind components, smooth with gaussian_filter
hght_500 = gaussian_filter(hght.sel(isobaric=500).data[0], sigma=3.0)
uwnd_500 = gaussian_filter(uwnd.sel(isobaric=500).data[0], sigma=3.0) * units('m/s')
vwnd_500 = gaussian_filter(vwnd.sel(isobaric=500).data[0], sigma=3.0) * units('m/s')
# Use MetPy to calculate the wind speed for colorfill plot, change units to knots from m/s
sped_500 = mpcalc.wind_speed(uwnd_500, vwnd_500).to('kt')
# Create a clean datetime object for plotting based on time of Geopotential heights
vtime = datetime.strptime(str(ds.time.data[0].astype('datetime64[ms]')),
'%Y-%m-%dT%H:%M:%S.%f')
# Set up the projection that will be used for plotting
mapcrs = ccrs.LambertConformal(central_longitude=-100,
central_latitude=35,
standard_parallels=(30, 60))
# Set up the projection of the data; if lat/lon then PlateCarree is what you want
datacrs = ccrs.PlateCarree()
# Start the figure and create plot axes with proper projection
fig = plt.figure(1, figsize=(14, 12))
ax = plt.subplot(111, projection=mapcrs)
ax.set_extent([-130, -72, 20, 55], ccrs.PlateCarree())
# Add geopolitical boundaries for map reference
ax.add_feature(cfeature.COASTLINE.with_scale('50m'))
ax.add_feature(cfeature.STATES.with_scale('50m'))
# Plot 500-hPa Colorfill Wind Speeds in knots
clevs_500_sped = np.arange(30, 150, 20)
cf = ax.contourf(lons, lats, sped_500, clevs_500_sped, cmap=plt.cm.BuPu,
transform=datacrs)
plt.colorbar(cf, orientation='horizontal', pad=0, aspect=50)
# Plot 500-hPa Geopotential Heights in meters
clevs_500_hght = np.arange(0, 8000, 60)
cs = ax.contour(lons, lats, hght_500, clevs_500_hght, colors='black',
transform=datacrs)
plt.clabel(cs, fmt='%d')
# Plot 500-hPa wind barbs in knots, regrid to reduce number of barbs
ax.barbs(lons, lats, uwnd_500.to('kt').m, vwnd_500.to('kt').m, pivot='middle',
color='black', regrid_shape=20, transform=datacrs)
# Make some nice titles for the plot (one right, one left)
plt.title('500-hPa NAM Geopotential Heights (m), Wind Speed (kt),'
' and Wind Barbs (kt)', loc='left')
plt.title('Valid Time: {}'.format(vtime), loc='right')
# Adjust image and show
plt.subplots_adjust(bottom=0, top=1)
plt.show()
| 0.827236 | 0.967747 |
```
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error, mean_squared_error, mean_absolute_error
from sklearn.model_selection import cross_val_score
from rfpimp import *
from sklearn.model_selection import train_test_split
from sklearn.base import clone
df_all = pd.read_csv("data/rent-cls.csv")
num_features = ['bathrooms','bedrooms','latitude','longitude','price']
target = 'interest_level'
df = df_all[num_features + [target]]
# compute median per num bedrooms
df = df.copy()
df_median_price_per_bedrooms = df.groupby(by='bedrooms')['price'].median().reset_index()
beds_to_median = df_median_price_per_bedrooms.to_dict(orient='dict')['price']
df['median_price_per_bedrooms'] = df['bedrooms'].map(beds_to_median)
# compute ratio of price to median price for that num of bedrooms
df['price_to_median_beds'] = df['price'] / df['median_price_per_bedrooms']
# ratio of num bedrooms to price
df["beds_per_price"] = df["bedrooms"] / df["price"]
# total rooms (bed, bath)
df["beds_baths"] = df["bedrooms"]+df["bathrooms"]
del df['median_price_per_bedrooms'] # don't need after computation
df_train, df_test = train_test_split(df, test_size=0.15)
from rfpimp import plot_corr_heatmap
viz = plot_corr_heatmap(df_train, figsize=(7,5))
viz.save('../article/images/corrheatmap.svg')
viz
X_train, y_train = df_train.drop('interest_level',axis=1), df_train['interest_level']
X_test, y_test = df_test.drop('interest_level',axis=1), df_test['interest_level']
rf = RandomForestClassifier(n_estimators=100, n_jobs=-1,
# max_features=X_train.shape[1]-1,
max_features=1.0,
min_samples_leaf=10, oob_score=True)
rf.fit(X_train, y_train)
```
Without specifying a feature list, the default is to give you each column as an individual feature:
```
I = importances(rf, X_test, y_test)
viz = plot_importances(I)
viz.save('../article/images/imp.svg')
viz
```
But you can specify a subset if you like:
```
I = importances(rf, X_test, y_test, features=['price',['latitude','longitude']])
viz = plot_importances(I, vscale=1.2)
viz.save('../article/images/subset_imp.svg')
viz
I = importances(rf, X_test, y_test, features=[['latitude','longitude']])
viz = plot_importances(I, vscale=1.2)
viz.save('../article/images/latlong_imp.svg')
viz
```
By specifying a list of lists, you can provide groups that should be treated together.
```
features = ['bathrooms', 'bedrooms',
['latitude', 'longitude'],
['price_to_median_beds', 'beds_baths', 'price'],
['beds_per_price','bedrooms']]
I = importances(rf, X_test, y_test, features=features)
viz = plot_importances(I, vscale=1.3)
viz.save('../article/images/grouped_imp.svg')
viz
```
Features can be duplicated in multiple groups:
```
features = [['latitude', 'longitude'],
['price_to_median_beds', 'beds_baths', 'beds_per_price', 'bedrooms'],
['price','beds_per_price','bedrooms']]
I = importances(rf, X_test, y_test, features=features)
viz = plot_importances(I, vscale=1.2)
viz.save('../article/images/grouped_dup_imp.svg')
viz
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error, mean_squared_error, mean_absolute_error
from sklearn.model_selection import cross_val_score
from rfpimp import *
from sklearn.model_selection import train_test_split
from sklearn.base import clone
df_all = pd.read_csv("data/rent-cls.csv")
num_features = ['bathrooms','bedrooms','latitude','longitude','price']
target = 'interest_level'
df = df_all[num_features + [target]]
# compute median per num bedrooms
df = df.copy()
df_median_price_per_bedrooms = df.groupby(by='bedrooms')['price'].median().reset_index()
beds_to_median = df_median_price_per_bedrooms.to_dict(orient='dict')['price']
df['median_price_per_bedrooms'] = df['bedrooms'].map(beds_to_median)
# compute ratio of price to median price for that num of bedrooms
df['price_to_median_beds'] = df['price'] / df['median_price_per_bedrooms']
# ratio of num bedrooms to price
df["beds_per_price"] = df["bedrooms"] / df["price"]
# total rooms (bed, bath)
df["beds_baths"] = df["bedrooms"]+df["bathrooms"]
del df['median_price_per_bedrooms'] # don't need after computation
df_train, df_test = train_test_split(df, test_size=0.15)
from rfpimp import plot_corr_heatmap
viz = plot_corr_heatmap(df_train, figsize=(7,5))
viz.save('../article/images/corrheatmap.svg')
viz
X_train, y_train = df_train.drop('interest_level',axis=1), df_train['interest_level']
X_test, y_test = df_test.drop('interest_level',axis=1), df_test['interest_level']
rf = RandomForestClassifier(n_estimators=100, n_jobs=-1,
# max_features=X_train.shape[1]-1,
max_features=1.0,
min_samples_leaf=10, oob_score=True)
rf.fit(X_train, y_train)
I = importances(rf, X_test, y_test)
viz = plot_importances(I)
viz.save('../article/images/imp.svg')
viz
I = importances(rf, X_test, y_test, features=['price',['latitude','longitude']])
viz = plot_importances(I, vscale=1.2)
viz.save('../article/images/subset_imp.svg')
viz
I = importances(rf, X_test, y_test, features=[['latitude','longitude']])
viz = plot_importances(I, vscale=1.2)
viz.save('../article/images/latlong_imp.svg')
viz
features = ['bathrooms', 'bedrooms',
['latitude', 'longitude'],
['price_to_median_beds', 'beds_baths', 'price'],
['beds_per_price','bedrooms']]
I = importances(rf, X_test, y_test, features=features)
viz = plot_importances(I, vscale=1.3)
viz.save('../article/images/grouped_imp.svg')
viz
features = [['latitude', 'longitude'],
['price_to_median_beds', 'beds_baths', 'beds_per_price', 'bedrooms'],
['price','beds_per_price','bedrooms']]
I = importances(rf, X_test, y_test, features=features)
viz = plot_importances(I, vscale=1.2)
viz.save('../article/images/grouped_dup_imp.svg')
viz
| 0.457137 | 0.762601 |
# Protein MD Setup tutorial using BioExcel Building Blocks (biobb)
**Based on the official GROMACS tutorial:** [http://www.mdtutorials.com/gmx/lysozyme/index.html](http://www.mdtutorials.com/gmx/lysozyme/index.html)
***
This tutorial aims to illustrate the process of **setting up a simulation system** containing a **protein**, step by step, using the **BioExcel Building Blocks library (biobb)**. The particular example used is the **Lysozyme** protein (PDB code 1AKI).
***
## Settings
### Biobb modules used
- [biobb_io](https://github.com/bioexcel/biobb_io): Tools to fetch biomolecular data from public databases.
- [biobb_model](https://github.com/bioexcel/biobb_model): Tools to model macromolecular structures.
- [biobb_md](https://github.com/bioexcel/biobb_md): Tools to setup and run Molecular Dynamics simulations.
- [biobb_analysis](https://github.com/bioexcel/biobb_analysis): Tools to analyse Molecular Dynamics trajectories.
### Auxiliar libraries used
- [nb_conda_kernels](https://github.com/Anaconda-Platform/nb_conda_kernels): Enables a Jupyter Notebook or JupyterLab application in one conda environment to access kernels for Python, R, and other languages found in other environments.
- [nglview](http://nglviewer.org/#nglview): Jupyter/IPython widget to interactively view molecular structures and trajectories in notebooks.
- [ipywidgets](https://github.com/jupyter-widgets/ipywidgets): Interactive HTML widgets for Jupyter notebooks and the IPython kernel.
- [plotly](https://plot.ly/python/offline/): Python interactive graphing library integrated in Jupyter notebooks.
- [simpletraj](https://github.com/arose/simpletraj): Lightweight coordinate-only trajectory reader based on code from GROMACS, MDAnalysis and VMD.
### Conda Installation and Launch
```console
git clone https://github.com/bioexcel/biobb_wf_md_setup.git
cd biobb_wf_md_setup
conda env create -f conda_env/environment.yml
conda activate biobb_MDsetup_tutorial
jupyter-nbextension enable --py --user widgetsnbextension
jupyter-nbextension enable --py --user nglview
jupyter-notebook biobb_wf_md_setup/notebooks/biobb_MDsetup_tutorial.ipynb
```
***
## Pipeline steps
1. [Input Parameters](#input)
2. [Fetching PDB Structure](#fetch)
3. [Fix Protein Structure](#fix)
4. [Create Protein System Topology](#top)
5. [Create Solvent Box](#box)
6. [Fill the Box with Water Molecules](#water)
7. [Adding Ions](#ions)
8. [Energetically Minimize the System](#min)
9. [Equilibrate the System (NVT)](#nvt)
10. [Equilibrate the System (NPT)](#npt)
11. [Free Molecular Dynamics Simulation](#free)
12. [Post-processing and Visualizing Resulting 3D Trajectory](#post)
13. [Output Files](#output)
14. [Questions & Comments](#questions)
***
<img src="https://bioexcel.eu/wp-content/uploads/2019/04/Bioexcell_logo_1080px_transp.png" alt="Bioexcel2 logo"
title="Bioexcel2 logo" width="400" />
***
<a id="input"></a>
## Input parameters
**Input parameters** needed:
- **pdbCode**: PDB code of the protein structure (e.g. 1AKI)
```
import nglview
import ipywidgets
pdbCode = "1AKI"
```
<a id="fetch"></a>
***
## Fetching PDB structure
Downloading **PDB structure** with the **protein molecule** from the RCSB PDB database.<br>
Alternatively, a **PDB file** can be used as starting structure. <br>
***
**Building Blocks** used:
- [Pdb](https://biobb-io.readthedocs.io/en/latest/api.html#module-api.pdb) from **biobb_io.api.pdb**
***
```
# Downloading desired PDB file
# Import module
from biobb_io.api.pdb import pdb
# Create properties dict and inputs/outputs
downloaded_pdb = pdbCode+'.pdb'
prop = {
'pdb_code': pdbCode
}
#Create and launch bb
pdb(output_pdb_path=downloaded_pdb,
properties=prop)
```
<a id="vis3D"></a>
### Visualizing 3D structure
Visualizing the downloaded/given **PDB structure** using **NGL**:
```
# Show protein
view = nglview.show_structure_file(downloaded_pdb)
view.add_representation(repr_type='ball+stick', selection='all')
view._remote_call('setSize', target='Widget', args=['','600px'])
view
```
<img src='ngl1.png'></img>
<a id="fix"></a>
***
## Fix protein structure
**Checking** and **fixing** (if needed) the protein structure:<br>
- **Modeling** **missing side-chain atoms**, modifying incorrect **amide assignments**, choosing **alternative locations**.<br>
- **Checking** for missing **backbone atoms**, **heteroatoms**, **modified residues** and possible **atomic clashes**.
***
**Building Blocks** used:
- [FixSideChain](https://biobb-model.readthedocs.io/en/latest/model.html#module-model.fix_side_chain) from **biobb_model.model.fix_side_chain**
***
```
# Check & Fix PDB
# Import module
from biobb_model.model.fix_side_chain import fix_side_chain
# Create prop dict and inputs/outputs
fixed_pdb = pdbCode + '_fixed.pdb'
# Create and launch bb
fix_side_chain(input_pdb_path=downloaded_pdb,
output_pdb_path=fixed_pdb)
```
### Visualizing 3D structure
Visualizing the fixed **PDB structure** using **NGL**. In this particular example, the checking step didn't find any issue to be solved, so there is no difference between the original structure and the fixed one.
```
# Show protein
view = nglview.show_structure_file(fixed_pdb)
view.add_representation(repr_type='ball+stick', selection='all')
view._remote_call('setSize', target='Widget', args=['','600px'])
view.camera='orthographic'
view
```
<img src='ngl2.png'></img>
<a id="top"></a>
***
## Create protein system topology
**Building GROMACS topology** corresponding to the protein structure.<br>
Force field used in this tutorial is [**amber99sb-ildn**](https://dx.doi.org/10.1002%2Fprot.22711): AMBER **parm99** force field with **corrections on backbone** (sb) and **side-chain torsion potentials** (ildn). Water molecules type used in this tutorial is [**spc/e**](https://pubs.acs.org/doi/abs/10.1021/j100308a038).<br>
Adding **hydrogen atoms** if missing. Automatically identifying **disulfide bridges**. <br>
Generating two output files:
- **GROMACS structure** (gro file)
- **GROMACS topology** ZIP compressed file containing:
- *GROMACS topology top file* (top file)
- *GROMACS position restraint file/s* (itp file/s)
***
**Building Blocks** used:
- [Pdb2gmx](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.pdb2gmx) from **biobb_md.gromacs.pdb2gmx**
***
```
# Create system topology
# Import module
from biobb_md.gromacs.pdb2gmx import pdb2gmx
# Create inputs/outputs
output_pdb2gmx_gro = pdbCode+'_pdb2gmx.gro'
output_pdb2gmx_top_zip = pdbCode+'_pdb2gmx_top.zip'
# Create and launch bb
pdb2gmx(input_pdb_path=fixed_pdb,
output_gro_path=output_pdb2gmx_gro,
output_top_zip_path=output_pdb2gmx_top_zip)
```
### Visualizing 3D structure
Visualizing the generated **GRO structure** using **NGL**. Note that **hydrogen atoms** were added to the structure by the **pdb2gmx GROMACS tool** when generating the **topology**.
```
# Show protein
view = nglview.show_structure_file(output_pdb2gmx_gro)
view.add_representation(repr_type='ball+stick', selection='all')
view._remote_call('setSize', target='Widget', args=['','600px'])
view.camera='orthographic'
view
```
<img src='ngl3.png'></img>
<a id="box"></a>
***
## Create solvent box
Define the unit cell for the **protein structure MD system** to fill it with water molecules.<br>
A **cubic box** is used to define the unit cell, with a **distance from the protein to the box edge of 1.0 nm**. The protein is **centered in the box**.
***
**Building Blocks** used:
- [Editconf](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.editconf) from **biobb_md.gromacs.editconf**
***
```
# Editconf: Create solvent box
# Import module
from biobb_md.gromacs.editconf import editconf
# Create prop dict and inputs/outputs
output_editconf_gro = pdbCode+'_editconf.gro'
prop = {
'box_type': 'cubic',
'distance_to_molecule': 1.0
}
#Create and launch bb
editconf(input_gro_path=output_pdb2gmx_gro,
output_gro_path=output_editconf_gro,
properties=prop)
```
<a id="water"></a>
***
## Fill the box with water molecules
Fill the unit cell for the **protein structure system** with water molecules.<br>
The solvent type used is the default **Simple Point Charge water (SPC)**, a generic equilibrated 3-point solvent model.
***
**Building Blocks** used:
- [Solvate](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.solvate) from **biobb_md.gromacs.solvate**
***
```
# Solvate: Fill the box with water molecules
from biobb_md.gromacs.solvate import solvate
# Create prop dict and inputs/outputs
output_solvate_gro = pdbCode+'_solvate.gro'
output_solvate_top_zip = pdbCode+'_solvate_top.zip'
# Create and launch bb
solvate(input_solute_gro_path=output_editconf_gro,
output_gro_path=output_solvate_gro,
input_top_zip_path=output_pdb2gmx_top_zip,
output_top_zip_path=output_solvate_top_zip)
```
### Visualizing 3D structure
Visualizing the **protein system** with the newly added **solvent box** using **NGL**.<br> Note the **cubic box** filled with **water molecules** surrounding the **protein structure**, which is **centered** right in the middle of the cube.
```
# Show protein
view = nglview.show_structure_file(output_solvate_gro)
view.clear_representations()
view.add_representation(repr_type='cartoon', selection='solute', color='green')
view.add_representation(repr_type='ball+stick', selection='SOL')
view._remote_call('setSize', target='Widget', args=['','600px'])
view.camera='orthographic'
view
```
<img src='ngl4.png'></img>
<a id="ions"></a>
***
## Adding ions
Add ions to neutralize the **protein structure** charge
- [Step 1](#ionsStep1): Creating portable binary run file for ion generation
- [Step 2](#ionsStep2): Adding ions to **neutralize** the system
***
**Building Blocks** used:
- [Grompp](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.grompp) from **biobb_md.gromacs.grompp**
- [Genion](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.genion) from **biobb_md.gromacs.genion**
***
<a id="ionsStep1"></a>
### Step 1: Creating portable binary run file for ion generation
A simple **energy minimization** molecular dynamics parameters (mdp) properties will be used to generate the portable binary run file for **ion generation**, although **any legitimate combination of parameters** could be used in this step.
```
# Grompp: Creating portable binary run file for ion generation
from biobb_md.gromacs.grompp import grompp
# Create prop dict and inputs/outputs
output_gppion_tpr = pdbCode+'_gppion.tpr'
prop = {
'simulation_type': 'minimization'
}
# Create and launch bb
grompp(input_gro_path=output_solvate_gro,
input_top_zip_path=output_solvate_top_zip,
output_tpr_path=output_gppion_tpr,
properties=prop)
```
<a id="ionsStep2"></a>
### Step 2: Adding ions to neutralize the system
Replace **solvent molecules** with **ions** to **neutralize** the system.
```
# Genion: Adding ions to neutralize the system
from biobb_md.gromacs.genion import genion
# Create prop dict and inputs/outputs
output_genion_gro = pdbCode+'_genion.gro'
output_genion_top_zip = pdbCode+'_genion_top.zip'
prop={
'neutral':True
}
# Create and launch bb
genion(input_tpr_path=output_gppion_tpr,
output_gro_path=output_genion_gro,
input_top_zip_path=output_solvate_top_zip,
output_top_zip_path=output_genion_top_zip,
properties=prop)
```
### Visualizing 3D structure
Visualizing the **neutralized protein system** with the newly added **ions** using **NGL**
```
# Show protein
view = nglview.show_structure_file(output_genion_gro)
view.clear_representations()
view.add_representation(repr_type='cartoon', selection='solute', color='sstruc')
view.add_representation(repr_type='ball+stick', selection='NA')
view.add_representation(repr_type='ball+stick', selection='CL')
view._remote_call('setSize', target='Widget', args=['','600px'])
view.camera='orthographic'
view
```
<img src='ngl5.png'></img>
<a id="min"></a>
***
## Energetically minimize the system
Energetically minimize the **protein system** till reaching a desired potential energy.
- [Step 1](#emStep1): Creating portable binary run file for energy minimization
- [Step 2](#emStep2): Energetically minimize the **system** till reaching a force of 500 kJ mol-1 nm-1.
- [Step 3](#emStep3): Checking **energy minimization** results. Plotting energy by time during the **minimization** process.
***
**Building Blocks** used:
- [Grompp](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.grompp) from **biobb_md.gromacs.grompp**
- [Mdrun](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.mdrun) from **biobb_md.gromacs.mdrun**
- [GMXEnergy](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_energy) from **biobb_analysis.gromacs.gmx_energy**
***
<a id="emStep1"></a>
### Step 1: Creating portable binary run file for energy minimization
The **minimization** type of the **molecular dynamics parameters (mdp) property** contains the main default parameters to run an **energy minimization**:
- integrator = steep ; Algorithm (steep = steepest descent minimization)
- emtol = 1000.0 ; Stop minimization when the maximum force < 1000.0 kJ/mol/nm
- emstep = 0.01 ; Minimization step size (nm)
- nsteps = 50000 ; Maximum number of (minimization) steps to perform
In this particular example, the method used to run the **energy minimization** is the default **steepest descent**, but the **maximum force** is placed at **500 KJ/mol\*nm^2**, and the **maximum number of steps** to perform (if the maximum force is not reached) to **5,000 steps**.
```
# Grompp: Creating portable binary run file for mdrun
from biobb_md.gromacs.grompp import grompp
# Create prop dict and inputs/outputs
output_gppmin_tpr = pdbCode+'_gppmin.tpr'
prop = {
'mdp':{
'emtol':'500',
'nsteps':'5000'
},
'simulation_type': 'minimization'
}
# Create and launch bb
grompp(input_gro_path=output_genion_gro,
input_top_zip_path=output_genion_top_zip,
output_tpr_path=output_gppmin_tpr,
properties=prop)
```
<a id="emStep2"></a>
### Step 2: Running Energy Minimization
Running **energy minimization** using the **tpr file** generated in the previous step.
```
# Mdrun: Running minimization
from biobb_md.gromacs.mdrun import mdrun
# Create prop dict and inputs/outputs
output_min_trr = pdbCode+'_min.trr'
output_min_gro = pdbCode+'_min.gro'
output_min_edr = pdbCode+'_min.edr'
output_min_log = pdbCode+'_min.log'
# Create and launch bb
mdrun(input_tpr_path=output_gppmin_tpr,
output_trr_path=output_min_trr,
output_gro_path=output_min_gro,
output_edr_path=output_min_edr,
output_log_path=output_min_log)
```
<a id="emStep3"></a>
### Step 3: Checking Energy Minimization results
Checking **energy minimization** results. Plotting **potential energy** by time during the minimization process.
```
# GMXEnergy: Getting system energy by time
from biobb_analysis.gromacs.gmx_energy import gmx_energy
# Create prop dict and inputs/outputs
output_min_ene_xvg = pdbCode+'_min_ene.xvg'
prop = {
'terms': ["Potential"]
}
# Create and launch bb
gmx_energy(input_energy_path=output_min_edr,
output_xvg_path=output_min_ene_xvg,
properties=prop)
import plotly
import plotly.graph_objs as go
#Read data from file and filter energy values higher than 1000 Kj/mol^-1
with open(output_min_ene_xvg,'r') as energy_file:
x,y = map(
list,
zip(*[
(float(line.split()[0]),float(line.split()[1]))
for line in energy_file
if not line.startswith(("#","@"))
if float(line.split()[1]) < 1000
])
)
plotly.offline.init_notebook_mode(connected=True)
fig = {
"data": [go.Scatter(x=x, y=y)],
"layout": go.Layout(title="Energy Minimization",
xaxis=dict(title = "Energy Minimization Step"),
yaxis=dict(title = "Potential Energy KJ/mol-1")
)
}
plotly.offline.iplot(fig)
```
<img src='plot1.png'></img>
<a id="nvt"></a>
***
## Equilibrate the system (NVT)
Equilibrate the **protein system** in **NVT ensemble** (constant Number of particles, Volume and Temperature). Protein **heavy atoms** will be restrained using position restraining forces: movement is permitted, but only after overcoming a substantial energy penalty. The utility of position restraints is that they allow us to equilibrate our solvent around our protein, without the added variable of structural changes in the protein.
- [Step 1](#eqNVTStep1): Creating portable binary run file for system equilibration
- [Step 2](#eqNVTStep2): Equilibrate the **protein system** with **NVT** ensemble.
- [Step 3](#eqNVTStep3): Checking **NVT Equilibration** results. Plotting **system temperature** by time during the **NVT equilibration** process.
***
**Building Blocks** used:
- [Grompp](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.grompp) from **biobb_md.gromacs.grompp**
- [Mdrun](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.mdrun) from **biobb_md.gromacs.mdrun**
- [GMXEnergy](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_energy) from **biobb_analysis.gromacs.gmx_energy**
***
<a id="eqNVTStep1"></a>
### Step 1: Creating portable binary run file for system equilibration (NVT)
The **nvt** type of the **molecular dynamics parameters (mdp) property** contains the main default parameters to run an **NVT equilibration** with **protein restraints** (see [GROMACS mdp options](http://manual.gromacs.org/documentation/2018/user-guide/mdp-options.html)):
- Define = -DPOSRES
- integrator = md
- dt = 0.002
- nsteps = 5000
- pcoupl = no
- gen_vel = yes
- gen_temp = 300
- gen_seed = -1
In this particular example, the default parameters will be used: **md** integrator algorithm, a **step size** of **2fs**, **5,000 equilibration steps** with the protein **heavy atoms restrained**, and a temperature of **300K**.
*Please note that for the sake of time this tutorial is only running 10ps of NVT equilibration, whereas in the [original example](http://www.mdtutorials.com/gmx/lysozyme/06_equil.html) the simulated time was 100ps.*
```
# Grompp: Creating portable binary run file for NVT Equilibration
from biobb_md.gromacs.grompp import grompp
# Create prop dict and inputs/outputs
output_gppnvt_tpr = pdbCode+'_gppnvt.tpr'
prop = {
'mdp':{
'nsteps': 5000,
'dt': 0.002,
'Define': '-DPOSRES',
#'tc_grps': "DNA Water_and_ions" # NOTE: uncomment this line if working with DNA
},
'simulation_type': 'nvt'
}
# Create and launch bb
grompp(input_gro_path=output_min_gro,
input_top_zip_path=output_genion_top_zip,
output_tpr_path=output_gppnvt_tpr,
properties=prop)
```
<a id="eqNVTStep2"></a>
### Step 2: Running NVT equilibration
```
# Mdrun: Running Equilibration NVT
from biobb_md.gromacs.mdrun import mdrun
# Create prop dict and inputs/outputs
output_nvt_trr = pdbCode+'_nvt.trr'
output_nvt_gro = pdbCode+'_nvt.gro'
output_nvt_edr = pdbCode+'_nvt.edr'
output_nvt_log = pdbCode+'_nvt.log'
output_nvt_cpt = pdbCode+'_nvt.cpt'
# Create and launch bb
mdrun(input_tpr_path=output_gppnvt_tpr,
output_trr_path=output_nvt_trr,
output_gro_path=output_nvt_gro,
output_edr_path=output_nvt_edr,
output_log_path=output_nvt_log,
output_cpt_path=output_nvt_cpt)
```
<a id="eqNVTStep3"></a>
### Step 3: Checking NVT Equilibration results
Checking **NVT Equilibration** results. Plotting **system temperature** by time during the NVT equilibration process.
```
# GMXEnergy: Getting system temperature by time during NVT Equilibration
from biobb_analysis.gromacs.gmx_energy import gmx_energy
# Create prop dict and inputs/outputs
output_nvt_temp_xvg = pdbCode+'_nvt_temp.xvg'
prop = {
'terms': ["Temperature"]
}
# Create and launch bb
gmx_energy(input_energy_path=output_nvt_edr,
output_xvg_path=output_nvt_temp_xvg,
properties=prop)
import plotly
import plotly.graph_objs as go
# Read temperature data from file
with open(output_nvt_temp_xvg,'r') as temperature_file:
x,y = map(
list,
zip(*[
(float(line.split()[0]),float(line.split()[1]))
for line in temperature_file
if not line.startswith(("#","@"))
])
)
plotly.offline.init_notebook_mode(connected=True)
fig = {
"data": [go.Scatter(x=x, y=y)],
"layout": go.Layout(title="Temperature during NVT Equilibration",
xaxis=dict(title = "Time (ps)"),
yaxis=dict(title = "Temperature (K)")
)
}
plotly.offline.iplot(fig)
```
<img src='plot2.png'></img>
<a id="npt"></a>
***
## Equilibrate the system (NPT)
Equilibrate the **protein system** in **NPT** ensemble (constant Number of particles, Pressure and Temperature).
- [Step 1](#eqNPTStep1): Creating portable binary run file for system equilibration
- [Step 2](#eqNPTStep2): Equilibrate the **protein system** with **NPT** ensemble.
- [Step 3](#eqNPTStep3): Checking **NPT Equilibration** results. Plotting **system pressure and density** by time during the **NPT equilibration** process.
***
**Building Blocks** used:
- [Grompp](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.grompp) from **biobb_md.gromacs.grompp**
- [Mdrun](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.mdrun) from **biobb_md.gromacs.mdrun**
- [GMXEnergy](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_energy) from **biobb_analysis.gromacs.gmx_energy**
***
<a id="eqNPTStep1"></a>
### Step 1: Creating portable binary run file for system equilibration (NPT)
The **npt** type of the **molecular dynamics parameters (mdp) property** contains the main default parameters to run an **NPT equilibration** with **protein restraints** (see [GROMACS mdp options](http://manual.gromacs.org/documentation/2018/user-guide/mdp-options.html)):
- Define = -DPOSRES
- integrator = md
- dt = 0.002
- nsteps = 5000
- pcoupl = Parrinello-Rahman
- pcoupltype = isotropic
- tau_p = 1.0
- ref_p = 1.0
- compressibility = 4.5e-5
- refcoord_scaling = com
- gen_vel = no
In this particular example, the default parameters will be used: **md** integrator algorithm, a **time step** of **2fs**, **5,000 equilibration steps** with the protein **heavy atoms restrained**, and a Parrinello-Rahman **pressure coupling** algorithm.
*Please note that for the sake of time this tutorial is only running 10ps of NPT equilibration, whereas in the [original example](http://www.mdtutorials.com/gmx/lysozyme/07_equil2.html) the simulated time was 100ps.*
```
# Grompp: Creating portable binary run file for NPT System Equilibration
from biobb_md.gromacs.grompp import grompp
# Create prop dict and inputs/outputs
output_gppnpt_tpr = pdbCode+'_gppnpt.tpr'
prop = {
'mdp':{
'nsteps':'5000',
#'tc_grps': "DNA Water_and_ions" # NOTE: uncomment this line if working with DNA
},
'simulation_type': 'npt'
}
# Create and launch bb
grompp(input_gro_path=output_nvt_gro,
input_top_zip_path=output_genion_top_zip,
output_tpr_path=output_gppnpt_tpr,
input_cpt_path=output_nvt_cpt,
properties=prop)
```
<a id="eqNPTStep2"></a>
### Step 2: Running NPT equilibration
```
# Mdrun: Running NPT System Equilibration
from biobb_md.gromacs.mdrun import mdrun
# Create prop dict and inputs/outputs
output_npt_trr = pdbCode+'_npt.trr'
output_npt_gro = pdbCode+'_npt.gro'
output_npt_edr = pdbCode+'_npt.edr'
output_npt_log = pdbCode+'_npt.log'
output_npt_cpt = pdbCode+'_npt.cpt'
# Create and launch bb
mdrun(input_tpr_path=output_gppnpt_tpr,
output_trr_path=output_npt_trr,
output_gro_path=output_npt_gro,
output_edr_path=output_npt_edr,
output_log_path=output_npt_log,
output_cpt_path=output_npt_cpt)
```
<a id="eqNPTStep3"></a>
### Step 3: Checking NPT Equilibration results
Checking **NPT Equilibration** results. Plotting **system pressure and density** by time during the **NPT equilibration** process.
```
# GMXEnergy: Getting system pressure and density by time during NPT Equilibration
from biobb_analysis.gromacs.gmx_energy import gmx_energy
# Create prop dict and inputs/outputs
output_npt_pd_xvg = pdbCode+'_npt_PD.xvg'
prop = {
'terms': ["Pressure","Density"]
}
# Create and launch bb
gmx_energy(input_energy_path=output_npt_edr,
output_xvg_path=output_npt_pd_xvg,
properties=prop)
import plotly
from plotly import subplots
import plotly.graph_objs as go
# Read pressure and density data from file
with open(output_npt_pd_xvg,'r') as pd_file:
x,y,z = map(
list,
zip(*[
(float(line.split()[0]),float(line.split()[1]),float(line.split()[2]))
for line in pd_file
if not line.startswith(("#","@"))
])
)
plotly.offline.init_notebook_mode(connected=True)
trace1 = go.Scatter(
x=x,y=y
)
trace2 = go.Scatter(
x=x,y=z
)
fig = subplots.make_subplots(rows=1, cols=2, print_grid=False)
fig.append_trace(trace1, 1, 1)
fig.append_trace(trace2, 1, 2)
fig['layout']['xaxis1'].update(title='Time (ps)')
fig['layout']['xaxis2'].update(title='Time (ps)')
fig['layout']['yaxis1'].update(title='Pressure (bar)')
fig['layout']['yaxis2'].update(title='Density (Kg*m^-3)')
fig['layout'].update(title='Pressure and Density during NPT Equilibration')
fig['layout'].update(showlegend=False)
plotly.offline.iplot(fig)
```
<img src='plot3.png'></img>
<a id="free"></a>
***
## Free Molecular Dynamics Simulation
Upon completion of the **two equilibration phases (NVT and NPT)**, the system is now well-equilibrated at the desired temperature and pressure. The **position restraints** can now be released. The last step of the **protein** MD setup is a short, **free MD simulation**, to ensure the robustness of the system.
- [Step 1](#mdStep1): Creating portable binary run file to run a **free MD simulation**.
- [Step 2](#mdStep2): Run short MD simulation of the **protein system**.
- [Step 3](#mdStep3): Checking results for the final step of the setup process, the **free MD run**. Plotting **Root Mean Square deviation (RMSd)** and **Radius of Gyration (Rgyr)** by time during the **free MD run** step.
***
**Building Blocks** used:
- [Grompp](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.grompp) from **biobb_md.gromacs.grompp**
- [Mdrun](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.mdrun) from **biobb_md.gromacs.mdrun**
- [GMXRms](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_rms) from **biobb_analysis.gromacs.gmx_rms**
- [GMXRgyr](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_rgyr) from **biobb_analysis.gromacs.gmx_rgyr**
***
<a id="mdStep1"></a>
### Step 1: Creating portable binary run file to run a free MD simulation
The **free** type of the **molecular dynamics parameters (mdp) property** contains the main default parameters to run an **free MD simulation** (see [GROMACS mdp options](http://manual.gromacs.org/documentation/2018/user-guide/mdp-options.html)):
- integrator = md
- dt = 0.002 (ps)
- nsteps = 50000
In this particular example, the default parameters will be used: **md** integrator algorithm, a **time step** of **2fs**, and a total of **50,000 md steps** (100ps).
*Please note that for the sake of time this tutorial is only running 100ps of free MD, whereas in the [original example](http://www.mdtutorials.com/gmx/lysozyme/08_MD.html) the simulated time was 1ns (1000ps).*
```
# Grompp: Creating portable binary run file for mdrun
from biobb_md.gromacs.grompp import grompp
# Create prop dict and inputs/outputs
output_gppmd_tpr = pdbCode+'_gppmd.tpr'
prop = {
'mdp':{
'nsteps':'50000',
#'tc_grps': "DNA Water_and_ions" # NOTE: uncomment this line if working with DNA
},
'simulation_type': 'free'
}
# Create and launch bb
grompp(input_gro_path=output_npt_gro,
input_top_zip_path=output_genion_top_zip,
output_tpr_path=output_gppmd_tpr,
input_cpt_path=output_npt_cpt,
properties=prop)
```
<a id="mdStep2"></a>
### Step 2: Running short free MD simulation
```
# Mdrun: Running free dynamics
from biobb_md.gromacs.mdrun import mdrun
# Create prop dict and inputs/outputs
output_md_trr = pdbCode+'_md.trr'
output_md_gro = pdbCode+'_md.gro'
output_md_edr = pdbCode+'_md.edr'
output_md_log = pdbCode+'_md.log'
output_md_cpt = pdbCode+'_md.cpt'
# Create and launch bb
mdrun(input_tpr_path=output_gppmd_tpr,
output_trr_path=output_md_trr,
output_gro_path=output_md_gro,
output_edr_path=output_md_edr,
output_log_path=output_md_log,
output_cpt_path=output_md_cpt)
```
<a id="mdStep3"></a>
### Step 3: Checking free MD simulation results
Checking results for the final step of the setup process, the **free MD run**. Plotting **Root Mean Square deviation (RMSd)** and **Radius of Gyration (Rgyr)** by time during the **free MD run** step. **RMSd** against the **experimental structure** (input structure of the pipeline) and against the **minimized and equilibrated structure** (output structure of the NPT equilibration step).
```
# GMXRms: Computing Root Mean Square deviation to analyse structural stability
# RMSd against minimized and equilibrated snapshot (backbone atoms)
from biobb_analysis.gromacs.gmx_rms import gmx_rms
# Create prop dict and inputs/outputs
output_rms_first = pdbCode+'_rms_first.xvg'
prop = {
'selection': 'Backbone',
#'selection': 'non-Water'
}
# Create and launch bb
gmx_rms(input_structure_path=output_gppmd_tpr,
input_traj_path=output_md_trr,
output_xvg_path=output_rms_first,
properties=prop)
# GMXRms: Computing Root Mean Square deviation to analyse structural stability
# RMSd against experimental structure (backbone atoms)
from biobb_analysis.gromacs.gmx_rms import gmx_rms
# Create prop dict and inputs/outputs
output_rms_exp = pdbCode+'_rms_exp.xvg'
prop = {
'selection': 'Backbone',
#'selection': 'non-Water'
}
# Create and launch bb
gmx_rms(input_structure_path=output_gppmin_tpr,
input_traj_path=output_md_trr,
output_xvg_path=output_rms_exp,
properties=prop)
import plotly
import plotly.graph_objs as go
# Read RMS vs first snapshot data from file
with open(output_rms_first,'r') as rms_first_file:
x,y = map(
list,
zip(*[
(float(line.split()[0]),float(line.split()[1]))
for line in rms_first_file
if not line.startswith(("#","@"))
])
)
# Read RMS vs experimental structure data from file
with open(output_rms_exp,'r') as rms_exp_file:
x2,y2 = map(
list,
zip(*[
(float(line.split()[0]),float(line.split()[1]))
for line in rms_exp_file
if not line.startswith(("#","@"))
])
)
trace1 = go.Scatter(
x = x,
y = y,
name = 'RMSd vs first'
)
trace2 = go.Scatter(
x = x,
y = y2,
name = 'RMSd vs exp'
)
data = [trace1, trace2]
plotly.offline.init_notebook_mode(connected=True)
fig = {
"data": data,
"layout": go.Layout(title="RMSd during free MD Simulation",
xaxis=dict(title = "Time (ps)"),
yaxis=dict(title = "RMSd (nm)")
)
}
plotly.offline.iplot(fig)
```
<img src='plot4.png'></img>
```
# GMXRgyr: Computing Radius of Gyration to measure the protein compactness during the free MD simulation
from biobb_analysis.gromacs.gmx_rgyr import gmx_rgyr
# Create prop dict and inputs/outputs
output_rgyr = pdbCode+'_rgyr.xvg'
prop = {
'selection': 'Backbone'
}
# Create and launch bb
gmx_rgyr(input_structure_path=output_gppmin_tpr,
input_traj_path=output_md_trr,
output_xvg_path=output_rgyr,
properties=prop)
import plotly
import plotly.graph_objs as go
# Read Rgyr data from file
with open(output_rgyr,'r') as rgyr_file:
x,y = map(
list,
zip(*[
(float(line.split()[0]),float(line.split()[1]))
for line in rgyr_file
if not line.startswith(("#","@"))
])
)
plotly.offline.init_notebook_mode(connected=True)
fig = {
"data": [go.Scatter(x=x, y=y)],
"layout": go.Layout(title="Radius of Gyration",
xaxis=dict(title = "Time (ps)"),
yaxis=dict(title = "Rgyr (nm)")
)
}
plotly.offline.iplot(fig)
```
<img src='plot5.png'></img>
<a id="post"></a>
***
## Post-processing and Visualizing resulting 3D trajectory
Post-processing and Visualizing the **protein system** MD setup **resulting trajectory** using **NGL**
- [Step 1](#ppStep1): *Imaging* the resulting trajectory, **stripping out water molecules and ions** and **correcting periodicity issues**.
- [Step 2](#ppStep2): Generating a *dry* structure, **removing water molecules and ions** from the final snapshot of the MD setup pipeline.
- [Step 3](#ppStep3): Visualizing the *imaged* trajectory using the *dry* structure as a **topology**.
***
**Building Blocks** used:
- [GMXImage](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_image) from **biobb_analysis.gromacs.gmx_image**
- [GMXTrjConvStr](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_trjconv_str) from **biobb_analysis.gromacs.gmx_trjconv_str**
***
<a id="ppStep1"></a>
### Step 1: *Imaging* the resulting trajectory.
Stripping out **water molecules and ions** and **correcting periodicity issues**
```
# GMXImage: "Imaging" the resulting trajectory
# Removing water molecules and ions from the resulting structure
from biobb_analysis.gromacs.gmx_image import gmx_image
# Create prop dict and inputs/outputs
output_imaged_traj = pdbCode+'_imaged_traj.trr'
prop = {
'center_selection': 'Protein',
'output_selection': 'Protein',
'pbc' : 'mol',
'center' : True
}
# Create and launch bb
gmx_image(input_traj_path=output_md_trr,
input_top_path=output_gppmd_tpr,
output_traj_path=output_imaged_traj,
properties=prop)
```
<a id="ppStep2"></a>
### Step 2: Generating the output *dry* structure.
**Removing water molecules and ions** from the resulting structure
```
# GMXTrjConvStr: Converting and/or manipulating a structure
# Removing water molecules and ions from the resulting structure
# The "dry" structure will be used as a topology to visualize
# the "imaged dry" trajectory generated in the previous step.
from biobb_analysis.gromacs.gmx_trjconv_str import gmx_trjconv_str
# Create prop dict and inputs/outputs
output_dry_gro = pdbCode+'_md_dry.gro'
prop = {
'selection': 'Protein'
}
# Create and launch bb
gmx_trjconv_str(input_structure_path=output_md_gro,
input_top_path=output_gppmd_tpr,
output_str_path=output_dry_gro,
properties=prop)
```
<a id="ppStep3"></a>
### Step 3: Visualizing the generated dehydrated trajectory.
Using the **imaged trajectory** (output of the [Post-processing step 1](#ppStep1)) with the **dry structure** (output of the [Post-processing step 2](#ppStep2)) as a topology.
```
# Show trajectory
view = nglview.show_simpletraj(nglview.SimpletrajTrajectory(output_imaged_traj, output_dry_gro), gui=True)
view
```
<img src='trajectory.gif'></img>
<a id="output"></a>
## Output files
Important **Output files** generated:
- {{output_md_gro}}: **Final structure** (snapshot) of the MD setup protocol.
- {{output_md_trr}}: **Final trajectory** of the MD setup protocol.
- {{output_md_cpt}}: **Final checkpoint file**, with information about the state of the simulation. It can be used to **restart** or **continue** a MD simulation.
- {{output_gppmd_tpr}}: **Final tpr file**, GROMACS portable binary run input file. This file contains the starting structure of the **MD setup free MD simulation step**, together with the molecular topology and all the simulation parameters. It can be used to **extend** the simulation.
- {{output_genion_top_zip}}: **Final topology** of the MD system. It is a compressed zip file including a **topology file** (.top) and a set of auxiliar **include topology** files (.itp).
**Analysis** (MD setup check) output files generated:
- {{output_rms_first}}: **Root Mean Square deviation (RMSd)** against **minimized and equilibrated structure** of the final **free MD run step**.
- {{output_rms_exp}}: **Root Mean Square deviation (RMSd)** against **experimental structure** of the final **free MD run step**.
- {{output_rgyr}}: **Radius of Gyration** of the final **free MD run step** of the **setup pipeline**.
***
<a id="questions"></a>
## Questions & Comments
Questions, issues, suggestions and comments are really welcome!
* GitHub issues:
* [https://github.com/bioexcel/biobb](https://github.com/bioexcel/biobb)
* BioExcel forum:
* [https://ask.bioexcel.eu/c/BioExcel-Building-Blocks-library](https://ask.bioexcel.eu/c/BioExcel-Building-Blocks-library)
|
github_jupyter
|
git clone https://github.com/bioexcel/biobb_wf_md_setup.git
cd biobb_wf_md_setup
conda env create -f conda_env/environment.yml
conda activate biobb_MDsetup_tutorial
jupyter-nbextension enable --py --user widgetsnbextension
jupyter-nbextension enable --py --user nglview
jupyter-notebook biobb_wf_md_setup/notebooks/biobb_MDsetup_tutorial.ipynb
```
***
## Pipeline steps
1. [Input Parameters](#input)
2. [Fetching PDB Structure](#fetch)
3. [Fix Protein Structure](#fix)
4. [Create Protein System Topology](#top)
5. [Create Solvent Box](#box)
6. [Fill the Box with Water Molecules](#water)
7. [Adding Ions](#ions)
8. [Energetically Minimize the System](#min)
9. [Equilibrate the System (NVT)](#nvt)
10. [Equilibrate the System (NPT)](#npt)
11. [Free Molecular Dynamics Simulation](#free)
12. [Post-processing and Visualizing Resulting 3D Trajectory](#post)
13. [Output Files](#output)
14. [Questions & Comments](#questions)
***
<img src="https://bioexcel.eu/wp-content/uploads/2019/04/Bioexcell_logo_1080px_transp.png" alt="Bioexcel2 logo"
title="Bioexcel2 logo" width="400" />
***
<a id="input"></a>
## Input parameters
**Input parameters** needed:
- **pdbCode**: PDB code of the protein structure (e.g. 1AKI)
<a id="fetch"></a>
***
## Fetching PDB structure
Downloading **PDB structure** with the **protein molecule** from the RCSB PDB database.<br>
Alternatively, a **PDB file** can be used as starting structure. <br>
***
**Building Blocks** used:
- [Pdb](https://biobb-io.readthedocs.io/en/latest/api.html#module-api.pdb) from **biobb_io.api.pdb**
***
<a id="vis3D"></a>
### Visualizing 3D structure
Visualizing the downloaded/given **PDB structure** using **NGL**:
<img src='ngl1.png'></img>
<a id="fix"></a>
***
## Fix protein structure
**Checking** and **fixing** (if needed) the protein structure:<br>
- **Modeling** **missing side-chain atoms**, modifying incorrect **amide assignments**, choosing **alternative locations**.<br>
- **Checking** for missing **backbone atoms**, **heteroatoms**, **modified residues** and possible **atomic clashes**.
***
**Building Blocks** used:
- [FixSideChain](https://biobb-model.readthedocs.io/en/latest/model.html#module-model.fix_side_chain) from **biobb_model.model.fix_side_chain**
***
### Visualizing 3D structure
Visualizing the fixed **PDB structure** using **NGL**. In this particular example, the checking step didn't find any issue to be solved, so there is no difference between the original structure and the fixed one.
<img src='ngl2.png'></img>
<a id="top"></a>
***
## Create protein system topology
**Building GROMACS topology** corresponding to the protein structure.<br>
Force field used in this tutorial is [**amber99sb-ildn**](https://dx.doi.org/10.1002%2Fprot.22711): AMBER **parm99** force field with **corrections on backbone** (sb) and **side-chain torsion potentials** (ildn). Water molecules type used in this tutorial is [**spc/e**](https://pubs.acs.org/doi/abs/10.1021/j100308a038).<br>
Adding **hydrogen atoms** if missing. Automatically identifying **disulfide bridges**. <br>
Generating two output files:
- **GROMACS structure** (gro file)
- **GROMACS topology** ZIP compressed file containing:
- *GROMACS topology top file* (top file)
- *GROMACS position restraint file/s* (itp file/s)
***
**Building Blocks** used:
- [Pdb2gmx](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.pdb2gmx) from **biobb_md.gromacs.pdb2gmx**
***
### Visualizing 3D structure
Visualizing the generated **GRO structure** using **NGL**. Note that **hydrogen atoms** were added to the structure by the **pdb2gmx GROMACS tool** when generating the **topology**.
<img src='ngl3.png'></img>
<a id="box"></a>
***
## Create solvent box
Define the unit cell for the **protein structure MD system** to fill it with water molecules.<br>
A **cubic box** is used to define the unit cell, with a **distance from the protein to the box edge of 1.0 nm**. The protein is **centered in the box**.
***
**Building Blocks** used:
- [Editconf](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.editconf) from **biobb_md.gromacs.editconf**
***
<a id="water"></a>
***
## Fill the box with water molecules
Fill the unit cell for the **protein structure system** with water molecules.<br>
The solvent type used is the default **Simple Point Charge water (SPC)**, a generic equilibrated 3-point solvent model.
***
**Building Blocks** used:
- [Solvate](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.solvate) from **biobb_md.gromacs.solvate**
***
### Visualizing 3D structure
Visualizing the **protein system** with the newly added **solvent box** using **NGL**.<br> Note the **cubic box** filled with **water molecules** surrounding the **protein structure**, which is **centered** right in the middle of the cube.
<img src='ngl4.png'></img>
<a id="ions"></a>
***
## Adding ions
Add ions to neutralize the **protein structure** charge
- [Step 1](#ionsStep1): Creating portable binary run file for ion generation
- [Step 2](#ionsStep2): Adding ions to **neutralize** the system
***
**Building Blocks** used:
- [Grompp](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.grompp) from **biobb_md.gromacs.grompp**
- [Genion](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.genion) from **biobb_md.gromacs.genion**
***
<a id="ionsStep1"></a>
### Step 1: Creating portable binary run file for ion generation
A simple **energy minimization** molecular dynamics parameters (mdp) properties will be used to generate the portable binary run file for **ion generation**, although **any legitimate combination of parameters** could be used in this step.
<a id="ionsStep2"></a>
### Step 2: Adding ions to neutralize the system
Replace **solvent molecules** with **ions** to **neutralize** the system.
### Visualizing 3D structure
Visualizing the **neutralized protein system** with the newly added **ions** using **NGL**
<img src='ngl5.png'></img>
<a id="min"></a>
***
## Energetically minimize the system
Energetically minimize the **protein system** till reaching a desired potential energy.
- [Step 1](#emStep1): Creating portable binary run file for energy minimization
- [Step 2](#emStep2): Energetically minimize the **system** till reaching a force of 500 kJ mol-1 nm-1.
- [Step 3](#emStep3): Checking **energy minimization** results. Plotting energy by time during the **minimization** process.
***
**Building Blocks** used:
- [Grompp](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.grompp) from **biobb_md.gromacs.grompp**
- [Mdrun](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.mdrun) from **biobb_md.gromacs.mdrun**
- [GMXEnergy](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_energy) from **biobb_analysis.gromacs.gmx_energy**
***
<a id="emStep1"></a>
### Step 1: Creating portable binary run file for energy minimization
The **minimization** type of the **molecular dynamics parameters (mdp) property** contains the main default parameters to run an **energy minimization**:
- integrator = steep ; Algorithm (steep = steepest descent minimization)
- emtol = 1000.0 ; Stop minimization when the maximum force < 1000.0 kJ/mol/nm
- emstep = 0.01 ; Minimization step size (nm)
- nsteps = 50000 ; Maximum number of (minimization) steps to perform
In this particular example, the method used to run the **energy minimization** is the default **steepest descent**, but the **maximum force** is placed at **500 KJ/mol\*nm^2**, and the **maximum number of steps** to perform (if the maximum force is not reached) to **5,000 steps**.
<a id="emStep2"></a>
### Step 2: Running Energy Minimization
Running **energy minimization** using the **tpr file** generated in the previous step.
<a id="emStep3"></a>
### Step 3: Checking Energy Minimization results
Checking **energy minimization** results. Plotting **potential energy** by time during the minimization process.
<img src='plot1.png'></img>
<a id="nvt"></a>
***
## Equilibrate the system (NVT)
Equilibrate the **protein system** in **NVT ensemble** (constant Number of particles, Volume and Temperature). Protein **heavy atoms** will be restrained using position restraining forces: movement is permitted, but only after overcoming a substantial energy penalty. The utility of position restraints is that they allow us to equilibrate our solvent around our protein, without the added variable of structural changes in the protein.
- [Step 1](#eqNVTStep1): Creating portable binary run file for system equilibration
- [Step 2](#eqNVTStep2): Equilibrate the **protein system** with **NVT** ensemble.
- [Step 3](#eqNVTStep3): Checking **NVT Equilibration** results. Plotting **system temperature** by time during the **NVT equilibration** process.
***
**Building Blocks** used:
- [Grompp](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.grompp) from **biobb_md.gromacs.grompp**
- [Mdrun](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.mdrun) from **biobb_md.gromacs.mdrun**
- [GMXEnergy](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_energy) from **biobb_analysis.gromacs.gmx_energy**
***
<a id="eqNVTStep1"></a>
### Step 1: Creating portable binary run file for system equilibration (NVT)
The **nvt** type of the **molecular dynamics parameters (mdp) property** contains the main default parameters to run an **NVT equilibration** with **protein restraints** (see [GROMACS mdp options](http://manual.gromacs.org/documentation/2018/user-guide/mdp-options.html)):
- Define = -DPOSRES
- integrator = md
- dt = 0.002
- nsteps = 5000
- pcoupl = no
- gen_vel = yes
- gen_temp = 300
- gen_seed = -1
In this particular example, the default parameters will be used: **md** integrator algorithm, a **step size** of **2fs**, **5,000 equilibration steps** with the protein **heavy atoms restrained**, and a temperature of **300K**.
*Please note that for the sake of time this tutorial is only running 10ps of NVT equilibration, whereas in the [original example](http://www.mdtutorials.com/gmx/lysozyme/06_equil.html) the simulated time was 100ps.*
<a id="eqNVTStep2"></a>
### Step 2: Running NVT equilibration
<a id="eqNVTStep3"></a>
### Step 3: Checking NVT Equilibration results
Checking **NVT Equilibration** results. Plotting **system temperature** by time during the NVT equilibration process.
<img src='plot2.png'></img>
<a id="npt"></a>
***
## Equilibrate the system (NPT)
Equilibrate the **protein system** in **NPT** ensemble (constant Number of particles, Pressure and Temperature).
- [Step 1](#eqNPTStep1): Creating portable binary run file for system equilibration
- [Step 2](#eqNPTStep2): Equilibrate the **protein system** with **NPT** ensemble.
- [Step 3](#eqNPTStep3): Checking **NPT Equilibration** results. Plotting **system pressure and density** by time during the **NPT equilibration** process.
***
**Building Blocks** used:
- [Grompp](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.grompp) from **biobb_md.gromacs.grompp**
- [Mdrun](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.mdrun) from **biobb_md.gromacs.mdrun**
- [GMXEnergy](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_energy) from **biobb_analysis.gromacs.gmx_energy**
***
<a id="eqNPTStep1"></a>
### Step 1: Creating portable binary run file for system equilibration (NPT)
The **npt** type of the **molecular dynamics parameters (mdp) property** contains the main default parameters to run an **NPT equilibration** with **protein restraints** (see [GROMACS mdp options](http://manual.gromacs.org/documentation/2018/user-guide/mdp-options.html)):
- Define = -DPOSRES
- integrator = md
- dt = 0.002
- nsteps = 5000
- pcoupl = Parrinello-Rahman
- pcoupltype = isotropic
- tau_p = 1.0
- ref_p = 1.0
- compressibility = 4.5e-5
- refcoord_scaling = com
- gen_vel = no
In this particular example, the default parameters will be used: **md** integrator algorithm, a **time step** of **2fs**, **5,000 equilibration steps** with the protein **heavy atoms restrained**, and a Parrinello-Rahman **pressure coupling** algorithm.
*Please note that for the sake of time this tutorial is only running 10ps of NPT equilibration, whereas in the [original example](http://www.mdtutorials.com/gmx/lysozyme/07_equil2.html) the simulated time was 100ps.*
<a id="eqNPTStep2"></a>
### Step 2: Running NPT equilibration
<a id="eqNPTStep3"></a>
### Step 3: Checking NPT Equilibration results
Checking **NPT Equilibration** results. Plotting **system pressure and density** by time during the **NPT equilibration** process.
<img src='plot3.png'></img>
<a id="free"></a>
***
## Free Molecular Dynamics Simulation
Upon completion of the **two equilibration phases (NVT and NPT)**, the system is now well-equilibrated at the desired temperature and pressure. The **position restraints** can now be released. The last step of the **protein** MD setup is a short, **free MD simulation**, to ensure the robustness of the system.
- [Step 1](#mdStep1): Creating portable binary run file to run a **free MD simulation**.
- [Step 2](#mdStep2): Run short MD simulation of the **protein system**.
- [Step 3](#mdStep3): Checking results for the final step of the setup process, the **free MD run**. Plotting **Root Mean Square deviation (RMSd)** and **Radius of Gyration (Rgyr)** by time during the **free MD run** step.
***
**Building Blocks** used:
- [Grompp](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.grompp) from **biobb_md.gromacs.grompp**
- [Mdrun](https://biobb-md.readthedocs.io/en/latest/gromacs.html#module-gromacs.mdrun) from **biobb_md.gromacs.mdrun**
- [GMXRms](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_rms) from **biobb_analysis.gromacs.gmx_rms**
- [GMXRgyr](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_rgyr) from **biobb_analysis.gromacs.gmx_rgyr**
***
<a id="mdStep1"></a>
### Step 1: Creating portable binary run file to run a free MD simulation
The **free** type of the **molecular dynamics parameters (mdp) property** contains the main default parameters to run an **free MD simulation** (see [GROMACS mdp options](http://manual.gromacs.org/documentation/2018/user-guide/mdp-options.html)):
- integrator = md
- dt = 0.002 (ps)
- nsteps = 50000
In this particular example, the default parameters will be used: **md** integrator algorithm, a **time step** of **2fs**, and a total of **50,000 md steps** (100ps).
*Please note that for the sake of time this tutorial is only running 100ps of free MD, whereas in the [original example](http://www.mdtutorials.com/gmx/lysozyme/08_MD.html) the simulated time was 1ns (1000ps).*
<a id="mdStep2"></a>
### Step 2: Running short free MD simulation
<a id="mdStep3"></a>
### Step 3: Checking free MD simulation results
Checking results for the final step of the setup process, the **free MD run**. Plotting **Root Mean Square deviation (RMSd)** and **Radius of Gyration (Rgyr)** by time during the **free MD run** step. **RMSd** against the **experimental structure** (input structure of the pipeline) and against the **minimized and equilibrated structure** (output structure of the NPT equilibration step).
<img src='plot4.png'></img>
<img src='plot5.png'></img>
<a id="post"></a>
***
## Post-processing and Visualizing resulting 3D trajectory
Post-processing and Visualizing the **protein system** MD setup **resulting trajectory** using **NGL**
- [Step 1](#ppStep1): *Imaging* the resulting trajectory, **stripping out water molecules and ions** and **correcting periodicity issues**.
- [Step 2](#ppStep2): Generating a *dry* structure, **removing water molecules and ions** from the final snapshot of the MD setup pipeline.
- [Step 3](#ppStep3): Visualizing the *imaged* trajectory using the *dry* structure as a **topology**.
***
**Building Blocks** used:
- [GMXImage](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_image) from **biobb_analysis.gromacs.gmx_image**
- [GMXTrjConvStr](https://biobb-analysis.readthedocs.io/en/latest/gromacs.html#module-gromacs.gmx_trjconv_str) from **biobb_analysis.gromacs.gmx_trjconv_str**
***
<a id="ppStep1"></a>
### Step 1: *Imaging* the resulting trajectory.
Stripping out **water molecules and ions** and **correcting periodicity issues**
<a id="ppStep2"></a>
### Step 2: Generating the output *dry* structure.
**Removing water molecules and ions** from the resulting structure
<a id="ppStep3"></a>
### Step 3: Visualizing the generated dehydrated trajectory.
Using the **imaged trajectory** (output of the [Post-processing step 1](#ppStep1)) with the **dry structure** (output of the [Post-processing step 2](#ppStep2)) as a topology.
| 0.879043 | 0.986111 |
# PR-028 DenseNet
김성훈 교수님의 [DenseNet 강의](https://www.youtube.com/watch?v=fe2Vn0mwALI) 감사드립니다.
CIFAR10 데이터를 DenseNet 으로 학습시킵니다.
논문: https://arxiv.org/abs/1608.06993
### Prerequisite
<code> pip install opencv-python</code>
<code> pip install scikit-learn</code>
### Data Set
케라스에서는 손쉽게 CIFAR-10 데이터를 쓸 수 있도록 api를 제공해 줍니다.
또한 [data generator](https://keras.io/preprocessing/image/)를 이용하여 augmentation을 쉽게 사용할 수 있습니다.
```
import numpy as np
from keras.datasets import cifar10
from keras.preprocessing.image import ImageDataGenerator
from keras.utils.np_utils import to_categorical
from sklearn.model_selection import train_test_split
(x_train, y_train), (x_test, y_test) = cifar10.load_data()
# normalization
x_train = np.array(x_train) / 127.5 - 1
x_test = np.array(x_test) / 127.5 - 1
# one-hot encoding
y_train = np.array(y_train)
y_test = np.array(y_test)
y_train = to_categorical(y_train)
y_test = to_categorical(y_test)
# train data의 1/4을 validation set으로 활용합니다.
x_train, x_val, y_train, y_val = train_test_split(x_train, y_train, test_size=0.2)
datagen = ImageDataGenerator(
rotation_range=20,
width_shift_range=0.1,
height_shift_range=0.1,
zoom_range=0.1,
horizontal_flip=True
)
```
### Build Model
논문에서는 크게 아래 사진과 같이 4개의 구조를 만들었습니다.

DenseNet-121 을 조금만 변형해서 사용해 봅니다.
input_shape 를 (32, 32, 3) 으로 입력시켜 주고
끝에 붙는 fully connected layer 의 크기를 10 으로 사용합니다.
```
from keras.models import Model
from keras.regularizers import l2
from keras.layers.normalization import BatchNormalization
from keras.layers.merge import concatenate
from keras.layers import Conv2D, Activation, Input, Dense
from keras.layers.pooling import MaxPooling2D, AveragePooling2D, GlobalAveragePooling2D
from keras.optimizers import Adam
# hyperparameters
K = 32
init_ch = 64 # 가장 첫 layer의 채널 수 입니다.
compression = 0.5 # transition layer 에서의 channel을 얼마나 줄일 것인지 비율입니다. 논문에서도 0.5를 사용하였습니다.
inp_shape = (32, 32, 3)
# conv_layer를 간편하게 쓰기 위해 wrapper 입니다.
def conv_layer(x, filters, k, s):
return Conv2D(filters,
kernel_size=(k, k),
strides=(s, s),
padding='same',
kernel_regularizer=l2(0.01))(x)
def dense_block(x, dense_block_size):
for i in range(dense_block_size):
tmp = x
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = conv_layer(x, 4*K, 1, 1)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = conv_layer(x, K, 3, 1)
x = concatenate([x, tmp])
return x
def transition_block(x):
shape = x.get_shape().as_list() # output 의 shape을 가져옵니다.
compressed_channels = int(shape[3] * compression) # compression 비율만큼 channel을 줄여 줍니다.
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = conv_layer(x, compressed_channels, 1, 1)
x = AveragePooling2D(pool_size=(2, 2), strides=(2, 2))(x)
return x
input_img = Input(inp_shape)
x = conv_layer(input_img, init_ch, 7, 2)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = MaxPooling2D(pool_size=(3, 3), strides=(2, 2), padding='same')(x)
x = dense_block(x, 6)
x = transition_block(x)
x = dense_block(x, 12)
x = transition_block(x)
x = dense_block(x, 24)
x = transition_block(x)
x = dense_block(x, 16)
x = GlobalAveragePooling2D()(x)
x = Dense(10, activation='softmax')(x)
dense121 = Model(input_img, x)
```
### Train Model
```
# callbacks
from keras.callbacks import ModelCheckpoint, EarlyStopping, LearningRateScheduler
import math
def lr_schedule(epoch):
return max(5e-5, 5e-4 * math.pow(0.7, epoch // 5))
lr_scheduler = LearningRateScheduler(lr_schedule)
early_stopping = EarlyStopping(monitor='val_loss', patience=10)
checkpoint = ModelCheckpoint(filepath='model_{epoch:02d}.h5',
save_best_only=True,
save_weights_only=True)
dense121.compile(optimizer=Adam(0.0), loss='categorical_crossentropy', metrics=['accuracy'])
history = dense121.fit_generator(datagen.flow(x_train, y_train, batch_size=32),
steps_per_epoch=x_train.shape[0] // 32,
validation_data=datagen.flow(x_val, y_val, batch_size=8),
validation_steps=x_val.shape[0] // 8,
epochs = 100,
callbacks=[early_stopping, checkpoint, lr_scheduler])
import matplotlib.pyplot as plt
%matplotlib inline
plt.subplot(211)
plt.title("accuracy")
plt.plot(history.history["acc"], color="r", label="train")
plt.plot(history.history["val_acc"], color="b", label="val")
plt.legend(loc="best")
plt.subplot(212)
plt.title("loss")
plt.plot(history.history["loss"], color="r", label="train")
plt.plot(history.history["val_loss"], color="b", label="val")
plt.legend(loc="best")
plt.tight_layout()
plt.show()
```
### Evaluate Model
```
score = dense121.evaluate(x_test, y_test)
print('test score:', score[0])
print('test accuracy:', score[1])
```
### Tip!
[keras application](https://keras.io/applications/#densenet)의 densenet을 사용하면 매우 편리하게 imagenet으로 학습된 모델을 가져올 수 있습니다.
```
from keras.applications import densenet
model = densenet.DenseNet121()
```
## Contact me
케라스를 사랑하는 개발자 입니다.
질문, 조언, contribtuion 등 소통은 언제나 환영합니다.
Anthony Kim(김동현) : artit.anthony@gmail.com
|
github_jupyter
|
import numpy as np
from keras.datasets import cifar10
from keras.preprocessing.image import ImageDataGenerator
from keras.utils.np_utils import to_categorical
from sklearn.model_selection import train_test_split
(x_train, y_train), (x_test, y_test) = cifar10.load_data()
# normalization
x_train = np.array(x_train) / 127.5 - 1
x_test = np.array(x_test) / 127.5 - 1
# one-hot encoding
y_train = np.array(y_train)
y_test = np.array(y_test)
y_train = to_categorical(y_train)
y_test = to_categorical(y_test)
# train data의 1/4을 validation set으로 활용합니다.
x_train, x_val, y_train, y_val = train_test_split(x_train, y_train, test_size=0.2)
datagen = ImageDataGenerator(
rotation_range=20,
width_shift_range=0.1,
height_shift_range=0.1,
zoom_range=0.1,
horizontal_flip=True
)
from keras.models import Model
from keras.regularizers import l2
from keras.layers.normalization import BatchNormalization
from keras.layers.merge import concatenate
from keras.layers import Conv2D, Activation, Input, Dense
from keras.layers.pooling import MaxPooling2D, AveragePooling2D, GlobalAveragePooling2D
from keras.optimizers import Adam
# hyperparameters
K = 32
init_ch = 64 # 가장 첫 layer의 채널 수 입니다.
compression = 0.5 # transition layer 에서의 channel을 얼마나 줄일 것인지 비율입니다. 논문에서도 0.5를 사용하였습니다.
inp_shape = (32, 32, 3)
# conv_layer를 간편하게 쓰기 위해 wrapper 입니다.
def conv_layer(x, filters, k, s):
return Conv2D(filters,
kernel_size=(k, k),
strides=(s, s),
padding='same',
kernel_regularizer=l2(0.01))(x)
def dense_block(x, dense_block_size):
for i in range(dense_block_size):
tmp = x
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = conv_layer(x, 4*K, 1, 1)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = conv_layer(x, K, 3, 1)
x = concatenate([x, tmp])
return x
def transition_block(x):
shape = x.get_shape().as_list() # output 의 shape을 가져옵니다.
compressed_channels = int(shape[3] * compression) # compression 비율만큼 channel을 줄여 줍니다.
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = conv_layer(x, compressed_channels, 1, 1)
x = AveragePooling2D(pool_size=(2, 2), strides=(2, 2))(x)
return x
input_img = Input(inp_shape)
x = conv_layer(input_img, init_ch, 7, 2)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = MaxPooling2D(pool_size=(3, 3), strides=(2, 2), padding='same')(x)
x = dense_block(x, 6)
x = transition_block(x)
x = dense_block(x, 12)
x = transition_block(x)
x = dense_block(x, 24)
x = transition_block(x)
x = dense_block(x, 16)
x = GlobalAveragePooling2D()(x)
x = Dense(10, activation='softmax')(x)
dense121 = Model(input_img, x)
# callbacks
from keras.callbacks import ModelCheckpoint, EarlyStopping, LearningRateScheduler
import math
def lr_schedule(epoch):
return max(5e-5, 5e-4 * math.pow(0.7, epoch // 5))
lr_scheduler = LearningRateScheduler(lr_schedule)
early_stopping = EarlyStopping(monitor='val_loss', patience=10)
checkpoint = ModelCheckpoint(filepath='model_{epoch:02d}.h5',
save_best_only=True,
save_weights_only=True)
dense121.compile(optimizer=Adam(0.0), loss='categorical_crossentropy', metrics=['accuracy'])
history = dense121.fit_generator(datagen.flow(x_train, y_train, batch_size=32),
steps_per_epoch=x_train.shape[0] // 32,
validation_data=datagen.flow(x_val, y_val, batch_size=8),
validation_steps=x_val.shape[0] // 8,
epochs = 100,
callbacks=[early_stopping, checkpoint, lr_scheduler])
import matplotlib.pyplot as plt
%matplotlib inline
plt.subplot(211)
plt.title("accuracy")
plt.plot(history.history["acc"], color="r", label="train")
plt.plot(history.history["val_acc"], color="b", label="val")
plt.legend(loc="best")
plt.subplot(212)
plt.title("loss")
plt.plot(history.history["loss"], color="r", label="train")
plt.plot(history.history["val_loss"], color="b", label="val")
plt.legend(loc="best")
plt.tight_layout()
plt.show()
score = dense121.evaluate(x_test, y_test)
print('test score:', score[0])
print('test accuracy:', score[1])
from keras.applications import densenet
model = densenet.DenseNet121()
| 0.87142 | 0.906073 |
# Brain network exploration with `neurolib`
In this example, we will run a parameter exploration of a whole-brain model that we load using the brain simulation framework `neurolib`. Please visit the [Github repo](https://github.com/neurolib-dev/neurolib) to learn more about this library or read the [gentle introduction to `neurolib`](https://caglorithm.github.io/notebooks/neurolib-intro/) to learn more about the neuroscience background of neural mass models and whole-brain simulations.
```
# change into the root directory of the project
import os
if os.getcwd().split("/")[-1] == "examples":
os.chdir('..')
%load_ext autoreload
%autoreload 2
import logging
logger = logging.getLogger()
logger.setLevel(logging.INFO)
!pip install matplotlib
import matplotlib.pyplot as plt
import numpy as np
# a nice color map
plt.rcParams['image.cmap'] = 'plasma'
!pip install neurolib
from neurolib.models.aln import ALNModel
from neurolib.utils.loadData import Dataset
import neurolib.utils.functions as func
ds = Dataset("hcp")
import mopet
```
We load a model with parameters that generate interesting dynamics.
```
model = ALNModel(Cmat = ds.Cmat, Dmat = ds.Dmat)
model.params['duration'] = 0.2*60*1000
model.params['mue_ext_mean'] = 1.57
model.params['mui_ext_mean'] = 1.6
# We set an appropriate level of noise
model.params['sigma_ou'] = 0.09
# And turn on adaptation with a low value of spike-triggered adaptation currents.
model.params['b'] = 5.0
```
Let's run it to see what kind of output it produces!
```
model.run(bold=True, chunkwise=True)
plt.plot(model.output.T);
```
We simualted the model with BOLD output, so let's compute the functional connectivity (fc) matrix:
```
plt.imshow(func.fc(model.BOLD.BOLD[:, model.BOLD.t_BOLD > 5000]))
```
This is our multi-stage evaluation function.
```
def evaluateSimulation(params):
model.params.update(params)
defaultDuration = model.params['duration']
invalid_result = {"fc" : [0]* len(ds.BOLDs)}
logging.info("Running stage 1")
# -------- stage wise simulation --------
# Stage 1 : simulate for a few seconds to see if there is any activity
# ---------------------------------------
model.params['duration'] = 3*1000.
model.run()
# check if stage 1 was successful
amplitude = np.max(model.output[:, model.t > 500]) - np.min(model.output[:, model.t > 500])
if amplitude < 0.05:
invalid_result = {"fc" : 0}
return invalid_result
logging.info("Running stage 2")
# Stage 2: simulate BOLD for a few seconds to see if it moves
# ---------------------------------------
model.params['duration'] = 20*1000.
model.run(bold = True, chunkwise=True)
if np.std(model.BOLD.BOLD[:, 5:10]) < 0.0001:
invalid_result = {"fc" : -1}
return invalid_result
logging.info("Running stage 3")
# Stage 3: full and final simulation
# ---------------------------------------
model.params['duration'] = defaultDuration
model.run(bold = True, chunkwise=True)
# -------- evaluation here --------
scores = []
for i, fc in enumerate(ds.FCs):#range(len(ds.FCs)):
fc_score = func.matrix_correlation(func.fc(model.BOLD.BOLD[:, 5:]), fc)
scores.append(fc_score)
meanScore = np.mean(scores)
result_dict = {"fc" : meanScore}
return result_dict
```
We test run the evaluation function.
```
model.params['duration'] = 20*1000.
evaluateSimulation(model.params)
# NOTE: These values are low for testing
model.params['duration'] = 10*1000.
explore_params = {"a": np.linspace(0, 40.0, 2)
,"K_gl": np.linspace(100, 400, 2)
,"sigma_ou" : np.linspace(0.1, 0.5, 2)
}
# we need this random filename to avoid testing clashes
hdf_filename = f"exploration-{np.random.randint(99999)}.h5"
ex = mopet.Exploration(evaluateSimulation, explore_params, default_params=model.params, hdf_filename=hdf_filename)
ex.run()
ex.load_results(as_dict=True)
ex.results
ex.params
ex.df
sigma_selectors = np.unique(ex.df.sigma_ou)
for s in sigma_selectors:
df = ex.df[(ex.df.sigma_ou == s)]
pivotdf = df.pivot_table(values='fc', index = 'K_gl', columns='a')
plt.imshow(pivotdf, \
extent = [min(df.a), max(df.a),
min(df.K_gl), max(df.K_gl)], origin='lower', aspect='auto')
plt.colorbar(label='Mean correlation to empirical rs-FC')
plt.xlabel("a")
plt.ylabel("K_gl")
plt.title("$\sigma_{ou}$" + "={}".format(s))
plt.show()
```
|
github_jupyter
|
# change into the root directory of the project
import os
if os.getcwd().split("/")[-1] == "examples":
os.chdir('..')
%load_ext autoreload
%autoreload 2
import logging
logger = logging.getLogger()
logger.setLevel(logging.INFO)
!pip install matplotlib
import matplotlib.pyplot as plt
import numpy as np
# a nice color map
plt.rcParams['image.cmap'] = 'plasma'
!pip install neurolib
from neurolib.models.aln import ALNModel
from neurolib.utils.loadData import Dataset
import neurolib.utils.functions as func
ds = Dataset("hcp")
import mopet
model = ALNModel(Cmat = ds.Cmat, Dmat = ds.Dmat)
model.params['duration'] = 0.2*60*1000
model.params['mue_ext_mean'] = 1.57
model.params['mui_ext_mean'] = 1.6
# We set an appropriate level of noise
model.params['sigma_ou'] = 0.09
# And turn on adaptation with a low value of spike-triggered adaptation currents.
model.params['b'] = 5.0
model.run(bold=True, chunkwise=True)
plt.plot(model.output.T);
plt.imshow(func.fc(model.BOLD.BOLD[:, model.BOLD.t_BOLD > 5000]))
def evaluateSimulation(params):
model.params.update(params)
defaultDuration = model.params['duration']
invalid_result = {"fc" : [0]* len(ds.BOLDs)}
logging.info("Running stage 1")
# -------- stage wise simulation --------
# Stage 1 : simulate for a few seconds to see if there is any activity
# ---------------------------------------
model.params['duration'] = 3*1000.
model.run()
# check if stage 1 was successful
amplitude = np.max(model.output[:, model.t > 500]) - np.min(model.output[:, model.t > 500])
if amplitude < 0.05:
invalid_result = {"fc" : 0}
return invalid_result
logging.info("Running stage 2")
# Stage 2: simulate BOLD for a few seconds to see if it moves
# ---------------------------------------
model.params['duration'] = 20*1000.
model.run(bold = True, chunkwise=True)
if np.std(model.BOLD.BOLD[:, 5:10]) < 0.0001:
invalid_result = {"fc" : -1}
return invalid_result
logging.info("Running stage 3")
# Stage 3: full and final simulation
# ---------------------------------------
model.params['duration'] = defaultDuration
model.run(bold = True, chunkwise=True)
# -------- evaluation here --------
scores = []
for i, fc in enumerate(ds.FCs):#range(len(ds.FCs)):
fc_score = func.matrix_correlation(func.fc(model.BOLD.BOLD[:, 5:]), fc)
scores.append(fc_score)
meanScore = np.mean(scores)
result_dict = {"fc" : meanScore}
return result_dict
model.params['duration'] = 20*1000.
evaluateSimulation(model.params)
# NOTE: These values are low for testing
model.params['duration'] = 10*1000.
explore_params = {"a": np.linspace(0, 40.0, 2)
,"K_gl": np.linspace(100, 400, 2)
,"sigma_ou" : np.linspace(0.1, 0.5, 2)
}
# we need this random filename to avoid testing clashes
hdf_filename = f"exploration-{np.random.randint(99999)}.h5"
ex = mopet.Exploration(evaluateSimulation, explore_params, default_params=model.params, hdf_filename=hdf_filename)
ex.run()
ex.load_results(as_dict=True)
ex.results
ex.params
ex.df
sigma_selectors = np.unique(ex.df.sigma_ou)
for s in sigma_selectors:
df = ex.df[(ex.df.sigma_ou == s)]
pivotdf = df.pivot_table(values='fc', index = 'K_gl', columns='a')
plt.imshow(pivotdf, \
extent = [min(df.a), max(df.a),
min(df.K_gl), max(df.K_gl)], origin='lower', aspect='auto')
plt.colorbar(label='Mean correlation to empirical rs-FC')
plt.xlabel("a")
plt.ylabel("K_gl")
plt.title("$\sigma_{ou}$" + "={}".format(s))
plt.show()
| 0.5564 | 0.962603 |
# string
Os objetos do tipo `string` são cadeias de caracteres, armazenadas em sequência na memória do computador. Para criar um string, podemos utilizar a função `str` ou escrever valores entre aspas.
Os strings funcionam como listas de caracteres, onde podemos acessar valores específicos com base em seu índice e fatiá-los.
```
print("Indexação: string[início:fim:salto]\n")
string = "ABC123"
print("String: ", string)
print("Primeira posição: ", string[0])
print("Última posição: ", string[-1])
print("Substring [1:3]:", string[1:3])
print("String invertido:", string[::-1])
print("Caracteres ímpares:", string[::2])
```
## Operações com strings
Os strings são imutáveis, logo não é possível modificar qualquer caractere diretamente com base em seu índice. A solução geralmente envolve fatiar o string ou utilizar os métodos de formatação disponibilizados para sua classe. Porém, é permitido somar (concatenar) e multiplicar (repetir) strings.
```
string_1 = "ABCDEF"
string_2 = "123456"
print("Strings: ", string_1, "e", string_2)
print("Soma: ", string_1 + string_2)
print("Produto: ", string_1 * 2)
```
O operador `in` permite verificar se um substring faz parte de um string
```
print("String: ", string)
print("3" in string)
print("C" not in string)
```
## Funções aplicáveis a strings
#### len( )
Permite encontrar o tamanho de um string
```
print("Tamanho: ", len(string_1), "caracteres")
```
#### input( )
A função `input()` permite que ao rodar o programa, o processo de leitura do código é interrompido até que o usuário digite algo diretamente no console do Python. O valor é lido como string e pode ser atribuído a uma variável.
```
entrada = input("Digite um valor: ")
print("Valor imputado: ", entrada)
```
## Métodos Importantes
#### Alfanuméricos
* string.isalnum() -> verifica se um string só contêm carcateres alfabéticos e numéricos
* string.isalpha() -> verifica se um string só contêm carcateres alfabéticos
* string.isdigit() -> verifica se um string só contêm os dígitos de 0 a 9
* string.isnumeric() -> verifica se um string só contêm carcateres numéricos e representações Unicode de outros dígitos. Ex: "\u2153"
* string.lower() -> retorna novo string com os caracteres em formato maiúsculo
* string.upper() -> retorna novo string com os caracteres em formato minúsculo
```
string = "ABCD1234"
print(string.isalnum())
print(string.isalpha())
print(string.isdigit())
print(string.isnumeric())
print(string.lower())
```
#### Alinhamento e ajuste
* string.center(quantidade, caracter) -> alinha o texto no centro do string e preenche o entorno com o caractere especificado
* string.ljust(quantidade, caracter) -> alinha o texto à esquerda do string e preenche com o caractere especificado
* string.rjust(quantidade, caracter) -> alinha o texto à direita do string e preenche com o caractere especificado
* string.strip(caracteres) -> remove todos os espaços em branco à esquerda e à direita do início e fim do string, respectivamente. Se passarmos caracteres como parâmetro, remove as suas ocorrências.
* string.lstrip() -> remove todos os espaços em branco à esquerda do início do string
* string.rstrip() -> remove todos os espaços em branco à direita do fim do string
```
string = "string"
print(string.center(10))
print(string.center(10, "*"))
print(string.ljust(10, "*"))
print(string.rjust(10, "*"))
string = " ***string*** "
print(string.strip())
print(string.strip(" *")) # remove tanto os espaços quanto o caractere *
print(string.lstrip(" *"))
print(string.rstrip(" *"))
```
#### Localização
* string.startswith("sub") -> verifica se o string começa com determinado substring
* string.endswith("sub") -> verifica se o string termina com determinado substring
* string.count(sub) -> retorna o número de ocorrências de um substring dentro do string original
* string.find(sub, inicio, fim) -> retorna a posição da primeira ocorrência do substring procurado, da esquerda para a direita (se não encontrar, retorna -1). Inicio e Fim são as posições a partir das quais a função inicia e termina a procura.
* string.rfind() -> retorna a posição da primeira ocorrência do substring procurado, da direita para a esquerda (se não encontrar, retorna -1). Inicio e Fim são as posições a partir das quais a função inicia e termina a procura.
```
string = "ABCDABCD"
print(string.startswith("ABC"))
print(string.endswith("ABC"))
string = "ABCDABCD"
print(string.count("ABC"))
print(string.find("BCD"))
print(string.rfind("BCD"))
```
#### Substituição
* string.replace(sub_antes, sub_depois, numero) -> substitui as ocorrências de um substring por outro, preservando o string original. O parâmetro numérico indica a quantidade máxima de reposições que devem ser feitas, da esquerda para a direita
* string.split(caracter) -> divide o string com base em um caractere, criando uma lista de substrings. Para criar uma lista separando cada caractere, usamos a função `list()`.
* string.splitlines(numero) -> divide o string com base no substring "\n", que ao ser printado pula uma linha. Se adicionarmos um número maior que zero como parâmetro, ele inclui o caractere ao final de cada divisão. Retorna uma lista de substrings
* sep.join(lista) -> concatena os substrings contidos em uma lista ou tupla, com base em um separador
```
string_1 = "ABCDEABCD"
print("string:", string_1)
print(string_1.replace("A", "F", 2))
print(string_1.split("E"))
lista = list(string_1)
print("string:", lista)
print(" ".join(lista))
string_2 = "ABCDE\nABCD"
print(string_2.splitlines())
print(string_2.splitlines(1))
```
#### Formatação de strings
##### string.format( )
O método `string.format()` indica os valores do string a serem substituídos, com um inteiro entre chaves indicando onde serão encaixados os parâmetros a serem passados ao método format(), que podem ser tanto substrings quanto valores numéricos.
Caso sejam colocados números entre as chaves, eles se referirão a qual parâmetro deverá ser encaixado naquele local. Ex: {0} -> O número 0 se refere ao primeiro parâmetro.
```
"ABC{1}DEF12{0}3456".format(" par_0 ", " par_1 ")
```
Se incluirmos uma lista como parâmetro, podemos indicar o índice do elemento a ser colocado dentro do string original. Ex: {0[0]}
```
print("String original: ABCD1234")
"ABC{1}D12{0[1]}34".format(["1", "2"], "3")
```
Dentro das chaves também pode ser especificado o tamanho desejado para o string que vamos colocar no lugar das chaves. Isso é feito com `:` após indicar a qual parâmetro nos referimos, se desejado. Se indicarmos um valor maior do que a quantidade de caracteres contida no parâmetro, o restante será preenchido à sua direita. Ex: {0:10}.
```
"{0:12}".format("string")
```
Os valores utilizados como preenchimento podem ser especificados após os dois-pontos, e depois deles podemos indicar também se o preenchimento ocorrerá à direita `<` do substring, à esquerda `>` ou ambos `^` (um a um, da direita para a esquerda). Ex: {:.^10}
```
"{0:*>12}".format("string")
```
Formatação exclusiva de números:
* {0:03} -> especifica a quantidade de zeros a serem incluídos à esquerda do número, para preenchimento do tamanho desejado
* {0:,} -> define agruplamento por milhar
* {0:+} -> mostra o sinal existente
* {0:d} -> para inserir o número em outra base, utilizamos letras antes do tamanho.
* d: base decimal
* b: binário
* o: octal
* x: hexadecimal minúsculo
* X: hexadecimal maiúsculo
* c: caractere UNICODE
* {0:.5f} -> impressão de número com 5 casas decimais
* f: float
* e: notação científica com `e` minúsculo
* E: notação científica com `e` maiúsculo
* %: converte o valor para porcentagem. Ex: 9,87 = 987%
```
print("Preenchimento com 10 zeros:", "{0:010}".format(123456))
print("Separador de milhar:", "{0:,}".format(123456))
print("Mostrar o sinal:", "{0:+}".format(123456))
print("Base Binária:", "{0:b}".format(123456))
print("Notação científica com 5 casas decimais:", "{0:.5e}".format(123456))
```
##### f-strings
Uma forma mais rápida de formatar strings ocorre utilizando `f` atrás da string. Isso possibilita que sejam incluídos os valores desejados dentro das chaves como se fossem os parâmetros da função format. Os valores dentro das chaves podem ser chamadas de funções, operações matemáticas, nomes de variáveis já criadas etc.
```
string = "texto"
print(f"{string}")
```
|
github_jupyter
|
print("Indexação: string[início:fim:salto]\n")
string = "ABC123"
print("String: ", string)
print("Primeira posição: ", string[0])
print("Última posição: ", string[-1])
print("Substring [1:3]:", string[1:3])
print("String invertido:", string[::-1])
print("Caracteres ímpares:", string[::2])
string_1 = "ABCDEF"
string_2 = "123456"
print("Strings: ", string_1, "e", string_2)
print("Soma: ", string_1 + string_2)
print("Produto: ", string_1 * 2)
print("String: ", string)
print("3" in string)
print("C" not in string)
print("Tamanho: ", len(string_1), "caracteres")
entrada = input("Digite um valor: ")
print("Valor imputado: ", entrada)
string = "ABCD1234"
print(string.isalnum())
print(string.isalpha())
print(string.isdigit())
print(string.isnumeric())
print(string.lower())
string = "string"
print(string.center(10))
print(string.center(10, "*"))
print(string.ljust(10, "*"))
print(string.rjust(10, "*"))
string = " ***string*** "
print(string.strip())
print(string.strip(" *")) # remove tanto os espaços quanto o caractere *
print(string.lstrip(" *"))
print(string.rstrip(" *"))
string = "ABCDABCD"
print(string.startswith("ABC"))
print(string.endswith("ABC"))
string = "ABCDABCD"
print(string.count("ABC"))
print(string.find("BCD"))
print(string.rfind("BCD"))
string_1 = "ABCDEABCD"
print("string:", string_1)
print(string_1.replace("A", "F", 2))
print(string_1.split("E"))
lista = list(string_1)
print("string:", lista)
print(" ".join(lista))
string_2 = "ABCDE\nABCD"
print(string_2.splitlines())
print(string_2.splitlines(1))
"ABC{1}DEF12{0}3456".format(" par_0 ", " par_1 ")
print("String original: ABCD1234")
"ABC{1}D12{0[1]}34".format(["1", "2"], "3")
"{0:12}".format("string")
"{0:*>12}".format("string")
print("Preenchimento com 10 zeros:", "{0:010}".format(123456))
print("Separador de milhar:", "{0:,}".format(123456))
print("Mostrar o sinal:", "{0:+}".format(123456))
print("Base Binária:", "{0:b}".format(123456))
print("Notação científica com 5 casas decimais:", "{0:.5e}".format(123456))
string = "texto"
print(f"{string}")
| 0.149159 | 0.92779 |
# Basic RoBERTa Fine-tuning Example
In this notebook, we will:
* Train a RoBERTa base model on MRPC and evaluate its performance
## Setup
#### Install dependencies
First, we will install libraries we need for this code.
```
%%capture
!git clone https://github.com/nyu-mll/jiant.git
%cd jiant
!pip install -r requirements-no-torch.txt
!pip install --no-deps -e ./
```
#### Download data
Next, we will download task data.
```
%%capture
# Download MRPC data
!PYTHONPATH=/content/jiant python jiant/jiant/scripts/download_data/runscript.py \
download \
--tasks mrpc \
--output_path=/content/tasks/
```
## `jiant` Pipeline
```
import sys
sys.path.insert(0, "/content/jiant")
import jiant.proj.main.tokenize_and_cache as tokenize_and_cache
import jiant.proj.main.export_model as export_model
import jiant.proj.main.scripts.configurator as configurator
import jiant.proj.main.runscript as main_runscript
import jiant.shared.caching as caching
import jiant.utils.python.io as py_io
import jiant.utils.display as display
import os
```
#### Download model
Next, we will download a `roberta-base` model. This also includes the tokenizer.
```
export_model.lookup_and_export_model(
model_type="roberta-base",
output_base_path="./models/roberta-base",
)
```
#### Tokenize and cache
With the model and data ready, we can now tokenize and cache the inputs features for our task. This converts the input examples to tokenized features ready to be consumed by the model, and saved them to disk in chunks.
```
# Tokenize and cache each task
task_name = "mrpc"
tokenize_and_cache.main(tokenize_and_cache.RunConfiguration(
task_config_path=f"./tasks/configs/{task_name}_config.json",
model_type="roberta-base",
model_tokenizer_path="./models/roberta-base/tokenizer",
output_dir=f"./cache/{task_name}",
phases=["train", "val"],
))
```
We can inspect the first examples of the first chunk of each task.
```
row = caching.ChunkedFilesDataCache("./cache/mrpc/train").load_chunk(0)[0]["data_row"]
print(row.input_ids)
print(row.tokens)
```
#### Writing a run config
Here we are going to write what we call a `jiant_task_container_config`. This configuration file basically defines a lot of the subtleties of our training pipeline, such as what tasks we will train on, do evaluation on, batch size for each task. The new version of `jiant` leans heavily toward explicitly specifying everything, for the purpose of inspectability and leaving minimal surprises for the user, even as the cost of being more verbose.
We use a helper "Configurator" to write out a `jiant_task_container_config`, since most of our setup is pretty standard.
**Depending on what GPU your Colab session is assigned to, you may need to lower the train batch size.**
```
jiant_run_config = configurator.SimpleAPIMultiTaskConfigurator(
task_config_base_path="./tasks/configs",
task_cache_base_path="./cache",
train_task_name_list=["mrpc"],
val_task_name_list=["mrpc"],
train_batch_size=8,
eval_batch_size=16,
epochs=3,
num_gpus=1,
).create_config()
os.makedirs("./run_configs/", exist_ok=True)
py_io.write_json(jiant_run_config, "./run_configs/mrpc_run_config.json")
display.show_json(jiant_run_config)
```
To briefly go over the major components of the `jiant_task_container_config`:
* `task_config_path_dict`: The paths to the task config files we wrote above.
* `task_cache_config_dict`: The paths to the task features caches we generated above.
* `sampler_config`: Determines how to sample from different tasks during training.
* `global_train_config`: The number of total steps and warmup steps during training.
* `task_specific_configs_dict`: Task-specific arguments for each task, such as training batch size and gradient accumulation steps.
* `taskmodels_config`: Task-model specific arguments for each task-model, including what tasks use which model.
* `metric_aggregator_config`: Determines how to weight/aggregate the metrics across multiple tasks.
#### Start training
Finally, we can start our training run.
Before starting training, the script also prints out the list of parameters in our model.
```
run_args = main_runscript.RunConfiguration(
jiant_task_container_config_path="./run_configs/mrpc_run_config.json",
output_dir="./runs/mrpc",
model_type="roberta-base",
model_path="./models/roberta-base/model/roberta-base.p",
model_config_path="./models/roberta-base/model/roberta-base.json",
model_tokenizer_path="./models/roberta-base/tokenizer",
learning_rate=1e-5,
eval_every_steps=500,
do_train=True,
do_val=True,
do_save=True,
force_overwrite=True,
)
main_runscript.run_loop(run_args)
```
At the end, you should see the evaluation scores for MRPC.
|
github_jupyter
|
%%capture
!git clone https://github.com/nyu-mll/jiant.git
%cd jiant
!pip install -r requirements-no-torch.txt
!pip install --no-deps -e ./
%%capture
# Download MRPC data
!PYTHONPATH=/content/jiant python jiant/jiant/scripts/download_data/runscript.py \
download \
--tasks mrpc \
--output_path=/content/tasks/
import sys
sys.path.insert(0, "/content/jiant")
import jiant.proj.main.tokenize_and_cache as tokenize_and_cache
import jiant.proj.main.export_model as export_model
import jiant.proj.main.scripts.configurator as configurator
import jiant.proj.main.runscript as main_runscript
import jiant.shared.caching as caching
import jiant.utils.python.io as py_io
import jiant.utils.display as display
import os
export_model.lookup_and_export_model(
model_type="roberta-base",
output_base_path="./models/roberta-base",
)
# Tokenize and cache each task
task_name = "mrpc"
tokenize_and_cache.main(tokenize_and_cache.RunConfiguration(
task_config_path=f"./tasks/configs/{task_name}_config.json",
model_type="roberta-base",
model_tokenizer_path="./models/roberta-base/tokenizer",
output_dir=f"./cache/{task_name}",
phases=["train", "val"],
))
row = caching.ChunkedFilesDataCache("./cache/mrpc/train").load_chunk(0)[0]["data_row"]
print(row.input_ids)
print(row.tokens)
jiant_run_config = configurator.SimpleAPIMultiTaskConfigurator(
task_config_base_path="./tasks/configs",
task_cache_base_path="./cache",
train_task_name_list=["mrpc"],
val_task_name_list=["mrpc"],
train_batch_size=8,
eval_batch_size=16,
epochs=3,
num_gpus=1,
).create_config()
os.makedirs("./run_configs/", exist_ok=True)
py_io.write_json(jiant_run_config, "./run_configs/mrpc_run_config.json")
display.show_json(jiant_run_config)
run_args = main_runscript.RunConfiguration(
jiant_task_container_config_path="./run_configs/mrpc_run_config.json",
output_dir="./runs/mrpc",
model_type="roberta-base",
model_path="./models/roberta-base/model/roberta-base.p",
model_config_path="./models/roberta-base/model/roberta-base.json",
model_tokenizer_path="./models/roberta-base/tokenizer",
learning_rate=1e-5,
eval_every_steps=500,
do_train=True,
do_val=True,
do_save=True,
force_overwrite=True,
)
main_runscript.run_loop(run_args)
| 0.29584 | 0.855127 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.