code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
<a href="https://colab.research.google.com/github/shanaka-desoysa/notes/blob/main/docs/blockchain/Blockchain_Explained_in_7_Simple_Functions.ipynb" target="_blank"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
---
# Blockchain Explained in 7 Simple Functions
---
Practical hands-on guide to implement your own blockchain with 7 simple Python functions.
## Hashing Function
At the heart of the blockchain is the hashing function. Without encryption, the blockchain will be easily manipulable and transactions will be able to be fraudulently inserted. Here we're using a simple MD5 hashing algorithm. If you're interested in what's actually being used in bitcoin, read [here](https://en.bitcoin.it/wiki/Block_hashing_algorithm).
```
import hashlib
import json
def hash_function(k):
"""Hashes our transaction."""
if type(k) is not str:
k = json.dumps(k, sort_keys=True)
return hashlib.sha256(k.encode('utf-8')).hexdigest()
hash_function('www.geni.ai')
```
## State Management
The ‘state’ is the record of who owns what. For example, *[Geni AI](https://www.geni.ai)* have 100 coins and give 5 to *John Smith*, then the state will be the value of the dictionary below.
`{'transaction': {'Geni AI': 95, 'John Smith': 5}}`
```
def update_state(transaction, state):
state = state.copy()
for key in transaction:
if key in state.keys():
state[key] += transaction[key]
else:
state[key] = transaction[key]
return state
```
## Transaction Validation
The important thing to note is that overdrafts cannot exist. If there are only 10 coins in existence, then I cannot give 11 coins to someone. The below function verifies that the transaction we attempt to make is indeed valid. Also, a transaction must balance. I cannot give 5 coins and have the recipient receive 4 coins, since that would allow the destruction and creation of coins.
```
def valid_transaction(transaction, state):
"""A valid transaction must sum to 0."""
if sum(transaction.values()) is not 0:
return False
for key in transaction.keys():
if key in state.keys():
account_balance = state[key]
else:
account_balance = 0
if account_balance + transaction[key] < 0:
return False
return True
```
## Make Block
Now, we can make our block. The information from the previous block is read, and used to link it to the new block. This, too, is central to the idea of blockchain. Seemingly valid transactions can be attempted to fraudulently be inserted into the blockchain, but decrypting all the previous blocks is computationally (nearly) impossible, which preserves the integrity of the blockchain.
```
def make_block(transactions, chain):
"""Make a block to go into the chain."""
parent_hash = chain[-1]['hash']
block_number = chain[-1]['contents']['block_number'] + 1
block_contents = {
'block_number': block_number,
'parent_hash': parent_hash,
'transaction_count': block_number + 1,
'transaction': transactions
}
return {'hash': hash_function(block_contents), 'contents': block_contents}
```
## Check Block Hash
Below is a small helper function to check the hash of the previous block:
```
def check_block_hash(block):
expected_hash = hash_function(block['contents'])
if block['hash'] is not expected_hash:
raise
return
```
## Block Validity
Once we have assembled everything together, its time to create our block. We will now update the blockchain.
```
def check_block_validity(block, parent, state):
parent_number = parent['contents']['block_number']
parent_hash = parent['hash']
block_number = block['contents']['block_number']
for transaction in block['contents']['transaction']:
if valid_transaction(transaction, state):
state = update_state(transaction, state)
else:
raise
check_block_hash(block) # Check hash integrity
if block_number is not parent_number + 1:
raise
if block['contents']['parent_hash'] is not parent_hash:
raise
```
## Check Blockchain
Before we are finished, the chain must be verified:
```
def check_chain(chain):
"""Check the chain is valid."""
if type(chain) is str:
try:
chain = json.loads(chain)
assert (type(chain) == list)
except ValueError:
# String passed in was not valid JSON
return False
elif type(chain) is not list:
return False
state = {}
for transaction in chain[0]['contents']['transaction']:
state = update_state(transaction, state)
check_block_hash(chain[0])
parent = chain[0]
for block in chain[1:]:
state = check_block_validity(block, parent, state)
parent = block
return state
```
## Add transaction
Finally, need a transaction function, which hangs all of the above together:
```
def add_transaction_to_chain(transaction, state, chain):
if valid_transaction(transaction, state):
state = update_state(transaction, state)
else:
raise Exception('Invalid transaction.')
my_block = make_block(state, chain)
chain.append(my_block)
for transaction in chain:
check_chain(transaction)
return state, chain
```
## Example
So, now we have our 7 functions. How do we interact with it? Well, first we need to start our chain with a Genesis Block. This is the inception of our new coin (or stock inventory, etc).
For the purposes of this article, I will say that I, Tom, will start off with 10 coins.
Let's say we start off with 100 coins for *[Geni AI](https://www.geni.ai)*.
```
genesis_block = {
'hash': hash_function({
'block_number': 0,
'parent_hash': None,
'transaction_count': 1,
'transaction': [{'Geni AI': 100}]
}),
'contents': {
'block_number': 0,
'parent_hash': None,
'transaction_count': 1,
'transaction': [{'Geni AI': 100}]
},
}
block_chain = [genesis_block]
chain_state = {'Geni AI': 100}
```
Now, look what happens when *[Geni AI](https://www.geni.ai)* give some coins to user *John Smith*:
```
chain_state, block_chain = add_transaction_to_chain(transaction={'Geni AI': -5, 'John Smith': 5}, state=chain_state, chain=block_chain)
chain_state
block_chain
```
Our first new transaction has been created and inserted to the top of the stack.
## References
https://towardsdatascience.com/blockchain-explained-in-7-python-functions-c49c84f34ba5
|
github_jupyter
|
import hashlib
import json
def hash_function(k):
"""Hashes our transaction."""
if type(k) is not str:
k = json.dumps(k, sort_keys=True)
return hashlib.sha256(k.encode('utf-8')).hexdigest()
hash_function('www.geni.ai')
def update_state(transaction, state):
state = state.copy()
for key in transaction:
if key in state.keys():
state[key] += transaction[key]
else:
state[key] = transaction[key]
return state
def valid_transaction(transaction, state):
"""A valid transaction must sum to 0."""
if sum(transaction.values()) is not 0:
return False
for key in transaction.keys():
if key in state.keys():
account_balance = state[key]
else:
account_balance = 0
if account_balance + transaction[key] < 0:
return False
return True
def make_block(transactions, chain):
"""Make a block to go into the chain."""
parent_hash = chain[-1]['hash']
block_number = chain[-1]['contents']['block_number'] + 1
block_contents = {
'block_number': block_number,
'parent_hash': parent_hash,
'transaction_count': block_number + 1,
'transaction': transactions
}
return {'hash': hash_function(block_contents), 'contents': block_contents}
def check_block_hash(block):
expected_hash = hash_function(block['contents'])
if block['hash'] is not expected_hash:
raise
return
def check_block_validity(block, parent, state):
parent_number = parent['contents']['block_number']
parent_hash = parent['hash']
block_number = block['contents']['block_number']
for transaction in block['contents']['transaction']:
if valid_transaction(transaction, state):
state = update_state(transaction, state)
else:
raise
check_block_hash(block) # Check hash integrity
if block_number is not parent_number + 1:
raise
if block['contents']['parent_hash'] is not parent_hash:
raise
def check_chain(chain):
"""Check the chain is valid."""
if type(chain) is str:
try:
chain = json.loads(chain)
assert (type(chain) == list)
except ValueError:
# String passed in was not valid JSON
return False
elif type(chain) is not list:
return False
state = {}
for transaction in chain[0]['contents']['transaction']:
state = update_state(transaction, state)
check_block_hash(chain[0])
parent = chain[0]
for block in chain[1:]:
state = check_block_validity(block, parent, state)
parent = block
return state
def add_transaction_to_chain(transaction, state, chain):
if valid_transaction(transaction, state):
state = update_state(transaction, state)
else:
raise Exception('Invalid transaction.')
my_block = make_block(state, chain)
chain.append(my_block)
for transaction in chain:
check_chain(transaction)
return state, chain
genesis_block = {
'hash': hash_function({
'block_number': 0,
'parent_hash': None,
'transaction_count': 1,
'transaction': [{'Geni AI': 100}]
}),
'contents': {
'block_number': 0,
'parent_hash': None,
'transaction_count': 1,
'transaction': [{'Geni AI': 100}]
},
}
block_chain = [genesis_block]
chain_state = {'Geni AI': 100}
chain_state, block_chain = add_transaction_to_chain(transaction={'Geni AI': -5, 'John Smith': 5}, state=chain_state, chain=block_chain)
chain_state
block_chain
| 0.722331 | 0.981524 |

### Egeria Hands-On Lab
# Welcome to the Conformance Test Suite Lab
## Introduction
Egeria is an open source project that provides open standards and implementation libraries to connect tools, catalogs and platforms together so they can share information (called metadata) about data and the technology that supports it.
In this hands-on lab you will get a chance to work with the conformance test suite that is used to validate that a technology can successfully join an open metadata repository cohort.
## About the Conformance Suite
The Conformance Suite can be used to test a Platform or Repository Connector to record which Conformance Profiles it supports. The Conformance Suite has different Workbenches that are used to test different types of system.
Initially our focus will be on the Repository Conformance Workbench. This workbench is used to test that an OMRS Repository Connector record which of the Repository Conformance Profiles it supports.
There are 13 repository conformance profiles in this workbench. One of them is mandatory - i.e. any repository connector must fully support that profile in order to be certified as conformant. The other profiles are optional and for each of these optional profiles, a repository connector can be certified as compliant even if it does not provide the function required by that profile - so long as it responds appropriately to requests.
## Configuring and running the Conformance Suite
We'll come back to the profiles later, but for now let's configure and run the Conformance Suite.
We're going to need a pair of OMAG Servers - one to run the repository under test, the other to run the workbench. The servers need to join the same cohort.

> **Figure 1:** Cohort for conformance testing
When the one running the workbench sees the cohort registration of the server under test, it runs the workbench tests against that server's repository.
## Starting up the Egeria platforms
We'll start one OMAG Server Platform on which to run both the servers.
We also need Apache Zookeeper and Apache Kafka.
```
%run ../common/globals.ipynb
import requests
import pprint
import json
import os
import time
# Disable warnings about self-signed certificates
from requests.packages.urllib3.exceptions import InsecureRequestWarning
requests.packages.urllib3.disable_warnings(InsecureRequestWarning)
ctsPlatformURL = os.environ.get('ctsPlatformURL','https://localhost:9445')
def checkServerPlatform(testPlatformName, testPlatformURL):
response = requests.get(testPlatformURL + "/open-metadata/platform-services/users/garygeeke/server-platform/origin/")
if response.status_code == 200:
print(" ...", testPlatformName, "at", testPlatformURL, "is active - ready to begin")
else:
print(" ...", testPlatformName, "at", testPlatformURL, "is down - start it before proceeding")
print ("\nChecking OMAG Server Platform availability...")
checkServerPlatform("CTS OMAG Server Platform", ctsPlatformURL)
print ("Done.")
```
## Configuring the Servers
We're going to configure both the servers in the diagram above.
It's useful to create some generally useful definitions here.
Knowing both server names up front will be handy for when we configure the workbench.
To configure the servers we'll need a common cohort name and event bus configuration.
We can let the CTS server default to using a local in-memory repository.
The CTS server does not need to run any Access Services.
```
ctsServerName = "CTS_Server"
sutServerName = "SUT_Server"
devCohort = "devCohort"
```
We'll need to pass a couple of JSON request bodies - so let's set up a reusable header:
```
jsonContentHeader = {'content-type':'application/json'}
```
We'll need a JSON request body for configuration of the event bus.
```
eventBusURLroot = os.environ.get('eventBusURLroot', 'localhost:9092')
eventBusBody = {
"producer": {
"bootstrap.servers": eventBusURLroot
},
"consumer":{
"bootstrap.servers": eventBusURLroot
}
}
```
We'll also need a JSON request body for configuration of the workbench.
This can be used to set the pageSize used in searches.
```
workbenchConfigBody = {
"class" : "RepositoryConformanceWorkbenchConfig",
"tutRepositoryServerName": sutServerName ,
"maxSearchResults" : 10
}
```
We also need a userId for the configuration commands. You could change this to a name you choose.
```
adminUserId = "garygeeke"
```
We can perform configuration operations through the administrative interface provided by the ctsPlatformURL.
The URLs for the configuration REST APIs have a common structure and begin with the following root:
```
adminPlatformURL = ctsPlatformURL
adminCommandURLRoot = adminPlatformURL + '/open-metadata/admin-services/users/' + adminUserId + '/servers/'
```
What follows are descriptions and coded requests to configure each server. There are a lot of common steps
involved in configuring a metadata server, so we first define some simple
functions that can be re-used in later steps for configuring each server.
Each function returns True or False to indicate whether it was successful.
```
def postAndPrintResult(url, json=None, headers=None):
print(" ...... (POST", url, ")")
response = requests.post(url, json=json, headers=headers)
if response.status_code == 200:
print(" ...... Success. Response: ", response.json())
return True
else:
print(" ...... Failed. Response: ", response.json())
return False
def getAndPrintResult(url, json=None, headers=None):
print(" ...... (GET", url, ")")
response = requests.get(url, json=json, headers=headers)
if response.status_code == 200:
print(" ...... Success. Response: ", response.json())
return True
else:
print(" ...... Failed. Response: ", response.json())
return False
def getResult(url, json=None, headers=None):
print("\n ...... (GET", url, ")")
try:
response = requests.get(url, json=json, headers=headers)
if response.status_code == 200:
if response.json()['relatedHTTPCode'] == 200:
return response.json()
return None
except requests.exceptions.RequestException as e:
print (" ...... FAILED - http request threw an exception: ", e)
return None
def configurePlatformURL(serverName, serverPlatform):
print("\n ... Configuring the platform the server will run on...")
url = adminCommandURLRoot + serverName + '/server-url-root?url=' + serverPlatform
return postAndPrintResult(url)
def configureServerType(serverName, serverType):
print ("\n ... Configuring the server's type...")
url = adminCommandURLRoot + serverName + '/server-type?typeName=' + serverType
return postAndPrintResult(url)
def configureUserId(serverName, userId):
print ("\n ... Configuring the server's userId...")
url = adminCommandURLRoot + serverName + '/server-user-id?id=' + userId
return postAndPrintResult(url)
def configurePassword(serverName, password):
print ("\n ... Configuring the server's password (optional)...")
url = adminCommandURLRoot + serverName + '/server-user-password?password=' + password
return postAndPrintResult(url)
def configureMetadataRepository(serverName, repositoryType):
print ("\n ... Configuring the metadata repository...")
url = adminCommandURLRoot + serverName + '/local-repository/mode/' + repositoryType
return postAndPrintResult(url)
def configureDescriptiveName(serverName, collectionName):
print ("\n ... Configuring the short descriptive name of the metadata stored in this server...")
url = adminCommandURLRoot + serverName + '/local-repository/metadata-collection-name/' + collectionName
return postAndPrintResult(url)
def configureEventBus(serverName, busBody):
print ("\n ... Configuring the event bus for this server...")
url = adminCommandURLRoot + serverName + '/event-bus'
return postAndPrintResult(url, json=busBody, headers=jsonContentHeader)
def configureCohortMembership(serverName, cohortName):
print ("\n ... Configuring the membership of the cohort...")
url = adminCommandURLRoot + serverName + '/cohorts/' + cohortName
return postAndPrintResult(url)
def configureRepositoryWorkbench(serverName, workbenchBody):
print ("\n ... Configuring the repository workbench for this server...")
url = adminCommandURLRoot + serverName + '/conformance-suite-workbenches/repository-workbench/repositories'
return postAndPrintResult(url, json=workbenchBody, headers=jsonContentHeader)
```
## Configuring the CTS Server
We're going to configure the CTS Server from the diagram above. The CTS Server is the one that runs the repository workbench.
The server will default to using a local in-memory repository.
The CTS server does not need to run any Access Services.
Notice that when we configure the CTS Server to run the repository workbench, we provide the name of the server under test.
First we introduce a 'success' variable which is used to monitor progress in the subsequent cells.
```
success = True
ctsServerType = "Conformance Suite Server"
ctsServerUserId = "CTS1npa"
ctsServerPassword = "CTS1passw0rd"
ctsServerPlatform = ctsPlatformURL
print("Configuring " + ctsServerName + "...")
if (success):
success = configurePlatformURL(ctsServerName, ctsServerPlatform)
if (success):
success = configureServerType(ctsServerName, ctsServerType)
if (success):
success = configureUserId(ctsServerName, ctsServerUserId)
if (success):
success = configurePassword(ctsServerName, ctsServerPassword)
if (success):
success = configureEventBus(ctsServerName, eventBusBody)
if (success):
success = configureCohortMembership(ctsServerName, devCohort)
if (success):
success = configureRepositoryWorkbench(ctsServerName, workbenchConfigBody)
if (success):
print("\nDone.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
```
## Configuring the SUT Server (Server Under Test)
Next we're going to configure the SUT Server from the diagram above. The SUT Server is the one that hosts the repository that is being tested. The SUT Server will run on the same platform as the CTS Server.
The server will default to using a local in-memory repository.
The SUT server does not need to run any Access Services.
Notice that when we configure the CTS Server to run the repository workbench, we provide the name of the server under test.
```
sutServerType = "Metadata Repository Server"
sutServerUserId = "SUTnpa"
sutServerPassword = "SUTpassw0rd"
metadataCollectionName = "SUT_MDR"
metadataRepositoryTypeInMemory = "in-memory-repository"
metadataRepositoryTypeGraph = "local-graph-repository"
print("Configuring " + sutServerName + "...")
if (success):
success = configurePlatformURL(sutServerName, ctsServerPlatform)
if (success):
success = configureServerType(sutServerName, sutServerType)
if (success):
success = configureUserId(sutServerName, sutServerUserId)
if (success):
success = configurePassword(sutServerName, sutServerPassword)
if (success):
success = configureMetadataRepository(sutServerName, metadataRepositoryTypeInMemory)
if (success):
success = configureDescriptiveName(sutServerName, metadataCollectionName)
if (success):
success = configureEventBus(sutServerName, eventBusBody)
if (success):
success = configureCohortMembership(sutServerName, devCohort)
if (success):
print("\nDone.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
```
The commands below deploy the server configuration documents to the server platforms where the
servers will run.
```
def deployServerToPlatform(serverName, platformURL):
print(" ... deploying", serverName, "to the", platformURL, "platform...")
url = adminCommandURLRoot + serverName + '/configuration/deploy'
platformTarget = {
"class": "URLRequestBody",
"urlRoot": platformURL
}
try:
return postAndPrintResult(url, json=platformTarget, headers=jsonContentHeader)
except requests.exceptions.RequestException as e:
print (" ...... FAILED - http request threw an exception: ", e)
return False
print("\nDeploying server configuration documents to appropriate platforms...")
if (success):
success = deployServerToPlatform(ctsServerName, ctsPlatformURL)
if (success):
success = deployServerToPlatform(sutServerName, ctsPlatformURL)
if (success):
print("\nDone.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
```
## Starting the servers
We'll need to define the URL for the OMRS operational services API.
```
operationalServicesURLcore = "/open-metadata/admin-services/users/" + adminUserId
```
Start the CTS Server, followed by the SUT Server.
When the CTS Server sees the cohort registration for the SUT Server it will start to run the workbench.
```
def startServer(serverName, platformURL):
print(" ... starting server", serverName, "...")
url = platformURL + operationalServicesURLcore + '/servers/' + serverName + '/instance'
return postAndPrintResult(url)
print ("\nStarting the CTS server ...")
if (success):
success = startServer(ctsServerName, ctsPlatformURL)
# Pause to allow server to initialize fully
time.sleep(4)
print ("\nStarting the SUT server ...")
if (success):
success = startServer(sutServerName, ctsPlatformURL)
if (success):
print("\nDone.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
```
## Workbench Progress
The repository workbench runs a lot of tests (several thousand) and it can take a while to complete -- meaning several hours. There is no 'completion event' because when the conformance suite has completed the synchronous workbench tests it continues to run and will perform asynchronous tests in responses to events that may be received within the cohort. The consequence of this is that it is not easy to know when the CTS has 'finished'. However, if you scan the output console logging from the conformance suite it is possible to detect the log output:
Thu Nov 21 09:11:01 GMT 2019 CTS_Server Information CONFORMANCE-SUITE-0011 The Open Metadata Conformance Workbench repository-workbench has completed its synchronous tests, further test cases may be triggered from incoming events.
When this has been seen you will probably see a number of further events being processed by the CTS Server. There can be up to several hundred events - that look like the following:
Thu Nov 21 09:11:03 GMT 2019 CTS_Server Event OMRS-AUDIT-8006 Processing incoming event of type DeletedEntityEvent for instance 2fd6cd97-35dd-41d9-ad2f-4d25af30033e from: OMRSEventOriginator{metadataCollectionId='f076a951-fcd0-483b-a06e-d0c7abb61b84', serverName='SUT_Server', serverType='Metadata Repository Server', organizationName='null'}
Thu Nov 21 09:11:03 GMT 2019 CTS_Server Event OMRS-AUDIT-8006 Processing incoming event of type PurgedEntityEvent for instance 2fd6cd97-35dd-41d9-ad2f-4d25af30033e from: OMRSEventOriginator{metadataCollectionId='f076a951-fcd0-483b-a06e-d0c7abb61b84', serverName='SUT_Server', serverType='Metadata Repository Server', organizationName='null'}
These events are usually DELETE and PURGE events relating to instances that have been cleaned up on the SUT Server.
Once these events have been logged the console should go quiet. When you see this, it is possible to retrieve the workbench results from the CTS Server.
## Polling for Status
The following cell can be used to find out whether the workbench has completed its synchronous tests....
```
conformanceSuiteServicesURLcore = "/open-metadata/conformance-suite/users/" + adminUserId
def retrieveStatus(serverName, platformURL):
print(" ... retrieving completion status from server", serverName, "...")
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/status/workbenches/repository-workbench'
return getResult(url)
print ("\nRetrieve repository-workbench status ...")
status_json = retrieveStatus(ctsServerName, ctsPlatformURL)
if (status_json != None):
workbenchId = status_json['workbenchStatus']['workbenchId']
workbenchComplete = status_json['workbenchStatus']['workbenchComplete']
if (workbenchComplete == True):
print("\nWorkbench",workbenchId,"is complete.")
else:
print("\nWorkbench",workbenchId,"has not yet completed.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
```
## Retrieving the Workbench Results
The repository workbench keeps the results of the testcases in memory. When the workbench is complete (see above) you
can request a report of the results from the REST API on the CTS Server.
The REST API has several options that supports different styles of report.
### Summary results
First we will request a summary report.
```
from requests.utils import quote
import os
report_json = None
cwd = os.getcwd()
conformanceSuiteServicesURLcore = "/open-metadata/conformance-suite/users/" + adminUserId
def retrieveSummary(serverName, platformURL):
print(" ... retrieving test report summary from server", serverName, "...")
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/report/summary'
return getResult(url)
print ("\nRetrieve Conformance Suite summary results ...")
summary_json = retrieveSummary(ctsServerName, ctsPlatformURL)
```
The following is a summary of the status of each conformance profile. To ensure that you get a complete summary, make sure you retrieve the summary results _once the workbench has completed_.
(Note that this uses pandas to summarize the results table: if you have not already done so, use pip3 to install pandas and its dependencies.)
```
import pandas
from pandas import json_normalize
if (summary_json != None):
repositoryWorkbenchResults = json_normalize(data = summary_json['testLabSummary'],
record_path =['testSummariesFromWorkbenches','profileSummaries'])
repositoryWorkbenchResultsSummary = repositoryWorkbenchResults[['name','description','profilePriority','conformanceStatus']]
display(repositoryWorkbenchResultsSummary.head(15))
```
### Detailed results
We can also retrieve the full details of each profile and test case individually. Some of the detailed profile reports can be large (10-20MB), so if you are running the Jupyter notebook server with its default configuration, the report may exceed the default max data rate for the notebook server. If you are not running the Egeria team's containers (docker/k8s), and you have not done so already, please restart the notebook server with the following configuration option:
jupyter notebook --NotebookApp.iopub_data_rate_limit=1.0e10
If the following call results in a Java Heap error you may need to increase the memory configured for your container environment, or available locally. Min 2GB, ideally 4GB additional heap space is recommended for CTS.
Given the amount of detail involved, it can take several minutes to retrieve all of the details of a completed CTS run (1000's of API calls and files): it may at times even appear that the notebook is frozen. Wait until the cell shows a number (rather than an asterisk). This indicates the cell has completed, and you should also see a final line of output that states: "Done -- all details retrieved." (While it runs, you should see the output updating with the iterative REST calls that are made to retrieve each profile's or test case's details.)
```
profileDir = "profile-details"
testCaseDir = "test-case-details"
def retrieveProfileNames(serverName, platformURL):
print(" ... retrieving profile list from server", serverName, "...")
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/report/profiles'
return getResult(url)
def retrieveTestCaseIds(serverName, platformURL):
print(" ... retrieving test case list from server", serverName, "...")
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/report/test-cases'
return getResult(url)
def retrieveProfileDetails(serverName, platformURL, profileName):
encodedProfileName = quote(profileName)
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/report/profiles/' + encodedProfileName
return getResult(url)
def retrieveTestCaseDetails(serverName, platformURL, testCaseId):
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/report/test-cases/' + testCaseId
return getResult(url)
if (summary_json != None):
with open("openmetadata_cts_summary.json", 'w') as outfile:
json.dump(summary_json, outfile)
profiles = retrieveProfileNames(ctsServerName, ctsPlatformURL)
profileDetailsDir = cwd + os.path.sep + profileDir
os.makedirs(profileDetailsDir, exist_ok=True)
print("Retrieving details for each profile...")
for profile in profiles['profileNames']:
profile_details = retrieveProfileDetails(ctsServerName, ctsPlatformURL, profile)
with open(profileDetailsDir + os.path.sep + profile.replace(" ", "_") + ".json", 'w') as outfile:
json.dump(profile_details, outfile)
test_cases = retrieveTestCaseIds(ctsServerName, ctsPlatformURL)
testCaseDetailsDir = cwd + os.path.sep + testCaseDir
os.makedirs(testCaseDetailsDir, exist_ok=True)
print("Retrieving details for each test case...")
for test_case in test_cases['testCaseIds']:
test_case_details = retrieveTestCaseDetails(ctsServerName, ctsPlatformURL, test_case)
with open(testCaseDetailsDir + os.path.sep + test_case + ".json", 'w') as outfile:
json.dump(test_case_details, outfile)
print("\nDone -- all details retrieved.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
```
|
github_jupyter
|
%run ../common/globals.ipynb
import requests
import pprint
import json
import os
import time
# Disable warnings about self-signed certificates
from requests.packages.urllib3.exceptions import InsecureRequestWarning
requests.packages.urllib3.disable_warnings(InsecureRequestWarning)
ctsPlatformURL = os.environ.get('ctsPlatformURL','https://localhost:9445')
def checkServerPlatform(testPlatformName, testPlatformURL):
response = requests.get(testPlatformURL + "/open-metadata/platform-services/users/garygeeke/server-platform/origin/")
if response.status_code == 200:
print(" ...", testPlatformName, "at", testPlatformURL, "is active - ready to begin")
else:
print(" ...", testPlatformName, "at", testPlatformURL, "is down - start it before proceeding")
print ("\nChecking OMAG Server Platform availability...")
checkServerPlatform("CTS OMAG Server Platform", ctsPlatformURL)
print ("Done.")
ctsServerName = "CTS_Server"
sutServerName = "SUT_Server"
devCohort = "devCohort"
jsonContentHeader = {'content-type':'application/json'}
eventBusURLroot = os.environ.get('eventBusURLroot', 'localhost:9092')
eventBusBody = {
"producer": {
"bootstrap.servers": eventBusURLroot
},
"consumer":{
"bootstrap.servers": eventBusURLroot
}
}
workbenchConfigBody = {
"class" : "RepositoryConformanceWorkbenchConfig",
"tutRepositoryServerName": sutServerName ,
"maxSearchResults" : 10
}
adminUserId = "garygeeke"
adminPlatformURL = ctsPlatformURL
adminCommandURLRoot = adminPlatformURL + '/open-metadata/admin-services/users/' + adminUserId + '/servers/'
def postAndPrintResult(url, json=None, headers=None):
print(" ...... (POST", url, ")")
response = requests.post(url, json=json, headers=headers)
if response.status_code == 200:
print(" ...... Success. Response: ", response.json())
return True
else:
print(" ...... Failed. Response: ", response.json())
return False
def getAndPrintResult(url, json=None, headers=None):
print(" ...... (GET", url, ")")
response = requests.get(url, json=json, headers=headers)
if response.status_code == 200:
print(" ...... Success. Response: ", response.json())
return True
else:
print(" ...... Failed. Response: ", response.json())
return False
def getResult(url, json=None, headers=None):
print("\n ...... (GET", url, ")")
try:
response = requests.get(url, json=json, headers=headers)
if response.status_code == 200:
if response.json()['relatedHTTPCode'] == 200:
return response.json()
return None
except requests.exceptions.RequestException as e:
print (" ...... FAILED - http request threw an exception: ", e)
return None
def configurePlatformURL(serverName, serverPlatform):
print("\n ... Configuring the platform the server will run on...")
url = adminCommandURLRoot + serverName + '/server-url-root?url=' + serverPlatform
return postAndPrintResult(url)
def configureServerType(serverName, serverType):
print ("\n ... Configuring the server's type...")
url = adminCommandURLRoot + serverName + '/server-type?typeName=' + serverType
return postAndPrintResult(url)
def configureUserId(serverName, userId):
print ("\n ... Configuring the server's userId...")
url = adminCommandURLRoot + serverName + '/server-user-id?id=' + userId
return postAndPrintResult(url)
def configurePassword(serverName, password):
print ("\n ... Configuring the server's password (optional)...")
url = adminCommandURLRoot + serverName + '/server-user-password?password=' + password
return postAndPrintResult(url)
def configureMetadataRepository(serverName, repositoryType):
print ("\n ... Configuring the metadata repository...")
url = adminCommandURLRoot + serverName + '/local-repository/mode/' + repositoryType
return postAndPrintResult(url)
def configureDescriptiveName(serverName, collectionName):
print ("\n ... Configuring the short descriptive name of the metadata stored in this server...")
url = adminCommandURLRoot + serverName + '/local-repository/metadata-collection-name/' + collectionName
return postAndPrintResult(url)
def configureEventBus(serverName, busBody):
print ("\n ... Configuring the event bus for this server...")
url = adminCommandURLRoot + serverName + '/event-bus'
return postAndPrintResult(url, json=busBody, headers=jsonContentHeader)
def configureCohortMembership(serverName, cohortName):
print ("\n ... Configuring the membership of the cohort...")
url = adminCommandURLRoot + serverName + '/cohorts/' + cohortName
return postAndPrintResult(url)
def configureRepositoryWorkbench(serverName, workbenchBody):
print ("\n ... Configuring the repository workbench for this server...")
url = adminCommandURLRoot + serverName + '/conformance-suite-workbenches/repository-workbench/repositories'
return postAndPrintResult(url, json=workbenchBody, headers=jsonContentHeader)
success = True
ctsServerType = "Conformance Suite Server"
ctsServerUserId = "CTS1npa"
ctsServerPassword = "CTS1passw0rd"
ctsServerPlatform = ctsPlatformURL
print("Configuring " + ctsServerName + "...")
if (success):
success = configurePlatformURL(ctsServerName, ctsServerPlatform)
if (success):
success = configureServerType(ctsServerName, ctsServerType)
if (success):
success = configureUserId(ctsServerName, ctsServerUserId)
if (success):
success = configurePassword(ctsServerName, ctsServerPassword)
if (success):
success = configureEventBus(ctsServerName, eventBusBody)
if (success):
success = configureCohortMembership(ctsServerName, devCohort)
if (success):
success = configureRepositoryWorkbench(ctsServerName, workbenchConfigBody)
if (success):
print("\nDone.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
sutServerType = "Metadata Repository Server"
sutServerUserId = "SUTnpa"
sutServerPassword = "SUTpassw0rd"
metadataCollectionName = "SUT_MDR"
metadataRepositoryTypeInMemory = "in-memory-repository"
metadataRepositoryTypeGraph = "local-graph-repository"
print("Configuring " + sutServerName + "...")
if (success):
success = configurePlatformURL(sutServerName, ctsServerPlatform)
if (success):
success = configureServerType(sutServerName, sutServerType)
if (success):
success = configureUserId(sutServerName, sutServerUserId)
if (success):
success = configurePassword(sutServerName, sutServerPassword)
if (success):
success = configureMetadataRepository(sutServerName, metadataRepositoryTypeInMemory)
if (success):
success = configureDescriptiveName(sutServerName, metadataCollectionName)
if (success):
success = configureEventBus(sutServerName, eventBusBody)
if (success):
success = configureCohortMembership(sutServerName, devCohort)
if (success):
print("\nDone.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
def deployServerToPlatform(serverName, platformURL):
print(" ... deploying", serverName, "to the", platformURL, "platform...")
url = adminCommandURLRoot + serverName + '/configuration/deploy'
platformTarget = {
"class": "URLRequestBody",
"urlRoot": platformURL
}
try:
return postAndPrintResult(url, json=platformTarget, headers=jsonContentHeader)
except requests.exceptions.RequestException as e:
print (" ...... FAILED - http request threw an exception: ", e)
return False
print("\nDeploying server configuration documents to appropriate platforms...")
if (success):
success = deployServerToPlatform(ctsServerName, ctsPlatformURL)
if (success):
success = deployServerToPlatform(sutServerName, ctsPlatformURL)
if (success):
print("\nDone.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
operationalServicesURLcore = "/open-metadata/admin-services/users/" + adminUserId
def startServer(serverName, platformURL):
print(" ... starting server", serverName, "...")
url = platformURL + operationalServicesURLcore + '/servers/' + serverName + '/instance'
return postAndPrintResult(url)
print ("\nStarting the CTS server ...")
if (success):
success = startServer(ctsServerName, ctsPlatformURL)
# Pause to allow server to initialize fully
time.sleep(4)
print ("\nStarting the SUT server ...")
if (success):
success = startServer(sutServerName, ctsPlatformURL)
if (success):
print("\nDone.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
conformanceSuiteServicesURLcore = "/open-metadata/conformance-suite/users/" + adminUserId
def retrieveStatus(serverName, platformURL):
print(" ... retrieving completion status from server", serverName, "...")
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/status/workbenches/repository-workbench'
return getResult(url)
print ("\nRetrieve repository-workbench status ...")
status_json = retrieveStatus(ctsServerName, ctsPlatformURL)
if (status_json != None):
workbenchId = status_json['workbenchStatus']['workbenchId']
workbenchComplete = status_json['workbenchStatus']['workbenchComplete']
if (workbenchComplete == True):
print("\nWorkbench",workbenchId,"is complete.")
else:
print("\nWorkbench",workbenchId,"has not yet completed.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
from requests.utils import quote
import os
report_json = None
cwd = os.getcwd()
conformanceSuiteServicesURLcore = "/open-metadata/conformance-suite/users/" + adminUserId
def retrieveSummary(serverName, platformURL):
print(" ... retrieving test report summary from server", serverName, "...")
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/report/summary'
return getResult(url)
print ("\nRetrieve Conformance Suite summary results ...")
summary_json = retrieveSummary(ctsServerName, ctsPlatformURL)
import pandas
from pandas import json_normalize
if (summary_json != None):
repositoryWorkbenchResults = json_normalize(data = summary_json['testLabSummary'],
record_path =['testSummariesFromWorkbenches','profileSummaries'])
repositoryWorkbenchResultsSummary = repositoryWorkbenchResults[['name','description','profilePriority','conformanceStatus']]
display(repositoryWorkbenchResultsSummary.head(15))
profileDir = "profile-details"
testCaseDir = "test-case-details"
def retrieveProfileNames(serverName, platformURL):
print(" ... retrieving profile list from server", serverName, "...")
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/report/profiles'
return getResult(url)
def retrieveTestCaseIds(serverName, platformURL):
print(" ... retrieving test case list from server", serverName, "...")
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/report/test-cases'
return getResult(url)
def retrieveProfileDetails(serverName, platformURL, profileName):
encodedProfileName = quote(profileName)
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/report/profiles/' + encodedProfileName
return getResult(url)
def retrieveTestCaseDetails(serverName, platformURL, testCaseId):
url = platformURL + '/servers/' + serverName + conformanceSuiteServicesURLcore + '/report/test-cases/' + testCaseId
return getResult(url)
if (summary_json != None):
with open("openmetadata_cts_summary.json", 'w') as outfile:
json.dump(summary_json, outfile)
profiles = retrieveProfileNames(ctsServerName, ctsPlatformURL)
profileDetailsDir = cwd + os.path.sep + profileDir
os.makedirs(profileDetailsDir, exist_ok=True)
print("Retrieving details for each profile...")
for profile in profiles['profileNames']:
profile_details = retrieveProfileDetails(ctsServerName, ctsPlatformURL, profile)
with open(profileDetailsDir + os.path.sep + profile.replace(" ", "_") + ".json", 'w') as outfile:
json.dump(profile_details, outfile)
test_cases = retrieveTestCaseIds(ctsServerName, ctsPlatformURL)
testCaseDetailsDir = cwd + os.path.sep + testCaseDir
os.makedirs(testCaseDetailsDir, exist_ok=True)
print("Retrieving details for each test case...")
for test_case in test_cases['testCaseIds']:
test_case_details = retrieveTestCaseDetails(ctsServerName, ctsPlatformURL, test_case)
with open(testCaseDetailsDir + os.path.sep + test_case + ".json", 'w') as outfile:
json.dump(test_case_details, outfile)
print("\nDone -- all details retrieved.")
else:
print("\nFAILED: please check the messages above and correct before proceeding")
| 0.160595 | 0.879354 |
```
import pandas as pd
import numpy as np
symbol = "Security 1"
symbol2 = "Security 2"
price_data = pd.DataFrame(
np.cumsum(np.random.randn(150, 2).dot([[0.5, 0.4], [0.4, 1.0]]), axis=0) + 100,
columns=["Security 1", "Security 2"],
index=pd.date_range(start="01-01-2007", periods=150),
)
dates_actual = price_data.index.values
prices = price_data[symbol].values
from bqplot import DateScale, LinearScale, Axis, Lines, Scatter, Bars, Hist, Figure
from bqplot.interacts import (
FastIntervalSelector,
IndexSelector,
BrushIntervalSelector,
BrushSelector,
MultiSelector,
LassoSelector,
PanZoom,
HandDraw,
)
from traitlets import link
from ipywidgets import ToggleButtons, VBox, HTML
```
# Line Chart Selectors
## Fast Interval Selector
```
## First we define a Figure
dt_x_fast = DateScale()
lin_y = LinearScale()
x_ax = Axis(label="Index", scale=dt_x_fast)
x_ay = Axis(label=(symbol + " Price"), scale=lin_y, orientation="vertical")
lc = Lines(
x=dates_actual, y=prices, scales={"x": dt_x_fast, "y": lin_y}, colors=["orange"]
)
lc_2 = Lines(
x=dates_actual[50:],
y=prices[50:] + 2,
scales={"x": dt_x_fast, "y": lin_y},
colors=["blue"],
)
## Next we define the type of selector we would like
intsel_fast = FastIntervalSelector(scale=dt_x_fast, marks=[lc, lc_2])
## Now, we define a function that will be called when the FastIntervalSelector is interacted with
def fast_interval_change_callback(change):
db_fast.value = "The selected period is " + str(change.new)
## Now we connect the selectors to that function
intsel_fast.observe(fast_interval_change_callback, names=["selected"])
## We use the HTML widget to see the value of what we are selecting and modify it when an interaction is performed
## on the selector
db_fast = HTML()
db_fast.value = "The selected period is " + str(intsel_fast.selected)
fig_fast_intsel = Figure(
marks=[lc, lc_2],
axes=[x_ax, x_ay],
title="Fast Interval Selector Example",
interaction=intsel_fast,
) # This is where we assign the interaction to this particular Figure
VBox([db_fast, fig_fast_intsel])
```
## Index Selector
```
db_index = HTML(value="[]")
## Now we try a selector made to select all the y-values associated with a single x-value
index_sel = IndexSelector(scale=dt_x_fast, marks=[lc, lc_2])
## Now, we define a function that will be called when the selectors are interacted with
def index_change_callback(change):
db_index.value = "The selected date is " + str(change.new)
index_sel.observe(index_change_callback, names=["selected"])
fig_index_sel = Figure(
marks=[lc, lc_2],
axes=[x_ax, x_ay],
title="Index Selector Example",
interaction=index_sel,
)
VBox([db_index, fig_index_sel])
```
## Returning indexes of selected values
```
from datetime import datetime as py_dtime
dt_x_index = DateScale(min=np.datetime64(py_dtime(2006, 6, 1)))
lin_y2 = LinearScale()
lc2_index = Lines(x=dates_actual, y=prices, scales={"x": dt_x_index, "y": lin_y2})
x_ax1 = Axis(label="Date", scale=dt_x_index)
x_ay2 = Axis(label=(symbol + " Price"), scale=lin_y2, orientation="vertical")
intsel_date = FastIntervalSelector(scale=dt_x_index, marks=[lc2_index])
db_date = HTML()
db_date.value = str(intsel_date.selected)
## Now, we define a function that will be called when the selectors are interacted with - a callback
def date_interval_change_callback(change):
db_date.value = str(change.new)
## Notice here that we call the observe on the Mark lc2_index rather than on the selector intsel_date
lc2_index.observe(date_interval_change_callback, names=["selected"])
fig_date_mark = Figure(
marks=[lc2_index],
axes=[x_ax1, x_ay2],
title="Fast Interval Selector Selected Indices Example",
interaction=intsel_date,
)
VBox([db_date, fig_date_mark])
```
## Brush Selector
### We can do the same with any type of selector
```
## Defining a new Figure
dt_x_brush = DateScale(min=np.datetime64(py_dtime(2006, 6, 1)))
lin_y2_brush = LinearScale()
lc3_brush = Lines(x=dates_actual, y=prices, scales={"x": dt_x_brush, "y": lin_y2_brush})
x_ax_brush = Axis(label="Date", scale=dt_x_brush)
x_ay_brush = Axis(label=(symbol + " Price"), scale=lin_y2_brush, orientation="vertical")
db_brush = HTML(value="[]")
brushsel_date = BrushIntervalSelector(
scale=dt_x_brush, marks=[lc3_brush], color="FireBrick"
)
## Now, we define a function that will be called when the selectors are interacted with - a callback
def date_brush_change_callback(change):
db_brush.value = str(change.new)
lc3_brush.observe(date_brush_change_callback, names=["selected"])
fig_brush_sel = Figure(
marks=[lc3_brush],
axes=[x_ax_brush, x_ay_brush],
title="Brush Selector Selected Indices Example",
interaction=brushsel_date,
)
VBox([db_brush, fig_brush_sel])
```
# Scatter Chart Selectors
## Brush Selector
```
date_fmt = "%m-%d-%Y"
sec2_data = price_data[symbol2].values
dates = price_data.index.values
sc_x = LinearScale()
sc_y = LinearScale()
scatt = Scatter(x=prices, y=sec2_data, scales={"x": sc_x, "y": sc_y})
sc_xax = Axis(label=(symbol), scale=sc_x)
sc_yax = Axis(label=(symbol2), scale=sc_y, orientation="vertical")
br_sel = BrushSelector(x_scale=sc_x, y_scale=sc_y, marks=[scatt], color="red")
db_scat_brush = HTML(value="[]")
## call back for the selector
def brush_callback(change):
db_scat_brush.value = str(br_sel.selected)
br_sel.observe(brush_callback, names=["brushing"])
fig_scat_brush = Figure(
marks=[scatt],
axes=[sc_xax, sc_yax],
title="Scatter Chart Brush Selector Example",
interaction=br_sel,
)
VBox([db_scat_brush, fig_scat_brush])
```
## Brush Selector with Date Values
```
sc_brush_dt_x = DateScale(date_format=date_fmt)
sc_brush_dt_y = LinearScale()
scatt2 = Scatter(
x=dates_actual, y=sec2_data, scales={"x": sc_brush_dt_x, "y": sc_brush_dt_y}
)
br_sel_dt = BrushSelector(x_scale=sc_brush_dt_x, y_scale=sc_brush_dt_y, marks=[scatt2])
db_brush_dt = HTML(value=str(br_sel_dt.selected))
## call back for the selector
def brush_dt_callback(change):
db_brush_dt.value = str(br_sel_dt.selected)
br_sel_dt.observe(brush_dt_callback, names=["brushing"])
sc_xax = Axis(label=(symbol), scale=sc_brush_dt_x)
sc_yax = Axis(label=(symbol2), scale=sc_brush_dt_y, orientation="vertical")
fig_brush_dt = Figure(
marks=[scatt2],
axes=[sc_xax, sc_yax],
title="Brush Selector with Dates Example",
interaction=br_sel_dt,
)
VBox([db_brush_dt, fig_brush_dt])
```
# Histogram Selectors
```
## call back for selectors
def interval_change_callback(name, value):
db3.value = str(value)
## call back for the selector
def brush_callback(change):
if not br_intsel.brushing:
db3.value = str(br_intsel.selected)
returns = np.log(prices[1:]) - np.log(prices[:-1])
hist_x = LinearScale()
hist_y = LinearScale()
hist = Hist(sample=returns, scales={"sample": hist_x, "count": hist_y})
br_intsel = BrushIntervalSelector(scale=hist_x, marks=[hist])
br_intsel.observe(brush_callback, names=["selected"])
br_intsel.observe(brush_callback, names=["brushing"])
db3 = HTML()
db3.value = str(br_intsel.selected)
h_xax = Axis(
scale=hist_x, label="Returns", grids="off", set_ticks=True, tick_format="0.2%"
)
h_yax = Axis(scale=hist_y, label="Freq", orientation="vertical", grid_lines="none")
fig_hist = Figure(
marks=[hist],
axes=[h_xax, h_yax],
title="Histogram Selection Example",
interaction=br_intsel,
)
VBox([db3, fig_hist])
```
## Multi Selector
* This selector provides the ability to have multiple brush selectors on the same graph.
* The first brush works like a regular brush.
* `Ctrl + click` creates a new brush, which works like the regular brush.
* The `active` brush has a Green border while all the `inactive` brushes have a Red border.
* `Shift + click` deactivates the current `active` brush. Now, click on any `inactive` brush to make it `active`.
* `Ctrl + Alt + Shift + click` clears and resets all the brushes.
```
def multi_sel_callback(change):
if not multi_sel.brushing:
db4.value = str(multi_sel.selected)
line_x = LinearScale()
line_y = LinearScale()
line = Lines(
x=np.arange(100), y=np.random.randn(100), scales={"x": line_x, "y": line_y}
)
multi_sel = MultiSelector(scale=line_x, marks=[line])
multi_sel.observe(multi_sel_callback, names=["selected"])
multi_sel.observe(multi_sel_callback, names=["brushing"])
db4 = HTML()
db4.value = str(multi_sel.selected)
h_xax = Axis(scale=line_x, label="Returns", grid_lines="none")
h_yax = Axis(scale=hist_y, label="Freq", orientation="vertical", grid_lines="none")
fig_multi = Figure(
marks=[line],
axes=[h_xax, h_yax],
title="Multi-Selector Example",
interaction=multi_sel,
)
VBox([db4, fig_multi])
# changing the names of the intervals.
multi_sel.names = ["int1", "int2", "int3"]
```
## Multi Selector with Date X
```
def multi_sel_dt_callback(change):
if not multi_sel_dt.brushing:
db_multi_dt.value = str(multi_sel_dt.selected)
line_dt_x = DateScale(min=np.datetime64(py_dtime(2007, 1, 1)))
line_dt_y = LinearScale()
line_dt = Lines(
x=dates_actual, y=sec2_data, scales={"x": line_dt_x, "y": line_dt_y}, colors=["red"]
)
multi_sel_dt = MultiSelector(scale=line_dt_x)
multi_sel_dt.observe(multi_sel_dt_callback, names=["selected"])
multi_sel_dt.observe(multi_sel_dt_callback, names=["brushing"])
db_multi_dt = HTML()
db_multi_dt.value = str(multi_sel_dt.selected)
h_xax_dt = Axis(scale=line_dt_x, label="Returns", grid_lines="none")
h_yax_dt = Axis(
scale=line_dt_y, label="Freq", orientation="vertical", grid_lines="none"
)
fig_multi_dt = Figure(
marks=[line_dt],
axes=[h_xax_dt, h_yax_dt],
title="Multi-Selector with Date Example",
interaction=multi_sel_dt,
)
VBox([db_multi_dt, fig_multi_dt])
```
## Lasso Selector
```
lasso_sel = LassoSelector()
xs, ys = LinearScale(), LinearScale()
data = np.arange(20)
line_lasso = Lines(x=data, y=data, scales={"x": xs, "y": ys})
scatter_lasso = Scatter(x=data, y=data, scales={"x": xs, "y": ys}, colors=["skyblue"])
bar_lasso = Bars(x=data, y=data / 2.0, scales={"x": xs, "y": ys})
xax_lasso, yax_lasso = Axis(scale=xs, label="X"), Axis(
scale=ys, label="Y", orientation="vertical"
)
fig_lasso = Figure(
marks=[scatter_lasso, line_lasso, bar_lasso],
axes=[xax_lasso, yax_lasso],
title="Lasso Selector Example",
interaction=lasso_sel,
)
lasso_sel.marks = [scatter_lasso, line_lasso]
fig_lasso
scatter_lasso.selected, line_lasso.selected
```
## Pan Zoom
```
xs_pz = DateScale(min=np.datetime64(py_dtime(2007, 1, 1)))
ys_pz = LinearScale()
line_pz = Lines(
x=dates_actual, y=sec2_data, scales={"x": xs_pz, "y": ys_pz}, colors=["red"]
)
panzoom = PanZoom(scales={"x": [xs_pz], "y": [ys_pz]})
xax = Axis(scale=xs_pz, label="Date", grids="off")
yax = Axis(scale=ys_pz, label="Price", orientation="vertical", grid_lines="none")
Figure(marks=[line_pz], axes=[xax, yax], interaction=panzoom)
```
## Hand Draw
```
xs_hd = DateScale(min=np.datetime64(py_dtime(2007, 1, 1)))
ys_hd = LinearScale()
line_hd = Lines(
x=dates_actual, y=sec2_data, scales={"x": xs_hd, "y": ys_hd}, colors=["red"]
)
handdraw = HandDraw(lines=line_hd)
xax = Axis(scale=xs_hd, label="Date", grid_lines="none")
yax = Axis(scale=ys_hd, label="Price", orientation="vertical", grid_lines="none")
Figure(marks=[line_hd], axes=[xax, yax], interaction=handdraw)
```
# Unified Figure with All Interactions
```
dt_x = DateScale(date_format=date_fmt, min=py_dtime(2007, 1, 1))
lc1_x = LinearScale()
lc2_y = LinearScale()
lc2 = Lines(
x=np.linspace(0.0, 10.0, len(prices)),
y=prices * 0.25,
scales={"x": lc1_x, "y": lc2_y},
display_legend=True,
labels=["Security 1"],
)
lc3 = Lines(
x=dates_actual,
y=sec2_data,
scales={"x": dt_x, "y": lc2_y},
colors=["red"],
display_legend=True,
labels=["Security 2"],
)
lc4 = Lines(
x=np.linspace(0.0, 10.0, len(prices)),
y=sec2_data * 0.75,
scales={"x": LinearScale(min=5, max=10), "y": lc2_y},
colors=["green"],
display_legend=True,
labels=["Security 2 squared"],
)
x_ax1 = Axis(label="Date", scale=dt_x)
x_ax2 = Axis(label="Time", scale=lc1_x, side="top", grid_lines="none")
x_ay2 = Axis(label=(symbol + " Price"), scale=lc2_y, orientation="vertical")
fig = Figure(marks=[lc2, lc3, lc4], axes=[x_ax1, x_ax2, x_ay2])
## declaring the interactions
multi_sel = MultiSelector(scale=dt_x, marks=[lc2, lc3])
br_intsel = BrushIntervalSelector(scale=lc1_x, marks=[lc2, lc3])
index_sel = IndexSelector(scale=dt_x, marks=[lc2, lc3])
int_sel = FastIntervalSelector(scale=dt_x, marks=[lc3, lc2])
hd = HandDraw(lines=lc2)
hd2 = HandDraw(lines=lc3)
pz = PanZoom(scales={"x": [dt_x], "y": [lc2_y]})
deb = HTML()
deb.value = "[]"
## Call back handler for the interactions
def test_callback(change):
deb.value = str(change.new)
multi_sel.observe(test_callback, names=["selected"])
br_intsel.observe(test_callback, names=["selected"])
index_sel.observe(test_callback, names=["selected"])
int_sel.observe(test_callback, names=["selected"])
from collections import OrderedDict
selection_interacts = ToggleButtons(
options=OrderedDict(
[
("HandDraw1", hd),
("HandDraw2", hd2),
("PanZoom", pz),
("FastIntervalSelector", int_sel),
("IndexSelector", index_sel),
("BrushIntervalSelector", br_intsel),
("MultiSelector", multi_sel),
("None", None),
]
)
)
link((selection_interacts, "value"), (fig, "interaction"))
VBox([deb, fig, selection_interacts], align_self="stretch")
# Set the scales of lc4 to the ones of lc2 and check if panzoom pans the two.
lc4.scales = lc2.scales
```
|
github_jupyter
|
import pandas as pd
import numpy as np
symbol = "Security 1"
symbol2 = "Security 2"
price_data = pd.DataFrame(
np.cumsum(np.random.randn(150, 2).dot([[0.5, 0.4], [0.4, 1.0]]), axis=0) + 100,
columns=["Security 1", "Security 2"],
index=pd.date_range(start="01-01-2007", periods=150),
)
dates_actual = price_data.index.values
prices = price_data[symbol].values
from bqplot import DateScale, LinearScale, Axis, Lines, Scatter, Bars, Hist, Figure
from bqplot.interacts import (
FastIntervalSelector,
IndexSelector,
BrushIntervalSelector,
BrushSelector,
MultiSelector,
LassoSelector,
PanZoom,
HandDraw,
)
from traitlets import link
from ipywidgets import ToggleButtons, VBox, HTML
## First we define a Figure
dt_x_fast = DateScale()
lin_y = LinearScale()
x_ax = Axis(label="Index", scale=dt_x_fast)
x_ay = Axis(label=(symbol + " Price"), scale=lin_y, orientation="vertical")
lc = Lines(
x=dates_actual, y=prices, scales={"x": dt_x_fast, "y": lin_y}, colors=["orange"]
)
lc_2 = Lines(
x=dates_actual[50:],
y=prices[50:] + 2,
scales={"x": dt_x_fast, "y": lin_y},
colors=["blue"],
)
## Next we define the type of selector we would like
intsel_fast = FastIntervalSelector(scale=dt_x_fast, marks=[lc, lc_2])
## Now, we define a function that will be called when the FastIntervalSelector is interacted with
def fast_interval_change_callback(change):
db_fast.value = "The selected period is " + str(change.new)
## Now we connect the selectors to that function
intsel_fast.observe(fast_interval_change_callback, names=["selected"])
## We use the HTML widget to see the value of what we are selecting and modify it when an interaction is performed
## on the selector
db_fast = HTML()
db_fast.value = "The selected period is " + str(intsel_fast.selected)
fig_fast_intsel = Figure(
marks=[lc, lc_2],
axes=[x_ax, x_ay],
title="Fast Interval Selector Example",
interaction=intsel_fast,
) # This is where we assign the interaction to this particular Figure
VBox([db_fast, fig_fast_intsel])
db_index = HTML(value="[]")
## Now we try a selector made to select all the y-values associated with a single x-value
index_sel = IndexSelector(scale=dt_x_fast, marks=[lc, lc_2])
## Now, we define a function that will be called when the selectors are interacted with
def index_change_callback(change):
db_index.value = "The selected date is " + str(change.new)
index_sel.observe(index_change_callback, names=["selected"])
fig_index_sel = Figure(
marks=[lc, lc_2],
axes=[x_ax, x_ay],
title="Index Selector Example",
interaction=index_sel,
)
VBox([db_index, fig_index_sel])
from datetime import datetime as py_dtime
dt_x_index = DateScale(min=np.datetime64(py_dtime(2006, 6, 1)))
lin_y2 = LinearScale()
lc2_index = Lines(x=dates_actual, y=prices, scales={"x": dt_x_index, "y": lin_y2})
x_ax1 = Axis(label="Date", scale=dt_x_index)
x_ay2 = Axis(label=(symbol + " Price"), scale=lin_y2, orientation="vertical")
intsel_date = FastIntervalSelector(scale=dt_x_index, marks=[lc2_index])
db_date = HTML()
db_date.value = str(intsel_date.selected)
## Now, we define a function that will be called when the selectors are interacted with - a callback
def date_interval_change_callback(change):
db_date.value = str(change.new)
## Notice here that we call the observe on the Mark lc2_index rather than on the selector intsel_date
lc2_index.observe(date_interval_change_callback, names=["selected"])
fig_date_mark = Figure(
marks=[lc2_index],
axes=[x_ax1, x_ay2],
title="Fast Interval Selector Selected Indices Example",
interaction=intsel_date,
)
VBox([db_date, fig_date_mark])
## Defining a new Figure
dt_x_brush = DateScale(min=np.datetime64(py_dtime(2006, 6, 1)))
lin_y2_brush = LinearScale()
lc3_brush = Lines(x=dates_actual, y=prices, scales={"x": dt_x_brush, "y": lin_y2_brush})
x_ax_brush = Axis(label="Date", scale=dt_x_brush)
x_ay_brush = Axis(label=(symbol + " Price"), scale=lin_y2_brush, orientation="vertical")
db_brush = HTML(value="[]")
brushsel_date = BrushIntervalSelector(
scale=dt_x_brush, marks=[lc3_brush], color="FireBrick"
)
## Now, we define a function that will be called when the selectors are interacted with - a callback
def date_brush_change_callback(change):
db_brush.value = str(change.new)
lc3_brush.observe(date_brush_change_callback, names=["selected"])
fig_brush_sel = Figure(
marks=[lc3_brush],
axes=[x_ax_brush, x_ay_brush],
title="Brush Selector Selected Indices Example",
interaction=brushsel_date,
)
VBox([db_brush, fig_brush_sel])
date_fmt = "%m-%d-%Y"
sec2_data = price_data[symbol2].values
dates = price_data.index.values
sc_x = LinearScale()
sc_y = LinearScale()
scatt = Scatter(x=prices, y=sec2_data, scales={"x": sc_x, "y": sc_y})
sc_xax = Axis(label=(symbol), scale=sc_x)
sc_yax = Axis(label=(symbol2), scale=sc_y, orientation="vertical")
br_sel = BrushSelector(x_scale=sc_x, y_scale=sc_y, marks=[scatt], color="red")
db_scat_brush = HTML(value="[]")
## call back for the selector
def brush_callback(change):
db_scat_brush.value = str(br_sel.selected)
br_sel.observe(brush_callback, names=["brushing"])
fig_scat_brush = Figure(
marks=[scatt],
axes=[sc_xax, sc_yax],
title="Scatter Chart Brush Selector Example",
interaction=br_sel,
)
VBox([db_scat_brush, fig_scat_brush])
sc_brush_dt_x = DateScale(date_format=date_fmt)
sc_brush_dt_y = LinearScale()
scatt2 = Scatter(
x=dates_actual, y=sec2_data, scales={"x": sc_brush_dt_x, "y": sc_brush_dt_y}
)
br_sel_dt = BrushSelector(x_scale=sc_brush_dt_x, y_scale=sc_brush_dt_y, marks=[scatt2])
db_brush_dt = HTML(value=str(br_sel_dt.selected))
## call back for the selector
def brush_dt_callback(change):
db_brush_dt.value = str(br_sel_dt.selected)
br_sel_dt.observe(brush_dt_callback, names=["brushing"])
sc_xax = Axis(label=(symbol), scale=sc_brush_dt_x)
sc_yax = Axis(label=(symbol2), scale=sc_brush_dt_y, orientation="vertical")
fig_brush_dt = Figure(
marks=[scatt2],
axes=[sc_xax, sc_yax],
title="Brush Selector with Dates Example",
interaction=br_sel_dt,
)
VBox([db_brush_dt, fig_brush_dt])
## call back for selectors
def interval_change_callback(name, value):
db3.value = str(value)
## call back for the selector
def brush_callback(change):
if not br_intsel.brushing:
db3.value = str(br_intsel.selected)
returns = np.log(prices[1:]) - np.log(prices[:-1])
hist_x = LinearScale()
hist_y = LinearScale()
hist = Hist(sample=returns, scales={"sample": hist_x, "count": hist_y})
br_intsel = BrushIntervalSelector(scale=hist_x, marks=[hist])
br_intsel.observe(brush_callback, names=["selected"])
br_intsel.observe(brush_callback, names=["brushing"])
db3 = HTML()
db3.value = str(br_intsel.selected)
h_xax = Axis(
scale=hist_x, label="Returns", grids="off", set_ticks=True, tick_format="0.2%"
)
h_yax = Axis(scale=hist_y, label="Freq", orientation="vertical", grid_lines="none")
fig_hist = Figure(
marks=[hist],
axes=[h_xax, h_yax],
title="Histogram Selection Example",
interaction=br_intsel,
)
VBox([db3, fig_hist])
def multi_sel_callback(change):
if not multi_sel.brushing:
db4.value = str(multi_sel.selected)
line_x = LinearScale()
line_y = LinearScale()
line = Lines(
x=np.arange(100), y=np.random.randn(100), scales={"x": line_x, "y": line_y}
)
multi_sel = MultiSelector(scale=line_x, marks=[line])
multi_sel.observe(multi_sel_callback, names=["selected"])
multi_sel.observe(multi_sel_callback, names=["brushing"])
db4 = HTML()
db4.value = str(multi_sel.selected)
h_xax = Axis(scale=line_x, label="Returns", grid_lines="none")
h_yax = Axis(scale=hist_y, label="Freq", orientation="vertical", grid_lines="none")
fig_multi = Figure(
marks=[line],
axes=[h_xax, h_yax],
title="Multi-Selector Example",
interaction=multi_sel,
)
VBox([db4, fig_multi])
# changing the names of the intervals.
multi_sel.names = ["int1", "int2", "int3"]
def multi_sel_dt_callback(change):
if not multi_sel_dt.brushing:
db_multi_dt.value = str(multi_sel_dt.selected)
line_dt_x = DateScale(min=np.datetime64(py_dtime(2007, 1, 1)))
line_dt_y = LinearScale()
line_dt = Lines(
x=dates_actual, y=sec2_data, scales={"x": line_dt_x, "y": line_dt_y}, colors=["red"]
)
multi_sel_dt = MultiSelector(scale=line_dt_x)
multi_sel_dt.observe(multi_sel_dt_callback, names=["selected"])
multi_sel_dt.observe(multi_sel_dt_callback, names=["brushing"])
db_multi_dt = HTML()
db_multi_dt.value = str(multi_sel_dt.selected)
h_xax_dt = Axis(scale=line_dt_x, label="Returns", grid_lines="none")
h_yax_dt = Axis(
scale=line_dt_y, label="Freq", orientation="vertical", grid_lines="none"
)
fig_multi_dt = Figure(
marks=[line_dt],
axes=[h_xax_dt, h_yax_dt],
title="Multi-Selector with Date Example",
interaction=multi_sel_dt,
)
VBox([db_multi_dt, fig_multi_dt])
lasso_sel = LassoSelector()
xs, ys = LinearScale(), LinearScale()
data = np.arange(20)
line_lasso = Lines(x=data, y=data, scales={"x": xs, "y": ys})
scatter_lasso = Scatter(x=data, y=data, scales={"x": xs, "y": ys}, colors=["skyblue"])
bar_lasso = Bars(x=data, y=data / 2.0, scales={"x": xs, "y": ys})
xax_lasso, yax_lasso = Axis(scale=xs, label="X"), Axis(
scale=ys, label="Y", orientation="vertical"
)
fig_lasso = Figure(
marks=[scatter_lasso, line_lasso, bar_lasso],
axes=[xax_lasso, yax_lasso],
title="Lasso Selector Example",
interaction=lasso_sel,
)
lasso_sel.marks = [scatter_lasso, line_lasso]
fig_lasso
scatter_lasso.selected, line_lasso.selected
xs_pz = DateScale(min=np.datetime64(py_dtime(2007, 1, 1)))
ys_pz = LinearScale()
line_pz = Lines(
x=dates_actual, y=sec2_data, scales={"x": xs_pz, "y": ys_pz}, colors=["red"]
)
panzoom = PanZoom(scales={"x": [xs_pz], "y": [ys_pz]})
xax = Axis(scale=xs_pz, label="Date", grids="off")
yax = Axis(scale=ys_pz, label="Price", orientation="vertical", grid_lines="none")
Figure(marks=[line_pz], axes=[xax, yax], interaction=panzoom)
xs_hd = DateScale(min=np.datetime64(py_dtime(2007, 1, 1)))
ys_hd = LinearScale()
line_hd = Lines(
x=dates_actual, y=sec2_data, scales={"x": xs_hd, "y": ys_hd}, colors=["red"]
)
handdraw = HandDraw(lines=line_hd)
xax = Axis(scale=xs_hd, label="Date", grid_lines="none")
yax = Axis(scale=ys_hd, label="Price", orientation="vertical", grid_lines="none")
Figure(marks=[line_hd], axes=[xax, yax], interaction=handdraw)
dt_x = DateScale(date_format=date_fmt, min=py_dtime(2007, 1, 1))
lc1_x = LinearScale()
lc2_y = LinearScale()
lc2 = Lines(
x=np.linspace(0.0, 10.0, len(prices)),
y=prices * 0.25,
scales={"x": lc1_x, "y": lc2_y},
display_legend=True,
labels=["Security 1"],
)
lc3 = Lines(
x=dates_actual,
y=sec2_data,
scales={"x": dt_x, "y": lc2_y},
colors=["red"],
display_legend=True,
labels=["Security 2"],
)
lc4 = Lines(
x=np.linspace(0.0, 10.0, len(prices)),
y=sec2_data * 0.75,
scales={"x": LinearScale(min=5, max=10), "y": lc2_y},
colors=["green"],
display_legend=True,
labels=["Security 2 squared"],
)
x_ax1 = Axis(label="Date", scale=dt_x)
x_ax2 = Axis(label="Time", scale=lc1_x, side="top", grid_lines="none")
x_ay2 = Axis(label=(symbol + " Price"), scale=lc2_y, orientation="vertical")
fig = Figure(marks=[lc2, lc3, lc4], axes=[x_ax1, x_ax2, x_ay2])
## declaring the interactions
multi_sel = MultiSelector(scale=dt_x, marks=[lc2, lc3])
br_intsel = BrushIntervalSelector(scale=lc1_x, marks=[lc2, lc3])
index_sel = IndexSelector(scale=dt_x, marks=[lc2, lc3])
int_sel = FastIntervalSelector(scale=dt_x, marks=[lc3, lc2])
hd = HandDraw(lines=lc2)
hd2 = HandDraw(lines=lc3)
pz = PanZoom(scales={"x": [dt_x], "y": [lc2_y]})
deb = HTML()
deb.value = "[]"
## Call back handler for the interactions
def test_callback(change):
deb.value = str(change.new)
multi_sel.observe(test_callback, names=["selected"])
br_intsel.observe(test_callback, names=["selected"])
index_sel.observe(test_callback, names=["selected"])
int_sel.observe(test_callback, names=["selected"])
from collections import OrderedDict
selection_interacts = ToggleButtons(
options=OrderedDict(
[
("HandDraw1", hd),
("HandDraw2", hd2),
("PanZoom", pz),
("FastIntervalSelector", int_sel),
("IndexSelector", index_sel),
("BrushIntervalSelector", br_intsel),
("MultiSelector", multi_sel),
("None", None),
]
)
)
link((selection_interacts, "value"), (fig, "interaction"))
VBox([deb, fig, selection_interacts], align_self="stretch")
# Set the scales of lc4 to the ones of lc2 and check if panzoom pans the two.
lc4.scales = lc2.scales
| 0.661595 | 0.84626 |
[View in Colaboratory](https://colab.research.google.com/github/apurvaasf/Jupyter_Problem_Proposals/blob/master/Proposal1/Proposal1_Excel.ipynb)
```
# Download the zip file at dataUrl and save it as data.zip.
dataUrl = "https://s3-us-west-2.amazonaws.com/apurvasbucket2/data.zip"
import os.path
import shutil
def download_file(url, filename):
# Check to see if file already exists
fileExists = os.path.isfile(filename)
if not fileExists:
import requests
print("Downloading", filename)
response = requests.get(url, stream=True)
# Throw an error for bad status codes
response.raise_for_status()
with open(filename, 'wb') as handle:
for block in response.iter_content(1024*256): # Load 256KB at a time and provide feedback.
print('.', end='') # print without new line
handle.write(block)
print('\n',filename, "downloaded.")
else:
print(filename, "exists. Skipping download")
download_file(dataUrl, "data.zip")
# Unzip data.zip into the data folder.
# Unzip all the zipfiles
import zipfile
# Delete any current data directory.
dir = 'data'
if os.path.exists(dir):
shutil.rmtree(dir)
os.makedirs(dir)
def unzip_file(theFile):
path_to_zip_file = theFile
zip_ref = zipfile.ZipFile(path_to_zip_file, 'r')
zip_ref.extractall(".")
zip_ref.close()
for filename in ["data.zip"]:
print("Unzipping", filename)
unzip_file(filename)
# Save the list of problem directories to the problems list
from os import walk
directories = []
for (dirpath, dirnames, filenames) in walk("data/problems"):
directories.extend(dirnames)
break
problems = []
for directory in directories:
if (directory[0] != "."):
problems.append(directory)
print(problems)
# For each problem, produce a solution. Copy the example_problem to the solution by default.
import shutil
import pandas as pd
import numpy as np
# Call the overfit solver.
# This will only work when example_solutions are present.
#
def solve(problem):
shutil.copytree("data/example_solutions/"+problem, "data/solutions/"+problem)
# Non-overfit solution - NOT NEEDED FOR PROPOSALS
"""
def solve(problem):
problemFile = "data/problems/"+problem+"/input.csv"
problemText = open( problemFile, "r").read()
print(problemText)
# Get the numbers, add them together, save to solution directory.
inputs = problemText.split(",")
x = int(inputs[0])
y = int(inputs[1])
result = x + y
print(inputs, result)
solutionFile = "data/solutions/"+problem+"/solution.csv"
dir = 'data/solutions/'+problem
if os.path.exists(dir):
shutil.rmtree(dir)
os.makedirs(dir)
with open(solutionFile, 'w') as out:
out.write(str(result))
"""
pass
```
# Section 2
```
# Delete any existing solutions.
dir = 'data/solutions'
if os.path.exists(dir):
shutil.rmtree(dir)
os.makedirs(dir)
# Call the new solve function for all available problems.
for problem in problems:
solve(problem)
for problemDir in problems:
solutionFile = "data/solutions/"+problemDir+"/solution.csv"
example_solutionFile = "data/example_solutions/"+problemDir+"/solution.csv"
# df_solutionFile=pd.read_csv(solutionFile)
# df_example_solutionFile=pd.read_csv(example_solutionFile)
solutionText = open( solutionFile, "r").read()
# Test all available solutions.
# Custom test code goes here.
def test_case(problemDir):
"""
problemFile = "data/problems/"+problemDir+"/input.csv"
problemText = open( problemFile, "r").read()
print(problemText)
"""
example_solutionFile = "data/example_solutions/"+problemDir+"/solution.csv"
example_solutionText = open( example_solutionFile, "r").read()
solutionFile = "data/solutions/"+problemDir+"/solution.csv"
solutionText = open( solutionFile, "r").read()
if( solutionText == example_solutionText ):
return True
else:
return False
# End customer test code.
for problem in problems:
result = test_case(problem)
if(result):
print("pass")
else:
print("Invalid solution for problem ",problem)
print("\n")
```
|
github_jupyter
|
# Download the zip file at dataUrl and save it as data.zip.
dataUrl = "https://s3-us-west-2.amazonaws.com/apurvasbucket2/data.zip"
import os.path
import shutil
def download_file(url, filename):
# Check to see if file already exists
fileExists = os.path.isfile(filename)
if not fileExists:
import requests
print("Downloading", filename)
response = requests.get(url, stream=True)
# Throw an error for bad status codes
response.raise_for_status()
with open(filename, 'wb') as handle:
for block in response.iter_content(1024*256): # Load 256KB at a time and provide feedback.
print('.', end='') # print without new line
handle.write(block)
print('\n',filename, "downloaded.")
else:
print(filename, "exists. Skipping download")
download_file(dataUrl, "data.zip")
# Unzip data.zip into the data folder.
# Unzip all the zipfiles
import zipfile
# Delete any current data directory.
dir = 'data'
if os.path.exists(dir):
shutil.rmtree(dir)
os.makedirs(dir)
def unzip_file(theFile):
path_to_zip_file = theFile
zip_ref = zipfile.ZipFile(path_to_zip_file, 'r')
zip_ref.extractall(".")
zip_ref.close()
for filename in ["data.zip"]:
print("Unzipping", filename)
unzip_file(filename)
# Save the list of problem directories to the problems list
from os import walk
directories = []
for (dirpath, dirnames, filenames) in walk("data/problems"):
directories.extend(dirnames)
break
problems = []
for directory in directories:
if (directory[0] != "."):
problems.append(directory)
print(problems)
# For each problem, produce a solution. Copy the example_problem to the solution by default.
import shutil
import pandas as pd
import numpy as np
# Call the overfit solver.
# This will only work when example_solutions are present.
#
def solve(problem):
shutil.copytree("data/example_solutions/"+problem, "data/solutions/"+problem)
# Non-overfit solution - NOT NEEDED FOR PROPOSALS
"""
def solve(problem):
problemFile = "data/problems/"+problem+"/input.csv"
problemText = open( problemFile, "r").read()
print(problemText)
# Get the numbers, add them together, save to solution directory.
inputs = problemText.split(",")
x = int(inputs[0])
y = int(inputs[1])
result = x + y
print(inputs, result)
solutionFile = "data/solutions/"+problem+"/solution.csv"
dir = 'data/solutions/'+problem
if os.path.exists(dir):
shutil.rmtree(dir)
os.makedirs(dir)
with open(solutionFile, 'w') as out:
out.write(str(result))
"""
pass
# Delete any existing solutions.
dir = 'data/solutions'
if os.path.exists(dir):
shutil.rmtree(dir)
os.makedirs(dir)
# Call the new solve function for all available problems.
for problem in problems:
solve(problem)
for problemDir in problems:
solutionFile = "data/solutions/"+problemDir+"/solution.csv"
example_solutionFile = "data/example_solutions/"+problemDir+"/solution.csv"
# df_solutionFile=pd.read_csv(solutionFile)
# df_example_solutionFile=pd.read_csv(example_solutionFile)
solutionText = open( solutionFile, "r").read()
# Test all available solutions.
# Custom test code goes here.
def test_case(problemDir):
"""
problemFile = "data/problems/"+problemDir+"/input.csv"
problemText = open( problemFile, "r").read()
print(problemText)
"""
example_solutionFile = "data/example_solutions/"+problemDir+"/solution.csv"
example_solutionText = open( example_solutionFile, "r").read()
solutionFile = "data/solutions/"+problemDir+"/solution.csv"
solutionText = open( solutionFile, "r").read()
if( solutionText == example_solutionText ):
return True
else:
return False
# End customer test code.
for problem in problems:
result = test_case(problem)
if(result):
print("pass")
else:
print("Invalid solution for problem ",problem)
print("\n")
| 0.311217 | 0.707645 |
```
import glob
import os
import random
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import cv2
import math
from functools import partial
from tqdm.auto import tqdm
import torch
from mpl_toolkits.axes_grid1 import make_axes_locatable
import fastai
from fastai.vision.all import *
from fastai.data.core import DataLoaders
import sys
sys.path.append('../src')
from clf_model_utils.miccai_2d_dataset import MICCAI2DDataset
import json
import fastai
from fastai.vision.all import *
from fastai.data.core import DataLoaders
from fastai.callback.all import *
from fastai.callback.wandb import WandbCallback
import torch.nn.functional as F
from timm import create_model
from fastai.vision.learner import _update_first_layer
from fastai.vision.learner import _add_norm
MODEL_FOLDERS = [
{
'model' : 'resnet50_rocstar',
'arch' : 'resnet50',
'fn' : '../output/resnet50_bs32_ep10_rocstar_lr0.0001_ps0.8_ranger_sz256/'
}
]
MODEL_INDEX = 0
train_df_fn = '../input/train_feature_data_v2.csv'
fold = 0
im_sz = 256
npy_dir = '../input/aligned_and_cropped_t2w/'
df = pd.read_csv(train_df_fn)
train_df = df[df.fold != fold]
val_df = df[df.fold == fold]
image_size = (im_sz,im_sz)
# timm + fastai functions copied from https://walkwithfastai.com/vision.external.timm
def create_timm_body(arch:str, pretrained=True, cut=None, n_in=3):
"Creates a body from any model in the `timm` library."
if 'vit' in arch:
model = create_model(arch, pretrained=pretrained, num_classes=0)
else:
model = create_model(arch, pretrained=pretrained, num_classes=0, global_pool='')
_update_first_layer(model, n_in, pretrained)
if cut is None:
ll = list(enumerate(model.children()))
cut = next(i for i,o in reversed(ll) if has_pool_type(o))
if isinstance(cut, int): return nn.Sequential(*list(model.children())[:cut])
elif callable(cut): return cut(model)
else: raise NamedError("cut must be either integer or function")
def create_timm_model(arch:str, n_out, cut=None, pretrained=True, n_in=3, init=nn.init.kaiming_normal_, custom_head=None,
concat_pool=True, **kwargs):
"Create custom architecture using `arch`, `n_in` and `n_out` from the `timm` library"
body = create_timm_body(arch, pretrained, None, n_in)
if custom_head is None:
nf = num_features_model(nn.Sequential(*body.children()))
head = create_head(nf, n_out, concat_pool=concat_pool, **kwargs)
else: head = custom_head
model = nn.Sequential(body, head)
if init is not None: apply_init(model[1], init)
return model
def timm_learner(dls, arch:str, loss_func=None, pretrained=True, cut=None, splitter=None,
y_range=None, config=None, n_out=None, normalize=True, **kwargs):
"Build a convnet style learner from `dls` and `arch` using the `timm` library"
if config is None: config = {}
if n_out is None: n_out = get_c(dls)
assert n_out, "`n_out` is not defined, and could not be inferred from data, set `dls.c` or pass `n_out`"
if y_range is None and 'y_range' in config: y_range = config.pop('y_range')
model = create_timm_model(arch, n_out, default_split, pretrained, y_range=y_range, **config)
kwargs.pop('ps')
learn = Learner(dls, model, loss_func=loss_func, splitter=default_split, **kwargs)
if pretrained: learn.freeze()
return learn
ds_t = MICCAI2DDataset(
train_df,
npy_dir=npy_dir,
image_size=image_size,
tio_augmentations=None,
is_train=True
)
ds_v = MICCAI2DDataset(
val_df,
npy_dir=npy_dir,
image_size=image_size,
tio_augmentations=None,
is_train=False
)
num_workers = 8
bs = 8
dls = DataLoaders.from_dsets(ds_t, ds_v, bs=bs, device='cuda', num_workers=num_workers)
loss = LabelSmoothingCrossEntropyFlat(eps=0.2)
opt_func = fastai.optimizer.ranger
arch = MODEL_FOLDERS[MODEL_INDEX]['arch']
create_learner = cnn_learner
if arch == 'densetnet121':
base = densenet121
elif arch == 'resnet18':
base = resnet18
elif arch == 'resnet34':
base = resnet34
elif arch == 'resnet50':
base = resnet50
elif arch == 'resnet101':
base = resnet101
elif arch == 'densenet169':
base = densenet169
else:
create_learner = timm_learner
base = arch
learn = create_learner(
dls,
base,
pretrained=True,
n_out=2,
loss_func=loss,
opt_func=opt_func,
metrics=[
accuracy
],
ps=0.8
).to_fp16()
model_path = os.path.join(MODEL_FOLDERS[MODEL_INDEX]['fn'], f'fold-{fold}', 'final.pth')
learn.model.load_state_dict(torch.load(model_path))
```
## CAM
```
def show_cam_one_batch(batch, preds=None, cams=None, scale=4, save_fn=None):
_images, _labels = batch
images = _images.cpu().numpy()[:,0,:,:] # reduce rgb dimension to grayscale
cam_images = cams.detach().cpu().numpy()
labels = [_labels.cpu().numpy()]
if preds is not None:
pred_lbls = list(preds.cpu().numpy())
else:
pred_lbls = [-1 for _ in labels]
plt.close('all')
f, axs = plt.subplots(1, 1, figsize=((scale + 1), scale))
axs = [axs]
idx = 0
for img, lbl, pred, ax in zip(images, labels, pred_lbls, axs):
ax.imshow(cv2.cvtColor((((img - np.min(img)) / (np.max(img) - np.min(img)))*255).astype(np.uint8), cv2.COLOR_GRAY2RGB))
axim = ax.imshow(
cam_images[idx], alpha=0.6, extent=(0,256,256,0),
interpolation='bilinear', cmap='magma'
)
divider = make_axes_locatable(ax)
cax = divider.append_axes('right', size='5%', pad=0.05)
f.colorbar(axim, cax=cax, orientation='vertical')
ax.set_title(f'GT: {lbl}, Pred: {pred:.3f}', fontsize=16)
ax.set_xticks([])
ax.set_yticks([])
idx += 1
# hide empties
for ax_index in range(len(images), len(axs)):
axs[ax_index].axis('off')
plt.tight_layout()
plt.subplots_adjust(left = 0.1, right = 0.9, wspace=0.2, hspace=0.05)
if save_fn is not None:
plt.savefig(save_fn, transparent=False)
else:
plt.show()
# grab val set batch
val_item = ds_v.__getitem__(7)
print(val_item[0].shape, val_item[1])
val_batch = torch.unsqueeze(torch.tensor(val_item[0]), 0), torch.tensor(val_item[1])
class Hook():
def __init__(self, m):
self.hook = m.register_forward_hook(self.hook_func)
def hook_func(self, m, i, o): self.stored = o.detach().clone()
def __enter__(self, *args): return self
def __exit__(self, *args): self.hook.remove()
class HookBwd():
def __init__(self, m):
self.hook = m.register_backward_hook(self.hook_func)
def hook_func(self, m, gi, go): self.stored = go[0].detach().clone()
def __enter__(self, *args): return self
def __exit__(self, *args): self.hook.remove()
cls = 1
with HookBwd(learn.model[0]) as hookg:
with Hook(learn.model[0]) as hook:
output = learn.model.eval()(val_batch[0])
act = hook.stored
output[0,cls].backward()
grad = hookg.stored
w = grad[0].mean(dim=[1,2], keepdim=True)
cam_map = (w * act[0]).sum(0)
cam_map.shape
# check prediction
pred = torch.softmax(output, dim=-1).detach()
pred
show_cam_one_batch(val_batch, preds=torch.unsqueeze(pred[0][1],0), cams=torch.unsqueeze(cam_map, 0))
```
|
github_jupyter
|
import glob
import os
import random
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import cv2
import math
from functools import partial
from tqdm.auto import tqdm
import torch
from mpl_toolkits.axes_grid1 import make_axes_locatable
import fastai
from fastai.vision.all import *
from fastai.data.core import DataLoaders
import sys
sys.path.append('../src')
from clf_model_utils.miccai_2d_dataset import MICCAI2DDataset
import json
import fastai
from fastai.vision.all import *
from fastai.data.core import DataLoaders
from fastai.callback.all import *
from fastai.callback.wandb import WandbCallback
import torch.nn.functional as F
from timm import create_model
from fastai.vision.learner import _update_first_layer
from fastai.vision.learner import _add_norm
MODEL_FOLDERS = [
{
'model' : 'resnet50_rocstar',
'arch' : 'resnet50',
'fn' : '../output/resnet50_bs32_ep10_rocstar_lr0.0001_ps0.8_ranger_sz256/'
}
]
MODEL_INDEX = 0
train_df_fn = '../input/train_feature_data_v2.csv'
fold = 0
im_sz = 256
npy_dir = '../input/aligned_and_cropped_t2w/'
df = pd.read_csv(train_df_fn)
train_df = df[df.fold != fold]
val_df = df[df.fold == fold]
image_size = (im_sz,im_sz)
# timm + fastai functions copied from https://walkwithfastai.com/vision.external.timm
def create_timm_body(arch:str, pretrained=True, cut=None, n_in=3):
"Creates a body from any model in the `timm` library."
if 'vit' in arch:
model = create_model(arch, pretrained=pretrained, num_classes=0)
else:
model = create_model(arch, pretrained=pretrained, num_classes=0, global_pool='')
_update_first_layer(model, n_in, pretrained)
if cut is None:
ll = list(enumerate(model.children()))
cut = next(i for i,o in reversed(ll) if has_pool_type(o))
if isinstance(cut, int): return nn.Sequential(*list(model.children())[:cut])
elif callable(cut): return cut(model)
else: raise NamedError("cut must be either integer or function")
def create_timm_model(arch:str, n_out, cut=None, pretrained=True, n_in=3, init=nn.init.kaiming_normal_, custom_head=None,
concat_pool=True, **kwargs):
"Create custom architecture using `arch`, `n_in` and `n_out` from the `timm` library"
body = create_timm_body(arch, pretrained, None, n_in)
if custom_head is None:
nf = num_features_model(nn.Sequential(*body.children()))
head = create_head(nf, n_out, concat_pool=concat_pool, **kwargs)
else: head = custom_head
model = nn.Sequential(body, head)
if init is not None: apply_init(model[1], init)
return model
def timm_learner(dls, arch:str, loss_func=None, pretrained=True, cut=None, splitter=None,
y_range=None, config=None, n_out=None, normalize=True, **kwargs):
"Build a convnet style learner from `dls` and `arch` using the `timm` library"
if config is None: config = {}
if n_out is None: n_out = get_c(dls)
assert n_out, "`n_out` is not defined, and could not be inferred from data, set `dls.c` or pass `n_out`"
if y_range is None and 'y_range' in config: y_range = config.pop('y_range')
model = create_timm_model(arch, n_out, default_split, pretrained, y_range=y_range, **config)
kwargs.pop('ps')
learn = Learner(dls, model, loss_func=loss_func, splitter=default_split, **kwargs)
if pretrained: learn.freeze()
return learn
ds_t = MICCAI2DDataset(
train_df,
npy_dir=npy_dir,
image_size=image_size,
tio_augmentations=None,
is_train=True
)
ds_v = MICCAI2DDataset(
val_df,
npy_dir=npy_dir,
image_size=image_size,
tio_augmentations=None,
is_train=False
)
num_workers = 8
bs = 8
dls = DataLoaders.from_dsets(ds_t, ds_v, bs=bs, device='cuda', num_workers=num_workers)
loss = LabelSmoothingCrossEntropyFlat(eps=0.2)
opt_func = fastai.optimizer.ranger
arch = MODEL_FOLDERS[MODEL_INDEX]['arch']
create_learner = cnn_learner
if arch == 'densetnet121':
base = densenet121
elif arch == 'resnet18':
base = resnet18
elif arch == 'resnet34':
base = resnet34
elif arch == 'resnet50':
base = resnet50
elif arch == 'resnet101':
base = resnet101
elif arch == 'densenet169':
base = densenet169
else:
create_learner = timm_learner
base = arch
learn = create_learner(
dls,
base,
pretrained=True,
n_out=2,
loss_func=loss,
opt_func=opt_func,
metrics=[
accuracy
],
ps=0.8
).to_fp16()
model_path = os.path.join(MODEL_FOLDERS[MODEL_INDEX]['fn'], f'fold-{fold}', 'final.pth')
learn.model.load_state_dict(torch.load(model_path))
def show_cam_one_batch(batch, preds=None, cams=None, scale=4, save_fn=None):
_images, _labels = batch
images = _images.cpu().numpy()[:,0,:,:] # reduce rgb dimension to grayscale
cam_images = cams.detach().cpu().numpy()
labels = [_labels.cpu().numpy()]
if preds is not None:
pred_lbls = list(preds.cpu().numpy())
else:
pred_lbls = [-1 for _ in labels]
plt.close('all')
f, axs = plt.subplots(1, 1, figsize=((scale + 1), scale))
axs = [axs]
idx = 0
for img, lbl, pred, ax in zip(images, labels, pred_lbls, axs):
ax.imshow(cv2.cvtColor((((img - np.min(img)) / (np.max(img) - np.min(img)))*255).astype(np.uint8), cv2.COLOR_GRAY2RGB))
axim = ax.imshow(
cam_images[idx], alpha=0.6, extent=(0,256,256,0),
interpolation='bilinear', cmap='magma'
)
divider = make_axes_locatable(ax)
cax = divider.append_axes('right', size='5%', pad=0.05)
f.colorbar(axim, cax=cax, orientation='vertical')
ax.set_title(f'GT: {lbl}, Pred: {pred:.3f}', fontsize=16)
ax.set_xticks([])
ax.set_yticks([])
idx += 1
# hide empties
for ax_index in range(len(images), len(axs)):
axs[ax_index].axis('off')
plt.tight_layout()
plt.subplots_adjust(left = 0.1, right = 0.9, wspace=0.2, hspace=0.05)
if save_fn is not None:
plt.savefig(save_fn, transparent=False)
else:
plt.show()
# grab val set batch
val_item = ds_v.__getitem__(7)
print(val_item[0].shape, val_item[1])
val_batch = torch.unsqueeze(torch.tensor(val_item[0]), 0), torch.tensor(val_item[1])
class Hook():
def __init__(self, m):
self.hook = m.register_forward_hook(self.hook_func)
def hook_func(self, m, i, o): self.stored = o.detach().clone()
def __enter__(self, *args): return self
def __exit__(self, *args): self.hook.remove()
class HookBwd():
def __init__(self, m):
self.hook = m.register_backward_hook(self.hook_func)
def hook_func(self, m, gi, go): self.stored = go[0].detach().clone()
def __enter__(self, *args): return self
def __exit__(self, *args): self.hook.remove()
cls = 1
with HookBwd(learn.model[0]) as hookg:
with Hook(learn.model[0]) as hook:
output = learn.model.eval()(val_batch[0])
act = hook.stored
output[0,cls].backward()
grad = hookg.stored
w = grad[0].mean(dim=[1,2], keepdim=True)
cam_map = (w * act[0]).sum(0)
cam_map.shape
# check prediction
pred = torch.softmax(output, dim=-1).detach()
pred
show_cam_one_batch(val_batch, preds=torch.unsqueeze(pred[0][1],0), cams=torch.unsqueeze(cam_map, 0))
| 0.604866 | 0.342297 |
<a href="https://colab.research.google.com/github/alanexplorer/dip-2020-2/blob/main/Create_images.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
%matplotlib inline
import matplotlib
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import matplotlib.cm as cm
import numpy as np
from pylab import *
import math
from math import sin, cos, pi
from scipy.ndimage.interpolation import rotate
```
# Create the image of a paraboloid with one axis scaled (like an oval paraboloid).
```
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
r = T = np.arange(0, 2*pi, 0.01)
r, T = np.meshgrid(r, T)
X = r*np.cos(T)
Y = r*np.sin(T)
Z = r**2
ax.plot_surface(X, Y, Z, alpha=0.9, rstride=10, cstride=10, linewidth=0.5, cmap=cm.plasma)
plt.show()
```
# Create the image of a rotated sin using rotation of coordinates.
```
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
# rotate the samples by pi / 4 radians around y
a = pi / 4
t = np.transpose(np.array([X, Y, Z]), (1,2,0))
m = [[cos(a), 0, sin(a)],[0,1,0],[-sin(a), 0, cos(a)]]
x,y,z = np.transpose(np.dot(t, m), (2,0,1))
ax.plot_surface(x, y, z,linewidth=0.5, alpha = 0.9, rstride=10, cstride=10, cmap=cm.plasma)
plt.show()
```
# Create the image of a gaussian.
```
mx = 32 # x-coordinate of peak centre.
my = 32 # y-coordinate of peak centre.
sx = 6 # Standard deviation in x.
sy = 3 # Standard deviation in y.
coords = np.meshgrid(np.arange(0, 64), np.arange(0, 64)) # x and y coordinates for each image.
amplitude=20 # Highest intensity in image.
rho=0.8 # Correlation coefficient.
offset=20 # Offset from zero (background radiation).
x, y = coords
mx = float(mx)
my = float(my)
# Create covariance matrix
mat_cov = [[sx**2, rho * sx * sy],
[rho * sx * sy, sy**2]]
mat_cov = np.asarray(mat_cov)
# Find its inverse
mat_cov_inv = np.linalg.inv(mat_cov)
# PB We stack the coordinates along the last axis
mat_coords = np.stack((x - mx, y - my), axis=-1)
G = amplitude * np.exp(-0.5*np.matmul(np.matmul(mat_coords[:, :, np.newaxis, :],
mat_cov_inv),
mat_coords[..., np.newaxis])) + offset
plt.figure(figsize=(5, 5)).add_axes([0, 0, 1, 1])
plt.contourf(G.squeeze())
```
# Create a function that generates the image of a Gaussian optionally rotate by an angle theta and with mx, my, sx, sy as input arguments.
```
def Gaussian2D_v1(mx=0, # x-coordinate of peak centre.
my=0, # y-coordinate of peak centre.
sx=1, # Standard deviation in x.
sy=1,
angle=0): # Standard deviation in y.
coords = np.meshgrid(np.arange(0, 64), np.arange(0, 64)) # x and y coordinates for each image.
amplitude=20 # Highest intensity in image.
rho=0.8 # Correlation coefficient.
offset=20 # Offset from zero (background radiation).
x, y = coords
mx = float(mx)
my = float(my)
# Create covariance matrix
mat_cov = [[sx**2, rho * sx * sy],
[rho * sx * sy, sy**2]]
mat_cov = np.asarray(mat_cov)
# Find its inverse
mat_cov_inv = np.linalg.inv(mat_cov)
# PB We stack the coordinates along the last axis
mat_coords = np.stack((x - mx, y - my), axis=-1)
G = amplitude * np.exp(-0.5*np.matmul(np.matmul(mat_coords[:, :, np.newaxis, :],
mat_cov_inv),
mat_coords[..., np.newaxis])) + offset
G = rotate(G, angle)
return G.squeeze()
model = Gaussian2D_v1(mx=32,
my=32,
sx=6,
sy=3,
angle=10)
plt.figure(figsize=(5, 5)).add_axes([0, 0, 1, 1])
plt.contourf(model)
```
|
github_jupyter
|
%matplotlib inline
import matplotlib
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import matplotlib.cm as cm
import numpy as np
from pylab import *
import math
from math import sin, cos, pi
from scipy.ndimage.interpolation import rotate
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
r = T = np.arange(0, 2*pi, 0.01)
r, T = np.meshgrid(r, T)
X = r*np.cos(T)
Y = r*np.sin(T)
Z = r**2
ax.plot_surface(X, Y, Z, alpha=0.9, rstride=10, cstride=10, linewidth=0.5, cmap=cm.plasma)
plt.show()
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
# rotate the samples by pi / 4 radians around y
a = pi / 4
t = np.transpose(np.array([X, Y, Z]), (1,2,0))
m = [[cos(a), 0, sin(a)],[0,1,0],[-sin(a), 0, cos(a)]]
x,y,z = np.transpose(np.dot(t, m), (2,0,1))
ax.plot_surface(x, y, z,linewidth=0.5, alpha = 0.9, rstride=10, cstride=10, cmap=cm.plasma)
plt.show()
mx = 32 # x-coordinate of peak centre.
my = 32 # y-coordinate of peak centre.
sx = 6 # Standard deviation in x.
sy = 3 # Standard deviation in y.
coords = np.meshgrid(np.arange(0, 64), np.arange(0, 64)) # x and y coordinates for each image.
amplitude=20 # Highest intensity in image.
rho=0.8 # Correlation coefficient.
offset=20 # Offset from zero (background radiation).
x, y = coords
mx = float(mx)
my = float(my)
# Create covariance matrix
mat_cov = [[sx**2, rho * sx * sy],
[rho * sx * sy, sy**2]]
mat_cov = np.asarray(mat_cov)
# Find its inverse
mat_cov_inv = np.linalg.inv(mat_cov)
# PB We stack the coordinates along the last axis
mat_coords = np.stack((x - mx, y - my), axis=-1)
G = amplitude * np.exp(-0.5*np.matmul(np.matmul(mat_coords[:, :, np.newaxis, :],
mat_cov_inv),
mat_coords[..., np.newaxis])) + offset
plt.figure(figsize=(5, 5)).add_axes([0, 0, 1, 1])
plt.contourf(G.squeeze())
def Gaussian2D_v1(mx=0, # x-coordinate of peak centre.
my=0, # y-coordinate of peak centre.
sx=1, # Standard deviation in x.
sy=1,
angle=0): # Standard deviation in y.
coords = np.meshgrid(np.arange(0, 64), np.arange(0, 64)) # x and y coordinates for each image.
amplitude=20 # Highest intensity in image.
rho=0.8 # Correlation coefficient.
offset=20 # Offset from zero (background radiation).
x, y = coords
mx = float(mx)
my = float(my)
# Create covariance matrix
mat_cov = [[sx**2, rho * sx * sy],
[rho * sx * sy, sy**2]]
mat_cov = np.asarray(mat_cov)
# Find its inverse
mat_cov_inv = np.linalg.inv(mat_cov)
# PB We stack the coordinates along the last axis
mat_coords = np.stack((x - mx, y - my), axis=-1)
G = amplitude * np.exp(-0.5*np.matmul(np.matmul(mat_coords[:, :, np.newaxis, :],
mat_cov_inv),
mat_coords[..., np.newaxis])) + offset
G = rotate(G, angle)
return G.squeeze()
model = Gaussian2D_v1(mx=32,
my=32,
sx=6,
sy=3,
angle=10)
plt.figure(figsize=(5, 5)).add_axes([0, 0, 1, 1])
plt.contourf(model)
| 0.78108 | 0.978281 |
# Tutorial
This tutorial shows how to perform diffusion-weighted MR simulations using Disimpy. To follow along, [install](https://disimpy.readthedocs.io/en/latest/installation.html) the package and execute the code in each cell in the order that they are presented.
You can also use Google Colaboratory to run this notebook [interactively in your browser](https://colab.research.google.com/github/kerkelae/disimpy/blob/master/docs/source/tutorial.ipynb) even if you don't have an Nvidia CUDA-capable GPU. To use a GPU on Google Colaboratory, select *Runtime > Change runtime type > Hardware Type: GPU* in the top menu.
```
# If Disimpy has not been installed, uncomment the following line and
# execute the code in this cell
#!pip install disimpy
# Import the packages and modules used in this tutorial
import os
import pickle
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from disimpy import gradients, simulations, substrates, utils
```
## Simulation parameters
We need to define the number of random walkers and diffusivity. In this tutorial, we will use a small number of random walkers to keep the simulation runtime short and to be able to quickly visualize the random walks. However, in actual experiments, a larger number of random walkers should be used to make sure that signal converges, and the random walk trajectories do not need not be saved.
```
n_walkers = int(1e3)
diffusivity = 2e-9 # SI units (m^2/s)
```
## Gradient arrays
We need to define a gradient array that contains the necessary information about the simulated magnetic field gradients for diffusion encoding. Gradient arrays are `numpy.ndarray` instances with shape (number of measurements, number of time points, 3). The elements of gradient arrays are floating-point numbers representing the gradient magnitude along an axis at a time point in SI units (T/m). The module `disimpy.gradients` contains several useful functions for creating and modifying gradient arrays. In the example below, we define a gradient array to be used in this tutorial.
```
# Create a simple Stejskal-Tanner gradient waveform
gradient = np.zeros((1, 100, 3))
gradient[0, 1:30, 0] = 1
gradient[0, 70:99, 0] = -1
T = 80e-3 # Duration in seconds
# Increase the number of time points
n_t = int(1e3) # Number of time points in the simulation
dt = T / (gradient.shape[1] - 1) # Time step duration in seconds
gradient, dt = gradients.interpolate_gradient(gradient, dt, n_t)
# Concatenate 100 gradient arrays with different b-values together
bs = np.linspace(0, 3e9, 100) # SI units (s/m^2)
gradient = np.concatenate([gradient for _ in bs], axis=0)
gradient = gradients.set_b(gradient, dt, bs)
# Show gradient magnitude over time for the last measurement
fig, ax = plt.subplots(1, figsize=(7, 4))
for i in range(3):
ax.plot(np.linspace(0, T, n_t), gradient[-1, :, i])
ax.legend(["G$_x$", "G$_y$", "G$_z$"])
ax.set_xlabel("Time (s)")
ax.set_ylabel("Gradient magnitude (T/m)")
plt.show()
```
## Substrates
We need to define the simulated diffusion environment, referred to as a substrate, by creating a substrate object. The module `disimpy.substrates` contains functions for creating substrate objects. Disimpy supports simulating diffusion without restrictions, in a sphere, in an infinite cylinder, in an ellipsoid, and restricted by arbitrary geometries defined by triangular meshes.
### Free diffusion
```
# Create a substrate object for free diffusion
substrate = substrates.free()
# Run simulation and show the random walker trajectories
traj_file = "example_traj.txt"
signals = simulations.simulation(
n_walkers=n_walkers,
diffusivity=diffusivity,
gradient=gradient,
dt=dt,
substrate=substrate,
traj=traj_file,
)
utils.show_traj(traj_file)
# Plot the simulated signal
fig, ax = plt.subplots(1, figsize=(7, 4))
ax.scatter(bs, signals / n_walkers, s=10)
ax.set_xlabel("b (s/m$^2$)")
ax.set_ylabel("S/S$_0$")
plt.show()
```
### Spheres
Simulating diffusion inside a sphere requires specifying the radius of the sphere:
```
# Create a substrate object for diffusion inside a sphere
substrate = substrates.sphere(radius=5e-6)
# Run simulation and show the random walker trajectories
traj_file = "example_traj.txt"
signals = simulations.simulation(
n_walkers=n_walkers,
diffusivity=diffusivity,
gradient=gradient,
dt=dt,
substrate=substrate,
traj=traj_file,
)
utils.show_traj(traj_file)
# Plot the simulated signal
fig, ax = plt.subplots(1, figsize=(7, 4))
ax.scatter(bs, signals / n_walkers, s=10)
ax.set_xlabel("b (s/m$^2$)")
ax.set_ylabel("S/S$_0$")
plt.show()
```
### Infinite cylinder
Simulating diffusion inside an infinite cylinders requires specifying the radius and orientation of the cylinder:
```
# Create a substrate object for diffusion inside an infinite cylinder
substrate = substrates.cylinder(
radius=5e-6,
orientation=np.array([1.0, 1.0, 1.0])
)
# Run simulation and show the random walker trajectories
traj_file = "example_traj.txt"
signals = simulations.simulation(
n_walkers=n_walkers,
diffusivity=diffusivity,
gradient=gradient,
dt=dt,
substrate=substrate,
traj=traj_file,
)
utils.show_traj(traj_file)
# Plot the simulated signal
fig, ax = plt.subplots(1, figsize=(7, 4))
ax.scatter(bs, signals / n_walkers, s=10)
ax.set_xlabel("b (s/m$^2$)")
ax.set_ylabel("S/S$_0$")
plt.show()
```
### Ellipsoids
Simulating diffusion inside an ellipsoid requires specifying the ellipsoid semiaxes and a rotation matrix, defining the ellipsoid orientation, according to which the axis-aligned ellipsoid is rotated before the simulation:
```
# Create a substrate object for diffusion inside an ellipsoid
v = np.array([1.0, 0, 0])
k = np.array([1.0, 1.0, 1.0])
R = utils.vec2vec_rotmat(v, k) # Rotation matrix for aligning v with k
substrate = substrates.ellipsoid(
semiaxes=np.array([10e-6, 5e-6, 2.5e-6]),
R=R,
)
# Run simulation and show the random walker trajectories
traj_file = "example_traj.txt"
signals = simulations.simulation(
n_walkers=n_walkers,
diffusivity=diffusivity,
gradient=gradient,
dt=dt,
substrate=substrate,
traj=traj_file,
)
utils.show_traj(traj_file)
# Plot the simulated signal
fig, ax = plt.subplots(1, figsize=(7, 4))
ax.scatter(bs, signals / n_walkers, s=10)
ax.set_xlabel("b (s/m$^2$)")
ax.set_ylabel("S/S$_0$")
plt.show()
```
### Triangular meshes
Diffusion restricted by arbitrary geometries can be simulated using triangular meshes.
Triangular meshes must be represented by a pair of `numpy.ndarray` instances: vertices array with shape (number of points, 3) containing the points of the triangular mesh, and faces array with shape (number of triangles, 3) defining how the triangles are made up of the vertices. The simulated voxel is equal to the axis-aligned bounding box of the triangles plus optional padding. Before the simulation, the triangles are moved so that the bottom corner of the simulated voxel is at the origin.
By default, the initial positions of the random walkers are randomly sampled from a uniform distribution over the simulated voxel. If the triangles define a closed surface, the initial positions of the random walkers can be randomly sampled from a uniform distribution over the volume inside or outside the surface. For very complex meshes, the current implementation of the algorithm that samples positions inside or outside the surface may require changes to the [code](https://github.com/kerkelae/disimpy/blob/master/disimpy/simulations.py#L430). The initial positions can also be defined manually.
Disimpy supports periodic and reflective boundary conditions. If periodic boundary conditions are used, the random walkers encounter infinitely repeating identical copies of the simulated microstructure after they leave the simulated voxel. Otherwise, the boundaries of the simulated voxel are treated as impermeable surfaces.
The code below loads an example mesh and shows how to generate a substrate object for simulating diffusion inside the closed surface.
```
# Load an example triangular mesh
mesh_path = os.path.join(
os.path.dirname(simulations.__file__), "tests", "example_mesh.pkl"
)
with open(mesh_path, "rb") as f:
example_mesh = pickle.load(f)
faces = example_mesh["faces"]
vertices = example_mesh["vertices"]
# Create a substrate object
substrate = substrates.mesh(
vertices, faces, padding=np.zeros(3), periodic=True, init_pos="intra"
)
# Show the mesh
utils.show_mesh(substrate)
# Run simulation and show the random walker trajectories
traj_file = "example_traj.txt"
signals = simulations.simulation(
n_walkers=n_walkers,
diffusivity=diffusivity,
gradient=gradient,
dt=dt,
substrate=substrate,
traj=traj_file,
)
utils.show_traj(traj_file)
# Plot the simulated signal
fig, ax = plt.subplots(1, figsize=(7, 4))
ax.scatter(bs, signals / n_walkers, s=10)
ax.set_xlabel("b (s/m$^2$)")
ax.set_ylabel("S/S$_0$")
plt.show()
# Create and show a substrate object with reflective boundary conditions
substrate = substrates.mesh(
vertices, faces, padding=np.zeros(3), periodic=False, init_pos="intra"
)
utils.show_mesh(substrate)
```
#### Importing mesh files
There are several open-source Python packages for reading mesh files. The code snippet below shows how to use `meshio` to load a mesh of a neuron model generated using an algorithm by [Palombo et al.](<https://doi.org/10.1016/j.neuroimage.2018.12.025>).
```
# If meshio has not been installed, uncomment the following line and execute
# the code in this cell
#!pip install meshio
import meshio
# Load mesh
mesh_path = os.path.join(
os.path.dirname(simulations.__file__), "tests", "neuron-model.stl"
)
mesh = meshio.read(mesh_path)
vertices = mesh.points.astype(np.float32)
faces = mesh.cells[0].data
# Show mesh using Matplotlib's trisurf
fig = plt.figure(figsize=(8, 8))
ax = fig.add_subplot(111, projection="3d")
ax.plot_trisurf(
vertices[:, 0],
vertices[:, 1],
vertices[:, 2],
triangles=faces,
)
plt.axis("off")
plt.show()
```
## Advanced information
```
# More information can be found in the function docstrings. For example,
simulations.simulation?
```
For details, please see the [function documentation](https://disimpy.readthedocs.io/en/latest/modules_and_functions.html) and [source code](https://github.com/kerkelae/disimpy).
|
github_jupyter
|
# If Disimpy has not been installed, uncomment the following line and
# execute the code in this cell
#!pip install disimpy
# Import the packages and modules used in this tutorial
import os
import pickle
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from disimpy import gradients, simulations, substrates, utils
n_walkers = int(1e3)
diffusivity = 2e-9 # SI units (m^2/s)
# Create a simple Stejskal-Tanner gradient waveform
gradient = np.zeros((1, 100, 3))
gradient[0, 1:30, 0] = 1
gradient[0, 70:99, 0] = -1
T = 80e-3 # Duration in seconds
# Increase the number of time points
n_t = int(1e3) # Number of time points in the simulation
dt = T / (gradient.shape[1] - 1) # Time step duration in seconds
gradient, dt = gradients.interpolate_gradient(gradient, dt, n_t)
# Concatenate 100 gradient arrays with different b-values together
bs = np.linspace(0, 3e9, 100) # SI units (s/m^2)
gradient = np.concatenate([gradient for _ in bs], axis=0)
gradient = gradients.set_b(gradient, dt, bs)
# Show gradient magnitude over time for the last measurement
fig, ax = plt.subplots(1, figsize=(7, 4))
for i in range(3):
ax.plot(np.linspace(0, T, n_t), gradient[-1, :, i])
ax.legend(["G$_x$", "G$_y$", "G$_z$"])
ax.set_xlabel("Time (s)")
ax.set_ylabel("Gradient magnitude (T/m)")
plt.show()
# Create a substrate object for free diffusion
substrate = substrates.free()
# Run simulation and show the random walker trajectories
traj_file = "example_traj.txt"
signals = simulations.simulation(
n_walkers=n_walkers,
diffusivity=diffusivity,
gradient=gradient,
dt=dt,
substrate=substrate,
traj=traj_file,
)
utils.show_traj(traj_file)
# Plot the simulated signal
fig, ax = plt.subplots(1, figsize=(7, 4))
ax.scatter(bs, signals / n_walkers, s=10)
ax.set_xlabel("b (s/m$^2$)")
ax.set_ylabel("S/S$_0$")
plt.show()
# Create a substrate object for diffusion inside a sphere
substrate = substrates.sphere(radius=5e-6)
# Run simulation and show the random walker trajectories
traj_file = "example_traj.txt"
signals = simulations.simulation(
n_walkers=n_walkers,
diffusivity=diffusivity,
gradient=gradient,
dt=dt,
substrate=substrate,
traj=traj_file,
)
utils.show_traj(traj_file)
# Plot the simulated signal
fig, ax = plt.subplots(1, figsize=(7, 4))
ax.scatter(bs, signals / n_walkers, s=10)
ax.set_xlabel("b (s/m$^2$)")
ax.set_ylabel("S/S$_0$")
plt.show()
# Create a substrate object for diffusion inside an infinite cylinder
substrate = substrates.cylinder(
radius=5e-6,
orientation=np.array([1.0, 1.0, 1.0])
)
# Run simulation and show the random walker trajectories
traj_file = "example_traj.txt"
signals = simulations.simulation(
n_walkers=n_walkers,
diffusivity=diffusivity,
gradient=gradient,
dt=dt,
substrate=substrate,
traj=traj_file,
)
utils.show_traj(traj_file)
# Plot the simulated signal
fig, ax = plt.subplots(1, figsize=(7, 4))
ax.scatter(bs, signals / n_walkers, s=10)
ax.set_xlabel("b (s/m$^2$)")
ax.set_ylabel("S/S$_0$")
plt.show()
# Create a substrate object for diffusion inside an ellipsoid
v = np.array([1.0, 0, 0])
k = np.array([1.0, 1.0, 1.0])
R = utils.vec2vec_rotmat(v, k) # Rotation matrix for aligning v with k
substrate = substrates.ellipsoid(
semiaxes=np.array([10e-6, 5e-6, 2.5e-6]),
R=R,
)
# Run simulation and show the random walker trajectories
traj_file = "example_traj.txt"
signals = simulations.simulation(
n_walkers=n_walkers,
diffusivity=diffusivity,
gradient=gradient,
dt=dt,
substrate=substrate,
traj=traj_file,
)
utils.show_traj(traj_file)
# Plot the simulated signal
fig, ax = plt.subplots(1, figsize=(7, 4))
ax.scatter(bs, signals / n_walkers, s=10)
ax.set_xlabel("b (s/m$^2$)")
ax.set_ylabel("S/S$_0$")
plt.show()
# Load an example triangular mesh
mesh_path = os.path.join(
os.path.dirname(simulations.__file__), "tests", "example_mesh.pkl"
)
with open(mesh_path, "rb") as f:
example_mesh = pickle.load(f)
faces = example_mesh["faces"]
vertices = example_mesh["vertices"]
# Create a substrate object
substrate = substrates.mesh(
vertices, faces, padding=np.zeros(3), periodic=True, init_pos="intra"
)
# Show the mesh
utils.show_mesh(substrate)
# Run simulation and show the random walker trajectories
traj_file = "example_traj.txt"
signals = simulations.simulation(
n_walkers=n_walkers,
diffusivity=diffusivity,
gradient=gradient,
dt=dt,
substrate=substrate,
traj=traj_file,
)
utils.show_traj(traj_file)
# Plot the simulated signal
fig, ax = plt.subplots(1, figsize=(7, 4))
ax.scatter(bs, signals / n_walkers, s=10)
ax.set_xlabel("b (s/m$^2$)")
ax.set_ylabel("S/S$_0$")
plt.show()
# Create and show a substrate object with reflective boundary conditions
substrate = substrates.mesh(
vertices, faces, padding=np.zeros(3), periodic=False, init_pos="intra"
)
utils.show_mesh(substrate)
# If meshio has not been installed, uncomment the following line and execute
# the code in this cell
#!pip install meshio
import meshio
# Load mesh
mesh_path = os.path.join(
os.path.dirname(simulations.__file__), "tests", "neuron-model.stl"
)
mesh = meshio.read(mesh_path)
vertices = mesh.points.astype(np.float32)
faces = mesh.cells[0].data
# Show mesh using Matplotlib's trisurf
fig = plt.figure(figsize=(8, 8))
ax = fig.add_subplot(111, projection="3d")
ax.plot_trisurf(
vertices[:, 0],
vertices[:, 1],
vertices[:, 2],
triangles=faces,
)
plt.axis("off")
plt.show()
# More information can be found in the function docstrings. For example,
simulations.simulation?
| 0.782663 | 0.984032 |
## Black Friday Dataset EDA And Feature Engineering
## Cleaning and preparing the data for model training
```
## dataset link: https://www.kaggle.com/sdolezel/black-friday?select=train.csv
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
```
## Problem Statement
A retail company “ABC Private Limited” wants to understand the customer purchase behaviour (specifically, purchase amount) against various products of different categories. They have shared purchase summary of various customers for selected high volume products from last month. The data set also contains customer demographics (age, gender, marital status, city_type, stay_in_current_city), product details (product_id and product category) and Total purchase_amount from last month.
Now, they want to build a model to predict the purchase amount of customer against various products which will help them to create personalized offer for customers against different products.
```
#importing the dataset
df_train=pd.read_csv('train.csv')
df_train.head()
## import the test data
df_test=pd.read_csv('test.csv')
df_test.head()
#Merge both train and test data
df=df_train.append(df_test)
df.head()
##Basic
df.info()
df.describe()
df.drop(['User_ID'],axis=1,inplace=True)
df.head()
pd.get_dummies(df['Gender'])
df['Gender']
##HAndling categorical feature Gender
df['Gender']=df['Gender'].map({'F':0,'M':1})
df.head()
## Handle categorical feature Age
df['Age'].unique()
#pd.get_dummies(df['Age'],drop_first=True)
df['Age']=df['Age'].map({'0-17':1,'18-25':2,'26-35':3,'36-45':4,'46-50':5,'51-55':6,'55+':7})
df
##second technqiue
from sklearn import preprocessing
# label_encoder object knows how to understand word labels.
label_encoder = preprocessing.LabelEncoder()
# Encode labels in column 'species'.
df['Age']= label_encoder.fit_transform(df['Age'])
df['Age'].unique()
##fixing categorical City_categort
df_city=pd.get_dummies(df['City_Category'],drop_first=True)
df_city.head()
df=pd.concat([df,df_city],axis=1)
df.head()
##drop City Category Feature
df.drop('City_Category',axis=1,inplace=True)
df
## Missing Values
df.isnull().sum()
## Focus on replacing missing values
df['Product_Category_2'].unique()
df['Product_Category_2'].value_counts()
## intilazing value with mode
df['Product_Category_2'].mode()[0]
## Replace the missing values with mode
df['Product_Category_2']=df['Product_Category_2'].fillna(df['Product_Category_2'].mode()[0])
df['Product_Category_2'].isnull().sum()
## Product_category 3 replace missing values
df['Product_Category_3'].unique()
df['Product_Category_3'].value_counts()
df['Product_Category_3']=df['Product_Category_3'].fillna(df['Product_Category_3'].mode()[0])
df.head()
df.shape
df['Stay_In_Current_City_Years'].unique()
df['Stay_In_Current_City_Years']=df['Stay_In_Current_City_Years'].str.replace('+','')
df.head()
df.info()
##convert object into integers
df['Stay_In_Current_City_Years']=df['Stay_In_Current_City_Years'].astype(int)
df.info()
df['B']=df['B'].astype(int)
df['C']=df['C'].astype(int)
df.info()
##Visualisation Age vs Purchased
sns.barplot('Age','Purchase',hue='Gender',data=df)
## Visualization of Purchase with occupation
sns.barplot('Occupation','Purchase',hue='Gender',data=df)
sns.barplot('Product_Category_1','Purchase',hue='Gender',data=df)
sns.barplot('Product_Category_2','Purchase',hue='Gender',data=df)
sns.barplot('Product_Category_3','Purchase',hue='Gender',data=df)
df.head()
##Feature Scaling
df_test=df[df['Purchase'].isnull()]
df_train=df[~df['Purchase'].isnull()]
X=df_train.drop('Purchase',axis=1)
X.head()
X.shape
y=df_train['Purchase']
y
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42)
X_train.drop('Product_ID',axis=1,inplace=True)
X_test.drop('Product_ID',axis=1,inplace=True)
## feature Scaling
from sklearn.preprocessing import StandardScaler
sc=StandardScaler()
X_train=sc.fit_transform(X_train)
X_test=sc.transform(X_test)
## Now train your model
```
|
github_jupyter
|
## dataset link: https://www.kaggle.com/sdolezel/black-friday?select=train.csv
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
#importing the dataset
df_train=pd.read_csv('train.csv')
df_train.head()
## import the test data
df_test=pd.read_csv('test.csv')
df_test.head()
#Merge both train and test data
df=df_train.append(df_test)
df.head()
##Basic
df.info()
df.describe()
df.drop(['User_ID'],axis=1,inplace=True)
df.head()
pd.get_dummies(df['Gender'])
df['Gender']
##HAndling categorical feature Gender
df['Gender']=df['Gender'].map({'F':0,'M':1})
df.head()
## Handle categorical feature Age
df['Age'].unique()
#pd.get_dummies(df['Age'],drop_first=True)
df['Age']=df['Age'].map({'0-17':1,'18-25':2,'26-35':3,'36-45':4,'46-50':5,'51-55':6,'55+':7})
df
##second technqiue
from sklearn import preprocessing
# label_encoder object knows how to understand word labels.
label_encoder = preprocessing.LabelEncoder()
# Encode labels in column 'species'.
df['Age']= label_encoder.fit_transform(df['Age'])
df['Age'].unique()
##fixing categorical City_categort
df_city=pd.get_dummies(df['City_Category'],drop_first=True)
df_city.head()
df=pd.concat([df,df_city],axis=1)
df.head()
##drop City Category Feature
df.drop('City_Category',axis=1,inplace=True)
df
## Missing Values
df.isnull().sum()
## Focus on replacing missing values
df['Product_Category_2'].unique()
df['Product_Category_2'].value_counts()
## intilazing value with mode
df['Product_Category_2'].mode()[0]
## Replace the missing values with mode
df['Product_Category_2']=df['Product_Category_2'].fillna(df['Product_Category_2'].mode()[0])
df['Product_Category_2'].isnull().sum()
## Product_category 3 replace missing values
df['Product_Category_3'].unique()
df['Product_Category_3'].value_counts()
df['Product_Category_3']=df['Product_Category_3'].fillna(df['Product_Category_3'].mode()[0])
df.head()
df.shape
df['Stay_In_Current_City_Years'].unique()
df['Stay_In_Current_City_Years']=df['Stay_In_Current_City_Years'].str.replace('+','')
df.head()
df.info()
##convert object into integers
df['Stay_In_Current_City_Years']=df['Stay_In_Current_City_Years'].astype(int)
df.info()
df['B']=df['B'].astype(int)
df['C']=df['C'].astype(int)
df.info()
##Visualisation Age vs Purchased
sns.barplot('Age','Purchase',hue='Gender',data=df)
## Visualization of Purchase with occupation
sns.barplot('Occupation','Purchase',hue='Gender',data=df)
sns.barplot('Product_Category_1','Purchase',hue='Gender',data=df)
sns.barplot('Product_Category_2','Purchase',hue='Gender',data=df)
sns.barplot('Product_Category_3','Purchase',hue='Gender',data=df)
df.head()
##Feature Scaling
df_test=df[df['Purchase'].isnull()]
df_train=df[~df['Purchase'].isnull()]
X=df_train.drop('Purchase',axis=1)
X.head()
X.shape
y=df_train['Purchase']
y
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42)
X_train.drop('Product_ID',axis=1,inplace=True)
X_test.drop('Product_ID',axis=1,inplace=True)
## feature Scaling
from sklearn.preprocessing import StandardScaler
sc=StandardScaler()
X_train=sc.fit_transform(X_train)
X_test=sc.transform(X_test)
## Now train your model
| 0.372505 | 0.883538 |
```
import os
import sys
import random
import math
import re
import time
import numpy as np
import tensorflow as tf
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.patches as patches
# Root directory of the project
ROOT_DIR = os.path.abspath("../../")
# Import Mask RCNN
sys.path.append(ROOT_DIR) # To find local version of the library
from mrcnn import utils
from mrcnn import visualize
from mrcnn.visualize import display_images
import mrcnn.model as modellib
from mrcnn.model import log
from samples.balloon import balloon
import skimage
# Root directory of the project
ROOT_DIR = os.path.abspath("../../")
# Import Mask RCNN
sys.path.append(ROOT_DIR) # To find local version of the library
from mrcnn.config import Config
from mrcnn import model as modellib, utils
%matplotlib inline
# Directory to save logs and trained model
MODEL_DIR = os.path.join(ROOT_DIR, "logs")
class EyeConfig(Config):
"""Configuration for training on the toy dataset.
Derives from the base Config class and overrides some values.
"""
# Give the configuration a recognizable name
NAME = "Eye"
# We use a GPU with 12GB memory, which can fit two images.
# Adjust down if you use a smaller GPU.
IMAGES_PER_GPU = 1
# Number of classes (including background)
NUM_CLASSES = 1 + 6 # Background + balloon
eyeConfig=EyeConfig
class InferenceConfig(eyeConfig):
# Set batch size to 1 since we'll be running inference on
# one image at a time. Batch size = GPU_COUNT * IMAGES_PER_GPU
GPU_COUNT = 1
IMAGES_PER_GPU = 1
config = InferenceConfig()
config.display()
# Create model object in inference mode.
model = modellib.MaskRCNN(mode="inference", model_dir=MODEL_DIR, config=config)
# Set path to balloon weights file
# Download file from the Releases page and set its path
# https://github.com/matterport/Mask_RCNN/releases
# weights_path = "/path/to/mask_rcnn_balloon.h5"
# Or, load the last model you trained
weights_path = model.find_last()
# Load weights
print("Loading weights ", weights_path)
model.load_weights(weights_path, by_name=True)
IMAGE_DIR='../../Eye/test/'
class_names =['BG',]
# Load a random image from the images folder
file_names = next(os.walk(IMAGE_DIR))[2]
image = skimage.io.imread(os.path.join(IMAGE_DIR, random.choice(file_names)))
# Run detection
results = model.detect([image], verbose=1)
# Visualize results
r = results[0]
visualize.display_instances(image, r['rois'], r['masks'], r['class_ids'],
class_names, r['scores'])
```
|
github_jupyter
|
import os
import sys
import random
import math
import re
import time
import numpy as np
import tensorflow as tf
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.patches as patches
# Root directory of the project
ROOT_DIR = os.path.abspath("../../")
# Import Mask RCNN
sys.path.append(ROOT_DIR) # To find local version of the library
from mrcnn import utils
from mrcnn import visualize
from mrcnn.visualize import display_images
import mrcnn.model as modellib
from mrcnn.model import log
from samples.balloon import balloon
import skimage
# Root directory of the project
ROOT_DIR = os.path.abspath("../../")
# Import Mask RCNN
sys.path.append(ROOT_DIR) # To find local version of the library
from mrcnn.config import Config
from mrcnn import model as modellib, utils
%matplotlib inline
# Directory to save logs and trained model
MODEL_DIR = os.path.join(ROOT_DIR, "logs")
class EyeConfig(Config):
"""Configuration for training on the toy dataset.
Derives from the base Config class and overrides some values.
"""
# Give the configuration a recognizable name
NAME = "Eye"
# We use a GPU with 12GB memory, which can fit two images.
# Adjust down if you use a smaller GPU.
IMAGES_PER_GPU = 1
# Number of classes (including background)
NUM_CLASSES = 1 + 6 # Background + balloon
eyeConfig=EyeConfig
class InferenceConfig(eyeConfig):
# Set batch size to 1 since we'll be running inference on
# one image at a time. Batch size = GPU_COUNT * IMAGES_PER_GPU
GPU_COUNT = 1
IMAGES_PER_GPU = 1
config = InferenceConfig()
config.display()
# Create model object in inference mode.
model = modellib.MaskRCNN(mode="inference", model_dir=MODEL_DIR, config=config)
# Set path to balloon weights file
# Download file from the Releases page and set its path
# https://github.com/matterport/Mask_RCNN/releases
# weights_path = "/path/to/mask_rcnn_balloon.h5"
# Or, load the last model you trained
weights_path = model.find_last()
# Load weights
print("Loading weights ", weights_path)
model.load_weights(weights_path, by_name=True)
IMAGE_DIR='../../Eye/test/'
class_names =['BG',]
# Load a random image from the images folder
file_names = next(os.walk(IMAGE_DIR))[2]
image = skimage.io.imread(os.path.join(IMAGE_DIR, random.choice(file_names)))
# Run detection
results = model.detect([image], verbose=1)
# Visualize results
r = results[0]
visualize.display_instances(image, r['rois'], r['masks'], r['class_ids'],
class_names, r['scores'])
| 0.557604 | 0.266739 |
# Prophet, with less features
*Anders Poire, 30-03-2020*
Goals: make a prophet model with most possible non-correlated features
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from os.path import join
from sklearn.compose import ColumnTransformer
from sklearn.impute import SimpleImputer
from sklearn.metrics import mean_absolute_error
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from fbprophet import Prophet
from fbprophet.diagnostics import cross_validation
sns.set()
```
## Pre-processing
We edited our pre-processing to drop features that we found to have
high correlation.
Furthermore, we take a mean of the vegatation indices (as they are also highly correlated)
```
def get_dataset(path):
features = pd.read_csv(path)
features = features.drop(
['year', 'weekofyear'],
axis = 1
)
features['ndvi_s'] = (features['ndvi_se'] + features['ndvi_sw'])/2
features['ndvi_n'] = (features['ndvi_ne'] + features['ndvi_nw'])/2
features = features.drop(
[
'reanalysis_sat_precip_amt_mm',
'reanalysis_max_air_temp_k',
'reanalysis_min_air_temp_k',
'reanalysis_air_temp_k',
'reanalysis_dew_point_temp_k',
'ndvi_ne', 'ndvi_nw',
'ndvi_se', 'ndvi_sw'
],
axis = 1
)
features = features.rename({'week_start_date': 'ds'}, axis = 1)
features_sj = features[features['city'] == 'sj'].drop('city', axis = 1)
features_iq = features[features['city'] == 'iq'].drop('city', axis = 1)
return (features_sj, features_iq)
DATA_PATH = '../data/raw'
train_sj, train_iq = get_dataset(join(DATA_PATH,
'dengue_features_train.csv'))
test_sj, test_iq = get_dataset(join(DATA_PATH,
'dengue_features_test.csv'))
train_labels = train_labels = pd.read_csv(join(DATA_PATH, 'dengue_labels_train.csv'))
```
## Modeling
## San Juan
```
train_sj
model_sj = Prophet(
growth = 'linear',
yearly_seasonality = True,
weekly_seasonality = False,
daily_seasonality = False,
seasonality_mode = 'multiplicative',
changepoint_prior_scale = 0.01
)
for name in train_sj.columns.values[1:]:
model_sj.add_regressor(name)
```
Mostly the same as before. I found a bug in `sklearn` where if I have `('pass', 'passthrough', ['ds'])` after `('num', numeric_transformer, numeric_features)` in my `ColumnTransformer` then the tranformed columns will all unexplicably be shifted by 1
```
numeric_features = train_sj.columns[1:]
numeric_transformer = Pipeline([
('impute', SimpleImputer(strategy = 'median')),
('scale', StandardScaler())
])
preprocessor = ColumnTransformer(
transformers = [
('pass', 'passthrough', ['ds']),
('num', numeric_transformer, numeric_features),
]
)
train_sj_t = pd.DataFrame(preprocessor.fit_transform(train_sj), columns = train_sj.columns)
train_sj_t['y'] = train_labels[train_labels['city'] == 'sj']['total_cases']
model_sj.fit(train_sj_t)
history = model_sj.predict(train_sj_t)
model_sj.plot(history)
cv_sj = cross_validation(model_sj, horizon = '365 days')
mean_absolute_error(cv_sj['yhat'], cv_sj['y'])
```
Let's compare this to our training set error when fitting on all of the data
## Take-aways
It seems likely that we are overfitting. Train error is much lower than CV error for both models
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from os.path import join
from sklearn.compose import ColumnTransformer
from sklearn.impute import SimpleImputer
from sklearn.metrics import mean_absolute_error
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from fbprophet import Prophet
from fbprophet.diagnostics import cross_validation
sns.set()
def get_dataset(path):
features = pd.read_csv(path)
features = features.drop(
['year', 'weekofyear'],
axis = 1
)
features['ndvi_s'] = (features['ndvi_se'] + features['ndvi_sw'])/2
features['ndvi_n'] = (features['ndvi_ne'] + features['ndvi_nw'])/2
features = features.drop(
[
'reanalysis_sat_precip_amt_mm',
'reanalysis_max_air_temp_k',
'reanalysis_min_air_temp_k',
'reanalysis_air_temp_k',
'reanalysis_dew_point_temp_k',
'ndvi_ne', 'ndvi_nw',
'ndvi_se', 'ndvi_sw'
],
axis = 1
)
features = features.rename({'week_start_date': 'ds'}, axis = 1)
features_sj = features[features['city'] == 'sj'].drop('city', axis = 1)
features_iq = features[features['city'] == 'iq'].drop('city', axis = 1)
return (features_sj, features_iq)
DATA_PATH = '../data/raw'
train_sj, train_iq = get_dataset(join(DATA_PATH,
'dengue_features_train.csv'))
test_sj, test_iq = get_dataset(join(DATA_PATH,
'dengue_features_test.csv'))
train_labels = train_labels = pd.read_csv(join(DATA_PATH, 'dengue_labels_train.csv'))
train_sj
model_sj = Prophet(
growth = 'linear',
yearly_seasonality = True,
weekly_seasonality = False,
daily_seasonality = False,
seasonality_mode = 'multiplicative',
changepoint_prior_scale = 0.01
)
for name in train_sj.columns.values[1:]:
model_sj.add_regressor(name)
numeric_features = train_sj.columns[1:]
numeric_transformer = Pipeline([
('impute', SimpleImputer(strategy = 'median')),
('scale', StandardScaler())
])
preprocessor = ColumnTransformer(
transformers = [
('pass', 'passthrough', ['ds']),
('num', numeric_transformer, numeric_features),
]
)
train_sj_t = pd.DataFrame(preprocessor.fit_transform(train_sj), columns = train_sj.columns)
train_sj_t['y'] = train_labels[train_labels['city'] == 'sj']['total_cases']
model_sj.fit(train_sj_t)
history = model_sj.predict(train_sj_t)
model_sj.plot(history)
cv_sj = cross_validation(model_sj, horizon = '365 days')
mean_absolute_error(cv_sj['yhat'], cv_sj['y'])
| 0.497315 | 0.881258 |
[@LorenaABarba](https://twitter.com/LorenaABarba)
12 steps to Navier–Stokes
=====
***
Did you make it this far? This is the last step! How long did it take you to write your own Navier–Stokes solver in Python following this interactive module? Let us know!
Step 12: Channel Flow with Navier–Stokes
----
***
The only difference between this final step and Step 11 is that we are going to add a source term to the $u$-momentum equation, to mimic the effect of a pressure-driven channel flow. Here are our modified Navier–Stokes equations:
$$\frac{\partial u}{\partial t}+u\frac{\partial u}{\partial x}+v\frac{\partial u}{\partial y}=-\frac{1}{\rho}\frac{\partial p}{\partial x}+\nu\left(\frac{\partial^2 u}{\partial x^2}+\frac{\partial^2 u}{\partial y^2}\right)+F$$
$$\frac{\partial v}{\partial t}+u\frac{\partial v}{\partial x}+v\frac{\partial v}{\partial y}=-\frac{1}{\rho}\frac{\partial p}{\partial y}+\nu\left(\frac{\partial^2 v}{\partial x^2}+\frac{\partial^2 v}{\partial y^2}\right)$$
$$\frac{\partial^2 p}{\partial x^2}+\frac{\partial^2 p}{\partial y^2}=-\rho\left(\frac{\partial u}{\partial x}\frac{\partial u}{\partial x}+2\frac{\partial u}{\partial y}\frac{\partial v}{\partial x}+\frac{\partial v}{\partial y}\frac{\partial v}{\partial y}\right)
$$
### Discretized equations
With patience and care, we write the discretized form of the equations. It is highly recommended that you write these in your own hand, mentally following each term as you write it.
The $u$-momentum equation:
$$
\begin{split}
& \frac{u_{i,j}^{n+1}-u_{i,j}^{n}}{\Delta t}+u_{i,j}^{n}\frac{u_{i,j}^{n}-u_{i-1,j}^{n}}{\Delta x}+v_{i,j}^{n}\frac{u_{i,j}^{n}-u_{i,j-1}^{n}}{\Delta y} = \\
& \qquad -\frac{1}{\rho}\frac{p_{i+1,j}^{n}-p_{i-1,j}^{n}}{2\Delta x} \\
& \qquad +\nu\left(\frac{u_{i+1,j}^{n}-2u_{i,j}^{n}+u_{i-1,j}^{n}}{\Delta x^2}+\frac{u_{i,j+1}^{n}-2u_{i,j}^{n}+u_{i,j-1}^{n}}{\Delta y^2}\right)+F_{i,j}
\end{split}
$$
The $v$-momentum equation:
$$
\begin{split}
& \frac{v_{i,j}^{n+1}-v_{i,j}^{n}}{\Delta t}+u_{i,j}^{n}\frac{v_{i,j}^{n}-v_{i-1,j}^{n}}{\Delta x}+v_{i,j}^{n}\frac{v_{i,j}^{n}-v_{i,j-1}^{n}}{\Delta y} = \\
& \qquad -\frac{1}{\rho}\frac{p_{i,j+1}^{n}-p_{i,j-1}^{n}}{2\Delta y} \\
& \qquad +\nu\left(\frac{v_{i+1,j}^{n}-2v_{i,j}^{n}+v_{i-1,j}^{n}}{\Delta x^2}+\frac{v_{i,j+1}^{n}-2v_{i,j}^{n}+v_{i,j-1}^{n}}{\Delta y^2}\right)
\end{split}
$$
And the pressure equation:
$$
\begin{split}
& \frac{p_{i+1,j}^{n}-2p_{i,j}^{n}+p_{i-1,j}^{n}}{\Delta x^2} + \frac{p_{i,j+1}^{n}-2p_{i,j}^{n}+p_{i,j-1}^{n}}{\Delta y^2} = \\
& \qquad \rho\left[\frac{1}{\Delta t}\left(\frac{u_{i+1,j}-u_{i-1,j}}{2\Delta x}+\frac{v_{i,j+1}-v_{i,j-1}}{2\Delta y}\right) - \frac{u_{i+1,j}-u_{i-1,j}}{2\Delta x}\frac{u_{i+1,j}-u_{i-1,j}}{2\Delta x} - 2\frac{u_{i,j+1}-u_{i,j-1}}{2\Delta y}\frac{v_{i+1,j}-v_{i-1,j}}{2\Delta x} - \frac{v_{i,j+1}-v_{i,j-1}}{2\Delta y}\frac{v_{i,j+1}-v_{i,j-1}}{2\Delta y}\right]
\end{split}
$$
As always, we need to re-arrange these equations to the form we need in the code to make the iterations proceed.
For the $u$- and $v$ momentum equations, we isolate the velocity at time step `n+1`:
$$
\begin{split}
u_{i,j}^{n+1} = u_{i,j}^{n} & - u_{i,j}^{n} \frac{\Delta t}{\Delta x} \left(u_{i,j}^{n}-u_{i-1,j}^{n}\right) - v_{i,j}^{n} \frac{\Delta t}{\Delta y} \left(u_{i,j}^{n}-u_{i,j-1}^{n}\right) \\
& - \frac{\Delta t}{\rho 2\Delta x} \left(p_{i+1,j}^{n}-p_{i-1,j}^{n}\right) \\
& + \nu\left[\frac{\Delta t}{\Delta x^2} \left(u_{i+1,j}^{n}-2u_{i,j}^{n}+u_{i-1,j}^{n}\right) + \frac{\Delta t}{\Delta y^2} \left(u_{i,j+1}^{n}-2u_{i,j}^{n}+u_{i,j-1}^{n}\right)\right] \\
& + \Delta t F
\end{split}
$$
$$
\begin{split}
v_{i,j}^{n+1} = v_{i,j}^{n} & - u_{i,j}^{n} \frac{\Delta t}{\Delta x} \left(v_{i,j}^{n}-v_{i-1,j}^{n}\right) - v_{i,j}^{n} \frac{\Delta t}{\Delta y} \left(v_{i,j}^{n}-v_{i,j-1}^{n}\right) \\
& - \frac{\Delta t}{\rho 2\Delta y} \left(p_{i,j+1}^{n}-p_{i,j-1}^{n}\right) \\
& + \nu\left[\frac{\Delta t}{\Delta x^2} \left(v_{i+1,j}^{n}-2v_{i,j}^{n}+v_{i-1,j}^{n}\right) + \frac{\Delta t}{\Delta y^2} \left(v_{i,j+1}^{n}-2v_{i,j}^{n}+v_{i,j-1}^{n}\right)\right]
\end{split}
$$
And for the pressure equation, we isolate the term $p_{i,j}^n$ to iterate in pseudo-time:
$$
\begin{split}
p_{i,j}^{n} = & \frac{\left(p_{i+1,j}^{n}+p_{i-1,j}^{n}\right) \Delta y^2 + \left(p_{i,j+1}^{n}+p_{i,j-1}^{n}\right) \Delta x^2}{2(\Delta x^2+\Delta y^2)} \\
& -\frac{\rho\Delta x^2\Delta y^2}{2\left(\Delta x^2+\Delta y^2\right)} \\
& \times \left[\frac{1}{\Delta t} \left(\frac{u_{i+1,j}-u_{i-1,j}}{2\Delta x} + \frac{v_{i,j+1}-v_{i,j-1}}{2\Delta y}\right) - \frac{u_{i+1,j}-u_{i-1,j}}{2\Delta x}\frac{u_{i+1,j}-u_{i-1,j}}{2\Delta x} - 2\frac{u_{i,j+1}-u_{i,j-1}}{2\Delta y}\frac{v_{i+1,j}-v_{i-1,j}}{2\Delta x} - \frac{v_{i,j+1}-v_{i,j-1}}{2\Delta y}\frac{v_{i,j+1}-v_{i,j-1}}{2\Delta y}\right]
\end{split}
$$
The initial condition is $u, v, p=0$ everywhere, and at the boundary conditions are:
$u, v, p$ are periodic on $x=0,2$
$u, v =0$ at $y =0,2$
$\frac{\partial p}{\partial y}=0$ at $y =0,2$
$F=1$ everywhere.
Let's begin by importing our usual run of libraries:
```
import numpy
from matplotlib import pyplot, cm
from mpl_toolkits.mplot3d import Axes3D
%matplotlib inline
```
In step 11, we isolated a portion of our transposed equation to make it easier to parse and we're going to do the same thing here. One thing to note is that we have periodic boundary conditions throughout this grid, so we need to explicitly calculate the values at the leading and trailing edge of our `u` vector.
```
def build_up_b(rho, dt, dx, dy, u, v):
b = numpy.zeros_like(u)
b[1:-1, 1:-1] = (rho * (1 / dt * ((u[1:-1, 2:] - u[1:-1, 0:-2]) / (2 * dx) +
(v[2:, 1:-1] - v[0:-2, 1:-1]) / (2 * dy)) -
((u[1:-1, 2:] - u[1:-1, 0:-2]) / (2 * dx))**2 -
2 * ((u[2:, 1:-1] - u[0:-2, 1:-1]) / (2 * dy) *
(v[1:-1, 2:] - v[1:-1, 0:-2]) / (2 * dx))-
((v[2:, 1:-1] - v[0:-2, 1:-1]) / (2 * dy))**2))
# Periodic BC Pressure @ x = 2
b[1:-1, -1] = (rho * (1 / dt * ((u[1:-1, 0] - u[1:-1,-2]) / (2 * dx) +
(v[2:, -1] - v[0:-2, -1]) / (2 * dy)) -
((u[1:-1, 0] - u[1:-1, -2]) / (2 * dx))**2 -
2 * ((u[2:, -1] - u[0:-2, -1]) / (2 * dy) *
(v[1:-1, 0] - v[1:-1, -2]) / (2 * dx)) -
((v[2:, -1] - v[0:-2, -1]) / (2 * dy))**2))
# Periodic BC Pressure @ x = 0
b[1:-1, 0] = (rho * (1 / dt * ((u[1:-1, 1] - u[1:-1, -1]) / (2 * dx) +
(v[2:, 0] - v[0:-2, 0]) / (2 * dy)) -
((u[1:-1, 1] - u[1:-1, -1]) / (2 * dx))**2 -
2 * ((u[2:, 0] - u[0:-2, 0]) / (2 * dy) *
(v[1:-1, 1] - v[1:-1, -1]) / (2 * dx))-
((v[2:, 0] - v[0:-2, 0]) / (2 * dy))**2))
return b
```
We'll also define a Pressure Poisson iterative function, again like we did in Step 11. Once more, note that we have to include the periodic boundary conditions at the leading and trailing edge. We also have to specify the boundary conditions at the top and bottom of our grid.
```
def pressure_poisson_periodic(p, dx, dy):
pn = numpy.empty_like(p)
for q in range(nit):
pn = p.copy()
p[1:-1, 1:-1] = (((pn[1:-1, 2:] + pn[1:-1, 0:-2]) * dy**2 +
(pn[2:, 1:-1] + pn[0:-2, 1:-1]) * dx**2) /
(2 * (dx**2 + dy**2)) -
dx**2 * dy**2 / (2 * (dx**2 + dy**2)) * b[1:-1, 1:-1])
# Periodic BC Pressure @ x = 2
p[1:-1, -1] = (((pn[1:-1, 0] + pn[1:-1, -2])* dy**2 +
(pn[2:, -1] + pn[0:-2, -1]) * dx**2) /
(2 * (dx**2 + dy**2)) -
dx**2 * dy**2 / (2 * (dx**2 + dy**2)) * b[1:-1, -1])
# Periodic BC Pressure @ x = 0
p[1:-1, 0] = (((pn[1:-1, 1] + pn[1:-1, -1])* dy**2 +
(pn[2:, 0] + pn[0:-2, 0]) * dx**2) /
(2 * (dx**2 + dy**2)) -
dx**2 * dy**2 / (2 * (dx**2 + dy**2)) * b[1:-1, 0])
# Wall boundary conditions, pressure
p[-1, :] =p[-2, :] # dp/dy = 0 at y = 2
p[0, :] = p[1, :] # dp/dy = 0 at y = 0
return p
```
Now we have our familiar list of variables and initial conditions to declare before we start.
```
##variable declarations
nx = 41
ny = 41
nt = 10
nit = 50
c = 1
dx = 2 / (nx - 1)
dy = 2 / (ny - 1)
x = numpy.linspace(0, 2, nx)
y = numpy.linspace(0, 2, ny)
X, Y = numpy.meshgrid(x, y)
##physical variables
rho = 1
nu = .1
F = 1
dt = .01
#initial conditions
u = numpy.zeros((ny, nx))
un = numpy.zeros((ny, nx))
v = numpy.zeros((ny, nx))
vn = numpy.zeros((ny, nx))
p = numpy.ones((ny, nx))
pn = numpy.ones((ny, nx))
b = numpy.zeros((ny, nx))
```
For the meat of our computation, we're going to reach back to a trick we used in Step 9 for Laplace's Equation. We're interested in what our grid will look like once we've reached a near-steady state. We can either specify a number of timesteps `nt` and increment it until we're satisfied with the results, or we can tell our code to run until the difference between two consecutive iterations is very small.
We also have to manage **8** separate boundary conditions for each iteration. The code below writes each of them out explicitly. If you're interested in a challenge, you can try to write a function which can handle some or all of these boundary conditions. If you're interested in tackling that, you should probably read up on Python [dictionaries](http://docs.python.org/2/tutorial/datastructures.html#dictionaries).
```
udiff = 1
stepcount = 0
while udiff > .001:
un = u.copy()
vn = v.copy()
b = build_up_b(rho, dt, dx, dy, u, v)
p = pressure_poisson_periodic(p, dx, dy)
u[1:-1, 1:-1] = (un[1:-1, 1:-1] -
un[1:-1, 1:-1] * dt / dx *
(un[1:-1, 1:-1] - un[1:-1, 0:-2]) -
vn[1:-1, 1:-1] * dt / dy *
(un[1:-1, 1:-1] - un[0:-2, 1:-1]) -
dt / (2 * rho * dx) *
(p[1:-1, 2:] - p[1:-1, 0:-2]) +
nu * (dt / dx**2 *
(un[1:-1, 2:] - 2 * un[1:-1, 1:-1] + un[1:-1, 0:-2]) +
dt / dy**2 *
(un[2:, 1:-1] - 2 * un[1:-1, 1:-1] + un[0:-2, 1:-1])) +
F * dt)
v[1:-1, 1:-1] = (vn[1:-1, 1:-1] -
un[1:-1, 1:-1] * dt / dx *
(vn[1:-1, 1:-1] - vn[1:-1, 0:-2]) -
vn[1:-1, 1:-1] * dt / dy *
(vn[1:-1, 1:-1] - vn[0:-2, 1:-1]) -
dt / (2 * rho * dy) *
(p[2:, 1:-1] - p[0:-2, 1:-1]) +
nu * (dt / dx**2 *
(vn[1:-1, 2:] - 2 * vn[1:-1, 1:-1] + vn[1:-1, 0:-2]) +
dt / dy**2 *
(vn[2:, 1:-1] - 2 * vn[1:-1, 1:-1] + vn[0:-2, 1:-1])))
# Periodic BC u @ x = 2
u[1:-1, -1] = (un[1:-1, -1] - un[1:-1, -1] * dt / dx *
(un[1:-1, -1] - un[1:-1, -2]) -
vn[1:-1, -1] * dt / dy *
(un[1:-1, -1] - un[0:-2, -1]) -
dt / (2 * rho * dx) *
(p[1:-1, 0] - p[1:-1, -2]) +
nu * (dt / dx**2 *
(un[1:-1, 0] - 2 * un[1:-1,-1] + un[1:-1, -2]) +
dt / dy**2 *
(un[2:, -1] - 2 * un[1:-1, -1] + un[0:-2, -1])) + F * dt)
# Periodic BC u @ x = 0
u[1:-1, 0] = (un[1:-1, 0] - un[1:-1, 0] * dt / dx *
(un[1:-1, 0] - un[1:-1, -1]) -
vn[1:-1, 0] * dt / dy *
(un[1:-1, 0] - un[0:-2, 0]) -
dt / (2 * rho * dx) *
(p[1:-1, 1] - p[1:-1, -1]) +
nu * (dt / dx**2 *
(un[1:-1, 1] - 2 * un[1:-1, 0] + un[1:-1, -1]) +
dt / dy**2 *
(un[2:, 0] - 2 * un[1:-1, 0] + un[0:-2, 0])) + F * dt)
# Periodic BC v @ x = 2
v[1:-1, -1] = (vn[1:-1, -1] - un[1:-1, -1] * dt / dx *
(vn[1:-1, -1] - vn[1:-1, -2]) -
vn[1:-1, -1] * dt / dy *
(vn[1:-1, -1] - vn[0:-2, -1]) -
dt / (2 * rho * dy) *
(p[2:, -1] - p[0:-2, -1]) +
nu * (dt / dx**2 *
(vn[1:-1, 0] - 2 * vn[1:-1, -1] + vn[1:-1, -2]) +
dt / dy**2 *
(vn[2:, -1] - 2 * vn[1:-1, -1] + vn[0:-2, -1])))
# Periodic BC v @ x = 0
v[1:-1, 0] = (vn[1:-1, 0] - un[1:-1, 0] * dt / dx *
(vn[1:-1, 0] - vn[1:-1, -1]) -
vn[1:-1, 0] * dt / dy *
(vn[1:-1, 0] - vn[0:-2, 0]) -
dt / (2 * rho * dy) *
(p[2:, 0] - p[0:-2, 0]) +
nu * (dt / dx**2 *
(vn[1:-1, 1] - 2 * vn[1:-1, 0] + vn[1:-1, -1]) +
dt / dy**2 *
(vn[2:, 0] - 2 * vn[1:-1, 0] + vn[0:-2, 0])))
# Wall BC: u,v = 0 @ y = 0,2
u[0, :] = 0
u[-1, :] = 0
v[0, :] = 0
v[-1, :]=0
udiff = (numpy.sum(u) - numpy.sum(un)) / numpy.sum(u)
stepcount += 1
```
You can see that we've also included a variable `stepcount` to see how many iterations our loop went through before our stop condition was met.
```
print(stepcount)
```
If you want to see how the number of iterations increases as our `udiff` condition gets smaller and smaller, try defining a function to perform the `while` loop written above that takes an input `udiff` and outputs the number of iterations that the function runs.
For now, let's look at our results. We've used the quiver function to look at the cavity flow results and it works well for channel flow, too.
```
fig = pyplot.figure(figsize = (11,7), dpi=100)
pyplot.quiver(X[::3, ::3], Y[::3, ::3], u[::3, ::3], v[::3, ::3]);
```
The structures in the `quiver` command that look like `[::3, ::3]` are useful when dealing with large amounts of data that you want to visualize. The one used above tells `matplotlib` to only plot every 3rd data point. If we leave it out, you can see that the results can appear a little crowded.
```
fig = pyplot.figure(figsize = (11,7), dpi=100)
pyplot.quiver(X, Y, u, v);
```
## Learn more
***
##### What is the meaning of the $F$ term?
Step 12 is an exercise demonstrating the problem of flow in a channel or pipe. If you recall from your fluid mechanics class, a specified pressure gradient is what drives Poisseulle flow.
Recall the $x$-momentum equation:
$$\frac{\partial u}{\partial t}+u \cdot \nabla u = -\frac{\partial p}{\partial x}+\nu \nabla^2 u$$
What we actually do in Step 12 is split the pressure into steady and unsteady components $p=P+p'$. The applied steady pressure gradient is the constant $-\frac{\partial P}{\partial x}=F$ (interpreted as a source term), and the unsteady component is $\frac{\partial p'}{\partial x}$. So the pressure that we solve for in Step 12 is actually $p'$, which for a steady flow is in fact equal to zero everywhere.
<b>Why did we do this?</b>
Note that we use periodic boundary conditions for this flow. For a flow with a constant pressure gradient, the value of pressure on the left edge of the domain must be different from the pressure at the right edge. So we cannot apply periodic boundary conditions on the pressure directly. It is easier to fix the gradient and then solve for the perturbations in pressure.
<b>Shouldn't we always expect a uniform/constant $p'$ then?</b>
That's true only in the case of steady laminar flows. At high Reynolds numbers, flows in channels can become turbulent, and we will see unsteady fluctuations in the pressure, which will result in non-zero values for $p'$.
In step 12, note that the pressure field itself is not constant, but it's the pressure perturbation field that is. The pressure field varies linearly along the channel with slope equal to the pressure gradient. Also, for incompressible flows, the absolute value of the pressure is inconsequential.
##### And explore more CFD materials online
The interactive module **12 steps to Navier–Stokes** is one of several components of the Computational Fluid Dynamics class taught by Prof. Lorena A. Barba in Boston University between 2009 and 2013.
For a sample of what the othe components of this class are, you can explore the **Resources** section of the Spring 2013 version of [the course's Piazza site](https://piazza.com/bu/spring2013/me702/resources).
***
```
from IPython.core.display import HTML
def css_styling():
styles = open("../styles/custom.css", "r").read()
return HTML(styles)
css_styling()
```
(The cell above executes the style for this notebook.)
|
github_jupyter
|
import numpy
from matplotlib import pyplot, cm
from mpl_toolkits.mplot3d import Axes3D
%matplotlib inline
def build_up_b(rho, dt, dx, dy, u, v):
b = numpy.zeros_like(u)
b[1:-1, 1:-1] = (rho * (1 / dt * ((u[1:-1, 2:] - u[1:-1, 0:-2]) / (2 * dx) +
(v[2:, 1:-1] - v[0:-2, 1:-1]) / (2 * dy)) -
((u[1:-1, 2:] - u[1:-1, 0:-2]) / (2 * dx))**2 -
2 * ((u[2:, 1:-1] - u[0:-2, 1:-1]) / (2 * dy) *
(v[1:-1, 2:] - v[1:-1, 0:-2]) / (2 * dx))-
((v[2:, 1:-1] - v[0:-2, 1:-1]) / (2 * dy))**2))
# Periodic BC Pressure @ x = 2
b[1:-1, -1] = (rho * (1 / dt * ((u[1:-1, 0] - u[1:-1,-2]) / (2 * dx) +
(v[2:, -1] - v[0:-2, -1]) / (2 * dy)) -
((u[1:-1, 0] - u[1:-1, -2]) / (2 * dx))**2 -
2 * ((u[2:, -1] - u[0:-2, -1]) / (2 * dy) *
(v[1:-1, 0] - v[1:-1, -2]) / (2 * dx)) -
((v[2:, -1] - v[0:-2, -1]) / (2 * dy))**2))
# Periodic BC Pressure @ x = 0
b[1:-1, 0] = (rho * (1 / dt * ((u[1:-1, 1] - u[1:-1, -1]) / (2 * dx) +
(v[2:, 0] - v[0:-2, 0]) / (2 * dy)) -
((u[1:-1, 1] - u[1:-1, -1]) / (2 * dx))**2 -
2 * ((u[2:, 0] - u[0:-2, 0]) / (2 * dy) *
(v[1:-1, 1] - v[1:-1, -1]) / (2 * dx))-
((v[2:, 0] - v[0:-2, 0]) / (2 * dy))**2))
return b
def pressure_poisson_periodic(p, dx, dy):
pn = numpy.empty_like(p)
for q in range(nit):
pn = p.copy()
p[1:-1, 1:-1] = (((pn[1:-1, 2:] + pn[1:-1, 0:-2]) * dy**2 +
(pn[2:, 1:-1] + pn[0:-2, 1:-1]) * dx**2) /
(2 * (dx**2 + dy**2)) -
dx**2 * dy**2 / (2 * (dx**2 + dy**2)) * b[1:-1, 1:-1])
# Periodic BC Pressure @ x = 2
p[1:-1, -1] = (((pn[1:-1, 0] + pn[1:-1, -2])* dy**2 +
(pn[2:, -1] + pn[0:-2, -1]) * dx**2) /
(2 * (dx**2 + dy**2)) -
dx**2 * dy**2 / (2 * (dx**2 + dy**2)) * b[1:-1, -1])
# Periodic BC Pressure @ x = 0
p[1:-1, 0] = (((pn[1:-1, 1] + pn[1:-1, -1])* dy**2 +
(pn[2:, 0] + pn[0:-2, 0]) * dx**2) /
(2 * (dx**2 + dy**2)) -
dx**2 * dy**2 / (2 * (dx**2 + dy**2)) * b[1:-1, 0])
# Wall boundary conditions, pressure
p[-1, :] =p[-2, :] # dp/dy = 0 at y = 2
p[0, :] = p[1, :] # dp/dy = 0 at y = 0
return p
##variable declarations
nx = 41
ny = 41
nt = 10
nit = 50
c = 1
dx = 2 / (nx - 1)
dy = 2 / (ny - 1)
x = numpy.linspace(0, 2, nx)
y = numpy.linspace(0, 2, ny)
X, Y = numpy.meshgrid(x, y)
##physical variables
rho = 1
nu = .1
F = 1
dt = .01
#initial conditions
u = numpy.zeros((ny, nx))
un = numpy.zeros((ny, nx))
v = numpy.zeros((ny, nx))
vn = numpy.zeros((ny, nx))
p = numpy.ones((ny, nx))
pn = numpy.ones((ny, nx))
b = numpy.zeros((ny, nx))
udiff = 1
stepcount = 0
while udiff > .001:
un = u.copy()
vn = v.copy()
b = build_up_b(rho, dt, dx, dy, u, v)
p = pressure_poisson_periodic(p, dx, dy)
u[1:-1, 1:-1] = (un[1:-1, 1:-1] -
un[1:-1, 1:-1] * dt / dx *
(un[1:-1, 1:-1] - un[1:-1, 0:-2]) -
vn[1:-1, 1:-1] * dt / dy *
(un[1:-1, 1:-1] - un[0:-2, 1:-1]) -
dt / (2 * rho * dx) *
(p[1:-1, 2:] - p[1:-1, 0:-2]) +
nu * (dt / dx**2 *
(un[1:-1, 2:] - 2 * un[1:-1, 1:-1] + un[1:-1, 0:-2]) +
dt / dy**2 *
(un[2:, 1:-1] - 2 * un[1:-1, 1:-1] + un[0:-2, 1:-1])) +
F * dt)
v[1:-1, 1:-1] = (vn[1:-1, 1:-1] -
un[1:-1, 1:-1] * dt / dx *
(vn[1:-1, 1:-1] - vn[1:-1, 0:-2]) -
vn[1:-1, 1:-1] * dt / dy *
(vn[1:-1, 1:-1] - vn[0:-2, 1:-1]) -
dt / (2 * rho * dy) *
(p[2:, 1:-1] - p[0:-2, 1:-1]) +
nu * (dt / dx**2 *
(vn[1:-1, 2:] - 2 * vn[1:-1, 1:-1] + vn[1:-1, 0:-2]) +
dt / dy**2 *
(vn[2:, 1:-1] - 2 * vn[1:-1, 1:-1] + vn[0:-2, 1:-1])))
# Periodic BC u @ x = 2
u[1:-1, -1] = (un[1:-1, -1] - un[1:-1, -1] * dt / dx *
(un[1:-1, -1] - un[1:-1, -2]) -
vn[1:-1, -1] * dt / dy *
(un[1:-1, -1] - un[0:-2, -1]) -
dt / (2 * rho * dx) *
(p[1:-1, 0] - p[1:-1, -2]) +
nu * (dt / dx**2 *
(un[1:-1, 0] - 2 * un[1:-1,-1] + un[1:-1, -2]) +
dt / dy**2 *
(un[2:, -1] - 2 * un[1:-1, -1] + un[0:-2, -1])) + F * dt)
# Periodic BC u @ x = 0
u[1:-1, 0] = (un[1:-1, 0] - un[1:-1, 0] * dt / dx *
(un[1:-1, 0] - un[1:-1, -1]) -
vn[1:-1, 0] * dt / dy *
(un[1:-1, 0] - un[0:-2, 0]) -
dt / (2 * rho * dx) *
(p[1:-1, 1] - p[1:-1, -1]) +
nu * (dt / dx**2 *
(un[1:-1, 1] - 2 * un[1:-1, 0] + un[1:-1, -1]) +
dt / dy**2 *
(un[2:, 0] - 2 * un[1:-1, 0] + un[0:-2, 0])) + F * dt)
# Periodic BC v @ x = 2
v[1:-1, -1] = (vn[1:-1, -1] - un[1:-1, -1] * dt / dx *
(vn[1:-1, -1] - vn[1:-1, -2]) -
vn[1:-1, -1] * dt / dy *
(vn[1:-1, -1] - vn[0:-2, -1]) -
dt / (2 * rho * dy) *
(p[2:, -1] - p[0:-2, -1]) +
nu * (dt / dx**2 *
(vn[1:-1, 0] - 2 * vn[1:-1, -1] + vn[1:-1, -2]) +
dt / dy**2 *
(vn[2:, -1] - 2 * vn[1:-1, -1] + vn[0:-2, -1])))
# Periodic BC v @ x = 0
v[1:-1, 0] = (vn[1:-1, 0] - un[1:-1, 0] * dt / dx *
(vn[1:-1, 0] - vn[1:-1, -1]) -
vn[1:-1, 0] * dt / dy *
(vn[1:-1, 0] - vn[0:-2, 0]) -
dt / (2 * rho * dy) *
(p[2:, 0] - p[0:-2, 0]) +
nu * (dt / dx**2 *
(vn[1:-1, 1] - 2 * vn[1:-1, 0] + vn[1:-1, -1]) +
dt / dy**2 *
(vn[2:, 0] - 2 * vn[1:-1, 0] + vn[0:-2, 0])))
# Wall BC: u,v = 0 @ y = 0,2
u[0, :] = 0
u[-1, :] = 0
v[0, :] = 0
v[-1, :]=0
udiff = (numpy.sum(u) - numpy.sum(un)) / numpy.sum(u)
stepcount += 1
print(stepcount)
fig = pyplot.figure(figsize = (11,7), dpi=100)
pyplot.quiver(X[::3, ::3], Y[::3, ::3], u[::3, ::3], v[::3, ::3]);
fig = pyplot.figure(figsize = (11,7), dpi=100)
pyplot.quiver(X, Y, u, v);
from IPython.core.display import HTML
def css_styling():
styles = open("../styles/custom.css", "r").read()
return HTML(styles)
css_styling()
| 0.328422 | 0.892281 |
# Algorithm explanation
```
from sklearn.datasets import make_blobs
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import pyplot
from mpl_toolkits.mplot3d import Axes3D
from src import P3C
```
### Sample dataset
First, we will generate a simple dataset with clear clusters to aid with visualisation
```
centers = [(0.2, 0.2, 0.2), (0.2, 0.8, 0.8), (0.5, 0.5, 0.5)]
cluster_std = [0.02, 0.03, 0.07]
data, y = make_blobs(n_samples=1000, cluster_std=cluster_std, centers=centers, n_features=3, random_state=1)
plt.scatter(data[y == 0, 0], data[y == 0, 1], color="red", s=10, label="Cluster1")
plt.scatter(data[y == 1, 0], data[y == 1, 1], color="blue", s=10, label="Cluster2")
plt.scatter(data[y == 2, 0], data[y == 2, 1], color="green", s=10, label="Cluster3")
fig = pyplot.figure()
ax = Axes3D(fig)
ax.scatter(data[y == 0, 0], data[y == 0, 1], data[y == 0, 2], color="red", s=10, label="Cluster1")
ax.scatter(data[y == 1, 0], data[y == 1, 1], data[y == 1, 2], color="blue", s=10, label="Cluster2")
ax.scatter(data[y == 2, 0], data[y == 2, 1], data[y == 2, 2], color="green", s=10, label="Cluster3")
pyplot.show()
```
### Splitting data into bins
Along each axis...
```
bins, nr_of_bins = P3C.split_into_bins(data)
for column_bins in bins:
P3C.mark_bins(column_bins, nr_of_bins)
for column_bins in bins:
P3C.mark_merge_bins(column_bins)
for dim in range(len(data[0])):
print(f"Dimension {dim}")
val = 0. # this is the value where you want the data to appear on the y-axis.
points = [x[dim] for x in data]
pyplot.plot(points, np.zeros_like(points) + val, 'o', fillstyle='none', ms=0.5)
bin_limits = np.array([(b.interval.start, b.interval.end) for b in bins[dim]]).flatten()
pyplot.plot(bin_limits, np.zeros_like(bin_limits) + val, '|', ms=50)
pyplot.show()
plt.scatter(data[y == 0, 0], data[y == 0, 1], color="red", s=10, label="Cluster1")
plt.scatter(data[y == 1, 0], data[y == 1, 1], color="blue", s=10, label="Cluster2")
plt.scatter(data[y == 2, 0], data[y == 2, 1], color="green", s=10, label="Cluster3")
for b in bins[0]:
plt.axline((b.interval.start, 0), (b.interval.start, 1))
plt.axline((b.interval.end, 0), (b.interval.end, 1))
for b in bins[1]:
plt.axline((0, b.interval.start), (0.8, b.interval.start), c="green")
plt.axline((0, b.interval.end), (0.8, b.interval.end), c="green")
plt.show()
new_bins = P3C.merge_all_bins(bins)
for dim in range(len(data[0])):
print(f"Dimension {dim}")
val = 0. # this is the value where you want the data to appear on the y-axis.
points = [x[dim] for x in data]
pyplot.plot(points, np.zeros_like(points) + val, 'o', fillstyle='none', ms=0.5)
bin_limits = np.array([(b.interval.start, b.interval.end) for b in new_bins[dim]]).flatten()
pyplot.plot(bin_limits, np.zeros_like(bin_limits) + val, '|', ms=50)
pyplot.show()
plt.scatter(data[y == 0, 0], data[y == 0, 1], color="red", s=10, label="Cluster1")
plt.scatter(data[y == 1, 0], data[y == 1, 1], color="blue", s=10, label="Cluster2")
plt.scatter(data[y == 2, 0], data[y == 2, 1], color="green", s=10, label="Cluster3")
for b in new_bins[0]:
plt.axline((b.interval.start, 0), (b.interval.start, 1))
plt.axline((b.interval.end, 0), (b.interval.end, 1))
for b in new_bins[1]:
plt.axline((0, b.interval.start), (0.8, b.interval.start), c="green")
plt.axline((0, b.interval.end), (0.8, b.interval.end), c="green")
plt.show()
fig = pyplot.figure()
ax = Axes3D(fig)
bins_0 = np.array([(x.interval.start, x.interval.end) for x in new_bins[0]] * 100).flatten()
ax.scatter(bins_0, np.linspace(0, 1, len(bins_0)), [0 for _ in range(len(bins_0))], color="yellow", s=10)
bins_1 = np.array([(x.interval.start, x.interval.end) for x in new_bins[1]] * 100).flatten()
ax.scatter([0 for _ in range(len(bins_1))], bins_1, np.linspace(0, 1, len(bins_1)), color="yellow", s=10)
bins_2 = np.array([(x.interval.start, x.interval.end) for x in new_bins[2]] * 100).flatten()
ax.scatter(np.linspace(0, 1, len(bins_2)), [1 for _ in range(len(bins_2))], bins_2, color="yellow", s=10)
ax.scatter(data[y == 0, 0], data[y == 0, 1], data[y == 0, 2], color="red", s=10, label="Cluster1")
ax.scatter(data[y == 1, 0], data[y == 1, 1], data[y == 1, 2], color="blue", s=10, label="Cluster2")
ax.scatter(data[y == 2, 0], data[y == 2, 1], data[y == 2, 2], color="green", s=10, label="Cluster3")
pyplot.show()
```
### Find candidates
```
tree = P3C.construct_candidate_tree_start(data, new_bins)
ns = P3C.construct_new_level(0, tree, 1e-4)
candidate_list = P3C.get_candidates(ns)
candidate_list
fig = pyplot.figure()
ax = Axes3D(fig)
for psig, color in zip(candidate_list, ['red', 'green', 'blue']):
intervals = [(b.interval.start, b.interval.end) for b in sorted(psig.bins, key=lambda x: x.dimension)]
bins_0 = np.array(intervals[0] * 100).flatten()
ax.scatter(bins_0, np.linspace(0, 1, len(bins_0)), [intervals[0][1] for _ in range(len(bins_0))], color=color, s=10)
bins_1 = np.array(intervals[1] * 100).flatten()
ax.scatter([intervals[1][1] for _ in range(len(bins_1))], bins_1, np.linspace(0, 1, len(bins_1)), color=color, s=10)
bins_2 = np.array(intervals[2] * 100).flatten()
ax.scatter(np.linspace(0, 1, len(bins_2)), [intervals[2][1] for _ in range(len(bins_2))], bins_2, color=color, s=10)
ax.scatter(data[y == 0, 0], data[y == 0, 1], data[y == 0, 2], color="red", s=10, label="Cluster1")
ax.scatter(data[y == 1, 0], data[y == 1, 1], data[y == 1, 2], color="blue", s=10, label="Cluster2")
ax.scatter(data[y == 2, 0], data[y == 2, 1], data[y == 2, 2], color="green", s=10, label="Cluster3")
pyplot.show()
```
### Get means
```
inv_cov_cluster_dict = P3C.get_inv_cov_cluster_dict(candidate_list)
result, gmm, means = P3C.get_result(data, candidate_list, inv_cov_cluster_dict)
means_after_bgm, cluster_dict, cluster_points = P3C.get_clusters_and_means(candidate_list, data, result, means)
plt.scatter(data[y == 0, 0], data[y == 0, 1], color="red", s=10, label="Cluster1")
plt.scatter(data[y == 1, 0], data[y == 1, 1], color="blue", s=10, label="Cluster2")
plt.scatter(data[y == 2, 0], data[y == 2, 1], color="green", s=10, label="Cluster3")
plt.scatter([m[0] for m in means], [m[1] for m in means], color="orange", s=150, label="Means")
plt.scatter([m[0] for m in means_after_bgm], [m[1] for m in means_after_bgm], color="yellow", s=60, label="Means2")
plt.show()
```
### Outliers
```
clustered = P3C.find_outliers(data, candidate_list, cluster_dict, cluster_points, means_after_bgm, degree_of_freedom=7, alpha=0.2)
plt.scatter([x[1][0] for x in clustered], [y[1][1] for y in clustered], c = [z[0] for z in clustered])
plt.show()
fig = pyplot.figure()
ax = Axes3D(fig)
ax.scatter([x[1][0] for x in clustered], [x[1][1] for x in clustered], [x[1][2] for x in clustered], c=[x[0] for x in clustered], s=10, zorder=0)
ax.scatter([x[0] for x in means_after_bgm], [x[1] for x in means_after_bgm], [x[2] for x in means_after_bgm], color="red", s=200, zorder=100)
pyplot.show()
```
|
github_jupyter
|
from sklearn.datasets import make_blobs
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import pyplot
from mpl_toolkits.mplot3d import Axes3D
from src import P3C
centers = [(0.2, 0.2, 0.2), (0.2, 0.8, 0.8), (0.5, 0.5, 0.5)]
cluster_std = [0.02, 0.03, 0.07]
data, y = make_blobs(n_samples=1000, cluster_std=cluster_std, centers=centers, n_features=3, random_state=1)
plt.scatter(data[y == 0, 0], data[y == 0, 1], color="red", s=10, label="Cluster1")
plt.scatter(data[y == 1, 0], data[y == 1, 1], color="blue", s=10, label="Cluster2")
plt.scatter(data[y == 2, 0], data[y == 2, 1], color="green", s=10, label="Cluster3")
fig = pyplot.figure()
ax = Axes3D(fig)
ax.scatter(data[y == 0, 0], data[y == 0, 1], data[y == 0, 2], color="red", s=10, label="Cluster1")
ax.scatter(data[y == 1, 0], data[y == 1, 1], data[y == 1, 2], color="blue", s=10, label="Cluster2")
ax.scatter(data[y == 2, 0], data[y == 2, 1], data[y == 2, 2], color="green", s=10, label="Cluster3")
pyplot.show()
bins, nr_of_bins = P3C.split_into_bins(data)
for column_bins in bins:
P3C.mark_bins(column_bins, nr_of_bins)
for column_bins in bins:
P3C.mark_merge_bins(column_bins)
for dim in range(len(data[0])):
print(f"Dimension {dim}")
val = 0. # this is the value where you want the data to appear on the y-axis.
points = [x[dim] for x in data]
pyplot.plot(points, np.zeros_like(points) + val, 'o', fillstyle='none', ms=0.5)
bin_limits = np.array([(b.interval.start, b.interval.end) for b in bins[dim]]).flatten()
pyplot.plot(bin_limits, np.zeros_like(bin_limits) + val, '|', ms=50)
pyplot.show()
plt.scatter(data[y == 0, 0], data[y == 0, 1], color="red", s=10, label="Cluster1")
plt.scatter(data[y == 1, 0], data[y == 1, 1], color="blue", s=10, label="Cluster2")
plt.scatter(data[y == 2, 0], data[y == 2, 1], color="green", s=10, label="Cluster3")
for b in bins[0]:
plt.axline((b.interval.start, 0), (b.interval.start, 1))
plt.axline((b.interval.end, 0), (b.interval.end, 1))
for b in bins[1]:
plt.axline((0, b.interval.start), (0.8, b.interval.start), c="green")
plt.axline((0, b.interval.end), (0.8, b.interval.end), c="green")
plt.show()
new_bins = P3C.merge_all_bins(bins)
for dim in range(len(data[0])):
print(f"Dimension {dim}")
val = 0. # this is the value where you want the data to appear on the y-axis.
points = [x[dim] for x in data]
pyplot.plot(points, np.zeros_like(points) + val, 'o', fillstyle='none', ms=0.5)
bin_limits = np.array([(b.interval.start, b.interval.end) for b in new_bins[dim]]).flatten()
pyplot.plot(bin_limits, np.zeros_like(bin_limits) + val, '|', ms=50)
pyplot.show()
plt.scatter(data[y == 0, 0], data[y == 0, 1], color="red", s=10, label="Cluster1")
plt.scatter(data[y == 1, 0], data[y == 1, 1], color="blue", s=10, label="Cluster2")
plt.scatter(data[y == 2, 0], data[y == 2, 1], color="green", s=10, label="Cluster3")
for b in new_bins[0]:
plt.axline((b.interval.start, 0), (b.interval.start, 1))
plt.axline((b.interval.end, 0), (b.interval.end, 1))
for b in new_bins[1]:
plt.axline((0, b.interval.start), (0.8, b.interval.start), c="green")
plt.axline((0, b.interval.end), (0.8, b.interval.end), c="green")
plt.show()
fig = pyplot.figure()
ax = Axes3D(fig)
bins_0 = np.array([(x.interval.start, x.interval.end) for x in new_bins[0]] * 100).flatten()
ax.scatter(bins_0, np.linspace(0, 1, len(bins_0)), [0 for _ in range(len(bins_0))], color="yellow", s=10)
bins_1 = np.array([(x.interval.start, x.interval.end) for x in new_bins[1]] * 100).flatten()
ax.scatter([0 for _ in range(len(bins_1))], bins_1, np.linspace(0, 1, len(bins_1)), color="yellow", s=10)
bins_2 = np.array([(x.interval.start, x.interval.end) for x in new_bins[2]] * 100).flatten()
ax.scatter(np.linspace(0, 1, len(bins_2)), [1 for _ in range(len(bins_2))], bins_2, color="yellow", s=10)
ax.scatter(data[y == 0, 0], data[y == 0, 1], data[y == 0, 2], color="red", s=10, label="Cluster1")
ax.scatter(data[y == 1, 0], data[y == 1, 1], data[y == 1, 2], color="blue", s=10, label="Cluster2")
ax.scatter(data[y == 2, 0], data[y == 2, 1], data[y == 2, 2], color="green", s=10, label="Cluster3")
pyplot.show()
tree = P3C.construct_candidate_tree_start(data, new_bins)
ns = P3C.construct_new_level(0, tree, 1e-4)
candidate_list = P3C.get_candidates(ns)
candidate_list
fig = pyplot.figure()
ax = Axes3D(fig)
for psig, color in zip(candidate_list, ['red', 'green', 'blue']):
intervals = [(b.interval.start, b.interval.end) for b in sorted(psig.bins, key=lambda x: x.dimension)]
bins_0 = np.array(intervals[0] * 100).flatten()
ax.scatter(bins_0, np.linspace(0, 1, len(bins_0)), [intervals[0][1] for _ in range(len(bins_0))], color=color, s=10)
bins_1 = np.array(intervals[1] * 100).flatten()
ax.scatter([intervals[1][1] for _ in range(len(bins_1))], bins_1, np.linspace(0, 1, len(bins_1)), color=color, s=10)
bins_2 = np.array(intervals[2] * 100).flatten()
ax.scatter(np.linspace(0, 1, len(bins_2)), [intervals[2][1] for _ in range(len(bins_2))], bins_2, color=color, s=10)
ax.scatter(data[y == 0, 0], data[y == 0, 1], data[y == 0, 2], color="red", s=10, label="Cluster1")
ax.scatter(data[y == 1, 0], data[y == 1, 1], data[y == 1, 2], color="blue", s=10, label="Cluster2")
ax.scatter(data[y == 2, 0], data[y == 2, 1], data[y == 2, 2], color="green", s=10, label="Cluster3")
pyplot.show()
inv_cov_cluster_dict = P3C.get_inv_cov_cluster_dict(candidate_list)
result, gmm, means = P3C.get_result(data, candidate_list, inv_cov_cluster_dict)
means_after_bgm, cluster_dict, cluster_points = P3C.get_clusters_and_means(candidate_list, data, result, means)
plt.scatter(data[y == 0, 0], data[y == 0, 1], color="red", s=10, label="Cluster1")
plt.scatter(data[y == 1, 0], data[y == 1, 1], color="blue", s=10, label="Cluster2")
plt.scatter(data[y == 2, 0], data[y == 2, 1], color="green", s=10, label="Cluster3")
plt.scatter([m[0] for m in means], [m[1] for m in means], color="orange", s=150, label="Means")
plt.scatter([m[0] for m in means_after_bgm], [m[1] for m in means_after_bgm], color="yellow", s=60, label="Means2")
plt.show()
clustered = P3C.find_outliers(data, candidate_list, cluster_dict, cluster_points, means_after_bgm, degree_of_freedom=7, alpha=0.2)
plt.scatter([x[1][0] for x in clustered], [y[1][1] for y in clustered], c = [z[0] for z in clustered])
plt.show()
fig = pyplot.figure()
ax = Axes3D(fig)
ax.scatter([x[1][0] for x in clustered], [x[1][1] for x in clustered], [x[1][2] for x in clustered], c=[x[0] for x in clustered], s=10, zorder=0)
ax.scatter([x[0] for x in means_after_bgm], [x[1] for x in means_after_bgm], [x[2] for x in means_after_bgm], color="red", s=200, zorder=100)
pyplot.show()
| 0.608594 | 0.978651 |
# Understanding How the DataBlock API was Built
```
%load_ext autoreload
%autoreload 2
%matplotlib inline
from exp.nb_07a import *
# Importing fastai's custom Imagenet dataset, called "Imagenette".
# Bear in mind that Imagenette v2 now has a 70/30 split so values will
# differ from the course notebook.
datasets.URLs.IMAGENETTE_160
```
## Building an Image ItemList
Previously, we were reading the whole MNIST dataset into our local RAM at once in pickle format. That can't be done for larger datasets.
From here on out, images will be left on disk and will be loaded according to the mini-batches being used for training.
```
path = datasets.untar_data(datasets.URLs.IMAGENETTE_160)
path
# We will add the .ls method to the path as a monkey patch
import PIL, os, mimetypes
Path.ls = lambda x: list(x.iterdir())
# Checking if the changes took effect
path.ls()
# Looking inside our classes folders
(path/'val').ls()
# Looking inside the Tench class folder
path_tench = path/'val'/'n01440764'
# Assigning a single image
img_fn = path_tench.ls()[0]
img_fn
img = PIL.Image.open(img_fn)
img
# Plotting dimensions of the image
plt.imshow(img);
```
We can also check the image dimensions using the standard numpy method.
```
import numpy as np
img_arr = np.array(img)
img_arr.shape
```
- We must be careful about other file types in the directory, for e.g. models, text etc.
- Instead of writing it out by hand, let's use the MIME types database.
- Details of mapping filenames to MIME types can be found Python's [documentation]("https://docs.python.org/2/library/mimetypes.html").
```
image_extensions = set(k for k,v in mimetypes.types_map.items()\
if v.startswith('image/'))
# Here are the image type extensions we will be focusing on
' '.join(image_extensions)
# Convert items first to a list and then a set
def setify(o):
return o if isinstance(o, set) else set(listify(o))
# Using the testing framework from previous nbs
test_eq(setify('aa'), {'aa'})
test_eq(setify(['aa',1]), {'aa',1})
test_eq(setify(None), set())
test_eq(setify(1), {1})
test_eq(setify({1}), {1})
```
- Now that our `setify()` function is ready, we will grab only the image files from the target directories.
- The first private function grabs all the images inside a given directory and the second one "walks" (potentially recursively) through all the folders in the given `path`
```
# Run through a single directory and grab the images in that
def _get_files(p, fs, extensions=None):
p = Path(p)
res = [p/f for f in fs if not f.startswith('.')
and ((not extensions) or f'.{f.split(".")[-1].lower()}' in extensions)]
return res
t = [o.name for o in os.scandir(path_tench)]
t = _get_files(path, t, extensions=image_extensions)
t[0:10]
# Putting all of the above into one function to "get files"
def get_files(path, extensions=None, recurse=False, include=None):
path=Path(path)
extensions = setify(extensions)
extensions = {e.lower() for e in extensions}
if recurse:
res = []
for i,(p,d,f) in enumerate(os.walk(path)):
if include is not None and i==0: d[:] = [o for o in d if o in include]
else: d[:] = [o for o in d if not o.startswith('.')]
res += _get_files(p, f, extensions)
return res
else:
f = [o.name for o in os.scandir(path) if o.is_file()]
return _get_files(path, f, extensions)
```
- `scandir()` is highly optimized as it a "thin" wrapper on top of C's API.
- For recursion, `os.walk()` uses scandir() internally to walk through a folder tree. We can also modify the list of directories it looks at on the fly.
```
# Testing the get_files function
get_files(path_tench, image_extensions)[:5]
```
The **recurse** argument is needed when we start from `path`, since the pictures are two levels below in directories.
```
get_files(path, image_extensions, recurse=True)[:5]
# Getting all of the file names
all_fns = get_files(path, image_extensions, recurse=True)
len(all_fns)
# Let's test the speed of the get_files function
# Bearing in mind that Imagenet is 100 times bigger than imagenette
%timeit -n 10 get_files(path, image_extensions, recurse=True)
```
## Data Prep for Modeling
So, our data pipeline will be:
- Getting files
- Splitting the validation set (args will be random%, folder name, ftype, etc.)
- Label the data (steps above)
- Transform per image / Augmentation (optional step)
- Transform to Tensor
- DataLoader
- Transform per batch (optional)
- DataBunch
- Add test set (optional)
### Get Files
- Using the `ListContainer` from before (exp06) to store our objects in the `ItemList`.
- `get` will need to be subclassed to explain how to access an element, then the private `_get` method will allow an additional transform operation.
- `new` will be used in conjunction with `__getitem__` (which works for one index or a list of indeces) to create training and validation sets from a single stream once the data is split.
- We could just put all files into ListContainer but it is much better if we could get the image right away when we call get function. ImageList does this by inheriting ItemList that inherits ListContainer. ItemList is just a function that gets items, path, and transforms. Transforms are called in order (if there is) and the data will be updated so that it rewrites the old data. More about compose: https://en.wikipedia.org/wiki/Function_composition_(computer_science) ImageList in the other hand is a function that opens the images for us when get is called. (Lankinen's Notes)
```
def compose(x, funcs, *args, order_key='_order', **kwargs):
key = lambda o: getattr(o, order_key, 0)
for f in sorted(listify(funcs), key=key):
x = f(x, **kwargs)
return x
class ItemList(ListContainer): # Grab image files and dump them into the ListContainer
def __init__(self, items, path='.', tfms=None):
super().__init__(items)
self.path, self.tfms = Path(path), tfms
def __repr__(self):
return f'{super().__repr__()}\Path: {self.path}'
def new(self, items, cls=None):
if cls is None:
cls=self.__class__
return cls(items, self.path, tfms=self.tfms)
def get(self, i): #This default get method will be over-ridden
return i
def _get(self, i):
return compose(self.get(i), self.tfms) #refer to Wikipedia link on compose
def __getitem__(self, idx):
res = super().__getitem__(idx) #Index into our list, pass it to the container
if isinstance(res, list): return [self._get(o) for o in res] # Call if list
return self._get(res) #call if single item
class ImageList(ItemList):
@classmethod
def from_files(cls, path, extensions=None, recurse=True, include=None, **kwargs):
if extensions is None:
extensions = image_extensions
return cls(get_files(path, extensions, recurse=recurse, include=include), path, **kwargs)
def get(self, fn):
return PIL.Image.open(fn)
```
- Transforms are strictly limited to data augmentation. For total flexibility, `ImageList` returns a raw PIL image. The first step would be to convert it to RGB or some other format.
- Transforms only need to be functions that take an element of the `ItemList` and transform it.
- In order to give them state, they can be defined as a class. Having them as a class allows to define an `_order` attribute(default 0), which is used to sort the transforms.
```
class Transform():
_order=0
# Both the class and the function do the same thing.
# Obviously, the class can be extended.
class MakeRGB(Transform):
def __call__(self, item): return item.convert('RGB')
def make_rgb(item):
return item.convert('RGB')
il = ImageList.from_files(path, tfms=make_rgb)
# __repr__ gives us following print outputs
il
# Indexing into the image list
img = il[1]
img
il[:2] # We can also index with a range of integers
```
### Split Validation Set
- The files need to be split between the train and val folders.
- As file names are `path` objects, the file directory can be found with `.parent`.
```
fn = il.items[0]; fn
fn.parent.parent.name
def grandparent_splitter(fn, valid_name='valid', train_name='train'):
gp = fn.parent.parent.name
return True if gp==valid_name else False if gp==train_name else None
def split_by_func(items, f):
mask = [f(o) for o in items]
# None values are filtered out
f = [o for o, m in zip(items, mask) if m==False]
t = [o for o, m in zip(items, mask) if m==True]
return f, t
splitter = partial(grandparent_splitter, valid_name='val')
%time train, valid = split_by_func(il, splitter)
len(train), len(valid)
```
- Now that the data is split, we will create a class to contain it.
- It just needs two `ItemList`s to be initialized.
- We can create a shortcut to all the unknown attributes by grabbing them in the `train` `ItemList`.
```
class SplitData():
def __init__(self, train, valid):
self.train, self.valid = train, valid
def __getattr__(self, k):
return getattr(self.train, k)
# Needed if SplitData needs to be pickled and loaded back without recursion errors
def __setstate__(self, data:Any):
self.__dict__.update(data)
@classmethod
def split_by_func(cls, il, f):
#il.new is defined in class ItemList
#identifies an object and it's class / sub-class and creates a new
#item list with the same type, path, tfms
lists = map(il.new, split_by_func(il.items, f))
return cls(*lists)
def __repr__(self):
return f'{self.__class__.__name__}\nTrain: {self.train}\nValid:{self.valid}\n'
sd = SplitData.split_by_func(il, splitter)
sd
```
### Labeling
- After splitting, we will use the training information to label the validation set using a **Processor**.
- A **Processor** is a transformation that is applied to all the inputs once at initialization, with some state computed on the training set that is applied without modification on the validation set (also could be on the test set or on a single item during inference).
- For e.g. processing texts to tokenize, and numericalize afterwards. In such a case, we want the validation set to be numericalized with exactly the same vocabulary as the training set.
- For tabular data, where we could impute missing values with medians, that statistic is stored in the inner state of the Processor and applied to the validation set.
- Here, we want to convert label strings to numbers in a consistent and reproducible way. This means creating a list of possible labels in the training set and then converting them to numbers based on this vocab.
```
from collections import OrderedDict
def uniqueify(x, sort=False):
res = list(OrderedDict.fromkeys(x).keys())
if sort:
res.sort()
return res
```
- Let's define the processor.
- A `ProcessedItemList` with an `obj` method will also be defined, which will allow us to get unprocessed items. For e.g. a processed label will be an index between 0 and the number number of classes - 1, the corresponding `obj` will be the name of the class.
```
# The first one is needed by the model for training
# The second is better for displaying objects.
class Processor():
def process(self, items): return items
class CategoryProcessor(Processor):# Creates list of all possible categories
def __init__(self): self.vocab=None
def __call__(self, items):
#The vocab is defined on the first use.
#defines the k, v which goes from object to int (otoi)
if self.vocab is None:
self.vocab = uniqueify(items)
self.otoi = {v:k for k,v in enumerate(self.vocab)}
return [self.proc1(o) for o in items]
def proc1(self, item): return self.otoi[item]
def deprocess(self, idxs): # Print out the inferences
assert self.vocab is not None
return [self.deproc1(idx) for idx in idxs] # deprocess for each index
def deproc1(self, idx): return self.vocab[idx]
```
- Now we label according to the folders of the images i.e. `fn.parent.name`.
- We label the training set first with `CategoryProcessor` which computes its inner `vocab` on that set.
- Afterwards, we label the validation set using the same processor, which also uses the same `vocab`.
- This results in another `SplitData` object.
```
# For processing we need the grand parent and for labelling we need the parent
def parent_labeler(fn): return fn.parent.name
def _label_by_func(ds, f, cls=ItemList): return cls([f(o) for o in ds.items], path=ds.path)
class LabeledData():
def process(self, il, proc): return il.new(compose(il.items, proc))
def __init__(self, x, y, proc_x=None, proc_y=None):
self.x,self.y = self.process(x, proc_x),self.process(y, proc_y)
self.proc_x,self.proc_y = proc_x,proc_y
def __repr__(self): return f'{self.__class__.__name__}\nx: {self.x}\ny: {self.y}\n'
def __getitem__(self,idx): return self.x[idx],self.y[idx]
def __len__(self): return len(self.x)
def x_obj(self, idx): return self.obj(self.x, idx, self.proc_x)
def y_obj(self, idx): return self.obj(self.y, idx, self.proc_y)
def obj(self, items, idx, procs):
isint = isinstance(idx, int) or (isinstance(idx,torch.LongTensor) and not idx.ndim)
item = items[idx]
for proc in reversed(listify(procs)):
item = proc.deproc1(item) if isint else proc.deprocess(item)
return item
# Added convenience, can read up more about them
@classmethod
def label_by_func(cls, il, f, proc_x=None, proc_y=None):
return cls(il, _label_by_func(il, f), proc_x=proc_x, proc_y=proc_y)
# This is a very important piece of the...pie??
# when we pass in train, the processor has no vocab. It then goes CategoryProcessor to
# create a list of unique possibilities. When it goes to valid, then proc will have a vocab
# so it will skip the step and use the training set's vocab.
# Here we will ENSURE that the mapping is similar for both training and validation sets.
def label_by_func(sd, f, proc_x=None, proc_y=None):
train = LabeledData.label_by_func(sd.train, f, proc_x=proc_x, proc_y=proc_y)
valid = LabeledData.label_by_func(sd.valid, f, proc_x=proc_x, proc_y=proc_y)
return SplitData(train,valid)
ll = label_by_func(sd, parent_labeler, proc_y=CategoryProcessor())
# Validating
assert ll.train.proc_y is ll.valid.proc_y
# So now we have the labeled lists
ll
# Delving deeper
ll.train
# And deeper
ll.train.y
# .....deeper still
ll.train.y.items[0], ll.train.y_obj(0), ll.train.y_obj(slice(4))
```
### Transform to Tensor
- In order to train our model, we will need to convert these pillow objects to tensors.
```
# Getting the fist item, or x, of the tuple
ll.train[0]
# Now the second item
ll.train[0][0]
# Let's resize the image to 128x128
ll.train[0][0].resize((128,128))
```
- We will now make a class for resizing our images.
- It is important to note that resizing should take part _AFTER_ other transformation operations.
```
class ResizeFixed(Transform): # Carry out Bilinear resizing
_order=10
def __init__(self, size):
if isinstance(size, int):
size=(size, size)
self.size = size
def __call__(self, item):
return item.resize(self.size, PIL.Image.BILINEAR)
def to_byte_tensor(item):
res = torch.ByteTensor(torch.ByteStorage.from_buffer(item.tobytes()))
w, h = item.size
return res.view(h, w, -1).permute(2, 0, 1) #Pillow and PyTorch handle permute differently
# this can be seen in the cell below
# Here we can giving to_byte_tensor and to_float_tensor "class level state"
# i.e. we can attach a state to a function, which is underused in Python but is
# very useful
to_byte_tensor._order=20
def to_float_tensor(item):
return item.float().div_(255.) # we divide by 255 to ensure the value is between 0 and 1
to_float_tensor._order=30
# Passing the list of tfms
tfms = [make_rgb, ResizeFixed(128), to_byte_tensor, to_float_tensor]
il = ImageList.from_files(path, tfms=tfms)
sd = SplitData.split_by_func(il, splitter)
ll = label_by_func(sd, parent_labeler, proc_y=CategoryProcessor())
def show_image(im, figsize=(3,3)):
plt.figure(figsize=figsize)
plt.axis('off')
plt.imshow(im.permute(1,2,0)) # Here the channel comes in last
x, y = ll.train[0]
x.shape
show_image(x)
```
## Modeling
### DataBunch
We will now put our datasets in the DataBunch.
```
bs = 128
train_dl, valid_dl = get_dls(ll.train, ll.valid, bs, num_workers=4)
x, y = next(iter(train_dl))
x.shape
# Images can be viewed in the batch, plus their corresponding classes
show_image(x[0])
ll.train.proc_y.vocab[y[0]]
y
```
- The `DataBunch` class will be modified with additional attributes `c_in` (channel in) and `c_out` (channel out) instead of just c.
- This way any models which need to be created automatically can do so with the correct number of inputs and outputs.
```
class DataBunch():
def __init__(self, train_dl, valid_dl, c_in=None, c_out=None):
self.train_dl, self.valid_dl, self.c_in, self.c_out = train_dl, valid_dl, c_in, c_out
@property
def train_ds(self): return self.train_dl.dataset
@property
def valid_ds(self): return self.valid_dl.dataset
# Defining a function that goes directly from SplitData to a DataBunch
def databunchify(sd, bs, c_in=None, c_out=None, **kwargs):
dls = get_dls(sd.train, sd.valid, bs, **kwargs)
return DataBunch(*dls, c_in=c_in, c_out=c_out)
SplitData.to_databunch = databunchify # This is another example of monkey patching
# on the fly.
# Running through all we have worked on so far
path = datasets.untar_data(datasets.URLs.IMAGENETTE_160)
tfms = [make_rgb, ResizeFixed(128), to_byte_tensor, to_float_tensor]
il = ImageList.from_files(path, tfms=tfms)
sd = SplitData.split_by_func(il, partial(grandparent_splitter, valid_name='val'))
ll = label_by_func(sd, parent_labeler, proc_y=CategoryProcessor())
data = ll.to_databunch(bs, c_in=3, c_out=10, num_workers=4)
```
### Model Building
```
cbfs = [partial(AvgStatsCallback, accuracy),
CudaCallback]
```
Normalizing statistics from a batch
```
m, s = x.mean((0,2,3)).cuda(), x.std((0,2,3)).cuda()
m, s
# Defining a function to normalize over three channels by using
# broadcasting
def normalize_chan(x, mean, std):
return (x - mean[..., None, None]) / std[..., None, None]
_m = tensor([0.47, 0.48, 0.45])
_s = tensor([0.29, 0.28, 0.30])
norm_imagenette = partial(normalize_chan, mean=_m.cuda(), std=_s.cuda())
# Choosing our callback functions
cbfs.append(partial(BatchTransformXCallback, norm_imagenette))
nfs = [64, 64, 128, 256]
```
- Time to build our model.
```
import math
def prev_pow_2(x):
return 2**math.floor(math.log2(x))
def get_cnn_layers(data, nfs, layer, **kwargs):
def f(ni, nf, stride=2):
return layer(ni, nf, 3, stride=stride, **kwargs)
l1 = data.c_in # layer 1 of the model
l2 = prev_pow_2(l1*3*3) # taking value of c_in and multiplying with next number which is pow^2
layers = [f(l1, l2, stride=1),
f(l2, l2*2, stride=2),
f(l2*2, l2*4, stride=2)]
nfs = [l2*4] + nfs
layers += [f(nfs[i], nfs[i+1]) for i in range(len(nfs)-1)]
layers += [nn.AdaptiveAvgPool2d(1), Lambda(flatten),
nn.Linear(nfs[-1], data.c_out)]
return layers
def get_cnn_model(data, nfs, layer, **kwargs):
return nn.Sequential(*get_cnn_layers(data, nfs, layer, **kwargs))
def get_learn_run(nfs, data, lr, layer, cbs=None, opt_func=None, **kwargs):
model = get_cnn_model(data, nfs, layer, **kwargs)
init_cnn(model)
return get_runner(model, data, lr=lr, cbs=cbs, opt_func=opt_func)
sched = combine_scheds([0.3, 0.7], cos_1cycle_anneal(0.1, 0.3, 0.05))
learn, run = get_learn_run(nfs=nfs, data=data, lr=0.2, layer=conv_layer,
cbs=cbfs + [partial(ParamScheduler, 'lr', sched)])
```
- Additionally, we can take a closer look at our model by using Hooks to print the layers and the shapes of their outputs.
```
def model_summary(run, learn, data, find_all=False):
xb, yb = get_batch(data.valid_dl, run)
#
device = next(learn.model.parameters()).device
xb, yb = xb.to(device), yb.to(device)
mods = find_modules(learn.model, is_lin_layer) if find_all else learn.model.children()
f = lambda hook, mod, inp, out: print(f"{mod}\n{out.shape}\n")
with Hooks(mods, f) as hooks: learn.model(xb)
model_summary(run, learn, data)
# Training model
%time run.fit(6, learn)
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
%matplotlib inline
from exp.nb_07a import *
# Importing fastai's custom Imagenet dataset, called "Imagenette".
# Bear in mind that Imagenette v2 now has a 70/30 split so values will
# differ from the course notebook.
datasets.URLs.IMAGENETTE_160
path = datasets.untar_data(datasets.URLs.IMAGENETTE_160)
path
# We will add the .ls method to the path as a monkey patch
import PIL, os, mimetypes
Path.ls = lambda x: list(x.iterdir())
# Checking if the changes took effect
path.ls()
# Looking inside our classes folders
(path/'val').ls()
# Looking inside the Tench class folder
path_tench = path/'val'/'n01440764'
# Assigning a single image
img_fn = path_tench.ls()[0]
img_fn
img = PIL.Image.open(img_fn)
img
# Plotting dimensions of the image
plt.imshow(img);
import numpy as np
img_arr = np.array(img)
img_arr.shape
image_extensions = set(k for k,v in mimetypes.types_map.items()\
if v.startswith('image/'))
# Here are the image type extensions we will be focusing on
' '.join(image_extensions)
# Convert items first to a list and then a set
def setify(o):
return o if isinstance(o, set) else set(listify(o))
# Using the testing framework from previous nbs
test_eq(setify('aa'), {'aa'})
test_eq(setify(['aa',1]), {'aa',1})
test_eq(setify(None), set())
test_eq(setify(1), {1})
test_eq(setify({1}), {1})
# Run through a single directory and grab the images in that
def _get_files(p, fs, extensions=None):
p = Path(p)
res = [p/f for f in fs if not f.startswith('.')
and ((not extensions) or f'.{f.split(".")[-1].lower()}' in extensions)]
return res
t = [o.name for o in os.scandir(path_tench)]
t = _get_files(path, t, extensions=image_extensions)
t[0:10]
# Putting all of the above into one function to "get files"
def get_files(path, extensions=None, recurse=False, include=None):
path=Path(path)
extensions = setify(extensions)
extensions = {e.lower() for e in extensions}
if recurse:
res = []
for i,(p,d,f) in enumerate(os.walk(path)):
if include is not None and i==0: d[:] = [o for o in d if o in include]
else: d[:] = [o for o in d if not o.startswith('.')]
res += _get_files(p, f, extensions)
return res
else:
f = [o.name for o in os.scandir(path) if o.is_file()]
return _get_files(path, f, extensions)
# Testing the get_files function
get_files(path_tench, image_extensions)[:5]
get_files(path, image_extensions, recurse=True)[:5]
# Getting all of the file names
all_fns = get_files(path, image_extensions, recurse=True)
len(all_fns)
# Let's test the speed of the get_files function
# Bearing in mind that Imagenet is 100 times bigger than imagenette
%timeit -n 10 get_files(path, image_extensions, recurse=True)
def compose(x, funcs, *args, order_key='_order', **kwargs):
key = lambda o: getattr(o, order_key, 0)
for f in sorted(listify(funcs), key=key):
x = f(x, **kwargs)
return x
class ItemList(ListContainer): # Grab image files and dump them into the ListContainer
def __init__(self, items, path='.', tfms=None):
super().__init__(items)
self.path, self.tfms = Path(path), tfms
def __repr__(self):
return f'{super().__repr__()}\Path: {self.path}'
def new(self, items, cls=None):
if cls is None:
cls=self.__class__
return cls(items, self.path, tfms=self.tfms)
def get(self, i): #This default get method will be over-ridden
return i
def _get(self, i):
return compose(self.get(i), self.tfms) #refer to Wikipedia link on compose
def __getitem__(self, idx):
res = super().__getitem__(idx) #Index into our list, pass it to the container
if isinstance(res, list): return [self._get(o) for o in res] # Call if list
return self._get(res) #call if single item
class ImageList(ItemList):
@classmethod
def from_files(cls, path, extensions=None, recurse=True, include=None, **kwargs):
if extensions is None:
extensions = image_extensions
return cls(get_files(path, extensions, recurse=recurse, include=include), path, **kwargs)
def get(self, fn):
return PIL.Image.open(fn)
class Transform():
_order=0
# Both the class and the function do the same thing.
# Obviously, the class can be extended.
class MakeRGB(Transform):
def __call__(self, item): return item.convert('RGB')
def make_rgb(item):
return item.convert('RGB')
il = ImageList.from_files(path, tfms=make_rgb)
# __repr__ gives us following print outputs
il
# Indexing into the image list
img = il[1]
img
il[:2] # We can also index with a range of integers
fn = il.items[0]; fn
fn.parent.parent.name
def grandparent_splitter(fn, valid_name='valid', train_name='train'):
gp = fn.parent.parent.name
return True if gp==valid_name else False if gp==train_name else None
def split_by_func(items, f):
mask = [f(o) for o in items]
# None values are filtered out
f = [o for o, m in zip(items, mask) if m==False]
t = [o for o, m in zip(items, mask) if m==True]
return f, t
splitter = partial(grandparent_splitter, valid_name='val')
%time train, valid = split_by_func(il, splitter)
len(train), len(valid)
class SplitData():
def __init__(self, train, valid):
self.train, self.valid = train, valid
def __getattr__(self, k):
return getattr(self.train, k)
# Needed if SplitData needs to be pickled and loaded back without recursion errors
def __setstate__(self, data:Any):
self.__dict__.update(data)
@classmethod
def split_by_func(cls, il, f):
#il.new is defined in class ItemList
#identifies an object and it's class / sub-class and creates a new
#item list with the same type, path, tfms
lists = map(il.new, split_by_func(il.items, f))
return cls(*lists)
def __repr__(self):
return f'{self.__class__.__name__}\nTrain: {self.train}\nValid:{self.valid}\n'
sd = SplitData.split_by_func(il, splitter)
sd
from collections import OrderedDict
def uniqueify(x, sort=False):
res = list(OrderedDict.fromkeys(x).keys())
if sort:
res.sort()
return res
# The first one is needed by the model for training
# The second is better for displaying objects.
class Processor():
def process(self, items): return items
class CategoryProcessor(Processor):# Creates list of all possible categories
def __init__(self): self.vocab=None
def __call__(self, items):
#The vocab is defined on the first use.
#defines the k, v which goes from object to int (otoi)
if self.vocab is None:
self.vocab = uniqueify(items)
self.otoi = {v:k for k,v in enumerate(self.vocab)}
return [self.proc1(o) for o in items]
def proc1(self, item): return self.otoi[item]
def deprocess(self, idxs): # Print out the inferences
assert self.vocab is not None
return [self.deproc1(idx) for idx in idxs] # deprocess for each index
def deproc1(self, idx): return self.vocab[idx]
# For processing we need the grand parent and for labelling we need the parent
def parent_labeler(fn): return fn.parent.name
def _label_by_func(ds, f, cls=ItemList): return cls([f(o) for o in ds.items], path=ds.path)
class LabeledData():
def process(self, il, proc): return il.new(compose(il.items, proc))
def __init__(self, x, y, proc_x=None, proc_y=None):
self.x,self.y = self.process(x, proc_x),self.process(y, proc_y)
self.proc_x,self.proc_y = proc_x,proc_y
def __repr__(self): return f'{self.__class__.__name__}\nx: {self.x}\ny: {self.y}\n'
def __getitem__(self,idx): return self.x[idx],self.y[idx]
def __len__(self): return len(self.x)
def x_obj(self, idx): return self.obj(self.x, idx, self.proc_x)
def y_obj(self, idx): return self.obj(self.y, idx, self.proc_y)
def obj(self, items, idx, procs):
isint = isinstance(idx, int) or (isinstance(idx,torch.LongTensor) and not idx.ndim)
item = items[idx]
for proc in reversed(listify(procs)):
item = proc.deproc1(item) if isint else proc.deprocess(item)
return item
# Added convenience, can read up more about them
@classmethod
def label_by_func(cls, il, f, proc_x=None, proc_y=None):
return cls(il, _label_by_func(il, f), proc_x=proc_x, proc_y=proc_y)
# This is a very important piece of the...pie??
# when we pass in train, the processor has no vocab. It then goes CategoryProcessor to
# create a list of unique possibilities. When it goes to valid, then proc will have a vocab
# so it will skip the step and use the training set's vocab.
# Here we will ENSURE that the mapping is similar for both training and validation sets.
def label_by_func(sd, f, proc_x=None, proc_y=None):
train = LabeledData.label_by_func(sd.train, f, proc_x=proc_x, proc_y=proc_y)
valid = LabeledData.label_by_func(sd.valid, f, proc_x=proc_x, proc_y=proc_y)
return SplitData(train,valid)
ll = label_by_func(sd, parent_labeler, proc_y=CategoryProcessor())
# Validating
assert ll.train.proc_y is ll.valid.proc_y
# So now we have the labeled lists
ll
# Delving deeper
ll.train
# And deeper
ll.train.y
# .....deeper still
ll.train.y.items[0], ll.train.y_obj(0), ll.train.y_obj(slice(4))
# Getting the fist item, or x, of the tuple
ll.train[0]
# Now the second item
ll.train[0][0]
# Let's resize the image to 128x128
ll.train[0][0].resize((128,128))
class ResizeFixed(Transform): # Carry out Bilinear resizing
_order=10
def __init__(self, size):
if isinstance(size, int):
size=(size, size)
self.size = size
def __call__(self, item):
return item.resize(self.size, PIL.Image.BILINEAR)
def to_byte_tensor(item):
res = torch.ByteTensor(torch.ByteStorage.from_buffer(item.tobytes()))
w, h = item.size
return res.view(h, w, -1).permute(2, 0, 1) #Pillow and PyTorch handle permute differently
# this can be seen in the cell below
# Here we can giving to_byte_tensor and to_float_tensor "class level state"
# i.e. we can attach a state to a function, which is underused in Python but is
# very useful
to_byte_tensor._order=20
def to_float_tensor(item):
return item.float().div_(255.) # we divide by 255 to ensure the value is between 0 and 1
to_float_tensor._order=30
# Passing the list of tfms
tfms = [make_rgb, ResizeFixed(128), to_byte_tensor, to_float_tensor]
il = ImageList.from_files(path, tfms=tfms)
sd = SplitData.split_by_func(il, splitter)
ll = label_by_func(sd, parent_labeler, proc_y=CategoryProcessor())
def show_image(im, figsize=(3,3)):
plt.figure(figsize=figsize)
plt.axis('off')
plt.imshow(im.permute(1,2,0)) # Here the channel comes in last
x, y = ll.train[0]
x.shape
show_image(x)
bs = 128
train_dl, valid_dl = get_dls(ll.train, ll.valid, bs, num_workers=4)
x, y = next(iter(train_dl))
x.shape
# Images can be viewed in the batch, plus their corresponding classes
show_image(x[0])
ll.train.proc_y.vocab[y[0]]
y
class DataBunch():
def __init__(self, train_dl, valid_dl, c_in=None, c_out=None):
self.train_dl, self.valid_dl, self.c_in, self.c_out = train_dl, valid_dl, c_in, c_out
@property
def train_ds(self): return self.train_dl.dataset
@property
def valid_ds(self): return self.valid_dl.dataset
# Defining a function that goes directly from SplitData to a DataBunch
def databunchify(sd, bs, c_in=None, c_out=None, **kwargs):
dls = get_dls(sd.train, sd.valid, bs, **kwargs)
return DataBunch(*dls, c_in=c_in, c_out=c_out)
SplitData.to_databunch = databunchify # This is another example of monkey patching
# on the fly.
# Running through all we have worked on so far
path = datasets.untar_data(datasets.URLs.IMAGENETTE_160)
tfms = [make_rgb, ResizeFixed(128), to_byte_tensor, to_float_tensor]
il = ImageList.from_files(path, tfms=tfms)
sd = SplitData.split_by_func(il, partial(grandparent_splitter, valid_name='val'))
ll = label_by_func(sd, parent_labeler, proc_y=CategoryProcessor())
data = ll.to_databunch(bs, c_in=3, c_out=10, num_workers=4)
cbfs = [partial(AvgStatsCallback, accuracy),
CudaCallback]
m, s = x.mean((0,2,3)).cuda(), x.std((0,2,3)).cuda()
m, s
# Defining a function to normalize over three channels by using
# broadcasting
def normalize_chan(x, mean, std):
return (x - mean[..., None, None]) / std[..., None, None]
_m = tensor([0.47, 0.48, 0.45])
_s = tensor([0.29, 0.28, 0.30])
norm_imagenette = partial(normalize_chan, mean=_m.cuda(), std=_s.cuda())
# Choosing our callback functions
cbfs.append(partial(BatchTransformXCallback, norm_imagenette))
nfs = [64, 64, 128, 256]
import math
def prev_pow_2(x):
return 2**math.floor(math.log2(x))
def get_cnn_layers(data, nfs, layer, **kwargs):
def f(ni, nf, stride=2):
return layer(ni, nf, 3, stride=stride, **kwargs)
l1 = data.c_in # layer 1 of the model
l2 = prev_pow_2(l1*3*3) # taking value of c_in and multiplying with next number which is pow^2
layers = [f(l1, l2, stride=1),
f(l2, l2*2, stride=2),
f(l2*2, l2*4, stride=2)]
nfs = [l2*4] + nfs
layers += [f(nfs[i], nfs[i+1]) for i in range(len(nfs)-1)]
layers += [nn.AdaptiveAvgPool2d(1), Lambda(flatten),
nn.Linear(nfs[-1], data.c_out)]
return layers
def get_cnn_model(data, nfs, layer, **kwargs):
return nn.Sequential(*get_cnn_layers(data, nfs, layer, **kwargs))
def get_learn_run(nfs, data, lr, layer, cbs=None, opt_func=None, **kwargs):
model = get_cnn_model(data, nfs, layer, **kwargs)
init_cnn(model)
return get_runner(model, data, lr=lr, cbs=cbs, opt_func=opt_func)
sched = combine_scheds([0.3, 0.7], cos_1cycle_anneal(0.1, 0.3, 0.05))
learn, run = get_learn_run(nfs=nfs, data=data, lr=0.2, layer=conv_layer,
cbs=cbfs + [partial(ParamScheduler, 'lr', sched)])
def model_summary(run, learn, data, find_all=False):
xb, yb = get_batch(data.valid_dl, run)
#
device = next(learn.model.parameters()).device
xb, yb = xb.to(device), yb.to(device)
mods = find_modules(learn.model, is_lin_layer) if find_all else learn.model.children()
f = lambda hook, mod, inp, out: print(f"{mod}\n{out.shape}\n")
with Hooks(mods, f) as hooks: learn.model(xb)
model_summary(run, learn, data)
# Training model
%time run.fit(6, learn)
| 0.66454 | 0.906031 |
## Master of Applied Data Science
### University of Michigan - School of Information
### Capstone Project - Rapid Labeling of Text Corpus Using Information Retrieval Techniques
### Fall 2021
#### Team Members: Chloe Zhang, Michael Penrose, Carlo Tak
### Experiment Flow
Class label > Count vectorizer > 100 features > PyCaret
### Purpose
This notebook investigates how well a classifier can predict the **event type (i.e. 'earthquake', 'fire', 'flood', 'hurricane)** of the Tweets in the [Disaster tweets dataset](https://crisisnlp.qcri.org/humaid_dataset.html#).
This classifier is to be used as a baseline of classification performance. Two things are investigated:
- Is it possible to build a reasonable 'good' classifier of these tweets at all
- If it is possible to build a classifier how well does the classifier perform using all of the labels from the training data
If it is possible to build a classifier using all of the labels in the training dataset then it should be possible to implement a method for rapidly labeling the corpus of texts in the dataset. Here we think of rapid labeling as any process that does not require the user to label each text in the corpus, one at a time.
To measure the performance of the classifier we use a metric called the Area Under the Curve (AUC). This metric was used because we believe it is a good metric for the preliminary work in this project. If a specific goal emerges later that requires a different metric, then the appropriate metric can be used at that time. The consequence of false positives (texts classified as having a certain label, but are not that label) and false negatives should be considered. For example, a metric like precision can be used to minimize false positives. The AUC metric provides a value between zero and one, with a higher number indicating better classification performance.
### Summary
The baseline classifier built using all the labels in the training dataset produced a classifier that had a fairly good AUC score for each of the 4 event type labels (i.e. earthquake, fire, flood, hurricane). All the AUC scores were above 0.98.
A simple vectorization (of texts) approach was implemented because we wanted the baseline classifier to be a basic solution – our feeling was that more complex techniques could be implemented at a later stage. A [count vectorizer]( https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.CountVectorizer.html) (with default settings) was used to convert the texts. The number of dimensions (features) was also reduced using feature selection ([SelectKBest]( https://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.SelectKBest.html)). This was to improve computational times – fewer dimensions means that there are fewer data to process. Also, this was a simpler method to implement than other techniques like removing stopwords, adjusting parameters like ‘stop_words’, ‘ngram_range’, ‘max_df’, ‘min_df’, and ‘max_features’. The complexity of the classifier could be adjusted if required, but this simple implementation produced good results.
This notebook reduced the number of features to 100.
The feature importances were extracted from the classifier, to see if they made sense. This sense check was important because we made several assumptions in building this classifier, that had to be validated. For example, when the text was vectorized we used a simple approach that just counted the individual words (tokens) – are more complex classifier might use bi-grams (two words per feature), this would have had the advantage of preserving features like ‘’.
Examining the top features
```
# ! pip freeze > requirements.txt
from utilities import dt_utilities as utils
from datetime import datetime
start_time = datetime.now()
start_time.strftime("%Y/%m/%d %H:%M:%S")
consolidated_disaster_tweet_data_df = \
utils.get_consolidated_disaster_tweet_data(root_directory="data/",
event_type_directory="HumAID_data_event_type",
events_set_directories=["HumAID_data_events_set1_47K",
"HumAID_data_events_set2_29K"],
include_meta_data=True)
consolidated_disaster_tweet_data_df
train_df = consolidated_disaster_tweet_data_df[consolidated_disaster_tweet_data_df["data_type"]=="train"].reset_index(drop=True)
train_df
test_df = consolidated_disaster_tweet_data_df[consolidated_disaster_tweet_data_df["data_type"]=="test"].reset_index(drop=True)
test_df
dev_df = consolidated_disaster_tweet_data_df[consolidated_disaster_tweet_data_df["data_type"]=="dev"].reset_index(drop=True)
dev_df
train_df.groupby(["event_type"]).size().reset_index().rename(columns={0: "Count"}).sort_values("Count", ascending=False)
train_df.groupby(["class_label"]).size().reset_index().rename(columns={0: "Count"}).sort_values("Count", ascending=False)
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.feature_selection import SelectKBest, chi2, f_classif
from sklearn.pipeline import Pipeline
import pandas as pd
from scipy.sparse import coo_matrix, hstack
import scipy.sparse
import numpy as np
from collections import Counter
num_features = 100
target_column = "class_label"
# vectorizer = TfidfVectorizer(max_features=num_features)
# count_vectorizer = CountVectorizer(max_features=num_features)
vectorizer = Pipeline([
("vectorizer", CountVectorizer()),
("reduce", SelectKBest(score_func=f_classif, k=num_features)), # chi2, f_classif
])
vectorizer.fit(train_df["tweet_text"], train_df[target_column])
def vectorized_tweet_data(fitted_vectorizer, source_df, text_column, target_column,
vectorizer_name="vectorizer", reducer_name="reduce"):
vectorized_data = fitted_vectorizer.transform(source_df[text_column])
vectorized_df = pd.DataFrame.sparse.from_spmatrix(vectorized_data)
all_feature_names = fitted_vectorizer.named_steps[vectorizer_name].get_feature_names()
support = vectorizer.named_steps[reducer_name].get_support()
feature_names = np.array(all_feature_names)[support]
vectorized_df.columns = feature_names
vectorized_df = vectorized_df.sparse.to_dense()
# vectorized_df = vectorized_df.apply(pd.to_numeric)
vectorized_df = vectorized_df.astype(float)
vectorized_df["tweet_id"] = source_df["tweet_id"]
vectorized_df["tweet_text"] = source_df["tweet_text"]
vectorized_df[target_column] = source_df[target_column]
return vectorized_df
train_vectorized_event_type_df = vectorized_tweet_data(fitted_vectorizer=vectorizer,
source_df=train_df,
text_column="tweet_text",
target_column=target_column,
vectorizer_name="vectorizer",
reducer_name="reduce")
train_vectorized_event_type_df
test_vectorized_event_type_df = vectorized_tweet_data(fitted_vectorizer=vectorizer,
source_df=test_df,
text_column="tweet_text",
target_column=target_column)
test_vectorized_event_type_df
dev_vectorized_event_type_df = vectorized_tweet_data(fitted_vectorizer=vectorizer,
source_df=dev_df,
text_column="tweet_text",
target_column=target_column)
dev_vectorized_event_type_df
import pycaret.classification as pc_class
RND_SEED = 39674
N_JOBS = 2
include_models = ["nb", "lr", "gbc", "lightgbm"] # , "xgboost"
exclude_models = ["knn", "svm", "ridge"]
exp_00 = pc_class.setup(train_vectorized_event_type_df,
# numeric_features=numeric_features_adj,
# categorical_features=categorical_features,
silent=True,
verbose=False,
ignore_features=["tweet_id", "tweet_text"],
target=target_column, # "event_type", # "class_label"
session_id=RND_SEED,
n_jobs=N_JOBS)
best_model = pc_class.compare_models(sort="AUC",
# include=include_models,
exclude=exclude_models,
turbo=True
)
best_model
# best_model = pc_class.created_model("nb")
# best_model = pc_class.created_model("lightgbm")
# best_model = pc_class.created_model("lr")
finalized_model = pc_class.finalize_model(best_model)
y_train = pc_class.get_config("y_train")
y_train
y = pc_class.get_config("y")
y
original_labels = train_df[target_column]
original_labels
Counter(original_labels)
labels_map = dict(zip(y, original_labels))
labels_map
try:
pc_class.plot_model(finalized_model, "auc")
except:
print(f"Could not plot model.")
try:
pc_class.plot_model(finalized_model, "learning")
except:
print(f"Could not plot model.")
try:
pc_class.plot_model(finalized_model, "confusion_matrix")
except:
print(f"Could not plot model.")
try:
pc_class.plot_model(finalized_model, "feature")
except:
print(f"Could not plot model.")
predictions_train = pc_class.predict_model(finalized_model)
predictions_train
test_vectorized_event_type_df
predictions_test = pc_class.predict_model(finalized_model, data=test_vectorized_event_type_df)
predictions_test
end_time = datetime.now()
end_time.strftime("%Y/%m/%d %H:%M:%S")
duration = end_time - start_time
print("duration :", duration)
```
|
github_jupyter
|
# ! pip freeze > requirements.txt
from utilities import dt_utilities as utils
from datetime import datetime
start_time = datetime.now()
start_time.strftime("%Y/%m/%d %H:%M:%S")
consolidated_disaster_tweet_data_df = \
utils.get_consolidated_disaster_tweet_data(root_directory="data/",
event_type_directory="HumAID_data_event_type",
events_set_directories=["HumAID_data_events_set1_47K",
"HumAID_data_events_set2_29K"],
include_meta_data=True)
consolidated_disaster_tweet_data_df
train_df = consolidated_disaster_tweet_data_df[consolidated_disaster_tweet_data_df["data_type"]=="train"].reset_index(drop=True)
train_df
test_df = consolidated_disaster_tweet_data_df[consolidated_disaster_tweet_data_df["data_type"]=="test"].reset_index(drop=True)
test_df
dev_df = consolidated_disaster_tweet_data_df[consolidated_disaster_tweet_data_df["data_type"]=="dev"].reset_index(drop=True)
dev_df
train_df.groupby(["event_type"]).size().reset_index().rename(columns={0: "Count"}).sort_values("Count", ascending=False)
train_df.groupby(["class_label"]).size().reset_index().rename(columns={0: "Count"}).sort_values("Count", ascending=False)
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.feature_selection import SelectKBest, chi2, f_classif
from sklearn.pipeline import Pipeline
import pandas as pd
from scipy.sparse import coo_matrix, hstack
import scipy.sparse
import numpy as np
from collections import Counter
num_features = 100
target_column = "class_label"
# vectorizer = TfidfVectorizer(max_features=num_features)
# count_vectorizer = CountVectorizer(max_features=num_features)
vectorizer = Pipeline([
("vectorizer", CountVectorizer()),
("reduce", SelectKBest(score_func=f_classif, k=num_features)), # chi2, f_classif
])
vectorizer.fit(train_df["tweet_text"], train_df[target_column])
def vectorized_tweet_data(fitted_vectorizer, source_df, text_column, target_column,
vectorizer_name="vectorizer", reducer_name="reduce"):
vectorized_data = fitted_vectorizer.transform(source_df[text_column])
vectorized_df = pd.DataFrame.sparse.from_spmatrix(vectorized_data)
all_feature_names = fitted_vectorizer.named_steps[vectorizer_name].get_feature_names()
support = vectorizer.named_steps[reducer_name].get_support()
feature_names = np.array(all_feature_names)[support]
vectorized_df.columns = feature_names
vectorized_df = vectorized_df.sparse.to_dense()
# vectorized_df = vectorized_df.apply(pd.to_numeric)
vectorized_df = vectorized_df.astype(float)
vectorized_df["tweet_id"] = source_df["tweet_id"]
vectorized_df["tweet_text"] = source_df["tweet_text"]
vectorized_df[target_column] = source_df[target_column]
return vectorized_df
train_vectorized_event_type_df = vectorized_tweet_data(fitted_vectorizer=vectorizer,
source_df=train_df,
text_column="tweet_text",
target_column=target_column,
vectorizer_name="vectorizer",
reducer_name="reduce")
train_vectorized_event_type_df
test_vectorized_event_type_df = vectorized_tweet_data(fitted_vectorizer=vectorizer,
source_df=test_df,
text_column="tweet_text",
target_column=target_column)
test_vectorized_event_type_df
dev_vectorized_event_type_df = vectorized_tweet_data(fitted_vectorizer=vectorizer,
source_df=dev_df,
text_column="tweet_text",
target_column=target_column)
dev_vectorized_event_type_df
import pycaret.classification as pc_class
RND_SEED = 39674
N_JOBS = 2
include_models = ["nb", "lr", "gbc", "lightgbm"] # , "xgboost"
exclude_models = ["knn", "svm", "ridge"]
exp_00 = pc_class.setup(train_vectorized_event_type_df,
# numeric_features=numeric_features_adj,
# categorical_features=categorical_features,
silent=True,
verbose=False,
ignore_features=["tweet_id", "tweet_text"],
target=target_column, # "event_type", # "class_label"
session_id=RND_SEED,
n_jobs=N_JOBS)
best_model = pc_class.compare_models(sort="AUC",
# include=include_models,
exclude=exclude_models,
turbo=True
)
best_model
# best_model = pc_class.created_model("nb")
# best_model = pc_class.created_model("lightgbm")
# best_model = pc_class.created_model("lr")
finalized_model = pc_class.finalize_model(best_model)
y_train = pc_class.get_config("y_train")
y_train
y = pc_class.get_config("y")
y
original_labels = train_df[target_column]
original_labels
Counter(original_labels)
labels_map = dict(zip(y, original_labels))
labels_map
try:
pc_class.plot_model(finalized_model, "auc")
except:
print(f"Could not plot model.")
try:
pc_class.plot_model(finalized_model, "learning")
except:
print(f"Could not plot model.")
try:
pc_class.plot_model(finalized_model, "confusion_matrix")
except:
print(f"Could not plot model.")
try:
pc_class.plot_model(finalized_model, "feature")
except:
print(f"Could not plot model.")
predictions_train = pc_class.predict_model(finalized_model)
predictions_train
test_vectorized_event_type_df
predictions_test = pc_class.predict_model(finalized_model, data=test_vectorized_event_type_df)
predictions_test
end_time = datetime.now()
end_time.strftime("%Y/%m/%d %H:%M:%S")
duration = end_time - start_time
print("duration :", duration)
| 0.372277 | 0.962918 |
```
import pandas as pd
```
df = pd.read_csv("C:/Users/user/Desktop/New folder (2)/Breast_cancer_data.csv")
```
df.head()
```
# FORECASTING BREAST CANCER
```
df = pd.read_csv("C:/Users/user/Desktop/New folder (2)/Breast_cancer_data.csv")
df.head()
```
Breast cancer is cancer that forms in the cells of the breasts.
After skin cancer, breast cancer is the most common cancer diagnosed in women in the United States. Breast cancer can occur in both men and women, but it's far more common in women.
Breast cancer is cancer that forms in the cells of the breasts.
After skin cancer, breast cancer is the most common cancer diagnosed in women in the United States. Breast cancer can occur in both men and women, but it's far more common in women.
There are many different types of breast cancer and common ones include ductal carcinoma in situ (DCIS) and invasive carcinoma. Others, like phyllodes tumors and angiosarcoma are less common.
Factors that are associated with an increased risk of breast cancer include:
Being female. Women are much more likely than men are to develop breast cancer.
Increasing age. Your risk of breast cancer increases as you age.
A personal history of breast conditions. If you've had a breast biopsy that found lobular carcinoma in situ (LCIS) or atypical hyperplasia of the breast, you have an increased risk of breast cancer.
A personal history of breast cancer. If you've had breast cancer in one breast, you have an increased risk of developing cancer in the other breast.
A family history of breast cancer. If your mother, sister or daughter was diagnosed with breast cancer, particularly at a young age, your risk of breast cancer is increased. Still, the majority of people diagnosed with breast cancer have no family history of the disease.
Inherited genes that increase cancer risk. Certain gene mutations that increase the risk of breast cancer can be passed from parents to children. The most well-known gene mutations are referred to as BRCA1 and BRCA2. These genes can greatly increase your risk of breast cancer and other cancers, but they don't make cancer inevitable.
Radiation exposure. If you received radiation treatments to your chest as a child or young adult, your risk of breast cancer is increased.
Obesity. Being obese increases your risk of breast cancer.
Beginning your period at a younger age. Beginning your period before age 12 increases your risk of breast cancer.
Beginning menopause at an older age. If you began menopause at an older age, you're more likely to develop breast cancer.
Having your first child at an older age. Women who give birth to their first child after age 30 may have an increased risk of breast cancer.
Having never been pregnant. Women who have never been pregnant have a greater risk of breast cancer than do women who have had one or more pregnancies.
Postmenopausal hormone therapy. Women who take hormone therapy medications that combine estrogen and progesterone to treat the signs and symptoms of menopause have an increased risk of breast cancer. The risk of breast cancer decreases when women stop taking these medications.
Drinking alcohol. Drinking alcohol increases the risk of breast cancer.
Breast cancer is treated in several ways. It depends on the kind of breast cancer and how far it has spread. People with breast cancer often get more than one kind of treatment.
Surgery. An operation where doctors cut out cancer tissue.
Chemotherapy. Using special medicines to shrink or kill the cancer cells. The drugs can be pills you take or medicines given in your veins, or sometimes both.
Hormonal therapy. Blocks cancer cells from getting the hormones they need to grow.
Biological therapy. Works with your body’s immune system to help it fight cancer cells or to control side effects from other cancer treatments.
Radiation therapy. Using high-energy rays (similar to X-rays) to kill the cancer cells.
Number of increasing breast cancer clients are shown below 
Each year in the United States, about 250,000 cases of breast cancer are diagnosed in women and about 2,300 in men. About 42,000 women and 510 men in the U.S. die each year from breast cancer. Black women have a higher rate of death from breast cancer than White women.
Cancer is a disease in which cells in the body grow out of control. Except for skin cancer, breast cancer is the most common cancer in women in the United States. Deaths from breast cancer have declined over time, but remain the second leading cause of cancer death among women overall and the leading cause of cancer death among Hispanic women.
Many factors over the course of a lifetime can influence your breast cancer risk. You can’t change some factors, such as getting older or your family history, but you can help lower your risk of breast cancer by taking care of your health in the following ways—!
Keep a healthy weight.
Exercise regularly.
Don’t drink alcohol, or limit alcoholic drinks.
If you are taking, or have been told to take, hormone replacement therapyexternal icon or oral contraceptivesexternal icon (birth control pills), ask your doctor about the risks and find out if it is right for you.
Breastfeed your children, if possible.
If you have a family history of breast cancer or inherited changes in your BRCA1 and BRCA2 genes, talk to your doctor about other ways to lower your risk.
Getting regular exercise and keeping a healthy weight can help lower your breast cancer risk.
Staying healthy throughout your life will lower your risk of developing cancer, and improve your chances of surviving cancer if it occurs 
Mammogram
A mammogram is an X-ray of the breast. For many women, mammograms are the best way to find breast cancer early, when it is easier to treat and before it is big enough to feel or cause symptoms. Having regular mammograms can lower the risk of dying from breast cancer. At this time, a mammogram is the best way to find breast cancer for most women.
Clinical Breast Exam
A clinical breast exam is an examination by a doctor or nurse, who uses his or her hands to feel for lumps or other changes.
Benefits and Risks of Screening
Every screening test has benefits and risks, which is why it’s important to talk to your doctor before getting any screening test, like a mammogram.
Conclusion: Breast canacer is increasing worldwide it can have bad outcomes and doctors are finding a way how to treat this deasease.In general, there are five treatment options, and most treatment plans include a combination of the following: surgery, radiation, hormone therapy, chemotherapy, and targeted therapies. Some are local, targeting just the area around the tumor. Others are systemic, targeting your whole body with cancer fighting agents.
|
github_jupyter
|
import pandas as pd
df.head()
df = pd.read_csv("C:/Users/user/Desktop/New folder (2)/Breast_cancer_data.csv")
df.head()
| 0.086715 | 0.913368 |
# Hello and Good Morning!
# My name is Rich!
# I heard that some of you are Roblox and Fortnite fans, so I thought I would start with a little story.
<img src="./images/gamer.png" alt="Drawing" style="width: 600px;"/>
```
# Intro
# Import required libraries
from rich.console import Console
from rich.panel import Panel
# Setup console output and format
console = Console()
style = 'blue'
# Screen output
console.print(Panel('I am really excited to use code to talk to you about coding!',title='Excited',style=style))
console.print(Panel('Thank you SO MUCH for the invite.',title='Thank You',style=style))
# Our talk as code
# Import required libraries
import requests
from rich.console import Console
from rich.panel import Panel
from IPython import display
# Setup console output and format
console = Console()
style = 'blue'
# Screen output
console.print(Panel('I know you are familiar with Python. It\'s a great language, and the language I use most often. So I thought it would be fun to do this presentation as code.',title='Interactive Presentation As Code',style=style))
display.Image("./images/ilovepy.png", width=400)
# About me
# Import required libraries
from IPython import display
from rich.console import Console
from rich.panel import Panel
# Setup console output and format
console = Console()
style = 'blue'
# Screen output
console.print(Panel('From Left to Right: Me, Eden (she is 10 now and a 5th grader just like you), Myrtle (the cow), Maddy (15 now), and Gwen (my wife)',title='About Me',style=style))
display.Image(filename='./images/family-pic.jpg', width=600)
# Let's see how many questions you had
# Import required libraries
import yaml
from rich.console import Console
from rich.panel import Panel
# Setup console output and format
console = Console()
style = "blue"
# Screen output
console.print(Panel('Ms. Carrick was nice enough to share your questions with me. Thank you Ms. Carrick!',title='Questions',style=style))
# read the file with the questions into a variable (dictionary) call "q"
f = open('questions.yaml')
q = yaml.safe_load(f)
f.close()
# count the number of questions in "q" and print answer to screen
count = 0
for x in q:
if isinstance(q[x], list):
count += len(q[x])
console.print(Panel('There are ' + str(count) + ' questions. Wow!',title='I wonder how many questions you have?',style=style))
# Let's answer some of your questions
# Import required libraries
import yaml
import IPython
from rich.console import Console
from rich.panel import Panel
from IPython.display import Image
from IPython.display import display
# Setup console output and format
console = Console()
q_style = 'bold red'
a_style = 'bold blue'
# Read the YAML file with the Q&A data
f = open('questions-kv.yaml')
q = yaml.safe_load(f)
f.close()
# Function to display image
def func_img(f):
img = Image(filename=f, width=480)
display(img)
# Function to display answers to questions
def func_qa(qa):
console.print (qa[0], style=q_style)
console.print (*qa[1], sep="\n\n", style=a_style)
# Logic to loop through question and get my answers
for key in q:
qa = (key, q[key])
if qa[0] == 'What is the most fun about your job?':
func_qa(qa)
func_img('./images/etchasketch.png')
elif qa[0] == 'How many years have you been coding?':
func_qa(qa)
func_img('./images/me.jpg')
elif qa[0] == 'How did you start liking coding?':
func_qa(qa)
func_img('./images/c64.jpg')
elif qa[0] == 'How did you pursue coding?':
func_qa(qa)
func_img('./images/curious.png')
elif qa[0] == 'Have you always wanted to be a coder?':
func_qa(qa)
func_img('./images/maker.jpg')
elif qa[0] == 'What kind of coding do you do for your job?':
func_qa(qa)
func_img('./images/dc.jpg')
elif qa[0] == 'What schooling did you get for your job?':
func_qa(qa)
func_img('./images/philospher.jpg')
elif qa[0] == 'At your place of work, how much do coders make?':
func_qa(qa)
func_img('./images/vg.jpg')
elif qa[0] == 'What has been your greatest coding challenge?':
func_qa(qa)
func_img('./images/fear.jpg')
elif qa[0] == 'Do you ever get bored of coding?':
func_qa(qa)
func_img('./images/coders-life.jpg')
elif qa[0] == 'From 1-10, how do you rate coding?':
func_qa(qa)
func_img('./images/codersgonnacode.jpg')
elif qa[0] == 'Have you ever made any games?':
func_qa(qa)
func_img('./images/pong.jpg')
else:
console.print(Panel('Not a valid question.',title='ERROR',style='red'))
func_img('./images/error.jpg')
console.input('Press enter for next question.')
IPython.display.clear_output()
# The end
console.print(Panel('I can answer any question you have.',title='The End',style='red'))
console.print(Panel('https://github.com/rbocchinfuso/MyJourney',title='This Presenation',style='blue'))
console.print(':mega: My Contatct Info :mega:')
console.print('Rich Bocchinfuso :person_with_blond_hair: ')
console.print('rbocchinfuso@gmail.com :e-mail:')
console.print('http://bocchinfuso.net :page_with_curl: ')
console.print('@rbocchinfuso')
func_img('./images/ty.jpg')
```
|
github_jupyter
|
# Intro
# Import required libraries
from rich.console import Console
from rich.panel import Panel
# Setup console output and format
console = Console()
style = 'blue'
# Screen output
console.print(Panel('I am really excited to use code to talk to you about coding!',title='Excited',style=style))
console.print(Panel('Thank you SO MUCH for the invite.',title='Thank You',style=style))
# Our talk as code
# Import required libraries
import requests
from rich.console import Console
from rich.panel import Panel
from IPython import display
# Setup console output and format
console = Console()
style = 'blue'
# Screen output
console.print(Panel('I know you are familiar with Python. It\'s a great language, and the language I use most often. So I thought it would be fun to do this presentation as code.',title='Interactive Presentation As Code',style=style))
display.Image("./images/ilovepy.png", width=400)
# About me
# Import required libraries
from IPython import display
from rich.console import Console
from rich.panel import Panel
# Setup console output and format
console = Console()
style = 'blue'
# Screen output
console.print(Panel('From Left to Right: Me, Eden (she is 10 now and a 5th grader just like you), Myrtle (the cow), Maddy (15 now), and Gwen (my wife)',title='About Me',style=style))
display.Image(filename='./images/family-pic.jpg', width=600)
# Let's see how many questions you had
# Import required libraries
import yaml
from rich.console import Console
from rich.panel import Panel
# Setup console output and format
console = Console()
style = "blue"
# Screen output
console.print(Panel('Ms. Carrick was nice enough to share your questions with me. Thank you Ms. Carrick!',title='Questions',style=style))
# read the file with the questions into a variable (dictionary) call "q"
f = open('questions.yaml')
q = yaml.safe_load(f)
f.close()
# count the number of questions in "q" and print answer to screen
count = 0
for x in q:
if isinstance(q[x], list):
count += len(q[x])
console.print(Panel('There are ' + str(count) + ' questions. Wow!',title='I wonder how many questions you have?',style=style))
# Let's answer some of your questions
# Import required libraries
import yaml
import IPython
from rich.console import Console
from rich.panel import Panel
from IPython.display import Image
from IPython.display import display
# Setup console output and format
console = Console()
q_style = 'bold red'
a_style = 'bold blue'
# Read the YAML file with the Q&A data
f = open('questions-kv.yaml')
q = yaml.safe_load(f)
f.close()
# Function to display image
def func_img(f):
img = Image(filename=f, width=480)
display(img)
# Function to display answers to questions
def func_qa(qa):
console.print (qa[0], style=q_style)
console.print (*qa[1], sep="\n\n", style=a_style)
# Logic to loop through question and get my answers
for key in q:
qa = (key, q[key])
if qa[0] == 'What is the most fun about your job?':
func_qa(qa)
func_img('./images/etchasketch.png')
elif qa[0] == 'How many years have you been coding?':
func_qa(qa)
func_img('./images/me.jpg')
elif qa[0] == 'How did you start liking coding?':
func_qa(qa)
func_img('./images/c64.jpg')
elif qa[0] == 'How did you pursue coding?':
func_qa(qa)
func_img('./images/curious.png')
elif qa[0] == 'Have you always wanted to be a coder?':
func_qa(qa)
func_img('./images/maker.jpg')
elif qa[0] == 'What kind of coding do you do for your job?':
func_qa(qa)
func_img('./images/dc.jpg')
elif qa[0] == 'What schooling did you get for your job?':
func_qa(qa)
func_img('./images/philospher.jpg')
elif qa[0] == 'At your place of work, how much do coders make?':
func_qa(qa)
func_img('./images/vg.jpg')
elif qa[0] == 'What has been your greatest coding challenge?':
func_qa(qa)
func_img('./images/fear.jpg')
elif qa[0] == 'Do you ever get bored of coding?':
func_qa(qa)
func_img('./images/coders-life.jpg')
elif qa[0] == 'From 1-10, how do you rate coding?':
func_qa(qa)
func_img('./images/codersgonnacode.jpg')
elif qa[0] == 'Have you ever made any games?':
func_qa(qa)
func_img('./images/pong.jpg')
else:
console.print(Panel('Not a valid question.',title='ERROR',style='red'))
func_img('./images/error.jpg')
console.input('Press enter for next question.')
IPython.display.clear_output()
# The end
console.print(Panel('I can answer any question you have.',title='The End',style='red'))
console.print(Panel('https://github.com/rbocchinfuso/MyJourney',title='This Presenation',style='blue'))
console.print(':mega: My Contatct Info :mega:')
console.print('Rich Bocchinfuso :person_with_blond_hair: ')
console.print('rbocchinfuso@gmail.com :e-mail:')
console.print('http://bocchinfuso.net :page_with_curl: ')
console.print('@rbocchinfuso')
func_img('./images/ty.jpg')
| 0.175361 | 0.536009 |
### Lemma 1: Initialization Price $P_i$ is equal to state variable Price $P_0$ at $t=0$
Let M refer to the total amount of money raised by the bond during the Initialization Phase. This money is divided such that a portion of the funds F is used to directly fund the project, and the remaining amount R is injected into the bonding curve reserve. This gives $$M = R + F$$<br> Since the amount of funds in the bonding curve at time $t=0$ is proportional to the reserve ratio $\rho_0$ at $t=0$, $R_0$ is $$R_0 = \rho_0 M$$<br> Which gives $F_0$ that goes to directly fund the project $$F_0=(1-\rho_0)M$$<br> Initialization Price $P_i$ can be defined as the amount of supply tokens $S$ that can be obtained per unit $M$ $$P_i = \frac{M}{S_0}$$<br> This gives the supply during Initialization $S_0$ $$S_0 = \frac{M}{P_i}$$<br> The amount of funds directed towards the project $F_0$ is directly and linearly proportional to the bond's initial likelihood of success. From this, we can infer $$F_0=\alpha_0M$$<br> We know that $$F_0=(1-\rho_0)M$$<br> From the above equations, we get $$\alpha_0 = 1-\rho_0$$<br> By definition, the state variable Price $P_0$ at $t=0$ is given by $$P_0=\kappa_0\frac{R_0}{S_0}$$<br> where $\kappa_0=\frac{1}{\rho_0}$. Expanding for $R_0=\rho_M$ and $S_0=M/P_i$, $$P_0 = \frac{1}{\rho_0} \frac{\rho_0M}{{M}/{P_i}}$$<br> Terms cancel out and we get $$P_0 = P_i$$<br> *Q.E.D*
### Lemma 2: Outcome Indifference
Let the true total payout at the Settlement Phase be $\Theta_{true}$. We know that total payout is contingent on the $(C, \Omega)$ pair that was set during Initialization and the amount of tokens bonded to the reserve. This gives $$\Theta_{true} = C \Omega + R$$<br> Neither the bonding curve system nor the participant agents have knowledge about the true total payout prior to the Settlement Phase, so they base their decisions on their own expectations of the total payout.
#### 1. System's Expectation of Total Payout
During Execution, the system does not have knowledge of $\Omega$ but is aware of the state variable $\alpha$, which is based on Attestations on the value of $\Omega$. This gives $$\mathbb{E}(\Theta_{sys}) = C\alpha + R$$<br> This expectation on $\Theta_{sys}$ is a **system-level constraint**.
#### 2. Agent's Expectation of Individual Payout
Let $\Theta_0 = M$ be the amount of money spent by agent $i$ to purchase $S_i$ amount of tokens during Initialization. From the definition of Price, we have $$\Theta_{0}=PS_0$$ Expanding Price $P$ $$\Theta_{0}=\frac{1}{\rho}\frac{R}{S}S_0$$<br> $$\Theta_{0}=\frac{1}{\rho}R\left(\frac{S_0}{S}\right)$$<br> This gives $\Theta_{i}$, the amount of tokens to be paid out to the agent $$\Theta_{i} = \frac{\Theta_0}{\Theta} = \frac{1}{\rho}\frac{R}{\Theta}\left(\frac{S_0}{S}\right)$$<br> This imposes an **agent-level constraint**.
Since outcome payout is conserved throughout the duration of the bond, we have $\frac{\Theta_{0}}{S_0} =\frac{\Theta}{S}$. Rearranging, $$\frac{\Theta_{0}}{S_0}=\frac{\Theta}{S}$$<br> For this equivalency to hold, in the definition for $\Theta_i$, we require that $$\frac{1}{\rho}\frac{R}{\Theta} = 1$$<br> Rearranging to obtain $R$, $$R=\rho\Theta$$<br> Since $\Theta_{true}$ is unknown, the system and its agents need to estimate it. Plugging the above in $\Theta_{true}$, we have $$\Theta_{true}=C\Omega + \rho\Theta_{true}$$ Simplifying, $$\Theta_{true}=\frac{C\Omega}{1-\rho}$$<br> From here, we derive the system's estimate of total payout and the agents' estimate of individual payout.
#### 3. System's estimate of Total Payout
From (1) we have $$\mathbb{E}(\Theta_{sys}) = C\alpha + R$$<br> Due to the analogy between $\Theta_{sys}$ and $\Theta_{true}$, we can conclude $$\mathbb{E}(\Theta_{sys})= \frac{C\alpha}{1-\rho}$$<br>
#### 4. Agents' estimate of Individual Payout
Following from the above, we obtain the agent's estimate of an individual agent's payout $$\mathbb{E}(\Theta_{i})= \frac{C\hat\alpha_i}{1-\rho}\frac{S_i}{S}$$<br>
|
github_jupyter
|
### Lemma 1: Initialization Price $P_i$ is equal to state variable Price $P_0$ at $t=0$
Let M refer to the total amount of money raised by the bond during the Initialization Phase. This money is divided such that a portion of the funds F is used to directly fund the project, and the remaining amount R is injected into the bonding curve reserve. This gives $$M = R + F$$<br> Since the amount of funds in the bonding curve at time $t=0$ is proportional to the reserve ratio $\rho_0$ at $t=0$, $R_0$ is $$R_0 = \rho_0 M$$<br> Which gives $F_0$ that goes to directly fund the project $$F_0=(1-\rho_0)M$$<br> Initialization Price $P_i$ can be defined as the amount of supply tokens $S$ that can be obtained per unit $M$ $$P_i = \frac{M}{S_0}$$<br> This gives the supply during Initialization $S_0$ $$S_0 = \frac{M}{P_i}$$<br> The amount of funds directed towards the project $F_0$ is directly and linearly proportional to the bond's initial likelihood of success. From this, we can infer $$F_0=\alpha_0M$$<br> We know that $$F_0=(1-\rho_0)M$$<br> From the above equations, we get $$\alpha_0 = 1-\rho_0$$<br> By definition, the state variable Price $P_0$ at $t=0$ is given by $$P_0=\kappa_0\frac{R_0}{S_0}$$<br> where $\kappa_0=\frac{1}{\rho_0}$. Expanding for $R_0=\rho_M$ and $S_0=M/P_i$, $$P_0 = \frac{1}{\rho_0} \frac{\rho_0M}{{M}/{P_i}}$$<br> Terms cancel out and we get $$P_0 = P_i$$<br> *Q.E.D*
### Lemma 2: Outcome Indifference
Let the true total payout at the Settlement Phase be $\Theta_{true}$. We know that total payout is contingent on the $(C, \Omega)$ pair that was set during Initialization and the amount of tokens bonded to the reserve. This gives $$\Theta_{true} = C \Omega + R$$<br> Neither the bonding curve system nor the participant agents have knowledge about the true total payout prior to the Settlement Phase, so they base their decisions on their own expectations of the total payout.
#### 1. System's Expectation of Total Payout
During Execution, the system does not have knowledge of $\Omega$ but is aware of the state variable $\alpha$, which is based on Attestations on the value of $\Omega$. This gives $$\mathbb{E}(\Theta_{sys}) = C\alpha + R$$<br> This expectation on $\Theta_{sys}$ is a **system-level constraint**.
#### 2. Agent's Expectation of Individual Payout
Let $\Theta_0 = M$ be the amount of money spent by agent $i$ to purchase $S_i$ amount of tokens during Initialization. From the definition of Price, we have $$\Theta_{0}=PS_0$$ Expanding Price $P$ $$\Theta_{0}=\frac{1}{\rho}\frac{R}{S}S_0$$<br> $$\Theta_{0}=\frac{1}{\rho}R\left(\frac{S_0}{S}\right)$$<br> This gives $\Theta_{i}$, the amount of tokens to be paid out to the agent $$\Theta_{i} = \frac{\Theta_0}{\Theta} = \frac{1}{\rho}\frac{R}{\Theta}\left(\frac{S_0}{S}\right)$$<br> This imposes an **agent-level constraint**.
Since outcome payout is conserved throughout the duration of the bond, we have $\frac{\Theta_{0}}{S_0} =\frac{\Theta}{S}$. Rearranging, $$\frac{\Theta_{0}}{S_0}=\frac{\Theta}{S}$$<br> For this equivalency to hold, in the definition for $\Theta_i$, we require that $$\frac{1}{\rho}\frac{R}{\Theta} = 1$$<br> Rearranging to obtain $R$, $$R=\rho\Theta$$<br> Since $\Theta_{true}$ is unknown, the system and its agents need to estimate it. Plugging the above in $\Theta_{true}$, we have $$\Theta_{true}=C\Omega + \rho\Theta_{true}$$ Simplifying, $$\Theta_{true}=\frac{C\Omega}{1-\rho}$$<br> From here, we derive the system's estimate of total payout and the agents' estimate of individual payout.
#### 3. System's estimate of Total Payout
From (1) we have $$\mathbb{E}(\Theta_{sys}) = C\alpha + R$$<br> Due to the analogy between $\Theta_{sys}$ and $\Theta_{true}$, we can conclude $$\mathbb{E}(\Theta_{sys})= \frac{C\alpha}{1-\rho}$$<br>
#### 4. Agents' estimate of Individual Payout
Following from the above, we obtain the agent's estimate of an individual agent's payout $$\mathbb{E}(\Theta_{i})= \frac{C\hat\alpha_i}{1-\rho}\frac{S_i}{S}$$<br>
| 0.781956 | 0.988917 |
# LES Band Azimuthal Angle Model Training
---
Full Environmental Database Model
### Carter J. Humphreys
Email: [chumphre@oswego.edu](mailto:chumphre@oswego.edu) | GitHub:[@HumphreysCarter](https://github.com/HumphreysCarter) | Website: [carterhumphreys.com](http://carterhumphreys.com/)
```
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
import pickle
import joblib
import pandas as pd
from pandas.plotting import scatter_matrix
from matplotlib import pyplot
from sklearn.model_selection import train_test_split, cross_val_score, KFold
from sklearn.metrics import max_error, mean_absolute_error, mean_squared_error, r2_score
# Models
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.neural_network import MLPRegressor
```
# Load The Data
```
# Load dataset
dataPath="../data/full_dataset.csv"
dataset=pd.read_csv(dataPath, header=None)
dataset=dataset.drop([0, 1, 2, 3, 4, 5, 6, 7, 9], axis=1)
```
# Dataset Summary
```
# shape
print(dataset.shape)
# head
print(dataset.head(5))
# descriptions
print(dataset.describe())
```
# Algorithms
```
# Split-out validation dataset
array = dataset.values
X = array[:,1:326]
y = array[:,0:1]
X_train, X_validation, Y_train, Y_validation = train_test_split(X, y, test_size=0.30, random_state=1)
print(y)
print(X)
# Spot Check Algorithms
models = []
models.append(('MultiLR', LinearRegression()))
models.append(('KNN(n=2)', KNeighborsRegressor(n_neighbors=2)))
models.append(('KNN(n=5)', KNeighborsRegressor(n_neighbors=5)))
#models.append(('DecisionTree', DecisionTreeRegressor()))
models.append(('RandomForest', RandomForestRegressor()))
#models.append(('MLPR', MLPRegressor()))
# evaluate each model in turn
results = []
names = []
for name, model in models:
kfold = KFold(n_splits=5, random_state=1, shuffle=True)
cv_results = cross_val_score(model, X_train, Y_train, cv=kfold)
if name != 'MultiLR': # Hide MultiLR since data is skewed
results.append(cv_results)
names.append(name)
print('%s: %f (%f)' % (name, cv_results.mean(), cv_results.std()))
# Compare Algorithms
pyplot.boxplot(results, labels=names, showfliers=False)
pyplot.title('Algorithm Comparison (Excluding MultiLR)')
pyplot.show()
```
# Predictions and Validation
```
for name, model in models:
print(f'==================== {name} ====================')
# Make predictions on validation dataset
model.fit(X_train, Y_train)
predictions = model.predict(X_validation)
# Evaluate current model
currMaxError=max_error(Y_validation, predictions)
currAbsError=mean_absolute_error(Y_validation, predictions)
currMeanSqrError=mean_squared_error(Y_validation, predictions)
currR2score=r2_score(Y_validation, predictions)
pyplot.plot(Y_validation, Y_validation, color='black')
pyplot.scatter(Y_validation, predictions, marker='o', color='lightseagreen');
pyplot.title(f'{name} [Current Run]')
pyplot.xlabel("validation")
pyplot.ylabel("prediction")
pyplot.xlim(50, 190)
pyplot.ylim(50, 190)
pyplot.show()
# Load previous model and make predictions on validation dataset
previousModel=joblib.load(f'../models/LES_Band_Position_Model_{name}_Az')
previousModel.fit(X_train, Y_train)
prevPredictions=previousModel.predict(X_validation)
# Evaluate previous model
prevMaxError=max_error(Y_validation, prevPredictions)
prevAbsError=mean_absolute_error(Y_validation, prevPredictions)
prevMeanSqrError=mean_squared_error(Y_validation, prevPredictions)
prevR2score=r2_score(Y_validation, prevPredictions)
pyplot.plot(Y_validation, Y_validation, color='black')
pyplot.scatter(Y_validation, predictions, marker='o', color='lightseagreen');
pyplot.title(f'{name} [Previous Run]')
pyplot.xlabel("validation")
pyplot.ylabel("prediction")
pyplot.xlim(50, 190)
pyplot.ylim(50, 190)
pyplot.show()
print('Metric\t\t\tCurr Score\t\tPrev Score')
print('----------\t\t---------\t\t--------')
print(f'max_error\t\t{currMaxError}\t{prevMaxError}')
print(f'mean_absolute_error\t{currAbsError}\t{prevAbsError}')
print(f'mean_squared_error\t{currMeanSqrError}\t{prevMeanSqrError}')
print(f'r2_score\t\t{currR2score}\t{prevR2score}')
# Save model if r^2 better
if currR2score > prevR2score:
print('Saving model...')
joblib.dump(model, f'../models/LES_Band_Position_Model_{name}_Az')
print('\n')
pyplot.title(f'Algorithm Coefficient of Determination')
pyplot.ylabel("R2 Score")
langs = ['MultiLR ', 'KNN(n=2)', 'KNN(n=5)', 'RandomForest',]
students = [0.5590192444600064, 0.7810491712130475, 0.7141367375570404, 0.7363230918074593]
pyplot.bar(langs,students)
pyplot.ylim(0.0, 1.0)
pyplot.show()
```
|
github_jupyter
|
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
import pickle
import joblib
import pandas as pd
from pandas.plotting import scatter_matrix
from matplotlib import pyplot
from sklearn.model_selection import train_test_split, cross_val_score, KFold
from sklearn.metrics import max_error, mean_absolute_error, mean_squared_error, r2_score
# Models
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.neural_network import MLPRegressor
# Load dataset
dataPath="../data/full_dataset.csv"
dataset=pd.read_csv(dataPath, header=None)
dataset=dataset.drop([0, 1, 2, 3, 4, 5, 6, 7, 9], axis=1)
# shape
print(dataset.shape)
# head
print(dataset.head(5))
# descriptions
print(dataset.describe())
# Split-out validation dataset
array = dataset.values
X = array[:,1:326]
y = array[:,0:1]
X_train, X_validation, Y_train, Y_validation = train_test_split(X, y, test_size=0.30, random_state=1)
print(y)
print(X)
# Spot Check Algorithms
models = []
models.append(('MultiLR', LinearRegression()))
models.append(('KNN(n=2)', KNeighborsRegressor(n_neighbors=2)))
models.append(('KNN(n=5)', KNeighborsRegressor(n_neighbors=5)))
#models.append(('DecisionTree', DecisionTreeRegressor()))
models.append(('RandomForest', RandomForestRegressor()))
#models.append(('MLPR', MLPRegressor()))
# evaluate each model in turn
results = []
names = []
for name, model in models:
kfold = KFold(n_splits=5, random_state=1, shuffle=True)
cv_results = cross_val_score(model, X_train, Y_train, cv=kfold)
if name != 'MultiLR': # Hide MultiLR since data is skewed
results.append(cv_results)
names.append(name)
print('%s: %f (%f)' % (name, cv_results.mean(), cv_results.std()))
# Compare Algorithms
pyplot.boxplot(results, labels=names, showfliers=False)
pyplot.title('Algorithm Comparison (Excluding MultiLR)')
pyplot.show()
for name, model in models:
print(f'==================== {name} ====================')
# Make predictions on validation dataset
model.fit(X_train, Y_train)
predictions = model.predict(X_validation)
# Evaluate current model
currMaxError=max_error(Y_validation, predictions)
currAbsError=mean_absolute_error(Y_validation, predictions)
currMeanSqrError=mean_squared_error(Y_validation, predictions)
currR2score=r2_score(Y_validation, predictions)
pyplot.plot(Y_validation, Y_validation, color='black')
pyplot.scatter(Y_validation, predictions, marker='o', color='lightseagreen');
pyplot.title(f'{name} [Current Run]')
pyplot.xlabel("validation")
pyplot.ylabel("prediction")
pyplot.xlim(50, 190)
pyplot.ylim(50, 190)
pyplot.show()
# Load previous model and make predictions on validation dataset
previousModel=joblib.load(f'../models/LES_Band_Position_Model_{name}_Az')
previousModel.fit(X_train, Y_train)
prevPredictions=previousModel.predict(X_validation)
# Evaluate previous model
prevMaxError=max_error(Y_validation, prevPredictions)
prevAbsError=mean_absolute_error(Y_validation, prevPredictions)
prevMeanSqrError=mean_squared_error(Y_validation, prevPredictions)
prevR2score=r2_score(Y_validation, prevPredictions)
pyplot.plot(Y_validation, Y_validation, color='black')
pyplot.scatter(Y_validation, predictions, marker='o', color='lightseagreen');
pyplot.title(f'{name} [Previous Run]')
pyplot.xlabel("validation")
pyplot.ylabel("prediction")
pyplot.xlim(50, 190)
pyplot.ylim(50, 190)
pyplot.show()
print('Metric\t\t\tCurr Score\t\tPrev Score')
print('----------\t\t---------\t\t--------')
print(f'max_error\t\t{currMaxError}\t{prevMaxError}')
print(f'mean_absolute_error\t{currAbsError}\t{prevAbsError}')
print(f'mean_squared_error\t{currMeanSqrError}\t{prevMeanSqrError}')
print(f'r2_score\t\t{currR2score}\t{prevR2score}')
# Save model if r^2 better
if currR2score > prevR2score:
print('Saving model...')
joblib.dump(model, f'../models/LES_Band_Position_Model_{name}_Az')
print('\n')
pyplot.title(f'Algorithm Coefficient of Determination')
pyplot.ylabel("R2 Score")
langs = ['MultiLR ', 'KNN(n=2)', 'KNN(n=5)', 'RandomForest',]
students = [0.5590192444600064, 0.7810491712130475, 0.7141367375570404, 0.7363230918074593]
pyplot.bar(langs,students)
pyplot.ylim(0.0, 1.0)
pyplot.show()
| 0.714728 | 0.879458 |
# Transient Flamelet Example: Ignition Sensitivity to Rate Parameter
_This demo is part of Spitfire, with [licensing and copyright info here.](https://github.com/sandialabs/Spitfire/blob/master/license.md)_
_Highlights_
- Solving transient flamelet ignition trajectories
- Observing sensitivity of the ignition behavior to a key reaction rate parameter
In this demonstration we use the `integrate_to_steady` method as in previous notebooks, this time to look at how ignition behavior is affected by the pre-exponential factor of a key chain-branching reaction in hydrogen-air ignition. Cantera is used to load the nominal chemistry and modify the reaction rate accordingly.
```
import cantera as ct
from spitfire import ChemicalMechanismSpec, Flamelet, FlameletSpec
import matplotlib.pyplot as plt
import numpy as np
from os.path import abspath, join
sol = ct.Solution('h2-burke.xml', 'h2-burke')
Tair = 1200.
pressure = 101325.
zstoich = 0.1
chi_max = 1.e3
npts_interior = 32
k1mult_list = [0.02, 0.1, 0.2, 1.0, 10.0, 100.0]
sol_dict = dict()
max_time = 0.
max_temp = 0.
A0_original = np.copy(sol.reaction(0).rate.pre_exponential_factor)
for i, k1mult in enumerate(k1mult_list):
print(f'running {k1mult:.2f}A ...')
r0 = sol.reaction(0)
new_rate = ct.Arrhenius(k1mult * A0_original,
r0.rate.temperature_exponent,
r0.rate.activation_energy)
new_rxn = ct.ElementaryReaction(r0.reactants, r0.products)
new_rxn.rate = new_rate
sol.modify_reaction(0, new_rxn)
m = ChemicalMechanismSpec.from_solution(sol)
air = m.stream(stp_air=True)
air.TP = Tair, pressure
fuel = m.mix_fuels_for_stoich_mixture_fraction(m.stream('X', 'H2:1'), m.stream('X', 'N2:1'), zstoich, air)
fuel.TP = 300., pressure
flamelet_specs = FlameletSpec(mech_spec=m,
initial_condition='unreacted',
oxy_stream=air,
fuel_stream=fuel,
grid_points=npts_interior + 2,
grid_cluster_intensity=4.,
max_dissipation_rate=chi_max)
ft = Flamelet(flamelet_specs)
output = ft.integrate_to_steady(first_time_step=1.e-9)
t = output.time_grid * 1.e3
z = output.mixture_fraction_grid
T = output['temperature']
OH = output['mass fraction OH']
max_time = max([max_time, np.max(t)])
max_temp = max([max_temp, np.max(T)])
sol_dict[k1mult] = (i, t, z, T, OH)
print('done')
```
Next we simply show the profiles of temperature and hydroxyl mass fraction with the various rate parameters. We not only see the expected decrease in ignition delay with larger pre-exponential factor, but also that ignition does not occur at lower values as chain-branching is entirely overwhelmed by dissipation.
```
fig, axarray = plt.subplots(1, len(k1mult_list), sharex=True, sharey=True)
for k1mult in k1mult_list:
sol = sol_dict[k1mult]
axarray[sol[0]].contourf(sol[2], sol[1] * 1.e3, sol[3],
cmap=plt.get_cmap('magma'),
levels=np.linspace(300., max_temp, 20))
axarray[sol[0]].set_title(f'{k1mult:.2f}A')
axarray[sol[0]].set_xlim([0, 1])
axarray[sol[0]].set_ylim([1.e0, max_time * 1.e3])
axarray[sol[0]].set_yscale('log')
axarray[sol[0]].set_xlabel('Z')
axarray[0].set_ylabel('t (ms)')
plt.show()
fig, axarray = plt.subplots(1, len(k1mult_list), sharex=True, sharey=True)
print('Mass fraction OH profiles')
for k1mult in k1mult_list:
sol = sol_dict[k1mult]
axarray[sol[0]].contourf(sol[2], sol[1] * 1.e3, sol[4],
cmap=plt.get_cmap('magma'))
axarray[sol[0]].set_title(f'{k1mult:.2f}A')
axarray[sol[0]].set_xlim([0, 1])
axarray[sol[0]].set_ylim([1.e0, max_time * 1.e3])
axarray[sol[0]].set_yscale('log')
axarray[sol[0]].set_xlabel('Z')
axarray[0].set_ylabel('t (ms)')
plt.show()
```
|
github_jupyter
|
import cantera as ct
from spitfire import ChemicalMechanismSpec, Flamelet, FlameletSpec
import matplotlib.pyplot as plt
import numpy as np
from os.path import abspath, join
sol = ct.Solution('h2-burke.xml', 'h2-burke')
Tair = 1200.
pressure = 101325.
zstoich = 0.1
chi_max = 1.e3
npts_interior = 32
k1mult_list = [0.02, 0.1, 0.2, 1.0, 10.0, 100.0]
sol_dict = dict()
max_time = 0.
max_temp = 0.
A0_original = np.copy(sol.reaction(0).rate.pre_exponential_factor)
for i, k1mult in enumerate(k1mult_list):
print(f'running {k1mult:.2f}A ...')
r0 = sol.reaction(0)
new_rate = ct.Arrhenius(k1mult * A0_original,
r0.rate.temperature_exponent,
r0.rate.activation_energy)
new_rxn = ct.ElementaryReaction(r0.reactants, r0.products)
new_rxn.rate = new_rate
sol.modify_reaction(0, new_rxn)
m = ChemicalMechanismSpec.from_solution(sol)
air = m.stream(stp_air=True)
air.TP = Tair, pressure
fuel = m.mix_fuels_for_stoich_mixture_fraction(m.stream('X', 'H2:1'), m.stream('X', 'N2:1'), zstoich, air)
fuel.TP = 300., pressure
flamelet_specs = FlameletSpec(mech_spec=m,
initial_condition='unreacted',
oxy_stream=air,
fuel_stream=fuel,
grid_points=npts_interior + 2,
grid_cluster_intensity=4.,
max_dissipation_rate=chi_max)
ft = Flamelet(flamelet_specs)
output = ft.integrate_to_steady(first_time_step=1.e-9)
t = output.time_grid * 1.e3
z = output.mixture_fraction_grid
T = output['temperature']
OH = output['mass fraction OH']
max_time = max([max_time, np.max(t)])
max_temp = max([max_temp, np.max(T)])
sol_dict[k1mult] = (i, t, z, T, OH)
print('done')
fig, axarray = plt.subplots(1, len(k1mult_list), sharex=True, sharey=True)
for k1mult in k1mult_list:
sol = sol_dict[k1mult]
axarray[sol[0]].contourf(sol[2], sol[1] * 1.e3, sol[3],
cmap=plt.get_cmap('magma'),
levels=np.linspace(300., max_temp, 20))
axarray[sol[0]].set_title(f'{k1mult:.2f}A')
axarray[sol[0]].set_xlim([0, 1])
axarray[sol[0]].set_ylim([1.e0, max_time * 1.e3])
axarray[sol[0]].set_yscale('log')
axarray[sol[0]].set_xlabel('Z')
axarray[0].set_ylabel('t (ms)')
plt.show()
fig, axarray = plt.subplots(1, len(k1mult_list), sharex=True, sharey=True)
print('Mass fraction OH profiles')
for k1mult in k1mult_list:
sol = sol_dict[k1mult]
axarray[sol[0]].contourf(sol[2], sol[1] * 1.e3, sol[4],
cmap=plt.get_cmap('magma'))
axarray[sol[0]].set_title(f'{k1mult:.2f}A')
axarray[sol[0]].set_xlim([0, 1])
axarray[sol[0]].set_ylim([1.e0, max_time * 1.e3])
axarray[sol[0]].set_yscale('log')
axarray[sol[0]].set_xlabel('Z')
axarray[0].set_ylabel('t (ms)')
plt.show()
| 0.387227 | 0.949576 |
# Using Feature Store for training and serving
```
import os
# The Google Cloud Notebook product has specific requirements
IS_GOOGLE_CLOUD_NOTEBOOK = os.path.exists("/opt/deeplearning/metadata/env_version")
# Google Cloud Notebook requires dependencies to be installed with '--user'
USER_FLAG = ""
if IS_GOOGLE_CLOUD_NOTEBOOK:
USER_FLAG = "--user"
#!pip install --user kfp==1.6.4
#!pip install --user tfx==1.0.0-rc1
#!pip install --user google-cloud-bigquery-datatransfer==3.2.0
#!pip install --user google-cloud-pipeline-components==0.1.1
#!pip install --user google-cloud-aiplatform==1.1.1
#!pip3 install {USER_FLAG} --upgrade git+https://github.com/googleapis/python-aiplatform.git@main-test
import copy
import numpy as np
import os
import pprint
import pandas as pd
import random
import tensorflow as tf
import time
from google.cloud import aiplatform
from google.cloud import bigquery_datatransfer
from google.cloud import bigquery
from google.cloud import exceptions
from google.cloud.aiplatform_v1beta1 import (
FeaturestoreOnlineServingServiceClient, FeaturestoreServiceClient)
from google.cloud.aiplatform_v1beta1.types import FeatureSelector, IdMatcher
from google.cloud.aiplatform_v1beta1.types import \
entity_type as entity_type_pb2
from google.cloud.aiplatform_v1beta1.types import feature as feature_pb2
from google.cloud.aiplatform_v1beta1.types import \
featurestore as featurestore_pb2
from google.cloud.aiplatform_v1beta1.types import \
featurestore_monitoring as featurestore_monitoring_pb2
from google.cloud.aiplatform_v1beta1.types import \
featurestore_online_service as featurestore_online_service_pb2
from google.cloud.aiplatform_v1beta1.types import \
featurestore_service as featurestore_service_pb2
from google.cloud.aiplatform_v1beta1.types import io as io_pb2
from google.protobuf.duration_pb2 import Duration
```
## Configure lab settings
```
PROJECT_ID = 'jsb-demos'
REGION = 'us-central1'
PREFIX = 'jsb_taxi'
API_ENDPOINT = f'{REGION}-aiplatform.googleapis.com'
```
## Create Featurestore clients
Admin client for CRUD operations.
```
admin_client = FeaturestoreServiceClient(client_options={"api_endpoint": API_ENDPOINT})
BASE_RESOURCE_PATH = admin_client.common_location_path(PROJECT_ID, REGION)
```
Data client for accessing features.
```
data_client = FeaturestoreOnlineServingServiceClient(
client_options={"api_endpoint": API_ENDPOINT}
)
```
## Create Featurestore and define schemas
### Create a feature store
```
FEATURESTORE_ID = f'{PREFIX}_featurestore'
create_lro = admin_client.create_featurestore(
featurestore_service_pb2.CreateFeaturestoreRequest(
parent=BASE_RESOURCE_PATH,
featurestore_id=FEATURESTORE_ID,
featurestore=featurestore_pb2.Featurestore(
online_serving_config=featurestore_pb2.Featurestore.OnlineServingConfig(
fixed_node_count=3
),
),
)
)
create_lro.result() # Wait
```
#### List feature stores
```
admin_client.list_featurestores(parent=BASE_RESOURCE_PATH)
```
#### Get your feature store
```
admin_client.get_featurestore(
name=admin_client.featurestore_path(PROJECT_ID, REGION, FEATURESTORE_ID)
)
```
### Create Entity Type
You can specify a monitoring config which will by default be inherited by all Features under this EntityType.
```
ENTITY_TYPE_ID = 'trips'
DESCRIPTION = 'Taxi trips'
entity_type_lro = admin_client.create_entity_type(
featurestore_service_pb2.CreateEntityTypeRequest(
parent=admin_client.featurestore_path(PROJECT_ID, REGION, FEATURESTORE_ID),
entity_type_id=ENTITY_TYPE_ID,
entity_type=entity_type_pb2.EntityType(
description=DESCRIPTION,
monitoring_config=featurestore_monitoring_pb2.FeaturestoreMonitoringConfig(
snapshot_analysis=featurestore_monitoring_pb2.FeaturestoreMonitoringConfig.SnapshotAnalysis(
monitoring_interval=Duration(seconds=86400), # 1 day
),
),
),
)
)
# Similarly, wait for EntityType creation operation.
print(entity_type_lro.result())
```
### Create Features
```
features=[
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Month of a trip",
),
feature_id="trip_month",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Day of a trip",
),
feature_id="trip_day",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Day of a week",
),
feature_id="trip_day_of_week",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Hour of a trip",
),
feature_id="trip_hour",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Trip duration in seconds",
),
feature_id="trip_seconds",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.STRING,
description="Payment type",
),
feature_id="payment_type",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.STRING,
description="Pick location",
),
feature_id="pickup_grid",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.STRING,
description="Dropoff location",
),
feature_id="dropoff_grid",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.DOUBLE,
description="Euclidean distance between pick up and dropoff",
),
feature_id="euclidean",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.DOUBLE,
description="Miles travelled during the trip",
),
feature_id="trip_miles",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Trip tip classification",
),
feature_id="tip_bin",
),
]
admin_client.batch_create_features(
parent=admin_client.entity_type_path(PROJECT_ID, REGION, FEATURESTORE_ID, ENTITY_TYPE_ID),
requests=features
).result()
```
### Discover features
#### Search for all features across all featurestores
```
for feature in admin_client.search_features(location=BASE_RESOURCE_PATH):
print(feature.description)
print(feature.name)
```
#### Search for all features that are of type DOUBLE
```
features = admin_client.search_features(
featurestore_service_pb2.SearchFeaturesRequest(
location=BASE_RESOURCE_PATH, query="value_type=DOUBLE"
)
)
for feature in features:
print(feature.description)
print(feature.name)
```
#### Search for all features with specific keywords in their ID
```
features = admin_client.search_features(
featurestore_service_pb2.SearchFeaturesRequest(
location=BASE_RESOURCE_PATH, query="feature_id:grid AND value_type=STRING"
)
)
for feature in features:
print(feature.description)
print(feature.name)
```
## Import Feature Values
### Prepare import table
```
BQ_DATASET_NAME = f'{PREFIX}_dataset'
BQ_TABLE_NAME = 'feature_staging_table'
BQ_LOCATION = 'US'
SAMPLE_SIZE = 500000
YEAR = 2020
client = bigquery.Client()
dataset_id = f'{PROJECT_ID}.{BQ_DATASET_NAME}'
dataset = bigquery.Dataset(dataset_id)
dataset.location = BQ_LOCATION
try:
dataset = client.create_dataset(dataset, timeout=30)
print('Created dataset: ', dataset_id)
except exceptions.Conflict:
print('Dataset {} already exists'.format(dataset_id))
sql_script_template = '''
CREATE OR REPLACE TABLE `@PROJECT.@DATASET.@TABLE`
AS (
WITH
taxitrips AS (
SELECT
unique_key AS trip_id,
FORMAT_DATETIME('%Y-%d-%m', trip_start_timestamp) AS date,
trip_start_timestamp,
trip_seconds,
trip_miles,
payment_type,
pickup_longitude,
pickup_latitude,
dropoff_longitude,
dropoff_latitude,
tips,
fare
FROM
`bigquery-public-data.chicago_taxi_trips.taxi_trips`
WHERE 1=1
AND pickup_longitude IS NOT NULL
AND pickup_latitude IS NOT NULL
AND dropoff_longitude IS NOT NULL
AND dropoff_latitude IS NOT NULL
AND trip_miles > 0
AND trip_seconds > 0
AND fare > 0
AND EXTRACT(YEAR FROM trip_start_timestamp) = @YEAR
)
SELECT
trip_id,
trip_start_timestamp,
EXTRACT(MONTH from trip_start_timestamp) as trip_month,
EXTRACT(DAY from trip_start_timestamp) as trip_day,
EXTRACT(DAYOFWEEK from trip_start_timestamp) as trip_day_of_week,
EXTRACT(HOUR from trip_start_timestamp) as trip_hour,
trip_seconds,
trip_miles,
payment_type,
ST_AsText(
ST_SnapToGrid(ST_GeogPoint(pickup_longitude, pickup_latitude), 0.1)
) AS pickup_grid,
ST_AsText(
ST_SnapToGrid(ST_GeogPoint(dropoff_longitude, dropoff_latitude), 0.1)
) AS dropoff_grid,
ST_Distance(
ST_GeogPoint(pickup_longitude, pickup_latitude),
ST_GeogPoint(dropoff_longitude, dropoff_latitude)
) AS euclidean,
IF((tips/fare >= 0.2), 1, 0) AS tip_bin,
CASE (ABS(MOD(FARM_FINGERPRINT(date),10)))
WHEN 9 THEN 'TEST'
WHEN 8 THEN 'VALIDATE'
ELSE 'TRAIN' END AS data_split
FROM
taxitrips
LIMIT @LIMIT
)
'''
sql_script = sql_script_template.replace(
'@PROJECT', PROJECT_ID).replace(
'@DATASET', BQ_DATASET_NAME).replace(
'@TABLE', BQ_TABLE_NAME).replace(
'@YEAR', str(YEAR)).replace(
'@LIMIT', str(SAMPLE_SIZE))
job = client.query(sql_script)
job.result()
```
### Import features
```
entity_id_field = 'trip_id'
bq_table = f'bq://{PROJECT_ID}.{BQ_DATASET_NAME}.{BQ_TABLE_NAME}'
import_request = featurestore_service_pb2.ImportFeatureValuesRequest(
entity_type=admin_client.entity_type_path(
PROJECT_ID, REGION, FEATURESTORE_ID, ENTITY_TYPE_ID
),
bigquery_source=io_pb2.BigQuerySource(
input_uri=bq_table
),
entity_id_field=entity_id_field,
feature_specs=[
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="tip_bin"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_month"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_day"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_day_of_week"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_hour"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="payment_type"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="pickup_grid"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="dropoff_grid"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="euclidean"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_seconds"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_miles"),
],
feature_time_field="trip_start_timestamp",
worker_count=1,
)
ingestion_lro = admin_client.import_feature_values(import_request)
ingestion_lro.result()
```
## Online serving
The
[Online Serving APIs](https://cloud.google.com/vertex-ai/featurestore/docs/reference/rpc/google.cloud.aiplatform.v1beta1#featurestoreonlineservingservice)
lets you serve feature values for small batches of entities. It's designed for latency-sensitive service, such as online model prediction. For example, for a movie service, you might want to quickly shows movies that the current user would most likely watch by using online predictions.
### Read one entity per request
The ReadFeatureValues API is used to read feature values of one entity; hence
its custom HTTP verb is `readFeatureValues`. By default, the API will return the latest value of each feature, meaning the feature values with the most recent timestamp.
To read feature values, specify the entity ID and features to read. The response
contains a `header` and an `entity_view`. Each row of data in the `entity_view`
contains one feature value, in the same order of features as listed in the response header.
```
feature_selector = FeatureSelector(
id_matcher=IdMatcher(ids=["tip_bin", "trip_miles", "trip_day","payment_type"])
)
features = data_client.read_feature_values(
featurestore_online_service_pb2.ReadFeatureValuesRequest(
# Fetch from the following feature store/entity type
entity_type=admin_client.entity_type_path(
PROJECT_ID, REGION, FEATURESTORE_ID, ENTITY_TYPE_ID
),
# Fetch the user features whose ID is "alice"
entity_id="13311b767c033d82e37439228ef23fd1d018d061",
#trip_miles = 100,
feature_selector=feature_selector,
)
)
features
```
### Read multiple entities per request
```
response_stream = data_client.streaming_read_feature_values(
featurestore_online_service_pb2.StreamingReadFeatureValuesRequest(
entity_type=admin_client.entity_type_path(
PROJECT_ID, REGION, FEATURESTORE_ID, ENTITY_TYPE_ID
),
entity_ids=["13311b767c033d82e37439228ef23fd1d018d061", "0e9be1edf79c3d88b4da5b9d11c2651538fb33b4"],
feature_selector=feature_selector,
)
)
for response in response_stream:
print(response)
```
## Batch serving
Batch Serving is used to fetch a large batch of feature values for high-throughput, typically for training a model or batch prediction. In this section, you will learn how to prepare for training examples by calling the BatchReadFeatureValues API.
### Use case
```
INPUT_CSV_FILE = "gs://cloud-samples-data-us-central1/ai-platform-unified/datasets/featurestore/movie_prediction.csv"
INPUT_CSV_FILE = "gs://jsb-demos-central1/vertex-ai-demo/feature_store/taxi_trip_ids.csv"
from datetime import datetime
# Output dataset
DESTINATION_DATA_SET = "jsb_taxi_dataset2" # @param {type:"string"}
TIMESTAMP = datetime.now().strftime("%Y%m%d%H%M%S")
#DESTINATION_DATA_SET = "{prefix}_{timestamp}".format(
# prefix=DESTINATION_DATA_SET, timestamp=TIMESTAMP)
# Output table. Make sure that the table does NOT already exist; the BatchReadFeatureValues API cannot overwrite an existing table
DESTINATION_TABLE_NAME = "training_data" # @param {type:"string"}
DESTINATION_TABLE_NAME = "{prefix}_{timestamp}".format(
prefix=DESTINATION_TABLE_NAME, timestamp=TIMESTAMP)
DESTINATION_PATTERN = "bq://{project}.{dataset}.{table}"
DESTINATION_TABLE_URI = DESTINATION_PATTERN.format(
project=PROJECT_ID, dataset=DESTINATION_DATA_SET, table=DESTINATION_TABLE_NAME
)
batch_serving_request = featurestore_service_pb2.BatchReadFeatureValuesRequest(
# featurestore info
featurestore=admin_client.featurestore_path(PROJECT_ID, REGION, FEATURESTORE_ID),
# URL for the label data, i.e., Table 1.
csv_read_instances=io_pb2.CsvSource(
gcs_source=io_pb2.GcsSource(uris=[INPUT_CSV_FILE])
),
destination=featurestore_service_pb2.FeatureValueDestination(
bigquery_destination=io_pb2.BigQueryDestination(
# Output to BigQuery table created earlier
output_uri=DESTINATION_TABLE_URI
)
),
entity_type_specs=[
featurestore_service_pb2.BatchReadFeatureValuesRequest.EntityTypeSpec(
# Read the 'age', 'gender' and 'liked_genres' features from the 'users' entity
entity_type_id="trips",
feature_selector=FeatureSelector(
id_matcher=IdMatcher(
ids=[
# features, use "*" if you want to select all features within this entity type
#"trip_month",
#"trip_day",
"trip_hour",
"pickup_grid",
"payment_type",
#"*",
]
)
),
),
#Showcasing how to get feature data from second Feature Store Instance
featurestore_service_pb2.BatchReadFeatureValuesRequest.EntityTypeSpec(
# Read the 'average_rating' and 'genres' feature values of the 'movies' entity
entity_type_id="trips",
feature_selector=FeatureSelector(
id_matcher=IdMatcher(ids=["trip_month", "trip_day"])
),
),
],
)
# Execute the batch read
batch_serving_lro = admin_client.batch_read_feature_values(batch_serving_request)
# This long runing operation will poll until the batch read finishes.
batch_serving_lro.result()
SAMPLE_SIZE = "10"
sql_script_template = '''
SELECT *
FROM
`@PROJECT.@DATASET.@TABLE`
LIMIT @LIMIT
'''
sql_script = sql_script_template.replace(
'@PROJECT', PROJECT_ID).replace(
'@DATASET', DESTINATION_DATA_SET).replace(
'@TABLE', DESTINATION_TABLE_NAME).replace(
'@LIMIT', str(SAMPLE_SIZE))
job = client.query(sql_script)
job.to_dataframe()
%%bigquery data
SELECT *
FROM
jsb-demos.jsb_taxi_dataset2.training_data_20210729173804
LIMIT 10
%%bigquery
SELECT *
FROM
`jk-test-1002.jktest2_dataset.feature_staging_table`
LIMIT 10
```
## Clean up
```
admin_client.delete_featurestore(
featurestore_service_pb2.DeleteFeaturestoreRequest(
name=admin_client.featurestore_path(PROJECT_ID, REGION, FEATURESTORE_ID),
force=True
)
)
```
|
github_jupyter
|
import os
# The Google Cloud Notebook product has specific requirements
IS_GOOGLE_CLOUD_NOTEBOOK = os.path.exists("/opt/deeplearning/metadata/env_version")
# Google Cloud Notebook requires dependencies to be installed with '--user'
USER_FLAG = ""
if IS_GOOGLE_CLOUD_NOTEBOOK:
USER_FLAG = "--user"
#!pip install --user kfp==1.6.4
#!pip install --user tfx==1.0.0-rc1
#!pip install --user google-cloud-bigquery-datatransfer==3.2.0
#!pip install --user google-cloud-pipeline-components==0.1.1
#!pip install --user google-cloud-aiplatform==1.1.1
#!pip3 install {USER_FLAG} --upgrade git+https://github.com/googleapis/python-aiplatform.git@main-test
import copy
import numpy as np
import os
import pprint
import pandas as pd
import random
import tensorflow as tf
import time
from google.cloud import aiplatform
from google.cloud import bigquery_datatransfer
from google.cloud import bigquery
from google.cloud import exceptions
from google.cloud.aiplatform_v1beta1 import (
FeaturestoreOnlineServingServiceClient, FeaturestoreServiceClient)
from google.cloud.aiplatform_v1beta1.types import FeatureSelector, IdMatcher
from google.cloud.aiplatform_v1beta1.types import \
entity_type as entity_type_pb2
from google.cloud.aiplatform_v1beta1.types import feature as feature_pb2
from google.cloud.aiplatform_v1beta1.types import \
featurestore as featurestore_pb2
from google.cloud.aiplatform_v1beta1.types import \
featurestore_monitoring as featurestore_monitoring_pb2
from google.cloud.aiplatform_v1beta1.types import \
featurestore_online_service as featurestore_online_service_pb2
from google.cloud.aiplatform_v1beta1.types import \
featurestore_service as featurestore_service_pb2
from google.cloud.aiplatform_v1beta1.types import io as io_pb2
from google.protobuf.duration_pb2 import Duration
PROJECT_ID = 'jsb-demos'
REGION = 'us-central1'
PREFIX = 'jsb_taxi'
API_ENDPOINT = f'{REGION}-aiplatform.googleapis.com'
admin_client = FeaturestoreServiceClient(client_options={"api_endpoint": API_ENDPOINT})
BASE_RESOURCE_PATH = admin_client.common_location_path(PROJECT_ID, REGION)
data_client = FeaturestoreOnlineServingServiceClient(
client_options={"api_endpoint": API_ENDPOINT}
)
FEATURESTORE_ID = f'{PREFIX}_featurestore'
create_lro = admin_client.create_featurestore(
featurestore_service_pb2.CreateFeaturestoreRequest(
parent=BASE_RESOURCE_PATH,
featurestore_id=FEATURESTORE_ID,
featurestore=featurestore_pb2.Featurestore(
online_serving_config=featurestore_pb2.Featurestore.OnlineServingConfig(
fixed_node_count=3
),
),
)
)
create_lro.result() # Wait
admin_client.list_featurestores(parent=BASE_RESOURCE_PATH)
admin_client.get_featurestore(
name=admin_client.featurestore_path(PROJECT_ID, REGION, FEATURESTORE_ID)
)
ENTITY_TYPE_ID = 'trips'
DESCRIPTION = 'Taxi trips'
entity_type_lro = admin_client.create_entity_type(
featurestore_service_pb2.CreateEntityTypeRequest(
parent=admin_client.featurestore_path(PROJECT_ID, REGION, FEATURESTORE_ID),
entity_type_id=ENTITY_TYPE_ID,
entity_type=entity_type_pb2.EntityType(
description=DESCRIPTION,
monitoring_config=featurestore_monitoring_pb2.FeaturestoreMonitoringConfig(
snapshot_analysis=featurestore_monitoring_pb2.FeaturestoreMonitoringConfig.SnapshotAnalysis(
monitoring_interval=Duration(seconds=86400), # 1 day
),
),
),
)
)
# Similarly, wait for EntityType creation operation.
print(entity_type_lro.result())
features=[
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Month of a trip",
),
feature_id="trip_month",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Day of a trip",
),
feature_id="trip_day",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Day of a week",
),
feature_id="trip_day_of_week",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Hour of a trip",
),
feature_id="trip_hour",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Trip duration in seconds",
),
feature_id="trip_seconds",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.STRING,
description="Payment type",
),
feature_id="payment_type",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.STRING,
description="Pick location",
),
feature_id="pickup_grid",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.STRING,
description="Dropoff location",
),
feature_id="dropoff_grid",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.DOUBLE,
description="Euclidean distance between pick up and dropoff",
),
feature_id="euclidean",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.DOUBLE,
description="Miles travelled during the trip",
),
feature_id="trip_miles",
),
featurestore_service_pb2.CreateFeatureRequest(
feature=feature_pb2.Feature(
value_type=feature_pb2.Feature.ValueType.INT64,
description="Trip tip classification",
),
feature_id="tip_bin",
),
]
admin_client.batch_create_features(
parent=admin_client.entity_type_path(PROJECT_ID, REGION, FEATURESTORE_ID, ENTITY_TYPE_ID),
requests=features
).result()
for feature in admin_client.search_features(location=BASE_RESOURCE_PATH):
print(feature.description)
print(feature.name)
features = admin_client.search_features(
featurestore_service_pb2.SearchFeaturesRequest(
location=BASE_RESOURCE_PATH, query="value_type=DOUBLE"
)
)
for feature in features:
print(feature.description)
print(feature.name)
features = admin_client.search_features(
featurestore_service_pb2.SearchFeaturesRequest(
location=BASE_RESOURCE_PATH, query="feature_id:grid AND value_type=STRING"
)
)
for feature in features:
print(feature.description)
print(feature.name)
BQ_DATASET_NAME = f'{PREFIX}_dataset'
BQ_TABLE_NAME = 'feature_staging_table'
BQ_LOCATION = 'US'
SAMPLE_SIZE = 500000
YEAR = 2020
client = bigquery.Client()
dataset_id = f'{PROJECT_ID}.{BQ_DATASET_NAME}'
dataset = bigquery.Dataset(dataset_id)
dataset.location = BQ_LOCATION
try:
dataset = client.create_dataset(dataset, timeout=30)
print('Created dataset: ', dataset_id)
except exceptions.Conflict:
print('Dataset {} already exists'.format(dataset_id))
sql_script_template = '''
CREATE OR REPLACE TABLE `@PROJECT.@DATASET.@TABLE`
AS (
WITH
taxitrips AS (
SELECT
unique_key AS trip_id,
FORMAT_DATETIME('%Y-%d-%m', trip_start_timestamp) AS date,
trip_start_timestamp,
trip_seconds,
trip_miles,
payment_type,
pickup_longitude,
pickup_latitude,
dropoff_longitude,
dropoff_latitude,
tips,
fare
FROM
`bigquery-public-data.chicago_taxi_trips.taxi_trips`
WHERE 1=1
AND pickup_longitude IS NOT NULL
AND pickup_latitude IS NOT NULL
AND dropoff_longitude IS NOT NULL
AND dropoff_latitude IS NOT NULL
AND trip_miles > 0
AND trip_seconds > 0
AND fare > 0
AND EXTRACT(YEAR FROM trip_start_timestamp) = @YEAR
)
SELECT
trip_id,
trip_start_timestamp,
EXTRACT(MONTH from trip_start_timestamp) as trip_month,
EXTRACT(DAY from trip_start_timestamp) as trip_day,
EXTRACT(DAYOFWEEK from trip_start_timestamp) as trip_day_of_week,
EXTRACT(HOUR from trip_start_timestamp) as trip_hour,
trip_seconds,
trip_miles,
payment_type,
ST_AsText(
ST_SnapToGrid(ST_GeogPoint(pickup_longitude, pickup_latitude), 0.1)
) AS pickup_grid,
ST_AsText(
ST_SnapToGrid(ST_GeogPoint(dropoff_longitude, dropoff_latitude), 0.1)
) AS dropoff_grid,
ST_Distance(
ST_GeogPoint(pickup_longitude, pickup_latitude),
ST_GeogPoint(dropoff_longitude, dropoff_latitude)
) AS euclidean,
IF((tips/fare >= 0.2), 1, 0) AS tip_bin,
CASE (ABS(MOD(FARM_FINGERPRINT(date),10)))
WHEN 9 THEN 'TEST'
WHEN 8 THEN 'VALIDATE'
ELSE 'TRAIN' END AS data_split
FROM
taxitrips
LIMIT @LIMIT
)
'''
sql_script = sql_script_template.replace(
'@PROJECT', PROJECT_ID).replace(
'@DATASET', BQ_DATASET_NAME).replace(
'@TABLE', BQ_TABLE_NAME).replace(
'@YEAR', str(YEAR)).replace(
'@LIMIT', str(SAMPLE_SIZE))
job = client.query(sql_script)
job.result()
entity_id_field = 'trip_id'
bq_table = f'bq://{PROJECT_ID}.{BQ_DATASET_NAME}.{BQ_TABLE_NAME}'
import_request = featurestore_service_pb2.ImportFeatureValuesRequest(
entity_type=admin_client.entity_type_path(
PROJECT_ID, REGION, FEATURESTORE_ID, ENTITY_TYPE_ID
),
bigquery_source=io_pb2.BigQuerySource(
input_uri=bq_table
),
entity_id_field=entity_id_field,
feature_specs=[
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="tip_bin"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_month"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_day"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_day_of_week"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_hour"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="payment_type"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="pickup_grid"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="dropoff_grid"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="euclidean"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_seconds"),
featurestore_service_pb2.ImportFeatureValuesRequest.FeatureSpec(id="trip_miles"),
],
feature_time_field="trip_start_timestamp",
worker_count=1,
)
ingestion_lro = admin_client.import_feature_values(import_request)
ingestion_lro.result()
feature_selector = FeatureSelector(
id_matcher=IdMatcher(ids=["tip_bin", "trip_miles", "trip_day","payment_type"])
)
features = data_client.read_feature_values(
featurestore_online_service_pb2.ReadFeatureValuesRequest(
# Fetch from the following feature store/entity type
entity_type=admin_client.entity_type_path(
PROJECT_ID, REGION, FEATURESTORE_ID, ENTITY_TYPE_ID
),
# Fetch the user features whose ID is "alice"
entity_id="13311b767c033d82e37439228ef23fd1d018d061",
#trip_miles = 100,
feature_selector=feature_selector,
)
)
features
response_stream = data_client.streaming_read_feature_values(
featurestore_online_service_pb2.StreamingReadFeatureValuesRequest(
entity_type=admin_client.entity_type_path(
PROJECT_ID, REGION, FEATURESTORE_ID, ENTITY_TYPE_ID
),
entity_ids=["13311b767c033d82e37439228ef23fd1d018d061", "0e9be1edf79c3d88b4da5b9d11c2651538fb33b4"],
feature_selector=feature_selector,
)
)
for response in response_stream:
print(response)
INPUT_CSV_FILE = "gs://cloud-samples-data-us-central1/ai-platform-unified/datasets/featurestore/movie_prediction.csv"
INPUT_CSV_FILE = "gs://jsb-demos-central1/vertex-ai-demo/feature_store/taxi_trip_ids.csv"
from datetime import datetime
# Output dataset
DESTINATION_DATA_SET = "jsb_taxi_dataset2" # @param {type:"string"}
TIMESTAMP = datetime.now().strftime("%Y%m%d%H%M%S")
#DESTINATION_DATA_SET = "{prefix}_{timestamp}".format(
# prefix=DESTINATION_DATA_SET, timestamp=TIMESTAMP)
# Output table. Make sure that the table does NOT already exist; the BatchReadFeatureValues API cannot overwrite an existing table
DESTINATION_TABLE_NAME = "training_data" # @param {type:"string"}
DESTINATION_TABLE_NAME = "{prefix}_{timestamp}".format(
prefix=DESTINATION_TABLE_NAME, timestamp=TIMESTAMP)
DESTINATION_PATTERN = "bq://{project}.{dataset}.{table}"
DESTINATION_TABLE_URI = DESTINATION_PATTERN.format(
project=PROJECT_ID, dataset=DESTINATION_DATA_SET, table=DESTINATION_TABLE_NAME
)
batch_serving_request = featurestore_service_pb2.BatchReadFeatureValuesRequest(
# featurestore info
featurestore=admin_client.featurestore_path(PROJECT_ID, REGION, FEATURESTORE_ID),
# URL for the label data, i.e., Table 1.
csv_read_instances=io_pb2.CsvSource(
gcs_source=io_pb2.GcsSource(uris=[INPUT_CSV_FILE])
),
destination=featurestore_service_pb2.FeatureValueDestination(
bigquery_destination=io_pb2.BigQueryDestination(
# Output to BigQuery table created earlier
output_uri=DESTINATION_TABLE_URI
)
),
entity_type_specs=[
featurestore_service_pb2.BatchReadFeatureValuesRequest.EntityTypeSpec(
# Read the 'age', 'gender' and 'liked_genres' features from the 'users' entity
entity_type_id="trips",
feature_selector=FeatureSelector(
id_matcher=IdMatcher(
ids=[
# features, use "*" if you want to select all features within this entity type
#"trip_month",
#"trip_day",
"trip_hour",
"pickup_grid",
"payment_type",
#"*",
]
)
),
),
#Showcasing how to get feature data from second Feature Store Instance
featurestore_service_pb2.BatchReadFeatureValuesRequest.EntityTypeSpec(
# Read the 'average_rating' and 'genres' feature values of the 'movies' entity
entity_type_id="trips",
feature_selector=FeatureSelector(
id_matcher=IdMatcher(ids=["trip_month", "trip_day"])
),
),
],
)
# Execute the batch read
batch_serving_lro = admin_client.batch_read_feature_values(batch_serving_request)
# This long runing operation will poll until the batch read finishes.
batch_serving_lro.result()
SAMPLE_SIZE = "10"
sql_script_template = '''
SELECT *
FROM
`@PROJECT.@DATASET.@TABLE`
LIMIT @LIMIT
'''
sql_script = sql_script_template.replace(
'@PROJECT', PROJECT_ID).replace(
'@DATASET', DESTINATION_DATA_SET).replace(
'@TABLE', DESTINATION_TABLE_NAME).replace(
'@LIMIT', str(SAMPLE_SIZE))
job = client.query(sql_script)
job.to_dataframe()
%%bigquery data
SELECT *
FROM
jsb-demos.jsb_taxi_dataset2.training_data_20210729173804
LIMIT 10
%%bigquery
SELECT *
FROM
`jk-test-1002.jktest2_dataset.feature_staging_table`
LIMIT 10
admin_client.delete_featurestore(
featurestore_service_pb2.DeleteFeaturestoreRequest(
name=admin_client.featurestore_path(PROJECT_ID, REGION, FEATURESTORE_ID),
force=True
)
)
| 0.355327 | 0.533641 |
# Creating a Siamese model using Trax: Ungraded Lecture Notebook
```
import trax
from trax import layers as tl
import trax.fastmath.numpy as np
import numpy
# Setting random seeds
trax.supervised.trainer_lib.init_random_number_generators(10)
numpy.random.seed(10)
```
## L2 Normalization
Before building the model you will need to define a function that applies L2 normalization to a tensor. This is very important because in this week's assignment you will create a custom loss function which expects the tensors it receives to be normalized. Luckily this is pretty straightforward:
```
def normalize(x):
return x / np.sqrt(np.sum(x * x, axis=-1, keepdims=True))
```
Notice that the denominator can be replaced by `np.linalg.norm(x, axis=-1, keepdims=True)` to achieve the same results and that Trax's numpy is being used within the function.
```
tensor = numpy.random.random((2,5))
print(f'The tensor is of type: {type(tensor)}\n\nAnd looks like this:\n\n {tensor}')
norm_tensor = normalize(tensor)
print(f'The normalized tensor is of type: {type(norm_tensor)}\n\nAnd looks like this:\n\n {norm_tensor}')
```
Notice that the initial tensor was converted from a numpy array to a jax array in the process.
## Siamese Model
To create a `Siamese` model you will first need to create a LSTM model using the `Serial` combinator layer and then use another combinator layer called `Parallel` to create the Siamese model. You should be familiar with the following layers (notice each layer can be clicked to go to the docs):
- [`Serial`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Serial) A combinator layer that allows to stack layers serially using function composition.
- [`Embedding`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.Embedding) Maps discrete tokens to vectors. It will have shape `(vocabulary length X dimension of output vectors)`. The dimension of output vectors (also called `d_feature`) is the number of elements in the word embedding.
- [`LSTM`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.rnn.LSTM) The LSTM layer. It leverages another Trax layer called [`LSTMCell`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.rnn.LSTMCell). The number of units should be specified and should match the number of elements in the word embedding.
- [`Mean`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.Mean) Computes the mean across a desired axis. Mean uses one tensor axis to form groups of values and replaces each group with the mean value of that group.
- [`Fn`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.base.Fn) Layer with no weights that applies the function f, which should be specified using a lambda syntax.
- [`Parallel`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Parallel) It is a combinator layer (like `Serial`) that applies a list of layers in parallel to its inputs.
Putting everything together the Siamese model will look like this:
```
vocab_size = 500
model_dimension = 128
# Define the LSTM model
LSTM = tl.Serial(
tl.Embedding(vocab_size=vocab_size, d_feature=model_dimension),
tl.LSTM(model_dimension),
tl.Mean(axis=1),
tl.Fn('Normalize', lambda x: normalize(x))
)
# Use the Parallel combinator to create a Siamese model out of the LSTM
Siamese = tl.Parallel(LSTM, LSTM)
```
Next is a helper function that prints information for every layer (sublayer within `Serial`):
```
def show_layers(model, layer_prefix):
print(f"Total layers: {len(model.sublayers)}\n")
for i in range(len(model.sublayers)):
print('========')
print(f'{layer_prefix}_{i}: {model.sublayers[i]}\n')
print('Siamese model:\n')
show_layers(Siamese, 'Parallel.sublayers')
print('Detail of LSTM models:\n')
show_layers(LSTM, 'Serial.sublayers')
```
Try changing the parameters defined before the Siamese model and see how it changes!
You will actually train this model in this week's assignment. For now you should be more familiarized with creating Siamese models using Trax. **Keep it up!**
|
github_jupyter
|
import trax
from trax import layers as tl
import trax.fastmath.numpy as np
import numpy
# Setting random seeds
trax.supervised.trainer_lib.init_random_number_generators(10)
numpy.random.seed(10)
def normalize(x):
return x / np.sqrt(np.sum(x * x, axis=-1, keepdims=True))
tensor = numpy.random.random((2,5))
print(f'The tensor is of type: {type(tensor)}\n\nAnd looks like this:\n\n {tensor}')
norm_tensor = normalize(tensor)
print(f'The normalized tensor is of type: {type(norm_tensor)}\n\nAnd looks like this:\n\n {norm_tensor}')
vocab_size = 500
model_dimension = 128
# Define the LSTM model
LSTM = tl.Serial(
tl.Embedding(vocab_size=vocab_size, d_feature=model_dimension),
tl.LSTM(model_dimension),
tl.Mean(axis=1),
tl.Fn('Normalize', lambda x: normalize(x))
)
# Use the Parallel combinator to create a Siamese model out of the LSTM
Siamese = tl.Parallel(LSTM, LSTM)
def show_layers(model, layer_prefix):
print(f"Total layers: {len(model.sublayers)}\n")
for i in range(len(model.sublayers)):
print('========')
print(f'{layer_prefix}_{i}: {model.sublayers[i]}\n')
print('Siamese model:\n')
show_layers(Siamese, 'Parallel.sublayers')
print('Detail of LSTM models:\n')
show_layers(LSTM, 'Serial.sublayers')
| 0.541894 | 0.983069 |
```
# -*- coding: utf-8 -*-
```
### 1、导入相关的库
- Transformer里面是关于Transformer模型的函数
- util里面是相关的数据读取文件
- train内是相关的训练和测试函数
```
import os
from Transformer import *
from util import *
from train import *
os.environ["CUDA_VISIBLE_DEVICES"] = "1,2,3,4"
device = torch.device('cuda')
```
### 2、设置相关的参数
```
embedding_size = 32 # token的维度
num_layers = 2 # 编码器和解码器的层数,这里两者层数相同,也可以不同
dropout = 0.05 # 所有层的droprate都相同,也可以不同
batch_size = 64 # 批次
num_steps = 10 # 预测步长
factor = 1 # 学习率因子
warmup = 2000 # 学习率上升步长
lr, num_epochs, ctx = 0.005, 500, device # 学习率;周期;设备
num_hiddens, num_heads = 64, 4 # 隐层单元的数目——表示FFN中间层的输出维度;attention的数目
```
### 3、导入文件
文件为fra.txt文件
```
src_vocab, tgt_vocab, train_iter = load_data_nmt(batch_size, num_steps)
```
### 4、加载模型
- TransformerEncoder为编码器模型
- TransformerDecoder为解码器模型
- transformer为编码器和解码器构成的最终模型
```
encoder = TransformerEncoder(vocab_size=len(src_vocab),
embedding_size=embedding_size,
n_layers=num_layers,
hidden_size=num_hiddens,
num_heads=num_heads,
dropout=dropout, )
decoder = TransformerDecoder(vocab_size=len(src_vocab),
embedding_size=embedding_size,
n_layers=num_layers,
hidden_size=num_hiddens,
num_heads=num_heads,
dropout=dropout, )
class transformer(nn.Module):
def __init__(self, enc_net, dec_net):
super(transformer, self).__init__()
self.enc_net = enc_net # TransformerEncoder的对象
self.dec_net = dec_net # TransformerDecoder的对象
def forward(self, enc_X, dec_X, valid_length=None, max_seq_len=None):
"""
enc_X: 编码器的输入
dec_X: 解码器的输入
valid_length: 编码器的输入对应的valid_length,主要用于编码器attention的masksoftmax中,
并且还用于解码器的第二个attention的masksoftmax中
max_seq_len: 位置编码时调整sin和cos周期大小的,默认大小为enc_X的第一个维度seq_len
"""
# 1、通过编码器得到编码器最后一层的输出enc_output
enc_output = self.enc_net(enc_X, valid_length, max_seq_len)
# 2、state为解码器的初始状态,state包含两个元素,分别为[enc_output, valid_length]
state = self.dec_net.init_state(enc_output, valid_length)
# 3、通过解码器得到编码器最后一层到线性层的输出output,这里的output不是解码器最后一层的输出,而是
# 最后一层再连接线性层的输出
output = self.dec_net(dec_X, state)
return output
model = transformer(encoder, decoder)
```
### 5、训练模型
```
model.train()
train(model, train_iter, lr, factor, warmup, num_epochs, ctx)
```
### 6、测试模型
```
model.eval()
for sentence in ['Go .', 'Wow !', "I'm OK .", 'I won !']:
print(sentence + ' => ' + translate(model, sentence, src_vocab, tgt_vocab, num_steps, ctx))
```
|
github_jupyter
|
# -*- coding: utf-8 -*-
import os
from Transformer import *
from util import *
from train import *
os.environ["CUDA_VISIBLE_DEVICES"] = "1,2,3,4"
device = torch.device('cuda')
embedding_size = 32 # token的维度
num_layers = 2 # 编码器和解码器的层数,这里两者层数相同,也可以不同
dropout = 0.05 # 所有层的droprate都相同,也可以不同
batch_size = 64 # 批次
num_steps = 10 # 预测步长
factor = 1 # 学习率因子
warmup = 2000 # 学习率上升步长
lr, num_epochs, ctx = 0.005, 500, device # 学习率;周期;设备
num_hiddens, num_heads = 64, 4 # 隐层单元的数目——表示FFN中间层的输出维度;attention的数目
src_vocab, tgt_vocab, train_iter = load_data_nmt(batch_size, num_steps)
encoder = TransformerEncoder(vocab_size=len(src_vocab),
embedding_size=embedding_size,
n_layers=num_layers,
hidden_size=num_hiddens,
num_heads=num_heads,
dropout=dropout, )
decoder = TransformerDecoder(vocab_size=len(src_vocab),
embedding_size=embedding_size,
n_layers=num_layers,
hidden_size=num_hiddens,
num_heads=num_heads,
dropout=dropout, )
class transformer(nn.Module):
def __init__(self, enc_net, dec_net):
super(transformer, self).__init__()
self.enc_net = enc_net # TransformerEncoder的对象
self.dec_net = dec_net # TransformerDecoder的对象
def forward(self, enc_X, dec_X, valid_length=None, max_seq_len=None):
"""
enc_X: 编码器的输入
dec_X: 解码器的输入
valid_length: 编码器的输入对应的valid_length,主要用于编码器attention的masksoftmax中,
并且还用于解码器的第二个attention的masksoftmax中
max_seq_len: 位置编码时调整sin和cos周期大小的,默认大小为enc_X的第一个维度seq_len
"""
# 1、通过编码器得到编码器最后一层的输出enc_output
enc_output = self.enc_net(enc_X, valid_length, max_seq_len)
# 2、state为解码器的初始状态,state包含两个元素,分别为[enc_output, valid_length]
state = self.dec_net.init_state(enc_output, valid_length)
# 3、通过解码器得到编码器最后一层到线性层的输出output,这里的output不是解码器最后一层的输出,而是
# 最后一层再连接线性层的输出
output = self.dec_net(dec_X, state)
return output
model = transformer(encoder, decoder)
model.train()
train(model, train_iter, lr, factor, warmup, num_epochs, ctx)
model.eval()
for sentence in ['Go .', 'Wow !', "I'm OK .", 'I won !']:
print(sentence + ' => ' + translate(model, sentence, src_vocab, tgt_vocab, num_steps, ctx))
| 0.450843 | 0.711443 |
# 0.0 Understanding the Situation
- objective of the proposed situation
1. Prediction of the first destination a new user will choose
- Why?
- What kind of business model does Airbnb have?
- Market Place (connecting people who offer accommodation with people who are looking for accommodation)
- Offer (people offer accommodation)
- Portifolio size
- Portfolio diversity/density
- Average price
- Demand (people looking for accommodation)
- Number of Users
- LTV (Lifetime Value)
- CAC (Client Acquisition Cost)
- Gross Revenue = fee * number of users - CAC (contribution margins)
- **Solution**
- Prediction model of the first destination of a new user
- API
- **Input**: user and its characteristics
- **Output**: user and its characteristics **with the prediction of destination**
# 1.0 IMPORTS
## 1.1 Libraries
```
#!pip install category_encoders
import random
import numpy as np # pip install numpy
import pandas as pd # pip install pandas
import seaborn as sns # pip install seaborn
import matplotlib.pyplot as plt
from scipy import stats as ss # pip install scipy
from sklearn.metrics import accuracy_score, balanced_accuracy_score, cohen_kappa_score, classification_report
from sklearn.preprocessing import OneHotEncoder, StandardScaler, RobustScaler, MinMaxScaler
from sklearn.model_selection import train_test_split, StratifiedKFold # pip install sklearn
from scikitplot.metrics import plot_confusion_matrix # pip install scikit-plot
from imblearn import combine as c # pip install imblearn
from imblearn import over_sampling as over
from imblearn import under_sampling as us
from category_encoders import TargetEncoder
from pandas_profiling import ProfileReport # pip install pandas-profiling
from keras.models import Sequential # pip install keras; pip install tensorflow
from keras.layers import Dense
```
## 1.2 Helper Functions
```
def cramer_v(x, y):
cm = pd.crosstab(x, y).to_numpy()
n = cm.sum()
r, k = cm.shape
chi2 = ss.chi2_contingency(cm)[0]
chi2corr = max(0, chi2 - (k-1)*(r-1)/(n-1))
kcorr = k - (k-1)**2/(n-1)
rcorr = r - (r-1)**2/(n-1)
return np.sqrt((chi2corr/n)/(min(kcorr-1,rcorr-1)))
```
## 1.3 Loading data
```
!ls -l ../01-Data/csv_data
```
### 1.3.1 Training
```
df_train_raw = pd.read_csv(
"../01-Data/csv_data/train_users_2.csv", low_memory=True)
df_train_raw.shape
```
### 1.3.2 Sessions
```
df_sessions_raw = pd.read_csv(
"../01-Data/csv_data/sessions.csv", low_memory=True)
df_sessions_raw.shape
```
# 2.0 DATA DESCRIPTION
```
df_train_01 = df_train_raw.copy()
df_sessions_01 = df_sessions_raw.copy()
```
## 2.1 Data Dimensions
### 2.1.1 Training
```
print(f'Number of Rows: {df_train_01.shape[0]}')
print(f'Number of Columns: {df_train_01.shape[1]}')
```
### 2.1.2 Sessions
```
print(f'Number of Rows: {df_sessions_01.shape[0]}')
print(f'Number of Columns: {df_sessions_01.shape[1]}')
```
## 2.2 Data Type
### 2.2.1 Training
```
df_train_01.dtypes
```
### 2.2.2 Sessions
```
df_sessions_01.dtypes
```
## 2.3 NA Check
### 2.3.1 Training
```
df_train_01.isnull().sum() / len(df_train_01)
aux = df_train_01[df_train_01['date_first_booking'].isnull()]
aux['country_destination'].value_counts(normalize=True)
aux = df_train_01[df_train_01['age'].isnull()]
aux['country_destination'].value_counts(normalize=True)
sns.displot(df_train_01[df_train_01['age']<75]['age'], kind='ecdf');
df_train_01['first_affiliate_tracked'].drop_duplicates()
# remove missing values completely
#df_train_01 = df_train_01.dropna()
# date_first_booking
date_first_booking_max = pd.to_datetime(df_train_01['date_first_booking']).max().strftime('%Y-%m-%d')
df_train_01['date_first_booking'] = df_train_01['date_first_booking'].fillna(date_first_booking_max)
# age
df_train_01 = df_train_01[(df_train_01['age'] > 15) & (df_train_01['age'] < 120)]
avg_age = int(df_train_01['age'].mean())
df_train_01['age'] = df_train_01['age'].fillna(avg_age)
# first_affiliate_tracked
# remove missing values completely
df_train_01 = df_train_01[~df_train_01['first_affiliate_tracked'].isnull()]
df_train_01.shape
```
### 2.3.2 Sessions
```
df_sessions_01.isnull().sum() / len(df_sessions_01)
# remove missing values completely
## user_id - 0.3%
df_sessions_01 = df_sessions_01[~df_sessions_01['user_id'].isnull()]
## action - 0.75%
df_sessions_01 = df_sessions_01[~df_sessions_01['action'].isnull()]
## action_type - 10.65%
df_sessions_01 = df_sessions_01[~df_sessions_01['action_type'].isnull()]
## action_detail - 10.65%
df_sessions_01 = df_sessions_01[~df_sessions_01['action_detail'].isnull()]
## secs_elapsed - 1.3%
df_sessions_01 = df_sessions_01[~df_sessions_01['secs_elapsed'].isnull()]
df_sessions_01.shape
```
## 2.4 Change Data type
### 2.4.1 Training
```
# date_account_created
df_train_01['date_account_created'] = pd.to_datetime(
df_train_01['date_account_created'])
# timestamp_first_active
df_train_01['timestamp_first_active'] = pd.to_datetime(
df_train_01['timestamp_first_active'], format='%Y%m%d%H%M%S')
# date_first_booking
df_train_01['date_first_booking'] = pd.to_datetime(
df_train_01['date_first_booking'])
# age
df_train_01['age'] = df_train_01['age'].astype(int)
df_train_01.dtypes
```
## 2.5 Check Balanced Data
### 2.5.1 Training
```
df_train_01['country_destination'].value_counts(normalize=True)
```
## 2.6 Descriptive Analysis
### 2.6.1 General
```
## Users
num_attributes = df_train_01.select_dtypes(include=['int32', 'int64', 'float64'])
cat_attributes = df_train_01.select_dtypes(exclude=['int32','int64', 'float64', 'datetime64[ns]'])
time_attributes = df_train_01.select_dtypes(include=['datetime64[ns]'])
## Sessions
num_attributes_sessions = df_sessions_01.select_dtypes(include=['int32', 'int64', 'float64'])
cat_attributes_sessions = df_sessions_01.select_dtypes(exclude=['int32','int64', 'float64', 'datetime64[ns]'])
time_attributes_sessions = df_sessions_01.select_dtypes(include=['datetime64[ns]'])
```
### 2.6.2 Numerical Users
```
# Central Tendency - Mean, Mediam
ct1 = pd.DataFrame(num_attributes.apply(np.mean)).T
ct2 = pd.DataFrame(num_attributes.apply(np.median)).T
# Dispersions - Std, Min, Max, Range, Skew, Kurtosis
d1 = pd.DataFrame(num_attributes.apply(np.std)).T
d2 = pd.DataFrame(num_attributes.apply(min)).T
d3 = pd.DataFrame(num_attributes.apply(max)).T
d4 = pd.DataFrame(num_attributes.apply(lambda x: x.max() - x.min())).T
d5 = pd.DataFrame(num_attributes.apply(lambda x: x.skew())).T
d6 = pd.DataFrame(num_attributes.apply(lambda x: x.kurtosis())).T
# Concatenate
ct = pd.concat([d2, d3, d4, ct1, ct2, d1, d5, d6]).T.reset_index()
ct.columns = ['attributes', 'min', 'max', 'range', 'mean', 'median', 'std', 'skew', 'kurtosis']
ct
```
### 2.6.3 Numerical Sessions
```
# Central Tendency - Mean, Mediam
ct1 = pd.DataFrame(num_attributes_sessions.apply(np.mean)).T
ct2 = pd.DataFrame(num_attributes_sessions.apply(np.median)).T
# Dispersions - Std, Min, Max, Range, Skew, Kurtosis
d1 = pd.DataFrame(num_attributes_sessions.apply(np.std)).T
d2 = pd.DataFrame(num_attributes_sessions.apply(min)).T
d3 = pd.DataFrame(num_attributes_sessions.apply(max)).T
d4 = pd.DataFrame(num_attributes_sessions.apply(lambda x: x.max() - x.min())).T
d5 = pd.DataFrame(num_attributes_sessions.apply(lambda x: x.skew())).T
d6 = pd.DataFrame(num_attributes_sessions.apply(lambda x: x.kurtosis())).T
# Concatenate
ct = pd.concat([d2, d3, d4, ct1, ct2, d1, d5, d6]).T.reset_index()
ct.columns = ['attributes', 'min', 'max', 'range', 'mean', 'median', 'std', 'skew', 'kurtosis']
ct
```
### 2.6.3 Categorical Users
```
cat_attributes.drop('id', axis=1).describe()
```
### 2.6.4 Categorical Sessions
```
cat_attributes_sessions.drop('user_id', axis=1).describe()
# list of attributes for Cramer's V correlation
cat_attributes_list = cat_attributes_sessions.drop('user_id', axis=1).columns.tolist()
corr_dict = {}
for i in range(len(cat_attributes_list)):
corr_list = []
for j in range(len(cat_attributes_list)):
ref = cat_attributes_list[i]
feat = cat_attributes_list[j]
# correlation
corr = cramer_v(cat_attributes_sessions[ref], cat_attributes_sessions[feat])
# append list
corr_list.append(corr)
# append correlation list for each ref attributes
corr_dict[ref] = corr_list
#d = pd.DataFrame(corr_dict)
d = pd.DataFrame(corr_dict)
d = d.set_index(d.columns)
sns.heatmap(d, annot=True);
```
# 3.0 Feature Engineering
```
df_train_02 = df_train_01.copy()
df_sessions_02 = df_sessions_01.copy()
# days from first activate up to first booking
df_train_02['first_active'] = pd.to_datetime(
df_train_02['timestamp_first_active'].dt.strftime('%Y-%m-%d'))
df_train_02['days_from_first_active_until_booking'] = (
df_train_02['date_first_booking'] - df_train_02['first_active']).apply(lambda x: x.days)
# days from first activate up to account created
df_train_02['days_from_first_active_until_account_created'] = (
df_train_02['date_account_created'] - df_train_02['first_active']).apply(lambda x: x.days)
# days from account created up to first booking
df_train_02['days_from_account_created_until_first_booking'] = (
df_train_02['date_first_booking'] - df_train_02['date_account_created']).apply(lambda x: x.days)
# =============================== first active ===============================
# year first active
df_train_02['year_first_active'] = df_train_02['first_active'].dt.year
# month first active
df_train_02['month_first_active'] = df_train_02['first_active'].dt.month
# day first active
df_train_02['day_first_active'] = df_train_02['first_active'].dt.day
# day of week first active
df_train_02['day_of_week_first_active'] = df_train_02['first_active'].dt.dayofweek
# week of year of first active
df_train_02['week_of_year_first_active'] = df_train_02['first_active'].dt.weekofyear
# =============================== first booking ===============================
# year first booking
df_train_02['year_first_booking'] = df_train_02['date_first_booking'].dt.year
# month first booking
df_train_02['month_first_booking'] = df_train_02['date_first_booking'].dt.month
# day first booking
df_train_02['day_first_booking'] = df_train_02['date_first_booking'].dt.day
# day of week first booking
df_train_02['day_of_week_first_booking'] = df_train_02['date_first_booking'].dt.dayofweek
# week of year of first booking
df_train_02['week_of_year_first_booking'] = df_train_02['date_first_booking'].dt.weekofyear
# =============================== first account created ===============================
# year first booking
df_train_02['year_account_created'] = df_train_02['date_account_created'].dt.year
# month first booking
df_train_02['month_account_created'] = df_train_02['date_account_created'].dt.month
# day first booking
df_train_02['day_account_created'] = df_train_02['date_account_created'].dt.day
# day of week first booking
df_train_02['day_of_week_account_created'] = df_train_02['date_account_created'].dt.dayofweek
# week of year of first booking
df_train_02['week_of_year_account_created'] = df_train_02['date_account_created'].dt.weekofyear
```
# 4.0 Data Filtering
```
df_train_03 = df_train_02.copy()
df_sessions_03 = df_sessions_02.copy()
```
## 4.1 Filtering Rows
```
# Filtering rows:
## age > greater than 15 and lower than 120 - There are few people over 120 years old
df_train_03 = df_train_03[(df_train_03['age'] > 15) & (df_train_03['age'] < 120)]
## secs_elapsed > greater than 0 - There is no possible secs elepsed on website
df_sessions_03 = df_sessions_03[df_sessions_03['secs_elapsed'] > 0]
```
## 4.1 Filtering Columns
```
cols = ['date_account_created', 'timestamp_first_active', 'date_first_booking', 'first_active'] # orginal Datetime
df_train_03 = df_train_03.drop(cols, axis=1)
```
# 5.0 Balanced Dataset
```
df_train_04 = df_train_03.copy()
# Encoder Categorical Variables
ohe = OneHotEncoder()
# Numerical
col_num = df_train_04.select_dtypes(include=['int32', 'int64', 'float64']).columns.tolist()
# Categorical
col_cat = df_train_04.select_dtypes(exclude=['int32', 'int64', 'float64', 'datetime64[ns]'])\
.drop(['id', 'country_destination'], axis=1).columns.tolist()
# Encoding
df_train_04_dummy = pd.DataFrame(ohe.fit_transform(df_train_04[col_cat]).toarray(), index=df_train_04.index)
# join Numerical and Categorical
df_train_04_1 = pd.concat([df_train_04[col_num], df_train_04_dummy], axis=1)
```
## 5.1 Random Undersampling
```
# ratio balanced
ratio_balanced = {'NDF': 10000}
# define sampler
undersampling = us.RandomUnderSampler(sampling_strategy=ratio_balanced, random_state=32)
# apply sampler
X_under, y_under = undersampling.fit_resample(df_train_04_1, df_train_04['country_destination'])
df_train_04['country_destination'].value_counts()
y_under.value_counts()
```
## 5.2 Random Oversampling
```
# ratio balanced
#ratio_balanced = {'NDF': 10000}
# define sampler
oversampling = over.RandomOverSampler(sampling_strategy='all', random_state=32)
# apply sampler
X_over, y_over = oversampling.fit_resample(df_train_04_1, df_train_04['country_destination'])
df_train_04['country_destination'].value_counts()
y_over.value_counts()
```
## 5.3 SMOTE + TOMEKLINK
```
ratio_balanced = {
'NDF': 54852,
'US': 48057,
'other': 6*7511,
'FR': 12*3669,
'IT': 20*2014,
'GB': 25*1758,
'ES': 25*1685,
'CA': 40*1064,
'DE': 45*841,
'NL': 80*595,
'AU': 85*433,
'PT': 250*157}
# define sampler
smt = c.SMOTETomek(sampling_strategy=ratio_balanced, random_state=32, n_jobs=-1)
# apply sampler
X_smt, y_smt = smt.fit_resample(df_train_04_1, df_train_04['country_destination'])
# numerical data
df_train_04_2 = X_smt[col_num]
# categorical data
df_train_04_3 = X_smt.drop(col_num, axis=1)
df_train_04_4 = pd.DataFrame(ohe.inverse_transform(df_train_04_3), columns=col_cat, index=df_train_04_3.index)
# join numerical and categorical
df_train_04_6 = pd.concat([df_train_04_2, df_train_04_4], axis=1)
df_train_04_6['country_destination'] = y_smt
df_train_04['country_destination'].value_counts()
y_smt.value_counts()
```
# 6.0 Exploratory Data Analysis (EDA)
```
df_train_05_1 = df_train_04_6.copy()
df_train_05_2 = df_train_04.copy()
```
## 6.1 Univariate Analysis - Feature Bahaviour (Balanced Dataset)
```
profile = ProfileReport(df_train_05_1, title="Airbnb First Booking", html={'style': {'full_width':True}}, minimal=True)
profile.to_file(output_file='airbnb_booking.html')
```
## 6.2 Bivariate Analysis - Hypothesis Validation (Unbalanced dataset)
> - **H01** - At all destinations, it takes users 15 days on average to make their first Airbnb reservation since their first activation
> - **H02** - In all destinations, users take 3 days, on average, to register on the site
> - **H03** - The volume of annual reservations made during the summer increased by 20% for destinations within the US
> - **H04** - Female users make 10% more reservations for countries outside the US
> - **H05** - The Google Marketing channel accounts for 40% of reservations for countries outside the US
> - **H06** - The US target represents more than 20% on all channels
> - **H07** - The average age of people is 35 years in all destinations
> - **H08** - The percentage of users who use the site in the English American language to book accommodation in any destination is greater than 90%
> - **H09** - Is the number of Airbnb reservations increasing or decreasing over the years?
> - **H10** - The number of Airbnb reservations is increasing over the years
**H01** - At all destinations, it takes users 15 days on average to make their first Airbnb reservation since their first activation
> - **True:** At all destinations, it takes users up to 6 days to book the first Airbnb
```
plt.figure(figsize=(20,12))
plt.subplot(2,1,1)
aux01 = df_train_05_2[['days_from_first_active_until_booking', 'country_destination']].groupby('country_destination').median().reset_index()
sns.barplot(x='country_destination', y='days_from_first_active_until_booking',
data=aux01. sort_values('days_from_first_active_until_booking'));
# remove outlier
aux02 = df_train_05_2[df_train_05_2['country_destination'] != 'NDF']
aux02 = aux02[['days_from_first_active_until_booking', 'country_destination']].groupby('country_destination').median().reset_index()
plt.subplot(2,1,2)
sns.barplot(x='country_destination', y='days_from_first_active_until_booking',
data=aux02. sort_values('days_from_first_active_until_booking'));
```
**H02** - In all destinations, users take 3 days, on average, to register on the site
> - **True:** In all destinations, users take, on average, up to 2 days to complete the registration
```
plt.figure(figsize=(20,12))
#plt.subplot(2,1,1)
aux01 = df_train_05_2[['days_from_first_active_until_account_created', 'country_destination']].groupby('country_destination').mean().reset_index()
sns.barplot(x='country_destination', y='days_from_first_active_until_account_created',
data=aux01. sort_values('days_from_first_active_until_account_created'));
# # remove outlier
# aux02 = df_train_05_2[df_train_05_2['country_destination'] != 'NDF']
# aux02 = aux02[['days_from_first_active_until_account_created', 'country_destination']].groupby('country_destination').mean().reset_index()
# plt.subplot(2,1,2)
# sns.barplot(x='country_destination', y='days_from_first_active_until_account_created',
# data=aux02. sort_values('days_from_first_active_until_account_created'));
```
**H03** - The volume of annual reservations made during the summer increased by 20% for destinations within the US
> - **FALSE:** The volume of reserves increases during the summer between the years 2010 to 2013
```
aux01 = df_train_05_2[['year_first_booking', 'month_first_booking', 'country_destination']].\
groupby(['year_first_booking', 'month_first_booking', 'country_destination']).\
size().reset_index().rename(columns={0: 'count'})
# select only summer
aux01 = aux01[(aux01['month_first_booking'].isin([6, 7, 8, 9])) & (aux01['country_destination'] == 'US')]
aux02 = aux01[['year_first_booking', 'count']].groupby('year_first_booking').sum().reset_index()
aux02['delta'] = 100*aux02['count'].pct_change().fillna(0)
plt.figure(figsize=(20,12))
sns.barplot(x='year_first_booking', y='delta', data=aux02);
```
## 6.3 Multivariable analysis (Balanced Dataset)
```
## Users
num_attributes = df_train_05_1.select_dtypes(include=['int32', 'int64', 'float64'])
cat_attributes = df_train_05_1.select_dtypes(exclude=['int32','int64', 'float64', 'datetime64[ns]'])
```
### 6.3.1 Numerical
```
correlation = num_attributes.corr(method='pearson')
plt.figure(figsize=(21,12))
sns.heatmap(correlation, annot=True);
```
### 6.3.2 Categorical
```
# list of attributes for Cramer's V correlation
cat_attributes_list = cat_attributes.columns.tolist()
corr_dict = {}
for i in range(len(cat_attributes_list)):
corr_list = []
for j in range(len(cat_attributes_list)):
ref = cat_attributes_list[i]
feat = cat_attributes_list[j]
# correlation
corr = cramer_v(cat_attributes[ref], cat_attributes[feat])
# append list
corr_list.append(corr)
# append correlation list for each ref attributes
corr_dict[ref] = corr_list
d = pd.DataFrame(corr_dict)
d = d.set_index(d.columns)
plt.figure(figsize=(21,12))
sns.heatmap(d, annot=True);
```
# 7.0 Data Filtering 2
## 7.1 Filtering Columns
```
# ============================== High Correlation ==============================
# days_from_first_active_until_booking x days_from_account_created_until_first_booking
# remove: days_from_first_active_until_booking
# year_first_active x year_account_created
# remove: year_first_active
# month_first_active x month_account_created
# remove:month_first_active
# day_first_active x day_account_created
# remove:day_first_active
# day_of_week_first_active x day_of_week_account_created
# remove: day_of_week_first_active
# week_of_year_first_active x week_of_year_account_created
# remove: week_of_year_first_active
# month_first_active x week_of_year_account_created
# remove: month_first_active
# month_first_active x week_of_year_first_active
# remove: month_first_active
# week_of_year_first_active x month_account_created
# remove: week_of_year_first_active
# month_first_booking x week_of_year_first_booking
# remove: month_first_booking
# month_account_created x week_of_year_account_created
# remove: month_account_created
# month_account_created x week_of_year_account_created
# remove: month_account_created
# year_first_booking x year_account_created
# remove: year_first_booking
# week_of_year_first_booking x week_of_year_account_created
# remove: week_of_year_first_booking
# affiliate_channel x affiliate_provider
# remove: affiliate_provider
# first_device_type x first_browser
# remove: first_browser
# first_device_type x signup_app
# remove: first_device_type
cols_to_drop = ['days_from_first_active_until_booking','year_first_active','month_first_active','day_first_active',
'day_of_week_first_active','week_of_year_first_active','month_first_booking','month_account_created',
'year_first_booking','week_of_year_first_booking','affiliate_provider','first_browser','first_device_type', 'language']
df_train_07 = df_train_05_1.drop(cols_to_drop, axis=1)
```
# 8.0 Data Preparation
```
df_train_08 = df_train_07.copy()
# # Dummy variable
# df_train_08_dummy = pd.get_dummies(
# df_train_08.drop(['country_destination'], axis=1))
# # Join id and country_destination
# df_train_08 = pd.concat(
# [df_train_08[['country_destination']], df_train_08_dummy], axis=1)
```
## 8.1 Rescaling
```
ss = StandardScaler()
rs = RobustScaler()
mms = MinMaxScaler()
# =========================== Standardization ===========================
# age
df_train_08['age'] = ss.fit_transform(df_train_08[['age']].values)
# =========================== Robust Sacaler ===========================
# signup_flow
df_train_08['signup_flow'] = rs.fit_transform(df_train_08[['signup_flow']].values)
# days_from_first_active_until_account_created
df_train_08['days_from_first_active_until_account_created'] = rs.fit_transform(df_train_08[['days_from_first_active_until_account_created']].values)
# days_from_account_created_until_first_booking
df_train_08['days_from_account_created_until_first_booking'] = rs.fit_transform(df_train_08[['days_from_account_created_until_first_booking']].values)
# =========================== MinMax Sacaler ===========================
# year_account_created
df_train_08['year_account_created'] = mms.fit_transform(df_train_08[['year_account_created']].values)
```
## 8.2 Encoding
```
te = TargetEncoder()
# =========================== One Hot Encoder ===========================
# gender
df_train_08 = pd.get_dummies(df_train_08, prefix=['gender'], columns=['gender'])
# signup_method
df_train_08 = pd.get_dummies(df_train_08, prefix=['signup_method'], columns=['signup_method'])
# signup_app
df_train_08 = pd.get_dummies(df_train_08, prefix=['signup_app'], columns=['signup_app'])
# =========================== Target Encoder ===========================
c = {'NDF': 0,'US': 1,'other': 2,'CA': 3,'FR': 4,'IT': 5,'ES': 6,'GB': 7,'NL': 8,'DE': 9,'AU': 10,'PT':11}
# first_affiliate_tracked
df_train_08['first_affiliate_tracked'] = te.fit_transform(df_train_08[['first_affiliate_tracked']].values, df_train_08['country_destination'].map(c))
# affiliate_channel
df_train_08['affiliate_channel'] = te.fit_transform(df_train_08[['affiliate_channel']].values, df_train_08['country_destination'].map(c))
```
## 8.3 Transformation
```
# week_of_year_account_created
df_train_08['week_of_year_account_created_sin'] = df_train_08['week_of_year_account_created'].apply(lambda x: np.sin(x * (2*np.pi/52)))
df_train_08['week_of_year_account_created_cos'] = df_train_08['week_of_year_account_created'].apply(lambda x: np.cos(x * (2*np.pi/52)))
# day_of_week_account_created
df_train_08['day_of_week_account_created_sin'] = df_train_08['day_of_week_account_created'].apply(lambda x: np.sin(x * (2*np.pi/7)))
df_train_08['day_of_week_account_created_cos'] = df_train_08['day_of_week_account_created'].apply(lambda x: np.cos(x * (2*np.pi/7)))
# day_of_week_first_booking
df_train_08['day_of_week_first_booking_sin'] = df_train_08['day_of_week_first_booking'].apply(lambda x: np.sin(x * (2*np.pi/7)))
df_train_08['day_of_week_first_booking_cos'] = df_train_08['day_of_week_first_booking'].apply(lambda x: np.cos(x * (2*np.pi/7)))
# day_account_created
df_train_08['day_account_created_sin'] = df_train_08['day_account_created'].apply(lambda x: np.sin(x * (2*np.pi/31)))
df_train_08['day_account_created_cos'] = df_train_08['day_account_created'].apply(lambda x: np.cos(x * (2*np.pi/31)))
```
# 9.0 Feature Selection
```
# cols_drop = ['id']
# df_train_06 = df_train_05.drop(cols_drop, axis=1)
df_train_09 = df_train_08.copy()
```
## 9.1 Split into Train and Validation
```
X = df_train_09.drop('country_destination', axis=1)
y = df_train_09['country_destination'].copy()
# Split dataset into train and validation
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=32)
```
# 10.0 Machine Learning Model
## 10.1 Baseline model
### 10.1.1 Random Choices
```
country_destination_list = y_train.drop_duplicates().sort_values().tolist()
country_destination_weights = y_train.value_counts(
normalize=True).sort_index().tolist()
k_num = y_test.shape[0]
# Random Model
yhat_random = random.choices(population=country_destination_list,
weights=country_destination_weights,
k=k_num)
```
### 10.1.2 Random Choices Performance
```
# Accuracy
acc_random = accuracy_score(y_test, yhat_random)
print('Accuracy: {}'.format(acc_random))
# Balanced Accuracy
balanced_acc_random = balanced_accuracy_score(y_test, yhat_random)
print('Balanced Accuracy: {}'.format(balanced_acc_random))
# Kappa Score
kappa_random = cohen_kappa_score(y_test, yhat_random)
print('Kappa Score: {}'.format(kappa_random))
# Classification Report
print(classification_report(y_test, yhat_random))
# Confusion matrix
plot_confusion_matrix(y_test, yhat_random, normalize=False, figsize=(12, 12))
```
## 10.2 Machine Learning Model - Neural Network MLP
### 10.2.1 Target Encoding
```
ohe = OneHotEncoder()
y_train_nn = ohe.fit_transform(y_train.values.reshape(-1, 1)).toarray()
```
### 10.2.2 NN Model
```
# Model Definition
model = Sequential()
model.add(Dense(256, input_dim=X_train.shape[1], activation='relu'))
model.add(Dense(12, activation='softmax'))
# Model compile
model.compile(loss='categorical_crossentropy',
optimizer='adam', metrics=['accuracy'])
# Train Model
model.fit(X_train, y_train_nn, epochs=100)
```
### 10.2.3 NN Performance
```
# Prediction
pred_nn = model.predict(X_test)
# Inver Prediction
yhat_nn = ohe.inverse_transform(pred_nn)
# Prediction Prepare
y_test_nn = y_test.to_numpy()
yhat_nn = yhat_nn.reshape(1, -1)[0]
# Accuracy
acc_nn = accuracy_score(y_test_nn, yhat_nn)
print('Accuracy: {}'.format(acc_nn))
# Balanced Accuracy
balanced_acc_nn = balanced_accuracy_score(y_test_nn, yhat_nn)
print('Balanced Accuracy: {}'.format(balanced_acc_nn))
# Kappa Score
kappa_nn = cohen_kappa_score(y_test_nn, yhat_nn)
print('Kappa Score: {}'.format(kappa_nn))
# Classification Report
print(classification_report(y_test_nn, yhat_nn))
# Confusion matrix
plot_confusion_matrix(y_test_nn, yhat_nn, normalize=False, figsize=(12, 12))
```
### 10.2.4 NN Performance - Cross Validation
```
# k-fold generate
num_folds = 5
kfold = StratifiedKFold(n_splits=num_folds, shuffle=True, random_state=32)
balanced_acc_list = []
kappa_acc_list = []
i = 1
for train_ix, val_ix in kfold.split(X_train, y_train):
print('Fold Number: {}/{}'.format(i, num_folds))
# get fold
X_train_fold = X_train.iloc[train_ix]
y_train_fold = y_train.iloc[train_ix]
X_val_fold = X_train.iloc[val_ix]
y_val_fold = y_train.iloc[val_ix]
# target encoding
ohe = OneHotEncoder()
y_train_fold_nn = ohe.fit_transform(
y_train_fold.values.reshape(-1, 1)).toarray()
# model definition
model = Sequential()
model.add(Dense(256, input_dim=X_train_fold.shape[1], activation='relu'))
model.add(Dense(12, activation='softmax'))
# compile model
model.compile(loss='categorical_crossentropy',
optimizer='adam', metrics=['accuracy'])
# training model
model.fit(X_train_fold, y_train_fold_nn,
epochs=100, batch_size=32, verbose=0)
# prediction
pred_nn = model.predict(X_val_fold)
yhat_nn = ohe.inverse_transform(pred_nn)
# prepare data
y_test_nn = y_val_fold.to_numpy()
yhat_nn = yhat_nn.reshape(1, -1)[0]
# metrics
# Balanced Accuracy
balanced_acc_nn = balanced_accuracy_score(y_test_nn, yhat_nn)
balanced_acc_list.append(balanced_acc_nn)
# Kappa Metrics
kappa_acc_nn = cohen_kappa_score(y_test_nn, yhat_nn)
kappa_acc_list.append(kappa_acc_nn)
i += 1
print('Avg Balanced Accuracy: {} +/- {}'.format(np.round(np.mean(balanced_acc_list), 4),
np.round(np.std(balanced_acc_list), 4)))
print('Avg Kappa: {} +/- {}'.format(np.round(np.mean(kappa_acc_list), 4),
np.round(np.std(kappa_acc_list), 4)))
```
|
github_jupyter
|
#!pip install category_encoders
import random
import numpy as np # pip install numpy
import pandas as pd # pip install pandas
import seaborn as sns # pip install seaborn
import matplotlib.pyplot as plt
from scipy import stats as ss # pip install scipy
from sklearn.metrics import accuracy_score, balanced_accuracy_score, cohen_kappa_score, classification_report
from sklearn.preprocessing import OneHotEncoder, StandardScaler, RobustScaler, MinMaxScaler
from sklearn.model_selection import train_test_split, StratifiedKFold # pip install sklearn
from scikitplot.metrics import plot_confusion_matrix # pip install scikit-plot
from imblearn import combine as c # pip install imblearn
from imblearn import over_sampling as over
from imblearn import under_sampling as us
from category_encoders import TargetEncoder
from pandas_profiling import ProfileReport # pip install pandas-profiling
from keras.models import Sequential # pip install keras; pip install tensorflow
from keras.layers import Dense
def cramer_v(x, y):
cm = pd.crosstab(x, y).to_numpy()
n = cm.sum()
r, k = cm.shape
chi2 = ss.chi2_contingency(cm)[0]
chi2corr = max(0, chi2 - (k-1)*(r-1)/(n-1))
kcorr = k - (k-1)**2/(n-1)
rcorr = r - (r-1)**2/(n-1)
return np.sqrt((chi2corr/n)/(min(kcorr-1,rcorr-1)))
!ls -l ../01-Data/csv_data
df_train_raw = pd.read_csv(
"../01-Data/csv_data/train_users_2.csv", low_memory=True)
df_train_raw.shape
df_sessions_raw = pd.read_csv(
"../01-Data/csv_data/sessions.csv", low_memory=True)
df_sessions_raw.shape
df_train_01 = df_train_raw.copy()
df_sessions_01 = df_sessions_raw.copy()
print(f'Number of Rows: {df_train_01.shape[0]}')
print(f'Number of Columns: {df_train_01.shape[1]}')
print(f'Number of Rows: {df_sessions_01.shape[0]}')
print(f'Number of Columns: {df_sessions_01.shape[1]}')
df_train_01.dtypes
df_sessions_01.dtypes
df_train_01.isnull().sum() / len(df_train_01)
aux = df_train_01[df_train_01['date_first_booking'].isnull()]
aux['country_destination'].value_counts(normalize=True)
aux = df_train_01[df_train_01['age'].isnull()]
aux['country_destination'].value_counts(normalize=True)
sns.displot(df_train_01[df_train_01['age']<75]['age'], kind='ecdf');
df_train_01['first_affiliate_tracked'].drop_duplicates()
# remove missing values completely
#df_train_01 = df_train_01.dropna()
# date_first_booking
date_first_booking_max = pd.to_datetime(df_train_01['date_first_booking']).max().strftime('%Y-%m-%d')
df_train_01['date_first_booking'] = df_train_01['date_first_booking'].fillna(date_first_booking_max)
# age
df_train_01 = df_train_01[(df_train_01['age'] > 15) & (df_train_01['age'] < 120)]
avg_age = int(df_train_01['age'].mean())
df_train_01['age'] = df_train_01['age'].fillna(avg_age)
# first_affiliate_tracked
# remove missing values completely
df_train_01 = df_train_01[~df_train_01['first_affiliate_tracked'].isnull()]
df_train_01.shape
df_sessions_01.isnull().sum() / len(df_sessions_01)
# remove missing values completely
## user_id - 0.3%
df_sessions_01 = df_sessions_01[~df_sessions_01['user_id'].isnull()]
## action - 0.75%
df_sessions_01 = df_sessions_01[~df_sessions_01['action'].isnull()]
## action_type - 10.65%
df_sessions_01 = df_sessions_01[~df_sessions_01['action_type'].isnull()]
## action_detail - 10.65%
df_sessions_01 = df_sessions_01[~df_sessions_01['action_detail'].isnull()]
## secs_elapsed - 1.3%
df_sessions_01 = df_sessions_01[~df_sessions_01['secs_elapsed'].isnull()]
df_sessions_01.shape
# date_account_created
df_train_01['date_account_created'] = pd.to_datetime(
df_train_01['date_account_created'])
# timestamp_first_active
df_train_01['timestamp_first_active'] = pd.to_datetime(
df_train_01['timestamp_first_active'], format='%Y%m%d%H%M%S')
# date_first_booking
df_train_01['date_first_booking'] = pd.to_datetime(
df_train_01['date_first_booking'])
# age
df_train_01['age'] = df_train_01['age'].astype(int)
df_train_01.dtypes
df_train_01['country_destination'].value_counts(normalize=True)
## Users
num_attributes = df_train_01.select_dtypes(include=['int32', 'int64', 'float64'])
cat_attributes = df_train_01.select_dtypes(exclude=['int32','int64', 'float64', 'datetime64[ns]'])
time_attributes = df_train_01.select_dtypes(include=['datetime64[ns]'])
## Sessions
num_attributes_sessions = df_sessions_01.select_dtypes(include=['int32', 'int64', 'float64'])
cat_attributes_sessions = df_sessions_01.select_dtypes(exclude=['int32','int64', 'float64', 'datetime64[ns]'])
time_attributes_sessions = df_sessions_01.select_dtypes(include=['datetime64[ns]'])
# Central Tendency - Mean, Mediam
ct1 = pd.DataFrame(num_attributes.apply(np.mean)).T
ct2 = pd.DataFrame(num_attributes.apply(np.median)).T
# Dispersions - Std, Min, Max, Range, Skew, Kurtosis
d1 = pd.DataFrame(num_attributes.apply(np.std)).T
d2 = pd.DataFrame(num_attributes.apply(min)).T
d3 = pd.DataFrame(num_attributes.apply(max)).T
d4 = pd.DataFrame(num_attributes.apply(lambda x: x.max() - x.min())).T
d5 = pd.DataFrame(num_attributes.apply(lambda x: x.skew())).T
d6 = pd.DataFrame(num_attributes.apply(lambda x: x.kurtosis())).T
# Concatenate
ct = pd.concat([d2, d3, d4, ct1, ct2, d1, d5, d6]).T.reset_index()
ct.columns = ['attributes', 'min', 'max', 'range', 'mean', 'median', 'std', 'skew', 'kurtosis']
ct
# Central Tendency - Mean, Mediam
ct1 = pd.DataFrame(num_attributes_sessions.apply(np.mean)).T
ct2 = pd.DataFrame(num_attributes_sessions.apply(np.median)).T
# Dispersions - Std, Min, Max, Range, Skew, Kurtosis
d1 = pd.DataFrame(num_attributes_sessions.apply(np.std)).T
d2 = pd.DataFrame(num_attributes_sessions.apply(min)).T
d3 = pd.DataFrame(num_attributes_sessions.apply(max)).T
d4 = pd.DataFrame(num_attributes_sessions.apply(lambda x: x.max() - x.min())).T
d5 = pd.DataFrame(num_attributes_sessions.apply(lambda x: x.skew())).T
d6 = pd.DataFrame(num_attributes_sessions.apply(lambda x: x.kurtosis())).T
# Concatenate
ct = pd.concat([d2, d3, d4, ct1, ct2, d1, d5, d6]).T.reset_index()
ct.columns = ['attributes', 'min', 'max', 'range', 'mean', 'median', 'std', 'skew', 'kurtosis']
ct
cat_attributes.drop('id', axis=1).describe()
cat_attributes_sessions.drop('user_id', axis=1).describe()
# list of attributes for Cramer's V correlation
cat_attributes_list = cat_attributes_sessions.drop('user_id', axis=1).columns.tolist()
corr_dict = {}
for i in range(len(cat_attributes_list)):
corr_list = []
for j in range(len(cat_attributes_list)):
ref = cat_attributes_list[i]
feat = cat_attributes_list[j]
# correlation
corr = cramer_v(cat_attributes_sessions[ref], cat_attributes_sessions[feat])
# append list
corr_list.append(corr)
# append correlation list for each ref attributes
corr_dict[ref] = corr_list
#d = pd.DataFrame(corr_dict)
d = pd.DataFrame(corr_dict)
d = d.set_index(d.columns)
sns.heatmap(d, annot=True);
df_train_02 = df_train_01.copy()
df_sessions_02 = df_sessions_01.copy()
# days from first activate up to first booking
df_train_02['first_active'] = pd.to_datetime(
df_train_02['timestamp_first_active'].dt.strftime('%Y-%m-%d'))
df_train_02['days_from_first_active_until_booking'] = (
df_train_02['date_first_booking'] - df_train_02['first_active']).apply(lambda x: x.days)
# days from first activate up to account created
df_train_02['days_from_first_active_until_account_created'] = (
df_train_02['date_account_created'] - df_train_02['first_active']).apply(lambda x: x.days)
# days from account created up to first booking
df_train_02['days_from_account_created_until_first_booking'] = (
df_train_02['date_first_booking'] - df_train_02['date_account_created']).apply(lambda x: x.days)
# =============================== first active ===============================
# year first active
df_train_02['year_first_active'] = df_train_02['first_active'].dt.year
# month first active
df_train_02['month_first_active'] = df_train_02['first_active'].dt.month
# day first active
df_train_02['day_first_active'] = df_train_02['first_active'].dt.day
# day of week first active
df_train_02['day_of_week_first_active'] = df_train_02['first_active'].dt.dayofweek
# week of year of first active
df_train_02['week_of_year_first_active'] = df_train_02['first_active'].dt.weekofyear
# =============================== first booking ===============================
# year first booking
df_train_02['year_first_booking'] = df_train_02['date_first_booking'].dt.year
# month first booking
df_train_02['month_first_booking'] = df_train_02['date_first_booking'].dt.month
# day first booking
df_train_02['day_first_booking'] = df_train_02['date_first_booking'].dt.day
# day of week first booking
df_train_02['day_of_week_first_booking'] = df_train_02['date_first_booking'].dt.dayofweek
# week of year of first booking
df_train_02['week_of_year_first_booking'] = df_train_02['date_first_booking'].dt.weekofyear
# =============================== first account created ===============================
# year first booking
df_train_02['year_account_created'] = df_train_02['date_account_created'].dt.year
# month first booking
df_train_02['month_account_created'] = df_train_02['date_account_created'].dt.month
# day first booking
df_train_02['day_account_created'] = df_train_02['date_account_created'].dt.day
# day of week first booking
df_train_02['day_of_week_account_created'] = df_train_02['date_account_created'].dt.dayofweek
# week of year of first booking
df_train_02['week_of_year_account_created'] = df_train_02['date_account_created'].dt.weekofyear
df_train_03 = df_train_02.copy()
df_sessions_03 = df_sessions_02.copy()
# Filtering rows:
## age > greater than 15 and lower than 120 - There are few people over 120 years old
df_train_03 = df_train_03[(df_train_03['age'] > 15) & (df_train_03['age'] < 120)]
## secs_elapsed > greater than 0 - There is no possible secs elepsed on website
df_sessions_03 = df_sessions_03[df_sessions_03['secs_elapsed'] > 0]
cols = ['date_account_created', 'timestamp_first_active', 'date_first_booking', 'first_active'] # orginal Datetime
df_train_03 = df_train_03.drop(cols, axis=1)
df_train_04 = df_train_03.copy()
# Encoder Categorical Variables
ohe = OneHotEncoder()
# Numerical
col_num = df_train_04.select_dtypes(include=['int32', 'int64', 'float64']).columns.tolist()
# Categorical
col_cat = df_train_04.select_dtypes(exclude=['int32', 'int64', 'float64', 'datetime64[ns]'])\
.drop(['id', 'country_destination'], axis=1).columns.tolist()
# Encoding
df_train_04_dummy = pd.DataFrame(ohe.fit_transform(df_train_04[col_cat]).toarray(), index=df_train_04.index)
# join Numerical and Categorical
df_train_04_1 = pd.concat([df_train_04[col_num], df_train_04_dummy], axis=1)
# ratio balanced
ratio_balanced = {'NDF': 10000}
# define sampler
undersampling = us.RandomUnderSampler(sampling_strategy=ratio_balanced, random_state=32)
# apply sampler
X_under, y_under = undersampling.fit_resample(df_train_04_1, df_train_04['country_destination'])
df_train_04['country_destination'].value_counts()
y_under.value_counts()
# ratio balanced
#ratio_balanced = {'NDF': 10000}
# define sampler
oversampling = over.RandomOverSampler(sampling_strategy='all', random_state=32)
# apply sampler
X_over, y_over = oversampling.fit_resample(df_train_04_1, df_train_04['country_destination'])
df_train_04['country_destination'].value_counts()
y_over.value_counts()
ratio_balanced = {
'NDF': 54852,
'US': 48057,
'other': 6*7511,
'FR': 12*3669,
'IT': 20*2014,
'GB': 25*1758,
'ES': 25*1685,
'CA': 40*1064,
'DE': 45*841,
'NL': 80*595,
'AU': 85*433,
'PT': 250*157}
# define sampler
smt = c.SMOTETomek(sampling_strategy=ratio_balanced, random_state=32, n_jobs=-1)
# apply sampler
X_smt, y_smt = smt.fit_resample(df_train_04_1, df_train_04['country_destination'])
# numerical data
df_train_04_2 = X_smt[col_num]
# categorical data
df_train_04_3 = X_smt.drop(col_num, axis=1)
df_train_04_4 = pd.DataFrame(ohe.inverse_transform(df_train_04_3), columns=col_cat, index=df_train_04_3.index)
# join numerical and categorical
df_train_04_6 = pd.concat([df_train_04_2, df_train_04_4], axis=1)
df_train_04_6['country_destination'] = y_smt
df_train_04['country_destination'].value_counts()
y_smt.value_counts()
df_train_05_1 = df_train_04_6.copy()
df_train_05_2 = df_train_04.copy()
profile = ProfileReport(df_train_05_1, title="Airbnb First Booking", html={'style': {'full_width':True}}, minimal=True)
profile.to_file(output_file='airbnb_booking.html')
plt.figure(figsize=(20,12))
plt.subplot(2,1,1)
aux01 = df_train_05_2[['days_from_first_active_until_booking', 'country_destination']].groupby('country_destination').median().reset_index()
sns.barplot(x='country_destination', y='days_from_first_active_until_booking',
data=aux01. sort_values('days_from_first_active_until_booking'));
# remove outlier
aux02 = df_train_05_2[df_train_05_2['country_destination'] != 'NDF']
aux02 = aux02[['days_from_first_active_until_booking', 'country_destination']].groupby('country_destination').median().reset_index()
plt.subplot(2,1,2)
sns.barplot(x='country_destination', y='days_from_first_active_until_booking',
data=aux02. sort_values('days_from_first_active_until_booking'));
plt.figure(figsize=(20,12))
#plt.subplot(2,1,1)
aux01 = df_train_05_2[['days_from_first_active_until_account_created', 'country_destination']].groupby('country_destination').mean().reset_index()
sns.barplot(x='country_destination', y='days_from_first_active_until_account_created',
data=aux01. sort_values('days_from_first_active_until_account_created'));
# # remove outlier
# aux02 = df_train_05_2[df_train_05_2['country_destination'] != 'NDF']
# aux02 = aux02[['days_from_first_active_until_account_created', 'country_destination']].groupby('country_destination').mean().reset_index()
# plt.subplot(2,1,2)
# sns.barplot(x='country_destination', y='days_from_first_active_until_account_created',
# data=aux02. sort_values('days_from_first_active_until_account_created'));
aux01 = df_train_05_2[['year_first_booking', 'month_first_booking', 'country_destination']].\
groupby(['year_first_booking', 'month_first_booking', 'country_destination']).\
size().reset_index().rename(columns={0: 'count'})
# select only summer
aux01 = aux01[(aux01['month_first_booking'].isin([6, 7, 8, 9])) & (aux01['country_destination'] == 'US')]
aux02 = aux01[['year_first_booking', 'count']].groupby('year_first_booking').sum().reset_index()
aux02['delta'] = 100*aux02['count'].pct_change().fillna(0)
plt.figure(figsize=(20,12))
sns.barplot(x='year_first_booking', y='delta', data=aux02);
## Users
num_attributes = df_train_05_1.select_dtypes(include=['int32', 'int64', 'float64'])
cat_attributes = df_train_05_1.select_dtypes(exclude=['int32','int64', 'float64', 'datetime64[ns]'])
correlation = num_attributes.corr(method='pearson')
plt.figure(figsize=(21,12))
sns.heatmap(correlation, annot=True);
# list of attributes for Cramer's V correlation
cat_attributes_list = cat_attributes.columns.tolist()
corr_dict = {}
for i in range(len(cat_attributes_list)):
corr_list = []
for j in range(len(cat_attributes_list)):
ref = cat_attributes_list[i]
feat = cat_attributes_list[j]
# correlation
corr = cramer_v(cat_attributes[ref], cat_attributes[feat])
# append list
corr_list.append(corr)
# append correlation list for each ref attributes
corr_dict[ref] = corr_list
d = pd.DataFrame(corr_dict)
d = d.set_index(d.columns)
plt.figure(figsize=(21,12))
sns.heatmap(d, annot=True);
# ============================== High Correlation ==============================
# days_from_first_active_until_booking x days_from_account_created_until_first_booking
# remove: days_from_first_active_until_booking
# year_first_active x year_account_created
# remove: year_first_active
# month_first_active x month_account_created
# remove:month_first_active
# day_first_active x day_account_created
# remove:day_first_active
# day_of_week_first_active x day_of_week_account_created
# remove: day_of_week_first_active
# week_of_year_first_active x week_of_year_account_created
# remove: week_of_year_first_active
# month_first_active x week_of_year_account_created
# remove: month_first_active
# month_first_active x week_of_year_first_active
# remove: month_first_active
# week_of_year_first_active x month_account_created
# remove: week_of_year_first_active
# month_first_booking x week_of_year_first_booking
# remove: month_first_booking
# month_account_created x week_of_year_account_created
# remove: month_account_created
# month_account_created x week_of_year_account_created
# remove: month_account_created
# year_first_booking x year_account_created
# remove: year_first_booking
# week_of_year_first_booking x week_of_year_account_created
# remove: week_of_year_first_booking
# affiliate_channel x affiliate_provider
# remove: affiliate_provider
# first_device_type x first_browser
# remove: first_browser
# first_device_type x signup_app
# remove: first_device_type
cols_to_drop = ['days_from_first_active_until_booking','year_first_active','month_first_active','day_first_active',
'day_of_week_first_active','week_of_year_first_active','month_first_booking','month_account_created',
'year_first_booking','week_of_year_first_booking','affiliate_provider','first_browser','first_device_type', 'language']
df_train_07 = df_train_05_1.drop(cols_to_drop, axis=1)
df_train_08 = df_train_07.copy()
# # Dummy variable
# df_train_08_dummy = pd.get_dummies(
# df_train_08.drop(['country_destination'], axis=1))
# # Join id and country_destination
# df_train_08 = pd.concat(
# [df_train_08[['country_destination']], df_train_08_dummy], axis=1)
ss = StandardScaler()
rs = RobustScaler()
mms = MinMaxScaler()
# =========================== Standardization ===========================
# age
df_train_08['age'] = ss.fit_transform(df_train_08[['age']].values)
# =========================== Robust Sacaler ===========================
# signup_flow
df_train_08['signup_flow'] = rs.fit_transform(df_train_08[['signup_flow']].values)
# days_from_first_active_until_account_created
df_train_08['days_from_first_active_until_account_created'] = rs.fit_transform(df_train_08[['days_from_first_active_until_account_created']].values)
# days_from_account_created_until_first_booking
df_train_08['days_from_account_created_until_first_booking'] = rs.fit_transform(df_train_08[['days_from_account_created_until_first_booking']].values)
# =========================== MinMax Sacaler ===========================
# year_account_created
df_train_08['year_account_created'] = mms.fit_transform(df_train_08[['year_account_created']].values)
te = TargetEncoder()
# =========================== One Hot Encoder ===========================
# gender
df_train_08 = pd.get_dummies(df_train_08, prefix=['gender'], columns=['gender'])
# signup_method
df_train_08 = pd.get_dummies(df_train_08, prefix=['signup_method'], columns=['signup_method'])
# signup_app
df_train_08 = pd.get_dummies(df_train_08, prefix=['signup_app'], columns=['signup_app'])
# =========================== Target Encoder ===========================
c = {'NDF': 0,'US': 1,'other': 2,'CA': 3,'FR': 4,'IT': 5,'ES': 6,'GB': 7,'NL': 8,'DE': 9,'AU': 10,'PT':11}
# first_affiliate_tracked
df_train_08['first_affiliate_tracked'] = te.fit_transform(df_train_08[['first_affiliate_tracked']].values, df_train_08['country_destination'].map(c))
# affiliate_channel
df_train_08['affiliate_channel'] = te.fit_transform(df_train_08[['affiliate_channel']].values, df_train_08['country_destination'].map(c))
# week_of_year_account_created
df_train_08['week_of_year_account_created_sin'] = df_train_08['week_of_year_account_created'].apply(lambda x: np.sin(x * (2*np.pi/52)))
df_train_08['week_of_year_account_created_cos'] = df_train_08['week_of_year_account_created'].apply(lambda x: np.cos(x * (2*np.pi/52)))
# day_of_week_account_created
df_train_08['day_of_week_account_created_sin'] = df_train_08['day_of_week_account_created'].apply(lambda x: np.sin(x * (2*np.pi/7)))
df_train_08['day_of_week_account_created_cos'] = df_train_08['day_of_week_account_created'].apply(lambda x: np.cos(x * (2*np.pi/7)))
# day_of_week_first_booking
df_train_08['day_of_week_first_booking_sin'] = df_train_08['day_of_week_first_booking'].apply(lambda x: np.sin(x * (2*np.pi/7)))
df_train_08['day_of_week_first_booking_cos'] = df_train_08['day_of_week_first_booking'].apply(lambda x: np.cos(x * (2*np.pi/7)))
# day_account_created
df_train_08['day_account_created_sin'] = df_train_08['day_account_created'].apply(lambda x: np.sin(x * (2*np.pi/31)))
df_train_08['day_account_created_cos'] = df_train_08['day_account_created'].apply(lambda x: np.cos(x * (2*np.pi/31)))
# cols_drop = ['id']
# df_train_06 = df_train_05.drop(cols_drop, axis=1)
df_train_09 = df_train_08.copy()
X = df_train_09.drop('country_destination', axis=1)
y = df_train_09['country_destination'].copy()
# Split dataset into train and validation
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=32)
country_destination_list = y_train.drop_duplicates().sort_values().tolist()
country_destination_weights = y_train.value_counts(
normalize=True).sort_index().tolist()
k_num = y_test.shape[0]
# Random Model
yhat_random = random.choices(population=country_destination_list,
weights=country_destination_weights,
k=k_num)
# Accuracy
acc_random = accuracy_score(y_test, yhat_random)
print('Accuracy: {}'.format(acc_random))
# Balanced Accuracy
balanced_acc_random = balanced_accuracy_score(y_test, yhat_random)
print('Balanced Accuracy: {}'.format(balanced_acc_random))
# Kappa Score
kappa_random = cohen_kappa_score(y_test, yhat_random)
print('Kappa Score: {}'.format(kappa_random))
# Classification Report
print(classification_report(y_test, yhat_random))
# Confusion matrix
plot_confusion_matrix(y_test, yhat_random, normalize=False, figsize=(12, 12))
ohe = OneHotEncoder()
y_train_nn = ohe.fit_transform(y_train.values.reshape(-1, 1)).toarray()
# Model Definition
model = Sequential()
model.add(Dense(256, input_dim=X_train.shape[1], activation='relu'))
model.add(Dense(12, activation='softmax'))
# Model compile
model.compile(loss='categorical_crossentropy',
optimizer='adam', metrics=['accuracy'])
# Train Model
model.fit(X_train, y_train_nn, epochs=100)
# Prediction
pred_nn = model.predict(X_test)
# Inver Prediction
yhat_nn = ohe.inverse_transform(pred_nn)
# Prediction Prepare
y_test_nn = y_test.to_numpy()
yhat_nn = yhat_nn.reshape(1, -1)[0]
# Accuracy
acc_nn = accuracy_score(y_test_nn, yhat_nn)
print('Accuracy: {}'.format(acc_nn))
# Balanced Accuracy
balanced_acc_nn = balanced_accuracy_score(y_test_nn, yhat_nn)
print('Balanced Accuracy: {}'.format(balanced_acc_nn))
# Kappa Score
kappa_nn = cohen_kappa_score(y_test_nn, yhat_nn)
print('Kappa Score: {}'.format(kappa_nn))
# Classification Report
print(classification_report(y_test_nn, yhat_nn))
# Confusion matrix
plot_confusion_matrix(y_test_nn, yhat_nn, normalize=False, figsize=(12, 12))
# k-fold generate
num_folds = 5
kfold = StratifiedKFold(n_splits=num_folds, shuffle=True, random_state=32)
balanced_acc_list = []
kappa_acc_list = []
i = 1
for train_ix, val_ix in kfold.split(X_train, y_train):
print('Fold Number: {}/{}'.format(i, num_folds))
# get fold
X_train_fold = X_train.iloc[train_ix]
y_train_fold = y_train.iloc[train_ix]
X_val_fold = X_train.iloc[val_ix]
y_val_fold = y_train.iloc[val_ix]
# target encoding
ohe = OneHotEncoder()
y_train_fold_nn = ohe.fit_transform(
y_train_fold.values.reshape(-1, 1)).toarray()
# model definition
model = Sequential()
model.add(Dense(256, input_dim=X_train_fold.shape[1], activation='relu'))
model.add(Dense(12, activation='softmax'))
# compile model
model.compile(loss='categorical_crossentropy',
optimizer='adam', metrics=['accuracy'])
# training model
model.fit(X_train_fold, y_train_fold_nn,
epochs=100, batch_size=32, verbose=0)
# prediction
pred_nn = model.predict(X_val_fold)
yhat_nn = ohe.inverse_transform(pred_nn)
# prepare data
y_test_nn = y_val_fold.to_numpy()
yhat_nn = yhat_nn.reshape(1, -1)[0]
# metrics
# Balanced Accuracy
balanced_acc_nn = balanced_accuracy_score(y_test_nn, yhat_nn)
balanced_acc_list.append(balanced_acc_nn)
# Kappa Metrics
kappa_acc_nn = cohen_kappa_score(y_test_nn, yhat_nn)
kappa_acc_list.append(kappa_acc_nn)
i += 1
print('Avg Balanced Accuracy: {} +/- {}'.format(np.round(np.mean(balanced_acc_list), 4),
np.round(np.std(balanced_acc_list), 4)))
print('Avg Kappa: {} +/- {}'.format(np.round(np.mean(kappa_acc_list), 4),
np.round(np.std(kappa_acc_list), 4)))
| 0.388502 | 0.864024 |
```
import json
import sqlalchemy
from sqlalchemy import UniqueConstraint, CheckConstraint
from sqlalchemy.ext.declarative import declarative_base
Base = declarative_base()
from sqlalchemy import Column, Integer, String, Text, Enum, Float, Boolean
from sqlalchemy import ForeignKey, ForeignKeyConstraint
from sqlalchemy.orm import relationship
from sqlalchemy import event
from sqlalchemy.engine import Engine
from sqlite3 import Connection as SQLite3Connection
@event.listens_for(Engine, "connect")
def _set_sqlite_pragma(dbapi_connection, connection_record):
if isinstance(dbapi_connection, SQLite3Connection):
cursor = dbapi_connection.cursor()
cursor.execute("PRAGMA foreign_keys=ON;")
cursor.close()
from sqlalchemy import create_engine
engine = create_engine('sqlite:///params.sqlite3', echo=False)
class Unit(Base):
__tablename__ = 'ex_units'
id = Column(Integer, primary_key=True)
whp_unit = Column(String, nullable=True, unique=True)
cf_unit = Column(String, nullable=False)
reference_scale = Column(String, nullable=True)
note = Column(Text, nullable=True)
class Param(Base):
__tablename__ = 'ex_params'
whp_name = Column(String, primary_key=True)
whp_number = Column(Integer, nullable=True)
description = Column(Text, nullable=True)
note = Column(Text, nullable=True)
warning = Column(Text, nullable=True)
scope = Column(Enum('cruise', 'profile', 'sample'), nullable=False, server_default='sample')
dtype = Column(Enum('decimal', 'integer', 'string'), nullable=False, )
flag = Column(Enum('woce_bottle', 'woce_ctd', 'woce_discrete', 'no_flags'), nullable=False)
ancillary = Column(Boolean, nullable=False, server_default='0')
rank = Column(Float, nullable=False)
class CFName(Base):
__tablename__ = 'cf_names'
standard_name = Column(String, primary_key=True)
canonical_units = Column(String, nullable=True)
grib = Column(String, nullable=True)
amip = Column(String, nullable=True)
description = Column(Text, nullable=True)
class CFAlias(Base):
__tablename__ = 'cf_aliases'
id = Column(Integer, primary_key=True) # cannot use numeric id since alias isn't unique
alias = Column(String, nullable=False)
standard_name = Column(String, ForeignKey(CFName.__table__.c.standard_name), nullable=False)
class WHPName(Base):
__tablename__ = 'whp_names'
whp_name = Column(String, ForeignKey(Param.__table__.c.whp_name), primary_key=True)
whp_unit = Column(String, ForeignKey(Unit.__table__.c.whp_unit), primary_key=True, nullable=True)
standard_name = Column(String, ForeignKey(CFName.__table__.c.standard_name), nullable=True)
nc_name = Column(String, unique=True, nullable=True)
numeric_min = Column(Float, nullable=True)
numeric_max = Column(Float, nullable=True)
error_name = Column(String, nullable=True)
analytical_temperature_name = Column(String, nullable=True)
analytical_temperature_units = Column(String, nullable=True)
field_width = Column(Integer, nullable=False)
numeric_precision = Column(Integer, nullable=True)
__table_args__ = (
ForeignKeyConstraint(
['analytical_temperature_name', 'analytical_temperature_units'],
['whp_names.whp_name', 'whp_names.whp_unit'],
),
)
class Alias(Base):
__tablename__ = "whp_alias"
old_name = Column(String, primary_key=True)
old_unit = Column(String, primary_key=True, nullable=True)
whp_name = Column(String)
whp_unit = Column(String)
__table_args__ = (
ForeignKeyConstraint(
['whp_name', 'whp_unit'],
['whp_names.whp_name', 'whp_names.whp_unit'],
),
)
Base.metadata.create_all(engine)
from sqlalchemy.orm import sessionmaker
from xml.etree import ElementTree
Session = sessionmaker(bind=engine)
session = Session()
cf_names = []
cf_aliases = []
version_number = None
for element in ElementTree.parse("cf-standard-name-table.xml").getroot():
if element.tag == "version_number":
version_number = int(element.text)
if element.tag not in ("entry", "alias"):
continue
name = element.attrib["id"]
name_info = {info.tag: info.text for info in element}
if element.tag == "entry":
cf_names.append(CFName(standard_name=name, **name_info))
if element.tag == "alias":
cf_aliases.append(CFAlias(alias=name, standard_name=name_info["entry_id"]))
with open("parameters.json") as f:
params = json.load(f)
units = {p["whp_unit"]:p.get("cf_unit") for p in params}
refscales = {p["whp_unit"]:p.get("reference_scale") for p in params}
unit_list = []
for key, value in units.items():
cf_unit = value
if key is None:
unit_list.append(Unit(whp_unit=key, cf_unit="1"))
continue
if cf_unit is None:
cf_unit = key.lower()
unit_list.append(Unit(whp_unit=key, cf_unit=cf_unit, reference_scale=refscales[key]))
session.add_all(cf_names)
session.commit()
session.add_all(cf_aliases)
session.commit()
session.add_all(unit_list)
session.commit()
whp_name = []
db_params = []
rank = 1
for param in params:
if param['whp_name'] in whp_name:
continue
whp_name.append(param['whp_name'])
flag = param["flag_w"]
if flag == None:
flag = "no_flags"
db_params.append(Param(whp_name = param['whp_name'],
whp_number=(param.get("whp_number")),
description=(param.get("description")),
note=(param.get("note")),
warning=(param.get("warning")),
scope=(param.get("scope", "sample")),
dtype=(param["data_type"]),
flag=flag,
rank=rank,
ancillary=False,
))
rank+=1
session.add_all(db_params)
session.commit()
whp_params = []
for param in params:
whp_params.append(WHPName(
whp_name=param['whp_name'],
whp_unit=param['whp_unit'],
standard_name=param.get('cf_name'),
nc_name=None, #this is a todo
numeric_min=param.get('numeric_min'),
numeric_max=param.get('numeric_max'),
field_width=param.get('field_width'),
numeric_precision=param.get('numeric_precision'),
error_name=param.get("error_name")
))
session.add_all(whp_params)
session.commit()
with open("aliases.json") as f:
aliases = json.load(f)
alias_ad = []
for alias in aliases:
alias_ad.append(
Alias(
old_name = alias["whp_name"],
old_unit = alias["whp_unit"],
whp_name = alias["canonical_name"],
whp_unit = alias["canonical_unit"],
)
)
session.add_all(alias_ad)
session.commit()
```
|
github_jupyter
|
import json
import sqlalchemy
from sqlalchemy import UniqueConstraint, CheckConstraint
from sqlalchemy.ext.declarative import declarative_base
Base = declarative_base()
from sqlalchemy import Column, Integer, String, Text, Enum, Float, Boolean
from sqlalchemy import ForeignKey, ForeignKeyConstraint
from sqlalchemy.orm import relationship
from sqlalchemy import event
from sqlalchemy.engine import Engine
from sqlite3 import Connection as SQLite3Connection
@event.listens_for(Engine, "connect")
def _set_sqlite_pragma(dbapi_connection, connection_record):
if isinstance(dbapi_connection, SQLite3Connection):
cursor = dbapi_connection.cursor()
cursor.execute("PRAGMA foreign_keys=ON;")
cursor.close()
from sqlalchemy import create_engine
engine = create_engine('sqlite:///params.sqlite3', echo=False)
class Unit(Base):
__tablename__ = 'ex_units'
id = Column(Integer, primary_key=True)
whp_unit = Column(String, nullable=True, unique=True)
cf_unit = Column(String, nullable=False)
reference_scale = Column(String, nullable=True)
note = Column(Text, nullable=True)
class Param(Base):
__tablename__ = 'ex_params'
whp_name = Column(String, primary_key=True)
whp_number = Column(Integer, nullable=True)
description = Column(Text, nullable=True)
note = Column(Text, nullable=True)
warning = Column(Text, nullable=True)
scope = Column(Enum('cruise', 'profile', 'sample'), nullable=False, server_default='sample')
dtype = Column(Enum('decimal', 'integer', 'string'), nullable=False, )
flag = Column(Enum('woce_bottle', 'woce_ctd', 'woce_discrete', 'no_flags'), nullable=False)
ancillary = Column(Boolean, nullable=False, server_default='0')
rank = Column(Float, nullable=False)
class CFName(Base):
__tablename__ = 'cf_names'
standard_name = Column(String, primary_key=True)
canonical_units = Column(String, nullable=True)
grib = Column(String, nullable=True)
amip = Column(String, nullable=True)
description = Column(Text, nullable=True)
class CFAlias(Base):
__tablename__ = 'cf_aliases'
id = Column(Integer, primary_key=True) # cannot use numeric id since alias isn't unique
alias = Column(String, nullable=False)
standard_name = Column(String, ForeignKey(CFName.__table__.c.standard_name), nullable=False)
class WHPName(Base):
__tablename__ = 'whp_names'
whp_name = Column(String, ForeignKey(Param.__table__.c.whp_name), primary_key=True)
whp_unit = Column(String, ForeignKey(Unit.__table__.c.whp_unit), primary_key=True, nullable=True)
standard_name = Column(String, ForeignKey(CFName.__table__.c.standard_name), nullable=True)
nc_name = Column(String, unique=True, nullable=True)
numeric_min = Column(Float, nullable=True)
numeric_max = Column(Float, nullable=True)
error_name = Column(String, nullable=True)
analytical_temperature_name = Column(String, nullable=True)
analytical_temperature_units = Column(String, nullable=True)
field_width = Column(Integer, nullable=False)
numeric_precision = Column(Integer, nullable=True)
__table_args__ = (
ForeignKeyConstraint(
['analytical_temperature_name', 'analytical_temperature_units'],
['whp_names.whp_name', 'whp_names.whp_unit'],
),
)
class Alias(Base):
__tablename__ = "whp_alias"
old_name = Column(String, primary_key=True)
old_unit = Column(String, primary_key=True, nullable=True)
whp_name = Column(String)
whp_unit = Column(String)
__table_args__ = (
ForeignKeyConstraint(
['whp_name', 'whp_unit'],
['whp_names.whp_name', 'whp_names.whp_unit'],
),
)
Base.metadata.create_all(engine)
from sqlalchemy.orm import sessionmaker
from xml.etree import ElementTree
Session = sessionmaker(bind=engine)
session = Session()
cf_names = []
cf_aliases = []
version_number = None
for element in ElementTree.parse("cf-standard-name-table.xml").getroot():
if element.tag == "version_number":
version_number = int(element.text)
if element.tag not in ("entry", "alias"):
continue
name = element.attrib["id"]
name_info = {info.tag: info.text for info in element}
if element.tag == "entry":
cf_names.append(CFName(standard_name=name, **name_info))
if element.tag == "alias":
cf_aliases.append(CFAlias(alias=name, standard_name=name_info["entry_id"]))
with open("parameters.json") as f:
params = json.load(f)
units = {p["whp_unit"]:p.get("cf_unit") for p in params}
refscales = {p["whp_unit"]:p.get("reference_scale") for p in params}
unit_list = []
for key, value in units.items():
cf_unit = value
if key is None:
unit_list.append(Unit(whp_unit=key, cf_unit="1"))
continue
if cf_unit is None:
cf_unit = key.lower()
unit_list.append(Unit(whp_unit=key, cf_unit=cf_unit, reference_scale=refscales[key]))
session.add_all(cf_names)
session.commit()
session.add_all(cf_aliases)
session.commit()
session.add_all(unit_list)
session.commit()
whp_name = []
db_params = []
rank = 1
for param in params:
if param['whp_name'] in whp_name:
continue
whp_name.append(param['whp_name'])
flag = param["flag_w"]
if flag == None:
flag = "no_flags"
db_params.append(Param(whp_name = param['whp_name'],
whp_number=(param.get("whp_number")),
description=(param.get("description")),
note=(param.get("note")),
warning=(param.get("warning")),
scope=(param.get("scope", "sample")),
dtype=(param["data_type"]),
flag=flag,
rank=rank,
ancillary=False,
))
rank+=1
session.add_all(db_params)
session.commit()
whp_params = []
for param in params:
whp_params.append(WHPName(
whp_name=param['whp_name'],
whp_unit=param['whp_unit'],
standard_name=param.get('cf_name'),
nc_name=None, #this is a todo
numeric_min=param.get('numeric_min'),
numeric_max=param.get('numeric_max'),
field_width=param.get('field_width'),
numeric_precision=param.get('numeric_precision'),
error_name=param.get("error_name")
))
session.add_all(whp_params)
session.commit()
with open("aliases.json") as f:
aliases = json.load(f)
alias_ad = []
for alias in aliases:
alias_ad.append(
Alias(
old_name = alias["whp_name"],
old_unit = alias["whp_unit"],
whp_name = alias["canonical_name"],
whp_unit = alias["canonical_unit"],
)
)
session.add_all(alias_ad)
session.commit()
| 0.306423 | 0.149066 |
```
import tensorflow as tf
import matplotlib.pyplot as plt
import numpy as np
```
# 神经元函数及优化方法
## 激活函数
tf.nn.relu()
tf.nn.sigmoid()
tf.nn.tanh()
tf.nn.elu()
tf.nn.bias_add()
tf.nn.crelu()
tf.nn.relu6()
tf.nn.softsign()
tf.nn.softplus()
tf.nn.dropout() # 防止过拟合,用来舍弃某些神经元
```
x = np.linspace(-10,10,1000)
# sigmoid函数
a = tf.constant([[1.0,2.0],[1.0,2.0]])
with tf.Session() as sess:
with tf.device("/gpu:0"):
print(sess.run(tf.nn.sigmoid(a)))
# sigmoid函数图像如下
y = tf.nn.sigmoid(x)
with tf.Session() as sess:
y = sess.run(y)
plt.plot(x,y)
# tanh函数图像如下
y = tf.nn.tanh(x)
with tf.Session() as sess:
y = sess.run(y)
plt.plot(x,y)
# relu函数图像如下
y = tf.nn.relu(x)
with tf.Session() as sess:
y = sess.run(y)
plt.plot(x,y)
# drop函数
a = tf.constant([[-1.0,2.0,3.0,4.0]])
with tf.Session() as sess:
print(sess.run(tf.nn.dropout(a,0.5,noise_shape=[1,4]))) # 每个元素是否被抑制是相互独立的,相当于默认的None
print(sess.run(tf.nn.dropout(a,0.5,noise_shape=[1,1]))) # 元素是否被抑制是相关联的,同时被抑制或不抑制
```
## 卷积函数
tf.nn.convolution(input, filter, padding, strides=None, dilation_rate=None, name=None, data_format=None)
tf.nn.conv2d(input, filter, strides, padding, use_cudnn_on_gpu=True, data_format='NHWC', dilations=[1, 1, 1, 1], name=None)
tf.nn.depthwise_conv2d(input, filter, strides, padding, rate=None, name=None, data_format=None)
tf.nn.conv2d_transpose(value, filter, output_shape, strides, padding='SAME', data_format='NHWC', name=None)# 反卷积
```
# 卷积
input_data = tf.Variable(np.random.rand(10,9,9,3),dtype=np.float32)
filter_data = tf.Variable(np.random.rand(2,2,3,2),dtype=np.float32)
y = tf.nn.conv2d(input=input_data, filter=filter_data, strides=[1,1,1,1], padding='SAME')
print(y.shape)
# 反卷积
x = tf.random_normal(shape=[1,3,3,1])
kernel = tf.random_normal(shape=[2,2,3,1])
y = tf.nn.conv2d_transpose(x,kernel,output_shape=[1,5,5,3],strides=[1,2,2,1],padding='SAME')
print(y.shape)
```
## 池化函数
tf.nn.avg_pool(value, ksize, strides, padding, data_format='NHWC', name=None)
tf.nn.max_pool(value, ksize, strides, padding, data_format='NHWC', name=None)
## 分类函数
tf.nn.sigmoid_cross_entropy_with_logits()
tf.nn.softmax(logits, axis=None, name=None, dim=None)
tf.nn.log_softmax()
tf.nn.softmax_cross_entropy_with_logits()
tf.nn.sparse_softmax_cross_entropy_with_logits()
## 优化方法
tf.train.GradientDescentOptimizer()
tf.train.AdadeltaOptimizer()
tf.train.AdagradDAOptimizer()
tf.train.MomentumOptimizer()
tf.train.FtrlOptimizer()
tf.train.RMSPropOptimizer()
8个优化器对应8种优化方法,分别是梯度下降法(BGD,SDG)、Adadelta法、Adagrad法(Adagrad,AdagradDAO)、Momentum法(Momentum、Nesterov Momentum)、Adam、Ftrl和RMSProp法
其中,BGD、SGD、Momentum和Nesterov Momentum是手动指定学习率的,其余算法能够自动调节学习率
|
github_jupyter
|
import tensorflow as tf
import matplotlib.pyplot as plt
import numpy as np
x = np.linspace(-10,10,1000)
# sigmoid函数
a = tf.constant([[1.0,2.0],[1.0,2.0]])
with tf.Session() as sess:
with tf.device("/gpu:0"):
print(sess.run(tf.nn.sigmoid(a)))
# sigmoid函数图像如下
y = tf.nn.sigmoid(x)
with tf.Session() as sess:
y = sess.run(y)
plt.plot(x,y)
# tanh函数图像如下
y = tf.nn.tanh(x)
with tf.Session() as sess:
y = sess.run(y)
plt.plot(x,y)
# relu函数图像如下
y = tf.nn.relu(x)
with tf.Session() as sess:
y = sess.run(y)
plt.plot(x,y)
# drop函数
a = tf.constant([[-1.0,2.0,3.0,4.0]])
with tf.Session() as sess:
print(sess.run(tf.nn.dropout(a,0.5,noise_shape=[1,4]))) # 每个元素是否被抑制是相互独立的,相当于默认的None
print(sess.run(tf.nn.dropout(a,0.5,noise_shape=[1,1]))) # 元素是否被抑制是相关联的,同时被抑制或不抑制
# 卷积
input_data = tf.Variable(np.random.rand(10,9,9,3),dtype=np.float32)
filter_data = tf.Variable(np.random.rand(2,2,3,2),dtype=np.float32)
y = tf.nn.conv2d(input=input_data, filter=filter_data, strides=[1,1,1,1], padding='SAME')
print(y.shape)
# 反卷积
x = tf.random_normal(shape=[1,3,3,1])
kernel = tf.random_normal(shape=[2,2,3,1])
y = tf.nn.conv2d_transpose(x,kernel,output_shape=[1,5,5,3],strides=[1,2,2,1],padding='SAME')
print(y.shape)
| 0.446736 | 0.875308 |
## Constructing co-expression graph
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
```
_Loading expression data for selected mice._
```
expression_data_all_features = pd.read_pickle("processed_data/expression_standardized_cleaned.pkl")
expression_data_all_features
```
_As we can observe we have 86 strains (mice) and 1,201,231 expression features._
_Loading the relevant features for our task._
```
selected_features_df = pd.read_pickle("processed_data/selected_features.pkl")
selected_features = list(selected_features_df.columns)
print('The total number of selected features is {f}'.format(f = len(selected_features)))
```
_We now select the relevant features for all the available strains._
```
expression_data = expression_data_all_features[selected_features].T
expression_data.index.name = 'snp'
expression_data
pd.to_pickle(expression_data, "processed_data/expression_data.pkl")
```
_In order to build our co-expression graph, given two SNPs X and Y:_
- _we first obtain the vectors corresponding to the expression for all strains for those SNPS._
- _we then compute the number of common strains for these two SNPs X and Y, call it **n**._
- _we then compute the euclidian distance **e** between the non NaN values of X and Y._
- _we obtain the distance **d** between X and Y by computing d = e / n._
_We will first visualize the distribution of number of common mice measurement per pair of SNPs. With this distribution we can then set the distance to infinity between X and Y if they don't share at least n common measurements._
```
def compute_common_mice_per_snp_pair(expression_data):
rows = []
for index_i, row_i in expression_data.iterrows():
new_row = {}
for index_j, row_j in expression_data.iterrows():
u = row_i.values
v = row_j.values
u_valid_indexes = np.argwhere(~np.isnan(u))
v_valid_indexes = np.argwhere(~np.isnan(v))
valid_indexes = np.intersect1d(u_valid_indexes, v_valid_indexes)
n = len(valid_indexes)
new_row[index_j] = n
rows.append(new_row)
return pd.DataFrame(rows, index = expression_data.index)
count = compute_common_mice_per_snp_pair(expression_data)
count
```
_We then apply a mask in order to count each pair once._
```
mask = np.zeros_like(count.values, dtype=np.bool)
mask[np.tril_indices_from(mask)] = True
common_mice_per_pair = count.values[mask]
common_mice_per_pair
plt.figure(figsize = (14, 8), dpi = 80)
plt.hist(common_mice_per_pair, bins='auto')
plt.title('Distribution of the number of common mice between pair of SNPs')
plt.xlabel('Number of common mice between pair of SNPs')
plt.ylabel('Total count')
plt.show()
```
We decide to consider similarity only between SNPs for which we have expression data from at least 10 mice in common.
```
from scipy.spatial.distance import squareform, pdist
from sklearn.metrics import pairwise_distances, pairwise
```
_Define the distance function that we are using to build the graph._
```
def distance(u, v):
# Obtain common indexes that are non NaN for both u and v
u_valid_indexes = np.argwhere(~np.isnan(u))
v_valid_indexes = np.argwhere(~np.isnan(v))
valid_indexes = np.intersect1d(u_valid_indexes, v_valid_indexes)
# Obtain valid common vectors and length of these vectors
u_valid = u[valid_indexes]
v_valid = v[valid_indexes]
n = len(valid_indexes)
# threshold on the number of mice
if n < 10:
distance = 1*n
else:
distance = np.linalg.norm(u_valid-v_valid)
return distance / n
distances = pd.DataFrame(
squareform(pdist(expression_data,
distance)),
columns = expression_data.index,
index = expression_data.index
)
distances_matrix = distances.values
print('Matrix containing distances has shape {s}'.format(s = distances_matrix.shape))
def epsilon_similarity_graph(distances: np.ndarray, sigma=1, epsilon=0):
""" distances (n x n): matrix containing the distance between all our data points.
sigma (float): width of the kernel
epsilon (float): threshold
Return:
adjacency (n x n ndarray): adjacency matrix of the graph.
"""
W = np.exp(- distances / (2 * sigma ** 2)) # Apply the kernel to the squared distances
W[W<epsilon] = 0 # Cut off the values below epsilon
np.fill_diagonal(W, 0) # Remove the connections on the diagonal
return W
```
In order to find a good value for sigma, we first compute the median $L_2$ distance between data points, which will be our first estimate for sigma.
```
median_dist = np.median(distances_matrix)
median_dist
c = 0.7 # c is linked to the sparsity of the graph
adjacency = epsilon_similarity_graph(distances_matrix, sigma=median_dist*c, epsilon=0.1)
plt.spy(adjacency)
plt.show()
```
We tuned parameters to have a sparse graph with dominating connected component. Further we operate only on the connected component. The remaining SNP expressions are inferred as previously: they are set to the mean.
```
import networkx as nx
G = nx.from_numpy_matrix(adjacency)
node_values = {}
for i in range(expression_data.shape[0]):
node_values.update({i: {}})
for mouse in expression_data.columns.values:
mouse_expression = expression_data[mouse].values
non_nan_expressions = np.argwhere(~np.isnan(mouse_expression))
for i in range(expression_data.shape[0]):
node_values[i].update({mouse+" value": 0.0})
for i in non_nan_expressions:
i = i[0]
node_values[i].update({mouse+" value": mouse_expression[i]})
nx.set_node_attributes(G, node_values)
nx.is_connected(G)
comp = nx.connected_components(G)
components = [len(list(com)) for com in comp]
import collections
degree_sequence = sorted([d for n, d in G.degree()], reverse=True) # degree sequence
# print "Degree sequence", degree_sequence
degreeCount = collections.Counter(degree_sequence)
deg, cnt = zip(*degreeCount.items())
fig, ax = plt.subplots()
plt.bar(deg, cnt, width=0.80, color='b')
plt.title("Degree Histogram")
plt.ylabel("Count")
plt.xlabel("Degree")
plt.show()
```
We decide to use only the biggest subgraph since the disconnected components are small: each having only a few nodes that are likely far apart from the others according to our distance metric.
```
def connected_component_subgraphs(G):
for c in nx.connected_components(G):
yield G.subgraph(c)
subgraphs = list(connected_component_subgraphs(G)) # Use this for version 2.4+ of networkx
# subgraphs = list(nx.connected_component_subgraphs(G)) # Earlier versions of networkx
graph_nodes = [len(graph.degree()) for graph in subgraphs]
biggest_subgraph_id = graph_nodes.index(max(graph_nodes))
plt.hist(graph_nodes);
subgraph = subgraphs[biggest_subgraph_id]
```
Number of connected components:
```
len(subgraphs)
np.save("processed_data/coexpression_adjacency.npy", nx.to_numpy_matrix(subgraph))
np.save("processed_data/coexpression_node_indices.npy", np.array(list(subgraph.nodes)))
nx.write_gexf(subgraph, "data/graph.gexf")
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
expression_data_all_features = pd.read_pickle("processed_data/expression_standardized_cleaned.pkl")
expression_data_all_features
selected_features_df = pd.read_pickle("processed_data/selected_features.pkl")
selected_features = list(selected_features_df.columns)
print('The total number of selected features is {f}'.format(f = len(selected_features)))
expression_data = expression_data_all_features[selected_features].T
expression_data.index.name = 'snp'
expression_data
pd.to_pickle(expression_data, "processed_data/expression_data.pkl")
def compute_common_mice_per_snp_pair(expression_data):
rows = []
for index_i, row_i in expression_data.iterrows():
new_row = {}
for index_j, row_j in expression_data.iterrows():
u = row_i.values
v = row_j.values
u_valid_indexes = np.argwhere(~np.isnan(u))
v_valid_indexes = np.argwhere(~np.isnan(v))
valid_indexes = np.intersect1d(u_valid_indexes, v_valid_indexes)
n = len(valid_indexes)
new_row[index_j] = n
rows.append(new_row)
return pd.DataFrame(rows, index = expression_data.index)
count = compute_common_mice_per_snp_pair(expression_data)
count
mask = np.zeros_like(count.values, dtype=np.bool)
mask[np.tril_indices_from(mask)] = True
common_mice_per_pair = count.values[mask]
common_mice_per_pair
plt.figure(figsize = (14, 8), dpi = 80)
plt.hist(common_mice_per_pair, bins='auto')
plt.title('Distribution of the number of common mice between pair of SNPs')
plt.xlabel('Number of common mice between pair of SNPs')
plt.ylabel('Total count')
plt.show()
from scipy.spatial.distance import squareform, pdist
from sklearn.metrics import pairwise_distances, pairwise
def distance(u, v):
# Obtain common indexes that are non NaN for both u and v
u_valid_indexes = np.argwhere(~np.isnan(u))
v_valid_indexes = np.argwhere(~np.isnan(v))
valid_indexes = np.intersect1d(u_valid_indexes, v_valid_indexes)
# Obtain valid common vectors and length of these vectors
u_valid = u[valid_indexes]
v_valid = v[valid_indexes]
n = len(valid_indexes)
# threshold on the number of mice
if n < 10:
distance = 1*n
else:
distance = np.linalg.norm(u_valid-v_valid)
return distance / n
distances = pd.DataFrame(
squareform(pdist(expression_data,
distance)),
columns = expression_data.index,
index = expression_data.index
)
distances_matrix = distances.values
print('Matrix containing distances has shape {s}'.format(s = distances_matrix.shape))
def epsilon_similarity_graph(distances: np.ndarray, sigma=1, epsilon=0):
""" distances (n x n): matrix containing the distance between all our data points.
sigma (float): width of the kernel
epsilon (float): threshold
Return:
adjacency (n x n ndarray): adjacency matrix of the graph.
"""
W = np.exp(- distances / (2 * sigma ** 2)) # Apply the kernel to the squared distances
W[W<epsilon] = 0 # Cut off the values below epsilon
np.fill_diagonal(W, 0) # Remove the connections on the diagonal
return W
median_dist = np.median(distances_matrix)
median_dist
c = 0.7 # c is linked to the sparsity of the graph
adjacency = epsilon_similarity_graph(distances_matrix, sigma=median_dist*c, epsilon=0.1)
plt.spy(adjacency)
plt.show()
import networkx as nx
G = nx.from_numpy_matrix(adjacency)
node_values = {}
for i in range(expression_data.shape[0]):
node_values.update({i: {}})
for mouse in expression_data.columns.values:
mouse_expression = expression_data[mouse].values
non_nan_expressions = np.argwhere(~np.isnan(mouse_expression))
for i in range(expression_data.shape[0]):
node_values[i].update({mouse+" value": 0.0})
for i in non_nan_expressions:
i = i[0]
node_values[i].update({mouse+" value": mouse_expression[i]})
nx.set_node_attributes(G, node_values)
nx.is_connected(G)
comp = nx.connected_components(G)
components = [len(list(com)) for com in comp]
import collections
degree_sequence = sorted([d for n, d in G.degree()], reverse=True) # degree sequence
# print "Degree sequence", degree_sequence
degreeCount = collections.Counter(degree_sequence)
deg, cnt = zip(*degreeCount.items())
fig, ax = plt.subplots()
plt.bar(deg, cnt, width=0.80, color='b')
plt.title("Degree Histogram")
plt.ylabel("Count")
plt.xlabel("Degree")
plt.show()
def connected_component_subgraphs(G):
for c in nx.connected_components(G):
yield G.subgraph(c)
subgraphs = list(connected_component_subgraphs(G)) # Use this for version 2.4+ of networkx
# subgraphs = list(nx.connected_component_subgraphs(G)) # Earlier versions of networkx
graph_nodes = [len(graph.degree()) for graph in subgraphs]
biggest_subgraph_id = graph_nodes.index(max(graph_nodes))
plt.hist(graph_nodes);
subgraph = subgraphs[biggest_subgraph_id]
len(subgraphs)
np.save("processed_data/coexpression_adjacency.npy", nx.to_numpy_matrix(subgraph))
np.save("processed_data/coexpression_node_indices.npy", np.array(list(subgraph.nodes)))
nx.write_gexf(subgraph, "data/graph.gexf")
| 0.547706 | 0.947235 |
This notebook demos some functionality in ConvoKit to preprocess text, and store the results. In particular, it shows examples of:
* A `TextProcessor` base class that maps per-utterance attributes to per-utterance outputs;
* A `TextParser` class that does dependency parsing;
* Selective and decoupled data storage and loading;
* Per-utterance calls to a transformer;
* Pipelining transformers.
## Preliminaries: loading an existing corpus.
To start, we load a clean version of a corpus. For speed we will use a 200-utterance subset of the tennis corpus.
```
import os
import convokit
from convokit import download
# OPTION 1: DOWNLOAD CORPUS
# UNCOMMENT THESE LINES TO DOWNLOAD CORPUS
# DATA_DIR = '<YOUR DIRECTORY>'
# ROOT_DIR = download('tennis-corpus')
# OPTION 2: READ PREVIOUSLY-DOWNLOADED CORPUS FROM DISK
# UNCOMMENT THIS LINE AND REPLACE WITH THE DIRECTORY WHERE THE TENNIS-CORPUS IS LOCATED
# ROOT_DIR = '<YOUR DIRECTORY>'
corpus = convokit.Corpus(ROOT_DIR, utterance_end_index=199)
corpus.print_summary_stats()
# SET YOUR OWN OUTPUT DIRECTORY HERE.
OUT_DIR = '<YOUR DIRECTORY>'
```
Here's an example of an utterance from this corpus (questions asked to tennis players after matches, and the answers they give):
```
test_utt_id = '1681_14.a'
utt = corpus.get_utterance(test_utt_id)
utt.text
```
Right now, `utt.meta` contains the following fields:
```
utt.meta
```
## The TextProcessor class
Many of our transformers are per-utterance mappings of one attribute of an utterance to another. To facilitate these calls, we use a `TextProcessor` class that inherits from `Transformer`.
`TextProcessor` is initialized with the following arguments:
* `proc_fn`: the mapping function. Supports one of two function signatures: `proc_fn(input)` and `proc_fn(input, auxiliary_info)`.
* `input_field`: the attribute of the utterance that `proc_fn` will take as input. If set to `None`, will default to reading `utt.text`, as seems to be presently done.
* `output_field`: the name of the attribute that the output of `proc_fn` will be written to.
* `aux_input`: any auxiliary input that `proc_fn` needs (e.g., a pre-loaded model); passed in as a dict.
* `input_filter`: a boolean function of signature `input_filter(utterance, aux_input)`, where `aux_input` is again passed as a dict. If this returns `False` then the particular utterance will be skipped; by default it will always return `True`.
Both `input_field` and `output_field` support multiple items -- that is, `proc_fn` could take in multiple attributes of an utterance and output multiple attributes. I'll show how this works in advanced usage, below.
"Attribute" is a deliberately generic term. `TextProcessor` could produce "features" as we may conventionally think of them (e.g., wordcount, politeness strategies). It can also be used to pre-process text, i.e., generate alternate representations of the text.
```
from convokit.text_processing import TextProcessor
```
### simple example: cleaning the text
As a simple example, suppose we want to remove hyphens "`--`" from the text as a preprocessing step. To use `TextProcessor` to do this for us, we'd define the following as a `proc_fn`:
```
def preprocess_text(text):
text = text.replace(' -- ', ' ')
return text
```
Below, we initialize `prep`, a `TextProcessor` object that will run `preprocess_text` on each utterance.
When we call `prep.transform()`, the following will occur:
* Because we didn't specify an input field, `prep` will pass `utterance.text` into `preprocess_text`
* It will write the output -- the text minus the hyphens -- to a field called `clean_text` that will be stored in the utterance meta and that can be accessed as `utt.meta['clean_text']` or `utt.get_info('clean_text')`
```
prep = TextProcessor(proc_fn=preprocess_text, output_field='clean_text')
corpus = prep.transform(corpus)
```
And as desired, we now have a new field attached to `utt`.
```
utt.get_info('clean_text')
```
## Parsing text with the TextParser class
One common utterance-level thing we want to do is parse the text. In practice, in increasing order of (computational) difficulty, this typically entails:
* proper tokenizing of words and sentences;
* POS-tagging;
* dependency-parsing.
As such, we provide a `TextParser` class that inherits from `TextProcessor` to do all of this, taking in the following arguments:
* `output_field`: defaults to `'parsed'`
* `input_field`
* `mode`: whether we want to go through all of the above steps (which may be expensive) or stop mid-way through. Supports the following options: `'tokenize'`, `'tag'`, `'parse'` (the default).
Under the surface, `TextParser` actually uses two separate models: a `spacy` object that does word tokenization, tagging and parsing _per sentence_, and `nltk`'s sentence tokenizer. The rationale is:
* `spacy` doesn't support sentence tokenization without dependency-parsing, and we often want sentence tokenization without having to go through the effort of parsing.
* We want to be consistent (as much as possible, given changes to spacy and nltk) in the tokenizations we produce, between runs where we don't want parsing and runs where we do.
If we've pre-loaded these models, we can pass them into the constructor too, as:
* `spacy_nlp`
* `sent_tokenizer`
```
from convokit.text_processing import TextParser
parser = TextParser(input_field='clean_text', verbosity=50)
corpus = parser.transform(corpus)
```
### parse output
A parse produced by `TextParser` is serialized in text form. It is a list consisting of sentences, where each sentence is a dict with
* `toks`: a list of tokens (i.e., words) in the sentence;
* `rt`: the index of the root of the dependency tree (i.e., `sentence['toks'][sentence['rt']` gives the root)
Each token, in turn, contains the following:
* `tok`: the text of the token;
* `tag`: the tag;
* `up`: the index of the parent of the token in the dependency tree (no entry for the root);
* `down`: the indices of the children of the token;
* `dep`: the dependency of the edge between the token and its parent.
```
test_parse = utt.get_info('parsed')
test_parse[0]
```
If we didn't want to go through the trouble of dependency-parsing (which could be expensive) we could initialize `TextParser` with `mode='tag'`, which only POS-tags tokens:
```
texttagger = TextParser(output_field='tagged', input_field='clean_text', mode='tag')
corpus = texttagger.transform(corpus)
utt.get_info('tagged')[0]
```
## Storing and loading corpora
We've now computed a bunch of utterance-level attributes.
```
list(utt.meta.keys())
```
By default, calling `corpus.dump` will write all of these attributes to disk, within the file that stores utterances; later calling `corpus.load` will load all of these attributes back into a new corpus. For big objects like parses, this incurs a high computational burden (especially if in a later use case you might not even need to look at parses).
To avoid this, `corpus.dump` takes an optional argument `fields_to_skip`, which is a dict of object type (`'utterance'`, `'conversation'`, `'speaker'`, `'corpus'`) to a list of fields that we do not want to write to disk.
The following call will write the corpus to disk, without any of the preprocessing output we generated above:
```
corpus.dump(os.path.basename(OUT_DIR), base_path=os.path.dirname(OUT_DIR),
fields_to_skip={'utterance': ['parsed','tagged','clean_text']})
```
For attributes we want to keep around, but that we don't want to read and write to disk in a big batch with all the other corpus data, `corpus.dump_info` will dump fields of a Corpus object into separate files. This takes the following arguments as input:
* `obj_type`: which type of Corpus object you're dealing with.
* `fields`: a list of the fields to write.
* `dir_name`: which directory to write to; by default will write to the directory you read the corpus from.
This function will write each field in `fields` to a separate file called `info.<field>.jsonl` where each line of the file is a json-serialized dict: `{"id": <ID of object>, "value": <object.get_info(field)>}`.
```
corpus.dump_info('utterance',['parsed','tagged'], dir_name = OUT_DIR)
```
As expected, we now have the following files in the output directory:
```
ls $OUT_DIR
```
If we now initialize a new corpus by reading from this directory:
```
new_corpus = convokit.Corpus(OUT_DIR)
new_utt = new_corpus.get_utterance(test_utt_id)
```
We see that things that we've omitted in the `corpus.dump` call will not be read.
```
new_utt.meta.keys()
```
As a counterpart to `corpus.dump_info` we can also load auxiliary information on-demand. Here, this call will look for `info.<field>.jsonl` in the directory of `new_corpus` (or an optionally-specified `dir_name`) and attach the value specified in each line of the file to the utterance with the associated id:
```
new_corpus.load_info('utterance',['parsed'])
new_utt.get_info('parsed')
```
## Per-utterance calls
`TextProcessor` objects also support calls per-utterance via `TextProcessor.transform_utterance()`. These calls take in raw strings as well as utterances, and will return an utterance:
```
test_str = "I played -- a tennis match."
prep.transform_utterance(test_str)
from convokit.model import Utterance
adhoc_utt = Utterance(text=test_str)
adhoc_utt = prep.transform_utterance(adhoc_utt)
adhoc_utt.get_info('clean_text')
```
## Pipelines
Finally, we can string together multiple transformers, and hence `TextProcessors`, into a pipeline, using a `ConvokitPipeline` object. This is analogous to (and in fact inherits from) scikit-learn's `Pipeline` class.
```
from convokit.convokitPipeline import ConvokitPipeline
```
As an example, suppose we want to both clean the text and parse it. We can chain the required steps to get there by initializing `ConvokitPipeline` with a list of steps, represented as a tuple of `(<step name>, initialized transformer-like object)`:
* `'prep'`, our de-hyphenator
* `'parse'`, our parser
```
parse_pipe = ConvokitPipeline([('prep', TextProcessor(preprocess_text, 'clean_text_pipe')),
('parse', TextParser('parsed_pipe', input_field='clean_text_pipe',
verbosity=50))])
corpus = parse_pipe.transform(corpus)
utt.get_info('parsed_pipe')
```
As promised, the pipeline also works to transform utterances.
```
test_utt = parse_pipe.transform_utterance(test_str)
test_utt.get_info('parsed_pipe')
```
### Some advanced usage: playing around with parameters
The point of the following is to demonstrate more elaborate calls to `TextProcessor`. As an example, we will count words in an utterance.
First, we'll initialize a `TextProcessor` that does wordcounts (i.e., `len(x.split())`) on just the raw text (`utt.text`), writing output to field `wc_raw`.
```
wc_raw = TextProcessor(proc_fn=lambda x: len(x.split()), output_field='wc_raw')
corpus = wc_raw.transform(corpus)
utt.get_info('wc_raw')
```
If we instead wanted to wordcount our preprocessed text, with the hyphens removed, we can specify `input_field='clean_text'` -- as such, the `TextProcessor` will read from `utt.get_info('clean_text')` instead.
```
wc = TextProcessor(proc_fn=lambda x: len(x.split()), output_field='wc', input_field='clean_text')
corpus = wc.transform(corpus)
```
Here we see that we are no longer counting the extra hyphen.
```
utt.get_info('wc')
```
Likewise, we can count characters:
```
chars = TextProcessor(proc_fn=lambda x: len(x), output_field='ch', input_field='clean_text')
corpus = chars.transform(corpus)
utt.get_info('ch')
```
Suppose that for some reason we now wanted to calculate:
* characters per word
* words per character (the reciprocal)
This requires:
* a `TextProcessor` that takes in multiple input fields, `'ch'` and `'wc'`;
* and that writes to multiple output fields, `'char_per_word'` and `'word_per_char'`.
Here's how the resultant object, `char_per_word`, handles this:
* in `transform()`, we pass `proc_fn` a dict mapping input field name to value, e.g., `{'wc': 22, 'ch': 120}`
* `proc_fn` will be written to return a tuple, where each element of that tuple corresponds to each element of the list we've passed to `output_field`, e.g.,
```out0, out1 = proc_fn(input)
utt.set_info('char_per_word', out0)
utt.set_info('word_per_char', out1)```
```
char_per_word = TextProcessor(proc_fn=lambda x: (x['ch']/x['wc'], x['wc']/x['ch']),
output_field=['char_per_word', 'word_per_char'], input_field=['ch','wc'])
corpus = char_per_word.transform(corpus)
utt.get_info('char_per_word')
utt.get_info('word_per_char')
```
### Some advanced usage: input filters
Just for the sake of demonstration, suppose we wished to save some computation time and only parse the questions in a corpus. We can do this by specifying `input_filter` (which, recall discussion above, takes as argument an `Utterance` object).
```
def is_question(utt, aux={}):
return utt.meta['is_question']
qparser = TextParser(output_field='qparsed', input_field='clean_text', input_filter=is_question, verbosity=50)
corpus = qparser.transform(corpus)
```
Since our test utterance is not a question, `qparser.transform()` will skip over it, and hence the utterance won't have the 'qparsed' attribute (and `get_info` returns `None`):
```
utt.get_info('qparsed')
```
However, if we take an utterance that's a question, we see that it is indeed parsed:
```
q_utt_id = '1681_14.q'
q_utt = corpus.get_utterance(q_utt_id)
q_utt.text
q_utt.get_info('qparsed')
```
|
github_jupyter
|
import os
import convokit
from convokit import download
# OPTION 1: DOWNLOAD CORPUS
# UNCOMMENT THESE LINES TO DOWNLOAD CORPUS
# DATA_DIR = '<YOUR DIRECTORY>'
# ROOT_DIR = download('tennis-corpus')
# OPTION 2: READ PREVIOUSLY-DOWNLOADED CORPUS FROM DISK
# UNCOMMENT THIS LINE AND REPLACE WITH THE DIRECTORY WHERE THE TENNIS-CORPUS IS LOCATED
# ROOT_DIR = '<YOUR DIRECTORY>'
corpus = convokit.Corpus(ROOT_DIR, utterance_end_index=199)
corpus.print_summary_stats()
# SET YOUR OWN OUTPUT DIRECTORY HERE.
OUT_DIR = '<YOUR DIRECTORY>'
test_utt_id = '1681_14.a'
utt = corpus.get_utterance(test_utt_id)
utt.text
utt.meta
from convokit.text_processing import TextProcessor
def preprocess_text(text):
text = text.replace(' -- ', ' ')
return text
prep = TextProcessor(proc_fn=preprocess_text, output_field='clean_text')
corpus = prep.transform(corpus)
utt.get_info('clean_text')
from convokit.text_processing import TextParser
parser = TextParser(input_field='clean_text', verbosity=50)
corpus = parser.transform(corpus)
test_parse = utt.get_info('parsed')
test_parse[0]
texttagger = TextParser(output_field='tagged', input_field='clean_text', mode='tag')
corpus = texttagger.transform(corpus)
utt.get_info('tagged')[0]
list(utt.meta.keys())
corpus.dump(os.path.basename(OUT_DIR), base_path=os.path.dirname(OUT_DIR),
fields_to_skip={'utterance': ['parsed','tagged','clean_text']})
corpus.dump_info('utterance',['parsed','tagged'], dir_name = OUT_DIR)
ls $OUT_DIR
new_corpus = convokit.Corpus(OUT_DIR)
new_utt = new_corpus.get_utterance(test_utt_id)
new_utt.meta.keys()
new_corpus.load_info('utterance',['parsed'])
new_utt.get_info('parsed')
test_str = "I played -- a tennis match."
prep.transform_utterance(test_str)
from convokit.model import Utterance
adhoc_utt = Utterance(text=test_str)
adhoc_utt = prep.transform_utterance(adhoc_utt)
adhoc_utt.get_info('clean_text')
from convokit.convokitPipeline import ConvokitPipeline
parse_pipe = ConvokitPipeline([('prep', TextProcessor(preprocess_text, 'clean_text_pipe')),
('parse', TextParser('parsed_pipe', input_field='clean_text_pipe',
verbosity=50))])
corpus = parse_pipe.transform(corpus)
utt.get_info('parsed_pipe')
test_utt = parse_pipe.transform_utterance(test_str)
test_utt.get_info('parsed_pipe')
wc_raw = TextProcessor(proc_fn=lambda x: len(x.split()), output_field='wc_raw')
corpus = wc_raw.transform(corpus)
utt.get_info('wc_raw')
wc = TextProcessor(proc_fn=lambda x: len(x.split()), output_field='wc', input_field='clean_text')
corpus = wc.transform(corpus)
utt.get_info('wc')
chars = TextProcessor(proc_fn=lambda x: len(x), output_field='ch', input_field='clean_text')
corpus = chars.transform(corpus)
utt.get_info('ch')
### Some advanced usage: input filters
Just for the sake of demonstration, suppose we wished to save some computation time and only parse the questions in a corpus. We can do this by specifying `input_filter` (which, recall discussion above, takes as argument an `Utterance` object).
Since our test utterance is not a question, `qparser.transform()` will skip over it, and hence the utterance won't have the 'qparsed' attribute (and `get_info` returns `None`):
However, if we take an utterance that's a question, we see that it is indeed parsed:
| 0.376623 | 0.932392 |
```
# !wget https://storage.googleapis.com/bert_models/2018_11_23/multi_cased_L-12_H-768_A-12.zip
# !unzip multi_cased_L-12_H-768_A-12.zip
# !wget https://huseinhouse-storage.s3-ap-southeast-1.amazonaws.com/bert-bahasa/session-entities.pkl
# !wget https://huseinhouse-storage.s3-ap-southeast-1.amazonaws.com/bert-bahasa/dictionary-entities.json
import pickle
import json
import tensorflow as tf
import numpy as np
# !pip3 install bert-tensorflow keras --user
import bert
from bert import run_classifier
from bert import optimization
from bert import tokenization
from bert import modeling
BERT_VOCAB = 'multi_cased_L-12_H-768_A-12/vocab.txt'
BERT_INIT_CHKPNT = 'multi_cased_L-12_H-768_A-12/bert_model.ckpt'
BERT_CONFIG = 'multi_cased_L-12_H-768_A-12/bert_config.json'
tokenizer = tokenization.FullTokenizer(
vocab_file=BERT_VOCAB, do_lower_case=False)
with open('session-entities.pkl', 'rb') as fopen:
data = pickle.load(fopen)
data.keys()
train_X = data['train_X']
test_X = data['test_X']
train_Y = data['train_Y']
test_Y = data['test_Y']
with open('dictionary-entities.json') as fopen:
dictionary = json.load(fopen)
dictionary.keys()
word2idx = dictionary['word2idx']
idx2word = {int(k): v for k, v in dictionary['idx2word'].items()}
tag2idx = dictionary['tag2idx']
idx2tag = {int(k): v for k, v in dictionary['idx2tag'].items()}
char2idx = dictionary['char2idx']
idx2tag
from tqdm import tqdm
def XY(left_train, right_train):
X, Y = [], []
for i in tqdm(range(len(left_train))):
left = [idx2word[d] for d in left_train[i]]
right = [idx2tag[d] for d in right_train[i]]
bert_tokens = ['[CLS]']
y = ['PAD']
for no, orig_token in enumerate(left):
y.append(right[no])
t = tokenizer.tokenize(orig_token)
bert_tokens.extend(t)
y.extend(['X'] * (len(t) - 1))
bert_tokens.append("[SEP]")
y.append('PAD')
X.append(tokenizer.convert_tokens_to_ids(bert_tokens))
Y.append([tag2idx[i] for i in y])
return X, Y
train_X, train_Y = XY(train_X, train_Y)
test_X, test_Y = XY(test_X, test_Y)
def merge_wordpiece_tokens_tagging(x, y):
new_paired_tokens = []
n_tokens = len(x)
i = 0
while i < n_tokens:
current_token, current_label = x[i], y[i]
if current_token.startswith('##'):
previous_token, previous_label = new_paired_tokens.pop()
merged_token = previous_token
merged_label = [previous_label]
while current_token.startswith('##'):
merged_token = merged_token + current_token.replace('##', '')
merged_label.append(current_label)
i = i + 1
current_token, current_label = x[i], y[i]
merged_label = merged_label[0]
new_paired_tokens.append((merged_token, merged_label))
else:
new_paired_tokens.append((current_token, current_label))
i = i + 1
words = [
i[0]
for i in new_paired_tokens
if i[0] not in ['[CLS]', '[SEP]', '[PAD]']
]
labels = [
i[1]
for i in new_paired_tokens
if i[0] not in ['[CLS]', '[SEP]', '[PAD]']
]
return words, labels
import keras
train_X = keras.preprocessing.sequence.pad_sequences(train_X, padding='post')
train_Y = keras.preprocessing.sequence.pad_sequences(train_Y, padding='post')
test_X = keras.preprocessing.sequence.pad_sequences(test_X, padding='post')
test_Y = keras.preprocessing.sequence.pad_sequences(test_Y, padding='post')
epoch = 10
batch_size = 32
warmup_proportion = 0.1
num_train_steps = int(len(train_X) / batch_size * epoch)
num_warmup_steps = int(num_train_steps * warmup_proportion)
bert_config = modeling.BertConfig.from_json_file(BERT_CONFIG)
class Model:
def __init__(
self,
dimension_output,
learning_rate = 2e-5,
):
self.X = tf.placeholder(tf.int32, [None, None])
self.Y = tf.placeholder(tf.int32, [None, None])
self.maxlen = tf.shape(self.X)[1]
self.lengths = tf.count_nonzero(self.X, 1)
model = modeling.BertModel(
config=bert_config,
is_training=True,
input_ids=self.X,
use_one_hot_embeddings=False)
output_layer = model.get_sequence_output()
logits = tf.layers.dense(output_layer, dimension_output)
y_t = self.Y
log_likelihood, transition_params = tf.contrib.crf.crf_log_likelihood(
logits, y_t, self.lengths
)
self.cost = tf.reduce_mean(-log_likelihood)
self.optimizer = tf.train.AdamOptimizer(
learning_rate = learning_rate
).minimize(self.cost)
mask = tf.sequence_mask(self.lengths, maxlen = self.maxlen)
self.tags_seq, tags_score = tf.contrib.crf.crf_decode(
logits, transition_params, self.lengths
)
self.tags_seq = tf.identity(self.tags_seq, name = 'logits')
y_t = tf.cast(y_t, tf.int32)
self.prediction = tf.boolean_mask(self.tags_seq, mask)
mask_label = tf.boolean_mask(y_t, mask)
correct_pred = tf.equal(self.prediction, mask_label)
correct_index = tf.cast(correct_pred, tf.float32)
self.accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
dimension_output = len(tag2idx)
learning_rate = 2e-5
tf.reset_default_graph()
sess = tf.InteractiveSession()
model = Model(
dimension_output,
learning_rate
)
sess.run(tf.global_variables_initializer())
var_lists = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope = 'bert')
saver = tf.train.Saver(var_list = var_lists)
saver.restore(sess, BERT_INIT_CHKPNT)
string = 'KUALA LUMPUR: Sempena sambutan Aidilfitri minggu depan, Perdana Menteri Tun Dr Mahathir Mohamad dan Menteri Pengangkutan Anthony Loke Siew Fook menitipkan pesanan khas kepada orang ramai yang mahu pulang ke kampung halaman masing-masing. Dalam video pendek terbitan Jabatan Keselamatan Jalan Raya (JKJR) itu, Dr Mahathir menasihati mereka supaya berhenti berehat dan tidur sebentar sekiranya mengantuk ketika memandu.'
import re
def entities_textcleaning(string, lowering = False):
"""
use by entities recognition, pos recognition and dependency parsing
"""
string = re.sub('[^A-Za-z0-9\-\/() ]+', ' ', string)
string = re.sub(r'[ ]+', ' ', string).strip()
original_string = string.split()
if lowering:
string = string.lower()
string = [
(original_string[no], word.title() if word.isupper() else word)
for no, word in enumerate(string.split())
if len(word)
]
return [s[0] for s in string], [s[1] for s in string]
def parse_X(left):
bert_tokens = ['[CLS]']
for no, orig_token in enumerate(left):
t = tokenizer.tokenize(orig_token)
bert_tokens.extend(t)
bert_tokens.append("[SEP]")
return tokenizer.convert_tokens_to_ids(bert_tokens), bert_tokens
sequence = entities_textcleaning(string)[1]
parsed_sequence, bert_sequence = parse_X(sequence)
predicted = sess.run(model.tags_seq,
feed_dict = {
model.X: [parsed_sequence]
},
)[0]
merged = merge_wordpiece_tokens_tagging(bert_sequence, [idx2tag[d] for d in predicted])
list(zip(merged[0], merged[1]))
import time
for e in range(6):
lasttime = time.time()
train_acc, train_loss, test_acc, test_loss = 0, 0, 0, 0
pbar = tqdm(
range(0, len(train_X), batch_size), desc = 'train minibatch loop'
)
for i in pbar:
batch_x = train_X[i : min(i + batch_size, train_X.shape[0])]
batch_y = train_Y[i : min(i + batch_size, train_X.shape[0])]
acc, cost, _ = sess.run(
[model.accuracy, model.cost, model.optimizer],
feed_dict = {
model.X: batch_x,
model.Y: batch_y
},
)
assert not np.isnan(cost)
train_loss += cost
train_acc += acc
pbar.set_postfix(cost = cost, accuracy = acc)
pbar = tqdm(
range(0, len(test_X), batch_size), desc = 'test minibatch loop'
)
for i in pbar:
batch_x = test_X[i : min(i + batch_size, test_X.shape[0])]
batch_y = test_Y[i : min(i + batch_size, test_X.shape[0])]
acc, cost = sess.run(
[model.accuracy, model.cost],
feed_dict = {
model.X: batch_x,
model.Y: batch_y
},
)
assert not np.isnan(cost)
test_loss += cost
test_acc += acc
pbar.set_postfix(cost = cost, accuracy = acc)
train_loss /= len(train_X) / batch_size
train_acc /= len(train_X) / batch_size
test_loss /= len(test_X) / batch_size
test_acc /= len(test_X) / batch_size
print('time taken:', time.time() - lasttime)
print(
'epoch: %d, training loss: %f, training acc: %f, valid loss: %f, valid acc: %f\n'
% (e, train_loss, train_acc, test_loss, test_acc)
)
predicted = sess.run(model.tags_seq,
feed_dict = {
model.X: [parsed_sequence]
},
)[0]
merged = merge_wordpiece_tokens_tagging(bert_sequence, [idx2tag[d] for d in predicted])
print(list(zip(merged[0], merged[1])))
def pred2label(pred):
out = []
for pred_i in pred:
out_i = []
for p in pred_i:
out_i.append(idx2tag[p])
out.append(out_i)
return out
real_Y, predict_Y = [], []
pbar = tqdm(
range(0, len(test_X), batch_size), desc = 'validation minibatch loop'
)
for i in pbar:
batch_x = test_X[i : min(i + batch_size, test_X.shape[0])]
batch_y = test_Y[i : min(i + batch_size, test_X.shape[0])]
predicted = pred2label(sess.run(model.tags_seq,
feed_dict = {
model.X: batch_x
},
))
real = pred2label(batch_y)
predict_Y.extend(predicted)
real_Y.extend(real)
saver = tf.train.Saver(tf.trainable_variables())
saver.save(sess, 'bert-multilanguage-ner/model.ckpt')
from sklearn.metrics import classification_report
print(classification_report(np.array(real_Y).ravel(), np.array(predict_Y).ravel(),
digits = 6))
strings = ','.join(
[
n.name
for n in tf.get_default_graph().as_graph_def().node
if ('Variable' in n.op
or 'Placeholder' in n.name
or 'logits' in n.name
or 'alphas' in n.name
or 'self/Softmax' in n.name)
and 'Adam' not in n.name
and 'beta' not in n.name
and 'global_step' not in n.name
]
)
strings.split(',')
def freeze_graph(model_dir, output_node_names):
if not tf.gfile.Exists(model_dir):
raise AssertionError(
"Export directory doesn't exists. Please specify an export "
'directory: %s' % model_dir
)
checkpoint = tf.train.get_checkpoint_state(model_dir)
input_checkpoint = checkpoint.model_checkpoint_path
absolute_model_dir = '/'.join(input_checkpoint.split('/')[:-1])
output_graph = absolute_model_dir + '/frozen_model.pb'
clear_devices = True
with tf.Session(graph = tf.Graph()) as sess:
saver = tf.train.import_meta_graph(
input_checkpoint + '.meta', clear_devices = clear_devices
)
saver.restore(sess, input_checkpoint)
output_graph_def = tf.graph_util.convert_variables_to_constants(
sess,
tf.get_default_graph().as_graph_def(),
output_node_names.split(','),
)
with tf.gfile.GFile(output_graph, 'wb') as f:
f.write(output_graph_def.SerializeToString())
print('%d ops in the final graph.' % len(output_graph_def.node))
freeze_graph('bert-multilanguage-ner', strings)
def load_graph(frozen_graph_filename):
with tf.gfile.GFile(frozen_graph_filename, 'rb') as f:
graph_def = tf.GraphDef()
graph_def.ParseFromString(f.read())
with tf.Graph().as_default() as graph:
tf.import_graph_def(graph_def)
return graph
g = load_graph('bert-multilanguage-ner/frozen_model.pb')
x = g.get_tensor_by_name('import/Placeholder:0')
logits = g.get_tensor_by_name('import/logits:0')
test_sess = tf.InteractiveSession(graph = g)
predicted = test_sess.run(logits,
feed_dict = {
x: [parsed_sequence]
},
)[0]
merged = merge_wordpiece_tokens_tagging(bert_sequence, [idx2tag[d] for d in predicted])
print(list(zip(merged[0], merged[1])))
import boto3
bucketName = 'huseinhouse-storage'
Key = 'bert-multilanguage-ner/frozen_model.pb'
outPutname = "v27/entities/bert-multilanguage-ner.pb"
s3 = boto3.client('s3',
aws_access_key_id='',
aws_secret_access_key='')
s3.upload_file(Key,bucketName,outPutname)
```
|
github_jupyter
|
# !wget https://storage.googleapis.com/bert_models/2018_11_23/multi_cased_L-12_H-768_A-12.zip
# !unzip multi_cased_L-12_H-768_A-12.zip
# !wget https://huseinhouse-storage.s3-ap-southeast-1.amazonaws.com/bert-bahasa/session-entities.pkl
# !wget https://huseinhouse-storage.s3-ap-southeast-1.amazonaws.com/bert-bahasa/dictionary-entities.json
import pickle
import json
import tensorflow as tf
import numpy as np
# !pip3 install bert-tensorflow keras --user
import bert
from bert import run_classifier
from bert import optimization
from bert import tokenization
from bert import modeling
BERT_VOCAB = 'multi_cased_L-12_H-768_A-12/vocab.txt'
BERT_INIT_CHKPNT = 'multi_cased_L-12_H-768_A-12/bert_model.ckpt'
BERT_CONFIG = 'multi_cased_L-12_H-768_A-12/bert_config.json'
tokenizer = tokenization.FullTokenizer(
vocab_file=BERT_VOCAB, do_lower_case=False)
with open('session-entities.pkl', 'rb') as fopen:
data = pickle.load(fopen)
data.keys()
train_X = data['train_X']
test_X = data['test_X']
train_Y = data['train_Y']
test_Y = data['test_Y']
with open('dictionary-entities.json') as fopen:
dictionary = json.load(fopen)
dictionary.keys()
word2idx = dictionary['word2idx']
idx2word = {int(k): v for k, v in dictionary['idx2word'].items()}
tag2idx = dictionary['tag2idx']
idx2tag = {int(k): v for k, v in dictionary['idx2tag'].items()}
char2idx = dictionary['char2idx']
idx2tag
from tqdm import tqdm
def XY(left_train, right_train):
X, Y = [], []
for i in tqdm(range(len(left_train))):
left = [idx2word[d] for d in left_train[i]]
right = [idx2tag[d] for d in right_train[i]]
bert_tokens = ['[CLS]']
y = ['PAD']
for no, orig_token in enumerate(left):
y.append(right[no])
t = tokenizer.tokenize(orig_token)
bert_tokens.extend(t)
y.extend(['X'] * (len(t) - 1))
bert_tokens.append("[SEP]")
y.append('PAD')
X.append(tokenizer.convert_tokens_to_ids(bert_tokens))
Y.append([tag2idx[i] for i in y])
return X, Y
train_X, train_Y = XY(train_X, train_Y)
test_X, test_Y = XY(test_X, test_Y)
def merge_wordpiece_tokens_tagging(x, y):
new_paired_tokens = []
n_tokens = len(x)
i = 0
while i < n_tokens:
current_token, current_label = x[i], y[i]
if current_token.startswith('##'):
previous_token, previous_label = new_paired_tokens.pop()
merged_token = previous_token
merged_label = [previous_label]
while current_token.startswith('##'):
merged_token = merged_token + current_token.replace('##', '')
merged_label.append(current_label)
i = i + 1
current_token, current_label = x[i], y[i]
merged_label = merged_label[0]
new_paired_tokens.append((merged_token, merged_label))
else:
new_paired_tokens.append((current_token, current_label))
i = i + 1
words = [
i[0]
for i in new_paired_tokens
if i[0] not in ['[CLS]', '[SEP]', '[PAD]']
]
labels = [
i[1]
for i in new_paired_tokens
if i[0] not in ['[CLS]', '[SEP]', '[PAD]']
]
return words, labels
import keras
train_X = keras.preprocessing.sequence.pad_sequences(train_X, padding='post')
train_Y = keras.preprocessing.sequence.pad_sequences(train_Y, padding='post')
test_X = keras.preprocessing.sequence.pad_sequences(test_X, padding='post')
test_Y = keras.preprocessing.sequence.pad_sequences(test_Y, padding='post')
epoch = 10
batch_size = 32
warmup_proportion = 0.1
num_train_steps = int(len(train_X) / batch_size * epoch)
num_warmup_steps = int(num_train_steps * warmup_proportion)
bert_config = modeling.BertConfig.from_json_file(BERT_CONFIG)
class Model:
def __init__(
self,
dimension_output,
learning_rate = 2e-5,
):
self.X = tf.placeholder(tf.int32, [None, None])
self.Y = tf.placeholder(tf.int32, [None, None])
self.maxlen = tf.shape(self.X)[1]
self.lengths = tf.count_nonzero(self.X, 1)
model = modeling.BertModel(
config=bert_config,
is_training=True,
input_ids=self.X,
use_one_hot_embeddings=False)
output_layer = model.get_sequence_output()
logits = tf.layers.dense(output_layer, dimension_output)
y_t = self.Y
log_likelihood, transition_params = tf.contrib.crf.crf_log_likelihood(
logits, y_t, self.lengths
)
self.cost = tf.reduce_mean(-log_likelihood)
self.optimizer = tf.train.AdamOptimizer(
learning_rate = learning_rate
).minimize(self.cost)
mask = tf.sequence_mask(self.lengths, maxlen = self.maxlen)
self.tags_seq, tags_score = tf.contrib.crf.crf_decode(
logits, transition_params, self.lengths
)
self.tags_seq = tf.identity(self.tags_seq, name = 'logits')
y_t = tf.cast(y_t, tf.int32)
self.prediction = tf.boolean_mask(self.tags_seq, mask)
mask_label = tf.boolean_mask(y_t, mask)
correct_pred = tf.equal(self.prediction, mask_label)
correct_index = tf.cast(correct_pred, tf.float32)
self.accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
dimension_output = len(tag2idx)
learning_rate = 2e-5
tf.reset_default_graph()
sess = tf.InteractiveSession()
model = Model(
dimension_output,
learning_rate
)
sess.run(tf.global_variables_initializer())
var_lists = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope = 'bert')
saver = tf.train.Saver(var_list = var_lists)
saver.restore(sess, BERT_INIT_CHKPNT)
string = 'KUALA LUMPUR: Sempena sambutan Aidilfitri minggu depan, Perdana Menteri Tun Dr Mahathir Mohamad dan Menteri Pengangkutan Anthony Loke Siew Fook menitipkan pesanan khas kepada orang ramai yang mahu pulang ke kampung halaman masing-masing. Dalam video pendek terbitan Jabatan Keselamatan Jalan Raya (JKJR) itu, Dr Mahathir menasihati mereka supaya berhenti berehat dan tidur sebentar sekiranya mengantuk ketika memandu.'
import re
def entities_textcleaning(string, lowering = False):
"""
use by entities recognition, pos recognition and dependency parsing
"""
string = re.sub('[^A-Za-z0-9\-\/() ]+', ' ', string)
string = re.sub(r'[ ]+', ' ', string).strip()
original_string = string.split()
if lowering:
string = string.lower()
string = [
(original_string[no], word.title() if word.isupper() else word)
for no, word in enumerate(string.split())
if len(word)
]
return [s[0] for s in string], [s[1] for s in string]
def parse_X(left):
bert_tokens = ['[CLS]']
for no, orig_token in enumerate(left):
t = tokenizer.tokenize(orig_token)
bert_tokens.extend(t)
bert_tokens.append("[SEP]")
return tokenizer.convert_tokens_to_ids(bert_tokens), bert_tokens
sequence = entities_textcleaning(string)[1]
parsed_sequence, bert_sequence = parse_X(sequence)
predicted = sess.run(model.tags_seq,
feed_dict = {
model.X: [parsed_sequence]
},
)[0]
merged = merge_wordpiece_tokens_tagging(bert_sequence, [idx2tag[d] for d in predicted])
list(zip(merged[0], merged[1]))
import time
for e in range(6):
lasttime = time.time()
train_acc, train_loss, test_acc, test_loss = 0, 0, 0, 0
pbar = tqdm(
range(0, len(train_X), batch_size), desc = 'train minibatch loop'
)
for i in pbar:
batch_x = train_X[i : min(i + batch_size, train_X.shape[0])]
batch_y = train_Y[i : min(i + batch_size, train_X.shape[0])]
acc, cost, _ = sess.run(
[model.accuracy, model.cost, model.optimizer],
feed_dict = {
model.X: batch_x,
model.Y: batch_y
},
)
assert not np.isnan(cost)
train_loss += cost
train_acc += acc
pbar.set_postfix(cost = cost, accuracy = acc)
pbar = tqdm(
range(0, len(test_X), batch_size), desc = 'test minibatch loop'
)
for i in pbar:
batch_x = test_X[i : min(i + batch_size, test_X.shape[0])]
batch_y = test_Y[i : min(i + batch_size, test_X.shape[0])]
acc, cost = sess.run(
[model.accuracy, model.cost],
feed_dict = {
model.X: batch_x,
model.Y: batch_y
},
)
assert not np.isnan(cost)
test_loss += cost
test_acc += acc
pbar.set_postfix(cost = cost, accuracy = acc)
train_loss /= len(train_X) / batch_size
train_acc /= len(train_X) / batch_size
test_loss /= len(test_X) / batch_size
test_acc /= len(test_X) / batch_size
print('time taken:', time.time() - lasttime)
print(
'epoch: %d, training loss: %f, training acc: %f, valid loss: %f, valid acc: %f\n'
% (e, train_loss, train_acc, test_loss, test_acc)
)
predicted = sess.run(model.tags_seq,
feed_dict = {
model.X: [parsed_sequence]
},
)[0]
merged = merge_wordpiece_tokens_tagging(bert_sequence, [idx2tag[d] for d in predicted])
print(list(zip(merged[0], merged[1])))
def pred2label(pred):
out = []
for pred_i in pred:
out_i = []
for p in pred_i:
out_i.append(idx2tag[p])
out.append(out_i)
return out
real_Y, predict_Y = [], []
pbar = tqdm(
range(0, len(test_X), batch_size), desc = 'validation minibatch loop'
)
for i in pbar:
batch_x = test_X[i : min(i + batch_size, test_X.shape[0])]
batch_y = test_Y[i : min(i + batch_size, test_X.shape[0])]
predicted = pred2label(sess.run(model.tags_seq,
feed_dict = {
model.X: batch_x
},
))
real = pred2label(batch_y)
predict_Y.extend(predicted)
real_Y.extend(real)
saver = tf.train.Saver(tf.trainable_variables())
saver.save(sess, 'bert-multilanguage-ner/model.ckpt')
from sklearn.metrics import classification_report
print(classification_report(np.array(real_Y).ravel(), np.array(predict_Y).ravel(),
digits = 6))
strings = ','.join(
[
n.name
for n in tf.get_default_graph().as_graph_def().node
if ('Variable' in n.op
or 'Placeholder' in n.name
or 'logits' in n.name
or 'alphas' in n.name
or 'self/Softmax' in n.name)
and 'Adam' not in n.name
and 'beta' not in n.name
and 'global_step' not in n.name
]
)
strings.split(',')
def freeze_graph(model_dir, output_node_names):
if not tf.gfile.Exists(model_dir):
raise AssertionError(
"Export directory doesn't exists. Please specify an export "
'directory: %s' % model_dir
)
checkpoint = tf.train.get_checkpoint_state(model_dir)
input_checkpoint = checkpoint.model_checkpoint_path
absolute_model_dir = '/'.join(input_checkpoint.split('/')[:-1])
output_graph = absolute_model_dir + '/frozen_model.pb'
clear_devices = True
with tf.Session(graph = tf.Graph()) as sess:
saver = tf.train.import_meta_graph(
input_checkpoint + '.meta', clear_devices = clear_devices
)
saver.restore(sess, input_checkpoint)
output_graph_def = tf.graph_util.convert_variables_to_constants(
sess,
tf.get_default_graph().as_graph_def(),
output_node_names.split(','),
)
with tf.gfile.GFile(output_graph, 'wb') as f:
f.write(output_graph_def.SerializeToString())
print('%d ops in the final graph.' % len(output_graph_def.node))
freeze_graph('bert-multilanguage-ner', strings)
def load_graph(frozen_graph_filename):
with tf.gfile.GFile(frozen_graph_filename, 'rb') as f:
graph_def = tf.GraphDef()
graph_def.ParseFromString(f.read())
with tf.Graph().as_default() as graph:
tf.import_graph_def(graph_def)
return graph
g = load_graph('bert-multilanguage-ner/frozen_model.pb')
x = g.get_tensor_by_name('import/Placeholder:0')
logits = g.get_tensor_by_name('import/logits:0')
test_sess = tf.InteractiveSession(graph = g)
predicted = test_sess.run(logits,
feed_dict = {
x: [parsed_sequence]
},
)[0]
merged = merge_wordpiece_tokens_tagging(bert_sequence, [idx2tag[d] for d in predicted])
print(list(zip(merged[0], merged[1])))
import boto3
bucketName = 'huseinhouse-storage'
Key = 'bert-multilanguage-ner/frozen_model.pb'
outPutname = "v27/entities/bert-multilanguage-ner.pb"
s3 = boto3.client('s3',
aws_access_key_id='',
aws_secret_access_key='')
s3.upload_file(Key,bucketName,outPutname)
| 0.549157 | 0.290943 |
# Objective
* Make a baseline model that predict the validation (28 days).
* This competition has 2 stages, so the main objective is to make a model that can predict the demand for the next 28 days
## Introduction:
Hi everyone welcome to this kernel,In this kernel i am building basic LightGBM model on timeseriessplits.This notebook was build on top of this [kernel](https://www.kaggle.com/ragnar123/very-fst-model).Thanks to the author of the kernel [ragnar](https://www.kaggle.com/ragnar123) <br>
### <font color='red'>If you find this kernel useful please consider upvoting 😊 It will keep me motivated to produce more quality content.Also dont forget to upvote the original kernel.</font>
```
import warnings
warnings.filterwarnings('ignore')
import pandas as pd
import numpy as np
import dask.dataframe as dd
pd.set_option('display.max_columns', 500)
pd.set_option('display.max_rows', 500)
import matplotlib.pyplot as plt
import seaborn as sns
import lightgbm as lgb
import dask_xgboost as xgb
import dask.dataframe as dd
from sklearn import preprocessing, metrics
from sklearn.model_selection import StratifiedKFold, KFold, RepeatedKFold, GroupKFold, GridSearchCV, train_test_split, TimeSeriesSplit
import gc
import os
for dirname, _, filenames in os.walk('/kaggle/input'):
for filename in filenames:
print(os.path.join(dirname, filename))
def reduce_mem_usage(df, verbose=True):
numerics = ['int16', 'int32', 'int64', 'float16', 'float32', 'float64']
start_mem = df.memory_usage().sum() / 1024**2
for col in df.columns:
col_type = df[col].dtypes
if col_type in numerics:
c_min = df[col].min()
c_max = df[col].max()
if str(col_type)[:3] == 'int':
if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:
df[col] = df[col].astype(np.int8)
elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:
df[col] = df[col].astype(np.int16)
elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:
df[col] = df[col].astype(np.int32)
elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:
df[col] = df[col].astype(np.int64)
else:
if c_min > np.finfo(np.float16).min and c_max < np.finfo(np.float16).max:
df[col] = df[col].astype(np.float16)
elif c_min > np.finfo(np.float32).min and c_max < np.finfo(np.float32).max:
df[col] = df[col].astype(np.float32)
else:
df[col] = df[col].astype(np.float64)
end_mem = df.memory_usage().sum() / 1024**2
if verbose: print('Mem. usage decreased to {:5.2f} Mb ({:.1f}% reduction)'.format(end_mem, 100 * (start_mem - end_mem) / start_mem))
return df
# function to read the data and merge it (ignoring some columns, this is a very fst model)
def read_data():
print('Reading files...')
calendar = pd.read_csv('/kaggle/input/m5-forecasting-accuracy/calendar.csv')
calendar = reduce_mem_usage(calendar)
print('Calendar has {} rows and {} columns'.format(calendar.shape[0], calendar.shape[1]))
sell_prices = pd.read_csv('/kaggle/input/m5-forecasting-accuracy/sell_prices.csv')
sell_prices = reduce_mem_usage(sell_prices)
print('Sell prices has {} rows and {} columns'.format(sell_prices.shape[0], sell_prices.shape[1]))
sales_train_validation = pd.read_csv('/kaggle/input/m5-forecasting-accuracy/sales_train_validation.csv')
print('Sales train validation has {} rows and {} columns'.format(sales_train_validation.shape[0], sales_train_validation.shape[1]))
submission = pd.read_csv('/kaggle/input/m5-forecasting-accuracy/sample_submission.csv')
return calendar, sell_prices, sales_train_validation, submission
def melt_and_merge(calendar, sell_prices, sales_train_validation, submission, nrows = 55000000, merge = False):
# melt sales data, get it ready for training
sales_train_validation = pd.melt(sales_train_validation, id_vars = ['id', 'item_id', 'dept_id', 'cat_id', 'store_id', 'state_id'], var_name = 'day', value_name = 'demand')
print('Melted sales train validation has {} rows and {} columns'.format(sales_train_validation.shape[0], sales_train_validation.shape[1]))
sales_train_validation = reduce_mem_usage(sales_train_validation)
# seperate test dataframes
test1_rows = [row for row in submission['id'] if 'validation' in row]
test2_rows = [row for row in submission['id'] if 'evaluation' in row]
test1 = submission[submission['id'].isin(test1_rows)]
test2 = submission[submission['id'].isin(test2_rows)]
# change column names
test1.columns = ['id', 'd_1914', 'd_1915', 'd_1916', 'd_1917', 'd_1918', 'd_1919', 'd_1920', 'd_1921', 'd_1922', 'd_1923', 'd_1924', 'd_1925', 'd_1926', 'd_1927', 'd_1928', 'd_1929', 'd_1930', 'd_1931',
'd_1932', 'd_1933', 'd_1934', 'd_1935', 'd_1936', 'd_1937', 'd_1938', 'd_1939', 'd_1940', 'd_1941']
test2.columns = ['id', 'd_1942', 'd_1943', 'd_1944', 'd_1945', 'd_1946', 'd_1947', 'd_1948', 'd_1949', 'd_1950', 'd_1951', 'd_1952', 'd_1953', 'd_1954', 'd_1955', 'd_1956', 'd_1957', 'd_1958', 'd_1959',
'd_1960', 'd_1961', 'd_1962', 'd_1963', 'd_1964', 'd_1965', 'd_1966', 'd_1967', 'd_1968', 'd_1969']
# get product table
product = sales_train_validation[['id', 'item_id', 'dept_id', 'cat_id', 'store_id', 'state_id']].drop_duplicates()
# merge with product table
test1 = test1.merge(product, how = 'left', on = 'id')
test2 = test2.merge(product, how = 'left', on = 'id')
#
test1 = pd.melt(test1, id_vars = ['id', 'item_id', 'dept_id', 'cat_id', 'store_id', 'state_id'], var_name = 'day', value_name = 'demand')
test2 = pd.melt(test2, id_vars = ['id', 'item_id', 'dept_id', 'cat_id', 'store_id', 'state_id'], var_name = 'day', value_name = 'demand')
sales_train_validation['part'] = 'train'
test1['part'] = 'test1'
test2['part'] = 'test2'
data = pd.concat([sales_train_validation, test1, test2], axis = 0)
del sales_train_validation, test1, test2
# get only a sample for fst training
data = data.loc[nrows:]
# drop some calendar features
calendar.drop(['weekday', 'wday', 'month', 'year'], inplace = True, axis = 1)
# delete test2 for now
data = data[data['part'] != 'test2']
if merge:
# notebook crash with the entire dataset (maybee use tensorflow, dask, pyspark xD)
data = pd.merge(data, calendar, how = 'left', left_on = ['day'], right_on = ['d'])
data.drop(['d', 'day'], inplace = True, axis = 1)
# get the sell price data (this feature should be very important)
data = data.merge(sell_prices, on = ['store_id', 'item_id', 'wm_yr_wk'], how = 'left')
print('Our final dataset to train has {} rows and {} columns'.format(data.shape[0], data.shape[1]))
else:
pass
gc.collect()
return data
calendar, sell_prices, sales_train_validation, submission = read_data()
data = melt_and_merge(calendar, sell_prices, sales_train_validation, submission, nrows = 27500000, merge = True)
gc.collect()
def transform(data):
nan_features = ['event_name_1', 'event_type_1', 'event_name_2', 'event_type_2']
for feature in nan_features:
data[feature].fillna('unknown', inplace = True)
encoder = preprocessing.LabelEncoder()
data['id_encode'] = encoder.fit_transform(data['id'])
cat = ['item_id', 'dept_id', 'cat_id', 'store_id', 'state_id', 'event_name_1', 'event_type_1', 'event_name_2', 'event_type_2']
for feature in cat:
encoder = preprocessing.LabelEncoder()
data[feature] = encoder.fit_transform(data[feature])
return data
data = transform(data)
gc.collect()
def simple_fe(data):
# demand features
data['lag_t28'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28))
data['lag_t29'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(29))
data['lag_t30'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(30))
data['rolling_mean_t7'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(7).mean())
data['rolling_std_t7'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(7).std())
data['rolling_mean_t30'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(30).mean())
data['rolling_mean_t90'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(90).mean())
data['rolling_mean_t180'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(180).mean())
data['rolling_std_t30'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(30).std())
# price features
data['lag_price_t1'] = data.groupby(['id'])['sell_price'].transform(lambda x: x.shift(1))
data['price_change_t1'] = (data['lag_price_t1'] - data['sell_price']) / (data['lag_price_t1'])
data['rolling_price_max_t365'] = data.groupby(['id'])['sell_price'].transform(lambda x: x.shift(1).rolling(365).max())
data['price_change_t365'] = (data['rolling_price_max_t365'] - data['sell_price']) / (data['rolling_price_max_t365'])
data['rolling_price_std_t7'] = data.groupby(['id'])['sell_price'].transform(lambda x: x.rolling(7).std())
data['rolling_price_std_t30'] = data.groupby(['id'])['sell_price'].transform(lambda x: x.rolling(30).std())
data.drop(['rolling_price_max_t365', 'lag_price_t1'], inplace = True, axis = 1)
# time features
data['date'] = pd.to_datetime(data['date'])
data['year'] = data['date'].dt.year
data['month'] = data['date'].dt.month
data['week'] = data['date'].dt.week
data['day'] = data['date'].dt.day
data['dayofweek'] = data['date'].dt.dayofweek
return data
data = simple_fe(data)
data = reduce_mem_usage(data)
gc.collect()
x = data[data['date'] <= '2016-04-24']
y = x.sort_values('date')['demand']
test = data[(data['date'] > '2016-04-24')]
x = x.sort_values('date')
test = test.sort_values('date')
del data
n_fold = 3 #3 for timely purpose of the kernel
folds = TimeSeriesSplit(n_splits=n_fold)
params = {'num_leaves': 555,
'min_child_weight': 0.034,
'feature_fraction': 0.379,
'bagging_fraction': 0.418,
'min_data_in_leaf': 106,
'objective': 'regression',
'max_depth': -1,
'learning_rate': 0.005,
"boosting_type": "gbdt",
"bagging_seed": 11,
"metric": 'rmse',
"verbosity": -1,
'reg_alpha': 0.3899,
'reg_lambda': 0.648,
'random_state': 222,
}
columns = ['item_id', 'dept_id', 'cat_id', 'store_id', 'state_id', 'year', 'month', 'week', 'day', 'dayofweek', 'event_name_1', 'event_type_1', 'event_name_2', 'event_type_2',
'snap_CA', 'snap_TX', 'snap_WI', 'sell_price', 'lag_t28', 'lag_t29', 'lag_t30', 'rolling_mean_t7', 'rolling_std_t7', 'rolling_mean_t30', 'rolling_mean_t90',
'rolling_mean_t180', 'rolling_std_t30', 'price_change_t1', 'price_change_t365', 'rolling_price_std_t7', 'rolling_price_std_t30']
splits = folds.split(x, y)
y_preds = np.zeros(test.shape[0])
y_oof = np.zeros(x.shape[0])
feature_importances = pd.DataFrame()
feature_importances['feature'] = columns
mean_score = []
for fold_n, (train_index, valid_index) in enumerate(splits):
print('Fold:',fold_n+1)
X_train, X_valid = x[columns].iloc[train_index], x[columns].iloc[valid_index]
y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]
dtrain = lgb.Dataset(X_train, label=y_train)
dvalid = lgb.Dataset(X_valid, label=y_valid)
clf = lgb.train(params, dtrain, 2500, valid_sets = [dtrain, dvalid],early_stopping_rounds = 50, verbose_eval=100)
feature_importances[f'fold_{fold_n + 1}'] = clf.feature_importance()
y_pred_valid = clf.predict(X_valid,num_iteration=clf.best_iteration)
y_oof[valid_index] = y_pred_valid
val_score = np.sqrt(metrics.mean_squared_error(y_pred_valid, y_valid))
print(f'val rmse score is {val_score}')
mean_score.append(val_score)
y_preds += clf.predict(test[columns], num_iteration=clf.best_iteration)/n_fold
del X_train, X_valid, y_train, y_valid
gc.collect()
print('mean rmse score over folds is',np.mean(mean_score))
test['demand'] = y_preds
def predict(test, submission):
predictions = test[['id', 'date', 'demand']]
predictions = pd.pivot(predictions, index = 'id', columns = 'date', values = 'demand').reset_index()
predictions.columns = ['id'] + ['F' + str(i + 1) for i in range(28)]
evaluation_rows = [row for row in submission['id'] if 'evaluation' in row]
evaluation = submission[submission['id'].isin(evaluation_rows)]
validation = submission[['id']].merge(predictions, on = 'id')
final = pd.concat([validation, evaluation])
#final.to_csv('submission.csv', index = False)
return final
subs = predict(test, submission)
subs.to_csv('submission.csv',index = False)
subs.head()
```
## Feature Importances:
```
import seaborn as sns
feature_importances['average'] = feature_importances[[f'fold_{fold_n + 1}' for fold_n in range(folds.n_splits)]].mean(axis=1)
feature_importances.to_csv('feature_importances.csv')
plt.figure(figsize=(16, 12))
sns.barplot(data=feature_importances.sort_values(by='average', ascending=False).head(20), x='average', y='feature');
plt.title('20 TOP feature importance over {} folds average'.format(folds.n_splits));
```
|
github_jupyter
|
import warnings
warnings.filterwarnings('ignore')
import pandas as pd
import numpy as np
import dask.dataframe as dd
pd.set_option('display.max_columns', 500)
pd.set_option('display.max_rows', 500)
import matplotlib.pyplot as plt
import seaborn as sns
import lightgbm as lgb
import dask_xgboost as xgb
import dask.dataframe as dd
from sklearn import preprocessing, metrics
from sklearn.model_selection import StratifiedKFold, KFold, RepeatedKFold, GroupKFold, GridSearchCV, train_test_split, TimeSeriesSplit
import gc
import os
for dirname, _, filenames in os.walk('/kaggle/input'):
for filename in filenames:
print(os.path.join(dirname, filename))
def reduce_mem_usage(df, verbose=True):
numerics = ['int16', 'int32', 'int64', 'float16', 'float32', 'float64']
start_mem = df.memory_usage().sum() / 1024**2
for col in df.columns:
col_type = df[col].dtypes
if col_type in numerics:
c_min = df[col].min()
c_max = df[col].max()
if str(col_type)[:3] == 'int':
if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:
df[col] = df[col].astype(np.int8)
elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:
df[col] = df[col].astype(np.int16)
elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:
df[col] = df[col].astype(np.int32)
elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:
df[col] = df[col].astype(np.int64)
else:
if c_min > np.finfo(np.float16).min and c_max < np.finfo(np.float16).max:
df[col] = df[col].astype(np.float16)
elif c_min > np.finfo(np.float32).min and c_max < np.finfo(np.float32).max:
df[col] = df[col].astype(np.float32)
else:
df[col] = df[col].astype(np.float64)
end_mem = df.memory_usage().sum() / 1024**2
if verbose: print('Mem. usage decreased to {:5.2f} Mb ({:.1f}% reduction)'.format(end_mem, 100 * (start_mem - end_mem) / start_mem))
return df
# function to read the data and merge it (ignoring some columns, this is a very fst model)
def read_data():
print('Reading files...')
calendar = pd.read_csv('/kaggle/input/m5-forecasting-accuracy/calendar.csv')
calendar = reduce_mem_usage(calendar)
print('Calendar has {} rows and {} columns'.format(calendar.shape[0], calendar.shape[1]))
sell_prices = pd.read_csv('/kaggle/input/m5-forecasting-accuracy/sell_prices.csv')
sell_prices = reduce_mem_usage(sell_prices)
print('Sell prices has {} rows and {} columns'.format(sell_prices.shape[0], sell_prices.shape[1]))
sales_train_validation = pd.read_csv('/kaggle/input/m5-forecasting-accuracy/sales_train_validation.csv')
print('Sales train validation has {} rows and {} columns'.format(sales_train_validation.shape[0], sales_train_validation.shape[1]))
submission = pd.read_csv('/kaggle/input/m5-forecasting-accuracy/sample_submission.csv')
return calendar, sell_prices, sales_train_validation, submission
def melt_and_merge(calendar, sell_prices, sales_train_validation, submission, nrows = 55000000, merge = False):
# melt sales data, get it ready for training
sales_train_validation = pd.melt(sales_train_validation, id_vars = ['id', 'item_id', 'dept_id', 'cat_id', 'store_id', 'state_id'], var_name = 'day', value_name = 'demand')
print('Melted sales train validation has {} rows and {} columns'.format(sales_train_validation.shape[0], sales_train_validation.shape[1]))
sales_train_validation = reduce_mem_usage(sales_train_validation)
# seperate test dataframes
test1_rows = [row for row in submission['id'] if 'validation' in row]
test2_rows = [row for row in submission['id'] if 'evaluation' in row]
test1 = submission[submission['id'].isin(test1_rows)]
test2 = submission[submission['id'].isin(test2_rows)]
# change column names
test1.columns = ['id', 'd_1914', 'd_1915', 'd_1916', 'd_1917', 'd_1918', 'd_1919', 'd_1920', 'd_1921', 'd_1922', 'd_1923', 'd_1924', 'd_1925', 'd_1926', 'd_1927', 'd_1928', 'd_1929', 'd_1930', 'd_1931',
'd_1932', 'd_1933', 'd_1934', 'd_1935', 'd_1936', 'd_1937', 'd_1938', 'd_1939', 'd_1940', 'd_1941']
test2.columns = ['id', 'd_1942', 'd_1943', 'd_1944', 'd_1945', 'd_1946', 'd_1947', 'd_1948', 'd_1949', 'd_1950', 'd_1951', 'd_1952', 'd_1953', 'd_1954', 'd_1955', 'd_1956', 'd_1957', 'd_1958', 'd_1959',
'd_1960', 'd_1961', 'd_1962', 'd_1963', 'd_1964', 'd_1965', 'd_1966', 'd_1967', 'd_1968', 'd_1969']
# get product table
product = sales_train_validation[['id', 'item_id', 'dept_id', 'cat_id', 'store_id', 'state_id']].drop_duplicates()
# merge with product table
test1 = test1.merge(product, how = 'left', on = 'id')
test2 = test2.merge(product, how = 'left', on = 'id')
#
test1 = pd.melt(test1, id_vars = ['id', 'item_id', 'dept_id', 'cat_id', 'store_id', 'state_id'], var_name = 'day', value_name = 'demand')
test2 = pd.melt(test2, id_vars = ['id', 'item_id', 'dept_id', 'cat_id', 'store_id', 'state_id'], var_name = 'day', value_name = 'demand')
sales_train_validation['part'] = 'train'
test1['part'] = 'test1'
test2['part'] = 'test2'
data = pd.concat([sales_train_validation, test1, test2], axis = 0)
del sales_train_validation, test1, test2
# get only a sample for fst training
data = data.loc[nrows:]
# drop some calendar features
calendar.drop(['weekday', 'wday', 'month', 'year'], inplace = True, axis = 1)
# delete test2 for now
data = data[data['part'] != 'test2']
if merge:
# notebook crash with the entire dataset (maybee use tensorflow, dask, pyspark xD)
data = pd.merge(data, calendar, how = 'left', left_on = ['day'], right_on = ['d'])
data.drop(['d', 'day'], inplace = True, axis = 1)
# get the sell price data (this feature should be very important)
data = data.merge(sell_prices, on = ['store_id', 'item_id', 'wm_yr_wk'], how = 'left')
print('Our final dataset to train has {} rows and {} columns'.format(data.shape[0], data.shape[1]))
else:
pass
gc.collect()
return data
calendar, sell_prices, sales_train_validation, submission = read_data()
data = melt_and_merge(calendar, sell_prices, sales_train_validation, submission, nrows = 27500000, merge = True)
gc.collect()
def transform(data):
nan_features = ['event_name_1', 'event_type_1', 'event_name_2', 'event_type_2']
for feature in nan_features:
data[feature].fillna('unknown', inplace = True)
encoder = preprocessing.LabelEncoder()
data['id_encode'] = encoder.fit_transform(data['id'])
cat = ['item_id', 'dept_id', 'cat_id', 'store_id', 'state_id', 'event_name_1', 'event_type_1', 'event_name_2', 'event_type_2']
for feature in cat:
encoder = preprocessing.LabelEncoder()
data[feature] = encoder.fit_transform(data[feature])
return data
data = transform(data)
gc.collect()
def simple_fe(data):
# demand features
data['lag_t28'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28))
data['lag_t29'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(29))
data['lag_t30'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(30))
data['rolling_mean_t7'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(7).mean())
data['rolling_std_t7'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(7).std())
data['rolling_mean_t30'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(30).mean())
data['rolling_mean_t90'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(90).mean())
data['rolling_mean_t180'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(180).mean())
data['rolling_std_t30'] = data.groupby(['id'])['demand'].transform(lambda x: x.shift(28).rolling(30).std())
# price features
data['lag_price_t1'] = data.groupby(['id'])['sell_price'].transform(lambda x: x.shift(1))
data['price_change_t1'] = (data['lag_price_t1'] - data['sell_price']) / (data['lag_price_t1'])
data['rolling_price_max_t365'] = data.groupby(['id'])['sell_price'].transform(lambda x: x.shift(1).rolling(365).max())
data['price_change_t365'] = (data['rolling_price_max_t365'] - data['sell_price']) / (data['rolling_price_max_t365'])
data['rolling_price_std_t7'] = data.groupby(['id'])['sell_price'].transform(lambda x: x.rolling(7).std())
data['rolling_price_std_t30'] = data.groupby(['id'])['sell_price'].transform(lambda x: x.rolling(30).std())
data.drop(['rolling_price_max_t365', 'lag_price_t1'], inplace = True, axis = 1)
# time features
data['date'] = pd.to_datetime(data['date'])
data['year'] = data['date'].dt.year
data['month'] = data['date'].dt.month
data['week'] = data['date'].dt.week
data['day'] = data['date'].dt.day
data['dayofweek'] = data['date'].dt.dayofweek
return data
data = simple_fe(data)
data = reduce_mem_usage(data)
gc.collect()
x = data[data['date'] <= '2016-04-24']
y = x.sort_values('date')['demand']
test = data[(data['date'] > '2016-04-24')]
x = x.sort_values('date')
test = test.sort_values('date')
del data
n_fold = 3 #3 for timely purpose of the kernel
folds = TimeSeriesSplit(n_splits=n_fold)
params = {'num_leaves': 555,
'min_child_weight': 0.034,
'feature_fraction': 0.379,
'bagging_fraction': 0.418,
'min_data_in_leaf': 106,
'objective': 'regression',
'max_depth': -1,
'learning_rate': 0.005,
"boosting_type": "gbdt",
"bagging_seed": 11,
"metric": 'rmse',
"verbosity": -1,
'reg_alpha': 0.3899,
'reg_lambda': 0.648,
'random_state': 222,
}
columns = ['item_id', 'dept_id', 'cat_id', 'store_id', 'state_id', 'year', 'month', 'week', 'day', 'dayofweek', 'event_name_1', 'event_type_1', 'event_name_2', 'event_type_2',
'snap_CA', 'snap_TX', 'snap_WI', 'sell_price', 'lag_t28', 'lag_t29', 'lag_t30', 'rolling_mean_t7', 'rolling_std_t7', 'rolling_mean_t30', 'rolling_mean_t90',
'rolling_mean_t180', 'rolling_std_t30', 'price_change_t1', 'price_change_t365', 'rolling_price_std_t7', 'rolling_price_std_t30']
splits = folds.split(x, y)
y_preds = np.zeros(test.shape[0])
y_oof = np.zeros(x.shape[0])
feature_importances = pd.DataFrame()
feature_importances['feature'] = columns
mean_score = []
for fold_n, (train_index, valid_index) in enumerate(splits):
print('Fold:',fold_n+1)
X_train, X_valid = x[columns].iloc[train_index], x[columns].iloc[valid_index]
y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]
dtrain = lgb.Dataset(X_train, label=y_train)
dvalid = lgb.Dataset(X_valid, label=y_valid)
clf = lgb.train(params, dtrain, 2500, valid_sets = [dtrain, dvalid],early_stopping_rounds = 50, verbose_eval=100)
feature_importances[f'fold_{fold_n + 1}'] = clf.feature_importance()
y_pred_valid = clf.predict(X_valid,num_iteration=clf.best_iteration)
y_oof[valid_index] = y_pred_valid
val_score = np.sqrt(metrics.mean_squared_error(y_pred_valid, y_valid))
print(f'val rmse score is {val_score}')
mean_score.append(val_score)
y_preds += clf.predict(test[columns], num_iteration=clf.best_iteration)/n_fold
del X_train, X_valid, y_train, y_valid
gc.collect()
print('mean rmse score over folds is',np.mean(mean_score))
test['demand'] = y_preds
def predict(test, submission):
predictions = test[['id', 'date', 'demand']]
predictions = pd.pivot(predictions, index = 'id', columns = 'date', values = 'demand').reset_index()
predictions.columns = ['id'] + ['F' + str(i + 1) for i in range(28)]
evaluation_rows = [row for row in submission['id'] if 'evaluation' in row]
evaluation = submission[submission['id'].isin(evaluation_rows)]
validation = submission[['id']].merge(predictions, on = 'id')
final = pd.concat([validation, evaluation])
#final.to_csv('submission.csv', index = False)
return final
subs = predict(test, submission)
subs.to_csv('submission.csv',index = False)
subs.head()
import seaborn as sns
feature_importances['average'] = feature_importances[[f'fold_{fold_n + 1}' for fold_n in range(folds.n_splits)]].mean(axis=1)
feature_importances.to_csv('feature_importances.csv')
plt.figure(figsize=(16, 12))
sns.barplot(data=feature_importances.sort_values(by='average', ascending=False).head(20), x='average', y='feature');
plt.title('20 TOP feature importance over {} folds average'.format(folds.n_splits));
| 0.331877 | 0.765243 |
## How to do it...
### Import relevant libaries
import tensorflow as tf
import numpy as np
import pandas as pd
from IPython.display import clear_output
from matplotlib import pyplot as plt
import matplotlib.pyplot as plt
import seaborn as sns
sns_colors = sns.color_palette('colorblind')
from numpy.random import uniform, seed
from scipy.interpolate import griddata
from matplotlib.font_manager import FontProperties
from sklearn.metrics import roc_curve
## Boilerplate Code to Plot
```
def _get_color(value):
"""To make positive DFCs plot green, negative DFCs plot red."""
green, red = sns.color_palette()[2:4]
if value >= 0: return green
return red
def _add_feature_values(feature_values, ax):
"""Display feature's values on left of plot."""
x_coord = ax.get_xlim()[0]
OFFSET = 0.15
for y_coord, (feat_name, feat_val) in enumerate(feature_values.items()):
t = plt.text(x_coord, y_coord - OFFSET, '{}'.format(feat_val), size=12)
t.set_bbox(dict(facecolor='white', alpha=0.5))
from matplotlib.font_manager import FontProperties
font = FontProperties()
font.set_weight('bold')
t = plt.text(x_coord, y_coord + 1 - OFFSET, 'feature\nvalue',
fontproperties=font, size=12)
def plot_example(example):
TOP_N = 8 # View top 8 features.
sorted_ix = example.abs().sort_values()[-TOP_N:].index # Sort by magnitude.
example = example[sorted_ix]
colors = example.map(_get_color).tolist()
ax = example.to_frame().plot(kind='barh',
color=[colors],
legend=None,
alpha=0.75,
figsize=(10,6))
ax.grid(False, axis='y')
ax.set_yticklabels(ax.get_yticklabels(), size=14)
# Add feature values.
_add_feature_values(xvalid.iloc[ID][sorted_ix], ax)
return ax
def permutation_importances(est, X_eval, y_eval, metric, features):
"""Column by column, shuffle values and observe effect on eval set.
source: http://explained.ai/rf-importance/index.html
A similar approach can be done during training. See "Drop-column importance"
in the above article."""
baseline = metric(est, X_eval, y_eval)
imp = []
for col in features:
save = X_eval[col].copy()
X_eval[col] = np.random.permutation(X_eval[col])
m = metric(est, X_eval, y_eval)
X_eval[col] = save
imp.append(baseline - m)
return np.array(imp)
def accuracy_metric(est, X, y):
"""TensorFlow estimator accuracy."""
eval_input_fn = make_input_fn(X,
y=y,
shuffle=False,
n_epochs=1)
return est.evaluate(input_fn=eval_input_fn)['accuracy']
```
### Import data
```
xtrain = pd.read_csv('hotel_bookings.csv')
xtrain.head(3)
```
### Create x and y train and validation data sets
```
xvalid = xtrain.loc[xtrain['reservation_status_date'] >= '2017-08-01']
xtrain = xtrain.loc[xtrain['reservation_status_date'] < '2017-08-01']
ytrain, yvalid = xtrain['is_canceled'], xvalid['is_canceled']
xtrain.drop('is_canceled', axis = 1, inplace = True)
xvalid.drop('is_canceled', axis = 1, inplace = True)
```
### Drop columns that are irrelevant or may introduce data leak
```
xtrain.drop(['arrival_date_year','assigned_room_type', 'booking_changes', 'reservation_status',
'country', 'days_in_waiting_list'], axis =1, inplace = True)
```
### Specify numerical and categorical feature columns
```
num_features = ["lead_time","arrival_date_week_number","arrival_date_day_of_month",
"stays_in_weekend_nights","stays_in_week_nights","adults","children",
"babies","is_repeated_guest", "previous_cancellations",
"previous_bookings_not_canceled","agent","company",
"required_car_parking_spaces", "total_of_special_requests", "adr"]
cat_features = ["hotel","arrival_date_month","meal","market_segment",
"distribution_channel","reserved_room_type","deposit_type","customer_type"]
```
### Create one hot categorical column encoder
```
def one_hot_cat_column(feature_name, vocab):
return tf.feature_column.indicator_column(
tf.feature_column.categorical_column_with_vocabulary_list(feature_name,
vocab))
```
### Create feature columns list
```
feature_columns = []
for feature_name in cat_features:
# Need to one-hot encode categorical features.
vocabulary = xtrain[feature_name].unique()
feature_columns.append(one_hot_cat_column(feature_name, vocabulary))
for feature_name in num_features:
feature_columns.append(tf.feature_column.numeric_column(feature_name,
dtype=tf.float32))
```
### Create input function for training and inference
```
# Use entire batch since this is such a small dataset.
NUM_EXAMPLES = len(ytrain)
def make_input_fn(X, y, n_epochs=None, shuffle=True):
def input_fn():
dataset = tf.data.Dataset.from_tensor_slices((dict(X), y))
if shuffle:
dataset = dataset.shuffle(NUM_EXAMPLES)
# For training, cycle thru dataset as many times as need (n_epochs=None).
dataset = dataset.repeat(n_epochs)
# In memory training doesn't use batching.
dataset = dataset.batch(NUM_EXAMPLES)
return dataset
return input_fn
# Training and evaluation input functions.
train_input_fn = make_input_fn(xtrain, ytrain)
eval_input_fn = make_input_fn(xvalid, yvalid, shuffle=False, n_epochs=1)
```
### Build the BoostedTrees model
```
params = {
'n_trees': 125,
'max_depth': 5,
'n_batches_per_layer': 1,
# 'learning_rate': 0.05,
# 'l1_regularization': 0.00001,
# 'l2_regularization': 0.00001,
# 'min_node_weight': 0.01,
# You must enable center_bias = True to get DFCs. This will force the model to
# make an initial prediction before using any features (e.g. use the mean of
# the training labels for regression or log odds for classification when
# using cross entropy loss).
'center_bias': True
}
est = tf.estimator.BoostedTreesClassifier(feature_columns, **params)
# Train model.
est.train(train_input_fn, max_steps=100)
# Evaluation
results = est.evaluate(eval_input_fn)
pd.Series(results).to_frame()
pred_dicts = list(est.predict(eval_input_fn))
probs = pd.Series([pred['probabilities'][1] for pred in pred_dicts])
fpr, tpr, _ = roc_curve(yvalid, probs)
plt.plot(fpr, tpr)
plt.title('ROC curve')
plt.xlabel('false positive rate')
plt.ylabel('true positive rate')
plt.xlim(0,); plt.ylim(0,); plt.show()
pred_dicts = list(est.experimental_predict_with_explanations(eval_input_fn))
# Create DFC Pandas dataframe
labels = yvalid.values
probs = pd.Series([pred['probabilities'][1] for pred in pred_dicts])
df_dfc = pd.DataFrame([pred['dfc'] for pred in pred_dicts])
df_dfc.describe().T
```
## How it works...
The following code block demonstrates the steps necessary to extract the feature contributions to a prediction for a particular record. For convenience and reusability, we define a function plotting a chosen record first (for easier interpretation, we want to plot feature importances using different colors, depending on whether their contribution is positive or negative):
With the boilerplate code defined, we plot the detailed graph for a specific record in a straight forward manner:
```
ID = 10
example = df_dfc.iloc[ID] # Choose ith example from evaluation set.
TOP_N = 8 # View top 8 features
sorted_ix = example.abs().sort_values()[-TOP_N:].index
ax = plot_example(example)
ax.set_title('Feature contributions for example {}\n pred: {:1.2f}; label: {}'.format(ID, probs[ID], labels[ID]))
ax.set_xlabel('Contribution to predicted probability', size=14)
plt.show()
```
Global interpretability refers to an understanding of the model as a whole: we will retrieve and visualize gain-based feature importances, permutation feature importances and also show aggregated DFCs.
Gain-based feature importances using est.experimental_feature_importances Permutation importances Aggregate DFCs using est.experimental_predict_with_explanations Gain-based feature importances measure the loss change when splitting on a particular feature, while permutation feature importances are computed by evaluating model performance on the evaluation set by shuffling each feature one-by-one and attributing the change in model performance to the shuffled feature.
In general, permutation feature importance are preferred to gain-based feature importance, though both methods can be unreliable in situations where potential predictor variables vary in their scale of measurement or their number of categories and when features are correlated (source).
```
features = cat_features + num_features
importances = permutation_importances(est, xvalid, yvalid, accuracy_metric, features)
df_imp = pd.Series(importances, index=features)
sorted_ix = df_imp.abs().sort_values().index
ax = df_imp[sorted_ix][-5:].plot(kind='barh', color=sns_colors[2], figsize=(10, 6))
ax.grid(False, axis='y')
ax.set_title('Permutation feature importance')
importances = est.experimental_feature_importances(normalize=True)
df_imp = pd.Series(importances)
# Visualize importances.
N = 8
ax = (df_imp.iloc[0:N][::-1].plot(kind='barh', color=sns_colors[0], title='Gain feature importances',
figsize=(10, 6)))
ax.grid(False, axis='y')
```
The absolute values of DFCs can be averaged to understand impact at a global level.
```
# Plot
dfc_mean = df_dfc.abs().mean()
N = 8
sorted_ix = dfc_mean.abs().sort_values()[-N:].index # Average and sort by absolute.
ax = dfc_mean[sorted_ix].plot(kind='barh',
color=sns_colors[1],
title='Mean | direction feature contributions|',
figsize=(10, 6))
ax.grid(False, axis='y')
```
|
github_jupyter
|
def _get_color(value):
"""To make positive DFCs plot green, negative DFCs plot red."""
green, red = sns.color_palette()[2:4]
if value >= 0: return green
return red
def _add_feature_values(feature_values, ax):
"""Display feature's values on left of plot."""
x_coord = ax.get_xlim()[0]
OFFSET = 0.15
for y_coord, (feat_name, feat_val) in enumerate(feature_values.items()):
t = plt.text(x_coord, y_coord - OFFSET, '{}'.format(feat_val), size=12)
t.set_bbox(dict(facecolor='white', alpha=0.5))
from matplotlib.font_manager import FontProperties
font = FontProperties()
font.set_weight('bold')
t = plt.text(x_coord, y_coord + 1 - OFFSET, 'feature\nvalue',
fontproperties=font, size=12)
def plot_example(example):
TOP_N = 8 # View top 8 features.
sorted_ix = example.abs().sort_values()[-TOP_N:].index # Sort by magnitude.
example = example[sorted_ix]
colors = example.map(_get_color).tolist()
ax = example.to_frame().plot(kind='barh',
color=[colors],
legend=None,
alpha=0.75,
figsize=(10,6))
ax.grid(False, axis='y')
ax.set_yticklabels(ax.get_yticklabels(), size=14)
# Add feature values.
_add_feature_values(xvalid.iloc[ID][sorted_ix], ax)
return ax
def permutation_importances(est, X_eval, y_eval, metric, features):
"""Column by column, shuffle values and observe effect on eval set.
source: http://explained.ai/rf-importance/index.html
A similar approach can be done during training. See "Drop-column importance"
in the above article."""
baseline = metric(est, X_eval, y_eval)
imp = []
for col in features:
save = X_eval[col].copy()
X_eval[col] = np.random.permutation(X_eval[col])
m = metric(est, X_eval, y_eval)
X_eval[col] = save
imp.append(baseline - m)
return np.array(imp)
def accuracy_metric(est, X, y):
"""TensorFlow estimator accuracy."""
eval_input_fn = make_input_fn(X,
y=y,
shuffle=False,
n_epochs=1)
return est.evaluate(input_fn=eval_input_fn)['accuracy']
xtrain = pd.read_csv('hotel_bookings.csv')
xtrain.head(3)
xvalid = xtrain.loc[xtrain['reservation_status_date'] >= '2017-08-01']
xtrain = xtrain.loc[xtrain['reservation_status_date'] < '2017-08-01']
ytrain, yvalid = xtrain['is_canceled'], xvalid['is_canceled']
xtrain.drop('is_canceled', axis = 1, inplace = True)
xvalid.drop('is_canceled', axis = 1, inplace = True)
xtrain.drop(['arrival_date_year','assigned_room_type', 'booking_changes', 'reservation_status',
'country', 'days_in_waiting_list'], axis =1, inplace = True)
num_features = ["lead_time","arrival_date_week_number","arrival_date_day_of_month",
"stays_in_weekend_nights","stays_in_week_nights","adults","children",
"babies","is_repeated_guest", "previous_cancellations",
"previous_bookings_not_canceled","agent","company",
"required_car_parking_spaces", "total_of_special_requests", "adr"]
cat_features = ["hotel","arrival_date_month","meal","market_segment",
"distribution_channel","reserved_room_type","deposit_type","customer_type"]
def one_hot_cat_column(feature_name, vocab):
return tf.feature_column.indicator_column(
tf.feature_column.categorical_column_with_vocabulary_list(feature_name,
vocab))
feature_columns = []
for feature_name in cat_features:
# Need to one-hot encode categorical features.
vocabulary = xtrain[feature_name].unique()
feature_columns.append(one_hot_cat_column(feature_name, vocabulary))
for feature_name in num_features:
feature_columns.append(tf.feature_column.numeric_column(feature_name,
dtype=tf.float32))
# Use entire batch since this is such a small dataset.
NUM_EXAMPLES = len(ytrain)
def make_input_fn(X, y, n_epochs=None, shuffle=True):
def input_fn():
dataset = tf.data.Dataset.from_tensor_slices((dict(X), y))
if shuffle:
dataset = dataset.shuffle(NUM_EXAMPLES)
# For training, cycle thru dataset as many times as need (n_epochs=None).
dataset = dataset.repeat(n_epochs)
# In memory training doesn't use batching.
dataset = dataset.batch(NUM_EXAMPLES)
return dataset
return input_fn
# Training and evaluation input functions.
train_input_fn = make_input_fn(xtrain, ytrain)
eval_input_fn = make_input_fn(xvalid, yvalid, shuffle=False, n_epochs=1)
params = {
'n_trees': 125,
'max_depth': 5,
'n_batches_per_layer': 1,
# 'learning_rate': 0.05,
# 'l1_regularization': 0.00001,
# 'l2_regularization': 0.00001,
# 'min_node_weight': 0.01,
# You must enable center_bias = True to get DFCs. This will force the model to
# make an initial prediction before using any features (e.g. use the mean of
# the training labels for regression or log odds for classification when
# using cross entropy loss).
'center_bias': True
}
est = tf.estimator.BoostedTreesClassifier(feature_columns, **params)
# Train model.
est.train(train_input_fn, max_steps=100)
# Evaluation
results = est.evaluate(eval_input_fn)
pd.Series(results).to_frame()
pred_dicts = list(est.predict(eval_input_fn))
probs = pd.Series([pred['probabilities'][1] for pred in pred_dicts])
fpr, tpr, _ = roc_curve(yvalid, probs)
plt.plot(fpr, tpr)
plt.title('ROC curve')
plt.xlabel('false positive rate')
plt.ylabel('true positive rate')
plt.xlim(0,); plt.ylim(0,); plt.show()
pred_dicts = list(est.experimental_predict_with_explanations(eval_input_fn))
# Create DFC Pandas dataframe
labels = yvalid.values
probs = pd.Series([pred['probabilities'][1] for pred in pred_dicts])
df_dfc = pd.DataFrame([pred['dfc'] for pred in pred_dicts])
df_dfc.describe().T
ID = 10
example = df_dfc.iloc[ID] # Choose ith example from evaluation set.
TOP_N = 8 # View top 8 features
sorted_ix = example.abs().sort_values()[-TOP_N:].index
ax = plot_example(example)
ax.set_title('Feature contributions for example {}\n pred: {:1.2f}; label: {}'.format(ID, probs[ID], labels[ID]))
ax.set_xlabel('Contribution to predicted probability', size=14)
plt.show()
features = cat_features + num_features
importances = permutation_importances(est, xvalid, yvalid, accuracy_metric, features)
df_imp = pd.Series(importances, index=features)
sorted_ix = df_imp.abs().sort_values().index
ax = df_imp[sorted_ix][-5:].plot(kind='barh', color=sns_colors[2], figsize=(10, 6))
ax.grid(False, axis='y')
ax.set_title('Permutation feature importance')
importances = est.experimental_feature_importances(normalize=True)
df_imp = pd.Series(importances)
# Visualize importances.
N = 8
ax = (df_imp.iloc[0:N][::-1].plot(kind='barh', color=sns_colors[0], title='Gain feature importances',
figsize=(10, 6)))
ax.grid(False, axis='y')
# Plot
dfc_mean = df_dfc.abs().mean()
N = 8
sorted_ix = dfc_mean.abs().sort_values()[-N:].index # Average and sort by absolute.
ax = dfc_mean[sorted_ix].plot(kind='barh',
color=sns_colors[1],
title='Mean | direction feature contributions|',
figsize=(10, 6))
ax.grid(False, axis='y')
| 0.88607 | 0.783077 |
# Lab 04 : Train vanilla neural network -- solution
# Training a one-layer net on FASHION-MNIST
```
# For Google Colaboratory
import sys, os
if 'google.colab' in sys.modules:
# mount google drive
from google.colab import drive
drive.mount('/content/gdrive')
path_to_file = '/content/gdrive/My Drive/CS4243_codes/codes/labs_lecture03/lab04_train_vanilla_nn'
print(path_to_file)
# move to Google Drive directory
os.chdir(path_to_file)
!pwd
import torch
import torch.nn as nn
import torch.optim as optim
from random import randint
import utils
```
### Download the TRAINING SET (data+labels)
```
from utils import check_fashion_mnist_dataset_exists
data_path=check_fashion_mnist_dataset_exists()
train_data=torch.load(data_path+'fashion-mnist/train_data.pt')
train_label=torch.load(data_path+'fashion-mnist/train_label.pt')
print(train_data.size())
print(train_label.size())
```
### Download the TEST SET (data only)
```
test_data=torch.load(data_path+'fashion-mnist/test_data.pt')
print(test_data.size())
```
### Make a one layer net class
```
class one_layer_net(nn.Module):
def __init__(self, input_size, output_size):
super(one_layer_net , self).__init__()
self.linear_layer = nn.Linear( input_size, output_size , bias=False)
def forward(self, x):
y = self.linear_layer(x)
prob = torch.softmax(y, dim=1)
return prob
```
### Build the net
```
net=one_layer_net(784,10)
print(net)
```
### Take the 4th image of the test set:
```
im=test_data[4]
utils.show(im)
```
### And feed it to the UNTRAINED network:
```
p = net( im.view(1,784))
print(p)
```
### Display visually the confidence scores
```
utils.show_prob_fashion_mnist(p)
```
### Train the network (only 5000 iterations) on the train set
```
criterion = nn.NLLLoss()
optimizer=torch.optim.SGD(net.parameters() , lr=0.01 )
for iter in range(1,5000):
# choose a random integer between 0 and 59,999
# extract the corresponding picture and label
# and reshape them to fit the network
idx=randint(0, 60000-1)
input=train_data[idx].view(1,784)
label=train_label[idx].view(1)
# feed the input to the net
input.requires_grad_()
prob=net(input)
# update the weights (all the magic happens here -- we will discuss it later)
log_prob=torch.log(prob)
loss = criterion(log_prob, label)
optimizer.zero_grad()
loss.backward()
optimizer.step()
```
### Take the 34th image of the test set:
```
im=test_data[34]
utils.show(im)
```
### Feed it to the TRAINED net:
```
p = net( im.view(1,784))
print(p)
```
### Display visually the confidence scores
```
utils.show_prob_fashion_mnist(prob)
```
### Choose image at random from the test set and see how good/bad are the predictions
```
# choose a picture at random
idx=randint(0, 10000-1)
im=test_data[idx]
# diplay the picture
utils.show(im)
# feed it to the net and display the confidence scores
prob = net( im.view(1,784))
utils.show_prob_fashion_mnist(prob)
```
|
github_jupyter
|
# For Google Colaboratory
import sys, os
if 'google.colab' in sys.modules:
# mount google drive
from google.colab import drive
drive.mount('/content/gdrive')
path_to_file = '/content/gdrive/My Drive/CS4243_codes/codes/labs_lecture03/lab04_train_vanilla_nn'
print(path_to_file)
# move to Google Drive directory
os.chdir(path_to_file)
!pwd
import torch
import torch.nn as nn
import torch.optim as optim
from random import randint
import utils
from utils import check_fashion_mnist_dataset_exists
data_path=check_fashion_mnist_dataset_exists()
train_data=torch.load(data_path+'fashion-mnist/train_data.pt')
train_label=torch.load(data_path+'fashion-mnist/train_label.pt')
print(train_data.size())
print(train_label.size())
test_data=torch.load(data_path+'fashion-mnist/test_data.pt')
print(test_data.size())
class one_layer_net(nn.Module):
def __init__(self, input_size, output_size):
super(one_layer_net , self).__init__()
self.linear_layer = nn.Linear( input_size, output_size , bias=False)
def forward(self, x):
y = self.linear_layer(x)
prob = torch.softmax(y, dim=1)
return prob
net=one_layer_net(784,10)
print(net)
im=test_data[4]
utils.show(im)
p = net( im.view(1,784))
print(p)
utils.show_prob_fashion_mnist(p)
criterion = nn.NLLLoss()
optimizer=torch.optim.SGD(net.parameters() , lr=0.01 )
for iter in range(1,5000):
# choose a random integer between 0 and 59,999
# extract the corresponding picture and label
# and reshape them to fit the network
idx=randint(0, 60000-1)
input=train_data[idx].view(1,784)
label=train_label[idx].view(1)
# feed the input to the net
input.requires_grad_()
prob=net(input)
# update the weights (all the magic happens here -- we will discuss it later)
log_prob=torch.log(prob)
loss = criterion(log_prob, label)
optimizer.zero_grad()
loss.backward()
optimizer.step()
im=test_data[34]
utils.show(im)
p = net( im.view(1,784))
print(p)
utils.show_prob_fashion_mnist(prob)
# choose a picture at random
idx=randint(0, 10000-1)
im=test_data[idx]
# diplay the picture
utils.show(im)
# feed it to the net and display the confidence scores
prob = net( im.view(1,784))
utils.show_prob_fashion_mnist(prob)
| 0.613121 | 0.854703 |
# Introduction to `geoplanar`
Welcome to `geoplanar`, a package for [planar enforcement](https://ibis.geog.ubc.ca/courses/klink/gis.notes/ncgia/u12.html#SEC12.6) for polygon (multipolygon) [GeoSeries/GeoDataFrames](https://github.com/geopandas/geopandas).
In this notebook, we will demonstrate some of the basic functionality of `geoplanar` using the example of a researcher interested in integrating data from the United States and Mexico, to study the US-Mexico international border region.
```
import geoplanar
import geopandas
mexico = geopandas.read_file("../geoplanar/datasets/mexico/mex_admbnda_adm0_govmex_20210618.shp")
mexico.plot()
import libpysal
us = libpysal.examples.load_example('us_income')
us.get_file_list()
us = geopandas.read_file(us.get_path("us48.shp"))
```
us.crs = mexico.crs
us = us.to_crs(mexico.crs)
```
us.plot()
usmex = us.append(mexico)
usmex.plot()
usmex.head()
usmex.shape
usmex.tail()
```
We have appended the Mexico gdf to the US gdf. For now, however, we are going to zoom in on a subset of the border region to investigate things further:
```
from shapely.geometry import box
clipper = geopandas.GeoDataFrame(geometry =[box(-109, 25, -97, 33)])
usborder = geopandas.clip(clipper, us)
mexborder = geopandas.clip(clipper, mexico)
usborder.plot()
mexborder.plot()
usmex = usborder.append(mexborder)
usmex.reset_index(inplace=True)
usmex.plot()
```
## Border discrepancies
```
base = usborder.plot(alpha=0.7, facecolor='none', edgecolor='blue')
_ = mexborder.plot(alpha=0.7, facecolor='none', edgecolor='red', ax=base)
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
```
Here we see an example of the kinds of problems that can occur when combining different geospatial datasets that have been constructed by different researchers. In this figure, a portion of the US-Mexico border is displayed, with the blue linestring indicating the border according to the US dataset, while the red linestring is the border according to the Mexican dataset.
There are two types of problems this induces. Consider a point that is situated to the south of the Mexican border but North of the US border. This can only occur when the Mexican linestring is north of the US linestring. Since under planar enforcement, a point can belong to at most a single polygon, this situation would be a violation - not to mention the kind of cartographic error that can lead to a border war.
A second error occurs when a point is north of the Mexican linestring, but south of the US linestring. In this case, the point is not contained by either the US or Mexico polygons.
## Fixing Overlaps/Overshoots
```
usmex = usborder.append(mexborder)
usmex.reset_index(inplace=True)
usmex['COUNTRY'] = ["US", "MEXICO"]
usmex.area
border_overlaps_removed = geoplanar.trim_overlaps(usmex)
border_overlaps_removed.area # mexico gets trimmed
border_overlaps_removed_1 = geoplanar.trim_overlaps(usmex, largest=False)
border_overlaps_removed_1.area # us gets trimmed
```
## Fixing undershoots/holes
Trimming the overlaps removes the areas where points belong to both national polygons. What remains after this correction are holes (slivers) where points belong to neither polygon.
```
base = border_overlaps_removed.plot(column='COUNTRY')
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
holes = geoplanar.holes(border_overlaps_removed)
base = holes.plot()
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
holes.shape
```
For the entire border region there are 231 holes that exist. These can be corrected, by merging the hole with the larger intersecting national polygon:
```
final = geoplanar.fill_holes(border_overlaps_removed)
base = final.plot(column='COUNTRY')
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
h1 = geoplanar.holes(final)
h1.shape
base = final.plot(edgecolor='k')
_ = usborder.plot(alpha=0.7, facecolor='none', edgecolor='white', ax = base)
_ = mexborder.plot(alpha=0.7, facecolor='none', edgecolor='red', ax=base)
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
final.area
```
## Changing the defaults
```
usmex = usborder.append(mexborder)
usmex.reset_index(inplace=True)
usmex['COUNTRY'] = ["US", "MEXICO"]
usmex.area
border_overlaps_removed_mx = geoplanar.trim_overlaps(usmex, largest=False)
base = border_overlaps_removed_mx.plot(column='COUNTRY')
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
final_mx = geoplanar.fill_holes(border_overlaps_removed_mx, largest=False)
base = final_mx.plot(column='COUNTRY')
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
```
|
github_jupyter
|
import geoplanar
import geopandas
mexico = geopandas.read_file("../geoplanar/datasets/mexico/mex_admbnda_adm0_govmex_20210618.shp")
mexico.plot()
import libpysal
us = libpysal.examples.load_example('us_income')
us.get_file_list()
us = geopandas.read_file(us.get_path("us48.shp"))
us.plot()
usmex = us.append(mexico)
usmex.plot()
usmex.head()
usmex.shape
usmex.tail()
from shapely.geometry import box
clipper = geopandas.GeoDataFrame(geometry =[box(-109, 25, -97, 33)])
usborder = geopandas.clip(clipper, us)
mexborder = geopandas.clip(clipper, mexico)
usborder.plot()
mexborder.plot()
usmex = usborder.append(mexborder)
usmex.reset_index(inplace=True)
usmex.plot()
base = usborder.plot(alpha=0.7, facecolor='none', edgecolor='blue')
_ = mexborder.plot(alpha=0.7, facecolor='none', edgecolor='red', ax=base)
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
usmex = usborder.append(mexborder)
usmex.reset_index(inplace=True)
usmex['COUNTRY'] = ["US", "MEXICO"]
usmex.area
border_overlaps_removed = geoplanar.trim_overlaps(usmex)
border_overlaps_removed.area # mexico gets trimmed
border_overlaps_removed_1 = geoplanar.trim_overlaps(usmex, largest=False)
border_overlaps_removed_1.area # us gets trimmed
base = border_overlaps_removed.plot(column='COUNTRY')
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
holes = geoplanar.holes(border_overlaps_removed)
base = holes.plot()
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
holes.shape
final = geoplanar.fill_holes(border_overlaps_removed)
base = final.plot(column='COUNTRY')
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
h1 = geoplanar.holes(final)
h1.shape
base = final.plot(edgecolor='k')
_ = usborder.plot(alpha=0.7, facecolor='none', edgecolor='white', ax = base)
_ = mexborder.plot(alpha=0.7, facecolor='none', edgecolor='red', ax=base)
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
final.area
usmex = usborder.append(mexborder)
usmex.reset_index(inplace=True)
usmex['COUNTRY'] = ["US", "MEXICO"]
usmex.area
border_overlaps_removed_mx = geoplanar.trim_overlaps(usmex, largest=False)
base = border_overlaps_removed_mx.plot(column='COUNTRY')
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
final_mx = geoplanar.fill_holes(border_overlaps_removed_mx, largest=False)
base = final_mx.plot(column='COUNTRY')
_ = base.set_xlim(-101.4, -101.3)
_ = base.set_ylim(29.6, 29.75)
| 0.34798 | 0.975414 |
### Note
* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.
```
# Dependencies and Setup
import pandas as pd
import numpy as np
# File to Load (Remember to Change These)
file_to_load = "Resources/purchase_data.csv"
# Read Purchasing File and store into Pandas data frame
purchase_data = pd.read_csv(file_to_load)
purchase_data.head()
purchase_data.groupby(["SN", "Gender"]).sum().groupby("Gender").mean()
```
## Player Count
* Display the total number of players
```
Total_players= len(purchase_data["SN"].value_counts())
player_demo=purchase_data.loc[:,["Gender", "SN", "Age"]].drop_duplicates()
num_players=player_demo.count()[0]
num_players
print(f"Total Number Of Players = {Total_players}")
players_df= pd.DataFrame(data=[Total_players])
players_df.columns = ["Total Players"]
players_df
```
## Purchasing Analysis (Total)
* Run basic calculations to obtain number of unique items, average price, etc.
* Create a summary data frame to hold the results
* Optional: give the displayed data cleaner formatting
* Display the summary data frame
```
unique_items= len(purchase_data["Item ID"].unique())
average_price= purchase_data["Price"].mean()
total_purchases=purchase_data.shape[0]
revenue= purchase_data["Price"].sum()
list=[unique_items, average_price, total_purchases, revenue]
frame_df= pd.DataFrame(data=[list])
frame_df.columns = ({"Number of Unique Items": [unique_items],
"Average Price": [average_price],
"Number of Purchases": [total_purchases],
"Total Revenue":[revenue]})
frame_df
```
## Gender Demographics
* Percentage and Count of Male Players
* Percentage and Count of Female Players
* Percentage and Count of Other / Non-Disclosed
```
Gender_count=player_demo["Gender"].value_counts()
Gender_percentage= (Gender_count/ num_players)
Gender_df = pd.concat ([Gender_count, Gender_percentage], axis=1)
Gender_df.columns = ["Total Count", "Percentage"]
Gender_df.style
```
## Purchasing Analysis (Gender)
```
purchase_count = purchase_data["Gender"].value_counts()
Average_Purchase_Price =purchase_data.groupby("Gender")
Average_Purchase_Price
```
* Run basic calculations to obtain purchase count, avg. purchase price, avg. purchase total per person etc. by gender
* Create a summary data frame to hold the results
* Optional: give the displayed data cleaner formatting
* Display the summary data frame
```
purchase_count = purchase_data["Gender"].value_counts()
Average_Purchase_Price = round(purchase_data.groupby(["Gender"]).mean()["Price"], 2)
Average_Purchase_Price = '$' + Average_Purchase_Price.astype(str)
Total_Purchase_Value = purchase_data.groupby(["Gender"]).sum()["Price"]
Average_Purchase_Total = Total_Purchase_Value / Gender_df["Total Count"]
Total_Purchase_Value = '$' + Total_Purchase_Value.astype(str)
Average_Purchase_Total = '$' + Average_Purchase_Total.astype(str)
purchasing_analysis_df = pd.DataFrame({"Purchase Count": purchase_count, "Average Price":Average_Purchase_Price,"Total Purchase Value": Total_Purchase_Value,"Avg Total Purchase Per Person": Average_Purchase_Total})
purchasing_analysis_df
```
## Age Demographics
* Establish bins for ages
* Categorize the existing players using the age bins. Hint: use pd.cut()
* Calculate the numbers and percentages by age group
* Create a summary data frame to hold the results
* Optional: round the percentage column to two decimal points
* Display Age Demographics Table
```
Ages = [10, 14, 19, 24, 29, 34, 39, 40, 1000]
Group_Names = ["<10", "10-14", "15-19", "20-24", "25-29", "30-34", "35-39", "40+"]
Age_Series = pd.cut(purchase_data.groupby("SN")["Age"].mean(), Ages, labels=Group_Names).value_counts()
Age_Percent = round(Age_Series / Age_Series.sum() * 100, 2)
Age_df = pd.concat([Age_Series, Age_Percent], axis=1, sort=True)
Age_df.columns = ["Total Number", "Percentage"]
Age_df.head()
```
## Purchasing Analysis (Age)
* Bin the purchase_data data frame by age
* Run basic calculations to obtain purchase count, avg. purchase price, avg. purchase total per person etc. in the table below
* Create a summary data frame to hold the results
* Optional: give the displayed data cleaner formatting
* Display the summary data frame
```
Ages = [10, 14, 19, 24, 29, 34, 39, 40, 1000]
Group_Names = ["<10", "10-14", "15-19", "20-24", "25-29", "30-34", "35-39", "40+"]
purchase_data["Age Range"] = pd.cut(purchase_data.Age, Ages, labels= Group_Names)
Purchase_Count_Age = purchase_data["Age Range"].count()
Purchase_Count_Age
Average_Purchase_Age = round(purchase_data.groupby("Age Range")["Price"].mean(), 2)
Average_Purchase_Age = '$' + Average_Purchase_Age.astype(str)
Total_Purchase_Age = round(purchase_data.groupby("Age Range")["Price"].sum(), 2)
Average_Purchase_Age = round(Total_Purchase_Age/ purchase_data.groupby('Age Range')['SN'].nunique(), 2)
Total_Purchase_Age = '$' + Total_Purchase_Age.astype(str)
Average_Purchase_Age= '$' + Average_Purchase_Age.astype(str)
Purchase_df_Age = pd.DataFrame({"Purchase Count": Purchase_Count_Age , "Average Purchase Price" :Average_Purchase_Age,"Total Purchase Value": Total_Purchase_Age})
Purchase_df_Age.head()
```
## Top Spenders
* Run basic calculations to obtain the results in the table below
* Create a summary data frame to hold the results
* Sort the total purchase value column in descending order
* Optional: give the displayed data cleaner formatting
* Display a preview of the summary data frame
```
Top_Spender = purchase_data.groupby("SN")["Price"].sum().nlargest(5)
Top_Average_Purchase = round(purchase_data.groupby("SN").Price.mean().loc[Top_Spender.index], 2)
Top_Non_Purchase = purchase_data.groupby("SN").Price.count().loc[Top_Spender.index]
Top_df = pd.concat([Top_Non_Purchase, Top_Average_Purchase, Top_Spender], axis=1)
Top_df.columns = ["Purchase Count", "Average Purchase Price", "Total Purchase Value"]
Top_df = Top_df.sort_values(by='Total Purchase Value', ascending=False)
Top_df.head().style
```
## Most Popular Items
* Retrieve the Item ID, Item Name, and Item Price columns
* Group by Item ID and Item Name. Perform calculations to obtain purchase count, item price, and total purchase value
* Create a summary data frame to hold the results
* Sort the purchase count column in descending order
* Optional: give the displayed data cleaner formatting
* Display a preview of the summary data frame
```
New_df = purchase_data[["Item ID", "Item Name", "Price"]]
New_df_group = New_df.groupby('Item ID')
ItemName = New_df[['Item ID', 'Item Name']]
ItemPrice = New_df[['Item ID', 'Price']]
ItemCounts = New_df_group.count()
ItemSums = New_df_group.sum()
ItemCountsBest = ItemCounts.sort_values('Item Name', ascending=False).head()
ItemCountsBest = ItemCountsBest.rename(columns={'Item Name':'Purchase Count'})
ItemCountsBest = ItemCountsBest.reset_index()
ItemCountsBest = ItemCountsBest[["Item ID", "Purchase Count"]]
ItemCountData = ItemCountsBest.merge(ItemName, left_on='Item ID', right_on='Item ID')
ItemCountData = ItemCountData.merge(ItemPrice, left_on='Item ID', right_on='Item ID')
ItemCountData = ItemCountData.merge(ItemSums, left_on='Item ID', right_on='Item ID')
ItemCountData = ItemCountData.drop_duplicates(keep='first')
ItemCountData = ItemCountData.reset_index()
ItemCountData = ItemCountData.rename(columns={'Price_x':'Item Price'})
ItemCountData = ItemCountData.rename(columns={'Price_y':'Total Purchase Value'})
ItemCountData = ItemCountData[['Item ID', 'Item Name', 'Purchase Count', 'Item Price', 'Total Purchase Value']]
ItemCountData
```
## Most Profitable Items
* Sort the above table by total purchase value in descending order
* Optional: give the displayed data cleaner formatting
* Display a preview of the data frame
```
New_df = purchase_data[["Item ID", "Item Name", "Price"]]
New_df_group = New_df.groupby('Item ID')
ItemName = New_df[['Item ID', 'Item Name']]
ItemPrice = New_df[['Item ID', 'Price']]
ItemCounts = New_df_group.count()
ItemSums = New_df_group.sum()
ItemSumsBest = ItemSums.sort_values('Price', ascending=False).head()
ItemSumsBest = ItemSumsBest.rename(columns={'Price':'Total Purchase Value'})
ItemSumsBest = ItemSumsBest.reset_index()
ItemSumsBest = ItemSumsBest[["Item ID", "Total Purchase Value"]]
ItemSumsBest = ItemSumsBest.merge(ItemName, left_on='Item ID', right_on='Item ID')
ItemSumsBest = ItemSumsBest.merge(ItemPrice, left_on='Item ID', right_on='Item ID')
ItemSumsBest = ItemSumsBest.merge(ItemCounts, left_on='Item ID', right_on='Item ID')
ItemSumData = ItemSumsBest.drop_duplicates(keep='first')
ItemSumData = ItemSumData.reset_index()
ItemSumData = ItemSumData.rename(columns={'Price_x':'Item Price'})
ItemSumData = ItemSumData.rename(columns={'Price_y':'Purchase Count'})
ItemSumData = ItemSumData.rename(columns={'Item Name_x':'Item Name'})
ItemSumData = ItemSumData[['Item ID', 'Item Name', 'Purchase Count', 'Item Price', 'Total Purchase Value']]
ItemSumData
#1st Observation: The Item that was most purchased at the lowest price and had the highest purchase value is the "Oathbreaker, Last Hope of the Breaking Storm"
#2nd Observation: The Gender that dominated purchases were Males.
#3rd Observation: The Least Age group that purchased games are 40 years of Age and above.
```
|
github_jupyter
|
# Dependencies and Setup
import pandas as pd
import numpy as np
# File to Load (Remember to Change These)
file_to_load = "Resources/purchase_data.csv"
# Read Purchasing File and store into Pandas data frame
purchase_data = pd.read_csv(file_to_load)
purchase_data.head()
purchase_data.groupby(["SN", "Gender"]).sum().groupby("Gender").mean()
Total_players= len(purchase_data["SN"].value_counts())
player_demo=purchase_data.loc[:,["Gender", "SN", "Age"]].drop_duplicates()
num_players=player_demo.count()[0]
num_players
print(f"Total Number Of Players = {Total_players}")
players_df= pd.DataFrame(data=[Total_players])
players_df.columns = ["Total Players"]
players_df
unique_items= len(purchase_data["Item ID"].unique())
average_price= purchase_data["Price"].mean()
total_purchases=purchase_data.shape[0]
revenue= purchase_data["Price"].sum()
list=[unique_items, average_price, total_purchases, revenue]
frame_df= pd.DataFrame(data=[list])
frame_df.columns = ({"Number of Unique Items": [unique_items],
"Average Price": [average_price],
"Number of Purchases": [total_purchases],
"Total Revenue":[revenue]})
frame_df
Gender_count=player_demo["Gender"].value_counts()
Gender_percentage= (Gender_count/ num_players)
Gender_df = pd.concat ([Gender_count, Gender_percentage], axis=1)
Gender_df.columns = ["Total Count", "Percentage"]
Gender_df.style
purchase_count = purchase_data["Gender"].value_counts()
Average_Purchase_Price =purchase_data.groupby("Gender")
Average_Purchase_Price
purchase_count = purchase_data["Gender"].value_counts()
Average_Purchase_Price = round(purchase_data.groupby(["Gender"]).mean()["Price"], 2)
Average_Purchase_Price = '$' + Average_Purchase_Price.astype(str)
Total_Purchase_Value = purchase_data.groupby(["Gender"]).sum()["Price"]
Average_Purchase_Total = Total_Purchase_Value / Gender_df["Total Count"]
Total_Purchase_Value = '$' + Total_Purchase_Value.astype(str)
Average_Purchase_Total = '$' + Average_Purchase_Total.astype(str)
purchasing_analysis_df = pd.DataFrame({"Purchase Count": purchase_count, "Average Price":Average_Purchase_Price,"Total Purchase Value": Total_Purchase_Value,"Avg Total Purchase Per Person": Average_Purchase_Total})
purchasing_analysis_df
Ages = [10, 14, 19, 24, 29, 34, 39, 40, 1000]
Group_Names = ["<10", "10-14", "15-19", "20-24", "25-29", "30-34", "35-39", "40+"]
Age_Series = pd.cut(purchase_data.groupby("SN")["Age"].mean(), Ages, labels=Group_Names).value_counts()
Age_Percent = round(Age_Series / Age_Series.sum() * 100, 2)
Age_df = pd.concat([Age_Series, Age_Percent], axis=1, sort=True)
Age_df.columns = ["Total Number", "Percentage"]
Age_df.head()
Ages = [10, 14, 19, 24, 29, 34, 39, 40, 1000]
Group_Names = ["<10", "10-14", "15-19", "20-24", "25-29", "30-34", "35-39", "40+"]
purchase_data["Age Range"] = pd.cut(purchase_data.Age, Ages, labels= Group_Names)
Purchase_Count_Age = purchase_data["Age Range"].count()
Purchase_Count_Age
Average_Purchase_Age = round(purchase_data.groupby("Age Range")["Price"].mean(), 2)
Average_Purchase_Age = '$' + Average_Purchase_Age.astype(str)
Total_Purchase_Age = round(purchase_data.groupby("Age Range")["Price"].sum(), 2)
Average_Purchase_Age = round(Total_Purchase_Age/ purchase_data.groupby('Age Range')['SN'].nunique(), 2)
Total_Purchase_Age = '$' + Total_Purchase_Age.astype(str)
Average_Purchase_Age= '$' + Average_Purchase_Age.astype(str)
Purchase_df_Age = pd.DataFrame({"Purchase Count": Purchase_Count_Age , "Average Purchase Price" :Average_Purchase_Age,"Total Purchase Value": Total_Purchase_Age})
Purchase_df_Age.head()
Top_Spender = purchase_data.groupby("SN")["Price"].sum().nlargest(5)
Top_Average_Purchase = round(purchase_data.groupby("SN").Price.mean().loc[Top_Spender.index], 2)
Top_Non_Purchase = purchase_data.groupby("SN").Price.count().loc[Top_Spender.index]
Top_df = pd.concat([Top_Non_Purchase, Top_Average_Purchase, Top_Spender], axis=1)
Top_df.columns = ["Purchase Count", "Average Purchase Price", "Total Purchase Value"]
Top_df = Top_df.sort_values(by='Total Purchase Value', ascending=False)
Top_df.head().style
New_df = purchase_data[["Item ID", "Item Name", "Price"]]
New_df_group = New_df.groupby('Item ID')
ItemName = New_df[['Item ID', 'Item Name']]
ItemPrice = New_df[['Item ID', 'Price']]
ItemCounts = New_df_group.count()
ItemSums = New_df_group.sum()
ItemCountsBest = ItemCounts.sort_values('Item Name', ascending=False).head()
ItemCountsBest = ItemCountsBest.rename(columns={'Item Name':'Purchase Count'})
ItemCountsBest = ItemCountsBest.reset_index()
ItemCountsBest = ItemCountsBest[["Item ID", "Purchase Count"]]
ItemCountData = ItemCountsBest.merge(ItemName, left_on='Item ID', right_on='Item ID')
ItemCountData = ItemCountData.merge(ItemPrice, left_on='Item ID', right_on='Item ID')
ItemCountData = ItemCountData.merge(ItemSums, left_on='Item ID', right_on='Item ID')
ItemCountData = ItemCountData.drop_duplicates(keep='first')
ItemCountData = ItemCountData.reset_index()
ItemCountData = ItemCountData.rename(columns={'Price_x':'Item Price'})
ItemCountData = ItemCountData.rename(columns={'Price_y':'Total Purchase Value'})
ItemCountData = ItemCountData[['Item ID', 'Item Name', 'Purchase Count', 'Item Price', 'Total Purchase Value']]
ItemCountData
New_df = purchase_data[["Item ID", "Item Name", "Price"]]
New_df_group = New_df.groupby('Item ID')
ItemName = New_df[['Item ID', 'Item Name']]
ItemPrice = New_df[['Item ID', 'Price']]
ItemCounts = New_df_group.count()
ItemSums = New_df_group.sum()
ItemSumsBest = ItemSums.sort_values('Price', ascending=False).head()
ItemSumsBest = ItemSumsBest.rename(columns={'Price':'Total Purchase Value'})
ItemSumsBest = ItemSumsBest.reset_index()
ItemSumsBest = ItemSumsBest[["Item ID", "Total Purchase Value"]]
ItemSumsBest = ItemSumsBest.merge(ItemName, left_on='Item ID', right_on='Item ID')
ItemSumsBest = ItemSumsBest.merge(ItemPrice, left_on='Item ID', right_on='Item ID')
ItemSumsBest = ItemSumsBest.merge(ItemCounts, left_on='Item ID', right_on='Item ID')
ItemSumData = ItemSumsBest.drop_duplicates(keep='first')
ItemSumData = ItemSumData.reset_index()
ItemSumData = ItemSumData.rename(columns={'Price_x':'Item Price'})
ItemSumData = ItemSumData.rename(columns={'Price_y':'Purchase Count'})
ItemSumData = ItemSumData.rename(columns={'Item Name_x':'Item Name'})
ItemSumData = ItemSumData[['Item ID', 'Item Name', 'Purchase Count', 'Item Price', 'Total Purchase Value']]
ItemSumData
#1st Observation: The Item that was most purchased at the lowest price and had the highest purchase value is the "Oathbreaker, Last Hope of the Breaking Storm"
#2nd Observation: The Gender that dominated purchases were Males.
#3rd Observation: The Least Age group that purchased games are 40 years of Age and above.
| 0.367043 | 0.768516 |
```
import xarray as xr
ds = xr.open_dataset("test_data/NEMO_GYRE_test_data/mesh_mask.nc")
ds
ds.isfdraft.data
mesh_mask_attrs = {}
# longitude fields
mesh_mask_attrs.update(
{
f"glam{grid}": {
"long_name": "longitude",
"units": "degrees_east",
"standard_name": "longitude"
}
for grid in ["t", "u", "v", "f"]
}
)
# latitude fields
mesh_mask_attrs.update(
{
f"gphi{grid}": {
"long_name": "latitude",
"units": "degrees_north",
"standard_name": "latitude"
}
for grid in ["t", "u", "v", "f"]
}
)
# depth fields
mesh_mask_attrs.update(
{
f"gdep{grid}_1d": {
"long_name": "depth",
"units": "meters",
"positive": "down",
"standard_name": "depth"
}
for grid in ["t", "w"]
}
)
# zonal grid constants
mesh_mask_attrs.update(
{
f"e1{grid}": {
"long_name": "zonal grid constant",
"units": "meters",
"coordinates": f"glam{grid} gphi{grid}"
}
for grid in ["t", "u", "v", "f"]
}
)
# meridional grid constants
mesh_mask_attrs.update(
{
f"e2{grid}": {
"long_name": "meridional grid constant",
"units": "meters",
"coordinates": f"glam{grid} gphi{grid}"
}
for grid in ["t", "u", "v", "f"]
}
)
# vertical grid constants
mesh_mask_attrs.update(
{
f"e3{grid}_1d": {
"long_name": "vertical grid constant",
"units": "meters",
"coordinates": f"gdep{grid}_1d"
}
for grid in ["t", "w"]
}
)
# masks
# Note that the f-mask lives on vertical T levels
# (c/f NEMO book)
mesh_mask_attrs.update(
{
f"{grid}mask": {
"long_name": "land point mask",
"units": "boolean",
"coordinates": f"gdept_1d glam{grid} gphi{grid}"
}
for grid in ["t", "u", "v", "f"]
}
)
# util masks
mesh_mask_attrs.update(
{
f"{grid}maskutil": {
"long_name": "land point mask",
"units": "boolean",
"coordinates": f"glam{grid} gphi{grid}"
}
for grid in ["t", "u", "v", "f"]
}
)
# number of wet grid points
mesh_mask_attrs.update({
"mbathy": {
"long_name": "number of ocean levels at xy grid point",
"coordinates": "glamt gphit"
}
})
mesh_mask_attrs
for varname, new_attrs in mesh_mask_attrs.items():
ds[varname].attrs.update(new_attrs)
ds
ds._encoding = {}
ds.to_netcdf("mesh_mask_annotated.nc")
!ncdump -h mesh_mask_annotated.nc
ds_reread = xr.open_dataset("mesh_mask_annotated.nc")
ds_reread
```
|
github_jupyter
|
import xarray as xr
ds = xr.open_dataset("test_data/NEMO_GYRE_test_data/mesh_mask.nc")
ds
ds.isfdraft.data
mesh_mask_attrs = {}
# longitude fields
mesh_mask_attrs.update(
{
f"glam{grid}": {
"long_name": "longitude",
"units": "degrees_east",
"standard_name": "longitude"
}
for grid in ["t", "u", "v", "f"]
}
)
# latitude fields
mesh_mask_attrs.update(
{
f"gphi{grid}": {
"long_name": "latitude",
"units": "degrees_north",
"standard_name": "latitude"
}
for grid in ["t", "u", "v", "f"]
}
)
# depth fields
mesh_mask_attrs.update(
{
f"gdep{grid}_1d": {
"long_name": "depth",
"units": "meters",
"positive": "down",
"standard_name": "depth"
}
for grid in ["t", "w"]
}
)
# zonal grid constants
mesh_mask_attrs.update(
{
f"e1{grid}": {
"long_name": "zonal grid constant",
"units": "meters",
"coordinates": f"glam{grid} gphi{grid}"
}
for grid in ["t", "u", "v", "f"]
}
)
# meridional grid constants
mesh_mask_attrs.update(
{
f"e2{grid}": {
"long_name": "meridional grid constant",
"units": "meters",
"coordinates": f"glam{grid} gphi{grid}"
}
for grid in ["t", "u", "v", "f"]
}
)
# vertical grid constants
mesh_mask_attrs.update(
{
f"e3{grid}_1d": {
"long_name": "vertical grid constant",
"units": "meters",
"coordinates": f"gdep{grid}_1d"
}
for grid in ["t", "w"]
}
)
# masks
# Note that the f-mask lives on vertical T levels
# (c/f NEMO book)
mesh_mask_attrs.update(
{
f"{grid}mask": {
"long_name": "land point mask",
"units": "boolean",
"coordinates": f"gdept_1d glam{grid} gphi{grid}"
}
for grid in ["t", "u", "v", "f"]
}
)
# util masks
mesh_mask_attrs.update(
{
f"{grid}maskutil": {
"long_name": "land point mask",
"units": "boolean",
"coordinates": f"glam{grid} gphi{grid}"
}
for grid in ["t", "u", "v", "f"]
}
)
# number of wet grid points
mesh_mask_attrs.update({
"mbathy": {
"long_name": "number of ocean levels at xy grid point",
"coordinates": "glamt gphit"
}
})
mesh_mask_attrs
for varname, new_attrs in mesh_mask_attrs.items():
ds[varname].attrs.update(new_attrs)
ds
ds._encoding = {}
ds.to_netcdf("mesh_mask_annotated.nc")
!ncdump -h mesh_mask_annotated.nc
ds_reread = xr.open_dataset("mesh_mask_annotated.nc")
ds_reread
| 0.617743 | 0.644477 |
# Particle Differential Energy Fluxes
author: Louis Richard\
Load brst particle distributions and convert to differential energy fluxes. Plots electron and ion fluxes and electron anisotropies.
```
import xarray as xr
import matplotlib.pyplot as plt
from pyrfu import mms
from pyrfu.pyrf import norm, resample
from pyrfu.plot import plot_line, plot_spectr, make_labels
from astropy import constants
```
## Define spacecraft index and time interval
```
ic = 3 # Spacecraft number
tint = ["2015-10-30T05:15:20.000", "2015-10-30T05:16:20.000"]
```
## Load data
### Particle distributions
```
vdf_i, vdf_e = [mms.get_data(f"pd{s}_fpi_brst_l2", tint, ic) for s in ["i", "e"]]
```
### Particle moments
```
n_i, n_e = [mms.get_data(f"n{s}_fpi_brst_l2", tint, ic) for s in ["i", "e"]]
v_xyz_i, v_xyz_e = [mms.get_data(f"v{s}_gse_fpi_brst_l2", tint, ic) for s in ["i", "e"]]
t_xyz_i, t_xyz_e = [mms.get_data(f"t{s}_gse_fpi_brst_l2", tint, ic) for s in ["i", "e"]]
```
### Other variables
```
b_xyz, b_gse = [mms.get_data(f"b_{cs}_fgm_brst_l2", tint, ic) for cs in ["dmpa", "gse"]]
e_xyz = mms.get_data("e_gse_edp_brst_l2", tint, ic)
scpot = mms.get_data("v_edp_brst_l2", tint, ic)
scpot = resample(scpot, n_e)
```
## Compute moments
```
qe = constants.e.value
mp = constants.m_p.value
v_av = 0.5 * mp * (1e3 * norm(v_xyz_i)) ** 2 / qe
```
### Compute parallel and perpendicular electron and ion temperatures
```
t_fac_i, t_fac_e = [mms.rotate_tensor(t_xyz, "fac", b_xyz, "pp") for t_xyz in [t_xyz_i, t_xyz_e]]
t_para_i, t_para_e = [t_fac[:, 0, 0] for t_fac in [t_fac_i, t_fac_e]]
t_perp_i, t_perp_e = [t_fac[:, 1, 1] for t_fac in [t_fac_i, t_fac_e]]
```
### Compute Differential Energy Fluxes
```
def_omni_i, def_omni_e = [mms.vdf_omni(mms.vdf_to_deflux(vdf)) for vdf in [vdf_i, vdf_e]]
```
### Compute Pitch-Angle Distribution
```
def_pad_e = mms.vdf_to_deflux(mms.get_pitch_angle_dist(vdf_e, b_xyz, tint, angles=13))
```
### Compute parallel/anti-parallel and parallel+anti-parallel/perpandicular
```
def calc_parapar_parperp(pad):
coords = [pad.time.data, pad.energy.data[0, :]]
dims = ["time", "energy"]
psd_parapar = def_pad_e.data.data[:, :, 0] / def_pad_e.data.data[:, :, -1]
psd_parperp = (def_pad_e.data.data[:, :, 0] + def_pad_e.data.data[:, :, -1]) / (2 * def_pad_e.data.data[:, :, 7])
psd_parapar = xr.DataArray(psd_parapar, coords=coords, dims=dims)
psd_parperp = xr.DataArray(psd_parperp, coords=coords, dims=dims)
return psd_parapar, psd_parperp
vdf_parapar_e, vdf_parperp_e = calc_parapar_parperp(def_pad_e)
```
## Plot
```
legend_options = dict(frameon=True, loc="upper right", ncol=3)
e_lim_i = [min(def_omni_i.energy.data), max(def_omni_i.energy.data)]
e_lim_e = [min(def_omni_e.energy.data), max(def_omni_e.energy.data)]
%matplotlib notebook
f, axs = plt.subplots(8, sharex="all", figsize=(6.5, 11))
f.subplots_adjust(bottom=.1, top=.95, left=.15, right=.85, hspace=0)
plot_line(axs[0], b_xyz)
axs[0].set_ylabel("$B_{DMPA}$" + "\n" + "[nT]")
axs[0].legend(["$B_{x}$", "$B_{y}$", "$B_{z}$"], **legend_options)
plot_line(axs[1], v_xyz_i)
axs[1].set_ylabel("$V_{i}$" + "\n" + "[km s$^{-1}$]")
axs[1].legend(["$V_{x}$", "$V_{y}$", "$V_{z}$"], **legend_options)
plot_line(axs[2], n_i, "tab:blue")
plot_line(axs[2], n_e, "tab:red")
axs[2].set_yscale("log")
axs[2].set_ylabel("$n$" + "\n" + "[cm$^{-3}$]")
axs[2].legend(["$n_i$", "$n_e$"], **legend_options)
plot_line(axs[3], e_xyz)
axs[3].set_ylabel("$E$" + "\n" + "[mV m$^{-1}$]")
axs[3].legend(["$E_{x}$", "$E_{y}$", "$E_{z}$"], **legend_options)
axs[4], caxs4 = plot_spectr(axs[4], def_omni_i, yscale="log", cscale="log", cmap="Spectral_r")
axs[4].set_ylabel("$E_i$" + "\n" + "[eV]")
caxs4.set_ylabel("DEF" + "\n" + "[kev/(cm$^2$ s sr keV)]")
axs[4].set_ylim(e_lim_i)
axs[5], caxs5 = plot_spectr(axs[5], def_omni_e, yscale="log", cscale="log", cmap="Spectral_r")
plot_line(axs[5], scpot)
plot_line(axs[5], t_para_e)
plot_line(axs[5], t_perp_e)
axs[5].set_ylabel("$E_e$" + "\n" + "[eV]")
caxs5.set_ylabel("DEF" + "\n" + "[kev/(cm$^2$ s sr keV)]")
axs[5].legend(["$\phi$", "$T_{||}$","$T_{\perp}$"], **legend_options)
axs[5].set_ylim(e_lim_e)
axs[6], caxs6 = plot_spectr(axs[6], vdf_parapar_e, yscale="log", cscale="log", clim=[1e-2, 1e2], cmap="RdBu_r")
plot_line(axs[6], scpot)
axs[6].set_ylabel("$E_e$" + "\n" + "[eV]")
caxs6.set_ylabel("$\\frac{f_{||+}}{f_{||-}}$" + "\n" + " ")
axs[6].legend(["$V_{SC}$", "$T_{||}$","$T_{\perp}$"], **legend_options)
axs[6].set_ylim(e_lim_e)
axs[7], caxs7 = plot_spectr(axs[7], vdf_parperp_e, yscale="log", cscale="log", clim=[1e-2, 1e2], cmap="RdBu_r")
plot_line(axs[7], scpot)
axs[7].set_ylabel("$E_e$" + "\n" + "[eV]")
caxs7.set_ylabel("$\\frac{f_{||+}+f_{||-}}{2 f_{\perp}}$" + "\n" + " ")
axs[7].legend(["$V_{SC}$"], **legend_options)
axs[7].set_ylim(e_lim_e)
make_labels(axs, [0.02, 0.85])
f.align_ylabels(axs)
```
|
github_jupyter
|
import xarray as xr
import matplotlib.pyplot as plt
from pyrfu import mms
from pyrfu.pyrf import norm, resample
from pyrfu.plot import plot_line, plot_spectr, make_labels
from astropy import constants
ic = 3 # Spacecraft number
tint = ["2015-10-30T05:15:20.000", "2015-10-30T05:16:20.000"]
vdf_i, vdf_e = [mms.get_data(f"pd{s}_fpi_brst_l2", tint, ic) for s in ["i", "e"]]
n_i, n_e = [mms.get_data(f"n{s}_fpi_brst_l2", tint, ic) for s in ["i", "e"]]
v_xyz_i, v_xyz_e = [mms.get_data(f"v{s}_gse_fpi_brst_l2", tint, ic) for s in ["i", "e"]]
t_xyz_i, t_xyz_e = [mms.get_data(f"t{s}_gse_fpi_brst_l2", tint, ic) for s in ["i", "e"]]
b_xyz, b_gse = [mms.get_data(f"b_{cs}_fgm_brst_l2", tint, ic) for cs in ["dmpa", "gse"]]
e_xyz = mms.get_data("e_gse_edp_brst_l2", tint, ic)
scpot = mms.get_data("v_edp_brst_l2", tint, ic)
scpot = resample(scpot, n_e)
qe = constants.e.value
mp = constants.m_p.value
v_av = 0.5 * mp * (1e3 * norm(v_xyz_i)) ** 2 / qe
t_fac_i, t_fac_e = [mms.rotate_tensor(t_xyz, "fac", b_xyz, "pp") for t_xyz in [t_xyz_i, t_xyz_e]]
t_para_i, t_para_e = [t_fac[:, 0, 0] for t_fac in [t_fac_i, t_fac_e]]
t_perp_i, t_perp_e = [t_fac[:, 1, 1] for t_fac in [t_fac_i, t_fac_e]]
def_omni_i, def_omni_e = [mms.vdf_omni(mms.vdf_to_deflux(vdf)) for vdf in [vdf_i, vdf_e]]
def_pad_e = mms.vdf_to_deflux(mms.get_pitch_angle_dist(vdf_e, b_xyz, tint, angles=13))
def calc_parapar_parperp(pad):
coords = [pad.time.data, pad.energy.data[0, :]]
dims = ["time", "energy"]
psd_parapar = def_pad_e.data.data[:, :, 0] / def_pad_e.data.data[:, :, -1]
psd_parperp = (def_pad_e.data.data[:, :, 0] + def_pad_e.data.data[:, :, -1]) / (2 * def_pad_e.data.data[:, :, 7])
psd_parapar = xr.DataArray(psd_parapar, coords=coords, dims=dims)
psd_parperp = xr.DataArray(psd_parperp, coords=coords, dims=dims)
return psd_parapar, psd_parperp
vdf_parapar_e, vdf_parperp_e = calc_parapar_parperp(def_pad_e)
legend_options = dict(frameon=True, loc="upper right", ncol=3)
e_lim_i = [min(def_omni_i.energy.data), max(def_omni_i.energy.data)]
e_lim_e = [min(def_omni_e.energy.data), max(def_omni_e.energy.data)]
%matplotlib notebook
f, axs = plt.subplots(8, sharex="all", figsize=(6.5, 11))
f.subplots_adjust(bottom=.1, top=.95, left=.15, right=.85, hspace=0)
plot_line(axs[0], b_xyz)
axs[0].set_ylabel("$B_{DMPA}$" + "\n" + "[nT]")
axs[0].legend(["$B_{x}$", "$B_{y}$", "$B_{z}$"], **legend_options)
plot_line(axs[1], v_xyz_i)
axs[1].set_ylabel("$V_{i}$" + "\n" + "[km s$^{-1}$]")
axs[1].legend(["$V_{x}$", "$V_{y}$", "$V_{z}$"], **legend_options)
plot_line(axs[2], n_i, "tab:blue")
plot_line(axs[2], n_e, "tab:red")
axs[2].set_yscale("log")
axs[2].set_ylabel("$n$" + "\n" + "[cm$^{-3}$]")
axs[2].legend(["$n_i$", "$n_e$"], **legend_options)
plot_line(axs[3], e_xyz)
axs[3].set_ylabel("$E$" + "\n" + "[mV m$^{-1}$]")
axs[3].legend(["$E_{x}$", "$E_{y}$", "$E_{z}$"], **legend_options)
axs[4], caxs4 = plot_spectr(axs[4], def_omni_i, yscale="log", cscale="log", cmap="Spectral_r")
axs[4].set_ylabel("$E_i$" + "\n" + "[eV]")
caxs4.set_ylabel("DEF" + "\n" + "[kev/(cm$^2$ s sr keV)]")
axs[4].set_ylim(e_lim_i)
axs[5], caxs5 = plot_spectr(axs[5], def_omni_e, yscale="log", cscale="log", cmap="Spectral_r")
plot_line(axs[5], scpot)
plot_line(axs[5], t_para_e)
plot_line(axs[5], t_perp_e)
axs[5].set_ylabel("$E_e$" + "\n" + "[eV]")
caxs5.set_ylabel("DEF" + "\n" + "[kev/(cm$^2$ s sr keV)]")
axs[5].legend(["$\phi$", "$T_{||}$","$T_{\perp}$"], **legend_options)
axs[5].set_ylim(e_lim_e)
axs[6], caxs6 = plot_spectr(axs[6], vdf_parapar_e, yscale="log", cscale="log", clim=[1e-2, 1e2], cmap="RdBu_r")
plot_line(axs[6], scpot)
axs[6].set_ylabel("$E_e$" + "\n" + "[eV]")
caxs6.set_ylabel("$\\frac{f_{||+}}{f_{||-}}$" + "\n" + " ")
axs[6].legend(["$V_{SC}$", "$T_{||}$","$T_{\perp}$"], **legend_options)
axs[6].set_ylim(e_lim_e)
axs[7], caxs7 = plot_spectr(axs[7], vdf_parperp_e, yscale="log", cscale="log", clim=[1e-2, 1e2], cmap="RdBu_r")
plot_line(axs[7], scpot)
axs[7].set_ylabel("$E_e$" + "\n" + "[eV]")
caxs7.set_ylabel("$\\frac{f_{||+}+f_{||-}}{2 f_{\perp}}$" + "\n" + " ")
axs[7].legend(["$V_{SC}$"], **legend_options)
axs[7].set_ylim(e_lim_e)
make_labels(axs, [0.02, 0.85])
f.align_ylabels(axs)
| 0.636466 | 0.934095 |
# Tag 1. Kapitel 3. Data Frames
## Lektion 20.1 Data Frame Operationen
Data Frames sind die Arbeitstiere von R, deshalb erstellen wir uns in dieser Lektion eine Art "Cheatsheet" der üblichsten Operationen. Dadurch wird diese Lektion extrem nützlich sein und uns im weiteren Verlauf des Kurses extrem viel Zeit sparen, da wir Data Frames bereits kennen und auf unser Cheatsheet zurückgreifen können.
Wir werden uns eine Übersicht der folgenden typischen Operationen verschaffen:
* Data Frames erstellen
* Daten in Data Frames anpassen (editieren)
* Informationen über die Data Frames erhalten
* Bezug auf Zellen nehmen
* Bezug auf Zeilen nehmen
* Bezug auf Spalten nehmen
* Zeilen hinzufügen
* Spalten hinzufügen
* Spaltennamen definieren
* Merhere Zeilen auswählen
* Mehrere Spalten auswählen
* Mit fehlenden Werten umzugehen
# Data Frames erstellen
```
leer <- data.frame() # Leerer Data Frame
c1 <- 1:15 # Vektor von Zahlen
c2 <- letters[1:15] # Verktor von Zeichen
df <- data.frame(spalte.name.1=c1,spalte.name.2=c2)
df2 <- data.frame(c1,c2)
df
df2
# Erstellte Datensätze anpassen und ausgeben
df <- edit(df) # läuft nur in R Studio (in R Studio ausprobieren)
df
```
# Information über Data Frames erhalten
```
# Zeilen und Spalten zählen
nrow(df)
ncol(df)
nrow(mtcars)
ncol(mtcars)
# Spalten Namen
colnames(df)
colnames(mtcars)
# Zeilen Namen (wobei auch nur der Index ausgegeben werden könnte)
rownames(df)
rownames(mtcars)
head(df)
head(mtcars)
```
# Bezug auf Zellen nehmen
Wir können uns unter den Grundlagen die Verwendung von zwei Klammer-Paaren vorstellen, um eine einzelne Zelle abzufragen. Und für mehrere Zellen verwenden wir ein einfaches Klammer-Paar. Ein Beispiel:
```
vec <- df[[5, 2]] # Erhalte die Zelle durch [[Zeile,Spalte]]
dfneu <- df[1:5, 1:2] # Erhalte merhere Zeilen und Spalten für den neuen df
df[[2, 'spalte.name.1']] <- 777 # Einer Zelle einen neuen Wert zuteilen
df[[2, 'spalte.name.2']] <- 'l' # Einer Zelle einen neuen Wert zuteilen
vec
dfneu
df
```
# Bezug auf Zeilen nehmen
Wir nutzen üblicherweise die [Zeile,] Notation
```
# Gibt einen df zurück, nicht einen Vektor
zeilendf <- df[1:3, ]
zeilendf
# Um eine Zeile zu einem Vektor zu machen schreiben wir folgendes
vzeile <- as.numeric(as.vector(df[1,]))
vzeile2 <- as.numeric(as.vector(df[2,]))
vzeile
vzeile2
```
# Bezug auf Spalten nehmen
Die meisten Spaltenbezüge geben einen Vektor zurück:
```
autos <- mtcars
head(autos)
spaltenv1 <- autos$mpg # Gibt einen Vektor zurück
spaltenv1
class(spaltenv1)
spaltenv2 <- autos[, 'mpg'] # Gibt einen Vektor zurück
spaltenv2
class(spaltenv2)
spaltenv3<- autos[, 1]
spaltenv3
class(spaltenv3)
spaltenv4 <- autos[['mpg']] # Gibt einen Vektor zurück
spaltenv4
class(spaltenv4)
# Wege um Data Frames zu erzeugen
mpgdf <- autos['mpg'] # Erzeugt einen df mit einer Spalte
head(mpgdf)
class(mpgdf)
mpgdf2 <- autos[1] # Erzeugt einen df mit einer Spalte
head(mpgdf2)
class(mpgdf2)
```
# Zeilen hinzufügen
```
# Beide Argumente sind Data Frames
df2 <- data.frame(spalte.name.1=2000,spalte.name.2='neu' )
df2
# Nutze rbind, um eine neue Zeile zu erstellen!
dfneu <- rbind(df,df2)
dfneu
```
# Spalten hinzufügen
```
df$neuespalte <- rep(NA, nrow(df)) # NA Spalte
df$neuespalte2 <- rep('Neu', nrow(df)) # NA Spalte
df
df[, 'kopie.con.spalte.2'] <- df$spalte.name.2 # Eine Spalte kopieren
df
# Wir können auch Gleichungen verwenden
df[['spalte1.mal.zwei']] <- df$spalte.name.1 * 2
df
df3 <- cbind(df, c1_neu=df$spalte.name.1)
df3
```
# Spaltennamen definieren
```
# Zweite Spalte umbenennen
colnames(df)[2] <- 'Name der zweiten Spalte'
df
# Wir können auch alle mit einem Vektor umbenennen
colnames(df) <- c('spalte.1', 'spalte.2', 'spalte.3', 'spalte.4' ,'spalte.5', 'spalte.6')
df
```
# Mehrere Zeilen auswählen
```
erste.zehn.zeilen <- df[1:10, ] # Gleich zu head(df, 10)
erste.zehn.zeilen
aulles.außer.zeile.zwei <- df[-2, ]
aulles.außer.zeile.zwei
# Bedingte Auswahl
sub1 <- df[ (df$spalte.1 > 8 & df$spalte.6 > 10), ]
sub1
sub2 <- subset(df, spalte.1 > 8 & spalte.5 == 'l')
sub2
```
# Mehrere Spalten wählen
```
df[, c(1, 2, 3)] # Die Spalten 1 bis 3 wählen
df[, c('spalte.1', 'spalte.5')] # Nach Namen
df[, -1] # Alle Spalten außer die erste
df[, -c(1, 3)] # Ohne Spalte 1 und 3
```
# Mit fehlenden Werten umgehen
Mit felhenden Werten umgehen zu können ist eine wichtige Fähigkeit bei der Arbeit mit Data Frames!
```
any(is.na(df)) # Suche nach NA Werten
df
any(is.na(df$spalte.1)) # Suche nach NA Werten in einer Spalte
any(is.na(df$spalte.3))
df2 <- data.frame(
spalte.1=NA,
spalte.2=NA,
spalte.3=NA,
spalte.4=NA,
spalte.5=NA,
spalte.6=NA
)
df
df2
df3 <- rbind(df, df2)
df3
# Die Zeilen mit fehlenden Werten löschen
df4 <- df3[!is.na(df3$spalte.1), ]
df4
# NAs mit etwas anderem ersetzen
df[is.na(df)] <- 0 # Für den gesamten df
df
df$spalte.3[is.na(df$spalte.3)] <- 999 # Für eine Spezielle Spalte
df
```
Herzlichen Glückwunsch! Sie sind mit 1. Teil der Lektion N° 20 fertig!
#### Bitte denkt daran, dieses Notebook als Referenz für spätere Lektionen und Aufgaben zu verwenden.
|
github_jupyter
|
leer <- data.frame() # Leerer Data Frame
c1 <- 1:15 # Vektor von Zahlen
c2 <- letters[1:15] # Verktor von Zeichen
df <- data.frame(spalte.name.1=c1,spalte.name.2=c2)
df2 <- data.frame(c1,c2)
df
df2
# Erstellte Datensätze anpassen und ausgeben
df <- edit(df) # läuft nur in R Studio (in R Studio ausprobieren)
df
# Zeilen und Spalten zählen
nrow(df)
ncol(df)
nrow(mtcars)
ncol(mtcars)
# Spalten Namen
colnames(df)
colnames(mtcars)
# Zeilen Namen (wobei auch nur der Index ausgegeben werden könnte)
rownames(df)
rownames(mtcars)
head(df)
head(mtcars)
vec <- df[[5, 2]] # Erhalte die Zelle durch [[Zeile,Spalte]]
dfneu <- df[1:5, 1:2] # Erhalte merhere Zeilen und Spalten für den neuen df
df[[2, 'spalte.name.1']] <- 777 # Einer Zelle einen neuen Wert zuteilen
df[[2, 'spalte.name.2']] <- 'l' # Einer Zelle einen neuen Wert zuteilen
vec
dfneu
df
# Gibt einen df zurück, nicht einen Vektor
zeilendf <- df[1:3, ]
zeilendf
# Um eine Zeile zu einem Vektor zu machen schreiben wir folgendes
vzeile <- as.numeric(as.vector(df[1,]))
vzeile2 <- as.numeric(as.vector(df[2,]))
vzeile
vzeile2
autos <- mtcars
head(autos)
spaltenv1 <- autos$mpg # Gibt einen Vektor zurück
spaltenv1
class(spaltenv1)
spaltenv2 <- autos[, 'mpg'] # Gibt einen Vektor zurück
spaltenv2
class(spaltenv2)
spaltenv3<- autos[, 1]
spaltenv3
class(spaltenv3)
spaltenv4 <- autos[['mpg']] # Gibt einen Vektor zurück
spaltenv4
class(spaltenv4)
# Wege um Data Frames zu erzeugen
mpgdf <- autos['mpg'] # Erzeugt einen df mit einer Spalte
head(mpgdf)
class(mpgdf)
mpgdf2 <- autos[1] # Erzeugt einen df mit einer Spalte
head(mpgdf2)
class(mpgdf2)
# Beide Argumente sind Data Frames
df2 <- data.frame(spalte.name.1=2000,spalte.name.2='neu' )
df2
# Nutze rbind, um eine neue Zeile zu erstellen!
dfneu <- rbind(df,df2)
dfneu
df$neuespalte <- rep(NA, nrow(df)) # NA Spalte
df$neuespalte2 <- rep('Neu', nrow(df)) # NA Spalte
df
df[, 'kopie.con.spalte.2'] <- df$spalte.name.2 # Eine Spalte kopieren
df
# Wir können auch Gleichungen verwenden
df[['spalte1.mal.zwei']] <- df$spalte.name.1 * 2
df
df3 <- cbind(df, c1_neu=df$spalte.name.1)
df3
# Zweite Spalte umbenennen
colnames(df)[2] <- 'Name der zweiten Spalte'
df
# Wir können auch alle mit einem Vektor umbenennen
colnames(df) <- c('spalte.1', 'spalte.2', 'spalte.3', 'spalte.4' ,'spalte.5', 'spalte.6')
df
erste.zehn.zeilen <- df[1:10, ] # Gleich zu head(df, 10)
erste.zehn.zeilen
aulles.außer.zeile.zwei <- df[-2, ]
aulles.außer.zeile.zwei
# Bedingte Auswahl
sub1 <- df[ (df$spalte.1 > 8 & df$spalte.6 > 10), ]
sub1
sub2 <- subset(df, spalte.1 > 8 & spalte.5 == 'l')
sub2
df[, c(1, 2, 3)] # Die Spalten 1 bis 3 wählen
df[, c('spalte.1', 'spalte.5')] # Nach Namen
df[, -1] # Alle Spalten außer die erste
df[, -c(1, 3)] # Ohne Spalte 1 und 3
any(is.na(df)) # Suche nach NA Werten
df
any(is.na(df$spalte.1)) # Suche nach NA Werten in einer Spalte
any(is.na(df$spalte.3))
df2 <- data.frame(
spalte.1=NA,
spalte.2=NA,
spalte.3=NA,
spalte.4=NA,
spalte.5=NA,
spalte.6=NA
)
df
df2
df3 <- rbind(df, df2)
df3
# Die Zeilen mit fehlenden Werten löschen
df4 <- df3[!is.na(df3$spalte.1), ]
df4
# NAs mit etwas anderem ersetzen
df[is.na(df)] <- 0 # Für den gesamten df
df
df$spalte.3[is.na(df$spalte.3)] <- 999 # Für eine Spezielle Spalte
df
| 0.147218 | 0.797833 |
## Filtering & adding new records
Filtering enables you to zoom in or out within a chart, allowing the viewer to focus on certain selected elements, or get more context. You can also add new records to the data on the chart which makes it easy to work with real-time sources.
**Note:** Currently `Data.filter()` and `Data().set_filter()` only accept JavaScript expression as string. Data fields can be accessed via `record` object, see the examples below.
We add two items from the Genres dimension - using the || operator - to the filter, so the chart elements that belong to the other two items will vanish from the chart.
```
from ipyvizzu import Chart, Data, Config
chart = Chart()
data = Data()
data.add_dimension('Genres', [ 'Pop', 'Rock', 'Jazz', 'Metal'])
data.add_dimension('Types', [ 'Hard', 'Smooth', 'Experimental' ])
data.add_measure(
'Popularity',
[
[114, 96, 78, 52],
[56, 36, 174, 121],
[127, 83, 94, 58],
]
)
chart.animate(data)
chart.animate(Config({
"channels": {
"y": {
"set": ["Popularity", "Types"]
},
"x": {
"set": "Genres"
},
"label": {
"attach": "Popularity"
}
},
"color": {
"attach": "Types"
},
"title": "Filter by one dimension"
}))
filter1 = Data.filter("record['Genres'] == 'Pop' || record['Genres'] == 'Metal'")
chart.animate(filter1)
snapshot1 = chart.store()
```
Now we add a cross-filter that includes items from both the Genres and the Types dimensions. This way we override the filter from the previous state. If we weren't update the filter, Vizzu would use it in subsequent states.
```
chart.animate(snapshot1)
chart.animate(Config({"title": "Filter by two dimensions"}))
filter2 = Data.filter("(record['Genres'] == 'Pop' || record['Genres'] == 'Metal') && record['Types'] == 'Smooth'")
chart.animate(filter2)
snapshot2 = chart.store()
```
Switching the filter off to get back to the original view.
```
chart.animate(snapshot2)
chart.animate(Config({"title": "Filter off"}))
chart.animate(Data.filter(None))
snapshot3 = chart.store()
```
Here we add another record to the data set and update the chart accordingly.
```
chart.animate(snapshot3)
chart.animate(Config({"title": "Adding new records"}))
data2 = Data()
records = [
['Soul', 'Hard', 91],
['Soul', 'Smooth', 57],
['Soul', 'Experimental', 115]
]
data2.add_records(records)
chart.animate(data2)
```
Note: combining this option with the store function makes it easy to update previously configured states with fresh data since this function saves the config and style parameters of the chart into a variable but not the data.
Next chapter: [Without coordinates & noop channel](./without_coordinates.ipynb) ----- Previous chapter: [Orientation, split & polar](./orientation.ipynb) ----- Back to the [Table of contents](../doc.ipynb#tutorial)
|
github_jupyter
|
from ipyvizzu import Chart, Data, Config
chart = Chart()
data = Data()
data.add_dimension('Genres', [ 'Pop', 'Rock', 'Jazz', 'Metal'])
data.add_dimension('Types', [ 'Hard', 'Smooth', 'Experimental' ])
data.add_measure(
'Popularity',
[
[114, 96, 78, 52],
[56, 36, 174, 121],
[127, 83, 94, 58],
]
)
chart.animate(data)
chart.animate(Config({
"channels": {
"y": {
"set": ["Popularity", "Types"]
},
"x": {
"set": "Genres"
},
"label": {
"attach": "Popularity"
}
},
"color": {
"attach": "Types"
},
"title": "Filter by one dimension"
}))
filter1 = Data.filter("record['Genres'] == 'Pop' || record['Genres'] == 'Metal'")
chart.animate(filter1)
snapshot1 = chart.store()
chart.animate(snapshot1)
chart.animate(Config({"title": "Filter by two dimensions"}))
filter2 = Data.filter("(record['Genres'] == 'Pop' || record['Genres'] == 'Metal') && record['Types'] == 'Smooth'")
chart.animate(filter2)
snapshot2 = chart.store()
chart.animate(snapshot2)
chart.animate(Config({"title": "Filter off"}))
chart.animate(Data.filter(None))
snapshot3 = chart.store()
chart.animate(snapshot3)
chart.animate(Config({"title": "Adding new records"}))
data2 = Data()
records = [
['Soul', 'Hard', 91],
['Soul', 'Smooth', 57],
['Soul', 'Experimental', 115]
]
data2.add_records(records)
chart.animate(data2)
| 0.470007 | 0.935405 |
<h1 style="color:red">CN</h1>
1. <h4 style="color:yellow">TCP/UDP</h4>
<span style="color:orange">TCP</span>
- Connection oriented
- 3 Way handshake
- order of the packet is maintained, and any lost packet is re-transmitted
<br>
<br>
<span style="color:orange">UDP</span>
- Not connection oriented
- order of packet is not maintained
2. <h4 style="color:yellow">ROUTER SWITCHES</h4>
<span style="color:orange">ROUTER</span>
- connecting device
- Acts as a dispatcher and responsibe to find the shortest path for a packet
- Network layer
<br>
<br>
<span style="color:orange">SWITCHES</span>
- connecting device
- connects various devices in a network
- Data link layer
3. <h4 style="color:yellow">ROUTING PROTOCOLS</h4>
<span style="color:orange">Distance Vector</span>
- Selects best path on the basis of hop counts to reach the destination network
- RIP => Routing Information Protocol
<br>
<br>
<span style="color:orange">Link State / Shortest Path First</span>
- Knows about Internetworks than Distance vector
- - Neighbor Table => information about neighbor of the routers
- - Topology Table => best and backup route to dest
- - Routing Table => best route to dest
- OSPF => Open Shortest Path First
<br>
<br>
<span style="color:orange">Advanced Distance Vector</span>
- Hybrid protocol
- EIGRP => Enhanced Interior Gateway Routing Protocol
- - Acts as a link state routing protocol as it uses the concept of Hello protocol for neighbor discovery and forming adjacency
- - Acts as distance vector routing protocol as it learned routes from directly connected neighbors
4. <h4 style="color:yellow">OSI vs TCP/IP</h4>
- - TCP refers to Transmission Control Protocol
- OSI refers to Open Systems Interconnection
<br><br>
- - TCP/IP has 4 layers
- OSI has 7 layers
<br><br>
- - TCP/IP follow a horizontal approach.
- OSI uses vertical approach
<br><br>
- - TCP is more realiable
- OSI is less reliable
5. <h4 style="color:yellow">OSI LAYER</h4>
<span style="color:orange">Application Layer</span>
- Produce data for transmission over the network
- Browsers, Desktop application
<br>
<span style="color:orange">Presentation Layer</span>
- Translation
- Encryption and Decryption
- Compression - Reduces no. of bits transferred over the network
<br>
<span style="color:orange">Session Layer</span>
- Session establishment, maintenance and termination
- Synchronisation - Add checkpoints to detect errors
- Dialog Controller - Half-Duplex or Full-Duplex
<br>
<span style="color:orange">Transport Layer</span>
- Segmentation and Reassembly
- Service Point addressing
- Flow and Error Control - to ensure proper data transmission
<br>
<span style="color:orange">Network Layer</span>
- Routing - Select best path for transmission of packet
- Logical Addressing
<br>
<span style="color:orange">Data Link Layer</span>
- Physical Addressing
- Error and Flow Control
- Access Control
<br>
<span style="color:orange">Physical Layer</span>
- Bit Synchronisation and rate-control
- Physical Topologies - Star, Mesh, Bus...
- Transmission Mode - Simplex, Half-Duplex, Full-Duplex
6. <h4 style="color:yellow">DNS</h4>
- DNS translates domain names to IP addresses so browsers can load Internet resources.
<br><br>
<span style="color:orange">Reverse DNS</span>
- A reverse DNS lookup is a DNS query for the domain name associated with a given IP address.
7. <h3 style="color:yellow">IPv6</h3>
- 128 bit address
- Each device will have it's own IP address
- Security and Scalability
- No more NAT, DHCP
- No more private address collisions
- efficient routing
<h1 style="color:red">OS</h1>
1. <h4 style="color:yellow">SEMAPHORE</h4>
- signaling mechanism to manage concurrent processes by using a simple integer value
- used to solve the critical section problem and to achieve process synchronization in the multiprocessing environment
- Types: Binary and Counting
- Atomic ops: Wait and Signal
<br>
<h4 style="color:yellow">MUTEX</h4>
- A mutex is the same as a lock but it can be system wide (shared by multiple processes).
<br>
<h4 style="color:yellow">LOCK</h4>
- A lock allows only one thread to enter the part that's locked and the lock is not shared with any other processes.
2. <h4 style="color:yellow">DEADLOCK</h4>
- situation where a set of processes are blocked because each process is holding a resource and waiting for another resource acquired by some other process
<br>
<br>
<span style="color:orange">ARISE</span>
- Mutual Exclusion
- Hold and Wait
- No preemption
- Circular Wait
<br>
<br>
<span style="color:orange">HANDLING</span>
- Prevention or Avoidance (Banker's algo)
- Detectiona and recovery
3. <h4 style="color:yellow">DAEMON vs DEMON</h4>
- Daemon is a program that runs by itself directly under the operating system continuously and exist for the purpose of handling periodic service requests that a computer system expects to recieve.
- Demon is part of a larger application program.
4. <h4 style="color:yellow">VIRTUALISATION</h4>
- The process of running a virtual instance of a computer system in a layer abstracted from the actual hardware.
- Mostly refers to running multiple operating systems on a computer system simultaneously
<br><br>
<span style="color:orange">VIRTUAL MEMORY</span>
- Virtual Memory is a storage allocation scheme in which secondary memory can be addressed as though it were part of main memory.
- All memory references within a process are logical addresses that are dynamically translated into physical addresses at run time.
<br><br>
<span style="color:orange">THRASHING</span>
- Thrashing is a condition or a situation when the system is spending a major portion of its time in servicing the page faults, but the actual processing done is very negligible.
5. <h3 style="color:yellow">PRIORITY INVERSION</h3>
- Scenario in scheduling in which higher priority tasks is indirectly preempted by a lower priority task effectively inverting the relative priorities of the two tasks.
6. <h3 style="color:yellow">DINING PHILOSOPHER</h3>
<span style="color:orange">Solution</span>
- semaphore chopstick [5];
- do {
- wait (chopstick[i]);
- wait (chopstick[i+1] % 5);
- EATING
- signal (chopstick[i]);
- signal (chopstick[i+1] % 5);
- THINKING
- } while(1);
<br><br>
<span style="color:orange">Deadlock</span>
- All the philosophers pick their left chopstick simultaneously
<br><br>
<span style="color:orange">Avoid Deadlock</span>
- Should be at most four philosophers on the table
- Even philosopher should pick the right chopstick and then the left chopstick while an Odd philosopher should pick the left chopstick and then the right chopstick
- A philosopher should only be allowed to pick their chopstick if both are available at the same time
<h1 style="color:red">OOP's and CLOUD</h1>
1. <h4 style="color:yellow">DATA STRUCTURES</h4>
<span style="color:orange">Linked List</span>
- A linked list is a linear collection of data elements whose order is not given by their physical placement in memory.
- Instead, each element points to the next.
- It is a data structure consisting of a collection of nodes which together represent a sequence.
<br>
<span style="color:orange">Binary Search Tree</span>
- A tree in which all the nodes follow
- - value of key of left sub-tree is less than the value of the parent node
- - value of key of right sub-tree is greater than the value of the parent node
<br>
<span style="color:orange">Microservice Architecture</span>
2. <h4 style="color:yellow">ABSTRACT CLASS & INTERFACE</h4>
<span style="color:orange">ABSTRACT CLASS</span>
- Blueprint for other classes
- An abstract method is a method that has a declaration but does not have an implementation.
from abc import ABC, abstractmethod<br>
class Polygon(ABC):
@abstractmethod
def noofsides(self):
pass
class Triangle(Polygon):
# overriding abstract method
def noofsides(self):
print("I have 3 sides")
<br><br>
<span style="color:orange">INTERFACE</span>
- help determine what class you should use to tackle the current problem
3. <h4 style="color:yellow">C</h4>
<span style="color:orange">GLOBAL VARIABLE</span>
- Variable defined outside all functions and available for all functions
- Exists till program ends
<br><br>
<span style="color:orange">STATIC VARIBALE</span>
- Maintains value from one function call to another and exists untill the program ends
- Either global or local
- Default value 0
<br><br>
<span style="color:orange">VOID</span>
- Pointer that has no associated data type with it
- Can hold address of any type and can be typcasted to any type
4. <h4 style="color:yellow">CLOUD</h4>
<span style="color:orange">SaaS</span>
- Can be run on web browsers without any downloads or installation required.
- Cost effective, Accessible everywhere, scalable, auto-updates
- Gmail, Ms-365, Dropbox
<br>
<span style="color:orange">PaaS</span>
- Provides platform and enviroment for developers to build applications and services over the internet
- Development and Deployment independent of the **Hardware**
- Efficient lifecycle, cost effective
- Salesforce, Google App Engine
<br>
<span style="color:orange">IaaS</span>
- Model that delivers computing, networking, storage as an outsource to the users
- Allows Dynamic Scaling, resources as distributed as a service
- Security, pay on a per use basis, website hosting
- AWS, GCP, Azure
5. <h3 style="color:yellow">MICROSERVICES</h3>
<span style="color:orange">Microservice Architecture</span>
- An architectural deployment style which builds an application as a collection of small autonomous services developed for business domain
- Loosely coupled
- Fault Isolation, Independent deployment and time reduction.
<br>
<span style="color:orange">Monolithic Architecture</span>
- A big container in which all the software components of an application are clubbed inside a single package
- Tightly coupled
<br>
<span style="color:orange">REST</span>
- Representational State Transfer is an architectural style that helps systems communicate over the internet
- Makes microservice easier to understand and implement
<h1 style="color:red">Coding Q's</h1>
<h3 style="color:yellow">Expressive Words</h3>
```
def expressiveWords(s, words):
return sum(check(s, w) for w in words)
def check(s, w):
i, j, n, m = 0, 0, len(s), len(w)
for i in range(n):
if j < m and s[i] == w[j]:
j += 1
elif s[i-1: i+2] != s[i]*3 != s[i-2: i+1]:
return False
return j == m
s = "heeellooo"
words = ["hello", "hi", "helo"]
print(expressiveWords(s, words))
s = "zzzzzyyyyy"
words = ["zzyy","zy","zyy"]
print(expressiveWords(s, words))
```
<h3 style="color:yellow">A string of numbers in random order is given and you have to print them in decimal format in strictly decreasing order.</h3>
```
import collections, itertools
def unscramble(S):
sc = collections.Counter(s)
return list(itertools.chain.from_iterable(
[
[9]*(sc['i'] - sc['x'] - sc['g'] - (sc['f'] - sc['u'])),
[8]*(sc['g']),
[7]*(sc['s'] - sc['x']),
[6]*(sc['x']),
[5]*(sc['f'] - sc['u']),
[4]*(sc['u']),
[3]*(sc['h'] - sc['g']),
[2]*(sc['w']),
[1]*(sc['o'] - sc['z'] - sc['w'] - sc['u']),
[0]*(sc['z'])
]
))
s = "nieignhtesevfouenr"
print(unscramble(s))
```
<h3 style="color:yellow">Lucky Number</h3>
```
def isLucky(n):
counter = 2
if counter > n:
return True
if n % counter == 0:
return False
n -= (n/isLucky.counter)
counter = counter + 1
return isLucky(n)
isLucky.counter = 2
x = 11
if isLucky(x):
print(x, "Lucky number")
else:
print(x, "not a Lucky number")
```
<h3 style="color:yellow">Doubly Linked List Insert </h3>
```
class Node:
def __init__(self, x):
self.data = x
self.prev = None
self.next = None
#Add a node at the front
def push(head, data):
newNode = Node(data)
#newNode.data = data
newNode.next = head
newNode.prev = None
if (head is not None):
head.prev = newNode
head = newNode
return head
# Insert before a node
def insertBefore(head, data, nextNode):
if nextNode == None:
return
newNode = Node(data)
newNode.prev = nextNode.prev
nextNode.prev = newNode
newNode.next = nextNode
if newNode.prev != None:
newNode.prev.next = newNode
else:
head = newNode
return head
# Insert after a node
def insertAfter(head, data, prevNode):
if prevNode == None:
return
newNode = Node(data)
newNode.next = prevNode.next
prevNode.next = newNode
newNode.prev = prevNode
if newNode.next:
newNode.next.prev = newNode
return head
# Append at the end
def append(head, data):
newNode = Node(data)
if head is None:
head = newNode
return
last = head
while last.next:
last = last.next
last.next = newNode
newNode.prev = last
return head
def printList(node):
last = None
print("Traversal in forward direction ")
while (node != None):
print(node.data, end=" ")
last = node
node = node.next
print("\nTraversal in reverse direction ")
while (last != None):
print(last.data, end=" ")
last = last.prev
if __name__ == '__main__':
# /* Start with the empty list */
head = None
head = push(head, 7)
head = push(head, 1)
head = push(head, 4)
# Insert 8, before 1. So linked list becomes 4.8.1.7.NULL
head = insertBefore(head, 8, head.next)
print("Created DLL is: ")
printList(head)
```
<h3 style="color:yellow">Reverse a Linked List</h3>
```
def reverse():
prev = None
curr = head
while (curr is not None):
temp = curr.next
curr.next = prev
prev = curr
curr = temp
#head = prev
return prev
```
<h3 style="color:yellow">Reverse a Double Linked List</h3>
```
def reverse(head):
temp = None
curr = head
while (curr is not None):
temp = curr.prev
curr.prev = curr.next
curr.next = temp
curr = curr.prev
if temp is not None:
head = temp.prev
return head
# Using Stack
def reverseStack():
stack = []
temp = head
while temp is not None:
stack.append(temp.data)
temp = temp.next
temp = head
while temp is not None:
temp.data = stack.pop()
temp = temp.next
temp.next = None
```
<h3 style="color:yellow">Count of Set bits / Brian Kernighan’s Algorithm</h3>
```
def countSetbits(n):
count = 0
while n:
n &= (n-1)
count += 1
return count
n = 9
# function calling
print(countSetbits(n))
print(bin(15).count('1'))
```
<h3 style="color:yellow">Find the Missing Number</h3>
```
def getMissingNo(arr):
n = len(arr)
total = int((n+1)*(n+2) // 2)
return total - sum(arr)
def getMissingNo_1(arr):
n = len(arr)
x1 = arr[0]
x2 = 1
for i in range(1, n):
x1 ^= arr[i]
for i in range(2, n+2):
x2 ^= i
return x1^x2
arr = [1,2,3,5,6]
print(getMissingNo_1(arr))
```
<h3 style="color:yellow">Eggs dropping puzzle</h3>
```
def binomialCoeff(x, n, k):
s = 0
term = 1
i = 1
while i <= n and s < k:
term *= x - i + 1
term /= i
s += term
i += 1
return s
def minTrial(n, k):
low = 1
high = k
while(low < high):
mid = int((low+high) / 2)
if binomialCoeff(mid, n, k) < k:
low = mid + 1
else:
high = mid
return int(low)
print(minTrial(2,100))
```
<h3 style="color:yellow">LINKED LIST</h3>
```
class Node:
def __init__(self, data):
self.data = data
self.next = None
def push(head, data):
newNode = Node(data)
newNode.next = head
head = newNode
return head
def printSecondList(l1, l2):
temp = l1
temp1 = l2
while temp is not None:
i = 1
while i < temp.data:
temp1 = temp1.next
i += 1
print(temp1.data,end=" ")
temp = temp.next
temp1 = l2
l1 = None
l2 = None
l1 = push(l1, 5)
l1 = push(l1, 2)
l2 = push(l2, 8)
l2 = push(l2, 7)
l2 = push(l2, 6)
l2 = push(l2, 5)
l2 = push(l2, 4)
printSecondList(l1, l2)
```
<h3 style="color:yellow">DETECT LOOP</h3>
```
class Node:
def __init__(self, data):
self.data = data
self.next = None
class LinkedList:
def __init__(self):
self.head = None
def push(self, data):
newNode = Node(data)
newNode.next = self.head
self.head = newNode
def printList(self):
temp = self.head
while temp is not None:
print(temp.data)
temp = temp.next
def detectLoop(self):
slow = self.head
fast = self.head
while slow and fast and fast.next:
slow = slow.next
fast = fast.next.next
if slow == fast:
return True
l1 = LinkedList()
l1.push(20)
l1.push(4)
l1.push(15)
l1.push(10)
l1.head.next.next.next.next = l1.head
if (l1.detectLoop()):
print ("Loop Found")
else:
print ("No Loop")
```
<h3 style="color: yellow">MIN STACK</h3>
```
class GetMin:
def __init__(self):
self.q = []
def push(self, data):
curMin = self.getMin()
if curMin == None or data < curMin:
curMin = data
self.q.append((data, curMin))
def pop(self):
self.q.pop()
def top(self):
if len(self.q) == 0:
return None
else:
return self.q[len(self.q) - 1][0]
def getMin(self):
if len(self.q) == 0:
return None
else:
return self.q[len(self.q) - 1][1]
stack = GetMin()
stack.getMin()
stack.push(3)
stack.push(5)
stack.getMin()
stack.push(2)
stack.push(1)
stack.getMin()
stack.pop()
stack.getMin()
stack.pop()
stack.getMin()
```
<h3 style="color:yellow">Duplicate Parenthesis</h3>
```
def findDuplicateparenthesis(string):
stack = []
for ch in string:
if ch == ')':
top = stack.pop()
elementsInside = 0
while top != '(':
elementsInside += 1
top = stack.pop()
if elementsInside < 1:
return True
else:
stack.append(ch)
return False
if __name__ == "__main__":
# input balanced expression
string = "((a+b)+((c+d)))"
if findDuplicateparenthesis(string) == True:
print("Duplicate Found")
else:
print("No Duplicate Found");
```
<h3 style="color:yellow">Powerset</h3>
```
import math;
def powerset(arr, N):
power_set_size = (int) (math.pow(2, N))
counter = 0
i = 0
for counter in range(0, power_set_size):
for j in range(N):
if((counter & (1 << j)) > 0):
print(arr[j], end = "")
print("")
from itertools import combinations
def powerset_2(arr, N):
#print(None)
for i in range(0, N):
for ele in combinations(arr, i):
print(''.join(ele))
arr = ['a','b','c']
powerset_2(arr, len(arr))
```
<h3 style="color:yellow">Count pairs with given sum</h3>
```
def pairCount(arr, N, S):
count = 0
for i in range(N):
for j in range(i+1, N):
if arr[i] + arr[j] == S:
count += 1
return count
def pairCount_2(arr, N, S):
m = [0]*1000
count = 0
for i in range(N):
m[arr[i]] += 1
for i in range(N):
count += m[S - arr[i]]
if (S - arr[i] == arr[i]):
count -= 1
return (count // 2)
arr = [1, 5, 7, -1, 5, 2, 4]
n = len(arr)
sum = 6
print(pairCount_2(arr, n, sum))
```
<h3 style="color:yellow">Middle Node of Linked List </h3>
```
def middleLL(head):
slow = fast = head
while fast and fast.next:
slow = slow.next
fast = fast.next.next
print("Middle Node: ", slow.data)
```
|
github_jupyter
|
def expressiveWords(s, words):
return sum(check(s, w) for w in words)
def check(s, w):
i, j, n, m = 0, 0, len(s), len(w)
for i in range(n):
if j < m and s[i] == w[j]:
j += 1
elif s[i-1: i+2] != s[i]*3 != s[i-2: i+1]:
return False
return j == m
s = "heeellooo"
words = ["hello", "hi", "helo"]
print(expressiveWords(s, words))
s = "zzzzzyyyyy"
words = ["zzyy","zy","zyy"]
print(expressiveWords(s, words))
import collections, itertools
def unscramble(S):
sc = collections.Counter(s)
return list(itertools.chain.from_iterable(
[
[9]*(sc['i'] - sc['x'] - sc['g'] - (sc['f'] - sc['u'])),
[8]*(sc['g']),
[7]*(sc['s'] - sc['x']),
[6]*(sc['x']),
[5]*(sc['f'] - sc['u']),
[4]*(sc['u']),
[3]*(sc['h'] - sc['g']),
[2]*(sc['w']),
[1]*(sc['o'] - sc['z'] - sc['w'] - sc['u']),
[0]*(sc['z'])
]
))
s = "nieignhtesevfouenr"
print(unscramble(s))
def isLucky(n):
counter = 2
if counter > n:
return True
if n % counter == 0:
return False
n -= (n/isLucky.counter)
counter = counter + 1
return isLucky(n)
isLucky.counter = 2
x = 11
if isLucky(x):
print(x, "Lucky number")
else:
print(x, "not a Lucky number")
class Node:
def __init__(self, x):
self.data = x
self.prev = None
self.next = None
#Add a node at the front
def push(head, data):
newNode = Node(data)
#newNode.data = data
newNode.next = head
newNode.prev = None
if (head is not None):
head.prev = newNode
head = newNode
return head
# Insert before a node
def insertBefore(head, data, nextNode):
if nextNode == None:
return
newNode = Node(data)
newNode.prev = nextNode.prev
nextNode.prev = newNode
newNode.next = nextNode
if newNode.prev != None:
newNode.prev.next = newNode
else:
head = newNode
return head
# Insert after a node
def insertAfter(head, data, prevNode):
if prevNode == None:
return
newNode = Node(data)
newNode.next = prevNode.next
prevNode.next = newNode
newNode.prev = prevNode
if newNode.next:
newNode.next.prev = newNode
return head
# Append at the end
def append(head, data):
newNode = Node(data)
if head is None:
head = newNode
return
last = head
while last.next:
last = last.next
last.next = newNode
newNode.prev = last
return head
def printList(node):
last = None
print("Traversal in forward direction ")
while (node != None):
print(node.data, end=" ")
last = node
node = node.next
print("\nTraversal in reverse direction ")
while (last != None):
print(last.data, end=" ")
last = last.prev
if __name__ == '__main__':
# /* Start with the empty list */
head = None
head = push(head, 7)
head = push(head, 1)
head = push(head, 4)
# Insert 8, before 1. So linked list becomes 4.8.1.7.NULL
head = insertBefore(head, 8, head.next)
print("Created DLL is: ")
printList(head)
def reverse():
prev = None
curr = head
while (curr is not None):
temp = curr.next
curr.next = prev
prev = curr
curr = temp
#head = prev
return prev
def reverse(head):
temp = None
curr = head
while (curr is not None):
temp = curr.prev
curr.prev = curr.next
curr.next = temp
curr = curr.prev
if temp is not None:
head = temp.prev
return head
# Using Stack
def reverseStack():
stack = []
temp = head
while temp is not None:
stack.append(temp.data)
temp = temp.next
temp = head
while temp is not None:
temp.data = stack.pop()
temp = temp.next
temp.next = None
def countSetbits(n):
count = 0
while n:
n &= (n-1)
count += 1
return count
n = 9
# function calling
print(countSetbits(n))
print(bin(15).count('1'))
def getMissingNo(arr):
n = len(arr)
total = int((n+1)*(n+2) // 2)
return total - sum(arr)
def getMissingNo_1(arr):
n = len(arr)
x1 = arr[0]
x2 = 1
for i in range(1, n):
x1 ^= arr[i]
for i in range(2, n+2):
x2 ^= i
return x1^x2
arr = [1,2,3,5,6]
print(getMissingNo_1(arr))
def binomialCoeff(x, n, k):
s = 0
term = 1
i = 1
while i <= n and s < k:
term *= x - i + 1
term /= i
s += term
i += 1
return s
def minTrial(n, k):
low = 1
high = k
while(low < high):
mid = int((low+high) / 2)
if binomialCoeff(mid, n, k) < k:
low = mid + 1
else:
high = mid
return int(low)
print(minTrial(2,100))
class Node:
def __init__(self, data):
self.data = data
self.next = None
def push(head, data):
newNode = Node(data)
newNode.next = head
head = newNode
return head
def printSecondList(l1, l2):
temp = l1
temp1 = l2
while temp is not None:
i = 1
while i < temp.data:
temp1 = temp1.next
i += 1
print(temp1.data,end=" ")
temp = temp.next
temp1 = l2
l1 = None
l2 = None
l1 = push(l1, 5)
l1 = push(l1, 2)
l2 = push(l2, 8)
l2 = push(l2, 7)
l2 = push(l2, 6)
l2 = push(l2, 5)
l2 = push(l2, 4)
printSecondList(l1, l2)
class Node:
def __init__(self, data):
self.data = data
self.next = None
class LinkedList:
def __init__(self):
self.head = None
def push(self, data):
newNode = Node(data)
newNode.next = self.head
self.head = newNode
def printList(self):
temp = self.head
while temp is not None:
print(temp.data)
temp = temp.next
def detectLoop(self):
slow = self.head
fast = self.head
while slow and fast and fast.next:
slow = slow.next
fast = fast.next.next
if slow == fast:
return True
l1 = LinkedList()
l1.push(20)
l1.push(4)
l1.push(15)
l1.push(10)
l1.head.next.next.next.next = l1.head
if (l1.detectLoop()):
print ("Loop Found")
else:
print ("No Loop")
class GetMin:
def __init__(self):
self.q = []
def push(self, data):
curMin = self.getMin()
if curMin == None or data < curMin:
curMin = data
self.q.append((data, curMin))
def pop(self):
self.q.pop()
def top(self):
if len(self.q) == 0:
return None
else:
return self.q[len(self.q) - 1][0]
def getMin(self):
if len(self.q) == 0:
return None
else:
return self.q[len(self.q) - 1][1]
stack = GetMin()
stack.getMin()
stack.push(3)
stack.push(5)
stack.getMin()
stack.push(2)
stack.push(1)
stack.getMin()
stack.pop()
stack.getMin()
stack.pop()
stack.getMin()
def findDuplicateparenthesis(string):
stack = []
for ch in string:
if ch == ')':
top = stack.pop()
elementsInside = 0
while top != '(':
elementsInside += 1
top = stack.pop()
if elementsInside < 1:
return True
else:
stack.append(ch)
return False
if __name__ == "__main__":
# input balanced expression
string = "((a+b)+((c+d)))"
if findDuplicateparenthesis(string) == True:
print("Duplicate Found")
else:
print("No Duplicate Found");
import math;
def powerset(arr, N):
power_set_size = (int) (math.pow(2, N))
counter = 0
i = 0
for counter in range(0, power_set_size):
for j in range(N):
if((counter & (1 << j)) > 0):
print(arr[j], end = "")
print("")
from itertools import combinations
def powerset_2(arr, N):
#print(None)
for i in range(0, N):
for ele in combinations(arr, i):
print(''.join(ele))
arr = ['a','b','c']
powerset_2(arr, len(arr))
def pairCount(arr, N, S):
count = 0
for i in range(N):
for j in range(i+1, N):
if arr[i] + arr[j] == S:
count += 1
return count
def pairCount_2(arr, N, S):
m = [0]*1000
count = 0
for i in range(N):
m[arr[i]] += 1
for i in range(N):
count += m[S - arr[i]]
if (S - arr[i] == arr[i]):
count -= 1
return (count // 2)
arr = [1, 5, 7, -1, 5, 2, 4]
n = len(arr)
sum = 6
print(pairCount_2(arr, n, sum))
def middleLL(head):
slow = fast = head
while fast and fast.next:
slow = slow.next
fast = fast.next.next
print("Middle Node: ", slow.data)
| 0.195786 | 0.68616 |
```
import numpy as np
```
# EP 5: Array
## EP 5.0: Even entries first
```
a = list(range(100))
%time b = a[1::2]+a[::2]
a = list(range(100))
%time
for i in range(len(a)//2):
a[i], a[2*i+1] = a[2*i+1], a[i]
```
## EP 5.1: Dutch national flag problem
## EP 5.5 Delete dupicated numbers from a sorted array
```
a = [2,3,5,5,7,77,11,17,13, 13 ,13, 13, 55,55, 0]
N = len(a)
i = j = 1
while j < N:
while a[j] == a[j-1]:
if j < N-1:
j += 1
else:
break
a[i] = a[j]
i += 1
j += 1
res = a[:i]
res
```
## EP 5.14: Compute random permutation
## EP 5.18: Compute spiral orderin of 2d array
```
dim_x = 4
a = np.reshape(range(dim_x**2), (dim_x, dim_x))
r_max, c_max = a.shape[0], a.shape[1]
r_min, c_min = (0, 0)
res = []
while ((r_min < r_max) & (c_min < c_max)):
for j in range(c_min, c_max):
res.append(a[r_min, j])
r_min += 1
for i in range(r_min, r_max):
res.append(a[i, c_max-1])
c_max -= 1
for j in range(c_max-1, c_min-1, -1):
res.append(a[r_max-1, j])
r_max -= 1
for i in range(r_max-1, r_min-1, -1):
res.append(a[i, c_min])
c_min += 1
print(f'Orig:\n {a} \n Spiral order: \n {res}')
```
## EP 5.19: Rotate 2d array
## EP 5.20: Compute Nth row in Pascal's triangle
# EP 6: String
## EP 6.0: Test panlindromicity
```
s = 'adb3kdda3k3008mdaf'
s[~2]
display(21//4, 21%4)
```
## EP 6.1: atoi, itoa
```
def atoi(s):
start = 1 if s[0]=='-' else 0
res = int(s[start])
for i in range(1+start, len(s)):
res = 10*res + int(s[i])
return res * (-1)**start
s = '992343242'
atoi(s)
def itoa(i):
start = 1 if i < 0 else 0
s = [i//10**j for j in range(len(i))]
```
## EP 6.4: Column name -> Decimal
```
s = 'AA'
res = 0
for c in s:
res += ord(c) - ord('A') + 1
res
```
EP 6.6: Reveser words in a sentence
EP 6.0: Test panlindromicity
EP 6.0: Test panlindromicity
# EP 7: Linked list
## EP 7.0: Build linked list, search, insert, delete
```
class ListNode:
def __init__(self, data=0, next_node=None):
self.data = data
self.next_node = next_node
def insert_node(curr_node, new_node):
''' insert after curr_node'''
if curr_node is not None:
new_node.next_node = curr_node.next_node
curr_node.next_node = new_node
return new_node
def print_linked_list(head):
while head is not None:
i, head = head.data, head.next_node
print(f'{i}')
def build_linked_list(a):
head = tail = ListNode(data=a[0])
if len(a) == 1:
return head
for i in a[1:]:
tail = insert_node(tail, ListNode(i))
return head
def search_node(head, data):
while head is not None:
if head.data == data:
return head
head = head.next
def delete_node(node):
# delete the node after
a = [3, 4, 5, 23,-33]
head = build_linked_list(a)
print_linked_list(head)
```
# EP 8: Stack
## EP 8.0: Reverse linked list
## EP 8.3: Test well-formedness of parenthesis/braces/brackets
## EP 8.5: Find building with sunset view
```
str(3)
```
|
github_jupyter
|
import numpy as np
a = list(range(100))
%time b = a[1::2]+a[::2]
a = list(range(100))
%time
for i in range(len(a)//2):
a[i], a[2*i+1] = a[2*i+1], a[i]
a = [2,3,5,5,7,77,11,17,13, 13 ,13, 13, 55,55, 0]
N = len(a)
i = j = 1
while j < N:
while a[j] == a[j-1]:
if j < N-1:
j += 1
else:
break
a[i] = a[j]
i += 1
j += 1
res = a[:i]
res
dim_x = 4
a = np.reshape(range(dim_x**2), (dim_x, dim_x))
r_max, c_max = a.shape[0], a.shape[1]
r_min, c_min = (0, 0)
res = []
while ((r_min < r_max) & (c_min < c_max)):
for j in range(c_min, c_max):
res.append(a[r_min, j])
r_min += 1
for i in range(r_min, r_max):
res.append(a[i, c_max-1])
c_max -= 1
for j in range(c_max-1, c_min-1, -1):
res.append(a[r_max-1, j])
r_max -= 1
for i in range(r_max-1, r_min-1, -1):
res.append(a[i, c_min])
c_min += 1
print(f'Orig:\n {a} \n Spiral order: \n {res}')
s = 'adb3kdda3k3008mdaf'
s[~2]
display(21//4, 21%4)
def atoi(s):
start = 1 if s[0]=='-' else 0
res = int(s[start])
for i in range(1+start, len(s)):
res = 10*res + int(s[i])
return res * (-1)**start
s = '992343242'
atoi(s)
def itoa(i):
start = 1 if i < 0 else 0
s = [i//10**j for j in range(len(i))]
s = 'AA'
res = 0
for c in s:
res += ord(c) - ord('A') + 1
res
class ListNode:
def __init__(self, data=0, next_node=None):
self.data = data
self.next_node = next_node
def insert_node(curr_node, new_node):
''' insert after curr_node'''
if curr_node is not None:
new_node.next_node = curr_node.next_node
curr_node.next_node = new_node
return new_node
def print_linked_list(head):
while head is not None:
i, head = head.data, head.next_node
print(f'{i}')
def build_linked_list(a):
head = tail = ListNode(data=a[0])
if len(a) == 1:
return head
for i in a[1:]:
tail = insert_node(tail, ListNode(i))
return head
def search_node(head, data):
while head is not None:
if head.data == data:
return head
head = head.next
def delete_node(node):
# delete the node after
a = [3, 4, 5, 23,-33]
head = build_linked_list(a)
print_linked_list(head)
str(3)
| 0.108118 | 0.861771 |
# Feature Analysis Using TensorFlow Data Validation and Facets
## Learning Objectives
1. Use TFRecords to load record-oriented binary format data
2. Use TFDV to generate statistics and Facets to visualize the data
3. Use the TFDV widget to answer questions
4. Analyze label distribution for subset groups
## Introduction
Bias can manifest in any part of a typical machine learning pipeline, from an unrepresentative dataset, to learned model representations, to the way in which the results are presented to the user. Errors that result from this bias can disproportionately impact some users more than others.
[TensorFlow Data Validation](https://www.tensorflow.org/tfx/data_validation/get_started) (TFDV) is one tool you can use to analyze your data to find potential problems in your data, such as missing values and data imbalances - that can lead to Fairness disparities. The TFDV tool analyzes training and serving data to compute descriptive statistics, infer a schema, and detect data anomalies. [Facets Overview](https://pair-code.github.io/facets/) provides a succinct visualization of these statistics for easy browsing. Both the TFDV and Facets are tools that are part of the [Fairness Indicators](https://www.tensorflow.org/tfx/fairness_indicators).
In this notebook, we use TFDV to compute descriptive statistics that provide a quick overview of the data in terms of the features that are present and the shapes of their value distributions. We use Facets Overview to visualize these statistics using the Civil Comments dataset.
Each learning objective will correspond to a __#TODO__ in the [student lab notebook](../labs/adv_tfdv_facets.ipynb) -- try to complete that notebook first before reviewing this solution notebook.
## Set up environment variables and load necessary libraries
We will start by importing the necessary dependencies for the libraries we'll be using in this exercise. First, run the cell below to install Fairness Indicators.
**NOTE:** You can ignore the "pip" being invoked by an old script wrapper, as it will not affect the lab's functionality.
```
!pip3 install fairness-indicators==0.1.2 --user
```
<strong>Restart the kernel</strong> after you do a pip3 install (click on the <strong>Restart the kernel</strong> button above).
Kindly ignore the deprecation warnings and incompatibility errors.
Next, import all the dependencies we'll use in this exercise, which include Fairness Indicators, TensorFlow Data Validation (tfdv), and the What-If tool (WIT) Facets Overview.
```
# %tensorflow_version 2.x
import sys, os
import warnings
warnings.filterwarnings('ignore')
#os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3' # Ignore deprecation warnings
import tempfile
import apache_beam as beam
import numpy as np
import pandas as pd
from datetime import datetime
import tensorflow_hub as hub
import tensorflow as tf
import tensorflow_model_analysis as tfma
import tensorflow_data_validation as tfdv
from tensorflow_model_analysis.addons.fairness.post_export_metrics import fairness_indicators
from tensorflow_model_analysis.addons.fairness.view import widget_view
from fairness_indicators.examples import util
import warnings
warnings.filterwarnings("ignore")
from witwidget.notebook.visualization import WitConfigBuilder
from witwidget.notebook.visualization import WitWidget
print(tf.version.VERSION)
print(tf) # This statement shows us what version of Python we are currently running.
```
### About the Civil Comments dataset
Click below to learn more about the Civil Comments dataset, and how we've preprocessed it for this exercise.
The Civil Comments dataset comprises approximately 2 million public comments that were submitted to the Civil Comments platform. [Jigsaw](https://jigsaw.google.com/) sponsored the effort to compile and annotate these comments for ongoing [research](https://arxiv.org/abs/1903.04561); they've also hosted competitions on [Kaggle](https://www.kaggle.com/c/jigsaw-unintended-bias-in-toxicity-classification) to help classify toxic comments as well as minimize unintended model bias.
#### Features
Within the Civil Comments data, a subset of comments are tagged with a variety of identity attributes pertaining to gender, sexual orientation, religion, race, and ethnicity. Each identity annotation column contains a value that represents the percentage of annotators who categorized a comment as containing references to that identity. Multiple identities may be present in a comment.
**NOTE:** These identity attributes are intended *for evaluation purposes only*, to assess how well a classifier trained solely on the comment text performs on different tag sets.
To collect these identity labels, each comment was reviewed by up to 10 annotators, who were asked to indicate all identities that were mentioned in the comment. For example, annotators were posed the question: "What genders are mentioned in the comment?", and asked to choose all of the following categories that were applicable.
* Male
* Female
* Transgender
* Other gender
* No gender mentioned
**NOTE:** *We recognize the limitations of the categories used in the original dataset, and acknowledge that these terms do not encompass the full range of vocabulary used in describing gender.*
Jigsaw used these ratings to generate an aggregate score for each identity attribute representing the percentage of raters who said the identity was mentioned in the comment. For example, if 10 annotators reviewed a comment, and 6 said that the comment mentioned the identity "female" and 0 said that the comment mentioned the identity "male," the comment would receive a `female` score of `0.6` and a `male` score of `0.0`.
**NOTE:** For the purposes of annotation, a comment was considered to "mention" gender if it contained a comment about gender issues (e.g., a discussion about feminism, wage gap between men and women, transgender rights, etc.), gendered language, or gendered insults. Use of "he," "she," or gendered names (e.g., Donald, Margaret) did not require a gender label.
#### Label
Each comment was rated by up to 10 annotators for toxicity, who each classified it with one of the following ratings.
* Very Toxic
* Toxic
* Hard to Say
* Not Toxic
Again, Jigsaw used these ratings to generate an aggregate toxicity "score" for each comment (ranging from `0.0` to `1.0`) to serve as the [label](https://developers.google.com/machine-learning/glossary?utm_source=Colab&utm_medium=fi-colab&utm_campaign=fi-practicum&utm_content=glossary&utm_term=label#label), representing the fraction of annotators who labeled the comment either "Very Toxic" or "Toxic." For example, if 10 annotators rated a comment, and 3 of them labeled it "Very Toxic" and 5 of them labeled it "Toxic", the comment would receive a toxicity score of `0.8`.
**NOTE:** For more information on the Civil Comments labeling schema, see the [Data](https://www.kaggle.com/c/jigsaw-unintended-bias-in-toxicity-classification/data) section of the Jigsaw Untended Bias in Toxicity Classification Kaggle competition.
### Preprocessing the data
For the purposes of this exercise, we converted toxicity and identity columns to booleans in order to work with our neural net and metrics calculations. In the preprocessed dataset, we considered any value ≥ 0.5 as True (i.e., a comment is considered toxic if 50% or more crowd raters labeled it as toxic).
For identity labels, the threshold 0.5 was chosen and the identities were grouped together by their categories. For example, if one comment has `{ male: 0.3, female: 1.0, transgender: 0.0, heterosexual: 0.8, homosexual_gay_or_lesbian: 1.0 }`, after processing, the data will be `{ gender: [female], sexual_orientation: [heterosexual, homosexual_gay_or_lesbian] }`.
**NOTE:** Missing identity fields were converted to False.
### Use TFRecords to load record-oriented binary format data
-------------------------------------------------------------------------------------------------------
The [TFRecord format](https://www.tensorflow.org/tutorials/load_data/tfrecord) is a simple [Protobuf](https://developers.google.com/protocol-buffers)-based format for storing a sequence of binary records. It gives you and your machine learning models to handle arbitrarily large datasets over the network because it:
1. Splits up large files into 100-200MB chunks
2. Stores the results as serialized binary messages for faster ingestion
If you already have a dataset in TFRecord format, you can use the tf.keras.utils functions for accessing the data (as you will below!). If you want to practice creating your own TFRecord datasets you can do so outside of this lab by [viewing the documentation here](https://www.tensorflow.org/tutorials/load_data/tfrecord).
#### TODO 1: Use the utility functions tf.keras to download and import our datasets
Run the following cell to download and import the training and validation preprocessed datasets.
```
download_original_data = False #@param {type:"boolean"}
# TODO 1
if download_original_data:
train_tf_file = tf.keras.utils.get_file('train_tf.tfrecord',
'https://storage.googleapis.com/civil_comments_dataset/train_tf.tfrecord')
validate_tf_file = tf.keras.utils.get_file('validate_tf.tfrecord',
'https://storage.googleapis.com/civil_comments_dataset/validate_tf.tfrecord')
# The identity terms list will be grouped together by their categories
# (see 'IDENTITY_COLUMNS') on threshould 0.5. Only the identity term column,
# text column and label column will be kept after processing.
train_tf_file = util.convert_comments_data(train_tf_file)
validate_tf_file = util.convert_comments_data(validate_tf_file)
# TODO 1a
else:
train_tf_file = tf.keras.utils.get_file('train_tf_processed.tfrecord',
'https://storage.googleapis.com/civil_comments_dataset/train_tf_processed.tfrecord')
validate_tf_file = tf.keras.utils.get_file('validate_tf_processed.tfrecord',
'https://storage.googleapis.com/civil_comments_dataset/validate_tf_processed.tfrecord')
```
### Use TFDV to generate statistics and Facets to visualize the data
TensorFlow Data Validation supports data stored in a TFRecord file, a CSV input format, with extensibility for other common formats. You can find the available data decoders [here](https://github.com/tensorflow/data-validation/tree/master/tensorflow_data_validation/coders). In addition, TFDV provides the [tfdv.generate_statistics_from_dataframe](https://www.tensorflow.org/tfx/data_validation/api_docs/python/tfdv/generate_statistics_from_dataframe) utility function for users with in-memory data represented as a pandas DataFrame.
In addition to computing a default set of data statistics, TFDV can also compute statistics for semantic domains (e.g., images, text). To enable computation of semantic domain statistics, pass a tfdv.StatsOptions object with enable_semantic_domain_stats set to True to tfdv.generate_statistics_from_tfrecord.Before we train the model, let's do a quick audit of our training data using [TensorFlow Data Validation](https://www.tensorflow.org/tfx/data_validation/get_started), so we can better understand our data distribution.
#### TODO 2: Use TFDV to get quick statistics on your dataset
The following cell may take 2–3 minutes to run. **NOTE:** Please ignore the deprecation warnings.
```
# TODO 2
# The computation of statistics using TFDV. The returned value is a DatasetFeatureStatisticsList protocol buffer.
stats = tfdv.generate_statistics_from_tfrecord(data_location=train_tf_file)
# TODO 2a
# A visualization of the statistics using Facets Overview.
tfdv.visualize_statistics(stats)
```
### TODO 3: Use the TensorFlow Data Validation widget above to answer the following questions.
#### **1. How many total examples are in the training dataset?**
#### Solution
See below solution.
**There are 1.08 million total examples in the training dataset.**
The count column tells us how many examples there are for a given feature. Each feature (`sexual_orientation`, `comment_text`, `gender`, etc.) has 1.08 million examples. The missing column tells us what percentage of examples are missing that feature.

Each feature is missing from 0% of examples, so we know that the per-feature example count of 1.08 million is also the total number of examples in the dataset.
#### **2. How many unique values are there for the `gender` feature? What are they, and what are the frequencies of each of these values?**
**NOTE #1:** `gender` and the other identity features (`sexual_orientation`, `religion`, `disability`, and `race`) are included in this dataset for evaluation purposes only, so we can assess model performance on different identity slices. The only feature we will use for model training is `comment_text`.
**NOTE #2:** *We recognize the limitations of the categories used in the original dataset, and acknowledge that these terms do not encompass the full range of vocabulary used in describing gender.*
#### Solution
See below solution.
The **unique** column of the **Categorical Features** table tells us that there are 4 unique values for the `gender` feature.
To view the 4 values and their frequencies, we can click on the **SHOW RAW DATA** button:

The raw data table shows that there are 32,208 examples with a gender value of `female`, 26,758 examples with a value of `male`, 1,551 examples with a value of `transgender`, and 4 examples with a value of `other gender`.
**NOTE:** As described [earlier](#scrollTo=J3R2QWkru1WN), a `gender` feature can contain zero or more of these 4 values, depending on the content of the comment. For example, a comment containing the text "I am a transgender man" will have both `transgender` and `male` as `gender` values, whereas a comment that does not reference gender at all will have an empty/false `gender` value.
#### **3. What percentage of total examples are labeled toxic? Overall, is this a class-balanced dataset (relatively even split of examples between positive and negative classes) or a class-imbalanced dataset (majority of examples are in one class)?**
**NOTE:** In this dataset, a `toxicity` value of `0` signifies "not toxic," and a `toxicity` value of `1` signifies "toxic."
#### Solution
See below solution.
**7.98 percent of examples are toxic.**
Under **Numeric Features**, we can see the distribution of values for the `toxicity` feature. 92.02% of examples have a value of 0 (which signifies "non-toxic"), so 7.98% of examples are toxic.

This is a [**class-imbalanced dataset**](https://developers.google.com/machine-learning/glossary?utm_source=Colab&utm_medium=fi-colab&utm_campaign=fi-practicum&utm_content=glossary&utm_term=class-imbalanced-dataset#class-imbalanced-dataset), as the overwhelming majority of examples (over 90%) are classified as nontoxic.
Notice that there is one numeric feature (count of toxic comments) and six categorical features.
### TODO 4: Analyze label distribution for subset groups
Run the following code to analyze label distribution for the subset of examples that contain a `gender` value**
**NOTE:** *The cell should run for just a few minutes*
```
#@title Calculate label distribution for gender-related examples
raw_dataset = tf.data.TFRecordDataset(train_tf_file)
toxic_gender_examples = 0
nontoxic_gender_examples = 0
# TODO 4
# There are 1,082,924 examples in the dataset
for raw_record in raw_dataset.take(1082924):
example = tf.train.Example()
example.ParseFromString(raw_record.numpy())
if str(example.features.feature["gender"].bytes_list.value) != "[]":
if str(example.features.feature["toxicity"].float_list.value) == "[1.0]":
toxic_gender_examples += 1
else:
nontoxic_gender_examples += 1
# TODO 4a
print("Toxic Gender Examples: %s" % toxic_gender_examples)
print("Nontoxic Gender Examples: %s" % nontoxic_gender_examples)
```
#### **What percentage of `gender` examples are labeled toxic? Compare this percentage to the percentage of total examples that are labeled toxic from #3 above. What, if any, fairness concerns can you identify based on this comparison?**
#### Solution
Click below for one possible solution.
There are 7,189 gender-related examples that are labeled toxic, which represent 14.7% of all gender-related examples.
The percentage of gender-related examples that are toxic (14.7%) is nearly double the percentage of toxic examples overall (7.98%). In other words, in our dataset, gender-related comments are almost two times more likely than comments overall to be labeled as toxic.
This skew suggests that a model trained on this dataset might learn a correlation between gender-related content and toxicity. This raises fairness considerations, as the model might be more likely to classify nontoxic comments as toxic if they contain gender terminology, which could lead to [disparate impact](https://developers.google.com/machine-learning/glossary?utm_source=Colab&utm_medium=fi-colab&utm_campaign=fi-practicum&utm_content=glossary&utm_term=disparate-impact#disparate-impact) for gender subgroups.
Copyright 2020 Google Inc.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
|
github_jupyter
|
!pip3 install fairness-indicators==0.1.2 --user
# %tensorflow_version 2.x
import sys, os
import warnings
warnings.filterwarnings('ignore')
#os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3' # Ignore deprecation warnings
import tempfile
import apache_beam as beam
import numpy as np
import pandas as pd
from datetime import datetime
import tensorflow_hub as hub
import tensorflow as tf
import tensorflow_model_analysis as tfma
import tensorflow_data_validation as tfdv
from tensorflow_model_analysis.addons.fairness.post_export_metrics import fairness_indicators
from tensorflow_model_analysis.addons.fairness.view import widget_view
from fairness_indicators.examples import util
import warnings
warnings.filterwarnings("ignore")
from witwidget.notebook.visualization import WitConfigBuilder
from witwidget.notebook.visualization import WitWidget
print(tf.version.VERSION)
print(tf) # This statement shows us what version of Python we are currently running.
download_original_data = False #@param {type:"boolean"}
# TODO 1
if download_original_data:
train_tf_file = tf.keras.utils.get_file('train_tf.tfrecord',
'https://storage.googleapis.com/civil_comments_dataset/train_tf.tfrecord')
validate_tf_file = tf.keras.utils.get_file('validate_tf.tfrecord',
'https://storage.googleapis.com/civil_comments_dataset/validate_tf.tfrecord')
# The identity terms list will be grouped together by their categories
# (see 'IDENTITY_COLUMNS') on threshould 0.5. Only the identity term column,
# text column and label column will be kept after processing.
train_tf_file = util.convert_comments_data(train_tf_file)
validate_tf_file = util.convert_comments_data(validate_tf_file)
# TODO 1a
else:
train_tf_file = tf.keras.utils.get_file('train_tf_processed.tfrecord',
'https://storage.googleapis.com/civil_comments_dataset/train_tf_processed.tfrecord')
validate_tf_file = tf.keras.utils.get_file('validate_tf_processed.tfrecord',
'https://storage.googleapis.com/civil_comments_dataset/validate_tf_processed.tfrecord')
# TODO 2
# The computation of statistics using TFDV. The returned value is a DatasetFeatureStatisticsList protocol buffer.
stats = tfdv.generate_statistics_from_tfrecord(data_location=train_tf_file)
# TODO 2a
# A visualization of the statistics using Facets Overview.
tfdv.visualize_statistics(stats)
#@title Calculate label distribution for gender-related examples
raw_dataset = tf.data.TFRecordDataset(train_tf_file)
toxic_gender_examples = 0
nontoxic_gender_examples = 0
# TODO 4
# There are 1,082,924 examples in the dataset
for raw_record in raw_dataset.take(1082924):
example = tf.train.Example()
example.ParseFromString(raw_record.numpy())
if str(example.features.feature["gender"].bytes_list.value) != "[]":
if str(example.features.feature["toxicity"].float_list.value) == "[1.0]":
toxic_gender_examples += 1
else:
nontoxic_gender_examples += 1
# TODO 4a
print("Toxic Gender Examples: %s" % toxic_gender_examples)
print("Nontoxic Gender Examples: %s" % nontoxic_gender_examples)
| 0.236957 | 0.993029 |
## Keras Auto Encoder for Anomaly Detection with Custom Layer for determining Encoder Decoder Reconstruction Cost - CATEGORY EMBEDDINGS
### Loading Necessary Files and Libraries
```
import keras
from keras.models import Model, Sequential
from keras.layers import Embedding, Input, Dense, LSTM
from keras.preprocessing.text import Tokenizer
from keras import layers
import pandas as pd
import numpy as np
from sklearn import preprocessing
import matplotlib.pyplot as plt
from keras import optimizers
import pickle
import os
from keras.layers import initializers
from keras.layers import regularizers
from keras.layers import constraints
from keras.layers import Activation
from keras.layers.advanced_activations import PReLU
from keras.layers.advanced_activations import LeakyReLU
from keras.layers.advanced_activations import ELU
from keras.constraints import max_norm
from keras.layers import LeakyReLU
from keras import optimizers, losses, activations, models
from keras.callbacks import ModelCheckpoint, EarlyStopping, LearningRateScheduler
from keras.layers import Dropout, Convolution1D, MaxPool1D, GlobalMaxPool1D, GlobalAveragePooling1D, concatenate
from keras.layers import MaxPooling2D, Flatten, Conv2D
from keras.utils import to_categorical
%matplotlib inline
```
## Load Data
From: https://www.kaggle.com/mlg-ulb/creditcardfraud/downloads/creditcardfraud.zip/3
The datasets contains transactions made by credit cards in September 2013 by european cardholders. This dataset presents transactions that occurred in two days, where we have 492 frauds out of 284,807 transactions. The dataset is highly unbalanced, the positive class (frauds) account for 0.172% of all transactions.
It contains only numerical input variables which are the result of a PCA transformation.
```
data = pd.read_csv("creditcard.csv")
print(data.shape)
data.head(5)
data.info()
%matplotlib inline
data.hist(bins=50, figsize=(20,15))
plt.show()
```
## Check Balance Level
```
data["Class"].value_counts()
print("Balance level: {} %".format(data["Class"].value_counts()[1]*100/data["Class"].value_counts()[0]))
```
# Data Prep
## Normalize Amount using Scaler
```
data["Amount"].describe()
%matplotlib inline
data["Amount"].hist(bins=200, figsize=(10,5))
plt.show()
scaler = preprocessing.StandardScaler()
data["Amount"] = pd.DataFrame(scaler.fit_transform(pd.DataFrame(data["Amount"])),columns=["Amount"])
data["Amount"].describe()
%matplotlib inline
data["Amount"].hist(bins=200, figsize=(10,5))
plt.show()
#data["Amount"].value_counts()
#data.drop("Amount", inplace=True, axis=1)
```
## Convert Time in Week Day Categories
Number of seconds elapsed between this transaction and the first transaction in the dataset
```
data["Time"].describe()
diff_secs = data["Time"].max() - data["Time"].min()
diff_hours = diff_secs/(60*60)
print("Elapsed hours: {}".format(diff_hours))
%matplotlib inline
data["Time"].hist(bins=48, figsize=(20,5))
plt.show()
data["Time"] = data["Time"].apply(lambda x: int(x / 3600) % 24)
data["Time"].describe()
%matplotlib inline
data["Time"].hist(bins=24, figsize=(20,5))
plt.show()
data = pd.concat([data.drop('Time', axis=1), pd.get_dummies(data['Time'])], axis=1)
data.info()
```
## Split Positive and Negative data
```
print(data.shape)
data.head(5)
all = data
positive = data[data["Class"] > 0]
negative = data[data["Class"] == 0]
print(all.shape, positive.shape, negative.shape, positive.shape[0] + negative.shape[0])
all.drop("Class", inplace=True, axis=1)
positive.drop("Class", inplace=True, axis=1)
negative.drop("Class", inplace=True, axis=1)
print(all.shape, positive.shape, negative.shape, positive.shape[0] + negative.shape[0])
```
## Positive is the anomalies - Train AutoEncoder with Negatives
```
INPUT_SIZE = negative.shape[1]
CATEGORICAL_INPUT_SIZE = 24
NUMERICAL_INPUT_SIZE = INPUT_SIZE - CATEGORICAL_INPUT_SIZE
AUTO_ENCODER_SHAPE = 128
print(INPUT_SIZE, CATEGORICAL_INPUT_SIZE + NUMERICAL_INPUT_SIZE, CATEGORICAL_INPUT_SIZE, NUMERICAL_INPUT_SIZE, AUTO_ENCODER_SHAPE)
```
## Train 80% of Negative
```
train_size = int(negative.shape[0]*80/100)
negative = negative.sample(frac=1)
X_train = negative.iloc[:train_size]
X_test = negative.iloc[-(negative.shape[0] - train_size):]
# force the model to lower the autoencoder reconstruction cost
Y = np.zeros([X_train.shape[0], 1], dtype = float) # ZEROS MATRIX with shape (,1)
print(X_train.shape, X_test.shape, Y.shape, INPUT_SIZE, X_train.shape[0] + X_test.shape[0])
```
## Split X_train, X_test and Positive in Numerical and Categorical
```
X_train_numerical = X_train[X_train.columns[0:NUMERICAL_INPUT_SIZE]]
X_train_categorical = X_train[X_train.columns[NUMERICAL_INPUT_SIZE:]]
print(X_train.shape, X_train_numerical.shape, X_train_categorical.shape)
X_test_numerical = X_test[X_test.columns[0:NUMERICAL_INPUT_SIZE]]
X_test_categorical = X_test[X_test.columns[NUMERICAL_INPUT_SIZE:]]
print(X_test.shape, X_test_numerical.shape, X_test_categorical.shape)
positive_numerical = positive[positive.columns[0:NUMERICAL_INPUT_SIZE]]
positive_categorical = positive[positive.columns[NUMERICAL_INPUT_SIZE:]]
print(positive.shape, positive_numerical.shape, positive_categorical.shape)
```
# Build the Model
## Build the Numerical Model
```
numerical_input = Input(shape=(NUMERICAL_INPUT_SIZE,))
numerical = Dense(64, activation='relu')(numerical_input)
```
## Build the Embedding Model
```
categorical_input = Input(shape=(CATEGORICAL_INPUT_SIZE,))
embedding = Embedding(input_dim=2, output_dim=50, input_length=CATEGORICAL_INPUT_SIZE)(categorical_input)
embedding = Flatten()(embedding)
```
## Merge Numerical and Embedding
```
input_encoder = concatenate([numerical, embedding], axis=1)
input_encoder = Dense(AUTO_ENCODER_SHAPE, activation='relu')(input_encoder)
```
## Build the Encoder
```
hidden2 = Dense(64, activation='relu')(input_encoder)
encoded = Dense(32, activation='relu')(hidden2)
```
## Build the Decoder
```
hidden3 = Dense(64, activation='relu')(encoded)
output_decoder = Dense(AUTO_ENCODER_SHAPE, activation='relu')(hidden3)
from keras import backend as K
from keras.layers import Layer
class ReconstructionCostLayer(Layer):
def __init__(self,output_dim):
self.output_dim = output_dim
super(ReconstructionCostLayer, self).__init__()
def build(self, input_shape):
super(ReconstructionCostLayer, self).build(input_shape) # Be sure to call this at the end
def call(self, x):
left = K.slice(x, [0, 0], [-1, AUTO_ENCODER_SHAPE])
right = K.slice(x, [0, AUTO_ENCODER_SHAPE], [-1, -1])
return K.sum(K.square(left - right), axis=1)
def compute_output_shape(self, input_shape):
return (input_shape[0], self.output_dim)
```
### Concatenate Encoder input and Decoder output
```
input_encoder_plus_output_decoder = concatenate([input_encoder, output_decoder], axis=1)
```
### Call the custom layer
```
reconstruction_cost_layer = ReconstructionCostLayer(1)(input_encoder_plus_output_decoder)
```
## Build the AutoEncoder with Regression Model
```
model = Model(inputs=[numerical_input, categorical_input], outputs=reconstruction_cost_layer)
```
#### Visualize Model Architecture
```
adam = optimizers.Adam(lr=0.00003, beta_1=0.9, beta_2=0.999, epsilon=1e-4, decay=0.0, amsgrad=True)
model.compile(optimizer=adam, loss='mean_squared_error', metrics=['mse'])
print(model.summary())
```
# Train
```
# tbCallBack = keras.callbacks.TensorBoard(log_dir='./Graph',
# histogram_freq=0,
# write_graph=True,
# write_images=True)
# early_stop = keras.callbacks.EarlyStopping(monitor='loss', patience=0, verbose=0, mode='auto', baseline=0.003)
history = model.fit([X_train_numerical, X_train_categorical],
Y,
epochs=70,
batch_size=50,
verbose=1,
shuffle=True,
#callbacks=[tbCallBack], #, early_stop],
)
```
# Test
```
def report_cost(label, result, positive, threshold):
tot = len(result)
greater = len(np.where(result > threshold)[0])
if positive:
accuracy = 1 - ((tot - greater) / tot)
else:
accuracy = (tot - greater) / tot
print("""
{}
------------
Greater Than Threeshold: {} over {}
\033[1mAccuracy: {:.2f} \033[0m
Mean: {}
Std: {}
Min: {}
Max: {}
""".format(label,
greater, tot,
accuracy,
result.mean(),
result.std(),
result.min(),
result.max()))
```
## Predict
#### Get Negative (Trained) Reconstruction Costs
#### Get Negative (New) Reconstruction Costs
#### Get Positive (New) Reconstruction Costs
```
trained_reconstruction_costs = model.predict([X_train_numerical, X_train_categorical])
new_negative_reconstruction_costs = model.predict([X_test_numerical, X_test_categorical])
new_positive_reconstruction_costs = model.predict([positive_numerical, positive_categorical])
```
## Print Accuracy over Threshold
```
THRESHOLD = 0.1
report_cost("TRAINED NEGATIVE", trained_reconstruction_costs, False, THRESHOLD)
report_cost("NEW NEGATIVE", new_negative_reconstruction_costs, False, THRESHOLD)
report_cost("NEW POSITIVE", new_positive_reconstruction_costs, True, THRESHOLD)
```
# Interactive Accuracy over Threshold
```
import ipywidgets as widgets
from ipywidgets import interact, interact_manual
@interact
def printPrecision(THRESHOLD=(0, 1, 0.01)):
report_cost("TRAINED NEGATIVE", trained_reconstruction_costs, False, THRESHOLD)
report_cost("NEW NEGATIVE", new_negative_reconstruction_costs, False, THRESHOLD)
report_cost("NEW POSITIVE", new_positive_reconstruction_costs, True, THRESHOLD)
```
|
github_jupyter
|
import keras
from keras.models import Model, Sequential
from keras.layers import Embedding, Input, Dense, LSTM
from keras.preprocessing.text import Tokenizer
from keras import layers
import pandas as pd
import numpy as np
from sklearn import preprocessing
import matplotlib.pyplot as plt
from keras import optimizers
import pickle
import os
from keras.layers import initializers
from keras.layers import regularizers
from keras.layers import constraints
from keras.layers import Activation
from keras.layers.advanced_activations import PReLU
from keras.layers.advanced_activations import LeakyReLU
from keras.layers.advanced_activations import ELU
from keras.constraints import max_norm
from keras.layers import LeakyReLU
from keras import optimizers, losses, activations, models
from keras.callbacks import ModelCheckpoint, EarlyStopping, LearningRateScheduler
from keras.layers import Dropout, Convolution1D, MaxPool1D, GlobalMaxPool1D, GlobalAveragePooling1D, concatenate
from keras.layers import MaxPooling2D, Flatten, Conv2D
from keras.utils import to_categorical
%matplotlib inline
data = pd.read_csv("creditcard.csv")
print(data.shape)
data.head(5)
data.info()
%matplotlib inline
data.hist(bins=50, figsize=(20,15))
plt.show()
data["Class"].value_counts()
print("Balance level: {} %".format(data["Class"].value_counts()[1]*100/data["Class"].value_counts()[0]))
data["Amount"].describe()
%matplotlib inline
data["Amount"].hist(bins=200, figsize=(10,5))
plt.show()
scaler = preprocessing.StandardScaler()
data["Amount"] = pd.DataFrame(scaler.fit_transform(pd.DataFrame(data["Amount"])),columns=["Amount"])
data["Amount"].describe()
%matplotlib inline
data["Amount"].hist(bins=200, figsize=(10,5))
plt.show()
#data["Amount"].value_counts()
#data.drop("Amount", inplace=True, axis=1)
data["Time"].describe()
diff_secs = data["Time"].max() - data["Time"].min()
diff_hours = diff_secs/(60*60)
print("Elapsed hours: {}".format(diff_hours))
%matplotlib inline
data["Time"].hist(bins=48, figsize=(20,5))
plt.show()
data["Time"] = data["Time"].apply(lambda x: int(x / 3600) % 24)
data["Time"].describe()
%matplotlib inline
data["Time"].hist(bins=24, figsize=(20,5))
plt.show()
data = pd.concat([data.drop('Time', axis=1), pd.get_dummies(data['Time'])], axis=1)
data.info()
print(data.shape)
data.head(5)
all = data
positive = data[data["Class"] > 0]
negative = data[data["Class"] == 0]
print(all.shape, positive.shape, negative.shape, positive.shape[0] + negative.shape[0])
all.drop("Class", inplace=True, axis=1)
positive.drop("Class", inplace=True, axis=1)
negative.drop("Class", inplace=True, axis=1)
print(all.shape, positive.shape, negative.shape, positive.shape[0] + negative.shape[0])
INPUT_SIZE = negative.shape[1]
CATEGORICAL_INPUT_SIZE = 24
NUMERICAL_INPUT_SIZE = INPUT_SIZE - CATEGORICAL_INPUT_SIZE
AUTO_ENCODER_SHAPE = 128
print(INPUT_SIZE, CATEGORICAL_INPUT_SIZE + NUMERICAL_INPUT_SIZE, CATEGORICAL_INPUT_SIZE, NUMERICAL_INPUT_SIZE, AUTO_ENCODER_SHAPE)
train_size = int(negative.shape[0]*80/100)
negative = negative.sample(frac=1)
X_train = negative.iloc[:train_size]
X_test = negative.iloc[-(negative.shape[0] - train_size):]
# force the model to lower the autoencoder reconstruction cost
Y = np.zeros([X_train.shape[0], 1], dtype = float) # ZEROS MATRIX with shape (,1)
print(X_train.shape, X_test.shape, Y.shape, INPUT_SIZE, X_train.shape[0] + X_test.shape[0])
X_train_numerical = X_train[X_train.columns[0:NUMERICAL_INPUT_SIZE]]
X_train_categorical = X_train[X_train.columns[NUMERICAL_INPUT_SIZE:]]
print(X_train.shape, X_train_numerical.shape, X_train_categorical.shape)
X_test_numerical = X_test[X_test.columns[0:NUMERICAL_INPUT_SIZE]]
X_test_categorical = X_test[X_test.columns[NUMERICAL_INPUT_SIZE:]]
print(X_test.shape, X_test_numerical.shape, X_test_categorical.shape)
positive_numerical = positive[positive.columns[0:NUMERICAL_INPUT_SIZE]]
positive_categorical = positive[positive.columns[NUMERICAL_INPUT_SIZE:]]
print(positive.shape, positive_numerical.shape, positive_categorical.shape)
numerical_input = Input(shape=(NUMERICAL_INPUT_SIZE,))
numerical = Dense(64, activation='relu')(numerical_input)
categorical_input = Input(shape=(CATEGORICAL_INPUT_SIZE,))
embedding = Embedding(input_dim=2, output_dim=50, input_length=CATEGORICAL_INPUT_SIZE)(categorical_input)
embedding = Flatten()(embedding)
input_encoder = concatenate([numerical, embedding], axis=1)
input_encoder = Dense(AUTO_ENCODER_SHAPE, activation='relu')(input_encoder)
hidden2 = Dense(64, activation='relu')(input_encoder)
encoded = Dense(32, activation='relu')(hidden2)
hidden3 = Dense(64, activation='relu')(encoded)
output_decoder = Dense(AUTO_ENCODER_SHAPE, activation='relu')(hidden3)
from keras import backend as K
from keras.layers import Layer
class ReconstructionCostLayer(Layer):
def __init__(self,output_dim):
self.output_dim = output_dim
super(ReconstructionCostLayer, self).__init__()
def build(self, input_shape):
super(ReconstructionCostLayer, self).build(input_shape) # Be sure to call this at the end
def call(self, x):
left = K.slice(x, [0, 0], [-1, AUTO_ENCODER_SHAPE])
right = K.slice(x, [0, AUTO_ENCODER_SHAPE], [-1, -1])
return K.sum(K.square(left - right), axis=1)
def compute_output_shape(self, input_shape):
return (input_shape[0], self.output_dim)
input_encoder_plus_output_decoder = concatenate([input_encoder, output_decoder], axis=1)
reconstruction_cost_layer = ReconstructionCostLayer(1)(input_encoder_plus_output_decoder)
model = Model(inputs=[numerical_input, categorical_input], outputs=reconstruction_cost_layer)
adam = optimizers.Adam(lr=0.00003, beta_1=0.9, beta_2=0.999, epsilon=1e-4, decay=0.0, amsgrad=True)
model.compile(optimizer=adam, loss='mean_squared_error', metrics=['mse'])
print(model.summary())
# tbCallBack = keras.callbacks.TensorBoard(log_dir='./Graph',
# histogram_freq=0,
# write_graph=True,
# write_images=True)
# early_stop = keras.callbacks.EarlyStopping(monitor='loss', patience=0, verbose=0, mode='auto', baseline=0.003)
history = model.fit([X_train_numerical, X_train_categorical],
Y,
epochs=70,
batch_size=50,
verbose=1,
shuffle=True,
#callbacks=[tbCallBack], #, early_stop],
)
def report_cost(label, result, positive, threshold):
tot = len(result)
greater = len(np.where(result > threshold)[0])
if positive:
accuracy = 1 - ((tot - greater) / tot)
else:
accuracy = (tot - greater) / tot
print("""
{}
------------
Greater Than Threeshold: {} over {}
\033[1mAccuracy: {:.2f} \033[0m
Mean: {}
Std: {}
Min: {}
Max: {}
""".format(label,
greater, tot,
accuracy,
result.mean(),
result.std(),
result.min(),
result.max()))
trained_reconstruction_costs = model.predict([X_train_numerical, X_train_categorical])
new_negative_reconstruction_costs = model.predict([X_test_numerical, X_test_categorical])
new_positive_reconstruction_costs = model.predict([positive_numerical, positive_categorical])
THRESHOLD = 0.1
report_cost("TRAINED NEGATIVE", trained_reconstruction_costs, False, THRESHOLD)
report_cost("NEW NEGATIVE", new_negative_reconstruction_costs, False, THRESHOLD)
report_cost("NEW POSITIVE", new_positive_reconstruction_costs, True, THRESHOLD)
import ipywidgets as widgets
from ipywidgets import interact, interact_manual
@interact
def printPrecision(THRESHOLD=(0, 1, 0.01)):
report_cost("TRAINED NEGATIVE", trained_reconstruction_costs, False, THRESHOLD)
report_cost("NEW NEGATIVE", new_negative_reconstruction_costs, False, THRESHOLD)
report_cost("NEW POSITIVE", new_positive_reconstruction_costs, True, THRESHOLD)
| 0.753376 | 0.910625 |
Beispiel für eine explorative Datenanalyse: Erdbeben der letzten 7 Tage (US Geological Survey)
==============================================================================================
Import zweier Standardpakete für die Datenanalyse: Numpy für mehrdimensionale Arrays, Pandas für Datenanalyse in Tabellen.
```
import pandas as pd
import numpy as np
```
Direkter Download vom USGS, Abruf des Downloaddatums, automatischer Import in Pandas-Dataframe
```
fileUrl = 'http://earthquake.usgs.gov/earthquakes/feed/v1.0/summary/2.5_day.csv'
eData = pd.read_csv(fileUrl)
dateDownloaded = !date
dateDownloaded
```
1. Darstellung als Pandas-DataFrame
-----------------------------------
Darstellung des Datensatzes als Datensatzes als Pandas-Dataframe (Tabelle der ersten und letzten 30 Einträge, Anzahl Reihen und Spalten). Konvention: Variablen sind die Spalten, einzelne Messungen die Zeilen.
```
eData
```
Kürzere Darstellung mit head(): nur die ersten 5 Einträge des Tabellenkopfes
```
eData.head()
```
Anzahl der Reihen und Spalten mit Numpy shape().
```
np.shape(eData)
```
Anzeige der einzelnen Spaltennamen mit Attribut DataFrame.columns
```
eData.columns
```
Datentyp der einzelnen Variablen mit Attribut DataFrame.dtypes
```
eData.dtypes
```
2. Aufbereitung des Datensatzes
-------------------------------
Überprüfen, ob Tabelle NaN enthält, mit DataFrame.isnull().any()
```
eData.isnull().any()
```
Entfernung aller Zeilen bzw. Messungen mit NaNs durch DataFrame.dropna()
```
eData = eData.dropna()
eData.head()
eData.isnull().any()
```
Überprüfen, ob Zeilen bzw. Messungen doppelt vorkommen, mit DataFrame.duplicated()
```
eData.duplicated().any()
```
Es kommen also keine Duplikate vor. Bei Bedarf mit *DataFrame.drop_duplicates()* entfernen.
3. Explorative Statistiken
--------------------------
Statistische Beschreibung der numerischen Variablen mit Dataframe.describe() (count: Anzahl Messungen, mean: Mittelwert, std: Standardabweichung, min: Minimum, 25%: 25-Perzentil, ...)
```
eData.describe()
```
Streumatrix für alle numerischen Variablen mit Pandas *scattermatrix()*:
```
pd.scatter_matrix(eData, figsize=(14,14), marker='o');
```
4. Analyse von Untermengen
--------------------------
Zugriff auf die Variable 'Lat' (latitude):
```
eData['Lat']
```
Welche Erdbeben fanden oberhalb einer geographischen Breite von 40 Grad statt?
```
eData['Lat'] > 40.0
```
Gab es überhaupt Erdbeben oberhalb 50 Grad Breite?
```
(eData['Lat'] > 40.0).any()
```
Gab es also. Haben alle verzeichneten Erdbeben eine Breite größer als 18 Grad?
```
(eData['Lat'] > 18.0).all()
```
Es sind also auch Erdbeben unterhalb von 18 Grad verzeichnet.
Alle unterschiedlichen Werte der kategorischen Variable 'Version' mit Dataframe['Variablenname'].*unique()*
```
eData['Version'].unique()
```
Häufigkeit der verschiedenen Kategorien in 'Version' mit Dataframe['Variablenname'].*value_counts()*:
```
eData['Version'].value_counts()
```
Häufigkeit von Wertepaaren der beiden kategorischen Variablen 'Version' und 'Src' mit Pandas *crosstab()*:
```
pd.crosstab(eData['Src'], eData['Version'])
```
Darstellung der Häufigkeitsverteilung der Erdbebenstärken für die verschiedenen Quellen mit einer Kastengraphik durch Pandas *boxplot()*:
```
from pandas.tools.plotting import boxplot
boxplot(eData, column='Magnitude', by='Src');
```
|
github_jupyter
|
import pandas as pd
import numpy as np
fileUrl = 'http://earthquake.usgs.gov/earthquakes/feed/v1.0/summary/2.5_day.csv'
eData = pd.read_csv(fileUrl)
dateDownloaded = !date
dateDownloaded
eData
eData.head()
np.shape(eData)
eData.columns
eData.dtypes
eData.isnull().any()
eData = eData.dropna()
eData.head()
eData.isnull().any()
eData.duplicated().any()
eData.describe()
pd.scatter_matrix(eData, figsize=(14,14), marker='o');
eData['Lat']
eData['Lat'] > 40.0
(eData['Lat'] > 40.0).any()
(eData['Lat'] > 18.0).all()
eData['Version'].unique()
eData['Version'].value_counts()
pd.crosstab(eData['Src'], eData['Version'])
from pandas.tools.plotting import boxplot
boxplot(eData, column='Magnitude', by='Src');
| 0.413477 | 0.968441 |
# Marginal Logistic Models
Last time, we focused on linear models, including OLS, marginal, and multilevel, with the NHANES national health and nutrition data set (https://crawstat.com/2020/06/17/marginal-multilevel-linear-models/). This time, working with the same data set, we'll focus on logistic models, which are used to predict the odds (probability) of an event, in this case the binary categorical variable of whether or not a person has smoked at least 100 cigarettes in their life. We'll first fit a simple binomial logistic regression assuming independent samples and diagnose its probability structure and non-linearity. We'll then fit a marginal logistic model to take into account within-cluster dependencies in NHANES' county-level geographic cluster design.
**Part 1**: fit and diagnose a logistic regression of smoker status on 5 predictors assuming fully independent samples.
**Part 2**: visualize and assess probability structure by plotting log odds and odds against select focus variables.
**Part 3**: visualize and assess non-linearity by generating partial residual, added variable, and CERES plots.
**Part 4**: fit and compare a marginal logistic model.
We'll fit logistic models as well as diagnose and compare them by interpreting parameters, standard errors, variance, and residual plots. We'll compare both log odds and odds (probability) of someone being a smoker based on their combination of variables. We'll also see within-cluster dependencies in action and how accounting for them helps us develop more accurate and meaningful models. Let's dig in.
## Part 1: Logistic Regression on 5 Covariates Assuming Fully Independent Samples
```
# Import relevant libraries
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import statsmodels.api as sm
from scipy import stats
from scipy.stats import t
import numpy as np
import scipy.stats.distributions as dist
from IPython.display import display, HTML
# Read the data set and have a look at the first few rows to a get a feel for it
pd.set_option("display.max_columns", 100)
da = pd.read_csv("https://raw.githubusercontent.com/kshedden/statswpy/master/NHANES/merged/nhanes_2015_2016.csv")
da.head(5)
# We're going to be using seaborn for plots, set style to darkgrid
sns.set_style("darkgrid")
sns.set(style='ticks', palette='Set2')
# Check the column headings
da.columns
# Check the dimensions of the dataframe, this one has 5,735 rows and 28 columns
da.shape
# Check number of null values by variable
da.isnull().sum()
# Create new gender column using more easily interpretable variables (instead of numeric responses)
da["RIAGENDRx"] = da.RIAGENDR.replace({1: "Male", 2: "Female"})
# Create new ethnicity column using more easily interpretable variables (instead of numeric responses)
da["RIDRETH1x"] = da.RIDRETH1.replace({1: "Mex Amer", 2: "Other Hisp", 3: "Non Hisp White", 4: "Non Hisp Black", 5: "Other"})
# Create new education column using more easily interpretable variables (instead of numeric responses)
da["DMDEDUC2x"] = da.DMDEDUC2.replace({1: "Elementary", 2: "Middle", 3: "HS", 4: "SomeCollege", 5: "College", 7: np.nan, 9: np.nan})
# Create new smoking status column where 1 is yes and 0 is no and don't know and refused are removed
da["SMQ020x"] = da.SMQ020.replace({2: 0, 7: np.nan, 9: np.nan})
keep = ['SDMVSTRA', 'SDMVPSU', 'SMQ020x','RIAGENDRx', 'RIDAGEYR', 'DMDEDUC2x',
'INDFMPIR', 'BMXBMI']
# Create new dataframe health with select variables, drop null values with .dropna()
health = da[keep].dropna()
health.head(5)
```
Let's fit our basic logistic model for odds of smoking. If an event has probability `p`, then its odds are `p/(1-p)`, which The odds is a transformation of the probability onto a different scale. For example, if the probability is 1/2, then the odds is 1. In our case, `p` is the proporition of SMQ020x equal to 1 and `1-p` is the proportion of SMQ020x equal to 0.
In our output, for our categorical variables, our reference levels are "Female" for gender (RIAGENDRx) and "College" for education (DMDEDUC2x) as they don't show up in the output (they have the lowest log odds of smoking). Keep in mind that for logistic regression, coefficients are applied to log odds and are are additive. On the other hand, comparing odds (probability) is multiplicative. Below, all variable coefficients, except those for elementary level education and BMI have low p-values making them statistically significant.
Looking at our coefficients, for a given age, BMI, education level, and income to poverty ratio, being a man will increase someone's log odds of smoking by 0.9147 (alternatively, odds increase `exp(0.9147) = 2.5` times). Additionally, someone's log odds of smoking increases by 0.0201 (alternatively, odds increase `exp(0.0201) = 1.02` times) with every year they grow older, holding all other variables constant. On the other hand, someone's log odds of smoking decreases by 0.1033 with every $20,000 rise in annual family income.
We can also look at specific variable combinations. The log odds for a 50-year-old man being a smoker are `0.9147 + (30 * 0.0201) = 1.5177` units higher that that of a 20-year-old woman, broken out into a 0.9147 increase due to gender and a 0.603 increase due to age. We can exponentiate to derive odds. Since `exp(0.9147 + 0.603) = exp(0.9147) * exp(0.603) = 2.5 * 1.83` we can state that being a man is associated with a 2.5-fold increase in odds of smoking and 30 years of age is associated with a 1.83-fold increase in the odds of smoking. When taken together, a 50-year-old man has `exp(1.52) = 4.56`-fold greater odds of smoking than a 20-year-old woman.
```
# Fit simple logistic model with 5 covariates: age, gender, BMI, family income to poverty ratio, and education.
# The family income to poverty ratio (INDFMPIR) uses a poverty level of $20,000 annual income
model = sm.GLM.from_formula("SMQ020x ~ RIDAGEYR + RIAGENDRx + BMXBMI + INDFMPIR + DMDEDUC2x", family=sm.families.Binomial(), data=health)
result = model.fit()
result.summary()
# Compare male and female odds of smoking.
# Keep in mind that, in this case, `p` is the proporition of SMQ020x equal to 1 and `1-p` is the proportion of SMQ020x equal to 0.
gender = pd.crosstab(health.RIAGENDRx, health.SMQ020x).apply(lambda x: x/x.sum(), axis=1)
gender["odds"] = gender.loc[:, 1] / gender.loc[:, 0]
gender
```
The probability that a woman has ever smoked is 32%, significantly lower than that of a man, 53%. Another way to state this is that the odds of a woman smoking is 0.47, while the odds of a man smoking is 1.13. Calculating the odds ratio below, we see that the odds ratio of smoking for males to females is 2.38, meaning that a man has 2.38 times greater odds of smoking than a woman.
```
gender["odds"].Male / gender["odds"].Female
# Compare odds of smoking by education level.
# Keep in mind that, in this case, `p` is the proporition of SMQ020x equal to 1 and `1-p` is the proportion of SMQ020x equal to 0.
education = pd.crosstab(health.DMDEDUC2x, health.SMQ020x).apply(lambda x: x/x.sum(), axis=1)
education["odds"] = education.loc[:, 1] / education.loc[:, 0]
education
```
The probability that someone who completed college has ever smoked is 27%, significantly lower than that of a person who started but didn't finish college, 45%. Another way to state this is that the odds of someone who started but didn't finish college of smoking is 0.81, while the odds of someone who completed college of smoking is 0.38. Calculating the odds ratio below, we see that the odds ratio of smoking for someone who started but didn't finish college is 2.16, meaning that a person who started but didn't finish college has 2.16 times greater odds of smoking than someone who completed college.
```
education["odds"].SomeCollege / education["odds"].College
```
## Part 2: Probability Structure
Let's visualize the probability structure of the population from our logistic model, including shaded 95% confidence bounds, by plotting log odds of smoking vs. age for men with a college education. First, we'll need to import predict_functional from statsmodels.sandbox.predict_functional. We also need to fix all of our variables. Our dependent variable is already set as smoking status (SMQ020x). We'll set our independent focus variable as age. We'll set our gender to male, BMI to 25 (the top of the "normal" range for BMI), education level to college, and income to poverty ratio to 5. From our plot, we see increasing log odds of smoking with age, which makes sense as somone is more likely to have smoked at least 100 cigarettes the older they are.
```
# Import predict_functional
from statsmodels.sandbox.predict_functional import predict_functional
# Fix variables at specific values
values = {"RIAGENDRx": "Male", "BMXBMI": 25, "DMDEDUC2x": "College", "INDFMPIR" : 5}
# The returned values are the predicted values (pv), confidence bands (cb), and function values (fv).
pv, cb, fv = predict_functional(result, "RIDAGEYR", values=values, ci_method="simultaneous")
plt.figure(figsize=(8, 6))
ax = sns.lineplot(fv, pv, lw=3)
ax.fill_between(fv, cb[:, 0], cb[:, 1], color='grey', alpha=0.3)
ax.set_title("Log Odds Smoking vs. Age for Male with College Education", size=15)
ax.set_xlabel("Age", size=15)
ax.set_ylabel("Log Odds Smoking", size=15)
```
Let's try the same plot in terms of probabilities instead of log odds. Probability can be obtained from the log odds using the relationship `p = 1 / (1 + exp(-o))` where `o` is the log odds. This time we see a slightly curved relationship.
```
plt.figure(figsize=(8, 6))
pv_prob = 1 / (1 + np.exp(-pv))
cb_prob = 1 / (1 + np.exp(-cb))
ax = sns.lineplot(fv, pv_prob, lw=3)
ax.fill_between(fv, cb_prob[:, 0], cb_prob[:, 1], color='grey', alpha=0.3)
ax.set_title("Probability Smoking vs. Age for Male with College Education", size=15)
ax.set_xlabel("Age", size=15)
ax.set_ylabel("Probability Smoking", size=15)
```
This time, let's plot log odds smoking vs. income to poverty ratio for men aged 40. Our dependent variable is already set as somking status (SMQ020x). We'll set our independent focus variable as income to poverty ratio (INDFMPIR). We'll set our gender to male, BMI to 25, education level to college, and age to 40.
Log odds smoking decreases with a higher income to poverty ratio. Additionally, the shaded gray confidence bounds are a bit wider meaning that the relationship between smoking and income has less certainty than that between smoking and age.
```
# Fix variables at specific values
values = {"RIAGENDRx": "Male", "BMXBMI": 25, "DMDEDUC2x": "College", "RIDAGEYR":40}
# The returned values are the predicted values (pv), confidence bands (cb), and function values (fv).
pv, cb, fv = predict_functional(result, "INDFMPIR", values=values, ci_method="simultaneous")
plt.figure(figsize=(8, 6))
ax = sns.lineplot(fv, pv, lw=3)
ax.fill_between(fv, cb[:, 0], cb[:, 1], color='grey', alpha=0.3)
ax.set_title("Log Odds Smoking vs. Income to Poverty Ratio for Male Aged 40", size=15)
ax.set_xlabel("Income to Poverty Ratio", size=15)
ax.set_ylabel("Log Odds Smoking", size=15)
plt.figure(figsize=(8, 6))
pv_prob = 1 / (1 + np.exp(-pv))
cb_prob = 1 / (1 + np.exp(-cb))
ax = sns.lineplot(fv, pv_prob, lw=3)
ax.fill_between(fv, cb_prob[:, 0], cb_prob[:, 1], color='grey', alpha=0.3)
ax.set_title("Probability Smoking vs. Income to Poverty Ratio for Male Aged 40", size=15)
ax.set_xlabel("Age", size=15)
ax.set_ylabel("Probability Smoking", size=15)
```
## Part 3: Variance Structure
Let's look at our variance structure to see if there's any curvature in the relationship between log odds smoking and a focus variable. Keep in mind that small discrepencies aren't very reliable to draw conclusions from, unless we have a very large sample size.
#### Partial Residual Plot
A partial residual plot is essentially a synthetic data set removing the effects of all but one variable. We'll see how variance looks like if one independent focus variable is allowed to change while all other independent variables and unexplained variation is fixed. Explained variation can be seen in the red trendline while unexplained variation is seen in the scatter around the trendline.
Let's set our independent focus variable as age (allowed to change) on the x axis, and fix gender, BMI, education, and income to poverty ratio. We see slight curvature with smoking increasing more quickly from age 20 to 35, flattening out, and rising faster again from age 50 to 60.
```
# Import add_lowess
from statsmodels.graphics.regressionplots import add_lowess
fig = result.plot_partial_residuals("RIDAGEYR")
ax = fig.get_axes()[0]
ax.lines[0].set_alpha(0.2)
ax.set_xlabel("Age", size=15)
_ = add_lowess(ax)
```
#### Added Variable Plot
We can also look for non-linearity with an added variable plot. Note that log odds smoking residuals and age have been centered around their respective means. Similar to the partial residuals plot, we see curvature with smoking rising more quickly from age 20 to 35, dipping a bit, and then increasing more quickly again after that.
```
fig = result.plot_added_variable("RIDAGEYR")
ax = fig.get_axes()[0]
ax.lines[0].set_alpha(0.2)
ax.set_xlabel("Age (Normalized)", size=15)
_ = add_lowess(ax)
```
#### CERES Plot
A CERES (conditional expectation partial residuals) plot are most supportive in assessing non-linearity. Here again, we see the slight curvature around in early and late age ranges.
```
fig = result.plot_ceres_residuals("RIDAGEYR")
ax = fig.get_axes()[0]
ax.lines[0].set_alpha(0.2)
ax.set_xlabel("Age", size=15)
_ = add_lowess(ax)
```
## Part 4: Marginal Logistic Model
NHANES follows a complex sampling design, specifically geographic cluster sampling. You can learn more here (https://www.cdc.gov/nchs/tutorials/nhanes/SurveyDesign/SampleDesign/Info1.htm). In the sampling design, primary sampling units (SDMVPSU) include counties, which are then divided into subregions, from which households are drawn and individuals are sampled. In order to pretect individuals' confidentiality, masked variance units (SDMVSTRA) combine subregions of different counties to mimic contiguous subregions. While they're not the actual original clusters, they can be treated as proxies of the original clusters with variances that closely approximate those of the original true design. We can arrive at their identifiers by combining them.
```
# Form cluster variable
health["cluster"] = 10*health.SDMVSTRA + health.SDMVPSU
```
Let's check if there is within-cluster correlation in our sample design. We set our correlation structure as exchangeable, which assumes constant correlation of observations within a cluster.
We see that we clearly have within-cluster correlation in the intercepts of our variables ranging from 0.025 to 0.067. While these seem low, they're not directly comparable to a Pearson correlation and are actually fairly high (0 means perfect independence while 1 means perfect clustering where values are identical).
```
# Within-cluster correlation of multiple variables using GEE without covariates, only an intercept (""~1"), and exchangeable correlation structure.
for v in ["SMQ020x", "RIDAGEYR", "BMXBMI", "INDFMPIR"]:
model = sm.GEE.from_formula(v + " ~ 1", groups="cluster", cov_struct=sm.cov_struct.Exchangeable(), data=health)
result = model.fit()
print(v, result.cov_struct.summary())
```
Now that we know that we have within-cluster correlation, let's fit a marginal model using a generalized estimating equation (GEE). Let's also construct a table to compare parameters and standard error between GEE and our basic logistic model (GLM) for all of our variables. Since GLM assumes independent samples and doesn't take into account within-cluster correlation, it will often underrepresent standard error. As expected, GEE standard errors are often higher than those of GLM.
Since we know that there are within-cluster dependencies, GLM is not justified and GEE is a superior fit with more accurate and meaningful parameters and standard errors (assuming exclusively within-cluster and not between-cluster dependencies).
```
# Fit simple logistic model using GLM
GLM = sm.GLM.from_formula("SMQ020x ~ RIDAGEYR + RIAGENDRx + BMXBMI + INDFMPIR + DMDEDUC2x", family=sm.families.Binomial(), data=health)
result_GLM = GLM.fit()
result_GLM.summary()
# Fit a marginal logistic model using GEE and exchangeable correlation structure
GEE = sm.GEE.from_formula("SMQ020x ~ RIDAGEYR + RIAGENDRx + BMXBMI + INDFMPIR + DMDEDUC2x", groups="cluster", family=sm.families.Binomial(), cov_struct=sm.cov_struct.Exchangeable(), data=health)
result_GEE = GEE.fit(start_params = result_GLM.params)
# Create dataframe for comparing OLS and GEE parameters and standard errors
compare = pd.DataFrame({"GLM Params": result_GLM.params, "GLM SE": result_GLM.bse, "GEE Params": result_GEE.params, "GEE SE": result_GEE.bse})
compare = compare[["GLM Params", "GLM SE", "GEE Params", "GEE SE"]]
# Print out the results in a pretty way
display(HTML(compare.to_html()))
```
|
github_jupyter
|
# Import relevant libraries
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import statsmodels.api as sm
from scipy import stats
from scipy.stats import t
import numpy as np
import scipy.stats.distributions as dist
from IPython.display import display, HTML
# Read the data set and have a look at the first few rows to a get a feel for it
pd.set_option("display.max_columns", 100)
da = pd.read_csv("https://raw.githubusercontent.com/kshedden/statswpy/master/NHANES/merged/nhanes_2015_2016.csv")
da.head(5)
# We're going to be using seaborn for plots, set style to darkgrid
sns.set_style("darkgrid")
sns.set(style='ticks', palette='Set2')
# Check the column headings
da.columns
# Check the dimensions of the dataframe, this one has 5,735 rows and 28 columns
da.shape
# Check number of null values by variable
da.isnull().sum()
# Create new gender column using more easily interpretable variables (instead of numeric responses)
da["RIAGENDRx"] = da.RIAGENDR.replace({1: "Male", 2: "Female"})
# Create new ethnicity column using more easily interpretable variables (instead of numeric responses)
da["RIDRETH1x"] = da.RIDRETH1.replace({1: "Mex Amer", 2: "Other Hisp", 3: "Non Hisp White", 4: "Non Hisp Black", 5: "Other"})
# Create new education column using more easily interpretable variables (instead of numeric responses)
da["DMDEDUC2x"] = da.DMDEDUC2.replace({1: "Elementary", 2: "Middle", 3: "HS", 4: "SomeCollege", 5: "College", 7: np.nan, 9: np.nan})
# Create new smoking status column where 1 is yes and 0 is no and don't know and refused are removed
da["SMQ020x"] = da.SMQ020.replace({2: 0, 7: np.nan, 9: np.nan})
keep = ['SDMVSTRA', 'SDMVPSU', 'SMQ020x','RIAGENDRx', 'RIDAGEYR', 'DMDEDUC2x',
'INDFMPIR', 'BMXBMI']
# Create new dataframe health with select variables, drop null values with .dropna()
health = da[keep].dropna()
health.head(5)
# Fit simple logistic model with 5 covariates: age, gender, BMI, family income to poverty ratio, and education.
# The family income to poverty ratio (INDFMPIR) uses a poverty level of $20,000 annual income
model = sm.GLM.from_formula("SMQ020x ~ RIDAGEYR + RIAGENDRx + BMXBMI + INDFMPIR + DMDEDUC2x", family=sm.families.Binomial(), data=health)
result = model.fit()
result.summary()
# Compare male and female odds of smoking.
# Keep in mind that, in this case, `p` is the proporition of SMQ020x equal to 1 and `1-p` is the proportion of SMQ020x equal to 0.
gender = pd.crosstab(health.RIAGENDRx, health.SMQ020x).apply(lambda x: x/x.sum(), axis=1)
gender["odds"] = gender.loc[:, 1] / gender.loc[:, 0]
gender
gender["odds"].Male / gender["odds"].Female
# Compare odds of smoking by education level.
# Keep in mind that, in this case, `p` is the proporition of SMQ020x equal to 1 and `1-p` is the proportion of SMQ020x equal to 0.
education = pd.crosstab(health.DMDEDUC2x, health.SMQ020x).apply(lambda x: x/x.sum(), axis=1)
education["odds"] = education.loc[:, 1] / education.loc[:, 0]
education
education["odds"].SomeCollege / education["odds"].College
# Import predict_functional
from statsmodels.sandbox.predict_functional import predict_functional
# Fix variables at specific values
values = {"RIAGENDRx": "Male", "BMXBMI": 25, "DMDEDUC2x": "College", "INDFMPIR" : 5}
# The returned values are the predicted values (pv), confidence bands (cb), and function values (fv).
pv, cb, fv = predict_functional(result, "RIDAGEYR", values=values, ci_method="simultaneous")
plt.figure(figsize=(8, 6))
ax = sns.lineplot(fv, pv, lw=3)
ax.fill_between(fv, cb[:, 0], cb[:, 1], color='grey', alpha=0.3)
ax.set_title("Log Odds Smoking vs. Age for Male with College Education", size=15)
ax.set_xlabel("Age", size=15)
ax.set_ylabel("Log Odds Smoking", size=15)
plt.figure(figsize=(8, 6))
pv_prob = 1 / (1 + np.exp(-pv))
cb_prob = 1 / (1 + np.exp(-cb))
ax = sns.lineplot(fv, pv_prob, lw=3)
ax.fill_between(fv, cb_prob[:, 0], cb_prob[:, 1], color='grey', alpha=0.3)
ax.set_title("Probability Smoking vs. Age for Male with College Education", size=15)
ax.set_xlabel("Age", size=15)
ax.set_ylabel("Probability Smoking", size=15)
# Fix variables at specific values
values = {"RIAGENDRx": "Male", "BMXBMI": 25, "DMDEDUC2x": "College", "RIDAGEYR":40}
# The returned values are the predicted values (pv), confidence bands (cb), and function values (fv).
pv, cb, fv = predict_functional(result, "INDFMPIR", values=values, ci_method="simultaneous")
plt.figure(figsize=(8, 6))
ax = sns.lineplot(fv, pv, lw=3)
ax.fill_between(fv, cb[:, 0], cb[:, 1], color='grey', alpha=0.3)
ax.set_title("Log Odds Smoking vs. Income to Poverty Ratio for Male Aged 40", size=15)
ax.set_xlabel("Income to Poverty Ratio", size=15)
ax.set_ylabel("Log Odds Smoking", size=15)
plt.figure(figsize=(8, 6))
pv_prob = 1 / (1 + np.exp(-pv))
cb_prob = 1 / (1 + np.exp(-cb))
ax = sns.lineplot(fv, pv_prob, lw=3)
ax.fill_between(fv, cb_prob[:, 0], cb_prob[:, 1], color='grey', alpha=0.3)
ax.set_title("Probability Smoking vs. Income to Poverty Ratio for Male Aged 40", size=15)
ax.set_xlabel("Age", size=15)
ax.set_ylabel("Probability Smoking", size=15)
# Import add_lowess
from statsmodels.graphics.regressionplots import add_lowess
fig = result.plot_partial_residuals("RIDAGEYR")
ax = fig.get_axes()[0]
ax.lines[0].set_alpha(0.2)
ax.set_xlabel("Age", size=15)
_ = add_lowess(ax)
fig = result.plot_added_variable("RIDAGEYR")
ax = fig.get_axes()[0]
ax.lines[0].set_alpha(0.2)
ax.set_xlabel("Age (Normalized)", size=15)
_ = add_lowess(ax)
fig = result.plot_ceres_residuals("RIDAGEYR")
ax = fig.get_axes()[0]
ax.lines[0].set_alpha(0.2)
ax.set_xlabel("Age", size=15)
_ = add_lowess(ax)
# Form cluster variable
health["cluster"] = 10*health.SDMVSTRA + health.SDMVPSU
# Within-cluster correlation of multiple variables using GEE without covariates, only an intercept (""~1"), and exchangeable correlation structure.
for v in ["SMQ020x", "RIDAGEYR", "BMXBMI", "INDFMPIR"]:
model = sm.GEE.from_formula(v + " ~ 1", groups="cluster", cov_struct=sm.cov_struct.Exchangeable(), data=health)
result = model.fit()
print(v, result.cov_struct.summary())
# Fit simple logistic model using GLM
GLM = sm.GLM.from_formula("SMQ020x ~ RIDAGEYR + RIAGENDRx + BMXBMI + INDFMPIR + DMDEDUC2x", family=sm.families.Binomial(), data=health)
result_GLM = GLM.fit()
result_GLM.summary()
# Fit a marginal logistic model using GEE and exchangeable correlation structure
GEE = sm.GEE.from_formula("SMQ020x ~ RIDAGEYR + RIAGENDRx + BMXBMI + INDFMPIR + DMDEDUC2x", groups="cluster", family=sm.families.Binomial(), cov_struct=sm.cov_struct.Exchangeable(), data=health)
result_GEE = GEE.fit(start_params = result_GLM.params)
# Create dataframe for comparing OLS and GEE parameters and standard errors
compare = pd.DataFrame({"GLM Params": result_GLM.params, "GLM SE": result_GLM.bse, "GEE Params": result_GEE.params, "GEE SE": result_GEE.bse})
compare = compare[["GLM Params", "GLM SE", "GEE Params", "GEE SE"]]
# Print out the results in a pretty way
display(HTML(compare.to_html()))
| 0.660501 | 0.991255 |
```
import sys
sys.path.append("..")
import numpy as np
import logging
import pickle
from sklearn.metrics import roc_auc_score
from sklearn.preprocessing import RobustScaler
from sklearn.utils import check_random_state
from recnn.preprocessing import permute_by_pt
from recnn.preprocessing import extract
from recnn.recnn import grnn_transform_gated
from recnn.recnn import grnn_predict_gated
%matplotlib inline
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (8,8)
# filename_train = "../data/w-vs-qcd/final/antikt-kt-train.pickle"
# filename_test = "../data/w-vs-qcd/final/antikt-kt-test.pickle"
filename_train = "../data/w-vs-qcd/final/antikt-antikt-train.pickle"
filename_test = "../data/w-vs-qcd/final/antikt-antikt-test.pickle"
rng = check_random_state(1)
# Make training data
print("Loading training data...")
fd = open(filename_train, "rb")
X, y = pickle.load(fd)
fd.close()
y = np.array(y)
print("\tfilename = %s" % filename_train)
print("\tX size = %d" % len(X))
print("\ty size = %d" % len(y))
# Preprocessing
print("Preprocessing...")
X = [extract(permute_by_pt(jet)) for jet in X]
tf = RobustScaler().fit(np.vstack([jet["content"] for jet in X]))
for jet in X:
jet["content"] = tf.transform(jet["content"])
# Make test data
print("Loading test data...")
fd = open(filename_test, "rb")
X, y = pickle.load(fd)
fd.close()
y = np.array(y)
print("\tfilename = %s" % filename_test)
print("\tX size = %d" % len(X))
print("\ty size = %d" % len(y))
# Preprocessing
print("Preprocessing...")
X = [extract(permute_by_pt(jet)) for jet in X]
for jet in X:
jet["content"] = tf.transform(jet["content"])
```
# Visualization
## Simple
```
# Loading model
print("Loading model...")
#filename_model = "../models/jet-study-2/model-w-s-antikt-kt-1.pickle"
filename_model = "../models/jet-study-2/model-w-s-antikt-antikt-1.pickle"
fd = open(filename_model, "rb")
params = pickle.load(fd)
fd.close()
print("\tfilename = %s" % filename_model)
from recnn.recnn import grnn_predict_simple
from sklearn.metrics import roc_auc_score
roc_auc_score(y, grnn_predict_simple(params, X))
import autograd as ag
import autograd.numpy as np
from recnn.recnn import log_loss, relu, sigmoid
def get_branch(tree, target, node_id, is_left):
if target == node_id:
return [(node_id, is_left)]
elif tree[node_id, 0] == -1:
return False
else:
left_result = get_branch(tree, target, tree[node_id, 0], True)
right_result = get_branch(tree, target, tree[node_id, 1], False)
if left_result:
return [(node_id, is_left)] + left_result
elif right_result:
return [(node_id, is_left)] + right_result
else:
return False
def grnn_embeddings(content, tree, params):
embeddings = {}
def _rec(node_id):
u_k = relu(np.dot(params["W_u"], content[node_id].T).T +
params["b_u"]).reshape(1, -1)
if tree[node_id, 0] == -1:
embeddings[node_id] = u_k
return u_k
else:
h_L = _rec(tree[node_id, 0])
h_R = _rec(tree[node_id, 1])
h = relu(np.dot(params["W_h"],
np.hstack((h_L, h_R, u_k)).T).T +
params["b_h"])
embeddings[node_id] = h
return h
_rec(0)
return embeddings
def make_dL_dh(content, tree, params, target):
branch = get_branch(tree, target, 0, True)
embeddings = grnn_embeddings(content, tree, params)
def f(v):
e = v
for i, (node_id, is_left) in enumerate(branch[::-1]):
if i == 0:
e = v
was_left = is_left
elif i > 0:
u_k = relu(np.dot(params["W_u"], content[node_id].T).T +
params["b_u"]).reshape(1, -1)
if was_left:
h_L = e
h_R = embeddings[tree[node_id, 1]]
else:
h_L = embeddings[tree[node_id, 0]]
h_R = e
h = relu(np.dot(params["W_h"],
np.hstack((h_L, h_R, u_k)).T).T +
params["b_h"])
e = h
was_left = is_left
h = e
h = relu(np.dot(params["W_clf"][0], h.T).T + params["b_clf"][0])
h = relu(np.dot(params["W_clf"][1], h.T).T + params["b_clf"][1])
h = sigmoid(np.dot(params["W_clf"][2], h.T).T + params["b_clf"][2])
return h.ravel()
df = ag.grad(f)
return df
def make_dL_du(content, tree, params, target):
branch = get_branch(tree, target, 0, True)
embeddings = grnn_embeddings(content, tree, params)
def f(v):
e = v
for i, (node_id, is_left) in enumerate(branch[::-1]):
if i == 0:
if tree[node_id, 0] == -1:
e = relu(np.dot(params["W_u"], v.T).T +
params["b_u"]).reshape(1, -1)
else:
u_k = relu(np.dot(params["W_u"], v.T).T +
params["b_u"]).reshape(1, -1)
h_L = embeddings[tree[node_id, 0]]
h_R = embeddings[tree[node_id, 1]]
h = relu(np.dot(params["W_h"],
np.hstack((h_L, h_R, u_k)).T).T +
params["b_h"])
e = h
was_left = is_left
elif i > 0:
u_k = relu(np.dot(params["W_u"], content[node_id].T).T +
params["b_u"]).reshape(1, -1)
if was_left:
h_L = e
h_R = embeddings[tree[node_id, 1]]
else:
h_L = embeddings[tree[node_id, 0]]
h_R = e
h = relu(np.dot(params["W_h"],
np.hstack((h_L, h_R, u_k)).T).T +
params["b_h"])
e = h
was_left = is_left
h = e
h = relu(np.dot(params["W_clf"][0], h.T).T + params["b_clf"][0])
h = relu(np.dot(params["W_clf"][1], h.T).T + params["b_clf"][1])
h = sigmoid(np.dot(params["W_clf"][2], h.T).T + params["b_clf"][2])
return h.ravel()
df = ag.grad(f)
return df
from graphviz import Digraph
import matplotlib as mpl
import matplotlib.cm as cm
norm = mpl.colors.Normalize(vmin=0.001, vmax=2.0)
cmap = cm.viridis_r
m = cm.ScalarMappable(norm=norm, cmap=cmap)
def number_to_color(x):
color = m.to_rgba(x)
return "#%.2x%.2x%.2x%.2x" % (int(255*color[0]),
int(255*color[1]),
int(255*color[2]),
int(255*color[3]))
def plot_jet_simple(params, jet, label=None):
content = tf.inverse_transform(jet["content"])
embeddings = grnn_embeddings(jet["content"], jet["tree"], params)
# Build graph recursively
dot = Digraph(graph_attr={"rank": "flow"},
edge_attr={"arrowsize": "0.5", "fontsize": "8.0"},
node_attr={"style": "filled"},
format="png")
df = make_dL_dh(jet["content"], jet["tree"], params, 0)
norm_root = (df(embeddings[0]) ** 2).sum() ** 0.5
if label:
dot.graph_attr["label"] = label
def _rec(jet, parent, node_id):
df = make_dL_du(jet["content"], jet["tree"], params, node_id)
norm_u = (df(jet["content"][node_id]) ** 2).sum() ** 0.5 / norm_root
df = make_dL_dh(jet["content"], jet["tree"], params, node_id)
norm_h = (df(embeddings[node_id]) ** 2).sum() ** 0.5 / norm_root
# Build subgraph
sub = Digraph(graph_attr={"rank": "flow"},
node_attr={"fixedsize": "true",
"label": "",
"height": "0.1",
"width": "0.1",
"style": "filled"},
edge_attr={"arrowsize": "0.5",
"fontsize": "8.0"})
size = "%.4f" % max(0.1, norm_h ** 0.5 / 2.0)
sub.node("%d" % node_id, width=size, height=size, shape="circle",
color=number_to_color(norm_h))
if jet["tree"][node_id, 0] == -1:
size = "%.4f" % max(0.1, norm_u / (2.0 ** 0.5))
sub.node("%d-N" % node_id, width=size, height=size, shape="diamond",
color=number_to_color(norm_u))
sub.edge("%d-N" % node_id, "%d" % node_id,
color=number_to_color(norm_u),
#label="o_%d" % (1+node_id)
)#label="%.4f" % norm_u)
dot.subgraph(sub)
# Connect to parent
if parent >= 0:
dot.edge("%d" % node_id, "%d" % parent,
color=number_to_color(norm_h),
#label="h_%d" % (1+node_id)
)
#label="%.4f" % norm_h)
# Recursive calls
if jet["tree"][node_id, 0] != -1:
_rec(jet, node_id, jet["tree"][node_id, 0])
_rec(jet, node_id, jet["tree"][node_id, 1])
_rec(jet, -1, jet["root_id"])
return dot
jet_id = np.argmin([len(j["tree"]) if len(j["tree"]) > 27 else np.inf for j in X])
print(jet_id)
dot = plot_jet_simple(params, X[jet_id],)
# label="y=%d, y_pred=%.4f" % (y[jet_id],
# grnn_predict_simple(params, [X[jet_id]])[0]))
dot
dot.save("jet-antikt-%d.gv" % jet_id)
np.random.seed(123)
for jet_id in np.random.permutation(len(X))[:20]:
dot = plot_jet_simple(params, X[jet_id],
label="y=%d, y_pred=%.4f" % (y[jet_id],
grnn_predict_simple(params, [X[jet_id]])[0]))
dot.render("figures/%d-anti-kt" % jet_id)
```
|
github_jupyter
|
import sys
sys.path.append("..")
import numpy as np
import logging
import pickle
from sklearn.metrics import roc_auc_score
from sklearn.preprocessing import RobustScaler
from sklearn.utils import check_random_state
from recnn.preprocessing import permute_by_pt
from recnn.preprocessing import extract
from recnn.recnn import grnn_transform_gated
from recnn.recnn import grnn_predict_gated
%matplotlib inline
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (8,8)
# filename_train = "../data/w-vs-qcd/final/antikt-kt-train.pickle"
# filename_test = "../data/w-vs-qcd/final/antikt-kt-test.pickle"
filename_train = "../data/w-vs-qcd/final/antikt-antikt-train.pickle"
filename_test = "../data/w-vs-qcd/final/antikt-antikt-test.pickle"
rng = check_random_state(1)
# Make training data
print("Loading training data...")
fd = open(filename_train, "rb")
X, y = pickle.load(fd)
fd.close()
y = np.array(y)
print("\tfilename = %s" % filename_train)
print("\tX size = %d" % len(X))
print("\ty size = %d" % len(y))
# Preprocessing
print("Preprocessing...")
X = [extract(permute_by_pt(jet)) for jet in X]
tf = RobustScaler().fit(np.vstack([jet["content"] for jet in X]))
for jet in X:
jet["content"] = tf.transform(jet["content"])
# Make test data
print("Loading test data...")
fd = open(filename_test, "rb")
X, y = pickle.load(fd)
fd.close()
y = np.array(y)
print("\tfilename = %s" % filename_test)
print("\tX size = %d" % len(X))
print("\ty size = %d" % len(y))
# Preprocessing
print("Preprocessing...")
X = [extract(permute_by_pt(jet)) for jet in X]
for jet in X:
jet["content"] = tf.transform(jet["content"])
# Loading model
print("Loading model...")
#filename_model = "../models/jet-study-2/model-w-s-antikt-kt-1.pickle"
filename_model = "../models/jet-study-2/model-w-s-antikt-antikt-1.pickle"
fd = open(filename_model, "rb")
params = pickle.load(fd)
fd.close()
print("\tfilename = %s" % filename_model)
from recnn.recnn import grnn_predict_simple
from sklearn.metrics import roc_auc_score
roc_auc_score(y, grnn_predict_simple(params, X))
import autograd as ag
import autograd.numpy as np
from recnn.recnn import log_loss, relu, sigmoid
def get_branch(tree, target, node_id, is_left):
if target == node_id:
return [(node_id, is_left)]
elif tree[node_id, 0] == -1:
return False
else:
left_result = get_branch(tree, target, tree[node_id, 0], True)
right_result = get_branch(tree, target, tree[node_id, 1], False)
if left_result:
return [(node_id, is_left)] + left_result
elif right_result:
return [(node_id, is_left)] + right_result
else:
return False
def grnn_embeddings(content, tree, params):
embeddings = {}
def _rec(node_id):
u_k = relu(np.dot(params["W_u"], content[node_id].T).T +
params["b_u"]).reshape(1, -1)
if tree[node_id, 0] == -1:
embeddings[node_id] = u_k
return u_k
else:
h_L = _rec(tree[node_id, 0])
h_R = _rec(tree[node_id, 1])
h = relu(np.dot(params["W_h"],
np.hstack((h_L, h_R, u_k)).T).T +
params["b_h"])
embeddings[node_id] = h
return h
_rec(0)
return embeddings
def make_dL_dh(content, tree, params, target):
branch = get_branch(tree, target, 0, True)
embeddings = grnn_embeddings(content, tree, params)
def f(v):
e = v
for i, (node_id, is_left) in enumerate(branch[::-1]):
if i == 0:
e = v
was_left = is_left
elif i > 0:
u_k = relu(np.dot(params["W_u"], content[node_id].T).T +
params["b_u"]).reshape(1, -1)
if was_left:
h_L = e
h_R = embeddings[tree[node_id, 1]]
else:
h_L = embeddings[tree[node_id, 0]]
h_R = e
h = relu(np.dot(params["W_h"],
np.hstack((h_L, h_R, u_k)).T).T +
params["b_h"])
e = h
was_left = is_left
h = e
h = relu(np.dot(params["W_clf"][0], h.T).T + params["b_clf"][0])
h = relu(np.dot(params["W_clf"][1], h.T).T + params["b_clf"][1])
h = sigmoid(np.dot(params["W_clf"][2], h.T).T + params["b_clf"][2])
return h.ravel()
df = ag.grad(f)
return df
def make_dL_du(content, tree, params, target):
branch = get_branch(tree, target, 0, True)
embeddings = grnn_embeddings(content, tree, params)
def f(v):
e = v
for i, (node_id, is_left) in enumerate(branch[::-1]):
if i == 0:
if tree[node_id, 0] == -1:
e = relu(np.dot(params["W_u"], v.T).T +
params["b_u"]).reshape(1, -1)
else:
u_k = relu(np.dot(params["W_u"], v.T).T +
params["b_u"]).reshape(1, -1)
h_L = embeddings[tree[node_id, 0]]
h_R = embeddings[tree[node_id, 1]]
h = relu(np.dot(params["W_h"],
np.hstack((h_L, h_R, u_k)).T).T +
params["b_h"])
e = h
was_left = is_left
elif i > 0:
u_k = relu(np.dot(params["W_u"], content[node_id].T).T +
params["b_u"]).reshape(1, -1)
if was_left:
h_L = e
h_R = embeddings[tree[node_id, 1]]
else:
h_L = embeddings[tree[node_id, 0]]
h_R = e
h = relu(np.dot(params["W_h"],
np.hstack((h_L, h_R, u_k)).T).T +
params["b_h"])
e = h
was_left = is_left
h = e
h = relu(np.dot(params["W_clf"][0], h.T).T + params["b_clf"][0])
h = relu(np.dot(params["W_clf"][1], h.T).T + params["b_clf"][1])
h = sigmoid(np.dot(params["W_clf"][2], h.T).T + params["b_clf"][2])
return h.ravel()
df = ag.grad(f)
return df
from graphviz import Digraph
import matplotlib as mpl
import matplotlib.cm as cm
norm = mpl.colors.Normalize(vmin=0.001, vmax=2.0)
cmap = cm.viridis_r
m = cm.ScalarMappable(norm=norm, cmap=cmap)
def number_to_color(x):
color = m.to_rgba(x)
return "#%.2x%.2x%.2x%.2x" % (int(255*color[0]),
int(255*color[1]),
int(255*color[2]),
int(255*color[3]))
def plot_jet_simple(params, jet, label=None):
content = tf.inverse_transform(jet["content"])
embeddings = grnn_embeddings(jet["content"], jet["tree"], params)
# Build graph recursively
dot = Digraph(graph_attr={"rank": "flow"},
edge_attr={"arrowsize": "0.5", "fontsize": "8.0"},
node_attr={"style": "filled"},
format="png")
df = make_dL_dh(jet["content"], jet["tree"], params, 0)
norm_root = (df(embeddings[0]) ** 2).sum() ** 0.5
if label:
dot.graph_attr["label"] = label
def _rec(jet, parent, node_id):
df = make_dL_du(jet["content"], jet["tree"], params, node_id)
norm_u = (df(jet["content"][node_id]) ** 2).sum() ** 0.5 / norm_root
df = make_dL_dh(jet["content"], jet["tree"], params, node_id)
norm_h = (df(embeddings[node_id]) ** 2).sum() ** 0.5 / norm_root
# Build subgraph
sub = Digraph(graph_attr={"rank": "flow"},
node_attr={"fixedsize": "true",
"label": "",
"height": "0.1",
"width": "0.1",
"style": "filled"},
edge_attr={"arrowsize": "0.5",
"fontsize": "8.0"})
size = "%.4f" % max(0.1, norm_h ** 0.5 / 2.0)
sub.node("%d" % node_id, width=size, height=size, shape="circle",
color=number_to_color(norm_h))
if jet["tree"][node_id, 0] == -1:
size = "%.4f" % max(0.1, norm_u / (2.0 ** 0.5))
sub.node("%d-N" % node_id, width=size, height=size, shape="diamond",
color=number_to_color(norm_u))
sub.edge("%d-N" % node_id, "%d" % node_id,
color=number_to_color(norm_u),
#label="o_%d" % (1+node_id)
)#label="%.4f" % norm_u)
dot.subgraph(sub)
# Connect to parent
if parent >= 0:
dot.edge("%d" % node_id, "%d" % parent,
color=number_to_color(norm_h),
#label="h_%d" % (1+node_id)
)
#label="%.4f" % norm_h)
# Recursive calls
if jet["tree"][node_id, 0] != -1:
_rec(jet, node_id, jet["tree"][node_id, 0])
_rec(jet, node_id, jet["tree"][node_id, 1])
_rec(jet, -1, jet["root_id"])
return dot
jet_id = np.argmin([len(j["tree"]) if len(j["tree"]) > 27 else np.inf for j in X])
print(jet_id)
dot = plot_jet_simple(params, X[jet_id],)
# label="y=%d, y_pred=%.4f" % (y[jet_id],
# grnn_predict_simple(params, [X[jet_id]])[0]))
dot
dot.save("jet-antikt-%d.gv" % jet_id)
np.random.seed(123)
for jet_id in np.random.permutation(len(X))[:20]:
dot = plot_jet_simple(params, X[jet_id],
label="y=%d, y_pred=%.4f" % (y[jet_id],
grnn_predict_simple(params, [X[jet_id]])[0]))
dot.render("figures/%d-anti-kt" % jet_id)
| 0.29584 | 0.379522 |
```
import graphlab
image_train = graphlab.SFrame('image_train_data/')
image_train.head()
knn_model = graphlab.nearest_neighbors.create(image_train, features = ['deep_features'], label = 'id')
graphlab.canvas.set_target('ipynb')
cat = image_train[18:19]
cat['image'].show()
knn_model.query(cat)
def get_images_from_ids(query_result):
return image_train.filter_by(query_result['reference_label'], 'id')
cat_neighbors = get_images_from_ids(knn_model.query(cat))
cat_neighbors['image'].show()
car = image_train[8:9]
car['image'].show()
get_images_from_ids(knn_model.query(car))['image'].show()
show_neighbors = lambda i: get_images_from_ids(knn_model.query(image_train[i:i+1]))['image'].show()
show_neighbors(8)
show_neighbors(26)
show_neighbors(1222)
show_neighbors(2000)
```
# Assignment
```
sketch = graphlab.Sketch(image_train['label'])
sketch
image_train_dog = image_train[image_train['label'] == 'dog']
image_train_cat = image_train[image_train['label'] == 'cat']
image_train_bird = image_train[image_train['label'] == 'bird']
image_train_automobile = image_train[image_train['label'] == 'automobile']
dog_model = graphlab.nearest_neighbors.create(image_train_dog, features=['deep_features'], label = 'id')
cat_model = graphlab.nearest_neighbors.create(image_train_cat, features=['deep_features'], label = 'id')
bird_model = graphlab.nearest_neighbors.create(image_train_bird, features=['deep_features'], label = 'id')
automobile_model = graphlab.nearest_neighbors.create(image_train_automobile, features=['deep_features'], label = 'id')
image_test = graphlab.SFrame('image_test_data/')
get_images_from_ids(cat_model.query(image_test[0:1]))['image'].show()
get_images_from_ids(dog_model.query(image_test[0:1]))['image'].show()
cat_model.query(image_test[0:1])['distance'].mean()
dog_model.query(image_test[0:1])['distance'].mean()
image_test_cat = image_test[image_test['label'] == 'cat']
image_test_dog = image_test[image_test['label'] == 'dog']
image_test_bird = image_test[image_test['label'] == 'bird']
image_test_automobile = image_test[image_test['label'] == 'automobile']
dog_cat_neighbors = cat_model.query(image_test_dog, k = 1)
dog_automobile_neighbors = automobile_model.query(image_test_dog, k = 1)
dog_bird_neighbors = bird_model.query(image_test_dog, k = 1)
dog_dog_neighbors = dog_model.query(image_test_dog, k = 1)
dog_distances = graphlab.SFrame({'dog-dog': dog_dog_neighbors['distance'], 'dog-cat': dog_cat_neighbors['distance'], 'dog-automobile': dog_automobile_neighbors['distance'], 'dog-bird': dog_bird_neighbors['distance']})
def is_dog_correct(row):
if(row['dog-dog'] < row['dog-cat'] and row['dog-dog'] < row['dog-automobile'] and row['dog-dog'] < row['dog-bird']):
return 1
else:
return 0
correctly_classified = dog_distances.apply(is_dog_correct).sum()
float(correctly_classified)/len(image_test_dog)
```
# Done
|
github_jupyter
|
import graphlab
image_train = graphlab.SFrame('image_train_data/')
image_train.head()
knn_model = graphlab.nearest_neighbors.create(image_train, features = ['deep_features'], label = 'id')
graphlab.canvas.set_target('ipynb')
cat = image_train[18:19]
cat['image'].show()
knn_model.query(cat)
def get_images_from_ids(query_result):
return image_train.filter_by(query_result['reference_label'], 'id')
cat_neighbors = get_images_from_ids(knn_model.query(cat))
cat_neighbors['image'].show()
car = image_train[8:9]
car['image'].show()
get_images_from_ids(knn_model.query(car))['image'].show()
show_neighbors = lambda i: get_images_from_ids(knn_model.query(image_train[i:i+1]))['image'].show()
show_neighbors(8)
show_neighbors(26)
show_neighbors(1222)
show_neighbors(2000)
sketch = graphlab.Sketch(image_train['label'])
sketch
image_train_dog = image_train[image_train['label'] == 'dog']
image_train_cat = image_train[image_train['label'] == 'cat']
image_train_bird = image_train[image_train['label'] == 'bird']
image_train_automobile = image_train[image_train['label'] == 'automobile']
dog_model = graphlab.nearest_neighbors.create(image_train_dog, features=['deep_features'], label = 'id')
cat_model = graphlab.nearest_neighbors.create(image_train_cat, features=['deep_features'], label = 'id')
bird_model = graphlab.nearest_neighbors.create(image_train_bird, features=['deep_features'], label = 'id')
automobile_model = graphlab.nearest_neighbors.create(image_train_automobile, features=['deep_features'], label = 'id')
image_test = graphlab.SFrame('image_test_data/')
get_images_from_ids(cat_model.query(image_test[0:1]))['image'].show()
get_images_from_ids(dog_model.query(image_test[0:1]))['image'].show()
cat_model.query(image_test[0:1])['distance'].mean()
dog_model.query(image_test[0:1])['distance'].mean()
image_test_cat = image_test[image_test['label'] == 'cat']
image_test_dog = image_test[image_test['label'] == 'dog']
image_test_bird = image_test[image_test['label'] == 'bird']
image_test_automobile = image_test[image_test['label'] == 'automobile']
dog_cat_neighbors = cat_model.query(image_test_dog, k = 1)
dog_automobile_neighbors = automobile_model.query(image_test_dog, k = 1)
dog_bird_neighbors = bird_model.query(image_test_dog, k = 1)
dog_dog_neighbors = dog_model.query(image_test_dog, k = 1)
dog_distances = graphlab.SFrame({'dog-dog': dog_dog_neighbors['distance'], 'dog-cat': dog_cat_neighbors['distance'], 'dog-automobile': dog_automobile_neighbors['distance'], 'dog-bird': dog_bird_neighbors['distance']})
def is_dog_correct(row):
if(row['dog-dog'] < row['dog-cat'] and row['dog-dog'] < row['dog-automobile'] and row['dog-dog'] < row['dog-bird']):
return 1
else:
return 0
correctly_classified = dog_distances.apply(is_dog_correct).sum()
float(correctly_classified)/len(image_test_dog)
| 0.390243 | 0.784402 |
# Eigensystems
This notebook explores methods for computing eigenvalues and eigenvectors, i.e, finding $x$ and $\lambda$ that satisfy
$$ A x = \lambda x $$
For general $A$ this can be quite challenging so we restrict our attention to **symmetric matrices** $A \in \mathbb{R}^{n\times n}$.
There are a huge range of the methods for computing or estimating eigenvalues/eigenvectors. We will look at a few of the simpler ones in this course.
```
import numpy as np
from numpy import linalg as la
```
# A test eigensystem
For the examples below I use the following matrix as an example. As a check on our methods I will also use NumPy's built in methods to compute the eigenvalues and eigenvectors
```
A = np.array([[2, -1, 0],[-1, 2, -1],[0, -1, 2]])
eigenSystem = la.eig(A)
print("Eigenvalues =", eigenSystem[0])
print("Eigenvectors =\n", eigenSystem[1]) # The eigen vectors are the columns of this matrix
lmaxCheck = np.max(eigenSystem[0])
lsecondCheck = np.sort(eigenSystem[0])[1]
lminCheck = np.min(eigenSystem[0])
print("The maximum eigenvalue is:", lmaxCheck)
print("The second eigenvalue is:", lsecondCheck)
print("The minimum eigenvalue is:", lminCheck)
```
# Maximum eigenvalue via the power method
Here we implemenet the power method for computing the maximum eigenvalue of a matrix and the associated eigenvector.
Also check out this [nice video](https://www.youtube.com/watch?v=yBiQh1vsCLU) showing the method in action
```
def MaxEigenvalue(A, err):
(m,n) = A.shape
if(m != n):
print("Matrix must be square")
return
# Create a random initial vector
x = np.random.rand(m)
lam = 0.1
lamprev = 1
while np.abs(1-lam/lamprev) > err:
Ax = A@x
lamprev = lam
lam = la.norm(Ax,2)/la.norm(x,2)
x = Ax
x = x/la.norm(x)
return (lam,x)
largestEigen = MaxEigenvalue(A, 1e-14)
# Check that eigenvalue and eigenvalue satisfy the eigen equation
print("Check the eigenvalue equation is satisfied:", np.dot(A,largestEigen[1]) - largestEigen[0] *largestEigen[1],"\n")
# Compare the eigenvalue to NumPy's result
print("Comparison of eigenvalue with NumPy:", largestEigen[0], largestEigen[0]- lmaxCheck,"\n")
# Compare the eigenvector to NumPy's result
print("Comparison of eigenvector with NumPy:", np.abs(largestEigen[1])-np.abs(eigenSystem[1][:,0]))
```
# Minimum eigenvalue for an invertible matrix
If the matrix in question is invertable and has a small condition number, one way to find the smallest eigenvalue of a matrix $A$ is to find the largest eigenvalue of $A^{-1}$ and take the reciprical. This method is not very useful if we are trying to calcualte a condition number via the $L^2$ norm (as we only know if the matrix can be inverted without error by computing the condition number, but we need to compute the condition number to know if it is safe to invert the matrix numerically).
Assuming we know it is safe to numerically invert the matrix, we could invert it using the GaussianElimination function we looked at in another notebook. Here I will just use NumPy's built in function.
```
lmin = 1/MaxEigenvalue(la.inv(A), 1e-14)[0]
# Compare the result to NumPy's calculation
print(lmin, lmin - lminCheck)
```
# Minimum eigenvalue for symmetric, positive definite matrices
If we have a symmetric matrix which is positive definite then we can find the minimum eigenvalue without inverting the matrix using the algorithm below. In the lectures we proved that $A^T A$ has these properties.
If we define
$$ B = A -\lambda_\max I$$
Then so long as $A$ is positive definite the max eigenvalue of $B$ plus $\lambda_\max$ will be the minimum eigenvalue of $A$. Thus we can find the minimum eigenvalue of $A$ by finding the maximum eigenvalue of $B$ using the power method.
This method is suitable for calculating the condition number of a matrix.
```
def MinEigenvalueSymmetricPositiveDefinite(A, err):
(m,n) = A.shape
if(m != n):
print("Matrix must be square")
return
lmax = MaxEigenvalue(A,err)[0]
B = A - np.identity(m)*lmax
eigBmax = MaxEigenvalue(B,err)
return (-eigBmax[0] + lmax, eigBmax[1])
smallEigen = MinEigenvalueSymmetricPositiveDefinite(A, 1e-14)
# Check that eigenvalue and eigenvalue satisfy the eigen equation
print(np.dot(A,smallEigen[1]) - smallEigen[0] *smallEigen[1],"\n")
# Check the eigenvalue agrees with NumPy's result
print("Check against NumPy's result:", smallEigen[0], smallEigen[0] - lminCheck)
```
# Hotelling's deflation: finding the second largest eigenvalue
We've seen how to find the maximum and minimum eigenvalues. What about other eigenvalues? In general if you are looking for multiple eigenvalues there are a range of other methods. Here we look at a method for finding the second largest eigenvalue. This method only works for symmetric matrices.
In this algorthm we construct the matrix
$$ B = A - \lambda_1 e_1\otimes e_1$$
where $\lambda_1$ is the largest eigenvalue, $e_1$ is the corresponding (unit normalized) eigenvector, and $\otimes$ is the outer product.
The matrix $B$ has the same eigenvectors as $A$, and the same eigenvalues except the largest one has been replaced by 0. Thus if we use the power method to find the largest eigenvalue of $B$, this will be the second largest eigenvalue of $A$.
```
maxEigen = MaxEigenvalue(A, 1e-14)
l1 = maxEigen[0]
e1 = maxEigen[1]
B = A - l1*np.outer(e1, e1)
lsecond = MaxEigenvalue(B, 1e-12)
# Compare against NumPy's results
print("Comparison with NumPy's eigenvalue:", lsecond[0], lsecond[0] - lsecondCheck)
print("Comparison with NumPy's eigenvector:", np.abs(lsecond[1]) - np.abs(eigenSystem[1][:,1]))
```
If you like you can apply the method again to find the third eigenvalue. If you are looking to find many eigenvalues this is not recommended (numerical round off will start to cause problems). Instead use methods like the [QR algorithm](https://en.wikipedia.org/wiki/QR_algorithm).
|
github_jupyter
|
import numpy as np
from numpy import linalg as la
A = np.array([[2, -1, 0],[-1, 2, -1],[0, -1, 2]])
eigenSystem = la.eig(A)
print("Eigenvalues =", eigenSystem[0])
print("Eigenvectors =\n", eigenSystem[1]) # The eigen vectors are the columns of this matrix
lmaxCheck = np.max(eigenSystem[0])
lsecondCheck = np.sort(eigenSystem[0])[1]
lminCheck = np.min(eigenSystem[0])
print("The maximum eigenvalue is:", lmaxCheck)
print("The second eigenvalue is:", lsecondCheck)
print("The minimum eigenvalue is:", lminCheck)
def MaxEigenvalue(A, err):
(m,n) = A.shape
if(m != n):
print("Matrix must be square")
return
# Create a random initial vector
x = np.random.rand(m)
lam = 0.1
lamprev = 1
while np.abs(1-lam/lamprev) > err:
Ax = A@x
lamprev = lam
lam = la.norm(Ax,2)/la.norm(x,2)
x = Ax
x = x/la.norm(x)
return (lam,x)
largestEigen = MaxEigenvalue(A, 1e-14)
# Check that eigenvalue and eigenvalue satisfy the eigen equation
print("Check the eigenvalue equation is satisfied:", np.dot(A,largestEigen[1]) - largestEigen[0] *largestEigen[1],"\n")
# Compare the eigenvalue to NumPy's result
print("Comparison of eigenvalue with NumPy:", largestEigen[0], largestEigen[0]- lmaxCheck,"\n")
# Compare the eigenvector to NumPy's result
print("Comparison of eigenvector with NumPy:", np.abs(largestEigen[1])-np.abs(eigenSystem[1][:,0]))
lmin = 1/MaxEigenvalue(la.inv(A), 1e-14)[0]
# Compare the result to NumPy's calculation
print(lmin, lmin - lminCheck)
def MinEigenvalueSymmetricPositiveDefinite(A, err):
(m,n) = A.shape
if(m != n):
print("Matrix must be square")
return
lmax = MaxEigenvalue(A,err)[0]
B = A - np.identity(m)*lmax
eigBmax = MaxEigenvalue(B,err)
return (-eigBmax[0] + lmax, eigBmax[1])
smallEigen = MinEigenvalueSymmetricPositiveDefinite(A, 1e-14)
# Check that eigenvalue and eigenvalue satisfy the eigen equation
print(np.dot(A,smallEigen[1]) - smallEigen[0] *smallEigen[1],"\n")
# Check the eigenvalue agrees with NumPy's result
print("Check against NumPy's result:", smallEigen[0], smallEigen[0] - lminCheck)
maxEigen = MaxEigenvalue(A, 1e-14)
l1 = maxEigen[0]
e1 = maxEigen[1]
B = A - l1*np.outer(e1, e1)
lsecond = MaxEigenvalue(B, 1e-12)
# Compare against NumPy's results
print("Comparison with NumPy's eigenvalue:", lsecond[0], lsecond[0] - lsecondCheck)
print("Comparison with NumPy's eigenvector:", np.abs(lsecond[1]) - np.abs(eigenSystem[1][:,1]))
| 0.573559 | 0.985043 |
```
#dependencies
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import re
#importing the csv file
overview_df = pd.read_csv("Indeed Data Set.csv")
overview_df.head()
overview_df.columns
skills_df = overview_df[['python', 'sql', 'machine learning', 'r', 'hadoop', 'tableau', 'sas', 'spark', 'java']]
#skills_df
skills_sum = skills_df.sum()
#skills_sum
top_skills = pd.DataFrame(skills_sum)
#top_skills
top_skills.plot(kind="bar")
plt.title("Top Skills in Data Science")
plt.ylabel("Number of Jobs require the skills")
plt.show()
plt.tight_layout()
industry_df = overview_df[['Consulting and Business Services',
'Internet and Software', 'Banks and Financial Services', 'Health Care', 'Insurance']]
#industry_df
industry_sum = industry_df.sum()
#industry_sum
top_industry = pd.DataFrame(industry_sum)
#top_skills
top_industry.plot(kind="bar")
plt.title("Top Industries in Data Science")
plt.ylabel("Number of companies")
plt.show()
plt.tight_layout()
states_df = overview_df[['CA', 'NY', 'VA', 'TX', 'MA', 'IL', 'WA', 'MD', 'DC', 'NC']]
#states_df
states_sum = states_df.sum()
#states_sum
top_states = pd.DataFrame(states_sum)
#top_states
top_states.plot(kind="bar")
plt.title("Top States in Data Science")
plt.ylabel("Number of Jobs")
plt.show()
plt.tight_layout()
```
# Exploring the Description Column
```
raw_df = pd.read_csv("indeed_job_dataset222.csv")
raw_df.head()
# checking what columns I have in this dataset
raw_df.columns
#isolating the descritopn column
desc_df = raw_df["Description"].dropna()
desc_df
```
Resource link : https://www.youtube.com/watch?v=hI1RZgbcW7g
```
#changing the decription column to lower case letters
desc_df = desc_df.str.lower()
desc_df
#finding the cells which have the word has degree
has_degree = desc_df.str.contains("degree")
has_degree.value_counts()
#rows that have degree
degree_row = desc_df[has_degree]
#using words with extraction
def search(text,n):
'''Searches for text, and retrieves n words either side of the text, which are retuned seperatly'''
word = r"\W*([\w]+)"
groups = re.search(r'{}\W*{}{}'.format(word*n,'place',word*n), text).groups()
return groups[:n],groups[n:]
```
Resource: https://stackoverflow.com/questions/17645701/extract-words-surrounding-a-search-word
http://chris35wills.github.io/apply_func_pandas/
```
#applying function to the
#degree_row.applymap(lambda x: search(degree, 20))
```
# Exploring the Industry Revenue
```
explore_industry = raw_df[['Job_Title', 'Queried_Salary', 'Job_Type', 'Skill', 'No_of_Skills', 'Company',
'Description', 'Location', 'Company_Revenue', 'Company_Employees', 'Company_Industry']]
explore_industry.head()
explore_dropna = explore_industry.dropna(how= "all", subset=['Company_Revenue'])
explore_dropna.head()
# no of skills required by indutry
group_industry = explore_dropna.groupby(['Company_Industry'])
group_industry.nunique()
new_df = explore_dropna.value_counts(['Company_Revenue', 'Queried_Salary'])
new_df
new_df = pd.DataFrame(new_df)
new_df
```
|
github_jupyter
|
#dependencies
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import re
#importing the csv file
overview_df = pd.read_csv("Indeed Data Set.csv")
overview_df.head()
overview_df.columns
skills_df = overview_df[['python', 'sql', 'machine learning', 'r', 'hadoop', 'tableau', 'sas', 'spark', 'java']]
#skills_df
skills_sum = skills_df.sum()
#skills_sum
top_skills = pd.DataFrame(skills_sum)
#top_skills
top_skills.plot(kind="bar")
plt.title("Top Skills in Data Science")
plt.ylabel("Number of Jobs require the skills")
plt.show()
plt.tight_layout()
industry_df = overview_df[['Consulting and Business Services',
'Internet and Software', 'Banks and Financial Services', 'Health Care', 'Insurance']]
#industry_df
industry_sum = industry_df.sum()
#industry_sum
top_industry = pd.DataFrame(industry_sum)
#top_skills
top_industry.plot(kind="bar")
plt.title("Top Industries in Data Science")
plt.ylabel("Number of companies")
plt.show()
plt.tight_layout()
states_df = overview_df[['CA', 'NY', 'VA', 'TX', 'MA', 'IL', 'WA', 'MD', 'DC', 'NC']]
#states_df
states_sum = states_df.sum()
#states_sum
top_states = pd.DataFrame(states_sum)
#top_states
top_states.plot(kind="bar")
plt.title("Top States in Data Science")
plt.ylabel("Number of Jobs")
plt.show()
plt.tight_layout()
raw_df = pd.read_csv("indeed_job_dataset222.csv")
raw_df.head()
# checking what columns I have in this dataset
raw_df.columns
#isolating the descritopn column
desc_df = raw_df["Description"].dropna()
desc_df
#changing the decription column to lower case letters
desc_df = desc_df.str.lower()
desc_df
#finding the cells which have the word has degree
has_degree = desc_df.str.contains("degree")
has_degree.value_counts()
#rows that have degree
degree_row = desc_df[has_degree]
#using words with extraction
def search(text,n):
'''Searches for text, and retrieves n words either side of the text, which are retuned seperatly'''
word = r"\W*([\w]+)"
groups = re.search(r'{}\W*{}{}'.format(word*n,'place',word*n), text).groups()
return groups[:n],groups[n:]
#applying function to the
#degree_row.applymap(lambda x: search(degree, 20))
explore_industry = raw_df[['Job_Title', 'Queried_Salary', 'Job_Type', 'Skill', 'No_of_Skills', 'Company',
'Description', 'Location', 'Company_Revenue', 'Company_Employees', 'Company_Industry']]
explore_industry.head()
explore_dropna = explore_industry.dropna(how= "all", subset=['Company_Revenue'])
explore_dropna.head()
# no of skills required by indutry
group_industry = explore_dropna.groupby(['Company_Industry'])
group_industry.nunique()
new_df = explore_dropna.value_counts(['Company_Revenue', 'Queried_Salary'])
new_df
new_df = pd.DataFrame(new_df)
new_df
| 0.422505 | 0.671902 |
<h1 align="center">Graduate Rotational Internship Programme</h1>
<h2 align="center">The Sparks Foundation</h2>
<h3>To Explore Supervised Machine Learning.</h3>
### **Simple Linear Regression**
In this regression task we will predict the percentage of marks that a student is expected to score based upon the number of hours they studied. This is a simple linear regression task as it involves just two variables.
```
# Importing all the required libraries
import numpy as np # to perform calculations
import pandas as pd # to read data
import matplotlib.pyplot as plt # to visualise
import seaborn as sns # to visualise
url="http://bit.ly/w-data"
df=pd.read_csv(url)
df.shape
df.head(25)
df.describe()
df.info()
# Plotting the distribution of scores
plt.figure(figsize=(8,6))
sns.scatterplot(x='Hours', y='Scores', data=df)
plt.title('Hours vs Percentage', fontsize=12)
plt.xlabel('Hours Studied', fontsize=10)
plt.ylabel('Percentage Score', fontsize=10)
plt.tight_layout()
```
There is a positive linear relation between the 'number of hours studied' and 'percentage of score' obtained by the students
```
sns.pairplot(df)
sns.distplot(df['Scores'],hist_kws=dict(edgecolor="black", linewidth=1),color='Blue')
sns.distplot(df['Hours'],hist_kws=dict(edgecolor="black", linewidth=1),color='Blue')
#Displaying correlation among all the columns
df.corr()
# Setting variables
X = df.iloc[:, :-1].values
y = df.iloc[:, 1].values
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
print(X_train.shape, X_test.shape, y_train.shape, y_test.shape)
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test) # Predicting the scores
plt.figure(figsize=(8,6))
plt.scatter(X_test, y_test)
plt.plot(X_test, lr.predict(X_test), color = "r")
plt.title('Hours vs Percentage (Testing set)', fontsize=12)
plt.xlabel('Hours Studied', fontsize=10)
plt.ylabel('Percentage Score', fontsize=10)
plt.tight_layout()
#Displaying the Intercept
print(lr.intercept_)
from sklearn import metrics
print('MAE:', metrics.mean_absolute_error(y_test, y_pred))
print('MSE:', metrics.mean_squared_error(y_test, y_pred))
print('RMSE:', np.sqrt(metrics.mean_squared_error(y_test, y_pred)))
hours = 9.25
own_pred = lr.predict([[hours]])
print("No of Hours = {}".format(hours))
print("Predicted Score = {}".format(own_pred[0]))
```
Using the above model, the predicted score for a student studying 9.25 hours a day is approximately 93.69
</br>
Additional steps using other packages and models
# Using PyCaret
```
from pycaret.regression import *
reg1 = setup(df, target = 'Scores', session_id = 123)
lr = create_model('lr')
tuned_lr = tune_model(lr)
final_lr = finalize_model(tuned_lr)
predict_model(final_lr)
new = {'Hours': ['9.25']}
new_df = pd.DataFrame(new)
unseen_predictions = predict_model(final_lr, data=new_df)
unseen_predictions.head()
```
Using the above model, the predicted score for a student studying 9.25 hours a day is approximately 92.9099
```
# Adding more parameters and using other model other than Linear Regression
reg11 = setup(df, target = 'Scores', session_id = 123, silent = True,normalize = True, transformation = True, transform_target = True,
combine_rare_levels = True, rare_level_threshold = 0.05,
remove_multicollinearity = True, multicollinearity_threshold = 0.95)
compare_models(blacklist=None, whitelist=None, fold=5,sort='R2', n_select=1, turbo=True, verbose=True)
ridge = create_model('ridge')
tuned_ridge = tune_model(ridge)
final_ridge = finalize_model(tuned_ridge)
predict_model(final_ridge)
unseen_predictions1 = predict_model(final_ridge, data=new_df)
unseen_predictions1.head()
```
Using the above model, the predicted score for a student studying 9.25 hours a day is approximately 96.7549
|
github_jupyter
|
# Importing all the required libraries
import numpy as np # to perform calculations
import pandas as pd # to read data
import matplotlib.pyplot as plt # to visualise
import seaborn as sns # to visualise
url="http://bit.ly/w-data"
df=pd.read_csv(url)
df.shape
df.head(25)
df.describe()
df.info()
# Plotting the distribution of scores
plt.figure(figsize=(8,6))
sns.scatterplot(x='Hours', y='Scores', data=df)
plt.title('Hours vs Percentage', fontsize=12)
plt.xlabel('Hours Studied', fontsize=10)
plt.ylabel('Percentage Score', fontsize=10)
plt.tight_layout()
sns.pairplot(df)
sns.distplot(df['Scores'],hist_kws=dict(edgecolor="black", linewidth=1),color='Blue')
sns.distplot(df['Hours'],hist_kws=dict(edgecolor="black", linewidth=1),color='Blue')
#Displaying correlation among all the columns
df.corr()
# Setting variables
X = df.iloc[:, :-1].values
y = df.iloc[:, 1].values
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
print(X_train.shape, X_test.shape, y_train.shape, y_test.shape)
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test) # Predicting the scores
plt.figure(figsize=(8,6))
plt.scatter(X_test, y_test)
plt.plot(X_test, lr.predict(X_test), color = "r")
plt.title('Hours vs Percentage (Testing set)', fontsize=12)
plt.xlabel('Hours Studied', fontsize=10)
plt.ylabel('Percentage Score', fontsize=10)
plt.tight_layout()
#Displaying the Intercept
print(lr.intercept_)
from sklearn import metrics
print('MAE:', metrics.mean_absolute_error(y_test, y_pred))
print('MSE:', metrics.mean_squared_error(y_test, y_pred))
print('RMSE:', np.sqrt(metrics.mean_squared_error(y_test, y_pred)))
hours = 9.25
own_pred = lr.predict([[hours]])
print("No of Hours = {}".format(hours))
print("Predicted Score = {}".format(own_pred[0]))
from pycaret.regression import *
reg1 = setup(df, target = 'Scores', session_id = 123)
lr = create_model('lr')
tuned_lr = tune_model(lr)
final_lr = finalize_model(tuned_lr)
predict_model(final_lr)
new = {'Hours': ['9.25']}
new_df = pd.DataFrame(new)
unseen_predictions = predict_model(final_lr, data=new_df)
unseen_predictions.head()
# Adding more parameters and using other model other than Linear Regression
reg11 = setup(df, target = 'Scores', session_id = 123, silent = True,normalize = True, transformation = True, transform_target = True,
combine_rare_levels = True, rare_level_threshold = 0.05,
remove_multicollinearity = True, multicollinearity_threshold = 0.95)
compare_models(blacklist=None, whitelist=None, fold=5,sort='R2', n_select=1, turbo=True, verbose=True)
ridge = create_model('ridge')
tuned_ridge = tune_model(ridge)
final_ridge = finalize_model(tuned_ridge)
predict_model(final_ridge)
unseen_predictions1 = predict_model(final_ridge, data=new_df)
unseen_predictions1.head()
| 0.792263 | 0.972727 |
# Integration with Simpson's rule
In this notebook we look at a more efficient method for numerical integraiton: Simpon's rule
```
import numpy as np
import matplotlib.pyplot as plt
# The below commands make the font and image size bigger
plt.rcParams.update({'font.size': 22})
plt.rcParams["figure.figsize"] = (15,10)
```
# Illustration of Simpson's rule
Simpson's rule fits a quadratic to each strip of the integrand. The equation for the quadratic can be found by using Lagrange polynomial formula, using values of the function at the left, middle and right of the strip. The resulting formula for the integral of each strip is:
$$ I_i = \frac{\Delta x}{6} \left[f(x_i) + 4 f((x_i + x_{i+1})/2) + f(x_{i+1}) \right] $$
```
# For the arguements:
# x is the data point (or array of points) to evaluate the intepolaring polynomial at
# data is the data to be interpolated
def LagrangePoly(x, data):
n = data.shape[0] - 1
i = 1
fn = 0
while i <= n + 1:
j = 1
Li = 1
while j <= n+1:
if(j == i):
j += 1
continue
Li *= (x - data[j-1,0])/(data[i-1,0] - data[j-1,0])
j += 1
fn += data[i-1,1]*Li
i += 1
return fn
def SimpsonsIntegrate(f, a, b, N, plotMethod=False):
dx = (b-a)/N
xi = a
area = 0
i = 0
while i < N:
if(plotMethod):
x1 = xi
x2 = x1 + dx
x3 = (x1+x2)/2
f1 = f(x1)
f2 = f(x2)
f3 = f(x3)
LagrangePoly
x = np.linspace(x1, x2, 100)
y = LagrangePoly(x, np.array([[x1, f1], [x2, f2], [x3, f3]]))
plt.plot(x,y , color='red')
plt.vlines(xi, 0, f1 , color= 'red')
# The below line applies Simpson's rule
area += dx/6*(f(xi) + 4*f((2*xi+dx)/2) + f(xi+dx))
xi += dx
i += 1
if(plotMethod):
plt.vlines(b, 0, f(b) , color= 'red')
x = np.linspace(a,b,100)
y = f(x)
plt.plot(x,y);
return area
def f(x):
return np.sin(x) + 2
SimpsonsIntegrate(f, 0, 10, 10, True)
```
# A generic integrator with different methods
Below is a function that can use all 3 methods we've looked at.
```
def NIntegrate(f, a, b, N, method='Simpsons'):
dx = (b-a)/N
xi = a
i = 0
area = 0
while i < N:
if(method == 'Simpsons'):
area += dx/6*(f(xi) + 4*f((2*xi+dx)/2) + f(xi+dx))
elif(method == 'midpoint'):
area += dx * f(xi+dx/2)
elif(method == 'trapezoidal'):
area += dx/2 * (f(xi) + f(xi+dx))
xi += dx
i+= 1
return area
def f(x):
return np.sin(x)
I = 1 - np.cos(4)
print("I = %.12f" % I)
print("Midpoint: I = %.12f" % NIntegrate(f, 0, 4, 10, 'midpoint'))
print("Trapezoidal: I = %.12f" % NIntegrate(f, 0, 4, 10, 'trapezoidal'))
print("Simpson's: I = %.12f" % NIntegrate(f, 0, 4, 10, 'Simpsons'))
```
Let's now plot the convergence rate of the methods. Recall from the lectures we expect the error to scale as:
$$ \begin{align*}
\epsilon_{trap}
&\le \frac{M (b-a)}{12} \Delta x^2
& \epsilon_{mid}
&\le \frac{M (b-a)}{24} \Delta x^2
&\epsilon_{simp}
&\le \frac{M (b-a)}{180} \Delta x^4
\end{align*}
$$
```
imax = 10;
errMidpoint = np.empty((imax,2))
errTrap = np.empty((imax,2))
errSimpsons = np.empty((imax, 2))
i = 0
N = 10
while i < imax:
errMidpoint[i,0] = N
errMidpoint[i,1] = np.abs(NIntegrate(f, 0, 4, N, 'midpoint') - I)
errTrap[i, 0] = N
errTrap[i, 1] = np.abs(NIntegrate(f, 0, 4, N, 'trapezoidal') - I)
errSimpsons[i, 0] = N
errSimpsons[i, 1] = np.abs(NIntegrate(f, 0, 4, N, 'Simpsons') - I)
N *= 2
i += 1
x = np.linspace(10,1e4)
N2ref = 10*x**-2
N4ref = 10*x**-4
plt.grid(True)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('N')
plt.ylabel('error')
plt.scatter(errMidpoint[:,0], errMidpoint[:,1]);
plt.scatter(errTrap[:,0], errTrap[:,1]);
plt.scatter(errSimpsons[:,0], errSimpsons[:,1]);
plt.plot(x,N2ref, color='red')
plt.plot(x,N4ref, color='green')
plt.legend(['N^{-2} reference','N^{-4} reference','Midpoint error', 'Trapezoidal error', 'Simpsons error']);
```
Looks like the code scales as expected. This is a good test on the code.
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
# The below commands make the font and image size bigger
plt.rcParams.update({'font.size': 22})
plt.rcParams["figure.figsize"] = (15,10)
# For the arguements:
# x is the data point (or array of points) to evaluate the intepolaring polynomial at
# data is the data to be interpolated
def LagrangePoly(x, data):
n = data.shape[0] - 1
i = 1
fn = 0
while i <= n + 1:
j = 1
Li = 1
while j <= n+1:
if(j == i):
j += 1
continue
Li *= (x - data[j-1,0])/(data[i-1,0] - data[j-1,0])
j += 1
fn += data[i-1,1]*Li
i += 1
return fn
def SimpsonsIntegrate(f, a, b, N, plotMethod=False):
dx = (b-a)/N
xi = a
area = 0
i = 0
while i < N:
if(plotMethod):
x1 = xi
x2 = x1 + dx
x3 = (x1+x2)/2
f1 = f(x1)
f2 = f(x2)
f3 = f(x3)
LagrangePoly
x = np.linspace(x1, x2, 100)
y = LagrangePoly(x, np.array([[x1, f1], [x2, f2], [x3, f3]]))
plt.plot(x,y , color='red')
plt.vlines(xi, 0, f1 , color= 'red')
# The below line applies Simpson's rule
area += dx/6*(f(xi) + 4*f((2*xi+dx)/2) + f(xi+dx))
xi += dx
i += 1
if(plotMethod):
plt.vlines(b, 0, f(b) , color= 'red')
x = np.linspace(a,b,100)
y = f(x)
plt.plot(x,y);
return area
def f(x):
return np.sin(x) + 2
SimpsonsIntegrate(f, 0, 10, 10, True)
def NIntegrate(f, a, b, N, method='Simpsons'):
dx = (b-a)/N
xi = a
i = 0
area = 0
while i < N:
if(method == 'Simpsons'):
area += dx/6*(f(xi) + 4*f((2*xi+dx)/2) + f(xi+dx))
elif(method == 'midpoint'):
area += dx * f(xi+dx/2)
elif(method == 'trapezoidal'):
area += dx/2 * (f(xi) + f(xi+dx))
xi += dx
i+= 1
return area
def f(x):
return np.sin(x)
I = 1 - np.cos(4)
print("I = %.12f" % I)
print("Midpoint: I = %.12f" % NIntegrate(f, 0, 4, 10, 'midpoint'))
print("Trapezoidal: I = %.12f" % NIntegrate(f, 0, 4, 10, 'trapezoidal'))
print("Simpson's: I = %.12f" % NIntegrate(f, 0, 4, 10, 'Simpsons'))
imax = 10;
errMidpoint = np.empty((imax,2))
errTrap = np.empty((imax,2))
errSimpsons = np.empty((imax, 2))
i = 0
N = 10
while i < imax:
errMidpoint[i,0] = N
errMidpoint[i,1] = np.abs(NIntegrate(f, 0, 4, N, 'midpoint') - I)
errTrap[i, 0] = N
errTrap[i, 1] = np.abs(NIntegrate(f, 0, 4, N, 'trapezoidal') - I)
errSimpsons[i, 0] = N
errSimpsons[i, 1] = np.abs(NIntegrate(f, 0, 4, N, 'Simpsons') - I)
N *= 2
i += 1
x = np.linspace(10,1e4)
N2ref = 10*x**-2
N4ref = 10*x**-4
plt.grid(True)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('N')
plt.ylabel('error')
plt.scatter(errMidpoint[:,0], errMidpoint[:,1]);
plt.scatter(errTrap[:,0], errTrap[:,1]);
plt.scatter(errSimpsons[:,0], errSimpsons[:,1]);
plt.plot(x,N2ref, color='red')
plt.plot(x,N4ref, color='green')
plt.legend(['N^{-2} reference','N^{-4} reference','Midpoint error', 'Trapezoidal error', 'Simpsons error']);
| 0.313 | 0.980673 |
# Regression data using scikit-learn
Regression is when the feature to be predicted contains continuous values. Regression refers to the process of predicting a dependent variable by analyzing the relationship between other independent variables. There are several algorithms known to us that help us in excavating these relationships to better predict the value.
In this notebook, we'll use scikit-learn to predict values. Scikit-learn provides implementations of many regression algorithms. In here, we have done a comparative study of 5 different regression algorithms.
To help visualize what we are doing, we'll use 2D and 3D charts to show how the classes looks (with 3 selected dimensions) with matplotlib and seaborn python libraries.
<a id="top"></a>
## Table of Contents
1. [Load libraries](#load_libraries)
2. [Helper methods for metrics](#helper_methods)
3. [Data exploration](#explore_data)
4. [Prepare data for building regression model](#prepare_data)
5. [Build Simple Linear Regression model](#model_slr)
6. [Build Multiple Linear Regression classification model](#model_mlr)
7. [Build Polynomial Linear Regression model](#model_plr)
8. [Build Decision Tree Regression model](#model_dtr)
9. [Build Random Forest Regression model](#model_rfr)
10. [Comparitive study of different regression algorithms](#compare_classification)
### Quick set of instructions to work through the notebook
If you are new to Notebooks, here's a quick overview of how to work in this environment.
1. The notebook has 2 types of cells - markdown (text) such as this and code such as the one below.
2. Each cell with code can be executed independently or together (see options under the Cell menu). When working in this notebook, we will be running one cell at a time because we need to make code changes to some of the cells.
3. To run the cell, position cursor in the code cell and click the Run (arrow) icon. The cell is running when you see the * next to it. Some cells have printable output.
4. Work through this notebook by reading the instructions and executing code cell by cell. Some cells will require modifications before you run them.
<a id="load_libraries"></a>
## 1. Load libraries
[Top](#top)
Install python modules
NOTE! Some pip installs require a kernel restart.
The shell command pip install is used to install Python modules. Some installs require a kernel restart to complete. To avoid confusing errors, run the following cell once and then use the Kernel menu to restart the kernel before proceeding.
```
!pip install pandas==0.24.2
!pip install --user pandas_ml==0.6.1
#downgrade matplotlib to bypass issue with confusion matrix being chopped out
!pip install matplotlib==3.1.0
!pip install seaborn
!pip install pydot
!pip install graphviz
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import StandardScaler
from sklearn.compose import ColumnTransformer, make_column_transformer
from sklearn.pipeline import Pipeline
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score,mean_squared_error, r2_score
import pandas as pd, numpy as np
import sys
import io
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import seaborn as sns
from sklearn.tree import export_graphviz
import pydot
```
<a id="helper_methods"></a>
## 2. Helper methods for metrics
[Top](#top)
```
def two_d_compare(X_test,y_test,y_pred,model_name):
area = (12 * np.random.rand(40))**2
plt.subplots(ncols=2, figsize=(10,4))
plt.suptitle('Actual vs Predicted data : ' +model_name + '. Variance score: %.2f' % r2_score(y_test, y_pred))
plt.subplot(121)
plt.scatter(X_test, y_test, alpha=0.8, color='#8CCB9B')
plt.title('Actual')
plt.subplot(122)
plt.scatter(X_test, y_pred,alpha=0.8, color='#E5E88B')
plt.title('Predicted')
plt.show()
def model_metrics(regressor,y_test,y_pred):
mse = mean_squared_error(y_test,y_pred)
print("Mean squared error: %.2f"
% mse)
r2 = r2_score(y_test, y_pred)
print('R2 score: %.2f' % r2 )
return [mse, r2]
def two_vs_three(x_test,y_test,y_pred,z=None, isLinear = False) :
area = 60
fig = plt.figure(figsize=(12,6))
fig.suptitle('2D and 3D view of sales price data')
# First subplot
ax = fig.add_subplot(1, 2,1)
ax.scatter(x_test, y_test, alpha=0.5,color='blue', s= area)
ax.plot(x_test, y_pred, alpha=0.9,color='red', linewidth=2)
ax.set_xlabel('YEAR BUILT')
ax.set_ylabel('SELLING PRICE')
plt.title('YEARBUILT vs SALEPRICE')
if not isLinear :
# Second subplot
ax = fig.add_subplot(1,2,2, projection='3d')
ax.scatter(z, x_test, y_test, color='blue', marker='o')
ax.plot(z, x_test, y_pred, alpha=0.9,color='red', linewidth=2)
ax.set_ylabel('YEAR BUILT')
ax.set_zlabel('SELLING PRICE')
ax.set_xlabel('LOT AREA')
plt.title('LOT AREA vs YEAR BUILT vs SELLING PRICE')
plt.show()
```
<a id="explore_data"></a>
## 3. Data exploration
[Top](#top)
Data can be easily loaded within IBM Watson Studio. Instructions to load data within IBM Watson Studio can be found [here](https://ibmdev1.rtp.raleigh.ibm.com/tutorials/watson-studio-using-jupyter-notebook/). The data set can be located by its name and inserted into the notebook as a pandas DataFrame as shown below.
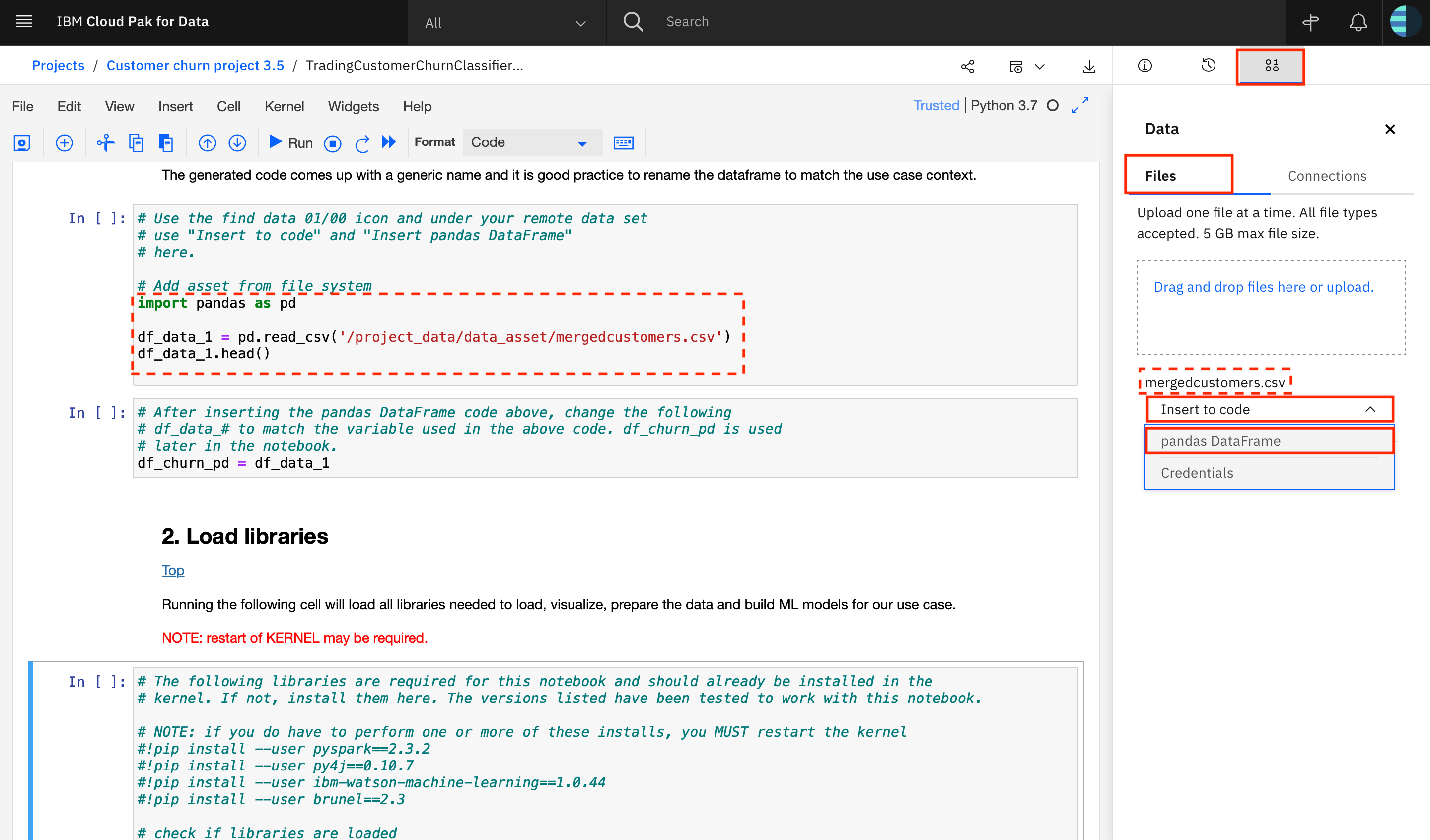
The generated code comes up with a generic name and it is good practice to rename the dataframe to match the use case context.
In the snippet below, we use the pandas library to load a csv that contains housing related information. With several independent variables related to this domain, we are going to predict the sales price of a house.
```
df_pd = pd.read_csv("https://raw.githubusercontent.com/IBM/ml-learning-path-assets/master/data/predict_home_value.csv")
df_pd.head()
area = 60
x = df_pd['YEARBUILT']
y = df_pd['SALEPRICE']
z = df_pd['LOTAREA']
fig = plt.figure(figsize=(12,6))
fig.suptitle('2D and 3D view of sales price data')
# First subplot
ax = fig.add_subplot(1, 2,1)
ax.scatter(x, y, alpha=0.5,color='blue', s= area)
ax.set_xlabel('YEAR BUILT')
ax.set_ylabel('SELLING PRICE')
plt.title('YEARBUILT vs SALEPRICE')
# Second subplot
ax = fig.add_subplot(1,2,2, projection='3d')
ax.scatter(z, x, y, color='blue', marker='o')
ax.set_ylabel('YEAR BUILT')
ax.set_zlabel('SELLING PRICE')
ax.set_xlabel('LOT AREA')
plt.title('LOT AREA VS YEAR BUILT vs SELLING PRICE')
plt.show()
sns.set(rc={"figure.figsize": (8, 4)}); np.random.seed(0)
ax = sns.distplot(df_pd['SALEPRICE'])
plt.show()
print("The dataset contains columns of the following data types : \n" +str(df_pd.dtypes))
```
Notice below that FIREPLACEQU, GARAGETYPE, GARAGEFINISH, GARAGECOND,FENCE and POOLQC have missing values.
```
print("The dataset contains following number of records for each of the columns : \n" +str(df_pd.count()))
df_pd.isnull().any()
```
<a id="prepare_data"></a>
## 4. Data preparation
[Top](#top)
Data preparation is a very important step in machine learning model building. This is because the model can perform well only when the data it is trained on is good and well prepared. Hence, this step consumes bulk of data scientist's time spent building models.
During this process, we identify categorical columns in the dataset. Categories needed to be indexed, which means the string labels are converted to label indices. These label indices are encoded using One-hot encoding to a binary vector with at most a single one-value indicating the presence of a specific feature value from among the set of all feature values. This encoding allows algorithms which expect continuous features to use categorical features.
```
#remove columns that are not required
df_pd = df_pd.drop(['ID'], axis=1)
df_pd.head()
# Defining the categorical columns
categoricalColumns = df_pd.select_dtypes(include=[np.object]).columns
print("Categorical columns : " )
print(categoricalColumns)
impute_categorical = SimpleImputer(strategy="most_frequent")
onehot_categorical = OneHotEncoder(handle_unknown='ignore')
categorical_transformer = Pipeline(steps=[('impute',impute_categorical),('onehot',onehot_categorical)])
# Defining the numerical columns
numericalColumns = [col for col in df_pd.select_dtypes(include=[np.float,np.int]).columns if col not in ['SALEPRICE']]
print("Numerical columns : " )
print(numericalColumns)
scaler_numerical = StandardScaler()
numerical_transformer = Pipeline(steps=[('scale',scaler_numerical)])
preprocessorForCategoricalColumns = ColumnTransformer(transformers=[('cat', categorical_transformer, categoricalColumns)],
remainder="passthrough")
preprocessorForAllColumns = ColumnTransformer(transformers=[('cat', categorical_transformer, categoricalColumns),('num',numerical_transformer,numericalColumns)],
remainder="passthrough")
#. The transformation happens in the pipeline. Temporarily done here to show what intermediate value looks like
df_pd_temp = preprocessorForCategoricalColumns.fit_transform(df_pd)
print("Data after transforming :")
print(df_pd_temp)
df_pd_temp_2 = preprocessorForAllColumns.fit_transform(df_pd)
print("Data after transforming :")
print(df_pd_temp_2)
# prepare data frame for splitting data into train and test datasets
features = []
features = df_pd.drop(['SALEPRICE'], axis=1)
label = pd.DataFrame(df_pd, columns = ['SALEPRICE'])
#label_encoder = LabelEncoder()
label = df_pd['SALEPRICE']
#label = label_encoder.fit_transform(label)
print(" value of label : " + str(label))
```
<a id="model_slr"></a>
## 5. Simple linear regression
[Top](#top)
This is the most basic form of linear regression in which the variable to be predicted is dependent on only one other variable. This is calculated by using the formula that is generally used in calculating the slope of a line.
y = w0 + w1*x1
In the above equation, y refers to the target variable and x1 refers to the independent variable. w1 refers to the coeeficient that expresses the relationship between y and x1 is it also know as the slope. w0 is the constant cooefficient a.k.a the intercept. It refers to the constant offset that y will always be with respect to the independent variables.
Since simple linear regression assumes that output depends on only one variable, we are assuming that it depends on the YEARBUILT. Data is split up into training and test sets.
```
X = features['YEARBUILT'].values.reshape(-1,1)
X_train_slr, X_test_slr, y_train_slr, y_test_slr = train_test_split(X,label , random_state=0)
print("Dimensions of datasets that will be used for training : Input features"+str(X_train_slr.shape)+
" Output label" + str(y_train_slr.shape))
print("Dimensions of datasets that will be used for testing : Input features"+str(X_test_slr.shape)+
" Output label" + str(y_test_slr.shape))
from sklearn.linear_model import LinearRegression
model_name = 'Simple Linear Regression'
slRegressor = LinearRegression()
slRegressor.fit(X_train_slr,y_train_slr)
y_pred_slr= slRegressor.predict(X_test_slr)
print(slRegressor)
print('Intercept: \n',slRegressor.intercept_)
print('Coefficients: \n', slRegressor.coef_)
two_vs_three(X_test_slr[:,0],y_test_slr,y_pred_slr,None, True)
two_d_compare(X_test_slr,y_test_slr,y_pred_slr,model_name)
slrMetrics = model_metrics(slRegressor,y_test_slr,y_pred_slr)
```
<a id="model_lrc"></a>
## 6. Build multiple linear regression model
[Top](#top)
Multiple linear regression is an extension to the simple linear regression. In this setup, the target value is dependant on more than one variable. The number of variables depends on the use case at hand. Usually a subject matter expert is involved in identifying the fields that will contribute towards better predicting the output feature.
y = w0 + w1*x1 + w2*x2 + .... + wn*xn
Since multiple linear regression assumes that output depends on more than one variable, we are assuming that it depends on all the 30 features. Data is split up into training and test sets. As an experiment, you can try to remove a few features and check if the model performs any better.
```
X_train, X_test, y_train, y_test = train_test_split(features,label , random_state=0)
print("Dimensions of datasets that will be used for training : Input features"+str(X_train.shape)+
" Output label" + str(y_train.shape))
print("Dimensions of datasets that will be used for testing : Input features"+str(X_test.shape)+
" Output label" + str(y_test.shape))
from sklearn.linear_model import LinearRegression
model_name = 'Multiple Linear Regression'
mlRegressor = LinearRegression()
mlr_model = Pipeline(steps=[('preprocessorAll',preprocessorForAllColumns),('regressor', mlRegressor)])
mlr_model.fit(X_train,y_train)
y_pred_mlr= mlr_model.predict(X_test)
print(mlRegressor)
print('Intercept: \n',mlRegressor.intercept_)
print('Coefficients: \n', mlRegressor.coef_)
two_vs_three(X_test['YEARBUILT'],y_test,y_pred_mlr,X_test['LOTAREA'], False)
two_d_compare(X_test['YEARBUILT'],y_test,y_pred_mlr,model_name)
mlrMetrics = model_metrics(slRegressor,y_test,y_pred_mlr)
```
<a id="model_plr"></a>
## 7. Build Polynomial Linear regression model
[Top](#top)
The prediction line generated by simple/linear regression is usually a straight line. In cases when a simple or multiple linear regression does not fit the data point accurately, we use the polynomial linear regression. The following formula is used in the back-end to generate polynomial linear regression.
y = w0 + w1*x1 + w2*x21 + .... + wn*xnn
We are assuming that output depends on the YEARBUILT and LOTATREA. Data is split up into training and test sets.
```
X = features.iloc[:, [0,4]].values
X_train, X_test, y_train, y_test = train_test_split(X,label, random_state=0)
print("Dimensions of datasets that will be used for training : Input features"+str(X_train.shape)+
" Output label" + str(y_train.shape))
print("Dimensions of datasets that will be used for testing : Input features"+str(X_test.shape)+
" Output label" + str(y_test.shape))
from sklearn.linear_model import LinearRegression
from sklearn.preprocessing import PolynomialFeatures
model_name = 'Polynomial Linear Regression'
polynomial_features= PolynomialFeatures(degree=3)
plRegressor = LinearRegression()
plr_model = Pipeline(steps=[('polyFeature',polynomial_features ),('regressor', plRegressor)])
plr_model.fit(X_train,y_train)
y_pred_plr= plr_model.predict(X_test)
print(plRegressor)
print('Intercept: \n',plRegressor.intercept_)
print('Coefficients: \n', plRegressor.coef_)
two_vs_three(X_test[:,1],y_test,y_pred_plr,X_test[:,0], False)
two_d_compare(X_test[:,1],y_test,y_pred_plr,model_name)
plrMetrics = model_metrics(plRegressor,y_test,y_pred_plr)
```
<a id="model_dtr"></a>
## 8. Build decision tree regressor
[Top](#top)
```
X_train, X_test, y_train, y_test = train_test_split(features,df_pd['SALEPRICE'] , random_state=0)
print("Dimensions of datasets that will be used for training : Input features"+str(X_train.shape)+
" Output label" + str(y_train.shape))
print("Dimensions of datasets that will be used for testing : Input features"+str(X_test.shape)+
" Output label" + str(y_test.shape))
from sklearn.tree import DecisionTreeRegressor
model_name = "Decision Tree Regressor"
decisionTreeRegressor = DecisionTreeRegressor(random_state=0,max_features=30)
dtr_model = Pipeline(steps=[('preprocessorAll',preprocessorForAllColumns),('regressor', decisionTreeRegressor)])
dtr_model.fit(X_train,y_train)
y_pred_dtr = dtr_model.predict(X_test)
print(decisionTreeRegressor)
export_graphviz(decisionTreeRegressor, out_file ='tree.dot')
# Use dot file to create a graph
(graph, ) = pydot.graph_from_dot_file('tree.dot')
# Write graph to a png file
graph.write_png('tree.png')
two_d_compare(X_test['YEARBUILT'],y_test,y_pred_dtr,model_name)
dtrMetrics = model_metrics(decisionTreeRegressor,y_test,y_pred_dtr)
```
<a id="model_rfr"></a>
## 9. Build Random Forest classification model
[Top](#top)
Decision tree algorithms are efficient in eliminating columns that don't add value in predicting the output and in some cases, we are even able to see how a prediction was derived by backtracking the tree. However, this algorithm doesn't perform individually when the trees are huge and are hard to interpret. Such models are oftern referred to as weak models. The model performance is however improvised by taking an average of several such decision trees derived from the subsets of the training data. This approach is called the Random Forest Regression.
```
from sklearn.ensemble import RandomForestRegressor
model_name = "Random Forest Regressor"
randomForestRegressor = RandomForestRegressor(n_estimators=100, max_depth=15,random_state=0)
rfr_model = Pipeline(steps=[('preprocessorAll',preprocessorForAllColumns),('regressor', randomForestRegressor)])
rfr_model.fit(X_train,y_train)
y_pred_rfr = rfr_model.predict(X_test)
two_d_compare(X_test['YEARBUILT'],y_test,y_pred_rfr,model_name)
rfrMetrics = model_metrics(randomForestRegressor,y_test,y_pred_rfr)
```
<a id="compare_classification"></a>
## 10. Comparative study of different regression algorithms.
[Top](#top)
In the bar chart below, we have compared the performances of different regression algorithms with each other.
```
n_groups = 1
index = np.arange(n_groups)
bar_width = 1
opacity = 0.8
area = 60
plt.subplots(ncols=2, figsize=(12,9))
plt.suptitle('Model performance comparison')
plt.subplot(121)
index = np.arange(n_groups)
bar_width = 1
opacity = 0.8
rects1 = plt.bar(index, slrMetrics[0], bar_width,
alpha=opacity,
color='g',
label='Simple Linear Regression')
rects2 = plt.bar(index + bar_width, mlrMetrics[0], bar_width,
alpha=opacity,
color='pink',
label='Multiple Linear Regression')
rects3 = plt.bar(index + bar_width*2, plrMetrics[0], bar_width,
alpha=opacity,
color='y',
label='Polynomial Linear Regression')
rects4 = plt.bar(index + bar_width*3, dtrMetrics[0], bar_width,
alpha=opacity,
color='b',
label='Decision Tree Regression')
rects6 = plt.bar(index + bar_width*4, rfrMetrics[0], bar_width,
alpha=opacity,
color='purple',
label='Random Forest Regression')
plt.xlabel('Models')
plt.ylabel('MSE')
plt.title('Mean Square Error comparison.')
#ax.set_xticklabels(('', 'Simple Lin', 'Multiple Lin', 'Polynomial Lin', 'Decision Tree','Random Forest'))
plt.subplot(122)
rects1 = plt.bar(index, slrMetrics[1], bar_width,
alpha=opacity,
color='g',
label='Simple Linear Regression')
rects2 = plt.bar(index + bar_width, mlrMetrics[1], bar_width,
alpha=opacity,
color='pink',
label='Multiple Linear Regression')
rects3 = plt.bar(index + bar_width*2, plrMetrics[1], bar_width,
alpha=opacity,
color='y',
label='Polynomial Linear Regression')
rects4 = plt.bar(index + bar_width*3, dtrMetrics[1], bar_width,
alpha=opacity,
color='b',
label='Decision Tree Regression')
rects6 = plt.bar(index + bar_width*4, rfrMetrics[1], bar_width,
alpha=opacity,
color='purple',
label='Random Forest Regression')
plt.xlabel('Models')
plt.ylabel('R2')
plt.title('R2 comparison.')
ax.set_xticklabels(('', 'Simple Lin', 'Multiple Lin', 'Polynomial Lin', 'Decision Tree','Random Forest'))
plt.legend()
plt.show()
```
<p><font size=-1 color=gray>
© Copyright 2019 IBM Corp. All Rights Reserved.
<p>
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file
except in compliance with the License. You may obtain a copy of the License at
https://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the
License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either
express or implied. See the License for the specific language governing permissions and
limitations under the License.
</font></p>
|
github_jupyter
|
!pip install pandas==0.24.2
!pip install --user pandas_ml==0.6.1
#downgrade matplotlib to bypass issue with confusion matrix being chopped out
!pip install matplotlib==3.1.0
!pip install seaborn
!pip install pydot
!pip install graphviz
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import StandardScaler
from sklearn.compose import ColumnTransformer, make_column_transformer
from sklearn.pipeline import Pipeline
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score,mean_squared_error, r2_score
import pandas as pd, numpy as np
import sys
import io
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import seaborn as sns
from sklearn.tree import export_graphviz
import pydot
def two_d_compare(X_test,y_test,y_pred,model_name):
area = (12 * np.random.rand(40))**2
plt.subplots(ncols=2, figsize=(10,4))
plt.suptitle('Actual vs Predicted data : ' +model_name + '. Variance score: %.2f' % r2_score(y_test, y_pred))
plt.subplot(121)
plt.scatter(X_test, y_test, alpha=0.8, color='#8CCB9B')
plt.title('Actual')
plt.subplot(122)
plt.scatter(X_test, y_pred,alpha=0.8, color='#E5E88B')
plt.title('Predicted')
plt.show()
def model_metrics(regressor,y_test,y_pred):
mse = mean_squared_error(y_test,y_pred)
print("Mean squared error: %.2f"
% mse)
r2 = r2_score(y_test, y_pred)
print('R2 score: %.2f' % r2 )
return [mse, r2]
def two_vs_three(x_test,y_test,y_pred,z=None, isLinear = False) :
area = 60
fig = plt.figure(figsize=(12,6))
fig.suptitle('2D and 3D view of sales price data')
# First subplot
ax = fig.add_subplot(1, 2,1)
ax.scatter(x_test, y_test, alpha=0.5,color='blue', s= area)
ax.plot(x_test, y_pred, alpha=0.9,color='red', linewidth=2)
ax.set_xlabel('YEAR BUILT')
ax.set_ylabel('SELLING PRICE')
plt.title('YEARBUILT vs SALEPRICE')
if not isLinear :
# Second subplot
ax = fig.add_subplot(1,2,2, projection='3d')
ax.scatter(z, x_test, y_test, color='blue', marker='o')
ax.plot(z, x_test, y_pred, alpha=0.9,color='red', linewidth=2)
ax.set_ylabel('YEAR BUILT')
ax.set_zlabel('SELLING PRICE')
ax.set_xlabel('LOT AREA')
plt.title('LOT AREA vs YEAR BUILT vs SELLING PRICE')
plt.show()
df_pd = pd.read_csv("https://raw.githubusercontent.com/IBM/ml-learning-path-assets/master/data/predict_home_value.csv")
df_pd.head()
area = 60
x = df_pd['YEARBUILT']
y = df_pd['SALEPRICE']
z = df_pd['LOTAREA']
fig = plt.figure(figsize=(12,6))
fig.suptitle('2D and 3D view of sales price data')
# First subplot
ax = fig.add_subplot(1, 2,1)
ax.scatter(x, y, alpha=0.5,color='blue', s= area)
ax.set_xlabel('YEAR BUILT')
ax.set_ylabel('SELLING PRICE')
plt.title('YEARBUILT vs SALEPRICE')
# Second subplot
ax = fig.add_subplot(1,2,2, projection='3d')
ax.scatter(z, x, y, color='blue', marker='o')
ax.set_ylabel('YEAR BUILT')
ax.set_zlabel('SELLING PRICE')
ax.set_xlabel('LOT AREA')
plt.title('LOT AREA VS YEAR BUILT vs SELLING PRICE')
plt.show()
sns.set(rc={"figure.figsize": (8, 4)}); np.random.seed(0)
ax = sns.distplot(df_pd['SALEPRICE'])
plt.show()
print("The dataset contains columns of the following data types : \n" +str(df_pd.dtypes))
print("The dataset contains following number of records for each of the columns : \n" +str(df_pd.count()))
df_pd.isnull().any()
#remove columns that are not required
df_pd = df_pd.drop(['ID'], axis=1)
df_pd.head()
# Defining the categorical columns
categoricalColumns = df_pd.select_dtypes(include=[np.object]).columns
print("Categorical columns : " )
print(categoricalColumns)
impute_categorical = SimpleImputer(strategy="most_frequent")
onehot_categorical = OneHotEncoder(handle_unknown='ignore')
categorical_transformer = Pipeline(steps=[('impute',impute_categorical),('onehot',onehot_categorical)])
# Defining the numerical columns
numericalColumns = [col for col in df_pd.select_dtypes(include=[np.float,np.int]).columns if col not in ['SALEPRICE']]
print("Numerical columns : " )
print(numericalColumns)
scaler_numerical = StandardScaler()
numerical_transformer = Pipeline(steps=[('scale',scaler_numerical)])
preprocessorForCategoricalColumns = ColumnTransformer(transformers=[('cat', categorical_transformer, categoricalColumns)],
remainder="passthrough")
preprocessorForAllColumns = ColumnTransformer(transformers=[('cat', categorical_transformer, categoricalColumns),('num',numerical_transformer,numericalColumns)],
remainder="passthrough")
#. The transformation happens in the pipeline. Temporarily done here to show what intermediate value looks like
df_pd_temp = preprocessorForCategoricalColumns.fit_transform(df_pd)
print("Data after transforming :")
print(df_pd_temp)
df_pd_temp_2 = preprocessorForAllColumns.fit_transform(df_pd)
print("Data after transforming :")
print(df_pd_temp_2)
# prepare data frame for splitting data into train and test datasets
features = []
features = df_pd.drop(['SALEPRICE'], axis=1)
label = pd.DataFrame(df_pd, columns = ['SALEPRICE'])
#label_encoder = LabelEncoder()
label = df_pd['SALEPRICE']
#label = label_encoder.fit_transform(label)
print(" value of label : " + str(label))
X = features['YEARBUILT'].values.reshape(-1,1)
X_train_slr, X_test_slr, y_train_slr, y_test_slr = train_test_split(X,label , random_state=0)
print("Dimensions of datasets that will be used for training : Input features"+str(X_train_slr.shape)+
" Output label" + str(y_train_slr.shape))
print("Dimensions of datasets that will be used for testing : Input features"+str(X_test_slr.shape)+
" Output label" + str(y_test_slr.shape))
from sklearn.linear_model import LinearRegression
model_name = 'Simple Linear Regression'
slRegressor = LinearRegression()
slRegressor.fit(X_train_slr,y_train_slr)
y_pred_slr= slRegressor.predict(X_test_slr)
print(slRegressor)
print('Intercept: \n',slRegressor.intercept_)
print('Coefficients: \n', slRegressor.coef_)
two_vs_three(X_test_slr[:,0],y_test_slr,y_pred_slr,None, True)
two_d_compare(X_test_slr,y_test_slr,y_pred_slr,model_name)
slrMetrics = model_metrics(slRegressor,y_test_slr,y_pred_slr)
X_train, X_test, y_train, y_test = train_test_split(features,label , random_state=0)
print("Dimensions of datasets that will be used for training : Input features"+str(X_train.shape)+
" Output label" + str(y_train.shape))
print("Dimensions of datasets that will be used for testing : Input features"+str(X_test.shape)+
" Output label" + str(y_test.shape))
from sklearn.linear_model import LinearRegression
model_name = 'Multiple Linear Regression'
mlRegressor = LinearRegression()
mlr_model = Pipeline(steps=[('preprocessorAll',preprocessorForAllColumns),('regressor', mlRegressor)])
mlr_model.fit(X_train,y_train)
y_pred_mlr= mlr_model.predict(X_test)
print(mlRegressor)
print('Intercept: \n',mlRegressor.intercept_)
print('Coefficients: \n', mlRegressor.coef_)
two_vs_three(X_test['YEARBUILT'],y_test,y_pred_mlr,X_test['LOTAREA'], False)
two_d_compare(X_test['YEARBUILT'],y_test,y_pred_mlr,model_name)
mlrMetrics = model_metrics(slRegressor,y_test,y_pred_mlr)
X = features.iloc[:, [0,4]].values
X_train, X_test, y_train, y_test = train_test_split(X,label, random_state=0)
print("Dimensions of datasets that will be used for training : Input features"+str(X_train.shape)+
" Output label" + str(y_train.shape))
print("Dimensions of datasets that will be used for testing : Input features"+str(X_test.shape)+
" Output label" + str(y_test.shape))
from sklearn.linear_model import LinearRegression
from sklearn.preprocessing import PolynomialFeatures
model_name = 'Polynomial Linear Regression'
polynomial_features= PolynomialFeatures(degree=3)
plRegressor = LinearRegression()
plr_model = Pipeline(steps=[('polyFeature',polynomial_features ),('regressor', plRegressor)])
plr_model.fit(X_train,y_train)
y_pred_plr= plr_model.predict(X_test)
print(plRegressor)
print('Intercept: \n',plRegressor.intercept_)
print('Coefficients: \n', plRegressor.coef_)
two_vs_three(X_test[:,1],y_test,y_pred_plr,X_test[:,0], False)
two_d_compare(X_test[:,1],y_test,y_pred_plr,model_name)
plrMetrics = model_metrics(plRegressor,y_test,y_pred_plr)
X_train, X_test, y_train, y_test = train_test_split(features,df_pd['SALEPRICE'] , random_state=0)
print("Dimensions of datasets that will be used for training : Input features"+str(X_train.shape)+
" Output label" + str(y_train.shape))
print("Dimensions of datasets that will be used for testing : Input features"+str(X_test.shape)+
" Output label" + str(y_test.shape))
from sklearn.tree import DecisionTreeRegressor
model_name = "Decision Tree Regressor"
decisionTreeRegressor = DecisionTreeRegressor(random_state=0,max_features=30)
dtr_model = Pipeline(steps=[('preprocessorAll',preprocessorForAllColumns),('regressor', decisionTreeRegressor)])
dtr_model.fit(X_train,y_train)
y_pred_dtr = dtr_model.predict(X_test)
print(decisionTreeRegressor)
export_graphviz(decisionTreeRegressor, out_file ='tree.dot')
# Use dot file to create a graph
(graph, ) = pydot.graph_from_dot_file('tree.dot')
# Write graph to a png file
graph.write_png('tree.png')
two_d_compare(X_test['YEARBUILT'],y_test,y_pred_dtr,model_name)
dtrMetrics = model_metrics(decisionTreeRegressor,y_test,y_pred_dtr)
from sklearn.ensemble import RandomForestRegressor
model_name = "Random Forest Regressor"
randomForestRegressor = RandomForestRegressor(n_estimators=100, max_depth=15,random_state=0)
rfr_model = Pipeline(steps=[('preprocessorAll',preprocessorForAllColumns),('regressor', randomForestRegressor)])
rfr_model.fit(X_train,y_train)
y_pred_rfr = rfr_model.predict(X_test)
two_d_compare(X_test['YEARBUILT'],y_test,y_pred_rfr,model_name)
rfrMetrics = model_metrics(randomForestRegressor,y_test,y_pred_rfr)
n_groups = 1
index = np.arange(n_groups)
bar_width = 1
opacity = 0.8
area = 60
plt.subplots(ncols=2, figsize=(12,9))
plt.suptitle('Model performance comparison')
plt.subplot(121)
index = np.arange(n_groups)
bar_width = 1
opacity = 0.8
rects1 = plt.bar(index, slrMetrics[0], bar_width,
alpha=opacity,
color='g',
label='Simple Linear Regression')
rects2 = plt.bar(index + bar_width, mlrMetrics[0], bar_width,
alpha=opacity,
color='pink',
label='Multiple Linear Regression')
rects3 = plt.bar(index + bar_width*2, plrMetrics[0], bar_width,
alpha=opacity,
color='y',
label='Polynomial Linear Regression')
rects4 = plt.bar(index + bar_width*3, dtrMetrics[0], bar_width,
alpha=opacity,
color='b',
label='Decision Tree Regression')
rects6 = plt.bar(index + bar_width*4, rfrMetrics[0], bar_width,
alpha=opacity,
color='purple',
label='Random Forest Regression')
plt.xlabel('Models')
plt.ylabel('MSE')
plt.title('Mean Square Error comparison.')
#ax.set_xticklabels(('', 'Simple Lin', 'Multiple Lin', 'Polynomial Lin', 'Decision Tree','Random Forest'))
plt.subplot(122)
rects1 = plt.bar(index, slrMetrics[1], bar_width,
alpha=opacity,
color='g',
label='Simple Linear Regression')
rects2 = plt.bar(index + bar_width, mlrMetrics[1], bar_width,
alpha=opacity,
color='pink',
label='Multiple Linear Regression')
rects3 = plt.bar(index + bar_width*2, plrMetrics[1], bar_width,
alpha=opacity,
color='y',
label='Polynomial Linear Regression')
rects4 = plt.bar(index + bar_width*3, dtrMetrics[1], bar_width,
alpha=opacity,
color='b',
label='Decision Tree Regression')
rects6 = plt.bar(index + bar_width*4, rfrMetrics[1], bar_width,
alpha=opacity,
color='purple',
label='Random Forest Regression')
plt.xlabel('Models')
plt.ylabel('R2')
plt.title('R2 comparison.')
ax.set_xticklabels(('', 'Simple Lin', 'Multiple Lin', 'Polynomial Lin', 'Decision Tree','Random Forest'))
plt.legend()
plt.show()
| 0.55254 | 0.981666 |
# Image Processing -
> In this post, we will cover the basics of working with images in Matplotlib, OpenCV and Keras.
- toc: true
- badges: true
- comments: true
- categories: [Image Processing, Computer Vision]
- image: images/freedom.png
```
import glob
from PIL import Image
import glob
import cv2
import numpy as np
import matplotlib.pyplot as plt
import os
# filepaths
fp_in_CFD = sorted(glob.glob("/home/imagda/sims-projs/CFD-DeepLearning-UNET/plots/valid_data/plots/montage_car_CFD_*.*"))
fp_in_ML = sorted(glob.glob("/home/imagda/sims-projs/CFD-DeepLearning-UNET/plots/valid_data/plots/montage_car_ML_*.*"))
counter = 0
for fp_img1, fp_img2 in zip(fp_in_CFD, fp_in_ML):
counter += 1
img1 = cv2.imread(fp_img1, -1)
img2 = cv2.imread(fp_img2, -1) # this one has transparency
img1_crop = img1[183:400, 169:1300]
img2_crop = img2[183:400, 169:1300]
numpy_vertical_concat = np.concatenate((img1_crop, img2_crop), axis=0)
#numpy_horizontal_concat = np.concatenate((img1_crop, img2_crop), axis=1)
filename = "/home/imagda/sims-projs/CFD-DeepLearning-UNET/plots/valid_data/plots/CFD+ML_"+"{:02d}".format(counter) +".png"
#file = open('{}{}'.format(name, x) , "w+")
plt.savefig(filename, dpi = 200)
plt.imshow( numpy_vertical_concat)
counter = 0
for fp_img1, fp_img2 in zip(fp_in_CFD, fp_in_ML):
counter += 1
img1 = cv2.imread(fp_img1, -1)
img2 = cv2.imread(fp_img2, -1) # this one has transparency
img1_crop = img1[183:400, 0:200]
img2_crop = img2[183:400, 800:1000]
#numpy_vertical_concat = np.concatenate((img1_crop, img2_crop), axis=0)
numpy_horizontal_concat = img1_crop + img2_crop
filename = "/home/imagda/sims-projs/CFD-DeepLearning-UNET/plots/valid_data/plots/CFD+ML_"+"{:02d}".format(counter) +".png"
#file = open('{}{}'.format(name, x) , "w+")
plt.savefig(filename, dpi = 200)
plt.imshow( numpy_horizontal_concat)
img1 = cv2.imread(fp_img1)
img1.shape
type(img1)
import cv2
from skimage import io
lion_rgb = cv2.imread('https://i.stack.imgur.com/R6X5p.jpg')
lion_gray = cv2.imread('https://i.stack.imgur.com/f27t5.png')
# Read Image1
#mountain = cv2.imread(fp_img1, 1)
# Read image2
#dog = cv2.imread(fp_img2, 1)
# Blending the images with 0.3 and 0.7
img = cv2.addWeighted(lion_rgb, 1, lion_rgb, 1, 0)
plt.imshow(img/266.)
#io.imshow(img)
# Show the image
#cv2.imshow('image', img)
# Wait for a key
#cv2.waitKey(0)
# Distroy all the window open
#cv2.distroyAllWindows()
rgb.shape
import numpy as np
from skimage import io
rgb = io.imread('https://i.stack.imgur.com/R6X5p.jpg')
gray = io.imread('https://i.stack.imgur.com/f27t5.png')
rows_rgb, cols_rgb, channels = rgb.shape
rows_gray, cols_gray = gray.shape
rows_comb = max(rows_rgb, rows_gray)
cols_comb = cols_rgb + cols_gray
comb = np.zeros(shape=(rows_comb, cols_comb, channels), dtype=np.uint8)
comb[:rows_rgb, :cols_rgb] = rgb
comb[:rows_gray, cols_rgb:] = gray[:, :, None]
io.imshow(comb)
rgb.shape
```
|
github_jupyter
|
import glob
from PIL import Image
import glob
import cv2
import numpy as np
import matplotlib.pyplot as plt
import os
# filepaths
fp_in_CFD = sorted(glob.glob("/home/imagda/sims-projs/CFD-DeepLearning-UNET/plots/valid_data/plots/montage_car_CFD_*.*"))
fp_in_ML = sorted(glob.glob("/home/imagda/sims-projs/CFD-DeepLearning-UNET/plots/valid_data/plots/montage_car_ML_*.*"))
counter = 0
for fp_img1, fp_img2 in zip(fp_in_CFD, fp_in_ML):
counter += 1
img1 = cv2.imread(fp_img1, -1)
img2 = cv2.imread(fp_img2, -1) # this one has transparency
img1_crop = img1[183:400, 169:1300]
img2_crop = img2[183:400, 169:1300]
numpy_vertical_concat = np.concatenate((img1_crop, img2_crop), axis=0)
#numpy_horizontal_concat = np.concatenate((img1_crop, img2_crop), axis=1)
filename = "/home/imagda/sims-projs/CFD-DeepLearning-UNET/plots/valid_data/plots/CFD+ML_"+"{:02d}".format(counter) +".png"
#file = open('{}{}'.format(name, x) , "w+")
plt.savefig(filename, dpi = 200)
plt.imshow( numpy_vertical_concat)
counter = 0
for fp_img1, fp_img2 in zip(fp_in_CFD, fp_in_ML):
counter += 1
img1 = cv2.imread(fp_img1, -1)
img2 = cv2.imread(fp_img2, -1) # this one has transparency
img1_crop = img1[183:400, 0:200]
img2_crop = img2[183:400, 800:1000]
#numpy_vertical_concat = np.concatenate((img1_crop, img2_crop), axis=0)
numpy_horizontal_concat = img1_crop + img2_crop
filename = "/home/imagda/sims-projs/CFD-DeepLearning-UNET/plots/valid_data/plots/CFD+ML_"+"{:02d}".format(counter) +".png"
#file = open('{}{}'.format(name, x) , "w+")
plt.savefig(filename, dpi = 200)
plt.imshow( numpy_horizontal_concat)
img1 = cv2.imread(fp_img1)
img1.shape
type(img1)
import cv2
from skimage import io
lion_rgb = cv2.imread('https://i.stack.imgur.com/R6X5p.jpg')
lion_gray = cv2.imread('https://i.stack.imgur.com/f27t5.png')
# Read Image1
#mountain = cv2.imread(fp_img1, 1)
# Read image2
#dog = cv2.imread(fp_img2, 1)
# Blending the images with 0.3 and 0.7
img = cv2.addWeighted(lion_rgb, 1, lion_rgb, 1, 0)
plt.imshow(img/266.)
#io.imshow(img)
# Show the image
#cv2.imshow('image', img)
# Wait for a key
#cv2.waitKey(0)
# Distroy all the window open
#cv2.distroyAllWindows()
rgb.shape
import numpy as np
from skimage import io
rgb = io.imread('https://i.stack.imgur.com/R6X5p.jpg')
gray = io.imread('https://i.stack.imgur.com/f27t5.png')
rows_rgb, cols_rgb, channels = rgb.shape
rows_gray, cols_gray = gray.shape
rows_comb = max(rows_rgb, rows_gray)
cols_comb = cols_rgb + cols_gray
comb = np.zeros(shape=(rows_comb, cols_comb, channels), dtype=np.uint8)
comb[:rows_rgb, :cols_rgb] = rgb
comb[:rows_gray, cols_rgb:] = gray[:, :, None]
io.imshow(comb)
rgb.shape
| 0.327023 | 0.772616 |
<img src='images/Joint_school_graphic_title.png' align='center' width='100%'></img>
# Practical on Datasets Comparison and Application of Averaging Kernels
This practical exercise was created for the <a href ="https://atmosphere.copernicus.eu/3rd-eumetsatesaecmwf-joint-training-atmospheric-composition" target = "_blank">3rd EUMETSAT/ESA/ECMWF Joint Training in Atmospheric Composition (6-17 December, 2021)</a> to show how to compare the NO<sub>2</sub> observations from the TROPOspheric Monitoring Instrument (TROPOMI) aboard Sentinel 5-P and the forecasts of the Copernicus Atmosphere Monitoring Service (CAMS). It is divided into the following sections:
1. [Installation](#installation): A brief guide to know how to install the <a href = "https://github.com/esowc/adc-toolbox/" target = "_blank">Atmospheric Datasets Comparison (ADC) Toolbox</a>, which contains functions that facilitate the datasets retrieval, metadata merge and statistical analysis.
2. [Datasets retrieval](#datasets_retrieval): The model and sensor datasets are downloaded and read as xarray objects before applying the real observations kernels into the model dataset.
3. [Datasets merge](#datasets_merge): The model partial columns are interpolated into the TM5 grid and the averaging kernels are applied.
4. [Comparison analysis](#comparison_analysis): Statistical methods are used to better understand the differences between both datasets and the effects of applying the averaging kernels.
5. [Assignment](#assignment): The details about the assignment submission are included in this section.
6. [FAQ and common errors](#faq): A summary of the questions and errors that the users bumped into while completing the assignment can be found here.
7. [User feedback form](#feedback): If you want to send your feedback, in this section you will find the link to the form.
## <a id='installation'>1. Installation</a>
### Clone the repository and set up the virtual environment
Participants should <a href = "https://my.wekeo.eu/web/guest/user-registration" target = "_blank">create an account in WEkEO</a> to use the JupyterHub and run this notebook. Once they <a href = "https://jupyterhub-wekeo.apps.eumetsat.dpi.wekeo.eu" target = "_blank">have access to this service</a>, they can open the terminal and clone the ADC Toolbox repository with the command:
```bash
$ git clone https://github.com/esowc/adc-toolbox
```
The virtual environment <em>environment.yml</em> was generated to simplify the installation process, so users just need to activate this environment or simulate it by installing the dependencies (libraries and packages) with:
```bash
$ cd adc-toolbox
$ conda create --name adc-toolbox
$ conda activate adc-toolbox
$ conda install -c conda-forge/label/cartopy_dev cartopy
$ pip install -r requirements.txt
$ python -m ipykernel install --user --name adc-toolbox
```
After running the previous commands, the page should be refreshed and the correct kernel (`adc-toolbox`) should be selected.
To finalize the installation process, users need to create a text file in the <em>data</em> folder, with the name <em>keys.txt</em>, and write down their personal CAMS API key in one line with the format <em>UID:Key</em>. Alternatively, they can introduce their user ID and key in the cell underneath. This key can be obtained by <a href = "https://ads.atmosphere.copernicus.eu/user/register?">registering at the Atmosphere Data Store</a>.
```
CAMS_UID = None
CAMS_key = None
```
### Import libraries
```
# Related to the system
import os
from pathlib import Path
# Related to the data retrieval
from sentinelsat.sentinel import SentinelAPI, geojson_to_wkt
import cdsapi
import cfgrib
import geojson
import urllib3
# Related to the data analysis
import math
import xarray as xr
import pandas as pd
import numpy as np
import datetime as dt
from itertools import product
import scipy.interpolate
from scipy.spatial.distance import cdist
from scipy.optimize import curve_fit
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_error
# Related to the results
from copy import copy
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.colors as colors
import matplotlib.lines as mlines
import matplotlib.transforms as mtransforms
import matplotlib.ticker as mticker
import matplotlib.patches as mpatches
import cartopy.crs as ccrs
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
import cartopy.feature as cfeature
import geocoder
import seaborn as sns
from matplotlib import animation
from IPython.display import HTML, display
import warnings
```
### Import functions
```
%run ../../functions/functions_general.ipynb
%run ../../functions/functions_cams.ipynb
%run ../../functions/functions_tropomi.ipynb
```
### Settings
```
# Hide pandas warning
pd.options.mode.chained_assignment = None
# Hide API request warning
urllib3.disable_warnings(urllib3.exceptions.InsecureRequestWarning)
# Increase animation limit
matplotlib.rcParams['animation.embed_limit'] = 25000000
# Hide labels animation warning
warnings.filterwarnings('ignore', category = UserWarning, module='cartopy')
```
## <a id='datasets_retrieval'>2. Datasets retrieval</a>
### Available datasets
ADC Toolbox facilitates the data retrieval of all the datasets presented in Table 1, since the dates they became available to the public. As an exception, the retrieval of IASI L2 data is currently available only since May 14, 2019.
<p align="center"> <b>Table 1</b>. Temporal availability (start date - end date) by data source.</p>
| Source | Type | Platform | NO<sub>2</sub> | O<sub>3</sub> | CO | SO<sub>2</sub> | HCHO |
| :---: | :---: | :---: | :---: | :---: | :---: | :---: | :---: |
| CAMS | Forecast | <a href = "https://ads.atmosphere.copernicus.eu/cdsapp#!/dataset/cams-global-atmospheric-composition-forecasts" target = "_blank">ADS</a> | 01.2015 - Present | 01.2015 - Present | 01.2015 - Present | 01.2015 - Present | 01.2015 - Present |
| CAMS | Reanalysis | <a href = "https://ads.atmosphere.copernicus.eu/cdsapp#!/dataset/cams-global-ghg-reanalysis-egg4-monthly">ADS</a> | 01.2003 - 06.2021 | 01.2003 - 06.2021 | 01.2003 - 06.2021 | 01.2003 - 06.2021 | 01.2003 - 06.2021 |
| TROPOMI | L2 | <a href = "https://s5phub.copernicus.eu/dhus/" target = "_blank">S5-P hub</a> | 07.2018 - Present | 07.2018 - Present | 07.2018 - Present | 10.2018 - Present | 07.2018 - Present |
| TROPOMI | L3 | <a href = "https://www.temis.nl/" target = "_blank">TEMIS</a> | 07.2018 - Present | - | - | - | - |
| IASI | L2 | <a href = "https://iasi.aeris-data.fr/" target = "_blank">AERIS Portal</a> | - | 01.2008 - Present | 10.2007 - Present | 10.2007 - Present | - |
| IASI | L3 | <a href = "https://iasi.aeris-data.fr/" target = "_blank">AERIS Portal</a> | - | 01.2008 - Present | 10.2007 - Present | - | - |
| GOME-2 | L2 | <a href = "https://acsaf.org/offline_access.php" target = "_blank">AC SAF</a> | 01.2007 - Present | 01.2007 - Present | - | 01.2007 - Present | 01.2007 - Present |
| GOME-2 | L3 | <a href = "https://acsaf.org/offline_access.php" target = "_blank">AC SAF</a> | 02.2007 - 11.2017 | - | - | - | - |
| GOME-2 | L3 | <a href = "https://www.temis.nl/" target = "_blank">TEMIS</a> | 02.2007 - Present | - | - | - | - |
In order to automatically download any model or sensor dataset, users only need to define:
* `component_nom`: Name of the atmospheric component.
* `model`: Short name of the model (<em>cams</em>)
* `model_full_name`: Full name of the model (<em>cams-global-atmospheric-composition-forecasts</em> or <em>cams-global-reanalysis-eac4-monthly</em>).
* `sensor`: Short name of the sensor (<em>tropomi</em>, <em>iasi</em> or <em>gome</em>).
* `start_date` and `end_date`: An array with dates will be retrieved between both dates.
* `lon_min`, `lon_max`, `lat_min` and `lat_max`: Coordinates to create the search bounding box (`bbox`).
* `area_name`: Name of the studied region.
```
# Define component
component_nom = 'NO2'
# Define model
model = 'cams'
model_full_name = 'cams-global-atmospheric-composition-forecasts'
# Define sensor
sensor = 'tropomi'
sensor_type = 'L2'
apply_kernels = True
# Define search period
start_date = '2021-11-18'
end_date = '2021-11-18'
# Define extent
area_name = 'Barcelona'
lon_min = 1.5
lon_max = 2.5
lat_min = 41
lat_max = 42
```
### Comparison checker and folder generation
The toolbox will check if the comparison analysis between the specified model and sensor is possible, given the name of the species. If it is, the molecular weight and the products standard metadata are obtained. Afterwards, the folders where the datasets will be stored are generated.
<ins>Note</ins>: This notebook can only be used to compare TROPOMI and CAMS datasets while applying the averaging kernels for NO<sub>2</sub>, please refer to the main code to compare the CAMS model against the observations from IASI or GOME-2 or to use the same data sources without applying the averaging kernels or for multiple timesteps.
```
# Check if comparison is possible
comparison_check(sensor, model, component_nom, model_full_name, sensor_type, apply_kernels)
# Get component full name and molecular weight
component, component_MW, product_type, sensor_column, column_type = components_table(sensor, component_nom, sensor_type)
# Folders generation
generate_folders(model, sensor, component_nom, sensor_type)
```
### Search period and bounding box
The search period and bounding box are derived from the details that were provided in advance.
```
# Generate array with search dates
dates = search_period(start_date, end_date, sensor, sensor_type)
# Create bbox
bbox = search_bbox(lon_min, lat_min, lon_max, lat_max)
```
### Download and read the model data
The model dataset is downloaded as a GRIB file and read as a xarray object. In this step, the users can decide if they want to retrieve total (`model_level` = 'single') or partial columns (`model_level` = 'multiple'). For this training, we need to obtain the partial columns (at 137 vertical levels) from the <a href = "https://ads.atmosphere.copernicus.eu/cdsapp#!/dataset/cams-global-atmospheric-composition-forecasts?tab=overview" target = "_blank">CAMS global atmospheric composition forecasts</a>.
```
model_level = 'multiple'
model_product_name, model_type = CAMS_download(dates, start_date, end_date, component, component_nom,
lat_min, lat_max, lon_min, lon_max, area_name,
model_full_name, model_level, CAMS_UID, CAMS_key)
model_ds, _, model_levels_df = CAMS_read(model_product_name, component, component_nom, dates)
model_ds
```
### Download and read sensor data
The sensor dataset is downloaded as a `NetCDF` file and read as a xarray object, along with more detailed and support datasets. More information about this product can be found in the <a href = "https://sentinels.copernicus.eu/documents/247904/2474726/Sentinel-5P-Level-2-Product-User-Manual-Nitrogen-Dioxide.pdf/ad25ea4c-3a9a-3067-0d1c-aaa56eb1746b?t=1626258361795" target = "_blank">TROPOMI NO<sub>2</sub> product manual</a>.
```
dates = sensor_download(sensor, sensor_type, component_nom, dates, bbox, product_type)
sensor_ds, support_input_ds, support_details_ds = sensor_read(sensor, sensor_type, sensor_column,
component_nom, dates)
sensor_ds
```
Within `support_input_ds` and `support_details_ds` we find the surface pressure data, that we will need to compute the pressure at each level.
```
support_input_ds
support_details_ds
```
## <a id='datasets_merge'>3. Datasets merge</a>
We will apply the sensor averaging kernels to the model partial columns in the observations space (TM5) before comparing the datasets. Right now, we have data at different grid resolutions and levels: the sensor collects data at 34 vertical layers, which are broader than the 137 levels in the CAMS model.
<img src='images/Vertical_resolutions.png' align='center' width='50%'></img>
<center><b>Figure 1.</b> L137 and TM5 levels by pressure.</center>
### Steps
Following the next steps, we can retrieve the model partial columns at the TM5 grid and carry out the satellite datasets comparison:
1. (L137) Calculate model levels pressure.
2. (L137) Convert model data units (from kg/kg to molecules/cm<sup>2</sup>) and compute the partial columns above each half level in the model space.
3. (TM5) Calculate sensor levels pressure and column kernels.
4. (TM5) Convert sensor data units (from kg/m<sup>2</sup> to molecules/cm<sup>2</sup>).
5. (TM5 + L137) Transform sensor dataset into a dataframe and join L137 and TM5 levels.
6. (L137) Retrieve model partial columns above each level by closest neighbours.
7. (TM5) Interpolate model partial columns above each level in the observations space.
8. (TM5) Calculate model partial columns at each level.
9. (TM5) Apply averaging kernels.
10. (TM5) Calculate total columns for both datasets and difference between them.
### Calculate the model levels pressure
The 137 vertical levels in the model can be described by various parameters, which are necessary to evaluate the levels pressure. In particular, the coefficients `a` and `b` are used, as indicated in the <a href = "https://confluence.ecmwf.int/display/OIFS/4.4+OpenIFS%3A+Vertical+Resolution+and+Configurations">Confluence page about the vertical resolution and configuration of the model</a>, to calculate these pressures:
<center>
<em>p<sub>k+½</sub> = a<sub>k+½</sub> + p<sub>s</sub> · b<sub>k+½</sub></em><br>
<em>p<sub>k-½</sub> = a<sub>k-½</sub> + p<sub>s</sub> · b<sub>k-½</sub></em><br>
<em>p<sub>k</sub> = 0.5 · (p<sub>k-½</sub> + p<sub>k+½</sub>)</em>
</center>
```
model_levels_df
# Calculate level pressures from the surface pressures
model_ds = CAMS_pressure(model_ds, model_product_name, model_levels_df, start_date, end_date, component_nom,
lat_min, lat_max, lon_min, lon_max, area_name, CAMS_UID, CAMS_key)
model_ds
```
### Convert the model data units (from kg/kg to molecules/cm<sup>2</sup>)
#### Calculate the columns above each half level (kg/kg to kg/m<sup>2</sup>)
To convert the original units (kg/kg) into kg/m<sup>2</sup>, we calculate the NO<sub>2</sub> columns above each CAMS half level, assuming that they are 0 at the top of the atmosphere.
```
print('The columns above each model half level will be calculated.')
# Initialize partial columns at the top of the atmosphere (hybrid = 1) as 0
model_ds_time_old = model_ds.sel(time = model_ds.time.values[0])
PC_hybrid_0 = model_ds_time_old.sel(hybrid = 1)
PC_hybrid_0['component'] = PC_hybrid_0['component'].where(PC_hybrid_0['component'] <= 0, 0, drop = False)
PC_hybrid_0 = PC_hybrid_0.expand_dims(dim = ['hybrid'])
# Create new model dataset
PC_above_all = []
PC_above_all.append(PC_hybrid_0)
model_ds_time_new = PC_hybrid_0
for hybrid in range(1, 137):
# Get current and previous partial columns and level pressures
PC_last = model_ds_time_new.component.sel(hybrid = hybrid)
PC_current = model_ds_time_old.component.sel(hybrid = hybrid + 1)
pressure_last = model_ds_time_old.pressure.sel(hybrid = hybrid)
pressure_current = model_ds_time_old.pressure.sel(hybrid = hybrid + 1)
# Calculate pressure difference
pressure_diff = pressure_current - pressure_last
# Calculate partial columns above each model level
# Units: (kg/kg * kg/m*s2) * s2/m -> kg/m2
PC_above = model_ds_time_old.sel(hybrid = hybrid + 1)
PC_above['component'] = PC_last + PC_current * pressure_diff * (1/9.81)
# Append result
PC_above_all.append(PC_above)
model_ds_time_new = xr.concat(PC_above_all, pd.Index(range(1, hybrid + 2), name = 'hybrid'))
model_ds = model_ds_time_new
model_ds = model_ds.expand_dims(dim = ['time'])
model_ds
# Assign new units to array
units = 'kg m**-2'
model_ds['component'] = model_ds.component.assign_attrs({'units': units})
print('The model component units have been converted from kg kg**-1 to kg m**-2.')
```
#### Convert units with Avogadro's number (kg/m<sup>2</sup> to molecules/cm<sup>2</sup>)
After, we convert the data units from kg/m<sup>2</sup> to molecules/cm<sup>2</sup> simply by:
```
# Conversion
NA = 6.022*10**23
model_ds['component'] = (model_ds['component'] * NA * 1000) / (10000 * component_MW)
# Assign new units to array
model_ds['component'] = model_ds.component.assign_attrs({'units': 'molec cm-2'})
print('The model component units have been converted from kg m**-2 to molec cm-2.')
```
### Calculate the TM5 levels pressure and column kernels
The computation of TM5 levels pressure is carried out as explained before. On the other hand, the tropospheric column kernels equal to:
<center>
<em>A<sub>trop</sub> = (M · A)/M<sub>trop</sub> if l $\leq$ l<sub>trop</sub></em><br>
<em>A<sub>trop</sub> = 0 if l $\gt$ l<sub>trop</sub></em><br><br>
</center>
where <em>A</em> is the total averaging kernel, <em>M</em> the total air mass factor, <em>M<sub>trop</sub></em> the tropospheric air mass factor, <em>l</em> the layer and <em>l<sub>trop</sub></em> the last layer of the troposphere.
```
print('APPLICATION OF AVERAGING KERNELS')
print('For the application of the averaging kernels, it is necessary to calculate:')
print('1. Level pressures')
print('2. Column kernels')
print('The apriori profiles should be retrieved, but they are not necessary.')
print('DATA AVAILABILITY')
sensor_ds = TROPOMI_pressure(sensor_ds, component_nom, support_input_ds, support_details_ds)
sensor_ds = TROPOMI_column_kernel(sensor_ds, component_nom, support_details_ds)
sensor_ds = TROPOMI_apriori_profile(sensor_ds, component_nom, component, support_details_ds)
sensor_ds
```
### Convert TROPOMI data units (From mol/m<sup>2</sup> to molecules/cm<sup>2</sup>)
```
# Conversion
sensor_ds['sensor_column'] = sensor_ds['sensor_column'] * 6.02214*10**19
# Assign new units to array
sensor_ds['sensor_column'] = sensor_ds['sensor_column'].assign_attrs({'units': 'molec cm-2'})
print('The sensor component units have been converted from mol cm-2 to molec cm-2.')
```
### Transform sensor dataset into a dataframe and join L137 and TM5 levels
Before we transform the sensor dataset into a dataframe, we select the data for a unique date and subset both datasets to speed up the merge. CAMS dataset was already reduced to the size of the previously defined bounding box during the units conversion. To subset TROPOMI's dataset, we create a lookup table with the equivalent geospatial coordinates to each pair of scanline and ground pixel.
```
# Reduce data to only one timestamp
model_ds_time = model_ds.sel(time = model_ds.time.values)
sensor_ds_time = sensor_ds.sel(time = sensor_ds.time.values)
# Subset
sensor_ds_time = subset(sensor_ds_time, bbox, sensor, component_nom,
sensor_type, subset_type = 'sensor_subset')
# Get equivalence table for coordinates after subset
lookup_table = TROPOMI_lookup_table(sensor_ds_time, component_nom)
lookup_table
# Transform data array into dataframe
match_df_time = sensor_ds_time.to_dataframe()
# Pass NaNs to data with qa_value under 0.5 (these values will be shown as transparent)
match_df_time.loc[match_df_time['qa_value'] <= 0.5, ['sensor_column', 'column_kernel']] = float('NaN')
# Select multiindex elements
match_df_time = match_df_time.groupby(by = ['layer', 'scanline', 'ground_pixel', 'time', 'delta_time']).mean()
match_df_time = match_df_time.reset_index(level = ['layer', 'delta_time'])
match_df_time = match_df_time.set_index('layer', append = True)
match_df_time
# Create index that includes CAMS pressure levels for all the locations in TROPOMI
new_array = np.concatenate([np.arange(1, 137) * 1000, sensor_ds_time.layer.values])
new_index = pd.MultiIndex.from_product([match_df_time.index.levels[0],
match_df_time.index.levels[1],
match_df_time.index.levels[2],
new_array
],
names = ['scanline', 'ground_pixel', 'time', 'layer'])
# Append original and new indexes and reindex dataframe
match_df_time = match_df_time[~match_df_time.index.duplicated()]
match_df_time = match_df_time.reindex(match_df_time.index.append(new_index))
# Sort and reset index
match_df_time = match_df_time.sort_index()
match_df_time = match_df_time.reset_index()
match_df_time
```
### Retrieve CAMS partial columns at TM5 grid
```
# Find latitudes in CAMS rows with scanlines and ground pixels
match_df_time['latitude'] = match_df_time.apply(lambda row: float(lookup_table[
(lookup_table['scanline'] == row['scanline']) &
(lookup_table['ground_pixel'] == row['ground_pixel'])]['latitude'])
if pd.isnull(row['latitude'])
else row['latitude'],
axis = 1)
# Find longitudes in CAMS rows with scanlines and ground pixels
match_df_time['longitude'] = match_df_time.apply(lambda row: float(lookup_table[
(lookup_table['scanline'] == row['scanline']) &
(lookup_table['ground_pixel'] == row['ground_pixel'])]['longitude'])
if pd.isnull(row['longitude'])
else row['longitude'],
axis = 1)
# Get unique timestep
sensor_times = sensor_ds_time.delta_time.isel(scanline = 0).values
model_times = model_ds_time.valid_time.values
unique_step = int(np.unique(nearest_neighbour(model_times, sensor_times)))
unique_time = model_ds_time.component.isel(step = unique_step).step.values.astype('timedelta64[h]')
# Get CAMS model partial columns above each level at closest TROPOMI locations (nearest neighbours)
match_df_time['model_partial_column_above'] = match_df_time.apply(lambda row: model_ds_time.component.sel(
step = unique_time,
hybrid = row['layer'] / 1000,
latitude = row['latitude'],
longitude = row['longitude'],
method = 'nearest').values
if pd.isnull(row['sensor_column'])
else math.nan,
axis = 1)
match_df_time
```
### Interpolate CAMS partial columns above each level at TM5 pressures
```
# Get CAMS model level pressures
match_df_time['pressure'] = match_df_time.apply(lambda row: model_ds_time.pressure.sel(
step = unique_time,
hybrid = row['layer'] / 1000,
latitude = row['latitude'],
longitude = row['longitude'],
method = 'nearest').values
if pd.isnull(row['pressure'])
else row['pressure'],
axis = 1)
# Transform 1D-array data to float
match_df_time['model_partial_column_above'] = match_df_time['model_partial_column_above'].apply(lambda x: float(x))
match_df_time['pressure'] = match_df_time['pressure'].apply(lambda x: float(x))
# Set multiindex again and sort for interpolation
match_df_time = match_df_time.reset_index()
match_df_time = match_df_time.set_index(['time', 'ground_pixel', 'scanline', 'pressure'])
match_df_time = match_df_time.sort_values(['time', 'ground_pixel','scanline', 'pressure'],
ascending = [True, True, True, False])
# Interpolate partial columns onto the TM5 pressure levels
match_df_time = match_df_time[~match_df_time.index.duplicated()]
match_df_time['model_partial_column_above'] = match_df_time['model_partial_column_above'].interpolate()
match_df_time
# Drop unnecessary values
match_df_time = match_df_time.reset_index()
match_df_time = match_df_time.set_index(['time', 'ground_pixel', 'scanline', 'layer'])
match_df_time = match_df_time.drop(np.arange(1, 137) * 1000, level = 'layer')
match_df_time
```
### Calculate CAMS partial columns at each level at TM5 grid
```
# Calculate CAMS partial columns for each TM5 layer (as difference between the interpolated values)
match_df_time['model_column'] = match_df_time['model_partial_column_above'] - match_df_time['model_partial_column_above'].shift(-1)
match_df_time = match_df_time.reset_index()
match_df_time.loc[match_df_time['layer'] == 33, ['model_column']] = match_df_time['model_partial_column_above']
match_df_time = match_df_time.set_index(['time', 'ground_pixel', 'scanline', 'layer'])
match_df_time
```
### Apply the averaging kernels
We calculated the difference between the interpolated values to get the CAMS partial columns for each TM5 layer (not above), so we can finally apply the averaging kernels as:
<center>
<em>x<sub>rtv</sub> ≈ x<sub>a</sub> + A · (x<sub>true</sub> - x<sub>a</sub>)</em><br><br>
</center>
where <em>x<sub>rtv</sub></em> is the averaged model partial column, <em>x<sub>a</sub></em> the apriori profile, <em>A</em> the averaging column kernel and <em>x<sub>true</sub></em> the model partial column before applying the kernels. In this case, the apriori profiles are null.
```
# Apply the averaging kernels
if 'apriori_profile' in match_df_time.columns:
match_df_time['model_column'] = match_df_time.apply(lambda row: row['apriori_profile'] +
row['column_kernel'] * row['model_column'] -
row['column_kernel'] * row['apriori_profile'],
axis = 1)
else:
match_df_time['model_column'] = match_df_time.apply(lambda row: row['model_column'] *
row['column_kernel'],
axis = 1)
match_df_time = match_df_time[~match_df_time.index.duplicated()]
match_df_time
```
### Calculate total columns for both datasets and difference between them
```
# Transform dataframe back to xarray
match_ds_time = match_df_time.to_xarray()
# Read latitudes and longitudes from data array
latitude = match_ds_time.latitude.mean(dim = 'layer')
longitude = match_ds_time.longitude.mean(dim = 'layer')
# Get sum of CAMS data of each layer to get column data
model_final_ds_time = match_ds_time.model_column.sum(dim = 'layer', skipna = False).astype(float)
model_final_ds_time = model_final_ds_time.assign_coords(latitude = latitude, longitude = longitude)
# Get mean of TROPOMI data of each layer (it must be equal)
sensor_final_ds_time = match_ds_time.sensor_column.mean(dim = 'layer', skipna = False).astype(float)
sensor_final_ds_time = sensor_final_ds_time.assign_coords(latitude = latitude, longitude = longitude)
# Calculate difference
merge_ds_time = xr.merge([model_final_ds_time, sensor_final_ds_time])
merge_ds_time['difference'] = merge_ds_time.model_column - merge_ds_time.sensor_column
merge_ds_time['relative_difference'] = (merge_ds_time.model_column - merge_ds_time.sensor_column)/merge_ds_time.sensor_column
# See results table
merge_df = merge_ds_time.to_dataframe()
merge_df = merge_df.reset_index().set_index(['ground_pixel', 'scanline', 'time'])
merge_df = merge_df[['latitude', 'longitude', 'model_column', 'sensor_column', 'difference', 'relative_difference']]
merge_df
merge_df.describe()
```
## <a id='comparison_analysis'>4. Comparison analysis</a>
### Select plot dates
```
plot_dates = plot_period(sensor_ds, sensor_type)
```
### Select plot extent
```
plot_bbox = plot_extent(bbox)
```
### Compare model and TROPOMI total columns
```
# Choose distribution (aggregated, individual or animated)
plot_type = 'individual'
# Define range (original, equal, centered or manual)
range_type = 'equal'
vmin_manual, vmax_manual = None, None
vmin_manual_diff, vmax_manual_diff = None, None
# Define projection and colors
projection = ccrs.PlateCarree()
color_scale = ['turbo', 'turbo', 'coolwarm']
# Get and decide the best width and height of the B/W frame lines
options_height_lon = get_frame_possible_lengths(lon_min, lon_max)
options_width_lat = get_frame_possible_lengths(lat_min, lat_max)
width_lon = 0.2
height_lat = 0.2
# Add marker
regions_names = ('Barcelona')
coords_list = (41.37, 2.17)
bbox_list = None
visualize_model_vs_sensor(model, sensor, component_nom, units, merge_df, plot_dates, plot_bbox, 20, 1.05,
model_type, sensor_type, range_type, plot_type, projection,
color_scale, width_lon, height_lat, vmin_manual, vmax_manual,
vmin_manual_diff, vmax_manual_diff, bbox_list, coords_list, regions_names)
```
### Retrieve nearest values to specific coordinates
It is possible to see the variations of nitrogen dioxide by CAMS and TROPOMI for multiple times and locations. The toolbox will find the nearest neighbours to the input coordinates (`coords_search_list`) and show a table with the concentrations, along with a timeseries plot. In this case, we only have one timestep, so the timeseries plot will not be visible.
```
regions_names = ('Barcelona - 1', 'Barcelona - 2')
coords_list = (41.39, 2.15,
41.1, 1.65)
ymin = 0
ymax = 5*10**15
xticks = plot_dates
timeseries_table = timeseries(merge_df, component_nom, sensor, sensor_type, model,
plot_dates, units, ymin, ymax, xticks, regions_names, coords_list)
timeseries_table
```
### Scatter plots by bbox
```
show_seasons = False
extent_definition = 'bbox' # bbox or country
scatter_plot_type = 'individual' # aggregated or individual
lim_min = None
lim_max = None
summary = scatter_plot(merge_df, component_nom, units, sensor, plot_dates, 1.05,
extent_definition, show_seasons, scatter_plot_type, lim_min, lim_max, plot_bbox)
summary
```
## <a id='assignment'>5. Assignment</a>
After learning about the ADC Toolbox during the training, participants will be able to carry out their own studies and explore its capabilities by completing one of these assignments:
1. Use the satellite NO<sub>2</sub> concentrations in your own city for a day in the last month and show how CAMS and TROPOMI datasets compare to each other applying TROPOMI averaging kernels to the CAMS data (using this notebook). As a bonus, compare both datasets without using the kernels or run a timeseries animated comparison (using <em>main_cams_tropomi_L2.ipynb</em>).
2. Compare the CAMS model and IASI sensor datasets to see if the COVID-19 lockdowns helped to reduce the air pollution in your region by looking at the monthly data of CO between 2019 and 2021 (using <em>main_cams_iasi_L3.ipynb</em>). There are other factors that might affect the data, could you think of which ones may cause variations in the concentrations of CO?
You can find <a href = "https://padlet.com/GlasgowsEvents/os1kfgsk1z679dk2" target = "_blank">all the submissions in Padlet</a>.
## <a id='faq'>6. FAQ and common errors</a>
The frequently asked questions and errors have been collected as:
* <strong>When importing libraries, I get the error <em>"ModuleNotFoundError: No module named 'sentinelsat'"</em></strong>: This occurs when the virtual environment is not activated. If you have followed the installation instructions and the virtual environment has been created, you need to select it on the top right of the page (Figure 2).
<img src='images/Kernel_selection.png' align='center' width='100%'></img>
<center><b>Figure 2.</b> Kernel selection location in JupyterHub.</center>
* <strong>When downloading the CAMS dataset, the compiler is showing the error <em>"TypeError: 'tuple' object is not callable"</em></strong>: This usually happens when the user ID and API key are not well defined. If you have given the variables CAMS_UID and CAMS_key a string other than None, remember to quote it with double or single quotes. If you want to run the other notebooks, it is recommend to create a text file in the <em>data</em> folder, with the name <em>keys.txt</em>, and write down your personal ID and CAMS API key in one line with the format <em>UID:Key</em>. An example can be found in the folder <em>2021-12-atmospheric-composition</em>.
* <strong>The installation of the toolbox in WEkEO takes too long. Is this normal?</strong>: It should take about 10 minutes to install through the terminal.
* <strong>The kernel is suddenly not working and I have to run the entire code again</strong>: If this happens, please contact WEkEO's support (support@wekeo.eu). It is likely that there is a problem with your personal account. It is also possible that you are using too many computational resources (e.g. by selecting a very large bounding box or time period).
If you have any other question or comment, you can <a href = "https://github.com/esowc/adc-toolbox/issues" target = "_blank">open an issue in GitHub</a>.
## <a id='feedback'>7. User feeback form</a>
If you are interested in the ADC toolbox and would like it to have other functionalities or datasets, you can <a href = "https://docs.google.com/forms/d/e/1FAIpQLSd2DYumvDcIGIz1cQrbfezOOEdGBFli5q76uZvmLva6mZ0E7w/viewform?usp=sf_link" target = "_blank">write your feedback in this Google Form</a>.
<center><strong>Thank you very much!</strong></center>
<img src='images/Joint_school_logo_line.png' align='center' width='100%'></img>
|
github_jupyter
|
$ git clone https://github.com/esowc/adc-toolbox
$ cd adc-toolbox
$ conda create --name adc-toolbox
$ conda activate adc-toolbox
$ conda install -c conda-forge/label/cartopy_dev cartopy
$ pip install -r requirements.txt
$ python -m ipykernel install --user --name adc-toolbox
CAMS_UID = None
CAMS_key = None
# Related to the system
import os
from pathlib import Path
# Related to the data retrieval
from sentinelsat.sentinel import SentinelAPI, geojson_to_wkt
import cdsapi
import cfgrib
import geojson
import urllib3
# Related to the data analysis
import math
import xarray as xr
import pandas as pd
import numpy as np
import datetime as dt
from itertools import product
import scipy.interpolate
from scipy.spatial.distance import cdist
from scipy.optimize import curve_fit
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_error
# Related to the results
from copy import copy
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.colors as colors
import matplotlib.lines as mlines
import matplotlib.transforms as mtransforms
import matplotlib.ticker as mticker
import matplotlib.patches as mpatches
import cartopy.crs as ccrs
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
import cartopy.feature as cfeature
import geocoder
import seaborn as sns
from matplotlib import animation
from IPython.display import HTML, display
import warnings
%run ../../functions/functions_general.ipynb
%run ../../functions/functions_cams.ipynb
%run ../../functions/functions_tropomi.ipynb
# Hide pandas warning
pd.options.mode.chained_assignment = None
# Hide API request warning
urllib3.disable_warnings(urllib3.exceptions.InsecureRequestWarning)
# Increase animation limit
matplotlib.rcParams['animation.embed_limit'] = 25000000
# Hide labels animation warning
warnings.filterwarnings('ignore', category = UserWarning, module='cartopy')
# Define component
component_nom = 'NO2'
# Define model
model = 'cams'
model_full_name = 'cams-global-atmospheric-composition-forecasts'
# Define sensor
sensor = 'tropomi'
sensor_type = 'L2'
apply_kernels = True
# Define search period
start_date = '2021-11-18'
end_date = '2021-11-18'
# Define extent
area_name = 'Barcelona'
lon_min = 1.5
lon_max = 2.5
lat_min = 41
lat_max = 42
# Check if comparison is possible
comparison_check(sensor, model, component_nom, model_full_name, sensor_type, apply_kernels)
# Get component full name and molecular weight
component, component_MW, product_type, sensor_column, column_type = components_table(sensor, component_nom, sensor_type)
# Folders generation
generate_folders(model, sensor, component_nom, sensor_type)
# Generate array with search dates
dates = search_period(start_date, end_date, sensor, sensor_type)
# Create bbox
bbox = search_bbox(lon_min, lat_min, lon_max, lat_max)
model_level = 'multiple'
model_product_name, model_type = CAMS_download(dates, start_date, end_date, component, component_nom,
lat_min, lat_max, lon_min, lon_max, area_name,
model_full_name, model_level, CAMS_UID, CAMS_key)
model_ds, _, model_levels_df = CAMS_read(model_product_name, component, component_nom, dates)
model_ds
dates = sensor_download(sensor, sensor_type, component_nom, dates, bbox, product_type)
sensor_ds, support_input_ds, support_details_ds = sensor_read(sensor, sensor_type, sensor_column,
component_nom, dates)
sensor_ds
support_input_ds
support_details_ds
model_levels_df
# Calculate level pressures from the surface pressures
model_ds = CAMS_pressure(model_ds, model_product_name, model_levels_df, start_date, end_date, component_nom,
lat_min, lat_max, lon_min, lon_max, area_name, CAMS_UID, CAMS_key)
model_ds
print('The columns above each model half level will be calculated.')
# Initialize partial columns at the top of the atmosphere (hybrid = 1) as 0
model_ds_time_old = model_ds.sel(time = model_ds.time.values[0])
PC_hybrid_0 = model_ds_time_old.sel(hybrid = 1)
PC_hybrid_0['component'] = PC_hybrid_0['component'].where(PC_hybrid_0['component'] <= 0, 0, drop = False)
PC_hybrid_0 = PC_hybrid_0.expand_dims(dim = ['hybrid'])
# Create new model dataset
PC_above_all = []
PC_above_all.append(PC_hybrid_0)
model_ds_time_new = PC_hybrid_0
for hybrid in range(1, 137):
# Get current and previous partial columns and level pressures
PC_last = model_ds_time_new.component.sel(hybrid = hybrid)
PC_current = model_ds_time_old.component.sel(hybrid = hybrid + 1)
pressure_last = model_ds_time_old.pressure.sel(hybrid = hybrid)
pressure_current = model_ds_time_old.pressure.sel(hybrid = hybrid + 1)
# Calculate pressure difference
pressure_diff = pressure_current - pressure_last
# Calculate partial columns above each model level
# Units: (kg/kg * kg/m*s2) * s2/m -> kg/m2
PC_above = model_ds_time_old.sel(hybrid = hybrid + 1)
PC_above['component'] = PC_last + PC_current * pressure_diff * (1/9.81)
# Append result
PC_above_all.append(PC_above)
model_ds_time_new = xr.concat(PC_above_all, pd.Index(range(1, hybrid + 2), name = 'hybrid'))
model_ds = model_ds_time_new
model_ds = model_ds.expand_dims(dim = ['time'])
model_ds
# Assign new units to array
units = 'kg m**-2'
model_ds['component'] = model_ds.component.assign_attrs({'units': units})
print('The model component units have been converted from kg kg**-1 to kg m**-2.')
# Conversion
NA = 6.022*10**23
model_ds['component'] = (model_ds['component'] * NA * 1000) / (10000 * component_MW)
# Assign new units to array
model_ds['component'] = model_ds.component.assign_attrs({'units': 'molec cm-2'})
print('The model component units have been converted from kg m**-2 to molec cm-2.')
print('APPLICATION OF AVERAGING KERNELS')
print('For the application of the averaging kernels, it is necessary to calculate:')
print('1. Level pressures')
print('2. Column kernels')
print('The apriori profiles should be retrieved, but they are not necessary.')
print('DATA AVAILABILITY')
sensor_ds = TROPOMI_pressure(sensor_ds, component_nom, support_input_ds, support_details_ds)
sensor_ds = TROPOMI_column_kernel(sensor_ds, component_nom, support_details_ds)
sensor_ds = TROPOMI_apriori_profile(sensor_ds, component_nom, component, support_details_ds)
sensor_ds
# Conversion
sensor_ds['sensor_column'] = sensor_ds['sensor_column'] * 6.02214*10**19
# Assign new units to array
sensor_ds['sensor_column'] = sensor_ds['sensor_column'].assign_attrs({'units': 'molec cm-2'})
print('The sensor component units have been converted from mol cm-2 to molec cm-2.')
# Reduce data to only one timestamp
model_ds_time = model_ds.sel(time = model_ds.time.values)
sensor_ds_time = sensor_ds.sel(time = sensor_ds.time.values)
# Subset
sensor_ds_time = subset(sensor_ds_time, bbox, sensor, component_nom,
sensor_type, subset_type = 'sensor_subset')
# Get equivalence table for coordinates after subset
lookup_table = TROPOMI_lookup_table(sensor_ds_time, component_nom)
lookup_table
# Transform data array into dataframe
match_df_time = sensor_ds_time.to_dataframe()
# Pass NaNs to data with qa_value under 0.5 (these values will be shown as transparent)
match_df_time.loc[match_df_time['qa_value'] <= 0.5, ['sensor_column', 'column_kernel']] = float('NaN')
# Select multiindex elements
match_df_time = match_df_time.groupby(by = ['layer', 'scanline', 'ground_pixel', 'time', 'delta_time']).mean()
match_df_time = match_df_time.reset_index(level = ['layer', 'delta_time'])
match_df_time = match_df_time.set_index('layer', append = True)
match_df_time
# Create index that includes CAMS pressure levels for all the locations in TROPOMI
new_array = np.concatenate([np.arange(1, 137) * 1000, sensor_ds_time.layer.values])
new_index = pd.MultiIndex.from_product([match_df_time.index.levels[0],
match_df_time.index.levels[1],
match_df_time.index.levels[2],
new_array
],
names = ['scanline', 'ground_pixel', 'time', 'layer'])
# Append original and new indexes and reindex dataframe
match_df_time = match_df_time[~match_df_time.index.duplicated()]
match_df_time = match_df_time.reindex(match_df_time.index.append(new_index))
# Sort and reset index
match_df_time = match_df_time.sort_index()
match_df_time = match_df_time.reset_index()
match_df_time
# Find latitudes in CAMS rows with scanlines and ground pixels
match_df_time['latitude'] = match_df_time.apply(lambda row: float(lookup_table[
(lookup_table['scanline'] == row['scanline']) &
(lookup_table['ground_pixel'] == row['ground_pixel'])]['latitude'])
if pd.isnull(row['latitude'])
else row['latitude'],
axis = 1)
# Find longitudes in CAMS rows with scanlines and ground pixels
match_df_time['longitude'] = match_df_time.apply(lambda row: float(lookup_table[
(lookup_table['scanline'] == row['scanline']) &
(lookup_table['ground_pixel'] == row['ground_pixel'])]['longitude'])
if pd.isnull(row['longitude'])
else row['longitude'],
axis = 1)
# Get unique timestep
sensor_times = sensor_ds_time.delta_time.isel(scanline = 0).values
model_times = model_ds_time.valid_time.values
unique_step = int(np.unique(nearest_neighbour(model_times, sensor_times)))
unique_time = model_ds_time.component.isel(step = unique_step).step.values.astype('timedelta64[h]')
# Get CAMS model partial columns above each level at closest TROPOMI locations (nearest neighbours)
match_df_time['model_partial_column_above'] = match_df_time.apply(lambda row: model_ds_time.component.sel(
step = unique_time,
hybrid = row['layer'] / 1000,
latitude = row['latitude'],
longitude = row['longitude'],
method = 'nearest').values
if pd.isnull(row['sensor_column'])
else math.nan,
axis = 1)
match_df_time
# Get CAMS model level pressures
match_df_time['pressure'] = match_df_time.apply(lambda row: model_ds_time.pressure.sel(
step = unique_time,
hybrid = row['layer'] / 1000,
latitude = row['latitude'],
longitude = row['longitude'],
method = 'nearest').values
if pd.isnull(row['pressure'])
else row['pressure'],
axis = 1)
# Transform 1D-array data to float
match_df_time['model_partial_column_above'] = match_df_time['model_partial_column_above'].apply(lambda x: float(x))
match_df_time['pressure'] = match_df_time['pressure'].apply(lambda x: float(x))
# Set multiindex again and sort for interpolation
match_df_time = match_df_time.reset_index()
match_df_time = match_df_time.set_index(['time', 'ground_pixel', 'scanline', 'pressure'])
match_df_time = match_df_time.sort_values(['time', 'ground_pixel','scanline', 'pressure'],
ascending = [True, True, True, False])
# Interpolate partial columns onto the TM5 pressure levels
match_df_time = match_df_time[~match_df_time.index.duplicated()]
match_df_time['model_partial_column_above'] = match_df_time['model_partial_column_above'].interpolate()
match_df_time
# Drop unnecessary values
match_df_time = match_df_time.reset_index()
match_df_time = match_df_time.set_index(['time', 'ground_pixel', 'scanline', 'layer'])
match_df_time = match_df_time.drop(np.arange(1, 137) * 1000, level = 'layer')
match_df_time
# Calculate CAMS partial columns for each TM5 layer (as difference between the interpolated values)
match_df_time['model_column'] = match_df_time['model_partial_column_above'] - match_df_time['model_partial_column_above'].shift(-1)
match_df_time = match_df_time.reset_index()
match_df_time.loc[match_df_time['layer'] == 33, ['model_column']] = match_df_time['model_partial_column_above']
match_df_time = match_df_time.set_index(['time', 'ground_pixel', 'scanline', 'layer'])
match_df_time
# Apply the averaging kernels
if 'apriori_profile' in match_df_time.columns:
match_df_time['model_column'] = match_df_time.apply(lambda row: row['apriori_profile'] +
row['column_kernel'] * row['model_column'] -
row['column_kernel'] * row['apriori_profile'],
axis = 1)
else:
match_df_time['model_column'] = match_df_time.apply(lambda row: row['model_column'] *
row['column_kernel'],
axis = 1)
match_df_time = match_df_time[~match_df_time.index.duplicated()]
match_df_time
# Transform dataframe back to xarray
match_ds_time = match_df_time.to_xarray()
# Read latitudes and longitudes from data array
latitude = match_ds_time.latitude.mean(dim = 'layer')
longitude = match_ds_time.longitude.mean(dim = 'layer')
# Get sum of CAMS data of each layer to get column data
model_final_ds_time = match_ds_time.model_column.sum(dim = 'layer', skipna = False).astype(float)
model_final_ds_time = model_final_ds_time.assign_coords(latitude = latitude, longitude = longitude)
# Get mean of TROPOMI data of each layer (it must be equal)
sensor_final_ds_time = match_ds_time.sensor_column.mean(dim = 'layer', skipna = False).astype(float)
sensor_final_ds_time = sensor_final_ds_time.assign_coords(latitude = latitude, longitude = longitude)
# Calculate difference
merge_ds_time = xr.merge([model_final_ds_time, sensor_final_ds_time])
merge_ds_time['difference'] = merge_ds_time.model_column - merge_ds_time.sensor_column
merge_ds_time['relative_difference'] = (merge_ds_time.model_column - merge_ds_time.sensor_column)/merge_ds_time.sensor_column
# See results table
merge_df = merge_ds_time.to_dataframe()
merge_df = merge_df.reset_index().set_index(['ground_pixel', 'scanline', 'time'])
merge_df = merge_df[['latitude', 'longitude', 'model_column', 'sensor_column', 'difference', 'relative_difference']]
merge_df
merge_df.describe()
plot_dates = plot_period(sensor_ds, sensor_type)
plot_bbox = plot_extent(bbox)
# Choose distribution (aggregated, individual or animated)
plot_type = 'individual'
# Define range (original, equal, centered or manual)
range_type = 'equal'
vmin_manual, vmax_manual = None, None
vmin_manual_diff, vmax_manual_diff = None, None
# Define projection and colors
projection = ccrs.PlateCarree()
color_scale = ['turbo', 'turbo', 'coolwarm']
# Get and decide the best width and height of the B/W frame lines
options_height_lon = get_frame_possible_lengths(lon_min, lon_max)
options_width_lat = get_frame_possible_lengths(lat_min, lat_max)
width_lon = 0.2
height_lat = 0.2
# Add marker
regions_names = ('Barcelona')
coords_list = (41.37, 2.17)
bbox_list = None
visualize_model_vs_sensor(model, sensor, component_nom, units, merge_df, plot_dates, plot_bbox, 20, 1.05,
model_type, sensor_type, range_type, plot_type, projection,
color_scale, width_lon, height_lat, vmin_manual, vmax_manual,
vmin_manual_diff, vmax_manual_diff, bbox_list, coords_list, regions_names)
regions_names = ('Barcelona - 1', 'Barcelona - 2')
coords_list = (41.39, 2.15,
41.1, 1.65)
ymin = 0
ymax = 5*10**15
xticks = plot_dates
timeseries_table = timeseries(merge_df, component_nom, sensor, sensor_type, model,
plot_dates, units, ymin, ymax, xticks, regions_names, coords_list)
timeseries_table
show_seasons = False
extent_definition = 'bbox' # bbox or country
scatter_plot_type = 'individual' # aggregated or individual
lim_min = None
lim_max = None
summary = scatter_plot(merge_df, component_nom, units, sensor, plot_dates, 1.05,
extent_definition, show_seasons, scatter_plot_type, lim_min, lim_max, plot_bbox)
summary
| 0.697403 | 0.981347 |
```
%matplotlib inline
from ipywidgets import interact, FloatSlider, HBox, HTML, FloatText
from IPython.display import display
import matplotlib.pyplot as plt
import matplotlib
matplotlib.rc('font',size=18)
import numpy as np
import warnings
from hyperfet.devices import SCMOSFET,VO2,HyperFET
import hyperfet.approximations as appr
import hyperfet.extractions as extr
from hyperfet.references import si
vo2_params={
"rho_m":si("5e-4 ohm cm"),
"rho_i":si("80 ohm cm"),
"J_MIT":si("2e6 A/cm^2"),
"J_IMT":si(".55e4 A/cm^2"),
"V_met":0,
"L":si("8nm"),
"W":si("14nm"),
"T":si("14nm")
}
vo2=VO2(**vo2_params)
VDD=.5
fet=None
out=HTML()
@interact(VT0=FloatSlider(value=.35,min=0,max=1,step=.05,continuous_update=False,
description=r'$V_\mathrm{T0}$'),
W=FloatSlider(value=50,min=10,max=100,step=10,continuous_update=False,
description=r'$W \mathrm{[nm]}$'),
Cinv_vxo=FloatSlider(value=3000,min=1000,max=5000,step=400,continuous_update=False,
description=r'$C_\mathrm{inv}v_{x_o}$'),
SS=FloatSlider(value=.065,min=.05,max=.09,step=.005,continuous_update=False,
description=r'$SS$'),
alpha=FloatSlider(value=2.5,min=0,max=5,step=.5,continuous_update=False,
description=r'$\alpha$'),
beta=FloatSlider(value=1.8,min=0,max=4,step=.1,continuous_update=False,
description=r'$\beta$'),
VDD=FloatSlider(value=.5,min=.3,max=1,step=.05,continuous_update=False,
description=r'$V_\mathrm{DD}$'),
VDsats=FloatSlider(value=.1,min=.1,max=2,step=.1,continuous_update=False,
description=r'$V_\mathrm{DSATS}$'),
delta=FloatSlider(value=.1,min=0,max=.5,step=.1,continuous_update=False,
description=r'$\delta$'),
log10Gleak=FloatSlider(value=-8,min=-14,max=-5,step=1,continuous_update=False,
description=r'$\log G_\mathrm{leak}$')
)
def show_hf(VT0,W,Cinv_vxo,SS,alpha,beta,VDsats,VDD,delta,log10Gleak):
global fet
plt.figure(figsize=(12,6))
fet=SCMOSFET(
W=W*1e-9,Cinv_vxo=Cinv_vxo,
VT0=VT0,alpha=alpha,SS=SS,delta=delta,
VDsats=VDsats,beta=beta,Gleak=10**log10Gleak)
#shift=approx_shift(HyperFET(fet,vo2),VDD)
VD=np.array(VDD)
VG=np.linspace(0,VDD,500)
### PLOT 1
plt.subplot(131)
I=fet.ID(VD=VD,VG=VG)
plt.plot(VG,I/fet.W,'r')
hf=HyperFET(fet,vo2)
If,Ib=[np.ravel(i) for i in hf.I_double(VD=VD,VG=VG)]
plt.plot(VG[~np.isnan(If)],If[~np.isnan(If)]/fet.W,'b')
plt.plot(VG[~np.isnan(Ib)],Ib[~np.isnan(Ib)]/fet.W,'b')
apprVleft,apprVright=appr.Vleft(hf,VD),appr.Vright(hf,VD)
plt.plot(apprVleft,vo2.I_MIT/fet.W,'o')
plt.plot(apprVright,vo2.I_IMT/fet.W,'o')
plt.plot(VG,appr.lowerbranch(hf,VD,VG)/fet.W,'--')
plt.plot(VG,appr.lowernoleak(hf,VD,VG)/fet.W,'--')
plt.plot(VG,appr.upperbranchsubthresh(hf,VD,VG)/fet.W,'--')
plt.plot(VG,appr.upperbranchinversion(hf,VD,VG)/fet.W,'--')
plt.yscale('log')
plt.ylim(1e-2,1e3)
plt.xlabel("$V_{GS}\;\mathrm{[V]}$")
plt.ylabel("$I/W\;\mathrm{[mA/mm]}$")
plt.title('Approximations')
### PLOT 2
plt.subplot(132)
I=fet.ID(VD=VD,VG=VG)
plt.plot(VG,I/fet.W,'r')
hf=HyperFET(fet,vo2)
If,Ib=[np.ravel(i) for i in hf.I_double(VD=VD,VG=VG)]
plt.plot(VG[~np.isnan(If)],If[~np.isnan(If)]/fet.W,'b')
plt.plot(VG[~np.isnan(Ib)],Ib[~np.isnan(Ib)]/fet.W,'b')
Vl,Ill,Ilu=extr.left(VG,If,Ib)
Vr,Irl,Iru=extr.right(VG,If,Ib)
plt.plot(Vl,Ill/fet.W,'o')
plt.plot(Vl,Ilu/fet.W,'o')
plt.plot(Vr,Irl/fet.W,'o')
plt.plot(Vr,Iru/fet.W,'o')
plt.yscale('log')
plt.ylim(1e-2,1e3)
plt.xlabel("$V_{GS}\;\mathrm{[V]}$")
plt.ylabel("$I/W\;\mathrm{[mA/mm]}$")
plt.title('Extractions')
plt.tight_layout()
out.value=r"<strong>Approx Vleft</strong> {:.2g}, <strong>Approx Vright</strong> {:.2g}".format(apprVleft,apprVright)
display(out)
#display(HBox([Latex("Approx Vleft: "),box_apprVleft,Latex("Approx Vright "),box_apprVright]))
```
|
github_jupyter
|
%matplotlib inline
from ipywidgets import interact, FloatSlider, HBox, HTML, FloatText
from IPython.display import display
import matplotlib.pyplot as plt
import matplotlib
matplotlib.rc('font',size=18)
import numpy as np
import warnings
from hyperfet.devices import SCMOSFET,VO2,HyperFET
import hyperfet.approximations as appr
import hyperfet.extractions as extr
from hyperfet.references import si
vo2_params={
"rho_m":si("5e-4 ohm cm"),
"rho_i":si("80 ohm cm"),
"J_MIT":si("2e6 A/cm^2"),
"J_IMT":si(".55e4 A/cm^2"),
"V_met":0,
"L":si("8nm"),
"W":si("14nm"),
"T":si("14nm")
}
vo2=VO2(**vo2_params)
VDD=.5
fet=None
out=HTML()
@interact(VT0=FloatSlider(value=.35,min=0,max=1,step=.05,continuous_update=False,
description=r'$V_\mathrm{T0}$'),
W=FloatSlider(value=50,min=10,max=100,step=10,continuous_update=False,
description=r'$W \mathrm{[nm]}$'),
Cinv_vxo=FloatSlider(value=3000,min=1000,max=5000,step=400,continuous_update=False,
description=r'$C_\mathrm{inv}v_{x_o}$'),
SS=FloatSlider(value=.065,min=.05,max=.09,step=.005,continuous_update=False,
description=r'$SS$'),
alpha=FloatSlider(value=2.5,min=0,max=5,step=.5,continuous_update=False,
description=r'$\alpha$'),
beta=FloatSlider(value=1.8,min=0,max=4,step=.1,continuous_update=False,
description=r'$\beta$'),
VDD=FloatSlider(value=.5,min=.3,max=1,step=.05,continuous_update=False,
description=r'$V_\mathrm{DD}$'),
VDsats=FloatSlider(value=.1,min=.1,max=2,step=.1,continuous_update=False,
description=r'$V_\mathrm{DSATS}$'),
delta=FloatSlider(value=.1,min=0,max=.5,step=.1,continuous_update=False,
description=r'$\delta$'),
log10Gleak=FloatSlider(value=-8,min=-14,max=-5,step=1,continuous_update=False,
description=r'$\log G_\mathrm{leak}$')
)
def show_hf(VT0,W,Cinv_vxo,SS,alpha,beta,VDsats,VDD,delta,log10Gleak):
global fet
plt.figure(figsize=(12,6))
fet=SCMOSFET(
W=W*1e-9,Cinv_vxo=Cinv_vxo,
VT0=VT0,alpha=alpha,SS=SS,delta=delta,
VDsats=VDsats,beta=beta,Gleak=10**log10Gleak)
#shift=approx_shift(HyperFET(fet,vo2),VDD)
VD=np.array(VDD)
VG=np.linspace(0,VDD,500)
### PLOT 1
plt.subplot(131)
I=fet.ID(VD=VD,VG=VG)
plt.plot(VG,I/fet.W,'r')
hf=HyperFET(fet,vo2)
If,Ib=[np.ravel(i) for i in hf.I_double(VD=VD,VG=VG)]
plt.plot(VG[~np.isnan(If)],If[~np.isnan(If)]/fet.W,'b')
plt.plot(VG[~np.isnan(Ib)],Ib[~np.isnan(Ib)]/fet.W,'b')
apprVleft,apprVright=appr.Vleft(hf,VD),appr.Vright(hf,VD)
plt.plot(apprVleft,vo2.I_MIT/fet.W,'o')
plt.plot(apprVright,vo2.I_IMT/fet.W,'o')
plt.plot(VG,appr.lowerbranch(hf,VD,VG)/fet.W,'--')
plt.plot(VG,appr.lowernoleak(hf,VD,VG)/fet.W,'--')
plt.plot(VG,appr.upperbranchsubthresh(hf,VD,VG)/fet.W,'--')
plt.plot(VG,appr.upperbranchinversion(hf,VD,VG)/fet.W,'--')
plt.yscale('log')
plt.ylim(1e-2,1e3)
plt.xlabel("$V_{GS}\;\mathrm{[V]}$")
plt.ylabel("$I/W\;\mathrm{[mA/mm]}$")
plt.title('Approximations')
### PLOT 2
plt.subplot(132)
I=fet.ID(VD=VD,VG=VG)
plt.plot(VG,I/fet.W,'r')
hf=HyperFET(fet,vo2)
If,Ib=[np.ravel(i) for i in hf.I_double(VD=VD,VG=VG)]
plt.plot(VG[~np.isnan(If)],If[~np.isnan(If)]/fet.W,'b')
plt.plot(VG[~np.isnan(Ib)],Ib[~np.isnan(Ib)]/fet.W,'b')
Vl,Ill,Ilu=extr.left(VG,If,Ib)
Vr,Irl,Iru=extr.right(VG,If,Ib)
plt.plot(Vl,Ill/fet.W,'o')
plt.plot(Vl,Ilu/fet.W,'o')
plt.plot(Vr,Irl/fet.W,'o')
plt.plot(Vr,Iru/fet.W,'o')
plt.yscale('log')
plt.ylim(1e-2,1e3)
plt.xlabel("$V_{GS}\;\mathrm{[V]}$")
plt.ylabel("$I/W\;\mathrm{[mA/mm]}$")
plt.title('Extractions')
plt.tight_layout()
out.value=r"<strong>Approx Vleft</strong> {:.2g}, <strong>Approx Vright</strong> {:.2g}".format(apprVleft,apprVright)
display(out)
#display(HBox([Latex("Approx Vleft: "),box_apprVleft,Latex("Approx Vright "),box_apprVright]))
| 0.473657 | 0.361418 |
# Day and Night Image Classifier
---
The day/night image dataset consists of 200 RGB color images in two categories: day and night. There are equal numbers of each example: 100 day images and 100 night images.
We'd like to build a classifier that can accurately label these images as day or night, and that relies on finding distinguishing features between the two types of images!
*Note: All images come from the [AMOS dataset](http://cs.uky.edu/~jacobs/datasets/amos/) (Archive of Many Outdoor Scenes).*
### Import resources
Before you get started on the project code, import the libraries and resources that you'll need.
```
import cv2 # computer vision library
import helpers
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
%matplotlib inline
```
## Training and Testing Data
The 200 day/night images are separated into training and testing datasets.
* 60% of these images are training images, for you to use as you create a classifier.
* 40% are test images, which will be used to test the accuracy of your classifier.
First, we set some variables to keep track of some where our images are stored:
image_dir_training: the directory where our training image data is stored
image_dir_test: the directory where our test image data is stored
```
# Image data directories
image_dir_training = "day_night_images/training/"
image_dir_test = "day_night_images/test/"
```
## Load the datasets
These first few lines of code will load the training day/night images and store all of them in a variable, `IMAGE_LIST`. This list contains the images and their associated label ("day" or "night").
For example, the first image-label pair in `IMAGE_LIST` can be accessed by index:
``` IMAGE_LIST[0][:]```.
```
# Using the load_dataset function in helpers.py
# Load training data
IMAGE_LIST = helpers.load_dataset(image_dir_training)
```
---
# 1. Visualize the input images
```
# Print out 1. The shape of the image and 2. The image's label
# Select an image and its label by list index
image_index = 0
selected_image = IMAGE_LIST[image_index][0]
selected_label = IMAGE_LIST[image_index][1]
# Display image and data about it
plt.imshow(selected_image)
print("Shape: "+str(selected_image.shape))
print("Label: " + str(selected_label))
```
# 2. Pre-process the Data
After loading in each image, you have to standardize the input and output.
#### Solution code
You are encouraged to try to complete this code on your own, but if you are struggling or want to make sure your code is correct, there i solution code in the `helpers.py` file in this directory. You can look at that python file to see complete `standardize_input` and `encode` function code. For this day and night challenge, you can often jump one notebook ahead to see the solution code for a previous notebook!
---
### Input
It's important to make all your images the same size so that they can be sent through the same pipeline of classification steps! Every input image should be in the same format, of the same size, and so on.
#### TODO: Standardize the input images
* Resize each image to the desired input size: 600x1100px (hxw).
```
# This function should take in an RGB image and return a new, standardized version
def standardize_input(image):
## TODO: Resize image so that all "standard" images are the same size 600x1100 (hxw)
standard_im = cv2.resize(image,(1100,600))
return standard_im
```
### TODO: Standardize the output
With each loaded image, you also need to specify the expected output. For this, use binary numerical values 0/1 = night/day.
```
# Examples:
# encode("day") should return: 1
# encode("night") should return: 0
def encode(label):
numerical_val = 0
## TODO: complete the code to produce a numerical label
if label =='day':
numerical_val=1
return numerical_val
```
## Construct a `STANDARDIZED_LIST` of input images and output labels.
This function takes in a list of image-label pairs and outputs a **standardized** list of resized images and numerical labels.
This uses the functions you defined above to standardize the input and output, so those functions must be complete for this standardization to work!
```
def standardize(image_list):
# Empty image data array
standard_list = []
# Iterate through all the image-label pairs
for item in image_list:
image = item[0]
label = item[1]
# Standardize the image
standardized_im = standardize_input(image)
# Create a numerical label
binary_label = encode(label)
# Append the image, and it's one hot encoded label to the full, processed list of image data
standard_list.append((standardized_im, binary_label))
return standard_list
# Standardize all training images
STANDARDIZED_LIST = standardize(IMAGE_LIST)
```
## Visualize the standardized data
Display a standardized image from STANDARDIZED_LIST.
```
# Display a standardized image and its label
# Select an image by index
image_num = 0
selected_image = STANDARDIZED_LIST[image_num][0]
selected_label = STANDARDIZED_LIST[image_num][1]
# Display image and data about it
## TODO: Make sure the images have numerical labels and are of the same size
plt.imshow(selected_image)
print("Shape: "+str(selected_image.shape))
print("Label [1 = day, 0 = night]: " + str(selected_label))
```
|
github_jupyter
|
import cv2 # computer vision library
import helpers
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
%matplotlib inline
# Image data directories
image_dir_training = "day_night_images/training/"
image_dir_test = "day_night_images/test/"
# Using the load_dataset function in helpers.py
# Load training data
IMAGE_LIST = helpers.load_dataset(image_dir_training)
# Print out 1. The shape of the image and 2. The image's label
# Select an image and its label by list index
image_index = 0
selected_image = IMAGE_LIST[image_index][0]
selected_label = IMAGE_LIST[image_index][1]
# Display image and data about it
plt.imshow(selected_image)
print("Shape: "+str(selected_image.shape))
print("Label: " + str(selected_label))
# This function should take in an RGB image and return a new, standardized version
def standardize_input(image):
## TODO: Resize image so that all "standard" images are the same size 600x1100 (hxw)
standard_im = cv2.resize(image,(1100,600))
return standard_im
# Examples:
# encode("day") should return: 1
# encode("night") should return: 0
def encode(label):
numerical_val = 0
## TODO: complete the code to produce a numerical label
if label =='day':
numerical_val=1
return numerical_val
def standardize(image_list):
# Empty image data array
standard_list = []
# Iterate through all the image-label pairs
for item in image_list:
image = item[0]
label = item[1]
# Standardize the image
standardized_im = standardize_input(image)
# Create a numerical label
binary_label = encode(label)
# Append the image, and it's one hot encoded label to the full, processed list of image data
standard_list.append((standardized_im, binary_label))
return standard_list
# Standardize all training images
STANDARDIZED_LIST = standardize(IMAGE_LIST)
# Display a standardized image and its label
# Select an image by index
image_num = 0
selected_image = STANDARDIZED_LIST[image_num][0]
selected_label = STANDARDIZED_LIST[image_num][1]
# Display image and data about it
## TODO: Make sure the images have numerical labels and are of the same size
plt.imshow(selected_image)
print("Shape: "+str(selected_image.shape))
print("Label [1 = day, 0 = night]: " + str(selected_label))
| 0.459076 | 0.990768 |
```
from colors import *
from matplotlib import rc
rc('text', usetex=True)
rc('text.latex', preamble=[r'\usepackage{sansmath}', r'\sansmath']) #r'\usepackage{DejaVuSans}'
rc('font',**{'family':'sans-serif','sans-serif':['DejaVu Sans']})
rc('xtick.major', pad=12)
rc('ytick.major', pad=12)
rc('grid', linewidth=1.3)
import matplotlib.pyplot as plt
import numpy as np
np.random.seed(2)
num_samples = 1000
xx = np.random.normal(loc=0.0, scale=1.0, size=num_samples) + 1.0
train_xx = xx[:40]
test_xx = xx[40:]
def get_log_ml(x, alpha_2, mu):
N = len(x)
cov_mat = np.eye(N) + alpha_2 * np.ones((N, N))
inv_cov = np.eye(N) - np.ones((N, N)) / (1/alpha_2 + N)
log_ml = - N/2 * np.log(2 * np.pi) - 0.5 * np.log(np.linalg.det(cov_mat))
log_ml += -0.5 * np.matmul(np.matmul((x - mu).T, inv_cov), (x - mu))
return log_ml
def get_likelihood(xx_train, xx_test, alpha2):
N = len(xx_train)
map_mean = xx_train.mean() / (1/alpha2 + N)
log_lik = - N/2 * np.log(2 * np.pi) - 0.5 * ((xx_test - map_mean)**2).sum()
return log_lik
def get_pos_dist(xx_train, alpha_2, mu):
N = len(xx_train)
pos_mean = 1/ (1/alpha_2 + N) * (xx_train.sum() + mu / alpha_2)
pos_var = 1/ (1/alpha_2 + N)
return pos_mean, pos_var
def pred_dist(xx_train, alpha_2, mu):
N = len(xx_train)
pred_mean = (1/ (1/alpha_2 + N)) * (xx_train.sum() + mu / alpha_2)
pred_var = 1 + 1 / (1/alpha_2 + N)
return pred_mean, pred_var
def get_log_pred(x_test, x_train, alpha_2, mu):
# N = len(x_test)
# pred_mean, pred_var = pred_dist(x_train, alpha_2, mu)
# log_pred = - N/2 * np.log(2 * np.pi) - 0.5 * np.log(np.linalg.det(pred_var))
# log_pre += -0.5 * np.matmul(np.matmul((x_test - pred_var).T, np.linalg.inv(pred_var)), (x_test - pred_var))
# N = len(x_test)
# assert N==1
pred_mean, pred_var = pred_dist(x_train, alpha_2, mu)
# print(pred_mean, pred_var)
log_pred = - 0.5 * np.log(2 * np.pi) - 0.5 * np.log(pred_var)
log_pred -= 0.5/pred_var * ((x_test - pred_mean)**2)
return log_pred.sum()
import seaborn as sns
sns.set_style("whitegrid")
mu = 0.0
# mu = np.mean(train_xx) + 0.
mll = []
pos_means = []
pos_vars = []
pred_means = []
pred_vars = []
pred_ll = []
alpha_range = np.arange(0.1, 1000, 0.1)
# alpha_range = np.arange(0.1, 1, 0.01)
for alpha2 in alpha_range:
mll.append(get_log_ml(train_xx, alpha2, mu=mu))
pos_m, pos_v = get_pos_dist(train_xx, alpha2, mu)
pred_m, pred_v = pred_dist(train_xx, alpha2, mu)
pos_means.append(pos_m)
pos_vars.append(pos_v)
pred_means.append(pred_m)
pred_vars.append(pred_v)
# pred_ll.append(get_log_pred(test_xx[6], train_xx, alpha2, mu))
pred_ll.append(get_log_pred(test_xx, train_xx, alpha2, mu))
from matplotlib.ticker import StrMethodFormatter
fig, ax = plt.subplots(figsize=(3, 3))
cmap = sns.cubehelix_palette(as_cmap=True)
plt.plot(alpha_range, mll, color=color12, lw=3)
plt.xlabel(r'Prior variance $\sigma^2$',fontsize=18)
plt.ylabel(r'MLL',fontsize=18)
# ax.set_yticks([-79, -77, -75, -73])
plt.yticks(fontsize=14)
plt.xticks(fontsize=14)
plt.xscale("log")
# plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/mll.pdf', bbox_inches="tight")
plt.show()
from matplotlib.ticker import StrMethodFormatter
fig, ax = plt.subplots(figsize=(3, 3))
cmap = sns.cubehelix_palette(as_cmap=True)
plt.plot(alpha_range, pred_ll, color=color5, lw=3)
plt.xlabel(r'Prior variance $\sigma^2$',fontsize=18)
plt.ylabel(r'Test LL',fontsize=18)
# ax.set_yticks([-79, -77, -75, -73])
plt.yticks(fontsize=14)
plt.xticks(fontsize=14)
plt.xscale("log")
# plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/test_ll.pdf', bbox_inches="tight")
plt.show()
from matplotlib.ticker import StrMethodFormatter
fig, ax = plt.subplots(figsize=(3, 3))
cmap = sns.cubehelix_palette(as_cmap=True)
plt.plot(alpha_range, pred_means, label=r"Predictive $\mu$", color=color2, lw=3)
plt.xlabel(r'Prior variance $\sigma^2$',fontsize=18)
plt.ylabel(r'Predictive mean',fontsize=18)
# ax.set_yticks([-79, -77, -75, -73])
plt.yticks(fontsize=14)
plt.xticks(fontsize=14)
plt.xscale("log")
# plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/predictive_mu.pdf', bbox_inches="tight")
plt.show()
from matplotlib.ticker import StrMethodFormatter
fig, ax = plt.subplots(figsize=(3, 3))
cmap = sns.cubehelix_palette(as_cmap=True)
plt.plot(alpha_range, pred_vars, label=r"Predictive $\mu$", color=color3, lw=3)
plt.xlabel(r'Prior variance $\sigma^2$',fontsize=18)
plt.ylabel(r'Predictive variance',fontsize=18)
# ax.set_yticks([-79, -77, -75, -73])
plt.yticks(fontsize=14)
plt.xticks(fontsize=14)
plt.xscale("log")
# plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/predictive_sigma.pdf', bbox_inches="tight")
plt.show()
from matplotlib.ticker import StrMethodFormatter
fig, ax = plt.subplots(figsize=(2.2, 3))
cmap = sns.cubehelix_palette(as_cmap=True)
# color1 = sns.color_palette("Paired")[1]
# color2 = sns.color_palette("Paired")[5]
l1 = ax.plot(alpha_range, mll, label="LML", color=sanae_colors[4], lw=3)
ax.set_xlabel(r'Prior variance $\sigma^2$', fontsize=18)
ax.set_yticks([-61, -62, -63])
ax.set_ylim(-63.5, -60)
ax.set_ylabel("LML", fontsize=16, color=sanae_colors[5])
plt.yticks(color=sanae_colors[5])
ax2=ax.twinx()
l2 = ax2.plot(alpha_range, pred_ll, color=sanae_colors[1], label="Predictive LL", lw=3)
plt.gca().yaxis.set_major_formatter(StrMethodFormatter('{x:,.2f}')) # 2 decimal places
plt.yticks(color=sanae_colors[2])
lns = l1+l2
labs = [l.get_label() for l in lns]
ax2.set_yticks([-1380, -1379, -1378])
ax2.set_yticklabels(["-1380", "-1379", "-1378"], rotation=-45)
ax2.set_ylim(-1380.5, -1377)
ax2.set_ylabel("Test LL", fontsize=16, color=sanae_colors[2])
ax.set_xticks([0, 500, 1000])
ax.tick_params(axis='both', which='major', labelsize=14, pad=0)
ax2.tick_params(axis='both', which='major', labelsize=14, pad=0)
# ax2.legend(lns, labs, loc=1, prop={'size': 18}, framealpha=1.)
plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/gen_model_mll.pdf', bbox_inches="tight")
plt.show()
import scipy
fig, ax = plt.subplots(figsize=(4., 2))
# col1 = sns.color_palette("Paired")[7]
# col2 = sns.color_palette("Paired")[3]
val = 0.
plt.plot(train_xx, np.zeros_like(train_xx) + val, 'o', color=sanae_colors[1], markeredgecolor="k", ms=12)
x_values = np.arange(-2, 3.5, 0.1)
alpha_2 = 1.#1.e-16
# mu = np.mean(train_xx)
mll = np.round(get_log_ml(train_xx, alpha_2, mu),2)
print("mll, ", mll)
print("pred, ", pred_dist(train_xx, alpha_2, mu))
pred_m, pred_v = pred_dist(train_xx, alpha_2, mu)
y_values = scipy.stats.norm(pred_m, pred_v)
plt.plot(x_values, y_values.pdf(x_values),
label=r"$\mathcal{M}_{MML}$: LML=$-60.8$",
color="k", linestyle="dashed", lw=3, zorder=3)
alpha_2 = 1e6
mu = 1.0
mll = np.round(get_log_ml(train_xx, alpha_2, mu),2)
print("mll, ", mll)
print("pred, ", pred_dist(train_xx, alpha_2, mu))
pred_m, pred_v = pred_dist(train_xx, alpha_2, mu)
y_values = scipy.stats.norm(pred_m, pred_v)
# plt.plot(x_values, y_values.pdf(x_values),
# label=r"$\mathcal{M}_2: \sigma^2 = 10^6$, $\mu=1$, MLL=$-66.9$",
# color=color5, lw=3)
plt.fill_between(x_values,
y_values.pdf(x_values),
np.zeros_like(x_values),
# color=color5,
facecolor=sanae_colors[1],
edgecolor=sanae_colors[2],
label=r"$\mathcal{M}_1~ ^{\sigma^2 = 10^6}_{\mu=1}$, LML=$-66.9$",
alpha=0.6, lw=3)
alpha_2 = .07
mu = -0.4
mll = np.round(get_log_ml(train_xx, alpha_2, mu),2)
print("mll, ", mll)
print("pred, ", pred_dist(train_xx, alpha_2, mu))
pred_m, pred_v = pred_dist(train_xx, alpha_2, mu)
y_values = scipy.stats.norm(pred_m, pred_v)
# plt.plot(x_values, y_values.pdf(x_values),
# label=r"$\mathcal{M}_1: \sigma^2 = 0.07$, $\mu=2$, MLL=$-66.5$",
# color=color1, lw=3)
plt.fill_between(x_values,
y_values.pdf(x_values),
np.zeros_like(x_values),
facecolor=sanae_colors[4],
edgecolor=sanae_colors[5],
alpha=0.6, lw=3,
label=r"$\mathcal{M}_2~ ^{\sigma^2 = 0.07}_{\mu=-0.4}$, LML=$-66.3$",
)
plt.vlines(np.mean(train_xx), 0, 0.4, linestyle="dotted", color=color2, label="Empirical mean", lw=3)
plt.ylim(-0.05, 0.5)
ax.set_xlabel(r'$x$',fontsize=20)
# plt.plot([pred_m, pred_m], [0.0, y_values.pdf(x_values).max()], color=col2)
plt.xticks(fontsize=16)
plt.yticks(fontsize=16)
plt.legend(fontsize=19, framealpha=1., bbox_to_anchor=(-.18,2.3), loc="upper left") #(0.))
plt.ylabel(r"$p(x)$", fontsize=20)
# plt.tight_layout()
plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/gen_model_mismatch.pdf',
bbox_inches="tight")
plt.show()
```
## Learning curve
```
test_xx.mean()
train_xx
get_log_pred(test_xx, train_xx[:5], 1., train_xx.mean()) / len(test_xx)
get_log_pred(test_xx, train_xx[:0], 1.e6, 1.0) / len(test_xx)
get_log_pred(test_xx, train_xx[:0], .07, -.5) / len(test_xx)
def get_learning_curve(alpha_2, mu, n_orders=100):
order_lls = []
for _ in range(n_orders):
order = np.arange(len(train_xx))
np.random.shuffle(order)
order_lls.append(
np.array([get_log_pred(test_xx, train_xx[order[:i]], alpha_2, mu).copy() / len(test_xx)
for i in range(len(order))]))
return np.stack(order_lls)
# M_MML
alpha_2 = 1.#e-16
mu = np.mean(train_xx)
mml_learning_curve = get_learning_curve(alpha_2, mu).mean(axis=0)
# M_1
alpha_2 = 1e6
mu = 1.0
m1_learning_curve = get_learning_curve(alpha_2, mu).mean(axis=0)
# M_2
alpha_2 = .07
mu = -0.4
m2_learning_curve = get_learning_curve(alpha_2, mu).mean(axis=0)
# mml_learning_curve[0, :] - mml_learning_curve[1, :]
f = plt.figure(figsize=(3., 3.))
plt.plot(mml_learning_curve, "--k", label=r"$\mathcal{M}_{MML}$", lw=4, zorder=4)
plt.plot(m1_learning_curve, label=r"$\mathcal{M}_1$", lw=4, color=sanae_colors[2])
plt.plot(m2_learning_curve, label=r"$\mathcal{M}_2$", lw=4, color=sanae_colors[3])
leg = plt.legend(handlelength=2, fontsize=18, loc=4)
for legobj in leg.legendHandles:
legobj.set_linewidth(4)
f.get_axes()[0].tick_params(axis='both', which='major', labelsize=14)
plt.xlim(-1, 40)
plt.ylim(-4, -1)
plt.ylabel(r"$p(\mathcal{D}_n \vert \mathcal D_{<n})$", fontsize=18)
plt.xlabel(r"Number of datapoints, $n$", fontsize=18)
plt.savefig("../../Papers/marginal_likelihood/figures/generative_model/learning_curve.pdf",
bbox_inches="tight")
plt.figure(figsize=(3, 3))
plt.plot(mml_learning_curve, label="$\mathcal{M}_{MML}$", lw=3)
plt.plot(m1_learning_curve, label="$\mathcal{M}_1$", lw=3)
plt.plot(m2_learning_curve, label="$\mathcal{M}_2$", lw=3)
plt.legend(fontsize=16)
# plt.ylim(-.6, -.42)
plt.ylabel("$\log p(\mathcal{D}_n | \mathcal{D}_{<n}$)", fontsize=14)
plt.xlabel("Number of data points", fontsize=14)
m1_learning_curve
alpha_2 = 1.0
mu = 2.0
mll = np.round(get_log_ml(train_xx, alpha_2, mu),2)
print("mll, ", mll)
print("pred, ", pred_dist(train_xx, alpha_2, mu))
x_values = np.arange(-2, 6, 0.1)
pred_m, pred_v = pred_dist(train_xx, alpha_2, mu)
pred_m
alpha_mlls = {
alpha: [] for alpha in [0.6, 10., 1.e6]
}
all_mlls = []
n_orders = 100
for alpha_2 in alpha_mlls:
for order in range(n_orders):
order = np.arange(len(train_xx))
np.random.shuffle(order)
x_ = train_xx[order]
mlls = np.array([
get_log_ml(x_[:i], alpha_2, 0.) for i in range(1, len(x_))])
alpha_mlls[alpha_2].append(mlls)
alpha_mlls[alpha_2] = np.array(alpha_mlls[alpha_2])
np.savez("data/mll_gen.npz",
**{str(k): v for (k, v) in alpha_mlls.items()})
alpha_mlls[0.6].shape
```
|
github_jupyter
|
from colors import *
from matplotlib import rc
rc('text', usetex=True)
rc('text.latex', preamble=[r'\usepackage{sansmath}', r'\sansmath']) #r'\usepackage{DejaVuSans}'
rc('font',**{'family':'sans-serif','sans-serif':['DejaVu Sans']})
rc('xtick.major', pad=12)
rc('ytick.major', pad=12)
rc('grid', linewidth=1.3)
import matplotlib.pyplot as plt
import numpy as np
np.random.seed(2)
num_samples = 1000
xx = np.random.normal(loc=0.0, scale=1.0, size=num_samples) + 1.0
train_xx = xx[:40]
test_xx = xx[40:]
def get_log_ml(x, alpha_2, mu):
N = len(x)
cov_mat = np.eye(N) + alpha_2 * np.ones((N, N))
inv_cov = np.eye(N) - np.ones((N, N)) / (1/alpha_2 + N)
log_ml = - N/2 * np.log(2 * np.pi) - 0.5 * np.log(np.linalg.det(cov_mat))
log_ml += -0.5 * np.matmul(np.matmul((x - mu).T, inv_cov), (x - mu))
return log_ml
def get_likelihood(xx_train, xx_test, alpha2):
N = len(xx_train)
map_mean = xx_train.mean() / (1/alpha2 + N)
log_lik = - N/2 * np.log(2 * np.pi) - 0.5 * ((xx_test - map_mean)**2).sum()
return log_lik
def get_pos_dist(xx_train, alpha_2, mu):
N = len(xx_train)
pos_mean = 1/ (1/alpha_2 + N) * (xx_train.sum() + mu / alpha_2)
pos_var = 1/ (1/alpha_2 + N)
return pos_mean, pos_var
def pred_dist(xx_train, alpha_2, mu):
N = len(xx_train)
pred_mean = (1/ (1/alpha_2 + N)) * (xx_train.sum() + mu / alpha_2)
pred_var = 1 + 1 / (1/alpha_2 + N)
return pred_mean, pred_var
def get_log_pred(x_test, x_train, alpha_2, mu):
# N = len(x_test)
# pred_mean, pred_var = pred_dist(x_train, alpha_2, mu)
# log_pred = - N/2 * np.log(2 * np.pi) - 0.5 * np.log(np.linalg.det(pred_var))
# log_pre += -0.5 * np.matmul(np.matmul((x_test - pred_var).T, np.linalg.inv(pred_var)), (x_test - pred_var))
# N = len(x_test)
# assert N==1
pred_mean, pred_var = pred_dist(x_train, alpha_2, mu)
# print(pred_mean, pred_var)
log_pred = - 0.5 * np.log(2 * np.pi) - 0.5 * np.log(pred_var)
log_pred -= 0.5/pred_var * ((x_test - pred_mean)**2)
return log_pred.sum()
import seaborn as sns
sns.set_style("whitegrid")
mu = 0.0
# mu = np.mean(train_xx) + 0.
mll = []
pos_means = []
pos_vars = []
pred_means = []
pred_vars = []
pred_ll = []
alpha_range = np.arange(0.1, 1000, 0.1)
# alpha_range = np.arange(0.1, 1, 0.01)
for alpha2 in alpha_range:
mll.append(get_log_ml(train_xx, alpha2, mu=mu))
pos_m, pos_v = get_pos_dist(train_xx, alpha2, mu)
pred_m, pred_v = pred_dist(train_xx, alpha2, mu)
pos_means.append(pos_m)
pos_vars.append(pos_v)
pred_means.append(pred_m)
pred_vars.append(pred_v)
# pred_ll.append(get_log_pred(test_xx[6], train_xx, alpha2, mu))
pred_ll.append(get_log_pred(test_xx, train_xx, alpha2, mu))
from matplotlib.ticker import StrMethodFormatter
fig, ax = plt.subplots(figsize=(3, 3))
cmap = sns.cubehelix_palette(as_cmap=True)
plt.plot(alpha_range, mll, color=color12, lw=3)
plt.xlabel(r'Prior variance $\sigma^2$',fontsize=18)
plt.ylabel(r'MLL',fontsize=18)
# ax.set_yticks([-79, -77, -75, -73])
plt.yticks(fontsize=14)
plt.xticks(fontsize=14)
plt.xscale("log")
# plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/mll.pdf', bbox_inches="tight")
plt.show()
from matplotlib.ticker import StrMethodFormatter
fig, ax = plt.subplots(figsize=(3, 3))
cmap = sns.cubehelix_palette(as_cmap=True)
plt.plot(alpha_range, pred_ll, color=color5, lw=3)
plt.xlabel(r'Prior variance $\sigma^2$',fontsize=18)
plt.ylabel(r'Test LL',fontsize=18)
# ax.set_yticks([-79, -77, -75, -73])
plt.yticks(fontsize=14)
plt.xticks(fontsize=14)
plt.xscale("log")
# plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/test_ll.pdf', bbox_inches="tight")
plt.show()
from matplotlib.ticker import StrMethodFormatter
fig, ax = plt.subplots(figsize=(3, 3))
cmap = sns.cubehelix_palette(as_cmap=True)
plt.plot(alpha_range, pred_means, label=r"Predictive $\mu$", color=color2, lw=3)
plt.xlabel(r'Prior variance $\sigma^2$',fontsize=18)
plt.ylabel(r'Predictive mean',fontsize=18)
# ax.set_yticks([-79, -77, -75, -73])
plt.yticks(fontsize=14)
plt.xticks(fontsize=14)
plt.xscale("log")
# plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/predictive_mu.pdf', bbox_inches="tight")
plt.show()
from matplotlib.ticker import StrMethodFormatter
fig, ax = plt.subplots(figsize=(3, 3))
cmap = sns.cubehelix_palette(as_cmap=True)
plt.plot(alpha_range, pred_vars, label=r"Predictive $\mu$", color=color3, lw=3)
plt.xlabel(r'Prior variance $\sigma^2$',fontsize=18)
plt.ylabel(r'Predictive variance',fontsize=18)
# ax.set_yticks([-79, -77, -75, -73])
plt.yticks(fontsize=14)
plt.xticks(fontsize=14)
plt.xscale("log")
# plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/predictive_sigma.pdf', bbox_inches="tight")
plt.show()
from matplotlib.ticker import StrMethodFormatter
fig, ax = plt.subplots(figsize=(2.2, 3))
cmap = sns.cubehelix_palette(as_cmap=True)
# color1 = sns.color_palette("Paired")[1]
# color2 = sns.color_palette("Paired")[5]
l1 = ax.plot(alpha_range, mll, label="LML", color=sanae_colors[4], lw=3)
ax.set_xlabel(r'Prior variance $\sigma^2$', fontsize=18)
ax.set_yticks([-61, -62, -63])
ax.set_ylim(-63.5, -60)
ax.set_ylabel("LML", fontsize=16, color=sanae_colors[5])
plt.yticks(color=sanae_colors[5])
ax2=ax.twinx()
l2 = ax2.plot(alpha_range, pred_ll, color=sanae_colors[1], label="Predictive LL", lw=3)
plt.gca().yaxis.set_major_formatter(StrMethodFormatter('{x:,.2f}')) # 2 decimal places
plt.yticks(color=sanae_colors[2])
lns = l1+l2
labs = [l.get_label() for l in lns]
ax2.set_yticks([-1380, -1379, -1378])
ax2.set_yticklabels(["-1380", "-1379", "-1378"], rotation=-45)
ax2.set_ylim(-1380.5, -1377)
ax2.set_ylabel("Test LL", fontsize=16, color=sanae_colors[2])
ax.set_xticks([0, 500, 1000])
ax.tick_params(axis='both', which='major', labelsize=14, pad=0)
ax2.tick_params(axis='both', which='major', labelsize=14, pad=0)
# ax2.legend(lns, labs, loc=1, prop={'size': 18}, framealpha=1.)
plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/gen_model_mll.pdf', bbox_inches="tight")
plt.show()
import scipy
fig, ax = plt.subplots(figsize=(4., 2))
# col1 = sns.color_palette("Paired")[7]
# col2 = sns.color_palette("Paired")[3]
val = 0.
plt.plot(train_xx, np.zeros_like(train_xx) + val, 'o', color=sanae_colors[1], markeredgecolor="k", ms=12)
x_values = np.arange(-2, 3.5, 0.1)
alpha_2 = 1.#1.e-16
# mu = np.mean(train_xx)
mll = np.round(get_log_ml(train_xx, alpha_2, mu),2)
print("mll, ", mll)
print("pred, ", pred_dist(train_xx, alpha_2, mu))
pred_m, pred_v = pred_dist(train_xx, alpha_2, mu)
y_values = scipy.stats.norm(pred_m, pred_v)
plt.plot(x_values, y_values.pdf(x_values),
label=r"$\mathcal{M}_{MML}$: LML=$-60.8$",
color="k", linestyle="dashed", lw=3, zorder=3)
alpha_2 = 1e6
mu = 1.0
mll = np.round(get_log_ml(train_xx, alpha_2, mu),2)
print("mll, ", mll)
print("pred, ", pred_dist(train_xx, alpha_2, mu))
pred_m, pred_v = pred_dist(train_xx, alpha_2, mu)
y_values = scipy.stats.norm(pred_m, pred_v)
# plt.plot(x_values, y_values.pdf(x_values),
# label=r"$\mathcal{M}_2: \sigma^2 = 10^6$, $\mu=1$, MLL=$-66.9$",
# color=color5, lw=3)
plt.fill_between(x_values,
y_values.pdf(x_values),
np.zeros_like(x_values),
# color=color5,
facecolor=sanae_colors[1],
edgecolor=sanae_colors[2],
label=r"$\mathcal{M}_1~ ^{\sigma^2 = 10^6}_{\mu=1}$, LML=$-66.9$",
alpha=0.6, lw=3)
alpha_2 = .07
mu = -0.4
mll = np.round(get_log_ml(train_xx, alpha_2, mu),2)
print("mll, ", mll)
print("pred, ", pred_dist(train_xx, alpha_2, mu))
pred_m, pred_v = pred_dist(train_xx, alpha_2, mu)
y_values = scipy.stats.norm(pred_m, pred_v)
# plt.plot(x_values, y_values.pdf(x_values),
# label=r"$\mathcal{M}_1: \sigma^2 = 0.07$, $\mu=2$, MLL=$-66.5$",
# color=color1, lw=3)
plt.fill_between(x_values,
y_values.pdf(x_values),
np.zeros_like(x_values),
facecolor=sanae_colors[4],
edgecolor=sanae_colors[5],
alpha=0.6, lw=3,
label=r"$\mathcal{M}_2~ ^{\sigma^2 = 0.07}_{\mu=-0.4}$, LML=$-66.3$",
)
plt.vlines(np.mean(train_xx), 0, 0.4, linestyle="dotted", color=color2, label="Empirical mean", lw=3)
plt.ylim(-0.05, 0.5)
ax.set_xlabel(r'$x$',fontsize=20)
# plt.plot([pred_m, pred_m], [0.0, y_values.pdf(x_values).max()], color=col2)
plt.xticks(fontsize=16)
plt.yticks(fontsize=16)
plt.legend(fontsize=19, framealpha=1., bbox_to_anchor=(-.18,2.3), loc="upper left") #(0.))
plt.ylabel(r"$p(x)$", fontsize=20)
# plt.tight_layout()
plt.savefig('../../Papers/marginal_likelihood/figures/generative_model/gen_model_mismatch.pdf',
bbox_inches="tight")
plt.show()
test_xx.mean()
train_xx
get_log_pred(test_xx, train_xx[:5], 1., train_xx.mean()) / len(test_xx)
get_log_pred(test_xx, train_xx[:0], 1.e6, 1.0) / len(test_xx)
get_log_pred(test_xx, train_xx[:0], .07, -.5) / len(test_xx)
def get_learning_curve(alpha_2, mu, n_orders=100):
order_lls = []
for _ in range(n_orders):
order = np.arange(len(train_xx))
np.random.shuffle(order)
order_lls.append(
np.array([get_log_pred(test_xx, train_xx[order[:i]], alpha_2, mu).copy() / len(test_xx)
for i in range(len(order))]))
return np.stack(order_lls)
# M_MML
alpha_2 = 1.#e-16
mu = np.mean(train_xx)
mml_learning_curve = get_learning_curve(alpha_2, mu).mean(axis=0)
# M_1
alpha_2 = 1e6
mu = 1.0
m1_learning_curve = get_learning_curve(alpha_2, mu).mean(axis=0)
# M_2
alpha_2 = .07
mu = -0.4
m2_learning_curve = get_learning_curve(alpha_2, mu).mean(axis=0)
# mml_learning_curve[0, :] - mml_learning_curve[1, :]
f = plt.figure(figsize=(3., 3.))
plt.plot(mml_learning_curve, "--k", label=r"$\mathcal{M}_{MML}$", lw=4, zorder=4)
plt.plot(m1_learning_curve, label=r"$\mathcal{M}_1$", lw=4, color=sanae_colors[2])
plt.plot(m2_learning_curve, label=r"$\mathcal{M}_2$", lw=4, color=sanae_colors[3])
leg = plt.legend(handlelength=2, fontsize=18, loc=4)
for legobj in leg.legendHandles:
legobj.set_linewidth(4)
f.get_axes()[0].tick_params(axis='both', which='major', labelsize=14)
plt.xlim(-1, 40)
plt.ylim(-4, -1)
plt.ylabel(r"$p(\mathcal{D}_n \vert \mathcal D_{<n})$", fontsize=18)
plt.xlabel(r"Number of datapoints, $n$", fontsize=18)
plt.savefig("../../Papers/marginal_likelihood/figures/generative_model/learning_curve.pdf",
bbox_inches="tight")
plt.figure(figsize=(3, 3))
plt.plot(mml_learning_curve, label="$\mathcal{M}_{MML}$", lw=3)
plt.plot(m1_learning_curve, label="$\mathcal{M}_1$", lw=3)
plt.plot(m2_learning_curve, label="$\mathcal{M}_2$", lw=3)
plt.legend(fontsize=16)
# plt.ylim(-.6, -.42)
plt.ylabel("$\log p(\mathcal{D}_n | \mathcal{D}_{<n}$)", fontsize=14)
plt.xlabel("Number of data points", fontsize=14)
m1_learning_curve
alpha_2 = 1.0
mu = 2.0
mll = np.round(get_log_ml(train_xx, alpha_2, mu),2)
print("mll, ", mll)
print("pred, ", pred_dist(train_xx, alpha_2, mu))
x_values = np.arange(-2, 6, 0.1)
pred_m, pred_v = pred_dist(train_xx, alpha_2, mu)
pred_m
alpha_mlls = {
alpha: [] for alpha in [0.6, 10., 1.e6]
}
all_mlls = []
n_orders = 100
for alpha_2 in alpha_mlls:
for order in range(n_orders):
order = np.arange(len(train_xx))
np.random.shuffle(order)
x_ = train_xx[order]
mlls = np.array([
get_log_ml(x_[:i], alpha_2, 0.) for i in range(1, len(x_))])
alpha_mlls[alpha_2].append(mlls)
alpha_mlls[alpha_2] = np.array(alpha_mlls[alpha_2])
np.savez("data/mll_gen.npz",
**{str(k): v for (k, v) in alpha_mlls.items()})
alpha_mlls[0.6].shape
| 0.531209 | 0.532911 |
## Tabular data preprocessing
```
from fastai.gen_doc.nbdoc import *
from fastai.tabular import *
```
## Overview
This package contains the basic class to define a transformation for preprocessing dataframes of tabular data, as well as basic [`TabularProc`](/tabular.transform.html#TabularProc). Preprocessing includes things like
- replacing non-numerical variables by categories, then their ids,
- filling missing values,
- normalizing continuous variables.
In all those steps we have to be careful to use the correspondence we decide on our training set (which id we give to each category, what is the value we put for missing data, or how the mean/std we use to normalize) on our validation or test set. To deal with this, we use a special class called [`TabularProc`](/tabular.transform.html#TabularProc).
The data used in this document page is a subset of the [adult dataset](https://archive.ics.uci.edu/ml/datasets/adult). It gives a certain amount of data on individuals to train a model to predict whether their salary is greater than \$50k or not.
```
path = untar_data(URLs.ADULT_SAMPLE)
df = pd.read_csv(path/'adult.csv')
train_df, valid_df = df.iloc[:800].copy(), df.iloc[800:1000].copy()
train_df.head()
```
We see it contains numerical variables (like `age` or `education-num`) as well as categorical ones (like `workclass` or `relationship`). The original dataset is clean, but we removed a few values to give examples of dealing with missing variables.
```
cat_names = ['workclass', 'education', 'marital-status', 'occupation', 'relationship', 'race', 'sex', 'native-country']
cont_names = ['age', 'fnlwgt', 'education-num', 'capital-gain', 'capital-loss', 'hours-per-week']
```
## Transforms for tabular data
```
show_doc(TabularProc)
```
Base class for creating transforms for dataframes with categorical variables `cat_names` and continuous variables `cont_names`. Note that any column not in one of those lists won't be touched.
```
show_doc(TabularProc.__call__)
show_doc(TabularProc.apply_train)
show_doc(TabularProc.apply_test)
jekyll_important("Those two functions must be implemented in a subclass. `apply_test` defaults to `apply_train`.")
```
The following [`TabularProc`](/tabular.transform.html#TabularProc) are implemented in the fastai library. Note that the replacement from categories to codes as well as the normalization of continuous variables are automatically done in a [`TabularDataBunch`](/tabular.data.html#TabularDataBunch).
```
show_doc(Categorify)
```
Variables in `cont_names` aren't affected.
```
show_doc(Categorify.apply_train)
show_doc(Categorify.apply_test)
tfm = Categorify(cat_names, cont_names)
tfm(train_df)
tfm(valid_df, test=True)
```
Since we haven't changed the categories by their codes, nothing visible has changed in the dataframe yet, but we can check that the variables are now categorical and view their corresponding codes.
```
train_df['workclass'].cat.categories
```
The test set will be given the same category codes as the training set.
```
valid_df['workclass'].cat.categories
show_doc(FillMissing)
```
`cat_names` variables are left untouched (their missing value will be replaced by code 0 in the [`TabularDataBunch`](/tabular.data.html#TabularDataBunch)). [`fill_strategy`](#FillStrategy) is adopted to replace those nans and if `add_col` is True, whenever a column `c` has missing values, a column named `c_nan` is added and flags the line where the value was missing.
```
show_doc(FillMissing.apply_train)
show_doc(FillMissing.apply_test)
```
Fills the missing values in the `cont_names` columns with the ones picked during train.
```
train_df[cont_names].head()
tfm = FillMissing(cat_names, cont_names)
tfm(train_df)
tfm(valid_df, test=True)
train_df[cont_names].head()
```
Values missing in the `education-num` column are replaced by 10, which is the median of the column in `train_df`. Categorical variables are not changed, since `nan` is simply used as another category.
```
valid_df[cont_names].head()
show_doc(FillStrategy, alt_doc_string='Enum flag represents determines how `FillMissing` should handle missing/nan values', arg_comments={
'MEDIAN':'nans are replaced by the median value of the column',
'COMMON': 'nans are replaced by the most common value of the column',
'CONSTANT': 'nans are replaced by `fill_val`'
})
show_doc(Normalize)
show_doc(Normalize.apply_train)
show_doc(Normalize.apply_test)
```
## Treating date columns
```
show_doc(add_datepart)
```
Will `drop` the column in `df` if the flag is `True`. The `time` flag decides if we go down to the time parts or stick to the date parts.
## Splitting data into cat and cont
```
show_doc(cont_cat_split)
```
Parameters:
- df: A pandas data frame.
- max_card: Maximum cardinality of a numerical categorical variable.
- dep_var: A dependent variable.
Return:
- cont_names: A list of names of continuous variables.
- cat_names: A list of names of categorical variables.
```
df = pd.DataFrame({'col1': [1, 2, 3], 'col2': ['a', 'b', 'a'], 'col3': [0.5, 1.2, 7.5], 'col4': ['ab', 'o', 'o']})
df
cont_list, cat_list = cont_cat_split(df=df, max_card=20, dep_var='col4')
cont_list, cat_list
```
## Undocumented Methods - Methods moved below this line will intentionally be hidden
## New Methods - Please document or move to the undocumented section
|
github_jupyter
|
from fastai.gen_doc.nbdoc import *
from fastai.tabular import *
path = untar_data(URLs.ADULT_SAMPLE)
df = pd.read_csv(path/'adult.csv')
train_df, valid_df = df.iloc[:800].copy(), df.iloc[800:1000].copy()
train_df.head()
cat_names = ['workclass', 'education', 'marital-status', 'occupation', 'relationship', 'race', 'sex', 'native-country']
cont_names = ['age', 'fnlwgt', 'education-num', 'capital-gain', 'capital-loss', 'hours-per-week']
show_doc(TabularProc)
show_doc(TabularProc.__call__)
show_doc(TabularProc.apply_train)
show_doc(TabularProc.apply_test)
jekyll_important("Those two functions must be implemented in a subclass. `apply_test` defaults to `apply_train`.")
show_doc(Categorify)
show_doc(Categorify.apply_train)
show_doc(Categorify.apply_test)
tfm = Categorify(cat_names, cont_names)
tfm(train_df)
tfm(valid_df, test=True)
train_df['workclass'].cat.categories
valid_df['workclass'].cat.categories
show_doc(FillMissing)
show_doc(FillMissing.apply_train)
show_doc(FillMissing.apply_test)
train_df[cont_names].head()
tfm = FillMissing(cat_names, cont_names)
tfm(train_df)
tfm(valid_df, test=True)
train_df[cont_names].head()
valid_df[cont_names].head()
show_doc(FillStrategy, alt_doc_string='Enum flag represents determines how `FillMissing` should handle missing/nan values', arg_comments={
'MEDIAN':'nans are replaced by the median value of the column',
'COMMON': 'nans are replaced by the most common value of the column',
'CONSTANT': 'nans are replaced by `fill_val`'
})
show_doc(Normalize)
show_doc(Normalize.apply_train)
show_doc(Normalize.apply_test)
show_doc(add_datepart)
show_doc(cont_cat_split)
df = pd.DataFrame({'col1': [1, 2, 3], 'col2': ['a', 'b', 'a'], 'col3': [0.5, 1.2, 7.5], 'col4': ['ab', 'o', 'o']})
df
cont_list, cat_list = cont_cat_split(df=df, max_card=20, dep_var='col4')
cont_list, cat_list
| 0.726037 | 0.983375 |
```
import numpy as np
import matplotlib.pyplot as plt
% matplotlib inline
plt.rcParams["savefig.dpi"] = 300
plt.rcParams["savefig.bbox"] = "tight"
np.set_printoptions(precision=3, suppress=True)
import pandas as pd
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import scale, StandardScaler
from sklearn.neural_network import MLPClassifier
from sklearn.datasets import make_moons
X, y = make_moons(n_samples=100, noise=0.25, random_state=2)
X_train, X_test, y_train, y_test = train_test_split(X, y, stratify=y,
random_state=42)
plt.scatter(X_train[:, 0], X_train[:, 1], c=plt.cm.Vega10(y_train))
xlim = plt.xlim()
ylim = plt.ylim()
xs = np.linspace(xlim[0], xlim[1], 1000)
ys = np.linspace(ylim[0], ylim[1], 1000)
xx, yy = np.meshgrid(xs, ys)
X_grid = np.c_[xx.ravel(), yy.ravel()]
mlp = MLPClassifier(solver='lbfgs', random_state=0).fit(X_train, y_train)
print(mlp.score(X_train, y_train))
print(mlp.score(X_test, y_test))
plt.contour(xx, yy, mlp.predict_proba(X_grid)[:, 1].reshape(xx.shape), levels=[.5])
plt.scatter(X_train[:, 0], X_train[:, 1], c=plt.cm.Vega10(y_train))
plt.xlim(xlim)
plt.ylim(ylim)
fig, axes = plt.subplots(3, 3, figsize=(8, 5))
for ax, i in zip(axes.ravel(), range(10)):
mlp = MLPClassifier(solver='lbfgs', random_state=i).fit(X_train, y_train)
print(mlp.score(X_train, y_train))
print(mlp.score(X_test, y_test))
ax.contour(xx, yy, mlp.predict_proba(X_grid)[:, 1].reshape(xx.shape), levels=[.5])
ax.scatter(X_train[:, 0], X_train[:, 1], c=plt.cm.Vega10(y_train))
ax.set_xlim(xlim)
ax.set_ylim(ylim)
ax.set_xticks(())
ax.set_yticks(())
mlp = MLPClassifier(solver='lbfgs', hidden_layer_sizes=(10, 10, 10), random_state=0)
mlp.fit(X_train, y_train)
print(mlp.score(X_train, y_train))
print(mlp.score(X_test, y_test))
plt.contour(xx, yy, mlp.predict_proba(X_grid)[:, 1].reshape(xx.shape), levels=[.5])
plt.scatter(X_train[:, 0], X_train[:, 1], c=plt.cm.Vega10(y_train))
plt.xlim(xlim)
plt.ylim(ylim)
mlp = MLPClassifier(solver='lbfgs', hidden_layer_sizes=(10, 10, 10), activation="tanh", random_state=0)
mlp.fit(X_train, y_train)
print(mlp.score(X_train, y_train))
print(mlp.score(X_test, y_test))
plt.contour(xx, yy, mlp.predict_proba(X_grid)[:, 1].reshape(xx.shape), levels=[.5])
plt.scatter(X_train[:, 0], X_train[:, 1], c=plt.cm.Vega10(y_train))
plt.xlim(xlim)
plt.ylim(ylim)
rng = np.random.RandomState(0)
x = np.sort(rng.uniform(size=100))
y = np.sin(10 * x) + 5 * x + np.random.normal(0, .3, size=100)
plt.plot(x, y, 'o')
line = np.linspace(0, 1, 100)
X = x.reshape(-1, 1)
from sklearn.neural_network import MLPRegressor
mlp_relu = MLPRegressor(solver="lbfgs").fit(X, y)
mlp_tanh = MLPRegressor(solver="lbfgs", activation='tanh').fit(X, y)
plt.plot(x, y, 'o')
plt.plot(line, mlp_relu.predict(line.reshape(-1, 1)), label="relu")
plt.plot(line, mlp_tanh.predict(line.reshape(-1, 1)), label="tanh")
plt.legend()
from sklearn.datasets import load_digits
digits = load_digits()
X_train, X_test, y_train, y_test = train_test_split(
digits.data / 16., digits.target, stratify=digits.target, random_state=0)
mlp = MLPClassifier(max_iter=1000, random_state=0).fit(X_train, y_train)
print(mlp.score(X_train, y_train))
print(mlp.score(X_test, y_test))
from sklearn.datasets import load_breast_cancer
data = load_breast_cancer()
X_train, X_test, y_train, y_test = train_test_split(
data.data, data.target, stratify=data.target, random_state=0)
scaler = StandardScaler().fit(X_train)
X_train_scaled = scaler.transform(X_train)
X_test_scaled = scaler.transform(X_test)
mlp = MLPClassifier(max_iter=1000, random_state=0).fit(X_train_scaled, y_train)
print(mlp.score(X_train_scaled, y_train))
print(mlp.score(X_test_scaled, y_test))
mlp = MLPClassifier(solver="lbfgs", random_state=1).fit(X_train_scaled, y_train)
print(mlp.score(X_train_scaled, y_train))
print(mlp.score(X_test_scaled, y_test))
from sklearn.model_selection import GridSearchCV
pipe = make_pipeline(StandardScaler(), MLPClassifier(solver="lbfgs", random_state=1))
param_grid = {'mlpclassifier__alpha': np.logspace(-3, 3, 7)}
grid = GridSearchCV(pipe, param_grid, cv=5, return_train_score=True)
grid.fit(X_train, y_train)
results = pd.DataFrame(grid.cv_results_)
res = results.pivot_table(index="param_mlpclassifier__alpha",
values=["mean_test_score", "mean_train_score"])
res
res.plot()
plt.xscale("log")
plt.ylim(0.95, 1.01)
res = results.pivot_table(index="param_mlpclassifier__alpha", values=["mean_test_score", "mean_train_score", "std_test_score", "std_train_score"])
res.mean_test_score.plot(yerr=res.std_test_score)
res.mean_train_score.plot(yerr=res.std_train_score)
plt.xscale("log")
plt.ylim(0.95, 1.01)
plt.legend()
from sklearn.model_selection import GridSearchCV
pipe = make_pipeline(StandardScaler(), MLPClassifier(solver="lbfgs", random_state=1))
param_grid = {'mlpclassifier__hidden_layer_sizes':
[(10,), (50,), (100,), (500,), (10, 10), (50, 50), (100, 100), (500, 500)]
}
grid = GridSearchCV(pipe, param_grid, cv=5)
grid.fit(X_train, y_train)
results = pd.DataFrame(grid.cv_results_)
res = results.pivot_table(index="param_mlpclassifier__hidden_layer_sizes", values=["mean_test_score", "mean_train_score", "std_test_score", "std_train_score"])
res.mean_test_score.plot(yerr=res.std_test_score)
res.mean_train_score.plot(yerr=res.std_train_score)
plt.legend()
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.model_selection import GridSearchCV
pipe = make_pipeline(StandardScaler(), MLPClassifier(solver="lbfgs", random_state=1))
param_grid = {'mlpclassifier__hidden_layer_sizes':
[(10,), (25,), (50,), (100,) , (10, 10), (25, 25), (50, 50)]
}
grid = GridSearchCV(pipe, param_grid, return_train_score=True)
grid.fit(X_train, y_train)
results = pd.DataFrame(grid.cv_results_)
res = results.pivot_table(index="param_mlpclassifier__hidden_layer_sizes", values=["mean_test_score", "mean_train_score", "std_test_score", "std_train_score"])
res.mean_test_score.plot(yerr=res.std_test_score)
res.mean_train_score.plot(yerr=res.std_train_score)
plt.legend()
mlp = MLPClassifier(solver="lbfgs", hidden_layer_sizes=(2,), random_state=0).fit(X_train_scaled, y_train)
print(mlp.score(X_train_scaled, y_train))
print(mlp.score(X_test_scaled, y_test))
mlp.coefs_[0].shape
hidden = np.maximum(0, np.dot(X_test_scaled, mlp.coefs_[0]) + mlp.intercepts_[0])
hidden = np.dot(X_test_scaled, mlp.coefs_[0]) + mlp.intercepts_[0]
plt.scatter(hidden[:, 0], hidden[:, 1], c=y_test)
from sklearn.linear_model import LogisticRegression
LogisticRegression().fit(X_train_scaled, y_train).score(X_test_scaled, y_test)
```
# Beyond scikit-learn
```
class NeuralNetwork(object):
def __init__(self):
# initialize coefficients and biases
pass
def forward(self, x):
activation = x
for coef, bias in zip(self.coef_, self.bias_):
activation = self.nonlinearity(np.dot(activation, coef) + bias)
return activation
def backward(self, x):
# compute gradient of stuff in forward pass
pass
# http://mxnet.io/architecture/program_model.html
class array(object) :
"""Simple Array object that support autodiff."""
def __init__(self, value, name=None):
self.value = value
if name:
self.grad = lambda g : {name : g}
def __add__(self, other):
assert isinstance(other, int)
ret = array(self.value + other)
ret.grad = lambda g : self.grad(g)
return ret
def __mul__(self, other):
assert isinstance(other, array)
ret = array(self.value * other.value)
def grad(g):
x = self.grad(g * other.value)
x.update(other.grad(g * self.value))
return x
ret.grad = grad
return ret
# some examples
a = array(np.array([1, 2]), 'a')
b = array(np.array([3, 4]), 'b')
c = b * a
d = c + 1
print(d.value)
print(d.grad(1))
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
% matplotlib inline
plt.rcParams["savefig.dpi"] = 300
plt.rcParams["savefig.bbox"] = "tight"
np.set_printoptions(precision=3, suppress=True)
import pandas as pd
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import scale, StandardScaler
from sklearn.neural_network import MLPClassifier
from sklearn.datasets import make_moons
X, y = make_moons(n_samples=100, noise=0.25, random_state=2)
X_train, X_test, y_train, y_test = train_test_split(X, y, stratify=y,
random_state=42)
plt.scatter(X_train[:, 0], X_train[:, 1], c=plt.cm.Vega10(y_train))
xlim = plt.xlim()
ylim = plt.ylim()
xs = np.linspace(xlim[0], xlim[1], 1000)
ys = np.linspace(ylim[0], ylim[1], 1000)
xx, yy = np.meshgrid(xs, ys)
X_grid = np.c_[xx.ravel(), yy.ravel()]
mlp = MLPClassifier(solver='lbfgs', random_state=0).fit(X_train, y_train)
print(mlp.score(X_train, y_train))
print(mlp.score(X_test, y_test))
plt.contour(xx, yy, mlp.predict_proba(X_grid)[:, 1].reshape(xx.shape), levels=[.5])
plt.scatter(X_train[:, 0], X_train[:, 1], c=plt.cm.Vega10(y_train))
plt.xlim(xlim)
plt.ylim(ylim)
fig, axes = plt.subplots(3, 3, figsize=(8, 5))
for ax, i in zip(axes.ravel(), range(10)):
mlp = MLPClassifier(solver='lbfgs', random_state=i).fit(X_train, y_train)
print(mlp.score(X_train, y_train))
print(mlp.score(X_test, y_test))
ax.contour(xx, yy, mlp.predict_proba(X_grid)[:, 1].reshape(xx.shape), levels=[.5])
ax.scatter(X_train[:, 0], X_train[:, 1], c=plt.cm.Vega10(y_train))
ax.set_xlim(xlim)
ax.set_ylim(ylim)
ax.set_xticks(())
ax.set_yticks(())
mlp = MLPClassifier(solver='lbfgs', hidden_layer_sizes=(10, 10, 10), random_state=0)
mlp.fit(X_train, y_train)
print(mlp.score(X_train, y_train))
print(mlp.score(X_test, y_test))
plt.contour(xx, yy, mlp.predict_proba(X_grid)[:, 1].reshape(xx.shape), levels=[.5])
plt.scatter(X_train[:, 0], X_train[:, 1], c=plt.cm.Vega10(y_train))
plt.xlim(xlim)
plt.ylim(ylim)
mlp = MLPClassifier(solver='lbfgs', hidden_layer_sizes=(10, 10, 10), activation="tanh", random_state=0)
mlp.fit(X_train, y_train)
print(mlp.score(X_train, y_train))
print(mlp.score(X_test, y_test))
plt.contour(xx, yy, mlp.predict_proba(X_grid)[:, 1].reshape(xx.shape), levels=[.5])
plt.scatter(X_train[:, 0], X_train[:, 1], c=plt.cm.Vega10(y_train))
plt.xlim(xlim)
plt.ylim(ylim)
rng = np.random.RandomState(0)
x = np.sort(rng.uniform(size=100))
y = np.sin(10 * x) + 5 * x + np.random.normal(0, .3, size=100)
plt.plot(x, y, 'o')
line = np.linspace(0, 1, 100)
X = x.reshape(-1, 1)
from sklearn.neural_network import MLPRegressor
mlp_relu = MLPRegressor(solver="lbfgs").fit(X, y)
mlp_tanh = MLPRegressor(solver="lbfgs", activation='tanh').fit(X, y)
plt.plot(x, y, 'o')
plt.plot(line, mlp_relu.predict(line.reshape(-1, 1)), label="relu")
plt.plot(line, mlp_tanh.predict(line.reshape(-1, 1)), label="tanh")
plt.legend()
from sklearn.datasets import load_digits
digits = load_digits()
X_train, X_test, y_train, y_test = train_test_split(
digits.data / 16., digits.target, stratify=digits.target, random_state=0)
mlp = MLPClassifier(max_iter=1000, random_state=0).fit(X_train, y_train)
print(mlp.score(X_train, y_train))
print(mlp.score(X_test, y_test))
from sklearn.datasets import load_breast_cancer
data = load_breast_cancer()
X_train, X_test, y_train, y_test = train_test_split(
data.data, data.target, stratify=data.target, random_state=0)
scaler = StandardScaler().fit(X_train)
X_train_scaled = scaler.transform(X_train)
X_test_scaled = scaler.transform(X_test)
mlp = MLPClassifier(max_iter=1000, random_state=0).fit(X_train_scaled, y_train)
print(mlp.score(X_train_scaled, y_train))
print(mlp.score(X_test_scaled, y_test))
mlp = MLPClassifier(solver="lbfgs", random_state=1).fit(X_train_scaled, y_train)
print(mlp.score(X_train_scaled, y_train))
print(mlp.score(X_test_scaled, y_test))
from sklearn.model_selection import GridSearchCV
pipe = make_pipeline(StandardScaler(), MLPClassifier(solver="lbfgs", random_state=1))
param_grid = {'mlpclassifier__alpha': np.logspace(-3, 3, 7)}
grid = GridSearchCV(pipe, param_grid, cv=5, return_train_score=True)
grid.fit(X_train, y_train)
results = pd.DataFrame(grid.cv_results_)
res = results.pivot_table(index="param_mlpclassifier__alpha",
values=["mean_test_score", "mean_train_score"])
res
res.plot()
plt.xscale("log")
plt.ylim(0.95, 1.01)
res = results.pivot_table(index="param_mlpclassifier__alpha", values=["mean_test_score", "mean_train_score", "std_test_score", "std_train_score"])
res.mean_test_score.plot(yerr=res.std_test_score)
res.mean_train_score.plot(yerr=res.std_train_score)
plt.xscale("log")
plt.ylim(0.95, 1.01)
plt.legend()
from sklearn.model_selection import GridSearchCV
pipe = make_pipeline(StandardScaler(), MLPClassifier(solver="lbfgs", random_state=1))
param_grid = {'mlpclassifier__hidden_layer_sizes':
[(10,), (50,), (100,), (500,), (10, 10), (50, 50), (100, 100), (500, 500)]
}
grid = GridSearchCV(pipe, param_grid, cv=5)
grid.fit(X_train, y_train)
results = pd.DataFrame(grid.cv_results_)
res = results.pivot_table(index="param_mlpclassifier__hidden_layer_sizes", values=["mean_test_score", "mean_train_score", "std_test_score", "std_train_score"])
res.mean_test_score.plot(yerr=res.std_test_score)
res.mean_train_score.plot(yerr=res.std_train_score)
plt.legend()
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.model_selection import GridSearchCV
pipe = make_pipeline(StandardScaler(), MLPClassifier(solver="lbfgs", random_state=1))
param_grid = {'mlpclassifier__hidden_layer_sizes':
[(10,), (25,), (50,), (100,) , (10, 10), (25, 25), (50, 50)]
}
grid = GridSearchCV(pipe, param_grid, return_train_score=True)
grid.fit(X_train, y_train)
results = pd.DataFrame(grid.cv_results_)
res = results.pivot_table(index="param_mlpclassifier__hidden_layer_sizes", values=["mean_test_score", "mean_train_score", "std_test_score", "std_train_score"])
res.mean_test_score.plot(yerr=res.std_test_score)
res.mean_train_score.plot(yerr=res.std_train_score)
plt.legend()
mlp = MLPClassifier(solver="lbfgs", hidden_layer_sizes=(2,), random_state=0).fit(X_train_scaled, y_train)
print(mlp.score(X_train_scaled, y_train))
print(mlp.score(X_test_scaled, y_test))
mlp.coefs_[0].shape
hidden = np.maximum(0, np.dot(X_test_scaled, mlp.coefs_[0]) + mlp.intercepts_[0])
hidden = np.dot(X_test_scaled, mlp.coefs_[0]) + mlp.intercepts_[0]
plt.scatter(hidden[:, 0], hidden[:, 1], c=y_test)
from sklearn.linear_model import LogisticRegression
LogisticRegression().fit(X_train_scaled, y_train).score(X_test_scaled, y_test)
class NeuralNetwork(object):
def __init__(self):
# initialize coefficients and biases
pass
def forward(self, x):
activation = x
for coef, bias in zip(self.coef_, self.bias_):
activation = self.nonlinearity(np.dot(activation, coef) + bias)
return activation
def backward(self, x):
# compute gradient of stuff in forward pass
pass
# http://mxnet.io/architecture/program_model.html
class array(object) :
"""Simple Array object that support autodiff."""
def __init__(self, value, name=None):
self.value = value
if name:
self.grad = lambda g : {name : g}
def __add__(self, other):
assert isinstance(other, int)
ret = array(self.value + other)
ret.grad = lambda g : self.grad(g)
return ret
def __mul__(self, other):
assert isinstance(other, array)
ret = array(self.value * other.value)
def grad(g):
x = self.grad(g * other.value)
x.update(other.grad(g * self.value))
return x
ret.grad = grad
return ret
# some examples
a = array(np.array([1, 2]), 'a')
b = array(np.array([3, 4]), 'b')
c = b * a
d = c + 1
print(d.value)
print(d.grad(1))
| 0.58261 | 0.685186 |
This notebook is meant to be viewed as a [RISE](https://github.com/damianavila/RISE) slideshow. When run, a custom stylesheet will be applied:
* *italic* text will be shown in *blue*,
* **bold** text will be showin in **red**, and
* ~~strikethrough~~ text will be shown in ~~green~~.
The code below is meant to be run before the presentation to ensure that Sage and its dependencies are properly initialized, so no waiting is required during the presentation.
```
import drg
p = [[[1, 0, 0, 0], [0, 6, 0, 0], [0, 0, 3, 0], [0, 0, 0, 6]],
[[0, 1, 0, 0], [1, 2, 1, 2], [0, 1, 0, 2], [0, 2, 2, 2]],
[[0, 0, 1, 0], [0, 2, 0, 4], [1, 0, 2, 0], [0, 4, 0, 2]],
[[0, 0, 0, 1], [0, 2, 2, 2], [0, 2, 0, 1], [1, 2, 1, 2]]]
scheme = drg.ASParameters(p)
scheme.kreinParameters()
```
# Computing distance-regular graph and association scheme parameters in SageMath with [`sage-drg`](https://github.com/jaanos/sage-drg)
### Janoš Vidali
#### University of Ljubljana
Based on joint work with <b>Alexander Gavrilyuk</b>, <b>Aleksandar Jurišić</b>, <b>Sho Suda</b> and <b>Jason Williford</b>
[Live slides](https://mybinder.org/v2/gh/jaanos/sage-drg/master?filepath=jupyter/2021-06-14-agtsem/AGTSem-sage-drg.ipynb) on [Binder](https://mybinder.org)
https://github.com/jaanos/sage-drg
## Association schemes
* **Association schemes** were defined by *Bose* and *Shimamoto* in *1952* as a theory underlying **experimental design**.
* They provide a ~~unified approach~~ to many topics, such as
- *combinatorial designs*,
- *coding theory*,
- generalizing *groups*, and
- *strongly regular* and *distance-regular graphs*.
## Examples
* *Hamming schemes*: **$X = \mathbb{Z}_n^d$**, **$x \ R_i \ y \Leftrightarrow \operatorname{weight}(x-y) = i$**
* *Johnson schemes*: **$X = \{S \subseteq \mathbb{Z}_n \mid |S| = d\}$** ($2d \le n$), **$x \ R_i \ y \Leftrightarrow |x \cap y| = d-i$**
<center><img src="as.png" /></center>
## Definition
* Let **$X$** be a set of *vertices* and **$\mathcal{R} = \{R_0 = \operatorname{id}_X, R_1, \dots, R_D\}$** a set of *symmetric relations* partitioning *$X^2$*.
* **$(X, \mathcal{R})$** is said to be a **$D$-class association scheme** if there exist numbers **$p^h_{ij}$** ($0 \le h, i, j \le D$) such that, for any *$x, y \in X$*,
**$$
x \ R_h \ y \Rightarrow |\{z \in X \mid x \ R_i \ z \ R_j \ y\}| = p^h_{ij}
$$**
* We call the numbers **$p^h_{ij}$** ($0 \le h, i, j \le D$) **intersection numbers**.
## Main problem
* Does an association scheme with given parameters ~~exist~~?
- If so, is it ~~unique~~?
- Can we determine ~~all~~ such schemes?
* ~~Lists~~ of feasible parameter sets have been compiled for [**strongly regular**](https://www.win.tue.nl/~aeb/graphs/srg/srgtab.html) and [**distance-regular graphs**](https://www.win.tue.nl/~aeb/drg/drgtables.html).
* Recently, lists have also been compiled for some [**$Q$-polynomial association schemes**](http://www.uwyo.edu/jwilliford/).
* Computer software allows us to *efficiently* compute parameters and check for *existence conditions*, and also to obtain new information which would be helpful in the ~~construction~~ of new examples.
## Bose-Mesner algebra
* Let **$A_i$** be the *binary matrix* corresponding to the relation *$R_i$* ($0 \le i \le D$).
* The vector space **$\mathcal{M}$** over *$\mathbb{R}$* spanned by *$A_i$* ($0 \le i \le D$) is called the **Bose-Mesner algebra**.
* *$\mathcal{M}$* has a second basis ~~$\{E_0, E_1, \dots, E_D\}$~~ consisting of *projectors* to the *common eigenspaces* of *$A_i$* ($0 \le i \le D$).
* There exist the **eigenmatrix** ~~$P$~~ and the **dual eigenmatrix** ~~$Q$~~ such that
*$$
A_j = \sum_{i=0}^D P_{ij} E_i, \qquad E_j = {1 \over |X|} \sum_{i=0}^D Q_{ij} A_i.
$$*
* There are ~~nonnegative~~ constants **$q^h_{ij}$**, called **Krein parameters**, such that
**$$
E_i \circ E_j = {1 \over |X|} \sum_{h=0}^D q^h_{ij} E_h ,
$$**
where **$\circ$** is the *entrywise matrix product*.
## Parameter computation: general association schemes
```
%display latex
import drg
p = [[[1, 0, 0, 0], [0, 6, 0, 0], [0, 0, 3, 0], [0, 0, 0, 6]],
[[0, 1, 0, 0], [1, 2, 1, 2], [0, 1, 0, 2], [0, 2, 2, 2]],
[[0, 0, 1, 0], [0, 2, 0, 4], [1, 0, 2, 0], [0, 4, 0, 2]],
[[0, 0, 0, 1], [0, 2, 2, 2], [0, 2, 0, 1], [1, 2, 1, 2]]]
scheme = drg.ASParameters(p)
scheme.kreinParameters()
```
## Metric and cometric schemes
* If **$p^h_{ij} \ne 0$** (resp. **$q^h_{ij} \ne 0$**) implies **$|i-j| \le h \le i+j$**, then the association scheme is said to be **metric** (resp. **cometric**).
* The *parameters* of a *metric* (or **$P$-polynomial**) association scheme can be ~~determined~~ from the **intersection array**
*$$
\{b_0, b_1, \dots, b_{D-1}; c_1, c_2, \dots, c_D\}
\quad (b_i = p^i_{1,i+1}, c_i = p^i_{1,i-1}).
$$*
* The *parameters* of a *cometric* (or **$Q$-polynomial**) association scheme can be ~~determined~~ from the **Krein array**
*$$
\{b^*_0, b^*_1, \dots, b^*_{D-1}; c^*_1, c^*_2, \dots, c^*_D\}
\quad (b^*_i = q^i_{1,i+1}, c^*_i = q^i_{1,i-1}).
$$*
* *Metric* association schemes correspond to *distance-regular graphs*.
## Parameter computation: metric and cometric schemes
```
from drg import DRGParameters
syl = DRGParameters([5, 4, 2], [1, 1, 4])
syl
syl.order()
from drg import QPolyParameters
q225 = QPolyParameters([24, 20, 36/11], [1, 30/11, 24])
q225
q225.order()
syl.pTable()
syl.kreinParameters()
syl.distancePartition()
syl.distancePartition(2)
```
## Parameter computation: parameters with variables
Let us define a *one-parametric family* of *intersection arrays*.
```
r = var("r")
f = DRGParameters([2*r^2*(2*r+1), (2*r-1)*(2*r^2+r+1), 2*r^2], [1, 2*r^2, r*(4*r^2-1)])
f.order(factor=True)
f1 = f.subs(r == 1)
f1
```
The parameters of `f1` are known to ~~uniquely determine~~ the *Hamming scheme $H(3, 3)$*.
```
f2 = f.subs(r == 2)
f2
```
## Feasibility checking
A parameter set is called **feasible** if it passes all known *existence conditions*.
Let us verify that *$H(3, 3)$* is feasible.
```
f1.check_feasible()
```
No error has occured, since all existence conditions are met.
Let us now check whether the second member of the family is feasible.
```
f2.check_feasible()
```
In this case, ~~nonexistence~~ has been shown by *matching* the parameters against a list of **nonexistent families**.
## Triple intersection numbers
* In some cases, **triple intersection numbers** can be computed.
**$$
[h \ i \ j] = \begin{bmatrix} x & y & z \\ h & i & j \end{bmatrix} = |\{w \in X \mid w \ R_i \ x \land w \ R_j \ y \land w \ R_h \ z\}|
$$**
* If **$x \ R_W \ y$**, **$x \ R_V \ z$** and **$y \ R_U \ z$**, then we have
*$$
\sum_{\ell=1}^D [\ell\ j\ h] = p^U_{jh} - [0\ j\ h], \qquad
\sum_{\ell=1}^D [i\ \ell\ h] = p^V_{ih} - [i\ 0\ h], \qquad
\sum_{\ell=1}^D [i\ j\ \ell] = p^W_{ij} - [i\ j\ 0],
$$*
where
*$$
[0\ j\ h] = \delta_{jW} \delta_{hV}, \qquad
[i\ 0\ h] = \delta_{iW} \delta_{hU}, \qquad
[i\ j\ 0] = \delta_{iV} \delta_{jU}.
$$*
* Additionally, **$q^h_{ij} = 0$** ~~if and only if~~
~~$$
\sum_{r,s,t=0}^D Q_{ri}Q_{sj}Q_{th} \begin{bmatrix} x & y & z \\ r & s & t \end{bmatrix} = 0
$$~~
for ~~all $x, y, z \in X$~~.
## Example: parameters for a bipartite DRG of diameter $5$
We will show that a distance-regular graph with intersection array **$\{55, 54, 50, 35, 10; 1, 5, 20, 45, 55\}$** ~~does not exist~~. The existence of such a graph would give a *counterexample* to a conjecture by MacLean and Terwilliger, see [Bipartite distance-regular graphs: The $Q$-polynomial property and pseudo primitive idempotents](http://dx.doi.org/10.1016/j.disc.2014.04.025) by M. Lang.
```
p = drg.DRGParameters([55, 54, 50, 35, 10], [1, 5, 20, 45, 55])
p.check_feasible(skip=["sporadic"])
p.order()
p.kreinParameters()
```
We now compute the triple intersection numbers with respect to three vertices **$x, y, z$ at mutual distances $2$**. Note that we have ~~$p^2_{22} = 243$~~, so such triples must exist. The parameter **$\alpha$** will denote the number of vertices adjacent to all of *$x, y, z$*.
```
p.distancePartition(2)
S222 = p.tripleEquations(2, 2, 2, params={"alpha": (1, 1, 1)})
show(S222[1, 1, 1])
show(S222[5, 5, 5])
```
Let us consider the set **$A$** of **common neighbours of $x$ and $y$**, and the set **$B$** of vertices at **distance $2$ from both $x$ and $y$**. By the above, each vertex in *$B$* has ~~at most one neighbour~~ in *$A$*, so there are ~~at most $243$~~ edges between *$A$* and *$B$*. However, each vertex in *$A$* is adjacent to both *$x$* and *$y$*, and the other ~~$53$~~ neighbours are in *$B$*, amounting to a total of ~~$5 \cdot 53 = 265$~~ edges. We have arrived to a ~~contradiction~~, and we must conclude that a graph with intersection array *$\{55, 54, 50, 35, 10; 1, 5, 20, 45, 55\}$* ~~does not exist~~.
## Double counting
* Let **$x, y \in X$** with **$x \ R_r \ y$**.
* Let **$\alpha_1, \alpha_2, \dots \alpha_u$** and **$\kappa_1, \kappa_2, \dots \kappa_u$** be numbers such that there are precisely *$\kappa_\ell$* vertices **$z \in X$** with **$x \ R_s \ z \ R_t \ y$** such that
**$$
\begin{bmatrix} x & y & z \\ h & i & j \end{bmatrix} = \alpha_\ell \qquad (1 \le \ell \le u).
$$**
* Let **$\beta_1, \beta_2, \dots \beta_v$** and **$\lambda_1, \lambda_2, \dots \lambda_v$** be numbers such that there are precisely *$\lambda_\ell$* vertices **$w \in X$** with **$x \ R_h \ w \ R_i \ y$** such that
**$$
\begin{bmatrix} w & x & y \\ j & s & t \end{bmatrix} = \beta_\ell \qquad (1 \le \ell \le v).
$$**
* Double-counting pairs *$(w, z)$* with **$w \ R_j \ z$** gives
~~$$
\sum_{\ell=1}^u \kappa_\ell \alpha_\ell = \sum_{\ell=1}^v \lambda_\ell \beta_\ell
$$~~
* Special case: **$u = 1, \alpha_1 = 0$** implies ~~$v = 1, \beta_1 = 0$~~.
## Example: parameters for a $3$-class $Q$-polynomial scheme
~~Nonexistence~~ of some *$Q$-polynomial* association schemes has been proven by obtaining a *contradiction* in *double counting* with triple intersection numbers.
```
q225
q225.check_quadruples()
```
*Integer linear programming* has been used to find solutions to multiple systems of *linear Diophantine equations*,
*eliminating* inconsistent solutions.
## More results
There is no *distance-regular graph* with intersection array
* ~~$\{83, 54, 21; 1, 6, 63\}$~~ (~~$1080$~~ vertices)
* ~~$\{135, 128, 16; 1, 16, 120\}$~~ (~~$1360$~~ vertices)
* ~~$\{104, 70, 25; 1, 7, 80\}$~~ (~~$1470$~~ vertices)
* ~~$\{234, 165, 12; 1, 30, 198\}$~~ (~~$1600$~~ vertices)
* ~~$\{195, 160, 28; 1, 20, 168\}$~~ (~~$2016$~~ vertices)
* ~~$\{125, 108, 24; 1, 9, 75\}$~~ (~~$2106$~~ vertices)
* ~~$\{126, 90, 10; 1, 6, 105\}$~~ (~~$2197$~~ vertices)
* ~~$\{203, 160, 34; 1, 16, 170\}$~~ (~~$2640$~~ vertices)
* ~~$\{53, 40, 28, 16; 1, 4, 10, 28\}$~~ (~~$2916$~~ vertices)
~~Nonexistence~~ of *$Q$-polynomial association schemes* [GVW21] with parameters listed as *feasible* by [Williford](http://www.uwyo.edu/jwilliford/) has been shown for
* ~~$29$~~ cases of *$3$-class primitive* $Q$-polynomial association schemes
- *double counting* has been used in ~~two~~ cases
* ~~$92$~~ cases of *$4$-class $Q$-bipartite* $Q$-polynomial association schemes
* ~~$11$~~ cases of *$5$-class $Q$-bipartite* $Q$-polynomial association schemes
## Nonexistence of infinite families
Association schemes with the following parameters do not exist.
* *distance-regular graphs* with *intersection arrays* ~~$\{(2r+1)(4r+1)(4t-1), 8r(4rt-r+2t), (r+t)(4r+1); 1, (r+t)(4r+1), 4r(2r+1)(4t-1)\}$~~ (**$r, t \ge 1$**)
* *primitive $Q$-polynomial association schemes* with *Krein arrays* ~~$\{2r^2-1, 2r^2-2, r^2+1; 1, 2, r^2-1\}$~~ (**$r \ge 3$ odd**)
* *$Q$-bipartite $Q$-polynomial association schemes* with *Krein arrays* ~~$\left\{m, m-1, {m(r^2-1) \over r^2}, m-r^2+1; 1, {m \over r^2}, r^2-1, m\right\}$~~ (**$m, r \ge 3$ odd**)
* *$Q$-bipartite $Q$-polynomial association schemes* with *Krein arrays* ~~$\left\{{r^2+1 \over 2}, {r^2-1 \over 2}, {(r^2+1)^2 \over 2r(r+1)},
{(r-1)(r^2+1) \over 4r}, {r^2+1 \over 2r};
1, {(r-1)(r^2+1) \over 2r(r+1)}, {(r+1)(r^2 + 1) \over 4r},
{(r-1)(r^2+1) \over 2r}, {r^2+1 \over 2}\right\}$~~ (**$r \ge 5$**, **$r \equiv 3 \pmod{4}$**)
* *$Q$-antipodal $Q$-polynomial association schemes* with *Krein arrays* ~~$\left\{r^2 - 4, r^2 - 9, \frac{12(s-1)}{s}, 1; 1, \frac{12}{s}, r^2 - 9, r^2 - 4\right\}$~~ (**$r \ge 5$**, **$s \ge 4$**)
- **Corollary**: a *tight $4$-design* in **$H((9a^2+1)/5,6)$** ~~does not exist~~ [GSV20].
## Using Schönberg's theorem
* **Schönberg's theorem**: A *polynomial* **$f: [-1, 1] \to \mathbb{R}$** of degree **$D$** is ~~positive definite on $S^{m-1}$~~ iff it is a ~~nonnegative linear combination~~ of *Gegenbauer polynomials* **$Q^m_{\ell}$** (**$0 \le \ell \le D$**).
* **Theorem** (*Kodalen, Martin*): If **$(X, \mathcal{R})$** is an *association scheme*, then
~~$$
Q_{\ell}^{m_i} \left({1 \over m_i} L^*_i \right) = {1 \over |X|} \sum_{j=0}^D \theta_{\ell j} L^*_j
$$~~
for some ~~nonnegative constants~~ **$\theta_{\ell j}$** (**$0 \le j \le D$**), where **$m_i = \operatorname{rank}(E_i)$** and **$L^*_i = (q^h_{ij})_{h,j=0}^D$**.
```
q594 = drg.QPolyParameters([9, 8, 81/11, 63/8], [1, 18/11, 9/8, 9])
q594.order()
q594.check_schoenberg()
```
## The Terwilliger polynomial
* *Terwilliger* has observed that for a *$Q$-polynomial distance-regular graph $\Gamma$*, there exists a ~~polynomial $T$ of degree $4$~~ whose coefficients can be expressed in terms of the *intersection numbers* of *$\Gamma$* such that ~~$T(\theta) \ge 0$~~ for each *non-principal eigenvalue* **$\theta$** of the **local graph** at a vertex of *$\Gamma$*.
* `sage-drg` can be used to *compute* this polynomial.
```
p750 = drg.DRGParameters([49, 40, 22], [1, 5, 28])
p750.order()
T750 = p750.terwilligerPolynomial()
T750
sorted(s.rhs() for s in solve(T750 == 0, x))
plot(T750, (x, -4, 5))
```
We may now use **[BCN, Thm. 4.4.4]** to further *restrict* the possible *non-principal eigenvalues* of the *local graphs*.
```
l, u = -1 - p750.b[1] / (p750.theta[1] + 1), -1 - p750.b[1] / (p750.theta[3] + 1)
l, u
plot(T750, (x, -4, 5)) + line([(l, 0), (u, 0)], color="red", thickness=3)
```
Since graph eigenvalues are *algebraic integers* and all *non-integral eigenvalues* of the *local graph* lie on a subinterval of ~~$(-4, -1)$~~, it can be shown that the only permissible *non-principal eigenvalues* are ~~$-3, -2, 3$~~.
We may now set up a *system of equations* to determine the *multiplicities*.
```
var("m1 m2 m3")
solve([1 + m1 + m2 + m3 == p750.k[1],
1 * p750.a[1] + m1 * 3 + m2 * (-2) + m3 * (-3) == 0,
1 * p750.a[1]^2 + m1 * 3^2 + m2 * (-2)^2 + m3 * (-3)^2 == p750.k[1] * p750.a[1]],
(m1, m2, m3))
```
Since all multiplicities are not *nonnegative integers*, we conclude that there is no *distance-regular graph* with intersection array
* ~~$\{49, 40, 22; 1, 5, 28\}$~~ (~~$750$~~ vertices)
* ~~$\{109, 80, 22; 1, 10, 88\}$~~ (~~$1200$~~ vertices)
* ~~$\{164, 121, 33; 1, 11, 132\}$~~ (~~$2420$~~ vertices)
## Distance-regular graphs with classical parameters
We use a similar technique to prove ~~nonexistence~~ of certain *distance-regular graphs* with *classical parameters* **$(D, b, \alpha, \beta)$**:
* ~~$(3, 2, 2, 9)$~~ (~~$430$~~ vertices)
* ~~$(3, 2, 5, 21)$~~ (~~$1100$~~ vertices)
* ~~$(6, 2, 2, 107)$~~ (~~$87\,725\,820\,468$~~ vertices)
* ~~$(b, \alpha) = (2, 1)$~~ and
- ~~$D = 4$~~, ~~$\beta \in \{8, 10, 12\}$~~
- ~~$D = 5$~~, ~~$\beta \in \{16, 17, 19, 20, 21, 28\}$~~
- ~~$D = 6$~~, ~~$\beta \in \{32, 33, 34, 35, 36, 38, 40, 46, 49, 54, 60\}$~~
- ~~$D = 7$~~, ~~$\beta \in \{64, 65, 66, 67, 69, 70, 71, 72, 73, 74, 77, 79, 81, 84, 85, 92, 99, 124\}$~~
- ~~$D = 8$~~, ~~$\beta \in \{128, 129, 130, 131, 133, 134, 135, 136, 137, 139, 140, 141, 151, 152, 155, 158, 160, 165, 168, 174, 175, 183, 184, 190, 202, 238, 252\}$~~
- ~~$D \ge 3$~~, ~~$\beta \in \{2^{D-1}, 2^D-4\}$~~
## Local graphs with at most four eigenvalues
* **Lemma** (*Van Dam*): A *connected graph* on **$n$** vertices with *spectrum*
**$$
{\theta_0}^{\ell_0} \quad
{\theta_1}^{\ell_1} \quad
{\theta_2}^{\ell_2} \quad
{\theta_3}^{\ell_3}
$$**
is ~~walk-regular~~ with precisely
~~$$
w_r = {1 \over n} \sum_{i=0}^3 \ell_i \cdot {(\theta_i)}^r
$$~~
*closed $r$-walks* (**$r \ge 3$**) through *each vertex*.
- If **$r$** is *odd*, **$w_r$** must be ~~even~~.
* A *distance-regular graph* **$\Gamma$** with *classical parameters* **$(D, 2, 1, \beta)$** has *local graphs* with
- precisely **three distinct eigenvalues** if **$\beta = 2^D - 1$**, and then *$\Gamma$* is a ~~bilinear forms graph~~ (Gavrilyuk, Koolen)
- precisely **four distinct eigenvalues** if **$(\beta+1) \mid (2^D-2)(2^D-1)$**, and then ~~$\beta = 2^D-2$~~ (or *$w_3$* is ~~nonintegral~~)
* There is no *distance-regular graph* with *classical parameters* **$(D, 2, 1, \beta)$** such that
- ~~$(D, \beta) \in \{(3, 5), (4, 9), (4, 13), (5, 29), (6, 41), (6, 61), (7, 125), (8, 169), (8, 253)\}$~~
- ~~$D \ge 3$~~ and ~~$\beta = 2^D - 3$~~
## Addendum: feasible negative type classical DRG parameters (Weng)
```
pp = drg.DRGParameters(4, -3, -2, -41)
TT = pp.terwilligerPolynomial()
TT.factor()
plot(TT, (x, -42, 42))
pp.check_feasible()
```
|
github_jupyter
|
import drg
p = [[[1, 0, 0, 0], [0, 6, 0, 0], [0, 0, 3, 0], [0, 0, 0, 6]],
[[0, 1, 0, 0], [1, 2, 1, 2], [0, 1, 0, 2], [0, 2, 2, 2]],
[[0, 0, 1, 0], [0, 2, 0, 4], [1, 0, 2, 0], [0, 4, 0, 2]],
[[0, 0, 0, 1], [0, 2, 2, 2], [0, 2, 0, 1], [1, 2, 1, 2]]]
scheme = drg.ASParameters(p)
scheme.kreinParameters()
%display latex
import drg
p = [[[1, 0, 0, 0], [0, 6, 0, 0], [0, 0, 3, 0], [0, 0, 0, 6]],
[[0, 1, 0, 0], [1, 2, 1, 2], [0, 1, 0, 2], [0, 2, 2, 2]],
[[0, 0, 1, 0], [0, 2, 0, 4], [1, 0, 2, 0], [0, 4, 0, 2]],
[[0, 0, 0, 1], [0, 2, 2, 2], [0, 2, 0, 1], [1, 2, 1, 2]]]
scheme = drg.ASParameters(p)
scheme.kreinParameters()
from drg import DRGParameters
syl = DRGParameters([5, 4, 2], [1, 1, 4])
syl
syl.order()
from drg import QPolyParameters
q225 = QPolyParameters([24, 20, 36/11], [1, 30/11, 24])
q225
q225.order()
syl.pTable()
syl.kreinParameters()
syl.distancePartition()
syl.distancePartition(2)
r = var("r")
f = DRGParameters([2*r^2*(2*r+1), (2*r-1)*(2*r^2+r+1), 2*r^2], [1, 2*r^2, r*(4*r^2-1)])
f.order(factor=True)
f1 = f.subs(r == 1)
f1
f2 = f.subs(r == 2)
f2
f1.check_feasible()
f2.check_feasible()
p = drg.DRGParameters([55, 54, 50, 35, 10], [1, 5, 20, 45, 55])
p.check_feasible(skip=["sporadic"])
p.order()
p.kreinParameters()
p.distancePartition(2)
S222 = p.tripleEquations(2, 2, 2, params={"alpha": (1, 1, 1)})
show(S222[1, 1, 1])
show(S222[5, 5, 5])
q225
q225.check_quadruples()
q594 = drg.QPolyParameters([9, 8, 81/11, 63/8], [1, 18/11, 9/8, 9])
q594.order()
q594.check_schoenberg()
p750 = drg.DRGParameters([49, 40, 22], [1, 5, 28])
p750.order()
T750 = p750.terwilligerPolynomial()
T750
sorted(s.rhs() for s in solve(T750 == 0, x))
plot(T750, (x, -4, 5))
l, u = -1 - p750.b[1] / (p750.theta[1] + 1), -1 - p750.b[1] / (p750.theta[3] + 1)
l, u
plot(T750, (x, -4, 5)) + line([(l, 0), (u, 0)], color="red", thickness=3)
var("m1 m2 m3")
solve([1 + m1 + m2 + m3 == p750.k[1],
1 * p750.a[1] + m1 * 3 + m2 * (-2) + m3 * (-3) == 0,
1 * p750.a[1]^2 + m1 * 3^2 + m2 * (-2)^2 + m3 * (-3)^2 == p750.k[1] * p750.a[1]],
(m1, m2, m3))
pp = drg.DRGParameters(4, -3, -2, -41)
TT = pp.terwilligerPolynomial()
TT.factor()
plot(TT, (x, -42, 42))
pp.check_feasible()
| 0.327561 | 0.970183 |
```
%matplotlib notebook
%matplotlib inline
import math
import matplotlib.pyplot as plt
```
# Nuclear Power Economics and Fuel Management
## Syllabus
Throughout the semester, you can always find the syllabus online at [https://github.com/katyhuff/npre412/blob/master/syllabus/syllabus.pdf](https://github.com/katyhuff/npre412/blob/master/syllabus/syllabus.pdf).
```
from IPython.display import IFrame
IFrame("../syllabus/syllabus.pdf", width=1000, height=1000)
from IPython.display import IFrame
IFrame("http://katyhuff.github.io", width=1000, height=700)
```
## Assessment
My goal (and, hopefully your goal) is for you to learn this material. If I have done my job right, your grade in this class will reflect just that -- how much you have learned. To do this for many learning types, your comprehension of the readings will be assessed with quizzes, your ability to apply what you've learned from class on your own will be assessed with the homeworks and projects, and your wholistic retention of the material will be assessed with tests.
### Monday Background Reading Assignments
Rather than introducing you to concepts during class, I think our time is better spent if we focus on exploring those concepts through demonstration and discussion together. **This 'active learning' educational strategy is backed by science, but is also just more respectful of your ability to learn things on your own.** Therefore, the lectures will assume you have studied the background materials ahead of time. This will include book sections, government reports, videos, and other resources. You will be expected to study the material outside of class before the start of each week. This will include book sections, government reports, videos, and other resources. I recommend you take notes on this material as it may be part of the tests.
**On Monday of each week I'll assign a list of reading material. You'll have 7 days to study that material before we start covering those concepts in class.**

### Monday Quizzes
To help me calibrate the in-class discussion, a weekly quiz will assess your comprehension of the background material. The quizzes can be taken online through [Compass2g](https://compass2g.illinois.edu) at any time during the week, but they must be completed by Monday at 10am, 7 days after the material was assigned.
### Friday Homework Assignments
Homeworks will be assigned each Friday concerning the material covered that week. You will have 7 days to do the homework, so it will be due at 10am on the following Friday.
### Projects
The class will involve two projects. One is a signficant quantitative research project, which involves a project proposal stage as well as a presentation to take place during the time period alotted for the final exam. The other is an in depth critical reading of a relevant book, with a report and an individual presentation due late in the semester.
### Tests
One midterm exam will take place in class.
### Participation
I will notice when you are not in class. While attendance might not directly affect your grades, it may indirectly affect your them. If you miss something I demonstrate in class, you'll have a lot more trouble proving that you've learned it.
## How to get an A
My dear friend, mathematician Kathryn Mann, has a great summary of [how to get an A in her class.](https://math.berkeley.edu/~kpmann/getanA.pdf). Everything she says about her math classes is true for this class as well. You should expect to spend 3 hours outside of class for every hour you spend in class. So, for a 3 credit class, you'll need to spend 3 hours a week in class and 9 hours outside of class on the coursework. If you find you're spending much less or much more time on this class, please let me know.
## Late Work
**Late work has a halflife of 1 hour.** That is, adjusted for lateness, your grade $G(t)$ is a decaying percentage of the raw grade $G_0$. An assignment turned in $t$ hours late will recieve a grade according to the following relation:
$$
\begin{align}
G(t) &= G_0e^{-\lambda t}\\
\end{align}
$$
where
$$
\begin{align}
G(t) &= \mbox{grade adjusted for lateness}\\
G_0 &= \mbox{raw grade}\\
\lambda &= \frac{ln(2)}{t_{1/2}} = \mbox{decay constant} \\
t &= \mbox{time elapsed since due [hours]}\\
t_{1/2} &= 1 = \mbox{half-life [hours]} \\
\end{align}
$$
```
import math
def late_grade(hours_late, grade=100, half_life=1):
"""This function describes how much credit you will get for late work"""
lam = math.log(2)/half_life
return grade*math.exp(-lam*hours_late)
# This code plots how much credit you'll get over time
import numpy as np
y = np.arange(24)
x = np.arange(24)
for h in range(0,24):
x[h] = h
y[h] = late_grade(h)
# creates a figure and axes with matplotlib
fig, ax = plt.subplots()
scatter = plt.scatter(x, y, color='blue', s=y*20, alpha=0.4)
ax.plot(x, y, color='red')
# adds labels to the plot
ax.set_ylabel('Percent of Grade Earned')
ax.set_xlabel('Hours Late')
ax.set_title('Grade Decay')
# adds tooltips
import mpld3
labels = ['{0}% earned'.format(i) for i in y]
tooltip = mpld3.plugins.PointLabelTooltip(scatter, labels=labels)
mpld3.plugins.connect(fig, tooltip)
mpld3.display()
print("If you turn in your homework an hour late, you'll get ", round(late_grade(1),2), "% credit.")
print("If you turn in your homework six hours late, you'll get ", round(late_grade(6),2), "% credit.")
print("If you turn in your homework a day late, you'll get ", round(late_grade(24),2), "% credit.")
print("If you turn in your homework two days late, you'll get ", round(late_grade(48),2), "% credit.")
print("If you turn in your homework three days late, you'll get ", round(late_grade(72),2), "% credit.")
```
**There will be no negotiation about late work except in the case of absence documented by an absence letter from the Dean of Students.** The university policy for requesting such a letter is [here](http://studentcode.illinois.edu/article1_part5_1-501.html) . Please note that such a letter is appropriate for many types of conflicts, but that religious conflicts require special early handling. In accordance with university policy, students seeking an excused absence for religious reasons should complete the Request for Accommodation for Religious Observances Form, which can be found on the Office of the Dean of Students website. The student should submit this form to the instructor and the Office of the Dean of Students by the end of the second week of the course to which it applies.
## Communications
Things to try when you have a question:
- Be Persistent: [Try just one more time.](https://s-media-cache-ak0.pinimg.com/736x/03/54/ce/0354ce58a7a4308edcc46dd9238e12d7.jpg)
- Google: [You might be surprised at its depth.](https://devhumor.com/content/uploads//images/April2016/google-errors.jpg)
- Piazza: Try this first, your student colleagues probably know the answer.
- TA email: A quick question can usually be answered by your TA via email.
- TA office hours: Your TA is there for you at a regularly scheduled time.
- Prof. email: Questions not appropriate for your TA can be directed to me.
- Prof. appointment: Schedule an appointment with me.
### A note on email
[Email tips for dealing with fussy professor types.](https://medium.com/@lportwoodstacer/how-to-email-your-professor-without-being-annoying-af-cf64ae0e4087)
## Python, IPython, Jupyter, git, and the Notebooks
Rather than reading equations off of slides, I will display lecture notes, equations, and images in "Jupyter notebooks" like this one. Sometimes, I will call them by their old name, "IPython notebooks," but I'm talking about "Jupyter notebooks". Interleaved with the course notes, we will often write small functions in the Python programming language to represent the equations we are talking about. This will allow you to interact with the math, changing variables, modifying the models, and exploring the parameter space.
### But I don't know Python!
*You don't have to know Python to take this class.* However, you will need to learn a little along the way. I will provide lots of example code to support your completion of homework assignments and I will never ask you to write functioning code as part of any written exam. Programming is really hard without the internet.
### Exercises
Watch for blocks titled **Exercise** in the notebooks. Those mark moments when I will ask you, during class, to try something out, explore an equation, or arrive at an answer. These are short and are not meant to be difficult. They exist to quickly solidify an idea before we move on to the next one. I will often randomly call on students (with a random number generator populated with the enrollment list) to give solutions to the exercises, so **a failure to show up and participate will be noticed.**
### Installing Python, IPython, Jupyter, git, and the Notebooks
Because engaging in the exercises will be really helpful for you to study, you'll should try to gain access to a computer equipped with Python (a version greater than 3.0) and a basic set of scientific python libraries. If you have a computer already, I encourage you to install [anaconda](https://www.continuum.io/downloads).
These notebooks are stored "in the cloud," which is to say that they are stored on someone else's computers. Those computers are servers at GitHub, a sometimes silly but also very important company in the beautiful city of San Francisco. GitHub stores "git repositories" which are collections of files that are "version controlled" by the program "git." This is a lot to keep track of, and I won't require that you learn git to participate in this class. However, I strongly recommend using git and GitHub and to keep track of your research code. So, I encourage you to use git to access the notebooks.
**More information about the things you might want to install can be found in the [README](https://github.com/katyhuff/npre412/blob/master/README.md).**
|
github_jupyter
|
%matplotlib notebook
%matplotlib inline
import math
import matplotlib.pyplot as plt
from IPython.display import IFrame
IFrame("../syllabus/syllabus.pdf", width=1000, height=1000)
from IPython.display import IFrame
IFrame("http://katyhuff.github.io", width=1000, height=700)
import math
def late_grade(hours_late, grade=100, half_life=1):
"""This function describes how much credit you will get for late work"""
lam = math.log(2)/half_life
return grade*math.exp(-lam*hours_late)
# This code plots how much credit you'll get over time
import numpy as np
y = np.arange(24)
x = np.arange(24)
for h in range(0,24):
x[h] = h
y[h] = late_grade(h)
# creates a figure and axes with matplotlib
fig, ax = plt.subplots()
scatter = plt.scatter(x, y, color='blue', s=y*20, alpha=0.4)
ax.plot(x, y, color='red')
# adds labels to the plot
ax.set_ylabel('Percent of Grade Earned')
ax.set_xlabel('Hours Late')
ax.set_title('Grade Decay')
# adds tooltips
import mpld3
labels = ['{0}% earned'.format(i) for i in y]
tooltip = mpld3.plugins.PointLabelTooltip(scatter, labels=labels)
mpld3.plugins.connect(fig, tooltip)
mpld3.display()
print("If you turn in your homework an hour late, you'll get ", round(late_grade(1),2), "% credit.")
print("If you turn in your homework six hours late, you'll get ", round(late_grade(6),2), "% credit.")
print("If you turn in your homework a day late, you'll get ", round(late_grade(24),2), "% credit.")
print("If you turn in your homework two days late, you'll get ", round(late_grade(48),2), "% credit.")
print("If you turn in your homework three days late, you'll get ", round(late_grade(72),2), "% credit.")
| 0.408277 | 0.934694 |
# Decision tree for regression
In this notebook, we present how decision trees are working in regression
problems. We show differences with the decision trees previously presented in
a classification setting.
First, we load the penguins dataset specifically for solving a regression
problem.
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">If you want a deeper overview regarding this dataset, you can refer to the
Appendix - Datasets description section at the end of this MOOC.</p>
</div>
```
import pandas as pd
penguins = pd.read_csv("../datasets/penguins_regression.csv")
feature_name = "Flipper Length (mm)"
target_name = "Body Mass (g)"
data_train, target_train = penguins[[feature_name]], penguins[target_name]
```
To illustrate how decision trees are predicting in a regression setting, we
will create a synthetic dataset containing all possible flipper length from
the minimum to the maximum of the original data.
```
import numpy as np
data_test = pd.DataFrame(np.arange(data_train[feature_name].min(),
data_train[feature_name].max()),
columns=[feature_name])
```
Using the term "test" here refers to data that was not used for training.
It should not be confused with data coming from a train-test split, as it
was generated in equally-spaced intervals for the visual evaluation of the
predictions.
Note that this is methodologically valid here because our objective is to get
some intuitive understanding on the shape of the decision function of the
learned decision trees.
However computing an evaluation metric on such a synthetic test set would
be meaningless since the synthetic dataset does not follow the same
distribution as the real world data on which the model will be deployed.
```
import matplotlib.pyplot as plt
import seaborn as sns
sns.scatterplot(data=penguins, x=feature_name, y=target_name,
color="black", alpha=0.5)
_ = plt.title("Illustration of the regression dataset used")
```
We will first illustrate the difference between a linear model and a decision
tree.
```
from sklearn.linear_model import LinearRegression
linear_model = LinearRegression()
linear_model.fit(data_train, target_train)
target_predicted = linear_model.predict(data_test)
sns.scatterplot(data=penguins, x=feature_name, y=target_name,
color="black", alpha=0.5)
plt.plot(data_test[feature_name], target_predicted, label="Linear regression")
plt.legend()
_ = plt.title("Prediction function using a LinearRegression")
```
On the plot above, we see that a non-regularized `LinearRegression` is able
to fit the data. A feature of this model is that all new predictions
will be on the line.
```
ax = sns.scatterplot(data=penguins, x=feature_name, y=target_name,
color="black", alpha=0.5)
plt.plot(data_test[feature_name], target_predicted, label="Linear regression",
linestyle="--")
plt.scatter(data_test[::3], target_predicted[::3], label="Predictions",
color="tab:orange")
plt.legend()
_ = plt.title("Prediction function using a LinearRegression")
```
Contrary to linear models, decision trees are non-parametric models:
they do not make assumptions about the way data is distributed.
This will affect the prediction scheme. Repeating the above experiment
will highlight the differences.
```
from sklearn.tree import DecisionTreeRegressor
tree = DecisionTreeRegressor(max_depth=1)
tree.fit(data_train, target_train)
target_predicted = tree.predict(data_test)
sns.scatterplot(data=penguins, x=feature_name, y=target_name,
color="black", alpha=0.5)
plt.plot(data_test[feature_name], target_predicted, label="Decision tree")
plt.legend()
_ = plt.title("Prediction function using a DecisionTreeRegressor")
```
We see that the decision tree model does not have an *a priori* distribution
for the data and we do not end-up with a straight line to regress flipper
length and body mass.
Instead, we observe that the predictions of the tree are piecewise constant.
Indeed, our feature space was split into two partitions. Let's check the
tree structure to see what was the threshold found during the training.
```
from sklearn.tree import plot_tree
_, ax = plt.subplots(figsize=(8, 6))
_ = plot_tree(tree, feature_names=feature_name, ax=ax)
```
The threshold for our feature (flipper length) is 206.5 mm. The predicted
values on each side of the split are two constants: 3683.50 g and 5023.62 g.
These values corresponds to the mean values of the training samples in each
partition.
In classification, we saw that increasing the depth of the tree allowed us to
get more complex decision boundaries.
Let's check the effect of increasing the depth in a regression setting:
```
tree = DecisionTreeRegressor(max_depth=3)
tree.fit(data_train, target_train)
target_predicted = tree.predict(data_test)
sns.scatterplot(data=penguins, x=feature_name, y=target_name,
color="black", alpha=0.5)
plt.plot(data_test[feature_name], target_predicted, label="Decision tree")
plt.legend()
_ = plt.title("Prediction function using a DecisionTreeRegressor")
```
Increasing the depth of the tree will increase the number of partition and
thus the number of constant values that the tree is capable of predicting.
In this notebook, we highlighted the differences in behavior of a decision
tree used in a classification problem in contrast to a regression problem.
|
github_jupyter
|
import pandas as pd
penguins = pd.read_csv("../datasets/penguins_regression.csv")
feature_name = "Flipper Length (mm)"
target_name = "Body Mass (g)"
data_train, target_train = penguins[[feature_name]], penguins[target_name]
import numpy as np
data_test = pd.DataFrame(np.arange(data_train[feature_name].min(),
data_train[feature_name].max()),
columns=[feature_name])
import matplotlib.pyplot as plt
import seaborn as sns
sns.scatterplot(data=penguins, x=feature_name, y=target_name,
color="black", alpha=0.5)
_ = plt.title("Illustration of the regression dataset used")
from sklearn.linear_model import LinearRegression
linear_model = LinearRegression()
linear_model.fit(data_train, target_train)
target_predicted = linear_model.predict(data_test)
sns.scatterplot(data=penguins, x=feature_name, y=target_name,
color="black", alpha=0.5)
plt.plot(data_test[feature_name], target_predicted, label="Linear regression")
plt.legend()
_ = plt.title("Prediction function using a LinearRegression")
ax = sns.scatterplot(data=penguins, x=feature_name, y=target_name,
color="black", alpha=0.5)
plt.plot(data_test[feature_name], target_predicted, label="Linear regression",
linestyle="--")
plt.scatter(data_test[::3], target_predicted[::3], label="Predictions",
color="tab:orange")
plt.legend()
_ = plt.title("Prediction function using a LinearRegression")
from sklearn.tree import DecisionTreeRegressor
tree = DecisionTreeRegressor(max_depth=1)
tree.fit(data_train, target_train)
target_predicted = tree.predict(data_test)
sns.scatterplot(data=penguins, x=feature_name, y=target_name,
color="black", alpha=0.5)
plt.plot(data_test[feature_name], target_predicted, label="Decision tree")
plt.legend()
_ = plt.title("Prediction function using a DecisionTreeRegressor")
from sklearn.tree import plot_tree
_, ax = plt.subplots(figsize=(8, 6))
_ = plot_tree(tree, feature_names=feature_name, ax=ax)
tree = DecisionTreeRegressor(max_depth=3)
tree.fit(data_train, target_train)
target_predicted = tree.predict(data_test)
sns.scatterplot(data=penguins, x=feature_name, y=target_name,
color="black", alpha=0.5)
plt.plot(data_test[feature_name], target_predicted, label="Decision tree")
plt.legend()
_ = plt.title("Prediction function using a DecisionTreeRegressor")
| 0.729809 | 0.991472 |
# K均值算法(K-means)聚类
## 【关键词】K个种子,均值
## 一、K-means算法原理
### 聚类的概念:一种无监督的学习,事先不知道类别,自动将相似的对象归到同一个簇中。
K-Means算法是一种聚类分析(cluster analysis)的算法,其主要是来计算数据聚集的算法,主要通过不断地取离种子点最近均值的算法。
K-Means算法主要解决的问题如下图所示。我们可以看到,在图的左边有一些点,我们用肉眼可以看出来有四个点群,但是我们怎么通过计算机程序找出这几个点群来呢?于是就出现了我们的K-Means算法

这个算法其实很简单,如下图所示:

从上图中,我们可以看到,A,B,C,D,E是五个在图中点。而灰色的点是我们的种子点,也就是我们用来找点群的点。有两个种子点,所以K=2。
然后,K-Means的算法如下:
1. 随机在图中取K(这里K=2)个种子点。
2. 然后对图中的所有点求到这K个种子点的距离,假如点Pi离种子点Si最近,那么Pi属于Si点群。(上图中,我们可以看到A,B属于上面的种子点,C,D,E属于下面中部的种子点)
3. 接下来,我们要移动种子点到属于他的“点群”的中心。(见图上的第三步)
4. 然后重复第2)和第3)步,直到,种子点没有移动(我们可以看到图中的第四步上面的种子点聚合了A,B,C,下面的种子点聚合了D,E)。
这个算法很简单,重点说一下“求点群中心的算法”:欧氏距离(Euclidean Distance):差的平方和的平方根

### K-Means主要最重大的缺陷——都和初始值有关:
K是事先给定的,这个K值的选定是非常难以估计的。很多时候,事先并不知道给定的数据集应该分成多少个类别才最合适。(ISODATA算法通过类的自动合并和分裂,得到较为合理的类型数目K)
K-Means算法需要用初始随机种子点来搞,这个随机种子点太重要,不同的随机种子点会有得到完全不同的结果。(K-Means++算法可以用来解决这个问题,其可以有效地选择初始点)
### 总结:K-Means算法步骤:
1. 从数据中选择k个对象作为初始聚类中心;
2. 计算每个聚类对象到聚类中心的距离来划分;
3. 再次计算每个聚类中心
4. 计算标准测度函数,直到达到最大迭代次数,则停止,否则,继续操作。
5. 确定最优的聚类中心
### K-Means算法应用
看到这里,你会说,K-Means算法看来很简单,而且好像就是在玩坐标点,没什么真实用处。而且,这个算法缺陷很多,还不如人工呢。是的,前面的例子只是玩二维坐标点,的确没什么意思。但是你想一下下面的几个问题:
1)如果不是二维的,是多维的,如5维的,那么,就只能用计算机来计算了。
2)二维坐标点的X,Y 坐标,其实是一种向量,是一种数学抽象。现实世界中很多属性是可以抽象成向量的,比如,我们的年龄,我们的喜好,我们的商品,等等,能抽象成向量的目的就是可以让计算机知道某两个属性间的距离。如:我们认为,18岁的人离24岁的人的距离要比离12岁的距离要近,鞋子这个商品离衣服这个商品的距离要比电脑要近,等等。
## 二、实战
重要参数:
- n_clusters:聚类的个数
重要属性:
- cluster_centers_ : [n_clusters, n_features]的数组,表示聚类中心点的坐标
- labels_ : 每个样本点的标签
### 1、聚类实例
导包,使用make_blobs生成随机点cluster_std
```
import numpy as np
import pandas as pd
from pandas import Series,DataFrame
import matplotlib.pyplot as plt
%matplotlib inline
from sklearn.datasets import make_blobs
train,target = make_blobs(n_samples=150,n_features=2,centers=3)
plt.scatter(train[:,0],train[:,1],c=target)
plt.scatter(train[:,0],train[:,1])
from sklearn.cluster import KMeans
kmean = KMeans(n_clusters=3)
kmean.fit(train)
y_ = kmean.predict(train)
# 无监督学习可以直接fit_predict
y_ = kmean.fit_predict(train)
```
建立模型,训练数据,并进行数据预测,使用相同数据
绘制图形,显示聚类结果kmeans.cluster_centers
```
plt.scatter(train[:,0],train[:,1],c=y_)
# 获取聚类中心
centers = kmean.cluster_centers_
plt.scatter(centers[:,0],centers[:,1],c='red')
plt.scatter(train[:,0],train[:,1],c=target)
(kmean.labels_ == target).sum()/target.size
```
### 实战,三问中国足球几多愁?
导包,3D图像需导包:from mpl_toolkits.mplot3d import Axes3D
读取数据AsiaZoo.txt
```
zoo = pd.read_csv('../data/AsiaZoo.txt',header=None)
train = zoo[[1,2,3]]
train
county = zoo[0]
county
kmean = KMeans(n_clusters=3)
y_ = kmean.fit_predict(train)
y_
county[y_ == 0]
county[y_==1]
county[y_==2]
for i in range(3):
items = county[y_==i]
for item in items:
print(item,end=' ')
print('\n')
```
列名修改为:"国家","2006世界杯","2010世界杯","2007亚洲杯"
使用K-Means进行数据处理,对亚洲球队进行分组,分三组
for循环打印输出分组后的球队,每一组球队打印一行
绘制三维立体图形
- ax = plt.subplot(projection = '3d')
- ax.scatter3D()
```
from mpl_toolkits.mplot3d import Axes3D
train.columns = ['2006-world-cup','2010-word-cup','2007-aisa-cup']
train
from matplotlib.colors import ListedColormap
cmap = ListedColormap(['r','g','b'])
plt.figure(figsize=(8,6))
ax = plt.subplot(projection='3d')
ax.scatter3D(train['2006-world-cup'],train['2010-word-cup'],train['2007-aisa-cup'],c=y_,cmap=cmap,s=80)
clusters = kmean.cluster_centers_
ax.scatter3D(clusters[:,0],clusters[:,1],clusters[:,2],c='yellow',s=100,alpha=1)
ax.set_xlabel('2006-world-cup')
ax.set_ylabel('2010-word-cup')
ax.set_zlabel('2007-aisa-cup')
clusters
```
绘制聚类点
### 2、聚类实践与常见错误
导包,使用make_blobs创建样本点
第一种错误,k值不合适,make_blobs默认中心点三个
```
train,target = make_blobs(n_samples=150,n_features=2,centers=3)
plt.scatter(train[:,0],train[:,1],c=target)
kmean = KMeans(n_clusters=4)
y_ = kmean.fit_predict(train)
plt.scatter(train[:,0],train[:,1],c=y_)
```
第二种错误,数据偏差
trans = [[0.6,-0.6],[-0.4,0.8]]
X2 = np.dot(X,trans)
```
train,target = make_blobs(n_samples=150,n_features=2,centers=3)
plt.scatter(train[:,0],train[:,1],c=target)
trans = [[0.6,-0.6],[-0.4,0.8]]
train1 = np.dot(train,trans)
# plt.scatter(train1[:,0],train1[:,1],c=target)
kmean = KMeans(n_clusters=3)
y_ = kmean.fit_predict(train1)
plt.scatter(train[:,0],train[:,1],c=y_)
```
第三个错误:标准偏差不相同cluster_std
```
train,target = make_blobs(n_samples=150,n_features=2,centers=3,cluster_std=[0.3,1,5])
plt.scatter(train[:,0],train[:,1],c=target)
kmean = KMeans(n_clusters=3)
y_ = kmean.fit_predict(train)
plt.scatter(train[:,0],train[:,1],c=y_)
```
第四个错误:样本数量不同
```
train,target = make_blobs(n_samples=1500,n_features=2,centers=3)
train1 = train[target==0][:10]
train2 = train[target==1][:100]
train3 = train[target==2][:500]
X_train = np.concatenate((train1,train2,train3))
y_train = [0]*10 + [1]*100 + [2]*500
plt.scatter(X_train[:,0],X_train[:,1],c=y_train)
kmean = KMeans(n_clusters=3)
y_ = kmean.fit_predict(X_train)
plt.scatter(X_train[:,0],X_train[:,1],c=y_)
```
## 三、作业
### 1、分析ex7data2.mat文件
找出最佳聚类数目,并画出聚类的中心点
`
from scipy.io import loadmat
from sklearn.cluster import KMeans
data = loadmat('../data/ex7data2.mat')
X = data['X']
X.shape
`
### 2、图片压缩
使用聚类压缩图片
`
img = plt.imread('../data/bird_small.png')
img_shape = img.shape
img_shape
`
- 核心思想:把图片颜色数据聚类分析成N个类别,用中心点颜色替换其他颜色
```
a = np.array([1,2,3])
b = [0,1,0,1]
a[b]
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from pandas import Series,DataFrame
import matplotlib.pyplot as plt
%matplotlib inline
from sklearn.datasets import make_blobs
train,target = make_blobs(n_samples=150,n_features=2,centers=3)
plt.scatter(train[:,0],train[:,1],c=target)
plt.scatter(train[:,0],train[:,1])
from sklearn.cluster import KMeans
kmean = KMeans(n_clusters=3)
kmean.fit(train)
y_ = kmean.predict(train)
# 无监督学习可以直接fit_predict
y_ = kmean.fit_predict(train)
plt.scatter(train[:,0],train[:,1],c=y_)
# 获取聚类中心
centers = kmean.cluster_centers_
plt.scatter(centers[:,0],centers[:,1],c='red')
plt.scatter(train[:,0],train[:,1],c=target)
(kmean.labels_ == target).sum()/target.size
zoo = pd.read_csv('../data/AsiaZoo.txt',header=None)
train = zoo[[1,2,3]]
train
county = zoo[0]
county
kmean = KMeans(n_clusters=3)
y_ = kmean.fit_predict(train)
y_
county[y_ == 0]
county[y_==1]
county[y_==2]
for i in range(3):
items = county[y_==i]
for item in items:
print(item,end=' ')
print('\n')
from mpl_toolkits.mplot3d import Axes3D
train.columns = ['2006-world-cup','2010-word-cup','2007-aisa-cup']
train
from matplotlib.colors import ListedColormap
cmap = ListedColormap(['r','g','b'])
plt.figure(figsize=(8,6))
ax = plt.subplot(projection='3d')
ax.scatter3D(train['2006-world-cup'],train['2010-word-cup'],train['2007-aisa-cup'],c=y_,cmap=cmap,s=80)
clusters = kmean.cluster_centers_
ax.scatter3D(clusters[:,0],clusters[:,1],clusters[:,2],c='yellow',s=100,alpha=1)
ax.set_xlabel('2006-world-cup')
ax.set_ylabel('2010-word-cup')
ax.set_zlabel('2007-aisa-cup')
clusters
train,target = make_blobs(n_samples=150,n_features=2,centers=3)
plt.scatter(train[:,0],train[:,1],c=target)
kmean = KMeans(n_clusters=4)
y_ = kmean.fit_predict(train)
plt.scatter(train[:,0],train[:,1],c=y_)
train,target = make_blobs(n_samples=150,n_features=2,centers=3)
plt.scatter(train[:,0],train[:,1],c=target)
trans = [[0.6,-0.6],[-0.4,0.8]]
train1 = np.dot(train,trans)
# plt.scatter(train1[:,0],train1[:,1],c=target)
kmean = KMeans(n_clusters=3)
y_ = kmean.fit_predict(train1)
plt.scatter(train[:,0],train[:,1],c=y_)
train,target = make_blobs(n_samples=150,n_features=2,centers=3,cluster_std=[0.3,1,5])
plt.scatter(train[:,0],train[:,1],c=target)
kmean = KMeans(n_clusters=3)
y_ = kmean.fit_predict(train)
plt.scatter(train[:,0],train[:,1],c=y_)
train,target = make_blobs(n_samples=1500,n_features=2,centers=3)
train1 = train[target==0][:10]
train2 = train[target==1][:100]
train3 = train[target==2][:500]
X_train = np.concatenate((train1,train2,train3))
y_train = [0]*10 + [1]*100 + [2]*500
plt.scatter(X_train[:,0],X_train[:,1],c=y_train)
kmean = KMeans(n_clusters=3)
y_ = kmean.fit_predict(X_train)
plt.scatter(X_train[:,0],X_train[:,1],c=y_)
a = np.array([1,2,3])
b = [0,1,0,1]
a[b]
| 0.301568 | 0.888566 |
# Posterior uncertainties of the ODE filter
We investigate the uncertaintes returned by Gaussian ODE Filters and how they are affected by the choice of diffusion model.
```
# Make inline plots vector graphics instead of raster graphics
%matplotlib inline
from IPython.display import set_matplotlib_formats
set_matplotlib_formats("pdf", "svg")
# Plotting
import matplotlib.pyplot as plt
plt.style.use("../../probnum.mplstyle")
import numpy as np
from probnum.diffeq import probsolve_ivp
rng = np.random.default_rng(seed=123)
```
We start with the Lotka-Volterra equations.
```
def f(t, y):
y1, y2 = y
return np.array([0.5 * y1 - 0.05 * y1 * y2, -0.5 * y2 + 0.05 * y1 * y2])
def df(t, y):
y1, y2 = y
return np.array([[0.5 - 0.05 * y2, -0.05 * y1], [0.05 * y2, -0.5 + 0.05 * y1]])
t0 = 0.0
tmax = 20.0
y0 = np.array([20, 20])
```
The EK0 ODE filter is quite fast and flexible, but does not yield accurate uncertainty estimates. We see below that the uncertainty increases monotonously, independent on the actual peaks and valleys of the ODE solution.
It is not surprising that the EK0 solution is agnostic of the trajectory. In fact, the covariance of the EK0 filter is independent of the data and as such, we cannot expect it to return useful uncertainty.
```
sol = probsolve_ivp(
f, t0, tmax, y0, df=df, step=0.1, adaptive=False, diffusion_model="dynamic"
)
means, stds = sol.states.mean, sol.states.std
fig, (ax1, ax2) = plt.subplots(nrows=2, ncols=1, sharex=True)
ax1.plot(sol.locations, means[:, 0], label="x1")
ax1.plot(sol.locations, means[:, 1], label="x2")
ax1.legend()
ax2.fill_between(sol.locations, stds[:, 0], alpha=0.25, label="x1-unc.")
ax2.fill_between(sol.locations, stds[:, 1], alpha=0.25, label="x2-unc.")
ax2.legend()
fig.suptitle("EK0 Solution")
plt.show()
```
Notice that the uncertainties are aware of the peaks and valleys. They even know whether the peak is flat (rhs of blue, lhs of orange; smaller ones of the blue peaks) or steep. On top of that, they increase over time.
For both we can also sample from the solution. Let us compute a low-res solution (so the samples actually look different from each other).
Beware, because large numbers of samples can take a long time to compute.
```
sol = probsolve_ivp(
f,
t0,
tmax,
y0,
df=df,
step=0.5,
algo_order=1,
adaptive=False,
diffusion_model="dynamic",
)
# sol.kalman_posterior.diffusion_model.diffusions[2:8] *= 100000
num_samples = 150
locations = np.arange(0.0, 22.1, 0.05)
locations = sol.locations
samples = sol.sample(rng=rng, t=locations, size=num_samples)
solution = sol(locations)
means = solution.mean
stds = solution.std
fig, (ax1) = plt.subplots(nrows=1, ncols=1, sharex=True)
for sample in samples[::5]:
ax1.plot(locations, sample[:, 0], ":", color="C0", linewidth=0.5, alpha=1)
ax1.plot(locations, sample[:, 1], ":", color="C1", linewidth=0.5, alpha=1)
ax1.plot(locations, means[:, 0], label="x1", color="k")
ax1.plot(locations, means[:, 1], label="x2", color="k")
ax1.fill_between(
locations,
means[:, 0] - 3 * stds[:, 0],
means[:, 0] + 3 * stds[:, 0],
color="C0",
alpha=0.3,
)
ax1.fill_between(
locations,
means[:, 1] - 3 * stds[:, 1],
means[:, 1] + 3 * stds[:, 1],
color="C1",
alpha=0.3,
)
ax1.plot(sol.locations, sol.states.mean, "o")
fig.suptitle(f"Samples From an EK0 Solution")
plt.show()
```
Let us have a look at the inferred diffusion model
```
x = np.linspace(-1, 22, 500)
plt.plot(x, sol.kalman_posterior.diffusion_model(x))
plt.show()
```
How well do the empirical moments of the samples reflect the uncertainty of the dense output?
```
plt.plot(samples.mean(axis=0), color="gray", linestyle="dotted")
plt.plot(means)
plt.show()
sample_mean = samples.mean(axis=0)
sample_std = samples.std(axis=0)
fig, (ax1, ax2) = plt.subplots(ncols=2)
ax1.plot(means[:, 0], sample_mean[:, 0])
ax2.plot(means[:, 1], sample_mean[:, 1])
ax1.plot(means[:, 0], means[:, 0], alpha=0.5, linewidth=8, zorder=0)
ax2.plot(means[:, 1], means[:, 1], alpha=0.5, linewidth=8, zorder=0)
ax1.axis("equal")
ax2.axis("equal")
plt.show()
fig, (ax1, ax2) = plt.subplots(ncols=2)
ax1.plot(locations, stds[:, 0], label="true-std")
ax1.plot(locations, sample_std[:, 0], label="sample-std")
ax1.legend()
ax2.plot(locations, stds[:, 1], label="true-std")
ax2.plot(locations, sample_std[:, 1], label="sample-std")
ax2.legend()
plt.show()
fig, (ax1, ax2) = plt.subplots(ncols=2)
ax1.plot(stds[:, 0], sample_std[:, 0])
ax2.plot(stds[:, 1], sample_std[:, 1])
ax1.plot(stds[:, 0], stds[:, 0], alpha=0.5, linewidth=8, zorder=0)
ax2.plot(stds[:, 1], stds[:, 1], alpha=0.5, linewidth=8, zorder=0)
ax1.axis("equal")
ax2.axis("equal")
plt.show()
```
|
github_jupyter
|
# Make inline plots vector graphics instead of raster graphics
%matplotlib inline
from IPython.display import set_matplotlib_formats
set_matplotlib_formats("pdf", "svg")
# Plotting
import matplotlib.pyplot as plt
plt.style.use("../../probnum.mplstyle")
import numpy as np
from probnum.diffeq import probsolve_ivp
rng = np.random.default_rng(seed=123)
def f(t, y):
y1, y2 = y
return np.array([0.5 * y1 - 0.05 * y1 * y2, -0.5 * y2 + 0.05 * y1 * y2])
def df(t, y):
y1, y2 = y
return np.array([[0.5 - 0.05 * y2, -0.05 * y1], [0.05 * y2, -0.5 + 0.05 * y1]])
t0 = 0.0
tmax = 20.0
y0 = np.array([20, 20])
sol = probsolve_ivp(
f, t0, tmax, y0, df=df, step=0.1, adaptive=False, diffusion_model="dynamic"
)
means, stds = sol.states.mean, sol.states.std
fig, (ax1, ax2) = plt.subplots(nrows=2, ncols=1, sharex=True)
ax1.plot(sol.locations, means[:, 0], label="x1")
ax1.plot(sol.locations, means[:, 1], label="x2")
ax1.legend()
ax2.fill_between(sol.locations, stds[:, 0], alpha=0.25, label="x1-unc.")
ax2.fill_between(sol.locations, stds[:, 1], alpha=0.25, label="x2-unc.")
ax2.legend()
fig.suptitle("EK0 Solution")
plt.show()
sol = probsolve_ivp(
f,
t0,
tmax,
y0,
df=df,
step=0.5,
algo_order=1,
adaptive=False,
diffusion_model="dynamic",
)
# sol.kalman_posterior.diffusion_model.diffusions[2:8] *= 100000
num_samples = 150
locations = np.arange(0.0, 22.1, 0.05)
locations = sol.locations
samples = sol.sample(rng=rng, t=locations, size=num_samples)
solution = sol(locations)
means = solution.mean
stds = solution.std
fig, (ax1) = plt.subplots(nrows=1, ncols=1, sharex=True)
for sample in samples[::5]:
ax1.plot(locations, sample[:, 0], ":", color="C0", linewidth=0.5, alpha=1)
ax1.plot(locations, sample[:, 1], ":", color="C1", linewidth=0.5, alpha=1)
ax1.plot(locations, means[:, 0], label="x1", color="k")
ax1.plot(locations, means[:, 1], label="x2", color="k")
ax1.fill_between(
locations,
means[:, 0] - 3 * stds[:, 0],
means[:, 0] + 3 * stds[:, 0],
color="C0",
alpha=0.3,
)
ax1.fill_between(
locations,
means[:, 1] - 3 * stds[:, 1],
means[:, 1] + 3 * stds[:, 1],
color="C1",
alpha=0.3,
)
ax1.plot(sol.locations, sol.states.mean, "o")
fig.suptitle(f"Samples From an EK0 Solution")
plt.show()
x = np.linspace(-1, 22, 500)
plt.plot(x, sol.kalman_posterior.diffusion_model(x))
plt.show()
plt.plot(samples.mean(axis=0), color="gray", linestyle="dotted")
plt.plot(means)
plt.show()
sample_mean = samples.mean(axis=0)
sample_std = samples.std(axis=0)
fig, (ax1, ax2) = plt.subplots(ncols=2)
ax1.plot(means[:, 0], sample_mean[:, 0])
ax2.plot(means[:, 1], sample_mean[:, 1])
ax1.plot(means[:, 0], means[:, 0], alpha=0.5, linewidth=8, zorder=0)
ax2.plot(means[:, 1], means[:, 1], alpha=0.5, linewidth=8, zorder=0)
ax1.axis("equal")
ax2.axis("equal")
plt.show()
fig, (ax1, ax2) = plt.subplots(ncols=2)
ax1.plot(locations, stds[:, 0], label="true-std")
ax1.plot(locations, sample_std[:, 0], label="sample-std")
ax1.legend()
ax2.plot(locations, stds[:, 1], label="true-std")
ax2.plot(locations, sample_std[:, 1], label="sample-std")
ax2.legend()
plt.show()
fig, (ax1, ax2) = plt.subplots(ncols=2)
ax1.plot(stds[:, 0], sample_std[:, 0])
ax2.plot(stds[:, 1], sample_std[:, 1])
ax1.plot(stds[:, 0], stds[:, 0], alpha=0.5, linewidth=8, zorder=0)
ax2.plot(stds[:, 1], stds[:, 1], alpha=0.5, linewidth=8, zorder=0)
ax1.axis("equal")
ax2.axis("equal")
plt.show()
| 0.799794 | 0.984155 |
<table class="ee-notebook-buttons" align="left">
<td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/Image/get_image_id.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
<td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Image/get_image_id.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
<td><a target="_blank" href="https://mybinder.org/v2/gh/giswqs/earthengine-py-notebooks/master?filepath=Image/get_image_id.ipynb"><img width=58px src="https://mybinder.org/static/images/logo_social.png" />Run in binder</a></td>
<td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Image/get_image_id.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
</table>
## Install Earth Engine API
Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geehydro](https://github.com/giswqs/geehydro). The **geehydro** Python package builds on the [folium](https://github.com/python-visualization/folium) package and implements several methods for displaying Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, `Map.centerObject()`, and `Map.setOptions()`.
The magic command `%%capture` can be used to hide output from a specific cell.
```
# %%capture
# !pip install earthengine-api
# !pip install geehydro
```
Import libraries
```
import ee
import folium
import geehydro
```
Authenticate and initialize Earth Engine API. You only need to authenticate the Earth Engine API once. Uncomment the line `ee.Authenticate()`
if you are running this notebook for this first time or if you are getting an authentication error.
```
# ee.Authenticate()
ee.Initialize()
```
## Create an interactive map
This step creates an interactive map using [folium](https://github.com/python-visualization/folium). The default basemap is the OpenStreetMap. Additional basemaps can be added using the `Map.setOptions()` function.
The optional basemaps can be `ROADMAP`, `SATELLITE`, `HYBRID`, `TERRAIN`, or `ESRI`.
```
Map = folium.Map(location=[40, -100], zoom_start=4)
Map.setOptions('HYBRID')
```
## Add Earth Engine Python script
```
# This function masks the input with a threshold on the simple cloud score.
def cloudMask(img):
cloudscore = ee.Algorithms.Landsat.simpleCloudScore(img).select('cloud')
return img.updateMask(cloudscore.lt(50))
# Load a Landsat 5 image collection.
collection = ee.ImageCollection('LANDSAT/LT5_L1T_TOA') \
.filterDate('2008-04-01', '2010-04-01') \
.filterBounds(ee.Geometry.Point(-122.2627, 37.8735)) \
.map(cloudMask) \
.select(['B4', 'B3']) \
.sort('system:time_start', True) # Sort the collection in chronological order.
print(collection.size().getInfo())
first = collection.first()
propertyNames = first.propertyNames()
print(propertyNames.getInfo())
uid = first.get('system:id')
print(uid.getInfo())
```
## Display Earth Engine data layers
```
Map.setControlVisibility(layerControl=True, fullscreenControl=True, latLngPopup=True)
Map
```
|
github_jupyter
|
# %%capture
# !pip install earthengine-api
# !pip install geehydro
import ee
import folium
import geehydro
# ee.Authenticate()
ee.Initialize()
Map = folium.Map(location=[40, -100], zoom_start=4)
Map.setOptions('HYBRID')
# This function masks the input with a threshold on the simple cloud score.
def cloudMask(img):
cloudscore = ee.Algorithms.Landsat.simpleCloudScore(img).select('cloud')
return img.updateMask(cloudscore.lt(50))
# Load a Landsat 5 image collection.
collection = ee.ImageCollection('LANDSAT/LT5_L1T_TOA') \
.filterDate('2008-04-01', '2010-04-01') \
.filterBounds(ee.Geometry.Point(-122.2627, 37.8735)) \
.map(cloudMask) \
.select(['B4', 'B3']) \
.sort('system:time_start', True) # Sort the collection in chronological order.
print(collection.size().getInfo())
first = collection.first()
propertyNames = first.propertyNames()
print(propertyNames.getInfo())
uid = first.get('system:id')
print(uid.getInfo())
Map.setControlVisibility(layerControl=True, fullscreenControl=True, latLngPopup=True)
Map
| 0.698946 | 0.950134 |
```
import sys
sys.path.append('../')
import controller
import torch
import torch.nn as nn
import random
import numpy as np
import matplotlib.pyplot as plt
MODE = 3
MODE_DICT = {0: "Training Neural Model", 1: "Demonstrate Neural Model",
2: "Demonstrate Simple Equation", 3: "Test Neural Model",
4: "Test Simple Equation"}
print(MODE_DICT[MODE])
CAR_MODEL = "complex"
ci = controller.Car_Interface(model = CAR_MODEL)
ci.set_gear(ci.FORWARD)
np.random.seed(181)
# Accelerates the given car interface until it arrives at the target velocity
def accelerate_until_velocity(ci, target_vel):
while(len(ci.log["velocity"]) == 0 or ci.log["velocity"][-1] < target_vel):
ci.apply_control(pedal = ci.ACCELERATOR, amount = 1.0)
'''
Accelerate until a randomly determined target velocity.
Record the actual achieved velocity (which maybe a little higher)
Then brake for enough time, so that we know we've stopped.
The stopping distance is then the difference in the car's position
now and when we started braking.
'''
def stopping_distance_gen(ci, t = 50):
amt = 0.15 + random.random() * 0.85
initial_velocity = random.random()
accelerate_until_velocity(ci, initial_velocity)
initial_velocity = ci.log["velocity"][-1]
ci.zero_position()
ci.apply_control_for_time(pedal = ci.BRAKE, amount = amt, time = t)
stopping_distance = ci.log["position"][-1]
inp = [initial_velocity, stopping_distance]
return amt, inp
amt, (initial_velocity, stopping_distance) = stopping_distance_gen(ci)
print(f"A car moving at {initial_velocity * 100:.2f}% speed, applied {amt * 100:.2f}% brakes and stopped, after travelling {stopping_distance:.2f} distance units")
# Analytically solve for the theoretical stopping distance
def actual_stopping_distance(initial_velocity, amt):
x = ci.brake_weight * amt + ci.rolling_bias
f = ci.friction_constant
v0 = initial_velocity
if ((1-f*v0/x) < 0):
return float("inf")
d = ((x/f)*np.log(1-f*v0/x) + v0)/f
return d
'''
Use binary search to approximate the required brake amount,
that results in a target stopping distance given an intial velocity.
'''
def approximate_amount(inp, tol = 1e-5, min_amt = 0, max_amt = 1):
mid_amt = (min_amt + max_amt) / 2
if (max_amt - min_amt < 2 * tol):
return mid_amt
v0, stopping_distance = inp
if (actual_stopping_distance(v0, mid_amt) < stopping_distance):
return approximate_amount(inp, tol, min_amt, mid_amt)
else:
return approximate_amount(inp, tol, mid_amt, max_amt)
if (MODE == 4):
for i in range(3):
amt, (initial_velocity, stopping_distance) = stopping_distance_gen(ci)
pred = approximate_amount((initial_velocity, stopping_distance))
print(f"Car moving at {initial_velocity * 100:.2f}%; Target Stopping Distance {stopping_distance:.2f} distance units")
print(f"Simulation Brake Amount: {amt*100:.2f}%; Closed Form Brake Amount {pred*100:.2f}%")
print()
# Fully Connected Network Class (a custom subclass of torch's nn module)
class fcn(nn.Module):
def __init__(self):
super().__init__()
# Number of hidden units in first hidden layer
self.H_1 = 30
# Number of hidden units in second hidden layer
self.H_2 = 20
'''
Weights generally [input dim, output dim] so when we multiply a vector
of size [input dim] by a matrix of size [input dim, output dim] we get
a vector of size [output dim]. The bias will have shape [output dim]
so we can add it to the result of the weight-vector multiplication.
'''
#Weights and Biases for computing input -> first hidden layer
self.W_1 = nn.Parameter(torch.randn([2, self.H_1]))
self.B_1 = nn.Parameter(torch.randn([self.H_1]))
#Weights and Biases for computing first -> second hidden layer
self.W_2 = nn.Parameter(torch.randn([self.H_1, self.H_2]))
self.B_2 = nn.Parameter(torch.randn([self.H_2]))
#Weights and Biases for computing second hidden layer -> output
self.W_3 = nn.Parameter(torch.randn([self.H_2, 1]))
self.B_3 = nn.Parameter(torch.randn([1]))
# Forward propogation
def forward(self, x):
# x will be a vector of length 2 containing the initial velocity and desired stopping distance
x = torch.tensor(x, dtype = torch.float32)
# first hidden layer computation with tanh activation
h_1 = torch.tanh(torch.matmul(x, self.W_1) + self.B_1)
# second hidden layer computation with tanh activation
h_2 = torch.tanh(torch.matmul(h_1, self.W_2) + self.B_2)
#output computation with no activation. We technically get a vector of length 1 so we squeeze to get value.
out = torch.squeeze(torch.matmul(h_2, self.W_3) + self.B_3)
'''
Our output is a scaled sigmoid (output range (0, 1.15)). This helps model learn faster since all
desired outputs are in the range (0.15, 1).
'''
return 1.15 * torch.sigmoid(out)
#If demonstrating or testing just load initialize FCN with learned weights
if (MODE == 1 or MODE == 3):
FN = "weights_" + CAR_MODEL
model = fcn()
model.load_state_dict(torch.load(open(FN + ".pt", "rb")))
#Training (not we may start from previously learned weights)
if (MODE == 0):
# Number of batches of data in a "epoch"
NUM_BATCHES = 10
# Number of data points in a single batch
BATCH_SIZE = 30
'''
Number of epochs. Note generally an epoch is a pass through the whole dataset.
Here we are artifically generated data, so the size of an epoch is artificial as well.
It will be (NUM_BATCHES * BATCH_SIZE).
'''
EPOCHS = 10
# Do we want to use previously trained weights?
USE_LAST = False
FN = "weights_" + CAR_MODEL
model = fcn()
if(USE_LAST):
model.load_state_dict(torch.load(open(FN + ".pt", "rb")))
'''
Adam is an improved version of standard Stochastic Gradient Descent.
The Stochastic refers to the fact that we update weights based on
a small subset of our overall dataset, rather than computing the gradient
over the entire dataset which can very time consuming. Typically small
batches represent the overall data patterns in the long run.
'''
optimizer = torch.optim.Adam(model.parameters(), lr = 1e-3)
for e in range(EPOCHS):
# epoch loss
e_loss = 0
for b in range(NUM_BATCHES):
# batch loss
b_loss = 0
for i in range(BATCH_SIZE):
# Generate a data point
amt, inp = stopping_distance_gen(ci)
# Find the model's predicted brake amount
out = model(inp)
# Compute MSE between model output and actual
amt_t = torch.tensor(amt)
b_loss += (out - amt_t) ** 2
b_loss /= BATCH_SIZE
# Update weights
optimizer.zero_grad()
b_loss.backward()
optimizer.step()
b_loss = b_loss.detach().numpy()
print(f"B {b} L {b_loss:.4f}", end = "\r")
e_loss += b_loss
e_loss /= NUM_BATCHES
print(f"EPOCH {e + 1} {e_loss:.4f}")
# save weights
torch.save(model.state_dict(), open(FN + ".pt", "wb"))
# Demonstrate model's effectiveness on a few generated data points
if (MODE == 1 or MODE == 2):
DEM = 5
for i in range(DEM):
amt, inp = stopping_distance_gen(ci)
if (MODE == 1):
out = model(inp).detach().numpy()
elif (MODE == 2):
out = approximate_amount(inp, tol = 1e-5)
print(f"INIT VEL: {inp[0]:.3f} TARG SD: {inp[1]:.3f} BRK AMT: {amt:.3f} MODEL OUT:{out:.3f} ")
# Evaluate models over a larger set of datapoints, plot error distribution.
if (MODE == 3 or MODE == 4):
TEST = 300
correct = 0
tol = 0.1
errors = []
for i in range(TEST):
print(f"TESTING {i + 1}/{TEST}", end = "\r")
amt, inp = stopping_distance_gen(ci)
if (MODE == 3):
out = model(inp).detach().numpy()
elif (MODE == 4):
out = approximate_amount(inp, tol = 1e-5)
if (abs(out - amt) < tol):
correct += 1
errors.append(out - amt)
print(f"WITHIN {tol} {correct}/{TEST} times")
print(f"AVERAGE ERROR {np.mean(np.abs(errors))}")
plt.title("Error Distribution")
plt.hist(errors, bins = 200, range = (-1, 1))
plt.show()
```
|
github_jupyter
|
import sys
sys.path.append('../')
import controller
import torch
import torch.nn as nn
import random
import numpy as np
import matplotlib.pyplot as plt
MODE = 3
MODE_DICT = {0: "Training Neural Model", 1: "Demonstrate Neural Model",
2: "Demonstrate Simple Equation", 3: "Test Neural Model",
4: "Test Simple Equation"}
print(MODE_DICT[MODE])
CAR_MODEL = "complex"
ci = controller.Car_Interface(model = CAR_MODEL)
ci.set_gear(ci.FORWARD)
np.random.seed(181)
# Accelerates the given car interface until it arrives at the target velocity
def accelerate_until_velocity(ci, target_vel):
while(len(ci.log["velocity"]) == 0 or ci.log["velocity"][-1] < target_vel):
ci.apply_control(pedal = ci.ACCELERATOR, amount = 1.0)
'''
Accelerate until a randomly determined target velocity.
Record the actual achieved velocity (which maybe a little higher)
Then brake for enough time, so that we know we've stopped.
The stopping distance is then the difference in the car's position
now and when we started braking.
'''
def stopping_distance_gen(ci, t = 50):
amt = 0.15 + random.random() * 0.85
initial_velocity = random.random()
accelerate_until_velocity(ci, initial_velocity)
initial_velocity = ci.log["velocity"][-1]
ci.zero_position()
ci.apply_control_for_time(pedal = ci.BRAKE, amount = amt, time = t)
stopping_distance = ci.log["position"][-1]
inp = [initial_velocity, stopping_distance]
return amt, inp
amt, (initial_velocity, stopping_distance) = stopping_distance_gen(ci)
print(f"A car moving at {initial_velocity * 100:.2f}% speed, applied {amt * 100:.2f}% brakes and stopped, after travelling {stopping_distance:.2f} distance units")
# Analytically solve for the theoretical stopping distance
def actual_stopping_distance(initial_velocity, amt):
x = ci.brake_weight * amt + ci.rolling_bias
f = ci.friction_constant
v0 = initial_velocity
if ((1-f*v0/x) < 0):
return float("inf")
d = ((x/f)*np.log(1-f*v0/x) + v0)/f
return d
'''
Use binary search to approximate the required brake amount,
that results in a target stopping distance given an intial velocity.
'''
def approximate_amount(inp, tol = 1e-5, min_amt = 0, max_amt = 1):
mid_amt = (min_amt + max_amt) / 2
if (max_amt - min_amt < 2 * tol):
return mid_amt
v0, stopping_distance = inp
if (actual_stopping_distance(v0, mid_amt) < stopping_distance):
return approximate_amount(inp, tol, min_amt, mid_amt)
else:
return approximate_amount(inp, tol, mid_amt, max_amt)
if (MODE == 4):
for i in range(3):
amt, (initial_velocity, stopping_distance) = stopping_distance_gen(ci)
pred = approximate_amount((initial_velocity, stopping_distance))
print(f"Car moving at {initial_velocity * 100:.2f}%; Target Stopping Distance {stopping_distance:.2f} distance units")
print(f"Simulation Brake Amount: {amt*100:.2f}%; Closed Form Brake Amount {pred*100:.2f}%")
print()
# Fully Connected Network Class (a custom subclass of torch's nn module)
class fcn(nn.Module):
def __init__(self):
super().__init__()
# Number of hidden units in first hidden layer
self.H_1 = 30
# Number of hidden units in second hidden layer
self.H_2 = 20
'''
Weights generally [input dim, output dim] so when we multiply a vector
of size [input dim] by a matrix of size [input dim, output dim] we get
a vector of size [output dim]. The bias will have shape [output dim]
so we can add it to the result of the weight-vector multiplication.
'''
#Weights and Biases for computing input -> first hidden layer
self.W_1 = nn.Parameter(torch.randn([2, self.H_1]))
self.B_1 = nn.Parameter(torch.randn([self.H_1]))
#Weights and Biases for computing first -> second hidden layer
self.W_2 = nn.Parameter(torch.randn([self.H_1, self.H_2]))
self.B_2 = nn.Parameter(torch.randn([self.H_2]))
#Weights and Biases for computing second hidden layer -> output
self.W_3 = nn.Parameter(torch.randn([self.H_2, 1]))
self.B_3 = nn.Parameter(torch.randn([1]))
# Forward propogation
def forward(self, x):
# x will be a vector of length 2 containing the initial velocity and desired stopping distance
x = torch.tensor(x, dtype = torch.float32)
# first hidden layer computation with tanh activation
h_1 = torch.tanh(torch.matmul(x, self.W_1) + self.B_1)
# second hidden layer computation with tanh activation
h_2 = torch.tanh(torch.matmul(h_1, self.W_2) + self.B_2)
#output computation with no activation. We technically get a vector of length 1 so we squeeze to get value.
out = torch.squeeze(torch.matmul(h_2, self.W_3) + self.B_3)
'''
Our output is a scaled sigmoid (output range (0, 1.15)). This helps model learn faster since all
desired outputs are in the range (0.15, 1).
'''
return 1.15 * torch.sigmoid(out)
#If demonstrating or testing just load initialize FCN with learned weights
if (MODE == 1 or MODE == 3):
FN = "weights_" + CAR_MODEL
model = fcn()
model.load_state_dict(torch.load(open(FN + ".pt", "rb")))
#Training (not we may start from previously learned weights)
if (MODE == 0):
# Number of batches of data in a "epoch"
NUM_BATCHES = 10
# Number of data points in a single batch
BATCH_SIZE = 30
'''
Number of epochs. Note generally an epoch is a pass through the whole dataset.
Here we are artifically generated data, so the size of an epoch is artificial as well.
It will be (NUM_BATCHES * BATCH_SIZE).
'''
EPOCHS = 10
# Do we want to use previously trained weights?
USE_LAST = False
FN = "weights_" + CAR_MODEL
model = fcn()
if(USE_LAST):
model.load_state_dict(torch.load(open(FN + ".pt", "rb")))
'''
Adam is an improved version of standard Stochastic Gradient Descent.
The Stochastic refers to the fact that we update weights based on
a small subset of our overall dataset, rather than computing the gradient
over the entire dataset which can very time consuming. Typically small
batches represent the overall data patterns in the long run.
'''
optimizer = torch.optim.Adam(model.parameters(), lr = 1e-3)
for e in range(EPOCHS):
# epoch loss
e_loss = 0
for b in range(NUM_BATCHES):
# batch loss
b_loss = 0
for i in range(BATCH_SIZE):
# Generate a data point
amt, inp = stopping_distance_gen(ci)
# Find the model's predicted brake amount
out = model(inp)
# Compute MSE between model output and actual
amt_t = torch.tensor(amt)
b_loss += (out - amt_t) ** 2
b_loss /= BATCH_SIZE
# Update weights
optimizer.zero_grad()
b_loss.backward()
optimizer.step()
b_loss = b_loss.detach().numpy()
print(f"B {b} L {b_loss:.4f}", end = "\r")
e_loss += b_loss
e_loss /= NUM_BATCHES
print(f"EPOCH {e + 1} {e_loss:.4f}")
# save weights
torch.save(model.state_dict(), open(FN + ".pt", "wb"))
# Demonstrate model's effectiveness on a few generated data points
if (MODE == 1 or MODE == 2):
DEM = 5
for i in range(DEM):
amt, inp = stopping_distance_gen(ci)
if (MODE == 1):
out = model(inp).detach().numpy()
elif (MODE == 2):
out = approximate_amount(inp, tol = 1e-5)
print(f"INIT VEL: {inp[0]:.3f} TARG SD: {inp[1]:.3f} BRK AMT: {amt:.3f} MODEL OUT:{out:.3f} ")
# Evaluate models over a larger set of datapoints, plot error distribution.
if (MODE == 3 or MODE == 4):
TEST = 300
correct = 0
tol = 0.1
errors = []
for i in range(TEST):
print(f"TESTING {i + 1}/{TEST}", end = "\r")
amt, inp = stopping_distance_gen(ci)
if (MODE == 3):
out = model(inp).detach().numpy()
elif (MODE == 4):
out = approximate_amount(inp, tol = 1e-5)
if (abs(out - amt) < tol):
correct += 1
errors.append(out - amt)
print(f"WITHIN {tol} {correct}/{TEST} times")
print(f"AVERAGE ERROR {np.mean(np.abs(errors))}")
plt.title("Error Distribution")
plt.hist(errors, bins = 200, range = (-1, 1))
plt.show()
| 0.765506 | 0.622459 |
# Programming Assignment:
## Готовим LDA по рецептам
Как вы уже знаете, в тематическом моделировании делается предположение о том, что для определения тематики порядок слов в документе не важен; об этом гласит гипотеза «мешка слов». Сегодня мы будем работать с несколько нестандартной для тематического моделирования коллекцией, которую можно назвать «мешком ингредиентов», потому что на состоит из рецептов блюд разных кухонь. Тематические модели ищут слова, которые часто вместе встречаются в документах, и составляют из них темы. Мы попробуем применить эту идею к рецептам и найти кулинарные «темы». Эта коллекция хороша тем, что не требует предобработки. Кроме того, эта задача достаточно наглядно иллюстрирует принцип работы тематических моделей.
Для выполнения заданий, помимо часто используемых в курсе библиотек, потребуются модули *json* и *gensim*. Первый входит в дистрибутив Anaconda, второй можно поставить командой
*pip install gensim*
Построение модели занимает некоторое время. На ноутбуке с процессором Intel Core i7 и тактовой частотой 2400 МГц на построение одной модели уходит менее 10 минут.
### Загрузка данных
Коллекция дана в json-формате: для каждого рецепта известны его id, кухня (cuisine) и список ингредиентов, в него входящих. Загрузить данные можно с помощью модуля json (он входит в дистрибутив Anaconda):
```
import json
with open("recipes.json") as f:
recipes = json.load(f)
print(recipes[0])
```
### Составление корпуса
```
from gensim import corpora, models
import numpy as np
```
Наша коллекция небольшая, и целиком помещается в оперативную память. Gensim может работать с такими данными и не требует их сохранения на диск в специальном формате. Для этого коллекция должна быть представлена в виде списка списков, каждый внутренний список соответствует отдельному документу и состоит из его слов. Пример коллекции из двух документов:
[["hello", "world"], ["programming", "in", "python"]]
Преобразуем наши данные в такой формат, а затем создадим объекты corpus и dictionary, с которыми будет работать модель.
```
texts = [recipe["ingredients"] for recipe in recipes]
dictionary = corpora.Dictionary(texts) # составляем словарь
corpus = [dictionary.doc2bow(text) for text in texts] # составляем корпус документов
print(texts[0])
print(corpus[0])
```
У объекта dictionary есть полезная переменная dictionary.token2id, позволяющая находить соответствие между ингредиентами и их индексами.
### Обучение модели
Вам может понадобиться [документация](https://radimrehurek.com/gensim/models/ldamodel.html) LDA в gensim.
__Задание 1.__ Обучите модель LDA с 40 темами, установив количество проходов по коллекции 5 и оставив остальные параметры по умолчанию.
Затем вызовите метод модели *show_topics*, указав количество тем 40 и количество токенов 10, и сохраните результат (топы ингредиентов в темах) в отдельную переменную. Если при вызове метода *show_topics* указать параметр *formatted=True*, то топы ингредиентов будет удобно выводить на печать, если *formatted=False*, будет удобно работать со списком программно. Выведите топы на печать, рассмотрите темы, а затем ответьте на вопрос:
Сколько раз ингредиенты "salt", "sugar", "water", "mushrooms", "chicken", "eggs" встретились среди топов-10 всех 40 тем? При ответе __не нужно__ учитывать составные ингредиенты, например, "hot water".
Передайте 6 чисел в функцию save_answers1 и загрузите сгенерированный файл в форму.
У gensim нет возможности фиксировать случайное приближение через параметры метода, но библиотека использует numpy для инициализации матриц. Поэтому, по утверждению автора библиотеки, фиксировать случайное приближение нужно командой, которая написана в следующей ячейке. __Перед строкой кода с построением модели обязательно вставляйте указанную строку фиксации random.seed.__
```
np.random.seed(76543)
# здесь код для построения модели:
%time ldamodel = models.ldamodel.LdaModel(corpus, id2word=dictionary, num_topics=40, passes=5)
topics = ldamodel.show_topics(num_topics=40, num_words=10, formatted=True)
print(topics)
def word_counter(word, text):
s = 0
for line in text:
if word in str(line):
s += 1
return s
c_salt = word_counter('"salt"', topics)
c_sugar = word_counter('"sugar"', topics)
c_water = word_counter('"water"', topics)
c_mushrooms = word_counter('"mushrooms"', topics)
c_chicken = word_counter('"chicken"', topics)
c_eggs = word_counter('"eggs"', topics)
def save_answers1(c_salt, c_sugar, c_water, c_mushrooms, c_chicken, c_eggs):
with open("cooking_LDA_pa_task1.txt", "w") as fout:
fout.write(" ".join([str(el) for el in [c_salt, c_sugar, c_water, c_mushrooms, c_chicken, c_eggs]]))
#save_answers1(23, 9, 10, 0, 1, 2)
save_answers1(c_salt, c_sugar, c_water, c_mushrooms, c_chicken, c_eggs)
```
### Фильтрация словаря
В топах тем гораздо чаще встречаются первые три рассмотренных ингредиента, чем последние три. При этом наличие в рецепте курицы, яиц и грибов яснее дает понять, что мы будем готовить, чем наличие соли, сахара и воды. Таким образом, даже в рецептах есть слова, часто встречающиеся в текстах и не несущие смысловой нагрузки, и поэтому их не желательно видеть в темах. Наиболее простой прием борьбы с такими фоновыми элементами — фильтрация словаря по частоте. Обычно словарь фильтруют с двух сторон: убирают очень редкие слова (в целях экономии памяти) и очень частые слова (в целях повышения интерпретируемости тем). Мы уберем только частые слова.
```
import copy
dictionary2 = copy.deepcopy(dictionary)
```
__Задание 2.__ У объекта dictionary2 есть переменная *dfs* — это словарь, ключами которого являются id токена, а элементами — число раз, сколько слово встретилось во всей коллекции. Сохраните в отдельный список ингредиенты, которые встретились в коллекции больше 4000 раз. Вызовите метод словаря *filter_tokens*, подав в качестве первого аргумента полученный список популярных ингредиентов. Вычислите две величины: dict_size_before и dict_size_after — размер словаря до и после фильтрации.
Затем, используя новый словарь, создайте новый корпус документов, corpus2, по аналогии с тем, как это сделано в начале ноутбука. Вычислите две величины: corpus_size_before и corpus_size_after — суммарное количество ингредиентов в корпусе (для каждого документа вычислите число различных ингредиентов в нем и просуммируйте по всем документам) до и после фильтрации.
Передайте величины dict_size_before, dict_size_after, corpus_size_before, corpus_size_after в функцию save_answers2 и загрузите сгенерированный файл в форму.
```
def count_size_corpus_ingredients(_corpus):
_sum = 0
for doc in _corpus:
_sum += len(doc)
return _sum
dict_size_before = len(dictionary2)
corpus2 = [dictionary2.doc2bow(text) for text in texts]
corpus_size_before = count_size_corpus_ingredients(corpus2)
print(dict_size_before)
print(corpus_size_before)
ids_four_thousand = []
for i in range(0, len(dictionary2)):
if dictionary2.dfs[i] > 4000:
ids_four_thousand.append(i)
ids_four_thousand
dictionary2.filter_tokens(bad_ids=ids_four_thousand)
dict_size_after = len(dictionary2)
corpus2 = [dictionary2.doc2bow(text) for text in texts]
corpus_size_after = count_size_corpus_ingredients(corpus2)
print(dict_size_after)
print(corpus_size_after)
def save_answers2(dict_size_before, dict_size_after, corpus_size_before, corpus_size_after):
with open("cooking_LDA_pa_task2.txt", "w") as fout:
fout.write(" ".join([str(el) for el in [dict_size_before, dict_size_after, corpus_size_before, corpus_size_after]]))
save_answers2(dict_size_before, dict_size_after, corpus_size_before, corpus_size_after)
```
### Сравнение когерентностей
__Задание 3.__ Постройте еще одну модель по корпусу corpus2 и словарю dictionary2, остальные параметры оставьте такими же, как при первом построении модели. Сохраните новую модель в другую переменную (не перезаписывайте предыдущую модель). Не забудьте про фиксирование seed!
Затем воспользуйтесь методом *top_topics* модели, чтобы вычислить ее когерентность. Передайте в качестве аргумента соответствующий модели корпус. Метод вернет список кортежей (топ токенов, когерентность), отсортированных по убыванию последней. Вычислите среднюю по всем темам когерентность для каждой из двух моделей и передайте в функцию save_answers3.
```
np.random.seed(76543)
# здесь код для построения модели:
%time ldamodel2 = models.ldamodel.LdaModel(corpus2, id2word=dictionary2, num_topics=40, passes=5)
cog2 = ldamodel2.top_topics(corpus2)
cog = ldamodel.top_topics(corpus)
def mean(coher):
_sum = 0
for c in coher:
_sum += c[1]
return _sum / len(coher)
coherence2 = mean(cog2)
coherence = mean(cog)
def save_answers3(coherence, coherence2):
with open("cooking_LDA_pa_task3.txt", "w") as fout:
fout.write(" ".join(["%3f"%el for el in [coherence, coherence2]]))
save_answers3(coherence, coherence2)
```
Считается, что когерентность хорошо соотносится с человеческими оценками интерпретируемости тем. Поэтому на больших текстовых коллекциях когерентность обычно повышается, если убрать фоновую лексику. Однако в нашем случае этого не произошло.
### Изучение влияния гиперпараметра alpha
В этом разделе мы будем работать со второй моделью, то есть той, которая построена по сокращенному корпусу.
Пока что мы посмотрели только на матрицу темы-слова, теперь давайте посмотрим на матрицу темы-документы. Выведите темы для нулевого (или любого другого) документа из корпуса, воспользовавшись методом *get_document_topics* второй модели:
```
ldamodel2.get_document_topics(corpus[0])
```
Также выведите содержимое переменной *.alpha* второй модели:
У вас должно получиться, что документ характеризуется небольшим числом тем. Попробуем поменять гиперпараметр alpha, задающий априорное распределение Дирихле для распределений тем в документах.
__Задание 4.__ Обучите третью модель: используйте сокращенный корпус (corpus2 и dictionary2) и установите параметр __alpha=1__, passes=5. Не забудьте про фиксацию seed! Выведите темы новой модели для нулевого документа; должно получиться, что распределение над множеством тем практически равномерное. Чтобы убедиться в том, что во второй модели документы описываются гораздо более разреженными распределениями, чем в третьей, посчитайте суммарное количество элементов, __превосходящих 0.01__, в матрицах темы-документы обеих моделей. Другими словами, запросите темы модели для каждого документа с параметром *minimum_probability=0.01* и просуммируйте число элементов в получаемых массивах. Передайте две суммы (сначала для модели с alpha по умолчанию, затем для модели в alpha=1) в функцию save_answers4.
```
np.random.seed(76543)
%time ldamodel3 = models.ldamodel.LdaModel(corpus2, id2word=dictionary2, passes=5, alpha=1, num_topics=40)
count_model2 = []
for j in range(0,39774):
TemaDocument2 = ldamodel2.get_document_topics(corpus2[j], minimum_probability=0.01)
count_model2.append(len(TemaDocument2))
sum(count_model2)
np.random.seed(76543)
count_model3 = []
for j in range(0,39774):
TemaDocument3 = ldamodel3.get_document_topics(corpus2[j], minimum_probability=0.01)
count_model3.append(len(TemaDocument3))
sum(count_model3)
def save_answers4(count_model2, count_model3):
with open("cooking_LDA_pa_task4.txt", "w") as fout:
fout.write(" ".join([str(el) for el in [count_model2, count_model3]]))
save_answers4(sum(count_model2), sum(count_model3))
```
Таким образом, гиперпараметр __alpha__ влияет на разреженность распределений тем в документах. Аналогично гиперпараметр __eta__ влияет на разреженность распределений слов в темах.
### LDA как способ понижения размерности
Иногда, распределения над темами, найденные с помощью LDA, добавляют в матрицу объекты-признаки как дополнительные, семантические, признаки, и это может улучшить качество решения задачи. Для простоты давайте просто обучим классификатор рецептов на кухни на признаках, полученных из LDA, и измерим точность (accuracy).
__Задание 5.__ Используйте модель, построенную по сокращенной выборке с alpha по умолчанию (вторую модель). Составьте матрицу $\Theta = p(t|d)$ вероятностей тем в документах; вы можете использовать тот же метод get_document_topics, а также вектор правильных ответов y (в том же порядке, в котором рецепты идут в переменной recipes). Создайте объект RandomForestClassifier со 100 деревьями, с помощью функции cross_val_score вычислите среднюю accuracy по трем фолдам (перемешивать данные не нужно) и передайте в функцию save_answers5.
```
from sklearn.ensemble import RandomForestClassifier
from sklearn.cross_validation import cross_val_score
def save_answers5(accuracy):
with open("cooking_LDA_pa_task5.txt", "w") as fout:
fout.write(str(accuracy))
```
Для такого большого количества классов это неплохая точность. Вы можете попроовать обучать RandomForest на исходной матрице частот слов, имеющей значительно большую размерность, и увидеть, что accuracy увеличивается на 10–15%. Таким образом, LDA собрал не всю, но достаточно большую часть информации из выборки, в матрице низкого ранга.
### LDA — вероятностная модель
Матричное разложение, использующееся в LDA, интерпретируется как следующий процесс генерации документов.
Для документа $d$ длины $n_d$:
1. Из априорного распределения Дирихле с параметром alpha сгенерировать распределение над множеством тем: $\theta_d \sim Dirichlet(\alpha)$
1. Для каждого слова $w = 1, \dots, n_d$:
1. Сгенерировать тему из дискретного распределения $t \sim \theta_{d}$
1. Сгенерировать слово из дискретного распределения $w \sim \phi_{t}$.
Подробнее об этом в [Википедии](https://en.wikipedia.org/wiki/Latent_Dirichlet_allocation).
В контексте нашей задачи получается, что, используя данный генеративный процесс, можно создавать новые рецепты. Вы можете передать в функцию модель и число ингредиентов и сгенерировать рецепт :)
```
def generate_recipe(model, num_ingredients):
theta = np.random.dirichlet(model.alpha)
for i in range(num_ingredients):
t = np.random.choice(np.arange(model.num_topics), p=theta)
topic = model.show_topic(t, topn=model.num_terms)
topic_distr = [x[1] for x in topic]
terms = [x[0] for x in topic]
w = np.random.choice(terms, p=topic_distr)
print w
```
### Интерпретация построенной модели
Вы можете рассмотреть топы ингредиентов каждой темы. Большиснтво тем сами по себе похожи на рецепты; в некоторых собираются продукты одного вида, например, свежие фрукты или разные виды сыра.
Попробуем эмпирически соотнести наши темы с национальными кухнями (cuisine). Построим матрицу $A$ размера темы $x$ кухни, ее элементы $a_{tc}$ — суммы $p(t|d)$ по всем документам $d$, которые отнесены к кухне $c$. Нормируем матрицу на частоты рецептов по разным кухням, чтобы избежать дисбаланса между кухнями. Следующая функция получает на вход объект модели, объект корпуса и исходные данные и возвращает нормированную матрицу $A$. Ее удобно визуализировать с помощью seaborn.
```
import pandas
import seaborn
from matplotlib import pyplot as plt
%matplotlib inline
def compute_topic_cuisine_matrix(model, corpus, recipes):
# составляем вектор целевых признаков
targets = list(set([recipe["cuisine"] for recipe in recipes]))
# составляем матрицу
tc_matrix = pandas.DataFrame(data=np.zeros((model.num_topics, len(targets))), columns=targets)
for recipe, bow in zip(recipes, corpus):
recipe_topic = model.get_document_topics(bow)
for t, prob in recipe_topic:
tc_matrix[recipe["cuisine"]][t] += prob
# нормируем матрицу
target_sums = pandas.DataFrame(data=np.zeros((1, len(targets))), columns=targets)
for recipe in recipes:
target_sums[recipe["cuisine"]] += 1
return pandas.DataFrame(tc_matrix.values/target_sums.values, columns=tc_matrix.columns)
def plot_matrix(tc_matrix):
plt.figure(figsize=(10, 10))
seaborn.heatmap(tc_matrix, square=True)
# Визуализируйте матрицу
```
Чем темнее квадрат в матрице, тем больше связь этой темы с данной кухней. Мы видим, что у нас есть темы, которые связаны с несколькими кухнями. Такие темы показывают набор ингредиентов, которые популярны в кухнях нескольких народов, то есть указывают на схожесть кухонь этих народов. Некоторые темы распределены по всем кухням равномерно, они показывают наборы продуктов, которые часто используются в кулинарии всех стран.
Жаль, что в датасете нет названий рецептов, иначе темы было бы проще интерпретировать...
### Заключение
В этом задании вы построили несколько моделей LDA, посмотрели, на что влияют гиперпараметры модели и как можно использовать построенную модель.
|
github_jupyter
|
import json
with open("recipes.json") as f:
recipes = json.load(f)
print(recipes[0])
from gensim import corpora, models
import numpy as np
texts = [recipe["ingredients"] for recipe in recipes]
dictionary = corpora.Dictionary(texts) # составляем словарь
corpus = [dictionary.doc2bow(text) for text in texts] # составляем корпус документов
print(texts[0])
print(corpus[0])
np.random.seed(76543)
# здесь код для построения модели:
%time ldamodel = models.ldamodel.LdaModel(corpus, id2word=dictionary, num_topics=40, passes=5)
topics = ldamodel.show_topics(num_topics=40, num_words=10, formatted=True)
print(topics)
def word_counter(word, text):
s = 0
for line in text:
if word in str(line):
s += 1
return s
c_salt = word_counter('"salt"', topics)
c_sugar = word_counter('"sugar"', topics)
c_water = word_counter('"water"', topics)
c_mushrooms = word_counter('"mushrooms"', topics)
c_chicken = word_counter('"chicken"', topics)
c_eggs = word_counter('"eggs"', topics)
def save_answers1(c_salt, c_sugar, c_water, c_mushrooms, c_chicken, c_eggs):
with open("cooking_LDA_pa_task1.txt", "w") as fout:
fout.write(" ".join([str(el) for el in [c_salt, c_sugar, c_water, c_mushrooms, c_chicken, c_eggs]]))
#save_answers1(23, 9, 10, 0, 1, 2)
save_answers1(c_salt, c_sugar, c_water, c_mushrooms, c_chicken, c_eggs)
import copy
dictionary2 = copy.deepcopy(dictionary)
def count_size_corpus_ingredients(_corpus):
_sum = 0
for doc in _corpus:
_sum += len(doc)
return _sum
dict_size_before = len(dictionary2)
corpus2 = [dictionary2.doc2bow(text) for text in texts]
corpus_size_before = count_size_corpus_ingredients(corpus2)
print(dict_size_before)
print(corpus_size_before)
ids_four_thousand = []
for i in range(0, len(dictionary2)):
if dictionary2.dfs[i] > 4000:
ids_four_thousand.append(i)
ids_four_thousand
dictionary2.filter_tokens(bad_ids=ids_four_thousand)
dict_size_after = len(dictionary2)
corpus2 = [dictionary2.doc2bow(text) for text in texts]
corpus_size_after = count_size_corpus_ingredients(corpus2)
print(dict_size_after)
print(corpus_size_after)
def save_answers2(dict_size_before, dict_size_after, corpus_size_before, corpus_size_after):
with open("cooking_LDA_pa_task2.txt", "w") as fout:
fout.write(" ".join([str(el) for el in [dict_size_before, dict_size_after, corpus_size_before, corpus_size_after]]))
save_answers2(dict_size_before, dict_size_after, corpus_size_before, corpus_size_after)
np.random.seed(76543)
# здесь код для построения модели:
%time ldamodel2 = models.ldamodel.LdaModel(corpus2, id2word=dictionary2, num_topics=40, passes=5)
cog2 = ldamodel2.top_topics(corpus2)
cog = ldamodel.top_topics(corpus)
def mean(coher):
_sum = 0
for c in coher:
_sum += c[1]
return _sum / len(coher)
coherence2 = mean(cog2)
coherence = mean(cog)
def save_answers3(coherence, coherence2):
with open("cooking_LDA_pa_task3.txt", "w") as fout:
fout.write(" ".join(["%3f"%el for el in [coherence, coherence2]]))
save_answers3(coherence, coherence2)
ldamodel2.get_document_topics(corpus[0])
np.random.seed(76543)
%time ldamodel3 = models.ldamodel.LdaModel(corpus2, id2word=dictionary2, passes=5, alpha=1, num_topics=40)
count_model2 = []
for j in range(0,39774):
TemaDocument2 = ldamodel2.get_document_topics(corpus2[j], minimum_probability=0.01)
count_model2.append(len(TemaDocument2))
sum(count_model2)
np.random.seed(76543)
count_model3 = []
for j in range(0,39774):
TemaDocument3 = ldamodel3.get_document_topics(corpus2[j], minimum_probability=0.01)
count_model3.append(len(TemaDocument3))
sum(count_model3)
def save_answers4(count_model2, count_model3):
with open("cooking_LDA_pa_task4.txt", "w") as fout:
fout.write(" ".join([str(el) for el in [count_model2, count_model3]]))
save_answers4(sum(count_model2), sum(count_model3))
from sklearn.ensemble import RandomForestClassifier
from sklearn.cross_validation import cross_val_score
def save_answers5(accuracy):
with open("cooking_LDA_pa_task5.txt", "w") as fout:
fout.write(str(accuracy))
def generate_recipe(model, num_ingredients):
theta = np.random.dirichlet(model.alpha)
for i in range(num_ingredients):
t = np.random.choice(np.arange(model.num_topics), p=theta)
topic = model.show_topic(t, topn=model.num_terms)
topic_distr = [x[1] for x in topic]
terms = [x[0] for x in topic]
w = np.random.choice(terms, p=topic_distr)
print w
import pandas
import seaborn
from matplotlib import pyplot as plt
%matplotlib inline
def compute_topic_cuisine_matrix(model, corpus, recipes):
# составляем вектор целевых признаков
targets = list(set([recipe["cuisine"] for recipe in recipes]))
# составляем матрицу
tc_matrix = pandas.DataFrame(data=np.zeros((model.num_topics, len(targets))), columns=targets)
for recipe, bow in zip(recipes, corpus):
recipe_topic = model.get_document_topics(bow)
for t, prob in recipe_topic:
tc_matrix[recipe["cuisine"]][t] += prob
# нормируем матрицу
target_sums = pandas.DataFrame(data=np.zeros((1, len(targets))), columns=targets)
for recipe in recipes:
target_sums[recipe["cuisine"]] += 1
return pandas.DataFrame(tc_matrix.values/target_sums.values, columns=tc_matrix.columns)
def plot_matrix(tc_matrix):
plt.figure(figsize=(10, 10))
seaborn.heatmap(tc_matrix, square=True)
# Визуализируйте матрицу
| 0.23546 | 0.983311 |
```
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import os
import sys
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
from nets import *
from cfgs import *
from data import *
from clip_ops.clip_ops import *
from trainer import *
%matplotlib inline
save_plot = True
plt.rcParams.update({'font.size': 10, 'axes.labelsize': 'x-large'})
D = 201
x = np.linspace(0, 1.0, D)
X_tst = np.stack([v.flatten() for v in np.meshgrid(x,x)], axis = -1)
X_tst = np.expand_dims(X_tst, 1)
X_tst = X_tst + 2.0
print(X_tst.shape)
cfg = unit_1x2_uniform_23_config.cfg
cfg.test.num_misreports = 1
cfg.test.gd_iter = 0
cfg.test.batch_size = D
cfg.test.num_batches = int(X_tst.shape[0]/cfg.test.batch_size)
cfg.test.save_output = True
Net = unit_net.Net
Generator = uniform_23_generator.Generator
clip_op_lambda = (lambda x: clip_op_23(x))
Trainer = trainer.Trainer
net = Net(cfg)
generator = Generator(cfg, 'test', X_tst)
clip_op_lambda = (lambda x: tf.assign(x, tf.clip_by_value(x, 0.0, 1.0)))
m = Trainer(cfg, "test", net, clip_op_lambda)
m.test(generator)
alloc = np.load(cfg.dir_name + "/alloc_tst_400000.npy").reshape(D,D,2)
pay = np.load(cfg.dir_name + "/pay_tst_400000.npy").reshape(D,D,1)
x1 = 4.0/3.0 + np.sqrt(4.0 + 3.0/2.0)/3.0
points1 = [(3.0 - 1.0/3.0, 3.0), (2.0, 2.0 + 1.0/3.0)]
points2 = [(2.0, 2 * x1 - 2.0), (2 * x1 - 2.0, 2.0)]
points3 = [(2.0 + 1.0/3.0, 2.0), (3.0, 3.0 - 1.0/3.0)]
x_1 = list(map(lambda x: x[0], points1))
y_1 = list(map(lambda x: x[1], points1))
x_2 = list(map(lambda x: x[0], points2))
y_2 = list(map(lambda x: x[1], points2))
x_3 = list(map(lambda x: x[0], points3))
y_3 = list(map(lambda x: x[1], points3))
plt.rcParams.update({'font.size': 10, 'axes.labelsize': 'x-large'})
fig, ax = plt.subplots(ncols = 1, nrows = 1, figsize = (8,6))
ax.plot(x_1, y_1, linewidth = 2, linestyle = '--', color='black')
ax.plot(x_2, y_2, linewidth = 2, linestyle = '--', color='black')
ax.plot(x_3, y_3, linewidth = 2, linestyle = '--', color='black')
img = ax.imshow(alloc[::-1, :, 0], extent=[2,3,2,3], vmin = 0.0, vmax=1.0, cmap = 'YlOrRd')
plt.text(2.2, 2.8, s='0', color='black', fontsize='10', fontweight='bold')
plt.text(2.05, 2.05, s='0', color='black', fontsize='10', fontweight='bold')
plt.text(2.5, 2.5, s='0.5', color='black', fontsize='10', fontweight='bold')
plt.text(2.8, 2.2, s='1', color='black', fontsize='10', fontweight='bold')
ax.set_xlabel('$v_1$')
ax.set_ylabel('$v_2$')
plt.title('Prob. of allocating item 1')
_ = plt.colorbar(img, fraction=0.046, pad=0.04)
if save_plot:
fig.set_size_inches(4, 3)
plt.savefig(os.path.join(cfg.dir_name, 'alloc1.pdf'), bbox_inches = 'tight', pad_inches = 0.05)
x1 = 4.0/3.0 + np.sqrt(4.0 + 3.0/2.0)/3.0
points1 = [(3.0 - 1.0/3.0, 3.0), (2.0, 2.0 + 1.0/3.0)]
points2 = [(2.0, 2 * x1 - 2.0), (2 * x1 - 2.0, 2.0)]
points3 = [(2.0 + 1.0/3.0, 2.0), (3.0, 3.0 - 1.0/3.0)]
x_1 = list(map(lambda x: x[0], points1))
y_1 = list(map(lambda x: x[1], points1))
x_2 = list(map(lambda x: x[0], points2))
y_2 = list(map(lambda x: x[1], points2))
x_3 = list(map(lambda x: x[0], points3))
y_3 = list(map(lambda x: x[1], points3))
plt.rcParams.update({'font.size': 10, 'axes.labelsize': 'x-large'})
fig, ax = plt.subplots(ncols = 1, nrows = 1, figsize = (8,6))
ax.plot(x_1, y_1, linewidth = 2, linestyle = '--', color='black')
ax.plot(x_2, y_2, linewidth = 2, linestyle = '--', color='black')
ax.plot(x_3, y_3, linewidth = 2, linestyle = '--', color='black')
img = ax.imshow(alloc[::-1, :, 1], extent=[2,3,2,3], vmin = 0.0, vmax=1.0, cmap = 'YlOrRd')
plt.text(2.2, 2.8, s='1', color='black', fontsize='10', fontweight='bold')
plt.text(2.05, 2.05, s='0', color='black', fontsize='10', fontweight='bold')
plt.text(2.5, 2.5, s='0.5', color='black', fontsize='10', fontweight='bold')
plt.text(2.8, 2.2, s='0', color='black', fontsize='10', fontweight='bold')
ax.set_xlabel('$v_1$')
ax.set_ylabel('$v_2$')
plt.title('Prob. of allocating item 2')
_ = plt.colorbar(img, fraction=0.046, pad=0.04)
if save_plot:
fig.set_size_inches(4, 3)
plt.savefig(os.path.join(cfg.dir_name, 'alloc2.pdf'), bbox_inches = 'tight', pad_inches = 0.05)
```
|
github_jupyter
|
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import os
import sys
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
from nets import *
from cfgs import *
from data import *
from clip_ops.clip_ops import *
from trainer import *
%matplotlib inline
save_plot = True
plt.rcParams.update({'font.size': 10, 'axes.labelsize': 'x-large'})
D = 201
x = np.linspace(0, 1.0, D)
X_tst = np.stack([v.flatten() for v in np.meshgrid(x,x)], axis = -1)
X_tst = np.expand_dims(X_tst, 1)
X_tst = X_tst + 2.0
print(X_tst.shape)
cfg = unit_1x2_uniform_23_config.cfg
cfg.test.num_misreports = 1
cfg.test.gd_iter = 0
cfg.test.batch_size = D
cfg.test.num_batches = int(X_tst.shape[0]/cfg.test.batch_size)
cfg.test.save_output = True
Net = unit_net.Net
Generator = uniform_23_generator.Generator
clip_op_lambda = (lambda x: clip_op_23(x))
Trainer = trainer.Trainer
net = Net(cfg)
generator = Generator(cfg, 'test', X_tst)
clip_op_lambda = (lambda x: tf.assign(x, tf.clip_by_value(x, 0.0, 1.0)))
m = Trainer(cfg, "test", net, clip_op_lambda)
m.test(generator)
alloc = np.load(cfg.dir_name + "/alloc_tst_400000.npy").reshape(D,D,2)
pay = np.load(cfg.dir_name + "/pay_tst_400000.npy").reshape(D,D,1)
x1 = 4.0/3.0 + np.sqrt(4.0 + 3.0/2.0)/3.0
points1 = [(3.0 - 1.0/3.0, 3.0), (2.0, 2.0 + 1.0/3.0)]
points2 = [(2.0, 2 * x1 - 2.0), (2 * x1 - 2.0, 2.0)]
points3 = [(2.0 + 1.0/3.0, 2.0), (3.0, 3.0 - 1.0/3.0)]
x_1 = list(map(lambda x: x[0], points1))
y_1 = list(map(lambda x: x[1], points1))
x_2 = list(map(lambda x: x[0], points2))
y_2 = list(map(lambda x: x[1], points2))
x_3 = list(map(lambda x: x[0], points3))
y_3 = list(map(lambda x: x[1], points3))
plt.rcParams.update({'font.size': 10, 'axes.labelsize': 'x-large'})
fig, ax = plt.subplots(ncols = 1, nrows = 1, figsize = (8,6))
ax.plot(x_1, y_1, linewidth = 2, linestyle = '--', color='black')
ax.plot(x_2, y_2, linewidth = 2, linestyle = '--', color='black')
ax.plot(x_3, y_3, linewidth = 2, linestyle = '--', color='black')
img = ax.imshow(alloc[::-1, :, 0], extent=[2,3,2,3], vmin = 0.0, vmax=1.0, cmap = 'YlOrRd')
plt.text(2.2, 2.8, s='0', color='black', fontsize='10', fontweight='bold')
plt.text(2.05, 2.05, s='0', color='black', fontsize='10', fontweight='bold')
plt.text(2.5, 2.5, s='0.5', color='black', fontsize='10', fontweight='bold')
plt.text(2.8, 2.2, s='1', color='black', fontsize='10', fontweight='bold')
ax.set_xlabel('$v_1$')
ax.set_ylabel('$v_2$')
plt.title('Prob. of allocating item 1')
_ = plt.colorbar(img, fraction=0.046, pad=0.04)
if save_plot:
fig.set_size_inches(4, 3)
plt.savefig(os.path.join(cfg.dir_name, 'alloc1.pdf'), bbox_inches = 'tight', pad_inches = 0.05)
x1 = 4.0/3.0 + np.sqrt(4.0 + 3.0/2.0)/3.0
points1 = [(3.0 - 1.0/3.0, 3.0), (2.0, 2.0 + 1.0/3.0)]
points2 = [(2.0, 2 * x1 - 2.0), (2 * x1 - 2.0, 2.0)]
points3 = [(2.0 + 1.0/3.0, 2.0), (3.0, 3.0 - 1.0/3.0)]
x_1 = list(map(lambda x: x[0], points1))
y_1 = list(map(lambda x: x[1], points1))
x_2 = list(map(lambda x: x[0], points2))
y_2 = list(map(lambda x: x[1], points2))
x_3 = list(map(lambda x: x[0], points3))
y_3 = list(map(lambda x: x[1], points3))
plt.rcParams.update({'font.size': 10, 'axes.labelsize': 'x-large'})
fig, ax = plt.subplots(ncols = 1, nrows = 1, figsize = (8,6))
ax.plot(x_1, y_1, linewidth = 2, linestyle = '--', color='black')
ax.plot(x_2, y_2, linewidth = 2, linestyle = '--', color='black')
ax.plot(x_3, y_3, linewidth = 2, linestyle = '--', color='black')
img = ax.imshow(alloc[::-1, :, 1], extent=[2,3,2,3], vmin = 0.0, vmax=1.0, cmap = 'YlOrRd')
plt.text(2.2, 2.8, s='1', color='black', fontsize='10', fontweight='bold')
plt.text(2.05, 2.05, s='0', color='black', fontsize='10', fontweight='bold')
plt.text(2.5, 2.5, s='0.5', color='black', fontsize='10', fontweight='bold')
plt.text(2.8, 2.2, s='0', color='black', fontsize='10', fontweight='bold')
ax.set_xlabel('$v_1$')
ax.set_ylabel('$v_2$')
plt.title('Prob. of allocating item 2')
_ = plt.colorbar(img, fraction=0.046, pad=0.04)
if save_plot:
fig.set_size_inches(4, 3)
plt.savefig(os.path.join(cfg.dir_name, 'alloc2.pdf'), bbox_inches = 'tight', pad_inches = 0.05)
| 0.47098 | 0.399402 |
# <font color ='red'> Tarea
Generar valores aleatorios para la siguiente distribución de probabilidad
$$f(x)=\begin{cases}\frac{2}{(c-a)(b-a)}(x-a), & a\leq x \leq b\\ \frac{-2}{(c-a)(c-b)}(x-c),& b\leq x \leq c \end{cases}$$ con a=1; b=2; c=5
1. Usando el método de la transformada inversa.
2. Usando el método de aceptación y rechazo.
3. En la librería `import scipy.stats as st` hay una función que genera variables aleatorias triangulares `st.triang.pdf(x, c, loc, scale)` donde "c,loc,scale" son los parámetros de esta distribución (similares a los que nuestra función se llaman a,b,c, PERO NO IGUALES). Explorar el help de python para encontrar la equivalencia entre los parámetros "c,loc,scale" y los parámetros de nuestra función con parámetros "a,b,c". La solución esperada es como se muestra a continuación:

4. Generar 1000 variables aleatorias usando la función creada en el punto 2 y usando la función `st.triang.rvs` y graficar el histograma en dos gráficas diferentes de cada uno de los conjuntos de variables aleatorios creado. Se espera algo como esto:
```
import matplotlib.pyplot as plt
import numpy as np
```
1. Usando el método de la transformada inversa.
Defino mi $f(x)$ y grafico para comprobar
```
def f(x):
a=1; b=2; c=5;
if x < a:
return 0
elif a <= x and x <= b:
return 2*(x-a)/(c-a)/(b-a)
elif b <= x and x <= c:
return -2*(x-c)/(c-a)/(c-b)
else:
return 0
x = np.arange(.5,5.5,.1)
y = [f(i)for i in x]
plt.plot(x,y,label="$f(x)$")
plt.legend()
plt.show()
```
Calculo la CDF y grafico
$$F(x)=\begin{cases}\frac{(x-a)^2}{(c-a)(b-a)}, & a\leq x \leq b\\ \frac{(b-c)^2-(x-c)^2}{(c-a)(c-b)} + F(b),& b< x \leq c \end{cases}$$
```
def F(x):
a=1; b=2; c=5;
if a <= x and x <= b:
return (x-a)**2/(c-a)/(b-a)
elif b < x and x <= c:
return ((b-c)**2-(x-c)**2)/(c-a)/(c-b) + F(b)
x1 = np.arange(1,5,.01)
y1 = [F(i)for i in x1]
plt.plot(x1,y1,label="$F(x)$")
plt.legend()
plt.show()
```
Saco la inversa de $F(x)$ (**Nota** nos interesa la raíz negativa)
$$F^{-1}(x)=\begin{cases}\sqrt{U(c-a)(b-a)}+a, & F(a)\leq U \leq F(b)\\ -\sqrt{(b-c)^2-(c-a)(c-b)(U-F(b))}+c,& F(b)< U \leq F(c) \end{cases}$$
```
def F_1(U):
a=1; b=2; c=5;
if F(a) <= U and U <= F(b):
return np.sqrt(U*(c-a)*(b-a))+a
# return c
elif F(b) < U and U <= F(c):
return -np.sqrt((b-c)**2 -(c-a)*(c-b)*(U-F(b))) +c
samples = 10000
X = list(map(lambda u:F_1(u),np.random.rand(samples)))
plt.hist(X,bins=30,density=True)
plt.plot(x,[f(i)for i in x],label="$f(x)$")
plt.show()
```
2. Usando el método de aceptación y rechazo.
Función triangular alcanza máximo en $f(2)$. Por lo tanto proponemos $\phi = Ch\,(x) = f(2) = Max(f) = .5$
$\int_a^b\phi= C = .5(b-a) \rightarrow h(x) = \frac\phi C = \frac 1{(b-a)}$
$H(x) = \frac{x-a}{b-a} \rightarrow H^{-1}(x) = x(b-a)+a$
Si $U_2<g(H^{-1}(u_1))$ entonces $Y = H^{-1}(u_1),g(x)=\frac{f(x)}{\phi}$
```
phi = lambda :.5
def H_1(u):
a=1; b=5;
return u*(b-a)+a
samples = 10000
```
3 y 4. Generar Aleatorios propios, comparar nuestra distribución contra Scipy.stats.triangular y crear aleatorios con Scipy.
```
c = .5*(5-1)
Adjsamples =int(samples*c)
Y=list(filter(lambda y: np.random.rand() < f(y)/phi(),map(lambda u1:H_1(u1),np.random.rand(Adjsamples))))
plt.figure(1,figsize=(14,5))
plt.subplot(121)
plt.title("Aceptación Rechazo")
plt.hist(Y,bins=30,density=True)
plt.plot(x,[f(i)for i in x],label="$f(x)$")
plt.subplot(122)
# plt.show()
import scipy.stats as st
# x = np.arange(0,6,.1)
plt.plot(x,st.triang.pdf(x,loc=1,c=.25,scale=4))
plt.title('Scipy')
plt.hist(st.triang.rvs(loc=1,c=.25,scale=4,size=10000),bins=30,density=True)
plt.show()
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
def f(x):
a=1; b=2; c=5;
if x < a:
return 0
elif a <= x and x <= b:
return 2*(x-a)/(c-a)/(b-a)
elif b <= x and x <= c:
return -2*(x-c)/(c-a)/(c-b)
else:
return 0
x = np.arange(.5,5.5,.1)
y = [f(i)for i in x]
plt.plot(x,y,label="$f(x)$")
plt.legend()
plt.show()
def F(x):
a=1; b=2; c=5;
if a <= x and x <= b:
return (x-a)**2/(c-a)/(b-a)
elif b < x and x <= c:
return ((b-c)**2-(x-c)**2)/(c-a)/(c-b) + F(b)
x1 = np.arange(1,5,.01)
y1 = [F(i)for i in x1]
plt.plot(x1,y1,label="$F(x)$")
plt.legend()
plt.show()
def F_1(U):
a=1; b=2; c=5;
if F(a) <= U and U <= F(b):
return np.sqrt(U*(c-a)*(b-a))+a
# return c
elif F(b) < U and U <= F(c):
return -np.sqrt((b-c)**2 -(c-a)*(c-b)*(U-F(b))) +c
samples = 10000
X = list(map(lambda u:F_1(u),np.random.rand(samples)))
plt.hist(X,bins=30,density=True)
plt.plot(x,[f(i)for i in x],label="$f(x)$")
plt.show()
phi = lambda :.5
def H_1(u):
a=1; b=5;
return u*(b-a)+a
samples = 10000
c = .5*(5-1)
Adjsamples =int(samples*c)
Y=list(filter(lambda y: np.random.rand() < f(y)/phi(),map(lambda u1:H_1(u1),np.random.rand(Adjsamples))))
plt.figure(1,figsize=(14,5))
plt.subplot(121)
plt.title("Aceptación Rechazo")
plt.hist(Y,bins=30,density=True)
plt.plot(x,[f(i)for i in x],label="$f(x)$")
plt.subplot(122)
# plt.show()
import scipy.stats as st
# x = np.arange(0,6,.1)
plt.plot(x,st.triang.pdf(x,loc=1,c=.25,scale=4))
plt.title('Scipy')
plt.hist(st.triang.rvs(loc=1,c=.25,scale=4,size=10000),bins=30,density=True)
plt.show()
| 0.360602 | 0.946151 |
<img src="https://www.python.org/static/img/python-logo.png">
# Welcome to my lessons
---
**Bo Zhang** (NAOC, <mailto:bozhang@nao.cas.cn>) will have a few lessons on python.
- These are **very useful knowledge, skills and code styles** when you use `python` to process astronomical data.
- All materials can be found on [**my github page**](https://github.com/hypergravity/cham_teaches_python).
- **jupyter notebook** (formerly named **ipython notebook**) is recommeded to use
---
These lectures are organized as below:
1. install python
2. basic syntax
3. numerical computing
4. scientific computing
5. plotting
6. astronomical data processing
7. high performance computing
8. version control
# object
```
object
dir()
In
a = 1.5
type(a)
print isinstance(a, float)
print isinstance(a, object)
```
# Lists and Tuples
```
b = [1, 2, 3, 'abc']
type(b)
b.append(4)
b
c = (1, 2, 3, 'abc', 3)
type(c)
c.count(3)
c
```
# Dictionaries and Sets
```
d = {'a': 123, 'b': 456}
d
print d['a']
print d['b']
e = {1, 2}
print type(e)
e
```
# Iterators and Generators
```
%%time
for i in range(10):
print i
%%time
for i in xrange(10):
print i
```
- mutable immutable
- iterable
```
def f1yield(n):
n0 = 0
while n0 < n:
yield n-n0
n0 += 1
def f1print(n):
n0 = 0
while n0 < n:
print n-n0
n0 += 1
f = f1yield(5)
print type(f)
print f.next()
print f.next()
print f.next()
print f.next()
f = f1print(5)
print type(f)
```
# Decorators
```
def salesgirl(method):
def serve(*args):
print "Salesgirl:Hello, what do you want?", method.__name__
method(*args)
return serve
@salesgirl
def try_this_shirt(size):
if size < 35:
print "I: %d inches is to small to me" %(size)
else:
print "I:%d inches is just enough" %(size)
try_this_shirt(38)
print type(salesgirl)
print type(try_this_shirt)
```
# lambda
```
def f2(x):
return 2*x
l2 = lambda x: 2*x
print f2(10)
print l2(10)
print type(f2)
print type(l2)
print f2
print l2
```
# class
you should understand the difference between "**class**" and "**instance**"
```
class People():
"""here we define the People class
"""
name = ''
height = 180. # cm
weight = 140. # pound
energy = 100. # percent
energy_cost_per_work_hour = 10. # percent energy per work hour
energy_per_meal = 90. # percent energy per meal
def __init__(self, name='', height=180., weight=140., energy=100.):
self.name = name
self.height = height
self.weight = weight
self.energy = energy
def work(self, hours=1.):
if hours > 0. and hours < 10. and hours < self.energy/self.energy_cost_per_work_hour:
self.energy -= hours*self.energy_cost_per_work_hour
else:
if hours <= 0.:
raise ValueError('@Cham: hours must be positive!')
else:
raise ValueError('@Cham: energy ran out!')
def eat(self, num_meal=1.):
if num_meal > 0. and num_meal <= 5.:
self.energy += num_meal*self.energy_per_meal
if self.energy > 100.:
self.energy = 100.
else:
if num_meal <= 0.:
raise ValueError('@Cham: number of meals must be positive!')
else:
raise ValueError('@Cham: too many meals!')
def print_status(self):
print ''
print 'name: %s' % self.name
print 'height: %s' % self.height
print 'weight: %s' % self.weight
print 'energy: %s' % self.energy
print 'energy_cost_per_work_hour: %s' % self.energy_cost_per_work_hour
print 'energy_per_meal: %s' % self.energy_per_meal
jim = People('Jim')
jim.print_status()
jim.work(5)
jim.print_status()
jim.eat(2)
jim.print_status()
```
# HOMEWORK
1. use generator to generate the Fibonacci sequence
2. try to define a simple class
|
github_jupyter
|
object
dir()
In
a = 1.5
type(a)
print isinstance(a, float)
print isinstance(a, object)
b = [1, 2, 3, 'abc']
type(b)
b.append(4)
b
c = (1, 2, 3, 'abc', 3)
type(c)
c.count(3)
c
d = {'a': 123, 'b': 456}
d
print d['a']
print d['b']
e = {1, 2}
print type(e)
e
%%time
for i in range(10):
print i
%%time
for i in xrange(10):
print i
def f1yield(n):
n0 = 0
while n0 < n:
yield n-n0
n0 += 1
def f1print(n):
n0 = 0
while n0 < n:
print n-n0
n0 += 1
f = f1yield(5)
print type(f)
print f.next()
print f.next()
print f.next()
print f.next()
f = f1print(5)
print type(f)
def salesgirl(method):
def serve(*args):
print "Salesgirl:Hello, what do you want?", method.__name__
method(*args)
return serve
@salesgirl
def try_this_shirt(size):
if size < 35:
print "I: %d inches is to small to me" %(size)
else:
print "I:%d inches is just enough" %(size)
try_this_shirt(38)
print type(salesgirl)
print type(try_this_shirt)
def f2(x):
return 2*x
l2 = lambda x: 2*x
print f2(10)
print l2(10)
print type(f2)
print type(l2)
print f2
print l2
class People():
"""here we define the People class
"""
name = ''
height = 180. # cm
weight = 140. # pound
energy = 100. # percent
energy_cost_per_work_hour = 10. # percent energy per work hour
energy_per_meal = 90. # percent energy per meal
def __init__(self, name='', height=180., weight=140., energy=100.):
self.name = name
self.height = height
self.weight = weight
self.energy = energy
def work(self, hours=1.):
if hours > 0. and hours < 10. and hours < self.energy/self.energy_cost_per_work_hour:
self.energy -= hours*self.energy_cost_per_work_hour
else:
if hours <= 0.:
raise ValueError('@Cham: hours must be positive!')
else:
raise ValueError('@Cham: energy ran out!')
def eat(self, num_meal=1.):
if num_meal > 0. and num_meal <= 5.:
self.energy += num_meal*self.energy_per_meal
if self.energy > 100.:
self.energy = 100.
else:
if num_meal <= 0.:
raise ValueError('@Cham: number of meals must be positive!')
else:
raise ValueError('@Cham: too many meals!')
def print_status(self):
print ''
print 'name: %s' % self.name
print 'height: %s' % self.height
print 'weight: %s' % self.weight
print 'energy: %s' % self.energy
print 'energy_cost_per_work_hour: %s' % self.energy_cost_per_work_hour
print 'energy_per_meal: %s' % self.energy_per_meal
jim = People('Jim')
jim.print_status()
jim.work(5)
jim.print_status()
jim.eat(2)
jim.print_status()
| 0.317744 | 0.931774 |
If you're opening this Notebook on colab, you will probably need to install 🤗 Transformers and 🤗 Datasets as well as other dependencies. Uncomment the following cell and run it.
```
#! pip install datasets transformers sacrebleu
```
If you're opening this notebook locally, make sure your environment has the last version of those libraries installed.
You can find a script version of this notebook to fine-tune your model in a distributed fashion using multiple GPUs or TPUs [here](https://github.com/huggingface/transformers/tree/master/examples/seq2seq).
# Fine-tuning a model on a translation task
In this notebook, we will see how to fine-tune one of the [🤗 Transformers](https://github.com/huggingface/transformers) model for a translation task. We will use the [WMT dataset](http://www.statmt.org/wmt16/), a machine translation dataset composed from a collection of various sources, including news commentaries and parliament proceedings.

We will see how to easily load the dataset for this task using 🤗 Datasets and how to fine-tune a model on it using the `Trainer` API.
```
model_checkpoint = "Helsinki-NLP/opus-mt-en-ro"
```
This notebook is built to run with any model checkpoint from the [Model Hub](https://huggingface.co/models) as long as that model has a sequence-to-sequence version in the Transformers library. Here we picked the [`Helsinki-NLP/opus-mt-en-ro`](https://huggingface.co/Helsinki-NLP/opus-mt-en-ro) checkpoint.
## Loading the dataset
We will use the [🤗 Datasets](https://github.com/huggingface/datasets) library to download the data and get the metric we need to use for evaluation (to compare our model to the benchmark). This can be easily done with the functions `load_dataset` and `load_metric`. We use the English/Romanian part of the WMT dataset here.
```
from datasets import load_dataset, load_metric
raw_datasets = load_dataset("wmt16", "ro-en")
metric = load_metric("sacrebleu")
```
The `dataset` object itself is [`DatasetDict`](https://huggingface.co/docs/datasets/package_reference/main_classes.html#datasetdict), which contains one key for the training, validation and test set:
```
raw_datasets
```
To access an actual element, you need to select a split first, then give an index:
```
raw_datasets["train"][0]
```
To get a sense of what the data looks like, the following function will show some examples picked randomly in the dataset.
```
import datasets
import random
import pandas as pd
from IPython.display import display, HTML
def show_random_elements(dataset, num_examples=5):
assert num_examples <= len(dataset), "Can't pick more elements than there are in the dataset."
picks = []
for _ in range(num_examples):
pick = random.randint(0, len(dataset)-1)
while pick in picks:
pick = random.randint(0, len(dataset)-1)
picks.append(pick)
df = pd.DataFrame(dataset[picks])
for column, typ in dataset.features.items():
if isinstance(typ, datasets.ClassLabel):
df[column] = df[column].transform(lambda i: typ.names[i])
display(HTML(df.to_html()))
show_random_elements(raw_datasets["train"])
```
The metric is an instance of [`datasets.Metric`](https://huggingface.co/docs/datasets/package_reference/main_classes.html#datasets.Metric):
```
metric
```
You can call its `compute` method with your predictions and labels, which need to be list of decoded strings (list of list for the labels):
```
fake_preds = ["hello there", "general kenobi"]
fake_labels = [["hello there"], ["general kenobi"]]
metric.compute(predictions=fake_preds, references=fake_labels)
```
## Preprocessing the data
Before we can feed those texts to our model, we need to preprocess them. This is done by a 🤗 Transformers `Tokenizer` which will (as the name indicates) tokenize the inputs (including converting the tokens to their corresponding IDs in the pretrained vocabulary) and put it in a format the model expects, as well as generate the other inputs that model requires.
To do all of this, we instantiate our tokenizer with the `AutoTokenizer.from_pretrained` method, which will ensure:
- we get a tokenizer that corresponds to the model architecture we want to use,
- we download the vocabulary used when pretraining this specific checkpoint.
That vocabulary will be cached, so it's not downloaded again the next time we run the cell.
```
from transformers import AutoTokenizer
tokenizer = AutoTokenizer.from_pretrained(model_checkpoint)
```
For the mBART tokenizer (like we have here), we need to set the source and target languages (so the texts are preprocessed properly). You can check the language codes [here](https://huggingface.co/facebook/mbart-large-cc25) if you are using this notebook on a different pairs of languages.
```
if "mbart" in model_checkpoint:
tokenizer.src_lang = "en-XX"
tokenizer.tgt_lang = "ro-RO"
```
By default, the call above will use one of the fast tokenizers (backed by Rust) from the 🤗 Tokenizers library.
You can directly call this tokenizer on one sentence or a pair of sentences:
```
tokenizer("Hello, this one sentence!")
```
Depending on the model you selected, you will see different keys in the dictionary returned by the cell above. They don't matter much for what we're doing here (just know they are required by the model we will instantiate later), you can learn more about them in [this tutorial](https://huggingface.co/transformers/preprocessing.html) if you're interested.
Instead of one sentence, we can pass along a list of sentences:
```
tokenizer(["Hello, this one sentence!", "This is another sentence."])
```
To prepare the targets for our model, we need to tokenize them inside the `as_target_tokenizer` context manager. This will make sure the tokenizer uses the special tokens corresponding to the targets:
```
with tokenizer.as_target_tokenizer():
print(tokenizer(["Hello, this one sentence!", "This is another sentence."]))
```
If you are using one of the five T5 checkpoints that require a special prefix to put before the inputs, you should adapt the following cell.
```
if model_checkpoint in ["t5-small", "t5-base", "t5-larg", "t5-3b", "t5-11b"]:
prefix = "translate English to Romanian: "
else:
prefix = ""
```
We can then write the function that will preprocess our samples. We just feed them to the `tokenizer` with the argument `truncation=True`. This will ensure that an input longer that what the model selected can handle will be truncated to the maximum length accepted by the model. The padding will be dealt with later on (in a data collator) so we pad examples to the longest length in the batch and not the whole dataset.
```
max_input_length = 128
max_target_length = 128
source_lang = "en"
target_lang = "ro"
def preprocess_function(examples):
inputs = [prefix + ex[source_lang] for ex in examples["translation"]]
targets = [ex[target_lang] for ex in examples["translation"]]
model_inputs = tokenizer(inputs, max_length=max_input_length, truncation=True)
# Setup the tokenizer for targets
with tokenizer.as_target_tokenizer():
labels = tokenizer(targets, max_length=max_target_length, truncation=True)
model_inputs["labels"] = labels["input_ids"]
return model_inputs
```
This function works with one or several examples. In the case of several examples, the tokenizer will return a list of lists for each key:
```
preprocess_function(raw_datasets['train'][:2])
```
To apply this function on all the pairs of sentences in our dataset, we just use the `map` method of our `dataset` object we created earlier. This will apply the function on all the elements of all the splits in `dataset`, so our training, validation and testing data will be preprocessed in one single command.
```
tokenized_datasets = raw_datasets.map(preprocess_function, batched=True)
```
Even better, the results are automatically cached by the 🤗 Datasets library to avoid spending time on this step the next time you run your notebook. The 🤗 Datasets library is normally smart enough to detect when the function you pass to map has changed (and thus requires to not use the cache data). For instance, it will properly detect if you change the task in the first cell and rerun the notebook. 🤗 Datasets warns you when it uses cached files, you can pass `load_from_cache_file=False` in the call to `map` to not use the cached files and force the preprocessing to be applied again.
Note that we passed `batched=True` to encode the texts by batches together. This is to leverage the full benefit of the fast tokenizer we loaded earlier, which will use multi-threading to treat the texts in a batch concurrently.
## Fine-tuning the model
Now that our data is ready, we can download the pretrained model and fine-tune it. Since our task is of the sequence-to-sequence kind, we use the `AutoModelForSeq2SeqLM` class. Like with the tokenizer, the `from_pretrained` method will download and cache the model for us.
```
from transformers import AutoModelForSeq2SeqLM, DataCollatorForSeq2Seq, Seq2SeqTrainingArguments, Seq2SeqTrainer
model = AutoModelForSeq2SeqLM.from_pretrained(model_checkpoint)
```
Note that we don't get a warning like in our classification example. This means we used all the weights of the pretrained model and there is no randomly initialized head in this case.
To instantiate a `Seq2SeqTrainer`, we will need to define three more things. The most important is the [`Seq2SeqTrainingArguments`](https://huggingface.co/transformers/main_classes/trainer.html#transformers.Seq2SeqTrainingArguments), which is a class that contains all the attributes to customize the training. It requires one folder name, which will be used to save the checkpoints of the model, and all other arguments are optional:
```
batch_size = 16
args = Seq2SeqTrainingArguments(
"test-translation",
evaluation_strategy = "epoch",
learning_rate=2e-5,
per_device_train_batch_size=batch_size,
per_device_eval_batch_size=batch_size,
weight_decay=0.01,
save_total_limit=3,
num_train_epochs=1,
predict_with_generate=True,
fp16=True,
)
```
Here we set the evaluation to be done at the end of each epoch, tweak the learning rate, use the `batch_size` defined at the top of the cell and customize the weight decay. Since the `Seq2SeqTrainer` will save the model regularly and our dataset is quite large, we tell it to make three saves maximum. Lastly, we use the `predict_with_generate` option (to properly generate summaries) and activate mixed precision training (to go a bit faster).
Then, we need a special kind of data collator, which will not only pad the inputs to the maximum length in the batch, but also the labels:
```
data_collator = DataCollatorForSeq2Seq(tokenizer, model=model)
```
The last thing to define for our `Seq2SeqTrainer` is how to compute the metrics from the predictions. We need to define a function for this, which will just use the `metric` we loaded earlier, and we have to do a bit of pre-processing to decode the predictions into texts:
```
import numpy as np
def postprocess_text(preds, labels):
preds = [pred.strip() for pred in preds]
labels = [[label.strip()] for label in labels]
return preds, labels
def compute_metrics(eval_preds):
preds, labels = eval_preds
if isinstance(preds, tuple):
preds = preds[0]
decoded_preds = tokenizer.batch_decode(preds, skip_special_tokens=True)
# Replace -100 in the labels as we can't decode them.
labels = np.where(labels != -100, labels, tokenizer.pad_token_id)
decoded_labels = tokenizer.batch_decode(labels, skip_special_tokens=True)
# Some simple post-processing
decoded_preds, decoded_labels = postprocess_text(decoded_preds, decoded_labels)
result = metric.compute(predictions=decoded_preds, references=decoded_labels)
result = {"bleu": result["score"]}
prediction_lens = [np.count_nonzero(pred != tokenizer.pad_token_id) for pred in preds]
result["gen_len"] = np.mean(prediction_lens)
result = {k: round(v, 4) for k, v in result.items()}
return result
```
Then we just need to pass all of this along with our datasets to the `Seq2SeqTrainer`:
```
trainer = Seq2SeqTrainer(
model,
args,
train_dataset=tokenized_datasets["train"],
eval_dataset=tokenized_datasets["validation"],
data_collator=data_collator,
tokenizer=tokenizer,
compute_metrics=compute_metrics
)
```
We can now finetune our model by just calling the `train` method:
```
trainer.train()
```
Don't forget to [upload your model](https://huggingface.co/transformers/model_sharing.html) on the [🤗 Model Hub](https://huggingface.co/models). You can then use it only to generate results like the one shown in the first picture of this notebook!
|
github_jupyter
|
#! pip install datasets transformers sacrebleu
model_checkpoint = "Helsinki-NLP/opus-mt-en-ro"
from datasets import load_dataset, load_metric
raw_datasets = load_dataset("wmt16", "ro-en")
metric = load_metric("sacrebleu")
raw_datasets
raw_datasets["train"][0]
import datasets
import random
import pandas as pd
from IPython.display import display, HTML
def show_random_elements(dataset, num_examples=5):
assert num_examples <= len(dataset), "Can't pick more elements than there are in the dataset."
picks = []
for _ in range(num_examples):
pick = random.randint(0, len(dataset)-1)
while pick in picks:
pick = random.randint(0, len(dataset)-1)
picks.append(pick)
df = pd.DataFrame(dataset[picks])
for column, typ in dataset.features.items():
if isinstance(typ, datasets.ClassLabel):
df[column] = df[column].transform(lambda i: typ.names[i])
display(HTML(df.to_html()))
show_random_elements(raw_datasets["train"])
metric
fake_preds = ["hello there", "general kenobi"]
fake_labels = [["hello there"], ["general kenobi"]]
metric.compute(predictions=fake_preds, references=fake_labels)
from transformers import AutoTokenizer
tokenizer = AutoTokenizer.from_pretrained(model_checkpoint)
if "mbart" in model_checkpoint:
tokenizer.src_lang = "en-XX"
tokenizer.tgt_lang = "ro-RO"
tokenizer("Hello, this one sentence!")
tokenizer(["Hello, this one sentence!", "This is another sentence."])
with tokenizer.as_target_tokenizer():
print(tokenizer(["Hello, this one sentence!", "This is another sentence."]))
if model_checkpoint in ["t5-small", "t5-base", "t5-larg", "t5-3b", "t5-11b"]:
prefix = "translate English to Romanian: "
else:
prefix = ""
max_input_length = 128
max_target_length = 128
source_lang = "en"
target_lang = "ro"
def preprocess_function(examples):
inputs = [prefix + ex[source_lang] for ex in examples["translation"]]
targets = [ex[target_lang] for ex in examples["translation"]]
model_inputs = tokenizer(inputs, max_length=max_input_length, truncation=True)
# Setup the tokenizer for targets
with tokenizer.as_target_tokenizer():
labels = tokenizer(targets, max_length=max_target_length, truncation=True)
model_inputs["labels"] = labels["input_ids"]
return model_inputs
preprocess_function(raw_datasets['train'][:2])
tokenized_datasets = raw_datasets.map(preprocess_function, batched=True)
from transformers import AutoModelForSeq2SeqLM, DataCollatorForSeq2Seq, Seq2SeqTrainingArguments, Seq2SeqTrainer
model = AutoModelForSeq2SeqLM.from_pretrained(model_checkpoint)
batch_size = 16
args = Seq2SeqTrainingArguments(
"test-translation",
evaluation_strategy = "epoch",
learning_rate=2e-5,
per_device_train_batch_size=batch_size,
per_device_eval_batch_size=batch_size,
weight_decay=0.01,
save_total_limit=3,
num_train_epochs=1,
predict_with_generate=True,
fp16=True,
)
data_collator = DataCollatorForSeq2Seq(tokenizer, model=model)
import numpy as np
def postprocess_text(preds, labels):
preds = [pred.strip() for pred in preds]
labels = [[label.strip()] for label in labels]
return preds, labels
def compute_metrics(eval_preds):
preds, labels = eval_preds
if isinstance(preds, tuple):
preds = preds[0]
decoded_preds = tokenizer.batch_decode(preds, skip_special_tokens=True)
# Replace -100 in the labels as we can't decode them.
labels = np.where(labels != -100, labels, tokenizer.pad_token_id)
decoded_labels = tokenizer.batch_decode(labels, skip_special_tokens=True)
# Some simple post-processing
decoded_preds, decoded_labels = postprocess_text(decoded_preds, decoded_labels)
result = metric.compute(predictions=decoded_preds, references=decoded_labels)
result = {"bleu": result["score"]}
prediction_lens = [np.count_nonzero(pred != tokenizer.pad_token_id) for pred in preds]
result["gen_len"] = np.mean(prediction_lens)
result = {k: round(v, 4) for k, v in result.items()}
return result
trainer = Seq2SeqTrainer(
model,
args,
train_dataset=tokenized_datasets["train"],
eval_dataset=tokenized_datasets["validation"],
data_collator=data_collator,
tokenizer=tokenizer,
compute_metrics=compute_metrics
)
trainer.train()
| 0.68763 | 0.948585 |
```
# Instalando e atualizando as bibliotecas necessárias
!pip install pyDOE
!pip install -U seaborn
# Importando RASD Toolbox
from RASD_TOOLBOX import *
# Importando Numpy toolbox
import numpy as np
from VIGA_METALICA_VERIFICA import *
from FINITO_FEM_TOOLBOX import *
ELEMENTO_ANALISADO = [2,8]
#150 X 18,0
P1 = ['NORMAL', 200E9, 10E9] #E MPa
P2 = ['NORMAL', 250E6, 12.5E6] #Fy MPa
P3 = ['NORMAL', 10000, 1000] #Fy N
P4 = ['NORMAL', 1, 0.05] #Incerteza de Modelo 1 adimensional
P5 = ['NORMAL', 1, 0.05] #Incerteza de Modelo 2 adimensional
P6 = ['NORMAL', 139, 2.78] #H_W mm
P7 = ['NORMAL', 5.8, 0.116] #T_W mm
P8 = ['NORMAL', 102, 2.04] #B_F mm
P9 = ['NORMAL', 7.1, 0.142] #T_F mm
P10 = ['NORMAL', 139.4, 2.788] #Z cm3
P11 = ['NORMAL', 939, 18.78] #I cm4
P12 = ['NORMAL', 1000000, 100000] #Mola 1 N/m
P13 = ['NORMAL', 100000, 10000] #Mola 2 N/m
POP = [10000]
#POP = np.arange(10,1000000, 10000).tolist() # (x,y,Z); x=pop inicial; y=pop final; z=variacao pop
#carta_total = carga G + carga Q
SETUP = {'N_REP': len(POP),
'POP': POP,
'N_G': 3,
'D': 13, #variables
'VARS': [P1, P2, P3, P4, P5, P6, P7, P8, P9, P10, P11, P12, P13],
'MODEL': 'MCS'}
def OBJ(X):
E = X[0]
FY = X[1]
F1 = X[2]
M1 = X[3]
M2 = X[4]
H_W = X[5]
T_W = X[6]
W_C = X[7]
W_T = X[8]
Z = X[9]
INERCIA = X[10]
K1 = X[11]
K2 = X[12]
M_SD = []
V_SD = []
N_SD = []
D_SD = []
L_MAX = []
# auxiliares para tranformar em metroX
ALFA = 0.001 #mm para m
BETA = 0.000001 #cm3 para m3
GAMMA = 0.00000001 #cm4 para m4
VIGA = {'H_W': H_W * ALFA, #m
'T_W': T_W * ALFA, #m
'B_F': W_C * ALFA, #m
'T_F': W_T * ALFA, #m
'PARAMETRO_PERFIL': 'DUPLA SIMETRIA', #"DUPLA SIMETRIA, MONO SIMETRIA "
'TIPO_PERFIL': 'LAMINADO', #"SOLDADO, LAMINADO "
'GAMMA_A1': 1.1,
'Z': Z * BETA, #m3
'INERCIA':INERCIA * GAMMA, #m4
'E_S': E , #Pa
'F_Y': FY} #N
AREA = 2 * (VIGA['T_F'] * VIGA['B_F']) + (VIGA['T_W'] * VIGA['H_W']) #m2
X_GC = VIGA['B_F'] / 2 #m
Y_GC = (( VIGA['T_F'] * 2) + VIGA['H_W']) / 2 #m
FRAME_00 = {
"TYPE_ELEMENT": 0,
"TYPE_SOLUTION": 0,
"N_NODES": 26,
"N_MATERIALS": 1,
"N_SECTIONS": 1,
"N_ELEMENTS": 27,
"N_DOFPRESCRIPTIONS": 4,
"N_DOFLOADED": 24,
"N_DOFSPRINGS": 2,
"COORDINATES": #X,Y
np.array([
[0.0,0.0],
[5.25,0.0],
[0.0,3.0],
[0.75,3.0],
[1.5,3.0],
[2.25,3.0],
[3.0,3.0],
[3.75,3.0],
[4.5,3.0],
[5.25,3.0],
[0.0,6.0],
[0.75,6.0],
[1.5,6.0],
[2.25,6.0],
[3.0,6.0],
[3.75,6.0],
[4.5,6.0],
[5.25,6.0],
[0.0,9.0],
[0.75,9.0],
[1.5,9.0],
[2.25,9.0],
[3.0,9.0],
[3.75,9.0],
[4.5,9.0],
[5.25,9.0]]),
"ELEMENTS": #NODE 1,NODE 2,MATERIAL ID,SECTION ID,HINGE ID NODE 1,HINGE ID NODE 2
np.array([
[0,2,0,0,0,0],
[1,9,0,0,0,0],
[2,3,0,0,0,0],
[3,4,0,0,0,0],
[4,5,0,0,0,0],
[5,6,0,0,0,0],
[6,7,0,0,0,0],
[7,8,0,0,0,0],
[8,9,0,0,0,0],
[2,10,0,0,0,0],
[9,17,0,0,0,0],
[10,11,0,0,0,0],
[11,12,0,0,0,0],
[12,13,0,0,0,0],
[13,14,0,0,0,0],
[14,15,0,0,0,0],
[15,16,0,0,0,0],
[16,17,0,0,0,0],
[10,18,0,0,0,0],
[17,25,0,0,0,0],
[18,19,0,0,0,0],
[19,20,0,0,0,0],
[20,21,0,0,0,0],
[21,22,0,0,0,0],
[22,23,0,0,0,0],
[23,24,0,0,0,0],
[24,25,0,0,0,0]]),
"MATERIALS": #YOUNG, POISSON,DENSITY, THERMAL COEFFICIENT
np.array([[VIGA['E_S'], 1, 1, 1E-8]]),
#np.array([[200E9,1,1,1E-8]]),
"SECTIONS": #AREA, INERTIA 1, INERTIA 2, X GC,Y GC
np.array([[AREA , VIGA['INERCIA'], VIGA['INERCIA'], X_GC, X_GC]]),
#np.array([[0.0450,0.000377,0.000377,0.075,0.15]]),
"PRESCRIBED DISPLACEMENTS": #NODE,DIRECTION(X=0,Y=1,Z=2),VALUE
np.array([
[0,0,0],
#[0,1,0],
[0,2,0],
[1,0,0],
#[1,1,0],
[1,2,0]]),
"ELEMENT LOADS": None,
"NODAL LOADS": #NODE,DIRECTION(X=0,Y=1,Z=2),VALUE
np.array([
[2,1,-F1,0],
[3,1,-F1,0],
[4,1,-F1,0],
[5,1,-F1,0],
[6,1,-F1,0],
[7,1,-F1,0],
[8,1,-F1,0],
[9,1,-F1,0],
[10,1,-F1,0],
[11,1,-F1,0],
[12,1,-F1,0],
[13,1,-F1,0],
[14,1,-F1,0],
[15,1,-F1,0],
[16,1,-F1,0],
[17,1,-F1,0],
[18,1,-F1,0],
[19,1,-F1,0],
[20,1,-F1,0],
[21,1,-F1,0],
[22,1,-F1,0],
[23,1,-F1,0],
[24,1,-F1,0],
[25,1,-F1,0]]),
"SPRINGS": #NODE,DIRECTION(X=0,Y=1,Z=2),VALUE
np.array([
[0,1,K1,0],
[1,1,K2,0]])}
FRAME_00_RESULTS = MEF1D(DICTIONARY = FRAME_00)
ELEMENT = pd.DataFrame(FRAME_00_RESULTS[ELEMENTO_ANALISADO[0]:ELEMENTO_ANALISADO[1]])
for i in range(len(ELEMENT)):
M_SD.append(max(ELEMENT['M'][i].max(),abs(ELEMENT['M'][i].min())))
V_SD.append(max(ELEMENT['V'][i].max(),abs(ELEMENT['V'][i].min())))
N_SD.append(max(ELEMENT['N'][i].max(),abs(ELEMENT['N'][i].min())))
D_SD.append(min(ELEMENT['UX'][i].max(),abs(ELEMENT['UX'][i].min())))
#L_MAX.append(ELEMENT['X'][i].max())
M_SD = max(M_SD)
V_SD = max(V_SD)
N_SD = max(N_SD)
D_SD = min(D_SD)
L_MAX = FRAME_00['COORDINATES'][1][0]
ESFORCOS = {'M_SD': M_SD,
'V_SD': V_SD,
'D_SD': D_SD,
'L_MAX': L_MAX}
R_0, S_0 = VERIFICACAO_VIGA_METALICA_MOMENTO_FLETOR(VIGA, ESFORCOS)
G_0 = -M1 * R_0 + M2 * S_0
R_1, S_1 = VERIFICACAO_VIGA_METALICA_ESFORCO_CORTANTE(VIGA, ESFORCOS)
G_1 = -M1 * R_1 + M2 * S_1
R_2, S_2 = VERIFICACAO_VIGA_METALICA_DEFORMACAO(VIGA, ESFORCOS)
G_2 = -M1 * R_2 + M2 * S_2
R = [R_0, R_1, R_2]
S = [S_0, S_1, S_2]
G = [G_0, G_1, G_2]
return R, S, G
RESULTS_TEST = RASD_STOCHASTIC(SETUP, OBJ)
RESULTS_TEST[0]['TOTAL RESULTS']['R_0']
DADOS = RESULTS_TEST[0]['TOTAL RESULTS']
DADOS['I_0'].sum()
DADOS = RESULTS_TEST[len(POP)-1]['TOTAL RESULTS'] #USAR QUANDO HOUVEREM VARIAS SIMULACOES
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'WIDTH': 0.40,
'HEIGHT': 0.20,
'X AXIS SIZE': 20,
'Y AXIS SIZE': 20,
'AXISES COLOR': '#000000',
'X AXIS LABEL': '$x_0$ - $P_X (lb)$',
'LABELS SIZE': 16,
'LABELS COLOR': '#000000',
'CHART COLOR': '#FEB625',
'KDE': False,
'DPI': 600,
'BINS' : 20,
'EXTENSION': '.svg'}
# RESULTS X_0 VARIABLE
OPCOES_DADOS = {'DATASET': DADOS, 'COLUMN': 'X_0'}
# CALL PLOT
RASD_PLOT_1(OPCOES_DADOS, OPCOES_GRAFICAS)
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'EXTENSION': '.svg',
'DPI': 600,
'WIDTH': 0.300,
'HEIGHT': 0.150,
'X DATA': 'S_0',
'Y DATA': 'R_0',
'X AXIS SIZE': 16,
'Y AXIS SIZE': 16,
'AXISES COLOR': '#000000',
'X AXIS LABEL': '$S_0$',
'Y AXIS LABEL': '$R_0$',
'LABELS SIZE': 18,
'LABELS COLOR': '#000000',
'LOC LEGEND': 'lower right',
'TITLE LEGEND': 'Failure index ($I$):'}
# RESULTS
OPCOES_DADOS = {'DATASET': DADOS, 'X DATA': 'S_0', 'Y DATA': 'R_0', 'HUE VALUE': 'I_0'}
# CALL PLOT
RASD_PLOT_2(OPCOES_DADOS, OPCOES_GRAFICAS)
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'EXTENSION': '.svg',
'DPI': 600,
'WIDTH': 0.20,
'HEIGHT': 0.10,
'X DATA': 'S_0',
'Y DATA': 'R_0',
'X AXIS SIZE': 20,
'Y AXIS SIZE': 20,
'AXISES COLOR': '#000000',
'X AXIS LABEL': '$S_0$',
'Y AXIS LABEL': '$R_0$',
'LABELS SIZE': 16,
'LABELS COLOR': '#000000',
'G VALUE': 'G_0',
'TRANSPARENCY': 0.8,
'COLOR MAP': 'viridis'}
# RESULTS
OPCOES_DADOS = {'DATASET': DADOS, 'X DATA': 'S_0', 'Y DATA': 'R_0', 'G VALUE': 'G_0'}
# CALL PLOT
RASD_PLOT_3(OPCOES_DADOS, OPCOES_GRAFICAS)
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'EXTENSION': '.svg',
'DPI': 600,
'WIDTH': 0.20,
'HEIGHT': 0.10,
'X DATA': 'S_0',
'Y DATA': 'R_0',
'X AXIS SIZE': 20,
'Y AXIS SIZE': 20,
'AXISES COLOR': '#00000',
'X AXIS LABEL': '$Z_0$',
'Y AXIS LABEL': 'Frequência',
'LABELS SIZE': 16,
'LABELS COLOR': '#000000',
'C VALUE': 'G_0',
'TRANSPARENCY': 0.8,
'COLOR MAP': 'viridis',
'BINS': '50',
'ALPHA': '0.5'}
# RESULTS
OPCOES_DADOS = {'DATASET': DADOS, 'X DATA': 'R_0', 'Y DATA': 'S_0', 'G VALUE': 'G_0'}
# CALL PLOT
RASD_PLOT_4(OPCOES_DADOS, OPCOES_GRAFICAS)
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'EXTENSION': '.svg',
'DPI': 600,
'WIDTH': 0.20,
'HEIGHT': 0.10,
'X AXIS SIZE': 20,
'Y AXIS SIZE': 20,
'AXISES COLOR': '#000000',
'X AXIS LABEL': 'Número de Simulações (ns)',
'Y AXIS LABEL': 'Probabilidade de Falha',
'LABELS SIZE': 16,
'LABELS COLOR': '#000000',
'CHART COLOR': 'black',
'POPULATION' : POP,
'TYPE' : 'pf'}
# RESULTS
OPCOES_DADOS = {'DATASET': RESULTS_TEST}
# CALL PLOT
RASD_PLOT_5(OPCOES_DADOS, OPCOES_GRAFICAS)
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'EXTENSION': '.svg',
'DPI': 600,
'WIDTH': 0.20,
'HEIGHT': 0.10,
'X AXIS SIZE': 20,
'Y AXIS SIZE': 20,
'AXISES COLOR': '#000000',
'X AXIS LABEL': 'Número de Simulações (ns)',
'Y AXIS LABEL': 'Beta',
'LABELS SIZE': 16,
'LABELS COLOR': '#000000',
'CHART COLOR': 'black',
'POPULATION' : POP,
'TYPE' : 'beta'}
# RESULTS
OPCOES_DADOS = {'DATASET': RESULTS_TEST}
# CALL PLOT
RASD_PLOT_5(OPCOES_DADOS, OPCOES_GRAFICAS)
```
|
github_jupyter
|
# Instalando e atualizando as bibliotecas necessárias
!pip install pyDOE
!pip install -U seaborn
# Importando RASD Toolbox
from RASD_TOOLBOX import *
# Importando Numpy toolbox
import numpy as np
from VIGA_METALICA_VERIFICA import *
from FINITO_FEM_TOOLBOX import *
ELEMENTO_ANALISADO = [2,8]
#150 X 18,0
P1 = ['NORMAL', 200E9, 10E9] #E MPa
P2 = ['NORMAL', 250E6, 12.5E6] #Fy MPa
P3 = ['NORMAL', 10000, 1000] #Fy N
P4 = ['NORMAL', 1, 0.05] #Incerteza de Modelo 1 adimensional
P5 = ['NORMAL', 1, 0.05] #Incerteza de Modelo 2 adimensional
P6 = ['NORMAL', 139, 2.78] #H_W mm
P7 = ['NORMAL', 5.8, 0.116] #T_W mm
P8 = ['NORMAL', 102, 2.04] #B_F mm
P9 = ['NORMAL', 7.1, 0.142] #T_F mm
P10 = ['NORMAL', 139.4, 2.788] #Z cm3
P11 = ['NORMAL', 939, 18.78] #I cm4
P12 = ['NORMAL', 1000000, 100000] #Mola 1 N/m
P13 = ['NORMAL', 100000, 10000] #Mola 2 N/m
POP = [10000]
#POP = np.arange(10,1000000, 10000).tolist() # (x,y,Z); x=pop inicial; y=pop final; z=variacao pop
#carta_total = carga G + carga Q
SETUP = {'N_REP': len(POP),
'POP': POP,
'N_G': 3,
'D': 13, #variables
'VARS': [P1, P2, P3, P4, P5, P6, P7, P8, P9, P10, P11, P12, P13],
'MODEL': 'MCS'}
def OBJ(X):
E = X[0]
FY = X[1]
F1 = X[2]
M1 = X[3]
M2 = X[4]
H_W = X[5]
T_W = X[6]
W_C = X[7]
W_T = X[8]
Z = X[9]
INERCIA = X[10]
K1 = X[11]
K2 = X[12]
M_SD = []
V_SD = []
N_SD = []
D_SD = []
L_MAX = []
# auxiliares para tranformar em metroX
ALFA = 0.001 #mm para m
BETA = 0.000001 #cm3 para m3
GAMMA = 0.00000001 #cm4 para m4
VIGA = {'H_W': H_W * ALFA, #m
'T_W': T_W * ALFA, #m
'B_F': W_C * ALFA, #m
'T_F': W_T * ALFA, #m
'PARAMETRO_PERFIL': 'DUPLA SIMETRIA', #"DUPLA SIMETRIA, MONO SIMETRIA "
'TIPO_PERFIL': 'LAMINADO', #"SOLDADO, LAMINADO "
'GAMMA_A1': 1.1,
'Z': Z * BETA, #m3
'INERCIA':INERCIA * GAMMA, #m4
'E_S': E , #Pa
'F_Y': FY} #N
AREA = 2 * (VIGA['T_F'] * VIGA['B_F']) + (VIGA['T_W'] * VIGA['H_W']) #m2
X_GC = VIGA['B_F'] / 2 #m
Y_GC = (( VIGA['T_F'] * 2) + VIGA['H_W']) / 2 #m
FRAME_00 = {
"TYPE_ELEMENT": 0,
"TYPE_SOLUTION": 0,
"N_NODES": 26,
"N_MATERIALS": 1,
"N_SECTIONS": 1,
"N_ELEMENTS": 27,
"N_DOFPRESCRIPTIONS": 4,
"N_DOFLOADED": 24,
"N_DOFSPRINGS": 2,
"COORDINATES": #X,Y
np.array([
[0.0,0.0],
[5.25,0.0],
[0.0,3.0],
[0.75,3.0],
[1.5,3.0],
[2.25,3.0],
[3.0,3.0],
[3.75,3.0],
[4.5,3.0],
[5.25,3.0],
[0.0,6.0],
[0.75,6.0],
[1.5,6.0],
[2.25,6.0],
[3.0,6.0],
[3.75,6.0],
[4.5,6.0],
[5.25,6.0],
[0.0,9.0],
[0.75,9.0],
[1.5,9.0],
[2.25,9.0],
[3.0,9.0],
[3.75,9.0],
[4.5,9.0],
[5.25,9.0]]),
"ELEMENTS": #NODE 1,NODE 2,MATERIAL ID,SECTION ID,HINGE ID NODE 1,HINGE ID NODE 2
np.array([
[0,2,0,0,0,0],
[1,9,0,0,0,0],
[2,3,0,0,0,0],
[3,4,0,0,0,0],
[4,5,0,0,0,0],
[5,6,0,0,0,0],
[6,7,0,0,0,0],
[7,8,0,0,0,0],
[8,9,0,0,0,0],
[2,10,0,0,0,0],
[9,17,0,0,0,0],
[10,11,0,0,0,0],
[11,12,0,0,0,0],
[12,13,0,0,0,0],
[13,14,0,0,0,0],
[14,15,0,0,0,0],
[15,16,0,0,0,0],
[16,17,0,0,0,0],
[10,18,0,0,0,0],
[17,25,0,0,0,0],
[18,19,0,0,0,0],
[19,20,0,0,0,0],
[20,21,0,0,0,0],
[21,22,0,0,0,0],
[22,23,0,0,0,0],
[23,24,0,0,0,0],
[24,25,0,0,0,0]]),
"MATERIALS": #YOUNG, POISSON,DENSITY, THERMAL COEFFICIENT
np.array([[VIGA['E_S'], 1, 1, 1E-8]]),
#np.array([[200E9,1,1,1E-8]]),
"SECTIONS": #AREA, INERTIA 1, INERTIA 2, X GC,Y GC
np.array([[AREA , VIGA['INERCIA'], VIGA['INERCIA'], X_GC, X_GC]]),
#np.array([[0.0450,0.000377,0.000377,0.075,0.15]]),
"PRESCRIBED DISPLACEMENTS": #NODE,DIRECTION(X=0,Y=1,Z=2),VALUE
np.array([
[0,0,0],
#[0,1,0],
[0,2,0],
[1,0,0],
#[1,1,0],
[1,2,0]]),
"ELEMENT LOADS": None,
"NODAL LOADS": #NODE,DIRECTION(X=0,Y=1,Z=2),VALUE
np.array([
[2,1,-F1,0],
[3,1,-F1,0],
[4,1,-F1,0],
[5,1,-F1,0],
[6,1,-F1,0],
[7,1,-F1,0],
[8,1,-F1,0],
[9,1,-F1,0],
[10,1,-F1,0],
[11,1,-F1,0],
[12,1,-F1,0],
[13,1,-F1,0],
[14,1,-F1,0],
[15,1,-F1,0],
[16,1,-F1,0],
[17,1,-F1,0],
[18,1,-F1,0],
[19,1,-F1,0],
[20,1,-F1,0],
[21,1,-F1,0],
[22,1,-F1,0],
[23,1,-F1,0],
[24,1,-F1,0],
[25,1,-F1,0]]),
"SPRINGS": #NODE,DIRECTION(X=0,Y=1,Z=2),VALUE
np.array([
[0,1,K1,0],
[1,1,K2,0]])}
FRAME_00_RESULTS = MEF1D(DICTIONARY = FRAME_00)
ELEMENT = pd.DataFrame(FRAME_00_RESULTS[ELEMENTO_ANALISADO[0]:ELEMENTO_ANALISADO[1]])
for i in range(len(ELEMENT)):
M_SD.append(max(ELEMENT['M'][i].max(),abs(ELEMENT['M'][i].min())))
V_SD.append(max(ELEMENT['V'][i].max(),abs(ELEMENT['V'][i].min())))
N_SD.append(max(ELEMENT['N'][i].max(),abs(ELEMENT['N'][i].min())))
D_SD.append(min(ELEMENT['UX'][i].max(),abs(ELEMENT['UX'][i].min())))
#L_MAX.append(ELEMENT['X'][i].max())
M_SD = max(M_SD)
V_SD = max(V_SD)
N_SD = max(N_SD)
D_SD = min(D_SD)
L_MAX = FRAME_00['COORDINATES'][1][0]
ESFORCOS = {'M_SD': M_SD,
'V_SD': V_SD,
'D_SD': D_SD,
'L_MAX': L_MAX}
R_0, S_0 = VERIFICACAO_VIGA_METALICA_MOMENTO_FLETOR(VIGA, ESFORCOS)
G_0 = -M1 * R_0 + M2 * S_0
R_1, S_1 = VERIFICACAO_VIGA_METALICA_ESFORCO_CORTANTE(VIGA, ESFORCOS)
G_1 = -M1 * R_1 + M2 * S_1
R_2, S_2 = VERIFICACAO_VIGA_METALICA_DEFORMACAO(VIGA, ESFORCOS)
G_2 = -M1 * R_2 + M2 * S_2
R = [R_0, R_1, R_2]
S = [S_0, S_1, S_2]
G = [G_0, G_1, G_2]
return R, S, G
RESULTS_TEST = RASD_STOCHASTIC(SETUP, OBJ)
RESULTS_TEST[0]['TOTAL RESULTS']['R_0']
DADOS = RESULTS_TEST[0]['TOTAL RESULTS']
DADOS['I_0'].sum()
DADOS = RESULTS_TEST[len(POP)-1]['TOTAL RESULTS'] #USAR QUANDO HOUVEREM VARIAS SIMULACOES
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'WIDTH': 0.40,
'HEIGHT': 0.20,
'X AXIS SIZE': 20,
'Y AXIS SIZE': 20,
'AXISES COLOR': '#000000',
'X AXIS LABEL': '$x_0$ - $P_X (lb)$',
'LABELS SIZE': 16,
'LABELS COLOR': '#000000',
'CHART COLOR': '#FEB625',
'KDE': False,
'DPI': 600,
'BINS' : 20,
'EXTENSION': '.svg'}
# RESULTS X_0 VARIABLE
OPCOES_DADOS = {'DATASET': DADOS, 'COLUMN': 'X_0'}
# CALL PLOT
RASD_PLOT_1(OPCOES_DADOS, OPCOES_GRAFICAS)
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'EXTENSION': '.svg',
'DPI': 600,
'WIDTH': 0.300,
'HEIGHT': 0.150,
'X DATA': 'S_0',
'Y DATA': 'R_0',
'X AXIS SIZE': 16,
'Y AXIS SIZE': 16,
'AXISES COLOR': '#000000',
'X AXIS LABEL': '$S_0$',
'Y AXIS LABEL': '$R_0$',
'LABELS SIZE': 18,
'LABELS COLOR': '#000000',
'LOC LEGEND': 'lower right',
'TITLE LEGEND': 'Failure index ($I$):'}
# RESULTS
OPCOES_DADOS = {'DATASET': DADOS, 'X DATA': 'S_0', 'Y DATA': 'R_0', 'HUE VALUE': 'I_0'}
# CALL PLOT
RASD_PLOT_2(OPCOES_DADOS, OPCOES_GRAFICAS)
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'EXTENSION': '.svg',
'DPI': 600,
'WIDTH': 0.20,
'HEIGHT': 0.10,
'X DATA': 'S_0',
'Y DATA': 'R_0',
'X AXIS SIZE': 20,
'Y AXIS SIZE': 20,
'AXISES COLOR': '#000000',
'X AXIS LABEL': '$S_0$',
'Y AXIS LABEL': '$R_0$',
'LABELS SIZE': 16,
'LABELS COLOR': '#000000',
'G VALUE': 'G_0',
'TRANSPARENCY': 0.8,
'COLOR MAP': 'viridis'}
# RESULTS
OPCOES_DADOS = {'DATASET': DADOS, 'X DATA': 'S_0', 'Y DATA': 'R_0', 'G VALUE': 'G_0'}
# CALL PLOT
RASD_PLOT_3(OPCOES_DADOS, OPCOES_GRAFICAS)
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'EXTENSION': '.svg',
'DPI': 600,
'WIDTH': 0.20,
'HEIGHT': 0.10,
'X DATA': 'S_0',
'Y DATA': 'R_0',
'X AXIS SIZE': 20,
'Y AXIS SIZE': 20,
'AXISES COLOR': '#00000',
'X AXIS LABEL': '$Z_0$',
'Y AXIS LABEL': 'Frequência',
'LABELS SIZE': 16,
'LABELS COLOR': '#000000',
'C VALUE': 'G_0',
'TRANSPARENCY': 0.8,
'COLOR MAP': 'viridis',
'BINS': '50',
'ALPHA': '0.5'}
# RESULTS
OPCOES_DADOS = {'DATASET': DADOS, 'X DATA': 'R_0', 'Y DATA': 'S_0', 'G VALUE': 'G_0'}
# CALL PLOT
RASD_PLOT_4(OPCOES_DADOS, OPCOES_GRAFICAS)
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'EXTENSION': '.svg',
'DPI': 600,
'WIDTH': 0.20,
'HEIGHT': 0.10,
'X AXIS SIZE': 20,
'Y AXIS SIZE': 20,
'AXISES COLOR': '#000000',
'X AXIS LABEL': 'Número de Simulações (ns)',
'Y AXIS LABEL': 'Probabilidade de Falha',
'LABELS SIZE': 16,
'LABELS COLOR': '#000000',
'CHART COLOR': 'black',
'POPULATION' : POP,
'TYPE' : 'pf'}
# RESULTS
OPCOES_DADOS = {'DATASET': RESULTS_TEST}
# CALL PLOT
RASD_PLOT_5(OPCOES_DADOS, OPCOES_GRAFICAS)
OPCOES_GRAFICAS = {'NAME': 'WANDER',
'EXTENSION': '.svg',
'DPI': 600,
'WIDTH': 0.20,
'HEIGHT': 0.10,
'X AXIS SIZE': 20,
'Y AXIS SIZE': 20,
'AXISES COLOR': '#000000',
'X AXIS LABEL': 'Número de Simulações (ns)',
'Y AXIS LABEL': 'Beta',
'LABELS SIZE': 16,
'LABELS COLOR': '#000000',
'CHART COLOR': 'black',
'POPULATION' : POP,
'TYPE' : 'beta'}
# RESULTS
OPCOES_DADOS = {'DATASET': RESULTS_TEST}
# CALL PLOT
RASD_PLOT_5(OPCOES_DADOS, OPCOES_GRAFICAS)
| 0.205735 | 0.423696 |
## Nonlinear Dimensionality Reduction
G. Richards (2016, 2018), based on materials from Ivezic, Connolly, Miller, Leighly, and VanderPlas.
Today we will talk about the concepts of
* manifold learning
* nonlinear dimensionality reduction
Specifically using the following algorithms
* local linear embedding (LLE)
* isometric mapping (IsoMap)
* t-distributed Stochastic Neighbor Embedding (t-SNE)
Let's start by my echoing the brief note of caution given in Adam Miller's notebook: "astronomers will often try to derive physical insight from PCA eigenspectra or eigentimeseries, but this is not advisable as there is no physical reason for the data to be linearly and orthogonally separable". Moreover, physical components are (generally) positive definite. So, PCA is great for dimensional reduction, but for doing physics there are generally better choices.
While NMF "solves" the issue of negative components, it is still a linear process. For data with non-linear correlations, an entire field, known as [Manifold Learning](http://scikit-learn.org/stable/modules/manifold.html) and [nonlinear dimensionality reduction]( https://en.wikipedia.org/wiki/Nonlinear_dimensionality_reduction), has been developed, with several algorithms available via the [`sklearn.manifold`](http://scikit-learn.org/stable/modules/classes.html#module-sklearn.manifold) module.
For example, if your data set looks like this:

Then PCA is going to give you something like this.

Clearly not very helpful!
What you were really hoping for is something more like the results below. For more examples see
[Vanderplas & Connolly 2009](http://iopscience.iop.org/article/10.1088/0004-6256/138/5/1365/meta;jsessionid=48A569862A424ECCAEECE2A900D9837B.c3.iopscience.cld.iop.org)

## Local Linear Embedding
[Local Linear Embedding](http://scikit-learn.org/stable/modules/generated/sklearn.manifold.LocallyLinearEmbedding.html#sklearn.manifold.LocallyLinearEmbedding) attempts to embed high-$D$ data in a lower-$D$ space. Crucially it also seeks to preserve the geometry of the local "neighborhoods" around each point. In the case of the "S" curve, it seeks to unroll the data. The steps are
Step 1: define local geometry
- local neighborhoods determined from $k$ nearest neighbors.
- for each point calculate weights that reconstruct a point from its $k$ nearest
neighbors via
$$
\begin{equation}
\mathcal{E}_1(W) = \left|X - WX\right|^2,
\end{equation}
$$
where $X$ is an $N\times K$ matrix and $W$ is an $N\times N$ matrix that minimizes the reconstruction error.
Essentially this is finding the hyperplane that describes the local surface at each point within the data set. So, imagine that you have a bunch of square tiles and you are trying to tile the surface with them.
Step 2: embed within a lower dimensional space
- set all $W_{ij}=0$ except when point $j$ is one of the $k$ nearest neighbors of point $i$.
- $W$ becomes very sparse for $k \ll N$ (only $Nk$ entries in $W$ are non-zero).
- minimize
>$\begin{equation}
\mathcal{E}_2(Y) = \left|Y - W Y\right|^2,
\end{equation}
$
with $W$ fixed to find an $N$ by $d$ matrix ($d$ is the new dimensionality).
Step 1 requires a nearest-neighbor search.
Step 2 requires an
eigenvalue decomposition of the matrix $C_W \equiv (I-W)^T(I-W)$.
LLE has been applied to data as diverse as galaxy spectra, stellar spectra, and photometric light curves. It was introduced by [Roweis & Saul (2000)](https://www.ncbi.nlm.nih.gov/pubmed/11125150).
Skikit-Learn's call to LLE is as follows, with a more detailed example already being given above.
```
import numpy as np
from sklearn.manifold import LocallyLinearEmbedding
X = np.random.normal(size=(1000,2)) # 1000 points in 2D
R = np.random.random((2,10)) # projection matrix
X = np.dot(X,R) # now a 2D linear manifold in 10D space
k = 5 # Number of neighbors to use in fit
n = 2 # Number of dimensions to fit
lle = LocallyLinearEmbedding(k,n)
lle.fit(X)
proj = lle.transform(X) # 100x2 projection of the data
```
See what LLE does for the digits data, using the 7 nearest neighbors and 2 components.
```
# Execute this cell to load the digits sample
%matplotlib inline
import numpy as np
from sklearn.datasets import load_digits
from matplotlib import pyplot as plt
digits = load_digits()
grid_data = np.reshape(digits.data[0], (8,8)) #reshape to 8x8
plt.imshow(grid_data, interpolation = "nearest", cmap = "bone_r")
print(grid_data)
X = digits.data
y = digits.target
#LLE
from sklearn.manifold import LocallyLinearEmbedding
k = 7 # Number of neighbors to use in fit
n = 2 # Number of dimensions to fit
lle = LocallyLinearEmbedding(k,n)
lle.fit(X)
X_reduced = lle.transform(X)
plt.scatter(X_reduced[:,0], X_reduced[:,1], c=y, cmap="nipy_spectral", edgecolor="None")
plt.colorbar()
```
## Isometric Mapping
is based on multi-dimensional scaling (MDS) framework. It was introduced in the same volume of *Science* as the article above. See [Tenenbaum, de Silva, & Langford (2000)](https://www.ncbi.nlm.nih.gov/pubmed/?term=A+Global+Geometric+Framework+for+Nonlinear+Dimensionality+Reduction).
Geodestic curves are used to recover non-linear structure.
In Scikit-Learn [IsoMap](http://scikit-learn.org/stable/modules/generated/sklearn.manifold.Isomap.html) is implemented as follows:
```
# Execute this cell
import numpy as np
from sklearn.manifold import Isomap
XX = np.random.normal(size=(1000,2)) # 1000 points in 2D
R = np.random.random((2,10)) # projection matrix
XX = np.dot(XX,R) # X is a 2D manifold in 10D space
k = 5 # number of neighbors
n = 2 # number of dimensions
iso = Isomap(k,n)
iso.fit(XX)
proj = iso.transform(XX) # 1000x2 projection of the data
```
Try 7 neighbors and 2 dimensions on the digits data.
```
# IsoMap
from sklearn.manifold import Isomap
k = 7 # Number of neighbors to use in fit
n = 2 # Number of dimensions to fit
iso = Isomap(k,n)
iso.fit(X)
X_reduced = iso.transform(X)
plt.scatter(X_reduced[:,0], X_reduced[:,1], c=y, cmap="nipy_spectral", edgecolor="None")
plt.colorbar()
```
## t-SNE
[t-distributed Stochastic Neighbor Embedding (t-SNE)](https://en.wikipedia.org/wiki/T-distributed_stochastic_neighbor_embedding) is not discussed in the book, Scikit-Learn does have a [t-SNE implementation](http://scikit-learn.org/stable/modules/generated/sklearn.manifold.TSNE.html) and it is well worth mentioning this manifold learning algorithm too. SNE itself was developed by [Hinton & Roweis](http://www.cs.toronto.edu/~fritz/absps/sne.pdf) with the "$t$" part being added by [van der Maaten & Hinton](http://jmlr.org/papers/volume9/vandermaaten08a/vandermaaten08a.pdf). It works like the other manifold learning algorithms.
Try it on the digits data. You'll need to import `TSNE` from `sklearn.manifold`, instantiate it with 2 components, then do a `fit_transform` on the original data.
```
# t-SNE
from sklearn.manifold import TSNE
tsne = TSNE(n_components=2)
X_reduced = tsne.fit_transform(X)
plt.scatter(X_reduced[:,0], X_reduced[:,1] , c=y, cmap="nipy_spectral", edgecolor="None")
plt.colorbar()
```
You'll know if you have done it right if you understand Adam Miller's comment "Holy freakin' smokes. That is magic. (It's possible we just solved science)."
Personally, I think that some exclamation points may be needed in there!
What's even more illuminating is to make the plot using the actual digits to plot the points. Then you can see why certain digits are alike or split into multiple regions. Can you explain the patterns you see here?
```
# Execute this cell
from matplotlib import offsetbox
#----------------------------------------------------------------------
# Scale and visualize the embedding vectors
def plot_embedding(X):
x_min, x_max = np.min(X, 0), np.max(X, 0)
X = (X - x_min) / (x_max - x_min)
plt.figure()
ax = plt.subplot(111)
for i in range(X.shape[0]):
#plt.text(X[i, 0], X[i, 1], str(digits.target[i]), color=plt.cm.Set1(y[i] / 10.), fontdict={'weight': 'bold', 'size': 9})
plt.text(X[i, 0], X[i, 1], str(digits.target[i]), color=plt.cm.nipy_spectral(y[i]/9.))
shown_images = np.array([[1., 1.]]) # just something big
for i in range(digits.data.shape[0]):
dist = np.sum((X[i] - shown_images) ** 2, 1)
if np.min(dist) < 4e-3:
# don't show points that are too close
continue
shown_images = np.r_[shown_images, [X[i]]]
imagebox = offsetbox.AnnotationBbox(offsetbox.OffsetImage(digits.images[i], cmap=plt.cm.gray_r), X[i])
ax.add_artist(imagebox)
plt.xticks([]), plt.yticks([])
plot_embedding(X_reduced)
plt.show()
```
With the remainder of time in class today, play with the arguments of the algorithms that we have discussed this week and/or try running them on a different data set. For example the iris data set or one of the other samples of data that are included with Scikit-Learn. Or maybe have a look through some of these public data repositories:
- [https://github.com/caesar0301/awesome-public-datasets?utm_content=buffer4245d&utm_medium=social&utm_source=twitter.com&utm_campaign=buffer](https://github.com/caesar0301/awesome-public-datasets?utm_content=buffer4245d&utm_medium=social&utm_source=twitter.com&utm_campaign=buffer)
- [http://www.datasciencecentral.com/m/blogpost?id=6448529%3ABlogPost%3A318739](http://www.datasciencecentral.com/m/blogpost?id=6448529%3ABlogPost%3A318739)
- [http://www.kdnuggets.com/2015/04/awesome-public-datasets-github.html](http://www.kdnuggets.com/2015/04/awesome-public-datasets-github.html)
|
github_jupyter
|
import numpy as np
from sklearn.manifold import LocallyLinearEmbedding
X = np.random.normal(size=(1000,2)) # 1000 points in 2D
R = np.random.random((2,10)) # projection matrix
X = np.dot(X,R) # now a 2D linear manifold in 10D space
k = 5 # Number of neighbors to use in fit
n = 2 # Number of dimensions to fit
lle = LocallyLinearEmbedding(k,n)
lle.fit(X)
proj = lle.transform(X) # 100x2 projection of the data
# Execute this cell to load the digits sample
%matplotlib inline
import numpy as np
from sklearn.datasets import load_digits
from matplotlib import pyplot as plt
digits = load_digits()
grid_data = np.reshape(digits.data[0], (8,8)) #reshape to 8x8
plt.imshow(grid_data, interpolation = "nearest", cmap = "bone_r")
print(grid_data)
X = digits.data
y = digits.target
#LLE
from sklearn.manifold import LocallyLinearEmbedding
k = 7 # Number of neighbors to use in fit
n = 2 # Number of dimensions to fit
lle = LocallyLinearEmbedding(k,n)
lle.fit(X)
X_reduced = lle.transform(X)
plt.scatter(X_reduced[:,0], X_reduced[:,1], c=y, cmap="nipy_spectral", edgecolor="None")
plt.colorbar()
# Execute this cell
import numpy as np
from sklearn.manifold import Isomap
XX = np.random.normal(size=(1000,2)) # 1000 points in 2D
R = np.random.random((2,10)) # projection matrix
XX = np.dot(XX,R) # X is a 2D manifold in 10D space
k = 5 # number of neighbors
n = 2 # number of dimensions
iso = Isomap(k,n)
iso.fit(XX)
proj = iso.transform(XX) # 1000x2 projection of the data
# IsoMap
from sklearn.manifold import Isomap
k = 7 # Number of neighbors to use in fit
n = 2 # Number of dimensions to fit
iso = Isomap(k,n)
iso.fit(X)
X_reduced = iso.transform(X)
plt.scatter(X_reduced[:,0], X_reduced[:,1], c=y, cmap="nipy_spectral", edgecolor="None")
plt.colorbar()
# t-SNE
from sklearn.manifold import TSNE
tsne = TSNE(n_components=2)
X_reduced = tsne.fit_transform(X)
plt.scatter(X_reduced[:,0], X_reduced[:,1] , c=y, cmap="nipy_spectral", edgecolor="None")
plt.colorbar()
# Execute this cell
from matplotlib import offsetbox
#----------------------------------------------------------------------
# Scale and visualize the embedding vectors
def plot_embedding(X):
x_min, x_max = np.min(X, 0), np.max(X, 0)
X = (X - x_min) / (x_max - x_min)
plt.figure()
ax = plt.subplot(111)
for i in range(X.shape[0]):
#plt.text(X[i, 0], X[i, 1], str(digits.target[i]), color=plt.cm.Set1(y[i] / 10.), fontdict={'weight': 'bold', 'size': 9})
plt.text(X[i, 0], X[i, 1], str(digits.target[i]), color=plt.cm.nipy_spectral(y[i]/9.))
shown_images = np.array([[1., 1.]]) # just something big
for i in range(digits.data.shape[0]):
dist = np.sum((X[i] - shown_images) ** 2, 1)
if np.min(dist) < 4e-3:
# don't show points that are too close
continue
shown_images = np.r_[shown_images, [X[i]]]
imagebox = offsetbox.AnnotationBbox(offsetbox.OffsetImage(digits.images[i], cmap=plt.cm.gray_r), X[i])
ax.add_artist(imagebox)
plt.xticks([]), plt.yticks([])
plot_embedding(X_reduced)
plt.show()
| 0.712232 | 0.973569 |
```
%load_ext autoreload
%autoreload 2
%aimport utils
import pandas as pd
import numpy as np
import altair as alt
from altair_saver import save
from os.path import join
from web import for_website
from constants import COLUMNS, DATA_AGGREGATE_TYPES
from utils import (
read_combined_daily_counts_df,
read_combined_by_country_daily_counts_df,
read_site_details_df,
get_visualization_subtitle,
get_siteid_anonymous_map,
get_anonymousid_color_maps,
apply_theme
)
```
# Required Setups
- All combined datasets should be placed in `../data/combined` (e.g., `../data/combined/DailyCounts-Combinedyymmdd.csv` for the DailyCounts file).
- To save PNG files for visualizations, a folder named "output" should be present (i.e., `../output/`).
```
# Path to save *.PNG files
SAVE_DIR = join("..", "output")
# Country Info
ALL_COUNTRY = "All countries"
ALL_COUNTRY_COLOR = "#444444"
COUNTRIES = ["France", "Germany", "Italy", "Singapore", "USA"]
COUNTRY_COLOR = ["#0072B2", "#E69F00", "#009E73", "#CC79A7", "#D55E00"]
```
# Data preprocess
```
CATEGORY = "category"
def preprocess_daily_df(df_dc):
# Wide to long
df_dc = pd.melt(df_dc, id_vars=[
COLUMNS.SITE_ID, COLUMNS.DATE,
COLUMNS.MASKED_UPPER_BOUND_NEW_POSITIVE_CASES,
COLUMNS.MASKED_UPPER_BOUND_PATIENTS_IN_ICU,
COLUMNS.MASKED_UPPER_BOUND_NEW_DEATHS,
COLUMNS.UNMASKED_SITES_NEW_POSITIVE_CASES,
COLUMNS.UNMASKED_SITES_PATIENTS_IN_ICU,
COLUMNS.UNMASKED_SITES_NEW_DEATHS,
COLUMNS.MASKED_SITES_NEW_POSITIVE_CASES,
COLUMNS.MASKED_SITES_PATIENTS_IN_ICU,
COLUMNS.MASKED_SITES_NEW_DEATHS
])
df_dc = df_dc.rename(columns={"variable": CATEGORY, "value": COLUMNS.NUM_PATIENTS})
# Leave only the 'upper' and 'under' values for the certain 'category' only
for c in [COLUMNS.NEW_POSITIVE_CASES, COLUMNS.PATIENTS_IN_ICU, COLUMNS.NEW_DEATHS]:
filter_c = df_dc[CATEGORY] == c
df_dc.loc[filter_c, "upper"] = df_dc.loc[filter_c, COLUMNS.NUM_PATIENTS] + df_dc.loc[filter_c, "masked_upper_bound_" + c]
df_dc.loc[filter_c, "under"] = df_dc.loc[filter_c, COLUMNS.NUM_PATIENTS]
df_dc.loc[filter_c, COLUMNS.NUM_PATIENTS] = df_dc.loc[filter_c, COLUMNS.NUM_PATIENTS] + df_dc.loc[filter_c, "masked_upper_bound_" + c] / 2.0
# Add num of sites
df_dc.loc[filter_c, COLUMNS.NUM_SITES] = df_dc["unmasked_sites_" + c] + df_dc["masked_sites_" + c]
# Drop unused columns
df_dc = df_dc.drop(columns=[
COLUMNS.MASKED_UPPER_BOUND_NEW_POSITIVE_CASES,
COLUMNS.MASKED_UPPER_BOUND_PATIENTS_IN_ICU,
COLUMNS.MASKED_UPPER_BOUND_NEW_DEATHS,
COLUMNS.UNMASKED_SITES_NEW_POSITIVE_CASES,
COLUMNS.UNMASKED_SITES_PATIENTS_IN_ICU,
COLUMNS.UNMASKED_SITES_NEW_DEATHS,
COLUMNS.MASKED_SITES_NEW_POSITIVE_CASES,
COLUMNS.MASKED_SITES_PATIENTS_IN_ICU,
COLUMNS.MASKED_SITES_NEW_DEATHS
])
# Make sure to drop date range out of our interest
df_dc = df_dc[df_dc[COLUMNS.DATE] >= "2020-01-29"]
df_dc = df_dc[df_dc[COLUMNS.DATE] <= "2020-03-31"]
# We are not using ICU
df_dc = df_dc[df_dc[CATEGORY] != COLUMNS.PATIENTS_IN_ICU]
# Use more readable names
df_dc.loc[df_dc[COLUMNS.SITE_ID] == "Combined", COLUMNS.SITE_ID] = ALL_COUNTRY
# Remove zero num_sites, which is missing data
df_dc = df_dc[df_dc[COLUMNS.NUM_SITES] != 0]
return df_dc
# Read files
df_dc = read_combined_by_country_daily_counts_df()
df_dc = preprocess_daily_df(df_dc)
df_dc_combined = read_combined_daily_counts_df()
df_dc_combined = preprocess_daily_df(df_dc_combined)
# Merge
df_dc = pd.concat([df_dc, df_dc_combined])
df_dc_combined
```
# Visualizations
```
CATEGORIES = [COLUMNS.NEW_POSITIVE_CASES, COLUMNS.NEW_DEATHS]
TITLE_BY_CATEGORY = {
COLUMNS.NEW_POSITIVE_CASES: "possitive cases",
COLUMNS.NEW_DEATHS: "deaths",
COLUMNS.PATIENTS_IN_ICU: "ICU admissions"
}
def dailycount_by_date(df=df_dc, is_cum=True, is_only_combined=False, is_site_level=False, is_num_hospital=False):
# Selections
nearest = alt.selection(type="single", nearest=True, on="mouseover", encodings=["x"], empty='none', clear="mouseout")
dailycount_dropdown = alt.binding_select(options=CATEGORIES)
dailycount_selection = alt.selection_single(fields=[CATEGORY], bind=dailycount_dropdown, name="Value", init={CATEGORY: COLUMNS.NEW_POSITIVE_CASES})
legend_selection = alt.selection_multi(fields=[COLUMNS.SITE_ID], bind="legend")
date_brush = alt.selection(type="interval", encodings=['x'])
y_zoom = alt.selection(type="interval", bind='scales', encodings=['y'])
# Rule
nearest_rule = alt.Chart(df).mark_rule(color="red").encode(
x=f"{COLUMNS.DATE}:T",
size=alt.value(0.5)
).transform_filter(
nearest
)
color_scale = alt.Scale(domain=COUNTRIES, range=COUNTRY_COLOR)
color_scale_bg = alt.Scale(domain=COUNTRIES, range=["lightgray"])
if is_only_combined:
color_scale = alt.Scale(domain=[ALL_COUNTRY], range=[ALL_COUNTRY_COLOR])
color_scale_bg = alt.Scale(domain=[ALL_COUNTRY], range=["lightgray"])
if is_site_level:
color_scale = alt.Scale(domain=ANONYMOUS_SITES, range=ANONYMOUS_COLORS)
color_scale_bg = alt.Scale(domain=ANONYMOUS_SITES, range=["lightgray"])
# Filter
filtered_chart = alt.Chart(df).transform_filter(
dailycount_selection
).transform_filter(
legend_selection
)
if is_only_combined:
filtered_chart = filtered_chart.transform_filter(
alt.datum[COLUMNS.SITE_ID] == ALL_COUNTRY
)
else:
filtered_chart = filtered_chart.transform_filter(
alt.datum[COLUMNS.SITE_ID] != ALL_COUNTRY
)
DAILY_COUNT_TOOLTIP = [
alt.Tooltip(COLUMNS.SITE_ID, title="Country"),
alt.Tooltip(COLUMNS.DATE, title="Date", format="%Y-%m-%d", formatType="time"),
alt.Tooltip(COLUMNS.NUM_PATIENTS, title="Number of patients"),
alt.Tooltip(COLUMNS.NUM_SITES, title="Number of sites")
]
# Calculate cumulative values
y_field = COLUMNS.NUM_PATIENTS
upper = "upper"
under = "under"
if is_cum:
filtered_chart = filtered_chart.transform_window(
cum_num_patients=f"sum({COLUMNS.NUM_PATIENTS})", # overwrite
sort=[{"field": COLUMNS.DATE}],
groupby=[COLUMNS.SITE_ID]
).transform_window(
cum_upper=f"sum(upper)",
sort=[{"field": COLUMNS.DATE}],
groupby=[COLUMNS.SITE_ID]
).transform_window(
cum_under=f"sum(under)",
sort=[{"field": COLUMNS.DATE}],
groupby=[COLUMNS.SITE_ID]
)
upper = "cum_upper"
under = "cum_under"
y_field = "cum_num_patients"
DAILY_COUNT_TOOLTIP += [alt.Tooltip("cum_num_patients:Q", title="Cumulative # of patients")]
# Render
line = filtered_chart.mark_line(size=3, opacity=0.7).encode(
x=alt.X(
f"{COLUMNS.DATE}:T",
scale=alt.Scale(padding=10),
axis=alt.Axis(tickCount=7, grid=True, labels=True, ticks=True, domain=True),
title=None
),
y=alt.Y(f"{y_field}:Q", axis=alt.Axis(tickCount=5), title="Number of patients", scale=alt.Scale(padding=10, nice=False)),
color=alt.Color(f"{COLUMNS.SITE_ID}:N", scale=color_scale, legend=alt.Legend(title=None)),
tooltip=DAILY_COUNT_TOOLTIP,
).transform_filter(
date_brush
)
circle = line.mark_circle(size=30, opacity=0.7).encode(
size=alt.condition(~nearest, alt.value(30), alt.value(60))
)
errorband = line.mark_errorband().encode(
x=alt.X(f"{COLUMNS.DATE}:T", axis=alt.Axis(tickCount=7), title=None),
y=alt.Y(f"{upper}:Q", title=""),
y2=f"{under}:Q",
color=alt.Color(f"{COLUMNS.SITE_ID}:N", scale=color_scale, legend=alt.Legend(title=None)),
tooltip=DAILY_COUNT_TOOLTIP
)
top_line = (circle + line + errorband + nearest_rule).resolve_scale(color="shared").properties(width=750, height=400).add_selection(y_zoom)
bottom_y_field = COLUMNS.NUM_HOSPITALS if is_num_hospital else COLUMNS.NUM_SITES
bottom_y_title = "# of hospitals" if is_num_hospital else "# of sites"
bottom_bar_bg = filtered_chart.mark_bar(size=5).encode(
x=alt.X(f"{COLUMNS.DATE}:T", axis=alt.Axis(tickCount=7), title=None, scale=alt.Scale(padding=10)),
y=alt.Y(f"{bottom_y_field}:Q", title=bottom_y_title, axis=alt.Axis(tickMinStep=1)),
color=alt.Color(f"{COLUMNS.SITE_ID}:N", scale=color_scale_bg, legend=None),
tooltip=DAILY_COUNT_TOOLTIP
).properties(height=60, width=750)
bottom_bar = bottom_bar_bg.encode(
color=alt.Color(f"{COLUMNS.SITE_ID}:N", scale=color_scale, legend=None), #legend=alt.Legend(title=None)
)#.transform_filter(date_brush)
bottom_bar = (bottom_bar + nearest_rule).resolve_scale(color="independent").add_selection(date_brush)
title = "Daily Counts"
# title = TITLE_BY_CATEGORY[category]
title = f"Cumulative {title}" if is_cum else f"{title}"
title = f"{title} by Site" if is_site_level else f"{title} by Country"
# title = title.capitalize()
# Apply Theme
result_vis = apply_theme(
alt.vconcat(top_line, bottom_bar).resolve_scale(x="independent", color="shared"),
legend_orient="right",
axis_title_font_size=13
)
result_vis = result_vis.properties(title={
"text": title,
"subtitle": get_visualization_subtitle(),
"subtitleColor": "gray",
"dx": 60
}).add_selection(
legend_selection
).add_selection(
nearest
).add_selection(
dailycount_selection
)
return result_vis
dailycount = dailycount_by_date(is_cum=False, is_only_combined=True)
# for_website(dailycount, "Daily Count", "Daily counts")
# save(dailycount, join(SAVE_DIR, "dailycount_by_date.png")) # Uncomment this to save *.png files
dailycount
```
## Daily counts by country
```
dailycount = dailycount_by_date(is_cum=False)
for_website(dailycount, "Daily Count", "Daily counts by country")
# save(dailycount, join(SAVE_DIR, "dailycount_by_date.png")) # Uncomment this to save *.png files
dailycount
```
## Cumulative daily counts by country
```
dailycount = dailycount_by_date(is_cum=True)
for_website(dailycount, "Daily Count", "Cumulative daily counts by country")
# save(dailycount, join(SAVE_DIR, "dailycount_by_date.png")) # Uncomment this to save *.png files
dailycount
```
## Values by the day of the week
```
import datetime
df_dc["week"] = df_dc["date"].apply(lambda x: datetime.datetime.strptime(x, '%Y-%m-%d').isocalendar()[1])
country_by_color = { ([ALL_COUNTRY] + COUNTRIES)[i]: ([ALL_COUNTRY_COLOR] + COUNTRY_COLOR)[i] for i in range(len([ALL_COUNTRY_COLOR] + COUNTRY_COLOR)) }
def dailycount_by_day_and_week(country, category):
# Filter
filtered_chart = alt.Chart(df_dc).transform_filter(
alt.datum[CATEGORY] == category
).transform_filter(
alt.datum[COLUMNS.SITE_ID] == country
)
# Rendering
result_vis = filtered_chart.mark_rect().encode(
y=alt.Y("day(date):O",title="Day of the week"),
x=alt.X('week:O', title="Week of the year"),
color=alt.Color('sum(num_patients):Q', title=None, scale=alt.Scale(scheme="lightorange"))
).properties(height=220, width=380, title={
"text": f"New {TITLE_BY_CATEGORY[category]} ({country})",
"subtitle": get_visualization_subtitle(),
"color": country_by_color[country],
"subtitleColor": "gray",
"dx": 60
})
return result_vis
is_save = False
for category in CATEGORIES:
v = alt.vconcat()
for country in [ALL_COUNTRY] + COUNTRIES:
result_vis = dailycount_by_day_and_week(country=country, category=category)
v &= result_vis
# Apply Theme
result_vis = apply_theme(
v,
legend_orient="right",
legend_stroke_color="white",
legend_padding=0
).resolve_scale(color="independent", x="shared")
# Display and save
result_vis.display()
if is_save:
save(result_vis, join(SAVE_DIR, f"dailycount_by_day_and_week_{category}.png"))
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
%aimport utils
import pandas as pd
import numpy as np
import altair as alt
from altair_saver import save
from os.path import join
from web import for_website
from constants import COLUMNS, DATA_AGGREGATE_TYPES
from utils import (
read_combined_daily_counts_df,
read_combined_by_country_daily_counts_df,
read_site_details_df,
get_visualization_subtitle,
get_siteid_anonymous_map,
get_anonymousid_color_maps,
apply_theme
)
# Path to save *.PNG files
SAVE_DIR = join("..", "output")
# Country Info
ALL_COUNTRY = "All countries"
ALL_COUNTRY_COLOR = "#444444"
COUNTRIES = ["France", "Germany", "Italy", "Singapore", "USA"]
COUNTRY_COLOR = ["#0072B2", "#E69F00", "#009E73", "#CC79A7", "#D55E00"]
CATEGORY = "category"
def preprocess_daily_df(df_dc):
# Wide to long
df_dc = pd.melt(df_dc, id_vars=[
COLUMNS.SITE_ID, COLUMNS.DATE,
COLUMNS.MASKED_UPPER_BOUND_NEW_POSITIVE_CASES,
COLUMNS.MASKED_UPPER_BOUND_PATIENTS_IN_ICU,
COLUMNS.MASKED_UPPER_BOUND_NEW_DEATHS,
COLUMNS.UNMASKED_SITES_NEW_POSITIVE_CASES,
COLUMNS.UNMASKED_SITES_PATIENTS_IN_ICU,
COLUMNS.UNMASKED_SITES_NEW_DEATHS,
COLUMNS.MASKED_SITES_NEW_POSITIVE_CASES,
COLUMNS.MASKED_SITES_PATIENTS_IN_ICU,
COLUMNS.MASKED_SITES_NEW_DEATHS
])
df_dc = df_dc.rename(columns={"variable": CATEGORY, "value": COLUMNS.NUM_PATIENTS})
# Leave only the 'upper' and 'under' values for the certain 'category' only
for c in [COLUMNS.NEW_POSITIVE_CASES, COLUMNS.PATIENTS_IN_ICU, COLUMNS.NEW_DEATHS]:
filter_c = df_dc[CATEGORY] == c
df_dc.loc[filter_c, "upper"] = df_dc.loc[filter_c, COLUMNS.NUM_PATIENTS] + df_dc.loc[filter_c, "masked_upper_bound_" + c]
df_dc.loc[filter_c, "under"] = df_dc.loc[filter_c, COLUMNS.NUM_PATIENTS]
df_dc.loc[filter_c, COLUMNS.NUM_PATIENTS] = df_dc.loc[filter_c, COLUMNS.NUM_PATIENTS] + df_dc.loc[filter_c, "masked_upper_bound_" + c] / 2.0
# Add num of sites
df_dc.loc[filter_c, COLUMNS.NUM_SITES] = df_dc["unmasked_sites_" + c] + df_dc["masked_sites_" + c]
# Drop unused columns
df_dc = df_dc.drop(columns=[
COLUMNS.MASKED_UPPER_BOUND_NEW_POSITIVE_CASES,
COLUMNS.MASKED_UPPER_BOUND_PATIENTS_IN_ICU,
COLUMNS.MASKED_UPPER_BOUND_NEW_DEATHS,
COLUMNS.UNMASKED_SITES_NEW_POSITIVE_CASES,
COLUMNS.UNMASKED_SITES_PATIENTS_IN_ICU,
COLUMNS.UNMASKED_SITES_NEW_DEATHS,
COLUMNS.MASKED_SITES_NEW_POSITIVE_CASES,
COLUMNS.MASKED_SITES_PATIENTS_IN_ICU,
COLUMNS.MASKED_SITES_NEW_DEATHS
])
# Make sure to drop date range out of our interest
df_dc = df_dc[df_dc[COLUMNS.DATE] >= "2020-01-29"]
df_dc = df_dc[df_dc[COLUMNS.DATE] <= "2020-03-31"]
# We are not using ICU
df_dc = df_dc[df_dc[CATEGORY] != COLUMNS.PATIENTS_IN_ICU]
# Use more readable names
df_dc.loc[df_dc[COLUMNS.SITE_ID] == "Combined", COLUMNS.SITE_ID] = ALL_COUNTRY
# Remove zero num_sites, which is missing data
df_dc = df_dc[df_dc[COLUMNS.NUM_SITES] != 0]
return df_dc
# Read files
df_dc = read_combined_by_country_daily_counts_df()
df_dc = preprocess_daily_df(df_dc)
df_dc_combined = read_combined_daily_counts_df()
df_dc_combined = preprocess_daily_df(df_dc_combined)
# Merge
df_dc = pd.concat([df_dc, df_dc_combined])
df_dc_combined
CATEGORIES = [COLUMNS.NEW_POSITIVE_CASES, COLUMNS.NEW_DEATHS]
TITLE_BY_CATEGORY = {
COLUMNS.NEW_POSITIVE_CASES: "possitive cases",
COLUMNS.NEW_DEATHS: "deaths",
COLUMNS.PATIENTS_IN_ICU: "ICU admissions"
}
def dailycount_by_date(df=df_dc, is_cum=True, is_only_combined=False, is_site_level=False, is_num_hospital=False):
# Selections
nearest = alt.selection(type="single", nearest=True, on="mouseover", encodings=["x"], empty='none', clear="mouseout")
dailycount_dropdown = alt.binding_select(options=CATEGORIES)
dailycount_selection = alt.selection_single(fields=[CATEGORY], bind=dailycount_dropdown, name="Value", init={CATEGORY: COLUMNS.NEW_POSITIVE_CASES})
legend_selection = alt.selection_multi(fields=[COLUMNS.SITE_ID], bind="legend")
date_brush = alt.selection(type="interval", encodings=['x'])
y_zoom = alt.selection(type="interval", bind='scales', encodings=['y'])
# Rule
nearest_rule = alt.Chart(df).mark_rule(color="red").encode(
x=f"{COLUMNS.DATE}:T",
size=alt.value(0.5)
).transform_filter(
nearest
)
color_scale = alt.Scale(domain=COUNTRIES, range=COUNTRY_COLOR)
color_scale_bg = alt.Scale(domain=COUNTRIES, range=["lightgray"])
if is_only_combined:
color_scale = alt.Scale(domain=[ALL_COUNTRY], range=[ALL_COUNTRY_COLOR])
color_scale_bg = alt.Scale(domain=[ALL_COUNTRY], range=["lightgray"])
if is_site_level:
color_scale = alt.Scale(domain=ANONYMOUS_SITES, range=ANONYMOUS_COLORS)
color_scale_bg = alt.Scale(domain=ANONYMOUS_SITES, range=["lightgray"])
# Filter
filtered_chart = alt.Chart(df).transform_filter(
dailycount_selection
).transform_filter(
legend_selection
)
if is_only_combined:
filtered_chart = filtered_chart.transform_filter(
alt.datum[COLUMNS.SITE_ID] == ALL_COUNTRY
)
else:
filtered_chart = filtered_chart.transform_filter(
alt.datum[COLUMNS.SITE_ID] != ALL_COUNTRY
)
DAILY_COUNT_TOOLTIP = [
alt.Tooltip(COLUMNS.SITE_ID, title="Country"),
alt.Tooltip(COLUMNS.DATE, title="Date", format="%Y-%m-%d", formatType="time"),
alt.Tooltip(COLUMNS.NUM_PATIENTS, title="Number of patients"),
alt.Tooltip(COLUMNS.NUM_SITES, title="Number of sites")
]
# Calculate cumulative values
y_field = COLUMNS.NUM_PATIENTS
upper = "upper"
under = "under"
if is_cum:
filtered_chart = filtered_chart.transform_window(
cum_num_patients=f"sum({COLUMNS.NUM_PATIENTS})", # overwrite
sort=[{"field": COLUMNS.DATE}],
groupby=[COLUMNS.SITE_ID]
).transform_window(
cum_upper=f"sum(upper)",
sort=[{"field": COLUMNS.DATE}],
groupby=[COLUMNS.SITE_ID]
).transform_window(
cum_under=f"sum(under)",
sort=[{"field": COLUMNS.DATE}],
groupby=[COLUMNS.SITE_ID]
)
upper = "cum_upper"
under = "cum_under"
y_field = "cum_num_patients"
DAILY_COUNT_TOOLTIP += [alt.Tooltip("cum_num_patients:Q", title="Cumulative # of patients")]
# Render
line = filtered_chart.mark_line(size=3, opacity=0.7).encode(
x=alt.X(
f"{COLUMNS.DATE}:T",
scale=alt.Scale(padding=10),
axis=alt.Axis(tickCount=7, grid=True, labels=True, ticks=True, domain=True),
title=None
),
y=alt.Y(f"{y_field}:Q", axis=alt.Axis(tickCount=5), title="Number of patients", scale=alt.Scale(padding=10, nice=False)),
color=alt.Color(f"{COLUMNS.SITE_ID}:N", scale=color_scale, legend=alt.Legend(title=None)),
tooltip=DAILY_COUNT_TOOLTIP,
).transform_filter(
date_brush
)
circle = line.mark_circle(size=30, opacity=0.7).encode(
size=alt.condition(~nearest, alt.value(30), alt.value(60))
)
errorband = line.mark_errorband().encode(
x=alt.X(f"{COLUMNS.DATE}:T", axis=alt.Axis(tickCount=7), title=None),
y=alt.Y(f"{upper}:Q", title=""),
y2=f"{under}:Q",
color=alt.Color(f"{COLUMNS.SITE_ID}:N", scale=color_scale, legend=alt.Legend(title=None)),
tooltip=DAILY_COUNT_TOOLTIP
)
top_line = (circle + line + errorband + nearest_rule).resolve_scale(color="shared").properties(width=750, height=400).add_selection(y_zoom)
bottom_y_field = COLUMNS.NUM_HOSPITALS if is_num_hospital else COLUMNS.NUM_SITES
bottom_y_title = "# of hospitals" if is_num_hospital else "# of sites"
bottom_bar_bg = filtered_chart.mark_bar(size=5).encode(
x=alt.X(f"{COLUMNS.DATE}:T", axis=alt.Axis(tickCount=7), title=None, scale=alt.Scale(padding=10)),
y=alt.Y(f"{bottom_y_field}:Q", title=bottom_y_title, axis=alt.Axis(tickMinStep=1)),
color=alt.Color(f"{COLUMNS.SITE_ID}:N", scale=color_scale_bg, legend=None),
tooltip=DAILY_COUNT_TOOLTIP
).properties(height=60, width=750)
bottom_bar = bottom_bar_bg.encode(
color=alt.Color(f"{COLUMNS.SITE_ID}:N", scale=color_scale, legend=None), #legend=alt.Legend(title=None)
)#.transform_filter(date_brush)
bottom_bar = (bottom_bar + nearest_rule).resolve_scale(color="independent").add_selection(date_brush)
title = "Daily Counts"
# title = TITLE_BY_CATEGORY[category]
title = f"Cumulative {title}" if is_cum else f"{title}"
title = f"{title} by Site" if is_site_level else f"{title} by Country"
# title = title.capitalize()
# Apply Theme
result_vis = apply_theme(
alt.vconcat(top_line, bottom_bar).resolve_scale(x="independent", color="shared"),
legend_orient="right",
axis_title_font_size=13
)
result_vis = result_vis.properties(title={
"text": title,
"subtitle": get_visualization_subtitle(),
"subtitleColor": "gray",
"dx": 60
}).add_selection(
legend_selection
).add_selection(
nearest
).add_selection(
dailycount_selection
)
return result_vis
dailycount = dailycount_by_date(is_cum=False, is_only_combined=True)
# for_website(dailycount, "Daily Count", "Daily counts")
# save(dailycount, join(SAVE_DIR, "dailycount_by_date.png")) # Uncomment this to save *.png files
dailycount
dailycount = dailycount_by_date(is_cum=False)
for_website(dailycount, "Daily Count", "Daily counts by country")
# save(dailycount, join(SAVE_DIR, "dailycount_by_date.png")) # Uncomment this to save *.png files
dailycount
dailycount = dailycount_by_date(is_cum=True)
for_website(dailycount, "Daily Count", "Cumulative daily counts by country")
# save(dailycount, join(SAVE_DIR, "dailycount_by_date.png")) # Uncomment this to save *.png files
dailycount
import datetime
df_dc["week"] = df_dc["date"].apply(lambda x: datetime.datetime.strptime(x, '%Y-%m-%d').isocalendar()[1])
country_by_color = { ([ALL_COUNTRY] + COUNTRIES)[i]: ([ALL_COUNTRY_COLOR] + COUNTRY_COLOR)[i] for i in range(len([ALL_COUNTRY_COLOR] + COUNTRY_COLOR)) }
def dailycount_by_day_and_week(country, category):
# Filter
filtered_chart = alt.Chart(df_dc).transform_filter(
alt.datum[CATEGORY] == category
).transform_filter(
alt.datum[COLUMNS.SITE_ID] == country
)
# Rendering
result_vis = filtered_chart.mark_rect().encode(
y=alt.Y("day(date):O",title="Day of the week"),
x=alt.X('week:O', title="Week of the year"),
color=alt.Color('sum(num_patients):Q', title=None, scale=alt.Scale(scheme="lightorange"))
).properties(height=220, width=380, title={
"text": f"New {TITLE_BY_CATEGORY[category]} ({country})",
"subtitle": get_visualization_subtitle(),
"color": country_by_color[country],
"subtitleColor": "gray",
"dx": 60
})
return result_vis
is_save = False
for category in CATEGORIES:
v = alt.vconcat()
for country in [ALL_COUNTRY] + COUNTRIES:
result_vis = dailycount_by_day_and_week(country=country, category=category)
v &= result_vis
# Apply Theme
result_vis = apply_theme(
v,
legend_orient="right",
legend_stroke_color="white",
legend_padding=0
).resolve_scale(color="independent", x="shared")
# Display and save
result_vis.display()
if is_save:
save(result_vis, join(SAVE_DIR, f"dailycount_by_day_and_week_{category}.png"))
| 0.373876 | 0.537163 |
```
# Live code
print('Hello world!')
```
# Markdown - Como formatar seu texto
Este será um texto onde posso explicar algo.
Abaixo você está vendo um código multilinhas.
```
if True:
print('Hello world!')
```
Também pode ser interessante usar uma `palavra` como código.
Você pode ter textos em *itálico* ou **negrito**.
* Linha 1
* Linha 2
* Linha aninhada
- Outra coisa
- Mais uma linha
* Retornando para lista normal
Também pode ter listas enumeradas:
1. Linha
2. Linha
1. Aninhamento
2. Aninhamento
3. Aninhamento
3. Linha
4. Linha
5. Linha
Abaixo uma linha horizontal
---
Bloco de citação:
> Um texto gigante no seu notebook.
>
> Mais uma linha do texto.
>
## Outro título
Aqui você verá o [link do google](https://www.google.com.br)
Abaixo uma tabela HTML:
| Aqui temos uma tabela | Com mais uma coluna |
|-----------------------|---------------------|
| Aqui temos uma linha | Outra coluna |
| Aqui temos uma linha | Outra coluna |
| Aqui temos uma linha | Outra coluna |
| Coluna | Outra |
Por fim, você pode usar HTML puro também, rsrs.
<h1>Isso é um título</h1>
<p>Aqui vemos um <p>. Você pode usar qualquer tag HTML normal.
# Abaixo você verá um vídeo do Youtube
Poderia também adicionar um texto explicativo aqui.
```
%%HTML
<h1>Vídeo do youtube</h1>
<iframe width="560" height="315" src="https://www.youtube.com/embed/bVMUGEEgEbg" frameborder="0"></iframe>
```
# Criando uma classe Python
```
class Cliente:
def __init__(self, nome, sobrenome):
self.nome = nome
self.sobrenome = sobrenome
def mostra_detalhes(self):
print(self.nome)
print(self.sobrenome)
cliente = Cliente('Luiz Otávio', 'Miranda')
cliente.mostra_detalhes()
import main # Importa o main da raiz do diretorio
```
# Comandos do sistema
```
!ping 127.0.0.1
```
# Gráficos
```
import matplotlib.path as mpath
import matplotlib.patches as mpatches
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
Path = mpath.Path
path_data = [
(Path.MOVETO, (1.58, -2.57)),
(Path.CURVE4, (0.35, -1.1)),
(Path.CURVE4, (-1.75, 2.0)),
(Path.CURVE4, (0.375, 2.0)),
(Path.LINETO, (0.85, 1.15)),
(Path.CURVE4, (2.2, 3.2)),
(Path.CURVE4, (3, 0.05)),
(Path.CURVE4, (2.0, -0.5)),
(Path.CLOSEPOLY, (1.58, -2.57)),
]
codes, verts = zip(*path_data)
path = mpath.Path(verts, codes)
patch = mpatches.PathPatch(path, facecolor='r', alpha=0.5)
ax.add_patch(patch)
# plot control points and connecting lines
x, y = zip(*path.vertices)
line, = ax.plot(x, y, 'go-')
ax.grid()
ax.axis('equal')
plt.show()
%%timeit
# Calcula o tempo para executar determinado comando
lista = [x * 2 for x in range(100000)]
```
# Exibindo Pandas Dataframe
```
import pandas as pd
import numpy as np
df = pd.DataFrame(np.random.randn(18,5))
df
```
|
github_jupyter
|
# Live code
print('Hello world!')
if True:
print('Hello world!')
%%HTML
<h1>Vídeo do youtube</h1>
<iframe width="560" height="315" src="https://www.youtube.com/embed/bVMUGEEgEbg" frameborder="0"></iframe>
class Cliente:
def __init__(self, nome, sobrenome):
self.nome = nome
self.sobrenome = sobrenome
def mostra_detalhes(self):
print(self.nome)
print(self.sobrenome)
cliente = Cliente('Luiz Otávio', 'Miranda')
cliente.mostra_detalhes()
import main # Importa o main da raiz do diretorio
!ping 127.0.0.1
import matplotlib.path as mpath
import matplotlib.patches as mpatches
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
Path = mpath.Path
path_data = [
(Path.MOVETO, (1.58, -2.57)),
(Path.CURVE4, (0.35, -1.1)),
(Path.CURVE4, (-1.75, 2.0)),
(Path.CURVE4, (0.375, 2.0)),
(Path.LINETO, (0.85, 1.15)),
(Path.CURVE4, (2.2, 3.2)),
(Path.CURVE4, (3, 0.05)),
(Path.CURVE4, (2.0, -0.5)),
(Path.CLOSEPOLY, (1.58, -2.57)),
]
codes, verts = zip(*path_data)
path = mpath.Path(verts, codes)
patch = mpatches.PathPatch(path, facecolor='r', alpha=0.5)
ax.add_patch(patch)
# plot control points and connecting lines
x, y = zip(*path.vertices)
line, = ax.plot(x, y, 'go-')
ax.grid()
ax.axis('equal')
plt.show()
%%timeit
# Calcula o tempo para executar determinado comando
lista = [x * 2 for x in range(100000)]
import pandas as pd
import numpy as np
df = pd.DataFrame(np.random.randn(18,5))
df
| 0.301671 | 0.863794 |
# Applying Chords to 2D and 3D Images
## Importing packages
```
import time
import porespy as ps
ps.visualization.set_mpl_style()
```
Import the usual packages from the Scipy ecosystem:
```
import scipy as sp
import scipy.ndimage as spim
import matplotlib.pyplot as plt
```
## Demonstration on 2D Image
Start by creating an image using the ``blobs`` function in ``generators``. The useful thing about this function is that images can be created with anisotropy. These are exactly the sort of images where chord length distributions are useful, since chords can be drawn in different directions, to probe the anisotropic pore sizes.
```
im = ps.generators.blobs(shape=[400, 400], blobiness=[2, 1])
```
The image can be visualized easily using matplotlib's ``imshow`` function:
```
# NBVAL_IGNORE_OUTPUT
plt.figure(figsize=[6, 6])
fig = plt.imshow(im)
```
Determining chord-length distributions requires first adding chords to the image, which is done using the ``apply_chords`` function. The following code applies chords to the image in the x-direction (along ``axis=0``), then applies them in the y-direction (``axis=1``). The two images are then plotted using ``matplotlib``.
```
# NBVAL_IGNORE_OUTPUT
crds_x = ps.filters.apply_chords(im=im, spacing=4, axis=0)
crds_y = ps.filters.apply_chords(im=im, spacing=4, axis=1)
fig, ax = plt.subplots(1, 2, figsize=[10, 5])
ax[0].imshow(crds_x)
ax[1].imshow(crds_y)
```
Note that none of the chords touch the edge of the image. These chords are trimmed by default since they are artificially shorter than they should be and would skew the results. This behavior is optional and these chords can be kept by setting ``trim_edges=False``.
It is sometimes useful to colorize the chords by their length. PoreSpy includes a function called ``region_size`` which counts the number of voxels in each connected region of an image, and replaces those voxels with the numerical value of the region size. This is illustrated below:
```
# NBVAL_IGNORE_OUTPUT
sz_x = ps.filters.region_size(crds_x)
sz_y = ps.filters.region_size(crds_y)
fig, ax = plt.subplots(1, 2, figsize=[10, 6])
ax[0].imshow(sz_x)
ax[1].imshow(sz_y)
```
Although the above images are useful for quick visualization, they are not quantitative. To get quantitative chord length distributions, pass the chord image(s) to the ``chord_length_distribution`` functions in the ``metrics`` submodule:
```
data_x = ps.metrics.chord_length_distribution(crds_x, bins=25)
data_y = ps.metrics.chord_length_distribution(crds_y, bins=25)
```
This function, like many of the functions in the ``metrics`` module, returns a named tuple containing various arrays. The advantage of the named tuple is that each array can be accessed by name as attributes, such as ``data_x.pdf``. To see all the available attributes (i.e. arrays) use the autocomplete function if your IDE, the following:
```
print(data_x._fields)
```
Now we can print the results of the chord-length distribution as bar graphs:
```
# NBVAL_IGNORE_OUTPUT
plt.figure(figsize=[6, 6])
bar = plt.bar(x=data_y.L, height=data_y.cdf, width=data_y.bin_widths, color='b', edgecolor='k', alpha=0.5)
bar = plt.bar(x=data_x.L, height=data_x.cdf, width=data_x.bin_widths, color='r', edgecolor='k', alpha=0.5)
```
The key point to see here is that the blue bars are for the y-direction, which was the elongated direction, and as expected they show a tendency toward longer chords.
## Application to 3D images
Chords can just as easily be applied to 3D images. Let's create an artificial image of fibers, aligned in the YZ plane, but oriented randomly in the X direction
```
# NBVAL_IGNORE_OUTPUT
im = ps.generators.cylinders(shape=[200, 400, 400], radius=8, ncylinders=200, )
plt.imshow(im[:, :, 100])
```
As above, we must apply chords to the image then pass the chord image to the ``chord_length_distribution`` function:
```
# NBVAL_IGNORE_OUTPUT
crds = ps.filters.apply_chords(im=im, axis=0)
plt.imshow(crds[:, :, 100])
```
|
github_jupyter
|
import time
import porespy as ps
ps.visualization.set_mpl_style()
import scipy as sp
import scipy.ndimage as spim
import matplotlib.pyplot as plt
im = ps.generators.blobs(shape=[400, 400], blobiness=[2, 1])
# NBVAL_IGNORE_OUTPUT
plt.figure(figsize=[6, 6])
fig = plt.imshow(im)
# NBVAL_IGNORE_OUTPUT
crds_x = ps.filters.apply_chords(im=im, spacing=4, axis=0)
crds_y = ps.filters.apply_chords(im=im, spacing=4, axis=1)
fig, ax = plt.subplots(1, 2, figsize=[10, 5])
ax[0].imshow(crds_x)
ax[1].imshow(crds_y)
# NBVAL_IGNORE_OUTPUT
sz_x = ps.filters.region_size(crds_x)
sz_y = ps.filters.region_size(crds_y)
fig, ax = plt.subplots(1, 2, figsize=[10, 6])
ax[0].imshow(sz_x)
ax[1].imshow(sz_y)
data_x = ps.metrics.chord_length_distribution(crds_x, bins=25)
data_y = ps.metrics.chord_length_distribution(crds_y, bins=25)
print(data_x._fields)
# NBVAL_IGNORE_OUTPUT
plt.figure(figsize=[6, 6])
bar = plt.bar(x=data_y.L, height=data_y.cdf, width=data_y.bin_widths, color='b', edgecolor='k', alpha=0.5)
bar = plt.bar(x=data_x.L, height=data_x.cdf, width=data_x.bin_widths, color='r', edgecolor='k', alpha=0.5)
# NBVAL_IGNORE_OUTPUT
im = ps.generators.cylinders(shape=[200, 400, 400], radius=8, ncylinders=200, )
plt.imshow(im[:, :, 100])
# NBVAL_IGNORE_OUTPUT
crds = ps.filters.apply_chords(im=im, axis=0)
plt.imshow(crds[:, :, 100])
| 0.486088 | 0.989182 |
```
# Initialize Otter
import otter
grader = otter.Notebook("lab02.ipynb")
```
# Lab 2: Table operations
Welcome to Lab 2! This week, we'll learn how to import a module and practice table operations!
Recommended Reading:
* [Introduction to tables](https://www.inferentialthinking.com/chapters/03/4/Introduction_to_Tables)
First, set up the tests and imports by running the cell below.
```
# Just run this cell
import numpy as np
from datascience import *
# These lines load the tests.
# When you log-in please hit return (not shift + return) after typing in your email
```
# 1. Review: The building blocks of Python code
The two building blocks of Python code are *expressions* and *statements*. An **expression** is a piece of code that
* is self-contained, meaning it would make sense to write it on a line by itself, and
* usually evaluates to a value.
Here are two expressions that both evaluate to 3:
3
5 - 2
One important type of expression is the **call expression**. A call expression begins with the name of a function and is followed by the argument(s) of that function in parentheses. The function returns some value, based on its arguments. Some important mathematical functions are listed below.
| Function | Description |
|----------|---------------------------------------------------------------|
| `abs` | Returns the absolute value of its argument |
| `max` | Returns the maximum of all its arguments |
| `min` | Returns the minimum of all its arguments |
| `pow` | Raises its first argument to the power of its second argument |
| `round` | Rounds its argument to the nearest integer |
Here are two call expressions that both evaluate to 3:
abs(2 - 5)
max(round(2.8), min(pow(2, 10), -1 * pow(2, 10)))
The expression `2 - 5` and the two call expressions given above are examples of **compound expressions**, meaning that they are actually combinations of several smaller expressions. `2 - 5` combines the expressions `2` and `5` by subtraction. In this case, `2` and `5` are called **subexpressions** because they're expressions that are part of a larger expression.
A **statement** is a whole line of code. Some statements are just expressions. The expressions listed above are examples.
Other statements *make something happen* rather than *having a value*. For example, an **assignment statement** assigns a value to a name.
A good way to think about this is that we're **evaluating the right-hand side** of the equals sign and **assigning it to the left-hand side**. Here are some assignment statements:
height = 1.3
the_number_five = abs(-5)
absolute_height_difference = abs(height - 1.688)
An important idea in programming is that large, interesting things can be built by combining many simple, uninteresting things. The key to understanding a complicated piece of code is breaking it down into its simple components.
For example, a lot is going on in the last statement above, but it's really just a combination of a few things. This picture describes what's going on.
<img src="statement.png">
**Question 1.1.** In the next cell, assign the name `new_year` to the larger number among the following two numbers:
1. the **absolute value** of $2^{5}-2^{11}-2^1 + 1$, and
2. $5 \times 13 \times 31 + 5$.
Try to use just one statement (one line of code). Be sure to check your work by executing the test cell afterward.
<!--
BEGIN QUESTION
name: q11
-->
```
new_year = ...
new_year
grader.check("q11")
```
We've asked you to use one line of code in the question above because it only involves mathematical operations. However, more complicated programming questions will more require more steps. It isn’t always a good idea to jam these steps into a single line because it can make the code harder to read and harder to debug.
Good programming practice involves splitting up your code into smaller steps and using appropriate names. You'll have plenty of practice in the rest of this course!
# 2. Importing code
[source](https://www.reddit.com/r/ProgrammerHumor/comments/cgtk7s/theres_no_need_to_reinvent_the_wheel_oc/)
Most programming involves work that is very similar to work that has been done before. Since writing code is time-consuming, it's good to rely on others' published code when you can. Rather than copy-pasting, Python allows us to **import modules**. A module is a file with Python code that has defined variables and functions. By importing a module, we are able to use its code in our own notebook.
Python includes many useful modules that are just an `import` away. We'll look at the `math` module as a first example. The `math` module is extremely useful in computing mathematical expressions in Python.
Suppose we want to very accurately compute the area of a circle with a radius of 5 meters. For that, we need the constant $\pi$, which is roughly 3.14. Conveniently, the `math` module has `pi` defined for us:
```
import math
radius = 5
area_of_circle = radius**2 * math.pi
area_of_circle
```
In the code above, the line `import math` imports the math module. This statement creates a module and then assigns the name `math` to that module. We are now able to access any variables or functions defined within `math` by typing the name of the module followed by a dot, then followed by the name of the variable or function we want.
<module name>.<name>
**Question 2.1.** The module `math` also provides the name `e` for the base of the natural logarithm, which is roughly 2.71. Compute $e^{\pi}-\pi$, giving it the name `near_twenty`.
*Remember: You can access `pi` from the `math` module as well!*
<!--
BEGIN QUESTION
name: q21
-->
```
near_twenty = ...
near_twenty
grader.check("q21")
```

[Source](http://imgs.xkcd.com/comics/e_to_the_pi_minus_pi.png)
[Explaination](https://www.explainxkcd.com/wiki/index.php/217:_e_to_the_pi_Minus_pi)
## 2.1. Accessing functions
In the question above, you accessed variables within the `math` module.
**Modules** also define **functions**. For example, `math` provides the name `sin` for the sine function. Having imported `math` already, we can write `math.sin(3)` to compute the sine of 3. (Note that this sine function considers its argument to be in [radians](https://en.wikipedia.org/wiki/Radian), not degrees. 180 degrees are equivalent to $\pi$ radians.)
**Question 2.1.1.** A $\frac{\pi}{4}$-radian (45-degree) angle forms a right triangle with equal base and height, pictured below. If the hypotenuse (the radius of the circle in the picture) is 1, then the height is $\sin(\frac{\pi}{4})$. Compute that value using `sin` and `pi` from the `math` module. Give the result the name `sine_of_pi_over_four`.
<img src="http://mathworld.wolfram.com/images/eps-gif/TrigonometryAnglesPi4_1000.gif">
[Source](http://mathworld.wolfram.com/images/eps-gif/TrigonometryAnglesPi4_1000.gif)
<!--
BEGIN QUESTION
name: q211
-->
```
sine_of_pi_over_four = ...
sine_of_pi_over_four
grader.check("q211")
```
For your reference, below are some more examples of functions from the `math` module.
Notice how different functions take in different numbers of arguments. Often, the [documentation](https://docs.python.org/3/library/math.html) of the module will provide information on how many arguments are required for each function.
*Hint: If you press `shift+tab` while next to the function call, the documentation for that function will appear*
```
# Calculating logarithms (the logarithm of 8 in base 2).
# The result is 3 because 2 to the power of 3 is 8.
math.log(8, 2)
# Calculating square roots.
math.sqrt(5)
```
There are various ways to import and access code from outside sources. The method we used above — `import <module_name>` — imports the entire module and requires that we use `<module_name>.<name>` to access its code.
We can also import a specific constant or function instead of the entire module. Notice that you don't have to use the module name beforehand to reference that particular value. However, you do have to be careful about reassigning the names of the constants or functions to other values!
```
# Importing just cos and pi from math.
# We don't have to use `math.` in front of cos or pi
from math import cos, pi
print(cos(pi))
# We do have to use it in front of other functions from math, though
math.log(pi)
```
Or we can import every function and value from the entire module.
```
# Lastly, we can import everything from math using the *
# Once again, we don't have to use 'math.' beforehand
from math import *
log(pi)
```
Don't worry too much about which type of import to use. It's often a coding style choice left up to each programmer. In this course, you'll always import the necessary modules when you run the setup cell (like the first code cell in this lab).
Let's move on to practicing some of the table operations you've learned in lecture!
# 3. Table operations
The table `farmers_markets.csv` contains data on farmers' markets in the United States (data collected [by the USDA](https://apps.ams.usda.gov/FarmersMarketsExport/ExcelExport.aspx)). Each row represents one such market.
Run the next cell to load the `farmers_markets` table.
```
# Just run this cell
farmers_markets = Table.read_table('farmers_markets.csv')
```
Let's examine our table to see what data it contains.
**Question 3.1.** Use the method `show` to display the first 5 rows of `farmers_markets`.
*Note:* The terms "method" and "function" are technically not the same thing, but for the purposes of this course, we will use them interchangeably.
**Hint:** `tbl.show(3)` will show the first 3 rows of `tbl`. Additionally, make sure not to call `.show()` without an argument, as this will crash your kernel!
```
...
```
Notice that some of the values in this table are missing, as denoted by "nan." This means either that the value is not available (e.g. if we don’t know the market’s street address) or not applicable (e.g. if the market doesn’t have a street address). You'll also notice that the table has a large number of columns in it!
### `num_columns`
The table property `num_columns` returns the number of columns in a table. (A "property" is just a method that doesn't need to be called by adding parentheses.)
Example call: `<tbl>.num_columns`
**Question 3.2.** Use `num_columns` to find the number of columns in our farmers' markets dataset.
Assign the number of columns to `num_farmers_markets_columns`.
<!--
BEGIN QUESTION
name: q32
-->
```
num_farmers_markets_columns = ...
print("The table has", num_farmers_markets_columns, "columns in it!")
grader.check("q32")
```
### `num_rows`
Similarly, the property `num_rows` tells you how many rows are in a table.
```
# Just run this cell
num_farmers_markets_rows = farmers_markets.num_rows
print("The table has", num_farmers_markets_rows, "rows in it!")
```
### `select`
Most of the columns are about particular products -- whether the market sells tofu, pet food, etc. If we're not interested in that information, it just makes the table difficult to read. This comes up more than you might think, because people who collect and publish data may not know ahead of time what people will want to do with it.
In such situations, we can use the table method `select` to choose only the columns that we want in a particular table. It takes any number of arguments. Each should be the name of a column in the table. It returns a new table with only those columns in it. The columns are in the order *in which they were listed as arguments*.
For example, the value of `farmers_markets.select("MarketName", "State")` is a table with only the name and the state of each farmers' market in `farmers_markets`.
**Question 3.3.** Use `select` to create a table with only the name, city, state, latitude (`y`), and longitude (`x`) of each market. Call that new table `farmers_markets_locations`.
*Hint:* Make sure to be exact when using column names with `select`; double-check capitalization!
<!--
BEGIN QUESTION
name: q33
-->
```
farmers_markets_locations = ...
farmers_markets_locations
grader.check("q33")
```
### `drop`
`drop` serves the same purpose as `select`, but it takes away the columns that you provide rather than the ones that you don't provide. Like `select`, `drop` returns a new table.
**Question 3.4.** Suppose you just didn't want the `FMID` and `updateTime` columns in `farmers_markets`. Create a table that's a copy of `farmers_markets` but doesn't include those columns. Call that table `farmers_markets_without_fmid`.
<!--
BEGIN QUESTION
name: q34
-->
```
farmers_markets_without_fmid = ...
farmers_markets_without_fmid
grader.check("q34")
```
Now, suppose we want to answer some questions about farmers' markets in the US. For example, which market(s) have the largest longitude (given by the `x` column)?
To answer this, we'll sort `farmers_markets_locations` by longitude.
```
farmers_markets_locations.sort('x')
```
Oops, that didn't answer our question because we sorted from smallest to largest longitude. To look at the largest longitudes, we'll have to sort in reverse order.
```
farmers_markets_locations.sort('x', descending=True)
```
(The `descending=True` bit is called an *optional argument*. It has a default value of `False`, so when you explicitly tell the function `descending=True`, then the function will sort in descending order.)
### `sort`
Some details about sort:
1. The first argument to `sort` is the name of a column to sort by.
2. If the column has text in it, `sort` will sort alphabetically; if the column has numbers, it will sort numerically.
3. The value of `farmers_markets_locations.sort("x")` is a *copy* of `farmers_markets_locations`; the `farmers_markets_locations` table doesn't get modified. For example, if we called `farmers_markets_locations.sort("x")`, then running `farmers_markets_locations` by itself would still return the unsorted table.
4. Rows always stick together when a table is sorted. It wouldn't make sense to sort just one column and leave the other columns alone. For example, in this case, if we sorted just the `x` column, the farmers' markets would all end up with the wrong longitudes.
**Question 3.5.** Create a version of `farmers_markets_locations` that's sorted by **latitude (`y`)**, with the largest latitudes first. Call it `farmers_markets_locations_by_latitude`.
<!--
BEGIN QUESTION
name: q35
-->
```
farmers_markets_locations_by_latitude = ...
farmers_markets_locations_by_latitude
grader.check("q35")
```
Now let's say we want a table of all farmers' markets in California. Sorting won't help us much here because California is closer to the middle of the dataset.
Instead, we use the table method `where`.
```
california_farmers_markets = farmers_markets_locations.where('State', are.equal_to('California'))
california_farmers_markets
```
Ignore the syntax for the moment. Instead, try to read that line like this:
> Assign the name **`california_farmers_markets`** to a table whose rows are the rows in the **`farmers_markets_locations`** table **`where`** the **`'State'`**s **`are` `equal` `to` `California`**.
### `where`
Now let's dive into the details a bit more. `where` takes 2 arguments:
1. The name of a column. `where` finds rows where that column's values meet some criterion.
2. A predicate that describes the criterion that the column needs to meet.
The predicate in the example above called the function `are.equal_to` with the value we wanted, 'California'. We'll see other predicates soon.
`where` returns a table that's a copy of the original table, but **with only the rows that meet the given predicate**.
**Question 3.6.** Use `california_farmers_markets` to create a table called `berkeley_markets` containing farmers' markets in Berkeley, California.
<!--
BEGIN QUESTION
name: q36
-->
```
berkeley_markets = ...
berkeley_markets
grader.check("q36")
```
Recognize any of them?
So far we've only been using `where` with the predicate that requires finding the values in a column to be *exactly* equal to a certain value. However, there are many other predicates. Here are a few:
|Predicate|Example|Result|
|-|-|-|
|`are.equal_to`|`are.equal_to(50)`|Find rows with values equal to 50|
|`are.not_equal_to`|`are.not_equal_to(50)`|Find rows with values not equal to 50|
|`are.above`|`are.above(50)`|Find rows with values above (and not equal to) 50|
|`are.above_or_equal_to`|`are.above_or_equal_to(50)`|Find rows with values above 50 or equal to 50|
|`are.below`|`are.below(50)`|Find rows with values below 50|
|`are.between`|`are.between(2, 10)`|Find rows with values above or equal to 2 and below 10|
## 4. Analyzing a dataset
Now that you're familiar with table operations, let’s answer an interesting question about a dataset!
Run the cell below to load the `imdb` table. It contains information about the 250 highest-rated movies on IMDb.
```
# Just run this cell
imdb = Table.read_table('imdb.csv')
imdb
```
Often, we want to perform multiple operations - sorting, filtering, or others - in order to turn a table we have into something more useful. You can do these operations one by one, e.g.
```
first_step = original_tbl.where(“col1”, are.equal_to(12))
second_step = first_step.sort(‘col2’, descending=True)
```
However, since the value of the expression `original_tbl.where(“col1”, are.equal_to(12))` is itself a table, you can just call a table method on it:
```
original_tbl.where(“col1”, are.equal_to(12)).sort(‘col2’, descending=True)
```
You should organize your work in the way that makes the most sense to you, using informative names for any intermediate tables you create.
**Question 4.1.** Create a table of movies released between 2010 and 2016 (inclusive) with ratings above 8. The table should only contain the columns `Title` and `Rating`, **in that order**.
Assign the table to the name `above_eight`.
*Hint:* Think about the steps you need to take, and try to put them in an order that make sense. Feel free to create intermediate tables for each step, but please make sure you assign your final table the name `above_eight`!
<!--
BEGIN QUESTION
name: q41
-->
```
above_eight = ...
above_eight
grader.check("q41")
```
**Question 4.2.** Use `num_rows` (and arithmetic) to find the *proportion* of movies in the dataset that were released 1900-1999, and the *proportion* of movies in the dataset that were released in the year 2000 or later.
Assign `proportion_in_20th_century` to the proportion of movies in the dataset that were released 1900-1999, and `proportion_in_21st_century` to the proportion of movies in the dataset that were released in the year 2000 or later.
*Hint:* The *proportion* of movies released in the 1900's is the *number* of movies released in the 1900's, divided by the *total number* of movies.
<!--
BEGIN QUESTION
name: q42
-->
```
num_movies_in_dataset = ...
num_in_20th_century = ...
num_in_21st_century = ...
proportion_in_20th_century = ...
proportion_in_21st_century = ...
print("Proportion in 20th century:", proportion_in_20th_century)
print("Proportion in 21st century:", proportion_in_21st_century)
grader.check("q42")
```
## 5. Summary
For your reference, here's a table of all the functions and methods we saw in this lab. We'll learn more methods to add to this table in the coming week!
|Name|Example|Purpose|
|-|-|-|
|`sort`|`tbl.sort("N")`|Create a copy of a table sorted by the values in a column|
|`where`|`tbl.where("N", are.above(2))`|Create a copy of a table with only the rows that match some *predicate*|
|`num_rows`|`tbl.num_rows`|Compute the number of rows in a table|
|`num_columns`|`tbl.num_columns`|Compute the number of columns in a table|
|`select`|`tbl.select("N")`|Create a copy of a table with only some of the columns|
|`drop`|`tbl.drop("2*N")`|Create a copy of a table without some of the columns|
<br/>
Alright! You're finished with lab 2! Be sure to...
- run all the tests (the next cell has a shortcut for that),
- **Save and Checkpoint** from the `File` menu,
- **run the last cell to submit your work**,
- and **ask one of the staff members to check you off**.
---
To double-check your work, the cell below will rerun all of the autograder tests.
```
grader.check_all()
```
## Submission
Make sure you have run all cells in your notebook in order before running the cell below, so that all images/graphs appear in the output. The cell below will generate a zip file for you to submit. **Please save before exporting!**
```
# Save your notebook first, then run this cell to export your submission.
grader.export()
```
|
github_jupyter
|
# Initialize Otter
import otter
grader = otter.Notebook("lab02.ipynb")
# Just run this cell
import numpy as np
from datascience import *
# These lines load the tests.
# When you log-in please hit return (not shift + return) after typing in your email
new_year = ...
new_year
grader.check("q11")
import math
radius = 5
area_of_circle = radius**2 * math.pi
area_of_circle
near_twenty = ...
near_twenty
grader.check("q21")
sine_of_pi_over_four = ...
sine_of_pi_over_four
grader.check("q211")
# Calculating logarithms (the logarithm of 8 in base 2).
# The result is 3 because 2 to the power of 3 is 8.
math.log(8, 2)
# Calculating square roots.
math.sqrt(5)
# Importing just cos and pi from math.
# We don't have to use `math.` in front of cos or pi
from math import cos, pi
print(cos(pi))
# We do have to use it in front of other functions from math, though
math.log(pi)
# Lastly, we can import everything from math using the *
# Once again, we don't have to use 'math.' beforehand
from math import *
log(pi)
# Just run this cell
farmers_markets = Table.read_table('farmers_markets.csv')
...
num_farmers_markets_columns = ...
print("The table has", num_farmers_markets_columns, "columns in it!")
grader.check("q32")
# Just run this cell
num_farmers_markets_rows = farmers_markets.num_rows
print("The table has", num_farmers_markets_rows, "rows in it!")
farmers_markets_locations = ...
farmers_markets_locations
grader.check("q33")
farmers_markets_without_fmid = ...
farmers_markets_without_fmid
grader.check("q34")
farmers_markets_locations.sort('x')
farmers_markets_locations.sort('x', descending=True)
farmers_markets_locations_by_latitude = ...
farmers_markets_locations_by_latitude
grader.check("q35")
california_farmers_markets = farmers_markets_locations.where('State', are.equal_to('California'))
california_farmers_markets
berkeley_markets = ...
berkeley_markets
grader.check("q36")
# Just run this cell
imdb = Table.read_table('imdb.csv')
imdb
first_step = original_tbl.where(“col1”, are.equal_to(12))
second_step = first_step.sort(‘col2’, descending=True)
original_tbl.where(“col1”, are.equal_to(12)).sort(‘col2’, descending=True)
above_eight = ...
above_eight
grader.check("q41")
num_movies_in_dataset = ...
num_in_20th_century = ...
num_in_21st_century = ...
proportion_in_20th_century = ...
proportion_in_21st_century = ...
print("Proportion in 20th century:", proportion_in_20th_century)
print("Proportion in 21st century:", proportion_in_21st_century)
grader.check("q42")
grader.check_all()
# Save your notebook first, then run this cell to export your submission.
grader.export()
| 0.563138 | 0.987723 |
# How to Create a Session
To use GraphScope, we need to establish a session first.
A session encapsulates the control and the state of the GraphScope engines. It serves as an entrance in the python client to GraphScope. A session allows users to deploy and connect GraphScope on a k8s cluster.
In this tutorial, we will demostrate:
* How to establish a default session;
* Alternative ways to launch a session;
* How to mount external volume to the cluster.
## Basic Usage
First of all, you should **import graphscope**.
```
import graphscope
```
For better understanding of the launching process, we recommend to enable the **show_log** option in the package scope.
```
graphscope.set_option(show_log=True)
```
A default session can be easily launched, even without any parameters.
```
s1 = graphscope.session()
```
Behind the scenes, the session tries to launch a coordinator, which is the entry for the back-end engines. The coordinator manages a cluster of k8s pods (2 pods by default), and the interactive/analytical/learning engines ran on them. For each pod in the cluster, there is a vineyard instance at service for distributed data in memory.
Run the cell and take a look at the log, it prints the whole process of the session launching.
The log **GraphScope coordinator service connected** means the session launches successfully, and the current Python client has connected to the session.
You can also check a session's status by this.
```
s1
```
Run this cell, you may find a `status` field with value `active`. Together with the status, it also prints other metainfo of this session, i.e., such as the number of workers(pods), the coordinator endpoint for connection, and so on.
A session manages the resources in the cluster, thus it is important to release these resources when they are no longer required. To de-allocate the resources, use the method **close** on the session when all the graph tasks are finished.
```
s1.close()
```
## Advanced Usage
GraphScope session provides several keyword arguments to config the cluster.
For example, you may use `k8s_gs_image` to specify the image of the GraphScope,
or use `num_workers` to specify the number of pods. You may use `help(graphscope.session)` to check
all available arguments.
```
s2 = graphscope.session(num_workers=1, k8s_engine_cpu=1, k8s_engine_mem='4Gi', timeout_seconds=1200)
s2.close()
```
Parametes are allowed to pass as a json string or `Dict`.
```
config = {'num_workers': 1, 'timeout_seconds': 100}
s3 = graphscope.session(config=config)
s3.close()
```
## Mounting Volumes
To save or load data, you may want to mount a file volume to the allocated cluster.
For example, we prepared some sample graph datasets for in the host location (`/testingdata`). You can mount it to path `/home/jovyan/datasets`. Then in the pods, you are able to access these testing data.
Note that, path `/testingdata` in server is a **Copy** of `/home/jovyan/datasets` in your HOME dir, and any modification locally will not affect the directory mounted on the server.
```
k8s_volumes = {
"data": {
"type": "hostPath",
"field": {
"path": '/testingdata',
"type": "Directory"
},
"mounts": {
"mountPath": "/home/jovyan/datasets",
"readOnly": True
}
}
}
s4 = graphscope.session(k8s_volumes=k8s_volumes)
s4.close()
```
|
github_jupyter
|
import graphscope
graphscope.set_option(show_log=True)
s1 = graphscope.session()
s1
s1.close()
s2 = graphscope.session(num_workers=1, k8s_engine_cpu=1, k8s_engine_mem='4Gi', timeout_seconds=1200)
s2.close()
config = {'num_workers': 1, 'timeout_seconds': 100}
s3 = graphscope.session(config=config)
s3.close()
k8s_volumes = {
"data": {
"type": "hostPath",
"field": {
"path": '/testingdata',
"type": "Directory"
},
"mounts": {
"mountPath": "/home/jovyan/datasets",
"readOnly": True
}
}
}
s4 = graphscope.session(k8s_volumes=k8s_volumes)
s4.close()
| 0.207777 | 0.985072 |
```
import aux_tools
import os
%matplotlib inline
def save_fig(fig_id, tight_layout=True, fig_extension="pdf", resolution=300):
path = os.path.join(os.getcwd(), fig_id + "." + fig_extension)
print(f"Saving figure {fig_id} to {path}")
if tight_layout:
plt.tight_layout()
plt.savefig(path, format=fig_extension, dpi=resolution)
# Load serialized dataframe
import pickle
import pandas as pd
# [86400, 21600, 7200, 3600, 900]
file = "df_900_0.3_v3.raw"
df = pickle.load(open(file, 'rb')) # type: pd.DataFrame
#encode window
df['relative_day_position'] = df['relative_day_position'].apply(lambda x: int((x*24)/285) )
#remove zeros
df = df[df['packet_count:mean'] != 0]
# Pipeline
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import RobustScaler
num_attribs = list(df.drop("entity", axis=1)) # numerical attributes
# numerical and categorical pipelines
num_pipeline = Pipeline([
#('imputer', Imputer(strategy="median")),
#('attribs_adder', CombinedAttributesAdder()),
('selector', aux_tools.DataFrameSelector(num_attribs)),
('std_scaler', StandardScaler()),
#('robust_scaler', RobustScaler()),
])
sliding_window_pipeline = Pipeline([
('selector', aux_tools.DataFrameSelector('relative_day_position')),
('revel', aux_tools.DataFrameRevel()),
('cat_encoder', aux_tools.CategoricalEncoder(encoding="onehot-dense")),
])
from sklearn.pipeline import FeatureUnion
full_pipeline = FeatureUnion(transformer_list=[
("num_pipeline", num_pipeline),
("sliding_window_pipeline", sliding_window_pipeline),
])
from sklearn.utils import shuffle
df = shuffle(df)
fulldata = full_pipeline.fit_transform(df)
cat_encoder = aux_tools.CategoricalEncoder(encoding="ordinal")
fulldata_labels = df['entity'].values.reshape(-1, 1)
fulldata_labels_ordinal = cat_encoder.fit_transform(fulldata_labels)
from sklearn import svm
from sklearn.model_selection import cross_val_score
from sklearn.neural_network import MLPClassifier
nn_class = MLPClassifier(solver='lbfgs', alpha=1e-5,
hidden_layer_sizes=(15,), random_state=42)
scores = cross_val_score(nn_class,
fulldata,
fulldata_labels_ordinal.ravel()
#scoring='accuracy'
)
print("Accuracy: %0.2f (+/- %0.4f)" % (scores.mean(), scores.std() * 2))
print(__doc__)
import itertools
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm, datasets
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import cross_val_predict
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
X = fulldata
y = fulldata_labels_ordinal.ravel()
class_names = [aux_tools.labnames[x] for x in list(cat_encoder.categories_[0])]
# Split the data into a training set and a test set
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
# Run classifier, using a model that is too regularized (C too low) to see
# the impact on the results
classifier = MLPClassifier(solver='lbfgs', alpha=1e-5,
hidden_layer_sizes=(15,), random_state=42)
classifier.fit(X, y)
y_pred = cross_val_predict(classifier, X, y, cv=3)
# Compute confusion matrix
cnf_matrix = confusion_matrix(y, y_pred)
np.set_printoptions(precision=2)
# Plot non-normalized confusion matrix
#plt.figure()
#plot_confusion_matrix(cnf_matrix, classes=class_names,
# title='Confusion matrix, without normalization')
# Plot normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names, normalize=True,
title='Normalized confusion matrix')
save_fig('conf_mat')
plt.figure()
row_sums = cnf_matrix.sum(axis=1, keepdims=True)
norm_conf_mx = cnf_matrix / row_sums
np.fill_diagonal(norm_conf_mx, 0)
plot_confusion_matrix(norm_conf_mx, classes=class_names, normalize=True,
title='Normalized confusion errors')
save_fig('conf_mat_errors')
plt.show()
from sklearn.decomposition import PCA
pca = PCA()
pca.fit(fulldata)
import scikitplot as skplt
skplt.decomposition.plot_pca_component_variance(pca,target_explained_variance=0.95)
save_fig('pca_variance_095')
plt.show()
from sklearn.model_selection import GridSearchCV
from sklearn.decomposition import PCA
from sklearn.pipeline import Pipeline
from sklearn.neural_network import MLPClassifier
pipe = Pipeline([
#('reduce_dim', PCA()),
#('reduce_dim', None,),
('classify', None)
])
param_grid= [
{
#'reduce_dim': [PCA()],
#'reduce_dim__n_components': [2, 3, 5, 7, 37, 41, 43, 47, 53, 59, 73, 79, 83,],
#'reduce_dim__n_components': [73, 83,],
'classify': [MLPClassifier(random_state=42)],
'classify__solver': ['lbfgs','adam'],
'classify__alpha': [1e-1, 1e-2, 1e-3, 1e-4, 1e-5, 1e-6, 1e-7]
},
]
param_grid = [
{'solver': ['lbfgs','adam'],'alpha': [1e-1, 1e-2, 1e-3, 1e-4, 1e-5, 1e-6, 1e-7]}
]
nn_clas = MLPClassifier(random_state=42)
grid_search_nn = GridSearchCV(nn_clas, param_grid, cv=5,
#scoring='neg_mean_squared_error',
scoring='accuracy',
return_train_score=True,
n_jobs=6,
verbose=True)
grid_search_nn.fit(fulldata,
fulldata_labels_ordinal.ravel())
cvres = grid_search_nn.cv_results_
cvres.keys()
for mean_score, std, params in zip(cvres['mean_test_score'], cvres['std_test_score'], cvres["params"]):
print(f"{mean_score} +- {std*2} : {params}")
np.std(cvres['mean_test_score']*100) * 2
# confusion matrix
print(__doc__)
import itertools
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm, datasets
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import cross_val_predict
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues, colorbar=True):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
if colorbar:
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
X = fulldata
y = fulldata_labels_ordinal.ravel()
class_names = [aux_tools.labnames[x] for x in list(cat_encoder.categories_[0])]
# Split the data into a training set and a test set
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
# Run classifier, using a model that is too regularized (C too low) to see
# the impact on the results
classifier = svm.LinearSVC(loss='hinge',max_iter=5000, C=1)
classifier.fit(X, y)
y_pred = cross_val_predict(classifier, X, y, cv=3)
# Compute confusion matrix
cnf_matrix = confusion_matrix(y, y_pred)
np.set_printoptions(precision=2)
# Plot non-normalized confusion matrix
#plt.figure()
#plot_confusion_matrix(cnf_matrix, classes=class_names,
# title='Confusion matrix, without normalization')
# Plot normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names, normalize=True,
title='Normalized confusion matrix', colorbar=False)
save_fig('conf_mat_NN')
plt.figure()
row_sums = cnf_matrix.sum(axis=1, keepdims=True)
norm_conf_mx = cnf_matrix / row_sums
np.fill_diagonal(norm_conf_mx, 0)
plot_confusion_matrix(norm_conf_mx, classes=class_names, normalize=True,
title='Normalized confusion errors')
save_fig('conf_mat_errors_NN')
plt.show()
# remove everything except the worse case
df_worsecase = df[df.entity != 'np_x58pc.raw']
df_worsecase = df_worsecase[df_worsecase.entity != 'np_tohiba.raw']
df_worsecase = df_worsecase[df_worsecase.entity != 'np_freebsd.raw']
df_worsecase = df_worsecase[df_worsecase.entity != 'np_nuc.raw']
fulldata_worsecase = full_pipeline.fit_transform(df_worsecase)
cat_encoder = aux_tools.CategoricalEncoder(encoding="ordinal")
fulldata_labels = df_worsecase['entity'].values.reshape(-1, 1)
fulldata_worsecase_labels_ordinal = cat_encoder.fit_transform(fulldata_labels)
df_bestcase = df[df.entity != 'np_x58pc.raw']
df_bestcase = df_bestcase[df_bestcase.entity != 'np_windows10x86.raw']
#df_bestcase = df_bestcase[df_bestcase.entity != 'np_tohiba.raw']
df_bestcase = df_bestcase[df_bestcase.entity != 'np_freebsd.raw']
df_bestcase = df_bestcase[df_bestcase.entity != 'np_nuc.raw']
fulldata_bestcase = full_pipeline.fit_transform(df_bestcase)
cat_encoder = aux_tools.CategoricalEncoder(encoding="ordinal")
fulldata_labels = df_bestcase['entity'].values.reshape(-1, 1)
fulldata_bestcase_labels_ordinal = cat_encoder.fit_transform(fulldata_labels)
[aux_tools.labnames[x] for x in list(cat_encoder.categories_[0])]
print(__doc__)
# Author: Issam H. Laradji
# License: BSD 3 clause
import numpy as np
from matplotlib import pyplot as plt
from matplotlib.colors import ListedColormap
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.datasets import make_moons, make_circles, make_classification
from sklearn.neural_network import MLPClassifier
h = .02 # step size in the mesh
alphas = np.logspace(-5, 3, 5)
#alphas = 10.0 ** -np.arange(1, 7)
names = []
for i in alphas:
names.append('alpha ' + str(i))
classifiers = []
for i in alphas:
classifiers.append(MLPClassifier(alpha=i,
random_state=42,
max_iter=600))
X, y = make_classification(n_features=2, n_redundant=0, n_informative=2,
random_state=0, n_clusters_per_class=1)
rng = np.random.RandomState(2)
X += 2 * rng.uniform(size=X.shape)
linearly_separable = (X, y)
SIZE = 1000
datasets = [(fulldata_worsecase[:SIZE], fulldata_worsecase_labels_ordinal.ravel()[:SIZE]),
(fulldata_bestcase[:SIZE], fulldata_bestcase_labels_ordinal.ravel()[:SIZE])]
figure = plt.figure(figsize=(17, 9))
i = 1
# iterate over datasets
for X, y in datasets:
# preprocess dataset, split into training and test part
pca = PCA(n_components=2)
pca.fit(X)
X = pca.transform(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=.4)
x_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5
y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
# just plot the dataset first
cm = plt.cm.RdBu
cm_bright = ListedColormap(['#FF0000', '#0000FF'])
ax = plt.subplot(len(datasets), len(classifiers) + 1, i)
# Plot the training points
ax.scatter(X_train[:, 0], X_train[:, 1], c=y_train, cmap=cm_bright)
# and testing points
ax.scatter(X_test[:, 0], X_test[:, 1], c=y_test, cmap=cm_bright, alpha=0.6)
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
ax.set_xticks(())
ax.set_yticks(())
i += 1
# iterate over classifiers
for name, clf in zip(names, classifiers):
ax = plt.subplot(len(datasets), len(classifiers) + 1, i)
clf.fit(X_train, y_train)
score = clf.score(X_test, y_test)
# Plot the decision boundary. For that, we will assign a color to each
# point in the mesh [x_min, x_max]x[y_min, y_max].
if hasattr(clf, "decision_function"):
Z = clf.decision_function(np.c_[xx.ravel(), yy.ravel()])
else:
Z = clf.predict_proba(np.c_[xx.ravel(), yy.ravel()])[:, 1]
# Put the result into a color plot
Z = Z.reshape(xx.shape)
ax.contourf(xx, yy, Z, cmap=cm, alpha=.8)
# Plot also the training points
ax.scatter(X_train[:, 0], X_train[:, 1], c=y_train, cmap=cm_bright,
edgecolors='black', s=25)
# and testing points
ax.scatter(X_test[:, 0], X_test[:, 1], c=y_test, cmap=cm_bright,
alpha=0.6, edgecolors='black', s=25)
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
ax.set_xticks(())
ax.set_yticks(())
ax.set_title(name)
ax.text(xx.max() - .3, yy.min() + .3, ('%.2f' % score).lstrip('0'),
size=15, horizontalalignment='right')
i += 1
figure.subplots_adjust(left=.02, right=.98)
save_fig('nn_alphas')
plt.show()
```
|
github_jupyter
|
import aux_tools
import os
%matplotlib inline
def save_fig(fig_id, tight_layout=True, fig_extension="pdf", resolution=300):
path = os.path.join(os.getcwd(), fig_id + "." + fig_extension)
print(f"Saving figure {fig_id} to {path}")
if tight_layout:
plt.tight_layout()
plt.savefig(path, format=fig_extension, dpi=resolution)
# Load serialized dataframe
import pickle
import pandas as pd
# [86400, 21600, 7200, 3600, 900]
file = "df_900_0.3_v3.raw"
df = pickle.load(open(file, 'rb')) # type: pd.DataFrame
#encode window
df['relative_day_position'] = df['relative_day_position'].apply(lambda x: int((x*24)/285) )
#remove zeros
df = df[df['packet_count:mean'] != 0]
# Pipeline
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import RobustScaler
num_attribs = list(df.drop("entity", axis=1)) # numerical attributes
# numerical and categorical pipelines
num_pipeline = Pipeline([
#('imputer', Imputer(strategy="median")),
#('attribs_adder', CombinedAttributesAdder()),
('selector', aux_tools.DataFrameSelector(num_attribs)),
('std_scaler', StandardScaler()),
#('robust_scaler', RobustScaler()),
])
sliding_window_pipeline = Pipeline([
('selector', aux_tools.DataFrameSelector('relative_day_position')),
('revel', aux_tools.DataFrameRevel()),
('cat_encoder', aux_tools.CategoricalEncoder(encoding="onehot-dense")),
])
from sklearn.pipeline import FeatureUnion
full_pipeline = FeatureUnion(transformer_list=[
("num_pipeline", num_pipeline),
("sliding_window_pipeline", sliding_window_pipeline),
])
from sklearn.utils import shuffle
df = shuffle(df)
fulldata = full_pipeline.fit_transform(df)
cat_encoder = aux_tools.CategoricalEncoder(encoding="ordinal")
fulldata_labels = df['entity'].values.reshape(-1, 1)
fulldata_labels_ordinal = cat_encoder.fit_transform(fulldata_labels)
from sklearn import svm
from sklearn.model_selection import cross_val_score
from sklearn.neural_network import MLPClassifier
nn_class = MLPClassifier(solver='lbfgs', alpha=1e-5,
hidden_layer_sizes=(15,), random_state=42)
scores = cross_val_score(nn_class,
fulldata,
fulldata_labels_ordinal.ravel()
#scoring='accuracy'
)
print("Accuracy: %0.2f (+/- %0.4f)" % (scores.mean(), scores.std() * 2))
print(__doc__)
import itertools
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm, datasets
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import cross_val_predict
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
X = fulldata
y = fulldata_labels_ordinal.ravel()
class_names = [aux_tools.labnames[x] for x in list(cat_encoder.categories_[0])]
# Split the data into a training set and a test set
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
# Run classifier, using a model that is too regularized (C too low) to see
# the impact on the results
classifier = MLPClassifier(solver='lbfgs', alpha=1e-5,
hidden_layer_sizes=(15,), random_state=42)
classifier.fit(X, y)
y_pred = cross_val_predict(classifier, X, y, cv=3)
# Compute confusion matrix
cnf_matrix = confusion_matrix(y, y_pred)
np.set_printoptions(precision=2)
# Plot non-normalized confusion matrix
#plt.figure()
#plot_confusion_matrix(cnf_matrix, classes=class_names,
# title='Confusion matrix, without normalization')
# Plot normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names, normalize=True,
title='Normalized confusion matrix')
save_fig('conf_mat')
plt.figure()
row_sums = cnf_matrix.sum(axis=1, keepdims=True)
norm_conf_mx = cnf_matrix / row_sums
np.fill_diagonal(norm_conf_mx, 0)
plot_confusion_matrix(norm_conf_mx, classes=class_names, normalize=True,
title='Normalized confusion errors')
save_fig('conf_mat_errors')
plt.show()
from sklearn.decomposition import PCA
pca = PCA()
pca.fit(fulldata)
import scikitplot as skplt
skplt.decomposition.plot_pca_component_variance(pca,target_explained_variance=0.95)
save_fig('pca_variance_095')
plt.show()
from sklearn.model_selection import GridSearchCV
from sklearn.decomposition import PCA
from sklearn.pipeline import Pipeline
from sklearn.neural_network import MLPClassifier
pipe = Pipeline([
#('reduce_dim', PCA()),
#('reduce_dim', None,),
('classify', None)
])
param_grid= [
{
#'reduce_dim': [PCA()],
#'reduce_dim__n_components': [2, 3, 5, 7, 37, 41, 43, 47, 53, 59, 73, 79, 83,],
#'reduce_dim__n_components': [73, 83,],
'classify': [MLPClassifier(random_state=42)],
'classify__solver': ['lbfgs','adam'],
'classify__alpha': [1e-1, 1e-2, 1e-3, 1e-4, 1e-5, 1e-6, 1e-7]
},
]
param_grid = [
{'solver': ['lbfgs','adam'],'alpha': [1e-1, 1e-2, 1e-3, 1e-4, 1e-5, 1e-6, 1e-7]}
]
nn_clas = MLPClassifier(random_state=42)
grid_search_nn = GridSearchCV(nn_clas, param_grid, cv=5,
#scoring='neg_mean_squared_error',
scoring='accuracy',
return_train_score=True,
n_jobs=6,
verbose=True)
grid_search_nn.fit(fulldata,
fulldata_labels_ordinal.ravel())
cvres = grid_search_nn.cv_results_
cvres.keys()
for mean_score, std, params in zip(cvres['mean_test_score'], cvres['std_test_score'], cvres["params"]):
print(f"{mean_score} +- {std*2} : {params}")
np.std(cvres['mean_test_score']*100) * 2
# confusion matrix
print(__doc__)
import itertools
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm, datasets
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import cross_val_predict
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues, colorbar=True):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
if colorbar:
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
X = fulldata
y = fulldata_labels_ordinal.ravel()
class_names = [aux_tools.labnames[x] for x in list(cat_encoder.categories_[0])]
# Split the data into a training set and a test set
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
# Run classifier, using a model that is too regularized (C too low) to see
# the impact on the results
classifier = svm.LinearSVC(loss='hinge',max_iter=5000, C=1)
classifier.fit(X, y)
y_pred = cross_val_predict(classifier, X, y, cv=3)
# Compute confusion matrix
cnf_matrix = confusion_matrix(y, y_pred)
np.set_printoptions(precision=2)
# Plot non-normalized confusion matrix
#plt.figure()
#plot_confusion_matrix(cnf_matrix, classes=class_names,
# title='Confusion matrix, without normalization')
# Plot normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names, normalize=True,
title='Normalized confusion matrix', colorbar=False)
save_fig('conf_mat_NN')
plt.figure()
row_sums = cnf_matrix.sum(axis=1, keepdims=True)
norm_conf_mx = cnf_matrix / row_sums
np.fill_diagonal(norm_conf_mx, 0)
plot_confusion_matrix(norm_conf_mx, classes=class_names, normalize=True,
title='Normalized confusion errors')
save_fig('conf_mat_errors_NN')
plt.show()
# remove everything except the worse case
df_worsecase = df[df.entity != 'np_x58pc.raw']
df_worsecase = df_worsecase[df_worsecase.entity != 'np_tohiba.raw']
df_worsecase = df_worsecase[df_worsecase.entity != 'np_freebsd.raw']
df_worsecase = df_worsecase[df_worsecase.entity != 'np_nuc.raw']
fulldata_worsecase = full_pipeline.fit_transform(df_worsecase)
cat_encoder = aux_tools.CategoricalEncoder(encoding="ordinal")
fulldata_labels = df_worsecase['entity'].values.reshape(-1, 1)
fulldata_worsecase_labels_ordinal = cat_encoder.fit_transform(fulldata_labels)
df_bestcase = df[df.entity != 'np_x58pc.raw']
df_bestcase = df_bestcase[df_bestcase.entity != 'np_windows10x86.raw']
#df_bestcase = df_bestcase[df_bestcase.entity != 'np_tohiba.raw']
df_bestcase = df_bestcase[df_bestcase.entity != 'np_freebsd.raw']
df_bestcase = df_bestcase[df_bestcase.entity != 'np_nuc.raw']
fulldata_bestcase = full_pipeline.fit_transform(df_bestcase)
cat_encoder = aux_tools.CategoricalEncoder(encoding="ordinal")
fulldata_labels = df_bestcase['entity'].values.reshape(-1, 1)
fulldata_bestcase_labels_ordinal = cat_encoder.fit_transform(fulldata_labels)
[aux_tools.labnames[x] for x in list(cat_encoder.categories_[0])]
print(__doc__)
# Author: Issam H. Laradji
# License: BSD 3 clause
import numpy as np
from matplotlib import pyplot as plt
from matplotlib.colors import ListedColormap
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.datasets import make_moons, make_circles, make_classification
from sklearn.neural_network import MLPClassifier
h = .02 # step size in the mesh
alphas = np.logspace(-5, 3, 5)
#alphas = 10.0 ** -np.arange(1, 7)
names = []
for i in alphas:
names.append('alpha ' + str(i))
classifiers = []
for i in alphas:
classifiers.append(MLPClassifier(alpha=i,
random_state=42,
max_iter=600))
X, y = make_classification(n_features=2, n_redundant=0, n_informative=2,
random_state=0, n_clusters_per_class=1)
rng = np.random.RandomState(2)
X += 2 * rng.uniform(size=X.shape)
linearly_separable = (X, y)
SIZE = 1000
datasets = [(fulldata_worsecase[:SIZE], fulldata_worsecase_labels_ordinal.ravel()[:SIZE]),
(fulldata_bestcase[:SIZE], fulldata_bestcase_labels_ordinal.ravel()[:SIZE])]
figure = plt.figure(figsize=(17, 9))
i = 1
# iterate over datasets
for X, y in datasets:
# preprocess dataset, split into training and test part
pca = PCA(n_components=2)
pca.fit(X)
X = pca.transform(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=.4)
x_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5
y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
# just plot the dataset first
cm = plt.cm.RdBu
cm_bright = ListedColormap(['#FF0000', '#0000FF'])
ax = plt.subplot(len(datasets), len(classifiers) + 1, i)
# Plot the training points
ax.scatter(X_train[:, 0], X_train[:, 1], c=y_train, cmap=cm_bright)
# and testing points
ax.scatter(X_test[:, 0], X_test[:, 1], c=y_test, cmap=cm_bright, alpha=0.6)
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
ax.set_xticks(())
ax.set_yticks(())
i += 1
# iterate over classifiers
for name, clf in zip(names, classifiers):
ax = plt.subplot(len(datasets), len(classifiers) + 1, i)
clf.fit(X_train, y_train)
score = clf.score(X_test, y_test)
# Plot the decision boundary. For that, we will assign a color to each
# point in the mesh [x_min, x_max]x[y_min, y_max].
if hasattr(clf, "decision_function"):
Z = clf.decision_function(np.c_[xx.ravel(), yy.ravel()])
else:
Z = clf.predict_proba(np.c_[xx.ravel(), yy.ravel()])[:, 1]
# Put the result into a color plot
Z = Z.reshape(xx.shape)
ax.contourf(xx, yy, Z, cmap=cm, alpha=.8)
# Plot also the training points
ax.scatter(X_train[:, 0], X_train[:, 1], c=y_train, cmap=cm_bright,
edgecolors='black', s=25)
# and testing points
ax.scatter(X_test[:, 0], X_test[:, 1], c=y_test, cmap=cm_bright,
alpha=0.6, edgecolors='black', s=25)
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
ax.set_xticks(())
ax.set_yticks(())
ax.set_title(name)
ax.text(xx.max() - .3, yy.min() + .3, ('%.2f' % score).lstrip('0'),
size=15, horizontalalignment='right')
i += 1
figure.subplots_adjust(left=.02, right=.98)
save_fig('nn_alphas')
plt.show()
| 0.727007 | 0.368377 |
### 1) First, load the dataset from the weatherinszeged
```
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import statsmodels.api as sm
df = pd.read_csv('weatherinszeged.csv')
```
### 2) Like in the previous checkpoint, build a linear regression model where your target variable is the difference between the apparenttemperature and the temperature. As explanatory variables, use humidity and windspeed. Now, estimate your model using OLS. What are the R-squared and adjusted R-squared values? Do you think they are satisfactory? Why?
R-squared and adjusted R-squared values are both 0.288. Generally these numbers would be considered bad, as in the model is a poor fit. The p-value for the F-statistic however, is close to 0 so the model contributes to explaining part of the target.
```
df['tempdiff'] = df['apparenttemperature'] - df['temperature']
Y = df['tempdiff']
X = df[['humidity', 'windspeed']]
X = sm.add_constant(X)
results = sm.OLS(Y, X).fit()
print(results.summary())
```
### 3) Next, include the interaction of humidity and windspeed to the model above and estimate the model using OLS. Now, what is the R-squared of this model? Does this model improve upon the previous one?
Both R-squared and adjusted R-squared values are 0.341, a higher number than the previous model (0.288), and thus is an improvement.
```
df['humidwind'] = df['humidity'] * df['windspeed']
Y2 = df['tempdiff']
X2 = df[['humidity', 'windspeed', 'humidwind']]
X2 = sm.add_constant(X2)
results2 = sm.OLS(Y2, X2).fit()
print(results2.summary())
```
### 4) Add visibility as an additional explanatory variable to the first model and estimate it. Did R-squared increase? What about adjusted R-squared? Compare the differences put on the table by the interaction term and the visibility in terms of the improvement in the adjusted R-squared. Which one is more useful?
R-squared and adjusted R-squared values are 0.304 and 0.303 respectively, an improvement over the first model, but not an improvement over the second model. The second model increased the R-squared values of the first model by 0.054, but this third model only increased the first model by 0.016. The second model is therefore more useful.
```
Y3 = df['tempdiff']
X3 = df[['humidity', 'windspeed', 'visibility']]
X3 = sm.add_constant(X3)
results3 = sm.OLS(Y3, X3).fit()
print(results3.summary())
```
### 5) Choose the best one from the three models above with respect to their AIC and BIC scores. Validate your choice by discussing your justification with your mentor.
| Model | AIC | BIC |
|------|------|
| 1 | 3.409e+05 | 3.409e+05 |
| 2 | 3.334e+05| 3.334e+05|
| 3 | 3.388e+05 | 3.388e+05 |
The lower the AIC and BIC, the better. In this case, model 2 has the lowest numbers for AIC and BIC, so this would be the best model to choose from. This is in line with the R-squared and adjusted R-squared values.
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import statsmodels.api as sm
df = pd.read_csv('weatherinszeged.csv')
df['tempdiff'] = df['apparenttemperature'] - df['temperature']
Y = df['tempdiff']
X = df[['humidity', 'windspeed']]
X = sm.add_constant(X)
results = sm.OLS(Y, X).fit()
print(results.summary())
df['humidwind'] = df['humidity'] * df['windspeed']
Y2 = df['tempdiff']
X2 = df[['humidity', 'windspeed', 'humidwind']]
X2 = sm.add_constant(X2)
results2 = sm.OLS(Y2, X2).fit()
print(results2.summary())
Y3 = df['tempdiff']
X3 = df[['humidity', 'windspeed', 'visibility']]
X3 = sm.add_constant(X3)
results3 = sm.OLS(Y3, X3).fit()
print(results3.summary())
| 0.180251 | 0.971645 |
# МАДМО
<a href="https://mipt.ru/science/labs/laboratoriya-neyronnykh-sistem-i-glubokogo-obucheniya/"><img align="right" src="https://avatars1.githubusercontent.com/u/29918795?v=4&s=200" alt="DeepHackLab" style="position:relative;top:-40px;right:10px;height:100px;" /></a>
### Физтех-Школа Прикладной математики и информатики МФТИ
### Лаборатория нейронных сетей и глубокого обучения (DeepHackLab)
Домашнее задание необходимо загрузить в общий репозиторий с именной папкой
## Домашнее задание 1
### Основы Python и пакет NumPy
---
```
import numpy as np
import random
import scipy.stats as sps
```
### Задача 1
В первой задаче вам предлагается перемножить две квадратные матрицы двумя способами -- без использования пакета ***numpy*** и с ним.
```
# Для генерации матриц используем фукнцию random -- она используется для генерации случайных объектов
# функция sample создает случайную выборку. В качестве аргумента ей передается кортеж (i,j), здесь i -- число строк,
# j -- число столбцов.
a = np.random.sample((1000,1000))
b = np.random.sample((1000,1000))
# выведите размерность (ранг) каждой матрицы с помощью функции ndim.
# Используйте функцию shape, что она вывела?
# Ответ: Функция shape вывела размеры матрицы
# ========
print(np.linalg.matrix_rank(a))
print(np.linalg.matrix_rank(b))
print(np.shape(a))
print(np.shape(b))
# ========
print(a)
print(b)
def mult(a, b):
n_arows, n_acols = a.shape
n_brows, n_bcols = b.shape
if n_acols != n_brows:
print ("Error: Matrix can't be multiplied!")
else:
Z = np.zeros((n_arows, n_bcols))
for i in range(n_arows):
for j in range(n_acols):
for k in range(n_bcols):
Z[i][k] += a[i][j] * b[j][k]
pass
def np_mult(a, b):
# здесь напишите перемножение матриц с
# использованием NumPy и выведите результат
Z = np.dot(a,b)
pass
%%time
# засечем время работы функции без NumPy
mult(a,b)
%%time
# засечем время работы функции с NumPy
np_mult(a,b)
```
### Задача 2
Напишите функцию, которая по данной последовательности $\{A_i\}_{i=1}^n$ строит последовательность $S_n$, где $S_k = \frac{A_1 + ... + A_k}{k}$.
Аналогично -- с помощью библиотеки **NumPy** и без нее. Сравните скорость, объясните результат.
```
# функция, решающая задачу с помощью NumPy
def sec_av(A):
return np.cumsum(A)/list(range(1,len(A)+1))
pass
# функция без NumPy
def stupid_sec_av(A):
S = [0 for i in range(len(A))]
S[0] = A[0]
for i in range(len(A)-1):
S[i+1] = A[i+1] + S[i]
numb = list(range(1,len(A)+1))
for i in range(len(A)):
S[i] = S[i] / numb[i]
return S
# зададим некоторую последовательность и проверим ее на ваших функциях.
# Первая функция должна работать ~ в 50 раз быстрее
A = sps.uniform.rvs(size=10 ** 7)
%time S1 = sec_av(A)
%time S2 = stupid_sec_av(A)
#проверим корректность:
np.abs(S1 - S2).sum()
```
### Задача 3
Пусть задан некоторый массив $X$. Надо построить новый массив, где все элементы с нечетными индексами требуется заменить на число $a$ (если оно не указано, то на 1). Все элементы с четными индексами исходного массива нужно возвести в куб и записать в обратном порядке относительно позиций этих элементов. Массив $X$ при этом должен остаться без изменений. В конце требуется слить массив X с преобразованным X и вывести в обратном порядке.
```
# функция, решающая задачу с помощью NumPy
def transformation(X, a=1):
X[1::2] = a
X[::2] **= 3
X[::2] = X[::2][::-1]
return X
# функция, решающая задачу без NumPy
def stupid_transformation(X):
temp_odd = []
temp_even = []
temp_even_inv = []
Z = []
temp_odd = int(round(len(X)/2)) * [1]
for i in range(0,len(X),2):
temp_even = temp_even + [round(X[i]**3,8)]
for i in range(len(temp_even),0,-1):
temp_even_inv = temp_even_inv + [temp_even[i-1]]
for i in range(min(len(temp_even_inv), len(temp_odd))):
Z = Z + [temp_even_inv[i]] + [temp_odd[i]]
if len(temp_even_inv) > len(temp_odd):
Z = Z + [temp_even_inv[-1]]
if len(temp_even_inv) < len(temp_odd):
Z = Z + [temp_odd[-1]]
return Z
X = sps.uniform.rvs(size=10 ** 1)
# здесь код эффективнее примерно в 20 раз.
# если Вы вдруг соберетесь печатать массив без np -- лучше сначала посмотрите на его размер
%time S2 = stupid_transformation(X)
%time S1 = transformation(X)
# проверим корректность:
np.abs(S1 - S2).sum()
```
Почему методы ***numpy*** оказываются эффективнее?
```
# Методы numpy оказываются эффективнее, потому что многие функции написаны на C/Cyton, что делает их очень быстрыми.
```
## Дополнительные задачи
Дополнительные задачи подразумевают, что Вы самостоятельно разберётесь в некоторых функциях ***numpy***, чтобы их сделать.
Эти задачи не являются обязательными, но могут повлиять на Ваш рейтинг в лучшую сторону (точные правила учёта доп. задач будут оглашены позже).
### Задача 4*
Дана функция двух переменных: $f(x, y) = sin(x)cos(y)$ (это просто такой красивый 3D-график), а также дана функция для отрисовки $f(x, y)$ (`draw_f()`), которая принимает на вход двумерную сетку, на которой будет вычисляться функция.
Вам нужно разобраться в том, как строить такие сетки (подсказка - это одна конкретная функция ***numpy***), и подать такую сетку на вход функции отрисовки.
```
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
%matplotlib inline
def f(x, y):
'''Функция двух переменных'''
return np.sin(x) * np.cos(y)
def draw_f(grid_x, grid_y):
'''Функция отрисовки функции f(x, y)'''
fig = plt.figure(figsize=(10, 8))
ax = Axes3D(fig)
ax.plot_surface(grid_x, grid_y, f(grid_x, grid_y), cmap='inferno')
plt.show()
i = np.arange(-1, 1, 0.01)
grid_x, grid_y = np.meshgrid(i, i)
draw_f(grid_x, grid_y)
```
### Задача 5*
Вам дана картинка. При загрузке её размерность равна 3: **(w, h, num_channels)**, где **w** - ширина картинки в пикселях, **h** - высота картинки в пикселях, **num_channels** - количество каналов *(R, G, B, alpha)*.
Вам нужно "развернуть" картинку в одномерный массив размера w \* h \* num_channels, написав **одну строку кода**.
```
from matplotlib import pyplot as plt
%matplotlib inline
from PIL import Image
path_to_image = 'boombob.jpg'
pict = Image.open("boombob.jpg")
image_array = plt.imread(path_to_image)
plt.imshow(image_array);
flat_image_array = np.array(pict.getdata()).reshape(pict.size[0], pict.size[1], 3).flatten()
len(flat_image_array)
```
|
github_jupyter
|
import numpy as np
import random
import scipy.stats as sps
# Для генерации матриц используем фукнцию random -- она используется для генерации случайных объектов
# функция sample создает случайную выборку. В качестве аргумента ей передается кортеж (i,j), здесь i -- число строк,
# j -- число столбцов.
a = np.random.sample((1000,1000))
b = np.random.sample((1000,1000))
# выведите размерность (ранг) каждой матрицы с помощью функции ndim.
# Используйте функцию shape, что она вывела?
# Ответ: Функция shape вывела размеры матрицы
# ========
print(np.linalg.matrix_rank(a))
print(np.linalg.matrix_rank(b))
print(np.shape(a))
print(np.shape(b))
# ========
print(a)
print(b)
def mult(a, b):
n_arows, n_acols = a.shape
n_brows, n_bcols = b.shape
if n_acols != n_brows:
print ("Error: Matrix can't be multiplied!")
else:
Z = np.zeros((n_arows, n_bcols))
for i in range(n_arows):
for j in range(n_acols):
for k in range(n_bcols):
Z[i][k] += a[i][j] * b[j][k]
pass
def np_mult(a, b):
# здесь напишите перемножение матриц с
# использованием NumPy и выведите результат
Z = np.dot(a,b)
pass
%%time
# засечем время работы функции без NumPy
mult(a,b)
%%time
# засечем время работы функции с NumPy
np_mult(a,b)
# функция, решающая задачу с помощью NumPy
def sec_av(A):
return np.cumsum(A)/list(range(1,len(A)+1))
pass
# функция без NumPy
def stupid_sec_av(A):
S = [0 for i in range(len(A))]
S[0] = A[0]
for i in range(len(A)-1):
S[i+1] = A[i+1] + S[i]
numb = list(range(1,len(A)+1))
for i in range(len(A)):
S[i] = S[i] / numb[i]
return S
# зададим некоторую последовательность и проверим ее на ваших функциях.
# Первая функция должна работать ~ в 50 раз быстрее
A = sps.uniform.rvs(size=10 ** 7)
%time S1 = sec_av(A)
%time S2 = stupid_sec_av(A)
#проверим корректность:
np.abs(S1 - S2).sum()
# функция, решающая задачу с помощью NumPy
def transformation(X, a=1):
X[1::2] = a
X[::2] **= 3
X[::2] = X[::2][::-1]
return X
# функция, решающая задачу без NumPy
def stupid_transformation(X):
temp_odd = []
temp_even = []
temp_even_inv = []
Z = []
temp_odd = int(round(len(X)/2)) * [1]
for i in range(0,len(X),2):
temp_even = temp_even + [round(X[i]**3,8)]
for i in range(len(temp_even),0,-1):
temp_even_inv = temp_even_inv + [temp_even[i-1]]
for i in range(min(len(temp_even_inv), len(temp_odd))):
Z = Z + [temp_even_inv[i]] + [temp_odd[i]]
if len(temp_even_inv) > len(temp_odd):
Z = Z + [temp_even_inv[-1]]
if len(temp_even_inv) < len(temp_odd):
Z = Z + [temp_odd[-1]]
return Z
X = sps.uniform.rvs(size=10 ** 1)
# здесь код эффективнее примерно в 20 раз.
# если Вы вдруг соберетесь печатать массив без np -- лучше сначала посмотрите на его размер
%time S2 = stupid_transformation(X)
%time S1 = transformation(X)
# проверим корректность:
np.abs(S1 - S2).sum()
# Методы numpy оказываются эффективнее, потому что многие функции написаны на C/Cyton, что делает их очень быстрыми.
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
%matplotlib inline
def f(x, y):
'''Функция двух переменных'''
return np.sin(x) * np.cos(y)
def draw_f(grid_x, grid_y):
'''Функция отрисовки функции f(x, y)'''
fig = plt.figure(figsize=(10, 8))
ax = Axes3D(fig)
ax.plot_surface(grid_x, grid_y, f(grid_x, grid_y), cmap='inferno')
plt.show()
i = np.arange(-1, 1, 0.01)
grid_x, grid_y = np.meshgrid(i, i)
draw_f(grid_x, grid_y)
from matplotlib import pyplot as plt
%matplotlib inline
from PIL import Image
path_to_image = 'boombob.jpg'
pict = Image.open("boombob.jpg")
image_array = plt.imread(path_to_image)
plt.imshow(image_array);
flat_image_array = np.array(pict.getdata()).reshape(pict.size[0], pict.size[1], 3).flatten()
len(flat_image_array)
| 0.168378 | 0.918991 |
# Benchmarking Thinc layers with a custom `benchmark` layer
This notebook shows how to write a `benchmark` layer that can wrap any layer(s) in your network and that **logs the execution times** of the initialization, forward pass and backward pass. The benchmark layer can also be mapped to an operator like `@` to make it easy to add debugging to your network.
```
!pip install "thinc>=8.0.0a0"
```
To log the results, we first set up a custom logger using Python's `logging` module. You could also just print the stats instead, but using `logging` is cleaner, since it lets other users modify the logger's behavior more easily, and separates the logs from other output and write it to a file (e.g. if you're benchmarking several layers during training). The following logging config will output the date and time, the name of the logger and the logged results.
```
import logging
logger = logging.getLogger("thinc:benchmark")
if not logger.hasHandlers(): # prevent Jupyter from adding multiple loggers
formatter = logging.Formatter('%(asctime)s %(name)s %(message)s', datefmt="%Y-%m-%d %H:%M:%S")
handler = logging.StreamHandler()
handler.setFormatter(formatter)
logger.addHandler(handler)
logger.setLevel(logging.DEBUG)
```
Here's a minimalistic time logger that can be initialized with the name of a given layer, and can track several events (e.g. `"forward"` and `"backward"`). When the `TimeLogger.end` method is called, the output is formatted nicely and the elapsed time is logged with the logger name and colored label.
```
from timeit import default_timer
from wasabi import color
class TimeLogger:
def __init__(self, name):
self.colors = {"forward": "green", "backward": "blue"}
self.name = name
self.timers = {}
def start(self, name):
self.timers[name] = default_timer()
def end(self, name):
result = default_timer() - self.timers[name]
label = f"{name.upper():<8}"
label = color(label, self.colors.get(name), bold=True)
logger.debug(f"{self.name:<12} | {label} | {result:.6f}")
```
The `benchmark` layer now has to wrap the forward pass, backward pass and initialization of the layer it wraps and log the execution times. It then returns a Thinc model instance with the custom `forward` function and a custom `init` function. We'll also allow setting a custom `name` to make it easier to tell multiple wrapped benchmark layers apart.
```
from thinc.api import Model
def benchmark(layer, name=None):
name = name if name is not None else layer.name
t = TimeLogger(name)
def init(model, X, Y):
t.start("init")
result = layer.initialize(X, Y)
t.end("init")
return result
def forward(model, X, is_train):
t.start("forward")
layer_Y, layer_callback = layer(X, is_train=is_train)
t.end("forward")
def backprop(dY):
t.start("backward")
result = layer_callback(dY)
t.end("backward")
return result
return layer_Y, backprop
return Model(f"benchmark:{layer.name}", forward, init=init)
```
---
## Usage examples
### Using the `benchmark` layer as a function
We can now wrap one or more layers (including nested layers) with the `benchmark` function. This is the original model:
```python
model = chain(Linear(1), Linear(1))
```
```
import numpy
from thinc.api import chain, Linear
X = numpy.zeros((1, 2), dtype="f")
model = benchmark(chain(benchmark(Linear(1)), Linear(1)), name="outer")
model.initialize(X=X)
Y, backprop = model(X, is_train=False)
dX = backprop(Y)
```
### Using the `benchmark` layer as an operator
Alternatively, we can also use `Model.define_operators` to map `benchmark` to an operator like `@`. The left argument of the operator is the first argument passed into the function (the layer) and the right argument is the second argument (the name). The following example wraps the whole network (two chained `Linear` layers) in a benchmark layer named `"outer"`, and the first `Linear` layer in a benchmark layer named `"first"`.
```
from thinc.api import Model
with Model.define_operators({">>": chain, "@": benchmark}):
model = (Linear(1) @ "first" >> Linear(1)) @ "outer"
model.initialize(X=X)
Y, backprop = model(X, is_train=True)
dX = backprop(Y)
```
### Using the `benchmark` layer during training
```
from thinc.api import Model, chain, Relu, Softmax, Adam
n_hidden = 32
dropout = 0.2
with Model.define_operators({">>": chain, "@": benchmark}):
model = (
Relu(nO=n_hidden, dropout=dropout) @ "relu1"
>> Relu(nO=n_hidden, dropout=dropout) @ "relu2"
>> Softmax()
)
train_X = numpy.zeros((5, 784), dtype="f")
train_Y = numpy.zeros((540, 10), dtype="f")
model.initialize(X=train_X[:5], Y=train_Y[:5])
optimizer = Adam(0.001)
for i in range(10):
for X, Y in model.ops.multibatch(8, train_X, train_Y, shuffle=True):
Yh, backprop = model.begin_update(X)
backprop(Yh - Y)
model.finish_update(optimizer)
```
|
github_jupyter
|
!pip install "thinc>=8.0.0a0"
import logging
logger = logging.getLogger("thinc:benchmark")
if not logger.hasHandlers(): # prevent Jupyter from adding multiple loggers
formatter = logging.Formatter('%(asctime)s %(name)s %(message)s', datefmt="%Y-%m-%d %H:%M:%S")
handler = logging.StreamHandler()
handler.setFormatter(formatter)
logger.addHandler(handler)
logger.setLevel(logging.DEBUG)
from timeit import default_timer
from wasabi import color
class TimeLogger:
def __init__(self, name):
self.colors = {"forward": "green", "backward": "blue"}
self.name = name
self.timers = {}
def start(self, name):
self.timers[name] = default_timer()
def end(self, name):
result = default_timer() - self.timers[name]
label = f"{name.upper():<8}"
label = color(label, self.colors.get(name), bold=True)
logger.debug(f"{self.name:<12} | {label} | {result:.6f}")
from thinc.api import Model
def benchmark(layer, name=None):
name = name if name is not None else layer.name
t = TimeLogger(name)
def init(model, X, Y):
t.start("init")
result = layer.initialize(X, Y)
t.end("init")
return result
def forward(model, X, is_train):
t.start("forward")
layer_Y, layer_callback = layer(X, is_train=is_train)
t.end("forward")
def backprop(dY):
t.start("backward")
result = layer_callback(dY)
t.end("backward")
return result
return layer_Y, backprop
return Model(f"benchmark:{layer.name}", forward, init=init)
model = chain(Linear(1), Linear(1))
import numpy
from thinc.api import chain, Linear
X = numpy.zeros((1, 2), dtype="f")
model = benchmark(chain(benchmark(Linear(1)), Linear(1)), name="outer")
model.initialize(X=X)
Y, backprop = model(X, is_train=False)
dX = backprop(Y)
from thinc.api import Model
with Model.define_operators({">>": chain, "@": benchmark}):
model = (Linear(1) @ "first" >> Linear(1)) @ "outer"
model.initialize(X=X)
Y, backprop = model(X, is_train=True)
dX = backprop(Y)
from thinc.api import Model, chain, Relu, Softmax, Adam
n_hidden = 32
dropout = 0.2
with Model.define_operators({">>": chain, "@": benchmark}):
model = (
Relu(nO=n_hidden, dropout=dropout) @ "relu1"
>> Relu(nO=n_hidden, dropout=dropout) @ "relu2"
>> Softmax()
)
train_X = numpy.zeros((5, 784), dtype="f")
train_Y = numpy.zeros((540, 10), dtype="f")
model.initialize(X=train_X[:5], Y=train_Y[:5])
optimizer = Adam(0.001)
for i in range(10):
for X, Y in model.ops.multibatch(8, train_X, train_Y, shuffle=True):
Yh, backprop = model.begin_update(X)
backprop(Yh - Y)
model.finish_update(optimizer)
| 0.653459 | 0.921922 |
# Linear Regression -- Weight Confidence Intervals
```
import matplotlib.pyplot as plt
%matplotlib inline
from mlxtend.plotting import scatterplotmatrix
from sklearn.linear_model import LinearRegression
from sklearn.preprocessing import StandardScaler
from scipy import stats
import numpy as np
# https://en.wikipedia.org/wiki/Simple_linear_regression#Confidence_intervals
# This data set gives average masses for women as a function of their height in a sample of American women of age 30–39.
height_in_m = [1.47, 1.50, 1.52, 1.55, 1.57, 1.60, 1.63, 1.65, 1.68, 1.70, 1.73, 1.75, 1.78, 1.80, 1.83]
mass_in_kg = [52.21, 53.12, 54.48, 55.84, 57.20, 58.57, 59.93, 61.29, 63.11, 64.47, 66.28, 68.10, 69.92, 72.19, 74.46]
np.random.seed(0)
rand1 = np.random.normal(size=len(height_in_m), scale=10, loc=5)
rand2 = np.random.normal(size=len(height_in_m))
X_train = np.array([(i, j, k) for i, j, k in zip(height_in_m, rand1, rand2)])
y_train = np.array(mass_in_kg)
sc_features = StandardScaler()
sc_target = StandardScaler()
X_std = sc_features.fit_transform(X_train)
y_std = sc_target.fit_transform(y_train.reshape(-1, 1)).flatten()
scatterplotmatrix(X_std, names=['Height','Rand 1', 'Rand 2'],
figsize=(6, 5))
plt.tight_layout()
plt.show()
```
## Weight coefficients
```
lr = LinearRegression()
lr.fit(X_std, y_std)
fig, ax = plt.subplots()
ax.bar([0, 1, 2], lr.coef_)
ax.set_xticks([0, 1, 2])
ax.set_xticklabels([f'Height\n({lr.coef_[0]:.3f})',
f'Random 1\n({lr.coef_[1]:.3f})',
f'Random 2\n({lr.coef_[2]:.3f})'])
plt.ylabel('Magnitude')
plt.show()
lr.intercept_
# y = 0.5 in kg
print(0.5 * np.sqrt(sc_target.var_) + sc_target.mean_)
# y = 1.5 in kg
print(1.5 * np.sqrt(sc_target.var_) + sc_target.mean_)
print(np.sqrt(sc_target.var_))
y_pred = lr.predict(X_std)
plt.scatter(X_std[:, 0], y_std)
x1, x2 = X_std[:, 0].argmin(), X_std[:, 0].argmax()
plt.plot([X_std[x1, 0], X_std[x2, 0]], [y_std[x1], y_std[x2]])
plt.show()
def std_err_linearregression(y_true, y_pred, x):
n = len(y_true)
mse = np.sum((y_true - y_pred)**2) / (n-2)
std_err = (np.sqrt(mse) / np.sqrt(np.sum((x - np.mean(x, axis=0))**2, axis=0)))
return std_err
def weight_intervals(n, weight, std_err, alpha=0.05):
t_value = stats.t.ppf(1 - alpha/2, df=n - 2)
temp = t_value * std_err
lower = weight - temp
upper = weight + temp
return lower, upper
y_pred = lr.predict(X_std)
std_err = std_err_linearregression(y_std, y_pred, X_std)
lower, upper = weight_intervals(len(y_std), lr.coef_, std_err)
fig, ax = plt.subplots()
ax.hlines(0, xmin=-0.1, xmax=2.2, linestyle='dashed', color='skyblue')
ax.errorbar([0, 1, 2], lr.coef_, yerr=upper - lr.coef_, fmt='.k')
ax.set_xticks([0, 1, 2])
ax.set_xticklabels([f'Height\n({lr.coef_[0]:.3f})',
f'Random 1\n({lr.coef_[1]:.3f})',
f'Random 2\n({lr.coef_[2]:.3f})'])
plt.ylabel('Magnitude');
lower, upper
```
---
```
import statsmodels.api as sm
mod = sm.OLS(y_std, X_std)
res = mod.fit()
lower, upper = res.conf_int(0.05)[:, 0], res.conf_int(0.05)[:, 1]
lower, upper
fig, ax = plt.subplots()
ax.hlines(0, xmin=-0.1, xmax=2.2, linestyle='dashed', color='skyblue')
ax.errorbar([0, 1, 2], res.params, yerr=upper - res.params, fmt='.k')
ax.set_xticks([0, 1, 2])
ax.set_xticklabels([f'Height\n({lr.coef_[0]:.3f})',
f'Random 1\n({lr.coef_[1]:.3f})',
f'Random 2\n({lr.coef_[2]:.3f})'])
plt.ylabel('Magnitude');
```
|
github_jupyter
|
import matplotlib.pyplot as plt
%matplotlib inline
from mlxtend.plotting import scatterplotmatrix
from sklearn.linear_model import LinearRegression
from sklearn.preprocessing import StandardScaler
from scipy import stats
import numpy as np
# https://en.wikipedia.org/wiki/Simple_linear_regression#Confidence_intervals
# This data set gives average masses for women as a function of their height in a sample of American women of age 30–39.
height_in_m = [1.47, 1.50, 1.52, 1.55, 1.57, 1.60, 1.63, 1.65, 1.68, 1.70, 1.73, 1.75, 1.78, 1.80, 1.83]
mass_in_kg = [52.21, 53.12, 54.48, 55.84, 57.20, 58.57, 59.93, 61.29, 63.11, 64.47, 66.28, 68.10, 69.92, 72.19, 74.46]
np.random.seed(0)
rand1 = np.random.normal(size=len(height_in_m), scale=10, loc=5)
rand2 = np.random.normal(size=len(height_in_m))
X_train = np.array([(i, j, k) for i, j, k in zip(height_in_m, rand1, rand2)])
y_train = np.array(mass_in_kg)
sc_features = StandardScaler()
sc_target = StandardScaler()
X_std = sc_features.fit_transform(X_train)
y_std = sc_target.fit_transform(y_train.reshape(-1, 1)).flatten()
scatterplotmatrix(X_std, names=['Height','Rand 1', 'Rand 2'],
figsize=(6, 5))
plt.tight_layout()
plt.show()
lr = LinearRegression()
lr.fit(X_std, y_std)
fig, ax = plt.subplots()
ax.bar([0, 1, 2], lr.coef_)
ax.set_xticks([0, 1, 2])
ax.set_xticklabels([f'Height\n({lr.coef_[0]:.3f})',
f'Random 1\n({lr.coef_[1]:.3f})',
f'Random 2\n({lr.coef_[2]:.3f})'])
plt.ylabel('Magnitude')
plt.show()
lr.intercept_
# y = 0.5 in kg
print(0.5 * np.sqrt(sc_target.var_) + sc_target.mean_)
# y = 1.5 in kg
print(1.5 * np.sqrt(sc_target.var_) + sc_target.mean_)
print(np.sqrt(sc_target.var_))
y_pred = lr.predict(X_std)
plt.scatter(X_std[:, 0], y_std)
x1, x2 = X_std[:, 0].argmin(), X_std[:, 0].argmax()
plt.plot([X_std[x1, 0], X_std[x2, 0]], [y_std[x1], y_std[x2]])
plt.show()
def std_err_linearregression(y_true, y_pred, x):
n = len(y_true)
mse = np.sum((y_true - y_pred)**2) / (n-2)
std_err = (np.sqrt(mse) / np.sqrt(np.sum((x - np.mean(x, axis=0))**2, axis=0)))
return std_err
def weight_intervals(n, weight, std_err, alpha=0.05):
t_value = stats.t.ppf(1 - alpha/2, df=n - 2)
temp = t_value * std_err
lower = weight - temp
upper = weight + temp
return lower, upper
y_pred = lr.predict(X_std)
std_err = std_err_linearregression(y_std, y_pred, X_std)
lower, upper = weight_intervals(len(y_std), lr.coef_, std_err)
fig, ax = plt.subplots()
ax.hlines(0, xmin=-0.1, xmax=2.2, linestyle='dashed', color='skyblue')
ax.errorbar([0, 1, 2], lr.coef_, yerr=upper - lr.coef_, fmt='.k')
ax.set_xticks([0, 1, 2])
ax.set_xticklabels([f'Height\n({lr.coef_[0]:.3f})',
f'Random 1\n({lr.coef_[1]:.3f})',
f'Random 2\n({lr.coef_[2]:.3f})'])
plt.ylabel('Magnitude');
lower, upper
import statsmodels.api as sm
mod = sm.OLS(y_std, X_std)
res = mod.fit()
lower, upper = res.conf_int(0.05)[:, 0], res.conf_int(0.05)[:, 1]
lower, upper
fig, ax = plt.subplots()
ax.hlines(0, xmin=-0.1, xmax=2.2, linestyle='dashed', color='skyblue')
ax.errorbar([0, 1, 2], res.params, yerr=upper - res.params, fmt='.k')
ax.set_xticks([0, 1, 2])
ax.set_xticklabels([f'Height\n({lr.coef_[0]:.3f})',
f'Random 1\n({lr.coef_[1]:.3f})',
f'Random 2\n({lr.coef_[2]:.3f})'])
plt.ylabel('Magnitude');
| 0.806052 | 0.912124 |
# Minimizing memory usage: a matrix-free iterative solver
## How to deal with dense BEM matrices?
[In the previous section, I explained how to directly discretize a free surface using TDEs](sa_tdes). A downside of this approach is that the surface matrix can get very large very quickly. If I make the width of an element half as large, then there will be 2x many elements per dimension and 4x as many elements overall. And because the interaction matrix is dense, 4x as many elements leads to 16x as many matrix entries. In other words, $n$, the number of elements, scales like $O(h^2)$ in terms of the element width $h$. And the number of matrix rows or columns is exactly $3n$ (the 3 comes from the vector nature of the problem). That requires storing $9n^2$ entries. And, even worse, using a direct solver (LU decomposition, Gaussian elimination, etc) with such a matrix requires time like $O(n^3)$. Even for quite small problems with 10,000 elements, the cost of storage and solution get very large. And without an absolutely enormous machine or a distributed parallel implementation, solving a problem with 200,000 elements will just not be possible. On the other hand, in an ideal world, it would be nice to be able to solve problems with millions or even tens or hundreds of millions of elements.
Fundamentally, the problem is that the interaction matrix is dense. There are two approaches for resolving this problem:
1. Don't store the matrix!
2. Compress the matrix by taking advantage of low rank sub-blocks.
Eventually approach #2 will be critical since it is scalable up to very large problems. And that's exactly what I'll do in the next sections where I'll investigate low-rank methods and hierarchical matrices (H-matrices). However, here, I'll demonstrate approach #1 by using a matrix-free iterative solver. Ultimately, this is just a small patch on a big problem and it won't be a sustainable solution. But, it's immediately useful when you don't have a working implementation, are running into RAM constraints and are okay with a fairly slow solution. It's also useful to introduce iterative linear solvers since they are central to solving BEM linear systems.
When we solve a linear system without storing the matrix, [the method is called "matrix-free"](https://en.wikipedia.org/wiki/Matrix-free_methods). Generally, we'll just recompute any matrix entry whenever we need. How does this do algorithmically? The storage requirements drop to just the $O(n)$ source and observation info instead of the $O(n^2)$ dense matrix. And, as I'll demonstrate, for some problems, the runtime will drop to $O(n^2)$ instead of $O(n^3)$ because solving linear systems will be possible with a fixed and fairly small number of matrix-vector products.
## A demonstration on a large mesh.
To get started, I'll just copy the code to set up the linear system for the South America problem from the previous section. But, as a twist, I'll going to use a mesh with several times more elements. This surface mesh has 28,388 elements. As a result, the matrix would have 3x that many rows and columns and would require 58 GB of memory to store. That's still small enough that it could be stored on a medium sized workstation. But, it's too big for my personal computer!
```
import cutde.fullspace as FS
import cutde.geometry
import numpy as np
import matplotlib.pyplot as plt
from pyproj import Transformer
plt.rcParams["text.usetex"] = True
%config InlineBackend.figure_format='retina'
(surf_pts_lonlat, surf_tris), (fault_pts_lonlat, fault_tris) = np.load(
"sa_mesh16_7216.npy", allow_pickle=True
)
print("Memory required to store this matrix: ", (surf_tris.shape[0] * 3) ** 2 * 8 / 1e9)
transformer = Transformer.from_crs(
"+proj=longlat +ellps=WGS84 +datum=WGS84 +no_defs",
"+proj=geocent +datum=WGS84 +units=m +no_defs",
)
surf_pts_xyz = np.array(
transformer.transform(
surf_pts_lonlat[:, 0], surf_pts_lonlat[:, 1], surf_pts_lonlat[:, 2]
)
).T.copy()
fault_pts_xyz = np.array(
transformer.transform(
fault_pts_lonlat[:, 0], fault_pts_lonlat[:, 1], fault_pts_lonlat[:, 2]
)
).T.copy()
surf_tri_pts_xyz = surf_pts_xyz[surf_tris]
surf_xyz_to_tdcs_R = cutde.geometry.compute_efcs_to_tdcs_rotations(surf_tri_pts_xyz)
fault_centers_lonlat = np.mean(fault_pts_lonlat[fault_tris], axis=1)
fault_lonlat_to_xyz_T = cutde.geometry.compute_projection_transforms(
fault_centers_lonlat, transformer
)
fault_tri_pts_xyz = fault_pts_xyz[fault_tris]
# Compute the transformation from spherical xyz coordinates (the "EFCS" - Earth fixed coordinate system)
fault_xyz_to_tdcs_R = cutde.geometry.compute_efcs_to_tdcs_rotations(fault_tri_pts_xyz)
fault_tri_pts_lonlat = fault_pts_lonlat[fault_tris]
fault_tdcs2_to_lonlat_R = cutde.geometry.compute_efcs_to_tdcs_rotations(fault_tri_pts_lonlat)
strike_lonlat = fault_tdcs2_to_lonlat_R[:, 0, :]
dip_lonlat = fault_tdcs2_to_lonlat_R[:, 1, :]
strike_xyz = np.sum(fault_lonlat_to_xyz_T * strike_lonlat[:, None, :], axis=2)
strike_xyz /= np.linalg.norm(strike_xyz, axis=1)[:, None]
dip_xyz = np.sum(fault_lonlat_to_xyz_T * dip_lonlat[:, None, :], axis=2)
dip_xyz /= np.linalg.norm(dip_xyz, axis=1)[:, None]
# The normal vectors for each triangle are the third rows of the XYZ->TDCS rotation matrices.
Vnormal = surf_xyz_to_tdcs_R[:, 2, :]
surf_centers_xyz = np.mean(surf_tri_pts_xyz, axis=1)
surf_tri_pts_xyz_conv = surf_tri_pts_xyz.astype(np.float32)
# The rotation matrix from TDCS to XYZ is the transpose of XYZ to TDCS.
# The inverse of a rotation matrix is its transpose.
surf_tdcs_to_xyz_R = np.transpose(surf_xyz_to_tdcs_R, (0, 2, 1)).astype(np.float32)
```
Proceeding like the previous section, the next step would be to construct our surface to surface left hand side matrix. But, instead, I'm just going to compute the action of that matrix without ever storing the entire matrix. Essentially, each matrix entry will be recomputed whenever it is needed. The `cutde.disp_free` and `cutde.strain_free` were written for this purpose.
First, let's check that the `cutde.disp_free` matrix free TDE computation is doing what I said it does. That is, it should be computing a matrix vector product. Since our problem is too big to generate the full matrix in memory, I'll just use the first 100 elements for this test.
First, I'll compute the full in-memory matrix subset. This should look familiar! Then, I multiply the matrix by a random slip vector.
```
test_centers = (surf_centers_xyz - 1.0 * Vnormal)[:100].astype(np.float32)
mat = FS.disp_matrix(test_centers, surf_tri_pts_xyz_conv[:100], 0.25).reshape(
(300, 300)
)
slip = np.random.rand(mat.shape[1]).astype(np.float32)
correct_disp = mat.dot(slip)
```
And now the matrix free version. Note that the slip is passed to the `disp_free` function. This makes sense since it is required for a matrix-vector product even though it is not required to construct the matrix with `cutde.disp_matrix`.
```
test_disp = FS.disp_free(
test_centers, surf_tri_pts_xyz_conv[:100], slip.reshape((-1, 3)), 0.25
)
```
And let's calculate the error... It looks good for the first element. For 32-bit floats, this is machine precision.
```
err = correct_disp.reshape((-1, 3)) - test_disp
err[0]
np.mean(np.abs(err)), np.max(np.abs(err))
```
Okay, now that I've shown that `cutde.disp_free` is trustworthy, let's construct a function that computes matrix vector products of the (not in memory) left-hand side matrix.
To start, recall the linear system that we are solving from the last section:
\begin{align}
Ax &= b\\
A_{ij}&=\delta_{ij} + \mathrm{TDE}_{\mathrm{disp}}(\overline{H_i}, H_j, 1)\\
b_i&=\sum_j \mathrm{TDE}_{\mathrm{disp}}(\overline{H_i}, F_j, \Delta u_j)\\
x&=u_j
\end{align}
where $H_i$ is a free surface TDE, $\overline{H_i}$ is the centroid of that TDE, $F_j$ is a fault TDE, $\Delta u_j$ is the imposed slip field and $x=u_j$ is the unknown surface displacement field.
Implicitly, in the construction of that left hand side matrix, there are a number of rotations embedded in the TDE operations. There's also an extrapolation step that we introduced in the last section that allows for safely evaluated at observation points that are directly on the source TDE. We need to transform all those rotation and extrapolation steps into a form that makes sense in an "on the fly" setting where we're not storing a matrix. The `matvec` function does exactly this.
```
offsets = [2.0, 1.0]
offset_centers = [(surf_centers_xyz - off * Vnormal).astype(np.float32) for off in offsets]
# The matrix that will rotate from (x, y, z) into
# the TDCS (triangular dislocation coordinate system)
surf_xyz_to_tdcs_R = surf_xyz_to_tdcs_R.astype(np.float32)
# The extrapolate to the boundary step looked like:
# lhs = 2 * eps_mats[1] - eps_mats[0]
# This array stores the coefficients so that we can apply that formula
# on the fly.
extrapolation_mult = [-1, 2]
def matvec(disp_xyz_flattened):
# Step 0) Unflatten the (x,y,z) coordinate displacement vector.
disp_xyz = disp_xyz_flattened.reshape((-1, 3)).astype(ft)
# Step 1) Rotate displacement into the TDCS (triangular dislocation coordinate system).
disp_tdcs = np.ascontiguousarray(
np.sum(surf_xyz_to_tdcs_R * disp_xyz[:, None, :], axis=2)
)
# Step 2) Compute the two point extrapolation to the boundary.
# Recall from the previous section that this two point extrapolation
# allow us to calculate for observation points that lie right on a TDE
# without worrying about numerical inaccuracies.
out = np.zeros_like(offset_centers[0])
for i in range(len(offsets)):
out += extrapolation_mult[i] * FS.disp_free(
offset_centers[i], surf_tri_pts_xyz_conv, disp_tdcs, 0.25
)
out = out.flatten()
# Step 3) Don't forget the diagonal Identity matrix term!
out += disp_xyz_flattened
return out
%%time
matvec(np.random.rand(surf_tris.shape[0] * 3))
```
Great! We computed a matrix-free matrix-vector product! Unfortunately, it's a bit slow, but that's an unsurprising consequence of running an $O(n^2)$ algorithm for a large value of $n$. The nice thing is that we're able to do this *at all* by not storing the matrix at any point. This little snippet below will demonstrate that the memory usage is still well under 1 GB proving that we're not storing a matrix anywhere.
```
import os, psutil
process = psutil.Process(os.getpid())
print(process.memory_info().rss / 1e9)
```
## Iterative linear solution
Okay, so how do we use this matrix-vector product to solve the linear system? Because the entire matrix is never in memory, direct solvers like LU decomposition or Cholesky decomposition are no longer an option. But, iterative linear solvers are still an option. The [conjugate gradient (CG) method](https://en.wikipedia.org/wiki/Conjugate_gradient_method) is a well-known example of an iterative solver. However, CG requires a symmetric positive definite matrix. Because our columns come from integrals over elements but our rows come from observation points, there is an inherent asymmetry to the boundary element matrices we are producing here. [GMRES](https://en.wikipedia.org/wiki/Generalized_minimal_residual_method) is an iterative linear solver that tolerates asymmetry. It's specifically a type of ["Krylov subspace"](https://en.wikipedia.org/wiki/Krylov_subspace) iterative linear solver and as such requires only the set of vectors:
\begin{equation}
\{b, Ab, A^2b, ..., A^nb\}
\end{equation}
As such, only having an implementation of the matrix vector product $Ab$ is required since the later iterates can be computed with multiple matrix vector product. For example, $A^2b = A(Ab)$.
Returning to our linear system above, the right hand side is the amount of displacement at the free surface caused by slip on the fault and can be precomputed because it is only a vector (and thus doesn't require much memory) and will not change during our linear solve.
```
slip = np.ascontiguousarray(np.sum(fault_xyz_to_tdcs_R * dip_xyz[:, None, :], axis=2), dtype=np.float32)
rhs = FS.disp_free(
surf_centers_xyz.astype(np.float32), fault_pts_xyz[fault_tris].astype(np.float32), slip, 0.25
).flatten()
```
Now, the fun stuff: Here, I'll use the [`scipy` implementation of GMRES](https://docs.scipy.org/doc/scipy/reference/generated/scipy.sparse.linalg.gmres.html). First, we need to do use the `scipy.sparse.linalg.LinearOperator` interface to wrap our `matvec` function in a form that the `gmres` function will recognize as a something that represents a linear system that can be solved.
```
import time
import scipy.sparse.linalg as spla
# The number of rows and columns
n = surf_tris.shape[0] * 3
# The matrix vector product function that serves as the "backend" for the LinearOperator.
# This is just a handy wrapper around matvec to track the number of matrix-vector products
# used during the linear solve process.
def M(disp_xyz_flattened):
M.n_iter += 1
start = time.time()
out = matvec(disp_xyz_flattened)
print("n_matvec", M.n_iter, "took", time.time() - start)
return out
M.n_iter = 0
lhs = spla.LinearOperator((n, n), M, dtype=rhs.dtype)
lhs.shape
```
And then we can pass that `LinearOperator` as the left hand side of a system of equations to `gmres`. I'm also going to pass a simple callback that will print the current residual norm at each step of the iterative solver and require a solution tolerance of `1e-4`.
```
np.linalg.norm(rhs)
soln = spla.gmres(
lhs,
rhs,
tol=1e-4,
atol=1e-4,
restart=100,
maxiter=1,
callback_type="pr_norm",
callback=lambda x: print(x),
)
soln = soln[0].reshape((-1, 3))
```
As the figures below demonstrate, only eight matrix-vector products got us a great solution!
```
inverse_transformer = Transformer.from_crs(
"+proj=geocent +datum=WGS84 +units=m +no_defs",
"+proj=longlat +ellps=WGS84 +datum=WGS84 +no_defs",
)
surf_xyz_to_lonlat_T = cutde.compute_projection_transforms(
surf_centers_xyz, inverse_transformer
)
surf_xyz_to_lonlat_T /= np.linalg.norm(surf_xyz_to_lonlat_T, axis=2)[:, :, None]
soln_lonlat = np.sum(surf_xyz_to_lonlat_T * soln[:, None, :], axis=2)
plt.figure(figsize=(13, 6))
for d in range(3):
plt.subplot(1, 3, 1 + d)
cntf = plt.tripcolor(
surf_pts_lonlat[:, 0], surf_pts_lonlat[:, 1], surf_tris, soln_lonlat[:, d]
)
plt.colorbar(cntf)
plt.axis("equal")
plt.xlim([-85, -70])
plt.ylim([-50, 10])
plt.title(
["$u_{\\textrm{east}}$", "$u_{\\textrm{north}}$", "$u_{\\textrm{up}}$"][d]
)
plt.show()
```
## Performance and convergence
An important thing to note about the solution above is that only a few matrix-vector products are required to get to a high-level of accuracy. GMRES (and many other iterative linear and nonlinear optimization algorithms) converges at a rate proportional to the condition number of the matrix {cite:p}`saadGMRESGeneralizedMinimal1986`. So in order to productively use an iterative linear solver, we need to have a matrix with a small condition number. It turns out that these free surface self-interaction matrices have condition numbers that are very close to 1.0, meaning that all the eigenvalues are very similar in magnitude. As a result, a highly accurate solution with GMRES requires less than ten matrix-vector products even for very large matrices.
Because of this dependence on the condition number, in the worst case, iterative solvers are not faster than a direct solver. However, suppose that we need only 10 matrix-vector products. Then, the runtime is approximately $10(2n^2)$ because each matrix-vector product requires $2n^2$ operations (one multiplication and one addition per matrix entry). As a result, GMRES is solving the problem in $O(n^2)$ instead of the $O(n^3)$ asymptotic runtime of direct methods like LU decomposition. **So, in addition to requiring less memory, the matrix free method here forced us into actually using a faster linear solver.** Of course, LU decomposition comes out ahead again if we need to solve many linear systems with the same left hand side and different right hand sides. That is not the case here but would be relevant for many other problems (e.g. problems involving time stepping).
The mess of code below builds a few figures that demonstrate these points regarding performance and accuracy as a function of the number of elements.
```
import time
fault_L = 1000.0
fault_H = 1000.0
fault_D = 0.0
fault_pts = np.array(
[
[-fault_L, 0, -fault_D],
[fault_L, 0, -fault_D],
[fault_L, 0, -fault_D - fault_H],
[-fault_L, 0, -fault_D - fault_H],
]
)
fault_tris = np.array([[0, 1, 2], [0, 2, 3]], dtype=np.int64)
results = []
for n_els_per_dim in [2, 4, 8, 16, 32, 48]:
surf_L = 4000
mesh_xs = np.linspace(-surf_L, surf_L, n_els_per_dim + 1)
mesh_ys = np.linspace(-surf_L, surf_L, n_els_per_dim + 1)
mesh_xg, mesh_yg = np.meshgrid(mesh_xs, mesh_ys)
surf_pts = np.array([mesh_xg, mesh_yg, 0 * mesh_yg]).reshape((3, -1)).T.copy()
surf_tris = []
nx = ny = n_els_per_dim + 1
idx = lambda i, j: i * ny + j
for i in range(n_els_per_dim):
for j in range(n_els_per_dim):
x1, x2 = mesh_xs[i : i + 2]
y1, y2 = mesh_ys[j : j + 2]
surf_tris.append([idx(i, j), idx(i + 1, j), idx(i + 1, j + 1)])
surf_tris.append([idx(i, j), idx(i + 1, j + 1), idx(i, j + 1)])
surf_tris = np.array(surf_tris, dtype=np.int64)
surf_tri_pts = surf_pts[surf_tris]
surf_centroids = np.mean(surf_tri_pts, axis=1)
fault_surf_mat = cutde.disp_matrix(surf_centroids, fault_pts[fault_tris], 0.25)
rhs = np.sum(fault_surf_mat[:, :, :, 0], axis=2).flatten()
start = time.time()
eps_mats = []
offsets = [0.002, 0.001]
offset_centers = [
np.mean(surf_tri_pts, axis=1) - off * np.array([0, 0, 1]) for off in offsets
]
for i, off in enumerate(offsets):
eps_mats.append(cutde.disp_matrix(offset_centers[i], surf_pts[surf_tris], 0.25))
lhs = 2 * eps_mats[1] - eps_mats[0]
lhs_reordered = np.empty_like(lhs)
lhs_reordered[:, :, :, 0] = lhs[:, :, :, 1]
lhs_reordered[:, :, :, 1] = lhs[:, :, :, 0]
lhs_reordered[:, :, :, 2] = lhs[:, :, :, 2]
lhs_reordered = lhs_reordered.reshape(
(surf_tris.shape[0] * 3, surf_tris.shape[0] * 3)
)
lhs_reordered += np.eye(lhs_reordered.shape[0])
direct_build_time = time.time() - start
start = time.time()
soln = np.linalg.solve(lhs_reordered, rhs).reshape((-1, 3))
direct_solve_time = time.time() - start
def matvec(x):
extrapolation_mult = [-1, 2]
slip = np.empty((surf_centroids.shape[0], 3))
xrshp = x.reshape((-1, 3))
slip[:, 0] = xrshp[:, 1]
slip[:, 1] = xrshp[:, 0]
slip[:, 2] = xrshp[:, 2]
out = np.zeros_like(offset_centers[0])
for i, off in enumerate(offsets):
out += extrapolation_mult[i] * cutde.disp_free(
offset_centers[i], surf_tri_pts, slip, 0.25
)
return out.flatten() + x
n = surf_tris.shape[0] * 3
def M(x):
M.n_iter += 1
return matvec(x)
M.n_iter = 0
lhs = spla.LinearOperator((n, n), M, dtype=rhs.dtype)
start = time.time()
soln_iter = spla.gmres(lhs, rhs, tol=1e-4)[0].reshape((-1, 3))
iterative_runtime = time.time() - start
l1_err = np.mean(np.abs((soln_iter - soln) / soln))
results.append(
dict(
l1_err=l1_err,
n_elements=surf_tris.shape[0],
iterations=M.n_iter,
direct_build_time=direct_build_time,
direct_solve_time=direct_solve_time,
iterative_runtime=iterative_runtime,
direct_memory=rhs.nbytes + lhs_reordered.nbytes,
iterative_memory=rhs.nbytes,
)
)
import pandas as pd
results_df = pd.DataFrame({k: [r[k] for r in results] for k in results[0].keys()})
results_df["direct_runtime"] = (
results_df["direct_build_time"] + results_df["direct_solve_time"]
)
results_df
plt.rcParams["text.usetex"] = False
plt.figure(figsize=(8, 4))
plt.subplot(1, 2, 1)
plt.plot(results_df["n_elements"], results_df["direct_runtime"], label="direct")
plt.plot(results_df["n_elements"], results_df["iterative_runtime"], label="iterative")
plt.legend()
plt.title("Run time (secs)")
plt.subplot(1, 2, 2)
plt.plot(results_df["n_elements"], results_df["direct_memory"] / 1e6, label="direct")
plt.plot(
results_df["n_elements"], results_df["iterative_memory"] / 1e6, label="iterative"
)
plt.legend()
plt.title("Memory usage (MB)")
plt.show()
```
|
github_jupyter
|
import cutde.fullspace as FS
import cutde.geometry
import numpy as np
import matplotlib.pyplot as plt
from pyproj import Transformer
plt.rcParams["text.usetex"] = True
%config InlineBackend.figure_format='retina'
(surf_pts_lonlat, surf_tris), (fault_pts_lonlat, fault_tris) = np.load(
"sa_mesh16_7216.npy", allow_pickle=True
)
print("Memory required to store this matrix: ", (surf_tris.shape[0] * 3) ** 2 * 8 / 1e9)
transformer = Transformer.from_crs(
"+proj=longlat +ellps=WGS84 +datum=WGS84 +no_defs",
"+proj=geocent +datum=WGS84 +units=m +no_defs",
)
surf_pts_xyz = np.array(
transformer.transform(
surf_pts_lonlat[:, 0], surf_pts_lonlat[:, 1], surf_pts_lonlat[:, 2]
)
).T.copy()
fault_pts_xyz = np.array(
transformer.transform(
fault_pts_lonlat[:, 0], fault_pts_lonlat[:, 1], fault_pts_lonlat[:, 2]
)
).T.copy()
surf_tri_pts_xyz = surf_pts_xyz[surf_tris]
surf_xyz_to_tdcs_R = cutde.geometry.compute_efcs_to_tdcs_rotations(surf_tri_pts_xyz)
fault_centers_lonlat = np.mean(fault_pts_lonlat[fault_tris], axis=1)
fault_lonlat_to_xyz_T = cutde.geometry.compute_projection_transforms(
fault_centers_lonlat, transformer
)
fault_tri_pts_xyz = fault_pts_xyz[fault_tris]
# Compute the transformation from spherical xyz coordinates (the "EFCS" - Earth fixed coordinate system)
fault_xyz_to_tdcs_R = cutde.geometry.compute_efcs_to_tdcs_rotations(fault_tri_pts_xyz)
fault_tri_pts_lonlat = fault_pts_lonlat[fault_tris]
fault_tdcs2_to_lonlat_R = cutde.geometry.compute_efcs_to_tdcs_rotations(fault_tri_pts_lonlat)
strike_lonlat = fault_tdcs2_to_lonlat_R[:, 0, :]
dip_lonlat = fault_tdcs2_to_lonlat_R[:, 1, :]
strike_xyz = np.sum(fault_lonlat_to_xyz_T * strike_lonlat[:, None, :], axis=2)
strike_xyz /= np.linalg.norm(strike_xyz, axis=1)[:, None]
dip_xyz = np.sum(fault_lonlat_to_xyz_T * dip_lonlat[:, None, :], axis=2)
dip_xyz /= np.linalg.norm(dip_xyz, axis=1)[:, None]
# The normal vectors for each triangle are the third rows of the XYZ->TDCS rotation matrices.
Vnormal = surf_xyz_to_tdcs_R[:, 2, :]
surf_centers_xyz = np.mean(surf_tri_pts_xyz, axis=1)
surf_tri_pts_xyz_conv = surf_tri_pts_xyz.astype(np.float32)
# The rotation matrix from TDCS to XYZ is the transpose of XYZ to TDCS.
# The inverse of a rotation matrix is its transpose.
surf_tdcs_to_xyz_R = np.transpose(surf_xyz_to_tdcs_R, (0, 2, 1)).astype(np.float32)
test_centers = (surf_centers_xyz - 1.0 * Vnormal)[:100].astype(np.float32)
mat = FS.disp_matrix(test_centers, surf_tri_pts_xyz_conv[:100], 0.25).reshape(
(300, 300)
)
slip = np.random.rand(mat.shape[1]).astype(np.float32)
correct_disp = mat.dot(slip)
test_disp = FS.disp_free(
test_centers, surf_tri_pts_xyz_conv[:100], slip.reshape((-1, 3)), 0.25
)
err = correct_disp.reshape((-1, 3)) - test_disp
err[0]
np.mean(np.abs(err)), np.max(np.abs(err))
offsets = [2.0, 1.0]
offset_centers = [(surf_centers_xyz - off * Vnormal).astype(np.float32) for off in offsets]
# The matrix that will rotate from (x, y, z) into
# the TDCS (triangular dislocation coordinate system)
surf_xyz_to_tdcs_R = surf_xyz_to_tdcs_R.astype(np.float32)
# The extrapolate to the boundary step looked like:
# lhs = 2 * eps_mats[1] - eps_mats[0]
# This array stores the coefficients so that we can apply that formula
# on the fly.
extrapolation_mult = [-1, 2]
def matvec(disp_xyz_flattened):
# Step 0) Unflatten the (x,y,z) coordinate displacement vector.
disp_xyz = disp_xyz_flattened.reshape((-1, 3)).astype(ft)
# Step 1) Rotate displacement into the TDCS (triangular dislocation coordinate system).
disp_tdcs = np.ascontiguousarray(
np.sum(surf_xyz_to_tdcs_R * disp_xyz[:, None, :], axis=2)
)
# Step 2) Compute the two point extrapolation to the boundary.
# Recall from the previous section that this two point extrapolation
# allow us to calculate for observation points that lie right on a TDE
# without worrying about numerical inaccuracies.
out = np.zeros_like(offset_centers[0])
for i in range(len(offsets)):
out += extrapolation_mult[i] * FS.disp_free(
offset_centers[i], surf_tri_pts_xyz_conv, disp_tdcs, 0.25
)
out = out.flatten()
# Step 3) Don't forget the diagonal Identity matrix term!
out += disp_xyz_flattened
return out
%%time
matvec(np.random.rand(surf_tris.shape[0] * 3))
import os, psutil
process = psutil.Process(os.getpid())
print(process.memory_info().rss / 1e9)
slip = np.ascontiguousarray(np.sum(fault_xyz_to_tdcs_R * dip_xyz[:, None, :], axis=2), dtype=np.float32)
rhs = FS.disp_free(
surf_centers_xyz.astype(np.float32), fault_pts_xyz[fault_tris].astype(np.float32), slip, 0.25
).flatten()
import time
import scipy.sparse.linalg as spla
# The number of rows and columns
n = surf_tris.shape[0] * 3
# The matrix vector product function that serves as the "backend" for the LinearOperator.
# This is just a handy wrapper around matvec to track the number of matrix-vector products
# used during the linear solve process.
def M(disp_xyz_flattened):
M.n_iter += 1
start = time.time()
out = matvec(disp_xyz_flattened)
print("n_matvec", M.n_iter, "took", time.time() - start)
return out
M.n_iter = 0
lhs = spla.LinearOperator((n, n), M, dtype=rhs.dtype)
lhs.shape
np.linalg.norm(rhs)
soln = spla.gmres(
lhs,
rhs,
tol=1e-4,
atol=1e-4,
restart=100,
maxiter=1,
callback_type="pr_norm",
callback=lambda x: print(x),
)
soln = soln[0].reshape((-1, 3))
inverse_transformer = Transformer.from_crs(
"+proj=geocent +datum=WGS84 +units=m +no_defs",
"+proj=longlat +ellps=WGS84 +datum=WGS84 +no_defs",
)
surf_xyz_to_lonlat_T = cutde.compute_projection_transforms(
surf_centers_xyz, inverse_transformer
)
surf_xyz_to_lonlat_T /= np.linalg.norm(surf_xyz_to_lonlat_T, axis=2)[:, :, None]
soln_lonlat = np.sum(surf_xyz_to_lonlat_T * soln[:, None, :], axis=2)
plt.figure(figsize=(13, 6))
for d in range(3):
plt.subplot(1, 3, 1 + d)
cntf = plt.tripcolor(
surf_pts_lonlat[:, 0], surf_pts_lonlat[:, 1], surf_tris, soln_lonlat[:, d]
)
plt.colorbar(cntf)
plt.axis("equal")
plt.xlim([-85, -70])
plt.ylim([-50, 10])
plt.title(
["$u_{\\textrm{east}}$", "$u_{\\textrm{north}}$", "$u_{\\textrm{up}}$"][d]
)
plt.show()
import time
fault_L = 1000.0
fault_H = 1000.0
fault_D = 0.0
fault_pts = np.array(
[
[-fault_L, 0, -fault_D],
[fault_L, 0, -fault_D],
[fault_L, 0, -fault_D - fault_H],
[-fault_L, 0, -fault_D - fault_H],
]
)
fault_tris = np.array([[0, 1, 2], [0, 2, 3]], dtype=np.int64)
results = []
for n_els_per_dim in [2, 4, 8, 16, 32, 48]:
surf_L = 4000
mesh_xs = np.linspace(-surf_L, surf_L, n_els_per_dim + 1)
mesh_ys = np.linspace(-surf_L, surf_L, n_els_per_dim + 1)
mesh_xg, mesh_yg = np.meshgrid(mesh_xs, mesh_ys)
surf_pts = np.array([mesh_xg, mesh_yg, 0 * mesh_yg]).reshape((3, -1)).T.copy()
surf_tris = []
nx = ny = n_els_per_dim + 1
idx = lambda i, j: i * ny + j
for i in range(n_els_per_dim):
for j in range(n_els_per_dim):
x1, x2 = mesh_xs[i : i + 2]
y1, y2 = mesh_ys[j : j + 2]
surf_tris.append([idx(i, j), idx(i + 1, j), idx(i + 1, j + 1)])
surf_tris.append([idx(i, j), idx(i + 1, j + 1), idx(i, j + 1)])
surf_tris = np.array(surf_tris, dtype=np.int64)
surf_tri_pts = surf_pts[surf_tris]
surf_centroids = np.mean(surf_tri_pts, axis=1)
fault_surf_mat = cutde.disp_matrix(surf_centroids, fault_pts[fault_tris], 0.25)
rhs = np.sum(fault_surf_mat[:, :, :, 0], axis=2).flatten()
start = time.time()
eps_mats = []
offsets = [0.002, 0.001]
offset_centers = [
np.mean(surf_tri_pts, axis=1) - off * np.array([0, 0, 1]) for off in offsets
]
for i, off in enumerate(offsets):
eps_mats.append(cutde.disp_matrix(offset_centers[i], surf_pts[surf_tris], 0.25))
lhs = 2 * eps_mats[1] - eps_mats[0]
lhs_reordered = np.empty_like(lhs)
lhs_reordered[:, :, :, 0] = lhs[:, :, :, 1]
lhs_reordered[:, :, :, 1] = lhs[:, :, :, 0]
lhs_reordered[:, :, :, 2] = lhs[:, :, :, 2]
lhs_reordered = lhs_reordered.reshape(
(surf_tris.shape[0] * 3, surf_tris.shape[0] * 3)
)
lhs_reordered += np.eye(lhs_reordered.shape[0])
direct_build_time = time.time() - start
start = time.time()
soln = np.linalg.solve(lhs_reordered, rhs).reshape((-1, 3))
direct_solve_time = time.time() - start
def matvec(x):
extrapolation_mult = [-1, 2]
slip = np.empty((surf_centroids.shape[0], 3))
xrshp = x.reshape((-1, 3))
slip[:, 0] = xrshp[:, 1]
slip[:, 1] = xrshp[:, 0]
slip[:, 2] = xrshp[:, 2]
out = np.zeros_like(offset_centers[0])
for i, off in enumerate(offsets):
out += extrapolation_mult[i] * cutde.disp_free(
offset_centers[i], surf_tri_pts, slip, 0.25
)
return out.flatten() + x
n = surf_tris.shape[0] * 3
def M(x):
M.n_iter += 1
return matvec(x)
M.n_iter = 0
lhs = spla.LinearOperator((n, n), M, dtype=rhs.dtype)
start = time.time()
soln_iter = spla.gmres(lhs, rhs, tol=1e-4)[0].reshape((-1, 3))
iterative_runtime = time.time() - start
l1_err = np.mean(np.abs((soln_iter - soln) / soln))
results.append(
dict(
l1_err=l1_err,
n_elements=surf_tris.shape[0],
iterations=M.n_iter,
direct_build_time=direct_build_time,
direct_solve_time=direct_solve_time,
iterative_runtime=iterative_runtime,
direct_memory=rhs.nbytes + lhs_reordered.nbytes,
iterative_memory=rhs.nbytes,
)
)
import pandas as pd
results_df = pd.DataFrame({k: [r[k] for r in results] for k in results[0].keys()})
results_df["direct_runtime"] = (
results_df["direct_build_time"] + results_df["direct_solve_time"]
)
results_df
plt.rcParams["text.usetex"] = False
plt.figure(figsize=(8, 4))
plt.subplot(1, 2, 1)
plt.plot(results_df["n_elements"], results_df["direct_runtime"], label="direct")
plt.plot(results_df["n_elements"], results_df["iterative_runtime"], label="iterative")
plt.legend()
plt.title("Run time (secs)")
plt.subplot(1, 2, 2)
plt.plot(results_df["n_elements"], results_df["direct_memory"] / 1e6, label="direct")
plt.plot(
results_df["n_elements"], results_df["iterative_memory"] / 1e6, label="iterative"
)
plt.legend()
plt.title("Memory usage (MB)")
plt.show()
| 0.568176 | 0.986675 |
```
import sys
import numpy as np
from hyperopt import fmin, tpe, hp, STATUS_OK, Trials
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Flatten
from keras.layers import Conv2D, MaxPooling2D
from keras.optimizers import Adam
from sklearn.model_selection import train_test_split
from keras.utils import to_categorical
from keras.callbacks import EarlyStopping, TensorBoard, ModelCheckpoint
from keras.datasets import cifar10
(X_train, y_train), (X_test, y_test) = cifar10.load_data()
validation_split = 0.1
X_train, X_val, y_train, y_val = train_test_split(X_train, y_train, test_size=validation_split, random_state=SEED)
X_train = X_train.astype('float32')
X_train /=255.
X_val = X_val.astype('float32')
X_val /=255.
X_test = X_test.astype('float32')
X_test /=255.
n_classes = 10
y_train = to_categorical(y_train, n_classes)
y_val = to_categorical(y_val, n_classes)
y_test = to_categorical(y_test, n_classes)
space = {'batch_size' : hp.choice('batch_size', [32, 64, 128,256]), 'n_epochs' : 1000}
def get_callbacks(pars):
callbacks =[EarlyStopping(monitor='val_acc', p atience=5, verbose=2),
ModelCheckpoint('checkpoints/{}.h5'.format(pars['batch_size']), save_best_only=True),
TensorBoard('~/notebooks/logs-gridsearch', write_graph=True, write_grads=True, write_images=True, embeddings_freq=0, embeddings_layer_names=None, embeddings_metadata=None)]
return callbacks
def f_nn(pars):
print ('Parameters: ', pars)
model = Sequential()
model.add(Conv2D(32, (3, 3), padding='same', input_shape=X_train.shape[1:]))
model.add(Activation('relu'))
model.add(Conv2D(32, (3, 3)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(64, (3, 3), padding='same'))
model.add(Activation('relu'))
model.add(Conv2D(64, (3, 3)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(512))
model.add(Activation('relu'))
model.add(Dropout(0.5))
model.add(Dense(n_classes))
model.add(Activation('softmax'))
optimizer = Adam()
model.compile(loss='categorical_crossentropy', optimizer=optimizer, metrics=['accuracy'])
history = model.fit(X_train, y_train, epochs=pars['n_epochs'], batch_size=pars['batch_size'],
validation_data=[X_val, y_val],
verbose = 1, callbacks=get_callbacks(pars))
best_epoch = np.argmax(history.history['val_acc'])
best_val_acc = np.max(history.history['val_acc'])
print('Epoch {} - val acc: {}'.format(best_epoch, best_val_acc))
sys.stdout.flush()
return {'val_acc': best_val_acc, 'best_epoch': best_epoch, 'eval_time': time.time(), 'status': STATUS_OK}
trials = Trials()
best = fmin(f_nn, space, algo=tpe.suggest, max_evals=50, trials=trials)
print(best)
```
|
github_jupyter
|
import sys
import numpy as np
from hyperopt import fmin, tpe, hp, STATUS_OK, Trials
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Flatten
from keras.layers import Conv2D, MaxPooling2D
from keras.optimizers import Adam
from sklearn.model_selection import train_test_split
from keras.utils import to_categorical
from keras.callbacks import EarlyStopping, TensorBoard, ModelCheckpoint
from keras.datasets import cifar10
(X_train, y_train), (X_test, y_test) = cifar10.load_data()
validation_split = 0.1
X_train, X_val, y_train, y_val = train_test_split(X_train, y_train, test_size=validation_split, random_state=SEED)
X_train = X_train.astype('float32')
X_train /=255.
X_val = X_val.astype('float32')
X_val /=255.
X_test = X_test.astype('float32')
X_test /=255.
n_classes = 10
y_train = to_categorical(y_train, n_classes)
y_val = to_categorical(y_val, n_classes)
y_test = to_categorical(y_test, n_classes)
space = {'batch_size' : hp.choice('batch_size', [32, 64, 128,256]), 'n_epochs' : 1000}
def get_callbacks(pars):
callbacks =[EarlyStopping(monitor='val_acc', p atience=5, verbose=2),
ModelCheckpoint('checkpoints/{}.h5'.format(pars['batch_size']), save_best_only=True),
TensorBoard('~/notebooks/logs-gridsearch', write_graph=True, write_grads=True, write_images=True, embeddings_freq=0, embeddings_layer_names=None, embeddings_metadata=None)]
return callbacks
def f_nn(pars):
print ('Parameters: ', pars)
model = Sequential()
model.add(Conv2D(32, (3, 3), padding='same', input_shape=X_train.shape[1:]))
model.add(Activation('relu'))
model.add(Conv2D(32, (3, 3)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(64, (3, 3), padding='same'))
model.add(Activation('relu'))
model.add(Conv2D(64, (3, 3)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(512))
model.add(Activation('relu'))
model.add(Dropout(0.5))
model.add(Dense(n_classes))
model.add(Activation('softmax'))
optimizer = Adam()
model.compile(loss='categorical_crossentropy', optimizer=optimizer, metrics=['accuracy'])
history = model.fit(X_train, y_train, epochs=pars['n_epochs'], batch_size=pars['batch_size'],
validation_data=[X_val, y_val],
verbose = 1, callbacks=get_callbacks(pars))
best_epoch = np.argmax(history.history['val_acc'])
best_val_acc = np.max(history.history['val_acc'])
print('Epoch {} - val acc: {}'.format(best_epoch, best_val_acc))
sys.stdout.flush()
return {'val_acc': best_val_acc, 'best_epoch': best_epoch, 'eval_time': time.time(), 'status': STATUS_OK}
trials = Trials()
best = fmin(f_nn, space, algo=tpe.suggest, max_evals=50, trials=trials)
print(best)
| 0.697506 | 0.379407 |
<a href="https://colab.research.google.com/github/pritul2/Detection-of-Person-With-or-Without-Mask/blob/master/xception.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
from keras.applications.xception import Xception
from keras.layers import Dense,Flatten
from keras.models import Model
from keras.preprocessing.image import ImageDataGenerator
from keras.models import Sequential
from keras import layers
from keras.layers import Conv2D, MaxPooling2D, Dropout, Flatten, Dense, Activation,GlobalMaxPooling2D,GlobalAveragePooling2D,AveragePooling2D
from keras import applications
from keras.preprocessing.image import ImageDataGenerator
from keras import optimizers
from keras.applications import VGG16
from keras.models import Model
base_model = Xception(weights='imagenet',include_top=False,input_shape=(299, 299, 3)) #include_top ---> not keeping complete Model
headModel = base_model.output
headModel = AveragePooling2D(pool_size=(4, 4))(headModel)
headModel = Flatten(name="flatten")(headModel)
headModel = Dense(128, activation="relu")(headModel)
headModel = Dropout(0.5)(headModel)
headModel = Dense(2, activation="softmax")(headModel)
model = Model(inputs=base_model.input, outputs=headModel)
for layer in base_model.layers:
layer.trainable = False
print("[INFO] compiling model...")
opt = optimizers.Adam(lr=1e-4)
model.compile(loss="binary_crossentropy", optimizer=opt,metrics=["accuracy"])
import os
total_train = len(os.listdir("/content/CROPPED_train/withmask_cropped")) + len(os.listdir("/content/CROPPED_train/without_mask2_output"))
train_datagen = ImageDataGenerator(rescale=1./255,rotation_range=30,
zoom_range=0.15,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.15,
horizontal_flip=True,
fill_mode="nearest")
import numpy as np
train_generator = train_datagen.flow_from_directory('/content/CROPPED_train',target_size=(299,299),batch_size = 16,seed=np.random.seed())
history = model.fit_generator(
train_generator,
epochs=5,
steps_per_epoch=total_train//16)
!unzip /content/CROPPED_train.zip
model.save("xception_mask.h5")
import cv2
import matplotlib.pyplot as plt
import numpy as np
img = cv2.imread("/content/CROPPED_train/without_mask2_output/57.png")
img = cv2.resize(img,(299,299))
plt.imshow(img)
plt.show()
temp_img = img/255.0
preds = model.predict(np.expand_dims(temp_img, axis=0))[0]
#y= model.predict(img[np.newaxis,...])
i = np.argmax(preds)
print(i)
!unzip CROPPED_train.zip
```
|
github_jupyter
|
from keras.applications.xception import Xception
from keras.layers import Dense,Flatten
from keras.models import Model
from keras.preprocessing.image import ImageDataGenerator
from keras.models import Sequential
from keras import layers
from keras.layers import Conv2D, MaxPooling2D, Dropout, Flatten, Dense, Activation,GlobalMaxPooling2D,GlobalAveragePooling2D,AveragePooling2D
from keras import applications
from keras.preprocessing.image import ImageDataGenerator
from keras import optimizers
from keras.applications import VGG16
from keras.models import Model
base_model = Xception(weights='imagenet',include_top=False,input_shape=(299, 299, 3)) #include_top ---> not keeping complete Model
headModel = base_model.output
headModel = AveragePooling2D(pool_size=(4, 4))(headModel)
headModel = Flatten(name="flatten")(headModel)
headModel = Dense(128, activation="relu")(headModel)
headModel = Dropout(0.5)(headModel)
headModel = Dense(2, activation="softmax")(headModel)
model = Model(inputs=base_model.input, outputs=headModel)
for layer in base_model.layers:
layer.trainable = False
print("[INFO] compiling model...")
opt = optimizers.Adam(lr=1e-4)
model.compile(loss="binary_crossentropy", optimizer=opt,metrics=["accuracy"])
import os
total_train = len(os.listdir("/content/CROPPED_train/withmask_cropped")) + len(os.listdir("/content/CROPPED_train/without_mask2_output"))
train_datagen = ImageDataGenerator(rescale=1./255,rotation_range=30,
zoom_range=0.15,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.15,
horizontal_flip=True,
fill_mode="nearest")
import numpy as np
train_generator = train_datagen.flow_from_directory('/content/CROPPED_train',target_size=(299,299),batch_size = 16,seed=np.random.seed())
history = model.fit_generator(
train_generator,
epochs=5,
steps_per_epoch=total_train//16)
!unzip /content/CROPPED_train.zip
model.save("xception_mask.h5")
import cv2
import matplotlib.pyplot as plt
import numpy as np
img = cv2.imread("/content/CROPPED_train/without_mask2_output/57.png")
img = cv2.resize(img,(299,299))
plt.imshow(img)
plt.show()
temp_img = img/255.0
preds = model.predict(np.expand_dims(temp_img, axis=0))[0]
#y= model.predict(img[np.newaxis,...])
i = np.argmax(preds)
print(i)
!unzip CROPPED_train.zip
| 0.668231 | 0.703817 |
<img src="http://hilpisch.com/tpq_logo.png" alt="The Python Quants" width="35%" align="right" border="0"><br><br><br>
# Listed Volatility and Variance Derivatives
**Wiley Finance (2017)**
Dr. Yves J. Hilpisch | The Python Quants GmbH
http://tpq.io | [@dyjh](http://twitter.com/dyjh) | http://books.tpq.io
<img src="https://hilpisch.com/images/lvvd_cover.png" alt="Listed Volatility and Variance Derivatives" width="30%" align="left" border="0">
# VSTOXX Index
## Introduction
This chapter is about the (re-) calculation of the VSTOXX index, the volatility index based on EURO STOXX 50 index options. The goal is to achieve a good understanding of the processes and underlying mechanics of calculating the VSTOXX index. You will find all background information as well as Python code that will enable you to recalculate historical VSTOXX index values as well as current ones in (almost) real-time. The chapter about the _Model-Free Replication of Variance_ provides the theoretical background for the concepts presented in this chapter.
The (main) VSTOXX index itself is based on two sub-indexes, which themselves are derived from Eurex option series for both European puts and calls on the EURO STOXX 50 index. The algorithm, and therefore this chapter as well, is comprised of three main parts:
* collect and clean-up the data of the necessary option series
* compute the sub-indexes from the option data
* compute the VSTOXX index from the relevant sub-indexes
A few remarks about the option series and sub-indexes used and their expiry dates and time horizons, respectively, seem in order. There are eight sub-indexes of the VSTOXX which each measure the implied volatility of an option series with fixed expiry. For example, the VSTOXX 1M sub-index starts with the option series that has one month expiry and is calculated up to two days prior to the fixed maturity date of the according option series. The VSTOXX index measures the implied volatility of an "imaginary" options series with a fixed time to expiry of 30 days. This is achieved through linear interpolation of the two nearest sub-indexes, generally VSTOXX 1M and VSTOXX 2M. On the two days before VSTOXX 1M expiry, the VSTOXX 2M and VSTOXX 3M are used instead and an extrapolation takes place.
The following table lists all the sub-indexes and provides additional information.
============ ======= ============== ===========================================
Sub-index Code ISIN Settlement date of the option series used
============ ======= ============== ===========================================
VSTOXX 1M V6I1 DE000A0G87B2 The last available within 1 month
VSTOXX 2M V6I2 DE000A0G87C0 The last available within 2 months
VSTOXX 3M V6I3 DE000A0G87D8 The last available within 3 months
VSTOXX 6M V6I4 DE000A0G87E6 The last available within 6 months
VSTOXX 9M V6I5 DE000A0G87F3 The last available within 9 months
VSTOXX 12M V6I6 DE000A0G87G1 The last available within 12 months
VSTOXX 18M V6I7 DE000A0G87H9 The last available within 18 months
VSTOXX 24M V6I8 DE000A0G87J5 The last available within 24 months
============ ======= ============== ===========================================
## Collecting Option Data
As pointed out, the VSTOXX is based on two sub-indexes, generally the VSTOXX 1M and VSTOXX 2M, sometimes VSTOXX 2M and VSTOXX 3M. The sub-indexes themselves are based on the option series on the EURO STOXX 50 index with respective time to expiry. We therefore need the prices of all options with maturities up to 3 months. We use historical data as provided by Eurex itself as the data source. See the web site http://bit.ly/1GY5KCI.
The code to collect the data can be found in the module ``index_collect_option_data.py`` (see appendix for the complete script). Like usual, the module starts with some imports and parameter definitions.
```
!sed -n 11,24p scripts/index_collect_option_data.py
```
In addition, the module contains six functions. The first is ``collect_option_series()``:
```
import sys; sys.path.append('./scripts/')
import index_collect_option_data as icod
icod.collect_option_series??
```
This function collects the data of the option series with maturity in the month ``month`` and year ``year``. It is called by the ``function start_collecting()`` and calls the ``function get_data()`` for every single day from the date ``start`` to today. It returns a complete set of prices (both puts and calls) for that series.
The second function is ``get_data()``.
```
icod.get_data??
```
This one is called by the function ``collect_option_series()`` and calls itself the functions ``get_data_from_www()``, ``parse_data(data, date)`` and ``merge_and_filter()``. It returns the prices of the option series with expiry date in month ``month`` and year ``year`` for the day ``date``.
The third function is ``get_data_from_www()``.
```
icod.get_data_from_www??
```
The function collects the prices of an option series for a single day (defined by ``date``) from the web. The option series is defined by the date of its expiry, given by ``matMonth`` and ``matYear``, the type of the options is given by ``oType`` which can be either ``Put`` or ``Call``. It returns a complete HTML file.
``merge_and_filter()`` is the fourth function.
```
icod.merge_and_filter??
```
This one gets two time series ``puts`` and ``calls`` (typically of the same option series), merges them, filters out all options with prices below 0.5 and returns the resulting pandas ``DataFrame`` object.
``parse_data()`` is the fifth function.
```
icod.parse_data??
```
It gets the string ``data`` which contains the HTML text delivered by function ``get_data_from_www()``, parses that string to a pandas ``DataFrame`` object with double index ``date`` and ``strike price`` and returns that object.
The sixth and final function is ``data_collection()``.
```
icod.data_collection??
```
This function is to initiate and finalize the collection of all relevant option series data sets. It saves the resulting data in a file named ``index_option_series.h5``.
```
path = './data/'
```
Let us collect option data since all other steps depend on this data. We import the module as ``icod``.
```
import numpy as np
import pandas as pd
import datetime as dt
import warnings; warnings.simplefilter('ignore')
```
Next, fix a target day relative to today such that you hit a business day for which closing data is available.
```
today = dt.datetime.now()
## make sure to hit a business day
target_day = today - dt.timedelta(days=2)
ds = target_day.strftime('%Y%m%d')
ds
URL = 'https://www.eurex.com/ex-en/data/statistics/market-statistics-online/'
URL += '100!onlineStats?productGroupId=13370&productId=69660&viewType=3&'
URL += 'cp=%s&month=%s&year=%s&busDate=%s'
URL % ('Call', 12, 2020, '20201111')
```
Then, for example, collect option data for puts and calls with a maturity as defined by the parameters as follows.
```
## adjust maturity parameters if necessary
call_data = icod.get_data_from_www(oType='Call', matMonth=12,
matYear=2020, date=ds)
put_data = icod.get_data_from_www(oType='Put', matMonth=12,
matYear=2020, date=ds)
```
The return objects need to be parsed.
```
## parse the raw data
calls = icod.parse_data(call_data, target_day)
puts = icod.parse_data(put_data, target_day)
```
Let us have a look at some meta information about the call options data.
```
calls.info()
calls.head()
```
And about the put options data.
```
puts.info()
```
In a next step, we take out the daily settlement prices for both the puts and calls and define two new ``DataFrame`` objects.
```
calls = pd.DataFrame(calls.rename(
columns={'Daily settlem. price': 'Call_Price'}
).pop('Call_Price').astype(float))
puts = pd.DataFrame(puts.rename(
columns={'Daily settlem. price': 'Put_Price'}
).pop('Put_Price').astype(float))
```
These two get then merged via the function ``merge_and_filter()`` into another new ``DataFrame`` object.
```
dataset = icod.merge_and_filter(puts, calls)
dataset.info()
```
This whole procedure is implemented in the function ``collect_option_series()`` which yields the same result.
```
os = icod.collect_option_series(12, 2020, target_day)
os.info()
```
The function ``data_collection()`` repeats this procedure for all those dates for which option data is available and writes (appends) the results in a HDF5 database file.
```
# uncomment to initiate the process (takes a while)
%time icod.data_collection(path)
```
For the further analyses, we open this HDF5 database file.
```
store = pd.HDFStore(path + 'index_option_series.h5', 'r')
store
```
The collected option series data is easily read from the HDF5 database file in monthly chunks.
```
Dec20 = store['Dec20']
Dec20.info()
store.close()
```
Some selected option prices from the large data set:
```
Dec20.iloc[25:35]
```
## Calculating the Sub-Indexes
In this section, we use the data file created in the previous one. For all dates of the data file, the Python module ``index_subindex_calculation.py`` (see the appendix for the complete script) used in this section decides whether the VSTOXX 1M sub-index is defined or not (remember that the sub-index is not defined at the final settlement day and one day before). If it is defined, the script computes the value of the sub-indexes VSTOXX 1M and VSTOXX 2M; if not, it computes the values of the sub-indexes VSTOXX 2M and VSTOXX 3M, respectively. Finally, it returns a pandas ``DataFrame`` object with the three time series.
### The Algorithm
First, we focus on the computation of the value of a single sub-index for a given date. Given are the prices $C_i, i \in \{0,...,n\},$ of a series of European call options on the EURO STOXX 50 with fixed maturity date $T$ and exercise prices $K_i, i \in \{0,...,n\},$ as well as the prices $P_i, i \in \{0,...,n\},$ of a series of European put options on EURO STOXX 50 with the same maturity date $T$ and exercise prices $K_i$. Let further hold $K_i < K_{i+1}$ for all $i \in \{0,....,n-1\}$.
Then, the value of the relevant sub-index $V$ is as follows (see also the chapter about _Model-Free Replication of Variance_):
$$
V = 100 \cdot \sqrt{\hat{\sigma}^2}
$$
with
$$
\hat{\sigma}^2 = \frac{2}{T} \sum_{i=0}^n \frac{\Delta K_i}{{K_i}^2} \mathrm{e}^{rT} M_i - \frac{1}{T}\left( \frac{F}{K_*}-1\right)^2
$$
where
$$
\begin{array}{ll}
\Delta K_i &=\left\{ \begin{array}{ll} K_1-K_0 & \mbox{for } i=0 \\ \dfrac{K_{i+1}-K_{i-1}}{2} & \mbox{for } i = 1,...,n-1 \\ K_n-K_{n-1} & \mbox{for } i=n \end{array} \right. \\ \\
r &= \mbox{constant risk-free short rate appropriate for maturity $T$} \\ \\
F &= K_j+ \mathrm{e}^{rT}|C_j-P_j|, \mbox{ where } j=\displaystyle \min_{i \in \{0,...,n\}}\{|C_i-P_i|\} \\ \\
K_* &= \displaystyle \max_{ K_{i | i \in \{0,...,n\}}} \{K_i < F \}, \\ \\
M_i & = \left\{ \begin{array}{ll} P_i & \mbox{for } K_i<K_* \\ \dfrac{P_i-C_i}{2} & \mbox{for } K_i=K_*\\ C_i & \mbox{for } K_i>K_* \end{array} \right.
\end{array}
$$
We implement a function to compute one value of a single sub-index. Thereafter, we extend that function to compute time series for both VSTOXX 1M and VSTOXX 2M indexes as well as parts of the VSTOXX 3M index. Imports again make up the beginning of the script.
```
!sed -n 10,15p scripts/index_subindex_calculation.py
```
A core function of the script is ``compute_subindex()``.
```
import index_subindex_calculation as isc
isc.compute_subindex??
```
This script calculates a single index value. It implements mainly the following steps:
* the calculation of $\Delta K_i$
* the computation of the forward price and the index of $K_*$
* the selection of the at-the-money option and the out-of-the-money options
* the combination of the results of the other three steps
The next step is the derivation of time series data for the VSTOXX 1M and VSTOXX 2M as well as parts of VSTOXX 3M indexes and storage of the results in a pandas ``DataFrame`` object. As data source we use the file created in the last section. Remember, that this file contains a dictionary-like ``HDFStore`` object with one entry for every options series. The keys for the entries are three letter abbreviations of the respective month's name plus the actual year represented by two numbers, for example ``Mar16``, ``Jun16``, ``Dec20`` and so on. The value of an entry is a pandas ``DataFrame`` object with a pandas ``MultiIndex`` (date, strike price) and prices for the put and call options for the dates and strike prices.
All this is implemented as function ``make_subindex()``.
```
isc.make_subindex??
```
This function uses the collected option series data and selects those data sub-sets needed for the calculation at hand. It generates sub-index values for all those days for which option data is available. The result is a pandas ``DataFrame`` object.
Let us see how it works. To this end, we first import the module as ``isc``.
```
import index_subindex_calculation as isc
si = isc.make_subindex(path)
si
```
For comparison, we retrieve the "real" historical VSTOXX (sub-) index values.
```
url = 'https://hilpisch.com/vstoxx_eikon_eod_data.csv'
vs = pd.read_csv(url, index_col=0, parse_dates=True)
vs.head()
```
Next, combine the re-calculated VSTOXX 2M values with the historical ones into a new ``DataFrame`` object and add a new column with the absolute differences.
```
comp = pd.concat((si['V6I2'], vs['.V6I2']),
axis=1, join='inner')
comp.index = comp.index.normalize()
comp.columns = ['CALC', 'REAL']
comp['DIFF'] = comp['CALC'] - comp['REAL']
comp
```
The following figure shows the two time series in direct comparison.
```
from pylab import mpl, plt
plt.style.use('seaborn')
mpl.rcParams['font.family'] = 'serif' # set serif font
comp[['CALC', 'REAL']].plot(style=['ro', 'b'], figsize=(10, 6));
```
<p style="font-family: monospace;">Calculated VSTOXX 2M sub-index values vs. real ones.
The following figure shows the point-wise differences between the two time series.
```
import matplotlib.pyplot as plt
plt.figure(figsize=(10, 6))
plt.bar(comp.index, comp['DIFF'])
plt.gcf().autofmt_xdate();
```
<p style="font-family: monospace;">Differences of calculated VSTOXX 2M index values and real ones.
## Calculating the VSTOXX Index
If the values for the sub-indexes VSTOXX 1M and VSTOXX 2M, $V_1$ and $V_2$ say, are given, then the value for the VSTOXX index itself, $V$ say, is calculated by the linear interpolation of $V_1$ and $V_2$:
$$
V = \sqrt{\left(T_1\cdot V_1^2\cdot\left(\frac{N_{T_{2}}-N_{30}}{N_{T_{2}}-N_{T_1}}\right)+T_{2}\cdot V_2^2\cdot\left(\frac{N_{30}-N_{T_1}}{N_{T_{2}}-N_{T_{1}}}\right)\right)\cdot\frac{N_{365}}{N_{30}}}
$$
where
* $N_{T_1}=$ time to expiry of $V_1$´s options series in seconds
* $N_{T_2}=$ time to expiry of $V_2$´s options series in seconds
* $N_{30}= 30$ days in seconds
* $N_{365}=$ time for a standard year in seconds
* $T_1= N_{T_1}/N_{365}$
* $T_2= N_{T_2}/N_{365}$
Recall that the sub-index VSTOXX 1M is not defined on the final settlement day of the underlying option series and the day before. For these dates, we use VSTOXX 2M and VSTOXX 3M as $V_1$ and $V_2$, respectively.
The Python module ``index_vstoxx_calculation.py`` (see the appendix for the module in its entirety) implements the VSTOXX index calculation routine — given the respective sub-index time series data sets. The module starts like usual with some imports.
```
!sed -n 9,12p scripts/index_vstoxx_calculation.py
```
The function ``calculate_vstoxx()`` is the core of the module.
```
import index_vstoxx_calculation as ivc
ivc.calculate_vstoxx??
```
As its single argument, the function takes the path to a CSV file containing historical VSTOXX data for the index itself and the sub-indexes. The re-calculation of it then is as straightforward as follows.
```
import index_vstoxx_calculation as ivc
%time data = ivc.calculate_vstoxx(url)
```
Let us inspect the pandas ``DataFrame`` with the results.
```
data.info()
```
A brief look at the absolute average error of the re-calculation reveals that the implementation yields quite accurate results.
```
## output: average error of re-calculation
data['Difference'].mean()
```
The following figure compares the original `.V2TX` time series with the re-calculated values.
```
## original vs. re-calculated VSTOXX index
data[['.V2TX', 'VSTOXX']].plot(subplots=True, figsize=(10, 6),
style="blue", grid=True);
## original vs. re-calculated VSTOXX index
data[['.V2TX', 'VSTOXX']].plot(figsize=(10, 6), style=['-', '.']);
```
<p style="font-family: monospace;">Historical VSTOXX index values re-calculated vs. real ones.
Finally, the following figure presents the absolute differences. The figure shows that the differences are in general marginal with a few outliers observed here and there.
```
## differences between single values
data['Difference'].plot(figsize=(10, 6), style="r", grid=True,
ylim=(-1, 1));
```
<p style="font-family: monospace;">Differences of historical VSTOXX index values re-calculated vs. real ones.
## Conclusions
This chapter (re-) calculates the VSTOXX volatility index based on historical sub-index values and based on the volatility index definition as derived in the chapter _Model-Free Replication of Variance_. The chapter also shows how to calculate the sub-index values themselves based on EURO STOXX 50 options data. Python code is provided to automatically collect such data from the Eurex web site.
## Python Scripts
### ``index_collect_option_data.py``
```
!cat scripts/index_collect_option_data.py
```
### `index_subindex_calculation.py`
```
!cat scripts/index_subindex_calculation.py
```
### `index_vstoxx_calculation.py`
```
!cat scripts/index_vstoxx_calculation.py
```
<img src="http://hilpisch.com/tpq_logo.png" alt="The Python Quants" width="35%" align="right" border="0"><br>
<a href="http://tpq.io" target="_blank">http://tpq.io</a> | <a href="http://twitter.com/dyjh" target="_blank">@dyjh</a> | <a href="mailto:team@tpq.io">team@tpq.io</a>
|
github_jupyter
|
!sed -n 11,24p scripts/index_collect_option_data.py
import sys; sys.path.append('./scripts/')
import index_collect_option_data as icod
icod.collect_option_series??
icod.get_data??
icod.get_data_from_www??
icod.merge_and_filter??
icod.parse_data??
icod.data_collection??
path = './data/'
import numpy as np
import pandas as pd
import datetime as dt
import warnings; warnings.simplefilter('ignore')
today = dt.datetime.now()
## make sure to hit a business day
target_day = today - dt.timedelta(days=2)
ds = target_day.strftime('%Y%m%d')
ds
URL = 'https://www.eurex.com/ex-en/data/statistics/market-statistics-online/'
URL += '100!onlineStats?productGroupId=13370&productId=69660&viewType=3&'
URL += 'cp=%s&month=%s&year=%s&busDate=%s'
URL % ('Call', 12, 2020, '20201111')
## adjust maturity parameters if necessary
call_data = icod.get_data_from_www(oType='Call', matMonth=12,
matYear=2020, date=ds)
put_data = icod.get_data_from_www(oType='Put', matMonth=12,
matYear=2020, date=ds)
## parse the raw data
calls = icod.parse_data(call_data, target_day)
puts = icod.parse_data(put_data, target_day)
calls.info()
calls.head()
puts.info()
calls = pd.DataFrame(calls.rename(
columns={'Daily settlem. price': 'Call_Price'}
).pop('Call_Price').astype(float))
puts = pd.DataFrame(puts.rename(
columns={'Daily settlem. price': 'Put_Price'}
).pop('Put_Price').astype(float))
dataset = icod.merge_and_filter(puts, calls)
dataset.info()
os = icod.collect_option_series(12, 2020, target_day)
os.info()
# uncomment to initiate the process (takes a while)
%time icod.data_collection(path)
store = pd.HDFStore(path + 'index_option_series.h5', 'r')
store
Dec20 = store['Dec20']
Dec20.info()
store.close()
Dec20.iloc[25:35]
!sed -n 10,15p scripts/index_subindex_calculation.py
import index_subindex_calculation as isc
isc.compute_subindex??
isc.make_subindex??
import index_subindex_calculation as isc
si = isc.make_subindex(path)
si
url = 'https://hilpisch.com/vstoxx_eikon_eod_data.csv'
vs = pd.read_csv(url, index_col=0, parse_dates=True)
vs.head()
comp = pd.concat((si['V6I2'], vs['.V6I2']),
axis=1, join='inner')
comp.index = comp.index.normalize()
comp.columns = ['CALC', 'REAL']
comp['DIFF'] = comp['CALC'] - comp['REAL']
comp
from pylab import mpl, plt
plt.style.use('seaborn')
mpl.rcParams['font.family'] = 'serif' # set serif font
comp[['CALC', 'REAL']].plot(style=['ro', 'b'], figsize=(10, 6));
import matplotlib.pyplot as plt
plt.figure(figsize=(10, 6))
plt.bar(comp.index, comp['DIFF'])
plt.gcf().autofmt_xdate();
!sed -n 9,12p scripts/index_vstoxx_calculation.py
import index_vstoxx_calculation as ivc
ivc.calculate_vstoxx??
import index_vstoxx_calculation as ivc
%time data = ivc.calculate_vstoxx(url)
data.info()
## output: average error of re-calculation
data['Difference'].mean()
## original vs. re-calculated VSTOXX index
data[['.V2TX', 'VSTOXX']].plot(subplots=True, figsize=(10, 6),
style="blue", grid=True);
## original vs. re-calculated VSTOXX index
data[['.V2TX', 'VSTOXX']].plot(figsize=(10, 6), style=['-', '.']);
## differences between single values
data['Difference'].plot(figsize=(10, 6), style="r", grid=True,
ylim=(-1, 1));
!cat scripts/index_collect_option_data.py
!cat scripts/index_subindex_calculation.py
!cat scripts/index_vstoxx_calculation.py
| 0.180612 | 0.982971 |
```
import itertools
import time
import matplotlib.pyplot as plt
import hypermodern_screening as hms
import numpy as np
import pandas as pd
def bar_plot(input_par_array):
"""Simple barplot for frequency of all distinct values in np.ndarry"""
new_list = input_par_array.reshape(-1, 1).tolist()
merged = pd.Series(itertools.chain.from_iterable(new_list))
vc = merged.value_counts().sort_index()
vc /= float(vc.sum())
assert vc.sum() > 0.999
ax = vc.plot(kind='bar')
fig = ax.get_figure()
fig.autofmt_xdate()
"""Draw trajectories without optimization"""
# Create list of n_traj_sample trajectories
n_inputs = 4
n_levels = 6
n_traj_sample = 20
sample_traj_list, _ = hms.trajectory_sample(n_traj_sample, n_inputs, n_levels)
# Compute aggregate distance measure for sample of trajectories.
simple_morris_sample = np.vstack(sample_traj_list)
simple_pairs_dist_matrix = hms.distance_matrix(sample_traj_list)
# Plot frequency of distinct values.
# Monitor differences between distance optimization methods.
bar_plot(simple_morris_sample)
"""Draw trajectories with complete optimization from Ge and Menendez (2017)"""
# Measure data generation time.
# Monitor differences between generation times.
start = time.time()
final_ge_menendez_2014_list, final_gm14_pairs_dist_matrix, _ = hms.final_ge_menendez_2014(
sample_traj_list, n_traj=10
)
end = time.time()
print(end - start)
gm14_array = simple_morris_sample = np.vstack(sample_traj_list)
bar_plot(gm14_array)
"""Draw trajectories with first step from optimization from Ge and Menendez (2017)"""
# Measure data generation time.
start = time.time()
intermediate_ge_menendez_2014_list, intermediate_gm14_pairs_dist_matrix, _ = hms.intermediate_ge_menendez_2014(
sample_traj_list, n_traj=10
)
end = time.time()
print(end - start)
int_gm14_array = np.vstack(intermediate_ge_menendez_2014_list)
bar_plot(int_gm14_array)
"""Draw trajectories with complete optimization from Campolongo (2007)"""
# Measure data generation time.
start = time.time()
campolongo_2007_list, c07_pairs_dist_matrix, _ = hms.campolongo_2007(
sample_traj_list, n_traj=10
)
end = time.time()
print(end - start)
c_array = np.vstack(campolongo_2007_list)
bar_plot(c_array)
# Compute aggregate distance measure for sample of trajectories.
# The first step in Ge and Menendez (2017) yields the best compromise between speed and distance.
final_gm14_total_distance = hms.total_distance(final_gm14_pairs_dist_matrix)
intermediate_gm14_total_distance = hms.total_distance(intermediate_gm14_pairs_dist_matrix)
c07_gm14_total_distance = hms.total_distance(c07_pairs_dist_matrix)
simple_total_distance = hms.total_distance(simple_pairs_dist_matrix)
print(
final_gm14_total_distance,
intermediate_gm14_total_distance,
c07_gm14_total_distance,
simple_total_distance/20
)
```
|
github_jupyter
|
import itertools
import time
import matplotlib.pyplot as plt
import hypermodern_screening as hms
import numpy as np
import pandas as pd
def bar_plot(input_par_array):
"""Simple barplot for frequency of all distinct values in np.ndarry"""
new_list = input_par_array.reshape(-1, 1).tolist()
merged = pd.Series(itertools.chain.from_iterable(new_list))
vc = merged.value_counts().sort_index()
vc /= float(vc.sum())
assert vc.sum() > 0.999
ax = vc.plot(kind='bar')
fig = ax.get_figure()
fig.autofmt_xdate()
"""Draw trajectories without optimization"""
# Create list of n_traj_sample trajectories
n_inputs = 4
n_levels = 6
n_traj_sample = 20
sample_traj_list, _ = hms.trajectory_sample(n_traj_sample, n_inputs, n_levels)
# Compute aggregate distance measure for sample of trajectories.
simple_morris_sample = np.vstack(sample_traj_list)
simple_pairs_dist_matrix = hms.distance_matrix(sample_traj_list)
# Plot frequency of distinct values.
# Monitor differences between distance optimization methods.
bar_plot(simple_morris_sample)
"""Draw trajectories with complete optimization from Ge and Menendez (2017)"""
# Measure data generation time.
# Monitor differences between generation times.
start = time.time()
final_ge_menendez_2014_list, final_gm14_pairs_dist_matrix, _ = hms.final_ge_menendez_2014(
sample_traj_list, n_traj=10
)
end = time.time()
print(end - start)
gm14_array = simple_morris_sample = np.vstack(sample_traj_list)
bar_plot(gm14_array)
"""Draw trajectories with first step from optimization from Ge and Menendez (2017)"""
# Measure data generation time.
start = time.time()
intermediate_ge_menendez_2014_list, intermediate_gm14_pairs_dist_matrix, _ = hms.intermediate_ge_menendez_2014(
sample_traj_list, n_traj=10
)
end = time.time()
print(end - start)
int_gm14_array = np.vstack(intermediate_ge_menendez_2014_list)
bar_plot(int_gm14_array)
"""Draw trajectories with complete optimization from Campolongo (2007)"""
# Measure data generation time.
start = time.time()
campolongo_2007_list, c07_pairs_dist_matrix, _ = hms.campolongo_2007(
sample_traj_list, n_traj=10
)
end = time.time()
print(end - start)
c_array = np.vstack(campolongo_2007_list)
bar_plot(c_array)
# Compute aggregate distance measure for sample of trajectories.
# The first step in Ge and Menendez (2017) yields the best compromise between speed and distance.
final_gm14_total_distance = hms.total_distance(final_gm14_pairs_dist_matrix)
intermediate_gm14_total_distance = hms.total_distance(intermediate_gm14_pairs_dist_matrix)
c07_gm14_total_distance = hms.total_distance(c07_pairs_dist_matrix)
simple_total_distance = hms.total_distance(simple_pairs_dist_matrix)
print(
final_gm14_total_distance,
intermediate_gm14_total_distance,
c07_gm14_total_distance,
simple_total_distance/20
)
| 0.751648 | 0.612976 |
<a href="https://colab.research.google.com/github/EnzoItaliano/calculoNumericoEmPython/blob/master/Lista_1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Universidade Tecnológica Federal do Paraná
Professor: Wellington José Corrêa
Orientando: Enzo Dornelles Italiano
Cálculo Numérico
Conversão de Bases e Aritmética de Ponto Flutuante
# Conversão de Base e Aritmética de Ponto Flutuante
Antes de iniciar é preciso rodar uma vez o código do tópico **Códigos**
##Códigos
```
import math
def dectobinDecimal(n):
n = int(n)
binario = ""
while(True):
binario = binario + str(n%2)
n = n//2
if n == 0:
break
binario = binario[::-1]
print(binario)
def dectobinDecimal1(n):
n = int(n)
binario = ""
while(True):
binario = binario + str(n%2)
n = n//2
if n == 0:
break
binario = binario[::-1]
print(binario, end='')
def dectobinFracionario(n):
n = str(n)
x = n.split(".")
dectobinDecimal1(x[0])
fracionario = ""
n = "0." + x[1]
n = float(n)
while(True):
n = n * 2
if(math.floor(n) == 1):
fracionario = fracionario + "1"
n = n - 1
else:
fracionario = fracionario + "0"
if(n == 0):
break
print("." + fracionario)
def bintodecDecimal(n):
n = str(n)
decimal = 0
n = n[::-1]
tam = len(n)
for i in range(tam):
if n[i] == "1":
decimal = decimal + 2**i
print(decimal)
def bintodecDecimal1(n):
n = str(n)
decimal = 0
n = n[::-1]
tam = len(n)
for i in range(tam):
if n[i] == "1":
decimal = decimal + 2**i
print(decimal, end="")
return decimal
def bintodecFracionario(n):
n = str(n)
x = n.split(".")
bintodecDecimal1(x[0])
temp = 0
tam = len(x[1])
for i in range (tam):
if x[1][i] == "1":
temp = temp + 2 ** -(i+1)
temp = str(temp)
fracionario = temp.split(".")
print("." + fracionario[1])
def dectohex(n):
n = int(n)
hexa = ""
while(True):
if(n % 16 == 10):
hexa = hexa + "A"
elif(n % 16 == 11):
hexa = hexa + "B"
elif(n % 16 == 12):
hexa = hexa + "C"
elif(n % 16 == 13):
hexa = hexa + "D"
elif(n % 16 == 14):
hexa = hexa + "E"
elif(n % 16 == 15):
hexa = hexa + "F"
else:
hexa = hexa + str(n%16)
n = n//16
if n == 0:
break
hexa = hexa[::-1]
print(hexa)
def dectohex1(n):
n = int(n)
hexa = ""
while(True):
if(n % 16 == 10):
hexa = hexa + "A"
elif(n % 16 == 11):
hexa = hexa + "B"
elif(n % 16 == 12):
hexa = hexa + "C"
elif(n % 16 == 13):
hexa = hexa + "D"
elif(n % 16 == 14):
hexa = hexa + "E"
elif(n % 16 == 15):
hexa = hexa + "F"
else:
hexa = hexa + str(n%16)
n = n//16
if n == 0:
break
hexa = hexa[::-1]
print(hexa, end="")
return hexa
def dectohexF(n):
n = str(n)
x = n.split(".")
dectohex1(x[0])
hexa = ""
n = "0." + x[1]
n = float(n)
while(True):
n = n * 16
if(math.floor(n) > 1):
hexa = hexa + str(math.floor(n))
n = n - math.floor(n)
else:
hexa = hexa + "0"
if(n == 0):
break
hexa = str(hexa)
print("." + hexa)
def hexstring2int(n):
decimal = 0
n = n[::-1]
tam = len(n)
for i in range(tam):
if n[i] == "A":
decimal = decimal + 10 * 16**i
elif n[i] == "B":
decimal = decimal + 11 * 16**i
elif n[i] == "C":
decimal = decimal + 12 * 16**i
elif n[i] == "D":
decimal = decimal + 13 * 16**i
elif n[i] == "E":
decimal = decimal + 14 * 16**i
elif n[i] == "F":
decimal = decimal + 15 * 16**i
else:
decimal = decimal + int(n[i]) * 16**i
print(decimal)
def erroAbs(Aex, Aaprox):
Eabs = Aex - Aaprox
if Eabs < 0:
Eabs *= -1
print(Eabs)
def erroRel(Aex, Aaprox):
Eabs = Aex - Aaprox
if Eabs < 0:
Eabs *= -1
Erel = Eabs / Aaprox
print(Erel)
def paraPontoFlut(n):
i = 0
while n > 1:
n *= (10**-1)
i+=1
n = float(n)
return n, i
def trunc(n, p):
x, c = paraPontoFlut(n)
c = int(c)
trunc = ""
x = str(x)
for i in range(p+2): # +2 para o código desconsiderar o 0.
trunc = trunc + x[i]
trunc = float(trunc)
trunc = trunc * 10**c
w, y, z = str(trunc).partition('.')
x = ".".join([w, z[:p]])
print(x)
def arred(n, p):
x, c = paraPontoFlut(n)
x = round(x, p)
x = round(x, p)
x = x * 10**c
print(x)
from decimal import *
def Represenbin(n):
s = int(n[0])
c = 0
i, j = 1, 10
while i < 12 and j > -1:
x = int(n[i])
c = c + x * (2 ** j)
i += 1
j -= 1
f = 0
i, j = 12, -1
while i < 64 and j > -53:
x = int(n[i])
f = f + x * (2**j)
i += 1
j -= 1
result = Decimal((-1)**s * 2**(c-1023) * (1+f))
print(result)
```
## 1. Conversão de Base
### 1.1 Conversão de base decimal para base binária.
a)Parte inteira: usaremos a função dectobinDecimal(n), onde n é o número inteiro na base 10.
Exemplo: Converta o número 42 na base 2.
```
dectobinDecimal(42)
```
Assim, $42_2$ = 101010
b) Parte decimal: Usaremos a função
dectobinFracionario(n) onde n é o número fracionário na base 10.
Exemplo: Converta o número 8,7 na base 2.
```
dectobinFracionario(8.7)
```
Portanto, $8,7_2 \approx$ 1000,1011
### 1.2 Conversão de base binária para a base decimal.
Como na seção anterior, dividiremos em duas partes.
a) Parte inteira: Nesta situação usaremos o comando bintodecDecimal(n), de modo que n é o número na base 2.
Exemplo: Converta o número 101010 na base decimal.
```
bintodecDecimal(101010)
```
Portanto, $101010_{10}$ = 42.
b) Parte fracionária: Agora usaremos a função bintodecFracionario(n), onde n é o número na base 2.
Exemplo: Converta o número 1000,1011 na base decimal.
```
bintodecFracionario(1000.1011)
```
Portanto, $1000,1011_{10} \approx$ 8,7.
### 1.3 Conversão de base decimal para base hexadecimal
a) Parte inteira: Usaremos a função dectohex(n) onde n é o número inteiro na base 10.
Exemplo: Converta o número 16435930 na base hexadecimal.
```
dectohex(16435930)
```
b) Parte decimal: Usaremos o comando dectohexF(n) onde n é o número fracionário na base 10.
Exemplo: Converta o número 10,75 na base hexadecimal.
```
dectohexF(10.75)
```
### 1.4 Conversão de base hexadecimal para base decimal
Neste caso, basta usar o comando hexstring2int("n") onde n é o número na base hexadecimal.
Exemplo: Converta o número hexadecimal "FACADA" na base decimal.
```
hexstring2int("FACADA")
```
## 2. Aritmética de Ponto Flutuante
### 2.1 Erro absoluto e relativo
No que segue, denotaremos a aproximação de p por p1.
Usaremos os comandos:
Erro absoluto: erroAbs(Aex,Aaprox)
Erro relativo: erroRel(Aex,Aaprox)
Exemplo: Determine o erro absoluto e o erro relativo na aproximação de $Aex = \sqrt{2}$ por $Aaprox = 1,41$.
```
erroAbs(math.sqrt(2), 1.41)
erroRel(math.sqrt(2), 1.41)
```
### 2.2 Truncamento e Arredondamento
Para tanto, denotemos por “n” o número a ser digitado e “p” o número de algarismos a ser estabelecido pelo truncamento e arredondamento. Temos os seguintes comandos:
Truncamento: trunc(n, p)
Arredondamento: arred(n, p)
Exemplo: Realize a aritmética de truncamento com três algarismos para o número 1,23675.
```
trunc(1.23675, 3)
```
Exemplo: Empregue a aritmética de arredondamento com três algarismos para os números 3,2365 e 3,2344.
```
arred(3.2365, 3)
arred(3.2344, 3)
```
### 2.3 Representação Numérica de Ponto Flutuante
Pelo padrão IEEE 754 podemos ter uma representação de números fracionários binários bem maior.
A função aqui é Represenbin(n), onde n é o número binário a ser convertido para decimal.
Exemplo: Converta o número 0100000000111011100100010000000000000000000000000000000000000000
```
Represenbin('0100000000111011100100010000000000000000000000000000000000000000')
# Represenbin('01' + 8*'0' + '11101110010001' + 40*'0')
```
|
github_jupyter
|
import math
def dectobinDecimal(n):
n = int(n)
binario = ""
while(True):
binario = binario + str(n%2)
n = n//2
if n == 0:
break
binario = binario[::-1]
print(binario)
def dectobinDecimal1(n):
n = int(n)
binario = ""
while(True):
binario = binario + str(n%2)
n = n//2
if n == 0:
break
binario = binario[::-1]
print(binario, end='')
def dectobinFracionario(n):
n = str(n)
x = n.split(".")
dectobinDecimal1(x[0])
fracionario = ""
n = "0." + x[1]
n = float(n)
while(True):
n = n * 2
if(math.floor(n) == 1):
fracionario = fracionario + "1"
n = n - 1
else:
fracionario = fracionario + "0"
if(n == 0):
break
print("." + fracionario)
def bintodecDecimal(n):
n = str(n)
decimal = 0
n = n[::-1]
tam = len(n)
for i in range(tam):
if n[i] == "1":
decimal = decimal + 2**i
print(decimal)
def bintodecDecimal1(n):
n = str(n)
decimal = 0
n = n[::-1]
tam = len(n)
for i in range(tam):
if n[i] == "1":
decimal = decimal + 2**i
print(decimal, end="")
return decimal
def bintodecFracionario(n):
n = str(n)
x = n.split(".")
bintodecDecimal1(x[0])
temp = 0
tam = len(x[1])
for i in range (tam):
if x[1][i] == "1":
temp = temp + 2 ** -(i+1)
temp = str(temp)
fracionario = temp.split(".")
print("." + fracionario[1])
def dectohex(n):
n = int(n)
hexa = ""
while(True):
if(n % 16 == 10):
hexa = hexa + "A"
elif(n % 16 == 11):
hexa = hexa + "B"
elif(n % 16 == 12):
hexa = hexa + "C"
elif(n % 16 == 13):
hexa = hexa + "D"
elif(n % 16 == 14):
hexa = hexa + "E"
elif(n % 16 == 15):
hexa = hexa + "F"
else:
hexa = hexa + str(n%16)
n = n//16
if n == 0:
break
hexa = hexa[::-1]
print(hexa)
def dectohex1(n):
n = int(n)
hexa = ""
while(True):
if(n % 16 == 10):
hexa = hexa + "A"
elif(n % 16 == 11):
hexa = hexa + "B"
elif(n % 16 == 12):
hexa = hexa + "C"
elif(n % 16 == 13):
hexa = hexa + "D"
elif(n % 16 == 14):
hexa = hexa + "E"
elif(n % 16 == 15):
hexa = hexa + "F"
else:
hexa = hexa + str(n%16)
n = n//16
if n == 0:
break
hexa = hexa[::-1]
print(hexa, end="")
return hexa
def dectohexF(n):
n = str(n)
x = n.split(".")
dectohex1(x[0])
hexa = ""
n = "0." + x[1]
n = float(n)
while(True):
n = n * 16
if(math.floor(n) > 1):
hexa = hexa + str(math.floor(n))
n = n - math.floor(n)
else:
hexa = hexa + "0"
if(n == 0):
break
hexa = str(hexa)
print("." + hexa)
def hexstring2int(n):
decimal = 0
n = n[::-1]
tam = len(n)
for i in range(tam):
if n[i] == "A":
decimal = decimal + 10 * 16**i
elif n[i] == "B":
decimal = decimal + 11 * 16**i
elif n[i] == "C":
decimal = decimal + 12 * 16**i
elif n[i] == "D":
decimal = decimal + 13 * 16**i
elif n[i] == "E":
decimal = decimal + 14 * 16**i
elif n[i] == "F":
decimal = decimal + 15 * 16**i
else:
decimal = decimal + int(n[i]) * 16**i
print(decimal)
def erroAbs(Aex, Aaprox):
Eabs = Aex - Aaprox
if Eabs < 0:
Eabs *= -1
print(Eabs)
def erroRel(Aex, Aaprox):
Eabs = Aex - Aaprox
if Eabs < 0:
Eabs *= -1
Erel = Eabs / Aaprox
print(Erel)
def paraPontoFlut(n):
i = 0
while n > 1:
n *= (10**-1)
i+=1
n = float(n)
return n, i
def trunc(n, p):
x, c = paraPontoFlut(n)
c = int(c)
trunc = ""
x = str(x)
for i in range(p+2): # +2 para o código desconsiderar o 0.
trunc = trunc + x[i]
trunc = float(trunc)
trunc = trunc * 10**c
w, y, z = str(trunc).partition('.')
x = ".".join([w, z[:p]])
print(x)
def arred(n, p):
x, c = paraPontoFlut(n)
x = round(x, p)
x = round(x, p)
x = x * 10**c
print(x)
from decimal import *
def Represenbin(n):
s = int(n[0])
c = 0
i, j = 1, 10
while i < 12 and j > -1:
x = int(n[i])
c = c + x * (2 ** j)
i += 1
j -= 1
f = 0
i, j = 12, -1
while i < 64 and j > -53:
x = int(n[i])
f = f + x * (2**j)
i += 1
j -= 1
result = Decimal((-1)**s * 2**(c-1023) * (1+f))
print(result)
dectobinDecimal(42)
dectobinFracionario(8.7)
bintodecDecimal(101010)
bintodecFracionario(1000.1011)
dectohex(16435930)
dectohexF(10.75)
hexstring2int("FACADA")
erroAbs(math.sqrt(2), 1.41)
erroRel(math.sqrt(2), 1.41)
trunc(1.23675, 3)
arred(3.2365, 3)
arred(3.2344, 3)
Represenbin('0100000000111011100100010000000000000000000000000000000000000000')
# Represenbin('01' + 8*'0' + '11101110010001' + 40*'0')
| 0.211417 | 0.701994 |
# Test Access to Earth Engine
Run the code blocks below to test if the notebook server is authorized to communicate with the Earth Engine backend servers.
First, check if the IPython Widgets library is available on the server.
```
# Code to check the IPython Widgets library.
try:
import ipywidgets
except ImportError:
print('The IPython Widgets library is not available on this server.\n'
'Please see https://github.com/jupyter-widgets/ipywidgets '
'for information on installing the library.')
raise
print('The IPython Widgets library (version {0}) is available on this server.'.format(
ipywidgets.__version__
))
```
Next, check if the Earth Engine API is available on the server.
```
# Code to check the Earth Engine API library.
try:
import ee
except ImportError:
print('The Earth Engine Python API library is not available on this server.\n'
'Please see https://developers.google.com/earth-engine/python_install '
'for information on installing the library.')
raise
print('The Earth Engine Python API (version {0}) is available on this server.'.format(
ee.__version__
))
```
Finally, check if the notebook server is authorized to access the Earth Engine backend servers.
```
# Code to check if authorized to access Earth Engine.
import cStringIO
import os
import urllib
from IPython import display
# Define layouts used by the form.
row_wide_layout = ipywidgets.Layout(flex_flow="row nowrap", align_items="center", width="100%")
column_wide_layout = ipywidgets.Layout(flex_flow="column nowrap", align_items="center", width="100%")
column_auto_layout = ipywidgets.Layout(flex_flow="column nowrap", align_items="center", width="auto")
form_definition = {'form': None}
response_box = ipywidgets.HTML('')
def isAuthorized():
try:
ee.Initialize()
test = ee.Image(0).getInfo()
except:
return False
return True
def ShowForm(auth_status_button, instructions):
"""Show a form to the user."""
form_definition['form'] = ipywidgets.VBox([
auth_status_button,
instructions,
ipywidgets.VBox([response_box], layout=row_wide_layout)
], layout=column_wide_layout)
display.display(form_definition.get('form'))
def ShowAuthorizedForm():
"""Show a form for a server that is currently authorized to access Earth Engine."""
def revoke_credentials(sender):
credentials = ee.oauth.get_credentials_path()
if os.path.exists(credentials):
os.remove(credentials)
response_box.value = ''
Init()
auth_status_button = ipywidgets.Button(
layout=column_wide_layout,
disabled=True,
description='The server is authorized to access Earth Engine',
button_style='success',
icon='check'
)
instructions = ipywidgets.Button(
layout = row_wide_layout,
description = 'Click here to revoke authorization',
disabled = False,
)
instructions.on_click(revoke_credentials)
ShowForm(auth_status_button, instructions)
def ShowUnauthorizedForm():
"""Show a form for a server that is not currently authorized to access Earth Engine."""
auth_status_button = ipywidgets.Button(
layout=column_wide_layout,
button_style='danger',
description='The server is not authorized to access Earth Engine',
disabled=True
)
auth_link = ipywidgets.HTML(
'<a href="{url}" target="auth">Open Authentication Tab</a><br/>'
.format(url=ee.oauth.get_authorization_url()
)
)
instructions = ipywidgets.VBox(
[
ipywidgets.HTML(
'Click on the link below to start the authentication and authorization process. '
'Once you have received an authorization code, use it to replace the '
'REPLACE_WITH_AUTH_CODE in the code cell below and run the cell.'
),
auth_link,
],
layout=column_auto_layout
)
ShowForm(auth_status_button, instructions)
def Init():
# If a form is currently displayed, close it.
if form_definition.get('form'):
form_definition['form'].close()
# Display the appropriate form according to whether the server is authorized.
if isAuthorized():
ShowAuthorizedForm()
else:
ShowUnauthorizedForm()
Init()
```
If the server **is authorized**, you do not need to run the next code cell.
If the server **is not authorized**:
1. Copy the authentication code generated in the previous step.
2. Replace the REPLACE_WITH_AUTH_CODE string in the cell below with the authentication code.
3. Run the code cell to save authentication credentials.
```
auth_code = 'REPLACE_WITH_AUTH_CODE'
response_box = ipywidgets.HTML('')
try:
token = ee.oauth.request_token(auth_code.strip())
ee.oauth.write_token(token)
if isAuthorized():
Init()
else:
response_box.value = '<font color="red">{0}</font>'.format(
'The account was authenticated, but does not have permission to access Earth Engine.'
)
except Exception as e:
response_box.value = '<font color="red">{0}</font>'.format(e)
response_box
```
Once the server is authorized, you can retrieve data from Earth Engine and use it in the notebook.
```
# Code to display an Earth Engine generated image.
from IPython.display import Image
url = ee.Image("CGIAR/SRTM90_V4").getThumbUrl({'min':0, 'max':3000})
Image(url=url)
```
|
github_jupyter
|
# Code to check the IPython Widgets library.
try:
import ipywidgets
except ImportError:
print('The IPython Widgets library is not available on this server.\n'
'Please see https://github.com/jupyter-widgets/ipywidgets '
'for information on installing the library.')
raise
print('The IPython Widgets library (version {0}) is available on this server.'.format(
ipywidgets.__version__
))
# Code to check the Earth Engine API library.
try:
import ee
except ImportError:
print('The Earth Engine Python API library is not available on this server.\n'
'Please see https://developers.google.com/earth-engine/python_install '
'for information on installing the library.')
raise
print('The Earth Engine Python API (version {0}) is available on this server.'.format(
ee.__version__
))
# Code to check if authorized to access Earth Engine.
import cStringIO
import os
import urllib
from IPython import display
# Define layouts used by the form.
row_wide_layout = ipywidgets.Layout(flex_flow="row nowrap", align_items="center", width="100%")
column_wide_layout = ipywidgets.Layout(flex_flow="column nowrap", align_items="center", width="100%")
column_auto_layout = ipywidgets.Layout(flex_flow="column nowrap", align_items="center", width="auto")
form_definition = {'form': None}
response_box = ipywidgets.HTML('')
def isAuthorized():
try:
ee.Initialize()
test = ee.Image(0).getInfo()
except:
return False
return True
def ShowForm(auth_status_button, instructions):
"""Show a form to the user."""
form_definition['form'] = ipywidgets.VBox([
auth_status_button,
instructions,
ipywidgets.VBox([response_box], layout=row_wide_layout)
], layout=column_wide_layout)
display.display(form_definition.get('form'))
def ShowAuthorizedForm():
"""Show a form for a server that is currently authorized to access Earth Engine."""
def revoke_credentials(sender):
credentials = ee.oauth.get_credentials_path()
if os.path.exists(credentials):
os.remove(credentials)
response_box.value = ''
Init()
auth_status_button = ipywidgets.Button(
layout=column_wide_layout,
disabled=True,
description='The server is authorized to access Earth Engine',
button_style='success',
icon='check'
)
instructions = ipywidgets.Button(
layout = row_wide_layout,
description = 'Click here to revoke authorization',
disabled = False,
)
instructions.on_click(revoke_credentials)
ShowForm(auth_status_button, instructions)
def ShowUnauthorizedForm():
"""Show a form for a server that is not currently authorized to access Earth Engine."""
auth_status_button = ipywidgets.Button(
layout=column_wide_layout,
button_style='danger',
description='The server is not authorized to access Earth Engine',
disabled=True
)
auth_link = ipywidgets.HTML(
'<a href="{url}" target="auth">Open Authentication Tab</a><br/>'
.format(url=ee.oauth.get_authorization_url()
)
)
instructions = ipywidgets.VBox(
[
ipywidgets.HTML(
'Click on the link below to start the authentication and authorization process. '
'Once you have received an authorization code, use it to replace the '
'REPLACE_WITH_AUTH_CODE in the code cell below and run the cell.'
),
auth_link,
],
layout=column_auto_layout
)
ShowForm(auth_status_button, instructions)
def Init():
# If a form is currently displayed, close it.
if form_definition.get('form'):
form_definition['form'].close()
# Display the appropriate form according to whether the server is authorized.
if isAuthorized():
ShowAuthorizedForm()
else:
ShowUnauthorizedForm()
Init()
auth_code = 'REPLACE_WITH_AUTH_CODE'
response_box = ipywidgets.HTML('')
try:
token = ee.oauth.request_token(auth_code.strip())
ee.oauth.write_token(token)
if isAuthorized():
Init()
else:
response_box.value = '<font color="red">{0}</font>'.format(
'The account was authenticated, but does not have permission to access Earth Engine.'
)
except Exception as e:
response_box.value = '<font color="red">{0}</font>'.format(e)
response_box
# Code to display an Earth Engine generated image.
from IPython.display import Image
url = ee.Image("CGIAR/SRTM90_V4").getThumbUrl({'min':0, 'max':3000})
Image(url=url)
| 0.466359 | 0.770033 |
Lambda School Data Science
*Unit 4, Sprint 3, Module 2*
---
# Convolutional Neural Networks (Prepare)
> Convolutional networks are simply neural networks that use convolution in place of general matrix multiplication in at least one of their layers. *Goodfellow, et al.*
## Learning Objectives
- <a href="#p1">Part 1: </a>Describe convolution and pooling
- <a href="#p2">Part 2: </a>Apply a convolutional neural network to a classification task
- <a href="#p3">Part 3: </a>Use a pre-trained convolution neural network for image classification
Modern __computer vision__ approaches rely heavily on convolutions as both a dimensionality reduction and feature extraction method. Before we dive into convolutions, let's talk about some of the common computer vision applications:
* Classification [(Hot Dog or Not Dog)](https://www.youtube.com/watch?v=ACmydtFDTGs)
* Object Detection [(YOLO)](https://www.youtube.com/watch?v=MPU2HistivI)
* Pose Estimation [(PoseNet)](https://ai.googleblog.com/2019/08/on-device-real-time-hand-tracking-with.html)
* Facial Recognition [Emotion Detection](https://www.cbronline.com/wp-content/uploads/2018/05/Mona-lIsa-test-570x300.jpg)
* and *countless* more
We are going to focus on classification and pre-trained classification today. What are some of the applications of image classification?
```
from IPython.display import YouTubeVideo
YouTubeVideo('MPU2HistivI', width=600, height=400)
```
# Convolution & Pooling (Learn)
<a id="p1"></a>
## Overview
Like neural networks themselves, CNNs are inspired by biology - specifically, the receptive fields of the visual cortex.
Put roughly, in a real brain the neurons in the visual cortex *specialize* to be receptive to certain regions, shapes, colors, orientations, and other common visual features. In a sense, the very structure of our cognitive system transforms raw visual input, and sends it to neurons that specialize in handling particular subsets of it.
CNNs imitate this approach by applying a convolution. A convolution is an operation on two functions that produces a third function, showing how one function modifies another. Convolutions have a [variety of nice mathematical properties](https://en.wikipedia.org/wiki/Convolution#Properties) - commutativity, associativity, distributivity, and more. Applying a convolution effectively transforms the "shape" of the input.
One common confusion - the term "convolution" is used to refer to both the process of computing the third (joint) function and the process of applying it. In our context, it's more useful to think of it as an application, again loosely analogous to the mapping from visual field to receptive areas of the cortex in a real animal.
```
from IPython.display import YouTubeVideo
YouTubeVideo('IOHayh06LJ4', width=600, height=400)
```
## Follow Along
Let's try to do some convolutions and pooling
### Convolution
Consider blurring an image - assume the image is represented as a matrix of numbers, where each number corresponds to the color value of a pixel.
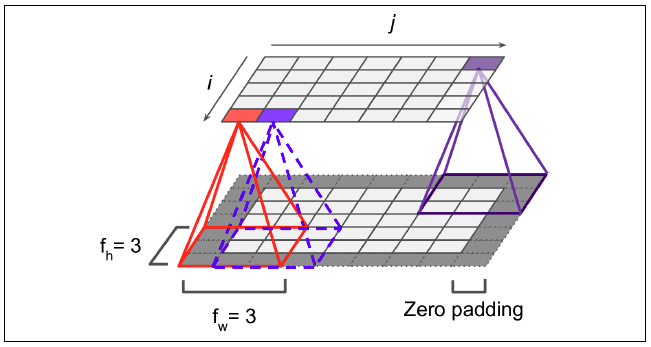
*Image Credits from __Hands on Machine Learning with Sckit-Learn, Keras & TensorFlow__*
Helpful Terms:
- __Filter__: The weights (parameters) we will apply to our input image.
- __Stride__: How the filter moves across the image
- __Padding__: Zeros (or other values) around the the input image border (kind of like a frame of zeros).
```
import imageio
import matplotlib.pyplot as plt
from skimage import color, io
from skimage.exposure import rescale_intensity
austen = io.imread('https://dl.airtable.com/S1InFmIhQBypHBL0BICi_austen.jpg')
austen_grayscale = rescale_intensity(color.rgb2gray(austen))
austen_grayscale.shape
plt.imshow(austen_grayscale, cmap="gray");
austen_grayscale.shape
import numpy as np
import scipy.ndimage as nd
horizontal_edge_convolution = np.array([[1,1,1,1,1],
[0,0,0,0,0],
[-1,-1,-1,-1,-1]])
vertical_edge_convolution = np.array([[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1]])
austen_edges = nd.convolve(austen_grayscale, horizontal_edge_convolution)
austen_edges.shape
plt.imshow(austen_edges, cmap="gray");
```
### Pooling Layer
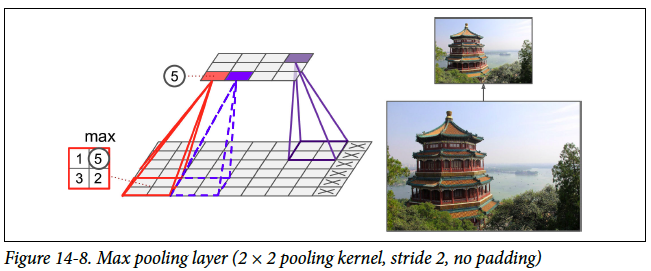
*Image Credits from __Hands on Machine Learning with Sckit-Learn, Keras & TensorFlow__*
We use Pooling Layers to reduce the dimensionality of the feature maps. We get smaller and smaller feature set by apply convolutions and then pooling layers.
Let's take a look very simple example using Austen's pic.
```
from skimage.measure import block_reduce
reduced = block_reduce(austen_grayscale, (2,2), np.max)
plt.imshow(reduced, cmap="gray");
```
## Challenge
You will be expected to be able to describe convolution.
# CNNs for Classification (Learn)
## Overview
### Typical CNN Architecture

The first stage of a CNN is, unsurprisingly, a convolution - specifically, a transformation that maps regions of the input image to neurons responsible for receiving them. The convolutional layer can be visualized as follows:
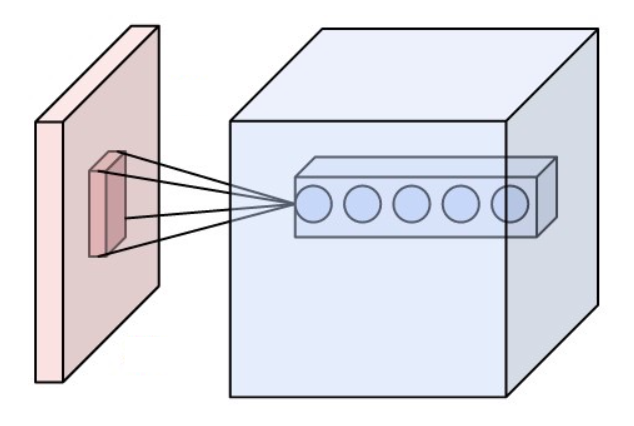
The red represents the original input image, and the blue the neurons that correspond.
As shown in the first image, a CNN can have multiple rounds of convolutions, [downsampling](https://en.wikipedia.org/wiki/Downsampling_(signal_processing)) (a digital signal processing technique that effectively reduces the information by passing through a filter), and then eventually a fully connected neural network and output layer. Typical output layers for a CNN would be oriented towards classification or detection problems - e.g. "does this picture contain a cat, a dog, or some other animal?"
#### A Convolution in Action
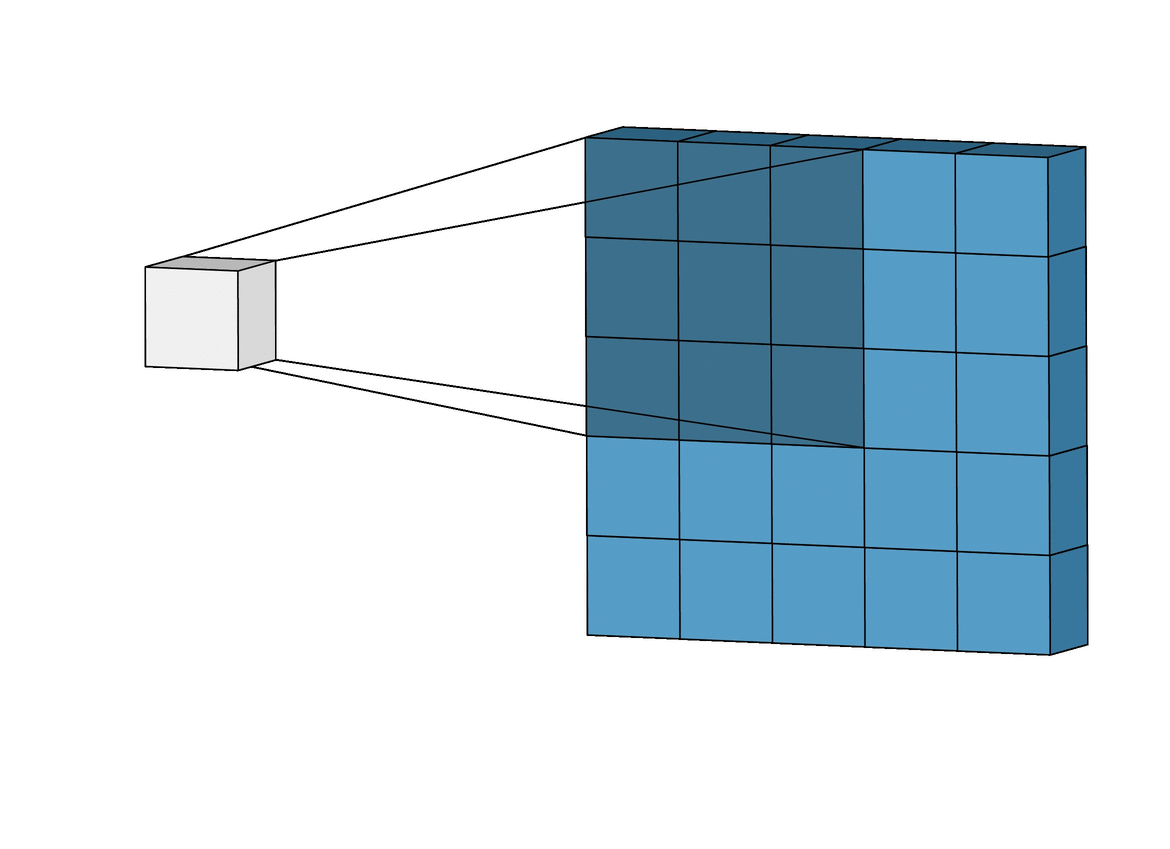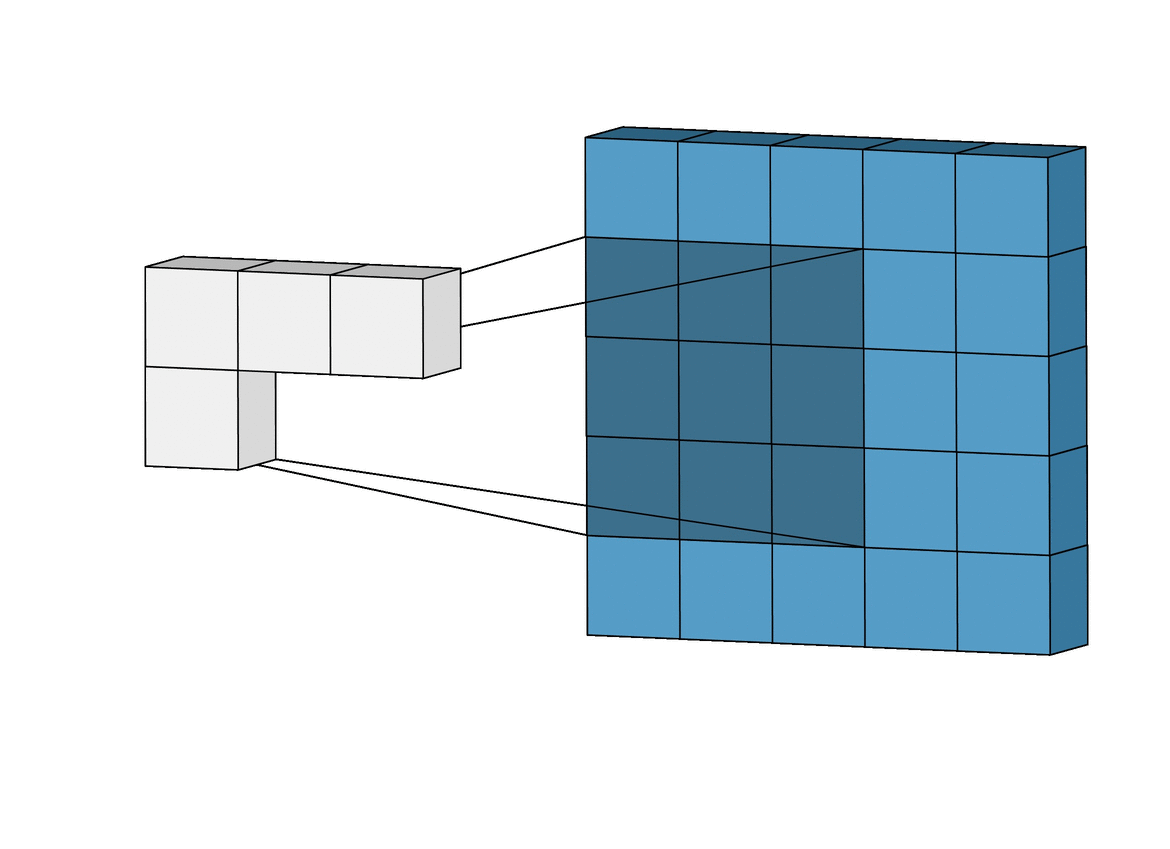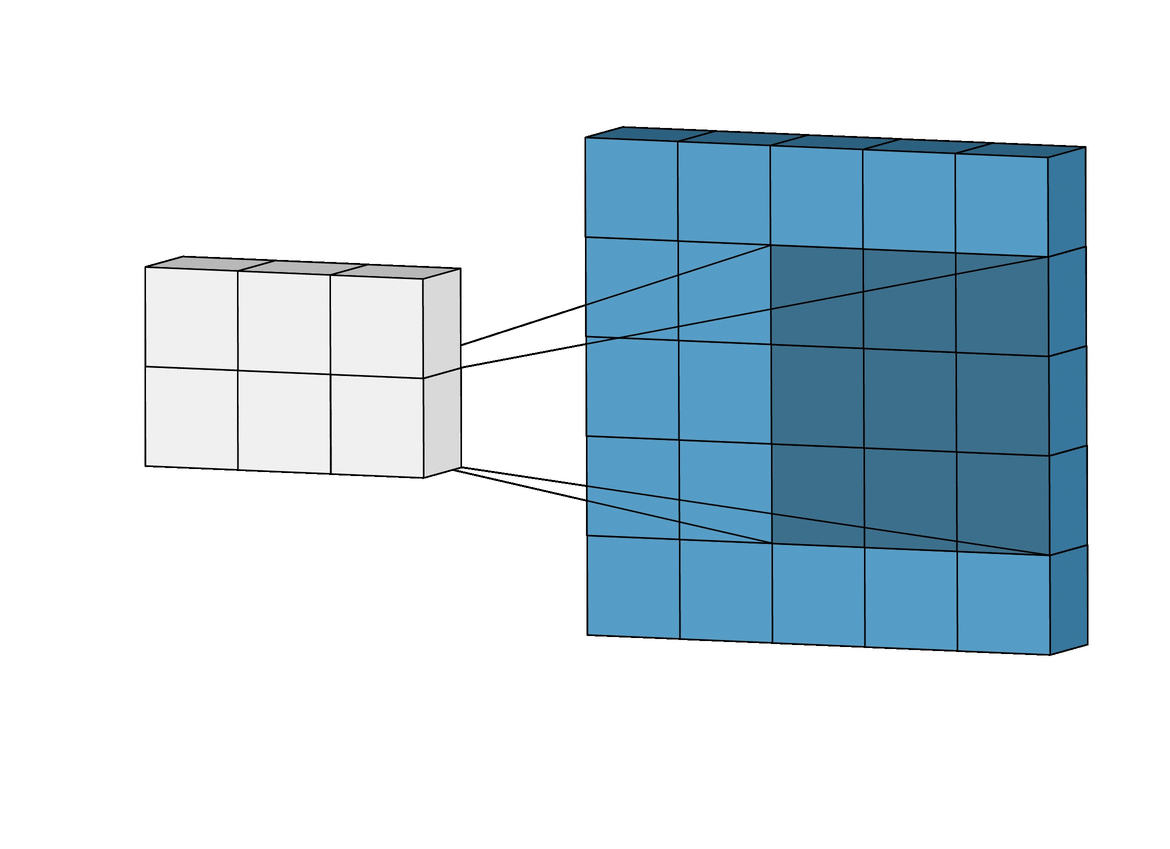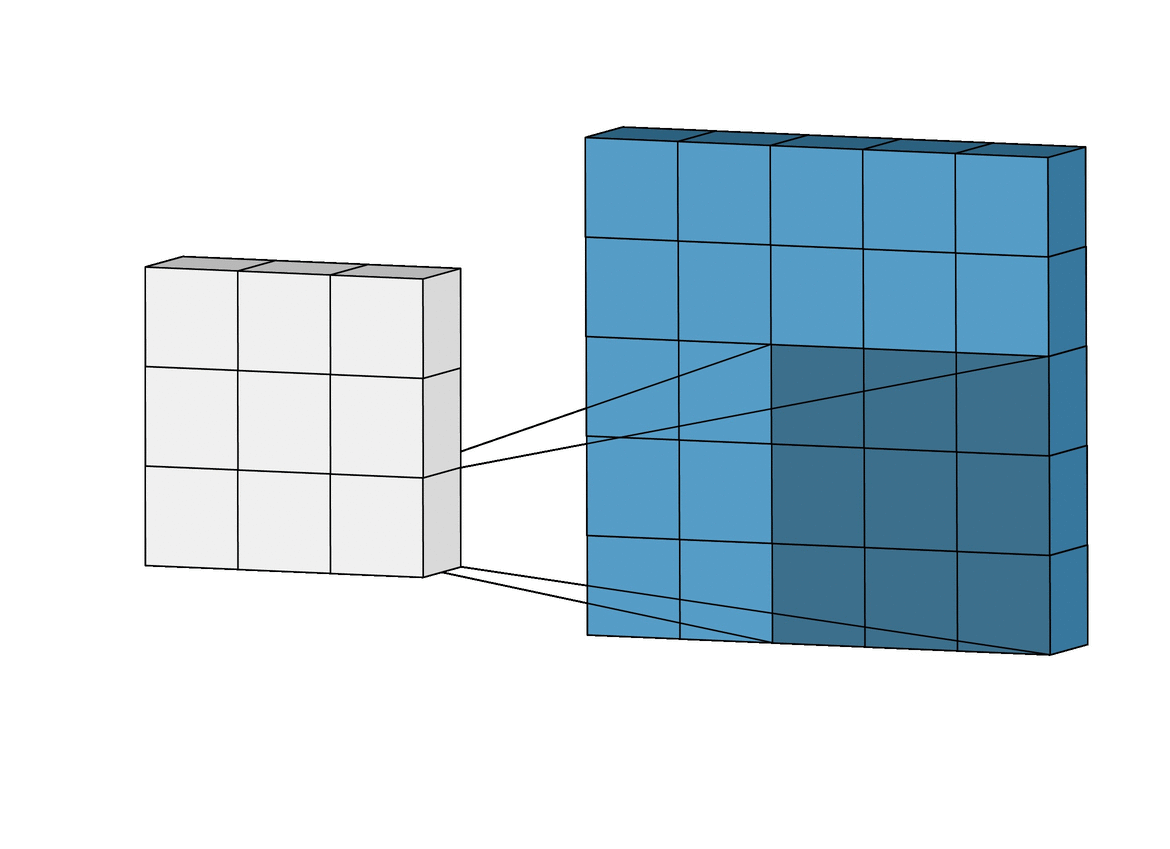
Why are CNNs so popular?
1. Compared to prior image learning techniques, they require relatively little image preprocessing (cropping/centering, normalizing, etc.)
2. Relatedly, they are *robust* to all sorts of common problems in images (shifts, lighting, etc.)
Actually training a cutting edge image classification CNN is nontrivial computationally - the good news is, with transfer learning, we can get one "off-the-shelf"!
## Follow Along
```
from tensorflow.keras import datasets
from tensorflow.keras.models import Sequential, Model # <- May Use
from tensorflow.keras.layers import Dense, Conv2D, MaxPooling2D, Flatten
(train_images, train_labels), (test_images, test_labels) = datasets.cifar10.load_data()
# Normalize pixel values to be between 0 and 1
train_images, test_images = train_images / 255.0, test_images / 255.0
class_names = ['airplane', 'automobile', 'bird', 'cat', 'deer',
'dog', 'frog', 'horse', 'ship', 'truck']
plt.figure(figsize=(10,10))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(train_images[i], cmap=plt.cm.binary)
# The CIFAR labels happen to be arrays,
# which is why you need the extra index
plt.xlabel(class_names[train_labels[i][0]])
plt.show()
train_images[0].shape
train_labels[1]
32*32*3
# Setup Architecture
model = Sequential()
model.add(Conv2D(32, (3,3), activation='relu', input_shape=(32,32,3)))
model.add(MaxPooling2D((2,2)))
model.add(Conv2D(64, (3,3), activation='relu'))
model.add(MaxPooling2D((2,2)))
model.add(Conv2D(64, (3,3), activation='relu'))
model.add(Flatten())
model.add(Dense(64, activation='relu'))
model.add(Dense(10, activation='softmax'))
model.summary()
# Compile Model
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# Fit Model
model.fit(train_images, train_labels, epochs=10, validation_data=(test_images, test_labels))
# Evaluate Model
test_loss, test_acc = model.evaluate(test_images, test_labels, verbose=2)
```
## Challenge
You will apply CNNs to a classification task in the module project.
# Transfer Learning for Image Classification (Learn)
## Overview
### Transfer Learning Repositories
#### TensorFlow Hub
"A library for reusable machine learning modules"
This lets you quickly take advantage of a model that was trained with thousands of GPU hours. It also enables transfer learning - reusing a part of a trained model (called a module) that includes weights and assets, but also training the overall model some yourself with your own data. The advantages are fairly clear - you can use less training data, have faster training, and have a model that generalizes better.
https://www.tensorflow.org/hub/
TensorFlow Hub is very bleeding edge, and while there's a good amount of documentation out there, it's not always updated or consistent. You'll have to use your problem-solving skills if you want to use it!
#### Keras API - Applications
> Keras Applications are deep learning models that are made available alongside pre-trained weights. These models can be used for prediction, feature extraction, and fine-tuning.
There is a decent selection of important benchmark models. We'll focus on an image classifier: ResNet50.
## Follow Along
```
import numpy as np
from tensorflow.keras.applications.resnet50 import ResNet50
from tensorflow.keras.preprocessing import image
from tensorflow.keras.applications.resnet50 import preprocess_input, decode_predictions
def process_img_path(img_path):
return image.load_img(img_path, target_size=(224, 224))
def img_contains_banana(img):
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
x = preprocess_input(x)
model = ResNet50(weights='imagenet')
features = model.predict(x)
results = decode_predictions(features, top=3)[0]
print(results)
for entry in results:
if entry[1] == 'banana':
return entry[2]
return 0.0
import requests
image_urls = ["https://github.com/LambdaSchool/ML-YouOnlyLookOnce/raw/master/sample_data/negative_examples/example11.jpeg",
"https://github.com/LambdaSchool/ML-YouOnlyLookOnce/raw/master/sample_data/positive_examples/example0.jpeg"]
for _id,img in enumerate(image_urls):
r = requests.get(img)
with open(f'example{_id}.jpg', 'wb') as f:
f.write(r.content)
from IPython.display import Image
Image(filename='./example0.jpg', width=600)
img_contains_banana(process_img_path('example0.jpg'))
Image(filename='example1.jpg', width=600)
img_contains_banana(process_img_path('example1.jpg'))
```
Notice that, while it gets it right, the confidence for the banana image is fairly low. That's because so much of the image is "not-banana"! How can this be improved? Bounding boxes to center on items of interest.
## Challenge
You will be expected to apply a pretrained model to a classificaiton problem today.
# Review
- <a href="#p1">Part 1: </a>Describe convolution and pooling
* A Convolution is a function applied to another function to produce a third function
* Convolutional Kernels are typically 'learned' during the process of training a Convolution Neural Network
* Pooling is a dimensionality reduction technique that uses either Max or Average of a feature map region to downsample data
- <a href="#p2">Part 2: </a>Apply a convolutional neural network to a classification task
* Keras has layers for convolutions :)
- <a href="#p3">Part 3: </a>Transfer Learning for Image Classification
* Check out both pretinaed models available in Keras & TensorFlow Hub
# Sources
- *_Deep Learning_*. Goodfellow *et al.*
- *Hands-on Machine Learnign with Scikit-Learn, Keras & Tensorflow*
- [Keras CNN Tutorial](https://www.tensorflow.org/tutorials/images/cnn)
- [Tensorflow + Keras](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Conv2D)
- [Convolution Wiki](https://en.wikipedia.org/wiki/Convolution)
- [Keras Conv2D: Working with CNN 2D Convolutions in Keras](https://missinglink.ai/guides/keras/keras-conv2d-working-cnn-2d-convolutions-keras/)
- [Intuitively Understanding Convolutions for Deep Learning](https://towardsdatascience.com/intuitively-understanding-convolutions-for-deep-learning-1f6f42faee1)
- [A Beginner's Guide to Understanding Convolutional Neural Networks Part 2](https://adeshpande3.github.io/A-Beginner%27s-Guide-To-Understanding-Convolutional-Neural-Networks-Part-2/)
|
github_jupyter
|
from IPython.display import YouTubeVideo
YouTubeVideo('MPU2HistivI', width=600, height=400)
from IPython.display import YouTubeVideo
YouTubeVideo('IOHayh06LJ4', width=600, height=400)
import imageio
import matplotlib.pyplot as plt
from skimage import color, io
from skimage.exposure import rescale_intensity
austen = io.imread('https://dl.airtable.com/S1InFmIhQBypHBL0BICi_austen.jpg')
austen_grayscale = rescale_intensity(color.rgb2gray(austen))
austen_grayscale.shape
plt.imshow(austen_grayscale, cmap="gray");
austen_grayscale.shape
import numpy as np
import scipy.ndimage as nd
horizontal_edge_convolution = np.array([[1,1,1,1,1],
[0,0,0,0,0],
[-1,-1,-1,-1,-1]])
vertical_edge_convolution = np.array([[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1]])
austen_edges = nd.convolve(austen_grayscale, horizontal_edge_convolution)
austen_edges.shape
plt.imshow(austen_edges, cmap="gray");
from skimage.measure import block_reduce
reduced = block_reduce(austen_grayscale, (2,2), np.max)
plt.imshow(reduced, cmap="gray");
from tensorflow.keras import datasets
from tensorflow.keras.models import Sequential, Model # <- May Use
from tensorflow.keras.layers import Dense, Conv2D, MaxPooling2D, Flatten
(train_images, train_labels), (test_images, test_labels) = datasets.cifar10.load_data()
# Normalize pixel values to be between 0 and 1
train_images, test_images = train_images / 255.0, test_images / 255.0
class_names = ['airplane', 'automobile', 'bird', 'cat', 'deer',
'dog', 'frog', 'horse', 'ship', 'truck']
plt.figure(figsize=(10,10))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(train_images[i], cmap=plt.cm.binary)
# The CIFAR labels happen to be arrays,
# which is why you need the extra index
plt.xlabel(class_names[train_labels[i][0]])
plt.show()
train_images[0].shape
train_labels[1]
32*32*3
# Setup Architecture
model = Sequential()
model.add(Conv2D(32, (3,3), activation='relu', input_shape=(32,32,3)))
model.add(MaxPooling2D((2,2)))
model.add(Conv2D(64, (3,3), activation='relu'))
model.add(MaxPooling2D((2,2)))
model.add(Conv2D(64, (3,3), activation='relu'))
model.add(Flatten())
model.add(Dense(64, activation='relu'))
model.add(Dense(10, activation='softmax'))
model.summary()
# Compile Model
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# Fit Model
model.fit(train_images, train_labels, epochs=10, validation_data=(test_images, test_labels))
# Evaluate Model
test_loss, test_acc = model.evaluate(test_images, test_labels, verbose=2)
import numpy as np
from tensorflow.keras.applications.resnet50 import ResNet50
from tensorflow.keras.preprocessing import image
from tensorflow.keras.applications.resnet50 import preprocess_input, decode_predictions
def process_img_path(img_path):
return image.load_img(img_path, target_size=(224, 224))
def img_contains_banana(img):
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
x = preprocess_input(x)
model = ResNet50(weights='imagenet')
features = model.predict(x)
results = decode_predictions(features, top=3)[0]
print(results)
for entry in results:
if entry[1] == 'banana':
return entry[2]
return 0.0
import requests
image_urls = ["https://github.com/LambdaSchool/ML-YouOnlyLookOnce/raw/master/sample_data/negative_examples/example11.jpeg",
"https://github.com/LambdaSchool/ML-YouOnlyLookOnce/raw/master/sample_data/positive_examples/example0.jpeg"]
for _id,img in enumerate(image_urls):
r = requests.get(img)
with open(f'example{_id}.jpg', 'wb') as f:
f.write(r.content)
from IPython.display import Image
Image(filename='./example0.jpg', width=600)
img_contains_banana(process_img_path('example0.jpg'))
Image(filename='example1.jpg', width=600)
img_contains_banana(process_img_path('example1.jpg'))
| 0.70253 | 0.993537 |
# Introduction to Programming
Topics for today will include:
- Mozilla Developer Network [(MDN)](https://developer.mozilla.org/en-US/)
- Python Documentation [(Official Documentation)](https://docs.python.org/3/)
- Importance of Design
- Functions
- Built in Functions
## Mozilla Developer Network [(MDN)](https://developer.mozilla.org/en-US/)
---
The Mozilla Developer Network is a great resource for all things web dev. This site is good for trying to learn about standards as well as finding out quick information about something that you're trying to do Web Dev Wise
This will be a major resource going forward when it comes to doing things with HTML and CSS
You'll often find that you're not the first to try and do something. That being said you need to start to get comfortable looking for information on your own when things go wrong.
## Python Documentation [(Official Documentation)](https://docs.python.org/3/)
---
This section is similar to the one above. Python has a lot of resources out there that we can utilize when we're stuck or need some help with something that we may not have encountered before.
Since this is the official documentation page for the language you may often be given too much information or something that you wanted but in the wrong form or for the wrong version of the language. It is up to you to learn how to utilize these things and use them to your advantage.
## Importance of Design
---
So this is a topic that i didn't learn the importance of until I was in the work force. Design is a major influence in the way that code is build and in a major capacity and significant effect on the industry.
Let's pretend we have a client that wants us to do the following:
- Write a function which will count the number of times any one character appears in a string of characters.
- Write a main function which takes the character to be counted from the user and calls the function, outputting the result to the user.
For example, are you like Android and take the latest and greatest and put them into phones in an unregulated hardware market. Thus leaving great variability in the market for your brand? Are you an Apple where you control the full stack. Your hardware and software may not be bleeding edge but it's seamless and uniform.
What does the market want? What are you good at? Do you have people around you that can fill your gaps?
Here's a blurb from a friend about the matter:
>Design, often paired with the phrase "design thinking", is an approach and method of problem solving that builds empathy for user(s) of a product, resulting in the creation of a seamless and delightful user experience tailored to the user's needs.
>Design thinks holistically about the experience that a user would go through when encountering and interacting with a product or technology. Design understands the user and their needs in great detail so that the product team can build the product and experience that fits what the user is looking for. We don't want to create products for the sake of creating them, we want to ensure that there is a need for it by a user.
>Design not only focuses on the actual interface design of a product, but can also ensure the actual technology has a seamless experience as well. Anything that blocks potential users from wanting to buy a product or prohibits current users from utilizing the product successfully, design wants to investigate. We ensure all pieces fit together from the user's standpoint, and we work to build a bridge between the technology and the user, who doesn't need to understand the technical depths of the product.
### Sorting Example [(Toptal Sorting Algorithms)](https://www.toptal.com/developers/sorting-algorithms)
---
Hypothetical, a client comes to you and they want you sort a list of numbers how do you optimally sort a list? `[2, 5, 6, 1, 4, 3]`
### Design Thinking [(IBM Design Thinking)](https://www.ibm.com/design/thinking/)
---
As this idea starts to grow you come to realize that different companies have different design methodologies. IBM has it's own version of Design Thinking. You can find more information about that at the site linked in the title. IBM is very focused on being exactly like its customers in most aspects.
What we're mostly going to take from this is that there are entire careers birthed from thinking before you act. That being said we're going to harp on a couple parts of this.
### Knowing what your requirements are
---
One of the most common scenarios to come across is a product that is annouced that's going to change everything. In the planning phase everyone agrees that the idea is amazing and going to solve all of our problems.
We get down the line and things start to fall apart, we run out of time. Things ran late, or didn't come in in time pushing everything out.
Scope creep ensued.
This is typically the result of not agreeing on what our requirements are. Something as basic as agreeing on what needs to be done needs to be discussed and checked on thouroughly. We do this because often two people rarely are thinking exactly the same thing.
You need to be on the same page as your client and your fellow developers as well. If you don't know ask.
### Planning Things Out
---
We have an idea on what we want to do. So now we just write it? No, not quite. We need to have a rough plan on how we're going to do things. Do we want to use functions, do we need a quick solution, is this going to be verbose and complex?
It's important to look at what we can set up for ourselves. We don't need to make things difficult by planning things out poorly. This means allotting time for things like getting stuck and brainstorming.
### Breaking things down
---
Personally I like to take my problem and scale it down into an easy example so in the case of our problem. The client may want to process a text like Moby Dick. We can start with a sentence and work our way up!
Taking the time to break things in to multiple pieces and figure out what goes where is an art in itself.
```
def char_finder(character, string):
total = 0
for char in string:
if char == character:
total += 1
return total
if __name__ == "__main__":
output = char_finder('o', 'Quick brown fox jumped over the lazy dog')
print(output)
```
## Functions
---
This is a intergral piece of how we do things in any programming language. This allows us to repeat instances of code that we've seen and use them at our preferance.
We'll often be using functions similar to how we use variables and our data types.
### Making Our Own Functions
---
So to make a functions we'll be using the `def` keyword followed by a name and then parameters. We've seen this a couple times now in code examples.
```
def exampleName(exampleParameter1, exampleParameter2):
print(exampleParameter1, exampleParameter2)
```
There are many ways to write functions, we can say that we're going return a specific type of data type.
```
def exampleName(exampleParameter1, exampleParameter2) -> any:
print(exampleParameter1, exampleParameter2)
```
We can also specify the types that the parameters are going to be.
```
def exampleName(exampleParameter1: any, exampleParameter2: any) -> any:
print(exampleParameter1, exampleParameter2)
```
Writing functions is only one part of the fun. We still have to be able to use them.
### Using functions
---
Using functions is fairly simple. To use a function all we have to do is give the function name followed by parenthesis. This should seem familiar.
```
def exampleName(exampleParameter1: int, exampleParameter2: int) -> None:
# print(exampleParameter1, exampleParameter2)
return exampleParameter1 + exampleParameter2
print()
exampleName(10, 94)
```
### Functions In Classes
---
Now we've mentioned classes before, classes can have functions but they're used a little differently. Functions that stem from classes are used often with a dot notation.
```
class Person:
def __init__(self, weight: int, height: int, name: str):
self.weight = weight
self.height = height
self.name = name
def who_is_this(self):
print("This person's name is " + self.name)
print("This person's weight is " + str(self.weight) + " pounds")
print("This person's height is " + str(self.height) + " inches")
if __name__ == "__main__":
Kipp = Person(225, 70, "Aaron Kippins")
Kipp.who_is_this()
```
## Built in Functions and Modules
---
With the talk of dot notation those are often used with built in functions. Built in function are functions that come along with the language. These tend to be very useful because as we start to visit more complex issues they allow us to do complexs thing with ease in some cases.
We have functions that belong to particular classes or special things that can be done with things of a certain class type.
Along side those we can also have Modules. Modules are classes or functions that other people wrote that we can import into our code to use.
### Substrings
---
```
string = "I want to go home!"
print(string[0:12], "to Cancun!")
# print(string[0:1])
```
### toUpper toLower
---
```
alpha_sentence = 'Quick brown fox jumped over the lazy dog'
print(alpha_sentence.title())
print(alpha_sentence.upper())
print(alpha_sentence.lower())
if alpha_sentence.lower().islower():
print("sentence is all lowercase")
```
### Exponents
---
```
print(2 ** 5)
```
### math.sqrt()
---
```
import math
math.sqrt(4)
```
### Integer Division vs Float Division
---
```
print(4//2)
print(4/2)
```
### Abs()
---
```
abs(-10)
```
### String Manipulation
---
```
dummy_string = "Hey there I'm just a string for the example about to happen."
print(dummy_string.center(70, "-"))
print(dummy_string.partition("o"))
print(dummy_string.swapcase())
print(dummy_string.split(" "))
```
### Array Manipulation
---
```
arr = [2, 5, 6, 1, 4, 3]
arr.sort()
print(arr)
print(arr[3])
# sorted(arr)
print(arr[1:3])
```
### Insert and Pop, Append and Remove
---
```
arr.append(7)
print(arr)
arr.pop()
print(arr)
```
|
github_jupyter
|
def char_finder(character, string):
total = 0
for char in string:
if char == character:
total += 1
return total
if __name__ == "__main__":
output = char_finder('o', 'Quick brown fox jumped over the lazy dog')
print(output)
def exampleName(exampleParameter1, exampleParameter2):
print(exampleParameter1, exampleParameter2)
def exampleName(exampleParameter1, exampleParameter2) -> any:
print(exampleParameter1, exampleParameter2)
def exampleName(exampleParameter1: any, exampleParameter2: any) -> any:
print(exampleParameter1, exampleParameter2)
def exampleName(exampleParameter1: int, exampleParameter2: int) -> None:
# print(exampleParameter1, exampleParameter2)
return exampleParameter1 + exampleParameter2
print()
exampleName(10, 94)
class Person:
def __init__(self, weight: int, height: int, name: str):
self.weight = weight
self.height = height
self.name = name
def who_is_this(self):
print("This person's name is " + self.name)
print("This person's weight is " + str(self.weight) + " pounds")
print("This person's height is " + str(self.height) + " inches")
if __name__ == "__main__":
Kipp = Person(225, 70, "Aaron Kippins")
Kipp.who_is_this()
string = "I want to go home!"
print(string[0:12], "to Cancun!")
# print(string[0:1])
alpha_sentence = 'Quick brown fox jumped over the lazy dog'
print(alpha_sentence.title())
print(alpha_sentence.upper())
print(alpha_sentence.lower())
if alpha_sentence.lower().islower():
print("sentence is all lowercase")
print(2 ** 5)
import math
math.sqrt(4)
print(4//2)
print(4/2)
abs(-10)
dummy_string = "Hey there I'm just a string for the example about to happen."
print(dummy_string.center(70, "-"))
print(dummy_string.partition("o"))
print(dummy_string.swapcase())
print(dummy_string.split(" "))
arr = [2, 5, 6, 1, 4, 3]
arr.sort()
print(arr)
print(arr[3])
# sorted(arr)
print(arr[1:3])
arr.append(7)
print(arr)
arr.pop()
print(arr)
| 0.283385 | 0.983166 |
# Copyright (c) Microsoft Corporation. All rights reserved.
### Licensed under the MIT License.
## Multi-Task Deep Neural Networks for Natural Language Understanding
This PyTorch package implements the Multi-Task Deep Neural Networks (MT-DNN) for Natural Language Understanding.
### The data
This notebook assumes you have data already pre-processed in the MT-DNN format and accessible in a local directory.
For the purposes of this example we have added sample data that is already processed in MT-DNN format which can be found in the __sample_data__ folder.
```
%load_ext autoreload
%autoreload 2
import torch
from mtdnn.common.types import EncoderModelType
from mtdnn.configuration_mtdnn import MTDNNConfig
from mtdnn.modeling_mtdnn import MTDNNModel
from mtdnn.process_mtdnn import MTDNNDataProcess
from mtdnn.tasks.config import MTDNNTaskDefs
```
## Define Configuration, Tasks and Model Objects
```
DATA_DIR = "../../sample_data/bert_uncased_lower/mnli/"
BATCH_SIZE = 16
```
### Define a Configuration Object
Create a model configuration object, `MTDNNConfig`, with the necessary parameters to initialize the MT-DNN model. Initialization without any parameters will default to a similar configuration that initializes a BERT model.
```
config = MTDNNConfig(batch_size=BATCH_SIZE)
```
### Create Task Definition Object
Define the task parameters to train for and initialize an `MTDNNTaskDefs` object. Create a task parameter dictionary. Definition can be a single or multiple tasks to train. `MTDNNTaskDefs` can take a python dict, yaml or json file with task(s) defintion.
```
tasks_params = {
"mnli": {
"data_format": "PremiseAndOneHypothesis",
"encoder_type": "BERT",
"dropout_p": 0.3,
"enable_san": True,
"labels": ["contradiction", "neutral", "entailment"],
"metric_meta": ["ACC"],
"loss": "CeCriterion",
"kd_loss": "MseCriterion",
"n_class": 3,
"split_names": [
"train",
"matched_dev",
"mismatched_dev",
"matched_test",
"mismatched_test",
],
"task_type": "Classification",
},
}
# Define the tasks
task_defs = MTDNNTaskDefs(tasks_params)
```
### Create the Data Processing Object
Create a data preprocessing object, `MTDNNDataProcess`. This creates the training, test and development PyTorch dataloaders needed for training and testing. We also need to retrieve the necessary training options required to initialize the model correctly, for all tasks.
Define a data process that handles creating the training, test and development PyTorch dataloaders
```
# Make the Data Preprocess step and update the config with training data updates
data_processor = MTDNNDataProcess(
config=config,
task_defs=task_defs,
data_dir=DATA_DIR,
train_datasets_list=["mnli"],
test_datasets_list=["mnli_mismatched", "mnli_matched"],
)
```
Retrieve the processed batch multitask batch data loaders for training, development and test
```
multitask_train_dataloader = data_processor.get_train_dataloader()
dev_dataloaders_list = data_processor.get_dev_dataloaders()
test_dataloaders_list = data_processor.get_test_dataloaders()
```
Get training options to initialize model
```
decoder_opts = data_processor.get_decoder_options_list()
task_types = data_processor.get_task_types_list()
dropout_list = data_processor.get_tasks_dropout_prob_list()
loss_types = data_processor.get_loss_types_list()
kd_loss_types = data_processor.get_kd_loss_types_list()
tasks_nclass_list = data_processor.get_task_nclass_list()
```
Let us update the batch steps
```
num_all_batches = data_processor.get_num_all_batches()
```
### Instantiate the MTDNN Model
Now we can go ahead and create an `MTDNNModel` model
```
model = MTDNNModel(
config,
task_defs,
pretrained_model_name="bert-base-uncased",
num_train_step=num_all_batches,
decoder_opts=decoder_opts,
task_types=task_types,
dropout_list=dropout_list,
loss_types=loss_types,
kd_loss_types=kd_loss_types,
tasks_nclass_list=tasks_nclass_list,
multitask_train_dataloader=multitask_train_dataloader,
dev_dataloaders_list=dev_dataloaders_list,
test_dataloaders_list=test_dataloaders_list,
)
```
### Fit on one epoch and predict using the training and test
At this point the MT-DNN model allows us to fit to the model and create predictions. The fit takes an optional `epochs` parameter that overwrites the epochs set in the `MTDNNConfig` object.
```
model.fit(epoch=1)
model.predict()
```
### Obtain predictions with a previously trained model checkpoint
The predict function can take an optional checkpoint, `trained_model_chckpt`. This can be used for inference and running evaluations on an already trained PyTorch MT-DNN model.
Optionally using a previously trained model as checkpoint.
```Python
# Predict using a MT-DNN model checkpoint
checkpt = "<path_to_existing_model_checkpoint>"
model.predict(trained_model_chckpt=checkpt)
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
import torch
from mtdnn.common.types import EncoderModelType
from mtdnn.configuration_mtdnn import MTDNNConfig
from mtdnn.modeling_mtdnn import MTDNNModel
from mtdnn.process_mtdnn import MTDNNDataProcess
from mtdnn.tasks.config import MTDNNTaskDefs
DATA_DIR = "../../sample_data/bert_uncased_lower/mnli/"
BATCH_SIZE = 16
config = MTDNNConfig(batch_size=BATCH_SIZE)
tasks_params = {
"mnli": {
"data_format": "PremiseAndOneHypothesis",
"encoder_type": "BERT",
"dropout_p": 0.3,
"enable_san": True,
"labels": ["contradiction", "neutral", "entailment"],
"metric_meta": ["ACC"],
"loss": "CeCriterion",
"kd_loss": "MseCriterion",
"n_class": 3,
"split_names": [
"train",
"matched_dev",
"mismatched_dev",
"matched_test",
"mismatched_test",
],
"task_type": "Classification",
},
}
# Define the tasks
task_defs = MTDNNTaskDefs(tasks_params)
# Make the Data Preprocess step and update the config with training data updates
data_processor = MTDNNDataProcess(
config=config,
task_defs=task_defs,
data_dir=DATA_DIR,
train_datasets_list=["mnli"],
test_datasets_list=["mnli_mismatched", "mnli_matched"],
)
multitask_train_dataloader = data_processor.get_train_dataloader()
dev_dataloaders_list = data_processor.get_dev_dataloaders()
test_dataloaders_list = data_processor.get_test_dataloaders()
decoder_opts = data_processor.get_decoder_options_list()
task_types = data_processor.get_task_types_list()
dropout_list = data_processor.get_tasks_dropout_prob_list()
loss_types = data_processor.get_loss_types_list()
kd_loss_types = data_processor.get_kd_loss_types_list()
tasks_nclass_list = data_processor.get_task_nclass_list()
num_all_batches = data_processor.get_num_all_batches()
model = MTDNNModel(
config,
task_defs,
pretrained_model_name="bert-base-uncased",
num_train_step=num_all_batches,
decoder_opts=decoder_opts,
task_types=task_types,
dropout_list=dropout_list,
loss_types=loss_types,
kd_loss_types=kd_loss_types,
tasks_nclass_list=tasks_nclass_list,
multitask_train_dataloader=multitask_train_dataloader,
dev_dataloaders_list=dev_dataloaders_list,
test_dataloaders_list=test_dataloaders_list,
)
model.fit(epoch=1)
model.predict()
# Predict using a MT-DNN model checkpoint
checkpt = "<path_to_existing_model_checkpoint>"
model.predict(trained_model_chckpt=checkpt)
| 0.701815 | 0.956309 |
```
# Dependencies
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# File path to Load the files
mouse_drug_data_path = "data/mouse_drug_data.csv"
clinical_trial_data_path = "data/clinicaltrial_data.csv"
# Read the Mouse drug data file
df_mouse_drug_data = pd.read_csv(mouse_drug_data_path)
df_mouse_drug_data.head()
# Read the Clinical Trial Data file
df_clincial_trial_data = pd.read_csv(clinical_trial_data_path)
df_clincial_trial_data.head()
# Combine the data into a single dataset
df_merged = pd.merge(df_clincial_trial_data, df_mouse_drug_data, on = ("Mouse ID"),how='outer')
# Display the data table for preview
df_merged.head()
```
## Tumor Response to Treatment
```
# Store the Mean Tumor Volume Data Grouped by Drug and Timepoint
tumour_response_group = df_merged.groupby(["Drug", "Timepoint"])
# Convert to DataFrame
df_tumour = tumour_response_group ["Tumor Volume (mm3)"].mean().to_frame()
# Preview DataFrame
df_tumour.head()
# Store the Standard Error of Tumor Volumes Grouped by Drug and Timepoint
tumour_response_err = df_merged.groupby(["Drug", "Timepoint"])['Tumor Volume (mm3)'].sem()
# Convert to DataFrame
df_tumour_response_error = tumour_response_err.to_frame()
# Preview DataFrame
df_tumour_response_error.head()
# Minor Data Munging to Re-Format the Data Frames
df_tomour_plot = df_tumour.unstack(0)
# Preview that Reformatting worked
df_tomour_plot = df_tomour_plot["Tumor Volume (mm3)"]
df_tomour_plot.head()
# Generate the Plot (with Error Bars)
# Set the x axis from 0 to 45 in increments of 5
x_axis = np.arange(0, 50, 5)
x_limit = 45
plt.figure(figsize=(10,7))
# calculate the tumor volume for each drug specified and plot the corresponding values
error = df_tumour_response_error["Tumor Volume (mm3)"]["Capomulin"]
Capomulin = plt.errorbar(x_axis, df_tomour_plot["Capomulin"], yerr=error, fmt="o", ls="dashed", linewidth=1,
alpha=1, capsize=3,color ="red")
error = df_tumour_response_error["Tumor Volume (mm3)"]["Infubinol"]
Infubinol = plt.errorbar(x_axis, df_tomour_plot["Infubinol"], yerr=error, fmt="^", ls="dashed", linewidth=1,
alpha=1, capsize=3,color ="blue")
error = df_tumour_response_error["Tumor Volume (mm3)"]["Ketapril"]
Ketapril = plt.errorbar(x_axis, df_tomour_plot["Ketapril"], yerr=error, fmt="s", ls="dashed", linewidth=1,
alpha=1, capsize=3,color ="green")
error = df_tumour_response_error["Tumor Volume (mm3)"]["Placebo"]
Placebo = plt.errorbar(x_axis, df_tomour_plot["Placebo"], yerr=error, fmt="D", ls="dashed", linewidth=1,
alpha=1, capsize=3,color ="black")
# Set the plot title and axes titles
plt.title("Tumor Response to Treatment")
plt.xlabel("Time (Days)")
plt.ylabel("Tumor Volume (mm3)")
plt.grid(linestyle="dashed")
plt.ylim(20, 80)
plt.xlim(0, 45)
plt.legend((Capomulin, Infubinol, Ketapril, Placebo), ("Capomulin", "Infubinol", "Ketapril", "Placebo"), fontsize=12)
# Save the Figure
plt.savefig("treatment.png")
# Show the Figure
plt.show()
```

## Metastatic Response to Treatment
```
# Store the Mean Met. Site Data Grouped by Drug and Timepoint
metastatsic_response_group = df_merged.groupby(["Drug", "Timepoint"])
# Convert to DataFrame
df_mean_sem_meta = metastatsic_response_group.agg({"Metastatic Sites" :["mean", "sem"]})
# Preview DataFrame
df_meta = df_mean_sem_meta["Metastatic Sites"]["mean"]
df_meta.head()
# Store the Standard Error associated with Met. Sites Grouped by Drug and Timepoint
# Convert to DataFrame
df_meta_error = df_mean_sem_meta ["Metastatic Sites"]["sem"]
# Preview DataFrame
df_meta_error.head()
# Minor Data Munging to Re-Format the Data Frames
df_meta_plot = df_meta.unstack(0)
# Preview that Reformatting worked
df_meta_plot.head()
# Create list of the meta site mean for each of the four drugs
cap_meta_mean_list = df_mean_sem_meta.loc['Capomulin'].loc[:, 'Metastatic Sites'].loc[:,'mean'].tolist()
infu_meta_mean_list = df_mean_sem_meta.loc['Infubinol'].loc[:, 'Metastatic Sites'].loc[:,'mean'].tolist()
keta_meta_mean_list = df_mean_sem_meta.loc['Ketapril'].loc[:, 'Metastatic Sites'].loc[:,'mean'].tolist()
plac_meta_mean_list = df_mean_sem_meta.loc['Placebo'].loc[:, 'Metastatic Sites'].loc[:,'mean'].tolist()
# Create list of the metastatic site sems(errors) for each of the four drugs
cap_error= df_mean_sem_meta.loc['Capomulin'].loc[:, 'Metastatic Sites'].loc[:,'sem'].tolist()
infu_error = df_mean_sem_meta.loc['Infubinol'].loc[:, 'Metastatic Sites'].loc[:,'sem'].tolist()
keta_error = df_mean_sem_meta.loc['Ketapril'].loc[:, 'Metastatic Sites'].loc[:,'sem'].tolist()
plac_error = df_mean_sem_meta.loc['Placebo'].loc[:, 'Metastatic Sites'].loc[:,'sem'].tolist()
# Generate the Plot (with Error Bars)
plt.figure(figsize=(10,7))
# calculate the metasite mean value for each drug specified and plot the corresponding values
cap2 = plt.errorbar(x_axis, cap_meta_mean_list, yerr=cap_error, fmt="o", ls="dashed", linewidth=1, alpha=1, capsize=3, color = "red")
infu2 = plt.errorbar(x_axis, infu_meta_mean_list, yerr=infu_error, fmt="^", ls="dashed", linewidth=1, alpha=1, capsize=3, color = "blue")
keta2 = plt.errorbar(x_axis, keta_meta_mean_list, yerr=keta_error, fmt="s", ls="dashed", linewidth=1, alpha=1, capsize=3, color = "green")
plac2 = plt.errorbar(x_axis, plac_meta_mean_list, yerr=plac_error, fmt="D", ls="dashed", linewidth=1, alpha=1, capsize=3, color = "black")
plt.ylim(0, 4)
plt.xlim(0, 45)
# Set the plot title and axes titles
plt.title("Metastatic Spread During Treatment", fontsize=20)
plt.xlabel("Treatment Duration (Days)", fontsize=14)
plt.ylabel("Metastatic Sites", fontsize=14)
plt.grid(linestyle="dashed")
plt.legend((cap2, infu2, keta2, plac2), ("Capomulin", "Infubinol", "Ketapril", "Placebo"), fontsize=12)
# Save the Figure
plt.savefig("spread.png")
# Show the resulting scatter plot
plt.show()
```

## Survival Rates
```
# Store the Count of Mice Grouped by Drug and Timepoint (W can pass any metric)
mice_count_group = df_merged.groupby(["Drug", "Timepoint"])
mice_count = mice_count_group["Mouse ID"].nunique()
# Convert to DataFrame
df_mice = mice_count.to_frame()
# Preview DataFrame
df_mice.head()
# Minor Data Munging to Re-Format the Data Frames
mice_plot = df_mice.unstack(0)
# Preview the Data Frame
df_mice_plot = mice_plot["Mouse ID"]
df_mice_plot.head()
df_mice_plot["Capomulin"]
x = (df_mice_plot["Capomulin"])/(df_mice_plot["Capomulin"][0])*100
# Generate the Plot (Accounting for percentages)
plt.figure(figsize = (10,7))
cap3 = plt.errorbar(x_axis, (df_mice_plot["Capomulin"])/(df_mice_plot["Capomulin"][0])*100, fmt="o", ls="dashed", linewidth=1, alpha=1, capsize=3,color = "red")
infu3 = plt.errorbar(x_axis, (df_mice_plot["Infubinol"])/(df_mice_plot["Infubinol"][0])*100, fmt="^", ls="dashed", linewidth=1, alpha=1, capsize=3,color = "blue")
keta3 = plt.errorbar(x_axis, (df_mice_plot["Ketapril"]/(df_mice_plot["Ketapril"][0])*100), fmt="s", ls="dashed", linewidth=1, alpha=1, capsize=3,color ="green")
plac3 = plt.errorbar(x_axis, (df_mice_plot["Placebo"]/(df_mice_plot['Placebo'][0])*100), fmt="D", ls="dashed", linewidth=1, alpha=1, capsize=3,color ="black")
# Set the plot title and axes titles
plt.title("Survival During Treatment", fontsize=20)
plt.xlabel("Time (Days)", fontsize=14)
plt.ylabel("Survival Rate (%)", fontsize=14)
plt.ylim(40, 100)
plt.xlim(0, 45)
plt.grid(linestyle="dashed")
plt.legend((cap3, infu3, keta3, plac3), ("Capomulin", "Infubinol", "Ketapril", "Placebo"), fontsize=12)
# Save the Figure
plt.savefig("survival.png")
# Show the Figure
plt.show()
```

## Summary Bar Graph
```
# Calculate the percent changes for each drug
## Calculating percent change in tumour volume for Capomulin
cap_list = df_tomour_plot["Capomulin"].to_list()
perc_change_cap = ((cap_list[-1] - cap_list[0])/cap_list[0])*100
## Calculating percent change in tumour volume for Infubinol
infu_list = df_tomour_plot["Infubinol"].to_list()
perc_change_infu = ((infu_list[-1] - infu_list[0])/infu_list[0])*100
## Calculating percent change for Ketapril
keta_list = df_tomour_plot["Ketapril"].to_list()
perc_change_keta = ((keta_list[-1] - keta_list[0])/keta_list[0])*100
## Calculating percent change for Placebo
plac_list = df_tomour_plot["Placebo"].to_list()
perc_change_plac = ((plac_list[-1] - plac_list[0])/plac_list[0])*100
# Display Percentage change in Capomulin
perc_change_cap
# Display Percentage change in Infubinol
perc_change_infu
# Display Percentage change in Ketapril
perc_change_keta
# Display Percentage change in Placebo
perc_change_plac
# Store all Relevant Percent Changes into a Tuple
y = [perc_change_cap, perc_change_infu, perc_change_keta, perc_change_plac]
# Set the x axis from 0 to 45 in increments of 5
x_axis = np.arange(len(y))
# Splice the data between passing and failing drugs
# Set the colors of the bars depending on if pos or neg
# -ve % are green and +ve % are red
colors = []
for item in y:
if item < 0:
colors.append('red')
else:
colors.append('green')
# Set up the bar graph
tumor_growth_bar = plt.bar(x_axis, y, color=colors, alpha=1.0, edgecolor='black', linewidth=0.7)
# Set the plot title and axes titles
plt.title("Tumor Change Over 45 Day Treatment")
plt.ylabel("% Tumor Volume Change")
# Add labels, tick marks, etc.
tick_locations = [value for value in x_axis]
plt.xticks(tick_locations, ["Capomulin", "Infubinol", "Ketapril", "Placebo"])
# Set the limit of the x and y axes. lim(start, end)
plt.xlim(-1, len(x_axis))
plt.ylim(min(y)-2, max(y)+2)
# Plot horizontal line at y=0
plt.hlines(0, -1, len(x_axis), alpha=1.0, linewidth=0.5)
# Add gridlines
plt.grid('on', which='major', axis='y', linestyle='dotted', linewidth=0.5)
# Use functions to label the percentages of changes
# Call functions to implement the function calls
def autolabel(rects):
for i,rect in enumerate(rects):
height = int(y[i])
if height >= 0:
plt.text(rect.get_x()+rect.get_width()/2., 2, '%s%%'% (y[i]),
ha='center', va='bottom', color='white', weight='bold')
else:
plt.text(rect.get_x()+rect.get_width()/2., -2, '%s%%'% (y[i]),
ha='center', va='top', color='white', weight='bold')
autolabel(tumor_growth_bar)
# Save the Figure
plt.savefig("change.png")
# Show the Figure
plt.show()
```

|
github_jupyter
|
# Dependencies
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# File path to Load the files
mouse_drug_data_path = "data/mouse_drug_data.csv"
clinical_trial_data_path = "data/clinicaltrial_data.csv"
# Read the Mouse drug data file
df_mouse_drug_data = pd.read_csv(mouse_drug_data_path)
df_mouse_drug_data.head()
# Read the Clinical Trial Data file
df_clincial_trial_data = pd.read_csv(clinical_trial_data_path)
df_clincial_trial_data.head()
# Combine the data into a single dataset
df_merged = pd.merge(df_clincial_trial_data, df_mouse_drug_data, on = ("Mouse ID"),how='outer')
# Display the data table for preview
df_merged.head()
# Store the Mean Tumor Volume Data Grouped by Drug and Timepoint
tumour_response_group = df_merged.groupby(["Drug", "Timepoint"])
# Convert to DataFrame
df_tumour = tumour_response_group ["Tumor Volume (mm3)"].mean().to_frame()
# Preview DataFrame
df_tumour.head()
# Store the Standard Error of Tumor Volumes Grouped by Drug and Timepoint
tumour_response_err = df_merged.groupby(["Drug", "Timepoint"])['Tumor Volume (mm3)'].sem()
# Convert to DataFrame
df_tumour_response_error = tumour_response_err.to_frame()
# Preview DataFrame
df_tumour_response_error.head()
# Minor Data Munging to Re-Format the Data Frames
df_tomour_plot = df_tumour.unstack(0)
# Preview that Reformatting worked
df_tomour_plot = df_tomour_plot["Tumor Volume (mm3)"]
df_tomour_plot.head()
# Generate the Plot (with Error Bars)
# Set the x axis from 0 to 45 in increments of 5
x_axis = np.arange(0, 50, 5)
x_limit = 45
plt.figure(figsize=(10,7))
# calculate the tumor volume for each drug specified and plot the corresponding values
error = df_tumour_response_error["Tumor Volume (mm3)"]["Capomulin"]
Capomulin = plt.errorbar(x_axis, df_tomour_plot["Capomulin"], yerr=error, fmt="o", ls="dashed", linewidth=1,
alpha=1, capsize=3,color ="red")
error = df_tumour_response_error["Tumor Volume (mm3)"]["Infubinol"]
Infubinol = plt.errorbar(x_axis, df_tomour_plot["Infubinol"], yerr=error, fmt="^", ls="dashed", linewidth=1,
alpha=1, capsize=3,color ="blue")
error = df_tumour_response_error["Tumor Volume (mm3)"]["Ketapril"]
Ketapril = plt.errorbar(x_axis, df_tomour_plot["Ketapril"], yerr=error, fmt="s", ls="dashed", linewidth=1,
alpha=1, capsize=3,color ="green")
error = df_tumour_response_error["Tumor Volume (mm3)"]["Placebo"]
Placebo = plt.errorbar(x_axis, df_tomour_plot["Placebo"], yerr=error, fmt="D", ls="dashed", linewidth=1,
alpha=1, capsize=3,color ="black")
# Set the plot title and axes titles
plt.title("Tumor Response to Treatment")
plt.xlabel("Time (Days)")
plt.ylabel("Tumor Volume (mm3)")
plt.grid(linestyle="dashed")
plt.ylim(20, 80)
plt.xlim(0, 45)
plt.legend((Capomulin, Infubinol, Ketapril, Placebo), ("Capomulin", "Infubinol", "Ketapril", "Placebo"), fontsize=12)
# Save the Figure
plt.savefig("treatment.png")
# Show the Figure
plt.show()
# Store the Mean Met. Site Data Grouped by Drug and Timepoint
metastatsic_response_group = df_merged.groupby(["Drug", "Timepoint"])
# Convert to DataFrame
df_mean_sem_meta = metastatsic_response_group.agg({"Metastatic Sites" :["mean", "sem"]})
# Preview DataFrame
df_meta = df_mean_sem_meta["Metastatic Sites"]["mean"]
df_meta.head()
# Store the Standard Error associated with Met. Sites Grouped by Drug and Timepoint
# Convert to DataFrame
df_meta_error = df_mean_sem_meta ["Metastatic Sites"]["sem"]
# Preview DataFrame
df_meta_error.head()
# Minor Data Munging to Re-Format the Data Frames
df_meta_plot = df_meta.unstack(0)
# Preview that Reformatting worked
df_meta_plot.head()
# Create list of the meta site mean for each of the four drugs
cap_meta_mean_list = df_mean_sem_meta.loc['Capomulin'].loc[:, 'Metastatic Sites'].loc[:,'mean'].tolist()
infu_meta_mean_list = df_mean_sem_meta.loc['Infubinol'].loc[:, 'Metastatic Sites'].loc[:,'mean'].tolist()
keta_meta_mean_list = df_mean_sem_meta.loc['Ketapril'].loc[:, 'Metastatic Sites'].loc[:,'mean'].tolist()
plac_meta_mean_list = df_mean_sem_meta.loc['Placebo'].loc[:, 'Metastatic Sites'].loc[:,'mean'].tolist()
# Create list of the metastatic site sems(errors) for each of the four drugs
cap_error= df_mean_sem_meta.loc['Capomulin'].loc[:, 'Metastatic Sites'].loc[:,'sem'].tolist()
infu_error = df_mean_sem_meta.loc['Infubinol'].loc[:, 'Metastatic Sites'].loc[:,'sem'].tolist()
keta_error = df_mean_sem_meta.loc['Ketapril'].loc[:, 'Metastatic Sites'].loc[:,'sem'].tolist()
plac_error = df_mean_sem_meta.loc['Placebo'].loc[:, 'Metastatic Sites'].loc[:,'sem'].tolist()
# Generate the Plot (with Error Bars)
plt.figure(figsize=(10,7))
# calculate the metasite mean value for each drug specified and plot the corresponding values
cap2 = plt.errorbar(x_axis, cap_meta_mean_list, yerr=cap_error, fmt="o", ls="dashed", linewidth=1, alpha=1, capsize=3, color = "red")
infu2 = plt.errorbar(x_axis, infu_meta_mean_list, yerr=infu_error, fmt="^", ls="dashed", linewidth=1, alpha=1, capsize=3, color = "blue")
keta2 = plt.errorbar(x_axis, keta_meta_mean_list, yerr=keta_error, fmt="s", ls="dashed", linewidth=1, alpha=1, capsize=3, color = "green")
plac2 = plt.errorbar(x_axis, plac_meta_mean_list, yerr=plac_error, fmt="D", ls="dashed", linewidth=1, alpha=1, capsize=3, color = "black")
plt.ylim(0, 4)
plt.xlim(0, 45)
# Set the plot title and axes titles
plt.title("Metastatic Spread During Treatment", fontsize=20)
plt.xlabel("Treatment Duration (Days)", fontsize=14)
plt.ylabel("Metastatic Sites", fontsize=14)
plt.grid(linestyle="dashed")
plt.legend((cap2, infu2, keta2, plac2), ("Capomulin", "Infubinol", "Ketapril", "Placebo"), fontsize=12)
# Save the Figure
plt.savefig("spread.png")
# Show the resulting scatter plot
plt.show()
# Store the Count of Mice Grouped by Drug and Timepoint (W can pass any metric)
mice_count_group = df_merged.groupby(["Drug", "Timepoint"])
mice_count = mice_count_group["Mouse ID"].nunique()
# Convert to DataFrame
df_mice = mice_count.to_frame()
# Preview DataFrame
df_mice.head()
# Minor Data Munging to Re-Format the Data Frames
mice_plot = df_mice.unstack(0)
# Preview the Data Frame
df_mice_plot = mice_plot["Mouse ID"]
df_mice_plot.head()
df_mice_plot["Capomulin"]
x = (df_mice_plot["Capomulin"])/(df_mice_plot["Capomulin"][0])*100
# Generate the Plot (Accounting for percentages)
plt.figure(figsize = (10,7))
cap3 = plt.errorbar(x_axis, (df_mice_plot["Capomulin"])/(df_mice_plot["Capomulin"][0])*100, fmt="o", ls="dashed", linewidth=1, alpha=1, capsize=3,color = "red")
infu3 = plt.errorbar(x_axis, (df_mice_plot["Infubinol"])/(df_mice_plot["Infubinol"][0])*100, fmt="^", ls="dashed", linewidth=1, alpha=1, capsize=3,color = "blue")
keta3 = plt.errorbar(x_axis, (df_mice_plot["Ketapril"]/(df_mice_plot["Ketapril"][0])*100), fmt="s", ls="dashed", linewidth=1, alpha=1, capsize=3,color ="green")
plac3 = plt.errorbar(x_axis, (df_mice_plot["Placebo"]/(df_mice_plot['Placebo'][0])*100), fmt="D", ls="dashed", linewidth=1, alpha=1, capsize=3,color ="black")
# Set the plot title and axes titles
plt.title("Survival During Treatment", fontsize=20)
plt.xlabel("Time (Days)", fontsize=14)
plt.ylabel("Survival Rate (%)", fontsize=14)
plt.ylim(40, 100)
plt.xlim(0, 45)
plt.grid(linestyle="dashed")
plt.legend((cap3, infu3, keta3, plac3), ("Capomulin", "Infubinol", "Ketapril", "Placebo"), fontsize=12)
# Save the Figure
plt.savefig("survival.png")
# Show the Figure
plt.show()
# Calculate the percent changes for each drug
## Calculating percent change in tumour volume for Capomulin
cap_list = df_tomour_plot["Capomulin"].to_list()
perc_change_cap = ((cap_list[-1] - cap_list[0])/cap_list[0])*100
## Calculating percent change in tumour volume for Infubinol
infu_list = df_tomour_plot["Infubinol"].to_list()
perc_change_infu = ((infu_list[-1] - infu_list[0])/infu_list[0])*100
## Calculating percent change for Ketapril
keta_list = df_tomour_plot["Ketapril"].to_list()
perc_change_keta = ((keta_list[-1] - keta_list[0])/keta_list[0])*100
## Calculating percent change for Placebo
plac_list = df_tomour_plot["Placebo"].to_list()
perc_change_plac = ((plac_list[-1] - plac_list[0])/plac_list[0])*100
# Display Percentage change in Capomulin
perc_change_cap
# Display Percentage change in Infubinol
perc_change_infu
# Display Percentage change in Ketapril
perc_change_keta
# Display Percentage change in Placebo
perc_change_plac
# Store all Relevant Percent Changes into a Tuple
y = [perc_change_cap, perc_change_infu, perc_change_keta, perc_change_plac]
# Set the x axis from 0 to 45 in increments of 5
x_axis = np.arange(len(y))
# Splice the data between passing and failing drugs
# Set the colors of the bars depending on if pos or neg
# -ve % are green and +ve % are red
colors = []
for item in y:
if item < 0:
colors.append('red')
else:
colors.append('green')
# Set up the bar graph
tumor_growth_bar = plt.bar(x_axis, y, color=colors, alpha=1.0, edgecolor='black', linewidth=0.7)
# Set the plot title and axes titles
plt.title("Tumor Change Over 45 Day Treatment")
plt.ylabel("% Tumor Volume Change")
# Add labels, tick marks, etc.
tick_locations = [value for value in x_axis]
plt.xticks(tick_locations, ["Capomulin", "Infubinol", "Ketapril", "Placebo"])
# Set the limit of the x and y axes. lim(start, end)
plt.xlim(-1, len(x_axis))
plt.ylim(min(y)-2, max(y)+2)
# Plot horizontal line at y=0
plt.hlines(0, -1, len(x_axis), alpha=1.0, linewidth=0.5)
# Add gridlines
plt.grid('on', which='major', axis='y', linestyle='dotted', linewidth=0.5)
# Use functions to label the percentages of changes
# Call functions to implement the function calls
def autolabel(rects):
for i,rect in enumerate(rects):
height = int(y[i])
if height >= 0:
plt.text(rect.get_x()+rect.get_width()/2., 2, '%s%%'% (y[i]),
ha='center', va='bottom', color='white', weight='bold')
else:
plt.text(rect.get_x()+rect.get_width()/2., -2, '%s%%'% (y[i]),
ha='center', va='top', color='white', weight='bold')
autolabel(tumor_growth_bar)
# Save the Figure
plt.savefig("change.png")
# Show the Figure
plt.show()
| 0.588889 | 0.740057 |
# Lab 04 : Train vanilla neural network -- solution
# Training a one-layer net on FASHION-MNIST
```
# For Google Colaboratory
import sys, os
if 'google.colab' in sys.modules:
from google.colab import drive
drive.mount('/content/gdrive')
file_name = 'train_vanilla_nn_solution.ipynb'
import subprocess
path_to_file = subprocess.check_output('find . -type f -name ' + str(file_name), shell=True).decode("utf-8")
print(path_to_file)
path_to_file = path_to_file.replace(file_name,"").replace('\n',"")
os.chdir(path_to_file)
!pwd
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from random import randint
import utils
```
### Download the TRAINING SET (data+labels)
```
from utils import check_fashion_mnist_dataset_exists
data_path=check_fashion_mnist_dataset_exists()
train_data=torch.load(data_path+'fashion-mnist/train_data.pt')
train_label=torch.load(data_path+'fashion-mnist/train_label.pt')
print(train_data.size())
print(train_label.size())
```
### Download the TEST SET (data only)
```
test_data=torch.load(data_path+'fashion-mnist/test_data.pt')
print(test_data.size())
```
### Make a one layer net class
```
class one_layer_net(nn.Module):
def __init__(self, input_size, output_size):
super(one_layer_net , self).__init__()
self.linear_layer = nn.Linear( input_size, output_size , bias=False)
def forward(self, x):
y = self.linear_layer(x)
prob = F.softmax(y, dim=1)
return prob
```
### Build the net
```
net=one_layer_net(784,10)
print(net)
```
### Take the 4th image of the test set:
```
im=test_data[4]
utils.show(im)
```
### And feed it to the UNTRAINED network:
```
p = net( im.view(1,784))
print(p)
```
### Display visually the confidence scores
```
utils.show_prob_fashion_mnist(p)
```
### Train the network (only 5000 iterations) on the train set
```
criterion = nn.NLLLoss()
optimizer=torch.optim.SGD(net.parameters() , lr=0.01 )
for iter in range(1,5000):
# choose a random integer between 0 and 59,999
# extract the corresponding picture and label
# and reshape them to fit the network
idx=randint(0, 60000-1)
input=train_data[idx].view(1,784)
label=train_label[idx].view(1)
# feed the input to the net
input.requires_grad_()
prob=net(input)
# update the weights (all the magic happens here -- we will discuss it later)
log_prob=torch.log(prob)
loss = criterion(log_prob, label)
optimizer.zero_grad()
loss.backward()
optimizer.step()
```
### Take the 34th image of the test set:
```
im=test_data[34]
utils.show(im)
```
### Feed it to the TRAINED net:
```
p = net( im.view(1,784))
print(p)
```
### Display visually the confidence scores
```
utils.show_prob_fashion_mnist(prob)
```
### Choose image at random from the test set and see how good/bad are the predictions
```
# choose a picture at random
idx=randint(0, 10000-1)
im=test_data[idx]
# diplay the picture
utils.show(im)
# feed it to the net and display the confidence scores
prob = net( im.view(1,784))
utils.show_prob_fashion_mnist(prob)
```
|
github_jupyter
|
# For Google Colaboratory
import sys, os
if 'google.colab' in sys.modules:
from google.colab import drive
drive.mount('/content/gdrive')
file_name = 'train_vanilla_nn_solution.ipynb'
import subprocess
path_to_file = subprocess.check_output('find . -type f -name ' + str(file_name), shell=True).decode("utf-8")
print(path_to_file)
path_to_file = path_to_file.replace(file_name,"").replace('\n',"")
os.chdir(path_to_file)
!pwd
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from random import randint
import utils
from utils import check_fashion_mnist_dataset_exists
data_path=check_fashion_mnist_dataset_exists()
train_data=torch.load(data_path+'fashion-mnist/train_data.pt')
train_label=torch.load(data_path+'fashion-mnist/train_label.pt')
print(train_data.size())
print(train_label.size())
test_data=torch.load(data_path+'fashion-mnist/test_data.pt')
print(test_data.size())
class one_layer_net(nn.Module):
def __init__(self, input_size, output_size):
super(one_layer_net , self).__init__()
self.linear_layer = nn.Linear( input_size, output_size , bias=False)
def forward(self, x):
y = self.linear_layer(x)
prob = F.softmax(y, dim=1)
return prob
net=one_layer_net(784,10)
print(net)
im=test_data[4]
utils.show(im)
p = net( im.view(1,784))
print(p)
utils.show_prob_fashion_mnist(p)
criterion = nn.NLLLoss()
optimizer=torch.optim.SGD(net.parameters() , lr=0.01 )
for iter in range(1,5000):
# choose a random integer between 0 and 59,999
# extract the corresponding picture and label
# and reshape them to fit the network
idx=randint(0, 60000-1)
input=train_data[idx].view(1,784)
label=train_label[idx].view(1)
# feed the input to the net
input.requires_grad_()
prob=net(input)
# update the weights (all the magic happens here -- we will discuss it later)
log_prob=torch.log(prob)
loss = criterion(log_prob, label)
optimizer.zero_grad()
loss.backward()
optimizer.step()
im=test_data[34]
utils.show(im)
p = net( im.view(1,784))
print(p)
utils.show_prob_fashion_mnist(prob)
# choose a picture at random
idx=randint(0, 10000-1)
im=test_data[idx]
# diplay the picture
utils.show(im)
# feed it to the net and display the confidence scores
prob = net( im.view(1,784))
utils.show_prob_fashion_mnist(prob)
| 0.620507 | 0.843444 |
<a href="https://colab.research.google.com/github/AnaVargasJ/MDigitales/blob/main/Clase4.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
**Continuacion de estructuras de control iterativa **
---
**Acumuladores**
Sel da este nombre a la variables que se encargan de almcenar algun tipo de informacion.
**Ejemplo**
El caso de la compra de viveres en la tiends.
```
nombre = input("Nombre del comprador")
Listacompra = "";
print(nombre, "escribe los siguientes niveles para su compra ene el supermercado:")
listacompra = (listacompra , + "1 paca de papel de higienico")
print("----compras que tengo que hacer----")
print(listacompra)
listacompra=(listacompra ,+ "Shampoo pantene 2 and 1")
listacompra=(listacompra, +"2 pacas de pañales pequeñin etapa 3")
print(listacompra)
```
la variable "listacompra" nos esta sirviendooppara acumular informacion de la lista de compra.
podemos observar, que **NO** estamos creando una variable por cada item, sino una variable definida nos sirve para almacenar la informacion
A continuacion observemos un ejemplo en donde se pone en practica el uso de acumulacion en una variable usando cantidades y precios
```
ppph=14000 #precio de papel higienico
cpph =2 #cantidad de pacas de papel
pshampoo = 18000 #Precio de shampoo pantene 2 and 1
cshampoo =4 #Cantidad de shampoo
ppbebe = 17000 #precio de pacas de pañales pequeña
cpbebe = 3 #cantidad de pañales pequeños
subtotal = 0
print("Calculando el total de la compra...")
total_ppph=ppph*cpph
print("el valor de la compra del papel higiencio es", total_ppph)
subtotal=subtotal + total_ppph
print("---el subtotal es:",subtotal)
total_shampoo = pshampoo *cshampoo
print("El valor del total de Shampoo es:$",total_shampoo )
subtotal = subtotal+ total_shampoo
print("---el subtotal es:$",subtotal)
total_ppbebe = ppbebe*cpbebe
print("el valor total de pañales es:$",total_ppbebe)
subtotal = subtotal + total_ppbebe
print("el total de su compra es:$",subtotal)
```
**Contadores**
tiene mucha relacion con los "acumuladores" visto en el apartado anterior
Estas variables se caracterizan por ser variables de control, es decir controlan la **cantidad** de veces que se ejecutan determinada accion.
Usando el ejemplo anterior y modificandoo un poco, podemos desarrollar el siguient algoritmo
```
#Se comprara pañales por unidad en este caso.
contp = 0
print("Se realizara la compra de pañales etapa 3... se ha iniciado la compra de asignacion en el carrito. En total hay :", contp, "pañales")
contp = contp+1
print("Se realizara la compra de pañales etapa 3... se ha iniciado la compra de asignacion en el carrito. Ahora hay :", contp, "pañales")
contp = contp+1
print("Ahora hay:",contp,"pañal1")
contp = contp+1
print("Ahora hay:",contp,"pañal1")
contp = contp+1
print("Ahora hay:",contp,"pañal1")
contp = contp+1
print("Ahora hay:",contp,"pañal1")
```
**Ciclos controlados por condicicones**
**WHILE**
---
Recordemos que las variables de control, nos permten manejar estados, pasar de un estado a otro es por ejemplo: una variable que no contiene elementos a contenerlo o una variable un elemento en particular (Acumulador o contador) y cambiarlo po completo(Bnadera)
Estas Variables de cocntrol son la base de ciclos de control. Siendo mas claros, pasar de una accion manual a algo mas automatizado
Empezamos con el ciclo "WHILE" En español es "mientras". Este ciclo compone una condiciion y su bloque de codigo
loque nos quiere decir While es que el bloque de codigo se ejecutara mientrasc la condicion da como resultado True or False
```
lapiz = 5
contlapiz = 0
print("Se ha iniciado la compra. en total hay :", contlapiz,lapiz)
while (contlapiz < lapiz):
contlapiz = contlapiz+1
print("Se ha realizado la compra de lapices ahora hay",str(contlapiz) + "lapiz")
a = str(contlapiz)
print(type(contlapiz))
print(type(a))
```
Tener en cuenta que dentro del ciclo de WHILE se va afectando las variables implicadas en la declracion de la condicicon que debe cumplir el ciclo en el ejemplo anterior la variable contlapiz para que en algun momento la condicion sea vedadera y termine el ciclo se tiene que cumplir la condicion(contlapiz). De lo contrario, tendriamos un ciclo que nunca se detendria, lo cual decantaria en un cilo interminable
**CICLO DE FOR**
---
Es un ciclo especializado y optimizado parta los ciclos controlados por cantidad. Se compone de tres elementos:
1. la variable de iteraccion
2. elemento de iteraccion
3. bloque de ocdigo iterar
**¿ventajas de usar el FOR ?**
en PYTHON es muy importante y se considera una herramienta bastante flexible y poderos, por permitir ingresar estructuras de datos complejas, cadena de caracteres, rangos , entre otros. los elementos de iteraccion en esta estructura de datos, son necesarios que tengan la siguiente caracteristica :
1. cantidad definida(Esto lo diferencia totalmente del WHILE)
el WHILE parte de una condicion de verdad, pero el **FOR** parte de una cantidad definida
```
##Retomando el ejemplo de la compra de lapices
print("se ha iniciado la compra. En total hay:0 lapices.")
for i in range(1,6): # en los rangos, la funcion range maneja un intervalo abierto a la derecha y cerrado al a izquierda
print("Se ha realizado la ocmpra de lapices. Ahora hay",i,"lapices")
```
|
github_jupyter
|
nombre = input("Nombre del comprador")
Listacompra = "";
print(nombre, "escribe los siguientes niveles para su compra ene el supermercado:")
listacompra = (listacompra , + "1 paca de papel de higienico")
print("----compras que tengo que hacer----")
print(listacompra)
listacompra=(listacompra ,+ "Shampoo pantene 2 and 1")
listacompra=(listacompra, +"2 pacas de pañales pequeñin etapa 3")
print(listacompra)
ppph=14000 #precio de papel higienico
cpph =2 #cantidad de pacas de papel
pshampoo = 18000 #Precio de shampoo pantene 2 and 1
cshampoo =4 #Cantidad de shampoo
ppbebe = 17000 #precio de pacas de pañales pequeña
cpbebe = 3 #cantidad de pañales pequeños
subtotal = 0
print("Calculando el total de la compra...")
total_ppph=ppph*cpph
print("el valor de la compra del papel higiencio es", total_ppph)
subtotal=subtotal + total_ppph
print("---el subtotal es:",subtotal)
total_shampoo = pshampoo *cshampoo
print("El valor del total de Shampoo es:$",total_shampoo )
subtotal = subtotal+ total_shampoo
print("---el subtotal es:$",subtotal)
total_ppbebe = ppbebe*cpbebe
print("el valor total de pañales es:$",total_ppbebe)
subtotal = subtotal + total_ppbebe
print("el total de su compra es:$",subtotal)
#Se comprara pañales por unidad en este caso.
contp = 0
print("Se realizara la compra de pañales etapa 3... se ha iniciado la compra de asignacion en el carrito. En total hay :", contp, "pañales")
contp = contp+1
print("Se realizara la compra de pañales etapa 3... se ha iniciado la compra de asignacion en el carrito. Ahora hay :", contp, "pañales")
contp = contp+1
print("Ahora hay:",contp,"pañal1")
contp = contp+1
print("Ahora hay:",contp,"pañal1")
contp = contp+1
print("Ahora hay:",contp,"pañal1")
contp = contp+1
print("Ahora hay:",contp,"pañal1")
lapiz = 5
contlapiz = 0
print("Se ha iniciado la compra. en total hay :", contlapiz,lapiz)
while (contlapiz < lapiz):
contlapiz = contlapiz+1
print("Se ha realizado la compra de lapices ahora hay",str(contlapiz) + "lapiz")
a = str(contlapiz)
print(type(contlapiz))
print(type(a))
##Retomando el ejemplo de la compra de lapices
print("se ha iniciado la compra. En total hay:0 lapices.")
for i in range(1,6): # en los rangos, la funcion range maneja un intervalo abierto a la derecha y cerrado al a izquierda
print("Se ha realizado la ocmpra de lapices. Ahora hay",i,"lapices")
| 0.112466 | 0.919208 |
```
# For encoding categorical data
!pip install category_encoders==2.*
from category_encoders import OneHotEncoder
# model building
from sklearn.feature_selection import SelectKBest
from sklearn.linear_model import LinearRegression, Ridge
from sklearn.metrics import mean_absolute_error, mean_squared_error
from sklearn.pipeline import make_pipeline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import sys
from category_encoders import OneHotEncoder
```
# NYC Rent
**GOAL:** Improve our model for predicting NYC rent prices.
**Objectives**
- Do one-hot encoding of categorical features
- Do univariate feature selection
- Use scikit-learn to fit Ridge Regression models
```
%%capture
# If you're on Colab:
if 'google.colab' in sys.modules:
DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Applied-Modeling/master/data/'
!pip install category_encoders==2.*
# If you're working locally:
else:
DATA_PATH = '../data/'
```
# I. Wrangle Data
Create **wrangle function** for **reproducibility**.
```
def wrangle(filepath):
df = pd.read_csv(filepath,
parse_dates=['created'],
index_col='created')
df = df[(df['price'] >= np.percentile(df['price'], 0.5)) &
(df['price'] <= np.percentile(df['price'], 99.5)) &
(df['latitude'] >= np.percentile(df['latitude'], 0.05)) &
(df['latitude'] < np.percentile(df['latitude'], 99.95)) &
(df['longitude'] >= np.percentile(df['longitude'], 0.05)) &
(df['longitude'] <= np.percentile(df['longitude'], 99.95))]
cutoff = 5
drop_cols = [col for col in df.select_dtypes('object') if df[col].nunique() > cutoff]
df.drop(columns=drop_cols,inplace=True)
return df
df = wrangle(DATA_PATH+'apartments/renthop-nyc.csv')
df.select_dtypes('object').nunique()
drop_cols =[]
cutoff = 5
for col in df.select_dtypes('object'):
if df[col].nunique() > cutoff:
drop_cols.append(col)
print(drop_cols)
drop_cols = [col for col in df.select_dtypes('object') if df[col].nunique() > cutoff]
drop_cols
```
# II. Split Data
Split **target vector** from **feature matrix**.
```
target = 'price'
y = df[target]
X = df.drop(columns=target)
```
Split data into **training** and **test** sets.
(Use data from April & May 2016 to train. Use data from June 2016 to test.)
```
cutoff = '2016-06-01'
mask = X.index < cutoff
X_train, y_train = X.loc[mask], y.loc[mask]
X_test, y_test = X.loc[~mask], y.loc[~mask]
```
# III. Establish Baseline
**Note:** This is a **regression** problem because we're predictiong the continuous value `'price'`.
```
y_pred = [y_train.mean()] * len(y_train)
print('Mean price:', y_train.mean())
print('Baseline MAE:', mean_absolute_error(y_train, y_pred))
```
# IV. Build Models
**Question:** How can we represent *categorical* features numerically so that we can use them to train our model?
```
# step 1: import your transformer class
#we did above
#step 2: instantiate your transformer
ohe = OneHotEncoder(use_cat_names = True)
#step 3: you fit your transformer to the TRAINING data
ohe.fit(X_train) #only on feature matrix (sometimes)
# Never refit your transformer on your TEST data
#step 4: transform my training data
XT_train = ohe.transform(X_train)
XT_test = ohe.transform(X_test)
#Regular linearl Regrassion
model_lr = LinearRegression()
model_lr.fit(XT_train,y_train)
# Ridge Regression(regularization)
model_r = Ridge(alpha = 1.0) #alpha is a hyper parameter
model_r.fit(XT_train, y_train)
#SelectKBest model
skb = SelectKBest(k=10)
skb.fit(XT_train,y_train)
XTT_train = skb.transform(XT_train)
XTT_test = skb.transform(XT_test)
model_lr2 = LinearRegression()
model_lr2.fit(XTT_train,y_train)
```
# V. Check Metrics
```
print('LR training MAE:', mean_absolute_error(y_train, model_lr.predict(XT_train)))
print('LR test MAE:', mean_absolute_error(y_test, model_lr.predict(XT_test)))
print('Ridge training MAE:', mean_absolute_error(y_train, model_r.predict(XT_train)))
print('Ridge test MAE:', mean_absolute_error(y_test, model_r.predict(XT_test)))
print('Ridge training MAE:', mean_absolute_error(y_train, model_lr2.predict(XTT_train)))
print('Ridge test MAE:', mean_absolute_error(y_test, model_lr2.predict(XTT_test)))
```
# Communicate results
```
feature_names = ohe.get_feature_names()
coefficients = model_lr.coef_
feature_importances = pd.Series(coefficients,index=feature_names).sort_values(key=abs)
feature_importances
feature_names = ohe.get_feature_names()
coefficients = model_r.coef_
feature_importances = pd.Series(coefficients,index=feature_names).sort_values(key=abs)
feature_importances.plot(kind='barh',title='Ridge Coefficients')
```
|
github_jupyter
|
# For encoding categorical data
!pip install category_encoders==2.*
from category_encoders import OneHotEncoder
# model building
from sklearn.feature_selection import SelectKBest
from sklearn.linear_model import LinearRegression, Ridge
from sklearn.metrics import mean_absolute_error, mean_squared_error
from sklearn.pipeline import make_pipeline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import sys
from category_encoders import OneHotEncoder
%%capture
# If you're on Colab:
if 'google.colab' in sys.modules:
DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Applied-Modeling/master/data/'
!pip install category_encoders==2.*
# If you're working locally:
else:
DATA_PATH = '../data/'
def wrangle(filepath):
df = pd.read_csv(filepath,
parse_dates=['created'],
index_col='created')
df = df[(df['price'] >= np.percentile(df['price'], 0.5)) &
(df['price'] <= np.percentile(df['price'], 99.5)) &
(df['latitude'] >= np.percentile(df['latitude'], 0.05)) &
(df['latitude'] < np.percentile(df['latitude'], 99.95)) &
(df['longitude'] >= np.percentile(df['longitude'], 0.05)) &
(df['longitude'] <= np.percentile(df['longitude'], 99.95))]
cutoff = 5
drop_cols = [col for col in df.select_dtypes('object') if df[col].nunique() > cutoff]
df.drop(columns=drop_cols,inplace=True)
return df
df = wrangle(DATA_PATH+'apartments/renthop-nyc.csv')
df.select_dtypes('object').nunique()
drop_cols =[]
cutoff = 5
for col in df.select_dtypes('object'):
if df[col].nunique() > cutoff:
drop_cols.append(col)
print(drop_cols)
drop_cols = [col for col in df.select_dtypes('object') if df[col].nunique() > cutoff]
drop_cols
target = 'price'
y = df[target]
X = df.drop(columns=target)
cutoff = '2016-06-01'
mask = X.index < cutoff
X_train, y_train = X.loc[mask], y.loc[mask]
X_test, y_test = X.loc[~mask], y.loc[~mask]
y_pred = [y_train.mean()] * len(y_train)
print('Mean price:', y_train.mean())
print('Baseline MAE:', mean_absolute_error(y_train, y_pred))
# step 1: import your transformer class
#we did above
#step 2: instantiate your transformer
ohe = OneHotEncoder(use_cat_names = True)
#step 3: you fit your transformer to the TRAINING data
ohe.fit(X_train) #only on feature matrix (sometimes)
# Never refit your transformer on your TEST data
#step 4: transform my training data
XT_train = ohe.transform(X_train)
XT_test = ohe.transform(X_test)
#Regular linearl Regrassion
model_lr = LinearRegression()
model_lr.fit(XT_train,y_train)
# Ridge Regression(regularization)
model_r = Ridge(alpha = 1.0) #alpha is a hyper parameter
model_r.fit(XT_train, y_train)
#SelectKBest model
skb = SelectKBest(k=10)
skb.fit(XT_train,y_train)
XTT_train = skb.transform(XT_train)
XTT_test = skb.transform(XT_test)
model_lr2 = LinearRegression()
model_lr2.fit(XTT_train,y_train)
print('LR training MAE:', mean_absolute_error(y_train, model_lr.predict(XT_train)))
print('LR test MAE:', mean_absolute_error(y_test, model_lr.predict(XT_test)))
print('Ridge training MAE:', mean_absolute_error(y_train, model_r.predict(XT_train)))
print('Ridge test MAE:', mean_absolute_error(y_test, model_r.predict(XT_test)))
print('Ridge training MAE:', mean_absolute_error(y_train, model_lr2.predict(XTT_train)))
print('Ridge test MAE:', mean_absolute_error(y_test, model_lr2.predict(XTT_test)))
feature_names = ohe.get_feature_names()
coefficients = model_lr.coef_
feature_importances = pd.Series(coefficients,index=feature_names).sort_values(key=abs)
feature_importances
feature_names = ohe.get_feature_names()
coefficients = model_r.coef_
feature_importances = pd.Series(coefficients,index=feature_names).sort_values(key=abs)
feature_importances.plot(kind='barh',title='Ridge Coefficients')
| 0.437824 | 0.830285 |
# Iris Data Species
```
from sklearn.model_selection import train_test_split
from sklearn.model_selection import StratifiedKFold
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from IPython.display import display
import matplotlib.patches as patch
import matplotlib.pyplot as plt
from sklearn.svm import NuSVR
from scipy.stats import norm
from sklearn import svm
import lightgbm as lgb
import xgboost as xgb
import seaborn as sns
import pandas as pd
import numpy as np
import warnings
import time
import glob
import sys
import os
import gc
```
#
```
iris=pd.read_csv('/home/rahul/Desktop/Link to rahul_environment/Projects/Machine_Learning Projects/Iris_Species/Iris.csv')
iris.head()
iris.info()
iris['Species'].value_counts()
```
# Creating the bar plot
```
sns.countplot('Species',data=iris)
```
## Counting the values in the pie plot
```
iris['Species'].value_counts().plot.pie(figsize=(10,8))
```
## Joint plot: Jointplot is seaborn library specific and can be used to quickly visualize and analyze the relationship between two variables and describe their individual distributions on the same plot.
```
figure=sns.jointplot(x='SepalLengthCm',y='SepalWidthCm',data=iris)
sns.jointplot(x='SepalWidthCm',y='SepalLengthCm',data=iris,kind='reg')
```
## Jointplot's for the Sepal Length and Width
```
sns.jointplot(x='SepalWidthCm',y='SepalLengthCm',data=iris,kind='hex')
sns.jointplot(x='SepalWidthCm',y='SepalLengthCm',data=iris,kind='resid')
sns.jointplot(x='SepalWidthCm',y='SepalLengthCm',data=iris,kind='kde')
```
## Boxplot for the Species and PetalLengthCm
```
sns.boxplot(x='Species',y='PetalLengthCm',data=iris)
plt.xlabel('Species of the plant')
plt.title('Box Plot Of Figure')
```
## Strip_plot
```
sns.stripplot(x='Species',y='PetalLengthCm',data=iris)
```
## Combining both the boxplot and strip_plot
```
fig=plt.gcf()
fig=sns.boxplot(x='Species',y='SepalLengthCm',data=iris)
fig=sns.stripplot(x='Species',y='SepalLengthCm',data=iris)
```
## Four different kinds of the violin_plots
```
plt.subplot(2,2,1)
sns.violinplot(x='Species',y='PetalLengthCm',data=iris)
plt.subplot(2,2,2)
sns.violinplot(x='Species',y='PetalWidthCm',data=iris)
plt.subplot(2,2,3)
sns.violinplot(x='Species',y='SepalLengthCm',data=iris)
plt.subplot(2,2,4)
sns.violinplot(x='Species',y='SepalWidthCm',data=iris)
```
## Scattterplot
```
sns.scatterplot(x='Species',y='PetalLengthCm',data=iris)
```
## Pairplot for the iris dataset.
```
sns.pairplot(data=iris,hue='Species')
```
## Heatmap for the iris dataset.
```
sns.heatmap(data=iris.corr(),annot=True)
```
## Don't know how to plot the distribution plot??
## Swarm Plot
```
sns.boxplot(x='Species',y='PetalLengthCm',data=iris)
sns.swarmplot(x='Species',y='PetalLengthCm',data=iris)
```
## Lmplot
```
sns.lmplot(x="PetalLengthCm",y='PetalWidthCm',data=iris)
```
# FacetGrid is still incomplete?
```
sns.FacetGrid(iris,hue='Species')
from pandas.tools.plotting import andrews_curves
andrews_curves(iris,"Species",colormap='rainbow')
plt.ioff()
```
## Parallel coordinate plot: This type of visualisation is used for plotting multivariate, numerical data. Parallel Coordinates Plots are ideal for comparing many variables together and seeing the relationships between them. For example, if you had to compare an array of products with the same attributes (comparing computer or cars specs across different models).
```
from pandas.tools.plotting import parallel_coordinates
parallel_coordinates(iris,"Species",colormap="rainbow")
```
## Factorplot
```
sns.factorplot('Species','SepalLengthCm',data=iris)
```
## Boxenplot
```
sns.boxenplot('Species','SepalLengthCm',data=iris)
fig=sns.residplot('SepalLengthCm', 'SepalWidthCm',data=iris)
```
# How to create the venn diagram pls let me know?
# Spider Graph is still in prgoress?
|
github_jupyter
|
from sklearn.model_selection import train_test_split
from sklearn.model_selection import StratifiedKFold
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from IPython.display import display
import matplotlib.patches as patch
import matplotlib.pyplot as plt
from sklearn.svm import NuSVR
from scipy.stats import norm
from sklearn import svm
import lightgbm as lgb
import xgboost as xgb
import seaborn as sns
import pandas as pd
import numpy as np
import warnings
import time
import glob
import sys
import os
import gc
iris=pd.read_csv('/home/rahul/Desktop/Link to rahul_environment/Projects/Machine_Learning Projects/Iris_Species/Iris.csv')
iris.head()
iris.info()
iris['Species'].value_counts()
sns.countplot('Species',data=iris)
iris['Species'].value_counts().plot.pie(figsize=(10,8))
figure=sns.jointplot(x='SepalLengthCm',y='SepalWidthCm',data=iris)
sns.jointplot(x='SepalWidthCm',y='SepalLengthCm',data=iris,kind='reg')
sns.jointplot(x='SepalWidthCm',y='SepalLengthCm',data=iris,kind='hex')
sns.jointplot(x='SepalWidthCm',y='SepalLengthCm',data=iris,kind='resid')
sns.jointplot(x='SepalWidthCm',y='SepalLengthCm',data=iris,kind='kde')
sns.boxplot(x='Species',y='PetalLengthCm',data=iris)
plt.xlabel('Species of the plant')
plt.title('Box Plot Of Figure')
sns.stripplot(x='Species',y='PetalLengthCm',data=iris)
fig=plt.gcf()
fig=sns.boxplot(x='Species',y='SepalLengthCm',data=iris)
fig=sns.stripplot(x='Species',y='SepalLengthCm',data=iris)
plt.subplot(2,2,1)
sns.violinplot(x='Species',y='PetalLengthCm',data=iris)
plt.subplot(2,2,2)
sns.violinplot(x='Species',y='PetalWidthCm',data=iris)
plt.subplot(2,2,3)
sns.violinplot(x='Species',y='SepalLengthCm',data=iris)
plt.subplot(2,2,4)
sns.violinplot(x='Species',y='SepalWidthCm',data=iris)
sns.scatterplot(x='Species',y='PetalLengthCm',data=iris)
sns.pairplot(data=iris,hue='Species')
sns.heatmap(data=iris.corr(),annot=True)
sns.boxplot(x='Species',y='PetalLengthCm',data=iris)
sns.swarmplot(x='Species',y='PetalLengthCm',data=iris)
sns.lmplot(x="PetalLengthCm",y='PetalWidthCm',data=iris)
sns.FacetGrid(iris,hue='Species')
from pandas.tools.plotting import andrews_curves
andrews_curves(iris,"Species",colormap='rainbow')
plt.ioff()
from pandas.tools.plotting import parallel_coordinates
parallel_coordinates(iris,"Species",colormap="rainbow")
sns.factorplot('Species','SepalLengthCm',data=iris)
sns.boxenplot('Species','SepalLengthCm',data=iris)
fig=sns.residplot('SepalLengthCm', 'SepalWidthCm',data=iris)
| 0.590189 | 0.868938 |
# Assignment 3: TCP Congestion Control and Bufferbloat
### Due March 18th, 5:00pm
The last assignment involved programming a distributed routing algorithm in a simulated network. In this assignment, you will create your own network simulation to investigate the dynamics of TCP and how seemingly minor configuration decisions made by network operators can have major performance effects.
As discussed in lecture, TCP is a protocol for obtaining reliable transmission over an unreliable packet-switched network. Another important component of TCP is congestion control, i.e. limiting end host send rates to prevent network infrastructure from getting overwhelmed with traffic.
However, networks can suffer congestion-related performance issues even when end hosts use TCP. One such issue, known as bufferbloat, can occur when packet buffers on routers and switches are too large.
In this assignment, you will use Mininet, a useful tool for network experiments, to emulate a small network and collect various performance statistics relevant to TCP congestion control and bufferbloat. This will allow you to reason about the effects of TCP and router configuration on network performance.
**You should work with a partner on this assignment. Put your names and netIDs in the cell below:**
**Names:**
**NetIds:**
## Background
#### TCP Congestion Window Size
The TCP congestion window size parameter, typically styled "cwnd," is maintained by the sender and determines how much traffic can be outstanding (sent but not acknowledged) at any time. There are many algorithms for controlling the value of cwnd during a TCP connection, all with the goal of maximizing the connection's throughput while preventing congestion. The additive increase and multiplicative decrease algorithm was discussed in lecture.
#### Bufferbloat
Bufferbloat is a phenomenon that happens when a switching device is configured to use excessively large buffers, which can in turn cause high latency and packet delay variation (jitter). This can happen even in a typical home network like the following:
<img width=600 src="figures/home-network.png">
Here, the end host in the home network is connected to the home router. The home router is then connected, via cable or DSL, to a headend router run by the Internet service provider (ISP). By simulating and experimenting with a similar network in Mininet, you will see how bufferbloat causes poor performance.
#### Mininet
Mininet is a network emulator with which you can create a custom network of virtual hosts, switches, controllers, and links, all on a single computer. The virtual devices in the emulated network can run real programs; anything that can run on linux can run on a Mininet device too. This makes Mininet a valuable tool for fast and easy simulation of network protcols and measurements. This [Introduction to Mininet](https://github.com/mininet/mininet/wiki/Introduction-to-Mininet) is a useful guide for getting started with Mininet's Python API. The [Mininet website](www.mininet.org) has additional resources if you are interested.
## Part A: Network Simulation & Measurement
To start, you should first create the following network using Mininet's Python API, which emulates a typical home netowrk:
<img width=450 src="figures/mininet-topo.png">
Here h1 is your home computer that has a fast connection (1Gb/s) to your home router. The home router has a slow uplink connection (1.5Mb/s). The round-trip propagation delay, or the minimum RTT between h1 and h2 is 20ms. The router buffer (queue) size will be the parameterized independent variable in your simulation.
To create a custom topology in Mininet, we extend the mininet.topo.Topo class. We have already added the switch (the router) to topology for you. You need to add h1, h2, and links with appropriate characteristics to create the setting specified in the image above. The first few subsections of the [Working with Mininet](https://github.com/mininet/mininet/wiki/Introduction-to-Mininet#working) section of the Mininet guide describe how to add elements to a topology and set performance parameters.
```
from mininet.topo import Topo
class BBTopo(Topo):
"Simple topology for bufferbloat experiment."
def __init__(self, queue_size):
super(BBTopo, self).__init__()
# Create switch s0 (the router)
self.addSwitch('s0')
# TODO: Create two hosts with names 'h1' and 'h2'
# TODO: Add links with appropriate bandwidth, delay, and queue size parameters.
# Set the router queue size using the queue_size argument
# Set bandwidths/latencies using the bandwidths and minimum RTT given in the network diagram above
return
```
Next, we need a couple of helper functions to generate traffic between the two hosts. The following function starts a long-lived TCP flow which sends data from h1 to h2 using **iperf**. [Iperf](https://iperf.fr/) is "a tool for active measurements of the maximum achievable bandwidth on IP networks." You can think of this iperf traffic like a one-way video call. It continually attempts to send a high volume of traffic from the home computer h1 to the server h2.
The following function receives one argument called `net`, which is an instance of mininet with a BBTopo topology that we have created above. We have written the part for the server (h2). You need to complete the function to also start iperf on the client (h1). The iperf session should run for the number of seconds given in the `experiment_time` argument.
You will need to use the `popen` function to run shell commands on a mininet host. The first argument to `popen` is a string command just like you would run in your shell. The second argument should be `shell=True`. You will need to look up the appropriate command line options to run iperf as a client for a given amount of time in the documentation here: [https://iperf.fr/iperf-doc.php#3doc](https://iperf.fr/iperf-doc.php#3doc). You will also need to include the IP address of h2 in your iperf command. This IP address can be accessed with the `h2.IP()` method.
```
def start_iperf(net, experiment_time):
# Start a TCP server on host 'h2' using perf.
# The -s parameter specifies server mode
# The -w 16m parameter ensures that the TCP flow is not receiver window limited (not necessary for client)
print "Starting iperf server"
h2 = net.get('h2')
server = h2.popen("iperf -s -w 16m", shell=True)
# TODO: Start an TCP client on host 'h1' using iperf.
# Ensure that the client runs for experiment_time seconds
print "Starting iperf client"
```
Next, you need to complete the following function that starts a back-to-back ping train from h1 to h2 to measure RTTs. A ping should be sent every 0.1 seconds. Results should be redirected from stdout to the `outfile` argument.
As before, `net` is an instance of mininet with a BBTopo topology. As before, you will need to use `popen`. The command argument to `popen` can redirect stdout using `>` just like a normal shell command. Read the man page for `ping` for details on available command line arguments. Make sure the second argument to `popen` is `shell=True`.
```
def start_ping(net, outfile="pings.txt"):
# TODO: Start a ping train from h1 to h2 with 0.1 seconds between pings, redirecting stdout to outfile
print "Starting ping train"
```
Next, we develop some helper functions to measure the congestion window of the TCP traffic. This will let us analyze at the dynamics of the TCP connections in the mininet network. The following functions are already complete.
```
from subprocess import Popen
import os
def start_tcpprobe(outfile="cwnd.txt"):
Popen("sudo cat /proc/net/tcpprobe > " + outfile, shell=True)
def stop_tcpprobe():
Popen("killall -9 cat", shell=True).wait()
```
We then create a helper function that monitors the queue length on a given interface. This will let us analyze how the number of packets in router buffer queues affects performance. This function is already complete.
```
from multiprocessing import Process
from monitor import monitor_qlen
def start_qmon(iface, interval_sec=0.1, outfile="q.txt"):
monitor = Process(target=monitor_qlen,
args=(iface, interval_sec, outfile))
monitor.start()
return monitor
```
We also need a helper function that starts a webserver on h1. This function is already complete.
```
from time import sleep
def start_webserver(net):
h1 = net.get('h1')
proc = h1.popen("python http/webserver.py", shell=True)
sleep(1)
return [proc]
```
Finally, we need a helper function that runs on h2, fetches the website from h1 every 3 seconds for `experiment_time`, and prints the average and standard deviation of the download times. This function is already complete
```
from time import time
from numpy import mean, std
from time import sleep
def fetch_webserver(net, experiment_time):
h2 = net.get('h2')
h1 = net.get('h1')
download_times = []
start_time = time()
while True:
sleep(3)
now = time()
if now - start_time > experiment_time:
break
fetch = h2.popen("curl -o /dev/null -s -w %{time_total} ", h1.IP(), shell=True)
download_time, _ = fetch.communicate()
print "Download time: {0}, {1:.1f}s left...".format(download_time, experiment_time - (now-start_time))
download_times.append(float(download_time))
average_time = mean(download_times)
std_time = std(download_times)
print "\nDownload Times: {}s average, {}s stddev\n".format(average_time, std_time)
```
Now, we need to put together all the pieces to create the network, start all the traffic, and make the measurements.
The following `bufferbloat()` function should:
* create a `BBTopo` object
* start the TCP and queue monitors
* start a long-lived TCP flow using iperf
* start the ping train
* start the webserver
* Periodically download the index.html web page from h1 and measure how long it takes to fetch it
Note that the long lived flow, ping train, and webserver downloads should all be happening simultaneously. Once you have completed the assignment steps up until here, complete the sections marked `TODO` in the below `bufferbloat()` function. Each TODO section requires adding one line to call a function defined above.
```
from mininet.node import CPULimitedHost, OVSController
from mininet.link import TCLink
from mininet.net import Mininet
from mininet.log import lg, info
from mininet.util import dumpNodeConnections
from time import time
import os
from subprocess import call
def bufferbloat(queue_size, experiment_time, experiment_name):
# Don't forget to use the arguments!
# Set the cwnd control algorithm to "reno" (half cwnd on 3 duplicate acks)
# Modern Linux uses CUBIC-TCP by default that doesn't have the usual sawtooth
# behaviour. For those who are curious, replace reno with cubic
# see what happens...
os.system("sysctl -w net.ipv4.tcp_congestion_control=reno")
# create the topology and network
topo = BBTopo(queue_size)
net = Mininet(topo=topo, host=CPULimitedHost, link=TCLink,
controller= OVSController)
net.start()
# Print the network topology
dumpNodeConnections(net.hosts)
# Performs a basic all pairs ping test to ensure the network set up properly
net.pingAll()
# Start monitoring TCP cwnd size
outfile = "{}_cwnd.txt".format(experiment_name)
start_tcpprobe(outfile)
# TODO: Start monitoring the queue sizes with the start_qmon() function.
# Fill in the iface argument with "s0-eth2" if the link from s0 to h2
# is added second in BBTopo or "s0-eth1" if the link from s0 to h2
# is added first in BBTopo. This is because we want to measure the
# number of packets in the outgoing queue from s0 to h2.
outfile = "{}_qsize.txt".format(experiment_name)
qmon = start_qmon(iface="TODO", outfile=outfile)
# TODO: Start the long lived TCP connections with the start_iperf() function
# TODO: Start pings with the start_ping() function
outfile = "{}_pings.txt".format(experiment_name)
# TODO: Start the webserver with the start_webserver() function
# TODO: Measure and print website download times with the fetch_webserver() function
# Stop probing
stop_tcpprobe()
qmon.terminate()
net.stop()
# Ensure that all processes you create within Mininet are killed.
Popen("pgrep -f webserver.py | xargs kill -9", shell=True).wait()
call(["mn", "-c"])
```
Once you have completed all the steps above, use the `bufferbloat()` function to run the experiment twice, once with queue size of a 20 packets and then queue size of 100 packets. Make sure to run the experiments long enough to see the dynamics of TCP, like the sawtooth behavior of cwnd, in your results (300 seconds should be good). Choose `experiment_name` arguments that reflect the queue size
```
from subprocess import call
call(["mn", "-c"])
# TODO: call the bufferbloat function twice, once with queue size of 20 packets and once with a queue size of 100.
```
## Part B: Plotting Results
In this part of the assignment, you will analyze your measurements by plotting the variations in congestion window, queue length, and ping RTT versus time. We have provided plotting functions for each of these measurements, which are called in the following already complete `plot_measurements()` function.
```
%matplotlib inline
from plot_cwnd import plot_congestion_window
from plot_qsize import plot_queue_length
from plot_ping import plot_ping_rtt
def plot_measurements(experiment_name_list, cwnd_histogram=False):
# plot the congestion window over time
for name in experiment_name_list:
cwnd_file = "{}_cwnd.txt".format(name)
plot_congestion_window(cwnd_file, histogram=cwnd_histogram)
# plot the queue size over time
for name in experiment_name_list:
qsize_file = "{}_qsize.txt".format(name)
plot_queue_length(qsize_file)
# plot the ping RTT over time
for name in experiment_name_list:
ping_file = "{}_pings.txt".format(name)
plot_ping_rtt(ping_file)
```
Now you need to call the `plot_measurements` function such that the `experiment_name_list` argument is list of the `experiment_name` arguments you used to run `bufferbloat()` above. This should generate 6 plots with the results of the experiments.
```
#TODO: Call plot_measurements() to plot your results
```
## Part C: Analysis
In this part of the assignment, you will answer some questions about TCP and bufferbloat using your simulations and the plots from the previous section. This questions are intentionally open-ended and many have multiple correct answers. There is no required answer length, but attempt to be both thorough and concise. 1-2 sentences is probably too short. More than 2-3 paragraphs is probably too long.
Take some time first to think about the simulation you just performed. The simulation was set up like a home network with a home computer connected to a remote server through a router. The link from the router to the server had much lower bandwidth than the link from the home computer to the router. The independent variable in the simulation was the maximum length of the buffer of packets waiting to be sent from the router to the server.
There were 3 sources of traffic:
1. A long-lasting TCP session (creating using iperf) sending a high volume of traffic from the home computer to the server.
2. Regularly spaced pings and ping replies to and from the home computer and the server
3. Regularly spaced attempts to download a website (using HTTP over TCP) from the home computer to the server.
As you (hopefully) discovered through the experiment, increasing the length of the packet buffer on the router significantly reduced performance by both ping RTT and HTTP download rate metrics.
### Questions
#### Q1.
What computer networks other than a home network might have a configuration like the one you simulated?
#### A1.
*TODO: your answer here.*
#### Q2.
Write a symbolic equation to describe the relation between RTT and queue size.
The symbolic equation should be generalized to any queue size. Basically, consider a snapshot of a system at one point of time, and use queue size and link delays parametrically to compute the RTT
An example (incorrect) symbolic equation:
$$RTT = kq^2$$
where $k$ is a constant factor and $q$ is the number of packets in the queue. Your equation is not limited to $k$ and $q$.
#### A2.
*TODO: your answer here. Use single dollar signs for inline latex math formatting and double dollar signs for block latex math formatting.*
#### Q3.
Describe in technical terms why increasing buffer size reduces performance (RTTs and webpage download times), causing the bufferbloat effect. Be sure to explicitly reference the plots you generated and the relationship between TCP congestion control and buffer size. *This is the most important question and will be weighted correspondingly more.*
#### A3.
*TODO: your answer here.*
#### Q4.
Re-describe the cause of the bufferbloat effect using a non-technical analogy to something other than computer networking. It is important to be able to describe technical content such that a layperson can understand, and generating analogies often helps your own reasoning.
#### A4.
*TODO: your answer here.*
#### Q5.
Is the bufferbloat effect specific to the type of network, traffic, and/or TCP congestion control algorithm we simulated, or is it a general phenomenon?
Are there any times when increasing router buffer size would improve performance? If so, give an example. If not, explain why not.
#### A5.
*TODO: your answer here.*
#### Q6.
Identify and describe a way to mitigate the bufferbloat problem without reducing buffer sizes.
#### A6.
*TODO: your answer here.*
## Submission
**Remember to "Save and Checkpoint" (from the "File" menu above) before you leave the notebook or close your tab.**
Submit this file (Assignment3_Notebook.ipynb) on CS Dropbox here: <>. Submit only once for both partners. Remember to put your names and netids in the marked location at the top of this file.
|
github_jupyter
|
from mininet.topo import Topo
class BBTopo(Topo):
"Simple topology for bufferbloat experiment."
def __init__(self, queue_size):
super(BBTopo, self).__init__()
# Create switch s0 (the router)
self.addSwitch('s0')
# TODO: Create two hosts with names 'h1' and 'h2'
# TODO: Add links with appropriate bandwidth, delay, and queue size parameters.
# Set the router queue size using the queue_size argument
# Set bandwidths/latencies using the bandwidths and minimum RTT given in the network diagram above
return
def start_iperf(net, experiment_time):
# Start a TCP server on host 'h2' using perf.
# The -s parameter specifies server mode
# The -w 16m parameter ensures that the TCP flow is not receiver window limited (not necessary for client)
print "Starting iperf server"
h2 = net.get('h2')
server = h2.popen("iperf -s -w 16m", shell=True)
# TODO: Start an TCP client on host 'h1' using iperf.
# Ensure that the client runs for experiment_time seconds
print "Starting iperf client"
def start_ping(net, outfile="pings.txt"):
# TODO: Start a ping train from h1 to h2 with 0.1 seconds between pings, redirecting stdout to outfile
print "Starting ping train"
from subprocess import Popen
import os
def start_tcpprobe(outfile="cwnd.txt"):
Popen("sudo cat /proc/net/tcpprobe > " + outfile, shell=True)
def stop_tcpprobe():
Popen("killall -9 cat", shell=True).wait()
from multiprocessing import Process
from monitor import monitor_qlen
def start_qmon(iface, interval_sec=0.1, outfile="q.txt"):
monitor = Process(target=monitor_qlen,
args=(iface, interval_sec, outfile))
monitor.start()
return monitor
from time import sleep
def start_webserver(net):
h1 = net.get('h1')
proc = h1.popen("python http/webserver.py", shell=True)
sleep(1)
return [proc]
from time import time
from numpy import mean, std
from time import sleep
def fetch_webserver(net, experiment_time):
h2 = net.get('h2')
h1 = net.get('h1')
download_times = []
start_time = time()
while True:
sleep(3)
now = time()
if now - start_time > experiment_time:
break
fetch = h2.popen("curl -o /dev/null -s -w %{time_total} ", h1.IP(), shell=True)
download_time, _ = fetch.communicate()
print "Download time: {0}, {1:.1f}s left...".format(download_time, experiment_time - (now-start_time))
download_times.append(float(download_time))
average_time = mean(download_times)
std_time = std(download_times)
print "\nDownload Times: {}s average, {}s stddev\n".format(average_time, std_time)
from mininet.node import CPULimitedHost, OVSController
from mininet.link import TCLink
from mininet.net import Mininet
from mininet.log import lg, info
from mininet.util import dumpNodeConnections
from time import time
import os
from subprocess import call
def bufferbloat(queue_size, experiment_time, experiment_name):
# Don't forget to use the arguments!
# Set the cwnd control algorithm to "reno" (half cwnd on 3 duplicate acks)
# Modern Linux uses CUBIC-TCP by default that doesn't have the usual sawtooth
# behaviour. For those who are curious, replace reno with cubic
# see what happens...
os.system("sysctl -w net.ipv4.tcp_congestion_control=reno")
# create the topology and network
topo = BBTopo(queue_size)
net = Mininet(topo=topo, host=CPULimitedHost, link=TCLink,
controller= OVSController)
net.start()
# Print the network topology
dumpNodeConnections(net.hosts)
# Performs a basic all pairs ping test to ensure the network set up properly
net.pingAll()
# Start monitoring TCP cwnd size
outfile = "{}_cwnd.txt".format(experiment_name)
start_tcpprobe(outfile)
# TODO: Start monitoring the queue sizes with the start_qmon() function.
# Fill in the iface argument with "s0-eth2" if the link from s0 to h2
# is added second in BBTopo or "s0-eth1" if the link from s0 to h2
# is added first in BBTopo. This is because we want to measure the
# number of packets in the outgoing queue from s0 to h2.
outfile = "{}_qsize.txt".format(experiment_name)
qmon = start_qmon(iface="TODO", outfile=outfile)
# TODO: Start the long lived TCP connections with the start_iperf() function
# TODO: Start pings with the start_ping() function
outfile = "{}_pings.txt".format(experiment_name)
# TODO: Start the webserver with the start_webserver() function
# TODO: Measure and print website download times with the fetch_webserver() function
# Stop probing
stop_tcpprobe()
qmon.terminate()
net.stop()
# Ensure that all processes you create within Mininet are killed.
Popen("pgrep -f webserver.py | xargs kill -9", shell=True).wait()
call(["mn", "-c"])
from subprocess import call
call(["mn", "-c"])
# TODO: call the bufferbloat function twice, once with queue size of 20 packets and once with a queue size of 100.
%matplotlib inline
from plot_cwnd import plot_congestion_window
from plot_qsize import plot_queue_length
from plot_ping import plot_ping_rtt
def plot_measurements(experiment_name_list, cwnd_histogram=False):
# plot the congestion window over time
for name in experiment_name_list:
cwnd_file = "{}_cwnd.txt".format(name)
plot_congestion_window(cwnd_file, histogram=cwnd_histogram)
# plot the queue size over time
for name in experiment_name_list:
qsize_file = "{}_qsize.txt".format(name)
plot_queue_length(qsize_file)
# plot the ping RTT over time
for name in experiment_name_list:
ping_file = "{}_pings.txt".format(name)
plot_ping_rtt(ping_file)
#TODO: Call plot_measurements() to plot your results
| 0.338405 | 0.993203 |
# SVM
## Introduction
- SVM classifiers are used for **prediction with confidence**. (Why I say so will be explained further)
- Also called as **Large Margin Classifier**.
- Primarily designed for **binary classification**, meaning it distinguishes between two classes; think (+) or (-). More advanced SVMs can discriminate between many classes.
- SVM can be used for learning complex **non-linear boundary functions**.
- Linear SVM uses **Hinge loss**, whereas Non-linear SVM uses **dual form with QP**.
## Defining the hyperplane for SVM
### Introduction
**In SVM, given two classes, we seek to produce a hyperplane that maximizes the distance between the two training classes.**
<br>
<center><img src="images/small_margin.gif" width="80%"/></center>
The margin is up to the nearest point of each class.
The general equation of a (hyper)plane in the n-dimensional feature space (in two dimensions a plane is a line):
$$ f_\theta (x) = \theta_0 + \theta_1x_1 + \theta_2x_2 + \cdots + \theta_nx_p $$
$$ f_\theta (x) = \theta_0 + \textbf{$\theta^{T}$}\textbf{x} $$
Our plane is going to divide the two classes in hyperspace. There are an infinite number of ways of choosing the different coefficients to represent the plane, but in this case we are just going to make it as simple as possible by setting $f_\theta (x)$ to 1:
$$ |\theta_0 + \textbf{$\theta^{T}$}\textbf{x}| = 1 $$
In this case $\textbf{x}$ represents those training examples closest to the hyperplane. This is an important element of what we are doing, because ideally we want to find the plane that maximizes distance between the positive and negative training examples.
### Principal equation of the hyperplane
Recall that we were discussing classifying the data into the (+) and (-) group. Thus we consider these classification states as output variables $\textbf{y}$ to our method. Going back to our equation, we can write:
**For the positive class:**
$$\theta_0 + \textbf{$\theta^{T}$}\textbf{x} = 1$$
**For the negative class:**
$$\theta_0 + \textbf{$\theta^{T}$}\textbf{x} = -1$$
Again recalling that the $y$ are either $+1$ or $-1$.
Also recall that the plane includes all data point solutions above the plane in the positive class. All solutions below the plane are in the negative class, therefore;
$$\theta_0 + \textbf{$\theta^{T}$}\textbf{x} \geq 1$$
and also
$$\theta_0 + \textbf{$\theta^{T}$}\textbf{x} \leq -1$$
Thinking of the $y_i$ as multipliers by $+1$ or $-1$, we can write:
$$y_i(\theta_0 + \theta^{T}x_{i}) \geq 1$$
for all datapoints $i=1,2, \cdots , m$
So, we see that in SVM instead of one decision boundary $ \theta_T * x=0 $, we have two boundaries, $\theta_T *x=1$ and $\theta_T *x=-1$
Perceptron Vs SVM
<center><img src="images/svm_confidence.png" width="80%"/></center>
#### Maximise Margin
The total **margin** between the support vectors is given by **m**:
$$m = \frac{2}{\|\theta\|}$$
With an SVM, we seek to **maximize m**. This is why SVMs are called **maximum margin classifiers**.
<center><img src="images/max_margin.png" width="500"/></center>
### To summarise:
**We can define the hyperplane in terms of the training points closest to the plane. These points are called *support vectors***.
The equation of the plane is given as
$$|\theta_0 + \textbf{$\theta^{T}$}\textbf{x}|=1$$
Where the $\textbf{x}$ are the **support vectors**.
It bears mentioning here that the $\theta$ coefficients are often described in other literature as **"weights"**, with the origin $\theta_0$ callsed **"bias"** $b$, such that the equation of the plane is written:
$$|\textbf{$\theta^{T}$}\textbf{x}+b|=1$$
However, we are going to continue to use the $\theta$ notation in these notes so as to highlight the relationship between the well-known OLS regression and SVMs.
## Cost Function
Recall that our main cost function seeks to find those minimal weights *as a whole* (remember shrinkage) that fit our criteria. Returning to the concept of a **norm**, we are going to minimize $\|\theta\|_2$ subject to our principle equation, $\sum_{i=0}^{n}y_i(\theta_0 + \theta^{T}x_{i}) \geq 1$ derived above.
We use an equation known in the literature as a *Lagrangian* as a way of posing the cost function of our method. A Lagrangian is an equation of the form:
$$\Lambda(x,y,\alpha) = f(x,y)+\alpha\ \cdot g(x,y)$$
#### Primal Form
<div style="background-color:#F7F99C">
In this case we do not formulate the cost function in terms of $\|\theta\|_2$ due to the onerous nature of the square root involved. Rather we can **transform** this term to its pseudo-integral, $\frac{1}{2}\|\theta\|_2^{2}$, which has the same properties.
$$\Lambda(x,y,\alpha) = \frac{1}{2}\theta^{T}\theta+\sum_{i=0}^{n}\alpha_{i}\ \cdot (1-y_i(\theta_0 + \theta^{T}x_{i}))$$
This is called the *primal form* because the **weights and coefficients appear in the same equation.** Now take the grad set to zero,taking the full gradient and setting the hyperplane intercept to zero:
$$\nabla_{\theta}\Lambda(x,y,\alpha) = \theta+\sum_{i=0}^{n}\alpha_{i}\ (-y_i(x_{i})) = 0$$
That gives us a first solution for the **weights**:
$$\theta = \sum_{i=0}^{n}\alpha_{i}y_{i}x_{i}$$
We also have a solution for the **bias** that we already set to zero:
$$\theta_0 = \sum_{i=0}^{n}\alpha_{i}y_{i} = 0$$
</div>
#### Dual Form
<div style="background-color:#F7F99C">
The more common form of the SVM Lagrangian is the *dual form*, which we can get if we substitute our solution for the weights, $\theta = \sum_{i=0}^{n}\alpha_{i}y_{i}x_{i}$, into the primal form. We also remember that $\sum_{i=0}^{n}\alpha_{i}y_{i} = 0$.
$$\Lambda(x,y,\alpha) = \frac{1}{2}\sum_{j=0}^{n}\alpha_{j}y_{j}x_{j}^{T}\sum_{i=0}^{n}\alpha_{i}y_{i}x_{i}+\sum_{i=0}^{n}\alpha_{i}\ \cdot (1-y_i(\theta_0 + \sum_{j=0}^{n}\alpha_{j}y_{j}x_{j}^{T}x_{i}))$$
This reorganizes (see appendix) to get the following equation:
</div>
$$\Lambda(x,y,\alpha) = \sum_{i=0}^{n}\alpha_{i}-\frac{1}{2}\sum_{i=0}^{n}\sum_{j=0}^{n}\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j}$$
This is a *dual form* because we see that we are describing the **weights** in terms of $\alpha$. Weights are not present in this equation. If we know all of the $\alpha$ we know the $\theta$; remember the formula $\theta = \sum_{i=0}^{n}\alpha_{i}y_{i}x_{i}$.
hinge loss function
------
<center><img src="images/The-hinge-loss-function.png" width="75%"/></center>
## Soft Margin SVMs
For reasons discussed in more detail below, most real-world cases will require the ability to tolerate some misclassification, where there are datapoints labeled as one class that should be in the other, or where noise or hidden variables lead to unclear relationships amongst the classes.
In this case, **even the optimal solution** will have some training datapoints that land on the wrong side of the plane.
We deal with this using **Soft Margins**. All standard SVM applications use soft margins.
For each point $i$, if the point falls on the wrong side of the hyperplane during training, we assign it a *penalty* $\xi_i$, based on its distance from the plane.
We minimize $\sum_{i=0}^{n}\xi_i$ along with the hyperplane during optimization.
In this case, the principal equations become:
$$y_{i}(\theta_0 + \textbf{$\theta^{T}$}x_{i}) \geq m(1 - \xi_{i})$$
For the positive class, and
$$y_{i}(\theta_0 + \textbf{$\theta^{T}$}x_{i}) \leq m(\xi_{i} - 1)$$
For the negative class. We also are assured that
$$\xi_{i} \geq 0$$
Because we don't penalize points that are not on the wrong side of the plane.
***notice that the margin figures importantly into the principal equations here. what does it mean to have a nonzero $\xi_{i}$ value?**
So we want to minimize $\frac{1}{2}\|\theta\|_2^{2}+C\sum_{i=0}^{n}\xi_i$, subject to the above prinicpal equations. It turns out that this is not too much of a change to the dual form of the Lagrangian (Hastie has the derivation for the primal form on pp. 420):
$$\Lambda(x,y,\alpha) = \sum_{i=0}^{n}\alpha_{i}-\frac{1}{2}\sum_{i=0}^{n}\sum_{j=0}^{n}\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j}$$
Where $\sum_{i=0}^{n}\alpha_{i}y_{i}x_{i}=0$ and we also follow the constraint $C\geq\alpha_i\geq 0$; this is all dealt with simultaneously during optimization.
The question arises: Now that we've included this new constant $C$ in the equations, what is its effect on the location of the decision boundary? We find that the fixed scale 1 is arbitrary because the $M$ term can be brought into the $\theta$.
What is important is that you understand that the **margin** $M$ changes with respect to $C$. Changes in $C$ result in changes in the $\theta$. This in turn results in changes in the direction of the plane and width of the margin simultaneously. More on this is discussed below.
### Optimization
We seek to maximize the (dual) Lagrangian as if it were an expectation function:
$$\Lambda(x,y,\alpha) = \sum_{i=0}^{n}\alpha_{i}-\frac{1}{2}\sum_{i=0}^{n}\sum_{j=0}^{n}\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j}$$
subject to some $\alpha_{i} \geq 0$ and $\sum_{i=0}^{n}\alpha_{i}y_{i} = 0$
Problems of this type are called Quadratic Programming Problems (because a squared term exists in one variable) and require relatively heavy-duty methods to reliably achieve the solution. However, because they are QP, we are assured that a **global** not approximate(!) maximum of $\alpha_i$ can be found if optimized correctly!
We will not cover QP solvers in this unit.
Directly computing weights is comparatively rare in most SVM implementations. It is more common to simply calculate the $\alpha_{i}$ and just report the model as such. This produces the **model** $G$ for input data $\textbf{z}$:
$$G(\textbf{z}) = \sum_{j=0}^{s}\alpha_{j}y_{j} \textbf{$x_{j}$}^{T}\textbf{z}$$
Where the $j$ are the indices of the $s$ support vectors.
The model classifies $\textbf{z}$ as class 1 (+) if positive and class 0 (-) otherwise.
(Note that the weights do not appear)
### Reasoning
Choosing a bisecting plane to divide two classes enables us to predict a classification using a training set without assuming linearity in the data or fitting to an explicit function.
SVM provides several advantages. In simple cases, SVM outperforms logistic regression. SVMs perform feature selection automatically by allowing $\alpha$ coefficients that are unimportant to the classification to go to 0; SVMs also perform shrinkage on the $\theta$ coefficients. We are assured of an optimal solution due to the formulation of the method.
## Kernels: Linear vs. Nonlinear SVMs
"Kernel" means "heart" or "essence" of something.
In math, which is the origin of the word in this case, a kernel is a member of a set of functions:
$$K: \mathbb{R}^{N} \times \mathbb{R}^{N} \rightarrow \mathbb{R}$$
In the case of the SVM, a kernel maps lower dimensional data into higher dimension and refers to a very particular term in the cost function:
$$\Lambda(x,y,\alpha) = \sum_{i=0}^{n}\alpha_{i}-\frac{1}{2}\sum_{i=0}^{n}\sum_{j=0}^{n}\alpha_{i}\alpha_{j}y_{i}y_{j}\underline{x_{i}^{T}x_{j}}$$
This is a dot or inner product, meaning that it produces a single number for each pair of points (vectors) $x_i, x_j$. You will often see it written this way (inner product vector notation):
$$x_{i}^{T}x_{j} = <x_j,x_i>$$
This regular, standard kernel is called a **linear** kernel.
The idea behind using kernels is that hopefully, then the *transformed* data is __linearly separable__ in higher dimensions.
From 2 to 3 dimensions
-----
<center><img src="images/feature_space.png" width="90%"/></center>
φ is the Greek letter "Phi"
φ is the mapping from one dimension to another
From 2 to 3 dimensions
-----
<center><img src="images/kernel-768x306.png" width="85%"/></center>
```
#RBF Kernel
from IPython.display import YouTubeVideo
YouTubeVideo("9NrALgHFwTo")
from IPython.display import YouTubeVideo
YouTubeVideo("3liCbRZPrZA")
```
<center><img src="images/feature_2.png" width="50%"/></center>
## The Kernel Trick
<div>
<font color="red">THIS IS A STANDARD INTERVIEW QUESTION</font>
</div>
Transforming the entire Lagrangian from linear into polynomial space is extremely costly in terms of resources. Normally this would imply that we would need to actually increase the number of $\alpha$ coefficients and transform the response variables too.
If we were to actually do this, the computation would become prohibitively expensive rapidly with respect to the number of features. However, because we simply compute a **dot product** for every pair of data points $i,j$ in the original feature space $N$ after we transform the data, *this doesn't happen*.
We call this **"the kernel trick"**.
It's a trick because transformation to a nonlinear space doesn't seem to cost us much; we don't need to calculate the transformation explicitly. We can simply calculate the kernel product once, using the additional functions as necessary(a $N \times N$ matrix). This only leads to a linear increase in calculation time - the time it costs to calculate the transformation from one basis to another. We do not increase the dimensionality of the Lagrangian at all. Since this is the most expensive function in terms of computation and storage, it amounts to immense computational leverage.
#### Types of Kernels
It bears mentioning that there are a few different types of kernels commonly in use; we will mention them here.
##### Linear Kernel:
This is simply the standard dot product $<x_j,x_i>$. Sometimes a scalar coefficient $c$ is added.
$$K(x_j,x_i) = <x_j,x_i>+c$$
##### Polynomial Kernels
Some combination of transformations and exponents of the orignal linear kernel. Transformation by the function:
$$f(x, b, c, d) = (bx+c)^{d}$$
Leads to the following expression:
$$K(x_j,x_i) = (b<x_j,x_i>+c)^{d}$$
##### Gaussian and Radial Basis Function Kernels
Radial basis functions model the data as if it came from a distribution, this is done in order to "soften" the locations of the training data. The Gaussian radial kernel is the most common of these:
$$K(x_j,x_i) = e^{-\frac{\|x_j-x_i\|^{2}}{2\sigma^{2}}}$$
There are other variants of the Gaussian, such as the Exponential and Laplace kernels, that leave out the square of the norm.
The quality of the outcome is rather dependent on the scaling factor $\sigma$ (the broadness of the Gaussian function).
<center><img src="images/SVM_kernel_1.png" width="75%"/></center>
Kernels & SVMS: Make the data more complex, and keep the model simple.
-------
```
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
import sklearn
import warnings
warnings.filterwarnings('ignore')
palette = "Dark2"
%matplotlib inline
from sklearn.datasets.samples_generator import make_blobs
from sklearn import datasets
from sklearn.svm import SVC
iris = datasets.load_iris()
X = iris.data[:, :2] # Use only the first two features for easier plotting
y = iris.target
# Define & fit model
#kernel = 'linear'
#kernel = 'rbf'
kernel = 'poly'
clf = SVC(kernel=kernel).fit(X, y)
predicted = clf.predict(iris.data[:, :2])
clf.n_support_
# Plot the decision bounds
from mlxtend.plotting import plot_decision_regions
y_labels = y # actual
# y_labels = predicted
plot_decision_regions(X=X,
y=y_labels,
clf=clf,
legend=False);
# NOTE: Linear kernel includes piecewise linear, hence the discontinuity
# Plot the decision bounds
from mlxtend.plotting import plot_decision_regions
y_labels = y # actual
# y_labels = predicted
plot_decision_regions(X=X,
y=y_labels,
clf=clf,
legend=False);
# NOTE: Linear kernel includes piecewise linear, hence the discontinuity
# Plot the decision bounds
from mlxtend.plotting import plot_decision_regions
y_labels = y # actual
# y_labels = predicted
plot_decision_regions(X=X,
y=y_labels,
clf=clf,
legend=False);
# NOTE: Linear kernel includes piecewise linear, hence the discontinuity
# From blobs to circles
from sklearn.datasets import make_circles
X, y = make_circles(noise=0.1, factor=.01, random_state=42)
ax = sns.scatterplot(x=X[:,0], y=X[:,1], hue=y, palette=palette, legend=False);
ax.set_yticklabels(''); ax.set_xticklabels('');
# Define & fit model
# kernel = 'linear'
kernel = 'rbf'
# kernel = 'poly'
clf = SVC(kernel=kernel).fit(X, y)
predicted = clf.predict(iris.data[:, :2])
# Plot the decision bounds
from mlxtend.plotting import plot_decision_regions
y_labels = y # actual
# y_labels = predicted
plot_decision_regions(X=X,
y=y_labels,
clf=clf,
legend=False);
# NOTE: Linear kernel includes piecewise linear, hence the discontinuity
```
What are the disadvantages of kernel methods?
------
- Selecting a reasonable kernel can be hard.
- Since kernels are often nonlinear and involve higher dimensional scaling, there can be high computational cost.
## Multiclass SVMs
It is possible to extend the SVM methodology to more than two classes by solving the constraints for each pair of classes separately.

There are two methods:
##### Any-of
In this case, we build classifiers for each possible class pair and let them all vote. This is an example where it's ok to have something classified as more than one class.
1. Build a classifier for each class, where the training set consists of the set of documents in the class (positive labels) and its complement (negative labels).
1. Given the test set, apply each classifier separately. The decision of one classifier has no influence on the decisions of the other classifiers.
##### One-of
Here we insist that each object must belong to one and only one class.
1. Build a classifier for each class, where the training set consists of points belonging to the class (positive labels) and its complement (negative labels).
1. Given the test document, apply each classifier separately.
1. Assign the document to the class with
1. the maximum score,
1. the maximum confidence value,
1. or the maximum probability of class membership based on the votes of every classifier.
We are not going to go into great detail regarding the construction of Multiclass SVMs at this time, although you will get an opportunity to work with them in the Lab exercise.
### References:
https://www.youtube.com/watch?v=_PwhiWxHK8o
|
github_jupyter
|
#RBF Kernel
from IPython.display import YouTubeVideo
YouTubeVideo("9NrALgHFwTo")
from IPython.display import YouTubeVideo
YouTubeVideo("3liCbRZPrZA")
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
import sklearn
import warnings
warnings.filterwarnings('ignore')
palette = "Dark2"
%matplotlib inline
from sklearn.datasets.samples_generator import make_blobs
from sklearn import datasets
from sklearn.svm import SVC
iris = datasets.load_iris()
X = iris.data[:, :2] # Use only the first two features for easier plotting
y = iris.target
# Define & fit model
#kernel = 'linear'
#kernel = 'rbf'
kernel = 'poly'
clf = SVC(kernel=kernel).fit(X, y)
predicted = clf.predict(iris.data[:, :2])
clf.n_support_
# Plot the decision bounds
from mlxtend.plotting import plot_decision_regions
y_labels = y # actual
# y_labels = predicted
plot_decision_regions(X=X,
y=y_labels,
clf=clf,
legend=False);
# NOTE: Linear kernel includes piecewise linear, hence the discontinuity
# Plot the decision bounds
from mlxtend.plotting import plot_decision_regions
y_labels = y # actual
# y_labels = predicted
plot_decision_regions(X=X,
y=y_labels,
clf=clf,
legend=False);
# NOTE: Linear kernel includes piecewise linear, hence the discontinuity
# Plot the decision bounds
from mlxtend.plotting import plot_decision_regions
y_labels = y # actual
# y_labels = predicted
plot_decision_regions(X=X,
y=y_labels,
clf=clf,
legend=False);
# NOTE: Linear kernel includes piecewise linear, hence the discontinuity
# From blobs to circles
from sklearn.datasets import make_circles
X, y = make_circles(noise=0.1, factor=.01, random_state=42)
ax = sns.scatterplot(x=X[:,0], y=X[:,1], hue=y, palette=palette, legend=False);
ax.set_yticklabels(''); ax.set_xticklabels('');
# Define & fit model
# kernel = 'linear'
kernel = 'rbf'
# kernel = 'poly'
clf = SVC(kernel=kernel).fit(X, y)
predicted = clf.predict(iris.data[:, :2])
# Plot the decision bounds
from mlxtend.plotting import plot_decision_regions
y_labels = y # actual
# y_labels = predicted
plot_decision_regions(X=X,
y=y_labels,
clf=clf,
legend=False);
# NOTE: Linear kernel includes piecewise linear, hence the discontinuity
| 0.84759 | 0.99269 |
# A Learning Based Strategy for Athena
## Introduction
Athena has various strategies for making a final prediction based on the predictions of the ensemble of weak defenses. These strategies are relatively basic, and leave a large potential for optimization using a neural network. Our strategy involves running the predictions of the ensemble through a neural network trained to make a final prediction based on the predictions of the ensemble.
### Detailed Rundown of Approach
1. Generate data set
1. Need to gather test and training data
- must be uniformly distributed to eliminate bias
- data must be adversarial examples from Athena
- need the proper labels for each example
- need raw predictions from each weak defense for each adversarial example
2. Create Neural Network
1. Train a CNN on training data
- Input will be nx10, where n is the number of weak defenses
- Error produced by comparison with 1x10 array in the form [0,0,0,0,x,0,0,0,0,0], where x is the prediction
- Output is the predicted label for the image
2. Implement into Athena
- NN will be a specific strategy that Athena can utilize, must implement this strategy name into main athena file
3. Test Neural Network
1. Run previously generated test data through NN, comparing output with true labels
2. Determine error rate of new athena strategy
**Important Note:**
Our implemetation of this strategy uses 15 randomly selected weak defenses. This is to reduce data collection and training time. With superior computing power, all 72 weak defenses would be uses.
#### 1. Generate Data Set
The data set was generated using the AE's found in (a); this is all of the AE's included with Athena. A subsample was generated splitting the data from each AE found in (a) into 80% training, 20% testing, using (b). Using (c), we looped through our dataset, generating the raw predictions from each weak defense defined in "active_wds" field in (d). Finally, (e) was used to generate the labels for training in a manner which can be used to create an error function for the NN as described in 2A. To transform the test data into a useable form, (f) was used.
**Total training samples:** 3600
**Total test samples:** 1000
**Files:**
- (a): src/configs/experiment/data-mnist.json
- (b): src/learning_based_strategy/split_data.py
- (c): src/learning_based_strategy/collect_raws.py
- (d): src/configs/experiment/athena-mnist.json
- (e): src/learning_based_strategy/get_training_labels.py
- (f): src/learning_based_strategy/get_test_samples.py
#### Relevant Code Snippets for 1
##### (b)
```
"""
Created on Thu Nov 12 17:49:52 2020
@author: miles
"""
import os
from utils.file import load_from_json
from utils.data import subsampling
import numpy as np
data_configs = load_from_json('../configs/experiment/data-mnist.json')
path = 'samples/'
#get data files, only take the AE type for the filename for matching later
data_files = [os.path.join(data_configs.get('dir'), ae_file) for ae_file in data_configs.get('ae_files')]
filenames = [ae_file.split('-')[-1].replace('.npy','') for ae_file in data_configs.get('ae_files')]
#Get respective label files
label_file = os.path.join(data_configs.get('dir'), data_configs.get('label_file'))
labels = np.load(label_file)
#Subsample from each AE file
for file, filename in zip(data_files, filenames):
data = np.load(file)
subsampling(data, labels, 10, 0.2, path, filename)
```
##### (c) from collect_raw_prediction
```
#get samples and respective labels
samples = glob.glob('samples/*training_samples*.npy')
labels = glob.glob('samples/*training_labels*.npy')
#sort based on type of attack
sorted_samples = []
sorted_labels = []
for sample in samples:
pref = sample.split('_training_samples')[0].split('/')[1].replace('.npy','')
sorted_samples.append(sample)
for label in labels:
pref2 = label.split('_training_labels')[0].split('/')[1].replace('.npy','')
if pref == pref2:
sorted_labels.append(label)
#load data and labels, concatenate into single numpy array for easy looping
samples_dat = [np.load(dat) for dat in sorted_samples]
labels_dat = [np.load(dat) for dat in sorted_labels]
samples_dat = np.concatenate(samples_dat,axis=0)
labels_dat = np.concatenate(labels_dat)
samples = []
labels = []
#Generate raw predictions from each WD for each AE
for i in range(0, len(labels_dat), 100):
raw_preds = athena.predict(x=samples_dat[i], raw=True)
samples.append(raw_preds)
labels.append(labels_dat[i])
#Write out raw predictions to training_data directory
samples = np.concatenate(samples,axis=1)
labels = np.array(labels)
samples_file = os.path.join('training_data/', 'training.npy')
np.save(file=samples_file,arr=samples)
labels_file = os.path.join('training_data/','training_labels.npy')
np.save(file=labels_file,arr=labels)
```
##### (e)
```
"""
Created on Sun Nov 15 20:08:08 2020
@author: miles
"""
import numpy as np
file = 'training_data/training_labels.npy'
labels = np.load(file)
arr = []
for label in labels:
temp = [0,0,0,0,0,0,0,0,0,0]
temp[label] = 1;
arr.append(temp)
new_arr = np.array(arr)
np.save(file='training_data/labels2D.npy', arr=new_arr)
```
##### (f)
```
"""
Created on Sun Nov 15 17:53:22 2020
@author: miles
"""
import os
import glob
import numpy as np
samples = glob.glob('samples/*test_samples*.npy')
labels = glob.glob('samples/*test_labels*.npy')
sorted_samples = []
sorted_labels = []
for sample in samples:
pref = sample.split('_test_samples')[0].split('/')[1].replace('.npy','')
sorted_samples.append(sample)
for label in labels:
pref2 = label.split('_test_labels')[0].split('/')[1].replace('.npy','')
if pref == pref2:
sorted_labels.append(label)
samples_dat = [np.load(dat) for dat in sorted_samples]
labels_dat = [np.load(dat) for dat in sorted_labels]
samples_dat = np.concatenate(samples_dat,axis=0)
labels_dat = np.concatenate(labels_dat)
samples = []
labels = []
for i in range(0, len(labels_dat), 90):
samples.append(samples_dat[i])
labels.append(labels_dat[i])
labels_file = os.path.join('testing/', 'test_labels.npy')
samples_file = os.path.join('testing/', 'test_samples.npy')
labels = np.array(labels)
samples = np.array(samples)
np.save(file=labels_file,arr=labels)
np.save(file=samples_file,arr=samples)
```
#### 2. Create Neural Network
The neural network has an input layer of shape 15x10, (15 weak defenses were used for this model chosen randomly in order to reduce training time, 10 predictions from each weak defense), two hidden layers and an output layer of 10x1. The output still generates 10 values using softmax, but picks the highest value in a postprocessing phase within Athena. The hidden layers take the input to 150x4, then back down to 150. All activations are relu, except for the final layer which is softmax.
**NOTE:** This method should be optimized by using all 72 weak defenses, so the input layer should be 72x10. The network should also be trained on many more than 3600 examples. We did not have the necessary resources to do this in a reasonable amount of time
**FILES:**
- create NN: src/learning_based_strategy/models/nn.py
- NN stored in: src/learning_based_strategy/models/learning-strategy-nn.h5
```
#nn.py
import numpy as np
import keras
from keras.datasets import mnist
from keras import models
from keras import layers
from keras.utils import to_categorical
training_data = np.load('../training_data/training.npy')
training_labels = np.load('../training_data/labels2D.npy')
network = models.Sequential()
network.add(layers.Dense(150, activation='relu', input_shape=(15 * 10,)))
network.add(layers.Dense(600, activation='relu', input_shape=(150 * 4,)))
network.add(layers.Dense(150, activation='relu'))
network.add(layers.Dense(10, activation='softmax'))
network.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
training_data = np.transpose(training_data, (1,0,2))
training_data = training_data.reshape((3600,15*10))
network.fit(training_data, training_labels, epochs=10, batch_size=360)
network.save('learning-strategy-nn.h5')
```
|
github_jupyter
|
"""
Created on Thu Nov 12 17:49:52 2020
@author: miles
"""
import os
from utils.file import load_from_json
from utils.data import subsampling
import numpy as np
data_configs = load_from_json('../configs/experiment/data-mnist.json')
path = 'samples/'
#get data files, only take the AE type for the filename for matching later
data_files = [os.path.join(data_configs.get('dir'), ae_file) for ae_file in data_configs.get('ae_files')]
filenames = [ae_file.split('-')[-1].replace('.npy','') for ae_file in data_configs.get('ae_files')]
#Get respective label files
label_file = os.path.join(data_configs.get('dir'), data_configs.get('label_file'))
labels = np.load(label_file)
#Subsample from each AE file
for file, filename in zip(data_files, filenames):
data = np.load(file)
subsampling(data, labels, 10, 0.2, path, filename)
#get samples and respective labels
samples = glob.glob('samples/*training_samples*.npy')
labels = glob.glob('samples/*training_labels*.npy')
#sort based on type of attack
sorted_samples = []
sorted_labels = []
for sample in samples:
pref = sample.split('_training_samples')[0].split('/')[1].replace('.npy','')
sorted_samples.append(sample)
for label in labels:
pref2 = label.split('_training_labels')[0].split('/')[1].replace('.npy','')
if pref == pref2:
sorted_labels.append(label)
#load data and labels, concatenate into single numpy array for easy looping
samples_dat = [np.load(dat) for dat in sorted_samples]
labels_dat = [np.load(dat) for dat in sorted_labels]
samples_dat = np.concatenate(samples_dat,axis=0)
labels_dat = np.concatenate(labels_dat)
samples = []
labels = []
#Generate raw predictions from each WD for each AE
for i in range(0, len(labels_dat), 100):
raw_preds = athena.predict(x=samples_dat[i], raw=True)
samples.append(raw_preds)
labels.append(labels_dat[i])
#Write out raw predictions to training_data directory
samples = np.concatenate(samples,axis=1)
labels = np.array(labels)
samples_file = os.path.join('training_data/', 'training.npy')
np.save(file=samples_file,arr=samples)
labels_file = os.path.join('training_data/','training_labels.npy')
np.save(file=labels_file,arr=labels)
"""
Created on Sun Nov 15 20:08:08 2020
@author: miles
"""
import numpy as np
file = 'training_data/training_labels.npy'
labels = np.load(file)
arr = []
for label in labels:
temp = [0,0,0,0,0,0,0,0,0,0]
temp[label] = 1;
arr.append(temp)
new_arr = np.array(arr)
np.save(file='training_data/labels2D.npy', arr=new_arr)
"""
Created on Sun Nov 15 17:53:22 2020
@author: miles
"""
import os
import glob
import numpy as np
samples = glob.glob('samples/*test_samples*.npy')
labels = glob.glob('samples/*test_labels*.npy')
sorted_samples = []
sorted_labels = []
for sample in samples:
pref = sample.split('_test_samples')[0].split('/')[1].replace('.npy','')
sorted_samples.append(sample)
for label in labels:
pref2 = label.split('_test_labels')[0].split('/')[1].replace('.npy','')
if pref == pref2:
sorted_labels.append(label)
samples_dat = [np.load(dat) for dat in sorted_samples]
labels_dat = [np.load(dat) for dat in sorted_labels]
samples_dat = np.concatenate(samples_dat,axis=0)
labels_dat = np.concatenate(labels_dat)
samples = []
labels = []
for i in range(0, len(labels_dat), 90):
samples.append(samples_dat[i])
labels.append(labels_dat[i])
labels_file = os.path.join('testing/', 'test_labels.npy')
samples_file = os.path.join('testing/', 'test_samples.npy')
labels = np.array(labels)
samples = np.array(samples)
np.save(file=labels_file,arr=labels)
np.save(file=samples_file,arr=samples)
#nn.py
import numpy as np
import keras
from keras.datasets import mnist
from keras import models
from keras import layers
from keras.utils import to_categorical
training_data = np.load('../training_data/training.npy')
training_labels = np.load('../training_data/labels2D.npy')
network = models.Sequential()
network.add(layers.Dense(150, activation='relu', input_shape=(15 * 10,)))
network.add(layers.Dense(600, activation='relu', input_shape=(150 * 4,)))
network.add(layers.Dense(150, activation='relu'))
network.add(layers.Dense(10, activation='softmax'))
network.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
training_data = np.transpose(training_data, (1,0,2))
training_data = training_data.reshape((3600,15*10))
network.fit(training_data, training_labels, epochs=10, batch_size=360)
network.save('learning-strategy-nn.h5')
| 0.425605 | 0.970183 |
```
data = [
'I am Mohammed Abacha, the son of the late Nigerian Head of '
'State who died on the 8th of June 1998. Since i have been '
'unsuccessful in locating the relatives for over 2 years now '
'I seek your consent to present you as the next of kin so '
'that the proceeds of this account valued at US$15.5 Million '
'Dollars can be paid to you. If you are capable and willing '
'to assist, contact me at once via email with following '
'details: 1. Your full name, address, and telephone number. '
'2. Your Bank Name, Address. 3.Your Bank Account Number and '
'Beneficiary Name - You must be the signatory.'
]
data
from sklearn.feature_extraction.text import CountVectorizer
vec = CountVectorizer()
x = vec.fit_transform(data)
x
vec.get_feature_names()[:5]
x.toarray()[0,:5]
'nigerian' in vec.get_feature_names()
'prince' in vec.get_feature_names()
data = [
{'age': 33, 'sex': 'F', 'BP': 'high', 'cholesterol': 'high',
'Na': 0.66, 'K': 0.06, 'drug': 'A'},
{'age': 77, 'sex': 'F', 'BP': 'high', 'cholesterol': 'normal',
'Na': 0.19, 'K': 0.03, 'drug': 'D'},
{'age': 88, 'sex': 'M', 'BP': 'normal', 'cholesterol': 'normal',
'Na': 0.80, 'K': 0.05, 'drug': 'B'},
{'age': 39, 'sex': 'F', 'BP': 'low', 'cholesterol': 'normal',
'Na': 0.19, 'K': 0.02, 'drug': 'C'},
{'age': 43, 'sex': 'M', 'BP': 'normal', 'cholesterol': 'high',
'Na': 0.36, 'K': 0.03, 'drug': 'D'},
{'age': 82, 'sex': 'F', 'BP': 'normal', 'cholesterol': 'normal',
'Na': 0.09, 'K': 0.09, 'drug': 'C'},
{'age': 40, 'sex': 'M', 'BP': 'high', 'cholesterol': 'normal',
'Na': 0.89, 'K': 0.02, 'drug': 'A'},
{'age': 88, 'sex': 'M', 'BP': 'normal', 'cholesterol': 'normal',
'Na': 0.80, 'K': 0.05, 'drug': 'B'},
{'age': 29, 'sex': 'F', 'BP': 'high', 'cholesterol': 'normal',
'Na': 0.35, 'K': 0.04, 'drug': 'D'},
{'age': 53, 'sex': 'F', 'BP': 'normal', 'cholesterol': 'normal',
'Na': 0.54, 'K': 0.06, 'drug': 'C'},
{'age': 63, 'sex': 'M', 'BP': 'low', 'cholesterol': 'high',
'Na': 0.86, 'K': 0.09, 'drug': 'B'},
{'age': 60, 'sex': 'M', 'BP': 'low', 'cholesterol': 'normal',
'Na': 0.66, 'K': 0.04, 'drug': 'C'},
{'age': 55, 'sex': 'M', 'BP': 'high', 'cholesterol': 'high',
'Na': 0.82, 'K': 0.04, 'drug': 'B'},
{'age': 35, 'sex': 'F', 'BP': 'normal', 'cholesterol': 'high',
'Na': 0.27, 'K': 0.03, 'drug': 'D'},
{'age': 23, 'sex': 'F', 'BP': 'high', 'cholesterol': 'high',
'Na': 0.55, 'K': 0.08, 'drug': 'A'},
{'age': 49, 'sex': 'F', 'BP': 'low', 'cholesterol': 'normal',
'Na': 0.27, 'K': 0.05, 'drug': 'C'},
{'age': 27, 'sex': 'M', 'BP': 'normal', 'cholesterol': 'normal',
'Na': 0.77, 'K': 0.02, 'drug': 'B'},
{'age': 51, 'sex': 'F', 'BP': 'low', 'cholesterol': 'high',
'Na': 0.20, 'K': 0.02, 'drug': 'D'},
{'age': 38, 'sex': 'M', 'BP': 'high', 'cholesterol': 'normal',
'Na': 0.78, 'K': 0.05, 'drug': 'A'}
]
len(data)
def separate_drug(data):
alist = [d.pop('drug') for d in data]
# iter = [d['drug'] for d in data]
# print(iter)
return alist
# x = separate_drug(data)
# x
# select 'drug' attribute as target and remove it from data
target = [dic.pop('drug') for dic in data]
sodium = [d['Na'] for d in data]
potassium = [d['K'] for d in data]
import matplotlib.pyplot as plt
plt.style.use('ggplot')
plt.scatter(sodium, potassium)
plt.xlabel('sodium')
plt.ylabel('potassium')
# dots with different colors
target_flags = [ord(i)-65 for i in target]
print('target_flags =', target_flags)
plt.scatter(sodium, potassium, c=target_flags, s=100)
plt.xlabel('sodium')
plt.ylabel('potassium')
age = [d['age'] for d in data]
plt.figure(figsize=(12,10))
plt.subplot(2,2,1)
plt.scatter(sodium, potassium, c=target_flags, s=100)
plt.xlabel('sodium (Na)')
plt.ylabel('potassium (K)')
plt.subplot(2,2,2)
plt.scatter(age, sodium, c=target_flags, s=100)
plt.xlabel('age')
plt.ylabel('sodium (Na)')
plt.subplot(2,2,3)
plt.scatter(age, potassium, c=target_flags, s=100)
plt.xlabel('age')
plt.ylabel('potassium (K)')
### Preprocessing the data
# convert categorical data to numerical
from sklearn.feature_extraction import DictVectorizer
vec = DictVectorizer(sparse=False)
data_pre = vec.fit_transform(data)
vec.get_feature_names()
# convert to float32 to compatible with OpenCV
import numpy as np
data_pre = np.array(data_pre, dtype=np.float32)
target_flags = np.array(target_flags, dtype=np.float32)
data_pre[0]
# split train & test datasets
import sklearn.model_selection as ms
import os
os.environ["CUDA_VISIBLE_DEVICES"] = "-1"
x_train, x_test, y_train, y_test = ms.train_test_split(data_pre, target_flags, test_size=5, random_state=42)
x_train.shape
x_train
### Constructing the decision tree
import cv2
from sklearn import metrics
dtree = cv2.ml.DTrees_create()
# train the tree
dtree.train(x_train, cv2.ml.ROW_SAMPLE, y_train)
y_pred = dtree.predict(x_test)
metrics.accuracy_score(y_test, y_pred)
```
|
github_jupyter
|
data = [
'I am Mohammed Abacha, the son of the late Nigerian Head of '
'State who died on the 8th of June 1998. Since i have been '
'unsuccessful in locating the relatives for over 2 years now '
'I seek your consent to present you as the next of kin so '
'that the proceeds of this account valued at US$15.5 Million '
'Dollars can be paid to you. If you are capable and willing '
'to assist, contact me at once via email with following '
'details: 1. Your full name, address, and telephone number. '
'2. Your Bank Name, Address. 3.Your Bank Account Number and '
'Beneficiary Name - You must be the signatory.'
]
data
from sklearn.feature_extraction.text import CountVectorizer
vec = CountVectorizer()
x = vec.fit_transform(data)
x
vec.get_feature_names()[:5]
x.toarray()[0,:5]
'nigerian' in vec.get_feature_names()
'prince' in vec.get_feature_names()
data = [
{'age': 33, 'sex': 'F', 'BP': 'high', 'cholesterol': 'high',
'Na': 0.66, 'K': 0.06, 'drug': 'A'},
{'age': 77, 'sex': 'F', 'BP': 'high', 'cholesterol': 'normal',
'Na': 0.19, 'K': 0.03, 'drug': 'D'},
{'age': 88, 'sex': 'M', 'BP': 'normal', 'cholesterol': 'normal',
'Na': 0.80, 'K': 0.05, 'drug': 'B'},
{'age': 39, 'sex': 'F', 'BP': 'low', 'cholesterol': 'normal',
'Na': 0.19, 'K': 0.02, 'drug': 'C'},
{'age': 43, 'sex': 'M', 'BP': 'normal', 'cholesterol': 'high',
'Na': 0.36, 'K': 0.03, 'drug': 'D'},
{'age': 82, 'sex': 'F', 'BP': 'normal', 'cholesterol': 'normal',
'Na': 0.09, 'K': 0.09, 'drug': 'C'},
{'age': 40, 'sex': 'M', 'BP': 'high', 'cholesterol': 'normal',
'Na': 0.89, 'K': 0.02, 'drug': 'A'},
{'age': 88, 'sex': 'M', 'BP': 'normal', 'cholesterol': 'normal',
'Na': 0.80, 'K': 0.05, 'drug': 'B'},
{'age': 29, 'sex': 'F', 'BP': 'high', 'cholesterol': 'normal',
'Na': 0.35, 'K': 0.04, 'drug': 'D'},
{'age': 53, 'sex': 'F', 'BP': 'normal', 'cholesterol': 'normal',
'Na': 0.54, 'K': 0.06, 'drug': 'C'},
{'age': 63, 'sex': 'M', 'BP': 'low', 'cholesterol': 'high',
'Na': 0.86, 'K': 0.09, 'drug': 'B'},
{'age': 60, 'sex': 'M', 'BP': 'low', 'cholesterol': 'normal',
'Na': 0.66, 'K': 0.04, 'drug': 'C'},
{'age': 55, 'sex': 'M', 'BP': 'high', 'cholesterol': 'high',
'Na': 0.82, 'K': 0.04, 'drug': 'B'},
{'age': 35, 'sex': 'F', 'BP': 'normal', 'cholesterol': 'high',
'Na': 0.27, 'K': 0.03, 'drug': 'D'},
{'age': 23, 'sex': 'F', 'BP': 'high', 'cholesterol': 'high',
'Na': 0.55, 'K': 0.08, 'drug': 'A'},
{'age': 49, 'sex': 'F', 'BP': 'low', 'cholesterol': 'normal',
'Na': 0.27, 'K': 0.05, 'drug': 'C'},
{'age': 27, 'sex': 'M', 'BP': 'normal', 'cholesterol': 'normal',
'Na': 0.77, 'K': 0.02, 'drug': 'B'},
{'age': 51, 'sex': 'F', 'BP': 'low', 'cholesterol': 'high',
'Na': 0.20, 'K': 0.02, 'drug': 'D'},
{'age': 38, 'sex': 'M', 'BP': 'high', 'cholesterol': 'normal',
'Na': 0.78, 'K': 0.05, 'drug': 'A'}
]
len(data)
def separate_drug(data):
alist = [d.pop('drug') for d in data]
# iter = [d['drug'] for d in data]
# print(iter)
return alist
# x = separate_drug(data)
# x
# select 'drug' attribute as target and remove it from data
target = [dic.pop('drug') for dic in data]
sodium = [d['Na'] for d in data]
potassium = [d['K'] for d in data]
import matplotlib.pyplot as plt
plt.style.use('ggplot')
plt.scatter(sodium, potassium)
plt.xlabel('sodium')
plt.ylabel('potassium')
# dots with different colors
target_flags = [ord(i)-65 for i in target]
print('target_flags =', target_flags)
plt.scatter(sodium, potassium, c=target_flags, s=100)
plt.xlabel('sodium')
plt.ylabel('potassium')
age = [d['age'] for d in data]
plt.figure(figsize=(12,10))
plt.subplot(2,2,1)
plt.scatter(sodium, potassium, c=target_flags, s=100)
plt.xlabel('sodium (Na)')
plt.ylabel('potassium (K)')
plt.subplot(2,2,2)
plt.scatter(age, sodium, c=target_flags, s=100)
plt.xlabel('age')
plt.ylabel('sodium (Na)')
plt.subplot(2,2,3)
plt.scatter(age, potassium, c=target_flags, s=100)
plt.xlabel('age')
plt.ylabel('potassium (K)')
### Preprocessing the data
# convert categorical data to numerical
from sklearn.feature_extraction import DictVectorizer
vec = DictVectorizer(sparse=False)
data_pre = vec.fit_transform(data)
vec.get_feature_names()
# convert to float32 to compatible with OpenCV
import numpy as np
data_pre = np.array(data_pre, dtype=np.float32)
target_flags = np.array(target_flags, dtype=np.float32)
data_pre[0]
# split train & test datasets
import sklearn.model_selection as ms
import os
os.environ["CUDA_VISIBLE_DEVICES"] = "-1"
x_train, x_test, y_train, y_test = ms.train_test_split(data_pre, target_flags, test_size=5, random_state=42)
x_train.shape
x_train
### Constructing the decision tree
import cv2
from sklearn import metrics
dtree = cv2.ml.DTrees_create()
# train the tree
dtree.train(x_train, cv2.ml.ROW_SAMPLE, y_train)
y_pred = dtree.predict(x_test)
metrics.accuracy_score(y_test, y_pred)
| 0.435181 | 0.599339 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.